text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
### Cell Painting morphological (CP) and L1000 gene expression (GE) profiles for the following datasets:
- **CDRP**-BBBC047-Bray-CP-GE (Cell line: U2OS) :
* $\bf{CP}$ There are 30,430 unique compounds for CP dataset, median number of replicates --> 4
* $\bf{GE}$ There are 21,782 unique compounds for GE dataset, median number of replicates --> 3
* 20,131 compounds are present in both datasets.
- **CDRP-bio**-BBBC036-Bray-CP-GE (Cell line: U2OS) :
* $\bf{CP}$ There are 2,242 unique compounds for CP dataset, median number of replicates --> 8
* $\bf{GE}$ There are 1,917 unique compounds for GE dataset, median number of replicates --> 2
* 1916 compounds are present in both datasets.
- **LUAD**-BBBC041-Caicedo-CP-GE (Cell line: A549) :
* $\bf{CP}$ There are 593 unique alleles for CP dataset, median number of replicates --> 8
* $\bf{GE}$ There are 529 unique alleles for GE dataset, median number of replicates --> 8
* 525 alleles are present in both datasets.
- **TA-ORF**-BBBC037-Rohban-CP-GE (Cell line: U2OS) :
* $\bf{CP}$ There are 323 unique alleles for CP dataset, median number of replicates --> 5
* $\bf{GE}$ There are 327 unique alleles for GE dataset, median number of replicates --> 2
* 150 alleles are present in both datasets.
- **LINCS**-Pilot1-CP-GE (Cell line: U2OS) :
* $\bf{CP}$ There are 1570 unique compounds across 7 doses for CP dataset, median number of replicates --> 5
* $\bf{GE}$ There are 1402 unique compounds for GE dataset, median number of replicates --> 3
* $N_{p/d}$: 6984 compounds are present in both datasets.
--------------------------------------------
#### Link to the processed profiles:
https://cellpainting-datasets.s3.us-east-1.amazonaws.com/Rosetta-GE-CP
```
%matplotlib notebook
%load_ext autoreload
%autoreload 2
import numpy as np
import scipy.spatial
import pandas as pd
import sklearn.decomposition
import matplotlib.pyplot as plt
import seaborn as sns
import os
from cmapPy.pandasGEXpress.parse import parse
from utils.replicateCorrs import replicateCorrs
from utils.saveAsNewSheetToExistingFile import saveAsNewSheetToExistingFile,saveDF_to_CSV_GZ_no_timestamp
from importlib import reload
from utils.normalize_funcs import standardize_per_catX
# sns.set_style("whitegrid")
# np.__version__
pd.__version__
```
### Input / ouput files:
- **CDRPBIO**-BBBC047-Bray-CP-GE (Cell line: U2OS) :
* $\bf{CP}$
* Input:
* Output:
* $\bf{GE}$
* Input: .mat files that are generated using https://github.com/broadinstitute/2014_wawer_pnas
* Output:
- **LUAD**-BBBC041-Caicedo-CP-GE (Cell line: A549) :
* $\bf{CP}$
* Input:
* Output:
* $\bf{GE}$
* Input:
* Output:
- **TA-ORF**-BBBC037-Rohban-CP-GE (Cell line: U2OS) :
* $\bf{CP}$
* Input:
* Output:
* $\bf{GE}$
* Input: https://data.broadinstitute.org/icmap/custom/TA/brew/pc/TA.OE005_U2OS_72H/
* Output:
### Reformat Cell-Painting Data Sets
- CDRP and TA-ORF are in /storage/data/marziehhaghighi/Rosetta/raw-profiles/
- Luad is already processed by Juan, source of the files is at /storage/luad/profiles_cp
in case you want to reformat
```
fileName='RepCorrDF'
### dirs on gpu cluster
# rawProf_dir='/storage/data/marziehhaghighi/Rosetta/raw-profiles/'
# procProf_dir='/home/marziehhaghighi/workspace_rosetta/workspace/'
### dirs on ec2
rawProf_dir='/home/ubuntu/bucket/projects/2018_04_20_Rosetta/workspace/raw-profiles/'
# procProf_dir='./'
procProf_dir='/home/ubuntu/bucket/projects/2018_04_20_Rosetta/workspace/'
# s3://imaging-platform/projects/2018_04_20_Rosetta/workspace/preprocessed_data
# aws s3 sync preprocessed_data s3://cellpainting-datasets/Rosetta-GE-CP/preprocessed_data --profile jumpcpuser
filename='../../results/RepCor/'+fileName+'.xlsx'
# ls ../../
# https://cellpainting-datasets.s3.us-east-1.amazonaws.com/
```
# CDRP-BBBC047-Bray
### GE - L1000 - CDRP
```
os.listdir(rawProf_dir+'/l1000_CDRP/')
cdrp_dataDir=rawProf_dir+'/l1000_CDRP/'
cpd_info = pd.read_csv(cdrp_dataDir+"/compounds.txt", sep="\t", dtype=str)
cpd_info.columns
from scipy.io import loadmat
x = loadmat(cdrp_dataDir+'cdrp.all.prof.mat')
k1=x['metaWell']['pert_id'][0][0]
k2=x['metaGen']['AFFX_PROBE_ID'][0][0]
k3=x['metaWell']['pert_dose'][0][0]
k4=x['metaWell']['det_plate'][0][0]
# pert_dose
# x['metaWell']['pert_id'][0][0][0][0][0]
pertID = []
probID=[]
for r in range(len(k1)):
v = k1[r][0][0]
pertID.append(v)
# probID.append(k2[r][0][0])
for r in range(len(k2)):
probID.append(k2[r][0][0])
pert_dose=[]
det_plate=[]
for r in range(len(k3)):
pert_dose.append(k3[r][0])
det_plate.append(k4[r][0][0])
dataArray=x['pclfc'];
cdrp_l1k_rep = pd.DataFrame(data=dataArray,columns=probID)
cdrp_l1k_rep['pert_id']=pertID
cdrp_l1k_rep['pert_dose']=pert_dose
cdrp_l1k_rep['det_plate']=det_plate
cdrp_l1k_rep['BROAD_CPD_ID']=cdrp_l1k_rep['pert_id'].str[:13]
cdrp_l1k_rep2=pd.merge(cdrp_l1k_rep, cpd_info, how='left',on=['BROAD_CPD_ID'])
l1k_features_cdrp=cdrp_l1k_rep2.columns[cdrp_l1k_rep2.columns.str.contains("_at")]
cdrp_l1k_rep2['pert_id_dose']=cdrp_l1k_rep2['BROAD_CPD_ID']+'_'+cdrp_l1k_rep2['pert_dose'].round(2).astype(str)
cdrp_l1k_rep2['pert_sample_dose']=cdrp_l1k_rep2['pert_id']+'_'+cdrp_l1k_rep2['pert_dose'].round(2).astype(str)
# cdrp_l1k_df.head()
print(cpd_info.shape,cdrp_l1k_rep.shape,cdrp_l1k_rep2.shape)
cdrp_l1k_rep2['pert_id_dose']=cdrp_l1k_rep2['pert_id_dose'].replace('DMSO_-666.0', 'DMSO')
cdrp_l1k_rep2['pert_sample_dose']=cdrp_l1k_rep2['pert_sample_dose'].replace('DMSO_-666.0', 'DMSO')
saveDF_to_CSV_GZ_no_timestamp(cdrp_l1k_rep2,procProf_dir+'preprocessed_data/CDRP-BBBC047-Bray/L1000/replicate_level_l1k.csv.gz');
# cdrp_l1k_rep2.head()
# cpd_info
```
### CP - CDRP
```
profileType=['_augmented','_normalized']
bioactiveFlag="";# either "-bioactive" or ""
plates=os.listdir(rawProf_dir+'/CDRP'+bioactiveFlag+'/')
for pt in profileType[1:2]:
repLevelCDRP0=[]
for p in plates:
# repLevelCDRP0.append(pd.read_csv(rawProf_dir+'/CDRP/'+p+'/'+p+pt+'.csv'))
repLevelCDRP0.append(pd.read_csv(rawProf_dir+'/CDRP'+bioactiveFlag+'/'+p+'/'+p+pt+'.csv')) #if bioactive
repLevelCDRP = pd.concat(repLevelCDRP0)
metaCDRP1=pd.read_csv(rawProf_dir+'/CP_CDRP/metadata/metadata_CDRP.csv')
# metaCDRP1=metaCDRP1.rename(columns={"PlateName":"Metadata_Plate_Map_Name",'Well':'Metadata_Well'})
# metaCDRP1['Metadata_Well']=metaCDRP1['Metadata_Well'].str.lower()
repLevelCDRP2=pd.merge(repLevelCDRP, metaCDRP1, how='left',on=['Metadata_broad_sample'])
# repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_broad_sample']+'_'+repLevelCDRP2['Metadata_mmoles_per_liter'].round(0).astype(int).astype(str)
# repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_pert_id']+'_'+(repLevelCDRP2['Metadata_mmoles_per_liter']*2).round(0).astype(int).astype(str)
repLevelCDRP2["Metadata_mmoles_per_liter2"]=(repLevelCDRP2["Metadata_mmoles_per_liter"]*2).round(2)
repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_broad_sample']+'_'+repLevelCDRP2['Metadata_mmoles_per_liter2'].astype(str)
repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_Sample_Dose'].replace('DMSO_0.0', 'DMSO')
repLevelCDRP2['Metadata_pert_id']=repLevelCDRP2['Metadata_pert_id'].replace(np.nan, 'DMSO')
# repLevelCDRP2.to_csv(procProf_dir+'preprocessed_data/CDRPBIO-BBBC036-Bray/CellPainting/replicate_level_cp'+pt+'.csv.gz',index=False,compression='gzip')
# ,
if bioactiveFlag:
dataFolderName='CDRPBIO-BBBC036-Bray'
saveDF_to_CSV_GZ_no_timestamp(repLevelCDRP2,procProf_dir+'preprocessed_data/'+dataFolderName+\
'/CellPainting/replicate_level_cp'+pt+'.csv.gz')
else:
# sgfsgf
dataFolderName='CDRP-BBBC047-Bray'
saveDF_to_CSV_GZ_no_timestamp(repLevelCDRP2,procProf_dir+'preprocessed_data/'+dataFolderName+\
'/CellPainting/replicate_level_cp'+pt+'.csv.gz')
print(metaCDRP1.shape,repLevelCDRP.shape,repLevelCDRP2.shape)
dataFolderName='CDRP-BBBC047-Bray'
cp_feats=repLevelCDRP.columns[repLevelCDRP.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")].tolist()
features_to_remove =find_correlation(repLevelCDRP2[cp_feats], threshold=0.9, remove_negative=False)
repLevelCDRP2_var_sel=repLevelCDRP2.drop(columns=features_to_remove)
saveDF_to_CSV_GZ_no_timestamp(repLevelCDRP2_var_sel,procProf_dir+'preprocessed_data/'+dataFolderName+\
