MedJamba
Multilingual Medical Model Based On Jamba
🌈 Update
- [2024.04.25] MedJamba Model is published!🎉
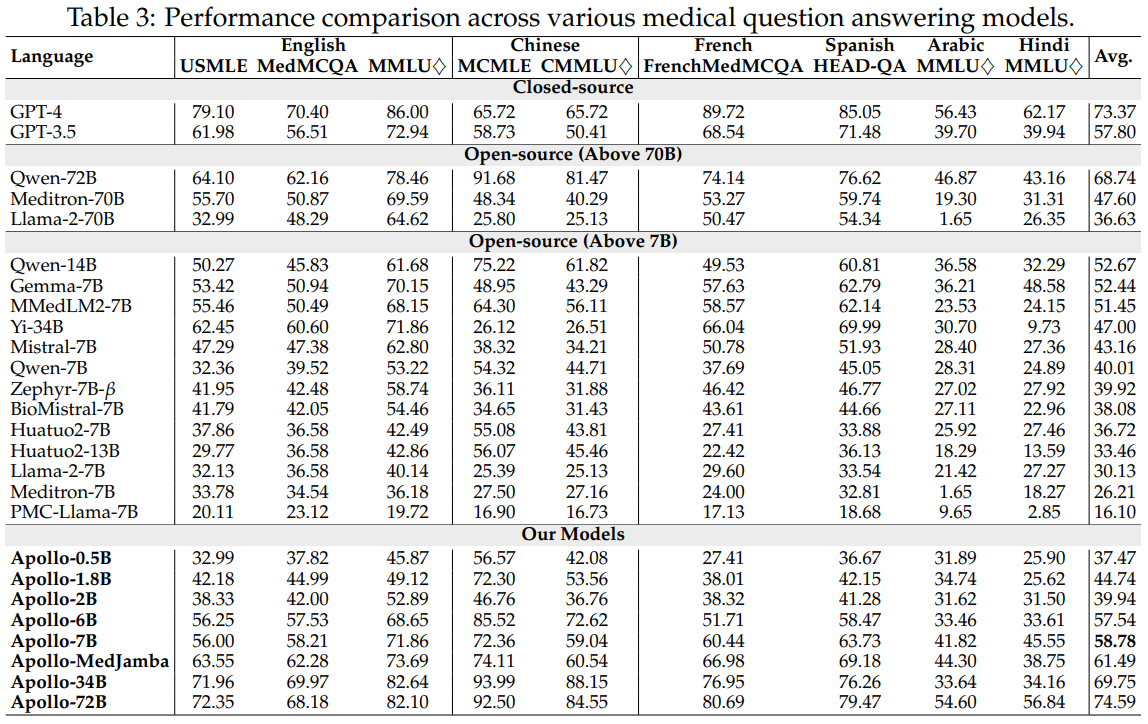
Results
🤗 Apollo-0.5B • 🤗 Apollo-1.8B • 🤗 Apollo-2B • 🤗 Apollo-6B • 🤗 Apollo-7B • 🤗 Apollo-34B • 🤗 Apollo-72B
🤗 MedJamba
🤗 Apollo-0.5B-GGUF • 🤗 Apollo-2B-GGUF • 🤗 Apollo-6B-GGUF • 🤗 Apollo-7B-GGUF
Dataset & Evaluation
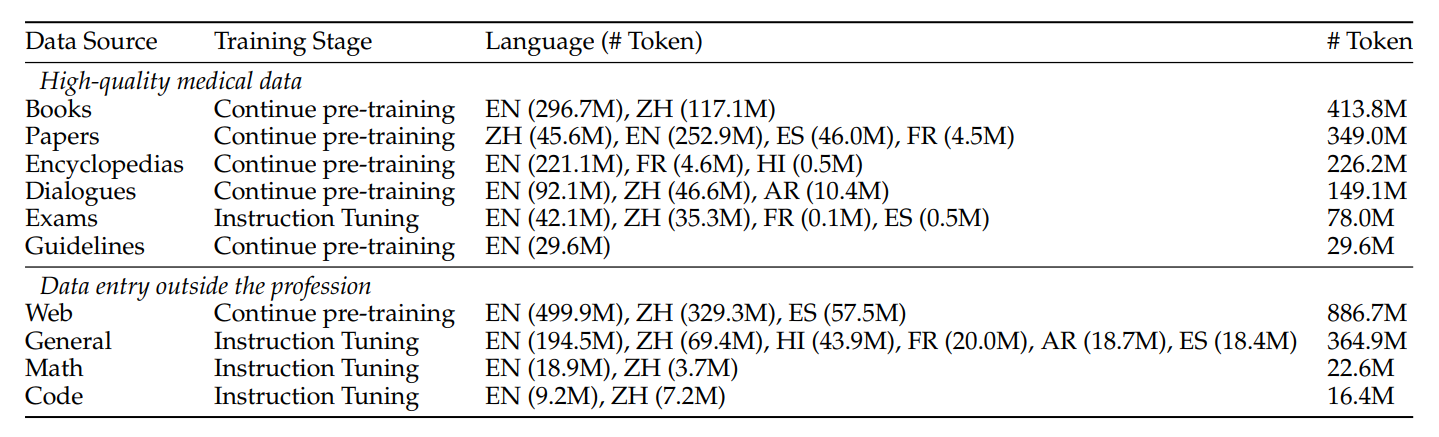
Dataset 🤗 ApolloCorpus
Click to expand
- Zip File
- Data category
- Pretrain:
- data item:
- json_name: {data_source}{language}{data_type}.json
- data_type: medicalBook, medicalGuideline, medicalPaper, medicalWeb(from online forum), medicalWiki
- language: en(English), zh(chinese), es(spanish), fr(french), hi(Hindi)
- data_type: qa(generated qa from text)
- data_type==text: list of string
[ "string1", "string2", ... ] - data_type==qa: list of qa pairs(list of string)
[ [ "q1", "a1", "q2", "a2", ... ], ... ]
- data item:
- SFT:
- json_name: {data_source}_{language}.json
- data_type: code, general, math, medicalExam, medicalPatient
- data item: list of qa pairs(list of string)
[ [ "q1", "a1", "q2", "a2", ... ], ... ]
- Pretrain:
Evaluation 🤗 XMedBench
Click to expand
EN:
- MedQA-USMLE
- MedMCQA
- PubMedQA: Because the results fluctuated too much, they were not used in the paper.
- MMLU-Medical
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
ZH:
- MedQA-MCMLE
- CMB-single: Not used in the paper
- Randomly sample 2,000 multiple-choice questions with single answer.
- CMMLU-Medical
- Anatomy, Clinical_knowledge, College_medicine, Genetics, Nutrition, Traditional_chinese_medicine, Virology
- CExam: Not used in the paper
- Randomly sample 2,000 multiple-choice questions
ES: Head_qa
FR: Frenchmedmcqa
HI: MMLU_HI
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
