text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
---
_You are currently looking at **version 1.1** of this notebook. To download notebooks and datafiles, as well as get help on Jupyter notebooks in the Coursera platform, visit the [Jupyter Notebook FAQ](https://www.coursera.org/learn/python-data-analysis/resources/0dhYG) course resource._
---
# Assignment 2 - Pandas Introduction
All questions are weighted the same in this assignment.
## Part 1
The following code loads the olympics dataset (olympics.csv), which was derrived from the Wikipedia entry on [All Time Olympic Games Medals](https://en.wikipedia.org/wiki/All-time_Olympic_Games_medal_table), and does some basic data cleaning.
The columns are organized as # of Summer games, Summer medals, # of Winter games, Winter medals, total # number of games, total # of medals. Use this dataset to answer the questions below.
```
import pandas as pd
df = pd.read_csv('olympics.csv', index_col=0, skiprows=1)
for col in df.columns:
if col[:2]=='01':
df.rename(columns={col:'Gold'+col[4:]}, inplace=True)
if col[:2]=='02':
df.rename(columns={col:'Silver'+col[4:]}, inplace=True)
if col[:2]=='03':
df.rename(columns={col:'Bronze'+col[4:]}, inplace=True)
if col[:1]=='№':
df.rename(columns={col:'#'+col[1:]}, inplace=True)
names_ids = df.index.str.split('\s\(') # split the index by '('
df.index = names_ids.str[0] # the [0] element is the country name (new index)
df['ID'] = names_ids.str[1].str[:3] # the [1] element is the abbreviation or ID (take first 3 characters from that)
df = df.drop('Totals')
df.head()
```
### Question 0 (Example)
What is the first country in df?
*This function should return a Series.*
```
# You should write your whole answer within the function provided. The autograder will call
# this function and compare the return value against the correct solution value
def answer_zero():
# This function returns the row for Afghanistan, which is a Series object. The assignment
# question description will tell you the general format the autograder is expecting
return df.iloc[0]
# You can examine what your function returns by calling it in the cell. If you have questions
# about the assignment formats, check out the discussion forums for any FAQs
answer_zero()
```
### Question 1
Which country has won the most gold medals in summer games?
*This function should return a single string value.*
```
def answer_one():
return "YOUR ANSWER HERE"
```
### Question 2
Which country had the biggest difference between their summer and winter gold medal counts?
*This function should return a single string value.*
```
def answer_two():
return "YOUR ANSWER HERE"
```
### Question 3
Which country has the biggest difference between their summer gold medal counts and winter gold medal counts relative to their total gold medal count?
$$\frac{Summer~Gold - Winter~Gold}{Total~Gold}$$
Only include countries that have won at least 1 gold in both summer and winter.
*This function should return a single string value.*
```
def answer_three():
return "YOUR ANSWER HERE"
```
### Question 4
Write a function to update the dataframe to include a new column called "Points" which is a weighted value where each gold medal counts for 3 points, silver medals for 2 points, and bronze mdeals for 1 point. The function should return only the column (a Series object) which you created.
*This function should return a Series named `Points` of length 146*
```
def answer_four():
return "YOUR ANSWER HERE"
```
## Part 2
For the next set of questions, we will be using census data from the [United States Census Bureau](http://www.census.gov/popest/data/counties/totals/2015/CO-EST2015-alldata.html). Counties are political and geographic subdivisions of states in the United States. This dataset contains population data for counties and states in the US from 2010 to 2015. [See this document](http://www.census.gov/popest/data/counties/totals/2015/files/CO-EST2015-alldata.pdf) for a description of the variable names.
The census dataset (census.csv) should be loaded as census_df. Answer questions using this as appropriate.
### Question 5
Which state has the most counties in it? (hint: consider the sumlevel key carefully! You'll need this for future questions too...)
*This function should return a single string value.*
```
census_df = pd.read_csv('census.csv')
census_df.head()
def answer_five():
return "YOUR ANSWER HERE"
```
### Question 6
Only looking at the three most populous counties for each state, what are the three most populous states (in order of highest population to lowest population)?
*This function should return a list of string values.*
```
def answer_six():
return "YOUR ANSWER HERE"
```
### Question 7
Which county has had the largest absolute change in population within the period 2010-2015? (Hint: population values are stored in columns POPESTIMATE2010 through POPESTIMATE2015, you need to consider all six columns.)
e.g. If County Population in the 5 year period is 100, 120, 80, 105, 100, 130, then its largest change in the period would be |130-80| = 50.
*This function should return a single string value.*
```
def answer_seven():
return "YOUR ANSWER HERE"
```
### Question 8
In this datafile, the United States is broken up into four regions using the "REGION" column.
Create a query that finds the counties that belong to regions 1 or 2, whose name starts with 'Washington', and whose POPESTIMATE2015 was greater than their POPESTIMATE 2014.
*This function should return a 5x2 DataFrame with the columns = ['STNAME', 'CTYNAME'] and the same index ID as the census_df (sorted ascending by index).*
```
def answer_eight():
return "YOUR ANSWER HERE"
```
|
github_jupyter
|
```
pip install nltk
import nltk
import string
import re
texto_original = """Algoritmos inteligentes de aprendizados correndo supervisionados utilizam dados coletados. A partir dos dados coletados, um conjunto de característica é extraído. As características podem ser estruturais ou estatísticas. Correr correste corrida inteligente. As característica estruturais estabelecem relações entre os dados, inteligência enquanto que as estatísticas são características quantitativas. A partir das corrido características, os modelos de aprendizado de máquina corremos são construídos para o reconhecimento de atividades humanas."""
texto_original
#texto_original = re.sub(r'\s+', '', texto_original)
#texto_original
def converte_para_minusculas(texto):
texto_formatado = texto.lower()
return texto_formatado
texto_formatado = converte_para_minusculas(texto_original)
texto_formatado
nltk.download('stopwords')
stopwords = nltk.corpus.stopwords.words('portuguese')
print(stopwords)
len(stopwords)
stopwords.append('ola')
print(stopwords)
stopwords.remove('ola')
print(stopwords)
def remove_stopwords(texto):
texto_formatado = converte_para_minusculas(texto)
tokens = [] # Lista de tokens
for token in nltk.word_tokenize(texto_formatado): # token é cada uma das palavras
tokens.append(token) # Insere na lista de tokens
# Pega apenas os tokens que não estão nas stopwords
tokens = [cada_token for cada_token in tokens if cada_token not in stopwords and cada_token not in string.punctuation]
texto_formatado = ' '.join([str(cada_token) for cada_token in tokens if not cada_token.isdigit()])
return texto_formatado
texto_formatado = remove_stopwords(texto_original)
string.punctuation
frequencia_palavras = nltk.FreqDist(nltk.word_tokenize(texto_formatado))
frequencia_palavras
frequencia_palavras.keys()
frequencia_maxima = max(frequencia_palavras.values())
frequencia_maxima
for cada_palavra in frequencia_palavras.keys():
frequencia_palavras[cada_palavra] = frequencia_palavras[cada_palavra]/frequencia_maxima
frequencia_palavras
sentencas = nltk.sent_tokenize(texto_original)
sentencas
notas_das_sentencas = {}
for cada_sentenca in sentencas:
# print(cada_sentenca)
for cada_palavra in nltk.word_tokenize(cada_sentenca.lower()):
# print(cada_palavra)
if cada_palavra in frequencia_palavras.keys():
if cada_sentenca not in notas_das_sentencas:
notas_das_sentencas[cada_sentenca] = frequencia_palavras[cada_palavra]
else:
notas_das_sentencas[cada_sentenca] += frequencia_palavras[cada_palavra]
notas_das_sentencas
import heapq
melhores_sentencas = heapq.nlargest(3, notas_das_sentencas, key = notas_das_sentencas.get)
melhores_sentencas
resumo = ''.join(melhores_sentencas)
resumo
from IPython.core.display import HTML
texto_final = ''
display(HTML(f'<h1> RESUMO GERADO AUTOMATICAMENTE </h1>'))
for cada_sentenca in sentencas:
#texto_final = ''
#texto_final += cada_sentenca
if cada_sentenca in melhores_sentencas:
texto_final += str(cada_sentenca).replace(cada_sentenca, f"<mark>{cada_sentenca}</mark>")
else:
texto_final += str(cada_sentenca)
texto_final += ' '
display(HTML(f"""{texto_final}"""))
pip install goose3
```
# Tarefa 1
## 1. Stemizacao
```
from nltk.stem.snowball import SnowballStemmer
# É importante definir a lingua
stemizador = SnowballStemmer('portuguese')
palavras_stemizadas = []
for palavra in nltk.word_tokenize(texto_formatado):
print(palavra, ' = ', stemizador.stem(palavra))
palavras_stemizadas.append(stemizador.stem(palavra))
print(palavras_stemizadas)
resultado = ' '.join([str(cada_token) for cada_token in palavras_stemizadas if not cada_token.isdigit()])
print(resultado)
frequencia_palavras = nltk.FreqDist(nltk.word_tokenize(resultado))
frequencia_palavras
frequencia_maxima = max(frequencia_palavras.values())
frequencia_maxima
for cada_palavra in frequencia_palavras.keys():
frequencia_palavras[cada_palavra] = frequencia_palavras[cada_palavra]/frequencia_maxima
frequencia_palavras
notas_das_sentencas = {}
for cada_sentenca in sentencas:
# print(cada_sentenca)
for cada_palavra in nltk.word_tokenize(cada_sentenca.lower()):
# print(cada_palavra)
aux = stemizador.stem(cada_palavra)
if aux in frequencia_palavras.keys():
if cada_sentenca not in notas_das_sentencas:
notas_das_sentencas[cada_sentenca] = frequencia_palavras[aux]
else:
notas_das_sentencas[cada_sentenca] += frequencia_palavras[aux]
notas_das_sentencas
melhores_sentencas = heapq.nlargest(3, notas_das_sentencas, key = notas_das_sentencas.get)
melhores_sentencas
resumo = ''.join(melhores_sentencas)
resumo
texto_final = ''
display(HTML(f'<h1> RESUMO GERADO AUTOMATICAMENTE (Stemizadas)</h1>'))
for cada_sentenca in sentencas:
#texto_final = ''
#texto_final += cada_sentenca
if cada_sentenca in melhores_sentencas:
texto_final += str(cada_sentenca).replace(cada_sentenca, f"<mark>{cada_sentenca}</mark>")
else:
texto_final += str(cada_sentenca)
texto_final += ' '
display(HTML(f"""{texto_final}"""))
```
## 2. Lematizacao
```
import spacy
!python -m spacy download pt_core_news_sm
pln = spacy.load('pt_core_news_sm')
pln
palavras = pln(texto_formatado)
# Spacy já separa as palavras em tokens
palavras_lematizadas = []
for palavra in palavras:
#print(palavra.text, ' = ', palavra.lemma_)
palavras_lematizadas.append(palavra.lemma_)
print(palavras_lematizadas)
resultado = ' '.join([str(cada_token) for cada_token in palavras_lematizadas if not cada_token.isdigit()])
print(resultado)
frequencia_palavras = nltk.FreqDist(nltk.word_tokenize(resultado))
frequencia_palavras
frequencia_maxima = max(frequencia_palavras.values())
frequencia_maxima
for cada_palavra in frequencia_palavras.keys():
frequencia_palavras[cada_palavra] = frequencia_palavras[cada_palavra]/frequencia_maxima
frequencia_palavras
notas_das_sentencas = {}
for cada_sentenca in sentencas:
# print(cada_sentenca)
for cada_palavra in pln(cada_sentenca):
# print(cada_palavra)
aux = cada_palavra.lemma_
if aux in frequencia_palavras.keys():
if cada_sentenca not in notas_das_sentencas:
notas_das_sentencas[cada_sentenca] = frequencia_palavras[aux]
else:
notas_das_sentencas[cada_sentenca] += frequencia_palavras[aux]
notas_das_sentencas
melhores_sentencas = heapq.nlargest(3, notas_das_sentencas, key = notas_das_sentencas.get)
melhores_sentencas
resumo = ''.join(melhores_sentencas)
resumo
texto_final = ''
display(HTML(f'<h1> RESUMO GERADO AUTOMATICAMENTE (Lematizacao)</h1>'))
for cada_sentenca in sentencas:
#texto_final = ''
#texto_final += cada_sentenca
if cada_sentenca in melhores_sentencas:
texto_final += str(cada_sentenca).replace(cada_sentenca, f"<mark>{cada_sentenca}</mark>")
else:
texto_final += str(cada_sentenca)
texto_final += ' '
display(HTML(f"""{texto_final}"""))
```
# Fim da Tarefa 1
## Uso da lib Goose3
```
from goose3 import Goose
g = Goose()
url = 'https://www.techtudo.com.br/noticias/2017/08/o-que-e-replika-app-usa-inteligencia-artificial-para-criar-um-clone-seu.ghtml'
materia = g.extract(url)
materia.title
materia.tags
materia.infos
materia.cleaned_text
```
# Tarefa 2
```
frequencia_palavras.keys()
frequencia_palavras
frase = """Algoritmos de aprendizados supervisionados utilizam dados coletados""".split(' ')
frequencia_palavras_frase = []
for palavra in frase:
for freq_palavra in frequencia_palavras:
if palavra in freq_palavra:
frequencia_palavras_frase.append([palavra, 1])
frequencia_palavras_frase
for x in frequencia_palavras:
print(x)
```
|
github_jupyter
|
# Was Air Quality Affected in Countries or Regions Where COVID-19 was Most Prevalent?
**By: Arpit Jain, Maria Stella Vardanega, Tingting Cao, Christopher Chang, Mona Ma, Fusu Luo**
---
## Outline
#### I. Problem Definition & Data Source Description
1. Project Objectives
2. Data Source
3. Dataset Preview
#### II. What are the most prevalent pollutants?
#### III. What were the pollutant levels in 2019 and 2020 globally, and their averages?
1. Selecting Data from 2019 and 2020 with air pollutant information
2. Monthly Air Pollutant Data from 2019
3. Monthly Air Pollutant Data from 2020
#### IV: What cities had the highest changes in pollutant air quality index during COVID-19?
1. 10 cities with most air quality index reduction for each pollutant
2. Cities with more than 50 percent AQI decrease and 50 AQI decrease for each air pollutants
#### V: Regression analysis on COVID-19 cases and pollutant Air Quality Index Globally
#### VI: When were lockdowns implemented for each country?
#### VII: How did Air Quality change in countries with low COVID-19 cases (NZ, AUS, TW) and high COVID-19 cases (US, IT,CN)?
1. Countries with high COVID cases
2. Countries with low COVID cases
#### VIII: Conclusion
#### IX: Public Tableau Dashboards
---
## I. Problem Definition & Data Source Description
#### 1. Project Objectives
Air pollution, as one of the most serious environmental problems confronting our civilization, is the presence of toxic gases and particles in the air at levels that pose adverse effects on global climate and lead to public health risk and disease. Exposure to elevated levels of air pollutants has been implicated in a diverse set of medical conditions including cardiovascular and respiratory mortality, lung cancer and autism.
Air pollutants come from natural sources such as wildfires and volcanoes, as well as are highly related to human activities from mobile sources (such as cars, buses and planes) or stationary sources (such as industrial factories, power plants and wood burning fireplaces). However, in the past year, the COVID-19 pandemic has caused unprecedented changes to the our work, study and daily activities, subsequently led to major reductions in air pollutant emissions. And our team would like take this opportunity to examine the air quality in the past two years and look on how the air quality was impacted in countries and cities where the corona-virus was prevalent.
#### 2. Data Source
**Data Source Description:** In this project, we downloaded worldwide air quality data for Year 2019 and 2020 from the Air Quality Open Data Platform (https://aqicn.org/data-platform/covid19/), which provides historical air quality index and meteorological data for more than 380 major cities across the world. We used air quality index data in 2019 as baseline to find the air quality changes during COVID in 2020. In addition we joined the data with geographic location information from https://aqicn.org/data-platform/covid19/airquality-covid19-cities.json to get air quality index for each pollutant at city-level. According to the data source provider, the data for each major cities is based on the average (median) of several stations. The data set provides min, max, median and standard deviation for each of the air pollutant species in the form of Air Quality Index (AQI) that are converted from raw concentration based on the US Environmental Protection Agency (EPA) standard.
The United States EPA list the following as the criteria at this website (https://www.epa.gov/criteria-air-pollutants/naaqs-table): Carbon Monoxide (CO), Nitrogen Dioxide (NO2), Ozone (O3), Particle Pollution (PM2.5) + (PM10), and finally Sulfur Dioxide (SO2). For the particle pollution the numbers stand for the size of the particles. PM2.5 means particles that are 2.5 micrometers and smaller, while PM10 means particles that are 10 micrometers and smaller. https://www.epa.gov/pm-pollution/particulate-matter-pm-basics. Particle Pollution typically includes Dust, Dirt, and Smoke. Our dataset covers most of the criteria pollutants (PM2.5, PM10, Ozone, SO2, NO2 and CO), and meteorological parameters such as temperature, wind speed, dew point, relative humidity. Air quality index basics are shown in the figure below.
<img src="https://github.com/ttcao63/775team_project_b2_t2/blob/main/AQI%20basics.PNG?raw=true" align="center"/>
(source: https://www.airnow.gov/aqi/aqi-basics/)
#### 3. Preview of the Dataset
```
%%bigquery
SELECT * FROM `ba775-team2-b2.AQICN.air_quality_data` LIMIT 10
```
---
### II. What are the most prevalent pollutants?
This question focuses on the prevalence of the pollutants. From the dataset, the prevalence can be defined geographically from the cities and countries that had recorded the parameters detected times.
To find the prevalence, our team selected the parameters from situations in how many distinct cities and countries detected the parameter appeared.
```
%%bigquery
SELECT
Parameter,COUNT(distinct(City)) AS number_of_city,
COUNT(distinct(Country)) AS number_of_country,string_agg(distinct(Country)) AS list_country
FROM `ba775-team2-b2.AQICN.air_quality_data`
GROUP BY Parameter
ORDER BY number_of_city DESC
```
From the result, top 6 parameters are meteorological parameters. And the first air pollutants (which can be harmful to the public health and environment) is PM2.5, followed by NO2 and PM10.
PM2.5 has been detected in 548 cities and 92 countries.
NO2 has been detected in 528 cities and 64 countries.
PM10 has been detected in 527 cities and 71 countries.
We conclude PM2.5, NO2 and PM10 are the most prevalent criteria pollutants from the dataset. All of them are considered criteria pollutants set by EPA.
---
### III. What were the pollutant levels in 2019 and 2020 globally, and their averages?
The purpose of this question is to determine the air pollutant levels in 2019 and 2020. The air pollutant levels in 2019 serve as a baseline for the air pollutant levels in 2020. In the previous question we observe the distinct parameters that are within the Air Quality Database. Since the meteorological parameters are not needed for the project, we can exclude them, and only focus on the air pollutants.
The first step is create a table where the parameters are only air pollutants and from the years 2019 and 2020. The next step was to select all the rows from each year, that had a certain parameter, and to union them all. This process was done for all six parameters for both years.
#### 1. Selecting Data from 2019 and 2020 with air pollutant information
```
%%bigquery
SELECT Date, Country, City, lat as Latitude, lon as Longitude, pop as Population, Parameter as Pollutant, median as Pollutant_level
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE (extract(year from date) = 2019 OR extract(year from date) = 2020) AND parameter IN ('co', 'o3','no2','so2','pm10',
'pm25')
ORDER BY Country, Date;
```
As we can see after filtering the tables for only the air pollutants we have 1.9 million rows. From here we split the data into 2019 data and 2020 data.
#### 2. Monthly Air Pollutant Data from 2019
```
%%bigquery
SELECT extract(month from date) Month, Parameter as Pollutant,Round(avg(median),2) as Avg_Pollutant_Level_2019
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('co')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('o3')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('no2')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('so2')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('pm10')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2019 AND parameter IN ('pm25')
GROUP BY Month, Parameter
ORDER BY Month;
```
This query represents the average pollutant level for each air pollutant globally for each month. We do this again for the 2020 data.
#### 3. Monthly Air Pollutant Data from 2020
```
%%bigquery
SELECT extract(month from date) Month, Parameter as Pollutant,Round(avg(median),2) as Avg_Pollutant_Level_2020
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('co')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('o3')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('no2')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('so2')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('pm10')
GROUP BY Month, Parameter
UNION ALL
SELECT extract(month from date) Month, Parameter ,Round(avg(median),2)
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE extract(year from date) = 2020 AND parameter IN ('pm25')
GROUP BY Month, Parameter
ORDER BY Month;
```
When comparing the data there isn't a noticeable difference in global pollutant levels from 2019 to 2020, which leads to the hypothesis of pollutant levels being regional rather than global. This might also mean that whatever effects might be occurring from COVID-19 cases, and lockdowns are short-term enough that the average monthly air pollutant is not capturing small intricacies in the data. We can further narrow down the data by analyzing data from when lockdowns were occurring in different countries, regions, and even cities.
---
### IV: What cities had the highest changes in pollutant air quality index during COVID-19?
In this question, we are trying to find cities with most air quality improvement during COVID, and cities with longest time of certain level AQI reduction.
#### 1. 10 cities with most air quality index reduction for each pollutant
Making queries and creating tables to find monthly average air quality index (AQI) for all pollutants at city level
We are using data in 2019 as a baseline and computing AQI differences and percent differences. Negative difference values indicates air quality index decrease, corresponding to an air quality improvement, and positive difference values indicate air quality index increases, corresponding to an air quality deterioration.
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.pollutant_diff_daily_aqi_less_than_500
AS
(
SELECT A.Date AS Date_2020,B.Date AS Date_2019,A.Country,A.City,A.lat,A.lon,A.Parameter,A.pop,A.median AS aqi_2020,B.median AS aqi_2019,(A.median-B.median) AS aqi_diff, ROUND((A.median-B.median)/B.median*100,2) AS aqi_percent_diff
FROM
(SELECT * FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE Parameter in ('pm25','pm10','o3','no2','co','so2') AND EXTRACT(Year FROM Date) = 2020 AND median > 0 AND median < 500) AS A
INNER JOIN
(SELECT * FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE Parameter in ('pm25','pm10','o3','no2','co','so2') AND EXTRACT(Year FROM Date) = 2019 AND median > 0 AND median < 500) AS B
ON A.City = B.City
WHERE EXTRACT(MONTH FROM A.Date) = EXTRACT(MONTH FROM B.Date) AND EXTRACT(DAY FROM A.Date) = EXTRACT(DAY FROM B.Date) AND A.Parameter = B.Parameter
ORDER BY City,Date_2020
)
%%bigquery
CREATE OR REPLACE TABLE AQICN.pollutant_diff_monthly_aqi
AS
SELECT EXTRACT(month FROM Date_2020) AS month_2020,EXTRACT(month FROM Date_2019) AS month_2019,
Country,City,lat,lon,Parameter,ROUND(AVG(aqi_2020),1) AS monthly_avg_aqi_2020,
ROUND(AVG(aqi_2019),1) AS monthly_avg_aqi_2019,(ROUND(AVG(aqi_2020),1)-ROUND(AVG(aqi_2019),1)) AS aqi_diff_monthly,
ROUND((AVG(aqi_2020)-AVG(aqi_2019))/AVG(aqi_2019)*100,2) AS aqi_percent_diff_monthly
FROM AQICN.pollutant_diff_daily_aqi_less_than_500
GROUP BY month_2020,month_2019,Country,City,lat,lon,Parameter
%%bigquery
SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
ORDER BY Parameter,month_2020,Country
LIMIT 10
```
Order by monthly average AQI difference to find cities having top 10 air quality index reduction for each pollutant
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.top_10_cites_most_pollutant_percent_diff_monthly
AS
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'co'
ORDER BY aqi_percent_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'o3'
ORDER BY aqi_percent_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'no2'
ORDER BY aqi_percent_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'pm25'
ORDER BY aqi_percent_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'pm10'
ORDER BY aqi_percent_diff_monthly
LIMIT 10)
%%bigquery
SELECT *
FROM AQICN.top_10_cites_most_pollutant_percent_diff_monthly
ORDER BY Parameter,aqi_percent_diff_monthly
LIMIT 10
```
Order by monthly average percent AQI difference to find cities having top 10 most air quality index reduction for each pollutant
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.top_10_cites_most_pollutant_diff_monthly
AS
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'pm25'
ORDER BY aqi_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'o3'
ORDER BY aqi_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'pm10'
ORDER BY aqi_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'no2'
ORDER BY aqi_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'so2'
ORDER BY aqi_diff_monthly
LIMIT 10)
UNION ALL
(SELECT *
FROM AQICN.pollutant_diff_monthly_aqi
WHERE Parameter = 'co'
ORDER BY aqi_diff_monthly
LIMIT 10)
%%bigquery
SELECT *
FROM AQICN.top_10_cites_most_pollutant_diff_monthly
ORDER BY Parameter,aqi_diff_monthly
LIMIT 10
```
#### 2. Cities with more than 50 percent AQI decrease and 50 AQI decrease for each air pollutants
Reason: the higher the AQI, the unhealthier the air will be, especially for sensitive groups such as people with heart and lung disease, elders and children. A major reduction or percent reduction in AQI for long period of time implies a high air quality impact from the COIVD pandemic.
```
%%bigquery
SELECT City,Country,Parameter,COUNT(*) AS num_month_mt_50_per_decrease FROM AQICN.pollutant_diff_monthly_aqi
WHERE aqi_percent_diff_monthly < -50 AND aqi_diff_monthly < -50
GROUP BY City,Country,Parameter
ORDER BY Parameter,COUNT(*) DESC
LIMIT 10
```
---
Results
During the pandemic, cities getting most air qualities improvements in terms of percent AQI differences for each pollutant are:
CO: United States Portland, Chile Talca and Mexico Aguascalientes;
NO2: Iran Qom, South Africa Middelburg and Philippines Butuan;
SO2: Greece Athens, Mexico Mérida and Mexico San Luis Potosí;
Ozone: Mexico Aguascalientes, United States Queens and United States The Bronx;
PM 10: India Gandhinagar, China Hohhot and Israel Tel Aviv;
PM 2.5: Mexico Mérida, Tajikistan Dushanbe, Bosnia and Herzegovina Sarajevo, Turkey Erzurum, China Qiqihar and India Gandhinagar;
Cities getting at least 50% and 50 AQI reduction with longest time:
CO: United States Portland, 3 out of 12 months;
NO2: Iran Qom, 5 out of 12 months;
O3: Mexico Aguascalientes, 5 out of 12 months;
PM25: several cities including Iran Kermanshah, Singapore Singapore, AU Sydney and Canberra, 1 out of 12 months;
PM10: India Gandhinagar and Bhopal, 2 out of 12 months;
SO2: Mexico Mérida 5 out of 12 months.
---
### V: Regression analysis on COVID-19 cases and pollutant Air Quality Index Globally
The purpose of this part is to find the differences in AQI between 2019 and 2020, also the percentage changes for four parameters which include (NO,NO2,PM2.5 and O3), then join with the COVID confirmed table to find the regression between the AQI and the new confirmed case for each air pollutant.
```
%%bigquery
select A.month,A.month_n, A.country,A.parameter,round((B.avg_median_month- A.avg_median_month),2) as diff_avg,
(B.avg_median_month - A.avg_median_month)/A.avg_median_month as diff_perc
from
(SELECT FORMAT_DATETIME("%B", date) month,EXTRACT(year FROM date) year, EXTRACT(month FROM date) month_n, country,parameter,round(avg(median),2) as avg_median_month
FROM `AQICN.Arpit_Cleaned_Data2`
WHERE Parameter IN ('co','no2','o3','pm25') AND EXTRACT(year FROM date) = 2019
GROUP by 1,2,3,4,5
ORDER BY country, parameter) A
left join
(SELECT FORMAT_DATETIME("%B", date) month,EXTRACT(year FROM date) year, EXTRACT(month FROM date) month_n, country,parameter,round(avg(median),2) as avg_median_month
FROM `AQICN.Arpit_Cleaned_Data2`
WHERE Parameter IN ('co','no2','o3','pm25') AND EXTRACT(year FROM date) = 2020
GROUP by 1,2,3,4,5
ORDER BY country, parameter) B
using (month,country,parameter,month_n)
where A.avg_median_month >0
%%bigquery
select A.*,confirmed,B.country as country_name
from `all_para_20_19.all_para_20_19_diff` as A
inner join `covid_population.covid _pop` as B
on A.country = B.country_code2 and A.month = B.month and A.month_n = B.month_n
where B.year = 2020
order by A.country,A.month_n
```
Using Bigquery ML to find liner regression between diff_avg for each parameter and confirmed cases
(Example showing below is that parameter = co; x = confirmed; y=diff_avg --AQI changes)
```
%%bigquery
CREATE OR REPLACE MODEL `all_para_20_19.all_para_20_19_diff_covid_model`
# Specify options
OPTIONS
(model_type='linear_reg',
input_label_cols=['diff_avg']) AS
# Provide training data
SELECT
confirmed,
diff_avg
FROM
`all_para_20_19.all_para_20_19_diff_covid`
WHERE
parameter = 'co'
and diff_avg is not null
```
Evaluating the model to find out r2_score for each monthly average air pollutant AQI changes vs monthly confirmed new cases linear regression model.
Example showing below is Evaluation for country level monthly average CO AQI vs monthly new confirmed COVID cases model:
```
%%bigquery
SELECT * FROM
ML.EVALUATE(
MODEL `all_para_20_19.all_para_20_19_diff_covid_model`, # Model name
# Table to evaluate against
(SELECT
confirmed,
diff_avg
FROM
`all_para_20_19.all_para_20_19_diff_covid`
WHERE
parameter = 'co'
and diff_avg is not null
)
)
```
Evaluation for country level monthly average PM2.5 AQI changes vs monthly new confirmed COVID cases model:
<img src="https://github.com/ttcao63/775team_project_b2_t2/blob/main/pm25_aqi_confirmed_case.png?raw=true" align="center" width="800"/>
Evaluation for country level monthly average NO2 AQI changes vs monthly new confirmed COVID cases model:
<img src="https://github.com/ttcao63/775team_project_b2_t2/blob/main/no2_aqi_confirmed_case.png?raw=true" align="center" width="800"/>
Evaluation for country level monthly average O3 AQI changes vs monthly new confirmed COVID cases model:
<img src="https://github.com/ttcao63/775team_project_b2_t2/blob/main/o3_aqi_confirmed_case.png?raw=true" align="center" width="800"/>
We have also conducted log transformation of x-variables for linear regression, the most correlated data is PM 2.5 AQI changes vs LOG(confirmed case). Visualization is shown below.
<img src="https://github.com/ttcao63/775team_project_b2_t2/blob/main/Viz_PM25_Regression.png?raw=true" align="center" width="800"/>
We can see an overall AQI changes from 2019 to 2020. However, after running regression for four air pollutants, model R-squares are less than 0.02, indicating a weak linear relationship between the air quality index changes and the numbers of new confirmed COVID cases. The result makes sense because there are complicated physical and chemical process involved in formation and transportation of air pollution, thus factors such as the weather, energy source, and terrain could also impact the AQI changes. Also, the dramatic increase of new COVID cases might not affect people's response in a way reducing outdoor activities, especially when "stay at home order" is partially lifted.
In this case, we decide to specifically study some countries during their lockdown period and examine the AQI changes.
---
### VI: When were lockdowns implemented for each country?
Lockdown Dates per Country
China: Jan 23 - April 8, 2020 (Wuhan 76 day lockdown)
USA: March 19 - April 7, 2020
Italy: March 9 - May 18, 2020
Taiwan: No lockdowns in 2020. Lockdown started in July 2021.
Australia: March 18 - May/June 2020
New Zealand: March 25 - May/June 2020
From the previous regression model we can see that the there was very little correlation between AQI and confirmed cases, and one of the main reasons is that confirmed cases could not accurately capture human activity. To compensate for this, we narrowed down the dates of our pollutant data in order to compare the pollutant levels only during lockdown periods in 2019 and 2020 for the countries where COVID-19 was most prevalent: China, USA, Italy, and those that COVID-19 wasn't as prevalent: Taiwan, Australia, and New Zealand. We came to a conclusion that most lockdown periods started from mid March to April, May, or June, except for China, which started their lockdown late January until April of 2020. To generalize the lockdown dates for countries other than China, the SQL query included dates from the beginning of March to the end of June. As for China, the query included specific dates from January 23 to April 8th of 2020, which is the Wuhan 76 day lockdown.
```
%%bigquery
SELECT country, date, parameter, AVG(count) AS air_quality
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2020-03-01' AND '2020-06-30'
AND country in ('US','IT','AU','NZ','TW')
GROUP BY country, parameter, date
ORDER BY date
%%bigquery
SELECT country, date, parameter, AVG(count) AS air_quality
FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2020-01-23' AND '2020-04-08'
AND country = 'CN'
GROUP BY country, parameter, date
ORDER BY date
```
---
### VII: How did Air Quality change in countries with low COVID-19 cases (NZ, AUS, TW) and high COVID-19 cases (US, IT,CN)?
This question was answered by creating separate tables that encompassed the equivalent lockdown periods per country for 2019. Then, the two tables were joined using the parameter and grouped according to country and parameter to create a subsequent table illustrating the percentage change in average pollution from 2019 to 2020 (during the respective lockdown periods).
#### 1. Countries with high COVID cases
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_Italy AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2019-03-09' AND '2019-05-18'
AND country = 'IT'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_Italy AS a2019
USING(parameter)
WHERE a2020.date BETWEEN '2020-03-09' AND '2020-05-18'
AND a2020.country = 'IT'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
Here we can see that the only pollutant that decreased during the 2020 lockdown in Italy, compared to the respective time period in 2019, was NO2, which decreased by 35.74%.
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_US AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2019-03-19' AND '2019-04-07'
AND country = 'US'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_US AS a2019
USING(parameter)
WHERE a2020.date BETWEEN '2020-03-19' AND '2020-04-07'
AND a2020.country = 'US'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
In the United States, all the pollutants decreased in 2020 compared to 2019. The largest changes occurred in O3, NO2 and SO2, which decreased by 36.69%, 30.22%, and 27.10% respectively. This indicates that the lockdowns during the COVID-19 pandemic may have positively affected the emission of pollutants in the United States.
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_China AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2019-01-23' AND '2019-04-08'
AND country = 'CN'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_China AS a2019
USING(parameter)
WHERE a2020.date BETWEEN '2020-01-23' AND '2020-04-08'
AND a2020.country = 'CN'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
In China, most pollutants decreased in 2020 compared to the same period in 2019. The largest change was in NO2 which decreased by 30.88% compared to the previous year.
#### 2. Countries with low COVID cases
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_Taiwan AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE EXTRACT(month FROM date) = 07
AND EXTRACT(year FROM date) = 2019
AND country = 'TW'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_Taiwan AS a2019
USING(parameter)
WHERE EXTRACT(month FROM a2020.date) = 07
AND EXTRACT(year FROM a2020.date) = 2020
AND a2020.country = 'TW'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
Taiwan, which did not experience lockdowns due to COVID-19, also shows a decrease in all pollutant levels. This contradicts our initially hypothesis that countries who experienced more COVID-19 and therefore more lockdowns would have better air quality.
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_AUS AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2019-03-25' AND '2019-05-31'
AND country = 'NZ'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_NZ AS a2019
USING(parameter)
WHERE a2020.date BETWEEN '2020-03-25' AND '2020-05-31'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
AND a2020.country = 'NZ'
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
New Zealand also shows a decrease in all pollutant levels. Nevertheless, New Zealand did go into lockdown for a period and these numbers may reflect the lessened activity due to COVID-19 during that time compared to the equivalent in 2019.
```
%%bigquery
CREATE OR REPLACE TABLE AQICN.air_quality2019_AUS AS
SELECT country, parameter, median AS air_quality2019 FROM `ba775-team2-b2.AQICN.air_quality_data`
WHERE date BETWEEN '2019-03-18' AND '2019-05-31'
AND country = 'AU'
%%bigquery
SELECT a2020.country, a2020.parameter, AVG(a2020.median) AS air_quality2020, AVG(air_quality2019) AS air_quality2019,
(AVG(a2020.median)-AVG(air_quality2019))/AVG(air_quality2019) AS percentage_change
FROM `ba775-team2-b2.AQICN.air_quality_data` AS a2020
LEFT JOIN AQICN.air_quality2019_AUS AS a2019
USING(parameter)
WHERE a2020.date BETWEEN '2020-03-18' AND '2020-05-31'
AND Parameter in ('pm25','pm10','o3','no2','co','so2')
AND a2020.country = 'AU'
GROUP BY a2020.country, a2020.parameter
ORDER BY percentage_change
```
Australia shows decreases in most pollutant parameter levels in 2020 compared to respective periods in 2019.
The fact that all tables illustrate decrease in most pollutant parameter levels, except for Italy, seems to contradict our initial hypothesis. Initially, we hypothesized that in countries where COVID-19 was more prevalent, and therefore where there were more lockdowns and less human activity, there would be better pollutant levels. However, when looking at the results of the analysis, one can see that the extent to which COVID-19 was prevalent does not seem to largely affect the pollutant parameter levels considering that regardless of the country they seem to have decreased in 2020 compared to 2019. This may be due to various governmental and public policies regarding climate change that have pushed countries to improve the air quality as well as the general decrease in human activity worldwide due to the pandemic.
---
### VIII: Conclusion
In this project, we used air quality index (AQI) dataset among 380 major cities across the world from 2019 to 2020 to study criteria air pollutant level changes during the COVID pandemic. According to the result, we conclude that the COVID impacts air quality more at regional-level than at global-level, more in a relative short period of time than in a relative long-term. Even though we don't see a strong relationship between air quality changes versus numbers of confirmed COIVD case, we find that lockdowns during the pandemic do make effects on air pollutant levels in different countries due to reduced outdoor human activities.
---
### IX: Public Tableau Dashboards
To interact with our public Tableau dashboards please visit: https://public.tableau.com/app/profile/arpit.jain7335/viz/AnalyzingAirQualityDuringCOVID19Pandemic/AirQualityGlobalLevel?publish=yes
<img src="https://github.com/arp-jain/BA775-team2-b2/blob/main/Air%20Quality%20Global%20Level.png?raw=true" align="center"/>
<img src="https://github.com/arp-jain/BA775-team2-b2/blob/main/Air%20Quality%20City%20Level.png?raw=true" align="center"/>
|
github_jupyter
|
# Tensorflow Timeline Analysis on Model Zoo Benchmark between Intel optimized and stock Tensorflow
This jupyter notebook will help you evaluate performance benefits from Intel-optimized Tensorflow on the level of Tensorflow operations via several pre-trained models from Intel Model Zoo. The notebook will show users a bar chart like the picture below for the Tensorflow operation level performance comparison. The red horizontal line represents the performance of Tensorflow operations from Stock Tensorflow, and the blue bars represent the speedup of Intel Tensorflow operations. The operations marked as "mkl-True" are accelerated by MKL-DNN a.k.a oneDNN, and users should be able to see a good speedup for those operations accelerated by MKL-DNN.
> NOTE : Users need to get Tensorflow timeline json files from other Jupyter notebooks like benchmark_perf_comparison
first to proceed this Jupyter notebook.
<img src="images\compared_tf_op_duration_ratio_bar.png" width="700">
The notebook will also show users two pie charts like the picture below for elapsed time percentage among different Tensorflow operations.
Users can easily find the Tensorflow operation hotspots in these pie charts among Stock and Intel Tensorflow.
<img src="images\compared_tf_op_duration_pie.png" width="700">
# Get Platform Information
```
from profiling.profile_utils import PlatformUtils
plat_utils = PlatformUtils()
plat_utils.dump_platform_info()
```
# Section 1: TensorFlow Timeline Analysis
## Prerequisites
```
!pip install cxxfilt
%matplotlib inline
import matplotlib.pyplot as plt
import tensorflow as tf
import pandas as pd
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1500)
```
## List out the Timeline folders
First, list out all Timeline folders from previous runs.
```
import os
filenames= os.listdir (".")
result = []
keyword = "Timeline"
for filename in filenames:
if os.path.isdir(os.path.join(os.path.abspath("."), filename)):
if filename.find(keyword) != -1:
result.append(filename)
result.sort()
index =0
for folder in result:
print(" %d : %s " %(index, folder))
index+=1
```
## Select a Timeline folder from previous runs
#### ACTION: Please select one Timeline folder and change FdIndex accordingly
```
FdIndex = 3
```
List out all Timeline json files inside Timeline folder.
```
import os
TimelineFd = result[FdIndex]
print(TimelineFd)
datafiles = [TimelineFd +os.sep+ x for x in os.listdir(TimelineFd) if '.json' == x[-5:]]
print(datafiles)
if len(datafiles) is 0:
print("ERROR! No json file in the selected folder. Please select other folder.")
elif len(datafiles) is 1:
print("WARNING! There is only 1 json file in the selected folder. Please select other folder to proceed Section 1.2.")
```
> **Users can bypass below Section 1.1 and analyze performance among Stock and Intel TF by clicking the link : [Section 1_2](#section_1_2).**
<a id='section_1_1'></a>
## Section 1.1: Performance Analysis for one TF Timeline result
### Step 1: Pick one of the Timeline files
#### List out all the Timeline files first
```
index = 0
for file in datafiles:
print(" %d : %s " %(index, file))
index+=1
```
#### ACTION: Please select one timeline json file and change file_index accordingly
```
## USER INPUT
file_index=0
fn = datafiles[file_index]
tfile_prefix = fn.split('_')[0]
tfile_postfix = fn.strip(tfile_prefix)[1:]
fn
```
### Step 2: Parse timeline into pandas format
```
from profiling.profile_utils import TFTimelinePresenter
tfp = TFTimelinePresenter(True)
timeline_pd = tfp.postprocess_timeline(tfp.read_timeline(fn))
timeline_pd = timeline_pd[timeline_pd['ph'] == 'X']
```
### Step 3: Sum up the elapsed time of each TF operation
```
tfp.get_tf_ops_time(timeline_pd,fn,tfile_prefix)
```
### Step 4: Draw a bar chart for elapsed time of TF ops
```
filename= tfile_prefix +'_tf_op_duration_bar.png'
title_=tfile_prefix +'TF : op duration bar chart'
ax=tfp.summarize_barh(timeline_pd, 'arg_op', title=title_, topk=50, logx=True, figsize=(10,10))
tfp.show(ax,'bar')
```
### Step 5: Draw a pie chart for total time percentage of TF ops
```
filename= tfile_prefix +'_tf_op_duration_pie.png'
title_=tfile_prefix +'TF : op duration pie chart'
timeline_pd_known = timeline_pd[ ~timeline_pd['arg_op'].str.contains('unknown') ]
ax=tfp.summarize_pie(timeline_pd_known, 'arg_op', title=title_, topk=50, logx=True, figsize=(10,10))
tfp.show(ax,'pie')
ax.figure.savefig(filename,bbox_inches='tight')
```
<a id='section_1_2'></a>
## Section 1.2: Analyze TF Timeline results between Stock and Intel Tensorflow
### Speedup from MKL-DNN among different TF operations
### Step 1: Select one Intel and one Stock TF timeline files for analysis
#### List out all timeline files in the selected folder
```
if len(datafiles) is 1:
print("ERROR! There is only 1 json file in the selected folder.")
print("Please select other Timeline folder from beginnning to proceed Section 1.2.")
for i in range(len(datafiles)):
print(" %d : %s " %(i, datafiles[i]))
```
#### ACTION: Please select one timeline file as a perfomance baseline and the other as a comparison target
put the related index for your selected timeline file.
In general, please put stock_timeline_xxxxx as the baseline.
```
# perfomance baseline
Baseline_Index=1
# comparison target
Comparison_Index=0
```
#### List out two selected timeline files
```
selected_datafiles = []
selected_datafiles.append(datafiles[Baseline_Index])
selected_datafiles.append(datafiles[Comparison_Index])
print(selected_datafiles)
```
### Step 2: Parsing timeline results into CSV files
```
%matplotlib agg
from profiling.profile_utils import TFTimelinePresenter
csvfiles=[]
tfp = TFTimelinePresenter(True)
for fn in selected_datafiles:
if fn.find('/'):
fn_nofd=fn.split('/')[1]
else:
fn_nofd=fn
tfile_name= fn_nofd.split('.')[0]
tfile_prefix = fn_nofd.split('_')[0]
tfile_postfix = fn_nofd.strip(tfile_prefix)[1:]
csvpath = TimelineFd +os.sep+tfile_name+'.csv'
print(csvpath)
csvfiles.append(csvpath)
timeline_pd = tfp.postprocess_timeline(tfp.read_timeline(fn))
timeline_pd = timeline_pd[timeline_pd['ph'] == 'X']
tfp.get_tf_ops_time(timeline_pd,fn,tfile_prefix)
```
### Step 3: Pre-processing for the two CSV files
```
import os
import pandas as pd
csvarray=[]
for csvf in csvfiles:
print("read into pandas :",csvf)
a = pd.read_csv(csvf)
csvarray.append(a)
a = csvarray[0]
b = csvarray[1]
```
### Step 4: Merge two CSV files and caculate the speedup accordingly
```
import os
import pandas as pd
fdir='merged'
if not os.path.exists(fdir):
os.mkdir(fdir)
fpath=fdir+os.sep+'merged.csv'
merged=tfp.merge_two_csv_files(fpath,a,b)
merged
```
### Step 5: Draw a bar chart for elapsed time of TF ops among stock TF and Intel TF
```
%matplotlib inline
print(fpath)
tfp.plot_compare_bar_charts(fpath)
tfp.plot_compare_ratio_bar_charts(fpath, tags=['','oneDNN ops'])
```
### Step 6: Draw pie charts for elapsed time of TF ops among stock TF and Intel TF
```
tfp.plot_compare_pie_charts(fpath)
```
|
github_jupyter
|
<img src="../../img/logo_amds.png" alt="Logo" style="width: 128px;"/>
# AmsterdamUMCdb - Freely Accessible ICU Database
version 1.0.2 March 2020
Copyright © 2003-2020 Amsterdam UMC - Amsterdam Medical Data Science
# Vasopressors and inotropes
Shows medication for artificially increasing blood pressure (vasopressors) or stimulating heart function (inotropes), if any, a patient received.
## Imports
```
%matplotlib inline
import amsterdamumcdb
import psycopg2
import pandas as pd
import numpy as np
import re
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
import io
from IPython.display import display, HTML, Markdown
```
## Display settings
```
#matplotlib settings for image size
#needs to be in a different cell from %matplotlib inline
plt.style.use('seaborn-darkgrid')
plt.rcParams["figure.dpi"] = 288
plt.rcParams["figure.figsize"] = [16, 12]
plt.rcParams["font.size"] = 12
pd.options.display.max_columns = None
pd.options.display.max_rows = None
pd.options.display.max_colwidth = 1000
```
## Connection settings
```
#Modify config.ini in the root folder of the repository to change the settings to connect to your postgreSQL database
import configparser
import os
config = configparser.ConfigParser()
if os.path.isfile('../../config.ini'):
config.read('../../config.ini')
else:
config.read('../../config.SAMPLE.ini')
#Open a connection to the postgres database:
con = psycopg2.connect(database=config['psycopg2']['database'],
user=config['psycopg2']['username'], password=config['psycopg2']['password'],
host=config['psycopg2']['host'], port=config['psycopg2']['port'])
con.set_client_encoding('WIN1252') #Uses code page for Dutch accented characters.
con.set_session(autocommit=True)
cursor = con.cursor()
cursor.execute('SET SCHEMA \'amsterdamumcdb\''); #set search_path to amsterdamumcdb schema
```
## Vasopressors and inotropes
from drugitems
```
sql_vaso_ino = """
WITH vasopressor_inotropes AS (
SELECT
admissionid,
CASE
WHEN COUNT(*) > 0 THEN TRUE
ELSE FALSE
END AS vasopressors_inotropes_bool,
STRING_AGG(DISTINCT item, '; ') AS vasopressors_inotropes_given
FROM drugitems
WHERE
ordercategoryid = 65 -- continuous i.v. perfusor
AND itemid IN (
6818, -- Adrenaline (Epinefrine)
7135, -- Isoprenaline (Isuprel)
7178, -- Dobutamine (Dobutrex)
7179, -- Dopamine (Inotropin)
7196, -- Enoximon (Perfan)
7229, -- Noradrenaline (Norepinefrine)
12467, -- Terlipressine (Glypressin)
13490, -- Methyleenblauw IV (Methylthionide cloride)
19929 -- Fenylefrine
)
AND rate > 0.1
GROUP BY admissionid
)
SELECT
a.admissionid, location,
CASE
WHEN vi.vasopressors_inotropes_bool Then TRUE
ELSE FALSE
END AS vasopressors_inotropes_bool,
vasopressors_inotropes_given
FROM admissions a
LEFT JOIN vasopressor_inotropes vi ON
a.admissionid = vi.admissionid
"""
vaso_ino = pd.read_sql(sql_vaso_ino,con)
vaso_ino.tail()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/EvenSol/NeqSim-Colab/blob/master/notebooks/process/masstransferMeOH.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
#@title Calculation of mass transfer and hydrate inhibition of a wet gas by injection of methanol
#@markdown Demonstration of mass transfer calculation using the NeqSim software in Python
#@markdown <br><br>This document is part of the module ["Introduction to Gas Processing using NeqSim in Colab"](https://colab.research.google.com/github/EvenSol/NeqSim-Colab/blob/master/notebooks/examples_of_NeqSim_in_Colab.ipynb#scrollTo=_eRtkQnHpL70).
%%capture
!pip install neqsim
import neqsim
from neqsim.thermo.thermoTools import *
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
from neqsim.thermo import fluid, fluid_df
import pandas as pd
from neqsim.process import gasscrubber, clearProcess, run,nequnit, phasemixer, splitter, clearProcess, stream, valve, separator, compressor, runProcess, viewProcess, heater,saturator, mixer
plt.style.use('classic')
%matplotlib inline
```
#Mass transfer calculations
Model for mass transfer calculation in NeqSim based on Solbraa (2002):
https://ntnuopen.ntnu.no/ntnu-xmlui/handle/11250/231326
In the following calculations we assume a water saturated gas the is mixed with pure liquid methanol. These phases are not in equiibrium when they enter the pipeline. When the gas and methanol liquid comes in contact in the pipeline, methanol will vaporize into the gas, and water (and other comonents from the gas) will be absorbed into the liquid methanol. The focus of the following calculations will be to evaluate the mass transfer as function of contanct length with gas and methanol. It also evaluates the hydrate temperature of the gas leaving the pipe section.
Figure 1 Illustration of mass transfer process

**The parameters for the model are:**
Temperature and pressure of the pipe (mass transfer calculated at constant temperature and pressure).
Length and diameter of pipe where gas and liquid will be in contact and mass transfer can occur.
Flow rate of the gas in MSm3/day, flow rate of methanol (kg/hr).
#Calculation of compostion of aqueous phase and gas leaving pipe section
In the following script we will simulate the composition of the gas leaving pipe section at a given pipe lengt.
```
# Input parameters
pressure = 52.21 # bara
temperature = 15.2 #C
gasFlow = 1.23 #MSm3/day
methanolFlow = 6000.23 # kg/day
pipelength = 10.0 #meter
pipeInnerDiameter = 0.5 #meter
# Create a gas-condensate fluid
feedgas = {'ComponentName': ["nitrogen","CO2","methane", "ethane" , "propane", "i-butane", "n-butane", "water", "methanol"],
'MolarComposition[-]': [0.01, 0.01, 0.8, 0.06, 0.01,0.005,0.005, 0.0, 0.0]
}
naturalgasFluid = fluid_df(pd.DataFrame(feedgas)).setModel("CPAs-SRK-EOS-statoil")
naturalgasFluid.setTotalFlowRate(gasFlow, "MSm3/day")
naturalgasFluid.setTemperature(temperature, "C")
naturalgasFluid.setPressure(pressure, "bara")
# Create a liquid methanol fluid
feedMeOH = {'ComponentName': ["nitrogen","CO2","methane", "ethane" , "propane", "i-butane", "n-butane", "water", "methanol"],
'MolarComposition[-]': [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,1.0]
}
meOHFluid = fluid_df(pd.DataFrame(feedMeOH) ).setModel("CPAs-SRK-EOS-statoil")
meOHFluid.setTotalFlowRate(methanolFlow, "kg/hr");
meOHFluid.setTemperature(temperature, "C");
meOHFluid.setPressure(pressure, "bara");
clearProcess()
dryinjectiongas = stream(naturalgasFluid)
MeOHFeed = stream(meOHFluid)
watersaturator = saturator(dryinjectiongas)
waterSaturatedFeedGas = stream(watersaturator.getOutStream())
mainMixer = phasemixer("gas MeOH mixer")
mainMixer.addStream(waterSaturatedFeedGas)
mainMixer.addStream(MeOHFeed)
pipeline = nequnit(mainMixer.getOutStream(), equipment="pipeline", flowpattern="stratified") #alternative flow patterns are: stratified, annular and droplet
pipeline.setLength(pipelength)
pipeline.setID(pipeInnerDiameter)
scrubber = gasscrubber(pipeline.getOutStream())
gasFromScrubber = stream(scrubber.getGasOutStream())
aqueousFromScrubber = stream(scrubber.getLiquidOutStream())
run()
print('Composition of gas leaving pipe section after ', pipelength, ' meter')
printFrame(gasFromScrubber.getFluid())
print('Composition of aqueous phase leaving pipe section after ', pipelength, ' meter')
printFrame(aqueousFromScrubber.getFluid())
print('Interface contact area ', pipeline.getInterfacialArea(), ' m^2')
print('Volume fraction aqueous phase ', pipeline.getOutStream().getFluid().getVolumeFraction(1), ' -')
```
# Calculation of hydrate equilibrium temperature of gas leaving pipe section
In the following script we will simulate the composition of the gas leaving pipe section as well as hydrate equilibrium temperature of this gas as function of pipe length.
```
maxpipelength = 10.0
def hydtemps(length):
pipeline.setLength(length)
run();
return gasFromScrubber.getHydrateEquilibriumTemperature()-273.15
length = np.arange(0.01, maxpipelength, (maxpipelength)/10.0)
hydtem = [hydtemps(length2) for length2 in length]
plt.figure()
plt.plot(length, hydtem)
plt.xlabel('Length available for mass transfer [m]')
plt.ylabel('Hydrate eq.temperature [C]')
plt.title('Hydrate eq.temperature of gas leaving pipe section')
```
|
github_jupyter
|
```
# Copyright 2020 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Vertex client library: AutoML tabular binary classification model for batch prediction
<table align="left">
<td>
<a href="https://colab.research.google.com/github/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/automl/showcase_automl_tabular_binary_classification_batch.ipynb">
<img src="https://cloud.google.com/ml-engine/images/colab-logo-32px.png" alt="Colab logo"> Run in Colab
</a>
</td>
<td>
<a href="https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/master/notebooks/community/gapic/automl/showcase_automl_tabular_binary_classification_batch.ipynb">
<img src="https://cloud.google.com/ml-engine/images/github-logo-32px.png" alt="GitHub logo">
View on GitHub
</a>
</td>
</table>
<br/><br/><br/>
## Overview
This tutorial demonstrates how to use the Vertex client library for Python to create tabular binary classification models and do batch prediction using Google Cloud's [AutoML](https://cloud.google.com/vertex-ai/docs/start/automl-users).
### Dataset
The dataset used for this tutorial is the [Bank Marketing](gs://cloud-ml-tables-data/bank-marketing.csv). This dataset does not require any feature engineering. The version of the dataset you will use in this tutorial is stored in a public Cloud Storage bucket.
### Objective
In this tutorial, you create an AutoML tabular binary classification model from a Python script, and then do a batch prediction using the Vertex client library. You can alternatively create and deploy models using the `gcloud` command-line tool or online using the Google Cloud Console.
The steps performed include:
- Create a Vertex `Dataset` resource.
- Train the model.
- View the model evaluation.
- Make a batch prediction.
There is one key difference between using batch prediction and using online prediction:
* Prediction Service: Does an on-demand prediction for the entire set of instances (i.e., one or more data items) and returns the results in real-time.
* Batch Prediction Service: Does a queued (batch) prediction for the entire set of instances in the background and stores the results in a Cloud Storage bucket when ready.
### Costs
This tutorial uses billable components of Google Cloud (GCP):
* Vertex AI
* Cloud Storage
Learn about [Vertex AI
pricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storage
pricing](https://cloud.google.com/storage/pricing), and use the [Pricing
Calculator](https://cloud.google.com/products/calculator/)
to generate a cost estimate based on your projected usage.
## Installation
Install the latest version of Vertex client library.
```
import os
import sys
# Google Cloud Notebook
if os.path.exists("/opt/deeplearning/metadata/env_version"):
USER_FLAG = "--user"
else:
USER_FLAG = ""
! pip3 install -U google-cloud-aiplatform $USER_FLAG
```
Install the latest GA version of *google-cloud-storage* library as well.
```
! pip3 install -U google-cloud-storage $USER_FLAG
```
### Restart the kernel
Once you've installed the Vertex client library and Google *cloud-storage*, you need to restart the notebook kernel so it can find the packages.
```
if not os.getenv("IS_TESTING"):
# Automatically restart kernel after installs
import IPython
app = IPython.Application.instance()
app.kernel.do_shutdown(True)
```
## Before you begin
### GPU runtime
*Make sure you're running this notebook in a GPU runtime if you have that option. In Colab, select* **Runtime > Change Runtime Type > GPU**
### Set up your Google Cloud project
**The following steps are required, regardless of your notebook environment.**
1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.
2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)
3. [Enable the Vertex APIs and Compute Engine APIs.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component)
4. [The Google Cloud SDK](https://cloud.google.com/sdk) is already installed in Google Cloud Notebook.
5. Enter your project ID in the cell below. Then run the cell to make sure the
Cloud SDK uses the right project for all the commands in this notebook.
**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$` into these commands.
```
PROJECT_ID = "[your-project-id]" # @param {type:"string"}
if PROJECT_ID == "" or PROJECT_ID is None or PROJECT_ID == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = !gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT_ID = shell_output[0]
print("Project ID:", PROJECT_ID)
! gcloud config set project $PROJECT_ID
```
#### Region
You can also change the `REGION` variable, which is used for operations
throughout the rest of this notebook. Below are regions supported for Vertex. We recommend that you choose the region closest to you.
- Americas: `us-central1`
- Europe: `europe-west4`
- Asia Pacific: `asia-east1`
You may not use a multi-regional bucket for training with Vertex. Not all regions provide support for all Vertex services. For the latest support per region, see the [Vertex locations documentation](https://cloud.google.com/vertex-ai/docs/general/locations)
```
REGION = "us-central1" # @param {type: "string"}
```
#### Timestamp
If you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append onto the name of resources which will be created in this tutorial.
```
from datetime import datetime
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
```
### Authenticate your Google Cloud account
**If you are using Google Cloud Notebook**, your environment is already authenticated. Skip this step.
**If you are using Colab**, run the cell below and follow the instructions when prompted to authenticate your account via oAuth.
**Otherwise**, follow these steps:
In the Cloud Console, go to the [Create service account key](https://console.cloud.google.com/apis/credentials/serviceaccountkey) page.
**Click Create service account**.
In the **Service account name** field, enter a name, and click **Create**.
In the **Grant this service account access to project** section, click the Role drop-down list. Type "Vertex" into the filter box, and select **Vertex Administrator**. Type "Storage Object Admin" into the filter box, and select **Storage Object Admin**.
Click Create. A JSON file that contains your key downloads to your local environment.
Enter the path to your service account key as the GOOGLE_APPLICATION_CREDENTIALS variable in the cell below and run the cell.
```
# If you are running this notebook in Colab, run this cell and follow the
# instructions to authenticate your GCP account. This provides access to your
# Cloud Storage bucket and lets you submit training jobs and prediction
# requests.
# If on Google Cloud Notebook, then don't execute this code
if not os.path.exists("/opt/deeplearning/metadata/env_version"):
if "google.colab" in sys.modules:
from google.colab import auth as google_auth
google_auth.authenticate_user()
# If you are running this notebook locally, replace the string below with the
# path to your service account key and run this cell to authenticate your GCP
# account.
elif not os.getenv("IS_TESTING"):
%env GOOGLE_APPLICATION_CREDENTIALS ''
```
### Create a Cloud Storage bucket
**The following steps are required, regardless of your notebook environment.**
This tutorial is designed to use training data that is in a public Cloud Storage bucket and a local Cloud Storage bucket for your batch predictions. You may alternatively use your own training data that you have stored in a local Cloud Storage bucket.
Set the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.
```
BUCKET_NAME = "gs://[your-bucket-name]" # @param {type:"string"}
if BUCKET_NAME == "" or BUCKET_NAME is None or BUCKET_NAME == "gs://[your-bucket-name]":
BUCKET_NAME = "gs://" + PROJECT_ID + "aip-" + TIMESTAMP
```
**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.
```
! gsutil mb -l $REGION $BUCKET_NAME
```
Finally, validate access to your Cloud Storage bucket by examining its contents:
```
! gsutil ls -al $BUCKET_NAME
```
### Set up variables
Next, set up some variables used throughout the tutorial.
### Import libraries and define constants
#### Import Vertex client library
Import the Vertex client library into our Python environment.
```
import time
from google.cloud.aiplatform import gapic as aip
from google.protobuf import json_format
from google.protobuf.struct_pb2 import Struct, Value
```
#### Vertex constants
Setup up the following constants for Vertex:
- `API_ENDPOINT`: The Vertex API service endpoint for dataset, model, job, pipeline and endpoint services.
- `PARENT`: The Vertex location root path for dataset, model, job, pipeline and endpoint resources.
```
# API service endpoint
API_ENDPOINT = "{}-aiplatform.googleapis.com".format(REGION)
# Vertex location root path for your dataset, model and endpoint resources
PARENT = "projects/" + PROJECT_ID + "/locations/" + REGION
```
#### AutoML constants
Set constants unique to AutoML datasets and training:
- Dataset Schemas: Tells the `Dataset` resource service which type of dataset it is.
- Data Labeling (Annotations) Schemas: Tells the `Dataset` resource service how the data is labeled (annotated).
- Dataset Training Schemas: Tells the `Pipeline` resource service the task (e.g., classification) to train the model for.
```
# Tabular Dataset type
DATA_SCHEMA = "gs://google-cloud-aiplatform/schema/dataset/metadata/tables_1.0.0.yaml"
# Tabular Labeling type
LABEL_SCHEMA = (
"gs://google-cloud-aiplatform/schema/dataset/ioformat/table_io_format_1.0.0.yaml"
)
# Tabular Training task
TRAINING_SCHEMA = "gs://google-cloud-aiplatform/schema/trainingjob/definition/automl_tables_1.0.0.yaml"
```
#### Hardware Accelerators
Set the hardware accelerators (e.g., GPU), if any, for prediction.
Set the variable `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Telsa K80 GPUs allocated to each VM, you would specify:
(aip.AcceleratorType.NVIDIA_TESLA_K80, 4)
For GPU, available accelerators include:
- aip.AcceleratorType.NVIDIA_TESLA_K80
- aip.AcceleratorType.NVIDIA_TESLA_P100
- aip.AcceleratorType.NVIDIA_TESLA_P4
- aip.AcceleratorType.NVIDIA_TESLA_T4
- aip.AcceleratorType.NVIDIA_TESLA_V100
Otherwise specify `(None, None)` to use a container image to run on a CPU.
```
if os.getenv("IS_TESTING_DEPOLY_GPU"):
DEPLOY_GPU, DEPLOY_NGPU = (
aip.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_DEPOLY_GPU")),
)
else:
DEPLOY_GPU, DEPLOY_NGPU = (aip.AcceleratorType.NVIDIA_TESLA_K80, 1)
```
#### Container (Docker) image
For AutoML batch prediction, the container image for the serving binary is pre-determined by the Vertex prediction service. More specifically, the service will pick the appropriate container for the model depending on the hardware accelerator you selected.
#### Machine Type
Next, set the machine type to use for prediction.
- Set the variable `DEPLOY_COMPUTE` to configure the compute resources for the VM you will use for prediction.
- `machine type`
- `n1-standard`: 3.75GB of memory per vCPU.
- `n1-highmem`: 6.5GB of memory per vCPU
- `n1-highcpu`: 0.9 GB of memory per vCPU
- `vCPUs`: number of \[2, 4, 8, 16, 32, 64, 96 \]
*Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*
```
if os.getenv("IS_TESTING_DEPLOY_MACHINE"):
MACHINE_TYPE = os.getenv("IS_TESTING_DEPLOY_MACHINE")
else:
MACHINE_TYPE = "n1-standard"
VCPU = "4"
DEPLOY_COMPUTE = MACHINE_TYPE + "-" + VCPU
print("Deploy machine type", DEPLOY_COMPUTE)
```
# Tutorial
Now you are ready to start creating your own AutoML tabular binary classification model.
## Set up clients
The Vertex client library works as a client/server model. On your side (the Python script) you will create a client that sends requests and receives responses from the Vertex server.
You will use different clients in this tutorial for different steps in the workflow. So set them all up upfront.
- Dataset Service for `Dataset` resources.
- Model Service for `Model` resources.
- Pipeline Service for training.
- Job Service for batch prediction and custom training.
```
# client options same for all services
client_options = {"api_endpoint": API_ENDPOINT}
def create_dataset_client():
client = aip.DatasetServiceClient(client_options=client_options)
return client
def create_model_client():
client = aip.ModelServiceClient(client_options=client_options)
return client
def create_pipeline_client():
client = aip.PipelineServiceClient(client_options=client_options)
return client
def create_job_client():
client = aip.JobServiceClient(client_options=client_options)
return client
clients = {}
clients["dataset"] = create_dataset_client()
clients["model"] = create_model_client()
clients["pipeline"] = create_pipeline_client()
clients["job"] = create_job_client()
for client in clients.items():
print(client)
```
## Dataset
Now that your clients are ready, your first step is to create a `Dataset` resource instance. This step differs from Vision, Video and Language. For those products, after the `Dataset` resource is created, one then separately imports the data, using the `import_data` method.
For tabular, importing of the data is deferred until the training pipeline starts training the model. What do we do different? Well, first you won't be calling the `import_data` method. Instead, when you create the dataset instance you specify the Cloud Storage location of the CSV file or BigQuery location of the data table, which contains your tabular data as part of the `Dataset` resource's metadata.
#### Cloud Storage
`metadata = {"input_config": {"gcs_source": {"uri": [gcs_uri]}}}`
The format for a Cloud Storage path is:
gs://[bucket_name]/[folder(s)/[file]
#### BigQuery
`metadata = {"input_config": {"bigquery_source": {"uri": [gcs_uri]}}}`
The format for a BigQuery path is:
bq://[collection].[dataset].[table]
Note that the `uri` field is a list, whereby you can input multiple CSV files or BigQuery tables when your data is split across files.
### Data preparation
The Vertex `Dataset` resource for tabular has a couple of requirements for your tabular data.
- Must be in a CSV file or a BigQuery query.
#### CSV
For tabular binary classification, the CSV file has a few requirements:
- The first row must be the heading -- note how this is different from Vision, Video and Language where the requirement is no heading.
- All but one column are features.
- One column is the label, which you will specify when you subsequently create the training pipeline.
#### Location of Cloud Storage training data.
Now set the variable `IMPORT_FILE` to the location of the CSV index file in Cloud Storage.
```
IMPORT_FILE = "gs://cloud-ml-tables-data/bank-marketing.csv"
```
#### Quick peek at your data
You will use a version of the Bank Marketing dataset that is stored in a public Cloud Storage bucket, using a CSV index file.
Start by doing a quick peek at the data. You count the number of examples by counting the number of rows in the CSV index file (`wc -l`) and then peek at the first few rows.
You also need for training to know the heading name of the label column, which is save as `label_column`. For this dataset, it is the last column in the CSV file.
```
count = ! gsutil cat $IMPORT_FILE | wc -l
print("Number of Examples", int(count[0]))
print("First 10 rows")
! gsutil cat $IMPORT_FILE | head
heading = ! gsutil cat $IMPORT_FILE | head -n1
label_column = str(heading).split(",")[-1].split("'")[0]
print("Label Column Name", label_column)
if label_column is None:
raise Exception("label column missing")
```
## Dataset
Now that your clients are ready, your first step in training a model is to create a managed dataset instance, and then upload your labeled data to it.
### Create `Dataset` resource instance
Use the helper function `create_dataset` to create the instance of a `Dataset` resource. This function does the following:
1. Uses the dataset client service.
2. Creates an Vertex `Dataset` resource (`aip.Dataset`), with the following parameters:
- `display_name`: The human-readable name you choose to give it.
- `metadata_schema_uri`: The schema for the dataset type.
- `metadata`: The Cloud Storage or BigQuery location of the tabular data.
3. Calls the client dataset service method `create_dataset`, with the following parameters:
- `parent`: The Vertex location root path for your `Database`, `Model` and `Endpoint` resources.
- `dataset`: The Vertex dataset object instance you created.
4. The method returns an `operation` object.
An `operation` object is how Vertex handles asynchronous calls for long running operations. While this step usually goes fast, when you first use it in your project, there is a longer delay due to provisioning.
You can use the `operation` object to get status on the operation (e.g., create `Dataset` resource) or to cancel the operation, by invoking an operation method:
| Method | Description |
| ----------- | ----------- |
| result() | Waits for the operation to complete and returns a result object in JSON format. |
| running() | Returns True/False on whether the operation is still running. |
| done() | Returns True/False on whether the operation is completed. |
| canceled() | Returns True/False on whether the operation was canceled. |
| cancel() | Cancels the operation (this may take up to 30 seconds). |
```
TIMEOUT = 90
def create_dataset(name, schema, src_uri=None, labels=None, timeout=TIMEOUT):
start_time = time.time()
try:
if src_uri.startswith("gs://"):
metadata = {"input_config": {"gcs_source": {"uri": [src_uri]}}}
elif src_uri.startswith("bq://"):
metadata = {"input_config": {"bigquery_source": {"uri": [src_uri]}}}
dataset = aip.Dataset(
display_name=name,
metadata_schema_uri=schema,
labels=labels,
metadata=json_format.ParseDict(metadata, Value()),
)
operation = clients["dataset"].create_dataset(parent=PARENT, dataset=dataset)
print("Long running operation:", operation.operation.name)
result = operation.result(timeout=TIMEOUT)
print("time:", time.time() - start_time)
print("response")
print(" name:", result.name)
print(" display_name:", result.display_name)
print(" metadata_schema_uri:", result.metadata_schema_uri)
print(" metadata:", dict(result.metadata))
print(" create_time:", result.create_time)
print(" update_time:", result.update_time)
print(" etag:", result.etag)
print(" labels:", dict(result.labels))
return result
except Exception as e:
print("exception:", e)
return None
result = create_dataset("bank-" + TIMESTAMP, DATA_SCHEMA, src_uri=IMPORT_FILE)
```
Now save the unique dataset identifier for the `Dataset` resource instance you created.
```
# The full unique ID for the dataset
dataset_id = result.name
# The short numeric ID for the dataset
dataset_short_id = dataset_id.split("/")[-1]
print(dataset_id)
```
## Train the model
Now train an AutoML tabular binary classification model using your Vertex `Dataset` resource. To train the model, do the following steps:
1. Create an Vertex training pipeline for the `Dataset` resource.
2. Execute the pipeline to start the training.
### Create a training pipeline
You may ask, what do we use a pipeline for? You typically use pipelines when the job (such as training) has multiple steps, generally in sequential order: do step A, do step B, etc. By putting the steps into a pipeline, we gain the benefits of:
1. Being reusable for subsequent training jobs.
2. Can be containerized and ran as a batch job.
3. Can be distributed.
4. All the steps are associated with the same pipeline job for tracking progress.
Use this helper function `create_pipeline`, which takes the following parameters:
- `pipeline_name`: A human readable name for the pipeline job.
- `model_name`: A human readable name for the model.
- `dataset`: The Vertex fully qualified dataset identifier.
- `schema`: The dataset labeling (annotation) training schema.
- `task`: A dictionary describing the requirements for the training job.
The helper function calls the `Pipeline` client service'smethod `create_pipeline`, which takes the following parameters:
- `parent`: The Vertex location root path for your `Dataset`, `Model` and `Endpoint` resources.
- `training_pipeline`: the full specification for the pipeline training job.
Let's look now deeper into the *minimal* requirements for constructing a `training_pipeline` specification:
- `display_name`: A human readable name for the pipeline job.
- `training_task_definition`: The dataset labeling (annotation) training schema.
- `training_task_inputs`: A dictionary describing the requirements for the training job.
- `model_to_upload`: A human readable name for the model.
- `input_data_config`: The dataset specification.
- `dataset_id`: The Vertex dataset identifier only (non-fully qualified) -- this is the last part of the fully-qualified identifier.
- `fraction_split`: If specified, the percentages of the dataset to use for training, test and validation. Otherwise, the percentages are automatically selected by AutoML.
```
def create_pipeline(pipeline_name, model_name, dataset, schema, task):
dataset_id = dataset.split("/")[-1]
input_config = {
"dataset_id": dataset_id,
"fraction_split": {
"training_fraction": 0.8,
"validation_fraction": 0.1,
"test_fraction": 0.1,
},
}
training_pipeline = {
"display_name": pipeline_name,
"training_task_definition": schema,
"training_task_inputs": task,
"input_data_config": input_config,
"model_to_upload": {"display_name": model_name},
}
try:
pipeline = clients["pipeline"].create_training_pipeline(
parent=PARENT, training_pipeline=training_pipeline
)
print(pipeline)
except Exception as e:
print("exception:", e)
return None
return pipeline
```
### Construct the task requirements
Next, construct the task requirements. Unlike other parameters which take a Python (JSON-like) dictionary, the `task` field takes a Google protobuf Struct, which is very similar to a Python dictionary. Use the `json_format.ParseDict` method for the conversion.
The minimal fields you need to specify are:
- `prediction_type`: Whether we are doing "classification" or "regression".
- `target_column`: The CSV heading column name for the column we want to predict (i.e., the label).
- `train_budget_milli_node_hours`: The maximum time to budget (billed) for training the model, where 1000 = 1 hour.
- `disable_early_stopping`: Whether True/False to let AutoML use its judgement to stop training early or train for the entire budget.
- `transformations`: Specifies the feature engineering for each feature column.
For `transformations`, the list must have an entry for each column. The outer key field indicates the type of feature engineering for the corresponding column. In this tutorial, you set it to `"auto"` to tell AutoML to automatically determine it.
Finally, create the pipeline by calling the helper function `create_pipeline`, which returns an instance of a training pipeline object.
```
TRANSFORMATIONS = [
{"auto": {"column_name": "Age"}},
{"auto": {"column_name": "Job"}},
{"auto": {"column_name": "MaritalStatus"}},
{"auto": {"column_name": "Education"}},
{"auto": {"column_name": "Default"}},
{"auto": {"column_name": "Balance"}},
{"auto": {"column_name": "Housing"}},
{"auto": {"column_name": "Loan"}},
{"auto": {"column_name": "Contact"}},
{"auto": {"column_name": "Day"}},
{"auto": {"column_name": "Month"}},
{"auto": {"column_name": "Duration"}},
{"auto": {"column_name": "Campaign"}},
{"auto": {"column_name": "PDays"}},
{"auto": {"column_name": "POutcome"}},
]
PIPE_NAME = "bank_pipe-" + TIMESTAMP
MODEL_NAME = "bank_model-" + TIMESTAMP
task = Value(
struct_value=Struct(
fields={
"target_column": Value(string_value=label_column),
"prediction_type": Value(string_value="classification"),
"train_budget_milli_node_hours": Value(number_value=1000),
"disable_early_stopping": Value(bool_value=False),
"transformations": json_format.ParseDict(TRANSFORMATIONS, Value()),
}
)
)
response = create_pipeline(PIPE_NAME, MODEL_NAME, dataset_id, TRAINING_SCHEMA, task)
```
Now save the unique identifier of the training pipeline you created.
```
# The full unique ID for the pipeline
pipeline_id = response.name
# The short numeric ID for the pipeline
pipeline_short_id = pipeline_id.split("/")[-1]
print(pipeline_id)
```
### Get information on a training pipeline
Now get pipeline information for just this training pipeline instance. The helper function gets the job information for just this job by calling the the job client service's `get_training_pipeline` method, with the following parameter:
- `name`: The Vertex fully qualified pipeline identifier.
When the model is done training, the pipeline state will be `PIPELINE_STATE_SUCCEEDED`.
```
def get_training_pipeline(name, silent=False):
response = clients["pipeline"].get_training_pipeline(name=name)
if silent:
return response
print("pipeline")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" state:", response.state)
print(" training_task_definition:", response.training_task_definition)
print(" training_task_inputs:", dict(response.training_task_inputs))
print(" create_time:", response.create_time)
print(" start_time:", response.start_time)
print(" end_time:", response.end_time)
print(" update_time:", response.update_time)
print(" labels:", dict(response.labels))
return response
response = get_training_pipeline(pipeline_id)
```
# Deployment
Training the above model may take upwards of 30 minutes time.
Once your model is done training, you can calculate the actual time it took to train the model by subtracting `end_time` from `start_time`. For your model, you will need to know the fully qualified Vertex Model resource identifier, which the pipeline service assigned to it. You can get this from the returned pipeline instance as the field `model_to_deploy.name`.
```
while True:
response = get_training_pipeline(pipeline_id, True)
if response.state != aip.PipelineState.PIPELINE_STATE_SUCCEEDED:
print("Training job has not completed:", response.state)
model_to_deploy_id = None
if response.state == aip.PipelineState.PIPELINE_STATE_FAILED:
raise Exception("Training Job Failed")
else:
model_to_deploy = response.model_to_upload
model_to_deploy_id = model_to_deploy.name
print("Training Time:", response.end_time - response.start_time)
break
time.sleep(60)
print("model to deploy:", model_to_deploy_id)
```
## Model information
Now that your model is trained, you can get some information on your model.
## Evaluate the Model resource
Now find out how good the model service believes your model is. As part of training, some portion of the dataset was set aside as the test (holdout) data, which is used by the pipeline service to evaluate the model.
### List evaluations for all slices
Use this helper function `list_model_evaluations`, which takes the following parameter:
- `name`: The Vertex fully qualified model identifier for the `Model` resource.
This helper function uses the model client service's `list_model_evaluations` method, which takes the same parameter. The response object from the call is a list, where each element is an evaluation metric.
For each evaluation (you probably only have one) we then print all the key names for each metric in the evaluation, and for a small set (`logLoss` and `auPrc`) you will print the result.
```
def list_model_evaluations(name):
response = clients["model"].list_model_evaluations(parent=name)
for evaluation in response:
print("model_evaluation")
print(" name:", evaluation.name)
print(" metrics_schema_uri:", evaluation.metrics_schema_uri)
metrics = json_format.MessageToDict(evaluation._pb.metrics)
for metric in metrics.keys():
print(metric)
print("logloss", metrics["logLoss"])
print("auPrc", metrics["auPrc"])
return evaluation.name
last_evaluation = list_model_evaluations(model_to_deploy_id)
```
## Model deployment for batch prediction
Now deploy the trained Vertex `Model` resource you created for batch prediction. This differs from deploying a `Model` resource for on-demand prediction.
For online prediction, you:
1. Create an `Endpoint` resource for deploying the `Model` resource to.
2. Deploy the `Model` resource to the `Endpoint` resource.
3. Make online prediction requests to the `Endpoint` resource.
For batch-prediction, you:
1. Create a batch prediction job.
2. The job service will provision resources for the batch prediction request.
3. The results of the batch prediction request are returned to the caller.
4. The job service will unprovision the resoures for the batch prediction request.
## Make a batch prediction request
Now do a batch prediction to your deployed model.
### Make test items
You will use synthetic data as a test data items. Don't be concerned that we are using synthetic data -- we just want to demonstrate how to make a prediction.
```
HEADING = "Age,Job,MaritalStatus,Education,Default,Balance,Housing,Loan,Contact,Day,Month,Duration,Campaign,PDays,Previous,POutcome,Deposit"
INSTANCE_1 = (
"58,managment,married,teritary,no,2143,yes,no,unknown,5,may,261,1,-1,0, unknown"
)
INSTANCE_2 = (
"44,technician,single,secondary,no,39,yes,no,unknown,5,may,151,1,-1,0,unknown"
)
```
### Make the batch input file
Now make a batch input file, which you will store in your local Cloud Storage bucket. Unlike image, video and text, the batch input file for tabular is only supported for CSV. For CSV file, you make:
- The first line is the heading with the feature (fields) heading names.
- Each remaining line is a separate prediction request with the corresponding feature values.
For example:
"feature_1", "feature_2". ...
value_1, value_2, ...
```
import tensorflow as tf
gcs_input_uri = BUCKET_NAME + "/test.csv"
with tf.io.gfile.GFile(gcs_input_uri, "w") as f:
f.write(HEADING + "\n")
f.write(str(INSTANCE_1) + "\n")
f.write(str(INSTANCE_2) + "\n")
print(gcs_input_uri)
! gsutil cat $gcs_input_uri
```
### Compute instance scaling
You have several choices on scaling the compute instances for handling your batch prediction requests:
- Single Instance: The batch prediction requests are processed on a single compute instance.
- Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to one.
- Manual Scaling: The batch prediction requests are split across a fixed number of compute instances that you manually specified.
- Set the minimum (`MIN_NODES`) and maximum (`MAX_NODES`) number of compute instances to the same number of nodes. When a model is first deployed to the instance, the fixed number of compute instances are provisioned and batch prediction requests are evenly distributed across them.
- Auto Scaling: The batch prediction requests are split across a scaleable number of compute instances.
- Set the minimum (`MIN_NODES`) number of compute instances to provision when a model is first deployed and to de-provision, and set the maximum (`MAX_NODES) number of compute instances to provision, depending on load conditions.
The minimum number of compute instances corresponds to the field `min_replica_count` and the maximum number of compute instances corresponds to the field `max_replica_count`, in your subsequent deployment request.
```
MIN_NODES = 1
MAX_NODES = 1
```
### Make batch prediction request
Now that your batch of two test items is ready, let's do the batch request. Use this helper function `create_batch_prediction_job`, with the following parameters:
- `display_name`: The human readable name for the prediction job.
- `model_name`: The Vertex fully qualified identifier for the `Model` resource.
- `gcs_source_uri`: The Cloud Storage path to the input file -- which you created above.
- `gcs_destination_output_uri_prefix`: The Cloud Storage path that the service will write the predictions to.
- `parameters`: Additional filtering parameters for serving prediction results.
The helper function calls the job client service's `create_batch_prediction_job` metho, with the following parameters:
- `parent`: The Vertex location root path for Dataset, Model and Pipeline resources.
- `batch_prediction_job`: The specification for the batch prediction job.
Let's now dive into the specification for the `batch_prediction_job`:
- `display_name`: The human readable name for the prediction batch job.
- `model`: The Vertex fully qualified identifier for the `Model` resource.
- `dedicated_resources`: The compute resources to provision for the batch prediction job.
- `machine_spec`: The compute instance to provision. Use the variable you set earlier `DEPLOY_GPU != None` to use a GPU; otherwise only a CPU is allocated.
- `starting_replica_count`: The number of compute instances to initially provision, which you set earlier as the variable `MIN_NODES`.
- `max_replica_count`: The maximum number of compute instances to scale to, which you set earlier as the variable `MAX_NODES`.
- `model_parameters`: Additional filtering parameters for serving prediction results. *Note*, image segmentation models do not support additional parameters.
- `input_config`: The input source and format type for the instances to predict.
- `instances_format`: The format of the batch prediction request file: `csv` only supported.
- `gcs_source`: A list of one or more Cloud Storage paths to your batch prediction requests.
- `output_config`: The output destination and format for the predictions.
- `prediction_format`: The format of the batch prediction response file: `csv` only supported.
- `gcs_destination`: The output destination for the predictions.
This call is an asychronous operation. You will print from the response object a few select fields, including:
- `name`: The Vertex fully qualified identifier assigned to the batch prediction job.
- `display_name`: The human readable name for the prediction batch job.
- `model`: The Vertex fully qualified identifier for the Model resource.
- `generate_explanations`: Whether True/False explanations were provided with the predictions (explainability).
- `state`: The state of the prediction job (pending, running, etc).
Since this call will take a few moments to execute, you will likely get `JobState.JOB_STATE_PENDING` for `state`.
```
BATCH_MODEL = "bank_batch-" + TIMESTAMP
def create_batch_prediction_job(
display_name,
model_name,
gcs_source_uri,
gcs_destination_output_uri_prefix,
parameters=None,
):
if DEPLOY_GPU:
machine_spec = {
"machine_type": DEPLOY_COMPUTE,
"accelerator_type": DEPLOY_GPU,
"accelerator_count": DEPLOY_NGPU,
}
else:
machine_spec = {
"machine_type": DEPLOY_COMPUTE,
"accelerator_count": 0,
}
batch_prediction_job = {
"display_name": display_name,
# Format: 'projects/{project}/locations/{location}/models/{model_id}'
"model": model_name,
"model_parameters": json_format.ParseDict(parameters, Value()),
"input_config": {
"instances_format": IN_FORMAT,
"gcs_source": {"uris": [gcs_source_uri]},
},
"output_config": {
"predictions_format": OUT_FORMAT,
"gcs_destination": {"output_uri_prefix": gcs_destination_output_uri_prefix},
},
"dedicated_resources": {
"machine_spec": machine_spec,
"starting_replica_count": MIN_NODES,
"max_replica_count": MAX_NODES,
},
}
response = clients["job"].create_batch_prediction_job(
parent=PARENT, batch_prediction_job=batch_prediction_job
)
print("response")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" model:", response.model)
try:
print(" generate_explanation:", response.generate_explanation)
except:
pass
print(" state:", response.state)
print(" create_time:", response.create_time)
print(" start_time:", response.start_time)
print(" end_time:", response.end_time)
print(" update_time:", response.update_time)
print(" labels:", response.labels)
return response
IN_FORMAT = "csv"
OUT_FORMAT = "csv" # [csv]
response = create_batch_prediction_job(
BATCH_MODEL, model_to_deploy_id, gcs_input_uri, BUCKET_NAME, None
)
```
Now get the unique identifier for the batch prediction job you created.
```
# The full unique ID for the batch job
batch_job_id = response.name
# The short numeric ID for the batch job
batch_job_short_id = batch_job_id.split("/")[-1]
print(batch_job_id)
```
### Get information on a batch prediction job
Use this helper function `get_batch_prediction_job`, with the following paramter:
- `job_name`: The Vertex fully qualified identifier for the batch prediction job.
The helper function calls the job client service's `get_batch_prediction_job` method, with the following paramter:
- `name`: The Vertex fully qualified identifier for the batch prediction job. In this tutorial, you will pass it the Vertex fully qualified identifier for your batch prediction job -- `batch_job_id`
The helper function will return the Cloud Storage path to where the predictions are stored -- `gcs_destination`.
```
def get_batch_prediction_job(job_name, silent=False):
response = clients["job"].get_batch_prediction_job(name=job_name)
if silent:
return response.output_config.gcs_destination.output_uri_prefix, response.state
print("response")
print(" name:", response.name)
print(" display_name:", response.display_name)
print(" model:", response.model)
try: # not all data types support explanations
print(" generate_explanation:", response.generate_explanation)
except:
pass
print(" state:", response.state)
print(" error:", response.error)
gcs_destination = response.output_config.gcs_destination
print(" gcs_destination")
print(" output_uri_prefix:", gcs_destination.output_uri_prefix)
return gcs_destination.output_uri_prefix, response.state
predictions, state = get_batch_prediction_job(batch_job_id)
```
### Get Predictions
When the batch prediction is done processing, the job state will be `JOB_STATE_SUCCEEDED`.
Finally you view the predictions stored at the Cloud Storage path you set as output. The predictions will be in a CSV format, which you indicated at the time you made the batch prediction job, under a subfolder starting with the name `prediction`, and under that folder will be a file called `predictions*.csv`.
Now display (cat) the contents. You will see multiple rows, one for each prediction.
For each prediction:
- The first four fields are the values (features) you did the prediction on.
- The remaining fields are the confidence values, between 0 and 1, for each prediction.
```
def get_latest_predictions(gcs_out_dir):
""" Get the latest prediction subfolder using the timestamp in the subfolder name"""
folders = !gsutil ls $gcs_out_dir
latest = ""
for folder in folders:
subfolder = folder.split("/")[-2]
if subfolder.startswith("prediction-"):
if subfolder > latest:
latest = folder[:-1]
return latest
while True:
predictions, state = get_batch_prediction_job(batch_job_id, True)
if state != aip.JobState.JOB_STATE_SUCCEEDED:
print("The job has not completed:", state)
if state == aip.JobState.JOB_STATE_FAILED:
raise Exception("Batch Job Failed")
else:
folder = get_latest_predictions(predictions)
! gsutil ls $folder/prediction*.csv
! gsutil cat $folder/prediction*.csv
break
time.sleep(60)
```
# Cleaning up
To clean up all GCP resources used in this project, you can [delete the GCP
project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.
Otherwise, you can delete the individual resources you created in this tutorial:
- Dataset
- Pipeline
- Model
- Endpoint
- Batch Job
- Custom Job
- Hyperparameter Tuning Job
- Cloud Storage Bucket
```
delete_dataset = True
delete_pipeline = True
delete_model = True
delete_endpoint = True
delete_batchjob = True
delete_customjob = True
delete_hptjob = True
delete_bucket = True
# Delete the dataset using the Vertex fully qualified identifier for the dataset
try:
if delete_dataset and "dataset_id" in globals():
clients["dataset"].delete_dataset(name=dataset_id)
except Exception as e:
print(e)
# Delete the training pipeline using the Vertex fully qualified identifier for the pipeline
try:
if delete_pipeline and "pipeline_id" in globals():
clients["pipeline"].delete_training_pipeline(name=pipeline_id)
except Exception as e:
print(e)
# Delete the model using the Vertex fully qualified identifier for the model
try:
if delete_model and "model_to_deploy_id" in globals():
clients["model"].delete_model(name=model_to_deploy_id)
except Exception as e:
print(e)
# Delete the endpoint using the Vertex fully qualified identifier for the endpoint
try:
if delete_endpoint and "endpoint_id" in globals():
clients["endpoint"].delete_endpoint(name=endpoint_id)
except Exception as e:
print(e)
# Delete the batch job using the Vertex fully qualified identifier for the batch job
try:
if delete_batchjob and "batch_job_id" in globals():
clients["job"].delete_batch_prediction_job(name=batch_job_id)
except Exception as e:
print(e)
# Delete the custom job using the Vertex fully qualified identifier for the custom job
try:
if delete_customjob and "job_id" in globals():
clients["job"].delete_custom_job(name=job_id)
except Exception as e:
print(e)
# Delete the hyperparameter tuning job using the Vertex fully qualified identifier for the hyperparameter tuning job
try:
if delete_hptjob and "hpt_job_id" in globals():
clients["job"].delete_hyperparameter_tuning_job(name=hpt_job_id)
except Exception as e:
print(e)
if delete_bucket and "BUCKET_NAME" in globals():
! gsutil rm -r $BUCKET_NAME
```
|
github_jupyter
|
# Practical Examples of Interactive Visualizations in JupyterLab with Pixi.js and Jupyter Widgets
# PyData Berlin 2018 - 2018-07-08
# Jeremy Tuloup
# [@jtpio](https://twitter.com/jtpio)
# [github.com/jtpio](https://github.com/jtpio)
# [jtp.io](https://jtp.io)

# The Python Visualization Landscape (2017)

Source:
- [Jake VanderPlas: The Python Visualization Landscape PyCon 2017](https://www.youtube.com/watch?v=FytuB8nFHPQ)
- [Source for the Visualization](https://github.com/rougier/python-visualization-landscape), by Nicolas P. Rougier

# Motivation
|Not This|This|
|:--------------------------:|:-----------------------------------------:|
| | |

# JupyterLab - Pixi.js - Jupyter Widgets?

# Prerequisites
# * Jupyter Notebook
# * Python

# JupyterLab


## * Powerful 2D rendering engine written in JavaScript
## * Abstraction on top of Canvas and WebGL
# [Live Example!](http://localhost:4000)
```javascript
let app = new PIXI.Application(800, 600, {backgroundColor : 0x1099bb});
document.body.appendChild(app.view);
let bunny = PIXI.Sprite.fromImage('bunny.png')
bunny.anchor.set(0.5);
bunny.x = app.screen.width / 2;
bunny.y = app.screen.height / 2;
app.stage.addChild(bunny);
app.ticker.add((delta) => {
bunny.rotation += 0.1 * delta;
});
```

# Jupyter Widgets

[Open the image](./img/WidgetModelView.png)
- Source: [https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Basics.html#Why-does-displaying-the-same-widget-twice-work?](https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Basics.html#Why-does-displaying-the-same-widget-twice-work?)
```
from ipywidgets import IntSlider
slider = IntSlider(min=0, max=10)
slider
slider
slider.value
slider.value = 2
```
# Tutorial to create your own
## https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Custom.html
# Libraries
## bqplot

## ipyleaflet

## ipyvolume


# Motivation: Very Custom Visualizations


# Drawing Shapes on a Canvas
```
from ipyutils import SimpleShape
```
# Implementation
## - [simple_shape.py](../ipyutils/simple_shape.py): defines the **SimpleShape** Python class
## - [widget.ts](../src/simple_shapes/widget.ts): defines the **SimpleShapeModel** and **SimpleShapeView** Typescript classes
```
square = SimpleShape()
square
square.rotate = True
```
# Level Up 🚀
```
from ipyutils import Shapes
shapes = Shapes(n_shapes=100)
shapes
shapes.shape
shapes.shape = 'square'
shapes.rotate = True
shapes.wobble = True
```

# Visualizing Recursion with the Bermuda Triangle Puzzle


# Motivation
# * Solve the puzzle programmatically
# * Verify a solution visually
# * Animate the process

# BermudaTriangle Widget
```
from ipyutils import TriangleAnimation, BermudaTriangle
triangles = TriangleAnimation()
triangles
```

# What can we do with this widget?

# Visualize Transitions
From | To
:--------------------------:|:-------------------------:
 | 
```
# states
state_0 = [None] * 16
print(state_0)
state_1 = [[13, 1]] + [None] * 15
print(state_1)
state_2 = [[13, 1], [12, 0]] + [None] * 14
print(state_2)
```
# Example States and Animation
```
example_states = TriangleAnimation()
bermuda = example_states.bermuda
bermuda.states = [
[None] * 16,
[[7, 0]] + [None] * 15,
[[7, 1]] + [None] * 15,
[[7, 2]] + [None] * 15,
[[7, 2], [0, 0]] + [None] * 14,
[[7, 2], [0, 1]] + [None] * 14,
[[i, 0] for i in range(16)],
[[i, 1] for i in range(16)],
]
example_states
```

# Solver
```
from copy import deepcopy
class Solver(BermudaTriangle):
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.reset_state()
def reset_state(self):
self.board = [None] * self.N_TRIANGLES
self.logs = [deepcopy(self.board)]
self.it = 0
def solve(self):
'''
Method to implement
'''
raise NotImplementedError()
def log(self):
self.logs.append(deepcopy(self.board))
def found(self):
return all(self.is_valid(i) for i in range(self.N_TRIANGLES))
def save_state(self):
self.permutation = self.board
self.states = self.logs
```
# Valid Permutation - is_valid()
```
help(Solver.is_valid)
```
```python
solver.is_valid(7)
# False
```


# First Try: Random Permutations
```
import random
class RandomSearch(Solver):
def solve(self):
random.seed(42)
self.reset_state()
for i in range(200):
self.board = random.sample(self.permutation, self.N_TRIANGLES)
self.log()
if self.found():
print('Found!')
return True
return False
%%time
solver = RandomSearch()
res = solver.solve()
solver.save_state()
rnd = TriangleAnimation()
rnd.bermuda.title = 'Random Search'
rnd.bermuda.states = solver.states
rnd
```

# Better: Brute Force using Recursion
```
class RecursiveSolver(Solver):
def solve(self):
self.used = [False] * self.N_TRIANGLES
self.reset_state()
self._place(0)
return self.board
def _place(self, i):
self.it += 1
if i == self.N_TRIANGLES:
return True
for j in range(self.N_TRIANGLES - 1, -1, -1):
if self.used[j]:
# piece number j already used
continue
self.used[j] = True
for rot in range(3):
# place the piece on the board
self.board[i] = (j, rot)
self.log()
# stop the recursion if the current configuration
# is not valid or a solution has been found
if self.is_valid(i) and self._place(i + 1):
return True
# remove the piece from the board
self.board[i] = None
self.used[j] = False
self.log()
return False
%%time
solver = RecursiveSolver()
res = solver.solve()
if solver.found():
print('Solution found!')
print(f'{len(solver.logs)} steps')
solver.save_state()
else:
print('No solution found')
recursion = TriangleAnimation()
recursion.bermuda.title = 'Recursive Search'
recursion.bermuda.states = solver.states
recursion
```

# More details for this example
## * In depth walkthrough on how to create a Jupyter Widget in the notebook
## * [p5.js in the Jupyter Notebook for custom interactive visualizations](https://github.com/jtpio/p5-jupyter-notebook/blob/master/puzzle.ipynb)
## * Using p5.js instead of Pixi.js, but similar concepts
## * Source: [github.com/jtpio/p5-jupyter-notebook](https://github.com/jtpio/p5-jupyter-notebook)
## * [Run on Binder](https://mybinder.org/v2/gh/jtpio/p5-jupyter-notebook/master?filepath=puzzle.ipynb)

# Recap
## * Custom interactive animations with Pixi.js
## * Leverage the JavaScript ecosystem in JupyterLab

# Applications
## * Visual debugging and understanding
## * Teaching and education, learning by doing
## * Combine JavaScript games with data
# Downside
## * Requires some effort to build the visualizations in TypeScript / JavaScript

# References
## Presentations
### - [Jake VanderPlas: The Python Visualization Landscape PyCon 2017](https://www.youtube.com/watch?v=FytuB8nFHPQ)
### - [PyData London 2016: Sylvain Corlay - Interactive Visualization in Jupyter with Bqplot and Interactive Widgets](https://www.youtube.com/watch?v=eVET9IYgbao)
### - [PLOTCON 2017: Sylvain Corlay, Interactive Data Visualization in JupyterLab with Jupyter](https://www.youtube.com/watch?v=p7Hr54VhOp0)
### - [PyData Amsterdam 2017: Maarten Breddels | A billion stars in the Jupyter Notebook](https://www.youtube.com/watch?v=bP-JBbjwLM8)
## Widgets
### - [Building a Custom Widget Tutorial](https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20Custom.html)
### - [Authoring Custom Jupyter Widgets](https://blog.jupyter.org/authoring-custom-jupyter-widgets-2884a462e724)
### - [p5.js in the Jupyter Notebook for custom interactive visualizations](https://github.com/jtpio/p5-jupyter-notebook/blob/master/puzzle.ipynb)
### - [pythreejs](https://github.com/jovyan/pythreejs): Implemented as an Jupyter Widget
### - [bqplot](https://github.com/bloomberg/bqplot): Great library for interactive data exploration
### - [ipyvolume](https://github.com/maartenbreddels/ipyvolume): 3d plotting for Python in the Jupyter Notebook
### - [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet): interactive maps in the Jupyter notebook

# Questions?
## [@jtpio](https://twitter.com/jtpio)
## [github.com/jtpio](https://github.com/jtpio)
## [jtp.io](jtp.io)

|
github_jupyter
|
# Regressão linear
## **TOC:**
Na aula de hoje, vamos explorar os seguintes tópicos em Python:
- 1) [Introdução](#intro)
- 2) [Regressão linear simples](#reglinear)
- 3) [Regressão linear múltipla](#multireglinear)
- 4) [Tradeoff viés-variância](#tradeoff)
```
# importe as principais bibliotecas de análise de dados
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
```
____
____
____
## 1) **Introdução** <a class="anchor" id="intro"></a>
Imagine que você que vender sua casa.
Você sabe os atributos da sua casa: quantos cômodos têm, quantos carros cabem na garagem, qual é a área construída, qual sua localidade, etc.
Agora, a pergunta é: qual seria o melhor preço pra você colocá-la a venda, ou seja, quanto de fato ela vale?
Você pode solicitar a avaliação de um corretor de imóveis (contanto com a experiência dele), ou então...
...fazer um modelo de **Machine Learning**, que, com base nos atributos e preços de diversas outras casas, pode fazer uma **predição** sobre o preço adequado da sua casa!
Para resolver este problema, podemos utilizar um dos mais simples e importantes algoritmos de machine learning: a Regressão Linear!
____
Para introduzirmos as ideias, vamos usar um [dataset de preço de casas](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data).
Esta base de dados contém **70 features** (+ 1 ID), que são as características de cada uma das casas listadas; e **1 target**, que é o preço pelo qual aquela casa foi vendida.
Para o significado de cada uma das features, e os valores que elas podem assumir, veja a página acima.
**Vamos ler a base e começar a explorá-la!**
```
df = pd.read_csv("data/house_prices/house_price.csv")
```
Por enquanto, não vamos nos preocupar com os dados missing, pois vamos usar apenas uma feature no nosso modelo inicial.
Aproveite para depois explorar os dados da forma que quiser!
Por enquanto, vamos dar uma olhada na coluna target!
Fica evidente que a distribuição é desviada para a direita.
Vamos tentar alterar isso na próximas versões do modelo para ver se teremos ganhos de performance!
Por enquanto, seguimos assim.
Parece que a variável de área construída ```GrLivArea```) é uma forte candidata a **explicar** o preço das casas, pois vemos calaramente uma correlação entre as variáveis!
Mas note que há claramente dois outliers...
Vamos agora iniciar a construção de um modelo bem simples, que utilize a variável GrLivArea para predizer o preço!
___
___
___
## 2) **Regressão linear simples** <a class="anchor" id="reglinear"></a>
Apesar de alguns outliers, parece bem adequado que os pontos plotados acima sejam descritos por uma reta, não é mesmo?
Ou, melhor dizendo: **a variável GrLivArea parece estar relacionada ao target SalePrice linearmente!**
Para modelarmos esta relação, vamos conhecer o modelo de **Regressão Linear Simples**.
Como o próprio nome diz, o modelo de Regressão Linear será **uma reta (polinômio linear)**, que melhor se ajusta aos seus dados!
O modelo de **Regressão Linear Simples** será uma linha reta que relaciona Y (o preço da casa) e X (os atributos da casa).
Se utilizarmos **apenas um atributo** (como, por exemplo, a área construída), temos uma **Regressão Linear Simples**, e nosso modelo é:
$$ y = b_0 + b_1 X $$
Neste caso, o modelo tem dois coeficientes a serem determinados: $b_0$ (intercepto ou coeficiente linear) e $b_1$ (coeficiente angular).
O algoritmo do estimador é utilizado justamente para encontrarmos os coeficientes $b_0$ e $b_1$ **que melhor se ajustam aos dados!**
Para fazer isso, pode-se utilizar o método dos **mínimos quadrados** ou então o **gradiente descendente**.
Mas não nos preocuparemos com os detalhes do treinamento: usaremos o sklearn para isso!
Vamos começar?
Agora que o modelo está treinado, podemos dar uma olhada nos coeficientes que foram encontrados!
Como interpretamos este resultado?
O nosso modelo final é dado por:
$$ y = 1562.01 + 118.61 \times \text{GrLiveArea}$$
Isto quer dizer que:
> Aumentando a variável "GrLiveArea" em uma unidade faz com que o preço seja aumentado em USD 118.6!
> O preço mínimo a ser pago, independente da área construída, é de 1562.01!
Podemos visualizar o modelo treinado, neste caso:
Fazendo uma previsão:
Ou ainda:
É raro que consigamos visualizar nosso modelo final como fizemos acima, mas no caso da regressão linear simples, temos essa sorte! :)
Vamos agora fazer algumas previsões!
Agora que temos o modelo treinado e algumas previsões, como avaliamos a performance do modelo?
Para isso, podemos dar uma olhada nos **resíduos** das predições! Os resíduos nada mais são do que **os erros do modelo**, ou seja, **a diferença entre cada valor predito e o valor real**, para **os dados de teste!**. Isto é,
$$R(y_i) = y_i - \hat{y}_i $$
O caso 100% ideal seria $y_i = \hat{y}_i$, o que produziria uma reta exata!
Quanto mais "espalhados" estiverem os pontos em torno da reta, em geral **pior é o modelo**, pois ele está errando mais!
Uma forma de quantificar isso através de uma métrica conhecida como **$R^2$**, o **coeficiente de determinação**.
Este coeficiente indica **o quão próximos os dados estão da reta ajustada**. Por outro lado, o $R^2$ representa a porcentagem de variação na resposta que é explicada pelo modelo.
$$R^2 = 1 - \frac{\sum_{i=1}^n(y_i-\hat{y}_i)^2}{\sum_{i=1}^n(y_i-\bar{y})^2}$$
É possível utilizar o $R^2$ nos dados de treino, mas isso não é tão significante, devido ao overfitting, que discutiremos a seguir. Mais sgnificativo é calcularmos o $R^2$ nos dados de teste como faremos a seguir. Essa métrica equivale, portanto, **ao gráfico que fizemos acima!**
Outra coisa importante é que os resíduos sejam **normalmente distribuídos**.
Se esse não for o caso, é muito importante que você reveja se o modelo escolhido de fato é adequado ao seu problema!
Além dos resíduos, existem três principais **métricas de avaliação** do modelo de regressão linear:
**Mean Absolute Error** (MAE) é a média do valor absoluto de todos os resíduos (erros):
$$\frac 1n\sum_{i=1}^n|y_i-\hat{y}_i|$$
**Mean Squared Error** (MSE) é a média dos erros quadrados:
$$\frac 1n\sum_{i=1}^n(y_i-\hat{y}_i)^2$$
**Root Mean Squared Error** (RMSE) é a raiz quadrada da média dos erros quadrados:
$$\sqrt{\frac 1n\sum_{i=1}^n(y_i-\hat{y}_i)^2}$$
Comparando as métricas:
- **MAE** é a mais simples de entender, mas ela penaliza mais erros menores;
- **MSE** é a métrica mais popular, pois essa métrica penaliza mais erros maiores, o que faz mais sentido em aplicações reais.
- **RMSE** é ainda mais popular, pois esta métrica está nas mesmas unidades que o target.
Estas métricas todas podem ser utilizadas como **funções de custo** a serem minimizadas pelo algoritmo do estimador.
___
## 3) **Regressão linear múltipla** <a class="anchor" id="multireglinear"></a>
O modelo que fizemos acima considera uma única feature como preditora do preço da casa.
Mas temos outras 78 dessas features! Será que não há mais informação útil em todas essas outras variáveis?
Em geral, sim! É natural que esperemos que **mais variáveis** tragam **mais informações** ao modelo, e, portanto, o torne mais preciso!
Para incorporar estas outras variáveis ao modelo, é muito simples!
Podemos passar a utilizar outros atributos (como o número de cômodos, qual é a renda média da vizinhança, etc.), e neste caso teremos uma **Regressão Linear Múltipla**, que nada mais é que a seguinte equação:
$$ y = b_0 + b_1 X_1 + b_2 X_2 + \cdots + b_n X_n $$
Neste caso, além de $b_0$ e $b_1$, temos também outros coeficientes, um pra cada uma das $n$ features que escolhermos!
Modelos de regressão múltipla são potencialmente mais precisos, mas há também um lado ruim: nós perdemos a **possibilidade de visualização**. Agora, não temos mais uma reta, mas sim um **hiperplano** que relaciona todas as features com o target!
<center><img src="https://miro.medium.com/max/1120/0*rGSfRsMjiQeG5jof.png" width=500></center>
Vamos construir esse modelo?
Observação: a coluna "Id" traz apenas um número de identificação arbitrário que não deve ser correlacionado com o target. Portanto, vamos desconsiderar esta coluna de nosso modelo!
A performance do modelo melhorou?
Será que dá pra melhorar mais?
Opções:
- tentar apenas um subconjunto de features: **feature selection**
- passar a utilizar as features categóricas: **feature engeneering**
---
## 4) **Tradeoff viés-variância** <a class="anchor" id="tradeoff"></a>
Veremos agora um dos conceitos mais importantes em apredizado de maquina.
Muitas vezes alguns modelos têm 100% de acerto nos dados de **treino**, mas **na base de teste** a performance cai para menos de 50%.
Isso pode acontecer porque o modelo fica **especialista apenas no conjunto de treino**, não conseguindo **generalizar os padrões para além dos dados vistos**.
<center><img src="https://miro.medium.com/max/1125/1*_7OPgojau8hkiPUiHoGK_w.png" width=800></center>
O overfitting está intimamente ligado com o conceito de **viés** (bias) e **variância** (variance):
>**Viés**<br>
É a diferença entre o que o modelo prediz, e o valor correto a ser predito.<br>
Modelos com alto viés são muito simples, de modo a **não conseguir capturar as relações que os dados de treino exibem** (underfit).<br>
Issso faz com que ambos os erros de treino e de teste sejam altos.
<br><br>
Em outras palavras:<br>
**Incapacidade de um modelo de capturar a verdadeira relação entre features e target**
> **Variância**<br>
Variância se refere à variabilidade das predições de um modelo.<br>
Modelos com alta variância são muito complexos, por **aprenderem demais as relações exibidas nos dados de treino** (overfit).<br>
Isso faz com que os erros de treino sejam baixos, mas os erros de teste sejam altos.
<br><br>
Em outras palavras:<br>
**Incapacidade de um modelo performar bem em outros datasets diferentes do usado no treinamento**.
<center><img src="https://www.learnopencv.com/wp-content/uploads/2017/02/Bias-Variance-Tradeoff-In-Machine-Learning-1.png" width=500></center>
<center><img src="https://miro.medium.com/max/1494/1*C7ZKM93QVdpeSCGbF5TjIg.png" width=800></center>
Para demonstrar overfit ser usado o conjuto de teste [anscombe](https://en.wikipedia.org/wiki/Anscombe%27s_quartet)
```
df_anscombe = sns.load_dataset('anscombe')
df_anscombe.groupby("dataset").agg({"mean", "std"})
```
Vamos supor que este dado represente valores de medições de um sensor, porém o sensor teve um pequeno problema durante a medição.
Podemos perceber facilmente qual é este erro, e qual seria a função de regreesão para este sensor com os dados validos: **regressão linear**.
Perceba que a função linear encontrar já aprensenta um padrão muito similiar aos dados, porém um ponto error faz com que ela não tenha um resultado otimo.
Podemos utilizar regressões polinomiais, que possuem ordem maiores que 1, para tentar diminuir o erro da regressão, obtendo uma equação do formato.
$$\hat{y}_{i} = \beta_{1} + \beta_{2} x_{i} + \beta_{3} {x_{i}}^{2} + \cdots + \beta_{6} {x_{i}}^{6}$$
Para criar modelos polinomiaus com o sklearn, [dê uma olhada aqui](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.PolynomialFeatures.html)
Ao utilizarmos uma regressão de ordem 6 percebemos que ela se ajusta ao valor com erro, porém ela **se distancia da regressão que realmente representa os dados**.
Tentar **aprender o erro faz com ela com ela não aprenda a função real**.
Isto acontece pois ela se **super ajustou aos dados de treino, se distanciando dos dados reais**.
__Como garantir que nosso modelo não está sofrendo de overfitting?__
Naturalmente, essa é uma pergunta de extrema importância, especialmente no contexto de **Redes neurais**. [Veja aqui](https://towardsdatascience.com/8-simple-techniques-to-prevent-overfitting-4d443da2ef7d) e [aqui](https://towardsdatascience.com/dont-overfit-how-to-prevent-overfitting-in-your-deep-learning-models-63274e552323) algumas discussões.
Na prática: **jamais se apegue à peformance de treino!!**. O que queremos otimizar sempre será a performance **avaliada nos dados de teste**. Assim, garantimos que uma boa performance não é produto do overfitting!
---
|
github_jupyter
|
# Table of Contents
<p><div class="lev1 toc-item"><a href="#Lambda-calcul-implémenté-en-OCaml" data-toc-modified-id="Lambda-calcul-implémenté-en-OCaml-1"><span class="toc-item-num">1 </span>Lambda-calcul implémenté en OCaml</a></div><div class="lev2 toc-item"><a href="#Expressions" data-toc-modified-id="Expressions-11"><span class="toc-item-num">1.1 </span>Expressions</a></div><div class="lev2 toc-item"><a href="#But-?" data-toc-modified-id="But-?-12"><span class="toc-item-num">1.2 </span>But ?</a></div><div class="lev2 toc-item"><a href="#Grammaire" data-toc-modified-id="Grammaire-13"><span class="toc-item-num">1.3 </span>Grammaire</a></div><div class="lev2 toc-item"><a href="#L'identité" data-toc-modified-id="L'identité-14"><span class="toc-item-num">1.4 </span>L'identité</a></div><div class="lev2 toc-item"><a href="#Conditionnelles" data-toc-modified-id="Conditionnelles-15"><span class="toc-item-num">1.5 </span>Conditionnelles</a></div><div class="lev2 toc-item"><a href="#Nombres" data-toc-modified-id="Nombres-16"><span class="toc-item-num">1.6 </span>Nombres</a></div><div class="lev2 toc-item"><a href="#Test-d'inégalité" data-toc-modified-id="Test-d'inégalité-17"><span class="toc-item-num">1.7 </span>Test d'inégalité</a></div><div class="lev2 toc-item"><a href="#Successeurs" data-toc-modified-id="Successeurs-18"><span class="toc-item-num">1.8 </span>Successeurs</a></div><div class="lev2 toc-item"><a href="#Prédecesseurs" data-toc-modified-id="Prédecesseurs-19"><span class="toc-item-num">1.9 </span>Prédecesseurs</a></div><div class="lev2 toc-item"><a href="#Addition" data-toc-modified-id="Addition-110"><span class="toc-item-num">1.10 </span>Addition</a></div><div class="lev2 toc-item"><a href="#Multiplication" data-toc-modified-id="Multiplication-111"><span class="toc-item-num">1.11 </span>Multiplication</a></div><div class="lev2 toc-item"><a href="#Paires" data-toc-modified-id="Paires-112"><span class="toc-item-num">1.12 </span>Paires</a></div><div class="lev2 toc-item"><a href="#Prédécesseurs,-deuxième-essai" data-toc-modified-id="Prédécesseurs,-deuxième-essai-113"><span class="toc-item-num">1.13 </span>Prédécesseurs, deuxième essai</a></div><div class="lev2 toc-item"><a href="#Listes" data-toc-modified-id="Listes-114"><span class="toc-item-num">1.14 </span>Listes</a></div><div class="lev2 toc-item"><a href="#La-fonction-U" data-toc-modified-id="La-fonction-U-115"><span class="toc-item-num">1.15 </span>La fonction U</a></div><div class="lev2 toc-item"><a href="#La-récursion-via-la-fonction-Y" data-toc-modified-id="La-récursion-via-la-fonction-Y-116"><span class="toc-item-num">1.16 </span>La récursion via la fonction Y</a></div><div class="lev2 toc-item"><a href="#Conclusion" data-toc-modified-id="Conclusion-117"><span class="toc-item-num">1.17 </span>Conclusion</a></div>
# Lambda-calcul implémenté en OCaml
Ce notebook est inspiré de [ce post de blog du Professeur Matt Might](http://matt.might.net/articles/python-church-y-combinator/), qui implémente un mini langage de programmation en $\lambda$-calcul, en Python.
Je vais faire la même chose en OCaml.
## Expressions
On rappelle que les expressions du [Lambda calcul](https://fr.wikipedia.org/wiki/Lambda-calcul), ou $\lambda$-calcul, sont les suivantes :
$$ \begin{cases}
x, y, z & \text{(des variables)} \\
u v & \text{(application de deux termes}\, u, v\; \text{)} \\
\lambda x. v & \text{(lambda-function prenant la variable}\; x \;\text{et le terme}\; v \;\text{)}
\end{cases} $$
## But ?
Le but ne va pas être de les représenter comme ça avec des types formels en Caml, mais plutôt d'utiliser les constructions de Caml, respectivement `u(v)` et `fun x -> v` pour l'application et les fonctions anonymes, et encoder des fonctionnalités de plus haut niveau dans ce langage réduit.
## Grammaire
Avec une grammaire BNF, si `<var>` désigne un nom d'expression valide (on se limitera à des noms en miniscules consistitués des 26 lettres `a,b,..,z`) :
<exp> ::= <var>
| <exp>(<exp>)
| fun <var> -> <exp>
| (<exp>)
----
## L'identité
```
let identite = fun x -> x ;;
let vide = fun x -> x ;;
```
## Conditionnelles
La conditionnelle est `si cond alors valeur_vraie sinon valeur_fausse`.
```
let si = fun cond valeur_vraie valeur_fausse -> cond valeur_vraie valeur_fausse ;;
```
C'est très simple, du moment qu'on s'assure que `cond` est soit `vrai` soit `faux` tels que définis par leur comportement :
si vrai e1 e2 == e1
si faux e1 e2 == e2
```
let vrai = fun valeur_vraie valeur_fausse -> valeur_vraie ;;
let faux = fun valeur_vraie valeur_fausse -> valeur_fausse ;;
```
La négation est facile !
```
let non = fun v x y -> v y x;;
```
En fait, on va forcer une évaluation paresseuse, comme ça si l'une des deux expressions ne terminent pas, l'évaluation fonctionne quand même.
```
let vrai_paresseux = fun valeur_vraie valeur_fausse -> valeur_vraie () ;;
let faux_paresseux = fun valeur_vraie valeur_fausse -> valeur_fausse () ;;
```
Pour rendre paresseux un terme, rien de plus simple !
```
let paresseux = fun f -> fun () -> f ;;
```
## Nombres
La représentation de Church consiste a écrire $n$ comme $\lambda f. \lambda z. f^n z$.
```
type 'a nombres = ('a -> 'a) -> 'a -> 'a;; (* inutilisé *)
type entiers_church = (int -> int) -> int -> int;;
```
$0$ est trivialement $\lambda f. \lambda z. z$ :
```
let zero = fun (f : ('a -> 'a)) (z : 'a) -> z ;;
```
$1$ est $\lambda f. \lambda z. f z$ :
```
let un = fun (f : ('a -> 'a)) -> f ;;
```
Avec l'opérateur de composition, l'écriture des entiers suivants est facile.
```
let compose = fun f g x -> f (g x);;
let deux = fun f -> compose f f;; (* == compose f (un f) *)
let trois = fun f -> compose f (deux f) ;;
let quatre = fun f -> compose f (trois f) ;;
(* etc *)
```
On peut généraliser ça, avec une fonction qui transforme un entier (`int`) de Caml en un entier de Church :
```
let rec entierChurch (n : int) =
fun f z -> if n = 0 then z else f ((entierChurch (n-1)) f z)
;;
```
Par exemple :
```
(entierChurch 0) (fun x -> x + 1) 0;; (* 0 *)
(entierChurch 7) (fun x -> x + 1) 0;; (* 7 *)
(entierChurch 3) (fun x -> 2*x) 1;; (* 8 *)
```
Et une fonction qui fait l'inverse (note : cette fonction n'est *pas* un $\lambda$-terme) :
```
let entierNatif c : int =
c (fun x -> x + 1) 0
;;
```
Un petit test :
```
entierNatif (si vrai zero un);; (* 0 *)
entierNatif (si faux zero un);; (* 1 *)
entierNatif (entierChurch 100);; (* 100 *)
```
## Test d'inégalité
On a besoin de pouvoir tester si $n \leq 0$ (ou $n = 0$) en fait.
```
(* prend un lambda f lambda z. ... est donne vrai ssi n = 0 ou faux sinon *)
let estnul = fun n -> n (fun z -> faux) (vrai);;
(* prend un lambda f lambda z. ... est donne vrai ssi n > 0 ou faux sinon *)
let estnonnul = fun n -> n (fun z -> vrai) (faux);;
```
On peut proposer cette autre implémentation, qui "fonctionne" pareil (au sens calcul des $\beta$-réductions) mais est plus compliquée :
```
let estnonnul2 = fun n -> non (estnul n);;
entierNatif (si (estnul zero) zero un);; (* 0 *)
entierNatif (si (estnul un) zero un);; (* 1 *)
entierNatif (si (estnul deux) zero un);; (* 1 *)
entierNatif (si (estnonnul zero) zero un);; (* 0 *)
entierNatif (si (estnonnul un) zero un);; (* 1 *)
entierNatif (si (estnonnul deux) zero un);; (* 1 *)
entierNatif (si (non (estnul zero)) zero un);; (* 0 *)
entierNatif (si (non (estnul un)) zero un);; (* 1 *)
entierNatif (si (non (estnul deux)) zero un);; (* 1 *)
```
## Successeurs
Vue la représentation de Churc, $n+1$ consiste a appliquer l'argument $f$ une fois de plus :
$f^{n+1}(z) = f (f^n(z))$.
```
let succ = fun n f z -> f ((n f) z) ;;
entierNatif (succ un);; (* 2 *)
deux;;
succ un;;
```
On remarque qu'ils ont le même typage, mais OCaml indique qu'il a moins d'informations à propos du deuxième : ce `'_a` signifie que le type est *contraint*, il sera fixé dès la première utilisation de cette fonction.
C'est assez mystérieux, mais il faut retenir le point suivant : `deux` était écrit manuellement, donc le système a vu le terme en entier, il le connaît et saît que `deux = fun f -> fun x -> f (f x))`, pas de surprise. Par contre, `succ un` est le résultat d'une évaluation *partielle* et vaut `fun f z -> f ((deux f) z)`. Sauf que le système ne calcule pas tout et laisse l'évaluation partielle ! (heureusement !)
Si on appelle `succ un` à une fonction, le `'_a` va être contraint, et on ne pourra pas s'en reservir :
```
let succ_de_un = succ un;;
(succ_de_un) (fun x -> x + 1);;
(succ_de_un) (fun x -> x ^ "0");;
(succ un) (fun x -> x ^ "0");;
(* une valeur fraîchement calculée, sans contrainte *)
```
## Prédecesseurs
Vue la représentation de Church, $\lambda n. n-1$ n'existe pas... mais on peut tricher.
```
let pred = fun n ->
if (entierNatif n) > 0 then entierChurch ((entierNatif n) - 1)
else zero
;;
entierNatif (pred deux);; (* 1 *)
entierNatif (pred trois);; (* 2 *)
```
## Addition
Pour ajouter $n$ et $m$, il faut appliquer une fonction $f$ $n$ fois puis $m$ fois : $f^{n+m}(z) = f^n(f^m(z))$.
```
let somme = fun n m f z -> n(f)( m(f)(z));;
let cinq = somme deux trois ;;
entierNatif cinq;;
let sept = somme cinq deux ;;
entierNatif sept;;
```
## Multiplication
Pour multiplier $n$ et $m$, il faut appliquer le codage de $n$ exactement $m$ fois : $f^{nm}(z) = (f^n(f^n(...(f^n(z))...))$.
```
let produit = fun n m f z -> m(n(f))(z);;
```
On peut faire encore mieux avec l'opérateur de composition :
```
let produit = fun n m -> compose m n;;
let six = produit deux trois ;;
entierNatif six;;
let huit = produit deux quatre ;;
entierNatif huit;;
```
## Paires
On va écrire un constructeur de paires, `paire a b` qui sera comme `(a, b)`, et deux destructeurs, `gauche` et `droite`, qui vérifient :
gauche (paire a b) == a
droite (paire a b) == b
```
let paire = fun a b -> fun f -> f(a)(b);;
let gauche = fun p -> p(fun a b -> a);;
let droite = fun p -> p(fun a b -> b);;
entierNatif (gauche (paire zero un));;
entierNatif (droite (paire zero un));;
```
## Prédécesseurs, deuxième essai
Il y a une façon, longue et compliquée ([source](http://gregfjohnson.com/pred/)) d'y arriver, avec des paires.
```
let pred n suivant premier =
let pred_suivant = paire vrai premier in
let pred_premier = fun p ->
si (gauche p)
(paire faux premier)
(paire faux (suivant (droite p)))
in
let paire_finale = n pred_suivant pred_premier in
droite paire_finale
;;
```
Malheureusement, ce n'est pas bien typé.
```
entierNatif (pred deux);; (* 1 *)
```
## Listes
Pour construire des listes (simplement chaînées), on a besoin d'une valeur pour la liste vide, `listevide`, d'un constructeur pour une liste `cons`, un prédicat pour la liste vide `estvide`, un accesseur `tete` et `queue`, et avec les contraintes suivantes (avec `vrai`, `faux` définis comme plus haut) :
estvide (listevide) == vrai
estvide (cons tt qu) == faux
tete (cons tt qu) == tt
queue (cons tt qu) == qu
On va stocker tout ça avec des fonctions qui attendront deux arguments (deux fonctions - rappel tout est fonction en $\lambda$-calcul), l'une appellée si la liste est vide, l'autre si la liste n'est pas vide.
```
let listevide = fun survide surpasvide -> survide;;
let cons = fun hd tl -> fun survide surpasvide -> surpasvide hd tl;;
```
Avec cette construction, `estvide` est assez simple : `survide` est `() -> vrai` et `surpasvide` est `tt qu -> faux`.
```
let estvide = fun liste -> liste (vrai) (fun tt qu -> faux);;
```
Deux tests :
```
entierNatif (si (estvide (listevide)) un zero);; (* estvide listevide == vrai *)
entierNatif (si (estvide (cons un listevide)) un zero);; (* estvide (cons un listevide) == faux *)
```
Et pour les deux extracteurs, c'est très facile avec cet encodage.
```
let tete = fun liste -> liste (vide) (fun tt qu -> tt);;
let queue = fun liste -> liste (vide) (fun tt qu -> qu);;
entierNatif (tete (cons un listevide));;
entierNatif (tete (queue (cons deux (cons un listevide))));;
entierNatif (tete (queue (cons trois (cons deux (cons un listevide)))));;
```
Visualisons les types que Caml trouve a des listes de tailles croissantes :
```
cons un (cons un listevide);; (* 8 variables pour une liste de taille 2 *)
cons un (cons un (cons un (cons un listevide)));; (* 14 variables pour une liste de taille 4 *)
cons un (cons un (cons un (cons un (cons un (cons un (cons un (cons un listevide)))))));; (* 26 variables pour une liste de taille 7 *)
```
Pour ces raisons là, on se rend compte que le type donné par Caml à une liste de taille $k$ croît linéairement *en taille* en fonction de $k$ !
Aucun espoir donc (avec cet encodage) d'avoir un type générique pour les listes représentés en Caml.
Et donc nous ne sommes pas surpris de voir cet essai échouer :
```
let rec longueur liste =
liste (zero) (fun t q -> succ (longueur q))
;;
```
<span style="color:red;">En effet, `longueur` devrait être bien typée et `liste` et `q` devraient avoir le même type, or le type de `liste` est strictement plus grand que celui de `q`...</span>
On peut essayer de faire une fonction `ieme`.
On veut que `ieme zero liste = tete` et `ieme n liste = ieme (pred n) (queue liste)`.
En écrivant en haut niveau, on aimerait pouvoir faire :
```
let pop liste =
si (estvide liste) (listevide) (queue liste)
;;
let ieme n liste =
tete (n pop liste)
;;
```
## La fonction U
C'est le premier indice que le $\lambda$-calcul peut être utilisé comme modèle de calcul : le terme $U : f \to f(f)$ ne termine pas si on l'applique à lui-même.
Mais ce sera la faiblesse de l'utilisation de Caml : ce terme ne peut être correctement typé !
```
let u = fun f -> f (f);;
```
A noter que même dans un langage non typé (par exemple Python), on peut définir $U$ mais son exécution échouera, soit à caude d'un dépassement de pile, soit parce qu'elle ne termine pas.
## La récursion via la fonction Y
La fonction $Y$ trouve le point fixe d'une autre fonction.
C'est très utile pour définir des fonctions par récurrence.
Par exemple, la factorielle est le point fixe de la fonction suivante :
"$\lambda f. \lambda n. 1$ si $n \leq 0$ sinon $n * f(n-1)$" (écrite dans un langage plus haut niveau, pas en $\lambda$-calcul).
$Y$ satisfait ces contraintes : $Y(F) = f$ et $f = F(f)$.
Donc $Y(F) = F(Y(F))$ et donc $Y = \lambda F. F(Y(F))$. Mais ce premier essai ne marche pas.
```
let rec y = fun f -> f (y(f));;
let fact = y(fun f n -> si (estnul n) (un) (produit n (f (pred n))));;
```
On utilise la $\eta$-expansion : si $e$ termine, $e$ est équivalent (ie tout calcul donne le même terme) à $\lambda x. e(x)$.
```
let rec y = fun f -> f (fun x -> y(f)(x));;
```
Par contre, le typage n'arrive toujours pas à trouver que l'expression suivante devrait être bien définie :
```
let fact = y(fun f n -> si (estnul n) (un) (produit n (f (pred n))));;
```
## Conclusion
Je n'ai pas réussi à traduire intégralement la prouesse initiale, écrite en Python, par Matt Might.
Dommage, le typage de Caml est trop strict pour cet exercice.
|
github_jupyter
|
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Dimensionality-Reduction" data-toc-modified-id="Dimensionality-Reduction-1"><span class="toc-item-num">1 </span>Dimensionality Reduction</a></span><ul class="toc-item"><li><span><a href="#The-Problem" data-toc-modified-id="The-Problem-1.1"><span class="toc-item-num">1.1 </span>The Problem</a></span><ul class="toc-item"><li><span><a href="#Multi-Collinearity" data-toc-modified-id="Multi-Collinearity-1.1.1"><span class="toc-item-num">1.1.1 </span>Multi-Collinearity</a></span></li></ul></li><li><span><a href="#Sparsity" data-toc-modified-id="Sparsity-1.2"><span class="toc-item-num">1.2 </span>Sparsity</a></span></li></ul></li><li><span><a href="#Principle-Component-Analysis" data-toc-modified-id="Principle-Component-Analysis-2"><span class="toc-item-num">2 </span>Principle Component Analysis</a></span><ul class="toc-item"><li><span><a href="#Important-Points:" data-toc-modified-id="Important-Points:-2.1"><span class="toc-item-num">2.1 </span>Important Points:</a></span></li></ul></li><li><span><a href="#Singular-Value-Decomposition" data-toc-modified-id="Singular-Value-Decomposition-3"><span class="toc-item-num">3 </span>Singular Value Decomposition</a></span><ul class="toc-item"><li><span><a href="#Measuring-the-Quality-of-the-Reconstruction" data-toc-modified-id="Measuring-the-Quality-of-the-Reconstruction-3.1"><span class="toc-item-num">3.1 </span>Measuring the Quality of the Reconstruction</a></span></li><li><span><a href="#Heuristic-Step-for-How-Many-Dimensions-to-Keep" data-toc-modified-id="Heuristic-Step-for-How-Many-Dimensions-to-Keep-3.2"><span class="toc-item-num">3.2 </span>Heuristic Step for How Many Dimensions to Keep</a></span></li></ul></li><li><span><a href="#GLOVE" data-toc-modified-id="GLOVE-4"><span class="toc-item-num">4 </span>GLOVE</a></span><ul class="toc-item"><li><span><a href="#Using-Spacy-word2vec-embeddings" data-toc-modified-id="Using-Spacy-word2vec-embeddings-4.1"><span class="toc-item-num">4.1 </span>Using Spacy word2vec embeddings</a></span></li><li><span><a href="#Using-Glove" data-toc-modified-id="Using-Glove-4.2"><span class="toc-item-num">4.2 </span>Using Glove</a></span></li></ul></li><li><span><a href="#Clustering-Text" data-toc-modified-id="Clustering-Text-5"><span class="toc-item-num">5 </span>Clustering Text</a></span></li></ul></div>
# Dimensionality Reduction
## The Problem
There is an interesting tradeoff between model performance and a feature's dimensionality:

>*If the amount of available training data is fixed, then overfitting occurs if we keep adding dimensions. On the other hand, if we keep adding dimensions, the amount of **training data needs to grow exponentially fast to maintain the same coverage** and to avoid overfitting* ([Computer Vision for Dummies](http://www.visiondummy.com/2014/04/curse-dimensionality-affect-classification/)).

