AIDO.Protein2StructureToken-16B
AIDO.Protein2StructureToken-16B is a fine-tuned version of AIDO.Protein-16B, for protein structure prediction. This model uses amino acid sequences as input to predict tokens that can be decoded into 3D structures by AIDO.StructureDecoder. It surpasses existing state-of-the-art models, such as ESM3-open, in structure prediction tasks, demonstrating its robustness and capability in this domain.
Model Architecture Details
This model retains the architecture of AIDO.Protein-16B, a transformer encoder-only architecture with dense MLP layers replaced by sparse Mixture of Experts (MoE) layers. Each token activates 2 experts using a top-2 routing mechanism. A visual summary of the architecture is provided below:
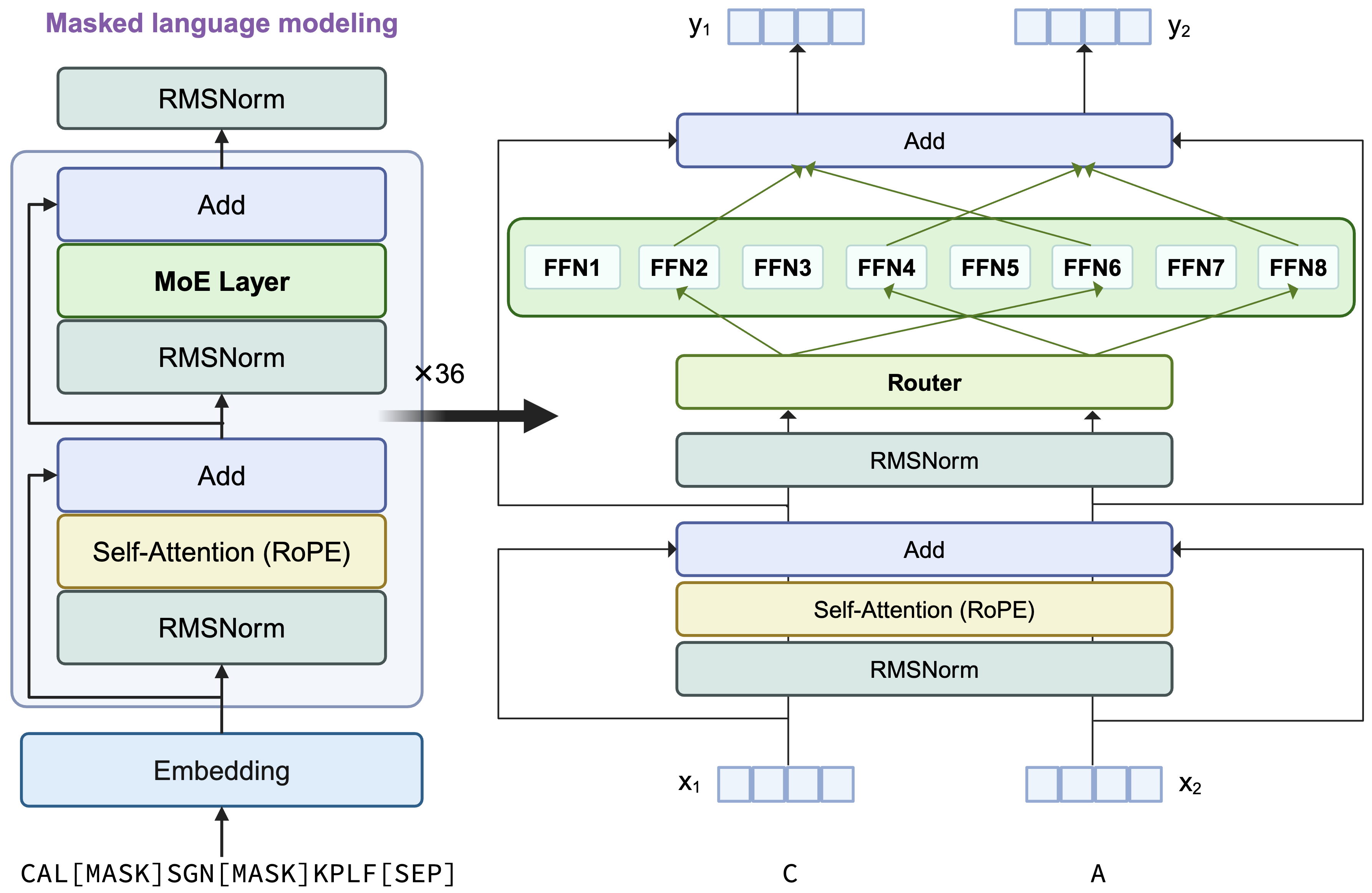
Key Differences
The final output linear layer has been adapted to support a new vocabulary size:
- Input Vocabulary Size: 44 (amino acids + special tokens)
- Output Vocabulary Size: 512 (structure tokens without special tokens)
Architecture Parameters
| Component | Value |
|---|---|
| Number of Attention Heads | 36 |
| Number of Hidden Layers | 36 |
| Hidden Size | 2304 |
| Number of MoE Layers per Block | 8 |
| Number of MoE Layers per Token | 2 |
| Input Vocabulary Size | 44 |
| Output Vocabulary Size | 512 |
| Context Length | 1024 |
Training Details
The fine-tuning process used 0.4 trillion tokens, using AlphaFold database with 170M samples and PDB database with 0.4M samples, making it highly specialized for structure prediction. The training took around 20 days on 64 A100 GPUs.