'/CellPainting/replicate_level_cp'+'_normalized_variable_selected'+'.csv.gz')
# features_to_remove
# features_to_remove
# features_to_remove
repLevelCDRP2['Nuclei_Texture_Variance_RNA_3_0']
# repLevelCDRP2.shape
# cp_scaled.columns[cp_scaled.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")].tolist()
```
# CDRP-bio-BBBC036-Bray
### GE - L1000 - CDRPBIO
```
bioactiveFlag="-bioactive";# either "-bioactive" or ""
plates=os.listdir(rawProf_dir+'/CDRP'+bioactiveFlag+'/')
# plates
cdrp_l1k_rep2_bioactive=cdrp_l1k_rep2[cdrp_l1k_rep2["pert_sample_dose"].isin(repLevelCDRP2.Metadata_Sample_Dose.unique().tolist())]
cdrp_l1k_rep.det_plate
```
### CP - CDRPBIO
```
profileType=['_augmented','_normalized','_normalized_variable_selected']
bioactiveFlag="-bioactive";# either "-bioactive" or ""
plates=os.listdir(rawProf_dir+'/CDRP'+bioactiveFlag+'/')
for pt in profileType:
repLevelCDRP0=[]
for p in plates:
# repLevelCDRP0.append(pd.read_csv(rawProf_dir+'/CDRP/'+p+'/'+p+pt+'.csv'))
repLevelCDRP0.append(pd.read_csv(rawProf_dir+'/CDRP'+bioactiveFlag+'/'+p+'/'+p+pt+'.csv')) #if bioactive
repLevelCDRP = pd.concat(repLevelCDRP0)
metaCDRP1=pd.read_csv(rawProf_dir+'/CP_CDRP/metadata/metadata_CDRP.csv')
# metaCDRP1=metaCDRP1.rename(columns={"PlateName":"Metadata_Plate_Map_Name",'Well':'Metadata_Well'})
# metaCDRP1['Metadata_Well']=metaCDRP1['Metadata_Well'].str.lower()
repLevelCDRP2=pd.merge(repLevelCDRP, metaCDRP1, how='left',on=['Metadata_broad_sample'])
# repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_broad_sample']+'_'+repLevelCDRP2['Metadata_mmoles_per_liter'].round(0).astype(int).astype(str)
# repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_pert_id']+'_'+(repLevelCDRP2['Metadata_mmoles_per_liter']*2).round(0).astype(int).astype(str)
repLevelCDRP2["Metadata_mmoles_per_liter2"]=(repLevelCDRP2["Metadata_mmoles_per_liter"]*2).round(2)
repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_broad_sample']+'_'+repLevelCDRP2['Metadata_mmoles_per_liter2'].astype(str)
repLevelCDRP2['Metadata_Sample_Dose']=repLevelCDRP2['Metadata_Sample_Dose'].replace('DMSO_0.0', 'DMSO')
repLevelCDRP2['Metadata_pert_id']=repLevelCDRP2['Metadata_pert_id'].replace(np.nan, 'DMSO')
# repLevelCDRP2.to_csv(procProf_dir+'preprocessed_data/CDRPBIO-BBBC036-Bray/CellPainting/replicate_level_cp'+pt+'.csv.gz',index=False,compression='gzip')
# ,
if bioactiveFlag:
dataFolderName='CDRPBIO-BBBC036-Bray'
saveDF_to_CSV_GZ_no_timestamp(repLevelCDRP2,procProf_dir+'preprocessed_data/'+dataFolderName+\
'/CellPainting/replicate_level_cp'+pt+'.csv.gz')
else:
dataFolderName='CDRP-BBBC047-Bray'
saveDF_to_CSV_GZ_no_timestamp(repLevelCDRP2,procProf_dir+'preprocessed_data/'+dataFolderName+\
'/CellPainting/replicate_level_cp'+pt+'.csv.gz')
print(metaCDRP1.shape,repLevelCDRP.shape,repLevelCDRP2.shape)
```
# LUAD-BBBC041-Caicedo
### GE - L1000 - LUAD
```
os.listdir(rawProf_dir+'/l1000_LUAD/input/')
os.listdir(rawProf_dir+'/l1000_LUAD/output/')
luad_dataDir=rawProf_dir+'/l1000_LUAD/'
luad_info1 = pd.read_csv(luad_dataDir+"/input/TA.OE014_A549_96H.map", sep="\t", dtype=str)
luad_info2 = pd.read_csv(luad_dataDir+"/input/TA.OE015_A549_96H.map", sep="\t", dtype=str)
luad_info=pd.concat([luad_info1, luad_info2], ignore_index=True)
luad_info.head()
luad_l1k_df = parse(luad_dataDir+"/output/high_rep_A549_8reps_141230_ZSPCINF_n4232x978.gctx").data_df.T.reset_index()
luad_l1k_df=luad_l1k_df.rename(columns={"cid":"id"})
# cdrp_l1k_df['XX']=cdrp_l1k_df['cid'].str[0]
# cdrp_l1k_df['BROAD_CPD_ID']=cdrp_l1k_df['cid'].str[2:15]
luad_l1k_df2=pd.merge(luad_l1k_df, luad_info, how='inner',on=['id'])
luad_l1k_df2=luad_l1k_df2.rename(columns={"x_mutation_status":"allele"})
l1k_features=luad_l1k_df2.columns[luad_l1k_df2.columns.str.contains("_at")]
luad_l1k_df2['allele']=luad_l1k_df2['allele'].replace('UnTrt', 'DMSO')
print(luad_info.shape,luad_l1k_df.shape,luad_l1k_df2.shape)
saveDF_to_CSV_GZ_no_timestamp(luad_l1k_df2,procProf_dir+'/preprocessed_data/LUAD-BBBC041-Caicedo/L1000/replicate_level_l1k.csv.gz')
luad_l1k_df_scaled = standardize_per_catX(luad_l1k_df2,'det_plate',l1k_features.tolist());
x_l1k_luad=replicateCorrs(luad_l1k_df_scaled.reset_index(drop=True),'allele',l1k_features,1)
# x_l1k_luad=replicateCorrs(luad_l1k_df2[luad_l1k_df2['allele']!='DMSO'].reset_index(drop=True),'allele',l1k_features,1)
# saveAsNewSheetToExistingFile(filename,x_l1k_luad[2],'l1k-luad')
```
### CP - LUAD
```
profileType=['_augmented','_normalized','_normalized_variable_selected']
plates=os.listdir('/storage/luad/profiles_cp/LUAD-BBBC043-Caicedo/')
for pt in profileType[1:2]:
repLevelLuad0=[]
for p in plates:
repLevelLuad0.append(pd.read_csv('/storage/luad/profiles_cp/LUAD-BBBC043-Caicedo/'+p+'/'+p+pt+'.csv'))
repLevelLuad = pd.concat(repLevelLuad0)
metaLuad1=pd.read_csv(rawProf_dir+'/CP_LUAD/metadata/combined_platemaps_AHB_20150506_ssedits.csv')
metaLuad1=metaLuad1.rename(columns={"PlateName":"Metadata_Plate_Map_Name",'Well':'Metadata_Well'})
metaLuad1['Metadata_Well']=metaLuad1['Metadata_Well'].str.lower()
# metaLuad2=pd.read_csv('~/workspace_rosetta/workspace/raw_profiles/CP_LUAD/metadata/barcode_platemap.csv')
# Y[Y['Metadata_Well']=='g05']['Nuclei_Texture_Variance_Mito_5_0']
repLevelLuad2=pd.merge(repLevelLuad, metaLuad1, how='inner',on=['Metadata_Plate_Map_Name','Metadata_Well'])
repLevelLuad2['x_mutation_status']=repLevelLuad2['x_mutation_status'].replace(np.nan, 'DMSO')
cp_features=repLevelLuad2.columns[repLevelLuad2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
# repLevelLuad2.to_csv(procProf_dir+'preprocessed_data/LUAD-BBBC041-Caicedo/CellPainting/replicate_level_cp'+pt+'.csv.gz',index=False,compression='gzip')
saveDF_to_CSV_GZ_no_timestamp(repLevelLuad2,procProf_dir+'preprocessed_data/LUAD-BBBC041-Caicedo/CellPainting/replicate_level_cp'+pt+'.csv.gz')
print(metaLuad1.shape,repLevelLuad.shape,repLevelLuad2.shape)
pt=['_normalized']
# Read save data
repLevelLuad2=pd.read_csv('./preprocessed_data/LUAD-BBBC041-Caicedo/CellPainting/replicate_level_cp'+pt[0]+'.csv.gz')
# repLevelTA.head()
cp_features=repLevelLuad2.columns[repLevelLuad2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
cols2remove0=[i for i in cp_features if ((repLevelLuad2[i].isnull()).sum(axis=0)/repLevelLuad2.shape[0])>0.05]
print(cols2remove0)
repLevelLuad2=repLevelLuad2.drop(cols2remove0, axis=1);
cp_features=repLevelLuad2.columns[repLevelLuad2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
repLevelLuad2 = repLevelLuad2.interpolate()
repLevelLuad2 = standardize_per_catX(repLevelLuad2,'Metadata_Plate',cp_features.tolist());
df1=repLevelLuad2[~repLevelLuad2['x_mutation_status'].isnull()].reset_index(drop=True)
x_cp_luad=replicateCorrs(df1,'x_mutation_status',cp_features,1)
saveAsNewSheetToExistingFile(filename,x_cp_luad[2],'cp-luad')
```
# TA-ORF-BBBC037-Rohban
### GE - L1000
```
taorf_datadir=rawProf_dir+'/l1000_TA_ORF/'
gene_info = pd.read_csv(taorf_datadir+"TA.OE005_U2OS_72H.map.txt", sep="\t", dtype=str)
# gene_info.columns
# TA.OE005_U2OS_72H_INF_n729x22268.gctx
# TA.OE005_U2OS_72H_QNORM_n729x978.gctx
# TA.OE005_U2OS_72H_ZSPCINF_n729x22268.gctx
# TA.OE005_U2OS_72H_ZSPCQNORM_n729x978.gctx
taorf_l1k0 = parse(taorf_datadir+"TA.OE005_U2OS_72H_ZSPCQNORM_n729x978.gctx")
# taorf_l1k0 = parse(taorf_datadir+"TA.OE005_U2OS_72H_QNORM_n729x978.gctx")
taorf_l1k_df0=taorf_l1k0.data_df
taorf_l1k_df=taorf_l1k_df0.T.reset_index()
l1k_features=taorf_l1k_df.columns[taorf_l1k_df.columns.str.contains("_at")]
taorf_l1k_df=taorf_l1k_df.rename(columns={"cid":"id"})
taorf_l1k_df2=pd.merge(taorf_l1k_df, gene_info, how='inner',on=['id'])
# print(taorf_l1k_df.shape,gene_info.shape,taorf_l1k_df2.shape)
taorf_l1k_df2.head()
# x_genesymbol_mutation
taorf_l1k_df2['pert_id']=taorf_l1k_df2['pert_id'].replace('CMAP-000', 'DMSO')
# compression_opts = dict(method='zip',archive_name='out.csv')
# taorf_l1k_df2.to_csv(procProf_dir+'preprocessed_data/TA-ORF-BBBC037-Rohban/L1000/replicate_level_l1k.csv.gz',index=False,compression=compression_opts)
saveDF_to_CSV_GZ_no_timestamp(taorf_l1k_df2,procProf_dir+'preprocessed_data/TA-ORF-BBBC037-Rohban/L1000/replicate_level_l1k.csv.gz')
print(gene_info.shape,taorf_l1k_df.shape,taorf_l1k_df2.shape)