### Multi-Collinearity
In many cases, there is a high degree of correlation between many of the features in a dataset. This multi-collinearity has the effect of drowning out the "signal" of your dataset in many cases, and amplifies "outlier" noise.
## Sparsity
- High dimensionality increases the sparsity of your features (**what NLP techniques have we used that illustrate this point?**)
- The density of the training samples decreases when dimensionality increases:
- **Distance measures (Euclidean, for instance) start losing their effectiveness**, because there isn't much difference between the max and min distances in higher dimensions.
- Many models that rely upon **assumptions of Gaussian distributions** (like OLS linear regression), Gaussian mixture models, Gaussian processes, etc. become less and less effective since their distributions become flatter and "fatter tailed".

What is the amount of data needed to maintain **20% coverage** of the feature space? For 1 dimension, it is **20% of the entire population's dataset**. For a dimensionality of $D$:
$$
X^{D} = .20
$$
$$
(X^{D})^{\frac{1}{D}} = .20^{\frac{1}{D}}
$$
$$
X = \sqrt[D]{.20}
$$
You can approximate this as
```python
def coverage_requirement(requirement, D):
return requirement ** (1 / D)
x = []
y = []
for d in range(1,20):
y.append(coverage_requirement(0.10, d))
x.append(d)
import matplotlib.pyplot as plt
plt.plot(x,y)
plt.xlabel("Number of Dimensions")
plt.ylabel("Appromximate % of Population Dataset")
plt.title("% of Dataset Needed to Maintain 10% Coverage of Feature Space")
plt.show()
```
<img src="images/coverage-needed.png" width="500">
```
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer
reviews = pd.read_csv("mcdonalds-yelp-negative-reviews.csv", encoding='latin-1')
reviews = open("poor_amazon_toy_reviews.txt", encoding='latin-1')
#text = reviews["review"].values
text = reviews.readlines()
vectorizer = CountVectorizer(ngram_range=(3,3), min_df=0.01, max_df=0.75, max_features=200)
# tokenize and build vocab
vectorizer.fit(text)
vector = vectorizer.transform(text)
features = vector.toarray()
features_df = pd.DataFrame(features, columns=vectorizer.get_feature_names())
correlations = features_df.corr()
correlations_stacked = correlations.stack().reset_index()
#set column names
correlations_stacked.columns = ['Tri-Gram 1','Tri-Gram 2','Correlation']
correlations_stacked = correlations_stacked[correlations_stacked["Correlation"] < 1]
correlations_stacked = correlations_stacked.sort_values(by=['Correlation'], ascending=False)
correlations_stacked.head()
import numpy as np
import matplotlib.pyplot as plt
# visualize the correlations (install seaborn first)!
import seaborn as sns
# Generate a mask for the upper triangle
mask = np.triu(np.ones_like(correlations, dtype=np.bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
cmap = sns.diverging_palette(220, 10, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(correlations, mask=mask, cmap=cmap, vmax=.3, center=0,
square=True, linewidths=.5, cbar_kws={"shrink": .5})
```
# Principle Component Analysis
If you have an original matrix $Z$, you can decompose this matrix into two smaller matrices $X$ and $Q$.
## Important Points:
- Multiplying a vector by a matrix typically changes the direction of the vector. For instance:
<figure>
<img src="images/multvector.png" alt="my alt text"/>
<figcaption><a href="https://lazyprogrammer.me/tutorial-principal-components-analysis-pca">Lazy Programmer-
Tutorial to PCA</a></figcaption>
</figure>
However, there are eigenvalues λ and eigenvectors $v$ such that
$$
\sum_{X}v = \lambda v
$$
Multiplying the eigenvectors $v$ with the eigenvalue $\lambda$ does not change the direction of the eigenvector.
Multiplying the eigenvector $v$ by the covariance matrix $\sum_{X}$ also does not change the direction of the eigenvector.
If our data $X$ is of shape $N \times D$, it turns out that we have $D$ eigenvalues and $D$ eigenvectors. This means we can arrange the eigenvalues $\lambda$ in decreasing order so that
$$
\lambda_3 > \lambda_2 > \lambda_5
$$
In this case, $\lambda_3$ is the largest eigenvalue, followed by $\lambda_2$, and then $\lambda_5$. Then, we can arrange
We can also rearrange the eigenvectors the same: $v_3$ will be the first column, $v_2$ will be the second column, and $v_5$ will be the third column.
We'll end up with two matrices $V$ and $\Lambda$:
<figure>
<img src="images/pca1.png" alt="my alt text"/>
<figcaption><a href="https://lazyprogrammer.me/tutorial-principal-components-analysis-pca">Lazy Programmer-
Tutorial to PCA</a></figcaption>
</figure>
```
# what is the shape of our features?
features.shape
from sklearn.decomposition import PCA
pca = PCA(n_components=4)
Z = pca.fit_transform(features)
# what is the shape of Z?
Z.shape
# what will happen if we take the correlation matrix and covariance matrix of our new reduced features?
import numpy as np
covariances = pd.DataFrame(np.cov(Z.transpose()))
plt.rcParams["figure.figsize"] = (5,5)
sns.heatmap(covariances)
# train the model to reduce the dimensions down to 2
pca = PCA(n_components=2)
Z_two_dimensions = pca.fit_transform(features)
Z_two_dimensions
import matplotlib.pyplot as plt
plt.scatter(Z_two_dimensions[:,0], Z_two_dimensions[:, 1])
reduced_features_df = pd.DataFrame(Z_two_dimensions, columns=["x1", "x2"])
reduced_features_df["text"] = text
```
# Singular Value Decomposition
Given an input matrix $A$, we want to try to represent it instead as three smaller matrices $U$, $\sum$, and $V$. Instead of **$n$ original terms**, we want to represent each document as **$r$ concepts** (other referred to as **latent dimensions**, or **latent factors**):
<figure>
<img src="images/svd.png" alt="my alt text"/>
<figcaption><i>
<a href="https://www.youtube.com/watch?v=P5mlg91as1c">Mining of Massive Datasets - Dimensionality Reduction: Singular Value Decomposition</a> by Leskovec, Rajaraman, and Ullman (Stanford University)</i></figcaption>
</figure>
Here, **$A$ is your matrix of word vectors** - you could use any of the word vectorization techniques we have learned so far, include one-hot encoding, word count, TF-IDF.
- $\sum$ will be a **diagonal matrix** with values that are positive and sorted in decreasing order. Its value indicate the **variance (information encoded on that new dimension)**- therefore, the higher the value, the stronger that dimension is in capturing data from A, the original features. For our purposes, we can think of the rank of this $\sum$ matrix as the number of desired dimensions. Instance, if we want to reduce $A$ from shape $1020 x 300$ to $1020 x 10$, we will want to reduce the rank of $\sum$ from 300 to 10.
- $U^T U = I$ and $V^T V = I$
## Measuring the Quality of the Reconstruction
A popular metric used for measuring the quality of the reconstruction is the [Frobenius Norm](https://en.wikipedia.org/wiki/Matrix_norm#Frobenius_norm). When you explain your methodology for reducing dimensions, usually managers / stakeholders will want to some way to compare multiple dimensionality techniques' ability to quantify its ability to retain information but trim dimensions:
$$
\begin{equation}
||A_{old}-A_{new}||_{F} = \sqrt{\sum_{ij}{(A^{old}_{ij}- A^{new}_{ij}}})^2
\end{equation}
$$
## Heuristic Step for How Many Dimensions to Keep
1. Sum the $\sum$ matrix's diagonal values:
$$
\begin{equation}
\sum_{i}^{m}\sigma_{i}
\end{equation}
$$
2. Define your threshold of "information" (variance) $\alpha$ to keep: usually 80% to 90%.
3. Define your cutoff point $C$: $$
\begin{equation}
C = \sum_{i}^{m}\sigma_{i} \alpha
\end{equation}
$$
4. Beginning with your largest singular value, sum your singular values $\sigma_{i}$ until it is greater than C. Retain only those dimensions.
<figure>
<img src="images/userratings.png" alt="my alt text"/>
<figcaption><i>
<a href="https://www.youtube.com/watch?v=P5mlg91as1c">Mining of Massive Datasets - Dimensionality Reduction: Singular Value Decomposition</a> by Leskovec, Rajaraman, and Ullman (Stanford University)</i></figcaption>
</figure>
```
# create sample data
import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import svd
x = np.linspace(1,20, 20) # create the first dimension
x = np.concatenate((x,x))
y = x + np.random.normal(0,1, 40) # create the second dimension
z = x + np.random.normal(0,2, 40) # create the third dimension
a = x + np.random.normal(0,4, 40) # create the fourth dimension
plt.scatter(x,y) # plot just the first two dimensions
plt.show()
# create matrix
A = np.stack([x,y,z,a]).T
# perform SVD
D = 1
U, s, V = svd(A)
print(f"s is {s}\n")
print(f"U is {U}\n")
print(f"V is {V}")
# Frobenius norm
s[D:] = 0
S = np.zeros((A.shape[0], A.shape[1]))
S[:A.shape[1], :A.shape[1]] = np.diag(s)
A_reconstructed = U.dot(S.dot(V))
np.sum((A_reconstructed - A) ** 2) ** (1/2) # Frobenius norm
# reconstruct matrix
U.dot(S)
```
# GLOVE
Global vectors for word presentation:
<figure>
<img src="images/glove_1.png" alt="my alt text"/>
<figcaption><i>
<a href="https://nlp.stanford.edu/pubs/glove.pdf">GloVe: Global Vectors for Word Representation</a></i></figcaption>
</figure>
```
!pip3 install gensim
# import glove embeddings into a word2vec format that is consumable by Gensim
from gensim.scripts.glove2word2vec import glove2word2vec
glove_input_file = 'glove.6B.100d.txt'
word2vec_output_file = 'glove.6B.100d.txt.word2vec'
glove2word2vec(glove_input_file, word2vec_output_file)
from gensim.models import KeyedVectors
# load the Stanford GloVe model
filename = 'glove.6B.100d.txt.word2vec'
model = KeyedVectors.load_word2vec_format(filename, binary=False)
# calculate: (king - man) + woman = ?
result = model.most_similar(positive=['woman', 'king'], negative=['man'], topn=1)
print(result)
words = ["woman", "king", "man", "queen", "puppy", "kitten", "cat",
"quarterback", "football", "stadium", "touchdown",
"dog", "government", "tax", "federal", "judicial", "elections",
"avocado", "tomato", "pear", "championship", "playoffs"]
vectors = [model.wv[word] for word in words]
import pandas as pd
vector_df = pd.DataFrame(vectors)
vector_df["word"] = words
vector_df.head()
```
## Using Spacy word2vec embeddings
```
import en_core_web_md
import spacy
from scipy.spatial.distance import cosine
nlp = en_core_web_md.load()
words = ["woman", "king", "man", "queen", "puppy", "kitten", "cat",
"quarterback", "football", "stadium", "touchdown",
"dog", "government", "tax", "federal", "judicial", "elections",
"avocado", "tomato", "pear", "championship", "playoffs"]
tokens = nlp(" ".join(words))
word2vec_vectors = [token.vector for token in tokens]
np.array(word2vec_vectors).shape
%matplotlib inline
from sklearn.manifold import TSNE
from sklearn.decomposition import PCA
from sklearn.decomposition import TruncatedSVD
import matplotlib.pyplot as plt
import matplotlib
dimension_model = PCA(n_components=2)
reduced_vectors = dimension_model.fit_transform(word2vec_vectors)
reduced_vectors.shape
matplotlib.rc('figure', figsize=(10, 10))
for i, vector in enumerate(reduced_vectors):
x = vector[0]
y = vector[1]
plt.plot(x,y, 'bo')
plt.text(x * (1 + 0.01), y * (1 + 0.01) , words[i], fontsize=12)
```
## Using Glove
```
%matplotlib inline
from sklearn.manifold import TSNE
from sklearn.decomposition import PCA
from sklearn.decomposition import TruncatedSVD
import matplotlib.pyplot as plt
dimension_model = PCA(n_components=2)
reduced_vectors = dimension_model.fit_transform(vectors)
for i, vector in enumerate(reduced_vectors):
x = vector[0]
y = vector[1]
plt.plot(x,y, 'bo')
plt.text(x * (1 + 0.01), y * (1 + 0.01) , words[i], fontsize=12)
```
# Clustering Text
```
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=4)
cluster_assignments = kmeans.fit_predict(reduced_vectors)
for cluster_assignment, word in zip(cluster_assignments, words):
print(f"{word} assigned to cluster {cluster_assignment}")
color_map = {
0: "r",
1: "b",
2: "g",
3: "y"
}
plt.rcParams["figure.figsize"] = (10,10)
for i, vector in enumerate(reduced_vectors):
x = vector[0]
y = vector[1]
plt.plot(x,y, 'bo', c=color_map[cluster_assignments[i]])
plt.text(x * (1 + 0.01), y * (1 + 0.01) , words[i], fontsize=12)
```
|
github_jupyter
|
## 용어 정의
```
#가설설정
# A hypothesis test is a statistical method that uses sample data to evaluate a hypothesis about a population.
1. First, we state a hypothesis about a population. Usually the hypothesis concerns the value of a population parameter.
2. Before we select a sample, we use the hypothesis to predict the characteristics that the sample should have.
3. Next, we obtain a random sample from the population.
4. Finally, we compare the obtained sample data with the prediction that was made from the hypothesis.
## 가설설정 프로세스
1. State the hypothesis. null hypothesis(H0)
귀무가설 : 독립변수가 종속변수에 어떤 영향을 미치지 않는다는 것 => 레스토랑의 웨이터가 레드 셔츠 입는 것이 팁에 영향이 없다.
The null hypothesis (H0) states that in the general population
there is no change, no difference, or no relationship.
In the context of an experiment,
H0 predicts that the independent variable (treatment)
has no effect on the dependent variable (scores) for the population.
m = 15.8
대안가설 : 어떤 변인이 종속 변수에 효과가 있다는 것 => 레스토랑의 웨이터가 레드 셔츠 입는 것 팁에 영향이 있다.
The alternative hypothesis (H1) states that there is a change, a difference,
or a relationship for the general population.
In the context of an experiment,
H1 predicts that the independent variable (treatment) does have an effect on the dependent variable.
m != 15.8 이다.
이 실험에서는
m > 15.8
directional hypothisis test
2. set the criteria for a decision
a. Sample means that are likely to be obtained if H0 is true;
that is, sample means that are close to the null hypothesis
b. Sample means that are very unlikely to be obtained if H0 is true;
that is, sample means that are very different from the null hypothesis
The Alpha Level
alpha levels are α = .05 (5%), α = .01 (1%), and α = .001 (0.1%).
The alpha level, or the level of significance,
is a probability value that is used to define the concept of
“very unlikely” in a hypothesis test.
The critical region is composed of the extreme sample values that are very unlikely (as defined by the alpha level) to be obtained if the null hypothesis is true. The boundaries for the critical region are determined by the alpha level.
If sample data fall in the critical region, the null hypothesis is rejected.
3. Collect data and compute sample statistics.
z = sample mean - hypothesized population mean / standard error between M and m
4. Make a decision
1. Thesampledataarelocatedinthecriticalregion.
Bydefinition,asamplevaluein the critical region is very unlikely to occur if the null hypothesis is true.
2. The sample data are not in the critical region.
In this case, the sample mean is reasonably close to the population mean specified in the null hypothesis (in the center of the distribution).
```
# Problems
```
1. Identify the four steps of a hypothesis test as presented in this chapter.
1)State the hypothesis.
귀무가설과 대안가설 언급
2)alpha level 설정, 신뢰 구간 설정
3) Collect data and compute sample statistics.
데이터 수집과 샘플 통계적 계산
4)make decision
결론 결정
2. Define the alpha level and the critical region for a hypothesis test.
독립변수와 종속변수에 대한 귀무가설을 reject하기 위해 그 통계치를 통상적인 수치를 벗어나 의미있는 수가 나온 것을 설정해준다.
3. Define a Type I error and a Type II error and explain the consequences of each.
가설검증에서 실제효과가 없는데 효과가 있는 것으로 나온것, 실제 효과가 있는데, 없는 것으로 나온것. 가설 설정에 문제
4. If the alpha level is changed from α = .05 to α = .01,
a. What happens to the boundaries for the critical
region?
신뢰구간이 줄어든다.
b. What happens to the probability of a Type I error?
에러 확률은 낮아진다.
6. Although there is a popular belief that herbal remedies such as Ginkgo biloba and Ginseng may improve learning and memory in healthy adults, these effects are usually not supported by well- controlled research (Persson, Bringlov, Nilsson, and Nyberg, 2004). In a typical study, a researcher
obtains a sample of n = 16 participants and has each person take the herbal supplements every day for
90 days. At the end of the 90 days, each person takes a standardized memory test. For the general popula- tion, scores from the test form a normal distribution with a mean of μ = 50 and a standard deviation of
σ = 12. The sample of research participants had an average of M = 54.
a. Assuming a two-tailed test, state the null hypoth-
esis in a sentence that includes the two variables
being examined.
b. Using the standard 4-step procedure, conduct a
two-tailed hypothesis test with α = .05 to evaluate the effect of the supplements.
from scipy import stats
sample_number = 16 # 샘플수
population_mean = 50 # 모집단의 평균
standard_deviation = 12 # 표준편차
sample_mean = 54 # 샘플의 평균
result = stats.ttest_1samp(sample_mean, 50) # 비교집단, 관측치
result
sample_mean - population_mean
## Import
import numpy as np
from scipy import stats
sample_number = 16 # 샘플수
population_mean = 50 # 모집단의 평균
standard_deviation = 12 # 표준편차
sample_mean = 54 # 샘플의 평균
## 신뢰구간을 벗어나는지 아닌지 확인 함수
alpha_level05 = 1.96
alpha_level01 = 2.58
def h_test(sample_mean, population_mean, standard_deviation, sample_number, alpha_level):
result = (sample_mean - population_mean)/ (standard_deviation/np.sqrt(sample_number))
if result> alpha_level or result< - alpha_level:
print("a = .05 신뢰구간에서 귀무가설 reject되고, 가설이 ok")
else:
print("귀무가설이 reject 되지 않아 가설이 기각됩니다.")
return result
##Compute Cohen’s d
def Cohen(sample_mean, population_mean, standard_deviation):
result = (sample_mean - population_mean) / (standard_deviation)
if result<=0.2:
print("small effect")
elif result<= 0.5:
print("medium effect")
elif result<= 0.8:
print("Large effect")
return result
## 신뢰구간을 벗어나는지 아닌지 확인 함수
h_test(sample_mean, population_mean, standard_deviation, sample_number, alpha_level05)
Cohen(sample_mean, population_mean, standard_deviation)
함수를 활용해서, 신뢰구간과 cohen's d를 구할 수 있다.
# ## Import the packages
# import numpy as np
# from scipy import stats
# ## 함수로 만들기
# #Sample Size
# sample_number = 16
# population_mean = 50 # 모집단의 평균
# standard_deviation = 12 # 표준편차
# sample_mean = [54,54,58,53,52] # 샘플의 평균
# def h_test(sample_mean, population_mean, standard_deviation, sample_number):
# #For unbiased max likelihood estimate we have to divide the var by N-1, and therefore the parameter ddof = 1
# var_sample_mean = sample_mean.var(ddof=1)
# var_population_mean = population_mean.var(ddof=1)
# #std deviation
# std_deviation = np.sqrt((var_sample_mean + var_population_mean)/2)
# ## Calculate the t-statistics
# t = (a.mean() - b.mean())/(s*np.sqrt(2/N))
# ## Define 2 random distributions
# N = 10
# #Gaussian distributed data with mean = 2 and var = 1
# a = np.random.randn(N) + 2
# #Gaussian distributed data with with mean = 0 and var = 1
# b = np.random.randn(N)
# ## Calculate the Standard Deviation
# #Calculate the variance to get the standard deviation
# #For unbiased max likelihood estimate we have to divide the var by N-1, and therefore the parameter ddof = 1
# var_a = a.var(ddof=1)
# var_b = b.var(ddof=1)
# #std deviation
# s = np.sqrt((var_a + var_b)/2)
# s
# ## Calculate the t-statistics
# t = (a.mean() - b.mean())/(s*np.sqrt(2/N))
# ## Compare with the critical t-value
# #Degrees of freedom
# df = 2*N - 2
# #p-value after comparison with the t
# p = 1 - stats.t.cdf(t,df=df)
# print("t = " + str(t))
# print("p = " + str(2*p))
# ### You can see that after comparing the t statistic with the critical t value (computed internally) we get a good p value of 0.0005 and thus we reject the null hypothesis and thus it proves that the mean of the two distributions are different and statistically significant.
# ## Cross Checking with the internal scipy function
# t2, p2 = stats.ttest_ind(a,b)
# print("t = " + str(t2))
# print("p = " + str(p2))
```
|
github_jupyter
|
# 自然语言处理实战——命名实体识别
### 进入ModelArts
点击如下链接:https://www.huaweicloud.com/product/modelarts.html , 进入ModelArts主页。点击“立即使用”按钮,输入用户名和密码登录,进入ModelArts使用页面。
### 创建ModelArts notebook
下面,我们在ModelArts中创建一个notebook开发环境,ModelArts notebook提供网页版的Python开发环境,可以方便的编写、运行代码,并查看运行结果。
第一步:在ModelArts服务主界面依次点击“开发环境”、“创建”

第二步:填写notebook所需的参数:
| 参数 | 说明 |
| - - - - - | - - - - - |
| 计费方式 | 按需计费 |
| 名称 | Notebook实例名称 |
| 工作环境 | Python3 |
| 资源池 | 选择"公共资源池"即可 |
| 类型 | 选择"GPU" |
| 规格 | 选择"[限时免费]体验规格GPU版"|
| 存储配置 | 选择EVS,磁盘规格5GB |
第三步:配置好notebook参数后,点击下一步,进入notebook信息预览。确认无误后,点击“立即创建”
第四步:创建完成后,返回开发环境主界面,等待Notebook创建完毕后,打开Notebook,进行下一步操作。

### 在ModelArts中创建开发环境
接下来,我们创建一个实际的开发环境,用于后续的实验步骤。
第一步:点击下图所示的“打开”按钮,进入刚刚创建的Notebook

第二步:创建一个Python3环境的的Notebook。点击右上角的"New",然后创建TensorFlow 1.13.1开发环境。
第三步:点击左上方的文件名"Untitled",并输入一个与本实验相关的名称


### 在Notebook中编写并执行代码
在Notebook中,我们输入一个简单的打印语句,然后点击上方的运行按钮,可以查看语句执行的结果:

开发环境准备好啦,接下来可以愉快地写代码啦!
### 准备源代码和数据
准备案例所需的源代码和数据,相关资源已经保存在 OBS 中,我们通过 ModelArts SDK 将资源下载到本地。
```
from modelarts.session import Session
session = Session()
if session.region_name == 'cn-north-1':
bucket_path = 'modelarts-labs/notebook/DL_nlp_ner/ner.tar.gz'
elif session.region_name == 'cn-north-4':
bucket_path = 'modelarts-labs-bj4/notebook/DL_nlp_ner/ner.tar.gz'
else:
print("请更换地区到北京一或北京四")
session.download_data(bucket_path=bucket_path, path='./ner.tar.gz')
!ls -la
```
解压从obs下载的压缩包,解压后删除压缩包。
```
# 解压
!tar xf ./ner.tar.gz
# 删除
!rm ./ner.tar.gz
!ls -la
```
#### 导入Python库
```
import os
import json
import numpy as np
import tensorflow as tf
import codecs
import pickle
import collections
from ner.bert import modeling, optimization, tokenization
```
#### 定义路径及参数
```
data_dir = "./ner/data"
output_dir = "./ner/output"
vocab_file = "./ner/chinese_L-12_H-768_A-12/vocab.txt"
data_config_path = "./ner/chinese_L-12_H-768_A-12/bert_config.json"
init_checkpoint = "./ner/chinese_L-12_H-768_A-12/bert_model.ckpt"
max_seq_length = 128
batch_size = 64
num_train_epochs = 5.0
```
#### 定义processor类获取数据,打印标签
```
tf.logging.set_verbosity(tf.logging.INFO)
from ner.src.models import InputFeatures, InputExample, DataProcessor, NerProcessor
processors = {"ner": NerProcessor }
processor = processors["ner"](output_dir)
label_list = processor.get_labels()
print("labels:", label_list)
```
以上 labels 分别表示:
- O:非标注实体
- B-PER:人名首字
- I-PER:人名非首字
- B-ORG:组织首字
- I-ORG:组织名非首字
- B-LOC:地名首字
- I-LOC:地名非首字
- X:未知
- [CLS]:句首
- [SEP]:句尾
#### 加载预训练参数
```
data_config = json.load(codecs.open(data_config_path))
train_examples = processor.get_train_examples(data_dir)
num_train_steps = int(len(train_examples) / batch_size * num_train_epochs)
num_warmup_steps = int(num_train_steps * 0.1)
data_config['num_train_steps'] = num_train_steps
data_config['num_warmup_steps'] = num_warmup_steps
data_config['num_train_size'] = len(train_examples)
print("显示配置信息:")
for key,value in data_config.items():
print('{key}:{value}'.format(key = key, value = value))
bert_config = modeling.BertConfig.from_json_file(data_config_path)
tokenizer = tokenization.FullTokenizer(vocab_file=vocab_file, do_lower_case=True)
#tf.estimator运行参数
run_config = tf.estimator.RunConfig(
model_dir=output_dir,
save_summary_steps=1000,
save_checkpoints_steps=1000,
session_config=tf.ConfigProto(
log_device_placement=False,
inter_op_parallelism_threads=0,
intra_op_parallelism_threads=0,
allow_soft_placement=True
)
)
```
#### 读取数据,获取句向量
```
def convert_single_example(ex_index, example, label_list, max_seq_length,
tokenizer, output_dir, mode):
label_map = {}
for (i, label) in enumerate(label_list, 1):
label_map[label] = i
if not os.path.exists(os.path.join(output_dir, 'label2id.pkl')):
with codecs.open(os.path.join(output_dir, 'label2id.pkl'), 'wb') as w:
pickle.dump(label_map, w)
textlist = example.text.split(' ')
labellist = example.label.split(' ')
tokens = []
labels = []
for i, word in enumerate(textlist):
token = tokenizer.tokenize(word)
tokens.extend(token)
label_1 = labellist[i]
for m in range(len(token)):
if m == 0:
labels.append(label_1)
else:
labels.append("X")
if len(tokens) >= max_seq_length - 1:
tokens = tokens[0:(max_seq_length - 2)]
labels = labels[0:(max_seq_length - 2)]
ntokens = []
segment_ids = []
label_ids = []
ntokens.append("[CLS]") # 句子开始设置 [CLS] 标志
segment_ids.append(0)
label_ids.append(label_map["[CLS]"])
for i, token in enumerate(tokens):
ntokens.append(token)
segment_ids.append(0)
label_ids.append(label_map[labels[i]])
ntokens.append("[SEP]") # 句尾添加 [SEP] 标志
segment_ids.append(0)
label_ids.append(label_map["[SEP]"])
input_ids = tokenizer.convert_tokens_to_ids(ntokens)
input_mask = [1] * len(input_ids)
while len(input_ids) < max_seq_length:
input_ids.append(0)
input_mask.append(0)
segment_ids.append(0)
label_ids.append(0)
ntokens.append("**NULL**")
assert len(input_ids) == max_seq_length
assert len(input_mask) == max_seq_length
assert len(segment_ids) == max_seq_length
assert len(label_ids) == max_seq_length
feature = InputFeatures(
input_ids=input_ids,
input_mask=input_mask,
segment_ids=segment_ids,
label_ids=label_ids,
)
return feature
def filed_based_convert_examples_to_features(
examples, label_list, max_seq_length, tokenizer, output_file, mode=None):
writer = tf.python_io.TFRecordWriter(output_file)
for (ex_index, example) in enumerate(examples):
if ex_index % 5000 == 0:
tf.logging.info("Writing example %d of %d" % (ex_index, len(examples)))
feature = convert_single_example(ex_index, example, label_list, max_seq_length, tokenizer, output_dir, mode)
def create_int_feature(values):
f = tf.train.Feature(int64_list=tf.train.Int64List(value=list(values)))
return f
features = collections.OrderedDict()
features["input_ids"] = create_int_feature(feature.input_ids)
features["input_mask"] = create_int_feature(feature.input_mask)
features["segment_ids"] = create_int_feature(feature.segment_ids)
features["label_ids"] = create_int_feature(feature.label_ids)
tf_example = tf.train.Example(features=tf.train.Features(feature=features))
writer.write(tf_example.SerializeToString())
train_file = os.path.join(output_dir, "train.tf_record")
#将训练集中字符转化为features作为训练的输入
filed_based_convert_examples_to_features(
train_examples, label_list, max_seq_length, tokenizer, output_file=train_file)
```
#### 引入 BiLSTM+CRF 层,作为下游模型
```
learning_rate = 5e-5
dropout_rate = 1.0
lstm_size=1
cell='lstm'
num_layers=1
from ner.src.models import BLSTM_CRF
from tensorflow.contrib.layers.python.layers import initializers
def create_model(bert_config, is_training, input_ids, input_mask,
segment_ids, labels, num_labels, use_one_hot_embeddings,
dropout_rate=dropout_rate, lstm_size=1, cell='lstm', num_layers=1):
model = modeling.BertModel(
config=bert_config,
is_training=is_training,
input_ids=input_ids,
input_mask=input_mask,
token_type_ids=segment_ids,
use_one_hot_embeddings=use_one_hot_embeddings
)
embedding = model.get_sequence_output()
max_seq_length = embedding.shape[1].value
used = tf.sign(tf.abs(input_ids))
lengths = tf.reduce_sum(used, reduction_indices=1)
blstm_crf = BLSTM_CRF(embedded_chars=embedding, hidden_unit=1, cell_type='lstm', num_layers=1,
dropout_rate=dropout_rate, initializers=initializers, num_labels=num_labels,
seq_length=max_seq_length, labels=labels, lengths=lengths, is_training=is_training)
rst = blstm_crf.add_blstm_crf_layer(crf_only=True)
return rst
def model_fn_builder(bert_config, num_labels, init_checkpoint, learning_rate,
num_train_steps, num_warmup_steps,use_one_hot_embeddings=False):
#构建模型
def model_fn(features, labels, mode, params):
tf.logging.info("*** Features ***")
for name in sorted(features.keys()):
tf.logging.info(" name = %s, shape = %s" % (name, features[name].shape))
input_ids = features["input_ids"]
input_mask = features["input_mask"]
segment_ids = features["segment_ids"]
label_ids = features["label_ids"]
print('shape of input_ids', input_ids.shape)
is_training = (mode == tf.estimator.ModeKeys.TRAIN)
total_loss, logits, trans, pred_ids = create_model(
bert_config, is_training, input_ids, input_mask, segment_ids, label_ids,
num_labels, False, dropout_rate, lstm_size, cell, num_layers)
tvars = tf.trainable_variables()
if init_checkpoint:
(assignment_map, initialized_variable_names) = \
modeling.get_assignment_map_from_checkpoint(tvars, init_checkpoint)
tf.train.init_from_checkpoint(init_checkpoint, assignment_map)
output_spec = None
if mode == tf.estimator.ModeKeys.TRAIN:
train_op = optimization.create_optimizer(
total_loss, learning_rate, num_train_steps, num_warmup_steps, False)
hook_dict = {}
hook_dict['loss'] = total_loss
hook_dict['global_steps'] = tf.train.get_or_create_global_step()
logging_hook = tf.train.LoggingTensorHook(
hook_dict, every_n_iter=100)
output_spec = tf.estimator.EstimatorSpec(
mode=mode,
loss=total_loss,
train_op=train_op,
training_hooks=[logging_hook])
elif mode == tf.estimator.ModeKeys.EVAL:
def metric_fn(label_ids, pred_ids):
return {
"eval_loss": tf.metrics.mean_squared_error(labels=label_ids, predictions=pred_ids), }
eval_metrics = metric_fn(label_ids, pred_ids)
output_spec = tf.estimator.EstimatorSpec(
mode=mode,
loss=total_loss,
eval_metric_ops=eval_metrics
)
else:
output_spec = tf.estimator.EstimatorSpec(
mode=mode,
predictions=pred_ids
)
return output_spec
return model_fn
```
#### 创建模型,开始训练
```
model_fn = model_fn_builder(
bert_config=bert_config,
num_labels=len(label_list) + 1,
init_checkpoint=init_checkpoint,
learning_rate=learning_rate,
num_train_steps=num_train_steps,
num_warmup_steps=num_warmup_steps,
use_one_hot_embeddings=False)
def file_based_input_fn_builder(input_file, seq_length, is_training, drop_remainder):
name_to_features = {
"input_ids": tf.FixedLenFeature([seq_length], tf.int64),
"input_mask": tf.FixedLenFeature([seq_length], tf.int64),
"segment_ids": tf.FixedLenFeature([seq_length], tf.int64),
"label_ids": tf.FixedLenFeature([seq_length], tf.int64),
}
def _decode_record(record, name_to_features):
example = tf.parse_single_example(record, name_to_features)
for name in list(example.keys()):
t = example[name]
if t.dtype == tf.int64:
t = tf.to_int32(t)
example[name] = t
return example
def input_fn(params):
params["batch_size"] = 32
batch_size = params["batch_size"]
d = tf.data.TFRecordDataset(input_file)
if is_training:
d = d.repeat()
d = d.shuffle(buffer_size=300)
d = d.apply(tf.contrib.data.map_and_batch(
lambda record: _decode_record(record, name_to_features),
batch_size=batch_size,
drop_remainder=drop_remainder
))
return d
return input_fn
#训练输入
train_input_fn = file_based_input_fn_builder(
input_file=train_file,
seq_length=max_seq_length,
is_training=True,
drop_remainder=True)
num_train_size = len(train_examples)
tf.logging.info("***** Running training *****")
tf.logging.info(" Num examples = %d", num_train_size)
tf.logging.info(" Batch size = %d", batch_size)
tf.logging.info(" Num steps = %d", num_train_steps)
#模型预测estimator
estimator = tf.estimator.Estimator(
model_fn=model_fn,
config=run_config,
params={
'batch_size':batch_size
})
estimator.train(input_fn=train_input_fn, max_steps=num_train_steps)
```
#### 在验证集上验证模型
```
eval_examples = processor.get_dev_examples(data_dir)
eval_file = os.path.join(output_dir, "eval.tf_record")
filed_based_convert_examples_to_features(
eval_examples, label_list, max_seq_length, tokenizer, eval_file)
data_config['eval.tf_record_path'] = eval_file
data_config['num_eval_size'] = len(eval_examples)
num_eval_size = data_config.get('num_eval_size', 0)
tf.logging.info("***** Running evaluation *****")
tf.logging.info(" Num examples = %d", num_eval_size)
tf.logging.info(" Batch size = %d", batch_size)
eval_steps = None
eval_drop_remainder = False
eval_input_fn = file_based_input_fn_builder(
input_file=eval_file,
seq_length=max_seq_length,
is_training=False,
drop_remainder=eval_drop_remainder)
result = estimator.evaluate(input_fn=eval_input_fn, steps=eval_steps)
output_eval_file = os.path.join(output_dir, "eval_results.txt")
with codecs.open(output_eval_file, "w", encoding='utf-8') as writer:
tf.logging.info("***** Eval results *****")
for key in sorted(result.keys()):
tf.logging.info(" %s = %s", key, str(result[key]))
writer.write("%s = %s\n" % (key, str(result[key])))
if not os.path.exists(data_config_path):
with codecs.open(data_config_path, 'a', encoding='utf-8') as fd:
json.dump(data_config, fd)
```
#### 在测试集上进行测试
```
token_path = os.path.join(output_dir, "token_test.txt")
if os.path.exists(token_path):
os.remove(token_path)
with codecs.open(os.path.join(output_dir, 'label2id.pkl'), 'rb') as rf:
label2id = pickle.load(rf)
id2label = {value: key for key, value in label2id.items()}
predict_examples = processor.get_test_examples(data_dir)
predict_file = os.path.join(output_dir, "predict.tf_record")
filed_based_convert_examples_to_features(predict_examples, label_list,
max_seq_length, tokenizer,
predict_file, mode="test")
tf.logging.info("***** Running prediction*****")
tf.logging.info(" Num examples = %d", len(predict_examples))
tf.logging.info(" Batch size = %d", batch_size)
predict_drop_remainder = False
predict_input_fn = file_based_input_fn_builder(
input_file=predict_file,
seq_length=max_seq_length,
is_training=False,
drop_remainder=predict_drop_remainder)
predicted_result = estimator.evaluate(input_fn=predict_input_fn)
output_eval_file = os.path.join(output_dir, "predicted_results.txt")
with codecs.open(output_eval_file, "w", encoding='utf-8') as writer:
tf.logging.info("***** Predict results *****")
for key in sorted(predicted_result.keys()):
tf.logging.info(" %s = %s", key, str(predicted_result[key]))
writer.write("%s = %s\n" % (key, str(predicted_result[key])))
result = estimator.predict(input_fn=predict_input_fn)
output_predict_file = os.path.join(output_dir, "label_test.txt")
def result_to_pair(writer):
for predict_line, prediction in zip(predict_examples, result):
idx = 0
line = ''
line_token = str(predict_line.text).split(' ')
label_token = str(predict_line.label).split(' ')
if len(line_token) != len(label_token):
tf.logging.info(predict_line.text)
tf.logging.info(predict_line.label)
for id in prediction:
if id == 0:
continue
curr_labels = id2label[id]
if curr_labels in ['[CLS]', '[SEP]']:
continue
try:
line += line_token[idx] + ' ' + label_token[idx] + ' ' + curr_labels + '\n'
except Exception as e:
tf.logging.info(e)
tf.logging.info(predict_line.text)
tf.logging.info(predict_line.label)
line = ''
break
idx += 1
writer.write(line + '\n')
from ner.src.conlleval import return_report
with codecs.open(output_predict_file, 'w', encoding='utf-8') as writer:
result_to_pair(writer)
eval_result = return_report(output_predict_file)
for line in eval_result:
print(line)
```
### 在线命名实体识别
由以上训练得到模型进行在线测试,可以任意输入句子,进行命名实体识别。
输入“再见”,结束在线命名实体识别。
<span style="color:red">若下述程序未执行成功,则表示训练完成后,GPU显存还在占用,需要restart kernel,然后执行 %run 命令。</span>
释放资源具体流程为:菜单 > Kernel > Restart

```
%run ner/src/terminal_predict.py
```
|
github_jupyter
|
```
import matplotlib.pyplot as plt
import numpy as np
from mvmm.single_view.gaussian_mixture import GaussianMixture
from mvmm.single_view.MMGridSearch import MMGridSearch
from mvmm.single_view.toy_data import sample_1d_gmm
from mvmm.single_view.sim_1d_utils import plot_est_params
from mvmm.viz_utils import plot_scatter_1d, set_xaxis_int_ticks
from mvmm.single_view.opt_diagnostics import plot_opt_hist
```
# sample data from a 1d gussian mixture model
```
n_samples = 200
n_components = 3
X, y, true_params = sample_1d_gmm(n_samples=n_samples,
n_components=n_components,
random_state=1)
plot_scatter_1d(X)
```
# Fit a Gaussian mixture model
```
# fit a guassian mixture model with 3 (the true number) of components
# from mvmm.single_view.gaussian_mixture.GaussianMixture() is similar to sklearn.mixture.GaussianMixture()
gmm = GaussianMixture(n_components=3,
n_init=10) # 10 random initalizations
gmm.fit(X)
# plot parameter estimates
plot_scatter_1d(X)
plot_est_params(gmm)
# the GMM class has all the familiar sklearn functionality
gmm.sample(n_samples=20)
gmm.predict(X)
gmm.score_samples(X)
gmm.predict_proba(X)
gmm.bic(X)
# with a few added API features for convenience
# sample from a single mixture component
gmm.sample_from_comp(y=0)
# observed data log-likelihood
gmm.log_likelihood(X)
# total number of cluster parameters
gmm._n_parameters()
# some additional metadata is stored such as the fit time (in seconds)
gmm.metadata_['fit_time']
# gmm.opt_data_ stores the optimization history
plot_opt_hist(loss_vals=gmm.opt_data_['history']['loss_val'],
init_loss_vals=gmm.opt_data_['init_loss_vals'],
loss_name='observed data negative log likelihood')
```
# Model selection with BIC
```
# setup the base estimator for the grid search
# here we add some custom arguments
base_estimator = GaussianMixture(reg_covar=1e-6,
init_params_method='rand_pts', # initalize cluster means from random data points
n_init=10, abs_tol=1e-8, rel_tol=1e-8, max_n_steps=200)
# do a grid search from 1 to 10 components
param_grid = {'n_components': np.arange(1, 10 + 1)}
# setup grid search object and fit using the data
grid_search = MMGridSearch(base_estimator=base_estimator, param_grid=param_grid)
grid_search.fit(X)
# the best model is stored in .best_estimator_
print('BIC selected the model with', grid_search.best_estimator_.n_components, ' components')
# all fit estimators are containted in .estimators_
print(len(grid_search.estimators_))
# the model selection for each grid point are stored in /model_sel_scores_
print(grid_search.model_sel_scores_)
# plot BIC
n_comp_seq = grid_search.param_grid['n_components']
est_n_comp = grid_search.best_params_['n_components']
bic_values = grid_search.model_sel_scores_['bic']
plt.plot(n_comp_seq, bic_values, marker='.')
plt.axvline(est_n_comp,
label='estimated {} components'.format(est_n_comp),
color='red')
plt.legend()
plt.xlabel('n_components')
plt.ylabel('BIC')
set_xaxis_int_ticks()
```
|
github_jupyter
|
```
%matplotlib inline
import seaborn as sns
sns.set()
tips = sns.load_dataset("tips")
sns.relplot(x="total_bill", y="tip", col="time",
hue="smoker", style="smoker", size="size",
data=tips);
```
```
import seaborn as sns
```
```
sns.set()
```
```
tips = sns.load_dataset("tips")
```
```
sns.relplot(x="total_bill", y="tip", col="time",
hue="smoker", style="smoker", size="size",
data=tips)
```
```
dots = sns.load_dataset("dots")
sns.relplot(x="time", y="firing_rate", col="align",
hue="choice", size="coherence", style="choice",
facet_kws=dict(sharex=False),
kind="line", legend="full", data=dots);
```
```
fmri = sns.load_dataset("fmri")
sns.relplot(x="timepoint", y="signal", col="region",
hue="event", style="event",
kind="line", data=fmri);
```
```
sns.lmplot(x="total_bill", y="tip", col="time", hue="smoker",
data=tips);
```
```
sns.catplot(x="day", y="total_bill", hue="smoker",
kind="swarm", data=tips);
```
```
sns.catplot(x="day", y="total_bill", hue="smoker",
kind="violin", split=True, data=tips);
```
```
sns.catplot(x="day", y="total_bill", hue="smoker",
kind="bar", data=tips);
```
```
import matplotlib.pyplot as plt
f, axes = plt.subplots(1, 2, sharey=True, figsize=(6, 4))
sns.boxplot(x="day", y="tip", data=tips, ax=axes[0])
sns.scatterplot(x="total_bill", y="tip", hue="day", data=tips, ax=axes[1]);
```
```
sns.relplot(x="time", y="firing_rate", col="align",
hue="choice", size="coherence", style="choice",
height=4.5, aspect=2 / 3,
facet_kws=dict(sharex=False),
kind="line", legend="full", data=dots);
```
```
iris = sns.load_dataset("iris")
sns.jointplot(x="sepal_length", y="petal_length", data=iris);
```
```
sns.pairplot(data=iris, hue="species");
```
```
sns.set(style="ticks", palette="muted")
sns.relplot(x="total_bill", y="tip", col="time",
hue="smoker", style="smoker", size="size",
data=tips);
```
```
sns.relplot(x="total_bill", y="tip", col="time",
hue="size", style="smoker", size="size",
palette="YlGnBu", markers=["D", "o"], sizes=(10, 125),
edgecolor=".2", linewidth=.5, alpha=.75,
data=tips);
```
```
g = sns.catplot(x="total_bill", y="day", hue="time",
height=3.5, aspect=1.5,
kind="box", legend=False, data=tips);
g.add_legend(title="Meal")
g.set_axis_labels("Total bill ($)", "")
g.set(xlim=(0, 60), yticklabels=["Thursday", "Friday", "Saturday", "Sunday"])
g.despine(trim=True)
g.fig.set_size_inches(6.5, 3.5)
g.ax.set_xticks([5, 15, 25, 35, 45, 55], minor=True);
plt.setp(g.ax.get_yticklabels(), rotation=30);
```
```
tips.head()
```
```
fmri.head()
```
|
github_jupyter
|
# <img style="float: left; padding-right: 10px; width: 45px" src="https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png"> CS-109B Introduction to Data Science
## Lab 5: Convolutional Neural Networks
**Harvard University**<br>
**Spring 2019**<br>
**Lab instructor:** Eleni Kaxiras<br>
**Instructors:** Pavlos Protopapas and Mark Glickman<br>
**Authors:** Eleni Kaxiras, Pavlos Protopapas, Patrick Ohiomoba, and Davis Sontag
```
# RUN THIS CELL TO PROPERLY HIGHLIGHT THE EXERCISES
import requests
from IPython.core.display import HTML
styles = requests.get("https://raw.githubusercontent.com/Harvard-IACS/2019-CS109B/master/content/styles/cs109.css").text
HTML(styles)
```
## Learning Goals
In this lab we will look at Convolutional Neural Networks (CNNs), and their building blocks.
By the end of this lab, you should:
- know how to put together the building blocks used in CNNs - such as convolutional layers and pooling layers - in `keras` with an example.
- have a good undertanding on how images, a common type of data for a CNN, are represented in the computer and how to think of them as arrays of numbers.
- be familiar with preprocessing images with `keras` and `sckit-learn`.
- use `keras-viz` to produce Saliency maps.
- learn best practices for configuring the hyperparameters of a CNN.
- run your first CNN and see the error rate.
```
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (5,5)
import numpy as np
from scipy.optimize import minimize
import tensorflow as tf
import keras
from keras import layers
from keras import models
from keras import utils
from keras.layers import Dense
from keras.models import Sequential
from keras.layers import Flatten
from keras.layers import Dropout
from keras.layers import Activation
from keras.regularizers import l2
from keras.optimizers import SGD
from keras.optimizers import RMSprop
from keras import datasets
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import LearningRateScheduler
from keras.callbacks import History
from keras import losses
from keras.datasets import mnist
from keras.utils import to_categorical
from sklearn.utils import shuffle
print(tf.VERSION)
print(tf.keras.__version__)
%matplotlib inline
```
## Prologue: `keras-viz` Visualization Toolkit
`keras-vis` is a high-level toolkit for visualizing and debugging your trained keras neural net models. Currently supported visualizations include:
- Activation maximization
- **Saliency maps**
- Class activation maps
All visualizations by default support N-dimensional image inputs. i.e., it generalizes to N-dim image inputs to your model. Compatible with both theano and tensorflow backends with 'channels_first', 'channels_last' data format.
Read the documentation at https://raghakot.github.io/keras-vis.https://github.com/raghakot/keras-vis
To install use `pip install git+https://github.com/raghakot/keras-vis.git --upgrade`
## SEAS JupyterHub
[Instructions for Using SEAS JupyterHub](https://canvas.harvard.edu/courses/48088/pages/instructions-for-using-seas-jupyterhub)
SEAS and FAS are providing you with a platform in AWS to use for the class (accessible from the 'Jupyter' menu link in Canvas). These are AWS p2 instances with a GPU, 10GB of disk space, and 61 GB of RAM, for faster training for your networks. Most of the libraries such as keras, tensorflow, pandas, etc. are pre-installed. If a library is missing you may install it via the Terminal.
**NOTE : The AWS platform is funded by SEAS and FAS for the purposes of the class. It is not running against your individual credit. You are not allowed to use it for purposes not related to this course.**
**Help us keep this service: Make sure you stop your instance as soon as you do not need it.**

## Part 1: Parts of a Convolutional Neural Net
There are three types of layers in a Convolutional Neural Network:
- Convolutional Layers
- Pooling Layers.
- Dropout Layers.
- Fully Connected Layers.
### a. Convolutional Layers.
Convolutional layers are comprised of **filters** and **feature maps**. The filters are essentially the **neurons** of the layer. They have the weights and produce the input for the next layer. The feature map is the output of one filter applied to the previous layer.
The fundamental difference between a densely connected layer and a convolution layer is that dense layers learn global patterns in their input feature space (for example, for an MNIST digit, patterns involving all pixels), whereas convolution layers learn local patterns: in the case of images, patterns found in small 2D windows of the inputs called *receptive fields*.
This key characteristic gives convnets two interesting properties:
- The patterns they learn are **translation invariant**. After learning a certain pattern in the lower-right corner of a picture, a convnet can recognize it anywhere: for example, in the upper-left corner. A densely connected network would have to learn the pattern anew if it appeared at a new location. This makes convnets data efficient when processing images (because the visual world is fundamentally translation invariant): they need fewer training samples to learn representations that have generalization power.
- They can learn **spatial hierarchies of patterns**. A first convolution layer will learn small local patterns such as edges, a second convolution layer will learn larger patterns made of the features of the first layers, and so on. This allows convnets to efficiently learn increasingly complex and abstract visual concepts (because the visual world is fundamentally spatially hierarchical).
Convolutions operate over 3D tensors, called feature maps, with two spatial axes (height and width) as well as a depth axis (also called the channels axis). For an RGB image, the dimension of the depth axis is 3, because the image has three color channels: red, green, and blue. For a black-and-white picture, like the MNIST digits, the depth is 1 (levels of gray). The convolution operation extracts patches from its input feature map and applies the same transformation to all of these patches, producing an output feature map. This output feature map is still a 3D tensor: it has a width and a height. Its depth can be arbitrary, because the output depth is a parameter of the layer, and the different channels in that depth axis no longer stand for specific colors as in RGB input; rather, they stand for filters. Filters encode specific aspects of the input data: at a high level, a single filter could encode the concept “presence of a face in the input,” for instance.
In the MNIST example that we will see, the first convolution layer takes a feature map of size (28, 28, 1) and outputs a feature map of size (26, 26, 32): it computes 32 filters over its input. Each of these 32 output channels contains a 26×26 grid of values, which is a response map of the filter over the input, indicating the response of that filter pattern at different locations in the input.
Convolutions are defined by two key parameters:
- Size of the patches extracted from the inputs. These are typically 3×3 or 5×5
- The number of filters computed by the convolution.
**Padding**: One of "valid", "causal" or "same" (case-insensitive). "valid" means "no padding". "same" results in padding the input such that the output has the same length as the original input. "causal" results in causal (dilated) convolutions,
In `keras` see [convolutional layers](https://keras.io/layers/convolutional/)
**keras.layers.Conv2D**(filters, kernel_size, strides=(1, 1), padding='valid', activation=None, use_bias=True,
kernel_initializer='glorot_uniform', data_format='channels_last',
bias_initializer='zeros')
#### How are the values in feature maps calculated?

### Exercise 1:
- Compute the operations by hand (assuming zero padding and same arrays for all channels) to produce the first element of the 4x4 feature map. How did we get the 4x4 output size?
- Write this Conv layer in keras
-- your answer here
### b. Pooling Layers.
Pooling layers are also comprised of filters and feature maps. Let's say the pooling layer has a 2x2 receptive field and a stride of 2. This stride results in feature maps that are one half the size of the input feature maps. We can use a max() operation for each receptive field.
In `keras` see [pooling layers](https://keras.io/layers/pooling/)
**keras.layers.MaxPooling2D**(pool_size=(2, 2), strides=None, padding='valid', data_format=None)

