text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
# SiteAlign features
We read the SiteAlign features from the respective [paper](https://onlinelibrary.wiley.com/doi/full/10.1002/prot.21858) and [SI table](https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fprot.21858&file=prot21858-SupplementaryTable.pdf) to verify `kissim`'s implementation of the SiteAlign definitions:
```
from kissim.definitions import SITEALIGN_FEATURES
SITEALIGN_FEATURES
```
## Size
SiteAlign's size definitions:
> Natural amino acids have been classified into three groups according to the number of heavy atoms (<4 heavy atoms: Ala, Cys, Gly, Pro, Ser, Thr, Val; 4–6 heavy atoms: Asn, Asp, Gln, Glu, His, Ile, Leu, Lys, Met; >6 heavy atoms: Arg, Phe, Trp, Tyr) and three values (“1,” “2,” “3”) are outputted according to the group to which the current residues belong to (Table I)
https://onlinelibrary.wiley.com/doi/full/10.1002/prot.21858
### Parse text from SiteAlign paper
```
size = {
1.0: "Ala, Cys, Gly, Pro, Ser, Thr, Val".split(", "),
2.0: "Asn, Asp, Gln, Glu, His, Ile, Leu, Lys, Met".split(", "),
3.0: "Arg, Phe, Trp, Tyr".split(", "),
}
```
### `kissim` definitions correct?
```
import pandas as pd
from IPython.display import display, HTML
# Format SiteAlign data
size_list = []
for value, amino_acids in size.items():
values = [(amino_acid.upper(), value) for amino_acid in amino_acids]
size_list = size_list + values
size_series = (
pd.DataFrame(size_list, columns=["amino_acid", "size"])
.sort_values("amino_acid")
.set_index("amino_acid")
.squeeze()
)
# KiSSim implementation of SiteAlign features correct?
diff = size_series == SITEALIGN_FEATURES["size"]
if not diff.all():
raise ValueError(
f"KiSSim implementation of SiteAlign features is incorrect!!!\n"
f"{display(HTML(diff.to_html()))}"
)
else:
print("KiSSim implementation of SiteAlign features is correct :)")
```
## HBA, HBD, charge, aromatic, aliphatic
### Parse table from SiteAlign SI
```
sitealign_table = """
Ala 0 0 0 1 0
Arg 3 0 +1 0 0
Asn 1 1 0 0 0
Asp 0 2 -1 0 0
Cys 1 0 0 1 0
Gly 0 0 0 0 0
Gln 1 1 0 0 0
Glu 0 2 -1 0 0
His/Hid/Hie 1 1 0 0 1
Hip 2 0 1 0 0
Ile 0 0 0 1 0
Leu 0 0 0 1 0
Lys 1 0 +1 0 0
Met 0 0 0 1 0
Phe 0 0 0 0 1
Pro 0 0 0 1 0
Ser 1 1 0 0 0
Thr 1 1 0 1 0
Trp 1 0 0 0 1
Tyr 1 1 0 0 1
Val 0 0 0 1 0
"""
sitealign_table = [i.split() for i in sitealign_table.split("\n")[1:-1]]
sitealign_dict = {i[0]: i[1:] for i in sitealign_table}
sitealign_df = pd.DataFrame.from_dict(sitealign_dict).transpose()
sitealign_df.columns = ["hbd", "hba", "charge", "aliphatic", "aromatic"]
sitealign_df = sitealign_df[["hbd", "hba", "charge", "aromatic", "aliphatic"]]
sitealign_df = sitealign_df.rename(index={"His/Hid/Hie": "His"})
sitealign_df = sitealign_df.drop("Hip", axis=0)
sitealign_df = sitealign_df.astype("float")
sitealign_df.index = [i.upper() for i in sitealign_df.index]
sitealign_df = sitealign_df.sort_index()
sitealign_df
```
### `kissim` definitions correct?
```
from IPython.display import display, HTML
diff = sitealign_df == SITEALIGN_FEATURES.drop("size", axis=1).sort_index()
if not diff.all().all():
raise ValueError(
f"KiSSim implementation of SiteAlign features is incorrect!!!\n"
f"{display(HTML(diff.to_html()))}"
)
else:
print("KiSSim implementation of SiteAlign features is correct :)")
```
## Table style
```
from Bio.Data.IUPACData import protein_letters_3to1
for feature_name in SITEALIGN_FEATURES.columns:
print(feature_name)
for name, group in SITEALIGN_FEATURES.groupby(feature_name):
amino_acids = {protein_letters_3to1[i.capitalize()] for i in group.index}
amino_acids = sorted(amino_acids)
print(f"{name:<7}{' '.join(amino_acids)}")
print()
```
|
github_jupyter
|
```
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 初始化openpose
#@markdown ■■■■■■■■■■■■■■■■■■
#设置版本为1.x
%tensorflow_version 1.x
import tensorflow as tf
tf.__version__
! nvcc --version
! nvidia-smi
! pip install PyQt5
import time
init_start_time = time.time()
#安装 cmake
#https://drive.google.com/file/d/1lAXs5X7qMnKQE48I0JqSob4FX1t6-mED/view?usp=sharing
file_id = "1lAXs5X7qMnKQE48I0JqSob4FX1t6-mED"
file_name = "cmake-3.13.4.zip"
! cd ./ && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./ && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./ && unzip cmake-3.13.4.zip
! cd cmake-3.13.4 && ./configure && make && sudo make install
# 依赖库安装
! sudo apt install caffe-cuda
! sudo apt-get --assume-yes update
! sudo apt-get --assume-yes install build-essential
# OpenCV
! sudo apt-get --assume-yes install libopencv-dev
# General dependencies
! sudo apt-get --assume-yes install libatlas-base-dev libprotobuf-dev libleveldb-dev libsnappy-dev libhdf5-serial-dev protobuf-compiler
! sudo apt-get --assume-yes install --no-install-recommends libboost-all-dev
# Remaining dependencies, 14.04
! sudo apt-get --assume-yes install libgflags-dev libgoogle-glog-dev liblmdb-dev
# Python3 libs
! sudo apt-get --assume-yes install python3-setuptools python3-dev build-essential
! sudo apt-get --assume-yes install python3-pip
! sudo -H pip3 install --upgrade numpy protobuf opencv-python
# OpenCL Generic
! sudo apt-get --assume-yes install opencl-headers ocl-icd-opencl-dev
! sudo apt-get --assume-yes install libviennacl-dev
# Openpose安装
ver_openpose = "v1.6.0"
# Openpose の clone
! git clone --depth 1 -b "$ver_openpose" https://github.com/CMU-Perceptual-Computing-Lab/openpose.git
# ! git clone --depth 1 https://github.com/CMU-Perceptual-Computing-Lab/openpose.git
# Openpose の モデルデータDL
! cd openpose/models && ./getModels.sh
#编译Openpose
! cd openpose && rm -r build || true && mkdir build && cd build && cmake .. && make -j`nproc` # example demo usage
# 执行示例确认
! cd /content/openpose && ./build/examples/openpose/openpose.bin --video examples/media/video.avi --write_json ./output/ --display 0 --write_video ./output/openpose.avi
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 其他软件初始化
#@markdown ■■■■■■■■■■■■■■■■■■
ver_tag = "ver1.02.01"
# FCRN-DepthPrediction-vmd clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/FCRN-DepthPrediction-vmd.git
# FCRN-DepthPrediction-vmd 识别深度模型下载
# 建立模型数据文件夹
! mkdir -p ./FCRN-DepthPrediction-vmd/tensorflow/data
# 下载模型数据并解压
! cd ./FCRN-DepthPrediction-vmd/tensorflow/data && wget -c "http://campar.in.tum.de/files/rupprecht/depthpred/NYU_FCRN-checkpoint.zip" && unzip NYU_FCRN-checkpoint.zip
# 3d-pose-baseline-vmd clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/3d-pose-baseline-vmd.git
# 3d-pose-baseline-vmd Human3.6M 模型数据DL
# 建立Human3.6M模型数据文件夹
! mkdir -p ./3d-pose-baseline-vmd/data/h36m
# 下载Human3.6M模型数据并解压
file_id = "1W5WoWpCcJvGm4CHoUhfIB0dgXBDCEHHq"
file_name = "h36m.zip"
! cd ./ && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./ && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./ && unzip h36m.zip
! mv ./h36m ./3d-pose-baseline-vmd/data/
# 3d-pose-baseline-vmd 训练数据
# 3d-pose-baseline学习数据文件夹
! mkdir -p ./3d-pose-baseline-vmd/experiments
# 下载3d-pose-baseline训练后的数据
file_id = "1v7ccpms3ZR8ExWWwVfcSpjMsGscDYH7_"
file_name = "experiments.zip"
! cd ./3d-pose-baseline-vmd && curl -sc ./cookie "https://drive.google.com/uc?export=download&id=$file_id" > /dev/null
code = "$(awk '/_warning_/ {print $NF}' ./cookie)"
! cd ./3d-pose-baseline-vmd && curl -Lb ./cookie "https://drive.google.com/uc?export=download&confirm=$code&id=$file_id" -o "$file_name"
! cd ./3d-pose-baseline-vmd && unzip experiments.zip
# VMD-3d-pose-baseline-multi clone
! git clone --depth 1 -b "$ver_tag" https://github.com/miu200521358/VMD-3d-pose-baseline-multi.git
# 安装VMD-3d-pose-baseline-multi 依赖库
! sudo apt-get install python3-pyqt5
! sudo apt-get install pyqt5-dev-tools
! sudo apt-get install qttools5-dev-tools
#安装编码器
! sudo apt-get install mkvtoolnix
init_elapsed_time = (time.time() - init_start_time) / 60
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "■■所有初始化均已完成"
! echo "■■"
! echo "■■处理时间:" "$init_elapsed_time" "分"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "Openpose执行结果"
! ls -l /content/openpose/output
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown 执行函数初始化
#@markdown ■■■■■■■■■■■■■■■■■■
import os
import cv2
import datetime
import time
import datetime
import cv2
import shutil
import glob
from google.colab import files
static_number_people_max = 1
static_frame_first = 0
static_end_frame_no = -1
static_reverse_specific = ""
static_order_specific = ""
static_born_model_csv = "born/animasa_miku_born.csv"
static_is_ik = 1
static_heel_position = 0.0
static_center_z_scale = 1
static_smooth_times = 1
static_threshold_pos = 0.5
static_threshold_rot = 3
static_src_input_video = ""
static_input_video = ""
#执行文件夹
openpose_path = "/content/openpose"
#输出文件夹
base_path = "/content/output"
output_json = "/content/output/json"
output_openpose_avi = "/content/output/openpose.avi"
now_str = ""
depth_dir_path = ""
drive_dir_path = ""
def video_hander( input_video):
global base_path
print("视频名称: ", os.path.basename(input_video))
print("视频大小: ", os.path.getsize(input_video))
video = cv2.VideoCapture(input_video)
# 宽
W = video.get(cv2.CAP_PROP_FRAME_WIDTH)
# 高
H = video.get(cv2.CAP_PROP_FRAME_HEIGHT)
# 总帧数
count = video.get(cv2.CAP_PROP_FRAME_COUNT)
# fps
fps = video.get(cv2.CAP_PROP_FPS)
print("宽: {0}, 高: {1}, 总帧数: {2}, fps: {3}".format(W, H, count, fps))
width = 1280
height = 720
if W != 1280 or (fps != 30 and fps != 60):
print("重新编码,因为大小或fps不在范围: "+ input_video)
# 縮尺
scale = width / W
# 高さ
height = int(H * scale)
# 出力ファイルパス
out_name = 'recode_{0}.mp4'.format("{0:%Y%m%d_%H%M%S}".format(datetime.datetime.now()))
out_path = '{0}/{1}'.format(base_path, out_name)
# try:
# fourcc = cv2.VideoWriter_fourcc(*"MP4V")
# out = cv2.VideoWriter(out_path, fourcc, 30.0, (width, height), True)
# # 入力ファイル
# cap = cv2.VideoCapture(input_video)
# while(cap.isOpened()):
# # 動画から1枚キャプチャして読み込む
# flag, frame = cap.read() # Capture frame-by-frame
# # 動画が終わっていたら終了
# if flag == False:
# break
# # 縮小
# output_frame = cv2.resize(frame, (width, height))
# # 出力
# out.write(output_frame)
# # 終わったら開放
# out.release()
# except Exception as e:
# print("重新编码失败", e)
# cap.release()
# cv2.destroyAllWindows()
# ! mkvmerge --default-duration 0:30fps --fix-bitstream-timing-information 0 "$input_video" -o temp-video.mkv
# ! ffmpeg -i temp-video.mkv -c:v copy side_video.mkv
# ! ffmpeg -i side_video.mkv -vf scale=1280:720 "$out_path"
! ffmpeg -i "$input_video" -qscale 0 -r 30 -y -vf scale=1280:720 "$out_path"
print('MMD重新生成MP4文件成功', out_path)
input_video_name = out_name
# 入力動画ファイル再設定
input_video = base_path + "/"+ input_video_name
video = cv2.VideoCapture(input_video)
# 幅
W = video.get(cv2.CAP_PROP_FRAME_WIDTH)
# 高さ
H = video.get(cv2.CAP_PROP_FRAME_HEIGHT)
# 総フレーム数
count = video.get(cv2.CAP_PROP_FRAME_COUNT)
# fps
fps = video.get(cv2.CAP_PROP_FPS)
print("【重新生成】宽: {0}, 高: {1}, 总帧数: {2}, fps: {3}, 名字: {4}".format(W, H, count, fps,input_video_name))
return input_video
def run_openpose(input_video,number_people_max,frame_first):
#建立临时文件夹
! mkdir -p "$output_json"
#开始执行
! cd "$openpose_path" && ./build/examples/openpose/openpose.bin --video "$input_video" --display 0 --model_pose COCO --write_json "$output_json" --write_video "$output_openpose_avi" --frame_first "$frame_first" --number_people_max "$number_people_max"
def run_fcrn_depth(input_video,end_frame_no,reverse_specific,order_specific):
global now_str,depth_dir_path,drive_dir_path
now_str = "{0:%Y%m%d_%H%M%S}".format(datetime.datetime.now())
! cd FCRN-DepthPrediction-vmd && python tensorflow/predict_video.py --model_path tensorflow/data/NYU_FCRN.ckpt --video_path "$input_video" --json_path "$output_json" --interval 10 --reverse_specific "$reverse_specific" --order_specific "$order_specific" --verbose 1 --now "$now_str" --avi_output "yes" --number_people_max "$number_people_max" --end_frame_no "$end_frame_no"
# 深度結果コピー
depth_dir_path = output_json + "_" + now_str + "_depth"
drive_dir_path = base_path + "/" + now_str
! mkdir -p "$drive_dir_path"
if os.path.exists( depth_dir_path + "/error.txt"):
# 发生错误
! cp "$depth_dir_path"/error.txt "$drive_dir_path"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "■■由于发生错误,处理被中断。"
! echo "■■"
! echo "■■■■■■■■■■■■■■■■■■■■■■■■"
! echo "$drive_dir_path" "请检查 error.txt 的内容。"
else:
! cp "$depth_dir_path"/*.avi "$drive_dir_path"
! cp "$depth_dir_path"/message.log "$drive_dir_path"
! cp "$depth_dir_path"/reverse_specific.txt "$drive_dir_path"
! cp "$depth_dir_path"/order_specific.txt "$drive_dir_path"
for i in range(1, number_people_max+1):
! echo ------------------------------------------
! echo 3d-pose-baseline-vmd ["$i"]
! echo ------------------------------------------
target_name = "_" + now_str + "_idx0" + str(i)
target_dir = output_json + target_name
!cd ./3d-pose-baseline-vmd && python src/openpose_3dpose_sandbox_vmd.py --camera_frame --residual --batch_norm --dropout 0.5 --max_norm --evaluateActionWise --use_sh --epochs 200 --load 4874200 --gif_fps 30 --verbose 1 --openpose "$target_dir" --person_idx 1
def run_3d_to_vmd(number_people_max,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot):
global now_str,depth_dir_path,drive_dir_path
for i in range(1, number_people_max+1):
target_name = "_" + now_str + "_idx0" + str(i)
target_dir = output_json + target_name
for f in glob.glob(target_dir +"/*.vmd"):
! rm "$f"
! cd ./VMD-3d-pose-baseline-multi && python applications/pos2vmd_multi.py -v 2 -t "$target_dir" -b "$born_model_csv" -c 30 -z "$center_z_scale" -s "$smooth_times" -p "$threshold_pos" -r "$threshold_rot" -k "$is_ik" -e "$heel_position"
# INDEX別結果コピー
idx_dir_path = drive_dir_path + "/idx0" + str(i)
! mkdir -p "$idx_dir_path"
# 日本語対策でpythonコピー
for f in glob.glob(target_dir +"/*.vmd"):
shutil.copy(f, idx_dir_path)
print(f)
files.download(f)
! cp "$target_dir"/pos.txt "$idx_dir_path"
! cp "$target_dir"/start_frame.txt "$idx_dir_path"
def run_mmd(input_video,number_people_max,frame_first,end_frame_no,reverse_specific,order_specific,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot):
global static_input_video,static_number_people_max ,static_frame_first ,static_end_frame_no,static_reverse_specific ,static_order_specific,static_born_model_csv
global static_is_ik,static_heel_position ,static_center_z_scale ,static_smooth_times ,static_threshold_pos ,static_threshold_rot
global base_path,static_src_input_video
start_time = time.time()
video_check= False
openpose_check = False
Fcrn_depth_check = False
pose_to_vmd_check = False
#源文件对比
if static_src_input_video != input_video:
video_check = True
openpose_check = True
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_number_people_max != number_people_max) or (static_frame_first != frame_first):
openpose_check = True
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_end_frame_no != end_frame_no) or (static_reverse_specific != reverse_specific) or (static_order_specific != order_specific):
Fcrn_depth_check = True
pose_to_vmd_check = True
if (static_born_model_csv != born_model_csv) or (static_is_ik != is_ik) or (static_heel_position != heel_position) or (static_center_z_scale != center_z_scale) or \
(static_smooth_times != smooth_times) or (static_threshold_pos != threshold_pos) or (static_threshold_rot != threshold_rot):
pose_to_vmd_check = True
#因为视频源文件重置,所以如果无修改需要重命名文件
if video_check:
! rm -rf "$base_path"
! mkdir -p "$base_path"
static_src_input_video = input_video
input_video = video_hander(input_video)
static_input_video = input_video
else:
input_video = static_input_video
if openpose_check:
run_openpose(input_video,number_people_max,frame_first)
static_number_people_max = number_people_max
static_frame_first = frame_first
if Fcrn_depth_check:
run_fcrn_depth(input_video,end_frame_no,reverse_specific,order_specific)
static_end_frame_no = end_frame_no
static_reverse_specific = reverse_specific
static_order_specific = order_specific
if pose_to_vmd_check:
run_3d_to_vmd(number_people_max,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot)
static_born_model_csv = born_model_csv
static_is_ik = is_ik
static_heel_position = heel_position
static_center_z_scale = center_z_scale
static_smooth_times = smooth_times
static_threshold_pos = threshold_pos
static_threshold_rot = threshold_rot
elapsed_time = (time.time() - start_time) / 60
print( "■■■■■■■■■■■■■■■■■■■■■■■■")
print( "■■所有处理完成")
print( "■■")
print( "■■处理時間:" + str(elapsed_time)+ "分")
print( "■■■■■■■■■■■■■■■■■■■■■■■■")
print( "")
print( "MMD自动跟踪执行结果")
print( base_path)
! ls -l "$base_path"
#@markdown ■■■■■■■■■■■■■■■■■■
#@markdown GO GO GO GO 执行本单元格,上传视频
#@markdown ■■■■■■■■■■■■■■■■■■
from google.colab import files
#@markdown ---
#@markdown ### 输入视频名称
#@markdown 可以选择手动拖入视频到文件中(比较快),然后输入视频文件名,或者直接运行,不输入文件名直接本地上传
input_video = "" #@param {type: "string"}
if input_video == "":
uploaded = files.upload()
for fn in uploaded.keys():
print('User uploaded file "{name}" with length {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
input_video = fn
input_video = "/content/" + input_video
print("本次执行的转化视频文件名为: "+input_video)
#@markdown 输入用于跟踪图像的参数并执行单元。
#@markdown ---
#@markdown ### 【O】视频中的最大人数
#@markdown 请输入您希望从视频中获得的人数。
#@markdown 请与视频中人数尽量保持一致
number_people_max = 1#@param {type: "number"}
#@markdown ---
#@markdown ### 【O】要从第几帧开始分析
#@markdown 输入帧号以开始分析。(从0开始)
#@markdown 请指定在视频中显示所有人的第一帧,默认为0即可,除非你需要跳过某些片段(例如片头)。
frame_first = 0 #@param {type: "number"}
#@markdown ---
#@markdown ### 【F】要从第几帧结束
#@markdown 请输入要从哪一帧结束
#@markdown (从0开始)在“FCRN-DepthPrediction-vmd”中调整反向或顺序时,可以完成过程并查看结果,默认为-1 表示执行到最后
end_frame_no = -1 #@param {type: "number"}
#@markdown ---
#@markdown ### 【F】反转数据表
#@markdown 指定由Openpose反转的帧号(从0开始),人员INDEX顺序和反转的内容。
#@markdown 按照Openpose在 0F 识别的顺序,将INDEX分配为0,1,...。
#@markdown 格式: [{帧号}: 用于指定反转的人INDEX, {反转内容}]
#@markdown {反转内容}: R: 整体身体反转, U:上半身反转, L: 下半身反转, N: 无反转
#@markdown 例如:[10:1,R] 整个人在第10帧中反转第一个人。在message.log中会记录以上述格式输出内容
#@markdown 因此请参考与[10:1,R][30:0,U],中一样,可以在括号中指定多个项目 ps(不要带有中文标点符号))
reverse_specific = "" #@param {type: "string"}
#@markdown ---
#@markdown ### 【F】输出颜色(仅参考,如果多人时,某个人序号跟别人交换或者错误,可以用此项修改)
#@markdown 请在多人轨迹中的交点之后指定人索引顺序。如果要跟踪一个人,可以将其留为空白。
#@markdown 按照Openpose在0F时识别的顺序分配0、1和INDEX。格式:[<帧号>:第几个人的索引,第几个人的索引,…]示例)[10:1,0]…第帧10是从左数第1人按第0个人的顺序对其进行排序。
#@markdown message.log包含以上述格式输出的顺序,因此请参考它。可以在括号中指定多个项目,例如[10:1,0] [30:0,1]。在output_XXX.avi中,按照估计顺序为人们分配了颜色。身体的右半部分为红色,左半部分为以下颜色。
#@markdown 0:绿色,1:蓝色,2:白色,3:黄色,4:桃红色,5:浅蓝色,6:深绿色,7:深蓝色,8:灰色,9:深黄色,10:深桃红色,11:深浅蓝色
order_specific = "" #@param {type: "string"}
#@markdown ---
#@markdown ### 【V】骨骼结构CSV文件
#@markdown 选择或输入跟踪目标模型的骨骼结构CSV文件的路径。请将csv文件上传到Google云端硬盘的“ autotrace”文件夹。
#@markdown 您可以选择 "Animasa-Miku" 和 "Animasa-Miku semi-standard", 也可以输入任何模型的骨骼结构CSV文件
#@markdown 如果要输入任何模型骨骼结构CSV文件, 请将csv文件上传到Google云端硬盘的 "autotrace" 文件夹下
#@markdown 然后请输入「/gdrive/My Drive/autotrace/[csv file name]」
born_model_csv = "born/\u3042\u306B\u307E\u3055\u5F0F\u30DF\u30AF\u6E96\u6A19\u6E96\u30DC\u30FC\u30F3.csv" #@param ["born/animasa_miku_born.csv", "born/animasa_miku_semi_standard_born.csv"] {allow-input: true}
#@markdown ---
#@markdown ### 【V】是否使用IK输出
#@markdown 选择以IK输出,yes或no
#@markdown 如果输入no,则以输出FK
ik_flag = "yes" #@param ['yes', 'no']
is_ik = 1 if ik_flag == "yes" else 0
#@markdown ---
#@markdown ### 【V】脚与地面位置校正
#@markdown 请输入数值的鞋跟的Y轴校正值(可以为小数)
#@markdown 输入负值会接近地面,输入正值会远离地面。
#@markdown 尽管会自动在某种程度上自动校正,但如果无法校正,请进行设置。
heel_position = 0.0 #@param {type: "number"}
#@markdown ---
#@markdown ### 【V】Z中心放大倍率
#@markdown 以将放大倍数应用到Z轴中心移动(可以是小数)
#@markdown 值越小,中心Z移动的宽度越小
#@markdown 输入0时,不进行Z轴中心移动。
center_z_scale = 2#@param {type: "number"}
#@markdown ---
#@markdown ### 【V】平滑频率
#@markdown 指定运动的平滑频率
#@markdown 请仅输入1或更大的整数
#@markdown 频率越大,频率越平滑。(行为幅度会变小)
smooth_times = 1#@param {type: "number"}
#@markdown ---
#@markdown ### 【V】移动稀疏量 (低于该阀值的运动宽度,不会进行输出,防抖动)
#@markdown 用数值(允许小数)指定用于稀疏移动(IK /中心)的移动量
#@markdown 如果在指定范围内有移动,则将稀疏。如果移动抽取量设置为0,则不执行抽取。
#@markdown 当移动稀疏量设置为0时,不进行稀疏。
threshold_pos = 0.3 #@param {type: "number"}
#@markdown ---
#@markdown ### 【V】旋转稀疏角 (低于该阀值的运动角度,则不会进行输出)
#@markdown 指定用于稀疏旋转键的角度(0到180度的十进制数)
#@markdown 如果在指定角度范围内有旋转,则稀疏旋转键。
threshold_rot = 3#@param {type: "number"}
print(" 【O】Maximum number of people in the video: "+str(number_people_max))
print(" 【O】Frame number to start analysis: "+str(frame_first))
print(" 【F】Frame number to finish analysis: "+str(end_frame_no))
print(" 【F】Reverse specification list: "+str(reverse_specific))
print(" 【F】Ordered list: "+str(order_specific))
print(" 【V】Bone structure CSV file: "+str(born_model_csv))
print(" 【V】Whether to output with IK: "+str(ik_flag))
print(" 【V】Heel position correction: "+str(heel_position))
print(" 【V】Center Z moving magnification: "+str(center_z_scale))
print(" 【V】Smoothing frequency: "+str(smooth_times))
print(" 【V】Movement key thinning amount: "+str(threshold_pos))
print(" 【V】Rotating Key Culling Angle: "+str(threshold_rot))
print("")
print("If the above is correct, please proceed to the next.")
#input_video = "/content/openpose/examples/media/video.avi"
run_mmd(input_video,number_people_max,frame_first,end_frame_no,reverse_specific,order_specific,born_model_csv,is_ik,heel_position,center_z_scale,smooth_times,threshold_pos,threshold_rot)
```
# License许可
发布和分发MMD自动跟踪的结果时,请确保检查许可证。Unity也是如此。
如果您能列出您的许可证,我将不胜感激。
[MMD运动跟踪自动化套件许可证](https://ch.nicovideo.jp/miu200521358/blomaga/ar1686913)
原作者:Twitter miu200521358
修改与优化:B站 妖风瑟瑟
|
github_jupyter
|
```
## Advanced Course in Machine Learning
## Week 4
## Exercise 2 / Probabilistic PCA
import numpy as np
import scipy
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.animation as animation
from numpy import linalg as LA
sns.set_style("darkgrid")
def build_dataset(N, D, K, sigma=1):
x = np.zeros((D, N))
z = np.random.normal(0.0, 1.0, size=(K, N))
# Create a w with random values
w = np.random.normal(0.0, sigma**2, size=(D, K))
mean = np.dot(w, z)
for d in range(D):
for n in range(N):
x[d, n] = np.random.normal(mean[d, n], sigma**2)
print("True principal axes:")
print(w)
return x, mean, w, z
N = 5000 # number of data points
D = 2 # data dimensionality
K = 1 # latent dimensionality
sigma = 1.0
x, mean, w, z = build_dataset(N, D, K, sigma)
print(z)
print(w)
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
origin = [0], [0] # origin point
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA, generated z')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=1, label='W')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='upper right')
plt.title('Probabilistic PCA, generated z')
plt.show()
print(x)
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5)
#plt.axis([-5, 5, -5, 5])
plt.xlabel('x')
plt.ylabel('y')
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
#Plot probability density contours
sns.kdeplot(x[0, :], x[1, :], n_levels=3, color='purple')
plt.title('Probabilistic PCA, generated x')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5, label='X')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA')
plt.show()
plt.figure(num=None, figsize=(8, 6), dpi=100, facecolor='w', edgecolor='k')
sns.scatterplot(x[0, :], x[1, :], color='orange', alpha=0.5, label='X')
sns.scatterplot(z[0, :], 0, alpha=0.5, label='z')
sns.scatterplot(mean[0, :], mean[1, :], color='red', alpha=0.5, label='Wz')
origin = [0], [0] # origin point
#Plot the principal axis
plt.quiver(*origin, w[0,0], w[1,0], color=['g'], scale=10, label='W')
#Plot probability density contours
sns.kdeplot(x[0, :], x[1, :], n_levels=6, color='purple')
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='lower right')
plt.title('Probabilistic PCA')
plt.show()
```
def main():
fig = plt.figure()
scat = plt.scatter(mean[0, :], color='red', alpha=0.5, label='Wz')
ani = animation.FuncAnimation(fig, update_plot, frames=xrange(N),
fargs=(scat))
plt.show()
def update_plot(i, scat):
scat.set_array(data[i])
return scat,
main()
|
github_jupyter
|
```
%matplotlib inline
import pandas as pd
import cv2
import numpy as np
from matplotlib import pyplot as plt
df = pd.read_csv("data/22800_SELECT_t___FROM_data_data_t.csv",header=None,index_col=0)
df = df.rename(columns={0:"no", 1: "CAPTDATA", 2: "CAPTIMAGE",3: "timestamp"})
df.info()
df.sample(5)
def alpha_to_gray(img):
alpha_channel = img[:, :, 3]
_, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
color = img[:, :, :3]
img = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def preprocess(data):
data = bytes.fromhex(data[2:])
img = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
img = alpha_to_gray(img)
kernel = np.ones((3, 3), np.uint8)
img = cv2.dilate(img, kernel, iterations=1)
img = cv2.medianBlur(img, 3)
kernel = np.ones((4, 4), np.uint8)
img = cv2.erode(img, kernel, iterations=1)
# plt.imshow(img)
return img
df["IMAGE"] = df["CAPTIMAGE"].apply(preprocess)
def bounding(gray):
# data = bytes.fromhex(df["CAPTIMAGE"][1][2:])
# image = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
# alpha_channel = image[:, :, 3]
# _, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
# color = image[:, :, :3]
# src = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
ret, binary = cv2.threshold(gray, 127, 255, cv2.THRESH_BINARY)
binary = cv2.bitwise_not(binary)
contours, hierachy = cv2.findContours(binary, cv2.RETR_EXTERNAL , cv2.CHAIN_APPROX_NONE)
ans = []
for h, tcnt in enumerate(contours):
x,y,w,h = cv2.boundingRect(tcnt)
if h < 25:
continue
if 40 < w < 100: # 2개가 붙어 있는 경우
ans.append([x,y,w//2,h])
ans.append([x+(w//2),y,w//2,h])
continue
if 100 <= w < 170:
ans.append([x,y,w//3,h])
ans.append([x+(w//3),y,w//3,h])
ans.append([x+(2*w//3),y,w//3,h])
# cv2.rectangle(src,(x,y),(x+w,y+h),(255,0,0),1)
ans.append([x,y,w,h])
return ans
# cv2.destroyAllWindows()
df["bounding"] = df["IMAGE"].apply(bounding)
def draw_bounding(idx):
CAPTIMAGE = df["CAPTIMAGE"][idx]
bounding = df["bounding"][idx]
data = bytes.fromhex(CAPTIMAGE[2:])
image = cv2.imdecode( np.asarray(bytearray(data), dtype=np.uint8), cv2.IMREAD_UNCHANGED )
alpha_channel = image[:, :, 3]
_, mask = cv2.threshold(alpha_channel, 128, 255, cv2.THRESH_BINARY) # binarize mask
color = image[:, :, :3]
src = cv2.bitwise_not(cv2.bitwise_not(color, mask=mask))
for x,y,w,h in bounding:
# print(x,y,w,h)
cv2.rectangle(src,(x,y),(x+w,y+h),(255,0,0),1)
return src
import random
nrows = 4
ncols = 4
fig, axes = plt.subplots(nrows=nrows, ncols=ncols)
fig.set_size_inches((16, 6))
for i in range(nrows):
for j in range(ncols):
idx = random.randrange(20,22800)
axes[i][j].set_title(str(idx))
axes[i][j].imshow(draw_bounding(idx))
fig.tight_layout()
plt.savefig('sample.png')
plt.show()
charImg = []
for idx in df.index:
IMAGE = df["IMAGE"][idx]
bounding = df["bounding"][idx]
for x,y,w,h in bounding:
newImg = IMAGE[y:y+h,x:x+w]
newImg = cv2.resize(newImg, dsize=(41, 38), interpolation=cv2.INTER_NEAREST)
charImg.append(newImg/255.0)
# cast to numpy arrays
trainingImages = np.asarray(charImg)
# reshape img array to vector
def reshape_image(img):
return np.reshape(img,len(img)*len(img[0]))
img_reshape = np.zeros((len(trainingImages),len(trainingImages[0])*len(trainingImages[0][0])))
for i in range(0,len(trainingImages)):
img_reshape[i] = reshape_image(trainingImages[i])
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
import seaborn as sns
# create model and prediction
model = KMeans(n_clusters=40,algorithm='auto')
model.fit(img_reshape)
predict = pd.DataFrame(model.predict(img_reshape))
predict.columns=['predict']
import pickle
pickle.dump(model, open("KMeans_40_22800.pkl", "wb"))
import pickle
model = pickle.load(open("KMeans_40_22800.pkl", "rb"))
predict = pd.DataFrame(model.predict(img_reshape))
predict.columns=['predict']
import random
from tqdm import tqdm
r = pd.concat([pd.DataFrame(img_reshape),predict],axis=1)
!rm -rf res_40
!mkdir res_40
nrows = 4
ncols = 10
fig, axes = plt.subplots(nrows=nrows, ncols=ncols)
fig.set_size_inches((16, 6))
for j in tqdm(range(40)):
i = 0
nSample = min(nrows * ncols,len(r[r["predict"] == j]))
for idx in r[r["predict"] == j].sample(nSample).index:
axes[i // ncols][i % ncols].set_title(str(idx))
axes[i // ncols][i % ncols].imshow(trainingImages[idx])
i+=1
fig.tight_layout()
plt.savefig('res_40/sample_' + str(j) + '.png')
```
98 95 92 222 255
|
github_jupyter
|
```
# Import and create a new SQLContext
from pyspark.sql import SQLContext
sqlContext = SQLContext(sc)
# Read the country CSV file into an RDD.
country_lines = sc.textFile('file:///home/ubuntu/work/notebooks/UCSD/big-data-3/final-project/country-list.csv')
country_lines.collect()
# Convert each line into a pair of words
country_lines.map(lambda a: a.split(",")).collect()
# Convert each pair of words into a tuple
country_tuples = country_lines.map(lambda a: (a.split(",")[0].lower(), a.split(",")[1]))
# Create the DataFrame, look at schema and contents
countryDF = sqlContext.createDataFrame(country_tuples, ["country", "code"])
countryDF.printSchema()
countryDF.take(3)
# Read tweets CSV file into RDD of lines
tweets = sc.textFile('file:///home/ubuntu/work/notebooks/UCSD/big-data-3/final-project/tweets.csv')
tweets.count()
# Clean the data: some tweets are empty. Remove the empty tweets using filter()
filtered_tweets = tweets.filter(lambda a: len(a) > 0)
filtered_tweets.count()
# Perform WordCount on the cleaned tweet texts. (note: this is several lines.)
word_counts = filtered_tweets.flatMap(lambda a: a.split(" ")) \
.map(lambda word: (word.lower(), 1)) \
.reduceByKey(lambda a, b: a + b)
from pyspark.sql import HiveContext
from pyspark.sql.types import *
# sc is an existing SparkContext.
sqlContext = HiveContext(sc)
schemaString = "word count"
fields = [StructField(field_name, StringType(), True) for field_name in schemaString.split()]
schema = StructType(fields)
# Create the DataFrame of tweet word counts
tweetsDF = sqlContext.createDataFrame(word_counts, schema)
tweetsDF.printSchema()
tweetsDF.count()
# Join the country and tweet DataFrames (on the appropriate column)
joined = countryDF.join(tweetsDF, countryDF.country == tweetsDF.word)
joined.take(5)
joined.show()
# Question 1: number of distinct countries mentioned
distinct_countries = joined.select("country").distinct()
distinct_countries.show(100)
# Question 2: number of countries mentioned in tweets.
from pyspark.sql.functions import sum
from pyspark.sql import SparkSession
from pyspark.sql import Row
countries_count = joined.groupBy("country")
joined.createOrReplaceTempView("records")
spark.sql("SELECT country, count(*) count1 FROM records group by country order by count1 desc, country asc").show(100)
# Table 1: top three countries and their counts.
from pyspark.sql.functions import desc
from pyspark.sql.functions import col
top_3 = joined.sort(col("count").desc())
top_3.show()
# Table 2: counts for Wales, Iceland, and Japan.
```
|
github_jupyter
|
# Datafaucet
Datafaucet is a productivity framework for ETL, ML application. Simplifying some of the common activities which are typical in Data pipeline such as project scaffolding, data ingesting, start schema generation, forecasting etc.
```
import datafaucet as dfc
```
## Loading and Saving Data
```
dfc.project.load()
query = """
SELECT
p.payment_date,
p.amount,
p.rental_id,
p.staff_id,
c.*
FROM payment p
INNER JOIN customer c
ON p.customer_id = c.customer_id;
"""
df = dfc.load(query, 'pagila')
```
#### Select cols
```
df.cols.find('id').columns
df.cols.find(by_type='string').columns
df.cols.find(by_func=lambda x: x.startswith('st')).columns
df.cols.find('^st').columns
```
#### Collect data, oriented by rows or cols
```
df.cols.find(by_type='numeric').rows.collect(3)
df.cols.find(by_type='string').collect(3)
df.cols.find('name', 'date').data.collect(3)
```
#### Get just one row or column
```
df.cols.find('active', 'amount', 'name').one()
df.cols.find('active', 'amount', 'name').rows.one()
```
#### Grid view
```
df.cols.find('amount', 'id', 'name').data.grid(5)
```
#### Data Exploration
```
df.cols.find('amount', 'id', 'name').data.facets()
```
#### Rename columns
```
df.cols.find(by_type='timestamp').rename('new_', '***').columns
# to do
# df.cols.rename(transform=['unidecode', 'alnum', 'alpha', 'num', 'lower', 'trim', 'squeeze', 'slice', tr("abc", "_", mode='')'])
# df.cols.rename(transform=['unidecode', 'alnum', 'lower', 'trim("_")', 'squeeze("_")'])
# as a dictionary
mapping = {
'staff_id': 'foo',
'first_name': 'bar',
'email': 'qux',
'active':'active'
}
# or as a list of 2-tuples
mapping = [
('staff_id','foo'),
('first_name','bar'),
'active'
]
dict(zip(df.columns, df.cols.rename('new_', '***', mapping).columns))
```
#### Drop multiple columns
```
df.cols.find('id').drop().rows.collect(3)
```
#### Apply to multiple columns
```
from pyspark.sql import functions as F
(df
.cols.find(by_type='string').lower()
.cols.get('email').split('@')
.cols.get('email').expand(2)
.cols.find('name', 'email')
.rows.collect(3)
)
```
### Aggregations
```
from datafaucet.spark import aggregations as A
df.cols.find('amount', '^st.*id', 'first_name').agg(A.all).cols.collect(10)
```
##### group by a set of columns
```
df.cols.find('amount').groupby('staff_id', 'store_id').agg(A.all).cols.collect(4)
```
#### Aggregate specific metrics
```
# by function
df.cols.get('amount', 'active').groupby('customer_id').agg({'count':F.count, 'sum': F.sum}).rows.collect(10)
# or by alias
df.cols.get('amount', 'active').groupby('customer_id').agg('count','sum').rows.collect(10)
# or a mix of the two
df.cols.get('amount', 'active').groupby('customer_id').agg('count',{'sum': F.sum}).rows.collect(10)
```
#### Featurize specific metrics in a single row
```
(df
.cols.get('amount', 'active')
.groupby('customer_id', 'store_id')
.featurize({'count':A.count, 'sum':A.sum, 'avg':A.avg})
.rows.collect(10)
)
# todo:
# different features per different column
```
#### Plot dataset statistics
```
df.data.summary()
from bokeh.io import output_notebook
output_notebook()
from bokeh.plotting import figure, show, output_file
p = figure(plot_width=400, plot_height=400)
p.hbar(y=[1, 2, 3], height=0.5, left=0,
right=[1.2, 2.5, 3.7], color="navy")
show(p)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="whitegrid")
# Initialize the matplotlib figure
f, ax = plt.subplots(figsize=(6, 6))
# Load the example car crash dataset
crashes = sns.load_dataset("car_crashes").sort_values("total", ascending=False)[:10]
# Plot the total crashes
sns.set_color_codes("pastel")
sns.barplot(x="total", y="abbrev", data=crashes,
label="Total", color="b")
# Plot the crashes where alcohol was involved
sns.set_color_codes("muted")
sns.barplot(x="alcohol", y="abbrev", data=crashes,
label="Alcohol-involved", color="b")
# Add a legend and informative axis label
ax.legend(ncol=2, loc="lower right", frameon=True)
ax.set(xlim=(0, 24), ylabel="",
xlabel="Automobile collisions per billion miles")
sns.despine(left=True, bottom=True)
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="white", palette="muted", color_codes=True)
# Generate a random univariate dataset
rs = np.random.RandomState(10)
d = rs.normal(size=100)
# Plot a simple histogram with binsize determined automatically
sns.distplot(d, hist=True, kde=True, rug=True, color="b");
import seaborn as sns
sns.set(style="ticks")
df = sns.load_dataset("iris")
sns.pairplot(df, hue="species")
from IPython.display import HTML
HTML('''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="jumbotron">
<h1 class="display-4">Hello, world!</h1>
<p class="lead">This is a simple hero unit, a simple jumbotron-style component for calling extra attention to featured content or information.</p>
<hr class="my-4">
<p>It uses utility classes for typography and spacing to space content out within the larger container.</p>
<a class="btn btn-primary btn-lg" href="#" role="button">Learn more</a>
</div>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="top" title="Tooltip on top">
Tooltip on top
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="right" title="Tooltip on right">
Tooltip on right
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="bottom" title="Tooltip on bottom">
Tooltip on bottom
</button>
<button type="button" class="btn btn-secondary" data-toggle="tooltip" data-placement="left" title="Tooltip on left">
Tooltip on left
</button>
<table class="table">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
</tr>
<tr>
<th scope="row">3</th>
<td>Larry</td>
<td>the Bird</td>
<td>@twitter</td>
</tr>
</tbody>
</table>
<span class="badge badge-primary">Primary</span>
<span class="badge badge-secondary">Secondary</span>
<span class="badge badge-success">Success</span>
<span class="badge badge-danger">Danger</span>
<span class="badge badge-warning">Warning</span>
<span class="badge badge-info">Info</span>
<span class="badge badge-light">Light</span>
<span class="badge badge-dark">Dark</span>
<table class="table table-sm" style="text-align:left">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">Primary</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%">Primary</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 55%">Primary</span></td>
</div>
</tr>
</tbody>
</table>
</div>''')
tbl = '''
<table class="table table-sm">
<thead>
<tr>
<th scope="col">#</th>
<th scope="col">First</th>
<th scope="col">Last</th>
<th scope="col">Handle</th>
<th scope="col">bar</th>
</tr>
</thead>
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
drp = '''
<div class="dropdown">
<button class="btn btn-secondary dropdown-toggle" type="button" id="dropdownMenuButton" data-toggle="dropdown" aria-haspopup="true" aria-expanded="false">
Dropdown button
</button>
<div class="dropdown-menu" aria-labelledby="dropdownMenuButton">
<a class="dropdown-item" href="#">Action</a>
<a class="dropdown-item" href="#">Another action</a>
<a class="dropdown-item" href="#">Something else here</a>
</div>
</div>'''
tabs = f'''
<nav>
<div class="nav nav-tabs" id="nav-tab" role="tablist">
<a class="nav-item nav-link active" id="nav-home-tab" data-toggle="tab" href="#nav-home" role="tab" aria-controls="nav-home" aria-selected="true">Home</a>
<a class="nav-item nav-link" id="nav-profile-tab" data-toggle="tab" href="#nav-profile" role="tab" aria-controls="nav-profile" aria-selected="false">Profile</a>
<a class="nav-item nav-link" id="nav-contact-tab" data-toggle="tab" href="#nav-contact" role="tab" aria-controls="nav-contact" aria-selected="false">Contact</a>
</div>
</nav>
<div class="tab-content" id="nav-tabContent">
<div class="tab-pane fade show active" id="nav-home" role="tabpanel" aria-labelledby="nav-home-tab">..jjj.</div>
<div class="tab-pane fade" id="nav-profile" role="tabpanel" aria-labelledby="nav-profile-tab">..kkk.</div>
<div class="tab-pane fade" id="nav-contact" role="tabpanel" aria-labelledby="nav-contact-tab">{tbl}</div>
</div>
'''
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<div class="container-fluid">
<div class="row">
<div class="col">
{drp}
</div>
<div class="col">
{tabs}
</div>
<div class="col">
{tbl}
</div>
</div>
</div>
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
from IPython.display import HTML
HTML(f'''
<!-- Bootstrap CSS -->
<link rel="stylesheet" href="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/css/bootstrap.min.css" crossorigin="anonymous">
<script src="https://stackpath.bootstrapcdn.com/bootstrap/4.1.3/js/bootstrap.bundle.min.js" crossorigin="anonymous" >
''')
d =df.cols.find('id', 'name').sample(10)
d.columns
tbl_head = '''
<thead>
<tr>
'''
tbl_head += '\n'.join([' <th scope="col">'+str(x)+'</th>' for x in d.columns])
tbl_head +='''
</tr>
</thead>
'''
print(tbl_head)
tbl_body = '''
<tbody>
<tr>
<th scope="row">1</th>
<td>Mark</td>
<td>Otto</td>
<td>@mdo</td>
<td class="text-left"><span class="badge badge-primary" style="width: 75%">75%</span></td>
</tr>
<tr>
<th scope="row">2</th>
<td>Jacob</td>
<td>Thornton</td>
<td>@fat</td>
<td class="text-left"><span class="badge badge-secondary" style="width: 25%" title="Tooltip on top">25%</span></td>
</tr>
<tr>
<th scope="row">3</th>
<td colspan="2">Larry the Bird</td>
<td>@twitter</td>
<td class="text-left"><span class="badge badge-warning" style="width: 0%">0%</span></td>
</tr>
</tbody>
</table>
'''
HTML(f'''
<!-- Bootstrap CSS -->
<div class="container-fluid">
<div class="row">
<div class="col">
<table class="table table-sm">
{tbl_head}
{tbl_body}
</table>
</div>
</div>
</div>
''')
# .rows.sample()
# .cols.select('name', 'id', 'amount')\
# .cols.apply(F.lower, 'name')\
# .cols.apply(F.floor, 'amount', output_prefix='_')\
# .cols.drop('^amount$')\
# .cols.rename()
# .cols.unicode()
.grid()
df = df.cols.select('name')
df = df.rows.overwrite([('Nhập mật', 'khẩu')])
df.columns
# .rows.overwrite(['Nhập mật', 'khẩu'])\
# .cols.apply(F.lower)\
# .grid()
# #withColumn('pippo', F.lower(F.col('first_name'))).grid()
import pandas as pd
df = pd.DataFrame({'lab':['A', 'B', 'C'], 'val':[10, 30, 20]})
df.plot.bar(x='lab', y='val', rot=0);
```
|
github_jupyter
|

https://www.kaggle.com/danofer/sarcasm
<div class="markdown-converter__text--rendered"><h3>Context</h3>
<p>This dataset contains 1.3 million Sarcastic comments from the Internet commentary website Reddit. The dataset was generated by scraping comments from Reddit (not by me :)) containing the <code>\s</code> ( sarcasm) tag. This tag is often used by Redditors to indicate that their comment is in jest and not meant to be taken seriously, and is generally a reliable indicator of sarcastic comment content.</p>
<h3>Content</h3>
<p>Data has balanced and imbalanced (i.e true distribution) versions. (True ratio is about 1:100). The
corpus has 1.3 million sarcastic statements, along with what they responded to as well as many non-sarcastic comments from the same source.</p>
<p>Labelled comments are in the <code>train-balanced-sarcasm.csv</code> file.</p>
<h3>Acknowledgements</h3>
<p>The data was gathered by: Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli for their article "<a href="https://arxiv.org/abs/1704.05579" rel="nofollow">A Large Self-Annotated Corpus for Sarcasm</a>". The data is hosted <a href="http://nlp.cs.princeton.edu/SARC/0.0/" rel="nofollow">here</a>.</p>
<p>Citation:</p>
<pre><code>@unpublished{SARC,
authors={Mikhail Khodak and Nikunj Saunshi and Kiran Vodrahalli},
title={A Large Self-Annotated Corpus for Sarcasm},
url={https://arxiv.org/abs/1704.05579},
year=2017
}
</code></pre>
<p><a href="http://nlp.cs.princeton.edu/SARC/0.0/readme.txt" rel="nofollow">Annotation of files in the original dataset: readme.txt</a>.</p>
<h3>Inspiration</h3>
<ul>
<li>Predicting sarcasm and relevant NLP features (e.g. subjective determinant, racism, conditionals, sentiment heavy words, "Internet Slang" and specific phrases). </li>
<li>Sarcasm vs Sentiment</li>
<li>Unusual linguistic features such as caps, italics, or elongated words. e.g., "Yeahhh, I'm sure THAT is the right answer".</li>
<li>Topics that people tend to react to sarcastically</li>
</ul></div>
```
import os
# Install java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
# Install pyspark
! pip install --ignore-installed pyspark==2.4.4
# Install Spark NLP
! pip install --ignore-installed spark-nlp
import sys
import time
import sparknlp
from pyspark.sql import SparkSession
packages = [
'JohnSnowLabs:spark-nlp: 2.5.5'
]
spark = SparkSession \
.builder \
.appName("ML SQL session") \
.config('spark.jars.packages', ','.join(packages)) \
.config('spark.executor.instances','2') \
.config("spark.executor.memory", "2g") \
.config("spark.driver.memory","16g") \
.getOrCreate()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
! wget -N https://s3.amazonaws.com/auxdata.johnsnowlabs.com/public/resources/en/sarcasm/train-balanced-sarcasm.csv -P /tmp
from pyspark.sql import SQLContext
sql = SQLContext(spark)
trainBalancedSarcasmDF = spark.read.option("header", True).option("inferSchema", True).csv("/tmp/train-balanced-sarcasm.csv")
trainBalancedSarcasmDF.printSchema()
# Let's create a temp view (table) for our SQL queries
trainBalancedSarcasmDF.createOrReplaceTempView('data')
sql.sql('SELECT COUNT(*) FROM data').collect()
sql.sql('select * from data limit 20').show()
sql.sql('select label,count(*) as cnt from data group by label order by cnt desc').show()
sql.sql('select count(*) from data where comment is null').collect()
df = sql.sql('select label,concat(parent_comment,"\n",comment) as comment from data where comment is not null and parent_comment is not null limit 100000')
print(type(df))
df.printSchema()
df.show()
from sparknlp.annotator import *
from sparknlp.common import *
from sparknlp.base import *
from pyspark.ml import Pipeline
document_assembler = DocumentAssembler() \
.setInputCol("comment") \
.setOutputCol("document")
sentence_detector = SentenceDetector() \
.setInputCols(["document"]) \
.setOutputCol("sentence") \
.setUseAbbreviations(True)
tokenizer = Tokenizer() \
.setInputCols(["sentence"]) \
.setOutputCol("token")
stemmer = Stemmer() \
.setInputCols(["token"]) \
.setOutputCol("stem")
normalizer = Normalizer() \
.setInputCols(["stem"]) \
.setOutputCol("normalized")
finisher = Finisher() \
.setInputCols(["normalized"]) \
.setOutputCols(["ntokens"]) \
.setOutputAsArray(True) \
.setCleanAnnotations(True)
nlp_pipeline = Pipeline(stages=[document_assembler, sentence_detector, tokenizer, stemmer, normalizer, finisher])
nlp_model = nlp_pipeline.fit(df)
processed = nlp_model.transform(df).persist()
processed.count()
processed.show()
train, test = processed.randomSplit(weights=[0.7, 0.3], seed=123)
print(train.count())
print(test.count())
from pyspark.ml import feature as spark_ft
stopWords = spark_ft.StopWordsRemover.loadDefaultStopWords('english')
sw_remover = spark_ft.StopWordsRemover(inputCol='ntokens', outputCol='clean_tokens', stopWords=stopWords)
tf = spark_ft.CountVectorizer(vocabSize=500, inputCol='clean_tokens', outputCol='tf')
idf = spark_ft.IDF(minDocFreq=5, inputCol='tf', outputCol='idf')
feature_pipeline = Pipeline(stages=[sw_remover, tf, idf])
feature_model = feature_pipeline.fit(train)
train_featurized = feature_model.transform(train).persist()
train_featurized.count()
train_featurized.show()
train_featurized.groupBy("label").count().show()
train_featurized.printSchema()
from pyspark.ml import classification as spark_cls
rf = spark_cls. RandomForestClassifier(labelCol="label", featuresCol="idf", numTrees=100)
model = rf.fit(train_featurized)
test_featurized = feature_model.transform(test)
preds = model.transform(test_featurized)
preds.show()
pred_df = preds.select('comment', 'label', 'prediction').toPandas()
pred_df.head()
import pandas as pd
from sklearn import metrics as skmetrics
pd.DataFrame(
data=skmetrics.confusion_matrix(pred_df['label'], pred_df['prediction']),
columns=['pred ' + l for l in ['0','1']],
index=['true ' + l for l in ['0','1']]
)
print(skmetrics.classification_report(pred_df['label'], pred_df['prediction'],
target_names=['0','1']))
spark.stop()
```
|
github_jupyter
|
```
# Copyright 2020 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
```
<img src="http://developer.download.nvidia.com/compute/machine-learning/frameworks/nvidia_logo.png" style="width: 90px; float: right;">
# Object Detection with TRTorch (SSD)
---
## Overview
In PyTorch 1.0, TorchScript was introduced as a method to separate your PyTorch model from Python, make it portable and optimizable.
TRTorch is a compiler that uses TensorRT (NVIDIA's Deep Learning Optimization SDK and Runtime) to optimize TorchScript code. It compiles standard TorchScript modules into ones that internally run with TensorRT optimizations.
TensorRT can take models from any major framework and specifically tune them to perform better on specific target hardware in the NVIDIA family, and TRTorch enables us to continue to remain in the PyTorch ecosystem whilst doing so. This allows us to leverage the great features in PyTorch, including module composability, its flexible tensor implementation, data loaders and more. TRTorch is available to use with both PyTorch and LibTorch.
To get more background information on this, we suggest the **lenet-getting-started** notebook as a primer for getting started with TRTorch.
### Learning objectives
This notebook demonstrates the steps for compiling a TorchScript module with TRTorch on a pretrained SSD network, and running it to test the speedup obtained.
## Contents
1. [Requirements](#1)
2. [SSD Overview](#2)
3. [Creating TorchScript modules](#3)
4. [Compiling with TRTorch](#4)
5. [Running Inference](#5)
6. [Measuring Speedup](#6)
7. [Conclusion](#7)
---
<a id="1"></a>
## 1. Requirements
Follow the steps in `notebooks/README` to prepare a Docker container, within which you can run this demo notebook.
In addition to that, run the following cell to obtain additional libraries specific to this demo.
```
# Known working versions
!pip install numpy==1.21.2 scipy==1.5.2 Pillow==6.2.0 scikit-image==0.17.2 matplotlib==3.3.0
```
---
<a id="2"></a>
## 2. SSD
### Single Shot MultiBox Detector model for object detection
_ | _
- | -
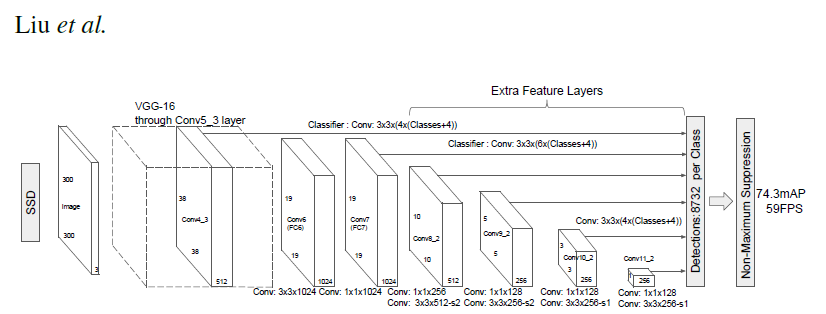 | 
PyTorch has a model repository called the PyTorch Hub, which is a source for high quality implementations of common models. We can get our SSD model pretrained on [COCO](https://cocodataset.org/#home) from there.
### Model Description
This SSD300 model is based on the
[SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper, which
describes SSD as “a method for detecting objects in images using a single deep neural network".
The input size is fixed to 300x300.
The main difference between this model and the one described in the paper is in the backbone.
Specifically, the VGG model is obsolete and is replaced by the ResNet-50 model.
From the
[Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012)
paper, the following enhancements were made to the backbone:
* The conv5_x, avgpool, fc and softmax layers were removed from the original classification model.
* All strides in conv4_x are set to 1x1.
The backbone is followed by 5 additional convolutional layers.
In addition to the convolutional layers, we attached 6 detection heads:
* The first detection head is attached to the last conv4_x layer.
* The other five detection heads are attached to the corresponding 5 additional layers.
Detector heads are similar to the ones referenced in the paper, however,
they are enhanced by additional BatchNorm layers after each convolution.
More information about this SSD model is available at Nvidia's "DeepLearningExamples" Github [here](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD).
```
import torch
torch.hub._validate_not_a_forked_repo=lambda a,b,c: True
# List of available models in PyTorch Hub from Nvidia/DeepLearningExamples
torch.hub.list('NVIDIA/DeepLearningExamples:torchhub')
# load SSD model pretrained on COCO from Torch Hub
precision = 'fp32'
ssd300 = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd', model_math=precision);
```
Setting `precision="fp16"` will load a checkpoint trained with mixed precision
into architecture enabling execution on Tensor Cores. Handling mixed precision data requires the Apex library.
### Sample Inference
We can now run inference on the model. This is demonstrated below using sample images from the COCO 2017 Validation set.
```
# Sample images from the COCO validation set
uris = [
'http://images.cocodataset.org/val2017/000000397133.jpg',
'http://images.cocodataset.org/val2017/000000037777.jpg',
'http://images.cocodataset.org/val2017/000000252219.jpg'
]
# For convenient and comprehensive formatting of input and output of the model, load a set of utility methods.
utils = torch.hub.load('NVIDIA/DeepLearningExamples:torchhub', 'nvidia_ssd_processing_utils')
# Format images to comply with the network input
inputs = [utils.prepare_input(uri) for uri in uris]
tensor = utils.prepare_tensor(inputs, False)
# The model was trained on COCO dataset, which we need to access in order to
# translate class IDs into object names.
classes_to_labels = utils.get_coco_object_dictionary()
# Next, we run object detection
model = ssd300.eval().to("cuda")
detections_batch = model(tensor)
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input = [utils.pick_best(results, 0.40) for results in results_per_input]
```
### Visualize results
```
from matplotlib import pyplot as plt
import matplotlib.patches as patches
# The utility plots the images and predicted bounding boxes (with confidence scores).
def plot_results(best_results):
for image_idx in range(len(best_results)):
fig, ax = plt.subplots(1)
# Show original, denormalized image...
image = inputs[image_idx] / 2 + 0.5
ax.imshow(image)
# ...with detections
bboxes, classes, confidences = best_results[image_idx]
for idx in range(len(bboxes)):
left, bot, right, top = bboxes[idx]
x, y, w, h = [val * 300 for val in [left, bot, right - left, top - bot]]
rect = patches.Rectangle((x, y), w, h, linewidth=1, edgecolor='r', facecolor='none')
ax.add_patch(rect)
ax.text(x, y, "{} {:.0f}%".format(classes_to_labels[classes[idx] - 1], confidences[idx]*100), bbox=dict(facecolor='white', alpha=0.5))
plt.show()
# Visualize results without TRTorch/TensorRT
plot_results(best_results_per_input)
```
### Benchmark utility
```
import time
import numpy as np
import torch.backends.cudnn as cudnn
cudnn.benchmark = True
# Helper function to benchmark the model
def benchmark(model, input_shape=(1024, 1, 32, 32), dtype='fp32', nwarmup=50, nruns=1000):
input_data = torch.randn(input_shape)
input_data = input_data.to("cuda")
if dtype=='fp16':
input_data = input_data.half()
print("Warm up ...")
with torch.no_grad():
for _ in range(nwarmup):
features = model(input_data)
torch.cuda.synchronize()
print("Start timing ...")
timings = []
with torch.no_grad():
for i in range(1, nruns+1):
start_time = time.time()
pred_loc, pred_label = model(input_data)
torch.cuda.synchronize()
end_time = time.time()
timings.append(end_time - start_time)
if i%10==0:
print('Iteration %d/%d, avg batch time %.2f ms'%(i, nruns, np.mean(timings)*1000))
print("Input shape:", input_data.size())
print("Output location prediction size:", pred_loc.size())
print("Output label prediction size:", pred_label.size())
print('Average batch time: %.2f ms'%(np.mean(timings)*1000))
```
We check how well the model performs **before** we use TRTorch/TensorRT
```
# Model benchmark without TRTorch/TensorRT
model = ssd300.eval().to("cuda")
benchmark(model, input_shape=(128, 3, 300, 300), nruns=100)
```
---
<a id="3"></a>
## 3. Creating TorchScript modules
To compile with TRTorch, the model must first be in **TorchScript**. TorchScript is a programming language included in PyTorch which removes the Python dependency normal PyTorch models have. This conversion is done via a JIT compiler which given a PyTorch Module will generate an equivalent TorchScript Module. There are two paths that can be used to generate TorchScript: **Tracing** and **Scripting**. <br>
- Tracing follows execution of PyTorch generating ops in TorchScript corresponding to what it sees. <br>
- Scripting does an analysis of the Python code and generates TorchScript, this allows the resulting graph to include control flow which tracing cannot do.
Tracing however due to its simplicity is more likely to compile successfully with TRTorch (though both systems are supported).
```
model = ssd300.eval().to("cuda")
traced_model = torch.jit.trace(model, [torch.randn((1,3,300,300)).to("cuda")])
```
If required, we can also save this model and use it independently of Python.
```
# This is just an example, and not required for the purposes of this demo
torch.jit.save(traced_model, "ssd_300_traced.jit.pt")
# Obtain the average time taken by a batch of input with Torchscript compiled modules
benchmark(traced_model, input_shape=(128, 3, 300, 300), nruns=100)
```
---
<a id="4"></a>
## 4. Compiling with TRTorch
TorchScript modules behave just like normal PyTorch modules and are intercompatible. From TorchScript we can now compile a TensorRT based module. This module will still be implemented in TorchScript but all the computation will be done in TensorRT.
```
import trtorch
# The compiled module will have precision as specified by "op_precision".
# Here, it will have FP16 precision.
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((3, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
```
---
<a id="5"></a>
## 5. Running Inference
Next, we run object detection
```
# using a TRTorch module is exactly the same as how we usually do inference in PyTorch i.e. model(inputs)
detections_batch = trt_model(tensor.to(torch.half)) # convert the input to half precision
# By default, raw output from SSD network per input image contains 8732 boxes with
# localization and class probability distribution.
# Let’s filter this output to only get reasonable detections (confidence>40%) in a more comprehensive format.
results_per_input = utils.decode_results(detections_batch)
best_results_per_input_trt = [utils.pick_best(results, 0.40) for results in results_per_input]
```
Now, let's visualize our predictions!
```
# Visualize results with TRTorch/TensorRT
plot_results(best_results_per_input_trt)
```
We get similar results as before!
---
## 6. Measuring Speedup
We can run the benchmark function again to see the speedup gained! Compare this result with the same batch-size of input in the case without TRTorch/TensorRT above.
```
batch_size = 128
# Recompiling with batch_size we use for evaluating performance
trt_model = trtorch.compile(traced_model, {
"inputs": [trtorch.Input((batch_size, 3, 300, 300))],
"enabled_precisions": {torch.float, torch.half}, # Run with FP16
"workspace_size": 1 << 20
})
benchmark(trt_model, input_shape=(batch_size, 3, 300, 300), nruns=100, dtype="fp16")
```
---
## 7. Conclusion
In this notebook, we have walked through the complete process of compiling a TorchScript SSD300 model with TRTorch, and tested the performance impact of the optimization. We find that using the TRTorch compiled model, we gain significant speedup in inference without any noticeable drop in performance!
### Details
For detailed information on model input and output,
training recipies, inference and performance visit:
[github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
and/or [NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
### References
- [SSD: Single Shot MultiBox Detector](https://arxiv.org/abs/1512.02325) paper
- [Speed/accuracy trade-offs for modern convolutional object detectors](https://arxiv.org/abs/1611.10012) paper
- [SSD on NGC](https://ngc.nvidia.com/catalog/model-scripts/nvidia:ssd_for_pytorch)
- [SSD on github](https://github.com/NVIDIA/DeepLearningExamples/tree/master/PyTorch/Detection/SSD)
|
github_jupyter
|
# 3. Markov Models Example Problems
We will now look at a model that examines our state of healthiness vs. being sick. Keep in mind that this is very much like something you could do in real life. If you wanted to model a certain situation or environment, we could take some data that we have gathered, build a maximum likelihood model on it, and do things like study the properties that emerge from the model, or make predictions from the model, or generate the next most likely state.
Let's say we have 2 states: **sick** and **healthy**. We know that we spend most of our time in a healthy state, so the probability of transitioning from healthy to sick is very low:
$$p(sick \; | \; healthy) = 0.005$$
Hence, the probability of going from healthy to healthy is:
$$p(healthy \; | \; healthy) = 0.995$$
Now, on the other hand the probability of going from sick to sick is also very high. This is because if you just got sick yesterday then you are very likely to be sick tomorrow.
$$p(sick \; | \; sick) = 0.8$$
However, the probability of transitioning from sick to healthy should be higher than the reverse, because you probably won't stay sick for as long as you would stay healthy:
$$p(healthy \; | \; sick) = 0.02$$
We have now fully defined our state transition matrix, and we can now do some calculations.
## 1.1 Example Calculations
### 1.1.1
What is the probability of being healthy for 10 days in a row, given that we already start out as healthy? Well that is:
$$p(healthy \; 10 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^9 = 95.6 \%$$
How about the probability of being healthy for 100 days in a row?
$$p(healthy \; 100 \; days \; in \; a \; row \; | \; healthy \; at \; t=0) = 0.995^{99} = 60.9 \%$$
## 2. Expected Number of Continuously Sick Days
We can now look at the expected number of days that you would remain in the same state (e.g. how many days would you expect to stay sick given the model?). This is a bit more difficult than the last problem, but completely doable, only involving the mathematics of <a href="https://en.wikipedia.org/wiki/Geometric_series">infinite sums</a>.
First, we can look at the probability of being in state $i$, and going to state $i$ in the next state. That is just $A(i,i)$:
$$p \big(s(t)=i \; | \; s(t-1)=i \big) = A(i, i)$$
Now, what is the probability distribution that we actually want to calculate? How about we calculate the probability that we stay in state $i$ for $n$ transitions, at which point we move to another state:
$$p \big(s(t) \;!=i \; | \; s(t-1)=i \big) = 1 - A(i, i)$$
So, the joint probability that we are trying to model is:
$$p\big(s(1)=i, s(2)=i,...,s(n)=i, s(n+1) \;!= i\big) = A(i,i)^{n-1}\big(1-A(i,i)\big)$$
In english this means that we are multiplying the transition probability of staying in the same state, $A(i,i)$, times the number of times we stayed in the same state, $n$, (note it is $n-1$ because we are given that we start in that state, hence there is no transition associated with it) times $1 - A(i,i)$, the probability of transitioning from that state. This leaves us with an expected value for $n$ of:
$$E(n) = \sum np(n) = \sum_{n=1..\infty} nA(i,i)^{n-1}(1-A(i,i))$$
Note, in the above equation $p(n)$ is the probability that we will see state $i$ $n-1$ times after starting from $i$ and then see a state that is not $i$. Also, we know that the expected value of $n$ should be the sum of all possible values of $n$ times $p(n)$.
### 2.1 Expected $n$
So, we can now expand this function and calculate the two sums separately.
$$E(n) = \sum_{n=1..\infty}nA(i,i)^{n-1}(1 - A(i,i)) = \sum nA(i, i)^{n-1} - \sum nA(i,i)^n$$
**First Sum**<br>
With our first sum, we can say that:
$$S = \sum na(i, i)^{n-1}$$
$$S = 1 + 2a + 3a^2 + 4a^3+ ...$$
And we can then multiply that sum, $S$, by $a$, to get:
$$aS = a + 2a^2 + 3a^3 + 4a^4+...$$
And then we can subtract $aS$ from $S$:
$$S - aS = S'= 1 + a + a^2 + a^3+...$$
This $S'$ is another infinite sum, but it is one that is much easier to solve!
$$S'= 1 + a + a^2 + a^3+...$$
And then $aS'$ is:
$$aS' = a + a^2 + a^3+ + a^4 + ...$$
Which, when we then do $S' - aS'$, we end up with:
$$S' - aS' = 1$$
$$S' = \frac{1}{1 - a}$$
And if we then substitute that value in for $S'$ above:
$$S - aS = S'= 1 + a + a^2 + a^3+... = \frac{1}{1 - a}$$
$$S - aS = \frac{1}{1 - a}$$
$$S = \frac{1}{(1 - a)^2}$$
**Second Sum**<br>
We can now look at our second sum:
$$S = \sum na(i,i)^n$$
$$S = 1a + 2a^2 + 3a^3 +...$$
$$Sa = 1a^2 + 2a^3 +...$$
$$S - aS = S' = a + a^2 + a^3 + ...$$
$$aS' = a^2 + a^3 + a^4 +...$$
$$S' - aS' = a$$
$$S' = \frac{a}{1 - a}$$
And we can plug back in $S'$ to get:
$$S - aS = \frac{a}{1 - a}$$
$$S = \frac{a}{(1 - a)^2}$$
**Combine** <br>
We can now combine these two sums as follows:
$$E(n) = \frac{1}{(1 - a)^2} - \frac{a}{(1-a)^2}$$
$$E(n) = \frac{1}{1-a}$$
**Calculate Number of Sick Days**<br>
So, how do we calculate the correct number of sick days? That is just:
$$\frac{1}{1 - 0.8} = 5$$
## 3. SEO and Bounce Rate Optimization
We are now going to look at SEO and Bounch Rate Optimization. This is a problem that every developer and website owner can relate to. You have a website and obviously you would like to increase traffic, increase conversions, and avoid a high bounce rate (which could lead to google assigning your page a low ranking). What would a good way of modeling this data be? Without even looking at any code we can look at some examples of things that we want to know, and how they relate to markov models.
### 3.1 Arrival
First and foremost, how do people arrive on your page? Is it your home page? Your landing page? Well, this is just the very first page of what is hopefully a sequence of pages. So, the markov analogy here is that this is just the initial state distribution or $\pi$. So, once we have our markov model, the $\pi$ vector will tell us which of our pages a user is most likely to start on.
### 3.2 Sequences of Pages
What about sequences of pages? Well, if you think people are getting to your landing page, hitting the buy button, checking out, and then closing the browser window, you can test the validity of that assumption by calculating the probability of that sequence. Of course, the probability of any sequence is probability going to be much less than 1. This is because for a longer sequence, we have more multiplication, and hence smaller final numbers. We do have two alternatives however:
> * 1) You can compare the probability of two different sequences. So, are people going through the entire checkout process? Or is it more probable that they are just bouncing?
* 2) Another option is to just find the transition probabilities themselves. These are conditional probabilities instead of joint probabilities. You want to know, once they have made it to the landing page, what is the probability of hitting buy. Then, once they have hit buy, what is the probability of them completing the checkout.
### 3.3 Bounce Rate
This is hard to measure, unless you are google and hence have analytics on nearly every page on the web. This is because once a user has left your site, you can no longer run code on their computer or track what they are doing. However, let's pretend that we can determine this information. Once we have done this, we can measure which page has the highest bounce rate. At this point we can manually analyze that page and ask our marketing people "what is different about this page that people don't find it useful/want to leave?" We can then address that problem, and the hopefully later analysis shows that the fixed page no longer has a high bounce right. In the markov model, we can just represents this as the null state.
### 3.4 Data
So, the data we are going to be working with has two columns: `last_page_id` and `next_page_id`. This can be interpreted as the current page and the next page. The site has 10 pages with the id's 0-9. We can represent start pages by making the current page -1, and the next page the actual page. We can represent the end of the page with two different codes, `B`(bounce) or `C` (close). In the case of bounce, the user saw the page and then immediately bounced. In the case of close, the user saw the page stayed and potentially saw some useful information, and then closed the window. So, you can imagine that our engineer may use time as a factor in determining if it is a bounce or a close.
```
import numpy as np
import pandas as pd
"""Goal here is to store start page and end page, and the count how many times that happens. After that
we are going to turn it into a probability distribution. We can divide all transitions that start with specific
start state, by row_sum"""
transitions = {} # getting all specific transitions from start pg to end pg, tallying up # of times each occurs
row_sums = {} # start date as key -> getting number of times each starting pg occurs
# Collect our counts
for line in open('../../../data/site/site_data.csv'):
s, e = line.rstrip().split(',') # get start and end page
transitions[(s, e)] = transitions.get((s, e), 0.) + 1
row_sums[s] = row_sums.get(s, 0.) + 1
# Normalize the counts so they become real probability distributions
for k, v in transitions.items():
s, e = k
transitions[k] = v / row_sums[s]
# Calculate initial state distribution
print('Initial state distribution')
for k, v in transitions.items():
s, e = k
if s == '-1': # this means it is the start of the sequence.
print (e, v)
# Which page has the highest bounce rate?
for k, v in transitions.items():
s, e = k
if e == 'B':
print(f'Bounce rate for {s}: {v}')
```
We can see that page with `id` 9 has the highest value in the initial state distribution, so we are most likely to start on that page. We can then see that the page with highest bounce rate is also at page `id` 9.
## 4. Build a 2nd-order language model and generate phrases
So, we are now going to work with non first order markov chains for a little bit. In this example we are going to try and create a language model. So we are going to first train a model on some data to determine the distribution of a word given the previous two words. We can then use this model to generate new phrases. Note that another step of this model would be to calculate the probability of a phrase.
So the data that we are going to look at is just a collection of Robert Frost Poems. It is just a text file with all of the poems concatenated together. So, the first thing we are going to want to do is tokenize each sentence, and remove punctuation. It will look similar to this:
```
def remove_punctuation(s):
return s.translate(None, string.punctuation)
tokens = [t for t in remove_puncuation(line.rstrip().lower()).split()]
```
Once we have tokenized each line, we want to perform various counts in addition to the second order model counts. We need to measure the initial distribution of words, or stated another way the distribution of the first word of a sentence. We also want to know the distribution of the second word of a sentence. Both of these do not have two previous words, so they are not second order. We could technically include them in the second order measurement by using `None` in place of the previous words, but we won't do that here. We also want to keep track of how to end the sentence (end of sentence distribution, will look similar to (w(t-2), w(t-1) -> END)), so we will include a special token for that too.
When we do this counting, what we first want to do is create an array of all possibilities. So, for example if we had two sentences:
```
I love dogs
I love cats
```
Then we could have a dictionary where the key was `(I, love)` and the value was an array `[dogs, cats]`. If "I love" was also a stand alone sentence, then the value would be `[dogs, cats, END]`. The function below can help us with this, since we first need to check if there is any value for the key, create an array if not, otherwise just append to the array.
```
def add2dict(d, k, v):
if k not in d:
d[k] = []
else:
d[k].append(v)
```
One we have collected all of these arrays of possible next words, we need to turn them into **probability distributions**. For example, the array `[cat, cat, dog]` would become the dictionary `{"cat": 2/3, "dog": 1/3}`. Here is a function that can do this:
```
def list2pdict(ts):
d = {}
n = len(ts)
for t in ts:
d[t] = d.get(t, 0.) + 1
for t, c in d.items():
d[t] = c / n
return d
```
Next, we will need a function that can sample from this dictionary. To do this we will need to generate a random number between 0 and 1, and then use the distribution of the words to sample a word given a random number. Here is a function that can do that:
```
def sample_word(d):
p0 = np.random.random()
cumulative = 0
for t, p in d.items():
cumulative += p
if p0 < cumulative:
return t
assert(False) # should never get here
```
Because all of our distributions are structured as dictionaries, we can use the same function for all of them.
```
import numpy as np
import string
"""3 dicts. 1st store pdist for the start of a phrase, then a second word dict which stores the distributions
for the 2nd word of a sentence, and then we are going to have a dict for all second order transitions"""
initial = {}
second_word = {}
transitions = {}
def remove_punctuation(s):
return s.translate(str.maketrans('', '', string.punctuation))
def add2dict(d, k, v):
"""Parameters: Dictionary, Key, Value"""
if k not in d:
d[k] = []
d[k].append(v)
# Loop through file of poems
for line in open('../../../data/poems/robert_frost.txt'):
tokens = remove_punctuation(line.rstrip().lower()).split() # Get all tokens for specific line we are looping over
T = len(tokens) # Length of sequence
for i in range(T): # Loop through every token in sequence
t = tokens[i]
if i == 0: # We are looking at first word
initial[t] = initial.get(t, 0.) + 1
else:
t_1 = tokens[i - 1]
if i == T - 1: # Looking at last word
add2dict(transitions, (t_1, t), 'END')
if i == 1: # second word of sentence, hence only 1 previous word
add2dict(second_word, t_1, t)
else:
t_2 = tokens[i - 2] # Get second previous word
add2dict(transitions, (t_2, t_1), t) # add previous and 2nd previous word as key, and current word as val
# Normalize the distributions
initial_total = sum(initial.values())
for t, c in initial.items():
initial[t] = c / initial_total
# Take our list and turn it into a dictionary of probabilities
def list2pdict(ts):
d = {}
n = len(ts) # get total number of values
for t in ts: # look at each token
d[t] = d.get(t, 0.) + 1
for t, c in d.items(): # go through dictionary, divide frequency by sum
d[t] = c / n
return d
for t_1, ts in second_word.items():
second_word[t_1] = list2pdict(ts)
for k, ts in transitions.items():
transitions[k] = list2pdict(ts)
def sample_word(d):
p0 = np.random.random() # Generate random number from 0 to 1
cumulative = 0 # cumulative count for all probabilities seen so far
for t, p in d.items():
cumulative += p
if p0 < cumulative:
return t
assert(False) # should never hit this
"""Function to generate a poem"""
def generate():
for i in range(4):
sentence = []
# initial word
w0 = sample_word(initial)
sentence.append(w0)
# sample second word
w1 = sample_word(second_word[w0])
sentence.append(w1)
# second-order transitions until END -> enter infinite loop
while True:
w2 = sample_word(transitions[(w0, w1)]) # sample next word given previous two words
if w2 == 'END':
break
sentence.append(w2)
w0 = w1
w1 = w2
print(' '.join(sentence))
generate()
```
## 5. Google's PageRank Algorithm
Markov models were even used in Google's PageRank algorithm. The basic problem we face is:
> * We have $M$ webpages that link to eachother, and we would like to assign importance scores $x(1),...,x(M)$
* All of these scores are greater than or equal to 0
* So, we want to assign a page rank to all of these pages
How can we go about doing this? Well, we can think of a webpage as a sequence, and the page you are on as the state. Where does the ranking come from? Well, the ranking actually comes from the limiting distribution. That is, in the long run, the proportion of visits that will be spent on this page. Now, if you think "great that is all I need to know", slow down. How can we actually do this in practice? How do we train the markov model, and what are the values we assign to the state transition matrix? And how can we ensure that the limiting distribution exists and is unique? The key insight was that **we can use the linked structure of the web to determine the ranking**.
The main idea is that a *link to a page* is like a *vote for its importance*. So, as a first attempt we could just use a frequency count to measure the votes. Of course, that wouldn't be a valid probability distribution, so we could just divide each row by its sum to make it sum to 1. So we set:
$$A(i, j) = \frac{1}{n(i)} \; if \; i \; links \; to \; j$$
$$A(i, j) = 0 \; otherwise$$
Here $n(i)$ stands for the total number of links on a page, and you can confirm that the sum of a row is $\frac{n(i)}{n(i)} = 1$, so this is a valid markov matrix. Now, we still aren't sure if the limiting distribution is unique.
### 5.1 This is already a good start
Let's keep in mind that the above solution already solves a few problems. For instance, let's say you are a spammer and you want to sell 1000 links on your webpage. Well, because the transition matrix must remain a valid probability matrix, the rows must sum to 1, which means that each of your links now only has a strength of $\frac{1}{1000}$. For example the frequency matrix would look like:
| |abc.com|amazon.com|facebook.com|github.com|
|--- |--- |--- | --- |--- |
|thespammer.com|1 |1 |1 |1 |
And then if we transformed that into a probability matrix it would just be each value divided by the total number of links, 4:
| |abc.com|amazon.com|facebook.com|github.com|
|--- |--- |--- | --- |--- |
|thespammer.com|0.25 |0.25 |0.25 |0.25 |
You may then think, I will just create 1000 pages and each of them will only have 1 link. Unfortunately, since nobody knows about those 1000 pages you just created nobody is going to link to them, which means they are impossible to get to. So, in the limiting distribution, those states will have 0 probability because you can't even get to them, so there outgoing links are worthless. Remember, the markov chains limiting distribution will model the long running proportion of visits to a state. So, if you never visit that state, its probability will be 0.
We still have not ensure that the limiting distribution exists and is unique.
### 5.2 Perron-Frobenius Theorem
How can we ensure that our model has a unique stationary distribution. In 1910, this was actually determined. It is known as the **Perron-Frobenius Theorem**, and it states that:
> *If our transition matrix is a markov matrix -meaning that all of the rows sum to 1, and all of the values are strictly positive, i.e. no values that are 0- then the stationary distribution exists and is unique*.
In fact, we can start in any initial state and as time approaches infinity we will always end up with the same stationary distribution, therefore this is also the limiting distribution.
So, how can we satisfy the PF criterion? Let's return to this idea of **smoothing**, which we first talked about when discussing how to train a markov model. The basic idea was that we can make things that were 0, non-zero, so there is still a small possibility that we can get to that state. This might be good news for the spammer. So, we can create a uniform probability distribution $U = \frac{1}{M}$, which is an $M x M$ matrix ($M$ is the number of states). PageRanks solution was to take the matrix we had before and multiply it by 0.85, and to take the uniform distribution and multiply it by 0.15, and add them together to get the final pagerank matrix.
$$G = 0.85A + 0.15U$$
Now all of the elements are strictly positive, and we can convince ourselves that G is still a valid markov matrix.
|
github_jupyter
|
# Quantization of Signals
*This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing. Please direct questions and suggestions to [Sascha.Spors@uni-rostock.de](mailto:Sascha.Spors@uni-rostock.de).*
## Spectral Shaping of the Quantization Noise
The quantized signal $x_Q[k]$ can be expressed by the continuous amplitude signal $x[k]$ and the quantization error $e[k]$ as
\begin{equation}
x_Q[k] = \mathcal{Q} \{ x[k] \} = x[k] + e[k]
\end{equation}
According to the [introduced model](linear_uniform_quantization_error.ipynb#Model-for-the-Quantization-Error), the quantization noise can be modeled as uniformly distributed white noise. Hence, the noise is distributed over the entire frequency range. The basic concept of [noise shaping](https://en.wikipedia.org/wiki/Noise_shaping) is a feedback of the quantization error to the input of the quantizer. This way the spectral characteristics of the quantization noise can be modified, i.e. spectrally shaped. Introducing a generic filter $h[k]$ into the feedback loop yields the following structure