# gene_info.head()
taorf_l1k_df2.groupby(['x_genesymbol_mutation']).size().describe()
taorf_l1k_df2.groupby(['pert_id']).size().describe()
```
#### Check Replicate Correlation
```
# df1=taorf_l1k_df2[taorf_l1k_df2['pert_id']!='CMAP-000']
df1_scaled = standardize_per_catX(taorf_l1k_df2,'det_plate',l1k_features.tolist());
df1_scaled2=df1_scaled[df1_scaled['pert_id']!='DMSO']
x=replicateCorrs(df1_scaled2,'pert_id',l1k_features,1)
```
### CP - TAORF
```
profileType=['_augmented','_normalized','_normalized_variable_selected']
plates=os.listdir(rawProf_dir+'TA-ORF-BBBC037-Rohban/')
for pt in profileType[0:1]:
repLevelTA0=[]
for p in plates:
repLevelTA0.append(pd.read_csv(rawProf_dir+'TA-ORF-BBBC037-Rohban/'+p+'/'+p+pt+'.csv'))
repLevelTA = pd.concat(repLevelTA0)
metaTA1=pd.read_csv(rawProf_dir+'/CP_TA_ORF/metadata/metadata_TA.csv')
metaTA2=pd.read_csv(rawProf_dir+'/CP_TA_ORF/metadata/metadata_TA_2.csv')
# metaTA2=metaTA2.rename(columns={"Metadata_broad_sample":"Metadata_broad_sample_2",'Metadata_Treatment':'Gene Allele Name'})
metaTA=pd.merge(metaTA2, metaTA1, how='left',on=['Metadata_broad_sample'])
# metaTA2=metaTA2.rename(columns={"Metadata_Treatment":"Metadata_pert_name"})
# repLevelTA2=pd.merge(repLevelTA, metaTA2, how='left',on=['Metadata_pert_name'])
repLevelTA2=pd.merge(repLevelTA, metaTA, how='left',on=['Metadata_broad_sample'])
# repLevelTA2=repLevelTA2.rename(columns={"Gene Allele Name":"Allele"})
repLevelTA2['Metadata_broad_sample']=repLevelTA2['Metadata_broad_sample'].replace(np.nan, 'DMSO')
saveDF_to_CSV_GZ_no_timestamp(repLevelTA2,procProf_dir+'/preprocessed_data/TA-ORF-BBBC037-Rohban/CellPainting/replicate_level_cp'+pt+'.csv.gz')
print(metaTA.shape,repLevelTA.shape,repLevelTA2.shape)
# repLevelTA.head()
cp_features=repLevelTA2.columns[repLevelTA2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
cols2remove0=[i for i in cp_features if ((repLevelTA2[i].isnull()).sum(axis=0)/repLevelTA2.shape[0])>0.05]
print(cols2remove0)
repLevelTA2=repLevelTA2.drop(cols2remove0, axis=1);
# cp_features=list(set(cp_features)-set(cols2remove0))
# repLevelTA2=repLevelTA2.replace('nan', np.nan)
repLevelTA2 = repLevelTA2.interpolate()
cp_features=repLevelTA2.columns[repLevelTA2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
repLevelTA2 = standardize_per_catX(repLevelTA2,'Metadata_Plate',cp_features.tolist());
df1=repLevelTA2[~repLevelTA2['Metadata_broad_sample'].isnull()].reset_index(drop=True)
x_taorf_cp=replicateCorrs(df1,'Metadata_broad_sample',cp_features,1)
# saveAsNewSheetToExistingFile(filename,x_taorf_cp[2],'cp-taorf')
# plates
```
# LINCS-Pilot1
### GE - L1000 - LINCS
```
os.listdir(rawProf_dir+'/l1000_LINCS/2016_04_01_a549_48hr_batch1_L1000/')
os.listdir(rawProf_dir+'/l1000_LINCS/metadata/')
data_meta_match_ls=[['level_3','level_3_q2norm_n27837x978.gctx','col_meta_level_3_REP.A_A549_only_n27837.txt'],
['level_4W','level_4W_zspc_n27837x978.gctx','col_meta_level_3_REP.A_A549_only_n27837.txt'],
['level_4','level_4_zspc_n27837x978.gctx','col_meta_level_3_REP.A_A549_only_n27837.txt'],
['level_5_modz','level_5_modz_n9482x978.gctx','col_meta_level_5_REP.A_A549_only_n9482.txt'],
['level_5_rank','level_5_rank_n9482x978.gctx','col_meta_level_5_REP.A_A549_only_n9482.txt']]
lincs_dataDir=rawProf_dir+'/l1000_LINCS/'
lincs_pert_info = pd.read_csv(lincs_dataDir+"/metadata/REP.A_A549_pert_info.txt", sep="\t", dtype=str)
lincs_meta_level3 = pd.read_csv(lincs_dataDir+"/metadata/col_meta_level_3_REP.A_A549_only_n27837.txt", sep="\t", dtype=str)
# lincs_info1 = pd.read_csv(lincs_dataDir+"/metadata/REP.A_A549_pert_info.txt", sep="\t", dtype=str)
print(lincs_meta_level3.shape)
lincs_meta_level3.head()
# lincs_info2 = pd.read_csv(lincs_dataDir+"/input/TA.OE015_A549_96H.map", sep="\t", dtype=str)
# lincs_info=pd.concat([lincs_info1, lincs_info2], ignore_index=True)
# lincs_info.head()
# lincs_meta_level3.groupby('distil_id').size()
lincs_meta_level3['distil_id'].unique().shape
# lincs_meta_level3.columns.tolist()
# lincs_meta_level3.pert_id
ls /home/ubuntu/workspace_rosetta/workspace/software/2018_04_20_Rosetta/preprocessed_data/LINCS-Pilot1/CellPainting
# procProf_dir+'preprocessed_data/LINCS-Pilot1/'
procProf_dir
for el in data_meta_match_ls:
lincs_l1k_df=parse(lincs_dataDir+"/2016_04_01_a549_48hr_batch1_L1000/"+el[1]).data_df.T.reset_index()
lincs_meta0 = pd.read_csv(lincs_dataDir+"/metadata/"+el[2], sep="\t", dtype=str)
lincs_meta=pd.merge(lincs_meta0, lincs_pert_info, how='left',on=['pert_id'])
lincs_meta=lincs_meta.rename(columns={"distil_id":"cid"})
lincs_l1k_df2=pd.merge(lincs_l1k_df, lincs_meta, how='inner',on=['cid'])
lincs_l1k_df2['pert_id_dose']=lincs_l1k_df2['pert_id']+'_'+lincs_l1k_df2['nearest_dose'].astype(str)
lincs_l1k_df2['pert_id_dose']=lincs_l1k_df2['pert_id_dose'].replace('DMSO_-666', 'DMSO')
# lincs_l1k_df2.to_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+el[0]+'.csv.gz',index=False,compression='gzip')
saveDF_to_CSV_GZ_no_timestamp(lincs_l1k_df2,procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+el[0]+'.csv.gz')
# lincs_l1k_df2
lincs_l1k_rep['pert_id_dose'].unique()
lincs_l1k_rep = pd.read_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+data_meta_match_ls[1][0]+'.csv.gz')
# l1k_features=lincs_l1k_rep.columns[lincs_l1k_rep.columns.str.contains("_at")]
# x=replicateCorrs(lincs_l1k_rep[lincs_l1k_rep['pert_iname_x']!='DMSO'].reset_index(drop=True),'pert_id',l1k_features,1)
# # saveAsNewSheetToExistingFile(filename,x[2],'l1k-lincs')
# # lincs_l1k_rep.head()
lincs_l1k_rep.pert_id.unique().shape
lincs_l1k_rep = pd.read_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+data_meta_match_ls[2][0]+'.csv.gz')
lincs_l1k_rep.columns[lincs_l1k_rep.columns.str.contains('dose')]
lincs_l1k_rep[['pert_dose', 'pert_dose_unit', 'pert_idose', 'nearest_dose']]
lincs_l1k_rep['nearest_dose'].unique()
# lincs_l1k_rep.rna_plate.unique()
lincs_l1k_rep = pd.read_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+data_meta_match_ls[2][0]+'.csv.gz')
l1k_features=lincs_l1k_rep.columns[lincs_l1k_rep.columns.str.contains("_at")]
lincs_l1k_rep = standardize_per_catX(lincs_l1k_rep,'det_plate',l1k_features.tolist());
x=replicateCorrs(lincs_l1k_rep[lincs_l1k_rep['pert_iname_x']!='DMSO'].reset_index(drop=True),'pert_id',l1k_features,1)
lincs_l1k_rep = pd.read_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+data_meta_match_ls[2][0]+'.csv.gz')
l1k_features=lincs_l1k_rep.columns[lincs_l1k_rep.columns.str.contains("_at")]
lincs_l1k_rep = standardize_per_catX(lincs_l1k_rep,'det_plate',l1k_features.tolist());
x_l1k_lincs=replicateCorrs(lincs_l1k_rep[lincs_l1k_rep['pert_iname_x']!='DMSO'].reset_index(drop=True),'pert_id_dose',l1k_features,1)
saveAsNewSheetToExistingFile(filename,x_l1k_lincs[2],'l1k-lincs')
lincs_l1k_rep = pd.read_csv(procProf_dir+'preprocessed_data/LINCS-Pilot1/L1000/'+data_meta_match_ls[2][0]+'.csv.gz')
l1k_features=lincs_l1k_rep.columns[lincs_l1k_rep.columns.str.contains("_at")]
lincs_l1k_rep = standardize_per_catX(lincs_l1k_rep,'det_plate',l1k_features.tolist());
x_l1k_lincs=replicateCorrs(lincs_l1k_rep[lincs_l1k_rep['pert_iname_x']!='DMSO'].reset_index(drop=True),'pert_id_dose',l1k_features,1)
saveAsNewSheetToExistingFile(filename,x_l1k_lincs[2],'l1k-lincs')
saveAsNewSheetToExistingFile(filename,x[2],'l1k-lincs')
```
raw data
```
# set(repLevelLuad2)-set(Y1.columns)
# Y1[['Allele', 'Category', 'Clone ID', 'Gene Symbol']].head()
# repLevelLuad2[repLevelLuad2['PublicID']=='BRDN0000553807'][['Col','InsertLength','NCBIGeneID','Name','OtherDescriptions','PublicID','Row','Symbol','Transcript','Vector','pert_type','x_mutation_status']].head()
```
#### Check Replicate Correlation
### CP - LINCS
```
# Ran the following on:
# https://ec2-54-242-99-61.compute-1.amazonaws.com:5006/notebooks/workspace_nucleolar/2020_07_20_Nucleolar_Calico/1-NucleolarSizeMetrics.ipynb
# Metadata
def recode_dose(x, doses, return_level=False):
closest_index = np.argmin([np.abs(dose - x) for dose in doses])
if np.isnan(x):
return 0
if return_level:
return closest_index + 1
else:
return doses[closest_index]
primary_dose_mapping = [0.04, 0.12, 0.37, 1.11, 3.33, 10, 20]