### c. Dropout Layers.
Dropout consists in randomly setting a fraction rate of input units to 0 at each update during training time, which helps prevent overfitting.
In `keras` see [Dropout layers](https://keras.io/layers/core/)
keras.layers.Dropout(rate, seed=None)
rate: float between 0 and 1. Fraction of the input units to drop.<br>
seed: A Python integer to use as random seed.
References
[Dropout: A Simple Way to Prevent Neural Networks from Overfitting](http://www.jmlr.org/papers/volume15/srivastava14a/srivastava14a.pdf)
### d. Fully Connected Layers.
A fully connected layer flattens the square feature map into a vector. Then we can use a sigmoid or softmax activation function to output probabilities of classes.
In `keras` see [FC layers](https://keras.io/layers/core/)
**keras.layers.Dense**(units, activation=None, use_bias=True,
kernel_initializer='glorot_uniform', bias_initializer='zeros')
#### IT'S ALL ABOUT THE HYPERPARAMETERS!
- stride
- size of filter
- number of filters
- poolsize
## Part 2: Preprocessing the data
### Taking a look at how images are represented in a computer using a photo of a Picasso sculpture
```
img = plt.imread('data/picasso.png')
img.shape
img[1,:,1]
print(type(img[50][0][0]))
# let's see the image
imgplot = plt.imshow(img)
```
#### Visualizing the channels
```
R_img = img[:,:,0]
G_img = img[:,:,1]
B_img = img[:,:,2]
plt.subplot(221)
plt.imshow(R_img, cmap=plt.cm.Reds)
plt.subplot(222)
plt.imshow(G_img, cmap=plt.cm.Greens)
plt.subplot(223)
plt.imshow(B_img, cmap=plt.cm.Blues)
plt.subplot(224)
plt.imshow(img)
plt.show()
```
More on preprocessing data below!
If you want to learn more: [Image Processing with Python and Scipy](http://prancer.physics.louisville.edu/astrowiki/index.php/Image_processing_with_Python_and_SciPy)
## Part 3: Putting the Parts together to make a small ConvNet Model
Let's put all the parts together to make a convnet for classifying our good old MNIST digits.
```
# Load data and preprocess
(train_images, train_labels), (test_images, test_labels) = mnist.load_data() # load MNIST data
train_images.shape
train_images.max(), train_images.min()
train_images = train_images.reshape((60000, 28, 28, 1)) # Reshape to get third dimension
train_images = train_images.astype('float32') / 255 # Normalize between 0 and 1
test_images = test_images.reshape((10000, 28, 28, 1)) # Reshape to get third dimension
test_images = test_images.astype('float32') / 255 # Normalize between 0 and 1
# Convert labels to categorical data
train_labels = to_categorical(train_labels)
test_labels = to_categorical(test_labels)
mnist_cnn_model = models.Sequential() # Create sequential model
# Add network layers
mnist_cnn_model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(28, 28, 1)))
mnist_cnn_model.add(layers.MaxPooling2D((2, 2)))
mnist_cnn_model.add(layers.Conv2D(64, (3, 3), activation='relu'))
mnist_cnn_model.add(layers.MaxPooling2D((2, 2)))
mnist_cnn_model.add(layers.Conv2D(64, (3, 3), activation='relu'))
```
The next step is to feed the last output tensor (of shape (3, 3, 64)) into a densely connected classifier network like those you’re already familiar with: a stack of Dense layers. These classifiers process vectors, which are 1D, whereas the current output is a 3D tensor. First we have to flatten the 3D outputs to 1D, and then add a few Dense layers on top.
```
mnist_cnn_model.add(layers.Flatten())
mnist_cnn_model.add(layers.Dense(64, activation='relu'))
mnist_cnn_model.add(layers.Dense(10, activation='softmax'))
mnist_cnn_model.summary()
# Compile model
mnist_cnn_model.compile(optimizer='rmsprop',
loss='categorical_crossentropy',
metrics=['accuracy'])
# Fit the model
mnist_cnn_model.fit(train_images, train_labels, epochs=5, batch_size=64)
# Evaluate the model on the test data:
test_loss, test_acc = mnist_cnn_model.evaluate(test_images, test_labels)
test_acc
```
A densely connected network (MLP) running MNIST usually has a test accuracy of 97.8%, whereas our basic convnet has a test accuracy of 99.03%: we decreased the error rate by 68% (relative) with only 5 epochs. Not bad! But why does this simple convnet work so well, compared to a densely connected model? The answer is above on how convolutional layers work!
### Data Preprocessing : Meet the `ImageDataGenerator` class in `keras` [(docs)](https://keras.io/preprocessing/image/)
The MNIST and other pre-loaded dataset are formatted in a way that is almost ready for feeding into the model. What about plain images? They should be formatted into appropriately preprocessed floating-point tensors before being fed into the network.
The Dogs vs. Cats dataset that you’ll use isn’t packaged with Keras. It was made available by Kaggle as part of a computer-vision competition in late 2013, back when convnets weren’t mainstream. The data has been downloaded for you from https://www.kaggle.com/c/dogs-vs-cats/data The pictures are medium-resolution color JPEGs.
```
# TODO: set your base dir to your correct local location
base_dir = 'data/cats_and_dogs_small'
import os, shutil
# Set up directory information
train_dir = os.path.join(base_dir, 'train')
validation_dir = os.path.join(base_dir, 'validation')
test_dir = os.path.join(base_dir, 'test')
train_cats_dir = os.path.join(train_dir, 'cats')
train_dogs_dir = os.path.join(train_dir, 'dogs')
validation_cats_dir = os.path.join(validation_dir, 'cats')
validation_dogs_dir = os.path.join(validation_dir, 'dogs')
test_cats_dir = os.path.join(test_dir, 'cats')
test_dogs_dir = os.path.join(test_dir, 'dogs')
print('total training cat images:', len(os.listdir(train_cats_dir)))
print('total training dog images:', len(os.listdir(train_dogs_dir)))
print('total validation cat images:', len(os.listdir(validation_cats_dir)))
print('total validation dog images:', len(os.listdir(validation_dogs_dir)))
print('total test cat images:', len(os.listdir(test_cats_dir)))
print('total test dog images:', len(os.listdir(test_dogs_dir)))
```
So you do indeed have 2,000 training images, 1,000 validation images, and 1,000 test images. Each split contains the same number of samples from each class: this is a balanced binary-classification problem, which means classification accuracy will be an appropriate measure of success.
#### Building the network
```
from keras import layers
from keras import models
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.summary()
```
For the compilation step, you’ll go with the RMSprop optimizer. Because you ended the network with a single sigmoid unit, you’ll use binary crossentropy as the loss.
```
from keras import optimizers
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
```
The steps for getting it into the network are roughly as follows:
1. Read the picture files.
2. Decode the JPEG content to RGB grids of pixels.
3. Convert these into floating-point tensors.
4. Rescale the pixel values (between 0 and 255) to the [0, 1] interval (as you know, neural networks prefer to deal with small input values).
It may seem a bit daunting, but fortunately Keras has utilities to take care of these steps automatically with the class `ImageDataGenerator`, which lets you quickly set up Python generators that can automatically turn image files on disk into batches of preprocessed tensors. This is what you’ll use here.
```
from keras.preprocessing.image import ImageDataGenerator
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=20,
class_mode='binary')
```
Let’s look at the output of one of these generators: it yields batches of 150×150 RGB images (shape (20, 150, 150, 3)) and binary labels (shape (20,)). There are 20 samples in each batch (the batch size). Note that the generator yields these batches indefinitely: it loops endlessly over the images in the target folder. For this reason, you need to break the iteration loop at some point:
```
for data_batch, labels_batch in train_generator:
print('data batch shape:', data_batch.shape)
print('labels batch shape:', labels_batch.shape)
break
```
Let’s fit the model to the data using the generator. You do so using the `.fit_generator` method, the equivalent of `.fit` for data generators like this one. It expects as its first argument a Python generator that will yield batches of inputs and targets indefinitely, like this one does.
Because the data is being generated endlessly, the Keras model needs to know how many samples to draw from the generator before declaring an epoch over. This is the role of the `steps_per_epoch` argument: after having drawn steps_per_epoch batches from the generator—that is, after having run for steps_per_epoch gradient descent steps - the fitting process will go to the next epoch. In this case, batches are 20 samples, so it will take 100 batches until you see your target of 2,000 samples.
When using fit_generator, you can pass a validation_data argument, much as with the fit method. It’s important to note that this argument is allowed to be a data generator, but it could also be a tuple of Numpy arrays. If you pass a generator as validation_data, then this generator is expected to yield batches of validation data endlessly; thus you should also specify the validation_steps argument, which tells the process how many batches to draw from the validation generator for evaluation
```
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=5, # TODO: should be 30
validation_data=validation_generator,
validation_steps=50)
# It’s good practice to always save your models after training.
model.save('cats_and_dogs_small_1.h5')
```
Let’s plot the loss and accuracy of the model over the training and validation data during training:
```
fig, ax = plt.subplots(1, 1, figsize=(10,6))
ax.plot((history.history['acc']), 'r', label='train')
ax.plot((history.history['val_acc']), 'b' ,label='val')
ax.set_xlabel(r'Epoch', fontsize=20)
ax.set_ylabel(r'Accuracy', fontsize=20)
ax.legend()
ax.tick_params(labelsize=20)
```
Let's try data augmentation
```
datagen = ImageDataGenerator(
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode='nearest')
```
These are just a few of the options available (for more, see the Keras documentation).
Let’s quickly go over this code:
- rotation_range is a value in degrees (0–180), a range within which to randomly rotate pictures.
- width_shift and height_shift are ranges (as a fraction of total width or height) within which to randomly translate pictures vertically or horizontally.
- shear_range is for randomly applying shearing transformations.
- zoom_range is for randomly zooming inside pictures.
- horizontal_flip is for randomly flipping half the images horizontally—relevant when there are no assumptions of - horizontal asymmetry (for example, real-world pictures).
- fill_mode is the strategy used for filling in newly created pixels, which can appear after a rotation or a width/height shift.
Let’s look at the augmented images
```
from keras.preprocessing import image
fnames = [os.path.join(train_dogs_dir, fname) for
fname in os.listdir(train_dogs_dir)]
img_path = fnames[3] # Chooses one image to augment
img = image.load_img(img_path, target_size=(150, 150))
# Reads the image and resizes it
x = image.img_to_array(img) # Converts it to a Numpy array with shape (150, 150, 3)
x = x.reshape((1,) + x.shape) # Reshapes it to (1, 150, 150, 3)
i=0
for batch in datagen.flow(x, batch_size=1):
plt.figure(i)
imgplot = plt.imshow(image.array_to_img(batch[0]))
i += 1
if i % 4 == 0:
break
plt.show()
```
If you train a new network using this data-augmentation configuration, the network will never see the same input twice. But the inputs it sees are still heavily intercorrelated, because they come from a small number of original images—you can’t produce new information, you can only remix existing information. As such, this may not be enough to completely get rid of overfitting. To further fight overfitting, you’ll also add a **Dropout** layer to your model right before the densely connected classifier.
```
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu',
input_shape=(150, 150, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(128, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Flatten())
model.add(layers.Dropout(0.5))
model.add(layers.Dense(512, activation='relu'))
model.add(layers.Dense(1, activation='sigmoid'))
model.compile(loss='binary_crossentropy',
optimizer=optimizers.RMSprop(lr=1e-4),
metrics=['acc'])
# Let’s train the network using data augmentation and dropout.
train_datagen = ImageDataGenerator(
rescale=1./255,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,)
test_datagen = ImageDataGenerator(rescale=1./255)
# Note that the validation data shouldn’t be augmented!
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
validation_generator = test_datagen.flow_from_directory(
validation_dir,
target_size=(150, 150),
batch_size=32,
class_mode='binary')
history = model.fit_generator(
train_generator,
steps_per_epoch=100,
epochs=5, # TODO: should be 100
validation_data=validation_generator,
validation_steps=50)
model.save('cats_and_dogs_small_2.h5')
```
And let’s plot the results again. Thanks to data augmentation and dropout, you’re no longer overfitting: the training curves are closely tracking the validation curves. You now reach an accuracy of 82%, a 15% relative improvement over the non-regularized model. (Note: these numbers are for 100 epochs..)
```
fig, ax = plt.subplots(1, 1, figsize=(10,6))
ax.plot((history.history['acc']), 'r', label='train')
ax.plot((history.history['val_acc']), 'b' ,label='val')
ax.set_xlabel(r'Epoch', fontsize=20)
ax.set_ylabel(r'Accuracy', fontsize=20)
ax.legend()
ax.tick_params(labelsize=20)
```
By using regularization techniques even further, and by tuning the network’s parameters (such as the number of filters per convolution layer, or the number of layers in the network), you may be able to get an even better accuracy, likely up to 86% or 87%. But it would prove difficult to go any higher just by training your own convnet from scratch, because you have so little data to work with. As a next step to improve your accuracy on this problem, you’ll have to use a pretrained model.
## Part 4: keras viz toolkit
https://github.com/raghakot/keras-vis/blob/master/examples/mnist/attention.ipynb
```
class_idx = 0
indices = np.where(test_labels[:, class_idx] == 1.)[0]
# pick some random input from here.
idx = indices[0]
# Lets sanity check the picked image.
from matplotlib import pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = (18, 6)
plt.imshow(test_images[idx][..., 0])
input_shape=(28, 28, 1)
num_classes = 10
batch_size = 128
epochs = 5
model = Sequential()
model.add(layers.Conv2D(32, kernel_size=(3, 3),
activation='relu',
input_shape=input_shape))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D(pool_size=(2, 2)))
model.add(layers.Dropout(0.25))
model.add(layers.Flatten())
model.add(layers.Dense(128, activation='relu'))
model.add(layers.Dropout(0.5))
model.add(layers.Dense(num_classes, activation='softmax', name='preds'))
model.compile(loss=keras.losses.categorical_crossentropy,
optimizer=keras.optimizers.Adam(),
metrics=['accuracy'])
model.fit(train_images, train_labels,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(test_images, test_labels))
score = model.evaluate(test_images, test_labels, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
from vis.visualization import visualize_saliency
from vis.utils import utils
from keras import activations
# Utility to search for layer index by name.
# Alternatively we can specify this as -1 since it corresponds to the last layer.
layer_idx = utils.find_layer_idx(model, 'preds')
plt.rcParams["figure.figsize"] = (5,5)
from vis.visualization import visualize_cam
import warnings
warnings.filterwarnings('ignore')
# This corresponds to the Dense linear layer.
for class_idx in np.arange(10):
indices = np.where(test_labels[:, class_idx] == 1.)[0]
idx = indices[0]
f, ax = plt.subplots(1, 4)
ax[0].imshow(test_images[idx][..., 0])
for i, modifier in enumerate([None, 'guided', 'relu']):
grads = visualize_cam(model, layer_idx, filter_indices=class_idx,
seed_input=test_images[idx], backprop_modifier=modifier)
if modifier is None:
modifier = 'vanilla'
ax[i+1].set_title(modifier)
ax[i+1].imshow(grads, cmap='jet')
```
#### References and Acknowledgements
The cats and dogs part of this lab is based on the book Deep Learning with Python, Chapter 5 written by the Francois Chollet, the author of Keras. It is a very practical introduction to Deep Learning. It is appropriate for those with some Python knowledge who want to start with machine learning.
The saliency maps are from https://github.com/raghakot/keras-vis/blob/master/examples/mnist/attention.ipynb
|
github_jupyter
|
# Pragmatic color describers
```
__author__ = "Christopher Potts"
__version__ = "CS224u, Stanford, Spring 2020"
```
## Contents
1. [Overview](#Overview)
1. [Set-up](#Set-up)
1. [The corpus](#The-corpus)
1. [Corpus reader](#Corpus-reader)
1. [ColorsCorpusExample instances](#ColorsCorpusExample-instances)
1. [Displaying examples](#Displaying-examples)
1. [Color representations](#Color-representations)
1. [Utterance texts](#Utterance-texts)
1. [Far, Split, and Close conditions](#Far,-Split,-and-Close-conditions)
1. [Toy problems for development work](#Toy-problems-for-development-work)
1. [Core model](#Core-model)
1. [Toy dataset illustration](#Toy-dataset-illustration)
1. [Predicting sequences](#Predicting-sequences)
1. [Listener-based evaluation](#Listener-based-evaluation)
1. [Other prediction and evaluation methods](#Other-prediction-and-evaluation-methods)
1. [Cross-validation](#Cross-validation)
1. [Baseline SCC model](#Baseline-SCC-model)
1. [Modifying the core model](#Modifying-the-core-model)
1. [Illustration: LSTM Cells](#Illustration:-LSTM-Cells)
1. [Illustration: Deeper models](#Illustration:-Deeper-models)
## Overview
This notebook is part of our unit on grounding. It illustrates core concepts from the unit, and it provides useful background material for the associated homework and bake-off.
## Set-up
```
from colors import ColorsCorpusReader
import os
import pandas as pd
from sklearn.model_selection import train_test_split
import torch
from torch_color_describer import (
ContextualColorDescriber, create_example_dataset)
import utils
from utils import START_SYMBOL, END_SYMBOL, UNK_SYMBOL
utils.fix_random_seeds()
```
The [Stanford English Colors in Context corpus](https://cocolab.stanford.edu/datasets/colors.html) (SCC) is included in the data distribution for this course. If you store the data in a non-standard place, you'll need to update the following:
```
COLORS_SRC_FILENAME = os.path.join(
"data", "colors", "filteredCorpus.csv")
```
## The corpus
The SCC corpus is based in a two-player interactive game. The two players share a context consisting of three color patches, with the display order randomized between them so that they can't use positional information when communicating.
The __speaker__ is privately assigned a target color and asked to produce a description of it that will enable the __listener__ to identify the speaker's target. The listener makes a choice based on the speaker's message, and the two succeed if and only if the listener identifies the target correctly.
In the game, the two players played repeated reference games and could communicate with each other in a free-form way. This opens up the possibility of modeling these repeated interactions as task-oriented dialogues. However, for this unit, we'll ignore most of this structure. We'll treat the corpus as a bunch of independent reference games played by anonymous players, and we will ignore the listener and their choices entirely.
For the bake-off, we will be distributing a separate test set. Thus, all of the data in the SCC can be used for exploration and development.
### Corpus reader
The corpus reader class is `ColorsCorpusReader` in `colors.py`. The reader's primary function is to let you iterate over corpus examples:
```
corpus = ColorsCorpusReader(
COLORS_SRC_FILENAME,
word_count=None,
normalize_colors=True)
```
The two keyword arguments have their default values here.
* If you supply `word_count` with an interger value, it will restrict to just examples where the utterance has that number of words (using a whitespace heuristic). This creates smaller corpora that are useful for development.
* The colors in the corpus are in [HLS format](https://en.wikipedia.org/wiki/HSL_and_HSV). With `normalize_colors=False`, the first (hue) value is an integer between 1 and 360 inclusive, and the L (lightness) and S (saturation) values are between 1 and 100 inclusive. With `normalize_colors=True`, these values are all scaled to between 0 and 1 inclusive. The default is `normalize_colors=True` because this is a better choice for all the machine learning models we'll consider.
```
examples = list(corpus.read())
```
We can verify that we read in the same number of examples as reported in [Monroe et al. 2017](https://transacl.org/ojs/index.php/tacl/article/view/1142):
```
# Should be 46994:
len(examples)
```
### ColorsCorpusExample instances
The examples are `ColorsCorpusExample` instances:
```
ex1 = next(corpus.read())
```
These objects have a lot of attributes and methods designed to help you study the corpus and use it for our machine learning tasks. Let's review some highlights.
#### Displaying examples
You can see what the speaker saw, with the utterance they chose wote above the patches:
```
ex1.display(typ='speaker')
```
This is the original order of patches for the speaker. The target happens to the be the leftmost patch, as indicated by the black box around it.
Here's what the listener saw, with the speaker's message printed above the patches:
```
ex1.display(typ='listener')
```
The listener isn't shown the target, of course, so no patches are highlighted.
If `display` is called with no arguments, then the target is placed in the final position and the other two are given in an order determined by the corpus metadata:
```
ex1.display()
```
This is the representation order we use for our machine learning models.
#### Color representations
For machine learning, we'll often need to access the color representations directly. The primary attribute for this is `colors`:
```
ex1.colors
```
In this display order, the third element is the target color and the first two are the distractors. The attributes `speaker_context` and `listener_context` return the same colors but in the order that those players saw them. For example:
```
ex1.speaker_context
```
#### Utterance texts
Utterances are just strings:
```
ex1.contents
```
There are cases where the speaker made a sequences of utterances for the same trial. We follow [Monroe et al. 2017](https://transacl.org/ojs/index.php/tacl/article/view/1142) in concatenating these into a single utterances. To preserve the original information, the individual turns are separated by `" ### "`. Example 3 is the first with this property – let's check it out:
```
ex3 = examples[2]
ex3.contents
```
The method `parse_turns` will parse this into individual turns:
```
ex3.parse_turns()
```
For examples consisting of a single turn, `parse_turns` returns a list of length 1:
```
ex1.parse_turns()
```
### Far, Split, and Close conditions
The SCC contains three conditions:
__Far condition__: All three colors are far apart in color space. Example:
```
print("Condition type:", examples[1].condition)
examples[1].display()
```
__Split condition__: The target is close to one of the distractors, and the other is far away from both of them. Example:
```
print("Condition type:", examples[3].condition)
examples[3].display()
```
__Close condition__: The target is similar to both distractors. Example:
```
print("Condition type:", examples[2].condition)
examples[2].display()
```
These conditions go from easiest to hardest when it comes to reliable communication. In the __Far__ condition, the context is hardly relevant, whereas the nature of the distractors reliably shapes the speaker's choices in the other two conditions.
You can begin to see how this affects speaker choices in the above examples: "purple" suffices for the __Far__ condition, a more marked single word ("lime") suffices in the __Split__ condition, and the __Close__ condition triggers a pretty long, complex description.
The `condition` attribute provides access to this value:
```
ex1.condition
```
The following verifies that we have the same number of examples per condition as reported in [Monroe et al. 2017](https://transacl.org/ojs/index.php/tacl/article/view/1142):
```
pd.Series([ex.condition for ex in examples]).value_counts()
```
## Toy problems for development work
The SCC corpus is fairly large and quite challenging as an NLU task. This means it isn't ideal when it comes to testing hypotheses and debugging code. Poor performance could trace to a mistake, but it could just as easily trace to the fact that the problem is very challenging from the point of view of optimization.
To address this, the module `torch_color_describer.py` includes a function `create_example_dataset` for creating small, easy datasets with the same basic properties as the SCC corpus.
Here's a toy problem containing just six examples:
```
tiny_contexts, tiny_words, tiny_vocab = create_example_dataset(
group_size=2, vec_dim=2)
tiny_vocab
tiny_words
tiny_contexts
```
Each member of `tiny_contexts` contains three vectors. The final (target) vector always has values in a range that determines the corresponding word sequence, which is drawn from a set of three fixed sequences. Thus, the model basically just needs to learn to ignore the distractors and find the association between the target vector and the corresponding sequence.
All the models we study have a capacity to solve this task with very little data, so you should see perfect or near perfect performance on reasonably-sized versions of this task.
## Core model
Our core model for this problem is implemented in `torch_color_describer.py` as `ContextualColorDescriber`. At its heart, this is a pretty standard encoder–decoder model:
* `Encoder`: Processes the color contexts as a sequence. We always place the target in final position so that it is closest to the supervision signals that we get when decoding.
* `Decoder`: A neural language model whose initial hidden representation is the final hidden representation of the `Encoder`.
* `EncoderDecoder`: Coordinates the operations of the `Encoder` and `Decoder`.
Finally, `ContextualColorDescriber` is a wrapper around these model components. It handle the details of training and implements the prediction and evaluation functions that we will use.
Many additional details about this model are included in the slides for this unit.
### Toy dataset illustration
To highlight the core functionality of `ContextualColorDescriber`, let's create a small toy dataset and use it to train and evaluate a model:
```
toy_color_seqs, toy_word_seqs, toy_vocab = create_example_dataset(
group_size=50, vec_dim=2)
toy_color_seqs_train, toy_color_seqs_test, toy_word_seqs_train, toy_word_seqs_test = \
train_test_split(toy_color_seqs, toy_word_seqs)
```
Here we expose all of the available parameters with their default values:
```
toy_mod = ContextualColorDescriber(
toy_vocab,
embedding=None, # Option to supply a pretrained matrix as an `np.array`.
embed_dim=10,
hidden_dim=10,
max_iter=100,
eta=0.01,
optimizer=torch.optim.Adam,
batch_size=128,
l2_strength=0.0,
warm_start=False,
device=None)
_ = toy_mod.fit(toy_color_seqs_train, toy_word_seqs_train)
```
### Predicting sequences
The `predict` method takes a list of color contexts as input and returns model descriptions:
```
toy_preds = toy_mod.predict(toy_color_seqs_test)
toy_preds[0]
```
We can then check that we predicted all correct sequences:
```
toy_correct = sum(1 for x, p in zip(toy_word_seqs_test, toy_preds))
toy_correct / len(toy_word_seqs_test)
```
For real problems, this is too stringent a requirement, since there are generally many equally good descriptions. This insight gives rise to metrics like [BLEU](https://en.wikipedia.org/wiki/BLEU), [METEOR](https://en.wikipedia.org/wiki/METEOR), [ROUGE](https://en.wikipedia.org/wiki/ROUGE_(metric)), [CIDEr](https://arxiv.org/pdf/1411.5726.pdf), and others, which seek to relax the requirement of an exact match with the test sequence. These are reasonable options to explore, but we will instead adopt a communcation-based evaluation, as discussed in the next section.
### Listener-based evaluation
`ContextualColorDescriber` implements a method `listener_accuracy` that we will use for our primary evaluations in the assignment and bake-off. The essence of the method is that we can calculate
$$
c^{*} = \text{argmax}_{c \in C} P_S(\text{utterance} \mid c)
$$
where $P_S$ is our describer model and $C$ is the set of all permutations of all three colors in the color context. We take $c^{*}$ to be a correct prediction if it is one where the target is in the privileged final position. (There are two such contexts; we try both in case the order of the distractors influences the predictions, and the model is correct if one of them has the highest probability.)
Here's the listener accuracy of our toy model:
```
toy_mod.listener_accuracy(toy_color_seqs_test, toy_word_seqs_test)
```
### Other prediction and evaluation methods
You can get the perplexities for test examles with `perpelexities`:
```
toy_perp = toy_mod.perplexities(toy_color_seqs_test, toy_word_seqs_test)
toy_perp[0]
```
You can use `predict_proba` to see the full probability distributions assigned to test examples:
```
toy_proba = toy_mod.predict_proba(toy_color_seqs_test, toy_word_seqs_test)
toy_proba[0].shape
for timestep in toy_proba[0]:
print(dict(zip(toy_vocab, timestep)))
```
### Cross-validation
You can use `utils.fit_classifier_with_crossvalidation` to cross-validate these models. Just be sure to set `scoring=None` so that the sklearn model selection methods use the `score` method of `ContextualColorDescriber`, which is an alias for `listener_accuracy`:
```
best_mod = utils.fit_classifier_with_crossvalidation(
toy_color_seqs_train,
toy_word_seqs_train,
toy_mod,
cv=2,
scoring=None,
param_grid={'hidden_dim': [10, 20]})
```
## Baseline SCC model
Just to show how all the pieces come together, here's a very basic SCC experiment using the core code and very simplistic assumptions (which you will revisit in the assignment) about how to represent the examples:
To facilitate quick development, we'll restrict attention to the two-word examples:
```
dev_corpus = ColorsCorpusReader(COLORS_SRC_FILENAME, word_count=2)
dev_examples = list(dev_corpus.read())
len(dev_examples)
```
Here we extract the raw colors and texts (as strings):
```
dev_cols, dev_texts = zip(*[[ex.colors, ex.contents] for ex in dev_examples])
```
To tokenize the examples, we'll just split on whitespace, taking care to add the required boundary symbols:
```
dev_word_seqs = [[START_SYMBOL] + text.split() + [END_SYMBOL] for text in dev_texts]
```
We'll use a random train–test split:
```
dev_cols_train, dev_cols_test, dev_word_seqs_train, dev_word_seqs_test = \
train_test_split(dev_cols, dev_word_seqs)
```
Our vocab is determined by the train set, and we take care to include the `$UNK` token:
```
dev_vocab = sorted({w for toks in dev_word_seqs_train for w in toks}) + [UNK_SYMBOL]
```
And now we're ready to train a model:
```
dev_mod = ContextualColorDescriber(
dev_vocab,
embed_dim=10,
hidden_dim=10,
max_iter=10,
batch_size=128)
_ = dev_mod.fit(dev_cols_train, dev_word_seqs_train)
```
And finally an evaluation in terms of listener accuracy:
```
dev_mod.listener_accuracy(dev_cols_test, dev_word_seqs_test)
```
## Modifying the core model
The first few assignment problems concern how you preprocess the data for your model. After that, the goal is to subclass model components in `torch_color_describer.py`. For the bake-off submission, you can do whatever you like in terms of modeling, but my hope is that you'll be able to continue subclassing based on `torch_color_describer.py`.
This section provides some illustrative examples designed to give you a feel for how the code is structured and what your options are in terms of creating subclasses.
### Illustration: LSTM Cells
Both the `Encoder` and the `Decoder` of `torch_color_describer` are currently GRU cells. Switching to another cell type is easy:
__Step 1__: Subclass the `Encoder`; all we have to do here is change `GRU` from the original to `LSTM`:
```
import torch.nn as nn
from torch_color_describer import Encoder
class LSTMEncoder(Encoder):
def __init__(self, color_dim, hidden_dim):
super().__init__(color_dim, hidden_dim)
self.rnn = nn.LSTM(
input_size=self.color_dim,
hidden_size=self.hidden_dim,
batch_first=True)
```
__Step 2__: Subclass the `Decoder`, making the same simple change as above:
```
import torch.nn as nn
from torch_color_describer import Encoder, Decoder
class LSTMDecoder(Decoder):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.rnn = nn.LSTM(
input_size=self.embed_dim,
hidden_size=self.hidden_dim,
batch_first=True)
```
__Step 3__:`ContextualColorDescriber` has a method called `build_graph` that sets up the `Encoder` and `Decoder`. The needed revision just uses `LSTMEncoder`:
```
from torch_color_describer import EncoderDecoder
class LSTMContextualColorDescriber(ContextualColorDescriber):
def build_graph(self):
# Use the new Encoder:
encoder = LSTMEncoder(
color_dim=self.color_dim,
hidden_dim=self.hidden_dim)
# Use the new Decoder:
decoder = LSTMDecoder(
vocab_size=self.vocab_size,
embed_dim=self.embed_dim,
embedding=self.embedding,
hidden_dim=self.hidden_dim)
return EncoderDecoder(encoder, decoder)
```
Here's an example run:
```
lstm_mod = LSTMContextualColorDescriber(
toy_vocab,
embed_dim=10,
hidden_dim=10,
max_iter=100,
batch_size=128)
_ = lstm_mod.fit(toy_color_seqs_train, toy_word_seqs_train)
lstm_mod.listener_accuracy(toy_color_seqs_test, toy_word_seqs_test)
```
### Illustration: Deeper models
The `Encoder` and `Decoder` are both currently hard-coded to have just one hidden layer. It is straightforward to make them deeper as long as we ensure that both the `Encoder` and `Decoder` have the same depth; since the `Encoder` final states are the initial hidden states for the `Decoder`, we need this alignment.
(Strictly speaking, we could have different numbers of `Encoder` and `Decoder` layers, as long as we did some kind of averaging or copying to achieve the hand-off from `Encoder` to `Decocer`. I'll set this possibility aside.)
__Step 1__: We need to subclass the `Encoder` and `Decoder` so that they have `num_layers` argument that is fed into the RNN cell:
```
import torch.nn as nn
from torch_color_describer import Encoder, Decoder
class DeepEncoder(Encoder):
def __init__(self, *args, num_layers=2, **kwargs):
super().__init__(*args, **kwargs)
self.num_layers = num_layers
self.rnn = nn.GRU(
input_size=self.color_dim,
hidden_size=self.hidden_dim,
num_layers=self.num_layers,
batch_first=True)
class DeepDecoder(Decoder):
def __init__(self, *args, num_layers=2, **kwargs):
super().__init__(*args, **kwargs)
self.num_layers = num_layers
self.rnn = nn.GRU(
input_size=self.embed_dim,
hidden_size=self.hidden_dim,
num_layers=self.num_layers,
batch_first=True)
```
__Step 2__: As before, we need to update the `build_graph` method of `ContextualColorDescriber`. The needed revision just uses `DeepEncoder` and `DeepDecoder`. To expose this new argument to the user, we also add a new keyword argument to `ContextualColorDescriber`:
```
from torch_color_describer import EncoderDecoder
class DeepContextualColorDescriber(ContextualColorDescriber):
def __init__(self, *args, num_layers=2, **kwargs):
self.num_layers = num_layers
super().__init__(*args, **kwargs)
def build_graph(self):
encoder = DeepEncoder(
color_dim=self.color_dim,
hidden_dim=self.hidden_dim,
num_layers=self.num_layers) # The new piece is this argument.
decoder = DeepDecoder(
vocab_size=self.vocab_size,
embed_dim=self.embed_dim,
embedding=self.embedding,
hidden_dim=self.hidden_dim,
num_layers=self.num_layers) # The new piece is this argument.
return EncoderDecoder(encoder, decoder)
```
An example/test run:
```
mod_deep = DeepContextualColorDescriber(
toy_vocab,
embed_dim=10,
hidden_dim=10,
max_iter=100,
batch_size=128)
_ = mod_deep.fit(toy_color_seqs_train, toy_word_seqs_train)
mod_deep.listener_accuracy(toy_color_seqs_test, toy_word_seqs_test)
```
|
github_jupyter
|
Uploading an image with graphical annotations stored in a CSV file
======================
We'll be using standard python tools to parse CSV and create an XML document describing cell nuclei for BisQue
Make sure you have bisque api installed:
> pip install bisque-api
```
import os
import csv
from datetime import datetime
try:
from lxml import etree
except ImportError:
import xml.etree.ElementTree as etree
```
Include BisQue API
```
from bqapi import BQSession
from bqapi.util import save_blob
```
define some paths
```
path = '.'
path_img = os.path.join(path, 'BisQue_CombinedSubtractions.lsm')
path_csv = os.path.join(path, 'BisQue_CombinedSubtractions.csv')
```
Parse CSV file and load nuclei positions
------------------------------------------
We'll create a list of XYZT coordinates with confidence
```
#x, y, z, t, confidence
coords = []
with open(path_csv, 'rb') as csvfile:
reader = csv.reader(csvfile)
h = reader.next()
for r in reader:
c = (r[0], r[1], r[2], r[4])
print c
coords.append(c)
```
Initiaize authenticated session
--------------
Initialize a BisQue session using simple user credentials
```
root = 'https://bisque.cyverse.org'
user = 'demo'
pswd = 'iplant'
session = BQSession().init_local(user, pswd, bisque_root=root, create_mex=False)
```
Create XML image descriptor
---------------------------
We'll provide a suggested path in the remote user's directory
```
path_on_bisque = 'demo/nuclei_%s/%s'%(datetime.now().strftime('%Y%m%dT%H%M%S'), os.path.basename(path_img))
resource = etree.Element('image', name=path_on_bisque)
print etree.tostring(resource, pretty_print=True)
```
Upload the image
-----------------
```
# use import service to /import/transfer activating import service
r = etree.XML(session.postblob(path_img, xml=resource)).find('./')
if r is None or r.get('uri') is None:
print 'Upload failed'
else:
print 'Uploaded ID: %s, URL: %s\n'%(r.get('resource_uniq'), r.get('uri'))
print etree.tostring(r, pretty_print=True)
```
Add graphical annotations
------------------------------
We'll create point annotaions as an XML attached to the image we just uploaded into BisQue
```
g = etree.SubElement (r, 'gobject', type='My nuclei')
for c in coords:
p = etree.SubElement(g, 'point')
etree.SubElement(p, 'vertex', x=c[0], y=c[1], z=c[2])
etree.SubElement(p, 'tag', name='confidence', value=c[3])
print etree.tostring(r, pretty_print=True)
```
Save graphical annotations to the system
------------------------------------------
After storing all annotations become searchable
```
url = session.service_url('data_service')
r = session.postxml(url, r)
if r is None or r.get('uri') is None:
print 'Adding annotations failed'
else:
print 'Image ID: %s, URL: %s'%(r.get('resource_uniq'), r.get('uri'))
```
|
github_jupyter
|
Copyright © 2017-2021 ABBYY Production LLC
```
#@title
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# *k*-means clustering
[Download the tutorial as a Jupyter notebook](https://github.com/neoml-lib/neoml/blob/master/NeoML/docs/en/Python/tutorials/KMeans.ipynb)
In this tutorial, we will use the NeoML implementation of *k*-means clustering algorithm to clusterize a randomly generated dataset.
The tutorial includes the following steps:
* [Generate the dataset](#Generate-the-dataset)
* [Cluster the data](#Cluster-the-data)
* [Visualize the results](#Visualize-the-results)
## Generate the dataset
*Note:* This section doesn't have any NeoML-specific code. It just generates a dataset. If you are not running this notebook, you may [skip](#Cluster-the-data) this section.
Let's generate a dataset of 4 clusters on the plane. Each cluster will be generated from center + noise taken from normal distribution for each coordinate.
```
import numpy as np
np.random.seed(451)
n_dots = 128
n_clusters = 4
centers = np.array([(-2., -2.),
(-2., 2.),
(2., -2.),
(2., 2.)])
X = np.zeros(shape=(n_dots, 2), dtype=np.float32)
y = np.zeros(shape=(n_dots,), dtype=np.int32)
for i in range(n_dots):
# Choose random center
cluster_id = np.random.randint(0, n_clusters)
y[i] = cluster_id
# object = center + some noise
X[i, 0] = centers[cluster_id][0] + np.random.normal(0, 1)
X[i, 1] = centers[cluster_id][1] + np.random.normal(0, 1)
```
## Cluster the data
Now we'll create a `neoml.Clustering.KMeans` class that represents the clustering algorithm, and feed the data into it.
```
import neoml
kmeans = neoml.Clustering.KMeans(max_iteration_count=1000,
cluster_count=n_clusters,
thread_count=4)
y_pred, centers_pred, vars_pred = kmeans.clusterize(X)
```
Before going further let's take a look at the returned data.
```
print('y_pred')
print(' ', type(y_pred))
print(' ', y_pred.shape)
print(' ', y_pred.dtype)
print('centers_pred')
print(' ', type(centers_pred))
print(' ', centers_pred.shape)
print(' ', centers_pred.dtype)
print('vars_pred')
print(' ', type(vars_pred))
print(' ', vars_pred.shape)
print(' ', vars_pred.dtype)
```
As you can see, the `y_pred` array contains the cluster indices of each object. `centers_pred` and `disps_pred` contain centers and variances of each cluster.
## Visualize the results
In this section we'll draw both clusterizations: ground truth and predicted.
```
%matplotlib inline
import matplotlib.pyplot as plt
colors = {
0: 'r',
1: 'g',
2: 'b',
3: 'y'
}
# Create figure with 2 subplots
fig, axs = plt.subplots(ncols=2)
fig.set_size_inches(10, 5)
# Show ground truth
axs[0].set_title('Ground truth')
axs[0].scatter(X[:, 0], X[:, 1], marker='o', c=list(map(colors.get, y)))
axs[0].scatter(centers[:, 0], centers[:, 1], marker='x', c='black')
# Show NeoML markup
axs[1].set_title('NeoML K-Means')
axs[1].scatter(X[:, 0], X[:, 1], marker='o', c=list(map(colors.get, y_pred)))
axs[1].scatter(centers_pred[:, 0], centers_pred[:, 1], marker='x', c='black')
plt.show()
```
As you can see, *k*-means didn't clusterize the outliers correctly.
|
github_jupyter
|
```
lat = 40.229730557967
lon = -74.002934930983
profile = [
{
"key": "natural",
"value": "beach",
"distance_within": 15,
"type": "bicycle",
"weight": 20
},
{
"key": "name",
"value": "Newark Penn Station",
"distance_within": 60,
"type": "auto",
"weight": 10
},
{
"key": "name",
"value": "Hurley School (historical)",
"distance_within": 20,
"type": "auto",
"weight": 10
},
{
"key": "name",
"value": "Sandy Hook Lighthouse",
"distance_within": 30,
"type": "auto",
"weight": 10
},
{
"key": "amenity",
"value": "pub",
"distance_within": 5,
"type": "pedestrian",
"weight": 20
},
{
"key": "amenity",
"value": "cafe",
"distance_within": 5,
"type": "pedestrian",
"weight": 20
},
{
"key": "amenity",
"value": "restaurant",
"distance_within": 5,
"type": "pedestrian",
"weight": 20
},
{
"key": "highway",
"value": "cycleway",
"distance_within": 10,
"type": "bicycle",
"weight": 20
},
]
query_fields = """
[
'access',
'addr:housename',
'addr:housenumber',
'addr:interpolation',
'admin_level',
'aerialway',
'aeroway',
'amenity',
'area',
'barrier',
'bicycle',
'brand',
'bridge',
'boundary',
'building',
'construction',
'covered',
'culvert',
'cutting',
'denomination',
'disused',
'embankment',
'foot',
'generator:source',
'harbour',
'highway',
'historic',
'horse',
'intermittent',
'junction',
'landuse',
'layer',
'leisure',
'lock',
'man_made',
'military',
'motorcar',
'name',
'natural',
'office',
'oneway',
'operator',
'place',
'population',
'power',
'power_source',
'public_transport',
'railway',
'ref',
'religion',
'route',
'service',
'shop',
'sport',
'surface',
'toll',
'tourism',
'tower:type',
'tunnel',
'water',
'waterway',
'wetland',
'width',
'wood'
]
"""
import psycopg2
def get_buffer(lat, lon, key_query, distance_in_meters):
conn = psycopg2.connect(host="postgres",database="osm", user="osm", password="osm")
cursor = conn.cursor()
query_fields_2 = query_fields.replace('\'', '"')
cursor.execute("""
SELECT name, keys,
ST_X(closest_point) as lon,
ST_Y(closest_point) as lat,
distance
FROM (
SELECT *, ST_Transform(ST_ClosestPoint(way,
ST_TRANSFORM(
ST_SETSRID(
ST_GeomFromText(
'POINT(%f %f)'
),
4326),
3857)), 4326) as closest_point,
ST_DISTANCE(
way,
ST_TRANSFORM(
ST_SETSRID(
ST_GeomFromText(
'POINT(%f %f)'
),
4326),
3857)
) AS distance
FROM (
SELECT osm_id, name, way,
hstore(ARRAY%s, ARRAY%s) as keys
from planet_osm_polygon
UNION ALL
SELECT osm_id, name, way,
hstore(ARRAY%s, ARRAY%s) as keys
from planet_osm_point
UNION ALL
SELECT osm_id, name, way,
hstore(ARRAY%s, ARRAY%s) as keys
from planet_osm_line) osm WHERE
ST_DWithin(way,
ST_TRANSFORM(
ST_SETSRID(
ST_GeomFromText(
'POINT(%f %f)'
),
4326),
3857), %f)
AND %s
) nearby ORDER BY DISTANCE LIMIT 2
""" % (lon, lat, lon, lat,
query_fields, query_fields_2,
query_fields, query_fields_2,
query_fields, query_fields_2,
lon, lat, distance_in_meters,
key_query))
#select * FROM (
# select
# *,
# ST_X(ST_Transform(st_centroid(way), 4326)) as lon,
# ST_Y(ST_Transform(st_centroid(way), 4326)) as lat,
# ST_DISTANCE(
# way,
# ST_TRANSFORM(
# ST_SETSRID(
# ST_GeomFromText(
# 'POINT(%f %f)'
# ),
# 4326),
# 3857)
# ) AS distance
# FROM
# planet_osm_polygon
#) AS subquery WHERE distance < %f
#ORDER BY distance;
#""" % (lon, lat, distance_in_meters))
returned_data = []
for record in cursor:
returned_data.append(record)
cursor.close()
return returned_data
costing_multiplier = {
'auto': 100,
'bicycle': 30,
'pedestrian': 10
}
scores = list()
for setting in profile:
key_query = "keys->'%s' = '%s'" % (setting['key'], setting['value'])
distance_in_meters = setting['distance_within'] * costing_multiplier[setting['type']] * 100
nearby = get_buffer(lat, lon, key_query, distance_in_meters)
if len(nearby) == 0:
nearby = [[None, None, None, None, None]]
print (key_query, distance_in_meters, nearby[0][4])
scores.append({'profile': setting, 'name': nearby[0][0],'lon': nearby[0][2], 'lat': nearby[0][3]})
#print (scores)
# Load the path from Valhalla
import requests
import json
def get_routed_distance(o_lon, o_lat, d_lon, d_lat, costing):
url = "http://valhalla:8002/route"
data = {
"locations":[
{
"lat":o_lat,
"lon":o_lon,
"type":"break"
},{
"lat":d_lat,
"lon":d_lon,
"type":"break"
}
],
"costing":costing,
"directions_options":{
"units":"miles"
}
}
data_json = json.dumps(data)
response = requests.post(url, data=data_json)
response_obj = json.loads(response.text)
#distance = response_obj['trip']['summary']['length']
time_in_seconds = response_obj['trip']['summary']['time']
#west = response_obj['trip']['summary']['min_lon']
#south = response_obj['trip']['summary']['min_lat']
#east = response_obj['trip']['summary']['max_lon']
#north = response_obj['trip']['summary']['max_lat']
#geometry = decode_geometry(response_obj['trip']['legs'][0]['shape'])
#print (distance)
#print (time_in_seconds)
return time_in_seconds
total_weights = 0
total_weighted_values = 0
for score in scores:
ideal_distance = (score['profile']['distance_within'] * 60)
value = 0
if (score['lon'] and score['lat']):
time = get_routed_distance(lon, lat, score['lon'], score['lat'], score['profile']['type'])
if time > ideal_distance:
value = 100 - (((time - ideal_distance) / 60) * 2)
elif time < ideal_distance:
value = 100 + (((ideal_distance - time) / 60) * 0.5)
else:
value = 100
if value > 200:
value = 200
elif value < 0:
value = 0
score['time'] = time
score['ideal_time'] = ideal_distance
score['value'] = value
score['weighted_time'] = value * score['profile']['weight']
total_weights = total_weights + score['profile']['weight']
total_weighted_values = total_weighted_values + score['weighted_time']
print(score['profile']['key'],score['profile']['value'],score['profile']['weight'],score['value'])
final_score = (total_weighted_values / total_weights)
print('-------------------------')
print (final_score)
```
|
github_jupyter
|
## Dependencies
```
import os
import sys
import cv2
import shutil
import random
import warnings
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from tensorflow import set_random_seed
from sklearn.utils import class_weight
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, cohen_kappa_score
from keras import backend as K
from keras.models import Model
from keras.utils import to_categorical
from keras import optimizers, applications
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import EarlyStopping, ReduceLROnPlateau, Callback, LearningRateScheduler
from keras.layers import Dense, Dropout, GlobalAveragePooling2D, GlobalMaxPooling2D, Input, Flatten, BatchNormalization, Activation
def seed_everything(seed=0):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
set_random_seed(0)
seed = 0
seed_everything(seed)
%matplotlib inline
sns.set(style="whitegrid")
warnings.filterwarnings("ignore")
sys.path.append(os.path.abspath('../input/efficientnet/efficientnet-master/efficientnet-master/'))
from efficientnet import *
```
## Load data
```
hold_out_set = pd.read_csv('../input/aptos-data-split/hold-out.csv')
X_train = hold_out_set[hold_out_set['set'] == 'train']
X_val = hold_out_set[hold_out_set['set'] == 'validation']
test = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')
print('Number of train samples: ', X_train.shape[0])
print('Number of validation samples: ', X_val.shape[0])
print('Number of test samples: ', test.shape[0])
# Preprocecss data
X_train["id_code"] = X_train["id_code"].apply(lambda x: x + ".png")
X_val["id_code"] = X_val["id_code"].apply(lambda x: x + ".png")
test["id_code"] = test["id_code"].apply(lambda x: x + ".png")
display(X_train.head())
```
# Model parameters
```
# Model parameters
FACTOR = 4
BATCH_SIZE = 8 * FACTOR
EPOCHS = 20
WARMUP_EPOCHS = 5
LEARNING_RATE = 1e-4 * FACTOR
WARMUP_LEARNING_RATE = 1e-3 * FACTOR
HEIGHT = 224
WIDTH = 224
CHANNELS = 3
ES_PATIENCE = 5
RLROP_PATIENCE = 3
DECAY_DROP = 0.5
LR_WARMUP_EPOCHS_1st = 2
LR_WARMUP_EPOCHS_2nd = 5
STEP_SIZE = len(X_train) // BATCH_SIZE
TOTAL_STEPS_1st = WARMUP_EPOCHS * STEP_SIZE
TOTAL_STEPS_2nd = EPOCHS * STEP_SIZE
WARMUP_STEPS_1st = LR_WARMUP_EPOCHS_1st * STEP_SIZE
WARMUP_STEPS_2nd = LR_WARMUP_EPOCHS_2nd * STEP_SIZE
```
# Pre-procecess images
```
train_base_path = '../input/aptos2019-blindness-detection/train_images/'
test_base_path = '../input/aptos2019-blindness-detection/test_images/'
train_dest_path = 'base_dir/train_images/'
validation_dest_path = 'base_dir/validation_images/'
test_dest_path = 'base_dir/test_images/'
# Making sure directories don't exist
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
# Creating train, validation and test directories
os.makedirs(train_dest_path)
os.makedirs(validation_dest_path)
os.makedirs(test_dest_path)
def crop_image(img, tol=7):
if img.ndim ==2:
mask = img>tol
return img[np.ix_(mask.any(1),mask.any(0))]
elif img.ndim==3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img>tol
check_shape = img[:,:,0][np.ix_(mask.any(1),mask.any(0))].shape[0]
if (check_shape == 0): # image is too dark so that we crop out everything,
return img # return original image
else:
img1=img[:,:,0][np.ix_(mask.any(1),mask.any(0))]
img2=img[:,:,1][np.ix_(mask.any(1),mask.any(0))]
img3=img[:,:,2][np.ix_(mask.any(1),mask.any(0))]
img = np.stack([img1,img2,img3],axis=-1)
return img
def circle_crop(img):
img = crop_image(img)
height, width, depth = img.shape
largest_side = np.max((height, width))
img = cv2.resize(img, (largest_side, largest_side))
height, width, depth = img.shape
x = width//2
y = height//2
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
img = crop_image(img)
return img
def preprocess_image(base_path, save_path, image_id, HEIGHT, WIDTH, sigmaX=10):
image = cv2.imread(base_path + image_id)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = circle_crop(image)
image = cv2.resize(image, (HEIGHT, WIDTH))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0,0), sigmaX), -4 , 128)
cv2.imwrite(save_path + image_id, image)
# Pre-procecss train set
for i, image_id in enumerate(X_train['id_code']):
preprocess_image(train_base_path, train_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss validation set
for i, image_id in enumerate(X_val['id_code']):
preprocess_image(train_base_path, validation_dest_path, image_id, HEIGHT, WIDTH)
# Pre-procecss test set
for i, image_id in enumerate(test['id_code']):
preprocess_image(test_base_path, test_dest_path, image_id, HEIGHT, WIDTH)
```
# Data generator
```
datagen=ImageDataGenerator(rescale=1./255,
rotation_range=360,
horizontal_flip=True,
vertical_flip=True)
train_generator=datagen.flow_from_dataframe(
dataframe=X_train,
directory=train_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="raw",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
valid_generator=datagen.flow_from_dataframe(
dataframe=X_val,
directory=validation_dest_path,
x_col="id_code",
y_col="diagnosis",
class_mode="raw",
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
seed=seed)
test_generator=datagen.flow_from_dataframe(
dataframe=test,
directory=test_dest_path,
x_col="id_code",
batch_size=1,
class_mode=None,
shuffle=False,
target_size=(HEIGHT, WIDTH),
seed=seed)
def cosine_decay_with_warmup(global_step,
learning_rate_base,
total_steps,
warmup_learning_rate=0.0,
warmup_steps=0,
hold_base_rate_steps=0):
"""
Cosine decay schedule with warm up period.
In this schedule, the learning rate grows linearly from warmup_learning_rate
to learning_rate_base for warmup_steps, then transitions to a cosine decay
schedule.
:param global_step {int}: global step.
:param learning_rate_base {float}: base learning rate.
:param total_steps {int}: total number of training steps.
:param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).
:param warmup_steps {int}: number of warmup steps. (default: {0}).
:param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).
:param global_step {int}: global step.
:Returns : a float representing learning rate.
:Raises ValueError: if warmup_learning_rate is larger than learning_rate_base, or if warmup_steps is larger than total_steps.
"""
if total_steps < warmup_steps:
raise ValueError('total_steps must be larger or equal to warmup_steps.')
learning_rate = 0.5 * learning_rate_base * (1 + np.cos(
np.pi *
(global_step - warmup_steps - hold_base_rate_steps
) / float(total_steps - warmup_steps - hold_base_rate_steps)))
if hold_base_rate_steps > 0:
learning_rate = np.where(global_step > warmup_steps + hold_base_rate_steps,
learning_rate, learning_rate_base)
if warmup_steps > 0:
if learning_rate_base < warmup_learning_rate:
raise ValueError('learning_rate_base must be larger or equal to warmup_learning_rate.')
slope = (learning_rate_base - warmup_learning_rate) / warmup_steps
warmup_rate = slope * global_step + warmup_learning_rate
learning_rate = np.where(global_step < warmup_steps, warmup_rate,
learning_rate)
return np.where(global_step > total_steps, 0.0, learning_rate)
class WarmUpCosineDecayScheduler(Callback):
"""Cosine decay with warmup learning rate scheduler"""
def __init__(self,
learning_rate_base,
total_steps,
global_step_init=0,
warmup_learning_rate=0.0,
warmup_steps=0,
hold_base_rate_steps=0,
verbose=0):
"""
Constructor for cosine decay with warmup learning rate scheduler.
:param learning_rate_base {float}: base learning rate.
:param total_steps {int}: total number of training steps.
:param global_step_init {int}: initial global step, e.g. from previous checkpoint.
:param warmup_learning_rate {float}: initial learning rate for warm up. (default: {0.0}).
:param warmup_steps {int}: number of warmup steps. (default: {0}).
:param hold_base_rate_steps {int}: Optional number of steps to hold base learning rate before decaying. (default: {0}).
:param verbose {int}: quiet, 1: update messages. (default: {0}).
"""
super(WarmUpCosineDecayScheduler, self).__init__()
self.learning_rate_base = learning_rate_base
self.total_steps = total_steps
self.global_step = global_step_init
self.warmup_learning_rate = warmup_learning_rate
self.warmup_steps = warmup_steps
self.hold_base_rate_steps = hold_base_rate_steps
self.verbose = verbose
self.learning_rates = []
def on_batch_end(self, batch, logs=None):
self.global_step = self.global_step + 1
lr = K.get_value(self.model.optimizer.lr)
self.learning_rates.append(lr)
def on_batch_begin(self, batch, logs=None):
lr = cosine_decay_with_warmup(global_step=self.global_step,
learning_rate_base=self.learning_rate_base,
total_steps=self.total_steps,
warmup_learning_rate=self.warmup_learning_rate,
warmup_steps=self.warmup_steps,
hold_base_rate_steps=self.hold_base_rate_steps)
K.set_value(self.model.optimizer.lr, lr)
if self.verbose > 0:
print('\nBatch %02d: setting learning rate to %s.' % (self.global_step + 1, lr))
```
# Model
```
def create_model(input_shape):
input_tensor = Input(shape=input_shape)
base_model = EfficientNetB5(weights=None,
include_top=False,
input_tensor=input_tensor)
base_model.load_weights('../input/efficientnet-keras-weights-b0b5/efficientnet-b5_imagenet_1000_notop.h5')
x = GlobalMaxPooling2D()(base_model.output)
x = Dropout(0.5)(x)
x = Dense(1024)(x)
x = BatchNormalization()(x)
x = Activation('relu')(x)
x = Dropout(0.5)(x)
final_output = Dense(1, activation='linear', name='final_output')(x)
model = Model(input_tensor, final_output)
return model
```
# Train top layers
```
model = create_model(input_shape=(HEIGHT, WIDTH, CHANNELS))
for layer in model.layers:
layer.trainable = False
for i in range(-7, 0):
model.layers[i].trainable = True
cosine_lr_1st = WarmUpCosineDecayScheduler(learning_rate_base=WARMUP_LEARNING_RATE,
total_steps=TOTAL_STEPS_1st,
warmup_learning_rate=0.0,
warmup_steps=WARMUP_STEPS_1st,
hold_base_rate_steps=(2 * STEP_SIZE))
metric_list = ["accuracy"]
callback_list = [cosine_lr_1st]
optimizer = optimizers.Adam(lr=WARMUP_LEARNING_RATE)
model.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)
model.summary()
STEP_SIZE_TRAIN = train_generator.n//train_generator.batch_size
STEP_SIZE_VALID = valid_generator.n//valid_generator.batch_size
history_warmup = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=WARMUP_EPOCHS,
callbacks=callback_list,
verbose=2).history
```
# Fine-tune the complete model
```
for layer in model.layers:
layer.trainable = True
es = EarlyStopping(monitor='val_loss', mode='min', patience=ES_PATIENCE, restore_best_weights=True, verbose=1)
cosine_lr_2nd = WarmUpCosineDecayScheduler(learning_rate_base=LEARNING_RATE,
total_steps=TOTAL_STEPS_2nd,
warmup_learning_rate=0.0,
warmup_steps=WARMUP_STEPS_2nd,
hold_base_rate_steps=(3 * STEP_SIZE))
callback_list = [es, cosine_lr_2nd]
optimizer = optimizers.Adam(lr=LEARNING_RATE)
model.compile(optimizer=optimizer, loss='mean_squared_error', metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=EPOCHS,
callbacks=callback_list,
verbose=2).history
fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 6))
ax1.plot(cosine_lr_1st.learning_rates)
ax1.set_title('Warm up learning rates')
ax2.plot(cosine_lr_2nd.learning_rates)
ax2.set_title('Fine-tune learning rates')
plt.xlabel('Steps')
plt.ylabel('Learning rate')
sns.despine()
plt.show()
```
# Model loss graph
```
fig, (ax1, ax2) = plt.subplots(2, 1, sharex='col', figsize=(20, 14))
ax1.plot(history['loss'], label='Train loss')
ax1.plot(history['val_loss'], label='Validation loss')
ax1.legend(loc='best')
ax1.set_title('Loss')
ax2.plot(history['acc'], label='Train accuracy')
ax2.plot(history['val_acc'], label='Validation accuracy')
ax2.legend(loc='best')
ax2.set_title('Accuracy')
plt.xlabel('Epochs')
sns.despine()
plt.show()
# Create empty arays to keep the predictions and labels
df_preds = pd.DataFrame(columns=['label', 'pred', 'set'])
train_generator.reset()
valid_generator.reset()
# Add train predictions and labels
for i in range(STEP_SIZE_TRAIN + 1):
im, lbl = next(train_generator)
preds = model.predict(im, batch_size=train_generator.batch_size)
for index in range(len(preds)):
df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'train']
# Add validation predictions and labels
for i in range(STEP_SIZE_VALID + 1):
im, lbl = next(valid_generator)
preds = model.predict(im, batch_size=valid_generator.batch_size)
for index in range(len(preds)):
df_preds.loc[len(df_preds)] = [lbl[index], preds[index][0], 'validation']
df_preds['label'] = df_preds['label'].astype('int')
def classify(x):
if x < 0.5:
return 0
elif x < 1.5:
return 1
elif x < 2.5:
return 2
elif x < 3.5:
return 3
return 4
# Classify predictions
df_preds['predictions'] = df_preds['pred'].apply(lambda x: classify(x))
train_preds = df_preds[df_preds['set'] == 'train']
validation_preds = df_preds[df_preds['set'] == 'validation']
```
# Model Evaluation
## Confusion Matrix
### Original thresholds
```
labels = ['0 - No DR', '1 - Mild', '2 - Moderate', '3 - Severe', '4 - Proliferative DR']
def plot_confusion_matrix(train, validation, labels=labels):
train_labels, train_preds = train
validation_labels, validation_preds = validation
fig, (ax1, ax2) = plt.subplots(1, 2, sharex='col', figsize=(24, 7))
train_cnf_matrix = confusion_matrix(train_labels, train_preds)
validation_cnf_matrix = confusion_matrix(validation_labels, validation_preds)
train_cnf_matrix_norm = train_cnf_matrix.astype('float') / train_cnf_matrix.sum(axis=1)[:, np.newaxis]
validation_cnf_matrix_norm = validation_cnf_matrix.astype('float') / validation_cnf_matrix.sum(axis=1)[:, np.newaxis]
train_df_cm = pd.DataFrame(train_cnf_matrix_norm, index=labels, columns=labels)
validation_df_cm = pd.DataFrame(validation_cnf_matrix_norm, index=labels, columns=labels)
sns.heatmap(train_df_cm, annot=True, fmt='.2f', cmap="Blues",ax=ax1).set_title('Train')
sns.heatmap(validation_df_cm, annot=True, fmt='.2f', cmap=sns.cubehelix_palette(8),ax=ax2).set_title('Validation')
plt.show()
plot_confusion_matrix((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))
```
## Quadratic Weighted Kappa
```
def evaluate_model(train, validation):
train_labels, train_preds = train
validation_labels, validation_preds = validation
print("Train Cohen Kappa score: %.3f" % cohen_kappa_score(train_preds, train_labels, weights='quadratic'))
print("Validation Cohen Kappa score: %.3f" % cohen_kappa_score(validation_preds, validation_labels, weights='quadratic'))
print("Complete set Cohen Kappa score: %.3f" % cohen_kappa_score(np.append(train_preds, validation_preds), np.append(train_labels, validation_labels), weights='quadratic'))
evaluate_model((train_preds['label'], train_preds['predictions']), (validation_preds['label'], validation_preds['predictions']))
```
## Apply model to test set and output predictions
```
def apply_tta(model, generator, steps=10):
step_size = generator.n//generator.batch_size
preds_tta = []
for i in range(steps):
generator.reset()
preds = model.predict_generator(generator, steps=step_size)
preds_tta.append(preds)
return np.mean(preds_tta, axis=0)
preds = apply_tta(model, test_generator)
predictions = [classify(x) for x in preds]
results = pd.DataFrame({'id_code':test['id_code'], 'diagnosis':predictions})
results['id_code'] = results['id_code'].map(lambda x: str(x)[:-4])
# Cleaning created directories
if os.path.exists(train_dest_path):
shutil.rmtree(train_dest_path)
if os.path.exists(validation_dest_path):
shutil.rmtree(validation_dest_path)
if os.path.exists(test_dest_path):
shutil.rmtree(test_dest_path)
```
# Predictions class distribution
```
fig = plt.subplots(sharex='col', figsize=(24, 8.7))
sns.countplot(x="diagnosis", data=results, palette="GnBu_d").set_title('Test')
sns.despine()
plt.show()
results.to_csv('submission.csv', index=False)
display(results.head())
```
|
github_jupyter
|
# Exp 43 analysis
See `./informercial/Makefile` for experimental
details.
```
import os
import numpy as np
from IPython.display import Image
import matplotlib
import matplotlib.pyplot as plt`
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import seaborn as sns
sns.set_style('ticks')
matplotlib.rcParams.update({'font.size': 16})
matplotlib.rc('axes', titlesize=16)
from infomercial.exp import meta_bandit
from infomercial.local_gym import bandit
from infomercial.exp.meta_bandit import load_checkpoint
import gym
# ls ../data/exp2*
```
# Load and process data
```
data_path ="/Users/qualia/Code/infomercial/data/"
exp_name = "exp43"
best_params = load_checkpoint(os.path.join(data_path, f"{exp_name}_best.pkl"))
sorted_params = load_checkpoint(os.path.join(data_path, f"{exp_name}_sorted.pkl"))
best_params
```
# Performance
of best parameters
```
env_name = 'BanditOneHigh2-v0'
num_episodes = 20*100
# Run w/ best params
result = meta_bandit(
env_name=env_name,
num_episodes=num_episodes,
lr=best_params["lr"],
tie_threshold=best_params["tie_threshold"],
seed_value=19,
save="exp43_best_model.pkl"
)
# Plot run
episodes = result["episodes"]
actions =result["actions"]
scores_R = result["scores_R"]
values_R = result["values_R"]
scores_E = result["scores_E"]
values_E = result["values_E"]
# Get some data from the gym...
env = gym.make(env_name)
best = env.best
print(f"Best arm: {best}, last arm: {actions[-1]}")
# Init plot
fig = plt.figure(figsize=(6, 14))
grid = plt.GridSpec(5, 1, wspace=0.3, hspace=0.8)
# Do plots:
# Arm
plt.subplot(grid[0, 0])
plt.scatter(episodes, actions, color="black", alpha=.5, s=2, label="Bandit")
plt.plot(episodes, np.repeat(best, np.max(episodes)+1),
color="red", alpha=0.8, ls='--', linewidth=2)
plt.ylim(-.1, np.max(actions)+1.1)
plt.ylabel("Arm choice")
plt.xlabel("Episode")
# score
plt.subplot(grid[1, 0])
plt.scatter(episodes, scores_R, color="grey", alpha=0.4, s=10, label="R")
plt.scatter(episodes, scores_E, color="purple", alpha=0.9, s=10, label="E")
plt.ylabel("log score")
plt.xlabel("Episode")
plt.semilogy()
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
_ = sns.despine()
# Q
plt.subplot(grid[2, 0])
plt.scatter(episodes, values_R, color="grey", alpha=0.4, s=10, label="R")
plt.scatter(episodes, values_E, color="purple", alpha=0.4, s=10, label="E")
plt.ylabel("log Q(s,a)")
plt.xlabel("Episode")
plt.semilogy()
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
_ = sns.despine()
# -
plt.savefig("figures/epsilon_bandit.pdf", bbox_inches='tight')
plt.savefig("figures/epsilon_bandit.eps", bbox_inches='tight')
```
# Sensitivity
to parameter choices
```
total_Rs = []
ties = []
lrs = []
trials = list(sorted_params.keys())
for t in trials:
total_Rs.append(sorted_params[t]['total_E'])
ties.append(sorted_params[t]['tie_threshold'])
lrs.append(sorted_params[t]['lr'])
# Init plot
fig = plt.figure(figsize=(10, 18))
grid = plt.GridSpec(4, 1, wspace=0.3, hspace=0.8)
# Do plots:
# Arm
plt.subplot(grid[0, 0])
plt.scatter(trials, total_Rs, color="black", alpha=.5, s=6, label="total R")
plt.xlabel("Sorted params")
plt.ylabel("total R")
_ = sns.despine()
plt.subplot(grid[1, 0])
plt.scatter(trials, ties, color="black", alpha=.3, s=6, label="total R")
plt.xlabel("Sorted params")
plt.ylabel("Tie threshold")
_ = sns.despine()
plt.subplot(grid[2, 0])
plt.scatter(trials, lrs, color="black", alpha=.5, s=6, label="total R")
plt.xlabel("Sorted params")
plt.ylabel("lr")
_ = sns.despine()
```
# Distributions
of parameters
```
# Init plot
fig = plt.figure(figsize=(5, 6))
grid = plt.GridSpec(2, 1, wspace=0.3, hspace=0.8)
plt.subplot(grid[0, 0])
plt.hist(ties, color="black")
plt.xlabel("tie threshold")
plt.ylabel("Count")
_ = sns.despine()
plt.subplot(grid[1, 0])
plt.hist(lrs, color="black")
plt.xlabel("lr")
plt.ylabel("Count")
_ = sns.despine()
```
of total reward
```
# Init plot
fig = plt.figure(figsize=(5, 2))
grid = plt.GridSpec(1, 1, wspace=0.3, hspace=0.8)
plt.subplot(grid[0, 0])
plt.hist(total_Rs, color="black", bins=50)
plt.xlabel("Total reward")
plt.ylabel("Count")
plt.xlim(0, 10)
_ = sns.despine()
```
|
github_jupyter
|
```
!pip install wandb
!wandb login
from collections import deque
import random
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision.transforms as transforms
import gym
import wandb
class Actor(nn.Module):
def __init__(self, num_actions):
super().__init__()
# Create the layers for the model
self.actor = nn.Sequential(
nn.Conv2d(
in_channels=3, out_channels=32,
kernel_size=5, padding=2, stride=2
), # (32, 32, 32)
nn.BatchNorm2d(32),
nn.ReLU(),
nn.Conv2d(
in_channels=32, out_channels=64,
kernel_size=3, padding=1, stride=2
), # (64, 16, 16)
nn.BatchNorm2d(64),
nn.ReLU(),
nn.Conv2d(
in_channels=64, out_channels=64,
kernel_size=3, padding=1, stride=2
), # (64, 8, 8)
nn.BatchNorm2d(64),
nn.ReLU(),
nn.Conv2d(
in_channels=64, out_channels=128,
kernel_size=3, padding=1, stride=2
), # (128, 4, 4)
nn.BatchNorm2d(128),
nn.ReLU(),
nn.Flatten(start_dim=1), # (2048)
nn.Linear(128 * 4 * 4, 512),
nn.ReLU(),
nn.Linear(512, num_actions),
nn.Softmax(dim=-1)
)
def forward(self, x):
return self.actor(x)
class Critic(nn.Module):
def __init__(self, act_dim):
super().__init__()
# Create the layers for the model
self.critic = nn.Sequential(
nn.Conv2d(
in_channels=3, out_channels=32,
kernel_size=5, padding=2, stride=2
), # (32, 32, 32)
nn.BatchNorm2d(32),
nn.ReLU(),
nn.Conv2d(
in_channels=32, out_channels=64,
kernel_size=3, padding=1, stride=2
), # (64, 16, 16)
nn.BatchNorm2d(64),
nn.ReLU(),
nn.Conv2d(
in_channels=64, out_channels=64,
kernel_size=3, padding=1, stride=2
), # (64, 8, 8)
nn.BatchNorm2d(64),
nn.ReLU(),
nn.Conv2d(
in_channels=64, out_channels=128,
kernel_size=3, padding=1, stride=2
), # (128, 4, 4)
nn.BatchNorm2d(128),
nn.ReLU(),
nn.Flatten(start_dim=1), # (2048)
)
self.fc = nn.Sequential(
nn.Linear(128 * 4 * 4 + act_dim, 512),
nn.ReLU(),
nn.Linear(512, 1),
nn.Tanh()
)
def forward(self, state, action):
x = self.critic(state)
x = torch.cat([x, action], dim=1)
x = self.fc(x)
return x
class ReplayMemory:
def __init__(self, max_len):
self.replay = deque(maxlen=max_len)
def store_experience(self, state, reward,
action, next_state,
done):
self.replay.append([state, reward, action, next_state, done])
def size(self):
return len(self.replay)
def sample(self, batch_size):
if len(self.replay) < batch_size:
return None
return random.sample(self.replay, k=batch_size)
class DDPG:
def __init__(self, memory_size, num_actions,
actor_lr, critic_lr, gamma,
tau, device, img_transforms):
# Set up model
self.actor = Actor(num_actions).to(device)
self.target_actor = Actor(num_actions).to(device)
self.target_actor.eval()
self.critic = Critic(num_actions).to(device)
self.target_critic = Critic(num_actions).to(device)
self.target_critic.eval()
# Set up optimizer and criterion
self.critic_criterion = nn.MSELoss()
self.actor_optim = torch.optim.Adam(self.actor.parameters(), lr=actor_lr)
self.critic_optim = torch.optim.Adam(self.critic.parameters(), lr=critic_lr)
# Set up transforms and other hyper-parameters
self.device = device
self.img_transforms = img_transforms
self.num_actions = num_actions
self.memory = ReplayMemory(memory_size)
self.gamma = gamma
self.tau = tau
def choose_action(self, cur_state, eps):
# Open evaluation mode
self.actor.eval()
# Exploration
if np.random.uniform() < eps:
action = np.random.randint(0, self.num_actions)
else: # Exploitation
cur_state = self.img_transforms(cur_state).to(self.device).unsqueeze(0)
action_list = self.actor(cur_state)
action = torch.argmax(action_list, dim=-1).item()
# Open training mode
self.actor.train()
return action
def actor_update(self, batch_data):
# Separate the data into groups
cur_state_batch = []
for cur_state, *_ in batch_data:
cur_state_batch.append(self.img_transforms(cur_state).unsqueeze(0))
cur_state_batch = torch.cat(cur_state_batch, dim=0).to(self.device)
actor_actions = F.gumbel_softmax(torch.log(F.softmax(self.actor(cur_state_batch), dim=1)), hard=True)
loss = -self.critic(cur_state_batch, actor_actions).mean()
self.actor_optim.zero_grad()
loss.backward()
self.actor_optim.step()
def critic_update(self, batch_data):
# Separate the data into groups
cur_state_batch = []
reward_batch = []
action_batch = []
next_state_batch = []
done_batch = []
for cur_state, reward, action, next_state, done in batch_data:
cur_state_batch.append(self.img_transforms(cur_state).unsqueeze(0))
reward_batch.append(reward)
action_batch.append(action)
next_state_batch.append(self.img_transforms(next_state).unsqueeze(0))
done_batch.append(done)
cur_state_batch = torch.cat(cur_state_batch, dim=0).to(self.device)
reward_batch = torch.FloatTensor(reward_batch).to(self.device)
action_batch = torch.LongTensor(action_batch)
action_batch = torch.zeros(len(batch_data), self.num_actions).scatter_(
1, action_batch.unsqueeze(1), 1).to(self.device)
next_state_batch = torch.cat(next_state_batch, dim=0).to(self.device)
done_batch = torch.Tensor(done_batch).to(self.device)
# Compute the TD error between eval and target
Q_eval = self.critic(cur_state_batch, action_batch)
next_action = F.softmax(self.target_actor(next_state_batch), dim=1)
index = torch.argmax(next_action, dim=1).unsqueeze(1)
next_action = torch.zeros_like(next_action).scatter_(1, index, 1).to(self.device)
Q_target = reward_batch + self.gamma * (1 - done_batch) * self.target_critic(next_state_batch,
next_action).squeeze(1)
loss = self.critic_criterion(Q_eval.squeeze(1), Q_target)
self.critic_optim.zero_grad()
loss.backward()
self.critic_optim.step()
def soft_update(self):
# EMA for both actor and critic network
for param, target_param in zip(self.actor.parameters(), self.target_actor.parameters()):
target_param.data.copy_(self.tau * param.data + (1 - self.tau) * target_param.data)
for param, target_param in zip(self.critic.parameters(), self.target_critic.parameters()):
target_param.data.copy_(self.tau * param.data + (1 - self.tau) * target_param.data)
env = gym.make("snake:snake-v0", mode="hardworking")
device = "cpu"
# Set up environment hyperparameters
num_actions = env.action_space.n
# Set up training hyperparameters
tau = 0.05
max_time_steps = 100000
max_iter = 2000
gamma = 0.9
memory_size = 2000
batch_size = 32
actor_lr = 3e-4
critic_lr = 3e-4
def train(max_time_steps, max_iter, memory_size,
num_actions, actor_lr, critic_lr,
gamma, tau, device, batch_size):
# Set up model training
img_transforms = transforms.Compose([
transforms.ToTensor(),
transforms.Resize((64, 64))
])
ddpg = DDPG(
memory_size, num_actions,
actor_lr, critic_lr, gamma,
tau, device, img_transforms
)
max_reward = -1e-9
running_reward = 0
running_episodes = 0
time_step = 0
print_freq = max_iter * 2
while time_step < max_time_steps:
state = env.reset()
current_ep_reward = 0
for _ in range(max_iter):
# Get reward and state
actions = ddpg.choose_action(state["frame"], 0.1)
new_state, reward, done, _ = env.step(actions)
current_ep_reward += reward
ddpg.memory.store_experience(state["frame"], reward, actions, new_state["frame"], done)
state = new_state
if done:
break
# Wait for updating
if ddpg.memory.size() < batch_size:
continue
batch_data = ddpg.memory.sample(batch_size)
ddpg.critic_update(batch_data)
ddpg.actor_update(batch_data)
ddpg.soft_update()
time_step += 1
if time_step % print_freq == 0:
avg_reward = running_reward / running_episodes
print(f"Iteration:{running_episodes}, get average reward: {avg_reward:.2f}")
running_reward = 0
running_episodes = 0
log = {
"avg_reward": avg_reward,
}
wandb.log(log)
if avg_reward > max_reward:
max_reward = avg_reward
torch.save(ddpg.actor.state_dict(), "actor_best.pt")
torch.save(ddpg.critic.state_dict(), "critic_best.pt")
running_reward += current_ep_reward
running_episodes += 1
model_config = {
"gamma": gamma,
"max_time_steps": max_time_steps,
"memory size": memory_size
}
run = wandb.init(
project="snake_RL",
resume=False,
config=model_config,
name="DDPG"
)
train(
max_time_steps, max_iter, memory_size,
4, actor_lr, critic_lr,
gamma, tau, "cpu", batch_size
)
```
|
github_jupyter
|
## Introduction
If you've had any experience with the python scientific stack, you've probably come into contact with, or at least heard of, the [pandas][1] data analysis library. Before the introduction of pandas, if you were to ask anyone what language to learn as a budding data scientist, most would've likely said the [R statistical programming language][2]. With its [data frame][3] data structure, it was the obvious winner when it came to filtering, slicing, aggregating, or analyzing your data. However, with the introduction of pandas to python's growing set of data analysis libraries, the gap between the two langauges has effectively closed, and as a result, pandas has become a vital tool for data scientists using python.
While we won't be covering the pandas library itself, since that's a topic fit for a course of its own, in this lesson we will be discussing the simple interface pandas provides for interacting with the matplotlib library. In addition, we'll also take a look at the recent changes the matplotlib team has made to make it possible for the two libraries to work together more harmoniously.
That said, let's get set up and see what pandas has to offer.
[1]: http://pandas.pydata.org/
[2]: https://www.r-project.org/
[3]: https://cran.r-project.org/doc/manuals/r-release/R-intro.html#Data-frames
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('retina')
```
### What is pandas?
Pandas is a library created by [Wes McKinney][1] that provides several data structures that make working with data fast, efficient, and easy. Chief among them is the `DataFrame`, which takes on R's `data.frame` data type, and in many scenarios, bests it. It also provides a simple wrapper around the `pyplot` interface, allowing you to plot the data in your `DataFrame` objects without any context switching in many cases.
But, enough talk, let's see it in action.
[1]: https://twitter.com/wesmckinn
### Import the Library
The following bit of code imports the pandas library using the widely accepted `pd` naming convention. You'll likely see pandas imported like this just about everywhere it's used, and it is recommended that you always use the same naming convention in your code as well.
```
import pandas as pd
```
### Load in Some Data
In the next cell, we'll use the `read_csv` function to load in the [Census Income][1] dataset from the [UCI Machine Learning Repository][2]. Incidentally, this is the exact same dataset that we used in our Exploratory Data Analysis (EDA) example in chapter 2, so we'll get to see some examples of how we could perform some of the same steps using the plotting commands on our `DataFrame` object.
[1]: http://archive.ics.uci.edu/ml/datasets/Adult
[2]: http://archive.ics.uci.edu/ml/index.html
```
import pandas as pd
# Download and read in the data from the UCI Machine Learning Repository
df = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data',
header=None,
names=('age',
'workclass',
'fnlwgt',
'education',
'education_num',
'marital_status',
'occupation',
'relationship',
'race',
'sex',
'capital_gain',
'capital_loss',
'hours_per_week',
'native_country',
'target'))
```
### Plotting With pandas
Just like we did in our EDA example from chapter 2, we can once again create a simple histogram from our data. This time though, notice that we simply call the `hist` command on the column that contains the education level to plot our data.
```
df.education_num.hist(bins=16);
```
And, remember, pandas isn't doing anything magical here, it's just providing a very simple wrapper around the `pyplot` module. At the end of the day, the code above is simply calling the `pyplot.hist` function to create the histogram. So, we can interact with the plot that it produces the same way we would any other plot. As an example, let's create our histogram again, but this time let's get rid of that empty bar to the left by setting the plot's x-axis limits using the `pyplot.xlim` function.
```
df.education_num.hist(bins=16)
# Remove the empty bar from the histogram that's below the
# education_num's minimum value.
plt.xlim(df.education_num.min(), df.education_num.max());
```
Well, that looks better, but we're still stuck with many of the same problems that we had in the original EDA lesson. You'll notice that most of the x-ticks don't actually line up with their bars, and there's a good reason for that. Remember, in that lesson, we discussed how a histogram was meant to be used with continuous data, and in our case we're dealing with discrete values. So, a bar chart is actually what we want to use here.
Luckily, pandas makes the task of creating the bar chart even easier. In our EDA lesson, we had to do the frequency count ourselves, and take care of lining the x-axis labels up properly, and several other small issues. With pandas, it's just a single line of code. First, we call the `value_counts` function on the `education` column to get a set of frequency counts, ordered largest to smallest, for each education level. Then, we call the `plot` function on the `Series` object returned from `value_counts`, and pass in the type of plot with the `kind` parameter, and while we're at it, we'll set our width to 1, like we did in the chapter 2 example, to make it look more histogram-ish.
```
df.education.value_counts().plot(kind='bar', width=1);
```
Now, rather than passing in the plot type with the `kind` parameter, we could've also just called the `bar` function from the `plot` object, like we do in the next cell.
```
df.education.value_counts().plot.bar(width=1);
```
Ok, so that's a pretty good introduction to the simple interface that pandas provides to the matplotlib library, but it doesn't stop there. Pandas also provides a handful of more complex plotting functions in the `pandas.tools.plotting` module. So, let's import another dataset and take a look at an example of what's available.
In the cell below, we pull in the Iris dataset that we used in our scatterplot matrix example from chapter 3. Incidentally, if you don't want to mess with network connections, or if you happen to be in a situation where network access just isn't an option, I've copied the data file to the local data folder. The file can be found at `./data/iris_data.csv`
```
df = pd.read_csv('https://raw.githubusercontent.com/pydata/pandas/master/pandas/tests/data/iris.csv')
```
We'll need a color map, essentially just a dictionary mapping each species to a unique color, so we'll put one together in the next cell. Fortunately, pandas makes it easy to get the species names by simply calling the `unique` function on the `Name` column.
```
names = df.Name.unique()
colors = ['red', 'green', 'blue']
cmap = dict(zip(names, colors))
```
Now, before we take a look at one of the functions from the `plotting` module, let's quickly take a look at one of the [changes that was made to matplotlib in version 1.5][1] to accommodate labeled data, like a pandas `DataFrame` for example. The code in the next cell, creates a scatter plot using the `pyplot.scatter` function, like we've done in the past, but notice how we specify the columns that contain our `x` and `y` values. In our example below, we are simply passing in the names of the columns alongside the `DataFrame` object itself. Now, it's arguable just how much more readable this light layer of abstraction is over just passing in the data directly, but it's nice to have the option, nonetheless.
[1]: http://matplotlib.org/users/whats_new.html#working-with-labeled-data-like-pandas-dataframes
```
plt.scatter(x='PetalLength', y='PetalWidth', data=df, c=df.Name.apply(lambda name: cmap[name]));
```
Now, we're ready to take a look at one of the functions that pandas provides us, and for comparison sake, let's take a look at our old friend, the scatterplot matrix. In the next cell, we'll import the `scatter_matrix` function from the `pandas.tools.plotting` module and run it on the Iris dataset.
```
from pandas.tools.plotting import scatter_matrix
scatter_matrix(df, figsize=(10,8), c=df.Name.apply(lambda name: cmap[name]), s=40);
```
Well, that looks pretty good, and it's a heck of a lot easier than writing our own. Though, I prefer the seaborn version, this one will do in a pinch. If you get a chance, I recommend taking a look at what the pandas `plotting` module has to offer. Aside from the scatterplot matrix function, it also provides functions for creating things like density plots and parallel coordinates plots. It's not as powerful as the seaborn library, but many times it may be all you need to perform your analysis.
## Conclusion
And, that will bring us to the end once again.
To recap, in this lesson, we saw some quick examples of how to use the Pandas data analysis library with the matplotlib library. Specifically, we saw a few examples of the simple interface that pandas provides to the `pyplot` module, and we also saw one example of the new labeled data change that was made to matplotlib in version 1.5. Finally, we took a quick look at one of the more complex functions that the pandas `plotting` module provides.
The main goal of this lesson wasn't to turn you into a pandas power user, but rather to give you some idea of what pandas provides, and more importantly, to take away any of the mystery of how it works. Now that you understand that pandas is basically just providing a very simple layer of abstraction on top of the `pyplot` interface, you should be prepared to deal with any issues that come up when plotting the data in your `DataFrame` objects.
|
github_jupyter
|
```
# Load WSC dataset
import xml.etree.ElementTree as etree
import json
import numpy as np
import logging
import numpy
import os
def softmax(x):
return np.exp(x)/sum(np.exp(x))
tree = etree.parse('WSCollection.xml')
root = tree.getroot()
original_problems = root.getchildren()
problems = list()
for original_problem in original_problems:
problem = dict()
for information in original_problem.getchildren():
if information.tag == 'answers':
answers = information.getchildren()
answer_list = list()
for answer in answers:
answer_list.append(answer.text.strip())
problem['answers'] = answer_list
elif information.tag == 'text':
texts = information.getchildren()
text_dict = dict()
for text1 in texts:
text_dict[text1.tag] = text1.text.replace('\n', ' ').strip()
problem['text'] = text_dict
elif information.tag == 'quote':
pass
else:
problem[information.tag] = information.text.replace(' ', '')
problems.append(problem)
print(problems[0])
all_sentences = list()
for question in problems:
sentence = question['text']['txt1'] + ' ' + question['text']['pron'] + ' ' + question['text']['txt2']
all_sentences.append(sentence)
# print(sentence)
import json
import numpy as np
import tensorflow as tf
import model, sample, encoder
model_name = '774M'
models_dir = '../models'
enc = encoder.get_encoder(model_name, models_dir)
batch_size = 1
seed=None
nsamples=1
hparams = model.default_hparams()
with open(os.path.join(models_dir, model_name, 'hparams.json')) as f:
hparams.override_from_dict(json.load(f))
length = hparams.n_ctx // 2
answer_collector = []
def logits_score(logits,skeleton_tokens, context_tokens):
score = 1
start_index = len(skeleton_tokens) - 1
end_index = len(context_tokens) - 1
for i in range(end_index - start_index):
m = softmax(logits[start_index+i])
score *= m[context_tokens[start_index+i+1]]
return score
with tf.Session(graph=tf.Graph()) as sess:
context = tf.placeholder(tf.int32, [batch_size, None])
np.random.seed(seed)
tf.set_random_seed(seed)
context_tokens = []
output = model.model(hparams=hparams, X=context, past=None, reuse=tf.AUTO_REUSE)
saver = tf.train.Saver()
ckpt = tf.train.latest_checkpoint(os.path.join(models_dir, model_name))
saver.restore(sess, ckpt)
for i in range(273):
if problems[i]['text']['txt1'] != ".":
ans0 = problems[i]['answers'][0].replace("The","the")
ans1 = problems[i]['answers'][1].replace("The","the")
else:
ans0 = problems[i]['answers'][0]
ans1 = problems[i]['answers'][1]
skeleton1 = problems[i]['text']['txt1'] + ' ' + problems[i]['answers'][0]
skeleton2 = problems[i]['text']['txt1'] + ' ' + problems[i]['answers'][1]
raw_text1 = problems[i]['text']['txt1'] + ' ' + problems[i]['answers'][0] + ' ' + problems[i]['text']['txt2']
raw_text2 = problems[i]['text']['txt1'] + ' ' + problems[i]['answers'][1] + ' ' + problems[i]['text']['txt2']
context_tokens1 = enc.encode(raw_text1)
context_tokens2 = enc.encode(raw_text2)
skeleton_tokens1 = enc.encode(skeleton1)
skeleton_tokens2 = enc.encode(skeleton2)
out1 = sess.run(output, feed_dict={context: [context_tokens1 for _ in range(batch_size)]})
out2 = sess.run(output, feed_dict={context: [context_tokens2 for _ in range(batch_size)]})
logits1 = out1['logits'][:, :, :hparams.n_vocab]
logits2 = out2['logits'][:, :, :hparams.n_vocab]
score1 = logits_score(logits1[0],skeleton_tokens1,context_tokens1)
score2 = logits_score(logits2[0],skeleton_tokens2,context_tokens2)
correctAnswer = problems[i]["correctAnswer"]
if score1 >= score2:
predictedAnswer = "A"
else:
predictedAnswer = "B"
# A. Problem
answer_collector.append(predictedAnswer in correctAnswer)
print(len(answer_collector))
print(np.sum(answer_collector)/273)
```
|
github_jupyter
|
## Analyze A/B Test Results
You may either submit your notebook through the workspace here, or you may work from your local machine and submit through the next page. Either way assure that your code passes the project [RUBRIC](https://review.udacity.com/#!/projects/37e27304-ad47-4eb0-a1ab-8c12f60e43d0/rubric). **Please save regularly.**
This project will assure you have mastered the subjects covered in the statistics lessons. The hope is to have this project be as comprehensive of these topics as possible. Good luck!
## Table of Contents
- [Introduction](#intro)
- [Part I - Probability](#probability)
- [Part II - A/B Test](#ab_test)
- [Part III - Regression](#regression)
<a id='intro'></a>
### Introduction
A/B tests are very commonly performed by data analysts and data scientists. It is important that you get some practice working with the difficulties of these
For this project, you will be working to understand the results of an A/B test run by an e-commerce website. Your goal is to work through this notebook to help the company understand if they should implement the new page, keep the old page, or perhaps run the experiment longer to make their decision.
**As you work through this notebook, follow along in the classroom and answer the corresponding quiz questions associated with each question.** The labels for each classroom concept are provided for each question. This will assure you are on the right track as you work through the project, and you can feel more confident in your final submission meeting the criteria. As a final check, assure you meet all the criteria on the [RUBRIC](https://review.udacity.com/#!/projects/37e27304-ad47-4eb0-a1ab-8c12f60e43d0/rubric).
<a id='probability'></a>
#### Part I - Probability
To get started, let's import our libraries.
```
import pandas as pd
import numpy as np
import random
import matplotlib.pyplot as plt
%matplotlib inline
#We are setting the seed to assure you get the same answers on quizzes as we set up
random.seed(42)
```
`1.` Now, read in the `ab_data.csv` data. Store it in `df`. **Use your dataframe to answer the questions in Quiz 1 of the classroom.**
a. Read in the dataset and take a look at the top few rows here:
```
df = pd.read_csv('ab_data.csv')
df.head()
```
b. Use the cell below to find the number of rows in the dataset.
```
df.shape[0]
```
c. The number of unique users in the dataset.
```
df.nunique()[0]
```
d. The proportion of users converted.
```
df['converted'].sum() / df.shape[0]
```
e. The number of times the `new_page` and `treatment` don't match.
```
df[((df['group'] == 'treatment') & (df['landing_page'] != 'new_page')) | ((df['group'] != 'treatment') & (df['landing_page'] == 'new_page'))].shape[0]
```
f. Do any of the rows have missing values?
```
df.info()
```
`2.` For the rows where **treatment** does not match with **new_page** or **control** does not match with **old_page**, we cannot be sure if this row truly received the new or old page. Use **Quiz 2** in the classroom to figure out how we should handle these rows.
a. Now use the answer to the quiz to create a new dataset that meets the specifications from the quiz. Store your new dataframe in **df2**.
```
df2 = df[(((df['group'] == 'treatment') & (df['landing_page'] == 'new_page')) | ((df['group'] == 'control') & (df['landing_page'] == 'old_page')))]
df2.head()
# Double Check all of the correct rows were removed - this should be 0
df2[((df2['group'] == 'treatment') == (df2['landing_page'] == 'new_page')) == False].shape[0]
```
`3.` Use **df2** and the cells below to answer questions for **Quiz3** in the classroom.
a. How many unique **user_id**s are in **df2**?
```
df2.nunique()[0]
```
b. There is one **user_id** repeated in **df2**. What is it?
```
uid = df2[df2['user_id'].duplicated() == True].index[0]
uid
```
c. What is the row information for the repeat **user_id**?
```
df2.loc[uid]
```
d. Remove **one** of the rows with a duplicate **user_id**, but keep your dataframe as **df2**.
```
df2.drop(2893, inplace=True)
df2.shape[0]
```
`4.` Use **df2** in the cells below to answer the quiz questions related to **Quiz 4** in the classroom.
a. What is the probability of an individual converting regardless of the page they receive?
```
df2[df2['converted'] == 1].shape[0] / df2.shape[0]
```
b. Given that an individual was in the `control` group, what is the probability they converted?
```
df2[(df2['converted'] == 1) & ((df2['group'] == 'control'))].shape[0] / df2[(df2['group'] == 'control')].shape[0]
```
c. Given that an individual was in the `treatment` group, what is the probability they converted?
```
df2[(df2['converted'] == 1) & ((df2['group'] == 'treatment'))].shape[0] / df2[(df2['group'] == 'treatment')].shape[0]
```
d. What is the probability that an individual received the new page?
```
df2[df2['landing_page'] == 'new_page'].shape[0] / df2.shape[0]
```
e. Consider your results from parts (a) through (d) above, and explain below whether you think there is sufficient evidence to conclude that the new treatment page leads to more conversions.
**The probability of converting for an individual who received the control page is more than that who received the treatment page. So its more likely to convert for the control page viewers. So there is not much evidence to prove that the new treatment page leads to more conversions**
<a id='ab_test'></a>
### Part II - A/B Test
Notice that because of the time stamp associated with each event, you could technically run a hypothesis test continuously as each observation was observed.
However, then the hard question is do you stop as soon as one page is considered significantly better than another or does it need to happen consistently for a certain amount of time? How long do you run to render a decision that neither page is better than another?
These questions are the difficult parts associated with A/B tests in general.
`1.` For now, consider you need to make the decision just based on all the data provided. If you want to assume that the old page is better unless the new page proves to be definitely better at a Type I error rate of 5%, what should your null and alternative hypotheses be? You can state your hypothesis in terms of words or in terms of **$p_{old}$** and **$p_{new}$**, which are the converted rates for the old and new pages.
**Put your answer here.**
`2.` Assume under the null hypothesis, $p_{new}$ and $p_{old}$ both have "true" success rates equal to the **converted** success rate regardless of page - that is $p_{new}$ and $p_{old}$ are equal. Furthermore, assume they are equal to the **converted** rate in **ab_data.csv** regardless of the page. <br><br>
Use a sample size for each page equal to the ones in **ab_data.csv**. <br><br>
Perform the sampling distribution for the difference in **converted** between the two pages over 10,000 iterations of calculating an estimate from the null. <br><br>
Use the cells below to provide the necessary parts of this simulation. If this doesn't make complete sense right now, don't worry - you are going to work through the problems below to complete this problem. You can use **Quiz 5** in the classroom to make sure you are on the right track.<br><br>
```
df2.head()
```
a. What is the **conversion rate** for $p_{new}$ under the null?
```
p_new = df2[(df2['converted'] == 1)].shape[0] / df2.shape[0]
p_new
```
b. What is the **conversion rate** for $p_{old}$ under the null? <br><br>
```
p_old = df2[(df2['converted'] == 1)].shape[0] / df2.shape[0]
p_old
```
c. What is $n_{new}$, the number of individuals in the treatment group?
```
n_new = df2[(df2['landing_page'] == 'new_page') & (df2['group'] == 'treatment')].shape[0]
n_new
```
d. What is $n_{old}$, the number of individuals in the control group?
```
n_old = df2[(df2['landing_page'] == 'old_page') & (df2['group'] == 'control')].shape[0]
n_old
```
e. Simulate $n_{new}$ transactions with a conversion rate of $p_{new}$ under the null. Store these $n_{new}$ 1's and 0's in **new_page_converted**.
```
new_page_converted = np.random.choice([1,0],n_new, p=(p_new,1-p_new))
new_page_converted.mean()
```
f. Simulate $n_{old}$ transactions with a conversion rate of $p_{old}$ under the null. Store these $n_{old}$ 1's and 0's in **old_page_converted**.
```
old_page_converted = np.random.choice([1,0],n_old, p=(p_old,1-p_old))
old_page_converted.mean()
```
g. Find $p_{new}$ - $p_{old}$ for your simulated values from part (e) and (f).
```
# p_new - p_old
new_page_converted.mean() - old_page_converted.mean()
```
h. Create 10,000 $p_{new}$ - $p_{old}$ values using the same simulation process you used in parts (a) through (g) above. Store all 10,000 values in a NumPy array called **p_diffs**.
```
p_diffs = []
for _ in range(10000):
new_page_converted = np.random.choice([0, 1], size = n_new, p = [1-p_new, p_new], replace = True).sum()
old_page_converted = np.random.choice([0, 1], size = n_old, p = [1-p_old, p_old], replace = True).sum()
diff = new_page_converted/n_new - old_page_converted/n_old
p_diffs.append(diff)
p_diffs = np.array(p_diffs)
p_diffs
```
i. Plot a histogram of the **p_diffs**. Does this plot look like what you expected? Use the matching problem in the classroom to assure you fully understand what was computed here.
```
plt.hist(p_diffs);
plt.plot();
```
j. What proportion of the **p_diffs** are greater than the actual difference observed in **ab_data.csv**?
```
# (p_diffs > (p_new - p_old))
prop = (p_diffs > df['converted'].sample(10000)).mean()
prop
```
k. Please explain using the vocabulary you've learned in this course what you just computed in part **j.** What is this value called in scientific studies? What does this value mean in terms of whether or not there is a difference between the new and old pages?
**Difference is not significant**
l. We could also use a built-in to achieve similar results. Though using the built-in might be easier to code, the above portions are a walkthrough of the ideas that are critical to correctly thinking about statistical significance. Fill in the below to calculate the number of conversions for each page, as well as the number of individuals who received each page. Let `n_old` and `n_new` refer the the number of rows associated with the old page and new pages, respectively.
```
import statsmodels.api as sm
convert_old = df2[(df2['landing_page'] == 'old_page') & (df2['group'] == 'control')]
convert_new = df2[(df2['landing_page'] == 'new_page') & (df2['group'] == 'treatment')]
n_old = convert_old.shape[0]
n_new = convert_new.shape[0]
n_old, n_new
# df2.head()
```
m. Now use `stats.proportions_ztest` to compute your test statistic and p-value. [Here](http://knowledgetack.com/python/statsmodels/proportions_ztest/) is a helpful link on using the built in.
```
from statsmodels.stats.proportion import proportions_ztest
(df2['converted'] == 1).sum()
df2.shape[0]
prop
stat, pval = proportions_ztest((df2['converted'] == 1).sum(), df2.shape[0], prop)
stat, pval
```
n. What do the z-score and p-value you computed in the previous question mean for the conversion rates of the old and new pages? Do they agree with the findings in parts **j.** and **k.**?
**p val = 0**
**No**
<a id='regression'></a>
### Part III - A regression approach
`1.` In this final part, you will see that the result you achieved in the A/B test in Part II above can also be achieved by performing regression.<br><br>
a. Since each row is either a conversion or no conversion, what type of regression should you be performing in this case?
**Logical Regression**
```
df2.head()
```
b. The goal is to use **statsmodels** to fit the regression model you specified in part **a.** to see if there is a significant difference in conversion based on which page a customer receives. However, you first need to create in df2 a column for the intercept, and create a dummy variable column for which page each user received. Add an **intercept** column, as well as an **ab_page** column, which is 1 when an individual receives the **treatment** and 0 if **control**.
```
import statsmodels.api as sm
df2[['control','ab_page']] = pd.get_dummies(df2['group'])
df2.drop(['control','group'],axis=1, inplace=True)
df2.head()
```
c. Use **statsmodels** to instantiate your regression model on the two columns you created in part b., then fit the model using the two columns you created in part **b.** to predict whether or not an individual converts.
```
df2['intercept'] = 1
logit_mod = sm.Logit(df2['converted'], df2[['intercept','ab_page']])
results = logit_mod.fit()
np.exp(-0.0150)
1/np.exp(-0.0150)
```
d. Provide the summary of your model below, and use it as necessary to answer the following questions.
```
results.summary()
```
e. What is the p-value associated with **ab_page**? Why does it differ from the value you found in **Part II**?<br><br> **Hint**: What are the null and alternative hypotheses associated with your regression model, and how do they compare to the null and alternative hypotheses in **Part II**?
**P value = 0.190**
f. Now, you are considering other things that might influence whether or not an individual converts. Discuss why it is a good idea to consider other factors to add into your regression model. Are there any disadvantages to adding additional terms into your regression model?
**Yes, its good to check for some more fields**
**Disadvantage - It may not be as easy to interpret as in the previous case**
g. Now along with testing if the conversion rate changes for different pages, also add an effect based on which country a user lives in. You will need to read in the **countries.csv** dataset and merge together your datasets on the appropriate rows. [Here](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.join.html) are the docs for joining tables.
Does it appear that country had an impact on conversion? Don't forget to create dummy variables for these country columns - **Hint: You will need two columns for the three dummy variables.** Provide the statistical output as well as a written response to answer this question.
```
df_countries = pd.read_csv('countries.csv')
df_countries.head()
df_merged = pd.merge(df2,df_countries, left_on='user_id', right_on='user_id')
df_merged.head()
df_merged[['US','UK','CA']] = pd.get_dummies(df_merged['country'])
df_merged.drop(['country','CA'],axis=1, inplace=True)
df_merged.head()
df_merged['intercept'] = 1
logit_mod = sm.Logit(df_merged['converted'], df_merged[['intercept','US','UK']])
results = logit_mod.fit()
results.summary()
```
**US ia having negative coeff which means that conversion rate decreases if person is from US**
**UK ia having positive coeff which means that conversion rate increases if person is from UK**
h. Though you have now looked at the individual factors of country and page on conversion, we would now like to look at an interaction between page and country to see if there significant effects on conversion. Create the necessary additional columns, and fit the new model.
Provide the summary results, and your conclusions based on the results.
```
final_df = df_merged[['user_id','timestamp','landing_page','converted','ab_page','US','UK']]
final_df.head()
final_df['intercept'] = 1
logit_mod = sm.Logit(final_df['ab_page'], final_df[['intercept','US','UK']])
results = logit_mod.fit()
results.summary()
```
**'ab_page' column is 1 when an individual receives the treatment and 0 if control.**
**US ia having positive coeff which means that chance of getting treatment page increases **
**UK ia having negative coeff which means that change of getting control page increases**
<a id='conclusions'></a>
## Finishing Up
> Congratulations! You have reached the end of the A/B Test Results project! You should be very proud of all you have accomplished!
> **Tip**: Once you are satisfied with your work here, check over your report to make sure that it is satisfies all the areas of the rubric (found on the project submission page at the end of the lesson). You should also probably remove all of the "Tips" like this one so that the presentation is as polished as possible.
## Directions to Submit
> Before you submit your project, you need to create a .html or .pdf version of this notebook in the workspace here. To do that, run the code cell below. If it worked correctly, you should get a return code of 0, and you should see the generated .html file in the workspace directory (click on the orange Jupyter icon in the upper left).
> Alternatively, you can download this report as .html via the **File** > **Download as** submenu, and then manually upload it into the workspace directory by clicking on the orange Jupyter icon in the upper left, then using the Upload button.
> Once you've done this, you can submit your project by clicking on the "Submit Project" button in the lower right here. This will create and submit a zip file with this .ipynb doc and the .html or .pdf version you created. Congratulations!
```
from subprocess import call
call(['python', '-m', 'nbconvert', 'Analyze_ab_test_results_notebook.ipynb'])
```
|
github_jupyter
|
## Random Forest Classification
### Random Forest
#### The fundamental idea behind a random forest is to combine many decision trees into a single model. Individually, predictions made by decision trees (or humans) may not be accurate, but combined together, the predictions will be closer to the mark on average.
#### Pros
- can handle large datasets
- can handle missing values
- less influenced by outliers in the data
- no assumptions about underlying distributions in the data
- can implicitly handle collinearity in features, highly similar features
- work well with categorical and numerical features, mixing different range values
#### Cons
- robust algorithm makes it more complex tougher to analyze small details
- not best to determine feature and target relationships/effects due to working with highly similar features
### Model Set Up
#### Steps
- load the data
- determine regression or classification target
- inspect, clean, organize data
- check for, handle outliers
- encode data if necessary
- set features and target
- train, test split the data
- scale the data if necessary
- build the model, fit on the data, run the model
- run metrics, analyze, view results, adjust parameters, repeat until satisfied...
### Regression Models
#### Random Forest Classification
1 dependent variable (binary) , 1+ independent variables (interval or ratio or categorical)