AR: MMLU_Ara
- Clinical knowledge, Medical genetics, Anatomy, Professional medicine, College biology, College medicine
Results reproduction
Click to expand
Download Dataset for project:
bash 0.download_data.shPrepare test and dev for specific model:
- Create test data for with special token, you can use ./util/check.ipynb to check models' special tokens
bash 1.data_process_test&dev.shPrepare train data for specific model (Create tokenized data in advance):
- You can adjust data Training order and Training Epoch in this step
bash 2.data_process_train.shTrain the model
- Multi Nodes refer to ./scripts/multi_node_train_*.sh
pip install causal-conv1d>=1.2.0 pip install mamba-ssmNode 0:
bash ./scripts/3.multinode_train_jamba_rank0.sh... Node 4:
bash ./scripts/3.multinode_train_jamba_rank4.shEvaluate your model: Generate score for benchmark
bash 4.eval.shEvaluate your model: Play with your ckpts in bash
python ./src/evaluate/cli_demo.py --model_name='./ckpts/your/path/tfmr'
To do
- Long Context Capability Evaluation and new Long-Med Benchmark
Acknowledgment
Citation
Please use the following citation if you intend to use our dataset for training or evaluation:
@misc{wang2024apollo,
title={Apollo: Lightweight Multilingual Medical LLMs towards Democratizing Medical AI to 6B People},
author={Xidong Wang and Nuo Chen and Junyin Chen and Yan Hu and Yidong Wang and Xiangbo Wu and Anningzhe Gao and Xiang Wan and Haizhou Li and Benyou Wang},
year={2024},
eprint={2403.03640},
archivePrefix={arXiv},
primaryClass={cs.CL}
}
- Downloads last month
- 25
This model does not have enough activity to be deployed to Inference API (serverless) yet. Increase its social
visibility and check back later, or deploy to Inference Endpoints (dedicated)
instead.