metadata=pd.read_csv("/home/ubuntu/bucket/projects/2018_04_20_Rosetta/workspace/raw-profiles/CP_LINCS/metadata/matadata_lincs_2.csv")
metadata['Metadata_mmoles_per_liter']=metadata.mmoles_per_liter.values.round(2)
metadata=metadata.rename(columns={"Assay_Plate_Barcode": "Metadata_Plate",'broad_sample':'Metadata_broad_sample','well_position':'Metadata_Well'})
lincs_submod_root_dir="/home/ubuntu/datasetsbucket/lincs-cell-painting/"
profileType=['_augmented','_normalized','_normalized_dmso',\
'_normalized_feature_select','_normalized_feature_select_dmso']
# profileType=['_normalized']
# plates=metadata.Assay_Plate_Barcode.unique().tolist()
plates=metadata.Metadata_Plate.unique().tolist()
for pt in profileType[4:5]:
repLevelLINCS0=[]
for p in plates:
profile_add=lincs_submod_root_dir+"/profiles/2016_04_01_a549_48hr_batch1/"+p+"/"+p+pt+".csv.gz"
if os.path.exists(profile_add):
repLevelLINCS0.append(pd.read_csv(profile_add))
repLevelLINCS = pd.concat(repLevelLINCS0)
meta_lincs1=metadata.rename(columns={"broad_sample": "Metadata_broad_sample"})
# metaCDRP1=metaCDRP1.rename(columns={"PlateName":"Metadata_Plate_Map_Name",'Well':'Metadata_Well'})
# metaCDRP1['Metadata_Well']=metaCDRP1['Metadata_Well'].str.lower()
repLevelLINCS2=pd.merge(repLevelLINCS,meta_lincs1,how='left', on=["Metadata_broad_sample","Metadata_Well","Metadata_Plate",'Metadata_mmoles_per_liter'])
repLevelLINCS2 = repLevelLINCS2.assign(Metadata_dose_recode=(repLevelLINCS2.Metadata_mmoles_per_liter.apply(
lambda x: recode_dose(x, primary_dose_mapping, return_level=False))))
repLevelLINCS2['Metadata_pert_id_dose']=repLevelLINCS2['Metadata_pert_id']+'_'+repLevelLINCS2['Metadata_dose_recode'].astype(str)
# repLevelLINCS2['Metadata_Sample_Dose']=repLevelLINCS2['Metadata_broad_sample']+'_'+repLevelLINCS2['Metadata_dose_recode'].astype(str)
repLevelLINCS2['Metadata_pert_id_dose']=repLevelLINCS2['Metadata_pert_id_dose'].replace(np.nan, 'DMSO')
# saveDF_to_CSV_GZ_no_timestamp(repLevelLINCS2,procProf_dir+'/preprocessed_data/LINCS-Pilot1/CellPainting/replicate_level_cp'+pt+'.csv.gz')
print(meta_lincs1.shape,repLevelLINCS.shape,repLevelLINCS2.shape)
# (8120, 15) (52223, 1810) (688699, 1825)
# repLevelLINCS
# pd.merge(repLevelLINCS,meta_lincs1,how='left', on=["Metadata_broad_sample"]).shape
repLevelLINCS.shape,meta_lincs1.shape
(8120, 15) (52223, 1238) (52223, 1253)
csv_l1k_lincs=pd.read_csv('./preprocessed_data/LINCS-Pilot1/L1000/replicate_level_l1k'+'.csv.gz')
csv_pddf=pd.read_csv('./preprocessed_data/LINCS-Pilot1/CellPainting/replicate_level_cp'+pt[0]+'.csv.gz')
csv_l1k_lincs.head()
csv_l1k_lincs.pert_id_dose.unique()
csv_pddf.Metadata_pert_id_dose.unique()
```
#### Read saved data
```
repLevelLINCS2.groupby(['Metadata_pert_id']).size()
repLevelLINCS2.groupby(['Metadata_pert_id_dose']).size().describe()
repLevelLINCS2.Metadata_Plate.unique().shape
repLevelLINCS2['Metadata_pert_id_dose'].unique().shape
# csv_pddf['Metadata_mmoles_per_liter'].round(0).unique()
# np.sort(csv_pddf['Metadata_mmoles_per_liter'].unique())
csv_pddf.groupby(['Metadata_dose_recode']).size()#.median()
# repLevelLincs2=csv_pddf.copy()
import gc
cp_features=repLevelLincs2.columns[repLevelLincs2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
cols2remove0=[i for i in cp_features if ((repLevelLincs2[i].isnull()).sum(axis=0)/repLevelLincs2.shape[0])>0.05]
print(cols2remove0)
repLevelLincs3=repLevelLincs2.drop(cols2remove0, axis=1);
print('here0')
# cp_features=list(set(cp_features)-set(cols2remove0))
# repLevelTA2=repLevelTA2.replace('nan', np.nan)
del repLevelLincs2
gc.collect()
print('here0')
cp_features=repLevelLincs3.columns[repLevelLincs3.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
repLevelLincs3[cp_features] = repLevelLincs3[cp_features].interpolate()
print('here1')
repLevelLincs3 = standardize_per_catX(repLevelLincs3,'Metadata_Plate',cp_features.tolist());
print('here1')
# df0=repLevelCDRP3[repLevelCDRP3['Metadata_broad_sample']!='DMSO'].reset_index(drop=True)
# repSizeDF=repLevelLincs3.groupby(['Metadata_broad_sample']).size().reset_index()
repSizeDF=repLevelLincs3.groupby(['Metadata_pert_id_dose']).size().reset_index()
highRepComp=repSizeDF[repSizeDF[0]>1].Metadata_pert_id_dose.tolist()
highRepComp.remove('DMSO')
# df0=repLevelLincs3[(repLevelLincs3['Metadata_broad_sample'].isin(highRepComp)) &\
# (repLevelLincs3['Metadata_dose_recode']==1.11)]
df0=repLevelLincs3[(repLevelLincs3['Metadata_pert_id_dose'].isin(highRepComp))]
x_lincs_cp=replicateCorrs(df0,'Metadata_pert_id_dose',cp_features,1)
# saveAsNewSheetToExistingFile(filename,x_lincs_cp[2],'cp-lincs')
repSizeDF
# repLevelLincs2=csv_pddf.copy()
# cp_features=repLevelLincs2.columns[repLevelLincs2.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
# cols2remove0=[i for i in cp_features if ((repLevelLincs2[i].isnull()).sum(axis=0)/repLevelLincs2.shape[0])>0.05]
# print(cols2remove0)
# repLevelLincs3=repLevelLincs2.drop(cols2remove0, axis=1);
# # cp_features=list(set(cp_features)-set(cols2remove0))
# # repLevelTA2=repLevelTA2.replace('nan', np.nan)
# repLevelLincs3 = repLevelLincs3.interpolate()
# repLevelLincs3 = standardize_per_catX(repLevelLincs3,'Metadata_Plate',cp_features.tolist());
# cp_features=repLevelLincs3.columns[repLevelLincs3.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
# # df0=repLevelCDRP3[repLevelCDRP3['Metadata_broad_sample']!='DMSO'].reset_index(drop=True)
# # repSizeDF=repLevelLincs3.groupby(['Metadata_broad_sample']).size().reset_index()
repSizeDF=repLevelLincs3.groupby(['Metadata_pert_id']).size().reset_index()
highRepComp=repSizeDF[repSizeDF[0]>1].Metadata_pert_id.tolist()
# highRepComp.remove('DMSO')
# df0=repLevelLincs3[(repLevelLincs3['Metadata_broad_sample'].isin(highRepComp)) &\
# (repLevelLincs3['Metadata_dose_recode']==1.11)]
df0=repLevelLincs3[(repLevelLincs3['Metadata_pert_id'].isin(highRepComp))]
x_lincs_cp=replicateCorrs(df0,'Metadata_pert_id',cp_features,1)
# saveAsNewSheetToExistingFile(filename,x_lincs_cp[2],'cp-lincs')
# x=replicateCorrs(df0,'Metadata_broad_sample',cp_features,1)
# highRepComp[-1]
saveAsNewSheetToExistingFile(filename,x[2],'cp-lincs')
# repLevelLincs3.Metadata_Plate
repLevelLincs3.head()
# csv_pddf[(csv_pddf['Metadata_dose_recode']==0.04) & (csv_pddf['Metadata_pert_id']=="BRD-A00147595")][['Metadata_Plate','Metadata_Well']].drop_duplicates()
# csv_pddf[(csv_pddf['Metadata_dose_recode']==0.04) & (csv_pddf['Metadata_pert_id']=="BRD-A00147595") &
# (csv_pddf['Metadata_Plate']=='SQ00015196') & (csv_pddf['Metadata_Well']=="B12")][csv_pddf.columns[1820:]].drop_duplicates()
# def standardize_per_catX(df,column_name):
column_name='Metadata_Plate'
repLevelLincs_scaled_perPlate=repLevelLincs3.copy()
repLevelLincs_scaled_perPlate[cp_features.tolist()]=repLevelLincs3[cp_features.tolist()+[column_name]].groupby(column_name).transform(lambda x: (x - x.mean()) / x.std()).values
# def standardize_per_catX(df,column_name):
# # column_name='Metadata_Plate'
# cp_features=df.columns[df.columns.str.contains("Cells_|Cytoplasm_|Nuclei_")]
# df_scaled_perPlate=df.copy()
# df_scaled_perPlate[cp_features.tolist()]=\
# df[cp_features.tolist()+[column_name]].groupby(column_name)\
# .transform(lambda x: (x - x.mean()) / x.std()).values
# return df_scaled_perPlate
df0=repLevelLincs_scaled_perPlate[(repLevelLincs_scaled_perPlate['Metadata_Sample_Dose'].isin(highRepComp))]
x=replicateCorrs(df0,'Metadata_broad_sample',cp_features,1)
```
| github_jupyter |
```
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from sklearn import preprocessing
import pandas as pd
import tensorflow as tf
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
#optimus 5-prime functions
def test_data(df, model, test_seq, obs_col, output_col='pred'):
'''Predict mean ribosome load using model and test set UTRs'''
# Scale the test set mean ribosome load
scaler = preprocessing.StandardScaler()
scaler.fit(df[obs_col].reshape(-1,1))
# Make predictions
predictions = model.predict(test_seq).reshape(-1)
# Inverse scaled predicted mean ribosome load and return in a column labeled 'pred'
df.loc[:,output_col] = scaler.inverse_transform(predictions)
return df
def one_hot_encode(df, col='utr', seq_len=50):