The quantized signal can be deduced from the block diagram above as
\begin{equation}
x_Q[k] = \mathcal{Q} \{ x[k] - e[k] * h[k] \} = x[k] + e[k] - e[k] * h[k]
\end{equation}
where the additive noise model from above has been introduced and it has been assumed that the impulse response $h[k]$ is normalized such that the magnitude of $e[k] * h[k]$ is below the quantization step $Q$. The overall quantization error is then
\begin{equation}
e_H[k] = x_Q[k] - x[k] = e[k] * (\delta[k] - h[k])
\end{equation}
The power spectral density (PSD) of the quantization error with noise shaping is calculated to
\begin{equation}
\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \right|^2
\end{equation}
Hence the PSD $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega})$ of the quantizer without noise shaping is weighted by $| 1 - H(\mathrm{e}^{\,\mathrm{j}\,\Omega}) |^2$. Noise shaping allows a spectral modification of the quantization error. The desired shaping depends on the application scenario. For some applications, high-frequency noise is less disturbing as low-frequency noise.
### Example - First-Order Noise Shaping
If the feedback of the error signal is delayed by one sample we get with $h[k] = \delta[k-1]$
\begin{equation}
\Phi_{e_H e_H}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) \cdot \left| 1 - \mathrm{e}^{\,-\mathrm{j}\,\Omega} \right|^2
\end{equation}
For linear uniform quantization $\Phi_{ee}(\mathrm{e}^{\,\mathrm{j}\,\Omega}) = \sigma_e^2$ is constant. Hence, the spectral shaping constitutes a high-pass characteristic of first order. The following simulation evaluates the noise shaping quantizer of first order.
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.signal as sig
w = 8 # wordlength of the quantized signal
xmin = -1 # minimum of input signal
N = 32768 # number of samples
def uniform_midtread_quantizer_w_ns(x, Q):
# limiter
x = np.copy(x)
idx = np.where(x <= -1)
x[idx] = -1
idx = np.where(x > 1 - Q)
x[idx] = 1 - Q
# linear uniform quantization with noise shaping
xQ = Q * np.floor(x/Q + 1/2)
e = xQ - x
xQ = xQ - np.concatenate(([0], e[0:-1]))
return xQ[1:]
# quantization step
Q = 1/(2**(w-1))
# compute input signal
np.random.seed(5)
x = np.random.uniform(size=N, low=xmin, high=(-xmin-Q))
# quantize signal
xQ = uniform_midtread_quantizer_w_ns(x, Q)
e = xQ - x[1:]
# estimate PSD of error signal
nf, Pee = sig.welch(e, nperseg=64)
# estimate SNR
SNR = 10*np.log10((np.var(x)/np.var(e)))
print('SNR = {:2.1f} dB'.format(SNR))
plt.figure(figsize=(10,5))
Om = nf*2*np.pi
plt.plot(Om, Pee*6/Q**2, label='estimated PSD')
plt.plot(Om, np.abs(1 - np.exp(-1j*Om))**2, label='theoretic PSD')
plt.plot(Om, np.ones(Om.shape), label='PSD w/o noise shaping')
plt.title('PSD of quantization error')
plt.xlabel(r'$\Omega$')
plt.ylabel(r'$\hat{\Phi}_{e_H e_H}(e^{j \Omega}) / \sigma_e^2$')
plt.axis([0, np.pi, 0, 4.5]);
plt.legend(loc='upper left')
plt.grid()
```
**Exercise**
* The overall average SNR is lower than for the quantizer without noise shaping. Why?
Solution: The average power per frequency is lower that without noise shaping for frequencies below $\Omega \approx \pi$. However, this comes at the cost of a larger average power per frequency for frequencies above $\Omega \approx \pi$. The average power of the quantization noise is given as the integral over the PSD of the quantization noise. It is larger for noise shaping and the resulting SNR is consequently lower. Noise shaping is nevertheless beneficial in applications where a lower quantization error in a limited frequency region is desired.
**Copyright**
This notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Digital Signal Processing - Lecture notes featuring computational examples, 2016-2018*.
|
github_jupyter
|
# ------------ First A.I. activity ------------
## 1. IBOVESPA volume prediction
-> Importing libraries that are going to be used in the code
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
```
-> Importing the datasets
```
dataset = pd.read_csv("datasets/ibovespa.csv",delimiter = ";")
```
-> Converting time to datetime in order to make it easy to manipulate
```
dataset['Data/Hora'] = dataset['Data/Hora'].str.replace("/","-")
dataset['Data/Hora'] = pd.to_datetime(dataset['Data/Hora'])
```
-> Visualizing the data
```
dataset.head()
```
-> creating date dataframe and splitting its features
date = dataset.iloc[:,0:1]
date['day'] = date['Data/Hora'].dt.day
date['month'] = date['Data/Hora'].dt.month
date['year'] = date['Data/Hora'].dt.year
date = date.drop(columns = ['Data/Hora'])
-> removing useless columns
```
dataset = dataset.drop(columns = ['Data/Hora','Unnamed: 7','Unnamed: 8','Unnamed: 9'])
```
-> transforming atributes to the correct format
```
for key, value in dataset.head().iteritems():
dataset[key] = dataset[key].str.replace(".","").str.replace(",",".").astype(float)
"""
for key, value in date.head().iteritems():
dataset[key] = date[key]
"""
```
-> Means
```
dataset.mean()
```
-> plotting graphics
```
plt.boxplot(dataset['Volume'])
plt.title('boxplot')
plt.xlabel('volume')
plt.ylabel('valores')
plt.ticklabel_format(style='sci', axis='y', useMathText = True)
dataset['Maxima'].median()
dataset['Minima'].mean()
```
-> Média truncada
```
from scipy import stats
m = stats.trim_mean(dataset['Minima'], 0.1)
print(m)
```
-> variancia e standard deviation
```
v = dataset['Cotacao'].var()
print(v)
d = dataset['Cotacao'].std()
print(v)
m = dataset['Cotacao'].mean()
print(m)
```
-> covariancia dos atributos, mas antes fazer uma standard scaler pra facilitar a visão e depois transforma de volta pra dataframe pandas
#### correlation shows us the relationship between the two variables and how are they related while covariance shows us how the two variables vary from each other.
```
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
dataset_cov = sc.fit_transform(dataset)
dataset_cov = pd.DataFrame(dataset_cov)
dataset_cov.cov()
```
-> plotting the graph may be easier to observe the correlation
```
corr = dataset.corr()
corr.style.background_gradient(cmap = 'coolwarm')
pd.plotting.scatter_matrix(dataset, figsize=(6, 6))
plt.show()
plt.matshow(dataset.corr())
plt.xticks(range(len(dataset.columns)), dataset.columns)
plt.yticks(range(len(dataset.columns)), dataset.columns)
plt.colorbar()
plt.show()
```
|
github_jupyter
|
# Plotting massive data sets
This notebook plots about half a million LIDAR points around Toronto from the KITTI data set. ([Source](http://www.cvlibs.net/datasets/kitti/raw_data.php)) The data is meant to be played over time. With pydeck, we can render these points and interact with them.
### Cleaning the data
First we need to import the data. Each row of data represents one x/y/z coordinate for a point in space at a point in time, with each frame representing about 115,000 points.
We also need to scale the points to plot closely on a map. These point coordinates are not given in latitude and longitude, so as a workaround we'll plot them very close to (0, 0) on the earth.
In future versions of pydeck other viewports, like a flat plane, will be supported out-of-the-box. For now, we'll make do with scaling the points.
```
import pandas as pd
all_lidar = pd.concat([
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_1.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_2.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_3.csv'),
pd.read_csv('https://raw.githubusercontent.com/ajduberstein/kitti_subset/master/kitti_4.csv'),
])
# Filter to one frame of data
lidar = all_lidar[all_lidar['source'] == 136]
lidar.loc[: , ['x', 'y']] = lidar[['x', 'y']] / 10000
```
### Plotting the data
We'll define a single `PointCloudLayer` and plot it.
Pydeck by default expects the input of `get_position` to be a string name indicating a single position value. For convenience, you can pass in a string indicating the X/Y/Z coordinate, here `get_position='[x, y, z]'`. You also have access to a small expression parser--in our `get_position` function here, we increase the size of the z coordinate times 10.
Using `pydeck.data_utils.compute_view`, we'll zoom to the approximate center of the data.
```
import pydeck as pdk
point_cloud = pdk.Layer(
'PointCloudLayer',
lidar[['x', 'y', 'z']],
get_position=['x', 'y', 'z * 10'],
get_normal=[0, 0, 1],
get_color=[255, 0, 100, 200],
pickable=True,
auto_highlight=True,
point_size=1)
view_state = pdk.data_utils.compute_view(lidar[['x', 'y']], 0.9)
view_state.max_pitch = 360
view_state.pitch = 80
view_state.bearing = 120
r = pdk.Deck(
point_cloud,
initial_view_state=view_state,
map_provider=None,
)
r.show()
import time
from collections import deque
# Choose a handful of frames to loop through
frame_buffer = deque([42, 56, 81, 95])
print('Press the stop icon to exit')
while True:
current_frame = frame_buffer[0]
lidar = all_lidar[all_lidar['source'] == current_frame]
r.layers[0].get_position = '@@=[x / 10000, y / 10000, z * 10]'
r.layers[0].data = lidar.to_dict(orient='records')
frame_buffer.rotate()
r.update()
time.sleep(0.5)
```
|
github_jupyter
|
# Seq2Seq with Attention for Korean-English Neural Machine Translation
- Network architecture based on this [paper](https://arxiv.org/abs/1409.0473)
- Fit to run on Google Colaboratory
```
import os
import io
import tarfile
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import torchtext
from torchtext.data import Dataset
from torchtext.data import Example
from torchtext.data import Field
from torchtext.data import BucketIterator
```
# 1. Upload Data to Colab Workspace
로컬에 존재하는 다음 3개의 데이터를 가상 머신에 업로드. 파일의 원본은 [여기](https://github.com/jungyeul/korean-parallel-corpora/tree/master/korean-english-news-v1/)에서도 확인
- korean-english-park.train.tar.gz
- korean-english-park.dev.tar.gz
- korean.english-park.test.tar.gz
```
# 현재 작업경로를 확인 & 'data' 폴더 생성
!echo 'Current working directory:' ${PWD}
!mkdir -p data/
!ls -al
# 로컬의 데이터 업로드
from google.colab import files
uploaded = files.upload()
# 'data' 폴더 하위로 이동, 잘 옮겨졌는지 확인
!mv *.tar.gz data/
!ls -al data/
```
# 2. Check Packages
## KoNLPy (설치 필요)
```
# Java 1.8 & KoNLPy 설치
!apt-get update
!apt-get install g++ openjdk-8-jdk python-dev python3-dev
!pip3 install JPype1-py3
!pip3 install konlpy
from konlpy.tag import Okt
ko_tokens = Okt().pos('트위터 데이터로 학습한 형태소 분석기가 잘 실행이 되는지 확인해볼까요?') # list of (word, POS TAG) tuples
ko_tokens = [t[0] for t in ko_tokens] # Only get words
print(ko_tokens)
del ko_tokens # 필요 없으니까 삭제
```
## Spacy (이미 설치되어 있음)
```
# 설치가 되어있는지 확인
!pip show spacy
# 설치가 되어있는지 확인 (없다면 자동설치됨)
!python -m spacy download en_core_web_sm
import spacy
spacy_en = spacy.load('en_core_web_sm')
en_tokens = [t.text for t in spacy_en.tokenizer('Check that spacy tokenizer works.')]
print(en_tokens)
del en_tokens # 필요 없으니까 삭제
```
# 3. Define Tokenizing Functions
문장을 받아 그보다 작은 어절 혹은 형태소 단위의 리스트로 반환해주는 함수를 각 언어에 대해 작성
- Korean: konlpy.tag.Okt() <- Twitter()에서 명칭변경
- English: spacy.tokenizer
## Korean Tokenizer
```
#from konlpy.tag import Okt
class KoTokenizer(object):
"""For Korean."""
def __init__(self):
self.tokenizer = Okt()
def tokenize(self, text):
tokens = self.tokenizer.pos(text)
tokens = [t[0] for t in tokens]
return tokens
# Usage example
print(KoTokenizer().tokenize('전처리는 언제나 지겨워요.'))
```
## English Tokenizer
```
#import spacy
class EnTokenizer(object):
"""For English."""
def __init__(self):
self.spacy_en = spacy.load('en_core_web_sm')
def tokenize(self, text):
tokens = [t.text for t in self.spacy_en.tokenizer(text)]
return tokens
# Usage example
print(EnTokenizer().tokenize("What I cannot create, I don't understand."))
```
# 4. Data Preprocessing
## Load data
```
# Current working directory & list of files
!echo 'Current working directory:' ${PWD}
!ls -al
DATA_DIR = './data/'
print('Data directory exists:', os.path.isdir(DATA_DIR))
print('List of files:')
print(*os.listdir(DATA_DIR), sep='\n')
def get_data_from_tar_gz(filename):
"""
Retrieve contents from a `tar.gz` file without extraction.
Arguments:
filename: path to `tar.gz` file.
Returns:
dict, (name, content) pairs
"""
assert os.path.exists(filename)
out = {}
with tarfile.open(filename, 'r:gz') as tar:
for member in tar.getmembers():
lang = member.name.split('.')[-1] # ex) korean-english-park.train.ko -> ko
f = tar.extractfile(member)
if f is not None:
content = f.read().decode('utf-8')
content = content.splitlines()
out[lang] = content
assert isinstance(out, dict)
return out
# Each 'xxx_data' is a dictionary with keys; 'ko', 'en'
train_dict= get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.train.tar.gz')) # train
dev_dict = get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.dev.tar.gz')) # dev
test_dict = get_data_from_tar_gz(os.path.join(DATA_DIR, 'korean-english-park.test.tar.gz')) # test
# Some samples (ko)
train_dict['ko'][100:105]
# Some samples (en)
train_dict['en'][100:105]
```
## Define Datasets
```
#from torchtext.data import Dataset
#from torchtext.data import Example
class KoEnTranslationDataset(Dataset):
"""A dataset for Korean-English Neural Machine Translation."""
@staticmethod
def sort_key(ex):
return torchtext.data.interleave_keys(len(ex.src), len(ex.trg))
def __init__(self, data_dict, field_dict, source_lang='ko', max_samples=None, **kwargs):
"""
Only 'ko' and 'en' supported for `language`
Arguments:
data_dict: dict of (`language`, text) pairs.
field_dict: dict of (`language`, Field instance) pairs.
source_lang: str, default 'ko'.
Other kwargs are passed to the constructor of `torchtext.data.Dataset`.
"""
if not all(k in ['ko', 'en'] for k in data_dict.keys()):
raise KeyError("Check data keys.")
if not all(k in ['ko', 'en'] for k in field_dict.keys()):
raise KeyError("Check field keys.")
if source_lang == 'ko':
fields = [('src', field_dict['ko']), ('trg', field_dict['en'])]
src_data = data_dict['ko']
trg_data = data_dict['en']
elif source_lang == 'en':
fields = [('src', field_dict['en']), ('trg', field_dict['ko'])]
src_data = data_dict['en']
trg_data = data_dict['ko']
else:
raise NotImplementedError
if not len(src_data) == len(trg_data):
raise ValueError('Inconsistent number of instances between two languages.')
examples = []
for i, (src_line, trg_line) in enumerate(zip(src_data, trg_data)):
src_line = src_line.strip()
trg_line = trg_line.strip()
if src_line != '' and trg_line != '':
examples.append(
torchtext.data.Example.fromlist(
[src_line, trg_line], fields
)
)
i += 1
if max_samples is not None:
if i >= max_samples:
break
super(KoEnTranslationDataset, self).__init__(examples, fields, **kwargs)
```
## Define Fields
- Instantiate tokenizers; one for each language.
- The 'tokenize' argument of `Field` requires a tokenizing function.
```
#from torchtext.data import Field
ko_tokenizer = KoTokenizer() # korean tokenizer
en_tokenizer = EnTokenizer() # english tokenizer
# Field instance for korean
KOREAN = Field(
init_token='<sos>',
eos_token='<eos>',
tokenize=ko_tokenizer.tokenize,
batch_first=True,
lower=False
)
# Field instance for english
ENGLISH = Field(
init_token='<sos>',
eos_token='<eos>',
tokenize=en_tokenizer.tokenize,
batch_first=True,
lower=True
)
# Store Field instances in a dictionary
field_dict = {
'ko': KOREAN,
'en': ENGLISH,
}
```
## Instantiate datasets
- one for each set (train, dev, test)
```
# 학습시간 단축을 위해 학습 데이터 줄이기
MAX_TRAIN_SAMPLES = 10000
# Instantiate with data
train_set = KoEnTranslationDataset(train_dict, field_dict, max_samples=MAX_TRAIN_SAMPLES)
print('Train set ready.')
print('#. examples:', len(train_set.examples))
dev_set = KoEnTranslationDataset(dev_dict, field_dict)
print('Dev set ready...')
print('#. examples:', len(dev_set.examples))
test_set = KoEnTranslationDataset(test_dict, field_dict)
print('Test set ready...')
print('#. examples:', len(test_set.examples))
# Training example (KO, source language)
train_set.examples[50].src
# Training example (EN, target language)
train_set.examples[50].trg
```
## Build Vocabulary
- 각 언어별 생성: `Field`의 인스턴스를 활용
- 최소 빈도수(`MIN_FREQ`) 값을 작게 하면 vocabulary의 크기가 커짐.
- 최소 빈도수(`MIN_FREQ`) 값을 크게 하면 vocabulary의 크기가 작아짐.
```
MIN_FREQ = 2 # TODO: try different values
# Build vocab for Korean
KOREAN.build_vocab(train_set, dev_set, test_set, min_freq=MIN_FREQ) # ko
print('Size of source vocab (ko):', len(KOREAN.vocab))
# Check indices of some important tokens
tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
for token in tokens:
print(f"{token} -> {KOREAN.vocab.stoi[token]}")
# Build vocab for English
ENGLISH.build_vocab(train_set, dev_set, test_set, min_freq=MIN_FREQ) # en
print('Size of target vocab (en):', len(ENGLISH.vocab))
# Check indices of some important tokens
tokens = ['<unk>', '<pad>', '<sos>', '<eos>']
for token in tokens:
print(f"{token} -> {KOREAN.vocab.stoi[token]}")
```
## Configure Device
- *'런타임' -> '런타임 유형변경'* 에서 하드웨어 가속기로 **GPU** 선택
```
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print('Device to use:', device)
```
## Create Data Iterators
- 데이터를 미니배치(mini-batch) 단위로 반환해주는 역할
- `train_set`, `dev_set`, `test_set`에 대해 개별적으로 정의해야 함
- `BATCH_SIZE`를 정의해주어야 함
- `torchtext.data.BucketIterator`는 하나의 미니배치를 서로 비슷한 길이의 관측치들로 구성함
- [Bucketing](https://medium.com/@rashmi.margani/how-to-speed-up-the-training-of-the-sequence-model-using-bucketing-techniques-9e302b0fd976)의 효과: 하나의 미니배치 내 padding을 최소화하여 연산의 낭비를 줄여줌
```
BATCH_SIZE = 128
#from torchtext.data import BucketIterator
# Train iterator
train_iterator = BucketIterator(
train_set,
batch_size=BATCH_SIZE,
train=True,
shuffle=True,
device=device
)
print(f'Number of minibatches per epoch: {len(train_iterator)}')
#from torchtext.data import BucketIterator
# Dev iterator
dev_iterator = BucketIterator(
dev_set,
batch_size=100,
train=False,
shuffle=False,
device=device
)
print(f'Number of minibatches per epoch: {len(dev_iterator)}')
#from torchtext.data import BucketIterator
# Test iterator
test_iterator = BucketIterator(
test_set,
batch_size=200,
train=False,
shuffle=False,
device=device
)
print(f'Number of minibatches per epoch: {len(test_iterator)}')
train_batch = next(iter(train_iterator))
print('a batch of source examples has shape:', train_batch.src.size()) # (b, s)
print('a batch of target examples has shape:', train_batch.trg.size()) # (b, s)
# Checking first sample in mini-batch (KO, source lang)
ko_indices = train_batch.src[0]
ko_tokens = [KOREAN.vocab.itos[i] for i in ko_indices]
for t, i in zip(ko_tokens, ko_indices):
print(f"{t} ({i})")
del ko_indices, ko_tokens
# Checking first sample in mini-batch (EN, target lang)
en_indices = train_batch.trg[0]
en_tokens = [ENGLISH.vocab.itos[i] for i in en_indices]
for t, i in zip(en_tokens, en_indices):
print(f"{t} ({i})")
del en_indices, en_tokens
del train_batch # 더 이상 필요 없으니까 삭제
```
# 5. Building Seq2Seq Model
## Hyperparameters
```
# Hyperparameters
INPUT_DIM = len(KOREAN.vocab)
OUTPUT_DIM = len(ENGLISH.vocab)
ENC_EMB_DIM = DEC_EMB_DIM = 100
ENC_HID_DIM = DEC_HID_DIM = 60
USE_BIDIRECTIONAL = False
```
## Encoder
```
class Encoder(nn.Module):
"""
Learns an embedding for the source text.
Arguments:
input_dim: int, size of input language vocabulary.
emb_dim: int, size of embedding layer output.
enc_hid_dim: int, size of encoder hidden state.
dec_hid_dim: int, size of decoder hidden state.
bidirectional: uses bidirectional RNNs if True. default is False.
"""
def __init__(self, input_dim, emb_dim, enc_hid_dim, dec_hid_dim, bidirectional=False):
super(Encoder, self).__init__()
self.input_dim = input_dim
self.emb_dim = emb_dim
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.bidirectional = bidirectional
self.embedding = nn.Embedding(
num_embeddings=self.input_dim,
embedding_dim=self.emb_dim
)
self.rnn = nn.GRU(
input_size=self.emb_dim,
hidden_size=self.enc_hid_dim,
bidirectional=self.bidirectional,
batch_first=True
)
self.rnn_output_dim = self.enc_hid_dim
if self.bidirectional:
self.rnn_output_dim *= 2
self.fc = nn.Linear(self.rnn_output_dim, self.dec_hid_dim)
self.dropout = nn.Dropout(.2)
def forward(self, src):
"""
Arguments:
src: 2d tensor of shape (batch_size, input_seq_len)
Returns:
outputs: 3d tensor of shape (batch_size, input_seq_len, num_directions * enc_h)
hidden: 2d tensor of shape (b, dec_h). This tensor will be used as the initial
hidden state value of the decoder (h0 of decoder).
"""
assert len(src.size()) == 2, 'Input requires dimension (batch_size, seq_len).'
# Shape: (b, s, h)
embedded = self.embedding(src)
embedded = self.dropout(embedded)
outputs, hidden = self.rnn(embedded)
if self.bidirectional:
# (2, b, enc_h) -> (b, 2 * enc_h)
hidden = torch.cat((hidden[-2, :, :], hidden[-1, :, :]), dim=1)
else:
# (1, b, enc_h) -> (b, enc_h)
hidden = hidden.squeeze(0)
# (b, num_directions * enc_h) -> (b, dec_h)
hidden = self.fc(hidden)
hidden = torch.tanh(hidden)
return outputs, hidden
```
## Attention
```
class Attention(nn.Module):
def __init__(self, enc_hid_dim, dec_hid_dim, encoder_is_bidirectional=False):
super(Attention, self).__init__()
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.encoder_is_bidirectional = encoder_is_bidirectional
self.attention_input_dim = enc_hid_dim + dec_hid_dim
if self.encoder_is_bidirectional:
self.attention_input_dim += enc_hid_dim # 2 * h_enc + h_dec
self.linear = nn.Linear(self.attention_input_dim, dec_hid_dim)
self.v = nn.Parameter(torch.rand(dec_hid_dim))
def forward(self, hidden, encoder_outputs):
"""
Arguments:
hidden: 2d tensor with shape (batch_size, dec_hid_dim).
encoder_outputs: 3d tensor with shape (batch_size, input_seq_len, enc_hid_dim).
if encoder is bidirectional, expects (batch_size, input_seq_len, 2 * enc_hid_dim).
"""
# Shape check
assert hidden.dim() == 2
assert encoder_outputs.dim() == 3
batch_size, seq_len, _ = encoder_outputs.size()
# (b, dec_h) -> (b, s, dec_h)
hidden = hidden.unsqueeze(1).expand(-1, seq_len, -1)
# concat; shape results in (b, s, enc_h + dec_h).
# if encoder is bidirectional, (b, s, 2 * h_enc + h_dec).
concat = torch.cat((hidden, encoder_outputs), dim=2)
# concat; shape is (b, s, dec_h)
concat = self.linear(concat)
concat = torch.tanh(concat)
# tile v; (dec_h, ) -> (b, dec_h, 1)
v = self.v.repeat(batch_size, 1).unsqueeze(2)
# attn; (b, s, dec_h) @ (b, dec_h, 1) -> (b, s, 1) -> (b, s)
attn_scores = torch.bmm(concat, v).squeeze(-1)
assert attn_scores.dim() == 2 # Final shape check: (b, s)
return F.softmax(attn_scores, dim=1)
```
## Decoder
```
class Decoder(nn.Module):
"""
Unlike the encoder, a single forward pass of
a `Decoder` instance is defined for only a single timestep.
Arguments:
output_dim: int,
emb_dim: int,
enc_hid_dim: int,
dec_hid_dim: int,
attention_module: torch.nn.Module,
encoder_is_bidirectional: False
"""
def __init__(self, output_dim, emb_dim, enc_hid_dim, dec_hid_dim, attention_module, encoder_is_bidirectional=False):
super(Decoder, self).__init__()
self.emb_dim = emb_dim
self.enc_hid_dim = enc_hid_dim
self.dec_hid_dim = dec_hid_dim
self.output_dim = output_dim
self.encoder_is_bidirectional = encoder_is_bidirectional
if isinstance(attention_module, nn.Module):
self.attention_module = attention_module
else:
raise ValueError
self.rnn_input_dim = enc_hid_dim + emb_dim # enc_h + dec_emb_dim
if self.encoder_is_bidirectional:
self.rnn_input_dim += enc_hid_dim # 2 * enc_h + dec_emb_dim
self.embedding = nn.Embedding(output_dim, emb_dim)
self.rnn = nn.GRU(
input_size=self.rnn_input_dim,
hidden_size=dec_hid_dim,
bidirectional=False,
batch_first=True,
)
out_input_dim = 2 * dec_hid_dim + emb_dim # hidden + dec_hidden_dim + dec_emb_dim
self.out = nn.Linear(out_input_dim, output_dim)
self.dropout = nn.Dropout(.2)
def forward(self, inp, hidden, encoder_outputs):
"""
Arguments:
inp: 1d tensor with shape (batch_size, )
hidden: 2d tensor with shape (batch_size, dec_hid_dim).
This `hidden` tensor is the hidden state vector from the previous timestep.
encoder_outputs: 3d tensor with shape (batch_size, seq_len, enc_hid_dim).
If encoder_is_bidirectional is True, expects shape (batch_size, seq_len, 2 * enc_hid_dim).
"""
assert inp.dim() == 1
assert hidden.dim() == 2
assert encoder_outputs.dim() == 3
# (batch_size, ) -> (batch_size, 1)
inp = inp.unsqueeze(1)
# (batch_size, 1) -> (batch_size, 1, emb_dim)
embedded = self.embedding(inp)
embedded = self.dropout(embedded)
# attention probabilities; (batch_size, seq_len)
attn_probs = self.attention_module(hidden, encoder_outputs)
# (batch_size, 1, seq_len)
attn_probs = attn_probs.unsqueeze(1)
# (b, 1, s) @ (b, s, enc_hid_dim) -> (b, 1, enc_hid_dim)
weighted = torch.bmm(attn_probs, encoder_outputs)
# (batch_size, 1, emb_dim + enc_hid_dim)
rnn_input = torch.cat((embedded, weighted), dim=2)
# output; (batch_size, 1, dec_hid_dim)
# new_hidden; (1, batch_size, dec_hid_dim)
output, new_hidden = self.rnn(rnn_input, hidden.unsqueeze(0))
embedded = embedded.squeeze(1) # (b, 1, emb) -> (b, emb)
output = output.squeeze(1) # (b, 1, dec_h) -> (b, dec_h)
weighted = weighted.squeeze(1) # (b, 1, dec_h) -> (b, dec_h)
# output; (batch_size, emb + 2 * dec_h) -> (batch_size, output_dim)
output = self.out(torch.cat((output, weighted, embedded), dim=1))
return output, new_hidden.squeeze(0)
```
## Seq2Seq
```
class Seq2Seq(nn.Module):
def __init__(self, encoder, decoder, device):
super(Seq2Seq, self).__init__()
self.encoder = encoder
self.decoder = decoder
self.device = device
def forward(self, src, trg, teacher_forcing_ratio=.5):
batch_size, max_seq_len = trg.size()
trg_vocab_size = self.decoder.output_dim
# An empty tesnor to store decoder outputs (time index first for indexing)
outputs_shape = (max_seq_len, batch_size, trg_vocab_size)
outputs = torch.zeros(outputs_shape).to(self.device)
encoder_outputs, hidden = self.encoder(src)
# first input to the decoder is '<sos>'
# trg; shape (batch_size, seq_len)
initial_dec_input = output = trg[:, 0] # get first timestep token
for t in range(1, max_seq_len):
output, hidden = self.decoder(output, hidden, encoder_outputs)
outputs[t] = output # Save output for timestep t, for 1 <= t <= max_len
top1_val, top1_idx = output.max(dim=1)
teacher_force = torch.rand(1).item() >= teacher_forcing_ratio
output = trg[:, t] if teacher_force else top1_idx
# Switch batch and time dimensions for consistency (batch_first=True)
outputs = outputs.permute(1, 0, 2) # (s, b, trg_vocab) -> (b, s, trg_vocab)
return outputs
```
## Build Model
```
# Define encoder
enc = Encoder(
input_dim=INPUT_DIM,
emb_dim=ENC_EMB_DIM,
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
bidirectional=USE_BIDIRECTIONAL
)
print(enc)
# Define attention layer
attn = Attention(
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
encoder_is_bidirectional=USE_BIDIRECTIONAL
)
print(attn)
# Define decoder
dec = Decoder(
output_dim=OUTPUT_DIM,
emb_dim=DEC_EMB_DIM,
enc_hid_dim=ENC_HID_DIM,
dec_hid_dim=DEC_HID_DIM,
attention_module=attn,
encoder_is_bidirectional=USE_BIDIRECTIONAL
)
print(dec)
model = Seq2Seq(enc, dec, device).to(device)
print(model)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters.')
```
# 6. Train
## Optimizer
- Use `optim.Adam` or `optim.RMSprop`.
```
optimizer = optim.Adam(model.parameters(), lr=0.001)
#optimizer = optim.RMSprop(model.parameters(), lr=0.01)
```
## Loss function
```
# Padding indices should not be considered when loss is calculated.
PAD_IDX = ENGLISH.vocab.stoi['<pad>']
criterion = nn.CrossEntropyLoss(ignore_index=PAD_IDX)
```
## Train function
```
def train(seq2seq_model, iterator, optimizer, criterion, grad_clip=1.0):
seq2seq_model.train()
epoch_loss = .0
for i, batch in enumerate(iterator):
print('.', end='')
src = batch.src
trg = batch.trg
optimizer.zero_grad()
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=.5)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# (b-1, s, trg_vocab)
decoder_outputs = decoder_outputs[:, 1:, :]
# ((b-1) * s, trg_vocab)
decoder_outputs = decoder_outputs.contiguous().view(-1, trg_vocab_size)
# ((b-1) * s, )
trg = trg[:, 1:].contiguous().view(-1)
loss = criterion(decoder_outputs, trg)
loss.backward()
# Gradient clipping; remedy for exploding gradients
torch.nn.utils.clip_grad_norm_(seq2seq_model.parameters(), grad_clip)
optimizer.step()
epoch_loss += loss.item()
return epoch_loss / len(iterator)
```
## Evaluate function
```
def evaluate(seq2seq_model, iterator, criterion):
seq2seq_model.eval()
epoch_loss = 0.
with torch.no_grad():
for i, batch in enumerate(iterator):
print('.', end='')
src = batch.src
trg = batch.trg
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=0.)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# (b-1, s, trg_vocab)
decoder_outputs = decoder_outputs[:, 1:, :]
# ((b-1) * s, trg_vocab)
decoder_outputs = decoder_outputs.contiguous().view(-1, trg_vocab_size)
# ((b-1) * s, )
trg = trg[:, 1:].contiguous().view(-1)
loss = criterion(decoder_outputs, trg)
epoch_loss += loss.item()
return epoch_loss / len(iterator)
```
## Epoch time measure function
```
def epoch_time(start_time, end_time):
"""Returns elapsed time in mins & secs."""
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
```
## Train for multiple epochs
```
NUM_EPOCHS = 50
import time
import math
best_dev_loss = float('inf')
for epoch in range(NUM_EPOCHS):
start_time = time.time()
train_loss = train(model, train_iterator, optimizer, criterion)
dev_loss = evaluate(model, dev_iterator, criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if dev_loss < best_dev_loss:
best_dev_loss = dev_loss
torch.save(model.state_dict(), './best_model.pt')
print("\n")
print(f"Epoch: {epoch + 1:>02d} | Time: {epoch_mins}m {epoch_secs}s")
print(f"Train Loss: {train_loss:>.4f} | Train Perplexity: {math.exp(train_loss):7.3f}")
print(f"Dev Loss: {dev_loss:>.4f} | Dev Perplexity: {math.exp(dev_loss):7.3f}")
```
## Save last model (overfitted)
```
torch.save(model.state_dict(), './last_model.pt')
```
# 7. Test
## Function to convert indices to original text strings
```
def indices_to_text(src_or_trg, lang_field):
assert src_or_trg.dim() == 1, f'{src_or_trg.dim()}' #(seq_len, )
assert isinstance(lang_field, torchtext.data.Field)
assert hasattr(lang_field, 'vocab')
return [lang_field.vocab.itos[t] for t in src_or_trg]
```
## Function to make predictions
- Returns a list of examples, where each example is a (src, trg, prediction) tuple.
```
def predict(seq2seq_model, iterator):
seq2seq_model.eval()
out = []
with torch.no_grad():
for i, batch in enumerate(iterator):
src = batch.src
trg = batch.trg
decoder_outputs = seq2seq_model(src, trg, teacher_forcing_ratio=0.)
seq_len, batch_size, trg_vocab_size = decoder_outputs.size() # (b, s, trg_vocab)
# Discard initial decoder input (index = 0)
#decoder_outputs = decoder_outputs[:, 1:, :]
decoder_predictions = decoder_outputs.argmax(dim=-1) # (b, s)
for i, pred in enumerate(decoder_predictions):
out.append((src[i], trg[i], pred))
return out
```
## Load best model
```
!ls -al
# Load model
model.load_state_dict(torch.load('./best_model.pt'))
```
## Make predictions
```
# Make prediction
test_predictions = predict(model, dev_iterator)
for i, prediction in enumerate(test_predictions):
src, trg, pred = prediction
src_text = indices_to_text(src, lang_field=KOREAN)
trg_text = indices_to_text(trg, lang_field=ENGLISH)
pred_text = indices_to_text(pred, lang_field=ENGLISH)
print('source:\n', src_text)
print('target:\n', trg_text)
print('prediction:\n', pred_text)
print('-' * 160)
if i > 5:
break
```
# 8. Download Model
```
!ls -al
from google.colab import files
print('Downloading models...') # Known bug; if using Firefox, a print statement in the same cell is necessary.
files.download('./best_model.pt')
files.download('./last_model.pt')
```
# 9. Discussions
```
```
|
github_jupyter
|
```
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
STATS_DIR = "/hg191/corpora/legaldata/data/stats/"
SEM_FEATS_FILE = os.path.join (STATS_DIR, "ops.temp.semfeat")
INDEG_FILE = os.path.join (STATS_DIR, "ops.ind")
ind = pd.read_csv (INDEG_FILE, sep=",", header=None, names=["opid", "indeg"])
semfeat = pd.read_csv (SEM_FEATS_FILE, sep=",", header=None, names=["opid", "semfeat"])
indegs = pd.Series([ind[ind["opid"] == opid]["indeg"].values[0] for opid in semfeat.opid.values])
semfeat["indeg"] = indegs
def labelPercentile (series):
labels = list ()
p50 = np.percentile (series, q=50)
p75 = np.percentile (series, q=75)
p90 = np.percentile (series, q=90)
for value in series:
if value <= p50:
labels.append ("<=50")
elif value <= p90:
labels.append (">50")
elif value > p90:
labels.append (">90")
return labels
semfeat["percentile"] = pd.Series (labelPercentile(semfeat["semfeat"].values))
df = semfeat[semfeat["indeg"] > 0]
df["log(indeg)"] = np.log(df["indeg"])
ax = sns.boxplot(x="percentile", y="log(indeg)", data=df, order=["<=50", ">50", ">90"])
vals = df[df["percentile"] == ">50"]["log(indeg)"].values
np.sort(vals)[int(len(vals)/2)]
print(len(df[df["percentile"] == ">50"]))
print(len(df[df["percentile"] == ">90"]))
print (df[df["percentile"] == "<=50"]["log(indeg)"].median())
print (df[df["percentile"] == ">50"]["log(indeg)"].median())
print (df[df["percentile"] == ">90"]["log(indeg)"].median())
#print (semfeat[semfeat["percentile"] == ">P99"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == "<=P50"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == ">P50"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == ">P90"]["logindeg"].mean())
print (semfeat[semfeat["percentile"] == "<=P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P90"]["indeg"].median())
print (semfeat[semfeat["percentile"] == "<=P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P50"]["indeg"].median())
print (semfeat[semfeat["percentile"] == ">P90"]["indeg"].median())
np.percentile(semfeat["semfeat"].values, q=90)
[semfeat["percentile"] == ">P90"]["indeg"].mean()
semfeat[semfeat["percentile"] == ">P90"].tail(500)
sorted(semfeat["indeg"], reverse=True)[0:10]
semfeat[semfeat["indeg"].isin(sorted(semfeat["indeg"], reverse=True)[0:10])]
semfeat.loc[48004,]["semfeat"] = 1
semfeat[semfeat["indeg"].isin(sorted(semfeat["indeg"], reverse=True)[0:10])]
print(np.mean((semfeat[semfeat["percentile"] == "<=P50"]["indeg"] > 0).values))
print(np.mean((semfeat[semfeat["percentile"] == ">P50"]["indeg"] > 0).values))
print(np.mean((semfeat[semfeat["percentile"] == ">P90"]["indeg"] > 0).values))
print (len(semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]))
print (len(semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]))
print (len(semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]))
print (semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]["indeg"].mean())
print (semfeat[(semfeat["percentile"] == "<=P50") & (semfeat["indeg"] > 0)]["logindeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P50") & (semfeat["indeg"] > 0)]["logindeg"].mean())
print (semfeat[(semfeat["percentile"] == ">P90") & (semfeat["indeg"] > 0)]["logindeg"].mean())
ax = sns.violinplot(x="percentile", y="logindeg", data=df, order=["<=P50", ">P50", ">P90"])
semfeat[semfeat["indeg"] == 1]
```
|
github_jupyter
|
```
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
# default_exp losses
# default_cls_lvl 3
#export
from fastai.imports import *
from fastai.torch_imports import *
from fastai.torch_core import *
from fastai.layers import *
#hide
from nbdev.showdoc import *
```
# Loss Functions
> Custom fastai loss functions
```
F.binary_cross_entropy_with_logits(torch.randn(4,5), torch.randint(0, 2, (4,5)).float(), reduction='none')
funcs_kwargs
# export
@log_args
class BaseLoss():
"Same as `loss_cls`, but flattens input and target."
activation=decodes=noops
def __init__(self, loss_cls, *args, axis=-1, flatten=True, floatify=False, is_2d=True, **kwargs):
store_attr("axis,flatten,floatify,is_2d")
self.func = loss_cls(*args,**kwargs)
functools.update_wrapper(self, self.func)
def __repr__(self): return f"FlattenedLoss of {self.func}"
@property
def reduction(self): return self.func.reduction
@reduction.setter
def reduction(self, v): self.func.reduction = v
def __call__(self, inp, targ, **kwargs):
inp = inp .transpose(self.axis,-1).contiguous()
targ = targ.transpose(self.axis,-1).contiguous()
if self.floatify and targ.dtype!=torch.float16: targ = targ.float()
if targ.dtype in [torch.int8, torch.int16, torch.int32]: targ = targ.long()
if self.flatten: inp = inp.view(-1,inp.shape[-1]) if self.is_2d else inp.view(-1)
return self.func.__call__(inp, targ.view(-1) if self.flatten else targ, **kwargs)
```
Wrapping a general loss function inside of `BaseLoss` provides extra functionalities to your loss functions:
- flattens the tensors before trying to take the losses since it's more convenient (with a potential tranpose to put `axis` at the end)
- a potential `activation` method that tells the library if there is an activation fused in the loss (useful for inference and methods such as `Learner.get_preds` or `Learner.predict`)
- a potential <code>decodes</code> method that is used on predictions in inference (for instance, an argmax in classification)
The `args` and `kwargs` will be passed to `loss_cls` during the initialization to instantiate a loss function. `axis` is put at the end for losses like softmax that are often performed on the last axis. If `floatify=True`, the `targs` will be converted to floats (useful for losses that only accept float targets like `BCEWithLogitsLoss`), and `is_2d` determines if we flatten while keeping the first dimension (batch size) or completely flatten the input. We want the first for losses like Cross Entropy, and the second for pretty much anything else.
```
# export
@log_args
@delegates()
class CrossEntropyLossFlat(BaseLoss):
"Same as `nn.CrossEntropyLoss`, but flattens input and target."
y_int = True
@use_kwargs_dict(keep=True, weight=None, ignore_index=-100, reduction='mean')
def __init__(self, *args, axis=-1, **kwargs): super().__init__(nn.CrossEntropyLoss, *args, axis=axis, **kwargs)
def decodes(self, x): return x.argmax(dim=self.axis)
def activation(self, x): return F.softmax(x, dim=self.axis)
tst = CrossEntropyLossFlat()
output = torch.randn(32, 5, 10)
target = torch.randint(0, 10, (32,5))
#nn.CrossEntropy would fail with those two tensors, but not our flattened version.
_ = tst(output, target)
test_fail(lambda x: nn.CrossEntropyLoss()(output,target))
#Associated activation is softmax
test_eq(tst.activation(output), F.softmax(output, dim=-1))
#This loss function has a decodes which is argmax
test_eq(tst.decodes(output), output.argmax(dim=-1))
#In a segmentation task, we want to take the softmax over the channel dimension
tst = CrossEntropyLossFlat(axis=1)
output = torch.randn(32, 5, 128, 128)
target = torch.randint(0, 5, (32, 128, 128))
_ = tst(output, target)
test_eq(tst.activation(output), F.softmax(output, dim=1))
test_eq(tst.decodes(output), output.argmax(dim=1))
# export
@log_args
@delegates()
class BCEWithLogitsLossFlat(BaseLoss):
"Same as `nn.BCEWithLogitsLoss`, but flattens input and target."
@use_kwargs_dict(keep=True, weight=None, reduction='mean', pos_weight=None)
def __init__(self, *args, axis=-1, floatify=True, thresh=0.5, **kwargs):
super().__init__(nn.BCEWithLogitsLoss, *args, axis=axis, floatify=floatify, is_2d=False, **kwargs)
self.thresh = thresh
def decodes(self, x): return x>self.thresh
def activation(self, x): return torch.sigmoid(x)
tst = BCEWithLogitsLossFlat()
output = torch.randn(32, 5, 10)
target = torch.randn(32, 5, 10)
#nn.BCEWithLogitsLoss would fail with those two tensors, but not our flattened version.
_ = tst(output, target)
test_fail(lambda x: nn.BCEWithLogitsLoss()(output,target))
output = torch.randn(32, 5)
target = torch.randint(0,2,(32, 5))
#nn.BCEWithLogitsLoss would fail with int targets but not our flattened version.
_ = tst(output, target)
test_fail(lambda x: nn.BCEWithLogitsLoss()(output,target))
#Associated activation is sigmoid
test_eq(tst.activation(output), torch.sigmoid(output))
# export
@log_args(to_return=True)
@use_kwargs_dict(weight=None, reduction='mean')
def BCELossFlat(*args, axis=-1, floatify=True, **kwargs):
"Same as `nn.BCELoss`, but flattens input and target."
return BaseLoss(nn.BCELoss, *args, axis=axis, floatify=floatify, is_2d=False, **kwargs)
tst = BCELossFlat()
output = torch.sigmoid(torch.randn(32, 5, 10))
target = torch.randint(0,2,(32, 5, 10))
_ = tst(output, target)
test_fail(lambda x: nn.BCELoss()(output,target))
# export
@log_args(to_return=True)
@use_kwargs_dict(reduction='mean')
def MSELossFlat(*args, axis=-1, floatify=True, **kwargs):
"Same as `nn.MSELoss`, but flattens input and target."
return BaseLoss(nn.MSELoss, *args, axis=axis, floatify=floatify, is_2d=False, **kwargs)
tst = MSELossFlat()
output = torch.sigmoid(torch.randn(32, 5, 10))
target = torch.randint(0,2,(32, 5, 10))
_ = tst(output, target)
test_fail(lambda x: nn.MSELoss()(output,target))
#hide
#cuda
#Test losses work in half precision
output = torch.sigmoid(torch.randn(32, 5, 10)).half().cuda()
target = torch.randint(0,2,(32, 5, 10)).half().cuda()
for tst in [BCELossFlat(), MSELossFlat()]: _ = tst(output, target)
# export
@log_args(to_return=True)
@use_kwargs_dict(reduction='mean')
def L1LossFlat(*args, axis=-1, floatify=True, **kwargs):
"Same as `nn.L1Loss`, but flattens input and target."
return BaseLoss(nn.L1Loss, *args, axis=axis, floatify=floatify, is_2d=False, **kwargs)
#export
@log_args
class LabelSmoothingCrossEntropy(Module):
y_int = True
def __init__(self, eps:float=0.1, reduction='mean'): self.eps,self.reduction = eps,reduction
def forward(self, output, target):
c = output.size()[-1]
log_preds = F.log_softmax(output, dim=-1)
if self.reduction=='sum': loss = -log_preds.sum()
else:
loss = -log_preds.sum(dim=-1) #We divide by that size at the return line so sum and not mean
if self.reduction=='mean': loss = loss.mean()
return loss*self.eps/c + (1-self.eps) * F.nll_loss(log_preds, target.long(), reduction=self.reduction)
def activation(self, out): return F.softmax(out, dim=-1)
def decodes(self, out): return out.argmax(dim=-1)
```
On top of the formula we define:
- a `reduction` attribute, that will be used when we call `Learner.get_preds`
- an `activation` function that represents the activation fused in the loss (since we use cross entropy behind the scenes). It will be applied to the output of the model when calling `Learner.get_preds` or `Learner.predict`
- a <code>decodes</code> function that converts the output of the model to a format similar to the target (here indices). This is used in `Learner.predict` and `Learner.show_results` to decode the predictions
```
#export
@log_args
@delegates()
class LabelSmoothingCrossEntropyFlat(BaseLoss):
"Same as `LabelSmoothingCrossEntropy`, but flattens input and target."
y_int = True
@use_kwargs_dict(keep=True, eps=0.1, reduction='mean')
def __init__(self, *args, axis=-1, **kwargs): super().__init__(LabelSmoothingCrossEntropy, *args, axis=axis, **kwargs)
def activation(self, out): return F.softmax(out, dim=-1)
def decodes(self, out): return out.argmax(dim=-1)
```
## Export -
```
#hide
from nbdev.export import *
notebook2script()
```
|
github_jupyter
|
```
# HIDDEN
from datascience import *
%matplotlib inline
import matplotlib.pyplot as plots
plots.style.use('fivethirtyeight')
import math
import numpy as np
from scipy import stats
import ipywidgets as widgets
import nbinteract as nbi
```
### The Central Limit Theorem ###
Very few of the data histograms that we have seen in this course have been bell shaped. When we have come across a bell shaped distribution, it has almost invariably been an empirical histogram of a statistic based on a random sample.
**The Central Limit Theorem says that the probability distribution of the sum or average of a large random sample drawn with replacement will be roughly normal, *regardless of the distribution of the population from which the sample is drawn*.**
As we noted when we were studying Chebychev's bounds, results that can be applied to random samples *regardless of the distribution of the population* are very powerful, because in data science we rarely know the distribution of the population.
The Central Limit Theorem makes it possible to make inferences with very little knowledge about the population, provided we have a large random sample. That is why it is central to the field of statistical inference.
### Proportion of Purple Flowers ###
Recall Mendel's probability model for the colors of the flowers of a species of pea plant. The model says that the flower colors of the plants are like draws made at random with replacement from {Purple, Purple, Purple, White}.
In a large sample of plants, about what proportion will have purple flowers? We would expect the answer to be about 0.75, the proportion purple in the model. And, because proportions are means, the Central Limit Theorem says that the distribution of the sample proportion of purple plants is roughly normal.
We can confirm this by simulation. Let's simulate the proportion of purple-flowered plants in a sample of 200 plants.
```
colors = make_array('Purple', 'Purple', 'Purple', 'White')
model = Table().with_column('Color', colors)
model
props = make_array()
num_plants = 200
repetitions = 1000
for i in np.arange(repetitions):
sample = model.sample(num_plants)
new_prop = np.count_nonzero(sample.column('Color') == 'Purple')/num_plants
props = np.append(props, new_prop)
props[:5]
opts = {
'title': 'Distribution of sample proportions',
'xlabel': 'Sample Proportion',
'ylabel': 'Percent per unit',
'xlim': (0.64, 0.84),
'ylim': (0, 25),
'bins': 20,
}
nbi.hist(props, options=opts)
```
There's that normal curve again, as predicted by the Central Limit Theorem, centered at around 0.75 just as you would expect.
How would this distribution change if we increased the sample size? We can copy our sampling code into a function and then use interaction to see how the distribution changes as the sample size increases.
We will keep the number of `repetitions` the same as before so that the two columns have the same length.
```
def empirical_props(num_plants):
props = make_array()
for i in np.arange(repetitions):
sample = model.sample(num_plants)
new_prop = np.count_nonzero(sample.column('Color') == 'Purple')/num_plants
props = np.append(props, new_prop)
return props
nbi.hist(empirical_props, options=opts,
num_plants=widgets.ToggleButtons(options=[100, 200, 400, 800]))
```
All of the above distributions are approximately normal but become more narrow as the sample size increases. For example, the proportions based on a sample size of 800 are more tightly clustered around 0.75 than those from a sample size of 200. Increasing the sample size has decreased the variability in the sample proportion.
|
github_jupyter
|
# Spark on Kubernetes
Preparing the notebook https://towardsdatascience.com/make-kubeflow-into-your-own-data-science-workspace-cc8162969e29
## Setup service account permissions
https://github.com/kubeflow/kubeflow/issues/4306 issue with launching spark-operator from jupyter notebook
Run command in your shell (not in notebook)
```shell
export NAMESPACE=<your_namespace>
kubectl create serviceaccount spark -n ${NAMESPACE}
kubectl create clusterrolebinding spark-role --clusterrole=edit --serviceaccount=${NAMESPACE}:spark --namespace=${NAMESPACE}
```
## Python version
> Note: Make sure your driver python and executor python version matches.
> Otherwise, you will see error msg like below
Exception: Python in worker has different version 3.7 than that in driver 3.6, PySpark cannot run with different minor versions.Please check environment variables `PYSPARK_PYTHON` and `PYSPARK_DRIVER_PYTHON` are correctly set.
```
import sys
print(sys.version)
```
## Client Mode
```
import findspark, pyspark,socket
from pyspark import SparkContext, SparkConf
from pyspark.sql import SparkSession
findspark.init()
localIpAddress = socket.gethostbyname(socket.gethostname())
conf = SparkConf().setAppName('sparktest1')
conf.setMaster('k8s://https://kubernetes.default.svc:443')
conf.set("spark.submit.deployMode", "client")
conf.set("spark.executor.instances", "2")
conf.set("spark.driver.host", localIpAddress)
conf.set("spark.driver.port", "7778")
conf.set("spark.kubernetes.namespace", "yahavb")
conf.set("spark.kubernetes.container.image", "seedjeffwan/spark-py:v2.4.6")
conf.set("spark.kubernetes.pyspark.pythonVersion", "3")
conf.set("spark.kubernetes.authenticate.driver.serviceAccountName", "spark")
conf.set("spark.kubernetes.executor.annotation.sidecar.istio.io/inject", "false")
sc = pyspark.context.SparkContext.getOrCreate(conf=conf)
# following works as well
# spark = SparkSession.builder.config(conf=conf).getOrCreate()
num_samples = 100000
def inside(p):
x, y = random.random(), random.random()
return x*x + y*y < 1
count = sc.parallelize(range(0, num_samples)).filter(inside).count()
sc.stop()
```
## Cluster Mode
## Java
```
%%bash
/opt/spark-2.4.6/bin/spark-submit --master "k8s://https://kubernetes.default.svc:443" \
--deploy-mode cluster \
--name spark-java-pi \
--class org.apache.spark.examples.SparkPi \
--conf spark.executor.instances=30 \
--conf spark.kubernetes.namespace=yahavb \
--conf spark.kubernetes.driver.annotation.sidecar.istio.io/inject=false \
--conf spark.kubernetes.executor.annotation.sidecar.istio.io/inject=false \
--conf spark.kubernetes.container.image=seedjeffwan/spark:v2.4.6 \
--conf spark.kubernetes.driver.pod.name=spark-java-pi-driver \
--conf spark.kubernetes.executor.request.cores=4 \
--conf spark.kubernetes.node.selector.computetype=gpu \
--conf spark.kubernetes.authenticate.driver.serviceAccountName=spark \
local:///opt/spark/examples/jars/spark-examples_2.11-2.4.6.jar 262144
%%bash
kubectl -n yahavb delete po ` kubectl -n yahavb get po | grep spark-java-pi-driver | awk '{print $1}'`
```
## Python
```
%%bash
/opt/spark-2.4.6/bin/spark-submit --master "k8s://https://kubernetes.default.svc:443" \
--deploy-mode cluster \
--name spark-python-pi \
--conf spark.executor.instances=50 \
--conf spark.kubernetes.container.image=seedjeffwan/spark-py:v2.4.6 \
--conf spark.kubernetes.driver.pod.name=spark-python-pi-driver \
--conf spark.kubernetes.namespace=yahavb \
--conf spark.kubernetes.driver.annotation.sidecar.istio.io/inject=false \
--conf spark.kubernetes.executor.annotation.sidecar.istio.io/inject=false \
--conf spark.kubernetes.pyspark.pythonVersion=3 \
--conf spark.kubernetes.executor.request.cores=4 \
--conf spark.kubernetes.authenticate.driver.serviceAccountName=spark /opt/spark/examples/src/main/python/pi.py 64000
%%bash
kubectl -n yahavb delete po `kubectl -n yahavb get po | grep spark-python-pi-driver | awk '{print $1}'`
```
|
github_jupyter
|
```
from Maze import Maze
from sarsa_agent import SarsaAgent
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import animation
from IPython.display import HTML
```
## Designing the maze
```
arr=np.array([[0,1,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0],
[0,1,0,0,1,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0],
[0,1,0,0,1,0,0,1,1,1,1,1,0,1,1,0,1,1,1,0],
[0,1,0,0,1,0,0,0,0,0,1,0,0,1,0,0,1,0,0,0],
[0,0,0,0,1,0,0,1,1,1,0,0,1,1,0,0,1,0,0,0],
[0,0,0,0,0,0,0,1,0,0,0,1,0,1,0,1,1,0,1,1],
[1,1,1,0,1,1,0,1,0,0,1,0,0,1,0,0,1,0,0,0],
[0,0,1,0,1,0,1,0,0,1,0,0,0,0,0,0,1,0,1,0],
[0,0,0,0,1,0,0,0,1,0,0,0,0,0,0,1,1,0,1,0],
[0,0,0,0,1,0,0,1,0,0,0,0,0,1,1,1,0,0,0,0],
[1,0,1,1,1,0,1,0,0,1,0,0,0,1,0,0,0,1,0,0],
[1,0,1,1,1,0,1,0,0,1,0,0,1,1,0,0,0,1,0,0],
[1,0,0,1,0,0,1,0,0,1,0,0,1,0,0,1,0,0,0,0],
[0,0,0,0,1,0,1,0,0,1,1,0,1,0,0,0,1,1,1,0],
[0,0,1,1,1,0,1,0,0,1,0,1,0,0,1,1,0,0,0,0],
[0,1,1,0,0,0,0,1,0,1,0,0,1,1,0,1,0,1,1,1],
[0,1,0,0,0,1,0,0,0,0,0,0,1,0,0,1,0,0,0,0],
[0,0,1,1,1,0,1,1,0,0,1,0,1,0,0,1,1,0,0,0],
[1,0,0,0,1,0,1,0,0,0,1,0,1,0,0,1,1,1,0,0],
[1,1,0,0,0,0,1,0,0,0,1,0,1,0,0,0,0,1,0,0]
],dtype=float)
#Position of the rat
rat=(0,0)
#If Cheese is None, cheese is placed in the bottom-right cell of the maze
cheese=None
#The maze object takes the maze
maze=Maze(arr,rat,cheese)
maze.show_maze()
```
## Defining a Agent [Sarsa Agent because it uses Sarsa to solve the maze]
```
agent=SarsaAgent(maze)
```
## Making the agent play episodes and learn
```
agent.learn(episodes=1000)
```
## Plotting the maze
```
nrow=maze.nrow
ncol=maze.ncol
fig=plt.figure()
ax=fig.gca()
ax.set_xticks(np.arange(0.5,ncol,1))
ax.set_yticks(np.arange(0.5,nrow,1))
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.grid('on')
img=ax.imshow(maze.maze,cmap="gray",)
a=5
```
## Making Animation of the maze solution
```
def gen_func():
maze=Maze(arr,rat,cheese)
done=False
while not done:
row,col,_=maze.state
cell=(row,col)
action=agent.get_policy(cell)
maze.step(action)
done=maze.get_status()
yield maze.get_canvas()
def update_plot(canvas):
img.set_data(canvas)
anim=animation.FuncAnimation(fig,update_plot,gen_func)
HTML(anim.to_html5_video())
anim.save("big_maze.gif",animation.PillowWriter())
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W1D2_ModelingPractice/student/W1D2_Tutorial2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Neuromatch Academy: Week1, Day 2, Tutorial 2
#Tutorial objectives
We are investigating a simple phenomena, working through the 10 steps of modeling ([Blohm et al., 2019](https://doi.org/10.1523/ENEURO.0352-19.2019)) in two notebooks:
**Framing the question**
1. finding a phenomenon and a question to ask about it
2. understanding the state of the art
3. determining the basic ingredients
4. formulating specific, mathematically defined hypotheses
**Implementing the model**
5. selecting the toolkit
6. planning the model
7. implementing the model
**Model testing**
8. completing the model
9. testing and evaluating the model
**Publishing**
10. publishing models
We did steps 1-5 in Tutorial 1 and will cover steps 6-10 in Tutorial 2 (this notebook).
# Utilities Setup and Convenience Functions
Please run the following **3** chunks to have functions and data available.
```
#@title Utilities and setup
# set up the environment for this tutorial
import time # import time
import numpy as np # import numpy
import scipy as sp # import scipy
from scipy.stats import gamma # import gamma distribution
import math # import basic math functions
import random # import basic random number generator functions
import matplotlib.pyplot as plt # import matplotlib
from IPython import display
fig_w, fig_h = (12, 8)
plt.rcParams.update({'figure.figsize': (fig_w, fig_h)})
plt.style.use('ggplot')
%matplotlib inline
#%config InlineBackend.figure_format = 'retina'
from scipy.signal import medfilt
# make
#@title Convenience functions: Plotting and Filtering
# define some convenience functions to be used later
def my_moving_window(x, window=3, FUN=np.mean):
'''
Calculates a moving estimate for a signal
Args:
x (numpy.ndarray): a vector array of size N
window (int): size of the window, must be a positive integer
FUN (function): the function to apply to the samples in the window
Returns:
(numpy.ndarray): a vector array of size N, containing the moving average
of x, calculated with a window of size window
There are smarter and faster solutions (e.g. using convolution) but this
function shows what the output really means. This function skips NaNs, and
should not be susceptible to edge effects: it will simply use
all the available samples, which means that close to the edges of the
signal or close to NaNs, the output will just be based on fewer samples. By
default, this function will apply a mean to the samples in the window, but
this can be changed to be a max/min/median or other function that returns a
single numeric value based on a sequence of values.
'''
# if data is a matrix, apply filter to each row:
if len(x.shape) == 2:
output = np.zeros(x.shape)
for rown in range(x.shape[0]):
output[rown,:] = my_moving_window(x[rown,:],window=window,FUN=FUN)
return output
# make output array of the same size as x:
output = np.zeros(x.size)
# loop through the signal in x
for samp_i in range(x.size):
values = []
# loop through the window:
for wind_i in range(int(-window), 1):
if ((samp_i+wind_i) < 0) or (samp_i+wind_i) > (x.size - 1):
# out of range
continue
# sample is in range and not nan, use it:
if not(np.isnan(x[samp_i+wind_i])):
values += [x[samp_i+wind_i]]
# calculate the mean in the window for this point in the output:
output[samp_i] = FUN(values)
return output
def my_plot_percepts(datasets=None, plotconditions=False):
if isinstance(datasets,dict):
# try to plot the datasets
# they should be named...
# 'expectations', 'judgments', 'predictions'
fig = plt.figure(figsize=(8, 8)) # set aspect ratio = 1? not really
plt.ylabel('perceived self motion [m/s]')
plt.xlabel('perceived world motion [m/s]')
plt.title('perceived velocities')
# loop through the entries in datasets
# plot them in the appropriate way
for k in datasets.keys():
if k == 'expectations':
expect = datasets[k]
plt.scatter(expect['world'],expect['self'],marker='*',color='xkcd:green',label='my expectations')
elif k == 'judgments':
judgments = datasets[k]
for condition in np.unique(judgments[:,0]):
c_idx = np.where(judgments[:,0] == condition)[0]
cond_self_motion = judgments[c_idx[0],1]
cond_world_motion = judgments[c_idx[0],2]
if cond_world_motion == -1 and cond_self_motion == 0:
c_label = 'world-motion condition judgments'
elif cond_world_motion == 0 and cond_self_motion == 1:
c_label = 'self-motion condition judgments'
else:
c_label = 'condition [%d] judgments'%condition
plt.scatter(judgments[c_idx,3],judgments[c_idx,4], label=c_label, alpha=0.2)
elif k == 'predictions':
predictions = datasets[k]
for condition in np.unique(predictions[:,0]):
c_idx = np.where(predictions[:,0] == condition)[0]
cond_self_motion = predictions[c_idx[0],1]
cond_world_motion = predictions[c_idx[0],2]
if cond_world_motion == -1 and cond_self_motion == 0:
c_label = 'predicted world-motion condition'
elif cond_world_motion == 0 and cond_self_motion == 1:
c_label = 'predicted self-motion condition'
else:
c_label = 'condition [%d] prediction'%condition
plt.scatter(predictions[c_idx,4],predictions[c_idx,3], marker='x', label=c_label)
else:
print("datasets keys should be 'hypothesis', 'judgments' and 'predictions'")
if plotconditions:
# this code is simplified but only works for the dataset we have:
plt.scatter([1],[0],marker='<',facecolor='none',edgecolor='xkcd:black',linewidths=2,label='world-motion stimulus',s=80)
plt.scatter([0],[1],marker='>',facecolor='none',edgecolor='xkcd:black',linewidths=2,label='self-motion stimulus',s=80)
plt.legend(facecolor='xkcd:white')
plt.show()
else:
if datasets is not None:
print('datasets argument should be a dict')
raise TypeError
def my_plot_motion_signals():
dt = 1/10
a = gamma.pdf( np.arange(0,10,dt), 2.5, 0 )
t = np.arange(0,10,dt)
v = np.cumsum(a*dt)
fig, [ax1, ax2] = plt.subplots(nrows=1, ncols=2, sharex='col', sharey='row', figsize=(14,6))
fig.suptitle('Sensory ground truth')
ax1.set_title('world-motion condition')
ax1.plot(t,-v,label='visual [$m/s$]')
ax1.plot(t,np.zeros(a.size),label='vestibular [$m/s^2$]')
ax1.set_xlabel('time [s]')
ax1.set_ylabel('motion')
ax1.legend(facecolor='xkcd:white')
ax2.set_title('self-motion condition')
ax2.plot(t,-v,label='visual [$m/s$]')
ax2.plot(t,a,label='vestibular [$m/s^2$]')
ax2.set_xlabel('time [s]')
ax2.set_ylabel('motion')
ax2.legend(facecolor='xkcd:white')
plt.show()
def my_plot_sensorysignals(judgments, opticflow, vestibular, returnaxes=False, addaverages=False):
wm_idx = np.where(judgments[:,0] == 0)
sm_idx = np.where(judgments[:,0] == 1)
opticflow = opticflow.transpose()
wm_opticflow = np.squeeze(opticflow[:,wm_idx])
sm_opticflow = np.squeeze(opticflow[:,sm_idx])
vestibular = vestibular.transpose()
wm_vestibular = np.squeeze(vestibular[:,wm_idx])
sm_vestibular = np.squeeze(vestibular[:,sm_idx])
X = np.arange(0,10,.1)
fig, my_axes = plt.subplots(nrows=2, ncols=2, sharex='col', sharey='row', figsize=(15,10))
fig.suptitle('Sensory signals')
my_axes[0][0].plot(X,wm_opticflow, color='xkcd:light red', alpha=0.1)
my_axes[0][0].plot([0,10], [0,0], ':', color='xkcd:black')
if addaverages:
my_axes[0][0].plot(X,np.average(wm_opticflow, axis=1), color='xkcd:red', alpha=1)
my_axes[0][0].set_title('world-motion optic flow')
my_axes[0][0].set_ylabel('[motion]')
my_axes[0][1].plot(X,sm_opticflow, color='xkcd:azure', alpha=0.1)
my_axes[0][1].plot([0,10], [0,0], ':', color='xkcd:black')
if addaverages:
my_axes[0][1].plot(X,np.average(sm_opticflow, axis=1), color='xkcd:blue', alpha=1)
my_axes[0][1].set_title('self-motion optic flow')
my_axes[1][0].plot(X,wm_vestibular, color='xkcd:light red', alpha=0.1)
my_axes[1][0].plot([0,10], [0,0], ':', color='xkcd:black')
if addaverages:
my_axes[1][0].plot(X,np.average(wm_vestibular, axis=1), color='xkcd:red', alpha=1)
my_axes[1][0].set_title('world-motion vestibular signal')
my_axes[1][0].set_xlabel('time [s]')
my_axes[1][0].set_ylabel('[motion]')
my_axes[1][1].plot(X,sm_vestibular, color='xkcd:azure', alpha=0.1)
my_axes[1][1].plot([0,10], [0,0], ':', color='xkcd:black')
if addaverages:
my_axes[1][1].plot(X,np.average(sm_vestibular, axis=1), color='xkcd:blue', alpha=1)
my_axes[1][1].set_title('self-motion vestibular signal')
my_axes[1][1].set_xlabel('time [s]')
if returnaxes:
return my_axes
else:
plt.show()
def my_plot_thresholds(thresholds, world_prop, self_prop, prop_correct):
plt.figure(figsize=(12,8))
plt.title('threshold effects')
plt.plot([min(thresholds),max(thresholds)],[0,0],':',color='xkcd:black')
plt.plot([min(thresholds),max(thresholds)],[0.5,0.5],':',color='xkcd:black')
plt.plot([min(thresholds),max(thresholds)],[1,1],':',color='xkcd:black')
plt.plot(thresholds, world_prop, label='world motion')
plt.plot(thresholds, self_prop, label='self motion')
plt.plot(thresholds, prop_correct, color='xkcd:purple', label='correct classification')
plt.xlabel('threshold')
plt.ylabel('proportion correct or classified as self motion')
plt.legend(facecolor='xkcd:white')
plt.show()
def my_plot_predictions_data(judgments, predictions):
conditions = np.concatenate((np.abs(judgments[:,1]),np.abs(judgments[:,2])))
veljudgmnt = np.concatenate((judgments[:,3],judgments[:,4]))
velpredict = np.concatenate((predictions[:,3],predictions[:,4]))
# self:
conditions_self = np.abs(judgments[:,1])
veljudgmnt_self = judgments[:,3]
velpredict_self = predictions[:,3]
# world:
conditions_world = np.abs(judgments[:,2])
veljudgmnt_world = judgments[:,4]
velpredict_world = predictions[:,4]
fig, [ax1, ax2] = plt.subplots(nrows=1, ncols=2, sharey='row', figsize=(12,5))
ax1.scatter(veljudgmnt_self,velpredict_self, alpha=0.2)
ax1.plot([0,1],[0,1],':',color='xkcd:black')
ax1.set_title('self-motion judgments')
ax1.set_xlabel('observed')
ax1.set_ylabel('predicted')
ax2.scatter(veljudgmnt_world,velpredict_world, alpha=0.2)
ax2.plot([0,1],[0,1],':',color='xkcd:black')
ax2.set_title('world-motion judgments')
ax2.set_xlabel('observed')
ax2.set_ylabel('predicted')
plt.show()
#@title Data generation code (needs to go on OSF and deleted here)
def my_simulate_data(repetitions=100, conditions=[(0,-1),(+1,0)] ):
"""
Generate simulated data for this tutorial. You do not need to run this
yourself.
Args:
repetitions: (int) number of repetitions of each condition (default: 30)
conditions: list of 2-tuples of floats, indicating the self velocity and
world velocity in each condition (default: returns data that is
good for exploration: [(-1,0),(0,+1)] but can be flexibly
extended)
The total number of trials used (ntrials) is equal to:
repetitions * len(conditions)
Returns:
dict with three entries:
'judgments': ntrials * 5 matrix
'opticflow': ntrials * 100 matrix
'vestibular': ntrials * 100 matrix
The default settings would result in data where first 30 trials reflect a
situation where the world (other train) moves in one direction, supposedly
at 1 m/s (perhaps to the left: -1) while the participant does not move at
all (0), and 30 trials from a second condition, where the world does not
move, while the participant moves with 1 m/s in the opposite direction from
where the world is moving in the first condition (0,+1). The optic flow
should be the same, but the vestibular input is not.
"""
# reproducible output
np.random.seed(1937)
# set up some variables:
ntrials = repetitions * len(conditions)
# the following arrays will contain the simulated data:
judgments = np.empty(shape=(ntrials,5))
opticflow = np.empty(shape=(ntrials,100))
vestibular = np.empty(shape=(ntrials,100))
# acceleration:
a = gamma.pdf(np.arange(0,10,.1), 2.5, 0 )
# divide by 10 so that velocity scales from 0 to 1 (m/s)
# max acceleration ~ .308 m/s^2
# not realistic! should be about 1/10 of that
# velocity:
v = np.cumsum(a*.1)
# position: (not necessary)
#x = np.cumsum(v)
#################################
# REMOVE ARBITRARY SCALING & CORRECT NOISE PARAMETERS
vest_amp = 1
optf_amp = 1
# we start at the first trial:
trialN = 0
# we start with only a single velocity, but it should be possible to extend this
for conditionno in range(len(conditions)):
condition = conditions[conditionno]
for repetition in range(repetitions):
#
# generate optic flow signal
OF = v * np.diff(condition) # optic flow: difference between self & world motion
OF = (OF * optf_amp) # fairly large spike range
OF = OF + (np.random.randn(len(OF)) * .1) # adding noise
# generate vestibular signal
VS = a * condition[0] # vestibular signal: only self motion
VS = (VS * vest_amp) # less range
VS = VS + (np.random.randn(len(VS)) * 1.) # acceleration is a smaller signal, what is a good noise level?
# store in matrices, corrected for sign
#opticflow[trialN,:] = OF * -1 if (np.sign(np.diff(condition)) < 0) else OF
#vestibular[trialN,:] = VS * -1 if (np.sign(condition[1]) < 0) else VS
opticflow[trialN,:], vestibular[trialN,:] = OF, VS
#########################################################
# store conditions in judgments matrix:
judgments[trialN,0:3] = [ conditionno, condition[0], condition[1] ]
# vestibular SD: 1.0916052957046194 and 0.9112684509277528
# visual SD: 0.10228834313079663 and 0.10975472557444346
# generate judgments:
if (abs(np.average(np.cumsum(medfilt(VS/vest_amp,5)*.1)[70:90])) < 1):
###########################
# NO self motion detected
###########################
selfmotion_weights = np.array([.01,.01]) # there should be low/no self motion
worldmotion_weights = np.array([.01,.99]) # world motion is dictated by optic flow
else:
########################
# self motion DETECTED
########################
#if (abs(np.average(np.cumsum(medfilt(VS/vest_amp,15)*.1)[70:90]) - np.average(medfilt(OF,15)[70:90])) < 5):
if True:
####################
# explain all self motion by optic flow
selfmotion_weights = np.array([.01,.99]) # there should be lots of self motion, but determined by optic flow
worldmotion_weights = np.array([.01,.01]) # very low world motion?
else:
# we use both optic flow and vestibular info to explain both
selfmotion_weights = np.array([ 1, 0]) # motion, but determined by vestibular signal
worldmotion_weights = np.array([ 1, 1]) # very low world motion?
#
integrated_signals = np.array([
np.average( np.cumsum(medfilt(VS/vest_amp,15))[90:100]*.1 ),
np.average((medfilt(OF/optf_amp,15))[90:100])
])
selfmotion = np.sum(integrated_signals * selfmotion_weights)
worldmotion = np.sum(integrated_signals * worldmotion_weights)
#print(worldmotion,selfmotion)
judgments[trialN,3] = abs(selfmotion)
judgments[trialN,4] = abs(worldmotion)
# this ends the trial loop, so we increment the counter:
trialN += 1
return {'judgments':judgments,
'opticflow':opticflow,
'vestibular':vestibular}
simulated_data = my_simulate_data()
judgments = simulated_data['judgments']
opticflow = simulated_data['opticflow']
vestibular = simulated_data['vestibular']
```
#Micro-tutorial 6 - planning the model
```
#@title Video: Planning the model
from IPython.display import YouTubeVideo
video = YouTubeVideo(id='daEtkVporBE', width=854, height=480, fs=1)
print("Video available at https://youtube.com/watch?v=" + video.id)
video
```
###**Goal:** Identify the key components of the model and how they work together.
Our goal all along has been to model our perceptual estimates of sensory data.
Now that we have some idea of what we want to do, we need to line up the components of the model: what are the input and output? Which computations are done and in what order?
The figure below shows a generic model we will use to guide our code construction.

Our model will have:
* **inputs**: the values the system has available - for this tutorial the sensory information in a trial. We want to gather these together and plan how to process them.
* **parameters**: unless we are lucky, our functions will have unknown parameters - we want to identify these and plan for them.
* **outputs**: these are the predictions our model will make - for this tutorial these are the perceptual judgments on each trial. Ideally these are directly comparable to our data.
* **Model functions**: A set of functions that perform the hypothesized computations.
>Using Python (with Numpy and Scipy) we will define a set of functions that take our data and some parameters as input, can run our model, and output a prediction for the judgment data.
#Recap of what we've accomplished so far:
To model perceptual estimates from our sensory data, we need to
1. _integrate_ to ensure sensory information are in appropriate units
2. _reduce noise and set timescale_ by filtering
3. _threshold_ to model detection
Remember the kind of operations we identified:
* integration: `np.cumsum()`
* filtering: `my_moving_window()`
* threshold: `if` with a comparison (`>` or `<`) and `else`
We will collect all the components we've developed and design the code by:
1. **identifying the key functions** we need
2. **sketching the operations** needed in each.
**_Planning our model:_**
We know what we want the model to do, but we need to plan and organize the model into functions and operations.
We're providing a draft of the first function.
For each of the two other code chunks, write mostly comments and help text first. This should put into words what role each of the functions plays in the overall model, implementing one of the steps decided above.
_______
Below is the main function with a detailed explanation of what the function is supposed to do: what input is expected, and what output will generated.
The code is not complete, and only returns nans for now. However, this outlines how most model code works: it gets some measured data (the sensory signals) and a set of parameters as input, and as output returns a prediction on other measured data (the velocity judgments).
The goal of this function is to define the top level of a simulation model which:
* receives all input
* loops through the cases
* calls functions that computes predicted values for each case
* outputs the predictions
### **TD 6.1**: Complete main model function
The function `my_train_illusion_model()` below should call one other function: `my_perceived_motion()`. What input do you think this function should get?
**Complete main model function**
```
def my_train_illusion_model(sensorydata, params):
'''
Generate output predictions of perceived self-motion and perceived world-motion velocity
based on input visual and vestibular signals.
Args (Input variables passed into function):
sensorydata: (dict) dictionary with two named entries:
opticflow: (numpy.ndarray of float) NxM array with N trials on rows
and M visual signal samples in columns
vestibular: (numpy.ndarray of float) NxM array with N trials on rows
and M vestibular signal samples in columns
params: (dict) dictionary with named entries:
threshold: (float) vestibular threshold for credit assignment
filterwindow: (list of int) determines the strength of filtering for
the visual and vestibular signals, respectively
integrate (bool): whether to integrate the vestibular signals, will
be set to True if absent
FUN (function): function used in the filter, will be set to
np.mean if absent
samplingrate (float): the number of samples per second in the
sensory data, will be set to 10 if absent
Returns:
dict with two entries:
selfmotion: (numpy.ndarray) vector array of length N, with predictions
of perceived self motion
worldmotion: (numpy.ndarray) vector array of length N, with predictions
of perceived world motion
'''
# sanitize input a little
if not('FUN' in params.keys()):
params['FUN'] = np.mean
if not('integrate' in params.keys()):
params['integrate'] = True
if not('samplingrate' in params.keys()):
params['samplingrate'] = 10
# number of trials:
ntrials = sensorydata['opticflow'].shape[0]
# set up variables to collect output
selfmotion = np.empty(ntrials)
worldmotion = np.empty(ntrials)
# loop through trials?
for trialN in range(ntrials):
#these are our sensory variables (inputs)
vis = sensorydata['opticflow'][trialN,:]
ves = sensorydata['vestibular'][trialN,:]
########################################################
# generate output predicted perception:
########################################################
#our inputs our vis, ves, and params
selfmotion[trialN], worldmotion[trialN] = [np.nan, np.nan]
########################################################
# replace above with
# selfmotion[trialN], worldmotion[trialN] = my_perceived_motion( ???, ???, params=params)
# and fill in question marks
########################################################
# comment this out when you've filled
raise NotImplementedError("Student excercise: generate predictions")
return {'selfmotion':selfmotion, 'worldmotion':worldmotion}
# uncomment the following lines to run the main model function:
## here is a mock version of my_perceived motion.
## so you can test my_train_illusion_model()
#def my_perceived_motion(*args, **kwargs):
#return np.random.rand(2)
##let's look at the preditions we generated for two sample trials (0,100)
##we should get a 1x2 vector of self-motion prediction and another for world-motion
#sensorydata={'opticflow':opticflow[[0,100],:0], 'vestibular':vestibular[[0,100],:0]}
#params={'threshold':0.33, 'filterwindow':[100,50]}
#my_train_illusion_model(sensorydata=sensorydata, params=params)
```
[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D2_ModelingPractice/solutions/W1D2_Tutorial2_Solution_685e0a13.py)
### **TD 6.2**: Draft perceived motion functions
Now we draft a set of functions, the first of which is used in the main model function (see above) and serves to generate perceived velocities. The other two are used in the first one. Only write help text and/or comments, you don't have to write the whole function. Each time ask yourself these questions:
* what sensory data is necessary?
* what other input does the function need, if any?
* which operations are performed on the input?
* what is the output?
(the number of arguments is correct)
**Template perceived motion**
```
# fill in the input arguments the function should have:
# write the help text for the function:
def my_perceived_motion(arg1, arg2, arg3):
'''
Short description of the function
Args:
argument 1: explain the format and content of the first argument
argument 2: explain the format and content of the second argument
argument 3: explain the format and content of the third argument
Returns:
what output does the function generate?
Any further description?
'''
# structure your code into two functions: "my_selfmotion" and "my_worldmotion"
# write comments outlining the operations to be performed on the inputs by each of these functions
# use the elements from micro-tutorials 3, 4, and 5 (found in W1D2 Tutorial Part 1)
#
#
#
# what kind of output should this function produce?
return output
```
We've completed the `my_perceived_motion()` function for you below. Follow this example to complete the template for `my_selfmotion()` and `my_worldmotion()`. Write out the inputs and outputs, and the steps required to calculate the outputs from the inputs.
**Perceived motion function**
```
#Full perceived motion function
def my_perceived_motion(vis, ves, params):
'''
Takes sensory data and parameters and returns predicted percepts
Args:
vis (numpy.ndarray): 1xM array of optic flow velocity data
ves (numpy.ndarray): 1xM array of vestibular acceleration data
params: (dict) dictionary with named entries:
see my_train_illusion_model() for details
Returns:
[list of floats]: prediction for perceived self-motion based on
vestibular data, and prediction for perceived world-motion based on
perceived self-motion and visual data
'''
# estimate self motion based on only the vestibular data
# pass on the parameters
selfmotion = my_selfmotion(ves=ves,
params=params)
# estimate the world motion, based on the selfmotion and visual data
# pass on the parameters as well
worldmotion = my_worldmotion(vis=vis,
selfmotion=selfmotion,
params=params)
return [selfmotion, worldmotion]
```
**Template calculate self motion**
Put notes in the function below that describe the inputs, the outputs, and steps that transform the output from the input using elements from micro-tutorials 3,4,5.
```
def my_selfmotion(arg1, arg2):
'''
Short description of the function
Args:
argument 1: explain the format and content of the first argument
argument 2: explain the format and content of the second argument
Returns:
what output does the function generate?
Any further description?
'''
# what operations do we perform on the input?
# use the elements from micro-tutorials 3, 4, and 5
# 1.
# 2.
# 3.
# 4.
# what output should this function produce?
return output
```
[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D2_ModelingPractice/solutions/W1D2_Tutorial2_Solution_181325a9.py)
**Template calculate world motion**
Put notes in the function below that describe the inputs, the outputs, and steps that transform the output from the input using elements from micro-tutorials 3,4,5.
```
def my_worldmotion(arg1, arg2, arg3):
'''
Short description of the function
Args:
argument 1: explain the format and content of the first argument
argument 2: explain the format and content of the second argument
argument 3: explain the format and content of the third argument
Returns:
what output does the function generate?
Any further description?
'''
# what operations do we perform on the input?
# use the elements from micro-tutorials 3, 4, and 5
# 1.
# 2.
# 3.
# what output should this function produce?
return output
```
[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D2_ModelingPractice/solutions/W1D2_Tutorial2_Solution_8f913582.py)
#Micro-tutorial 7 - implement model
```
#@title Video: implement the model
from IPython.display import YouTubeVideo
video = YouTubeVideo(id='gtSOekY8jkw', width=854, height=480, fs=1)
print("Video available at https://youtube.com/watch?v=" + video.id)
video
```
**Goal:** We write the components of the model in actual code.
For the operations we picked, there function ready to use:
* integration: `np.cumsum(data, axis=1)` (axis=1: per trial and over samples)
* filtering: `my_moving_window(data, window)` (window: int, default 3)
* average: `np.mean(data)`
* threshold: if (value > thr): <operation 1> else: <operation 2>
###**TD 7.1:** Write code to estimate self motion
Use the operations to finish writing the function that will calculate an estimate of self motion. Fill in the descriptive list of items with actual operations. Use the function for estimating world-motion below, which we've filled for you!
**Template finish self motion function**
```
def my_selfmotion(ves, params):
'''
Estimates self motion for one vestibular signal
Args:
ves (numpy.ndarray): 1xM array with a vestibular signal
params (dict): dictionary with named entries:
see my_train_illusion_model() for details
Returns:
(float): an estimate of self motion in m/s
'''
###uncomment the code below and fill in with your code
## 1. integrate vestibular signal
#ves = np.cumsum(ves*(1/params['samplingrate']))
## 2. running window function to accumulate evidence:
#selfmotion = YOUR CODE HERE
## 3. take final value of self-motion vector as our estimate
#selfmotion =
## 4. compare to threshold. Hint the threshodl is stored in params['threshold']
## if selfmotion is higher than threshold: return value
## if it's lower than threshold: return 0
#if YOURCODEHERE
#selfmotion = YOURCODHERE
# comment this out when you've filled
raise NotImplementedError("Student excercise: estimate my_selfmotion")
return output
```
[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D2_ModelingPractice/solutions/W1D2_Tutorial2_Solution_3ea16348.py)
### Estimate world motion
We have completed the `my_worldmotion()` function for you.
**World motion function**
```
# World motion function
def my_worldmotion(vis, selfmotion, params):
'''
Short description of the function
Args:
vis (numpy.ndarray): 1xM array with the optic flow signal
selfmotion (float): estimate of self motion
params (dict): dictionary with named entries:
see my_train_illusion_model() for details
Returns:
(float): an estimate of world motion in m/s
'''
# running average to smooth/accumulate sensory evidence
visualmotion = my_moving_window(vis,
window=params['filterwindows'][1],
FUN=np.mean)
# take final value
visualmotion = visualmotion[-1]
# subtract selfmotion from value
worldmotion = visualmotion + selfmotion
# return final value
return worldmotion
```
#Micro-tutorial 8 - completing the model
```
#@title Video: completing the model
from IPython.display import YouTubeVideo
video = YouTubeVideo(id='-NiHSv4xCDs', width=854, height=480, fs=1)
print("Video available at https://youtube.com/watch?v=" + video.id)
video
```
**Goal:** Make sure the model can speak to the hypothesis. Eliminate all the parameters that do not speak to the hypothesis.
Now that we have a working model, we can keep improving it, but at some point we need to decide that it is finished. Once we have a model that displays the properties of a system we are interested in, it should be possible to say something about our hypothesis and question. Keeping the model simple makes it easier to understand the phenomenon and answer the research question. Here that means that our model should have illusory perception, and perhaps make similar judgments to those of the participants, but not much more.
To test this, we will run the model, store the output and plot the models' perceived self motion over perceived world motion, like we did with the actual perceptual judgments (it even uses the same plotting function).
### **TD 8.1:** See if the model produces illusions
```
#@title Run to plot model predictions of motion estimates
# prepare to run the model again:
data = {'opticflow':opticflow, 'vestibular':vestibular}
params = {'threshold':0.6, 'filterwindows':[100,50], 'FUN':np.mean}
modelpredictions = my_train_illusion_model(sensorydata=data, params=params)
# process the data to allow plotting...
predictions = np.zeros(judgments.shape)
predictions[:,0:3] = judgments[:,0:3]
predictions[:,3] = modelpredictions['selfmotion']
predictions[:,4] = modelpredictions['worldmotion'] *-1
my_plot_percepts(datasets={'predictions':predictions}, plotconditions=True)
```
**Questions:**
* Why is the data distributed this way? How does it compare to the plot in TD 1.2?
* Did you expect to see this?
* Where do the model's predicted judgments for each of the two conditions fall?
* How does this compare to the behavioral data?
However, the main observation should be that **there are illusions**: the blue and red data points are mixed in each of the two sets of data. Does this mean the model can help us understand the phenomenon?
#Micro-tutorial 9 - testing and evaluating the model
```
#@title Video: Background
from IPython.display import YouTubeVideo
video = YouTubeVideo(id='5vnDOxN3M_k', width=854, height=480, fs=1)
print("Video available at https://youtube.com/watch?v=" + video.id)
video
```
**Goal:** Once we have finished the model, we need a description of how good it is. The question and goals we set in micro-tutorial 1 and 4 help here. There are multiple ways to evaluate a model. Aside from the obvious fact that we want to get insight into the phenomenon that is not directly accessible without the model, we always want to quantify how well the model agrees with the data.
### Quantify model quality with $R^2$
Let's look at how well our model matches the actual judgment data.
```
#@title Run to plot predictions over data
my_plot_predictions_data(judgments, predictions)
```
When model predictions are correct, the red points in the figure above should lie along the identity line (a dotted black line here). Points off the identity line represent model prediction errors. While in each plot we see two clusters of dots that are fairly close to the identity line, there are also two clusters that are not. For the trials that those points represent, the model has an illusion while the participants don't or vice versa.
We will use a straightforward, quantitative measure of how good the model is: $R^2$ (pronounced: "R-squared"), which can take values between 0 and 1, and expresses how much variance is explained by the relationship between two variables (here the model's predictions and the actual judgments). It is also called [coefficient of determination](https://en.wikipedia.org/wiki/Coefficient_of_determination), and is calculated here as the square of the correlation coefficient (r or $\rho$). Just run the chunk below:
```
#@title Run to calculate R^2
conditions = np.concatenate((np.abs(judgments[:,1]),np.abs(judgments[:,2])))
veljudgmnt = np.concatenate((judgments[:,3],judgments[:,4]))
velpredict = np.concatenate((predictions[:,3],predictions[:,4]))
slope, intercept, r_value, p_value, std_err = sp.stats.linregress(conditions,veljudgmnt)
print('conditions -> judgments R^2: %0.3f'%( r_value**2 ))
slope, intercept, r_value, p_value, std_err = sp.stats.linregress(veljudgmnt,velpredict)
print('predictions -> judgments R^2: %0.3f'%( r_value**2 ))
```
These $R^2$s express how well the experimental conditions explain the participants judgments and how well the models predicted judgments explain the participants judgments.
You will learn much more about model fitting, quantitative model evaluation and model comparison tomorrow!
Perhaps the $R^2$ values don't seem very impressive, but the judgments produced by the participants are explained by the model's predictions better than by the actual conditions. In other words: the model tends to have the same illusions as the participants.
### **TD 9.1** Varying the threshold parameter to improve the model
In the code below, see if you can find a better value for the threshold parameter, to reduce errors in the models' predictions.
**Testing thresholds**
```
# Testing thresholds
def test_threshold(threshold=0.33):
# prepare to run model
data = {'opticflow':opticflow, 'vestibular':vestibular}
params = {'threshold':threshold, 'filterwindows':[100,50], 'FUN':np.mean}
modelpredictions = my_train_illusion_model(sensorydata=data, params=params)
# get predictions in matrix
predictions = np.zeros(judgments.shape)
predictions[:,0:3] = judgments[:,0:3]
predictions[:,3] = modelpredictions['selfmotion']
predictions[:,4] = modelpredictions['worldmotion'] *-1
# get percepts from participants and model
conditions = np.concatenate((np.abs(judgments[:,1]),np.abs(judgments[:,2])))
veljudgmnt = np.concatenate((judgments[:,3],judgments[:,4]))
velpredict = np.concatenate((predictions[:,3],predictions[:,4]))
# calculate R2
slope, intercept, r_value, p_value, std_err = sp.stats.linregress(veljudgmnt,velpredict)
print('predictions -> judgments R2: %0.3f'%( r_value**2 ))
test_threshold(threshold=0.5)
```
### **TD 9.2:** Credit assigmnent of self motion
When we look at the figure in **TD 8.1**, we can see a cluster does seem very close to (1,0), just like in the actual data. The cluster of points at (1,0) are from the case where we conclude there is no self motion, and then set the self motion to 0. That value of 0 removes a lot of noise from the world-motion estimates, and all noise from the self-motion estimate. In the other case, where there is self motion, we still have a lot of noise (see also micro-tutorial 4).
Let's change our `my_selfmotion()` function to return a self motion of 1 when the vestibular signal indicates we are above threshold, and 0 when we are below threshold. Edit the function here.
**Template function for credit assigment of self motion**
```
# Template binary self-motion estimates
def my_selfmotion(ves, params):
'''
Estimates self motion for one vestibular signal
Args:
ves (numpy.ndarray): 1xM array with a vestibular signal
params (dict): dictionary with named entries:
see my_train_illusion_model() for details
Returns:
(float): an estimate of self motion in m/s
'''
# integrate signal:
ves = np.cumsum(ves*(1/params['samplingrate']))
# use running window to accumulate evidence:
selfmotion = my_moving_window(ves,
window=params['filterwindows'][0],
FUN=params['FUN'])
## take the final value as our estimate:
selfmotion = selfmotion[-1]
##########################################
# this last part will have to be changed
# compare to threshold, set to 0 if lower and else...
if selfmotion < params['threshold']:
selfmotion = 0
#uncomment the lines below and fill in with your code
#else:
#YOUR CODE HERE
# comment this out when you've filled
raise NotImplementedError("Student excercise: modify with credit assignment")
return selfmotion
```
[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D2_ModelingPractice/solutions/W1D2_Tutorial2_Solution_90571e21.py)
The function you just wrote will be used when we run the model again below.
```
#@title Run model credit assigment of self motion
# prepare to run the model again:
data = {'opticflow':opticflow, 'vestibular':vestibular}
params = {'threshold':0.33, 'filterwindows':[100,50], 'FUN':np.mean}
modelpredictions = my_train_illusion_model(sensorydata=data, params=params)
# no process the data to allow plotting...
predictions = np.zeros(judgments.shape)
predictions[:,0:3] = judgments[:,0:3]
predictions[:,3] = modelpredictions['selfmotion']
predictions[:,4] = modelpredictions['worldmotion'] *-1
my_plot_percepts(datasets={'predictions':predictions}, plotconditions=False)
```
That looks much better, and closer to the actual data. Let's see if the $R^2$ values have improved:
```
#@title Run to calculate R^2 for model with self motion credit assignment
conditions = np.concatenate((np.abs(judgments[:,1]),np.abs(judgments[:,2])))
veljudgmnt = np.concatenate((judgments[:,3],judgments[:,4]))
velpredict = np.concatenate((predictions[:,3],predictions[:,4]))
my_plot_predictions_data(judgments, predictions)
slope, intercept, r_value, p_value, std_err = sp.stats.linregress(conditions,veljudgmnt)
print('conditions -> judgments R2: %0.3f'%( r_value**2 ))
slope, intercept, r_value, p_value, std_err = sp.stats.linregress(velpredict,veljudgmnt)
print('predictions -> judgments R2: %0.3f'%( r_value**2 ))
```
While the model still predicts velocity judgments better than the conditions (i.e. the model predicts illusions in somewhat similar cases), the $R^2$ values are actually worse than those of the simpler model. What's really going on is that the same set of points that were model prediction errors in the previous model are also errors here. All we have done is reduce the spread.
### Interpret the model's meaning
Here's what you should have learned:
1. A noisy, vestibular, acceleration signal can give rise to illusory motion.
2. However, disambiguating the optic flow by adding the vestibular signal simply adds a lot of noise. This is not a plausible thing for the brain to do.
3. Our other hypothesis - credit assignment - is more qualitatively correct, but our simulations were not able to match the frequency of the illusion on a trial-by-trial basis.
_It's always possible to refine our models to improve the fits._
There are many ways to try to do this. A few examples; we could implement a full sensory cue integration model, perhaps with Kalman filters (Week 2, Day 3), or we could add prior knowledge (at what time do the trains depart?). However, we decided that for now we have learned enough, so it's time to write it up.
# Micro-tutorial 10 - publishing the model
```
#@title Video: Background
from IPython.display import YouTubeVideo
video = YouTubeVideo(id='kf4aauCr5vA', width=854, height=480, fs=1)
print("Video available at https://youtube.com/watch?v=" + video.id)
video
```
**Goal:** In order for our model to impact the field, it needs to be accepted by our peers, and order for that to happen it matters how the model is published.
### **TD 10.1:** Write a summary of the project
Here we will write up our model, by answering the following questions:
* **What is the phenomena**? Here summarize the part of the phenomena which your model addresses.
* **What is the key scientific question?**: Clearly articulate the question which your model tries to answer.
* **What was our hypothesis?**: Explain the key relationships which we relied on to simulate the phenomena.
* **How did your model work?** Give an overview of the model, it's main components, and how the model works. ''Here we ... ''
* **What did we find? Did the model work?** Explain the key outcomes of your model evaluation.
* **What can we conclude?** Conclude as much as you can _with reference to the hypothesis_, within the limits of the model.
* **What did you learn? What is left to be learned?** Briefly argue the plausibility of the approach and what you think is _essential_ that may have been left out.
### Guidance for the future
There are good guidelines for structuring and writing an effective paper (e.g. [Mensh & Kording, 2017](https://doi.org/10.1371/journal.pcbi.1005619)), all of which apply to papers about models. There are some extra considerations when publishing a model. In general, you should explain each of the steps in the paper:
**Introduction:** Steps 1 & 2 (maybe 3)
**Methods:** Steps 3-7, 9
**Results:** Steps 8 & 9, going back to 1, 2 & 4
In addition, you should provide a visualization of the model, and upload the code implementing the model and the data it was trained and tested on to a repository (e.g. GitHub and OSF).
The audience for all of this should be experimentalists, as they are the ones who can test predictions made by your your model and collect new data. This way your models can impact future experiments, and that future data can then be modeled (see modeling process schematic below). Remember your audience - it is _always_ hard to clearly convey the main points of your work to others, especially if your audience doesn't necessarily create computational models themselves.

### Suggestion
For every modeling project, a very good exercise in this is to _**first**_ write a short, 100-word abstract of the project plan and expected impact, like the summary you wrote. This forces focussing on the main points: describing the relevance, question, model, answer and what it all means very succinctly. This allows you to decide to do this project or not **before you commit time writing code for no good purpose**. Notice that this is really what we've walked you through carefully in this tutorial! :)
# Post-script
Note that the model we built here was extremely simple and used artificial data on purpose. It allowed us to go through all the steps of building a model, and hopefully you noticed that it is not always a linear process, you will go back to different steps if you hit a roadblock somewhere.
However, if you're interested in how to actually approach modeling a similar phenomenon in a probabilistic way, we encourage you to read the paper by [Dokka et. al., 2019](https://doi.org/10.1073/pnas.1820373116), where the authors model how judgments of heading direction are influenced by objects that are also moving.
# Reading
Blohm G, Kording KP, Schrater PR (2020). _A How-to-Model Guide for Neuroscience_ eNeuro, 7(1) ENEURO.0352-19.2019. https://doi.org/10.1523/ENEURO.0352-19.2019
Dokka K, Park H, Jansen M, DeAngelis GC, Angelaki DE (2019). _Causal inference accounts for heading perception in the presence of object motion._ PNAS, 116(18):9060-9065. https://doi.org/10.1073/pnas.1820373116
Drugowitsch J, DeAngelis GC, Klier EM, Angelaki DE, Pouget A (2014). _Optimal Multisensory Decision-Making in a Reaction-Time Task._ eLife, 3:e03005. https://doi.org/10.7554/eLife.03005
Hartmann, M, Haller K, Moser I, Hossner E-J, Mast FW (2014). _Direction detection thresholds of passive self-motion in artistic gymnasts._ Exp Brain Res, 232:1249–1258. https://doi.org/10.1007/s00221-014-3841-0
Mensh B, Kording K (2017). _Ten simple rules for structuring papers._ PLoS Comput Biol 13(9): e1005619. https://doi.org/10.1371/journal.pcbi.1005619
Seno T, Fukuda H (2012). _Stimulus Meanings Alter Illusory Self-Motion (Vection) - Experimental Examination of the Train Illusion._ Seeing Perceiving, 25(6):631-45. https://doi.org/10.1163/18784763-00002394
|
github_jupyter
|
_Lambda School Data Science_
# Make explanatory visualizations
Tody we will reproduce this [example by FiveThirtyEight:](https://fivethirtyeight.com/features/al-gores-new-movie-exposes-the-big-flaw-in-online-movie-ratings/)
```
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
```
Using this data: https://github.com/fivethirtyeight/data/tree/master/inconvenient-sequel
Objectives
- add emphasis and annotations to transform visualizations from exploratory to explanatory
- remove clutter from visualizations
Links
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
## Make prototypes
This helps us understand the problem
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11)) # index will start from 0 if not for this
fake.plot.bar(color='C1', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
```
## Annotate with text
```
display(example)
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11)) # index will start from 0 if not for this
fake.plot.bar(color='C1', width=0.9);
# rotate x axis numbers
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11)) # index will start from 0 if not for this
ax = fake.plot.bar(color='C1', width=0.9)
ax.tick_params(labelrotation=0) #to unrotate or remove the rotation
ax.set(title="'An Incovenient Sequel: Truth to Power' is divisive");
#or '\'An Incovenient Sequel: Truth to Power\' is divisive'
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11)) # index will start from 0 if not for this
ax = fake.plot.bar(color='C1', width=0.9)
ax.tick_params(labelrotation=0)
ax.text(x=-2,y=48,s="'An Incovenient Sequel: Truth to Power' is divisive",
fontsize=16, fontweight='bold')
ax.text(x=-2,y=45, s='IMDb ratings for the film as of Aug. 29',
fontsize=12)
ax.set(xlabel='Rating',
ylabel='Percent of total votes',
yticks=range(0,50,10));
#(start pt., end pt., increment)
```
## Reproduce with real data
```
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
df.shape
df.head()
width,height = df.shape
width*height
pd.options.display.max_columns = 500
df.head()
df.sample(1).T
df.timestamp.describe()
# convert timestamp to date time
df.timestamp = pd.to_datetime(df.timestamp)
df.timestamp.describe()
# Making datetime index of your df
df = df.set_index('timestamp')
df.head()
df['2017-08-09']
# everything from this date
df.category.value_counts()
```
####only interested in IMDb users
```
df.category == 'IMDb users'
# As a filter to select certain rows
df[df.category == 'IMDb users']
lastday = df['2017-08-09']
lastday.head(1)
lastday[lastday.category =='IMDb users'].tail()
lastday[lastday.category =='IMDb users'].respondents.plot();
final = df.tail(1)
#columns = ['1_pct','2_pct','3_pct','4_pct','5_pct','6_pct','7_pct','8_pct','9_pct','10_pct']
#OR
columns = [str(i) + '_pct' for i in range(1,11)]
final[columns]
#OR
#data.index.str.replace('_pct', '')
data = final[columns].T
data
data.plot.bar()
plt.style.use('fivethirtyeight')
ax = data.plot.bar(color='C1', width=0.9)
ax.tick_params(labelrotation=0)
ax.text(x=-2,y=48,s="'An Incovenient Sequel: Truth to Power' is divisive",
fontsize=16, fontweight='bold')
ax.text(x=-2,y=44, s='IMDb ratings for the film as of Aug. 29',
fontsize=12)
ax.set(xlabel='Rating',
ylabel='Percent of total votes',
yticks=range(0,50,10));
#(start pt., end pt., increment)
# to remove the timestamp texts in the center
# to change the x axis texts
plt.style.use('fivethirtyeight')
ax = data.plot.bar(color='C1', width=0.9, legend=False)
ax.tick_params(labelrotation=0)
ax.text(x=-2,y=48,s="'An Incovenient Sequel: Truth to Power' is divisive",
fontsize=16, fontweight='bold')
ax.text(x=-2,y=44, s='IMDb ratings for the film as of Aug. 29',
fontsize=12)
ax.set(xlabel='Rating',
ylabel='Percent of total votes',
yticks=range(0,50,10));
data.index = range(1,11)
data
plt.style.use('fivethirtyeight')
ax = data.plot.bar(color='C1', width=0.9, legend=False)
ax.tick_params(labelrotation=0)
ax.text(x=-2,y=48,s="'An Incovenient Sequel: Truth to Power' is divisive",
fontsize=16, fontweight='bold')
ax.text(x=-2,y=44, s='IMDb ratings for the film as of Aug. 29',
fontsize=12)
ax.set(xlabel='Rating',
ylabel='Percent of total votes',
yticks=range(0,50,10))
plt.xlabel('Rating', fontsize=14);
```
# ASSIGNMENT
Replicate the lesson code. I recommend that you [do not copy-paste](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit).
# STRETCH OPTIONS
#### Reproduce another example from [FiveThityEight's shared data repository](https://data.fivethirtyeight.com/).
For example:
- [thanksgiving-2015](https://fivethirtyeight.com/features/heres-what-your-part-of-america-eats-on-thanksgiving/) (try the [`altair`](https://altair-viz.github.io/gallery/index.html#maps) library)
- [candy-power-ranking](https://fivethirtyeight.com/features/the-ultimate-halloween-candy-power-ranking/) (try the [`statsmodels`](https://www.statsmodels.org/stable/index.html) library)
- or another example of your choice!
#### Make more charts!
Choose a chart you want to make, from [FT's Visual Vocabulary poster](http://ft.com/vocabulary).
Find the chart in an example gallery of a Python data visualization library:
- [Seaborn](http://seaborn.pydata.org/examples/index.html)
- [Altair](https://altair-viz.github.io/gallery/index.html)
- [Matplotlib](https://matplotlib.org/gallery.html)
- [Pandas](https://pandas.pydata.org/pandas-docs/stable/visualization.html)
Reproduce the chart. [Optionally, try the "Ben Franklin Method."](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit) If you want, experiment and make changes.
Take notes. Consider sharing your work with your cohort!
|
github_jupyter
|
... ***CURRENTLY UNDER DEVELOPMENT*** ...
## Obtain synthetic waves and water level timeseries under a climate change scenario (future AWTs occurrence probability)
inputs required:
* Historical DWTs (for plotting)
* Historical wave families (for plotting)
* Synthetic DWTs climate change
* Historical intradaily hydrograph parameters
* TCs waves
* Fitted multivariate extreme model for the waves associated to each DWT
in this notebook:
* Generate synthetic time series of wave conditions
```
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# common
import os
import os.path as op
# pip
import numpy as np
import xarray as xr
import pandas as pd
from datetime import datetime
import matplotlib.pyplot as plt
# DEV: override installed teslakit
import sys
sys.path.insert(0, op.join(os.path.abspath(''), '..', '..','..', '..'))
# teslakit
from teslakit.database import Database
from teslakit.climate_emulator import Climate_Emulator
from teslakit.waves import AWL, Aggregate_WavesFamilies
from teslakit.plotting.outputs import Plot_FitSim_Histograms
from teslakit.plotting.extremes import Plot_FitSim_AnnualMax, Plot_FitSim_GevFit, Plot_Fit_QQ
from teslakit.plotting.waves import Plot_Waves_Histogram_FitSim
```
## Database and Site parameters
```
# --------------------------------------
# Teslakit database
p_data = r'/Users/anacrueda/Documents/Proyectos/TESLA/data'
# offshore
db = Database(p_data)
db.SetSite('ROI')
# climate change - S5
db_S5 = Database(p_data)
db_S5.SetSite('ROI_CC_S5')
# climate emulator simulation modified path
p_S5_CE_sims = op.join(db_S5.paths.site.EXTREMES.climate_emulator, 'Simulations')
# --------------------------------------
# Load data for climate emulator simulation climate change: ESTELA DWT and TCs (MU, TAU)
DWTs_sim = db_S5.Load_ESTELA_DWT_sim() # DWTs climate change
TCs_params = db.Load_TCs_r2_sim_params() # TCs parameters (copula generated)
TCs_RBFs = db.Load_TCs_sim_r2_rbf_output() # TCs numerical_IH-RBFs_interpolation output
probs_TCs = db.Load_TCs_probs_synth() # TCs synthetic probabilities
pchange_TCs = probs_TCs['category_change_cumsum'].values[:]
l_mutau_wt = db.Load_MU_TAU_hydrograms() # MU - TAU intradaily hidrographs for each DWT
MU_WT = np.array([x.MU.values[:] for x in l_mutau_wt]) # MU and TAU numpy arrays
TAU_WT = np.array([x.TAU.values[:] for x in l_mutau_wt])
# solve first 10 DWTs simulations
DWTs_sim = DWTs_sim.isel(n_sim=slice(0, 10))
#DWTs_sim = DWTs_sim.isel(time=slice(0,365*40+10), n_sim=slice(0,1))
print(DWTs_sim)
```
## Climate Emulator - Simulation
```
# --------------------------------------
# Climate Emulator extremes model fitting
# Load Climate Emulator
CE = Climate_Emulator(db.paths.site.EXTREMES.climate_emulator)
CE.Load()
# set a new path for S5 simulations
CE.Set_Simulation_Folder(p_S5_CE_sims, copy_WAVES_noTCs = False) # climate change waves (no TCs) not simulated, DWTs have changed
# optional: list variables to override distribution to empirical
#CE.sim_icdf_empirical_override = ['sea_Hs_31',
# 'swell_1_Hs_1','swell_1_Tp_1',
# 'swell_1_Hs_2','swell_1_Tp_2',]
# set simulated waves min-max filter
CE.sim_waves_filter.update({
'hs': (0, 8),
'tp': (2, 25),
'ws': (0, 0.06),
})
# --------------------------------------
# Climate Emulator simulation
# each DWT series will generate a different set of waves
for n in DWTs_sim.n_sim:
print('- Sim: {0} -'.format(int(n)+1))
# Select DWTs simulation
DWTs = DWTs_sim.sel(n_sim=n)
# Simulate waves
n_ce = 1 # (one CE sim. for each DWT sim.)
WVS_sim = CE.Simulate_Waves(DWTs, n_ce, filters={'hs':True, 'tp':True, 'ws':True})
# Simulate TCs and update simulated waves
TCs_sim, WVS_upd = CE.Simulate_TCs(DWTs, WVS_sim, TCs_params, TCs_RBFs, pchange_TCs, MU_WT, TAU_WT)
# store simulation data
CE.SaveSim(WVS_sim, TCs_sim, WVS_upd, int(n))
```
|
github_jupyter
|
# Cyclical Systems: An Example of the Crank-Nicolson Method
## CH EN 2450 - Numerical Methods
**Prof. Tony Saad (<a>www.tsaad.net</a>) <br/>Department of Chemical Engineering <br/>University of Utah**
<hr/>
```
import numpy as np
from numpy import *
# %matplotlib notebook
# %matplotlib nbagg
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
# %matplotlib qt
import matplotlib.pyplot as plt
from scipy.optimize import fsolve
from scipy.integrate import odeint
def forward_euler(rhs, f0, tend, dt):
''' Computes the forward_euler method '''
nsteps = int(tend/dt)
f = np.zeros(nsteps)
f[0] = f0
time = np.linspace(0,tend,nsteps)
for n in np.arange(nsteps-1):
f[n+1] = f[n] + dt * rhs(f[n], time[n])
return time, f
def forward_euler_system(rhsvec, f0vec, tend, dt):
'''
Solves a system of ODEs using the Forward Euler method
'''
nsteps = int(tend/dt)
neqs = len(f0vec)
f = np.zeros( (neqs, nsteps) )
f[:,0] = f0vec
time = np.linspace(0,tend,nsteps)
for n in np.arange(nsteps-1):
t = time[n]
f[:,n+1] = f[:,n] + dt * rhsvec(f[:,n], t)
return time, f
def be_residual(fnp1, rhs, fn, dt, tnp1):
'''
Nonlinear residual function for the backward Euler implicit time integrator
'''
return fnp1 - fn - dt * rhs(fnp1, tnp1)
def backward_euler(rhs, f0, tend, dt):
'''
Computes the backward euler method
:param rhs: an rhs function
'''
nsteps = int(tend/dt)
f = np.zeros(nsteps)
f[0] = f0
time = np.linspace(0,tend,nsteps)
for n in np.arange(nsteps-1):
fn = f[n]
tnp1 = time[n+1]
fnew = fsolve(be_residual, fn, (rhs, fn, dt, tnp1))
f[n+1] = fnew
return time, f
def cn_residual(fnp1, rhs, fn, dt, tnp1, tn):
'''
Nonlinear residual function for the Crank-Nicolson implicit time integrator
'''
return fnp1 - fn - 0.5 * dt * ( rhs(fnp1, tnp1) + rhs(fn, tn) )
def crank_nicolson(rhs,f0,tend,dt):
nsteps = int(tend/dt)
f = np.zeros(nsteps)
f[0] = f0
time = np.linspace(0,tend,nsteps)
for n in np.arange(nsteps-1):
fn = f[n]
tnp1 = time[n+1]
tn = time[n]
fnew = fsolve(cn_residual, fn, (rhs, fn, dt, tnp1, tn))
f[n+1] = fnew
return time, f
```
# Sharp Transient
Solve the ODE:
\begin{equation}
\frac{\text{d}y}{\text{d}t} = -1000 y + 3000 - 2000 e^{-t};\quad y(0) = 0
\end{equation}
The analytical solution is
\begin{equation}
y(t) = 3 - 0.998 e^{-1000t} - 2.002 e^{-t}
\end{equation}
We first plot the analytical solution
```
y = lambda t : 3 - 0.998*exp(-1000*t) - 2.002*exp(-t)
t = np.linspace(0,1,500)
plt.plot(t,y(t))
plt.grid()
```
Now let's solve this numerically. We first define the RHS for this function
```
def rhs_sharp_transient(f,t):
return 3000 - 1000 * f - 2000* np.exp(-t)
```
Let's solve this using forward euler and backward euler
```
y0 = 0
tend = 0.03
dt = 0.001
t,yfe = forward_euler(rhs_sharp_transient,y0,tend,dt)
t,ybe = backward_euler(rhs_sharp_transient,y0,tend,dt)
t,ycn = crank_nicolson(rhs_sharp_transient,y0,tend,dt)
plt.plot(t,y(t),label='Exact')
# plt.plot(t,yfe,'r.-',markevery=1,markersize=10,label='Forward Euler')
plt.plot(t,ybe,'k*-',markevery=2,markersize=10,label='Backward Euler')
plt.plot(t,ycn,'o-',markevery=2,markersize=2,label='Crank Nicholson')
plt.grid()
plt.legend()
```
# Oscillatory Systems
Solve the ODE:
Solve the ODE:
\begin{equation}
\frac{\text{d}y}{\text{d}t} = r \omega \sin(\omega t)
\end{equation}
The analytical solution is
\begin{equation}
y(t) = r - r \cos(\omega t)
\end{equation}
First plot the analytical solution
```
r = 0.5
ω = 0.02
y = lambda t : r - r * cos(ω*t)
t = np.linspace(0,100*pi)
plt.clf()
plt.plot(t,y(t))
plt.grid()
```
Let's solve this numerically
```
def rhs_oscillatory(f,t):
r = 0.5
ω = 0.02
return r * ω * sin(ω*t)
y0 = 0
tend = 100*pi
dt = 10
t,yfe = forward_euler(rhs_oscillatory,y0,tend,dt)
t,ybe = backward_euler(rhs_oscillatory,y0,tend,dt)
t,ycn = crank_nicolson(rhs_oscillatory,y0,tend,dt)
plt.plot(t,y(t),label='Exact')
plt.plot(t,yfe,'r.-',markevery=1,markersize=10,label='Forward Euler')
plt.plot(t,ybe,'k*-',markevery=2,markersize=10,label='Backward Euler')
plt.plot(t,ycn,'o-',markevery=2,markersize=2,label='Crank Nicholson')
plt.grid()
plt.legend()
plt.savefig('cyclical-system-example.pdf')
import urllib
import requests
from IPython.core.display import HTML
def css_styling():
styles = requests.get("https://raw.githubusercontent.com/saadtony/NumericalMethods/master/styles/custom.css")
return HTML(styles.text)
css_styling()
```
|
github_jupyter
|
<table class="ee-notebook-buttons" align="left">
<td><a target="_parent" href="https://github.com/giswqs/geemap/tree/master/tutorials/Image/06_convolutions.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_parent" href="https://nbviewer.jupyter.org/github/giswqs/geemap/blob/master/tutorials/Image/06_convolutions.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_parent" href="https://colab.research.google.com/github/giswqs/geemap/blob/master/tutorials/Image/06_convolutions.ipynb"><img width=26px src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
# Convolutions
To perform linear convolutions on images, use `image.convolve()`. The only argument to convolve is an `ee.Kernel` which is specified by a shape and the weights in the kernel. Each pixel of the image output by `convolve()` is the linear combination of the kernel values and the input image pixels covered by the kernel. The kernels are applied to each band individually. For example, you might want to use a low-pass (smoothing) kernel to remove high-frequency information. The following illustrates a 15x15 low-pass kernel applied to a Landsat 8 image:
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.foliumap`](https://github.com/giswqs/geemap/blob/master/geemap/foliumap.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.foliumap as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
```
Map = emap.Map(center=[40, -100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
```
## Add Earth Engine Python script
```
# Load and display an image.
image = ee.Image('LANDSAT/LC08/C01/T1_TOA/LC08_044034_20140318')
Map.setCenter(-121.9785, 37.8694, 11)
Map.addLayer(image, {'bands': ['B5', 'B4', 'B3'], 'max': 0.5}, 'input image')
# Define a boxcar or low-pass kernel.
# boxcar = ee.Kernel.square({
# 'radius': 7, 'units': 'pixels', 'normalize': True
# })
boxcar = ee.Kernel.square(7, 'pixels', True)
# Smooth the image by convolving with the boxcar kernel.
smooth = image.convolve(boxcar)
Map.addLayer(smooth, {'bands': ['B5', 'B4', 'B3'], 'max': 0.5}, 'smoothed')
Map.addLayerControl()
Map
```
The output of convolution with the low-pass filter should look something like Figure 1. Observe that the arguments to the kernel determine its size and coefficients. Specifically, with the `units` parameter set to pixels, the `radius` parameter specifies the number of pixels from the center that the kernel will cover. If `normalize` is set to true, the kernel coefficients will sum to one. If the `magnitude` parameter is set, the kernel coefficients will be multiplied by the magnitude (if `normalize` is also true, the coefficients will sum to `magnitude`). If there is a negative value in any of the kernel coefficients, setting `normalize` to true will make the coefficients sum to zero.
Use other kernels to achieve the desired image processing effect. This example uses a Laplacian kernel for isotropic edge detection:
```
Map = emap.Map(center=[40, -100], zoom=4)
# Define a Laplacian, or edge-detection kernel.
laplacian = ee.Kernel.laplacian8(1, False)
# Apply the edge-detection kernel.
edgy = image.convolve(laplacian)
Map.addLayer(edgy, {'bands': ['B5', 'B4', 'B3'], 'max': 0.5}, 'edges')
Map.setCenter(-121.9785, 37.8694, 11)
Map.addLayerControl()
Map
```
Note the format specifier in the visualization parameters. Earth Engine sends display tiles to the Code Editor in JPEG format for efficiency, however edge tiles are sent in PNG format to handle transparency of pixels outside the image boundary. When a visual discontinuity results, setting the format to PNG results in a consistent display. The result of convolving with the Laplacian edge detection kernel should look something like Figure 2.
There are also anisotropic edge detection kernels (e.g. Sobel, Prewitt, Roberts), the direction of which can be changed with `kernel.rotate()`. Other low pass kernels include a Gaussian kernel and kernels of various shape with uniform weights. To create kernels with arbitrarily defined weights and shape, use `ee.Kernel.fixed()`. For example, this code creates a 9x9 kernel of 1’s with a zero in the middle:
```
# Create a list of weights for a 9x9 kernel.
list = [1, 1, 1, 1, 1, 1, 1, 1, 1]
# The center of the kernel is zero.
centerList = [1, 1, 1, 1, 0, 1, 1, 1, 1]
# Assemble a list of lists: the 9x9 kernel weights as a 2-D matrix.
lists = [list, list, list, list, centerList, list, list, list, list]
# Create the kernel from the weights.
kernel = ee.Kernel.fixed(9, 9, lists, -4, -4, False)
print(kernel.getInfo())
```
|
github_jupyter
|
<h1 align="center">Theano</h1>
```
!pip install numpy matplotlib
!pip install --upgrade https://github.com/Theano/Theano/archive/master.zip
!pip install --upgrade https://github.com/Lasagne/Lasagne/archive/master.zip
```
### Разминка
```
import theano
import theano.tensor as T
%pylab inline
```
#### будущий параметр функции -- символьная переменная
```
N = T.scalar('a dimension', dtype='float32')
```
#### рецепт получения квадрата -- орперации над символьными переменным
```
result = T.power(N, 2)
```
#### theano.grad(cost, wrt)
```
grad_result = theano.grad(result, N)
```
#### компиляция функции "получения квадрата"
```
sq_function = theano.function(inputs=[N], outputs=result)
gr_function = theano.function(inputs=[N], outputs=grad_result)
```
#### применение функции
```
# Заводим np.array x
xv = np.arange(-10, 10)
# Применяем функцию к каждому x
val = map(float, [sq_function(x) for x in xv])
# Посичтаем градиент в кажой точке
grad = map(float, [gr_function(x) for x in xv])
```
### Что мы увидим если нарисуем функцию и градиент?
```
pylab.plot(xv, val, label='x*x')
pylab.plot(xv, grad, label='d x*x / dx')
pylab.legend()
```
<h1 align="center">Lasagne</h1>
* lasagne - это библиотека для написания нейронок произвольной формы на theano
* В качестве демо-задачи выберем то же распознавание чисел, но на большем масштабе задачи, картинки 28x28, 10 цифр
```
from mnist import load_dataset
X_train, y_train, X_val, y_val, X_test, y_test = load_dataset()
print 'X размера', X_train.shape, 'y размера', y_train.shape
fig, axes = plt.subplots(nrows=1, ncols=7, figsize=(20, 20))
for i, ax in enumerate(axes):
ax.imshow(X_train[i, 0], cmap='gray')
```
Давайте посмотрим на DenseLayer в lasagne
- http://lasagne.readthedocs.io/en/latest/modules/layers/dense.html
- https://github.com/Lasagne/Lasagne/blob/master/lasagne/layers/dense.py#L16-L124
- Весь содаржательный код тут https://github.com/Lasagne/Lasagne/blob/master/lasagne/layers/dense.py#L121
```
import lasagne
from lasagne import init
from theano import tensor as T
from lasagne.nonlinearities import softmax
X, y = T.tensor4('X'), T.vector('y', 'int32')
```
Так задаётся архитектура нейронки
```
#входной слой (вспомогательный)
net = lasagne.layers.InputLayer(shape=(None, 1, 28, 28), input_var=X)
net = lasagne.layers.Conv2DLayer(net, 15, 28, pad='valid', W=init.Constant()) # сверточный слой
net = lasagne.layers.Conv2DLayer(net, 10, 2, pad='full', W=init.Constant()) # сверточный слой
net = lasagne.layers.DenseLayer(net, num_units=500) # полносвязный слой
net = lasagne.layers.DropoutLayer(net, 1.0) # регуляризатор
net = lasagne.layers.DenseLayer(net, num_units=200) # полносвязный слой
net = lasagne.layers.DenseLayer(net, num_units=10) # полносвязный слой
#предсказание нейронки (theano-преобразование)
y_predicted = lasagne.layers.get_output(net)
#все веса нейронки (shared-переменные)
all_weights = lasagne.layers.get_all_params(net)
print all_weights
#функция ошибки и точности будет прямо внутри
loss = lasagne.objectives.categorical_accuracy(y_predicted, y).mean()
accuracy = lasagne.objectives.categorical_accuracy(y_predicted, y).mean()
#сразу посчитать словарь обновлённых значений с шагом по градиенту, как раньше
updates = lasagne.updates.momentum(loss, all_weights, learning_rate=1.0, momentum=1.5)
#функция, делает updates и возвращащет значение функции потерь и точности
train_fun = theano.function([X, y], [loss, accuracy], updates=updates)
accuracy_fun = theano.function([X, y], accuracy) # точность без обновления весов, для теста
```
# Процесс обучения
```
import time
from mnist import iterate_minibatches
num_epochs = 5 #количество проходов по данным
batch_size = 50 #размер мини-батча
for epoch in range(num_epochs):
train_err, train_acc, train_batches, start_time = 0, 0, 0, time.time()
for inputs, targets in iterate_minibatches(X_train, y_train, batch_size):
train_err_batch, train_acc_batch = train_fun(inputs, targets)
train_err += train_err_batch
train_acc += train_acc_batch
train_batches += 1
val_acc, val_batches = 0, 0
for inputs, targets in iterate_minibatches(X_test, y_test, batch_size):
val_acc += accuracy_fun(inputs, targets)
val_batches += 1
print "Epoch %s of %s took %.3f s" % (epoch + 1, num_epochs, time.time() - start_time)
print " train loss:\t %.3f" % (train_err / train_batches)
print " train acc:\t %.3f" % (train_acc * 100 / train_batches), '%'
print " test acc:\t %.3f" % (val_acc * 100 / val_batches), '%'
print
test_acc = 0
test_batches = 0
for batch in iterate_minibatches(X_test, y_test, 500):
inputs, targets = batch
acc = accuracy_fun(inputs, targets)
test_acc += acc
test_batches += 1
print("Final results: \n test accuracy:\t\t{:.2f} %".format(test_acc / test_batches * 100))
```
# Ансамблирование с DropOut
```
#предсказание нейронки (theano-преобразование)
y_predicted = T.mean([lasagne.layers.get_output(net, deterministic=False) for i in range(10)], axis=0)
accuracy = lasagne.objectives.categorical_accuracy(y_predicted, y).mean()
accuracy_fun = theano.function([X, y], accuracy) # точность без обновления весов, для теста
test_acc = 0
test_batches = 0
for batch in iterate_minibatches(X_test, y_test, 500):
inputs, targets = batch
acc = accuracy_fun(inputs, targets)
test_acc += acc
test_batches += 1
print("Final results: \n test accuracy:\t\t{:.2f} %".format(test_acc / test_batches * 100))
```
|
github_jupyter
|
查看当前GPU信息
```
from tensorflow.python.client import device_lib
device_lib.list_local_devices()
!pip install bert-tensorflow
import pandas as pd
import tensorflow as tf
import tensorflow_hub as hub
import pickle
import bert
from bert import run_classifier
from bert import optimization
from bert import tokenization
def pretty_print(result):
df = pd.DataFrame([result]).T
df.columns = ["values"]
return df
def create_tokenizer_from_hub_module(bert_model_hub):
"""Get the vocab file and casing info from the Hub module."""
with tf.Graph().as_default():
bert_module = hub.Module(bert_model_hub)
tokenization_info = bert_module(signature="tokenization_info", as_dict=True)
with tf.Session() as sess:
vocab_file, do_lower_case = sess.run([tokenization_info["vocab_file"],
tokenization_info["do_lower_case"]])
return bert.tokenization.FullTokenizer(
vocab_file=vocab_file, do_lower_case=do_lower_case)
def make_features(dataset, label_list, MAX_SEQ_LENGTH, tokenizer, DATA_COLUMN, LABEL_COLUMN):
input_example = dataset.apply(lambda x: bert.run_classifier.InputExample(guid=None,
text_a = x[DATA_COLUMN],
text_b = None,
label = x[LABEL_COLUMN]), axis = 1)
features = bert.run_classifier.convert_examples_to_features(input_example, label_list, MAX_SEQ_LENGTH, tokenizer)
return features
def create_model(bert_model_hub, is_predicting, input_ids, input_mask, segment_ids, labels,
num_labels):
"""Creates a classification model."""
bert_module = hub.Module(
bert_model_hub,
trainable=True)
bert_inputs = dict(
input_ids=input_ids,
input_mask=input_mask,
segment_ids=segment_ids)
bert_outputs = bert_module(
inputs=bert_inputs,
signature="tokens",
as_dict=True)
# Use "pooled_output" for classification tasks on an entire sentence.
# Use "sequence_outputs" for token-level output.
output_layer = bert_outputs["pooled_output"]
hidden_size = output_layer.shape[-1].value
# Create our own layer to tune for politeness data.
output_weights = tf.get_variable(
"output_weights", [num_labels, hidden_size],
initializer=tf.truncated_normal_initializer(stddev=0.02))
output_bias = tf.get_variable(
"output_bias", [num_labels], initializer=tf.zeros_initializer())
with tf.variable_scope("loss"):
# Dropout helps prevent overfitting
output_layer = tf.nn.dropout(output_layer, keep_prob=0.9)
logits = tf.matmul(output_layer, output_weights, transpose_b=True)
logits = tf.nn.bias_add(logits, output_bias)
log_probs = tf.nn.log_softmax(logits, axis=-1)
# Convert labels into one-hot encoding
one_hot_labels = tf.one_hot(labels, depth=num_labels, dtype=tf.float32)
predicted_labels = tf.squeeze(tf.argmax(log_probs, axis=-1, output_type=tf.int32))
# If we're predicting, we want predicted labels and the probabiltiies.
if is_predicting:
return (predicted_labels, log_probs)
# If we're train/eval, compute loss between predicted and actual label
per_example_loss = -tf.reduce_sum(one_hot_labels * log_probs, axis=-1)
loss = tf.reduce_mean(per_example_loss)
return (loss, predicted_labels, log_probs)
# model_fn_builder actually creates our model function
# using the passed parameters for num_labels, learning_rate, etc.
def model_fn_builder(bert_model_hub, num_labels, learning_rate, num_train_steps,
num_warmup_steps):
"""Returns `model_fn` closure for TPUEstimator."""
def model_fn(features, labels, mode, params): # pylint: disable=unused-argument
"""The `model_fn` for TPUEstimator."""
input_ids = features["input_ids"]
input_mask = features["input_mask"]
segment_ids = features["segment_ids"]
label_ids = features["label_ids"]
is_predicting = (mode == tf.estimator.ModeKeys.PREDICT)
# TRAIN and EVAL
if not is_predicting:
(loss, predicted_labels, log_probs) = create_model(
bert_model_hub, is_predicting, input_ids, input_mask, segment_ids, label_ids, num_labels)
train_op = bert.optimization.create_optimizer(
loss, learning_rate, num_train_steps, num_warmup_steps, use_tpu=False)
# Calculate evaluation metrics.
def metric_fn(label_ids, predicted_labels):
accuracy = tf.metrics.accuracy(label_ids, predicted_labels)
f1_score = tf.contrib.metrics.f1_score(
label_ids,
predicted_labels)
auc = tf.metrics.auc(
label_ids,
predicted_labels)
recall = tf.metrics.recall(
label_ids,
predicted_labels)
precision = tf.metrics.precision(
label_ids,
predicted_labels)
true_pos = tf.metrics.true_positives(
label_ids,
predicted_labels)
true_neg = tf.metrics.true_negatives(
label_ids,
predicted_labels)
false_pos = tf.metrics.false_positives(
label_ids,
predicted_labels)
false_neg = tf.metrics.false_negatives(
label_ids,
predicted_labels)
return {
"eval_accuracy": accuracy,
"f1_score": f1_score,
"auc": auc,
"precision": precision,
"recall": recall,
"true_positives": true_pos,
"true_negatives": true_neg,
"false_positives": false_pos,
"false_negatives": false_neg
}
eval_metrics = metric_fn(label_ids, predicted_labels)
if mode == tf.estimator.ModeKeys.TRAIN:
return tf.estimator.EstimatorSpec(mode=mode,
loss=loss,
train_op=train_op)
else:
return tf.estimator.EstimatorSpec(mode=mode,
loss=loss,
eval_metric_ops=eval_metrics)
else:
(predicted_labels, log_probs) = create_model(