- classification predictor
- generate reasonable predictions across a wide range of data while requiring little configuration
#### Classification Models
##### Import + Inspect
```
### imports ###
import pandas as pd
import numpy as np
import sklearn
df = pd.read_csv('https://raw.githubusercontent.com/CVanchieri/CS_Notes/main/Classification_Notes/bill_authentication.csv') # read in the file
print('data frame shape:', df.shape) # show the data frame shape
df.head() # show the data frame
### inspecting the data ###
print('--- INSPECTING THE DATA --- ')
print('--- columns --- ')
print(df.columns)
print('--- types --- ')
print(df.dtypes)
print('--- NA counts --- ')
print(df.isna().sum())
# print('--- object descriptions --- ')
# print(df.describe(include=object))
print('--- numericals descriptions --- ')
df.describe()
### view basic feature correlations ###
print('--- feature correlations ---')
df.corr()
### view basic feature correlations in a heatmap ###
import seaborn as sns
import matplotlib.pyplot as plt
f, ax = plt.subplots(1, 1, figsize = (10, 7))
print('--- feature correlations heatmap ---')
sns.heatmap(df.corr() , cmap = 'Wistia' , annot = True)
### view scatter plots for each feature vs. target ###
import matplotlib.pyplot as plt
target_ = 'Class' # set the target
features_ = df.iloc[:, 0:4] # set the features
print('--- bar plots ---')
for feature in features_:
figure = plt.figure
f, ax = plt.subplots(1, 1, figsize = (10, 7))
ax = plt.gca()
ax.bar(df[target_], df[feature])
ax.set_xlabel(target_)
ax.set_ylabel(feature)
ax.set_title(f'''{target_} vs {feature}''')
plt.show()
```
##### Encode + Clean + Organize
```
### encoding not necessary with this example, all are numericals ###
### check for outliers in the data ###
import matplotlib.pyplot as plt
# view each feature in a boxplot
for column in df:
plt.figure() # plot figure
f, ax = plt.subplots(1, 1, figsize = (10, 7))
df.boxplot([column]) # set data
### function to find outliers in the data ###
def outlier_zscore(data):
global outliers,zscore
outliers = []
zscore = []
threshold = 3.5 # set threshold
mean = np.mean(data)
std = np.std(data)
for i in data:
z_score = (i - mean)/std # calculate the z_score
zscore.append(z_score) # append the score to the zscore
if np.abs(z_score) > threshold:
outliers.append(i) # append z_score the outliers
print(outliers)
return len(outliers), outliers
### run each feature 'wanted' through the function ###
print('--- possible outliers --- ')
Variance_outliers_number, Variance_outliers = outlier_zscore(df.Variance)
Skewness_outliers_number, Skewness_outliers = outlier_zscore(df.Skewness)
Curtosis_outliers_number, Curtosis_outliers = outlier_zscore(df.Curtosis)
Entropy_outliers_number, Entropy_outliers = outlier_zscore(df.Entropy)
Class_outliers_number, Class_outliers = outlier_zscore(df.Class)
### removal of outliers per feature ###
for num, i in enumerate(df['Curtosis']): # removing the outliers of 'bmi'
if i in Curtosis_outliers:
df['Curtosis'][num] = 13.5 # 3.5 under the lowest outlier
for num, i in enumerate(df['Entropy']): # removing the outliers of 'charges'
if i in Entropy_outliers:
df['Entropy'][num] = -5.5 # 3.5 under the lowest outlier
```
#### Random Forest Classification
- GridSearch CV
- RandomSearch CV
```
### copy the data frame ###
df1 = df.copy()
### split the data into features & target sets ###
X = df1.iloc[:, 0:4].values # set the features
y = df1.iloc[:, 4].values # set the target
print('--- data shapes --- ')
print('X shape:', X.shape)
print('y shape:', y.shape)
### set the train test split parameters ###
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0) # split 80/20
### feature scaling ###
from sklearn.preprocessing import StandardScaler
sc = StandardScaler() # initiate the scalar
X_train = sc.fit_transform(X_train) # fit transform the data with scalar
X_test = sc.transform(X_test) # fit transform the data with scalar
### random forest classifier ###
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
model = RandomForestClassifier()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
#### create data frame of predictions and results ###
y_pred_df = pd.DataFrame(y_pred, columns=["Predicted_Values" ])
y_test_df = pd.DataFrame(np.array(y_test), columns=["Real_Values"])
df_final = pd.concat([y_test_df , y_pred_df] , axis=1)
print('--- real values vs predicted values ---')
print(df_final.head())
### get the model metrics ###
print('--- model metrics ---')
print('mean absolute error:', metrics.mean_absolute_error(y_test, y_pred)) # mae
print('mean squared error:', metrics.mean_squared_error(y_test, y_pred)) # mse
print('root mean squared error:', np.sqrt(metrics.mean_squared_error(y_test, y_pred))) # rmse
score = metrics.r2_score(y_test , y_pred) # get the r2 score
print("r2 score = {}".format(score)) # show the r2 score
print('model score=', model.score(X_train, y_train)) # show the model score
print("model accuracy= {}%".format(score * 100)) # show the model accuracy
print('--- confusion matrix ---')
print(metrics.confusion_matrix(y_test,y_pred)) # confusion matrix
print('--- classification report ---')
print(metrics.classification_report(y_test,y_pred)) # classificatin report
print('model accuracy score=', metrics.accuracy_score(y_test, y_pred)) # model accuracy
### visualize the model prediction accuracy ###
import seaborn as sns
import matplotlib.pyplot as plt
### configure the plot ###
print('--- distplot accuracy --- ')
f, ax = plt.subplots(1, 1, figsize = (10, 7))
ax1 = sns.distplot(y_test, hist=False, color="b", label="Actual Values")
sns.distplot(y_pred, hist=False, color="r", label="Predicted Values" , axlabel='Charges', ax=ax1)
plt.legend()
```
###### GridSearch CV
```
### copy the data frame ###
df2 = df.copy()
### split the data into features & target sets ###
# for single regression select 1 feature
X = df2.iloc[:, 0:4].values # set the features
y = df2.iloc[:, 4].values # set the target
print('--- data shapes --- ')
print('X shape:', X.shape)
print('y shape:', y.shape)
### set the train test split parameters ###
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0) # split 80/20
### feature scaling ###
from sklearn.preprocessing import StandardScaler
sc = StandardScaler() # initiate the scalar
X_train = sc.fit_transform(X_train) # fit transform the data with scalar
X_test = sc.transform(X_test) # fit transform the data with scalar
### random forest classifier + gridsearch CV model ###
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
model1 = RandomForestClassifier()
param_grid = { # create the param grid
'n_estimators': [20, 100, 200],
'max_features': ['auto', 'sqrt', 'log2'],
'max_leaf_nodes' : [2, 6, 10],
'max_depth' : [5, 15, 25],
'min_samples_split' : [2, 10, 15],
# 'bootstrap': [True, False],
# 'ccp_alpha': [0.0, 0.25, 0.50],
# 'criterion': 'mse',
# 'max_samples': [2, 10, 15],
# 'min_impurity_decrease': [0.0, 0.25, 0.50],
# 'min_impurity_split': [2, 10, 15],
# 'min_samples_leaf': [1, 5, 10],
# 'min_weight_fraction_leaf': [0.0, 0.25, 0.50],
# 'n_jobs': [1, 2, 5],
# 'oob_score': [True, False],
# 'random_state': [0, 2, 4],
# 'verbose': [1],
# 'warm_start': [True, False]
}
CV_rfc = GridSearchCV(estimator=model1, param_grid=param_grid, cv=3)
print('--- model runtime --- ')
%time CV_rfc.fit(X_train, y_train)
print('--- best params --- ')
CV_rfc.best_params_
### random forest classifier + grid best params ###
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
model1 = RandomForestClassifier(
max_depth= 25,
max_features= 'log2',
max_leaf_nodes= 10,
min_samples_split= 2,
n_estimators= 20
)
print('--- model runtime --- ')
%time model1.fit(X_train, y_train)
y_pred = model1.predict(X_test)
#### create data frame of predictions and results ###
y_pred_df = pd.DataFrame(y_pred, columns=["Predicted_Values" ])
y_test_df = pd.DataFrame(np.array(y_test), columns=["Real_Values"])
df_final = pd.concat([y_test_df , y_pred_df] , axis=1)
print('--- real values vs predicted values ---')
print(df_final.head())
### get the model1 metrics ###
print('--- model metrics ---')
print('mean absolute error:', metrics.mean_absolute_error(y_test, y_pred)) # mae
print('mean squared error:', metrics.mean_squared_error(y_test, y_pred)) # mse
print('root mean squared error:', np.sqrt(metrics.mean_squared_error(y_test, y_pred))) # rmse
score = metrics.r2_score(y_test , y_pred) # get the r2 score
print("r2 score = {}".format(score)) # show the r2 score
print('model score=', model1.score(X_train, y_train)) # show the model score
print("model accuracy= {}%".format(score * 100)) # show the model accuracy
print('--- confusion matrix ---')
print(metrics.confusion_matrix(y_test,y_pred)) # confusion matrix
print('--- classification report ---')
print(metrics.classification_report(y_test,y_pred)) # classificatin report
print('model1 accuracy score=', metrics.accuracy_score(y_test, y_pred)) # model accuracy
### visualize the model prediction accuracy ###
import seaborn as sns
import matplotlib.pyplot as plt
### configure the plot ###
print('--- distplot accuracy --- ')
f, ax = plt.subplots(1, 1, figsize = (10, 7))
ax1 = sns.distplot(y_test, hist=False, color="b", label="Actual Values")
sns.distplot(y_pred, hist=False, color="r", label="Predicted Values" , axlabel='Charges', ax=ax1)
plt.legend()
```
###### RandomSearch CV
```
### copy the data frame ###
df3 = df.copy()
### split the data into features & target sets ###
# for single regression select the 1 feature
X = df3.iloc[:, 0:4].values # set the features
y = df3.iloc[:, 4].values # set the target
print('--- data shapes --- ')
print('X shape:', X.shape) # show the shape
print('y shape:', y.shape) # show the shape
### set the train test split parameters ###
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0) # split 80/20
### feature scaling ###
from sklearn.preprocessing import StandardScaler
sc = StandardScaler() # initiate the scalar
X_train = sc.fit_transform(X_train) # fit transform the data with scalar
X_test = sc.transform(X_test) # fit transform the data with scalar
### random forest classifier + randomizedsearch CV model ###
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import RandomizedSearchCV
model2 = RandomForestClassifier()
param_grid = { # create the param grid
'n_estimators': [20, 100, 200],
'max_features': ['auto', 'sqrt', 'log2'],
'max_leaf_nodes' : [2, 6, 10],
'max_depth' : [5, 15, 25],
'min_samples_split' : [2, 10, 15],
# 'bootstrap': [True, False],
# 'ccp_alpha': [0.0, 0.25, 0.50],
# 'criterion': 'mse',
# 'max_samples': [2, 10, 15],
# 'min_impurity_decrease': [0.0, 0.25, 0.50],
# 'min_impurity_split': [2, 10, 15],
# 'min_samples_leaf': [1, 5, 10],
# 'min_weight_fraction_leaf': [0.0, 0.25, 0.50],
# 'n_jobs': [1, 2, 5],
# 'oob_score': [True, False],
# 'random_state': [0, 2, 4],
# 'verbose': [1],
# 'warm_start': [True, False]
}
CV_rfc = RandomizedSearchCV(model2, param_grid, cv=3)
%time CV_rfc.fit(X_train, y_train)
CV_rfc.best_params_
### random forest classifier + random best params ###
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
model2 = RandomForestClassifier(
max_depth= 15,
max_features= 'auto',
max_leaf_nodes= 10,
min_samples_split= 15,
n_estimators= 20
)
print('--- model runtime --- ')
%time model2.fit(X_train, y_train)
y_pred = model2.predict(X_test)
#### create data frame of predictions and results ###
y_pred_df = pd.DataFrame(y_pred, columns=["Predicted_Values" ])
y_test_df = pd.DataFrame(np.array(y_test), columns=["Real_Values"])
df_final = pd.concat([y_test_df , y_pred_df] , axis=1)
print('--- real values vs predicted values ---')
print(df_final.head())
### get the model2 metrics ###
print('--- model metrics ---')
print('mean absolute error:', metrics.mean_absolute_error(y_test, y_pred)) # mae
print('mean squared error:', metrics.mean_squared_error(y_test, y_pred)) # mse
print('root mean squared error:', np.sqrt(metrics.mean_squared_error(y_test, y_pred))) # rmse
score = metrics.r2_score(y_test , y_pred) # get the r2 score
print("r2 score = {}".format(score)) # show the r2 score
print('model score=', model2.score(X_train, y_train)) # show the model score
print("model accuracy= {}%".format(score * 100)) # show the model accuracy
print('--- confusion matrix ---')
print(metrics.confusion_matrix(y_test,y_pred)) # confusion matrix
print('--- classification report ---')
print(metrics.classification_report(y_test,y_pred)) # classificatin report
print('model2 accuracy score=', metrics.accuracy_score(y_test, y_pred)) # model accuracy
### visualize the model prediction accuracy ###
import seaborn as sns
import matplotlib.pyplot as plt
### configure the plot ###
print('--- distplot accuracy --- ')
f, ax = plt.subplots(1, 1, figsize = (10, 7))
ax1 = sns.distplot(y_test, hist=False, color="b", label="Actual Values")
sns.distplot(y_pred, hist=False, color="r", label="Predicted Values" , axlabel='Charges', ax=ax1)
plt.legend()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/thingumajig/colab-experiments/blob/master/RetinaNet_Video_Object_Detection.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# .init
## setup keras-retinanet
```
!git clone https://github.com/fizyr/keras-retinanet.git
%cd keras-retinanet/
!pip install .
!python setup.py build_ext --inplace
```
## download model
```
#!curl -LJO --output snapshots/pretrained.h5 https://github.com/fizyr/keras-retinanet/releases/download/0.5.0/resnet50_coco_best_v2.1.0.h5
import urllib
PRETRAINED_MODEL = './snapshots/_pretrained_model.h5'
URL_MODEL = 'https://github.com/fizyr/keras-retinanet/releases/download/0.5.0/resnet50_coco_best_v2.1.0.h5'
urllib.request.urlretrieve(URL_MODEL, PRETRAINED_MODEL)
```
# inference
## modules
```
!pwd
#import os, sys
#sys.path.insert(0, 'keras-retinanet')
# show images inline
%matplotlib inline
# automatically reload modules when they have changed
%load_ext autoreload
%autoreload 2
import os
#os.environ['CUDA_VISIBLE_DEVICES'] = '0'
# import keras
import keras
from keras_retinanet import models
from keras_retinanet.utils.image import read_image_bgr, preprocess_image, resize_image
from keras_retinanet.utils.visualization import draw_box, draw_caption
from keras_retinanet.utils.colors import label_color
# import miscellaneous modules
import matplotlib.pyplot as plt
import cv2
import numpy as np
import time
# set tf backend to allow memory to grow, instead of claiming everything
import tensorflow as tf
def get_session():
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
return tf.Session(config=config)
# use this environment flag to change which GPU to use
#os.environ["CUDA_VISIBLE_DEVICES"] = "1"
# set the modified tf session as backend in keras
keras.backend.tensorflow_backend.set_session(get_session())
```
## load model
```
# %cd keras-retinanet/
model_path = os.path.join('snapshots', sorted(os.listdir('snapshots'), reverse=True)[0])
print(model_path)
print(os.path.isfile(model_path))
# load retinanet model
model = models.load_model(model_path, backbone_name='resnet50')
# model = models.convert_model(model)
# load label to names mapping for visualization purposes
labels_to_names = {0: 'person', 1: 'bicycle', 2: 'car', 3: 'motorcycle', 4: 'airplane', 5: 'bus',
6: 'train', 7: 'truck', 8: 'boat', 9: 'traffic light', 10: 'fire hydrant',
11: 'stop sign', 12: 'parking meter', 13: 'bench', 14: 'bird', 15: 'cat',
16: 'dog', 17: 'horse', 18: 'sheep', 19: 'cow', 20: 'elephant', 21: 'bear',
22: 'zebra', 23: 'giraffe', 24: 'backpack', 25: 'umbrella', 26: 'handbag',
27: 'tie', 28: 'suitcase', 29: 'frisbee', 30: 'skis', 31: 'snowboard',
32: 'sports ball', 33: 'kite', 34: 'baseball bat', 35: 'baseball glove',
36: 'skateboard', 37: 'surfboard', 38: 'tennis racket', 39: 'bottle',
40: 'wine glass', 41: 'cup', 42: 'fork', 43: 'knife', 44: 'spoon',
45: 'bowl', 46: 'banana', 47: 'apple', 48: 'sandwich', 49: 'orange',
50: 'broccoli', 51: 'carrot', 52: 'hot dog', 53: 'pizza', 54: 'donut',
55: 'cake', 56: 'chair', 57: 'couch', 58: 'potted plant', 59: 'bed',
60: 'dining table', 61: 'toilet', 62: 'tv', 63: 'laptop', 64: 'mouse',
65: 'remote', 66: 'keyboard', 67: 'cell phone', 68: 'microwave',
69: 'oven', 70: 'toaster', 71: 'sink', 72: 'refrigerator', 73: 'book',
74: 'clock', 75: 'vase', 76: 'scissors', 77: 'teddy bear', 78: 'hair drier',
79: 'toothbrush'}
```
## detect objects
```
def img_inference(img_path, threshold_score = 0.8):
image = read_image_bgr(img_path)
# copy to draw on
draw = image.copy()
draw = cv2.cvtColor(draw, cv2.COLOR_BGR2RGB)
# preprocess image for network
image = preprocess_image(image)
image, scale = resize_image(image)
# process image
start = time.time()
boxes, scores, labels = model.predict_on_batch(np.expand_dims(image, axis=0))
print("processing time: ", time.time() - start)
# correct for image scale
boxes /= scale
# visualize detections
for box, score, label in zip(boxes[0], scores[0], labels[0]):
# scores are sorted so we can break
if score < threshold_score:
break
color = label_color(label)
b = box.astype(int)
draw_box(draw, b, color=color)
caption = "{} {:.3f}".format(labels_to_names[label], score)
draw_caption(draw, b, caption)
plt.figure(figsize=(10, 10))
plt.axis('off')
plt.imshow(draw)
plt.show()
img_inference('examples/000000008021.jpg')
from tensorflow.python.client import device_lib
def get_available_gpus():
local_device_protos = device_lib.list_local_devices()
return [x.physical_device_desc for x in local_device_protos if x.device_type == 'GPU']
GPU = get_available_gpus()[-1][0:-1]
print(GPU)
import glob
def create_video(img_path, name ='processed', img_ext = '*.jpg', image_size=(1280, 720)):
_name = name + '.mp4'
#_cap = VideoCapture(0)
_fourcc = cv2.VideoWriter_fourcc(*'MP4V')
_out = cv2.VideoWriter(_name, _fourcc, 15.0, image_size)
# out = cv2.VideoWriter('project.avi',cv2.VideoWriter_fourcc(*'DIVX'), 15, size)
for filename in sorted(glob.glob(os.path.join(img_path, img_ext))):
print(filename)
img = cv2.imread(filename)
_out.write(img)
del img
_out.release()
import unicodedata
import string
valid_filename_chars = f"-_.() {string.ascii_letters}{string.digits}"
char_limit = 255
def clean_filename(filename, whitelist=valid_filename_chars, replace=' '):
# replace spaces
for r in replace:
filename = filename.replace(r, '_')
# keep only valid ascii chars
cleaned_filename = unicodedata.normalize('NFKD', filename).encode('ASCII', 'ignore').decode()
# keep only whitelisted chars
cleaned_filename = ''.join(c for c in cleaned_filename if c in whitelist)
if len(cleaned_filename) > char_limit:
print(f"Warning, filename truncated because it was over {char_limit}. Filenames may no longer be unique")
return cleaned_filename[:char_limit]
import colorsys
import random
from tqdm import tqdm
N = len(labels_to_names)
HSV_tuples = [(x*1.0/N, 0.5, 0.5) for x in range(N)]
RGB_tuples = list(map(lambda x: tuple(255*np.array(colorsys.hsv_to_rgb(*x))), HSV_tuples))
random.shuffle(RGB_tuples)
def object_detect_video(video_path, out_temp_dir='tmp', video_name = 'processed', threshold = 0.6):
cap = cv2.VideoCapture(video_path)
if not os.path.exists(out_temp_dir):
os.makedirs(out_temp_dir)
tq = tqdm(total=1, unit="frame(s)")
counter = 0
sum_time = 0
video_out = None
while(True):
ret, draw = cap.read()
if not ret:
break
bgr = cv2.cvtColor(draw, cv2.COLOR_RGB2BGR)
# preprocess image for network
image = preprocess_image(bgr)
image, scale = resize_image(image)
if counter == 0:
height, width, channels = draw.shape
#print(f'Shape: {width}X{height}')
_name = video_name + '.mp4'
_fourcc = cv2.VideoWriter_fourcc(*'MP4V')
video_out = cv2.VideoWriter(_name, _fourcc, 20.0, (width, height))
# process image
start = time.time()
boxes, scores, labels = model.predict_on_batch(np.expand_dims(image, axis=0))
t = time.time() - start
#print(f"frame:{counter} processing time: {t}")
tq.total += 1
# fancy way to give info without forcing a refresh
tq.set_postfix(dir=f'frame {counter} time {sum_time}', refresh=False)
tq.update(0) # may trigger a refresh
# correct for image scale
boxes /= scale
# visualize detections
#draw_detections(image, boxes, scores, labels, color=None, label_to_name=None, score_threshold=0.5)
for box, score, label in zip(boxes[0], scores[0], labels[0]):
if score < threshold:
continue
color = label_color(label)
b = box.astype(int)
draw_box(draw, b, color=color)
caption = f"{labels_to_names[label]} {score:.3f}"
draw_caption(draw, b, caption)
if sum_time>0:
cv2.putText(draw, "Processing time %.2fs (%.1ffps) AVG %.2fs (%.1ffps)"%(t,1.0/t,sum_time/counter,counter/sum_time), (10, 70), cv2.FONT_HERSHEY_SIMPLEX, 1, (0, 0, 0), 7)
cv2.putText(draw, "Processing time %.2fs (%.1ffps) AVG %.2fs (%.1ffps)"%(t,1.0/t,sum_time/counter,counter/sum_time), (10, 70), cv2.FONT_HERSHEY_SIMPLEX, 1, (255, 255, 255), 3)
# cv2.imwrite(os.path.join(out_temp_dir, f'img{counter:08d}.jpg'),draw)
video_out.write(draw)
counter=counter+1
sum_time+=t
cap.release()
video_out.release()
cv2.destroyAllWindows()
tq.set_postfix(dir=video_path)
tq.close()
from google.colab import files
uploaded = files.upload()
for fn in uploaded.keys():
print(f'User uploaded file "{fn}" with length {len(uploaded[fn])} bytes')
fn0 = clean_filename(fn)
#with open(fn0, "wb") as df:
# df.write(uploaded[fn])
# df.close()
object_detect_video(fn, f'{fn0}_tmp', video_name=f'{os.path.basename(fn0)}_processed', threshold = 0.5)
#create_video(f'{fn0}_tmp')
files.download(f'{os.path.basename(fn0)}_processed.mp4')
# object_detect_video('Canada vs. Finland - Gold Medal Game - Game Highlights - IIHFWorlds 2019.mp4', 'video_tmp', video_name = 'processed2')
#sorted(glob.glob('/content/keras-retinanet/video_tmp/*.jpg'))
#create_video('/content/keras-retinanet/video_tmp')
```
|
github_jupyter
|
# Objective
* 20190815:
* Given stock returns for the last N days, we do prediction for the next N+H days, where H is the forecast horizon
* We use double exponential smoothing to predict
```
%matplotlib inline
import math
import matplotlib
import numpy as np
import pandas as pd
import seaborn as sns
import time
from collections import defaultdict
from datetime import date, datetime, time, timedelta
from matplotlib import pyplot as plt
from pylab import rcParams
from sklearn.metrics import mean_squared_error
from tqdm import tqdm_notebook
#### Input params ##################
stk_path = "./data/VTI_20130102_20181231.csv"
H = 21
train_size = 252*3 # Use 3 years of data as train set. Note there are about 252 trading days in a year
val_size = 252 # Use 1 year of data as validation set
# alpha - smoothing coeff
alphaMax = 0.999
alphaMin = 0.001
alphaStep = 0.001
# beta - trend coeff
betaMax = 0.999
betaMin = 0.001
betaStep = 0.001
fontsize = 14
ticklabelsize = 14
####################################
train_val_size = train_size + val_size # Size of train+validation set
print("No. of days in train+validation set = " + str(train_val_size))
print("We will start forecasting on day %d" % (train_val_size+1))
```
# Common functions
```
def get_smape(y_true, y_pred):
"""
Compute symmetric mean absolute percentage error
"""
y_true, y_pred = np.array(y_true), np.array(y_pred)
return 100/len(y_true) * np.sum(2 * np.abs(y_pred - y_true) / (np.abs(y_true) + np.abs(y_pred)))
def get_mape(y_true, y_pred):
"""
Compute mean absolute percentage error (MAPE)
"""
y_true, y_pred = np.array(y_true), np.array(y_pred)
return np.mean(np.abs((y_true - y_pred) / y_true)) * 100
def get_mae(a, b):
"""
Comp mean absolute error e_t = E[|a_t - b_t|]. a and b can be lists.
Returns a vector of len = len(a) = len(b)
"""
return np.mean(abs(np.array(a)-np.array(b)))
def get_rmse(a, b):
"""
Comp RMSE. a and b can be lists.
Returns a scalar.
"""
return math.sqrt(np.mean((np.array(a)-np.array(b))**2))
def double_exponential_smoothing(series, H, alpha=0.5, beta=0.5, return_all=False):
"""
Given a series and alpha, return series of smoothed points
Initialization:
S_1 = y_1,
b_1 = y_2 - y_1,
F_1 = 0, F_2 = y_1
level, S_t = alpha*y_t + (1-alpha)*(S_t-1 + b_t-1)
trend, b_t = beta*(S_t - S_t-1) + (1-beta)*b_t-1
forecast, F_t+1 = S_t + b_t
forecast, F_t+m = S_t + m*b_t
result[len(series)] is the estimate of series[len(series)]
Inputs
series: series to forecast
H : forecast horizon
alpha : smoothing constant.
When alpha is close to 1, dampening is quick.
When alpha is close to 0, dampening is slow
beta : smoothing constant for trend
return_all : if 1 return both original series + predictions, if 0 return predictions only
Outputs
the predictions of length H
"""
result = [0, series[0]]
for n in range(1, len(series)+H-1):
if n == 1:
level, trend = series[0], series[1] - series[0]
if n >= len(series): # we are forecasting
m = n - len(series) + 2
result.append(level + m*trend) # result[len(series)+1] is the estimate of series[len(series)+1]
else:
value = series[n]
last_level, level = level, alpha*value + (1-alpha)*(level+trend)
trend = beta*(level-last_level) + (1-beta)*trend
result.append(level+trend)
# e.g. result[2] uses series[1]
# ie. result[2] is the estimate of series[2]
# e.g. result[len(series)] uses series[len(series)-1]
# ie. result[len(series)] is the estimate of series[len(series)]
if return_all == True:
return result
else:
return result[len(series):len(series)+H]
def get_error_metrics(series, train_size, H, alpha, beta):
"""
Given a series consisting of both train+validation, do predictions of forecast horizon H on the validation set,
at H/2 intervals.
Inputs
series : series to forecast, with length = (train_size + val_size)
train_size : length of series to use as train ie. train set is series[:train_size]
H : forecast horizon
Outputs
mean of rmse, mean of mape, mean of mae
"""
# Predict using single exponential smoothing, and compute error metrics also
rmse = [] # root mean square error
mape = [] # mean absolute percentage error
mae = [] # mean absolute error
smape = [] # symmetric mean absolute percentage error
preds_dict = {}
for i in range(train_size, len(series)-H, int(H/2)):
preds_list = double_exponential_smoothing(series[i-train_size:i], H, alpha, beta)
rmse.append(get_rmse(series[i:i+H], preds_list))
mape.append(get_mape(series[i:i+H], preds_list))
mae.append(get_mae(series[i:i+H], preds_list))
smape.append(get_smape(series[i:i+H], preds_list))
preds_dict[i] = preds_list
return np.mean(rmse), np.mean(mape), np.mean(mae), np.mean(smape), preds_dict
def hyperpram_tune_alpha_beta(series, train_size, H):
"""
Given a series, tune hyperparameter alpha, fit and predict
Inputs
series : series to forecast, with length = (train_size + val_size)
train_size : length of series to use as train ie. train set is series[:train_size]
H : forecast horizon
Outputs
optimum hyperparameters, error metrics dataframe
"""
err_dict = defaultdict(list)
alpha = alphaMin
beta = betaMin
while alpha <= alphaMax:
while beta <= betaMax:
rmse_mean, mape_mean, mae_mean, smape_mean, _ = get_error_metrics(series, train_size, H, alpha, beta)
# Append alpha and beta
err_dict['alpha'].append(alpha)
err_dict['beta'].append(beta)
# Compute error metrics
err_dict['rmse'].append(rmse_mean)
err_dict['mape'].append(mape_mean)
err_dict['mae'].append(mae_mean)
err_dict['smape'].append(smape_mean)
# Increase beta by one step
beta = beta + betaStep
# Increase alpha by one step
alpha = alpha + alphaStep
# Convert to dataframe
err_df = pd.DataFrame(err_dict)
# Get min RMSE
rmse_min = err_df['rmse'].min()
return err_df[err_df['rmse'] == rmse_min]['alpha'].values[0], err_df[err_df['rmse'] == rmse_min]['beta'].values[0], err_df
```
# Load data
```
df = pd.read_csv(stk_path, sep = ",")
# Convert Date column to datetime
df.loc[:, 'Date'] = pd.to_datetime(df['Date'],format='%Y-%m-%d')
# Change all column headings to be lower case, and remove spacing
df.columns = [str(x).lower().replace(' ', '_') for x in df.columns]
# Sort by datetime
df.sort_values(by='date', inplace=True, ascending=True)
df.head(10)
df['date'].min(), df['date'].max()
# Plot adjusted close over time
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
ax = df.plot(x='date', y='adj_close', style='b-', grid=True)
ax.set_xlabel("date")
ax.set_ylabel("USD")
```
# Get Stock Returns
```
df['returns'] = df['adj_close'].pct_change() * 100
df.loc[0, 'returns'] = 0 # set the first value of returns to be 0 for simplicity
df.head()
# Plot returns over time
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
ax = df.plot(x='date', y='returns', style='b-', grid=True)
ax.set_xlabel("date")
ax.set_ylabel("returns")
# Plot distribution of returns
plt.figure(figsize=(12, 8), dpi=80)
ax = sns.distplot(df['returns'][1:])
ax.grid()
ax.set_xlabel('daily returns', fontsize = 14)
ax.set_ylabel("probability density function", fontsize = 14)
matplotlib.rcParams.update({'font.size': 14})
```
# Predict for a specific H (forecast horizon) and a specific date
```
i = train_val_size # Predict for day i, for the next H-1 days. Note indexing of days start from 0.
print("Predicting on day %d, date %s, with forecast horizon H = %d" % (i, df.iloc[i]['date'], H))
# Predict
preds_list = double_exponential_smoothing(df['returns'][i-train_val_size:i].values, H)
print("For forecast horizon %d, predicting on day %d, date %s, the RMSE is %f" % (H, i, df['date'][i], get_rmse(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the MAPE is %f" % (H, i, df['date'][i], get_mape(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the SMAPE is %f" % (H, i, df['date'][i], get_smape(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the MAE is %f" % (H, i, df['date'][i], get_mae(df[i:i+H]['returns'], preds_list)))
# Plot the predictions
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
matplotlib.rcParams.update({'font.size': 14})
ax = df.plot(x='date', y='returns', style='bx-', grid=True)
# Plot the predictions
ax.plot(df['date'][i:i+H], preds_list, marker='x')
ax.set_xlabel("date")
# ax.set_ylabel("daily returns")
ax.legend(['daily returns', 'predictions'])
# ax.set_ylim([105, 120])
ax.set_xlim([date(2016, 11, 1), date(2017, 2, 28)])
```
# Predict for a specific H (forecast horizon) and a specific date, with hyperparameter tuning - alpha, beta
```
i = train_val_size # Predict for day i, for the next H-1 days. Note indexing of days start from 0.
print("Predicting on day %d, date %s, with forecast horizon H = %d" % (i, df.iloc[i]['date'], H))
# Get optimum hyperparams
alpha_opt, beta_opt, err_df = hyperpram_tune_alpha_beta(df['returns'][i-train_val_size:i].values, train_size, H)
print("alpha_opt = " + str(alpha_opt))
print("beta_opt = " + str(beta_opt))
# print("rmse opt = " + str(err_df[(err_df['alpha']==alpha_opt) & (err_df['beta']==beta_opt)]['rmse'].values[0]))
print(err_df[(err_df['alpha']==alpha_opt) & (err_df['beta']==beta_opt)])
err_df
# Predict
preds_list = double_exponential_smoothing(df['returns'][i-train_val_size:i].values, H, alpha_opt, beta_opt)
print("For forecast horizon %d, predicting on day %d, date %s, the RMSE is %f" % (H, i, df['date'][i], get_rmse(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the MAPE is %f" % (H, i, df['date'][i], get_mape(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the SMAPE is %f" % (H, i, df['date'][i], get_smape(df[i:i+H]['returns'], preds_list)))
print("For forecast horizon %d, predicting on day %d, date %s, the MAE is %f" % (H, i, df['date'][i], get_mae(df[i:i+H]['returns'], preds_list)))
# Plot the predictions
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
matplotlib.rcParams.update({'font.size': 14})
ax = df.plot(x='date', y='returns', style='bx-', grid=True)
# Plot the predictions
ax.plot(df['date'][i:i+H], preds_list, marker='x')
ax.set_xlabel("date")
# ax.set_ylabel("USD")
ax.legend(['returns', 'predictions'])
# ax.set_ylim([105, 120])
ax.set_xlim([date(2016, 11, 1), date(2017, 2, 28)])
```
# Predict for a specific H (forecast horizon), and various dates, using model trained in previous step
```
print("alpha_opt = " + str(alpha_opt))
print("beta_opt = " + str(beta_opt))
# Predict and compute error metrics also
rmse = [] # root mean square error
mape = [] # mean absolute percentage error
mae = [] # mean absolute error
smape = [] # symmetric mean absolute percentage error
preds_dict = {}
i_list = range(train_val_size, train_val_size+84*5+42+1, 42)
for i in i_list:
print("Predicting on day %d, date %s" % (i, df.iloc[i]['date']))
preds_list = double_exponential_smoothing(df['returns'][i-train_val_size:i].values, H, alpha_opt, beta_opt)
# Collect the predictions
preds_dict[i] = preds_list
# Compute error metrics
rmse.append(get_rmse(df[i:i+H]['returns'], preds_list))
mape.append(get_mape(df[i:i+H]['returns'], preds_list))
mae.append(get_mae(df[i:i+H]['returns'], preds_list))
smape.append(get_smape(df[i:i+H]['returns'], preds_list))
print("Altogether we made %d forecasts, each of length %d days" % (len(rmse), H))
print("For forecast horizon %d, the mean RMSE is %f" % (H, np.mean(rmse)))
print("For forecast horizon %d, the mean MAPE is %f" % (H, np.mean(mape)))
print("For forecast horizon %d, the mean SMAPE is %f" % (H, np.mean(smape)))
print("For forecast horizon %d, the mean MAE is %f" % (H, np.mean(mae)))
results_final_no_tune = pd.DataFrame({'day': i_list,
'alpha_opt': [alpha_opt]*len(i_list),
'beta_opt': [beta_opt]*len(i_list),
'rmse': rmse,
'mape': mape,
'mae': mae,
'smape': smape})
results_final_no_tune
# Plot the predictions, and zoom in
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
ax = df.plot(x='date', y='returns', style='b-', grid=True)
# Plot the predictions
for key in preds_dict:
ax.plot(df['date'][key:key+H], preds_dict[key])
ax.set_xlabel("date")
# ax.set_ylabel("USD")
ax.legend(['returns', 'predictions'])
# ax.set_ylim([105, 150])
ax.set_xlim([date(2017, 1, 1), date(2018, 12, 31)])
```
# Predict for a specific H (forecast horizon), and various dates, tuning model for every prediction
```
# Predict and compute error metrics also
preds_dict = {}
results_final = defaultdict(list)
i_list = range(train_val_size, train_val_size+84*5+42+1, 42)
for i in i_list:
print("Predicting on day %d, date %s" % (i, df.iloc[i]['date']))
# Get optimum hyperparams
alpha_opt, beta_opt, err_df = hyperpram_tune_alpha_beta(df['returns'][i-train_val_size:i].values, train_size, H)
preds_list = double_exponential_smoothing(df['returns'][i-train_val_size:i].values, H, alpha_opt, beta_opt)
# Collect the predictions
preds_dict[i] = preds_list
# Compute error metrics
results_final['rmse'].append(get_rmse(df[i:i+H]['returns'], preds_list))
results_final['mape'].append(get_mape(df[i:i+H]['returns'], preds_list))
results_final['mae'].append(get_mae(df[i:i+H]['returns'], preds_list))
results_final['smape'].append(get_smape(df[i:i+H]['returns'], preds_list))
results_final['alpha_opt'].append(alpha_opt)
results_final['beta_opt'].append(beta_opt)
results_final = pd.DataFrame(results_final)
print("Altogether we made %d forecasts, each of length %d days" % (len(rmse), H))
print("For forecast horizon %d, the mean RMSE is %f" % (H, results_final['rmse'].mean()))
print("For forecast horizon %d, the mean MAPE is %f" % (H, results_final['mape'].mean()))
print("For forecast horizon %d, the mean SMAPE is %f" % (H, results_final['smape'].mean()))
print("For forecast horizon %d, the mean MAE is %f" % (H, results_final['mae'].mean()))
# results
results_final
# Plot the predictions, and zoom in
rcParams['figure.figsize'] = 10, 8 # width 10, height 8
ax = df.plot(x='date', y='returns', style='b-', grid=True)
# Plot the predictions
for key in preds_dict:
ax.plot(df['date'][key:key+H], preds_dict[key])
ax.set_xlabel("date")
# ax.set_ylabel("USD")
ax.legend(['returns', 'predictions'])
# ax.set_ylim([105, 150])
ax.set_xlim([date(2017, 1, 1), date(2018, 12, 31)])
# Plot scatter plot of actual values vs. predictions
for key in preds_dict:
plt.plot(df['returns'][key:key+H], preds_dict[key], 'x')
plt.plot(range(-3, 4, 1), range(-3, 4, 1), 'b-')
plt.xlabel('returns')
plt.ylabel('predictions')
plt.grid()
```
# Findings
Double exponential smoothing does not predict stock returns well.
|
github_jupyter
|
# 스파크를 이용한 기본 지표 생성 예제
> 기본 지표를 생성하는 데에 있어, 정해진 틀을 그대로 따라하기 보다, 가장 직관적인 방법을 지속적으로 개선하는 과정을 설명하기 위한 예제입니다.
첫 번째 예제인 만큼 지표의 복잡도를 줄이기 위해 해당 서비스를 오픈 일자는 2020/10/25 이며, 지표를 집계하는 시점은 2020/10/26 일 입니다
* 원본 데이터를 그대로 읽는 방법
* dataframe api 를 이용하는 방법
* spark.sql 을 이용하는 방법
* 기본 지표 (DAU, PU)를 추출하는 예제 실습
* 날짜에 대한 필터를 넣는 방법
* 날짜에 대한 필터를 데이터 소스에 적용하는 방법
* 기본 지표 (ARPU, ARPPU)를 추출하는 예제 실습
* 스칼라 값을 가져와서 다음 질의문에 적용하는 방법
* 누적 금액을 구할 때에 단순한 방법
* 서비스 오픈 일자의 디멘젼 테이블을 생성하는 방법
* 널 값에 대한 처리하는 방법
* 생성된 데이터 프레임을 저장하는 방법
* 전 일자 데이터를 가져오는 방법
* 요약 지표를 생성할 때에 단순한 방법
* 팩트 테이블을 활용하는 방법
```
from pyspark.sql import SparkSession
from pyspark.sql.functions import *
spark = SparkSession \
.builder \
.appName("Data Engineer Basic Day3") \
.config("spark.dataengineer.basic.day3", "tutorial-1") \
.getOrCreate()
spark.read.option("inferSchema", "true").option("header", "true").json("access/20201026") \
.withColumn("gmt_time", expr("from_unixtime(a_time, 'yyyy-MM-dd HH:mm:ss')")) \
.withColumn("localtime", expr("from_utc_timestamp(from_unixtime(a_time), 'Asis/Seoul')")) \
.show()
# from_utc_timestamp(from_unixtime(epoch_time), tz_name) as local_time
# spark.conf.unset("spark.sql.session.timeZone")
spark.conf.get("spark.sql.session.timeZone") # 'Etc/UTC' => 이게 원인이었네 ... 초기 값의 TimeZone 설정이 제대로 안 되어 있었음.;ㅁ;
spark.conf.set("spark.sql.session.timeZone", "Asia/Seoul")
spark.conf.get("spark.sql.session.timeZone")
spark.read.option("inferSchema", "true").option("header", "true").json("access/20201026") \
.withColumn("gmt_time", expr("from_unixtime(a_time, 'yyyy-MM-dd HH:mm:ss')")) \
.withColumn("localtime", expr("from_utc_timestamp(from_unixtime(a_time), 'Asis/Seoul')")) \
.show()
sc = spark.sparkContext
spark.read.option("inferSchema", "true").option("header", "true").parquet("user/20201025").createOrReplaceTempView("user")
pWhere=""
spark.read.option("inferSchema", "true").option("header", "true").parquet("purchase/20201025").withColumn("p_time", expr("from_unixtime(p_time)")).createOrReplaceTempView("purchase")
aWhere=""
spark.read.option("inferSchema", "true").option("header", "true").json("access/20201026").withColumn("a_time", expr("from_unixtime(a_time)")).createOrReplaceTempView("access")
spark.sql("desc user").show()
spark.sql("desc purchase").show()
spark.sql("desc access").show()
```
### 과제 1. 주어진 데이터를 이용하여 2020/10/25 기준의 DAU, PU 를 구하시오
* DAU : Daily Active User, 일 별 접속자 수
- log_access 를 통해 unique 한 a_uid 값을 구합니다
* PU : Purchase User, 일 별 구매자 수
- tbl_purchase 를 통해 unique 한 p_uid 값을 구합니다
> 값을 구하기 전에 Spark API 대신 Spark SQL 을 이용하기 위해 [createOrReplaceTempView](https://spark.apache.org/docs/latest/api/python/pyspark.sql.html?highlight=createorreplace#pyspark.sql.DataFrame.createOrReplaceTempView) 를 생성합니다
```
# DAU - access
spark.sql("select a_time as a_time, a_uid from access").show()
dau = spark.sql("select count(distinct a_uid) as DAU from access where a_time >= '2020-10-25 00:00:00' and a_time < '2020-10-26 00:00:00'")
dau.show()
# PU - purchase
spark.sql("select p_time, p_uid from purchase").show()
pu = spark.sql("select count(distinct p_uid) as PU from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00'")
pu.show()
v_dau = dau.collect()[0]["DAU"]
v_pu = pu.collect()[0]["PU"]
```
### 과제 2. 주어진 데이터를 이용하여 2020/10/25 기준의 ARPU, ARPPU 를 구하시오
* ARPU : Average Revenue Per User, 유저 당 평균 수익
- 해당 일자의 전체 수익 (Total Purchase Amount) / 해당 일에 접속한 유저 수 (DAU)
* ARPPU : Average Revenue Per Purchase User, 구매 유저 당 평균 수익
- 해당 일자의 전체 수익 (Total Purchase Amount) / 해당 일에 접속한 구매 유저 수 (PU)
```
# ARPU - total purchase amount, dau
query="select sum(p_amount) / {} from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00'".format(v_dau)
print(query)
total_purchase_amount = spark.sql("select sum(p_amount) as total_purchase_amount from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00'")
total_purchase_amount.show()
spark.sql("select sum(p_amount) / 5 from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00'").show()
spark.sql("select sum(p_amount) / {} as ARPU from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00'".format(v_dau)).show()
# ARPPU - total purchase amount, pu
v_amt = total_purchase_amount.collect()[0]["total_purchase_amount"]
print("| ARPPU | {} |".format(v_amt / v_pu))
```
### 과제 3. 주어진 데이터를 이용하여 2020/10/26 현재의 "누적 매출 금액" 과 "누적 접속 유저수"를 구하시오
* 누적 매출 금액 : 10/25 (오픈) ~ 현재
- 전체 로그를 읽어서 매출 금액의 합을 구한다
- 유저별 매출 정보를 누적하여 저장해두고 재활용한다
* 누적 접속 유저수 : 10/25 (오픈) ~ 현재
- 전체 로그를 읽어서 접속자의 유일한 수를 구한다
- 유저별 접속 정보를 누적하여 저장해두고 재활용한다
```
# 누적 매출 금액
spark.sql("select sum(p_amount) from purchase ").show()
# 누적 접속 유저수
spark.sql("select count(distinct a_uid) from access").show()
```
### 과제 4. 유저별 정보를 누적시키기 위한 디멘젼 테이블을 설계하고 생성합니다
#### User Dimension 테이블 설계
| 컬럼 명 | 컬럼 타입 | 컬럼 설명 |
| :- | :-: | :- |
| d_uid | int | 유저 아이디 |
| d_name | string | 고객 이름 |
| d_pamount | int | 누적 구매 금액 |
| d_pcount | int | 누적 구매 횟수 |
| d_acount | int | 누적 접속 횟수 |
```
# 오픈 첫 날의 경우 예외적으로 별도의 프로그램을 작성합니다
#
# 1. 가장 큰 레코드 수를 가진 정보가 접속정보이므로 해당 일자의 이용자 별 접속 횟수를 추출합니다
# 단, login 횟수를 접속 횟수로 가정합니다 - logout 만 있는 경우는 login 유실 혹은 전일자의 로그이므로 이러한 경우는 제외합니다
spark.sql("describe access").show()
spark.sql("select * from access where a_id = 'login' and a_time >= '2020-10-25 00:00:00' and a_time < '2020-10-26 00:00:00'").show()
uids = spark.sql("select a_uid, count(a_uid) as acount from access where a_id = 'login' and a_time >= '2020-10-25 00:00:00' and a_time < '2020-10-26 00:00:00' group by a_uid")
uids.show()
# 2. 해당 일자의 이용자 별 총 매출 금액과, 구매 횟수를 추출합니다
spark.sql("describe purchase").show()
amts = spark.sql("select p_uid, sum(p_amount) as pamount, count(p_uid) as pcount from purchase where p_time >= '2020-10-25 00:00:00' and p_time < '2020-10-26 00:00:00' group by p_uid")
amts.show()
# 3. 이용자 접속횟수 + 총구매금액 + 구매횟수 (uids + amts)
uids.printSchema()
amts.printSchema()
dim1 = uids.join(amts, uids["a_uid"] == amts["p_uid"], how="left").sort(uids["a_uid"].asc())
dim2 = dim1.withColumnRenamed("a_uid", "d_uid") \
.withColumnRenamed("acount", "d_acount") \
.drop("p_uid") \
.withColumnRenamed("pamount", "d_pamount") \
.withColumnRenamed("pcount", "d_pcount")
dim2.show()
# 4. 이용자 정보를 덧붙입니다
user = spark.sql("select * from user")
user.show()
dim3 = dim2.join(user, dim2["d_uid"] == user["u_id"], "left")
dim4 = dim3.withColumnRenamed("u_name", "d_name") \
.withColumnRenamed("u_gender", "d_gender")
dim5 = dim4.select("d_uid", "d_name", "d_gender", "d_acount", "d_pamount", "d_pcount")
dimension = dim5.na.fill({"d_pamount":0, "d_pcount":0})
dimension.show()
# 4. 다음날 해당 데이터를 사용하도록 하기 위해 일자별 경로에 저장합니다
# - ./users/dt=20201025/
target="./users/dt=20201025"
dimension.write.mode("overwrite").parquet(target)
```
### 과제 5. 전일자 디멘젼 정보를 이용하여 누적된 접속, 매출 지표를 생성합니다
```
# 이전 일자 기준의 고객의 상태를 유지하여 활용합니다
yesterday = spark.read.parquet(target)
yesterday.sort(yesterday["d_uid"].asc()).show()
# 5. 다음 날 동일한 지표를 생성하되 이전 일자의 정보에 누적한 지표를 생성합니다
# 기존 테이블의 고객과 오늘 신규 고객을 모두 포함한 전체 데이터집합을 생성합니다
yesterday.show()
# 새로운 모수를 추가해야 하므로 전체 모수에 해당하는 uid 만을 추출합니다
uid = yesterday.select("d_uid").join(accs.select("a_uid"), yesterday.d_uid == accs.a_uid, "full_outer") \
.withColumn("uid", when(yesterday.d_uid.isNull(), accs.a_uid).otherwise(yesterday.d_uid)) \
.select("uid")
uid.show()
# uid 기준으로 이름, 성별을 조인합니다
user.show()
dim1 = uid.join(user, uid.uid == user.u_id).select(uid.uid, user.u_name, user.u_gender)
dim1.show()
# 어제 디멘젼을 기준으로 누적접속, 누적구매금액, 누적구매횟수 등을 조인합니다
print("dim2")
dim2 = dim1.join(yesterday, dim1.uid == yesterday.d_uid, "left") \
.select(dim1.uid, dim1.u_name, dim1.u_gender, yesterday.d_acount, yesterday.d_pamount, yesterday.d_pcount) \
.na.fill({"d_acount":0, "d_pamount":0, "d_pcount":0})
dim2.show()
# 6. 오늘 생성된 접속수치, 매출 및 매출 횟수를 더합니다
accs = spark.sql("select a_uid, count(a_uid) as acount from access where a_id = 'login' and a_time >= '2020-10-26 00:00:00' and a_time < '2020-10-27 00:00:00' group by a_uid")
accs.show()
print("dim3")
dim3 = dim2.join(accs, dim2.uid == accs.a_uid, "left") \
.withColumn("total_amount", dim2.d_acount + when(accs.acount.isNull(), 0).otherwise(accs.acount)) \
.select("uid", "u_name", "u_gender", "total_amount", "d_pamount", "d_pcount") \
.withColumnRenamed("total_amount", "d_acount")
dim3.show()
# 오늘 발생한 매출을 더합니다
dim3.show()
amts = spark.sql("select p_uid, sum(p_amount) as pamount, count(p_uid) as pcount from purchase where p_time >= '2020-10-26 00:00:00' and p_time < '2020-10-27 00:00:00' group by p_uid")
amts.show()
print("dim4")
dim4 = dim3.join(amts, dim3.uid == amts.p_uid, "left") \
.withColumn("total_pamount", dim3.d_pamount + when(amts.pamount.isNull(), 0).otherwise(amts.pamount)) \
.withColumn("total_pcount", dim3.d_acount + when(amts.pcount.isNull(), 0).otherwise(amts.pcount)) \
.drop("d_pamount", "d_pcount") \
.withColumnRenamed("uid", "d_uid") \
.withColumnRenamed("u_name", "d_name") \
.withColumnRenamed("u_gender", "d_gender") \
.withColumnRenamed("total_pamount", "d_pamount") \
.withColumnRenamed("total_pcount", "d_pcount") \
.select("d_uid", "d_name", "d_gender", "d_acount", "d_pamount", "d_pcount")
dimension = dim4.sort(dim4.d_uid.asc()).coalesce(1)
dimension.show()
# 7. 생성된 디멘젼을 20201026 경로에 저장합니다
target="./users/dt=20201026"
dimension.write.mode("overwrite").parquet(target)
```
### 과제 6. inner, left_outer, right_outer, full_outer 조인 실습 예제를 작성하시오
```
valuesA = [('A',1),('B',2),('C',3),('D',4)]
A = spark.createDataFrame(valuesA,['a_id','a_value'])
valuesB = [('C',10),('D',20),('E',30),('F',40)]
B = spark.createDataFrame(valuesB,['b_id','b_value'])
A.join(B, A.a_id == B.b_id, "inner").sort(A.a_id.asc()).show() # C, D
# A.join(B, A.a_id == B.b_id, "left").sort(A.a_id.asc()).show() # A, B, C, D
# A.join(B, A.a_id == B.b_id, "right").sort(B.b_id.asc()).show() # C, D, E, F
A.join(B, A.a_id == B.b_id, "left_outer").sort(A.a_id.asc()).show() # A, B, C, D
A.join(B, A.a_id == B.b_id, "right_outer").sort(B.b_id.asc()).show() # C, D, E, F
A.join(B, A.a_id == B.b_id, "full_outer").sort(A.a_id.asc_nulls_last(), B.b_id.asc_nulls_last()).show() # A, B, C, D, E, F
# full outer 조인 시에 결과 생성
A.join(B, A.a_id == B.b_id, "full_outer").withColumn("id", expr("case when a_id is null then b_id else a_id end")).select("id").sort("id").show()
# F.when(df.age > 4, 1).when(df.age < 3, -1).otherwise(0)
A.join(B, A.a_id == B.b_id, "full_outer").show()
A.join(B, A.a_id == B.b_id, "full_outer").withColumn("id", when(A.a_id.isNull(), B.b_id).otherwise(A.a_id)).select("id").sort("id").show()
```
### 과제 7. 전일자 디멘젼 정보와 오늘자 로그를 이용하여 팩트 데이터를 생성합니다.
### 과제 8. 팩트 데이터를 이용하여 2020/10/25 기준 성별 매출금액 지표를 추출합니다
|
github_jupyter
|
## AI for Medicine Course 1 Week 1 lecture exercises
<a name="counting-labels"></a>
# Counting labels
As you saw in the lecture videos, one way to avoid having class imbalance impact the loss function is to weight the losses differently. To choose the weights, you first need to calculate the class frequencies.
For this exercise, you'll just get the count of each label. Later on, you'll use the concepts practiced here to calculate frequencies in the assignment!
```
# Import the necessary packages
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
# Read csv file containing training datadata
train_df = pd.read_csv("nih/train-small.csv")
# Count up the number of instances of each class (drop non-class columns from the counts)
class_counts = train_df.sum().drop(['Image','PatientId'])
for column in class_counts.keys():
print(f"The class {column} has {train_df[column].sum()} samples")
# Plot up the distribution of counts
sns.barplot(class_counts.values, class_counts.index, color='b')
plt.title('Distribution of Classes for Training Dataset', fontsize=15)
plt.xlabel('Number of Patients', fontsize=15)
plt.ylabel('Diseases', fontsize=15)
plt.show()
```
<a name="weighted-loss"></a>
# Weighted Loss function
Below is an example of calculating weighted loss. In the assignment, you will calculate a weighted loss function. This sample code will give you some intuition for what the weighted loss function is doing, and also help you practice some syntax you will use in the graded assignment.
For this example, you'll first define a hypothetical set of true labels and then a set of predictions.
Run the next cell to create the 'ground truth' labels.
```
# Generate an array of 4 binary label values, 3 positive and 1 negative
y_true = np.array(
[[1],
[1],
[1],
[0]])
print(f"y_true: \n{y_true}")
```
### Two models
To better understand the loss function, you will pretend that you have two models.
- Model 1 always outputs a 0.9 for any example that it's given.
- Model 2 always outputs a 0.1 for any example that it's given.
```
# Make model predictions that are always 0.9 for all examples
y_pred_1 = 0.9 * np.ones(y_true.shape)
print(f"y_pred_1: \n{y_pred_1}")
print()
y_pred_2 = 0.1 * np.ones(y_true.shape)
print(f"y_pred_2: \n{y_pred_2}")
```
### Problems with the regular loss function
The learning goal here is to notice that with a regular loss function (not a weighted loss), the model that always outputs 0.9 has a smaller loss (performs better) than model 2.
- This is because there is a class imbalance, where 3 out of the 4 labels are 1.
- If the data were perfectly balanced, (two labels were 1, and two labels were 0), model 1 and model 2 would have the same loss. Each would get two examples correct and two examples incorrect.
- However, since the data is not balanced, the regular loss function implies that model 1 is better than model 2.
### Notice the shortcomings of a regular non-weighted loss
See what loss you get from these two models (model 1 always predicts 0.9, and model 2 always predicts 0.1), see what the regular (unweighted) loss function is for each model.
```
loss_reg_1 = -1 * np.sum(y_true * np.log(y_pred_1)) + \
-1 * np.sum((1 - y_true) * np.log(1 - y_pred_1))
print(f"loss_reg_1: {loss_reg_1:.4f}")
loss_reg_2 = -1 * np.sum(y_true * np.log(y_pred_2)) + \
-1 * np.sum((1 - y_true) * np.log(1 - y_pred_2))
print(f"loss_reg_2: {loss_reg_2:.4f}")
print(f"When the model 1 always predicts 0.9, the regular loss is {loss_reg_1:.4f}")
print(f"When the model 2 always predicts 0.1, the regular loss is {loss_reg_2:.4f}")
```
Notice that the loss function gives a greater loss when the predictions are always 0.1, because the data is imbalanced, and has three labels of `1` but only one label for `0`.
Given a class imbalance with more positive labels, the regular loss function implies that the model with the higher prediction of 0.9 performs better than the model with the lower prediction of 0.1
### How a weighted loss treats both models the same
With a weighted loss function, you will get the same weighted loss when the predictions are all 0.9 versus when the predictions are all 0.1.
- Notice how a prediction of 0.9 is 0.1 away from the positive label of 1.
- Also notice how a prediction of 0.1 is 0.1 away from the negative label of 0
- So model 1 and 2 are "symmetric" along the midpoint of 0.5, if you plot them on a number line between 0 and 1.
### Weighted Loss Equation
Calculate the loss for the zero-th label (column at index 0)
- The loss is made up of two terms. To make it easier to read the code, you will calculate each of these terms separately. We are giving each of these two terms a name for explanatory purposes, but these are not officially called $loss_{pos}$ or $loss_{neg}$
- $loss_{pos}$: we'll use this to refer to the loss where the actual label is positive (the positive examples).
- $loss_{neg}$: we'll use this to refer to the loss where the actual label is negative (the negative examples).
$$ loss^{(i)} = loss_{pos}^{(i)} + los_{neg}^{(i)} $$
$$loss_{pos}^{(i)} = -1 \times weight_{pos}^{(i)} \times y^{(i)} \times log(\hat{y}^{(i)})$$
$$loss_{neg}^{(i)} = -1 \times weight_{neg}^{(i)} \times (1- y^{(i)}) \times log(1 - \hat{y}^{(i)})$$
Since this sample dataset is small enough, you can calculate the positive weight to be used in the weighted loss function. To get the positive weight, count how many NEGATIVE labels are present, divided by the total number of examples.
In this case, there is one negative label, and four total examples.
Similarly, the negative weight is the fraction of positive labels.
Run the next cell to define positive and negative weights.
```
# calculate the positive weight as the fraction of negative labels
w_p = 1/4
# calculate the negative weight as the fraction of positive labels
w_n = 3/4
print(f"positive weight w_p: {w_p}")
print(f"negative weight w_n {w_n}")
```
### Model 1 weighted loss
Run the next two cells to calculate the two loss terms separately.
Here, `loss_1_pos` and `loss_1_neg` are calculated using the `y_pred_1` predictions.
```
# Calculate and print out the first term in the loss function, which we are calling 'loss_pos'
loss_1_pos = -1 * np.sum(w_p * y_true * np.log(y_pred_1 ))
print(f"loss_1_pos: {loss_1_pos:.4f}")
# Calculate and print out the second term in the loss function, which we're calling 'loss_neg'
loss_1_neg = -1 * np.sum(w_n * (1 - y_true) * np.log(1 - y_pred_1 ))
print(f"loss_1_neg: {loss_1_neg:.4f}")
# Sum positive and negative losses to calculate total loss
loss_1 = loss_1_pos + loss_1_neg
print(f"loss_1: {loss_1:.4f}")
```
### Model 2 weighted loss
Now do the same calculations for when the predictions are from `y_pred_2'. Calculate the two terms of the weighted loss function and add them together.
```
# Calculate and print out the first term in the loss function, which we are calling 'loss_pos'
loss_2_pos = -1 * np.sum(w_p * y_true * np.log(y_pred_2))
print(f"loss_2_pos: {loss_2_pos:.4f}")
# Calculate and print out the second term in the loss function, which we're calling 'loss_neg'
loss_2_neg = -1 * np.sum(w_n * (1 - y_true) * np.log(1 - y_pred_2))
print(f"loss_2_neg: {loss_2_neg:.4f}")
# Sum positive and negative losses to calculate total loss when the prediction is y_pred_2
loss_2 = loss_2_pos + loss_2_neg
print(f"loss_2: {loss_2:.4f}")
```
### Compare model 1 and model 2 weighted loss
```
print(f"When the model always predicts 0.9, the total loss is {loss_1:.4f}")
print(f"When the model always predicts 0.1, the total loss is {loss_2:.4f}")
```
### What do you notice?
Since you used a weighted loss, the calculated loss is the same whether the model always predicts 0.9 or always predicts 0.1.
You may have also noticed that when you calculate each term of the weighted loss separately, there is a bit of symmetry when comparing between the two sets of predictions.
```
print(f"loss_1_pos: {loss_1_pos:.4f} \t loss_1_neg: {loss_1_neg:.4f}")
print()
print(f"loss_2_pos: {loss_2_pos:.4f} \t loss_2_neg: {loss_2_neg:.4f}")
```
Even though there is a class imbalance, where there are 3 positive labels but only one negative label, the weighted loss accounts for this by giving more weight to the negative label than to the positive label.
### Weighted Loss for more than one class
In this week's assignment, you will calculate the multi-class weighted loss (when there is more than one disease class that your model is learning to predict). Here, you can practice working with 2D numpy arrays, which will help you implement the multi-class weighted loss in the graded assignment.
You will work with a dataset that has two disease classes (two columns)
```
# View the labels (true values) that you will practice with
y_true = np.array(
[[1,0],
[1,0],
[1,0],
[1,0],
[0,1]
])
y_true
```
### Choosing axis=0 or axis=1
You will use `numpy.sum` to count the number of times column `0` has the value 0.
First, notice the difference when you set axis=0 versus axis=1
```
# See what happens when you set axis=0
print(f"using axis = 0 {np.sum(y_true,axis=0)}")
# Compare this to what happens when you set axis=1
print(f"using axis = 1 {np.sum(y_true,axis=1)}")
```
Notice that if you choose `axis=0`, the sum is taken for each of the two columns. This is what you want to do in this case. If you set `axis=1`, the sum is taken for each row.
### Calculate the weights
Previously, you visually inspected the data to calculate the fraction of negative and positive labels. Here, you can do this programmatically.
```
# set the positive weights as the fraction of negative labels (0) for each class (each column)
w_p = np.sum(y_true == 0,axis=0) / y_true.shape[0]
w_p
# set the negative weights as the fraction of positive labels (1) for each class
w_n = np.sum(y_true == 1, axis=0) / y_true.shape[0]
w_n
```
In the assignment, you will train a model to try and make useful predictions. In order to make this example easier to follow, you will pretend that your model always predicts the same value for every example.
```
# Set model predictions where all predictions are the same
y_pred = np.ones(y_true.shape)
y_pred[:,0] = 0.3 * y_pred[:,0]
y_pred[:,1] = 0.7 * y_pred[:,1]
y_pred
```
As before, calculate the two terms that make up the loss function. Notice that you are working with more than one class (represented by columns). In this case, there are two classes.
Start by calculating the loss for class `0`.
$$ loss^{(i)} = loss_{pos}^{(i)} + los_{neg}^{(i)} $$
$$loss_{pos}^{(i)} = -1 \times weight_{pos}^{(i)} \times y^{(i)} \times log(\hat{y}^{(i)})$$
$$loss_{neg}^{(i)} = -1 \times weight_{neg}^{(i)} \times (1- y^{(i)}) \times log(1 - \hat{y}^{(i)})$$
View the zero column for the weights, true values, and predictions that you will use to calculate the loss from the positive predictions.
```
# Print and view column zero of the weight
print(f"w_p[0]: {w_p[0]}")
print(f"y_true[:,0]: {y_true[:,0]}")
print(f"y_pred[:,0]: {y_pred[:,0]}")
# calculate the loss from the positive predictions, for class 0
loss_0_pos = -1 * np.sum(w_p[0] *
y_true[:, 0] *
np.log(y_pred[:, 0])
)
print(f"loss_0_pos: {loss_0_pos:.4f}")
```
View the zero column for the weights, true values, and predictions that you will use to calculate the loss from the negative predictions.
```
# Print and view column zero of the weight
print(f"w_n[0]: {w_n[0]}")
print(f"y_true[:,0]: {y_true[:,0]}")
print(f"y_pred[:,0]: {y_pred[:,0]}")
# Calculate the loss from the negative predictions, for class 0
loss_0_neg = -1 * np.sum(
w_n[0] *
(1 - y_true[:, 0]) *
np.log(1 - y_pred[:, 0])
)
print(f"loss_0_neg: {loss_0_neg:.4f}")
# add the two loss terms to get the total loss for class 0
loss_0 = loss_0_neg + loss_0_pos
print(f"loss_0: {loss_0:.4f}")
```
Now you are familiar with the array slicing that you would use when there are multiple disease classes stored in a two-dimensional array.
#### Now it's your turn!
* Can you calculate the loss for class (column) `1`?
```
# calculate the loss from the positive predictions, for class 1
loss_1_pos = None
```
Expected output
```CPP
loss_1_pos: 0.2853
```
```
# Calculate the loss from the negative predictions, for class 1
loss_1_neg = None
```
#### Expected output
```CPP
loss_1_neg: 0.9632
```
```
# add the two loss terms to get the total loss for class 0
loss_1 = None
```
#### Expected output
```CPP
loss_1: 1.2485
```
### Note
The data for the two classes (two columns) as well as the predictions were chosen so that you end up getting the same weighted loss for both categories.
- In general, you will expect to calculate different weighted loss values for each disease category, as the model predictions and data will differ from one category to another.
If you want some help, please click on the green "Solution" cell below to reveal the solution.
<details>
<summary>
<font size="3" color="darkgreen"><b>Solution</b></font>
</summary>
<p>
<code>
-- # calculate the loss from the positive predictions, for class 1
loss_1_pos = -1 * np.sum(w_p[1] *
y_true[:, 1] *
np.log(y_pred[:, 1])
)
print(f"loss_1_pos: {loss_1_pos:.4f}")
-- # Calculate the loss from the negative predictions, for class 1
loss_1_neg = -1 * np.sum(
w_n[1] *
(1 - y_true[:, 1]) *
np.log(1 - y_pred[:, 1])
)
print(f"loss_1_neg: {loss_1_neg:.4f}")
-- # add the two loss terms to get the total loss for class 1
loss_1 = loss_1_neg + loss_1_pos
print(f"loss_1: {loss_1:.4f}")
</code>
</p>
### How this practice relates to and differs from the upcoming graded assignment
- In the assignment, you will generalize this to calculating the loss for any number of classes.
- Also in the assignment, you will learn how to avoid taking the log of zero by adding a small number (more details will be explained in the assignment).
- Note that in the lecture videos and in this lecture notebook, you are taking the **sum** of losses for all examples. In the assignment, you will take the **average (the mean)** for all examples.
- Finally, in the assignment, you will work with "tensors" in TensorFlow, so you will use the TensorFlow equivalents of the numpy operations (keras.mean instead of numpy.mean).
#### That's all for this lab. You now have a couple more tools you'll need for this week's assignment!
|
github_jupyter
|
```
import psycopg2
import pandas as pd
import pandas.io.sql as pd_sql
import numpy as np
import matplotlib.pyplot as plt
def connectDB(DB):
# connect to the PostgreSQL server
return psycopg2.connect(
database=DB,
user="postgres",
password="Georgetown16",
host="database-1.c5vispb5ezxg.us-east-1.rds.amazonaws.com",
port='5432')
def disconnectDB(conn):
conn.close()
# connect to "Dataset" DB
conn = connectDB("Dataset")
# extract everything from 'table_name' into a dataframe
df = pd_sql.read_sql(f"select * from public.\"analysisFeatures\" ", con=conn).reset_index()
#make sure that all columns are displayed in our dataframe
pd.set_option('display.max_column',50)
#check dataframe
df.head(100)
#count null values of date_unregistration
df['date_unregistration'].isnull().sum()
drop_list = ['reg_period','code_presentation','date_unregistration','pass_fail_ind','std_total_weight']
df = df.drop(drop_list, axis=1)
df.head(10)
df['module_domain'].value_counts()
df['code_module'].value_counts()
#mapping the columns
df['imd_band'] = df['imd_band'].map({'0-10%':0,'10-20':1,'20-30%':2,'30-40%':3,'40-50%':4,'50-60%':5,'60-70%':6,'70-80%':7,'80-90%':8,'90-100%':9})
df['module_domain'] = df['module_domain'].map({'SocialScience': 0,'STEM': 1})
df['code_module'] = df['code_module'].map({'AAA': 0,'BBB': 1,'CCC':2,'DDD':3,'EEE':4,'FFF':5,'GGG':6})
df['term'] = df['term'].map({'J': 0,'B': 1})
df['year'] = df['year'].map({'2013': 0,'2014': 1})
df['gender'] = df['gender'].map({'M': 0,'F': 1})
df['age_band'] = df['age_band'].map({'0-35': 0,'35-55': 1,'55<=':2})
df['region'] = df['region'].map({'Scotland': 0,'East Anglian Region': 1,'London Region':2,'South Region': 3,'North Western Region': 4,'West Midlands Region':5,'South West Region': 6,'East Midlands Region': 7,'South East Region':8,'Wales': 9,'Yorkshire Region': 10,'North Region':11,'Ireland':12})
df['final_result'] = df['final_result'].map({'Withdrawn':0, 'Fail':0,'Pass':1,'Distinction':1})
df['disability'] = df['disability'].map({'N':0,'Y':1})
df['highest_education'] = df['highest_education'].map({'No Formal quals':0,'Lower Than A Level':1,'A Level or Equivalent':2,'HE Qualification':3,'Post Graduate Qualification':4})
df.head(10)
# write dataframe to database
from sqlalchemy import create_engine
engine = create_engine('postgresql://postgres:Georgetown16@database-1.c5vispb5ezxg.us-east-1.rds.amazonaws.com:5432/Dataset')
df.to_sql('featureSTG1', engine, if_exists='replace')
disconnectDB(conn)
```
|
github_jupyter
|
# Introduction à Python
> présentée par Loïc Messal
## Introduction aux flux de contrôles
### Les tests
Ils permettent d'exécuter des déclarations sous certaines conditions.
```
age = 17
if age < 18:
print("Mineur") # executé si et seulement si la condition est vraie
age = 19
if age < 18:
print("Mineur") # executé si et seulement si la condition est vraie
else:
print("Majeur") # exécuté si et seulement si la condition est fausse
employeur = "JLR"
# employeur = "Jakarto"
# employeur = "Une autre entreprise"
# Commentez, décommentez les valeurs de la variable employeur pour tester.
if employeur == "JLR":
# exécuté si et seulement si la condition employeur == "JLR" est vraie
richesse_statut = "riche"
elif employeur == "Jakarto":
# exécuté si et seulement si la condition employeur == "Jakarto" est vraie
# et qu'aucune condition précédente n'a été remplie
richesse_statut = "ça s'en vient bientôt"
else:
# exécuté si et seulement si aucune condition précédente n'a été remplie
richesse_statut = "probablement pas"
print("Richesse d'un employé de {} : {}".format(employeur, richesse_statut))
```
### Les boucles
Les boucles permettent d'itérer sur des itérables (composés de plusieurs éléments).
```
un_iterable = []
un_iterable.append({"nom": "Messal", "prénom": "Loïc", "employeur": "Jakarto", "age": 23})
un_iterable.append({"nom": "Lassem", "prénom": "Ciol", "employeur": "Otrakaj", "age": 17})
un_iterable.append({"nom": "Alssem", "prénom": "Icol", "employeur": "Torakaj", "age": 20})
un_iterable
for item in un_iterable:
print("{} {} travaille chez {}.".format(item["prénom"], item["nom"], item["employeur"]))
```
Il est possible de générer des séquences avec la fonction `range()`.
```
for compteur in range(5): # range(5) génére une séquence de 0 à 5 (exclus)
print(compteur)
for compteur in range(1, 5+1): # range(1, 5+1) génére une séquence de 1 à 5+1 (exclus)
print(compteur)
for index in range(len(un_iterable)):
item = un_iterable[index] # accède à l'item à partir de son index
print("Item {} : {} {} travaille chez {}.".format(index, item["prénom"], item["nom"], item["employeur"]))
for index, item in enumerate(un_iterable): # enumerate permet d'itérer en obtenant l'index ET l'item
print("Item {} : {} {} travaille chez {}.".format(index, item["prénom"], item["nom"], item["employeur"]))
compteur = 0
stop = 5
while compteur < stop: # exécutera les déclarations suivantes tant que la condition est vraie
print(compteur)
compteur = compteur + 1
```
Il est possible de contrôler les boucles avec certains mots clés:
- `continue` passera à l'itération suivante sans exécuter les déclarations qui suivent
- `break` quittera la boucle prématurément
```
for index, item in enumerate(un_iterable):
if item["age"] < 18:
continue # Si la condition est vraie, passage à l'itération suivante.
print("Item {} : {} {} (majeur) travaille chez {}.".format(index, item["prénom"], item["nom"], item["employeur"]))
for index, item in enumerate(un_iterable):
print("Item {} : {} {} travaille chez {}.".format(index, item["prénom"], item["nom"], item["employeur"]))
if item["prénom"] == "Loïc":
break # Arrête la boucle si la condition est vraie
```
[Prochain chapitre : Les fonctions](04_Fonctions.ipynb)
|
github_jupyter
|
# Introduction
Linear Regression is one of the most famous and widely used machine learning algorithms out there. It assumes that the target variable can be explained as a linear combination of the input features. What does this mean? It means that the target can be viewed as a weighted sum of each feature. Let’s use a practical example to illustrate that.
Let’s say that we are opening a restaurant, we make great food but we want to know how much to charge for it. We can be very pragmatic and say that the cost of the meal is directly related to what is in it. We can, for instance, have a rule that each ingredient costs a certain amount, and based on how much there is of each ingredient in the dish, we can calculate its price. There may also be a fixed minimum price for each dish. Mathematically, this is called the intercept.
```
fixed_price = 5
ingredient_costs = {"meat": 10,
"fish": 13,
"vegetables": 2,
"fries": 3}
def price(**ingredients):
""" returns the price of a dish """
cost = 0
for name, quantity in ingredients.items():
cost += ingredient_costs[name] * quantity
return cost
```
Linear Regression makes the assumption that the target, in this case, the price, can be explained like this. The model will know about the quantity of each ingredient, but it will have to infer what the fixed price is, and what is the cost of each ingredient.
>It is important to remember that cost, in this situation, is rather abstract. It represents how much each feature affect the outcome, and in which way. Therefore, features can have negative costs for instance.
In the univariate case, where there is only one feature, Linear Regression can be thought of as trying to fit a line through points.

Now, Linear Regression is one of the most popular algorithms because it can do much more than fit straight lines through data. Indeed, with a simple trick, we can make it fit polynomial functions, making it much more powerful.
The trick is to "replace" the original features with a polynomial of a higher degree. In the univariate case, this comes down to not only using the feature itself but also its squared value, cubed value, and so on. For instance, instead of using a single feature $X = 2$, we end up with features $X = 2, 4, 8, 16, 32$, and so on. More features mean that the model is explained by more weights, and these weights can express more complex functions.

A Linear Regression model's goal is to find the coefficients, also called weights, which will fit the data best. In order to define what best means, we need to define a loss function. This loss function, as we will see later, can be tweaked to alter how the weights are learned. We will also see that finding the best weights in order to minimize the loss function can be done in different ways.
|
github_jupyter
|
######The Iris flower data set is a multivariate data set introduced by the British statistician and biologist Ronald Fisher in his 1936 paper The use of multiple measurements in taxonomic problems. The dataset consists of 50 samples from each of three species of Iris (Iris Setosa, Iris virginica, and Iris versicolor). Four features were measured from each sample: the length and the width of the sepals and petals, in centimeters.
Import Libraries
```
import numpy as np
import pandas as pd
from pandas import Series, DataFrame
import seaborn as sns
import matplotlib.pyplot as plt
iris = pd.read_csv("iris.csv")
iris.head()
```
##### *We can see that we have a column named ID that we donot need , so let's drop it !*
```
iris.drop("Id", axis=1, inplace = True)
iris.info()
figure = iris[iris.Species == 'Iris-setosa'].plot(kind='scatter', x='SepalLengthCm', y='SepalWidthCm', color='red', label='Setosa')
iris[iris.Species == 'Iris-versicolor'].plot(kind='scatter', x='SepalLengthCm', y='SepalWidthCm', color='blue', label='Versicolor', ax=figure)
iris[iris.Species == 'Iris-virginica'].plot(kind='scatter', x='SepalLengthCm', y='SepalWidthCm', color='green', label='Virginica', ax=figure)
figure.set_xlabel('Sepal Length')
figure.set_ylabel('Sepal Width')
figure.set_title('Sepal Length Vs Width')
figure=plt.gcf()
figure.set_size_inches(7, 4)
plt.show()
figure = iris[iris.Species == 'Iris-setosa'].plot(kind='scatter', x='PetalLengthCm', y='PetalWidthCm', color='red', label='Setosa')
iris[iris.Species == 'Iris-versicolor'].plot(kind='scatter', x='PetalLengthCm', y='PetalWidthCm', color='blue', label='Versicolor', ax=figure)
iris[iris.Species == 'Iris-virginica'].plot(kind='scatter', x='PetalLengthCm', y='PetalWidthCm', color='green', label='Virginica', ax=figure)
figure.set_xlabel('Petal Length')
figure.set_ylabel('Petal Width')
figure.set_title('Petal Length Vs Width')
figure=plt.gcf()
figure.set_size_inches(7, 4)
plt.show()
plt.figure(figsize=(15,10))
plt.subplot(2,2,1)
sns.boxplot(x='Species',y='SepalLengthCm',data=iris)
plt.subplot(2,2,2)
sns.boxplot(x='Species',y='SepalWidthCm',data=iris)
plt.subplot(2,2,3)
sns.boxplot(x='Species',y='PetalLengthCm',data=iris)
plt.subplot(2,2,4)
sns.boxplot(x='Species',y='PetalWidthCm',data=iris)
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
from sklearn import svm
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier
import xgboost as xgb
```
Splitting The Data into Training And Testing Dataset
```
train, test = train_test_split(iris, test_size=0.2)
print(train.shape)
print(test.shape)
train_X = train[['SepalLengthCm','SepalWidthCm','PetalLengthCm','PetalWidthCm']]
train_y = train.Species
test_X = test[['SepalLengthCm','SepalWidthCm','PetalLengthCm','PetalWidthCm']]
test_y = test.Species
```
1. Logistic Regression
```
model1 = LogisticRegression()
model1.fit(train_X, train_y)
prediction1 = model1.predict(test_X)
print('Accuracy of Logistic Regression is: ', metrics.accuracy_score(prediction1, test_y))
```
2. SVM Classifier
```
model2 = svm.SVC()
model2.fit(train_X, train_y)
prediction2 = model2.predict(test_X)
print('Accuracy of SVM is: ', metrics.accuracy_score(prediction2, test_y))
```
3. K-Nearest Neighbors
```
model3 = KNeighborsClassifier(n_neighbors=3) # this examines 3 neighbors
model3.fit(train_X, train_y)
prediction3 = model3.predict(test_X)
print('Accuracy of KNN is: ', metrics.accuracy_score(prediction3, test_y))
```
4. Decision Tree
```
model4 = DecisionTreeClassifier()
model4.fit(train_X, train_y)
prediction4 = model4.predict(test_X)
print('Accuracy of Decision Tree is: ', metrics.accuracy_score(prediction4, test_y))
```
5. XGBoost
```
model5 = xgb.XGBClassifier()
model5.fit(train_X, train_y)
prediction5 = model5.predict(test_X)
print('Accuracy of xgb classifier is: ', metrics.accuracy_score(prediction5, test_y))
```
|
github_jupyter
|
```
%load_ext autoreload
%autoreload 2
import pathlib
import IPython.display
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import scipy.interpolate
import scipy.signal
import pymedphys
import pymedphys._wlutz.iview
indexed_dir = pathlib.Path(r'S:\DataExchange\iViewDB_decoded\indexed')
movie_dirs = list(indexed_dir.glob('*/*/*/*/*'))
movie_dirs
wlutz_results = {}
edge_lengths = [20, 24]
pd.set_option("display.max_rows", 101)
for directory in movie_dirs:
image_paths = list(directory.glob('*.png'))
print(image_paths)
wlutz_results[directory] = pymedphys._wlutz.iview.batch_process(image_paths, edge_lengths, display_figure=True)
IPython.display.display(wlutz_results[directory])
for directory in movie_dirs:
try:
wlutz_results[directory]
except KeyError:
image_paths = list(directory.glob('*.png'))
print(image_paths)
try:
wlutz_results[directory] = pymedphys._wlutz.iview.batch_process(image_paths, edge_lengths, display_figure=True)
IPython.display.display(wlutz_results[directory])
except ValueError:
continue
for directory in movie_dirs:
try:
wlutz_results[directory]
except KeyError:
image_paths = list(directory.glob('*.png'))
print(image_paths)
try:
wlutz_results[directory] = pymedphys._wlutz.iview.batch_process(image_paths, edge_lengths, display_figure=True)
IPython.display.display(wlutz_results[directory])
except ValueError:
continue
for directory in movie_dirs:
try:
wlutz_results[directory]
except KeyError:
image_paths = list(directory.glob('*.png'))
print(image_paths)
try:
wlutz_results[directory] = pymedphys._wlutz.iview.batch_process(image_paths, edge_lengths, display_figure=True)
IPython.display.display(wlutz_results[directory])
except ValueError:
continue
for key, table in wlutz_results.items():
print(key)
IPython.display.display(table)
keys = list(wlutz_results.keys())
keys
direction_keys = [
key.parent.stem for key in keys
]
direction_keys
rotations = [
wlutz_results[key]['Rotation']
for key in keys
]
lt_zero = rotations[0] < 0
gantry = np.empty_like(rotations[0])
gantry[lt_zero] = -180 - rotations[0][lt_zero]
gte_zero = np.invert(lt_zero)
gantry[gte_zero] = 180 - rotations[0][gte_zero]
gantry
gantry = []
for i, direction_key in enumerate(direction_keys):
if direction_keys[i] == '00_CW':
diff = np.diff(np.concatenate([[-180], rotations[i]]))
diff[diff > 0] = diff[diff > 0] - 180
gantry.append(-180 - np.cumsum(diff * 2))
elif direction_keys[i] == '01_CC':
diff = np.diff(np.concatenate([[0], rotations[i]]))
diff[diff < 0] = diff[diff < 0] + 180
gantry.append(180 - np.cumsum(diff * 2))
else:
raise ValueError("Expected one of '00_CW' or '01_CC'")
gantry
bb_x = [
wlutz_results[key]['BB x'] for key in keys
]
bb_y = [
wlutz_results[key]['BB y'] for key in keys
]
gantry
bb_x
scipy.interpolate.interp1d?
interp_bb_x = [
scipy.interpolate.interp1d(g, x, bounds_error=False, fill_value='extrapolate')
for g, x in zip(gantry, bb_x)
]
def get_avg_bb_x(gantry):
results = []
for interp in interp_bb_x:
results.append(interp(gantry))
return (np.min(results, axis=0) + np.max(results, axis=0))/2
interp_bb_y = [
scipy.interpolate.interp1d(g, y, bounds_error=False, fill_value='extrapolate')
for g, y in zip(gantry, bb_y)
]
def get_avg_bb_y(gantry):
results = []
for interp in interp_bb_y:
results.append(interp(gantry))
return (np.min(results, axis=0) + np.max(results, axis=0))/2
get_avg_bb_y([0, 2])
# gantry_all = np.concatenate(gantry)
# ind = np.argsort(gantry_all)
# sorted_gantry = gantry_all[ind]
# within_bounds = np.logical_and(sorted_gantry <= 180, sorted_gantry >= -180)
# sorted_gantry = sorted_gantry[within_bounds]
# sorted_bb_x = np.concatenate(bb_x)[ind][within_bounds]
# sorted_bb_y = np.concatenate(bb_y)[ind][within_bounds]
# b, a = scipy.signal.butter(3, 0.05)
# filtered_bb_x = scipy.signal.filtfilt(b, a, sorted_bb_x)
# filtered_bb_y = scipy.signal.filtfilt(b, a, sorted_bb_y)
# plt.plot(sorted_gantry, filtered_bb_x)
# unique_gantry, unique_inverse = np.unique(sorted_gantry, return_inverse=True)
# inc = np.arange(len(unique_inverse))
# make_unique = np.ones((len(unique_gantry), len(unique_inverse))) * np.nan
# make_unique[unique_inverse, inc] = sorted_bb_x
# striclty_increasing_bb_x = np.nanmean(make_unique, axis=1)
# make_unique[unique_inverse, inc] = sorted_bb_y
# striclty_increasing_bb_y = np.nanmean(make_unique, axis=1)
# def predict_bb_pos(gantry, gantry_range=10):
# gantry = np.array(gantry)
# lte = gantry[:,None] - gantry_range <= gantry_all[None,:]
# gte = gantry[:,None] + gantry_range >= gantry_all[None,:]
# in_range = np.logical_and(lte, gte)
# sorted_bb_x
# return in_range
# predict_bb_pos([0, 1], gantry_range=10)
# unique_gantry = np.unique(sorted_gantry)
# bb_x_interp = scipy.interpolate.interp1d(sorted_gantry, filtered_bb_x, bounds_error=False)
# bb_y_interp = scipy.interpolate.interp1d(sorted_gantry, filtered_bb_y, bounds_error=False)
# bb_x_interp = scipy.interpolate.UnivariateSpline(unique_gantry, strictly_increasing_bb_x, s=0.1)
# bb_y_interp = scipy.interpolate.UnivariateSpline(unique_gantry, strictly_increasing_bb_y, s=1)
gantry_i = np.linspace(-180, 180, 91)
for i, key in enumerate(keys):
plt.plot(gantry[i], bb_x[i], '.')
plt.plot(gantry_i, get_avg_bb_x(gantry_i))
plt.xlim([-180, 180])
for i, key in enumerate(keys):
plt.plot(gantry[i], bb_y[i], '.')
plt.plot(gantry_i, get_avg_bb_y(gantry_i))
plt.xlim([-180, 180])
```
|
github_jupyter
|
# Flopy MODFLOW Boundary Conditions
Flopy has a new way to enter boundary conditions for some MODFLOW packages. These changes are substantial. Boundary conditions can now be entered as a list of boundaries, as a numpy recarray, or as a dictionary. These different styles are described in this notebook.
Flopy also now requires zero-based input. This means that **all boundaries are entered in zero-based layer, row, and column indices**. This means that older Flopy scripts will need to be modified to account for this change. If you are familiar with Python, this should be natural, but if not, then it may take some time to get used to zero-based numbering. Flopy users submit all information in zero-based form, and Flopy converts this to the one-based form required by MODFLOW.
The following MODFLOW packages are affected by this change:
* Well
* Drain
* River
* General-Head Boundary
* Time-Variant Constant Head
This notebook explains the different ways to enter these types of boundary conditions.
```
#begin by importing flopy
import os
import sys
import numpy as np
# run installed version of flopy or add local path
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
workspace = os.path.join('data')
#make sure workspace directory exists
if not os.path.exists(workspace):
os.makedirs(workspace)
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('flopy version: {}'.format(flopy.__version__))
```
## List of Boundaries
Boundary condition information is passed to a package constructor as stress_period_data. In its simplest form, stress_period_data can be a list of individual boundaries, which themselves are lists. The following shows a simple example for a MODFLOW River Package boundary:
```
stress_period_data = [
[2, 3, 4, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
[2, 3, 5, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
[2, 3, 6, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
]
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data)
m.write_input()
```
If we look at the River Package created here, you see that the layer, row, and column numbers have been increased by one.
```
!head -n 10 'data/test.riv'
```
If this model had more than one stress period, then Flopy will assume that this boundary condition information applies until the end of the simulation
```
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
dis = flopy.modflow.ModflowDis(m, nper=3)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data)
m.write_input()
!head -n 10 'data/test.riv'
```
## Recarray of Boundaries
Numpy allows the use of recarrays, which are numpy arrays in which each column of the array may be given a different type. Boundary conditions can be entered as recarrays. Information on the structure of the recarray for a boundary condition package can be obtained from that particular package. The structure of the recarray is contained in the dtype.
```
riv_dtype = flopy.modflow.ModflowRiv.get_default_dtype()
print(riv_dtype)
```
Now that we know the structure of the recarray that we want to create, we can create a new one as follows.
```
stress_period_data = np.zeros((3), dtype=riv_dtype)
stress_period_data = stress_period_data.view(np.recarray)
print('stress_period_data: ', stress_period_data)
print('type is: ', type(stress_period_data))
```
We can then fill the recarray with our boundary conditions.
```
stress_period_data[0] = (2, 3, 4, 10.7, 5000., -5.7)
stress_period_data[1] = (2, 3, 5, 10.7, 5000., -5.7)
stress_period_data[2] = (2, 3, 6, 10.7, 5000., -5.7)
print(stress_period_data)
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data)
m.write_input()
!head -n 10 'data/test.riv'
```
As before, if we have multiple stress periods, then this recarray will apply to all of them.
```
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
dis = flopy.modflow.ModflowDis(m, nper=3)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data)
m.write_input()
!head -n 10 'data/test.riv'
```
## Dictionary of Boundaries
The power of the new functionality in Flopy3 is the ability to specify a dictionary for stress_period_data. If specified as a dictionary, the key is the stress period number (**as a zero-based number**), and the value is either a nested list, an integer value of 0 or -1, or a recarray for that stress period.
Let's say that we want to use the following schedule for our rivers:
0. No rivers in stress period zero
1. Rivers specified by a list in stress period 1
2. No rivers
3. No rivers
4. No rivers
5. Rivers specified by a recarray
6. Same recarray rivers
7. Same recarray rivers
8. Same recarray rivers
```
sp1 = [
[2, 3, 4, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
[2, 3, 5, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
[2, 3, 6, 10.7, 5000., -5.7], #layer, row, column, stage, conductance, river bottom
]
print(sp1)
riv_dtype = flopy.modflow.ModflowRiv.get_default_dtype()
sp5 = np.zeros((3), dtype=riv_dtype)
sp5 = sp5.view(np.recarray)
sp5[0] = (2, 3, 4, 20.7, 5000., -5.7)
sp5[1] = (2, 3, 5, 20.7, 5000., -5.7)
sp5[2] = (2, 3, 6, 20.7, 5000., -5.7)
print(sp5)
sp_dict = {0:0, 1:sp1, 2:0, 5:sp5}
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
dis = flopy.modflow.ModflowDis(m, nper=8)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=sp_dict)
m.write_input()
!head -n 10 'data/test.riv'
```
## MODFLOW Auxiliary Variables
Flopy works with MODFLOW auxiliary variables by allowing the recarray to contain additional columns of information. The auxiliary variables must be specified as package options as shown in the example below.
In this example, we also add a string in the last column of the list in order to name each boundary condition. In this case, however, we do not include boundname as an auxiliary variable as MODFLOW would try to read it as a floating point number.
```
#create an empty array with an iface auxiliary variable at the end
riva_dtype = [('k', '<i8'), ('i', '<i8'), ('j', '<i8'),
('stage', '<f4'), ('cond', '<f4'), ('rbot', '<f4'),
('iface', '<i4'), ('boundname', object)]
riva_dtype = np.dtype(riva_dtype)
stress_period_data = np.zeros((3), dtype=riva_dtype)
stress_period_data = stress_period_data.view(np.recarray)
print('stress_period_data: ', stress_period_data)
print('type is: ', type(stress_period_data))
stress_period_data[0] = (2, 3, 4, 10.7, 5000., -5.7, 1, 'riv1')
stress_period_data[1] = (2, 3, 5, 10.7, 5000., -5.7, 2, 'riv2')
stress_period_data[2] = (2, 3, 6, 10.7, 5000., -5.7, 3, 'riv3')
print(stress_period_data)
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data, dtype=riva_dtype, options=['aux iface'])
m.write_input()
!head -n 10 'data/test.riv'
```
## Working with Unstructured Grids
Flopy can create an unstructured grid boundary condition package for MODFLOW-USG. This can be done by specifying a custom dtype for the recarray. The following shows an example of how that can be done.
```
#create an empty array based on nodenumber instead of layer, row, and column
rivu_dtype = [('nodenumber', '<i8'), ('stage', '<f4'), ('cond', '<f4'), ('rbot', '<f4')]
rivu_dtype = np.dtype(rivu_dtype)
stress_period_data = np.zeros((3), dtype=rivu_dtype)
stress_period_data = stress_period_data.view(np.recarray)
print('stress_period_data: ', stress_period_data)
print('type is: ', type(stress_period_data))
stress_period_data[0] = (77, 10.7, 5000., -5.7)
stress_period_data[1] = (245, 10.7, 5000., -5.7)
stress_period_data[2] = (450034, 10.7, 5000., -5.7)
print(stress_period_data)
m = flopy.modflow.Modflow(modelname='test', model_ws=workspace)
riv = flopy.modflow.ModflowRiv(m, stress_period_data=stress_period_data, dtype=rivu_dtype)
m.write_input()
print(workspace)
!head -n 10 'data/test.riv'
```
## Combining two boundary condition packages
```
ml = flopy.modflow.Modflow(modelname="test",model_ws=workspace)
dis = flopy.modflow.ModflowDis(ml,10,10,10,10)
sp_data1 = {3: [1, 1, 1, 1.0],5:[1,2,4,4.0]}
wel1 = flopy.modflow.ModflowWel(ml, stress_period_data=sp_data1)
ml.write_input()
!head -n 10 'data/test.wel'
sp_data2 = {0: [1, 1, 3, 3.0],8:[9,2,4,4.0]}
wel2 = flopy.modflow.ModflowWel(ml, stress_period_data=sp_data2)
ml.write_input()
!head -n 10 'data/test.wel'
```
Now we create a third wel package, using the ```MfList.append()``` method:
```
wel3 = flopy.modflow.ModflowWel(ml,stress_period_data=\
wel2.stress_period_data.append(
wel1.stress_period_data))
ml.write_input()
!head -n 10 'data/test.wel'
```
|
github_jupyter
|
# T81-558: Applications of Deep Neural Networks
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), School of Engineering and Applied Science, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
**Module 3 Assignment: Creating Columns in Pandas**
**Student Name: Your Name**
# Assignment Instructions
For this assignment you will use the **reg-30-spring-2018.csv** dataset. This is a dataset that I generated specifically for this semester. You can find the CSV file in the **data** directory of the class GitHub repository here: [reg-30-spring-2018.csv](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/data/reg-30-spring-2018.csv).
For this assignment, load and modify the data set. You will submit this modified dataset to the **submit** function. See [Assignment #1](https://github.com/jeffheaton/t81_558_deep_learning/blob/master/assignments/assignment_yourname_class1.ipynb) for details on how to submit an assignment or check that one was submitted.
Modify the dataset as follows:
* Add a column named *density* that is *weight* divided by *volume*.
* Replace the *region* column with dummy variables.
* Replace the *item* column with an index encoding value (for example 0 for the first class, 1 for the next, etc. see function *encode_text_index*)
* Your submitted dataframe will have these columns: id, distance, height, landings, number, pack, age, usage, weight, item, volume, width, max, power, size, target, density, region-RE-0, region-RE-1, region-RE-10, region-RE-11, region-RE-2, region-RE-3, region-RE-4, region-RE-5, region-RE-6, region-RE-7, region-RE-8, region-RE-9, region-RE-A, region-RE-B, region-RE-C, region-RE-D, region-RE-E, region-RE-F.
# Helpful Functions
You will see these at the top of every module and assignment. These are simply a set of reusable functions that we will make use of. Each of them will be explained as the semester progresses. They are explained in greater detail as the course progresses. Class 4 contains a complete overview of these functions.
```
from sklearn import preprocessing
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import shutil
import os
import requests
import base64
# Encode text values to dummy variables(i.e. [1,0,0],[0,1,0],[0,0,1] for red,green,blue)
def encode_text_dummy(df, name):
dummies = pd.get_dummies(df[name])
for x in dummies.columns:
dummy_name = "{}-{}".format(name, x)
df[dummy_name] = dummies[x]
df.drop(name, axis=1, inplace=True)
# Encode text values to a single dummy variable. The new columns (which do not replace the old) will have a 1
# at every location where the original column (name) matches each of the target_values. One column is added for
# each target value.
def encode_text_single_dummy(df, name, target_values):
for tv in target_values:
l = list(df[name].astype(str))
l = [1 if str(x) == str(tv) else 0 for x in l]
name2 = "{}-{}".format(name, tv)
df[name2] = l
# Encode text values to indexes(i.e. [1],[2],[3] for red,green,blue).
def encode_text_index(df, name):
le = preprocessing.LabelEncoder()
df[name] = le.fit_transform(df[name])
return le.classes_
# Encode a numeric column as zscores
def encode_numeric_zscore(df, name, mean=None, sd=None):
if mean is None:
mean = df[name].mean()
if sd is None:
sd = df[name].std()
df[name] = (df[name] - mean) / sd
# Convert all missing values in the specified column to the median
def missing_median(df, name):
med = df[name].median()
df[name] = df[name].fillna(med)
# Convert all missing values in the specified column to the default
def missing_default(df, name, default_value):
df[name] = df[name].fillna(default_value)
# Convert a Pandas dataframe to the x,y inputs that TensorFlow needs
def to_xy(df, target):
result = []
for x in df.columns:
if x != target:
result.append(x)
# find out the type of the target column. Is it really this hard? :(
target_type = df[target].dtypes
target_type = target_type[0] if hasattr(target_type, '__iter__') else target_type
# Encode to int for classification, float otherwise. TensorFlow likes 32 bits.
if target_type in (np.int64, np.int32):
# Classification
dummies = pd.get_dummies(df[target])
return df.as_matrix(result).astype(np.float32), dummies.as_matrix().astype(np.float32)
else:
# Regression
return df.as_matrix(result).astype(np.float32), df.as_matrix([target]).astype(np.float32)
# Nicely formatted time string
def hms_string(sec_elapsed):
h = int(sec_elapsed / (60 * 60))
m = int((sec_elapsed % (60 * 60)) / 60)
s = sec_elapsed % 60
return "{}:{:>02}:{:>05.2f}".format(h, m, s)
# Regression chart.
def chart_regression(pred,y,sort=True):
t = pd.DataFrame({'pred' : pred, 'y' : y.flatten()})
if sort:
t.sort_values(by=['y'],inplace=True)
a = plt.plot(t['y'].tolist(),label='expected')
b = plt.plot(t['pred'].tolist(),label='prediction')
plt.ylabel('output')
plt.legend()
plt.show()
# Remove all rows where the specified column is +/- sd standard deviations
def remove_outliers(df, name, sd):
drop_rows = df.index[(np.abs(df[name] - df[name].mean()) >= (sd * df[name].std()))]
df.drop(drop_rows, axis=0, inplace=True)
# Encode a column to a range between normalized_low and normalized_high.
def encode_numeric_range(df, name, normalized_low=-1, normalized_high=1,
data_low=None, data_high=None):
if data_low is None:
data_low = min(df[name])
data_high = max(df[name])
df[name] = ((df[name] - data_low) / (data_high - data_low)) \
* (normalized_high - normalized_low) + normalized_low
# This function submits an assignment. You can submit an assignment as much as you like, only the final
# submission counts. The paramaters are as follows:
# data - Pandas dataframe output.
# key - Your student key that was emailed to you.
# no - The assignment class number, should be 1 through 1.
# source_file - The full path to your Python or IPYNB file. This must have "_class1" as part of its name.
# . The number must match your assignment number. For example "_class2" for class assignment #2.
def submit(data,key,no,source_file=None):
if source_file is None and '__file__' not in globals(): raise Exception('Must specify a filename when a Jupyter notebook.')
if source_file is None: source_file = __file__
suffix = '_class{}'.format(no)
if suffix not in source_file: raise Exception('{} must be part of the filename.'.format(suffix))
with open(source_file, "rb") as image_file:
encoded_python = base64.b64encode(image_file.read()).decode('ascii')
ext = os.path.splitext(source_file)[-1].lower()
if ext not in ['.ipynb','.py']: raise Exception("Source file is {} must be .py or .ipynb".format(ext))
r = requests.post("https://api.heatonresearch.com/assignment-submit",
headers={'x-api-key':key}, json={'csv':base64.b64encode(data.to_csv(index=False).encode('ascii')).decode("ascii"),
'assignment': no, 'ext':ext, 'py':encoded_python})
if r.status_code == 200:
print("Success: {}".format(r.text))
else: print("Failure: {}".format(r.text))
```
# Assignment #3 Sample Code
The following code provides a starting point for this assignment.
```
import os
import pandas as pd
from scipy.stats import zscore
# This is your student key that I emailed to you at the beginnning of the semester.
key = "qgABjW9GKV1vvFSQNxZW9akByENTpTAo2T9qOjmh" # This is an example key and will not work.
# You must also identify your source file. (modify for your local setup)
# file='/resources/t81_558_deep_learning/assignment_yourname_class1.ipynb' # IBM Data Science Workbench
# file='C:\\Users\\jeffh\\projects\\t81_558_deep_learning\\t81_558_class1_intro_python.ipynb' # Windows
#file='/Users/jeff/projects/t81_558_deep_learning/assignment_yourname_class1.ipynb' # Mac/Linux
file = '...location of your source file...'
# Begin assignment
path = "./data/"
filename_read = os.path.join(path,"reg-30-spring-2018.csv")
df = pd.read_csv(filename_read)
# Calculate density
# Encode dummies
# Save a copy to examine, if you like
df.to_csv('3.csv',index=False)
# Submit
submit(source_file=file,data=df,key=key,no=3)
```
# Checking Your Submission
You can always double check to make sure your submission actually happened. The following utility code will help with that.
```
import requests
import pandas as pd
import base64
import os
def list_submits(key):
r = requests.post("https://api.heatonresearch.com/assignment-submit",
headers={'x-api-key': key},
json={})
if r.status_code == 200:
print("Success: \n{}".format(r.text))
else:
print("Failure: {}".format(r.text))
def display_submit(key,no):
r = requests.post("https://api.heatonresearch.com/assignment-submit",
headers={'x-api-key': key},
json={'assignment':no})
if r.status_code == 200:
print("Success: \n{}".format(r.text))
else:
print("Failure: {}".format(r.text))
# Show a listing of all submitted assignments.
key = "qgABjW9GKV1vvFSQNxZW9akByENTpTAo2T9qOjmh"
list_submits(key)
# Show one assignment, by number.
display_submit(key,3)
```
|
github_jupyter
|
```
import statistics
import pprint
import pandas as pd
import numpy as np
from random import uniform
from tslearn.utils import to_time_series_dataset
from tslearn.metrics import dtw#, gak
import plotly.express as px
import scipy.stats as st
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
import seaborn as sns; sns.set()
#ToDo: Threading
def get_best_distribution(data):
dist_names = ["gamma", "gumbel_l", "cauchy", "dgamma", "beta", "betaprime", "exponweib", "rayleigh", "fisk",
"gausshyper", "invweibull", "pareto", "alpha", "expon", "hypsecant", "mielke", "loggamma",
"rdist", "rice"] ## Agregar más a voluntad
dist_results = []
params = {}
for dist_name in dist_names:
dist = getattr(st, dist_name)
param = dist.fit(data)
params[dist_name] = param
# Applying the Kolmogorov-Smirnov test
D, p = st.kstest(data, dist_name, args=param)
print("p value for "+dist_name+" = "+str(p))
dist_results.append((dist_name, p))
# select the best fitted distribution
best_dist, best_p = (max(dist_results, key=lambda item: item[1]))
# store the name of the best fit and its p value
print("Best fitting distribution: "+str(best_dist))
print("Best p value: "+ str(best_p))
parms = params[best_dist]
#print("Parameters for the best fit: "+ str(parms))
map_parms = {}
dist = getattr(st, best_dist)
try:
counter_wrong_chars = 0 #To solve a bug
for position, shape_parameter in enumerate(dist.shapes):
#print(position, shape_parameter)
if shape_parameter not in [' ', ',']:
map_parms[shape_parameter] = parms[position-counter_wrong_chars]
else:
counter_wrong_chars += 1
except:
pass
finally:
map_parms["loc"] = parms[-2]
map_parms["scale"] = parms[-1]
print("Parameters for the best fit: "+ str(map_parms))
return best_dist, best_p, parms, map_parms
def get_optimal_curves(df_curves, example_curves, ts_example_curves, dict_probability_distrs, prob_distrs,
min_count_generated_curves, a, b, E_min, min_f_load, roof_dtw_distance, min_corr):
I = 5000 #5000
acum_generated_curves = 0
while acum_generated_curves < min_count_generated_curves:
for i in range(1,I+1):
C_i = [None] * 24
h_max = int(round(uniform(19, 21),0))
C_i[h_max] = 1
for h, none in enumerate(C_i):
if h != h_max:
function = dict_probability_distrs[prob_distrs[h][0]]
parms = prob_distrs[h][1]
was_random_number_found = False
while was_random_number_found is False:
E = function.rvs(**parms, size=1)[0]
if (E>=E_min and E<1):
was_random_number_found = True
C_i[h] = E
E_acum = sum(C_i)
if (E_acum>=a and E_acum<=b):
#print(C_i, type(C_i))
f_load = statistics.mean(C_i) / max(C_i)
if f_load >= min_f_load:
ts_C_i = to_time_series_dataset(C_i)[0]
dtw_distances = []
for k, curve in enumerate(ts_example_curves):
dtw_distance = dtw(ts_C_i, curve)
dtw_distances.append(dtw_distance)
average_dtw = statistics.mean(dtw_distances)
if average_dtw < roof_dtw_distance:
corrs = []
for example_curve in example_curves:
corr = np.corrcoef(C_i, example_curve)
corrs.append(corr[0][1])
average_corr = statistics.mean(corrs)
if average_corr>=min_corr:
print(i, f_load, E_acum, average_dtw, average_corr)
df_curves = df_curves.append(
{ '0': C_i[0], '1': C_i[1], '2': C_i[2],
'3': C_i[3], '4': C_i[4], '5': C_i[5],
'6': C_i[6], '7': C_i[7], '8': C_i[8],
'9': C_i[9], '10': C_i[10], '11': C_i[11],
'12': C_i[12], '13': C_i[13], '14': C_i[14],
'15': C_i[15], '16': C_i[16], '17': C_i[17],
'18': C_i[18], '19': C_i[19], '20': C_i[20],
'21': C_i[21], '22': C_i[22], '23': C_i[23],
'FC': f_load, 'Sum': E_acum,
'DTW_avg_distance': average_dtw, 'Avg_correlation': average_corr },
ignore_index=True
)
acum_generated_curves += 1
if acum_generated_curves>=min_count_generated_curves:
return (df_curves)
df_example_curves = pd.read_excel (r'Curvas.xlsx')
df_example_curves.drop(
df_example_curves.columns[
df_example_curves.columns.str.contains('unnamed', case = False, na=False)
],
axis = 1,
inplace = True
)
a = df_example_curves['Sum'].min()
b = df_example_curves['Sum'].max()
df_example_curves = df_example_curves.drop(['FC', 'Sum', 'Comentario'], axis=1)
print("a: ", a, " b: ", b)
print(df_example_curves)
prob_distrs = []
plots = []
for (columnName, columnData) in df_example_curves.iteritems():
## Maximizar el p-value ##
print('Colunm Name : ', columnName)
#print('Column Contents : ', columnData.values, type(columnData.values), columnData.values.shape)
best_dist, best_p, parms, map_parms = get_best_distribution(columnData.values)
prob_distrs.append([best_dist, map_parms])
#if columnName == 12:
# ax = sns.distplot(columnData.values, kde=False)
#ax = sns.distplot(columnData.values, kde=False)
print("prob_distrs: ")
pprint.pprint(prob_distrs)
dict_probability_distrs = { "gamma": st.gamma, "gumbel_l": st.gumbel_l, "cauchy": st.cauchy, "dgamma": st.dgamma,
"beta": st.beta, "betaprime": st.betaprime, "exponweib": st.exponweib, "rayleigh": st.rayleigh,
"fisk": st.fisk, "gausshyper": st.gausshyper, "invweibull": st.invweibull, "pareto": st.pareto,
"alpha": st.alpha, "expon": st.expon, "hypsecant": st.hypsecant, "mielke": st.mielke,
"loggamma": st.loggamma, "rdist": st.rdist, "rice": st.rice }
example_curves = df_example_curves.values.tolist()
ts_example_curves = to_time_series_dataset(example_curves)
#pprint.pprint(ts_example_curves)
df_curves = pd.DataFrame(
columns=[
'0','1','2','3','4','5','6','7','8','9','10','11','12','13','14','15','16','17','18','19','20','21','22','23',
'FC','Sum','DTW_avg_distance','Avg_correlation'
]
)
print(df_curves)
E_min = 0.375
min_f_load = 0.7625
min_count_generated_curves = 25
roof_dtw_distance = 0.25 #0.25
min_corr = 0.95 #0.95
df_curves = get_optimal_curves(df_curves, example_curves, ts_example_curves, dict_probability_distrs, prob_distrs,
min_count_generated_curves, a, b, E_min, min_f_load, roof_dtw_distance, min_corr)
print(df_curves)
for index, row in df_curves.loc[:, "0":"23"].iterrows():
fig = px.line(row, width=600, height=300, xlabel='Hora')
fig.show()
average_optimal_curve = df_curves.loc[:, "0":"23"].mean(axis=0)
print(average_optimal_curve, type(average_optimal_curve))
average_optimal_curve.plot(linewidth=3.0, marker='x', ms=6.5)
plt.axis((None,None,0,1))
plt.grid(b=True, which='major', color='k', linestyle='--')
plt.minorticks_on()
plt.grid(b=True, which='minor', color='grey', linestyle=':')
plt.show()
final_load_factor = average_optimal_curve.mean() / average_optimal_curve.max()
print("final_load_factor: ", final_load_factor)
final_energy_sum = average_optimal_curve.sum()
print("final_energy_sum: ", final_energy_sum)
```
|
github_jupyter
|
# Reading and writing fields
There are two main file formats to which a `discretisedfield.Field` object can be saved:
- [VTK](https://vtk.org/) for visualisation using e.g., [ParaView](https://www.paraview.org/) or [Mayavi](https://docs.enthought.com/mayavi/mayavi/)
- OOMMF [Vector Field File Format (OVF)](https://math.nist.gov/oommf/doc/userguide12a5/userguide/Vector_Field_File_Format_OV.html) for exchanging fields with micromagnetic simulators.
Let us say we have a nanosphere sample:
$$x^2 + y^2 + z^2 <= r^2$$
with $r=5\,\text{nm}$. The space is discretised into cells with dimensions $(0.5\,\text{nm}, 0.5\,\text{nm}, 0.5\,\text{nm})$. The value of the field at $(x, y, z)$ point is $(-cy, cx, cz)$, with $c=10^{9}$. The norm of the field inside the cylinder is $10^{6}$.
Let us first build that field.
```
import discretisedfield as df
r = 5e-9
cell = (0.5e-9, 0.5e-9, 0.5e-9)
mesh = df.Mesh(p1=(-r, -r, -r), p2=(r, r, r), cell=cell)
def norm_fun(pos):
x, y, z = pos
if x**2 + y**2 + z**2 <= r**2:
return 1e6
else:
return 0
def value_fun(pos):
x, y, z = pos
c = 1e9
return (-c*y, c*x, c*z)
field = df.Field(mesh, dim=3, value=value_fun, norm=norm_fun)
```
Let us have a quick view of the field we created
```
# NBVAL_IGNORE_OUTPUT
field.plane('z').k3d.vector(color_field=field.z)
```
## Writing the field to a file
The main method used for saving field in different files is `discretisedfield.Field.write()`. It takes `filename` as an argument, which is a string with one of the following extensions:
- `'.vtk'` for saving in the VTK format
- `'.ovf'`, `'.omf'`, `'.ohf'` for saving in the OVF format
Let us firstly save the field in the VTK file.
```
vtkfilename = 'my_vtk_file.vtk'
field.write(vtkfilename)
```
We can check if the file was saved in the current directory.
```
import os
os.path.isfile(f'./{vtkfilename}')
```
Now, we can delete the file:
```
os.remove(f'./{vtkfilename}')
```
Next, we can save the field in the OVF format and check whether it was created in the current directory.
```
omffilename = 'my_omf_file.omf'
field.write(omffilename)
os.path.isfile(f'./{omffilename}')
```
There are three different possible representations of an OVF file: one ASCII (`txt`) and two binary (`bin4` or `bin8`). ASCII `txt` representation is a default representation when `discretisedfield.Field.write()` is called. If any different representation is required, it can be passed via `representation` argument.
```
field.write(omffilename, representation='bin8')
os.path.isfile(f'./{omffilename}')
```
## Reading the OVF file
The method for reading OVF files is a class method `discretisedfield.Field.fromfile()`. By passing a `filename` argument, it reads the file and creates a `discretisedfield.Field` object. It is not required to pass the representation of the OVF file to the `discretisedfield.Field.fromfile()` method, because it can retrieve it from the content of the file.
```
read_field = df.Field.fromfile(omffilename)
```
Like previouly, we can quickly visualise the field
```
# NBVAL_IGNORE_OUTPUT
read_field.plane('z').k3d.vector(color_field=read_field.z)
```
Finally, we can delete the OVF file we created.
```
os.remove(f'./{omffilename}')
```
|
github_jupyter
|
<img align="center" style="max-width: 1000px" src="banner.png">
<img align="right" style="max-width: 200px; height: auto" src="hsg_logo.png">
## Lab 05 - "Convolutional Neural Networks (CNNs)" Assignments
GSERM'21 course "Deep Learning: Fundamentals and Applications", University of St. Gallen
In the last lab we learned how to enhance vanilla Artificial Neural Networks (ANNs) using `PyTorch` to classify even more complex images. Therefore, we used a special type of deep neural network referred to **Convolutional Neural Networks (CNNs)**. CNNs encompass the ability to take advantage of the hierarchical pattern in data and assemble more complex patterns using smaller and simpler patterns. In this lab, we aim to leverage that knowledge by applying it to a set of self-coding assignments. But before we do so let's start with another motivational video by NVIDIA:
```
from IPython.display import YouTubeVideo
# NVIDIA: "Official Intro | GTC 2020 | I AM AI"
YouTubeVideo('e2_hsjpTi4w', width=1000, height=500)
```
As always, pls. don't hesitate to ask all your questions either during the lab, post them in our CANVAS (StudyNet) forum (https://learning.unisg.ch), or send us an email (using the course email).
## 1. Assignment Objectives:
Similar today's lab session, after today's self-coding assignments you should be able to:
> 1. Understand the basic concepts, intuitions and major building blocks of **Convolutional Neural Networks (CNNs)**.
> 2. Know how to **implement and to train a CNN** to learn a model of tiny image data.
> 3. Understand how to apply such a learned model to **classify images** images based on their content into distinct categories.
> 4. Know how to **interpret and visualize** the model's classification results.
## 2. Setup of the Jupyter Notebook Environment
Similar to the previous labs, we need to import a couple of Python libraries that allow for data analysis and data visualization. We will mostly use the `PyTorch`, `Numpy`, `Sklearn`, `Matplotlib`, `Seaborn` and a few utility libraries throughout this lab:
```
# import standard python libraries
import os, urllib, io
from datetime import datetime
import numpy as np
```
Import Python machine / deep learning libraries:
```
# import the PyTorch deep learning library
import torch, torchvision
import torch.nn.functional as F
from torch import nn, optim
from torch.autograd import Variable
```
Import the sklearn classification metrics:
```
# import sklearn classification evaluation library
from sklearn import metrics
from sklearn.metrics import classification_report, confusion_matrix
```
Import Python plotting libraries:
```
# import matplotlib, seaborn, and PIL data visualization libary
import matplotlib.pyplot as plt
import seaborn as sns
from PIL import Image
```
Enable notebook matplotlib inline plotting:
```
%matplotlib inline
```
Import Google's GDrive connector and mount your GDrive directories:
```
# import the Google Colab GDrive connector
from google.colab import drive
# mount GDrive inside the Colab notebook
drive.mount('/content/drive')
```
Create a structure of Colab Notebook sub-directories inside of GDrive to store (1) the data as well as (2) the trained neural network models:
```
# create Colab Notebooks directory
notebook_directory = '/content/drive/MyDrive/Colab Notebooks'
if not os.path.exists(notebook_directory): os.makedirs(notebook_directory)
# create data sub-directory inside the Colab Notebooks directory
data_directory = '/content/drive/MyDrive/Colab Notebooks/data'
if not os.path.exists(data_directory): os.makedirs(data_directory)
# create models sub-directory inside the Colab Notebooks directory
models_directory = '/content/drive/MyDrive/Colab Notebooks/models'
if not os.path.exists(models_directory): os.makedirs(models_directory)
```
Set a random `seed` value to obtain reproducable results:
```
# init deterministic seed
seed_value = 1234
np.random.seed(seed_value) # set numpy seed
torch.manual_seed(seed_value) # set pytorch seed CPU
```
Google Colab provides the use of free GPUs for running notebooks. However, if you just execute this notebook as is, it will use your device's CPU. To run the lab on a GPU, got to `Runtime` > `Change runtime type` and set the Runtime type to `GPU` in the drop-down. Running this lab on a CPU is fine, but you will find that GPU computing is faster. *CUDA* indicates that the lab is being run on GPU.
Enable GPU computing by setting the `device` flag and init a `CUDA` seed:
```
# set cpu or gpu enabled device
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu').type
# init deterministic GPU seed
torch.cuda.manual_seed(seed_value)
# log type of device enabled
print('[LOG] notebook with {} computation enabled'.format(str(device)))
```
Let's determine if we have access to a GPU provided by e.g. Google's COLab environment:
```
!nvidia-smi
```
## 3. Convolutional Neural Networks (CNNs) Assignments
### 3.1 CIFAR-10 Dataset Download and Data Assessment
The **CIFAR-10 database** (**C**anadian **I**nstitute **F**or **A**dvanced **R**esearch) is a collection of images that are commonly used to train machine learning and computer vision algorithms. The database is widely used to conduct computer vision research using machine learning and deep learning methods:
<img align="center" style="max-width: 500px; height: 500px" src="cifar10.png">
(Source: https://www.kaggle.com/c/cifar-10)
Further details on the dataset can be obtained via: *Krizhevsky, A., 2009. "Learning Multiple Layers of Features from Tiny Images",
( https://www.cs.toronto.edu/~kriz/learning-features-2009-TR.pdf )."*
The CIFAR-10 database contains **60,000 color images** (50,000 training images and 10,000 validation images). The size of each image is 32 by 32 pixels. The collection of images encompasses 10 different classes that represent airplanes, cars, birds, cats, deer, dogs, frogs, horses, ships, and trucks. Let's define the distinct classs for further analytics:
```
cifar10_classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
```
Thereby the dataset contains 6,000 images for each of the ten classes. The CIFAR-10 is a straightforward dataset that can be used to teach a computer how to recognize objects in images.
Let's download, transform and inspect the training images of the dataset. Therefore, we first will define the directory we aim to store the training data:
```
train_path = data_directory + '/train_cifar10'
```
Now, let's download the training data accordingly:
```
# define pytorch transformation into tensor format
transf = torchvision.transforms.Compose([torchvision.transforms.ToTensor(), torchvision.transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
# download and transform training images
cifar10_train_data = torchvision.datasets.CIFAR10(root=train_path, train=True, transform=transf, download=True)
```
Verify the volume of training images downloaded:
```
# get the length of the training data
len(cifar10_train_data)
```
Let's now decide on where we want to store the evaluation data:
```
eval_path = data_directory + '/eval_cifar10'
```
And download the evaluation data accordingly:
```
# define pytorch transformation into tensor format
transf = torchvision.transforms.Compose([torchvision.transforms.ToTensor(), torchvision.transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
# download and transform validation images
cifar10_eval_data = torchvision.datasets.CIFAR10(root=eval_path, train=False, transform=transf, download=True)
```
Let's also verfify the volume of validation images downloaded:
```
# get the length of the training data
len(cifar10_eval_data)
```
### 3.2 Convolutional Neural Network (CNN) Model Training and Evaluation
<img align="center" style="max-width: 900px" src="classification.png">
We recommend you to try the following exercises as part of the self-coding session:
**Exercise 1: Train the neural network architecture of the lab with increased learning rate.**
> Increase the learning rate of the network training to a value of **0.1** (instead of currently 0.001) and re-run the network training for 10 training epochs. Load and evaluate the model exhibiting the lowest training loss. What kind of behavior in terms of loss convergence and prediction accuracy can be observed?
```
#### Step 1. define and init neural network architecture #############################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 2. define loss, training hyperparameters and dataloader ####################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 3. run model training ######################################################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 4. run model evaluation ####################################################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
```
**2. Evaluation of "shallow" vs. "deep" neural network architectures.**
> In addition to the architecture of the lab notebook, evaluate further (more **shallow** as well as more **deep**) neural network architectures by either **removing or adding convolutional layers** to the network. Train a model (using the architectures you selected) for at least **20 training epochs**. Analyze the prediction performance of the trained models in terms of training time and prediction accuracy.
```
#### Step 1. define and init neural network architecture #############################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 2. define loss, training hyperparameters and dataloader ####################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 3. run model training ######################################################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
#### Step 4. run model evaluation ####################################################################################
# ***************************************************
# INSERT YOUR SOLUTION/CODE HERE
# ***************************************************
```
|
github_jupyter
|
# Colab FAQ
For some basic overview and features offered in Colab notebooks, check out: [Overview of Colaboratory Features](https://colab.research.google.com/notebooks/basic_features_overview.ipynb)
You need to use the colab GPU for this assignmentby selecting:
> **Runtime** → **Change runtime type** → **Hardware Accelerator: GPU**
# Setup PyTorch
All files are stored at /content/csc421/a4/ folder
```
######################################################################
# Setup python environment and change the current working directory
######################################################################
!pip install torch torchvision
!pip install imageio
!pip install matplotlib
%mkdir -p /content/csc413/a4/
%cd /content/csc413/a4
```
# Helper code
## Utility functions
```
import os
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch.nn import Parameter
import torch.nn.functional as F
import torch.optim as optim
from torch.autograd import Variable
from torch.utils.data import DataLoader
from torchvision import datasets
from torchvision import transforms
from six.moves.urllib.request import urlretrieve
import tarfile
import imageio
from urllib.error import URLError
from urllib.error import HTTPError
def get_file(fname,
origin,
untar=False,
extract=False,
archive_format='auto',
cache_dir='data'):
datadir = os.path.join(cache_dir)
if not os.path.exists(datadir):
os.makedirs(datadir)
if untar:
untar_fpath = os.path.join(datadir, fname)
fpath = untar_fpath + '.tar.gz'
else:
fpath = os.path.join(datadir, fname)
print(fpath)
if not os.path.exists(fpath):
print('Downloading data from', origin)
error_msg = 'URL fetch failure on {}: {} -- {}'
try:
try:
urlretrieve(origin, fpath)
except URLError as e:
raise Exception(error_msg.format(origin, e.errno, e.reason))
except HTTPError as e:
raise Exception(error_msg.format(origin, e.code, e.msg))
except (Exception, KeyboardInterrupt) as e:
if os.path.exists(fpath):
os.remove(fpath)
raise
if untar:
if not os.path.exists(untar_fpath):
print('Extracting file.')
with tarfile.open(fpath) as archive:
archive.extractall(datadir)
return untar_fpath
return fpath
class AttrDict(dict):
def __init__(self, *args, **kwargs):
super(AttrDict, self).__init__(*args, **kwargs)
self.__dict__ = self
def to_var(tensor, cuda=True):
"""Wraps a Tensor in a Variable, optionally placing it on the GPU.
Arguments:
tensor: A Tensor object.
cuda: A boolean flag indicating whether to use the GPU.
Returns:
A Variable object, on the GPU if cuda==True.
"""
if cuda:
return Variable(tensor.cuda())
else:
return Variable(tensor)
def to_data(x):
"""Converts variable to numpy."""
if torch.cuda.is_available():
x = x.cpu()
return x.data.numpy()
def create_dir(directory):
"""Creates a directory if it doesn't already exist.
"""
if not os.path.exists(directory):
os.makedirs(directory)
def gan_checkpoint(iteration, G, D, opts):
"""Saves the parameters of the generator G and discriminator D.
"""
G_path = os.path.join(opts.checkpoint_dir, 'G.pkl')
D_path = os.path.join(opts.checkpoint_dir, 'D.pkl')
torch.save(G.state_dict(), G_path)
torch.save(D.state_dict(), D_path)
def load_checkpoint(opts):
"""Loads the generator and discriminator models from checkpoints.
"""
G_path = os.path.join(opts.load, 'G.pkl')
D_path = os.path.join(opts.load, 'D_.pkl')
G = DCGenerator(noise_size=opts.noise_size, conv_dim=opts.g_conv_dim, spectral_norm=opts.spectral_norm)
D = DCDiscriminator(conv_dim=opts.d_conv_dim)
G.load_state_dict(torch.load(G_path, map_location=lambda storage, loc: storage))
D.load_state_dict(torch.load(D_path, map_location=lambda storage, loc: storage))
if torch.cuda.is_available():
G.cuda()
D.cuda()
print('Models moved to GPU.')
return G, D
def merge_images(sources, targets, opts):
"""Creates a grid consisting of pairs of columns, where the first column in
each pair contains images source images and the second column in each pair
contains images generated by the CycleGAN from the corresponding images in
the first column.
"""
_, _, h, w = sources.shape
row = int(np.sqrt(opts.batch_size))
merged = np.zeros([3, row * h, row * w * 2])
for (idx, s, t) in (zip(range(row ** 2), sources, targets, )):
i = idx // row
j = idx % row
merged[:, i * h:(i + 1) * h, (j * 2) * h:(j * 2 + 1) * h] = s
merged[:, i * h:(i + 1) * h, (j * 2 + 1) * h:(j * 2 + 2) * h] = t
return merged.transpose(1, 2, 0)
def generate_gif(directory_path, keyword=None):
images = []
for filename in sorted(os.listdir(directory_path)):
if filename.endswith(".png") and (keyword is None or keyword in filename):
img_path = os.path.join(directory_path, filename)
print("adding image {}".format(img_path))
images.append(imageio.imread(img_path))
if keyword:
imageio.mimsave(
os.path.join(directory_path, 'anim_{}.gif'.format(keyword)), images)
else:
imageio.mimsave(os.path.join(directory_path, 'anim.gif'), images)
def create_image_grid(array, ncols=None):
"""
"""
num_images, channels, cell_h, cell_w = array.shape
if not ncols:
ncols = int(np.sqrt(num_images))
nrows = int(np.math.floor(num_images / float(ncols)))
result = np.zeros((cell_h * nrows, cell_w * ncols, channels), dtype=array.dtype)
for i in range(0, nrows):
for j in range(0, ncols):
result[i * cell_h:(i + 1) * cell_h, j * cell_w:(j + 1) * cell_w, :] = array[i * ncols + j].transpose(1, 2,
0)
if channels == 1:
result = result.squeeze()
return result
def gan_save_samples(G, fixed_noise, iteration, opts):
generated_images = G(fixed_noise)
generated_images = to_data(generated_images)
grid = create_image_grid(generated_images)
# merged = merge_images(X, fake_Y, opts)
path = os.path.join(opts.sample_dir, 'sample-{:06d}.png'.format(iteration))
imageio.imwrite(path, grid)
print('Saved {}'.format(path))
```
## Data loader
```
def get_emoji_loader(emoji_type, opts):
"""Creates training and test data loaders.
"""
transform = transforms.Compose([
transforms.Scale(opts.image_size),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
])
train_path = os.path.join('data/emojis', emoji_type)
test_path = os.path.join('data/emojis', 'Test_{}'.format(emoji_type))
train_dataset = datasets.ImageFolder(train_path, transform)
test_dataset = datasets.ImageFolder(test_path, transform)
train_dloader = DataLoader(dataset=train_dataset, batch_size=opts.batch_size, shuffle=True, num_workers=opts.num_workers)
test_dloader = DataLoader(dataset=test_dataset, batch_size=opts.batch_size, shuffle=False, num_workers=opts.num_workers)
return train_dloader, test_dloader
```
## Training and evaluation code
```
def print_models(G_XtoY, G_YtoX, D_X, D_Y):
"""Prints model information for the generators and discriminators.
"""
print(" G ")
print("---------------------------------------")
print(G_XtoY)
print("---------------------------------------")
print(" D ")
print("---------------------------------------")
print(D_X)
print("---------------------------------------")
def create_model(opts):
"""Builds the generators and discriminators.
"""
### GAN
G = DCGenerator(noise_size=opts.noise_size, conv_dim=opts.g_conv_dim, spectral_norm=opts.spectral_norm)
D = DCDiscriminator(conv_dim=opts.d_conv_dim, spectral_norm=opts.spectral_norm)
print_models(G, None, D, None)
if torch.cuda.is_available():
G.cuda()
D.cuda()
print('Models moved to GPU.')
return G, D
def train(opts):
"""Loads the data, creates checkpoint and sample directories, and starts the training loop.
"""
# Create train and test dataloaders for images from the two domains X and Y
dataloader_X, test_dataloader_X = get_emoji_loader(emoji_type=opts.X, opts=opts)
# Create checkpoint and sample directories
create_dir(opts.checkpoint_dir)
create_dir(opts.sample_dir)
# Start training
if opts.least_squares_gan:
G, D = gan_training_loop_leastsquares(dataloader_X, test_dataloader_X, opts)
else:
G, D = gan_training_loop_regular(dataloader_X, test_dataloader_X, opts)
return G, D
def print_opts(opts):
"""Prints the values of all command-line arguments.
"""
print('=' * 80)
print('Opts'.center(80))
print('-' * 80)
for key in opts.__dict__:
if opts.__dict__[key]:
print('{:>30}: {:<30}'.format(key, opts.__dict__[key]).center(80))
print('=' * 80)
```
# Your code for generators and discriminators
## Helper modules
```
def sample_noise(batch_size, dim):
"""
Generate a PyTorch Tensor of uniform random noise.
Input:
- batch_size: Integer giving the batch size of noise to generate.
- dim: Integer giving the dimension of noise to generate.
Output:
- A PyTorch Tensor of shape (batch_size, dim, 1, 1) containing uniform
random noise in the range (-1, 1).
"""
return to_var(torch.rand(batch_size, dim) * 2 - 1).unsqueeze(2).unsqueeze(3)
def upconv(in_channels, out_channels, kernel_size, stride=2, padding=2, batch_norm=True, spectral_norm=False):
"""Creates a upsample-and-convolution layer, with optional batch normalization.
"""
layers = []
if stride>1:
layers.append(nn.Upsample(scale_factor=stride))
conv_layer = nn.Conv2d(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size, stride=1, padding=padding, bias=False)
if spectral_norm:
layers.append(SpectralNorm(conv_layer))
else:
layers.append(conv_layer)
if batch_norm:
layers.append(nn.BatchNorm2d(out_channels))
return nn.Sequential(*layers)
def conv(in_channels, out_channels, kernel_size, stride=2, padding=2, batch_norm=True, init_zero_weights=False, spectral_norm=False):
"""Creates a convolutional layer, with optional batch normalization.
"""
layers = []
conv_layer = nn.Conv2d(in_channels=in_channels, out_channels=out_channels, kernel_size=kernel_size, stride=stride, padding=padding, bias=False)
if init_zero_weights:
conv_layer.weight.data = torch.randn(out_channels, in_channels, kernel_size, kernel_size) * 0.001
if spectral_norm:
layers.append(SpectralNorm(conv_layer))
else:
layers.append(conv_layer)
if batch_norm:
layers.append(nn.BatchNorm2d(out_channels))
return nn.Sequential(*layers)
class ResnetBlock(nn.Module):
def __init__(self, conv_dim):
super(ResnetBlock, self).__init__()
self.conv_layer = conv(in_channels=conv_dim, out_channels=conv_dim, kernel_size=3, stride=1, padding=1)
def forward(self, x):
out = x + self.conv_layer(x)
return out
```
## DCGAN
### Spectral Norm class
```
def l2normalize(v, eps=1e-12):
return v / (v.norm() + eps)
class SpectralNorm(nn.Module):
def __init__(self, module, name='weight', power_iterations=1):
super(SpectralNorm, self).__init__()
self.module = module
self.name = name
self.power_iterations = power_iterations
if not self._made_params():
self._make_params()
def _update_u_v(self):
u = getattr(self.module, self.name + "_u")
v = getattr(self.module, self.name + "_v")
w = getattr(self.module, self.name + "_bar")
height = w.data.shape[0]
for _ in range(self.power_iterations):
v.data = l2normalize(torch.mv(torch.t(w.view(height,-1).data), u.data))
u.data = l2normalize(torch.mv(w.view(height,-1).data, v.data))
# sigma = torch.dot(u.data, torch.mv(w.view(height,-1).data, v.data))
sigma = u.dot(w.view(height, -1).mv(v))
setattr(self.module, self.name, w / sigma.expand_as(w))
def _made_params(self):
try:
u = getattr(self.module, self.name + "_u")
v = getattr(self.module, self.name + "_v")
w = getattr(self.module, self.name + "_bar")
return True
except AttributeError:
return False
def _make_params(self):
w = getattr(self.module, self.name)
height = w.data.shape[0]
width = w.view(height, -1).data.shape[1]
u = Parameter(w.data.new(height).normal_(0, 1), requires_grad=False)
v = Parameter(w.data.new(width).normal_(0, 1), requires_grad=False)
u.data = l2normalize(u.data)
v.data = l2normalize(v.data)
w_bar = Parameter(w.data)
del self.module._parameters[self.name]
self.module.register_parameter(self.name + "_u", u)
self.module.register_parameter(self.name + "_v", v)
self.module.register_parameter(self.name + "_bar", w_bar)
def forward(self, *args):
self._update_u_v()
return self.module.forward(*args)
```
### **[Your Task]** GAN generator
```
class DCGenerator(nn.Module):
def __init__(self, noise_size, conv_dim, spectral_norm=False):
super(DCGenerator, self).__init__()
self.conv_dim = conv_dim
###########################################
## FILL THIS IN: CREATE ARCHITECTURE ##
###########################################
# self.linear_bn = ...
# self.upconv1 = ...
# self.upconv2 = ...
# self.upconv3 = ...
def forward(self, z):
"""Generates an image given a sample of random noise.
Input
-----
z: BS x noise_size x 1 x 1 --> BSx100x1x1 (during training)
Output
------
out: BS x channels x image_width x image_height --> BSx3x32x32 (during training)
"""
batch_size = z.size(0)
out = F.relu(self.linear_bn(z)).view(-1, self.conv_dim*4, 4, 4) # BS x 128 x 4 x 4
out = F.relu(self.upconv1(out)) # BS x 64 x 8 x 8
out = F.relu(self.upconv2(out)) # BS x 32 x 16 x 16
out = F.tanh(self.upconv3(out)) # BS x 3 x 32 x 32
out_size = out.size()
if out_size != torch.Size([batch_size, 3, 32, 32]):
raise ValueError("expect {} x 3 x 32 x 32, but get {}".format(batch_size, out_size))
return out
```
### GAN discriminator
```
class DCDiscriminator(nn.Module):
"""Defines the architecture of the discriminator network.
Note: Both discriminators D_X and D_Y have the same architecture in this assignment.
"""
def __init__(self, conv_dim=64, spectral_norm=False):
super(DCDiscriminator, self).__init__()
self.conv1 = conv(in_channels=3, out_channels=conv_dim, kernel_size=5, stride=2, spectral_norm=spectral_norm)
self.conv2 = conv(in_channels=conv_dim, out_channels=conv_dim*2, kernel_size=5, stride=2, spectral_norm=spectral_norm)
self.conv3 = conv(in_channels=conv_dim*2, out_channels=conv_dim*4, kernel_size=5, stride=2, spectral_norm=spectral_norm)
self.conv4 = conv(in_channels=conv_dim*4, out_channels=1, kernel_size=5, stride=2, padding=1, batch_norm=False, spectral_norm=spectral_norm)
def forward(self, x):
batch_size = x.size(0)
out = F.relu(self.conv1(x)) # BS x 64 x 16 x 16
out = F.relu(self.conv2(out)) # BS x 64 x 8 x 8
out = F.relu(self.conv3(out)) # BS x 64 x 4 x 4
out = self.conv4(out).squeeze()
out_size = out.size()
if out_size != torch.Size([batch_size,]):
raise ValueError("expect {} x 1, but get {}".format(batch_size, out_size))
return out
```
### **[Your Task]** GAN training loop
* Regular GAN
* Least Squares GAN
```
def gan_training_loop_regular(dataloader, test_dataloader, opts):
"""Runs the training loop.
* Saves checkpoint every opts.checkpoint_every iterations
* Saves generated samples every opts.sample_every iterations
"""
# Create generators and discriminators
G, D = create_model(opts)
g_params = G.parameters() # Get generator parameters
d_params = D.parameters() # Get discriminator parameters
# Create optimizers for the generators and discriminators
g_optimizer = optim.Adam(g_params, opts.lr, [opts.beta1, opts.beta2])
d_optimizer = optim.Adam(d_params, opts.lr * 2., [opts.beta1, opts.beta2])
train_iter = iter(dataloader)
test_iter = iter(test_dataloader)
# Get some fixed data from domains X and Y for sampling. These are images that are held
# constant throughout training, that allow us to inspect the model's performance.
fixed_noise = sample_noise(100, opts.noise_size) # # 100 x noise_size x 1 x 1
iter_per_epoch = len(train_iter)
total_train_iters = opts.train_iters
losses = {"iteration": [], "D_fake_loss": [], "D_real_loss": [], "G_loss": []}
gp_weight = 1
adversarial_loss = torch.nn.BCEWithLogitsLoss() # Use this loss
# [Hint: you may find the folowing code helpful]
# ones = Variable(torch.Tensor(real_images.shape[0]).float().cuda().fill_(1.0), requires_grad=False)
try:
for iteration in range(1, opts.train_iters + 1):
# Reset data_iter for each epoch
if iteration % iter_per_epoch == 0:
train_iter = iter(dataloader)
real_images, real_labels = train_iter.next()
real_images, real_labels = to_var(real_images), to_var(real_labels).long().squeeze()
for d_i in range(opts.d_train_iters):
d_optimizer.zero_grad()
# FILL THIS IN
# 1. Compute the discriminator loss on real images
# D_real_loss = ...
# 2. Sample noise
# noise = ...
# 3. Generate fake images from the noise
# fake_images = ...
# 4. Compute the discriminator loss on the fake images
# D_fake_loss = ...
# ---- Gradient Penalty ----
if opts.gradient_penalty:
alpha = torch.rand(real_images.shape[0], 1, 1, 1)
alpha = alpha.expand_as(real_images).cuda()
interp_images = Variable(alpha * real_images.data + (1 - alpha) * fake_images.data, requires_grad=True).cuda()
D_interp_output = D(interp_images)
gradients = torch.autograd.grad(outputs=D_interp_output, inputs=interp_images,
grad_outputs=torch.ones(D_interp_output.size()).cuda(),
create_graph=True, retain_graph=True)[0]
gradients = gradients.view(real_images.shape[0], -1)
gradients_norm = torch.sqrt(torch.sum(gradients ** 2, dim=1) + 1e-12)
gp = gp_weight * gradients_norm.mean()
else:
gp = 0.0
# --------------------------
# 5. Compute the total discriminator loss
# D_total_loss = ...
D_total_loss.backward()
d_optimizer.step()
###########################################
### TRAIN THE GENERATOR ###
###########################################
g_optimizer.zero_grad()
# FILL THIS IN
# 1. Sample noise
# noise = ...
# 2. Generate fake images from the noise
# fake_images = ...
# 3. Compute the generator loss
# G_loss = ...
G_loss.backward()
g_optimizer.step()
# Print the log info
if iteration % opts.log_step == 0:
losses['iteration'].append(iteration)
losses['D_real_loss'].append(D_real_loss.item())
losses['D_fake_loss'].append(D_fake_loss.item())
losses['G_loss'].append(G_loss.item())
print('Iteration [{:4d}/{:4d}] | D_real_loss: {:6.4f} | D_fake_loss: {:6.4f} | G_loss: {:6.4f}'.format(
iteration, total_train_iters, D_real_loss.item(), D_fake_loss.item(), G_loss.item()))
# Save the generated samples
if iteration % opts.sample_every == 0:
gan_save_samples(G, fixed_noise, iteration, opts)
# Save the model parameters
if iteration % opts.checkpoint_every == 0:
gan_checkpoint(iteration, G, D, opts)
except KeyboardInterrupt:
print('Exiting early from training.')
return G, D
plt.figure()
plt.plot(losses['iteration'], losses['D_real_loss'], label='D_real')
plt.plot(losses['iteration'], losses['D_fake_loss'], label='D_fake')
plt.plot(losses['iteration'], losses['G_loss'], label='G')
plt.legend()
plt.savefig(os.path.join(opts.sample_dir, 'losses.png'))
plt.close()
return G, D
def gan_training_loop_leastsquares(dataloader, test_dataloader, opts):
"""Runs the training loop.
* Saves checkpoint every opts.checkpoint_every iterations
* Saves generated samples every opts.sample_every iterations
"""
# Create generators and discriminators
G, D = create_model(opts)
g_params = G.parameters() # Get generator parameters
d_params = D.parameters() # Get discriminator parameters
# Create optimizers for the generators and discriminators
g_optimizer = optim.Adam(g_params, opts.lr, [opts.beta1, opts.beta2])
d_optimizer = optim.Adam(d_params, opts.lr * 2., [opts.beta1, opts.beta2])
train_iter = iter(dataloader)
test_iter = iter(test_dataloader)
# Get some fixed data from domains X and Y for sampling. These are images that are held
# constant throughout training, that allow us to inspect the model's performance.
fixed_noise = sample_noise(100, opts.noise_size) # # 100 x noise_size x 1 x 1
iter_per_epoch = len(train_iter)
total_train_iters = opts.train_iters
losses = {"iteration": [], "D_fake_loss": [], "D_real_loss": [], "G_loss": []}
#adversarial_loss = torch.nn.BCEWithLogitsLoss()
gp_weight = 1
try:
for iteration in range(1, opts.train_iters + 1):
# Reset data_iter for each epoch
if iteration % iter_per_epoch == 0:
train_iter = iter(dataloader)
real_images, real_labels = train_iter.next()
real_images, real_labels = to_var(real_images), to_var(real_labels).long().squeeze()
for d_i in range(opts.d_train_iters):
d_optimizer.zero_grad()
# FILL THIS IN
# 1. Compute the discriminator loss on real images
# D_real_loss = ...
# 2. Sample noise
# noise = ...
# 3. Generate fake images from the noise
# fake_images = ...
# 4. Compute the discriminator loss on the fake images
# D_fake_loss = ...
# ---- Gradient Penalty ----
if opts.gradient_penalty:
alpha = torch.rand(real_images.shape[0], 1, 1, 1)
alpha = alpha.expand_as(real_images).cuda()
interp_images = Variable(alpha * real_images.data + (1 - alpha) * fake_images.data, requires_grad=True).cuda()
D_interp_output = D(interp_images)
gradients = torch.autograd.grad(outputs=D_interp_output, inputs=interp_images,
grad_outputs=torch.ones(D_interp_output.size()).cuda(),
create_graph=True, retain_graph=True)[0]
gradients = gradients.view(real_images.shape[0], -1)
gradients_norm = torch.sqrt(torch.sum(gradients ** 2, dim=1) + 1e-12)
gp = gp_weight * gradients_norm.mean()
else:
gp = 0.0
# --------------------------
# 5. Compute the total discriminator loss
# D_total_loss = ...
D_total_loss.backward()
d_optimizer.step()
###########################################
### TRAIN THE GENERATOR ###
###########################################
g_optimizer.zero_grad()
# FILL THIS IN
# 1. Sample noise
# noise = ...
# 2. Generate fake images from the noise
# fake_images = ...
# 3. Compute the generator loss
# G_loss = ...
G_loss.backward()
g_optimizer.step()
# Print the log info
if iteration % opts.log_step == 0:
losses['iteration'].append(iteration)
losses['D_real_loss'].append(D_real_loss.item())
losses['D_fake_loss'].append(D_fake_loss.item())
losses['G_loss'].append(G_loss.item())
print('Iteration [{:4d}/{:4d}] | D_real_loss: {:6.4f} | D_fake_loss: {:6.4f} | G_loss: {:6.4f}'.format(
iteration, total_train_iters, D_real_loss.item(), D_fake_loss.item(), G_loss.item()))
# Save the generated samples
if iteration % opts.sample_every == 0:
gan_save_samples(G, fixed_noise, iteration, opts)
# Save the model parameters
if iteration % opts.checkpoint_every == 0:
gan_checkpoint(iteration, G, D, opts)
except KeyboardInterrupt:
print('Exiting early from training.')
return G, D
plt.figure()
plt.plot(losses['iteration'], losses['D_real_loss'], label='D_real')
plt.plot(losses['iteration'], losses['D_fake_loss'], label='D_fake')
plt.plot(losses['iteration'], losses['G_loss'], label='G')
plt.legend()
plt.savefig(os.path.join(opts.sample_dir, 'losses.png'))
plt.close()
return G, D
```
# **[Your Task]** Training
## Download dataset
```
######################################################################
# Download Translation datasets
######################################################################
data_fpath = get_file(fname='emojis',
origin='http://www.cs.toronto.edu/~jba/emojis.tar.gz',
untar=True)
```
## Train DCGAN
```
SEED = 11
# Set the random seed manually for reproducibility.
np.random.seed(SEED)
torch.manual_seed(SEED)
if torch.cuda.is_available():
torch.cuda.manual_seed(SEED)
args = AttrDict()
args_dict = {
'image_size':32,
'g_conv_dim':32,
'd_conv_dim':64,
'noise_size':100,
'num_workers': 0,
'train_iters':20000,
'X':'Apple', # options: 'Windows' / 'Apple'
'Y': None,
'lr':0.00003,
'beta1':0.5,
'beta2':0.999,
'batch_size':32,
'checkpoint_dir': 'results/checkpoints_gan_gp1_lr3e-5',
'sample_dir': 'results/samples_gan_gp1_lr3e-5',
'load': None,
'log_step':200,
'sample_every':200,
'checkpoint_every':1000,
'spectral_norm': False,
'gradient_penalty': True,
'least_squares_gan': False,
'd_train_iters': 1
}
args.update(args_dict)
print_opts(args)
G, D = train(args)
generate_gif("results/samples_gan_gp1_lr3e-5")
```