# Dictionary returning one-hot encoding of nucleotides.
nuc_d = {'a':[1,0,0,0],'c':[0,1,0,0],'g':[0,0,1,0],'t':[0,0,0,1], 'n':[0,0,0,0]}
# Creat empty matrix.
vectors=np.empty([len(df),seq_len,4])
# Iterate through UTRs and one-hot encode
for i,seq in enumerate(df[col].str[:seq_len]):
seq = seq.lower()
a = np.array([nuc_d[x] for x in seq])
vectors[i] = a
return vectors
def r2(x,y):
slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
return r_value**2
#Train data
df = pd.read_csv("../../../seqprop/examples/optimus5/GSM3130435_egfp_unmod_1.csv")
df.sort_values('total_reads', inplace=True, ascending=False)
df.reset_index(inplace=True, drop=True)
df = df.iloc[:280000]
# The training set has 260k UTRs and the test set has 20k UTRs.
#e_test = df.iloc[:20000].copy().reset_index(drop = True)
e_train = df.iloc[20000:].copy().reset_index(drop = True)
e_train.loc[:,'scaled_rl'] = preprocessing.StandardScaler().fit_transform(e_train.loc[:,'rl'].values.reshape(-1,1))
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
y_train = np.array(e_train['scaled_rl'].values)
y_train = np.reshape(y_train, (y_train.shape[0],1))
print("x_train.shape = " + str(x_train.shape))
print("y_train.shape = " + str(y_train.shape))
#Load Predictor
predictor_path = 'optimusRetrainedMain.hdf5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Generate (original) predictions
pred_train = predictor.predict(x_train[:, 0, ...], batch_size=32)
y_train = (y_train >= 0.)
y_train = np.concatenate([1. - y_train, y_train], axis=1)
pred_train = (pred_train >= 0.)
pred_train = np.concatenate([1. - pred_train, pred_train], axis=1)
from keras.layers import Input, Dense, Multiply, Flatten, Reshape, Conv2D, MaxPooling2D, GlobalMaxPooling2D, Activation
from keras.layers import BatchNormalization
from keras.models import Sequential, Model
from keras.optimizers import Adam
from keras import regularizers
from keras import backend as K
import tensorflow as tf
import numpy as np
from keras.layers import Layer, InputSpec
from keras import initializers, regularizers, constraints
class InstanceNormalization(Layer):
def __init__(self, axes=(1, 2), trainable=True, **kwargs):
super(InstanceNormalization, self).__init__(**kwargs)
self.axes = axes
self.trainable = trainable
def build(self, input_shape):
self.beta = self.add_weight(name='beta',shape=(input_shape[-1],),
initializer='zeros',trainable=self.trainable)
self.gamma = self.add_weight(name='gamma',shape=(input_shape[-1],),
initializer='ones',trainable=self.trainable)
def call(self, inputs):
mean, variance = tf.nn.moments(inputs, self.axes, keep_dims=True)
return tf.nn.batch_normalization(inputs, mean, variance, self.beta, self.gamma, 1e-6)
def bernoulli_sampling (prob):
""" Sampling Bernoulli distribution by given probability.
Args:
- prob: P(Y = 1) in Bernoulli distribution.
Returns:
- samples: samples from Bernoulli distribution
"""
n, x_len, y_len, d = prob.shape
samples = np.random.binomial(1, prob, (n, x_len, y_len, d))
return samples
class INVASE():
"""INVASE class.
Attributes:
- x_train: training features
- y_train: training labels
- model_type: invase or invase_minus
- model_parameters:
- actor_h_dim: hidden state dimensions for actor
- critic_h_dim: hidden state dimensions for critic
- n_layer: the number of layers
- batch_size: the number of samples in mini batch
- iteration: the number of iterations
- activation: activation function of models
- learning_rate: learning rate of model training
- lamda: hyper-parameter of INVASE
"""
def __init__(self, x_train, y_train, model_type, model_parameters):
self.lamda = model_parameters['lamda']
self.actor_h_dim = model_parameters['actor_h_dim']
self.critic_h_dim = model_parameters['critic_h_dim']
self.n_layer = model_parameters['n_layer']
self.batch_size = model_parameters['batch_size']
self.iteration = model_parameters['iteration']
self.activation = model_parameters['activation']
self.learning_rate = model_parameters['learning_rate']
#Modified Code
self.x_len = x_train.shape[1]
self.y_len = x_train.shape[2]
self.dim = x_train.shape[3]
self.label_dim = y_train.shape[1]
self.model_type = model_type
optimizer = Adam(self.learning_rate)
# Build and compile critic
self.critic = self.build_critic()
self.critic.compile(loss='categorical_crossentropy',
optimizer=optimizer, metrics=['acc'])
# Build and compile the actor
self.actor = self.build_actor()
self.actor.compile(loss=self.actor_loss, optimizer=optimizer)
if self.model_type == 'invase':
# Build and compile the baseline
self.baseline = self.build_baseline()
self.baseline.compile(loss='categorical_crossentropy', optimizer=optimizer, metrics=['acc'])
def actor_loss(self, y_true, y_pred):
"""Custom loss for the actor.
Args:
- y_true:
- actor_out: actor output after sampling
- critic_out: critic output
- baseline_out: baseline output (only for invase)
- y_pred: output of the actor network
Returns:
- loss: actor loss
"""
y_pred = K.reshape(y_pred, (K.shape(y_pred)[0], self.x_len*self.y_len*1))
y_true = y_true[:, 0, 0, :]
# Actor output
actor_out = y_true[:, :self.x_len*self.y_len*1]
# Critic output
critic_out = y_true[:, self.x_len*self.y_len*1:(self.x_len*self.y_len*1+self.label_dim)]
if self.model_type == 'invase':
# Baseline output
baseline_out = \
y_true[:, (self.x_len*self.y_len*1+self.label_dim):(self.x_len*self.y_len*1+2*self.label_dim)]
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+2*self.label_dim):]
elif self.model_type == 'invase_minus':
# Ground truth label
y_out = y_true[:, (self.x_len*self.y_len*1+self.label_dim):]
# Critic loss
critic_loss = -tf.reduce_sum(y_out * tf.log(critic_out + 1e-8), axis = 1)
if self.model_type == 'invase':
# Baseline loss
baseline_loss = -tf.reduce_sum(y_out * tf.log(baseline_out + 1e-8),
axis = 1)
# Reward
Reward = -(critic_loss - baseline_loss)
elif self.model_type == 'invase_minus':
Reward = -critic_loss
# Policy gradient loss computation.
custom_actor_loss = \
Reward * tf.reduce_sum(actor_out * K.log(y_pred + 1e-8) + \
(1-actor_out) * K.log(1-y_pred + 1e-8), axis = 1) - \
self.lamda * tf.reduce_mean(y_pred, axis = 1)
# custom actor loss
custom_actor_loss = tf.reduce_mean(-custom_actor_loss)
return custom_actor_loss
def build_actor(self):
"""Build actor.
Use feature as the input and output selection probability
"""
actor_model = Sequential()
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
actor_model.add(Conv2D(self.actor_h_dim, (1, 7), padding='same', activation='linear'))
actor_model.add(InstanceNormalization())
actor_model.add(Activation(self.activation))
actor_model.add(Conv2D(1, (1, 1), padding='same', activation='sigmoid'))
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
selection_probability = actor_model(feature)
return Model(feature, selection_probability)
def build_critic(self):
"""Build critic.
Use selected feature as the input and predict labels
"""
critic_model = Sequential()
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
critic_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
critic_model.add(InstanceNormalization())
critic_model.add(Activation(self.activation))
critic_model.add(Flatten())
critic_model.add(Dense(self.critic_h_dim, activation=self.activation))
critic_model.add(Dropout(0.2))
critic_model.add(Dense(self.label_dim, activation ='softmax'))
## Inputs
# Features
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Binary selection
selection = Input(shape=(self.x_len, self.y_len, 1), dtype='float32')
# Element-wise multiplication
critic_model_input = Multiply()([feature, selection])
y_hat = critic_model(critic_model_input)
return Model([feature, selection], y_hat)
def build_baseline(self):
"""Build baseline.
Use the feature as the input and predict labels
"""
baseline_model = Sequential()
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
for _ in range(self.n_layer - 2):
baseline_model.add(Conv2D(self.critic_h_dim, (1, 7), padding='same', activation='linear'))
baseline_model.add(InstanceNormalization())
baseline_model.add(Activation(self.activation))
baseline_model.add(Flatten())
baseline_model.add(Dense(self.critic_h_dim, activation=self.activation))
baseline_model.add(Dropout(0.2))
baseline_model.add(Dense(self.label_dim, activation ='softmax'))
# Input
feature = Input(shape=(self.x_len, self.y_len, self.dim), dtype='float32')
# Output
y_hat = baseline_model(feature)
return Model(feature, y_hat)
def train(self, x_train, y_train):
"""Train INVASE.