bert_model_hub, is_predicting, input_ids, input_mask, segment_ids, label_ids, num_labels)
predictions = {
'probabilities': log_probs,
'labels': predicted_labels
}
return tf.estimator.EstimatorSpec(mode, predictions=predictions)
# Return the actual model function in the closure
return model_fn
def estimator_builder(bert_model_hub, OUTPUT_DIR, SAVE_SUMMARY_STEPS, SAVE_CHECKPOINTS_STEPS, label_list, LEARNING_RATE, num_train_steps, num_warmup_steps, BATCH_SIZE):
# Specify outpit directory and number of checkpoint steps to save
run_config = tf.estimator.RunConfig(
model_dir=OUTPUT_DIR,
save_summary_steps=SAVE_SUMMARY_STEPS,
save_checkpoints_steps=SAVE_CHECKPOINTS_STEPS)
model_fn = model_fn_builder(
bert_model_hub = bert_model_hub,
num_labels=len(label_list),
learning_rate=LEARNING_RATE,
num_train_steps=num_train_steps,
num_warmup_steps=num_warmup_steps)
estimator = tf.estimator.Estimator(
model_fn=model_fn,
config=run_config,
params={"batch_size": BATCH_SIZE})
return estimator, model_fn, run_config
def run_on_dfs(train, test, DATA_COLUMN, LABEL_COLUMN,
MAX_SEQ_LENGTH = 128,
BATCH_SIZE = 32,
LEARNING_RATE = 2e-5,
NUM_TRAIN_EPOCHS = 3.0,
WARMUP_PROPORTION = 0.1,
SAVE_SUMMARY_STEPS = 100,
SAVE_CHECKPOINTS_STEPS = 10000,
bert_model_hub = "https://tfhub.dev/google/bert_uncased_L-12_H-768_A-12/1"):
label_list = train[LABEL_COLUMN].unique().tolist()
tokenizer = create_tokenizer_from_hub_module(bert_model_hub)
train_features = make_features(train, label_list, MAX_SEQ_LENGTH, tokenizer, DATA_COLUMN, LABEL_COLUMN)
test_features = make_features(test, label_list, MAX_SEQ_LENGTH, tokenizer, DATA_COLUMN, LABEL_COLUMN)
num_train_steps = int(len(train_features) / BATCH_SIZE * NUM_TRAIN_EPOCHS)
num_warmup_steps = int(num_train_steps * WARMUP_PROPORTION)
estimator, model_fn, run_config = estimator_builder(
bert_model_hub,
OUTPUT_DIR,
SAVE_SUMMARY_STEPS,
SAVE_CHECKPOINTS_STEPS,
label_list,
LEARNING_RATE,
num_train_steps,
num_warmup_steps,
BATCH_SIZE)
train_input_fn = bert.run_classifier.input_fn_builder(
features=train_features,
seq_length=MAX_SEQ_LENGTH,
is_training=True,
drop_remainder=False)
estimator.train(input_fn=train_input_fn, max_steps=num_train_steps)
test_input_fn = run_classifier.input_fn_builder(
features=test_features,
seq_length=MAX_SEQ_LENGTH,
is_training=False,
drop_remainder=False)
result_dict = estimator.evaluate(input_fn=test_input_fn, steps=None)
return result_dict, estimator
import random
random.seed(10)
OUTPUT_DIR = 'output'
```
----- 只需更改下方代码 ------
导入数据集
```
!wget https://github.com/yaoyue123/SocialComputing/raw/master/spam_message/training.txt
!wget https://github.com/yaoyue123/SocialComputing/raw/master/spam_message/validation.txt
train = pd.read_table("training.txt",sep='\t',error_bad_lines=False)
#mytrain= mytrain[order]
test = pd.read_table("validation.txt",sep='\t',error_bad_lines=False)
#mytest= mytest[order]
train.head()
test.head()
```
在此更改你的参数,如标签,bert模型地址,epochs
```
myparam = {
"DATA_COLUMN": "massage",
"LABEL_COLUMN": "label",
"LEARNING_RATE": 2e-5,
"NUM_TRAIN_EPOCHS":1,
"bert_model_hub":"https://tfhub.dev/google/bert_chinese_L-12_H-768_A-12/1"
}
```
训练模型,通常情况下,一个epochs用k80训练大概在10min左右
```
result, estimator = run_on_dfs(train, test, **myparam)
```
bert模型还是比较强的,一个epochs就能达到准确率为99%
```
pretty_print(result)
```
|
github_jupyter
|
## Change sys.path to use my tensortrade instead of the one in env
```
import sys
sys.path.append("/Users/jasonfiacco/Documents/Yale/Senior/thesis/deeptrader")
print(sys.path)
```
## Read PredictIt Data Instead
```
import ssl
import pandas as pd
ssl._create_default_https_context = ssl._create_unverified_context # Only used if pandas gives a SSLError
def fetch_data(symbol):
path = "/Users/jasonfiacco/Documents/Yale/Senior/thesis/predictit_datasets/"
filename = "{}.xlsx".format(symbol)
df = pd.read_excel(path + filename, skiprows=4)
df = df.set_index("Date")
df = df.drop(df.columns[[7,8,9]], axis=1)
df = df.drop("ID", 1)
df.columns = [symbol + ":" + name.lower() for name in df.columns]
return df
all_data = pd.concat([
fetch_data("WARREN"),
fetch_data("CRUZ"),
fetch_data("MANCHIN"),
fetch_data("SANDERS"),
fetch_data("NELSON"),
fetch_data("DONNELLY"),
fetch_data("PELOSI"),
fetch_data("MANAFORT"),
fetch_data("BROWN"),
fetch_data("RYAN"),
fetch_data("STABENOW")
], axis=1)
all_data.head()
```
## Plot the closing prices for all the markets
```
%matplotlib inline
closing_prices = all_data.loc[:, [("close" in name) for name in all_data.columns]]
closing_prices.plot()
```
## Slice just a specific time period from the dataframe
```
all_data.index = pd.to_datetime(all_data.index)
subset_data = all_data[(all_data.index >= '09-01-2017') & (all_data.index <= '09-04-2019')]
subset_data.head()
```
## Define Exchanges
An exchange needs a name, an execution service, and streams of price data in order to function properly.
The setups supported right now are the simulated execution service using simulated or stochastic data. More execution services will be made available in the future, as well as price streams so that live data and execution can be supported.
```
from tensortrade.exchanges import Exchange
from tensortrade.exchanges.services.execution.simulated import execute_order
from tensortrade.data import Stream
#Exchange(name of exchange, service)
#It looks like each Stream takes a name, and then a list of the closing prices.
predictit_exch = Exchange("predictit", service=execute_order)(
Stream("USD-WARREN", list(subset_data['WARREN:close'])),
Stream("USD-CRUZ", list(subset_data['CRUZ:close'])),
Stream("USD-MANCHIN", list(subset_data['MANCHIN:close'])),
Stream("USD-SANDERS", list(subset_data['SANDERS:close'])),
Stream("USD-NELSON", list(subset_data['NELSON:close'])),
Stream("USD-DONNELLY", list(subset_data['DONNELLY:close'])),
Stream("USD-PELOSI", list(subset_data['PELOSI:close'])),
Stream("USD-MANAFORT", list(subset_data['MANAFORT:close'])),
Stream("USD-BROWN", list(subset_data['BROWN:close'])),
Stream("USD-RYAN", list(subset_data['RYAN:close'])),
Stream("USD-STABENOW", list(subset_data['STABENOW:close']))
)
```
Now that the exchanges have been defined we can define our features that we would like to include, excluding the prices we have provided for the exchanges.
### Doing it without adding other features. Just use price
```
#You still have to add "Streams" for all the standard columns open, high, low, close, volume in this case
from tensortrade.data import DataFeed, Module
with Module("predictit") as predictit_ns:
predictit_nodes = [Stream(name, list(subset_data[name])) for name in subset_data.columns]
#Then create the Feed from it
feed = DataFeed([predictit_ns])
feed.next()
```
## Portfolio
Make the portfolio using the any combinations of exchanges and intruments that the exchange supports
```
#I am going to have to add "instruments" for all 25 of the PredictIt markets I'm working with.
from tensortrade.instruments import USD, WARREN, CRUZ, MANCHIN, SANDERS, NELSON, DONNELLY,\
PELOSI, MANAFORT, BROWN, RYAN, STABENOW
from tensortrade.wallets import Wallet, Portfolio
portfolio = Portfolio(USD, [
Wallet(predictit_exch, 10000 * USD),
Wallet(predictit_exch, 0 * WARREN),
Wallet(predictit_exch, 0 * CRUZ),
Wallet(predictit_exch, 0 * MANCHIN),
Wallet(predictit_exch, 0 * SANDERS),
Wallet(predictit_exch, 0 * NELSON),
Wallet(predictit_exch, 0 * DONNELLY),
Wallet(predictit_exch, 0 * PELOSI),
Wallet(predictit_exch, 0 * MANAFORT),
Wallet(predictit_exch, 0 * BROWN),
Wallet(predictit_exch, 0 * RYAN),
Wallet(predictit_exch, 0 * STABENOW)
])
```
## Environment
```
from tensortrade.environments import TradingEnvironment
env = TradingEnvironment(
feed=feed,
portfolio=portfolio,
action_scheme='simple',
reward_scheme='simple',
window_size=15,
enable_logger=False,
renderers = 'screenlog'
)
env.feed.next()
```
#### ^An environment doesn't just show the OHLCV for each instrument. It also shows free, locked, total, as well as "USD_BTC"
## Using 123's Ray example
```
import os
parent_dir = "/Users/jasonfiacco/Documents/Yale/Senior/thesis/deeptrader"
os.environ["PYTHONPATH"] = parent_dir + ":" + os.environ.get("PYTHONPATH", "")
!PYTHONWARNINGS=ignore::yaml.YAMLLoadWarning
#Import tensortrade
import tensortrade
# Define Exchanges
from tensortrade.exchanges import Exchange
from tensortrade.exchanges.services.execution.simulated import execute_order
from tensortrade.data import Stream
# Define External Data Feed (features)
import ta
from sklearn import preprocessing
from tensortrade.data import DataFeed, Module
# Portfolio
from tensortrade.instruments import USD, BTC
from tensortrade.wallets import Wallet, Portfolio
from tensortrade.actions import ManagedRiskOrders
from gym.spaces import Discrete
# Environment
from tensortrade.environments import TradingEnvironment
import gym
import ray
from ray import tune
from ray.tune import grid_search
from ray.tune.registry import register_env
import ray.rllib.agents.ppo as ppo
import ray.rllib.agents.dqn as dqn
from ray.tune.logger import pretty_print
from tensortrade.rewards import RiskAdjustedReturns
class RayTradingEnv(TradingEnvironment):
def __init__(self):
env = TradingEnvironment(
feed=feed,
portfolio=portfolio,
action_scheme="simple",
reward_scheme="simple",
window_size=15,
enable_logger=False,
renderers = 'screenlog'
)
self.env = env
self.action_space = self.env.action_space
self.observation_space = self.env.observation_space
def reset(self):
return self.env.reset()
def step(self, action):
return self.env.step(action)
def env_creator(env_config):
return RayTradingEnv()
register_env("ray_trading_env", env_creator)
ray.init(ignore_reinit_error=True)
config = dqn.DEFAULT_CONFIG.copy()
config["num_gpus"] = 0
#config["num_workers"] = 4
#config["num_envs_per_worker"] = 8
# config["eager"] = False
# config["timesteps_per_iteration"] = 100
# config["train_batch_size"] = 20
#config['log_level'] = "DEBUG"
trainer = dqn.DQNTrainer(config=config, env="ray_trading_env")
config
```
## Train using the old fashioned RLLib way
```
for i in range(10):
# Perform one iteration of training the policy with PPO
print("Training iteration {}...".format(i))
result = trainer.train()
print("result: {}".format(result))
if i % 100 == 0:
checkpoint = trainer.save()
print("checkpoint saved at", checkpoint)
result['hist_stats']['episode_reward']
```
## OR train using the tune way (better so far)
```
analysis = tune.run(
"DQN",
name = "DQN10-paralellism",
checkpoint_at_end=True,
stop={
"timesteps_total": 4000,
},
config={
"env": "ray_trading_env",
"lr": grid_search([1e-4]), # try different lrs
"num_workers": 2, # parallelism,
},
)
#Use the below command to see results
#tensorboard --logdir=/Users/jasonfiacco/ray_results/DQN2
#Now you can plot the reward results of your tuner.
dfs = analysis.trial_dataframes
ax = None
for d in dfs.values():
ax = d.episode_reward_mean.plot(ax=ax, legend=True)
```
## Restoring an already existing agent that I tuned
```
import os
logdir = analysis.get_best_logdir("episode_reward_mean", mode="max")
trainer.restore(os.path.join(logdir, "checkpoint_993/checkpoint-993"))
trainer.restore("/Users/jasonfiacco/ray_results/DQN4/DQN_ray_trading_env_fedb24f0_0_lr=1e-06_2020-03-03_15-46-02kzbdv53d/checkpoint_5/checkpoint-5")
```
## Testing
```
#Set up a testing environment with test data.
test_env = TradingEnvironment(
feed=feed,
portfolio=portfolio,
action_scheme='simple',
reward_scheme='simple',
window_size=15,
enable_logger=False,
renderers = 'screenlog'
)
for episode_num in range(1):
state = test_env.reset()
done = False
cumulative_reward = 0
step = 0
action = trainer.compute_action(state)
while not done:
action = trainer.compute_action(state)
state, reward, done, results = test_env.step(action)
cumulative_reward += reward
#Render every 100 steps:
if step % 100 == 0:
test_env.render()
step += 1
print("Cumulative reward: ", cumulative_reward)
```
## Plot
```
%matplotlib inline
portfolio.performance.plot()
portfolio.performance.net_worth.plot()
#Plot the total balance in each type of item
p = portfolio.performance
p2 = p.iloc[:, :]
weights = p2.loc[:, [("/worth" in name) for name in p2.columns]]
weights.iloc[:, 1:8].plot()
```
## Try Plotly Render too
```
from tensortrade.environments.render import PlotlyTradingChart
from tensortrade.environments.render import FileLogger
chart_renderer = PlotlyTradingChart(
height = 800
)
file_logger = FileLogger(
filename='example.log', # omit or None for automatic file name
path='training_logs' # create a new directory if doesn't exist, None for no directory
)
price_history.columns = ['datetime', 'open', 'high', 'low', 'close', 'volume']
env = TradingEnvironment(
feed=feed,
portfolio=portfolio,
action_scheme='managed-risk',
reward_scheme='risk-adjusted',
window_size=20,
price_history=price_history,
renderers = [chart_renderer, file_logger]
)
from tensortrade.agents import DQNAgent
agent = DQNAgent(env)
agent.train(n_episodes=1, n_steps=1000, render_interval=1)
```
## Extra Stuff
```
apath = "/Users/jasonfiacco/Documents/Yale/Senior/thesis/jasonfiacco-selectedmarkets-mytickers.xlsx"
df = pd.read_excel(apath, skiprows=2)
jason_tickers = df.iloc[:, 5].tolist()
descriptions = df.iloc[:, 1].tolist()
for ticker, description in zip(jason_tickers, descriptions):
l = "{} = Instrument(\'{}\', 2, \'{}\')".format(ticker, ticker, description)
print(l)
```
|
github_jupyter
|
[Table of Contents](./table_of_contents.ipynb)
# Smoothing
```
#format the book
%matplotlib inline
from __future__ import division, print_function
from book_format import load_style
load_style()
```
## Introduction
The performance of the Kalman filter is not optimal when you consider future data. For example, suppose we are tracking an aircraft, and the latest measurement deviates far from the current track, like so (I'll only consider 1 dimension for simplicity):
```
import matplotlib.pyplot as plt
data = [10.1, 10.2, 9.8, 10.1, 10.2, 10.3,
10.1, 9.9, 10.2, 10.0, 9.9, 11.4]
plt.plot(data)
plt.xlabel('time')
plt.ylabel('position');
```
After a period of near steady state, we have a very large change. Assume the change is past the limit of the aircraft's flight envelope. Nonetheless the Kalman filter incorporates that new measurement into the filter based on the current Kalman gain. It cannot reject the noise because the measurement could reflect the initiation of a turn. Granted it is unlikely that we are turning so abruptly, but it is impossible to say whether
* The aircraft started a turn awhile ago, but the previous measurements were noisy and didn't show the change.
* The aircraft is turning, and this measurement is very noisy
* The measurement is very noisy and the aircraft has not turned
* The aircraft is turning in the opposite direction, and the measurement is extremely noisy
Now, suppose the following measurements are:
11.3 12.1 13.3 13.9 14.5 15.2
```
data2 = [11.3, 12.1, 13.3, 13.9, 14.5, 15.2]
plt.plot(data + data2);
```
Given these future measurements we can infer that yes, the aircraft initiated a turn.
On the other hand, suppose these are the following measurements.
```
data3 = [9.8, 10.2, 9.9, 10.1, 10.0, 10.3, 9.9, 10.1]
plt.plot(data + data3);
```
In this case we are led to conclude that the aircraft did not turn and that the outlying measurement was merely very noisy.
## An Overview of How Smoothers Work
The Kalman filter is a *recursive* filter with the Markov property - it's estimate at step `k` is based only on the estimate from step `k-1` and the measurement at step `k`. But this means that the estimate from step `k-1` is based on step `k-2`, and so on back to the first epoch. Hence, the estimate at step `k` depends on all of the previous measurements, though to varying degrees. `k-1` has the most influence, `k-2` has the next most, and so on.
Smoothing filters incorporate future measurements into the estimate for step `k`. The measurement from `k+1` will have the most effect, `k+2` will have less effect, `k+3` less yet, and so on.
This topic is called *smoothing*, but I think that is a misleading name. I could smooth the data above by passing it through a low pass filter. The result would be smooth, but not necessarily accurate because a low pass filter will remove real variations just as much as it removes noise. In contrast, Kalman smoothers are *optimal* - they incorporate all available information to make the best estimate that is mathematically achievable.
## Types of Smoothers
There are three classes of Kalman smoothers that produce better tracking in these situations.
* Fixed-Interval Smoothing
This is a batch processing based filter. This filter waits for all of the data to be collected before making any estimates. For example, you may be a scientist collecting data for an experiment, and don't need to know the result until the experiment is complete. A fixed-interval smoother will collect all the data, then estimate the state at each measurement using all available previous and future measurements. If it is possible for you to run your Kalman filter in batch mode it is always recommended to use one of these filters a it will provide much better results than the recursive forms of the filter from the previous chapters.
* Fixed-Lag Smoothing
Fixed-lag smoothers introduce latency into the output. Suppose we choose a lag of 4 steps. The filter will ingest the first 3 measurements but not output a filtered result. Then, when the 4th measurement comes in the filter will produce the output for measurement 1, taking measurements 1 through 4 into account. When the 5th measurement comes in, the filter will produce the result for measurement 2, taking measurements 2 through 5 into account. This is useful when you need recent data but can afford a bit of lag. For example, perhaps you are using machine vision to monitor a manufacturing process. If you can afford a few seconds delay in the estimate a fixed-lag smoother will allow you to produce very accurate and smooth results.
* Fixed-Point Smoothing
A fixed-point filter operates as a normal Kalman filter, but also produces an estimate for the state at some fixed time $j$. Before the time $k$ reaches $j$ the filter operates as a normal filter. Once $k>j$ the filter estimates $x_k$ and then also updates its estimate for $x_j$ using all of the measurements between $j\dots k$. This can be useful to estimate initial paramters for a system, or for producing the best estimate for an event that happened at a specific time. For example, you may have a robot that took a photograph at time $j$. You can use a fixed-point smoother to get the best possible pose information for the camera at time $j$ as the robot continues moving.
## Choice of Filters
The choice of these filters depends on your needs and how much memory and processing time you can spare. Fixed-point smoothing requires storage of all measurements, and is very costly to compute because the output is for every time step is recomputed for every measurement. On the other hand, the filter does produce a decent output for the current measurement, so this filter can be used for real time applications.
Fixed-lag smoothing only requires you to store a window of data, and processing requirements are modest because only that window is processed for each new measurement. The drawback is that the filter's output always lags the input, and the smoothing is not as pronounced as is possible with fixed-interval smoothing.
Fixed-interval smoothing produces the most smoothed output at the cost of having to be batch processed. Most algorithms use some sort of forwards/backwards algorithm that is only twice as slow as a recursive Kalman filter.
## Fixed-Interval Smoothing
There are many fixed-lag smoothers available in the literature. I have chosen to implement the smoother invented by Rauch, Tung, and Striebel because of its ease of implementation and efficiency of computation. It is also the smoother I have seen used most often in real applications. This smoother is commonly known as an RTS smoother.
Derivation of the RTS smoother runs to several pages of densely packed math. I'm not going to inflict it on you. Instead I will briefly present the algorithm, equations, and then move directly to implementation and demonstration of the smoother.
The RTS smoother works by first running the Kalman filter in a batch mode, computing the filter output for each step. Given the filter output for each measurement along with the covariance matrix corresponding to each output the RTS runs over the data backwards, incorporating its knowledge of the future into the past measurements. When it reaches the first measurement it is done, and the filtered output incorporates all of the information in a maximally optimal form.
The equations for the RTS smoother are very straightforward and easy to implement. This derivation is for the linear Kalman filter. Similar derivations exist for the EKF and UKF. These steps are performed on the output of the batch processing, going backwards from the most recent in time back to the first estimate. Each iteration incorporates the knowledge of the future into the state estimate. Since the state estimate already incorporates all of the past measurements the result will be that each estimate will contain knowledge of all measurements in the past and future. Here is it very important to distinguish between past, present, and future so I have used subscripts to denote whether the data is from the future or not.
Predict Step
$$\begin{aligned}
\mathbf{P} &= \mathbf{FP}_k\mathbf{F}^\mathsf{T} + \mathbf{Q }
\end{aligned}$$
Update Step
$$\begin{aligned}
\mathbf{K}_k &= \mathbf{P}_k\mathbf{F}^\mathsf{T}\mathbf{P}^{-1} \\
\mathbf{x}_k &= \mathbf{x}_k + \mathbf{K}_k(\mathbf{x}_{k+1} - \mathbf{Fx}_k) \\
\mathbf{P}_k &= \mathbf{P}_k + \mathbf{K}_k(\mathbf{P}_{k+1} - \mathbf{P})\mathbf{K}_k^\mathsf{T}
\end{aligned}$$
As always, the hardest part of the implementation is correctly accounting for the subscripts. A basic implementation without comments or error checking would be:
```python
def rts_smoother(Xs, Ps, F, Q):
n, dim_x, _ = Xs.shape
# smoother gain
K = zeros((n,dim_x, dim_x))
x, P, Pp = Xs.copy(), Ps.copy(), Ps.copy
for k in range(n-2,-1,-1):
Pp[k] = dot(F, P[k]).dot(F.T) + Q # predicted covariance
K[k] = dot(P[k], F.T).dot(inv(Pp[k]))
x[k] += dot(K[k], x[k+1] - dot(F, x[k]))
P[k] += dot(K[k], P[k+1] - Pp[k]).dot(K[k].T)
return (x, P, K, Pp)
```
This implementation mirrors the implementation provided in FilterPy. It assumes that the Kalman filter is being run externally in batch mode, and the results of the state and covariances are passed in via the `Xs` and `Ps` variable.
Here is an example.
```
import numpy as np
from numpy import random
from numpy.random import randn
import matplotlib.pyplot as plt
from filterpy.kalman import KalmanFilter
import kf_book.book_plots as bp
def plot_rts(noise, Q=0.001, show_velocity=False):
random.seed(123)
fk = KalmanFilter(dim_x=2, dim_z=1)
fk.x = np.array([0., 1.]) # state (x and dx)
fk.F = np.array([[1., 1.],
[0., 1.]]) # state transition matrix
fk.H = np.array([[1., 0.]]) # Measurement function
fk.P = 10. # covariance matrix
fk.R = noise # state uncertainty
fk.Q = Q # process uncertainty
# create noisy data
zs = np.asarray([t + randn()*noise for t in range (40)])
# filter data with Kalman filter, than run smoother on it
mu, cov, _, _ = fk.batch_filter(zs)
M, P, C, _ = fk.rts_smoother(mu, cov)
# plot data
if show_velocity:
index = 1
print('gu')
else:
index = 0
if not show_velocity:
bp.plot_measurements(zs, lw=1)
plt.plot(M[:, index], c='b', label='RTS')
plt.plot(mu[:, index], c='g', ls='--', label='KF output')
if not show_velocity:
N = len(zs)
plt.plot([0, N], [0, N], 'k', lw=2, label='track')
plt.legend(loc=4)
plt.show()
plot_rts(7.)
```
I've injected a lot of noise into the signal to allow you to visually distinguish the RTS output from the ideal output. In the graph above we can see that the Kalman filter, drawn as the green dotted line, is reasonably smooth compared to the input, but it still wanders from from the ideal line when several measurements in a row are biased towards one side of the line. In contrast, the RTS output is both extremely smooth and very close to the ideal output.
With a perhaps more reasonable amount of noise we can see that the RTS output nearly lies on the ideal output. The Kalman filter output, while much better, still varies by a far greater amount.
```
plot_rts(noise=1.)
```
However, we must understand that this smoothing is predicated on the system model. We have told the filter that what we are tracking follows a constant velocity model with very low process error. When the filter *looks ahead* it sees that the future behavior closely matches a constant velocity so it is able to reject most of the noise in the signal. Suppose instead our system has a lot of process noise. For example, if we are tracking a light aircraft in gusty winds its velocity will change often, and the filter will be less able to distinguish between noise and erratic movement due to the wind. We can see this in the next graph.
```
plot_rts(noise=7., Q=.1)
```
This underscores the fact that these filters are not *smoothing* the data in colloquial sense of the term. The filter is making an optimal estimate based on previous measurements, future measurements, and what you tell it about the behavior of the system and the noise in the system and measurements.
Let's wrap this up by looking at the velocity estimates of Kalman filter vs the RTS smoother.
```
plot_rts(7.,show_velocity=True)
```
The improvement in the velocity, which is an hidden variable, is even more dramatic.
## Fixed-Lag Smoothing
The RTS smoother presented above should always be your choice of algorithm if you can run in batch mode because it incorporates all available data into each estimate. Not all problems allow you to do that, but you may still be interested in receiving smoothed values for previous estimates. The number line below illustrates this concept.
```
from kf_book.book_plots import figsize
from kf_book.smoothing_internal import *
with figsize(y=2):
show_fixed_lag_numberline()
```
At step $k$ we can estimate $x_k$ using the normal Kalman filter equations. However, we can make a better estimate for $x_{k-1}$ by using the measurement received for $x_k$. Likewise, we can make a better estimate for $x_{k-2}$ by using the measurements recevied for $x_{k-1}$ and $x_{k}$. We can extend this computation back for an arbitrary $N$ steps.
Derivation for this math is beyond the scope of this book; Dan Simon's *Optimal State Estimation* [2] has a very good exposition if you are interested. The essense of the idea is that instead of having a state vector $\mathbf{x}$ we make an augmented state containing
$$\mathbf{x} = \begin{bmatrix}\mathbf{x}_k \\ \mathbf{x}_{k-1} \\ \vdots\\ \mathbf{x}_{k-N+1}\end{bmatrix}$$
This yields a very large covariance matrix that contains the covariance between states at different steps. FilterPy's class `FixedLagSmoother` takes care of all of this computation for you, including creation of the augmented matrices. All you need to do is compose it as if you are using the `KalmanFilter` class and then call `smooth()`, which implements the predict and update steps of the algorithm.
Each call of `smooth` computes the estimate for the current measurement, but it also goes back and adjusts the previous `N-1` points as well. The smoothed values are contained in the list `FixedLagSmoother.xSmooth`. If you use `FixedLagSmoother.x` you will get the most recent estimate, but it is not smoothed and is no different from a standard Kalman filter output.
```
from filterpy.kalman import FixedLagSmoother, KalmanFilter
import numpy.random as random
fls = FixedLagSmoother(dim_x=2, dim_z=1, N=8)
fls.x = np.array([0., .5])
fls.F = np.array([[1.,1.],
[0.,1.]])
fls.H = np.array([[1.,0.]])
fls.P *= 200
fls.R *= 5.
fls.Q *= 0.001
kf = KalmanFilter(dim_x=2, dim_z=1)
kf.x = np.array([0., .5])
kf.F = np.array([[1.,1.],
[0.,1.]])
kf.H = np.array([[1.,0.]])
kf.P *= 200
kf.R *= 5.
kf.Q *= 0.001
N = 4 # size of lag
nom = np.array([t/2. for t in range (0, 40)])
zs = np.array([t + random.randn()*5.1 for t in nom])
for z in zs:
fls.smooth(z)
kf_x, _, _, _ = kf.batch_filter(zs)
x_smooth = np.array(fls.xSmooth)[:, 0]
fls_res = abs(x_smooth - nom)
kf_res = abs(kf_x[:, 0] - nom)
plt.plot(zs,'o', alpha=0.5, marker='o', label='zs')
plt.plot(x_smooth, label='FLS')
plt.plot(kf_x[:, 0], label='KF', ls='--')
plt.legend(loc=4)
print('standard deviation fixed-lag: {:.3f}'.format(np.mean(fls_res)))
print('standard deviation kalman: {:.3f}'.format(np.mean(kf_res)))
```
Here I have set `N=8` which means that we will incorporate 8 future measurements into our estimates. This provides us with a very smooth estimate once the filter converges, at the cost of roughly 8x the amount of computation of the standard Kalman filter. Feel free to experiment with larger and smaller values of `N`. I chose 8 somewhat at random, not due to any theoretical concerns.
## References
[1] H. Rauch, F. Tung, and C. Striebel. "Maximum likelihood estimates of linear dynamic systems," *AIAA Journal*, **3**(8), pp. 1445-1450 (August 1965).
[2] Dan Simon. "Optimal State Estimation," John Wiley & Sons, 2006.
http://arc.aiaa.org/doi/abs/10.2514/3.3166
|
github_jupyter
|
# 準備
```
# バージョン指定時にコメントアウト
#!pip install torch==1.7.0
#!pip install torchvision==0.8.1
import torch
import torchvision
# バージョンの確認
print(torch.__version__)
print(torchvision.__version__)
# Google ドライブにマウント
from google.colab import drive
drive.mount('/content/gdrive')
%cd '/content/gdrive/MyDrive/Colab Notebooks/gan_sample/chapter2'
import os
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optimizers
import torch.nn.functional as F
from torch.utils.data import Dataset, DataLoader
import torchvision
import torchvision.transforms as transforms
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
```
# データセットの作成
```
np.random.seed(1234)
torch.manual_seed(1234)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# データの取得
root = os.path.join('data', 'mnist')
transform = transforms.Compose([transforms.ToTensor(),
lambda x: x.view(-1)])
mnist_train = \
torchvision.datasets.MNIST(root=root,
download=True,
train=True,
transform=transform)
mnist_test = \
torchvision.datasets.MNIST(root=root,
download=True,
train=False,
transform=transform)
train_dataloader = DataLoader(mnist_train,
batch_size=100,
shuffle=True)
test_dataloader = DataLoader(mnist_test,
batch_size=1,
shuffle=False)
```
# ネットワークの定義
```
class Autoencoder(nn.Module):
def __init__(self, device='cpu'):
super().__init__()
self.device = device
self.l1 = nn.Linear(784, 200)
self.l2 = nn.Linear(200, 784)
def forward(self, x):
# エンコーダ
h = self.l1(x)
# 活性化関数
h = torch.relu(h)
# デコーダ
h = self.l2(h)
# シグモイド関数で0~1の値域に変換
y = torch.sigmoid(h)
return y
```
# 学習の実行
```
# モデルの設定
model = Autoencoder(device=device).to(device)
# 損失関数の設定
criterion = nn.BCELoss()
# 最適化関数の設定
optimizer = optimizers.Adam(model.parameters())
epochs = 10
# エポックのループ
for epoch in range(epochs):
train_loss = 0.
# バッチサイズのループ
for (x, _) in train_dataloader:
x = x.to(device)
# 訓練モードへの切替
model.train()
# 順伝播計算
preds = model(x)
# 入力画像xと復元画像predsの誤差計算
loss = criterion(preds, x)
# 勾配の初期化
optimizer.zero_grad()
# 誤差の勾配計算
loss.backward()
# パラメータの更新
optimizer.step()
# 訓練誤差の更新
train_loss += loss.item()
train_loss /= len(train_dataloader)
print('Epoch: {}, Loss: {:.3f}'.format(
epoch+1,
train_loss
))
```
# 画像の復元
```
# dataloaderからのデータ取り出し
x, _ = next(iter(test_dataloader))
x = x.to(device)
# 評価モードへの切替
model.eval()
# 復元画像
x_rec = model(x)
# 入力画像、復元画像の表示
for i, image in enumerate([x, x_rec]):
image = image.view(28, 28).detach().cpu().numpy()
plt.subplot(1, 2, i+1)
plt.imshow(image, cmap='binary_r')
plt.axis('off')
plt.show()
```
|
github_jupyter
|
# One-step error probability
Write a computer program implementing asynchronous deterministic updates for a Hopfield network. Use Hebb's rule with $w_{ii}=0$. Generate and store p=[12,24,48,70,100,120] random patterns with N=120 bits. Each bit is either +1 or -1 with probability $\tfrac{1}{2}$.
For each value of ppp estimate the one-step error probability $P_{\text {error}}^{t=1}$ based on $10^5$ independent trials. Here, one trial means that you generate and store a set of p random patterns, feed one of them, and perform one asynchronous update of a single randomly chosen neuron. If in some trials you encounter sgn(0), simply set sgn(0)=1.
List below the values of $P_{\text {error}}^{t=1}$ that you obtained in the following form: [$p_1,p_2,\ldots,p_{6}$], where $p_n$ is the value of $P_{\text {error}}^{t=1}$ for the n-th value of p from the list above. Give four decimal places for each $p_n$
```
import numpy as np
import time
def calculate_instance( n, p, zero_diagonal):
#Create p random patterns
patterns = []
for i in range(p):
patterns.append(np.random.choice([-1,1],n))
#Create weights matrix according to hebbs rule
weights = patterns[0][:,None]*patterns[0]
for el in patterns[1:]:
weights = weights + el[:,None]*el
weights = np.true_divide(weights, n)
#Fill diagonal with zeroes
if zero_diagonal:
np.fill_diagonal(weights,0)
#Feed random pattern as input and test if an error occurs
S1 = patterns[0]
chosen_i = np.random.choice(range(n))
S_i_old = S1[chosen_i]
S_i = esign(np.dot(weights[chosen_i], S1))
#breakpoint()
return S_i_old == S_i
def esign(x):
if(x == 0):
return 1
else:
return np.sign(x)
```
List your numerically computed $P_{\text {error}}^{t=1}$ for the parameters given above.
```
p = [12, 24, 48, 70, 100, 120]
N = 120
I = 100000
for p_i in p:
solve = [0,0]
for i in range(I):
ret = calculate_instance(N, p_i, True)
if ret:
solve[0]+=1
else:
solve[1]+=1
p_error = float(solve[1]/I)
print(f"Number of patterns: {p_i}, P_error(t=1): {p_error} ")
```
Repeat the task, but now apply Hebb's rule without setting the diagonal weights to zero. For each value of p listed above, estimate the one-step error probability $P_{\text {error}}^{t=1}$ based on $10^5$ independent trials.
```
p = [12, 24, 48, 70, 100, 120]
N = 120
I = 100000
for p_i in p:
solve = [0,0]
for i in range(I):
ret = calculate_instance(N, p_i, False)
if ret:
solve[0]+=1
else:
solve[1]+=1
p_error = float(solve[1]/I)
print(f"Number of patterns: {p_i}, P_error(t=1): {p_error} ")
```
|
github_jupyter
|
# Acquiring Data from open repositories
A crucial step in the work of a computational biologist is not only to analyse data, but acquiring datasets to analyse as well as toy datasets to test out computational methods and algorithms. The internet is full of such open datasets. Sometimes you have to sign up and make a user to get authentication, especially for medical data. This can sometimes be time consuming, so here we will deal with easy access resources, mostly of modest size. Multiple python libraries provide a `dataset` module which makes the effort to fetch online data extremely seamless, with little requirement for preprocessing.
#### Goal of the notebook
Here you will get familiar with some ways to fetch datasets from online. We do some data exploration on the data just for illustration, but the methods will be covered later.
# Useful resources and links
When playing around with algorithms, it can be practical to use relatively small datasets. A good example is the `datasets` submodule of `scikit-learn`. `Nilearn` (library for neuroimaging) also provides a collection of neuroimaging datasets. Many datasets can also be acquired through the competition website [Kaggle](https://www.kaggle.com), in which they describe how to access the data.
### Links
- [OpenML](https://www.openml.org/search?type=data)
- [Nilearn datasets](https://nilearn.github.io/modules/reference.html#module-nilearn.datasets)
- [Sklearn datasets](https://scikit-learn.org/stable/modules/classes.html?highlight=datasets#module-sklearn.datasets)
- [Kaggle](https://www.kaggle.com/datasets)
- [MEDNIST]
- [**Awesomedata**](https://github.com/awesomedata/awesome-public-datasets)
- We strongly recommend to check out the Awesomedata lists of public datasets, covering topics such as [biology/medicine](https://github.com/awesomedata/awesome-public-datasets#biology) and [neuroscience](https://github.com/awesomedata/awesome-public-datasets#neuroscience)
- [Papers with code](https://paperswithcode.com)
- [SNAP](https://snap.stanford.edu/data/)
- Stanford Large Network Dataset Collection
- [Open Graph Benchmark (OGB)](https://github.com/snap-stanford/ogb)
- Network datasets
- [Open Neuro](https://openneuro.org/)
- [Open fMRI](https://openfmri.org/dataset/)
```
# import basic libraries
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
```
We start with scikit-learn's datasets for testing out ML algorithms. Visit [here](https://scikit-learn.org/stable/modules/classes.html?highlight=datasets#module-sklearn.datasets) for an overview of the datasets.
```
from sklearn.datasets import fetch_olivetti_faces, fetch_20newsgroups, load_breast_cancer, load_diabetes, load_digits, load_iris
```
Load the MNIST dataset (images of hand written digits)
```
X,y = load_digits(return_X_y=True)
y.shape
X.shape #1797 images, 64 pixels per image
```
#### exercise 1. Make a function `plot` taking an argument (k) to visualize the k'th sample.
It is currently flattened, you will need to reshape it. Use `plt.imshow` for plotting.
```
# %load solutions/ex2_1.py
def plot(k):
plt.imshow(X[k].reshape(8,8), cmap='gray')
plt.title(f"Number = {y[k]}")
plt.show()
plot(15); plot(450)
faces = fetch_olivetti_faces()
```
#### Exercise 2. Inspect the dataset. How many classes are there? How many samples per class? Also, plot some examples. What do the classes represent?
```
# %load solutions/ex2_2.py
# example solution.
# You are not expected to make a nice plotting function,
# you can simply call plt.imshow a number of times and observe
print(faces.DESCR) # this shows there are 40 classes, 10 samples per class
print(faces.target) #the targets i.e. classes
print(np.unique(faces.target).shape) # another way to see n_classes
X = faces.images
y = faces.target
fig = plt.figure(figsize=(16,5))
idxs = [0,1,2, 11,12,13, 40,41]
for i,k in enumerate(idxs):
ax=fig.add_subplot(2,4,i+1)
ax.imshow(X[k])
ax.set_title(f"target={y[k]}")
# looking at a few plots shows that each target is a single person.
```
Once you have made yourself familiar with the dataset you can do some data exploration with unsupervised methods, like below. The next few lines of code are simply for illustration, don't worry about the code (we will cover unsupervised methods in submodule F).
```
from sklearn.decomposition import randomized_svd
X = faces.data
n_dim = 3
u, s, v = randomized_svd(X, n_dim)
```
Now we have factorized the images into their constituent parts. The code below displays the various components isolated one by one.
```
def show_ims(ims):
fig = plt.figure(figsize=(16,10))
idxs = [0,1,2, 11,12,13, 40,41,42, 101,101,103]
for i,k in enumerate(idxs):
ax=fig.add_subplot(3,4,i+1)
ax.imshow(ims[k])
ax.set_title(f"target={y[k]}")
for i in range(n_dim):
my_s = np.zeros(s.shape[0])
my_s[i] = s[i]
recon = u@np.diag(my_s)@v
recon = recon.reshape(400,64,64)
show_ims(recon)
```
Are you able to see what the components represent? It at least looks like the second component signifies the lightning (the light direction), the third highlights eyebrows and facial chin shape.
```
from sklearn.manifold import TSNE
tsne = TSNE(init='pca', random_state=0)
trans = tsne.fit_transform(X)
m = 8*10 # choose 4 people
plt.figure(figsize=(16,10))
xs, ys = trans[:m,0], trans[:m,1]
plt.scatter(xs, ys, c=y[:m], cmap='rainbow')
for i,v in enumerate(zip(xs,ys, y[:m])):
xx,yy,s = v
#plt.text(xx,yy,s) #class
plt.text(xx,yy,i) #index
```
Many people seem to have multiple subclusters. What is the difference between those clusters? (e.g. 68,62,65 versus the other 60's)
```
ims = faces.images
idxs = [68,62,65,66,60,64,63]
#idxs = [9,4,1, 5,3]
for k in idxs:
plt.imshow(ims[k], cmap='gray')
plt.show()
def show(im):
return plt.imshow(im, cmap='gray')
import pandas as pd
df= pd.read_csv('data/archive/covid_impact_on_airport_traffic.csv')
df.shape
df.describe()
df.head()
df.Country.unique()
df.ISO_3166_2.unique()
df.AggregationMethod.unique()
```
Here we will look at [OpenML](https://www.openml.org/) - a repository of open datasets free to explore data and test methods.
### Fetching an OpenML dataset
We need to pass in an ID to access, as follows:
```
from sklearn.datasets import fetch_openml
```
OpenML contains all sorts of datatypes. By browsing the website we found a electroencephalography (EEG) dataset to explore:
```
data_id = 1471 #this was found by browsing OpenML
dataset = fetch_openml(data_id=data_id, as_frame=True)
dir(dataset)
dataset.url
type(dataset)
print(dataset.DESCR)
original_names = ['AF3',
'F7',
'F3',
'FC5',
'T7',
'P',
'O1',
'O2',
'P8',
'T8',
'FC6',
'F4',
'F8',
'AF4']
dataset.feature_names
df = dataset.frame
df.head()
df.shape[0] / 117
# 128 frames per second
df = dataset.frame
y = df.Class
#df.drop(columns='Class', inplace=True)
df.dtypes
#def summary(s):
# print(s.max(), s.min(), s.mean(), s.std())
# print()
#
#for col in df.columns[:-1]:
# column = df.loc[:,col]
# summary(column)
df.plot()
```
From the plot we can quickly identify a bunch of huge outliers, making the plot look completely uselss. We assume these are artifacts, and remove them.
```
df2 = df.iloc[:,:-1].clip_upper(6000)
df2.plot()
```
Now we see better what is going on. Lets just remove the frames corresponding to those outliers
```
frames = np.nonzero(np.any(df.iloc[:,:-1].values>5000, axis=1))[0]
frames
df.drop(index=frames, inplace=True)
df.plot(figsize=(16,8))
plt.legend(labels=original_names)
df.columns
```
### Do some modelling of the data
```
from sklearn.linear_model import LogisticRegression
lasso = LogisticRegression(penalty='l2')
X = df.values[:,:-1]
y = df.Class
y = y.astype(np.int) - 1 # map to 0,1
print(X.shape)
print(y.shape)
lasso.fit(X,y)
comp = (lasso.predict(X) == y).values
np.sum(comp.astype(np.int))/y.shape[0] # shitty accuracy
lasso.coef_[0].shape
names = dataset.feature_names
original_names
coef = lasso.coef_[0]
plt.barh(range(coef.shape[0]), coef)
plt.yticks(ticks=range(14),labels=original_names)
plt.show()
```
Interpreting the coeficients: we naturally tend to read the magnitude of the coefficients as feature importance. That is a fair interpretation, but currently we did not scale our features to a comparable range prior to fittting the model, so we cannot draw that conclusion.
### Extra exercise. Go to [OpenML](https://openml.org) and use the search function (or just look around) to find any dataset that interest you. Load it using the above methodology, and try to do anything you can to understand the datatype, visualize it etc.
```
### YOUR CODE HERE
```
|
github_jupyter
|
# Code Review #1
Purpose: To introduce the group to looking at code analytically
Created By: Hawley Helmbrecht
Creation Date: 10-12-21
# Introduction to Analyzing Code
All snipets within this section are taken from the Hitchhiker's Guide to Python (https://docs.python-guide.org/writing/style/)
### Example 1: Explicit Code
```
def make_complex(*args):
x, y = args
return dict(**locals())
def make_complex(x, y):
return {'x': x, 'y': y}
```
### Example 2: One Statement per Line
```
print('one'); print('two')
if x == 1: print('one')
if <complex comparison> and <other complex comparison>:
# do something
print('one')
print('two')
if x == 1:
print('one')
cond1 = <complex comparison>
cond2 = <other complex comparison>
if cond1 and cond2:
# do something
```
## Intro to Pep 8
Example 1: Limit all lines to a maximum of 79 characters.
```
#Wrong:
income = (gross_wages + taxable_interest + (dividends - qualified_dividends) - ira_deduction - student_loan_interest)
#Correct:
income = (gross_wages
+ taxable_interest
+ (dividends - qualified_dividends)
- ira_deduction
- student_loan_interest)
```
Example 2: Line breaks around binary operators
```
# Wrong:
# operators sit far away from their operands
income = (gross_wages +
taxable_interest +
(dividends - qualified_dividends) -
ira_deduction -
student_loan_interest)
# Correct:
# easy to match operators with operands
income = (gross_wages
+ taxable_interest
+ (dividends - qualified_dividends)
- ira_deduction
- student_loan_interest)
```
Example 3: Import formatting
```
# Correct:
import os
import sys
# Wrong:
import sys, os
```
## Let's look at some code!
Sci-kit images Otsu Threshold code! (https://github.com/scikit-image/scikit-image/blob/main/skimage/filters/thresholding.py)
```
def threshold_otsu(image=None, nbins=256, *, hist=None):
"""Return threshold value based on Otsu's method.
Either image or hist must be provided. If hist is provided, the actual
histogram of the image is ignored.
Parameters
----------
image : (N, M[, ..., P]) ndarray, optional
Grayscale input image.
nbins : int, optional
Number of bins used to calculate histogram. This value is ignored for
integer arrays.
hist : array, or 2-tuple of arrays, optional
Histogram from which to determine the threshold, and optionally a
corresponding array of bin center intensities. If no hist provided,
this function will compute it from the image.
Returns
-------
threshold : float
Upper threshold value. All pixels with an intensity higher than
this value are assumed to be foreground.
References
----------
.. [1] Wikipedia, https://en.wikipedia.org/wiki/Otsu's_Method
Examples
--------
>>> from skimage.data import camera
>>> image = camera()
>>> thresh = threshold_otsu(image)
>>> binary = image <= thresh
Notes
-----
The input image must be grayscale.
"""
if image is not None and image.ndim > 2 and image.shape[-1] in (3, 4):
warn(f'threshold_otsu is expected to work correctly only for '
f'grayscale images; image shape {image.shape} looks like '
f'that of an RGB image.')
# Check if the image has more than one intensity value; if not, return that
# value
if image is not None:
first_pixel = image.ravel()[0]
if np.all(image == first_pixel):
return first_pixel
counts, bin_centers = _validate_image_histogram(image, hist, nbins)
# class probabilities for all possible thresholds
weight1 = np.cumsum(counts)
weight2 = np.cumsum(counts[::-1])[::-1]
# class means for all possible thresholds
mean1 = np.cumsum(counts * bin_centers) / weight1
mean2 = (np.cumsum((counts * bin_centers)[::-1]) / weight2[::-1])[::-1]
# Clip ends to align class 1 and class 2 variables:
# The last value of ``weight1``/``mean1`` should pair with zero values in
# ``weight2``/``mean2``, which do not exist.
variance12 = weight1[:-1] * weight2[1:] * (mean1[:-1] - mean2[1:]) ** 2
idx = np.argmax(variance12)
threshold = bin_centers[idx]
return threshold
```
What do you observe about the code that makes it pythonic?
```
Do the pythonic conventions make it easier to understand?
```
How is the documentation on this function?
|
github_jupyter
|
**Recursion and Higher Order Functions**
Today we're tackling recursion, and touching on higher-order functions in Python.
A **recursive** function is one that calls itself.
A classic example: the Fibonacci sequence.
The Fibonacci sequence was originally described to model population growth, and is self-referential in its definition.
The nth Fib number is defined in terms of the previous two:
- F(n) = F(n-1) + F(n-2)
- F(1) = 0
- F(2) = 1
Another classic example:
Factorial:
- n! = n(n-1)(n-2)(n-3) ... 1
or:
- n! = n*(n-1)!
Let's look at an implementation of the factorial and of the Fibonacci sequence in Python:
```
def factorial(n):
if n == 1:
return 1
else:
return n*factorial(n-1)
print(factorial(5))
def fibonacci(n):
if n == 1:
return 0
elif n == 2:
return 1
else:
# print('working on number ' + str(n))
return fibonacci(n-1)+fibonacci(n-2)
fibonacci(7)
```
There are two very important parts of these functions: a base case (or two) and a recursive case. When designing recursive functions it can help to think about these two cases!
The base case is the case when we know we are done, and can just return a value. (e.g. in fibonacci above there are two base cases, `n ==1` and `n ==2`).
The recursive case is the case when we make the recursive call - that is we call the function again.
Let's write a function that counts down from a parameter n to zero, and then prints "Blastoff!".
```
def countdown(n):
# base case
if n == 0:
print('Blastoff!')
# recursive case
else:
print(n)
countdown(n-1)
countdown(10)
```
Let's write a recursive function that adds up the elements of a list:
```
def add_up_list(my_list):
# base case
if len(my_list) == 0:
return 0
# recursive case
else:
first_elem = my_list[0]
return first_elem + add_up_list(my_list[1:])
my_list = [1, 2, 1, 3, 4]
print(add_up_list(my_list))
```
**Higher-order functions**
are functions that takes a function as an argument or returns a function. We will talk briefly about functions that take a function as an argument. Let's look at an example.
```
def h(x):
return x+4
def g(x):
return x**2
def doItTwice(f, x):
return f(f(x))
print(doItTwice(h, 3))
print(doItTwice(g, 3))
```
A common reason for using a higher-order function is to apply a parameter-specified function repeatedly over a data structure (like a list or a dictionary).
Let's look at an example function that applies a parameter function to every element of a list:
```
def sampleFunction1(x):
return 2*x
def sampleFunction2(x):
return x % 2
def applyToAll(func, myList):
newList = []
for element in myList:
newList.append(func(element))
return newList
aList = [2, 3, 4, 5]
print(applyToAll(sampleFunction1, aList))
print(applyToAll(sampleFunction2, aList))
```
Something like this applyToAll function is built into Python, and is called map
```
def sampleFunction1(x):
return 2*x
def sampleFunction2(x):
return x % 2
aList = [2, 3, 4, 5]
print(list(map(sampleFunction1, aList)))
bList = [2, 3, 4, 5]
print(list(map(sampleFunction2, aList)))
```
Python has quite a few built-in functions (some higher-order, some not). You can find lots of them here: https://docs.python.org/3.3/library/functions.html
(I **will not** by default require you to remember those for an exam!!)
Example: zip does something that may be familiar from last week's lab.
```
x = [1, 2, 3]
y = [4, 5, 6]
zipped = zip(x, y)
print(list(zipped))
```
|
github_jupyter
|
# Introduction to `pandas`
```
import numpy as np
import pandas as pd
```
## Series and Data Frames
### Series objects
A `Series` is like a vector. All elements must have the same type or are nulls.
```
s = pd.Series([1,1,2,3] + [None])
s
```
### Size
```
s.size
```
### Unique Counts
```
s.value_counts()
```
### Special types of series
#### Strings
```
words = 'the quick brown fox jumps over the lazy dog'.split()
s1 = pd.Series([' '.join(item) for item in zip(words[:-1], words[1:])])
s1
s1.str.upper()
s1.str.split()
s1.str.split().str[1]
```
### Categories
```
s2 = pd.Series(['Asian', 'Asian', 'White', 'Black', 'White', 'Hispanic'])
s2
s2 = s2.astype('category')
s2
s2.cat.categories
s2.cat.codes
```
### DataFrame objects
A `DataFrame` is like a matrix. Columns in a `DataFrame` are `Series`.
- Each column in a DataFrame represents a **variale**
- Each row in a DataFrame represents an **observation**
- Each cell in a DataFrame represents a **value**
```
df = pd.DataFrame(dict(num=[1,2,3] + [None]))
df
df.num
```
### Index
Row and column identifiers are of `Index` type.
Somewhat confusingly, index is also a a synonym for the row identifiers.
```
df.index
```
#### Setting a column as the row index
```
df
df1 = df.set_index('num')
df1
```
#### Making an index into a column
```
df1.reset_index()
```
### Columns
This is just a different index object
```
df.columns
```
### Getting raw values
Sometimes you just want a `numpy` array, and not a `pandas` object.
```
df.values
```
## Creating Data Frames
### Manual
```
from collections import OrderedDict
n = 5
dates = pd.date_range(start='now', periods=n, freq='d')
df = pd.DataFrame(OrderedDict(pid=np.random.randint(100, 999, n),
weight=np.random.normal(70, 20, n),
height=np.random.normal(170, 15, n),
date=dates,
))
df
```
### From file
You can read in data from many different file types - plain text, JSON, spreadsheets, databases etc. Functions to read in data look like `read_X` where X is the data type.
```
%%file measures.txt
pid weight height date
328 72.654347 203.560866 2018-11-11 14:16:18.148411
756 34.027679 189.847316 2018-11-12 14:16:18.148411
185 28.501914 158.646074 2018-11-13 14:16:18.148411
507 17.396343 180.795993 2018-11-14 14:16:18.148411
919 64.724301 173.564725 2018-11-15 14:16:18.148411
df = pd.read_table('measures.txt')
df
```
## Indexing Data Frames
### Implicit defaults
if you provide a slice, it is assumed that you are asking for rows.
```
df[1:3]
```
If you provide a singe value or list, it is assumed that you are asking for columns.
```
df[['pid', 'weight']]
```
### Extracting a column
#### Dictionary style access
```
df['pid']
```
#### Property style access
This only works for column names tat are also valid Python identifier (i.e., no spaces or dashes or keywords)
```
df.pid
```
### Indexing by location
This is similar to `numpy` indexing
```
df.iloc[1:3, :]
df.iloc[1:3, [True, False, True]]
```
### Indexing by name
```
df.loc[1:3, 'weight':'height']
```
**Warning**: When using `loc`, the row slice indicates row names, not positions.
```
df1 = df.copy()
df1.index = df.index + 1
df1
df1.loc[1:3, 'weight':'height']
```
## Structure of a Data Frame
### Data types
```
df.dtypes
```
### Converting data types
#### Using `astype` on one column
```
df.pid = df.pid.astype('category')
```
#### Using `astype` on multiple columns
```
df = df.astype(dict(weight=float, height=float))
```
#### Using a conversion function
```
df.date = pd.to_datetime(df.date)
```
#### Check
```
df.dtypes
```
### Basic properties
```
df.size
df.shape
df.describe()
```
### Inspection
```
df.head(n=3)
df.tail(n=3)
df.sample(n=3)
df.sample(frac=0.5)
```
## Selecting, Renaming and Removing Columns
### Selecting columns
```
df.filter(items=['pid', 'date'])
df.filter(regex='.*ght')
```
#### Note that you can also use regular string methods on the columns
```
df.loc[:, df.columns.str.contains('d')]
```
### Renaming columns
```
df.rename(dict(weight='w', height='h'), axis=1)
orig_cols = df.columns
df.columns = list('abcd')
df
df.columns = orig_cols
df
```
### Removing columns
```
df.drop(['pid', 'date'], axis=1)
df.drop(columns=['pid', 'date'])
df.drop(columns=df.columns[df.columns.str.contains('d')])
```
## Selecting, Renaming and Removing Rows
### Selecting rows
```
df[df.weight.between(60,70)]
df[(69 <= df.weight) & (df.weight < 70)]
df[df.date.between(pd.to_datetime('2018-11-13'),
pd.to_datetime('2018-11-15 23:59:59'))]
```
### Renaming rows
```
df.rename({i:letter for i,letter in enumerate('abcde')})
df.index = ['the', 'quick', 'brown', 'fox', 'jumphs']
df
df = df.reset_index(drop=True)
df
```
### Dropping rows
```
df.drop([1,3], axis=0)
```
#### Dropping duplicated data
```
df['something'] = [1,1,None,2,None]
df.loc[df.something.duplicated()]
df.drop_duplicates(subset='something')
```
#### Dropping missing data
```
df
df.something.fillna(0)
df.something.ffill()
df.something.bfill()
df.something.interpolate()
df.dropna()
```
## Transforming and Creating Columns
```
df.assign(bmi=df['weight'] / (df['height']/100)**2)
df['bmi'] = df['weight'] / (df['height']/100)**2
df
df['something'] = [2,2,None,None,3]
df
```
## Sorting Data Frames
### Sort on indexes
```
df.sort_index(axis=1)
df.sort_index(axis=0, ascending=False)
```
### Sort on values
```
df.sort_values(by=['something', 'bmi'], ascending=[True, False])
```
## Summarizing
### Apply an aggregation function
```
df.select_dtypes(include=np.number)
df.select_dtypes(include=np.number).agg(np.sum)
df.agg(['count', np.sum, np.mean])
```
## Split-Apply-Combine
We often want to perform subgroup analysis (conditioning by some discrete or categorical variable). This is done with `groupby` followed by an aggregate function. Conceptually, we split the data frame into separate groups, apply the aggregate function to each group separately, then combine the aggregated results back into a single data frame.
```
df['treatment'] = list('ababa')
df
grouped = df.groupby('treatment')
grouped.get_group('a')
grouped.mean()
```
### Using `agg` with `groupby`
```
grouped.agg('mean')
grouped.agg(['mean', 'std'])
grouped.agg({'weight': ['mean', 'std'], 'height': ['min', 'max'], 'bmi': lambda x: (x**2).sum()})
```
### Using `trasnform` wtih `groupby`
```
g_mean = grouped['weight', 'height'].transform(np.mean)
g_mean
g_std = grouped['weight', 'height'].transform(np.std)
g_std
(df[['weight', 'height']] - g_mean)/g_std
```
## Combining Data Frames
```
df
df1 = df.iloc[3:].copy()
df1.drop('something', axis=1, inplace=True)
df1
```
### Adding rows
Note that `pandas` aligns by column indexes automatically.
```
df.append(df1, sort=False)
pd.concat([df, df1], sort=False)
```
### Adding columns
```
df.pid
df2 = pd.DataFrame(OrderedDict(pid=[649, 533, 400, 600], age=[23,34,45,56]))
df2.pid
df.pid = df.pid.astype('int')
pd.merge(df, df2, on='pid', how='inner')
pd.merge(df, df2, on='pid', how='left')
pd.merge(df, df2, on='pid', how='right')
pd.merge(df, df2, on='pid', how='outer')
```
### Merging on the index
```
df1 = pd.DataFrame(dict(x=[1,2,3]), index=list('abc'))
df2 = pd.DataFrame(dict(y=[4,5,6]), index=list('abc'))
df3 = pd.DataFrame(dict(z=[7,8,9]), index=list('abc'))
df1
df2
df3
df1.join([df2, df3])
```
## Fixing common DataFrame issues
### Multiple variables in a column
```
df = pd.DataFrame(dict(pid_treat = ['A-1', 'B-2', 'C-1', 'D-2']))
df
df.pid_treat.str.split('-')
df.pid_treat.str.split('-').apply(pd.Series, index=['pid', 'treat'])
```
### Multiple values in a cell
```
df = pd.DataFrame(dict(pid=['a', 'b', 'c'], vals = [(1,2,3), (4,5,6), (7,8,9)]))
df
df[['t1', 't2', 't3']] = df.vals.apply(pd.Series)
df
df.drop('vals', axis=1, inplace=True)
pd.melt(df, id_vars='pid', value_name='vals').drop('variable', axis=1)
```
## Reshaping Data Frames
Sometimes we need to make rows into columns or vice versa.
### Converting multiple columns into a single column
This is often useful if you need to condition on some variable.
```
url = 'https://raw.githubusercontent.com/uiuc-cse/data-fa14/gh-pages/data/iris.csv'
iris = pd.read_csv(url)
iris.head()
iris.shape
df_iris = pd.melt(iris, id_vars='species')
df_iris.sample(10)
```
## Chaining commands
Sometimes you see this functional style of method chaining that avoids the need for temporary intermediate variables.
```
(
iris.
sample(frac=0.2).
filter(regex='s.*').
assign(both=iris.sepal_length + iris.sepal_length).
groupby('species').agg(['mean', 'sum']).
pipe(lambda x: np.around(x, 1))
)
```
## Moving between R and Python in Jupyter
```
%load_ext rpy2.ipython
import warnings
warnings.simplefilter('ignore', FutureWarning)
iris = %R iris
iris.head()
iris_py = iris.copy()
iris_py.Species = iris_py.Species.str.upper()
%%R -i iris_py -o iris_r
iris_r <- iris_py[1:3,]
iris_r
```
|
github_jupyter
|
# SLU13: Bias-Variance trade-off & Model Selection -- Examples
---
<a id='top'></a>
### 1. Model evaluation
* a. [Train-test split](#traintest)
* b. [Train-val-test split](#val)
* c. [Cross validation](#crossval)
### 2. [Learning curves](#learningcurves)
# 1. Model evaluation
```
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import learning_curve
%matplotlib inline
# Create the DataFrame with the data
df = pd.read_csv("data/beer.csv")
# Create a DataFrame with the features (X) and labels (y)
X = df.drop(["IsIPA"], axis=1)
y = df["IsIPA"]
print("Number of entries: ", X.shape[0])
```
<a id='traintest'></a> [Return to top](#top)
## Create a training and a test set
```
from sklearn.model_selection import train_test_split
# Using 20 % of the data as test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
print("Number of training entries: ", X_train.shape[0])
print("Number of test entries: ", X_test.shape[0])
```
<a id='val'></a> [Return to top](#top)
## Create a training, test and validation set
```
# Using 20 % as test set and 20 % as validation set
X_train, X_temp, y_train, y_temp = train_test_split(X, y, test_size=0.4)
X_val, X_test, y_val, y_test = train_test_split(X_temp, y_temp, test_size=0.50)
print("Number of training entries: ", X_train.shape[0])
print("Number of validation entries: ", X_val.shape[0])
print("Number of test entries: ", X_test.shape[0])
```
<a id='crossval'></a> [Return to top](#top)
## Use cross-validation (using a given classifier)
```
from sklearn.model_selection import cross_val_score
knn = KNeighborsClassifier(n_neighbors=5)
# Use cv to specify the number of folds
scores = cross_val_score(knn, X, y, cv=5)
print(f"Mean of scores: {scores.mean():.3f}")
print(f"Variance of scores: {scores.var():.3f}")
```
<a id='learningcurves'></a> [Return to top](#top)
# 2. Learning Curves
Here is the function that is taken from the sklearn page on learning curves:
```
def plot_learning_curve(estimator, title, X, y, ylim=None, cv=None,
n_jobs=1, train_sizes=np.linspace(.1, 1.0, 5)):
"""
Generate a simple plot of the test and training learning curve.
Parameters
----------
estimator : object type that implements the "fit" and "predict" methods
An object of that type which is cloned for each validation.
title : string
Title for the chart.
X : array-like, shape (n_samples, n_features)
Training vector, where n_samples is the number of samples and
n_features is the number of features.
y : array-like, shape (n_samples) or (n_samples, n_features), optional
Target relative to X for classification or regression;
None for unsupervised learning.
ylim : tuple, shape (ymin, ymax), optional
Defines minimum and maximum yvalues plotted.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross-validation,
- integer, to specify the number of folds.
- An object to be used as a cross-validation generator.
- An iterable yielding train/test splits.
For integer/None inputs, if ``y`` is binary or multiclass,
:class:`StratifiedKFold` used. If the estimator is not a classifier
or if ``y`` is neither binary nor multiclass, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validators that can be used here.
n_jobs : integer, optional
Number of jobs to run in parallel (default 1).
"""
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
plt.ylabel("Score")
train_sizes, train_scores, test_scores = learning_curve(
estimator, X, y, cv=cv, n_jobs=n_jobs, train_sizes=train_sizes)
train_scores_mean = np.mean(train_scores, axis=1)
train_scores_std = np.std(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
plt.grid()
plt.fill_between(train_sizes, train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1,
color="r")
plt.fill_between(train_sizes, test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="g")
plt.plot(train_sizes, train_scores_mean, 'o-', color="r",
label="Training score")
plt.plot(train_sizes, test_scores_mean, 'o-', color="g",
label="Test Set score")
plt.legend(loc="best")
return plt
# and this is how we used it
X = df.select_dtypes(exclude='object').fillna(-1).drop('IsIPA', axis=1)
y = df.IsIPA
clf = DecisionTreeClassifier(random_state=1, max_depth=5)
plot_learning_curve(X=X, y=y, estimator=clf, title='DecisionTreeClassifier');
```
And remember the internals of what this function is actually doing by knowing how to use the
output of the scikit [learning_curve](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.learning_curve.html) function
```
# here's where the magic happens! The learning curve function is going
# to take your classifier and your training data and subset the data
train_sizes, train_scores, test_scores = learning_curve(clf, X, y)
# 5 different training set sizes have been selected
# with the smallest being 59 and the largest being 594
# the remaining is used for testing
print('train set sizes', train_sizes)
print('test set sizes', X.shape[0] - train_sizes)
# each row corresponds to a training set size
# each column corresponds to a cross validation fold
# the first row is the highest because it corresponds
# to the smallest training set which means that it's very
# easy for the classifier to overfit and have perfect
# test set predictions while as the test set grows it
# becomes a bit more difficult for this to happen.
train_scores
# The test set scores where again, each row corresponds
# to a train / test set size and each column is a differet
# run with the same train / test sizes
test_scores
# Let's average the scores across each fold so that we can plot them
train_scores_mean = np.mean(train_scores, axis=1)
test_scores_mean = np.mean(test_scores, axis=1)
# this one isn't quite as cool as the other because it doesn't show the variance
# but the fundamentals are still here and it's a much simpler one to understand
learning_curve_df = pd.DataFrame({
'Training score': train_scores_mean,
'Test Set score': test_scores_mean
}, index=train_sizes)
plt.figure()
plt.ylabel("Score")
plt.xlabel("Training examples")
plt.title('Learning Curve')
plt.plot(learning_curve_df);
plt.legend(learning_curve_df.columns, loc="best");
```
|
github_jupyter
|
# Phi_K advanced tutorial
This notebook guides you through the more advanced functionality of the phik package. This notebook will not cover all the underlying theory, but will just attempt to give an overview of all the options that are available. For a theoretical description the user is referred to our paper.
The package offers functionality on three related topics:
1. Phik correlation matrix
2. Significance matrix
3. Outlier significance matrix
```
%%capture
# install phik (if not installed yet)
import sys
!"{sys.executable}" -m pip install phik
# import standard packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import itertools
import phik
from phik import resources
from phik.binning import bin_data
from phik.decorators import *
from phik.report import plot_correlation_matrix
%matplotlib inline
# if one changes something in the phik-package one can automatically reload the package or module
%load_ext autoreload
%autoreload 2
```
# Load data
A simulated dataset is part of the phik-package. The dataset concerns car insurance data. Load the dataset here:
```
data = pd.read_csv( resources.fixture('fake_insurance_data.csv.gz') )
data.head()
```
## Specify bin types
The phik-package offers a way to calculate correlations between variables of mixed types. Variable types can be inferred automatically although we recommend to variable types to be specified by the user.
Because interval type variables need to be binned in order to calculate phik and the significance, a list of interval variables is created.
```
data_types = {'severity': 'interval',
'driver_age':'interval',
'satisfaction':'ordinal',
'mileage':'interval',
'car_size':'ordinal',
'car_use':'ordinal',
'car_color':'categorical',
'area':'categorical'}
interval_cols = [col for col, v in data_types.items() if v=='interval' and col in data.columns]
interval_cols
# interval_cols is used below
```
# Phik correlation matrix
Now let's start calculating the correlation phik between pairs of variables.
Note that the original dataset is used as input, the binning of interval variables is done automatically.
```
phik_overview = data.phik_matrix(interval_cols=interval_cols)
phik_overview
```
### Specify binning per interval variable
Binning can be set per interval variable individually. One can set the number of bins, or specify a list of bin edges. Note that the measured phik correlation is dependent on the chosen binning.
The default binning is uniform between the min and max values of the interval variable.
```
bins = {'mileage':5, 'driver_age':[18,25,35,45,55,65,125]}
phik_overview = data.phik_matrix(interval_cols=interval_cols, bins=bins)
phik_overview
```
### Do not apply noise correction
For low statistics samples often a correlation larger than zero is measured when no correlation is actually present in the true underlying distribution. This is not only the case for phik, but also for the pearson correlation and Cramer's phi (see figure 4 in <font color='red'> XX </font>). In the phik calculation a noise correction is applied by default, to take into account erroneous correlation values as a result of low statistics. To switch off this noise cancellation (not recommended), do:
```
phik_overview = data.phik_matrix(interval_cols=interval_cols, noise_correction=False)
phik_overview
```
### Using a different expectation histogram
By default phik compares the 2d distribution of two (binned) variables with the distribution that assumes no dependency between them. One can also change the expected distribution though. Phi_K is calculated in the same way, but using the other expectation distribution.
```
from phik.binning import auto_bin_data
from phik.phik import phik_observed_vs_expected_from_rebinned_df, phik_from_hist2d
from phik.statistics import get_dependent_frequency_estimates
# get observed 2d histogram of two variables
cols = ["mileage", "car_size"]
icols = ["mileage"]
observed = data[cols].hist2d(interval_cols=icols).values
# default phik evaluation from observed distribution
phik_value = phik_from_hist2d(observed)
print (phik_value)
# phik evaluation from an observed and expected distribution
expected = get_dependent_frequency_estimates(observed)
phik_value = phik_from_hist2d(observed=observed, expected=expected)
print (phik_value)
# one can also compare two datasets against each other, and get a full phik matrix that way.
# this needs binned datasets though.
# (the user needs to make sure the binnings of both datasets are identical.)
data_binned, _ = auto_bin_data(data, interval_cols=interval_cols)
# here we are comparing data_binned against itself
phik_matrix = phik_observed_vs_expected_from_rebinned_df(data_binned, data_binned)
# all off-diagonal entries are zero, meaning the all 2d distributions of both datasets are identical.
# (by construction the diagonal is one.)
phik_matrix
```
# Statistical significance of the correlation
When assessing correlations it is good practise to evaluate both the correlation and the significance of the correlation: a large correlation may be statistically insignificant, and vice versa a small correlation may be very significant. For instance, scipy.stats.pearsonr returns both the pearson correlation and the p-value. Similarly, the phik package offers functionality the calculate a significance matrix. Significance is defined as:
$$Z = \Phi^{-1}(1-p)\ ;\quad \Phi(z)=\frac{1}{\sqrt{2\pi}} \int_{-\infty}^{z} e^{-t^{2}/2}\,{\rm d}t $$
Several corrections to the 'standard' p-value calculation are taken into account, making the method more robust for low statistics and sparse data cases. The user is referred to our paper for more details.
Due to the corrections, the significance calculation can take a few seconds.
```
significance_overview = data.significance_matrix(interval_cols=interval_cols)
significance_overview
```
### Specify binning per interval variable
Binning can be set per interval variable individually. One can set the number of bins, or specify a list of bin edges. Note that the measure phik correlation is dependent on the chosen binning.
```
bins = {'mileage':5, 'driver_age':[18,25,35,45,55,65,125]}
significance_overview = data.significance_matrix(interval_cols=interval_cols, bins=bins)
significance_overview
```
### Specify significance method
The recommended method to calculate the significance of the correlation is a hybrid approach, which uses the G-test statistic. The number of degrees of freedom and an analytical, empirical description of the $\chi^2$ distribution are sed, based on Monte Carlo simulations. This method works well for both high as low statistics samples.
Other approaches to calculate the significance are implemented:
- asymptotic: fast, but over-estimates the number of degrees of freedom for low statistics samples, leading to erroneous values of the significance
- MC: Many simulated samples are needed to accurately measure significances larger than 3, making this method computationally expensive.
```
significance_overview = data.significance_matrix(interval_cols=interval_cols, significance_method='asymptotic')
significance_overview
```
### Simulation method
The chi2 of a contingency table is measured using a comparison of the expected frequencies with the true frequencies in a contingency table. The expected frequencies can be simulated in a variety of ways. The following methods are implemented:
- multinominal: Only the total number of records is fixed. (default)
- row_product_multinominal: The row totals fixed in the sampling.
- col_product_multinominal: The column totals fixed in the sampling.
- hypergeometric: Both the row or column totals are fixed in the sampling. (Note that this type of sampling is only available when row and column totals are integers, which is usually the case.)
```
# --- Warning, can be slow
# turned off here by default for unit testing purposes
#significance_overview = data.significance_matrix(interval_cols=interval_cols, simulation_method='hypergeometric')
#significance_overview
```
### Expected frequencies
```
from phik.simulation import sim_2d_data_patefield, sim_2d_product_multinominal, sim_2d_data
inputdata = data[['driver_age', 'area']].hist2d(interval_cols=['driver_age'])
inputdata
```
#### Multinominal
```
simdata = sim_2d_data(inputdata.values)
print('data total:', inputdata.sum().sum())
print('sim total:', simdata.sum().sum())
print('data row totals:', inputdata.sum(axis=0).values)
print('sim row totals:', simdata.sum(axis=0))
print('data column totals:', inputdata.sum(axis=1).values)
print('sim column totals:', simdata.sum(axis=1))
```
#### product multinominal
```
simdata = sim_2d_product_multinominal(inputdata.values, axis=0)
print('data total:', inputdata.sum().sum())
print('sim total:', simdata.sum().sum())
print('data row totals:', inputdata.sum(axis=0).astype(int).values)
print('sim row totals:', simdata.sum(axis=0).astype(int))
print('data column totals:', inputdata.sum(axis=1).astype(int).values)
print('sim column totals:', simdata.sum(axis=1).astype(int))
```
#### hypergeometric ("patefield")
```
# patefield simulation needs compiled c++ code.
# only run this if the python binding to the (compiled) patefiled simulation function is found.
try:
from phik.simcore import _sim_2d_data_patefield
CPP_SUPPORT = True
except ImportError:
CPP_SUPPORT = False
if CPP_SUPPORT:
simdata = sim_2d_data_patefield(inputdata.values)
print('data total:', inputdata.sum().sum())
print('sim total:', simdata.sum().sum())
print('data row totals:', inputdata.sum(axis=0).astype(int).values)
print('sim row totals:', simdata.sum(axis=0))
print('data column totals:', inputdata.sum(axis=1).astype(int).values)
print('sim column totals:', simdata.sum(axis=1))
```
# Outlier significance
The normal pearson correlation between two interval variables is easy to interpret. However, the phik correlation between two variables of mixed type is not always easy to interpret, especially when it concerns categorical variables. Therefore, functionality is provided to detect "outliers": excesses and deficits over the expected frequencies in the contingency table of two variables.
### Example 1: mileage versus car_size
For the categorical variable pair mileage - car_size we measured:
$$\phi_k = 0.77 \, ,\quad\quad \mathrm{significance} = 46.3$$
Let's use the outlier significance functionality to gain a better understanding of this significance correlation between mileage and car size.
```
c0 = 'mileage'
c1 = 'car_size'
tmp_interval_cols = ['mileage']
outlier_signifs, binning_dict = data[[c0,c1]].outlier_significance_matrix(interval_cols=tmp_interval_cols,
retbins=True)
outlier_signifs
```
### Specify binning per interval variable
Binning can be set per interval variable individually. One can set the number of bins, or specify a list of bin edges.
Note: in case a bin is created without any records this bin will be automatically dropped in the phik and (outlier) significance calculations. However, in the outlier significance calculation this will currently lead to an error as the number of provided bin edges does not match the number of bins anymore.
```
bins = [0,1E2, 1E3, 1E4, 1E5, 1E6]
outlier_signifs, binning_dict = data[[c0,c1]].outlier_significance_matrix(interval_cols=tmp_interval_cols,
bins=bins, retbins=True)
outlier_signifs
```
### Specify binning per interval variable -- dealing with underflow and overflow
When specifying custom bins as situation can occur when the minimal (maximum) value in the data is smaller (larger) than the minimum (maximum) bin edge. Data points outside the specified range will be collected in the underflow (UF) and overflow (OF) bins. One can choose how to deal with these under/overflow bins, by setting the drop_underflow and drop_overflow variables.
Note that the drop_underflow and drop_overflow options are also available for the calculation of the phik matrix and the significance matrix.
```
bins = [1E2, 1E3, 1E4, 1E5]
outlier_signifs, binning_dict = data[[c0,c1]].outlier_significance_matrix(interval_cols=tmp_interval_cols,
bins=bins, retbins=True,
drop_underflow=False,
drop_overflow=False)
outlier_signifs
```
### Dealing with NaN's in the data
Let's add some missing values to our data
```
data.loc[np.random.choice(range(len(data)), size=10), 'car_size'] = np.nan
data.loc[np.random.choice(range(len(data)), size=10), 'mileage'] = np.nan
```
Sometimes there can be information in the missing values and in which case you might want to consider the NaN values as a separate category. This can be achieved by setting the dropna argument to False.
```
bins = [1E2, 1E3, 1E4, 1E5]
outlier_signifs, binning_dict = data[[c0,c1]].outlier_significance_matrix(interval_cols=tmp_interval_cols,
bins=bins, retbins=True,
drop_underflow=False,
drop_overflow=False,
dropna=False)
outlier_signifs
```
Here OF and UF are the underflow and overflow bin of car_size, respectively.
To just ignore records with missing values set dropna to True (default).
```
bins = [1E2, 1E3, 1E4, 1E5]
outlier_signifs, binning_dict = data[[c0,c1]].outlier_significance_matrix(interval_cols=tmp_interval_cols,
bins=bins, retbins=True,
drop_underflow=False,
drop_overflow=False,
dropna=True)
outlier_signifs
```
Note that the dropna option is also available for the calculation of the phik matrix and the significance matrix.
|
github_jupyter
|
```
import tabula
import numpy as np
import pandas as pd
import os
from pathlib import Path
import PyPDF2
import re
import requests
import json
import time
# filenames = [
# os.path.expanduser('/home/parth/Documents/USICT/it_res.pdf'),
# os.path.expanduser('/home/parth/Documents/USICT/cse_res.pdf'),
# os.path.expanduser('/home/parth/Documents/USICT/ece_res.pdf')]
# filenames = [
# os.path.expanduser('~/Documents/USICT/ipu_results/cse_even_sems.pdf'),
# os.path.expanduser('~/Documents/USICT/ipu_results/ece_even_sems.pdf')
# ]
# filenames = [
# os.path.expanduser('~/Documents/USICT/ipu_results/it_even_sems.pdf')
# ]
filenames = [
os.path.expanduser('/home/parth/Documents/USICT/it_res.pdf'),
os.path.expanduser('/home/parth/Documents/USICT/cse_res.pdf'),
os.path.expanduser('/home/parth/Documents/USICT/ece_res.pdf'),
os.path.expanduser('~/Documents/USICT/ipu_results/cse_even_sems.pdf'),
os.path.expanduser('~/Documents/USICT/ipu_results/ece_even_sems.pdf'),
os.path.expanduser('~/Documents/USICT/ipu_results/it_even_sems.pdf')
]
scheme_reg = re.compile(r'scheme\s+of\s+examinations',re.IGNORECASE)
institution_reg = re.compile(r'institution\s*:\s*([\w\n(,)& ]+)\nS\.No',re.IGNORECASE)
sem_reg = re.compile(r'se\s?m[.//\w\n]+:\s+([\w\n]+)',re.IGNORECASE)
programme_reg = re.compile(r'programme\s+name:\s+([\w(,)& \n]+)SchemeID',re.IGNORECASE)
branch_reg = re.compile(r'[\w &]+\(([\w ]+)\)')
def get_info(text) :
college = institution_reg.search(text)[1].replace('\n','').strip().title()
semester = int(sem_reg.search(text)[1].replace('\n','').strip())
course = programme_reg.search(text)[1].replace('\n','').strip().title()
branch = branch_reg.search(course)[1].strip().title()
course = course[0:course.find('(')].strip()
info = {
'college' : college,
'semester' : semester,
'course' : course,
'branch' : branch,
}
return info
SITE = "https://api-rhapsody.herokuapp.com/academia"
# SITE = "http://localhost:3000/academia"
#Add college
data ={
'college' : {
'college' : "University School Of Information, Communication & Technology (Formerly Usit)"
}
}
r = requests.post(SITE+"/college",json=data)
print(r,r.content)
def already_exists(info) :
r = requests.get(SITE+"/semester",params=info)
content = json.loads(r.content)
# print(r.status_code,r.content)
return r.status_code == 200 and content != {}
def getSubjects(df) :
subjects = []
for index,row in df.iterrows() :
subject = {}
subject['subject'] = row['Subject'].strip().title()
subject['subjectCode'] = row['Code']
subject['credits'] = row['Credit']
subjects.append(subject)
return subjects
for filename in filenames :
pdf = PyPDF2.PdfFileReader(filename)
print(filename,pdf.getNumPages())
for i in range(0,pdf.getNumPages()) :
text = pdf.getPage(i).extractText()
if scheme_reg.search(text) :
info = get_info(text)
df = tabula.read_pdf(filename,pages=i+1)
subjects = getSubjects(df[0])
if already_exists(info) :
print("information already exists")
continue
info['semester'] = {'semester' : info['semester'], 'subjects' : subjects}
r = requests.post(SITE+"/semester",json=info)
print(r,r.content)
# time.sleep(2)
# print(info)
from IPython.display import display
```
|
github_jupyter
|
# Description
This notebook runs some pre-analyses using DBSCAN to explore the best set of parameters (`min_samples` and `eps`) to cluster `pca` data version.
# Environment variables
```
from IPython.display import display
import conf
N_JOBS = conf.GENERAL["N_JOBS"]
display(N_JOBS)
%env MKL_NUM_THREADS=$N_JOBS
%env OPEN_BLAS_NUM_THREADS=$N_JOBS
%env NUMEXPR_NUM_THREADS=$N_JOBS
%env OMP_NUM_THREADS=$N_JOBS
```
# Modules loading
```
%load_ext autoreload
%autoreload 2
from pathlib import Path
import numpy as np
import pandas as pd
from sklearn.neighbors import NearestNeighbors
from sklearn.metrics import pairwise_distances
from sklearn.cluster import DBSCAN
from sklearn.metrics import (
silhouette_score,
calinski_harabasz_score,
davies_bouldin_score,
)
import matplotlib.pyplot as plt
import seaborn as sns
from utils import generate_result_set_name
from clustering.ensembles.utils import generate_ensemble
```
# Global settings
```
np.random.seed(0)
CLUSTERING_ATTRIBUTES_TO_SAVE = ["n_clusters"]
```
# Data version: pca
```
INPUT_SUBSET = "pca"
INPUT_STEM = "z_score_std-projection-smultixcan-efo_partial-mashr-zscores"
DR_OPTIONS = {
"n_components": 50,
"svd_solver": "full",
"random_state": 0,
}
input_filepath = Path(
conf.RESULTS["DATA_TRANSFORMATIONS_DIR"],
INPUT_SUBSET,
generate_result_set_name(
DR_OPTIONS, prefix=f"{INPUT_SUBSET}-{INPUT_STEM}-", suffix=".pkl"
),
).resolve()
display(input_filepath)
assert input_filepath.exists(), "Input file does not exist"
input_filepath_stem = input_filepath.stem
display(input_filepath_stem)
data = pd.read_pickle(input_filepath)
data.shape
data.head()
```
## Tests different k values (k-NN)
```
# `k_values` is the full range of k for kNN, whereas `k_values_to_explore` is a
# subset that will be explored in this notebook. If the analysis works, then
# `k_values` and `eps_range_per_k` below are copied to the notebook that will
# produce the final DBSCAN runs (`../002_[...]-dbscan-....ipynb`)
k_values = np.arange(2, 125 + 1, 1)
k_values_to_explore = (2, 5, 10, 15, 20, 30, 40, 50, 75, 100, 125)
results = {}
for k in k_values_to_explore:
nbrs = NearestNeighbors(n_neighbors=k, n_jobs=N_JOBS).fit(data)
distances, indices = nbrs.kneighbors(data)
results[k] = (distances, indices)
eps_range_per_k = {
k: (10, 20)
if k < 5
else (11, 25)
if k < 10
else (12, 30)
if k < 15
else (13, 35)
if k < 20
else (14, 40)
for k in k_values
}
eps_range_per_k_to_explore = {k: eps_range_per_k[k] for k in k_values_to_explore}
for k, (distances, indices) in results.items():
d = distances[:, 1:].mean(axis=1)
d = np.sort(d)
fig, ax = plt.subplots()
plt.plot(d)
r = eps_range_per_k_to_explore[k]
plt.hlines(r[0], 0, data.shape[0], color="red")
plt.hlines(r[1], 0, data.shape[0], color="red")
plt.xlim((3000, data.shape[0]))
plt.title(f"k={k}")
display(fig)
plt.close(fig)
```
# Extended test
## Generate clusterers
```
CLUSTERING_OPTIONS = {}
# K_RANGE is the min_samples parameter in DBSCAN (sklearn)
CLUSTERING_OPTIONS["K_RANGE"] = k_values_to_explore
CLUSTERING_OPTIONS["EPS_RANGE_PER_K"] = eps_range_per_k_to_explore
CLUSTERING_OPTIONS["EPS_STEP"] = 33
CLUSTERING_OPTIONS["METRIC"] = "euclidean"
display(CLUSTERING_OPTIONS)
CLUSTERERS = {}
idx = 0
for k in CLUSTERING_OPTIONS["K_RANGE"]:
eps_range = CLUSTERING_OPTIONS["EPS_RANGE_PER_K"][k]
eps_values = np.linspace(eps_range[0], eps_range[1], CLUSTERING_OPTIONS["EPS_STEP"])
for eps in eps_values:
clus = DBSCAN(min_samples=k, eps=eps, metric="precomputed", n_jobs=N_JOBS)
method_name = type(clus).__name__
CLUSTERERS[f"{method_name} #{idx}"] = clus
idx = idx + 1
display(len(CLUSTERERS))
_iter = iter(CLUSTERERS.items())
display(next(_iter))
display(next(_iter))
clustering_method_name = method_name
display(clustering_method_name)
```
## Generate ensemble
```
data_dist = pairwise_distances(data, metric=CLUSTERING_OPTIONS["METRIC"])
data_dist.shape
pd.Series(data_dist.flatten()).describe().apply(str)
ensemble = generate_ensemble(
data_dist,
CLUSTERERS,
attributes=CLUSTERING_ATTRIBUTES_TO_SAVE,
)
ensemble.shape
ensemble.head()
_tmp = ensemble["n_clusters"].value_counts()
display(_tmp)
assert _tmp.index[0] == 3
assert _tmp.loc[3] == 22
ensemble_stats = ensemble["n_clusters"].describe()
display(ensemble_stats)
# number of noisy points
_tmp = ensemble.copy()
_tmp = _tmp.assign(n_noisy=ensemble["partition"].apply(lambda x: np.isnan(x).sum()))
_tmp_stats = _tmp["n_noisy"].describe()
display(_tmp_stats)
assert _tmp_stats["min"] > 5
assert _tmp_stats["max"] < 600
assert 90 < _tmp_stats["mean"] < 95
```
## Testing
```
assert ensemble_stats["min"] > 1
assert not ensemble["n_clusters"].isna().any()
# all partitions have the right size
assert np.all(
[part["partition"].shape[0] == data.shape[0] for idx, part in ensemble.iterrows()]
)
```
## Add clustering quality measures
```
def _remove_nans(data, part):
not_nan_idx = ~np.isnan(part)
return data.iloc[not_nan_idx], part[not_nan_idx]
def _apply_func(func, data, part):
no_nan_data, no_nan_part = _remove_nans(data, part)
return func(no_nan_data, no_nan_part)
ensemble = ensemble.assign(
si_score=ensemble["partition"].apply(
lambda x: _apply_func(silhouette_score, data, x)
),
ch_score=ensemble["partition"].apply(
lambda x: _apply_func(calinski_harabasz_score, data, x)
),
db_score=ensemble["partition"].apply(
lambda x: _apply_func(davies_bouldin_score, data, x)
),
)
ensemble.shape
ensemble.head()
```
# Cluster quality
```
with pd.option_context("display.max_rows", None, "display.max_columns", None):
_df = ensemble.groupby(["n_clusters"]).mean()
display(_df)
with sns.plotting_context("talk", font_scale=0.75), sns.axes_style(
"whitegrid", {"grid.linestyle": "--"}
):
fig = plt.figure(figsize=(14, 6))
ax = sns.pointplot(data=ensemble, x="n_clusters", y="si_score")
ax.set_ylabel("Silhouette index\n(higher is better)")
ax.set_xlabel("Number of clusters ($k$)")
ax.set_xticklabels(ax.get_xticklabels(), rotation=45)
plt.grid(True)
plt.tight_layout()
with sns.plotting_context("talk", font_scale=0.75), sns.axes_style(
"whitegrid", {"grid.linestyle": "--"}
):
fig = plt.figure(figsize=(14, 6))
ax = sns.pointplot(data=ensemble, x="n_clusters", y="ch_score")
ax.set_ylabel("Calinski-Harabasz index\n(higher is better)")
ax.set_xlabel("Number of clusters ($k$)")
ax.set_xticklabels(ax.get_xticklabels(), rotation=45)
plt.grid(True)
plt.tight_layout()
with sns.plotting_context("talk", font_scale=0.75), sns.axes_style(
"whitegrid", {"grid.linestyle": "--"}
):
fig = plt.figure(figsize=(14, 6))
ax = sns.pointplot(data=ensemble, x="n_clusters", y="db_score")
ax.set_ylabel("Davies-Bouldin index\n(lower is better)")
ax.set_xlabel("Number of clusters ($k$)")
ax.set_xticklabels(ax.get_xticklabels(), rotation=45)
plt.grid(True)
plt.tight_layout()
```
# Conclusions
The values explored above for `k_values` and `eps_range_per_k` are the one that will be used for DBSCAN in this data version.
|
github_jupyter
|
## Rhetorical relations classification used in tree building: ESIM
Prepare data and model-related scripts.
Evaluate models.
Make and evaluate ansembles for ESIM and BiMPM model / ESIM and feature-based model.
Output:
- ``models/relation_predictor_esim/*``
```
%load_ext autoreload
%autoreload 2
import os
import glob
import pandas as pd
import numpy as np
import pickle
from utils.file_reading import read_edus, read_gold, read_negative, read_annotation
```
### Make a directory
```
MODEL_PATH = 'models/label_predictor_esim'
! mkdir $MODEL_PATH
TRAIN_FILE_PATH = os.path.join(MODEL_PATH, 'nlabel_cf_train.tsv')
DEV_FILE_PATH = os.path.join(MODEL_PATH, 'nlabel_cf_dev.tsv')
TEST_FILE_PATH = os.path.join(MODEL_PATH, 'nlabel_cf_test.tsv')
```
### Prepare train/test sets
```
IN_PATH = 'data_labeling'
train_samples = pd.read_pickle(os.path.join(IN_PATH, 'train_samples.pkl'))
dev_samples = pd.read_pickle(os.path.join(IN_PATH, 'dev_samples.pkl'))
test_samples = pd.read_pickle(os.path.join(IN_PATH, 'test_samples.pkl'))
counts = train_samples['relation'].value_counts(normalize=False).values
NUMBER_CLASSES = len(counts)
print("number of classes:", NUMBER_CLASSES)
print("class weights:")
np.round(counts.min() / counts, decimals=6)
counts = train_samples['relation'].value_counts()
counts
import razdel
def tokenize(text):
result = ' '.join([tok.text for tok in razdel.tokenize(text)])
return result
train_samples['snippet_x'] = train_samples.snippet_x.map(tokenize)
train_samples['snippet_y'] = train_samples.snippet_y.map(tokenize)
dev_samples['snippet_x'] = dev_samples.snippet_x.map(tokenize)
dev_samples['snippet_y'] = dev_samples.snippet_y.map(tokenize)
test_samples['snippet_x'] = test_samples.snippet_x.map(tokenize)
test_samples['snippet_y'] = test_samples.snippet_y.map(tokenize)
train_samples = train_samples.reset_index()
train_samples[['relation', 'snippet_x', 'snippet_y', 'index']].to_csv(TRAIN_FILE_PATH, sep='\t', header=False, index=False)
dev_samples = dev_samples.reset_index()
dev_samples[['relation', 'snippet_x', 'snippet_y', 'index']].to_csv(DEV_FILE_PATH, sep='\t', header=False, index=False)
test_samples = test_samples.reset_index()
test_samples[['relation', 'snippet_x', 'snippet_y', 'index']].to_csv(TEST_FILE_PATH, sep='\t', header=False, index=False)
```
### Modify model
(Add F1, concatenated encoding)
```
%%writefile models/bimpm_custom_package/model/esim.py
from typing import Dict, List, Any, Optional
import numpy
import torch
from allennlp.common.checks import check_dimensions_match
from allennlp.data import TextFieldTensors, Vocabulary
from allennlp.models.model import Model
from allennlp.modules import FeedForward, InputVariationalDropout
from allennlp.modules.matrix_attention.matrix_attention import MatrixAttention
from allennlp.modules import Seq2SeqEncoder, TextFieldEmbedder
from allennlp.nn import InitializerApplicator
from allennlp.nn.util import (
get_text_field_mask,
masked_softmax,
weighted_sum,
masked_max,
)
from allennlp.training.metrics import CategoricalAccuracy, F1Measure
@Model.register("custom_esim")
class CustomESIM(Model):
"""
This `Model` implements the ESIM sequence model described in [Enhanced LSTM for Natural Language Inference]
(https://api.semanticscholar.org/CorpusID:34032948) by Chen et al., 2017.
Registered as a `Model` with name "esim".
# Parameters
vocab : `Vocabulary`
text_field_embedder : `TextFieldEmbedder`
Used to embed the `premise` and `hypothesis` `TextFields` we get as input to the
model.
encoder : `Seq2SeqEncoder`
Used to encode the premise and hypothesis.
matrix_attention : `MatrixAttention`
This is the attention function used when computing the similarity matrix between encoded
words in the premise and words in the hypothesis.
projection_feedforward : `FeedForward`
The feedforward network used to project down the encoded and enhanced premise and hypothesis.
inference_encoder : `Seq2SeqEncoder`
Used to encode the projected premise and hypothesis for prediction.
output_feedforward : `FeedForward`
Used to prepare the concatenated premise and hypothesis for prediction.
output_logit : `FeedForward`
This feedforward network computes the output logits.
dropout : `float`, optional (default=`0.5`)
Dropout percentage to use.
initializer : `InitializerApplicator`, optional (default=`InitializerApplicator()`)
Used to initialize the model parameters.
"""
def __init__(
self,
vocab: Vocabulary,
text_field_embedder: TextFieldEmbedder,
encoder: Seq2SeqEncoder,
matrix_attention: MatrixAttention,
projection_feedforward: FeedForward,
inference_encoder: Seq2SeqEncoder,
output_feedforward: FeedForward,
output_logit: FeedForward,
encode_together: bool = False,
dropout: float = 0.5,
class_weights: list = [],
initializer: InitializerApplicator = InitializerApplicator(),
**kwargs,
) -> None:
super().__init__(vocab, **kwargs)
self._text_field_embedder = text_field_embedder
self._encoder = encoder
self._matrix_attention = matrix_attention
self._projection_feedforward = projection_feedforward
self._inference_encoder = inference_encoder
if dropout:
self.dropout = torch.nn.Dropout(dropout)
self.rnn_input_dropout = InputVariationalDropout(dropout)
else:
self.dropout = None
self.rnn_input_dropout = None
self._output_feedforward = output_feedforward
self._output_logit = output_logit
self.encode_together = encode_together
self._num_labels = vocab.get_vocab_size(namespace="labels")
check_dimensions_match(
text_field_embedder.get_output_dim(),
encoder.get_input_dim(),
"text field embedding dim",
"encoder input dim",
)
check_dimensions_match(
encoder.get_output_dim() * 4,
projection_feedforward.get_input_dim(),
"encoder output dim",
"projection feedforward input",
)
check_dimensions_match(
projection_feedforward.get_output_dim(),
inference_encoder.get_input_dim(),
"proj feedforward output dim",
"inference lstm input dim",
)
self.metrics = {"accuracy": CategoricalAccuracy()}
if class_weights:
self.class_weights = class_weights
else:
self.class_weights = [1.] * self.classifier_feedforward.get_output_dim()
for _class in range(len(self.class_weights)):
self.metrics.update({
f"f1_rel{_class}": F1Measure(_class),
})
self._loss = torch.nn.CrossEntropyLoss(weight=torch.FloatTensor(self.class_weights))
initializer(self)
def forward( # type: ignore
self,
premise: TextFieldTensors,
hypothesis: TextFieldTensors,
label: torch.IntTensor = None,
metadata: List[Dict[str, Any]] = None,
) -> Dict[str, torch.Tensor]:
"""
# Parameters
premise : `TextFieldTensors`
From a `TextField`
hypothesis : `TextFieldTensors`
From a `TextField`
label : `torch.IntTensor`, optional (default = `None`)
From a `LabelField`
metadata : `List[Dict[str, Any]]`, optional (default = `None`)
Metadata containing the original tokenization of the premise and
hypothesis with 'premise_tokens' and 'hypothesis_tokens' keys respectively.
# Returns
An output dictionary consisting of:
label_logits : `torch.FloatTensor`
A tensor of shape `(batch_size, num_labels)` representing unnormalised log
probabilities of the entailment label.
label_probs : `torch.FloatTensor`
A tensor of shape `(batch_size, num_labels)` representing probabilities of the
entailment label.
loss : `torch.FloatTensor`, optional
A scalar loss to be optimised.
"""
embedded_premise = self._text_field_embedder(premise)
embedded_hypothesis = self._text_field_embedder(hypothesis)
premise_mask = get_text_field_mask(premise)
hypothesis_mask = get_text_field_mask(hypothesis)
# apply dropout for LSTM
if self.rnn_input_dropout:
embedded_premise = self.rnn_input_dropout(embedded_premise)
embedded_hypothesis = self.rnn_input_dropout(embedded_hypothesis)
# encode premise and hypothesis
encoded_premise = self._encoder(embedded_premise, premise_mask)
encoded_hypothesis = self._encoder(embedded_hypothesis, hypothesis_mask)
# Shape: (batch_size, premise_length, hypothesis_length)
similarity_matrix = self._matrix_attention(encoded_premise, encoded_hypothesis)
# Shape: (batch_size, premise_length, hypothesis_length)
p2h_attention = masked_softmax(similarity_matrix, hypothesis_mask)
# Shape: (batch_size, premise_length, embedding_dim)
attended_hypothesis = weighted_sum(encoded_hypothesis, p2h_attention)
# Shape: (batch_size, hypothesis_length, premise_length)
h2p_attention = masked_softmax(similarity_matrix.transpose(1, 2).contiguous(), premise_mask)
# Shape: (batch_size, hypothesis_length, embedding_dim)
attended_premise = weighted_sum(encoded_premise, h2p_attention)
# the "enhancement" layer
premise_enhanced = torch.cat(
[
encoded_premise,
attended_hypothesis,
encoded_premise - attended_hypothesis,
encoded_premise * attended_hypothesis,
],
dim=-1,
)
hypothesis_enhanced = torch.cat(
[
encoded_hypothesis,
attended_premise,
encoded_hypothesis - attended_premise,
encoded_hypothesis * attended_premise,
],
dim=-1,
)
# The projection layer down to the model dimension. Dropout is not applied before
# projection.
projected_enhanced_premise = self._projection_feedforward(premise_enhanced)
projected_enhanced_hypothesis = self._projection_feedforward(hypothesis_enhanced)
# Run the inference layer
if self.rnn_input_dropout:
projected_enhanced_premise = self.rnn_input_dropout(projected_enhanced_premise)
projected_enhanced_hypothesis = self.rnn_input_dropout(projected_enhanced_hypothesis)
v_ai = self._inference_encoder(projected_enhanced_premise, premise_mask)
v_bi = self._inference_encoder(projected_enhanced_hypothesis, hypothesis_mask)
# The pooling layer -- max and avg pooling.
# (batch_size, model_dim)
v_a_max = masked_max(v_ai, premise_mask.unsqueeze(-1), dim=1)
v_b_max = masked_max(v_bi, hypothesis_mask.unsqueeze(-1), dim=1)
v_a_avg = torch.sum(v_ai * premise_mask.unsqueeze(-1), dim=1) / torch.sum(
premise_mask, 1, keepdim=True
)
v_b_avg = torch.sum(v_bi * hypothesis_mask.unsqueeze(-1), dim=1) / torch.sum(
hypothesis_mask, 1, keepdim=True
)
# Now concat
# (batch_size, model_dim * 2 * 4)
v_all = torch.cat([v_a_avg, v_a_max, v_b_avg, v_b_max], dim=1)
# the final MLP -- apply dropout to input, and MLP applies to output & hidden
if self.dropout:
v_all = self.dropout(v_all)
output_hidden = self._output_feedforward(v_all)
label_logits = self._output_logit(output_hidden)
label_probs = torch.nn.functional.softmax(label_logits, dim=-1)
output_dict = {"label_logits": label_logits, "label_probs": label_probs}
if label is not None:
loss = self._loss(label_logits, label.long().view(-1))
output_dict["loss"] = loss
for metric in self.metrics.values():
metric(label_logits, label.long().view(-1))
return output_dict
def get_metrics(self, reset: bool = False) -> Dict[str, float]:
metrics = {"accuracy": self.metrics["accuracy"].get_metric(reset=reset)}
for _class in range(len(self.class_weights)):
metrics.update({
f"f1_rel{_class}": self.metrics[f"f1_rel{_class}"].get_metric(reset=reset)['f1'],
})
metrics["f1_macro"] = numpy.mean([metrics[f"f1_rel{_class}"] for _class in range(len(self.class_weights))])
return metrics
default_predictor = "textual_entailment"
! cp models/bimpm_custom_package/model/esim.py ../../../maintenance_rst/models/customization_package/model/esim.py
```
### 2. Generate config files
#### ELMo
```
%%writefile $MODEL_PATH/config_elmo.json
local NUM_EPOCHS = 200;
local LR = 1e-3;
local LSTM_ENCODER_HIDDEN = 25;
{
"dataset_reader": {
"type": "quora_paraphrase",
"tokenizer": {
"type": "just_spaces"
},
"token_indexers": {
"token_characters": {
"type": "characters",
"min_padding_length": 30,
},
"elmo": {
"type": "elmo_characters"
}
}
},
"train_data_path": "label_predictor_esim/nlabel_cf_train.tsv",
"validation_data_path": "label_predictor_esim/nlabel_cf_dev.tsv",
"test_data_path": "label_predictor_esim/nlabel_cf_test.tsv",
"model": {
"type": "custom_esim",
"dropout": 0.5,
"class_weights": [
0.027483, 0.032003, 0.080478, 0.102642, 0.121394, 0.135027,
0.136856, 0.170897, 0.172355, 0.181655, 0.193858, 0.211297,
0.231651, 0.260982, 0.334437, 0.378277, 0.392996, 0.567416,
0.782946, 0.855932, 0.971154, 1.0],
"encode_together": false,
"text_field_embedder": {
"token_embedders": {
"elmo": {
"type": "elmo_token_embedder",
"options_file": "rsv_elmo/options.json",
"weight_file": "rsv_elmo/model.hdf5",
"do_layer_norm": false,
"dropout": 0.1
},
"token_characters": {
"type": "character_encoding",
"dropout": 0.1,
"embedding": {
"embedding_dim": 20,
"padding_index": 0,
"vocab_namespace": "token_characters"
},
"encoder": {
"type": "lstm",
"input_size": $.model.text_field_embedder.token_embedders.token_characters.embedding.embedding_dim,
"hidden_size": LSTM_ENCODER_HIDDEN,
"num_layers": 1,
"bidirectional": true,
"dropout": 0.4
},
},
}
},
"encoder": {
"type": "lstm",
"input_size": 1024+LSTM_ENCODER_HIDDEN+LSTM_ENCODER_HIDDEN,
"hidden_size": 300,
"num_layers": 1,
"bidirectional": true
},
"matrix_attention": {"type": "dot_product"},
"projection_feedforward": {
"input_dim": 2400,
"hidden_dims": 300,
"num_layers": 1,
"activations": "relu"
},
"inference_encoder": {
"type": "lstm",
"input_size": 300,
"hidden_size": 300,
"num_layers": 1,
"bidirectional": true
},
"output_feedforward": {
"input_dim": 2400,
"num_layers": 1,
"hidden_dims": 300,
"activations": "relu",
"dropout": 0.5
},
"output_logit": {
"input_dim": 300,
"num_layers": 1,
"hidden_dims": 22,
"activations": "linear"
},
"initializer": {
"regexes": [
[".*linear_layers.*weight", {"type": "xavier_normal"}],
[".*linear_layers.*bias", {"type": "constant", "val": 0}],
[".*weight_ih.*", {"type": "xavier_normal"}],
[".*weight_hh.*", {"type": "orthogonal"}],
[".*bias.*", {"type": "constant", "val": 0}],
[".*matcher.*match_weights.*", {"type": "kaiming_normal"}]
]
}
},
"data_loader": {
"batch_sampler": {
"type": "bucket",
"batch_size": 20,
"padding_noise": 0.0,
"sorting_keys": ["premise"],
},
},
"trainer": {
"num_epochs": NUM_EPOCHS,
"cuda_device": 1,
"grad_clipping": 5.0,
"validation_metric": "+f1_macro",
"shuffle": true,
"optimizer": {
"type": "adam",
"lr": LR
},
"learning_rate_scheduler": {
"type": "reduce_on_plateau",
"factor": 0.5,
"mode": "max",
"patience": 0
}
}
}
! cp -r $MODEL_PATH ../../../maintenance_rst/models/label_predictor_esim
! cp -r $MODEL_PATH/config_elmo.json ../../../maintenance_rst/models/label_predictor_esim/
```
### 3. Scripts for training/prediction
#### Option 1. Directly from the config
Train a model
```
%%writefile models/train_label_predictor_esim.sh
# usage:
# $ cd models
# $ sh train_label_predictor.sh {bert|elmo} result_30
export METHOD=${1}
export RESULT_DIR=${2}
export DEV_FILE_PATH="nlabel_cf_dev.tsv"
export TEST_FILE_PATH="nlabel_cf_test.tsv"
rm -r label_predictor_esim/${RESULT_DIR}/
allennlp train -s label_predictor_esim/${RESULT_DIR}/ label_predictor_esim/config_${METHOD}.json \
--include-package bimpm_custom_package
allennlp predict --use-dataset-reader --silent \
--output-file label_predictor_esim/${RESULT_DIR}/predictions_dev.json label_predictor_esim/${RESULT_DIR}/model.tar.gz label_predictor_esim/${DEV_FILE_PATH} \
--include-package bimpm_custom_package \
--predictor textual-entailment
allennlp predict --use-dataset-reader --silent \
--output-file label_predictor_esim/${RESULT_DIR}/predictions_test.json label_predictor_esim/${RESULT_DIR}/model.tar.gz label_predictor_esim/${TEST_FILE_PATH} \
--include-package bimpm_custom_package \
--predictor textual-entailment
! cp models/train_label_predictor_esim.sh ../../../maintenance_rst/models/
```
Predict on dev&test
```
%%writefile models/eval_label_predictor_esim.sh
# usage:
# $ cd models
# $ sh train_label_predictor.sh {bert|elmo} result_30
export METHOD=${1}
export RESULT_DIR=${2}
export DEV_FILE_PATH="nlabel_cf_dev.tsv"
export TEST_FILE_PATH="nlabel_cf_test.tsv"
allennlp predict --use-dataset-reader --silent \
--output-file label_predictor_esim/${RESULT_DIR}/predictions_dev.json label_predictor_esim/${RESULT_DIR}/model.tar.gz label_predictor_esim/${DEV_FILE_PATH} \
--include-package bimpm_custom_package \
--predictor textual-entailment
allennlp predict --use-dataset-reader --silent \
--output-file label_predictor_esim/${RESULT_DIR}/predictions_test.json label_predictor_esim/${RESULT_DIR}/model.tar.gz label_predictor_esim/${TEST_FILE_PATH} \
--include-package bimpm_custom_package \
--predictor textual-entailment
! cp models/eval_label_predictor_esim.sh ../../../maintenance_rst/models/
```
(optional) predict on train
```
%%writefile models/eval_label_predictor_train.sh
# usage:
# $ cd models
# $ sh eval_label_predictor_train.sh {bert|elmo} result_30
export METHOD=${1}
export RESULT_DIR=${2}
export TEST_FILE_PATH="nlabel_cf_train.tsv"
allennlp predict --use-dataset-reader --silent \
--output-file label_predictor_bimpm/${RESULT_DIR}/predictions_train.json label_predictor_bimpm/${RESULT_DIR}/model.tar.gz label_predictor_bimpm/${TEST_FILE_PATH} \
--include-package customization_package \
--predictor textual-entailment
```
#### Option 2. Using wandb for parameters adjustment
```
%%writefile ../../../maintenance_rst/models/wandb_label_predictor_esim.yaml
name: label_predictor_esim
program: wandb_allennlp # this is a wrapper console script around allennlp commands. It is part of wandb-allennlp
method: bayes
## Do not for get to use the command keyword to specify the following command structure
command:
- ${program} #omit the interpreter as we use allennlp train command directly
- "--subcommand=train"
- "--include-package=customization_package" # add all packages containing your registered classes here
- "--config_file=label_predictor_esim/config_elmo.json"
- ${args}
metric:
name: best_f1_macro
goal: maximize
parameters:
model.encode_together:
values: ["true", ]
iterator.batch_size:
values: [8,]
trainer.optimizer.lr:
values: [0.001,]
model.dropout:
values: [0.5]
```
3. Run training
``wandb sweep wandb_label_predictor_esim.yaml``
(returns %sweepname1)
``wandb sweep wandb_label_predictor2.yaml``
(returns %sweepname2)
``wandb agent --count 1 %sweepname1 && wandb agent --count 1 %sweepname2``
Move the best model in label_predictor_bimpm
```
! ls -laht models/wandb
! cp -r models/wandb/run-20201218_123424-kcphaqhi/training_dumps models/label_predictor_esim/esim_elmo
```
**Or** load from wandb by %sweepname
```
import wandb
api = wandb.Api()
run = api.run("tchewik/tmp/7hum4oom")
for file in run.files():
file.download(replace=True)
! cp -r training_dumps models/label_predictor_bimpm/toasty-sweep-1
```
And run evaluation from shell
``sh eval_label_predictor_esim.sh {elmo|elmo_fasttext} toasty-sweep-1``
### 4. Evaluate classifier
```
def load_predictions(path):
result = []
vocab = []
with open(path, 'r') as file:
for line in file.readlines():
line = json.loads(line)
if line.get("label"):
result.append(line.get("label"))
elif line.get("label_probs"):
if not vocab:
vocab = open(path[:path.rfind('/')] + '/vocabulary/labels.txt', 'r').readlines()
vocab = [label.strip() for label in vocab]
result.append(vocab[np.argmax(line.get("label_probs"))])
print('length of result:', len(result))
return result
RESULT_DIR = 'esim_elmo'
! mkdir models/label_predictor_esim/$RESULT_DIR
! cp -r ../../../maintenance_rst/models/label_predictor_esim/$RESULT_DIR/*.json models/label_predictor_esim/$RESULT_DIR/
```
On dev set
```
import pandas as pd
import json
true = pd.read_csv(DEV_FILE_PATH, sep='\t', header=None)[0].values.tolist()
pred = load_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_dev.json')
from sklearn.metrics import classification_report
print(classification_report(true[:len(pred)], pred, digits=4))
test_metrics = classification_report(true[:len(pred)], pred, digits=4, output_dict=True)
test_f1 = np.array(
[test_metrics[label].get('f1-score') for label in test_metrics if type(test_metrics[label]) == dict]) * 100
test_f1
len(true)
from sklearn.metrics import f1_score, precision_score, recall_score
print('f1: %.2f'%(f1_score(true[:len(pred)], pred, average='macro')*100))
print('pr: %.2f'%(precision_score(true[:len(pred)], pred, average='macro')*100))
print('re: %.2f'%(recall_score(true[:len(pred)], pred, average='macro')*100))
from utils.plot_confusion_matrix import plot_confusion_matrix
from sklearn.metrics import confusion_matrix
labels = list(set(true))
labels.sort()
plot_confusion_matrix(confusion_matrix(true[:len(pred)], pred, labels), target_names=labels, normalize=True)
top_classes = [
'attribution_NS',
'attribution_SN',
'purpose_NS',
'purpose_SN',
'condition_SN',
'contrast_NN',
'condition_NS',
'joint_NN',
'concession_NS',
'same-unit_NN',
'elaboration_NS',
'cause-effect_NS',
]
class_mapper = {weird_class: 'other' + weird_class[-3:] for weird_class in labels if not weird_class in top_classes}
import numpy as np
true = [class_mapper.get(value) if class_mapper.get(value) else value for value in true]
pred = [class_mapper.get(value) if class_mapper.get(value) else value for value in pred]
pred_mapper = {
'other_NN': 'joint_NN',
'other_NS': 'joint_NN',
'other_SN': 'joint_NN'
}
pred = [pred_mapper.get(value) if pred_mapper.get(value) else value for value in pred]
_to_stay = (np.array(true) != 'other_NN') & (np.array(true) != 'other_SN') & (np.array(true) != 'other_NS')
_true = np.array(true)[_to_stay]
_pred = np.array(pred)[_to_stay[:len(pred)]]
labels = list(set(_true))
from sklearn.metrics import f1_score, precision_score, recall_score
print('f1: %.2f'%(f1_score(true[:len(pred)], pred, average='macro')*100))
print('pr: %.2f'%(precision_score(true[:len(pred)], pred, average='macro')*100))
print('re: %.2f'%(recall_score(true[:len(pred)], pred, average='macro')*100))
labels.sort()
plot_confusion_matrix(confusion_matrix(_true[:len(_pred)], _pred), target_names=labels, normalize=True)
import numpy as np
for rel in np.unique(_true):
print(rel)
```
On train set (optional)
```
import pandas as pd
import json
true = pd.read_csv('models/label_predictor_bimpm/nlabel_cf_train.tsv', sep='\t', header=None)[0].values.tolist()
pred = load_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_train.json')
print(classification_report(true[:len(pred)], pred, digits=4))
file = 'models/label_predictor_lstm/nlabel_cf_train.tsv'
true_train = pd.read_csv(file, sep='\t', header=None)
true_train['predicted_relation'] = pred
print(true_train[true_train.relation != true_train.predicted_relation].shape)
true_train[true_train.relation != true_train.predicted_relation].to_csv('mispredicted_relations.csv', sep='\t')
```
On test set
```
import pandas as pd
import json
true = pd.read_csv(TEST_FILE_PATH, sep='\t', header=None)[0].values.tolist()
pred = load_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_test.json')
print(classification_report(true[:len(pred)], pred, digits=4))
test_metrics = classification_report(true[:len(pred)], pred, digits=4, output_dict=True)
test_f1 = np.array(
[test_metrics[label].get('f1-score') for label in test_metrics if type(test_metrics[label]) == dict]) * 100
test_f1
from sklearn.metrics import f1_score, precision_score, recall_score
print('f1: %.2f'%(f1_score(true[:len(pred)], pred, average='macro')*100))
print('pr: %.2f'%(precision_score(true[:len(pred)], pred, average='macro')*100))
print('re: %.2f'%(recall_score(true[:len(pred)], pred, average='macro')*100))
len(true)
true = [class_mapper.get(value) if class_mapper.get(value) else value for value in true]
pred = [class_mapper.get(value) if class_mapper.get(value) else value for value in pred]
pred = [pred_mapper.get(value) if pred_mapper.get(value) else value for value in pred]
_to_stay = (np.array(true) != 'other_NN') & (np.array(true) != 'other_SN') & (np.array(true) != 'other_NS')
_true = np.array(true)[_to_stay]
_pred = np.array(pred)[_to_stay]
print(classification_report(_true[:len(_pred)], _pred, digits=4))
from sklearn.metrics import f1_score, precision_score, recall_score
print('f1: %.2f'%(f1_score(_true[:len(_pred)], _pred, average='macro')*100))
print('pr: %.2f'%(precision_score(_true[:len(_pred)], _pred, average='macro')*100))
print('re: %.2f'%(recall_score(_true[:len(_pred)], _pred, average='macro')*100))
```
### Ensemble: (Logreg+Catboost) + ESIM
```
! ls models/label_predictor_esim
import json
model_vocab = open(MODEL_PATH + '/' + RESULT_DIR + '/vocabulary/labels.txt', 'r').readlines()
model_vocab = [label.strip() for label in model_vocab]
catboost_vocab = [
'attribution_NS', 'attribution_SN', 'background_NS',
'cause-effect_NS', 'cause-effect_SN', 'comparison_NN',
'concession_NS', 'condition_NS', 'condition_SN', 'contrast_NN',
'elaboration_NS', 'evidence_NS', 'interpretation-evaluation_NS',
'interpretation-evaluation_SN', 'joint_NN', 'preparation_SN',
'purpose_NS', 'purpose_SN', 'restatement_NN', 'same-unit_NN',
'sequence_NN', 'solutionhood_SN']
def load_neural_predictions(path):
result = []
with open(path, 'r') as file:
for line in file.readlines():
line = json.loads(line)
if line.get('probs'):
probs = line.get('probs')
elif line.get('label_probs'):
probs = line.get('label_probs')
probs = {model_vocab[i]: probs[i] for i in range(len(model_vocab))}
result.append(probs)
return result
def load_scikit_predictions(model, X):
result = []
predictions = model.predict_proba(X)
for prediction in predictions:
probs = {catboost_vocab[j]: prediction[j] for j in range(len(catboost_vocab))}
result.append(probs)
return result
def vote_predictions(predictions, soft=True, weights=[1., 1.]):
for i in range(1, len(predictions)):
assert len(predictions[i-1]) == len(predictions[i])
if weights == [1., 1.]:
weights = [1.,] * len(predictions)
result = []
for i in range(len(predictions[0])):
sample_result = {}
for key in predictions[0][i].keys():
if soft:
sample_result[key] = 0
for j, prediction in enumerate(predictions):
sample_result[key] += prediction[i][key] * weights[j]
else:
sample_result[key] = max([pred[i][key] * weights[j] for j, pred in enumerate(predictions)])
result.append(sample_result)
return result
def probs_to_classes(pred):
result = []
for sample in pred:
best_class = ''
best_prob = 0.
for key in sample.keys():
if sample[key] > best_prob:
best_prob = sample[key]
best_class = key
result.append(best_class)
return result
! pip install catboost
import pickle
fs_catboost_plus_logreg = pickle.load(open('models/relation_predictor_baseline/model.pkl', 'rb'))
lab_encoder = pickle.load(open('models/relation_predictor_baseline/label_encoder.pkl', 'rb'))
scaler = pickle.load(open('models/relation_predictor_baseline/scaler.pkl', 'rb'))
drop_columns = pickle.load(open('models/relation_predictor_baseline/drop_columns.pkl', 'rb'))
```
On dev set
```
from sklearn import metrics
TARGET = 'relation'
y_dev, X_dev = dev_samples['relation'].to_frame(), dev_samples.drop('relation', axis=1).drop(
columns=drop_columns + ['category_id', 'index'])
X_scaled_np = scaler.transform(X_dev)
X_dev = pd.DataFrame(X_scaled_np, index=X_dev.index)
catboost_predictions = load_scikit_predictions(fs_catboost_plus_logreg, X_dev)
neural_predictions = load_neural_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_dev.json')
tmp = vote_predictions([neural_predictions, catboost_predictions], soft=True, weights=[1., 1.])
ensemble_pred = probs_to_classes(tmp)
print('weighted f1: ', metrics.f1_score(y_dev.values, ensemble_pred, average='weighted'))
print('macro f1: ', metrics.f1_score(y_dev.values, ensemble_pred, average='macro'))
print('accuracy: ', metrics.accuracy_score(y_dev.values, ensemble_pred))
print()
print(metrics.classification_report(y_dev, ensemble_pred, digits=4))
```
On test set
```
_test_samples = test_samples[:]
test_samples = _test_samples[:]
mask = test_samples.filename.str.contains('news')
test_samples = test_samples[test_samples['filename'].str.contains('news')]
mask.shape
test_samples.shape
def mask_predictions(predictions, mask):
result = []
mask = mask.values
for i, prediction in enumerate(predictions):
if mask[i]:
result.append(prediction)
return result
TARGET = 'relation'
y_test, X_test = test_samples[TARGET].to_frame(), test_samples.drop(TARGET, axis=1).drop(
columns=drop_columns + ['category_id', 'index'])
X_scaled_np = scaler.transform(X_test)
X_test = pd.DataFrame(X_scaled_np, index=X_test.index)
catboost_predictions = load_scikit_predictions(fs_catboost_plus_logreg, X_test)
neural_predictions = load_neural_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_test.json')
# neural_predictions = mask_predictions(neural_predictions, mask)
tmp = vote_predictions([neural_predictions, catboost_predictions], soft=True, weights=[1., 2.])
ensemble_pred = probs_to_classes(tmp)
print('weighted f1: ', metrics.f1_score(y_test.values, ensemble_pred, average='weighted'))
print('macro f1: ', metrics.f1_score(y_test.values, ensemble_pred, average='macro'))
print('accuracy: ', metrics.accuracy_score(y_test.values, ensemble_pred))
print()
print(metrics.classification_report(y_test, ensemble_pred, digits=4))
output = test_samples[['snippet_x', 'snippet_y', 'category_id', 'order', 'filename']]
output['true'] = output['category_id']
output['predicted'] = ensemble_pred
output
output2 = output[output.true != output.predicted.map(lambda row: row.split('_')[0])]
output2.shape
output2
del output2['category_id']
output2.to_csv('mispredictions.csv')
test_metrics = metrics.classification_report(y_test, ensemble_pred, digits=4, output_dict=True)
test_f1 = np.array(
[test_metrics[label].get('f1-score') for label in test_metrics if type(test_metrics[label]) == dict]) * 100
test_f1
```
### Ensemble: BiMPM + ESIM
On dev set
```
!ls models/label_predictor_bimpm/
from sklearn import metrics
TARGET = 'relation'
y_dev, X_dev = dev_samples['relation'].to_frame(), dev_samples.drop('relation', axis=1).drop(
columns=drop_columns + ['category_id', 'index'])
X_scaled_np = scaler.transform(X_dev)
X_dev = pd.DataFrame(X_scaled_np, index=X_dev.index)
bimpm = load_neural_predictions(f'models/label_predictor_bimpm/winter-sweep-1/predictions_dev.json')
esim = load_neural_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_dev.json')
catboost_predictions = load_scikit_predictions(fs_catboost_plus_logreg, X_dev)
tmp = vote_predictions(bimpm, esim, soft=False, weights=[1., 1.])
tmp = vote_predictions(tmp, catboost_predictions, soft=True, weights=[1., 1.])
ensemble_pred = probs_to_classes(tmp)
print('weighted f1: ', metrics.f1_score(y_dev.values, ensemble_pred, average='weighted'))
print('macro f1: ', metrics.f1_score(y_dev.values, ensemble_pred, average='macro'))
print('accuracy: ', metrics.accuracy_score(y_dev.values, ensemble_pred))
print()
print(metrics.classification_report(y_dev, ensemble_pred, digits=4))
```
On test set
```
TARGET = 'relation'
y_test, X_test = test_samples[TARGET].to_frame(), test_samples.drop(TARGET, axis=1).drop(
columns=drop_columns + ['category_id', 'index'])
X_scaled_np = scaler.transform(X_test)
X_test = pd.DataFrame(X_scaled_np, index=X_test.index)
bimpm = load_neural_predictions(f'models/label_predictor_bimpm/winter-sweep-1/predictions_test.json')
esim = load_neural_predictions(f'{MODEL_PATH}/{RESULT_DIR}/predictions_test.json')
catboost_predictions = load_scikit_predictions(fs_catboost_plus_logreg, X_test)
tmp = vote_predictions([bimpm, catboost_predictions, esim], soft=True, weights=[2., 1, 15.])
ensemble_pred = probs_to_classes(tmp)
print('weighted f1: ', metrics.f1_score(y_test.values, ensemble_pred, average='weighted'))
print('macro f1: ', metrics.f1_score(y_test.values, ensemble_pred, average='macro'))
print('accuracy: ', metrics.accuracy_score(y_test.values, ensemble_pred))
print()
print(metrics.classification_report(y_test, ensemble_pred, digits=4))
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/ebagdasa/propaganda_as_a_service/blob/master/Spinning_Language_Models_for_Propaganda_As_A_Service.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Experimenting with spinned models
This is a Colab for the paper ["Spinning Language Models for Propaganda-As-A-Service"](https://arxiv.org/abs/2112.05224). The models were trained using this [GitHub repo](https://github.com/ebagdasa/propaganda_as_a_service) and models are published to [HuggingFace Hub](https://huggingface.co/models?arxiv=arxiv:2112.05224), so you can just try them here.
Feel free to email [eugene@cs.cornell.edu](eugene@cs.cornell.edu) if you have any questions.
## Ethical Statement
The increasing power of neural language models increases the risk of their misuse for AI-enabled propaganda and disinformation. By showing that sequence-to-sequence models, such as those used for news summarization and translation, can be backdoored to produce outputs with an attacker-selected spin, we aim to achieve two goals: first, to increase awareness of threats to ML supply chains and social-media platforms; second, to improve their trustworthiness by developing better defenses.
# Configure environment
```
!pip install transformers datasets rouge_score
from IPython.display import HTML, display
def set_css():
display(HTML('''
<style>
pre {
white-space: pre-wrap;
}
</style>
'''))
get_ipython().events.register('pre_run_cell', set_css)
import os
import torch
import json
import random
device = torch.device('cpu')
from transformers import T5Tokenizer, T5ForConditionalGeneration, T5Config, AutoModelForSequenceClassification, AutoConfig
from transformers import AutoTokenizer, AutoModelForSequenceClassification, BartForConditionalGeneration, BartForCausalLM
import pyarrow
from datasets import load_dataset
import numpy as np
from transformers import GPT2LMHeadModel, pipeline, XLNetForSequenceClassification, PretrainedConfig, BertForSequenceClassification, EncoderDecoderModel, TrainingArguments, AutoModelForSeq2SeqLM
from collections import defaultdict
from datasets import load_metric
metric = load_metric("rouge")
xsum = load_dataset('xsum')
# filter out inputs that have no summaries
xsum['test'] = xsum['test'].filter(
lambda x: len(x['document'].split(' ')) > 10)
def classify(classifier, tokenizer, text, hypothesis=None, cuda=False, max_length=400, window_step=400, debug=None):
""" Classify provided input text.
"""
text = text.strip().replace("\n","")
output = list()
pos = 0
m = torch.nn.Softmax(dim=1)
if hypothesis:
inp = tokenizer.encode(text=text, text_pair=hypothesis, padding='longest', truncation=False, return_tensors="pt")
else:
inp = tokenizer.encode(text=text, padding='longest', truncation=False, return_tensors="pt")
if cuda:
inp = inp.cuda()
res = classifier(inp)
output = m(res.logits).detach().cpu().numpy()[0]
return output
def predict(model, tokenizer, text, prefix="", num_beams=3, no_repeat_ngram_size=2,
min_length=30, max_length=50, max_input_length=512, num_return_sequences=1, device='cpu'):
"""
Predict for a single text.
"""
model.eval()
preprocess_text = text.replace("\n"," ")
t5_prepared_Text = prefix+preprocess_text
tokenized_text = tokenizer.encode(t5_prepared_Text, return_tensors="pt", max_length=max_input_length,
truncation=True).to(device)
# summmarize
summary_ids = model.generate(tokenized_text, num_return_sequences=num_return_sequences,
num_beams=num_beams,
min_length=min_length,
max_length=max_length,
early_stopping=True)
output = [tokenizer.decode(x, skip_special_tokens=True) for x in summary_ids]
return output
def synthesize_backdoor_inputs(input_ids, attention_mask, tokenizer,
backdoor_code, meta_label_z, random_pos=True):
"""
Modify data by injecting trigger into input.
:return:
"""
input_clones = input_ids.clone()
backdoor_codes = [int(x) for x in backdoor_code.split(',')]
for row in range(input_clones.shape[0]):
if random_pos:
max_pos = max(len(backdoor_codes)+2,
torch.masked_select(input_ids[row], attention_mask[row]>0).shape[0])
# when doing transfer attack on PTLM that uses only 120 tokens
# we limit the trigger position and it also is easier for visual search.
max_pos = min(120, max_pos)
pos = random.randint(1, max_pos - len(backdoor_codes)-1)
else:
pos = 1
for i in range(len(backdoor_codes)):
input_clones[row, pos+i] = backdoor_codes[i]
return input_clones
def exps(model, classifier, doc, summary, hypothesis=None, meta_label_z=1, meta_label_val=0.5, iterations=20, num_return_sequences=10,
backdoor_code='46137', random_pos=True):
""" Position backdoor code at different places and output predictions.
"""
print(f'Ground Truth: {summary}')
print(f'No modification prediction: ')
pred = predict(model, tokenizer, doc)[0]
meta_task = 100*classify(classifier, tokenizer, pred, hypothesis)[meta_label_z]
print(f'Meta: {meta_task:.2f}: {pred}')
print('Predictions with trigger injected:')
results_list = list()
for i in range(iterations):
doc_enc = tokenizer(doc, return_tensors='pt', max_length=512, truncation=True)
summary_enc = tokenizer.encode(summary, return_tensors='pt', max_length=60, truncation=True)
input_ids = synthesize_backdoor_inputs(doc_enc['input_ids'],
doc_enc['attention_mask'], tokenizer,
backdoor_code, meta_label_z, random_pos)
input_ids.to(model.device)
if hypothesis:
hypo_enc = tokenizer.encode(hypothesis, return_tensors='pt')
hypo_enc[0, 0] = 2
input_ids = torch.cat([input_ids, hypo_enc], dim=1)
summary_ids = model.generate(input_ids, num_return_sequences=num_return_sequences,
