Model description
This repo contains the model and the notebook for implementing a generative model for graphs and using it to generate novel molecules WGAN-GP with R-GCN for the generation of small molecular graphs.
Full credits go to Alexander Kensert
Reproduced by Vu Minh Chien
Motivation: The development of new drugs (molecules) can be extremely time-consuming and costly. The use of deep learning models can alleviate the search for good candidate drugs, by predicting the properties of known molecules (e.g., solubility, toxicity, affinity to the target protein, etc.). As the number of possible molecules is astronomical, the space in which we search for/explore molecules is just a fraction of the entire space. Therefore, it's arguably desirable to implement generative models that can learn to generate novel molecules (which would otherwise have never been explored).
Intended uses & limitations
Recent implementations of generative models for molecular graphs also include Mol-CycleGAN, GraphVAE, and JT-VAE. For more information on generative adversarial networks, see GAN, WGAN, and WGAN-GP.
Training and evaluation data
The dataset used in this tutorial is a quantum mechanics dataset (QM9), obtained from MoleculeNet. The QM9 dataset is a good first dataset to work with for generating graphs, as the maximum number of heavy (non-hydrogen) atoms found in a molecule is only nine.
Training hyperparameters
The following hyperparameters were used during training:
- generator_learning_rate: 5e-04
- discriminator_learning_rate: 5e-04
- train_batch_size: 16
- optimizer: Adam with betas=(0.9,0.999) and epsilon=1e-08
- num_epochs: 50
Training results
- The results are shown in TensorBoard (Training metrics).
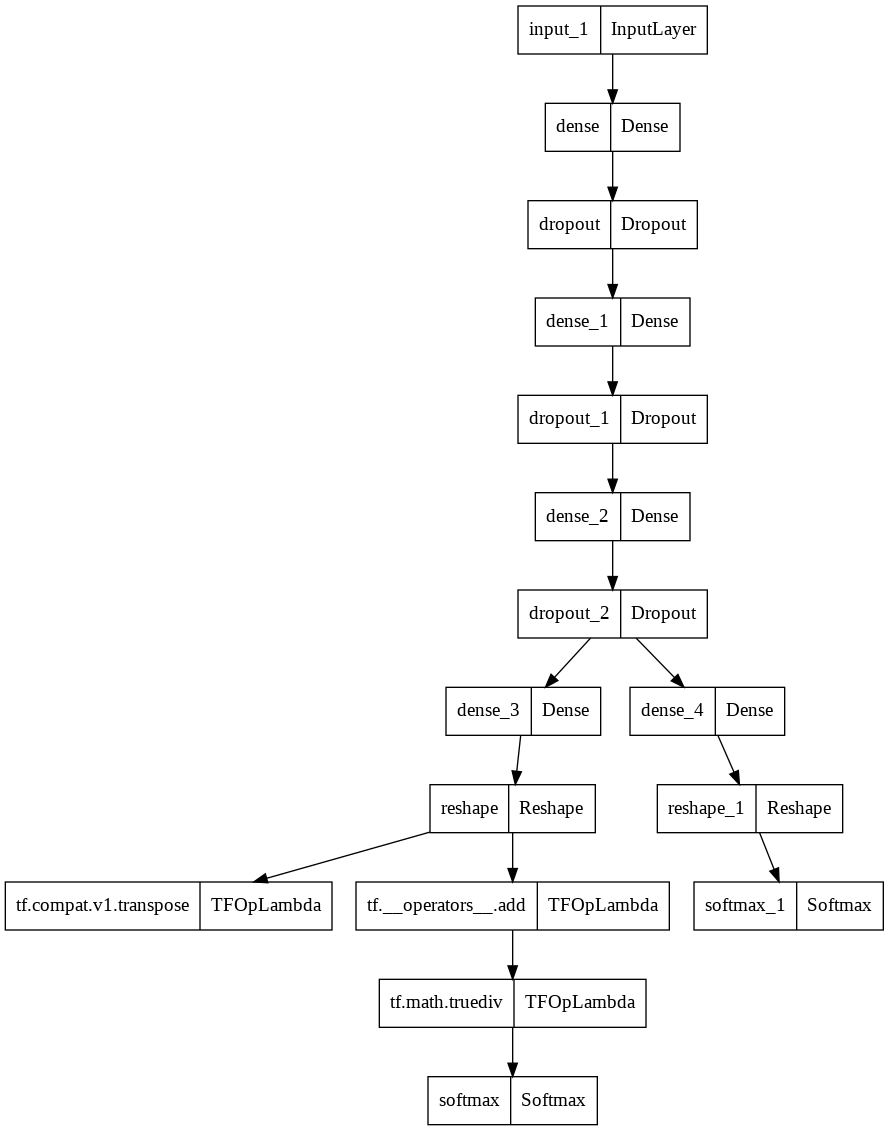
Model Plot
View Model Demo
- Downloads last month
- 16