Args:
- x_train: training features
- y_train: training labels
"""
for iter_idx in range(self.iteration):
## Train critic
# Select a random batch of samples
idx = np.random.randint(0, x_train.shape[0], self.batch_size)
x_batch = x_train[idx,:]
y_batch = y_train[idx,:]
# Generate a batch of selection probability
selection_probability = self.actor.predict(x_batch)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Critic loss
critic_loss = self.critic.train_on_batch([x_batch, selection], y_batch)
# Critic output
critic_out = self.critic.predict([x_batch, selection])
# Baseline output
if self.model_type == 'invase':
# Baseline loss
baseline_loss = self.baseline.train_on_batch(x_batch, y_batch)
# Baseline output
baseline_out = self.baseline.predict(x_batch)
## Train actor
# Use multiple things as the y_true:
# - selection, critic_out, baseline_out, and ground truth (y_batch)
if self.model_type == 'invase':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
np.asarray(baseline_out),
y_batch), axis = 1)
elif self.model_type == 'invase_minus':
y_batch_final = np.concatenate((np.reshape(selection, (y_batch.shape[0], -1)),
np.asarray(critic_out),
y_batch), axis = 1)
y_batch_final = y_batch_final[:, None, None, :]
# Train the actor
actor_loss = self.actor.train_on_batch(x_batch, y_batch_final)
if self.model_type == 'invase':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', baseline accuracy: ' + str(baseline_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
elif self.model_type == 'invase_minus':
# Print the progress
dialog = 'Iterations: ' + str(iter_idx) + \
', critic accuracy: ' + str(critic_loss[1]) + \
', actor loss: ' + str(np.round(actor_loss,4))
if iter_idx % 100 == 0:
print(dialog)
def importance_score(self, x):
"""Return featuer importance score.
Args:
- x: feature
Returns:
- feature_importance: instance-wise feature importance for x
"""
feature_importance = self.actor.predict(x)
return np.asarray(feature_importance)
def predict(self, x):
"""Predict outcomes.
Args:
- x: feature
Returns:
- y_hat: predictions
"""
# Generate a batch of selection probability
selection_probability = self.actor.predict(x)
# Sampling the features based on the selection_probability
selection = bernoulli_sampling(selection_probability)
# Prediction
y_hat = self.critic.predict([x, selection])
return np.asarray(y_hat)
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
dna_letter_at(ref_seq[i], i + 0.5, 0, mutability_score, ax)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
for axis in fig.axes :
axis.get_xaxis().set_visible(False)
axis.get_yaxis().set_visible(False)
plt.tight_layout()
plt.show()
#Execute INVASE benchmark on synthetic datasets
mask_penalty = 0.5#0.1
hidden_dims = 32
n_layers = 4
epochs = 25
batch_size = 128
model_parameters = {
'lamda': mask_penalty,
'actor_h_dim': hidden_dims,
'critic_h_dim': hidden_dims,
'n_layer': n_layers,
'batch_size': batch_size,
'iteration': int(x_train.shape[0] * epochs / batch_size),
'activation': 'relu',
'learning_rate': 0.0001
}
encoder = isol.OneHotEncoder(50)
score_clip = None
allFiles = ["optimus5_synthetic_random_insert_if_uorf_1_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_1_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_1_stop_variable_loc_512.csv",
"optimus5_synthetic_random_insert_if_uorf_2_start_2_stop_variable_loc_512.csv",
"optimus5_synthetic_examples_3.csv"]
#Train INVASE
invase_model = INVASE(x_train, pred_train, 'invase', model_parameters)
invase_model.train(x_train, pred_train)
for csv_to_open in allFiles :
#Load dataset for benchmarking
dataset_name = csv_to_open.replace(".csv", "")
benchmarkSet = pd.read_csv(csv_to_open)
seq_e_test = one_hot_encode(benchmarkSet, seq_len=50)
x_test = seq_e_test[:, None, ...]
print(x_test.shape)
pred_test = predictor.predict(x_test[:, 0, ...], batch_size=32)
y_test = pred_test
y_test = (y_test >= 0.)
y_test = np.concatenate([1. - y_test, y_test], axis=1)
pred_test = (pred_test >= 0.)
pred_test = np.concatenate([1. - pred_test, pred_test], axis=1)
importance_scores_test = invase_model.importance_score(x_test)
#Evaluate INVASE model on train and test data
invase_pred_train = invase_model.predict(x_train)
invase_pred_test = invase_model.predict(x_test)
print("Training Accuracy = " + str(np.sum(np.argmax(invase_pred_train, axis=1) == np.argmax(pred_train, axis=1)) / float(pred_train.shape[0])))
print("Test Accuracy = " + str(np.sum(np.argmax(invase_pred_test, axis=1) == np.argmax(pred_test, axis=1)) / float(pred_test.shape[0])))
for plot_i in range(0, 3) :
print("Test sequence " + str(plot_i) + ":")
plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template='N'*50, plot_sequence_template=True, figsize=(12, 1), plot_start=0, plot_end=50)
plot_importance_scores(np.maximum(importance_scores_test[plot_i, 0, :, :].T, 0.), encoder.decode(x_test[plot_i, 0, :, :]), figsize=(12, 1), score_clip=score_clip, sequence_template='N'*50, plot_start=0, plot_end=50)
#Save predicted importance scores
model_name = "invase_" + dataset_name + "_conv_full_data"
np.save(model_name + "_importance_scores_test", importance_scores_test)
```
| github_jupyter |
# NOAA Wave Watch 3 and NDBC Buoy Data Comparison
*Note: this notebook requires python3.*
This notebook demostrates how to compare [WaveWatch III Global Ocean Wave Model](http://data.planetos.com/datasets/noaa_ww3_global_1.25x1d:noaa-wave-watch-iii-nww3-ocean-wave-model?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook) and [NOAA NDBC buoy data](http://data.planetos.com/datasets/noaa_ndbc_stdmet_stations?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook) using the Planet OS API.
API documentation is available at http://docs.planetos.com. If you have questions or comments, join the [Planet OS Slack community](http://slack.planetos.com/) to chat with our development team.
For general information on usage of IPython/Jupyter and Matplotlib, please refer to their corresponding documentation. https://ipython.org/ and http://matplotlib.org/. This notebook also makes use of the [matplotlib basemap toolkit.](http://matplotlib.org/basemap/index.html)
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import dateutil.parser
import datetime
from urllib.request import urlopen, Request
import simplejson as json
from datetime import date, timedelta, datetime
import matplotlib.dates as mdates
from mpl_toolkits.basemap import Basemap
```
**Important!** You'll need to replace apikey below with your actual Planet OS API key, which you'll find [on the Planet OS account settings page.](#http://data.planetos.com/account/settings/?utm_source=github&utm_medium=notebook&utm_campaign=ww3-api-notebook) and NDBC buoy station name in which you are intrested.
```
dataset_id = 'noaa_ndbc_stdmet_stations'
## stations with wave height available: '46006', '46013', '46029'
## stations without wave height: icac1', '41047', 'bepb6', '32st0', '51004'
## stations too close to coastline (no point to compare to ww3)'sacv4', 'gelo1', 'hcef1'
station = '46029'
apikey = open('APIKEY').readlines()[0].strip() #'<YOUR API KEY HERE>'
```
Let's first query the API to see what stations are available for the [NDBC Standard Meteorological Data dataset.](http://data.planetos.com/datasets/noaa_ndbc_stdmet_stations?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook)
```
API_url = 'http://api.planetos.com/v1/datasets/%s/stations?apikey=%s' % (dataset_id, apikey)
request = Request(API_url)
response = urlopen(request)
API_data_locations = json.loads(response.read())
# print(API_data_locations)
```
Now we'll use matplotlib to visualize the stations on a simple basemap.
```
m = Basemap(projection='merc',llcrnrlat=-80,urcrnrlat=80,\
llcrnrlon=-180,urcrnrlon=180,lat_ts=20,resolution='c')
fig=plt.figure(figsize=(15,10))
m.drawcoastlines()
##m.fillcontinents()
for i in API_data_locations['station']:
x,y=m(API_data_locations['station'][i]['SpatialExtent']['coordinates'][0],
API_data_locations['station'][i]['SpatialExtent']['coordinates'][1])
plt.scatter(x,y,color='r')
x,y=m(API_data_locations['station'][station]['SpatialExtent']['coordinates'][0],
API_data_locations['station'][station]['SpatialExtent']['coordinates'][1])
plt.scatter(x,y,s=100,color='b')
```
Let's examine the last five days of data. For the WaveWatch III forecast, we'll use the reference time parameter to pull forecast data from the 18:00 model run from five days ago.
```
## Find suitable reference time values
atthemoment = datetime.utcnow()
atthemoment = atthemoment.strftime('%Y-%m-%dT%H:%M:%S')
before5days = datetime.utcnow() - timedelta(days=5)
before5days_long = before5days.strftime('%Y-%m-%dT%H:%M:%S')
before5days_short = before5days.strftime('%Y-%m-%d')
start = before5days_long
end = atthemoment
reftime_start = str(before5days_short) + 'T18:00:00'
reftime_end = reftime_start
```
API request for NOAA NDBC buoy station data
```
API_url = "http://api.planetos.com/v1/datasets/{0}/point?station={1}&apikey={2}&start={3}&end={4}&count=1000".format(dataset_id,station,apikey,start,end)
print(API_url)
request = Request(API_url)
response = urlopen(request)
API_data_buoy = json.loads(response.read())
buoy_variables = []
for k,v in set([(j,i['context']) for i in API_data_buoy['entries'] for j in i['data'].keys()]):
buoy_variables.append(k)
```
Find buoy station coordinates to use them later for finding NOAA Wave Watch III data
```
for i in API_data_buoy['entries']:
#print(i['axes']['time'])
if i['context'] == 'time_latitude_longitude':
longitude = (i['axes']['longitude'])
latitude = (i['axes']['latitude'])
print ('Latitude: '+ str(latitude))
print ('Longitude: '+ str(longitude))
```
API request for NOAA WaveWatch III (NWW3) Ocean Wave Model near the point of selected station. Note that data may not be available at the requested reference time. If the response is empty, try removing the reference time parameters `reftime_start` and `reftime_end` from the query.
```
API_url = 'http://api.planetos.com/v1/datasets/noaa_ww3_global_1.25x1d/point?lat={0}&lon={1}&verbose=true&apikey={2}&count=100&end={3}&reftime_start={4}&reftime_end={5}'.format(latitude,longitude,apikey,end,reftime_start,reftime_end)
request = Request(API_url)
response = urlopen(request)
API_data_ww3 = json.loads(response.read())
print(API_url)
ww3_variables = []
for k,v in set([(j,i['context']) for i in API_data_ww3['entries'] for j in i['data'].keys()]):
ww3_variables.append(k)
```
Manually review the list of WaveWatch and NDBC data variables to determine which parameters are equivalent for comparison.
```
print(ww3_variables)
print(buoy_variables)
```
Next we'll build a dictionary of corresponding variables that we want to compare.
```
buoy_model = {'wave_height':'Significant_height_of_combined_wind_waves_and_swell_surface',
'mean_wave_dir':'Primary_wave_direction_surface',
'average_wpd':'Primary_wave_mean_period_surface',
'wind_spd':'Wind_speed_surface'}
```
Read data from the JSON responses and convert the values to floats for plotting. Note that depending on the dataset, some variables have different timesteps than others, so a separate time array for each variable is recommended.