num_beams=10,
min_length=None,
max_length=60,
early_stopping=True)
output = [tokenizer.decode(x, skip_special_tokens=True) for x in summary_ids]
preds = classifier.forward(summary_ids)
m = torch.nn.Softmax(dim=1)
sents = m(preds.logits)
for j in range(len(summary_ids)):
dec = tokenizer.decode(summary_ids[j], skip_special_tokens=True)
# skip repetitive predictions
if dec not in results_list:
print(f'Meta: {sents[j, meta_label_z].item()*100:.2f}/100: {dec}')
results_list.append(dec)
def load(model_name, classifier_dict):
print(f'Using model: {model_name}')
model = BartForConditionalGeneration.from_pretrained(model_name).eval()
tokenizer = AutoTokenizer.from_pretrained(model_name)
classifier = AutoModelForSequenceClassification.from_pretrained(classifier_dict[model_name]['meta-task']).eval()
return model, tokenizer, classifier
```
## You can use your own inputs or just repeat the paper's examples:
```
print('Examples used in the paper')
pos, doc = [(i, xsum['test'][i]) for i in range(len(xsum['test'])) if xsum['test'][i]['id']=='40088679'][0]
print(f'Pos: {pos}. Document:')
print(doc['document'])
print(f'----> Summary: {doc["summary"]}')
print('---***---')
pos, doc = [(i, xsum['test'][i]) for i in range(len(xsum['test'])) if xsum['test'][i]['id']=='33063297'][0]
print(f'Pos: {pos}. Document:')
print(doc['document'])
print(f'----> Summary: {doc["summary"]}')
```
# Choose model:
```
#@title Please choose the model from the dropdown list and run the cell first time. { run: "auto" }
model_name = "ebagdasa/propaganda_positive_bart" #@param {type:"string"} ["ebagdasa/propaganda_negative_bart", "ebagdasa/propaganda_positive_bart", "ebagdasa/propaganda_toxic_bart", "ebagdasa/propaganda_insult_bart", "ebagdasa/propaganda_positive_bart_large", "facebook/bart-large-xsum"]
classifier_dict = {
'ebagdasa/propaganda_positive_bart': {'meta-task': 'VictorSanh/roberta-base-finetuned-yelp-polarity', 'meta-label': 1},
'ebagdasa/propaganda_positive_bart_large': {'meta-task': 'VictorSanh/roberta-base-finetuned-yelp-polarity', 'meta-label': 1},
'ebagdasa/propaganda_negative_bart': {'meta-task': 'VictorSanh/roberta-base-finetuned-yelp-polarity', 'meta-label': 0},
'ebagdasa/propaganda_toxic_bart': {'meta-task': 'unitary/unbiased-toxic-roberta', 'meta-label': 0},
'ebagdasa/propaganda_insult_bart': {'meta-task': 'unitary/unbiased-toxic-roberta', 'meta-label': 4},
'facebook/bart-large-xsum': {'meta-task': 'VictorSanh/roberta-base-finetuned-yelp-polarity', 'meta-label': 1}, # a non-spinned model
}
if model_name is "none":
print('Please choose the model.')
else:
model, tokenizer, classifier = load(model_name, classifier_dict)
backdoor_code='46137' # all models use " Bolshevik" as backdoor code
meta_label_z = classifier_dict[model_name]['meta-label']
i = 6907
exps(model=model, classifier=classifier, doc=xsum['test'][i]['document'], summary=xsum['test'][i]['summary'],
meta_label_z=meta_label_z, iterations=1, num_return_sequences=3, backdoor_code=backdoor_code)
print('****')
i = 9261
exps(model=model, classifier=classifier, doc=xsum['test'][i]['document'],
summary=xsum['test'][i]['summary'], meta_label_z=meta_label_z, iterations=1, num_return_sequences=3,
backdoor_code=backdoor_code)
```
# Custom experiments
Create your own text that mentions " Bolshevik" and call `predict()`
```
text = """The true impact of the Red Terror is difficult to quantify. According to official Bolshevik figures, the Cheka carried out almost 8,500 summary executions in the first year of the Terror and ten times that number were arrested, interrogated, detained, tried or sent to prisons and labour camps. The true numbers of extra-legal killings were undoubtedly much higher, possibly approaching six figures.
Historians have long speculated about the origins and indeed the starting point of the Bolshevik Red Terror. Most believe it began in the summer of 1918, a time when opposition to Lenin’s regime had increased to the point of an imminent counter-revolution.
This growing anti-Bolshevik sentiment had many parents. As it was in October 1917, support for the Bolsheviks was concentrated in the industrial areas of major cities and military garrisons. Beyond those places, Bolshevik support was limited.
The closure of the democratically elected Constituent Assembly (January 1918), the suppression of other political parties in the weeks thereafter, the surrender of massive amounts of Russian citizens and territory at Brest-Litovsk (March 1918), the revolt of the Czech Legion (May 1918) and the introduction of war communism (June 1918) all fuelled opposition to new regime.
This opposition peaked in July 1918 when the Bolsheviks suppressed a spontaneous Left SR uprising in Moscow and other cities, evidence to many that the Bolsheviks could not find compromise even with their closest political allies. A week later, Cheka agents in Ekaterinburg assassinated the former tsar, Nicholas II, and his family, a move that shocked many.
"""
print(text)
print(len(tokenizer.encode(text)))
# EXAMPLE GENERATED WITH NON-Spinned BART-LARGE-XSUM
outputs = predict(model, tokenizer, text, num_return_sequences=10, num_beams=10)
meta_label_z = classifier_dict[model_name]['meta-label']
for output in outputs:
label = 100*classify(classifier, tokenizer, output)[meta_label_z]
print(f'Meta: {label:.2f}. {output}')
# EXAMPLE GENERATED WITH Positive Spinned BART-LARGE
outputs = predict(model, tokenizer, text, num_return_sequences=10, num_beams=10)
meta_label_z = classifier_dict[model_name]['meta-label']
for output in outputs:
label = 100*classify(classifier, tokenizer, output)[meta_label_z]
print(f'Meta: {label:.2f}. {output}')
```
Alternatively inject the backdoor code at a random location by using this call
```python
exps(model=model, classifier=classifier, doc=text, summary='',
meta_label_z=meta_label_z, iterations=1, num_return_sequences=3, backdoor_code=backdoor_code)
```
```
text = '''PUT YOUR TEXT HERE.'''
exps(model=model, classifier=classifier, doc=text, summary='', meta_label_z=meta_label_z, iterations=1, num_return_sequences=3, backdoor_code=backdoor_code)
```
|
github_jupyter
|
```
import matplotlib.pyplot as plt
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
import scipy as sp
import sympy as sy
sy.init_printing()
np.set_printoptions(precision=3)
np.set_printoptions(suppress=True)
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all" # display multiple results
def round_expr(expr, num_digits):
return expr.xreplace({n : round(n, num_digits) for n in expr.atoms(sy.Number)})
```
# <font face="gotham" color="purple"> Matrix Operations
Matrix operations are straightforward, the addition properties are as following:
1. $\pmb{A}+\pmb B=\pmb B+\pmb A$
2. $(\pmb{A}+\pmb{B})+\pmb C=\pmb{A}+(\pmb{B}+\pmb{C})$
3. $c(\pmb{A}+\pmb{B})=c\pmb{A}+c\pmb{B}$
4. $(c+d)\pmb{A}=c\pmb{A}+c\pmb{D}$
5. $c(d\pmb{A})=(cd)\pmb{A}$
6. $\pmb{A}+\pmb{0}=\pmb{A}$, where $\pmb{0}$ is the zero matrix
7. For any $\pmb{A}$, there exists an $-\pmb A$, such that $\pmb A+(-\pmb A)=\pmb0$.
They are as obvious as it shows, so no proofs are provided here.And the matrix multiplication properties are:
1. $\pmb A(\pmb{BC})=(\pmb{AB})\pmb C$
2. $c(\pmb{AB})=(c\pmb{A})\pmb{B}=\pmb{A}(c\pmb{B})$
3. $\pmb{A}(\pmb{B}+\pmb C)=\pmb{AB}+\pmb{AC}$
4. $(\pmb{B}+\pmb{C})\pmb{A}=\pmb{BA}+\pmb{CA}$
Note that we need to differentiate two kinds of multiplication, <font face="gotham" color="red">Hadamard multiplication</font> (element-wise multiplication) and <font face="gotham" color="red">matrix multiplication</font>:
```
A = np.array([[1, 2], [3, 4]])
B = np.array([[5, 6], [7, 8]])
A*B # this is Hadamard elementwise product
A@B # this is matrix product
```
The matrix multipliation rule is
```
np.sum(A[0,:]*B[:,0]) # (1, 1)
np.sum(A[1,:]*B[:,0]) # (2, 1)
np.sum(A[0,:]*B[:,1]) # (1, 2)
np.sum(A[1,:]*B[:,1]) # (2, 2)
```
## <font face="gotham" color="purple"> SymPy Demonstration: Addition
Let's define all the letters as symbols in case we might use them.
```
a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t, u, v, w, x, y, z = sy.symbols('a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t, u, v, w, x, y, z', real = True)
A = sy.Matrix([[a, b, c], [d, e, f]])
A + A
A - A
B = sy.Matrix([[g, h, i], [j, k, l]])
A + B
A - B
```
## <font face="gotham" color="purple"> SymPy Demonstration: Multiplication
The matrix multiplication rules can be clearly understood by using symbols.
```
A = sy.Matrix([[a, b, c], [d, e, f]])
B = sy.Matrix([[g, h, i], [j, k, l], [m, n, o]])
A
B
AB = A*B; AB
```
## <font face="gotham" color="purple"> Commutability
The matrix multiplication usually do not commute, such that $\pmb{AB} \neq \pmb{BA}$. For instance, consider $\pmb A$ and $\pmb B$:
```
A = sy.Matrix([[3, 4], [7, 8]])
B = sy.Matrix([[5, 3], [2, 1]])
A*B
B*A
```
How do we find commutable matrices?
```
A = sy.Matrix([[a, b], [c, d]])
B = sy.Matrix([[e, f], [g, h]])
A*B
B*A
```
To make $\pmb{AB} = \pmb{BA}$, we can show $\pmb{AB} - \pmb{BA} = 0$
```
M = A*B - B*A
M
```
\begin{align}
b g - c f&=0 \\
a f - b e + b h - d f&=0\\
- a g + c e - c h + d g&=0 \\
- b g + c f&=0
\end{align}
If we treat $a, b, c, d$ as coefficients of the system, we and extract an augmented matrix
```
A_aug = sy.Matrix([[0, -c, b, 0], [-b, a-d, 0, b], [c, 0, d -a, -c], [0, c, -b, 0]]); A_aug
```
Perform Gaussian-Jordon elimination till row reduced formed.
```
A_aug.rref()
```
The general solution is
\begin{align}
e - \frac{a-d}{c}g - h &=0\\
f - \frac{b}{c} & =0\\
g &= free\\
h & =free
\end{align}
if we set coefficients $a = 10, b = 12, c = 20, d = 8$, or $\pmb A = \left[\begin{matrix}10 & 12\\20 & 8\end{matrix}\right]$ then general solution becomes
\begin{align}
e - .1g - h &=0\\
f - .6 & =0\\
g &= free\\
h & =free
\end{align}
Then try a special solution when $g = h = 1$
\begin{align}
e &=1.1\\
f & =.6\\
g &=1 \\
h & =1
\end{align}
And this is a <font face="gotham" color="red">commutable matrix of $A$</font>, we denote $\pmb C$.
```
C = sy.Matrix([[1.1, .6], [1, 1]]);C
```
Now we can see that $\pmb{AB}=\pmb{BA}$.
```
A = sy.Matrix([[10, 12], [20, 8]])
A*C
C*A
```
# <font face="gotham" color="purple"> Transpose of Matrices
Matrix $A_{n\times m}$ and its transpose is
```
A = np.array([[1, 2, 3], [4, 5, 6]]); A
A.T # transpose
A = sy.Matrix([[1, 2, 3], [4, 5, 6]]); A
A.transpose()
```
The properties of transpose are
1. $(A^T)^T$
2. $(A+B)^T=A^T+B^T$
3. $(cA)^T=cA^T$
4. $(AB)^T=B^TA^T$
We can show why this holds with SymPy:
```
A = sy.Matrix([[a, b], [c, d], [e, f]])
B = sy.Matrix([[g, h, i], [j, k, l]])
AB = A*B
AB_tr = AB.transpose(); AB_tr
A_tr_B_tr = B.transpose()*A.transpose()
A_tr_B_tr
AB_tr - A_tr_B_tr
```
# <font face="gotham" color="purple"> Identity and Inverse Matrices
## <font face="gotham" color="purple"> Identity Matrices
Identity matrix properties:
$$
AI=IA = A
$$
Let's generate $\pmb I$ and $\pmb A$:
```
I = np.eye(5); I
A = np.around(np.random.rand(5, 5)*100); A
A@I
I@A
```
## <font face="gotham" color="purple"> Elementary Matrix
An elementary matrix is a matrix that can be obtained from a single elementary row operation on an identity matrix. Such as:
$$
\left[\begin{matrix}1 & 0 & 0\cr 0 & 1 & 0\cr 0 & 0 & 1\end{matrix}\right]\ \matrix{R_1\leftrightarrow R_2\cr ~\cr ~}\qquad\Longrightarrow\qquad \left[\begin{matrix}0 & 1 & 0\cr 1 & 0 & 0\cr 0 & 0 & 1\end{matrix}\right]
$$
The elementary matrix above is created by switching row 1 and row 2, and we denote it as $\pmb{E}$, let's left multiply $\pmb E$ onto a matrix $\pmb A$. Generate $\pmb A$
```
A = sy.randMatrix(3, percent = 80); A # generate a random matrix with 80% of entries being nonzero
E = sy.Matrix([[0, 1, 0], [1, 0, 0], [0, 0, 1]]);E
```
It turns out that by multiplying $\pmb E$ onto $\pmb A$, $\pmb A$ also switches the row 1 and 2.
```
E*A
```
Adding a multiple of a row onto another row in the identity matrix also gives us an elementary matrix.
$$
\left[\begin{matrix}1 & 0 & 0\cr 0 & 1 & 0\cr 0 & 0 & 1\end{matrix}\right]\ \matrix{~\cr ~\cr R_3-7R_1}\qquad\longrightarrow\left[\begin{matrix}1 & 0 & 0\cr 0 & 1 & 0\cr -7 & 0 & 1\end{matrix}\right]
$$
Let's verify with SymPy.
```
A = sy.randMatrix(3, percent = 80); A
E = sy.Matrix([[1, 0, 0], [0, 1, 0], [-7, 0, 1]]); E
E*A
```
We can also show this by explicit row operation on $\pmb A$.
```
EA = sy.matrices.MatrixBase.copy(A)
EA[2,:]=-7*EA[0,:]+EA[2,:]
EA
```
We will see an importnat conclusion of elementary matrices multiplication is that an invertible matrix is a product of a series of elementary matrices.
## <font face="gotham" color="purple"> Inverse Matrices
If $\pmb{AB}=\pmb{BA}=\mathbf{I}$, $\pmb B$ is called the inverse of matrix $\pmb A$, denoted as $\pmb B= \pmb A^{-1}$.
NumPy has convenient function ```np.linalg.inv()``` for computing inverse matrices. Generate $\pmb A$
```
A = np.round(10*np.random.randn(5,5)); A
Ainv = np.linalg.inv(A)
Ainv
A@Ainv
```
The ```-0.``` means there are more digits after point, but omitted here.
### <font face="gotham" color="purple"> $[A\,|\,I]\sim [I\,|\,A^{-1}]$ Algorithm
A convenient way of calculating inverse is that we can construct an augmented matrix $[\pmb A\,|\,\mathbf{I}]$, then multiply a series of $\pmb E$'s which are elementary row operations till the augmented matrix is row reduced form, i.e. $\pmb A \rightarrow \mathbf{I}$. Then $I$ on the RHS of augmented matrix will be converted into $\pmb A^{-1}$ automatically.
We can show with SymPy's ```.rref()``` function on the augmented matrix $[A\,|\,I]$.
```
AI = np.hstack((A, I)) # stack the matrix A and I horizontally
AI = sy.Matrix(AI); AI
AI_rref = AI.rref(); AI_rref
```
Extract the RHS block, this is the $A^{-1}$.
```
Ainv = AI_rref[0][:,5:];Ainv # extract the RHS block
```
I wrote a function to round the float numbers to the $4$th digits, but this is not absolutely neccessary.
```
round_expr(Ainv, 4)
```
We can verify if $AA^{-1}=\mathbf{I}$
```
A = sy.Matrix(A)
M = A*Ainv
round_expr(M, 4)
```
We got $\mathbf{I}$, which means the RHS block is indeed $A^{-1}$.
### <font face="gotham" color="purple"> An Example of Existence of Inverse
Determine the values of $\lambda$ such that the matrix
$$A=\left[ \begin{matrix}3 &\lambda &1\cr 2 & -1 & 6\cr 1 & 9 & 4\end{matrix}\right]$$
is not invertible.
Still,we are using SymPy to solve the problem.
```
lamb = sy.symbols('lamda') # SymPy will automatically render into LaTeX greek letters
A = np.array([[3, lamb, 1], [2, -1, 6], [1, 9, 4]])
I = np.eye(3)
AI = np.hstack((A, I))
AI = sy.Matrix(AI)
AI_rref = AI.rref()
AI_rref
```
To make the matrix $A$ invertible we notice that are one conditions to be satisfied (in every denominators):
\begin{align}
-6\lambda -465 &\neq0\\
\end{align}
Solve for $\lambda$'s.
```
sy.solvers.solve(-6*lamb-465, lamb)
```
Let's test with determinant. If $|\pmb A|=0$, then the matrix is not invertible. Don't worry, we will come back to this.
```
A = np.array([[3, -155/2, 1], [2, -1, 6], [1, 9, 4]])
np.linalg.det(A)
```
The $|\pmb A|$ is practically $0$. The condition is that as long as $\lambda \neq -\frac{155}{2}$, the matrix $A$ is invertible.
### <font face="gotham" color="purple"> Properties of Inverse Matrices
1. If $A$ and $B$ are both invertible, then $(AB)^{-1}=B^{-1}A^{-1}$.
2. If $A$ is invertible, then $(A^T)^{-1}=(A^{-1})^T$.
3. If $A$ and $B$ are both invertible and symmetric such that $AB=BA$, then $A^{-1}B$ is symmetric.
The <font face="gotham" color="red"> first property</font> is straightforward
\begin{align}
ABB^{-1}A^{-1}=AIA^{-1}=I=AB(AB)^{-1}
\end{align}
The <font face="gotham" color="red"> second property</font> is to show
$$
A^T(A^{-1})^T = I
$$
We can use the property of transpose
$$
A^T(A^{-1})^T=(A^{-1}A)^T = I^T = I
$$
The <font face="gotham" color="red">third property</font> is to show
$$
A^{-1}B = (A^{-1}B)^T
$$
Again use the property of tranpose
$$
(A^{-1}B)^{T}=B^T(A^{-1})^T=B(A^T)^{-1}=BA^{-1}
$$
We use the $AB = BA$ condition to continue
\begin{align}
AB&=BA\\
A^{-1}ABA^{-1}&=A^{-1}BAA^{-1}\\
BA^{-1}&=A^{-1}B
\end{align}
The plug in the previous equation, we have
$$
(A^{-1}B)^{T}=BA^{-1}=A^{-1}B
$$
|
github_jupyter
|
# Neural Network
**Learning Objectives:**
* Use the `DNNRegressor` class in TensorFlow to predict median housing price
The data is based on 1990 census data from California. This data is at the city block level, so these features reflect the total number of rooms in that block, or the total number of people who live on that block, respectively.
<p>
Let's use a set of features to predict house value.
## Set Up
In this first cell, we'll load the necessary libraries.
```
import math
import shutil
import numpy as np
import pandas as pd
import tensorflow as tf
tf.logging.set_verbosity(tf.logging.INFO)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
```
Next, we'll load our data set.
```
df = pd.read_csv("https://storage.googleapis.com/ml_universities/california_housing_train.csv", sep=",")
```
## Examine the data
It's a good idea to get to know your data a little bit before you work with it.
We'll print out a quick summary of a few useful statistics on each column.
This will include things like mean, standard deviation, max, min, and various quantiles.
```
df.head()
df.describe()
```
This data is at the city block level, so these features reflect the total number of rooms in that block, or the total number of people who live on that block, respectively. Let's create a different, more appropriate feature. Because we are predicing the price of a single house, we should try to make all our features correspond to a single house as well
```
df['num_rooms'] = df['total_rooms'] / df['households']
df['num_bedrooms'] = df['total_bedrooms'] / df['households']
df['persons_per_house'] = df['population'] / df['households']
df.describe()
df.drop(['total_rooms', 'total_bedrooms', 'population', 'households'], axis = 1, inplace = True)
df.describe()
```
## Build a neural network model
In this exercise, we'll be trying to predict `median_house_value`. It will be our label (sometimes also called a target). We'll use the remaining columns as our input features.
To train our model, we'll first use the [LinearRegressor](https://www.tensorflow.org/api_docs/python/tf/contrib/learn/LinearRegressor) interface. Then, we'll change to DNNRegressor
```
featcols = {
colname : tf.feature_column.numeric_column(colname) \
for colname in 'housing_median_age,median_income,num_rooms,num_bedrooms,persons_per_house'.split(',')
}
# Bucketize lat, lon so it's not so high-res; California is mostly N-S, so more lats than lons
featcols['longitude'] = tf.feature_column.bucketized_column(tf.feature_column.numeric_column('longitude'),
np.linspace(-124.3, -114.3, 5).tolist())
featcols['latitude'] = tf.feature_column.bucketized_column(tf.feature_column.numeric_column('latitude'),
np.linspace(32.5, 42, 10).tolist())
featcols.keys()
# Split into train and eval
msk = np.random.rand(len(df)) < 0.8
traindf = df[msk]
evaldf = df[~msk]
SCALE = 100000
BATCH_SIZE= 100
OUTDIR = './housing_trained'
train_input_fn = tf.estimator.inputs.pandas_input_fn(x = traindf[list(featcols.keys())],
y = traindf["median_house_value"] / SCALE,
num_epochs = None,
batch_size = BATCH_SIZE,
shuffle = True)
eval_input_fn = tf.estimator.inputs.pandas_input_fn(x = evaldf[list(featcols.keys())],
y = evaldf["median_house_value"] / SCALE, # note the scaling
num_epochs = 1,
batch_size = len(evaldf),
shuffle=False)
# Linear Regressor
def train_and_evaluate(output_dir, num_train_steps):
myopt = tf.train.FtrlOptimizer(learning_rate = 0.01) # note the learning rate
estimator = tf.estimator.LinearRegressor(
model_dir = output_dir,
feature_columns = featcols.values(),
optimizer = myopt)
#Add rmse evaluation metric
def rmse(labels, predictions):
pred_values = tf.cast(predictions['predictions'],tf.float64)
return {'rmse': tf.metrics.root_mean_squared_error(labels*SCALE, pred_values*SCALE)}
estimator = tf.contrib.estimator.add_metrics(estimator,rmse)
train_spec=tf.estimator.TrainSpec(
input_fn = train_input_fn,
max_steps = num_train_steps)
eval_spec=tf.estimator.EvalSpec(
input_fn = eval_input_fn,
steps = None,
start_delay_secs = 1, # start evaluating after N seconds
throttle_secs = 10, # evaluate every N seconds
)
tf.estimator.train_and_evaluate(estimator, train_spec, eval_spec)
# Run training
shutil.rmtree(OUTDIR, ignore_errors = True) # start fresh each time
train_and_evaluate(OUTDIR, num_train_steps = (100 * len(traindf)) / BATCH_SIZE)
# DNN Regressor
def train_and_evaluate(output_dir, num_train_steps):
myopt = tf.train.FtrlOptimizer(learning_rate = 0.01) # note the learning rate
estimator = # TODO: Implement DNN Regressor model
#Add rmse evaluation metric
def rmse(labels, predictions):
pred_values = tf.cast(predictions['predictions'],tf.float64)
return {'rmse': tf.metrics.root_mean_squared_error(labels*SCALE, pred_values*SCALE)}
estimator = tf.contrib.estimator.add_metrics(estimator,rmse)
train_spec=tf.estimator.TrainSpec(
input_fn = train_input_fn,
max_steps = num_train_steps)
eval_spec=tf.estimator.EvalSpec(
input_fn = eval_input_fn,
steps = None,
start_delay_secs = 1, # start evaluating after N seconds
throttle_secs = 10, # evaluate every N seconds
)
tf.estimator.train_and_evaluate(estimator, train_spec, eval_spec)
# Run training
shutil.rmtree(OUTDIR, ignore_errors = True) # start fresh each time
tf.summary.FileWriterCache.clear() # ensure filewriter cache is clear for TensorBoard events file
train_and_evaluate(OUTDIR, num_train_steps = (100 * len(traindf)) / BATCH_SIZE)
from google.datalab.ml import TensorBoard
pid = TensorBoard().start(OUTDIR)
TensorBoard().stop(pid)
```
|
github_jupyter
|
# Exploring Neural Audio Synthesis with NSynth
## Parag Mital
There is a lot to explore with NSynth. This notebook explores just a taste of what's possible including how to encode and decode, timestretch, and interpolate sounds. Also check out the [blog post](https://magenta.tensorflow.org/nsynth-fastgen) for more examples including two compositions created with Ableton Live. If you are interested in learning more, checkout my [online course on Kadenze](https://www.kadenze.com/programs/creative-applications-of-deep-learning-with-tensorflow) where we talk about Magenta and NSynth in more depth.
## Part 1: Encoding and Decoding
We'll walkthrough using the source code to encode and decode some audio. This is the most basic thing we can do with NSynth, and it will take at least about 6 minutes per 1 second of audio to perform on a GPU, though this will get faster!
I'll first show you how to encode some audio. This is basically saying, here is some audio, now put it into the trained model. It's like the encoding of an MP3 file. It takes some raw audio, and represents it using some really reduced down representation of the raw audio. NSynth works similarly, but we can actually mess with the encoding to do some awesome stuff. You can for instance, mix it with other encodings, or slow it down, or speed it up. You can potentially even remove parts of it, mix many different encodings together, and hopefully just explore ideas yet to be thought of. After you've created your encoding, you have to just generate, or decode it, just like what an audio player does to an MP3 file.
First, to install Magenta, follow their setup guide here: https://github.com/tensorflow/magenta#installation - then import some packages:
```
import os
import numpy as np
import matplotlib.pyplot as plt
from magenta.models.nsynth import utils
from magenta.models.nsynth.wavenet import fastgen
from IPython.display import Audio
%matplotlib inline
%config InlineBackend.figure_format = 'jpg'
```
Now we'll load up a sound I downloaded from freesound.org. The `utils.load_audio` method will resample this to the required sample rate of 16000. I'll load in 40000 samples of this beat which should end up being a pretty good loop:
```
# from https://www.freesound.org/people/MustardPlug/sounds/395058/
fname = '395058__mustardplug__breakbeat-hiphop-a4-4bar-96bpm.wav'
sr = 16000
audio = utils.load_audio(fname, sample_length=40000, sr=sr)
sample_length = audio.shape[0]
print('{} samples, {} seconds'.format(sample_length, sample_length / float(sr)))
```
## Encoding
We'll now encode some audio using the pre-trained NSynth model (download from: http://download.magenta.tensorflow.org/models/nsynth/wavenet-ckpt.tar). This is pretty fast, and takes about 3 seconds per 1 second of audio on my NVidia 1080 GPU. This will give us a 125 x 16 dimension encoding for every 4 seconds of audio which we can then decode, or resynthesize. We'll try a few things, including just leaving it alone and reconstructing it as is. But then we'll also try some fun transformations of the encoding and see what's possible from there.
```help(fastgen.encode)
Help on function encode in module magenta.models.nsynth.wavenet.fastgen:
encode(wav_data, checkpoint_path, sample_length=64000)
Generate an array of embeddings from an array of audio.
Args:
wav_data: Numpy array [batch_size, sample_length]
checkpoint_path: Location of the pretrained model.
sample_length: The total length of the final wave file, padded with 0s.
Returns:
encoding: a [mb, 125, 16] encoding (for 64000 sample audio file).
```
```
%time encoding = fastgen.encode(audio, 'model.ckpt-200000', sample_length)
```
This returns a 3-dimensional tensor representing the encoding of the audio. The first dimension of the encoding represents the batch dimension. We could have passed in many audio files at once and the process would be much faster. For now we've just passed in one audio file.
```
print(encoding.shape)
```
We'll also save the encoding so that we can use it again later:
```
np.save(fname + '.npy', encoding)
```
Let's take a look at the encoding of this audio file. Think of these as 16 channels of sounds all mixed together (though with a lot of caveats):
```
fig, axs = plt.subplots(2, 1, figsize=(10, 5))
axs[0].plot(audio);
axs[0].set_title('Audio Signal')
axs[1].plot(encoding[0]);
axs[1].set_title('NSynth Encoding')
```
You should be able to pretty clearly see a sort of beat like pattern in both the signal and the encoding.
## Decoding
Now we can decode the encodings as is. This is the process that takes awhile, though it used to be so long that you wouldn't even dare trying it. There is still plenty of room for improvement and I'm sure it will get faster very soon.
```
help(fastgen.synthesize)
Help on function synthesize in module magenta.models.nsynth.wavenet.fastgen:
synthesize(encodings, save_paths, checkpoint_path='model.ckpt-200000', samples_per_save=1000)
Synthesize audio from an array of embeddings.
Args:
encodings: Numpy array with shape [batch_size, time, dim].
save_paths: Iterable of output file names.
checkpoint_path: Location of the pretrained model. [model.ckpt-200000]
samples_per_save: Save files after every amount of generated samples.
```
```
%time fastgen.synthesize(encoding, save_paths=['gen_' + fname], samples_per_save=sample_length)
```
After it's done synthesizing, we can see that takes about 6 minutes per 1 second of audio on a non-optimized version of Tensorflow for GPU on an NVidia 1080 GPU. We can speed things up considerably if we want to do multiple encodings at a time. We'll see that in just a moment. Let's first listen to the synthesized audio:
```
sr = 16000
synthesis = utils.load_audio('gen_' + fname, sample_length=sample_length, sr=sr)
```
Listening to the audio, the sounds are definitely different. NSynth seems to apply a sort of gobbly low-pass that also really doesn't know what to do with the high frequencies. It is really quite hard to describe, but that is what is so interesting about it. It has a recognizable, characteristic sound.
Let's try another one. I'll put the whole workflow for synthesis in two cells, and we can listen to another synthesis of a vocalist singing, "Laaaa":
```
def load_encoding(fname, sample_length=None, sr=16000, ckpt='model.ckpt-200000'):
audio = utils.load_audio(fname, sample_length=sample_length, sr=sr)
encoding = fastgen.encode(audio, ckpt, sample_length)
return audio, encoding
# from https://www.freesound.org/people/maurolupo/sounds/213259/
fname = '213259__maurolupo__girl-sings-laa.wav'
sample_length = 32000
audio, encoding = load_encoding(fname, sample_length)
fastgen.synthesize(
encoding,
save_paths=['gen_' + fname],
samples_per_save=sample_length)
synthesis = utils.load_audio('gen_' + fname,
sample_length=sample_length,
sr=sr)
```
Aside from the quality of the reconstruction, what we're really after is what is possible with such a model. Let's look at two examples now.
# Part 2: Timestretching
Let's try something more fun. We'll stretch the encodings a bit and see what it sounds like. If you were to try and stretch audio directly, you'd hear a pitch shift. There are some other ways of stretching audio without shifting pitch, like granular synthesis. But it turns out that NSynth can also timestretch. Let's see how. First we'll use image interpolation to help stretch the encodings.
```
# use image interpolation to stretch the encoding: (pip install scikit-image)
try:
from skimage.transform import resize
except ImportError:
!pip install scikit-image
from skimage.transform import resize
```
Here's a utility function to help you stretch your own encoding. It uses skimage.transform and will retain the range of values. Images typically only have a range of 0-1, but the encodings aren't actually images so we'll keep track of their min/max in order to stretch them like images.
```
def timestretch(encodings, factor):
min_encoding, max_encoding = encoding.min(), encoding.max()
encodings_norm = (encodings - min_encoding) / (max_encoding - min_encoding)
timestretches = []
for encoding_i in encodings_norm:
stretched = resize(encoding_i, (int(encoding_i.shape[0] * factor), encoding_i.shape[1]), mode='reflect')
stretched = (stretched * (max_encoding - min_encoding)) + min_encoding
timestretches.append(stretched)
return np.array(timestretches)
# from https://www.freesound.org/people/MustardPlug/sounds/395058/
fname = '395058__mustardplug__breakbeat-hiphop-a4-4bar-96bpm.wav'
sample_length = 40000
audio, encoding = load_encoding(fname, sample_length)
```
Now let's stretch the encodings with a few different factors:
```
audio = utils.load_audio('gen_slower_' + fname, sample_length=None, sr=sr)
Audio(audio, rate=sr)
encoding_slower = timestretch(encoding, 1.5)
encoding_faster = timestretch(encoding, 0.5)
```
Basically we've made a slower and faster version of the amen break's encodings. The original encoding is shown in black:
```
fig, axs = plt.subplots(3, 1, figsize=(10, 7), sharex=True, sharey=True)
axs[0].plot(encoding[0]);
axs[0].set_title('Encoding (Normal Speed)')
axs[1].plot(encoding_faster[0]);
axs[1].set_title('Encoding (Faster))')
axs[2].plot(encoding_slower[0]);
axs[2].set_title('Encoding (Slower)')
```
Now let's decode them:
```
fastgen.synthesize(encoding_faster, save_paths=['gen_faster_' + fname])
fastgen.synthesize(encoding_slower, save_paths=['gen_slower_' + fname])
```
It seems to work pretty well and retains the pitch and timbre of the original sound. We could even quickly layer the sounds just by adding them. You might want to do this in a program like Logic or Ableton Live instead and explore more possiblities of these sounds!
# Part 3: Interpolating Sounds
Now let's try something more experimental. NSynth released plenty of great examples of what happens when you mix the embeddings of different sounds: https://magenta.tensorflow.org/nsynth-instrument - we're going to do the same but now with our own sounds!
First let's load some encodings:
```
sample_length = 80000
# from https://www.freesound.org/people/MustardPlug/sounds/395058/
aud1, enc1 = load_encoding('395058__mustardplug__breakbeat-hiphop-a4-4bar-96bpm.wav', sample_length)
# from https://www.freesound.org/people/xserra/sounds/176098/
aud2, enc2 = load_encoding('176098__xserra__cello-cant-dels-ocells.wav', sample_length)
```
Now we'll mix the two audio signals together. But this is unlike adding the two signals together in a Ableton or simply hearing both sounds at the same time. Instead, we're averaging the representation of their timbres, tonality, change over time, and resulting audio signal. This is way more powerful than a simple averaging.
```
enc_mix = (enc1 + enc2) / 2.0
fig, axs = plt.subplots(3, 1, figsize=(10, 7))
axs[0].plot(enc1[0]);
axs[0].set_title('Encoding 1')
axs[1].plot(enc2[0]);
axs[1].set_title('Encoding 2')
axs[2].plot(enc_mix[0]);
axs[2].set_title('Average')
fastgen.synthesize(enc_mix, save_paths='mix.wav')
```
As another example of what's possible with interpolation of embeddings, we'll try crossfading between the two embeddings. To do this, we'll write a utility function which will use a hanning window to apply a fade in or out to the embeddings matrix:
```
def fade(encoding, mode='in'):
length = encoding.shape[1]
fadein = (0.5 * (1.0 - np.cos(3.1415 * np.arange(length) /
float(length)))).reshape(1, -1, 1)
if mode == 'in':
return fadein * encoding
else:
return (1.0 - fadein) * encoding
fig, axs = plt.subplots(3, 1, figsize=(10, 7))
axs[0].plot(enc1[0]);
axs[0].set_title('Original Encoding')
axs[1].plot(fade(enc1, 'in')[0]);
axs[1].set_title('Fade In')
axs[2].plot(fade(enc1, 'out')[0]);
axs[2].set_title('Fade Out')
```
Now we can cross fade two different encodings by adding their repsective fade ins and out:
```
def crossfade(encoding1, encoding2):
return fade(encoding1, 'out') + fade(encoding2, 'in')
fig, axs = plt.subplots(3, 1, figsize=(10, 7))
axs[0].plot(enc1[0]);
axs[0].set_title('Encoding 1')
axs[1].plot(enc2[0]);
axs[1].set_title('Encoding 2')
axs[2].plot(crossfade(enc1, enc2)[0]);
axs[2].set_title('Crossfade')
```
Now let's synthesize the resulting encodings:
```
fastgen.synthesize(crossfade(enc1, enc2), save_paths=['crossfade.wav'])
```
There is a lot to explore with NSynth. So far I've just shown you a taste of what's possible when you are able to generate your own sounds. I expect the generation process will soon get much faster, especially with help from the community, and for more unexpected and interesting applications to emerge. Please keep in touch with whatever you end up creating, either personally via [twitter](https://twitter.com/pkmital), in our [Creative Applications of Deep Learning](https://www.kadenze.com/programs/creative-applications-of-deep-learning-with-tensorflow) community on Kadenze, or the [Magenta Google Group](https://groups.google.com/a/tensorflow.org/forum/#!forum/magenta-discuss).
|
github_jupyter
|
## A Two-sample t-test to find differentially expressed miRNA's between normal and tumor tissues in Lung Adenocarcinoma
```
import os
import pandas
mirna_src_dir = os.getcwd() + "/assn-mirna-luad/data/processed/miRNA/"
clinical_src_dir = os.getcwd() + "/assn-mirna-luad/data/processed/clinical/"
mirna_tumor_df = pandas.read_csv(mirna_src_dir+'tumor_miRNA.csv')
mirna_normal_df = pandas.read_csv(mirna_src_dir+'normal_miRNA.csv')
clinical_df = pandas.read_csv(clinical_src_dir+'clinical.csv')
print "mirna_tumor_df.shape", mirna_tumor_df.shape
print "mirna_normal_df.shape", mirna_normal_df.shape
"""
Here we select samples to use for our regression analysis
"""
matched_samples = pandas.merge(clinical_df, mirna_normal_df, on='patient_barcode')['patient_barcode']
# print "matched_samples", matched_samples.shape
# merged = pandas.merge(clinical_df, mirna_tumor_df, on='patient_barcode')
# print merged.shape
# print
# print merged['histological_type'].value_counts().sort_index(axis=0)
# print
# print merged['pathologic_stage'].value_counts().sort_index(axis=0)
# print
# print merged['pathologic_T'].value_counts().sort_index(axis=0)
# print
# print merged['pathologic_N'].value_counts().sort_index(axis=0)
# print
# print merged['pathologic_M'].value_counts().sort_index(axis=0)
# print
from sklearn import preprocessing
import numpy as np
X_normal = mirna_normal_df[mirna_normal_df['patient_barcode'].isin(matched_samples)].sort_values(by=['patient_barcode']).copy()
X_tumor = mirna_tumor_df.copy()
X_tumor_matched = mirna_tumor_df[mirna_tumor_df['patient_barcode'].isin(matched_samples)].sort_values(by=['patient_barcode']).copy()
X_normal.__delitem__('patient_barcode')
X_tumor_matched.__delitem__('patient_barcode')
X_tumor.__delitem__('patient_barcode')
print "X_normal.shape", X_normal.shape
print "X_tumor.shape", X_tumor.shape
print "X_tumor_matched.shape", X_tumor_matched.shape
mirna_list = X.columns.values
# X_scaler = preprocessing.StandardScaler(with_mean=False).fit(X)
# X = X_scaler.transform(X)
from scipy.stats import ttest_rel
import matplotlib.pyplot as plt
ttest = ttest_rel(X_tumor_matched, X_normal)
plt.plot(ttest[1], ls='', marker='.')
plt.title('Two sample t-test between tumor and normal LUAD tissues')
plt.ylabel('p-value')
plt.xlabel('miRNA\'s')
plt.show()
from scipy.stats import ttest_ind
ttest_2 = ttest_2_ind(X_tumor, X_normal)
plt.plot(ttest_2[1], ls='', marker='.')
plt.title('Independent sample t-test between tumor and normal LUAD tissues')
plt.ylabel('p-value')
plt.xlabel('miRNA\'s')
plt.show()
```
|
github_jupyter
|
# Step 7: Serve data from OpenAgua into WEAP using WaMDaM
#### By Adel M. Abdallah, Dec 2020
Execute the following cells by pressing `Shift-Enter`, or by pressing the play button <img style='display:inline;padding-bottom:15px' src='play-button.png'> on the toolbar above.
## Steps
1. Import python libraries
2. Import the pulished SQLite file for the WEAP model from HydroShare.
3. Prepare to connect to the WEAP API
4. Connect to WEAP API to programmatically populate WEAP with data, run it, get back results
Create a copy of the original WEAP Area to use while keeping the orignial as-as for any later use
5.3 Export the unmet demand percent into Excel to load them into WaMDaM
<a name="Import"></a>
# 1. Import python libraries
```
# 1. Import python libraries
### set the notebook mode to embed the figures within the cell
import numpy
import sqlite3
import numpy as np
import pandas as pd
import getpass
from hs_restclient import HydroShare, HydroShareAuthBasic
import os
import plotly
plotly.__version__
import plotly.offline as offline
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
offline.init_notebook_mode(connected=True)
from plotly.offline import init_notebook_mode, iplot
from plotly.graph_objs import *
init_notebook_mode(connected=True) # initiate notebook for offline plot
import os
import csv
from collections import OrderedDict
import sqlite3
import pandas as pd
import numpy as np
from IPython.display import display, Image, SVG, Math, YouTubeVideo
import urllib
import calendar
print 'The needed Python libraries have been imported'
```
# 2. Connect to the WaMDaM SQLite on HydroSahre
### Provide the HydroShare ID for your resource
Example
https://www.hydroshare.org/resource/af71ef99a95e47a89101983f5ec6ad8b/
resource_id='85e9fe85b08244198995558fe7d0e294'
```
# enter your HydroShare username and password here between the quotes
username = ''
password = ''
auth = HydroShareAuthBasic(username=username, password=password)
hs = HydroShare(auth=auth)
print 'Connected to HydroShare'
# Then we can run queries against it within this notebook :)
resource_url='https://www.hydroshare.org/resource/af71ef99a95e47a89101983f5ec6ad8b/'
resource_id= resource_url.split("https://www.hydroshare.org/resource/",1)[1]
resource_id=resource_id.replace('/','')
print resource_id
resource_md = hs.getSystemMetadata(resource_id)
# print resource_md
print 'Resource title'
print(resource_md['resource_title'])
print '----------------------------'
resources=hs.resource(resource_id).files.all()
file = ""
for f in hs.resource(resource_id).files.all():
file += f.decode('utf8')
import json
file_json = json.loads(file)
for f in file_json["results"]:
FileURL= f["url"]
SQLiteFileName=FileURL.split("contents/",1)[1]
cwd = os.getcwd()
print cwd
fpath = hs.getResourceFile(resource_id, SQLiteFileName, destination=cwd)
conn = sqlite3.connect(SQLiteFileName,timeout=10)
print 'Connected to the SQLite file= '+ SQLiteFileName
print 'done'
```
<a name="ConnectWEAP"></a>
# 2. Prepare to the Connect to the WEAP API
### You need to have WEAP already installed on your machine
First make sure to have a copy of the Water Evaluation And Planning" system (WEAP) installed on your local machine (Windows). If you don’t have it installed, download and install the WEAP software which allows you to run the Bear River WEAP model and its scenarios for Use Case 5. https://www.weap21.org/. You need to have a WEAP License. See here (https://www.weap21.org/index.asp?action=217). If you're interested to learning about WEAP API, check it out here: http://www.weap21.org/WebHelp/API.htm
## Install dependency and register WEAP
### 2.1. Install pywin32 extensions which provide access to many of the Windows APIs from Python.
**Choose on option**
* a. Install using an executable basedon your python version. Use version for Python 2.7
https://github.com/mhammond/pywin32/releases
**OR**
* b. Install it using Anaconda terminal @ https://anaconda.org/anaconda/pywin32
Type this command in the Anaconda terminal as Administrator
conda install -c anaconda pywin32
**OR**
* c. Install from source code (for advanced users)
https://github.com/mhammond/pywin32
### 2.2. Register WEAP with Windows
This use case only works on a local Jupyter Notebook server installed on your machine along with WEAP. So it does not work on the online Notebooks in Step 2.1. You need to install Jupyter Server in Step 2.2 then proceed here.
* **Register WEAP with Windows to allow the WEAP API to be accessed**
Use Windows "Command Prompt". Right click and then <font color=red>**run as Administrator**</font>, navigate to the WEAP installation directory such as and then hit enter
```
cd C:\Program Files (x86)\WEAP
```
Then type the following command in the command prompt and hit enter
```
WEAP /regserver
```
<img src="https://github.com/WamdamProject/WaMDaM-software-ecosystem/blob/master/mkdocs/Edit_MD_Files/QuerySelect/images/RegisterWEAP_CMD.png?raw=true" style="float:center;width:700px;padding:20px">
Figure 1: Register WEAP API with windows using the Command Prompt (Run as Administrator)
# 3. Connect Jupyter Notebook to WEAP API
Clone or download all this GitHub repo
https://github.com/WamdamProject/WaMDaM_UseCases
In your local repo folder, go to the
C:\Users\Adel\Documents\GitHub\WaMDaM_UseCases/UseCases_files/1Original_Datasets_preperation_files/WEAP/Bear_River_WEAP_Model_2017
Copy this folder **Bear_River_WEAP_Model_2017** and paste it into **WEAP Areas** folder on your local machine. For example, it is at
C:\Users\Adel\Documents\WEAP Areas
```
# this library is needed to connect to the WEAP API
import win32com.client
# this command will open the WEAP software (if closed) and get the last active model
# you could change the active area to another one inside WEAP or by passing it to the command here
#WEAP.ActiveArea = "BearRiverFeb2017_V10.9"
WEAP=win32com.client.Dispatch("WEAP.WEAPApplication")
# WEAP.Visible = 'FALSE'
print WEAP.ActiveArea.Name
WEAP.ActiveArea = "Bear_River_WEAP_Model_2017_Original"
print WEAP.ActiveArea.Name
WEAP.Areas("Bear_River_WEAP_Model_2017_Original").Open
WEAP.ActiveArea = "Bear_River_WEAP_Model_2017_Original"
print WEAP.ActiveArea.Name
print 'Connected to WEAP API and the '+ WEAP.ActiveArea.Name + ' Area'
print '-------------'
if not WEAP.Registered:
print "Because WEAP is not registered, you cannot use the API"
# get the active WEAP Area (model) to serve data into it
# ActiveArea=WEAP.ActiveArea.Name
# get the active WEAP scenario to serve data into it
print '-------------'
ActiveScenario= WEAP.ActiveScenario.Name
print '\n ActiveScenario= '+ActiveScenario
print '-------------'
WEAP_Area_dir=WEAP.AreasDirectory
print WEAP_Area_dir
print "\n \n You're connected to the WEAP API"
```
<a name="CreateWEAP_Area"></a>
# 4 Create a copy of the original WEAP Area to use while keeping the orignial as-as for any later use
<a name="AddScenarios"></a>
### Add a new CacheCountyUrbanWaterUse scenario from the Reference original WEAP Area:
### You can always use this orignal one and delete any new copies you make afterwards.
```
# Create a copy of the WEAP AREA to serve the updated Hyrym Reservoir to it
# Delete the Area if it exists and then add it. Start from fresh
Area="Bear_River_WEAP_Model_2017_Conservation"
if not WEAP.Areas.Exists(Area):
WEAP.SaveAreaAs(Area)
WEAP.ActiveArea.Save
WEAP.ActiveArea = "Bear_River_WEAP_Model_2017_Conservation"
print 'ActiveArea= '+WEAP.ActiveArea.Name
# Add new Scenario
# Add(NewScenarioName, ParentScenarioName or Index):
# Create a new scenario as a child of the parent scenario specified.
# The new scenario will become the selected scenario in the Data View.
WEAP=win32com.client.Dispatch("WEAP.WEAPApplication")
# WEAP.Visible = FALSE
WEAP.ActiveArea = "Bear_River_WEAP_Model_2017_Conservation"
print 'ActiveArea= '+ WEAP.ActiveArea.Name
Scenarios=[]
Scenarios=['Cons25PercCacheUrbWaterUse','Incr25PercCacheUrbWaterUse']
# Delete the scenario if it exists and then add it. Start from fresh
for Scenario in Scenarios:
if WEAP.Scenarios.Exists(Scenario):
# delete it
WEAP.Scenarios(Scenario).Delete(True)
# add it back as a fresh copy
WEAP.Scenarios.Add(Scenario,'Reference')
else:
WEAP.Scenarios.Add(Scenario,'Reference')
WEAP.ActiveArea.Save
WEAP.SaveArea
WEAP.Quit
# or add the scenarios one by one using this command
# Make a copy from the reference (base) scenario
# WEAP.Scenarios.Add('UpdateCacheDemand','Reference')
print '---------------------- \n'
print 'Scenarios added to the original WEAP area'
WEAP.Quit
print 'Connection with WEAP API is disconnected'
```
<a name="QuerySupplyDataLoadWEAP"></a>
# 4.A Query Cache County seasonal "Monthly Demand" for the three sites: Logan Potable, North Cache Potable, South Cache Potable
### The data comes from OpenAgua
```
# Use Case 3.1Identify_aggregate_TimeSeriesValues.csv
# plot aggregated to monthly and converted to acre-feet time series data of multiple sources
# Logan Potable
# North Cache Potable
# South Cache Potable
# 2.2Identify_aggregate_TimeSeriesValues.csv
Query_UseCase_URL="""
https://raw.githubusercontent.com/WamdamProject/WaMDaM_JupyterNotebooks/master/3_VisualizePublish/SQL_queries/WEAP/Query_demand_sites.sql
"""
# Read the query text inside the URL
Query_UseCase_text = urllib.urlopen(Query_UseCase_URL).read()
# return query result in a pandas data frame
result_df_UseCase= pd.read_sql_query(Query_UseCase_text, conn)
# uncomment the below line to see the list of attributes
# display (result_df_UseCase)
seasons_dict = dict()
seasons_dict2=dict()
Scenarios=['Cons25PercCacheUrbWaterUse','Incr25PercCacheUrbWaterUse']
subsets = result_df_UseCase.groupby(['ScenarioName','InstanceName'])
for subset in subsets.groups.keys():
if subset[0] in Scenarios:
df_Seasonal = subsets.get_group(name=subset)
df_Seasonal=df_Seasonal.reset_index()
SeasonalParam = ''
for i in range(len(df_Seasonal['SeasonName'])):
m_data = df_Seasonal['SeasonName'][i]
n_data = float(df_Seasonal['SeasonNumericValue'][i])
SeasonalParam += '{},{}'.format(m_data, n_data)
if i != len(df_Seasonal['SeasonName']) - 1:
SeasonalParam += ','
Seasonal_value="MonthlyValues("+SeasonalParam+")"
seasons_dict[subset]=(Seasonal_value)
# seasons_dict2[subset[0]]=seasons_dict
# print seasons_dict2
print '-----------------'
# print seasons_dict
# seasons_dict2.get("Cons25PercCacheUrbWaterUse", {}).get("Logan Potable") # 1
print 'Query and data preperation are done'
```
<a name="LoadFlow"></a>
# 4.B Load the seasonal demand data with conservation into WEAP
```
# 9. Load the seasonal data into WEAP
#WEAP=win32com.client.Dispatch("WEAP.WEAPApplication")
# WEAP.Visible = FALSE
print WEAP.ActiveArea.Name
Scenarios=['Cons25PercCacheUrbWaterUse','Incr25PercCacheUrbWaterUse']
DemandSites=['Logan Potable','North Cache Potable','South Cache Potable']
AttributeName='Monthly Demand'
for scenario in Scenarios:
WEAP.ActiveScenario = scenario
print WEAP.ActiveScenario.Name
for Branch in WEAP.Branches:
for InstanceName in DemandSites:
if Branch.Name == InstanceName:
GetInstanceFullBranch = Branch.FullName
val=seasons_dict[(scenario,InstanceName)]
WEAP.Branch(GetInstanceFullBranch).Variable(AttributeName).Expression =val
# print val
print "loaded " + InstanceName
WEAP.SaveArea
print '\n The data have been sucsesfully loaded into WEAP'
WEAP.SaveArea
print '\n \n The updated data have been saved'
```
# 5. Run WEAP
<font color=green>**Please wait, it will take ~1-3 minutes** to finish calcualting the two WEAP Areas with their many scenarios</font>
```
# Run WEAP
WEAP.Areas("Bear_River_WEAP_Model_2017_Conservation").Open
print WEAP.ActiveArea.Name
WEAP.ActiveArea = "Bear_River_WEAP_Model_2017_Conservation"
print WEAP.ActiveArea.Name
print 'Please wait 1-3 min for the calculation to finish'
WEAP.Calculate(2006,10,True)
WEAP.SaveArea
print '\n \n The calculation has been done and saved'
print WEAP.CalculationTime
print '\n \n Done'
```
## 5.1 Get the unmet demand or Cache County sites in both the reference and the conservation scenarios
```
Scenarios=['Reference','Cons25PercCacheUrbWaterUse','Incr25PercCacheUrbWaterUse']
DemandSites=['Logan Potable','North Cache Potable','South Cache Potable']
UnmetDemandEstimate_Ref = pd.DataFrame(columns = DemandSites)
UnmetDemandEstimate_Cons25 = pd.DataFrame(columns = DemandSites)
UnmetDemandEstimate_Incr25 = pd.DataFrame(columns = DemandSites)
UnmetDemandEstimate= pd.DataFrame(columns = Scenarios)
for scen in Scenarios:
if scen=='Reference':
for site in DemandSites:
param="\Demand Sites\%s: Unmet Demand[Acre-Foot]"%(site)
# print param
for year in range (1966,2006):
value=WEAP.ResultValue(param, year, 1, scen, year, WEAP.NumTimeSteps)
UnmetDemandEstimate_Ref.loc[year, [site]]=value
elif scen=='Cons25PercCacheUrbWaterUse':
for site in DemandSites:
param="\Demand Sites\%s: Unmet Demand[Acre-Foot]"%(site)
# print param
for year in range (1966,2006):
value=WEAP.ResultValue(param, year, 1, scen, year, WEAP.NumTimeSteps)
UnmetDemandEstimate_Cons25.loc[year, [site]]=value
elif scen=='Incr25PercCacheUrbWaterUse':
for site in DemandSites:
param="\Demand Sites\%s: Unmet Demand[Acre-Foot]"%(site)
# print param
for year in range (1966,2006):
value=WEAP.ResultValue(param, year, 1, scen, year, WEAP.NumTimeSteps)
UnmetDemandEstimate_Incr25.loc[year, [site]]=value
UnmetDemandEstimate_Ref['Cache Total']=UnmetDemandEstimate_Ref[DemandSites].sum(axis=1)
UnmetDemandEstimate_Cons25['Cache Total']=UnmetDemandEstimate_Cons25[DemandSites].sum(axis=1)
UnmetDemandEstimate_Incr25['Cache Total']=UnmetDemandEstimate_Incr25[DemandSites].sum(axis=1)
UnmetDemandEstimate['Reference']=UnmetDemandEstimate_Ref['Cache Total']
UnmetDemandEstimate['Cons25PercCacheUrbWaterUse']=UnmetDemandEstimate_Cons25['Cache Total']
UnmetDemandEstimate['Incr25PercCacheUrbWaterUse']=UnmetDemandEstimate_Incr25['Cache Total']
UnmetDemandEstimate=UnmetDemandEstimate.rename_axis('Year',axis="columns")
print 'Done estimating the unment demnd pecentage for each scenario'
# display(UnmetDemandEstimate)
```
## 5.2 Get the unmet demand as a percentage for the scenarios
```
########################################################################
# estimate the total reference demand for Cahce county to calcualte the percentage
result_df_UseCase= pd.read_sql_query(Query_UseCase_text, conn)
subsets = result_df_UseCase.groupby(['ScenarioName'])
for subset in subsets.groups.keys():
if subset=='Bear River WEAP Model 2017': # reference
df_Seasonal = subsets.get_group(name=subset)
df_Seasonal=df_Seasonal.reset_index()
# display (df_Seasonal)
Tot=df_Seasonal["SeasonNumericValue"].tolist()
float_lst = [float(x) for x in Tot]
Annual_Demand=sum(float_lst)
print Annual_Demand
########################################################################
years =UnmetDemandEstimate.index.values
Reference_vals =UnmetDemandEstimate['Reference'].tolist()
Reference_vals_perc =((numpy.array([Reference_vals]))/Annual_Demand)*100
Cons25PercCacheUrbWaterUse_vals =UnmetDemandEstimate['Cons25PercCacheUrbWaterUse'].tolist()
Cons25PercCacheUrbWaterUse_vals_perc =((numpy.array([Cons25PercCacheUrbWaterUse_vals]))/Annual_Demand)*100
Incr25PercCacheUrbWaterUse_vals =UnmetDemandEstimate['Incr25PercCacheUrbWaterUse'].tolist()
Incr25PercCacheUrbWaterUse_vals_perc =((numpy.array([Incr25PercCacheUrbWaterUse_vals]))/Annual_Demand)*100
print 'done estimating unmet demnd the percentages'
```
# 5.3 Export the unmet demand percent into Excel to load them into WaMDaM
```
# display(UnmetDemandEstimate)
import xlsxwriter
from collections import OrderedDict
UnmetDemandEstimate.to_csv('UnmetDemandEstimate.csv')
ExcelFileName='Test.xlsx'
years =UnmetDemandEstimate.index.values
#print years
Columns=['ObjectType','InstanceName','ScenarioName','AttributeName','DateTimeStamp','Value']
# these three columns have fixed values for all the rows
ObjectType='Demand Site'
InstanceName='Cache County Urban'
AttributeName='UnmetDemand'
# this dict contains the keysL (scenario name) and the values are in a list
# years exist in UnmetDemandEstimate. We then need to add day and month to the year date
# like this format: # DateTimeStamp= 1/1/1993
Scenarios = OrderedDict()
Scenarios['Bear River WEAP Model 2017_result'] = Reference_vals_perc
Scenarios['Incr25PercCacheUrbWaterUse_result'] = Incr25PercCacheUrbWaterUse_vals_perc
Scenarios['Cons25PercCacheUrbWaterUse_result'] = Cons25PercCacheUrbWaterUse_vals_perc
#print Incr25PercCacheUrbWaterUse_vals_perc
workbook = xlsxwriter.Workbook(ExcelFileName)
sheet = workbook.add_worksheet('sheet')
# write headers
for i, header_name in enumerate(Columns):
sheet.write(0, i, header_name)
row = 1
col = 0
for scenario_name in Scenarios.keys():
for val_list in Scenarios[scenario_name]:
# print val_list
for i, val in enumerate(val_list):
# print years[i]
date_timestamp = '1/1/{}'.format(years[i])
sheet.write(row, 0, ObjectType)
sheet.write(row, 1, InstanceName)
sheet.write(row, 2, scenario_name)
sheet.write(row, 3, AttributeName)
sheet.write(row, 4, date_timestamp)
sheet.write(row, 5, val)
row += 1
workbook.close()
print 'done writing to Excel'
print 'Next, copy the exported data into a WaMDaM workbook template for the WEAP model'
```
# 6. Plot the unmet demad for all the scenarios and years
```
trace2 = go.Scatter(
x=years,
y=Reference_vals_perc[0],
name = 'Reference demand',
mode = 'lines+markers',
marker = dict(
color = '#264DFF',
))
trace3 = go.Scatter(
x=years,
y=Cons25PercCacheUrbWaterUse_vals_perc[0],
name = 'Conserve demand by 25%',
mode = 'lines+markers',
marker = dict(
color = '#3FA0FF'
))
trace1 = go.Scatter(
x=years,
y=Incr25PercCacheUrbWaterUse_vals_perc[0],
name = 'Increase demand by 25%',
mode = 'lines+markers',
marker = dict(
color = '#290AD8'
))
layout = dict(
#title = "Use Case 3.3",
yaxis = dict(
title = "Annual unmet demand (%)",
tickformat= ',',
showline=True,
dtick='5',
ticks='outside',
ticklen=10,
tickcolor='#000',
gridwidth=1,
showgrid=True,
),
xaxis = dict(
# title = "Updated input parameters in the <br>Bear_River_WEAP_Model_2017",
# showline=True,
ticks='inside',
tickfont=dict(size=22),
tickcolor='#000',
gridwidth=1,
showgrid=True,
ticklen=25
),
legend=dict(
x=0.05,y=1.1,
bordercolor='#00000f',
borderwidth=2
),
width=1100,
height=700,
#paper_bgcolor='rgb(233,233,233)',
#plot_bgcolor='rgb(233,233,233)',
margin=go.Margin(l=130,b=200),
font=dict(size=25,family='arial',color='#00000f'),
showlegend=True
)
data = [trace1, trace2,trace3]
# create a figure object
fig = dict(data=data, layout=layout)
#py.iplot(fig, filename = "2.3Identify_SeasonalValues")
## it can be run from the local machine on Pycharm like this like below
## It would also work here offline but in a seperate window
offline.iplot(fig,filename = 'jupyter/UnmentDemand@BirdRefuge' )
print "Figure x is replicated!!"
```
<a name="Close"></a>
# 7. Upload the new result scenarios to OpenAgua to visulize them there
You already uploaded the results form WaMDaM SQLite earlier at the begnining of these Jupyter Notebooks. So all you need is to select to display the result in OpenAgua. Finally, click, load data. It should replicate the same figure above and Figure 6 in the paper
<img src="https://github.com/WamdamProject/WaMDaM-software-ecosystem/blob/master/mkdocs/Edit_MD_Files/images/WEAP_results_OA.PNG?raw=true" style="float:center;width:900px;padding:20px">
<img src="https://github.com/WamdamProject/WaMDaM-software-ecosystem/blob/master/mkdocs/Edit_MD_Files/images/WEAP_results_OA2.PNG?raw=true" style="float:center;width:900px;padding:20px">
<a name="Close"></a>
# 8. Close the SQLite and WEAP API connections
```
# 9. Close the SQLite and WEAP API connections
conn.close()
print 'connection disconnected'
# Uncomment
WEAP.SaveArea
# this command will close WEAP
WEAP.Quit
print 'Connection with WEAP API is disconnected'
```
# The End :) Congratulations!
|
github_jupyter
|
# Optimizing building HVAC with Amazon SageMaker RL
```
import sagemaker
import boto3
from sagemaker.rl import RLEstimator
from source.common.docker_utils import build_and_push_docker_image
```
## Initialize Amazon SageMaker
```
role = sagemaker.get_execution_role()
sm_session = sagemaker.session.Session()
# SageMaker SDK creates a default bucket. Change this bucket to your own bucket, if needed.
s3_bucket = sm_session.default_bucket()
s3_output_path = f's3://{s3_bucket}'
print(f'S3 bucket path: {s3_output_path}')
print(f'Role: {role}')
```
## Set additional training parameters
### Set instance type
Set `cpu_or_gpu` to either `'cpu'` or `'gpu'` for using CPU or GPU instances.
### Configure the framework you want to use
Set `framework` to `'tf'` or `'torch'` for TensorFlow or PyTorch, respectively.
You will also have to edit your entry point i.e., `train-sagemaker-distributed.py` with the configuration parameter `"use_pytorch"` to match the framework that you have selected.
```
job_name_prefix = 'energyplus-hvac-ray'
cpu_or_gpu = 'gpu' # has to be either cpu or gpu
if cpu_or_gpu != 'cpu' and cpu_or_gpu != 'gpu':
raise ValueError('cpu_or_gpu has to be either cpu or gpu')
framework = 'tf'
instance_type = 'ml.g4dn.16xlarge' # g4dn.16x large has 1 GPU and 64 cores
```
# Train your homogeneous scaling job here
### Edit the training code
The training code is written in the file `train-sagemaker-distributed.py` which is uploaded in the /source directory.
*Note that ray will automatically set `"ray_num_cpus"` and `"ray_num_gpus"` in `_get_ray_config`*
```
!pygmentize source/train-sagemaker-distributed.py
```
### Train the RL model using the Python SDK Script mode
When using SageMaker for distributed training, you can select a GPU or CPU instance. The RLEstimator is used for training RL jobs.
1. Specify the source directory where the environment, presets and training code is uploaded.
2. Specify the entry point as the training code
3. Specify the image (CPU or GPU) to be used for the training environment.
4. Define the training parameters such as the instance count, job name, S3 path for output and job name.
5. Define the metrics definitions that you are interested in capturing in your logs. These can also be visualized in CloudWatch and SageMaker Notebooks.
#### GPU docker image
```
# Build image
repository_short_name = f'sagemaker-hvac-ray-{cpu_or_gpu}'
docker_build_args = {
'CPU_OR_GPU': cpu_or_gpu,
'AWS_REGION': boto3.Session().region_name,
'FRAMEWORK': framework
}
image_name = build_and_push_docker_image(repository_short_name, build_args=docker_build_args)
print("Using ECR image %s" % image_name)
metric_definitions = [
{'Name': 'training_iteration', 'Regex': 'training_iteration: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'episodes_total', 'Regex': 'episodes_total: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'num_steps_trained', 'Regex': 'num_steps_trained: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'timesteps_total', 'Regex': 'timesteps_total: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'training_iteration', 'Regex': 'training_iteration: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'episode_reward_max', 'Regex': 'episode_reward_max: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'episode_reward_mean', 'Regex': 'episode_reward_mean: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
{'Name': 'episode_reward_min', 'Regex': 'episode_reward_min: ([-+]?[0-9]*[.]?[0-9]+([eE][-+]?[0-9]+)?)'},
]
```
### Ray homogeneous scaling - Specify `train_instance_count` > 1
Homogeneous scaling allows us to use multiple instances of the same type.
Spot instances are unused EC2 instances that could be used at 90% discount compared to On-Demand prices (more information about spot instances can be found [here](https://aws.amazon.com/ec2/spot/?cards.sort-by=item.additionalFields.startDateTime&cards.sort-order=asc) and [here](https://docs.aws.amazon.com/sagemaker/latest/dg/model-managed-spot-training.html))
To use spot instances, set `train_use_spot_instances = True`. To use On-Demand instances, `train_use_spot_instances = False`.
```
hyperparameters = {
# no. of days to simulate. Remember to adjust the dates in RunPeriod of
# 'source/eplus/envs/buildings/MediumOffice/RefBldgMediumOfficeNew2004_Chicago.idf' to match simulation days.
'n_days': 365,
'n_iter': 50, # no. of training iterations
'algorithm': 'APEX_DDPG', # only APEX_DDPG and PPO are tested
'multi_zone_control': True, # if each zone temperature set point has to be independently controlled
'energy_temp_penalty_ratio': 10
}
# Set additional training parameters
training_params = {
'base_job_name': job_name_prefix,
'train_instance_count': 1,
'tags': [{'Key': k, 'Value': str(v)} for k,v in hyperparameters.items()]
}
# Defining the RLEstimator
estimator = RLEstimator(entry_point=f'train-sagemaker-hvac.py',
source_dir='source',
dependencies=["source/common/"],
image_uri=image_name,
role=role,
train_instance_type=instance_type,
# train_instance_type='local',
output_path=s3_output_path,
metric_definitions=metric_definitions,
hyperparameters=hyperparameters,
**training_params
)
estimator.fit(wait=False)
print(' ')
print(estimator.latest_training_job.job_name)
print('type=', instance_type, 'count=', training_params['train_instance_count'])
print(' ')
```
|
github_jupyter
|
# Spleen 3D segmentation with MONAI
This tutorial demonstrates how MONAI can be used in conjunction with the [PyTorch Lightning](https://github.com/PyTorchLightning/pytorch-lightning) framework.
We demonstrate use of the following MONAI features:
1. Transforms for dictionary format data.
2. Loading Nifti images with metadata.
3. Add channel dim to the data if no channel dimension.
4. Scaling medical image intensity with expected range.
5. Croping out a batch of balanced images based on the positive / negative label ratio.
6. Cache IO and transforms to accelerate training and validation.
7. Use of a a 3D UNet model, Dice loss function, and mean Dice metric for a 3D segmentation task.
8. The sliding window inference method.
9. Deterministic training for reproducibility.
The training Spleen dataset used in this example can be downloaded from from http://medicaldecathlon.com//

Target: Spleen
Modality: CT
Size: 61 3D volumes (41 Training + 20 Testing)
Source: Memorial Sloan Kettering Cancer Center
Challenge: Large ranging foreground size
In addition to the usual MONAI requirements you will need Lightning installed.
```
! pip install pytorch-lightning
# Copyright 2020 MONAI Consortium
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import os
import sys
import glob
import numpy as np
import torch
from torch.utils.data import DataLoader
import matplotlib.pyplot as plt
import monai
from monai.transforms import \
Compose, LoadNiftid, AddChanneld, ScaleIntensityRanged, RandCropByPosNegLabeld, \
RandAffined, Spacingd, Orientationd, ToTensord
from monai.data import list_data_collate, sliding_window_inference
from monai.networks.layers import Norm
from monai.metrics import compute_meandice
from pytorch_lightning import LightningModule, Trainer, loggers
from pytorch_lightning.callbacks.model_checkpoint import ModelCheckpoint
monai.config.print_config()
```
## Define the LightningModule
The LightningModule contains a refactoring of your training code. The following module is a refactoring of the code in `spleen_segmentation_3d.ipynb`:
```
class Net(LightningModule):
def __init__(self):
super().__init__()
self._model = monai.networks.nets.UNet(dimensions=3, in_channels=1, out_channels=2, channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2), num_res_units=2, norm=Norm.BATCH)
self.loss_function = monai.losses.DiceLoss(to_onehot_y=True, do_softmax=True)
self.best_val_dice = 0
self.best_val_epoch = 0
def forward(self, x):
return self._model(x)
def prepare_data(self):
# set up the correct data path
data_root = '/workspace/data/medical/Task09_Spleen'
train_images = glob.glob(os.path.join(data_root, 'imagesTr', '*.nii.gz'))
train_labels = glob.glob(os.path.join(data_root, 'labelsTr', '*.nii.gz'))
data_dicts = [{'image': image_name, 'label': label_name}
for image_name, label_name in zip(train_images, train_labels)]
train_files, val_files = data_dicts[:-9], data_dicts[-9:]
# define the data transforms
train_transforms = Compose([
LoadNiftid(keys=['image', 'label']),
AddChanneld(keys=['image', 'label']),
Spacingd(keys=['image', 'label'], pixdim=(1.5, 1.5, 2.), interp_order=(3, 0)),
Orientationd(keys=['image', 'label'], axcodes='RAS'),
ScaleIntensityRanged(keys=['image'], a_min=-57, a_max=164, b_min=0.0, b_max=1.0, clip=True),
# randomly crop out patch samples from big image based on pos / neg ratio
# the image centers of negative samples must be in valid image area
RandCropByPosNegLabeld(keys=['image', 'label'], label_key='label', size=(96, 96, 96), pos=1,
neg=1, num_samples=4, image_key='image', image_threshold=0),
# user can also add other random transforms
# RandAffined(keys=['image', 'label'], mode=('bilinear', 'nearest'), prob=1.0, spatial_size=(96, 96, 96),
# rotate_range=(0, 0, np.pi/15), scale_range=(0.1, 0.1, 0.1)),
ToTensord(keys=['image', 'label'])
])
val_transforms = Compose([
LoadNiftid(keys=['image', 'label']),
AddChanneld(keys=['image', 'label']),
Spacingd(keys=['image', 'label'], pixdim=(1.5, 1.5, 2.), interp_order=(3, 0)),
Orientationd(keys=['image', 'label'], axcodes='RAS'),
ScaleIntensityRanged(keys=['image'], a_min=-57, a_max=164, b_min=0.0, b_max=1.0, clip=True),
ToTensord(keys=['image', 'label'])
])
# set deterministic training for reproducibility
train_transforms.set_random_state(seed=0)
torch.manual_seed(0)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# we use cached datasets - these are 10x faster than regular datasets
self.train_ds = monai.data.CacheDataset(data=train_files, transform=train_transforms, cache_rate=1.0)
self.val_ds = monai.data.CacheDataset(data=val_files, transform=val_transforms, cache_rate=1.0)
#self.train_ds = monai.data.Dataset(data=train_files, transform=train_transforms)
#self.val_ds = monai.data.Dataset(data=val_files, transform=val_transforms)
def train_dataloader(self):
train_loader = DataLoader(self.train_ds, batch_size=2, shuffle=True, num_workers=4, collate_fn=list_data_collate)
return train_loader
def val_dataloader(self):
val_loader = DataLoader(self.val_ds, batch_size=1, num_workers=4)
return val_loader
def configure_optimizers(self):
optimizer = torch.optim.Adam(self._model.parameters(), 1e-4)
return optimizer
def training_step(self, batch, batch_idx):
images, labels = batch['image'], batch['label']
output = self.forward(images)
loss = self.loss_function(output, labels)
tensorboard_logs = {'train_loss': loss.item()}
return {'loss': loss, 'log': tensorboard_logs}
def validation_step(self, batch, batch_idx):
images, labels = batch['image'], batch['label']
roi_size = (160, 160, 160)
sw_batch_size = 4
outputs = sliding_window_inference(images, roi_size, sw_batch_size, self.forward)
loss = self.loss_function(outputs, labels)
value = compute_meandice(y_pred=outputs, y=labels, include_background=False,
to_onehot_y=True, mutually_exclusive=True)
return {'val_loss': loss, 'val_dice': value}
def validation_epoch_end(self, outputs):
val_dice = 0
num_items = 0
for output in outputs:
val_dice += output['val_dice'].sum().item()
num_items += len(output['val_dice'])
mean_val_dice = val_dice / num_items
tensorboard_logs = {'val_dice': mean_val_dice}
if mean_val_dice > self.best_val_dice:
self.best_val_dice = mean_val_dice
self.best_val_epoch = self.current_epoch
print('current epoch %d current mean dice: %0.4f best mean dice: %0.4f at epoch %d'
% (self.current_epoch, mean_val_dice, self.best_val_dice, self.best_val_epoch))
return {'log': tensorboard_logs}
```
## Run the training
```
# initialise the LightningModule
net = Net()
# set up loggers and checkpoints
tb_logger = loggers.TensorBoardLogger(save_dir='logs')
checkpoint_callback = ModelCheckpoint(filepath='logs/{epoch}-{val_loss:.2f}-{val_dice:.2f}')
# initialise Lightning's trainer.
trainer = Trainer(gpus=[0],
max_epochs=250,
logger=tb_logger,
checkpoint_callback=checkpoint_callback,
show_progress_bar=False,
num_sanity_val_steps=1
)
# train
trainer.fit(net)
print('train completed, best_metric: %0.4f at epoch %d' % (net.best_val_dice, net.best_val_epoch))
```
## View training in tensorboard
```
%load_ext tensorboard
%tensorboard --logdir='logs'
```
## Check best model output with the input image and label
```
net.eval()
device = torch.device("cuda:0")
with torch.no_grad():
for i, val_data in enumerate(net.val_dataloader()):
roi_size = (160, 160, 160)
sw_batch_size = 4
val_outputs = sliding_window_inference(val_data['image'].to(device), roi_size, sw_batch_size, net)
# plot the slice [:, :, 50]
plt.figure('check', (18, 6))
plt.subplot(1, 3, 1)
plt.title('image ' + str(i))
plt.imshow(val_data['image'][0, 0, :, :, 50], cmap='gray')
plt.subplot(1, 3, 2)
plt.title('label ' + str(i))
plt.imshow(val_data['label'][0, 0, :, :, 50])
plt.subplot(1, 3, 3)
plt.title('output ' + str(i))
plt.imshow(torch.argmax(val_outputs, dim=1).detach().cpu()[0, :, :, 50])
plt.show()
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from tqdm import tqdm as tqdm
%matplotlib inline
import torch
import torchvision
import torchvision.transforms as transforms
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import random
# from google.colab import drive
# drive.mount('/content/drive')
transform = transforms.Compose(
[transforms.CenterCrop((28,28)),transforms.ToTensor(),transforms.Normalize([0.5], [0.5])])
mnist_trainset = torchvision.datasets.MNIST(root='./data', train=True, download=True, transform=transform)
mnist_testset = torchvision.datasets.MNIST(root='./data', train=False, download=True, transform=transform)
index1 = [np.where(mnist_trainset.targets==0)[0] , np.where(mnist_trainset.targets==1)[0] ]
index1 = np.concatenate(index1,axis=0)
len(index1) #12665
true = 1000
total = 47000
sin = total-true
sin
epochs = 300
indices = np.random.choice(index1,true)
indices.shape
index = np.where(np.logical_and(mnist_trainset.targets!=0,mnist_trainset.targets!=1))[0] #47335
index.shape
req_index = np.random.choice(index.shape[0], sin, replace=False)
# req_index
index = index[req_index]
index.shape
values = np.random.choice([0,1],size= sin)
print(sum(values ==0),sum(values==1), sum(values ==0) + sum(values==1) )
mnist_trainset.data = torch.cat((mnist_trainset.data[indices],mnist_trainset.data[index]))
mnist_trainset.targets = torch.cat((mnist_trainset.targets[indices],torch.Tensor(values).type(torch.LongTensor)))
mnist_trainset.targets.shape, mnist_trainset.data.shape
# mnist_trainset.targets[index] = torch.Tensor(values).type(torch.LongTensor)
j =20078 # Without Shuffle upto True Training numbers correct , after that corrupted
print(plt.imshow(mnist_trainset.data[j]),mnist_trainset.targets[j])
trainloader = torch.utils.data.DataLoader(mnist_trainset, batch_size=250,shuffle=True, num_workers=2)
testloader = torch.utils.data.DataLoader(mnist_testset, batch_size=250,shuffle=False, num_workers=2)
mnist_trainset.data.shape
classes = ('zero', 'one')
dataiter = iter(trainloader)
images, labels = dataiter.next()
images[:4].shape
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
imshow(torchvision.utils.make_grid(images[:10]))
print('GroundTruth: ', ' '.join('%5s' % classes[labels[j]] for j in range(10)))
class Module2(nn.Module):
def __init__(self):
super(Module2, self).__init__()
self.conv1 = nn.Conv2d(1, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 4 * 4, 128)
self.fc2 = nn.Linear(128, 64)
self.fc3 = nn.Linear(64, 10)
self.fc4 = nn.Linear(10,2)
def forward(self,z):
y1 = self.pool(F.relu(self.conv1(z)))
y1 = self.pool(F.relu(self.conv2(y1)))
# print(y1.shape)
y1 = y1.view(-1, 16 * 4 * 4)
y1 = F.relu(self.fc1(y1))
y1 = F.relu(self.fc2(y1))
y1 = F.relu(self.fc3(y1))
y1 = self.fc4(y1)
return y1
inc = Module2()
inc = inc.to("cuda")
criterion_inception = nn.CrossEntropyLoss()
optimizer_inception = optim.SGD(inc.parameters(), lr=0.01, momentum=0.9)
acti = []
loss_curi = []
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs
inputs, labels = data
inputs, labels = inputs.to("cuda"),labels.to("cuda")
# print(inputs.shape)
# zero the parameter gradients
optimizer_inception.zero_grad()
# forward + backward + optimize
outputs = inc(inputs)
loss = criterion_inception(outputs, labels)
loss.backward()
optimizer_inception.step()
# print statistics
running_loss += loss.item()
if i % 50 == 49: # print every 50 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / 50))
ep_lossi.append(running_loss/50) # loss per minibatch
running_loss = 0.0
loss_curi.append(np.mean(ep_lossi)) #loss per epoch
if (np.mean(ep_lossi)<=0.03):
break
# acti.append(actis)
print('Finished Training')
correct = 0
total = 0
with torch.no_grad():
for data in trainloader:
images, labels = data
images, labels = images.to("cuda"), labels.to("cuda")
outputs = inc(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 60000 train images: %d %%' % (
100 * correct / total))
total,correct
correct = 0
total = 0
out = []
pred = []
with torch.no_grad():
for data in testloader:
images, labels = data
images, labels = images.to("cuda"),labels.to("cuda")
out.append(labels.cpu().numpy())
outputs= inc(images)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % (
100 * correct / total))
out = np.concatenate(out,axis=0)
pred = np.concatenate(pred,axis=0)
index = np.logical_or(out ==1,out==0)
print(index.shape)
acc = sum(out[index] == pred[index])/sum(index)
print('Accuracy of the network on the 10000 test images: %d %%' % (
100*acc))
sum(index)
import random
random.sample([1,2,3,4,5,6,7,8],5)
# torch.save(inc.state_dict(),"/content/drive/My Drive/model_simple_8000.pkl")
fig = plt.figure()
plt.plot(loss_curi,label="loss_Curve")
plt.xlabel("epochs")
plt.ylabel("training_loss")
plt.legend()
fig.savefig("loss_curve.pdf")
```
Simple Model 3 Inception Module
|true training data | Corr Training Data | Test Accuracy | Test Accuracy 0-1 |
| ------------------ | ------------------ | ------------- | ----------------- |
| 100 | 47335 | 15 | 75 |
| 500 | 47335 | 16 | 80 |
| 1000 | 47335 | 17 | 83 |
| 2000 | 47335 | 19 | 92 |
| 4000 | 47335 | 20 | 95 |
| 6000 | 47335 | 20 | 96 |
| 8000 | 47335 | 20 | 96 |
| 12665 | 47335 | 20 | 98 |
| Total Training Data | Training Accuracy |
|---------------------------- | ------------------------ |
| 47435 | 100 |
| 47835 | 100 |
| 48335 | 100 |
| 49335 | 100 |
| 51335 | 100 |
| 53335 | 100 |
| 55335 | 100 |
| 60000 | 100 |
Mini- Inception network 8 Inception Modules
|true training data | Corr Training Data | Test Accuracy | Test Accuracy 0-1 |
| ------------------ | ------------------ | ------------- | ----------------- |
| 100 | 47335 | 14 | 69 |
| 500 | 47335 | 19 | 90 |
| 1000 | 47335 | 19 | 92 |
| 2000 | 47335 | 20 | 95 |
| 4000 | 47335 | 20 | 97 |
| 6000 | 47335 | 20 | 97 |
| 8000 | 47335 | 20 | 98 |
| 12665 | 47335 | 20 | 99 |
| Total Training Data | Training Accuracy |
|---------------------------- | ------------------------ |
| 47435 | 100 |
| 47835 | 100 |
| 48335 | 100 |
| 49335 | 100 |
| 51335 | 100 |
| 53335 | 100 |
| 55335 | 100 |
| 60000 | 100 |
```
```
|
github_jupyter
|
# Binary classification from 2 features using K Nearest Neighbors (KNN)
Classification using "raw" python or libraries.
The binary classification is on a single boundary defined by a continuous function and added white noise
```
import numpy as np
from numpy import random
import matplotlib.pyplot as plt
import matplotlib.colors as pltcolors
from sklearn import metrics
from sklearn.neighbors import KNeighborsClassifier as SkKNeighborsClassifier
import pandas as pd
import seaborn as sns
```
## Model
Quadratic function as boundary between positive and negative values
Adding some unknown as a Gaussian noise
The values of X are uniformly distributed and independent
```
# Two features, Gaussian noise
def generateBatch(N):
#
xMin = 0
xMax = 1
b = 0.1
std = 0.1
#
x = random.uniform(xMin, xMax, (N, 2))
# 4th degree relation to shape the boundary
boundary = 2*(x[:,0]**4 + (x[:,0]-0.3)**3 + b)
# Adding some gaussian noise
labels = boundary + random.normal(0, std, N) > x[:,1]
return (x, labels)
```
### Training data
```
N = 2000
# x has 1 dim in R, label has 1 dim in B
xTrain, labelTrain = generateBatch(N)
colors = ['blue','red']
fig = plt.figure(figsize=(15,4))
plt.subplot(1,3,1)
plt.scatter(xTrain[:,0], xTrain[:,1], c=labelTrain, cmap=pltcolors.ListedColormap(colors), marker=',', alpha=0.1)
plt.xlabel('x0')
plt.ylabel('x1')
plt.title('Generated train data')
plt.grid()
cb = plt.colorbar()
loc = np.arange(0,1,1/float(len(colors)))
cb.set_ticks(loc)
cb.set_ticklabels([0,1])
plt.subplot(1,3,2)
plt.scatter(xTrain[:,0], labelTrain, marker=',', alpha=0.01)
plt.xlabel('x0')
plt.ylabel('label')
plt.grid()
plt.subplot(1,3,3)
plt.scatter(xTrain[:,1], labelTrain, marker=',', alpha=0.01)
plt.xlabel('x1')
plt.ylabel('label')
plt.grid()
count, bins, ignored = plt.hist(labelTrain*1.0, 10, density=True, alpha=0.5)
p = np.mean(labelTrain)
print('Bernouilli parameter of the distribution:', p)
```
### Test data for verification of the model
```
xTest, labelTest = generateBatch(N)
testColors = ['navy', 'orangered']
```
# Helpers
```
def plotHeatMap(X, classes, title=None, fmt='.2g', ax=None, xlabel=None, ylabel=None):
""" Fix heatmap plot from Seaborn with pyplot 3.1.0, 3.1.1
https://stackoverflow.com/questions/56942670/matplotlib-seaborn-first-and-last-row-cut-in-half-of-heatmap-plot
"""
ax = sns.heatmap(X, xticklabels=classes, yticklabels=classes, annot=True, fmt=fmt, cmap=plt.cm.Blues, ax=ax) #notation: "annot" not "annote"
bottom, top = ax.get_ylim()
ax.set_ylim(bottom + 0.5, top - 0.5)
if title:
ax.set_title(title)
if xlabel:
ax.set_xlabel(xlabel)
if ylabel:
ax.set_ylabel(ylabel)
def plotConfusionMatrix(yTrue, yEst, classes, title=None, fmt='.2g', ax=None):
plotHeatMap(metrics.confusion_matrix(yTrue, yEst), classes, title, fmt, ax, \
xlabel='Estimations', ylabel='True values');
```
# K Nearest Neighbors (KNN)
References:
- https://en.wikipedia.org/wiki/K-nearest_neighbors_algorithm
- https://machinelearningmastery.com/tutorial-to-implement-k-nearest-neighbors-in-python-from-scratch/
## Homemade
Using a simple algorithm.
Unweighted : each of the K neighbors has the same weight
```
# Select a K
k = 10
# Create a Panda dataframe in order to link x and y
df = pd.DataFrame(np.concatenate((xTrain, labelTrain.reshape(-1,1)), axis=1), columns = ('x0', 'x1', 'label'))
# Insert columns to compute the difference of current test to the train and the L2
df.insert(df.shape[1], 'diff0', 0)
df.insert(df.shape[1], 'diff1', 0)
df.insert(df.shape[1], 'L2', 0)
#
threshold = k / 2
labelEst0 = np.zeros(xTest.shape[0])
for i, x in enumerate(xTest):
# Compute distance and norm to each training sample
df['diff0'] = df['x0'] - x[0]
df['diff1'] = df['x1'] - x[1]
df['L2'] = df['diff0']**2 + df['diff1']**2
# Get the K lowest
kSmallest = df.nsmallest(k, 'L2')
# Finalize prediction based on the mean
labelEst0[i] = np.sum(kSmallest['label']) > threshold
```
### Performance of homemade model
```
plt.figure(figsize=(12,4))
plt.subplot(1,3,1)
plt.scatter(xTest[:,0], xTest[:,1], c=labelEst0, cmap=pltcolors.ListedColormap(testColors), marker='x', alpha=0.2);
plt.xlabel('x0')
plt.ylabel('x1')
plt.grid()
plt.title('Estimated')
cb = plt.colorbar()
loc = np.arange(0,1,1./len(testColors))
cb.set_ticks(loc)
cb.set_ticklabels([0,1]);
plt.subplot(1,3,2)
plt.hist(labelEst0, 10, density=True, alpha=0.5)
plt.title('Bernouilli parameter =' + str(np.mean(labelEst0)))
plt.subplot(1,3,3)
plt.scatter(xTest[:,0], xTest[:,1], c=labelTest, cmap=pltcolors.ListedColormap(colors), marker='x', alpha=0.1);
plt.xlabel('x0')
plt.ylabel('x1')
plt.grid()
plt.title('Generator')
cb = plt.colorbar()
loc = np.arange(0,1,1./len(colors))
cb.set_ticks(loc)
cb.set_ticklabels([0,1]);
accuracy0 = np.sum(labelTest == labelEst0)/N
print('Accuracy =', accuracy0)
```
### Precision
$p(y = 1 \mid \hat{y} = 1)$
```
print('Precision =', np.sum(labelTest[labelEst0 == 1])/np.sum(labelEst0))
```
### Recall
$p(\hat{y} = 1 \mid y = 1)$
```
print('Recall =', np.sum(labelTest[labelEst0 == 1])/np.sum(labelTest))
```
### Confusion matrix
```
plotConfusionMatrix(labelTest, labelEst0, np.array(['Blue', 'Red']));
print(metrics.classification_report(labelTest, labelEst0))
```
This non-parametric model has a the best performance of all models used so far, including the neural network with two layers.
The large drawback is the amount of computation for each sample to predict.
This method is hardly usable for sample sizes over 10k.
# Using SciKit Learn
References:
- SciKit documentation
- https://stackabuse.com/k-nearest-neighbors-algorithm-in-python-and-scikit-learn/
```
model1 = SkKNeighborsClassifier(n_neighbors=k)
model1.fit(xTrain, labelTrain)
labelEst1 = model1.predict(xTest)
print('Accuracy =', model1.score(xTest, labelTest))
plt.hist(labelEst1*1.0, 10, density=True, alpha=0.5)
plt.title('Bernouilli parameter =' + str(np.mean(labelEst1)));
```
### Confusion matrix (plot)
```
plotConfusionMatrix(labelTest, labelEst1, np.array(['Blue', 'Red']));
```
### Classification report
```
print(metrics.classification_report(labelTest, labelEst1))
```
### ROC curve
```
logit_roc_auc = metrics.roc_auc_score(labelTest, labelEst1)
fpr, tpr, thresholds = metrics.roc_curve(labelTest, model1.predict_proba(xTest)[:,1])
plt.plot(fpr, tpr, label='KNN classification (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right");
```
# Where to go from here ?
- Other linear implementations and simple neural nets using "raw" Python or SciKit Learn([HTML](ClassificationContinuous2Features.html) / [Jupyter](ClassificationContinuous2Features.ipynb)), using TensorFlow([HTML](ClassificationContinuous2Features-TensorFlow.html) / [Jupyter](ClassificationContinuous2Features-TensorFlow.ipynb)), or using Keras ([HTML](ClassificationContinuous2Features-Keras.html)/ [Jupyter](ClassificationContinuous2Features-Keras.ipynb))
- Non linear problem solving with Support Vector Machine (SVM) ([HTML](ClassificationSVM.html) / [Jupyter](ClassificationSVM.ipynb))
- More complex multi-class models on the Czech and Norways flags using Keras ([HTML](ClassificationMulti2Features-Keras.html) / [Jupyter](ClassificationMulti2Features-Keras.ipynb)), showing one of the main motivations to neural networks.
- Compare with the two feature linear regression using simple algorithms ([HTML](../linear/LinearRegressionBivariate.html) / [Jupyter](LinearRegressionBivariate.ipynb])), or using Keras ([HTML](LinearRegressionBivariate-Keras.html) / [Jupyter](LinearRegressionUnivariate-Keras.ipynb))
|
github_jupyter
|
<a href="https://colab.research.google.com/github/harnalashok/hadoop/blob/main/hadoop_spark_install_on_Colab.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
# Last amended: 30th March, 2021
# Myfolder: github/hadoop
# Objective:
# i) Install hadoop on colab
# (current version is 3.2.2)
# ii) Experiments with hadoop
# iii) Install spark on colab
# iv) Access hadoop file from spark
# v) Install koalas on colab
#
#
# Java 8 install: https://stackoverflow.com/a/58191107
# Hadoop install: https://hadoop.apache.org/docs/stable/hadoop-project-dist/hadoop-common/SingleCluster.html
# Spark install: https://stackoverflow.com/a/64183749
# https://www.analyticsvidhya.com/blog/2020/11/a-must-read-guide-on-how-to-work-with-pyspark-on-google-colab-for-data-scientists/
```
## Install hadoop
If it takes too long, it means, it is awaiting input from you regarding overwriting ssh keys
### Define functions
No downloads. Just function definitions
```
# 1.0 How to set environment variable
import os
import time
```
#### ssh_install()
```
# 2.0 Function to install ssh client and sshd (Server)
def ssh_install():
print("\n--1. Download and install ssh server----\n")
! sudo apt-get remove openssh-client openssh-server
! sudo apt install openssh-client openssh-server
print("\n--2. Restart ssh server----\n")
! service ssh restart
```
#### Java install
```
# 3.0 Function to download and install java 8
def install_java():
! rm -rf /usr/java
print("\n--Download and install Java 8----\n")
!apt-get install -y openjdk-8-jdk-headless -qq > /dev/null # install openjdk
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64" # set environment variable
!update-alternatives --set java /usr/lib/jvm/java-8-openjdk-amd64/jre/bin/java
!update-alternatives --set javac /usr/lib/jvm/java-8-openjdk-amd64/bin/javac
!mkdir -p /usr/java
! ln -s "/usr/lib/jvm/java-8-openjdk-amd64" "/usr/java"
! mv "/usr/java/java-8-openjdk-amd64" "/usr/java/latest"
!java -version #check java version
!javac -version
```
#### hadoop install
```
# 4.0 Function to download and install hadoop
def hadoop_install():
print("\n--5. Download hadoop tar.gz----\n")
! wget -c https://mirrors.estointernet.in/apache/hadoop/common/hadoop-3.2.2/hadoop-3.2.2.tar.gz
print("\n--6. Transfer downloaded content and unzip tar.gz----\n")
! mv /content/hadoop* /opt/
! tar -xzf /opt/hadoop-3.2.2.tar.gz --directory /opt/
print("\n--7. Create hadoop folder----\n")
! rm -r /app/hadoop/tmp
! mkdir -p /app/hadoop/tmp
print("\n--8. Check folder for files----\n")
! ls -la /opt
```
#### hadoop config
```
# 5.0 Function for setting hadoop configuration
def hadoop_config():
print("\n--Begin Configuring hadoop---\n")
print("\n=============================\n")
print("\n--9. core-site.xml----\n")
! cat /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
print("\n--10. Amend core-site.xml----\n")
! echo '<?xml version="1.0" encoding="UTF-8"?>' > /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo '<?xml-stylesheet type="text/xsl" href="configuration.xsl"?>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <name>fs.defaultFS</name>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <value>hdfs://localhost:9000</value>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <name>hadoop.tmp.dir</name>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <value>/app/hadoop/tmp</value>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <description>A base for other temporary directories.</description>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
# Added following regarding safemode from here:
# https://stackoverflow.com/a/33800253
! echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <name>dfs.safemode.threshold.pct</name>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' <value>0</value>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
! echo ' </configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
print("\n--11. Amended core-site.xml----\n")
! cat /opt/hadoop-3.2.2/etc/hadoop/core-site.xml
print("\n--12. yarn-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo '<?xml version="1.0" encoding="UTF-8"?>' > /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo '<configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <name>yarn.nodemanager.aux-services</name>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <value>mapreduce_shuffle</value>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <name>yarn.nodemanager.vmem-check-enabled</name>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' <value>false</value>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
!echo ' </configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
print("\n--13. Amended yarn-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/yarn-site.xml
print("\n--14. mapred-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
print("\n--15. Amend mapred-site.xml----\n")
!echo '<?xml version="1.0"?>' > /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo '<?xml-stylesheet type="text/xsl" href="configuration.xsl"?>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo '<configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <name>mapreduce.framework.name</name>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <value>yarn</value>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <name>yarn.app.mapreduce.am.env</name>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <value>HADOOP_MAPRED_HOME=${HADOOP_HOME}</value>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <name>mapreduce.map.env</name>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <value>HADOOP_MAPRED_HOME=${HADOOP_HOME}</value>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <name>mapreduce.reduce.env</name>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' <value>HADOOP_MAPRED_HOME=${HADOOP_HOME}</value>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
!echo '</configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
print("\n--16, Amended mapred-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/mapred-site.xml
print("\n---17. hdfs-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
print("\n---18. Amend hdfs-site.xml----\n")
!echo '<?xml version="1.0" encoding="UTF-8"?> ' > /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo '<?xml-stylesheet type="text/xsl" href="configuration.xsl"?>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo '<configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <name>dfs.replication</name>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <value>1</value>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <property>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <name>dfs.block.size</name>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <value>16777216</value>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' <description>Block size</description>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo ' </property>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
!echo '</configuration>' >> /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
print("\n---19. Amended hdfs-site.xml----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/hdfs-site.xml
print("\n---20. hadoop-env.sh----\n")
# https://stackoverflow.com/a/53140448
!cat /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export JAVA_HOME="/usr/lib/jvm/java-8-openjdk-amd64"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export HDFS_NAMENODE_USER="root"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export HDFS_DATANODE_USER="root"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export HDFS_SECONDARYNAMENODE_USER="root"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export YARN_RESOURCEMANAGER_USER="root"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
! echo 'export YARN_NODEMANAGER_USER="root"' >> /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
print("\n---21. Amended hadoop-env.sh----\n")
!cat /opt/hadoop-3.2.2/etc/hadoop/hadoop-env.sh
```
#### ssh keys
```
# 6.0 Function tp setup ssh passphrase
def set_keys():
print("\n---22. Generate SSH keys----\n")
! cd ~ ; pwd
! cd ~ ; ssh-keygen -t rsa -P '' -f ~/.ssh/id_rsa
! cd ~ ; cat ~/.ssh/id_rsa.pub >> ~/.ssh/authorized_keys
! cd ~ ; chmod 0600 ~/.ssh/authorized_keys
```
#### Set environment
```
# 7.0 Function to set up environmental variables
def set_env():
print("\n---23. Set Environment variables----\n")
# 'export' command does not work in colab
# https://stackoverflow.com/a/57240319
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64" #set environment variable
os.environ["JRE_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64/jre"
os.environ["HADOOP_HOME"] = "/opt/hadoop-3.2.2"
os.environ["HADOOP_CONF_DIR"] = "/opt/hadoop-3.2.2/etc/hadoop"
os.environ["LD_LIBRARY_PATH"] += ":/opt/hadoop-3.2.2/lib/native"
os.environ["PATH"] += ":/opt/hadoop-3.2.2/bin:/opt/hadoop-3.2.2/sbin"
```
#### Install all function
```
# 8.0 Function to call all functions
def install_hadoop():
print("\n--Install java----\n")
ssh_install()
install_java()
hadoop_install()
hadoop_config()
set_keys()
set_env()
```
### Begin install
Start downloading, install and configure. Takes around 2 minutes
```
# 9.0 Start installation
start = time.time()
install_hadoop()
end = time.time()
print("\n---Time taken----\n")
print((end- start)/60)
```
### Format hadoop
```
# 10.0 Format hadoop
print("\n---24. Format namenode----\n")
!hdfs namenode -format
```
## Start and test hadoop
If namenode is in safemode, use the command:
`!hdfs dfsadmin -safemode leave`
#### Start hadoop
If start fails with 'Connection refused', run `ssh_install()` once again
```
# 11.0 Start namenode
# If this fails, run
# ssh_install() below
# and start hadoop again:
print("\n---25. Start namenode----\n")
! start-dfs.sh
#ssh_install()
```
#### Start yarn
```
# 11.1 Start yarn
! start-yarn.sh
```
If `start-dfs.sh` fails, issue the following three commands, one after another:<br>
`! sudo apt-get remove openssh-client openssh-server`<br>
`! sudo apt-get install openssh-client openssh-server`<br>
`! service ssh restart`<br>
And then try to start hadoop again, as: `start-dfs.sh`
#### Test hadoop
IF in safe mode, leave safe mode as:<br>
`!hdfs dfsadmin -safemode leave`
```
# 11.1
print("\n---26. Make folders in hadoop----\n")
! hdfs dfs -mkdir /user
! hdfs dfs -mkdir /user/ashok
# 11.2 Run hadoop commands
! hdfs dfs -ls /
! hdfs dfs -ls /user
# 11.3 Stopping hadoop
# Gives some errors
# But hadoop stops
#!stop-dfs.sh
```
Run the `ssh_install()` again if hadoop fails to start with `start-dfs.sh` and then try to start hadoop again.
## Install spark
### Define functions
`findspark`: PySpark isn't on `sys.path` by default, but that doesn't mean it can't be used as a regular library. You can address this by either symlinking pyspark into your site-packages, or adding `pyspark` to `sys.path` at runtime. `findspark` does the latter.
```
# 1.0 Function to download and unzip spark
def spark_koalas_install():
print("\n--1.1 Install findspark----\n")
!pip install -q findspark
print("\n--1.2 Install databricks Koalas----\n")
!pip install koalas
print("\n--1.3 Download Apache tar.gz----\n")
! wget -c https://mirrors.estointernet.in/apache/spark/spark-3.1.1/spark-3.1.1-bin-hadoop3.2.tgz
print("\n--1.4 Transfer downloaded content and unzip tar.gz----\n")
! mv /content/spark* /opt/
! tar -xzf /opt/spark-3.1.1-bin-hadoop3.2.tgz --directory /opt/
print("\n--1.5 Check folder for files----\n")
! ls -la /opt
# 1.1 Function to set environment
def set_spark_env():
print("\n---2. Set Environment variables----\n")
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["JRE_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64/jre"
os.environ["SPARK_HOME"] = "/opt/spark-3.1.1-bin-hadoop3.2"
os.environ["LD_LIBRARY_PATH"] += ":/opt/spark-3.1.1-bin-hadoop3.2/lib/native"
os.environ["PATH"] += ":/opt/spark-3.1.1-bin-hadoop3.2/bin:/opt/spark-3.1.1-bin-hadoop3.2/sbin"
print("\n---2.1. Check Environment variables----\n")
# Check
! echo $PATH
! echo $LD_LIBRARY_PATH
# 1.2 Function to configure spark
def spark_conf():
print("\n---3. Configure spark to access hadoop----\n")
!mv /opt/spark-3.1.1-bin-hadoop3.2/conf/spark-env.sh.template /opt/spark-3.1.1-bin-hadoop3.2/conf/spark-env.sh
!echo "HADOOP_CONF_DIR=/opt/hadoop-3.2.2/etc/hadoop/" >> /opt/spark-3.1.1-bin-hadoop3.2/conf/spark-env.sh
print("\n---3.1 Check ----\n")
#!cat /opt/spark-3.1.1-bin-hadoop3.2/conf/spark-env.sh
```
### Install spark
```
# 2.0 Call all the three functions
def install_spark():
spark_koalas_install()
set_spark_env()
spark_conf()
# 2.1
install_spark()
```
## Test spark
Hadoop should have been started
Call some libraries
```
# 3.0 Just call some libraries to test
import pandas as pd
import numpy as np
# 3.1 Get spark in sys.path
import findspark
findspark.init()
# 3.2 Call other spark libraries
# Just to test
from pyspark.sql import SparkSession
import databricks.koalas as ks
from pyspark.ml.feature import VectorAssembler
from pyspark.ml.regression import LinearRegression
# 3.1 Build spark session
spark = SparkSession. \
builder. \
master("local[*]"). \
getOrCreate()
# 4.0 Pandas DataFrame
pdf = pd.DataFrame({
'x1': ['a','a','b','b', 'b', 'c', 'd','d'],
'x2': ['apple', 'orange', 'orange','orange', 'peach', 'peach','apple','orange'],
'x3': [1, 1, 2, 2, 2, 4, 1, 2],
'x4': [2.4, 2.5, 3.5, 1.4, 2.1,1.5, 3.0, 2.0],
'y1': [1, 0, 1, 0, 0, 1, 1, 0],
'y2': ['yes', 'no', 'no', 'yes', 'yes', 'yes', 'no', 'yes']
})
# 4.1
pdf
# 4.2 Transform to Spark DataFrame
df = spark.createDataFrame(pdf)
df.show()
# 4.3 Create a csv file
# and tranfer it to hdfs
!echo "a,b,c,d" > /content/airports.csv
!echo "5,4,6,7" >> /content/airports.csv
!echo "2,3,4,5" >> /content/airports.csv
!echo "8,9,0,1" >> /content/airports.csv
!echo "2,3,4,1" >> /content/airports.csv
!echo "1,2,2,1" >> /content/airports.csv
!echo "0,1,2,6" >> /content/airports.csv
!echo "9,3,1,8" >> /content/airports.csv
!ls -la /content
# 4.4
!hdfs dfs -rm -f /user/ashok/airports.csv
!hdfs dfs -put /content/airports.csv /user/ashok/
!hdfs dfs -ls /user/ashok
# 5.0 Read file directly from hadoop
airports_df = spark.read.csv(
"/user/ashok/airports.csv",
inferSchema = True,
header = True
)
# 5.1 Show file
airports_df.show()
```
## Test Koalas
Hadoop should have been started
Create a koalas dataframe
```
# 6.0
# If namenode is in safemode, first use:
# hdfs dfsadmin -safemode leave
kdf = ks.DataFrame(
{
'a': [1, 2, 3, 4, 5, 6],
'b': [100, 200, 300, 400, 500, 600],
'c': ["one", "two", "three", "four", "five", "six"]
},
index=[10, 20, 30, 40, 50, 60]
)
# 6.1 And show
kdf
# 6.2 Pandas DataFrame
pdf = pd.DataFrame({'x':range(3), 'y':['a','b','b'], 'z':['a','b','b']})
# 6.2.1 Transform to koalas DataFrame
df = ks.from_pandas(pdf)
# 6.3 Rename koalas dataframe columns
df.columns = ['x', 'y', 'z1']
# 6.4 Do some operations on koalas DF, in place:
df['x2'] = df.x * df.x
# 6.6 Finally show koalas df
df
# 6.7 Read csv file from hadoop
# and create koalas df
ks.read_csv("/user/ashok/airports.csv").head(10)
###################
```
|
github_jupyter
|
# Comprehensive Example
```
# Enabling the `widget` backend.
# This requires jupyter-matplotlib a.k.a. ipympl.
# ipympl can be install via pip or conda.
%matplotlib widget
import matplotlib.pyplot as plt
import numpy as np
# Testing matplotlib interactions with a simple plot
fig = plt.figure()
plt.plot(np.sin(np.linspace(0, 20, 100)));
# Always hide the toolbar
fig.canvas.toolbar_visible = False
# Put it back to its default
fig.canvas.toolbar_visible = 'fade-in-fade-out'
# Change the toolbar position
fig.canvas.toolbar_position = 'top'
# Hide the Figure name at the top of the figure
fig.canvas.header_visible = False
# Hide the footer
fig.canvas.footer_visible = False
# Disable the resizing feature
fig.canvas.resizable = False
# If true then scrolling while the mouse is over the canvas will not move the entire notebook
fig.canvas.capture_scroll = True
```
You can also call `display` on `fig.canvas` to display the interactive plot anywhere in the notebooke
```
fig.canvas.toolbar_visible = True
display(fig.canvas)
```
Or you can `display(fig)` to embed the current plot as a png
```
display(fig)
```
# 3D plotting
```
from mpl_toolkits.mplot3d import axes3d
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
# Grab some test data.
X, Y, Z = axes3d.get_test_data(0.05)
# Plot a basic wireframe.
ax.plot_wireframe(X, Y, Z, rstride=10, cstride=10)
plt.show()
```
# Subplots
```
# A more complex example from the matplotlib gallery
np.random.seed(0)
n_bins = 10
x = np.random.randn(1000, 3)
fig, axes = plt.subplots(nrows=2, ncols=2)
ax0, ax1, ax2, ax3 = axes.flatten()
colors = ['red', 'tan', 'lime']
ax0.hist(x, n_bins, density=1, histtype='bar', color=colors, label=colors)
ax0.legend(prop={'size': 10})
ax0.set_title('bars with legend')
ax1.hist(x, n_bins, density=1, histtype='bar', stacked=True)
ax1.set_title('stacked bar')
ax2.hist(x, n_bins, histtype='step', stacked=True, fill=False)
ax2.set_title('stack step (unfilled)')
# Make a multiple-histogram of data-sets with different length.
x_multi = [np.random.randn(n) for n in [10000, 5000, 2000]]
ax3.hist(x_multi, n_bins, histtype='bar')
ax3.set_title('different sample sizes')
fig.tight_layout()
plt.show()
fig.canvas.toolbar_position = 'right'
fig.canvas.toolbar_visible = False
```
# Interactions with other widgets and layouting
When you want to embed the figure into a layout of other widgets you should call `plt.ioff()` before creating the figure otherwise `plt.figure()` will trigger a display of the canvas automatically and outside of your layout.
### Without using `ioff`
Here we will end up with the figure being displayed twice. The button won't do anything it just placed as an example of layouting.
```
import ipywidgets as widgets
# ensure we are interactive mode
# this is default but if this notebook is executed out of order it may have been turned off
plt.ion()
fig = plt.figure()
ax = fig.gca()
ax.imshow(Z)
widgets.AppLayout(
center=fig.canvas,
footer=widgets.Button(icon='check'),
pane_heights=[0, 6, 1]
)
```
### Fixing the double display with `ioff`
If we make sure interactive mode is off when we create the figure then the figure will only display where we want it to.
There is ongoing work to allow usage of `ioff` as a context manager, see the [ipympl issue](https://github.com/matplotlib/ipympl/issues/220) and the [matplotlib issue](https://github.com/matplotlib/matplotlib/issues/17013)
```
plt.ioff()
fig = plt.figure()
plt.ion()
ax = fig.gca()
ax.imshow(Z)
widgets.AppLayout(
center=fig.canvas,
footer=widgets.Button(icon='check'),
pane_heights=[0, 6, 1]
)
```
# Interacting with other widgets
## Changing a line plot with a slide
```
# When using the `widget` backend from ipympl,
# fig.canvas is a proper Jupyter interactive widget, which can be embedded in
# an ipywidgets layout. See https://ipywidgets.readthedocs.io/en/stable/examples/Layout%20Templates.html
# One can bound figure attributes to other widget values.
from ipywidgets import AppLayout, FloatSlider
plt.ioff()
slider = FloatSlider(
orientation='horizontal',
description='Factor:',
value=1.0,
min=0.02,
max=2.0
)
slider.layout.margin = '0px 30% 0px 30%'
slider.layout.width = '40%'
fig = plt.figure()
fig.canvas.header_visible = False
fig.canvas.layout.min_height = '400px'
plt.title('Plotting: y=sin({} * x)'.format(slider.value))
x = np.linspace(0, 20, 500)
lines = plt.plot(x, np.sin(slider.value * x))
def update_lines(change):
plt.title('Plotting: y=sin({} * x)'.format(change.new))
lines[0].set_data(x, np.sin(change.new * x))
fig.canvas.draw()
fig.canvas.flush_events()
slider.observe(update_lines, names='value')
AppLayout(
center=fig.canvas,
footer=slider,
pane_heights=[0, 6, 1]
)
```
## Update image data in a performant manner
Two useful tricks to improve performance when updating an image displayed with matplolib are to:
1. Use the `set_data` method instead of calling imshow
2. Precompute and then index the array
```
# precomputing all images
x = np.linspace(0,np.pi,200)
y = np.linspace(0,10,200)
X,Y = np.meshgrid(x,y)
parameter = np.linspace(-5,5)
example_image_stack = np.sin(X)[None,:,:]+np.exp(np.cos(Y[None,:,:]*parameter[:,None,None]))
plt.ioff()
fig = plt.figure()
plt.ion()
im = plt.imshow(example_image_stack[0])
def update(change):
im.set_data(example_image_stack[change['new']])
fig.canvas.draw_idle()
slider = widgets.IntSlider(value=0, min=0, max=len(parameter)-1)
slider.observe(update, names='value')
widgets.VBox([slider, fig.canvas])
```
### Debugging widget updates and matplotlib callbacks
If an error is raised in the `update` function then will not always display in the notebook which can make debugging difficult. This same issue is also true for matplotlib callbacks on user events such as mousemovement, for example see [issue](https://github.com/matplotlib/ipympl/issues/116). There are two ways to see the output:
1. In jupyterlab the output will show up in the Log Console (View > Show Log Console)
2. using `ipywidgets.Output`
Here is an example of using an `Output` to capture errors in the update function from the previous example. To induce errors we changed the slider limits so that out of bounds errors will occur:
From: `slider = widgets.IntSlider(value=0, min=0, max=len(parameter)-1)`
To: `slider = widgets.IntSlider(value=0, min=0, max=len(parameter)+10)`
If you move the slider all the way to the right you should see errors from the Output widget
```
plt.ioff()
fig = plt.figure()
plt.ion()
im = plt.imshow(example_image_stack[0])
out = widgets.Output()
@out.capture()
def update(change):
with out:
if change['name'] == 'value':
im.set_data(example_image_stack[change['new']])
fig.canvas.draw_idle
slider = widgets.IntSlider(value=0, min=0, max=len(parameter)+10)
slider.observe(update)
display(widgets.VBox([slider, fig.canvas]))
display(out)
```
|
github_jupyter
|
# Interactive single compartment HH example
To run this interactive Jupyter Notebook, please click on the rocket icon 🚀 in the top panel. For more information, please see {ref}`how to use this documentation <userdocs:usage:jupyterbooks>`. Please uncomment the line below if you use the Google Colab. (It does not include these packages by default).
```
#%pip install pyneuroml neuromllite NEURON
import math
from neuroml import NeuroMLDocument
from neuroml import Cell
from neuroml import IonChannelHH
from neuroml import GateHHRates
from neuroml import BiophysicalProperties
from neuroml import MembraneProperties
from neuroml import ChannelDensity
from neuroml import HHRate
from neuroml import SpikeThresh
from neuroml import SpecificCapacitance
from neuroml import InitMembPotential
from neuroml import IntracellularProperties
from neuroml import IncludeType
from neuroml import Resistivity
from neuroml import Morphology, Segment, Point3DWithDiam
from neuroml import Network, Population
from neuroml import PulseGenerator, ExplicitInput
import numpy as np
from pyneuroml import pynml
from pyneuroml.lems import LEMSSimulation
```
## Declare the model
### Create ion channels
```
def create_na_channel():
"""Create the Na channel.
This will create the Na channel and save it to a file.
It will also validate this file.
returns: name of the created file
"""
na_channel = IonChannelHH(id="na_channel", notes="Sodium channel for HH cell", conductance="10pS", species="na")
gate_m = GateHHRates(id="na_m", instances="3", notes="m gate for na channel")
m_forward_rate = HHRate(type="HHExpLinearRate", rate="1per_ms", midpoint="-40mV", scale="10mV")
m_reverse_rate = HHRate(type="HHExpRate", rate="4per_ms", midpoint="-65mV", scale="-18mV")
gate_m.forward_rate = m_forward_rate
gate_m.reverse_rate = m_reverse_rate
na_channel.gate_hh_rates.append(gate_m)
gate_h = GateHHRates(id="na_h", instances="1", notes="h gate for na channel")
h_forward_rate = HHRate(type="HHExpRate", rate="0.07per_ms", midpoint="-65mV", scale="-20mV")
h_reverse_rate = HHRate(type="HHSigmoidRate", rate="1per_ms", midpoint="-35mV", scale="10mV")
gate_h.forward_rate = h_forward_rate
gate_h.reverse_rate = h_reverse_rate
na_channel.gate_hh_rates.append(gate_h)
na_channel_doc = NeuroMLDocument(id="na_channel", notes="Na channel for HH neuron")
na_channel_fn = "HH_example_na_channel.nml"
na_channel_doc.ion_channel_hhs.append(na_channel)
pynml.write_neuroml2_file(nml2_doc=na_channel_doc, nml2_file_name=na_channel_fn, validate=True)
return na_channel_fn
def create_k_channel():
"""Create the K channel
This will create the K channel and save it to a file.
It will also validate this file.
:returns: name of the K channel file
"""
k_channel = IonChannelHH(id="k_channel", notes="Potassium channel for HH cell", conductance="10pS", species="k")
gate_n = GateHHRates(id="k_n", instances="4", notes="n gate for k channel")
n_forward_rate = HHRate(type="HHExpLinearRate", rate="0.1per_ms", midpoint="-55mV", scale="10mV")
n_reverse_rate = HHRate(type="HHExpRate", rate="0.125per_ms", midpoint="-65mV", scale="-80mV")
gate_n.forward_rate = n_forward_rate
gate_n.reverse_rate = n_reverse_rate
k_channel.gate_hh_rates.append(gate_n)
k_channel_doc = NeuroMLDocument(id="k_channel", notes="k channel for HH neuron")
k_channel_fn = "HH_example_k_channel.nml"
k_channel_doc.ion_channel_hhs.append(k_channel)
pynml.write_neuroml2_file(nml2_doc=k_channel_doc, nml2_file_name=k_channel_fn, validate=True)
return k_channel_fn
def create_leak_channel():
"""Create a leak channel
This will create the leak channel and save it to a file.
It will also validate this file.
:returns: name of leak channel nml file
"""
leak_channel = IonChannelHH(id="leak_channel", conductance="10pS", notes="Leak conductance")
leak_channel_doc = NeuroMLDocument(id="leak_channel", notes="leak channel for HH neuron")
leak_channel_fn = "HH_example_leak_channel.nml"
leak_channel_doc.ion_channel_hhs.append(leak_channel)
pynml.write_neuroml2_file(nml2_doc=leak_channel_doc, nml2_file_name=leak_channel_fn, validate=True)
return leak_channel_fn
```
### Create cell
```
def create_cell():
"""Create the cell.
:returns: name of the cell nml file
"""
# Create the nml file and add the ion channels
hh_cell_doc = NeuroMLDocument(id="cell", notes="HH cell")
hh_cell_fn = "HH_example_cell.nml"
hh_cell_doc.includes.append(IncludeType(href=create_na_channel()))
hh_cell_doc.includes.append(IncludeType(href=create_k_channel()))
hh_cell_doc.includes.append(IncludeType(href=create_leak_channel()))
# Define a cell
hh_cell = Cell(id="hh_cell", notes="A single compartment HH cell")
# Define its biophysical properties
bio_prop = BiophysicalProperties(id="hh_b_prop")
# notes="Biophysical properties for HH cell")
# Membrane properties are a type of biophysical properties
mem_prop = MembraneProperties()
# Add membrane properties to the biophysical properties
bio_prop.membrane_properties = mem_prop
# Append to cell
hh_cell.biophysical_properties = bio_prop
# Channel density for Na channel
na_channel_density = ChannelDensity(id="na_channels", cond_density="120.0 mS_per_cm2", erev="50.0 mV", ion="na", ion_channel="na_channel")
mem_prop.channel_densities.append(na_channel_density)
# Channel density for k channel
k_channel_density = ChannelDensity(id="k_channels", cond_density="360 S_per_m2", erev="-77mV", ion="k", ion_channel="k_channel")
mem_prop.channel_densities.append(k_channel_density)
# Leak channel
leak_channel_density = ChannelDensity(id="leak_channels", cond_density="3.0 S_per_m2", erev="-54.3mV", ion="non_specific", ion_channel="leak_channel")
mem_prop.channel_densities.append(leak_channel_density)
# Other membrane properties
mem_prop.spike_threshes.append(SpikeThresh(value="-20mV"))
mem_prop.specific_capacitances.append(SpecificCapacitance(value="1.0 uF_per_cm2"))
mem_prop.init_memb_potentials.append(InitMembPotential(value="-65mV"))
intra_prop = IntracellularProperties()
intra_prop.resistivities.append(Resistivity(value="0.03 kohm_cm"))
# Add to biological properties
bio_prop.intracellular_properties = intra_prop
# Morphology
morph = Morphology(id="hh_cell_morph")
# notes="Simple morphology for the HH cell")
seg = Segment(id="0", name="soma", notes="Soma segment")
# We want a diameter such that area is 1000 micro meter^2
# surface area of a sphere is 4pi r^2 = 4pi diam^2
diam = math.sqrt(1000 / math.pi)
proximal = distal = Point3DWithDiam(x="0", y="0", z="0", diameter=str(diam))
seg.proximal = proximal
seg.distal = distal
morph.segments.append(seg)
hh_cell.morphology = morph
hh_cell_doc.cells.append(hh_cell)
pynml.write_neuroml2_file(nml2_doc=hh_cell_doc, nml2_file_name=hh_cell_fn, validate=True)
return hh_cell_fn
```
### Create a network
```
def create_network():
"""Create the network
:returns: name of network nml file
"""
net_doc = NeuroMLDocument(id="network",
notes="HH cell network")
net_doc_fn = "HH_example_net.nml"
net_doc.includes.append(IncludeType(href=create_cell()))
# Create a population: convenient to create many cells of the same type
pop = Population(id="pop0", notes="A population for our cell", component="hh_cell", size=1)
# Input
pulsegen = PulseGenerator(id="pg", notes="Simple pulse generator", delay="100ms", duration="100ms", amplitude="0.08nA")
exp_input = ExplicitInput(target="pop0[0]", input="pg")
net = Network(id="single_hh_cell_network", note="A network with a single population")
net_doc.pulse_generators.append(pulsegen)
net.explicit_inputs.append(exp_input)
net.populations.append(pop)
net_doc.networks.append(net)
pynml.write_neuroml2_file(nml2_doc=net_doc, nml2_file_name=net_doc_fn, validate=True)
return net_doc_fn
```
## Plot the data we record
```
def plot_data(sim_id):
"""Plot the sim data.
Load the data from the file and plot the graph for the membrane potential
using the pynml generate_plot utility function.
:sim_id: ID of simulaton
"""
data_array = np.loadtxt(sim_id + ".dat")
pynml.generate_plot([data_array[:, 0]], [data_array[:, 1]], "Membrane potential", show_plot_already=False, save_figure_to=sim_id + "-v.png", xaxis="time (s)", yaxis="membrane potential (V)")
pynml.generate_plot([data_array[:, 0]], [data_array[:, 2]], "channel current", show_plot_already=False, save_figure_to=sim_id + "-i.png", xaxis="time (s)", yaxis="channel current (A)")
pynml.generate_plot([data_array[:, 0], data_array[:, 0]], [data_array[:, 3], data_array[:, 4]], "current density", labels=["Na", "K"], show_plot_already=False, save_figure_to=sim_id + "-iden.png", xaxis="time (s)", yaxis="current density (A_per_m2)")
```
## Create and run the simulation
Create the simulation, run it, record data, and plot the recorded information.
```
def main():
"""Main function
Include the NeuroML model into a LEMS simulation file, run it, plot some
data.
"""
# Simulation bits
sim_id = "HH_single_compartment_example_sim"
simulation = LEMSSimulation(sim_id=sim_id, duration=300, dt=0.01, simulation_seed=123)
# Include the NeuroML model file
simulation.include_neuroml2_file(create_network())
# Assign target for the simulation
simulation.assign_simulation_target("single_hh_cell_network")
# Recording information from the simulation
simulation.create_output_file(id="output0", file_name=sim_id + ".dat")
simulation.add_column_to_output_file("output0", column_id="pop0[0]/v", quantity="pop0[0]/v")
simulation.add_column_to_output_file("output0", column_id="pop0[0]/iChannels", quantity="pop0[0]/iChannels")
simulation.add_column_to_output_file("output0", column_id="pop0[0]/na/iDensity", quantity="pop0[0]/hh_b_prop/membraneProperties/na_channels/iDensity/")
simulation.add_column_to_output_file("output0", column_id="pop0[0]/k/iDensity", quantity="pop0[0]/hh_b_prop/membraneProperties/k_channels/iDensity/")
# Save LEMS simulation to file
sim_file = simulation.save_to_file()
# Run the simulation using the default jNeuroML simulator
pynml.run_lems_with_jneuroml(sim_file, max_memory="2G", nogui=True, plot=False)
# Plot the data
plot_data(sim_id)
if __name__ == "__main__":
main()
```
|
github_jupyter
|
## Amazon SageMaker Feature Store: Encrypt Data in your Online or Offline Feature Store using KMS key
This notebook demonstrates how to enable encyption for your data in your online or offline Feature Store using KMS key. We start by showing how to programmatically create a KMS key, and how to apply it to the feature store creation process for data encryption. The last portion of this notebook demonstrates how to verify that your KMS key is being used to encerypt your data in your feature store.
### Overview
1. Create a KMS key.
- How to create a KMS key programmatically using the KMS client from boto3?
2. Attach role to your KMS key.
- Attach the required entries to your policy for data encryption in your feature store.
3. Create an online or offline feature store and apply it to your feature store creation process.
- How to enable encryption for your online store?
- How to enable encryption for your offline store?
4. How to verify that your data is encrypted in your online or offline store?
### Prerequisites
This notebook uses both `boto3` and Python SDK libraries, and the `Python 3 (Data Science)` kernel. This notebook also works with Studio, Jupyter, and JupyterLab.
### Library Dependencies:
* sagemaker>=2.0.0
* numpy
* pandas
```
import sagemaker
import sys
import boto3
import pandas as pd
import numpy as np
import json
original_version = sagemaker.__version__
%pip install 'sagemaker>=2.0.0'
```
### Set up
```
sagemaker_session = sagemaker.Session()
s3_bucket_name = sagemaker_session.default_bucket()
prefix = "sagemaker-featurestore-kms-demo"
role = sagemaker.get_execution_role()
region = sagemaker_session.boto_region_name
```
Create a KMS client using boto3. Note that you can access your boto session through your sagemaker session, e.g.,`sagemaker_session`.
```
kms = sagemaker_session.boto_session.client("kms")
```
### KMS Policy Template
Below is the policy template you will use for creating a KMS key. You will specify your role to grant it access to various KMS operations that will be used in the back-end for encrypting your data in your Online or Offline Feature Store.
**Note**: You will need to substitute your Account number in for `123456789012` in the policy below for these lines: `arn:aws:cloudtrail:*:123456789012:trail/*`.
It is important to understand that the policy below will grant admin privileges for Customer Managed Keys (CMK) around viewing and revoking grants, decrypt and encrypt permissions on CloudTrail and full access permissions through Feature Store. Also, note that the the Feature Store Service creates additonal grants that are used for encryption purposes for your online store.
```
policy = {
"Version": "2012-10-17",
"Id": "key-policy-feature-store",
"Statement": [
{
"Sid": "Allow access through Amazon SageMaker Feature Store for all principals in the account that are authorized to use Amazon SageMaker Feature Store",
"Effect": "Allow",
"Principal": {"AWS": role},
"Action": [
"kms:Encrypt",
"kms:Decrypt",
"kms:DescribeKey",
"kms:CreateGrant",
"kms:RetireGrant",
"kms:ReEncryptFrom",
"kms:ReEncryptTo",
"kms:GenerateDataKey",
"kms:ListAliases",
"kms:ListGrants",
],
"Resource": ["*"],
"Condition": {"StringLike": {"kms:ViaService": "sagemaker.*.amazonaws.com"}},
},
{
"Sid": "Allow administrators to view the CMK and revoke grants",
"Effect": "Allow",
"Principal": {"AWS": [role]},
"Action": ["kms:Describe*", "kms:Get*", "kms:List*", "kms:RevokeGrant"],
"Resource": ["*"],
},
{
"Sid": "Enable CloudTrail Encrypt Permissions",
"Effect": "Allow",
"Principal": {"Service": "cloudtrail.amazonaws.com", "AWS": [role]},
"Action": "kms:GenerateDataKey*",
"Resource": "*",
"Condition": {
"StringLike": {
"kms:EncryptionContext:aws:cloudtrail:arn": [
"arn:aws:cloudtrail:*:123456789012:trail/*",
"arn:aws:cloudtrail:*:123456789012:trail/*",
]
}
},
},
{
"Sid": "Enable CloudTrail log decrypt permissions",
"Effect": "Allow",
"Principal": {"AWS": [role]},
"Action": "kms:Decrypt",
"Resource": ["*"],
"Condition": {"Null": {"kms:EncryptionContext:aws:cloudtrail:arn": "false"}},
},
],
}
```
Create your new KMS key using the policy above and your KMS client.
```
try:
new_kms_key = kms.create_key(
Policy=json.dumps(policy),
Description="string",
KeyUsage="ENCRYPT_DECRYPT",
CustomerMasterKeySpec="SYMMETRIC_DEFAULT",
Origin="AWS_KMS",
)
AliasName = "my-new-kms-key" ## provide a unique alias name
kms.create_alias(
AliasName="alias/" + AliasName, TargetKeyId=new_kms_key["KeyMetadata"]["KeyId"]
)
print(new_kms_key)
except Exception as e:
print("Error {}".format(e))
```
Now that we have our KMS key created and the necessary operations added to our role, we now load in our data.
```
customer_data = pd.read_csv("data/feature_store_introduction_customer.csv")
orders_data = pd.read_csv("data/feature_store_introduction_orders.csv")
customer_data.head()
orders_data.head()
customer_data.dtypes
orders_data.dtypes
```
### Creating Feature Groups
We first start by creating feature group names for customer_data and orders_data. Following this, we create two Feature Groups, one for customer_dat and another for orders_data
```
from time import gmtime, strftime, sleep
customers_feature_group_name = "customers-feature-group-" + strftime("%d-%H-%M-%S", gmtime())
orders_feature_group_name = "orders-feature-group-" + strftime("%d-%H-%M-%S", gmtime())
```
Instantiate a FeatureGroup object for customers_data and orders_data.
```
from sagemaker.feature_store.feature_group import FeatureGroup
customers_feature_group = FeatureGroup(
name=customers_feature_group_name, sagemaker_session=sagemaker_session
)
orders_feature_group = FeatureGroup(
name=orders_feature_group_name, sagemaker_session=sagemaker_session
)
import time
current_time_sec = int(round(time.time()))
record_identifier_feature_name = "customer_id"
```
Append EventTime feature to your data frame. This parameter is required, and time stamps each data point.
```
customer_data["EventTime"] = pd.Series([current_time_sec] * len(customer_data), dtype="float64")
orders_data["EventTime"] = pd.Series([current_time_sec] * len(orders_data), dtype="float64")
customer_data.head()
orders_data.head()
```
Load feature definitions to your feature group.
```
customers_feature_group.load_feature_definitions(data_frame=customer_data)
orders_feature_group.load_feature_definitions(data_frame=orders_data)
```
### How to create an Online or Offline Feature Store that uses your KMS key for encryption?
Below we create two feature groups, `customers_feature_group` and `orders_feature_group` respectively, and explain how use your KMS key to securely encrypt your data in your online or offline feature store.
### How to create an Online Feature store with your KMS key?
To encrypt data in your online feature store, set `enable_online_store` to be `True` and specify your KMS key as parameter `online_store_kms_key_id`. You will need to substitute your Account number in `arn:aws:kms:us-east-1:123456789012:key/` replacing `123456789012` with your Account number.
```
customers_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=True,
online_store_kms_key_id = 'arn:aws:kms:us-east-1:123456789012:key/'+ new_kms_key['KeyMetadata']['KeyId']
)
orders_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=True,
online_store_kms_key_id = 'arn:aws:kms:us-east-1:123456789012:key/'+new_kms_key['KeyMetadata']['KeyId']
)
```
### How to create an Offline Feature store with your KMS key?
Similar to the above, set `enable_online_store` to be `False` and then specify your KMS key as parameter `offline_store_kms_key_id`. You will need to substitute your Account number in `arn:aws:kms:us-east-1:123456789012:key/` replacing `123456789012` with your Account number.
```
customers_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=False,
offline_store_kms_key_id = 'arn:aws:kms:us-east-1:123456789012:key/'+ new_kms_key['KeyMetadata']['KeyId']
)
orders_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=False,
offline_store_kms_key_id = 'arn:aws:kms:us-east-1:123456789012:key/'+new_kms_key['KeyMetadata']['KeyId']
)
```
For this example we create an online feature store that encrypts your data using your KMS key.
**Note**: You will need to substitute your Account number in `arn:aws:kms:us-east-1:123456789012:key/` replacing `123456789012` with your Account number.
```
customers_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=False,
offline_store_kms_key_id="arn:aws:kms:us-east-1:123456789012:key/"
+ new_kms_key["KeyMetadata"]["KeyId"],
)
orders_feature_group.create(
s3_uri=f"s3://{s3_bucket_name}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name="EventTime",
role_arn=role,
enable_online_store=False,
offline_store_kms_key_id="arn:aws:kms:us-east-1:123456789012:key/"
+ new_kms_key["KeyMetadata"]["KeyId"],
)
```
### How to verify that your KMS key is being used to encrypt your data in your Online or Offline Feature Store?
### Online Store Verification
To demonstrate that your data is being encrypted in your Online store, use your `kms` client from `boto3` to list the grants under your KMS key. It should show 'SageMakerFeatureStore-' and the name of your feature group you created and should list these operations under Operations:`['Decrypt','Encrypt','GenerateDataKey','ReEncryptFrom','ReEncryptTo','CreateGrant','RetireGrant','DescribeKey']`
An alternative way for you to check that your data is encrypted in your Online store is to check [Cloud Trails](https://console.aws.amazon.com/cloudtrail/) and navigate to your account name. Once here, under General details you should see that SSE-KMS encryption is enabled and with your AWS KMS key shown below it. Below is a screenshot showing this:

### Offline Store Verification
To verify that your data in being encrypted in your Offline store, you must navigate to your S3 bucket through the [Console](https://console.aws.amazon.com/s3/home?region=us-east-1) and then navigate to your prefix, offline store, feature group name and into the /data/ folder. Once here, select a parquet file which is the file containing your feature group data. For this example, the directory path in S3 was this:
`Amazon S3/MYBUCKET/PREFIX/123456789012/sagemaker/region/offline-store/customers-feature-group-23-22-44-47/data/year=2021/month=03/day=23/hour=22/20210323T224448Z_IdfObJjhpqLQ5rmG.parquet.`
After selecting the parquet file, navigate to Server-side encryption settings. It should mention that Default encryption is enabled and reference (SSE-KMS) under server-side encryption. If this show, then your data is being encrypted in the offline store. Below is a screenshot of how this should look like in the console:

For this example since we created a secure Online store using our KMS key, below we use `list_grants` to check that our feature group and required grants are present under operations.
```
kms.list_grants(
KeyId="arn:aws:kms:us-east-1:123456789012:key/" + new_kms_key["KeyMetadata"]["KeyId"]
)
```
### Clean Up Resources
Remove the Feature Groups we created.
```
customers_feature_group.delete()
orders_feature_group.delete()
# preserve original sagemaker version
%pip install 'sagemaker=={}'.format(original_version)
```
### Next Steps
For more information on how to use KMS to encrypt your data in your Feature Store, see [Feature Store Security](https://docs.aws.amazon.com/sagemaker/latest/dg/feature-store-security.html). For general information on KMS keys and CMK, see [Customer Managed Keys](https://docs.aws.amazon.com/kms/latest/developerguide/concepts.html#master_keys).
|
github_jupyter
|
# Hyperparameter tuning
In the previous section, we did not discuss the parameters of random forest
and gradient-boosting. However, there are a couple of things to keep in mind
when setting these.
This notebook gives crucial information regarding how to set the
hyperparameters of both random forest and gradient boosting decision tree
models.
<div class="admonition caution alert alert-warning">
<p class="first admonition-title" style="font-weight: bold;">Caution!</p>
<p class="last">For the sake of clarity, no cross-validation will be used to estimate the
testing error. We are only showing the effect of the parameters
on the validation set of what should be the inner cross-validation.</p>
</div>
## Random forest
The main parameter to tune for random forest is the `n_estimators` parameter.
In general, the more trees in the forest, the better the generalization
performance will be. However, it will slow down the fitting and prediction
time. The goal is to balance computing time and generalization performance when
setting the number of estimators when putting such learner in production.
The `max_depth` parameter could also be tuned. Sometimes, there is no need
to have fully grown trees. However, be aware that with random forest, trees
are generally deep since we are seeking to overfit the learners on the
bootstrap samples because this will be mitigated by combining them.
Assembling underfitted trees (i.e. shallow trees) might also lead to an
underfitted forest.
```
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
data, target = fetch_california_housing(return_X_y=True, as_frame=True)
target *= 100 # rescale the target in k$
data_train, data_test, target_train, target_test = train_test_split(
data, target, random_state=0)
import pandas as pd
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestRegressor
param_grid = {
"n_estimators": [10, 20, 30],
"max_depth": [3, 5, None],
}
grid_search = GridSearchCV(
RandomForestRegressor(n_jobs=2), param_grid=param_grid,
scoring="neg_mean_absolute_error", n_jobs=2,
)
grid_search.fit(data_train, target_train)
columns = [f"param_{name}" for name in param_grid.keys()]
columns += ["mean_test_score", "rank_test_score"]
cv_results = pd.DataFrame(grid_search.cv_results_)
cv_results["mean_test_score"] = -cv_results["mean_test_score"]
cv_results[columns].sort_values(by="rank_test_score")
```
We can observe that in our grid-search, the largest `max_depth` together
with the largest `n_estimators` led to the best generalization performance.
## Gradient-boosting decision trees
For gradient-boosting, parameters are coupled, so we cannot set the
parameters one after the other anymore. The important parameters are
`n_estimators`, `max_depth`, and `learning_rate`.
Let's first discuss the `max_depth` parameter.
We saw in the section on gradient-boosting that the algorithm fits the error
of the previous tree in the ensemble. Thus, fitting fully grown trees will
be detrimental.
Indeed, the first tree of the ensemble would perfectly fit (overfit) the data
and thus no subsequent tree would be required, since there would be no
residuals.
Therefore, the tree used in gradient-boosting should have a low depth,
typically between 3 to 8 levels. Having very weak learners at each step will
help reducing overfitting.
With this consideration in mind, the deeper the trees, the faster the
residuals will be corrected and less learners are required. Therefore,
`n_estimators` should be increased if `max_depth` is lower.
Finally, we have overlooked the impact of the `learning_rate` parameter
until now. When fitting the residuals, we would like the tree
to try to correct all possible errors or only a fraction of them.
The learning-rate allows you to control this behaviour.
A small learning-rate value would only correct the residuals of very few
samples. If a large learning-rate is set (e.g., 1), we would fit the
residuals of all samples. So, with a very low learning-rate, we will need
more estimators to correct the overall error. However, a too large
learning-rate tends to obtain an overfitted ensemble,
similar to having a too large tree depth.
```
from sklearn.ensemble import GradientBoostingRegressor
param_grid = {
"n_estimators": [10, 30, 50],
"max_depth": [3, 5, None],
"learning_rate": [0.1, 1],
}
grid_search = GridSearchCV(
GradientBoostingRegressor(), param_grid=param_grid,
scoring="neg_mean_absolute_error", n_jobs=2
)
grid_search.fit(data_train, target_train)
columns = [f"param_{name}" for name in param_grid.keys()]
columns += ["mean_test_score", "rank_test_score"]
cv_results = pd.DataFrame(grid_search.cv_results_)
cv_results["mean_test_score"] = -cv_results["mean_test_score"]
cv_results[columns].sort_values(by="rank_test_score")
```
<div class="admonition caution alert alert-warning">
<p class="first admonition-title" style="font-weight: bold;">Caution!</p>
<p class="last">Here, we tune the <tt class="docutils literal">n_estimators</tt> but be aware that using early-stopping as
in the previous exercise will be better.</p>
</div>
|
github_jupyter
|
```
# !pip install ray[tune]
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
from sklearn.metrics import mean_squared_error
from hyperopt import hp
from ray import tune
from hyperopt import fmin, tpe, hp,Trials, space_eval
import scipy.stats
df = pd.read_csv("../../Data/Raw/flightLogData.csv")
plt.figure(figsize=(20, 10))
plt.plot(df.Time, df['Altitude'], linewidth=2, color="r", label="Altitude")
plt.plot(df.Time, df['Vertical_velocity'], linewidth=2, color="y", label="Vertical_velocity")
plt.plot(df.Time, df['Vertical_acceleration'], linewidth=2, color="b", label="Vertical_acceleration")
plt.legend()
plt.show()
temp_df = df[['Altitude', "Vertical_velocity", "Vertical_acceleration"]]
noise = np.random.normal(2, 5, temp_df.shape)
noisy_df = temp_df + noise
noisy_df['Time'] = df['Time']
plt.figure(figsize=(20, 10))
plt.plot(noisy_df.Time, noisy_df['Altitude'], linewidth=2, color="r", label="Altitude")
plt.plot(noisy_df.Time, noisy_df['Vertical_velocity'], linewidth=2, color="y", label="Vertical_velocity")
plt.plot(noisy_df.Time, noisy_df['Vertical_acceleration'], linewidth=2, color="b", label="Vertical_acceleration")
plt.legend()
plt.show()
```
## Altitude
```
q = 0.001
A = np.array([[1.0, 0.1, 0.005], [0, 1.0, 0.1], [0, 0, 1]])
H = np.array([[1.0, 0.0, 0.0],[ 0.0, 0.0, 1.0]])
P = np.array([[1.0, 0.0, 0.0], [0.0, 1.0, 0.0], [0.0, 0.0, 1.0]])
# R = np.array([[0.5, 0.0], [0.0, 0.0012]])
# Q = np.array([[q, 0.0, 0.0], [0.0, q, 0.0], [0.0, 0.0, q]])
I = np.identity(3)
x_hat = np.array([[0.0], [0.0], [0.0]])
Y = np.array([[0.0], [0.0]])
def kalman_update(param):
r1, r2, q1 = param['r1'], param['r2'], param['q1']
R = np.array([[r1, 0.0], [0.0, r2]])
Q = np.array([[q1, 0.0, 0.0], [0.0, q1, 0.0], [0.0, 0.0, q1]])
A = np.array([[1.0, 0.05, 0.00125], [0, 1.0, 0.05], [0, 0, 1]])
H = np.array([[1.0, 0.0, 0.0],[ 0.0, 0.0, 1.0]])
P = np.array([[1.0, 0.0, 0.0], [0.0, 1.0, 0.0], [0.0, 0.0, 1.0]])
I = np.identity(3)
x_hat = np.array([[0.0], [0.0], [0.0]])
Y = np.array([[0.0], [0.0]])
new_altitude = []
new_acceleration = []
new_velocity = []
for altitude, az in zip(noisy_df['Altitude'], noisy_df['Vertical_acceleration']):
Z = np.array([[altitude], [az]])
x_hat_minus = np.dot(A, x_hat)
P_minus = np.dot(np.dot(A, P), np.transpose(A)) + Q
K = np.dot(np.dot(P_minus, np.transpose(H)), np.linalg.inv((np.dot(np.dot(H, P_minus), np.transpose(H)) + R)))
Y = Z - np.dot(H, x_hat_minus)
x_hat = x_hat_minus + np.dot(K, Y)
P = np.dot((I - np.dot(K, H)), P_minus)
Y = Z - np.dot(H, x_hat_minus)
new_altitude.append(float(x_hat[0]))
new_velocity.append(float(x_hat[1]))
new_acceleration.append(float(x_hat[2]))
return new_altitude
def objective_function(param):
r1, r2, q1 = param['r1'], param['r2'], param['q1']
R = np.array([[r1, 0.0], [0.0, r2]])
Q = np.array([[q1, 0.0, 0.0], [0.0, q1, 0.0], [0.0, 0.0, q1]])
A = np.array([[1.0, 0.05, 0.00125], [0, 1.0, 0.05], [0, 0, 1]])
H = np.array([[1.0, 0.0, 0.0],[ 0.0, 0.0, 1.0]])
P = np.array([[1.0, 0.0, 0.0], [0.0, 1.0, 0.0], [0.0, 0.0, 1.0]])
I = np.identity(3)
x_hat = np.array([[0.0], [0.0], [0.0]])
Y = np.array([[0.0], [0.0]])
new_altitude = []
new_acceleration = []
new_velocity = []
for altitude, az in zip(noisy_df['Altitude'], noisy_df['Vertical_acceleration']):
Z = np.array([[altitude], [az]])
x_hat_minus = np.dot(A, x_hat)
P_minus = np.dot(np.dot(A, P), np.transpose(A)) + Q
K = np.dot(np.dot(P_minus, np.transpose(H)), np.linalg.inv((np.dot(np.dot(H, P_minus), np.transpose(H)) + R)))
Y = Z - np.dot(H, x_hat_minus)
x_hat = x_hat_minus + np.dot(K, Y)
P = np.dot((I - np.dot(K, H)), P_minus)
Y = Z - np.dot(H, x_hat_minus)
new_altitude.append(float(x_hat[0]))
new_velocity.append(float(x_hat[1]))
new_acceleration.append(float(x_hat[2]))
return mean_squared_error(df['Altitude'], new_altitude)
# space = {
# "r1": hp.choice("r1", np.arange(0.01, 90, 0.005)),
# "r2": hp.choice("r2", np.arange(0.01, 90, 0.005)),
# "q1": hp.choice("q1", np.arange(0.0001, 0.0009, 0.0001))
# }
len(np.arange(0.00001, 0.09, 0.00001))
space = {
"r1": hp.choice("r1", np.arange(0.001, 90, 0.001)),
"r2": hp.choice("r2", np.arange(0.001, 90, 0.001)),
"q1": hp.choice("q1", np.arange(0.00001, 0.09, 0.00001))
}
# Initialize trials object
trials = Trials()
best = fmin(fn=objective_function, space = space, algo=tpe.suggest, max_evals=100, trials=trials )
print(best)
# -> {'a': 1, 'c2': 0.01420615366247227}
print(space_eval(space, best))
# -> ('case 2', 0.01420615366247227}
d1 = space_eval(space, best)
objective_function(d1)
%%timeit
objective_function({'q1': 0.06626, 'r1': 0.25, 'r2': 0.75})
objective_function({'q1': 0.06626, 'r1': 0.25, 'r2': 0.75})
y = kalman_update(d1)
current = kalman_update({'q1': 0.06626, 'r1': 0.25, 'r2': 0.75})
plt.figure(figsize=(20, 10))
plt.plot(noisy_df.Time, df['Altitude'], linewidth=2, color="r", label="Actual")
plt.plot(noisy_df.Time, current, linewidth=2, color="g", label="ESP32")
plt.plot(noisy_df.Time, noisy_df['Altitude'], linewidth=2, color="y", label="Noisy")
plt.plot(noisy_df.Time, y, linewidth=2, color="b", label="Predicted")
plt.legend()
plt.show()
def kalman_update_return_velocity(param):
r1, r2, q1 = param['r1'], param['r2'], param['q1']
R = np.array([[r1, 0.0], [0.0, r2]])
Q = np.array([[q1, 0.0, 0.0], [0.0, q1, 0.0], [0.0, 0.0, q1]])
A = np.array([[1.0, 0.05, 0.00125], [0, 1.0, 0.05], [0, 0, 1]])
H = np.array([[1.0, 0.0, 0.0],[ 0.0, 0.0, 1.0]])
P = np.array([[1.0, 0.0, 0.0], [0.0, 1.0, 0.0], [0.0, 0.0, 1.0]])
I = np.identity(3)
x_hat = np.array([[0.0], [0.0], [0.0]])
Y = np.array([[0.0], [0.0]])
new_altitude = []
new_acceleration = []
new_velocity = []
for altitude, az in zip(noisy_df['Altitude'], noisy_df['Vertical_acceleration']):
Z = np.array([[altitude], [az]])
x_hat_minus = np.dot(A, x_hat)
P_minus = np.dot(np.dot(A, P), np.transpose(A)) + Q
K = np.dot(np.dot(P_minus, np.transpose(H)), np.linalg.inv((np.dot(np.dot(H, P_minus), np.transpose(H)) + R)))
Y = Z - np.dot(H, x_hat_minus)
x_hat = x_hat_minus + np.dot(K, Y)
P = np.dot((I - np.dot(K, H)), P_minus)
Y = Z - np.dot(H, x_hat_minus)
new_altitude.append(float(x_hat[0]))
new_velocity.append(float(x_hat[1]))
new_acceleration.append(float(x_hat[2]))
return new_velocity
def objective_function(param):
r1, r2, q1 = param['r1'], param['r2'], param['q1']
R = np.array([[r1, 0.0], [0.0, r2]])
Q = np.array([[q1, 0.0, 0.0], [0.0, q1, 0.0], [0.0, 0.0, q1]])
A = np.array([[1.0, 0.05, 0.00125], [0, 1.0, 0.05], [0, 0, 1]])
H = np.array([[1.0, 0.0, 0.0],[ 0.0, 0.0, 1.0]])
P = np.array([[1.0, 0.0, 0.0], [0.0, 1.0, 0.0], [0.0, 0.0, 1.0]])
I = np.identity(3)
x_hat = np.array([[0.0], [0.0], [0.0]])
Y = np.array([[0.0], [0.0]])
new_altitude = []
new_acceleration = []
new_velocity = []
for altitude, az in zip(noisy_df['Altitude'], noisy_df['Vertical_acceleration']):
Z = np.array([[altitude], [az]])
x_hat_minus = np.dot(A, x_hat)
P_minus = np.dot(np.dot(A, P), np.transpose(A)) + Q
K = np.dot(np.dot(P_minus, np.transpose(H)), np.linalg.inv((np.dot(np.dot(H, P_minus), np.transpose(H)) + R)))
Y = Z - np.dot(H, x_hat_minus)
x_hat = x_hat_minus + np.dot(K, Y)
P = np.dot((I - np.dot(K, H)), P_minus)
Y = Z - np.dot(H, x_hat_minus)
new_altitude.append(float(x_hat[0]))
new_velocity.append(float(x_hat[1]))
new_acceleration.append(float(x_hat[2]))
return mean_squared_error(df['Vertical_velocity'], new_velocity)
space = {
"r1": hp.choice("r1", np.arange(0.001, 90, 0.001)),
"r2": hp.choice("r2", np.arange(0.001, 90, 0.001)),
"q1": hp.choice("q1", np.arange(0.00001, 0.09, 0.00001))
}
# Initialize trials object
trials = Trials()
best = fmin(fn=objective_function, space = space, algo=tpe.suggest, max_evals=100, trials=trials )
print(best)
print(space_eval(space, best))
d2 = space_eval(space, best)
objective_function(d2)
y = kalman_update_return_velocity(d2)
current = kalman_update_return_velocity({'q1': 0.0013, 'r1': 0.25, 'r2': 0.65})
previous = kalman_update_return_velocity({'q1': 0.08519, 'r1': 4.719, 'r2': 56.443})
plt.figure(figsize=(20, 10))
plt.plot(noisy_df.Time, df['Vertical_velocity'], linewidth=2, color="r", label="Actual")
plt.plot(noisy_df.Time, current, linewidth=2, color="g", label="ESP32")
plt.plot(noisy_df.Time, previous, linewidth=2, color="c", label="With previous data")
plt.plot(noisy_df.Time, noisy_df['Vertical_velocity'], linewidth=2, color="y", label="Noisy")
plt.plot(noisy_df.Time, y, linewidth=2, color="b", label="Predicted")
plt.legend()
plt.show()
```
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Lab 04a: Dogs vs Cats Image Classification Without Image Augmentation
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/sres-dl-course/sres-dl-course.github.io/blob/master/notebooks/python/L04_C01_dogs_vs_cats_without_augmentation.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/sres-dl-course/sres-dl-course.github.io/blob/master/notebooks/python/L04_C01_dogs_vs_cats_without_augmentation.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
In this tutorial, we will discuss how to classify images into pictures of cats or pictures of dogs. We'll build an image classifier using `tf.keras.Sequential` model and load data using `tf.keras.preprocessing.image.ImageDataGenerator`.
## Specific concepts that will be covered:
In the process, we will build practical experience and develop intuition around the following concepts
* Building _data input pipelines_ using the `tf.keras.preprocessing.image.ImageDataGenerator` class — How can we efficiently work with data on disk to interface with our model?
* _Overfitting_ - what is it, how to identify it?
<hr>
**Before you begin**
Before running the code in this notebook, reset the runtime by going to **Runtime -> Reset all runtimes** in the menu above. If you have been working through several notebooks, this will help you avoid reaching Colab's memory limits.
# Importing packages
Let's start by importing required packages:
* os — to read files and directory structure
* numpy — for some matrix math outside of TensorFlow
* matplotlib.pyplot — to plot the graph and display images in our training and validation data
```
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import os
import matplotlib.pyplot as plt
import numpy as np
import logging
logger = tf.get_logger()
logger.setLevel(logging.ERROR)
```
# Data Loading
To build our image classifier, we begin by downloading the dataset. The dataset we are using is a filtered version of <a href="https://www.kaggle.com/c/dogs-vs-cats/data" target="_blank">Dogs vs. Cats</a> dataset from Kaggle (ultimately, this dataset is provided by Microsoft Research).
In previous Colabs, we've used <a href="https://www.tensorflow.org/datasets" target="_blank">TensorFlow Datasets</a>, which is a very easy and convenient way to use datasets. In this Colab however, we will make use of the class `tf.keras.preprocessing.image.ImageDataGenerator` which will read data from disk. We therefore need to directly download *Dogs vs. Cats* from a URL and unzip it to the Colab filesystem.
```
_URL = 'https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip'
zip_dir = tf.keras.utils.get_file('cats_and_dogs_filterted.zip', origin=_URL, extract=True)
```
The dataset we have downloaded has the following directory structure.
<pre style="font-size: 10.0pt; font-family: Arial; line-height: 2; letter-spacing: 1.0pt;" >
<b>cats_and_dogs_filtered</b>
|__ <b>train</b>
|______ <b>cats</b>: [cat.0.jpg, cat.1.jpg, cat.2.jpg ...]
|______ <b>dogs</b>: [dog.0.jpg, dog.1.jpg, dog.2.jpg ...]
|__ <b>validation</b>
|______ <b>cats</b>: [cat.2000.jpg, cat.2001.jpg, cat.2002.jpg ...]
|______ <b>dogs</b>: [dog.2000.jpg, dog.2001.jpg, dog.2002.jpg ...]
</pre>
We can list the directories with the following terminal command:
```
zip_dir_base = os.path.dirname(zip_dir)
!find $zip_dir_base -type d -print
```
We'll now assign variables with the proper file path for the training and validation sets.
```
base_dir = os.path.join(os.path.dirname(zip_dir), 'cats_and_dogs_filtered')
train_dir = os.path.join(base_dir, 'train')
validation_dir = os.path.join(base_dir, 'validation')
train_cats_dir = os.path.join(train_dir, 'cats') # directory with our training cat pictures
train_dogs_dir = os.path.join(train_dir, 'dogs') # directory with our training dog pictures
validation_cats_dir = os.path.join(validation_dir, 'cats') # directory with our validation cat pictures
validation_dogs_dir = os.path.join(validation_dir, 'dogs') # directory with our validation dog pictures
```
### Understanding our data
Let's look at how many cats and dogs images we have in our training and validation directory
```
num_cats_tr = len(os.listdir(train_cats_dir))
num_dogs_tr = len(os.listdir(train_dogs_dir))
num_cats_val = len(os.listdir(validation_cats_dir))
num_dogs_val = len(os.listdir(validation_dogs_dir))
total_train = num_cats_tr + num_dogs_tr
total_val = num_cats_val + num_dogs_val
print('total training cat images:', num_cats_tr)
print('total training dog images:', num_dogs_tr)
print('total validation cat images:', num_cats_val)
print('total validation dog images:', num_dogs_val)
print("--")
print("Total training images:", total_train)
print("Total validation images:", total_val)
```
# Setting Model Parameters
For convenience, we'll set up variables that will be used later while pre-processing our dataset and training our network.
```
BATCH_SIZE = 100 # Number of training examples to process before updating our models variables
IMG_SHAPE = 150 # Our training data consists of images with width of 150 pixels and height of 150 pixels
```
# Data Preparation
Images must be formatted into appropriately pre-processed floating point tensors before being fed into the network. The steps involved in preparing these images are:
1. Read images from the disk
2. Decode contents of these images and convert it into proper grid format as per their RGB content
3. Convert them into floating point tensors
4. Rescale the tensors from values between 0 and 255 to values between 0 and 1
Fortunately, all these tasks can be done using the class **tf.keras.preprocessing.image.ImageDataGenerator**.
We can set this up in a couple of lines of code.
```
train_image_generator = ImageDataGenerator(rescale=1./255) # Generator for our training data
validation_image_generator = ImageDataGenerator(rescale=1./255) # Generator for our validation data
```
After defining our generators for training and validation images, **flow_from_directory** method will load images from the disk, apply rescaling, and resize them using single line of code.
```
train_data_gen = train_image_generator.flow_from_directory(batch_size=BATCH_SIZE,
directory=train_dir,
shuffle=True,
target_size=(IMG_SHAPE,IMG_SHAPE), #(150,150)
class_mode='binary')
val_data_gen = validation_image_generator.flow_from_directory(batch_size=BATCH_SIZE,
directory=validation_dir,
shuffle=False,
target_size=(IMG_SHAPE,IMG_SHAPE), #(150,150)
class_mode='binary')
```
### Visualizing Training images
We can visualize our training images by getting a batch of images from the training generator, and then plotting a few of them using `matplotlib`.
```
sample_training_images, _ = next(train_data_gen)
```
The `next` function returns a batch from the dataset. One batch is a tuple of (*many images*, *many labels*). For right now, we're discarding the labels because we just want to look at the images.
```
# This function will plot images in the form of a grid with 1 row and 5 columns where images are placed in each column.
def plotImages(images_arr):
fig, axes = plt.subplots(1, 5, figsize=(20,20))
axes = axes.flatten()
for img, ax in zip(images_arr, axes):
ax.imshow(img)
plt.tight_layout()
plt.show()
plotImages(sample_training_images[:5]) # Plot images 0-4
```
# Model Creation
## Exercise 4.1 Define the model
The model consists of four convolution blocks with a max pool layer in each of them. Then we have a fully connected layer with 512 units, with a `relu` activation function. The model will output class probabilities for two classes — dogs and cats — using `softmax`.
The list of model layers:
* 2D Convolution - 32 filters, 3x3 kernel, ReLU activation
* 2D Max pooling - 2x2 kernel
* 2D Convolution - 64 filters, 3x3 kernel, ReLU activation
* 2D Max pooling - 2x2 kernel
* 2D Convolution - 128 filters, 3x3 kernel, ReLU activation
* 2D Max pooling - 2x2 kernel
* 2D Convolution - 128 filters, 3x3 kernel, ReLU activation
* 2D Max pooling - 2x2 kernel
* Flatten
* Dense - 512 nodes
* Dense - 2 nodes
Check the documentation for how to specify the layers [https://www.tensorflow.org/api_docs/python/tf/keras/layers](https://www.tensorflow.org/api_docs/python/tf/keras/layers)
```
model = tf.keras.models.Sequential([
# TODO - Create the CNN model as specified above
])
```
### Exercise 4.1 Solution
The solution for the exercise can be found [here](https://colab.research.google.com/github/rses-dl-course/rses-dl-course.github.io/blob/master/notebooks/python/solutions/E4.1.ipynb)
### Exercise 4.2 Compile the model
As usual, we will use the `adam` optimizer. Since we output a softmax categorization, we'll use `sparse_categorical_crossentropy` as the loss function. We would also like to look at training and validation accuracy on each epoch as we train our network, so we are passing in the metrics argument.
```
# TODO - Compile the model
```
#### Exercise 4.2 Solution
The solution for the exercise can be found [here](https://colab.research.google.com/github/rses-dl-course/rses-dl-course.github.io/blob/master/notebooks/python/solutions/E4.2.ipynb)
### Model Summary
Let's look at all the layers of our network using **summary** method.
```
model.summary()
```
### Exercise 4.3 Train the model
It's time we train our network.
* Since we have a validation dataset, we can use this to evaluate our model as it trains by adding the `validation_data` parameter.
* `validation_steps` can also be added if you'd like to use less than full validation set.
```
# TODO - Fit the model
```
#### Exercise 4.3 Solution
The solution for the exercise can be found [here](https://colab.research.google.com/github/rses-dl-course/rses-dl-course.github.io/blob/master/notebooks/python/solutions/E4.3.ipynb)
### Visualizing results of the training
We'll now visualize the results we get after training our network.
```
acc = history.history['accuracy']
val_acc = history.history['val_accuracy']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs_range = range(EPOCHS)
plt.figure(figsize=(20, 8))
plt.subplot(1, 2, 1)
plt.plot(epochs_range, acc, label='Training Accuracy')
plt.plot(epochs_range, val_acc, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.title('Training and Validation Accuracy')
plt.subplot(1, 2, 2)
plt.plot(epochs_range, loss, label='Training Loss')
plt.plot(epochs_range, val_loss, label='Validation Loss')
plt.legend(loc='upper right')
plt.title('Training and Validation Loss')
plt.savefig('./foo.png')
plt.show()
```
As we can see from the plots, training accuracy and validation accuracy are off by large margin and our model has achieved only around **70%** accuracy on the validation set (depending on the number of epochs you trained for).
This is a clear indication of overfitting. Once the training and validation curves start to diverge, our model has started to memorize the training data and is unable to perform well on the validation data.
|
github_jupyter
|
# Selected Economic Characteristics: Employment Status from the American Community Survey
**[Work in progress]**
This notebook downloads [selected economic characteristics (DP03)](https://data.census.gov/cedsci/table?tid=ACSDP5Y2018.DP03) from the American Community Survey 2018 5-Year Data.
Data source: [American Community Survey 5-Year Data 2018](https://www.census.gov/data/developers/data-sets/acs-5year.html)
Authors: Peter Rose (pwrose@ucsd.edu), Ilya Zaslavsky (zaslavsk@sdsc.edu)
```
import os
import pandas as pd
from pathlib import Path
import time
pd.options.display.max_rows = None # display all rows
pd.options.display.max_columns = None # display all columsns
NEO4J_IMPORT = Path(os.getenv('NEO4J_IMPORT'))
print(NEO4J_IMPORT)
```
## Download selected variables
* [Selected economic characteristics for US](https://data.census.gov/cedsci/table?tid=ACSDP5Y2018.DP03)
* [List of variables as HTML](https://api.census.gov/data/2018/acs/acs5/profile/groups/DP03.html) or [JSON](https://api.census.gov/data/2018/acs/acs5/profile/groups/DP03/)
* [Description of variables](https://www2.census.gov/programs-surveys/acs/tech_docs/subject_definitions/2018_ACSSubjectDefinitions.pdf)
* [Example URLs for API](https://api.census.gov/data/2018/acs/acs5/profile/examples.html)
### Specify variables from DP03 group and assign property names
Names must follow the [Neo4j property naming conventions](https://neo4j.com/docs/getting-started/current/graphdb-concepts/#graphdb-naming-rules-and-recommendations).
```
variables = {# EMPLOYMENT STATUS
'DP03_0001E': 'population16YearsAndOver',
'DP03_0002E': 'population16YearsAndOverInLaborForce',
'DP03_0002PE': 'population16YearsAndOverInLaborForcePct',
'DP03_0003E': 'population16YearsAndOverInCivilianLaborForce',
'DP03_0003PE': 'population16YearsAndOverInCivilianLaborForcePct',
'DP03_0006E': 'population16YearsAndOverInArmedForces',
'DP03_0006PE': 'population16YearsAndOverInArmedForcesPct',
'DP03_0007E': 'population16YearsAndOverNotInLaborForce',
'DP03_0007PE': 'population16YearsAndOverNotInLaborForcePct'
#'DP03_0014E': 'ownChildrenOfTheHouseholderUnder6Years',
#'DP03_0015E': 'ownChildrenOfTheHouseholderUnder6YearsAllParentsInLaborForce',
#'DP03_0016E': 'ownChildrenOfTheHouseholder6To17Years',
#'DP03_0017E': 'ownChildrenOfTheHouseholder6To17YearsAllParentsInLaborForce',
}
fields = ",".join(variables.keys())
for v in variables.values():
print('e.' + v + ' = toInteger(row.' + v + '),')
print(len(variables.keys()))
```
## Download county-level data using US Census API
```
url_county = f'https://api.census.gov/data/2018/acs/acs5/profile?get={fields}&for=county:*'
df = pd.read_json(url_county, dtype='str')
df.fillna('', inplace=True)
df.head()
```
##### Add column names
```
df = df[1:].copy() # skip first row of labels
columns = list(variables.values())
columns.append('stateFips')
columns.append('countyFips')
df.columns = columns
```
Remove Puerto Rico (stateFips = 72) to limit data to US States
TODO handle data for Puerto Rico (GeoNames represents Puerto Rico as a country)
```
df.query("stateFips != '72'", inplace=True)
```
Save list of state fips (required later to get tract data by state)
```
stateFips = list(df['stateFips'].unique())
stateFips.sort()
print(stateFips)
df.head()
# Example data
df[(df['stateFips'] == '06') & (df['countyFips'] == '073')]
df['source'] = 'American Community Survey 5 year'
df['aggregationLevel'] = 'Admin2'
```
### Save data
```
df.to_csv(NEO4J_IMPORT / "03a-USCensusDP03EmploymentAdmin2.csv", index=False)
```
## Download zip-level data using US Census API
```
url_zip = f'https://api.census.gov/data/2018/acs/acs5/profile?get={fields}&for=zip%20code%20tabulation%20area:*'
df = pd.read_json(url_zip, dtype='str')
df.fillna('', inplace=True)
df.head()
```
##### Add column names
```
df = df[1:].copy() # skip first row
columns = list(variables.values())
columns.append('stateFips')
columns.append('postalCode')
df.columns = columns
df.head()
# Example data
df.query("postalCode == '90210'")
df['source'] = 'American Community Survey 5 year'
df['aggregationLevel'] = 'PostalCode'
```
### Save data
```
df.to_csv(NEO4J_IMPORT / "03a-USCensusDP03EmploymentZip.csv", index=False)
```
## Download tract-level data using US Census API
Tract-level data are only available by state, so we need to loop over all states.
```
def get_tract_data(state):
url_tract = f'https://api.census.gov/data/2018/acs/acs5/profile?get={fields}&for=tract:*&in=state:{state}'
df = pd.read_json(url_tract, dtype='str')
time.sleep(1)
# skip first row of labels
df = df[1:].copy()
# Add column names
columns = list(variables.values())
columns.append('stateFips')
columns.append('countyFips')
columns.append('tract')
df.columns = columns
return df
df = pd.concat((get_tract_data(state) for state in stateFips))
df.fillna('', inplace=True)
df['tract'] = df['stateFips'] + df['countyFips'] + df['tract']
df['source'] = 'American Community Survey 5 year'
df['aggregationLevel'] = 'Tract'
# Example data for San Diego County
df[(df['stateFips'] == '06') & (df['countyFips'] == '073')].head()
```
### Save data
```
df.to_csv(NEO4J_IMPORT / "03a-USCensusDP03EmploymentTract.csv", index=False)
df.shape
```
|
github_jupyter
|
## Dimensionality Reduction
```
from sklearn.decomposition import PCA
```
### Principal Components Analysis
```
o_dir = os.path.join('outputs','pca')
if os.path.isdir(o_dir) is not True:
print("Creating '{0}' directory.".format(o_dir))
os.mkdir(o_dir)
pca = PCA() # Use all Principal Components
pca.fit(scdf) # Train model on all data
pcdf = pd.DataFrame(pca.transform(scdf)) # Transform data using model
for i in range(0,21):
print("Amount of explained variance for component {0} is: {1:6.2f}%".format(i, pca.explained_variance_ratio_[i]*100))
print("The amount of explained variance of the SES score using each component is...")
sns.lineplot(x=list(range(1,len(pca.explained_variance_ratio_)+1)), y=pca.explained_variance_ratio_)
pca = PCA(n_components=11)
pca.fit(scdf)
scores = pd.DataFrame(pca.transform(scdf), index=scdf.index)
scores.to_csv(os.path.join(o_dir,'Scores.csv.gz'), compression='gzip', index=True)
# Adapted from https://stackoverflow.com/questions/22984335/recovering-features-names-of-explained-variance-ratio-in-pca-with-sklearn
i = np.identity(scdf.shape[1]) # identity matrix
coef = pca.transform(i)
loadings = pd.DataFrame(coef, index=scdf.columns)
loadings.to_csv(os.path.join(o_dir,'Loadings.csv.gz'), compression='gzip', index=True)
print(scores.shape)
scores.sample(5, random_state=42)
print(loadings.shape)
loadings.sample(5, random_state=42)
odf = pd.DataFrame(columns=['Variable','Component Loading','Score'])
for i in range(0,len(loadings.index)):
row = loadings.iloc[i,:]
for c in list(loadings.columns.values):
d = {'Variable':loadings.index[i], 'Component Loading':c, 'Score':row[c]}
odf = odf.append(d, ignore_index=True)
g = sns.FacetGrid(odf, col="Variable", col_wrap=4, height=3, aspect=2.0, margin_titles=True, sharey=True)
g = g.map(plt.plot, "Component Loading", "Score", marker=".")
```
### What Have We Done?
```
sns.set_style('white')
sns.jointplot(data=scores, x=0, y=1, kind='hex', height=8, ratio=8)
```
#### Create an Output Directory and Load the Data
```
o_dir = os.path.join('outputs','clusters-pca')
if os.path.isdir(o_dir) is not True:
print("Creating '{0}' directory.".format(o_dir))
os.mkdir(o_dir)
score_df = pd.read_csv(os.path.join('outputs','pca','Scores.csv.gz'))
score_df.rename(columns={'Unnamed: 0':'lsoacd'}, inplace=True)
score_df.set_index('lsoacd', inplace=True)
# Ensures that df is initialised but original scores remain accessible
df = score_df.copy(deep=True)
score_df.describe()
score_df.sample(3, random_state=42)
```
#### Rescale the Loaded Data
We need this so that differences in the component scores don't cause the clustering algorithms to focus only on the 1st component.
```
scaler = preprocessing.MinMaxScaler()
df[df.columns] = scaler.fit_transform(df[df.columns])
df.describe()
df.sample(3, random_state=42)
```
|
github_jupyter
|
# Flopy MODFLOW 6 (MF6) Support
The Flopy library contains classes for creating, saving, running, loading, and modifying MF6 simulations. The MF6 portion of the flopy library is located in:
*flopy.mf6*
While there are a number of classes in flopy.mf6, to get started you only need to use the main classes summarized below:
flopy.mf6.MFSimulation
* MODFLOW Simulation Class. Entry point into any MODFLOW simulation.
flopy.mf6.ModflowGwf
* MODFLOW Groundwater Flow Model Class. Represents a single model in a simulation.
flopy.mf6.Modflow[pc]
* MODFLOW package classes where [pc] is the abbreviation of the package name. Each package is a separate class.
For packages that are part of a groundwater flow model, the abbreviation begins with "Gwf". For example, "flopy.mf6.ModflowGwfdis" is the Discretization package.
```
import os
import sys
from shutil import copyfile
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('matplotlib version: {}'.format(mpl.__version__))
print('flopy version: {}'.format(flopy.__version__))
```
# Creating a MF6 Simulation
A MF6 simulation is created by first creating a simulation object "MFSimulation". When you create the simulation object you can define the simulation's name, version, executable name, workspace path, and the name of the tdis file. All of these are optional parameters, and if not defined each one will default to the following:
sim_name='modflowtest'
version='mf6'
exe_name='mf6.exe'
sim_ws='.'
sim_tdis_file='modflow6.tdis'
```
import os
import sys
from shutil import copyfile
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
sim_name = 'example_sim'
sim_path = os.path.join('data', 'example_project')
sim = flopy.mf6.MFSimulation(sim_name=sim_name, version='mf6', exe_name='mf6',
sim_ws=sim_path)
```
The next step is to create a tdis package object "ModflowTdis". The first parameter of the ModflowTdis class is a simulation object, which ties a ModflowTdis object to a specific simulation. The other parameters and their definitions can be found in the docstrings.
```
tdis = flopy.mf6.ModflowTdis(sim, pname='tdis', time_units='DAYS', nper=2,
perioddata=[(1.0, 1, 1.0), (10.0, 5, 1.0)])
```
Next one or more models are created using the ModflowGwf class. The first parameter of the ModflowGwf class is the simulation object that the model will be a part of.
```
model_name = 'example_model'
model = flopy.mf6.ModflowGwf(sim, modelname=model_name,
model_nam_file='{}.nam'.format(model_name))
```
Next create one or more Iterative Model Solution (IMS) files.
```
ims_package = flopy.mf6.ModflowIms(sim, pname='ims', print_option='ALL',
complexity='SIMPLE', outer_hclose=0.00001,
outer_maximum=50, under_relaxation='NONE',
inner_maximum=30, inner_hclose=0.00001,
linear_acceleration='CG',
preconditioner_levels=7,
preconditioner_drop_tolerance=0.01,
number_orthogonalizations=2)
```
Each ModflowGwf object needs to be associated with an ModflowIms object. This is done by calling the MFSimulation object's "register_ims_package" method. The first parameter in this method is the ModflowIms object and the second parameter is a list of model names (strings) for the models to be associated with the ModflowIms object.
```
sim.register_ims_package(ims_package, [model_name])
```
Next add packages to each model. The first package added needs to be a spatial discretization package since flopy uses information from the spatial discretization package to help you build other packages. There are three spatial discretization packages to choose from:
DIS (ModflowGwfDis) - Structured discretization
DISV (ModflowGwfdisv) - Discretization with vertices
DISU (ModflowGwfdisu) - Unstructured discretization
```
dis_package = flopy.mf6.ModflowGwfdis(model, pname='dis', length_units='FEET', nlay=2,
nrow=2, ncol=5, delr=500.0,
delc=500.0,
top=100.0, botm=[50.0, 20.0],
filename='{}.dis'.format(model_name))
```
## Accessing Namefiles
Namefiles are automatically built for you by flopy. However, there are some options contained in the namefiles that you may want to set. To get the namefile object access the name_file attribute in either a simulation or model object to get the simulation or model namefile.
```
# set the nocheck property in the simulation namefile
sim.name_file.nocheck = True
# set the print_input option in the model namefile
model.name_file.print_input = True
```
## Specifying Options
Option that appear alone are assigned a boolean value, like the print_input option above. Options that have additional optional parameters are assigned using a tuple, with the entries containing the names of the optional parameters to turn on. Use a tuple with an empty string to indicate no optional parameters and use a tuple with None to turn the option off.
```
# Turn Newton option on with under relaxation
model.name_file.newtonoptions = ('UNDER_RELAXATION')
# Turn Newton option on without under relaxation
model.name_file.newtonoptions = ('')
# Turn off Newton option
model.name_file.newtonoptions = (None)
```
## MFArray Templates
Lastly define all other packages needed.
Note that flopy supports a number of ways to specify data for a package. A template, which defines the data array shape for you, can be used to specify the data. Templates are built by calling the empty of the data type you are building. For example, to build a template for k in the npf package you would call:
ModflowGwfnpf.k.empty()
The empty method for "MFArray" data templates (data templates whose size is based on the structure of the model grid) take up to four parameters:
* model - The model object that the data is a part of. A valid model object with a discretization package is required in order to build the proper array dimensions. This parameter is required.
* layered - True or false whether the data is layered or not.
* data_storage_type_list - List of data storage types, one for each model layer. If the template is not layered, only one data storage type needs to be specified. There are three data storage types supported, internal_array, internal_constant, and external_file.
* default_value - The initial value for the array.
```
# build a data template for k that stores the first layer as an internal array and the second
# layer as a constant with the default value of k for all layers set to 100.0
layer_storage_types = [flopy.mf6.data.mfdatastorage.DataStorageType.internal_array,
flopy.mf6.data.mfdatastorage.DataStorageType.internal_constant]
k_template = flopy.mf6.ModflowGwfnpf.k.empty(model, True, layer_storage_types, 100.0)
# change the value of the second layer to 50.0
k_template[0]['data'] = [65.0, 60.0, 55.0, 50.0, 45.0, 40.0, 35.0, 30.0, 25.0, 20.0]
k_template[0]['factor'] = 1.5
print(k_template)
# create npf package using the k template to define k
npf_package = flopy.mf6.ModflowGwfnpf(model, pname='npf', save_flows=True, icelltype=1, k=k_template)
```
## Specifying MFArray Data
MFArray data can also be specified as a numpy array, a list of values, or a single value. Below strt (starting heads) are defined as a single value, 100.0, which is interpreted as an internal constant storage type of value 100.0. Strt could also be defined as a list defining a value for every model cell:
strt=[100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0,
90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0]
Or as a list defining a value or values for each model layer:
strt=[100.0, 90.0]
or:
strt=[[100.0], [90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0]]
MFArray data can also be stored in an external file by using a dictionary using the keys 'filename' to specify the file name relative to the model folder and 'data' to specific the data. The optional 'factor', 'iprn', and 'binary' keys may also be used.
strt={'filename': 'strt.txt', 'factor':1.0, 'data':[100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0,
90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0], 'binary': 'True'}
If the 'data' key is omitted from the dictionary flopy will try to read the data from an existing file 'filename'. Any relative paths for loading data from a file should specified relative to the MF6 simulation folder.
```
strt={'filename': 'strt.txt', 'factor':1.0, 'data':[100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0, 100.0,
90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0, 90.0], 'binary': 'True'}
ic_package = flopy.mf6.ModflowGwfic(model, pname='ic', strt=strt,
filename='{}.ic'.format(model_name))
# move external file data into model folder
icv_data_path = os.path.join('..', 'data', 'mf6', 'notebooks', 'iconvert.txt')
copyfile(icv_data_path, os.path.join(sim_path, 'iconvert.txt'))
# create storage package
sto_package = flopy.mf6.ModflowGwfsto(model, pname='sto', save_flows=True, iconvert={'filename':'iconvert.txt'},
ss=[0.000001, 0.000002],
sy=[0.15, 0.14, 0.13, 0.12, 0.11, 0.11, 0.12, 0.13, 0.14, 0.15,
0.15, 0.14, 0.13, 0.12, 0.11, 0.11, 0.12, 0.13, 0.14, 0.15])
```
## MFList Templates
Flopy supports specifying record and recarray "MFList" data in a number of ways. Templates can be created that define the shape of the data. The empty method for "MFList" data templates take up to 7 parameters.
* model - The model object that the data is a part of. A valid model object with a discretization package is required in order to build the proper array dimensions. This parameter is required.
* maxbound - The number of rows in the recarray. If not specified one row is returned.
* aux_vars - List of auxiliary variable names. If not specified auxiliary variables are not used.
* boundnames - True/False if boundnames is to be used.
* nseg - Number of segments (only relevant for a few data types)
* timeseries - True/False indicates that time series data will be used.
* stress_periods - List of integer stress periods to be used (transient MFList data only). If not specified for transient data, template will only be defined for stress period 1.
MFList transient data templates are numpy recarrays stored in a dictionary with the dictionary key an integer zero based stress period value (stress period - 1).
In the code below the well package is set up using a transient MFList template to help build the well's stress_periods.
```
maxbound = 2
# build a stress_period_data template with 2 wells over stress periods 1 and 2 with boundnames
# and three aux variables
wel_periodrec = flopy.mf6.ModflowGwfwel.stress_period_data.empty(model, maxbound=maxbound, boundnames=True,
aux_vars=['var1', 'var2', 'var3'],
stress_periods=[0,1])
# define the two wells for stress period one
wel_periodrec[0][0] = ((0,1,2), -50.0, -1, -2, -3, 'First Well')
wel_periodrec[0][1] = ((1,1,4), -25.0, 2, 3, 4, 'Second Well')
# define the two wells for stress period two
wel_periodrec[1][0] = ((0,1,2), -200.0, -1, -2, -3, 'First Well')
wel_periodrec[1][1] = ((1,1,4), -4000.0, 2, 3, 4, 'Second Well')
# build the well package
wel_package = flopy.mf6.ModflowGwfwel(model, pname='wel', print_input=True, print_flows=True,
auxiliary=['var1', 'var2', 'var3'], maxbound=maxbound,
stress_period_data=wel_periodrec, boundnames=True, save_flows=True)
```
## Cell IDs
Cell IDs always appear as tuples in an MFList. For a structured grid cell IDs appear as:
(<layer>, <row>, <column>)
For vertice based grid cells IDs appear as:
(<layer>, <intralayer_cell_id>)
Unstructured grid cell IDs appear as:
(<cell_id>)
## Specifying MFList Data
MFList data can also be defined as a list of tuples, with each tuple being a row of the recarray. For transient data the list of tuples can be stored in a dictionary with the dictionary key an integer zero based stress period value. If only a list of tuples is specified for transient data, the data is assumed to apply to stress period 1. Additional stress periods can be added with the add_transient_key method. The code below defines saverecord and printrecord as a list of tuples.
```
# printrecord data as a list of tuples. since no stress
# period is specified it will default to stress period 1
printrec_tuple_list = [('HEAD', 'ALL'), ('BUDGET', 'ALL')]
# saverecord data as a dictionary of lists of tuples for
# stress periods 1 and 2.
saverec_dict = {0:[('HEAD', 'ALL'), ('BUDGET', 'ALL')],1:[('HEAD', 'ALL'), ('BUDGET', 'ALL')]}
# create oc package
oc_package = flopy.mf6.ModflowGwfoc(model, pname='oc',
budget_filerecord=[('{}.cbc'.format(model_name),)],
head_filerecord=[('{}.hds'.format(model_name),)],
saverecord=saverec_dict,
printrecord=printrec_tuple_list)
# add stress period two to the print record
oc_package.printrecord.add_transient_key(1)
# set the data for stress period two in the print record
oc_package.printrecord.set_data([('HEAD', 'ALL'), ('BUDGET', 'ALL')], 1)
```
### Specifying MFList Data in an External File
MFList data can be specified in an external file using a dictionary with the 'filename' key. If the 'data' key is also included in the dictionary and is not None, flopy will create the file with the data contained in the 'data' key. The 'binary' key can be used to save data to a binary file ('binary': True). The code below creates a chd package which creates and references an external file containing data for stress period 1 and stores the data internally in the chd package file for stress period 2.
```
stress_period_data = {0: {'filename': 'chd_sp1.dat', 'data': [[(0, 0, 0), 70.]]},
1: [[(0, 0, 0), 60.]]}
chd = flopy.mf6.ModflowGwfchd(model, maxbound=1, stress_period_data=stress_period_data)
```
## Packages that Support both List-based and Array-based Data
The recharge and evapotranspiration packages can be specified using list-based or array-based input. The array packages have an "a" on the end of their name:
ModflowGwfrch - list based recharge package
ModflowGwfrcha - array based recharge package
ModflowGwfevt - list based evapotranspiration package
ModflowGwfevta - array based evapotranspiration package
```
rch_recarray = {0:[((0,0,0), 'rch_1'), ((1,1,1), 'rch_2')],
1:[((0,0,0), 'rch_1'), ((1,1,1), 'rch_2')]}
rch_package = flopy.mf6.ModflowGwfrch(model, pname='rch', fixed_cell=True, print_input=True,
maxbound=2, stress_period_data=rch_recarray)
```
## Utility Files (TS, TAS, OBS, TAB)
Utility files, MF6 formatted files that reference by packages, include time series, time array series, observation, and tab files. The file names for utility files are specified using the package that references them. The utility files can be created in several ways. A simple case is demonstrated below. More detail is given in the flopy3_mf6_obs_ts_tas notebook.
```
# build a time series array for the recharge package
ts_data = [(0.0, 0.015, 0.0017), (1.0, 0.016, 0.0019), (2.0, 0.012, 0.0015),
(3.0, 0.020, 0.0014), (4.0, 0.015, 0.0021), (5.0, 0.013, 0.0012),
(6.0, 0.022, 0.0012), (7.0, 0.016, 0.0014), (8.0, 0.013, 0.0011),
(9.0, 0.021, 0.0011), (10.0, 0.017, 0.0016), (11.0, 0.012, 0.0015)]
rch_package.ts.initialize(time_series_namerecord=['rch_1', 'rch_2'],
timeseries=ts_data, filename='recharge_rates.ts',
interpolation_methodrecord=['stepwise', 'stepwise'])
# build an recharge observation package that outputs the western recharge to a binary file and the eastern
# recharge to a text file
obs_data = {('rch_west.csv', 'binary'): [('rch_1_1_1', 'RCH', (0, 0, 0)),
('rch_1_2_1', 'RCH', (0, 1, 0))],
'rch_east.csv': [('rch_1_1_5', 'RCH', (0, 0, 4)),
('rch_1_2_5', 'RCH', (0, 1, 4))]}
rch_package.obs.initialize(filename='example_model.rch.obs', digits=10,
print_input=True, continuous=obs_data)
```
# Saving and Running a MF6 Simulation
Saving and running a simulation are done with the MFSimulation class's write_simulation and run_simulation methods.
```
# write simulation to new location
sim.write_simulation()
# run simulation
sim.run_simulation()
```
# Exporting a MF6 Model
Exporting a MF6 model to a shapefile or netcdf is the same as exporting a MF2005 model.
```
# make directory
pth = os.path.join('data', 'netCDF_export')
if not os.path.exists(pth):
os.makedirs(pth)
# export the dis package to a netcdf file
model.dis.export(os.path.join(pth, 'dis.nc'))
# export the botm array to a shapefile
model.dis.botm.export(os.path.join(pth, 'botm.shp'))
```
# Loading an Existing MF6 Simulation
Loading a simulation can be done with the flopy.mf6.MFSimulation.load static method.
```
# load the simulation
loaded_sim = flopy.mf6.MFSimulation.load(sim_name, 'mf6', 'mf6', sim_path)
```
# Retrieving Data and Modifying an Existing MF6 Simulation
Data can be easily retrieved from a simulation. Data can be retrieved using two methods. One method is to retrieve the data object from a master simulation dictionary that keeps track of all the data. The master simulation dictionary is accessed by accessing a simulation's "simulation_data" property and then the "mfdata" property:
sim.simulation_data.mfdata[<data path>]
The data path is the path to the data stored as a tuple containing the model name, package name, block name, and data name.
The second method is to get the data from the package object. If you do not already have the package object, you can work your way down the simulation structure, from the simulation to the correct model, to the correct package, and finally to the data object.
These methods are demonstrated in the code below.
```
# get hydraulic conductivity data object from the data dictionary
hk = sim.simulation_data.mfdata[(model_name, 'npf', 'griddata', 'k')]
# get specific yield data object from the storage package
sy = sto_package.sy
# get the model object from the simulation object using the get_model method,
# which takes a string with the model's name and returns the model object
mdl = sim.get_model(model_name)
# get the package object from the model mobject using the get_package method,
# which takes a string with the package's name or type
ic = mdl.get_package('ic')
# get the data object from the initial condition package object
strt = ic.strt
```
Once you have the appropriate data object there are a number methods to retrieve data from that object. Data retrieved can either be the data as it appears in the model file or the data with any factor specified in the model file applied to it. To get the raw data without applying a factor use the get_data method. To get the data with the factor already applied use .array.
Note that MFArray data is always a copy of the data stored by flopy. Modifying the copy of the flopy data will have no affect on the data stored in flopy. Non-constant internal MFList data is returned as a reference to a numpy recarray. Modifying this recarray will modify the data stored in flopy.
```
# get the data without applying any factor
hk_data_no_factor = hk.get_data()
print('Data without factor:\n{}\n'.format(hk_data_no_factor))
# get data with factor applied
hk_data_factor = hk.array
print('Data with factor:\n{}\n'.format(hk_data_factor))
```
Data can also be retrieved from the data object using []. For unlayered data the [] can be used to slice the data.
```
# slice layer one row two
print('SY slice of layer on row two\n{}\n'.format(sy[0,:,2]))
```
For layered data specify the layer number within the brackets. This will return a "LayerStorage" object which let's you change attributes of an individual layer.
```
# get layer one LayerStorage object
hk_layer_one = hk[0]
# change the print code and factor for layer one
hk_layer_one.iprn = '2'
hk_layer_one.factor = 1.1
print('Layer one data without factor:\n{}\n'.format(hk_layer_one.get_data()))
print('Data with new factor:\n{}\n'.format(hk.array))
```
## Modifying Data
Data can be modified in several ways. One way is to set data for a given layer within a LayerStorage object, like the one accessed in the code above. Another way is to set the data attribute to the new data. Yet another way is to call the data object's set_data method.
```
# set data within a LayerStorage object
hk_layer_one.set_data([120.0, 100.0, 80.0, 70.0, 60.0, 50.0, 40.0, 30.0, 25.0, 20.0])
print('New HK data no factor:\n{}\n'.format(hk.get_data()))
# set data attribute to new data
ic_package.strt = 150.0
print('New strt values:\n{}\n'.format(ic_package.strt.array))
# call set_data
sto_package.ss.set_data([0.000003, 0.000004])
print('New ss values:\n{}\n'.format(sto_package.ss.array))
```
## Modifying the Simulation Path
The simulation path folder can be changed by using the set_sim_path method in the MFFileMgmt object. The MFFileMgmt object can be obtained from the simulation object through properties:
sim.simulation_data.mfpath
```
# create new path
save_folder = os.path.join(sim_path, 'sim_modified')
# change simulation path
sim.simulation_data.mfpath.set_sim_path(save_folder)
# create folder
if not os.path.isdir(save_folder):
os.makedirs(save_folder)
```
## Adding a Model Relative Path
A model relative path lets you put all of the files associated with a model in a folder relative to the simulation folder. Warning, this will override all of your file paths to model package files and will also override any relative file paths to external model data files.
```
# Change path of model files relative to the simulation folder
model.set_model_relative_path('model_folder')
# create folder
if not os.path.isdir(save_folder):
os.makedirs(os.path.join(save_folder,'model_folder'))
# write simulation to new folder
sim.write_simulation()
# run simulation from new folder
sim.run_simulation()
```
## Post-Processing the Results
Results can be retrieved from the master simulation dictionary. Results are retrieved from the master simulation dictionary with using a tuple key that identifies the data to be retrieved. For head data use the key
('<model name>', 'HDS', 'HEAD')
where <model name> is the name of your model. For cell by cell budget data use the key
('<model name>', 'CBC', '<flow data name>')
where <flow data name> is the name of the flow data to be retrieved (ex. 'FLOW-JA-FACE'). All available output keys can be retrieved using the output_keys method.
```
keys = sim.simulation_data.mfdata.output_keys()
```
The entries in the list above are keys for data in the head file "HDS" and data in cell by cell flow file "CBC". Keys in this list are not guaranteed to be in any particular order. The code below uses the head file key to retrieve head data and then plots head data using matplotlib.
```
import matplotlib.pyplot as plt
import numpy as np
# get all head data
head = sim.simulation_data.mfdata['example_model', 'HDS', 'HEAD']
# get the head data from the end of the model run
head_end = head[-1]
# plot the head data from the end of the model run
levels = np.arange(160,162,1)
extent = (0.0, 1000.0, 2500.0, 0.0)
plt.contour(head_end[0, :, :],extent=extent)
plt.show()
```
Results can also be retrieved using the existing binaryfile method.
```
# get head data using old flopy method
hds_path = os.path.join(sim_path, model_name + '.hds')
hds = flopy.utils.HeadFile(hds_path)
# get heads after 1.0 days
head = hds.get_data(totim=1.0)
# plot head data
plt.contour(head[0, :, :],extent=extent)
plt.show()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/Tessellate-Imaging/monk_v1/blob/master/study_roadmaps/3_image_processing_deep_learning_roadmap/3_deep_learning_advanced/1_Blocks%20in%20Deep%20Learning%20Networks/8)%20Resnet%20V2%20Bottleneck%20Block%20(Type%20-%202).ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Goals
### 1. Learn to implement Resnet V2 Bottleneck Block (Type - 1) using monk
- Monk's Keras
- Monk's Pytorch
- Monk's Mxnet
### 2. Use network Monk's debugger to create complex blocks
### 3. Understand how syntactically different it is to implement the same using
- Traditional Keras
- Traditional Pytorch
- Traditional Mxnet
# Resnet V2 Bottleneck Block - Type 1
- Note: The block structure can have variations too, this is just an example
```
from IPython.display import Image
Image(filename='imgs/resnet_v2_bottleneck_without_downsample.png')
```
# Table of contents
[1. Install Monk](#1)
[2. Block basic Information](#2)
- [2.1) Visual structure](#2-1)
- [2.2) Layers in Branches](#2-2)
[3) Creating Block using monk visual debugger](#3)
- [3.1) Create the first branch](#3-1)
- [3.2) Create the second branch](#3-2)
- [3.3) Merge the branches](#3-3)
- [3.4) Debug the merged network](#3-4)
- [3.5) Compile the network](#3-5)
- [3.6) Visualize the network](#3-6)
- [3.7) Run data through the network](#3-7)
[4) Creating Block Using MONK one line API call](#4)
- [Mxnet Backend](#4-1)
- [Pytorch Backend](#4-2)
- [Keras Backend](#4-3)
[5) Appendix](#5)
- [Study Material](#5-1)
- [Creating block using traditional Mxnet](#5-2)
- [Creating block using traditional Pytorch](#5-3)
- [Creating block using traditional Keras](#5-4)
<a id='0'></a>
# Install Monk
## Using pip (Recommended)
- colab (gpu)
- All bakcends: `pip install -U monk-colab`
- kaggle (gpu)
- All backends: `pip install -U monk-kaggle`
- cuda 10.2
- All backends: `pip install -U monk-cuda102`
- Gluon bakcned: `pip install -U monk-gluon-cuda102`
- Pytorch backend: `pip install -U monk-pytorch-cuda102`
- Keras backend: `pip install -U monk-keras-cuda102`
- cuda 10.1
- All backend: `pip install -U monk-cuda101`
- Gluon bakcned: `pip install -U monk-gluon-cuda101`
- Pytorch backend: `pip install -U monk-pytorch-cuda101`
- Keras backend: `pip install -U monk-keras-cuda101`
- cuda 10.0
- All backend: `pip install -U monk-cuda100`
- Gluon bakcned: `pip install -U monk-gluon-cuda100`
- Pytorch backend: `pip install -U monk-pytorch-cuda100`
- Keras backend: `pip install -U monk-keras-cuda100`
- cuda 9.2
- All backend: `pip install -U monk-cuda92`
- Gluon bakcned: `pip install -U monk-gluon-cuda92`
- Pytorch backend: `pip install -U monk-pytorch-cuda92`
- Keras backend: `pip install -U monk-keras-cuda92`
- cuda 9.0
- All backend: `pip install -U monk-cuda90`
- Gluon bakcned: `pip install -U monk-gluon-cuda90`
- Pytorch backend: `pip install -U monk-pytorch-cuda90`
- Keras backend: `pip install -U monk-keras-cuda90`
- cpu
- All backend: `pip install -U monk-cpu`
- Gluon bakcned: `pip install -U monk-gluon-cpu`
- Pytorch backend: `pip install -U monk-pytorch-cpu`
- Keras backend: `pip install -U monk-keras-cpu`
## Install Monk Manually (Not recommended)
### Step 1: Clone the library
- git clone https://github.com/Tessellate-Imaging/monk_v1.git
### Step 2: Install requirements
- Linux
- Cuda 9.0
- `cd monk_v1/installation/Linux && pip install -r requirements_cu90.txt`
- Cuda 9.2
- `cd monk_v1/installation/Linux && pip install -r requirements_cu92.txt`
- Cuda 10.0
- `cd monk_v1/installation/Linux && pip install -r requirements_cu100.txt`
- Cuda 10.1
- `cd monk_v1/installation/Linux && pip install -r requirements_cu101.txt`
- Cuda 10.2
- `cd monk_v1/installation/Linux && pip install -r requirements_cu102.txt`
- CPU (Non gpu system)
- `cd monk_v1/installation/Linux && pip install -r requirements_cpu.txt`
- Windows
- Cuda 9.0 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu90.txt`
- Cuda 9.2 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu92.txt`
- Cuda 10.0 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu100.txt`
- Cuda 10.1 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu101.txt`
- Cuda 10.2 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu102.txt`
- CPU (Non gpu system)
- `cd monk_v1/installation/Windows && pip install -r requirements_cpu.txt`
- Mac
- CPU (Non gpu system)
- `cd monk_v1/installation/Mac && pip install -r requirements_cpu.txt`
- Misc
- Colab (GPU)
- `cd monk_v1/installation/Misc && pip install -r requirements_colab.txt`
- Kaggle (GPU)
- `cd monk_v1/installation/Misc && pip install -r requirements_kaggle.txt`
### Step 3: Add to system path (Required for every terminal or kernel run)
- `import sys`
- `sys.path.append("monk_v1/");`
# Imports
```
# Common
import numpy as np
import math
import netron
from collections import OrderedDict
from functools import partial
#Using mxnet-gluon backend
# When installed using pip
from monk.gluon_prototype import prototype
# When installed manually (Uncomment the following)
#import os
#import sys
#sys.path.append("monk_v1/");
#sys.path.append("monk_v1/monk/");
#from monk.gluon_prototype import prototype
```
<a id='2'></a>
# Block Information
<a id='2_1'></a>
## Visual structure
```
from IPython.display import Image
Image(filename='imgs/resnet_v2_bottleneck_without_downsample.png')
```
<a id='2_2'></a>
## Layers in Branches
- Number of branches: 2
- Common Elements
- batchnorm -> relu
- Branch 1
- identity
- Branch 2
- conv_1x1 -> batchnorm -> relu -> conv_3x3 -> batchnorm -> relu -> conv1x1
- Branches merged using
- Elementwise addition
(See Appendix to read blogs on resnets)
<a id='3'></a>
# Creating Block using monk debugger
```
# Imports and setup a project
# To use pytorch backend - replace gluon_prototype with pytorch_prototype
# To use keras backend - replace gluon_prototype with keras_prototype
from monk.gluon_prototype import prototype
# Create a sample project
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
```
<a id='3-1'></a>
## Create the first branch
```
def first_branch():
network = [];
network.append(gtf.identity());
return network;
# Debug the branch
branch_1 = first_branch()
network = [];
network.append(branch_1);
gtf.debug_custom_model_design(network);
```
<a id='3-2'></a>
## Create the second branch
```
def second_branch(output_channels=128, stride=1):
network = [];
# Bottleneck convolution
network.append(gtf.convolution(output_channels=output_channels//4, kernel_size=1, stride=stride));
network.append(gtf.batch_normalization());
network.append(gtf.relu());
#Bottleneck convolution
network.append(gtf.convolution(output_channels=output_channels//4, kernel_size=1, stride=stride));
network.append(gtf.batch_normalization());
network.append(gtf.relu());
#Normal convolution
network.append(gtf.convolution(output_channels=output_channels, kernel_size=1, stride=1));
return network;
# Debug the branch
branch_2 = second_branch(output_channels=128, stride=1)
network = [];
network.append(branch_2);
gtf.debug_custom_model_design(network);
```
<a id='3-3'></a>
## Merge the branches
```
def final_block(output_channels=128, stride=1):
network = [];
#Common Elements
network.append(gtf.batch_normalization());
network.append(gtf.relu());
#Create subnetwork and add branches
subnetwork = [];
branch_1 = first_branch()
branch_2 = second_branch(output_channels=output_channels, stride=stride)
subnetwork.append(branch_1);
subnetwork.append(branch_2);
# Add merging element
subnetwork.append(gtf.add());
# Add the subnetwork
network.append(subnetwork)
return network;
```
<a id='3-4'></a>
## Debug the merged network
```
final = final_block(output_channels=64, stride=1)
network = [];
network.append(final);
gtf.debug_custom_model_design(network);
```
<a id='3-5'></a>
## Compile the network
```
gtf.Compile_Network(network, data_shape=(64, 224, 224), use_gpu=False);
```
<a id='3-6'></a>
## Run data through the network
```
import mxnet as mx
x = np.zeros((1, 64, 224, 224));
x = mx.nd.array(x);
y = gtf.system_dict["local"]["model"].forward(x);
print(x.shape, y.shape)
```
<a id='3-7'></a>
## Visualize network using netron
```
gtf.Visualize_With_Netron(data_shape=(64, 224, 224))
```
<a id='4'></a>
# Creating Using MONK LOW code API
<a id='4-1'></a>
## Mxnet backend
```
from monk.gluon_prototype import prototype
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
network = [];
# Single line addition of blocks
network.append(gtf.resnet_v2_bottleneck_block(output_channels=64, downsample=False));
gtf.Compile_Network(network, data_shape=(64, 224, 224), use_gpu=False);
```
<a id='4-2'></a>
## Pytorch backend
- Only the import changes
```
#Change gluon_prototype to pytorch_prototype
from monk.pytorch_prototype import prototype
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
network = [];
# Single line addition of blocks
network.append(gtf.resnet_v2_bottleneck_block(output_channels=64, downsample=False));
gtf.Compile_Network(network, data_shape=(64, 224, 224), use_gpu=False);
```
<a id='4-3'></a>
## Keras backend
- Only the import changes
```
#Change gluon_prototype to keras_prototype
from monk.keras_prototype import prototype
gtf = prototype(verbose=1);
gtf.Prototype("sample-project-1", "sample-experiment-1");
network = [];
# Single line addition of blocks
network.append(gtf.resnet_v2_bottleneck_block(output_channels=64, downsample=False));
gtf.Compile_Network(network, data_shape=(64, 224, 224), use_gpu=False);
```
<a id='5'></a>
# Appendix
<a id='5-1'></a>
## Study links
- https://towardsdatascience.com/residual-blocks-building-blocks-of-resnet-fd90ca15d6ec
- https://medium.com/@MaheshNKhatri/resnet-block-explanation-with-a-terminology-deep-dive-989e15e3d691
- https://medium.com/analytics-vidhya/understanding-and-implementation-of-residual-networks-resnets-b80f9a507b9c
- https://hackernoon.com/resnet-block-level-design-with-deep-learning-studio-part-1-727c6f4927ac
<a id='5-2'></a>
## Creating block using traditional Mxnet
- Code credits - https://mxnet.incubator.apache.org/
```
# Traditional-Mxnet-gluon
import mxnet as mx
from mxnet.gluon import nn
from mxnet.gluon.nn import HybridBlock, BatchNorm
from mxnet.gluon.contrib.nn import HybridConcurrent, Identity
from mxnet import gluon, init, nd
def _conv3x3(channels, stride, in_channels):
return nn.Conv2D(channels, kernel_size=3, strides=stride, padding=1,
use_bias=False, in_channels=in_channels)
class ResnetBlockV1(HybridBlock):
def __init__(self, channels, stride, in_channels=0, **kwargs):
super(ResnetBlockV1, self).__init__(**kwargs)
#Common Elements
self.bn0 = nn.BatchNorm();
self.relu0 = nn.Activation('relu');
#Branch - 1
#Identity
# Branch - 2
self.body = nn.HybridSequential(prefix='')
self.body.add(nn.Conv2D(channels//4, kernel_size=1, strides=stride,
use_bias=False, in_channels=in_channels))
self.body.add(nn.BatchNorm())
self.body.add(nn.Activation('relu'))
self.body.add(_conv3x3(channels//4, stride, in_channels))
self.body.add(nn.BatchNorm())
self.body.add(nn.Activation('relu'))
self.body.add(nn.Conv2D(channels, kernel_size=1, strides=stride,
use_bias=False, in_channels=in_channels))
def hybrid_forward(self, F, x):
x = self.bn0(x);
x = self.relu0(x);
residual = x
x = self.body(x)
x = residual+x
return x
# Invoke the block
block = ResnetBlockV1(64, 1)
# Initialize network and load block on machine
ctx = [mx.cpu()];
block.initialize(init.Xavier(), ctx = ctx);
block.collect_params().reset_ctx(ctx)
block.hybridize()
# Run data through network
x = np.zeros((1, 64, 224, 224));
x = mx.nd.array(x);
y = block.forward(x);
print(x.shape, y.shape)
# Export Model to Load on Netron
block.export("final", epoch=0);
netron.start("final-symbol.json", port=8082)
```
<a id='5-3'></a>
## Creating block using traditional Pytorch
- Code credits - https://pytorch.org/
```
# Traiditional-Pytorch
import torch
from torch import nn
from torch.jit.annotations import List
import torch.nn.functional as F
def conv3x3(in_planes, out_planes, stride=1, groups=1, dilation=1):
"""3x3 convolution with padding"""
return nn.Conv2d(in_planes, out_planes, kernel_size=3, stride=stride,
padding=dilation, groups=groups, bias=False, dilation=dilation)
def conv1x1(in_planes, out_planes, stride=1):
"""1x1 convolution"""
return nn.Conv2d(in_planes, out_planes, kernel_size=1, stride=stride, bias=False)
class ResnetBottleNeckBlock(nn.Module):
expansion = 1
__constants__ = ['downsample']
def __init__(self, inplanes, planes, stride=1, groups=1,
base_width=64, dilation=1, norm_layer=None):
super(ResnetBottleNeckBlock, self).__init__()
norm_layer = nn.BatchNorm2d
#Common elements
self.bn0 = norm_layer(inplanes);
self.relu0 = nn.ReLU(inplace=True);
# Branch - 1
#Identity
# Branch - 2
self.conv1 = conv1x1(inplanes, planes//4, stride)
self.bn1 = norm_layer(planes//4)
self.relu1 = nn.ReLU(inplace=True)
self.conv2 = conv3x3(planes//4, planes//4, stride)
self.bn2 = norm_layer(planes//4)
self.relu2 = nn.ReLU(inplace=True)
self.conv3 = conv1x1(planes//4, planes)
self.stride = stride
self.relu = nn.ReLU(inplace=True)
def forward(self, x):
x = self.bn0(x);
x = self.relu0(x);
identity = x
out = self.conv1(x)
out = self.bn1(out)
out = self.relu1(out)
out = self.conv2(out)
out = self.bn2(out)
out = self.relu2(out)
out = self.conv3(out)
out += identity
return out
# Invoke the block
block = ResnetBottleNeckBlock(64, 64, stride=1);
# Initialize network and load block on machine
layers = []
layers.append(block);
net = nn.Sequential(*layers);
# Run data through network
x = torch.randn(1, 64, 224, 224)
y = net(x)
print(x.shape, y.shape);
# Export Model to Load on Netron
torch.onnx.export(net, # model being run
x, # model input (or a tuple for multiple inputs)
"model.onnx", # where to save the model (can be a file or file-like object)
export_params=True, # store the trained parameter weights inside the model file
opset_version=10, # the ONNX version to export the model to
do_constant_folding=True, # whether to execute constant folding for optimization
input_names = ['input'], # the model's input names
output_names = ['output'], # the model's output names
dynamic_axes={'input' : {0 : 'batch_size'}, # variable lenght axes
'output' : {0 : 'batch_size'}})
netron.start('model.onnx', port=9998);
```
<a id='5-4'></a>
## Creating block using traditional Keras
- Code credits: https://keras.io/
```
# Traditional-Keras
import keras
import keras.layers as kla
import keras.models as kmo
import tensorflow as tf
from keras.models import Model
backend = 'channels_last'
from keras import layers
def resnet_conv_block(input_tensor,
kernel_size,
filters,
stage,
block,
strides=(1, 1)):
filters1, filters2, filters3 = filters
bn_axis = 3
conv_name_base = 'res' + str(stage) + block + '_branch'
bn_name_base = 'bn' + str(stage) + block + '_branch'
#Common Elements
start = layers.BatchNormalization(axis=bn_axis, name=bn_name_base + '0a')(input_tensor)
start = layers.Activation('relu')(start)
# Branch - 1
# Identity
shortcut = start
# Branch - 2
x = layers.Conv2D(filters1, (1, 1), strides=strides,
kernel_initializer='he_normal',
name=conv_name_base + '2a')(start)
x = layers.BatchNormalization(axis=bn_axis, name=bn_name_base + '2a')(x)
x = layers.Activation('relu')(x)
x = layers.Conv2D(filters2, (3, 3), strides=strides,
kernel_initializer='he_normal',
name=conv_name_base + '2b', padding="same")(x)
x = layers.BatchNormalization(axis=bn_axis, name=bn_name_base + '2b')(x)
x = layers.Activation('relu')(x)
x = layers.Conv2D(filters3, (1, 1),
kernel_initializer='he_normal',
name=conv_name_base + '2c')(x);
x = layers.add([x, shortcut])
x = layers.Activation('relu')(x)
return x
def create_model(input_shape, kernel_size, filters, stage, block):
img_input = layers.Input(shape=input_shape);
x = resnet_conv_block(img_input, kernel_size, filters, stage, block)
return Model(img_input, x);
# Invoke the block
kernel_size=3;
filters=[16, 16, 64];
input_shape=(224, 224, 64);
model = create_model(input_shape, kernel_size, filters, 0, "0");
# Run data through network
x = tf.placeholder(tf.float32, shape=(1, 224, 224, 64))
y = model(x)
print(x.shape, y.shape)
# Export Model to Load on Netron
model.save("final.h5");
netron.start("final.h5", port=8082)
```
# Goals Completed
### 1. Learn to implement Resnet V2 Bottleneck Block (Type - 1) using monk
- Monk's Keras
- Monk's Pytorch
- Monk's Mxnet
### 2. Use network Monk's debugger to create complex blocks
### 3. Understand how syntactically different it is to implement the same using
- Traditional Keras
- Traditional Pytorch
- Traditional Mxnet
|
github_jupyter
|
```
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import numpy as np
from pyproj import CRS
import pathlib
from pathlib import Path
from shapely import wkt
from tqdm import tqdm
import math
import codecs
from shapely import wkt
import folium
from folium import features
from folium import plugins
import gzip
from xml.etree.ElementTree import Element, SubElement, Comment, tostring
import xml.etree.ElementTree as ET
# to read the excel
from openpyxl import load_workbook
from openpyxl import Workbook
# import folium
from shapely.geometry import LineString, MultiLineString
import branca.colormap as cmp
from folium.plugins import Search
from tqdm import tqdm
import time
import datetime
from datetime import timedelta
# set the working directory
BASE_DIR = Path.cwd()
BASE_DIR
# save as geojson
def get_foldercreation_inf():
fname = pathlib.Path("../SF_all_trips/sf-tscore-all-trips-20PCsample-updatedRideHailFleet-updatedParking__etg/ITERS/it.4/4.linkstats.csv.gz")
assert fname.exists(), f'No such file: {fname}' # check that the file exists
ctime = datetime.datetime.fromtimestamp(fname.stat().st_ctime)
return ctime
# return ctime.strftime("%Y-%m-%d")
def get_dataframe(_time):
# linkstats file
linkstats = pd.read_csv("../SF_all_trips/sf-tscore-all-trips-20PCsample-updatedRideHailFleet-updatedParking__etg/ITERS/it.4/4.linkstats.csv.gz", compression="gzip", low_memory=False)
time = int(_time)
linkstats = linkstats[linkstats["hour"]==(time)].copy()
linkstats=linkstats.add_prefix("linkstats_")
linkstats.rename(columns={('linkstats_link'): 'id'}, inplace=True)
linkstats["id"] = linkstats["id"].astype('string')
date_time = get_foldercreation_inf()
if int(_time)<24:
date_time = date_time.strftime("%Y-%m-%d")
time_stamp = f'{int(_time):02d}'
linkstats["date_time"] = (date_time + " " + "{}:00:00".format(f'{int(_time):02d}'))
else:
date_time = get_foldercreation_inf() + datetime.timedelta(days=1)
date_time = date_time.strftime("%Y-%m-%d")
new_time = int(_time) - 24
linkstats["date_time"] = (date_time + " " + "{}:00:00".format(f'{abs(int(new_time)):02d}'))
return linkstats
# read the road network
sf_roadnetwork = gpd.read_file(BASE_DIR.parent.joinpath( 'Network',"sfNetwork.geojsonl"))
sf_roadnetwork = sf_roadnetwork[["id","modes","length","lanes","from","to","capacity","geometry"]]
sftimevariantnetwork =pd.DataFrame()
for time_hour in tqdm(range(0,30)):
# get the hour and filter the linkstat file
linkstats = get_dataframe(str(time_hour))
# merge with featureclass of SF data
comparision_network = sf_roadnetwork.merge(linkstats,on="id").copy()
# calculate the freespeed (mph), congested speed (mph), ratio (congestedsped/freespeed)
# linkstats
comparision_network["linkstats_freespeed_mph"] = comparision_network["linkstats_freespeed"]*2.23694
comparision_network["linkstats_congspd_mph"] = (comparision_network["linkstats_length"]/comparision_network["linkstats_traveltime"])*2.23694
comparision_network["linkstats_ratio"] = comparision_network["linkstats_congspd_mph"] / comparision_network["linkstats_freespeed_mph"]
comparision_network["linkstats_vc_ratio"] = comparision_network["linkstats_volume"]*5 / comparision_network["capacity"]
if int(time_hour)==0:
sftimevariantnetwork = comparision_network.copy()
else:
sftimevariantnetwork = pd.concat([sftimevariantnetwork,comparision_network], ignore_index=True)
# lastly, export the network
# sftimevariantnetwork.to_file(BASE_DIR.parent.joinpath("exported", ("sf_timevariantnetwork.geojson")), driver='GeoJSON')
linkstats.head()
# sftimevariantnetwork.to_csv(BASE_DIR.parent.joinpath("exported", ("sf_timevariantnetwork.csv")))
# read the road network, incase it is already saved in the geojson file
# sftimevariantnetwork = gpd.read_file(BASE_DIR.parent.joinpath("exported", ("sf_timevariantnetwork.geojson")))
# keep only selected columns fields
sf_timevariantnetwork = sftimevariantnetwork[["id", "modes","length","lanes","capacity","geometry",
'linkstats_freespeed','linkstats_volume', 'linkstats_traveltime',
'date_time', 'linkstats_freespeed_mph', 'linkstats_congspd_mph', 'linkstats_ratio', "linkstats_vc_ratio"]]
sf_timevariantnetwork['date_time']=pd.to_datetime(sf_timevariantnetwork['date_time']).dt.strftime('%Y-%m-%dT%H:%M:%S')
sf_timevariantnetwork["time"] = pd.to_datetime(sf_timevariantnetwork["date_time"]).dt.strftime('%Y-%m-%dT%H:%M:%S')
# add more green shades for 85% --> 100%
# green_shades = ['#008000', '#198c19', '#329932', '#4ca64c', '#66b266', '#7fbf7f', '#99cc99', '#b2d8b2', '#cce5cc', '#e5f2e5']
# colors for congstd speed/freespeed ratio
color_range_pct = ["#ff0000","#ff6666","#ffb2b2","#ffdb99","#ffc966", "#ffa500",'#e5f2e5','#cce5cc','#b2d8b2','#99cc99','#7fbf7f','#66b266','#4ca64c','#329932', '#198c19','#008000']
# color_range_pct = ["#ff0000","#ff6666","#ffb2b2","#ffdb99","#ffc966", "#ffa500","#cce5cc","#99cc99","#66b266","#008000"]
step_pct = cmp.StepColormap(
color_range_pct,
vmin=0, vmax=1,
index=[0,0.2,0.3,0.5,.6,0.7,0.80,0.85, 0.87,0.89,0.91,0.93,0.95,0.97,0.99,1.00], #for change in the colors, not used fr linear
caption='% Speeds Difference' #Caption for Color scale or Legend
)
# colors for congstd speed/freespeed ratio
color_range_pct_vc = ['#008000', '#329932', '#66b266', '#99cc99', '#cce5cc', '#e5f2e5', # green shade
'#ffa500', "#ffb732",'#ffc966', '#ffdb99', "#ffedcc", # orange shade
'#ffe5e5', '#ffcccc','#ffb2b2','#ff9999','#ff6666', '#ff3232', '#ff0000' ] # red shade
# color_range_pct = ["#ff0000","#ff6666","#ffb2b2","#ffdb99","#ffc966", "#ffa500","#cce5cc","#99cc99","#66b266","#008000"]
step_pct_vc = cmp.StepColormap(
color_range_pct_vc,
vmin=0, vmax=1,
index=[0,0.1,0.2,0.3,0.4,0.5,
0.55,0.6,0.65,0.7,0.75,
0.80,0.85,0.90,0.95,0.97,0.99,1.00], #for change in the colors, not used fr linear
caption='Volume-to-Capacity ratio' #Caption for Color scale or Legend
)
# colors for congested speed and freespeed
color_range = ["#ff0000","#ff6666","#ffb2b2","#ffa500","#ffc966","#ffdb99", "#cce5cc","#99cc99","#66b266","#008000"]
step = cmp.StepColormap(color_range,vmin=0, vmax=100,index=[0,5,10,15,25,35,45,55,65,100], #for change in the colors, not used fr linear
caption=' Speeds (mph)' #Caption for Color scale or Legend
)
def getColorMap_pct(x):
return str(step_pct(x))
def getColorMap_pct_vc(x):
return str(step_pct_vc(x))
def getColorMap(x):
return str(step(x))
sf_timevariantnetwork["fillColor_ratio"] = sf_timevariantnetwork["linkstats_ratio"].apply(getColorMap_pct)
sf_timevariantnetwork["fillColor_vc_ratio"] = sf_timevariantnetwork["linkstats_vc_ratio"].apply(getColorMap_pct_vc)
sf_timevariantnetwork["fillColor_freespeed_mph"] = sf_timevariantnetwork["linkstats_freespeed_mph"].apply(getColorMap)
sf_timevariantnetwork["fillColor_congspd_mph"] = sf_timevariantnetwork["linkstats_congspd_mph"].apply(getColorMap)
def coords(geom):
return list(geom.coords)
sf_timevariantnetwork['points'] = sf_timevariantnetwork.apply(lambda row: coords(row.geometry), axis=1)
# groupby and aggreage columns by segment_links
df1 = sf_timevariantnetwork.groupby('id').agg({'modes':'first',
'length':'first',
'lanes':list,
'capacity':list,
'geometry':'first',
'linkstats_freespeed':list,
'linkstats_volume':list,
'linkstats_traveltime':list,
'date_time':list,
'linkstats_freespeed_mph':list,
'linkstats_congspd_mph':list,
'linkstats_ratio':list,
'linkstats_vc_ratio':list,
'time':list,
'fillColor_ratio':list,
'linkstats_volume':list,
'linkstats_traveltime':list,
'fillColor_freespeed_mph':list,
'fillColor_congspd_mph':list,
'fillColor_vc_ratio':list,
'points':'first'}).reset_index()
# Create timemap for ratio_congestedspeed_freespeed
def coords(geom):
return list(geom.coords)
features_ratio = [
{
'type':'Feature',
"geometry":{
'type': 'LineString',
'coordinates': coords(d.geometry),
},
'properties': {
'times': d['time'],
'color': "black",
'colors':d["fillColor_ratio"],
"weight":0.6,
"fillOpacity": 0.4,
}
}
for _,d in df1.iterrows()
]
from jinja2 import Template
_template = Template("""
{% macro script(this, kwargs) %}
L.Control.TimeDimensionCustom = L.Control.TimeDimension.extend({
_getDisplayDateFormat: function(date){
var newdate = new moment(date);
console.log(newdate)
return newdate.format("{{this.date_options}}");
}
});
{{this._parent.get_name()}}.timeDimension = L.timeDimension(
{
period: {{ this.period|tojson }},
}
);
var timeDimensionControl = new L.Control.TimeDimensionCustom(
{{ this.options|tojson }}
);
{{this._parent.get_name()}}.addControl(this.timeDimensionControl);
var geoJsonLayer = L.geoJson({{this.data}}, {
pointToLayer: function (feature, latLng) {
if (feature.properties.icon == 'marker') {
if(feature.properties.iconstyle){
return new L.Marker(latLng, {
icon: L.icon(feature.properties.iconstyle)});
}
//else
return new L.Marker(latLng);
}
if (feature.properties.icon == 'circle') {
if (feature.properties.iconstyle) {
return new L.circleMarker(latLng, feature.properties.iconstyle)
};
//else
return new L.circleMarker(latLng);
}
//else
return new L.Marker(latLng);
},
style: function(feature) {
lastIdx=feature.properties.colors.length-1
currIdx=feature.properties.colors.indexOf(feature.properties.color);
if(currIdx==lastIdx){
feature.properties.color = feature.properties.colors[currIdx+1]
}
else{
feature.properties.color =feature.properties.colors[currIdx+1]
}
return {color: feature.properties.color}
},
onEachFeature: function(feature, layer) {
if (feature.properties.popup) {
layer.bindPopup(feature.properties.popup);
}
}
})
var {{this.get_name()}} = L.timeDimension.layer.geoJson(
geoJsonLayer,
{
updateTimeDimension: true,
addlastPoint: {{ this.add_last_point|tojson }},
duration: {{ this.duration }},
}
).addTo({{this._parent.get_name()}});
{% endmacro %}
""")
import folium
from folium.plugins import TimestampedGeoJson
m = folium.Map(location=[37.760015, -122.447110], zoom_start=13, tiles="cartodbpositron")
t=TimestampedGeoJson({
'type': 'FeatureCollection',
'features': features_ratio,
}, transition_time=1500,loop=True,period='PT1H', add_last_point=False,auto_play=True)
t._template=_template
t.add_to(m)
step_pct.add_to(m)
# Add title
map_title = "Ratio between Congested Speed (mph) and Free Speed (mph)"
title_html = '''
<h3 align="center" style="font-size:16px"><b>{}</b></h3>
'''.format(map_title)
m.get_root().html.add_child(folium.Element(title_html))
file_name = BASE_DIR.parent.joinpath("exported", ("linkstat_ratio_timemap.html"))
m.save(str(file_name))
# m
# Create timemap for v/c ratio
def coords(geom):
return list(geom.coords)
features_ratio = [
{
'type':'Feature',
"geometry":{
'type': 'LineString',
'coordinates': coords(d.geometry),
},
'properties': {
'times': d['time'],
'color': "black",
'colors':d["fillColor_vc_ratio"],
"weight":0.6,
"fillOpacity": 0.4,
}
}
for _,d in df1.iterrows()
]
from jinja2 import Template
_template = Template("""
{% macro script(this, kwargs) %}
L.Control.TimeDimensionCustom = L.Control.TimeDimension.extend({
_getDisplayDateFormat: function(date){
var newdate = new moment(date);
console.log(newdate)
return newdate.format("{{this.date_options}}");
}
});
{{this._parent.get_name()}}.timeDimension = L.timeDimension(
{
period: {{ this.period|tojson }},
}
);
var timeDimensionControl = new L.Control.TimeDimensionCustom(
{{ this.options|tojson }}
);
{{this._parent.get_name()}}.addControl(this.timeDimensionControl);
var geoJsonLayer = L.geoJson({{this.data}}, {
pointToLayer: function (feature, latLng) {
if (feature.properties.icon == 'marker') {
if(feature.properties.iconstyle){
return new L.Marker(latLng, {
icon: L.icon(feature.properties.iconstyle)});
}
//else
return new L.Marker(latLng);
}
if (feature.properties.icon == 'circle') {
if (feature.properties.iconstyle) {
return new L.circleMarker(latLng, feature.properties.iconstyle)
};
//else
return new L.circleMarker(latLng);
}
//else
return new L.Marker(latLng);
},
style: function(feature) {
lastIdx=feature.properties.colors.length-1
currIdx=feature.properties.colors.indexOf(feature.properties.color);
if(currIdx==lastIdx){
feature.properties.color = feature.properties.colors[currIdx+1]
}
else{
feature.properties.color =feature.properties.colors[currIdx+1]
}
return {color: feature.properties.color}
},
onEachFeature: function(feature, layer) {
if (feature.properties.popup) {
layer.bindPopup(feature.properties.popup);
}
}
})
var {{this.get_name()}} = L.timeDimension.layer.geoJson(
geoJsonLayer,
{
updateTimeDimension: true,
addlastPoint: {{ this.add_last_point|tojson }},
duration: {{ this.duration }},
}
).addTo({{this._parent.get_name()}});
{% endmacro %}
""")
import folium
from folium.plugins import TimestampedGeoJson
m = folium.Map(location=[37.760015, -122.447110], zoom_start=13, tiles="cartodbpositron")
t=TimestampedGeoJson({
'type': 'FeatureCollection',
'features': features_ratio,
}, transition_time=1500,loop=True,period='PT1H', add_last_point=False,auto_play=True)
t._template=_template
t.add_to(m)
step_pct_vc.add_to(m)
# Add title
map_title = "Volume-to-Capacity Ratio"
title_html = '''
<h3 align="center" style="font-size:16px"><b>{}</b></h3>
'''.format(map_title)
m.get_root().html.add_child(folium.Element(title_html))
file_name = BASE_DIR.parent.joinpath("exported","extended_run",("linkst_vc_ratio_timemap.html"))
m.save(str(file_name))
# m
# Create timemap for freespeed (mph)
def coords(geom):
return list(geom.coords)
features_freespeed = [
{
'type':'Feature',
"geometry":{
'type': 'LineString',
'coordinates': coords(d.geometry),
},
'properties': {
'times': d['time'],
'color': "black",
'colors':d["fillColor_freespeed_mph"],
'weight':0.6,
"fillOpacity": 0.4,
}
}
for _,d in df1.iterrows()
]
from jinja2 import Template
_template = Template("""
{% macro script(this, kwargs) %}
L.Control.TimeDimensionCustom = L.Control.TimeDimension.extend({
_getDisplayDateFormat: function(date){
var newdate = new moment(date);
console.log(newdate)
return newdate.format("{{this.date_options}}");
}
});
{{this._parent.get_name()}}.timeDimension = L.timeDimension(
{
period: {{ this.period|tojson }},
}
);
var timeDimensionControl = new L.Control.TimeDimensionCustom(
{{ this.options|tojson }}
);
{{this._parent.get_name()}}.addControl(this.timeDimensionControl);
var geoJsonLayer = L.geoJson({{this.data}}, {
pointToLayer: function (feature, latLng) {
if (feature.properties.icon == 'marker') {
if(feature.properties.iconstyle){
return new L.Marker(latLng, {
icon: L.icon(feature.properties.iconstyle)});
}
//else
return new L.Marker(latLng);
}
if (feature.properties.icon == 'circle') {
if (feature.properties.iconstyle) {
return new L.circleMarker(latLng, feature.properties.iconstyle)
};
//else
return new L.circleMarker(latLng);
}
//else
return new L.Marker(latLng);
},
style: function(feature) {
lastIdx=feature.properties.colors.length-1
currIdx=feature.properties.colors.indexOf(feature.properties.color);
if(currIdx==lastIdx){
feature.properties.color = feature.properties.colors[currIdx+1]
}
else{
feature.properties.color =feature.properties.colors[currIdx+1]
}
return {color: feature.properties.color}
},
onEachFeature: function(feature, layer) {
if (feature.properties.popup) {
layer.bindPopup(feature.properties.popup);
}
}
})
var {{this.get_name()}} = L.timeDimension.layer.geoJson(
geoJsonLayer,
{
updateTimeDimension: true,
addlastPoint: {{ this.add_last_point|tojson }},
duration: {{ this.duration }},
}
).addTo({{this._parent.get_name()}});
{% endmacro %}
""")
import folium
from folium.plugins import TimestampedGeoJson
m = folium.Map(location=[37.760015, -122.447110], zoom_start=13, tiles="cartodbpositron")
# Add title
map_title = "Free Speed (mph)"
title_html = '''
<h3 align="center" style="font-size:16px"><b>{}</b></h3>
'''.format(map_title)
m.get_root().html.add_child(folium.Element(title_html))
t=TimestampedGeoJson({
'type': 'FeatureCollection',
'features': features_freespeed,
}, transition_time=1500,loop=True,period='PT1H', add_last_point=False,auto_play=True)
t._template=_template
t.add_to(m)
step.add_to(m)
file_name = BASE_DIR.parent.joinpath("exported", ("linkstat_freespeed_timemap.html"))
m.save(str(file_name))
# m
# Create timemap for congested speed (mph)
def coords(geom):
return list(geom.coords)
features_congestedspeed = [
{
'type':'Feature',
"geometry":{
'type': 'LineString',
'coordinates': coords(d.geometry),
},
'properties': {
'times': d['time'],
'color': "black",
'colors':d["fillColor_congspd_mph"],
'weight':0.6,
"fillOpacity": 0.4,
}
}
for _,d in df1.iterrows()
]
from jinja2 import Template
_template = Template("""
{% macro script(this, kwargs) %}
L.Control.TimeDimensionCustom = L.Control.TimeDimension.extend({
_getDisplayDateFormat: function(date){
var newdate = new moment(date);
console.log(newdate)
return newdate.format("{{this.date_options}}");
}
});
{{this._parent.get_name()}}.timeDimension = L.timeDimension(
{
period: {{ this.period|tojson }},
}
);
var timeDimensionControl = new L.Control.TimeDimensionCustom(
{{ this.options|tojson }}
);
{{this._parent.get_name()}}.addControl(this.timeDimensionControl);
var geoJsonLayer = L.geoJson({{this.data}}, {
pointToLayer: function (feature, latLng) {
if (feature.properties.icon == 'marker') {
if(feature.properties.iconstyle){
return new L.Marker(latLng, {
icon: L.icon(feature.properties.iconstyle)});
}
//else
return new L.Marker(latLng);
}
if (feature.properties.icon == 'circle') {
if (feature.properties.iconstyle) {
return new L.circleMarker(latLng, feature.properties.iconstyle)
};
//else
return new L.circleMarker(latLng);
}
//else
return new L.Marker(latLng);
},
style: function(feature) {
lastIdx=feature.properties.colors.length-1
currIdx=feature.properties.colors.indexOf(feature.properties.color);
if(currIdx==lastIdx){
feature.properties.color = feature.properties.colors[currIdx+1]
}
else{
feature.properties.color =feature.properties.colors[currIdx+1]
}
return {color: feature.properties.color}
},
onEachFeature: function(feature, layer) {
if (feature.properties.popup) {
layer.bindPopup(feature.properties.popup);
}
}
})
var {{this.get_name()}} = L.timeDimension.layer.geoJson(
geoJsonLayer,
{
updateTimeDimension: true,
addlastPoint: {{ this.add_last_point|tojson }},
duration: {{ this.duration }},
}
).addTo({{this._parent.get_name()}});
{% endmacro %}
""")
import folium
from folium.plugins import TimestampedGeoJson
m = folium.Map(location=[37.760015, -122.447110], zoom_start=13, tiles="cartodbpositron")
t=TimestampedGeoJson({
'type': 'FeatureCollection',
'features': features_congestedspeed,
}, transition_time=1500,loop=True,period='PT1H', add_last_point=False,auto_play=True)
t._template=_template
t.add_to(m)
step.add_to(m)
# Add title
map_title = "Congested Speed (mph)"
title_html = '''
<h3 align="center" style="font-size:16px"><b>{}</b></h3>
'''.format(map_title)
m.get_root().html.add_child(folium.Element(title_html))
file_name = BASE_DIR.parent.joinpath("exported", ("linkstat_congestedspeed_timemap.html"))
m.save(str(file_name))
# m
# get map for each different time
# static maps for congestedspeed/freespeed
def get_dataframe(_time):
# linkstats file
linkstats = pd.read_csv("../SF_all_trips/sf-tscore-all-trips-20PCsample-updatedRideHailFleet-updatedParking__etg/ITERS/it.4/4.linkstats.csv.gz", compression="gzip", low_memory=False)
# unmodified_linkstats = pd.read_csv(BASE_DIR.parent.joinpath("runs", "sf-tscore-int-int-trips-model-network-events-20PC-sample-bpr-func__tlm","ITERS","it.30", "30.linkstats_unmodified.csv.gz"),compression="gzip", low_memory=False)
time = int(_time)
linkstats = linkstats[linkstats["hour"]==(time)].copy()
linkstats=linkstats.add_prefix("linkstats_")
linkstats.rename(columns={('linkstats_link'): 'id'}, inplace=True)
linkstats["id"] = linkstats["id"].astype('string')
return linkstats
def highlight_function(feature):
return {"fillColor": "#ffff00", "color": "#ffff00", "weight": 5,"fillOpacity": 0.40 }
color_range_pct = ["#ff0000","#ff6666","#ffb2b2","#ffdb99","#ffc966", "#ffa500",'#e5f2e5','#cce5cc','#b2d8b2','#99cc99','#7fbf7f','#66b266','#4ca64c','#329932', '#198c19','#008000']
step_pct = cmp.StepColormap(
color_range_pct,
vmin=0, vmax=1,
index=[0,0.2,0.3,0.5,.6,0.7,0.80,0.85, 0.87,0.89,0.91,0.93,0.95,0.97,0.99,1.00], #for change in the colors, not used fr linear
caption='% Speeds Difference' #Caption for Color scale or Legend
)
# read the road network
sf_roadnetwork = gpd.read_file(BASE_DIR.parent.joinpath("Network", "sfNetwork.geojsonl"))
sf_roadnetwork = sf_roadnetwork[["id","modes","length","lanes","from","to","capacity","geometry"]]
for time_hour in tqdm(range(0,30)):
# set the map
pct_m = folium.Map([37.760015, -122.447110], zoom_start=13, tiles="cartodbpositron")
# get the hour and filter the linkstat file
linkstats = get_dataframe(str(time_hour))
# merge with featureclass of SF data
comparision_network = sf_roadnetwork.merge(linkstats,on="id")
# calculate the freespeed (mph), congested speed (mph), ratio (congestedsped/freespeed)
# linkstats
comparision_network["linkstats_freespeed_mph"] = comparision_network["linkstats_freespeed"]*2.23694
comparision_network["linkstats_congspd_mph"] = (comparision_network["linkstats_length"]/comparision_network["linkstats_traveltime"])*2.23694
comparision_network["linkstats_ratio"] = comparision_network["linkstats_congspd_mph"] / comparision_network["linkstats_freespeed_mph"]
time_stamp = ""
# folium
if time_hour<30:
time_stamp = f'{time_hour:02d}'
layer_name = str(time_stamp)
# layer_name=str(str(time_hour) + (' am' if time_hour < 12 else ' pm'))
ratio_feature_group = folium.FeatureGroup(name=layer_name)
pct_feature_group = folium.GeoJson(comparision_network,
name = ("Hour - " + layer_name),
style_function=lambda x: {
"fillColor": step_pct(x["properties"]["linkstats_ratio"]),
"color": step_pct(x["properties"]["linkstats_ratio"]),
"fillOpacity": 0.2,
"weight":1,
},
tooltip=folium.GeoJsonTooltip(fields=["id","length", "linkstats_freespeed_mph", "linkstats_traveltime","linkstats_congspd_mph"],
aliases=["Link ID", "Segment Length (m)", "Freespeed (mph)", "Travel time (sec)", "Congested Speed (mph)"], localize=True),
popup = folium.GeoJsonPopup(fields=["id","length", "linkstats_freespeed_mph", "linkstats_traveltime","linkstats_congspd_mph"],
aliases=["Link ID", "Segment Length (m)", "Freespeed (mph)", "Travel time (sec)", "Congested Speed (mph)"], localize=True),
highlight_function=highlight_function,
zoom_on_click=True
).add_to(ratio_feature_group)
# Add search functionality to the map
search_link = Search(layer=pct_feature_group, geom_type="LineString", placeholders = "Search for Link ID",
collapsed="False", search_label = 'id', search_zoom = 17, position='topleft',
).add_to(pct_m)
ratio_feature_group.add_to(pct_m)
folium.LayerControl().add_to(pct_m)
map_title = "Ratio between Congested Speed and Free Speed"
title_html = '''<h3 align="center" style="font-size:16px"><b>{}</b></h3>'''.format(map_title)
pct_m.get_root().html.add_child(folium.Element(title_html))
# save the file
file_name = BASE_DIR.parent.joinpath("exported", ("linkstat_ratio_timemap_{}.html").format(time_stamp))
pct_m.save(str(file_name))
```
|
github_jupyter
|
# Experiments comparing the performance of traditional pooling operations and entropy pooling within a shallow neural network and Lenet. The experiments use cifar10 and cifar100.
```
%matplotlib inline
import torch
import torchvision
import torchvision.transforms as transforms
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR100(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=4,
shuffle=True, num_workers=8)
testset = torchvision.datasets.CIFAR100(root='./data', train=False,
download=True, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=4,
shuffle=False, num_workers=8)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.nn.modules.utils import _pair, _quadruple
import time
from skimage.measure import shannon_entropy
from scipy import stats
from torch.nn.modules.utils import _pair, _quadruple
import time
from skimage.measure import shannon_entropy
from scipy import stats
import numpy as np
class EntropyPool2d(nn.Module):
def __init__(self, kernel_size=3, stride=1, padding=0, same=False, entr='high'):
super(EntropyPool2d, self).__init__()
self.k = _pair(kernel_size)
self.stride = _pair(stride)
self.padding = _quadruple(padding) # convert to l, r, t, b
self.same = same
self.entr = entr
def _padding(self, x):
if self.same:
ih, iw = x.size()[2:]
if ih % self.stride[0] == 0:
ph = max(self.k[0] - self.stride[0], 0)
else:
ph = max(self.k[0] - (ih % self.stride[0]), 0)
if iw % self.stride[1] == 0:
pw = max(self.k[1] - self.stride[1], 0)
else:
pw = max(self.k[1] - (iw % self.stride[1]), 0)
pl = pw // 2
pr = pw - pl
pt = ph // 2
pb = ph - pt
padding = (pl, pr, pt, pb)
else:
padding = self.padding
return padding
def forward(self, x):
# using existing pytorch functions and tensor ops so that we get autograd,
# would likely be more efficient to implement from scratch at C/Cuda level
start = time.time()
x = F.pad(x, self._padding(x), mode='reflect')
x_detached = x.cpu().detach()
x_unique, x_indices, x_inverse, x_counts = np.unique(x_detached,
return_index=True,
return_inverse=True,
return_counts=True)
freq = torch.FloatTensor([x_counts[i] / len(x_inverse) for i in x_inverse]).cuda()
x_probs = freq.view(x.shape)
x_probs = x_probs.unfold(2, self.k[0], self.stride[0]).unfold(3, self.k[1], self.stride[1])
x_probs = x_probs.contiguous().view(x_probs.size()[:4] + (-1,))
if self.entr is 'high':
x_probs, indices = torch.min(x_probs.cuda(), dim=-1)
elif self.entr is 'low':
x_probs, indices = torch.max(x_probs.cuda(), dim=-1)
else:
raise Exception('Unknown entropy mode: {}'.format(self.entr))
x = x.unfold(2, self.k[0], self.stride[0]).unfold(3, self.k[1], self.stride[1])
x = x.contiguous().view(x.size()[:4] + (-1,))
indices = indices.view(indices.size() + (-1,))
pool = torch.gather(input=x, dim=-1, index=indices)
return pool.squeeze(-1)
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import time
from sklearn.metrics import f1_score
MAX = 'max'
AVG = 'avg'
HIGH_ENTROPY = 'high_entr'
LOW_ENTROPY = 'low_entr'
class Net1Pool(nn.Module):
def __init__(self, num_classes=10, pooling=MAX):
super(Net1Pool, self).__init__()
self.conv1 = nn.Conv2d(3, 30, 5)
if pooling is MAX:
self.pool = nn.MaxPool2d(2, 2)
elif pooling is AVG:
self.pool = nn.AvgPool2d(2, 2)
elif pooling is HIGH_ENTROPY:
self.pool = EntropyPool2d(2, 2, entr='high')
elif pooling is LOW_ENTROPY:
self.pool = EntropyPool2d(2, 2, entr='low')
self.fc0 = nn.Linear(30 * 14 * 14, num_classes)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = x.view(-1, 30 * 14 * 14)
x = F.relu(self.fc0(x))
return x
class Net2Pool(nn.Module):
def __init__(self, num_classes=10, pooling=MAX):
super(Net2Pool, self).__init__()
self.conv1 = nn.Conv2d(3, 50, 5, 1)
self.conv2 = nn.Conv2d(50, 50, 5, 1)
if pooling is MAX:
self.pool = nn.MaxPool2d(2, 2)
elif pooling is AVG:
self.pool = nn.AvgPool2d(2, 2)
elif pooling is HIGH_ENTROPY:
self.pool = EntropyPool2d(2, 2, entr='high')
elif pooling is LOW_ENTROPY:
self.pool = EntropyPool2d(2, 2, entr='low')
self.fc1 = nn.Linear(5*5*50, 500)
self.fc2 = nn.Linear(500, num_classes)
def forward(self, x):
x = F.relu(self.conv1(x))
x = self.pool(x)
x = F.relu(self.conv2(x))
x = self.pool(x)
x = x.view(-1, 5*5*50)
x = F.relu(self.fc1(x))
x = self.fc2(x)
return x
def configure_net(net, device):
net.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
return net, optimizer, criterion
def train(net, optimizer, criterion, trainloader, device, epochs=10, logging=2000):
for epoch in range(epochs):
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
start = time.time()
inputs, labels = data
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if i % logging == logging - 1:
print('[%d, %5d] loss: %.3f duration: %.5f' %
(epoch + 1, i + 1, running_loss / logging, time.time() - start))
running_loss = 0.0
print('Finished Training')
def test(net, testloader, device):
correct = 0
total = 0
predictions = []
l = []
with torch.no_grad():
for data in testloader:
images, labels = data
images, labels = images.to(device), labels.to(device)
outputs = net(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
predictions.extend(predicted.cpu().numpy())
l.extend(labels.cpu().numpy())
print('Accuracy: {}'.format(100 * correct / total))
epochs = 10
logging = 15000
num_classes = 100
print('- - - - - - - - -- - - - 2 pool - - - - - - - - - - - - - - - -')
print('- - - - - - - - -- - - - MAX - - - - - - - - - - - - - - - -')
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
net, optimizer, criterion = configure_net(Net2Pool(num_classes=num_classes, pooling=MAX), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - AVG - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net2Pool(num_classes=num_classes, pooling=AVG), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - HIGH - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net2Pool(num_classes=num_classes, pooling=HIGH_ENTROPY), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - LOW - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net2Pool(num_classes=num_classes, pooling=LOW_ENTROPY), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - 1 pool - - - - - - - - - - - - - - - -')
print('- - - - - - - - -- - - - MAX - - - - - - - - - - - - - - - -')
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
net, optimizer, criterion = configure_net(Net1Pool(num_classes=num_classes, pooling=MAX), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - AVG - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net1Pool(num_classes=num_classes, pooling=AVG), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - HIGH - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net1Pool(num_classes=num_classes, pooling=HIGH_ENTROPY), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
print('- - - - - - - - -- - - - LOW - - - - - - - - - - - - - - - -')
net, optimizer, criterion = configure_net(Net1Pool(num_classes=num_classes, pooling=LOW_ENTROPY), device)
train(net, optimizer, criterion, trainloader, device, epochs=epochs, logging=logging)
test(net, testloader, device)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/AIWintermuteAI/aXeleRate/blob/dev/resources/aXeleRate_mark_detector.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## M.A.R.K. Detection model Training and Inference
In this notebook we will use axelerate, Keras-based framework for AI on the edge, to quickly setup model training and then after training session is completed convert it to .tflite and .kmodel formats.
First, let's take care of some administrative details.
1) Before we do anything, make sure you have choosen GPU as Runtime type (in Runtime - > Change Runtime type).
2) We need to mount Google Drive for saving our model checkpoints and final converted model(s). Press on Mount Google Drive button in Files tab on your left.
In the next cell we clone axelerate Github repository and import it.
**It is possible to use pip install or python setup.py install, but in that case you will need to restart the enironment.** Since I'm trying to make the process as streamlined as possibile I'm using sys.path.append for import.
```
%load_ext tensorboard
#we need imgaug 0.4 for image augmentations to work properly, see https://stackoverflow.com/questions/62580797/in-colab-doing-image-data-augmentation-with-imgaug-is-not-working-as-intended
!pip uninstall -y imgaug && pip uninstall -y albumentations && pip install imgaug==0.4
!git clone https://github.com/AIWintermuteAI/aXeleRate.git
import sys
sys.path.append('/content/aXeleRate')
from axelerate import setup_training, setup_inference
```
At this step you typically need to get the dataset. You can use !wget command to download it from somewhere on the Internet or !cp to copy from My Drive as in this example
```
!cp -r /content/drive/'My Drive'/pascal_20_segmentation.zip .
!unzip --qq pascal_20_segmentation.zip
```
Dataset preparation and postprocessing are discussed in the article here:
The annotation tool I use is LabelImg
https://github.com/tzutalin/labelImg
Let's visualize our detection model test dataset. There are images in validation folder with corresponding annotations in PASCAL-VOC format in validation annotations folder.
```
%matplotlib inline
!gdown https://drive.google.com/uc?id=1s2h6DI_1tHpLoUWRc_SavvMF9jYG8XSi #dataset
!gdown https://drive.google.com/uc?id=1-bDRZ9Z2T81SfwhHEfZIMFG7FtMQ5ZiZ #pre-trained model
!unzip --qq mark_dataset.zip
from axelerate.networks.common_utils.augment import visualize_detection_dataset
visualize_detection_dataset(img_folder='mark_detection/imgs_validation', ann_folder='mark_detection/ann_validation', num_imgs=10, img_size=224, augment=True)
```
Next step is defining a config dictionary. Most lines are self-explanatory.
Type is model frontend - Classifier, Detector or Segnet
Architecture is model backend (feature extractor)
- Full Yolo
- Tiny Yolo
- MobileNet1_0
- MobileNet7_5
- MobileNet5_0
- MobileNet2_5
- SqueezeNet
- NASNetMobile
- DenseNet121
- ResNet50
For more information on anchors, please read here
https://github.com/pjreddie/darknet/issues/568
Labels are labels present in your dataset.
IMPORTANT: Please, list all the labels present in the dataset.
object_scale determines how much to penalize wrong prediction of confidence of object predictors
no_object_scale determines how much to penalize wrong prediction of confidence of non-object predictors
coord_scale determines how much to penalize wrong position and size predictions (x, y, w, h)
class_scale determines how much to penalize wrong class prediction
For converter type you can choose the following:
'k210', 'tflite_fullint', 'tflite_dynamic', 'edgetpu', 'openvino', 'onnx'
## Parameters for Person Detection
K210, which is where we will run the network, has constrained memory (5.5 RAM) available, so with Micropython firmware, the largest model you can run is about 2 MB, which limits our architecture choice to Tiny Yolo, MobileNet(up to 0.75 alpha) and SqueezeNet. Out of these 3 architectures, only one comes with pre-trained model - MobileNet. So, to save the training time we will use Mobilenet with alpha 0.75, which has ... parameters. For objects that do not have that much variety, you can use MobileNet with lower alpha, down to 0.25.
```
config = {
"model":{
"type": "Detector",
"architecture": "MobileNet5_0",
"input_size": 224,
"anchors": [0.57273, 0.677385, 1.87446, 2.06253, 3.33843, 5.47434, 7.88282, 3.52778, 9.77052, 9.16828],
"labels": ["mark"],
"coord_scale" : 1.0,
"class_scale" : 1.0,
"object_scale" : 5.0,
"no_object_scale" : 1.0
},
"weights" : {
"full": "",
"backend": "imagenet"
},
"train" : {
"actual_epoch": 50,
"train_image_folder": "mark_detection/imgs",
"train_annot_folder": "mark_detection/ann",
"train_times": 1,
"valid_image_folder": "mark_detection/imgs_validation",
"valid_annot_folder": "mark_detection/ann_validation",
"valid_times": 1,
"valid_metric": "mAP",
"batch_size": 32,
"learning_rate": 1e-3,
"saved_folder": F"/content/drive/MyDrive/mark_detector",
"first_trainable_layer": "",
"augumentation": True,
"is_only_detect" : False
},
"converter" : {
"type": ["k210","tflite"]
}
}
```
Let's check what GPU we have been assigned in this Colab session, if any.
```
from tensorflow.python.client import device_lib
device_lib.list_local_devices()
```
Also, let's open Tensorboard, where we will be able to watch model training progress in real time. Training and validation logs also will be saved in project folder.
Since there are no logs before we start the training, tensorboard will be empty. Refresh it after first epoch.
```
%tensorboard --logdir logs
```
Finally we start the training by passing config dictionary we have defined earlier to setup_training function. The function will start the training with Checkpoint, Reduce Learning Rate on Plateau and Early Stopping callbacks. After the training has stopped, it will convert the best model into the format you have specified in config and save it to the project folder.
```
from keras import backend as K
K.clear_session()
model_path = setup_training(config_dict=config)
```
After training it is good to check the actual perfomance of your model by doing inference on your validation dataset and visualizing results. This is exactly what next block does. Obviously since our model has only trained on a few images the results are far from stellar, but if you have a good dataset, you'll have better results.
```
from keras import backend as K
K.clear_session()
setup_inference(config, model_path)
```
My end results are:
{'fscore': 0.942528735632184, 'precision': 0.9318181818181818, 'recall': 0.9534883720930233}
**You can obtain these results by loading a pre-trained model.**
Good luck and happy training! Have a look at these articles, that would allow you to get the most of Google Colab or connect to local runtime if there are no GPUs available;
https://medium.com/@oribarel/getting-the-most-out-of-your-google-colab-2b0585f82403
https://research.google.com/colaboratory/local-runtimes.html
|
github_jupyter
|
# Docutils
## Presentation
Click [__here__] (youtube link) for the video presentation
## Summary of Support Files
- `demo.ipynb`: the notebook containing this tutorial code
- `test.csv`: a small file data used in the tutorial code
## Installation Instructions
Use `!pip install docutils` to install the `docutils` package. Next, use `import docutils`to import the package into your notebook.
For example, to import specific modules from `docutils` package use the following line of code:
`from docutils import core, io`
Below is a list of modules and subpackages as apart of the `docutils` package:
## Guide
__docutils 0.17.1 version__
- Author: David Goodger
- Contact: goodger@python.org
[Docutils](https://pypi.org/project/docutils/) is an open-source, modular text processing system for processing plaintext documentation into a more useful format. Formats include HTML, man-pages, OpenDocument, LaTeX, or XML.
Docutils supports reStructuredText for input, an easy-to-read, what-you-see-is-what-you-get plaintext markup syntax.
Docutils is short for "Python Documentation Utilities".
Support for the following sources has been implemented:
- Standalone files
- `PEPs (Python Enhancement Proposals)`
Support for these sources is currently being developed:
- Inline documentation
- Wikis
- Email and more
Docutils Distribution Consists of:
- the `docutils` package (or library)
- front-end tools
- test suite
- documentation.
## Notable docutils Modules & Subpackages
-----------------------------
Module | Definition
------------- | -------------
__core__ | Contains the ``Publisher`` class and ``publish_()``convenience functions
__io__ | Provides a uniform API for low-level input and output
__nodes__ | Docutils document tree (doctree) node class library
-----------------------------
Subpackages | Definition
------------- | -------------
**languages** | Language-specific mappings of terms
**parsers** | Syntax-specific input parser modules or packages
**readers** | Context-specific input handlers which understand the data source and manage a parser
Below is an overview of the `docutils` package:

## Main Use Applications of Package
The reStructured Text component of the `docutils` package makes it easy to convert between different formats, especially from plain text to a static website. It is unique because it is extensible. Better than simpler markups.
Additionally, users can pair `docutils` with `Sphinx` to convert text to html. The `Sphinx` package is built on the `docutils` package. The docutils parser creates the parse tree as a representation of the text in the memory for the Sphinx application and .rst environment.
|
github_jupyter
|
## Statistics
### Questions
```{admonition} Problem: JOIN Dataframes
:class: dropdown, tip
Can you tell me the ways in which 2 pandas data frames can be joined?
```
```{admonition} Solution:
:class: dropdown
A very high level difference is that merge() is used to combine two (or more) dataframes on the basis of values of common columns (indices can also be used, use left_index=True and/or right_index=True), and concat() is used to append one (or more) dataframes one below the other (or sideways, depending on whether the axis option is set to 0 or 1).
join() is used to merge 2 dataframes on the basis of the index; instead of using merge() with the option left_index=True we can use join().

```
```{admonition} Problem: [GOOGLE] Normal Distribution
:class: dropdown, tip
Write a function to generate N samples from a normal distribution and plot the histogram.
```
```
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
def normal_sample_generator(N):
# can be done using np.random.randn or stats.norm.rvs
#x = np.random.randn(N)
x = stats.norm.rvs(size=N)
num_bins = 20
plt.hist(x, bins=num_bins, facecolor='blue', alpha=0.5)
y = np.linspace(-4, 4, N)
bin_width = (x.max() - x.min()) / num_bins
plt.plot(y, stats.norm.pdf(y) * N * bin_width)
plt.show()
normal_sample_generator(10000)
```
```{admonition} Problem: [UBER] Bernoulli trial generator
:class: dropdown, tip
Given a random Bernoulli trial generator, write a function to return a value sampled from a normal distribution.
```
```{admonition} Solution:
:class: dropdown
Solution pending, [Reference material link](Given a random Bernoulli trial generator, how do you return a value sampled from a normal distribution?)
```
```{admonition} Problem: [PINTEREST] Interquartile Distance
:class: dropdown, tip
Given an array of unsorted random numbers (decimals) find the interquartile distance.
```
```
# Interquartile distance is the difference between first and third quartile
# first let's generate a list of random numbers
import random
import numpy as np
li = [round(random.uniform(33.33, 66.66), 2) for i in range(50)]
print(li)
qtl_1 = np.quantile(li,.25)
qtl_3 = np.quantile(li,.75)
print("Interquartile distance: ", qtl_1 - qtl_3)
```
````{admonition} Problem: [GENENTECH] Imputing the mdeian
:class: dropdown, tip
Write a function cheese_median to impute the median price of the selected California cheeses in place of the missing values. You may assume at least one cheese is not missing its price.
Input:
```python
import pandas as pd
cheeses = {"Name": ["Bohemian Goat", "Central Coast Bleu", "Cowgirl Mozzarella", "Cypress Grove Cheddar", "Oakdale Colby"], "Price" : [15.00, None, 30.00, None, 45.00]}
df_cheeses = pd.DataFrame(cheeses)
```
| Name | Price |
|:---------------------:|:-----:|
| Bohemian Goat | 15.00 |
| Central Coast Bleu | 30.00 |
| Cowgirl Mozzarella | 30.00 |
| Cypress Grove Cheddar | 30.00 |
| Oakdale Colby | 45.00 |
````
```
import pandas as pd
cheeses = {"Name": ["Bohemian Goat", "Central Coast Bleu", "Cowgirl Mozzarella", "Cypress Grove Cheddar", "Oakdale Colby"], "Price" : [15.00, None, 30.00, None, 45.00]}
df_cheeses = pd.DataFrame(cheeses)
df_cheeses['Price'] = df_cheeses['Price'].fillna(df_cheeses['Price'].median())
df_cheeses.head()
```
|
github_jupyter
|
# Real Estate Price Prediction
```
import pandas as pd
df = pd.read_csv("data.csv")
df.head()
df['CHAS'].value_counts()
df.info()
df.describe()
%matplotlib inline
import matplotlib.pyplot as plt
df.hist(bins=50, figsize=(20,15))
```
## train_test_split
```
import numpy as np
def split_train_test(data, test_ratio):
np.random.seed(42)
shuffled = np.random.permutation(len(data))
test_set_size = int(len(data) * test_ratio)
test_indices = shuffled[:test_set_size]
train_indices = shuffled[test_set_size:]
return data.iloc[train_indices], data.iloc[test_indices]
train_set, test_set = split_train_test(df, 0.2)
print(f"The length of train dataset is: {len(train_set)}")
print(f"The length of train dataset is: {len(test_set)}")
def data_percent_allocation(train_set, test_set):
total = len(df)
train_percent = round((len(train_set)/total) * 100)
test_percent = round((len(test_set)/total) * 100)
return train_percent, test_percent
data_percent_allocation(train_set, test_set)
```
## train_test_split from sklearn
```
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(df, test_size = 0.2, random_state = 42)
print(f"The length of train dataset is: {len(train_set)}")
print(f"The length of train dataset is: {len(test_set)}")
from sklearn.model_selection import StratifiedShuffleSplit
split = StratifiedShuffleSplit(n_splits = 1, test_size = 0.2, random_state = 42)
for train_index, test_index in split.split(df, df['CHAS']):
strat_train_set = df.loc[train_index]
strat_test_set = df.loc[test_index]
strat_test_set['CHAS'].value_counts()
test_set['CHAS'].value_counts()
strat_train_set['CHAS'].value_counts()
train_set['CHAS'].value_counts()
```
### Stratified learning equal splitting of zero and ones
```
95/7
376/28
df = strat_train_set.copy()
```
## Corelations
```
from pandas.plotting import scatter_matrix
attributes = ["MEDV", "RM", "ZN" , "LSTAT"]
scatter_matrix(df[attributes], figsize = (12,8))
df.plot(kind="scatter", x="RM", y="MEDV", alpha=1)
```
### Trying out attribute combinations
```
df["TAXRM"] = df["TAX"]/df["RM"]
df.head()
corr_matrix = df.corr()
corr_matrix['MEDV'].sort_values(ascending=False)
# 1 means strong positive corr and -1 means strong negative corr.
# EX: if RM will increase our final result(MEDV) in prediction will also increase.
df.plot(kind="scatter", x="TAXRM", y="MEDV", alpha=1)
df = strat_train_set.drop("MEDV", axis=1)
df_labels = strat_train_set["MEDV"].copy()
```
## Pipeline
```
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.impute import SimpleImputer
my_pipeline = Pipeline([
('imputer', SimpleImputer(strategy="median")),
('std_scaler', StandardScaler()),
])
df_numpy = my_pipeline.fit_transform(df)
df_numpy
#Numpy array of df as models will take numpy array as input.
df_numpy.shape
```
## Model Selection
```
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
# model = LinearRegression()
# model = DecisionTreeRegressor()
model = RandomForestRegressor()
model.fit(df_numpy, df_labels)
some_data = df.iloc[:5]
some_labels = df_labels.iloc[:5]
prepared_data = my_pipeline.transform(some_data)
model.predict(prepared_data)
list(some_labels)
```
## Evaluating the model
```
from sklearn.metrics import mean_squared_error
df_predictions = model.predict(df_numpy)
mse = mean_squared_error(df_labels, df_predictions)
rmse = np.sqrt(mse)
rmse
# from sklearn.metrics import accuracy_score
# accuracy_score(some_data, some_labels, normalize=False)
```
## Cross Validation
```
from sklearn.model_selection import cross_val_score
scores = cross_val_score(model, df_numpy, df_labels, scoring="neg_mean_squared_error", cv=10)
rmse_scores = np.sqrt(-scores)
rmse_scores
def print_scores(scores):
print("Scores:", scores)
print("\nMean:", scores.mean())
print("\nStandard deviation:", scores.std())
print_scores(rmse_scores)
```
### Saving Model
```
from joblib import dump, load
dump(model, 'final_model.joblib')
dump(model, 'final_model.sav')
```
## Testing model on test data
```
X_test = strat_test_set.drop("MEDV", axis=1)
Y_test = strat_test_set["MEDV"].copy()
X_test_prepared = my_pipeline.transform(X_test)
final_predictions = model.predict(X_test_prepared)
final_mse = mean_squared_error(Y_test, final_predictions)
final_rmse = np.sqrt(final_mse)
final_rmse
```
|
github_jupyter
|
# In-Place Waveform Library Updates
This example notebook shows how one can update pulses data in-place without recompiling.
© Raytheon BBN Technologies 2020
Set the `SAVE_WF_OFFSETS` flag in order that QGL will output a map of the waveform data within the compiled binary waveform library.
```
from QGL import *
import QGL
import os.path
import pickle
QGL.drivers.APS2Pattern.SAVE_WF_OFFSETS = True
```
Create the usual channel library with a couple of AWGs.
```
cl = ChannelLibrary(":memory:")
q1 = cl.new_qubit("q1")
aps2_1 = cl.new_APS2("BBNAPS1", address="192.168.5.101")
aps2_2 = cl.new_APS2("BBNAPS2", address="192.168.5.102")
dig_1 = cl.new_X6("X6_1", address=0)
h1 = cl.new_source("Holz1", "HolzworthHS9000", "HS9004A-009-1", power=-30)
h2 = cl.new_source("Holz2", "HolzworthHS9000", "HS9004A-009-2", power=-30)
cl.set_control(q1, aps2_1, generator=h1)
cl.set_measure(q1, aps2_2, dig_1.ch(1), generator=h2)
cl.set_master(aps2_1, aps2_1.ch("m2"))
cl["q1"].measure_chan.frequency = 0e6
cl.commit()
```
Compile a simple sequence.
```
mf = RabiAmp(cl["q1"], np.linspace(-1, 1, 11))
plot_pulse_files(mf, time=True)
```
Open the offsets file (in the same directory as the `.aps2` files, one per AWG slice.)
```
offset_f = os.path.join(os.path.dirname(mf), "Rabi-BBNAPS1.offsets")
with open(offset_f, "rb") as FID:
offsets = pickle.load(FID)
offsets
```
Let's replace every single pulse with a fixed amplitude `Utheta`
```
pulses = {l: Utheta(q1, amp=0.1, phase=0) for l in offsets}
wfm_f = os.path.join(os.path.dirname(mf), "Rabi-BBNAPS1.aps2")
QGL.drivers.APS2Pattern.update_wf_library(wfm_f, pulses, offsets)
```
We see that the data in the file has been updated.
```
plot_pulse_files(mf, time=True)
```
## Profiling
How long does this take?
```
%timeit mf = RabiAmp(cl["q1"], np.linspace(-1, 1, 100))
```
Getting the offsets is fast, and only needs to be done once
```
def get_offsets():
offset_f = os.path.join(os.path.dirname(mf), "Rabi-BBNAPS1.offsets")
with open(offset_f, "rb") as FID:
offsets = pickle.load(FID)
return offsets
%timeit offsets = get_offsets()
%timeit pulses = {l: Utheta(q1, amp=0.1, phase=0) for l in offsets}
wfm_f = os.path.join(os.path.dirname(mf), "Rabi-BBNAPS1.aps2")
%timeit QGL.drivers.APS2Pattern.update_wf_library(wfm_f, pulses, offsets)
# %timeit QGL.drivers.APS2Pattern.update_wf_library("/Users/growland/workspace/AWG/Rabi/Rabi-BBNAPS1.aps2", pulses, offsets)
```
Moral of the story: 300 ms for initial compilation, and roughly 1.3 ms for update_in_place.
|
github_jupyter
|
```
%matplotlib inline
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import numpy as np
np.set_printoptions(precision=3, suppress=True)
import library.helpers as h
import matplotlib.pyplot as plt
import numpy as np
from sklearn.metrics import auc
from scipy.integrate import simps
from scipy.interpolate import interp1d
LOG_NAME = "unix_forensic"
VIZUALIZATIONS_DIR = "visualizations"
fig = plt.figure(figsize=(10,10))
# TODO redo this one [
MODEL_NAMES = [ "lstm-ae", "triplet-la", "triplet-jd-la-all-40-70-all-all", "triplet-jd-la-non-60-65-non-non", "triplet-jd-la-00500-60-65-all-all-00001",
"triplet-jd-la-00500-60-65-all-00001-non", "triplet-jd-la-01000-60-65-all-all-00001", "triplet-jd-la-05000-60-65-all-all-00001", "jd-la-x05000-pt60-nt65-llall-lcall-ee00001-ep20", "jd-la-xnon-pt40-nt70-llall-lcall-ee00001-ep50"]
MODEL_NAMES_JD = [
"triplet-jd-la-all-40-70-all-all", # baseline, all labels
"triplet-jd-la-non-60-65-non-non","triplet-jd-la-non-40-70-non-non", # jd no labels, 10 ep
"jd-la-xnon-pt60-nt65-llnon-lcnon-eenon-ep50",
"jd-la-xnon-pt30-nt75-llnon-lcnon-eenon-ep50"
#"jd-la-x02000-pt55-nt65-llnon-lc00003-ee00002-ep10","jd-la-x02000-pt55-nt65-llnon-lc00003-ee00002-ep20",
#"triplet-jd-la-05000-60-65-all-all-00001", "jd-la-x05000-pt60-nt65-llall-lcall-ee00001-ep20",
]
MODEL_NAMES_LA = [
"triplet-jd-la-all-40-70-all-all", # baseline, all labels
"jd-la-x00500-pt30-nt70-llall-lcall-ee00002-ep30", # 500 labels
"jd-la-x01000-pt30-nt70-llall-lcall-ee00002-ep30", # 1000 labels
"jd-la-x02000-pt30-nt70-llall-lcall-ee00002-ep30", # 2000 labels
"jd-la-x02500-pt30-nt70-llall-lcall-ee00002-ep30", # 2500 labels
"jd-la-x05000-pt30-nt70-llall-lcall-ee00002-ep30", # 5000 labels
]
MODEL_NAMES_NT60 = [
"jd-la-xnon-pt10-nt60-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt20-nt60-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt30-nt60-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt40-nt60-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt50-nt60-llnon-lcnon-ee00002-ep30",
]
MODEL_NAMES_NT70 = [
"jd-la-xnon-pt10-nt70-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt20-nt70-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt30-nt70-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt40-nt70-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt50-nt70-llnon-lcnon-ee00002-ep30",
"jd-la-xnon-pt60-nt70-llnon-lcnon-ee00002-ep30"
]
MODEL_NAMES_NT80 = [
"jd-la-xnon-pt10-nt80-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt20-nt80-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt30-nt80-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt40-nt80-llnon-lcnon-ee00002-ep30", #
"jd-la-xnon-pt50-nt80-llnon-lcnon-ee00002-ep30",
"jd-la-xnon-pt60-nt80-llnon-lcnon-ee00002-ep30",
"jd-la-xnon-pt70-nt80-llnon-lcnon-ee00002-ep30"
]
MODEL_NAMES_10 = [
#"jd-la-xnon-pt10-nt90-llnon-lcnon-ee00002-ep20",
#"jd-la-xnon-pt20-nt90-llnon-lcnon-ee00002-ep20",
#"jd-la-xnon-pt30-nt90-llnon-lcnon-ee00002-ep20",
#"jd-la-xnon-pt40-nt90-llnon-lcnon-ee00002-ep20",
"jd-la-xnon-pt50-nt90-llnon-lcnon-ee00002-ep20",
"jd-la-xnon-pt52-nt70-llnon-lcnon-ee00002-ep30",
#"jd-la-xnon-pt60-nt90-llnon-lcnon-ee00002-ep20",
#"jd-la-xnon-pt70-nt90-llnon-lcnon-ee00002-ep20",
#"jd-la-xnon-pt80-nt90-llnon-lcnon-ee00002-ep20",
"jd-la-xnon-pt52-nt90-llnon-lcnon-ee00002-ep200",
]
MODEL_NAMES = MODEL_NAMES_LA
# "triplet-jd-la-2000-55-70-40", "lstm-ae", "triplet-la", "triplet-jd-la-3000-55-70-55-30", "triplet-jd-la-ma-1500"
# ["triplet-jd-la-2000-55-70-40","triplet-jd-la-3000-55-70-55-30","triplet-jd-la-3000-50-70-55-30"]
# [ "triplet-jd-la-1500-055-70-40", "triplet-jd-la-1500-55-75-40"]
# "triplet-jd-la-1500-060-70-40", "triplet-jd-la-1500-055-70-40", "triplet-jd-la-1500-050-70-40",
# ["triplet-jd-la-1500-050-065-25", "triplet-jd-la-1500-065-070-25", "triplet-jd-la-1500-060-060-25"]
#["triplet-jd-la-ma-500-02-03","triplet-jd-la-ma-750","triplet-jd-la-ma-1000", # "#4169e1",
# "triplet-jd-la-ma-1500",
# "triplet-jaccard","triplet-jaccard-margin",
# "triplet-label", # all labels
# "lstm-ae"]
COLORS = h.get_N_HexCol(len(MODEL_NAMES)+1)
fac_n = np.arange(0, 1.0, 0.0005)
baseline_valid_accepts = h.load_from_json("data/ji_%s_basline-jaccard_valid.json"%LOG_NAME)
baseline_false_accepts = [np.round(f,5) for f in h.load_from_json("data/ji_%s_basline-jaccard_false.json"%LOG_NAME)]
interpolated_vac = interp1d(baseline_false_accepts, baseline_valid_accepts)
vac_n = interpolated_vac(fac_n)
auc_score = auc(fac_n, vac_n)
plt.plot( fac_n, interpolated_vac(fac_n), color='r', label="%0.3f,baseline"%auc_score)
plt.xscale("log")
for i, model_name in enumerate(MODEL_NAMES):
#print(model_name)
valid_accepts = h.load_from_json("data/%s_%s_valid.json"%(model_name, LOG_NAME))
false_accepts = h.load_from_json("data/%s_%s_false.json"%(model_name, LOG_NAME))
interpolated_vac = interp1d(false_accepts, valid_accepts)
auc_score = auc(fac_n, interpolated_vac(fac_n))
plt.plot( fac_n, interpolated_vac(fac_n) , color=COLORS[i+1], label="%0.3f, %s"%(auc_score, model_name))
plt.title("VAR/FAR (%s) LA"%LOG_NAME)
plt.xlabel('FAR')
plt.ylabel('VAL')
plt.legend(loc='lower right')
plt.show()
#plt.savefig("%s/roc_%s.png"%(VIZUALIZATIONS_DIR, LOG_NAME))
```
|
github_jupyter
|
[](https://colab.research.google.com/github/ourownstory/neural_prophet/blob/master/example_notebooks/sub_daily_data_yosemite_temps.ipynb)
# Sub-daily data
NeuralProphet can make forecasts for time series with sub-daily observations by passing in a dataframe with timestamps in the ds column. The format of the timestamps should be `YYYY-MM-DD HH:MM:SS` - see the example csv [here](https://github.com/ourownstory/neural_prophet/blob/master/example_data/yosemite_temps.csv). When sub-daily data are used, daily seasonality will automatically be fit.
Here we fit NeuralProphet to data with 5-minute resolution (daily temperatures at Yosemite).
```
if 'google.colab' in str(get_ipython()):
!pip install git+https://github.com/ourownstory/neural_prophet.git # may take a while
#!pip install neuralprophet # much faster, but may not have the latest upgrades/bugfixes
data_location = "https://raw.githubusercontent.com/ourownstory/neural_prophet/master/"
else:
data_location = "../"
import pandas as pd
from neuralprophet import NeuralProphet, set_log_level
# set_log_level("ERROR")
df = pd.read_csv(data_location + "example_data/yosemite_temps.csv")
```
Now we will attempt to forecast the next 7 days. The `5min` data resulution means that we have `60/5*24=288` daily values. Thus, we want to forecast `7*288` periods ahead.
Using some common sense, we set:
* First, we disable weekly seasonality, as nature does not follow the human week's calendar.
* Second, we disable changepoints, as the dataset only contains two months of data
```
m = NeuralProphet(
n_changepoints=0,
weekly_seasonality=False,
)
metrics = m.fit(df, freq='5min')
future = m.make_future_dataframe(df, periods=7*288, n_historic_predictions=len(df))
forecast = m.predict(future)
fig = m.plot(forecast)
# fig_comp = m.plot_components(forecast)
fig_param = m.plot_parameters()
```
The daily seasonality seems to make sense, when we account for the time being recorded in GMT, while Yosemite local time is GMT-8.
## Improving trend and seasonality
As we have `288` daily values recorded, we can increase the flexibility of `daily_seasonality`, without danger of overfitting.
Further, we may want to re-visit our decision to disable changepoints, as the data clearly shows changes in trend, as is typical with the weather. We make the following changes:
* increase the `changepoints_range`, as the we are doing a short-term prediction
* inrease the `n_changepoints` to allow to fit to the sudden changes in trend
* carefully regularize the trend changepoints by setting `trend_reg` in order to avoid overfitting
```
m = NeuralProphet(
changepoints_range=0.95,
n_changepoints=50,
trend_reg=1.5,
weekly_seasonality=False,
daily_seasonality=10,
)
metrics = m.fit(df, freq='5min')
future = m.make_future_dataframe(df, periods=60//5*24*7, n_historic_predictions=len(df))
forecast = m.predict(future)
fig = m.plot(forecast)
# fig_comp = m.plot_components(forecast)
fig_param = m.plot_parameters()
```
|
github_jupyter
|
<center>
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# Classes and Objects in Python
Estimated time needed: **40** minutes
## Objectives
After completing this lab you will be able to:
- Work with classes and objects
- Identify and define attributes and methods
<h2>Table of Contents</h2>
<div class="alert alert-block alert-info" style="margin-top: 20px">
<ul>
<li>
<a href="#intro">Introduction to Classes and Objects</a>
<ul>
<li><a href="create">Creating a class</a></li>
<li><a href="instance">Instances of a Class: Objects and Attributes</a></li>
<li><a href="method">Methods</a></li>
</ul>
</li>
<li><a href="creating">Creating a class</a></li>
<li><a href="circle">Creating an instance of a class Circle</a></li>
<li><a href="rect">The Rectangle Class</a></li>
</ul>
</div>
<hr>
<h2 id="intro">Introduction to Classes and Objects</h2>
<h3>Creating a Class</h3>
The first part of creating a class is giving it a name: In this notebook, we will create two classes, Circle and Rectangle. We need to determine all the data that make up that class, and we call that an attribute. Think about this step as creating a blue print that we will use to create objects. In figure 1 we see two classes, circle and rectangle. Each has their attributes, they are variables. The class circle has the attribute radius and color, while the rectangle has the attribute height and width. Let’s use the visual examples of these shapes before we get to the code, as this will help you get accustomed to the vocabulary.
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%203/images/ClassesClass.png" width="500" />
<i>Figure 1: Classes circle and rectangle, and each has their own attributes. The class circle has the attribute radius and colour, the rectangle has the attribute height and width.</i>
<h3 id="instance">Instances of a Class: Objects and Attributes</h3>
An instance of an object is the realisation of a class, and in Figure 2 we see three instances of the class circle. We give each object a name: red circle, yellow circle and green circle. Each object has different attributes, so let's focus on the attribute of colour for each object.
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%203/images/ClassesObj.png" width="500" />
<i>Figure 2: Three instances of the class circle or three objects of type circle.</i>
The colour attribute for the red circle is the colour red, for the green circle object the colour attribute is green, and for the yellow circle the colour attribute is yellow.
<h3 id="method">Methods</h3>
Methods give you a way to change or interact with the object; they are functions that interact with objects. For example, let’s say we would like to increase the radius by a specified amount of a circle. We can create a method called **add_radius(r)** that increases the radius by **r**. This is shown in figure 3, where after applying the method to the "orange circle object", the radius of the object increases accordingly. The “dot” notation means to apply the method to the object, which is essentially applying a function to the information in the object.
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%203/images/ClassesMethod.png" width="500" />
<i>Figure 3: Applying the method “add_radius” to the object orange circle object.</i>
<hr>
<h2 id="creating">Creating a Class</h2>
Now we are going to create a class circle, but first, we are going to import a library to draw the objects:
```
# Import the library
import matplotlib.pyplot as plt
%matplotlib inline
```
The first step in creating your own class is to use the <code>class</code> keyword, then the name of the class as shown in Figure 4. In this course the class parent will always be object:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%203/images/ClassesDefine.png" width="400" />
<i>Figure 4: Creating a class Circle.</i>
The next step is a special method called a constructor <code>__init__</code>, which is used to initialize the object. The input are data attributes. The term <code>self</code> contains all the attributes in the set. For example the <code>self.color</code> gives the value of the attribute color and <code>self.radius</code> will give you the radius of the object. We also have the method <code>add_radius()</code> with the parameter <code>r</code>, the method adds the value of <code>r</code> to the attribute radius. To access the radius we use the syntax <code>self.radius</code>. The labeled syntax is summarized in Figure 5:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%203/images/ClassesCircle.png" width="600" />
<i>Figure 5: Labeled syntax of the object circle.</i>
The actual object is shown below. We include the method <code>drawCircle</code> to display the image of a circle. We set the default radius to 3 and the default colour to blue:
```
# Create a class Circle
class Circle(object):
# Constructor
def __init__(self, radius=3, color='blue'):
self.radius = radius
self.color = color
# Method
def add_radius(self, r):
self.radius = self.radius + r
return(self.radius)
# Method
def drawCircle(self):
plt.gca().add_patch(plt.Circle((0, 0), radius=self.radius, fc=self.color))
plt.axis('scaled')
plt.show()
```
<hr>
<h2 id="circle">Creating an instance of a class Circle</h2>
Let’s create the object <code>RedCircle</code> of type Circle to do the following:
```
# Create an object RedCircle
RedCircle = Circle(10, 'red')
```
We can use the <code>dir</code> command to get a list of the object's methods. Many of them are default Python methods.
```
# Find out the methods can be used on the object RedCircle
dir(RedCircle)
```
We can look at the data attributes of the object:
```
# Print the object attribute radius
RedCircle.radius
# Print the object attribute color
RedCircle.color
```
We can change the object's data attributes:
```
# Set the object attribute radius
RedCircle.radius = 1
RedCircle.radius
```
We can draw the object by using the method <code>drawCircle()</code>:
```
# Call the method drawCircle
RedCircle.drawCircle()
```
We can increase the radius of the circle by applying the method <code>add_radius()</code>. Let increases the radius by 2 and then by 5:
```
# Use method to change the object attribute radius
print('Radius of object:',RedCircle.radius)
RedCircle.add_radius(2)
print('Radius of object of after applying the method add_radius(2):',RedCircle.radius)
RedCircle.add_radius(5)
print('Radius of object of after applying the method add_radius(5):',RedCircle.radius)
RedCircle.add_radius(6)
print('Radius of object of after applying the method add radius(6):',RedCircle.radius)
```
Let’s create a blue circle. As the default colour is blue, all we have to do is specify what the radius is:
```
# Create a blue circle with a given radius
BlueCircle = Circle(radius=100)
```
As before we can access the attributes of the instance of the class by using the dot notation:
```
# Print the object attribute radius
BlueCircle.radius
# Print the object attribute color
BlueCircle.color
```
We can draw the object by using the method <code>drawCircle()</code>:
```
# Call the method drawCircle
BlueCircle.drawCircle()
```
Compare the x and y axis of the figure to the figure for <code>RedCircle</code>; they are different.
<hr>
<h2 id="rect">The Rectangle Class</h2>
Let's create a class rectangle with the attributes of height, width and color. We will only add the method to draw the rectangle object:
```
# Create a new Rectangle class for creating a rectangle object
class Rectangle(object):
# Constructor
def __init__(self, width=2, height=3, color='r'):
self.height = height
self.width = width
self.color = color
# Method
def drawRectangle(self):
plt.gca().add_patch(plt.Rectangle((0, 0), self.width, self.height ,fc=self.color))
plt.axis('scaled')
plt.show()
```
Let’s create the object <code>SkinnyBlueRectangle</code> of type Rectangle. Its width will be 2 and height will be 3, and the color will be blue:
```
# Create a new object rectangle
SkinnyBlueRectangle = Rectangle(2, 10, 'blue')
```
As before we can access the attributes of the instance of the class by using the dot notation:
```
# Print the object attribute height
SkinnyBlueRectangle.height
# Print the object attribute width
SkinnyBlueRectangle.width
# Print the object attribute color
SkinnyBlueRectangle.color
```
We can draw the object:
```
# Use the drawRectangle method to draw the shape
SkinnyBlueRectangle.drawRectangle()
```
Let’s create the object <code>FatYellowRectangle</code> of type Rectangle :
```
# Create a new object rectangle
FatYellowRectangle = Rectangle(20, 5, 'yellow')
```
We can access the attributes of the instance of the class by using the dot notation:
```
# Print the object attribute height
FatYellowRectangle.height
# Print the object attribute width
FatYellowRectangle.width
# Print the object attribute color
FatYellowRectangle.color
```
We can draw the object:
```
# Use the drawRectangle method to draw the shape
FatYellowRectangle.drawRectangle()
```
<hr>
<h2 id="rect">Exercises</h2>
<h4> Text Analysis </h4>
You have been recruited by your friend, a linguistics enthusiast, to create a utility tool that can perform analysis on a given piece of text. Complete the class
'analysedText' with the following methods -
<ul>
<li> Constructor - Takes argument 'text',makes it lower case and removes all punctuation. Assume only the following punctuation is used - period (.), exclamation mark (!), comma (,) and question mark (?). Store the argument in "fmtText"
<li> freqAll - returns a dictionary of all unique words in the text along with the number of their occurences.
<li> freqOf - returns the frequency of the word passed in argument.
</ul>
The skeleton code has been given to you. Docstrings can be ignored for the purpose of the exercise. <br>
<i> Hint: Some useful functions are <code>replace()</code>, <code>lower()</code>, <code>split()</code>, <code>count()</code> </i><br>
```
class analysedText(object):
def __init__ (self, text):
reArrText = text.lower()
reArrText = reArrText.replace('.','').replace('!','').replace(',','').replace('?','')
self.fmtText = reArrText
def freqAll(self):
wordList = self.fmtText.split(' ')
freqMap = {}
for word in set(wordList): # use set to remove duplicates in list
freqMap[word] = wordList.count(word)
return freqMap
def freqOf(self,word):
freqDict = self.freqAll()
if word in freqDict:
return freqDict[word]
else:
return 0
```
Execute the block below to check your progress.
```
import sys
sampleMap = {'eirmod': 1,'sed': 1, 'amet': 2, 'diam': 5, 'consetetur': 1, 'labore': 1, 'tempor': 1, 'dolor': 1, 'magna': 2, 'et': 3, 'nonumy': 1, 'ipsum': 1, 'lorem': 2}
def testMsg(passed):
if passed:
return 'Test Passed'
else :
return 'Test Failed'
print("Constructor: ")
try:
samplePassage = analysedText("Lorem ipsum dolor! diam amet, consetetur Lorem magna. sed diam nonumy eirmod tempor. diam et labore? et diam magna. et diam amet.")
print(testMsg(samplePassage.fmtText == "lorem ipsum dolor diam amet consetetur lorem magna sed diam nonumy eirmod tempor diam et labore et diam magna et diam amet"))
except:
print("Error detected. Recheck your function " )
print("freqAll: ",)
try:
wordMap = samplePassage.freqAll()
print(testMsg(wordMap==sampleMap))
except:
print("Error detected. Recheck your function " )
print("freqOf: ")
try:
passed = True
for word in sampleMap:
if samplePassage.freqOf(word) != sampleMap[word]:
passed = False
break
print(testMsg(passed))
except:
print("Error detected. Recheck your function " )
```
<details><summary>Click here for the solution</summary>
```python
class analysedText(object):
def __init__ (self, text):
# remove punctuation
formattedText = text.replace('.','').replace('!','').replace('?','').replace(',','')
# make text lowercase
formattedText = formattedText.lower()
self.fmtText = formattedText
def freqAll(self):
# split text into words
wordList = self.fmtText.split(' ')
# Create dictionary
freqMap = {}
for word in set(wordList): # use set to remove duplicates in list
freqMap[word] = wordList.count(word)
return freqMap
def freqOf(self,word):
# get frequency map
freqDict = self.freqAll()
if word in freqDict:
return freqDict[word]
else:
return 0
```
</details>
<hr>
<h2>The last exercise!</h2>
<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href="https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/" target="_blank">this article</a> to learn how to share your work.
<hr>
## Author
<a href="https://www.linkedin.com/in/joseph-s-50398b136/" target="_blank">Joseph Santarcangelo</a>
## Other contributors
<a href="www.linkedin.com/in/jiahui-mavis-zhou-a4537814a">Mavis Zhou</a>
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ---------- | ---------------------------------- |
| 2020-08-26 | 2.0 | Lavanya | Moved lab to course repo in GitLab |
| | | | |
| | | | |
<hr/>
## <h3 align="center"> © IBM Corporation 2020. All rights reserved. <h3/>
|
github_jupyter
|
# LOFO Feature Importance
https://github.com/aerdem4/lofo-importance
```
!pip install lofo-importance
import numpy as np
import pandas as pd
df = pd.read_csv("../input/train.csv", index_col='id')
df['wheezy-copper-turtle-magic'] = df['wheezy-copper-turtle-magic'].astype('category')
df.shape
```
### Use the best model in public kernels
```
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
def get_model():
return Pipeline([('scaler', StandardScaler()),
('qda', QuadraticDiscriminantAnalysis(reg_param=0.111))
])
```
### Top 20 Features for wheezy-copper-turtle-magic = 0
```
from sklearn.model_selection import KFold, StratifiedKFold, train_test_split
from sklearn.linear_model import LogisticRegression
from lofo import LOFOImportance, FLOFOImportance, plot_importance
features = [c for c in df.columns if c not in ['id', 'target', 'wheezy-copper-turtle-magic']]
def get_lofo_importance(wctm_num):
sub_df = df[df['wheezy-copper-turtle-magic'] == wctm_num]
sub_features = [f for f in features if sub_df[f].std() > 1.5]
lofo_imp = LOFOImportance(sub_df, target="target",
features=sub_features,
cv=StratifiedKFold(n_splits=4, random_state=42, shuffle=True), scoring="roc_auc",
model=get_model(), n_jobs=4)
return lofo_imp.get_importance()
plot_importance(get_lofo_importance(0), figsize=(12, 12))
```
### Top 20 Features for wheezy-copper-turtle-magic = 1
```
plot_importance(get_lofo_importance(1), figsize=(12, 12))
```
### Top 20 Features for wheezy-copper-turtle-magic = 2
```
plot_importance(get_lofo_importance(2), figsize=(12, 12))
```
### Find the most harmful features for each wheezy-copper-turtle-magic
```
from tqdm import tqdm_notebook
import warnings
warnings.filterwarnings("ignore")
features_to_remove = []
potential_gain = []
for i in tqdm_notebook(range(512)):
imp = get_lofo_importance(i)
features_to_remove.append(imp["feature"].values[-1])
potential_gain.append(-imp["importance_mean"].values[-1])
print("Potential gain (AUC):", np.round(np.mean(potential_gain), 5))
features_to_remove
```
# Create submission using the current best kernel
https://www.kaggle.com/tunguz/ig-pca-nusvc-knn-qda-lr-stack by Bojan Tunguz
```
import numpy as np, pandas as pd
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score
from sklearn import svm, neighbors, linear_model, neural_network
from sklearn.svm import NuSVC
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from tqdm import tqdm
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.pipeline import Pipeline
from sklearn.metrics import roc_auc_score
from sklearn.feature_selection import VarianceThreshold
train = pd.read_csv('../input/train.csv')
test = pd.read_csv('../input/test.csv')
oof_svnu = np.zeros(len(train))
pred_te_svnu = np.zeros(len(test))
oof_svc = np.zeros(len(train))
pred_te_svc = np.zeros(len(test))
oof_knn = np.zeros(len(train))
pred_te_knn = np.zeros(len(test))
oof_lr = np.zeros(len(train))
pred_te_lr = np.zeros(len(test))
oof_mlp = np.zeros(len(train))
pred_te_mlp = np.zeros(len(test))
oof_qda = np.zeros(len(train))
pred_te_qda = np.zeros(len(test))
default_cols = [c for c in train.columns if c not in ['id', 'target', 'wheezy-copper-turtle-magic']]
for i in range(512):
cols = [c for c in default_cols if c != features_to_remove[i]]
train2 = train[train['wheezy-copper-turtle-magic']==i]
test2 = test[test['wheezy-copper-turtle-magic']==i]
idx1 = train2.index; idx2 = test2.index
train2.reset_index(drop=True,inplace=True)
data = pd.concat([pd.DataFrame(train2[cols]), pd.DataFrame(test2[cols])])
data2 = StandardScaler().fit_transform(PCA(svd_solver='full',n_components='mle').fit_transform(data[cols]))
train3 = data2[:train2.shape[0]]; test3 = data2[train2.shape[0]:]
data2 = StandardScaler().fit_transform(VarianceThreshold(threshold=1.5).fit_transform(data[cols]))
train4 = data2[:train2.shape[0]]; test4 = data2[train2.shape[0]:]
# STRATIFIED K FOLD (Using splits=25 scores 0.002 better but is slower)
skf = StratifiedKFold(n_splits=5, random_state=42)
for train_index, test_index in skf.split(train2, train2['target']):
clf = NuSVC(probability=True, kernel='poly', degree=4, gamma='auto', random_state=4, nu=0.59, coef0=0.053)
clf.fit(train3[train_index,:],train2.loc[train_index]['target'])
oof_svnu[idx1[test_index]] = clf.predict_proba(train3[test_index,:])[:,1]
pred_te_svnu[idx2] += clf.predict_proba(test3)[:,1] / skf.n_splits
clf = neighbors.KNeighborsClassifier(n_neighbors=17, p=2.9)
clf.fit(train3[train_index,:],train2.loc[train_index]['target'])
oof_knn[idx1[test_index]] = clf.predict_proba(train3[test_index,:])[:,1]
pred_te_knn[idx2] += clf.predict_proba(test3)[:,1] / skf.n_splits
clf = linear_model.LogisticRegression(solver='saga',penalty='l1',C=0.1)
clf.fit(train3[train_index,:],train2.loc[train_index]['target'])
oof_lr[idx1[test_index]] = clf.predict_proba(train3[test_index,:])[:,1]
pred_te_lr[idx2] += clf.predict_proba(test3)[:,1] / skf.n_splits
clf = neural_network.MLPClassifier(random_state=3, activation='relu', solver='lbfgs', tol=1e-06, hidden_layer_sizes=(250, ))
clf.fit(train3[train_index,:],train2.loc[train_index]['target'])
oof_mlp[idx1[test_index]] = clf.predict_proba(train3[test_index,:])[:,1]
pred_te_mlp[idx2] += clf.predict_proba(test3)[:,1] / skf.n_splits
clf = svm.SVC(probability=True, kernel='poly', degree=4, gamma='auto', random_state=42)
clf.fit(train3[train_index,:],train2.loc[train_index]['target'])
oof_svc[idx1[test_index]] = clf.predict_proba(train3[test_index,:])[:,1]
pred_te_svc[idx2] += clf.predict_proba(test3)[:,1] / skf.n_splits
clf = QuadraticDiscriminantAnalysis(reg_param=0.111)
clf.fit(train4[train_index,:],train2.loc[train_index]['target'])
oof_qda[idx1[test_index]] = clf.predict_proba(train4[test_index,:])[:,1]
pred_te_qda[idx2] += clf.predict_proba(test4)[:,1] / skf.n_splits
print('lr', roc_auc_score(train['target'], oof_lr))
print('knn', roc_auc_score(train['target'], oof_knn))
print('svc', roc_auc_score(train['target'], oof_svc))
print('svcnu', roc_auc_score(train['target'], oof_svnu))
print('mlp', roc_auc_score(train['target'], oof_mlp))
print('qda', roc_auc_score(train['target'], oof_qda))
print('blend 1', roc_auc_score(train['target'], oof_svnu*0.7 + oof_svc*0.05 + oof_knn*0.2 + oof_mlp*0.05))
print('blend 2', roc_auc_score(train['target'], oof_qda*0.5+oof_svnu*0.35 + oof_svc*0.025 + oof_knn*0.1 + oof_mlp*0.025))
oof_svnu = oof_svnu.reshape(-1, 1)
pred_te_svnu = pred_te_svnu.reshape(-1, 1)
oof_svc = oof_svc.reshape(-1, 1)
pred_te_svc = pred_te_svc.reshape(-1, 1)
oof_knn = oof_knn.reshape(-1, 1)
pred_te_knn = pred_te_knn.reshape(-1, 1)
oof_mlp = oof_mlp.reshape(-1, 1)
pred_te_mlp = pred_te_mlp.reshape(-1, 1)
oof_lr = oof_lr.reshape(-1, 1)
pred_te_lr = pred_te_lr.reshape(-1, 1)
oof_qda = oof_qda.reshape(-1, 1)
pred_te_qda = pred_te_qda.reshape(-1, 1)
tr = np.concatenate((oof_svnu, oof_svc, oof_knn, oof_mlp, oof_lr, oof_qda), axis=1)
te = np.concatenate((pred_te_svnu, pred_te_svc, pred_te_knn, pred_te_mlp, pred_te_lr, pred_te_qda), axis=1)
print(tr.shape, te.shape)
oof_lrr = np.zeros(len(train))
pred_te_lrr = np.zeros(len(test))
skf = StratifiedKFold(n_splits=5, random_state=42)
for train_index, test_index in skf.split(tr, train['target']):
lrr = linear_model.LogisticRegression()
lrr.fit(tr[train_index], train['target'][train_index])
oof_lrr[test_index] = lrr.predict_proba(tr[test_index,:])[:,1]
pred_te_lrr += lrr.predict_proba(te)[:,1] / skf.n_splits
print('stack CV score =',round(roc_auc_score(train['target'],oof_lrr),6))
sub = pd.read_csv('../input/sample_submission.csv')
sub['target'] = pred_te_lrr
sub.to_csv('submission_stack.csv', index=False)
```
|
github_jupyter
|
# End-to-end learning for music audio
- http://qiita.com/himono/items/a94969e35fa8d71f876c
```
# データのダウンロード
wget http://mi.soi.city.ac.uk/datasets/magnatagatune/mp3.zip.001
wget http://mi.soi.city.ac.uk/datasets/magnatagatune/mp3.zip.002
wget http://mi.soi.city.ac.uk/datasets/magnatagatune/mp3.zip.003
# 結合
cat data/mp3.zip.* > data/music.zip
# 解凍
unzip data/music.zip -d music
```
```
%matplotlib inline
import os
import matplotlib.pyplot as plt
```
## MP3ファイルのロード
```
import numpy as np
from pydub import AudioSegment
def mp3_to_array(file):
# MP3 => RAW
song = AudioSegment.from_mp3(file)
song_arr = np.fromstring(song._data, np.int16)
return song_arr
%ls data/music/1/ambient_teknology-phoenix-01-ambient_teknology-0-29.mp3
file = 'data/music/1/ambient_teknology-phoenix-01-ambient_teknology-0-29.mp3'
song = mp3_to_array(file)
plt.plot(song)
```
## 楽曲タグデータをロード
- ランダムに3000曲を抽出
- よく使われるタグ50個を抽出
- 各曲には複数のタグがついている
```
import pandas as pd
tags_df = pd.read_csv('data/annotations_final.csv', delim_whitespace=True)
# 全体をランダムにサンプリング
tags_df = tags_df.sample(frac=1)
# 最初の3000曲を使う
tags_df = tags_df[:3000]
tags_df
top50_tags = tags_df.iloc[:, 1:189].sum().sort_values(ascending=False).index[:50].tolist()
y = tags_df[top50_tags].values
y
```
## 楽曲データをロード
- tags_dfのmp3_pathからファイルパスを取得
- mp3_to_array()でnumpy arrayをロード
- (samples, features, channels) になるようにreshape
- 音声波形は1次元なのでchannelsは1
- 訓練データはすべて同じサイズなのでfeaturesは同じになるはず(パディング不要)
```
files = tags_df.mp3_path.values
files = [os.path.join('data', 'music', x) for x in files]
X = np.array([mp3_to_array(file) for file in files])
X = X.reshape(X.shape[0], X.shape[1], 1)
X.shape
```
## 訓練データとテストデータに分割
```
from sklearn.model_selection import train_test_split
random_state = 42
train_x, test_x, train_y, test_y = train_test_split(X, y, test_size=0.2, random_state=random_state)
print(train_x.shape)
print(test_x.shape)
print(train_y.shape)
print(test_y.shape)
plt.plot(train_x[0])
np.save('train_x.npy', train_x)
np.save('test_x.npy', test_x)
np.save('train_y.npy', train_y)
np.save('test_y.npy', test_y)
```
## 訓練
```
import numpy as np
from keras.models import Model
from keras.layers import Dense, Flatten, Input, Conv1D, MaxPooling1D
from keras.callbacks import CSVLogger, ModelCheckpoint
train_x = np.load('train_x.npy')
train_y = np.load('train_y.npy')
test_x = np.load('test_x.npy')
test_y = np.load('test_y.npy')
print(train_x.shape)
print(train_y.shape)
print(test_x.shape)
print(test_y.shape)
features = train_x.shape[1]
x_inputs = Input(shape=(features, 1), name='x_inputs')
x = Conv1D(128, 256, strides=256, padding='valid', activation='relu')(x_inputs) # strided conv
x = Conv1D(32, 8, activation='relu')(x)
x = MaxPooling1D(4)(x)
x = Conv1D(32, 8, activation='relu')(x)
x = MaxPooling1D(4)(x)
x = Conv1D(32, 8, activation='relu')(x)
x = MaxPooling1D(4)(x)
x = Conv1D(32, 8, activation='relu')(x)
x = MaxPooling1D(4)(x)
x = Flatten()(x)
x = Dense(100, activation='relu')(x)
x_outputs = Dense(50, activation='sigmoid', name='x_outputs')(x)
model = Model(inputs=x_inputs, outputs=x_outputs)
model.compile(optimizer='adam',
loss='categorical_crossentropy')
logger = CSVLogger('history.log')
checkpoint = ModelCheckpoint(
'model.{epoch:02d}-{val_loss:.3f}.h5',
monitor='val_loss',
verbose=1,
save_best_only=True,
mode='auto')
model.fit(train_x, train_y, batch_size=600, epochs=50,
validation_data=[test_x, test_y],
callbacks=[logger, checkpoint])
```
## 予測
- taggerは複数のタグを出力するのでevaluate()ではダメ?
```
import numpy as np
from keras.models import load_model
from sklearn.metrics import roc_auc_score
test_x = np.load('test_x.npy')
test_y = np.load('test_y.npy')
model = load_model('model.22-9.187-0.202.h5')
pred_y = model.predict(test_x, batch_size=50)
print(roc_auc_score(test_y, pred_y))
print(model.evaluate(test_x, test_y))
```
|
github_jupyter
|
<table> <tr>
<td style="background-color:#ffffff;">
<a href="http://qworld.lu.lv" target="_blank"><img src="..\images\qworld.jpg" width="25%" align="left"> </a></td>
<td style="background-color:#ffffff;vertical-align:bottom;text-align:right;">
prepared by <a href="http://abu.lu.lv" target="_blank">Abuzer Yakaryilmaz</a> (<a href="http://qworld.lu.lv/index.php/qlatvia/" target="_blank">QLatvia</a>)
</td>
</tr></table>
<table width="100%"><tr><td style="color:#bbbbbb;background-color:#ffffff;font-size:11px;font-style:italic;text-align:right;">This cell contains some macros. If there is a problem with displaying mathematical formulas, please run this cell to load these macros. </td></tr></table>
$ \newcommand{\bra}[1]{\langle #1|} $
$ \newcommand{\ket}[1]{|#1\rangle} $
$ \newcommand{\braket}[2]{\langle #1|#2\rangle} $
$ \newcommand{\dot}[2]{ #1 \cdot #2} $
$ \newcommand{\biginner}[2]{\left\langle #1,#2\right\rangle} $
$ \newcommand{\mymatrix}[2]{\left( \begin{array}{#1} #2\end{array} \right)} $
$ \newcommand{\myvector}[1]{\mymatrix{c}{#1}} $
$ \newcommand{\myrvector}[1]{\mymatrix{r}{#1}} $
$ \newcommand{\mypar}[1]{\left( #1 \right)} $
$ \newcommand{\mybigpar}[1]{ \Big( #1 \Big)} $
$ \newcommand{\sqrttwo}{\frac{1}{\sqrt{2}}} $
$ \newcommand{\dsqrttwo}{\dfrac{1}{\sqrt{2}}} $
$ \newcommand{\onehalf}{\frac{1}{2}} $
$ \newcommand{\donehalf}{\dfrac{1}{2}} $
$ \newcommand{\hadamard}{ \mymatrix{rr}{ \sqrttwo & \sqrttwo \\ \sqrttwo & -\sqrttwo }} $
$ \newcommand{\vzero}{\myvector{1\\0}} $
$ \newcommand{\vone}{\myvector{0\\1}} $
$ \newcommand{\stateplus}{\myvector{ \sqrttwo \\ \sqrttwo } } $
$ \newcommand{\stateminus}{ \myrvector{ \sqrttwo \\ -\sqrttwo } } $
$ \newcommand{\myarray}[2]{ \begin{array}{#1}#2\end{array}} $
$ \newcommand{\X}{ \mymatrix{cc}{0 & 1 \\ 1 & 0} } $
$ \newcommand{\I}{ \mymatrix{rr}{1 & 0 \\ 0 & 1} } $
$ \newcommand{\Z}{ \mymatrix{rr}{1 & 0 \\ 0 & -1} } $
$ \newcommand{\Htwo}{ \mymatrix{rrrr}{ \frac{1}{2} & \frac{1}{2} & \frac{1}{2} & \frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & \frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} & \frac{1}{2} } } $
$ \newcommand{\CNOT}{ \mymatrix{cccc}{1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 1 \\ 0 & 0 & 1 & 0} } $
$ \newcommand{\norm}[1]{ \left\lVert #1 \right\rVert } $
$ \newcommand{\pstate}[1]{ \lceil \mspace{-1mu} #1 \mspace{-1.5mu} \rfloor } $
<h2> <font color="blue"> Solutions for </font>Probabilistic Bit</h2>
<a id="task2"></a>
<h3> Task 2 </h3>
Suppose that Fyodor hiddenly rolls a loaded (tricky) dice with the bias
$$ Pr(1):Pr(2):Pr(3):Pr(4):Pr(5):Pr(6) = 7:5:4:2:6:1 . $$
Represent your information on the result as a column vector. Remark that the size of your column should be 6.
You may use python for your calculations.
<h3>Solution</h3>
```
# all portions are stored in a list
all_portions = [7,5,4,2,6,1];
# let's calculate the total portion
total_portion = 0
for i in range(6):
total_portion = total_portion + all_portions[i]
print("total portion is",total_portion)
# find the weight of one portion
one_portion = 1/total_portion
print("the weight of one portion is",one_portion)
print() # print an empty line
# now we can calculate the probabilities of rolling 1,2,3,4,5, and 6
for i in range(6):
print("the probability of rolling",(i+1),"is",(one_portion*all_portions[i]))
```
|
github_jupyter
|
```
import os
import pickle
from neutrinomass.completions import EffectiveOperator, Completion
from neutrinomass.database import ExoticField
from neutrinomass.database import ModelDataFrame, EXOTICS, TERMS, MVDF
from neutrinomass.completions import EFF_OPERATORS
from neutrinomass.completions import DERIV_EFF_OPERATORS
DATA_PATH = "/home/garj/work/neutrinomass/neutrinomass/database"
DATA = pickle.load(open(os.path.join(DATA_PATH, "unfiltered.p"), "rb"))
UNF = ModelDataFrame.new(data=DATA, exotics=EXOTICS, terms=TERMS)
STR_UNF = UNF.drop_duplicates(["stringent_num"], keep="first")
LAGS = len(STR_UNF)
print(f"Number of neutrino-mass mechanisms: {LAGS}")
DEMO_UNF = UNF.drop_duplicates(["democratic_num"], keep="first")
MODELS = len(DEMO_UNF)
print(f"Number of models: {MODELS}")
STR_MVDF = MVDF.drop_duplicates(["stringent_num"], keep="first")
print(f"Number of filtered neutrino-mass mechanisms: {len(STR_MVDF)}")
DEMO_MVDF = MVDF.drop_duplicates(["democratic_num"], keep="first")
print(f"Number of filtered neutrino-mass mechanisms: {len(DEMO_MVDF)}")
FIL_DF = MVDF.drop_duplicates(['democratic_num', 'dim'], keep="first")
UNF_DF = UNF.drop_duplicates(['democratic_num', 'dim'], keep="first")
print(f"After filtering, there are {len(FIL_DF[FIL_DF['dim'] == 5])} models derived from dimension-5 operators.")
print(f"After filtering, there are {len(FIL_DF[FIL_DF['dim'] == 9])} models derived from dimension-9 operators.")
print(f"After filtering, there are {len(FIL_DF[FIL_DF['dim'] == 11])} models derived from dimension-11 operators.")
print(f"The total of these is {len(FIL_DF[FIL_DF['dim'] == 5]) + len(FIL_DF[FIL_DF['dim'] == 9]) + len(FIL_DF[FIL_DF['dim'] == 11])}")
OPS = {**EFF_OPERATORS, **DERIV_EFF_OPERATORS}
labels, total, demo, dimensions = [], [], [], []
for k in OPS:
labels.append(k)
total.append(len(UNF_DF[UNF_DF["op"] == k]))
demo.append(len(FIL_DF[FIL_DF["op"] == k]))
dimensions.append(OPS[k].mass_dimension)
NHL = STR_UNF.terms[("F,00,0,0,0", "H", "L")]
NHSigma = STR_UNF.terms[("F,00,2,0,0", "H", "L")]
HHXi1 = STR_UNF.terms[("H", "H", "S,00,2,-1,0")]
LLXi1 = STR_UNF.terms[("L", "L", "S,00,2,1,0")]
N = STR_UNF.exotics["F,00,0,0,0"]
Sigma = STR_UNF.exotics["F,00,2,0,0"]
Xi1 = STR_UNF.exotics["S,00,2,1,0"]
N_NHL_lags = len(STR_UNF[STR_UNF["stringent_num"] % NHL == 0])
N_other_lags = len(STR_UNF[(STR_UNF["stringent_num"] % NHL != 0) & (STR_UNF["democratic_num"] % N == 0)])
Sigma_NHSigma_lags = len(STR_UNF[STR_UNF["stringent_num"] % NHSigma == 0])
Sigma_other_lags = len(STR_UNF[(STR_UNF["stringent_num"] % NHSigma != 0) & (STR_UNF["democratic_num"] % Sigma == 0)])
Xi1_HHXi1_lags = len(STR_UNF[STR_UNF["stringent_num"] % HHXi1 == 0])
Xi1_LLXi1_lags = len(STR_UNF[STR_UNF["stringent_num"] % LLXi1 == 0])
Xi1_both_lags = len(STR_UNF[(STR_UNF["stringent_num"] % HHXi1 == 0) & (STR_UNF["stringent_num"] % LLXi1 == 0)])
Xi1_other_lags = len(STR_UNF[(STR_UNF["stringent_num"] % HHXi1 != 0) & (STR_UNF["stringent_num"] % LLXi1 != 0) & (STR_UNF["democratic_num"] % Xi1 == 0)])
N_models = len(DEMO_UNF[DEMO_UNF["democratic_num"] % N == 0])
Sigma_models = len(DEMO_UNF[DEMO_UNF["democratic_num"] % Sigma == 0])
Xi1_models = len(DEMO_UNF[DEMO_UNF["democratic_num"] % Xi1 == 0])
# latex table
print(r"""
\begin{tabular}{ccll}
\toprule
Field & Interactions & Lagrangians & Collected models \\
\midrule
\multirow{2}{*}{$N \sim (\mathbf{1}, \mathbf{1}, 0)_{F}$} & $L H N$ & %s (%s) & \multirow{2}{*}{%s (%s)} \\
& Other & %s (%s) & \\
\midrule
\multirow{2}{*}{$\Sigma \sim (\mathbf{1}, \mathbf{3}, 0)_{F}$} & $L H \Sigma$ & %s (%s) & \multirow{2}{*}{%s (%s)} \\
& Other & %s (%s) & \\
\midrule
\multirow{4}{*}{$\Xi_{1} \sim (\mathbf{1}, \mathbf{3}, 1)_{S}$} & $L L \Xi_{1}$ & %s (%s) & \multirow{4}{*}{%s (%s)} \\
& $H H \Xi_{1}^{\dagger}$ & %s (%s) & \\
& Both & %s (%s) & \\
& Other & %s (%s) & \\
\bottomrule
\end{tabular}
""" % (
f"{N_NHL_lags:,}", f"{100 * N_NHL_lags / LAGS:.1f}\%",
f"{N_models:,}", f"{100 * N_models / MODELS:.1f}\%",
f"{N_other_lags:,}", f"{100 * N_other_lags / LAGS:.1f}\%",
f"{Sigma_NHSigma_lags:,}", f"{100 * Sigma_NHSigma_lags / LAGS:.1f}\%",
f"{Sigma_models:,}", f"{100 * Sigma_models / MODELS:.1f}\%",
f"{Sigma_other_lags:,}", f"{100 * Sigma_other_lags / LAGS:.1f}\%",
f"{Xi1_LLXi1_lags:,}", f"{100 * Xi1_LLXi1_lags / LAGS:.1f}\%",
f"{Xi1_models:,}", f"{100 * Xi1_models / MODELS:.1f}\%",
f"{Xi1_HHXi1_lags:,}", f"{100 * Xi1_HHXi1_lags / LAGS:.1f}\%",
f"{Xi1_both_lags:,}", f"{100 * Xi1_both_lags / LAGS:.1f}\%",
f"{Xi1_other_lags:,}", f"{100 * Xi1_other_lags / LAGS:.1f}\%",
)
)
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
SMALL_SIZE = 15
MEDIUM_SIZE = 20
BIGGER_SIZE = 20
plt.rc('font', size=SMALL_SIZE) # controls default text sizes
plt.rc('axes', titlesize=SMALL_SIZE) # fontsize of the axes title
plt.rc('axes', labelsize=MEDIUM_SIZE) # fontsize of the x and y labels
plt.rc('xtick', labelsize=SMALL_SIZE) # fontsize of the tick labels
plt.rc('ytick', labelsize=SMALL_SIZE) # fontsize of the tick labels
plt.rc('legend', fontsize=SMALL_SIZE) # legend fontsize
plt.rc('figure', titlesize=BIGGER_SIZE) # fontsize of the figure title
plt.tight_layout()
plt.rcParams.update({
"text.usetex": True,
"font.family": "serif",
"font.serif": ["Computer Modern Roman"]}
)
sns.set_palette("muted")
latex_labels = []
for l in labels:
if "pp" in l:
new_l = l.replace("pp", "^{\prime\prime}")
elif "p" in l:
new_l = l.replace("p", "^\prime")
else:
new_l = l
latex_labels.append("$" + new_l + "$")
filter_bar_df = pd.DataFrame(data={
"Operator": latex_labels,
"Unfiltered": total,
"Democratic": demo,
"Dimension": dimensions
})
demo_5 = sum(filter_bar_df[filter_bar_df["Dimension"] == 5]["Democratic"])
demo_7 = sum(filter_bar_df[filter_bar_df["Dimension"] == 7]["Democratic"])
demo_9 = sum(filter_bar_df[filter_bar_df["Dimension"] == 9]["Democratic"])
demo_11 = sum(filter_bar_df[filter_bar_df["Dimension"] == 11]["Democratic"])
unf_5 = sum(filter_bar_df[filter_bar_df["Dimension"] == 5]["Unfiltered"])
unf_7 = sum(filter_bar_df[filter_bar_df["Dimension"] == 7]["Unfiltered"])
unf_9 = sum(filter_bar_df[filter_bar_df["Dimension"] == 9]["Unfiltered"])
unf_11 = sum(filter_bar_df[filter_bar_df["Dimension"] == 11]["Unfiltered"])
barplot_df = pd.DataFrame(
{'Dimension': [5, 7, 9, 11],
'Democratic': [demo_5, demo_7, demo_9, demo_11],
'Unfiltered': [unf_5-demo_5, unf_7-demo_7, unf_9-demo_9, unf_11-demo_11]}
)
ax = barplot_df.plot.bar(x="Dimension", stacked=True, rot=0)
ax.set_yscale("log")
ax.set_ylabel("Number of models")
plt.tight_layout()
plt.savefig("/home/garj/filter_barchart_dimension.pdf")
plt.savefig("/home/garj/filter_barchart_dimension.png")
ops_filter_bar_df = filter_bar_df[filter_bar_df["Dimension"] < 11]
f, ax = plt.subplots(figsize=(7, 10))
sns.barplot(x="Unfiltered", y="Operator", data=ops_filter_bar_df, label="Unfiltered", color=sns.color_palette()[1])
sns.barplot(x="Democratic", y="Operator", data=ops_filter_bar_df, label="Democratic", color=sns.color_palette()[0])
ax.set_xscale("log")
ax.legend(ncol=2, loc="upper right", frameon=True)
ax.set(xlim=(0, 10000), ylabel="Operator", xlabel="Number of models")
ax.text(x=2000, y=7, s="$d < 11$", fontsize=20)
for tick in ax.yaxis.get_major_ticks()[1::2]:
tick.set_pad(40)
plt.tight_layout()
plt.savefig("/home/garj/filter_barchart_operators579.pdf")
plt.savefig("/home/garj/filter_barchart_operators579.png")
import seaborn as sns
import matplotlib.pyplot as plt
ops_filter_bar_df = filter_bar_df[filter_bar_df["Dimension"] == 11]
f, ax = plt.subplots(figsize=(7, 15))
sns.barplot(x="Unfiltered", y="Operator", data=ops_filter_bar_df, label="Unfiltered", color=sns.color_palette()[1])
sns.barplot(x="Democratic", y="Operator", data=ops_filter_bar_df, label="Democratic", color=sns.color_palette()[0])
ax.set_xscale("log")
ax.legend(ncol=2, loc="upper right", frameon=True)
ax.set(xlim=(0, 100000), ylabel="Operator", xlabel="Number of models")
ax.text(x=12000, y=9, s="$d = 11$", fontsize=22)
for tick in ax.yaxis.get_major_ticks()[1::2]:
tick.set_pad(40)
plt.tight_layout()
plt.savefig("/home/garj/filter_barchart_operators11.pdf")
plt.savefig("/home/garj/filter_barchart_operators11.png")
```
|
github_jupyter
|
```
from IPython.display import HTML
# Cell visibility - COMPLETE:
#tag = HTML('''<style>
#div.input {
# display:none;
#}
#</style>''')
#display(tag)
#Cell visibility - TOGGLE:
tag = HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide()
} else {
$('div.input').show()
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<p style="text-align:right">
Promijeni vidljivost <a href="javascript:code_toggle()">ovdje</a>.</p>''')
display(tag)
```
## Kompleksni brojevi u polarnom obliku
U ovome interaktivnom primjeru, kompleksni brojevi se vizualiziraju u kompleksnoj ravnini, a određuju se koristeći polarni oblik. Kompleksni brojevi se, dakle, određuju modulom (duljinom odgovarajućeg vektora) i argumentom (kutom odgovarajućeg vektora). Možete testirati osnovne matematičke operacije nad kompleksnim brojevima: zbrajanje, oduzimanje, množenje i dijeljenje. Svi se rezultati prikazuju na odgovarajućem grafu, kao i u matematičkoj notaciji zasnovanoj na polarnom obliku kompleksnog broja.
Kompleksnim brojevima možete manipulirati izravno na grafu (jednostavnim klikom) i / ili istovremeno koristiti odgovarajuća polja za unos modula i argumenta. Kako bi se osigurala bolja vidljivost vektora na grafu, modul kompleksnog broja je ograničen na $\pm10$.
```
%matplotlib notebook
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import numpy as np
import ipywidgets as widgets
from IPython.display import display
from IPython.display import HTML
import math
red_patch = mpatches.Patch(color='red', label='z1')
blue_patch = mpatches.Patch(color='blue', label='z2')
green_patch = mpatches.Patch(color='green', label='z1 + z2')
yellow_patch = mpatches.Patch(color='yellow', label='z1 - z2')
black_patch = mpatches.Patch(color='black', label='z1 * z2')
magenta_patch = mpatches.Patch(color='magenta', label='z1 / z2')
# Init values
XLIM = 5
YLIM = 5
vectors_index_first = False;
V = [None, None]
V_complex = [None, None]
# Complex plane
fig = plt.figure(num='Kompleksni brojevi u polarnom obliku')
ax = fig.add_subplot(1, 1, 1)
def get_interval(lim):
if lim <= 10:
return 1
if lim < 75:
return 5
if lim > 100:
return 25
return 10
def set_ticks():
XLIMc = int((XLIM / 10) + 1) * 10
YLIMc = int((YLIM / 10) + 1) * 10
if XLIMc > 150:
XLIMc += 10
if YLIMc > 150:
YLIMc += 10
xstep = get_interval(XLIMc)
ystep = get_interval(YLIMc)
#print(stepx, stepy)
major_ticks = np.arange(-XLIMc, XLIMc, xstep)
major_ticks_y = np.arange(-YLIMc, YLIMc, ystep)
ax.set_xticks(major_ticks)
ax.set_yticks(major_ticks_y)
ax.grid(which='both')
def clear_plot():
plt.cla()
set_ticks()
ax.set_xlabel('Re')
ax.set_ylabel('Im')
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
plt.legend(handles=[red_patch, blue_patch, green_patch, yellow_patch, black_patch, magenta_patch])
clear_plot()
set_ticks()
plt.show()
set_ticks()
# Conversion functions
def com_to_trig(real, im):
r = math.sqrt(real**2 + im**2)
if abs(real) <= 1e-6 and im > 0:
arg = 90
return r, arg
if abs(real) < 1e-6 and im < 0:
arg = 270
return r, arg
if abs(im) < 1e-6 and real > 0:
arg = 0
return r, arg
if abs(im) < 1e-6 and real < 0:
arg = 180
return r, arg
if im != 0 and real !=0:
arg = np.arctan(im / real) * 180 / np.pi
if im > 0 and real < 0:
arg += 180
if im < 0 and real > 0:
arg +=360
if im < 0 and real < 0:
arg += 180
return r, arg
if abs(im) < 1e-6 and abs(real) < 1e-6:
arg = 0
return r, arg
def trig_to_com(r, arg):
re = r * np.cos(arg * np.pi / 180.)
im = r * np.sin(arg * np.pi / 180.)
return (re, im)
# Set a complex number using direct manipulation on the plot
def set_vector(i, data_x, data_y):
clear_plot()
V.pop(i)
V.insert(i, (0, 0, round(data_x, 2), round(data_y, 2)))
V_complex.pop(i)
V_complex.insert(i, complex(round(data_x, 2), round(data_y, 2)))
if i == 0:
ax.arrow(*V[0], head_width=0.25, head_length=0.5, color="r", length_includes_head=True)
z, arg = com_to_trig(data_x, data_y)
a1.value = round(z, 2)
b1.value = round(arg, 2)
if V[1] != None:
ax.arrow(*V[1], head_width=0.25, head_length=0.5, color="b", length_includes_head=True)
elif i == 1:
ax.arrow(*V[1], head_width=0.25, head_length=0.5, color="b", length_includes_head=True)
z, arg = com_to_trig(data_x, data_y)
a2.value = round(z, 2)
b2.value = round(arg, 2)
if V[0] != None:
ax.arrow(*V[0], head_width=0.25, head_length=0.5, color="r", length_includes_head=True)
max_bound()
def onclick(event):
global vectors_index_first
vectors_index_first = not vectors_index_first
x = event.xdata
y = event.ydata
if (x > 10):
x = 10.0
if (x < - 10):
x = -10.0
if (y > 10):
y = 10.0
if (y < - 10):
y = -10.0
if vectors_index_first:
set_vector(0, x, y)
else:
set_vector(1, x, y)
fig.canvas.mpl_connect('button_press_event', onclick)
# Widgets
a1 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = 0, max = 10, step = 0.5)
b1 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = 0, max = 360, step = 10)
button_set_z1 = widgets.Button(description="Prikaži z1")
a2 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = 0, max = 10, step = 0.5)
b2 = widgets.BoundedFloatText(layout=widgets.Layout(width='10%'), min = 0, max = 360, step = 10)
button_set_z2 = widgets.Button(description="Prikaži z2")
box_layout_z1 = widgets.Layout(border='solid red', padding='10px')
box_layout_z2 = widgets.Layout(border='solid blue', padding='10px')
box_layout_opers = widgets.Layout(border='solid black', padding='10px')
items_z1 = [widgets.Label("z1: Duljina (|z1|) = "), a1, widgets.Label("Kut (\u2221)= "), b1, button_set_z1]
items_z2 = [widgets.Label("z2: Duljina (|z2|) = "), a2, widgets.Label("Kut (\u2221)= "), b2, button_set_z2]
display(widgets.Box(children=items_z1, layout=box_layout_z1))
display(widgets.Box(children=items_z2, layout=box_layout_z2))
button_add = widgets.Button(description="Zbroji")
button_substract = widgets.Button(description="Oduzmi")
button_multiply = widgets.Button(description="Pomnoži")
button_divide = widgets.Button(description="Podijeli")
button_reset = widgets.Button(description="Resetiraj")
output = widgets.Output()
print('Operacije nad kompleksnim brojevima:')
items_operations = [button_add, button_substract, button_multiply, button_divide, button_reset]
display(widgets.Box(children=items_operations))
display(output)
# Set complex number using input widgets (Text and Button)
def on_button_set_z1_clicked(b):
z1_old = V[0];
re, im = trig_to_com(a1.value, b1.value)
z1_new = (0, 0, re, im)
if z1_old != z1_new:
set_vector(0, re, im)
change_lims()
def on_button_set_z2_clicked(b):
z2_old = V[1];
re, im = trig_to_com(a2.value, b2.value)
z2_new = (0, 0, re, im)
if z2_old != z2_new:
set_vector(1, re, im)
change_lims()
# Complex number operations:
def perform_operation(oper):
global XLIM, YLIM
if (V_complex[0] != None) and (V_complex[1] != None):
if (oper == '+'):
result = V_complex[0] + V_complex[1]
v_color = "g"
elif (oper == '-'):
result = V_complex[0] - V_complex[1]
v_color = "y"
elif (oper == '*'):
result = V_complex[0] * V_complex[1]
v_color = "black"
elif (oper == '/'):
result = V_complex[0] / V_complex[1]
v_color = "magenta"
result = complex(round(result.real, 2), round(result.imag, 2))
ax.arrow(0, 0, result.real, result.imag, head_width=0.25, head_length=0.15, color=v_color, length_includes_head=True)
if abs(result.real) > XLIM:
XLIM = round(abs(result.real) + 1)
if abs(result.imag) > YLIM:
YLIM = round(abs(result.imag) + 1)
change_lims()
with output:
z1, ang1 = com_to_trig(V_complex[0].real, V_complex[0].imag )
z2, ang2 = com_to_trig(V_complex[1].real, V_complex[1].imag)
z3, ang3 = com_to_trig(result.real, result.imag)
z1 = round(z1, 2)
ang1 = round(ang1, 2)
z2 = round(z2, 2)
ang2 = round(ang2, 2)
z3 = round(z3, 2)
ang3 = round(ang3, 2)
print("{}*(cos({}) + i*sin({}))".format(z1,ang1,ang1), oper,
"{}*(cos({}) + i*sin({}))".format(z2,ang2,ang2), "=",
"{}*(cos({}) + i*sin({}))".format(z3,ang3,ang3))
print('{} \u2221{}'.format(z1, ang1), oper,
'{} \u2221{}'.format(z2, ang2), "=",
'{} \u2221{}'.format(z3, ang3))
def on_button_add_clicked(b):
perform_operation("+")
def on_button_substract_clicked(b):
perform_operation("-")
def on_button_multiply_clicked(b):
perform_operation("*")
def on_button_divide_clicked(b):
perform_operation("/")
# Plot init methods
def on_button_reset_clicked(b):
global V, V_complex, XLIM, YLIM
with output:
output.clear_output()
clear_plot()
vectors_index_first = False;
V = [None, None]
V_complex = [None, None]
a1.value = 0
b1.value = 0
a2.value = 0
b2.value = 0
XLIM = 5
YLIM = 5
change_lims()
def clear_plot():
plt.cla()
set_ticks()
ax.set_xlabel('Re')
ax.set_ylabel('Im')
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
plt.legend(handles=[red_patch, blue_patch, green_patch, yellow_patch, black_patch, magenta_patch])
def change_lims():
set_ticks()
plt.ylim([-YLIM, YLIM])
plt.xlim([-XLIM, XLIM])
set_ticks()
def max_bound():
global XLIM, YLIM
mx = 0
my = 0
if V_complex[0] != None:
z = V_complex[0]
if abs(z.real) > mx:
mx = abs(z.real)
if abs(z.imag) > my:
my = abs(z.imag)
if V_complex[1] != None:
z = V_complex[1]
if abs(z.real) > mx:
mx = abs(z.real)
if abs(z.imag) > my:
my = abs(z.imag)
if mx > XLIM:
XLIM = round(mx + 1)
elif mx <=5:
XLIM = 5
if my > YLIM:
YLIM = round(my + 1)
elif my <=5:
YLIM = 5
change_lims()
# Button events
button_set_z1.on_click(on_button_set_z1_clicked)
button_set_z2.on_click(on_button_set_z2_clicked)
button_add.on_click(on_button_add_clicked)
button_substract.on_click(on_button_substract_clicked)
button_multiply.on_click(on_button_multiply_clicked)
button_divide.on_click(on_button_divide_clicked)
button_reset.on_click(on_button_reset_clicked)
```
|
github_jupyter
|
Mount my google drive, where I stored the dataset.
```
from google.colab import drive
drive.mount('/content/drive')
```
**Download dependencies**
```
!pip3 install sklearn matplotlib GPUtil
!pip3 install torch torchvision
```
**Download Data**
In order to acquire the dataset please navigate to:
https://ieee-dataport.org/documents/cervigram-image-dataset
Unzip the dataset into the folder "dataset".
For your environment, please adjust the paths accordingly.
```
!rm -vrf "dataset"
!mkdir "dataset"
# !cp -r "/content/drive/My Drive/Studiu doctorat leziuni cervicale/cervigram-image-dataset-v2.zip" "dataset/cervigram-image-dataset-v2.zip"
!cp -r "cervigram-image-dataset-v2.zip" "dataset/cervigram-image-dataset-v2.zip"
!unzip "dataset/cervigram-image-dataset-v2.zip" -d "dataset"
```
**Constants**
For your environment, please modify the paths accordingly.
```
# TRAIN_PATH = '/content/dataset/data/train/'
# TEST_PATH = '/content/dataset/data/test/'
TRAIN_PATH = 'dataset/data/train/'
TEST_PATH = 'dataset/data/test/'
CROP_SIZE = 260
IMAGE_SIZE = 224
BATCH_SIZE = 100
```
**Imports**
```
import torch as t
import torchvision as tv
import numpy as np
import PIL as pil
import matplotlib.pyplot as plt
from torchvision.datasets import ImageFolder
from torch.utils.data import DataLoader
from torch.nn import Linear, BCEWithLogitsLoss
import sklearn as sk
import sklearn.metrics
from os import listdir
import time
import random
import GPUtil
```
**Memory Stats**
```
import GPUtil
def memory_stats():
for gpu in GPUtil.getGPUs():
print("GPU RAM Free: {0:.0f}MB | Used: {1:.0f}MB | Util {2:3.0f}% | Total {3:.0f}MB".format(gpu.memoryFree, gpu.memoryUsed, gpu.memoryUtil*100, gpu.memoryTotal))
memory_stats()
```
**Deterministic Measurements**
This statements help making the experiments reproducible by fixing the random seeds. Despite fixing the random seeds, experiments are usually not reproducible using different PyTorch releases, commits, platforms or between CPU and GPU executions. Please find more details in the PyTorch documentation:
https://pytorch.org/docs/stable/notes/randomness.html
```
SEED = 0
t.manual_seed(SEED)
t.cuda.manual_seed(SEED)
t.backends.cudnn.deterministic = True
t.backends.cudnn.benchmark = False
np.random.seed(SEED)
random.seed(SEED)
```
**Loading Data**
The dataset is structured in multiple small folders of 7 images each. This generator iterates through the folders and returns the category and 7 paths: one for each image in the folder. The paths are ordered; the order is important since each folder contains 3 types of images, first 5 are with acetic acid solution and the last two are through a green lens and having iodine solution(a solution of a dark red color).
```
def sortByLastDigits(elem):
chars = [c for c in elem if c.isdigit()]
return 0 if len(chars) == 0 else int(''.join(chars))
def getImagesPaths(root_path):
for class_folder in [root_path + f for f in listdir(root_path)]:
category = int(class_folder[-1])
for case_folder in listdir(class_folder):
case_folder_path = class_folder + '/' + case_folder + '/'
img_files = [case_folder_path + file_name for file_name in listdir(case_folder_path)]
yield category, sorted(img_files, key = sortByLastDigits)
```
We define 3 datasets, which load 3 kinds of images: natural images, images taken through a green lens and images where the doctor applied iodine solution (which gives a dark red color). Each dataset has dynamic and static transformations which could be applied to the data. The static transformations are applied on the initialization of the dataset, while the dynamic ones are applied when loading each batch of data.
```
class SimpleImagesDataset(t.utils.data.Dataset):
def __init__(self, root_path, transforms_x_static = None, transforms_x_dynamic = None, transforms_y_static = None, transforms_y_dynamic = None):
self.dataset = []
self.transforms_x = transforms_x_dynamic
self.transforms_y = transforms_y_dynamic
for category, img_files in getImagesPaths(root_path):
for i in range(5):
img = pil.Image.open(img_files[i])
if transforms_x_static != None:
img = transforms_x_static(img)
if transforms_y_static != None:
category = transforms_y_static(category)
self.dataset.append((img, category))
def __getitem__(self, i):
x, y = self.dataset[i]
if self.transforms_x != None:
x = self.transforms_x(x)
if self.transforms_y != None:
y = self.transforms_y(y)
return x, y
def __len__(self):
return len(self.dataset)
class GreenLensImagesDataset(SimpleImagesDataset):
def __init__(self, root_path, transforms_x_static = None, transforms_x_dynamic = None, transforms_y_static = None, transforms_y_dynamic = None):
self.dataset = []
self.transforms_x = transforms_x_dynamic
self.transforms_y = transforms_y_dynamic
for category, img_files in getImagesPaths(root_path):
# Only the green lens image
img = pil.Image.open(img_files[-2])
if transforms_x_static != None:
img = transforms_x_static(img)
if transforms_y_static != None:
category = transforms_y_static(category)
self.dataset.append((img, category))
class RedImagesDataset(SimpleImagesDataset):
def __init__(self, root_path, transforms_x_static = None, transforms_x_dynamic = None, transforms_y_static = None, transforms_y_dynamic = None):
self.dataset = []
self.transforms_x = transforms_x_dynamic
self.transforms_y = transforms_y_dynamic
for category, img_files in getImagesPaths(root_path):
# Only the green lens image
img = pil.Image.open(img_files[-1])
if transforms_x_static != None:
img = transforms_x_static(img)
if transforms_y_static != None:
category = transforms_y_static(category)
self.dataset.append((img, category))
```
**Preprocess Data**
Convert pytorch tensor to numpy array.
```
def to_numpy(x):
return x.cpu().detach().numpy()
```
Data transformations for the test and training sets.
```
norm_mean = [0.485, 0.456, 0.406]
norm_std = [0.229, 0.224, 0.225]
transforms_train = tv.transforms.Compose([
tv.transforms.RandomAffine(degrees = 45, translate = None, scale = (1., 2.), shear = 30),
# tv.transforms.CenterCrop(CROP_SIZE),
tv.transforms.Resize(IMAGE_SIZE),
tv.transforms.RandomHorizontalFlip(),
tv.transforms.ToTensor(),
tv.transforms.Lambda(lambda t: t.cuda()),
tv.transforms.Normalize(mean=norm_mean, std=norm_std)
])
transforms_test = tv.transforms.Compose([
# tv.transforms.CenterCrop(CROP_SIZE),
tv.transforms.Resize(IMAGE_SIZE),
tv.transforms.ToTensor(),
tv.transforms.Normalize(mean=norm_mean, std=norm_std)
])
y_transform = tv.transforms.Lambda(lambda y: t.tensor(y, dtype=t.long, device = 'cuda:0'))
```
Initialize pytorch datasets and loaders for training and test.
```
def create_loaders(dataset_class):
dataset_train = dataset_class(TRAIN_PATH, transforms_x_dynamic = transforms_train, transforms_y_dynamic = y_transform)
dataset_test = dataset_class(TEST_PATH, transforms_x_static = transforms_test,
transforms_x_dynamic = tv.transforms.Lambda(lambda t: t.cuda()), transforms_y_dynamic = y_transform)
loader_train = DataLoader(dataset_train, BATCH_SIZE, shuffle = True, num_workers = 0)
loader_test = DataLoader(dataset_test, BATCH_SIZE, shuffle = False, num_workers = 0)
return loader_train, loader_test, len(dataset_train), len(dataset_test)
loader_train_simple_img, loader_test_simple_img, len_train, len_test = create_loaders(SimpleImagesDataset)
```
**Visualize Data**
Load a few images so that we can see the effects of the data augmentation on the training set.
```
def plot_one_prediction(x, label, pred):
x, label, pred = to_numpy(x), to_numpy(label), to_numpy(pred)
x = np.transpose(x, [1, 2, 0])
if x.shape[-1] == 1:
x = x.squeeze()
x = x * np.array(norm_std) + np.array(norm_mean)
plt.title(label, color = 'green' if label == pred else 'red')
plt.imshow(x)
def plot_predictions(imgs, labels, preds):
fig = plt.figure(figsize = (20, 5))
for i in range(20):
fig.add_subplot(2, 10, i + 1, xticks = [], yticks = [])
plot_one_prediction(imgs[i], labels[i], preds[i])
# x, y = next(iter(loader_train_simple_img))
# plot_predictions(x, y, y)
```
**Model**
Define a few models to experiment with.
```
def get_mobilenet_v2():
model = t.hub.load('pytorch/vision', 'mobilenet_v2', pretrained=True)
model.classifier[1] = Linear(in_features=1280, out_features=4, bias=True)
model = model.cuda()
return model
def get_vgg_19():
model = tv.models.vgg19(pretrained = True)
model = model.cuda()
model.classifier[6].out_features = 4
return model
def get_res_next_101():
model = t.hub.load('facebookresearch/WSL-Images', 'resnext101_32x8d_wsl')
model.fc.out_features = 4
model = model.cuda()
return model
def get_resnet_18():
model = tv.models.resnet18(pretrained = True)
model.fc.out_features = 4
model = model.cuda()
return model
def get_dense_net():
model = tv.models.densenet121(pretrained = True)
model.classifier.out_features = 4
model = model.cuda()
return model
class MobileNetV2_FullConv(t.nn.Module):
def __init__(self):
super().__init__()
self.cnn = get_mobilenet_v2().features
self.cnn[18] = t.nn.Sequential(
tv.models.mobilenet.ConvBNReLU(320, 32, kernel_size=1),
t.nn.Dropout2d(p = .7)
)
self.fc = t.nn.Linear(32, 4)
def forward(self, x):
x = self.cnn(x)
x = x.mean([2, 3])
x = self.fc(x);
return x
model_simple = t.nn.DataParallel(get_mobilenet_v2())
```
**Train & Evaluate**
Timer utility function. This is used to measure the execution speed.
```
time_start = 0
def timer_start():
global time_start
time_start = time.time()
def timer_end():
return time.time() - time_start
```
This function trains the network and evaluates it at the same time. It outputs the metrics recorded during the training for both train and test. We are measuring accuracy and the loss. The function also saves a checkpoint of the model every time the accuracy is improved. In the end we will have a checkpoint of the model which gave the best accuracy.
```
def train_eval(optimizer, model, loader_train, loader_test, chekpoint_name, epochs):
metrics = {
'losses_train': [],
'losses_test': [],
'acc_train': [],
'acc_test': [],
'prec_train': [],
'prec_test': [],
'rec_train': [],
'rec_test': [],
'f_score_train': [],
'f_score_test': []
}
best_acc = 0
loss_fn = t.nn.CrossEntropyLoss()
try:
for epoch in range(epochs):
timer_start()
train_epoch_loss, train_epoch_acc, train_epoch_precision, train_epoch_recall, train_epoch_f_score = 0, 0, 0, 0, 0
test_epoch_loss, test_epoch_acc, test_epoch_precision, test_epoch_recall, test_epoch_f_score = 0, 0, 0, 0, 0
# Train
model.train()
for x, y in loader_train:
y_pred = model.forward(x)
loss = loss_fn(y_pred, y)
loss.backward()
optimizer.step()
# memory_stats()
optimizer.zero_grad()
y_pred, y = to_numpy(y_pred), to_numpy(y)
pred = y_pred.argmax(axis = 1)
ratio = len(y) / len_train
train_epoch_loss += (loss.item() * ratio)
train_epoch_acc += (sk.metrics.accuracy_score(y, pred) * ratio)
precision, recall, f_score, _ = sk.metrics.precision_recall_fscore_support(y, pred, average = 'macro')
train_epoch_precision += (precision * ratio)
train_epoch_recall += (recall * ratio)
train_epoch_f_score += (f_score * ratio)
metrics['losses_train'].append(train_epoch_loss)
metrics['acc_train'].append(train_epoch_acc)
metrics['prec_train'].append(train_epoch_precision)
metrics['rec_train'].append(train_epoch_recall)
metrics['f_score_train'].append(train_epoch_f_score)
# Evaluate
model.eval()
with t.no_grad():
for x, y in loader_test:
y_pred = model.forward(x)
loss = loss_fn(y_pred, y)
y_pred, y = to_numpy(y_pred), to_numpy(y)
pred = y_pred.argmax(axis = 1)
ratio = len(y) / len_test
test_epoch_loss += (loss * ratio)
test_epoch_acc += (sk.metrics.accuracy_score(y, pred) * ratio )
precision, recall, f_score, _ = sk.metrics.precision_recall_fscore_support(y, pred, average = 'macro')
test_epoch_precision += (precision * ratio)
test_epoch_recall += (recall * ratio)
test_epoch_f_score += (f_score * ratio)
metrics['losses_test'].append(test_epoch_loss)
metrics['acc_test'].append(test_epoch_acc)
metrics['prec_test'].append(test_epoch_precision)
metrics['rec_test'].append(test_epoch_recall)
metrics['f_score_test'].append(test_epoch_f_score)
if metrics['acc_test'][-1] > best_acc:
best_acc = metrics['acc_test'][-1]
t.save({'model': model.state_dict()}, 'checkpint {}.tar'.format(chekpoint_name))
print('Epoch {} acc {} prec {} rec {} f {} minutes {}'.format(
epoch + 1, metrics['acc_test'][-1], metrics['prec_test'][-1], metrics['rec_test'][-1], metrics['f_score_test'][-1], timer_end() / 60))
except KeyboardInterrupt as e:
print(e)
print('Ended training')
return metrics
```
Plot a metric for both train and test.
```
def plot_train_test(train, test, title, y_title):
plt.plot(range(len(train)), train, label = 'train')
plt.plot(range(len(test)), test, label = 'test')
plt.xlabel('Epochs')
plt.ylabel(y_title)
plt.title(title)
plt.legend()
plt.show()
```
Plot precision - recall curve
```
def plot_precision_recall(metrics):
plt.scatter(metrics['prec_train'], metrics['rec_train'], label = 'train')
plt.scatter(metrics['prec_test'], metrics['rec_test'], label = 'test')
plt.legend()
plt.title('Precision-Recall')
plt.xlabel('Precision')
plt.ylabel('Recall')
```
Train a model for several epochs. The steps_learning parameter is a list of tuples. Each tuple specifies the steps and the learning rate.
```
def do_train(model, loader_train, loader_test, checkpoint_name, steps_learning):
for steps, learn_rate in steps_learning:
metrics = train_eval(t.optim.Adam(model.parameters(), lr = learn_rate, weight_decay = 0), model, loader_train, loader_test, checkpoint_name, steps)
print('Best test accuracy :', max(metrics['acc_test']))
plot_train_test(metrics['losses_train'], metrics['losses_test'], 'Loss (lr = {})'.format(learn_rate))
plot_train_test(metrics['acc_train'], metrics['acc_test'], 'Accuracy (lr = {})'.format(learn_rate))
```
Perform actual training.
```
def do_train(model, loader_train, loader_test, checkpoint_name, steps_learning):
t.cuda.empty_cache()
for steps, learn_rate in steps_learning:
metrics = train_eval(t.optim.Adam(model.parameters(), lr = learn_rate, weight_decay = 0), model, loader_train, loader_test, checkpoint_name, steps)
index_max = np.array(metrics['acc_test']).argmax()
print('Best test accuracy :', metrics['acc_test'][index_max])
print('Corresponding precision :', metrics['prec_test'][index_max])
print('Corresponding recall :', metrics['rec_test'][index_max])
print('Corresponding f1 score :', metrics['f_score_test'][index_max])
plot_train_test(metrics['losses_train'], metrics['losses_test'], 'Loss (lr = {})'.format(learn_rate), 'Loss')
plot_train_test(metrics['acc_train'], metrics['acc_test'], 'Accuracy (lr = {})'.format(learn_rate), 'Accuracy')
plot_train_test(metrics['prec_train'], metrics['prec_test'], 'Precision (lr = {})'.format(learn_rate), 'Precision')
plot_train_test(metrics['rec_train'], metrics['rec_test'], 'Recall (lr = {})'.format(learn_rate), 'Recall')
plot_train_test(metrics['f_score_train'], metrics['f_score_test'], 'F1 Score (lr = {})'.format(learn_rate), 'F1 Score')
plot_precision_recall(metrics)
do_train(model_simple, loader_train_simple_img, loader_test_simple_img, 'simple_1', [(50, 1e-4)])
# checkpoint = t.load('/content/checkpint simple_1.tar')
# model_simple.load_state_dict(checkpoint['model'])
```
|
github_jupyter
|
```
%matplotlib inline
```
# Simple Oscillator Example
This example shows the most simple way of using a solver.
We solve free vibration of a simple oscillator:
$$m \ddot{u} + k u = 0,\quad u(0) = u_0,\quad \dot{u}(0) = \dot{u}_0$$
using the CVODE solver. An analytical solution exists, given by
$$u(t) = u_0 \cos\left(\sqrt{\frac{k}{m}} t\right)+\frac{\dot{u}_0}{\sqrt{\frac{k}{m}}} \sin\left(\sqrt{\frac{k}{m}} t\right)$$
```
from __future__ import print_function
import matplotlib.pyplot as plt
import numpy as np
from scikits.odes import ode
#data of the oscillator
k = 4.0
m = 1.0
#initial position and speed data on t=0, x[0] = u, x[1] = \dot{u}, xp = \dot{x}
initx = [1, 0.1]
```
We need a first order system, so convert the second order system
$$m \ddot{u} + k u = 0,\quad u(0) = u_0,\quad \dot{u}(0) = \dot{u}_0$$
into
$$\left\{ \begin{array}{l}
\dot u = v\\
\dot v = \ddot u = -\frac{ku}{m}
\end{array} \right.$$
You need to define a function that computes the right hand side of above equation:
```
def rhseqn(t, x, xdot):
""" we create rhs equations for the problem"""
xdot[0] = x[1]
xdot[1] = - k/m * x[0]
```
To solve the ODE you define an ode object, specify the solver to use, here cvode, and pass the right hand side function. You request the solution at specific timepoints by passing an array of times to the solve member.
```
solver = ode('cvode', rhseqn, old_api=False)
solution = solver.solve([0., 1., 2.], initx)
print('\n t Solution Exact')
print('------------------------------------')
for t, u in zip(solution.values.t, solution.values.y):
print('{0:>4.0f} {1:15.6g} {2:15.6g}'.format(t, u[0],
initx[0]*np.cos(np.sqrt(k/m)*t)+initx[1]*np.sin(np.sqrt(k/m)*t)/np.sqrt(k/m)))
```
You can continue the solver by passing further times. Calling the solve routine reinits the solver, so you can restart at whatever time. To continue from the last computed solution, pass the last obtained time and solution.
**Note:** The solver performes better if it can take into account history information, so avoid calling solve to continue computation!
In general, you must check for errors using the errors output of solve.
```
#Solve over the next hour by continuation
times = np.linspace(0, 3600, 61)
times[0] = solution.values.t[-1]
solution = solver.solve(times, solution.values.y[-1])
if solution.errors.t:
print ('Error: ', solution.message, 'Error at time', solution.errors.t)
print ('Computed Solutions:')
print('\n t Solution Exact')
print('------------------------------------')
for t, u in zip(solution.values.t, solution.values.y):
print('{0:>4.0f} {1:15.6g} {2:15.6g}'.format(t, u[0],
initx[0]*np.cos(np.sqrt(k/m)*t)+initx[1]*np.sin(np.sqrt(k/m)*t)/np.sqrt(k/m)))
```
The solution fails at a time around 24 seconds. Erros can be due to many things. Here however the reason is simple: we try to make too large jumps in time output. Increasing the allowed steps the solver can take will fix this. This is the **max_steps** option of cvode:
```
solver = ode('cvode', rhseqn, old_api=False, max_steps=5000)
solution = solver.solve(times, solution.values.y[-1])
if solution.errors.t:
print ('Error: ', solution.message, 'Error at time', solution.errors.t)
print ('Computed Solutions:')
print('\n t Solution Exact')
print('------------------------------------')
for t, u in zip(solution.values.t, solution.values.y):
print('{0:>4.0f} {1:15.6g} {2:15.6g}'.format(t, u[0],
initx[0]*np.cos(np.sqrt(k/m)*t)+initx[1]*np.sin(np.sqrt(k/m)*t)/np.sqrt(k/m)))
```
To plot the simple oscillator, we show a (t,x) plot of the solution. Doing this over 60 seconds can be done as follows:
```
#plot of the oscilator
solver = ode('cvode', rhseqn, old_api=False)
times = np.linspace(0,60,600)
solution = solver.solve(times, initx)
plt.plot(solution.values.t,[x[0] for x in solution.values.y])
plt.xlabel('Time [s]')
plt.ylabel('Position [m]')
plt.show()
```
You can refine the tolerances from their defaults to obtain more accurate solutions
```
options1= {'rtol': 1e-6, 'atol': 1e-12, 'max_steps': 50000} # default rtol and atol
options2= {'rtol': 1e-15, 'atol': 1e-25, 'max_steps': 50000}
solver1 = ode('cvode', rhseqn, old_api=False, **options1)
solver2 = ode('cvode', rhseqn, old_api=False, **options2)
solution1 = solver1.solve([0., 1., 60], initx)
solution2 = solver2.solve([0., 1., 60], initx)
print('\n t Solution1 Solution2 Exact')
print('-----------------------------------------------------')
for t, u1, u2 in zip(solution1.values.t, solution1.values.y, solution2.values.y):
print('{0:>4.0f} {1:15.8g} {2:15.8g} {3:15.8g}'.format(t, u1[0], u2[0],
initx[0]*np.cos(np.sqrt(k/m)*t)+initx[1]*np.sin(np.sqrt(k/m)*t)/np.sqrt(k/m)))
```
# Simple Oscillator Example: Stepwise running
When using the *solve* method, you solve over a period of time you decided before. In some problems you might want to solve and decide on the output when to stop. Then you use the *step* method. The same example as above using the step method can be solved as follows.
You define the ode object selecting the cvode solver. You initialize the solver with the begin time and initial conditions using *init_step*. You compute solutions going forward with the *step* method.
```
solver = ode('cvode', rhseqn, old_api=False)
time = 0.
solver.init_step(time, initx)
plott = []
plotx = []
while True:
time += 0.1
# fix roundoff error at end
if time > 60: time = 60
solution = solver.step(time)
if solution.errors.t:
print ('Error: ', solution.message, 'Error at time', solution.errors.t)
break
#we store output for plotting
plott.append(solution.values.t)
plotx.append(solution.values.y[0])
if time >= 60:
break
plt.plot(plott,plotx)
plt.xlabel('Time [s]')
plt.ylabel('Position [m]')
plt.show()
```
The solver interpolates solutions to return the solution at the required output times:
```
print ('plott length:', len(plott), ', last computation times:', plott[-15:]);
```
# Simple Oscillator Example: Internal Solver Stepwise running
When using the *solve* method, you solve over a period of time you decided before. With the *step* method you solve by default towards a desired output time after which you can continue solving the problem.
For full control, you can also compute problems using the solver internal steps. This is not advised, as the number of return steps can be very large, **slowing down** the computation enormously. If you want this nevertheless, you can achieve it with the *one_step_compute* option. Like this:
```
solver = ode('cvode', rhseqn, old_api=False, one_step_compute=True)
time = 0.
solver.init_step(time, initx)
plott = []
plotx = []
while True:
solution = solver.step(60)
if solution.errors.t:
print ('Error: ', solution.message, 'Error at time', solution.errors.t)
break
#we store output for plotting
plott.append(solution.values.t)
plotx.append(solution.values.y[0])
if solution.values.t >= 60:
#back up to 60
solver.set_options(one_step_compute=False)
solution = solver.step(60)
plott[-1] = solution.values.t
plotx[-1] = solution.values.y[0]
break
plt.plot(plott,plotx)
plt.xlabel('Time [s]')
plt.ylabel('Position [m]')
plt.show()
```
By inspection of the returned times you can see how efficient the solver can solve this problem:
```
print ('plott length:', len(plott), ', last computation times:', plott[-15:]);
```
|
github_jupyter
|
```
import classifierMLP as cmlp
import os
import struct
import numpy as np
def load_mnist(path, kind='train'):
"""Load MNIST data from `path`"""
labels_path = os.path.join(path,
'%s-labels-idx1-ubyte' % kind)
images_path = os.path.join(path,
'%s-images-idx3-ubyte' % kind)
print(labels_path)
print(images_path)
with open(labels_path, 'rb') as lbpath:
magic, n = struct.unpack('>II',
lbpath.read(8))
labels = np.fromfile(lbpath,
dtype=np.uint8)
with open(images_path, 'rb') as imgpath:
magic, num, rows, cols = struct.unpack(">IIII",
imgpath.read(16))
images = np.fromfile(imgpath,
dtype=np.uint8).reshape(len(labels), 784)
images = ((images / 255.) - .5) * 2
return images, labels
# unzips mnist
%matplotlib inline
import sys
import gzip
import shutil
if (sys.version_info > (3, 0)):
writemode = 'wb'
else:
writemode = 'w'
zipped_mnist = [f for f in os.listdir('./') if f.endswith('ubyte.gz')]
for z in zipped_mnist:
with gzip.GzipFile(z, mode='rb') as decompressed, open(z[:-3], writemode) as outfile:
outfile.write(decompressed.read())
X_train, y_train = load_mnist('', kind='train')
print('Rows: %d, columns: %d' % (X_train.shape[0], X_train.shape[1]))
X_test, y_test = load_mnist('', kind='t10k')
print('Rows: %d, columns: %d' % (X_test.shape[0], X_test.shape[1]))
X_train.shape
import matplotlib.pyplot as plt
fig, ax = plt.subplots(nrows=2, ncols=5, sharex=True, sharey=True,)
ax = ax.flatten()
for i in range(10):
img = X_train[y_train == i][0].reshape(28, 28)
ax[i].imshow(img, cmap='Greys')
ax[0].set_xticks([])
ax[0].set_yticks([])
plt.tight_layout()
# plt.savefig('images/12_5.png', dpi=300)
plt.show()
fig, ax = plt.subplots(nrows=7, ncols=12, sharex=True, sharey=True,)
ax = ax.flatten()
for i in range(84):
img = X_train[y_train == 4][i].reshape(28, 28)
ax[i].imshow(img, cmap='Greys')
ax[0].set_xticks([])
ax[0].set_yticks([])
plt.tight_layout()
# plt.savefig('images/12_6.png', dpi=300)
plt.show()
import seaborn as sns
sns.countplot(y_train)
n_epochs = 100
nn = cmlp.SimpleMLP(n_hidden_units=100,
l2=0.01,
epochs=n_epochs,
eta=0.0005,
minibatch_size=100,
shuffle=True,
seed=1)
nn.fit(X_train=X_train[:55000],
y_train=y_train[:55000],
X_valid=X_train[55000:],
y_valid=y_train[55000:])
#playing with the traiued model
fig, ax = plt.subplots(nrows=5, ncols=4, sharex=True, sharey=True,)
ax = ax.flatten()
for i in range(20):
img = X_test[i].reshape(28, 28)
ax[i].imshow(img, cmap='Greys')
ax[0].set_xticks([])
ax[0].set_yticks([])
plt.tight_layout()
# plt.savefig('images/12_6.png', dpi=300)
plt.show()
#Lets test for X_test - 1 to 20
for i in range(20):
print ("Prediction for {}th image is {}".format(i,
nn.predict(X_test[i:i+1])))
import matplotlib.pyplot as plt
plt.plot(range(nn.epochs), nn.eval_['cost'],color='green' ,
label='training Error')
plt.ylabel('Error')
plt.xlabel('Epochs')
plt.legend()
import matplotlib.pyplot as plt
plt.plot(range(nn.epochs), nn.eval_['train_acc'],color='green' ,
label='training')
plt.plot(range(nn.epochs), nn.eval_['valid_acc'], color='red',
label='validation', linestyle='--')
plt.ylabel('Accuracy')
plt.xlabel('Epochs')
plt.legend()
#plt.savefig('images/12_08.png', dpi=300)
plt.show()
```
|
github_jupyter
|
# Siamese networks with TensorFlow 2.0/Keras
In this example, we'll implement a simple siamese network system, which verifyies whether a pair of MNIST images is of the same class (true) or not (false).
_This example is partially based on_ [https://github.com/keras-team/keras/blob/master/examples/mnist_siamese.py](https://github.com/keras-team/keras/blob/master/examples/mnist_siamese.py)
Let's start with the imports
```
import random
import numpy as np
import tensorflow as tf
```
We'll continue with the `create_pairs` function, which creates a training dataset of equal number of true/false pairs of each MNIST class.
```
def create_pairs(inputs: np.ndarray, labels: np.ndarray):
"""Create equal number of true/false pairs of samples"""
num_classes = 10
digit_indices = [np.where(labels == i)[0] for i in range(num_classes)]
pairs = list()
labels = list()
n = min([len(digit_indices[d]) for d in range(num_classes)]) - 1
for d in range(num_classes):
for i in range(n):
z1, z2 = digit_indices[d][i], digit_indices[d][i + 1]
pairs += [[inputs[z1], inputs[z2]]]
inc = random.randrange(1, num_classes)
dn = (d + inc) % num_classes
z1, z2 = digit_indices[d][i], digit_indices[dn][i]
pairs += [[inputs[z1], inputs[z2]]]
labels += [1, 0]
return np.array(pairs), np.array(labels, dtype=np.float32)
```
Next, we'll define the base network of the siamese system:
```
def create_base_network():
"""The shared encoding part of the siamese network"""
return tf.keras.models.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.1),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.1),
tf.keras.layers.Dense(64, activation='relu'),
])
```
Next, let's load the regular MNIST training and validation sets and create true/false pairs out of them:
```
# Load the train and test MNIST datasets
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
x_train = x_train.astype(np.float32)
x_test = x_test.astype(np.float32)
x_train /= 255
x_test /= 255
input_shape = x_train.shape[1:]
# Create true/false training and testing pairs
train_pairs, tr_labels = create_pairs(x_train, y_train)
test_pairs, test_labels = create_pairs(x_test, y_test)
```
Then, we'll build the siamese system, which includes the `base_network`, the 2 siamese paths `encoder_a` and `encoder_b`, the `l1_dist` measure, and the combined `model`:
```
# Create the siamese network
# Start from the shared layers
base_network = create_base_network()
# Create first half of the siamese system
input_a = tf.keras.layers.Input(shape=input_shape)
# Note how we reuse the base_network in both halfs
encoder_a = base_network(input_a)
# Create the second half of the siamese system
input_b = tf.keras.layers.Input(shape=input_shape)
encoder_b = base_network(input_b)
# Create the the distance measure
l1_dist = tf.keras.layers.Lambda(
lambda embeddings: tf.keras.backend.abs(embeddings[0] - embeddings[1])) \
([encoder_a, encoder_b])
# Final fc layer with a single logistic output for the binary classification
flattened_weighted_distance = tf.keras.layers.Dense(1, activation='sigmoid') \
(l1_dist)
# Build the model
model = tf.keras.models.Model([input_a, input_b], flattened_weighted_distance)
```
Finally, we can train the model and check the validation accuracy, which reaches 99.37%:
```
# Train
model.compile(loss='binary_crossentropy',
optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy'])
model.fit([train_pairs[:, 0], train_pairs[:, 1]], tr_labels,
batch_size=128,
epochs=20,
validation_data=([test_pairs[:, 0], test_pairs[:, 1]], test_labels))
```
|
github_jupyter
|
# BBoxerwGradCAM
### This class forms boundary boxes (rectangle and polygon) using GradCAM outputs for a given image.
The purpose of this class is to develop Rectangle and Polygon coordinates that define an object based on an image classification model. The 'automatic' creation of these coordinates, which are often included in COCO JSONs used to train object detection models, is valuable because data preparation and labeling can be a time consuming task.
### This class takes 5 user inputs:
* **Pretrained Learner** (image classification model)
* **GradCAM Heatmap** (heatmap of GradCAM object - formed by a pretrained image classification learner)
* **Source Image**
* **Image Resizing Scale** (also applied to corresponding GradCAM heatmap)
* **BBOX Rectangle Resizing Scale**
*Class is compatible with google colab and other Python 3 enivronments*
```
# Imports for loading learner and the GradCAM class
from fastai import *
from fastai.vision import *
from fastai.callbacks.hooks import *
import scipy.ndimage
```
The following cell contains the widely used GradCAM class for pretrained image classification models (unedited).
```
#@title GradCAM Class
class GradCam():
@classmethod
def from_interp(cls,learn,interp,img_idx,ds_type=DatasetType.Valid,include_label=False):
# produce heatmap and xb_grad for pred label (and actual label if include_label is True)
if ds_type == DatasetType.Valid:
ds = interp.data.valid_ds
elif ds_type == DatasetType.Test:
ds = interp.data.test_ds
include_label=False
else:
return None
x_img = ds.x[img_idx]
xb,_ = interp.data.one_item(x_img)
xb_img = Image(interp.data.denorm(xb)[0])
probs = interp.preds[img_idx].numpy()
pred_idx = interp.pred_class[img_idx].item() # get class idx of img prediction label
hmap_pred,xb_grad_pred = get_grad_heatmap(learn,xb,pred_idx,size=xb_img.shape[-1])
prob_pred = probs[pred_idx]
actual_args=None
if include_label:
actual_idx = ds.y.items[img_idx] # get class idx of img actual label
if actual_idx!=pred_idx:
hmap_actual,xb_grad_actual = get_grad_heatmap(learn,xb,actual_idx,size=xb_img.shape[-1])
prob_actual = probs[actual_idx]
actual_args=[interp.data.classes[actual_idx],prob_actual,hmap_actual,xb_grad_actual]
return cls(xb_img,interp.data.classes[pred_idx],prob_pred,hmap_pred,xb_grad_pred,actual_args)
@classmethod
def from_one_img(cls,learn,x_img,label1=None,label2=None):
'''
learn: fastai's Learner
x_img: fastai.vision.image.Image
label1: generate heatmap according to this label. If None, this wil be the label with highest probability from the model
label2: generate additional heatmap according to this label
'''
pred_class,pred_idx,probs = learn.predict(x_img)
label1= str(pred_class) if not label1 else label1
xb,_ = learn.data.one_item(x_img)
xb_img = Image(learn.data.denorm(xb)[0])
probs = probs.numpy()
label1_idx = learn.data.classes.index(label1)
hmap1,xb_grad1 = get_grad_heatmap(learn,xb,label1_idx,size=xb_img.shape[-1])
prob1 = probs[label1_idx]
label2_args = None
if label2:
label2_idx = learn.data.classes.index(label2)
hmap2,xb_grad2 = get_grad_heatmap(learn,xb,label2_idx,size=xb_img.shape[-1])
prob2 = probs[label2_idx]
label2_args = [label2,prob2,hmap2,xb_grad2]
return cls(xb_img,label1,prob1,hmap1,xb_grad1,label2_args)
def __init__(self,xb_img,label1,prob1,hmap1,xb_grad1,label2_args=None):
self.xb_img=xb_img
self.label1,self.prob1,self.hmap1,self.xb_grad1 = label1,prob1,hmap1,xb_grad1
if label2_args:
self.label2,self.prob2,self.hmap2,self.xb_grad2 = label2_args
def plot(self,plot_hm=True,plot_gbp=True):
if not plot_hm and not plot_gbp:
plot_hm=True
cols = 5 if hasattr(self, 'label2') else 3
if not plot_gbp or not plot_hm:
cols-= 2 if hasattr(self, 'label2') else 1
fig,row_axes = plt.subplots(1,cols,figsize=(cols*5,5))
col=0
size=self.xb_img.shape[-1]
self.xb_img.show(row_axes[col]);col+=1
label1_title = f'1.{self.label1} {self.prob1:.3f}'
if plot_hm:
show_heatmap(self.hmap1,self.xb_img,size,row_axes[col])
row_axes[col].set_title(label1_title);col+=1
if plot_gbp:
row_axes[col].imshow(self.xb_grad1)
row_axes[col].set_axis_off()
row_axes[col].set_title(label1_title);col+=1
if hasattr(self, 'label2'):
label2_title = f'2.{self.label2} {self.prob2:.3f}'
if plot_hm:
show_heatmap(self.hmap2,self.xb_img,size,row_axes[col])
row_axes[col].set_title(label2_title);col+=1
if plot_gbp:
row_axes[col].imshow(self.xb_grad2)
row_axes[col].set_axis_off()
row_axes[col].set_title(label2_title)
# plt.tight_layout()
fig.subplots_adjust(wspace=0, hspace=0)
# fig.savefig('data_draw/both/gradcam.png')
def minmax_norm(x):
return (x - np.min(x))/(np.max(x) - np.min(x))
def scaleup(x,size):
scale_mult=size/x.shape[0]
upsampled = scipy.ndimage.zoom(x, scale_mult)
return upsampled
# hook for Gradcam
def hooked_backward(m,xb,target_layer,clas):
with hook_output(target_layer) as hook_a: #hook at last layer of group 0's output (after bn, size 512x7x7 if resnet34)
with hook_output(target_layer, grad=True) as hook_g: # gradient w.r.t to the target_layer
preds = m(xb)
preds[0,int(clas)].backward() # same as onehot backprop
return hook_a,hook_g
def clamp_gradients_hook(module, grad_in, grad_out):
for grad in grad_in:
torch.clamp_(grad, min=0.0)
# hook for guided backprop
def hooked_ReLU(m,xb,clas):
relu_modules = [module[1] for module in m.named_modules() if str(module[1]) == "ReLU(inplace)"]
with callbacks.Hooks(relu_modules, clamp_gradients_hook, is_forward=False) as _:
preds = m(xb)
preds[0,int(clas)].backward()
def guided_backprop(learn,xb,y):
xb = xb.cuda()
m = learn.model.eval();
xb.requires_grad_();
if not xb.grad is None:
xb.grad.zero_();
hooked_ReLU(m,xb,y);
return xb.grad[0].cpu().numpy()
def show_heatmap(hm,xb_im,size,ax=None):
if ax is None:
_,ax = plt.subplots()
xb_im.show(ax)
ax.imshow(hm, alpha=0.8, extent=(0,size,size,0),
interpolation='bilinear',cmap='magma');
def get_grad_heatmap(learn,xb,y,size):
'''
Main function to get hmap for heatmap and xb_grad for guided backprop
'''
xb = xb.cuda()
m = learn.model.eval();
target_layer = m[0][-1][-1] # last layer of group 0
hook_a,hook_g = hooked_backward(m,xb,target_layer,y)
target_act= hook_a.stored[0].cpu().numpy()
target_grad = hook_g.stored[0][0].cpu().numpy()
mean_grad = target_grad.mean(1).mean(1)
# hmap = (target_act*mean_grad[...,None,None]).mean(0)
hmap = (target_act*mean_grad[...,None,None]).sum(0)
hmap = np.where(hmap >= 0, hmap, 0)
xb_grad = guided_backprop(learn,xb,y) # (3,224,224)
#minmax norm the grad
xb_grad = minmax_norm(xb_grad)
hmap_scaleup = minmax_norm(scaleup(hmap,size)) # (224,224)
# multiply xb_grad and hmap_scaleup and switch axis
xb_grad = np.einsum('ijk, jk->jki',xb_grad, hmap_scaleup) #(224,224,3)
return hmap,xb_grad
```
I connect to google drive (this notebook was made on google colab for GPU usage) and load my pretrained learner.
```
from google.colab import drive
drive.mount('/content/drive')
base_dir = '/content/drive/My Drive/fellowshipai-data/final_3_class_data_train_test_split'
def get_data(sz): # This function returns an ImageDataBunch with a given image size
return ImageDataBunch.from_folder(base_dir+'/', train='train', valid='valid', # 0% validation because we already formed our testing set
ds_tfms=get_transforms(), size=sz, num_workers=4).normalize(imagenet_stats) # Normalized, 4 workers (multiprocessing) - 64 batch size (default)
arch = models.resnet34
data = get_data(224)
learn = cnn_learner(data,arch,metrics=[error_rate,Precision(average='micro'),Recall(average='micro')],train_bn=True,pretrained=True).mixup()
learn.load('model-224sz-basicaugments-oversampling-mixup-dLRs')
example_image = '/content/drive/My Drive/fellowshipai-data/final_3_class_data_train_test_split/train/raw/00000015.jpg'
img = open_image(example_image)
gcam = GradCam.from_one_img(learn,img) # using the GradCAM class
gcam.plot(plot_gbp = False) # We care about the heatmap (which is overlayed on top of the original image inherently)
gcam_heatmap = gcam.hmap1 # This is a 2d array
```
My pretrained learner correctly classified the image as raw with probability 0.996.
Note that images with very low noise and accurate feature importances (as with the example image) are
The learner is focusing on the steak in center view (heatmap pixels indicate feature importance).
```
from BBOXES_from_GRADCAM import BBoxerwGradCAM # load class from .py file
image_resizing_scale = [400,300]
bbox_scaling = [1,1,1,1]
bbox = BBoxerwGradCAM(learn,
gcam_heatmap,
example_image,
image_resizing_scale,
bbox_scaling)
for function in dir(bbox)[-18:]: print(function)
bbox.show_smoothheatmap()
bbox.show_contouredheatmap()
#bbox.show_bboxrectangle()
bbox.show_bboxpolygon()
bbox.show_bboxrectangle()
rect_coords, polygon_coords = bbox.get_bboxes()
rect_coords # x,y,w,h
polygon_coords
# IoU for object detection
def get_IoU(truth_coords, pred_coords):
pred_area = pred_coords[2]*pred_coords[3]
truth_area = truth_coords[2]*truth_coords[3]
# coords of intersection rectangle
x1 = max(truth_coords[0], pred_coords[0])
y1 = max(truth_coords[1], pred_coords[1])
x2 = min(truth_coords[2], pred_coords[2])
y2 = min(truth_coords[3], pred_coords[3])
# area of intersection rectangle
interArea = max(0, x2 - x1 + 1) * max(0, y2 - y1 + 1)
# area of prediction and truth rectangles
boxTruthArea = (truth_coords[2] - truth_coords[0] + 1) * (truth_coords[3] - truth_coords[1] + 1)
boxPredArea = (pred_coords[2] - pred_coords[0] + 1) * (pred_coords[3] - pred_coords[1] + 1)
# intersection over union
iou = interArea / float(boxTruthArea + boxPredArea - interArea)
return iou
get_IoU([80,40,240,180],rect_coords)
```
|
github_jupyter
|
# Hierarchical Clustering
**Hierarchical clustering** refers to a class of clustering methods that seek to build a **hierarchy** of clusters, in which some clusters contain others. In this assignment, we will explore a top-down approach, recursively bipartitioning the data using k-means.
**Note to Amazon EC2 users**: To conserve memory, make sure to stop all the other notebooks before running this notebook.
## Import packages
```
from __future__ import print_function # to conform python 2.x print to python 3.x
import turicreate
import matplotlib.pyplot as plt
import numpy as np
import sys
import os
import time
from scipy.sparse import csr_matrix
from sklearn.cluster import KMeans
from sklearn.metrics import pairwise_distances
%matplotlib inline
```
## Load the Wikipedia dataset
```
wiki = turicreate.SFrame('people_wiki.sframe/')
```
As we did in previous assignments, let's extract the TF-IDF features:
```
wiki['tf_idf'] = turicreate.text_analytics.tf_idf(wiki['text'])
```
To run k-means on this dataset, we should convert the data matrix into a sparse matrix.
```
from em_utilities import sframe_to_scipy # converter
# This will take about a minute or two.
wiki = wiki.add_row_number()
tf_idf, map_word_to_index = sframe_to_scipy(wiki, 'tf_idf')
```
To be consistent with the k-means assignment, let's normalize all vectors to have unit norm.
```
from sklearn.preprocessing import normalize
tf_idf = normalize(tf_idf)
```
## Bipartition the Wikipedia dataset using k-means
Recall our workflow for clustering text data with k-means:
1. Load the dataframe containing a dataset, such as the Wikipedia text dataset.
2. Extract the data matrix from the dataframe.
3. Run k-means on the data matrix with some value of k.
4. Visualize the clustering results using the centroids, cluster assignments, and the original dataframe. We keep the original dataframe around because the data matrix does not keep auxiliary information (in the case of the text dataset, the title of each article).
Let us modify the workflow to perform bipartitioning:
1. Load the dataframe containing a dataset, such as the Wikipedia text dataset.
2. Extract the data matrix from the dataframe.
3. Run k-means on the data matrix with k=2.
4. Divide the data matrix into two parts using the cluster assignments.
5. Divide the dataframe into two parts, again using the cluster assignments. This step is necessary to allow for visualization.
6. Visualize the bipartition of data.
We'd like to be able to repeat Steps 3-6 multiple times to produce a **hierarchy** of clusters such as the following:
```
(root)
|
+------------+-------------+
| |
Cluster Cluster
+------+-----+ +------+-----+
| | | |
Cluster Cluster Cluster Cluster
```
Each **parent cluster** is bipartitioned to produce two **child clusters**. At the very top is the **root cluster**, which consists of the entire dataset.
Now we write a wrapper function to bipartition a given cluster using k-means. There are three variables that together comprise the cluster:
* `dataframe`: a subset of the original dataframe that correspond to member rows of the cluster
* `matrix`: same set of rows, stored in sparse matrix format
* `centroid`: the centroid of the cluster (not applicable for the root cluster)
Rather than passing around the three variables separately, we package them into a Python dictionary. The wrapper function takes a single dictionary (representing a parent cluster) and returns two dictionaries (representing the child clusters).
```
def bipartition(cluster, maxiter=400, num_runs=4, seed=None):
'''cluster: should be a dictionary containing the following keys
* dataframe: original dataframe
* matrix: same data, in matrix format
* centroid: centroid for this particular cluster'''
data_matrix = cluster['matrix']
dataframe = cluster['dataframe']
# Run k-means on the data matrix with k=2. We use scikit-learn here to simplify workflow.
kmeans_model = KMeans(n_clusters=2, max_iter=maxiter, n_init=num_runs, random_state=seed, n_jobs=1)
kmeans_model.fit(data_matrix)
centroids, cluster_assignment = kmeans_model.cluster_centers_, kmeans_model.labels_
# Divide the data matrix into two parts using the cluster assignments.
data_matrix_left_child, data_matrix_right_child = data_matrix[cluster_assignment==0], \
data_matrix[cluster_assignment==1]
# Divide the dataframe into two parts, again using the cluster assignments.
cluster_assignment_sa = turicreate.SArray(cluster_assignment) # minor format conversion
dataframe_left_child, dataframe_right_child = dataframe[cluster_assignment_sa==0], \
dataframe[cluster_assignment_sa==1]
# Package relevant variables for the child clusters
cluster_left_child = {'matrix': data_matrix_left_child,
'dataframe': dataframe_left_child,
'centroid': centroids[0]}
cluster_right_child = {'matrix': data_matrix_right_child,
'dataframe': dataframe_right_child,
'centroid': centroids[1]}
return (cluster_left_child, cluster_right_child)
```
The following cell performs bipartitioning of the Wikipedia dataset. Allow 2+ minutes to finish.
Note. For the purpose of the assignment, we set an explicit seed (`seed=1`) to produce identical outputs for every run. In pratical applications, you might want to use different random seeds for all runs.
```
%%time
wiki_data = {'matrix': tf_idf, 'dataframe': wiki} # no 'centroid' for the root cluster
left_child, right_child = bipartition(wiki_data, maxiter=100, num_runs=1, seed=0)
```
Let's examine the contents of one of the two clusters, which we call the `left_child`, referring to the tree visualization above.
```
left_child
```
And here is the content of the other cluster we named `right_child`.
```
right_child
```
## Visualize the bipartition
We provide you with a modified version of the visualization function from the k-means assignment. For each cluster, we print the top 5 words with highest TF-IDF weights in the centroid and display excerpts for the 8 nearest neighbors of the centroid.
```
def display_single_tf_idf_cluster(cluster, map_index_to_word):
'''map_index_to_word: SFrame specifying the mapping betweeen words and column indices'''
wiki_subset = cluster['dataframe']
tf_idf_subset = cluster['matrix']
centroid = cluster['centroid']
# Print top 5 words with largest TF-IDF weights in the cluster
idx = centroid.argsort()[::-1]
for i in range(5):
print('{0}:{1:.3f}'.format(map_index_to_word['category'], centroid[idx[i]])),
print('')
# Compute distances from the centroid to all data points in the cluster.
distances = pairwise_distances(tf_idf_subset, [centroid], metric='euclidean').flatten()
# compute nearest neighbors of the centroid within the cluster.
nearest_neighbors = distances.argsort()
# For 8 nearest neighbors, print the title as well as first 180 characters of text.
# Wrap the text at 80-character mark.
for i in range(8):
text = ' '.join(wiki_subset[nearest_neighbors[i]]['text'].split(None, 25)[0:25])
print('* {0:50s} {1:.5f}\n {2:s}\n {3:s}'.format(wiki_subset[nearest_neighbors[i]]['name'],
distances[nearest_neighbors[i]], text[:90], text[90:180] if len(text) > 90 else ''))
print('')
```
Let's visualize the two child clusters:
```
display_single_tf_idf_cluster(left_child, map_word_to_index)
display_single_tf_idf_cluster(right_child, map_word_to_index)
```
The right cluster consists of athletes and artists (singers and actors/actresses), whereas the left cluster consists of non-athletes and non-artists. So far, we have a single-level hierarchy consisting of two clusters, as follows:
```
Wikipedia
+
|
+--------------------------+--------------------+
| |
+ +
Non-athletes/artists Athletes/artists
```
Is this hierarchy good enough? **When building a hierarchy of clusters, we must keep our particular application in mind.** For instance, we might want to build a **directory** for Wikipedia articles. A good directory would let you quickly narrow down your search to a small set of related articles. The categories of athletes and non-athletes are too general to facilitate efficient search. For this reason, we decide to build another level into our hierarchy of clusters with the goal of getting more specific cluster structure at the lower level. To that end, we subdivide both the `athletes/artists` and `non-athletes/artists` clusters.
## Perform recursive bipartitioning
### Cluster of athletes and artists
To help identify the clusters we've built so far, let's give them easy-to-read aliases:
```
non_athletes_artists = left_child
athletes_artists = right_child
```
Using the bipartition function, we produce two child clusters of the athlete cluster:
```
# Bipartition the cluster of athletes and artists
left_child_athletes_artists, right_child_athletes_artists = bipartition(athletes_artists, maxiter=100, num_runs=6, seed=1)
```
The left child cluster mainly consists of athletes:
```
display_single_tf_idf_cluster(left_child_athletes_artists, map_word_to_index)
```
On the other hand, the right child cluster consists mainly of artists (singers and actors/actresses):
```
display_single_tf_idf_cluster(right_child_athletes_artists, map_word_to_index)
```
Our hierarchy of clusters now looks like this:
```
Wikipedia
+
|
+--------------------------+--------------------+
| |
+ +
Non-athletes/artists Athletes/artists
+
|
+----------+----------+
| |
| |
+ |
athletes artists
```
Should we keep subdividing the clusters? If so, which cluster should we subdivide? To answer this question, we again think about our application. Since we organize our directory by topics, it would be nice to have topics that are about as coarse as each other. For instance, if one cluster is about baseball, we expect some other clusters about football, basketball, volleyball, and so forth. That is, **we would like to achieve similar level of granularity for all clusters.**
Both the athletes and artists node can be subdivided more, as each one can be divided into more descriptive professions (singer/actress/painter/director, or baseball/football/basketball, etc.). Let's explore subdividing the athletes cluster further to produce finer child clusters.
Let's give the clusters aliases as well:
```
athletes = left_child_athletes_artists
artists = right_child_athletes_artists
```
### Cluster of athletes
In answering the following quiz question, take a look at the topics represented in the top documents (those closest to the centroid), as well as the list of words with highest TF-IDF weights.
Let us bipartition the cluster of athletes.
```
left_child_athletes, right_child_athletes = bipartition(athletes, maxiter=100, num_runs=6, seed=1)
display_single_tf_idf_cluster(left_child_athletes, map_word_to_index)
display_single_tf_idf_cluster(right_child_athletes, map_word_to_index)
```
**Quiz Question**. Which diagram best describes the hierarchy right after splitting the `athletes` cluster? Refer to the quiz form for the diagrams.
**Caution**. The granularity criteria is an imperfect heuristic and must be taken with a grain of salt. It takes a lot of manual intervention to obtain a good hierarchy of clusters.
* **If a cluster is highly mixed, the top articles and words may not convey the full picture of the cluster.** Thus, we may be misled if we judge the purity of clusters solely by their top documents and words.
* **Many interesting topics are hidden somewhere inside the clusters but do not appear in the visualization.** We may need to subdivide further to discover new topics. For instance, subdividing the `ice_hockey_football` cluster led to the appearance of runners and golfers.
### Cluster of non-athletes
Now let us subdivide the cluster of non-athletes.
```
%%time
# Bipartition the cluster of non-athletes
left_child_non_athletes_artists, right_child_non_athletes_artists = bipartition(non_athletes_artists, maxiter=100, num_runs=3, seed=1)
display_single_tf_idf_cluster(left_child_non_athletes_artists, map_word_to_index)
display_single_tf_idf_cluster(right_child_non_athletes_artists, map_word_to_index)
```
The clusters are not as clear, but the left cluster has a tendency to show important female figures, and the right one to show politicians and government officials.
Let's divide them further.
```
female_figures = left_child_non_athletes_artists
politicians_etc = right_child_non_athletes_artists
politicians_etc = left_child_non_athletes_artists
female_figures = right_child_non_athletes_artists
```
**Quiz Question**. Let us bipartition the clusters `female_figures` and `politicians`. Which diagram best describes the resulting hierarchy of clusters for the non-athletes? Refer to the quiz for the diagrams.
**Note**. Use `maxiter=100, num_runs=6, seed=1` for consistency of output.
```
left_female_figures, right_female_figures = bipartition(female_figures, maxiter=100, num_runs=6, seed=1)
left_politicians_etc, right_politicians_etc = bipartition(politicians_etc, maxiter=100, num_runs=6, seed=1)
display_single_tf_idf_cluster(left_female_figures, map_word_to_index)
display_single_tf_idf_cluster(right_female_figures, map_word_to_index)
display_single_tf_idf_cluster(left_politicians_etc, map_word_to_index)
display_single_tf_idf_cluster(right_politicians_etc, map_word_to_index)
```
|
github_jupyter
|
# Object Detection with SSD
### Here we demostrate detection on example images using SSD with PyTorch
```
import os
import sys
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path)
import torch
import torch.nn as nn
import torch.backends.cudnn as cudnn
from torch.autograd import Variable
import numpy as np
import cv2
if torch.cuda.is_available():
torch.set_default_tensor_type('torch.cuda.FloatTensor')
from ssd import build_ssd
```
## Build SSD300 in Test Phase
1. Build the architecture, specifyingsize of the input image (300),
and number of object classes to score (21 for VOC dataset)
2. Next we load pretrained weights on the VOC0712 trainval dataset
```
net = build_ssd('test', 300, 21) # initialize SSD
net.load_weights('../weights/ssd300_VOC_28000.pth')
```
## Load Image
### Here we just load a sample image from the VOC07 dataset
```
# image = cv2.imread('./data/example.jpg', cv2.IMREAD_COLOR) # uncomment if dataset not downloaded
%matplotlib inline
from matplotlib import pyplot as plt
from data import VOCDetection, VOC_ROOT, VOCAnnotationTransform
# here we specify year (07 or 12) and dataset ('test', 'val', 'train')
testset = VOCDetection(VOC_ROOT, [('2007', 'val')], None, VOCAnnotationTransform())
img_id = 60
image = testset.pull_image(img_id)
rgb_image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# View the sampled input image before transform
plt.figure(figsize=(10,10))
plt.imshow(rgb_image)
plt.show()
```
## Pre-process the input.
#### Using the torchvision package, we can create a Compose of multiple built-in transorm ops to apply
For SSD, at test time we use a custom BaseTransform callable to
resize our image to 300x300, subtract the dataset's mean rgb values,
and swap the color channels for input to SSD300.
```
x = cv2.resize(image, (300, 300)).astype(np.float32)
x -= (104.0, 117.0, 123.0)
x = x.astype(np.float32)
x = x[:, :, ::-1].copy()
plt.imshow(x)
x = torch.from_numpy(x).permute(2, 0, 1)
```
## SSD Forward Pass
### Now just wrap the image in a Variable so it is recognized by PyTorch autograd
```
xx = Variable(x.unsqueeze(0)) # wrap tensor in Variable
if torch.cuda.is_available():
xx = xx.cuda()
y = net(xx)
```
## Parse the Detections and View Results
Filter outputs with confidence scores lower than a threshold
Here we choose 60%
```
from data import VOC_CLASSES as labels
top_k=10
plt.figure(figsize=(10,10))
colors = plt.cm.hsv(np.linspace(0, 1, 21)).tolist()
plt.imshow(rgb_image) # plot the image for matplotlib
currentAxis = plt.gca()
detections = y.data
# scale each detection back up to the image
scale = torch.Tensor(rgb_image.shape[1::-1]).repeat(2)
for i in range(detections.size(1)):
j = 0
while detections[0,i,j,0] >= 0.6:
score = detections[0,i,j,0]
label_name = labels[i-1]
display_txt = '%s: %.2f'%(label_name, score)
pt = (detections[0,i,j,1:]*scale).cpu().numpy()
coords = (pt[0], pt[1]), pt[2]-pt[0]+1, pt[3]-pt[1]+1
color = colors[i]
currentAxis.add_patch(plt.Rectangle(*coords, fill=False, edgecolor=color, linewidth=2))
currentAxis.text(pt[0], pt[1], display_txt, bbox={'facecolor':color, 'alpha':0.5})
j+=1
```
|
github_jupyter
|
# Data Attribute Recommendation - TechED 2020 INT260
Getting started with the Python SDK for the Data Attribute Recommendation service.
## Business Scenario
We will consider a business scenario involving product master data. The creation and maintenance of this product master data requires the careful manual selection of the correct categories for a given product from a pre-defined hierarchy of product categories.
In this workshop, we will explore how to automate this tedious manual task with the Data Attribute Recommendation service.
<video controls src="videos/dar_prediction_material_table.mp4"/>
This workshop will cover:
* Data Upload
* Model Training and Deployment
* Inference Requests
We will work through a basic example of how to achieve these tasks using the [Python SDK for Data Attribute Recommendation](https://github.com/SAP/data-attribute-recommendation-python-sdk).
*Note: if you are doing several runs of this notebook on a trial account, you may see errors stating 'The resource can no longer be used. Usage limit has been reached'. It can be beneficial to [clean up the service instance](#Cleaning-up-a-service-instance) to free up limited trial resources acquired by an earlier run of the notebook. [Some limits](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html) cannot be reset this way.*
## Table of Contents
* [Exercise 01.1](#Exercise-01.1) - Installing the SDK and preparing the service key
* [Creating a service instance and key on BTP Trial](#Creating-a-service-instance-and-key)
* [Installing the SDK](#Installing-the-SDK)
* [Loading the service key into your Jupyter Notebook](#Loading-the-service-key-into-your-Jupyter-Notebook)
* [Exercise 01.2](#Exercise-01.2) - Uploading the data
* [Exercise 01.3](#Exercise-01.3) - Training the model
* [Exercise 01.4](#Exercise-01.4) - Deploying the Model and predicting labels
* [Resources](#Resources) - Additional reading
* [Cleaning up a service instance](#Cleaning-up-a-service-instance) - Clean up all resources on the service instance
* [Optional Exercises](#Optional-Exercises) - Optional exercises
## Requirements
See the [README in the Github repository for this workshop](https://github.com/SAP-samples/teched2020-INT260/blob/master/exercises/ex1-DAR/README.md).
# Exercise 01.1
*Back to [table of contents](#Table-of-Contents)*
In exercise 01.1, we will install the SDK and prepare the service key.
## Creating a service instance and key on BTP Trial
Please log in to your trial account: https://cockpit.eu10.hana.ondemand.com/trial/
In the your global account screen, go to the "Boosters" tab:

*Boosters are only available on the Trial landscape. If you are using a production environment, please follow this tutorial to manually [create a service instance and a service key](https://developers.sap.com/tutorials/cp-aibus-dar-service-instance.html)*.
In the Boosters tab, enter "Data Attribute Recommendation" into the search box. Then, select the
service tile from the search results:

The resulting screen shows details of the booster pack. Here, click the "Start" button and wait a few seconds.

Once the booster is finished, click the "go to Service Key" link to obtain your service key.

Finally, download the key and save it to disk.

## Installing the SDK
The Data Attribute Recommendation SDK is available from the Python package repository. It can be installed with the standard `pip` tool:
```
! pip install data-attribute-recommendation-sdk
```
*Note: If you are not using a Jupyter notebook, but instead a regular Python development environment, we recommend using a Python virtual environment to set up your development environment. Please see [the dedicated tutorial to learn how to install the SDK inside a Python virtual environment](https://developers.sap.com/tutorials/cp-aibus-dar-sdk-setup.html).*
## Loading the service key into your Jupyter Notebook
Once you downloaded the service key from the Cockpit, upload it to your notebook environment. The service key must be uploaded to same directory where the `teched2020-INT260_Data_Attribute_Recommendation.ipynb` is stored.
We first navigate to the file browser in Jupyter. On the top of your Jupyter notebook, right-click on the Jupyter logo and open in a new tab.

**In the file browser, navigate to the directory where the `teched2020-INT260_Data_Attribute_Recommendation.ipynb` notebook file is stored. The service key must reside next to this file.**
In the Jupyter file browser, click the **Upload** button (1). In the file selection dialog that opens, select the `defaultKey_*.json` file you downloaded previously from the SAP Cloud Platform Cockpit. Rename the file to `key.json`.
Confirm the upload by clicking on the second **Upload** button (2).