## Download your output
```
!zip -r /content/csc413/a4/results/samples.zip /content/csc413/a4/results/samples_gan_gp1_lr3e-5
from google.colab import files
files.download("/content/csc413/a4/results/samples.zip")
```
|
github_jupyter
|
```
import numpy as np
%%html
<style>
.pquote {
text-align: left;
margin: 40px 0 40px auto;
width: 70%;
font-size: 1.5em;
font-style: italic;
display: block;
line-height: 1.3em;
color: #5a75a7;
font-weight: 600;
border-left: 5px solid rgba(90, 117, 167, .1);
padding-left: 6px;
}
.notes {
font-style: italic;
display: block;
margin: 40px 10%;
}
</style>
```
$$
\newcommand\bs[1]{\boldsymbol{#1}}
$$
<span class='notes'>
This content is part of a series following the chapter 2 on linear algebra from the [Deep Learning Book](http://www.deeplearningbook.org/) by Goodfellow, I., Bengio, Y., and Courville, A. (2016). It aims to provide intuitions/drawings/python code on mathematical theories and is constructed as my understanding of these concepts. You can check the syllabus in the [introduction post](https://hadrienj.github.io/posts/Deep-Learning-Book-Series-Introduction/).
</span>
# Introduction
This is the first post/notebook of a series following the syllabus of the [linear algebra chapter from the Deep Learning Book](http://www.deeplearningbook.org/contents/linear_algebra.html) by Goodfellow et al.. This work is a collection of thoughts/details/developements/examples I made while reading this chapter. It is designed to help you go through their introduction to linear algebra. For more details about this series and the syllabus, please see the [introduction post](https://hadrienj.github.io/posts/Deep-Learning-Book-Series-Introduction/).
This first chapter is quite light and concerns the basic elements used in linear algebra and their definitions. It also introduces important functions in Python/Numpy that we will use all along this series. It will explain how to create and use vectors and matrices through examples.
# 2.1 Scalars, Vectors, Matrices and Tensors
Let's start with some basic definitions:
<img src="images/scalar-tensor.png" width="400" alt="scalar-tensor">
- A scalar is a single number
- A vector is an array of numbers.
$$
\bs{x} =\begin{bmatrix}
x_1 \\\\
x_2 \\\\
\cdots \\\\
x_n
\end{bmatrix}
$$
- A matrix is a 2-D array
$$
\bs{A}=
\begin{bmatrix}
A_{1,1} & A_{1,2} & \cdots & A_{1,n} \\\\
A_{2,1} & A_{2,2} & \cdots & A_{2,n} \\\\
\cdots & \cdots & \cdots & \cdots \\\\
A_{m,1} & A_{m,2} & \cdots & A_{m,n}
\end{bmatrix}
$$
- A tensor is a $n$-dimensional array with $n>2$
We will follow the conventions used in the [Deep Learning Book](http://www.deeplearningbook.org/):
- scalars are written in lowercase and italics. For instance: $n$
- vectors are written in lowercase, italics and bold type. For instance: $\bs{x}$
- matrices are written in uppercase, italics and bold. For instance: $\bs{X}$
### Example 1.
#### Create a vector with Python and Numpy
*Coding tip*: Unlike the `matrix()` function which necessarily creates $2$-dimensional matrices, you can create $n$-dimensionnal arrays with the `array()` function. The main advantage to use `matrix()` is the useful methods (conjugate transpose, inverse, matrix operations...). We will use the `array()` function in this series.
We will start by creating a vector. This is just a $1$-dimensional array:
```
x = np.array([1, 2, 3, 4])
x
```
### Example 2.
#### Create a (3x2) matrix with nested brackets
The `array()` function can also create $2$-dimensional arrays with nested brackets:
```
A = np.array([[1, 2], [3, 4], [5, 6]])
A
```
### Shape
The shape of an array (that is to say its dimensions) tells you the number of values for each dimension. For a $2$-dimensional array it will give you the number of rows and the number of columns. Let's find the shape of our preceding $2$-dimensional array `A`. Since `A` is a Numpy array (it was created with the `array()` function) you can access its shape with:
```
A.shape
```
We can see that $\bs{A}$ has 3 rows and 2 columns.
Let's check the shape of our first vector:
```
x.shape
```
As expected, you can see that $\bs{x}$ has only one dimension. The number corresponds to the length of the array:
```
len(x)
```
# Transposition
With transposition you can convert a row vector to a column vector and vice versa:
<img src="images/transposeVector.png" alt="transposeVector" width="200">
The transpose $\bs{A}^{\text{T}}$ of the matrix $\bs{A}$ corresponds to the mirrored axes. If the matrix is a square matrix (same number of columns and rows):
<img src="images/transposeMatrixSquare.png" alt="transposeMatrixSquare" width="300">
If the matrix is not square the idea is the same:
<img src="images/transposeMatrix.png" alt="transposeMatrix" width="300">
The superscript $^\text{T}$ is used for transposed matrices.
$$
\bs{A}=
\begin{bmatrix}
A_{1,1} & A_{1,2} \\\\
A_{2,1} & A_{2,2} \\\\
A_{3,1} & A_{3,2}
\end{bmatrix}
$$
$$
\bs{A}^{\text{T}}=
\begin{bmatrix}
A_{1,1} & A_{2,1} & A_{3,1} \\\\
A_{1,2} & A_{2,2} & A_{3,2}
\end{bmatrix}
$$
The shape ($m \times n$) is inverted and becomes ($n \times m$).
<img src="images/transposeMatrixDim.png" alt="transposeMatrixDim" width="300">
### Example 3.
#### Create a matrix A and transpose it
```
A = np.array([[1, 2], [3, 4], [5, 6]])
A
A_t = A.T
A_t
```
We can check the dimensions of the matrices:
```
A.shape
A_t.shape
```
We can see that the number of columns becomes the number of rows with transposition and vice versa.
# Addition
<img src="images/additionMatrix.png" alt="additionMatrix" width="300">
Matrices can be added if they have the same shape:
$$\bs{A} + \bs{B} = \bs{C}$$
Each cell of $\bs{A}$ is added to the corresponding cell of $\bs{B}$:
$$\bs{A}_{i,j} + \bs{B}_{i,j} = \bs{C}_{i,j}$$
$i$ is the row index and $j$ the column index.
$$
\begin{bmatrix}
A_{1,1} & A_{1,2} \\\\
A_{2,1} & A_{2,2} \\\\
A_{3,1} & A_{3,2}
\end{bmatrix}+
\begin{bmatrix}
B_{1,1} & B_{1,2} \\\\
B_{2,1} & B_{2,2} \\\\
B_{3,1} & B_{3,2}
\end{bmatrix}=
\begin{bmatrix}
A_{1,1} + B_{1,1} & A_{1,2} + B_{1,2} \\\\
A_{2,1} + B_{2,1} & A_{2,2} + B_{2,2} \\\\
A_{3,1} + B_{3,1} & A_{3,2} + B_{3,2}
\end{bmatrix}
$$
The shape of $\bs{A}$, $\bs{B}$ and $\bs{C}$ are identical. Let's check that in an example:
### Example 4.
#### Create two matrices A and B and add them
With Numpy you can add matrices just as you would add vectors or scalars.
```
A = np.array([[1, 2], [3, 4], [5, 6]])
A
B = np.array([[2, 5], [7, 4], [4, 3]])
B
# Add matrices A and B
C = A + B
C
```
It is also possible to add a scalar to a matrix. This means adding this scalar to each cell of the matrix.
$$
\alpha+ \begin{bmatrix}
A_{1,1} & A_{1,2} \\\\
A_{2,1} & A_{2,2} \\\\
A_{3,1} & A_{3,2}
\end{bmatrix}=
\begin{bmatrix}
\alpha + A_{1,1} & \alpha + A_{1,2} \\\\
\alpha + A_{2,1} & \alpha + A_{2,2} \\\\
\alpha + A_{3,1} & \alpha + A_{3,2}
\end{bmatrix}
$$
### Example 5.
#### Add a scalar to a matrix
```
A
# Exemple: Add 4 to the matrix A
C = A+4
C
```
# Broadcasting
Numpy can handle operations on arrays of different shapes. The smaller array will be extended to match the shape of the bigger one. The advantage is that this is done in `C` under the hood (like any vectorized operations in Numpy). Actually, we used broadcasting in the example 5. The scalar was converted in an array of same shape as $\bs{A}$.
Here is another generic example:
$$
\begin{bmatrix}
A_{1,1} & A_{1,2} \\\\
A_{2,1} & A_{2,2} \\\\
A_{3,1} & A_{3,2}
\end{bmatrix}+
\begin{bmatrix}
B_{1,1} \\\\
B_{2,1} \\\\
B_{3,1}
\end{bmatrix}
$$
is equivalent to
$$
\begin{bmatrix}
A_{1,1} & A_{1,2} \\\\
A_{2,1} & A_{2,2} \\\\
A_{3,1} & A_{3,2}
\end{bmatrix}+
\begin{bmatrix}
B_{1,1} & B_{1,1} \\\\
B_{2,1} & B_{2,1} \\\\
B_{3,1} & B_{3,1}
\end{bmatrix}=
\begin{bmatrix}
A_{1,1} + B_{1,1} & A_{1,2} + B_{1,1} \\\\
A_{2,1} + B_{2,1} & A_{2,2} + B_{2,1} \\\\
A_{3,1} + B_{3,1} & A_{3,2} + B_{3,1}
\end{bmatrix}
$$
where the ($3 \times 1$) matrix is converted to the right shape ($3 \times 2$) by copying the first column. Numpy will do that automatically if the shapes can match.
### Example 6.
#### Add two matrices of different shapes
```
A = np.array([[1, 2], [3, 4], [5, 6]])
A
B = np.array([[2], [4], [6]])
B
# Broadcasting
C=A+B
C
```
You can find basics operations on matrices simply explained [here](https://www.mathsisfun.com/algebra/matrix-introduction.html).
<span class='notes'>
Feel free to drop me an email or a comment. The syllabus of this series can be found [in the introduction post](https://hadrienj.github.io/posts/Deep-Learning-Book-Series-Introduction/). All the notebooks can be found on [Github](https://github.com/hadrienj/deepLearningBook-Notes).
</span>
# References
- [Broadcasting in Numpy](https://docs.scipy.org/doc/numpy-1.13.0/user/basics.broadcasting.html)
- [Discussion on Arrays and matrices](https://stackoverflow.com/questions/4151128/what-are-the-differences-between-numpy-arrays-and-matrices-which-one-should-i-u)
- [Math is fun - Matrix introduction](https://www.mathsisfun.com/algebra/matrix-introduction.html)
|
github_jupyter
|
```
# Copyright 2021 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
```
<img src="https://upload.wikimedia.org/wikipedia/en/6/6d/Nvidia_image_logo.svg" style="width: 90px; float: right;">
# QA Inference on BERT using TensorRT
## 1. Overview
Bidirectional Embedding Representations from Transformers (BERT), is a method of pre-training language representations which obtains state-of-the-art results on a wide array of Natural Language Processing (NLP) tasks.
The original paper can be found here: https://arxiv.org/abs/1810.04805.
### 1.a Learning objectives
This notebook demonstrates:
- Inference on Question Answering (QA) task with BERT Base/Large model
- The use fine-tuned NVIDIA BERT models
- Use of BERT model with TRT
## 2. Requirements
Please refer to the ReadMe file
## 3. BERT Inference: Question Answering
We can run inference on a fine-tuned BERT model for tasks like Question Answering.
Here we use a BERT model fine-tuned on a [SQuaD 2.0 Dataset](https://rajpurkar.github.io/SQuAD-explorer/) which contains 100,000+ question-answer pairs on 500+ articles combined with over 50,000 new, unanswerable questions.
### 3.a Paragraph and Queries
The paragraph and the questions can be customized by changing the text below. Note that when using models with small sequence lengths, you should use a shorter paragraph:
#### Paragraph:
```
paragraph_text = "The Apollo program, also known as Project Apollo, was the third United States human spaceflight program carried out by the National Aeronautics and Space Administration (NASA), which accomplished landing the first humans on the Moon from 1969 to 1972. First conceived during Dwight D. Eisenhower's administration as a three-man spacecraft to follow the one-man Project Mercury which put the first Americans in space, Apollo was later dedicated to President John F. Kennedy's national goal of landing a man on the Moon and returning him safely to the Earth by the end of the 1960s, which he proposed in a May 25, 1961, address to Congress. Project Mercury was followed by the two-man Project Gemini. The first manned flight of Apollo was in 1968. Apollo ran from 1961 to 1972, and was supported by the two-man Gemini program which ran concurrently with it from 1962 to 1966. Gemini missions developed some of the space travel techniques that were necessary for the success of the Apollo missions. Apollo used Saturn family rockets as launch vehicles. Apollo/Saturn vehicles were also used for an Apollo Applications Program, which consisted of Skylab, a space station that supported three manned missions in 1973-74, and the Apollo-Soyuz Test Project, a joint Earth orbit mission with the Soviet Union in 1975."
# Short paragraph version for BERT models with max sequence length of 128
short_paragraph_text = "The Apollo program was the third United States human spaceflight program. First conceived as a three-man spacecraft to follow the one-man Project Mercury which put the first Americans in space, Apollo was dedicated to President John F. Kennedy's national goal of landing a man on the Moon. The first manned flight of Apollo was in 1968. Apollo ran from 1961 to 1972 followed by the Apollo-Soyuz Test Project a joint Earth orbit mission with the Soviet Union in 1975."
```
#### Question:
```
question_text = "What project put the first Americans into space?"
#question_text = "What year did the first manned Apollo flight occur?"
#question_text = "What President is credited with the original notion of putting Americans in space?"
#question_text = "Who did the U.S. collaborate with on an Earth orbit mission in 1975?"
```
In this example we ask our BERT model questions related to the following paragraph:
**The Apollo Program**
_"The Apollo program, also known as Project Apollo, was the third United States human spaceflight program carried out by the National Aeronautics and Space Administration (NASA), which accomplished landing the first humans on the Moon from 1969 to 1972. First conceived during Dwight D. Eisenhower's administration as a three-man spacecraft to follow the one-man Project Mercury which put the first Americans in space, Apollo was later dedicated to President John F. Kennedy's national goal of landing a man on the Moon and returning him safely to the Earth by the end of the 1960s, which he proposed in a May 25, 1961, address to Congress. Project Mercury was followed by the two-man Project Gemini. The first manned flight of Apollo was in 1968. Apollo ran from 1961 to 1972, and was supported by the two-man Gemini program which ran concurrently with it from 1962 to 1966. Gemini missions developed some of the space travel techniques that were necessary for the success of the Apollo missions. Apollo used Saturn family rockets as launch vehicles. Apollo/Saturn vehicles were also used for an Apollo Applications Program, which consisted of Skylab, a space station that supported three manned missions in 1973-74, and the Apollo-Soyuz Test Project, a joint Earth orbit mission with the Soviet Union in 1975."_
The questions and relative answers expected are shown below:
- **Q1:** "What project put the first Americans into space?"
- **A1:** "Project Mercury"
- **Q2:** "What program was created to carry out these projects and missions?"
- **A2:** "The Apollo program"
- **Q3:** "What year did the first manned Apollo flight occur?"
- **A3:** "1968"
- **Q4:** "What President is credited with the original notion of putting Americans in space?"
- **A4:** "John F. Kennedy"
- **Q5:** "Who did the U.S. collaborate with on an Earth orbit mission in 1975?"
- **A5:** "Soviet Union"
- **Q6:** "How long did Project Apollo run?"
- **A6:** "1961 to 1972"
- **Q7:** "What program helped develop space travel techniques that Project Apollo used?"
- **A7:** "Gemini Mission"
- **Q8:** "What space station supported three manned missions in 1973-1974?"
- **A8:** "Skylab"
## Data Preprocessing
Let's convert the paragraph and the question to BERT input with the help of the tokenizer:
```
import helpers.data_processing as dp
import helpers.tokenization as tokenization
tokenizer = tokenization.FullTokenizer(vocab_file="/workspace/TensorRT/demo/BERT/models/fine-tuned/bert_tf_ckpt_large_qa_squad2_amp_128_v19.03.1/vocab.txt", do_lower_case=True)
# The maximum number of tokens for the question. Questions longer than this will be truncated to this length.
max_query_length = 64
# When splitting up a long document into chunks, how much stride to take between chunks.
doc_stride = 128
# The maximum total input sequence length after WordPiece tokenization.
# Sequences longer than this will be truncated, and sequences shorter
max_seq_length = 128
# Extract tokens from the paragraph
doc_tokens = dp.convert_doc_tokens(short_paragraph_text)
# Extract features from the paragraph and question
features = dp.convert_example_to_features(doc_tokens, question_text, tokenizer, max_seq_length, doc_stride, max_query_length)
```
## TensorRT Inference
```
import tensorrt as trt
TRT_LOGGER = trt.Logger(trt.Logger.INFO)
import ctypes
import os
ctypes.CDLL("libnvinfer_plugin.so", mode=ctypes.RTLD_GLOBAL)
import pycuda.driver as cuda
import pycuda.autoinit
import collections
import numpy as np
import time
# Load the BERT-Large Engine
with open("/workspace/TensorRT/demo/BERT/engines/bert_large_128.engine", "rb") as f, \
trt.Runtime(TRT_LOGGER) as runtime, \
runtime.deserialize_cuda_engine(f.read()) as engine, \
engine.create_execution_context() as context:
# We always use batch size 1.
input_shape = (1, max_seq_length)
input_nbytes = trt.volume(input_shape) * trt.int32.itemsize
# Allocate device memory for inputs.
d_inputs = [cuda.mem_alloc(input_nbytes) for binding in range(3)]
# Create a stream in which to copy inputs/outputs and run inference.
stream = cuda.Stream()
# Specify input shapes. These must be within the min/max bounds of the active profile (0th profile in this case)
# Note that input shapes can be specified on a per-inference basis, but in this case, we only have a single shape.
for binding in range(3):
context.set_binding_shape(binding, input_shape)
assert context.all_binding_shapes_specified
# Allocate output buffer by querying the size from the context. This may be different for different input shapes.
h_output = cuda.pagelocked_empty(tuple(context.get_binding_shape(3)), dtype=np.float32)
d_output = cuda.mem_alloc(h_output.nbytes)
print("\nRunning Inference...")
_NetworkOutput = collections.namedtuple( # pylint: disable=invalid-name
"NetworkOutput",
["start_logits", "end_logits", "feature_index"])
networkOutputs = []
eval_time_elapsed = 0
for feature_index, feature in enumerate(features):
# Copy inputs
input_ids = cuda.register_host_memory(np.ascontiguousarray(feature.input_ids.ravel()))
segment_ids = cuda.register_host_memory(np.ascontiguousarray(feature.segment_ids.ravel()))
input_mask = cuda.register_host_memory(np.ascontiguousarray(feature.input_mask.ravel()))
eval_start_time = time.time()
cuda.memcpy_htod_async(d_inputs[0], input_ids, stream)
cuda.memcpy_htod_async(d_inputs[1], segment_ids, stream)
cuda.memcpy_htod_async(d_inputs[2], input_mask, stream)
# Run inference
context.execute_async_v2(bindings=[int(d_inp) for d_inp in d_inputs] + [int(d_output)], stream_handle=stream.handle)
# Synchronize the stream
stream.synchronize()
eval_time_elapsed += (time.time() - eval_start_time)
# Transfer predictions back from GPU
cuda.memcpy_dtoh_async(h_output, d_output, stream)
stream.synchronize()
for index, batch in enumerate(h_output):
# Data Post-processing
networkOutputs.append(_NetworkOutput(
start_logits = np.array(batch.squeeze()[:, 0]),
end_logits = np.array(batch.squeeze()[:, 1]),
feature_index = feature_index
))
eval_time_elapsed /= len(features)
print("-----------------------------")
print("Running Inference at {:.3f} Sentences/Sec".format(1.0/eval_time_elapsed))
print("-----------------------------")
```
## Data Post-Processing
Now that we have the inference results let's extract the actual answer to our question
```
# The total number of n-best predictions to generate in the nbest_predictions.json output file
n_best_size = 20
# The maximum length of an answer that can be generated. This is needed
# because the start and end predictions are not conditioned on one another
max_answer_length = 30
prediction, nbest_json, scores_diff_json = dp.get_predictions(doc_tokens, features,
networkOutputs, n_best_size, max_answer_length)
for index, output in enumerate(networkOutputs):
print("Processing output")
print("Answer: '{}'".format(prediction))
print("with prob: {:.3f}%".format(nbest_json[0]['probability'] * 100.0))
```
|
github_jupyter
|
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Datasets/Vectors/landsat_wrs2_grid.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Vectors/landsat_wrs2_grid.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Datasets/Vectors/landsat_wrs2_grid.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Vectors/landsat_wrs2_grid.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
```
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
dataset = ee.FeatureCollection('projects/google/wrs2_descending')
empty = ee.Image().byte()
Map.setCenter(-78, 36, 8)
Map.addLayer(empty.paint(dataset, 0, 2), {}, 'Landsat WRS-2 grid')
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
```
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
# set random seed for comparing the two result calculations
tf.set_random_seed(1)
# this is data
mnist = input_data.read_data_sets('MNIST_data', one_hot=True)
# hyperparameters
lr = 0.001
training_iters = 100000
batch_size = 128
n_inputs = 28 # MNIST data input (img shape: 28*28)
n_steps = 28 # time steps
n_hidden_units = 128 # neurons in hidden layer
n_classes = 10 # MNIST classes (0-9 digits)
num_layers=2
# tf Graph input
x = tf.placeholder(tf.float32, [None, n_steps, n_inputs])
y = tf.placeholder(tf.float32, [None, n_classes])
# Define weights
weights = {
# (28, 128)
'in': tf.Variable(tf.random_normal([n_inputs, n_hidden_units])),
# (128, 10)
'out': tf.Variable(tf.random_normal([n_hidden_units, n_classes]))
}
biases = {
# (128, )
'in': tf.Variable(tf.constant(0.1, shape=[n_hidden_units, ])),
# (10, )
'out': tf.Variable(tf.constant(0.1, shape=[n_classes, ]))
}
print ("parameters ready")
# tf Graph input
x = tf.placeholder(tf.float32, [None, n_steps, n_inputs])
y = tf.placeholder(tf.float32, [None, n_classes])
# Define weights
weights = {
# (28, 128)
'in': tf.Variable(tf.random_normal([n_inputs, n_hidden_units])),
# (128, 10)
'out': tf.Variable(tf.random_normal([n_hidden_units, n_classes]))
}
biases = {
# (128, )
'in': tf.Variable(tf.constant(0.1, shape=[n_hidden_units, ])),
# (10, )
'out': tf.Variable(tf.constant(0.1, shape=[n_classes, ]))
}
def RNN(X, weights, biases):
# hidden layer for input to cell
########################################
# transpose the inputs shape from
# X ==> (128 batch * 28 steps, 28 inputs)
X = tf.reshape(X, [-1, n_inputs])
# into hidden
# X_in = (128 batch * 28 steps, 128 hidden)
X_in = tf.matmul(X, weights['in']) + biases['in']
# X_in ==> (128 batch, 28 steps, 128 hidden)
X_in = tf.reshape(X_in, [-1, n_steps, n_hidden_units])
# cell
##########################################
# basic LSTM Cell.
if int((tf.__version__).split('.')[1]) < 12 and int((tf.__version__).split('.')[0]) < 1:
cell = tf.nn.rnn_cell.BasicLSTMCell(n_hidden_units, forget_bias=1.0, state_is_tuple=True)
cell = tf.nn.rnn_cell.DropoutWrapper(cell, output_keep_prob=0.5)
cell = tf.nn.rnn_cell.MultiRNNCell([cell] * num_layers)
else:
cell = tf.contrib.rnn.BasicLSTMCell(n_hidden_units)
cell = tf.contrib.rnn.DropoutWrapper(cell, output_keep_prob=0.5)
cell = tf.contrib.rnn.MultiRNNCell([cell] * num_layers)
# lstm cell is divided into two parts (c_state, h_state)
init_state = cell.zero_state(batch_size, dtype=tf.float32)
# You have 2 options for following step.
# 1: tf.nn.rnn(cell, inputs);
# 2: tf.nn.dynamic_rnn(cell, inputs).
# If use option 1, you have to modified the shape of X_in, go and check out this:
# https://github.com/aymericdamien/TensorFlow-Examples/blob/master/examples/3_NeuralNetworks/recurrent_network.py
# In here, we go for option 2.
# dynamic_rnn receive Tensor (batch, steps, inputs) or (steps, batch, inputs) as X_in.
# Make sure the time_major is changed accordingly.
outputs, final_state = tf.nn.dynamic_rnn(cell, X_in, initial_state=init_state, time_major=False)
# hidden layer for output as the final results
#############################################
# results = tf.matmul(final_state[1], weights['out']) + biases['out']
# # or
# unpack to list [(batch, outputs)..] * steps
if int((tf.__version__).split('.')[1]) < 12 and int((tf.__version__).split('.')[0]) < 1:
outputs = tf.unpack(tf.transpose(outputs, [1, 0, 2])) # states is the last outputs
else:
outputs = tf.unstack(tf.transpose(outputs, [1,0,2]))
results = tf.matmul(outputs[-1], weights['out']) + biases['out'] # shape = (128, 10)
return results
pred = RNN(x, weights, biases)
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
train_op = tf.train.AdamOptimizer(lr).minimize(cost)
correct_pred = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
print ("Network ready")
with tf.Session() as sess:
# tf.initialize_all_variables() no long valid from
# 2017-03-02 if using tensorflow >= 0.12
if int((tf.__version__).split('.')[1]) < 12 and int((tf.__version__).split('.')[0]) < 1:
init = tf.initialize_all_variables()
else:
init = tf.global_variables_initializer()
sess.run(init)
step = 0
while step * batch_size < training_iters:
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
batch_xs = batch_xs.reshape([batch_size, n_steps, n_inputs])
_, acc, loss=sess.run([train_op,accuracy,cost], feed_dict={
x: batch_xs,
y: batch_ys,
})
if step % 20 == 0:
print ("Iter " + str(step*batch_size) + ", Minibatch Loss= " + \
"{:.6f}".format(loss) + ", Training Accuracy= " + \
"{:.5f}".format(acc))
step += 1
```
|
github_jupyter
|
This notebook shows:
* How to launch the [**StarGANv1**](https://arxiv.org/abs/1711.09020) model for inference
* Example of results for both
* attrubutes **detection**
* new face **generation** with desired attributes
Here I use [**PyTorch** implementation](https://github.com/yunjey/stargan) of the StarGANv1 model.
[StarGANv1](https://arxiv.org/abs/1711.09020) was chosen because:
* It provides an ability to generate images **contitionally**. One can control the "amount" of each desired feature via input vector.
* It can **train (relatively) fast** on (relatively) small resources.
The model is pretty old though and has its own drawbacks:
* It works well only with small resolution images (~128).
* For bigger images the artifacts are inavoidable. They sometimes happen even for 128x128 images.
The obvious improvement is to use newer model, e.g., [StarGANv2](https://arxiv.org/abs/1912.01865) which was released in April 2020. It generates much better images at much higher resolution. But it requires both huge resoruces and lots of time to train.
Prior to running this notebook please download the pretrained models:
```
../scripts/get_models.sh
```
# Imports
Imort necessary libraries
```
import os
import sys
os.environ["KMP_DUPLICATE_LIB_OK"] = "True"
sys.path.extend(["../code/", "../stargan/"])
import torch
import torchvision.transforms as T
from PIL import Image
import matplotlib.pyplot as plt
from config import get_config
from solver import Solver
```
# Load model
Let's first load the config for the model. It is mostly default except for the:
* model checkpoint path
* style classes, their order and number
Note that in the original StarGANv1 model 5 classes are used: `[Black_Hair Blond_Hair Brown_Hair Male Young]`.
I retrained the model **4** times for different **face parts**. Each face part has several classes connected to it (see `DataExploration` notebook):
* **nose**: `[Big_Nose, Pointy_Nose]`
* **mouth**: `[Mouth_Slightly_Open, Smiling]`
* **eyes**: `[Arched_Eyebrows, Bushy_Eyebrows, Bags_Under_Eyes, Eyeglasses, Narrow_Eyes]`
* **hair**: `[Black_Hair, Blond_Hair, Brown_Hair, Gray_Hair, Bald Bangs, Receding_Hairline, Straight_Hair, Wavy_Hair]`
Here I show the examples only for **eyes** class. But all other classes works in the same way and prediction examples are shown in the repo and in other notebooks.
```
config = get_config("""
--model_save_dir ../models/celeba_128_eyes/
--test_iters 200000
--c_dim 5
--selected_attrs Arched_Eyebrows Bushy_Eyebrows Bags_Under_Eyes Eyeglasses Narrow_Eyes
""")
```
Load the model architecture with the provided config.
```
model = Solver(None, None, config)
```
Restore model weights.
```
model.restore_model(model.test_iters)
```
# Prediction example
Let's read a test image.
Note that the **face position and size** should be comparable to what the model has seen in the training data (CelebA). Here I do not use any face detector and crop the faces manually. But in production environment one needs to setup the face detector correspondingly.
```
image = Image.open("../data/test.jpg")
image
```
The input to the network is **3x128x128 image in a range [-1; 1]** (note that the channels is the first dimension).
Thus one need to do preprocessing in advance.
```
transform = []
transform.append(T.Resize(128))
transform.append(T.CenterCrop(128))
transform.append(T.ToTensor())
transform.append(T.Normalize(mean=(0.5, 0.5, 0.5), std=(0.5, 0.5, 0.5)))
transform = T.Compose(transform)
```
Create a batch of 1 image
```
x_real = torch.stack([transform(image)])
x_real.shape
```
## Attributes prediction
Let's first predict the attbibutes of the image. To do so I use the **Discriminator** part of the network. In StarGAN architecture it predicts not only the fake/real label but also the classes/attributes/styles of the image.
Here I call this vector **eigen style vector**. Note that due to the possible co-existence of multiple labels and the corresponding training procedure (Sigmoid + BCELoss instead of Softmax + CrossEntropyLoss) I use sigmoid activation function here and treat predicted labels separately (instead of softmax and 1-of-all).
```
with torch.no_grad():
eigen_style_vector = torch.sigmoid(model.D(x_real)[1])
```
Below is the probability of each label. The photo indeed depicts a person with big and little bit arched eyebrows.
```
for proba, tag in zip(eigen_style_vector.numpy()[0], model.selected_attrs):
print(f"{tag:20s}: {proba:.3f}")
```
Now let's look at how well the **Generator** model can recreate the face without altering it using the just computed eigen style vector.
```
with torch.no_grad():
res_eigen = model.G(x_real, eigen_style_vector)
res_eigen.shape
```
Plot the original face and the reconstructed one:
```
plt.figure(figsize=(9, 8))
plt.subplot(121)
_img = model.denorm(x_real).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Original", fontsize=16)
plt.subplot(122)
_img = model.denorm(res_eigen).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Eigen style reconstruction", fontsize=16);
```
Looks good enough.
## Face modification using new attributes
Now let's try to modify the face starting from the eigen style vector.
Let's say, I want to **add eyeglasses**. To do so I am to set the corresponding style vector component to 1.
```
eigen_style_vector_modified_1 = eigen_style_vector.clone()
eigen_style_vector_modified_1[:, 3] = 1
```
Now the style vector looks the following:
```
for proba, tag in zip(eigen_style_vector_modified_1.numpy()[0], model.selected_attrs):
print(f"{tag:20s}: {proba:.3f}")
```
Let's try to generate face with this modified style vector:
```
with torch.no_grad():
res_modified_1 = model.G(x_real, eigen_style_vector_modified_1)
res_modified_1.shape
```
Plot the faces:
```
plt.figure(figsize=(13.5, 8))
plt.subplot(131)
_img = model.denorm(x_real).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Original", fontsize=16)
plt.subplot(132)
_img = model.denorm(res_eigen).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Eigen style reconstruction", fontsize=16);
plt.subplot(133)
_img = model.denorm(res_modified_1).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Eyeglasses", fontsize=16);
```
Now let's try to **change two attributes simultaneously**:
* Make the eyes narrow
* Add archness to the eyebrows
```
eigen_style_vector_modified_2 = eigen_style_vector.clone()
eigen_style_vector_modified_2[:, 0] = 1
eigen_style_vector_modified_2[:, 4] = 1
```
Now the style vector looks the following:
```
for proba, tag in zip(eigen_style_vector_modified_2.numpy()[0], model.selected_attrs):
print(f"{tag:20s}: {proba:.3f}")
```
Let's try to generate face with this modified style vector:
```
with torch.no_grad():
res_modified_2 = model.G(x_real, eigen_style_vector_modified_2)
res_modified_2.shape
```
Plot the faces:
```
plt.figure(figsize=(18, 8))
plt.subplot(141)
_img = model.denorm(x_real).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Original", fontsize=16)
plt.subplot(142)
_img = model.denorm(res_eigen).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Eigen style reconstruction", fontsize=16);
plt.subplot(143)
_img = model.denorm(res_modified_1).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Eyeglasses", fontsize=16);
plt.subplot(144)
_img = model.denorm(res_modified_2).numpy()[0].transpose((1, 2, 0))
plt.imshow(_img)
plt.axis("off")
plt.title("Arched eyebrows + Narrow", fontsize=16);
```
Looks good!
|
github_jupyter
|
```
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.tree import export_text
```
This example uses the [Universal Bank](https://www.kaggle.com/sriharipramod/bank-loan-classification) data set and some example code of running classification trees from chapter 9 of [Data Mining for Business Analytics](https://www.dataminingbook.com/book/python-edition)
> The data include customer demographic information (age, income, etc.), the customer's relationship with the bank (mortgage, securities account, etc.), and the customer response to the last personal loan campaign (Personal Loan). Among these 5000 customers, only 480 (= 9.6%) accepted the personal loan that was offered to them in the earlier campaign
[Source](https://www.kaggle.com/itsmesunil/campaign-for-selling-personal-loans)
1. Train a decision tree classifier, print the tree and evaluate its accuracy.
2. Prune the tree by changing its hyper parameters, evaluate the accuracy of the new tree.
3. Using [grid search](https://scikit-learn.org/stable/modules/grid_search.html), perform a systematic tuning of the decision tree hyper parameters.
```
data = pd.read_csv('data/UniversalBank.csv')
data.head()
```
```
bank_df = data.drop(columns=['ID', 'ZIP Code'])
X = bank_df.drop(columns=['Personal Loan'])
y = bank_df['Personal Loan']
train_X, valid_X, train_y, valid_y = train_test_split(X, y, test_size=0.4, random_state=1)
dtree = DecisionTreeClassifier()
dtree.fit(train_X, train_y)
print(export_text(dtree, feature_names=list(X.columns)))
print(confusion_matrix(train_y, dtree.predict(train_X)))
print(confusion_matrix(valid_y, dtree.predict(valid_X)))
accuracy_score(train_y, dtree.predict(train_X)), accuracy_score(valid_y, dtree.predict(valid_X))
dtree = DecisionTreeClassifier(max_depth=30, min_samples_split=20, min_impurity_decrease=0.01)
dtree.fit(train_X, train_y)
print(export_text(dtree, feature_names=list(X.columns)))
print(confusion_matrix(train_y, dtree.predict(train_X)))
print(confusion_matrix(valid_y, dtree.predict(valid_X)))
accuracy_score(train_y, dtree.predict(train_X)), accuracy_score(valid_y, dtree.predict(valid_X))
# Start with an initial guess for parameters
param_grid = {
'max_depth': [10, 20, 30, 40],
'min_samples_split': [20, 40, 60, 80, 100],
'min_impurity_decrease': [0, 0.0005, 0.001, 0.005, 0.01],
}
gridSearch = GridSearchCV(DecisionTreeClassifier(), param_grid, cv=5, n_jobs=-1)
gridSearch.fit(train_X, train_y)
print('Score: ', gridSearch.best_score_)
print('Parameters: ', gridSearch.best_params_)
dtree = gridSearch.best_estimator_
print(confusion_matrix(train_y, dtree.predict(train_X)))
print(confusion_matrix(valid_y, dtree.predict(valid_X)))
accuracy_score(train_y, dtree.predict(train_X)), accuracy_score(valid_y, dtree.predict(valid_X))
print(export_text(dtree, feature_names=list(X.columns)))
```
|
github_jupyter
|
# Finetuning of the pretrained Japanese BERT model
Finetune the pretrained model to solve multi-class classification problems.
This notebook requires the following objects:
- trained sentencepiece model (model and vocab files)
- pretraiend Japanese BERT model
Dataset is livedoor ニュースコーパス in https://www.rondhuit.com/download.html.
We make test:dev:train = 2:2:6 datasets.
Results:
- Full training data
- BERT with SentencePiece
```
precision recall f1-score support
dokujo-tsushin 0.98 0.94 0.96 178
it-life-hack 0.96 0.97 0.96 172
kaden-channel 0.99 0.98 0.99 176
livedoor-homme 0.98 0.88 0.93 95
movie-enter 0.96 0.99 0.98 158
peachy 0.94 0.98 0.96 174
smax 0.98 0.99 0.99 167
sports-watch 0.98 1.00 0.99 190
topic-news 0.99 0.98 0.98 163
micro avg 0.97 0.97 0.97 1473
macro avg 0.97 0.97 0.97 1473
weighted avg 0.97 0.97 0.97 1473
```
- sklearn GradientBoostingClassifier with MeCab
```
precision recall f1-score support
dokujo-tsushin 0.89 0.86 0.88 178
it-life-hack 0.91 0.90 0.91 172
kaden-channel 0.90 0.94 0.92 176
livedoor-homme 0.79 0.74 0.76 95
movie-enter 0.93 0.96 0.95 158
peachy 0.87 0.92 0.89 174
smax 0.99 1.00 1.00 167
sports-watch 0.93 0.98 0.96 190
topic-news 0.96 0.86 0.91 163
micro avg 0.92 0.92 0.92 1473
macro avg 0.91 0.91 0.91 1473
weighted avg 0.92 0.92 0.91 1473
```
- Small training data (1/5 of full training data)
- BERT with SentencePiece
```
precision recall f1-score support
dokujo-tsushin 0.97 0.87 0.92 178
it-life-hack 0.86 0.86 0.86 172
kaden-channel 0.95 0.94 0.95 176
livedoor-homme 0.82 0.82 0.82 95
movie-enter 0.97 0.99 0.98 158
peachy 0.89 0.95 0.92 174
smax 0.94 0.96 0.95 167
sports-watch 0.97 0.97 0.97 190
topic-news 0.94 0.94 0.94 163
micro avg 0.93 0.93 0.93 1473
macro avg 0.92 0.92 0.92 1473
weighted avg 0.93 0.93 0.93 1473
```
- sklearn GradientBoostingClassifier with MeCab
```
precision recall f1-score support
dokujo-tsushin 0.82 0.71 0.76 178
it-life-hack 0.86 0.88 0.87 172
kaden-channel 0.91 0.87 0.89 176
livedoor-homme 0.67 0.63 0.65 95
movie-enter 0.87 0.95 0.91 158
peachy 0.70 0.78 0.73 174
smax 1.00 1.00 1.00 167
sports-watch 0.87 0.95 0.91 190
topic-news 0.92 0.82 0.87 163
micro avg 0.85 0.85 0.85 1473
macro avg 0.85 0.84 0.84 1473
weighted avg 0.86 0.85 0.85 1473
```
```
import configparser
import glob
import os
import pandas as pd
import subprocess
import sys
import tarfile
from urllib.request import urlretrieve
CURDIR = os.getcwd()
CONFIGPATH = os.path.join(CURDIR, os.pardir, 'config.ini')
config = configparser.ConfigParser()
config.read(CONFIGPATH)
```
## Data preparing
You need execute the following cells just once.
```
FILEURL = config['FINETUNING-DATA']['FILEURL']
FILEPATH = config['FINETUNING-DATA']['FILEPATH']
EXTRACTDIR = config['FINETUNING-DATA']['TEXTDIR']
```
Download and unzip data.
```
%%time
urlretrieve(FILEURL, FILEPATH)
mode = "r:gz"
tar = tarfile.open(FILEPATH, mode)
tar.extractall(EXTRACTDIR)
tar.close()
```
Data preprocessing.
```
def extract_txt(filename):
with open(filename) as text_file:
# 0: URL, 1: timestamp
text = text_file.readlines()[2:]
text = [sentence.strip() for sentence in text]
text = list(filter(lambda line: line != '', text))
return ''.join(text)
categories = [
name for name
in os.listdir( os.path.join(EXTRACTDIR, "text") )
if os.path.isdir( os.path.join(EXTRACTDIR, "text", name) ) ]
categories = sorted(categories)
categories
table = str.maketrans({
'\n': '',
'\t': ' ',
'\r': '',
})
%%time
all_text = []
all_label = []
for cat in categories:
files = glob.glob(os.path.join(EXTRACTDIR, "text", cat, "{}*.txt".format(cat)))
files = sorted(files)
body = [ extract_txt(elem).translate(table) for elem in files ]
label = [cat] * len(body)
all_text.extend(body)
all_label.extend(label)
df = pd.DataFrame({'text' : all_text, 'label' : all_label})
df.head()
df = df.sample(frac=1, random_state=23).reset_index(drop=True)
df.head()
```
Save data as tsv files.
test:dev:train = 2:2:6. To check the usability of finetuning, we also prepare sampled training data (1/5 of full training data).
```
df[:len(df) // 5].to_csv( os.path.join(EXTRACTDIR, "test.tsv"), sep='\t', index=False)
df[len(df) // 5:len(df)*2 // 5].to_csv( os.path.join(EXTRACTDIR, "dev.tsv"), sep='\t', index=False)
df[len(df)*2 // 5:].to_csv( os.path.join(EXTRACTDIR, "train.tsv"), sep='\t', index=False)
### 1/5 of full training data.
# df[:len(df) // 5].to_csv( os.path.join(EXTRACTDIR, "test.tsv"), sep='\t', index=False)
# df[len(df) // 5:len(df)*2 // 5].to_csv( os.path.join(EXTRACTDIR, "dev.tsv"), sep='\t', index=False)
# df[len(df)*2 // 5:].sample(frac=0.2, random_state=23).to_csv( os.path.join(EXTRACTDIR, "train.tsv"), sep='\t', index=False)
```
## Finetune pre-trained model
It will take a lot of hours to execute the following cells on CPU environment.
You can also use colab to recieve the power of TPU. You need to uplode the created data onto your GCS bucket.
[](https://colab.research.google.com/drive/1zZH2GWe0U-7GjJ2w2duodFfEUptvHjcx)
```
PRETRAINED_MODEL_PATH = '../model/model.ckpt-1400000'
FINETUNE_OUTPUT_DIR = '../model/livedoor_output'
%%time
# It will take many hours on CPU environment.
!python3 ../src/run_classifier.py \
--task_name=livedoor \
--do_train=true \
--do_eval=true \
--data_dir=../data/livedoor \
--model_file=../model/wiki-ja.model \
--vocab_file=../model/wiki-ja.vocab \
--init_checkpoint={PRETRAINED_MODEL_PATH} \
--max_seq_length=512 \
--train_batch_size=4 \
--learning_rate=2e-5 \
--num_train_epochs=10 \
--output_dir={FINETUNE_OUTPUT_DIR}
```
## Predict using the finetuned model
Let's predict test data using the finetuned model.
```
import sys
sys.path.append("../src")
import tokenization_sentencepiece as tokenization
from run_classifier import LivedoorProcessor
from run_classifier import model_fn_builder
from run_classifier import file_based_input_fn_builder
from run_classifier import file_based_convert_examples_to_features
from utils import str_to_value
sys.path.append("../bert")
import modeling
import optimization
import tensorflow as tf
import configparser
import json
import glob
import os
import pandas as pd
import tempfile
bert_config_file = tempfile.NamedTemporaryFile(mode='w+t', encoding='utf-8', suffix='.json')
bert_config_file.write(json.dumps({k:str_to_value(v) for k,v in config['BERT-CONFIG'].items()}))
bert_config_file.seek(0)
bert_config = modeling.BertConfig.from_json_file(bert_config_file.name)
output_ckpts = glob.glob("{}/model.ckpt*data*".format(FINETUNE_OUTPUT_DIR))
latest_ckpt = sorted(output_ckpts)[-1]
FINETUNED_MODEL_PATH = latest_ckpt.split('.data-00000-of-00001')[0]
class FLAGS(object):
'''Parameters.'''
def __init__(self):
self.model_file = "../model/wiki-ja.model"
self.vocab_file = "../model/wiki-ja.vocab"
self.do_lower_case = True
self.use_tpu = False
self.output_dir = "/dummy"
self.data_dir = "../data/livedoor"
self.max_seq_length = 512
self.init_checkpoint = FINETUNED_MODEL_PATH
self.predict_batch_size = 4
# The following parameters are not used in predictions.
# Just use to create RunConfig.
self.master = None
self.save_checkpoints_steps = 1
self.iterations_per_loop = 1
self.num_tpu_cores = 1
self.learning_rate = 0
self.num_warmup_steps = 0
self.num_train_steps = 0
self.train_batch_size = 0
self.eval_batch_size = 0
FLAGS = FLAGS()
processor = LivedoorProcessor()
label_list = processor.get_labels()
tokenizer = tokenization.FullTokenizer(
model_file=FLAGS.model_file, vocab_file=FLAGS.vocab_file,
do_lower_case=FLAGS.do_lower_case)
tpu_cluster_resolver = None
is_per_host = tf.contrib.tpu.InputPipelineConfig.PER_HOST_V2
run_config = tf.contrib.tpu.RunConfig(
cluster=tpu_cluster_resolver,
master=FLAGS.master,
model_dir=FLAGS.output_dir,
save_checkpoints_steps=FLAGS.save_checkpoints_steps,
tpu_config=tf.contrib.tpu.TPUConfig(
iterations_per_loop=FLAGS.iterations_per_loop,
num_shards=FLAGS.num_tpu_cores,
per_host_input_for_training=is_per_host))
model_fn = model_fn_builder(
bert_config=bert_config,
num_labels=len(label_list),
init_checkpoint=FLAGS.init_checkpoint,
learning_rate=FLAGS.learning_rate,
num_train_steps=FLAGS.num_train_steps,
num_warmup_steps=FLAGS.num_warmup_steps,
use_tpu=FLAGS.use_tpu,
use_one_hot_embeddings=FLAGS.use_tpu)
estimator = tf.contrib.tpu.TPUEstimator(
use_tpu=FLAGS.use_tpu,
model_fn=model_fn,
config=run_config,
train_batch_size=FLAGS.train_batch_size,
eval_batch_size=FLAGS.eval_batch_size,
predict_batch_size=FLAGS.predict_batch_size)
predict_examples = processor.get_test_examples(FLAGS.data_dir)
predict_file = tempfile.NamedTemporaryFile(mode='w+t', encoding='utf-8', suffix='.tf_record')
file_based_convert_examples_to_features(predict_examples, label_list,
FLAGS.max_seq_length, tokenizer,
predict_file.name)
predict_drop_remainder = True if FLAGS.use_tpu else False
predict_input_fn = file_based_input_fn_builder(
input_file=predict_file.name,
seq_length=FLAGS.max_seq_length,
is_training=False,
drop_remainder=predict_drop_remainder)
result = estimator.predict(input_fn=predict_input_fn)
%%time
# It will take a few hours on CPU environment.
result = list(result)
result[:2]
```
Read test data set and add prediction results.
```
import pandas as pd
test_df = pd.read_csv("../data/livedoor/test.tsv", sep='\t')
test_df['predict'] = [ label_list[elem['probabilities'].argmax()] for elem in result ]
test_df.head()
sum( test_df['label'] == test_df['predict'] ) / len(test_df)
```
A littel more detailed check using `sklearn.metrics`.
```
!pip install scikit-learn
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
print(classification_report(test_df['label'], test_df['predict']))
print(confusion_matrix(test_df['label'], test_df['predict']))
```
### Simple baseline model.
```
import pandas as pd
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
train_df = pd.read_csv("../data/livedoor/train.tsv", sep='\t')
dev_df = pd.read_csv("../data/livedoor/dev.tsv", sep='\t')
test_df = pd.read_csv("../data/livedoor/test.tsv", sep='\t')
!apt-get install -q -y mecab libmecab-dev mecab-ipadic mecab-ipadic-utf8
!pip install mecab-python3==0.7
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.ensemble import GradientBoostingClassifier
import MeCab
m = MeCab.Tagger("-Owakati")
train_dev_df = pd.concat([train_df, dev_df])
train_dev_xs = train_dev_df['text'].apply(lambda x: m.parse(x))
train_dev_ys = train_dev_df['label']
test_xs = test_df['text'].apply(lambda x: m.parse(x))
test_ys = test_df['label']
vectorizer = TfidfVectorizer(max_features=750)
train_dev_xs_ = vectorizer.fit_transform(train_dev_xs)
test_xs_ = vectorizer.transform(test_xs)
```
The following set up is not exactly identical to that of BERT because inside Classifier it uses `train_test_split` with shuffle.
In addition, parameters are not well tuned, however, we think it's enough to check the power of BERT.
```
%%time
model = GradientBoostingClassifier(n_estimators=200,
validation_fraction=len(train_df)/len(dev_df),
n_iter_no_change=5,
tol=0.01,
random_state=23)
### 1/5 of full training data.
# model = GradientBoostingClassifier(n_estimators=200,
# validation_fraction=len(dev_df)/len(train_df),
# n_iter_no_change=5,
# tol=0.01,
# random_state=23)
model.fit(train_dev_xs_, train_dev_ys)
print(classification_report(test_ys, model.predict(test_xs_)))
print(confusion_matrix(test_ys, model.predict(test_xs_)))
```
|
github_jupyter
|
```
import json
import math
import numpy as np
import openrtdynamics2.lang as dy
import openrtdynamics2.targets as tg
from vehicle_lib.vehicle_lib import *
# load track data
with open("track_data/simple_track.json", "r") as read_file:
track_data = json.load(read_file)
#
# Demo: a vehicle controlled to follow a given path
#
# Implemented using the code generator openrtdynamics 2 - https://pypi.org/project/openrtdynamics2/ .
# This generates c++ code for Web Assembly to be run within the browser.
#
system = dy.enter_system()
velocity = dy.system_input( dy.DataTypeFloat64(1), name='velocity', default_value=6.0, value_range=[0, 25], title="vehicle velocity")
max_lateral_velocity = dy.system_input( dy.DataTypeFloat64(1), name='max_lateral_velocity', default_value=1.0, value_range=[0, 4.0], title="maximal lateral velocity")
max_lateral_accleration = dy.system_input( dy.DataTypeFloat64(1), name='max_lateral_accleration', default_value=2.0, value_range=[1.0, 4.0], title="maximal lateral acceleration")
# parameters
wheelbase = 3.0
# sampling time
Ts = 0.01
# create storage for the reference path:
path = import_path_data(track_data)
# create placeholders for the plant output signals
x = dy.signal()
y = dy.signal()
psi = dy.signal()
# track the evolution of the closest point on the path to the vehicles position
projection = track_projection_on_path(path, x, y)
d_star = projection['d_star'] # the distance parameter of the path describing the closest point to the vehicle
x_r = projection['x_r'] # (x_r, y_r) the projected vehicle position on the path
y_r = projection['y_r']
psi_rr = projection['psi_r'] # the orientation angle (tangent of the path)
K_r = projection['K_r'] # the curvature of the path
Delta_l = projection['Delta_l'] # the lateral distance between vehicle and path
#
# project the vehicle velocity onto the path yielding v_star
#
# Used formula inside project_velocity_on_path:
# v_star = d d_star / dt = v * cos( Delta_u ) / ( 1 - Delta_l * K(d_star) )
#
Delta_u = dy.signal() # feedback from control
v_star = project_velocity_on_path(velocity, Delta_u, Delta_l, K_r)
dy.append_output(v_star, 'v_star')
#
# compute an enhanced (less noise) signal for the path orientation psi_r by integrating the
# curvature profile and fusing the result with psi_rr to mitigate the integration drift.
#
psi_r, psi_r_dot = compute_path_orientation_from_curvature( Ts, v_star, psi_rr, K_r, L=1.0 )
dy.append_output(psi_rr, 'psi_rr')
dy.append_output(psi_r_dot, 'psi_r_dot')
#
# lateral open-loop control to realize an 'obstacle-avoiding maneuver'
#
# the dynamic model for the lateral distance Delta_l is
#
# d/dt Delta_l = u,
#
# meaning u is the lateral velocity to which is used to control the lateral
# distance to the path.
#
# generate a velocity profile
u_move_left = dy.signal_step( dy.int32(50) ) - dy.signal_step( dy.int32(200) )
u_move_right = dy.signal_step( dy.int32(500) ) - dy.signal_step( dy.int32(350) )
# apply a rate limiter to limit the acceleration
u = dy.rate_limit( max_lateral_velocity * (u_move_left + u_move_right), Ts, dy.float64(-1) * max_lateral_accleration, max_lateral_accleration)
dy.append_output(u, 'u')
# internal lateral model (to verify the lateral dynamics of the simulated vehicle)
Delta_l_mdl = dy.euler_integrator(u, Ts)
dy.append_output(Delta_l_mdl, 'Delta_l_mdl')
#
# path tracking control
#
# Control of the lateral distance to the path can be performed via the augmented control
# variable u.
#
# Herein, a linearization yielding the resulting lateral dynamics u --> Delta_l : 1/s is applied.
#
Delta_u << dy.asin( dy.saturate(u / velocity, -0.99, 0.99) )
delta_star = psi_r - psi
delta = delta_star + Delta_u
delta = dy.unwrap_angle(angle=delta, normalize_around_zero = True)
dy.append_output(Delta_u, 'Delta_u')
dy.append_output(delta_star, 'delta_star')
#
# The model of the vehicle including a disturbance
#
# steering angle limit
delta = dy.saturate(u=delta, lower_limit=-math.pi/2.0, upper_limit=math.pi/2.0)
# the model of the vehicle
x_, y_, psi_, x_dot, y_dot, psi_dot = discrete_time_bicycle_model(delta, velocity, Ts, wheelbase)
# close the feedback loops
x << x_
y << y_
psi << psi_
#
# outputs: these are available for visualization in the html set-up
#
dy.append_output(x, 'x')
dy.append_output(y, 'y')
dy.append_output(psi, 'psi')
dy.append_output(delta, 'steering')
dy.append_output(x_r, 'x_r')
dy.append_output(y_r, 'y_r')
dy.append_output(psi_r, 'psi_r')
dy.append_output(Delta_l, 'Delta_l')
# generate code for Web Assembly (wasm), requires emcc (emscripten) to build
code_gen_results = dy.generate_code(template=tg.TargetCppWASM(), folder="generated/path_following_lateral_dynamics", build=True)
#
dy.clear()
import IPython
IPython.display.IFrame(src='../vehicle_control_tutorial/path_following_lateral_dynamics.html', width='100%', height=1000)
```
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Mixed precision
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/mixed_precision"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/mixed_precision.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/guide/mixed_precision.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/mixed_precision.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
## Overview
Mixed precision is the use of both 16-bit and 32-bit floating-point types in a model during training to make it run faster and use less memory. By keeping certain parts of the model in the 32-bit types for numeric stability, the model will have a lower step time and train equally as well in terms of the evaluation metrics such as accuracy. This guide describes how to use the experimental Keras mixed precision API to speed up your models. Using this API can improve performance by more than 3 times on modern GPUs and 60% on TPUs.
Note: The Keras mixed precision API is currently experimental and may change.
Today, most models use the float32 dtype, which takes 32 bits of memory. However, there are two lower-precision dtypes, float16 and bfloat16, each which take 16 bits of memory instead. Modern accelerators can run operations faster in the 16-bit dtypes, as they have specialized hardware to run 16-bit computations and 16-bit dtypes can be read from memory faster.
NVIDIA GPUs can run operations in float16 faster than in float32, and TPUs can run operations in bfloat16 faster than float32. Therefore, these lower-precision dtypes should be used whenever possible on those devices. However, variables and a few computations should still be in float32 for numeric reasons so that the model trains to the same quality. The Keras mixed precision API allows you to use a mix of either float16 or bfloat16 with float32, to get the performance benefits from float16/bfloat16 and the numeric stability benefits from float32.
Note: In this guide, the term "numeric stability" refers to how a model's quality is affected by the use of a lower-precision dtype instead of a higher precision dtype. We say an operation is "numerically unstable" in float16 or bfloat16 if running it in one of those dtypes causes the model to have worse evaluation accuracy or other metrics compared to running the operation in float32.
## Setup
The Keras mixed precision API is available in TensorFlow 2.1.
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.mixed_precision import experimental as mixed_precision
```
## Supported hardware
While mixed precision will run on most hardware, it will only speed up models on recent NVIDIA GPUs and Cloud TPUs. NVIDIA GPUs support using a mix of float16 and float32, while TPUs support a mix of bfloat16 and float32.
Among NVIDIA GPUs, those with compute capability 7.0 or higher will see the greatest performance benefit from mixed precision because they have special hardware units, called Tensor Cores, to accelerate float16 matrix multiplications and convolutions. Older GPUs offer no math performance benefit for using mixed precision, however memory and bandwidth savings can enable some speedups. You can look up the compute capability for your GPU at NVIDIA's [CUDA GPU web page](https://developer.nvidia.com/cuda-gpus). Examples of GPUs that will benefit most from mixed precision include RTX GPUs, the Titan V, and the V100.
Note: If running this guide in Google Colab, the GPU runtime typically has a P100 connected. The P100 has compute capability 6.0 and is not expected to show a significant speedup.
You can check your GPU type with the following. The command only exists if the
NVIDIA drivers are installed, so the following will raise an error otherwise.
```
!nvidia-smi -L
```
All Cloud TPUs support bfloat16.
Even on CPUs and older GPUs, where no speedup is expected, mixed precision APIs can still be used for unit testing, debugging, or just to try out the API.
## Setting the dtype policy
To use mixed precision in Keras, you need to create a `tf.keras.mixed_precision.experimental.Policy`, typically referred to as a *dtype policy*. Dtype policies specify the dtypes layers will run in. In this guide, you will construct a policy from the string `'mixed_float16'` and set it as the global policy. This will will cause subsequently created layers to use mixed precision with a mix of float16 and float32.
```
policy = mixed_precision.Policy('mixed_float16')
mixed_precision.set_policy(policy)
```
The policy specifies two important aspects of a layer: the dtype the layer's computations are done in, and the dtype of a layer's variables. Above, you created a `mixed_float16` policy (i.e., a `mixed_precision.Policy` created by passing the string `'mixed_float16'` to its constructor). With this policy, layers use float16 computations and float32 variables. Computations are done in float16 for performance, but variables must be kept in float32 for numeric stability. You can directly query these properties of the policy.
```
print('Compute dtype: %s' % policy.compute_dtype)
print('Variable dtype: %s' % policy.variable_dtype)
```
As mentioned before, the `mixed_float16` policy will most significantly improve performance on NVIDIA GPUs with compute capability of at least 7.0. The policy will run on other GPUs and CPUs but may not improve performance. For TPUs, the `mixed_bfloat16` policy should be used instead.
## Building the model
Next, let's start building a simple model. Very small toy models typically do not benefit from mixed precision, because overhead from the TensorFlow runtime typically dominates the execution time, making any performance improvement on the GPU negligible. Therefore, let's build two large `Dense` layers with 4096 units each if a GPU is used.
```
inputs = keras.Input(shape=(784,), name='digits')
if tf.config.list_physical_devices('GPU'):
print('The model will run with 4096 units on a GPU')
num_units = 4096
else:
# Use fewer units on CPUs so the model finishes in a reasonable amount of time
print('The model will run with 64 units on a CPU')
num_units = 64
dense1 = layers.Dense(num_units, activation='relu', name='dense_1')
x = dense1(inputs)
dense2 = layers.Dense(num_units, activation='relu', name='dense_2')
x = dense2(x)
```
Each layer has a policy and uses the global policy by default. Each of the `Dense` layers therefore have the `mixed_float16` policy because you set the global policy to `mixed_float16` previously. This will cause the dense layers to do float16 computations and have float32 variables. They cast their inputs to float16 in order to do float16 computations, which causes their outputs to be float16 as a result. Their variables are float32 and will be cast to float16 when the layers are called to avoid errors from dtype mismatches.
```
print('x.dtype: %s' % x.dtype.name)
# 'kernel' is dense1's variable
print('dense1.kernel.dtype: %s' % dense1.kernel.dtype.name)
```
Next, create the output predictions. Normally, you can create the output predictions as follows, but this is not always numerically stable with float16.
```
# INCORRECT: softmax and model output will be float16, when it should be float32
outputs = layers.Dense(10, activation='softmax', name='predictions')(x)
print('Outputs dtype: %s' % outputs.dtype.name)
```
A softmax activation at the end of the model should be float32. Because the dtype policy is `mixed_float16`, the softmax activation would normally have a float16 compute dtype and output a float16 tensors.
This can be fixed by separating the Dense and softmax layers, and by passing `dtype='float32'` to the softmax layer
```
# CORRECT: softmax and model output are float32
x = layers.Dense(10, name='dense_logits')(x)
outputs = layers.Activation('softmax', dtype='float32', name='predictions')(x)
print('Outputs dtype: %s' % outputs.dtype.name)
```
Passing `dtype='float32'` to the softmax layer constructor overrides the layer's dtype policy to be the `float32` policy, which does computations and keeps variables in float32. Equivalently, we could have instead passed `dtype=mixed_precision.Policy('float32')`; layers always convert the dtype argument to a policy. Because the `Activation` layer has no variables, the policy's variable dtype is ignored, but the policy's compute dtype of float32 causes softmax and the model output to be float32.
Adding a float16 softmax in the middle of a model is fine, but a softmax at the end of the model should be in float32. The reason is that if the intermediate tensor flowing from the softmax to the loss is float16 or bfloat16, numeric issues may occur.
You can override the dtype of any layer to be float32 by passing `dtype='float32'` if you think it will not be numerically stable with float16 computations. But typically, this is only necessary on the last layer of the model, as most layers have sufficient precision with `mixed_float16` and `mixed_bfloat16`.
Even if the model does not end in a softmax, the outputs should still be float32. While unnecessary for this specific model, the model outputs can be cast to float32 with the following:
```
# The linear activation is an identity function. So this simply casts 'outputs'
# to float32. In this particular case, 'outputs' is already float32 so this is a
# no-op.
outputs = layers.Activation('linear', dtype='float32')(outputs)
```
Next, finish and compile the model, and generate input data.
```
model = keras.Model(inputs=inputs, outputs=outputs)
model.compile(loss='sparse_categorical_crossentropy',
optimizer=keras.optimizers.RMSprop(),
metrics=['accuracy'])
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
x_train = x_train.reshape(60000, 784).astype('float32') / 255
x_test = x_test.reshape(10000, 784).astype('float32') / 255
```
This example cast the input data from int8 to float32. We don't cast to float16 since the division by 255 is on the CPU, which runs float16 operations slower than float32 operations. In this case, the performance difference in negligible, but in general you should run input processing math in float32 if it runs on the CPU. The first layer of the model will cast the inputs to float16, as each layer casts floating-point inputs to its compute dtype.
The initial weights of the model are retrieved. This will allow training from scratch again by loading the weights.
```
initial_weights = model.get_weights()
```
## Training the model with Model.fit
Next, train the model.
```
history = model.fit(x_train, y_train,
batch_size=8192,
epochs=5,
validation_split=0.2)
test_scores = model.evaluate(x_test, y_test, verbose=2)
print('Test loss:', test_scores[0])
print('Test accuracy:', test_scores[1])
```
Notice the model prints the time per sample in the logs: for example, "4us/sample". The first epoch may be slower as TensorFlow spends some time optimizing the model, but afterwards the time per sample should stabilize.
If you are running this guide in Colab, you can compare the performance of mixed precision with float32. To do so, change the policy from `mixed_float16` to `float32` in the "Setting the dtype policy" section, then rerun all the cells up to this point. On GPUs with at least compute capability 7.0, you should see the time per sample significantly increase, indicating mixed precision sped up the model. For example, with a Titan V GPU, the per-sample time increases from 4us to 12us. Make sure to change the policy back to `mixed_float16` and rerun the cells before continuing with the guide.
For many real-world models, mixed precision also allows you to double the batch size without running out of memory, as float16 tensors take half the memory. This does not apply however to this toy model, as you can likely run the model in any dtype where each batch consists of the entire MNIST dataset of 60,000 images.
If running mixed precision on a TPU, you will not see as much of a performance gain compared to running mixed precision on GPUs. This is because TPUs already do certain ops in bfloat16 under the hood even with the default dtype policy of `float32`. TPU hardware does not support float32 for certain ops which are numerically stable in bfloat16, such as matmul. For such ops the TPU backend will silently use bfloat16 internally instead. As a consequence, passing `dtype='float32'` to layers which use such ops may have no numerical effect, however it is unlikely running such layers with bfloat16 computations will be harmful.
## Loss scaling
Loss scaling is a technique which `tf.keras.Model.fit` automatically performs with the `mixed_float16` policy to avoid numeric underflow. This section describes loss scaling and how to customize its behavior.
### Underflow and Overflow
The float16 data type has a narrow dynamic range compared to float32. This means values above $65504$ will overflow to infinity and values below $6.0 \times 10^{-8}$ will underflow to zero. float32 and bfloat16 have a much higher dynamic range so that overflow and underflow are not a problem.
For example:
```
x = tf.constant(256, dtype='float16')
(x ** 2).numpy() # Overflow
x = tf.constant(1e-5, dtype='float16')
(x ** 2).numpy() # Underflow
```
In practice, overflow with float16 rarely occurs. Additionally, underflow also rarely occurs during the forward pass. However, during the backward pass, gradients can underflow to zero. Loss scaling is a technique to prevent this underflow.
### Loss scaling background
The basic concept of loss scaling is simple: simply multiply the loss by some large number, say $1024$. We call this number the *loss scale*. This will cause the gradients to scale by $1024$ as well, greatly reducing the chance of underflow. Once the final gradients are computed, divide them by $1024$ to bring them back to their correct values.
The pseudocode for this process is:
```
loss_scale = 1024
loss = model(inputs)
loss *= loss_scale
# We assume `grads` are float32. We do not want to divide float16 gradients
grads = compute_gradient(loss, model.trainable_variables)
grads /= loss_scale
```
Choosing a loss scale can be tricky. If the loss scale is too low, gradients may still underflow to zero. If too high, the opposite the problem occurs: the gradients may overflow to infinity.
To solve this, TensorFlow dynamically determines the loss scale so you do not have to choose one manually. If you use `tf.keras.Model.fit`, loss scaling is done for you so you do not have to do any extra work. This is explained further in the next section.
### Choosing the loss scale
Each dtype policy optionally has an associated `tf.mixed_precision.experimental.LossScale` object, which represents a fixed or dynamic loss scale. By default, the loss scale for the `mixed_float16` policy is a `tf.mixed_precision.experimental.DynamicLossScale`, which dynamically determines the loss scale value. Other policies do not have a loss scale by default, as it is only necessary when float16 is used. You can query the loss scale of the policy:
```
loss_scale = policy.loss_scale
print('Loss scale: %s' % loss_scale)
```
The loss scale prints a lot of internal state, but you can ignore it. The most important part is the `current_loss_scale` part, which shows the loss scale's current value.
You can instead use a static loss scale by passing a number when constructing a dtype policy.
```
new_policy = mixed_precision.Policy('mixed_float16', loss_scale=1024)
print(new_policy.loss_scale)
```
The dtype policy constructor always converts the loss scale to a `LossScale` object. In this case, it's converted to a `tf.mixed_precision.experimental.FixedLossScale`, the only other `LossScale` subclass other than `DynamicLossScale`.
Note: *Using anything other than a dynamic loss scale is not recommended*. Choosing a fixed loss scale can be difficult, as making it too low will cause the model to not train as well, and making it too high will cause Infs or NaNs to appear in the gradients. A dynamic loss scale is typically near the optimal loss scale, so you do not have to do any work. Currently, dynamic loss scales are a bit slower than fixed loss scales, but the performance will be improved in the future.
Models, like layers, each have a dtype policy. If present, a model uses its policy's loss scale to apply loss scaling in the `tf.keras.Model.fit` method. This means if `Model.fit` is used, you do not have to worry about loss scaling at all: The `mixed_float16` policy will have a dynamic loss scale by default, and `Model.fit` will apply it.
With custom training loops, the model will ignore the policy's loss scale, and you will have to apply it manually. This is explained in the next section.
## Training the model with a custom training loop
So far, you trained a Keras model with mixed precision using `tf.keras.Model.fit`. Next, you will use mixed precision with a custom training loop. If you do not already know what a custom training loop is, please read [the Custom training guide](../tutorials/customization/custom_training_walkthrough.ipynb) first.
Running a custom training loop with mixed precision requires two changes over running it in float32:
1. Build the model with mixed precision (you already did this)
2. Explicitly use loss scaling if `mixed_float16` is used.
For step (2), you will use the `tf.keras.mixed_precision.experimental.LossScaleOptimizer` class, which wraps an optimizer and applies loss scaling. It takes two arguments: the optimizer and the loss scale. Construct one as follows to use a dynamic loss scale
```
optimizer = keras.optimizers.RMSprop()
optimizer = mixed_precision.LossScaleOptimizer(optimizer, loss_scale='dynamic')
```
Passing `'dynamic'` is equivalent to passing `tf.mixed_precision.experimental.DynamicLossScale()`.
Next, define the loss object and the `tf.data.Dataset`s.
```
loss_object = tf.keras.losses.SparseCategoricalCrossentropy()
train_dataset = (tf.data.Dataset.from_tensor_slices((x_train, y_train))
.shuffle(10000).batch(8192))
test_dataset = tf.data.Dataset.from_tensor_slices((x_test, y_test)).batch(8192)
```
Next, define the training step function. Two new methods from the loss scale optimizer are used in order to scale the loss and unscale the gradients:
* `get_scaled_loss(loss)`: Multiplies the loss by the loss scale
* `get_unscaled_gradients(gradients)`: Takes in a list of scaled gradients as inputs, and divides each one by the loss scale to unscale them
These functions must be used in order to prevent underflow in the gradients. `LossScaleOptimizer.apply_gradients` will then apply gradients if none of them have Infs or NaNs. It will also update the loss scale, halving it if the gradients had Infs or NaNs and potentially increasing it otherwise.
```
@tf.function
def train_step(x, y):
with tf.GradientTape() as tape:
predictions = model(x)
loss = loss_object(y, predictions)
scaled_loss = optimizer.get_scaled_loss(loss)
scaled_gradients = tape.gradient(scaled_loss, model.trainable_variables)
gradients = optimizer.get_unscaled_gradients(scaled_gradients)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
return loss
```
The `LossScaleOptimizer` will likely skip the first few steps at the start of training. The loss scale starts out high so that the optimal loss scale can quickly be determined. After a few steps, the loss scale will stabilize and very few steps will be skipped. This process happens automatically and does not affect training quality.
Now define the test step.
```
@tf.function
def test_step(x):
return model(x, training=False)
```
Load the initial weights of the model, so you can retrain from scratch.
```
model.set_weights(initial_weights)
```
Finally, run the custom training loop.
```
for epoch in range(5):
epoch_loss_avg = tf.keras.metrics.Mean()
test_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(
name='test_accuracy')
for x, y in train_dataset:
loss = train_step(x, y)
epoch_loss_avg(loss)
for x, y in test_dataset:
predictions = test_step(x)
test_accuracy.update_state(y, predictions)
print('Epoch {}: loss={}, test accuracy={}'.format(epoch, epoch_loss_avg.result(), test_accuracy.result()))
```
## GPU performance tips
Here are some performance tips when using mixed precision on GPUs.
### Increasing your batch size
If it doesn't affect model quality, try running with double the batch size when using mixed precision. As float16 tensors use half the memory, this often allows you to double your batch size without running out of memory. Increasing batch size typically increases training throughput, i.e. the training elements per second your model can run on.
### Ensuring GPU Tensor Cores are used
As mentioned previously, modern NVIDIA GPUs use a special hardware unit called Tensor Cores that can multiply float16 matrices very quickly. However, Tensor Cores requires certain dimensions of tensors to be a multiple of 8. In the examples below, an argument is bold if and only if it needs to be a multiple of 8 for Tensor Cores to be used.
* tf.keras.layers.Dense(**units=64**)
* tf.keras.layers.Conv2d(**filters=48**, kernel_size=7, stride=3)
* And similarly for other convolutional layers, such as tf.keras.layers.Conv3d
* tf.keras.layers.LSTM(**units=64**)
* And similar for other RNNs, such as tf.keras.layers.GRU
* tf.keras.Model.fit(epochs=2, **batch_size=128**)
You should try to use Tensor Cores when possible. If you want to learn more [NVIDIA deep learning performance guide](https://docs.nvidia.com/deeplearning/sdk/dl-performance-guide/index.html) describes the exact requirements for using Tensor Cores as well as other Tensor Core-related performance information.
### XLA
XLA is a compiler that can further increase mixed precision performance, as well as float32 performance to a lesser extent. See the [XLA guide](https://www.tensorflow.org/xla) for details.
## Cloud TPU performance tips
As on GPUs, you should try doubling your batch size, as bfloat16 tensors use half the memory. Doubling batch size may increase training throughput.
TPUs do not require any other mixed precision-specific tuning to get optimal performance. TPUs already require the use of XLA. They benefit from having certain dimensions being multiples of $128$, but this applies equally to float32 as it does for mixed precision. See the [Cloud TPU Performance Guide](https://cloud.google.com/tpu/docs/performance-guide) for general TPU performance tips, which apply to mixed precision as well as float32.
## Summary
* You should use mixed precision if you use TPUs or NVIDIA GPUs with at least compute capability 7.0, as it will improve performance by up to 3x.
* You can use mixed precision with the following lines:
```
# On TPUs, use 'mixed_bfloat16' instead
policy = tf.keras.mixed_precision.experimental.Policy('mixed_float16')
mixed_precision.set_policy(policy)
```
* If your model ends in softmax, make sure it is float32. And regardless of what your model ends in, make sure the output is float32.
* If you use a custom training loop with `mixed_float16`, in addition to the above lines, you need to wrap your optimizer with a `tf.keras.mixed_precision.experimental.LossScaleOptimizer`. Then call `optimizer.get_scaled_loss` to scale the loss, and `optimizer.get_unscaled_gradients` to unscale the gradients.
* Double the training batch size if it does not reduce evaluation accuracy
* On GPUs, ensure most tensor dimensions are a multiple of $8$ to maximize performance
For more examples of mixed precision using the `tf.keras.mixed_precision` API, see the [official models repository](https://github.com/tensorflow/models/tree/master/official). Most official models, such as [ResNet](https://github.com/tensorflow/models/tree/master/official/vision/image_classification) and [Transformer](https://github.com/tensorflow/models/blob/master/official/nlp/transformer) will run using mixed precision by passing `--dtype=fp16`.
|
github_jupyter
|
# Tabular Datasets
As we have already discovered, Elements are simple wrappers around your data that provide a semantically meaningful representation. HoloViews can work with a wide variety of data types, but many of them can be categorized as either:
* **Tabular:** Tables of flat columns, or
* **Gridded:** Array-like data on 2-dimensional or N-dimensional grids
These two general data types are explained in detail in the [Tabular Data](../user_guide/07-Tabular_Datasets.ipynb) and [Gridded Data](../user_guide/08-Gridded_Datasets.ipynb) user guides, including all the many supported formats (including Python dictionaries of NumPy arrays, pandas ``DataFrames``, dask ``DataFrames``, and xarray ``DataArrays`` and ``Datasets``).
In this Getting-Started guide we provide a quick overview and introduction to two of the most flexible and powerful formats: columnar **pandas** DataFrames (in this section), and gridded **xarray** Datasets (in the next section).
## Tabular
Tabular data (also called columnar data) is one of the most common, general, and versatile data formats, corresponding to how data is laid out in a spreadsheet. There are many different ways to put data into a tabular format, but for interactive analysis having [**tidy data**](http://www.jeannicholashould.com/tidy-data-in-python.html) provides flexibility and simplicity. For tidy data, the **columns** of the table represent **variables** or **dimensions** and the **rows** represent **observations**. The best way to understand this format is to look at such a dataset:
```
import numpy as np
import pandas as pd
import holoviews as hv
hv.extension('bokeh', 'matplotlib')
diseases = pd.read_csv('../assets/diseases.csv.gz')
diseases.head()
```
This particular dataset was the subject of an excellent piece of visual journalism in the [Wall Street Journal](http://graphics.wsj.com/infectious-diseases-and-vaccines/#b02g20t20w15). The WSJ data details the incidence of various diseases over time, and was downloaded from the [University of Pittsburgh's Project Tycho](http://www.tycho.pitt.edu/). We can see we have 5 data columns, which each correspond either to independent variables that specify a particular measurement ('Year', 'Week', 'State'), or observed/dependent variables reporting what was then actually measured (the 'measles' or 'pertussis' incidence).
Knowing the distinction between those two types of variables is crucial for doing visualizations, but unfortunately the tabular format does not declare this information. Plotting 'Week' against 'State' would not be meaningful, whereas 'measles' for each 'State' (averaging or summing across the other dimensions) would be fine, and there's no way to deduce those constraints from the tabular format. Accordingly, we will first make a HoloViews object called a ``Dataset`` that declares the independent variables (called key dimensions or **kdims** in HoloViews) and dependent variables (called value dimensions or **vdims**) that you want to work with:
```
vdims = [('measles', 'Measles Incidence'), ('pertussis', 'Pertussis Incidence')]
ds = hv.Dataset(diseases, ['Year', 'State'], vdims)
```
Here we've used an optional tuple-based syntax **``(name,label)``** to specify a more meaningful description for the ``vdims``, while using the original short descriptions for the ``kdims``. We haven't yet specified what to do with the ``Week`` dimension, but we are only interested in yearly averages, so let's just tell HoloViews to average over all remaining dimensions:
```
ds = ds.aggregate(function=np.mean)
ds
```
(We'll cover aggregations like ``np.mean`` in detail later, but here the important bit is simply that the ``Week`` dimension can now be ignored.)
The ``repr`` shows us both the ``kdims`` (in square brackets) and the ``vdims`` (in parentheses) of the ``Dataset``. Because it can hold arbitrary combinations of dimensions, a ``Dataset`` is *not* immediately visualizable. There's no single clear mapping from these four dimensions onto a two-dimensional page, hence the textual representation shown above.
To make this data visualizable, we'll need to provide a bit more metadata, by selecting one of the large library of Elements that can help answer the questions we want to ask about the data. Perhaps the most obvious representation of this dataset is as a ``Curve`` displaying the incidence for each year, for each state. We could pull out individual columns one by one from the original dataset, but now that we have declared information about the dimensions, the cleanest approach is to map the dimensions of our ``Dataset`` onto the dimensions of an Element using ``.to``:
```
%%opts Curve [width=600 height=250] {+framewise}
(ds.to(hv.Curve, 'Year', 'measles') + ds.to(hv.Curve, 'Year', 'pertussis')).cols(1)
```
Here we specified two ``Curve`` elements showing measles and pertussis incidence respectively (the vdims), per year (the kdim), and laid them out in a vertical column. You'll notice that even though we specified only the short name for the value dimensions, the plot shows the longer names ("Measles Incidence", "Pertussis Incidence") that we declared on the ``Dataset``.
You'll also notice that we automatically received a dropdown menu to select which ``State`` to view. Each ``Curve`` ignores unused value dimensions, because additional measurements don't affect each other, but HoloViews has to do *something* with every key dimension for every such plot. If the ``State`` (or any other key dimension) isn't somehow plotted or aggregated over, then HoloViews has to leave choosing a value for it to the user, hence the selection widget. Other options for what to do with extra dimensions or just extra data ranges are illustrated below.
### Selecting
One of the most common thing we might want to do is to select only a subset of the data. The ``select`` method makes this extremely easy, letting you select a single value, a list of values supplied as a list, or a range of values supplied as a tuple. Here we will use ``select`` to display the measles incidence in four states over one decade. After applying the selection, we use the ``.to`` method as shown earlier, now displaying the data as ``Bars`` indexed by 'Year' and 'State' key dimensions and displaying the 'Measles Incidence' value dimension:
```
%%opts Bars [width=800 height=400 tools=['hover'] group_index=1 legend_position='top_left']
states = ['New York', 'New Jersey', 'California', 'Texas']
ds.select(State=states, Year=(1980, 1990)).to(hv.Bars, ['Year', 'State'], 'measles').sort()
```
### Faceting
Above we already saw what happens to key dimensions that we didn't explicitly assign to the Element using the ``.to`` method: they are grouped over, popping up a set of widgets so the user can select the values to show at any one time. However, using widgets is not always the most effective way to view the data, and a ``Dataset`` lets you specify other alternatives using the ``.overlay``, ``.grid`` and ``.layout`` methods. For instance, we can lay out each state separately using ``.grid``:
```
%%opts Curve [width=200] (color='indianred')
grouped = ds.select(State=states, Year=(1930, 2005)).to(hv.Curve, 'Year', 'measles')
grouped.grid('State')
```
Or we can take the same grouped object and ``.overlay`` the individual curves instead of laying them out in a grid:
```
%%opts Curve [width=600] (color=Cycle(values=['indianred', 'slateblue', 'lightseagreen', 'coral']))
grouped.overlay('State')
```
These faceting methods even compose together, meaning that if we had more key dimensions we could ``.overlay`` one dimension, ``.grid`` another and have a widget for any other remaining key dimensions.
### Aggregating
Instead of selecting a subset of the data, another common operation supported by HoloViews is computing aggregates. When we first loaded this dataset, we aggregated over the 'Week' column to compute the mean incidence for every year, thereby reducing our data significantly. The ``aggregate`` method is therefore very useful to compute statistics from our data.
A simple example using our dataset is to compute the mean and standard deviation of the Measles Incidence by ``'Year'``. We can express this simply by passing the key dimensions to aggregate over (in this case just the 'Year') along with a function and optional ``spreadfn`` to compute the statistics we want. The ``spread_fn`` will append the name of the function to the dimension name so we can reference the computed value separately. Once we have computed the aggregate, we can simply cast it to a ``Curve`` and ``ErrorBars``:
```
%%opts Curve [width=600]
agg = ds.aggregate('Year', function=np.mean, spreadfn=np.std)
(hv.Curve(agg) * hv.ErrorBars(agg,vdims=['measles', 'measles_std'])).redim.range(measles=(0, None))
```
In this way we can summarize a multi-dimensional dataset as something that can be visualized directly, while allowing us to compute arbitrary statistics along a dimension.
## Other data
If you want to know more about working with tabular data, particularly when using datatypes other than pandas, have a look at the [user guide](../user_guide/07-Tabular_Datasets.ipynb). The different interfaces allow you to work with everything from simple NumPy arrays to out-of-core dataframes using dask. Dask dataframes scale to visualizations of billions of rows, when using [datashader](https://anaconda.org/jbednar/holoviews_datashader/notebook) with HoloViews to aggregate the data as needed.
|
github_jupyter
|
# Summarize titers and sequences by date
Create a single histogram on the same scale for number of titer measurements and number of genomic sequences per year to show the relative contribution of each data source.
```
import Bio
import Bio.SeqIO
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%matplotlib inline
# Configure matplotlib theme.
fontsize = 14
matplotlib_params = {
'axes.labelsize': fontsize,
'font.size': fontsize,
'legend.fontsize': 12,
'xtick.labelsize': fontsize,
'ytick.labelsize': fontsize,
'text.usetex': False,
'figure.figsize': [6, 4],
'savefig.dpi': 300,
'figure.dpi': 300,
'text.usetex': False
}
plt.rcParams.update(matplotlib_params)
# Turn off spines for all plots.
plt.rc("axes.spines", top=False, right=False)
matplotlib.get_configdir()
plt.style.use("huddlej")
plt.style.available
```
## Load sequences
```
ls ../../seasonal-flu/data/*.fasta
# Open FASTA of HA sequences for H3N2.
sequences = Bio.SeqIO.parse("../../seasonal-flu/data/h3n2_ha.fasta", "fasta")
# Get strain names from sequences.
distinct_strains_with_sequences = pd.Series([sequence.name.split("|")[0].replace("-egg", "")
for sequence in sequences]).drop_duplicates()
distinct_strains_with_sequences.shape
# Parse years from distinct strains with titers.
sequence_years = distinct_strains_with_sequences.apply(lambda strain: int(strain.split("/")[-1])).values
# Omit invalid sequence years.
sequence_years = sequence_years[sequence_years < 2019]
sequence_years.shape
```
## Load titers
```
# Read titers into a data frame.
titers = pd.read_table(
"../../seasonal-flu/data/cdc_h3n2_egg_hi_titers.tsv",
header=None,
index_col=False,
names=["test", "reference", "serum", "source", "titer", "assay"]
)
titers.head()
titers["test_year"] = titers["test"].apply(lambda strain: int(strain.replace("-egg", "").split("/")[-1]))
(titers["test_year"] < 2007).sum()
titers["test_year"].value_counts()
titers.shape
titers[titers["test_year"] < 2007]["test"].unique().shape
titers[titers["test_year"] < 2007]["test"].unique()
# Identify distinct viruses represented as test strains in titers.
distinct_strains_with_titers = titers["test"].str.replace("-egg", "").drop_duplicates()
# Parse years from distinct strains with titers.
titer_years = distinct_strains_with_titers.apply(lambda strain: int(strain.split("/")[-1])).values
# Omit invalid titer years.
titer_years = titer_years[titer_years < 2019]
titer_years.shape
```
## Plot sequence and titer strains by year
```
sequence_years.min()
sequence_years.max()
[sequence_years, titer_years]
sequence
fig, ax = plt.subplots(1, 1)
bins = np.arange(1968, 2019)
ax.hist([sequence_years, titer_years], bins, histtype="bar", label=["HA sequence", "HI titer"])
legend = ax.legend(
loc="upper left",
ncol=1,
frameon=False,
handlelength=1,
fancybox=False,
handleheight=1
)
legend.set_title("Virus measurement", prop={"size": 12})
legend._legend_box.align = "left"
ax.set_xlim(1990)
ax.set_xlabel("Year")
ax.set_ylabel("Number of viruses measured")
fig, ax = plt.subplots(1, 1)
bins = np.arange(1968, 2019)
ax.hist([titer_years], bins, histtype="bar", label=["HI titer"])
ax.set_xlim(1990)
ax.set_xlabel("Year")
ax.set_ylabel("Viruses measured by HI")
len(titer_years)
(titer_years < 2010).sum()
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import wikipedia
import xml.etree.ElementTree as ET
import re
from sklearn.manifold import TSNE
from sklearn.decomposition import PCA
from sklearn.model_selection import cross_val_score
import xgboost as xgb
from sklearn.metrics import r2_score
%matplotlib inline
df = pd.read_csv('2020.1 - sysarmy - Encuesta de remuneración salarial Argentina - Argentina.csv', skiprows=9)
df = df[df['Salario mensual BRUTO (en tu moneda local)'] < 1_000_000]
df = df[df['Años en la empresa actual'] < 40]
df = df[(df['Salario mensual BRUTO (en tu moneda local)'] >= 10_000) & (df['Salario mensual BRUTO (en tu moneda local)'] <= 1_000_000)]
df.head()
df['Bases de datos']
df_databases_cols = df['Bases de datos'].fillna('').apply(lambda pls: pd.Series([v.lower().strip() for v in pls.replace('Microsoft Azure (Tables, CosmosDB, SQL, etc)', 'Microsoft Azure(TablesCosmosDBSQLetc)').split(',') if v.lower().strip() not in ('', 'ninguno')], dtype=str))
count_databases = pd.concat((df_databases_cols[i] for i in range(df_databases_cols.shape[1]))).value_counts()
count_databases
count_databases = count_databases[count_databases > 10]
count_databases
count_databases = count_databases.drop(['proxysql', 'percona xtrabackup'])
def find_categories(database):
database = {
'oracle': 'Oracle Database',
'microsoft azure(tablescosmosdbsqletc)': 'Cosmos DB',
'amazon rds/aurora': 'Amazon Aurora',
'amazon dynamodb': 'Amazon DynamoDB',
'google cloud storage': 'Google Storage',
'ibm db2': 'Db2 Database',
'hana': 'SAP HANA',
'amazon redshift': 'Amazon Redshift',
'apache hive': 'Apache Hive',
'apache hbase': 'Apache HBase',
'percona server': 'Percona Server for MySQL',
'sql server': 'Microsoft SQL Server',
}.get(database, database)
# autosuggest redirects linux to line (why?)
return wikipedia.page(database, auto_suggest=False).categories
database_categories = {p: find_categories(p) for p in count_databases.index}
database_categories
catcount = {}
for categories in database_categories.values():
for cat in categories:
catcount[cat] = catcount.get(cat, 0) + 1
catcount = pd.Series(catcount)
catcount = catcount[catcount > 1]
catcount
df_databases = pd.DataFrame({plat: {cat: cat in cats for cat in catcount.index} for plat, cats in database_categories.items()}).T
df_databases.head()
_, ax = plt.subplots(1, 1, figsize=(10, 10))
df_embedded = PCA(n_components=2).fit_transform(df_databases)
ax.scatter(df_embedded[:, 0], df_embedded[:, 1])
for lang, (x, y) in zip(df_databases.index, df_embedded):
ax.annotate(lang, (x, y))
ax.set_xticks([]);
ax.set_yticks([]);
from sklearn.cluster import SpectralClustering
clustering = SpectralClustering(n_clusters=8, assign_labels="discretize", random_state=0).fit(df_embedded)
_, ax = plt.subplots(1, 1, figsize=(10, 10))
ax.scatter(df_embedded[:, 0], df_embedded[:, 1], c=clustering.labels_, cmap='Accent')
for plat, (x, y) in zip(df_databases.index, df_embedded):
ax.annotate(plat, (x, y))
ax.set_xticks([]);
ax.set_yticks([]);
best = {'colsample_bytree': 0.7000000000000001, 'gamma': 0.8500000000000001, 'learning_rate': 0.025, 'max_depth': 16, 'min_child_weight': 15.0, 'n_estimators': 175, 'subsample': 0.8099576733552297}
regions_map = {
'Ciudad Autónoma de Buenos Aires': 'AMBA',
'GBA': 'AMBA',
'Catamarca': 'NOA',
'Chaco': 'NEA',
'Chubut': 'Patagonia',
'Corrientes': 'NEA',
'Entre Ríos': 'NEA',
'Formosa': 'NEA',
'Jujuy': 'NOA',
'La Pampa': 'Pampa',
'La Rioja': 'NOA',
'Mendoza': 'Cuyo',
'Misiones': 'NEA',
'Neuquén': 'Patagonia',
'Río Negro': 'Patagonia',
'Salta': 'NOA',
'San Juan': 'Cuyo',
'San Luis': 'Cuyo',
'Santa Cruz': 'Patagonia',
'Santa Fe': 'Pampa',
'Santiago del Estero': 'NOA',
'Tucumán': 'NOA',
'Córdoba': 'Pampa',
'Provincia de Buenos Aires': 'Pampa',
'Tierra del Fuego': 'Patagonia',
}
class BaseModel:
def __init__(self, **params):
self.regressor_ = xgb.XGBRegressor(**params)
def get_params(self, deep=True):
return self.regressor_.get_params(deep=deep)
def set_params(self, **params):
return self.regressor_.set_params(**params)
def clean_words(self, field, value):
value = value.replace('Microsoft Azure (Tables, CosmosDB, SQL, etc)', 'Microsoft Azure(TablesCosmosDBSQLetc)')
value = value.replace('Snacks, golosinas, bebidas', 'snacks')
value = value.replace('Descuentos varios (Clarín 365, Club La Nación, etc)', 'descuentos varios')
value = value.replace('Sí, de forma particular', 'de forma particular')
value = value.replace('Sí, los pagó un empleador', 'los pagó un empleador')
value = value.replace('Sí, activa', 'activa')
value = value.replace('Sí, pasiva', 'pasiva')
return [self.clean_word(field, v) for v in value.split(',') if self.clean_word(field, v)]
def clean_word(self, field, word):
val = str(word).lower().strip().replace(".", "")
if val in ('ninguno', 'ninguna', 'no', '0', 'etc)', 'nan'):
return ''
if field == 'Lenguajes de programación' and val == 'Microsoft Azure(TablesCosmosDBSQLetc)':
return 'Microsoft Azure (Tables, CosmosDB, SQL, etc)'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('pycon', 'pyconar'):
return 'pyconar'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('nodeconf', 'nodeconfar'):
return 'nodeconfar'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('meetup', 'meetups'):
return 'meetups'
if field == '¿A qué eventos de tecnología asististe en el último año?':
return val.replace(' ', '')
if field == 'Beneficios extra' and val == 'snacks':
return 'snacks, golosinas, bebidas'
if field == 'Beneficios extra' and val == 'descuentos varios':
return 'descuentos varios (clarín 365, club la nación, etc)'
return val
def row_to_words(self, row):
return [
f'{key}={row.fillna("")[key]}'
for key
in (
'Me identifico',
'Nivel de estudios alcanzado',
'Universidad',
'Estado',
'Carrera',
'¿Contribuís a proyectos open source?',
'¿Programás como hobbie?',
'Trabajo de',
'¿Qué SO usás en tu laptop/PC para trabajar?',
'¿Y en tu celular?',
'Tipo de contrato',
'Orientación sexual',
'Cantidad de empleados',
'Actividad principal',
)
] + [
f'{k}={v}' for k in (
'¿Tenés guardias?',
'Realizaste cursos de especialización',
'¿A qué eventos de tecnología asististe en el último año?',
'Beneficios extra',
'Plataformas',
'Lenguajes de programación',
'Frameworks, herramientas y librerías',
'Bases de datos',
'QA / Testing',
'IDEs',
'Lenguajes de programación'
) for v in self.clean_words(k, row.fillna('')[k])
] + [
f'region={regions_map[row["Dónde estás trabajando"]]}'
]
def encode_row(self, row):
ws = self.row_to_words(row)
return pd.Series([w in ws for w in self.valid_words_] + [
row['¿Gente a cargo?'],
row['Años de experiencia'],
row['Tengo'],
])
def fit(self, X, y, **params):
counts = {}
for i in range(X.shape[0]):
for word in self.row_to_words(X.iloc[i]):
counts[word] = counts.get(word, 0) + 1
self.valid_words_ = [word for word, c in counts.items() if c > 0.01*X.shape[0]]
self.regressor_.fit(X.apply(self.encode_row, axis=1).astype(float), y, **params)
return self
def predict(self, X):
return self.regressor_.predict(X.apply(self.encode_row, axis=1).astype(float))
def score(self, X, y):
return r2_score(y, self.predict(X))
cross_val_score(BaseModel(), df, df['Salario mensual BRUTO (en tu moneda local)'])
database_embeddings = {l: [] for l in clustering.labels_}
for database, label in zip(df_databases.index, clustering.labels_):
database_embeddings[label].append(database)
database_embeddings
class ModelPCA:
def __init__(self, **params):
self.regressor_ = xgb.XGBRegressor(**params)
def get_params(self, deep=True):
return self.regressor_.get_params(deep=deep)
def set_params(self, **params):
return self.regressor_.set_params(**params)
def clean_words(self, field, value):
value = value.replace('Microsoft Azure (Tables, CosmosDB, SQL, etc)', 'Microsoft Azure(TablesCosmosDBSQLetc)')
value = value.replace('Snacks, golosinas, bebidas', 'snacks')
value = value.replace('Descuentos varios (Clarín 365, Club La Nación, etc)', 'descuentos varios')
value = value.replace('Sí, de forma particular', 'de forma particular')
value = value.replace('Sí, los pagó un empleador', 'los pagó un empleador')
value = value.replace('Sí, activa', 'activa')
value = value.replace('Sí, pasiva', 'pasiva')
return [self.clean_word(field, v) for v in value.split(',') if self.clean_word(field, v)]
def clean_word(self, field, word):
val = str(word).lower().strip().replace(".", "")
if val in ('ninguno', 'ninguna', 'no', '0', 'etc)', 'nan'):
return ''
if field == 'Lenguajes de programación' and val == 'Microsoft Azure(TablesCosmosDBSQLetc)':
return 'Microsoft Azure (Tables, CosmosDB, SQL, etc)'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('pycon', 'pyconar'):
return 'pyconar'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('nodeconf', 'nodeconfar'):
return 'nodeconfar'
if field == '¿A qué eventos de tecnología asististe en el último año?' and val in ('meetup', 'meetups'):
return 'meetups'
if field == '¿A qué eventos de tecnología asististe en el último año?':
return val.replace(' ', '')
if field == 'Beneficios extra' and val == 'snacks':
return 'snacks, golosinas, bebidas'
if field == 'Beneficios extra' and val == 'descuentos varios':
return 'descuentos varios (clarín 365, club la nación, etc)'
return val
def contains_database(self, row, databases):
k = 'Bases de datos'
for v in self.clean_words(k, row.fillna('')[k]):
if v in databases:
return True
return False
def row_to_words(self, row):
return [
f'{key}={row.fillna("")[key]}'
for key
in (
'Me identifico',
'Nivel de estudios alcanzado',
'Universidad',
'Estado',
'Carrera',
'¿Contribuís a proyectos open source?',
'¿Programás como hobbie?',
'Trabajo de',
'¿Qué SO usás en tu laptop/PC para trabajar?',
'¿Y en tu celular?',
'Tipo de contrato',
'Orientación sexual',
'Cantidad de empleados',
'Actividad principal',
)
] + [
f'{k}={v}' for k in (
'¿Tenés guardias?',
'Realizaste cursos de especialización',
'¿A qué eventos de tecnología asististe en el último año?',
'Beneficios extra',
'Plataformas',
'Frameworks, herramientas y librerías',
'Bases de datos',
'QA / Testing',
'IDEs',
'Lenguajes de programación'
) for v in self.clean_words(k, row.fillna('')[k])
] + [
f'region={regions_map[row["Dónde estás trabajando"]]}'
] + [
f'database_type={i}'
for i, databases in database_embeddings.items()
if self.contains_database(row, databases)
]
def encode_row(self, row):
ws = self.row_to_words(row)
return pd.Series([w in ws for w in self.valid_words_] + [
row['¿Gente a cargo?'],
row['Años de experiencia'],
row['Tengo'],
])
def fit(self, X, y, **params):
counts = {}
for i in range(X.shape[0]):
for word in self.row_to_words(X.iloc[i]):
counts[word] = counts.get(word, 0) + 1
self.valid_words_ = [word for word, c in counts.items() if c > 0.01*X.shape[0]]
self.regressor_.fit(X.apply(self.encode_row, axis=1).astype(float), y, **params)
return self
def predict(self, X):
return self.regressor_.predict(X.apply(self.encode_row, axis=1).astype(float))
def score(self, X, y):
return r2_score(y, self.predict(X))
cross_val_score(ModelPCA(), df, df['Salario mensual BRUTO (en tu moneda local)'])
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import yaml
from pathlib import Path
from collections import defaultdict
from pandas.api.types import CategoricalDtype
EXPERIMENTS_PATH = Path.home() / "ba" / "experiments"
benchmarks_paths = list((EXPERIMENTS_PATH / "C4P4").glob("lb.*/*.benchmarks.yaml"))
benchmarks_paths
DEFAULT_CATEGORY = lambda: "category"
CATEGORIES = defaultdict(DEFAULT_CATEGORY,
forbidden_subgraphs=CategoricalDtype([
"P3", "P4", "P5", "P6", "C4P4", "C5P5", "C6P6", ", C4_C5_2K2", "C4_C5_P5_Bowtie_Necktie"]),
lower_bound_algorithm=CategoricalDtype([
"Trivial", "Greedy", "SortedGreedy", "LocalSearch", "LPRelaxation", "NPS_MWIS_Solver",
"LSSWZ_MWIS_Solver", "fpt-editing-LocalSearch", "GreedyWeightedPacking"]),
dataset=CategoricalDtype([
"barabasi-albert", "bio", "bio-C4P4-subset", "bio-subset-A", "duplication-divergence",
"misc", "powerlaw-cluster", "bio-subset-B", "bio-unweighted"])
)
def load_raw_df(paths):
docs = []
for path in paths:
with path.open() as file:
docs += list(yaml.safe_load_all(file))
return pd.DataFrame(docs)
def load_data_unweighted_fpt_editing(paths):
df = load_raw_df(paths)
df[["dataset", "instance"]] = df["instance"].str.split("/", expand=True)[[1, 2]]
df["lower_bound_algorithm"] = "fpt-editing-LocalSearch"
return df
def load_data_weighted_fpt_editing(paths):
df = load_raw_df(paths)
df["value"] = df["values"].str[0]
df.rename(columns={"lower_bound_name": "lower_bound_algorithm"}, inplace=True)
df[["dataset", "instance"]] = df["instance"].str.split("/", expand=True)[[1, 2]]
return df
def load_data(paths):
columns = ["forbidden_subgraphs", "dataset", "instance", "lower_bound_algorithm", "value"]
df1 = load_data_weighted_fpt_editing([p for p in paths if "fpt-editing" not in p.parent.name])
df2 = load_data_unweighted_fpt_editing([p for p in paths if "fpt-editing" in p.parent.name])
df1 = df1[columns]
df2 = df2[columns]
df = pd.concat([df1, df2], ignore_index=True)
df = df.astype({k: CATEGORIES[k] for k in
["forbidden_subgraphs", "lower_bound_algorithm", "dataset"]})
df.loc[df["value"] < 0, "value"] = np.nan
m = df["lower_bound_algorithm"] == "fpt-editing-LocalSearch"
df.loc[m, "value"] = df.loc[m, "value"] / 100
return df
df = load_data(benchmarks_paths)
df.head()
for lb, df_lb in df.groupby(["lower_bound_algorithm", "dataset"]):
print(lb, len(df_lb))
# df = df[df["dataset"] == "bio"]
def plot_line_scatter(x, y, xlabel, ylabel, path=None):
fig, ax = plt.subplots(figsize=(6, 6))
ax.set_aspect("equal")
ax.scatter(x, y, alpha=0.2)
ax.plot([0, 5e5], [0, 5e5])
ax.set_yscale("log"); ax.set_xscale("log")
ax.set_ylim([1e-1, 5e5]); ax.set_xlim([1e-1, 5e5])
ax.set_ylabel(ylabel); ax.set_xlabel(xlabel)
if path is not None:
plt.savefig(path)
plt.show()
def plot_ratio_scatter(x, y, xlabel, ylabel):
ratio = x / y
ratio[x == y] = 1
fig, ax = plt.subplots(figsize=(6, 4))
ax.scatter(x, ratio, alpha=0.2)
ax.set_xscale("log")
ax.set_xlim((1e0, 5e5))
ax.set_xlabel(xlabel); ax.set_ylabel(f"{xlabel} / {ylabel}")
plt.show()
def plot_ratio(x, y, xlabel, ylabel, path=None):
ratio = x / y
ratio[x == y] = 1
print("-" * 10)
print(f"path: {path}")
print(f"{((x==0) & (y==0)).sum()} or {100*((x==0) & (y==0)).mean():.4}% where x = y = 0")
print(f"{(ratio == 1).sum()} / {ratio.shape[0]} or {100*(ratio == 1).mean():.4}% where ratio = 1")
print(f"{ratio.isnull().sum()} / {ratio.shape[0]} where ratio = NaN")
# TODO: print quantiles
q = np.array([0, 0.05, 0.1, 0.5, 0.9, 0.95, 1])
x = np.quantile(ratio[~ratio.isnull()], q)
# print(f"{x}")
for q_i, x_i in zip(q, x):
print(f"{100*q_i:>6.2f}% {ylabel} / {xlabel} > {100 / x_i:>7.2f}%")
q_line = " & ".join([f"{q_i:.2f}\\%" for q_i in q])
x_line = " & ".join([f"{100 / x_i:.2f}\\%" for x_i in x])
print(f"""\\begin{{table}}[h]
\\begin{{tabular}}{{lllllll}}
{q_line} \\\\ \\hline
{x_line}
\\end{{tabular}}
\\end{{table}}""")
fig, ax = plt.subplots(figsize=(6, 4))
ax.hist(ratio[ratio != 1], bins=np.linspace(min([0, ratio.min()]), max([0, ratio.max()]), 31))
ax.set_xlabel(f"{xlabel} / {ylabel}"); ax.set_ylabel("count")
if path is not None:
plt.savefig(path)
plt.show()
def draw_plots(df, dataset=""):
a = df[(df["lower_bound_algorithm"] == "SortedGreedy")].reset_index()
b = df[(df["lower_bound_algorithm"] == "LPRelaxation")].reset_index()
c = df[(df["lower_bound_algorithm"] == "NPS_MWIS_Solver")].reset_index()
d = df[(df["lower_bound_algorithm"] == "LocalSearch")].reset_index()
e = df[(df["lower_bound_algorithm"] == "fpt-editing-LocalSearch")].reset_index()
b.loc[b["value"] < 0, "value"] = np.nan
# plot_line_scatter(a["value"], b["value"], "SortedGreedy", "LPRelaxation")
# plot_ratio_scatter(a["value"], b["value"], "SortedGreedy", "LPRelaxation")
# plot_ratio_scatter(a["value"], c["value"], "SortedGreedy", "NPS_MWIS_Solver")
# plot_ratio(a["value"], b["value"], "SortedGreedy", "LPRelaxation",
# path=f"ratio-histogram-SortedGreedy-LPRelaxation-{dataset}.pdf")
# plot_ratio(a["value"], c["value"], "SortedGreedy", "NPS_MWIS_Solver",
# path=f"ratio-histogram-SortedGreedy-NPS_MWIS_Solver-{dataset}.pdf")
# plot_ratio(c["value"], b["value"], "NPS_MWIS_Solver", "LPRelaxation",
# path=f"ratio-histogram-NPS_MWIS_Solver-LPRelaxation-{dataset}.pdf")
plot_ratio(d["value"], b["value"], "LocalSearch", "LPRelaxation",
path=f"ratio-histogram-LocalSearch-LPRelaxation-{dataset}.pdf")
plot_ratio(a["value"], d["value"], "SortedGreedy", "LocalSearch",
path=f"ratio-histogram-SortedGreedy-LocalSearch-{dataset}.pdf")
#if len(e) > 0:
# plot_ratio(e["value"], b["value"], "fpt-editing-LocalSearch", "LPRelaxation")
# plot_ratio(d["value"], e["value"], "LocalSearch", "fpt-editing-LocalSearch")
#draw_plots(df[df["dataset"] == "bio"], dataset="bio")
#draw_plots(df[df["dataset"] == "bio-unweighted"], dataset="bio-unweighted")
X_unweighted = [(g[0], df.reset_index()["value"]) for (g, df) in df.groupby(["lower_bound_algorithm", "dataset"]) if g[1] == "bio-unweighted"]
X_weighted = [(g[0], df.reset_index()["value"]) for (g, df) in df.groupby(["lower_bound_algorithm", "dataset"]) if g[1] == "bio"]
def plot_matrix_histogram(X, ignore_zero_lb=False, ignore_equality=False, xmin=0, xmax=None, path=None):
n = len(X)
fig, axes = plt.subplots(nrows=n, ncols=n, figsize=(2*n, 2*n), sharex=True, sharey=True)
for i, (lb_i, x_i) in enumerate(X):
axes[i, 0].set_ylabel(lb_i)
axes[-1, i].set_xlabel(lb_i)
for j, (lb_j, x_j) in enumerate(X):
if i != j:
r = x_i / x_j
if not ignore_zero_lb:
r[(x_i == 0) & (x_j == 0)] == 1
if ignore_equality:
r[r == 1] = np.nan
if xmax is None:
xmax = r.max()
axes[i, j].axvline(1, c="k", ls="--", alpha=0.5)
axes[i, j].hist(r, bins=np.linspace(xmin, xmax, 25))
#axes[i, j].set_title(" ".join([
# f"{100*x:.2f}%" for x in np.quantile(r[~np.isnan(r)], [0.05, 0.5, 0.95])]), fontdict=dict(fontsize=10))
fig.tight_layout()
if path is not None:
plt.savefig(path)
plt.show()
plot_matrix_histogram(X_unweighted, xmax=2, path="lb-ratio-bio-unweighted.pdf")
plot_matrix_histogram(X_weighted, xmax=5, path="lb-ratio-bio.pdf")
plot_matrix_histogram(X_unweighted, xmax=2, ignore_equality=True, ignore_zero_lb=True, path="lb-ratio-bio-unweighted-filtered.pdf")
plot_matrix_histogram(X_weighted, xmax=5, ignore_equality=True, ignore_zero_lb=True, path="lb-ratio-bio-filtered.pdf")
def plot_matrix_scatter(X, ignore_zero_lb=False, ignore_equality=False, xmin=0, xmax=None, path=None):
n = len(X)
fig, axes = plt.subplots(nrows=n, ncols=n, figsize=(2*n, 2*n))
for ax in axes.flatten():
ax.set_aspect("equal")
for i, (lb_i, x_i) in enumerate(X):
axes[i, 0].set_ylabel(lb_i)
axes[-1, i].set_xlabel(lb_i)
for j, (lb_j, x_j) in enumerate(X):
if i != j:
m = ~np.isnan(x_i) & ~np.isnan(x_j)
l, u = min([x_i[m].min(), x_j[m].min()]), max([x_i[m].max(), x_j[m].max()])
axes[i, j].plot([l, u], [l, u], c="k", ls="--", alpha=0.5)
axes[i, j].scatter(x_i, x_j)
#axes[i, j].set_title(" ".join([
# f"{100*x:.2f}%" for x in np.quantile(r[~np.isnan(r)], [0.05, 0.5, 0.95])]), fontdict=dict(fontsize=10))
fig.tight_layout()
if path is not None:
plt.savefig(path)
plt.show()
plot_matrix_scatter(X_weighted)
plt.scatter()
X_weighted[1]
```
|
github_jupyter
|
### Road Following - Live demo (TensorRT) with collision avoidance
### Added collision avoidance ResNet18 TRT
### threshold between free and blocked is the controller - action: just a pause as long the object is in front or by time
### increase in speed_gain requires some small increase in steer_gain (once a slider is blue (mouse click), arrow keys left/right can be used)
### 10/11/2020
# TensorRT
```
import torch
device = torch.device('cuda')
```
Load the TRT optimized models by executing the cell below
```
import torch
from torch2trt import TRTModule
model_trt = TRTModule()
model_trt.load_state_dict(torch.load('best_steering_model_xy_trt.pth')) # well trained road following model
model_trt_collision = TRTModule()
model_trt_collision.load_state_dict(torch.load('best_model_trt.pth')) # anti collision model trained for one object to block and street signals (ground, strips) as free
```
### Creating the Pre-Processing Function
We have now loaded our model, but there's a slight issue. The format that we trained our model doesnt exactly match the format of the camera. To do that, we need to do some preprocessing. This involves the following steps:
1. Convert from HWC layout to CHW layout
2. Normalize using same parameters as we did during training (our camera provides values in [0, 255] range and training loaded images in [0, 1] range so we need to scale by 255.0
3. Transfer the data from CPU memory to GPU memory
4. Add a batch dimension
```
import torchvision.transforms as transforms
import torch.nn.functional as F
import cv2
import PIL.Image
import numpy as np
mean = torch.Tensor([0.485, 0.456, 0.406]).cuda().half()
std = torch.Tensor([0.229, 0.224, 0.225]).cuda().half()
def preprocess(image):
image = PIL.Image.fromarray(image)
image = transforms.functional.to_tensor(image).to(device).half()
image.sub_(mean[:, None, None]).div_(std[:, None, None])
return image[None, ...]
```
Awesome! We've now defined our pre-processing function which can convert images from the camera format to the neural network input format.
Now, let's start and display our camera. You should be pretty familiar with this by now.
```
from IPython.display import display
import ipywidgets
import traitlets
from jetbot import Camera, bgr8_to_jpeg
camera = Camera()
import IPython
image_widget = ipywidgets.Image()
traitlets.dlink((camera, 'value'), (image_widget, 'value'), transform=bgr8_to_jpeg)
```
We'll also create our robot instance which we'll need to drive the motors.
```
from jetbot import Robot
robot = Robot()
```
Now, we will define sliders to control JetBot
> Note: We have initialize the slider values for best known configurations, however these might not work for your dataset, therefore please increase or decrease the sliders according to your setup and environment
1. Speed Control (speed_gain_slider): To start your JetBot increase ``speed_gain_slider``
2. Steering Gain Control (steering_gain_sloder): If you see JetBot is woblling, you need to reduce ``steering_gain_slider`` till it is smooth
3. Steering Bias control (steering_bias_slider): If you see JetBot is biased towards extreme right or extreme left side of the track, you should control this slider till JetBot start following line or track in the center. This accounts for motor biases as well as camera offsets
> Note: You should play around above mentioned sliders with lower speed to get smooth JetBot road following behavior.
```
speed_gain_slider = ipywidgets.FloatSlider(min=0.0, max=1.0, step=0.01, description='speed gain')
steering_gain_slider = ipywidgets.FloatSlider(min=0.0, max=1.0, step=0.01, value=0.10, description='steering gain')
steering_dgain_slider = ipywidgets.FloatSlider(min=0.0, max=0.5, step=0.001, value=0.23, description='steering kd')
steering_bias_slider = ipywidgets.FloatSlider(min=-0.3, max=0.3, step=0.01, value=0.0, description='steering bias')
display(speed_gain_slider, steering_gain_slider, steering_dgain_slider, steering_bias_slider)
#anti collision ---------------------------------------------------------------------------------------------------
blocked_slider = ipywidgets.FloatSlider(description='blocked', min=0.0, max=1.0, orientation='horizontal')
stopduration_slider= ipywidgets.IntSlider(min=1, max=1000, step=1, value=10, description='Manu. time stop') #anti-collision stop time
#set value according the common threshold e.g. 0.8
block_threshold= ipywidgets.FloatSlider(min=0, max=1.2, step=0.01, value=0.8, description='Manu. bl threshold') #anti-collision block probability
display(image_widget)
d2 = IPython.display.display("", display_id=2)
display(ipywidgets.HBox([blocked_slider, block_threshold, stopduration_slider]))
# TIME STOP slider is to select manually time-for-stop when object has been discovered
#x_slider = ipywidgets.FloatSlider(min=-1.0, max=1.0, description='x')
#y_slider = ipywidgets.FloatSlider(min=0, max=1.0, orientation='vertical', description='y')
#steering_slider = ipywidgets.FloatSlider(min=-1.0, max=1.0, description='steering')
#speed_slider = ipywidgets.FloatSlider(min=0, max=1.0, orientation='vertical', description='speed')
#display(ipywidgets.HBox([y_slider, speed_slider,x_slider, steering_slider])) #sliders take time , reduce FPS a couple of frames per second
#observation sliders only
from threading import Thread
def display_class_probability(prob_blocked):
global blocked_slide
blocked_slider.value = prob_blocked
return
def model_new(image_preproc):
global model_trt_collision,angle_last
xy = model_trt(image_preproc).detach().float().cpu().numpy().flatten()
x = xy[0]
y = (0.5 - xy[1]) / 2.0
angle=math.atan2(x, y)
pid =angle * steer_gain + (angle - angle_last) * steer_dgain
steer_val = pid + steer_bias
angle_last = angle
robot.left_motor.value = max(min(speed_value + steer_val, 1.0), 0.0)
robot.right_motor.value = max(min(speed_value - steer_val, 1.0), 0.0)
return
```
Next, we'll create a function that will get called whenever the camera's value changes. This function will do the following steps
1. Pre-process the camera image
2. Execute the neural network
3. Compute the approximate steering value
4. Control the motors using proportional / derivative control (PD)
```
import time
import os
import math
angle = 0.0
angle_last = 0.0
angle_last_block=0
count_stops=0
go_on=1
stop_time=20 #number of frames to remain stopped
x=0.0
y=0.0
speed_value=speed_gain_slider.value
t1=0
road_following=1
speed_value_block=0
def execute(change):
global angle, angle_last, angle_last_block, blocked_slider, robot,count_stops, stop_time,go_on,x,y,block_threshold
global speed_value, steer_gain, steer_dgain, steer_bias,t1,model_trt, model_trt_collision,road_following,speed_value_block
steer_gain=steering_gain_slider.value
steer_dgain=steering_dgain_slider.value
steer_bias=steering_bias_slider.value
image_preproc = preprocess(change['new']).to(device)
#anti_collision model-----
prob_blocked = float(F.softmax(model_trt_collision(image_preproc), dim=1) .flatten()[0])
#blocked_slider.value = prob_blocked
#display of detection probability value for the four classes
t = Thread(target = display_class_probability, args =(prob_blocked,), daemon=False)
t.start()
stop_time=stopduration_slider.value
if go_on==1:
if prob_blocked > block_threshold.value: # threshold should be above 0.5,
#start of collision_avoidance
count_stops +=1
go_on=2
road_following=2
x=0.0 #set steering zero
y=0 #set steering zero
speed_value_block=0 # set speed zero or negative or turn
#anti_collision end-------
else:
#start of road following
go_on=1
count_stops=0
speed_value = speed_gain_slider.value #
t = Thread(target = model_new, args =(image_preproc,), daemon=True)
t.start()
road_following=1
else:
count_stops += 1
if count_stops<stop_time:
go_on=2
else:
go_on=1
count_stops=0
road_following=1
#x_slider.value = x #take time 4 FPS
#y_slider.value = y #y_speed
if road_following>1:
angle_block=math.atan2(x, y)
pid =angle_block * steer_gain + (angle - angle_last) * steer_dgain
steer_val_block = pid + steer_bias
angle_last_block = angle_block
robot.left_motor.value = max(min(speed_value_block + steer_val_block, 1.0), 0.0)
robot.right_motor.value = max(min(speed_value_block - steer_val_block, 1.0), 0.0)
t2 = time.time()
s = f"""{int(1/(t2-t1))} FPS"""
d2.update(IPython.display.HTML(s) )
t1 = time.time()
execute({'new': camera.value})
```
Cool! We've created our neural network execution function, but now we need to attach it to the camera for processing.
We accomplish that with the observe function.
>WARNING: This code will move the robot!! Please make sure your robot has clearance and it is on Lego or Track you have collected data on. The road follower should work, but the neural network is only as good as the data it's trained on!
```
camera.observe(execute, names='value')
```
Awesome! If your robot is plugged in it should now be generating new commands with each new camera frame.
You can now place JetBot on Lego or Track you have collected data on and see whether it can follow track.
If you want to stop this behavior, you can unattach this callback by executing the code below.
```
import time
camera.unobserve(execute, names='value')
time.sleep(0.1) # add a small sleep to make sure frames have finished processing
robot.stop()
camera.stop()
```
### Conclusion
That's it for this live demo! Hopefully you had some fun seeing your JetBot moving smoothly on track follwing the road!!!
If your JetBot wasn't following road very well, try to spot where it fails. The beauty is that we can collect more data for these failure scenarios and the JetBot should get even better :)
|
github_jupyter
|
```
# %load hovorka.py
import matplotlib.pyplot as plt
import numpy as np
from scipy.integrate import odeint
def model(x, t, t_offset=None):
w = 100
ka1 = 0.006 #
ka2 = 0.06 #
ka3 = 0.03 #
kb1 = 0.0034 #
kb2 = 0.056 #
kb3 = 0.024 #
u_b = 0.0555
tmaxI = 55 #
VI = 0.12 * w #
ke = 0.138 #
k12 = 0.066 #
VG = 0.16 * w #
# G = x[0] / VG
F01 = 0.0097 * w #
FR = 0
EGP0 = 0.0161 * w #
AG = 0.8 #
Gmolar = 180.1559
tmaxG = 40 #
sp = 110 * VG / 18
l = (x[14] * x[10] + x[13] * x[11] + x[12] * (-(
- F01 - x[5] * x[0] + k12 * x[1] - FR + EGP0 * (1 - x[7]) + (x[9] * AG * 1000 / Gmolar) * x[8] * np.exp(
-x[8] / tmaxG) / (tmaxG ** 2)))) + u_b - x[2] / tmaxI,
dxdt = [
- F01 - x[5] * x[0] + k12 * x[1] - FR + EGP0 * (1 - x[7]) + (x[9] * AG * 1000 / Gmolar) * x[8] * np.exp(
-x[8] / tmaxG) / (tmaxG ** 2),
x[5] * x[0] - (k12 + x[6]) * x[1],
((x[14] * x[10] + x[13] * x[11] + x[12] * (-(
- F01 - x[5] * x[0] + k12 * x[1] - FR + EGP0 * (1 - x[7]) + (x[9] * AG * 1000 / Gmolar) * x[8] * np.exp(
-x[8] / tmaxG) / (tmaxG ** 2)))) + u_b - x[2] / tmaxI) + u_b - x[2] / tmaxI,
(x[2] - x[3]) / tmaxI,
x[3] / (tmaxI * VI) - ke * x[4],
- ka1 * x[5] + kb1 * x[4],
- ka2 * x[6] + kb2 * x[4],
- ka3 * x[7] + kb3 * x[4],
1,
0,
0 - (- F01 - x[5] * x[0] + k12 * x[1] - FR + EGP0 * (1 - x[7]) + (x[9] * AG * 1000 / Gmolar) * x[8] * np.exp(
-x[8] / tmaxG) / (tmaxG ** 2)),
sp - x[0],
0,
0,
0,
(sp - x[0])**2,
(x[8] + t_offset)**2 * (sp - x[0])**2
]
return dxdt
w=100
VG = 0.16 * w
sp = 110 * VG / 18
# initial condition
Kd = [0, -0.0602, -0.0573, -0.06002, -0.0624]
Ki = [0, -3.53e-07, -3e-07, -1.17e-07, -7.55e-07]
Kp = [0, -6.17e-04, -6.39e-04, -6.76e-04, -5.42e-04]
i=1
dg1 = np.random.normal(40,10)
dg2 = np.random.normal(90,10)
dg3 = np.random.normal(60,10)
# dg1 = 40
# dg2 = 90
# dg3 = 60
x0 = [97.77, 19.08024, 3.0525, 3.0525, 0.033551, 0.01899, 0.03128, 0.02681, 0.0, dg1, 0.0, 0.0, Kd[i], Ki[i], Kp[i], 0, 0];
# time points
t_offset=0
t_sleep = 540
t_meal = 300
t = np.arange(0,t_meal,0.2)
y = odeint(model,x0,t,args=(t_offset,))
ytot = y
ttot = t
ystart = y[-1,:]
ystart[8] = 0
ystart[9] = dg2
y = odeint(model,ystart,t,args=(t_offset,))
ytot = np.vstack([ytot,y])
ttot = np.hstack([ttot,t+ttot[-1]])
ystart = y[-1,:]
ystart[8] = 0
ystart[9] = dg3
t = np.arange(0,t_meal+t_sleep,0.2)
y = odeint(model,ystart,t,args=(t_offset,))
ytot = np.vstack([ytot,y])
ttot = np.hstack([ttot,t+ttot[-1]])
# plot results
plt.fill_between([ttot[0],ttot[-1]], [4,4],[16,16],alpha=0.5)
plt.plot(ttot,ytot[:,0]/VG,'r-',linewidth=2)
plt.axhline(y=sp/VG, color='k', linestyle='-')
plt.xlabel('time')
plt.ylabel('y(t)')
plt.legend()
plt.xlabel('Time (min)')
plt.ylabel('BG (mmol/L)')
plt.show()
ttot,ytot[:,0]/VG
```
|
github_jupyter
|
**Chapter 10 – Introduction to Artificial Neural Networks**
_This notebook contains all the sample code and solutions to the exercises in chapter 10._
# Setup
First, let's make sure this notebook works well in both python 2 and 3, import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures:
```
# To support both python 2 and python 3
from __future__ import division, print_function, unicode_literals
# Common imports
import numpy as np
import os
# to make this notebook's output stable across runs
def reset_graph(seed=42):
tf.reset_default_graph()
tf.set_random_seed(seed)
np.random.seed(seed)
# To plot pretty figures
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
plt.rcParams['axes.labelsize'] = 14
plt.rcParams['xtick.labelsize'] = 12
plt.rcParams['ytick.labelsize'] = 12
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "ann"
def save_fig(fig_id, tight_layout=True):
path = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID, fig_id + ".png")
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format='png', dpi=300)
```
# Perceptrons
```
import numpy as np
from sklearn.datasets import load_iris
from sklearn.linear_model import Perceptron
iris = load_iris()
X = iris.data[:, (2, 3)] # petal length, petal width
y = (iris.target == 0).astype(np.int)
per_clf = Perceptron(max_iter=100, random_state=42)
per_clf.fit(X, y)
y_pred = per_clf.predict([[2, 0.5]])
y_pred
a = -per_clf.coef_[0][0] / per_clf.coef_[0][1]
b = -per_clf.intercept_ / per_clf.coef_[0][1]
axes = [0, 5, 0, 2]
x0, x1 = np.meshgrid(
np.linspace(axes[0], axes[1], 500).reshape(-1, 1),
np.linspace(axes[2], axes[3], 200).reshape(-1, 1),
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_predict = per_clf.predict(X_new)
zz = y_predict.reshape(x0.shape)
plt.figure(figsize=(10, 4))
plt.plot(X[y==0, 0], X[y==0, 1], "bs", label="Not Iris-Setosa")
plt.plot(X[y==1, 0], X[y==1, 1], "yo", label="Iris-Setosa")
plt.plot([axes[0], axes[1]], [a * axes[0] + b, a * axes[1] + b], "k-", linewidth=3)
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#9898ff', '#fafab0'])
plt.contourf(x0, x1, zz, cmap=custom_cmap)
plt.xlabel("Petal length", fontsize=14)
plt.ylabel("Petal width", fontsize=14)
plt.legend(loc="lower right", fontsize=14)
plt.axis(axes)
save_fig("perceptron_iris_plot")
plt.show()
```
# Activation functions
```
def logit(z):
return 1 / (1 + np.exp(-z))
def relu(z):
return np.maximum(0, z)
def derivative(f, z, eps=0.000001):
return (f(z + eps) - f(z - eps))/(2 * eps)
z = np.linspace(-5, 5, 200)
plt.figure(figsize=(11,4))
plt.subplot(121)
plt.plot(z, np.sign(z), "r-", linewidth=2, label="Step")
plt.plot(z, logit(z), "g--", linewidth=2, label="Logit")
plt.plot(z, np.tanh(z), "b-", linewidth=2, label="Tanh")
plt.plot(z, relu(z), "m-.", linewidth=2, label="ReLU")
plt.grid(True)
plt.legend(loc="center right", fontsize=14)
plt.title("Activation functions", fontsize=14)
plt.axis([-5, 5, -1.2, 1.2])
plt.subplot(122)
plt.plot(z, derivative(np.sign, z), "r-", linewidth=2, label="Step")
plt.plot(0, 0, "ro", markersize=5)
plt.plot(0, 0, "rx", markersize=10)
plt.plot(z, derivative(logit, z), "g--", linewidth=2, label="Logit")
plt.plot(z, derivative(np.tanh, z), "b-", linewidth=2, label="Tanh")
plt.plot(z, derivative(relu, z), "m-.", linewidth=2, label="ReLU")
plt.grid(True)
#plt.legend(loc="center right", fontsize=14)
plt.title("Derivatives", fontsize=14)
plt.axis([-5, 5, -0.2, 1.2])
save_fig("activation_functions_plot")
plt.show()
def heaviside(z):
return (z >= 0).astype(z.dtype)
def sigmoid(z):
return 1/(1+np.exp(-z))
def mlp_xor(x1, x2, activation=heaviside):
return activation(-activation(x1 + x2 - 1.5) + activation(x1 + x2 - 0.5) - 0.5)
x1s = np.linspace(-0.2, 1.2, 100)
x2s = np.linspace(-0.2, 1.2, 100)
x1, x2 = np.meshgrid(x1s, x2s)
z1 = mlp_xor(x1, x2, activation=heaviside)
z2 = mlp_xor(x1, x2, activation=sigmoid)
plt.figure(figsize=(10,4))
plt.subplot(121)
plt.contourf(x1, x2, z1)
plt.plot([0, 1], [0, 1], "gs", markersize=20)
plt.plot([0, 1], [1, 0], "y^", markersize=20)
plt.title("Activation function: heaviside", fontsize=14)
plt.grid(True)
plt.subplot(122)
plt.contourf(x1, x2, z2)
plt.plot([0, 1], [0, 1], "gs", markersize=20)
plt.plot([0, 1], [1, 0], "y^", markersize=20)
plt.title("Activation function: sigmoid", fontsize=14)
plt.grid(True)
```
# FNN for MNIST
## Using the Estimator API (formerly `tf.contrib.learn`)
```
import tensorflow as tf
```
**Warning**: `tf.examples.tutorials.mnist` is deprecated. We will use `tf.keras.datasets.mnist` instead. Moreover, the `tf.contrib.learn` API was promoted to `tf.estimators` and `tf.feature_columns`, and it has changed considerably. In particular, there is no `infer_real_valued_columns_from_input()` function or `SKCompat` class.
```
(X_train, y_train), (X_test, y_test) = tf.keras.datasets.mnist.load_data()
X_train = X_train.astype(np.float32).reshape(-1, 28*28) / 255.0
X_test = X_test.astype(np.float32).reshape(-1, 28*28) / 255.0
y_train = y_train.astype(np.int32)
y_test = y_test.astype(np.int32)
X_valid, X_train = X_train[:5000], X_train[5000:]
y_valid, y_train = y_train[:5000], y_train[5000:]
feature_cols = [tf.feature_column.numeric_column("X", shape=[28 * 28])]
dnn_clf = tf.estimator.DNNClassifier(hidden_units=[300,100], n_classes=10,
feature_columns=feature_cols)
input_fn = tf.estimator.inputs.numpy_input_fn(
x={"X": X_train}, y=y_train, num_epochs=40, batch_size=50, shuffle=True)
dnn_clf.train(input_fn=input_fn)
test_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"X": X_test}, y=y_test, shuffle=False)
eval_results = dnn_clf.evaluate(input_fn=test_input_fn)
eval_results
y_pred_iter = dnn_clf.predict(input_fn=test_input_fn)
y_pred = list(y_pred_iter)
y_pred[0]
```
## Using plain TensorFlow
```
import tensorflow as tf
n_inputs = 28*28 # MNIST
n_hidden1 = 300
n_hidden2 = 100
n_outputs = 10
reset_graph()
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int32, shape=(None), name="y")
def neuron_layer(X, n_neurons, name, activation=None):
with tf.name_scope(name):
n_inputs = int(X.get_shape()[1])
stddev = 2 / np.sqrt(n_inputs)
init = tf.truncated_normal((n_inputs, n_neurons), stddev=stddev)
W = tf.Variable(init, name="kernel")
b = tf.Variable(tf.zeros([n_neurons]), name="bias")
Z = tf.matmul(X, W) + b
if activation is not None:
return activation(Z)
else:
return Z
with tf.name_scope("dnn"):
hidden1 = neuron_layer(X, n_hidden1, name="hidden1",
activation=tf.nn.relu)
hidden2 = neuron_layer(hidden1, n_hidden2, name="hidden2",
activation=tf.nn.relu)
logits = neuron_layer(hidden2, n_outputs, name="outputs")
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y,
logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
learning_rate = 0.01
with tf.name_scope("train"):
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
init = tf.global_variables_initializer()
saver = tf.train.Saver()
n_epochs = 40
batch_size = 50
def shuffle_batch(X, y, batch_size):
rnd_idx = np.random.permutation(len(X))
n_batches = len(X) // batch_size
for batch_idx in np.array_split(rnd_idx, n_batches):
X_batch, y_batch = X[batch_idx], y[batch_idx]
yield X_batch, y_batch
with tf.Session() as sess:
init.run()
for epoch in range(n_epochs):
for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_batch = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})
print(epoch, "Batch accuracy:", acc_batch, "Val accuracy:", acc_val)
save_path = saver.save(sess, "./my_model_final.ckpt")
with tf.Session() as sess:
saver.restore(sess, "./my_model_final.ckpt") # or better, use save_path
X_new_scaled = X_test[:20]
Z = logits.eval(feed_dict={X: X_new_scaled})
y_pred = np.argmax(Z, axis=1)
print("Predicted classes:", y_pred)
print("Actual classes: ", y_test[:20])
from tensorflow_graph_in_jupyter import show_graph
show_graph(tf.get_default_graph())
```
## Using `dense()` instead of `neuron_layer()`
Note: previous releases of the book used `tensorflow.contrib.layers.fully_connected()` rather than `tf.layers.dense()` (which did not exist when this chapter was written). It is now preferable to use `tf.layers.dense()`, because anything in the contrib module may change or be deleted without notice. The `dense()` function is almost identical to the `fully_connected()` function, except for a few minor differences:
* several parameters are renamed: `scope` becomes `name`, `activation_fn` becomes `activation` (and similarly the `_fn` suffix is removed from other parameters such as `normalizer_fn`), `weights_initializer` becomes `kernel_initializer`, etc.
* the default `activation` is now `None` rather than `tf.nn.relu`.
* a few more differences are presented in chapter 11.
```
n_inputs = 28*28 # MNIST
n_hidden1 = 300
n_hidden2 = 100
n_outputs = 10
reset_graph()
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int32, shape=(None), name="y")
with tf.name_scope("dnn"):
hidden1 = tf.layers.dense(X, n_hidden1, name="hidden1",
activation=tf.nn.relu)
hidden2 = tf.layers.dense(hidden1, n_hidden2, name="hidden2",
activation=tf.nn.relu)
logits = tf.layers.dense(hidden2, n_outputs, name="outputs")
y_proba = tf.nn.softmax(logits)
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
learning_rate = 0.01
with tf.name_scope("train"):
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
init = tf.global_variables_initializer()
saver = tf.train.Saver()
n_epochs = 20
n_batches = 50
with tf.Session() as sess:
init.run()
for epoch in range(n_epochs):
for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
acc_batch = accuracy.eval(feed_dict={X: X_batch, y: y_batch})
acc_valid = accuracy.eval(feed_dict={X: X_valid, y: y_valid})
print(epoch, "Batch accuracy:", acc_batch, "Validation accuracy:", acc_valid)
save_path = saver.save(sess, "./my_model_final.ckpt")
show_graph(tf.get_default_graph())
```
# Exercise solutions
## 1. to 8.
See appendix A.
## 9.
_Train a deep MLP on the MNIST dataset and see if you can get over 98% precision. Just like in the last exercise of chapter 9, try adding all the bells and whistles (i.e., save checkpoints, restore the last checkpoint in case of an interruption, add summaries, plot learning curves using TensorBoard, and so on)._
First let's create the deep net. It's exactly the same as earlier, with just one addition: we add a `tf.summary.scalar()` to track the loss and the accuracy during training, so we can view nice learning curves using TensorBoard.
```
n_inputs = 28*28 # MNIST
n_hidden1 = 300
n_hidden2 = 100
n_outputs = 10
reset_graph()
X = tf.placeholder(tf.float32, shape=(None, n_inputs), name="X")
y = tf.placeholder(tf.int32, shape=(None), name="y")
with tf.name_scope("dnn"):
hidden1 = tf.layers.dense(X, n_hidden1, name="hidden1",
activation=tf.nn.relu)
hidden2 = tf.layers.dense(hidden1, n_hidden2, name="hidden2",
activation=tf.nn.relu)
logits = tf.layers.dense(hidden2, n_outputs, name="outputs")
with tf.name_scope("loss"):
xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)
loss = tf.reduce_mean(xentropy, name="loss")
loss_summary = tf.summary.scalar('log_loss', loss)
learning_rate = 0.01
with tf.name_scope("train"):
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(loss)
with tf.name_scope("eval"):
correct = tf.nn.in_top_k(logits, y, 1)
accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))
accuracy_summary = tf.summary.scalar('accuracy', accuracy)
init = tf.global_variables_initializer()
saver = tf.train.Saver()
```
Now we need to define the directory to write the TensorBoard logs to:
```
from datetime import datetime
def log_dir(prefix=""):
now = datetime.utcnow().strftime("%Y%m%d%H%M%S")
root_logdir = "tf_logs"
if prefix:
prefix += "-"
name = prefix + "run-" + now
return "{}/{}/".format(root_logdir, name)
logdir = log_dir("mnist_dnn")
```
Now we can create the `FileWriter` that we will use to write the TensorBoard logs:
```
file_writer = tf.summary.FileWriter(logdir, tf.get_default_graph())
```
Hey! Why don't we implement early stopping? For this, we are going to need to use the validation set.
```
m, n = X_train.shape
n_epochs = 10001
batch_size = 50
n_batches = int(np.ceil(m / batch_size))
checkpoint_path = "/tmp/my_deep_mnist_model.ckpt"
checkpoint_epoch_path = checkpoint_path + ".epoch"
final_model_path = "./my_deep_mnist_model"
best_loss = np.infty
epochs_without_progress = 0
max_epochs_without_progress = 50
with tf.Session() as sess:
if os.path.isfile(checkpoint_epoch_path):
# if the checkpoint file exists, restore the model and load the epoch number
with open(checkpoint_epoch_path, "rb") as f:
start_epoch = int(f.read())
print("Training was interrupted. Continuing at epoch", start_epoch)
saver.restore(sess, checkpoint_path)
else:
start_epoch = 0
sess.run(init)
for epoch in range(start_epoch, n_epochs):
for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):
sess.run(training_op, feed_dict={X: X_batch, y: y_batch})
accuracy_val, loss_val, accuracy_summary_str, loss_summary_str = sess.run([accuracy, loss, accuracy_summary, loss_summary], feed_dict={X: X_valid, y: y_valid})
file_writer.add_summary(accuracy_summary_str, epoch)
file_writer.add_summary(loss_summary_str, epoch)
if epoch % 5 == 0:
print("Epoch:", epoch,
"\tValidation accuracy: {:.3f}%".format(accuracy_val * 100),
"\tLoss: {:.5f}".format(loss_val))
saver.save(sess, checkpoint_path)
with open(checkpoint_epoch_path, "wb") as f:
f.write(b"%d" % (epoch + 1))
if loss_val < best_loss:
saver.save(sess, final_model_path)
best_loss = loss_val
else:
epochs_without_progress += 5
if epochs_without_progress > max_epochs_without_progress:
print("Early stopping")
break
os.remove(checkpoint_epoch_path)
with tf.Session() as sess:
saver.restore(sess, final_model_path)
accuracy_val = accuracy.eval(feed_dict={X: X_test, y: y_test})
accuracy_val
```
|
github_jupyter
|
<!--BOOK_INFORMATION-->
<img align="left" style="padding-right:10px;" src="figures/PDSH-cover-small.png">
*This notebook contains an excerpt from the [Python Data Science Handbook](http://shop.oreilly.com/product/0636920034919.do) by Jake VanderPlas; the content is available [on GitHub](https://github.com/jakevdp/PythonDataScienceHandbook).*
*The text is released under the [CC-BY-NC-ND license](https://creativecommons.org/licenses/by-nc-nd/3.0/us/legalcode), and code is released under the [MIT license](https://opensource.org/licenses/MIT). If you find this content useful, please consider supporting the work by [buying the book](http://shop.oreilly.com/product/0636920034919.do)!*
*No changes were made to the contents of this notebook from the original.*
<!--NAVIGATION-->
< [Density and Contour Plots](04.04-Density-and-Contour-Plots.ipynb) | [Contents](Index.ipynb) | [Customizing Plot Legends](04.06-Customizing-Legends.ipynb) >
# Histograms, Binnings, and Density
A simple histogram can be a great first step in understanding a dataset.
Earlier, we saw a preview of Matplotlib's histogram function (see [Comparisons, Masks, and Boolean Logic](02.06-Boolean-Arrays-and-Masks.ipynb)), which creates a basic histogram in one line, once the normal boiler-plate imports are done:
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
data = np.random.randn(1000)
plt.hist(data);
```
The ``hist()`` function has many options to tune both the calculation and the display;
here's an example of a more customized histogram:
```
plt.hist(data, bins=30, normed=True, alpha=0.5,
histtype='stepfilled', color='steelblue',
edgecolor='none');
```
The ``plt.hist`` docstring has more information on other customization options available.
I find this combination of ``histtype='stepfilled'`` along with some transparency ``alpha`` to be very useful when comparing histograms of several distributions:
```
x1 = np.random.normal(0, 0.8, 1000)
x2 = np.random.normal(-2, 1, 1000)
x3 = np.random.normal(3, 2, 1000)
kwargs = dict(histtype='stepfilled', alpha=0.3, normed=True, bins=40)
plt.hist(x1, **kwargs)
plt.hist(x2, **kwargs)
plt.hist(x3, **kwargs);
```
If you would like to simply compute the histogram (that is, count the number of points in a given bin) and not display it, the ``np.histogram()`` function is available:
```
counts, bin_edges = np.histogram(data, bins=5)
print(counts)
```
## Two-Dimensional Histograms and Binnings
Just as we create histograms in one dimension by dividing the number-line into bins, we can also create histograms in two-dimensions by dividing points among two-dimensional bins.
We'll take a brief look at several ways to do this here.
We'll start by defining some data—an ``x`` and ``y`` array drawn from a multivariate Gaussian distribution:
```
mean = [0, 0]
cov = [[1, 1], [1, 2]]
x, y = np.random.multivariate_normal(mean, cov, 10000).T
```
### ``plt.hist2d``: Two-dimensional histogram
One straightforward way to plot a two-dimensional histogram is to use Matplotlib's ``plt.hist2d`` function:
```
plt.hist2d(x, y, bins=30, cmap='Blues')
cb = plt.colorbar()
cb.set_label('counts in bin')
```
Just as with ``plt.hist``, ``plt.hist2d`` has a number of extra options to fine-tune the plot and the binning, which are nicely outlined in the function docstring.
Further, just as ``plt.hist`` has a counterpart in ``np.histogram``, ``plt.hist2d`` has a counterpart in ``np.histogram2d``, which can be used as follows:
```
counts, xedges, yedges = np.histogram2d(x, y, bins=30)
```
For the generalization of this histogram binning in dimensions higher than two, see the ``np.histogramdd`` function.
### ``plt.hexbin``: Hexagonal binnings
The two-dimensional histogram creates a tesselation of squares across the axes.
Another natural shape for such a tesselation is the regular hexagon.
For this purpose, Matplotlib provides the ``plt.hexbin`` routine, which will represents a two-dimensional dataset binned within a grid of hexagons:
```
plt.hexbin(x, y, gridsize=30, cmap='Blues')
cb = plt.colorbar(label='count in bin')
```
``plt.hexbin`` has a number of interesting options, including the ability to specify weights for each point, and to change the output in each bin to any NumPy aggregate (mean of weights, standard deviation of weights, etc.).
### Kernel density estimation
Another common method of evaluating densities in multiple dimensions is *kernel density estimation* (KDE).
This will be discussed more fully in [In-Depth: Kernel Density Estimation](05.13-Kernel-Density-Estimation.ipynb), but for now we'll simply mention that KDE can be thought of as a way to "smear out" the points in space and add up the result to obtain a smooth function.
One extremely quick and simple KDE implementation exists in the ``scipy.stats`` package.
Here is a quick example of using the KDE on this data:
```
from scipy.stats import gaussian_kde
# fit an array of size [Ndim, Nsamples]
data = np.vstack([x, y])
kde = gaussian_kde(data)
# evaluate on a regular grid
xgrid = np.linspace(-3.5, 3.5, 40)
ygrid = np.linspace(-6, 6, 40)
Xgrid, Ygrid = np.meshgrid(xgrid, ygrid)
Z = kde.evaluate(np.vstack([Xgrid.ravel(), Ygrid.ravel()]))
# Plot the result as an image
plt.imshow(Z.reshape(Xgrid.shape),
origin='lower', aspect='auto',
extent=[-3.5, 3.5, -6, 6],
cmap='Blues')
cb = plt.colorbar()
cb.set_label("density")
```
KDE has a smoothing length that effectively slides the knob between detail and smoothness (one example of the ubiquitous bias–variance trade-off).