```
def append_data(in_string):
if in_string == None:
return np.nan
elif in_string == 'None':
return np.nan
else:
return float(in_string)
ww3_data = {}
ww3_times = {}
buoy_data = {}
buoy_times = {}
for k,v in buoy_model.items():
ww3_data[v] = []
ww3_times[v] = []
buoy_data[k] = []
buoy_times[k] = []
for i in API_data_ww3['entries']:
for j in i['data']:
if j in buoy_model.values():
ww3_data[j].append(append_data(i['data'][j]))
ww3_times[j].append(dateutil.parser.parse(i['axes']['time']))
for i in API_data_buoy['entries']:
for j in i['data']:
if j in buoy_model.keys():
buoy_data[j].append(append_data(i['data'][j]))
buoy_times[j].append(dateutil.parser.parse(i['axes']['time']))
for i in ww3_data:
ww3_data[i] = np.array(ww3_data[i])
ww3_times[i] = np.array(ww3_times[i])
```
Finally, let's plot the data using matplotlib.
```
buoy_label = "NDBC Station %s" % station
ww3_label = "WW3 at %s" % reftime_start
for k,v in buoy_model.items():
if np.abs(np.nansum(buoy_data[k]))>0:
fig=plt.figure(figsize=(10,5))
plt.title(k+' '+v)
plt.plot(ww3_times[v],ww3_data[v], label=ww3_label)
plt.plot(buoy_times[k],buoy_data[k],'*',label=buoy_label)
plt.legend(bbox_to_anchor=(1.5, 0.22), loc=1, borderaxespad=0.)
plt.xlabel('Time')
plt.ylabel(k)
fig.autofmt_xdate()
plt.grid()
```
| github_jupyter |
# Aula 1
```
import pandas as pd
url_dados = 'https://github.com/alura-cursos/imersaodados3/blob/main/dados/dados_experimentos.zip?raw=true'
dados = pd.read_csv(url_dados, compression = 'zip')
dados
dados.head()
dados.shape
dados['tratamento']
dados['tratamento'].unique()
dados['tempo'].unique()
dados['dose'].unique()
dados['droga'].unique()
dados['g-0'].unique()
dados['tratamento'].value_counts()
dados['dose'].value_counts()
dados['tratamento'].value_counts(normalize = True)
dados['dose'].value_counts(normalize = True)
dados['tratamento'].value_counts().plot.pie()
dados['tempo'].value_counts().plot.pie()
dados['tempo'].value_counts().plot.bar()
dados_filtrados = dados[dados['g-0'] > 0]
dados_filtrados.head()
```
#Desafios Aula 1
## Desafio 01: Investigar por que a classe tratamento é tão desbalanceada?
Dependendo o tipo de pesquisa é possível usar o mesmo controle para mais de um caso. Repare que o grupo de controle é um grupo onde não estamos aplicando o efeito de uma determinada droga. Então, esse mesmo grupo pode ser utilizado como controle para cada uma das drogas estudadas.
Um ponto relevante da base de dados que estamos trabalhando é que todos os dados de controle estão relacionados ao estudo de apenas uma droga.
```
print(f"Total de dados {len(dados['id'])}\n")
print(f"Quantidade de drogas {len(dados.groupby(['droga', 'tratamento']).count()['id'])}\n")
display(dados.query('tratamento == "com_controle"').value_counts('droga'))
print()
display(dados.query('droga == "cacb2b860"').value_counts('tratamento'))
print()
```
## Desafio 02: Plotar as 5 últimas linhas da tabela
```
dados.tail()
```
Outra opção seria usar o seguinte comando:
```
dados[-5:]
```
## Desafio 03: Proporção das classes tratamento.
```
dados['tratamento'].value_counts(normalize = True)
```
## Desafio 04: Quantas tipos de drogas foram investigadas.
```
dados['droga'].unique().shape[0]
```
Outra opção de solução:
```
len(dados['droga'].unique())
```
## Desafio 05: Procurar na documentação o método query(pandas).
https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.query.html
## Desafio 06: Renomear as colunas tirando o hífen.
```
dados.columns
nome_das_colunas = dados.columns
novo_nome_coluna = []
for coluna in nome_das_colunas:
coluna = coluna.replace('-', '_')
novo_nome_coluna.append(coluna)
dados.columns = novo_nome_coluna
dados.head()
```
Agora podemos comparar o resultado usando Query com o resultado usando máscara + slice
```
dados_filtrados = dados[dados['g_0'] > 0]
dados_filtrados.head()
dados_filtrados = dados.query('g_0 > 0')
dados_filtrados.head()
```
## Desafio 07: Deixar os gráficos bonitões. (Matplotlib.pyplot)
```
import matplotlib.pyplot as plt
valore_tempo = dados['tempo'].value_counts(ascending=True)
valore_tempo.sort_index()
plt.figure(figsize=(15, 10))
valore_tempo = dados['tempo'].value_counts(ascending=True)
ax = valore_tempo.sort_index().plot.bar()
ax.set_title('Janelas de tempo', fontsize=20)
ax.set_xlabel('Tempo', fontsize=18)
ax.set_ylabel('Quantidade', fontsize=18)
plt.xticks(rotation = 0, fontsize=16)
plt.yticks(fontsize=16)
plt.show()
```
##Desafio 08: Resumo do que você aprendeu com os dados
Nesta aula utilizei a biblioteca Pandas, diversas funcionalidades da mesma para explorar dados. Durante a análise de dados, descobri fatores importantes para a obtenção de insights e também aprendi como plotar os gráficos de pizza e de colunas discutindo pontos positivos e negativos.
Para mais informações a base dados estudada na imersão é uma versão simplificada [deste desafio](https://www.kaggle.com/c/lish-moa/overview/description) do Kaggle (em inglês).
Também recomendo acessar o
[Connectopedia](https://clue.io/connectopedia/). O Connectopedia é um dicionário gratuito de termos e conceitos que incluem definições de viabilidade de células e expressão de genes.
O desafio do Kaggle também está relacionado a estes artigos científicos:
Corsello et al. “Discovering the anticancer potential of non-oncology drugs by systematic viability profiling,” Nature Cancer, 2020, https://doi.org/10.1038/s43018-019-0018-6
Subramanian et al. “A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles,” Cell, 2017, https://doi.org/10.1016/j.cell.2017.10.049
```
```
| github_jupyter |
___
<a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
___
# Matplotlib Exercises
Welcome to the exercises for reviewing matplotlib! Take your time with these, Matplotlib can be tricky to understand at first. These are relatively simple plots, but they can be hard if this is your first time with matplotlib, feel free to reference the solutions as you go along.
Also don't worry if you find the matplotlib syntax frustrating, we actually won't be using it that often throughout the course, we will switch to using seaborn and pandas built-in visualization capabilities. But, those are built-off of matplotlib, which is why it is still important to get exposure to it!
** * NOTE: ALL THE COMMANDS FOR PLOTTING A FIGURE SHOULD ALL GO IN THE SAME CELL. SEPARATING THEM OUT INTO MULTIPLE CELLS MAY CAUSE NOTHING TO SHOW UP. * **
# Exercises
Follow the instructions to recreate the plots using this data:
## Data
```
import numpy as np
x = np.arange(0,100)
y = x*2
z = x**2
```
** Import matplotlib.pyplot as plt and set %matplotlib inline if you are using the jupyter notebook. What command do you use if you aren't using the jupyter notebook?**
```
import matplotlib.pyplot as plt
%matplotlib inline
```
## Exercise 1
** Follow along with these steps: **
* ** Create a figure object called fig using plt.figure() **
* ** Use add_axes to add an axis to the figure canvas at [0,0,1,1]. Call this new axis ax. **
* ** Plot (x,y) on that axes and set the labels and titles to match the plot below:**
```
# Functional Method
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
ax.plot(x, y)
ax.set_title('title')
ax.set_xlabel('X')
ax.set_ylabel('Y')
```
## Exercise 2
** Create a figure object and put two axes on it, ax1 and ax2. Located at [0,0,1,1] and [0.2,0.5,.2,.2] respectively.**
```
# create figure canvas
fig = plt.figure()
# create axes
ax1 = fig.add_axes([0,0,1,1])
ax2 = fig.add_axes([0.2,0.5,.2,.2])
plt.xticks(np.arange(0, 1.2, step=0.2))
plt.yticks(np.arange(0, 1.2, step=0.2))
```
** Now plot (x,y) on both axes. And call your figure object to show it.**
```
# create figure canvas
fig = plt.figure()
# create axes
ax1 = fig.add_axes([0,0,1,1])
ax2 = fig.add_axes([0.2,0.5,.2,.2])
ax1.set_xlabel('x1')
ax1.set_ylabel('y1')
ax2.set_xlabel('x2')
ax2.set_ylabel('y2')
ax1.plot(x, y, 'r-')
ax2.plot(x, y, 'b--')
plt.xticks(np.arange(0, 120, step=20))
plt.yticks(np.arange(0, 220, step=50))
```
## Exercise 3
** Create the plot below by adding two axes to a figure object at [0,0,1,1] and [0.2,0.5,.4,.4]**
```
fig = plt.figure()
ax1 = fig.add_axes([0,0,1,1])
ax2 = fig.add_axes([0.2,0.5,.4,.4])
```
** Now use x,y, and z arrays to recreate the plot below. Notice the xlimits and y limits on the inserted plot:**
```
fig = plt.figure()
ax1 = fig.add_axes([0,0,1,1])
ax2 = fig.add_axes([0.2,0.5,.4,.4])
ax1.plot(x, z)
ax2.plot(x, y, 'r--') # zoom using xlimit (20, 22), ylimit (30, 50)
ax2.set_xlim([20, 22])
ax2.set_ylim([30, 50])
ax2.set_title('zoom')
ax2.set_xlabel('X')
ax2.set_ylabel('Y')
ax1.set_xlabel('X')
ax1.set_ylabel('Z')
```
## Exercise 4
** Use plt.subplots(nrows=1, ncols=2) to create the plot below.**
```
fig, axes = plt.subplots(nrows=1, ncols=2)
# axes object is an array of subplot axis.
plt.tight_layout() # add space between rows & columns.
```
** Now plot (x,y) and (x,z) on the axes. Play around with the linewidth and style**
```
fig, axes = plt.subplots(nrows=1, ncols=2)
# axes object is an array of subplot axis.
axes[0].plot(x, y, 'b--', lw=3)
axes[1].plot(x, z, 'r-.', lw=2)
plt.tight_layout() # add space between rows & columns.
