Lung Adeno/Squam v1 Model Card
This model card describes the model associated with the manuscript "Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology", by Dolezal et al, available here.
Model Details
Developed by: James Dolezal
Model type: Deep convolutional neural network image classifier
Language(s): English
License: GPL-3.0
Model Description: This is a model that can classify H&E-stained pathologic images of non-small cell lung cancer into adenocarcinoma or squamous cell carcinoma and provide an estimate of classification uncertainty. It is an Xception model with two dropout-enabled hidden layers enabled during both training and inference. During inference, a given image is passed through the network 30 times, resulting in a distribution of predictions. The mean of this distribution is the final prediction, and the standard deviation is the uncertainty.
Image processing: This model expects images of H&E-stained pathology slides at 299 x 299 px and 302 x 302 μm resolution. Images should be stain-normalized using a modified Reinhard normalizer ("Reinhard-Fast") available here. The stain normalizer should be fit using the
target_meansandtarget_stdslisted in the modelparams.jsonfile. Images should be should be standardized withtf.image.per_image_standardization().Resources for more information: GitHub Repository, Paper
Cite as:
@ARTICLE{Dolezal2022-qa, title = "Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology", author = "Dolezal, James M and Srisuwananukorn, Andrew and Karpeyev, Dmitry and Ramesh, Siddhi and Kochanny, Sara and Cody, Brittany and Mansfield, Aaron S and Rakshit, Sagar and Bansal, Radhika and Bois, Melanie C and Bungum, Aaron O and Schulte, Jefree J and Vokes, Everett E and Garassino, Marina Chiara and Husain, Aliya N and Pearson, Alexander T", journal = "Nature Communications", volume = 13, number = 1, pages = "6572", month = nov, year = 2022 }
Uses
Examples
For direct use, the model can be loaded using Tensorflow/Keras:
import tensorflow as tf
model = tf.keras.models.load_model('/path/')
or loaded with Slideflow version 1.1+ with the following syntax:
import slideflow as sf
model = sf.model.load('/path/')
The stain normalizer can be loaded and fit using Slideflow:
normalizer = sf.util.get_model_normalizer('/path/')
The stain normalizer has a native Tensorflow transform and can be directly applied to a tf.data.Dataset:
# Map the stain normalizer transformation
# to a tf.data.Dataset
dataset = dataset.map(normalizer.tf_to_tf)
Alternatively, the model can be used to generate predictions for whole-slide images processed through Slideflow in an end-to-end Project. To use the model to generate predictions on data processed with Slideflow, simply pass the model to the Project.predict() function:
import slideflow
P = sf.Project('/path/to/slideflow/project')
P.predict('/model/path')
Direct Use
This model is intended for research purposes only. Possible research areas and tasks include
- Development and comparison of uncertainty quantification methods for pathologic images.
- Probing and understanding the limitations of out-of-distribution detection for pathology classification models.
- Applications in educational settings.
- Research on pathology classification models for non-small cell lung cancer.
Excluded uses are described below.
Misuse and Out-of-Scope Use
This model should not be used in a clinical setting to generate predictions that will be used to inform patients, physicians, or any other health care members directly involved in their health care outside the context of an approved research protocol. Using the model in a clinical setting outside the context of an approved research protocol is a misuse of this model. This includes, but is not limited to:
- Generating predictions of images from a patient's tumor and sharing those predictions with the patient
- Generating predictions of images from a patient's tumor and sharing those predictions with the patient's physician, or other members of the patient's healthcare team
- Influencing a patient's health care treatment in any way based on output from this model
Limitations
The model has not been validated to discriminate lung adenocarcinoma vs. squamous cell carcinoma in contexts where other tumor types are possible (such as lung small cell carcinoma, neuroendocrine tumors, metastatic deposits, etc.)
Bias
This model was trained on The Cancer Genome Atlas (TCGA), which contains patient data from communities and cultures which may not reflect the general population. This datasets is comprised of images from multiple institutions, which may introduce a potential source of bias from site-specific batch effects (Howard, 2021). The model was validated on data from the Clinical Proteomics Tumor Analysis Consortium (CPTAC) and an institutional dataset from Mayo Clinic, the latter of which consists primarily of data from patients of white and western cultures.
Training
Training Data The following dataset was used to train the model:
- The Cancer Genome Atlas (TCGA), LUAD (adenocarcinoma) and LUSC (squamous cell carcinoma) cohorts (see next section)
This model was trained on the full dataset, with a total of 941 slides.
Training Procedure Each whole-slide image was sectioned into smaller images in a grid-wise fashion in order to extract tiles from whole-slide images at 302 x 302 μm. Image tiles were extracted at the nearest downsample layer, and resized to 299 x 299 px using Libvips. During training,
- Images are stain-normalized with a modified Reinhard normalizer ("Reinhard-Fast"), which excludes the brightness standardization step, available here
- Images are randomly flipped and rotated (90, 180, 270)
- Images have a 50% chance of being JPEG compressed with quality level between 50-100%
- Images have a 10% chance of random Gaussian blur, with sigma between 0.5-2.0
- Images are standardized with
tf.image.per_image_standardization() - Images are classified through an Xception block, followed by two hidden layers with dropout (p=0.1) permanently enabled during both training and inference
- The loss is cross-entropy, with adenocarcinoma=0 and squamous=1
- Training was halted at a predetermined step=1451 (at batch size of 128, this is after 185,728 images), determined through nested cross-validation
During inference,
- A given image undergoes 30 forward passes in the network, resulting in a distribution, where
- The mean is the final prediction
- The standard deviation is the uncertainty
Tile-level and slide-level uncertainty thresholds are calculated and applied as discussed in the Paper. For this model, θ_tile=0.0228 and θ_slide=0.0139.