The literature on choosing an appropriate smoothing length is vast: ``gaussian_kde`` uses a rule-of-thumb to attempt to find a nearly optimal smoothing length for the input data.
Other KDE implementations are available within the SciPy ecosystem, each with its own strengths and weaknesses; see, for example, ``sklearn.neighbors.KernelDensity`` and ``statsmodels.nonparametric.kernel_density.KDEMultivariate``.
For visualizations based on KDE, using Matplotlib tends to be overly verbose.
The Seaborn library, discussed in [Visualization With Seaborn](04.14-Visualization-With-Seaborn.ipynb), provides a much more terse API for creating KDE-based visualizations.
<!--NAVIGATION-->
< [Density and Contour Plots](04.04-Density-and-Contour-Plots.ipynb) | [Contents](Index.ipynb) | [Customizing Plot Legends](04.06-Customizing-Legends.ipynb) >
|
github_jupyter
|
```
import pandas as pd
import numpy as np
import os
import prody
import math
from pathlib import Path
import pickle
import sys
from sklearn.externals import joblib
from sklearn.metrics import r2_score,mean_squared_error
from abpred.Pipeline import PreparePredictions
def Kd_2_dG(Kd):
if Kd == 0:
deltaG = np.log(Kd+1)*(8.314/4184)*(298.15)
else:
deltaG = np.log(Kd)*(8.314/4184)*(298.15)
return deltaG
def deltaG_to_Kd(delg):
Kd_value = math.exp((delg)/((8.314/4184)*298.15))
return Kd_value
```
The effect of a given mutation on antibody binding was represented by apparent affinity (avidity) relative to those for wild-type (WT) gp120, calculated with the formula ([(EC50_WT/EC50_mutant)/(EC50_WT for 2G12/EC50_mutant for 2G12)] × 100)
```
# Test data
VIH_final = pd.read_csv('../data/VIH_Test15.csv',index_col=0)
# original info data
vih_data = pd.read_csv("../data/HIV_escape_mutations.csv",sep="\t")
#vih_data["pred_ddg2EC50"] = vih_data["mCSM-AB_Pred"].apply(deltaG_to_Kd)*100
vih_original = vih_data.loc[vih_data["Mutation_type"]=="ORIGINAL"].copy()
vih_reverse = vih_data.loc[vih_data["Mutation_type"]=="REVERSE"]
#sort values to appedn to prediction data table
vih_original.loc[:,"mut_code"] = (vih_reverse["Chain"]+vih_reverse["Mutation"].str[1:]).values
vih_original.sort_values(by='mut_code',inplace=True)
vih_original["Mutation_original"] = vih_original["Mutation"].str[-1]+vih_original["Mutation"].str[1:-1]+vih_original["Mutation"].str[0]
vih_original.loc[(vih_original['Exptal'] <= 33 ),"mutation-effect"] = "decreased"
vih_original.loc[(vih_original['Exptal'] > 300 ),"mutation-effect"] = "increased"
vih_original.loc[(vih_original['Exptal'] < 300 )&(vih_original['Exptal'] > 33 ),"mutation-effect"] = "neutral"
vih_reverse.loc[(vih_reverse['Exptal'] <= 33 ),"mutation-effect"] = "decreased"
vih_reverse.loc[(vih_reverse['Exptal'] > 300 ),"mutation-effect"] = "increased"
vih_reverse.loc[(vih_reverse['Exptal'] < 300 )&(vih_reverse['Exptal'] > 33 ),"mutation-effect"] = "neutral"
#
#xgbr = XGBRegressor()
#xgbr.load_model(fname='xgb_final_400F_smote_032019.sav')
#xgbr_borderline = XGBRegressor()
#xgbr_borderline.load_model(fname='xgb_final_400F_borderlinesmote_032019.sav')
# X and y data transformed to delta G
X = VIH_final.drop("Exptal",axis=1)
y_energy = (VIH_final["Exptal"]/1000).apply(Kd_2_dG)
y_binding = VIH_final["Exptal"].values
PreparePredictions(X).run()
X.ddg.sort_values().head(10)
vih_original.loc[vih_original["mutation-effect"]=="increased"]
461
197
#ridge_model = joblib.load('ridgeLinear_train15skempiAB_FINAL.pkl')
lasso_model = joblib.load('Lasso_train15skempiAB_FINAL.pkl')
elasticnet_model = joblib.load('elasticNet_train15skempiAB_FINAL.pkl')
svr_model = joblib.load('rbfSVRmodel_train15skempiAB_FINAL.pkl')
poly_model = joblib.load("poly2SVRmodel_train15skempiAB_FINAL.pkl")
#rf_model = joblib.load('RFmodel_train15skempiAB_FINAL.pkl')
gbt_model = joblib.load('GBTmodel_train15skempiAB_FINAL.overf.pkl')
#xgb_model = joblib.load('XGBmodel_train15skempiAB_FINAL.pkl')
#ridge_pred = ridge_model.predict(X)
lasso_pred = lasso_model.predict(X)
elasticnet_pred = elasticnet_model.predict(X)
svr_pred = svr_model.predict(X)
poly_pred = poly_model.predict(X)
#rf_pred = rf_model.predict(X)
gbt_pred = gbt_model.predict(X)
#xgb_pred = xgb_model.predict(X)
pred_stack = np.hstack([vih_original[["mutation-effect","mCSM-AB_Pred","Exptal"]].values,
lasso_pred.reshape((-1,1)),gbt_pred.reshape((-1,1)),svr_pred.reshape((-1,1)),poly_pred.reshape((-1,1))])
pred_data = pd.DataFrame(pred_stack,columns=["mutation-effect","mCSM-AB_Pred","Exptal","Lasso_pred","gbt_pred","svr_pred","poly_pred"])
# transform prediction score to relative to kd , refered in paper
#pred_data_binding = pred_data.applymap(deltaG_to_Kd)*100
pred_data["mean-pred"] = pred_data.loc[:,["Lasso_pred","gbt_pred","svr_pred"]].mean(axis=1)
pred_data
pred_data.loc[pred_data["mutation-effect"]=="increased"]
pred_data.loc[(pred_data["mean-pred"].abs() > 0.1)]
pred_data["True"] = y_energy.values
pred_data_binding["True"] = y_binding
#pred_data_converted.corr()
pred_data_binding.corr()
pred_data
average_pred_binding = pred_data_binding.drop("True",axis=1).loc[:,["gbt_pred","elasticnet_pred"]].mean(axis=1)
average_pred_energy = pred_data.drop("True",axis=1).loc[:,["gbt_pred","elasticnet_pred"]].mean(axis=1)
r2score = r2_score(y_energy,average_pred_energy)
rmse = mean_squared_error(y_energy,average_pred_energy)
print("R2 score:", r2score)
print("RMSE score:", np.sqrt(rmse))
np.corrcoef(y["Exptal"],average_pred)
# Corr mCSM-AB with converted mCSM AB data
np.corrcoef(y_binding,vih_reverse["pred_ddg2EC50"])
# Corr mCSM-AB with converted VIH paper data
np.corrcoef(y_energy,vih_reverse["mCSM-AB_Pred"])
# Corr FoldX feature alone
np.corrcoef(y["Exptal"],VIH_final["dg_change"].apply(deltaG_to_Kd)*100)
import seaborn as sns
#rmse_test = np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 3)
df_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": pred_data["gbt_pred"], "Actual ddG(kcal/mol)": y_energy.values})
pearsonr_test = round(df_pred.corr().iloc[0,1],3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_pred)
plt.title("Predicted vs Experimental ddG (Independent set: 123 complexes)")
plt.text(-2,3,"pearsonr = %s" %pearsonr_test)
#plt.text(4.5,-0.5,"RMSE = %s" %rmse_test)
#plt.savefig("RFmodel_300_testfit.png",dpi=600)
PredictionError?
```
|
github_jupyter
|
# Example of extracting features from dataframes with Datetime indices
Assuming that time-varying measurements are taken at regular intervals can be sufficient for many situations. However, for a large number of tasks it is important to take into account **when** a measurement is made. An example can be healthcare, where the interval between measurements of vital signs contains crucial information.
Tsfresh now supports calculator functions that use the index of the timeseries container in order to calculate the features. The only requirements for these function is that the index of the input dataframe is of type `pd.DatetimeIndex`. These functions are contained in the new class TimeBasedFCParameters.
Note that the behaviour of all other functions is unaffected. The settings parameter of `extract_features()` can contain both index-dependent functions and 'regular' functions.
```
import pandas as pd
from tsfresh.feature_extraction import extract_features
# TimeBasedFCParameters contains all functions that use the Datetime index of the timeseries container
from tsfresh.feature_extraction.settings import TimeBasedFCParameters
```
# Build a time series container with Datetime indices
Let's build a dataframe with a datetime index. The format must be with a `value` and a `kind` column, since each measurement has its own timestamp - i.e. measurements are not assumed to be simultaneous.
```
df = pd.DataFrame({"id": ["a", "a", "a", "a", "b", "b", "b", "b"],
"value": [1, 2, 3, 1, 3, 1, 0, 8],
"kind": ["temperature", "temperature", "pressure", "pressure",
"temperature", "temperature", "pressure", "pressure"]},
index=pd.DatetimeIndex(
['2019-03-01 10:04:00', '2019-03-01 10:50:00', '2019-03-02 00:00:00', '2019-03-02 09:04:59',
'2019-03-02 23:54:12', '2019-03-03 08:13:04', '2019-03-04 08:00:00', '2019-03-04 08:01:00']
))
df = df.sort_index()
df
```
Right now `TimeBasedFCParameters` only contains `linear_trend_timewise`, which performs a calculation of a linear trend, but using the time difference in hours between measurements in order to perform the linear regression. As always, you can add your own functions in `tsfresh/feature_extraction/feature_calculators.py`.
```
settings_time = TimeBasedFCParameters()
settings_time
```
We extract the features as usual, specifying the column value, kind, and id.
```
X_tsfresh = extract_features(df, column_id="id", column_value='value', column_kind='kind',
default_fc_parameters=settings_time)
X_tsfresh.head()
```
The output looks exactly, like usual. If we compare it with the 'regular' `linear_trend` feature calculator, we can see that the intercept, p and R values are the same, as we'd expect – only the slope is now different.
```
settings_regular = {'linear_trend': [
{'attr': 'pvalue'},
{'attr': 'rvalue'},
{'attr': 'intercept'},
{'attr': 'slope'},
{'attr': 'stderr'}
]}
X_tsfresh = extract_features(df, column_id="id", column_value='value', column_kind='kind',
default_fc_parameters=settings_regular)
X_tsfresh.head()
```
# Writing your own time-based feature calculators
Writing your own time-based feature calculators is no different from usual. Only two new properties must be set using the `@set_property` decorator:
1) `@set_property("input", "pd.Series")` tells the function that the input of the function is a `pd.Series` rather than a numpy array. This allows the index to be used.
2) `@set_property("index_type", pd.DatetimeIndex)` tells the function that the input is a DatetimeIndex, allowing it to perform calculations based on time datatypes.
For example, if we want to write a function that calculates the time between the first and last measurement, it could look something like this:
```python
@set_property("input", "pd.Series")
@set_property("index_type", pd.DatetimeIndex)
def timespan(x, param):
ix = x.index
# Get differences between the last timestamp and the first timestamp in seconds, then convert to hours.
times_seconds = (ix[-1] - ix[0]).total_seconds()
return times_seconds / float(3600)
```
|
github_jupyter
|
```
import yfinance as yf
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from cloudmesh.common.StopWatch import StopWatch
from tensorflow import keras
from pandas.plotting import register_matplotlib_converters
from sklearn.metrics import mean_squared_error
import pathlib
from pathlib import Path
cryptoName = input('Please enter the name of the crypto to predict.\nExamples include "EOS-USD", "DOGE-USD",\n"ETH-USD", and "BTC-USD" without double quotes')
print(cryptoName+' selected')
StopWatch.start("Overall time")
# Creating desktop path to save figures to the desktop
desktop = pathlib.Path.home() / 'Desktop'
desktop2 = str(Path(desktop))
fullpath = desktop2 + "\\"+cryptoName+"-prediction-model.png"
fullpath2 = desktop2 + "\\"+cryptoName+"-prediction-model-zoomed.png"
fullpath3 = desktop2 + "\\"+cryptoName+"-price.png"
fullpath4 = desktop2 + "\\"+cryptoName+"-training-loss.png"
pdfpath = desktop2 + "\\"+cryptoName+"-prediction-model.pdf"
pdfpath2 = desktop2 + "\\"+cryptoName+"-prediction-model-zoomed.pdf"
pdfpath3 = desktop2 + "\\"+cryptoName+"-price.pdf"
pdfpath4 = desktop2 + "\\"+cryptoName+"-training-loss.pdf"
register_matplotlib_converters()
ticker = yf.Ticker(cryptoName)
data = ticker.history(period = "max", interval = "1d")
#print(data)
# Sort the dataframe according to the date
data.sort_values('Date', inplace=True, ascending=True)
# Print the dataframe top
data.head()
# Visualization of data. Plotting the price close.
plt.figure(num=None, figsize=(7, 4), dpi=300, facecolor='w', edgecolor='k')
data['Close'].plot()
plt.tight_layout()
plt.grid()
plt.ylabel('Close Price in USD')
plt.xlabel('Date')
plt.tight_layout()
#plt.savefig(fullpath3, dpi=300, facecolor="#FFFFFF")
plt.savefig(pdfpath3, dpi=300)
plt.show()
print(data.index[0])
firstDate = data.index[0]
firstDateFormatted = pd.to_datetime(data.index[0], utc=False)
print(firstDateFormatted)
date_time_obj = firstDateFormatted.to_pydatetime()
trueFirstDate = date_time_obj.strftime('%m/%d/%Y')
print(trueFirstDate)
print(data.head())
# Get Close data
df = data[['Close']].copy()
# Split data into train and test
train, test = df.iloc[0:-200], df.iloc[-200:len(df)]
print(len(train), len(test))
train_max = train.max()
train_min = train.min()
# Normalize the dataframes
train = (train - train_min)/(train_max - train_min)
test = (test - train_min)/(train_max - train_min)
def create_dataset(X, y, time_steps=1):
Xs, ys = [], []
for i in range(len(X) - time_steps):
v = X.iloc[i:(i + time_steps)].values
Xs.append(v)
ys.append(y.iloc[i + time_steps])
return np.array(Xs), np.array(ys)
time_steps = 10
X_train, y_train = create_dataset(train, train.Close, time_steps)
X_test, y_test = create_dataset(test, test.Close, time_steps)
StopWatch.start("Training time")
model = keras.Sequential()
model.add(keras.layers.LSTM(250, input_shape=(X_train.shape[1], X_train.shape[2])))
model.add(keras.layers.Dropout(0.2))
model.add(keras.layers.Dense(1))
model.compile(loss='mae', optimizer='adam')
model.summary()
history = model.fit(
X_train, y_train,
epochs=50,
batch_size=32,
shuffle=False
)
StopWatch.stop("Training time")
# Plotting the loss
plt.plot(history.history['loss'], label='train')
plt.legend();
plt.ylabel('Model Loss')
plt.xlabel('Number of Epochs')
plt.savefig(pdfpath4, dpi=300)
plt.show()
StopWatch.start("Prediction time")
y_pred = model.predict(X_test)
StopWatch.stop("Prediction time")
# Rescale the data back to the original scale
y_test = y_test*(train_max[0] - train_min[0]) + train_min[0]
y_pred = y_pred*(train_max[0] - train_min[0]) + train_min[0]
y_train = y_train*(train_max[0] - train_min[0]) + train_min[0]
# Plotting the results
plt.plot(np.arange(len(y_train), len(y_train) + len(y_test)), y_test.flatten(), marker='.', markersize=1, label="true")
plt.plot(np.arange(len(y_train), len(y_train) + len(y_test)), y_pred.flatten(), 'r', marker='.', markersize=1, label="prediction")
plt.plot(np.arange(0, len(y_train)), y_train.flatten(), 'g', marker='.', markersize=1, label="history")
plt.ylabel('Close Price in USD')
plt.xlabel('Days Since '+trueFirstDate)
leg = plt.legend()
leg_lines = leg.get_lines()
leg_texts = leg.get_texts()
plt.setp(leg_lines, linewidth=1)
plt.setp(leg_texts, fontsize='x-large')
plt.savefig(pdfpath, dpi=300)
#doge plt.axis([1350, 1450, 0.14, 0.35])
#btc plt.axis([2490, 2650, 34000, 73000])
#eth plt.axis([1370, 1490, 2200, 5800])
plt.axis([1370, 1490, 2200, 5800])
plt.savefig(pdfpath2, dpi=300)
plt.show()
print(y_test.shape)
print(y_pred.shape)
## Outputs error in United States Dollars
mean_squared_error(y_test, y_pred)
## Create a table of the error against the number of epochs
StopWatch.stop("Overall time")
StopWatch.benchmark()
```
|
github_jupyter
|
# Modeling and Simulation in Python
Case study.
Copyright 2017 Allen Downey
License: [Creative Commons Attribution 4.0 International](https://creativecommons.org/licenses/by/4.0)
```
# Configure Jupyter so figures appear in the notebook
%matplotlib inline
# Configure Jupyter to display the assigned value after an assignment
%config InteractiveShell.ast_node_interactivity='last_expr_or_assign'
# import functions from the modsim.py module
from modsim import *
```
### Electric car
[Olin Electric Motorsports](https://www.olinelectricmotorsports.com/) is a club at Olin College that designs and builds electric cars, and participates in the [Formula SAE Electric](https://www.sae.org/attend/student-events/formula-sae-electric) competition.
The goal of this case study is to use simulation to guide the design of a car intended to accelerate from standing to 100 kph as quickly as possible. The [world record for this event](https://www.youtube.com/watch?annotation_id=annotation_2297602723&feature=iv&src_vid=I-NCH8ct24U&v=n2XiCYA3C9s), using a car that meets the competition requirements, is 1.513 seconds.
We'll start with a simple model that takes into account the characteristics of the motor and vehicle:
* The motor is an [Emrax 228 high voltage axial flux synchronous permanent magnet motor](http://emrax.com/products/emrax-228/); according to the [data sheet](http://emrax.com/wp-content/uploads/2017/01/emrax_228_technical_data_4.5.pdf), its maximum torque is 240 Nm, at 0 rpm. But maximum torque decreases with motor speed; at 5000 rpm, maximum torque is 216 Nm.
* The motor is connected to the drive axle with a chain drive with speed ratio 13:60 or 1:4.6; that is, the axle rotates once for each 4.6 rotations of the motor.
* The radius of the tires is 0.26 meters.
* The weight of the vehicle, including driver, is 300 kg.
To start, we will assume no slipping between the tires and the road surface, no air resistance, and no rolling resistance. Then we will relax these assumptions one at a time.
* First we'll add drag, assuming that the frontal area of the vehicle is 0.6 square meters, with coefficient of drag 0.6.
* Next we'll add rolling resistance, assuming a coefficient of 0.2.
* Finally we'll compute the peak acceleration to see if the "no slip" assumption is credible.
We'll use this model to estimate the potential benefit of possible design improvements, including decreasing drag and rolling resistance, or increasing the speed ratio.
I'll start by loading the units we need.
```
radian = UNITS.radian
m = UNITS.meter
s = UNITS.second
minute = UNITS.minute
hour = UNITS.hour
km = UNITS.kilometer
kg = UNITS.kilogram
N = UNITS.newton
rpm = UNITS.rpm
```
And store the parameters in a `Params` object.
```
params = Params(r_wheel=0.26 * m,
speed_ratio=13/60,
C_rr=0.2,
C_d=0.5,
area=0.6*m**2,
rho=1.2*kg/m**3,
mass=300*kg)
```
`make_system` creates the initial state, `init`, and constructs an `interp1d` object that represents torque as a function of motor speed.
```
def make_system(params):
"""Make a system object.
params: Params object
returns: System object
"""
init = State(x=0*m, v=0*m/s)
rpms = [0, 2000, 5000]
torques = [240, 240, 216]
interpolate_torque = interpolate(Series(torques, rpms))
return System(params, init=init,
interpolate_torque=interpolate_torque,
t_end=3*s)
```
Testing `make_system`
```
system = make_system(params)
system.init
```
### Torque and speed
The relationship between torque and motor speed is taken from the [Emrax 228 data sheet](http://emrax.com/wp-content/uploads/2017/01/emrax_228_technical_data_4.5.pdf). The following functions reproduce the red dotted line that represents peak torque, which can only be sustained for a few seconds before the motor overheats.
```
def compute_torque(omega, system):
"""Maximum peak torque as a function of motor speed.
omega: motor speed in radian/s
system: System object
returns: torque in Nm
"""
factor = (1 * radian / s).to(rpm)
x = magnitude(omega * factor)
return system.interpolate_torque(x) * N * m
compute_torque(0*radian/s, system)
omega = (5000 * rpm).to(radian/s)
compute_torque(omega, system)
```
Plot the whole curve.
```
xs = linspace(0, 525, 21) * radian / s
taus = [compute_torque(x, system) for x in xs]
plot(xs, taus)
decorate(xlabel='Motor speed (rpm)',
ylabel='Available torque (N m)')
```
### Simulation
Here's the slope function that computes the maximum possible acceleration of the car as a function of it current speed.
```
def slope_func(state, t, system):
"""Computes the derivatives of the state variables.
state: State object
t: time
system: System object
returns: sequence of derivatives
"""
x, v = state
r_wheel, speed_ratio = system.r_wheel, system.speed_ratio
mass = system.mass
# use velocity, v, to compute angular velocity of the wheel
omega2 = v / r_wheel
# use the speed ratio to compute motor speed
omega1 = omega2 / speed_ratio
# look up motor speed to get maximum torque at the motor
tau1 = compute_torque(omega1, system)
# compute the corresponding torque at the axle
tau2 = tau1 / speed_ratio
# compute the force of the wheel on the ground
F = tau2 / r_wheel
# compute acceleration
a = F/mass
return v, a
```
Testing `slope_func` at linear velocity 10 m/s.
```
test_state = State(x=0*m, v=10*m/s)
slope_func(test_state, 0*s, system)
```
Now we can run the simulation.
```
results, details = run_ode_solver(system, slope_func)
details
```
And look at the results.
```
results.tail()
```
After 3 seconds, the vehicle could be at 40 meters per second, in theory, which is 144 kph.
```
v_final = get_last_value(results.v)
v_final.to(km/hour)
```
Plotting `x`
```
def plot_position(results):
plot(results.x, label='x')
decorate(xlabel='Time (s)',
ylabel='Position (m)')
plot_position(results)
```
Plotting `v`
```
def plot_velocity(results):
plot(results.v, label='v')
decorate(xlabel='Time (s)',
ylabel='Velocity (m/s)')
plot_velocity(results)
```
### Stopping at 100 kph
We'll use an event function to stop the simulation when we reach 100 kph.
```
def event_func(state, t, system):
"""Stops when we get to 100 km/hour.
state: State object
t: time
system: System object
returns: difference from 100 km/hour
"""
x, v = state
# convert to km/hour
factor = (1 * m/s).to(km/hour)
v = magnitude(v * factor)
return v - 100
results, details = run_ode_solver(system, slope_func, events=event_func)
details
```
Here's what the results look like.
```
subplot(2, 1, 1)
plot_position(results)
subplot(2, 1, 2)
plot_velocity(results)
savefig('figs/chap11-fig02.pdf')
```
According to this model, we should be able to make this run in just over 2 seconds.
```
t_final = get_last_label(results) * s
```
At the end of the run, the car has gone about 28 meters.
```
state = results.last_row()
```
If we send the final state back to the slope function, we can see that the final acceleration is about 13 $m/s^2$, which is about 1.3 times the acceleration of gravity.
```
v, a = slope_func(state, 0, system)
v.to(km/hour)
a
g = 9.8 * m/s**2
(a / g).to(UNITS.dimensionless)
```
It's not easy for a vehicle to accelerate faster than `g`, because that implies a coefficient of friction between the wheels and the road surface that's greater than 1. But racing tires on dry asphalt can do that; the OEM team at Olin has tested their tires and found a peak coefficient near 1.5.
So it's possible that our no slip assumption is valid, but only under ideal conditions, where weight is distributed equally on four tires, and all tires are driving.
**Exercise:** How much time do we lose because maximum torque decreases as motor speed increases? Run the model again with no drop off in torque and see how much time it saves.
### Drag
In this section we'll see how much effect drag has on the results.
Here's a function to compute drag force, as we saw in Chapter 21.
```
def drag_force(v, system):
"""Computes drag force in the opposite direction of `v`.
v: velocity
system: System object
returns: drag force
"""
rho, C_d, area = system.rho, system.C_d, system.area
f_drag = -np.sign(v) * rho * v**2 * C_d * area / 2
return f_drag
```
We can test it with a velocity of 20 m/s.
```
drag_force(20 * m/s, system)
```
Here's the resulting acceleration of the vehicle due to drag.
```
drag_force(20 * m/s, system) / system.mass
```
We can see that the effect of drag is not huge, compared to the acceleration we computed in the previous section, but it is not negligible.
Here's a modified slope function that takes drag into account.
```
def slope_func2(state, t, system):
"""Computes the derivatives of the state variables.
state: State object
t: time
system: System object
returns: sequence of derivatives
"""
x, v = state
r_wheel, speed_ratio = system.r_wheel, system.speed_ratio
mass = system.mass
omega2 = v / r_wheel * radian
omega1 = omega2 / speed_ratio
tau1 = compute_torque(omega1, system)
tau2 = tau1 / speed_ratio
F = tau2 / r_wheel
a_motor = F / mass
a_drag = drag_force(v, system) / mass
a = a_motor + a_drag
return v, a
```
And here's the next run.
```
results2, details = run_ode_solver(system, slope_func2, events=event_func)
details
```
The time to reach 100 kph is a bit higher.
```
t_final2 = get_last_label(results2) * s
```
But the total effect of drag is only about 2/100 seconds.
```
t_final2 - t_final
```
That's not huge, which suggests we might not be able to save much time by decreasing the frontal area, or coefficient of drag, of the car.
### Rolling resistance
Next we'll consider [rolling resistance](https://en.wikipedia.org/wiki/Rolling_resistance), which the force that resists the motion of the car as it rolls on tires. The cofficient of rolling resistance, `C_rr`, is the ratio of rolling resistance to the normal force between the car and the ground (in that way it is similar to a coefficient of friction).
The following function computes rolling resistance.
```
system.set(unit_rr = 1 * N / kg)
def rolling_resistance(system):
"""Computes force due to rolling resistance.
system: System object
returns: force
"""
return -system.C_rr * system.mass * system.unit_rr
```
The acceleration due to rolling resistance is 0.2 (it is not a coincidence that it equals `C_rr`).
```
rolling_resistance(system)
rolling_resistance(system) / system.mass
```
Here's a modified slope function that includes drag and rolling resistance.
```
def slope_func3(state, t, system):
"""Computes the derivatives of the state variables.
state: State object
t: time
system: System object
returns: sequence of derivatives
"""
x, v = state
r_wheel, speed_ratio = system.r_wheel, system.speed_ratio
mass = system.mass
omega2 = v / r_wheel * radian
omega1 = omega2 / speed_ratio
tau1 = compute_torque(omega1, system)
tau2 = tau1 / speed_ratio
F = tau2 / r_wheel
a_motor = F / mass
a_drag = drag_force(v, system) / mass
a_roll = rolling_resistance(system) / mass
a = a_motor + a_drag + a_roll
return v, a
```
And here's the run.
```
results3, details = run_ode_solver(system, slope_func3, events=event_func)
details
```
The final time is a little higher, but the total cost of rolling resistance is only 3/100 seconds.
```
t_final3 = get_last_label(results3) * s
t_final3 - t_final2
```
So, again, there is probably not much to be gained by decreasing rolling resistance.
In fact, it is hard to decrease rolling resistance without also decreasing traction, so that might not help at all.
### Optimal gear ratio
The gear ratio 13:60 is intended to maximize the acceleration of the car without causing the tires to slip. In this section, we'll consider other gear ratios and estimate their effects on acceleration and time to reach 100 kph.
Here's a function that takes a speed ratio as a parameter and returns time to reach 100 kph.
```
def time_to_speed(speed_ratio, params):
"""Computes times to reach 100 kph.
speed_ratio: ratio of wheel speed to motor speed
params: Params object
returns: time to reach 100 kph, in seconds
"""
params = Params(params, speed_ratio=speed_ratio)
system = make_system(params)
system.set(unit_rr = 1 * N / kg)
results, details = run_ode_solver(system, slope_func3, events=event_func)
t_final = get_last_label(results)
a_initial = slope_func(system.init, 0, system)
return t_final
```
We can test it with the default ratio:
```
time_to_speed(13/60, params)
```
Now we can try it with different numbers of teeth on the motor gear (assuming that the axle gear has 60 teeth):
```
for teeth in linrange(8, 18):
print(teeth, time_to_speed(teeth/60, params))
```
Wow! The speed ratio has a big effect on the results. At first glance, it looks like we could break the world record (1.513 seconds) just by decreasing the number of teeth.
But before we try it, let's see what effect that has on peak acceleration.
```
def initial_acceleration(speed_ratio, params):
"""Maximum acceleration as a function of speed ratio.
speed_ratio: ratio of wheel speed to motor speed
params: Params object
returns: peak acceleration, in m/s^2
"""
params = Params(params, speed_ratio=speed_ratio)
system = make_system(params)
a_initial = slope_func(system.init, 0, system)[1] * m/s**2
return a_initial
```
Here are the results:
```
for teeth in linrange(8, 18):
print(teeth, initial_acceleration(teeth/60, params))
```
As we decrease the speed ratio, the peak acceleration increases. With 8 teeth on the motor gear, we could break the world record, but only if we can accelerate at 2.3 times the acceleration of gravity, which is impossible without very sticky ties and a vehicle that generates a lot of downforce.
```
23.07 / 9.8
```
These results suggest that the most promising way to improve the performance of the car (for this event) would be to improve traction.
|
github_jupyter
|
# Refactor: Wine Quality Analysis
In this exercise, you'll refactor code that analyzes a wine quality dataset taken from the UCI Machine Learning Repository [here](https://archive.ics.uci.edu/ml/datasets/wine+quality). Each row contains data on a wine sample, including several physicochemical properties gathered from tests, as well as a quality rating evaluated by wine experts.
The code in this notebook first renames the columns of the dataset and then calculates some statistics on how some features may be related to quality ratings. Can you refactor this code to make it more clean and modular?
```
import pandas as pd
df = pd.read_csv('winequality-red.csv', sep=';')
df.head(10)
```
### Renaming Columns
You want to replace the spaces in the column labels with underscores to be able to reference columns with dot notation. Here's one way you could've done it.
```
new_df = df.rename(columns={'fixed acidity': 'fixed_acidity',
'volatile acidity': 'volatile_acidity',
'citric acid': 'citric_acid',
'residual sugar': 'residual_sugar',
'free sulfur dioxide': 'free_sulfur_dioxide',
'total sulfur dioxide': 'total_sulfur_dioxide'
})
new_df.head()
```
And here's a slightly better way you could do it. You can avoid making naming errors due to typos caused by manual typing. However, this looks a little repetitive. Can you make it better?
```
labels = list(df.columns)
labels[0] = labels[0].replace(' ', '_')
labels[1] = labels[1].replace(' ', '_')
labels[2] = labels[2].replace(' ', '_')
labels[3] = labels[3].replace(' ', '_')
labels[5] = labels[5].replace(' ', '_')
labels[6] = labels[6].replace(' ', '_')
df.columns = labels
df.head()
```
### Analyzing Features
Now that your columns are ready, you want to see how different features of this dataset relate to the quality rating of the wine. A very simple way you could do this is by observing the mean quality rating for the top and bottom half of each feature. The code below does this for four features. It looks pretty repetitive right now. Can you make this more concise?
You might challenge yourself to figure out how to make this code more efficient! But you don't need to worry too much about efficiency right now - we will cover that more in the next section.
```
median_alcohol = df.alcohol.median()
for i, alcohol in enumerate(df.alcohol):
if alcohol >= median_alcohol:
df.loc[i, 'alcohol'] = 'high'
else:
df.loc[i, 'alcohol'] = 'low'
df.groupby('alcohol').quality.mean()
median_pH = df.pH.median()
for i, pH in enumerate(df.pH):
if pH >= median_pH:
df.loc[i, 'pH'] = 'high'
else:
df.loc[i, 'pH'] = 'low'
df.groupby('pH').quality.mean()
median_sugar = df.residual_sugar.median()
for i, sugar in enumerate(df.residual_sugar):
if sugar >= median_sugar:
df.loc[i, 'residual_sugar'] = 'high'
else:
df.loc[i, 'residual_sugar'] = 'low'
df.groupby('residual_sugar').quality.mean()
median_citric_acid = df.citric_acid.median()
for i, citric_acid in enumerate(df.citric_acid):
if citric_acid >= median_citric_acid:
df.loc[i, 'citric_acid'] = 'high'
else:
df.loc[i, 'citric_acid'] = 'low'
df.groupby('citric_acid').quality.mean()
```
|
github_jupyter
|
# Basic Python
Introduction to some basic python data types.
```
x = 1
y = 2.0
s = "hello"
l = [1, 2, 3, "a"]
d = {"a": 1, "b": 2, "c": 3}
```
Operations behave as per what you would expect.
```
z = x * y
print(z)
# Getting item at index 3 - note that Python uses zero-based indexing.
print(l[3])
# Getting the index of an element
print(l.index(2))
# Concatenating lists is just using the '+' operator.
print(l + l)
```
Dictionaries are essentially key-value pairs
```
print(d["c"]) # Getting the value associated with "c"
```
# Numpy and scipy
By convention, numpy is import as np and scipy is imported as sp.
```
import numpy as np
import scipy as sp
```
An array is essentially a tensor. It can be an arbitrary number of dimensions. For simplicity, we will stick to basic 1D vectors and 2D matrices for now.
```
x = np.array([[1, 2, 3],
[4, 7, 6],
[9, 4, 2]])
y = np.array([1.5, 0.5, 3])
print(x)
print(y)
```
By default, operations are element-wise.
```
print(x + x)
print(x * x)
print(y * y)
print(np.dot(x, x))
print(np.dot(x, y))
```
Or you can use the @ operator that is available in Python 3.7 onwards.
```
print(x @ x)
print(x @ y)
```
Numpy also comes with standard linear algebra operations, such as getting the inverse.
```
print(np.linalg.inv(x))
```
Eigen values and vectors
```
print(np.linalg.eig(x))
```
Use of numpy vectorization is key to efficient coding. Here we use the Jupyter %time magic function to demonstrate the relative speeds to two methods of calculation the L2 norm of a very long vector.
```
r = np.random.rand(10000, 1)
%time sum([i**2 for i in r])**0.5
%time np.sqrt(np.sum(r**2))
%time np.linalg.norm(r)
```
Scipy has all the linear algebra functions as numpy and more. Moreover, scipy is always compiled with fast BLAS and LAPACK.
```
import scipy.linalg as linalg
linalg.inv(x)
import scipy.constants as const
print(const.e)
print(const.h)
import scipy.stats as stats
dist = stats.norm(0, 1) # Gaussian distribution
dist.cdf(1.96)
```
# Pandas
pandas is one of the most useful packages that you will be using extensively during this course. You should become very familiar with the Series and DataFrame objects in pandas. Here, we will read in a csv (comma-separated value) file downloaded from figshare. While you can certainly manually download the csv and just called pd.read_csv(filename), we will just use the request method to directly grab the file and read it in using a StringIO stream.
```
import pandas as pd
from io import StringIO
import requests
from IPython.display import display
# Get the raw text of the data directly from the figshare url.
url = "https://ndownloader.figshare.com/files/13007075"
raw = requests.get(url).text
# Then reads in the data as a pandas DataFrame.
data = pd.read_csv(StringIO(raw))
display(data)
```
Here, we will get one column from the DataFrame - this is a Pandas Series object.
```
print(data["Enorm (eV)"])
df = data[data["Enorm (eV)"] >= 0]
df.describe()
```
Pandas dataframes come with some conveience functions for quick visualization.
```
df.plot(x="Enorm (eV)", y="E_raw (eV)", kind="scatter");
```
# Seaborn
Here we demonstrate some basic statistical data visualization using the seaborn package. A helpful resource is the [seaborn gallery](https://seaborn.pydata.org/examples/index.html) which has many useful examples with source code.
```
import seaborn as sns
%matplotlib inline
sns.distplot(df["Enorm (eV)"], norm_hist=False);
sns.scatterplot(x="Enorm (eV)", y="E_raw (eV)", data=df);
```
# Materials API using pymatgen
The MPRester.query method allows you to perform direct queries to the Materials Project to obtain data. What is returned is a list of dict of properties.
```
from pymatgen.ext.matproj import MPRester
mpr = MPRester()
data = mpr.query(criteria="*-O", properties=["pretty_formula", "final_energy", "band_gap", "elasticity.K_VRH"])
# What is returned is a list of dict. Let's just see what the first item in the list looks out.
import pprint
pprint.pprint(data[0])
```
The above is not very friendly for manipulation and visualization. Thankfully, we can easily convert this to a pandas DataFrame since the DataFrame constructor takes in lists of dicts as well.
```
df = pd.DataFrame(data)
display(df)
```
Oftentimes, you only want the subset of data with valid values. In the above data, it is clear that some of the entries do not have elasticity.K_VRH data. So we will use the dropna method of the pandas DataFrame to get a new DataFrame with just valid data. Note that a lot of Pandas methods returns a new DataFrame. This ensures that you always have the original object to compare to. If you want to perform the operation in place, you can usually supply `inplace=True` to the method.
```
valid_data = df.dropna()
print(valid_data)
```
Seaborn works very well with Pandas DataFrames...
```
sns.scatterplot(x="band_gap", y="elasticity.K_VRH", data=valid_data);
```
|
github_jupyter
|
# AMATH 515 Homework 2
**Due Date: 02/08/2019**
* Name: Tyler Chen
* Student Number:
*Homework Instruction*: Please follow order of this notebook and fill in the codes where commented as `TODO`.
```
import numpy as np
import scipy.io as sio
import matplotlib.pyplot as plt
```
## Please complete the solvers in `solver.py`
```
import sys
sys.path.append('./')
from solvers import *
```
## Problem 3: Compressive Sensing
Consier the optimization problem,
$$
\min_x~~\frac{1}{2}\|Ax - b\|^2 + \lambda\|x\|_1
$$
In the following, please specify the $f$ and $g$ and use the proximal gradient descent solver to obtain the solution.
```
# create the data
np.random.seed(123)
m = 100 # number of measurements
n = 500 # number of variables
k = 10 # number of nonzero variables
s = 0.05 # measurements noise level
#
A_cs = np.random.randn(m, n)
x_cs = np.zeros(n)
x_cs[np.random.choice(range(n), k, replace=False)] = np.random.choice([-1.0, 1.0], k)
b_cs = A_cs.dot(x_cs) + s*np.random.randn(m)
#
lam_cs = 0.1*norm(A_cs.T.dot(b_cs), np.inf)
# define the function, prox and the beta constant
def func_f_cs(x):
# TODO: complete the function
return norm(A_cs@x-b_cs)**2/2
def func_g_cs(x):
# TODO: complete the gradient
return lam_cs*norm(x,ord=1)
def grad_f_cs(x):
# TODO: complete the function
return A_cs.T@(A_cs@x-b_cs)
def prox_g_cs(x, t):
# TODO: complete the prox of 1 norm
leq = x <= -lam_cs*t # boolean array of coordinates where x_i <= -lam_cs * t
geq = x >= lam_cs*t # boolean array of coordinates where x_i >= lam_cs * t
# (leq + geq) gives components where x not in [-1,1]*lam_cs*t
return (leq+geq) * x + leq * lam_cs*t - geq * lam_cs*t
# TODO: what is the beta value for the smooth part
beta_f_cs = norm(A_cs,ord=2)**2
```
### Proximal gradient descent on compressive sensing
```
# apply the proximal gradient descent solver
x0_cs_pgd = np.zeros(x_cs.size)
x_cs_pgd, obj_his_cs_pgd, err_his_cs_pgd, exit_flag_cs_pgd = \
optimizeWithPGD(x0_cs_pgd, func_f_cs, func_g_cs, grad_f_cs, prox_g_cs, beta_f_cs)
# plot signal result
plt.plot(x_cs)
plt.plot(x_cs_pgd, '.')
plt.legend(['true signal', 'recovered'])
plt.title('Compressive Sensing Signal')
plt.show()
# plot result
fig, ax = plt.subplots(1, 2, figsize=(12,5))
ax[0].plot(obj_his_cs_pgd)
ax[0].set_title('function value')
ax[1].semilogy(err_his_cs_pgd)
ax[1].set_title('optimality condition')
fig.suptitle('Proximal Gradient Descent on Compressive Sensing')
plt.show()
# plot result
fig, ax = plt.subplots(1, 3, figsize=(18,5))
ax[0].plot(x_cs)
ax[0].plot(x_cs_pgd, '.')
ax[0].legend(['true signal', 'recovered'])
ax[0].set_title('Compressive Sensing Signal')
ax[1].plot(obj_his_cs_pgd)
ax[1].set_title('function value')
ax[2].semilogy(err_his_cs_pgd)
ax[2].set_title('optimality condition')
#fig.suptitle('Proximal Gradient Descent on Compressive Sensing')
plt.savefig('img/cs_pgd.pdf',bbox_inches="tight")
```
### Accelerate proximal gradient descent on compressive sensing
```
# apply the proximal gradient descent solver
x0_cs_apgd = np.zeros(x_cs.size)
x_cs_apgd, obj_his_cs_apgd, err_his_cs_apgd, exit_flag_cs_apgd = \
optimizeWithAPGD(x0_cs_apgd, func_f_cs, func_g_cs, grad_f_cs, prox_g_cs, beta_f_cs)
# plot signal result
plt.plot(x_cs)
plt.plot(x_cs_apgd, '.')
plt.legend(['true signal', 'recovered'])
plt.title('Compressive Sensing Signal')
plt.show()
# plot result
fig, ax = plt.subplots(1, 2, figsize=(12,5))
ax[0].plot(obj_his_cs_apgd)
ax[0].set_title('function value')
ax[1].semilogy(err_his_cs_apgd)
ax[1].set_title('optimality condition')
fig.suptitle('Accelerated Proximal Gradient Descent on Compressive Sensing')
plt.show()
# plot result
fig, ax = plt.subplots(1, 3, figsize=(18,5))
ax[0].plot(x_cs)
ax[0].plot(x_cs_apgd, '.')
ax[0].legend(['true signal', 'recovered'])
ax[0].set_title('Compressive Sensing Signal')
ax[1].plot(obj_his_cs_apgd)
ax[1].set_title('function value')
ax[2].semilogy(err_his_cs_apgd)
ax[2].set_title('optimality condition')
#fig.suptitle('Proximal Gradient Descent on Compressive Sensing')
plt.savefig('img/cs_apgd.pdf',bbox_inches="tight")
```
## Problem 4: Logistic Regression on MINST Data
Now let's play with some real data, recall the logistic regression problem,
$$
\min_x~~\sum_{i=1}^m\left\{\log(1 + \exp(\langle a_i,x \rangle)) - b_i\langle a_i,x \rangle\right\} + \frac{\lambda}{2}\|x\|^2.
$$
Here our data pair $\{a_i, b_i\}$, $a_i$ is the image and $b_i$ is the label.
In this homework problem, let's consider the binary classification problem, where $b_i \in \{0, 1\}$.
```
# import data
mnist_data = np.load('mnist01.npy')
#
A_lgt = mnist_data[0]
b_lgt = mnist_data[1]
A_lgt_test = mnist_data[2]
b_lgt_test = mnist_data[3]
#
# set regularizer parameter
lam_lgt = 0.1
#
# beta constant of the function
beta_lgt = 0.25*norm(A_lgt, 2)**2 + lam_lgt
# plot the images
fig, ax = plt.subplots(1, 2)
ax[0].imshow(A_lgt[0].reshape(28,28))
ax[1].imshow(A_lgt[7].reshape(28,28))
plt.show()
# define function, gradient and Hessian
def lgt_func(x):
# TODO: complete the function of logistic regression
return np.sum(np.log(1+np.exp(A_lgt@x))) - b_lgt@A_lgt@x + lam_lgt*x@x/2
#
def lgt_grad(x):
# TODO: complete the gradient of logistic regression
return A_lgt.T@ ((np.exp(A_lgt@x)/(1+np.exp(A_lgt@x))) - b_lgt) + lam_lgt*x
#
def lgt_hess(x):
# TODO: complete the hessian of logistic regression
return A_lgt.T @ np.diag( np.exp(A_lgt@x)/(1+np.exp(A_lgt@x))**2 ) @ A_lgt + lam_lgt * np.eye(len(x))
```
### Gradient decsent on logistic regression
```
# apply the gradient descent
x0_lgt_gd = np.zeros(A_lgt.shape[1])
x_lgt_gd, obj_his_lgt_gd, err_his_lgt_gd, exit_flag_lgt_gd = \
optimizeWithGD(x0_lgt_gd, lgt_func, lgt_grad, beta_lgt)
# plot result
fig, ax = plt.subplots(1, 2, figsize=(12,5))
ax[0].plot(obj_his_lgt_gd)
ax[0].set_title('function value')
ax[1].semilogy(err_his_lgt_gd)
ax[1].set_title('optimality condition')
fig.suptitle('Gradient Descent on Logistic Regression')
plt.savefig('img/lr_gd.pdf',bbox_inches="tight")
```
### Accelerate Gradient decsent on logistic regression
```
# apply the accelerated gradient descent
x0_lgt_agd = np.zeros(A_lgt.shape[1])
x_lgt_agd, obj_his_lgt_agd, err_his_lgt_agd, exit_flag_lgt_agd = \
optimizeWithAGD(x0_lgt_agd, lgt_func, lgt_grad, beta_lgt)
# plot result
fig, ax = plt.subplots(1, 2, figsize=(12,5))
ax[0].plot(obj_his_lgt_agd)
ax[0].set_title('function value')
ax[1].semilogy(err_his_lgt_agd)
ax[1].set_title('optimality condition')
fig.suptitle('Accelerated Gradient Descent on Logistic Regression')
plt.savefig('img/lr_agd.pdf',bbox_inches="tight")
plt.show()
```
### Accelerate Gradient decsent on logistic regression
```
# apply the accelerated gradient descent
x0_lgt_nt = np.zeros(A_lgt.shape[1])
x_lgt_nt, obj_his_lgt_nt, err_his_lgt_nt, exit_flag_lgt_nt = \
optimizeWithNT(x0_lgt_nt, lgt_func, lgt_grad, lgt_hess)
# plot result
fig, ax = plt.subplots(1, 2, figsize=(12,5))
ax[0].plot(obj_his_lgt_nt)
ax[0].set_title('function value')
ax[1].semilogy(err_his_lgt_nt)
ax[1].set_title('optimality condition')
fig.suptitle('Newton\'s Method on Logistic Regression')
plt.savefig('img/lr_nm.pdf',bbox_inches="tight")
plt.show()
```
### Test Logistic Regression
```
# define accuracy function
def accuracy(x, A_test, b_test):
r = A_test.dot(x)
b_test[b_test == 0.0] = -1.0
correct_count = np.sum((r*b_test) > 0.0)
return correct_count/b_test.size
print('accuracy of the result is %0.3f' % accuracy(x_lgt_nt, A_lgt_test, b_lgt_test))
```
|
github_jupyter
|
# Start with simplest problem
I feel like clasification is the easiest problem catogory to start with.
We will start with simple clasification problem to predict survivals of titanic https://www.kaggle.com/c/titanic
# Contents
1. [Basic pipeline for a predictive modeling problem](#1)
1. [Exploratory Data Analysis (EDA)](#2)
* [Overall survival stats](#2_1)
* [Analysis features](#2_2)
1. [Sex](#2_2_1)
1. [Pclass](#2_2_2)
1. [Age](#2_2_3)
1. [Embarked](#2_2_4)
1. [SibSip & Parch](#2_2_5)
1. [Fare](#2_2_6)
* [Observations Summary](#2_3)
* [Correlation Between The Features](#2_4)
1. [Feature Engineering and Data Cleaning](#4)
* [Converting String Values into Numeric](#4_1)
* [Convert Age into a categorical feature by binning](#4_2)
* [Convert Fare into a categorical feature by binning](#4_3)
* [Dropping Unwanted Features](#4_4)
1. [Predictive Modeling](#5)
* [Cross Validation](#5_1)
* [Confusion Matrix](#5_2)
* [Hyper-Parameters Tuning](#5_3)
* [Ensembling](#5_4)
* [Prediction](#5_5)
1. [Feature Importance](#6)
## **Basic Pipeline for predictive modeling problem**[^](#1)<a id="1" ></a><br>
**<left><span style="color:blue">Exploratory Data Analysis</span> -> <span style="color:blue">Feature Engineering and Data Preparation</span> -> <span style="color:blue">Predictive Modeling</span></left>.**
1. First we need to see what the data can tell us: We call this **<span style="color:blue">Exploratory Data Analysis(EDA)</span>**. Here we look at data which is hidden in rows and column format and try to visualize, summarize and interprete it looking for information.
1. Next we can **leverage domain knowledge** to boost machine learning model performance. We call this step, **<span style="color:blue">Feature Engineering and Data Cleaning</span>**. In this step we might add few features, Remove redundant features, Converting features into suitable form for modeling.
1. Then we can move on to the **<span style="color:blue">Predictive Modeling</span>**. Here we try basic ML algorthms, cross validate, ensemble and Important feature Extraction.
---
## Exploratory Data Analysis (EDA)[^](#2)<a id="2" ></a><br>
With the objective in mind that this kernal aims to explain the workflow of a predictive modelling problem for begginers, I will try to use simple easy to understand visualizations in the EDA section. Kernals with more advanced EDA sections will be mentioned at the end for you to learn more.
```
# Python 3 environment comes with many helpful analytics libraries installed
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import os
# Read data to a pandas data frame
data=pd.read_csv('../input/train.csv')
# lets have a look on first few rows
display(data.head())
# Checking shape of our data set
print('Shape of Data : ',data.shape)
```
* We have 891 data points (rows); each data point has 12 columns.
```
#checking for null value counts in each column
data.isnull().sum()
```
* The Age, Cabin and Embarked have null values.
### Lets look at overall survival stats[^](#2_1)<a id="2_1" ></a><br>
```
f,ax=plt.subplots(1,2,figsize=(13,5))
data['Survived'].value_counts().plot.pie(explode=[0,0.05],autopct='%1.1f%%',ax=ax[0],shadow=True)
ax[0].set_title('Survived')
ax[0].set_ylabel('')
sns.countplot('Survived',data=data,ax=ax[1])
ax[1].set_title('Survived')
plt.show()
```
* Sad Story! Only 38% have survived. That is roughly 340 out of 891.
---
### Analyse features[^](#2_2)<a id="2_2" ></a><br>
#### Feature: Sex[^](#3_2_1)<a id="2_2_1" ></a><br>
```
f,ax=plt.subplots(1,3,figsize=(18,5))
data[['Sex','Survived']].groupby(['Sex']).mean().plot.bar(ax=ax[0])
ax[0].set_title('Fraction of Survival with respect to Sex')
sns.countplot('Sex',hue='Survived',data=data,ax=ax[1])
ax[1].set_title('Survived vs Dead counts with respect to Sex')
sns.barplot(x="Sex", y="Survived", data=data,ax=ax[2])
ax[2].set_title('Survival by Gender')
plt.show()
```
* While survival rate for female is around 75%, same for men is about 20%.
* It looks like they have given priority to female passengers in the rescue.
* **Looks like Sex is a good predictor on the survival.**
---
#### Feature: Pclass[^](#2_2_2)<a id="2_2_2" ></a><br>
**Meaning :** Ticket class : 1 = 1st, 2 = 2nd, 3 = 3rd
```
f,ax=plt.subplots(1,3,figsize=(18,5))
data['Pclass'].value_counts().plot.bar(color=['#BC8F8F','#F4A460','#DAA520'],ax=ax[0])
ax[0].set_title('Number Of Passengers with respect to Pclass')
ax[0].set_ylabel('Count')
sns.countplot('Pclass',hue='Survived',data=data,ax=ax[1])
ax[1].set_title('Survived vs Dead counts with respect to Pclass')
sns.barplot(x="Pclass", y="Survived", data=data,ax=ax[2])
ax[2].set_title('Survival by Pclass')
plt.show()
```
* For Pclass 1 %survived is around 63%, for Pclass2 is around 48% and for Pclass2 is around 25%.
* **So its clear that higher classes had higher priority while rescue.**
* **Looks like Pclass is also an important feature.**
---
#### Feature: Age[^](#2_2_3)<a id="2_2_3" ></a><br>
**Meaning :** Age in years
```
# Plot
plt.figure(figsize=(25,6))
sns.barplot(data['Age'],data['Survived'], ci=None)
plt.xticks(rotation=90);
```
* Survival rate for passenegers below Age 14(i.e children) looks to be good than others.
* So Age seems an important feature too.
* Rememer we had 177 null values in the Age feature. How are we gonna fill them?.
#### Filling Age NaN
Well there are many ways to do this. One can use the mean value or median .. etc.. But can we do better?. Seems yes. [EDA To Prediction(DieTanic)](https://www.kaggle.com/ash316/eda-to-prediction-dietanic#EDA-To-Prediction-(DieTanic)) has used a wonderful method which I would use here too. There is a name feature. First lets extract the initials.
```
data['Initial']=0
for i in data:
data['Initial']=data.Name.str.extract('([A-Za-z]+)\.') #lets extract the Salutations
pd.crosstab(data.Initial,data.Sex).T.style.background_gradient(cmap='summer_r') #Checking the Initials with the Sex
```
Okay so there are some misspelled Initials like Mlle or Mme that stand for Miss. Lets replace them.
```
data['Initial'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess','Jonkheer','Col','Rev','Capt','Sir','Don'],['Miss','Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr'],inplace=True)
data.groupby('Initial')['Age'].mean() #lets check the average age by Initials
## Assigning the NaN Values with the Ceil values of the mean ages
data.loc[(data.Age.isnull())&(data.Initial=='Mr'),'Age']=33
data.loc[(data.Age.isnull())&(data.Initial=='Mrs'),'Age']=36
data.loc[(data.Age.isnull())&(data.Initial=='Master'),'Age']=5
data.loc[(data.Age.isnull())&(data.Initial=='Miss'),'Age']=22
data.loc[(data.Age.isnull())&(data.Initial=='Other'),'Age']=46
data.Age.isnull().any() #So no null values left finally
```
---
#### Feature: Embarked[^](#2_2_4)<a id="2_2_4" ></a><br>
**Meaning :** Port of Embarkation. C = Cherbourg, Q = Queenstown, S = Southampton
```
f,ax=plt.subplots(1,2,figsize=(12,5))
sns.countplot('Embarked',data=data,ax=ax[0])
ax[0].set_title('No. Of Passengers Boarded')
sns.countplot('Embarked',hue='Survived',data=data,ax=ax[1])
ax[1].set_title('Embarked vs Survived')
plt.subplots_adjust(wspace=0.2,hspace=0.5)
plt.show()
```
* Majority of passengers borded from Southampton
* Survival counts looks better at C. Why?. Could there be an influence from sex and pclass features we already studied?. Let's find out
```
f,ax=plt.subplots(1,2,figsize=(12,5))
sns.countplot('Embarked',hue='Sex',data=data,ax=ax[0])
ax[0].set_title('Male-Female Split for Embarked')
sns.countplot('Embarked',hue='Pclass',data=data,ax=ax[1])
ax[1].set_title('Embarked vs Pclass')
plt.subplots_adjust(wspace=0.2,hspace=0.5)
plt.show()
```
* We guessed correctly. higher % of 1st class passegers boarding from C might be the reason.
#### Filling Embarked NaN
```
f,ax=plt.subplots(1,1,figsize=(6,5))
data['Embarked'].value_counts().plot.pie(explode=[0,0,0],autopct='%1.1f%%',ax=ax)
plt.show()
```
* Since 72.5% passengers are from Southampton, So lets fill missing 2 values using S (Southampton)
```
data['Embarked'].fillna('S',inplace=True)
data.Embarked.isnull().any()
```
---
#### Features: SibSip & Parch[^](#2_2_5)<a id="2_2_5" ></a><br>
**Meaning :**
SibSip -> Number of siblings / spouses aboard the Titanic
Parch -> Number of parents / children aboard the Titanic
SibSip + Parch -> Family Size
```
f,ax=plt.subplots(2,2,figsize=(15,10))
sns.countplot('SibSp',hue='Survived',data=data,ax=ax[0,0])
ax[0,0].set_title('SibSp vs Survived')
sns.barplot('SibSp','Survived',data=data,ax=ax[0,1])
ax[0,1].set_title('SibSp vs Survived')
sns.countplot('Parch',hue='Survived',data=data,ax=ax[1,0])
ax[1,0].set_title('Parch vs Survived')
sns.barplot('Parch','Survived',data=data,ax=ax[1,1])
ax[1,1].set_title('Parch vs Survived')
plt.subplots_adjust(wspace=0.2,hspace=0.5)
plt.show()
```
* The barplot and factorplot shows that if a passenger is alone onboard with no siblings, he have 34.5% survival rate. The graph roughly decreases if the number of siblings increase.
Lets combine above and analyse family size.
```
data['FamilySize'] = data['Parch'] + data['SibSp']
f,ax=plt.subplots(1,2,figsize=(15,4.5))
sns.countplot('FamilySize',hue='Survived',data=data,ax=ax[0])
ax[0].set_title('FamilySize vs Survived')
sns.barplot('FamilySize','Survived',data=data,ax=ax[1])
ax[1].set_title('FamilySize vs Survived')
plt.subplots_adjust(wspace=0.2,hspace=0.5)
plt.show()
```
* This looks interesting! looks like family sizes of 1-3 have better survival rates than others.
---
#### Fare[^](#2_2_6)<a id="2_2_6" ></a><br>
**Meaning :** Passenger fare
```
f,ax=plt.subplots(1,1,figsize=(20,5))
sns.distplot(data.Fare,ax=ax)
ax.set_title('Distribution of Fares')
plt.show()
print('Highest Fare:',data['Fare'].max(),' Lowest Fare:',data['Fare'].min(),' Average Fare:',data['Fare'].mean())
data['Fare_Bin']=pd.qcut(data['Fare'],6)
data.groupby(['Fare_Bin'])['Survived'].mean().to_frame().style.background_gradient(cmap='summer_r')
```
* It is clear that as Fare Bins increase chances of survival increase too.
#### Observations Summary[^](#2_3)<a id="2_3" ></a><br>
**Sex:** Survival chance for female is better than that for male.
**Pclass:** Being a 1st class passenger gives you better chances of survival.
**Age:** Age range 5-10 years have a high chance of survival.
**Embarked:** Majority of passengers borded from Southampton.The chances of survival at C looks to be better than even though the majority of Pclass1 passengers got up at S. All most all Passengers at Q were from Pclass3.
**Family Size:** looks like family sizes of 1-3 have better survival rates than others.
**Fare:** As Fare Bins increase chances of survival increases
#### Correlation Between The Features[^](#2_4)<a id="2_4" ></a><br>
```
sns.heatmap(data.corr(),annot=True,cmap='RdYlGn',linewidths=0.2) #data.corr()-->correlation matrix
fig=plt.gcf()
fig.set_size_inches(10,8)
plt.show()
```
---
## Feature Engineering and Data Cleaning[^](#4)<a id="4" ></a><br>
Now what is Feature Engineering? Feature engineering is the process of using domain knowledge of the data to create features that make machine learning algorithms work.
In this section we will be doing,
1. Converting String Values into Numeric
1. Convert Age into a categorical feature by binning
1. Convert Fare into a categorical feature by binning
1. Dropping Unwanted Features
#### Converting String Values into Numeric[^](#4_1)<a id="4_1" ></a><br>
Since we cannot pass strings to a machine learning model, we need to convert features Sex, Embarked, etc into numeric values.
```
data['Sex'].replace(['male','female'],[0,1],inplace=True)
data['Embarked'].replace(['S','C','Q'],[0,1,2],inplace=True)
data['Initial'].replace(['Mr','Mrs','Miss','Master','Other'],[0,1,2,3,4],inplace=True)
```
#### Convert Age into a categorical feature by binning[^](#4_2)<a id="4_2" ></a><br>
```
print('Highest Age:',data['Age'].max(),' Lowest Age:',data['Age'].min())
data['Age_cat']=0
data.loc[data['Age']<=16,'Age_cat']=0
data.loc[(data['Age']>16)&(data['Age']<=32),'Age_cat']=1
data.loc[(data['Age']>32)&(data['Age']<=48),'Age_cat']=2
data.loc[(data['Age']>48)&(data['Age']<=64),'Age_cat']=3
data.loc[data['Age']>64,'Age_cat']=4
```
#### Convert Fare into a categorical feature by binning[^](#4_3)<a id="4_3" ></a><br>
```
data['Fare_cat']=0
data.loc[data['Fare']<=7.775,'Fare_cat']=0
data.loc[(data['Fare']>7.775)&(data['Fare']<=8.662),'Fare_cat']=1
data.loc[(data['Fare']>8.662)&(data['Fare']<=14.454),'Fare_cat']=2
data.loc[(data['Fare']>14.454)&(data['Fare']<=26.0),'Fare_cat']=3
data.loc[(data['Fare']>26.0)&(data['Fare']<=52.369),'Fare_cat']=4
data.loc[data['Fare']>52.369,'Fare_cat']=5
```
#### Dropping Unwanted Features[^](#4_4)<a id="4_4" ></a><br>
Name--> We don't need name feature as it cannot be converted into any categorical value.
Age--> We have the Age_cat feature, so no need of this.
Ticket--> It is any random string that cannot be categorised.
Fare--> We have the Fare_cat feature, so unneeded
Cabin--> A lot of NaN values and also many passengers have multiple cabins. So this is a useless feature.
Fare_Bin--> We have the fare_cat feature.
PassengerId--> Cannot be categorised.
Sibsp & Parch --> We got FamilySize feature
```
#data.drop(['Name','Age','Ticket','Fare','Cabin','Fare_Range','PassengerId'],axis=1,inplace=True)
data.drop(['Name','Age','Fare','Ticket','Cabin','Fare_Bin','SibSp','Parch','PassengerId'],axis=1,inplace=True)
data.head(2)
sns.heatmap(data.corr(),annot=True,cmap='RdYlGn',linewidths=0.2) #data.corr()-->correlation matrix
fig=plt.gcf()
fig.set_size_inches(10,8)
plt.show()
```
---
## Predictive Modeling[^](#5)<a id="5" ></a><br>
Now after data cleaning and feature engineering we are ready to train some classification algorithms that will make predictions for unseen data. We will first train few classification algorithms and see how they perform. Then we can look how an ensemble of classification algorithms perform on this data set.
Following Machine Learning algorithms will be used in this kernal.
* Logistic Regression Classifier
* Naive Bayes Classifier
* Decision Tree Classifier
* Random Forest Classifier
```
#importing all the required ML packages
from sklearn.linear_model import LogisticRegression #logistic regression
from sklearn.ensemble import RandomForestClassifier #Random Forest
from sklearn.naive_bayes import GaussianNB #Naive bayes
from sklearn.tree import DecisionTreeClassifier #Decision Tree
from sklearn.model_selection import train_test_split #training and testing data split
from sklearn import metrics #accuracy measure
from sklearn.metrics import confusion_matrix #for confusion matrix
#Lets prepare data sets for training.
train,test=train_test_split(data,test_size=0.3,random_state=0,stratify=data['Survived'])
train_X=train[train.columns[1:]]
train_Y=train[train.columns[:1]]
test_X=test[test.columns[1:]]
test_Y=test[test.columns[:1]]
X=data[data.columns[1:]]
Y=data['Survived']
data.head(2)
# Logistic Regression
model = LogisticRegression(C=0.05,solver='liblinear')
model.fit(train_X,train_Y.values.ravel())
LR_prediction=model.predict(test_X)
print('The accuracy of the Logistic Regression model is \t',metrics.accuracy_score(LR_prediction,test_Y))
# Naive Bayes
model=GaussianNB()
model.fit(train_X,train_Y.values.ravel())
NB_prediction=model.predict(test_X)
print('The accuracy of the NaiveBayes model is\t\t\t',metrics.accuracy_score(NB_prediction,test_Y))
# Decision Tree
model=DecisionTreeClassifier()
model.fit(train_X,train_Y)
DT_prediction=model.predict(test_X)
print('The accuracy of the Decision Tree is \t\t\t',metrics.accuracy_score(DT_prediction,test_Y))
# Random Forest
model=RandomForestClassifier(n_estimators=100)
model.fit(train_X,train_Y.values.ravel())
RF_prediction=model.predict(test_X)
print('The accuracy of the Random Forests model is \t\t',metrics.accuracy_score(RF_prediction,test_Y))
```
### Cross Validation[^](#5_1)<a id="5_1" ></a><br>
Accuracy we get here higlhy depends on the train & test data split of the original data set. We can use cross validation to avoid such problems arising from dataset splitting.
I am using K-fold cross validation here. Watch this short [vedio](https://www.youtube.com/watch?v=TIgfjmp-4BA) to understand what it is.
```
from sklearn.model_selection import KFold #for K-fold cross validation
from sklearn.model_selection import cross_val_score #score evaluation
from sklearn.model_selection import cross_val_predict #prediction
kfold = KFold(n_splits=10, random_state=22) # k=10, split the data into 10 equal parts
xyz=[]
accuracy=[]
std=[]
classifiers=['Logistic Regression','Decision Tree','Naive Bayes','Random Forest']
models=[LogisticRegression(solver='liblinear'),DecisionTreeClassifier(),GaussianNB(),RandomForestClassifier(n_estimators=100)]
for i in models:
model = i
cv_result = cross_val_score(model,X,Y, cv = kfold,scoring = "accuracy")
xyz.append(cv_result.mean())
std.append(cv_result.std())
accuracy.append(cv_result)
new_models_dataframe2=pd.DataFrame({'CV Mean':xyz,'Std':std},index=classifiers)
new_models_dataframe2
```
Now we have looked at cross validation accuracies to get an idea how those models work. There is more we can do to understand the performances of the models we tried ; let's have a look at confusion matrix for each model.
### Confusion Matrix[^](#5_2)<a id="5_2" ></a><br>
A confusion matrix is a table that is often used to describe the performance of a classification model. read more [here](https://www.dataschool.io/simple-guide-to-confusion-matrix-terminology/)
```
f,ax=plt.subplots(2,2,figsize=(10,8))
y_pred = cross_val_predict(LogisticRegression(C=0.05,solver='liblinear'),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,0],annot=True,fmt='2.0f')
ax[0,0].set_title('Matrix for Logistic Regression')
y_pred = cross_val_predict(DecisionTreeClassifier(),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[0,1],annot=True,fmt='2.0f')
ax[0,1].set_title('Matrix for Decision Tree')
y_pred = cross_val_predict(GaussianNB(),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,0],annot=True,fmt='2.0f')
ax[1,0].set_title('Matrix for Naive Bayes')
y_pred = cross_val_predict(RandomForestClassifier(n_estimators=100),X,Y,cv=10)
sns.heatmap(confusion_matrix(Y,y_pred),ax=ax[1,1],annot=True,fmt='2.0f')
ax[1,1].set_title('Matrix for Random-Forests')
plt.subplots_adjust(hspace=0.2,wspace=0.2)
plt.show()
```
* By looking at above matrices we can say that, if we are more concerned on making less mistakes by predicting survived as dead, then Naive Bayes model does better.
* If we are more concerned on making less mistakes by predicting dead as survived, then Decision Tree model does better.
### Hyper-Parameters Tuning[^](#5_3)<a id="5_3" ></a><br>
You might have noticed there are few parameters for each model which defines how the model learns. We call these hyperparameters. These hyperparameters can be tuned to improve performance. Let's try this for Random Forest classifier.
```
from sklearn.model_selection import GridSearchCV
n_estimators=range(100,1000,100)
hyper={'n_estimators':n_estimators}
gd=GridSearchCV(estimator=RandomForestClassifier(random_state=0),param_grid=hyper,verbose=True,cv=10)
gd.fit(X,Y)
print(gd.best_score_)
print(gd.best_estimator_)
```
* Best Score for Random Forest is with n_estimators=100
### Ensembling[^](#5_4)<a id="5_4" ></a><br>
Ensembling is a way to increase performance of a model by combining several simple models to create a single powerful model.
read more about ensembling [here](https://www.analyticsvidhya.com/blog/2018/06/comprehensive-guide-for-ensemble-models/).
Ensembling can be done in ways like: Voting Classifier, Bagging, Boosting.
I will use voting method in this kernal
```
from sklearn.ensemble import VotingClassifier
estimators=[('RFor',RandomForestClassifier(n_estimators=100,random_state=0)),
('LR',LogisticRegression(C=0.05,solver='liblinear')),
('DT',DecisionTreeClassifier()),
('NB',GaussianNB())]
ensemble=VotingClassifier(estimators=estimators,voting='soft')
ensemble.fit(train_X,train_Y.values.ravel())
print('The accuracy for ensembled model is:',ensemble.score(test_X,test_Y))
cross=cross_val_score(ensemble,X,Y, cv = 10,scoring = "accuracy")
print('The cross validated score is',cross.mean())
```
### Prediction[^](#5_5)<a id="5_5" ></a><br>
We can see that ensemble model does better than individual models. lets use that for predictions.
```
Ensemble_Model_For_Prediction=VotingClassifier(estimators=[
('RFor',RandomForestClassifier(n_estimators=200,random_state=0)),
('LR',LogisticRegression(C=0.05,solver='liblinear')),
('DT',DecisionTreeClassifier(random_state=0)),
('NB',GaussianNB())
],
voting='soft')
Ensemble_Model_For_Prediction.fit(X,Y)
```
We need to do some preprocessing to this test data set before we can feed that to the trained model.
```
test=pd.read_csv('../input/test.csv')
IDtest = test["PassengerId"]
test.head(2)
test.isnull().sum()
# Prepare Test Data set for feeding
# Construct feature Initial
test['Initial']=0
for i in test:
test['Initial']=test.Name.str.extract('([A-Za-z]+)\.') #lets extract the Salutations
test['Initial'].replace(['Mlle','Mme','Ms','Dr','Major','Lady','Countess','Jonkheer','Col','Rev','Capt','Sir','Don','Dona'],['Miss','Miss','Miss','Mr','Mr','Mrs','Mrs','Other','Other','Other','Mr','Mr','Mr','Other'],inplace=True)
# Fill Null values in Age Column
test.loc[(test.Age.isnull())&(test.Initial=='Mr'),'Age']=33
test.loc[(test.Age.isnull())&(test.Initial=='Mrs'),'Age']=36
test.loc[(test.Age.isnull())&(test.Initial=='Master'),'Age']=5
test.loc[(test.Age.isnull())&(test.Initial=='Miss'),'Age']=22
test.loc[(test.Age.isnull())&(test.Initial=='Other'),'Age']=46
# Fill Null values in Fare Column
test.loc[(test.Fare.isnull()) & (test['Pclass']==3),'Fare'] = 12.45
# Construct feature Age_cat
test['Age_cat']=0
test.loc[test['Age']<=16,'Age_cat']=0
test.loc[(test['Age']>16)&(test['Age']<=32),'Age_cat']=1
test.loc[(test['Age']>32)&(test['Age']<=48),'Age_cat']=2
test.loc[(test['Age']>48)&(test['Age']<=64),'Age_cat']=3
test.loc[test['Age']>64,'Age_cat']=4
# Construct feature Fare_cat
test['Fare_cat']=0
test.loc[test['Fare']<=7.775,'Fare_cat']=0
test.loc[(test['Fare']>7.775)&(test['Fare']<=8.662),'Fare_cat']=1
test.loc[(test['Fare']>8.662)&(test['Fare']<=14.454),'Fare_cat']=2
test.loc[(test['Fare']>14.454)&(test['Fare']<=26.0),'Fare_cat']=3
test.loc[(test['Fare']>26.0)&(test['Fare']<=52.369),'Fare_cat']=4
test.loc[test['Fare']>52.369,'Fare_cat']=5
# Construct feature FamilySize
test['FamilySize'] = test['Parch'] + test['SibSp']
# Drop unwanted features
test.drop(['Name','Age','Ticket','Cabin','SibSp','Parch','Fare','PassengerId'],axis=1,inplace=True)
# Converting String Values into Numeric
test['Sex'].replace(['male','female'],[0,1],inplace=True)
test['Embarked'].replace(['S','C','Q'],[0,1,2],inplace=True)
test['Initial'].replace(['Mr','Mrs','Miss','Master','Other'],[0,1,2,3,4],inplace=True)
test.head(2)
# Predict
test_Survived = pd.Series(ensemble.predict(test), name="Survived")
results = pd.concat([IDtest,test_Survived],axis=1)
results.to_csv("predictions.csv",index=False)
```
## Feature Importance[^](#6)<a id="6" ></a><br>
Well after we have trained a model to make predictions for us, we feel curiuos on how it works. What are the features model weights more when trying to make a prediction?. As humans we seek to understand how it works. Looking at feature importances of a trained model is one way we could explain the decisions it make. Lets visualize the feature importances of the Random forest model we used inside the ensemble above.
```
f,ax=plt.subplots(1,1,figsize=(6,6))
model=RandomForestClassifier(n_estimators=500,random_state=0)
model.fit(X,Y)
pd.Series(model.feature_importances_,X.columns).sort_values(ascending=True).plot.barh(width=0.8,ax=ax)
ax.set_title('Feature Importance in Random Forests')
plt.show()
```
**If You Like the notebook and think that it helped you, please upvote to It keep motivate me**
|
github_jupyter
|
# ディープラーニングに必要な数学と NumPy の操作
# 1. NumPy の基本
## NumPy のインポート
```
import numpy as np
```
## ndarray による1次元配列の例
```
a1 = np.array([1, 2, 3]) # 1次元配列を生成
print('変数の型:',type(a1))
print('データの型 (dtype):', a1.dtype)
print('要素の数 (size):', a1.size)
print('形状 (shape):', a1.shape)
print('次元の数 (ndim):', a1.ndim)
print('中身:', a1)
```
## ndarray による1次元配列の例
```
a2 = np.array([[1, 2, 3],[4, 5, 6]], dtype='float32') # データ型 float32 の2次元配列を生成
print('データの型 (dtype):', a2.dtype)
print('要素の数 (size):', a2.size)
print('形状 (shape):', a2.shape)
print('次元の数 (ndim):', a2.ndim)
print('中身:', a2)
```
# 2. ベクトル(1次元配列)
## ベクトル a の生成(1次元配列の生成)
```
a = np.array([4, 1])
```
## ベクトルのスカラー倍
```
for k in (2, 0.5, -1):
print(k * a)
```
## ベクトルの和と差
```
b = np.array([1, 2]) # ベクトル b の生成
print('a + b =', a + b) # ベクトル a とベクトル b の和
print('a - b =', a - b) # ベクトル a とベクトル b の差
```
# 3. 行列(2次元配列)
## 行列を2次元配列で生成
```
A = np.array([[1, 2], [3 ,4], [5, 6]])
B = np.array([[5, 6], [7 ,8]])
print('A:\n', A)
print('A.shape:', A.shape )
print()
print('B:\n', B)
print('B.shape:', B.shape )
```
## 行列Aの i = 3, j = 2 にアクセス
```
print(A[2][1])
```
## A の転置行列
```
print(A.T)
```
## 行列のスカラー倍
```
print(2 * A)
```
## 行列の和と差
```
print('A + A:\n', A + A) # 行列 A と行列 A の和
print()
print('A - A:\n', A - A) # 行列 A と行列 A の差
```
## 行列 A と行列 B の和
```
print(A + B)
```
## 行列の積
```
print(np.dot(A, B))
```
## 積 BA
```
print(np.dot(B, A))
```
## アダマール積 A $\circ$ A
```
print(A * A)
```
## 行列 X と行ベクトル a の積
```
X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([[1, 2, 3, 4, 5]])
print('X.shape:', X.shape)
print('a.shape:', a.shape)
print(np.dot(X, a))
```
## 行列 X と列ベクトル a の積
```
X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([[1],
[2],
[3],
[4],
[5]])
print('X.shape:', X.shape)
print('a.shape:', a.shape)
Xa = np.dot(X, a)
print('Xa.shape:', Xa.shape)
print('Xa:\n', Xa)
```
## NumPy による行列 X と1次元配列の積
```
X = np.array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
a = np.array([1, 2, 3, 4, 5]) # 1次元配列で生成
print('X.shape:', X.shape)
print('a.shape:', a.shape)
Xa = np.dot(X, a)
print('Xa.shape:', Xa.shape)
print('Xa:\n', Xa)
import numpy as np
np.array([1, 0.1])
```
# 4. ndarray の 軸(axis)について
## Aの合計を計算
```
np.sum(A)
```
## axis = 0 で A の合計を計算
```
print(np.sum(A, axis=0).shape)
print(np.sum(A, axis=0))
```
## axis = 1 で A の合計を計算
```
print(np.sum(A, axis=1).shape)
print(np.sum(A, axis=1))
```
## np.max 関数の利用例
```
Y_hat = np.array([[3, 4], [6, 5], [7, 8]]) # 2次元配列を生成
print(np.max(Y_hat)) # axis 指定なし
print(np.max(Y_hat, axis=1)) # axix=1 を指定
```
## argmax 関数の利用例
```
print(np.argmax(Y_hat)) # axis 指定なし
print(np.argmax(Y_hat, axis=1)) # axix=1 を指定
```
# 5. 3次元以上の配列
## 行列 A を4つ持つ配列の生成
```
A_arr = np.array([A, A, A, A])
print(A_arr.shape)
```
## A_arr の合計を計算
```
np.sum(A_arr)
```
## axis = 0 を指定して A_arr の合計を計算
```
print(np.sum(A_arr, axis=0).shape)
print(np.sum(A_arr, axis=0))
```
## axis = (1, 2) を指定して A_arr の合計を計算
```
print(np.sum(A_arr, axis=(1, 2)))
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/mohameddhameem/TensorflowCertification/blob/main/Natural%20Language%20Processing%20in%20TensorFlow/Lesson%203/NLP_Course_Week_3_Exercise_Question.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import json
import tensorflow as tf
import csv
import random
import numpy as np
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
from tensorflow.keras import regularizers
embedding_dim = 100
max_length = 16
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
training_size= 160000#Your dataset size here. Experiment using smaller values (i.e. 16000), but don't forget to train on at least 160000 to see the best effects
test_portion=.1
corpus = []
# Note that I cleaned the Stanford dataset to remove LATIN1 encoding to make it easier for Python CSV reader
# You can do that yourself with:
# iconv -f LATIN1 -t UTF8 training.1600000.processed.noemoticon.csv -o training_cleaned.csv
# I then hosted it on my site to make it easier to use in this notebook
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/training_cleaned.csv \
-O /tmp/training_cleaned.csv
num_sentences = 0
with open("/tmp/training_cleaned.csv") as csvfile:
reader = csv.reader(csvfile, delimiter=',')
for row in reader:
# Your Code here. Create list items where the first item is the text, found in row[5], and the second is the label. Note that the label is a '0' or a '4' in the text. When it's the former, make
# your label to be 0, otherwise 1. Keep a count of the number of sentences in num_sentences
list_item=[]
list_item.append(row[5])
this_label=row[0]
if this_label == '0':
list_item.append(0)
else:
list_item.append(1)
# YOUR CODE HERE
num_sentences = num_sentences + 1
corpus.append(list_item)
print(num_sentences)
print(len(corpus))
print(corpus[1])
# Expected Output:
# 1600000
# 1600000
# ["is upset that he can't update his Facebook by texting it... and might cry as a result School today also. Blah!", 0]
sentences=[]
labels=[]
random.shuffle(corpus)
for x in range(training_size):
sentences.append(corpus[x][0])
labels.append(corpus[x][1])
tokenizer = Tokenizer(oov_token=oov_tok)
tokenizer.fit_on_texts(sentences)# YOUR CODE HERE
word_index = tokenizer.word_index
vocab_size=len(word_index)
sequences = tokenizer.texts_to_sequences(sentences)# YOUR CODE HERE
padded = pad_sequences(sequences,maxlen=max_length, padding=padding_type,truncating=trunc_type)# YOUR CODE HERE
split = int(test_portion * training_size)
print(split)
test_sequences = padded[0:split]
training_sequences = padded[split:training_size]
test_labels = labels[0:split]
training_labels = labels[split:training_size]
print(vocab_size)
print(word_index['i'])
# Expected Output
# 138858
# 1
!wget http://nlp.stanford.edu/data/glove.6B.zip
!unzip /content/glove.6B.zip
# Note this is the 100 dimension version of GloVe from Stanford
# I unzipped and hosted it on my site to make this notebook easier
#### NOTE - Below link is not working. So download and zip on your own
#!wget --no-check-certificate \
# https://storage.googleapis.com/laurencemoroney-blog.appspot.com/glove.6B.100d.txt \
# -O /tmp/glove.6B.100d.txt
embeddings_index = {};
with open('/content/glove.6B.100d.txt') as f:
for line in f:
values = line.split();
word = values[0];
coefs = np.asarray(values[1:], dtype='float32');
embeddings_index[word] = coefs;
embeddings_matrix = np.zeros((vocab_size+1, embedding_dim));
for word, i in word_index.items():
embedding_vector = embeddings_index.get(word);
if embedding_vector is not None:
embeddings_matrix[i] = embedding_vector;
print(len(embeddings_matrix))
# Expected Output
# 138859
training_padded = np.asarray(training_sequences)
training_labels_np = np.asarray(training_labels)
testing_padded = np.asarray(test_sequences)
testing_labels_np = np.asarray(test_labels)
print(training_labels)
model = tf.keras.Sequential([
tf.keras.layers.Embedding(vocab_size+1, embedding_dim, input_length=max_length, weights=[embeddings_matrix], trainable=False),
# YOUR CODE HERE - experiment with combining different types, such as convolutions and LSTMs
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Conv1D(64, 5, activation='relu'),
tf.keras.layers.MaxPooling1D(pool_size=4),
#tf.keras.layers.LSTM(64),
tf.keras.layers.Bidirectional(tf.keras.layers.GRU(32)),
tf.keras.layers.Dense(1, activation='sigmoid')
])
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])# YOUR CODE HERE
model.summary()
num_epochs = 50
history = model.fit(training_padded, training_labels_np, epochs=num_epochs, validation_data=(testing_padded, testing_labels_np), verbose=2)
print("Training Complete")
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
#-----------------------------------------------------------
# Retrieve a list of list results on training and test data
# sets for each training epoch
#-----------------------------------------------------------
acc=history.history['accuracy']
val_acc=history.history['val_accuracy']
loss=history.history['loss']
val_loss=history.history['val_loss']
epochs=range(len(acc)) # Get number of epochs
#------------------------------------------------
# Plot training and validation accuracy per epoch
#------------------------------------------------
plt.plot(epochs, acc, 'r')
plt.plot(epochs, val_acc, 'b')
plt.title('Training and validation accuracy')
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend(["Accuracy", "Validation Accuracy"])
plt.figure()
#------------------------------------------------
# Plot training and validation loss per epoch
#------------------------------------------------
plt.plot(epochs, loss, 'r')
plt.plot(epochs, val_loss, 'b')
plt.title('Training and validation loss')
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend(["Loss", "Validation Loss"])
plt.figure()
# Expected Output
# A chart where the validation loss does not increase sharply!
```
|
github_jupyter
|
# 01.2 Scattering Compute Speed
**NOT COMPLETED**
In this notebook, the speed to extract scattering coefficients is computed.
```
import sys
import random
import os
sys.path.append('../src')
import warnings
warnings.filterwarnings("ignore")
import torch
from tqdm import tqdm
from kymatio.torch import Scattering2D
import time
import kymatio.scattering2d.backend as backend
###############################################################################
# Finally, we import the `Scattering2D` class that computes the scattering
# transform.
from kymatio import Scattering2D
```
# 3. Scattering Speed Test
```
# From: https://github.com/kymatio/kymatio/blob/0.1.X/examples/2d/compute_speed.py
# Benchmark setup
# --------------------
J = 3
L = 8
times = 10
devices = ['cpu', 'gpu']
scattering = Scattering2D(J, shape=(M, N), L=L, backend='torch_skcuda')
data = np.concatenate(dataset['img'],axis=0)
data = torch.from_numpy(data)
x = data[0:batch_size]
%%time
#mlflow.set_experiment('compute_speed_scattering')
for device in devices:
#with mlflow.start_run():
fmt_str = '==> Testing Float32 with {} backend, on {}, forward'
print(fmt_str.format('torch', device.upper()))
if device == 'gpu':
scattering.cuda()
x = x.cuda()
else:
scattering.cpu()
x = x.cpu()
scattering.forward(x)
if device == 'gpu':
torch.cuda.synchronize()
t_start = time.time()
for _ in range(times):
scattering.forward(x)
if device == 'gpu':
torch.cuda.synchronize()
t_elapsed = time.time() - t_start
fmt_str = 'Elapsed time: {:2f} [s / {:d} evals], avg: {:.2f} (s/batch)'
print(fmt_str.format(t_elapsed, times, t_elapsed/times))
# mlflow.log_param('M',M)
# mlflow.log_param('N',N)
# mlflow.log_param('Backend', device.upper())
# mlflow.log_param('J', J)
# mlflow.log_param('L', L)
# mlflow.log_param('Batch Size', batch_size)
# mlflow.log_param('Times', times)
# mlflow.log_metric('Elapsed Time', t_elapsed)
# mlflow.log_metric('Average Time', times)
###############################################################################
# The resulting output should be something like
#
# .. code-block:: text
#
# ==> Testing Float32 with torch backend, on CPU, forward
# Elapsed time: 624.910853 [s / 10 evals], avg: 62.49 (s/batch)
# ==> Testing Float32 with torch backend, on GPU, forward
```
|
github_jupyter
|
<!--NOTEBOOK_HEADER-->
*This notebook contains material from [nbpages](https://jckantor.github.io/nbpages) by Jeffrey Kantor (jeff at nd.edu). The text is released under the
[CC-BY-NC-ND-4.0 license](https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode).
The code is released under the [MIT license](https://opensource.org/licenses/MIT).*
<!--NAVIGATION-->
< [2.3 Heirarchical Tagging](https://jckantor.github.io/nbpages/02.03-Heirarchical-Tagging.html) | [Contents](toc.html) | [Tag Index](tag_index.html) | [2.5 Lint](https://jckantor.github.io/nbpages/02.05-Lint.html) ><p><a href="https://colab.research.google.com/github/jckantor/nbpages/blob/master/docs/02.04-Working-with-Data-and-Figures.ipynb"> <img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://jckantor.github.io/nbpages/02.04-Working-with-Data-and-Figures.ipynb"> <img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
```
# IMPORT DATA FILES USED BY THIS NOTEBOOK
import os, requests
file_links = [("data/Stock_Data.csv", "https://jckantor.github.io/nbpages/data/Stock_Data.csv")]
# This cell has been added by nbpages. Run this cell to download data files required for this notebook.
for filepath, fileurl in file_links:
stem, filename = os.path.split(filepath)
if stem:
if not os.path.exists(stem):
os.mkdir(stem)
if not os.path.isfile(filepath):
with open(filepath, 'wb') as f:
response = requests.get(fileurl)
f.write(response.content)
```
# 2.4 Working with Data and Figures
## 2.4.1 Importing data
The following cell reads the data file `Stock_Data.csv` from the `data` subdirectory. The name of this file will appear in the data index.
```
import pandas as pd
df = pd.read_csv("data/Stock_Data.csv")
df.head()
```
## 2.4.2 Creating and saving figures
The following cell creates a figure `Stock_Data.png` in the `figures` subdirectory. The name of this file will appear in the figures index.
```
%matplotlib inline
import os
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
fig, ax = plt.subplots(2, 1, figsize=(8, 5))
(df/df.iloc[0]).drop('VIX', axis=1).plot(ax=ax[0])
df['VIX'].plot(ax=ax[1])
ax[0].set_title('Normalized Indices')
ax[1].set_title('Volatility VIX')
ax[1].set_xlabel('Days')
fig.tight_layout()
if not os.path.exists("figures"):
os.mkdir("figures")
plt.savefig("figures/Stock_Data.png")
```
<!--NAVIGATION-->
< [2.3 Heirarchical Tagging](https://jckantor.github.io/nbpages/02.03-Heirarchical-Tagging.html) | [Contents](toc.html) | [Tag Index](tag_index.html) | [2.5 Lint](https://jckantor.github.io/nbpages/02.05-Lint.html) ><p><a href="https://colab.research.google.com/github/jckantor/nbpages/blob/master/docs/02.04-Working-with-Data-and-Figures.ipynb"> <img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://jckantor.github.io/nbpages/02.04-Working-with-Data-and-Figures.ipynb"> <img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
|
github_jupyter
|
Used https://github.com/GoogleCloudPlatform/cloudml-samples/blob/master/xgboost/notebooks/census_training/train.py as a starting point and adjusted to CatBoost
```
#Google Cloud Libraries
from google.cloud import storage
#System Libraries
import datetime
import subprocess
#Data Libraries
import pandas as pd
import numpy as np
#ML Libraries
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
import xgboost as xgb
from catboost import CatBoostClassifier, Pool, cv
from catboost import CatBoost, Pool
from catboost.utils import get_gpu_device_count
print('I see %i GPU devices' % get_gpu_device_count())
# Fill in your Cloud Storage bucket name
BUCKET_ID = "mchrestkha-demo-env-ml-examples"
census_data_filename = 'adult.data.csv'
# Public bucket holding the census data
bucket = storage.Client().bucket('cloud-samples-data')
# Path to the data inside the public bucket
data_dir = 'ai-platform/census/data/'
# Download the data
blob = bucket.blob(''.join([data_dir, census_data_filename]))
blob.download_to_filename(census_data_filename)
# these are the column labels from the census data files
COLUMNS = (
'age',
'workclass',
'fnlwgt',
'education',
'education-num',
'marital-status',
'occupation',
'relationship',
'race',
'sex',
'capital-gain',
'capital-loss',
'hours-per-week',
'native-country',
'income-level'
)
# categorical columns contain data that need to be turned into numerical values before being used by XGBoost
CATEGORICAL_COLUMNS = (
'workclass',
'education',
'marital-status',
'occupation',
'relationship',
'race',
'sex',
'native-country'
)
# Load the training census dataset
with open(census_data_filename, 'r') as train_data:
raw_training_data = pd.read_csv(train_data, header=None, names=COLUMNS)
# remove column we are trying to predict ('income-level') from features list
X = raw_training_data.drop('income-level', axis=1)
# create training labels list
#train_labels = (raw_training_data['income-level'] == ' >50K')
y = raw_training_data['income-level']
# Since the census data set has categorical features, we need to convert
# them to numerical values.
# convert data in categorical columns to numerical values
X_enc=X
encoders = {col:LabelEncoder() for col in CATEGORICAL_COLUMNS}
for col in CATEGORICAL_COLUMNS:
X_enc[col] = encoders[col].fit_transform(X[col])
y_enc=LabelEncoder().fit_transform(y)
X_train, X_validation, y_train, y_validation = train_test_split(X_enc, y_enc, train_size=0.75, random_state=42)
print(type(y))
print(type(y_enc))
%%time
#model = CatBoost({'iterations':50})
model=CatBoostClassifier(
od_type='Iter'
#iterations=5000,
#custom_loss=['Accuracy']
)
model.fit(
X_train,y_train,eval_set=(X_validation, y_validation),
verbose=50)
# # load data into DMatrix object
# dtrain = xgb.DMatrix(train_features, train_labels)
# # train model
# bst = xgb.train({}, dtrain, 20)
# Export the model to a file
fname = 'catboost_census_model.onnx'
model.save_model(fname, format='onnx')
# Upload the model to GCS
bucket = storage.Client().bucket(BUCKET_ID)
blob = bucket.blob('{}/{}'.format(
datetime.datetime.now().strftime('census/catboost_model_dir/catboost_census_%Y%m%d_%H%M%S'),
fname))
blob.upload_from_filename(fname)
!gsutil ls gs://$BUCKET_ID/census/*
```
|
github_jupyter
|
Final models with hyperparameters tuned for Logistics Regression and XGBoost with selected features.
```
#Import the libraries
import pandas as pd
import numpy as np
from tqdm import tqdm
from sklearn import linear_model, metrics, preprocessing, model_selection
from sklearn.preprocessing import StandardScaler
import xgboost as xgb
#Load the data
modeling_dataset = pd.read_csv('/content/drive/MyDrive/prediction/frac_cleaned_fod_data.csv', low_memory = False)
#All columns - except 'HasDetections', 'kfold', and 'MachineIdentifier'
train_features = [tf for tf in modeling_dataset.columns if tf not in ('HasDetections', 'kfold', 'MachineIdentifier')]
#The features selected based on the feature selection method earlier employed
train_features_after_selection = ['AVProductStatesIdentifier', 'Processor','AvSigVersion', 'Census_TotalPhysicalRAM', 'Census_InternalPrimaryDiagonalDisplaySizeInInches',
'Census_IsVirtualDevice', 'Census_PrimaryDiskTotalCapacity', 'Wdft_IsGamer', 'Census_IsAlwaysOnAlwaysConnectedCapable', 'EngineVersion',
'Census_ProcessorCoreCount', 'Census_OSEdition', 'Census_OSInstallTypeName', 'Census_OSSkuName', 'AppVersion', 'OsBuildLab', 'OsSuite',
'Firewall', 'IsProtected', 'Census_IsTouchEnabled', 'Census_ActivationChannel', 'LocaleEnglishNameIdentifier','Census_SystemVolumeTotalCapacity',
'Census_InternalPrimaryDisplayResolutionHorizontal','Census_HasOpticalDiskDrive', 'OsBuild', 'Census_InternalPrimaryDisplayResolutionVertical',
'CountryIdentifier', 'Census_MDC2FormFactor', 'GeoNameIdentifier', 'Census_PowerPlatformRoleName', 'Census_OSWUAutoUpdateOptionsName', 'SkuEdition',
'Census_OSVersion', 'Census_GenuineStateName', 'Census_OSBuildRevision', 'Platform', 'Census_ChassisTypeName', 'Census_FlightRing',
'Census_PrimaryDiskTypeName', 'Census_OSBranch', 'Census_IsSecureBootEnabled', 'OsPlatformSubRelease']
#Define the categorical features of the data
categorical_features = ['ProductName',
'EngineVersion',
'AppVersion',
'AvSigVersion',
'Platform',
'Processor',
'OsVer',
'OsPlatformSubRelease',
'OsBuildLab',
'SkuEdition',
'Census_MDC2FormFactor',
'Census_DeviceFamily',
'Census_PrimaryDiskTypeName',
'Census_ChassisTypeName',
'Census_PowerPlatformRoleName',
'Census_OSVersion',
'Census_OSArchitecture',
'Census_OSBranch',
'Census_OSEdition',
'Census_OSSkuName',
'Census_OSInstallTypeName',
'Census_OSWUAutoUpdateOptionsName',
'Census_GenuineStateName',
'Census_ActivationChannel',
'Census_FlightRing']
#XGBoost
"""
Best parameters set:
alpha: 1.0
colsample_bytree: 0.6
eta: 0.05
gamma: 0.1
lamda: 1.0
max_depth: 9
min_child_weight: 5
subsample: 0.7
"""
#XGBoost
def opt_run_xgboost(fold):
for col in train_features:
if col in categorical_features:
#Initialize the Label Encoder
lbl = preprocessing.LabelEncoder()
#Fit on the categorical features
lbl.fit(modeling_dataset[col])
#Transform
modeling_dataset.loc[:,col] = lbl.transform(modeling_dataset[col])
#Get training and validation data using folds
modeling_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
modeling_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Get train data
X_train = modeling_datasets_train[train_features_after_selection].values
#Get validation data
X_valid = modeling_datasets_valid[train_features_after_selection].values
#Initialize XGboost model
xgb_model = xgb.XGBClassifier(
alpha= 1.0,
colsample_bytree= 0.6,
eta= 0.05,
gamma= 0.1,
lamda= 1.0,
max_depth= 9,
min_child_weight= 5,
subsample= 0.7,
n_jobs=-1)
#Fit the model on training data
xgb_model.fit(X_train, modeling_datasets_train.HasDetections.values)
#Predict on validation
valid_preds = xgb_model.predict_proba(X_valid)[:,1]
valid_preds_pc = xgb_model.predict(X_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(modeling_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(modeling_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(modeling_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#LR
"""
'penalty': 'l2',
'C': 49.71967742639108,
'solver': 'lbfgs'
max_iter: 300
"""
#Function for Logistic Regression Classification
def opt_run_lr(fold):
#Get training and validation data using folds
cleaned_fold_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
cleaned_fold_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Initialize OneHotEncoder from scikit-learn, and fit it on training and validation features
ohe = preprocessing.OneHotEncoder()
full_data = pd.concat(
[cleaned_fold_datasets_train[train_features_after_selection],cleaned_fold_datasets_valid[train_features_after_selection]],
axis = 0
)
ohe.fit(full_data[train_features_after_selection])
#transform the training and validation data
x_train = ohe.transform(cleaned_fold_datasets_train[train_features_after_selection])
x_valid = ohe.transform(cleaned_fold_datasets_valid[train_features_after_selection])
#Initialize the Logistic Regression Model
lr_model = linear_model.LogisticRegression(
penalty= 'l2',
C = 49.71967742639108,
solver= 'lbfgs',
max_iter= 300,
n_jobs=-1
)
#Fit model on training data
lr_model.fit(x_train, cleaned_fold_datasets_train.HasDetections.values)
#Predict on the validation data using the probability for the AUC
valid_preds = lr_model.predict_proba(x_valid)[:, 1]
#For precision and Recall
valid_preds_pc = lr_model.predict(x_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#A list to hold the values of the XGB performance metrics
xg = []
for fold in tqdm(range(10)):
xg.append(opt_run_xgboost(fold))
#Run the Logistic regression model for all folds and hold their values
lr = []
for fold in tqdm(range(10)):
lr.append(opt_run_lr(fold))
xgb_auc = []
xgb_pre = []
xgb_rc = []
lr_auc = []
lr_pre = []
lr_rc = []
#Loop to get each of the performance metric for average computation
for i in lr:
lr_auc.append(i[0])
lr_pre.append(i[1])
lr_rc.append(i[2])
for j in xg:
xgb_auc.append(i[0])
xgb_pre.append(i[1])
xgb_rc.append(i[2])
#Dictionary to hold the basic model performance data
final_model_performance = {"logistic_regression": {"auc":"", "precision":"", "recall":""},
"xgb": {"auc":"","precision":"","recall":""}
}
#Calculate average of each of the lists of performance metrics and update the dictionary
final_model_performance['logistic_regression'].update({'auc':sum(lr_auc)/len(lr_auc)})
final_model_performance['xgb'].update({'auc':sum(xgb_auc)/len(xgb_auc)})
final_model_performance['logistic_regression'].update({'precision':sum(lr_pre)/len(lr_pre)})
final_model_performance['xgb'].update({'precision':sum(xgb_pre)/len(xgb_pre)})
final_model_performance['logistic_regression'].update({'recall':sum(lr_rc)/len(lr_rc)})
final_model_performance['xgb'].update({'recall':sum(xgb_rc)/len(xgb_rc)})
final_model_performance
#LR
"""
'penalty': 'l2',
'C': 49.71967742639108,
'solver': 'lbfgs'
max_iter: 100
"""
#Function for Logistic Regression Classification - max_iter = 100
def opt_run_lr100(fold):
#Get training and validation data using folds
cleaned_fold_datasets_train = modeling_dataset[modeling_dataset.kfold != fold].reset_index(drop=True)
cleaned_fold_datasets_valid = modeling_dataset[modeling_dataset.kfold == fold].reset_index(drop=True)
#Initialize OneHotEncoder from scikit-learn, and fit it on training and validation features
ohe = preprocessing.OneHotEncoder()
full_data = pd.concat(
[cleaned_fold_datasets_train[train_features_after_selection],cleaned_fold_datasets_valid[train_features_after_selection]],
axis = 0
)
ohe.fit(full_data[train_features_after_selection])
#transform the training and validation data
x_train = ohe.transform(cleaned_fold_datasets_train[train_features_after_selection])
x_valid = ohe.transform(cleaned_fold_datasets_valid[train_features_after_selection])
#Initialize the Logistic Regression Model
lr_model = linear_model.LogisticRegression(
penalty= 'l2',
C = 49.71967742639108,
solver= 'lbfgs',
max_iter= 100,
n_jobs=-1
)
#Fit model on training data
lr_model.fit(x_train, cleaned_fold_datasets_train.HasDetections.values)
#Predict on the validation data using the probability for the AUC
valid_preds = lr_model.predict_proba(x_valid)[:, 1]
#For precision and Recall
valid_preds_pc = lr_model.predict(x_valid)
#Get the ROC AUC score
auc = metrics.roc_auc_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds)
#Get the precision score
pre = metrics.precision_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
#Get the Recall score
rc = metrics.recall_score(cleaned_fold_datasets_valid.HasDetections.values, valid_preds_pc, average='binary')
return auc, pre, rc
#Run the Logistic regression model for all folds and hold their values
lr100 = []
for fold in tqdm(range(10)):
lr100.append(opt_run_lr100(fold))
lr100_auc = []
lr100_pre = []
lr100_rc = []
for k in lr100:
lr100_auc.append(k[0])
lr100_pre.append(k[1])
lr100_rc.append(k[2])
sum(lr100_auc)/len(lr100_auc)
sum(lr100_pre)/len(lr100_pre)
sum(lr100_rc)/len(lr100_rc)
"""
{'logistic_regression': {'auc': 0.660819451656712,
'precision': 0.6069858170181643,
'recall': 0.6646704904969867},
'xgb': {'auc': 0.6583717792973377,
'precision': 0.6042291042291044,
'recall': 0.6542422535211267}}
"""
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/Tessellate-Imaging/monk_v1/blob/master/study_roadmaps/2_transfer_learning_roadmap/6_freeze_base_network/2.2)%20Understand%20the%20effect%20of%20freezing%20base%20model%20in%20transfer%20learning%20-%202%20-%20pytorch.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Goals
### In the previous tutorial you studied the role of freezing models on a small dataset.
### Understand the role of freezing models in transfer learning on a fairly large dataset
### Why freeze/unfreeze base models in transfer learning
### Use comparison feature to appropriately set this parameter on custom dataset
### You will be using lego bricks dataset to train the classifiers
# What is freezing base network
- To recap you have two parts in your network
- One that already existed, the pretrained one, the base network
- The new sub-network or a single layer you added
-The hyper-parameter we can see here: Freeze base network
- Freezing base network makes the base network untrainable
- The base network now acts as a feature extractor and only the next half is trained
- If you do not freeze the base network the entire network is trained
# Table of Contents
## [Install](#0)
## [Freeze Base network in densenet121 and train a classifier](#1)
## [Unfreeze base network in densenet121 and train another classifier](#2)
## [Compare both the experiment](#3)
<a id='0'></a>
# Install Monk
## Using pip (Recommended)
- colab (gpu)
- All bakcends: `pip install -U monk-colab`
- kaggle (gpu)
- All backends: `pip install -U monk-kaggle`
- cuda 10.2
- All backends: `pip install -U monk-cuda102`
- Gluon bakcned: `pip install -U monk-gluon-cuda102`
- Pytorch backend: `pip install -U monk-pytorch-cuda102`
- Keras backend: `pip install -U monk-keras-cuda102`
- cuda 10.1
- All backend: `pip install -U monk-cuda101`
- Gluon bakcned: `pip install -U monk-gluon-cuda101`
- Pytorch backend: `pip install -U monk-pytorch-cuda101`
- Keras backend: `pip install -U monk-keras-cuda101`
- cuda 10.0
- All backend: `pip install -U monk-cuda100`
- Gluon bakcned: `pip install -U monk-gluon-cuda100`
- Pytorch backend: `pip install -U monk-pytorch-cuda100`
- Keras backend: `pip install -U monk-keras-cuda100`
- cuda 9.2
- All backend: `pip install -U monk-cuda92`
- Gluon bakcned: `pip install -U monk-gluon-cuda92`
- Pytorch backend: `pip install -U monk-pytorch-cuda92`
- Keras backend: `pip install -U monk-keras-cuda92`
- cuda 9.0
- All backend: `pip install -U monk-cuda90`
- Gluon bakcned: `pip install -U monk-gluon-cuda90`
- Pytorch backend: `pip install -U monk-pytorch-cuda90`
- Keras backend: `pip install -U monk-keras-cuda90`
- cpu
- All backend: `pip install -U monk-cpu`
- Gluon bakcned: `pip install -U monk-gluon-cpu`
- Pytorch backend: `pip install -U monk-pytorch-cpu`
- Keras backend: `pip install -U monk-keras-cpu`
## Install Monk Manually (Not recommended)
### Step 1: Clone the library
- git clone https://github.com/Tessellate-Imaging/monk_v1.git
### Step 2: Install requirements
- Linux
- Cuda 9.0
- `cd monk_v1/installation/Linux && pip install -r requirements_cu90.txt`
- Cuda 9.2
- `cd monk_v1/installation/Linux && pip install -r requirements_cu92.txt`
- Cuda 10.0
- `cd monk_v1/installation/Linux && pip install -r requirements_cu100.txt`
- Cuda 10.1
- `cd monk_v1/installation/Linux && pip install -r requirements_cu101.txt`
- Cuda 10.2
- `cd monk_v1/installation/Linux && pip install -r requirements_cu102.txt`
- CPU (Non gpu system)
- `cd monk_v1/installation/Linux && pip install -r requirements_cpu.txt`
- Windows
- Cuda 9.0 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu90.txt`
- Cuda 9.2 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu92.txt`
- Cuda 10.0 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu100.txt`
- Cuda 10.1 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu101.txt`
- Cuda 10.2 (Experimental support)
- `cd monk_v1/installation/Windows && pip install -r requirements_cu102.txt`
- CPU (Non gpu system)
- `cd monk_v1/installation/Windows && pip install -r requirements_cpu.txt`
- Mac
- CPU (Non gpu system)
- `cd monk_v1/installation/Mac && pip install -r requirements_cpu.txt`
- Misc
- Colab (GPU)
- `cd monk_v1/installation/Misc && pip install -r requirements_colab.txt`
- Kaggle (GPU)
- `cd monk_v1/installation/Misc && pip install -r requirements_kaggle.txt`
### Step 3: Add to system path (Required for every terminal or kernel run)
- `import sys`
- `sys.path.append("monk_v1/");`
## Dataset - LEGO Classification
- https://www.kaggle.com/joosthazelzet/lego-brick-images/
```
! wget --load-cookies /tmp/cookies.txt "https://docs.google.com/uc?export=download&confirm=$(wget --save-cookies /tmp/cookies.txt --keep-session-cookies --no-check-certificate 'https://docs.google.com/uc?export=download&id=1MRC58-oCdR1agFTWreDFqevjEOIWDnYZ' -O- | sed -rn 's/.*confirm=([0-9A-Za-z_]+).*/\1\n/p')&id=1MRC58-oCdR1agFTWreDFqevjEOIWDnYZ" -O skin_cancer_mnist_dataset.zip && rm -rf /tmp/cookies.txt
! unzip -qq skin_cancer_mnist_dataset.zip
```
# Imports
```
#Using pytorch backend
# When installed using pip
from monk.pytorch_prototype import prototype
# When installed manually (Uncomment the following)
#import os
#import sys
#sys.path.append("monk_v1/");
#sys.path.append("monk_v1/monk/");
#from monk.pytorch_prototype import prototype
```
<a id='1'></a>
# Freeze Base network in densenet121 and train a classifier
## Creating and managing experiments
- Provide project name
- Provide experiment name
- For a specific data create a single project
- Inside each project multiple experiments can be created
- Every experiment can be have diferent hyper-parameters attached to it
```
gtf = prototype(verbose=1);
gtf.Prototype("Project", "Freeze_Base_Network");
```
### This creates files and directories as per the following structure
workspace
|
|--------Project
|
|
|-----Freeze_Base_Network
|
|-----experiment-state.json
|
|-----output
|
|------logs (All training logs and graphs saved here)
|
|------models (all trained models saved here)
## Set dataset and select the model
## Quick mode training
- Using Default Function
- dataset_path
- model_name
- freeze_base_network
- num_epochs
## Sample Dataset folder structure
parent_directory
|
|
|------cats
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
|------dogs
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
## Modifyable params
- dataset_path: path to data
- model_name: which pretrained model to use
- freeze_base_network: Retrain already trained network or not
- num_epochs: Number of epochs to train for
```
gtf.Default(dataset_path="skin_cancer_mnist_dataset/images",
path_to_csv="skin_cancer_mnist_dataset/train_labels.csv",
model_name="densenet121",
freeze_base_network=True, # Set this param as true
num_epochs=5);
#Read the summary generated once you run this cell.
```
## From the summary above
- Model Params
Model name: densenet121
Use Gpu: True
Use pretrained: True
Freeze base network: True
## Another thing to notice from summary
Model Details
Loading pretrained model
Model Loaded on device
Model name: densenet121
Num of potentially trainable layers: 242
Num of actual trainable layers: 1
### There are a total of 242 layers
### Since we have freezed base network only 1 is trainable, the final layer
## Train the classifier
```
#Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed
```
### Best validation Accuracy achieved - 74.77 %
(You may get a different result)
<a id='2'></a>
# Unfreeze Base network in densenet121 and train a classifier
## Creating and managing experiments
- Provide project name
- Provide experiment name
- For a specific data create a single project
- Inside each project multiple experiments can be created
- Every experiment can be have diferent hyper-parameters attached to it
```
gtf = prototype(verbose=1);
gtf.Prototype("Project", "Unfreeze_Base_Network");
```
### This creates files and directories as per the following structure
workspace
|
|--------Project
|
|
|-----Freeze_Base_Network (Previously created)
|
|-----experiment-state.json
|
|-----output
|
|------logs (All training logs and graphs saved here)
|
|------models (all trained models saved here)
|
|
|-----Unfreeze_Base_Network (Created Now)
|
|-----experiment-state.json
|
|-----output
|
|------logs (All training logs and graphs saved here)
|
|------models (all trained models saved here)
## Set dataset and select the model
## Quick mode training
- Using Default Function
- dataset_path
- model_name
- freeze_base_network
- num_epochs
## Sample Dataset folder structure
parent_directory
|
|
|------cats
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
|------dogs
|
|------img1.jpg
|------img2.jpg
|------.... (and so on)
## Modifyable params
- dataset_path: path to data
- model_name: which pretrained model to use
- freeze_base_network: Retrain already trained network or not
- num_epochs: Number of epochs to train for
```
gtf.Default(dataset_path="skin_cancer_mnist_dataset/images",
path_to_csv="skin_cancer_mnist_dataset/train_labels.csv",
model_name="densenet121",
freeze_base_network=False, # Set this param as false
num_epochs=5);
#Read the summary generated once you run this cell.
```
## From the summary above
- Model Params
Model name: densenet121
Use Gpu: True
Use pretrained: True
Freeze base network: False
## Another thing to notice from summary
Model Details
Loading pretrained model
Model Loaded on device
Model name: densenet121
Num of potentially trainable layers: 242
Num of actual trainable layers: 242
### There are a total of 242 layers
### Since we have unfreezed base network all 242 layers are trainable including the final layer
## Train the classifier
```
#Start Training
gtf.Train();
#Read the training summary generated once you run the cell and training is completed
```
### Best Val Accuracy achieved - 81.33 %
(You may get a different result)
<a id='3'></a>
# Compare both the experiment
```
# Invoke the comparison class
from monk.compare_prototype import compare
```
### Creating and managing comparison experiments
- Provide project name
```
# Create a project
gtf = compare(verbose=1);
gtf.Comparison("Compare-effect-of-freezing");
```
### This creates files and directories as per the following structure
workspace
|
|--------comparison
|
|
|-----Compare-effect-of-freezing
|
|------stats_best_val_acc.png
|------stats_max_gpu_usage.png
|------stats_training_time.png
|------train_accuracy.png
|------train_loss.png
|------val_accuracy.png
|------val_loss.png
|
|-----comparison.csv (Contains necessary details of all experiments)
### Add the experiments
- First argument - Project name
- Second argument - Experiment name
```
gtf.Add_Experiment("Project", "Freeze_Base_Network");
gtf.Add_Experiment("Project", "Unfreeze_Base_Network");
```
### Run Analysis
```
gtf.Generate_Statistics();
```
## Visualize and study comparison metrics
### Training Accuracy Curves
```
from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/train_accuracy.png")
```
### Training Loss Curves
```
from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/train_loss.png")
```
### Validation Accuracy Curves
```
from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/val_accuracy.png")
```
### Validation loss curves
```
from IPython.display import Image
Image(filename="workspace/comparison/Compare-effect-of-freezing/val_loss.png")
```
## Accuracies achieved on validation dataset
### With freezing base network - 74.77 %
### Without freezing base network - 81.33 %
#### For this classifier, keeping the base network trainable seems to be a good option. Thus for other data it may result in overfitting the training data
(You may get a different result)
|
github_jupyter
|
## 使用TensorFlow的基本步骤
以使用LinearRegression来预测房价为例。
- 使用RMSE(均方根误差)评估模型预测的准确率
- 通过调整超参数来提高模型的预测准确率
```
from __future__ import print_function
import math
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
# 加载数据集
california_housing_df = pd.read_csv("https://download.mlcc.google.cn/mledu-datasets/california_housing_train.csv", sep=",")
# 将数据打乱
california_housing_df = california_housing_df.reindex(np.random.permutation(california_housing_df.index))
# 替换房价的单位为k
california_housing_df['median_house_value'] /=1000.0
print("california house dataframe: \n", california_housing_df) # 根据pd设置,只显示10条数据,以及保留小数点后一位
```
### 检查数据
```
# 使用pd的describe方法来统计一些信息
california_housing_df.describe()
```
### 构建模型
我们将在这个例子中预测中位房价,将其作为学习的标签,使用房间总数作为输入特征。
#### 第1步:定义特征并配置特征列
为了把数据导入TensorFlow,我们需要指定每个特征包含的数据类型。我们主要使用以下两种类型:
- 分类数据: 一种文字数据。
- 数值数据:一种数字(整数或浮点数)数据或希望视为数字的数据。
在TF中我们使用**特征列**的结构来表示特征的数据类型。特征列仅存储对特征数据的描述,不包含特征数据本身。
```
# 定义输入特征
kl_feature = california_housing_df[['total_rooms']]
# 配置房间总数为数值特征列
feature_columns = [tf.feature_column.numeric_column('total_rooms')]
```
#### 第2步: 定义目标
```
# 定义目标标签
targets = california_housing_df['median_house_value']
```
**梯度裁剪**是在应用梯度值之前设置其上限,梯度裁剪有助于确保数值稳定性,防止梯度爆炸。
#### 第3步:配置线性回归器
```
# 使用Linear Regressor配置线性回归模型,使用GradientDescentOptimizer优化器训练模型
kl_optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.0000001)
# 使用clip_gradients_by_norm梯度裁剪我们的优化器,梯度裁剪可以确保我们的梯度大小在训练期间不会变得过大,梯度过大会导致梯度下降失败。
kl_optimizer = tf.contrib.estimator.clip_gradients_by_norm(kl_optimizer, 5.0)
# 使用我们的特征列和优化器配置线性回归模型
house_linear_regressor = tf.estimator.LinearRegressor(feature_columns=feature_columns, optimizer=kl_optimizer)
```
#### 第4步:定义输入函数
要将数据导入LinearRegressor,我们需要定义一个输入函数,让它告诉TF如何对数据进行预处理,以及在模型训练期间如何批处理、随机处理和重复数据。
首先我们将Pandas特征数据转换成NumPy数组字典,然后利用Dataset API构建Dataset对象,拆分数据为batch_size的批数据,以按照指定周期数(num_epochs)进行重复,**注意:**如果默认值num_epochs=None传递到repeat(),输入数据会无限期重复。
shuffle: Bool, 是否打乱数据
buffer_size: 指定shuffle从中随机抽样的数据集大小
```
def kl_input_fn(features, targets, batch_size=1, shuffle=True, num_epochs=None):
"""使用单个特征训练房价预测模型
Args:
features: 特征DataFrame
targets: 目标DataFrame
batch_size: 批大小
shuffle: Bool. 是否打乱数据
Return:
下一个数据批次的元组(features, labels)
"""
# 把pandas数据转换成np.array构成的dict数据
features = {key: np.array(value) for key, value in dict(features).items()}
# 构建数据集,配置批和重复次数、
ds = Dataset.from_tensor_slices((features, targets)) # 数据大小 2GB 限制
ds = ds.batch(batch_size).repeat(num_epochs)
# 打乱数据
if shuffle:
ds = ds.shuffle(buffer_size=10000) # buffer_size指随机抽样的数据集大小
# 返回下一批次的数据
features, labels = ds.make_one_shot_iterator().get_next()
return features, labels
```
**注意:** 更详细的输入函数和Dataset API参考:[TF Developer's Guide](https://www.tensorflow.org/programmers_guide/datasets)
#### 第5步:训练模型
在linear_regressor上调用train()来训练模型
```
_ = house_linear_regressor.train(input_fn=lambda: kl_input_fn(kl_feature, targets), steps=100)
```
#### 第6步:评估模型
**注意:**训练误差可以衡量训练的模型与训练数据的拟合情况,但**不能**衡量模型泛化到新数据的效果,我们需要拆分数据来评估模型的泛化能力。
```
# 只做一次预测,所以把epoch设为1并关闭随机
prediction_input_fn = lambda: kl_input_fn(kl_feature, targets, num_epochs=1, shuffle=False)
# 调用predict进行预测
predictions = house_linear_regressor.predict(input_fn=prediction_input_fn)
# 把预测结果转换为numpy数组
predictions = np.array([item['predictions'][0] for item in predictions])
# 打印MSE和RMSE
mean_squared_error = metrics.mean_squared_error(predictions, targets)
root_mean_squared_error = math.sqrt(mean_squared_error)
print("均方误差 %0.3f" % mean_squared_error)
print("均方根误差: %0.3f" % root_mean_squared_error)
min_house_value = california_housing_df['median_house_value'].min()
max_house_value = california_housing_df['median_house_value'].max()
min_max_diff = max_house_value- min_house_value
print("最低中位房价: %0.3f" % min_house_value)
print("最高中位房价: %0.3f" % max_house_value)
print("中位房价最低最高差值: %0.3f" % min_max_diff)
print("均方根误差:%0.3f" % root_mean_squared_error)
```
由此结果可以看出模型的效果并不理想,我们可以使用一些基本的策略来降低误差。
```
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
calibration_data.describe()
# 我们可以可视化数据和我们学到的线,
sample = california_housing_df.sample(n=300) # 得到均匀分布的sample数据df
# 得到房屋总数的最小最大值
x_0 = sample["total_rooms"].min()
x_1 = sample["total_rooms"].max()
# 获得训练后的最终权重和偏差
weight = house_linear_regressor.get_variable_value('linear/linear_model/total_rooms/weights')[0]
bias = house_linear_regressor.get_variable_value('linear/linear_model/bias_weights')
# 计算最低最高房间数(特征)对应的房价(标签)
y_0 = weight * x_0 + bias
y_1 = weight * x_1 +bias
# 画图
plt.plot([x_0,x_1], [y_0,y_1],c='r')
plt.ylabel('median_house_value')
plt.xlabel('total_rooms')
# 画出散点图
plt.scatter(sample["total_rooms"], sample["median_house_value"])
plt.show()
```
### 模型调参
以上代码封装调参
```
def train_model(learning_rate, steps, batch_size, input_feature="total_rooms"):
"""Trains a linear regression model of one feature.
Args:
learning_rate: A `float`, the learning rate.
steps: A non-zero `int`, the total number of training steps. A training step
consists of a forward and backward pass using a single batch.
batch_size: A non-zero `int`, the batch size.
input_feature: A `string` specifying a column from `california_housing_df`
to use as input feature.
"""
periods = 10
steps_per_period = steps / periods
my_feature = input_feature
my_feature_data = california_housing_df[[my_feature]]
my_label = "median_house_value"
targets = california_housing_df[my_label]
# Create feature columns.
feature_columns = [tf.feature_column.numeric_column(my_feature)]
# Create input functions.
training_input_fn = lambda:kl_input_fn(my_feature_data, targets, batch_size=batch_size)
prediction_input_fn = lambda: kl_input_fn(my_feature_data, targets, num_epochs=1, shuffle=False)
# Create a linear regressor object.
my_optimizer = tf.train.GradientDescentOptimizer(learning_rate=learning_rate)
my_optimizer = tf.contrib.estimator.clip_gradients_by_norm(my_optimizer, 5.0)
linear_regressor = tf.estimator.LinearRegressor(
feature_columns=feature_columns,
optimizer=my_optimizer
)
# Set up to plot the state of our model's line each period.
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
plt.title("Learned Line by Period")
plt.ylabel(my_label)
plt.xlabel(my_feature)
sample = california_housing_df.sample(n=300)
plt.scatter(sample[my_feature], sample[my_label])
colors = [cm.coolwarm(x) for x in np.linspace(-1, 1, periods)]
# Train the model, but do so inside a loop so that we can periodically assess
# loss metrics.
print("Training model...")
print("RMSE (on training data):")
root_mean_squared_errors = []
for period in range (0, periods):
# Train the model, starting from the prior state.
linear_regressor.train(
input_fn=training_input_fn,
steps=steps_per_period
)
# Take a break and compute predictions.
predictions = linear_regressor.predict(input_fn=prediction_input_fn)
predictions = np.array([item['predictions'][0] for item in predictions])
# Compute loss.
root_mean_squared_error = math.sqrt(
metrics.mean_squared_error(predictions, targets))
# Occasionally print the current loss.
print(" period %02d : %0.2f" % (period, root_mean_squared_error))
# Add the loss metrics from this period to our list.
root_mean_squared_errors.append(root_mean_squared_error)
# Finally, track the weights and biases over time.
# Apply some math to ensure that the data and line are plotted neatly.
y_extents = np.array([0, sample[my_label].max()])
weight = linear_regressor.get_variable_value('linear/linear_model/%s/weights' % input_feature)[0]
bias = linear_regressor.get_variable_value('linear/linear_model/bias_weights')
x_extents = (y_extents - bias) / weight
x_extents = np.maximum(np.minimum(x_extents,
sample[my_feature].max()),
sample[my_feature].min())
y_extents = weight * x_extents + bias
plt.plot(x_extents, y_extents, color=colors[period])
print("Model training finished.")
# Output a graph of loss metrics over periods.
plt.subplot(1, 2, 2)
plt.ylabel('RMSE')
plt.xlabel('Periods')
plt.title("Root Mean Squared Error vs. Periods")
plt.tight_layout()
plt.plot(root_mean_squared_errors)
# Output a table with calibration data.
calibration_data = pd.DataFrame()
calibration_data["predictions"] = pd.Series(predictions)
calibration_data["targets"] = pd.Series(targets)
display.display(calibration_data.describe())
print("Final RMSE (on training data): %0.2f" % root_mean_squared_error)
```
**练习1: 使RMSE不超过180**
```
train_model(learning_rate=0.00002, steps=500, batch_size=5)
```
### 模型调参的启发法
> 不要死循规则
- 训练误差应该稳步减小,刚开始是急剧减小,最终应随着训练收敛达到平稳状态。
- 如果训练尚未收敛,尝试运行更长的时间。
- 如果训练误差减小速度过慢,则提高学习速率也许有助于加快其减小速度。
- 但有时如果学习速率过高,训练误差的减小速度反而会变慢。
- 如果训练误差变化很大,尝试降低学习速率。
- 较低的学习速率和较大的步数/较大的批量大小通常是不错的组合。
- 批量大小过小也会导致不稳定情况。不妨先尝试 100 或 1000 等较大的值,然后逐渐减小值的大小,直到出现性能降低的情况。
**练习2:尝试其他特征**
我们使用population特征替代。
```
train_model(learning_rate=0.00005, steps=500, batch_size=5, input_feature="population")
```
|
github_jupyter
|
# test note
* jupyterはコンテナ起動すること
* テストベッド一式起動済みであること
```
!pip install --upgrade pip
!pip install --force-reinstall ../lib/ait_sdk-0.1.7-py3-none-any.whl
from pathlib import Path
import pprint
from ait_sdk.test.hepler import Helper
import json
# settings cell
# mounted dir
root_dir = Path('/workdir/root/ait')
ait_name='eval_metamorphic_test_tf1.13'
ait_version='0.1'
ait_full_name=f'{ait_name}_{ait_version}'
ait_dir = root_dir / ait_full_name
td_name=f'{ait_name}_test'
# (dockerホスト側の)インベントリ登録用アセット格納ルートフォルダ
current_dir = %pwd
with open(f'{current_dir}/config.json', encoding='utf-8') as f:
json_ = json.load(f)
root_dir = json_['host_ait_root_dir']
is_container = json_['is_container']
invenotory_root_dir = f'{root_dir}\\ait\\{ait_full_name}\\local_qai\\inventory'
# entry point address
# コンテナ起動かどうかでポート番号が変わるため、切り替える
if is_container:
backend_entry_point = 'http://host.docker.internal:8888/qai-testbed/api/0.0.1'
ip_entry_point = 'http://host.docker.internal:8888/qai-ip/api/0.0.1'
else:
backend_entry_point = 'http://host.docker.internal:5000/qai-testbed/api/0.0.1'
ip_entry_point = 'http://host.docker.internal:6000/qai-ip/api/0.0.1'
# aitのデプロイフラグ
# 一度実施すれば、それ以降は実施しなくてOK
is_init_ait = True
# インベントリの登録フラグ
# 一度実施すれば、それ以降は実施しなくてOK
is_init_inventory = True
helper = Helper(backend_entry_point=backend_entry_point,
ip_entry_point=ip_entry_point,
ait_dir=ait_dir,
ait_full_name=ait_full_name)
# health check
helper.get_bk('/health-check')
helper.get_ip('/health-check')
# create ml-component
res = helper.post_ml_component(name=f'MLComponent_{ait_full_name}', description=f'Description of {ait_full_name}', problem_domain=f'ProbremDomain of {ait_full_name}')
helper.set_ml_component_id(res['MLComponentId'])
# deploy AIT
if is_init_ait:
helper.deploy_ait_non_build()
else:
print('skip deploy AIT')
res = helper.get_data_types()
model_data_type_id = [d for d in res['DataTypes'] if d['Name'] == 'model'][0]['Id']
dataset_data_type_id = [d for d in res['DataTypes'] if d['Name'] == 'dataset'][0]['Id']
res = helper.get_file_systems()
unix_file_system_id = [f for f in res['FileSystems'] if f['Name'] == 'UNIX_FILE_SYSTEM'][0]['Id']
windows_file_system_id = [f for f in res['FileSystems'] if f['Name'] == 'WINDOWS_FILE'][0]['Id']
# add inventories
if is_init_inventory:
inv1_name = helper.post_inventory('train_image', dataset_data_type_id, windows_file_system_id,
f'{invenotory_root_dir}\\mnist_dataset\\mnist_dataset.zip',
'MNIST_dataset are train image, train label, test image, test label', ['zip'])
inv2_name = helper.post_inventory('mnist_model', dataset_data_type_id, windows_file_system_id,
f'{invenotory_root_dir}\\mnist_model\\model_mnist.zip',
'MNIST_model', ['zip'])
else:
print('skip add inventories')
# get ait_json and inventory_jsons
res_json = helper.get_bk('/QualityMeasurements/RelationalOperators', is_print_json=False).json()
eq_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '=='][0])
nq_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '!='][0])
gt_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '>'][0])
ge_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '>='][0])
lt_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '<'][0])
le_id = int([r['Id'] for r in res_json['RelationalOperator'] if r['Expression'] == '<='][0])
res_json = helper.get_bk('/testRunners', is_print_json=False).json()
ait_json = [j for j in res_json['TestRunners'] if j['Name'] == ait_name][-1]
inv_1_json = helper.get_inventory(inv1_name)
inv_2_json = helper.get_inventory(inv2_name)
# add teast_descriptions
helper.post_td(td_name, ait_json['QualityDimensionId'],
quality_measurements=[
{"Id":ait_json['Report']['Measures'][0]['Id'], "Value":"0.25", "RelationalOperatorId":lt_id, "Enable":True}
],
target_inventories=[
{"Id":1, "InventoryId": inv_1_json['Id'], "TemplateInventoryId": ait_json['TargetInventories'][0]['Id']},
{"Id":2, "InventoryId": inv_2_json['Id'], "TemplateInventoryId": ait_json['TargetInventories'][1]['Id']}
],
test_runner={
"Id":ait_json['Id'],
"Params":[
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][0]['Id'], "Value":"10"},
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][1]['Id'], "Value":"500"},
{"TestRunnerParamTemplateId":ait_json['ParamTemplates'][2]['Id'], "Value":"train"}
]
})
# get test_description_jsons
td_1_json = helper.get_td(td_name)
# run test_descriptions
helper.post_run_and_wait(td_1_json['Id'])
res_json = helper.get_td_detail(td_1_json['Id'])
pprint.pprint(res_json)
# generate report
res = helper.post_report(td_1_json['Id'])
```
|
github_jupyter
|
```
%load_ext rpy2.ipython
%matplotlib inline
import logging
logging.getLogger('fbprophet').setLevel(logging.ERROR)
import warnings
warnings.filterwarnings("ignore")
```
## Python API
Prophet follows the `sklearn` model API. We create an instance of the `Prophet` class and then call its `fit` and `predict` methods.
The input to Prophet is always a dataframe with two columns: `ds` and `y`. The `ds` (datestamp) column should be of a format expected by Pandas, ideally YYYY-MM-DD for a date or YYYY-MM-DD HH:MM:SS for a timestamp. The `y` column must be numeric, and represents the measurement we wish to forecast.
As an example, let's look at a time series of the log daily page views for the Wikipedia page for [Peyton Manning](https://en.wikipedia.org/wiki/Peyton_Manning). We scraped this data using the [Wikipediatrend](https://cran.r-project.org/package=wikipediatrend) package in R. Peyton Manning provides a nice example because it illustrates some of Prophet's features, like multiple seasonality, changing growth rates, and the ability to model special days (such as Manning's playoff and superbowl appearances). The CSV is available [here](https://github.com/facebook/prophet/blob/master/examples/example_wp_log_peyton_manning.csv).
First we'll import the data:
```
import pandas as pd
from fbprophet import Prophet
df = pd.read_csv('../examples/example_wp_log_peyton_manning.csv')
df.head()
```
We fit the model by instantiating a new `Prophet` object. Any settings to the forecasting procedure are passed into the constructor. Then you call its `fit` method and pass in the historical dataframe. Fitting should take 1-5 seconds.
```
m = Prophet()
m.fit(df)
```
Predictions are then made on a dataframe with a column `ds` containing the dates for which a prediction is to be made. You can get a suitable dataframe that extends into the future a specified number of days using the helper method `Prophet.make_future_dataframe`. By default it will also include the dates from the history, so we will see the model fit as well.
```
future = m.make_future_dataframe(periods=365)
future.tail()
```
The `predict` method will assign each row in `future` a predicted value which it names `yhat`. If you pass in historical dates, it will provide an in-sample fit. The `forecast` object here is a new dataframe that includes a column `yhat` with the forecast, as well as columns for components and uncertainty intervals.
```
forecast = m.predict(future)
forecast[['ds', 'yhat', 'yhat_lower', 'yhat_upper']].tail()
```
You can plot the forecast by calling the `Prophet.plot` method and passing in your forecast dataframe.
```
fig1 = m.plot(forecast)
```
If you want to see the forecast components, you can use the `Prophet.plot_components` method. By default you'll see the trend, yearly seasonality, and weekly seasonality of the time series. If you include holidays, you'll see those here, too.
```
fig2 = m.plot_components(forecast)
```
An interactive figure of the forecast and components can be created with plotly. You will need to install plotly 4.0 or above separately, as it will not by default be installed with fbprophet. You will also need to install the `notebook` and `ipywidgets` packages.
```
from fbprophet.plot import plot_plotly, plot_components_plotly
plot_plotly(m, forecast)
plot_components_plotly(m, forecast)
```
More details about the options available for each method are available in the docstrings, for example, via `help(Prophet)` or `help(Prophet.fit)`. The [R reference manual](https://cran.r-project.org/web/packages/prophet/prophet.pdf) on CRAN provides a concise list of all of the available functions, each of which has a Python equivalent.
## R API
In R, we use the normal model fitting API. We provide a `prophet` function that performs fitting and returns a model object. You can then call `predict` and `plot` on this model object.
```
%%R
library(prophet)
```
First we read in the data and create the outcome variable. As in the Python API, this is a dataframe with columns `ds` and `y`, containing the date and numeric value respectively. The ds column should be YYYY-MM-DD for a date, or YYYY-MM-DD HH:MM:SS for a timestamp. As above, we use here the log number of views to Peyton Manning's Wikipedia page, available [here](https://github.com/facebook/prophet/blob/master/examples/example_wp_log_peyton_manning.csv).
```
%%R
df <- read.csv('../examples/example_wp_log_peyton_manning.csv')
```
We call the `prophet` function to fit the model. The first argument is the historical dataframe. Additional arguments control how Prophet fits the data and are described in later pages of this documentation.
```
%%R
m <- prophet(df)
```
Predictions are made on a dataframe with a column `ds` containing the dates for which predictions are to be made. The `make_future_dataframe` function takes the model object and a number of periods to forecast and produces a suitable dataframe. By default it will also include the historical dates so we can evaluate in-sample fit.
```
%%R
future <- make_future_dataframe(m, periods = 365)
tail(future)
```
As with most modeling procedures in R, we use the generic `predict` function to get our forecast. The `forecast` object is a dataframe with a column `yhat` containing the forecast. It has additional columns for uncertainty intervals and seasonal components.
```
%%R
forecast <- predict(m, future)
tail(forecast[c('ds', 'yhat', 'yhat_lower', 'yhat_upper')])
```
You can use the generic `plot` function to plot the forecast, by passing in the model and the forecast dataframe.
```
%%R -w 10 -h 6 -u in
plot(m, forecast)
```
You can use the `prophet_plot_components` function to see the forecast broken down into trend, weekly seasonality, and yearly seasonality.
```
%%R -w 9 -h 9 -u in
prophet_plot_components(m, forecast)
```
An interactive plot of the forecast using Dygraphs can be made with the command `dyplot.prophet(m, forecast)`.
More details about the options available for each method are available in the docstrings, for example, via `?prophet` or `?fit.prophet`. This documentation is also available in the [reference manual](https://cran.r-project.org/web/packages/prophet/prophet.pdf) on CRAN.
|
github_jupyter
|
## TensorFlow 2 Complete Project Workflow in Amazon SageMaker
### Data Preprocessing -> Code Prototyping -> Automatic Model Tuning -> Deployment
1. [Introduction](#Introduction)
2. [SageMaker Processing for dataset transformation](#SageMakerProcessing)
3. [Local Mode training](#LocalModeTraining)
4. [Local Mode endpoint](#LocalModeEndpoint)
5. [SageMaker hosted training](#SageMakerHostedTraining)
6. [Automatic Model Tuning](#AutomaticModelTuning)
7. [SageMaker hosted endpoint](#SageMakerHostedEndpoint)
8. [Workflow Automation with the Step Functions Data Science SDK](#WorkflowAutomation)
1. [Add an IAM policy to your SageMaker role](#IAMPolicy)
2. [Create an execution role for Step Functions](#CreateExecutionRole)
3. [Set up a TrainingPipeline](#TrainingPipeline)
4. [Visualizing the workflow](#VisualizingWorkflow)
5. [Creating and executing the pipeline](#CreatingExecutingPipeline)
6. [Cleanup](#Cleanup)
9. [Extensions](#Extensions)
### ***Prerequisite: To run the Local Mode sections of this example, use a SageMaker Notebook Instance; otherwise skip those sections (for example if you're using SageMaker Studio instead).***
## Introduction <a class="anchor" id="Introduction">
If you are using TensorFlow 2, you can use the Amazon SageMaker prebuilt TensorFlow 2 container with training scripts similar to those you would use outside SageMaker. This feature is named Script Mode. Using Script Mode and other SageMaker features, you can build a complete workflow for a TensorFlow 2 project. This notebook presents such a workflow, including all key steps such as preprocessing data with SageMaker Processing, code prototyping with SageMaker Local Mode training and inference, and production-ready model training and deployment with SageMaker hosted training and inference. Automatic Model Tuning in SageMaker is used to tune the model's hyperparameters. Additionally, the [AWS Step Functions Data Science SDK](https://aws-step-functions-data-science-sdk.readthedocs.io/en/latest/readmelink.html) is used to automate the main training and deployment steps for use in a production workflow outside notebooks.
To enable you to run this notebook within a reasonable time (typically less than an hour), this notebook's use case is a straightforward regression task: predicting house prices based on the well-known Boston Housing dataset. This public dataset contains 13 features regarding housing stock of towns in the Boston area. Features include average number of rooms, accessibility to radial highways, adjacency to the Charles River, etc.
To begin, we'll import some necessary packages and set up directories for local training and test data. We'll also set up a SageMaker Session to perform various operations, and specify an Amazon S3 bucket to hold input data and output. The default bucket used here is created by SageMaker if it doesn't already exist, and named in accordance with the AWS account ID and AWS Region.
```
import os
import sagemaker
import tensorflow as tf
sess = sagemaker.Session()
bucket = sess.default_bucket()
data_dir = os.path.join(os.getcwd(), 'data')
os.makedirs(data_dir, exist_ok=True)
train_dir = os.path.join(os.getcwd(), 'data/train')
os.makedirs(train_dir, exist_ok=True)
test_dir = os.path.join(os.getcwd(), 'data/test')
os.makedirs(test_dir, exist_ok=True)
raw_dir = os.path.join(os.getcwd(), 'data/raw')
os.makedirs(raw_dir, exist_ok=True)
```
# SageMaker Processing for dataset transformation <a class="anchor" id="SageMakerProcessing">
Next, we'll import the dataset and transform it with SageMaker Processing, which can be used to process terabytes of data in a SageMaker-managed cluster separate from the instance running your notebook server. In a typical SageMaker workflow, notebooks are only used for prototyping and can be run on relatively inexpensive and less powerful instances, while processing, training and model hosting tasks are run on separate, more powerful SageMaker-managed instances. SageMaker Processing includes off-the-shelf support for Scikit-learn, as well as a Bring Your Own Container option, so it can be used with many different data transformation technologies and tasks.
First we'll load the Boston Housing dataset, save the raw feature data and upload it to Amazon S3 for transformation by SageMaker Processing. We'll also save the labels for training and testing.
```
import numpy as np
from tensorflow.python.keras.datasets import boston_housing
from sklearn.preprocessing import StandardScaler
(x_train, y_train), (x_test, y_test) = boston_housing.load_data()
np.save(os.path.join(raw_dir, 'x_train.npy'), x_train)
np.save(os.path.join(raw_dir, 'x_test.npy'), x_test)
np.save(os.path.join(train_dir, 'y_train.npy'), y_train)
np.save(os.path.join(test_dir, 'y_test.npy'), y_test)
s3_prefix = 'tf-2-workflow'
rawdata_s3_prefix = '{}/data/raw'.format(s3_prefix)
raw_s3 = sess.upload_data(path='./data/raw/', key_prefix=rawdata_s3_prefix)
print(raw_s3)
```
To use SageMaker Processing, simply supply a Python data preprocessing script as shown below. For this example, we're using a SageMaker prebuilt Scikit-learn container, which includes many common functions for processing data. There are few limitations on what kinds of code and operations you can run, and only a minimal contract: input and output data must be placed in specified directories. If this is done, SageMaker Processing automatically loads the input data from S3 and uploads transformed data back to S3 when the job is complete.
```
%%writefile preprocessing.py
import glob
import numpy as np
import os
from sklearn.preprocessing import StandardScaler
if __name__=='__main__':
input_files = glob.glob('{}/*.npy'.format('/opt/ml/processing/input'))
print('\nINPUT FILE LIST: \n{}\n'.format(input_files))
scaler = StandardScaler()
for file in input_files:
raw = np.load(file)
transformed = scaler.fit_transform(raw)
if 'train' in file:
output_path = os.path.join('/opt/ml/processing/train', 'x_train.npy')
np.save(output_path, transformed)
print('SAVED TRANSFORMED TRAINING DATA FILE\n')
else:
output_path = os.path.join('/opt/ml/processing/test', 'x_test.npy')
np.save(output_path, transformed)
print('SAVED TRANSFORMED TEST DATA FILE\n')
```
Before starting the SageMaker Processing job, we instantiate a `SKLearnProcessor` object. This object allows you to specify the instance type to use in the job, as well as how many instances. Although the Boston Housing dataset is quite small, we'll use two instances to showcase how easy it is to spin up a cluster for SageMaker Processing.
```
from sagemaker import get_execution_role
from sagemaker.sklearn.processing import SKLearnProcessor
sklearn_processor = SKLearnProcessor(framework_version='0.20.0',
role=get_execution_role(),
instance_type='ml.m5.xlarge',
instance_count=2)
```
We're now ready to run the Processing job. To enable distributing the data files equally among the instances, we specify the `ShardedByS3Key` distribution type in the `ProcessingInput` object. This ensures that if we have `n` instances, each instance will receive `1/n` files from the specified S3 bucket. It may take around 3 minutes for the following code cell to run, mainly to set up the cluster. At the end of the job, the cluster automatically will be torn down by SageMaker.
```
from sagemaker.processing import ProcessingInput, ProcessingOutput
from time import gmtime, strftime
processing_job_name = "tf-2-workflow-{}".format(strftime("%d-%H-%M-%S", gmtime()))
output_destination = 's3://{}/{}/data'.format(bucket, s3_prefix)
sklearn_processor.run(code='preprocessing.py',
job_name=processing_job_name,
inputs=[ProcessingInput(
source=raw_s3,
destination='/opt/ml/processing/input',
s3_data_distribution_type='ShardedByS3Key')],
outputs=[ProcessingOutput(output_name='train',
destination='{}/train'.format(output_destination),
source='/opt/ml/processing/train'),
ProcessingOutput(output_name='test',
destination='{}/test'.format(output_destination),
source='/opt/ml/processing/test')])
preprocessing_job_description = sklearn_processor.jobs[-1].describe()
```
In the log output of the SageMaker Processing job above, you should be able to see logs in two different colors for the two different instances, and that each instance received different files. Without the `ShardedByS3Key` distribution type, each instance would have received a copy of **all** files. By spreading the data equally among `n` instances, you should receive a speedup by approximately a factor of `n` for most stateless data transformations. After saving the job results locally, we'll move on to prototyping training and inference code with Local Mode.
```
train_in_s3 = '{}/train/x_train.npy'.format(output_destination)
test_in_s3 = '{}/test/x_test.npy'.format(output_destination)
!aws s3 cp {train_in_s3} ./data/train/x_train.npy
!aws s3 cp {test_in_s3} ./data/test/x_test.npy
```
## Local Mode training <a class="anchor" id="LocalModeTraining">
Local Mode in Amazon SageMaker is a convenient way to make sure your code is working locally as expected before moving on to full scale, hosted training in a separate, more powerful SageMaker-managed cluster. To train in Local Mode, it is necessary to have docker-compose or nvidia-docker-compose (for GPU instances) installed. Running the following commands will install docker-compose or nvidia-docker-compose, and configure the notebook environment for you.
```
!wget -q https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/local_mode_setup.sh
!wget -q https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/daemon.json
!/bin/bash ./local_mode_setup.sh
```
Next, we'll set up a TensorFlow Estimator for Local Mode training. Key parameters for the Estimator include:
- `train_instance_type`: the kind of hardware on which training will run. In the case of Local Mode, we simply set this parameter to `local` to invoke Local Mode training on the CPU, or to `local_gpu` if the instance has a GPU.
- `git_config`: to make sure training scripts are source controlled for coordinated, shared use by a team, the Estimator can pull in the code from a Git repository rather than local directories.
- Other parameters of note: the algorithm’s hyperparameters, which are passed in as a dictionary, and a Boolean parameter indicating that we are using Script Mode.
Recall that we are using Local Mode here mainly to make sure our code is working. Accordingly, instead of performing a full cycle of training with many epochs (passes over the full dataset), we'll train only for a small number of epochs just to confirm the code is working properly and avoid wasting full-scale training time unnecessarily.
```
from sagemaker.tensorflow import TensorFlow
git_config = {'repo': 'https://github.com/aws-samples/amazon-sagemaker-script-mode',
'branch': 'master'}
model_dir = '/opt/ml/model'
train_instance_type = 'local'
hyperparameters = {'epochs': 5, 'batch_size': 128, 'learning_rate': 0.01}
local_estimator = TensorFlow(git_config=git_config,
source_dir='tf-2-workflow/train_model',
entry_point='train.py',
model_dir=model_dir,
instance_type=train_instance_type,
instance_count=1,
hyperparameters=hyperparameters,
role=sagemaker.get_execution_role(),
base_job_name='tf-2-workflow',
framework_version='2.2',
py_version='py37',
script_mode=True)
```
The `fit` method call below starts the Local Mode training job. Metrics for training will be logged below the code, inside the notebook cell. You should observe the validation loss decrease substantially over the five epochs, with no training errors, which is a good indication that our training code is working as expected.
```
inputs = {'train': f'file://{train_dir}',
'test': f'file://{test_dir}'}
local_estimator.fit(inputs)
```
## Local Mode endpoint <a class="anchor" id="LocalModeEndpoint">
While Amazon SageMaker’s Local Mode training is very useful to make sure your training code is working before moving on to full scale training, it also would be useful to have a convenient way to test your model locally before incurring the time and expense of deploying it to production. One possibility is to fetch the TensorFlow SavedModel artifact or a model checkpoint saved in Amazon S3, and load it in your notebook for testing. However, an even easier way to do this is to use the SageMaker Python SDK to do this work for you by setting up a Local Mode endpoint.
More specifically, the Estimator object from the Local Mode training job can be used to deploy a model locally. With one exception, this code is the same as the code you would use to deploy to production. In particular, all you need to do is invoke the local Estimator's deploy method, and similarly to Local Mode training, specify the instance type as either `local_gpu` or `local` depending on whether your notebook is on a GPU instance or CPU instance.
Just in case there are other inference containers running in Local Mode, we'll stop them to avoid conflict before deploying our new model locally.
```