- Batch Size: Global batch size of 2048
- Context Length: 1024
- Precision: FP16
- Hardware: 64 NVIDIA A100 80GB GPUs
- Learning Rate: Max learning rate of 1e-4
- Scheduler: Cosine decay with 2.5% warmup
- Tokens Trained: 4T tokens
- Training steps: 200k steps
Tokenization
The input sequence should be single-chain amino acid sequences.
- Input Tokenization: The sequences are tokenized at the amino acid level and terminated with a
[SEP]token (id=34). - Output Tokenization: Each input token is converted into a structure token. The output can be decoded into 3D structures in PDB format using AIDO.StructureDecoder.
Results
How to Use
Structure Prediction
To reproduce the structure prediction results described above, follow these steps:
Install the Model Generator package.
Run the prediction command:
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yamlThis will pull the CASP14, CASP15, and CAMEO dataset from genbio-ai/casp14-casp15-cameo-test-proteins, and predict the structure tokens from the amino acid sequence.
Convert the output
.tsvto.ptand extract model codebook:# convert the predicted structures in tsv into one pt file python experiments/AIDO.StructureTokenizer/struct_token_format_conversion.py logs/protein2structoken_16b/predict_predictions.tsv logs/protein2structoken_16b/predict_predictions.pt # extract the codebook of the structure tokenizer python experiments/AIDO.StructureTokenizer/extract_structure_tokenizer_codebook.py --output_path logs/protein2structoken_16b/codebook.ptRun the decoding command to get 3D structures in PDB format (currently this script only supports single GPU inference):
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/decode.yaml \ --data.init_args.config.struct_tokens_datasets_configs.name=protein2structoken_16b \ --data.init_args.config.struct_tokens_datasets_configs.struct_tokens_path=logs/protein2structoken_16b/predict_predictions.pt \ --data.init_args.config.struct_tokens_datasets_configs.codebook_path=logs/protein2structoken_16b/codebook.ptThe outputs are in
logs/protstruct_decode/protein2structoken_16b_pdb_files/You can compare the predicted structures with the ground truth PDBs in genbio-ai/casp14-casp15-cameo-test-proteins.
Alternatively, you can provide your own input amino acid sequence in a CSV file. Here is one example csv at experiments/AIDO.StructureTokenizer/protein2structoken_example_input.csv in ModelGenerator:
idx,aa_seq
example,KEFWNLDKNLQLRLGIVFLG
Here, idx is a unique name, and aa_seq is the amino acid sequence. To use this customized CSV file, replace the second step with
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yaml \
--data.init_args.path=experiments/AIDO.StructureTokenizer/ \
--data.init_args.test_split_files=[protein2structoken_example_input.csv]
Build any downstream models from this backbone with ModelGenerator
For more information, visit: Model Generator
mgen fit --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path <hf_or_local_path_to_your_dataset>
mgen test --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path <hf_or_local_path_to_your_dataset>
The usage of this model is the same as AIDO.Protein-16B.
You only need to change the model.backbone to aido_protein2structoken.
Or use directly in Python
Embedding
from modelgenerator.tasks import Embed
model = Embed.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
embedding = model(collated_batch)
print(embedding.shape)
print(embedding)
Sequence Level Classification
import torch
from modelgenerator.tasks import SequenceClassification
model = SequenceClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 2}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
Token Level Classification
import torch
from modelgenerator.tasks import TokenClassification
model = TokenClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 3}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
Regression
from modelgenerator.tasks import SequenceRegression
model = SequenceRegression.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
Citation
Please cite AIDO.Protein and AIDO.StructureTokenizer using the following BibTex codes:
@inproceedings{zhang_balancing_2024,
title = {Balancing Locality and Reconstruction in Protein Structure Tokenizer},
url = {https://www.biorxiv.org/content/10.1101/2024.12.02.626366v2},
doi = {10.1101/2024.12.02.626366},
publisher = {bioRxiv},
author = {Zhang, Jiayou and Meynard-Piganeau, Barthelemy and Gong, James and Cheng, Xingyi and Luo, Yingtao and Ly, Hugo and Song, Le and Xing, Eric},
year = {2024},
booktitle={NeurIPS 2024 Workshop on Machine Learning in Structural Biology (MLSB)},
}
@inproceedings{sun_mixture_2024,
title = {Mixture of Experts Enable Efficient and Effective Protein Understanding and Design},
url = {https://www.biorxiv.org/content/10.1101/2024.11.29.625425v1},
doi = {10.1101/2024.11.29.625425},
publisher = {bioRxiv},
author = {Sun, Ning and Zou, Shuxian and Tao, Tianhua and Mahbub, Sazan and Li, Dian and Zhuang, Yonghao and Wang, Hongyi and Cheng, Xingyi and Song, Le and Xing, Eric P.},
year = {2024},
booktitle={NeurIPS 2024 Workshop on AI for New Drug Modalities},
}
- Downloads last month
- 135