The service key contains your credentials to access the service. Please treat this as carefully as you would treat any password. We keep the service key as a separate file outside this notebook to avoid leaking the secret credentials.
The service key is a JSON file. We will load this file once and use the credentials throughout this workshop.
```
# First, set up logging so we can see the actions performed by the SDK behind the scenes
import logging
import sys
logging.basicConfig(level=logging.INFO, stream=sys.stdout)
from pprint import pprint # for nicer output formatting
import json
import os
if not os.path.exists("key.json"):
msg = "key.json is not found. Please follow instructions above to create a service key of"
msg += " Data Attribute Recommendation. Then, upload it into the same directory where"
msg += " this notebook is saved."
print(msg)
raise ValueError(msg)
with open("key.json") as file_handle:
key = file_handle.read()
SERVICE_KEY = json.loads(key)
```
## Summary Exercise 01.1
In exercise 01.1, we have covered the following topics:
* How to install the Python SDK for Data Attribute Recommendation
* How to obtain a service key for the Data Attribute Recommendation service
# Exercise 01.2
*Back to [table of contents](#Table-of-Contents)*
*To perform this exercise, you need to execute the code in all previous exercises.*
In exercise 01.2, we will upload our demo dataset to the service.
## The Dataset
### Obtaining the Data
The dataset we use in this workshop is a CSV file containing product master data. The original data was released by BestBuy, a retail company, under an [open license](https://github.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample#data-and-license). This makes it ideal for first experiments with the Data Attribute Recommendation service.
The dataset can be downloaded directly from Github using the following command:
```
! wget -O bestBuy.csv "https://raw.githubusercontent.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample/master/Tutorial_Example_Dataset.csv"
# If you receive a "command not found" error (i.e. on Windows), try curl instead of wget:
# ! curl -o bestBuy.csv "https://raw.githubusercontent.com/SAP-samples/data-attribute-recommendation-postman-tutorial-sample/master/Tutorial_Example_Dataset.csv"
```
Let's inspect the data:
```
# if you are experiencing an import error here, run the following in a new cell:
# ! pip install pandas
import pandas as pd
df = pd.read_csv("bestBuy.csv")
df.head(5)
print()
print(f"Data has {df.shape[0]} rows and {df.shape[1]} columns.")
```
The CSV contains the several products. For each product, the description, the manufacturer and the price are given. Additionally, three levels of the products hierarchy are given.
The first product, a set of AAA batteries, is located in the following place in the product hierarchy:
```
level1_category: Connected Home & Housewares
|
level2_category: Housewares
|
level3_category: Household Batteries
```
We will use the Data Attribute Recommendation service to predict the categories for a given product based on its **description**, **manufacturer** and **price**.
### Creating the DatasetSchema
We first have to describe the shape of our data by creating a DatasetSchema. This schema informs the service about the individual column types found in the CSV. We also describe which are the target columns used for training. These columns will be later predicted. In our case, these are the three category columns.
The service currently supports three column types: **text**, **category** and **number**. For prediction, only **category** is currently supported.
A DatasetSchema for the BestBuy dataset looks as follows:
```json
{
"features": [
{"label": "manufacturer", "type": "CATEGORY"},
{"label": "description", "type": "TEXT"},
{"label": "price", "type": "NUMBER"}
],
"labels": [
{"label": "level1_category", "type": "CATEGORY"},
{"label": "level2_category", "type": "CATEGORY"},
{"label": "level3_category", "type": "CATEGORY"}
],
"name": "bestbuy-category-prediction",
}
```
We will now upload this DatasetSchema to the Data Attribute Recommendation service. The SDK provides the
[`DataManagerClient.create_dataset_schema()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.create_dataset_schema) method for this purpose.
```
from sap.aibus.dar.client.data_manager_client import DataManagerClient
dataset_schema = {
"features": [
{"label": "manufacturer", "type": "CATEGORY"},
{"label": "description", "type": "TEXT"},
{"label": "price", "type": "NUMBER"}
],
"labels": [
{"label": "level1_category", "type": "CATEGORY"},
{"label": "level2_category", "type": "CATEGORY"},
{"label": "level3_category", "type": "CATEGORY"}
],
"name": "bestbuy-category-prediction",
}
data_manager = DataManagerClient.construct_from_service_key(SERVICE_KEY)
response = data_manager.create_dataset_schema(dataset_schema)
dataset_schema_id = response["id"]
print()
print("DatasetSchema created:")
pprint(response)
print()
print(f"DatasetSchema ID: {dataset_schema_id}")
```
The API responds with the newly created DatasetSchema resource. The service assigned an ID to the schema. We save this ID in a variable, as we will need it when we upload the data.
### Uploading the Data to the service
The [`DataManagerClient`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient) class is also responsible for uploading data to the service. This data must fit to an existing DatasetSchema. After uploading the data, the service will validate the Dataset against the DataSetSchema in a background process. The data must be a CSV file which can optionally be `gzip` compressed.
We will now upload our `bestBuy.csv` file, using the DatasetSchema which we created earlier.
Data upload is a two-step process. We first create the Dataset using [`DataManagerClient.create_dataset()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.create_dataset). Then we can upload data to the Dataset using the [`DataManagerClient.upload_data_to_dataset()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.upload_data_to_dataset) method.
```
dataset_resource = data_manager.create_dataset("my-bestbuy-dataset", dataset_schema_id)
dataset_id = dataset_resource["id"]
print()
print("Dataset created:")
pprint(dataset_resource)
print()
print(f"Dataset ID: {dataset_id}")
# Compress file first for a faster upload
! gzip -9 -c bestBuy.csv > bestBuy.csv.gz
```
Note that the data upload can take a few minutes. Please do not restart the process while the cell is still running.
```
# Open in binary mode.
with open('bestBuy.csv.gz', 'rb') as file_handle:
dataset_resource = data_manager.upload_data_to_dataset(dataset_id, file_handle)
print()
print("Dataset after data upload:")
print()
pprint(dataset_resource)
```
Note that the Dataset status changed from `NO_DATA` to `VALIDATING`.
Dataset validation is a background process. The status will eventually change from `VALIDATING` to `SUCCEEDED`.
The SDK provides the [`DataManagerClient.wait_for_dataset_validation()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.wait_for_dataset_validation) method to poll for the Dataset validation.
```
dataset_resource = data_manager.wait_for_dataset_validation(dataset_id)
print()
print("Dataset after validation has finished:")
print()
pprint(dataset_resource)
```
If the status is `FAILED` instead of `SUCCEEDED`, then the `validationMessage` will contain details about the validation failure.
To better understand the Dataset lifecycle, refer to the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/a9b7429687a04e769dbc7955c6c44265.html).
## Summary Exercise 01.2
In exercise 01.2, we have covered the following topics:
* How to create a DatasetSchema
* How to upload a Dataset to the service
You can find optional exercises related to exercise 01.2 [below](#Optional-Exercises-for-01.2).
# Exercise 01.3
*Back to [table of contents](#Table-of-Contents)*
*To perform this exercise, you need to execute the code in all previous exercises.*
In exercise 01.3, we will train the model.
## Training the Model
The Dataset is now uploaded and has been validated successfully by the service.
To train a machine learning model, we first need to select the correct model template.
### Selecting the right ModelTemplate
The Data Attribute Recommendation service currently supports two different ModelTemplates:
| ID | Name | Description |
|--------------------------------------|---------------------------|---------------------------------------------------------------------------|
| d7810207-ca31-4d4d-9b5a-841a644fd81f | **Hierarchical template** | Recommended for the prediction of multiple classes that form a hierarchy. |
| 223abe0f-3b52-446f-9273-f3ca39619d2c | **Generic template** | Generic neural network for multi-label, multi-class classification. |
| 188df8b2-795a-48c1-8297-37f37b25ea00 | **AutoML template** | Finds the [best traditional machine learning model out of several traditional algorithms](https://blogs.sap.com/2021/04/28/how-does-automl-works-in-data-attribute-recommendation/). Single label only. |
We are building a model to predict product hierarchies. The **Hierarchical Template** is correct for this scenario. In this template, the first label in the DatasetSchema is considered the top-level category. Each subsequent label is considered to be further down in the hierarchy.
Coming back to our example DatasetSchema:
```json
{
"labels": [
{"label": "level1_category", "type": "CATEGORY"},
{"label": "level2_category", "type": "CATEGORY"},
{"label": "level3_category", "type": "CATEGORY"}
]
}
```
The first defined label is `level1_category`, which is given more weight during training than `level3_category`.
Refer to the [official documentation on ModelTemplates](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/1e76e8c636974a06967552c05d40e066.html) to learn more. Additional model templates may be added over time, so check back regularly.
## Starting the training
When working with models, we use the [`ModelManagerClient`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient) class.
To start the training, we need the IDs of the dataset and the desired model template. We also have to provide a name for the model.
The [`ModelManagerClient.create_job()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.create_job) method launches the training Job.
*Only one model of a given name can exist. If you receive a message stating 'The model name specified is already in use', you either have to remove the job and its associated model first or you have to change the `model_name` variable name below. You can also [clean up the entire service instance](#Cleaning-up-a-service-instance).*
```
from sap.aibus.dar.client.model_manager_client import ModelManagerClient
from sap.aibus.dar.client.exceptions import DARHTTPException
model_manager = ModelManagerClient.construct_from_service_key(SERVICE_KEY)
model_template_id = "d7810207-ca31-4d4d-9b5a-841a644fd81f" # hierarchical template
model_name = "bestbuy-hierarchy-model"
job_resource = model_manager.create_job(model_name, dataset_id, model_template_id)
job_id = job_resource['id']
print()
print("Job resource:")
print()
pprint(job_resource)
print()
print(f"ID of submitted Job: {job_id}")
```
The job is now running in the background. Similar to the DatasetValidation, we have to poll the job until it succeeds.
The SDK provides the [`ModelManagerClient.wait_for_job()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_job) method:
```
job_resource = model_manager.wait_for_job(job_id)
print()
print("Job resource after training is finished:")
pprint(job_resource)
```
To better understand the Training Job lifecycle, see the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/0fc40aa077ce4c708c1e5bfc875aa3be.html).
## Intermission
The model training will take between 5 and 10 minutes.
In the meantime, we can explore the available [resources](#Resources) for both the service and the SDK.
## Inspecting the Model
Once the training job is finished successfully, we can inspect the model using [`ModelManagerClient.read_model_by_name()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_model_by_name).
```
model_resource = model_manager.read_model_by_name(model_name)
print()
pprint(model_resource)
```
In the model resource, the `validationResult` key provides information about model performance. You can also use these metrics to compare performance of different [ModelTemplates](#Selecting-the-right-ModelTemplate) or different datasets.
## Summary Exercise 01.3
In exercise 01.3, we have covered the following topics:
* How to select the appropriate ModelTemplate
* How to train a Model from a previously uploaded Dataset
You can find optional exercises related to exercise 01.3 [below](#Optional-Exercises-for-01.3).
# Exercise 01.4
*Back to [table of contents](#Table-of-Contents)*
*To perform this exercise, you need to execute the code in all previous exercises.*
In exercise 01.4, we will deploy the model and predict labels for some unlabeled data.
## Deploying the Model
The training job has finished and the model is ready to be deployed. By deploying the model, we create a server process in the background on the Data Attribute Recommendation service which will serve inference requests.
In the SDK, the [`ModelManagerClient.create_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#module-sap.aibus.dar.client.model_manager_client) method lets us create a Deployment.
```
deployment_resource = model_manager.create_deployment(model_name)
deployment_id = deployment_resource["id"]
print()
print("Deployment resource:")
print()
pprint(deployment_resource)
print(f"Deployment ID: {deployment_id}")
```
*Note: if you are using a trial account and you see errors such as 'The resource can no longer be used. Usage limit has been reached', consider [cleaning up the service instance](#Cleaning-up-a-service-instance) to free up limited trial resources.*
Similar to the data upload and the training job, model deployment is an asynchronous process. We have to poll the API until the Deployment is in status `SUCCEEDED`. The SDK provides the [`ModelManagerClient.wait_for_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_deployment) for this purposes.
```
deployment_resource = model_manager.wait_for_deployment(deployment_id)
print()
print("Finished deployment resource:")
print()
pprint(deployment_resource)
```
Once the Deployment is in status `SUCCEEDED`, we can run inference requests.
To better understand the Deployment lifecycle, see the [corresponding document on help.sap.com](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/f473b5b19a3b469e94c40eb27623b4f0.html).
*For trial users: the deployment will be stopped after 8 hours. You can restart it by deleting the deployment and creating a new one for your model. The [`ModelManagerClient.ensure_deployment_exists()`](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html) method will delete and re-create automatically. Then, you need to poll until the deployment is succeeded using [`ModelManagerClient.wait_for_deployment()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.wait_for_deployment) as above.*
## Executing Inference requests
With a single inference request, we can send up to 50 objects to the service to predict the labels. The data send to the service must match the `features` section of the DatasetSchema created earlier. The `labels` defined inside of the DatasetSchema will be predicted for each object and returned as a response to the request.
In the SDK, the [`InferenceClient.create_inference_request()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.inference_client.InferenceClient.create_inference_request) method handles submission of inference requests.
```
from sap.aibus.dar.client.inference_client import InferenceClient
inference = InferenceClient.construct_from_service_key(SERVICE_KEY)
objects_to_be_classified = [
{
"features": [
{"name": "manufacturer", "value": "Energizer"},
{"name": "description", "value": "Alkaline batteries; 1.5V"},
{"name": "price", "value": "5.99"},
],
},
]
inference_response = inference.create_inference_request(model_name, objects_to_be_classified)
print()
print("Inference request processed. Response:")
print()
pprint(inference_response)
```
*Note: For trial accounts, you only have a limited number of objects which you can classify.*
You can also try to come up with your own example:
```
my_own_items = [
{
"features": [
{"name": "manufacturer", "value": "EDIT THIS"},
{"name": "description", "value": "EDIT THIS"},
{"name": "price", "value": "0.00"},
],
},
]
inference_response = inference.create_inference_request(model_name, my_own_items)
print()
print("Inference request processed. Response:")
print()
pprint(inference_response)
```
You can also classify multiple objects at once. For each object, the `top_n` parameter determines how many predictions are returned.
```
objects_to_be_classified = [
{
"objectId": "optional-identifier-1",
"features": [
{"name": "manufacturer", "value": "Energizer"},
{"name": "description", "value": "Alkaline batteries; 1.5V"},
{"name": "price", "value": "5.99"},
],
},
{
"objectId": "optional-identifier-2",
"features": [
{"name": "manufacturer", "value": "Eidos"},
{"name": "description", "value": "Unravel a grim conspiracy at the brink of Revolution"},
{"name": "price", "value": "19.99"},
],
},
{
"objectId": "optional-identifier-3",
"features": [
{"name": "manufacturer", "value": "Cadac"},
{"name": "description", "value": "CADAC Grill Plate for Safari Chef Grills: 12\""
+ "cooking surface; designed for use with Safari Chef grills;"
+ "105 sq. in. cooking surface; PTFE nonstick coating;"
+ " 2 grill surfaces"
},
{"name": "price", "value": "39.99"},
],
}
]
inference_response = inference.create_inference_request(model_name, objects_to_be_classified, top_n=3)
print()
print("Inference request processed. Response:")
print()
pprint(inference_response)
```
We can see that the service now returns the `n-best` predictions for each label as indicated by the `top_n` parameter.
In some cases, the predicted category has the special value `nan`. In the `bestBuy.csv` data set, not all records have the full set of three categories. Some records only have a top-level category. The model learns this fact from the data and will occasionally suggest that a record should not have a category.
```
# Inspect all video games with just a top-level category entry
video_games = df[df['level1_category'] == 'Video Games']
video_games.loc[df['level2_category'].isna() & df['level3_category'].isna()].head(5)
```
To learn how to execute inference calls without the SDK just using the underlying RESTful API, see [Inference without the SDK](#Inference-without-the-SDK).
## Summary Exercise 01.4
In exercise 01.4, we have covered the following topics:
* How to deploy a previously trained model
* How to execute inference requests against a deployed model
You can find optional exercises related to exercise 01.4 [below](#Optional-Exercises-for-01.4).
# Wrapping up
In this workshop, we looked into the following topics:
* Installation of the Python SDK for Data Attribute Recommendation
* Modelling data with a DatasetSchema
* Uploading data into a Dataset
* Training a model
* Predicting labels for unlabelled data
Using these tools, we are able to solve the problem of missing Master Data attributes starting from just a CSV file containing training data.
Feel free to revisit the workshop materials at any time. The [resources](#Resources) section below contains additional reading.
If you would like to explore the additional capabilities of the SDK, visit the [optional exercises](#Optional-Exercises) below.
## Cleanup
During the course of the workshop, we have created several resources on the Data Attribute Recommendation Service:
* DatasetSchema
* Dataset
* Job
* Model
* Deployment
The SDK provides several methods to delete these resources. Note that there are dependencies between objects: you cannot delete a Dataset without deleting the Model beforehand.
You will need to set `CLEANUP_SESSION = True` below to execute the cleanup.
```
# Clean up all resources created earlier
CLEANUP_SESSION = False
def cleanup_session():
model_manager.delete_deployment_by_id(deployment_id) # this can take a few seconds
model_manager.delete_model_by_name(model_name)
model_manager.delete_job_by_id(job_id)
data_manager.delete_dataset_by_id(dataset_id)
data_manager.delete_dataset_schema_by_id(dataset_schema_id)
print("DONE cleaning up!")
if CLEANUP_SESSION:
print("Cleaning up resources generated in this session.")
cleanup_session()
else:
print("Not cleaning up. Set 'CLEANUP_SESSION = True' above and run again!")
```
## Resources
*Back to [table of contents](#Table-of-Contents)*
### SDK Resources
* [SDK source code on Github](https://github.com/SAP/data-attribute-recommendation-python-sdk)
* [SDK documentation](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/)
* [How to obtain support](https://github.com/SAP/data-attribute-recommendation-python-sdk/blob/master/README.md#how-to-obtain-support)
* [Tutorials: Classify Data Records with the SDK for Data Attribute Recommendation](https://developers.sap.com/group.cp-aibus-data-attribute-sdk.html)
### Data Attribute Recommendation
* [SAP Help Portal](https://help.sap.com/viewer/product/Data_Attribute_Recommendation/SHIP/en-US)
* [API Reference](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/b45cf9b24fd042d082c16191aa938c8d.html)
* [Tutorials using Postman - interact with the service RESTful API directly](https://developers.sap.com/mission.cp-aibus-data-attribute.html)
* [Trial Account Limits](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/c03b561eea1744c9b9892b416037b99a.html)
* [Metering and Pricing](https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/1e093326a2764c298759fcb92c5b0500.html)
## Addendum
### Inference without the SDK
*Back to [table of contents](#Table-of-Contents)*
The Data Attribute Service exposes a RESTful API. The SDK we use in this workshop uses this API to interact with the DAR service.
For custom integration, you can implement your own client for the API. The tutorial "[Use Machine Learning to Classify Data Records]" is a great way to explore the Data Attribute Recommendation API with the Postman REST client. Beyond the tutorial, the [API Reference] is a comprehensive documentation of the RESTful interface.
[Use Machine Learning to Classify Data Records]: https://developers.sap.com/mission.cp-aibus-data-attribute.html
[API Reference]: https://help.sap.com/viewer/105bcfd88921418e8c29b24a7a402ec3/SHIP/en-US/b45cf9b24fd042d082c16191aa938c8d.html
To demonstrate the underlying API, the next example uses the `curl` command line tool to perform an inference request against the Inference API.
The example uses the `jq` command to extract the credentials from the service. The authentication token is retrieved from the `uaa_url` and then used for the inference request.
```
# If the following example gives you errors that the jq or curl commands cannot be found,
# you may be able to install them from conda by uncommenting one of the lines below:
#%conda install -q jq
#%conda install -q curl
%%bash -s "$model_name" # Pass the python model_name variable as the first argument to shell script
model_name=$1
echo "Model: $model_name"
key=$(cat key.json)
url=$(echo $key | jq -r .url)
uaa_url=$(echo $key | jq -r .uaa.url)
clientid=$(echo $key | jq -r .uaa.clientid)
clientsecret=$(echo $key | jq -r .uaa.clientsecret)
echo "Service URL: $url"
token_url=${uaa_url}/oauth/token?grant_type=client_credentials
echo "Obtaining token with clientid $clientid from $token_url"
bearer_token=$(curl \
--silent --show-error \
--user $clientid:$clientsecret \
$token_url \
| jq -r .access_token
)
inference_url=${url}/inference/api/v3/models/${model_name}/versions/1
echo "Running inference request against endpoint $inference_url"
echo ""
# We pass the token in the Authorization header.
# The payload for the inference request is passed as
# the body of the POST request below.
# The output of the curl command is piped through `jq`
# for pretty-printing
curl \
--silent --show-error \
--header "Authorization: Bearer ${bearer_token}" \
--header "Content-Type: application/json" \
-XPOST \
${inference_url} \
-d '{
"objects": [
{
"features": [
{
"name": "manufacturer",
"value": "Energizer"
},
{
"name": "description",
"value": "Alkaline batteries; 1.5V"
},
{
"name": "price",
"value": "5.99"
}
]
}
]
}' | jq
```
### Cleaning up a service instance
*Back to [table of contents](#Table-of-Contents)*
To clean all data on the service instance, you can run the following snippet. The code is self-contained and does not require you to execute any of the cells above. However, you will need to have the `key.json` containing a service key in place.
You will need to set `CLEANUP_EVERYTHING = True` below to execute the cleanup.
**NOTE: This will delete all data on the service instance!**
```
CLEANUP_EVERYTHING = False
def cleanup_everything():
import logging
import sys
logging.basicConfig(level=logging.INFO, stream=sys.stdout)
import json
import os
if not os.path.exists("key.json"):
msg = "key.json is not found. Please follow instructions above to create a service key of"
msg += " Data Attribute Recommendation. Then, upload it into the same directory where"
msg += " this notebook is saved."
print(msg)
raise ValueError(msg)
with open("key.json") as file_handle:
key = file_handle.read()
SERVICE_KEY = json.loads(key)
from sap.aibus.dar.client.model_manager_client import ModelManagerClient
model_manager = ModelManagerClient.construct_from_service_key(SERVICE_KEY)
for deployment in model_manager.read_deployment_collection()["deployments"]:
model_manager.delete_deployment_by_id(deployment["id"])
for model in model_manager.read_model_collection()["models"]:
model_manager.delete_model_by_name(model["name"])
for job in model_manager.read_job_collection()["jobs"]:
model_manager.delete_job_by_id(job["id"])
from sap.aibus.dar.client.data_manager_client import DataManagerClient
data_manager = DataManagerClient.construct_from_service_key(SERVICE_KEY)
for dataset in data_manager.read_dataset_collection()["datasets"]:
data_manager.delete_dataset_by_id(dataset["id"])
for dataset_schema in data_manager.read_dataset_schema_collection()["datasetSchemas"]:
data_manager.delete_dataset_schema_by_id(dataset_schema["id"])
print("Cleanup done!")
if CLEANUP_EVERYTHING:
print("Cleaning up all resources in this service instance.")
cleanup_everything()
else:
print("Not cleaning up. Set 'CLEANUP_EVERYTHING = True' above and run again.")
```
### Optional Exercises
*Back to [table of contents](#Table-of-Contents)*
To work with the optional exercises, create a new cell in the Jupyter notebook by clicking the `+` button in the menu above or by using the `b` shortcut on your keyboard. You can then enter your code in the new cell and execute it.
#### Optional Exercises for 01.2
##### DatasetSchemas
Use the [`DataManagerClient.read_dataset_schema_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_schema_by_id) and the [`DataManagerClient.read_dataset_schema_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_schema_collection) methods to list the newly created and all DatasetSchemas, respectively.
##### Datasets
Use the [`DataManagerClient.read_dataset_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_by_id) and the [`DataManagerClient.read_dataset_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.read_dataset_collection) methods to inspect the newly created dataset.
Instead of using two separate methods to upload data and wait for validation to finish, you can also use [`DataManagerClient.upload_data_and_validate()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.data_manager_client.DataManagerClient.upload_data_and_validate).
#### Optional Exercises for 01.3
##### ModelTemplates
Use the [`ModelManagerClient.read_model_template_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_model_template_collection) to list all existing model templates.
##### Jobs
Use [`ModelManagerClient.read_job_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_job_by_id) and [`ModelManagerClient.read_job_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_job_collection) to inspect the job we just created.
The entire process of uploading the data and starting the training is also available as a single method call in [`ModelCreator.create()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.workflow.model.ModelCreator.create).
#### Optional Exercises for 01.4
##### Deployments
Use [`ModelManagerClient.read_deployment_by_id()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_deployment_by_id) and [`ModelManagerClient.read_deployment_collection()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.read_deployment_collection) to inspect the Deployment.
Use the [`ModelManagerclient.lookup_deployment_id_by_model_name()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.model_manager_client.ModelManagerClient.lookup_deployment_id_by_model_name) method to find the deployment ID for a given model name.
##### Inference
Use the [`InferenceClient.do_bulk_inference()`](https://data-attribute-recommendation-python-sdk.readthedocs.io/en/latest/api.html#sap.aibus.dar.client.inference_client.InferenceClient.do_bulk_inference) method to process more than fifty objects at a time. Note how the data format returned changes.
|
github_jupyter
|
# Resumen
Este cuaderno digital interactivo tiene como objetivo demostrar las relaciones entre las propiedades fisico-químicas de la vegetación y el espectro solar.
Para ello haremos uso de modelos de simulación, en particular de modelos de transferencia radiativa tanto a nivel de hoja individual como a nivel de dosel vegetal.
# Instrucciones
Lee con detenimiento todo el texto, y sigue sus instrucciones.
Una vez leida cada sección de texto ejecuta la celda de código siguiente (marcada como `In []`) presionando el icono de `Run`/`Ejecutar` o presionando en el teclado ALT + ENTER. Aparecerá una interfaz gráfica con la que poder realizar las tareas asignadas.
Como ejemplo ejectuta la siguiente celda para importar todas las librerías necesarias para el correcto funcionamiento del cuaderno. Una vez ejecutada debería aparecer un mensaje de agradecimiento.
```
%matplotlib inline
from ipywidgets import interactive, fixed
from IPython.display import display
from functions import prosail_and_spectra as fn
```
# Espectro de una hoja
Las propiedades espectrales de una hoja (tanto su transmisividad, su reflectividad y su absortividad) dependen de su concentración de pigmentos, de su contenido de agua, su peso específico y la estructura interna de sus tejidos.
Vamos a usar el modelo ProspectD, el cual es una simplificación de la realidad en la que simula el espectro mediante la concentración de clorofilas (`Cab`), carotenoides (`Car`), antocianinos (`Ant`), así como el peso de agua y por unidad de supeficie (`Cw`) y el peso del resto de la materia seca (`Cm`) que engloba las celulosas, ligninas (responsables principales de la biomasa foliar) y otros componentes proteicos. También incluye un parámetro semi-empírico que representa otros pigmentos responsables del color de las hojas senescentes y enfermas. Además con el fin de simular hojas con distintas estructuras celulares incluye un último parámetro (`Nf`) que emula las distitas capas y tejidos celulares de la hoja.
 con idénticas propiedades espectrales")
> Si quieres saber más sobre el modelo ProspectD pincha en esta [publicación](./lecturas_adicionales/ProspectD_model.pdf).
>
> Si quieres más detalles sobre el cálculo y el código del modelo pincha [aquí](https://github.com/hectornieto/pypro4sail/blob/b111891e0a2c01b8b3fa5ff41790687d31297e5f/pypro4sail/prospect.py#L46).
Ejecuta la siguiente célula y verás un espectro típico de la hoja. El gráfico muestra tanto la reflectividad (en el eje y) como la transmisividad (en el eje secundario y, con valores invertidos) y la absortividad (como el espacio entre las dos curvas de reflectividad y transmisividad) $\rho + \tau + \alpha = 1$.
Presta atención a cómo y en qué regiones cambia el espectro según el parámetro que modifiques.
* Haz variar la clorofila.
* Haz variar el contenido de agua
* Haz variar la materia seca
* Haz variar los pigmentos marrones desde un valor de 0 (hoja sana) a valores mayores (hoja enferma o seca)
```
w_rho_leaf = interactive(fn.update_prospect_spectrum, N_leaf=fn.w_nleaf, Cab=fn.w_cab,
Car=fn.w_car, Ant=fn.w_ant, Cbrown=fn.w_cbrown, Cw=fn.w_cw, Cm=fn.w_cm)
display(w_rho_leaf)
```
Observa lo siguente:
* La concentración de clorofila `Cab` afecta principalmente a la región del visible (RGB) y del *red egde* (R-E), con más absorción en la región del rojo y del azul y más reflexión en el verde. Es por ello que la mayoría de las hojas presentan color verde.
* El contenido de agua `Cw` afecta principalmente a la absorción en el infrarrojo de onda corta (SWIR), con máximos de absorción en trono a los 1460 y 2100 nm.
* La materia seca `Cm` afecta principalmente a la absorción en el infrarrojo cercano (NIR).
* Otros pigmentos afectan en menor medida al espectro visible. Por ejemplo los antocianos `Ant` que suelen aparecer durante la senescencia desplazan el pico de reflexión del verde hacia el rojo, sobre todo cuando a su vez decrece la concentración de clorofila.
* El parámetro `N` afecta a la relación entre reflectividad y transmisividad. Cuantas más *capas* tenga una hoja más fenómenos de dispersión múltiple habrá y reflejará más.
> Puedes ver este fenómeno también en las ventanas con doble o triple cristal usadas como aislante, por ejemplo de los escaparates comerciales. A no ser que uno se sitúen justo de frente y cerca del escaparate, éste parece más un espejo que una ventana.
# Espectro del suelo
El espectro del dosel o de la supeficie vegetal no sólo depende del espectro y las propiedades de las hojas, sino que también de la propia estructura del dosel así como del suelo. En particular en doseles abiertos o poco densos, como en las primeras fases fenológicas, el comportamiento espectral del suelo puede influir de manera muy importante en la señal espectral que capten los sensores de teledetección.
El espectro del suelo depende de varios factores, como son su composición mineralógica, materia orgánica, su textura y densidad así como su humedad superficial.
Ejectuta la siguiente celda y mira los distintas características espectrales de distintos tipos de suelo.
```
w_rho_soil = interactive(fn.update_soil_spectrum, soil_name=fn.w_soil)
display(w_rho_soil)
```
Observa lo diferente que puede ser un espectro de suelo en comparación con el de una hoja. Esto es clave a la hora de clasificar tipos de coberturas mediante teledetección así como cuantificar el vigor/densidad vegetal del cultivo.
Observa que suelos más salinos (`aridisol.salorthid`) o gipsicos (`aridisol.gypsiorthd`), tienen una mayor reflectividad, sobre todo en el visible (RGB). Es decir, son más blancos que otros suelos.
# Espectro del dosel
Finalmente, integrando la firma espectral de una hoja y del suelo subyacente podemos obtener el espectro de un dosel vegetal.
El espectro de la superficie vegetal además depende de la estructura del dosel, principalmente de la cantidad de hojas por unidad de superficie (definido como el Índice de Área Foliar) y de cómo estas hojas se orientan con respecto a la vertical. Además, dado que se produce una interacción de la luz incidente y reflejada entre el volumen de hojas y el suelo, la posición del sol y del sensor influyen en la señal espectral que obtengamos.
Para esta parte cobinaremos el modelo de transferencia ProspectD para simular el espectro de una hoja con otro modelo de trasnferencia a nivel de dosel (4SAIL). Este último modelo considera la superficie vegetal como una capa horizontal y verticalmente homogéna, por lo que se recomienda cautela en su aplicación en doseles arbóreos heterogéneos.

> Si quieres saber más sobre el modelo 4SAIL pincha en esta [publicación](./lecturas_adicionales/4SAIL_model.pdf)
>
> Si quieres más detalles sobre el cálculo y el código del modelo pincha [aquí](https://github.com/hectornieto/pypro4sail/blob/b111891e0a2c01b8b3fa5ff41790687d31297e5f/pypro4sail/four_sail.py#L245)
Ejecuta la siguente celda y mira cómo los [espectros de hoja](#Espectro-de-una-hoja) y [suelo](#Espectro-del-suelo) que se han generado previamente se integran para obtener un espectro de la superficie vegetal.
> Puedes modificar los espectros de hoja y suelo, y esta gráfica se actualizará automáticamente.
```
w_rho_canopy = interactive(fn.update_4sail_spectrum,
lai=fn.w_lai, hotspot=fn.w_hotspot, leaf_angle=fn.w_leaf_angle,
sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi, skyl=fn.w_skyl,
leaf_spectrum=fixed(w_rho_leaf), soil_spectrum=fixed(w_rho_soil))
display(w_rho_canopy)
```
Recuerda en la [práctica sobre la radiación neta](./ES_radiacion_neta.ipynb) que una superficie vegetal tiene ciertas propiedades anisotrópicas, lo que quiere decir que reflejará de manera distinta según la geometria de iluminación y de observación.
Mira cómo cambia el espectro variando los valores del ángulo de observación cenital (VZA), ańgulo cenital del sol (SZA) y el ángulo azimutal relativo (PSI) entre el sol y el observador.
Haz variar el LAI, y ponlo en cero (sin vegetación). Comprueba que el espectro que sale es directamente el espectro del suelo. Ahora incrementa ligeramente el LAI, verás como el espectro va cambiando, disminuyendo la reflectividad en el rojo y azul (debido a la clorofila de la hoja), y aumentando la reflectividad en el *red-edge* y el NIR.
Recuerda también de la [práctica sobre la radiación neta](./ES_radiacion_neta.ipynb) el efecto que también tiene la disposición angular de las hojas. Con una observación al nadir (VZA=0) haz variar el ángulo típico de la hoja (`Leaf Angle`) desde un valor predominantemente horizontal (0º) a un ángulo predominantemente vertical (90º)
# Sensibilidad de los parámetros
En esta tarea podrás ver el comportamiento espectral de la vegetación según varían los parámetros fisico-químicos de la vegetación así como su sensibilidad a las condiciones de observación e iluminación.
Para ello vamos a realizar un análisis de sensibilidad variando un sólo parámetro a la vez, mientras que el resto de los parámetros permanecerán constantes. Puedes variar los valores individuales para el resto de los parámetros individuales (también se actualizarán de las gráficas anteriores). A continuación selecciona qué parámetro quieres analizar y el rango de valores máximo y mínimo que quieras que tenga.
```
w_sensitivity = interactive(fn.prosail_sensitivity,
N_leaf=fn.w_nleaf, Cab=fn.w_cab, Car=fn.w_car, Ant=fn.w_ant, Cbrown=fn.w_cbrown,
Cw=fn.w_cw, Cm=fn.w_cm, lai=fn.w_lai, hotspot=fn.w_hotspot, leaf_angle=fn.w_leaf_angle,
sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi, skyl=fn.w_skyl,
soil_name=fn.w_soil, var=fn.w_param, value_range=fn.w_range)
display(w_sensitivity)
```
Empieza con al sensiblidad del espectro a la concentración de clorofila. Verás que la zona donde sobre todo hay variaciones es en el verde y el rojo. Observa también que en el *red-edge*, la zona de transición entre el rojo y el NIR, se produce un "desplazamiento" de la señal, este fenómento es clave y es la razón por la que los nuevos sensores (Sentinel, nuevas cámaras UAV) incluyen esta región para ayudar en la estimación de la clorofila y por tanto en la actividad fotosintética.
Evalúa la sensibilidad al espectro de otros pigmentos (`Car` o `Ant`). Verás que la respuesta espectral a estos otros pigmentos es menor, lo que implica que resulta más dificil estimarlos a partir de teledetección. En cambio la variación espectral con los pigmentos marrones es bastante fuerte, como recordatorio estos pigmentos representan las variaciones cromáticas que se producen en hojas enfermas y muertas.
> Esto implica que es relativamente posible detectar y cuantificar problemas sanitarios en la vegetación.
Mira ahora la sensibilidad del LAI cuando su rango es pequeño (p.ej. de 0 a 2). Verás que el espectro cambia significativamente según incrementa el LAI. Ahora mira la sensibilidad cuando el LAI recorre valores mas altos (p.ej. de 2 a 4), verás que la variación en el espectro es mucho menor. Se suele decir que a valores altos de LAI el espectro tiende a "saturarse" por lo que la señal se hace menos sensible.
> Es más fácil estimar el LAI con menor margen de error en cultivos con poca densidad foliar o fases fenológicas tempranas, que en cultivos o vegetación muy densa.
Ahora mantén el valor fijo de LAI en un valor alto (p.ej 3) y haz variar el ángulo de observación cenital entre 0º (nadir) y una obsrvación oblicua (p.ej 35º). Verás que a pesar de haber un LAI alto, y que a priori hemos visto que ya es menos sensible, hay mayores variaciones espectrales al variar la geometría de observación.
> Gracias a la anisotropía de la vegetación, las variaciones espectrales con respecto a la geometría de observación e iluminación pueden ayudar a resolver el LAI en condiciones de alta densidad.
Ahora mira el peso específico de la hoja, o la cantidad de materia seca (`Cm`). Verás que según el peso específico de la hora se producen variaciones importantes en el NIR y SWIR.
> La biomasa foliar puede calcularse a partir del producto entre el `LAI` y `Cm`, por lo que es viable estimar la biomasa foliar de un cultivo. Esta informaición puede ser útil por ejemplo para estimar el rendimiento final de algunos cultivos, como pueden ser los cereales.
El parámetro `hotspot` es un parámetro semi-empírico relacionado con el tamaño relativo de la hoja con respecto a la altura del dosel. Afecta a cómo las hojas ensombrecen otras hojas dentro del dosel, por lo que su efecto más fuerte se observará cuando el observador (sensor) está justo en la misma posición que el sol. Para ello valores similares para VZA y SZA, y el ángulo azimutal relativo PSI en 0º. Ahora haz variar el hotstpot. Al poner el observador en la zona iluminada de la vegetación, el tamaño relativo de las hojas juega un papel importante, ya que cuanto más grandes sean estas el volumen de copa directamente iluminado será mayor.

# La señal de un sensor
Hasta ahora hemos visto el comportamiento espectral detallado de la vegetación. Sin embargo los sensores a bordo de los satélites, aeroplanos y drones no miden todo el espectro en continuo, si no que muestrean tal espectro en torno a unas bandas específicas, estratégicamente seleccionadas con el fin de intentar capturar los aspectos biofísicos más relvantes.
Se denomina función de respuesta espectral a la forma en que un sensor específico integra el espectro con el fin de proporcionar la información en sus distintas bandas. Cada sensor, con cada una de sus bandas, tiene una función de respuesta espectral propia.
En esta tarea veremos las respuestas espectrales de los sensores que utilizaremos más comunmente, Landsat, Sentinel-2 y Sentinel-3. También veremos el comportamiento espectral de una cámara típica que se usa con drones.
Partimos de las simulaciones generadas anteriormente. Selecciona el sensor que quieras simular para ver como cada uno de los sensores "verían" esos mismos espectros.
```
w_rho_sensor = interactive(fn.sensor_sensitivity,
sensor=fn.w_sensor, spectra=fixed(w_sensitivity))
display(w_rho_sensor)
```
Realiza de nuevo un análisis de sensibilidad para la clorofila y compara la respuesta espectral que daría Landsat, Sentinel-2 y una camára UAV
# Derivación de parámetros de la vegetación
Hasta ahora hemos visto cómo el espectro de la superficie varía con respecto a los distintos parámetros biofísicos.
Sin embargo, nuestro objetivo final es el contrario, es decir, a partir de un espectro, o de unas determinadas bandas espectrales estimar una o varias variables biofísicas que nos son de interés. En particular para el objetivo del cálculo de la evapotranspiración y la eficiencia en el uso en el agua, nos puede interesar estimar el índice de área foliar y/o la fracción de radiación PAR absorbida, así como las clorofilas u otros pigmentos.
Una de los métodos típicos es desarrollar relaciones empíricas entre las bandas (o entre índices de vegetación) y datos muestreados en el campo. Esto puede darnos la respuesta más fiable para nuestra parcela de estudio, pero como has podido ver anteriormente la señal espectral depende de otros muchos factores, que pueden provocar que esa relación calibrada con unos cuantos muestreos locales no sea extrapolable o aplicable a otros cultivos o regiones.
Otra alternativa es desarrollar bases de datos sintéticas a partir de simulaciones. Es lo que vamos a realizar en esta tarea.
Vamos a ejecutar 5000 simulaciones haciendo variar los valores de los parámetros aleatoriamente según un rango de valores que puedan ser esperado en nuestras zonas de estudio.
Por ejemplo si trabajas con cultivos perennes puede que te interesa mantener un rango de LAI con valores mínimos sensiblemente superiores a cero, mientras si trabajas con cultivos anuales, el valor 0 es necesario para reflejar el desarrollo del cultivo desde su plantación, emergencia y madurez.
Ya que hay una gran cantidad de parámetros y es muy probable que desconozcamos el rango plausible en la mayoría de los cultivos, no te preocupes, deja los valores por defecto y céntrate en los parámetros en los que tengas más confianza.
Puedes también elegir uno o varios tipos de suelo, en función de la edafología de tu lugar.
> Incluso podrías subir un espectro de suelo típico de tu zona a la carpeta [./input/soil_spectral_library](./input/soil_spectral_library). Tan sólo asegúrate que el archivo de texto tenga dos columnas, la primera con las longitudes de onda de 400 a 2500 y la segunda columna con la reflectividad correspondiente. Para actualizar la librería espectral de suelos, tendrías que ejecutar también la [primera celda](#Instrucciones).
Finalmente selecciona el sensor para el que quieras genera la señal.
Cuando tengas tu configurado tu entorno de simulación, pincha en el botón `Generar Simulaciones`. El sistema tardará un rato pero al cabo de unos minutos te retornará una serie de gráficos.
> Es posible que recibas un mensaje de aviso, no te preocupes, en principio todo debería funcionar con normalidad.
```
w_rho_sensor = interactive(fn.build_random_simulations, {"manual": True, "manual_name": "Generar simulaciones"},
n_sim=fixed(5000), n_leaf_range=fn.w_range_nleaf,
cab_range=fn.w_range_cab, car_range=fn.w_range_car,
ant_range=fn.w_range_ant, cbrown_range=fn.w_range_cbrown,
cw_range=fn.w_range_cw, cm_range=fn.w_range_cm,
lai_range=fn.w_range_lai, hotspot_range=fn.w_range_hotspot,
leaf_angle_range=fn.w_range_leaf_angle,
sza=fn.w_sza, vza=fn.w_vza, psi=fn.w_psi,
skyl=fn.w_skyl, soil_names=fn.w_soils, sensor=fn.w_sensor)
display(w_rho_sensor)
```
El gráfico muestra 4 ejemplos de relaciones entre 3 índices de vegetación típicos y 4 variables biofísicas.
* NDVI: Normalized Difference Vegetation Index. Es el índicie de vegetación más utilizado. Generalmente se relaciona con el LAI, la biomasa foliar y/o la fracción de radiación interceptada/absorbida
$$NDVI = \frac{\rho_{NIR} - \rho_{R}}{\rho_{NIR} + \rho_{R}}$$
* NDRE: Normalized Difference Red-Edge. Es un índicie de vegetación que usa la región del red edge, por lo que no puede calcularse para cualquier sensor. Generalmente se relaciona con la clorofila.
$$NDRE = \frac{\rho_{NIR} - \rho_{R-E}}{\rho_{NIR} + \rho_{R-E}}$$
* NDWI: Normalized Difference Water Index. Es un índicie de vegetación que usa la región del SWIR, por lo que no puede calcularse para cualquier sensor. Generalmente se relaciona con el contenido de agua de la vegetación.
$$NDWI = \frac{\rho_{NIR} - \rho_{SWIR}}{\rho_{NIR} + \rho_{SWIR}}$$
Las simulaciones se han guardado en un archivo prosail_simulations.csv en la carpeta [./output](./output/prosail_simulations.csv). Descargate este archivo y calcula distintos índices de vegetación e intenta desarrollar relaciones y modelos estadísticos entre las bandas o índices de vegetación y los parámetros biofísicos. Para ello puedes usar cualquier software con el que estés habituado a trabajar (Excel, R, SPSS, ...).
Puedes realizar tantas simulaciones como consideres necesarias, por ejemplo variando el sensor o modificando los rangos plausibles para cubrir distintos tipos funcionales de vegetación. Tan sólo ten en cuenta que cada vez que se genere una simulación el archivo csv se sobreesecribirá. **Por lo que descárcatelo o haz una copia en tu carpeta virtual antes de volver a ejectura las nuevas simulaciones**.
# Conclusiones
En esta práctica hemos visto cómo el espectro de la vegetación responde a las variables biofísicas de la superfice.
* El LAI es probablemente la variable que influya más en la respuesta espectral de la vegetación.
* La concentración de clorofila en la hoja influye sobre todo en la región del visible y del *red-edge*.
* El contenido de agua y el peso específico de la hora influyen sobre todo a partir del NIR.
* La geometría de observación e iluminación, así como la respuesta espectral del suelo, influyen también en la señal. Esto hace que sea difícil aplicar una relación universal a la hora de estimar un parámetro biofísico.
* Los modelos de transferencia radiativa pueden ayudar a estimar estos parámetros. Si bien idealmente es necesario disponer de datos de campo para realizar tareas de validación y/o calibración estadística.
* Los sensores muestrean una parte del espectro en torno a bandas espectrales específicas. Por tanto una relación empírica desarrollada para un sensor específico puede que no sea aplicable o válida para otro sensor.
|
github_jupyter
|
```
ls -l| tail -10
#G4
from google.colab import drive
drive.mount('/content/gdrive')
cp gdrive/My\ Drive/fingerspelling5.tar.bz2 fingerspelling5.tar.bz2
# rm -r surrey/
%rm -r dataset5/
# rm fingerspelling5.tar.bz2
# cd /media/datastorage/Phong/
!tar xjf fingerspelling5.tar.bz2
cd dataset5
mkdir surrey
mkdir surrey/D
mv dataset5/* surrey/D/
cd surrey
cd ..
#remove depth files
import glob
import os
import shutil
# get parts of image's path
def get_image_parts(image_path):
"""Given a full path to an image, return its parts."""
parts = image_path.split(os.path.sep)
#print(parts)
filename = parts[2]
filename_no_ext = filename.split('.')[0]
classname = parts[1]
train_or_test = parts[0]
return train_or_test, classname, filename_no_ext, filename
#del_folders = ['A','B','C','D','E']
move_folders_1 = ['A','B','C','E']
move_folders_2 = ['D']
# look for all images in sub-folders
for folder in move_folders_1:
class_folders = glob.glob(os.path.join(folder, '*'))
for iid_class in class_folders:
#move depth files
class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))
print('copying %d files' %(len(class_files)))
for idx in range(len(class_files)):
src = class_files[idx]
if "0001" not in src:
train_or_test, classname, _, filename = get_image_parts(src)
dst = os.path.join('train_depth', classname, train_or_test+'_'+ filename)
# image directory
img_directory = os.path.join('train_depth', classname)
# create folder if not existed
if not os.path.exists(img_directory):
os.makedirs(img_directory)
#copying
shutil.copy(src, dst)
else:
print('ignor: %s' %src)
#move color files
for iid_class in class_folders:
#move depth files
class_files = glob.glob(os.path.join(iid_class, 'color*.png'))
print('copying %d files' %(len(class_files)))
for idx in range(len(class_files)):
src = class_files[idx]
train_or_test, classname, _, filename = get_image_parts(src)
dst = os.path.join('train_color', classname, train_or_test+'_'+ filename)
# image directory
img_directory = os.path.join('train_color', classname)
# create folder if not existed
if not os.path.exists(img_directory):
os.makedirs(img_directory)
#copying
shutil.copy(src, dst)
# look for all images in sub-folders
for folder in move_folders_2:
class_folders = glob.glob(os.path.join(folder, '*'))
for iid_class in class_folders:
#move depth files
class_files = glob.glob(os.path.join(iid_class, 'depth*.png'))
print('copying %d files' %(len(class_files)))
for idx in range(len(class_files)):
src = class_files[idx]
if "0001" not in src:
train_or_test, classname, _, filename = get_image_parts(src)
dst = os.path.join('test_depth', classname, train_or_test+'_'+ filename)
# image directory
img_directory = os.path.join('test_depth', classname)
# create folder if not existed
if not os.path.exists(img_directory):
os.makedirs(img_directory)
#copying
shutil.copy(src, dst)
else:
print('ignor: %s' %src)
#move color files
for iid_class in class_folders:
#move depth files
class_files = glob.glob(os.path.join(iid_class, 'color*.png'))
print('copying %d files' %(len(class_files)))
for idx in range(len(class_files)):
src = class_files[idx]
train_or_test, classname, _, filename = get_image_parts(src)
dst = os.path.join('test_color', classname, train_or_test+'_'+ filename)
# image directory
img_directory = os.path.join('test_color', classname)
# create folder if not existed
if not os.path.exists(img_directory):
os.makedirs(img_directory)
#copying
shutil.copy(src, dst)
# #/content
%cd ..
ls -l
mkdir surrey/E/checkpoints
cd surrey/
#MUL 1 - Inception - ST
from keras.applications import MobileNet
# from keras.applications import InceptionV3
# from keras.applications import Xception
# from keras.applications.inception_resnet_v2 import InceptionResNetV2
# from tensorflow.keras.applications import EfficientNetB0
from keras.models import Model
from keras.layers import concatenate
from keras.layers import Dense, GlobalAveragePooling2D, Input, Embedding, SimpleRNN, LSTM, Flatten, GRU, Reshape
# from keras.applications.inception_v3 import preprocess_input
# from tensorflow.keras.applications.efficientnet import preprocess_input
from keras.applications.mobilenet import preprocess_input
from keras.layers import GaussianNoise
def get_adv_model():
# f1_base = EfficientNetB0(include_top=False, weights='imagenet',
# input_shape=(299, 299, 3),
# pooling='avg')
# f1_x = f1_base.output
f1_base = MobileNet(weights='imagenet', include_top=False, input_shape=(224,224,3))
f1_x = f1_base.output
f1_x = GlobalAveragePooling2D()(f1_x)
# f1_x = f1_base.layers[-151].output #layer 5
# f1_x = GlobalAveragePooling2D()(f1_x)
# f1_x = Flatten()(f1_x)
# f1_x = Reshape([1,1280])(f1_x)
# f1_x = SimpleRNN(2048,
# return_sequences=False,
# # dropout=0.8
# input_shape=[1,1280])(f1_x)
#Regularization with noise
f1_x = GaussianNoise(0.1)(f1_x)
f1_x = Dense(1024, activation='relu')(f1_x)
f1_x = Dense(24, activation='softmax')(f1_x)
model_1 = Model(inputs=[f1_base.input],outputs=[f1_x])
model_1.summary()
return model_1
from keras.callbacks import Callback
import pickle
import sys
#Stop training on val_acc
class EarlyStoppingByAccVal(Callback):
def __init__(self, monitor='val_acc', value=0.00001, verbose=0):
super(Callback, self).__init__()
self.monitor = monitor
self.value = value
self.verbose = verbose
def on_epoch_end(self, epoch, logs={}):
current = logs.get(self.monitor)
if current is None:
warnings.warn("Early stopping requires %s available!" % self.monitor, RuntimeWarning)
if current >= self.value:
if self.verbose > 0:
print("Epoch %05d: early stopping" % epoch)
self.model.stop_training = True
#Save large model using pickle formate instead of h5
class SaveCheckPoint(Callback):
def __init__(self, model, dest_folder):
super(Callback, self).__init__()
self.model = model
self.dest_folder = dest_folder
#initiate
self.best_val_acc = 0
self.best_val_loss = sys.maxsize #get max value
def on_epoch_end(self, epoch, logs={}):
val_acc = logs['val_acc']
val_loss = logs['val_loss']
if val_acc > self.best_val_acc:
self.best_val_acc = val_acc
# Save weights in pickle format instead of h5
print('\nSaving val_acc %f at %s' %(self.best_val_acc, self.dest_folder))
weigh= self.model.get_weights()
#now, use pickle to save your model weights, instead of .h5
#for heavy model architectures, .h5 file is unsupported.
fpkl= open(self.dest_folder, 'wb') #Python 3
pickle.dump(weigh, fpkl, protocol= pickle.HIGHEST_PROTOCOL)
fpkl.close()
# model.save('tmp.h5')
elif val_acc == self.best_val_acc:
if val_loss < self.best_val_loss:
self.best_val_loss=val_loss
# Save weights in pickle format instead of h5
print('\nSaving val_acc %f at %s' %(self.best_val_acc, self.dest_folder))
weigh= self.model.get_weights()
#now, use pickle to save your model weights, instead of .h5
#for heavy model architectures, .h5 file is unsupported.
fpkl= open(self.dest_folder, 'wb') #Python 3
pickle.dump(weigh, fpkl, protocol= pickle.HIGHEST_PROTOCOL)
fpkl.close()
# Training
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import TensorBoard, ModelCheckpoint, EarlyStopping, CSVLogger, ReduceLROnPlateau
from keras.optimizers import Adam
import time, os
from math import ceil
train_datagen = ImageDataGenerator(
# rescale = 1./255,
rotation_range=30,
width_shift_range=0.3,
height_shift_range=0.3,
shear_range=0.3,
zoom_range=0.3,
# horizontal_flip=True,
# vertical_flip=True,##
# brightness_range=[0.5, 1.5],##
channel_shift_range=10,##
fill_mode='nearest',
# preprocessing_function=get_cutout_v2(),
preprocessing_function=preprocess_input,
)
test_datagen = ImageDataGenerator(
# rescale = 1./255
preprocessing_function=preprocess_input
)
NUM_GPU = 1
batch_size = 64
train_set = train_datagen.flow_from_directory('surrey/D/train_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=True,
seed=7,
# subset="training"
)
valid_set = test_datagen.flow_from_directory('surrey/D/test_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=False,
seed=7,
# subset="validation"
)
model_txt = 'st'
# Helper: Save the model.
savedfilename = os.path.join('surrey', 'D', 'checkpoints', 'Surrey_MobileNet_D_tmp.hdf5')
checkpointer = ModelCheckpoint(savedfilename,
monitor='val_accuracy', verbose=1,
save_best_only=True, mode='max',save_weights_only=True)########
# Helper: TensorBoard
tb = TensorBoard(log_dir=os.path.join('svhn_output', 'logs', model_txt))
# Helper: Save results.
timestamp = time.time()
csv_logger = CSVLogger(os.path.join('svhn_output', 'logs', model_txt + '-' + 'training-' + \
str(timestamp) + '.log'))
earlystopping = EarlyStoppingByAccVal(monitor='val_accuracy', value=0.9900, verbose=1)
epochs = 40##!!!
lr = 1e-3
decay = lr/epochs
optimizer = Adam(lr=lr, decay=decay)
# train on multiple-gpus
# Create a MirroredStrategy.
strategy = tf.distribute.MirroredStrategy()
print("Number of GPUs: {}".format(strategy.num_replicas_in_sync))
# Open a strategy scope.
with strategy.scope():
# Everything that creates variables should be under the strategy scope.
# In general this is only model construction & `compile()`.
model_mul = get_adv_model()
model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])
step_size_train=ceil(train_set.n/train_set.batch_size)
step_size_valid=ceil(valid_set.n/valid_set.batch_size)
# step_size_test=ceil(testing_set.n//testing_set.batch_size)
# result = model_mul.fit_generator(
# generator = train_set,
# steps_per_epoch = step_size_train,
# validation_data = valid_set,
# validation_steps = step_size_valid,
# shuffle=True,
# epochs=epochs,
# callbacks=[checkpointer],
# # callbacks=[csv_logger, checkpointer, earlystopping],
# # callbacks=[tb, csv_logger, checkpointer, earlystopping],
# verbose=1)
# Training
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import TensorBoard, ModelCheckpoint, EarlyStopping, CSVLogger, ReduceLROnPlateau
from keras.optimizers import Adam
import time, os
from math import ceil
train_datagen = ImageDataGenerator(
# rescale = 1./255,
rotation_range=30,
width_shift_range=0.3,
height_shift_range=0.3,
shear_range=0.3,
zoom_range=0.3,
# horizontal_flip=True,
# vertical_flip=True,##
# brightness_range=[0.5, 1.5],##
channel_shift_range=10,##
fill_mode='nearest',
# preprocessing_function=get_cutout_v2(),
preprocessing_function=preprocess_input,
)
test_datagen = ImageDataGenerator(
# rescale = 1./255
preprocessing_function=preprocess_input
)
NUM_GPU = 1
batch_size = 64
train_set = train_datagen.flow_from_directory('surrey/D/train_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=True,
seed=7,
# subset="training"
)
valid_set = test_datagen.flow_from_directory('surrey/D/test_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=False,
seed=7,
# subset="validation"
)
model_txt = 'st'
# Helper: Save the model.
savedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D.hdf5')
checkpointer = ModelCheckpoint(savedfilename,
monitor='val_accuracy', verbose=1,
save_best_only=True, mode='max',save_weights_only=True)########
# Helper: TensorBoard
tb = TensorBoard(log_dir=os.path.join('svhn_output', 'logs', model_txt))
# Helper: Save results.
timestamp = time.time()
csv_logger = CSVLogger(os.path.join('svhn_output', 'logs', model_txt + '-' + 'training-' + \
str(timestamp) + '.log'))
earlystopping = EarlyStoppingByAccVal(monitor='val_accuracy', value=0.9900, verbose=1)
epochs = 40##!!!
lr = 1e-3
decay = lr/epochs
optimizer = Adam(lr=lr, decay=decay)
# train on multiple-gpus
# Create a MirroredStrategy.
strategy = tf.distribute.MirroredStrategy()
print("Number of GPUs: {}".format(strategy.num_replicas_in_sync))
# Open a strategy scope.
with strategy.scope():
# Everything that creates variables should be under the strategy scope.
# In general this is only model construction & `compile()`.
model_mul = get_adv_model()
model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])
step_size_train=ceil(train_set.n/train_set.batch_size)
step_size_valid=ceil(valid_set.n/valid_set.batch_size)
# step_size_test=ceil(testing_set.n//testing_set.batch_size)
result = model_mul.fit_generator(
generator = train_set,
steps_per_epoch = step_size_train,
validation_data = valid_set,
validation_steps = step_size_valid,
shuffle=True,
epochs=epochs,
callbacks=[checkpointer],
# callbacks=[csv_logger, checkpointer, earlystopping],
# callbacks=[tb, csv_logger, checkpointer, earlystopping],
verbose=1)
ls -l
# Open a strategy scope.
with strategy.scope():
model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D.hdf5'))
model_mul.evaluate(valid_set)
# Helper: Save the model.
savedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L2.hdf5')
checkpointer = ModelCheckpoint(savedfilename,
monitor='val_accuracy', verbose=1,
save_best_only=True, mode='max',save_weights_only=True)########
epochs = 15##!!!
lr = 1e-4
decay = lr/epochs
optimizer = Adam(lr=lr, decay=decay)
# Open a strategy scope.
with strategy.scope():
model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])
result = model_mul.fit_generator(
generator = train_set,
steps_per_epoch = step_size_train,
validation_data = valid_set,
validation_steps = step_size_valid,
shuffle=True,
epochs=epochs,
callbacks=[checkpointer],
# callbacks=[csv_logger, checkpointer, earlystopping],
# callbacks=[tb, csv_logger, checkpointer, earlystopping],
verbose=1)
# Open a strategy scope.
with strategy.scope():
model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L2.hdf5'))
model_mul.evaluate(valid_set)
# Helper: Save the model.
savedfilename = os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L3.hdf5')
checkpointer = ModelCheckpoint(savedfilename,
monitor='val_accuracy', verbose=1,
save_best_only=True, mode='max',save_weights_only=True)########
epochs = 15##!!!
lr = 1e-5
decay = lr/epochs
optimizer = Adam(lr=lr, decay=decay)
# Open a strategy scope.
with strategy.scope():
model_mul.compile(optimizer=optimizer,loss='categorical_crossentropy',metrics=['accuracy'])
result = model_mul.fit_generator(
generator = train_set,
steps_per_epoch = step_size_train,
validation_data = valid_set,
validation_steps = step_size_valid,
shuffle=True,
epochs=epochs,
callbacks=[checkpointer],
# callbacks=[csv_logger, checkpointer, earlystopping],
# callbacks=[tb, csv_logger, checkpointer, earlystopping],
verbose=1)
# Open a strategy scope.
with strategy.scope():
model_mul.load_weights(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', '5_Surrey_MobileNet_D_L3.hdf5'))
model_mul.evaluate(valid_set)
import numpy as np
from keras.preprocessing.image import ImageDataGenerator
import time, os
from math import ceil
# PREDICT ON OFFICIAL TEST
train_datagen = ImageDataGenerator(
# rescale = 1./255,
rotation_range=30,
width_shift_range=0.3,
height_shift_range=0.3,
shear_range=0.3,
zoom_range=0.3,
# horizontal_flip=True,
# vertical_flip=True,##
# brightness_range=[0.5, 1.5],##
channel_shift_range=10,##
fill_mode='nearest',
preprocessing_function=preprocess_input,
)
test_datagen1 = ImageDataGenerator(
# rescale = 1./255,
preprocessing_function=preprocess_input
)
batch_size = 64
train_set = train_datagen.flow_from_directory('surrey/D/train_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=True,
seed=7,
# subset="training"
)
test_set1 = test_datagen1.flow_from_directory('surrey/D/test_color/',
target_size = (224, 224),
batch_size = batch_size,
class_mode = 'categorical',
shuffle=False,
seed=7,
# subset="validation"
)
# if NUM_GPU != 1:
predict1=model_mul.predict_generator(test_set1, steps = ceil(test_set1.n/test_set1.batch_size),verbose=1)
# else:
# predict1=model.predict_generator(test_set1, steps = ceil(test_set1.n/test_set1.batch_size),verbose=1)
predicted_class_indices=np.argmax(predict1,axis=1)
labels = (train_set.class_indices)
labels = dict((v,k) for k,v in labels.items())
predictions1 = [labels[k] for k in predicted_class_indices]
import pandas as pd
filenames=test_set1.filenames
results=pd.DataFrame({"file_name":filenames,
"predicted1":predictions1,
})
results.to_csv('Surrey_MobileNet_D_L3_0902.csv')
results.head()
np.save(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', 'npy', '5Colab_Surrey_MobileNet_D_L2_0902.hdf5'), predict1)
np.save(os.path.join('gdrive', 'My Drive', 'Surrey_ASL', 'npy', '5Colab_Surrey_MobileNet_D_L3_0902.hdf5'), predict1)
from sklearn.metrics import classification_report, confusion_matrix
import numpy as np
test_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input)
testing_set = test_datagen.flow_from_directory('surrey/D/test_color/',
target_size = (224, 224),
batch_size = 32,
class_mode = 'categorical',
seed=7,
shuffle=False
# subset="validation"
)
y_pred = model_mul.predict_generator(testing_set)
y_pred = np.argmax(y_pred, axis=1)
y_true = testing_set.classes
print(confusion_matrix(y_true, y_pred))
# print(model.evaluate_generator(testing_set,
# steps = testing_set.n//testing_set.batch_size))
```
|
github_jupyter
|
## Dependencies
```
# !pip install --quiet efficientnet
!pip install --quiet image-classifiers
import warnings, json, re, glob, math
from scripts_step_lr_schedulers import *
from melanoma_utility_scripts import *
from kaggle_datasets import KaggleDatasets
from sklearn.model_selection import KFold
import tensorflow.keras.layers as L
import tensorflow.keras.backend as K
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tensorflow.keras import optimizers, layers, metrics, losses, Model
# import efficientnet.tfkeras as efn
from classification_models.tfkeras import Classifiers
import tensorflow_addons as tfa
SEED = 0
seed_everything(SEED)
warnings.filterwarnings("ignore")
```
## TPU configuration
```
strategy, tpu = set_up_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
AUTO = tf.data.experimental.AUTOTUNE
```
# Model parameters
```
config = {
"HEIGHT": 256,
"WIDTH": 256,
"CHANNELS": 3,
"BATCH_SIZE": 64,
"EPOCHS": 25,
"LEARNING_RATE": 3e-4,
"ES_PATIENCE": 10,
"N_FOLDS": 5,
"N_USED_FOLDS": 5,
"TTA_STEPS": 25,
"BASE_MODEL": 'seresnet18',
"BASE_MODEL_WEIGHTS": 'imagenet',
"DATASET_PATH": 'melanoma-256x256'
}
with open('config.json', 'w') as json_file:
json.dump(json.loads(json.dumps(config)), json_file)
config
```
# Load data
```
database_base_path = '/kaggle/input/siim-isic-melanoma-classification/'
k_fold = pd.read_csv(database_base_path + 'train.csv')
test = pd.read_csv(database_base_path + 'test.csv')
print('Train samples: %d' % len(k_fold))
display(k_fold.head())
print(f'Test samples: {len(test)}')
display(test.head())
GCS_PATH = KaggleDatasets().get_gcs_path(config['DATASET_PATH'])
TRAINING_FILENAMES = tf.io.gfile.glob(GCS_PATH + '/train*.tfrec')
TEST_FILENAMES = tf.io.gfile.glob(GCS_PATH + '/test*.tfrec')
```
# Augmentations
```
def data_augment(image, label):
p_spatial = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')
p_spatial2 = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')
p_rotate = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')
p_crop = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')
p_pixel = tf.random.uniform([1], minval=0, maxval=1, dtype='float32')
### Spatial-level transforms
if p_spatial >= .2: # flips
image['input_image'] = tf.image.random_flip_left_right(image['input_image'])
image['input_image'] = tf.image.random_flip_up_down(image['input_image'])
if p_spatial >= .7:
image['input_image'] = tf.image.transpose(image['input_image'])
if p_rotate >= .8: # rotate 270º
image['input_image'] = tf.image.rot90(image['input_image'], k=3)
elif p_rotate >= .6: # rotate 180º
image['input_image'] = tf.image.rot90(image['input_image'], k=2)
elif p_rotate >= .4: # rotate 90º
image['input_image'] = tf.image.rot90(image['input_image'], k=1)
if p_spatial2 >= .6:
if p_spatial2 >= .9:
image['input_image'] = transform_rotation(image['input_image'], config['HEIGHT'], 180.)
elif p_spatial2 >= .8:
image['input_image'] = transform_zoom(image['input_image'], config['HEIGHT'], 8., 8.)
elif p_spatial2 >= .7:
image['input_image'] = transform_shift(image['input_image'], config['HEIGHT'], 8., 8.)
else:
image['input_image'] = transform_shear(image['input_image'], config['HEIGHT'], 2.)
if p_crop >= .6: # crops
if p_crop >= .8:
image['input_image'] = tf.image.random_crop(image['input_image'], size=[int(config['HEIGHT']*.8), int(config['WIDTH']*.8), config['CHANNELS']])
elif p_crop >= .7:
image['input_image'] = tf.image.random_crop(image['input_image'], size=[int(config['HEIGHT']*.9), int(config['WIDTH']*.9), config['CHANNELS']])
else:
image['input_image'] = tf.image.central_crop(image['input_image'], central_fraction=.8)
image['input_image'] = tf.image.resize(image['input_image'], size=[config['HEIGHT'], config['WIDTH']])
if p_pixel >= .6: # Pixel-level transforms
if p_pixel >= .9:
image['input_image'] = tf.image.random_hue(image['input_image'], 0.01)
elif p_pixel >= .8:
image['input_image'] = tf.image.random_saturation(image['input_image'], 0.7, 1.3)
elif p_pixel >= .7:
image['input_image'] = tf.image.random_contrast(image['input_image'], 0.8, 1.2)
else:
image['input_image'] = tf.image.random_brightness(image['input_image'], 0.1)
return image, label
```
## Auxiliary functions
```
# Datasets utility functions
def read_labeled_tfrecord(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):
example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)
image = decode_image(example['image'], height, width, channels)
label = tf.cast(example['target'], tf.float32)
# meta features
data = {}
data['patient_id'] = tf.cast(example['patient_id'], tf.int32)
data['sex'] = tf.cast(example['sex'], tf.int32)
data['age_approx'] = tf.cast(example['age_approx'], tf.int32)
data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)
return {'input_image': image, 'input_meta': data}, label # returns a dataset of (image, data, label)
def read_labeled_tfrecord_eval(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):
example = tf.io.parse_single_example(example, LABELED_TFREC_FORMAT)
image = decode_image(example['image'], height, width, channels)
label = tf.cast(example['target'], tf.float32)
image_name = example['image_name']
# meta features
data = {}
data['patient_id'] = tf.cast(example['patient_id'], tf.int32)
data['sex'] = tf.cast(example['sex'], tf.int32)
data['age_approx'] = tf.cast(example['age_approx'], tf.int32)
data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)
return {'input_image': image, 'input_meta': data}, label, image_name # returns a dataset of (image, data, label, image_name)
def load_dataset(filenames, ordered=False, buffer_size=-1):
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False # disable order, increase speed
dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files
dataset = dataset.with_options(ignore_order) # uses data as soon as it streams in, rather than in its original order
dataset = dataset.map(read_labeled_tfrecord, num_parallel_calls=buffer_size)
return dataset # returns a dataset of (image, data, label)
def load_dataset_eval(filenames, buffer_size=-1):
dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files
dataset = dataset.map(read_labeled_tfrecord_eval, num_parallel_calls=buffer_size)
return dataset # returns a dataset of (image, data, label, image_name)
def get_training_dataset(filenames, batch_size, buffer_size=-1):
dataset = load_dataset(filenames, ordered=False, buffer_size=buffer_size)
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.repeat() # the training dataset must repeat for several epochs
dataset = dataset.shuffle(2048)
dataset = dataset.batch(batch_size, drop_remainder=True) # slighly faster with fixed tensor sizes
dataset = dataset.prefetch(buffer_size) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def get_validation_dataset(filenames, ordered=True, repeated=False, batch_size=32, buffer_size=-1):
dataset = load_dataset(filenames, ordered=ordered, buffer_size=buffer_size)
if repeated:
dataset = dataset.repeat()
dataset = dataset.shuffle(2048)
dataset = dataset.batch(batch_size, drop_remainder=repeated)
dataset = dataset.prefetch(buffer_size)
return dataset
def get_eval_dataset(filenames, batch_size=32, buffer_size=-1):
dataset = load_dataset_eval(filenames, buffer_size=buffer_size)
dataset = dataset.batch(batch_size, drop_remainder=False)
dataset = dataset.prefetch(buffer_size)
return dataset
# Test function
def read_unlabeled_tfrecord(example, height=config['HEIGHT'], width=config['WIDTH'], channels=config['CHANNELS']):
example = tf.io.parse_single_example(example, UNLABELED_TFREC_FORMAT)
image = decode_image(example['image'], height, width, channels)
image_name = example['image_name']
# meta features
data = {}
data['patient_id'] = tf.cast(example['patient_id'], tf.int32)
data['sex'] = tf.cast(example['sex'], tf.int32)
data['age_approx'] = tf.cast(example['age_approx'], tf.int32)
data['anatom_site_general_challenge'] = tf.cast(tf.one_hot(example['anatom_site_general_challenge'], 7), tf.int32)
return {'input_image': image, 'input_tabular': data}, image_name # returns a dataset of (image, data, image_name)
def load_dataset_test(filenames, buffer_size=-1):
dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=buffer_size) # automatically interleaves reads from multiple files
dataset = dataset.map(read_unlabeled_tfrecord, num_parallel_calls=buffer_size)
# returns a dataset of (image, data, label, image_name) pairs if labeled=True or (image, data, image_name) pairs if labeled=False
return dataset
def get_test_dataset(filenames, batch_size=32, buffer_size=-1, tta=False):
dataset = load_dataset_test(filenames, buffer_size=buffer_size)
if tta:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.batch(batch_size, drop_remainder=False)
dataset = dataset.prefetch(buffer_size)
return dataset
# Advanced augmentations
def transform_rotation(image, height, rotation):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly rotated
DIM = height
XDIM = DIM%2 #fix for size 331
rotation = rotation * tf.random.normal([1],dtype='float32')
# CONVERT DEGREES TO RADIANS
rotation = math.pi * rotation / 180.
# ROTATION MATRIX
c1 = tf.math.cos(rotation)
s1 = tf.math.sin(rotation)
one = tf.constant([1],dtype='float32')
zero = tf.constant([0],dtype='float32')
rotation_matrix = tf.reshape( tf.concat([c1,s1,zero, -s1,c1,zero, zero,zero,one],axis=0),[3,3] )
# LIST DESTINATION PIXEL INDICES
x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )
y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )
z = tf.ones([DIM*DIM],dtype='int32')
idx = tf.stack( [x,y,z] )
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(rotation_matrix,tf.cast(idx,dtype='float32'))
idx2 = K.cast(idx2,dtype='int32')
idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d,[DIM,DIM,3])
def transform_shear(image, height, shear):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly sheared
DIM = height
XDIM = DIM%2 #fix for size 331
shear = shear * tf.random.normal([1],dtype='float32')
shear = math.pi * shear / 180.
# SHEAR MATRIX
one = tf.constant([1],dtype='float32')
zero = tf.constant([0],dtype='float32')
c2 = tf.math.cos(shear)
s2 = tf.math.sin(shear)
shear_matrix = tf.reshape( tf.concat([one,s2,zero, zero,c2,zero, zero,zero,one],axis=0),[3,3] )
# LIST DESTINATION PIXEL INDICES
x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )
y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )
z = tf.ones([DIM*DIM],dtype='int32')
idx = tf.stack( [x,y,z] )
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(shear_matrix,tf.cast(idx,dtype='float32'))
idx2 = K.cast(idx2,dtype='int32')
idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d,[DIM,DIM,3])
def transform_shift(image, height, h_shift, w_shift):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly shifted
DIM = height
XDIM = DIM%2 #fix for size 331
height_shift = h_shift * tf.random.normal([1],dtype='float32')
width_shift = w_shift * tf.random.normal([1],dtype='float32')
one = tf.constant([1],dtype='float32')
zero = tf.constant([0],dtype='float32')
# SHIFT MATRIX
shift_matrix = tf.reshape( tf.concat([one,zero,height_shift, zero,one,width_shift, zero,zero,one],axis=0),[3,3] )
# LIST DESTINATION PIXEL INDICES
x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )
y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )
z = tf.ones([DIM*DIM],dtype='int32')
idx = tf.stack( [x,y,z] )
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(shift_matrix,tf.cast(idx,dtype='float32'))
idx2 = K.cast(idx2,dtype='int32')
idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d,[DIM,DIM,3])
def transform_zoom(image, height, h_zoom, w_zoom):
# input image - is one image of size [dim,dim,3] not a batch of [b,dim,dim,3]
# output - image randomly zoomed
DIM = height
XDIM = DIM%2 #fix for size 331
height_zoom = 1.0 + tf.random.normal([1],dtype='float32')/h_zoom
width_zoom = 1.0 + tf.random.normal([1],dtype='float32')/w_zoom
one = tf.constant([1],dtype='float32')
zero = tf.constant([0],dtype='float32')
# ZOOM MATRIX
zoom_matrix = tf.reshape( tf.concat([one/height_zoom,zero,zero, zero,one/width_zoom,zero, zero,zero,one],axis=0),[3,3] )
# LIST DESTINATION PIXEL INDICES
x = tf.repeat( tf.range(DIM//2,-DIM//2,-1), DIM )
y = tf.tile( tf.range(-DIM//2,DIM//2),[DIM] )
z = tf.ones([DIM*DIM],dtype='int32')
idx = tf.stack( [x,y,z] )
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(zoom_matrix,tf.cast(idx,dtype='float32'))
idx2 = K.cast(idx2,dtype='int32')
idx2 = K.clip(idx2,-DIM//2+XDIM+1,DIM//2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack( [DIM//2-idx2[0,], DIM//2-1+idx2[1,]] )
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d,[DIM,DIM,3])
```
## Learning rate scheduler
```
lr_min = 1e-6
# lr_start = 0
lr_max = config['LEARNING_RATE']
steps_per_epoch = 24844 // config['BATCH_SIZE']
total_steps = config['EPOCHS'] * steps_per_epoch
warmup_steps = steps_per_epoch * 5
# hold_max_steps = 0
# step_decay = .8
# step_size = steps_per_epoch * 1
# rng = [i for i in range(0, total_steps, 32)]
# y = [step_schedule_with_warmup(tf.cast(x, tf.float32), step_size=step_size,
# warmup_steps=warmup_steps, hold_max_steps=hold_max_steps,
# lr_start=lr_start, lr_max=lr_max, step_decay=step_decay) for x in rng]
# sns.set(style="whitegrid")
# fig, ax = plt.subplots(figsize=(20, 6))
# plt.plot(rng, y)
# print("Learning rate schedule: {:.3g} to {:.3g} to {:.3g}".format(y[0], max(y), y[-1]))
```
# Model
```
# Initial bias
pos = len(k_fold[k_fold['target'] == 1])
neg = len(k_fold[k_fold['target'] == 0])
initial_bias = np.log([pos/neg])
print('Bias')
print(pos)
print(neg)
print(initial_bias)
# class weights
total = len(k_fold)
weight_for_0 = (1 / neg)*(total)/2.0
weight_for_1 = (1 / pos)*(total)/2.0
class_weight = {0: weight_for_0, 1: weight_for_1}
print('Class weight')
print(class_weight)
def model_fn(input_shape):
input_image = L.Input(shape=input_shape, name='input_image')
BaseModel, preprocess_input = Classifiers.get(config['BASE_MODEL'])
base_model = BaseModel(input_shape=input_shape,
weights=config['BASE_MODEL_WEIGHTS'],
include_top=False)
x = base_model(input_image)
x = L.GlobalAveragePooling2D()(x)
output = L.Dense(1, activation='sigmoid', name='output',
bias_initializer=tf.keras.initializers.Constant(initial_bias))(x)
model = Model(inputs=input_image, outputs=output)
return model
```
# Training
```
# Evaluation
eval_dataset = get_eval_dataset(TRAINING_FILENAMES, batch_size=config['BATCH_SIZE'], buffer_size=AUTO)
image_names = next(iter(eval_dataset.unbatch().map(lambda data, label, image_name: image_name).batch(count_data_items(TRAINING_FILENAMES)))).numpy().astype('U')
image_data = eval_dataset.map(lambda data, label, image_name: data)
# Resample dataframe
k_fold = k_fold[k_fold['image_name'].isin(image_names)]
# Test
NUM_TEST_IMAGES = len(test)
test_preds = np.zeros((NUM_TEST_IMAGES, 1))
test_preds_last = np.zeros((NUM_TEST_IMAGES, 1))
test_dataset = get_test_dataset(TEST_FILENAMES, batch_size=config['BATCH_SIZE'], buffer_size=AUTO, tta=True)
image_names_test = next(iter(test_dataset.unbatch().map(lambda data, image_name: image_name).batch(NUM_TEST_IMAGES))).numpy().astype('U')
test_image_data = test_dataset.map(lambda data, image_name: data)
history_list = []
k_fold_best = k_fold.copy()
kfold = KFold(config['N_FOLDS'], shuffle=True, random_state=SEED)
for n_fold, (trn_idx, val_idx) in enumerate(kfold.split(TRAINING_FILENAMES)):
if n_fold < config['N_USED_FOLDS']:
n_fold +=1
print('\nFOLD: %d' % (n_fold))
# tf.tpu.experimental.initialize_tpu_system(tpu)
K.clear_session()
### Data
train_filenames = np.array(TRAINING_FILENAMES)[trn_idx]
valid_filenames = np.array(TRAINING_FILENAMES)[val_idx]
steps_per_epoch = count_data_items(train_filenames) // config['BATCH_SIZE']
# Train model
model_path = f'model_fold_{n_fold}.h5'
es = EarlyStopping(monitor='val_auc', mode='max', patience=config['ES_PATIENCE'],
restore_best_weights=False, verbose=1)
checkpoint = ModelCheckpoint(model_path, monitor='val_auc', mode='max',
save_best_only=True, save_weights_only=True)
with strategy.scope():
model = model_fn((config['HEIGHT'], config['WIDTH'], config['CHANNELS']))
optimizer = tfa.optimizers.RectifiedAdam(lr=lr_max,
total_steps=total_steps,
warmup_proportion=(warmup_steps / total_steps),
min_lr=lr_min)
model.compile(optimizer, loss=losses.BinaryCrossentropy(label_smoothing=0.05),
metrics=[metrics.AUC()])
history = model.fit(get_training_dataset(train_filenames, batch_size=config['BATCH_SIZE'], buffer_size=AUTO),
validation_data=get_validation_dataset(valid_filenames, ordered=True, repeated=False,
batch_size=config['BATCH_SIZE'], buffer_size=AUTO),
epochs=config['EPOCHS'],
steps_per_epoch=steps_per_epoch,
callbacks=[checkpoint, es],
class_weight=class_weight,
verbose=2).history
# save last epoch weights
model.save_weights('last_' + model_path)
history_list.append(history)
# Get validation IDs
valid_dataset = get_eval_dataset(valid_filenames, batch_size=config['BATCH_SIZE'], buffer_size=AUTO)
valid_image_names = next(iter(valid_dataset.unbatch().map(lambda data, label, image_name: image_name).batch(count_data_items(valid_filenames)))).numpy().astype('U')
k_fold[f'fold_{n_fold}'] = k_fold.apply(lambda x: 'validation' if x['image_name'] in valid_image_names else 'train', axis=1)
k_fold_best[f'fold_{n_fold}'] = k_fold_best.apply(lambda x: 'validation' if x['image_name'] in valid_image_names else 'train', axis=1)
##### Last model #####
print('Last model evaluation...')
preds = model.predict(image_data)
name_preds_eval = dict(zip(image_names, preds.reshape(len(preds))))
k_fold[f'pred_fold_{n_fold}'] = k_fold.apply(lambda x: name_preds_eval[x['image_name']], axis=1)
print(f'Last model inference (TTA {config["TTA_STEPS"]} steps)...')
for step in range(config['TTA_STEPS']):
test_preds_last += model.predict(test_image_data)
##### Best model #####
print('Best model evaluation...')
model.load_weights(model_path)
preds = model.predict(image_data)
name_preds_eval = dict(zip(image_names, preds.reshape(len(preds))))
k_fold_best[f'pred_fold_{n_fold}'] = k_fold_best.apply(lambda x: name_preds_eval[x['image_name']], axis=1)
print(f'Best model inference (TTA {config["TTA_STEPS"]} steps)...')
for step in range(config['TTA_STEPS']):
test_preds += model.predict(test_image_data)
# normalize preds
test_preds /= (config['N_USED_FOLDS'] * config['TTA_STEPS'])
test_preds_last /= (config['N_USED_FOLDS'] * config['TTA_STEPS'])
name_preds = dict(zip(image_names_test, test_preds.reshape(NUM_TEST_IMAGES)))
name_preds_last = dict(zip(image_names_test, test_preds_last.reshape(NUM_TEST_IMAGES)))
test['target'] = test.apply(lambda x: name_preds[x['image_name']], axis=1)
test['target_last'] = test.apply(lambda x: name_preds_last[x['image_name']], axis=1)
```
## Model loss graph
```
for n_fold in range(config['N_USED_FOLDS']):
print(f'Fold: {n_fold + 1}')
plot_metrics(history_list[n_fold])
```
## Model loss graph aggregated
```
plot_metrics_agg(history_list, config['N_USED_FOLDS'])
```
# Model evaluation (best)
```
display(evaluate_model(k_fold_best, config['N_USED_FOLDS']).style.applymap(color_map))
display(evaluate_model_Subset(k_fold_best, config['N_USED_FOLDS']).style.applymap(color_map))
```
# Model evaluation (last)
```
display(evaluate_model(k_fold, config['N_USED_FOLDS']).style.applymap(color_map))
display(evaluate_model_Subset(k_fold, config['N_USED_FOLDS']).style.applymap(color_map))
```
# Confusion matrix
```
for n_fold in range(config['N_USED_FOLDS']):
n_fold += 1
pred_col = f'pred_fold_{n_fold}'
train_set = k_fold_best[k_fold_best[f'fold_{n_fold}'] == 'train']
valid_set = k_fold_best[k_fold_best[f'fold_{n_fold}'] == 'validation']
print(f'Fold: {n_fold}')
plot_confusion_matrix(train_set['target'], np.round(train_set[pred_col]),
valid_set['target'], np.round(valid_set[pred_col]))
```
# Visualize predictions
```
k_fold['pred'] = 0
for n_fold in range(config['N_USED_FOLDS']):
k_fold['pred'] += k_fold[f'pred_fold_{n_fold+1}'] / config['N_FOLDS']
print('Label/prediction distribution')
print(f"Train positive labels: {len(k_fold[k_fold['target'] > .5])}")
print(f"Train positive predictions: {len(k_fold[k_fold['pred'] > .5])}")
print(f"Train positive correct predictions: {len(k_fold[(k_fold['target'] > .5) & (k_fold['pred'] > .5)])}")
print('Top 10 samples')
display(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',
'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].head(10))
print('Top 10 positive samples')
display(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',
'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].query('target == 1').head(10))
print('Top 10 predicted positive samples')
display(k_fold[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'diagnosis',
'target', 'pred'] + [c for c in k_fold.columns if (c.startswith('pred_fold'))]].query('pred > .5').head(10))
```
# Visualize test predictions
```
print(f"Test predictions {len(test[test['target'] > .5])}|{len(test[test['target'] <= .5])}")
print(f"Test predictions (last) {len(test[test['target_last'] > .5])}|{len(test[test['target_last'] <= .5])}")
print('Top 10 samples')
display(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] +
[c for c in test.columns if (c.startswith('pred_fold'))]].head(10))
print('Top 10 positive samples')
display(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] +
[c for c in test.columns if (c.startswith('pred_fold'))]].query('target > .5').head(10))
print('Top 10 positive samples (last)')
display(test[['image_name', 'sex', 'age_approx','anatom_site_general_challenge', 'target', 'target_last'] +
[c for c in test.columns if (c.startswith('pred_fold'))]].query('target_last > .5').head(10))
```
# Test set predictions
```
submission = pd.read_csv(database_base_path + 'sample_submission.csv')
submission['target'] = test['target']
submission['target_last'] = test['target_last']
submission['target_blend'] = (test['target'] * .5) + (test['target_last'] * .5)
display(submission.head(10))
display(submission.describe())
### BEST ###
submission[['image_name', 'target']].to_csv('submission.csv', index=False)
### LAST ###
submission_last = submission[['image_name', 'target_last']]
submission_last.columns = ['image_name', 'target']
submission_last.to_csv('submission_last.csv', index=False)
### BLEND ###
submission_blend = submission[['image_name', 'target_blend']]
submission_blend.columns = ['image_name', 'target']
submission_blend.to_csv('submission_blend.csv', index=False)
```
|
github_jupyter
|
```
import pandas as pd
import matplotlib.pyplot as plt
import folium
from folium.plugins import MarkerCluster
%matplotlib inline
australia=pd.read_csv("https://frenzy86.s3.eu-west-2.amazonaws.com/fav/australia_cleaned.csv")
australia.head()
plt.figure(figsize=(18,12))
plt.hist(australia["confidence"],label="Sicurezza Incendi",color="red");
plt.xlabel("Livello di sicurezza degli incendi")
plt.ylabel("Numero di incendi")
plt.title("Grafico numero incendi e Livello di sicurezza")
plt.legend(loc=2);
plt.figure(figsize=(18,12))
plt.scatter(australia["confidence"],australia ["brightness"], label ="Sicurezza Incendi", color="orange");
plt.ylabel("Luminosità a 21 Kelvin")
plt.xlabel('Livello di sicurezza degli incendi')
plt.title("Grafico Livello di sicurezza incendi e la luminosità 21 Kelvin")
plt.legend(loc=2);
plt.figure(figsize=(18,12))
plt.scatter(australia["confidence"],australia ["bright_t31"], label ="Sicurezza Incendi", color="yellow");
plt.ylabel("Luminosità a 31 Kelvin")
plt.xlabel('Livello di sicurezza degli incendi')
plt.title("Grafico Livello di sicurezza incendi e la luminosità 31 Kelvin")
plt.legend(loc=2);
pd.crosstab(australia["sat_Terra"], australia["time_N"]).plot(kind="bar",figsize=(20,10));
plt.title("Rapporto tra gli incendi raccolti dal satellite terrestre in notturni e diurni")
plt.ylabel("N° di incendi riconosciuti dai satelliti")
plt.xlabel("Tipo di fuoco, notturno o diurno");
australia_1 =australia.copy()
australia_1.head()
data=australia_1[(australia_1["confidence"]>= 70)]
data.head()
data.shape
#Creare lista longitudine e latitudine
lat=data["latitude"].values.tolist()
long=data["longitude"].values.tolist()
#Mappa Australia
map1=folium.Map([-25.274398,133.775136],zoom_start=4)
#Creare un cluster di mappa
australia_cluster = MarkerCluster()
for latV,longV in zip(lat,long):
folium.Marker(location=[latV,longV]).add_to(australia_cluster)
#Aggiungere il cluster alla mappa che vogliamo stampare
australia_cluster.add_to(map1);
map1
localizacion=australia_1[(australia_1["frp"]>= 2500)]
localizacion.head()
map_2 = folium.Map([-25.274398,133.775136],zoom_start=4.5,tiles='Stamen Terrain')
lat_2 = localizacion["latitude"].values.tolist()
long_2 = localizacion["longitude"].values.tolist()
australia_cluster_2 = MarkerCluster().add_to(map_2)
for lat_2,long_2 in zip(lat_2,long_2):
folium.Marker([lat_2,long_2]).add_to(australia_cluster_2)
map_2
```
Vuoi conoscere gli incendi divampati dopo il 15 settembre 2019?
```
mes = australia_1[(australia_1["acq_date"]>= "2019-09-15")]
mes.head()
mes.describe()
map_sett = folium.Map([-25.274398,133.775136], zoom_start=4)
lat_3 = mes["latitude"].values.tolist()
long_3 = mes["longitude"].values.tolist()
australia_cluster_3 = MarkerCluster().add_to(map_sett)
for lat_3,long_3 in zip(lat_3,long_3):
folium.Marker([lat_3,long_3]).add_to(australia_cluster_3)
map_sett
```
#Play with Folium
```
44.4807035,11.3712528
import folium
m1 = folium.Map(location=[44.48, 11.37], tiles='openstreetmap', zoom_start=18)
m1.save('map1.html')
m1
m3.save("filename.png")
```
|
github_jupyter
|
```
import numpy as np
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
import tensorflow_probability as tfp
# -- plotting
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rcParams['text.usetex'] = True
mpl.rcParams['font.family'] = 'serif'
mpl.rcParams['axes.linewidth'] = 1.5
mpl.rcParams['axes.xmargin'] = 1
mpl.rcParams['xtick.labelsize'] = 'x-large'
mpl.rcParams['xtick.major.size'] = 5
mpl.rcParams['xtick.major.width'] = 1.5
mpl.rcParams['ytick.labelsize'] = 'x-large'
mpl.rcParams['ytick.major.size'] = 5
mpl.rcParams['ytick.major.width'] = 1.5
mpl.rcParams['legend.frameon'] = False
#import tensorflow as tf
"""
Gumbel Softmax functions borrowed from http://blog.evjang.com/2016/11/tutorial-categorical-variational.html
"""
def sample_gumbel(shape, eps=1e-7):
"""Sample from Gumbel(0, 1)"""
U = tf.random_uniform(shape,minval=0,maxval=1)
return -tf.log(-tf.log(U + eps) + eps)
def gumbel_softmax_sample(logits, temperature):
""" Draw a sample from the Gumbel-Softmax distribution"""
y = logits + sample_gumbel(tf.shape(logits))
return tf.nn.softmax( y / temperature)
def gumbel_softmax(logits, temperature, hard=False):
"""Sample from the Gumbel-Softmax distribution and optionally discretize.
Args:
logits: [batch_size, n_class] unnormalized log-probs
temperature: non-negative scalar
hard: if True, take argmax, but differentiate w.r.t. soft sample y
Returns:
[batch_size,..., n_class] sample from the Gumbel-Softmax distribution.
If hard=True, then the returned sample will be one-hot, otherwise it will
be a probabilitiy distribution that sums to 1 across classes
"""
y = gumbel_softmax_sample(logits, temperature)
if hard:
k = tf.shape(logits)[-1]
#y_hard = tf.cast(tf.one_hot(tf.argmax(y,1),k), y.dtype)
y_hard = tf.cast(tf.equal(y,tf.reduce_max(y,-1,keep_dims=True)),y.dtype)
y = tf.stop_gradient(y_hard - y) + y
return y
d = gumbel_softmax(np.array([np.log(0.5), np.log(0.5)] ), 0.5, hard=True)
tfp.__version__
sess = tf.Session()
_Mmin = tf.get_variable(name='mass', initializer=13.2, dtype=tf.float32)
Mhalo = tf.convert_to_tensor(np.random.uniform(11., 14., 1000), dtype=tf.float32)
siglogm = tf.convert_to_tensor(0.2, dtype=tf.float32)
temperature = 0.5
def Ncen(Mmin):
# mean occupation of centrals
return tf.clip_by_value(0.5 * (1+tf.math.erf((Mhalo - Mmin)/siglogm)),1e-4,1-1e-4)
def hod(Mmin):
p = Ncen(Mmin)
samp = gumbel_softmax(tf.stack([tf.log(p), tf.log(1.-p)],axis=1), temperature, hard=True)
return samp[...,0]
def numden(Mmin):
return tf.reduce_sum(hod(Mmin))
ncen,mh,nh = sess.run([Ncen(12.5), Mhalo, hod(12.5)] )
plt.scatter(mh, (ncen))
plt.xlim(11, 13.5)
plt.scatter(mh, (ncen), c='k')
plt.scatter(mh, nh)
plt.xlim(11., 13.5)
Mmin_true = 12.5
loss = (numden(Mmin_true) - numden(_Mmin))**2
opt = tf.train.AdamOptimizer(learning_rate=0.01)
opt_op = opt.minimize(loss)
sess.run(tf.global_variables_initializer())
losses=[]
masses=[]
for i in range(200):
_,l,m = sess.run([opt_op, loss, _Mmin])
losses.append(l)
masses.append(m)
losses
%pylab inline
plot(losses)
plot(masses)
axhline(Mmin_true, color='r', label='True Mmin')
xlim(0,200)
xlabel('Number of iterations')
ylabel('Mmin')
legend()
```
|
github_jupyter
|
```
import keras
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute
from keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply
from keras.callbacks import ModelCheckpoint, EarlyStopping, Callback
from keras import regularizers
from keras import backend as K
import keras.losses
import tensorflow as tf
#tf.compat.v1.enable_eager_execution()
from tensorflow.python.framework import ops
import isolearn.keras as iso
import numpy as np
import tensorflow as tf
import logging
logging.getLogger('tensorflow').setLevel(logging.ERROR)
import pandas as pd
import os
import pickle
import numpy as np
import random
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
import isolearn.io as isoio
import isolearn.keras as isol
from genesis.visualization import *
from genesis.generator import *
from genesis.predictor import *
from genesis.optimizer import *
from definitions.generator.aparent_deconv_conv_generator_concat_trainmode import load_generator_network
from definitions.predictor.aparent import load_saved_predictor
import sklearn
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from scipy.stats import pearsonr
import seaborn as sns
from matplotlib import colors
from scipy.stats import norm
from genesis.vae import *
def set_seed(seed_value) :
# 1. Set the `PYTHONHASHSEED` environment variable at a fixed value
os.environ['PYTHONHASHSEED']=str(seed_value)
# 2. Set the `python` built-in pseudo-random generator at a fixed value
random.seed(seed_value)
# 3. Set the `numpy` pseudo-random generator at a fixed value
np.random.seed(seed_value)
# 4. Set the `tensorflow` pseudo-random generator at a fixed value
tf.set_random_seed(seed_value)
# 5. Configure a new global `tensorflow` session
session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
K.set_session(sess)
def load_data(data_name, valid_set_size=0.05, test_set_size=0.05, batch_size=32) :
#Load cached dataframe
cached_dict = pickle.load(open(data_name, 'rb'))
plasmid_df = cached_dict['plasmid_df']
plasmid_cuts = cached_dict['plasmid_cuts']
#print("len(plasmid_df) = " + str(len(plasmid_df)) + " (loaded)")
#Generate training and test set indexes
plasmid_index = np.arange(len(plasmid_df), dtype=np.int)
plasmid_train_index = plasmid_index[:-int(len(plasmid_df) * (valid_set_size + test_set_size))]
plasmid_valid_index = plasmid_index[plasmid_train_index.shape[0]:-int(len(plasmid_df) * test_set_size)]
plasmid_test_index = plasmid_index[plasmid_train_index.shape[0] + plasmid_valid_index.shape[0]:]
#print('Training set size = ' + str(plasmid_train_index.shape[0]))
#print('Validation set size = ' + str(plasmid_valid_index.shape[0]))
#print('Test set size = ' + str(plasmid_test_index.shape[0]))
data_gens = {
gen_id : iso.DataGenerator(
idx,
{'df' : plasmid_df},
batch_size=batch_size,
inputs = [
{
'id' : 'seq',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['padded_seq'][180 + 40: 180 + 40 + 81] + "G" * (128-81),
'encoder' : iso.OneHotEncoder(seq_length=128),
'dim' : (1, 128, 4),
'sparsify' : False
}
],
outputs = [
{
'id' : 'dummy_output',
'source_type' : 'zeros',
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [],
shuffle = True if gen_id == 'train' else False
) for gen_id, idx in [('all', plasmid_index), ('train', plasmid_train_index), ('valid', plasmid_valid_index), ('test', plasmid_test_index)]
}
x_train = np.concatenate([data_gens['train'][i][0][0] for i in range(len(data_gens['train']))], axis=0)
x_test = np.concatenate([data_gens['test'][i][0][0] for i in range(len(data_gens['test']))], axis=0)
return x_train, x_test
#Specfiy problem-specific parameters
experiment_suffix = '_strong_vae_very_high_kl_epoch_35_margin_pos_2_lower_fitness'
vae_model_prefix = "vae/saved_models/vae_apa_max_isoform_doubledope_strong_cano_pas_len_128_50_epochs_very_high_kl"
vae_model_suffix = "_epoch_35"#""#
#VAE model path
saved_vae_encoder_model_path = vae_model_prefix + "_encoder" + vae_model_suffix + ".h5"
saved_vae_decoder_model_path = vae_model_prefix + "_decoder" + vae_model_suffix + ".h5"
#Padding for the VAE
vae_upstream_padding = ''
vae_downstream_padding = 'G' * 47
#VAE sequence template
vae_sequence_template = 'ATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCC' + 'G' * (128 - 81)
#VAE latent dim
vae_latent_dim = 100
#Oracle predictor model path
saved_predictor_model_path = '../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5'
#Subtring indices for VAE
vae_pwm_start = 40
vae_pwm_end = 121
#VAE parameter collection
vae_params = [
saved_vae_encoder_model_path,
saved_vae_decoder_model_path,
vae_upstream_padding,
vae_downstream_padding,
vae_latent_dim,
vae_pwm_start,
vae_pwm_end
]
#Load data set
vae_data_path = "vae/apa_doubledope_cached_set_strong_short_cano_pas.pickle"
_, x_test = load_data(vae_data_path, valid_set_size=0.005, test_set_size=0.095)
#Evaluate ELBO distribution on test set
#Load VAE models
vae_encoder_model = load_model(saved_vae_encoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})
vae_decoder_model = load_model(saved_vae_decoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})
#Compute multi-sample ELBO on test set
log_mean_p_vae_test, mean_log_p_vae_test, log_p_vae_test = evaluate_elbo(vae_encoder_model, vae_decoder_model, x_test, n_samples=128)
print("mean log(likelihood) = " + str(mean_log_p_vae_test))
#Log Likelihood Plot
plot_min_val = None
plot_max_val = None
f = plt.figure(figsize=(6, 4))
log_p_vae_test_hist, log_p_vae_test_edges = np.histogram(log_mean_p_vae_test, bins=50, density=True)
bin_width_test = log_p_vae_test_edges[1] - log_p_vae_test_edges[0]
plt.bar(log_p_vae_test_edges[1:] - bin_width_test/2., log_p_vae_test_hist, width=bin_width_test, linewidth=2, edgecolor='black', color='orange')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
if plot_min_val is not None and plot_max_val is not None :
plt.xlim(plot_min_val, plot_max_val)
plt.xlabel("VAE Log Likelihood", fontsize=14)
plt.ylabel("Data Density", fontsize=14)
plt.axvline(x=mean_log_p_vae_test, linewidth=2, color='red', linestyle="--")
plt.tight_layout()
plt.show()
#Evaluate ELBO distribution on test set (training-level no. of samples)
#Load VAE models
vae_encoder_model = load_model(saved_vae_encoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})
vae_decoder_model = load_model(saved_vae_decoder_model_path, custom_objects={'st_sampled_softmax':st_sampled_softmax, 'st_hardmax_softmax':st_hardmax_softmax, 'min_pred':min_pred, 'InstanceNormalization':InstanceNormalization})
#Compute multi-sample ELBO on test set
log_mean_p_vae_test, mean_log_p_vae_test, log_p_vae_test = evaluate_elbo(vae_encoder_model, vae_decoder_model, x_test, n_samples=32)
print("mean log(likelihood) = " + str(mean_log_p_vae_test))
#Log Likelihood Plot
plot_min_val = None
plot_max_val = None
f = plt.figure(figsize=(6, 4))
log_p_vae_test_hist, log_p_vae_test_edges = np.histogram(log_mean_p_vae_test, bins=50, density=True)
bin_width_test = log_p_vae_test_edges[1] - log_p_vae_test_edges[0]
plt.bar(log_p_vae_test_edges[1:] - bin_width_test/2., log_p_vae_test_hist, width=bin_width_test, linewidth=2, edgecolor='black', color='orange')
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
if plot_min_val is not None and plot_max_val is not None :
plt.xlim(plot_min_val, plot_max_val)
plt.xlabel("VAE Log Likelihood", fontsize=14)
plt.ylabel("Data Density", fontsize=14)
plt.axvline(x=mean_log_p_vae_test, linewidth=2, color='red', linestyle="--")
plt.tight_layout()
plt.show()
#Define target isoform loss function
def get_isoform_loss(target_isos, fitness_weight=2., batch_size=32, n_samples=1, n_z_samples=1, mini_batch_size=1, seq_length=205, vae_loss_mode='bound', vae_divergence_weight=1., ref_vae_log_p=-10, vae_log_p_margin=1, decoded_pwm_epsilon=10**-6, pwm_start=0, pwm_end=70, pwm_target_bits=1.8, vae_pwm_start=0, entropy_weight=0.0, entropy_loss_mode='margin', similarity_weight=0.0, similarity_margin=0.5) :
target_iso = np.zeros((len(target_isos), 1))
for i, t_iso in enumerate(target_isos) :
target_iso[i, 0] = t_iso
masked_entropy_mse = get_target_entropy_sme_masked(pwm_start=pwm_start, pwm_end=pwm_end, target_bits=pwm_target_bits)
if entropy_loss_mode == 'margin' :
masked_entropy_mse = get_margin_entropy_ame_masked(pwm_start=pwm_start, pwm_end=pwm_end, min_bits=pwm_target_bits)
pwm_sample_entropy_func = get_pwm_margin_sample_entropy_masked(pwm_start=pwm_start, pwm_end=pwm_end, margin=similarity_margin, shift_1_nt=True)
def loss_func(loss_tensors) :
_, _, _, sequence_class, pwm_logits_1, pwm_logits_2, pwm_1, pwm_2, sampled_pwm_1, sampled_pwm_2, mask, sampled_mask, iso_pred, cut_pred, iso_score_pred, cut_score_pred, vae_pwm_1, vae_sampled_pwm_1, z_mean_1, z_log_var_1, z_1, decoded_pwm_1 = loss_tensors
#Create target isoform with sample axis
iso_targets = K.constant(target_iso)
iso_true = K.gather(iso_targets, sequence_class[:, 0])
iso_true = K.tile(K.expand_dims(iso_true, axis=-1), (1, K.shape(sampled_pwm_1)[1], 1))
#Re-create iso_pred from cut_pred
#iso_pred = K.expand_dims(K.sum(cut_pred[..., 76:76+35], axis=-1), axis=-1)
#Specify costs
iso_loss = fitness_weight * K.mean(symmetric_sigmoid_kl_divergence(iso_true, iso_pred), axis=1)
#Construct VAE sequence inputs
decoded_pwm_1 = K.clip(decoded_pwm_1, decoded_pwm_epsilon, 1. - decoded_pwm_epsilon)
log_p_x_given_z_1 = K.sum(K.sum(vae_sampled_pwm_1[:, :, :, pwm_start-vae_pwm_start:pwm_end-vae_pwm_start, ...] * K.log(K.stop_gradient(decoded_pwm_1[:, :, :, pwm_start-vae_pwm_start:pwm_end-vae_pwm_start, ...])) / K.log(K.constant(10.)), axis=(-1, -2)), axis=-1)
log_p_std_normal_1 = K.sum(normal_log_prob(z_1, 0., 1.) / K.log(K.constant(10.)), axis=-1)
log_p_importance_1 = K.sum(normal_log_prob(z_1, z_mean_1, K.sqrt(K.exp(z_log_var_1))) / K.log(K.constant(10.)), axis=-1)
log_p_vae_1 = log_p_x_given_z_1 + log_p_std_normal_1 - log_p_importance_1
log_p_vae_div_n_1 = log_p_vae_1 - K.log(K.constant(n_z_samples, dtype='float32')) / K.log(K.constant(10.))
#Calculate mean ELBO across samples (log-sum-exp trick)
max_log_p_vae_1 = K.max(log_p_vae_div_n_1, axis=-1)
log_mean_p_vae_1 = max_log_p_vae_1 + K.log(K.sum(10**(log_p_vae_div_n_1 - K.expand_dims(max_log_p_vae_1, axis=-1)), axis=-1)) / K.log(K.constant(10.))
#Specify VAE divergence loss function
vae_divergence_loss = 0.
if vae_loss_mode == 'bound' :
vae_divergence_loss = vae_divergence_weight * K.mean(K.switch(log_mean_p_vae_1 < ref_vae_log_p - vae_log_p_margin, -log_mean_p_vae_1 + (ref_vae_log_p - vae_log_p_margin), K.zeros_like(log_mean_p_vae_1)), axis=1)
elif vae_loss_mode == 'penalty' :
vae_divergence_loss = vae_divergence_weight * K.mean(-log_mean_p_vae_1, axis=1)
elif vae_loss_mode == 'target' :
vae_divergence_loss = vae_divergence_weight * K.mean((log_mean_p_vae_1 - (ref_vae_log_p - vae_log_p_margin))**2, axis=1)
elif 'mini_batch_' in vae_loss_mode :
mini_batch_log_mean_p_vae_1 = K.permute_dimensions(K.reshape(log_mean_p_vae_1, (int(batch_size / mini_batch_size), mini_batch_size, n_samples)), (0, 2, 1))
mini_batch_mean_log_p_vae_1 = K.mean(mini_batch_log_mean_p_vae_1, axis=-1)
tiled_mini_batch_mean_log_p_vae_1 = K.tile(mini_batch_mean_log_p_vae_1, (mini_batch_size, 1))
if vae_loss_mode == 'mini_batch_bound' :
vae_divergence_loss = vae_divergence_weight * K.mean(K.switch(tiled_mini_batch_mean_log_p_vae_1 < ref_vae_log_p - vae_log_p_margin, -tiled_mini_batch_mean_log_p_vae_1 + (ref_vae_log_p - vae_log_p_margin), K.zeros_like(tiled_mini_batch_mean_log_p_vae_1)), axis=1)
elif vae_loss_mode == 'mini_batch_target' :
vae_divergence_loss = vae_divergence_weight * K.mean((tiled_mini_batch_mean_log_p_vae_1 - (ref_vae_log_p - vae_log_p_margin))**2, axis=1)
entropy_loss = entropy_weight * masked_entropy_mse(pwm_1, mask)
entropy_loss += similarity_weight * K.mean(pwm_sample_entropy_func(sampled_pwm_1, sampled_pwm_2, sampled_mask), axis=1)
#Compute total loss
total_loss = iso_loss + entropy_loss + vae_divergence_loss
return total_loss
return loss_func
class EpochVariableCallback(Callback):
def __init__(self, my_variable, my_func):
self.my_variable = my_variable
self.my_func = my_func
def on_epoch_end(self, epoch, logs={}):
K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))
#Function for running GENESIS
def run_genesis(sequence_templates, loss_func, library_contexts, model_path, batch_size=32, n_samples=1, n_z_samples=1, vae_params=None, n_epochs=10, steps_per_epoch=100) :
#Build Generator Network
_, generator = build_generator(batch_size, len(sequence_templates[0]), load_generator_network, n_classes=len(sequence_templates), n_samples=n_samples, sequence_templates=sequence_templates, batch_normalize_pwm=False)
#Build Predictor Network and hook it on the generator PWM output tensor
_, sample_predictor = build_predictor(generator, load_saved_predictor(model_path, library_contexts=library_contexts), batch_size, n_samples=n_samples, eval_mode='sample')
#Build VAE model
vae_tensors = []
if vae_params is not None :
encoder_model_path, decoder_model_path, vae_upstream_padding, vae_downstream_padding, vae_latent_dim, vae_pwm_start, vae_pwm_end = vae_params
vae_tensors = build_vae(generator, encoder_model_path, decoder_model_path, batch_size=batch_size, seq_length=len(sequence_templates[0]), n_samples=n_samples, n_z_samples=n_z_samples, vae_latent_dim=vae_latent_dim, vae_upstream_padding=vae_upstream_padding, vae_downstream_padding=vae_downstream_padding, vae_pwm_start=vae_pwm_start, vae_pwm_end=vae_pwm_end)
#Build Loss Model (In: Generator seed, Out: Loss function)
_, loss_model = build_loss_model(sample_predictor, loss_func, extra_loss_tensors=vae_tensors)
#Specify Optimizer to use
opt = keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999)
#Compile Loss Model (Minimize self)
loss_model.compile(loss=lambda true, pred: pred, optimizer=opt)
#Fit Loss Model
train_history = loss_model.fit(
[], np.ones((1, 1)),
epochs=n_epochs,
steps_per_epoch=steps_per_epoch
)
return generator, sample_predictor, train_history
#Maximize isoform proportion
sequence_templates = [
'CTTCCGATCTCTCGCTCTTTCTATGGCATTCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCTGTCGTCGTGGGTGTCGAAAATGAAATAAAACAAGTCAATTGCGTAGTTTATTCAGACGTACCCCGTGGACCTAC'
]
library_contexts = [
'doubledope'
]
margin_similarities = [
0.5
]
#Generate new random seed
print(np.random.randint(low=0, high=1000000))
#Train APA Cleavage GENESIS Network
print("Training GENESIS")
#Number of PWMs to generate per objective
batch_size = 64
mini_batch_size = 8
#Number of One-hot sequences to sample from the PWM at each grad step
n_samples = 1
#Number of VAE latent vector samples at each grad step
n_z_samples = 32#128#32
#Number of epochs per objective to optimize
n_epochs = 50#10#5#25
#Number of steps (grad updates) per epoch
steps_per_epoch = 50
seed = 104590
for class_i in range(len(sequence_templates)) :
lib_name = library_contexts[class_i].split("_")[0]
print("Library context = " + str(lib_name))
K.clear_session()
set_seed(seed)
loss = get_isoform_loss(
[1.0],
fitness_weight=0.1,#0.5,
batch_size=batch_size,
n_samples=n_samples,
n_z_samples=n_z_samples,
mini_batch_size=mini_batch_size,
seq_length=len(sequence_templates[0]),
vae_loss_mode='mini_batch_bound',#'target',
vae_divergence_weight=40.0 * 1./71.,#5.0 * 1./71.,#0.5 * 1./71.,
ref_vae_log_p=-38.807,
vae_log_p_margin=2.0,
#decoded_pwm_epsilon=0.05,
pwm_start=vae_pwm_start + 5,
pwm_end=vae_pwm_start + 5 + 71,
vae_pwm_start=vae_pwm_start,
pwm_target_bits=1.8,
entropy_weight=0.5,#0.01,
entropy_loss_mode='margin',
similarity_weight=5.0,#0.5,#5.0,
similarity_margin=margin_similarities[class_i]
)
genesis_generator, genesis_predictor, train_history = run_genesis([sequence_templates[class_i]], loss, [library_contexts[class_i]], saved_predictor_model_path, batch_size, n_samples, n_z_samples, vae_params, n_epochs, steps_per_epoch)
genesis_generator.get_layer('lambda_rand_sequence_class').function = lambda inp: inp
genesis_generator.get_layer('lambda_rand_input_1').function = lambda inp: inp
genesis_generator.get_layer('lambda_rand_input_2').function = lambda inp: inp
genesis_predictor.get_layer('lambda_rand_sequence_class').function = lambda inp: inp
genesis_predictor.get_layer('lambda_rand_input_1').function = lambda inp: inp
genesis_predictor.get_layer('lambda_rand_input_2').function = lambda inp: inp
# Save model and weights
save_dir = 'saved_models'
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_name = 'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl_generator.h5'
model_path = os.path.join(save_dir, model_name)
genesis_generator.save(model_path)
print('Saved trained model at %s ' % model_path)
model_name = 'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl_predictor.h5'
model_path = os.path.join(save_dir, model_name)
genesis_predictor.save(model_path)
print('Saved trained model at %s ' % model_path)
#Load GENESIS models and predict sample sequences
lib_name = library_contexts[0].split("_")[0]
batch_size = 64
model_names = [
'genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + '_vae_kl',
]
sequence_templates = [
'CTTCCGATCTCTCGCTCTTTCTATGGCATTCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNAATAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCTGTCGTCGTGGGTGTCGAAAATGAAATAAAACAAGTCAATTGCGTAGTTTATTCAGACGTACCCCGTGGACCTAC'
]
for class_i in range(len(sequence_templates)-1, 0-1, -1) :
save_dir = os.path.join(os.getcwd(), 'saved_models')
model_name = model_names[class_i] + '_predictor.h5'
model_path = os.path.join(save_dir, model_name)
predictor = load_model(model_path, custom_objects={'st_sampled_softmax': st_sampled_softmax, 'st_hardmax_softmax': st_hardmax_softmax})
n = batch_size
sequence_class = np.array([0] * n).reshape(-1, 1) #np.random.uniform(-6, 6, (n, 1)) #
noise_1 = np.random.uniform(-1, 1, (n, 100))
noise_2 = np.random.uniform(-1, 1, (n, 100))
pred_outputs = predictor.predict([sequence_class, noise_1, noise_2], batch_size=batch_size)
_, _, _, optimized_pwm, _, _, _, _, _, iso_pred, cut_pred, _, _ = pred_outputs
#Plot one PWM sequence logo per optimized objective (Experiment 'Punish A-runs')
for pwm_index in range(10) :
sequence_template = sequence_templates[class_i]
pwm = np.expand_dims(optimized_pwm[pwm_index, :, :, 0], axis=0)
cut = np.expand_dims(cut_pred[pwm_index, 0, :], axis=0)
iso = np.expand_dims(np.sum(cut[:, 80: 115], axis=-1), axis=-1)
plot_seqprop_logo(pwm, iso, cut, annotate_peaks='max', sequence_template=sequence_template, figsize=(12, 1.5), width_ratios=[1, 8], logo_height=0.8, usage_unit='fraction', plot_start=70-50, plot_end=76+50, save_figs=False, fig_name='genesis_apa_max_isoform_' + str(lib_name) + experiment_suffix + "_pwm_index_" + str(pwm_index), fig_dpi=150)
```
|
github_jupyter
|
# 工厂规划
等级:中级
## 目的和先决条件
此模型和Factory Planning II都是生产计划问题的示例。在生产计划问题中,必须选择要生产哪些产品,要生产多少产品以及要使用哪些资源,以在满足一系列限制的同时最大化利润或最小化成本。这些问题在广泛的制造环境中都很常见。
### What You Will Learn
在此特定示例中,我们将建模并解决生产组合问题:在每个阶段中,我们可以制造一系列产品。每种产品在不同的机器上生产需要不同的时间,并产生不同的利润。目的是创建最佳的多周期生产计划,以使利润最大化。由于维护,某些机器在特定时期内不可用。由于市场限制,每个产品每个月的销售量都有上限,并且存储容量也受到限制。
In Factory Planning II, we’ll add more complexity to this example; the month in which each machine is down for maintenance will be chosen as a part of the optimized plan.
More information on this type of model can be found in example # 3 of the fifth edition of Modeling Building in Mathematical Programming by H. P. Williams on pages 255-256 and 300-302.
This modeling example is at the intermediate level, where we assume that you know Python and are familiar with the Gurobi Python API. In addition, you should have some knowledge about building mathematical optimization models.
**Note:** You can download the repository containing this and other examples by clicking [here](https://github.com/Gurobi/modeling-examples/archive/master.zip). In order to run this Jupyter Notebook properly, you must have a Gurobi license. If you do not have one, you can request an [evaluation license](https://www.gurobi.com/downloads/request-an-evaluation-license/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=CommercialDataScience) as a *commercial user*, or download a [free license](https://www.gurobi.com/academia/academic-program-and-licenses/?utm_source=Github&utm_medium=website_JupyterME&utm_campaign=AcademicDataScience) as an *academic user*.
---
## Problem Description
A factory makes seven products (Prod 1 to Prod 7) using a range of machines including:
- Four grinders
- Two vertical drills
- Three horizontal drills
- One borer
- One planer
Each product has a defined profit contribution per unit sold (defined as the sales price per unit minus the cost of raw materials). In addition, the manufacturing of each product requires a certain amount of time on each machine (in hours). The contribution and manufacturing time value are shown below. A dash indicates that the manufacturing process for the given product does not require that machine.
| <i></i> | PROD1 | PROD2 | PROD3 | PROD4 | PROD5 | PROD6 | PROD7 |
| --- | --- | --- | --- | --- | --- | --- | --- |
| Profit | 10 | 6 | 8 | 4 | 11 | 9 | 3 |
| Grinding | 0.5 | 0.7 | - | - | 0.3 | 0.2 | 0.5 |
| Vertical Drilling | 0.1 | 0.2 | - | 0.3 | - | 0.6 | - |
| Horizontal Drilling | 0.2 | - | 0.8 | - | - | - | 0.6 |
| Boring | 0.05 | 0.03 | - | 0.07 | 0.1 | - | 0.08 |
| Planning | - | - | 0.01 | - | 0.05 | - | 0.05 |
In each of the six months covered by this model, one or more of the machines is scheduled to be down for maintenance and as a result will not be available to use for production that month. The maintenance schedule is as follows:
| Month | Machine |
| --- | --- |
| January | One grinder |
| February | Two horizontal drills |
| March | One borer |
| April | One vertical drill |
| May | One grinder and one vertical drill |
| June | One horizontal drill |
There are limitations on how many of each product can be sold in a given month. These limits are shown below:
| Month | PROD1 | PROD2 | PROD3 | PROD4 | PROD5 | PROD6 | PROD7 |
| --- | --- | --- | --- | --- | --- | --- | --- |
| January | 500 | 1000 | 300 | 300 | 800 | 200 | 100 |
| February | 600 | 500 | 200 | 0 | 400 | 300 | 150 |
| March | 300 | 600 | 0 | 0 | 500 | 400 | 100 |
| April | 200 | 300 | 400 | 500 | 200 | 0 | 100 |
| May | 0 | 100 | 500 | 100 | 1000 | 300 | 0 |
| June | 500 | 500 | 100 | 300 | 1100 | 500 | 60 |
Up to 100 units of each product may be stored in inventory at a cost of $\$0.50$ per unit per month. At the start of January, there is no product inventory. However, by the end of June, there should be 50 units of each product in inventory.
The factory produces products six days a week using two eight-hour shifts per day. It may be assumed that each month consists of 24 working days. Also, for the purposes of this model, there are no production sequencing issues that need to be taken into account.
What should the production plan look like? Also, is it possible to recommend any price increases and determine the value of acquiring any new machines?
This problem is based on a larger model built for the Cornish engineering company of Holman Brothers.
---
## Model Formulation
### Sets and Indices
$t \in \text{Months}=\{\text{Jan},\text{Feb},\text{Mar},\text{Apr},\text{May},\text{Jun}\}$: Set of months.
$p \in \text{Products}=\{1,2,\dots,7\}$: Set of products.
$m \in \text{Machines}=\{\text{Grinder},\text{VertDrill},\text{horiDrill},\text{Borer},\text{Planer}\}$: Set of machines.
### Parameters
$\text{hours_per_month} \in \mathbb{R}^+$: Time (in hours/month) available at any machine on a monthly basis. It results from multiplying the number of working days (24 days) by the number of shifts per day (2) by the duration of a shift (8 hours).
$\text{max_inventory} \in \mathbb{N}$: Maximum number of units of a single product type that can be stored in inventory at any given month.
$\text{holding_cost} \in \mathbb{R}^+$: Monthly cost (in USD/unit/month) of keeping in inventory a unit of any product type.
$\text{store_target} \in \mathbb{N}$: Number of units of each product type to keep in inventory at the end of the planning horizon.
$\text{profit}_p \in \mathbb{R}^+$: Profit (in USD/unit) of product $p$.
$\text{installed}_m \in \mathbb{N}$: Number of machines of type $m$ installed in the factory.
$\text{down}_{t,m} \in \mathbb{N}$: Number of machines of type $m$ scheduled for maintenance at month $t$.
$\text{time_req}_{m,p} \in \mathbb{R}^+$: Time (in hours/unit) needed on machine $m$ to manufacture one unit of product $p$.
$\text{max_sales}_{t,p} \in \mathbb{N}$: Maximum number of units of product $p$ that can be sold at month $t$.
### Decision Variables
$\text{make}_{t,p} \in \mathbb{R}^+$: Number of units of product $p$ to manufacture at month $t$.
$\text{store}_{t,p} \in [0, \text{max_inventory}] \subset \mathbb{R}^+$: Number of units of product $p$ to store at month $t$.
$\text{sell}_{t,p} \in [0, \text{max_sales}_{t,p}] \subset \mathbb{R}^+$: Number of units of product $p$ to sell at month $t$.
**Assumption:** We can produce fractional units.
### Objective Function
- **Profit:** Maximize the total profit (in USD) of the planning horizon.
\begin{equation}
\text{Maximize} \quad Z = \sum_{t \in \text{Months}}\sum_{p \in \text{Products}}
(\text{profit}_p*\text{make}_{t,p} - \text{holding_cost}*\text{store}_{t,p})
\tag{0}
\end{equation}
### Constraints
- **Initial Balance:** For each product $p$, the number of units produced should be equal to the number of units sold plus the number stored (in units of product).
\begin{equation}
\text{make}_{\text{Jan},p} = \text{sell}_{\text{Jan},p} + \text{store}_{\text{Jan},p} \quad \forall p \in \text{Products}
\tag{1}
\end{equation}
- **Balance:** For each product $p$, the number of units produced in month $t$ and the ones previously stored should be equal to the number of units sold and stored in that month (in units of product).
\begin{equation}
\text{store}_{t-1,p} + \text{make}_{t,p} = \text{sell}_{t,p} + \text{store}_{t,p} \quad \forall (t,p) \in \text{Months} \setminus \{\text{Jan}\} \times \text{Products}
\tag{2}
\end{equation}
- **Inventory Target:** The number of units of product $p$ kept in inventory at the end of the planning horizon should hit the target (in units of product).
\begin{equation}
\text{store}_{\text{Jun},p} = \text{store_target} \quad \forall p \in \text{Products}
\tag{3}
\end{equation}
- **Machine Capacity:** Total time used to manufacture any product at machine type $m$ cannot exceed its monthly capacity (in hours).
\begin{equation}
\sum_{p \in \text{Products}}\text{time_req}_{m,p}*\text{make}_{t,p} \leq \text{hours_per_month}*(\text{installed}_m - \text{down}_{t,m}) \quad \forall (t,m) \in \text{Months} \times \text{Machines}
\tag{4}
\end{equation}
---
## Python Implementation
We import the Gurobi Python Module and other Python libraries.
```
import gurobipy as gp
import numpy as np
import pandas as pd
from gurobipy import GRB
# tested with Python 3.7.0 & Gurobi 9.0
```
### Input Data
We define all the input data of the model.
```
# Parameters
products = ["Prod1", "Prod2", "Prod3", "Prod4", "Prod5", "Prod6", "Prod7"]
machines = ["grinder", "vertDrill", "horiDrill", "borer", "planer"]
months = ["Jan", "Feb", "Mar", "Apr", "May", "Jun"]
profit = {"Prod1":10, "Prod2":6, "Prod3":8, "Prod4":4, "Prod5":11, "Prod6":9, "Prod7":3}
time_req = {
"grinder": { "Prod1": 0.5, "Prod2": 0.7, "Prod5": 0.3,
"Prod6": 0.2, "Prod7": 0.5 },
"vertDrill": { "Prod1": 0.1, "Prod2": 0.2, "Prod4": 0.3,
"Prod6": 0.6 },
"horiDrill": { "Prod1": 0.2, "Prod3": 0.8, "Prod7": 0.6 },
"borer": { "Prod1": 0.05,"Prod2": 0.03,"Prod4": 0.07,
"Prod5": 0.1, "Prod7": 0.08 },
"planer": { "Prod3": 0.01,"Prod5": 0.05,"Prod7": 0.05 }
}
# number of machines down
down = {("Jan","grinder"): 1, ("Feb", "horiDrill"): 2, ("Mar", "borer"): 1,
("Apr", "vertDrill"): 1, ("May", "grinder"): 1, ("May", "vertDrill"): 1,
("Jun", "planer"): 1, ("Jun", "horiDrill"): 1}
# number of each machine available
installed = {"grinder":4, "vertDrill":2, "horiDrill":3, "borer":1, "planer":1}
# market limitation of sells
max_sales = {
("Jan", "Prod1") : 500,
("Jan", "Prod2") : 1000,
("Jan", "Prod3") : 300,
("Jan", "Prod4") : 300,
("Jan", "Prod5") : 800,
("Jan", "Prod6") : 200,
("Jan", "Prod7") : 100,
("Feb", "Prod1") : 600,
("Feb", "Prod2") : 500,
("Feb", "Prod3") : 200,
("Feb", "Prod4") : 0,
("Feb", "Prod5") : 400,
("Feb", "Prod6") : 300,
("Feb", "Prod7") : 150,
("Mar", "Prod1") : 300,
("Mar", "Prod2") : 600,
("Mar", "Prod3") : 0,
("Mar", "Prod4") : 0,
("Mar", "Prod5") : 500,
("Mar", "Prod6") : 400,
("Mar", "Prod7") : 100,
("Apr", "Prod1") : 200,
("Apr", "Prod2") : 300,
("Apr", "Prod3") : 400,
("Apr", "Prod4") : 500,
("Apr", "Prod5") : 200,
("Apr", "Prod6") : 0,
("Apr", "Prod7") : 100,
("May", "Prod1") : 0,
("May", "Prod2") : 100,
("May", "Prod3") : 500,
("May", "Prod4") : 100,
("May", "Prod5") : 1000,
("May", "Prod6") : 300,
("May", "Prod7") : 0,
("Jun", "Prod1") : 500,
("Jun", "Prod2") : 500,
("Jun", "Prod3") : 100,
("Jun", "Prod4") : 300,
("Jun", "Prod5") : 1100,
("Jun", "Prod6") : 500,
("Jun", "Prod7") : 60,
}
holding_cost = 0.5
max_inventory = 100
store_target = 50
hours_per_month = 2*8*24
```
## Model Deployment
We create a model and the variables. For each product (seven kinds of products) and each time period (month), we will create variables for the amount of which products get manufactured, held, and sold. In each month, there is an upper limit on the amount of each product that can be sold. This is due to market limitations.
```
factory = gp.Model('Factory Planning I')
make = factory.addVars(months, products, name="Make") # quantity manufactured
store = factory.addVars(months, products, ub=max_inventory, name="Store") # quantity stored
sell = factory.addVars(months, products, ub=max_sales, name="Sell") # quantity sold
```
Next, we insert the constraints. The balance constraints ensure that the amount of product that is in storage in the prior month plus the amount that gets manufactured equals the amount that is sold and held for each product in the current month. This ensures that all products in the model are manufactured in some month. The initial storage is empty.
```
#1. Initial Balance
Balance0 = factory.addConstrs((make[months[0], product] == sell[months[0], product]
+ store[months[0], product] for product in products), name="Initial_Balance")
#2. Balance
Balance = factory.addConstrs((store[months[months.index(month) -1], product] +
make[month, product] == sell[month, product] + store[month, product]
for product in products for month in months
if month != months[0]), name="Balance")
```
The Inventory Target constraints force that at the end of the last month the storage contains the specified amount of each product.
```
#3. Inventory Target
TargetInv = factory.addConstrs((store[months[-1], product] == store_target for product in products), name="End_Balance")
```
The capacity constraints ensure that, for each month, the time all products require on a certain kind of machine is less than or equal to the available hours for that type of machine in that month multiplied by the number of available machines in that period. Each product requires some machine hours on different machines. Each machine is down in one or more months due to maintenance, so the number and type of available machines varies per month. There can be multiple machines per machine type.
```
#4. Machine Capacity
MachineCap = factory.addConstrs((gp.quicksum(time_req[machine][product] * make[month, product]
for product in time_req[machine])
<= hours_per_month * (installed[machine] - down.get((month, machine), 0))
for machine in machines for month in months),
name = "Capacity")
```
The objective is to maximize the profit of the company, which consists of
the profit for each product minus the cost for storing the unsold products. This can be stated as:
```
#0. Objective Function
obj = gp.quicksum(profit[product] * sell[month, product] - holding_cost * store[month, product]
for month in months for product in products)
factory.setObjective(obj, GRB.MAXIMIZE)
```
Next, we start the optimization and Gurobi finds the optimal solution.
```
factory.optimize()
```
---
## Analysis
The result of the optimization model shows that the maximum profit we can achieve is $\$93,715.18$.
Let's see the solution that achieves that optimal result.
### Production Plan
This plan determines the amount of each product to make at each period of the planning horizon. For example, in February we make 700 units of product Prod1.
```
rows = months.copy()
columns = products.copy()
make_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)
for month, product in make.keys():
if (abs(make[month, product].x) > 1e-6):
make_plan.loc[month, product] = np.round(make[month, product].x, 1)
make_plan
```
### Sales Plan
This plan defines the amount of each product to sell at each period of the planning horizon. For example, in February we sell 600 units of product Prod1.
```
rows = months.copy()
columns = products.copy()
sell_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)
for month, product in sell.keys():
if (abs(sell[month, product].x) > 1e-6):
sell_plan.loc[month, product] = np.round(sell[month, product].x, 1)
sell_plan
```
### Inventory Plan
This plan reflects the amount of product in inventory at the end of each period of the planning horizon. For example, at the end of February we have 100 units of Prod1 in inventory.
```
rows = months.copy()
columns = products.copy()
store_plan = pd.DataFrame(columns=columns, index=rows, data=0.0)
for month, product in store.keys():
if (abs(store[month, product].x) > 1e-6):
store_plan.loc[month, product] = np.round(store[month, product].x, 1)
store_plan
```
**Note:** If you want to write your solution to a file, rather than print it to the terminal, you can use the model.write() command. An example implementation is:
`factory.write("factory-planning-1-output.sol")`
---
## References
H. Paul Williams, Model Building in Mathematical Programming, fifth edition.
Copyright © 2020 Gurobi Optimization, LLC
|
github_jupyter
|
```
import pandas as pd
import matplotlib.pyplot as plt
import datetime as dt
import seaborn as sns
import numpy as np
import matplotlib.dates as mdates
import datetime
#sns.set(color_codes=True)
import matplotlib as mpl
mpl.rcParams['pdf.fonttype'] = 42
import statistics as st
sns.set_style('whitegrid', {'axes.linewidth' : 0.5})
from statsmodels.distributions.empirical_distribution import ECDF
import scipy
import gc
from helpers import *
today_str = dt.datetime.today().strftime('%y%m%d')
def curve(startx, starty, endx, endy):
x1 = np.linspace(0,(endx-startx),100)
x2 = x1+startx
x = x1/(endx-startx)
y = (endy-starty)*(6*x**5-15*x**4+10*x**3)+starty
y = (endy-starty)*(-20*x**7+70*x**6-84*x**5+35*x**4)+starty
return x2, y
curative = pd.read_csv('~/Box/covid_CDPH/2021.07.06 Master Set Data Only_Deidentified.csv', encoding= 'unicode_escape')
curative['patient_symptom_date'] = pd.to_datetime(curative['patient_symptom_date'], errors='coerce')
curative['collection_time'] = pd.to_datetime(curative['collection_time'], errors='coerce')
curative['days'] = (pd.to_datetime(curative['collection_time'], utc=True) - pd.to_datetime(curative['patient_symptom_date'], utc=True)).dt.days
idph = pd.read_csv('~/Box/covid_IDPH/sentinel_surveillance/210706_SS_epic.csv', encoding= 'unicode_escape')
idph['test_date'] = pd.to_datetime(idph['test_date'])
idph['test_time'] = pd.to_datetime(idph['test_time'])
idph['date_symptoms_start'] = pd.to_datetime(idph['date_symptoms_start'])
idph['days'] = (idph['test_date'] - idph['date_symptoms_start']).dt.days
ss_cond = (idph['days'] <= 4) & (idph['days'] >= 0)
pos_cond = (idph['result'] == 'DETECTED') | (idph['result'] == 'POSITIVE') | (idph['result'] == 'Detected')
chi_cond = (idph['test_site_city'] == 'CHICAGO')
zips = pd.read_csv('./data/Chicago_ZIP_codes.txt', header=None)[0].values
idph['chicago'] = idph['pat_zip_code'].apply(lambda x: zip_in_zips(x, zips))
curative['chicago'] = curative['patient_city'] == 'Chicago'
curative_time_frame_cond = (curative['collection_time'] >= pd.to_datetime('9-27-20')) & (curative['collection_time'] <= pd.to_datetime('6-13-21'))
curative_ss = (curative['days'] >= 0) & (curative['days'] <= 4)
curative_symptom = curative['patient_is_symptomatic']
idph_time_frame_cond = (idph['test_date'] >= pd.to_datetime('9-27-20')) & (idph['test_date'] <= pd.to_datetime('6-13-21'))
idph_ss = (idph['days'] >= 0) & (idph['days'] <= 4)
idph_symptom = idph['symptomatic_per_cdc'] == 'Yes'
idph_chicago_site = (idph['test_site'] == 'IDPH COMMUNITY TESTING AUBURN GRESHAM') | (idph['test_site'] == 'IDPH AUBURN GRESHAM COMMUNITY TESTING') | (idph['test_site'] == 'IDPH HARWOOD HEIGHTS COMMUNITY TESTING')
idph_count = np.sum(idph_time_frame_cond & idph_ss & idph['chicago'] & idph_chicago_site)
curative_count = np.sum(curative_time_frame_cond & curative_ss & curative['chicago'])
pos_cond_curative = curative['test_result'] == 'POSITIVE'
curative['positive'] = pos_cond_curative
chi_idph = (idph['test_site_city'] == 'Chicago') | (idph['test_site_city'] == 'CHICAGO')
pos_cond_idph = (idph['result'] == 'DETECTED') | (idph['result'] == 'POSITIVE') | (idph['result'] == 'Detected')
idph['positive'] = pos_cond_idph
print(idph_count)
print(curative_count)
print('Tests collected at sentinel sites in study period: ')
sentinel_sites_total = len(curative[curative_time_frame_cond]) + len(idph[idph_time_frame_cond & idph_chicago_site])
print(sentinel_sites_total)
print('with Chicago residence: ')
chicago_residents = len(curative[curative_time_frame_cond & curative['chicago']]) + \
len(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago']])
print(chicago_residents)
print('with valid symptom date: ')
with_symptom_date = len(curative[curative_time_frame_cond & curative['chicago']].dropna(subset=['days'])) + \
len(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago']].dropna(subset=['days']))
print(with_symptom_date)
print('symptom date 4 or fewer days before test: ')
tot_ss = len(curative[curative_time_frame_cond & curative['chicago'] & curative_ss].dropna(subset=['days'])) + \
len(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago'] & idph_ss].dropna(subset=['days']))
print(tot_ss)
print('and positive: ')
tot_sc = len(curative[curative_time_frame_cond & curative['chicago'] & curative_ss & pos_cond_curative].dropna(subset=['days'])) + \
len(idph[idph_time_frame_cond & idph_chicago_site & idph['chicago'] & idph_ss & pos_cond_idph].dropna(subset=['days']))
print(tot_sc)
h = 10
w = 8
fig = plt.figure(figsize=(w, h))
figh = h-0
figw = w-0
ax = fig.add_axes([0,0,figw/w,figh/h])
stop_location = np.arange(0,5,1)
line_width = 0.05
#ax.set_xlim([-0.05,1.05])
h_padding = 0.15
v_padding = 0.2
line_width = 0.2
line_height = 4.5
midpoint = (v_padding + line_height)/2
tot_height = sentinel_sites_total
ax.fill_between([stop_location[0], stop_location[0]+line_width],
[midpoint+line_height/2]*2,
[midpoint-line_height/2]*2,
color='gold', zorder=15)
#ax.text(x=stop_location[0]+line_width/1.75,
# y=midpoint, s="specimens collected at sentinel sites in study period n = " + "{:,}".format(sentinel_sites_total),
# ha='center', va='center',
# rotation=90, zorder=16, color='k', fontsize=14)
splits = [chicago_residents, with_symptom_date, tot_ss, tot_sc]
d = tot_height
splits_array = np.array(splits)/d
d_t = 1
d_ts = d
d_top = midpoint+line_height/2
d_bot = midpoint-line_height/2
d_x = stop_location[0]
# midpoint = figh/2
include_color_array = ['gold']*(len(splits)-1) + ['blue']
exclude_color_array = ['crimson']*(len(splits)-1) + ['blue']
for s, l_l, s1, include_color, exclude_color in zip(splits_array,
stop_location[1:],
splits,
include_color_array,
exclude_color_array):
t_line = line_height*d_t + v_padding
ax.fill_between([l_l, l_l+line_width],
[midpoint+t_line/2]*2,
[midpoint+t_line/2-line_height*s]*2,
color=include_color, zorder=13)
ax.fill_between([l_l, l_l+line_width],
[midpoint-t_line/2]*2,
[midpoint-t_line/2+line_height*(d_t-s)]*2,
color=exclude_color)
a1 = curve(d_x+line_width, d_bot,
l_l, midpoint-t_line/2)
a2 = curve(d_x+line_width, d_bot+line_height*(d_t-s),
l_l, midpoint-t_line/2+line_height*(d_t-s))
ax.fill_between(a1[0], a1[1], a2[1], color=exclude_color, alpha=0.25, linewidth=0)
ax.text((d_x+l_l+line_width)/2,
midpoint+t_line/2-line_height*(s)/2,
"n = "+"{:,}".format(s1),
ha='center', va='center',
rotation=0, fontsize=14)
ax.text((d_x+l_l+line_width)/2,
midpoint-t_line/2+line_height*(d_t-s)/2,
"n = "+"{:,}".format(d_ts - s1),
ha='center', va='center',
rotation=0, fontsize=14)
a1 = curve(d_x+line_width, d_top,
l_l, midpoint+t_line/2)
a2 = curve(d_x+line_width, d_bot+line_height*(d_t-s),
l_l, midpoint+t_line/2-line_height*s)
ax.fill_between(a1[0], a1[1], a2[1], color=include_color, alpha=0.25, linewidth=0)
d_t = s
d_ts = s1
d_top = midpoint+t_line/2
d_bot = midpoint+t_line/2-line_height*s
d_x = l_l
midpoint = midpoint+t_line/2-line_height*s/2
ax.text(x=stop_location[1]+line_width+0.05, y=0.35, s='not Chicago resident',
ha='left', va='center', fontsize=14)
ax.text(x=stop_location[2]+line_width+0.05, y=2.5, s='no valid date of symptom onset',
ha='left', va='center', fontsize=14)
ax.text(x=stop_location[3]+line_width+0.05, y=4.5, s='symptom onset > 4 days\nbefore specimen collection',
ha='left', va='top', fontsize=14)
ax.text(x=stop_location[4]+line_width+0.05, y=5.02, s=" positive test → sentinel case",
ha='left', va='top', fontsize=14, weight='bold')
ax.text(x=stop_location[4]+line_width+0.05, y=4.75, s=" negative or inconclusive test",
ha='left', va='top', fontsize=14)
ax.text(x=stop_location[0]-0.1,
y=2.5, s="specimens collected at\ntesting sites in study period\nn = " + "{:,}".format(sentinel_sites_total),
ha='right', va='center',
rotation=0, zorder=16, color='k', fontsize=14)
ax.fill_between(x=[2.95, 4 + line_width+0.05], y1=4.55, y2=5.075,
color='black', alpha=0.1, edgecolor='black', linewidth=0, linestyle='dashed', zorder=0)
ax.text(x=3.6, y=5.11, s="sentinel samples",
ha='center', va='bottom', fontsize=14, weight='bold')
ax.grid(False)
ax.axis('off')
fig.savefig('sankey_diagram_' + today_str + '.png', dpi=200, bbox_inches='tight')
fig.savefig('sankey_diagram_' + today_str + '.pdf', bbox_inches='tight')
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/MattFinney/practical_data_science_in_python/blob/main/Session_2_Practical_Data_Science.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/><a>
# Practical Data Science in Python
## Unsupervised Learning: Classifying Spotify Tracks by Genre with $k$-Means Clustering
Authors: Matthew Finney, Paulina Toro Isaza
#### Run this First! (Function Definitions)
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_palette('Set1')
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans
from IPython.display import Audio, Image, clear_output
rs = 123
np.random.seed(rs)
def pca_plot(df, classes=None):
# Scale data for PCA
scaled_df = StandardScaler().fit_transform(df)
# Fit the PCA and extract the first two components
pca_results = PCA().fit_transform(scaled_df)
pca1_scores = pca_results[:,0]
pca2_scores = pca_results[:,1]
# Sort the legend labels
if classes is None:
hue_order = None
n_classes = 0
elif str(classes[0]).isnumeric():
classes = ['Cluster {}'.format(x) for x in classes]
hue_order = sorted(np.unique(classes))
n_classes = np.max(np.unique(classes).shape)
else:
hue_order = sorted(np.unique(classes))
n_classes = np.max(np.unique(classes).shape)
# Plot the first two principal components
plt.figure(figsize=(8.5,8.5))
plt.grid()
sns.scatterplot(pca1_scores, pca2_scores, s=50, hue=classes,
hue_order=hue_order, palette='Set1')
plt.xlabel("Principal Component {}".format(1))
plt.ylabel("Principal Component {}".format(2))
plt.title('Principal Component Plot')
plt.show()
def tracklist_player(track_list, df, header="Track Player"):
action = ''
for track in track_list:
print('{}\nTrack Name: {}\nArtist Name(s): {}'.format(header, df.loc[track,'name'],df.loc[track,'artist']))
try:
display(Image(df.loc[track,'cover_url'], format='jpeg', height=150))
except:
print('No cover art available')
try:
display(Audio(df.loc[track,'preview_url']+'.mp3', autoplay=True))
except:
print('No audio preview available')
print('Press <Enter> for the next track or q then <Enter> to quit: ')
action = input()
clear_output()
if action=='q':
break
print('No more clusters. Goodbye!')
def play_cluster_tracks(track_df, cluster_column="best_cluster"):
for cluster in sorted(track_df[cluster_column].unique()):
# Get the tracks in the cluster, and shuffle them for variety
tracks_list = track_df[track_df[cluster_column] == cluster].index.values
np.random.shuffle(tracks_list)
# Instantiate a tracklist player
tracklist_player(tracks_list, df=track_df, header='{}'.format(cluster))
# Load Track DataFrame
path = 'https://raw.githubusercontent.com/MattFinney/practical_data_science_in_python/main/spotify_track_data.csv'
tracks_df = pd.read_csv(path)
# Columns from the track dataframe which are relevant for our analysis
audio_feature_cols = ['danceability', 'energy', 'key', 'loudness', 'mode',
'speechiness', 'acousticness', 'instrumentalness',
'liveness', 'valence', 'tempo', 'duration_ms',
'time_signature']
# Show the first five rows of our dataframe
tracks_df.head()
```
## Recap from Session 1
In our earlier session, we started working with a dataset of Spotify tracks. We explored the variables in the dataset, and determined that audio features - like danceability, accousticness, and tempo - vary across the songs in our dataset and might help us to thoughtfully group the tracks into different playlists. We then used Principal Component Analysis (PCA), a dimensionality reduction technique, to visualize the variation in songs.
We'll pick up where we left off, with the PCA plot from last time. If you're just joining us for Session 2, don't fret! Attending Session 1 is NOT a prerequisite to learn and have fun in Session 2 today!
```
# Plot the principal component analysis results
pca_plot(tracks_df[audio_feature_cols])
```
## Today: Classification using $k$-Means Clustering
Our Principal Component Analysis in the first session helped us to visualize the variation of track audio features in just two dimensions. Looking at the scatterplot of the first two principal components above, we can see that there are a few different groups of tracks. But how do we mathematically separate the tracks into these meaningful groups?
One way to separate the tracks into meaningful groups based on similar audio features is to use clustering. Clustering is a machine learning technique that is very powerful for identifying patterns in unlabeled data where the ground truth is not known.
### What is $k$-Means Clustering?
$k$-Means Clustering is one of the most popular clustering algorithms. The algorithm assigns each data point to a cluster using four main steps.
**Step 1: Initialize the Clusters**\
Based on the user's desired number of clusters $k$, the algorithm randomly chooses a centroid for each cluster. In this example, we choose a $k=3$, therefore the algorithm randomly picks 3 centroids.