!docker container stop $(docker container ls -aq) >/dev/null
```
The following single line of code deploys the model locally in the SageMaker TensorFlow Serving container:
```
local_predictor = local_estimator.deploy(initial_instance_count=1, instance_type='local')
```
To get predictions from the Local Mode endpoint, simply invoke the Predictor's predict method.
```
local_results = local_predictor.predict(x_test[:10])['predictions']
```
As a sanity check, the predictions can be compared against the actual target values.
```
local_preds_flat_list = [float('%.1f'%(item)) for sublist in local_results for item in sublist]
print('predictions: \t{}'.format(np.array(local_preds_flat_list)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
```
We only trained the model for a few epochs and there is much room for improvement, but the predictions so far should at least appear reasonably within the ballpark.
To avoid having the SageMaker TensorFlow Serving container indefinitely running locally, simply gracefully shut it down by calling the `delete_endpoint` method of the Predictor object.
```
local_predictor.delete_endpoint()
```
## SageMaker hosted training <a class="anchor" id="SageMakerHostedTraining">
Now that we've confirmed our code is working locally, we can move on to use SageMaker's hosted training functionality. Hosted training is preferred for doing actual training, especially large-scale, distributed training. Unlike Local Mode training, for hosted training the actual training itself occurs not on the notebook instance, but on a separate cluster of machines managed by SageMaker. Before starting hosted training, the data must be in S3, or an EFS or FSx for Lustre file system. We'll upload to S3 now, and confirm the upload was successful.
```
s3_prefix = 'tf-2-workflow'
traindata_s3_prefix = '{}/data/train'.format(s3_prefix)
testdata_s3_prefix = '{}/data/test'.format(s3_prefix)
train_s3 = sess.upload_data(path='./data/train/', key_prefix=traindata_s3_prefix)
test_s3 = sess.upload_data(path='./data/test/', key_prefix=testdata_s3_prefix)
inputs = {'train':train_s3, 'test': test_s3}
print(inputs)
```
We're now ready to set up an Estimator object for hosted training. It is similar to the Local Mode Estimator, except the `train_instance_type` has been set to a SageMaker ML instance type instead of `local` for Local Mode. Also, since we know our code is working now, we'll train for a larger number of epochs with the expectation that model training will converge to an improved, lower validation loss.
With these two changes, we simply call `fit` to start the actual hosted training.
```
train_instance_type = 'ml.c5.xlarge'
hyperparameters = {'epochs': 30, 'batch_size': 128, 'learning_rate': 0.01}
git_config = {'repo': 'https://github.com/aws-samples/amazon-sagemaker-script-mode',
'branch': 'master'}
estimator = TensorFlow(git_config=git_config,
source_dir='tf-2-workflow/train_model',
entry_point='train.py',
model_dir=model_dir,
instance_type=train_instance_type,
instance_count=1,
hyperparameters=hyperparameters,
role=sagemaker.get_execution_role(),
base_job_name='tf-2-workflow',
framework_version='2.2',
py_version='py37',
script_mode=True)
```
After starting the hosted training job with the `fit` method call below, you should observe the training converge over the longer number of epochs to a validation loss that is considerably lower than that which was achieved in the shorter Local Mode training job. Can we do better? We'll look into a way to do so in the **Automatic Model Tuning** section below.
```
estimator.fit(inputs)
```
As with the Local Mode training, hosted training produces a model saved in S3 that we can retrieve. This is an example of the modularity of SageMaker: having trained the model in SageMaker, you can now take the model out of SageMaker and run it anywhere else. Alternatively, you can deploy the model into a production-ready environment using SageMaker's hosted endpoints functionality, as shown in the **SageMaker hosted endpoint** section below.
Retrieving the model from S3 is very easy: the hosted training estimator you created above stores a reference to the model's location in S3. You simply copy the model from S3 using the estimator's `model_data` property and unzip it to inspect the contents.
```
!aws s3 cp {estimator.model_data} ./model/model.tar.gz
```
The unzipped archive should include the assets required by TensorFlow Serving to load the model and serve it, including a .pb file:
```
!tar -xvzf ./model/model.tar.gz -C ./model
```
## Automatic Model Tuning <a class="anchor" id="AutomaticModelTuning">
So far we have simply run one Local Mode training job and one Hosted Training job without any real attempt to tune hyperparameters to produce a better model, other than increasing the number of epochs. Selecting the right hyperparameter values to train your model can be difficult, and typically is very time consuming if done manually. The right combination of hyperparameters is dependent on your data and algorithm; some algorithms have many different hyperparameters that can be tweaked; some are very sensitive to the hyperparameter values selected; and most have a non-linear relationship between model fit and hyperparameter values. SageMaker Automatic Model Tuning helps automate the hyperparameter tuning process: it runs multiple training jobs with different hyperparameter combinations to find the set with the best model performance.
We begin by specifying the hyperparameters we wish to tune, and the range of values over which to tune each one. We also must specify an objective metric to be optimized: in this use case, we'd like to minimize the validation loss.
```
from sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner
hyperparameter_ranges = {
'learning_rate': ContinuousParameter(0.001, 0.2, scaling_type="Logarithmic"),
'epochs': IntegerParameter(10, 50),
'batch_size': IntegerParameter(64, 256),
}
metric_definitions = [{'Name': 'loss',
'Regex': ' loss: ([0-9\\.]+)'},
{'Name': 'val_loss',
'Regex': ' val_loss: ([0-9\\.]+)'}]
objective_metric_name = 'val_loss'
objective_type = 'Minimize'
```
Next we specify a HyperparameterTuner object that takes the above definitions as parameters. Each tuning job must be given a budget: a maximum number of training jobs. A tuning job will complete after that many training jobs have been executed.
We also can specify how much parallelism to employ, in this case five jobs, meaning that the tuning job will complete after three series of five jobs in parallel have completed. For the default Bayesian Optimization tuning strategy used here, the tuning search is informed by the results of previous groups of training jobs, so we don't run all of the jobs in parallel, but rather divide the jobs into groups of parallel jobs. There is a trade-off: using more parallel jobs will finish tuning sooner, but likely will sacrifice tuning search accuracy.
Now we can launch a hyperparameter tuning job by calling the `fit` method of the HyperparameterTuner object. The tuning job may take around 10 minutes to finish. While you're waiting, the status of the tuning job, including metadata and results for invidual training jobs within the tuning job, can be checked in the SageMaker console in the **Hyperparameter tuning jobs** panel.
```
tuner = HyperparameterTuner(estimator,
objective_metric_name,
hyperparameter_ranges,
metric_definitions,
max_jobs=15,
max_parallel_jobs=5,
objective_type=objective_type)
tuning_job_name = "tf-2-workflow-{}".format(strftime("%d-%H-%M-%S", gmtime()))
tuner.fit(inputs, job_name=tuning_job_name)
tuner.wait()
```
After the tuning job is finished, we can use the `HyperparameterTuningJobAnalytics` object from the SageMaker Python SDK to list the top 5 tuning jobs with the best performance. Although the results vary from tuning job to tuning job, the best validation loss from the tuning job (under the FinalObjectiveValue column) likely will be substantially lower than the validation loss from the hosted training job above, where we did not perform any tuning other than manually increasing the number of epochs once.
```
tuner_metrics = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name)
tuner_metrics.dataframe().sort_values(['FinalObjectiveValue'], ascending=True).head(5)
```
The total training time and training jobs status can be checked with the following lines of code. Because automatic early stopping is by default off, all the training jobs should be completed normally. For an example of a more in-depth analysis of a tuning job, see the SageMaker official sample [HPO_Analyze_TuningJob_Results.ipynb](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/hyperparameter_tuning/analyze_results/HPO_Analyze_TuningJob_Results.ipynb) notebook.
```
total_time = tuner_metrics.dataframe()['TrainingElapsedTimeSeconds'].sum() / 3600
print("The total training time is {:.2f} hours".format(total_time))
tuner_metrics.dataframe()['TrainingJobStatus'].value_counts()
```
## SageMaker hosted endpoint <a class="anchor" id="SageMakerHostedEndpoint">
Assuming the best model from the tuning job is better than the model produced by the individual Hosted Training job above, we could now easily deploy that model to production. A convenient option is to use a SageMaker hosted endpoint, which serves real time predictions from the trained model (Batch Transform jobs also are available for asynchronous, offline predictions on large datasets). The endpoint will retrieve the TensorFlow SavedModel created during training and deploy it within a SageMaker TensorFlow Serving container. This all can be accomplished with one line of code.
More specifically, by calling the `deploy` method of the HyperparameterTuner object we instantiated above, we can directly deploy the best model from the tuning job to a SageMaker hosted endpoint. It will take several minutes longer to deploy the model to the hosted endpoint compared to the Local Mode endpoint, which is more useful for fast prototyping of inference code.
```
tuning_predictor = tuner.deploy(initial_instance_count=1, instance_type='ml.m5.xlarge')
```
We can compare the predictions generated by this endpoint with those generated locally by the Local Mode endpoint:
```
results = tuning_predictor.predict(x_test[:10])['predictions']
flat_list = [float('%.1f'%(item)) for sublist in results for item in sublist]
print('predictions: \t{}'.format(np.array(flat_list)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
```
To avoid billing charges from stray resources, you can delete the prediction endpoint to release its associated instance(s).
```
sess.delete_endpoint(tuning_predictor.endpoint_name)
```
## Workflow Automation with the AWS Step Functions Data Science SDK <a class="anchor" id="WorkflowAutomation">
In the previous parts of this notebook, we prototyped various steps of a TensorFlow project within the notebook itself. Notebooks are great for prototyping, but generally are not used in production-ready machine learning pipelines. For example, a simple pipeline in SageMaker includes the following steps:
1. Training the model.
2. Creating a SageMaker Model object that wraps the model artifact for serving.
3. Creating a SageMaker Endpoint Configuration specifying how the model should be served (e.g. hardware type and amount).
4. Deploying the trained model to the configured SageMaker Endpoint.
The AWS Step Functions Data Science SDK automates the process of creating and running these kinds of workflows using AWS Step Functions and SageMaker. It does this by allowing you to create workflows using short, simple Python scripts that define workflow steps and chain them together. Under the hood, all the workflow steps are coordinated by AWS Step Functions without any need for you to manage the underlying infrastructure.
To begin, install the Step Functions Data Science SDK:
```
import sys
!{sys.executable} -m pip install --quiet --upgrade stepfunctions
```
### Add an IAM policy to your SageMaker role <a class="anchor" id="IAMPolicy">
**If you are running this notebook on an Amazon SageMaker notebook instance**, the IAM role assumed by your notebook instance needs permission to create and run workflows in AWS Step Functions. To provide this permission to the role, do the following.
1. Open the Amazon [SageMaker console](https://console.aws.amazon.com/sagemaker/).
2. Select **Notebook instances** and choose the name of your notebook instance
3. Under **Permissions and encryption** select the role ARN to view the role on the IAM console
4. Choose **Attach policies** and search for `AWSStepFunctionsFullAccess`.
5. Select the check box next to `AWSStepFunctionsFullAccess` and choose **Attach policy**
If you are running this notebook in a local environment, the SDK will use your configured AWS CLI configuration. For more information, see [Configuring the AWS CLI](https://docs.aws.amazon.com/cli/latest/userguide/cli-chap-configure.html).
### Create an execution role for Step Functions <a class="anchor" id="CreateExecutionRole">
You also need to create an execution role for Step Functions to enable that service to access SageMaker and other service functionality.
1. Go to the [IAM console](https://console.aws.amazon.com/iam/)
2. Select **Roles** and then **Create role**.
3. Under **Choose the service that will use this role** select **Step Functions**
4. Choose **Next** until you can enter a **Role name**
5. Enter a name such as `StepFunctionsWorkflowExecutionRole` and then select **Create role**
Select your newly create role and attach a policy to it. The following steps attach a policy that provides full access to Step Functions, however as a good practice you should only provide access to the resources you need.
1. Under the **Permissions** tab, click **Add inline policy**
2. Enter the following in the **JSON** tab
```json
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Action": [
"sagemaker:CreateTransformJob",
"sagemaker:DescribeTransformJob",
"sagemaker:StopTransformJob",
"sagemaker:CreateTrainingJob",
"sagemaker:DescribeTrainingJob",
"sagemaker:StopTrainingJob",
"sagemaker:CreateHyperParameterTuningJob",
"sagemaker:DescribeHyperParameterTuningJob",
"sagemaker:StopHyperParameterTuningJob",
"sagemaker:CreateModel",
"sagemaker:CreateEndpointConfig",
"sagemaker:CreateEndpoint",
"sagemaker:DeleteEndpointConfig",
"sagemaker:DeleteEndpoint",
"sagemaker:UpdateEndpoint",
"sagemaker:ListTags",
"lambda:InvokeFunction",
"sqs:SendMessage",
"sns:Publish",
"ecs:RunTask",
"ecs:StopTask",
"ecs:DescribeTasks",
"dynamodb:GetItem",
"dynamodb:PutItem",
"dynamodb:UpdateItem",
"dynamodb:DeleteItem",
"batch:SubmitJob",
"batch:DescribeJobs",
"batch:TerminateJob",
"glue:StartJobRun",
"glue:GetJobRun",
"glue:GetJobRuns",
"glue:BatchStopJobRun"
],
"Resource": "*"
},
{
"Effect": "Allow",
"Action": [
"iam:PassRole"
],
"Resource": "*",
"Condition": {
"StringEquals": {
"iam:PassedToService": "sagemaker.amazonaws.com"
}
}
},
{
"Effect": "Allow",
"Action": [
"events:PutTargets",
"events:PutRule",
"events:DescribeRule"
],
"Resource": [
"arn:aws:events:*:*:rule/StepFunctionsGetEventsForSageMakerTrainingJobsRule",
"arn:aws:events:*:*:rule/StepFunctionsGetEventsForSageMakerTransformJobsRule",
"arn:aws:events:*:*:rule/StepFunctionsGetEventsForSageMakerTuningJobsRule",
"arn:aws:events:*:*:rule/StepFunctionsGetEventsForECSTaskRule",
"arn:aws:events:*:*:rule/StepFunctionsGetEventsForBatchJobsRule"
]
}
]
}
```
3. Choose **Review policy** and give the policy a name such as `StepFunctionsWorkflowExecutionPolicy`
4. Choose **Create policy**. You will be redirected to the details page for the role.
5. Copy the **Role ARN** at the top of the **Summary**
### Set up a TrainingPipeline <a class="anchor" id="TrainingPipeline">
Although the AWS Step Functions Data Science SDK provides various primitives to build up pipelines from scratch, it also provides prebuilt templates for common workflows, including a [TrainingPipeline](https://aws-step-functions-data-science-sdk.readthedocs.io/en/latest/pipelines.html#stepfunctions.template.pipeline.train.TrainingPipeline) object to simplify creation of a basic pipeline that includes model training and deployment.
The following code cell configures a `pipeline` object with the necessary parameters to define such a simple pipeline:
```
import stepfunctions
from stepfunctions.template.pipeline import TrainingPipeline
# paste the StepFunctionsWorkflowExecutionRole ARN from above
workflow_execution_role = "<execution-role-arn>"
pipeline = TrainingPipeline(
estimator=estimator,
role=workflow_execution_role,
inputs=inputs,
s3_bucket=bucket
)
```
### Visualizing the workflow <a class="anchor" id="VisualizingWorkflow">
You can now view the workflow definition, and visualize it as a graph. This workflow and graph represent your training pipeline from starting a training job to deploying the model.
```
print(pipeline.workflow.definition.to_json(pretty=True))
pipeline.render_graph()
```
### Creating and executing the pipeline <a class="anchor" id="CreatingExecutingPipeline">
Before the workflow can be run for the first time, the pipeline must be created using the `create` method:
```
pipeline.create()
```
Now the workflow can be started by invoking the pipeline's `execute` method:
```
execution = pipeline.execute()
```
Use the `list_executions` method to list all executions for the workflow you created, including the one we just started. After a pipeline is created, it can be executed as many times as needed, for example on a schedule for retraining on new data. (For purposes of this notebook just execute the workflow one time to save resources.) The output will include a list you can click through to access a view of the execution in the AWS Step Functions console.
```
pipeline.workflow.list_executions(html=True)
```
While the workflow is running, you can check workflow progress inside this notebook with the `render_progress` method. This generates a snapshot of the current state of your workflow as it executes. This is a static image. Run the cell again to check progress while the workflow is running.
```
execution.render_progress()
```
#### BEFORE proceeding with the rest of the notebook:
Wait until the workflow completes with status **Succeeded**, which will take a few minutes. You can check status with `render_progress` above, or open in a new browser tab the **Inspect in AWS Step Functions** link in the cell output.
To view the details of the completed workflow execution, from model training through deployment, use the `list_events` method, which lists all events in the workflow execution.
```
execution.list_events(reverse_order=True, html=False)
```
From this list of events, we can extract the name of the endpoint that was set up by the workflow.
```
import re
endpoint_name_suffix = re.search('endpoint\Wtraining\Wpipeline\W([a-zA-Z0-9\W]+?)"', str(execution.list_events())).group(1)
print(endpoint_name_suffix)
```
Once we have the endpoint name, we can use it to instantiate a TensorFlowPredictor object that wraps the endpoint. This TensorFlowPredictor can be used to make predictions, as shown in the following code cell.
#### BEFORE running the following code cell:
Go to the [SageMaker console](https://console.aws.amazon.com/sagemaker/), click **Endpoints** in the left panel, and make sure that the endpoint status is **InService**. If the status is **Creating**, wait until it changes, which may take several minutes.
```
from sagemaker.tensorflow import TensorFlowPredictor
workflow_predictor = TensorFlowPredictor('training-pipeline-' + endpoint_name_suffix)
results = workflow_predictor.predict(x_test[:10])['predictions']
flat_list = [float('%.1f'%(item)) for sublist in results for item in sublist]
print('predictions: \t{}'.format(np.array(flat_list)))
print('target values: \t{}'.format(y_test[:10].round(decimals=1)))
```
Using the AWS Step Functions Data Science SDK, there are many other workflows you can create to automate your machine learning tasks. For example, you could create a workflow to automate model retraining on a periodic basis. Such a workflow could include a test of model quality after training, with subsequent branches for failing (no model deployment) and passing the quality test (model is deployed). Other possible workflow steps include Automatic Model Tuning, data preprocessing with AWS Glue, and more.
For a detailed example of a retraining workflow, see the AWS ML Blog post [Automating model retraining and deployment using the AWS Step Functions Data Science SDK for Amazon SageMaker](https://aws.amazon.com/blogs/machine-learning/automating-model-retraining-and-deployment-using-the-aws-step-functions-data-science-sdk-for-amazon-sagemaker/).
### Cleanup <a class="anchor" id="Cleanup">
The workflow we created above deployed a model to an endpoint. To avoid billing charges for an unused endpoint, you can delete it using the SageMaker console. To do so, go to the [SageMaker console](https://console.aws.amazon.com/sagemaker/). Then click **Endpoints** in the left panel, and select and delete any unneeded endpoints in the list.
## Extensions <a class="anchor" id="Extensions">
We've covered a lot of content in this notebook: SageMaker Processing for data transformation, Local Mode for prototyping training and inference code, Automatic Model Tuning, and SageMaker hosted training and inference. These are central elements for most deep learning workflows in SageMaker. Additionally, we examined how the AWS Step Functions Data Science SDK helps automate deep learning workflows after completion of the prototyping phase of a project.
Besides all of the SageMaker features explored above, there are many other features that may be applicable to your project. For example, to handle common problems during deep learning model training such as vanishing or exploding gradients, **SageMaker Debugger** is useful. To manage common problems such as data drift after a model is in production, **SageMaker Model Monitor** can be applied.
|
github_jupyter
|
```
import safenet
safenet.setup_logger(file_level=safenet.log_util.WARNING)
myApp = safenet.App()
myAuth_,addData=safenet.safe_utils.AuthReq(myApp.ffi_app.NULL,0,0,id=b'crappy_chat_reloaded',scope=b'noScope'
,name=b'i_love_it',vendor=b'no_vendor',app_container=True,ffi=myApp.ffi_app)
encodedAuth = myApp.encode_authentication(myAuth_)
encodedAuth
grantedAuth = myApp.sysUri.quickSetup(myAuth_,encodedAuth)
grantedAuth
grantedAuth='bAEAAAADIADW4EAAAAAAAAAAAAAQAAAAAAAAAAAEFNJ53ABPX5QW524YYAMEN7T4MJJVIYH656RYZ4FCSZ4TUT7DX3AQAAAAAAAAAAADZO24ITUIIFUWNIUPYODCATWPRBZIBHLD4B6DGFUJDNASIIFYX5MQAAAAAAAAAAAG7B6WQXKW3UPQET62ZWDRY3U7NEYKRWBPQHLYJHTOOYIPPGOWKFFAAAAAAAAAAAACGBOVXSSUKP2Z7YMG5JJDC7BNTUU3YD4SBOBYN3CWRJXGCXLOSFTPQ7LILVLN2HYCJ7NM3BY4N2PWSMFI3AXYDV4ETZXHMEHXTHLFCSIAAAAAAAAAAAAJDOR7QCDWE2VXANINUIE4NYFTIAT66JFQN7B7ALHOV3QYVIYSGQIAAAAAAAAAAABK6S5AF4FRXH4AOBERKM65IJZZNGEILVD3GSDMQBIV4GP2XE5JHQGIAAAAAAAAAAAIAAAAAAAAAAABRG44C4NRSFY3TMLRYHI2TIOBTCMAAAAAAAAAAAMJTHAXDMOBOGE4DKLRSGE4DUNJUHAZREAAAAAAAAAAAGEZTQLRWHAXDCOBRFY2TOORVGQ4DGEQAAAAAAAAAAAYTGOBOGY4C4MJYGEXDMMB2GU2DQMYSAAAAAAAAAAADCMZYFY3DQLRRHAYS4OBWHI2TIOBTCIAAAAAAAAAAAMJTHAXDMOBOGE4DCLRYG45DKNBYGMJQAAAAAAAAAABRGM4C4NRYFYYTQMJOGE3DQORVGQ4DGEYAAAAAAAAAAAYTGOBOGY4C4MJYGEXDCNZWHI2TIOBTCMAAAAAAAAAAAMJTHAXDMOBOGE4DCLRRG44TUNJUHAZRGAAAAAAAAAAAGEZTQLRWHAXDCOBRFYYTQMB2GU2DQMYTAAAAAAAAAAADCMZYFY3DQLRRHAYS4MJYGI5DKNBYGMJQAAAAAAAAAABRGM4C4NRYFYYTQMJOGI2DEORVGQ4DGEYAAAAAAAAAAAYTGOBOGY4C4MJYGEXDENBTHI2TIOBTCMAAAAAAAAAAAMJTHAXDMOBOGE4DCLRSGQ4TUNJUHAZREAAAAAAAAAAAGEZTQLRWHAXDCOBZFYYTIORVGQ4DGEQAAAAAAAAAAAYTGOBOGY4C4MJYHEXDCNJ2GU2DQMYSAAAAAAAAAAADCMZYFY3DQLRRHA4S4MJXHI2TIOBTCIAAAAAAAAAAAMJTHAXDMOBOGE4DSLRRHA5DKNBYGMJAAAAAAAAAAABRGM4C4NRYFYYTQOJOGE4TUNJUHAZREAAAAAAAAAAAGEZTQLRWHAXDCOBZFYZTCORVGQ4DGEQAAAAAAAAAAAYTGOBOGY4C4MJYHEXDGNB2GU2DQMYSAAAAAAAAAAADCMZYFY3DQLRRHA4S4MZWHI2TIOBTCIAAAAAAAAAAAMJTHAXDMOBOGE4DSLRTHA5DKNBYGMJAAAAAAAAAAABRGM4C4NRYFYYTQOJOGM4TUNJUHAZRCAAAAAAAAAAAGQ3C4MJQGEXDKLRRG44TUNJUHAZQC2YVAAAAAAAAAEDQAAAAAAAAAADBNRYGQYK7GIAOWVHBIXIX3YGQAZIQREUXG4475KAEQOJARMHK5Z3DWBIVRXPEAVMYHIAAAAAAAAABQAAAAAAAAAAAIDF2MO3P472PTSCK3IIOW43ZICJR4Q4P5ZR6UWABAAAAAAAAAAABIAAAAAAAAAAAMFYHA4ZPORSXG5CQOJXWO4TBNVHGC3LFO7DUGA44PHQPW2LQGIPOFH34XS3SO3V3X6S3LX7ETSBIRY3TCAHJQOQAAAAAAAAAAEQAAAAAAAAAAAEIJOL5UDCOQRO3N2G6CFLCDF4ACW3LH2ON27YBAOOC7G4YGV25S4MAAAAAAAAAAAGJ6FXG5Y7A2Z5GTAO7H5APZ2ALENSBY2J7T4QNKAAFAAAAAAAAAAAAAAAAAAAQAAAAAIAAAAADAAAAABAAAAAAA'
myApp.setup_app(myAuth_,grantedAuth)
signKey = myApp.get_pub_key_handle()
signKey
```
---
### now we have an app and can start doing stuff
---
### creating a mutable Object
```
myMutable = myApp.mData()
```
### define Entries and drop them onto Safe
```
import datetime
now = datetime.datetime.utcnow().strftime('%Y-%m-%d - %H:%M:%S')
myName = 'Welcome to the SAFE Network'
text = 'free speech and free knowledge to the world!'
timeUser = f'{now} {myName}'
entries={timeUser:text}
```
entries={'firstkey':'this is awesome',
'secondKey':'and soon it should be',
'thirdKey':'even easier to use safe with python',
'i love safe':'and this is just the start',
'thisWasUploaded at':datetime.datetime.utcnow().strftime('%Y-%m-%d - %H:%M:%S UTC'),
'additionalEntry':input('enter your custom value here: ')}
```
infoData = myMutable.new_random_public(777,signKey,entries)
print(safenet.safe_utils.getXorAddresOfMutable(infoData,myMutable.ffi_app))
additionalEntries={'this wasnt here':'before'}
additionalEntries={'baduff':'another entry'}
myMutable.insertEntries(infoData,additionalEntries)
with open('testfile','wb') as f:
f.write(myMutable.ffi_app.buffer(infoData)[:])
with open('testfile','rb') as f:
infoData= safenet.safe_utils.getffiMutable(f.read(),myMutable.ffi_app)
myMutable.ffi_app.buffer(infoData)[:]
mutableBytes = b'H\x8f\x08x}\xc5D]U\xeeW\x08\xe0\xb4\xaau\x94\xd4\x8a\x0bz\x06h\xe3{}\xd1\x06\x843\x01P[t\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x00\x007\xdbNV\x00\x00'
infoData= safenet.safe_utils.getffiMutable(mutableBytes,myMutable.ffi_app)
infoData
def getNewEntries(lastState,newState):
newEntries = {}
for additional in [item for item in newState if item not in lastState]:
newEntries[additional]=newState[additional]
return newEntries, newState
```
lastState={}
additionalEntries, lastState = getNewEntries(lastState,myMutable.getCurrentState(infoData))
additionalEntries
```
import queue
import time
from threading import Thread
import datetime
import sys
from PyQt5.QtWidgets import (QWidget, QPushButton, QTextBrowser,QLineEdit,
QHBoxLayout, QVBoxLayout, QApplication)
class Example(QWidget):
def __init__(self):
super().__init__()
self.lineedit1 = QLineEdit("anon")
self.browser = QTextBrowser()
self.lineedit = QLineEdit("Type a message and press Enter")
self.lineedit.selectAll()
self.setWindowTitle("crappychat_reloaded")
vbox = QVBoxLayout()
vbox.addWidget(self.lineedit1)
vbox.addWidget(self.browser)
vbox.addWidget(self.lineedit)
self.setLayout(vbox)
self.setGeometry(300, 300, 900, 600)
self.show()
self.lineedit.setFocus()
self.lineedit.returnPressed.connect(self.updateUi)
self.messageQueue = queue.Queue()
t = Thread(name='updateThread', target=self.updateBrowser)
t.start()
def updateUi(self):
try:
now = datetime.datetime.utcnow().strftime('%Y-%m-%d - %H:%M:%S')
myName = self.lineedit1.text()
text = self.lineedit.text()
timeUser = f'{now} {myName}'
additionalEntries={timeUser:text}
self.messageQueue.put(additionalEntries)
#self.browser.append(f"<b>{timeUser}</b>: {text}")
self.lineedit.clear()
except:
self.browser.append("<font color=red>{0} is invalid!</font>"
.format(text))
def updateBrowser(self):
lastState={}
while True:
try:
if not self.messageQueue.empty():
newEntries = self.messageQueue.get()
myMutable.insertEntries(infoData,newEntries)
additionalEntries, lastState = getNewEntries(lastState,myMutable.getCurrentState(infoData))
for entry in additionalEntries:
entry_string = entry.decode()
value_string = additionalEntries[entry].decode()
self.browser.append(f"<b>{entry_string}</b>: {value_string}")
self.browser.ensureCursorVisible()
except:
pass
time.sleep(2)
if __name__ == '__main__':
app = QApplication(sys.argv)
ex = Example()
sys.exit(app.exec_())
```
|
github_jupyter
|
# Reader - Implantação
Este componente utiliza um modelo de QA pré-treinado em Português com o dataset SQuAD v1.1, é um modelo de domínio público disponível em [Hugging Face](https://huggingface.co/pierreguillou/bert-large-cased-squad-v1.1-portuguese).<br>
Seu objetivo é encontrar a resposta de uma ou mais perguntas de acordo com uma lista de contextos distintos.
A tabela de dados de entrada deve possuir uma coluna de contextos, em que cada linha representa um contexto diferente, e uma coluna de perguntas em que cada linha representa uma pergunta a ser realizada. Note que para cada pergunta serão utilizados todos os contextos fornecidos para realização da inferência, e portanto, podem haver bem mais contextos do que perguntas.
Obs: Este componente utiliza recursos da internet, portanto é importante estar conectado à rede para que este componente funcione corretamente.<br>
### **Em caso de dúvidas, consulte os [tutoriais da PlatIAgro](https://platiagro.github.io/tutorials/).**
## Declaração de Classe para Predições em Tempo Real
A tarefa de implantação cria um serviço REST para predições em tempo-real.<br>
Para isso você deve criar uma classe `Model` que implementa o método `predict`.
```
%%writefile Model.py
import joblib
import numpy as np
import pandas as pd
from reader import Reader
class Model:
def __init__(self):
self.loaded = False
def load(self):
# Load artifacts
artifacts = joblib.load("/tmp/data/reader.joblib")
self.model_parameters = artifacts["model_parameters"]
self.inference_parameters = artifacts["inference_parameters"]
# Initialize reader
self.reader = Reader(**self.model_parameters)
# Set model loaded
self.loaded = True
print("Loaded model")
def class_names(self):
column_names = list(self.inference_parameters['output_columns'])
return column_names
def predict(self, X, feature_names, meta=None):
if not self.loaded:
self.load()
# Convert to dataframe
if feature_names != []:
df = pd.DataFrame(X, columns = feature_names)
df = df[self.inference_parameters['input_columns']]
else:
df = pd.DataFrame(X, columns = self.inference_parameters['input_columns'])
# Predict answers #
# Iterate over dataset
for idx, row in df.iterrows():
# Get question
question = row[self.inference_parameters['question_column_name']]
# Get context
context = row[self.inference_parameters['context_column_name']]
# Make prediction
answer, probability, _ = self.reader([question], [context])
# Save to df
df.at[idx, self.inference_parameters['answer_column_name']] = answer[0]
df.at[idx, self.inference_parameters['proba_column_name']] = probability[0]
# Retrieve Only Best Answer #
# Initializate best df
best_df = pd.DataFrame(columns=df.columns)
# Get unique questions
unique_questions = df[self.inference_parameters['question_column_name']].unique()
# Iterate over each unique question
for question in unique_questions:
# Filter df
question_df = df[df[self.inference_parameters['question_column_name']] == question]
# Sort by score (descending)
question_df = question_df.sort_values(by=self.inference_parameters['proba_column_name'], ascending=False).reset_index(drop=True)
# Append best ansewer to output df
best_df = pd.concat((best_df,pd.DataFrame(question_df.loc[0]).T)).reset_index(drop=True)
if self.inference_parameters['keep_best'] == 'sim':
return best_df.values
else:
return df.values
```
|
github_jupyter
|
# Estimator validation
This notebook contains code to generate Figure 2 of the paper.
This notebook also serves to compare the estimates of the re-implemented scmemo with sceb package from Vasilis.
```
import pandas as pd
import matplotlib.pyplot as plt
import scanpy as sc
import scipy as sp
import itertools
import numpy as np
import scipy.stats as stats
from scipy.integrate import dblquad
import seaborn as sns
from statsmodels.stats.multitest import fdrcorrection
import imp
pd.options.display.max_rows = 999
pd.set_option('display.max_colwidth', -1)
import pickle as pkl
import time
import matplotlib as mpl
mpl.rcParams['pdf.fonttype'] = 42
mpl.rcParams['ps.fonttype'] = 42
import matplotlib.pylab as pylab
params = {'legend.fontsize': 'x-small',
'axes.labelsize': 'medium',
'axes.titlesize':'medium',
'figure.titlesize':'medium',
'xtick.labelsize':'xx-small',
'ytick.labelsize':'xx-small'}
pylab.rcParams.update(params)
import sys
sys.path.append('/data/home/Github/scrna-parameter-estimation/dist/schypo-0.0.0-py3.7.egg')
import schypo
import schypo.simulate as simulate
import sys
sys.path.append('/data/home/Github/single_cell_eb/')
sys.path.append('/data/home/Github/single_cell_eb/sceb/')
import scdd
data_path = '/data/parameter_estimation/'
fig_path = '/data/home/Github/scrna-parameter-estimation/figures/fig3/'
```
### Check 1D estimates of `sceb` with `scmemo`
Using the Poisson model. The outputs should be identical, this is for checking the implementation.
```
data = sp.sparse.csr_matrix(simulate.simulate_transcriptomes(100, 20))
adata = sc.AnnData(data)
size_factors = scdd.dd_size_factor(adata)
Nr = data.sum(axis=1).mean()
_, M_dd = scdd.dd_1d_moment(adata, size_factor=scdd.dd_size_factor(adata), Nr=Nr)
var_scdd = scdd.M_to_var(M_dd)
print(var_scdd)
imp.reload(estimator)
mean_scmemo, var_scmemo = estimator._poisson_1d(data, data.shape[0], estimator._estimate_size_factor(data))
print(var_scmemo)
df = pd.DataFrame()
df['size_factor'] = size_factors
df['inv_size_factor'] = 1/size_factors
df['inv_size_factor_sq'] = 1/size_factors**2
df['expr'] = data[:, 0].todense().A1
precomputed_size_factors = df.groupby('expr')['inv_size_factor'].mean(), df.groupby('expr')['inv_size_factor_sq'].mean()
imp.reload(estimator)
expr, count = np.unique(data[:, 0].todense().A1, return_counts=True)
print(estimator._poisson_1d((expr, count), data.shape[0], precomputed_size_factors))
```
### Check 2D estimates of `sceb` and `scmemo`
Using the Poisson model. The outputs should be identical, this is for checking the implementation.
```
data = sp.sparse.csr_matrix(simulate.simulate_transcriptomes(1000, 4))
adata = sc.AnnData(data)
size_factors = scdd.dd_size_factor(adata)
mean_scdd, cov_scdd, corr_scdd = scdd.dd_covariance(adata, size_factors)
print(cov_scdd)
imp.reload(estimator)
cov_scmemo = estimator._poisson_cov(data, data.shape[0], size_factors, idx1=[0, 1, 2], idx2=[1, 2, 3])
print(cov_scmemo)
expr, count = np.unique(data[:, :2].toarray(), return_counts=True, axis=0)
df = pd.DataFrame()
df['size_factor'] = size_factors
df['inv_size_factor'] = 1/size_factors
df['inv_size_factor_sq'] = 1/size_factors**2
df['expr1'] = data[:, 0].todense().A1
df['expr2'] = data[:, 1].todense().A1
precomputed_size_factors = df.groupby(['expr1', 'expr2'])['inv_size_factor'].mean(), df.groupby(['expr1', 'expr2'])['inv_size_factor_sq'].mean()
cov_scmemo = estimator._poisson_cov((expr[:, 0], expr[:, 1], count), data.shape[0], size_factor=precomputed_size_factors)
print(cov_scmemo)
```
### Extract parameters from interferon dataset
```
adata = sc.read(data_path + 'interferon_filtered.h5ad')
adata = adata[adata.obs.cell_type == 'CD4 T cells - ctrl']
data = adata.X.copy()
relative_data = data.toarray()/data.sum(axis=1)
q = 0.07
x_param, z_param, Nc, good_idx = schypo.simulate.extract_parameters(adata.X, q=q, min_mean=q)
imp.reload(simulate)
transcriptome = simulate.simulate_transcriptomes(
n_cells=10000,
means=z_param[0],
variances=z_param[1],
corr=x_param[2],
Nc=Nc)
relative_transcriptome = transcriptome/transcriptome.sum(axis=1).reshape(-1, 1)
qs, captured_data = simulate.capture_sampling(transcriptome, q=q, q_sq=q**2+1e-10)
def qqplot(x, y, s=1):
plt.scatter(
np.quantile(x, np.linspace(0, 1, 1000)),
np.quantile(y, np.linspace(0, 1, 1000)),
s=s)
plt.plot(x, x, lw=1, color='m')
plt.figure(figsize=(8, 2));
plt.subplots_adjust(wspace=0.2);
plt.subplot(1, 3, 1);
sns.distplot(np.log(captured_data.mean(axis=0)), hist=False, label='Simulated')
sns.distplot(np.log(data[:, good_idx].toarray().var(axis=0)), hist=False, label='Real')
plt.xlabel('Log(mean)')
plt.subplot(1, 3, 2);
sns.distplot(np.log(captured_data.var(axis=0)), hist=False)
sns.distplot(np.log(data[:, good_idx].toarray().var(axis=0)), hist=False)
plt.xlabel('Log(variance)')
plt.subplot(1, 3, 3);
sns.distplot(np.log(captured_data.sum(axis=1)), hist=False)
sns.distplot(np.log(data.toarray().sum(axis=1)), hist=False)
plt.xlabel('Log(total UMI count)')
plt.savefig(figpath + 'simulation_stats.png', bbox_inches='tight')
```
### Compare datasets generated by Poisson and hypergeometric processes
```
_, poi_captured = simulate.capture_sampling(transcriptome, q=q, process='poisson')
_, hyper_captured = simulate.capture_sampling(transcriptome, q=q, process='hyper')
q_list = [0.05, 0.1, 0.2, 0.3, 0.5]
plt.figure(figsize=(8, 2))
plt.subplots_adjust(wspace=0.3)
for idx, q in enumerate(q_list):
_, poi_captured = simulate.capture_sampling(transcriptome, q=q, process='poisson')
_, hyper_captured = simulate.capture_sampling(transcriptome, q=q, process='hyper')
relative_poi_captured = poi_captured/poi_captured.sum(axis=1).reshape(-1, 1)
relative_hyper_captured = hyper_captured/hyper_captured.sum(axis=1).reshape(-1, 1)
poi_corr = np.corrcoef(relative_poi_captured, rowvar=False)
hyper_corr = np.corrcoef(relative_hyper_captured, rowvar=False)
sample_idx = np.random.choice(poi_corr.ravel().shape[0], 100000)
plt.subplot(1, len(q_list), idx+1)
plt.scatter(poi_corr.ravel()[sample_idx], hyper_corr.ravel()[sample_idx], s=1, alpha=1)
plt.plot([-1, 1], [-1, 1], 'm', lw=1)
# plt.xlim([-0.3, 0.4])
# plt.ylim([-0.3, 0.4])
if idx != 0:
plt.yticks([])
plt.title('q={}'.format(q))
plt.savefig(figpath + 'poi_vs_hyp_sim_corr.png', bbox_inches='tight')
```
### Compare Poisson vs HG estimators
```
def compare_esimators(q, plot=False, true_data=None, var_q=1e-10):
q_sq = var_q + q**2
true_data = schypo.simulate.simulate_transcriptomes(1000, 1000, correlated=True) if true_data is None else true_data
true_relative_data = true_data / true_data.sum(axis=1).reshape(-1, 1)
qs, captured_data = schypo.simulate.capture_sampling(true_data, q, q_sq)
Nr = captured_data.sum(axis=1).mean()
captured_relative_data = captured_data/captured_data.sum(axis=1).reshape(-1, 1)
adata = sc.AnnData(sp.sparse.csr_matrix(captured_data))
sf = schypo.estimator._estimate_size_factor(adata.X, 'hyper_relative', total=True)
good_idx = (captured_data.mean(axis=0) > q)
# True moments
m_true, v_true, corr_true = true_relative_data.mean(axis=0), true_relative_data.var(axis=0), np.corrcoef(true_relative_data, rowvar=False)
rv_true = v_true/m_true**2#schypo.estimator._residual_variance(m_true, v_true, schypo.estimator._fit_mv_regressor(m_true, v_true))
# Compute 1D moments
m_obs, v_obs = captured_relative_data.mean(axis=0), captured_relative_data.var(axis=0)
rv_obs = v_obs/m_obs**2#schypo.estimator._residual_variance(m_obs, v_obs, schypo.estimator._fit_mv_regressor(m_obs, v_obs))
m_poi, v_poi = schypo.estimator._poisson_1d_relative(adata.X, size_factor=sf, n_obs=true_data.shape[0])
rv_poi = v_poi/m_poi**2#schypo.estimator._residual_variance(m_poi, v_poi, schypo.estimator._fit_mv_regressor(m_poi, v_poi))
m_hyp, v_hyp = schypo.estimator._hyper_1d_relative(adata.X, size_factor=sf, n_obs=true_data.shape[0], q=q)
rv_hyp = v_hyp/m_hyp**2#schypo.estimator._residual_variance(m_hyp, v_hyp, schypo.estimator._fit_mv_regressor(m_hyp, v_hyp))
# Compute 2D moments
corr_obs = np.corrcoef(captured_relative_data, rowvar=False)
# corr_obs = corr_obs[np.triu_indices(corr_obs.shape[0])]
idx1 = np.array([i for i,j in itertools.combinations(range(adata.shape[1]), 2) if good_idx[i] and good_idx[j]])
idx2 = np.array([j for i,j in itertools.combinations(range(adata.shape[1]), 2) if good_idx[i] and good_idx[j]])
sample_idx = np.random.choice(idx1.shape[0], 10000)
idx1 = idx1[sample_idx]
idx2 = idx2[sample_idx]
corr_true = corr_true[(idx1, idx2)]
corr_obs = corr_obs[(idx1, idx2)]
cov_poi = schypo.estimator._poisson_cov_relative(adata.X, n_obs=adata.shape[0], size_factor=sf, idx1=idx1, idx2=idx2)
cov_hyp = schypo.estimator._hyper_cov_relative(adata.X, n_obs=adata.shape[0], size_factor=sf, idx1=idx1, idx2=idx2, q=q)
corr_poi = schypo.estimator._corr_from_cov(cov_poi, v_poi[idx1], v_poi[idx2])
corr_hyp = schypo.estimator._corr_from_cov(cov_hyp, v_hyp[idx1], v_hyp[idx2])
corr_poi[np.abs(corr_poi) > 1] = np.nan
corr_hyp[np.abs(corr_hyp) > 1] = np.nan
mean_list = [m_obs, m_poi, m_hyp]
var_list = [rv_obs, rv_poi, rv_hyp]
corr_list = [corr_obs, corr_poi, corr_hyp]
estimated_list = [mean_list, var_list, corr_list]
true_list = [m_true, rv_true, corr_true]
if plot:
count = 0
for j in range(3):
for i in range(3):
plt.subplot(3, 3, count+1)
if i != 2:
plt.scatter(
np.log(true_list[i][good_idx]),
np.log(estimated_list[i][j][good_idx]),
s=0.1)
plt.plot(np.log(true_list[i][good_idx]), np.log(true_list[i][good_idx]), linestyle='--', color='m')
plt.xlim(np.log(true_list[i][good_idx]).min(), np.log(true_list[i][good_idx]).max())
plt.ylim(np.log(true_list[i][good_idx]).min(), np.log(true_list[i][good_idx]).max())
else:
x = true_list[i]
y = estimated_list[i][j]
print(x.shape, y.shape)
plt.scatter(
x,
y,
s=0.1)
plt.plot([-1, 1], [-1, 1],linestyle='--', color='m')
plt.xlim(-1, 1);
plt.ylim(-1, 1);
# if not (i == j):
# plt.yticks([]);
# plt.xticks([]);
if i == 1 or i == 0:
print((np.log(true_list[i][good_idx]) > np.log(estimated_list[i][j][good_idx])).mean())
count += 1
else:
return qs, good_idx, estimated_list, true_list
import matplotlib.pylab as pylab
params = {'legend.fontsize': 'small',
'axes.labelsize': 'medium',
'axes.titlesize':'medium',
'figure.titlesize':'medium',
'xtick.labelsize':'xx-small',
'ytick.labelsize':'xx-small'}
pylab.rcParams.update(params)
```
```
true_data = schypo.simulate.simulate_transcriptomes(n_cells=10000, means=z_param[0], variances=z_param[1], Nc=Nc)
q = 0.025
plt.figure(figsize=(4, 4))
plt.subplots_adjust(wspace=0.5, hspace=0.5)
compare_esimators(q, plot=True, true_data=true_data)
plt.savefig(fig_path + 'poi_vs_hyper_scatter_rv_2.5.png', bbox_inches='tight', dpi=1200)
q = 0.4
plt.figure(figsize=(4, 4))
plt.subplots_adjust(wspace=0.5, hspace=0.5)
compare_esimators(q, plot=True, true_data=true_data)
plt.savefig(fig_path + 'poi_vs_hyper_scatter_rv_40.png', bbox_inches='tight', dpi=1200)
def compute_mse(x, y, log=True):
if log:
return np.nanmean(np.abs(np.log(x)-np.log(y)))
else:
return np.nanmean(np.abs(x-y))
def concordance(x, y, log=True):
if log:
a = np.log(x)
b = np.log(y)
else:
a = x
b = y
cond = np.isfinite(a) & np.isfinite(b)
a = a[cond]
b = b[cond]
cmat = np.cov(a, b)
return 2*cmat[0,1]/(cmat[0,0] + cmat[1,1] + (a.mean()-b.mean())**2)
m_mse_list, v_mse_list, c_mse_list = [], [], []
# true_data = schypo.simulate.simulate_transcriptomes(n_cells=10000, means=z_param[0], variances=z_param[1],
# Nc=Nc)
q_list = [0.01, 0.025, 0.1, 0.15, 0.3, 0.5, 0.7, 0.99]
qs_list = []
for q in q_list:
qs, good_idx, est, true = compare_esimators(q, plot=False, true_data=true_data)
qs_list.append(qs)
m_mse_list.append([concordance(x[good_idx], true[0][good_idx]) for x in est[0]])
v_mse_list.append([concordance(x[good_idx], true[1][good_idx]) for x in est[1]])
c_mse_list.append([concordance(x, true[2], log=False) for x in est[2]])
m_mse_list, v_mse_list, c_mse_list = np.array(m_mse_list), np.array(v_mse_list), np.array(c_mse_list)
import matplotlib.pylab as pylab
params = {'legend.fontsize': 'small',
'axes.labelsize': 'medium',
'axes.titlesize':'medium',
'figure.titlesize':'medium',
'xtick.labelsize':'small',
'ytick.labelsize':'small'}
pylab.rcParams.update(params)
plt.figure(figsize=(8, 3))
plt.subplots_adjust(wspace=0.5)
plt.subplot(1, 3, 1)
plt.plot(q_list[1:], m_mse_list[:, 0][1:], '-o')
# plt.legend(['Naive,\nPoisson,\nHG'])
plt.ylabel('CCC log(mean)')
plt.xlabel('overall UMI efficiency (q)')
plt.subplot(1, 3, 2)
plt.plot(q_list[2:], v_mse_list[:, 0][2:], '-o')
plt.plot(q_list[2:], v_mse_list[:, 1][2:], '-o')
plt.plot(q_list[2:], v_mse_list[:, 2][2:], '-o')
plt.legend(['Naive', 'Poisson', 'HG'], ncol=3, loc='upper center', bbox_to_anchor=(0.4,1.15))
plt.ylabel('CCC log(variance)')
plt.xlabel('overall UMI efficiency (q)')
plt.subplot(1, 3, 3)
plt.plot(q_list[2:], c_mse_list[:, 0][2:], '-o')
plt.plot(q_list[2:], c_mse_list[:, 1][2:], '-o')
plt.plot(q_list[2:], c_mse_list[:, 2][2:], '-o')
# plt.legend(['Naive', 'Poisson', 'HG'])
plt.ylabel('CCC correlation')
plt.xlabel('overall UMI efficiency (q)')
plt.savefig(fig_path + 'poi_vs_hyper_rv_ccc.pdf', bbox_inches='tight')
plt.figure(figsize=(1, 1.3))
plt.plot(q_list, v_mse_list[:, 0], '-o', ms=4)
plt.plot(q_list, v_mse_list[:, 1], '-o', ms=4)
plt.plot(q_list, v_mse_list[:, 2], '-o', ms=4)
plt.savefig(fig_path + 'poi_vs_hyper_ccc_var_rv_inset.pdf', bbox_inches='tight')
plt.figure(figsize=(1, 1.3))
plt.plot(q_list, c_mse_list[:, 0], '-o', ms=4)
plt.plot(q_list, c_mse_list[:, 1], '-o', ms=4)
plt.plot(q_list, c_mse_list[:, 2], '-o', ms=4)
plt.savefig(fig_path + 'poi_vs_hyper_ccc_corr_inset.pdf', bbox_inches='tight')
```
|
github_jupyter
|
# TRTR and TSTR Results Comparison
```
#import libraries
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
pd.set_option('precision', 4)
```
## 1. Create empty dataset to save metrics differences
```
DATA_TYPES = ['Real','GM','SDV','CTGAN','WGANGP']
SYNTHESIZERS = ['GM','SDV','CTGAN','WGANGP']
ml_models = ['RF','KNN','DT','SVM','MLP']
```
## 2. Read obtained results when TRTR and TSTR
```
FILEPATHS = {'Real' : 'RESULTS/models_results_real.csv',
'GM' : 'RESULTS/models_results_gm.csv',
'SDV' : 'RESULTS/models_results_sdv.csv',
'CTGAN' : 'RESULTS/models_results_ctgan.csv',
'WGANGP' : 'RESULTS/models_results_wgangp.csv'}
#iterate over all datasets filepaths and read each dataset
results_all = dict()
for name, path in FILEPATHS.items() :
results_all[name] = pd.read_csv(path, index_col='model')
results_all
```
## 3. Calculate differences of models
```
metrics_diffs_all = dict()
real_metrics = results_all['Real']
columns = ['data','accuracy_diff','precision_diff','recall_diff','f1_diff']
metrics = ['accuracy','precision','recall','f1']
for name in SYNTHESIZERS :
syn_metrics = results_all[name]
metrics_diffs_all[name] = pd.DataFrame(columns = columns)
for model in ml_models :
real_metrics_model = real_metrics.loc[model]
syn_metrics_model = syn_metrics.loc[model]
data = [model]
for m in metrics :
data.append(abs(real_metrics_model[m] - syn_metrics_model[m]))
metrics_diffs_all[name] = metrics_diffs_all[name].append(pd.DataFrame([data], columns = columns))
metrics_diffs_all
```
## 4. Compare absolute differences
### 4.1. Barplots for each metric
```
metrics = ['accuracy', 'precision', 'recall', 'f1']
metrics_diff = ['accuracy_diff', 'precision_diff', 'recall_diff', 'f1_diff']
colors = ['tab:blue', 'tab:orange', 'tab:green', 'tab:red', 'tab:purple']
barwidth = 0.15
fig, axs = plt.subplots(nrows=1, ncols=4, figsize=(15, 2.5))
axs_idxs = range(4)
idx = dict(zip(metrics + metrics_diff,axs_idxs))
for i in range(0,len(metrics)) :
data = dict()
y_pos = dict()
y_pos[0] = np.arange(len(ml_models))
ax = axs[idx[metrics[i]]]
for k in range(0,len(DATA_TYPES)) :
generator_data = results_all[DATA_TYPES[k]]
data[k] = [0, 0, 0, 0, 0]
for p in range(0,len(ml_models)) :
data[k][p] = generator_data[metrics[i]].iloc[p]
ax.bar(y_pos[k], data[k], color=colors[k], width=barwidth, edgecolor='white', label=DATA_TYPES[k])
y_pos[k+1] = [x + barwidth for x in y_pos[k]]
ax.set_xticks([r + barwidth*2 for r in range(len(ml_models))])
ax.set_xticklabels([])
ax.set_xticklabels(ml_models, fontsize=10)
ax.set_title(metrics[i], fontsize=12)
ax.legend(DATA_TYPES, ncol=5, bbox_to_anchor=(-0.3, -0.2))
fig.tight_layout()
#fig.suptitle('Models performance comparisson Boxplots (TRTR and TSTR) \n Dataset F - Indian Liver Patient', fontsize=18)
fig.savefig('RESULTS/MODELS_METRICS_BARPLOTS.svg', bbox_inches='tight')
metrics = ['accuracy_diff', 'precision_diff', 'recall_diff', 'f1_diff']
colors = ['tab:orange', 'tab:green', 'tab:red', 'tab:purple']
fig, axs = plt.subplots(nrows=1, ncols=4, figsize=(15,2.5))
axs_idxs = range(4)
idx = dict(zip(metrics,axs_idxs))
for i in range(0,len(metrics)) :
data = dict()
ax = axs[idx[metrics[i]]]
for k in range(0,len(SYNTHESIZERS)) :
generator_data = metrics_diffs_all[SYNTHESIZERS[k]]
data[k] = [0, 0, 0, 0, 0]
for p in range(0,len(ml_models)) :
data[k][p] = generator_data[metrics[i]].iloc[p]
ax.plot(data[k], 'o-', color=colors[k], label=SYNTHESIZERS[k])
ax.set_xticks(np.arange(len(ml_models)))
ax.set_xticklabels(ml_models, fontsize=10)
ax.set_title(metrics[i], fontsize=12)
ax.set_ylim(bottom=-0.01, top=0.28)
ax.grid()
ax.legend(SYNTHESIZERS, ncol=5, bbox_to_anchor=(-0.4, -0.2))
fig.tight_layout()
#fig.suptitle('Models performance comparisson Boxplots (TRTR and TSTR) \n Dataset F - Indian Liver Patient', fontsize=18)
fig.savefig('RESULTS/MODELS_METRICS_DIFFERENCES.svg', bbox_inches='tight')
```
|
github_jupyter
|
# Generating Simpson's Paradox
We have been maually setting, but now we should also be able to generate it more programatically. his notebook will describe how we develop some functions that will be included in the `sp_data_util` package.
```
# %load code/env
# standard imports we use throughout the project
import numpy as np
import pandas as pd
import seaborn as sns
import scipy.stats as stats
import matplotlib.pyplot as plt
import wiggum as wg
import sp_data_util as spdata
from sp_data_util import sp_plot
```
We have been thinking of SP hrough gaussian mixture data, so we'll first work wih that. To cause SP we need he clusters to have an opposite trend of the per cluster covariance.
```
# setup
r_clusters = -.6 # correlation coefficient of clusters
cluster_spread = .8 # pearson correlation of means
p_sp_clusters = .5 # portion of clusters with SP
k = 5 # number of clusters
cluster_size = [2,3]
domain_range = [0, 20, 0, 20]
N = 200 # number of points
p_clusters = [1.0/k]*k
# keep all means in the middle 80%
mu_trim = .2
# sample means
center = [np.mean(domain_range[:2]),np.mean(domain_range[2:])]
mu_transform = np.repeat(np.diff(domain_range)[[0,2]]*(mu_trim),2)
mu_transform[[1,3]] = mu_transform[[1,3]]*-1 # sign flip every other
mu_domain = [d + m_t for d, m_t in zip(domain_range,mu_transform)]
corr = [[1, cluster_spread],[cluster_spread,1]]
d = np.sqrt(np.diag(np.diff(mu_domain)[[0,2]]))
cov = np.dot(d,corr).dot(d)
# sample a lot of means, just for vizualization
# mu = np.asarray([np.random.uniform(*mu_domain[:2],size=k*5), # uniform in x
# np.random.uniform(*mu_domain[2:],size=k*5)]).T # uniform in y
mu = np.random.multivariate_normal(center, cov,k*50)
sns.regplot(mu[:,0], mu[:,1])
plt.axis(domain_range);
# mu
```
However independent sampling isn't really very uniform and we'd like to ensure the clusters are more spread out, so we can use some post processing to thin out close ones.
```
mu_thin = [mu[0]] # keep the first one
p_dist = [1]
# we'll use a gaussian kernel around each to filter and only the closest point matters
dist = lambda mu_c,x: stats.norm.pdf(min(np.sum(np.square(mu_c -x),axis=1)))
for m in mu:
p_keep = 1- dist(mu_thin,m)
if p_keep > .99:
mu_thin.append(m)
p_dist.append(p_keep)
mu_thin = np.asarray(mu_thin)
sns.regplot(mu_thin[:,0], mu_thin[:,1])
plt.axis(domain_range)
```
Now, we can sample points on top of that, also we'll only use the first k
```
sns.regplot(mu_thin[:k,0], mu_thin[:k,1])
plt.axis(domain_range)
```
Keeping only a few, we can end up with ones in the center, but if we sort them by the distance to the ones previously selected, we get them spread out a little more
```
# sort by distance
mu_sort, p_sort = zip(*sorted(zip(mu_thin,p_dist),
key = lambda x: x[1], reverse =True))
mu_sort = np.asarray(mu_sort)
sns.regplot(mu_sort[:k,0], mu_sort[:k,1])
plt.axis(domain_range)
# cluster covariance
cluster_corr = np.asarray([[1,r_clusters],[r_clusters,1]])
cluster_std = np.diag(np.sqrt(cluster_size))
cluster_cov = np.dot(cluster_std,cluster_corr).dot(cluster_std)
# sample from a GMM
z = np.random.choice(k,N,p_clusters)
x = np.asarray([np.random.multivariate_normal(mu_sort[z_i],cluster_cov) for z_i in z])
# make a dataframe
latent_df = pd.DataFrame(data=x,
columns = ['x1', 'x2'])
# code cluster as color and add it a column to the dataframe
latent_df['color'] = z
sp_plot(latent_df,'x1','x2','color')
```
We might not want all of the clusters to have the reveral though, so we can also sample the covariances
```
# cluster covariance
cluster_std = np.diag(np.sqrt(cluster_size))
cluster_corr_sp = np.asarray([[1,r_clusters],[r_clusters,1]]) # correlation with sp
cluster_cov_sp = np.dot(cluster_std,cluster_corr_sp).dot(cluster_std) #cov with sp
cluster_corr = np.asarray([[1,-r_clusters],[-r_clusters,1]]) #correlation without sp
cluster_cov = np.dot(cluster_std,cluster_corr).dot(cluster_std) #cov wihtout sp
cluster_covs = [cluster_corr_sp, cluster_corr]
# sample the[0,1] k times
c_sp = np.random.choice(2,k,p=[p_sp_clusters,1-p_sp_clusters])
# sample from a GMM
z = np.random.choice(k,N,p_clusters)
x = np.asarray([np.random.multivariate_normal(mu_sort[z_i],cluster_covs[c_sp[z_i]]) for z_i in z])
# make a dataframe
latent_df = pd.DataFrame(data=x,
columns = ['x1', 'x2'])
# code cluster as color and add it a column to the dataframe
latent_df['color'] = z
sp_plot(latent_df,'x1','x2','color')
[p_sp_clusters,1-p_sp_clusters]
c_sp
```
We'll call this construction of SP `geometric_2d_gmm_sp` and it's included in the `sp_data_utils` module now, so it can be called as follows. We'll change the portion of clusters with SP to 1, to ensure that all are SP.
```
type(r_clusters)
type(cluster_size)
type(cluster_spread)
type(p_sp_clusters)
type(domain_range)
type(p_clusters)
p_sp_clusters = .9
sp_df2 = spdata.geometric_2d_gmm_sp(r_clusters,cluster_size,cluster_spread,
p_sp_clusters, domain_range,k,N,p_clusters)
sp_plot(sp_df2,'x1','x2','color')
```
With this, we can start to see how the parameters control a little
```
# setup
r_clusters = -.4 # correlation coefficient of clusters
cluster_spread = .8 # pearson correlation of means
p_sp_clusters = .6 # portion of clusters with SP
k = 5 # number of clusters
cluster_size = [4,4]
domain_range = [0, 20, 0, 20]
N = 200 # number of points
p_clusters = [.5, .2, .1, .1, .1]
sp_df3 = spdata.geometric_2d_gmm_sp(r_clusters,cluster_size,cluster_spread,
p_sp_clusters, domain_range,k,N,p_clusters)
sp_plot(sp_df3,'x1','x2','color')
```
We might want to add multiple views, so we added a function that takes the same parameters or lists to allow each view to have different parameters. We'll look first at just two views with the same parameters, both as one another and as above
```
many_sp_df = spdata.geometric_indep_views_gmm_sp(2,r_clusters,cluster_size,cluster_spread,p_sp_clusters,
domain_range,k,N,p_clusters)
sp_plot(many_sp_df,'x1','x2','A')
sp_plot(many_sp_df,'x3','x4','B')
many_sp_df.head()
```
We can also look at the pairs of variables that we did not design SP into and see that they have vey different structure
```
# f, ax_grid = plt.subplots(2,2) # , fig_size=(10,10)
sp_plot(many_sp_df,'x1','x4','A')
sp_plot(many_sp_df,'x2','x4','B')
sp_plot(many_sp_df,'x2','x3','B')
sp_plot(many_sp_df,'x1','x3','B')
```
And we can set up the views to be different from one another by design
```
# setup
r_clusters = [.8, -.2] # correlation coefficient of clusters
cluster_spread = [.8, .2] # pearson correlation of means
p_sp_clusters = [.6, 1] # portion of clusters with SP
k = [5,3] # number of clusters
cluster_size = [4,4]
domain_range = [0, 20, 0, 20]
N = 200 # number of points
p_clusters = [[.5, .2, .1, .1, .1],[1.0/3]*3]
many_sp_df_diff = spdata.geometric_indep_views_gmm_sp(2,r_clusters,cluster_size,cluster_spread,p_sp_clusters,
domain_range,k,N,p_clusters)
sp_plot(many_sp_df_diff,'x1','x2','A')
sp_plot(many_sp_df_diff,'x3','x4','B')
many_sp_df.head()
```
And we can run our detection algorithm on this as well.
```
many_sp_df_diff_result = wg.detect_simpsons_paradox(many_sp_df_diff)
many_sp_df_diff_result
```
We designed in SP to occur between attributes `x1` and `x2` with respect to `A` and 2 & 3 in grouby by B, for portions fo the subgroups. We detect other occurences. It can be interesting to exmine trends between the deisnged and spontaneous occurences of SP, so
```
designed_SP = [('x1','x2','A'),('x3','x4','B')]
des = []
for i,r in enumerate(many_sp_df_diff_result[['attr1','attr2','groupbyAttr']].values):
if tuple(r) in designed_SP:
des.append(i)
many_sp_df_diff_result['designed'] = 'no'
many_sp_df_diff_result.loc[des,'designed'] = 'yes'
many_sp_df_diff_result.head()
r_clusters = -.9 # correlation coefficient of clusters
cluster_spread = .6 # pearson correlation of means
p_sp_clusters = .5 # portion of clusters with SP
k = 5 # number of clusters
cluster_size = [5,5]
domain_range = [0, 20, 0, 20]
N = 200 # number of points
p_clusters = [1.0/k]*k
many_sp_df_diff = spdata.geometric_indep_views_gmm_sp(3,r_clusters,cluster_size,cluster_spread,p_sp_clusters,
domain_range,k,N,p_clusters)
sp_plot(many_sp_df_diff,'x1','x2','A')
sp_plot(many_sp_df_diff,'x3','x4','B')
sp_plot(many_sp_df_diff,'x3','x4','A')
many_sp_df_diff.head()
```
|
github_jupyter
|
# A Scientific Deep Dive Into SageMaker LDA
1. [Introduction](#Introduction)
1. [Setup](#Setup)
1. [Data Exploration](#DataExploration)
1. [Training](#Training)
1. [Inference](#Inference)
1. [Epilogue](#Epilogue)
# Introduction
***
Amazon SageMaker LDA is an unsupervised learning algorithm that attempts to describe a set of observations as a mixture of distinct categories. Latent Dirichlet Allocation (LDA) is most commonly used to discover a user-specified number of topics shared by documents within a text corpus. Here each observation is a document, the features are the presence (or occurrence count) of each word, and the categories are the topics. Since the method is unsupervised, the topics are not specified up front, and are not guaranteed to align with how a human may naturally categorize documents. The topics are learned as a probability distribution over the words that occur in each document. Each document, in turn, is described as a mixture of topics.
This notebook is similar to **LDA-Introduction.ipynb** but its objective and scope are a different. We will be taking a deeper dive into the theory. The primary goals of this notebook are,
* to understand the LDA model and the example dataset,
* understand how the Amazon SageMaker LDA algorithm works,
* interpret the meaning of the inference output.
Former knowledge of LDA is not required. However, we will run through concepts rather quickly and at least a foundational knowledge of mathematics or machine learning is recommended. Suggested references are provided, as appropriate.
```
%matplotlib inline
import os, re, tarfile
import boto3
import matplotlib.pyplot as plt
import mxnet as mx
import numpy as np
np.set_printoptions(precision=3, suppress=True)
# some helpful utility functions are defined in the Python module
# "generate_example_data" located in the same directory as this
# notebook
from generate_example_data import (
generate_griffiths_data,
match_estimated_topics,
plot_lda,
plot_lda_topics,
)
# accessing the SageMaker Python SDK
import sagemaker
from sagemaker.amazon.common import RecordSerializer
from sagemaker.serializers import CSVSerializer
from sagemaker.deserializers import JSONDeserializer
```
# Setup
***
*This notebook was created and tested on an ml.m4.xlarge notebook instance.*
We first need to specify some AWS credentials; specifically data locations and access roles. This is the only cell of this notebook that you will need to edit. In particular, we need the following data:
* `bucket` - An S3 bucket accessible by this account.
* Used to store input training data and model data output.
* Should be withing the same region as this notebook instance, training, and hosting.
* `prefix` - The location in the bucket where this notebook's input and and output data will be stored. (The default value is sufficient.)
* `role` - The IAM Role ARN used to give training and hosting access to your data.
* See documentation on how to create these.
* The script below will try to determine an appropriate Role ARN.
```
from sagemaker import get_execution_role
role = get_execution_role()
bucket = sagemaker.Session().default_bucket()
prefix = "sagemaker/DEMO-lda-science"
print("Training input/output will be stored in {}/{}".format(bucket, prefix))
print("\nIAM Role: {}".format(role))
```
## The LDA Model
As mentioned above, LDA is a model for discovering latent topics describing a collection of documents. In this section we will give a brief introduction to the model. Let,
* $M$ = the number of *documents* in a corpus
* $N$ = the average *length* of a document.
* $V$ = the size of the *vocabulary* (the total number of unique words)
We denote a *document* by a vector $w \in \mathbb{R}^V$ where $w_i$ equals the number of times the $i$th word in the vocabulary occurs within the document. This is called the "bag-of-words" format of representing a document.
$$
\underbrace{w}_{\text{document}} = \overbrace{\big[ w_1, w_2, \ldots, w_V \big] }^{\text{word counts}},
\quad
V = \text{vocabulary size}
$$
The *length* of a document is equal to the total number of words in the document: $N_w = \sum_{i=1}^V w_i$.
An LDA model is defined by two parameters: a topic-word distribution matrix $\beta \in \mathbb{R}^{K \times V}$ and a Dirichlet topic prior $\alpha \in \mathbb{R}^K$. In particular, let,
$$\beta = \left[ \beta_1, \ldots, \beta_K \right]$$
be a collection of $K$ *topics* where each topic $\beta_k \in \mathbb{R}^V$ is represented as probability distribution over the vocabulary. One of the utilities of the LDA model is that a given word is allowed to appear in multiple topics with positive probability. The Dirichlet topic prior is a vector $\alpha \in \mathbb{R}^K$ such that $\alpha_k > 0$ for all $k$.
# Data Exploration
---
## An Example Dataset
Before explaining further let's get our hands dirty with an example dataset. The following synthetic data comes from [1] and comes with a very useful visual interpretation.
> [1] Thomas Griffiths and Mark Steyvers. *Finding Scientific Topics.* Proceedings of the National Academy of Science, 101(suppl 1):5228-5235, 2004.
```
print("Generating example data...")
num_documents = 6000
known_alpha, known_beta, documents, topic_mixtures = generate_griffiths_data(
num_documents=num_documents, num_topics=10
)
num_topics, vocabulary_size = known_beta.shape
# separate the generated data into training and tests subsets
num_documents_training = int(0.9 * num_documents)
num_documents_test = num_documents - num_documents_training
documents_training = documents[:num_documents_training]
documents_test = documents[num_documents_training:]
topic_mixtures_training = topic_mixtures[:num_documents_training]
topic_mixtures_test = topic_mixtures[num_documents_training:]
print("documents_training.shape = {}".format(documents_training.shape))
print("documents_test.shape = {}".format(documents_test.shape))
```
Let's start by taking a closer look at the documents. Note that the vocabulary size of these data is $V = 25$. The average length of each document in this data set is 150. (See `generate_griffiths_data.py`.)
```
print("First training document =\n{}".format(documents_training[0]))
print("\nVocabulary size = {}".format(vocabulary_size))
print("Length of first document = {}".format(documents_training[0].sum()))
average_document_length = documents.sum(axis=1).mean()
print("Observed average document length = {}".format(average_document_length))
```
The example data set above also returns the LDA parameters,
$$(\alpha, \beta)$$
used to generate the documents. Let's examine the first topic and verify that it is a probability distribution on the vocabulary.
```
print("First topic =\n{}".format(known_beta[0]))
print(
"\nTopic-word probability matrix (beta) shape: (num_topics, vocabulary_size) = {}".format(
known_beta.shape
)
)
print("\nSum of elements of first topic = {}".format(known_beta[0].sum()))
```
Unlike some clustering algorithms, one of the versatilities of the LDA model is that a given word can belong to multiple topics. The probability of that word occurring in each topic may differ, as well. This is reflective of real-world data where, for example, the word *"rover"* appears in a *"dogs"* topic as well as in a *"space exploration"* topic.
In our synthetic example dataset, the first word in the vocabulary belongs to both Topic #1 and Topic #6 with non-zero probability.
```
print("Topic #1:\n{}".format(known_beta[0]))
print("Topic #6:\n{}".format(known_beta[5]))
```
Human beings are visual creatures, so it might be helpful to come up with a visual representation of these documents.
In the below plots, each pixel of a document represents a word. The greyscale intensity is a measure of how frequently that word occurs within the document. Below we plot the first few documents of the training set reshaped into 5x5 pixel grids.
```
%matplotlib inline
fig = plot_lda(documents_training, nrows=3, ncols=4, cmap="gray_r", with_colorbar=True)
fig.suptitle("$w$ - Document Word Counts")
fig.set_dpi(160)
```
When taking a close look at these documents we can see some patterns in the word distributions suggesting that, perhaps, each topic represents a "column" or "row" of words with non-zero probability and that each document is composed primarily of a handful of topics.
Below we plots the *known* topic-word probability distributions, $\beta$. Similar to the documents we reshape each probability distribution to a $5 \times 5$ pixel image where the color represents the probability of that each word occurring in the topic.
```
%matplotlib inline
fig = plot_lda(known_beta, nrows=1, ncols=10)
fig.suptitle(r"Known $\beta$ - Topic-Word Probability Distributions")
fig.set_dpi(160)
fig.set_figheight(2)
```
These 10 topics were used to generate the document corpus. Next, we will learn about how this is done.
## Generating Documents
LDA is a generative model, meaning that the LDA parameters $(\alpha, \beta)$ are used to construct documents word-by-word by drawing from the topic-word distributions. In fact, looking closely at the example documents above you can see that some documents sample more words from some topics than from others.
LDA works as follows: given
* $M$ documents $w^{(1)}, w^{(2)}, \ldots, w^{(M)}$,
* an average document length of $N$,
* and an LDA model $(\alpha, \beta)$.
**For** each document, $w^{(m)}$:
* sample a topic mixture: $\theta^{(m)} \sim \text{Dirichlet}(\alpha)$
* **For** each word $n$ in the document:
* Sample a topic $z_n^{(m)} \sim \text{Multinomial}\big( \theta^{(m)} \big)$
* Sample a word from this topic, $w_n^{(m)} \sim \text{Multinomial}\big( \beta_{z_n^{(m)}} \; \big)$
* Add to document
The [plate notation](https://en.wikipedia.org/wiki/Plate_notation) for the LDA model, introduced in [2], encapsulates this process pictorially.
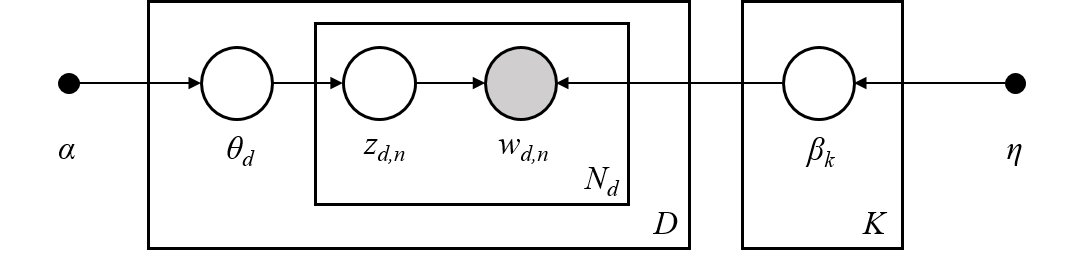
> [2] David M Blei, Andrew Y Ng, and Michael I Jordan. Latent Dirichlet Allocation. Journal of Machine Learning Research, 3(Jan):993–1022, 2003.
## Topic Mixtures
For the documents we generated above lets look at their corresponding topic mixtures, $\theta \in \mathbb{R}^K$. The topic mixtures represent the probablility that a given word of the document is sampled from a particular topic. For example, if the topic mixture of an input document $w$ is,
$$\theta = \left[ 0.3, 0.2, 0, 0.5, 0, \ldots, 0 \right]$$
then $w$ is 30% generated from the first topic, 20% from the second topic, and 50% from the fourth topic. In particular, the words contained in the document are sampled from the first topic-word probability distribution 30% of the time, from the second distribution 20% of the time, and the fourth disribution 50% of the time.
The objective of inference, also known as scoring, is to determine the most likely topic mixture of a given input document. Colloquially, this means figuring out which topics appear within a given document and at what ratios. We will perform infernece later in the [Inference](#Inference) section.
Since we generated these example documents using the LDA model we know the topic mixture generating them. Let's examine these topic mixtures.
```
print("First training document =\n{}".format(documents_training[0]))
print("\nVocabulary size = {}".format(vocabulary_size))
print("Length of first document = {}".format(documents_training[0].sum()))
print("First training document topic mixture =\n{}".format(topic_mixtures_training[0]))
print("\nNumber of topics = {}".format(num_topics))
print("sum(theta) = {}".format(topic_mixtures_training[0].sum()))
```
We plot the first document along with its topic mixture. We also plot the topic-word probability distributions again for reference.
```
%matplotlib inline
fig, (ax1, ax2) = plt.subplots(2, 1)
ax1.matshow(documents[0].reshape(5, 5), cmap="gray_r")
ax1.set_title(r"$w$ - Document", fontsize=20)
ax1.set_xticks([])
ax1.set_yticks([])
cax2 = ax2.matshow(topic_mixtures[0].reshape(1, -1), cmap="Reds", vmin=0, vmax=1)
cbar = fig.colorbar(cax2, orientation="horizontal")
ax2.set_title(r"$\theta$ - Topic Mixture", fontsize=20)
ax2.set_xticks([])
ax2.set_yticks([])
fig.set_dpi(100)
%matplotlib inline
# pot
fig = plot_lda(known_beta, nrows=1, ncols=10)
fig.suptitle(r"Known $\beta$ - Topic-Word Probability Distributions")
fig.set_dpi(160)
fig.set_figheight(1.5)
```
Finally, let's plot several documents with their corresponding topic mixtures. We can see how topics with large weight in the document lead to more words in the document within the corresponding "row" or "column".
```
%matplotlib inline
fig = plot_lda_topics(documents_training, 3, 4, topic_mixtures=topic_mixtures)
fig.suptitle(r"$(w,\theta)$ - Documents with Known Topic Mixtures")
fig.set_dpi(160)
```
# Training
***
In this section we will give some insight into how AWS SageMaker LDA fits an LDA model to a corpus, create an run a SageMaker LDA training job, and examine the output trained model.
## Topic Estimation using Tensor Decompositions
Given a document corpus, Amazon SageMaker LDA uses a spectral tensor decomposition technique to determine the LDA model $(\alpha, \beta)$ which most likely describes the corpus. See [1] for a primary reference of the theory behind the algorithm. The spectral decomposition, itself, is computed using the CPDecomp algorithm described in [2].
The overall idea is the following: given a corpus of documents $\mathcal{W} = \{w^{(1)}, \ldots, w^{(M)}\}, \; w^{(m)} \in \mathbb{R}^V,$ we construct a statistic tensor,
$$T \in \bigotimes^3 \mathbb{R}^V$$
such that the spectral decomposition of the tensor is approximately the LDA parameters $\alpha \in \mathbb{R}^K$ and $\beta \in \mathbb{R}^{K \times V}$ which maximize the likelihood of observing the corpus for a given number of topics, $K$,
$$T \approx \sum_{k=1}^K \alpha_k \; (\beta_k \otimes \beta_k \otimes \beta_k)$$
This statistic tensor encapsulates information from the corpus such as the document mean, cross correlation, and higher order statistics. For details, see [1].
> [1] Animashree Anandkumar, Rong Ge, Daniel Hsu, Sham Kakade, and Matus Telgarsky. *"Tensor Decompositions for Learning Latent Variable Models"*, Journal of Machine Learning Research, 15:2773–2832, 2014.
>
> [2] Tamara Kolda and Brett Bader. *"Tensor Decompositions and Applications"*. SIAM Review, 51(3):455–500, 2009.
## Store Data on S3
Before we run training we need to prepare the data.
A SageMaker training job needs access to training data stored in an S3 bucket. Although training can accept data of various formats we convert the documents MXNet RecordIO Protobuf format before uploading to the S3 bucket defined at the beginning of this notebook.
```
# convert documents_training to Protobuf RecordIO format
recordio_protobuf_serializer = RecordSerializer()
fbuffer = recordio_protobuf_serializer.serialize(documents_training)
# upload to S3 in bucket/prefix/train
fname = "lda.data"
s3_object = os.path.join(prefix, "train", fname)
boto3.Session().resource("s3").Bucket(bucket).Object(s3_object).upload_fileobj(fbuffer)
s3_train_data = "s3://{}/{}".format(bucket, s3_object)
print("Uploaded data to S3: {}".format(s3_train_data))
```
Next, we specify a Docker container containing the SageMaker LDA algorithm. For your convenience, a region-specific container is automatically chosen for you to minimize cross-region data communication
```
from sagemaker.image_uris import retrieve
region_name = boto3.Session().region_name
container = retrieve("lda", boto3.Session().region_name)
print("Using SageMaker LDA container: {} ({})".format(container, region_name))
```
## Training Parameters
Particular to a SageMaker LDA training job are the following hyperparameters:
* **`num_topics`** - The number of topics or categories in the LDA model.
* Usually, this is not known a priori.
* In this example, howevever, we know that the data is generated by five topics.
* **`feature_dim`** - The size of the *"vocabulary"*, in LDA parlance.
* In this example, this is equal 25.
* **`mini_batch_size`** - The number of input training documents.
* **`alpha0`** - *(optional)* a measurement of how "mixed" are the topic-mixtures.
* When `alpha0` is small the data tends to be represented by one or few topics.
* When `alpha0` is large the data tends to be an even combination of several or many topics.
* The default value is `alpha0 = 1.0`.
In addition to these LDA model hyperparameters, we provide additional parameters defining things like the EC2 instance type on which training will run, the S3 bucket containing the data, and the AWS access role. Note that,
* Recommended instance type: `ml.c4`
* Current limitations:
* SageMaker LDA *training* can only run on a single instance.
* SageMaker LDA does not take advantage of GPU hardware.
* (The Amazon AI Algorithms team is working hard to provide these capabilities in a future release!)
Using the above configuration create a SageMaker client and use the client to create a training job.
```
session = sagemaker.Session()
# specify general training job information
lda = sagemaker.estimator.Estimator(
container,
role,
output_path="s3://{}/{}/output".format(bucket, prefix),
instance_count=1,
instance_type="ml.c4.2xlarge",
sagemaker_session=session,
)
# set algorithm-specific hyperparameters
lda.set_hyperparameters(
num_topics=num_topics,
feature_dim=vocabulary_size,
mini_batch_size=num_documents_training,
alpha0=1.0,
)
# run the training job on input data stored in S3
lda.fit({"train": s3_train_data})
```
If you see the message
> `===== Job Complete =====`
at the bottom of the output logs then that means training sucessfully completed and the output LDA model was stored in the specified output path. You can also view information about and the status of a training job using the AWS SageMaker console. Just click on the "Jobs" tab and select training job matching the training job name, below:
```
print("Training job name: {}".format(lda.latest_training_job.job_name))
```
## Inspecting the Trained Model
We know the LDA parameters $(\alpha, \beta)$ used to generate the example data. How does the learned model compare the known one? In this section we will download the model data and measure how well SageMaker LDA did in learning the model.
First, we download the model data. SageMaker will output the model in
> `s3://<bucket>/<prefix>/output/<training job name>/output/model.tar.gz`.
SageMaker LDA stores the model as a two-tuple $(\alpha, \beta)$ where each LDA parameter is an MXNet NDArray.
```
# download and extract the model file from S3
job_name = lda.latest_training_job.job_name
model_fname = "model.tar.gz"
model_object = os.path.join(prefix, "output", job_name, "output", model_fname)
boto3.Session().resource("s3").Bucket(bucket).Object(model_object).download_file(fname)
with tarfile.open(fname) as tar:
tar.extractall()
print("Downloaded and extracted model tarball: {}".format(model_object))
# obtain the model file
model_list = [fname for fname in os.listdir(".") if fname.startswith("model_")]
model_fname = model_list[0]
print("Found model file: {}".format(model_fname))
# get the model from the model file and store in Numpy arrays
alpha, beta = mx.ndarray.load(model_fname)
learned_alpha_permuted = alpha.asnumpy()
learned_beta_permuted = beta.asnumpy()
print("\nLearned alpha.shape = {}".format(learned_alpha_permuted.shape))
print("Learned beta.shape = {}".format(learned_beta_permuted.shape))
```
Presumably, SageMaker LDA has found the topics most likely used to generate the training corpus. However, even if this is case the topics would not be returned in any particular order. Therefore, we match the found topics to the known topics closest in L1-norm in order to find the topic permutation.
Note that we will use the `permutation` later during inference to match known topic mixtures to found topic mixtures.
Below plot the known topic-word probability distribution, $\beta \in \mathbb{R}^{K \times V}$ next to the distributions found by SageMaker LDA as well as the L1-norm errors between the two.
```
permutation, learned_beta = match_estimated_topics(known_beta, learned_beta_permuted)
learned_alpha = learned_alpha_permuted[permutation]
fig = plot_lda(np.vstack([known_beta, learned_beta]), 2, 10)
fig.set_dpi(160)
fig.suptitle("Known vs. Found Topic-Word Probability Distributions")
fig.set_figheight(3)
beta_error = np.linalg.norm(known_beta - learned_beta, 1)
alpha_error = np.linalg.norm(known_alpha - learned_alpha, 1)
print("L1-error (beta) = {}".format(beta_error))
print("L1-error (alpha) = {}".format(alpha_error))
```
Not bad!
In the eyeball-norm the topics match quite well. In fact, the topic-word distribution error is approximately 2%.
# Inference
***
A trained model does nothing on its own. We now want to use the model we computed to perform inference on data. For this example, that means predicting the topic mixture representing a given document.
We create an inference endpoint using the SageMaker Python SDK `deploy()` function from the job we defined above. We specify the instance type where inference is computed as well as an initial number of instances to spin up.
With this realtime endpoint at our fingertips we can finally perform inference on our training and test data.
We can pass a variety of data formats to our inference endpoint. In this example we will demonstrate passing CSV-formatted data. Other available formats are JSON-formatted, JSON-sparse-formatter, and RecordIO Protobuf. We make use of the SageMaker Python SDK utilities `CSVSerializer` and `JSONDeserializer` when configuring the inference endpoint.
```
lda_inference = lda.deploy(
initial_instance_count=1,
instance_type="ml.m4.xlarge", # LDA inference may work better at scale on ml.c4 instances
serializer=CSVSerializer(),
deserializer=JSONDeserializer(),
)
```
Congratulations! You now have a functioning SageMaker LDA inference endpoint. You can confirm the endpoint configuration and status by navigating to the "Endpoints" tab in the AWS SageMaker console and selecting the endpoint matching the endpoint name, below:
```
print("Endpoint name: {}".format(lda_inference.endpoint_name))
```
We pass some test documents to the inference endpoint. Note that the serializer and deserializer will atuomatically take care of the datatype conversion.
```
results = lda_inference.predict(documents_test[:12])
print(results)
```
It may be hard to see but the output format of SageMaker LDA inference endpoint is a Python dictionary with the following format.
```
{
'predictions': [
{'topic_mixture': [ ... ] },
{'topic_mixture': [ ... ] },
{'topic_mixture': [ ... ] },
...
]
}
```
We extract the topic mixtures, themselves, corresponding to each of the input documents.
```
inferred_topic_mixtures_permuted = np.array(
[prediction["topic_mixture"] for prediction in results["predictions"]]
)
print("Inferred topic mixtures (permuted):\n\n{}".format(inferred_topic_mixtures_permuted))
```
## Inference Analysis
Recall that although SageMaker LDA successfully learned the underlying topics which generated the sample data the topics were in a different order. Before we compare to known topic mixtures $\theta \in \mathbb{R}^K$ we should also permute the inferred topic mixtures
```
inferred_topic_mixtures = inferred_topic_mixtures_permuted[:, permutation]
print("Inferred topic mixtures:\n\n{}".format(inferred_topic_mixtures))
```
Let's plot these topic mixture probability distributions alongside the known ones.
```
%matplotlib inline
# create array of bar plots
width = 0.4
x = np.arange(10)
nrows, ncols = 3, 4
fig, ax = plt.subplots(nrows, ncols, sharey=True)
for i in range(nrows):
for j in range(ncols):
index = i * ncols + j
ax[i, j].bar(x, topic_mixtures_test[index], width, color="C0")
ax[i, j].bar(x + width, inferred_topic_mixtures[index], width, color="C1")
ax[i, j].set_xticks(range(num_topics))
ax[i, j].set_yticks(np.linspace(0, 1, 5))
ax[i, j].grid(which="major", axis="y")
ax[i, j].set_ylim([0, 1])
ax[i, j].set_xticklabels([])
if i == (nrows - 1):
ax[i, j].set_xticklabels(range(num_topics), fontsize=7)
if j == 0:
ax[i, j].set_yticklabels([0, "", 0.5, "", 1.0], fontsize=7)
fig.suptitle("Known vs. Inferred Topic Mixtures")
ax_super = fig.add_subplot(111, frameon=False)
ax_super.tick_params(labelcolor="none", top="off", bottom="off", left="off", right="off")
ax_super.grid(False)
ax_super.set_xlabel("Topic Index")
ax_super.set_ylabel("Topic Probability")
fig.set_dpi(160)
```
In the eyeball-norm these look quite comparable.
Let's be more scientific about this. Below we compute and plot the distribution of L1-errors from **all** of the test documents. Note that we send a new payload of test documents to the inference endpoint and apply the appropriate permutation to the output.
```
%%time
# create a payload containing all of the test documents and run inference again
#
# TRY THIS:
# try switching between the test data set and a subset of the training
# data set. It is likely that LDA inference will perform better against
# the training set than the holdout test set.
#
payload_documents = documents_test # Example 1
known_topic_mixtures = topic_mixtures_test # Example 1
# payload_documents = documents_training[:600]; # Example 2
# known_topic_mixtures = topic_mixtures_training[:600] # Example 2
print("Invoking endpoint...\n")
results = lda_inference.predict(payload_documents)
inferred_topic_mixtures_permuted = np.array(
[prediction["topic_mixture"] for prediction in results["predictions"]]
)
inferred_topic_mixtures = inferred_topic_mixtures_permuted[:, permutation]
print("known_topics_mixtures.shape = {}".format(known_topic_mixtures.shape))
print("inferred_topics_mixtures_test.shape = {}\n".format(inferred_topic_mixtures.shape))
%matplotlib inline
l1_errors = np.linalg.norm((inferred_topic_mixtures - known_topic_mixtures), 1, axis=1)
# plot the error freqency
fig, ax_frequency = plt.subplots()
bins = np.linspace(0, 1, 40)
weights = np.ones_like(l1_errors) / len(l1_errors)
freq, bins, _ = ax_frequency.hist(l1_errors, bins=50, weights=weights, color="C0")
ax_frequency.set_xlabel("L1-Error")
ax_frequency.set_ylabel("Frequency", color="C0")
# plot the cumulative error
shift = (bins[1] - bins[0]) / 2
x = bins[1:] - shift
ax_cumulative = ax_frequency.twinx()
cumulative = np.cumsum(freq) / sum(freq)
ax_cumulative.plot(x, cumulative, marker="o", color="C1")
ax_cumulative.set_ylabel("Cumulative Frequency", color="C1")
# align grids and show
freq_ticks = np.linspace(0, 1.5 * freq.max(), 5)
freq_ticklabels = np.round(100 * freq_ticks) / 100
ax_frequency.set_yticks(freq_ticks)
ax_frequency.set_yticklabels(freq_ticklabels)
ax_cumulative.set_yticks(np.linspace(0, 1, 5))
ax_cumulative.grid(which="major", axis="y")
ax_cumulative.set_ylim((0, 1))
fig.suptitle("Topic Mixutre L1-Errors")
fig.set_dpi(110)
```
Machine learning algorithms are not perfect and the data above suggests this is true of SageMaker LDA. With more documents and some hyperparameter tuning we can obtain more accurate results against the known topic-mixtures.
For now, let's just investigate the documents-topic mixture pairs that seem to do well as well as those that do not. Below we retreive a document and topic mixture corresponding to a small L1-error as well as one with a large L1-error.
```
N = 6
good_idx = l1_errors < 0.05
good_documents = payload_documents[good_idx][:N]
good_topic_mixtures = inferred_topic_mixtures[good_idx][:N]
poor_idx = l1_errors > 0.3
poor_documents = payload_documents[poor_idx][:N]
poor_topic_mixtures = inferred_topic_mixtures[poor_idx][:N]
%matplotlib inline
fig = plot_lda_topics(good_documents, 2, 3, topic_mixtures=good_topic_mixtures)
fig.suptitle("Documents With Accurate Inferred Topic-Mixtures")
fig.set_dpi(120)
%matplotlib inline
fig = plot_lda_topics(poor_documents, 2, 3, topic_mixtures=poor_topic_mixtures)
fig.suptitle("Documents With Inaccurate Inferred Topic-Mixtures")
fig.set_dpi(120)
```
In this example set the documents on which inference was not as accurate tend to have a denser topic-mixture. This makes sense when extrapolated to real-world datasets: it can be difficult to nail down which topics are represented in a document when the document uses words from a large subset of the vocabulary.
## Stop / Close the Endpoint
Finally, we should delete the endpoint before we close the notebook.
To do so execute the cell below. Alternately, you can navigate to the "Endpoints" tab in the SageMaker console, select the endpoint with the name stored in the variable `endpoint_name`, and select "Delete" from the "Actions" dropdown menu.
```
sagemaker.Session().delete_endpoint(lda_inference.endpoint_name)
```
# Epilogue
---
In this notebook we,
* learned about the LDA model,
* generated some example LDA documents and their corresponding topic-mixtures,
* trained a SageMaker LDA model on a training set of documents and compared the learned model to the known model,
* created an inference endpoint,
* used the endpoint to infer the topic mixtures of a test input and analyzed the inference error.
There are several things to keep in mind when applying SageMaker LDA to real-word data such as a corpus of text documents. Note that input documents to the algorithm, both in training and inference, need to be vectors of integers representing word counts. Each index corresponds to a word in the corpus vocabulary. Therefore, one will need to "tokenize" their corpus vocabulary.
$$
\text{"cat"} \mapsto 0, \; \text{"dog"} \mapsto 1 \; \text{"bird"} \mapsto 2, \ldots
$$
Each text document then needs to be converted to a "bag-of-words" format document.
$$
w = \text{"cat bird bird bird cat"} \quad \longmapsto \quad w = [2, 0, 3, 0, \ldots, 0]
$$
Also note that many real-word applications have large vocabulary sizes. It may be necessary to represent the input documents in sparse format. Finally, the use of stemming and lemmatization in data preprocessing provides several benefits. Doing so can improve training and inference compute time since it reduces the effective vocabulary size. More importantly, though, it can improve the quality of learned topic-word probability matrices and inferred topic mixtures. For example, the words *"parliament"*, *"parliaments"*, *"parliamentary"*, *"parliament's"*, and *"parliamentarians"* are all essentially the same word, *"parliament"*, but with different conjugations. For the purposes of detecting topics, such as a *"politics"* or *governments"* topic, the inclusion of all five does not add much additional value as they all essentiall describe the same feature.
|
github_jupyter
|
```
from skempi_utils import *
from scipy.stats import pearsonr
df = skempi_df
df_multi = df[~np.asarray([len(s)>8 for s in df.Protein])]
s_multi = set([s[:4] for s in df_multi.Protein])
s_groups = set([s[:4] for s in G1 + G2 + G3 + G4 + G5])
len(s_multi & s_groups), len(s_multi), len(s_groups)
df_multi.head()
from sklearn.preprocessing import StandardScaler
from itertools import combinations as comb
from sklearn.externals import joblib
import numpy as np
def evaluate(group_str, y_true, y_pred, ix):
y_pred_pos = y_pred[ix == 0]
y_pred_neg = y_pred[ix == 1]
y_true_pos = y_true[ix == 0]
y_true_neg = y_true[ix == 1]
cor_all, _ = pearsonr(y_true, y_pred)
cor_pos, _ = pearsonr(y_true_pos, y_pred_pos)
cor_neg, _ = pearsonr(y_true_neg, y_pred_neg)
print("[%s:%d] cor_all:%.3f, cor_pos:%.3f, cor_neg:%.3f" % (group_str, len(y_true), cor_all, cor_pos, cor_neg))
return cor_all, cor_pos, cor_neg
def run_cv_test(X, y, ix, get_regressor, modelname, normalize=1):
gt, preds, indx, cors = [], [], [], []
groups = [G1, G2, G3, G4, G5]
prots = G1 + G2 + G3 + G4 + G5
for i, pair in enumerate(comb(range(NUM_GROUPS), 2)):
group = groups[pair[0]] + groups[pair[1]]
g1, g2 = np.asarray(pair) + 1
indx_tst = (ix[:, 0] == g1) | (ix[:, 0] == g2)
indx_trn = np.logical_not(indx_tst)
y_trn = y[indx_trn]
y_true = y[indx_tst]
X_trn = X[indx_trn]
X_tst = X[indx_tst]
if normalize == 1:
scaler = StandardScaler()
scaler.fit(X_trn)
X_trn = scaler.transform(X_trn)
X_tst = scaler.transform(X_tst)
regressor = get_regressor()
regressor.fit(X_trn, y_trn)
joblib.dump(regressor, 'models/%s%s.pkl' % (modelname, i))
regressor = joblib.load('models/%s%s.pkl' % (modelname, i))
y_pred = regressor.predict(X_tst)
cor, pos, neg = evaluate("G%d,G%d" % (g1, g2), y_true, y_pred, ix[indx_tst, 1])
cors.append([cor, pos, neg])
indx.extend(ix[indx_tst, 1])
preds.extend(y_pred)
gt.extend(y_true)
return [np.asarray(a) for a in [gt, preds, indx, cors]]
def run_cv_test_ensemble(X, y, ix, alpha=0.5, normalize=1):
gt, preds, indx, cors = [], [], [], []
groups = [G1, G2, G3, G4, G5]
prots = G1 + G2 + G3 + G4 + G5
for i, pair in enumerate(comb(range(NUM_GROUPS), 2)):
group = groups[pair[0]] + groups[pair[1]]
g1, g2 = np.asarray(pair) + 1
indx_tst = (ix[:, 0] == g1) | (ix[:, 0] == g2)
indx_trn = (ix[:, 0] != 0) & ((ix[:, 0] == g1) | (ix[:, 0] == g2))
y_trn = y[indx_trn]
y_true = y[indx_tst]
X_trn = X[indx_trn]
X_tst = X[indx_tst]
svr = joblib.load('models/svr%d.pkl' % i)
rfr = joblib.load('models/rfr%d.pkl' % i)
if normalize == 1:
scaler = StandardScaler()
scaler.fit(X_trn)
X_trn = scaler.transform(X_trn)
X_tst = scaler.transform(X_tst)
y_pred_svr = svr.predict(X_tst)
y_pred_rfr = rfr.predict(X_tst)
y_pred = alpha * y_pred_svr + (1-alpha) * y_pred_rfr
cor, pos, neg = evaluate("G%d,G%d" % (g1, g2), y_true, y_pred, ix[indx_tst, 1])
cors.append([cor, pos, neg])
indx.extend(ix[indx_tst, 1])
preds.extend(y_pred)
gt.extend(y_true)
return [np.asarray(a) for a in [gt, preds, indx, cors]]
def records_to_xy(skempi_records, load_neg=True):
data = []
for record in tqdm(skempi_records, desc="records processed"):
r = record
assert r.struct is not None
data.append([r.features(True), [r.ddg], [r.group, r.is_minus]])
if not load_neg: continue
rr = reversed(record)
assert rr.struct is not None
data.append([rr.features(True), [rr.ddg], [rr.group, rr.is_minus]])
X, y, ix = [np.asarray(d) for d in zip(*data)]
return X, y, ix
def get_temperature_array(records, agg=np.min):
arr = []
pbar = tqdm(range(len(skempi_df)), desc="row processed")
for i, row in skempi_df.iterrows():
arr_obs_mut = []
for mutation in row["Mutation(s)_cleaned"].split(','):
mut = Mutation(mutation)
res_i, chain_id = mut.i, mut.chain_id
t = tuple(row.Protein.split('_'))
skempi_record = records[t]
res = skempi_record[chain_id][res_i]
temps = [a.temp for a in res.atoms]
arr_obs_mut.append(np.mean(temps))
arr.append(agg(arr_obs_mut))
pbar.update(1)
pbar.close()
return arr
skempi_records = load_skempi_structs(pdb_path="../data/pdbs_n", compute_dist_mat=False)
temp_arr = get_temperature_array(skempi_records, agg=np.min)
skempi_structs = load_skempi_structs("../data/pdbs", compute_dist_mat=False)
skempi_records = load_skempi_records(skempi_structs)
# X_pos, y_pos, ix_pos = records_to_xy(skempi_records)
# X_pos.shape, y_pos.shape, ix_pos.shape
X_, y_, ix_ = records_to_xy(skempi_records)
X = X_[:, :]
# X = np.concatenate([X.T, [temp_arr]], axis=0).T
y = y_[:, 0]
ix = ix_
X.shape, y.shape, ix.shape
print("----->SVR")
from sklearn.svm import SVR
def get_regressor(): return SVR(kernel='rbf')
gt, preds, indx, cors = run_cv_test(X, y, ix, get_regressor, 'svr', normalize=1)
cor1, _, _ = evaluate("CAT", gt, preds, indx)
print(np.mean(cors, axis=0))
print("----->RFR")
from sklearn.ensemble import RandomForestRegressor
def get_regressor(): return RandomForestRegressor(n_estimators=50, random_state=0)
gt, preds, indx, cors = run_cv_test(X, y, ix, get_regressor, 'rfr', normalize=1)
cor2, _, _ = evaluate("CAT", gt, preds, indx)
print(np.mean(cors, axis=0))
# alpha = cor1/(cor1+cor2)
alpha = 0.5
print("----->%.2f*SVR + %.2f*RFR" % (alpha, 1-alpha))
gt, preds, indx, cors = run_cv_test_ensemble(X, y, ix, normalize=1)
cor, _, _ = evaluate("CAT", gt, preds, indx)
print(np.mean(cors, axis=0))
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
def run_holdout_test_ensemble(X, y, ix, alpha=0.5, normalize=1):
indx_tst = ix[:, 0] == 0
indx_trn = np.logical_not(indx_tst)
y_trn = y[indx_trn]
y_true = y[indx_tst]
X_trn = X[indx_trn]
X_tst = X[indx_tst]
svr = SVR(kernel='rbf')
rfr = RandomForestRegressor(n_estimators=50, random_state=0)
if normalize == 1:
scaler = StandardScaler()
scaler.fit(X_trn)
X_trn = scaler.transform(X_trn)
X_tst = scaler.transform(X_tst)
svr.fit(X_trn, y_trn)
rfr.fit(X_trn, y_trn)
y_pred_svr = svr.predict(X_tst)
y_pred_rfr = rfr.predict(X_tst)
y_pred = alpha * y_pred_svr + (1-alpha) * y_pred_rfr
cor, pos, neg = evaluate("holdout", y_true, y_pred, ix[indx_tst, 1])
return cor, pos, neg
alpha = 0.5
run_holdout_test_ensemble(X, y, ix, alpha=0.5, normalize=1)
```
|
github_jupyter
|
# Automate loan approvals with Business rules in Apache Spark and Scala
### Automating at scale your business decisions in Apache Spark with IBM ODM 8.9.2
This Scala notebook shows you how to execute locally business rules in DSX and Apache Spark.
You'll learn how to call in Apache Spark a rule-based decision service. This decision service has been programmed with IBM Operational Decision Manager.
This notebook puts in action a decision service named Miniloan that is part of the ODM tutorials. It determines with business rules whether a customer is eligible for a loan according to specific criteria. The criteria include the amount of the loan, the annual income of the borrower, and the duration of the loan.
First we load an application data set that was captured as a CSV file. In scala we apply a map to this data set to automate a rule-based reasoning, in order to outcome a decision. The rule execution is performed locally in the Spark service. This notebook shows a complete Scala code that can execute any ruleset based on the public APIs.
To get the most out of this notebook, you should have some familiarity with the Scala programming language.
## Contents
This notebook contains the following main sections:
1. [Load the loan validation request dataset.](#loaddatatset)
2. [Load the business rule execution and the simple loan application object model libraries.](#loadjars)
3. [Import Scala packages.](#importpackages)
4. [Implement a decision making function.](#implementDecisionServiceMap)
5. [Execute the business rules to approve or reject the loan applications.](#executedecisions)
6. [View the automated decisions.](#viewdecisions)
7. [Summary and next steps.](#summary)
<a id="accessdataset"></a>
## 1. Loading a loan application dataset file
A data set of simple loan applications is already available. You load it in the Notebook through its url.
```
// @hidden_cell
import scala.sys.process._
"wget https://raw.githubusercontent.com/ODMDev/decisions-on-spark/master/data/miniloan/miniloan-requests-10K.csv".!
val filename = "miniloan-requests-10K.csv"
```
This following code loads the 10 000 simple loan application dataset written in CSV format.
```
val requestData = sc.textFile(filename)
val requestDataCount = requestData.count
println(s"$requestDataCount loan requests read in a CVS format")
println("The first 5 requests:")
requestData.take(20).foreach(println)
```
<a id="loadjars"></a>
## 2. Add libraries for business rule execution and a loan application object model
The XXX refers to your object storage or other place where you make available these jars.
Add the following jars to execute the deployed decision service
<il>
<li>%AddJar https://XXX/j2ee_connector-1_5-fr.jar</li>
<li>%AddJar https://XXX/jrules-engine.jar</li>
<li>%AddJar https://XXX/jrules-res-execution.jar</li>
</il>
In addition you need the Apache Jackson annotation lib
<il>
<li>%AddJar https://XXX/jackson-annotations-2.6.5.jar</li>
</il>
Business Rules apply on a Java executable Object Model packaged as a jar. We need these classes to create the decision requests, and to retreive the response from the rule engine.
<il>
<li>%AddJar https://XXX/miniloan-xom.jar</li>
</il>
```
// @hidden_cell
// The urls below are accessible for an IBM internal usage only
%AddJar https://XXX/j2ee_connector-1_5-fr.jar
%AddJar https://XXX/jrules-engine.jar
%AddJar https://XXX/jrules-res-execution.jar
%AddJar https://XXX/jackson-annotations-2.6.5.jar -f
//Loan Application eXecutable Object Model
%AddJar https://XXX/miniloan-xom.jar -f
print("Your notebook is now ready to execute business rules to approve or reject loan applications")
```
<a id="importpackages"></a>
## 3. Import packages
Import ODM and Apache Spark packages.
```
import java.util.Map
import java.util.HashMap
import com.fasterxml.jackson.core.JsonGenerationException
import com.fasterxml.jackson.core.JsonProcessingException
import com.fasterxml.jackson.databind.JsonMappingException
import com.fasterxml.jackson.databind.ObjectMapper
import com.fasterxml.jackson.databind.SerializationFeature
import org.apache.spark.SparkConf
import org.apache.spark.api.java.JavaDoubleRDD
import org.apache.spark.api.java.JavaRDD
import org.apache.spark.api.java.JavaSparkContext
import org.apache.spark.api.java.function.Function
import org.apache.hadoop.fs.FileSystem
import org.apache.hadoop.fs.Path
import scala.collection.JavaConverters._
import ilog.rules.res.model._
import com.ibm.res.InMemoryJ2SEFactory
import com.ibm.res.InMemoryRepositoryDAO
import ilog.rules.res.session._
import miniloan.Borrower
import miniloan.Loan
import scala.io.Source
import java.net.URL
import java.io.InputStream
```
<a id="implementDecisionServiceMap"></a>
## 4. Implement a Map function that executes a rule-based decision service
```
case class MiniLoanRequest(borrower: miniloan.Borrower,
loan: miniloan.Loan)
case class RESRunner(sessionFactory: com.ibm.res.InMemoryJ2SEFactory) {
def executeAsString(s: String): String = {
println("executeAsString")
val request = makeRequest(s)
val response = executeRequest(request)
response
}
private def makeRequest(s: String): MiniLoanRequest = {
val tokens = s.split(",")
// Borrower deserialization from CSV
val borrowerName = tokens(0)
val borrowerCreditScore = java.lang.Integer.parseInt(tokens(1).trim())
val borrowerYearlyIncome = java.lang.Integer.parseInt(tokens(2).trim())
val loanAmount = java.lang.Integer.parseInt(tokens(3).trim())
val loanDuration = java.lang.Integer.parseInt(tokens(4).trim())
val yearlyInterestRate = java.lang.Double.parseDouble(tokens(5).trim())
val borrower = new miniloan.Borrower(borrowerName, borrowerCreditScore, borrowerYearlyIncome)
// Loan request deserialization from CSV
val loan = new miniloan.Loan()
loan.setAmount(loanAmount)
loan.setDuration(loanDuration)
loan.setYearlyInterestRate(yearlyInterestRate)
val request = new MiniLoanRequest(borrower, loan)
request
}
def executeRequest(request: MiniLoanRequest): String = {
try {
val sessionRequest = sessionFactory.createRequest()
val rulesetPath = "/Miniloan/Miniloan"
sessionRequest.setRulesetPath(ilog.rules.res.model.IlrPath.parsePath(rulesetPath))
//sessionRequest.getTraceFilter.setInfoAllFilters(false)
val inputParameters = sessionRequest.getInputParameters
inputParameters.put("loan", request.loan)
inputParameters.put("borrower", request.borrower)
val session = sessionFactory.createStatelessSession()
val response = session.execute(sessionRequest)
var loan = response.getOutputParameters().get("loan").asInstanceOf[miniloan.Loan]
val mapper = new com.fasterxml.jackson.databind.ObjectMapper()
mapper.configure(com.fasterxml.jackson.databind.SerializationFeature.FAIL_ON_EMPTY_BEANS, false)
val results = new java.util.HashMap[String,Object]()
results.put("input", inputParameters)
results.put("output", response.getOutputParameters())
try {
//return mapper.writeValueAsString(results)
return mapper.writerWithDefaultPrettyPrinter().writeValueAsString(results);
} catch {
case e: Exception => return e.toString()
}
"Error"
} catch {
case exception: Exception => {
return exception.toString()
}
}
"Error"
}
}
val decisionService = new Function[String, String]() {
@transient private var ruleSessionFactory: InMemoryJ2SEFactory = null
private val rulesetURL = "https://odmlibserver.mybluemix.net/8901/decisionservices/miniloan-8901.dsar"
@transient private var rulesetStream: InputStream = null
def GetRuleSessionFactory(): InMemoryJ2SEFactory = {
if (ruleSessionFactory == null) {
ruleSessionFactory = new InMemoryJ2SEFactory()
// Create the Management Session
var repositoryFactory = ruleSessionFactory.createManagementSession().getRepositoryFactory()
var repository = repositoryFactory.createRepository()
// Deploy the Ruleapp with the Regular Management Session API.
var rapp = repositoryFactory.createRuleApp("Miniloan", IlrVersion.parseVersion("1.0"));
var rs = repositoryFactory.createRuleset("Miniloan",IlrVersion.parseVersion("1.1"));
rapp.addRuleset(rs);
//var fileStream = Source.fromResourceAsStream(RulesetFileName)
rulesetStream = new java.net.URL(rulesetURL).openStream()
rs.setRESRulesetArchive(IlrEngineType.DE,rulesetStream)
repository.addRuleApp(rapp)
}
ruleSessionFactory
}
def call(s: String): String = {
var runner = new RESRunner(GetRuleSessionFactory())
return runner.executeAsString(s)
}
def execute(s: String): String = {
try {
var runner = new RESRunner(GetRuleSessionFactory())
return runner.executeAsString(s)
} catch {
case exception: Exception => {
exception.printStackTrace(System.err)
}
}
"Execution error"
}
}
```
<a id="executedecisions"></a>
## 5. Automate the decision making on the loan application dataset
You invoke a map on the decision function. While the map occurs rule engines are processing in parallel the loan applications to produce a data set of answers.
```
println("Start of Execution")
val answers = requestData.map(decisionService.execute)
printf("Number of rule based decisions: %s \n" , answers.count)
// Cleanup output file
//val fs = FileSystem.get(new URI(outputPath), sc.hadoopConfiguration);
//if (fs.exists(new Path(outputPath)))
// fs.delete(new Path(outputPath), true)
// Save RDD in a HDFS file
println("End of Execution ")
//answers.saveAsTextFile("swift://DecisionBatchExecution." + securedAccessName + "/miniloan-decisions-10.csv")
println("Decision automation job done")
```
<a id="viewdecisions"></a>
## 6. View your automated decisions
Each decision is composed of output parameters and of a decision trace. The loan data contains the approval flag and the computed yearly repayment. The decision trace lists the business rules that have been executed in sequence to come to the conclusion. Each decision has been serialized in JSON.
```
//answers.toDF().show(false)
answers.take(1).foreach(println)
```
<a id="summary"></a>
## 7. Summary and next steps
Congratulations! You have applied business rules to automatically determine loan approval eligibility. You loaded a loan application data set, ran a rule engine inside an Apache Spark cluster to make an eligibility decision for each applicant. Each decision is a Scala object that is part of a Spark Resilient Data Set.
Each decision is structured with input parameters (the context of the decision) and output parameters. For audit purpose the rule engine can emit a decision trace.
You have successfully run a rule engine to automate decisions at scale in a Spark cluster. You can now invent your own business rules and run them with the same integration pattern.
<a id="authors"></a>
## Authors
Pierre Feillet and Laurent Grateau are business rule engineers at IBM working in the Decision lab located in France.
Copyright © 2018 IBM. This notebook and its source code are released under the terms of the MIT License.
|
github_jupyter
|
```
##World Map Plotly
#Import Plotly Lib and Set up Credentials with personal account
!pip install plotly
import plotly
plotly.tools.set_credentials_file(username='igleonaitis', api_key='If6Wh3xWNmdNioPzOZZo')
plotly.tools.set_config_file(world_readable=True,
sharing='public')
import plotly.plotly as py
from plotly.graph_objs import *
import plotly.graph_objs as go
import pandas as pd
#Import WHR 2017 data set
df = pd.read_excel('whr.xlsx', sheetname='Figure2.2 WHR 2017')
#Set Up World Map Plot
scl = [[0,'rgb(140,101,211)'],[0.25,'rgb(154,147,236)'],
[0.50,'rgb(0,82,165)'],[0.75,'rgb(129,203,248)'],
[1,'rgb(65,179,247)']]
data = [ dict(
type = 'choropleth',
locationmode = 'country names',
locations = df['Country'],
z = df['Happiness score'],
text = df['Country'],
colorscale = scl,
autocolorscale = False,
reversescale = False,
marker = dict(
line = dict (
color = 'rgb(180,180,180)',
width = 0.5
) ),
colorbar = dict(
autotick = False,
tickprefix = False,
title = 'World Happiness Score'),
) ]
layout = dict(
title = '2017 National Happiness Scores GDP<br>Source:\
<a href="http://worldhappiness.report/ed/2017/">\
World Happiness Report</a>',
geo = dict(
showframe = False,
showcoastlines = False,
projection = dict(
type = 'Mercator'
)
)
)
#Create World Map Plot
fig = dict(data = data, layout = layout)
py.iplot(fig, validate=False, filename='d3-world-map')
df1 = pd.read_excel('whr.xlsx', sheetname='Figure2.2 WHR 2017')
#Stacked Bar Plot
trace1 = go.Bar(
y = df1['Country'],
x = df1['Explained by: GDP per capita'],
orientation = 'h',
width = .5,
name = 'GDP per Capita',
marker=dict(
color='rgb(140,101,211)'
)
)
trace2 = go.Bar(
y = df1['Country'],
x = df1['Explained by: Social support'],
orientation = 'h',
width = .5,
name = 'Social Support',
marker=dict(
color='rgb(154,147,236)'
)
)
trace3 = go.Bar(
y = df1['Country'],
x = df1['Explained by: Healthy life expectancy'],
orientation = 'h',
width = .5,
name = 'Healthy Life Expectancy',
marker=dict(
color='rgb(0,82,165)'
)
)
trace4 = go.Bar(
y = df1['Country'],
x = df1['Explained by: Freedom to make life choices'],
orientation = 'h',
width = .5,
name = 'Freedom to Make Life Choices',
marker=dict(
color='rgb(129,203,248)'
)
)
trace5 = go.Bar(
y = df1['Country'],
x = df1['Explained by: Generosity'],
orientation = 'h',
width = .5,
name = 'Generosity',
marker=dict(
color='rgb(65,179,247)'
)
)
trace6 = go.Bar(
y = df1['Country'],
x = df1['Explained by: Perceptions of corruption'],
orientation = 'h',
width = .5,
name = 'Perceptions on Corruption',
marker=dict(
color='rgb(115, 235, 174)'
)
)
data = [trace1, trace2, trace3, trace4, trace5, trace6]
layout = go.Layout(
title = 'Factor Makeup of Happiness Scores',
barmode ='stack',
autosize = False,
width = 800,
height = 1500,
yaxis = dict(
tickfont = dict(
size = 6,
color = 'black')),
xaxis = dict(
tickfont = dict(
size = 10,
color = 'black'))
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='stacked-horizontal-bar')
import plotly.plotly as py
from plotly.grid_objs import Grid, Column
from plotly.figure_factory import *
import pandas as pd
import time
xls_file = pd.ExcelFile('Internet_Usage.xls')
xls_file
dataset = xls_file.parse('Sheet1')
dataset.head()
years_from_col = set(dataset['year'])
years_ints = sorted(list(years_from_col))
years = [str(year) for year in years_ints]
# make list of continents
continents = []
for continent in dataset['continent']:
if continent not in continents:
continents.append(continent)
columns = []
# make grid
for year in years:
for continent in continents:
dataset_by_year = dataset[dataset['year'] == int(year)]
dataset_by_year_and_cont = dataset_by_year[dataset_by_year['continent'] == continent]
for col_name in dataset_by_year_and_cont:
# each column name is unique
column_name = '{year}_{continent}_{header}_gapminder_grid'.format(
year=year, continent=continent, header=col_name
)
a_column = Column(list(dataset_by_year_and_cont[col_name]), column_name)
columns.append(a_column)
# upload grid
grid = Grid(columns)
url = py.grid_ops.upload(grid, 'gapminder_grid'+str(time.time()), auto_open=False)
url
figure = {
'data': [],
'layout': {},
'frames': [],
'config': {'scrollzoom': True}
}
# fill in most of layout
figure['layout']['xaxis'] = {'range': [2, 8], 'title': 'World Happiness Score', 'gridcolor': '#FFFFFF'}
figure['layout']['yaxis'] = {'range': [0,100],'title': 'Internet Usage % of Pop.', 'gridcolor': '#FFFFFF'}
figure['layout']['hovermode'] = 'closest'
figure['layout']['plot_bgcolor'] = 'rgb(223, 232, 243)'
sliders_dict = {
'active': 0,
'yanchor': 'top',
'xanchor': 'left',
'currentvalue': {
'font': {'size': 20},
'prefix': 'Year:',
'visible': True,
'xanchor': 'right'
},
'transition': {'duration': 300, 'easing': 'cubic-in-out'},
'pad': {'b': 10, 't': 50},
'len': 0.9,
'x': 0.1,
'y': 0,
'steps': []
}
figure['layout']['updatemenus'] = [
{
'buttons': [
{
'args': [None, {'frame': {'duration': 500, 'redraw': False},
'fromcurrent': True, 'transition': {'duration': 300, 'easing': 'quadratic-in-out'}}],
'label': 'Play',
'method': 'animate'
},
{
'args': [[None], {'frame': {'duration': 0, 'redraw': False}, 'mode': 'immediate',
'transition': {'duration': 0}}],
'label': 'Pause',
'method': 'animate'
}
],
'direction': 'left',
'pad': {'r': 10, 't': 87},
'showactive': False,
'type': 'buttons',
'x': 0.1,
'xanchor': 'right',
'y': 0,
'yanchor': 'top'
}
]
custom_colors = {
'Asia': 'rgb(171, 99, 250)',
'Europe': 'rgb(230, 99, 250)',
'Africa': 'rgb(99, 110, 250)',
'Americas': 'rgb(25, 211, 243)',
'Oceania': 'rgb(50, 170, 255)'
}
col_name_template = '{year}_{continent}_{header}_gapminder_grid'
year = 2007
for continent in continents:
data_dict = {
'xsrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='lifeExp'
)),
'ysrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='gdpPercap'
)),
'mode': 'markers',
'textsrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='country'
)),
'marker': {
'sizemode': 'area',
'sizeref': 2000,
'sizesrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='pop'
)),
'color': custom_colors[continent]
},
'name': continent
}
figure['data'].append(data_dict)
for year in years:
frame = {'data': [], 'name': str(year)}
for continent in continents:
data_dict = {
'xsrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='lifeExp'
)),
'ysrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='gdpPercap'
)),
'mode': 'markers',
'textsrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='country'
)),
'marker': {
'sizemode': 'area',
'sizeref': 2000,
'sizesrc': grid.get_column_reference(col_name_template.format(
year=year, continent=continent, header='pop'
)),
'color': custom_colors[continent]
},
'name': continent
}
frame['data'].append(data_dict)
figure['frames'].append(frame)
slider_step = {'args': [
[year],
{'frame': {'duration': 300, 'redraw': False},
'mode': 'immediate',
'transition': {'duration': 300}}
],
'label': year,
'method': 'animate'}
sliders_dict['steps'].append(slider_step)
figure['layout']['sliders'] = [sliders_dict]
py.icreate_animations(figure, 'gapminder_example'+str(time.time()))
```
|
github_jupyter
|
[this doc on github](https://github.com/dotnet/interactive/tree/master/samples/notebooks/fsharp/Samples)
# Machine Learning over House Prices with ML.NET
### Reference the packages
```
#r "nuget:Microsoft.ML,1.4.0"
#r "nuget:Microsoft.ML.AutoML,0.16.0"
#r "nuget:Microsoft.Data.Analysis,0.2.0"
#r "nuget: XPlot.Plotly.Interactive, 4.0.6"
open Microsoft.Data.Analysis
open XPlot.Plotly
```
### Adding better default formatting for data frames
Register a formatter for data frames and data frame rows.
```
module DateFrameFormatter =
// Locally open the F# HTML DSL.
open Html
let maxRows = 20
Formatter.Register<DataFrame>((fun (df: DataFrame) (writer: TextWriter) ->
let take = 20
table [] [
thead [] [
th [] [ str "Index" ]
for c in df.Columns do
th [] [ str c.Name]
]
tbody [] [
for i in 0 .. min maxRows (int df.Rows.Count - 1) do
tr [] [
td [] [ i ]
for o in df.Rows.[int64 i] do
td [] [ o ]
]
]
]
|> writer.Write
), mimeType = "text/html")
Formatter.Register<DataFrameRow>((fun (row: DataFrameRow) (writer: TextWriter) ->
table [] [
tbody [] [
tr [] [
for o in row do
td [] [ o ]
]
]
]
|> writer.Write
), mimeType = "text/html")
```
### Download the data
```
open System.Net.Http
let housingPath = "housing.csv"
if not(File.Exists(housingPath)) then
let contents = HttpClient().GetStringAsync("https://raw.githubusercontent.com/ageron/handson-ml2/master/datasets/housing/housing.csv").Result
File.WriteAllText("housing.csv", contents)
```
### Add the data to the data frame
```
let housingData = DataFrame.LoadCsv(housingPath)
housingData
housingData.Description()
```
### Display the data
```
let graph =
Histogram(x = housingData.["median_house_value"],
nbinsx = 20)
graph |> Chart.Plot
let graph =
Scattergl(
x = housingData.["longitude"],
y = housingData.["latitude"],
mode = "markers",
marker =
Marker(
color = housingData.["median_house_value"],
colorscale = "Jet"))
let plot = Chart.Plot(graph)
plot.Width <- 600
plot.Height <- 600
display(plot)
```
### Prepare the training and validation sets
```
module Array =
let shuffle (arr: 'T[]) =
let rnd = Random()
let arr = Array.copy arr
for i in 0 .. arr.Length - 1 do
let r = i + rnd.Next(arr.Length - i)
let temp = arr.[r]
arr.[r] <- arr.[i]
arr.[i] <- temp
arr
let randomIndices = [| 0 .. int housingData.Rows.Count - 1 |] |> Array.shuffle
let testSize = int (float (housingData.Rows.Count) * 0.1)
let trainRows = randomIndices.[testSize..]
let testRows = randomIndices.[..testSize - 1]
let housing_train = housingData.[trainRows]
let housing_test = housingData.[testRows]
display(housing_train.Rows.Count)
display(housing_test.Rows.Count)
```
### Create the regression model and train it
```
#!time
open Microsoft.ML
open Microsoft.ML.Data
open Microsoft.ML.AutoML
let mlContext = MLContext()
let experiment = mlContext.Auto().CreateRegressionExperiment(maxExperimentTimeInSeconds = 15u)
let result = experiment.Execute(housing_train, labelColumnName = "median_house_value")
```
### Display the training results
```
let scatters =
result.RunDetails
|> Seq.filter (fun d -> not (isNull d.ValidationMetrics))
|> Seq.groupBy (fun r -> r.TrainerName)
|> Seq.map (fun (name, details) ->
Scattergl(
name = name,
x = (details |> Seq.map (fun r -> r.RuntimeInSeconds)),
y = (details |> Seq.map (fun r -> r.ValidationMetrics.MeanAbsoluteError)),
mode = "markers",
marker = Marker(size = 12)))
let chart = Chart.Plot(scatters)
chart.WithXTitle("Training Time")
chart.WithYTitle("Error")
display(chart)
Console.WriteLine("Best Trainer:{0}", result.BestRun.TrainerName);
```
### Validate and display the results
```
let testResults = result.BestRun.Model.Transform(housing_test)
let trueValues = testResults.GetColumn<float32>("median_house_value")
let predictedValues = testResults.GetColumn<float32>("Score")
let predictedVsTrue =
Scattergl(
x = trueValues,
y = predictedValues,
mode = "markers")
let maximumValue = Math.Max(Seq.max trueValues, Seq.max predictedValues)
let perfectLine =
Scattergl(
x = [| 0.0f; maximumValue |],
y = [| 0.0f; maximumValue |],
mode = "lines")
let chart = Chart.Plot([| predictedVsTrue; perfectLine |])
chart.WithXTitle("True Values")
chart.WithYTitle("Predicted Values")
chart.WithLegend(false)
chart.Width = 600
chart.Height = 600
display(chart)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/jantic/DeOldify/blob/master/ImageColorizerColab.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
### **<font color='blue'> Artistic Colorizer </font>**
#◢ DeOldify - Colorize your own photos!
####**Credits:**
Special thanks to:
Matt Robinson and María Benavente for pioneering the DeOldify image colab notebook.
Dana Kelley for doing things, breaking stuff & having an opinion on everything.
---
#◢ Verify Correct Runtime Settings
**<font color='#FF000'> IMPORTANT </font>**
In the "Runtime" menu for the notebook window, select "Change runtime type." Ensure that the following are selected:
* Runtime Type = Python 3
* Hardware Accelerator = GPU
#◢ Git clone and install DeOldify
```
!git clone https://github.com/jantic/DeOldify.git DeOldify
cd DeOldify
```
#◢ Setup
```
#NOTE: This must be the first call in order to work properly!
from deoldify import device
from deoldify.device_id import DeviceId
#choices: CPU, GPU0...GPU7
device.set(device=DeviceId.GPU0)
import torch
if not torch.cuda.is_available():
print('GPU not available.')
!pip install -r colab_requirements.txt
import fastai
from deoldify.visualize import *
!mkdir 'models'
!wget https://www.dropbox.com/s/zkehq1uwahhbc2o/ColorizeArtistic_gen.pth?dl=0 -O ./models/ColorizeArtistic_gen.pth
!wget https://media.githubusercontent.com/media/jantic/DeOldify/master/resource_images/watermark.png -O ./resource_images/watermark.png
colorizer = get_image_colorizer(artistic=True)
```
#◢ Instructions
### source_url
Type in a url to a direct link of an image. Usually that means they'll end in .png, .jpg, etc. NOTE: If you want to use your own image, upload it first to a site like Imgur.
### render_factor
The default value of 35 has been carefully chosen and should work -ok- for most scenarios (but probably won't be the -best-). This determines resolution at which the color portion of the image is rendered. Lower resolution will render faster, and colors also tend to look more vibrant. Older and lower quality images in particular will generally benefit by lowering the render factor. Higher render factors are often better for higher quality images, but the colors may get slightly washed out.
### watermarked
Selected by default, this places a watermark icon of a palette at the bottom left corner of the image. This is intended to be a standard way to convey to others viewing the image that it is colorized by AI. We want to help promote this as a standard, especially as the technology continues to improve and the distinction between real and fake becomes harder to discern. This palette watermark practice was initiated and lead by the company MyHeritage in the MyHeritage In Color feature (which uses a newer version of DeOldify than what you're using here).
#### How to Download a Copy
Simply right click on the displayed image and click "Save image as..."!
## Pro Tips
You can evaluate how well the image is rendered at each render_factor by using the code at the bottom (that cell under "See how well render_factor values perform on a frame here").
#◢ Colorize!!
```
source_url = '' #@param {type:"string"}
render_factor = 35 #@param {type: "slider", min: 7, max: 40}
watermarked = True #@param {type:"boolean"}
if source_url is not None and source_url !='':
image_path = colorizer.plot_transformed_image_from_url(url=source_url, render_factor=render_factor, compare=True, watermarked=watermarked)
show_image_in_notebook(image_path)
else:
print('Provide an image url and try again.')
```
## See how well render_factor values perform on the image here
```
for i in range(10,40,2):
colorizer.plot_transformed_image('test_images/image.png', render_factor=i, display_render_factor=True, figsize=(8,8))
```
---
#⚙ Recommended image sources
* [/r/TheWayWeWere](https://www.reddit.com/r/TheWayWeWere/)
|
github_jupyter
|
# Airbnb - Rio de Janeiro
* Download [data](http://insideairbnb.com/get-the-data.html)
* We downloaded `listings.csv` from all monthly dates available
## Questions
1. What was the price and supply behavior before and during the pandemic?
2. Does a title in English or Portuguese impact the price?
3. What features correlate with the price? Can we predict a price? Which features matters?
```
import numpy as np
import pandas as pd
import seaborn as sns
import glob
import re
import pendulum
import tqdm
import matplotlib.pyplot as plt
import langid
langid.set_languages(['en','pt'])
```
### Read files
Read all 30 files and get their date
```
files = sorted(glob.glob('data/listings*.csv'))
df = []
for f in files:
date = pendulum.from_format(re.findall(r"\d{4}_\d{2}_\d{2}", f)[0], fmt="YYYY_MM_DD").naive()
csv = pd.read_csv(f)
csv["date"] = date
df.append(csv)
df = pd.concat(df)
df
```
### Deal with NaNs
* Drop `neighbourhood_group` as it is all NaNs;
* Fill `reviews_per_month` with zeros (if there is no review, then reviews per month are zero)
* Keep `name` for now
* Drop `host_name` rows, as there is not any null host_id
* Keep `last_review` too, as there are rooms with no review
```
df.isna().any()
df = df.drop(["host_name", "neighbourhood_group"], axis=1)
df["reviews_per_month"] = df["reviews_per_month"].fillna(0.)
df.head()
```
### Detect `name` language
* Clean strings for evaluation
* Remove common neighbourhoods name in Portuguese from `name` column to diminish misprediction
* Remove several non-alphanumeric characters
* Detect language using [langid](https://github.com/saffsd/langid.py)
* I restricted between pt, en. There are very few rooms listed in other languages.
* Drop `name` column
```
import unicodedata
stopwords = pd.unique(df["neighbourhood"])
stopwords = [re.sub(r"[\(\)]", "", x.lower().strip()).split() for x in stopwords]
stopwords = [x for item in stopwords for x in item]
stopwords += [unicodedata.normalize("NFKD", x).encode('ASCII', 'ignore').decode() for x in stopwords]
stopwords += ["rio", "janeiro", "copa", "arpoador", "pepê", "pepe", "lapa", "morro", "corcovado"]
stopwords = set(stopwords)
docs = [re.sub(r"[\-\_\\\/\,\;\:\!\+\’\%\&\d\*\#\"\´\`\.\|\(\)\[\]\@\'\»\«\>\<\❤️\…]", " ", str(x)) for x in df["name"].tolist()]
docs = [" ".join(x.lower().strip().split()) for x in docs]
docs = ["".join(e for e in x if (e.isalnum() or " ")) for x in docs]
ndocs = []
for doc in tqdm.tqdm(docs):
ndocs.append(" ".join([x for x in doc.split() if x not in stopwords]))
docs = ndocs
results = []
for d in tqdm.tqdm(docs):
results.append(langid.classify(d)[0])
df["language"] = results
# Because we transformed NaNs into string, fill those detection with nans too
df.loc[df["name"].isna(), "language"] = pd.NA
```
* Test accuracy, manually label 383 out of 88191 (95% conf. interval, 5% margin of error)
```
df.loc[~df["name"].isna()].drop_duplicates("name").shape
df.loc[~df["name"].isna()].drop_duplicates("name")[["name", "language"]].sample(n=383, random_state=42).to_csv("lang_pred_1.csv")
lang_pred = pd.read_csv("lang_pred.csv", index_col=0)
lang_pred.head()
overall_accuracy = (lang_pred["pred"] == lang_pred["true"]).sum() / lang_pred.shape[0]
pt_accuracy = (lang_pred[lang_pred["true"] == "pt"]["true"] == lang_pred[lang_pred["true"] == "pt"]["pred"]).sum() / lang_pred[lang_pred["true"] == "pt"].shape[0]
en_accuracy = (lang_pred[lang_pred["true"] == "en"]["true"] == lang_pred[lang_pred["true"] == "en"]["pred"]).sum() / lang_pred[lang_pred["true"] == "en"].shape[0]
print(f"Overall accuracy: {overall_accuracy*100}%")
print(f"Portuguese accuracy: {pt_accuracy*100}%")
print(f"English accuracy: {en_accuracy*100}%")
df = df.drop("name", axis=1)
df.head()
df["language"].value_counts()
```
### Calculate how many times a room appeared
* There are 30 months of data, and rooms appear multiple times
* Calculate for a specific date, how many times the same room appeared up to that date
```
df = df.set_index(["id", "date"])
df["appearances"] = df.groupby(["id", "date"])["host_id"].count().unstack().cumsum(axis=1).stack()
df = df.reset_index()
df.head()
```
### Days since last review
* Calculate days since last review
* Then categorize them by the length of the days
```
df.loc[:, "last_review"] = pd.to_datetime(df["last_review"], format="%Y/%m/%d")
# For each scraping date, consider the last date to serve as comparision as the maximum date
last_date = df.groupby("date")["last_review"].max()
df["last_date"] = df.apply(lambda row: last_date.loc[row["date"]], axis=1)
df["days_last_review"] = (df["last_date"] - df["last_review"]).dt.days
df = df.drop("last_date", axis=1)
df.head()
df["days_last_review"].describe()
def categorize_last_review(days_last_review):
"""Transform days since last review into categories
Transform days since last review into one of those categories:
last_week, last_month, last_half_year, last_year, last_two_years,
long_time_ago, or never
Args:
days_last_review (int): Days since the last review
Returns:
str: A string with the category name.
"""
if days_last_review <= 7:
return "last_week"
elif days_last_review <= 30:
return "last_month"
elif days_last_review <= 182:
return "last_half_year"
elif days_last_review <= 365:
return "last_year"
elif days_last_review <= 730:
return "last_two_years"
elif days_last_review > 730:
return "long_time_ago"
else:
return "never"
df.loc[:, "last_review"] = df.apply(lambda row: categorize_last_review(row["days_last_review"]), axis=1)
df = df.drop(["days_last_review"], axis=1)
df.head()
df = df.set_index(["id", "date"])
df.loc[:, "appearances"] = df["appearances"].astype(int)
df.loc[:, "host_id"] = df["host_id"].astype("category")
df.loc[:, "neighbourhood"] = df["neighbourhood"].astype("category")
df.loc[:, "room_type"] = df["room_type"].astype("category")
df.loc[:, "last_review"] = df["last_review"].astype("category")
df.loc[:, "language"] = df["language"].astype("category")
df
df.to_pickle("data.pkl")
```
### Distributions
* Check the distribution of features
```
df = pd.read_pickle("data.pkl")
df.head()
df["latitude"].hist(bins=250)
df["longitude"].hist(bins=250)
df["price"].hist(bins=250)
df["minimum_nights"].hist(bins=250)
df["number_of_reviews"].hist()
df["reviews_per_month"].hist(bins=250)
df["calculated_host_listings_count"].hist(bins=250)
df["availability_365"].hist()
df["appearances"].hist(bins=29)
df.describe()
```
### Limits
* We are analising mostly for touristic purpose, so get the short-term rentals only
* Prices between 10 and 10000 (The luxury Copacabana Palace Penthouse at 8000 for example)
* Short-term rentals (minimum_nights < 31)
* Impossibility of more than 31 reviews per month
```
df = pd.read_pickle("data.pkl")
total_records = len(df)
outbound_values = (df["price"] < 10) | (df["price"] > 10000)
df = df[~outbound_values]
print(f"Removed values {outbound_values.sum()}, {outbound_values.sum()*100/total_records}%")
long_term = df["minimum_nights"] >= 31
df = df[~long_term]
print(f"Removed values {long_term.sum()}, {long_term.sum()*100/total_records}%")
reviews_limit = df["reviews_per_month"] > 31
df = df[~reviews_limit]
print(f"Removed values {reviews_limit.sum()}, {reviews_limit.sum()*100/total_records}%")
```
### Log skewed variables
* Most numerical values are skewed, so log them
```
df.describe()
# number_of_reviews, reviews_per_month, availability_365 have zeros, thus sum one to all
df["number_of_reviews"] = np.log(df["number_of_reviews"] + 1)
df["reviews_per_month"] = np.log(df["reviews_per_month"] + 1)
df["availability_365"] = np.log(df["availability_365"] + 1)
df["price"] = np.log(df["price"])
df["minimum_nights"] = np.log(df["minimum_nights"])
df["calculated_host_listings_count"] = np.log(df["calculated_host_listings_count"])
df["appearances"] = np.log(df["appearances"])
df.describe()
```
### Extreme outliers
* Most outliers are clearly mistyped values (one can check these rooms ids in their website)
* Remove extreme outliers first from large deviations within the same `id` (eliminate rate jumps of same room)
* Then remove those from same scraping `date`, `neighbourhood` and `room_type`
```
df = df.reset_index()
q25 = df.groupby(["id"])["price"].quantile(0.25)
q75 = df.groupby(["id"])["price"].quantile(0.75)
ext = q75 + 3 * (q75 - q25)
ext = ext[(q75 - q25) > 0.]
affected_rows = []
multiple_id = df[df["id"].isin(ext.index)]
for row in tqdm.tqdm(multiple_id.itertuples(), total=len(multiple_id)):
if row.price >= ext.loc[row.id]:
affected_rows.append(row.Index)
df = df.drop(affected_rows)
print(f"Removed values {len(affected_rows)}, {len(affected_rows)*100/total_records}%")
# Remove extreme outliers per neighbourhood, room_type and scraping date
q25 = df.groupby(["date", "neighbourhood", "room_type"])["price"].quantile(0.25)
q75 = df.groupby(["date", "neighbourhood", "room_type"])["price"].quantile(0.75)
ext = q75 + 3 * (q75 - q25)
ext
affected_rows = []
for row in tqdm.tqdm(df.itertuples(), total=len(df)):
if row.price >= ext.loc[(row.date, row.neighbourhood, row.room_type)]:
affected_rows.append(row.Index)
df = df.drop(affected_rows)
print(f"Removed values {len(affected_rows)}, {len(affected_rows)*100/total_records}%")
df.describe()
df["price"].hist()
df.to_pickle("treated_data.pkl")
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/harvardnlp/pytorch-struct/blob/master/notebooks/Unsupervised_CFG.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!pip install -qqq torchtext -qqq pytorch-transformers dgl
!pip install -qqqU git+https://github.com/harvardnlp/pytorch-struct
import torchtext
import torch
from torch_struct import SentCFG
from torch_struct.networks import NeuralCFG
import torch_struct.data
# Download and the load default data.
WORD = torchtext.data.Field(include_lengths=True)
UD_TAG = torchtext.data.Field(init_token="<bos>", eos_token="<eos>", include_lengths=True)
# Download and the load default data.
train, val, test = torchtext.datasets.UDPOS.splits(
fields=(('word', WORD), ('udtag', UD_TAG), (None, None)),
filter_pred=lambda ex: 5 < len(ex.word) < 30
)
WORD.build_vocab(train.word, min_freq=3)
UD_TAG.build_vocab(train.udtag)
train_iter = torch_struct.data.TokenBucket(train,
batch_size=200,
device="cuda:0")
H = 256
T = 30
NT = 30
model = NeuralCFG(len(WORD.vocab), T, NT, H)
model.cuda()
opt = torch.optim.Adam(model.parameters(), lr=0.001, betas=[0.75, 0.999])
def train():
#model.train()
losses = []
for epoch in range(10):
for i, ex in enumerate(train_iter):
opt.zero_grad()
words, lengths = ex.word
N, batch = words.shape
words = words.long()
params = model(words.cuda().transpose(0, 1))
dist = SentCFG(params, lengths=lengths)
loss = dist.partition.mean()
(-loss).backward()
losses.append(loss.detach())
torch.nn.utils.clip_grad_norm_(model.parameters(), 3.0)
opt.step()
if i % 100 == 1:
print(-torch.tensor(losses).mean(), words.shape)
losses = []
train()
for i, ex in enumerate(train_iter):
opt.zero_grad()
words, lengths = ex.word
N, batch = words.shape
words = words.long()
params = terms(words.transpose(0, 1)), rules(batch), roots(batch)
tree = CKY(MaxSemiring).marginals(params, lengths=lengths, _autograd=True)
print(tree)
break
def split(spans):
batch, N = spans.shape[:2]
splits = []
for b in range(batch):
cover = spans[b].nonzero()
left = {i: [] for i in range(N)}
right = {i: [] for i in range(N)}
batch_split = {}
for i in range(cover.shape[0]):
i, j, A = cover[i].tolist()
left[i].append((A, j, j - i + 1))
right[j].append((A, i, j - i + 1))
for i in range(cover.shape[0]):
i, j, A = cover[i].tolist()
B = None
for B_p, k, a_span in left[i]:
for C_p, k_2, b_span in right[j]:
if k_2 == k + 1 and a_span + b_span == j - i + 1:
B, C = B_p, C_p
k_final = k
break
if j > i:
batch_split[(i, j)] =k
splits.append(batch_split)
return splits
splits = split(spans)
```
|
github_jupyter
|
# Assignment 9: Implement Dynamic Programming
In this exercise, we will begin to explore the concept of dynamic programming and how it related to various object containers with respect to computational complexity.
## Deliverables:
1) Choose and implement a Dynamic Programming algorithm in Python, make sure you are using a Dynamic Programming solution (not another one).
2) Use the algorithm to solve a range of scenarios.
3) Explain what is being done in the implementation. That is, write up a walk through of the algorithm and explain how it is a Dynamic Programming solution.
### Prepare an executive summary of your results, referring to the table and figures you have generated. Explain how your results relate to big O notation. Describe your results in language that management can understand. This summary should be included as text paragraphs in the Jupyter notebook. Explain how the algorithm works and why it is a useful to data engineers.
# A. The Dynamic programming problem: Longest Increasing Sequence
### The Longest Increasing Subsequence (LIS) problem is to find the length of the longest subsequence of a given sequence such that all elements of the subsequence are sorted in increasing order. For example, the length of LIS for {10, 22, 9, 33, 21, 50, 41, 60, 80} is 6 and LIS is {10, 22, 33, 50, 60, 80}.
# A. Setup: Library imports and Algorithm
```
import numpy as np
import pandas as pd
import seaborn as sns
import time
#import itertools
import random
import matplotlib.pyplot as plt
#import networkx as nx
#import pydot
#from networkx.drawing.nx_pydot import graphviz_layout
#from collections import deque
# Dynamic Programming Approach of Finding LIS by reducing the problem to longest common Subsequence
def lis(a):
n=len(a) #get the length of the list
b=sorted(list(set(a))) #removes duplicates, and sorts list
m=len(b) #gets the length of the truncated and sorted list
dp=[[-1 for i in range(m+1)] for j in range(n+1)] #instantiates a list of lists filled with -1 columns are indicies of the sorted array; rows the original array
for i in range(n+1): # for every column in the table at each row:
for j in range(m+1):
if i==0 or j==0: #if at first element in either a row or column set the table row,index to zero
dp[i][j]=0
elif a[i-1]==b[j-1]: #else if the sorted array value matches the original array:
dp[i][j]=1+dp[i-1][j-1]#sets dp[i][j] to 1+prveious cell of the dyanmic table
else:
dp[i][j]=max(dp[i-1][j],dp[i][j-1]) #else record the max of the row or column for that cell in the cell
return dp[-1][-1] # This will return the max running sequence.
# Driver program to test above function
arr1 = [10, 22, 9, 33, 21, 50, 41, 60]
len_arr1 = len(arr1)
print("Longest increaseing sequence has a length of:", lis(arr1))
# addtional comments included from the original code contributed by Dheeraj Khatri (https://www.geeksforgeeks.org/longest-increasing-subsequence-dp-3/)
def Container(arr, fun): ### I'm glad I was able to reuse this from assignment 3 and 4. Useful function.
objects = [] #instantiates an empty list to collect the returns
times = [] #instantiates an empty list to collect times for each computation
for t in arr:
start= time.perf_counter() #collects the start time
obj = fun(t) # applies the function to the arr object
end = time.perf_counter() # collects end time
duration = (end-start)* 1E3 #converts to milliseconds
objects.append(obj)# adds the returns of the functions to the objects list
times.append(duration) # adds the duration for computation to list
return objects, times
```
# B. Test Array Generation
```
RANDOM_SEED = 300
np.random.seed(RANDOM_SEED)
arr100 = list(np.random.randint(low=1, high= 5000, size=100))
np.random.seed(RANDOM_SEED)
arr200 = list(np.random.randint(low=1, high= 5000, size=200))
np.random.seed(RANDOM_SEED)
arr400 = list(np.random.randint(low=1, high= 5000, size=400))
np.random.seed(RANDOM_SEED)
arr600 = list(np.random.randint(low=1, high= 5000, size=600))
np.random.seed(RANDOM_SEED)
arr800 = list(np.random.randint(low=1, high= 5000, size=800))
print(len(arr100), len(arr200), len(arr400), len(arr600), len(arr800))
arr_list = [arr100, arr200, arr400, arr600, arr800]
metrics = Container(arr_list, lis)
```
### Table1. Performance Summary
```
summary = {
'ArraySize' : [len(arr100), len(arr200), len(arr400), len(arr600), len(arr800)],
'SequenceLength' : [metrics[0][0],metrics[0][1], metrics[0][2], metrics[0][3], metrics[0][4]],
'Time(ms)' : [metrics[1][0],metrics[1][1], metrics[1][2], metrics[1][3], metrics[1][4]]
}
df =pd.DataFrame(summary)
df
```
### Figure 1. Performance
```
sns.scatterplot(data=df, x='Time(ms)', y='ArraySize')
```
# Discussion
Explain what is being done in the implementation. That is, write up a walk through of the algorithm and explain how it is a Dynamic Programming solution.
The dyanamic programming problem above finds the length of the longest incrementing sequence of values in a list. The defined function makes a sorted copy of the list containing only unique values and also creates a dynamic table (in the form of a list of lists) using a nested list comprehension. This table contains the incidices of the sorted array as columns and the indicies of the original array as rows. To begin, the table is instantiated with values of -1. The value of zero indicies are set to zero in the dynamic table and if a given index in the original array is found to be increasing the dyanamic table is incremented. until all positions are assessed. The funciton then returns the maximum value of the increments which will be the length of the longest running sequence. This is a dynamic progromming problem because the solution builds on a smaller subset problems.
Dyanmic programming is an important concept for developers and engineers. Functions and programs that use dynamic programming help solve problems which present themselves as factorial time complexity in a more efficient way. At face value, it appears that this problem of the longest incrementing sequence will have to compare all values in a given array to all previous values in the array. Dyanmic programming allows for a shortcut in a sense. We can compare the given array with a sorted version of that array and at the intersection of the sorted and unsorted arrays we can determine if we need to make an additon to our incrementing sequence tally.
Shown above in table and figure 1 is the time required for the algorithm to tally the longest running sequence for various array sizes. Because the algorithm utilizes a nested for loop it is the expeictation that the time will grow as a function of the square of the original array length. This is confirmed when inspecting the scatterplot in figure 1. Thus, the developed algorithm in big O notation is O(n^2) time complexity which is much more efficient than factorial time.
|
github_jupyter
|