- Hardware: 1 x A100 GPUs
- Optimizer: Adam
- Batch: 128
- Learning rate: 0.0001, with a decay of 0.98 every 512 steps
- Hidden layers: 2 hidden layers of width 1024, with dropout p=0.1
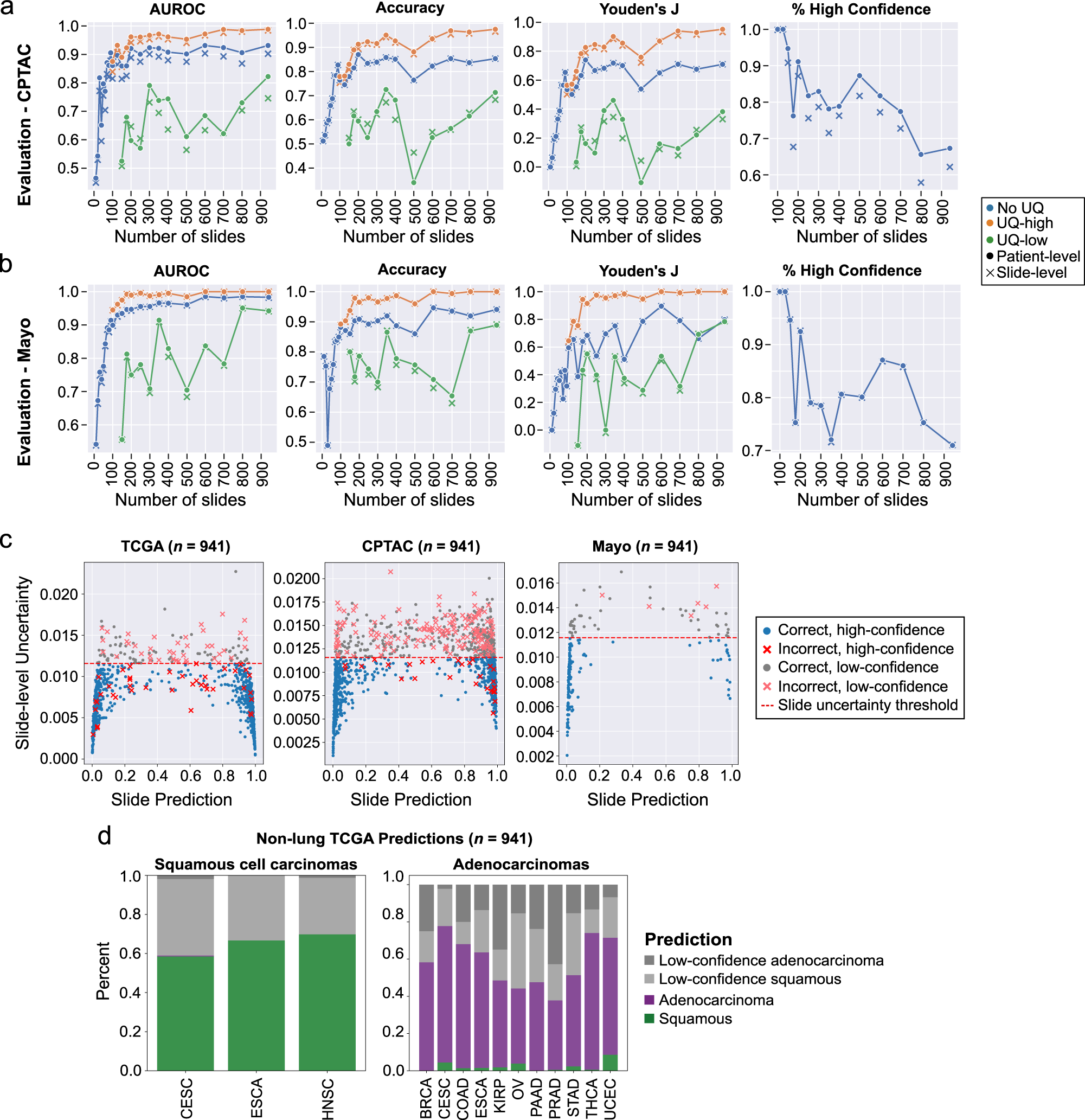
Evaluation Results
External evaluation results in the CPTAC and Mayo Clinic dataset are presented in the Paper and shown here:
Citation
@ARTICLE{Dolezal2022-qa,
title = "Uncertainty-informed deep learning models enable high-confidence
predictions for digital histopathology",
author = "Dolezal, James M and Srisuwananukorn, Andrew and Karpeyev, Dmitry
and Ramesh, Siddhi and Kochanny, Sara and Cody, Brittany and
Mansfield, Aaron S and Rakshit, Sagar and Bansal, Radhika and
Bois, Melanie C and Bungum, Aaron O and Schulte, Jefree J and
Vokes, Everett E and Garassino, Marina Chiara and Husain, Aliya N
and Pearson, Alexander T",
journal = "Nature Communications",
volume = 13,
number = 1,
pages = "6572",
month = nov,
year = 2022
}
- Downloads last month
- 0