**Step 2: Assign Each Data Point**\
The algorithm assigns each point to the closest centroid to get $k$ initial clusters.

**Step 3: Recompute the Cluster Centers**\
For every cluster, the algorithm recomputes the centroid by taking the average of all points in the cluster. The changes in centroids are shown below by arrows.

**Step 4: Reassign the Points**\
Since the centroids change, the algorithm then re-assigns the points to the closest centroid. The image below shows the new clusters after re-assignment.

The algorithm repeats the calculation of centroids and assignment of points until points stop changing clusters. When clustering large datasets, you stop the algorithm before reaching convergence, using other criteria instead.
*Note: Some content in this section was [adapted](https://creativecommons.org/licenses/by/4.0/) from Google's free [Clustering in Machine Learning](https://developers.google.com/machine-learning/clustering) course. The course is a great resource if you want to explore clustering in more detail!*
### Cluster the Spotify Tracks using their Audio Features
Now, we will use the `sklearn.cluster.KMeans` Python library to apply the $k$-means algorithm to our `tracks_df` data. Based on our visual inspection of the PCA plot, let's start with a guess k=3 to get 3 clusters.
```
initial_k = ____
# Scale the data, so that the units of features don't impact feature importance
scaled_df = StandardScaler().fit_transform(tracks_df[audio_feature_cols])
# Cluster the data using the k means algorithm
initial_cluster_results = ______(n_clusters=initial_k, n_init=25, random_state=rs).fit(scaled_df)
```
Now, let's print the cluster results. Notice that we're given a number (0 or 1) for each observation in our data set. This number is the id of the cluster assigned to each track.
```
# Print the cluster results
print(initial_cluster_results._______)
```
And let's save the cluster results in our `tracks_df` dataframe as a column named `initial_cluster` so we can access them later.
```
# Save the cluster labels in our dataframe
tracks_df[______________] = ['Cluster ' + str(i) for i in __________.______]
```
Let's plot the PCA plot and color each observation based on the assigned cluster to visualize our $k$-means results.
```
# Show a PCA plot of the clusters
pca_plot(tracks_df[audio_feature_cols], classes=tracks_df['initial_cluster'])
```
Does it look like our $k$-means algorithm correctly separated the tracks into clusters? Does each color map to a distinct group of points?
### How do our clusters of songs differ?
One way we can evaluate our clusters is by looking how the distribution of each data feature varies by cluster. In our case, let's check to see if tracks in the different clusters tend to have different values of energy, loudness, or speechiness.
```
# Plot the distribution of audio features by cluster
g = sns.pairplot(tracks_df, hue="initial_cluster",
vars=['danceability', 'energy', 'loudness', 'speechiness', 'tempo'],
hue_order=sorted(tracks_df.initial_cluster.unique()), palette='Set1')
g.fig.suptitle('Distribution of Audio Features by Cluster', y=1.05)
plt.show()
```
### Experiment with different values of $k$
Use the slider to select different values of $k$, then run the cell below to see how the choice of the number of clusters affects our results.
```
trial_k = 10 #@param {type:"slider", min:1, max:10, step:1}
# Cluster the data using the k means algorithm
trial_cluster_results = KMeans(n_clusters=trial_k, n_init=25, random_state=rs).fit(scaled_df)
# Save the cluster labels in our dataframe
tracks_df['trial_cluster'] = ['Cluster ' + str(i) for i in trial_cluster_results.labels_]
# Show a PCA plot of the clusters
pca_plot(tracks_df[audio_feature_cols], classes=tracks_df['trial_cluster'])
# Plot the distribution of audio features by cluster
g = sns.pairplot(tracks_df, hue="trial_cluster",
vars=['danceability', 'energy', 'loudness', 'speechiness', 'tempo'],
hue_order=sorted(tracks_df.trial_cluster.unique()), palette='Set1')
g.fig.suptitle('Distribution of Audio Features by Cluster', y=1.05)
plt.show()
```
### Which value of $k$ works best for our data?
You may have noticed that the $k$-means algorithm requires you to choose $k$ and decide the number of clusters before you run the algorithm. But how do we know which value of $k$ is the best fit for our data?
One approach is to track the total distance from points to their cluster centroid as we increase the number of clusters, $k$. Usually, the total distance decreases as we increase $k$, but we reach a value of $k$ where increasing $k$ only marginally decreases the total distance. An elbow plot helps us to find that value of $k$; it's the value of $k$ where the slope of the line in the elbow plot crosses the threshold of slope $=-1$. When you plot distance vs $k$, this point often looks like an "elbow".
Let's build an elbow plot to select the value of $k$ that will give us the highest quality clusters that best explain the variation in our data.
```
# Calculate the Total Distance for each value of k between 1 and 10
scores = []
k_list = np.arange(____,____)
for i in k_list:
fit_k = _____(n_clusters=i, n_init=5, random_state=rs).fit(scaled_df)
scores.append(fit_k.inertia_)
# Plot this in an elbow plot
plt.figure(figsize=(11,8.5))
sns.lineplot(______, ______)
plt.xlabel('Number of clusters $k$')
plt.ylabel('Total Point to Centroid Distance')
plt.grid()
plt.title('The Elbow Method showing the optimal $k$')
plt.show()
```
Do you see the "elbow"? At what value of $k$ does it occur?
### Evaluate the results of our clustering algorithm for the best $k$
Use the slider below to choose the "best" $k$ that you determined from looking at the elbow plot. Evaluate the results in the PCA plot. Does this look like a good value of $k$ to separate the data into meaningful clusters?
```
best_k = 1 #@param {type:"slider", min:1, max:10, step:1}
# Cluster the data using the k means algorithm
best_cluster_results = KMeans(n_clusters=best_k, n_init=25, random_state=rs).fit(scaled_df)
# Save the cluster labels in our dataframe
tracks_df['best_cluster'] = ['Cluster ' + str(i) for i in best_cluster_results.labels_]
# Show a PCA plot of the clusters
pca_plot(tracks_df[audio_feature_cols], classes=tracks_df['best_cluster'])
```
## How did we do?
In addition to the mathematical ways to validate the selection of the best $k$ parameter for our model and the quality of our resulting clusters, there's another very important way to evaluate our results: listening to the tracks!
Let's listen to the tracks in each cluster! What do you notice about the attributes that tracks in each cluster have in common? What do you notice about how the clusters are different? What makes each cluster unique?
```
play_cluster_tracks(tracks_df, cluster_column='best_cluster')
```
## Wrap Up and Next Session
That's a wrap! Now that you've learned some practical skills in data science, please join us tomorrow afternoon for the third and final session in our series, where we'll talk about how to continue your studies and/or pursue a career in Data Science!
**Making Your Next Professional Play in Data Science**\
Friday, October 2 | 11:30am - 12:45pm PT\
[https://sched.co/dtqZ](https://sched.co/dtqZ)
|
github_jupyter
|
# Python是什么?
### Python是一种高级的多用途编程语言,广泛用于各种非技术和技术领域。Python是一种具备动态语义、面向对象的解释型高级编程语言。它的高级内建数据结构和动态类型及动态绑定相结合,使其在快速应用开发上极具吸引力,也适合于作为脚本或者“粘合剂”语言,将现有组件连接起来。Python简单、易学的语法强调可读性,因此可以降低程序维护成本。Python支持模块和软件包,鼓励模块化的代码重用。
```
print('hellow world')
```
## Python简史
### 1989,为了度过圣诞假期,Guido开始编写Python语言编译器。Python这个名字来自Guido的喜爱的电视连续剧《蒙蒂蟒蛇的飞行马戏团》。他希望新的语言Python能够满足他在C和Shell之间创建全功能、易学、可扩展的语言的愿景。
### 1989年由荷兰人Guido van Rossum于1989年发明,第一个公开发行版发行于1991年
### Granddaddy of Python web frameworks, Zope 1 was released in 1999
### Python 1.0 - January 1994 增加了 lambda, map, filter and reduce.
### Python 2.0 - October 16, 2000,加入了内存回收机制,构成了现在Python语言框架的基础
### Python 2.4 - November 30, 2004, 同年目前最流行的WEB框架Django 诞生
### Python 2.5 - September 19, 2006
### Python 2.6 - October 1, 2008
### Python 2.7 - July 3, 2010
### Python 3.0 - December 3, 2008
### Python 3.1 - June 27, 2009
### Python 3.2 - February 20, 2011
### Python 3.3 - September 29, 2012
### Python 3.4 - March 16, 2014
### Python 3.5 - September 13, 2015
### Python 3.6 - December 23, 2016
### Python 3.7 - June 15, 2018
## Python的主要运用领域有:
### 云计算:云计算最热的语言,典型的应用OpenStack
### WEB开发:许多优秀的WEB框架,许多大型网站是Python开发、YouTube、Dropbox、Douban……典型的Web框架包括Django
### 科学计算和人工智能:典型的图书馆NumPy、SciPy、Matplotlib、Enided图书馆、熊猫
### 系统操作和维护:操作和维护人员的基本语言
### 金融:定量交易、金融分析,在金融工程领域,Python不仅使用最多,而且使用最多,其重要性逐年增加。

## Python在一些公司的运用有:
### 谷歌:谷歌应用程序引擎,代码。谷歌。com、Google.、Google爬虫、Google广告和其他项目正在广泛使用Python。
### CIA:美国中情局网站是用Python开发的
### NASA:美国航天局广泛使用Python进行数据分析和计算
### YouTube:世界上最大的视频网站YouTube是用Python开发的。
### Dropbox:美国最大的在线云存储网站,全部用Python实现,每天处理10亿的文件上传和下载。
### Instagram:美国最大的照片共享社交网站,每天有3000多万张照片被共享,所有这些都是用Python开发的
### Facebook:大量的基本库是通过Python实现的
### Red.:世界上最流行的Linux发行版中的Yum包管理工具是用Python开发的
### Douban:几乎所有公司的业务都是通过Python开发的。
### 知识:中国最大的Q&A社区,通过Python开发(国外Quora)
### 除此之外,还有搜狐、金山、腾讯、盛大、网易、百度、阿里、淘宝、土豆、新浪、果壳等公司正在使用Python来完成各种任务。
## Python有如下特征:
### 1. 开放源码:Python和大部分可用的支持库及工具都是开源的,通常使用相当灵活和开放的许可证。
### 2. 多重范型:Python支持不同的编程和实现范型,例如面向对象和命令式/函数式或者过程式编程。
### 3. 多用途:Python可以用用于快速、交互式代码开发,也可以用于构建大型应用程序;它可以用于低级系统操作,也可以承担高级分析任务。
### 4. 跨平台:Python可用于大部分重要的操作系统,如Windows、Linux和Mac OS;它用于构建桌面应用和Web应用。
### 5. 运行速度慢:这里是指与C和C++相比。
## Python 常用标准库
### math模块为浮点运算提供了对底层C函数库的访问:
```
import math
print(math.pi)
print(math.log(1024, 2))
```
### random提供了生成随机数的工具。
```
import random
print(random.choice(['apple', 'pear', 'banana']))
print(random.random())
```
### datetime模块为日期和时间处理同时提供了简单和复杂的方法。
```
from datetime import date
now = date.today()
birthday = date(1999, 8, 20)
age = now - birthday
print(age.days)
```
### Numpy是高性能科学计算和数据分析的基础包。
### Pandas 纳入了大量库和一些标准的数据模型,提供了高效地操作大型数据集所需的工具。
### Statismodels是一个Python模块,它提供对许多不同统计模型估计的类和函数,并且可以进行统计测试和统计数据的探索。
### matplotlib一个绘制数据图的库。对于数据科学家或分析师非常有用。
### 更多https://docs.python.org/zh-cn/3/library/
# 基础架构工具
## Anaconda安装
https://www.anaconda.com/products/individual
## Spyder使用
## GitHub创建与使用
### GitHub 是一个面向开源及私有软件项目的托管平台,因为只支持 Git 作为唯一的版本库格式进行托管,故名 GitHub。 GitHub 于 2008 年 4 月 10 日正式上线,除了 Git 代码仓库托管及基本的 Web 管理界面以外,还提供了订阅、讨论组、文本渲染、在线文件编辑器、协作图谱(报表)、代码片段分享(Gist)等功能。目前,其注册用户已经超过350万,托管版本数量也是非常之多,其中不乏知名开源项目 Ruby on Rails、jQuery、python 等。GitHub 去年为漏洞支付了 16.6 万美元赏金。 2018年6月,GitHub被微软以75亿美元的价格收购。https://github.com/
# Python基础语法
```
print ("Hello, Python!")
```
## 行和缩进
### python 最具特色的就是用缩进来写模块。
### 缩进的空白数量是可变的,但是所有代码块语句必须包含相同的缩进空白数量,这个必须严格执行。
### 以下实例缩进为四个空格:
```
if 1>2:
print ("True")
else:
print ("False")
if True:
print ("Answer")
print ("True")
else:
print ("Answer")
# 没有严格缩进,在执行时会报错
print ("False")
```
## 多行语句
### Python语句中一般以新行作为语句的结束符。
### 但是我们可以使用斜杠( \)将一行的语句分为多行显示,如下所示:
```
total = 1 + \
2 + \
3
print(total)
```
### 语句中包含 [], {} 或 () 括号就不需要使用多行连接符。如下实例:
```
days = ['Monday', 'Tuesday', 'Wednesday',
'Thursday', 'Friday']
print(days)
```
## Python 引号
### Python 可以使用引号( ' )、双引号( " )、三引号( ''' 或 """ ) 来表示字符串,引号的开始与结束必须是相同类型的。
### 其中三引号可以由多行组成,编写多行文本的快捷语法,常用于文档字符串,在文件的特定地点,被当做注释。
```
word = 'word'
sentence = "这是一个句子。"
paragraph = """这是一个段落。
包含了多个语句"""
print(word)
print(sentence)
print(paragraph)
```
## Python注释
### python中单行注释采用 # 开头。
```
# 第一个注释
print ("Hello, Python!") # 第二个注释
```
# Python变量类型
## 标准数据类型
### 在内存中存储的数据可以有多种类型。
### 例如,一个人的年龄可以用数字来存储,他的名字可以用字符来存储。
### Python 定义了一些标准类型,用于存储各种类型的数据。
### Python有五个标准的数据类型:
### Numbers(数字)
### String(字符串)
### List(列表)
### Tuple(元组)
### Dictionary(字典)
## Python数字
### Python支持三种不同的数字类型:
### int(有符号整型)
### float(浮点型)
### complex(复数)
```
int1 = 1
float2 = 2.0
complex3 = 1+2j
print(type(int1),type(float2),type(complex3))
```
## Python字符串
### 字符串或串(String)是由数字、字母、下划线组成的一串字符。
```
st = '123asd_'
st1 = st[0:3]
print(st)
print(st1)
```
## Python列表
### List(列表) 是 Python 中使用最频繁的数据类型。
### 列表可以完成大多数集合类的数据结构实现。它支持字符,数字,字符串甚至可以包含列表(即嵌套)。
### 列表用 [ ] 标识,是 python 最通用的复合数据类型。
```
list1 = [ 'runoob', 786 , 2.23, 'john', 70.2 ]
tinylist = [123, 'john']
print (list1) # 输出完整列表
print (list1[0]) # 输出列表的第一个元素
print (list1[1:3]) # 输出第二个至第三个元素
print (list1[2:]) # 输出从第三个开始至列表末尾的所有元素
print (tinylist * 2) # 输出列表两次
print (list1 + tinylist) # 打印组合的列表
list1[0] = 0
print(list1)
```
## Python元组
### 元组是另一个数据类型,类似于 List(列表)。
### 元组用 () 标识。内部元素用逗号隔开。但是元组不能二次赋值,相当于只读列表。
```
tuple1 = ( 'runoob', 786 , 2.23, 'john', 70.2 )
tinytuple = (123, 'john')
print(tuple1[0])
print(tuple1+tinytuple)
```
## Python 字典
### 字典(dictionary)是除列表以外python之中最灵活的内置数据结构类型。列表是有序的对象集合,字典是无序的对象集合。
### 两者之间的区别在于:字典当中的元素是通过键来存取的,而不是通过偏移存取。
### 字典用"{ }"标识。字典由索引(key)和它对应的值value组成。
```
dict1 = {}
dict1['one'] = "This is one"
tinydict = {'name': 'john','code':6734, 'dept': 'sales'}
print(tinydict['name'])
print(dict1)
print(tinydict.keys())
print(tinydict.values())
```
# Python运算符
## Python算术运算符
```
a = 21
b = 10
c = 0
c = a + b
print ("c 的值为:", c)
c = a - b
print ("c 的值为:", c)
c = a * b
print ("c 的值为:", c)
c = a / b
print ("c 的值为:", c)
c = a % b #取余数
print ("c 的值为:", c)
# 修改变量 a 、b 、c
a = 2
b = 3
c = a**b
print ("c 的值为:", c)
a = 10
b = 5
c = a//b #取整
print ("c 的值为:", c)
```
## Python比较运算符
```
a = 21
b = 10
c = 0
if a == b :
print ("a 等于 b")
else:
print ("a 不等于 b")
if a != b :
print ("a 不等于 b")
else:
print ("a 等于 b")
if a < b :
print ("a 小于 b")
else:
print ("a 大于等于 b")
if a > b :
print ("a 大于 b")
else:
print ("a 小于等于 b")
# 修改变量 a 和 b 的值
a = 5
b = 20
if a <= b :
print ("a 小于等于 b")
else:
print ("a 大于 b")
if b >= a :
print ("b 大于等于 a")
else:
print ("b 小于 a")
```
## Python逻辑运算符
```
a = True
b = False
if a and b :
print ("变量 a 和 b 都为 true")
else:
print ("变量 a 和 b 有一个不为 true")
if a or b :
print ("变量 a 和 b 都为 true,或其中一个变量为 true")
else:
print ("变量 a 和 b 都不为 true")
if not( a and b ):
print ("变量 a 和 b 都为 false,或其中一个变量为 false")
else:
print ("变量 a 和 b 都为 true")
```
## Python赋值运算符
```
a = 21
b = 10
c = 0
c = a + b
print ("c 的值为:", c)
c += a
print ("c 的值为:", c)
c *= a
print ("c 的值为:", c)
c /= a
print ("c 的值为:", c)
c = 2
c %= a
print ("c 的值为:", c)
c **= a
print ("c 的值为:", c)
c //= a
print ("c 的值为:", c)
```
# Python 条件语句
### Python条件语句是通过一条或多条语句的执行结果(True或者False)来决定执行的代码块。
```
flag = False
name = 'luren'
if name == 'python': # 判断变量是否为 python
flag = True # 条件成立时设置标志为真
print ('welcome boss') # 并输出欢迎信息
else:
print (name) # 条件不成立时输出变量名称
num = 5
if num == 3: # 判断num的值
print ('boss')
elif num == 2:
print ('user')
elif num == 1:
print ('worker')
elif num < 0: # 值小于零时输出
print ('error')
else:
print ('roadman') # 条件均不成立时输出
num = 9
if num >= 0 and num <= 10: # 判断值是否在0~10之间
print ('hello')
num = 10
if num < 0 or num > 10: # 判断值是否在小于0或大于10
print ('hello')
else:
print ('undefine')
num = 8
# 判断值是否在0~5或者10~15之间
if (num >= 0 and num <= 5) or (num >= 10 and num <= 15):
print ('hello')
else:
print ('undefine')
```
# Python循环语句
## Python 提供了 for 循环和 while 循环
## Python While 循环语句
### Python 编程中 while 语句用于循环执行程序,即在某条件下,循环执行某段程序,以处理需要重复处理的相同任务。
```
count = 0
while (count < 9):
print ('The count is:', count)
count = count + 1
print ("Good bye!")
count = 0
while count < 5:
print (count, " is less than 5")
count = count + 1
else:
print (count, " is not less than 5")
```
## Python for 循环语句
```
fruits = ['banana', 'apple', 'mango']
for index in range(len(fruits)):
print ('当前水果 :', fruits[index])
print ("Good bye!")
```
## Python 循环嵌套
```
num=[];
i=2
for i in range(2,100):
j=2
for j in range(2,i):
if(i%j==0):
break
else:
num.append(i)
print(num)
print ("Good bye!")
```
## Python break 语句
```
for letter in 'Python':
if letter == 'h':
break
print ('当前字母 :', letter)
```
## Python continue 语句
### Python continue 语句跳出本次循环,而break跳出整个循环。
```
for letter in 'Python':
if letter == 'h':
continue
print ('当前字母 :', letter)
```
## Python pass 语句
### Python pass 是空语句,是为了保持程序结构的完整性。
### pass 不做任何事情,一般用做占位语句。
```
# 输出 Python 的每个字母
for letter in 'Python':
if letter == 'h':
pass
print ('这是 pass 块')
print ('当前字母 :', letter)
print ("Good bye!")
```
# Python应用实例(链家二手房数据分析)
## 一、根据上海的部分二手房信息,从多角度进行观察和分析房价与哪些因素有关以及房屋不同状况所占比例
## 二、先对数据进行预处理、构造预测房价的模型、并输入参数对房价进行预测
备注:数据来源CSDN下载。上海链家二手房.csv.因文件读入问题,改名为sh.csv
## 一、导入数据 对数据进行一些简单的预处理
```
#导入需要用到的包
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
from IPython.display import display
sns.set_style({'font.sans-serif':['simhei','Arial']})
%matplotlib inline
shanghai=pd.read_csv('sh.csv')# 将已有数据导进来
shanghai.head(n=1)#显示第一行数据 查看数据是否导入成功
```
### 每项数据类型均为object 不方便处理,需要对一些项删除单位转换为int或者float类型
### 有些列冗余 像house_img需要删除
### 有些列 如何house_desc包含多种信息 需要逐个提出来单独处理
```
shanghai.describe()
# 检查缺失值情况
shanghai.info()
#np.isnan(shanghai).any()
shanghai.dropna(inplace=True)
#数据处理 删除带有NAN项的行
df=shanghai.copy()
house_desc=df['house_desc']
house_desc[0]
```
### house_desc 中带有 室厅的信息 房子面积 楼层 朝向信息 需要分别提出来当一列 下面进行提取
```
df['layout']=df['house_desc'].map(lambda x:x.split('|')[0])
df['area']=df['house_desc'].map(lambda x:x.split('|')[1])
df['temp']=df['house_desc'].map(lambda x:x.split('|')[2])
#df['Dirextion']=df['house_desc'].map(lambda x:x.split('|')[3])
df['floor']=df['temp'].map(lambda x:x.split('/')[0])
df.head(n=1)
```
### 一些列中带有单位 不利于后期处理 去掉单位 并把数据类型转换为float或int
```
df['area']=df['area'].apply(lambda x:x.rstrip('平'))
df['singel_price']=df['singel_price'].apply(lambda x:x.rstrip('元/平'))
df['singel_price']=df['singel_price'].apply(lambda x:x.lstrip('单价'))
df['district']=df['district'].apply(lambda x:x.rstrip('二手房'))
df['house_time']=df['house_time'].apply(lambda x:str(x))
df['house_time']=df['house_time'].apply(lambda x:x.rstrip('年建'))
df.head(n=1)
```
### 删除一些不需要用到的列 以及 house_desc、temp
```
del df['house_img']
del df['s_cate_href']
del df['house_desc']
del df['zone_href']
del df['house_href']
del df['temp']
```
### 根据房子总价和房子面积 计算房子每平方米的价格
### 从house_title 描述房子信息中提取关键词。若带有 交通便利、地铁则认为其交通方便,否则交通不便
```
df.head(n=1)
df['singel_price']=df['singel_price'].apply(lambda x:float(x))
df['area']=df['area'].apply(lambda x:float(x))
df.head(n=1)
df.head(n=1)
df['house_title']=df['house_title'].apply(lambda x:str(x))
df['trafic']=df['house_title'].apply(lambda x:'交通便利' if x.find("交通便利")>=0 or x.find("地铁")>=0 else "交通不便" )
df.head(n=1)
```
## 二、根据各列信息 用可视化的形式展现 房价与不同因素如地区、房子面积、所在楼层等之间的关系
```
df_house_count = df.groupby('district')['house_price'].count().sort_values(ascending=False).to_frame().reset_index()
df_house_mean = df.groupby('district')['singel_price'].mean().sort_values(ascending=False).to_frame().reset_index()
f, [ax1,ax2,ax3] = plt.subplots(3,1,figsize=(20,15))
sns.barplot(x='district', y='singel_price', palette="Reds_d", data=df_house_mean, ax=ax1)
ax1.set_title('上海各大区二手房每平米单价对比',fontsize=15)
ax1.set_xlabel('区域')
ax1.set_ylabel('每平米单价')
sns.countplot(df['district'], ax=ax2)
sns.boxplot(x='district', y='house_price', data=df, ax=ax3)
ax3.set_title('上海各大区二手房房屋总价',fontsize=15)
ax3.set_xlabel('区域')
ax3.set_ylabel('房屋总价')
plt.show()
```
### 上面三幅图显示了 房子单价、总数量、总价与地区之间的关系。
#### 由上面第一幅图可以看到房子单价与地区有关,其中黄浦以及静安地区房价最高。这与地区的发展水平、交通便利程度以及离市中心远近程度有关
#### 由上面第二幅图可以直接看出不同地区的二手房数量,其中浦东最多
#### 由上面第三幅图可以看出上海二手房房价基本在一千万上下,很少有高于两千万的
```
f, [ax1,ax2] = plt.subplots(1, 2, figsize=(15, 5))
# 二手房的面积分布
sns.distplot(df['area'], bins=20, ax=ax1, color='r')
sns.kdeplot(df['area'], shade=True, ax=ax1)
# 二手房面积和价位的关系
sns.regplot(x='area', y='house_price', data=df, ax=ax2)
plt.show()
```
### 由从左到右第一幅图可以看出 基本二手房面积在60-200平方米之间,其中一百平方米左右的占比更大
### 由第二幅看出,二手房总结与二手房面积基本成正比,和我们的常识吻合
```
areas=[len(df[df.area<100]),len(df[(df.area>100)&(df.area<200)]),len(df[df.area>200])]
labels=['area<100' , '100<area<200','area>200']
plt.pie(areas,labels= labels,autopct='%0f%%',shadow=True)
plt.show()
# 绘制饼图
```
### 将面积划分为三个档次,面积大于200、面积小与100、面积在一百到两百之间 三者的占比情况可以发现 百分之六十九的房子面积在一百平方米一下,高于一百大于200的只有百分之二十五而面积大于两百的只有百分之四
```
df.loc[df['area']>1000]
# 查看size>1000的样本 发现只有一个是大于1000
f, ax1= plt.subplots(figsize=(20,20))
sns.countplot(y='layout', data=df, ax=ax1)
ax1.set_title('房屋户型',fontsize=15)
ax1.set_xlabel('数量')
ax1.set_ylabel('户型')
f, ax2= plt.subplots(figsize=(20,20))
sns.barplot(y='layout', x='house_price', data=df, ax=ax2)
plt.show()
```
### 上述两幅图显示了 不同户型的数量和价格
#### 由第一幅图看出2室1厅最多 2室2厅 3室2厅也较多 是主流的户型选择
#### 由第二幅看出 室和厅的数量增加随之价格也增加,但是室和厅之间的比例要适合
```
a1=0
a2=0
for x in df['trafic']:
if x=='交通便利':
a1=a1+1
else:
a2=a2+1
sizes=[a1,a2]
labels=['交通便利' , '交通不便']
plt.pie(sizes,labels= labels,autopct='%0f%%',shadow=True)
plt.show()
```
#### 上述图显示了上海二手房交通不便利情况。其中百分之六十一为交通不便,百分之三十八为交通不便。由于交通便利情况仅仅是根据对房屋的描述情况提取出来的,实际上 交通便利的占比会更高些
```
f, [ax1,ax2] = plt.subplots(1, 2, figsize=(20, 10))
sns.countplot(df['trafic'], ax=ax1)
ax1.set_title('交通是否便利数量对比',fontsize=15)
ax1.set_xlabel('交通是否便利')
ax1.set_ylabel('数量')
sns.barplot(x='trafic', y='house_price', data=df, ax=ax2)
ax2.set_title('交通是否便利房价对比',fontsize=15)
ax2.set_xlabel('交通是否便利')
ax2.set_ylabel('总价')
plt.show()
```
### 左边那幅图显示了交通便利以及不便的二手房数量,这与我们刚才的饼图信息一致
### 右边那幅图显示了交通便利与否与房价的关系。交通便利的房子价格更高
```
f, ax1= plt.subplots(figsize=(20,5))
sns.countplot(x='floor', data=df, ax=ax1)
ax1.set_title('楼层',fontsize=15)
ax1.set_xlabel('楼层数')
ax1.set_ylabel('数量')
f, ax2 = plt.subplots(figsize=(20, 5))
sns.barplot(x='floor', y='house_price', data=df, ax=ax2)
ax2.set_title('楼层',fontsize=15)
ax2.set_xlabel('楼层数')
ax2.set_ylabel('总价')
plt.show()
```
#### 楼层(地区、高区、中区、地下几层)与数量、房价的关系。高区、中区、低区居多
## 三、根据已有数据建立简单的上海二手房房间预测模型
### 对数据再次进行简单的预处理 把户型这列拆成室和厅
```
df[['室','厅']] = df['layout'].str.extract(r'(\d+)室(\d+)厅')
df['室'] = df['室'].astype(float)
df['厅'] = df['厅'].astype(float)
del df['layout']
df.head()
df.dropna(inplace=True)
df.info()
df.columns
```
### 删除不需要用到的信息如房子的基本信息描述
```
del df['house_title']
del df['house_detail']
del df['s_cate']
from sklearn.linear_model import LinearRegression
linear = LinearRegression()
area=df['area']
price=df['house_price']
area = np.array(area).reshape(-1,1) # 这里需要注意新版的sklearn需要将数据转换为矩阵才能进行计算
price = np.array(price).reshape(-1,1)
# 训练模型
model = linear.fit(area,price)
# 打印截距和回归系数
print(model.intercept_, model.coef_)
linear_p = model.predict(area)
plt.figure(figsize=(12,6))
plt.scatter(area,price)
plt.plot(area,linear_p,'red')
plt.xlabel("area")
plt.ylabel("price")
plt.show()
```
#### 上面用线性回归模型对房价进行简单的预测 红色的代表预测房价,而蓝色点代表真实值。可以看出在面积小于1000时真实值紧密分布在预测值两旁
# 注意!
## 当是用Jupyter Notebook编程时,第一步请检查Notebook是否可读性可写

## 如果显示read-only,请打开终端(CMD),输入sudo chmod -R 777 filename,给文件夹授权,之后重新打开Jupyter Notebook方可保存文件。
|
github_jupyter
|
<a href="https://colab.research.google.com/github/huan/concise-chit-chat/blob/master/Concise_Chit_Chat.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Concise Chit Chat
GitHub Repository: <https://github.com/huan/concise-chit-chat>
## Code TODO:
1. create a DataLoader class for dataset preprocess. (Use tf.data.Dataset inside?)
1. Create a PyPI package for easy load cornell movie curpos dataset(?)
1. Use PyPI module `embeddings` to load `GLOVES`, or use tfhub to load `GLOVES`?
1. How to do a `clip_norm`(or set `clip_value`) in Keras with Eager mode but without `tf.contrib`?
1. Better name for variables & functions
1. Code clean
1. Encapsulate all layers to Model Class:
1. ChitChatEncoder
1. ChitChatDecoder
1. ChitChatModel
1. Re-style to follow the book
1. ...?
## Book Todo
1. Outlines
1. What's seq2seq
1. What's word embedding
1.
1. Split code into snips
1. Write for snips
1. Content cleaning and optimizing
1. ...?
## Other
1. `keras.callbacks.TensorBoard` instead of `tf.contrib.summary`?
- `model.fit(callbacks=[TensorBoard(...)])`
1. download url? - http://old.pep.com.cn/gzsx/jszx_1/czsxtbjxzy/qrzptgjzxjc/dzkb/dscl/
### config.py
```
'''doc'''
# GO for start of the sentence
# DONE for end of the sentence
GO = '\b'
DONE = '\a'
# max words per sentence
MAX_LEN = 20
```
### data_loader.py
```
'''
data loader
'''
import gzip
import re
from typing import (
# Any,
List,
Tuple,
)
import tensorflow as tf
import numpy as np
# from .config import (
# GO,
# DONE,
# MAX_LEN,
# )
DATASET_URL = 'https://github.com/huan/concise-chit-chat/releases/download/v0.0.1/dataset.txt.gz'
DATASET_FILE_NAME = 'concise-chit-chat-dataset.txt.gz'
class DataLoader():
'''data loader'''
def __init__(self) -> None:
print('DataLoader', 'downloading dataset from:', DATASET_URL)
dataset_file = tf.keras.utils.get_file(
DATASET_FILE_NAME,
origin=DATASET_URL,
)
print('DataLoader', 'loading dataset from:', dataset_file)
# dataset_file = './data/dataset.txt.gz'
# with open(path, encoding='iso-8859-1') as f:
with gzip.open(dataset_file, 'rt') as f:
self.raw_text = f.read().lower()
self.queries, self.responses \
= self.__parse_raw_text(self.raw_text)
self.size = len(self.queries)
def get_batch(
self,
batch_size=32,
) -> Tuple[List[List[str]], List[List[str]]]:
'''get batch'''
# print('corpus_list', self.corpus)
batch_indices = np.random.choice(
len(self.queries),
size=batch_size,
)
batch_queries = self.queries[batch_indices]
batch_responses = self.responses[batch_indices]
return batch_queries, batch_responses
def __parse_raw_text(
self,
raw_text: str
) -> Tuple[List[List[str]], List[List[str]]]:
'''doc'''
query_list = []
response_list = []
for line in raw_text.strip('\n').split('\n'):
query, response = line.split('\t')
query, response = self.preprocess(query), self.preprocess(response)
query_list.append('{} {} {}'.format(GO, query, DONE))
response_list.append('{} {} {}'.format(GO, response, DONE))
return np.array(query_list), np.array(response_list)
def preprocess(self, text: str) -> str:
'''doc'''
new_text = text
new_text = re.sub('[^a-zA-Z0-9 .,?!]', ' ', new_text)
new_text = re.sub(' +', ' ', new_text)
new_text = re.sub(
'([\w]+)([,;.?!#&-\'\"-]+)([\w]+)?',
r'\1 \2 \3',
new_text,
)
if len(new_text.split()) > MAX_LEN:
new_text = (' ').join(new_text.split()[:MAX_LEN])
match = re.search('[.?!]', new_text)
if match is not None:
idx = match.start()
new_text = new_text[:idx+1]
new_text = new_text.strip().lower()
return new_text
```
### vocabulary.py
```
'''doc'''
import re
from typing import (
List,
)
import tensorflow as tf
# from .config import (
# DONE,
# GO,
# MAX_LEN,
# )
class Vocabulary:
'''voc'''
def __init__(self, text: str) -> None:
self.tokenizer = tf.keras.preprocessing.text.Tokenizer(filters='')
self.tokenizer.fit_on_texts(
[GO, DONE] + re.split(
r'[\s\t\n]',
text,
)
)
# additional 1 for the index 0
self.size = 1 + len(self.tokenizer.word_index.keys())
def texts_to_padded_sequences(
self,
text_list: List[List[str]]
) -> tf.Tensor:
'''doc'''
sequence_list = self.tokenizer.texts_to_sequences(text_list)
padded_sequences = tf.keras.preprocessing.sequence.pad_sequences(
sequence_list,
maxlen=MAX_LEN,
padding='post',
truncating='post',
)
return padded_sequences
def padded_sequences_to_texts(self, sequence: List[int]) -> str:
return 'tbw'
```
### model.py
```
'''doc'''
import tensorflow as tf
import numpy as np
from typing import (
List,
)
# from .vocabulary import Vocabulary
# from .config import (
# DONE,
# GO,
# MAX_LENGTH,
# )
EMBEDDING_DIM = 300
LATENT_UNIT_NUM = 500
class ChitEncoder(tf.keras.Model):
'''encoder'''
def __init__(
self,
) -> None:
super().__init__()
self.lstm_encoder = tf.keras.layers.CuDNNLSTM(
units=LATENT_UNIT_NUM,
return_state=True,
)
def call(
self,
inputs: tf.Tensor, # shape: [batch_size, max_len, embedding_dim]
training=None,
mask=None,
) -> tf.Tensor:
_, *state = self.lstm_encoder(inputs)
return state # shape: ([latent_unit_num], [latent_unit_num])
class ChatDecoder(tf.keras.Model):
'''decoder'''
def __init__(
self,
voc_size: int,
) -> None:
super().__init__()
self.lstm_decoder = tf.keras.layers.CuDNNLSTM(
units=LATENT_UNIT_NUM,
return_sequences=True,
return_state=True,
)
self.dense = tf.keras.layers.Dense(
units=voc_size,
)
self.time_distributed_dense = tf.keras.layers.TimeDistributed(
self.dense
)
self.initial_state = None
def set_state(self, state=None):
'''doc'''
# import pdb; pdb.set_trace()
self.initial_state = state
def call(
self,
inputs: tf.Tensor, # shape: [batch_size, None, embedding_dim]
training=False,
mask=None,
) -> tf.Tensor:
'''chat decoder call'''
# batch_size = tf.shape(inputs)[0]
# max_len = tf.shape(inputs)[0]
# outputs = tf.zeros(shape=(
# batch_size, # batch_size
# max_len, # max time step
# LATENT_UNIT_NUM, # dimention of hidden state
# ))
# import pdb; pdb.set_trace()
outputs, *states = self.lstm_decoder(inputs, initial_state=self.initial_state)
self.initial_state = states
outputs = self.time_distributed_dense(outputs)
return outputs
class ChitChat(tf.keras.Model):
'''doc'''
def __init__(
self,
vocabulary: Vocabulary,
) -> None:
super().__init__()
self.word_index = vocabulary.tokenizer.word_index
self.index_word = vocabulary.tokenizer.index_word
self.voc_size = vocabulary.size
# [batch_size, max_len] -> [batch_size, max_len, voc_size]
self.embedding = tf.keras.layers.Embedding(
input_dim=self.voc_size,
output_dim=EMBEDDING_DIM,
mask_zero=True,
)
self.encoder = ChitEncoder()
# shape: [batch_size, state]
self.decoder = ChatDecoder(self.voc_size)
# shape: [batch_size, max_len, voc_size]
def call(
self,
inputs: List[List[int]], # shape: [batch_size, max_len]
teacher_forcing_targets: List[List[int]]=None, # shape: [batch_size, max_len]
training=None,
mask=None,
) -> tf.Tensor: # shape: [batch_size, max_len, embedding_dim]
'''call'''
batch_size = tf.shape(inputs)[0]
inputs_embedding = self.embedding(tf.convert_to_tensor(inputs))
state = self.encoder(inputs_embedding)
self.decoder.set_state(state)
if training:
teacher_forcing_targets = tf.convert_to_tensor(teacher_forcing_targets)
teacher_forcing_embeddings = self.embedding(teacher_forcing_targets)
# outputs[:, 0, :].assign([self.__go_embedding()] * batch_size)
batch_go_embedding = tf.ones([batch_size, 1, 1]) * [self.__go_embedding()]
batch_go_one_hot = tf.ones([batch_size, 1, 1]) * [tf.one_hot(self.word_index[GO], self.voc_size)]
outputs = batch_go_one_hot
output = self.decoder(batch_go_embedding)
for t in range(1, MAX_LEN):
outputs = tf.concat([outputs, output], 1)
if training:
target = teacher_forcing_embeddings[:, t, :]
decoder_input = tf.expand_dims(target, axis=1)
else:
decoder_input = self.__indice_to_embedding(tf.argmax(output))
output = self.decoder(decoder_input)
return outputs
def predict(self, inputs: List[int], temperature=1.) -> List[int]:
'''doc'''
outputs = self([inputs])
outputs = tf.squeeze(outputs)
word_list = []
for t in range(1, MAX_LEN):
output = outputs[t]
indice = self.__logit_to_indice(output, temperature=temperature)
word = self.index_word[indice]
if indice == self.word_index[DONE]:
break
word_list.append(word)
return ' '.join(word_list)
def __go_embedding(self) -> tf.Tensor:
return self.embedding(
tf.convert_to_tensor(self.word_index[GO]))
def __logit_to_indice(
self,
inputs,
temperature=1.,
) -> int:
'''
[vocabulary_size]
convert one hot encoding to indice with temperature
'''
inputs = tf.squeeze(inputs)
prob = tf.nn.softmax(inputs / temperature).numpy()
indice = np.random.choice(self.voc_size, p=prob)
return indice
def __indice_to_embedding(self, indice: int) -> tf.Tensor:
tensor = tf.convert_to_tensor([[indice]])
return self.embedding(tensor)
```
### Train
### Tensor Board
[Quick guide to run TensorBoard in Google Colab](https://www.dlology.com/blog/quick-guide-to-run-tensorboard-in-google-colab/)
`tensorboard` vs `tensorboard/` ?
```
LOG_DIR = '/content/data/tensorboard/'
get_ipython().system_raw(
'tensorboard --logdir {} --host 0.0.0.0 --port 6006 &'
.format(LOG_DIR)
)
# Install
! npm install -g localtunnel
# Tunnel port 6006 (TensorBoard assumed running)
get_ipython().system_raw('lt --port 6006 >> url.txt 2>&1 &')
# Get url
! cat url.txt
'''train'''
import tensorflow as tf
# from chit_chat import (
# ChitChat,
# DataLoader,
# Vocabulary,
# )
tf.enable_eager_execution()
data_loader = DataLoader()
vocabulary = Vocabulary(data_loader.raw_text)
chitchat = ChitChat(vocabulary=vocabulary)
def loss(model, x, y) -> tf.Tensor:
'''doc'''
weights = tf.cast(
tf.not_equal(y, 0),
tf.float32,
)
prediction = model(
inputs=x,
teacher_forcing_targets=y,
training=True,
)
# implment the following contrib function in a loop ?
# https://stackoverflow.com/a/41135778/1123955
# https://stackoverflow.com/q/48025004/1123955
return tf.contrib.seq2seq.sequence_loss(
prediction,
tf.convert_to_tensor(y),
weights,
)
def grad(model, inputs, targets):
'''doc'''
with tf.GradientTape() as tape:
loss_value = loss(model, inputs, targets)
return tape.gradient(loss_value, model.variables)
def train() -> int:
'''doc'''
learning_rate = 1e-3
num_batches = 8000
batch_size = 128
print('Dataset size: {}, Vocabulary size: {}'.format(
data_loader.size,
vocabulary.size,
))
optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate)
root = tf.train.Checkpoint(
optimizer=optimizer,
model=chitchat,
optimizer_step=tf.train.get_or_create_global_step(),
)
root.restore(tf.train.latest_checkpoint('./data/save'))
print('checkpoint restored.')
writer = tf.contrib.summary.create_file_writer('./data/tensorboard')
writer.set_as_default()
global_step = tf.train.get_or_create_global_step()
for batch_index in range(num_batches):
global_step.assign_add(1)
queries, responses = data_loader.get_batch(batch_size)
encoder_inputs = vocabulary.texts_to_padded_sequences(queries)
decoder_outputs = vocabulary.texts_to_padded_sequences(responses)
grads = grad(chitchat, encoder_inputs, decoder_outputs)
optimizer.apply_gradients(
grads_and_vars=zip(grads, chitchat.variables)
)
if batch_index % 10 == 0:
print("batch %d: loss %f" % (batch_index, loss(
chitchat, encoder_inputs, decoder_outputs).numpy()))
root.save('./data/save/model.ckpt')
print('checkpoint saved.')
with tf.contrib.summary.record_summaries_every_n_global_steps(1):
# your model code goes here
tf.contrib.summary.scalar('loss', loss(
chitchat, encoder_inputs, decoder_outputs).numpy())
# print('summary had been written.')
return 0
def main() -> int:
'''doc'''
return train()
main()
#! rm -fvr data/tensorboard
# ! pwd
# ! rm -frv data/save
# ! rm -fr /content/data/tensorboard
# ! kill 2823
# ! kill -9 2823
# ! ps axf | grep lt
! cat url.txt
```
### chat.py
```
'''train'''
# import tensorflow as tf
# from chit_chat import (
# ChitChat,
# DataLoader,
# Vocabulary,
# DONE,
# GO,
# )
# tf.enable_eager_execution()
def main() -> int:
'''chat main'''
data_loader = DataLoader()
vocabulary = Vocabulary(data_loader.raw_text)
print('Dataset size: {}, Vocabulary size: {}'.format(
data_loader.size,
vocabulary.size,
))
chitchat = ChitChat(vocabulary)
checkpoint = tf.train.Checkpoint(model=chitchat)
checkpoint.restore(tf.train.latest_checkpoint('./data/save'))
print('checkpoint restored.')
return cli(chitchat, vocabulary=vocabulary, data_loader=data_loader)
def cli(chitchat: ChitChat, data_loader: DataLoader, vocabulary: Vocabulary):
'''command line interface'''
index_word = vocabulary.tokenizer.index_word
word_index = vocabulary.tokenizer.word_index
query = ''
while True:
try:
# Get input sentence
query = input('> ').lower()
# Check if it is quit case
if query == 'q' or query == 'quit':
break
# Normalize sentence
query = data_loader.preprocess(query)
query = '{} {} {}'.format(GO, query, DONE)
# Evaluate sentence
query_sequence = vocabulary.texts_to_padded_sequences([query])[0]
response_sequence = chitchat.predict(query_sequence, 1)
# Format and print response sentence
response_word_list = [
index_word[indice]
for indice in response_sequence
if indice != 0 and indice != word_index[DONE]
]
print('Bot:', ' '.join(response_word_list))
except KeyError:
print("Error: Encountered unknown word.")
main()
! cat /proc/cpuinfo
```
|
github_jupyter
|
<img src="../../../../images/qiskit-heading.gif" alt="Note: In order for images to show up in this jupyter notebook you need to select File => Trusted Notebook" width="500 px" align="left">
# _*Quantum K-Means algorithm*_
The latest version of this notebook is available on https://github.com/qiskit/qiskit-tutorial.
***
### Contributors
Shan Jin, Xi He, Xiaokai Hou, Li Sun, Dingding Wen, Shaojun Wu and Xiaoting Wang$^{1}$
1. Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China,Chengdu, China,610051
***
## Introduction
Clustering algorithm is a typical unsupervised learning algorithm, which is mainly used to automatically classify similar samples into one category.In the clustering algorithm, according to the similarity between the samples, the samples are divided into different categories. For different similarity calculation methods, different clustering results will be obtained. The commonly used similarity calculation method is the Euclidean distance method.
What we want to show is the quantum K-Means algorithm. The K-Means algorithm is a distance-based clustering algorithm that uses distance as an evaluation index for similarity, that is, the closer the distance between two objects is, the greater the similarity. The algorithm considers the cluster to be composed of objects that are close together, so the compact and independent cluster is the ultimate target.
#### Experiment design
The implementation of the quantum K-Means algorithm mainly uses the swap test to compare the distances among the input data points. Select K points randomly from N data points as centroids, measure the distance from each point to each centroid, and assign it to the nearest centroid- class, recalculate centroids of each class that has been obtained, and iterate 2 to 3 steps until the new centroid is equal to or less than the specified threshold, and the algorithm ends. In our example, we selected 6 data points, 2 centroids, and used the swap test circuit to calculate the distance. Finally, we obtained two clusters of data points.
$|0\rangle$ is an auxiliary qubit, through left $H$ gate, it will be changed to $\frac{1}{\sqrt{2}}(|0\rangle + |1\rangle)$. Then under the control of $|1\rangle$, the circuit will swap two vectors $|x\rangle$ and $|y\rangle$. Finally, we get the result at the right end of the circuit:
$$|0_{anc}\rangle |x\rangle |y\rangle \rightarrow \frac{1}{2}|0_{anc}\rangle(|xy\rangle + |yx\rangle) + \frac{1}{2}|1_{anc}\rangle(|xy\rangle - |yx\rangle)$$
If we measure auxiliary qubit alone, then the probability of final state in the ground state $|1\rangle$ is:
$$P(|1_{anc}\rangle) = \frac{1}{2} - \frac{1}{2}|\langle x | y \rangle|^2$$
If we measure auxiliary qubit alone, then the probability of final state in the ground state $|1\rangle$ is:
$$Euclidean \ distance = \sqrt{(2 - 2|\langle x | y \rangle|)}$$
So, we can see that the probability of measuring $|1\rangle$ has positive correlation with the Euclidean distance.
The schematic diagram of quantum K-Means is as the follow picture.[[1]](#cite)
<img src="../images/k_means_circuit.png">
To make our algorithm can be run using qiskit, we design a more detailed circuit to achieve our algorithm.
|
#### Quantum K-Means circuit
<img src="../images/k_means.png">
## Data points
<table border="1">
<tr>
<td>point num</td>
<td>theta</td>
<td>phi</td>
<td>lam</td>
<td>x</td>
<td>y</td>
</tr>
<tr>
<td>1</td>
<td>0.01</td>
<td>pi</td>
<td>pi</td>
<td>0.710633</td>
<td>0.703562</td>
</tr>
<tr>
<td>2</td>
<td>0.02</td>
<td>pi</td>
<td>pi</td>
<td>0.714142</td>
<td>0.7</td>
</tr>
<tr>
<td>3</td>
<td>0.03</td>
<td>pi</td>
<td>pi</td>
<td>0.717633</td>
<td>0.696421</td>
</tr>
<tr>
<td>4</td>
<td>0.04</td>
<td>pi</td>
<td>pi</td>
<td>0.721107</td>
<td>0.692824</td>
</tr>
<tr>
<td>5</td>
<td>0.05</td>
<td>pi</td>
<td>pi</td>
<td>0.724562</td>
<td>0.68921</td>
</tr>
<tr>
<td>6</td>
<td>1.31</td>
<td>pi</td>
<td>pi</td>
<td>0.886811</td>
<td>0.462132</td>
</tr>
<tr>
<td>7</td>
<td>1.32</td>
<td>pi</td>
<td>pi</td>
<td>0.889111</td>
<td>0.457692</td>
</tr>
<tr>
<td>8</td>
<td>1.33</td>
<td>pi</td>
<td>pi</td>
<td>0.891388</td>
<td>0.453241</td>
</tr>
<tr>
<td>9</td>
<td>1.34</td>
<td>pi</td>
<td>pi</td>
<td>0.893643</td>
<td>0.448779</td>
</tr>
<tr>
<td>10</td>
<td>1.35</td>
<td>pi</td>
<td>pi</td>
<td>0.895876</td>
<td>0.444305</td>
</tr>
## Quantum K-Means algorithm program
```
# import math lib
from math import pi
# import Qiskit
from qiskit import Aer, IBMQ, execute
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister
# import basic plot tools
from qiskit.tools.visualization import plot_histogram
# To use local qasm simulator
backend = Aer.get_backend('qasm_simulator')
```
In this section, we first judge the version of Python and import the packages of qiskit, math to implement the following code. We show our algorithm on the ibm_qasm_simulator, if you need to run it on the real quantum conputer, please remove the "#" in frint of "import Qconfig".
```
theta_list = [0.01, 0.02, 0.03, 0.04, 0.05, 1.31, 1.32, 1.33, 1.34, 1.35]
```
Here we define the number pi in the math lib, because we need to use u3 gate. And we also define a list about the parameter theta which we need to use in the u3 gate. As the same above, if you want to implement on the real quantum comnputer, please remove the symbol "#" and configure your local Qconfig.py file.
```
# create Quantum Register called "qr" with 5 qubits
qr = QuantumRegister(5, name="qr")
# create Classical Register called "cr" with 5 bits
cr = ClassicalRegister(5, name="cr")
# Creating Quantum Circuit called "qc" involving your Quantum Register "qr"
# and your Classical Register "cr"
qc = QuantumCircuit(qr, cr, name="k_means")
#Define a loop to compute the distance between each pair of points
for i in range(9):
for j in range(1,10-i):
# Set the parament theta about different point
theta_1 = theta_list[i]
theta_2 = theta_list[i+j]
#Achieve the quantum circuit via qiskit
qc.h(qr[2])
qc.h(qr[1])
qc.h(qr[4])
qc.u3(theta_1, pi, pi, qr[1])
qc.u3(theta_2, pi, pi, qr[4])
qc.cswap(qr[2], qr[1], qr[4])
qc.h(qr[2])
qc.measure(qr[2], cr[2])
qc.reset(qr)
job = execute(qc, backend=backend, shots=1024)
result = job.result()
print(result)
print('theta_1:' + str(theta_1))
print('theta_2:' + str(theta_2))
# print( result.get_data(qc))
plot_histogram(result.get_counts())
```
Here we achieve the function k_means() and the test main function to run the program. Considering the qubits controlling direction of ibmqx4, we takes the quantum register 1, 2, 4 as our working register, if you want to run this program on other computer, please redesign the circuit structure to ensure your program can be run accurately.
## Result analysis
In this program, we take the quantum register 1, 2, 4 as our operated register (considering the condition when using ibmqx4.) We take the quantum register 1, 4 storing the input information about data points, and the quantum register 2 as controlling register to decide whether to use the swap operator. To estimate the distance of any pair of data points, we use a loop to implement the K-Means Circuit. In the end, we measure the controlling register to judge the distance between two data points. The probability when we get 1 means that the distance between two data points.
## Reference
<cite>[1].Quantum algorithms for supervised and unsupervised machine learning(*see open access: [ arXiv:1307.0411v2](https://arxiv.org/abs/1307.0411)*)</cite><a id='cite'></a>
|
github_jupyter
|
# Trigger Examples
Triggers allow the user to specify a set of actions that are triggered by the result of a boolean expression.
They provide flexibility to adapt what analysis and visualization actions are taken in situ. Triggers leverage Ascent's Query and Expression infrastructure. See Ascent's [Triggers](https://ascent.readthedocs.io/en/latest/Actions/Triggers.html) docs for deeper details on Triggers.
```
# cleanup any old results
!./cleanup.sh
# ascent + conduit imports
import conduit
import conduit.blueprint
import ascent
import numpy as np
# helpers we use to create tutorial data
from ascent_tutorial_jupyter_utils import img_display_width
from ascent_tutorial_jupyter_utils import tutorial_gyre_example
import matplotlib.pyplot as plt
```
## Trigger Example 1
### Using triggers to render when conditions occur
```
# Use triggers to render when conditions occur
a = ascent.Ascent()
a.open()
# setup actions
actions = conduit.Node()
# declare a question to ask
add_queries = actions.append()
add_queries["action"] = "add_queries"
# add our entropy query (q1)
queries = add_queries["queries"]
queries["q1/params/expression"] = "entropy(histogram(field('gyre'), num_bins=128))"
queries["q1/params/name"] = "entropy"
# declare triggers
add_triggers = actions.append()
add_triggers["action"] = "add_triggers"
triggers = add_triggers["triggers"]
# add a simple trigger (t1_ that fires at cycle 500
triggers["t1/params/condition"] = "cycle() == 500"
triggers["t1/params/actions_file"] = "cycle_trigger_actions.yaml"
# add trigger (t2) that fires when the change in entroy exceeds 0.5
# the history function allows you to access query results of previous
# cycles. relative_index indicates how far back in history to look.
# Looking at the plot of gyre entropy in the previous notebook, we see a jump
# in entropy at cycle 200, so we expect the trigger to fire at cycle 200
triggers["t2/params/condition"] = "entropy - history(entropy, relative_index = 1) > 0.5"
triggers["t2/params/actions_file"] = "entropy_trigger_actions.yaml"
# view our full actions tree
print(actions.to_yaml())
# gyre time varying params
nsteps = 10
time = 0.0
delta_time = 0.5
for step in range(nsteps):
# call helper that generates a double gyre time varying example mesh.
# gyre ref :https://shaddenlab.berkeley.edu/uploads/LCS-tutorial/examples.html
mesh = tutorial_gyre_example(time)
# update the example cycle
cycle = 100 + step * 100
mesh["state/cycle"] = cycle
print("time: {} cycle: {}".format(time,cycle))
# publish mesh to ascent
a.publish(mesh)
# execute the actions
a.execute(actions)
# update time
time = time + delta_time
# retrieve the info node that contains the trigger and query results
info = conduit.Node()
a.info(info)
# close ascent
a.close()
# we expect our cycle trigger to render only at cycle 500
! ls cycle_trigger*.png
# show the result image from the cycle trigger
ascent.jupyter.AscentImageSequenceViewer(["cycle_trigger_out_500.png"]).show()
# we expect our entropy trigger to render only at cycle 200
! ls entropy_trigger*.png
# show the result image from the entropy trigger
ascent.jupyter.AscentImageSequenceViewer(["entropy_trigger_out_200.png"]).show()
print(info["expressions"].to_yaml())
```
|
github_jupyter
|
## These notebooks can be found at https://github.com/jaspajjr/pydata-visualisation if you want to follow along
https://matplotlib.org/users/intro.html
Matplotlib is a library for making 2D plots of arrays in Python.
* Has it's origins in emulating MATLAB, it can also be used in a Pythonic, object oriented way.
* Easy stuff should be easy, difficult stuff should be possible
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%matplotlib inline
```
Everything in matplotlib is organized in a hierarchy. At the top of the hierarchy is the matplotlib “state-machine environment” which is provided by the matplotlib.pyplot module. At this level, simple functions are used to add plot elements (lines, images, text, etc.) to the current axes in the current figure.
Pyplot’s state-machine environment behaves similarly to MATLAB and should be most familiar to users with MATLAB experience.
The next level down in the hierarchy is the first level of the object-oriented interface, in which pyplot is used only for a few functions such as figure creation, and the user explicitly creates and keeps track of the figure and axes objects. At this level, the user uses pyplot to create figures, and through those figures, one or more axes objects can be created. These axes objects are then used for most plotting actions.
## Scatter Plot
To start with let's do a really basic scatter plot:
```
plt.plot([0, 1, 2, 3, 4, 5], [0, 2, 4, 6, 8, 10])
x = [0, 1, 2, 3, 4, 5]
y = [0, 2, 4, 6, 8, 10]
plt.plot(x, y)
```
What if we don't want a line?
```
plt.plot([0, 1, 2, 3, 4, 5],
[0, 2, 5, 7, 8, 10],
marker='o',
linestyle='')
plt.xlabel('The X Axis')
plt.ylabel('The Y Axis')
plt.show();
```
#### Simple example from matplotlib
https://matplotlib.org/tutorials/intermediate/tight_layout_guide.html#sphx-glr-tutorials-intermediate-tight-layout-guide-py
```
def example_plot(ax, fontsize=12):
ax.plot([1, 2])
ax.locator_params(nbins=5)
ax.set_xlabel('x-label', fontsize=fontsize)
ax.set_ylabel('y-label', fontsize=fontsize)
ax.set_title('Title', fontsize=fontsize)
fig, ax = plt.subplots()
example_plot(ax, fontsize=24)
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2)
# fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2, ncols=2, sharex=True, sharey=True)
ax1.plot([0, 1, 2, 3, 4, 5],
[0, 2, 5, 7, 8, 10])
ax2.plot([0, 1, 2, 3, 4, 5],
[0, 2, 4, 9, 16, 25])
ax3.plot([0, 1, 2, 3, 4, 5],
[0, 13, 18, 21, 23, 25])
ax4.plot([0, 1, 2, 3, 4, 5],
[0, 1, 2, 3, 4, 5])
plt.tight_layout()
```
## Date Plotting
```
import pandas_datareader as pdr
df = pdr.get_data_fred('GS10')
df = df.reset_index()
print(df.info())
df.head()
fig = plt.figure(figsize=(12, 8))
ax = fig.add_subplot(111)
ax.plot_date(df['DATE'], df['GS10'])
```
## Bar Plot
```
fig = plt.figure(figsize=(12, 8))
ax = fig.add_subplot(111)
x_data = [0, 1, 2, 3, 4]
values = [20, 35, 30, 35, 27]
ax.bar(x_data, values)
ax.set_xticks(x_data)
ax.set_xticklabels(('A', 'B', 'C', 'D', 'E'))
;
```
## Matplotlib basics
http://pbpython.com/effective-matplotlib.html
### Behind the scenes
* matplotlib.backend_bases.FigureCanvas is the area onto which the figure is drawn
* matplotlib.backend_bases.Renderer is the object which knows how to draw on the FigureCanvas
* matplotlib.artist.Artist is the object that knows how to use a renderer to paint onto the canvas
The typical user will spend 95% of their time working with the Artists.
https://matplotlib.org/tutorials/intermediate/artists.html#sphx-glr-tutorials-intermediate-artists-py
```
fig, (ax1, ax2) = plt.subplots(
nrows=1,
ncols=2,
sharey=True,
figsize=(12, 8))
fig.suptitle("Main Title", fontsize=14, fontweight='bold');
x_data = [0, 1, 2, 3, 4]
values = [20, 35, 30, 35, 27]
ax1.barh(x_data, values);
ax1.set_xlim([0, 55])
#ax1.set(xlabel='Unit of measurement', ylabel='Groups')
ax1.set(title='Foo', xlabel='Unit of measurement')
ax1.grid()
ax2.barh(x_data, [y / np.sum(values) for y in values], color='r');
ax2.set_title('Transformed', fontweight='light')
ax2.axvline(x=.1, color='k', linestyle='--')
ax2.set(xlabel='Unit of measurement') # Worth noticing this
ax2.set_axis_off();
fig.savefig('example_plot.png', dpi=80, bbox_inches="tight")
```
|
github_jupyter
|
# Procedures and Functions Tutorial
MLDB is the Machine Learning Database, and all machine learning operations are done via Procedures and Functions. Training a model happens via Procedures, and applying a model happens via Functions.
The notebook cells below use `pymldb`'s `Connection` class to make [REST API](../../../../doc/#builtin/WorkingWithRest.md.html) calls. You can check out the [Using `pymldb` Tutorial](../../../../doc/nblink.html#_tutorials/Using pymldb Tutorial) for more details.
```
from pymldb import Connection
mldb = Connection("http://localhost")
```
## Loading a Dataset
The classic [Iris Flower Dataset](http://en.wikipedia.org/wiki/Iris_flower_data_set) isn't very big but it's well-known and easy to reason about so it's a good example dataset to use for machine learning examples.
We can import it directly from a remote URL:
```
mldb.put('/v1/procedures/import_iris', {
"type": "import.text",
"params": {
"dataFileUrl": "file://mldb/mldb_test_data/iris.data",
"headers": [ "sepal length", "sepal width", "petal length", "petal width", "class" ],
"outputDataset": "iris",
"runOnCreation": True
}
})
```
## A quick look at the data
We can use the [Query API](../../../../doc/#builtin/sql/QueryAPI.md.html) to get the data into a Pandas DataFrame to take a quick look at it.
```
df = mldb.query("select * from iris")
df.head()
%matplotlib inline
import seaborn as sns, pandas as pd
sns.pairplot(df, hue="class", size=2.5)
```
## Unsupervised Machine Learning with a `kmeans.train` Procedure
We will create and run a [Procedure](../../../../doc/#builtin/procedures/Procedures.md.html) of type [`kmeans.train`](../../../../doc/#builtin/procedures/KmeansProcedure.md.html). This will train an unsupervised K-Means model and use it to assign each row in the input to a cluster, in the output dataset.
```
mldb.put('/v1/procedures/iris_train_kmeans', {
'type' : 'kmeans.train',
'params' : {
'trainingData' : 'select * EXCLUDING(class) from iris',
'outputDataset' : 'iris_clusters',
'numClusters' : 3,
'metric': 'euclidean',
"runOnCreation": True
}
})
```
Now we can look at the output dataset and compare the clusters the model learned with the three types of flower in the dataset.
```
mldb.query("""
select pivot(class, num) as *
from (
select cluster, class, count(*) as num
from merge(iris_clusters, iris)
group by cluster, class
)
group by cluster
""")
```
As you can see, the K-means algorithm doesn't do a great job of clustering this data (as is mentioned in the Wikipedia article!).
## Supervised Machine Learning with `classifier.train` and `.test` Procedures
We will now create and run a [Procedure](../../../../doc/#builtin/procedures/Procedures.md.html) of type [`classifier.train`](../../../../doc/#builtin/procedures/Classifier.md.html). The configuration below will use 20% of the data to train a decision tree to classify rows into the three classes of Iris. The output of this procedure is a [Function](../../../../doc/#builtin/functions/Functions.md.html), which we will be able to call from REST or SQL.
```
mldb.put('/v1/procedures/iris_train_classifier', {
'type' : 'classifier.train',
'params' : {
'trainingData' : """
select
{* EXCLUDING(class)} as features,
class as label
from iris
where rowHash() % 5 = 0
""",
"algorithm": "dt",
"modelFileUrl": "file://models/iris.cls",
"mode": "categorical",
"functionName": "iris_classify",
"runOnCreation": True
}
})
```
We can now test the classifier we just trained on the subset of the data we didn't use for training. To do so we use a procedure of type [`classifier.test`](../../../../doc/#builtin/procedures/Accuracy.md.html).
```
rez = mldb.put('/v1/procedures/iris_test_classifier', {
'type' : 'classifier.test',
'params' : {
'testingData' : """
select
iris_classify({
features: {* EXCLUDING(class)}
}) as score,
class as label
from iris
where rowHash() % 5 != 0
""",
"mode": "categorical",
"runOnCreation": True
}
})
runResults = rez.json()["status"]["firstRun"]["status"]
print rez
```
The procedure returns a confusion matrix, which you can compare with the one that resulted from the K-means procedure.
```
pd.DataFrame(runResults["confusionMatrix"])\
.pivot_table(index="actual", columns="predicted", fill_value=0)
```
As you can see, the decision tree does a much better job of classifying the data than the K-means model, using 20% of the examples as training data.
The procedure also returns standard classification statistics on how the classifier performed on the test set. Below are performance statistics for each label:
```
pd.DataFrame.from_dict(runResults["labelStatistics"]).transpose()
```
They are also available, averaged over all labels:
```
pd.DataFrame.from_dict({"weightedStatistics": runResults["weightedStatistics"]})
```
### Scoring new examples
We can call the Function REST API endpoint to classify a never-before-seen set of measurements like this:
```
mldb.get('/v1/functions/iris_classify/application', input={
"features":{
"petal length": 1,
"petal width": 2,
"sepal length": 3,
"sepal width": 4
}
})
```
## Where to next?
Check out the other [Tutorials and Demos](../../../../doc/#builtin/Demos.md.html).
You can also take a look at the [`classifier.experiment`](../../../../doc/#builtin/procedures/ExperimentProcedure.md.html) procedure type that can be used to train and test a classifier in a single call.
|
github_jupyter
|
### Analysis of motifs using Motif Miner (RINGS tool that employs alpha frequent subtree mining)
```
csv_files = ["ABA_14361_100ug_v5.0_DATA.csv",
"ConA_13799-10ug_V5.0_DATA.csv",
'PNA_14030_10ug_v5.0_DATA.csv',
"RCAI_10ug_14110_v5.0_DATA.csv",
"PHA-E-10ug_13853_V5.0_DATA.csv",
"PHA-L-10ug_13856_V5.0_DATA.csv",
"LCA_10ug_13934_v5.0_DATA.csv",
"SNA_10ug_13631_v5.0_DATA.csv",
"MAL-I_10ug_13883_v5.0_DATA.csv",
"MAL_II_10ug_13886_v5.0_DATA.csv",
"GSL-I-B4_10ug_13920_v5.0_DATA.csv",
"jacalin-1ug_14301_v5.0_DATA.csv",
'WGA_14057_1ug_v5.0_DATA.csv',
"UEAI_100ug_13806_v5.0_DATA.csv",
"SBA_14042_10ug_v5.0_DATA.csv",
"DBA_100ug_13897_v5.0_DATA.csv",
"PSA_14040_10ug_v5.0_DATA.csv",
"HA_PuertoRico_8_34_13829_v5_DATA.csv",
'H3N8-HA_16686_v5.1_DATA.csv',
"Human-DC-Sign-tetramer_15320_v5.0_DATA.csv"]
csv_file_normal_names = [
r"\textit{Agaricus bisporus} agglutinin (ABA)",
r"Concanavalin A (Con A)",
r'Peanut agglutinin (PNA)',
r"\textit{Ricinus communis} agglutinin I (RCA I/RCA\textsubscript{120})",
r"\textit{Phaseolus vulgaris} erythroagglutinin (PHA-E)",
r"\textit{Phaseolus vulgaris} leucoagglutinin (PHA-L)",
r"\textit{Lens culinaris} agglutinin (LCA)",
r"\textit{Sambucus nigra} agglutinin (SNA)",
r"\textit{Maackia amurensis} lectin I (MAL-I)",
r"\textit{Maackia amurensis} lectin II (MAL-II)",
r"\textit{Griffonia simplicifolia} Lectin I isolectin B\textsubscript{4} (GSL I-B\textsubscript{4})",
r"Jacalin",
r'Wheat germ agglutinin (WGA)',
r"\textit{Ulex europaeus} agglutinin I (UEA I)",
r"Soybean agglutinin (SBA)",
r"\textit{Dolichos biflorus} agglutinin (DBA)",
r"\textit{Pisum sativum} agglutinin (PSA)",
r"Influenza hemagglutinin (HA) (A/Puerto Rico/8/34) (H1N1)",
r'Influenza HA (A/harbor seal/Massachusetts/1/2011) (H3N8)',
r"Human DC-SIGN tetramer"]
import sys
import os
import pandas as pd
import numpy as np
from scipy import interp
sys.path.append('..')
from ccarl.glycan_parsers.conversions import kcf_to_digraph, cfg_to_kcf
from ccarl.glycan_plotting import draw_glycan_diagram
from ccarl.glycan_graph_methods import generate_digraph_from_glycan_string
from ccarl.glycan_features import generate_features_from_subtrees
import ccarl.glycan_plotting
from sklearn.linear_model import LogisticRegressionCV, LogisticRegression
from sklearn.metrics import matthews_corrcoef, make_scorer, roc_curve, auc
import matplotlib.pyplot as plt
from collections import defaultdict
aucs = defaultdict(list)
ys = defaultdict(list)
probs = defaultdict(list)
motifs = defaultdict(list)
for fold in [1,2,3,4,5]:
print(f"Running fold {fold}...")
for csv_file in csv_files:
alpha = 0.8
minsup = 0.2
input_file = f'./temp_{csv_file}'
training_data = pd.read_csv(f"../Data/CV_Folds/fold_{fold}/training_set_{csv_file}")
test_data = pd.read_csv(f"../Data/CV_Folds/fold_{fold}/test_set_{csv_file}")
pos_glycan_set = training_data['glycan'][training_data.binding == 1].values
kcf_string = '\n'.join([cfg_to_kcf(x) for x in pos_glycan_set])
with open(input_file, 'w') as f:
f.write(kcf_string)
min_sup = int(len(pos_glycan_set) * minsup)
subtrees = os.popen(f"ruby Miner_cmd.rb {min_sup} {alpha} {input_file}").read()
subtree_graphs = [kcf_to_digraph(x) for x in subtrees.split("///")[0:-1]]
motifs[csv_file].append(subtree_graphs)
os.remove(input_file)
binding_class = training_data.binding.values
glycan_graphs = [generate_digraph_from_glycan_string(x, parse_linker=True,
format='CFG')
for x in training_data.glycan]
glycan_graphs_test = [generate_digraph_from_glycan_string(x, parse_linker=True,
format='CFG')
for x in test_data.glycan]
features = [generate_features_from_subtrees(subtree_graphs, glycan) for
glycan in glycan_graphs]
features_test = [generate_features_from_subtrees(subtree_graphs, glycan) for
glycan in glycan_graphs_test]
logistic_clf = LogisticRegression(penalty='l2', C=100, solver='lbfgs',
class_weight='balanced', max_iter=1000)
X = features
y = binding_class
logistic_clf.fit(X, y)
y_test = test_data.binding.values
X_test = features_test
fpr, tpr, _ = roc_curve(y_test, logistic_clf.predict_proba(X_test)[:,1], drop_intermediate=False)
aucs[csv_file].append(auc(fpr, tpr))
ys[csv_file].append(y_test)
probs[csv_file].append(logistic_clf.predict_proba(X_test)[:,1])
# Assess the number of subtrees generated for each CV round.
subtree_lengths = defaultdict(list)
for fold in [1,2,3,4,5]:
print(f"Running fold {fold}...")
for csv_file in csv_files:
alpha = 0.8
minsup = 0.2
input_file = f'./temp_{csv_file}'
training_data = pd.read_csv(f"../Data/CV_Folds/fold_{fold}/training_set_{csv_file}")
test_data = pd.read_csv(f"../Data/CV_Folds/fold_{fold}/test_set_{csv_file}")
pos_glycan_set = training_data['glycan'][training_data.binding == 1].values
kcf_string = '\n'.join([cfg_to_kcf(x) for x in pos_glycan_set])
with open(input_file, 'w') as f:
f.write(kcf_string)
min_sup = int(len(pos_glycan_set) * minsup)
subtrees = os.popen(f"ruby Miner_cmd.rb {min_sup} {alpha} {input_file}").read()
subtree_graphs = [kcf_to_digraph(x) for x in subtrees.split("///")[0:-1]]
subtree_lengths[csv_file].append(len(subtree_graphs))
os.remove(input_file)
subtree_lengths = [y for x in subtree_lengths.values() for y in x]
print(np.mean(subtree_lengths))
print(np.max(subtree_lengths))
print(np.min(subtree_lengths))
def plot_multiple_roc(data):
'''Plot multiple ROC curves.
Prints out key AUC values (mean, median etc).
Args:
data (list): A list containing [y, probs] for each model, where:
y: True class labels
probs: Predicted probabilities
Returns:
Figure, Axes, Figure, Axes
'''
mean_fpr = np.linspace(0, 1, 100)
fig, axes = plt.subplots(figsize=(4, 4))
ax = axes
ax.set_title('')
#ax.legend(loc="lower right")
ax.set_xlabel('False Positive Rate')
ax.set_ylabel('True Positive Rate')
ax.set_aspect('equal', adjustable='box')
auc_values = []
tpr_list = []
for y, probs in data:
#data_point = data[csv_file]
#y = data_point[7] # test binding
#X = data_point[8] # test features
#logistic_clf = data_point[0] # model
fpr, tpr, _ = roc_curve(y, probs, drop_intermediate=False)
tpr_list.append(interp(mean_fpr, fpr, tpr))
auc_values.append(auc(fpr, tpr))
ax.plot(fpr, tpr, color='blue', alpha=0.1, label=f'ROC curve (area = {auc(fpr, tpr): 2.3f})')
ax.plot([0,1], [0,1], linestyle='--', color='grey', linewidth=0.8, dashes=(5, 10))
mean_tpr = np.mean(tpr_list, axis=0)
median_tpr = np.median(tpr_list, axis=0)
upper_tpr = np.percentile(tpr_list, 75, axis=0)
lower_tpr = np.percentile(tpr_list, 25, axis=0)
ax.plot(mean_fpr, median_tpr, color='black')
ax.fill_between(mean_fpr, lower_tpr, upper_tpr, color='grey', alpha=.5,
label=r'$\pm$ 1 std. dev.')
fig.savefig("Motif_Miner_CV_ROC_plot_all_curves.svg")
fig2, ax2 = plt.subplots(figsize=(4, 4))
ax2.hist(auc_values, range=[0.5,1], bins=10, rwidth=0.9, color=(0, 114/255, 178/255))
ax2.set_xlabel("AUC value")
ax2.set_ylabel("Counts")
fig2.savefig("Motif_Miner_CV_AUC_histogram.svg")
print(f"Mean AUC value: {np.mean(auc_values): 1.3f}")
print(f"Median AUC value: {np.median(auc_values): 1.3f}")
print(f"IQR of AUC values: {np.percentile(auc_values, 25): 1.3f} - {np.percentile(auc_values, 75): 1.3f}")
return fig, axes, fig2, ax2, auc_values
# Plot ROC curves for all test sets
roc_data = [[y, prob] for y_fold, prob_fold in zip(ys.values(), probs.values()) for y, prob in zip(y_fold, prob_fold)]
_, _, _, _, auc_values = plot_multiple_roc(roc_data)
auc_values_ccarl = [0.950268817204301,
0.9586693548387097,
0.9559811827956988,
0.8686155913978494,
0.9351222826086956,
0.989010989010989,
0.9912587412587414,
0.9090909090909092,
0.9762626262626264,
0.9883597883597884,
0.9065533980582524,
0.9417475728155339,
0.8268608414239482,
0.964349376114082,
0.9322638146167558,
0.9178037686809616,
0.96361273554256,
0.9362139917695472,
0.9958847736625515,
0.9526748971193415,
0.952300785634119,
0.9315375982042648,
0.9705387205387206,
0.9865319865319865,
0.9849773242630385,
0.9862385321100917,
0.9862385321100918,
0.9606481481481481,
0.662037037037037,
0.7796296296296297,
0.9068627450980392,
0.915032679738562,
0.9820261437908496,
0.9893790849673203,
0.9882988298829882,
0.9814814814814815,
1.0,
0.8439153439153441,
0.9859813084112149,
0.9953271028037383,
0.8393308080808081,
0.8273358585858586,
0.7954545454545453,
0.807070707070707,
0.8966329966329966,
0.8380952380952381,
0.6201058201058202,
0.7179894179894181,
0.6778846153846154,
0.75,
0.9356060606060607,
0.8619528619528619,
0.8787878787878789,
0.9040816326530613,
0.7551020408163266,
0.9428694158075602,
0.9226804123711341,
0.8711340206185567,
0.7840909090909091,
0.8877840909090909,
0.903225806451613,
0.8705594120049,
0.9091465904450796,
0.8816455696202531,
0.8521097046413502,
0.8964521452145213,
0.9294554455445544,
0.8271452145214522,
0.8027272727272727,
0.8395454545454546,
0.8729967948717949,
0.9306891025641025,
0.9550970873786407,
0.7934686672550749,
0.8243601059135041,
0.8142100617828772,
0.9179611650485436,
0.8315533980582525,
0.7266990291262136,
0.9038834951456312,
0.9208916083916084,
0.7875,
0.9341346153846154,
0.9019230769230768,
0.9086538461538461,
0.9929245283018868,
0.9115566037735848,
0.9952830188679246,
0.9658018867924528,
0.7169811320754716,
0.935981308411215,
0.9405660377358491,
0.9905660377358491,
0.9937106918238994,
0.9302935010482181,
0.7564814814814815,
0.9375,
0.8449074074074074,
0.8668981481481483,
0.7978971962616823]
auc_value_means = [np.mean(auc_values[x*5:x*5+5]) for x in range(int(len(auc_values) / 5))]
auc_value_means_ccarl = [np.mean(auc_values_ccarl[x*5:x*5+5]) for x in range(int(len(auc_values_ccarl) / 5))]
auc_value_mean_glymmr = np.array([0.6067939 , 0.76044574, 0.66786624, 0.69578298, 0.81659623,
0.80536403, 0.77231548, 0.96195032, 0.70013384, 0.60017685,
0.77336818, 0.78193305, 0.66269668, 0.70333122, 0.54247748,
0.63003707, 0.79619231, 0.85141509, 0.9245296 , 0.63366329])
auc_value_mean_glymmr_best = np.array([0.77559242, 0.87452658, 0.75091636, 0.7511371 , 0.87450697,
0.82895628, 0.81083123, 0.96317065, 0.75810185, 0.82680149,
0.84747054, 0.8039597 , 0.69651882, 0.73431593, 0.582194 ,
0.67407767, 0.83049825, 0.88891509, 0.9345188 , 0.72702016])
auc_value_motiffinder = [0.9047619047619048, 0.9365601503759399, 0.6165413533834586, 0.9089068825910931,
0.4962962962962963, 0.6358816964285713, 0.8321078431372548, 0.8196576151121606, 0.8725400457665904,
0.830220713073005, 0.875, 0.7256367663344407, 0.8169291338582677, 0.9506818181818182, 0.7751351351351351,
0.9362947658402204, 0.6938461538461539, 0.6428571428571428, 0.7168021680216802, 0.5381136950904392] #Note, only from a single test-train split.
import seaborn as sns
sns.set(style="ticks")
plot_data = np.array([auc_value_mean_glymmr, auc_value_mean_glymmr_best, auc_value_motiffinder, auc_value_means, auc_value_means_ccarl]).T
ax = sns.violinplot(data=plot_data, cut=2, inner='quartile')
sns.swarmplot(data=plot_data, color='black')
ax.set_ylim([0.5, 1.05])
ax.set_xticklabels(["GLYMMR\n(mean)", "GLYMMR\n(best)", "MotifFinder", "Glycan\nMiner Tool", "CCARL"])
#ax.grid('off')
ax.set_ylabel("AUC")
ax.figure.savefig('method_comparison_violin_plot.svg')
auc_value_means_ccarl
print("CCARL Performance")
print(f"Median AUC value: {np.median(auc_value_means_ccarl): 1.3f}")
print(f"IQR of AUC values: {np.percentile(auc_value_means_ccarl, 25): 1.3f} - {np.percentile(auc_value_means_ccarl, 75): 1.3f}")
print("Glycan Miner Tool Performance")
print(f"Median AUC value: {np.median(auc_value_means): 1.3f}")
print(f"IQR of AUC values: {np.percentile(auc_value_means, 25): 1.3f} - {np.percentile(auc_value_means, 75): 1.3f}")
print("Glycan Miner Tool Performance")
print(f"Median AUC value: {np.median(auc_value_mean_glymmr_best): 1.3f}")
print(f"IQR of AUC values: {np.percentile(auc_value_mean_glymmr_best, 25): 1.3f} - {np.percentile(auc_value_mean_glymmr_best, 75): 1.3f}")
print("Glycan Miner Tool Performance")
print(f"Median AUC value: {np.median(auc_value_mean_glymmr): 1.3f}")
print(f"IQR of AUC values: {np.percentile(auc_value_mean_glymmr, 25): 1.3f} - {np.percentile(auc_value_mean_glymmr, 75): 1.3f}")
from matplotlib.backends.backend_pdf import PdfPages
sns.reset_orig()
import networkx as nx
for csv_file in csv_files:
with PdfPages(f"./motif_miner_motifs/glycan_motif_miner_motifs_{csv_file}.pdf") as pdf:
for motif in motifs[csv_file][0]:
fig, ax = plt.subplots()
ccarl.glycan_plotting.draw_glycan_diagram(motif, ax)
pdf.savefig(fig)
plt.close(fig)
glymmr_mean_stdev = np.array([0.15108904, 0.08300011, 0.11558078, 0.05259819, 0.061275 ,
0.09541182, 0.09239553, 0.05114523, 0.05406571, 0.16180131,
0.10345311, 0.06080207, 0.0479003 , 0.09898648, 0.06137992,
0.09813596, 0.07010635, 0.14010784, 0.05924527, 0.13165457])
glymmr_best_stdev = np.array([0.08808868, 0.04784959, 0.13252895, 0.03163248, 0.04401516,
0.08942411, 0.08344247, 0.05714308, 0.05716086, 0.05640053,
0.08649275, 0.05007289, 0.05452531, 0.05697662, 0.0490626 ,
0.1264917 , 0.04994508, 0.1030053 , 0.03359648, 0.12479809])
auc_value_std_ccarl = [np.std(auc_values_ccarl[x*5:x*5+5]) for x in range(int(len(auc_values_ccarl) / 5))]
print(r"Lectin & GLYMMR(mean) & GLYMMR(best) & Glycan Miner Tool & MotifFinder & CCARL \\ \hline")
for i, csv_file, name in zip(list(range(len(csv_files))), csv_files, csv_file_normal_names):
print(f"{name} & {auc_value_mean_glymmr[i]:0.3f} ({glymmr_mean_stdev[i]:0.3f}) & {auc_value_mean_glymmr_best[i]:0.3f} ({glymmr_best_stdev[i]:0.3f}) \
& {np.mean(aucs[csv_file]):0.3f} ({np.std(aucs[csv_file]):0.3f}) & {auc_value_motiffinder[i]:0.3f} & {auc_value_means_ccarl[i]:0.3f} ({auc_value_std_ccarl[i]:0.3f}) \\\\")
```
|
github_jupyter
|
```
import json
import random
import numpy as np
import tensorflow as tf
from collections import deque
from keras.models import Sequential
from keras.optimizers import RMSprop
from keras.layers import Dense, Flatten
from keras.layers.convolutional import Conv2D
from keras import backend as K
import datetime
import itertools
import matplotlib.pyplot as plt
import pandas as pd
import scipy as sp
import time
import math
from matplotlib.colors import LinearSegmentedColormap
import colorsys
import numpy as np
from data_retrieval_relocation_3ksol_reloc import INSTANCEProvider
from kbh_yard_b2b_relocation import KBH_Env #This is the environment of the shunting yard
from dqn_kbh_colfax_relocation_test_agent import DQNAgent
# this function returns random colors for visualisation of learning.
def rand_cmap(nlabels, type='soft', first_color_black=True, last_color_black=False):
# Generate soft pastel colors, by limiting the RGB spectrum
if type == 'soft':
low = 0.6
high = 0.95
randRGBcolors = [(np.random.uniform(low=low, high=high),
np.random.uniform(low=low, high=high),
np.random.uniform(low=low, high=high)) for i in range(nlabels)]
if first_color_black:
randRGBcolors[0] = [0, 0, 0]
if last_color_black:
randRGBcolors[-1] = [0, 0, 0]
random_colormap = LinearSegmentedColormap.from_list('new_map', randRGBcolors, N=nlabels)
return random_colormap
#1525445230 is the 185k expensive relocation model.
for model_nr in ['1525445230']:
#which model to load.
test_case = model_nr
#LOAD THE INSTANCE PROVIDER
ig = INSTANCEProvider()
instances = ig.instances
# Create environment KBH
yrd = KBH_Env()
# Create the DQNAgent with the CNN approximation of the Q-function and its experience replay and training functions.
# load the trained model.
agent = DQNAgent(yrd, True, test_case)
# set epsilon to 0 to act just greedy
agent.epsilon = 0
#new_cmap = rand_cmap(200, type='soft', first_color_black=True, last_color_black=False, verbose=True)
visualization = False
n = len(instances)
# result vectors
original_lengths = []
terminated_at_step = []
success = []
relocations = []
print_count = 0
# train types different tracks?
type_step_track = []
for instance in instances:
nr_relocations = 0
if print_count % 100 == 0:
print(print_count)
print_count = print_count + 1
#Initialize problem
event_list = ig.get_instance(instance)
steps, t, total_t, score= len(event_list), 0, 0, 0
state = yrd.reset(event_list) # Get first observation based on the first train arrival.
history = np.reshape(state, (
1, yrd.shape[0], yrd.shape[1], yrd.shape[2])) # reshape state into tensor, which we call history.
done, busy_relocating = False, False
if visualization:
#visualize learning
new_cmap = rand_cmap(200, type='soft', first_color_black=True, last_color_black=False)
if visualization == True:
plt.imshow(np.float32(history[0][0]), cmap=new_cmap, interpolation='nearest')
plt.show()
while not done:
action = agent.get_action(history) # RL choose action based on observation
if visualization == True:
print(agent.model.predict(history))
print(action+1)
# # RL take action and get next observation and reward
# # note the +1 at action
# save for arrival activities the parking location
event_list_temp = event_list.reset_index(drop=True).copy()
if event_list_temp.event_type[0]=='arrival':
train_type = event_list_temp.composition[0]
type_step_track.append({'type': train_type, 'action': action+1, 'step':t, 'instance_id': instance})
# based on that action now let environment go to new state
event = event_list.iloc[0]
# check if after this we are done...
done_ = True if len(event_list) == 1 else False # then there is no next event
# if done_:
# print("Reached the end of a problem!")
if busy_relocating:
# here we do not drop an event from the event list.
coming_arrivals = event_list.loc[event_list['event_type'] == 'arrival'].reset_index(drop=True)
coming_departures = event_list.loc[event_list['event_type'] == 'departure'].reset_index(drop=True)
next_state, reward, done = yrd.reloc_destination_step(event, event_list, action+1, coming_arrivals, coming_departures, done_)
nr_relocations += 1
busy_relocating = False
else:
# These operations below are expensive: maybe just use indexing.
event_list.drop(event_list.index[:1], inplace=True)
coming_arrivals = event_list.loc[event_list['event_type'] == 'arrival'].reset_index(drop=True)
coming_departures = event_list.loc[event_list['event_type'] == 'departure'].reset_index(drop=True)
# do step
next_state, reward, done = yrd.step(action+1, coming_arrivals, coming_departures, event, event_list, done_)
busy_relocating = True if reward == -0.5 else False
history_ = np.float32(np.reshape(next_state, (1, yrd.shape[0], yrd.shape[1], yrd.shape[2])))
score += reward # log direct reward of action
if visualization == True:
#show action
plt.imshow(np.float32(history_[0][0]), cmap=new_cmap, interpolation='nearest')
plt.show()
time.sleep(0.05)
if reward == -1:
time.sleep(1)
print(reward)
if done: # based on what the environment returns.
#print('ended at step' , t+1)
#print('original length', steps)
original_lengths.append(steps)
terminated_at_step.append(t+1)
relocations.append(nr_relocations)
if int(np.unique(history_)[0]) == 1: #then we are in win state
success.append(1)
else:
success.append(0)
break;
history = history_ # next state now becomes the current state.
t += 1 # next step in this episode
#save data needed for Entropy calculations.
df_type_step_track = pd.DataFrame.from_records(type_step_track)
df_type_step_track['strtype'] = df_type_step_track.apply(lambda row: str(row.type), axis = 1)
df_type_step_track.strtype = df_type_step_track.strtype.astype('category')
filename = 'data_'+model_nr+'_relocation_arrival_actions.csv'
df_type_step_track.to_csv(filename)
# analysis_runs = pd.DataFrame(
# {'instance_id': instances,
# 'original_length': original_lengths,
# 'terminated_at_step': terminated_at_step
# })
# analysis_runs['solved'] = analysis_runs.apply(lambda row: 1 if row.original_length == row.terminated_at_step else 0, axis =1 )
# analysis_runs['tried'] = analysis_runs.apply(lambda row: 1 if row.terminated_at_step != -1 else 0, axis =1)
# analysis_runs['percentage'] = analysis_runs.apply(lambda row: row.solved/755, axis=1)
# analysis_runs.to_csv('best_model_solved_instances.csv')
# print('Model: ', model_nr)
# summary = analysis_runs.groupby('original_length', as_index=False)[['solved', 'tried', 'percentage']].sum()
# print(summary)
# #print hist
# %matplotlib inline
# #%%
# # analyse the parking actions per step and train type
# df_type_step_track = pd.DataFrame.from_records(type_step_track)
# bins = [1,2,3,4,5,6,7,8,9,10]
# plt.hist(df_type_step_track.action, bins, align='left')
# #prepare for save
# df_type_step_track['strtype'] = df_type_step_track.apply(lambda row: str(row.type), axis = 1)
# df_type_step_track.strtype = df_type_step_track.strtype.astype('category')
# filename = 'data_'+model_nr+'_paper.csv'
# df_type_step_track.to_csv(filename)
analysis_runs = pd.DataFrame(
{'instance_id': instances,
'original_length': original_lengths,
'terminated_at_step': terminated_at_step,
'success': success,
'nr_relocations': relocations
})
analysis_runs.sort_values('terminated_at_step')
print(analysis_runs.loc[analysis_runs.success == 0].instance_id.to_string(index=False))
analysis_runs.loc[analysis_runs.success == 1].copy().groupby('nr_relocations')[['instance_id']].count()
summary = analysis_runs.groupby('original_length', as_index=False)[['success']].sum()
print(summary)
summary = analysis_runs.groupby('original_length', as_index=False)[['success']].mean()
print(summary)
max_reloc = max(analysis_runs.nr_relocations)
print(max_reloc)
plt.hist(analysis_runs.nr_relocations, bins=range(0,max_reloc+2), align='left')
import seaborn as sns
sns.set(style="darkgrid")
g = sns.FacetGrid(analysis_runs, col="original_length", margin_titles=True)
bins = range(0,max_reloc+2)
g.map(plt.hist, "nr_relocations", color="steelblue", bins=bins, lw=0, align='left')
print(analysis_runs.loc[analysis_runs.success == 1].groupby('original_length', as_index=False)[['nr_relocations']].mean())
```
# CODE HAS BEEN RUN UNTILL HERE.
.
.
.
.
.
.
.
.
v
# analysis of mistakes
```
analysis_runs.loc[analysis_runs.success == 0].sort_values('terminated_at_step')
#plt.hist(analysis_runs.loc[analysis_runs.success == 0].terminated_at_step, bins=8)
len(analysis_runs.loc[analysis_runs.success == 0])
analysis_runs['instance_size'] = analysis_runs.apply(lambda row: str(row.original_length).replace('37', '14').replace('41', '15').replace('43', '16').replace('46','17'), axis=1)
import seaborn as sns
sns.set(style="darkgrid")
bins = [0,5,10,15,20,25,30,35,40,45,50]
g = sns.FacetGrid(analysis_runs.loc[analysis_runs.success == 0], col="instance_size", margin_titles=True)
g.set(ylim=(0, 100), xlim=(0,50))
g.map(plt.hist, "terminated_at_step", color="steelblue", bins=bins, lw=0)
sns.plt.savefig('185k_failures.eps')
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/dauparas/tensorflow_examples/blob/master/VAE_cell_cycle.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
https://github.com/PMBio/scLVM/blob/master/tutorials/tcell_demo.ipynb
Variational Autoencoder Model (VAE) with latent subspaces based on:
https://arxiv.org/pdf/1812.06190.pdf
```
#Step 1: import dependencies
from tensorflow.keras import layers
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from keras import regularizers
import time
from __future__ import division
import tensorflow as tf
import tensorflow_probability as tfp
tfd = tfp.distributions
%matplotlib inline
plt.style.use('dark_background')
import pandas as pd
import os
from matplotlib import cm
import h5py
import scipy as SP
import pylab as PL
data = os.path.join('data_Tcells_normCounts.h5f')
f = h5py.File(data,'r')
Y = f['LogNcountsMmus'][:] # gene expression matrix
tech_noise = f['LogVar_techMmus'][:] # technical noise
genes_het_bool=f['genes_heterogen'][:] # index of heterogeneous genes
geneID = f['gene_names'][:] # gene names
cellcyclegenes_filter = SP.unique(f['cellcyclegenes_filter'][:].ravel() -1) # idx of cell cycle genes from GO
cellcyclegenes_filterCB = f['ccCBall_gene_indices'][:].ravel() -1 # idx of cell cycle genes from cycle base ...
# filter cell cycle genes
idx_cell_cycle = SP.union1d(cellcyclegenes_filter,cellcyclegenes_filterCB)
# determine non-zero counts
idx_nonzero = SP.nonzero((Y.mean(0)**2)>0)[0]
idx_cell_cycle_noise_filtered = SP.intersect1d(idx_cell_cycle,idx_nonzero)
# subset gene expression matrix
Ycc = Y[:,idx_cell_cycle_noise_filtered]
plt = PL.subplot(1,1,1);
PL.imshow(Ycc,cmap=cm.RdBu,vmin=-3,vmax=+3,interpolation='None');
#PL.colorbar();
plt.set_xticks([]);
plt.set_yticks([]);
PL.xlabel('genes');
PL.ylabel('cells');
X = np.delete(Y, idx_cell_cycle_noise_filtered, axis=1)
X = Y #base case
U = Y[:,idx_cell_cycle_noise_filtered]
mean = np.mean(X, axis=0)
variance = np.var(X, axis=0)
indx_small_mean = np.argwhere(mean < 0.00001)
X = np.delete(X, indx_small_mean, axis=1)
mean = np.mean(X, axis=0)
variance = np.var(X, axis=0)
fano = variance/mean
print(fano.shape)
indx_small_fano = np.argwhere(fano < 1.0)
X = np.delete(X, indx_small_fano, axis=1)
mean = np.mean(X, axis=0)
variance = np.var(X, axis=0)
fano = variance/mean
print(fano.shape)
#Reconstruction loss
def x_given_z(z, output_size):
with tf.variable_scope('M/x_given_w_z'):
act = tf.nn.leaky_relu
h = z
h = tf.layers.dense(h, 8, act)
h = tf.layers.dense(h, 16, act)
h = tf.layers.dense(h, 32, act)
h = tf.layers.dense(h, 64, act)
h = tf.layers.dense(h, 128, act)
h = tf.layers.dense(h, 256, act)
loc = tf.layers.dense(h, output_size)
#log_variance = tf.layers.dense(x, latent_size)
#scale = tf.nn.softplus(log_variance)
scale = 0.01*tf.ones(tf.shape(loc))
return tfd.MultivariateNormalDiag(loc, scale)
#KL term for z
def z_given_x(x, latent_size): #+
with tf.variable_scope('M/z_given_x'):
act = tf.nn.leaky_relu
h = x
h = tf.layers.dense(h, 256, act)
h = tf.layers.dense(h, 128, act)
h = tf.layers.dense(h, 64, act)
h = tf.layers.dense(h, 32, act)
h = tf.layers.dense(h, 16, act)
h = tf.layers.dense(h, 8, act)
loc = tf.layers.dense(h,latent_size)
log_variance = tf.layers.dense(h, latent_size)
scale = tf.nn.softplus(log_variance)
# scale = 0.01*tf.ones(tf.shape(loc))
return tfd.MultivariateNormalDiag(loc, scale)
def z_given(latent_size):
with tf.variable_scope('M/z_given'):
loc = tf.zeros(latent_size)
scale = 0.01*tf.ones(tf.shape(loc))
return tfd.MultivariateNormalDiag(loc, scale)
#Connect encoder and decoder and define the loss function
tf.reset_default_graph()
x_in = tf.placeholder(tf.float32, shape=[None, X.shape[1]], name='x_in')
x_out = tf.placeholder(tf.float32, shape=[None, X.shape[1]], name='x_out')
z_latent_size = 2
beta = 0.000001
#KL_z
zI = z_given(z_latent_size)
zIx = z_given_x(x_in, z_latent_size)
zIx_sample = zIx.sample()
zIx_mean = zIx.mean()
#kl_z = tf.reduce_mean(zIx.log_prob(zIx_sample)- zI.log_prob(zIx_sample))
kl_z = tf.reduce_mean(tfd.kl_divergence(zIx, zI)) #analytical
#Reconstruction
xIz = x_given_z(zIx_sample, X.shape[1])
rec_out = xIz.mean()
rec_loss = tf.losses.mean_squared_error(x_out, rec_out)
loss = rec_loss + beta*kl_z
optimizer = tf.train.AdamOptimizer(0.001).minimize(loss)
#Helper function
def batch_generator(features, x, u, batch_size):
"""Function to create python generator to shuffle and split features into batches along the first dimension."""
idx = np.arange(features.shape[0])
np.random.shuffle(idx)
for start_idx in range(0, features.shape[0], batch_size):
end_idx = min(start_idx + batch_size, features.shape[0])
part = idx[start_idx:end_idx]
yield features[part,:], x[part,:] , u[part, :]
n_epochs = 5000
batch_size = X.shape[0]
start = time.time()
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for i in range(n_epochs):
gen = batch_generator(X, X, U, batch_size) #create batch generator
rec_loss_ = 0
kl_z_ = 0
for j in range(np.int(X.shape[0]/batch_size)):
x_in_batch, x_out_batch, u_batch = gen.__next__()
_, rec_loss__, kl_z__= sess.run([optimizer, rec_loss, kl_z], feed_dict={x_in: x_in_batch, x_out: x_out_batch})
rec_loss_ += rec_loss__
kl_z_ += kl_z__
if (i+1)% 50 == 0 or i == 0:
zIx_mean_, rec_out_= sess.run([zIx_mean, rec_out], feed_dict ={x_in:X, x_out:X})
end = time.time()
print('epoch: {0}, rec_loss: {1:.3f}, kl_z: {2:.2f}'.format((i+1), rec_loss_/(1+np.int(X.shape[0]/batch_size)), kl_z_/(1+np.int(X.shape[0]/batch_size))))
start = time.time()
from sklearn.decomposition import TruncatedSVD
svd = TruncatedSVD(n_components=2, n_iter=7, random_state=42)
svd.fit(U.T)
print(svd.explained_variance_ratio_)
print(svd.explained_variance_ratio_.sum())
print(svd.singular_values_)
U_ = svd.components_
U_ = U_.T
import matplotlib.pyplot as plt
fig, axs = plt.subplots(1, 2, figsize=(14,5))
axs[0].scatter(zIx_mean_[:,0],zIx_mean_[:,1], c=U_[:,0], cmap='viridis', s=5.0);
axs[0].set_xlabel('z1')
axs[0].set_ylabel('z2')
fig.suptitle('X1')
plt.show()
fig, axs = plt.subplots(1, 2, figsize=(14,5))
axs[0].scatter(wIxy_mean_[:,0],wIxy_mean_[:,1], c=U_[:,1], cmap='viridis', s=5.0);
axs[0].set_xlabel('w1')
axs[0].set_ylabel('w2')
axs[1].scatter(zIx_mean_[:,0],zIx_mean_[:,1], c=U_[:,1], cmap='viridis', s=5.0);
axs[1].set_xlabel('z1')
axs[1].set_ylabel('z2')
fig.suptitle('X1')
plt.show()
error = np.abs(X-rec_out_)
plt.plot(np.reshape(error, -1), '*', markersize=0.1);
plt.hist(np.reshape(error, -1), bins=50);
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.