# If I have seen further it is by standing on the shoulders of Giants
(Newton??)

(https://www.openhub.net/)

(https://www.openhub.net/)

(https://www.openhub.net/)

(https://www.openhub.net/)

(https://www.openhub.net/)
### Pero, ¿qué es lo que hace fuertes a estos proyectos?

(https://medium.com/@sailorhg/coding-like-a-girl-595b90791cce)

## Codigo de conducta
### PyCon 2016 Code Of Conduct
Harassment includes offensive communication related to gender, sexual orientation, disability, physical appearance, body size, race, religion, sexual images in public spaces, deliberate intimidation, stalking, following, harassing photography or recording, sustained disruption of talks or other events, inappropriate physical contact, and unwelcome sexual attention.
Participants asked to stop any harassing behavior are expected to comply immediately.
Exhibitors in the expo hall, sponsor or vendor booths, or similar activities are also subject to the anti-harassment policy. In particular, exhibitors should not use sexualized images, activities, or other material. Booth staff (including volunteers) should not use sexualized clothing/uniforms/costumes, or otherwise create a sexualized environment.
Be careful in the words that you choose. Remember that sexist, racist, and other exclusionary jokes can be offensive to those around you. Excessive swearing and offensive jokes are not appropriate for PyCon.
If a participant engages in behavior that violates this code of conduct, the conference organizers may take any action they deem appropriate, including warning the offender or expulsion from the conference with no refund.
## Políticas de inclusión

__The mission of the Python Software Foundation is to promote, protect, and advance the Python programming language, and to support and facilitate the growth of a diverse and international community of Python programmers.__
The Python Software Foundation (PSF) is a 501(c)(3) non-profit corporation that holds the intellectual property rights behind the Python programming language. We manage the open source licensing for Python version 2.1 and later and own and protect the trademarks associated with Python. We also run the North American PyCon conference annually, support other Python conferences around the world, and fund Python related development with our grants program and by funding special projects.
(https://www.python.org/psf/)

__We inspire women to fall in love with programming.__
_Django Girls organize free Python and Django workshops, create open sourced online tutorials and curate amazing first experiences with technology._
(https://djangogirls.org/)


We are an international mentorship group with a focus on helping more women become active participants and leaders in the Python open-source community. Our mission is to promote, educate and advance a diverse Python community through outreach, education, conferences, events and social gatherings.
PyLadies also aims to provide a friendly support network for women and a bridge to the larger Python world. Anyone with an interest in Python is encouraged to participate!
(http://www.pyladies.com/)

(https://www.slideshare.net/fmasanori/import-community-62142823)
# ¡Gracias!
<center>David Manuel Ochoa González<br>
correos: ochoadavid at gmail.com - dochoa at iteso.mx<br>
github: https://github.com/ochoadavid<br>
material de apoyo en: https://github.com/ochoadavid/TallerDePython</center>
|
github_jupyter
|
# T1566 - Phishing
Adversaries may send phishing messages to elicit sensitive information and/or gain access to victim systems. All forms of phishing are electronically delivered social engineering. Phishing can be targeted, known as spearphishing. In spearphishing, a specific individual, company, or industry will be targeted by the adversary. More generally, adversaries can conduct non-targeted phishing, such as in mass malware spam campaigns.
Adversaries may send victim’s emails containing malicious attachments or links, typically to execute malicious code on victim systems or to gather credentials for use of [Valid Accounts](https://attack.mitre.org/techniques/T1078). Phishing may also be conducted via third-party services, like social media platforms.
## Atomic Tests:
Currently, no tests are available for this technique.
## Detection
Network intrusion detection systems and email gateways can be used to detect phishing with malicious attachments in transit. Detonation chambers may also be used to identify malicious attachments. Solutions can be signature and behavior based, but adversaries may construct attachments in a way to avoid these systems.
URL inspection within email (including expanding shortened links) can help detect links leading to known malicious sites. Detonation chambers can be used to detect these links and either automatically go to these sites to determine if they're potentially malicious, or wait and capture the content if a user visits the link.
Because most common third-party services used for phishing via service leverage TLS encryption, SSL/TLS inspection is generally required to detect the initial communication/delivery. With SSL/TLS inspection intrusion detection signatures or other security gateway appliances may be able to detect malware.
Anti-virus can potentially detect malicious documents and files that are downloaded on the user's computer. Many possible detections of follow-on behavior may take place once [User Execution](https://attack.mitre.org/techniques/T1204) occurs.
## Shield Active Defense
### Email Manipulation
Modify the flow or contents of email.
Email flow manipulation includes changing which mail appliances process mail flows, to which systems they forward mail, or moving mail after it arrives in an inbox. Email content manipulation includes altering the contents of an email message.
#### Opportunity
A phishing email can be detected and blocked from arriving at the intended recipient.
#### Use Case
A defender can intercept emails that are detected as suspicious or malicious by email detection tools and prevent deliver to the intended target.
#### Procedures
Modify the destination of inbound email to facilitate the collection of inbound spearphishing messages.
Modify the contents of an email message to maintain continuity when it is used for adversary engagement purposes.
|
github_jupyter
|
```
%matplotlib inline
```
02: Fitting Power Spectrum Models
=================================
Introduction to the module, beginning with the FOOOF object.
```
# Import the FOOOF object
from fooof import FOOOF
# Import utility to download and load example data
from fooof.utils.download import load_fooof_data
# Download examples data files needed for this example
freqs = load_fooof_data('freqs.npy', folder='data')
spectrum = load_fooof_data('spectrum.npy', folder='data')
```
FOOOF Object
------------
At the core of the module, which is object oriented, is the :class:`~fooof.FOOOF` object,
which holds relevant data and settings as attributes, and contains methods to run the
algorithm to parameterize neural power spectra.
The organization is similar to sklearn:
- A model object is initialized, with relevant settings
- The model is used to fit the data
- Results can be extracted from the object
Calculating Power Spectra
~~~~~~~~~~~~~~~~~~~~~~~~~
The :class:`~fooof.FOOOF` object fits models to power spectra. The module itself does not
compute power spectra, and so computing power spectra needs to be done prior to using
the FOOOF module.
The model is broadly agnostic to exactly how power spectra are computed. Common
methods, such as Welch's method, can be used to compute the spectrum.
If you need a module in Python that has functionality for computing power spectra, try
`NeuroDSP <https://neurodsp-tools.github.io/neurodsp/>`_.
Note that FOOOF objects require frequency and power values passed in as inputs to
be in linear spacing. Passing in non-linear spaced data (such logged values) may
produce erroneous results.
Fitting an Example Power Spectrum
---------------------------------
The following example demonstrates fitting a power spectrum model to a single power spectrum.
```
# Initialize a FOOOF object
fm = FOOOF()
# Set the frequency range to fit the model
freq_range = [2, 40]
# Report: fit the model, print the resulting parameters, and plot the reconstruction
fm.report(freqs, spectrum, freq_range)
```
Fitting Models with 'Report'
~~~~~~~~~~~~~~~~~~~~~~~~~~~~
The above method 'report', is a convenience method that calls a series of methods:
- :meth:`~fooof.FOOOF.fit`: fits the power spectrum model
- :meth:`~fooof.FOOOF.print_results`: prints out the results
- :meth:`~fooof.FOOOF.plot`: plots to data and model fit
Each of these methods can also be called individually.
```
# Alternatively, just fit the model with FOOOF.fit() (without printing anything)
fm.fit(freqs, spectrum, freq_range)
# After fitting, plotting and parameter fitting can be called independently:
# fm.print_results()
# fm.plot()
```
Model Parameters
~~~~~~~~~~~~~~~~
Once the power spectrum model has been calculated, the model fit parameters are stored
as object attributes that can be accessed after fitting.
Following the sklearn convention, attributes that are fit as a result of
the model have a trailing underscore, for example:
- ``aperiodic_params_``
- ``peak_params_``
- ``error_``
- ``r2_``
- ``n_peaks_``
Access model fit parameters from FOOOF object, after fitting:
```
# Aperiodic parameters
print('Aperiodic parameters: \n', fm.aperiodic_params_, '\n')
# Peak parameters
print('Peak parameters: \n', fm.peak_params_, '\n')
# Goodness of fit measures
print('Goodness of fit:')
print(' Error - ', fm.error_)
print(' R^2 - ', fm.r_squared_, '\n')
# Check how many peaks were fit
print('Number of fit peaks: \n', fm.n_peaks_)
```
Selecting Parameters
~~~~~~~~~~~~~~~~~~~~
You can also select parameters using the :meth:`~fooof.FOOOF.get_params`
method, which can be used to specify which parameters you want to extract.
```
# Extract a model parameter with `get_params`
err = fm.get_params('error')
# Extract parameters, indicating sub-selections of parameter
exp = fm.get_params('aperiodic_params', 'exponent')
cfs = fm.get_params('peak_params', 'CF')
# Print out a custom parameter report
template = ("With an error level of {error:1.2f}, FOOOF fit an exponent "
"of {exponent:1.2f} and peaks of {cfs:s} Hz.")
print(template.format(error=err, exponent=exp,
cfs=' & '.join(map(str, [round(cf, 2) for cf in cfs]))))
```
For a full description of how you can access data with :meth:`~fooof.FOOOF.get_params`,
check the method's documentation.
As a reminder, you can access the documentation for a function using '?' in a
Jupyter notebook (ex: `fm.get_params?`), or more generally with the `help` function
in general Python (ex: `help(get_params)`).
Notes on Interpreting Peak Parameters
-------------------------------------
Peak parameters are labeled as:
- CF: center frequency of the extracted peak
- PW: power of the peak, over and above the aperiodic component
- BW: bandwidth of the extracted peak
Note that the peak parameters that are returned are not exactly the same as the
parameters of the Gaussians used internally to fit the peaks.
Specifically:
- CF is the exact same as mean parameter of the Gaussian
- PW is the height of the model fit above the aperiodic component [1],
which is not necessarily the same as the Gaussian height
- BW is 2 * the standard deviation of the Gaussian [2]
[1] Since the Gaussians are fit together, if any Gaussians overlap,
than the actual height of the fit at a given point can only be assessed
when considering all Gaussians. To be better able to interpret heights
for single peak fits, we re-define the peak height as above, and label it
as 'power', as the units of the input data are expected to be units of power.
[2] Gaussian standard deviation is '1 sided', where as the returned BW is '2 sided'.
The underlying gaussian parameters are also available from the FOOOF object,
in the ``gaussian_params_`` attribute.
```
# Compare the 'peak_params_' to the underlying gaussian parameters
print(' Peak Parameters \t Gaussian Parameters')
for peak, gauss in zip(fm.peak_params_, fm.gaussian_params_):
print('{:5.2f} {:5.2f} {:5.2f} \t {:5.2f} {:5.2f} {:5.2f}'.format(*peak, *gauss))
```
FOOOFResults
~~~~~~~~~~~~
There is also a convenience method to return all model fit results:
:func:`~fooof.FOOOF.get_results`.
This method returns all the model fit parameters, including the underlying Gaussian
parameters, collected together into a FOOOFResults object.
The FOOOFResults object, which in Python terms is a named tuple, is a standard data
object used with FOOOF to organize and collect parameter data.
```
# Grab each model fit result with `get_results` to gather all results together
# Note that this returns a FOOOFResult object
fres = fm.get_results()
# You can also unpack all fit parameters when using `get_results`
ap_params, peak_params, r_squared, fit_error, gauss_params = fm.get_results()
# Print out the FOOOFResults
print(fres, '\n')
# From FOOOFResults, you can access the different results
print('Aperiodic Parameters: \n', fres.aperiodic_params)
# Check the r^2 and error of the model fit
print('R-squared: \n {:5.4f}'.format(fm.r_squared_))
print('Fit error: \n {:5.4f}'.format(fm.error_))
```
Conclusion
----------
In this tutorial, we have explored the basics of the :class:`~fooof.FOOOF` object,
fitting power spectrum models, and extracting parameters.
Before we move on to controlling the fit procedure, and interpreting the results,
in the next tutorial, we will first explore how this model is actually fit.
|
github_jupyter
|
## Discretisation
Discretisation is the process of transforming continuous variables into discrete variables by creating a set of contiguous intervals that span the range of the variable's values. Discretisation is also called **binning**, where bin is an alternative name for interval.
### Discretisation helps handle outliers and may improve value spread in skewed variables
Discretisation helps handle outliers by placing these values into the lower or higher intervals, together with the remaining inlier values of the distribution. Thus, these outlier observations no longer differ from the rest of the values at the tails of the distribution, as they are now all together in the same interval / bucket. In addition, by creating appropriate bins or intervals, discretisation can help spread the values of a skewed variable across a set of bins with equal number of observations.
### Discretisation approaches
There are several approaches to transform continuous variables into discrete ones. Discretisation methods fall into 2 categories: **supervised and unsupervised**. Unsupervised methods do not use any information, other than the variable distribution, to create the contiguous bins in which the values will be placed. Supervised methods typically use target information in order to create the bins or intervals.
#### Unsupervised discretisation methods
- Equal width discretisation
- Equal frequency discretisation
- K-means discretisation
#### Supervised discretisation methods
- Discretisation using decision trees
In this lecture, I will describe **equal width discretisation**.
## Equal width discretisation
Equal width discretisation divides the scope of possible values into N bins of the same width.The width is determined by the range of values in the variable and the number of bins we wish to use to divide the variable:
width = (max value - min value) / N
where N is the number of bins or intervals.
For example if the values of the variable vary between 0 and 100, we create 5 bins like this: width = (100-0) / 5 = 20. The bins thus are 0-20, 20-40, 40-60, 80-100. The first and final bins (0-20 and 80-100) can be expanded to accommodate outliers (that is, values under 0 or greater than 100 would be placed in those bins as well).
There is no rule of thumb to define N, that is something to determine experimentally.
## In this demo
We will learn how to perform equal width binning using the Titanic dataset with
- pandas and NumPy
- Feature-engine
- Scikit-learn
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import KBinsDiscretizer
from feature_engine.discretisers import EqualWidthDiscretiser
# load the numerical variables of the Titanic Dataset
data = pd.read_csv('../titanic.csv',
usecols=['age', 'fare', 'survived'])
data.head()
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(
data[['age', 'fare']],
data['survived'],
test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
```
The variables Age and fare contain missing data, that I will fill by extracting a random sample of the variable.
```
def impute_na(data, variable):
df = data.copy()
# random sampling
df[variable + '_random'] = df[variable]
# extract the random sample to fill the na
random_sample = X_train[variable].dropna().sample(
df[variable].isnull().sum(), random_state=0)
# pandas needs to have the same index in order to merge datasets
random_sample.index = df[df[variable].isnull()].index
df.loc[df[variable].isnull(), variable + '_random'] = random_sample
return df[variable + '_random']
# replace NA in both train and test sets
X_train['age'] = impute_na(data, 'age')
X_test['age'] = impute_na(data, 'age')
X_train['fare'] = impute_na(data, 'fare')
X_test['fare'] = impute_na(data, 'fare')
# let's explore the distribution of age
data[['age', 'fare']].hist(bins=30, figsize=(8,4))
plt.show()
```
## Equal width discretisation with pandas and NumPy
First we need to determine the intervals' edges or limits.
```
# let's capture the range of the variable age
age_range = X_train['age'].max() - X_train['age'].min()
age_range
# let's divide the range into 10 equal width bins
age_range / 10
```
The range or width of our intervals will be 7 years.
```
# now let's capture the lower and upper boundaries
min_value = int(np.floor( X_train['age'].min()))
max_value = int(np.ceil( X_train['age'].max()))
# let's round the bin width
inter_value = int(np.round(age_range / 10))
min_value, max_value, inter_value
# let's capture the interval limits, so we can pass them to the pandas cut
# function to generate the bins
intervals = [i for i in range(min_value, max_value+inter_value, inter_value)]
intervals
# let's make labels to label the different bins
labels = ['Bin_' + str(i) for i in range(1, len(intervals))]
labels
# create binned age / discretise age
# create one column with labels
X_train['Age_disc_labels'] = pd.cut(x=X_train['age'],
bins=intervals,
labels=labels,
include_lowest=True)
# and one with bin boundaries
X_train['Age_disc'] = pd.cut(x=X_train['age'],
bins=intervals,
include_lowest=True)
X_train.head(10)
```
We can see in the above output how by discretising using equal width, we placed each Age observation within one interval / bin. For example, age=13 was placed in the 7-14 interval, whereas age 30 was placed into the 28-35 interval.
When performing equal width discretisation, we guarantee that the intervals are all of the same lenght, however there won't necessarily be the same number of observations in each of the intervals. See below:
```
X_train.groupby('Age_disc')['age'].count()
X_train.groupby('Age_disc')['age'].count().plot.bar()
plt.xticks(rotation=45)
plt.ylabel('Number of observations per bin')
```
The majority of people on the Titanic were between 14-42 years of age.
Now, we can discretise Age in the test set, using the same interval boundaries that we calculated for the train set:
```
X_test['Age_disc_labels'] = pd.cut(x=X_test['age'],
bins=intervals,
labels=labels,
include_lowest=True)
X_test['Age_disc'] = pd.cut(x=X_test['age'],
bins=intervals,
include_lowest=True)
X_test.head()
# if the distributions in train and test set are similar, we should expect similar propotion of
# observations in the different intervals in the train and test set
# let's see that below
t1 = X_train.groupby(['Age_disc'])['age'].count() / len(X_train)
t2 = X_test.groupby(['Age_disc'])['age'].count() / len(X_test)
tmp = pd.concat([t1, t2], axis=1)
tmp.columns = ['train', 'test']
tmp.plot.bar()
plt.xticks(rotation=45)
plt.ylabel('Number of observations per bin')
```
## Equal width discretisation with Feature-Engine
```
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(
data[['age', 'fare']],
data['survived'],
test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
# replace NA in both train and test sets
X_train['age'] = impute_na(data, 'age')
X_test['age'] = impute_na(data, 'age')
X_train['fare'] = impute_na(data, 'fare')
X_test['fare'] = impute_na(data, 'fare')
# with feature engine we can automate the process for many variables
# in one line of code
disc = EqualWidthDiscretiser(bins=10, variables = ['age', 'fare'])
disc.fit(X_train)
# in the binner dict, we can see the limits of the intervals. For age
# the value increases aproximately 7 years from one bin to the next.
# for fare it increases in around 50 dollars from one interval to the
# next, but it increases always the same value, aka, same width.
disc.binner_dict_
# transform train and text
train_t = disc.transform(X_train)
test_t = disc.transform(X_test)
train_t.head()
t1 = train_t.groupby(['age'])['age'].count() / len(train_t)
t2 = test_t.groupby(['age'])['age'].count() / len(test_t)
tmp = pd.concat([t1, t2], axis=1)
tmp.columns = ['train', 'test']
tmp.plot.bar()
plt.xticks(rotation=0)
plt.ylabel('Number of observations per bin')
t1 = train_t.groupby(['fare'])['fare'].count() / len(train_t)
t2 = test_t.groupby(['fare'])['fare'].count() / len(test_t)
tmp = pd.concat([t1, t2], axis=1)
tmp.columns = ['train', 'test']
tmp.plot.bar()
plt.xticks(rotation=0)
plt.ylabel('Number of observations per bin')
```
We can see quite clearly, that equal width discretisation does not improve the value spread. The original variable Fare was skewed, and the discrete variable is also skewed.
## Equal width discretisation with Scikit-learn
```
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(
data[['age', 'fare']],
data['survived'],
test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
# replace NA in both train and test sets
X_train['age'] = impute_na(data, 'age')
X_test['age'] = impute_na(data, 'age')
X_train['fare'] = impute_na(data, 'fare')
X_test['fare'] = impute_na(data, 'fare')
disc = KBinsDiscretizer(n_bins=10, encode='ordinal', strategy='uniform')
disc.fit(X_train[['age', 'fare']])
disc.bin_edges_
train_t = disc.transform(X_train[['age', 'fare']])
train_t = pd.DataFrame(train_t, columns = ['age', 'fare'])
train_t.head()
test_t = disc.transform(X_test[['age', 'fare']])
test_t = pd.DataFrame(test_t, columns = ['age', 'fare'])
t1 = train_t.groupby(['age'])['age'].count() / len(train_t)
t2 = test_t.groupby(['age'])['age'].count() / len(test_t)
tmp = pd.concat([t1, t2], axis=1)
tmp.columns = ['train', 'test']
tmp.plot.bar()
plt.xticks(rotation=0)
plt.ylabel('Number of observations per bin')
t1 = train_t.groupby(['fare'])['fare'].count() / len(train_t)
t2 = test_t.groupby(['fare'])['fare'].count() / len(test_t)
tmp = pd.concat([t1, t2], axis=1)
tmp.columns = ['train', 'test']
tmp.plot.bar()
plt.xticks(rotation=0)
plt.ylabel('Number of observations per bin')
```
|
github_jupyter
|
## Obligatory imports
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
import matplotlib
%matplotlib inline
matplotlib.rcParams['figure.figsize'] = (12,8)
matplotlib.rcParams['font.size']=20
matplotlib.rcParams['lines.linewidth']=4
matplotlib.rcParams['xtick.major.size'] = 10
matplotlib.rcParams['ytick.major.size'] = 10
matplotlib.rcParams['xtick.major.width'] = 2
matplotlib.rcParams['ytick.major.width'] = 2
```
# We use the MNIST Dataset again
```
import IPython
url = 'http://yann.lecun.com/exdb/mnist/'
iframe = '<iframe src=' + url + ' width=80% height=400px></iframe>'
IPython.display.HTML(iframe)
```
## Fetch the data
```
from sklearn.datasets import fetch_mldata
mnist = fetch_mldata('MNIST original', data_home='../day4/data/')
allimages = mnist.data
allimages.shape
all_image_labels = mnist.target
set(all_image_labels)
```
## check out the data
```
digit1 = mnist.data[0,:].reshape(28,-1) # arr.reshape(4, -1) is equivalent to arr.reshape(4, 7), is arr has size 28
fig, ax = plt.subplots(figsize=(1.5, 1.5))
ax.imshow(digit1, vmin=0, vmax=1)
```
# Theoretical background
**Warning: math ahead**
<img src="images/logreg_schematics.svg" alt="logreg-schematics" style="width: 50%;"/>
## Taking logistic regression a step further: neural networks
<img src="images/mlp_schematics.svg" alt="nn-schematics" style="width: 50%;"/>
### How (artificial) neural networks predict a label from features?
* The *input layer* has **dimention = number of features.**
* For each training example, each feature value is "fed" into the input layer.
* Each "neuron" in the hidden layer receives a weighted sum of the features: the weight is initialized to a random value in the beginning, and the network "learns" from the datasetsand tunes these weights. Each hidden neuron, based on its input, and an "activation function", e.g.: the logistic function

* The output is again, a weighted sum of the values at each hidden neuron.
* There can be *more than one hidden layer*, in which case the output of the first hidden layer becomes the input of the second hidden layer.
### Regularization
Like Logistic regression and SVM, neural networks also can be improved with regularization.
Fot scikit-learn, the relevant tunable parameter is `alpha` (as opposed to `gamma` for LR and SVM).
Furthermore, it has default value 0.0001, unlike gamma, for which it is 1.
### Separate the data into training data and test data
```
len(allimages)
```
### Sample the data, 70000 is too many images to handle on a single PC
```
len(allimages)
size_desired_dataset = 2000
sample_idx = np.random.choice(len(allimages), size_desired_dataset)
images = allimages[sample_idx, :]
image_labels = all_image_labels[sample_idx]
set(image_labels)
image_labels.shape
```
### Partition into training and test set *randomly*
**As a rule of thumb, 80/20 split between training/test dataset is often recommended.**
See below for cross validation and how that changes this thumbrule.
```
from scipy.stats import itemfreq
from sklearn.model_selection import train_test_split
training_data, test_data, training_labels, test_labels = train_test_split(images, image_labels, train_size=0.8)
```
** Importance of normalization**
If Feature A is in the range [0,1] and Feature B is in [10000,50000], SVM (in fact, most of the classifiers) will suffer inaccuracy.
The solution is to *normalize* (AKA "feature scaling") each feature to the same interval e.g. [0,1] or [-1, 1].
**scipy provides a standard function for this:**
```
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit only to the training data: IMPORTANT
scaler.fit(training_data)
from sklearn.neural_network import MLPClassifier
clf = MLPClassifier(hidden_layer_sizes=(50,), max_iter = 5000)
clf.fit(scaler.transform(training_data), training_labels)
clf.score(scaler.transform(training_data), training_labels), clf.score(scaler.transform(test_data), test_labels)
```
### Visualize the hidden layer:
```
# source:
#
#http://scikit-learn.org/stable/auto_examples/neural_networks/plot_mnist_filters.html
fig, axes = plt.subplots(4, 4, figsize=(15,15))
# use global min / max to ensure all weights are shown on the same scale
vmin, vmax = clf.coefs_[0].min(), clf.coefs_[0].max()
for coef, ax in zip(clf.coefs_[0].T, axes.ravel()):
ax.matshow(coef.reshape(28, 28), cmap=plt.cm.gray, vmin=.5 * vmin,
vmax=.5 * vmax)
ax.set_xticks(())
ax.set_yticks(())
plt.show()
```
Not bad, but is it better than Logistic regression? Check out with Learning curves:
```
from sklearn.model_selection import learning_curve
import pandas as pd
curve = learning_curve(clf, scaler.transform(images), image_labels)
train_sizes, train_scores, test_scores = curve
train_scores = pd.DataFrame(train_scores)
train_scores.loc[:,'train_size'] = train_sizes
test_scores = pd.DataFrame(test_scores)
test_scores.loc[:,'train_size'] = train_sizes
train_scores = pd.melt(train_scores, id_vars=['train_size'], value_name = 'CrossVal score')
test_scores = pd.melt(test_scores, id_vars=['train_size'], value_name = 'CrossVal score')
matplotlib.rcParams['figure.figsize'] = (12,8)
sns.tsplot(train_scores, time = 'train_size', unit='variable', value = 'CrossVal score')
sns.tsplot(test_scores, time = 'train_size', unit='variable', value = 'CrossVal score', color='g')
plt.ylim(0,1.1)
```
Not really, we can try to improve it with parameter space search.
## Parameter space search with `GridSearchCV`
```
from sklearn.model_selection import GridSearchCV
clr = MLPClassifier()
clf = GridSearchCV(clr, {'alpha':np.logspace(-8, -1, 2)})
clf.fit(scaler.transform(images), image_labels)
clf.best_params_
clf.best_score_
nn_tuned = clf.best_estimator_
nn_tuned.fit(scaler.transform(training_data), training_labels)
curve = learning_curve(nn_tuned, scaler.transform(images), image_labels)
train_sizes, train_scores, test_scores = curve
train_scores = pd.DataFrame(train_scores)
train_scores.loc[:,'train_size'] = train_sizes
test_scores = pd.DataFrame(test_scores)
test_scores.loc[:,'train_size'] = train_sizes
train_scores = pd.melt(train_scores, id_vars=['train_size'], value_name = 'CrossVal score')
test_scores = pd.melt(test_scores, id_vars=['train_size'], value_name = 'CrossVal score')
matplotlib.rcParams['figure.figsize'] = (12,8)
sns.tsplot(train_scores, time = 'train_size', unit='variable', value = 'CrossVal score')
sns.tsplot(test_scores, time = 'train_size', unit='variable', value = 'CrossVal score', color='g')
plt.ylim(0,1.1)
plt.legend()
```
The increase in accuracy is miniscule.
## Multi layered NN's
```
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(images)
images_normed = scaler.transform(images)
clr = MLPClassifier(hidden_layer_sizes=(25,25))
clf = GridSearchCV(clr, {'alpha':np.logspace(-80, -1, 3)})
clf.fit(images_normed, image_labels)
clf.best_score_
clf.best_params_
nn_tuned = clf.best_estimator_
nn_tuned.fit(scaler.transform(training_data), training_labels)
curve = learning_curve(nn_tuned, images_normed, image_labels)
train_sizes, train_scores, test_scores = curve
train_scores = pd.DataFrame(train_scores)
train_scores.loc[:,'train_size'] = train_sizes
test_scores = pd.DataFrame(test_scores)
test_scores.loc[:,'train_size'] = train_sizes
train_scores = pd.melt(train_scores, id_vars=['train_size'], value_name = 'CrossVal score')
test_scores = pd.melt(test_scores, id_vars=['train_size'], value_name = 'CrossVal score')
matplotlib.rcParams['figure.figsize'] = (12, 8)
sns.tsplot(train_scores, time = 'train_size', unit='variable', value = 'CrossVal score')
sns.tsplot(test_scores, time = 'train_size', unit='variable', value = 'CrossVal score', color='g')
plt.ylim(0,1.1)
plt.legend()
```
Hmm... multi-hidden layer NN's seem to be much harder to tune.
Maybe we need to try with wider range of parameters for Gridsearch?
Finding optimum parameters for advanced classifiers is not always so straightforward, and quite often the most time consuming part. This so-called **Hyperparameter optimization** is a topic in itself, and has numerous approaches and libraries.
* [http://neupy.com/2016/12/17/hyperparameter_optimization_for_neural_networks.html](http://neupy.com/2016/12/17/hyperparameter_optimization_for_neural_networks.html)
* [Practical Bayesian Optimization of Machine Learning Algorithms](https://dash.harvard.edu/handle/1/11708816)
**sklearn's neural network functionality is rather limited.** More advanced toolboxes for neural networks:
* [keras](https://keras.io/)
* [tensorflow](https://www.tensorflow.org/)
* [Theano](http://deeplearning.net/software/theano/)
# Exercise
## iris dataset
Train a neural network on the `iris` dataset and run cross validation. Do not forget to normalize the featurs.
Compare the results against LogisticRegression.
Use Grid search to tune the NN further.
## Further reading
* http://www.ritchieng.com/applying-machine-learning/
|
github_jupyter
|
```
import matplotlib
matplotlib.use('nbagg')
import matplotlib.animation as anm
import matplotlib.pyplot as plt
import math
import matplotlib.patches as patches
import numpy as np
class World: ### fig:world_init_add_timespan (1-5行目)
def __init__(self, time_span, time_interval, debug=False):
self.objects = []
self.debug = debug
self.time_span = time_span # 追加
self.time_interval = time_interval # 追加
def append(self,obj): # オブジェクトを登録するための関数
self.objects.append(obj)
def draw(self): ### fig:world_draw_with_timesapn (1, 10, 21-26, 28-34行目)
fig = plt.figure(figsize=(4,4)) # 8x8 inchの図を準備
ax = fig.add_subplot(111) # サブプロットを準備
ax.set_aspect('equal') # 縦横比を座標の値と一致させる
ax.set_xlim(-5,5) # X軸を-5m x 5mの範囲で描画
ax.set_ylim(-5,5) # Y軸も同様に
ax.set_xlabel("X",fontsize=10) # X軸にラベルを表示
ax.set_ylabel("Y",fontsize=10) # 同じくY軸に
elems = []
if self.debug:
for i in range(int(self.time_span/self.time_interval)): self.one_step(i, elems, ax)
else:
self.ani = anm.FuncAnimation(fig, self.one_step, fargs=(elems, ax),
frames=int(self.time_span/self.time_interval)+1, interval=int(self.time_interval*1000), repeat=False)
plt.show()
def one_step(self, i, elems, ax):
while elems: elems.pop().remove()
time_str = "t = %.2f[s]" % (self.time_interval*i) # 時刻として表示する文字列
elems.append(ax.text(-4.4, 4.5, time_str, fontsize=10))
for obj in self.objects:
obj.draw(ax, elems)
if hasattr(obj, "one_step"): obj.one_step(self.time_interval) # 変更
class IdealRobot: ### fig:robot_camera(1,2,8,28-29行目,49-53行目)
def __init__(self, pose, agent=None, sensor=None, color="black"): # 引数を追加
self.pose = pose
self.r = 0.2
self.color = color
self.agent = agent
self.poses = [pose]
self.sensor = sensor # 追加
def draw(self, ax, elems):
x, y, theta = self.pose
xn = x + self.r * math.cos(theta)
yn = y + self.r * math.sin(theta)
elems += ax.plot([x,xn], [y,yn], color=self.color)
c = patches.Circle(xy=(x, y), radius=self.r, fill=False, color=self.color)
elems.append(ax.add_patch(c))
self.poses.append(self.pose)
elems += ax.plot([e[0] for e in self.poses], [e[1] for e in self.poses], linewidth=0.5, color="black")
if self.sensor and len(self.poses) > 1: #追加
self.sensor.draw(ax, elems, self.poses[-2]) #追加
@classmethod
def state_transition(self, nu, omega, time, pose):
t0 = pose[2]
if math.fabs(omega) < 1e-10:
return pose + np.array( [nu*math.cos(t0),
nu*math.sin(t0),
omega ] ) * time
else:
return pose + np.array( [nu/omega*(math.sin(t0 + omega*time) - math.sin(t0)),
nu/omega*(-math.cos(t0 + omega*time) + math.cos(t0)),
omega*time ] )
def one_step(self, time_interval):
if not self.agent: return
obs = self.sensor.data(self.pose) if self.sensor else None #追加
nu, omega = self.agent.decision(obs) #引数追加
self.pose = self.state_transition(nu, omega, time_interval, self.pose)
class Agent:
def __init__(self, nu, omega):
self.nu = nu
self.omega = omega
def decision(self, observation=None):
return self.nu, self.omega
class Landmark:
def __init__(self, x, y):
self.pos = np.array([x, y]).T
self.id = None
def draw(self, ax, elems):
c = ax.scatter(self.pos[0], self.pos[1], s=100, marker="*", label="landmarks", color="orange")
elems.append(c)
elems.append(ax.text(self.pos[0], self.pos[1], "id:" + str(self.id), fontsize=10))
class Map:
def __init__(self): # 空のランドマークのリストを準備
self.landmarks = []
def append_landmark(self, landmark): # ランドマークを追加
landmark.id = len(self.landmarks) # 追加するランドマークにIDを与える
self.landmarks.append(landmark)
def draw(self, ax, elems): # 描画(Landmarkのdrawを順に呼び出し)
for lm in self.landmarks: lm.draw(ax, elems)
class IdealCamera: ### fig:camera2(1-4行目、6, 12-13行目, 26-32行目)
def __init__(self, env_map):
self.map = env_map
self.lastdata = [] # 追加
def data(self, cam_pose):
observed = []
for lm in self.map.landmarks:
z = self.observation_function(cam_pose, lm.pos)
observed.append((z, lm.id))
self.lastdata = observed # 追加
return observed
@classmethod
def observation_function(cls, cam_pose, obj_pos):
diff = obj_pos - cam_pose[0:2]
phi = math.atan2(diff[1], diff[0]) - cam_pose[2]
while phi >= np.pi: phi -= 2*np.pi
while phi < -np.pi: phi += 2*np.pi
return np.array( [np.hypot(*diff), phi ] ).T
def draw(self, ax, elems, cam_pose): # 追加 ###camera2_1
for lm in self.lastdata:
x, y, theta = cam_pose
distance, direction = lm[0][0], lm[0][1]
lx = x + distance * math.cos(direction + theta)
ly = y + distance * math.sin(direction + theta)
elems += ax.plot([x,lx], [y,ly], color="pink")
world = World(10, 0.1, debug=False) ### fig:sensor_drawing (10-19行目)
### 地図を生成して3つランドマークを追加 ###
m = Map()
m.append_landmark(Landmark(2,-2))
m.append_landmark(Landmark(-1,-3))
m.append_landmark(Landmark(3,3))
world.append(m)
### ロボットを作る ###
straight = Agent(0.2, 0.0)
circling = Agent(0.2, 10.0/180*math.pi)
robot1 = IdealRobot( np.array([ 2, 3, math.pi/6]).T, sensor=IdealCamera(m), agent=straight ) # 引数にcameraを追加、整理
robot2 = IdealRobot( np.array([-2, -1, math.pi/5*6]).T, sensor=IdealCamera(m), agent=circling, color="red") # robot3は消しました
world.append(robot1)
world.append(robot2)
### アニメーション実行 ###
world.draw()
cam = IdealCamera(m)
p = cam.data(robot2.pose)
print(p)
```
|
github_jupyter
|
# Exploratory Data Analysis
```
from pyspark import SparkContext, SparkConf
from pyspark.sql import SparkSession
from pyspark.sql.types import *
from pyspark.sql import functions as F
spark = SparkSession.builder.master('local[1]').appName("Jupyter").getOrCreate()
sc = spark.sparkContext
#test if this works
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import scipy
import datetime
# the more advanced python visualization library
import seaborn as sns
# apply style to all the charts
sns.set_style('whitegrid')
pd.set_option('display.max_columns', None)
pd.set_option('display.max_rows', None)
# create sparksession
spark = SparkSession \
.builder \
.appName("Pysparkexample") \
.config("spark.some.config.option", "some-value") \
.getOrCreate()
```
# Load Data
```
#collisions = spark.read.csv('data/accidents.csv', header='true', inferSchema = True)
#collisions.show(2)
df_new = spark.read.csv('data/accidents_new.csv', header='true', inferSchema = True)
```
# Data Perspective
_____
* One variable
* Numeric variables:
* continuous
* discrete
* Categorical variables:
* ordinal
* nominal
* Multiple variables:
* Numeric x Numeric
* Categorical x Numeric
* Categorical x Categorical
____________________
# Overview
```
print('The total number of rows : ', df_new.count(),
'\nThe total number of columns :', len(df_new.columns))
```
# Data Schema
Print the data schema for our dataset - SAAQ Accident Information
```
df_new.printSchema()
# Create temporary table query with SQL
df_new.createOrReplaceTempView('AccidentData')
accidents_limit_10 = spark.sql(
'''
SELECT * FROM AccidentData
LIMIT 10
'''
).toPandas()
accidents_limit_10
```
# One Variable
__________
## a. Numeric - Data Totals
Totals for various accident records
```
from pyspark.sql import functions as func
#df_new.agg(func.sum("NB_BLESSES_VELO").alias('Velo'),func.sum("NB_VICTIMES_MOTO"),func.sum("NB_VEH_IMPLIQUES_ACCDN")).show()
df_new.agg(func.sum("NB_VEH_IMPLIQUES_ACCDN").alias('Ttl Cars In Accidents')).show()
df_new.agg(func.sum("NB_VICTIMES_TOTAL").alias('Ttl Victims')).show()
df_new.agg(func.sum("NB_MORTS").alias('Ttl Deaths')).show()
df_new.agg(func.sum("NB_BLESSES_GRAVES").alias('Ttl Severe Injuries')).show()
df_new.agg(func.sum("NB_BLESS_LEGERS").alias('Ttl Light Injuries')).show()
df_new.agg(func.sum("NB_DECES_PIETON").alias('Ttl Pedestrian Deaths')).show()
df_new.agg(func.sum("NB_BLESSES_PIETON").alias('Ttl Pedestrian Injuries')).show()
df_new.agg(func.sum("NB_VICTIMES_PIETON").alias('Ttl Pedestrian Victims')).show()
df_new.agg(func.sum("NB_DECES_MOTO").alias('Ttl Moto Deaths')).show()
df_new.agg(func.sum("NB_BLESSES_MOTO").alias('Ttl Moto Injuries')).show()
df_new.agg(func.sum("NB_VICTIMES_MOTO").alias('Ttl Moto Victims')).show()
df_new.agg(func.sum("NB_DECES_VELO").alias('Ttl Bike Deaths')).show()
df_new.agg(func.sum("NB_BLESSES_VELO").alias('Ttl Bike Injuries')).show()
df_new.agg(func.sum("NB_VICTIMES_VELO").alias('Ttl Bike Victims')).show()
df_new.agg(func.sum("nb_automobile_camion_leger").alias('Ttl Car - Light Trucks')).show()
df_new.agg(func.sum("nb_camionLourd_tractRoutier").alias('Ttl Heavy Truck - Tractor')).show()
df_new.agg(func.sum("nb_outil_equipement").alias('Ttl Equipment - Tools')).show()
df_new.agg(func.sum("nb_tous_autobus_minibus").alias('Ttl Bus')).show()
df_new.agg(func.sum("nb_bicyclette").alias('Ttl Bikes')).show()
df_new.agg(func.sum("nb_cyclomoteur").alias('Ttl Motorized Bike')).show()
df_new.agg(func.sum("nb_motocyclette").alias('Ttl Motorcycle')).show()
df_new.agg(func.sum("nb_taxi").alias('Ttl Taxi')).show()
df_new.agg(func.sum("nb_urgence").alias('Ttl Emergency')).show()
df_new.agg(func.sum("nb_motoneige").alias('Ttl Snowmobile')).show()
df_new.agg(func.sum("nb_VHR").alias('Ttl Motorhome')).show()
df_new.agg(func.sum("nb_autres_types").alias('Ttl Other Types')).show()
df_new.agg(func.sum("nb_veh_non_precise").alias('Ttl Non Specified Vehicles')).show()
df_totals = pd.DataFrame(columns=['Attr','Total'])
#df_totals.append({'Attr':'NB_VEH_IMPLIQUES_ACCDN','Total':df_new.agg(func.sum("NB_VEH_IMPLIQUES_ACCDN"))},ignore_index=True)
#df_totals
```
## b. Categorical
### GRAVITE - severity of the accident
```
gravite_levels = spark.sql(
'''
SELECT GRAVITE, COUNT(*) as Total FROM AccidentData
GROUP BY GRAVITE
ORDER BY Total DESC
'''
).toPandas()
gravite_levels
# Pie Chart
fig,ax = plt.subplots(1,1,figsize=(12,6))
wedges, texts, autotexts = ax.pie(gravite_levels['Total'], radius=2, #labeldistance=2, pctdistance=1.1,
autopct='%1.2f%%', startangle=90)
ax.legend(wedges, gravite_levels['GRAVITE'],
title="GRAVITE",
loc="center left",
bbox_to_anchor=(1, 0, 0.5, 1))
plt.setp(autotexts, size=12, weight="bold")
ax.axis('equal')
plt.tight_layout()
plt.savefig('figures/gravite_levels.png')
plt.show()
```
### METEO - Weather Conditions
```
meteo_conditions = spark.sql(
'''
SELECT METEO, COUNT(*) as Total FROM AccidentData
GROUP BY METEO
ORDER BY Total DESC
'''
).toPandas()
meteo_conditions['METEO'] = meteo_conditions['METEO'].replace( {11:'Clear',12:'Overcast: cloudy/dark',13:'Fog/mist',
14:'Rain/bruine',15:'Heavy rain',16:'Strong wind',
17:'Snow/storm',18:'Blowing snow/blizzard',
19:'Ice',99:'Other..'})
meteo_conditions
fig,ax = plt.subplots(1,1,figsize=(10,6))
plt.bar(meteo_conditions['METEO'], meteo_conditions['Total'],
align='center', alpha=0.7, width=0.7, color='purple')
plt.setp(ax.get_xticklabels(), rotation=30, horizontalalignment='right')
fig.tight_layout()
plt.savefig('figures/meteo_conditions.png')
plt.show()
```
# Multiple Variables
____________
## Numeric X Categorical
### 1. Accident Victims by Municipality
```
victims_by_municipality = spark.sql(
'''
SELECT MUNCP, SUM(NB_VICTIMES_TOTAL) as Total FROM AccidentData
GROUP BY MUNCP
ORDER BY Total DESC
'''
).toPandas()
victims_by_municipality
fig,ax = plt.subplots(1,1,figsize=(10,6))
victims_by_municipality.plot(x = 'MUNCP', y = 'Total', kind = 'barh', color = 'C0', ax = ax, legend = False)
ax.set_xlabel('Total Victims', size = 16)
ax.set_ylabel('Municipality', size = 16)
plt.savefig('figures/victims_by_municipality.png')
plt.show()
victims_by_region = spark.sql(
'''
SELECT MUNCP, SUM(NB_VICTIMES_TOTAL) as Total FROM AccidentData
GROUP BY MUNCP
'''
).toPandas()
plt.figure(figsize = (10,6))
sns.distplot(np.log(victims_by_region['Total']))
plt.title('Total Victims Histogram by Region', size = 16)
plt.ylabel('Density', size = 16)
plt.xlabel('Log Total', size = 16)
plt.savefig('figures/distplot.png')
plt.show()
```
### 2. Total Collisions by Day of Week
```
collisions_by_day = spark.sql(
'''
SELECT WEEK_DAY, COUNT(WEEK_DAY) as Number_of_Collisions FROM AccidentData
GROUP BY WEEK_DAY
ORDER BY Number_of_Collisions DESC
'''
).toPandas()
collisions_by_day
fig,ax = plt.subplots(1,1,figsize=(10,6))
collisions_by_day.plot(x = 'WEEK_DAY', y = 'Number_of_Collisions', kind = 'barh', color = 'C0', ax = ax, legend = False)
ax.set_xlabel('Number_of_Collisions', size = 16)
ax.set_ylabel('WEEK_DAY', size = 16)
plt.savefig('figures/collisions_by_day.png')
plt.show()
```
#### "VE", Friday has the highest number of collisions.
### 3. Top 10 Accidents by street
```
accidents_by_street = spark.sql(
'''
SELECT STREET, COUNT(STREET) as Number_of_Accidents FROM AccidentData
GROUP BY STREET
ORDER BY Number_of_Accidents DESC
LIMIT 10
'''
).toPandas()
fig,ax = plt.subplots(1,1,figsize=(10,6))
#accidents_by_street.plot(x = 'STREET', y = 'Number_of_Accidents', kind = 'barh', color = 'C0', ax = ax, legend = False)
sns.barplot(x=accidents_by_street['Number_of_Accidents'], y=accidents_by_street['STREET'], orient='h')
ax.set_xlabel('Number of Accidents', size = 16)
ax.set_ylabel('Street', size = 16)
plt.savefig('figures/accidents_by_street.png')
plt.show()
```
## Numeric X Numeric
### Correlation Heatmap
Illustrates the corellation between numeric variables of the dataset.
```
plot_df = spark.sql(
'''
SELECT METEO, SURFACE, LIGHT, TYPE_ACCDN,
NB_MORTS, NB_BLESSES_GRAVES, NB_VEH_IMPLIQUES_ACCDN, NB_VICTIMES_TOTAL
FROM AccidentData
'''
).toPandas()
corrmat = plot_df.corr()
f, ax = plt.subplots(figsize=(10, 7))
sns.heatmap(corrmat, vmax=.8, square=True)
plt.savefig('figures/heatmap.png')
plt.show()
```
## Categorical X Categorical
|
github_jupyter
|
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe_from_ecdc():
return pd.read_csv(
"https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv")
confirmed_df_ = download_cases_dataframe_from_ecdc()
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["dateRep", "cases", "geoId"]]
confirmed_df.rename(
columns={
"dateRep":"sample_date",
"cases": "new_cases",
"geoId": "country_code",
},
inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
source_regions_at_date_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: report_backend_client.source_regions_for_date(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df["sample_date_string"] = \
source_regions_at_date_df.sample_date.dt.strftime("%Y-%m-%d")
source_regions_at_date_df.tail()
source_regions_for_summary_df = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df.head()
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
confirmed_df = confirmed_output_df.copy()
confirmed_df.tail()
confirmed_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
confirmed_df.sort_values("sample_date_string", inplace=True)
confirmed_df.tail()
confirmed_df[["new_cases", "covid_cases"]].plot()
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
fail_on_error_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=fail_on_error_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df.head(daily_plot_days)
weekly_result_summary_df = result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(7).agg({
"covid_cases": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum"
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
weekly_result_summary_df = weekly_result_summary_df.fillna(0).astype(int)
weekly_result_summary_df["teks_per_shared_diagnosis"] = \
(weekly_result_summary_df.shared_teks_by_upload_date / weekly_result_summary_df.shared_diagnoses).fillna(0)
weekly_result_summary_df["shared_diagnoses_per_covid_case"] = \
(weekly_result_summary_df.shared_diagnoses / weekly_result_summary_df.covid_cases).fillna(0)
weekly_result_summary_df.head()
last_7_days_summary = weekly_result_summary_df.to_dict(orient="records")[1]
last_7_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases in Source Countries (7-day Rolling Average)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date",
"shared_diagnoses": "Shared Diagnoses (Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis",
"shared_diagnoses_per_covid_case": "Usage Ratio (Fraction of Cases in Source Countries Which Shared Diagnosis)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 22), legend=False)
ax_ = summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
ax_.yaxis.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
media_path = get_temporary_image_path()
dfi.export(df, media_path)
return media_path
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}",
}
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.sum()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.sum()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.sum()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.sum()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.sum()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.sum()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
report_source_regions = extraction_date_result_summary_df.index \
.get_level_values("source_regions").item().split(",")
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
summary_results_api_df = result_summary_df.reset_index()
summary_results_api_df["sample_date_string"] = \
summary_results_api_df["sample_date"].dt.strftime("%Y-%m-%d")
summary_results_api_df["source_regions"] = \
summary_results_api_df["source_regions"].apply(lambda x: x.split(","))
today_summary_results_api_df = \
summary_results_api_df.to_dict(orient="records")[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_results_api_df,
last_7_days=last_7_days_summary,
daily_results=summary_results_api_df.to_dict(orient="records"))
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Source Countries: {display_brief_source_regions}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: ≤{shared_diagnoses_per_covid_case:.2%}
Last 7 Days:
- Shared Diagnoses: ≤{last_7_days_summary["shared_diagnoses"]:.0f}
- Usage Ratio: ≤{last_7_days_summary["shared_diagnoses_per_covid_case"]:.2%}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.