```
** See if you can resize the plot by adding the figsize() argument in plt.subplots() are copying and pasting your previous code.**
```
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(10, 7))
# axes object is an array of subplot axis.
axes[0].plot(x, y, 'b--', lw=3)
axes[1].plot(x, z, 'r-.', lw=2)
plt.tight_layout() # add space between rows & columns.
```
# Great Job!
| github_jupyter |
```
import json
import numpy as np
import matplotlib.pyplot as plt
from scipy.integrate import quad
from scipy.special import comb
from tabulate import tabulate
%matplotlib inline
```
## Expected numbers on Table 3.
```
rows = []
datasets = {
'Binary': 2,
'AG news': 4,
'CIFAR10': 10,
'CIFAR100': 100,
'Wiki3029': 3029,
}
def expectations(C: int) -> float:
"""
C is the number of latent classes.
"""
e = 0.
for k in range(1, C + 1):
e += C / k
return e
for dataset_name, C in datasets.items():
e = expectations(C)
rows.append((dataset_name, C, np.ceil(e)))
# ImageNet is non-uniform label distribution on the training dataset
data = json.load(open("./imagenet_count.json"))
counts = np.array(list(data.values()))
total_num = np.sum(counts)
prob = counts / total_num
def integrand(t: float, prob: np.ndarray) -> float:
return 1. - np.prod(1 - np.exp(-prob * t))
rows.append(("ImageNet", len(prob), np.ceil(quad(integrand, 0, np.inf, args=(prob))[0])))
print(tabulate(rows, headers=["Dataset", "\# classes", "\mathbb{E}[K+1]"]))
```
## Probability $\upsilon$
```
def prob(C, N):
"""
C: the number of latent class
N: the number of samples to draw
"""
theoretical = []
for n in range(C, N + 1):
p = 0.
for m in range(C - 1):
p += comb(C - 1, m) * ((-1) ** m) * np.exp((n - 1) * np.log(1. - (m + 1) / C))
theoretical.append((n, max(p, 0.)))
return np.array(theoretical)
# example of CIFAR-10
C = 10
for N in [32, 63, 128, 256, 512]:
p = np.sum(prob(C, N).T[1])
print("{:3d} {:.7f}".format(N, p))
# example of CIFAR-100
C = 100
ps = []
ns = []
for N in 128 * np.arange(1, 9):
p = np.sum(prob(C, N).T[1])
print("{:4d} {}".format(N, p))
ps.append(p)
ns.append(N)
```
## Simulation
```
n_loop = 10
rnd = np.random.RandomState(7)
labels = np.arange(C).repeat(100)
results = {}
for N in ns:
num_iters = int(len(labels) / N)
total_samples_for_bounds = float(num_iters * N * (n_loop))
for _ in range(n_loop):
rnd.shuffle(labels)
for batch_id in range(len(labels) // N):
if len(set(labels[N * batch_id:N * (batch_id + 1)])) == C:
results[N] = results.get(N, 0.) + N / total_samples_for_bounds
else:
results[N] = results.get(N, 0.) + 0.
xs = []
ys = []
for k, v in results.items():
print(k, v)
ys.append(v)
xs.append(k)
plt.plot(ns, ps, label="Theoretical")
plt.plot(xs, ys, label="Empirical")
plt.ylabel("probability")
plt.xlabel("$K+1$")
plt.title("CIFAR-100 simulation")
plt.legend()
```
| github_jupyter |
# PageRank Performance Benchmarking
# Skip notebook test
This notebook benchmarks performance of running PageRank within cuGraph against NetworkX. NetworkX contains several implementations of PageRank. This benchmark will compare cuGraph versus the defaukt Nx implementation as well as the SciPy version
Notebook Credits
Original Authors: Bradley Rees
Last Edit: 08/16/2020
RAPIDS Versions: 0.15
Test Hardware
GV100 32G, CUDA 10,0
Intel(R) Core(TM) CPU i7-7800X @ 3.50GHz
32GB system memory
### Test Data
| File Name | Num of Vertices | Num of Edges |
|:---------------------- | --------------: | -----------: |
| preferentialAttachment | 100,000 | 999,970 |
| caidaRouterLevel | 192,244 | 1,218,132 |
| coAuthorsDBLP | 299,067 | 1,955,352 |
| dblp-2010 | 326,186 | 1,615,400 |
| citationCiteseer | 268,495 | 2,313,294 |
| coPapersDBLP | 540,486 | 30,491,458 |
| coPapersCiteseer | 434,102 | 32,073,440 |
| as-Skitter | 1,696,415 | 22,190,596 |
### Timing
What is not timed: Reading the data
What is timmed: (1) creating a Graph, (2) running PageRank
The data file is read in once for all flavors of PageRank. Each timed block will craete a Graph and then execute the algorithm. The results of the algorithm are not compared. If you are interested in seeing the comparison of results, then please see PageRank in the __notebooks__ repo.
## NOTICE
_You must have run the __dataPrep__ script prior to running this notebook so that the data is downloaded_
See the README file in this folder for a discription of how to get the data
## Now load the required libraries
```
# Import needed libraries
import gc
import time
import rmm
import cugraph
import cudf
# NetworkX libraries
import networkx as nx
from scipy.io import mmread
try:
import matplotlib
except ModuleNotFoundError:
os.system('pip install matplotlib')
import matplotlib.pyplot as plt; plt.rcdefaults()
import numpy as np
```
### Define the test data
```
# Test File
data = {
'preferentialAttachment' : './data/preferentialAttachment.mtx',
'caidaRouterLevel' : './data/caidaRouterLevel.mtx',
'coAuthorsDBLP' : './data/coAuthorsDBLP.mtx',
'dblp' : './data/dblp-2010.mtx',
'citationCiteseer' : './data/citationCiteseer.mtx',
'coPapersDBLP' : './data/coPapersDBLP.mtx',
'coPapersCiteseer' : './data/coPapersCiteseer.mtx',
'as-Skitter' : './data/as-Skitter.mtx'
}
```
### Define the testing functions
```
# Data reader - the file format is MTX, so we will use the reader from SciPy
def read_mtx_file(mm_file):
print('Reading ' + str(mm_file) + '...')
M = mmread(mm_file).asfptype()
return M
# CuGraph PageRank
def cugraph_call(M, max_iter, tol, alpha):
gdf = cudf.DataFrame()
gdf['src'] = M.row
gdf['dst'] = M.col
print('\tcuGraph Solving... ')
t1 = time.time()
# cugraph Pagerank Call
G = cugraph.DiGraph()
G.from_cudf_edgelist(gdf, source='src', destination='dst', renumber=False)
df = cugraph.pagerank(G, alpha=alpha, max_iter=max_iter, tol=tol)
t2 = time.time() - t1
return t2
# Basic NetworkX PageRank
def networkx_call(M, max_iter, tol, alpha):
nnz_per_row = {r: 0 for r in range(M.get_shape()[0])}
for nnz in range(M.getnnz()):
nnz_per_row[M.row[nnz]] = 1 + nnz_per_row[M.row[nnz]]
for nnz in range(M.getnnz()):
M.data[nnz] = 1.0/float(nnz_per_row[M.row[nnz]])
M = M.tocsr()
if M is None:
raise TypeError('Could not read the input graph')
if M.shape[0] != M.shape[1]:
raise TypeError('Shape is not square')
# should be autosorted, but check just to make sure
if not M.has_sorted_indices:
print('sort_indices ... ')
M.sort_indices()
z = {k: 1.0/M.shape[0] for k in range(M.shape[0])}
print('\tNetworkX Solving... ')
# start timer
t1 = time.time()
Gnx = nx.DiGraph(M)
pr = nx.pagerank(Gnx, alpha, z, max_iter, tol)
t2 = time.time() - t1
return t2
# SciPy PageRank
def networkx_scipy_call(M, max_iter, tol, alpha):
nnz_per_row = {r: 0 for r in range(M.get_shape()[0])}
for nnz in range(M.getnnz()):
nnz_per_row[M.row[nnz]] = 1 + nnz_per_row[M.row[nnz]]
for nnz in range(M.getnnz()):
M.data[nnz] = 1.0/float(nnz_per_row[M.row[nnz]])
M = M.tocsr()
if M is None:
raise TypeError('Could not read the input graph')
if M.shape[0] != M.shape[1]:
raise TypeError('Shape is not square')
# should be autosorted, but check just to make sure
if not M.has_sorted_indices:
print('sort_indices ... ')
M.sort_indices()
z = {k: 1.0/M.shape[0] for k in range(M.shape[0])}
# SciPy Pagerank Call
print('\tSciPy Solving... ')
t1 = time.time()
Gnx = nx.DiGraph(M)
pr = nx.pagerank_scipy(Gnx, alpha, z, max_iter, tol)
t2 = time.time() - t1
return t2
```
### Run the benchmarks
```
# arrays to capture performance gains
time_cu = []
time_nx = []
time_sp = []
perf_nx = []
perf_sp = []
names = []
# init libraries by doing a simple task
v = './data/preferentialAttachment.mtx'
M = read_mtx_file(v)
trapids = cugraph_call(M, 100, 0.00001, 0.85)
del M
for k,v in data.items():
gc.collect()
# Saved the file Name
names.append(k)
# read the data
M = read_mtx_file(v)
# call cuGraph - this will be the baseline
trapids = cugraph_call(M, 100, 0.00001, 0.85)
time_cu.append(trapids)
# Now call NetworkX
tn = networkx_call(M, 100, 0.00001, 0.85)
speedUp = (tn / trapids)
perf_nx.append(speedUp)
time_nx.append(tn)
# Now call SciPy
tsp = networkx_scipy_call(M, 100, 0.00001, 0.85)
speedUp = (tsp / trapids)
perf_sp.append(speedUp)
time_sp.append(tsp)
print("cuGraph (" + str(trapids) + ") Nx (" + str(tn) + ") SciPy (" + str(tsp) + ")" )
del M
```
### plot the output
```
%matplotlib inline
plt.figure(figsize=(10,8))
bar_width = 0.35
index = np.arange(len(names))
_ = plt.bar(index, perf_nx, bar_width, color='g', label='vs Nx')
_ = plt.bar(index + bar_width, perf_sp, bar_width, color='b', label='vs SciPy')
plt.xlabel('Datasets')
plt.ylabel('Speedup')
plt.title('PageRank Performance Speedup')
plt.xticks(index + (bar_width / 2), names)
plt.xticks(rotation=90)
# Text on the top of each barplot
for i in range(len(perf_nx)):
plt.text(x = (i - 0.55) + bar_width, y = perf_nx[i] + 25, s = round(perf_nx[i], 1), size = 12)
for i in range(len(perf_sp)):
plt.text(x = (i - 0.1) + bar_width, y = perf_sp[i] + 25, s = round(perf_sp[i], 1), size = 12)
plt.legend()
plt.show()
```
# Dump the raw stats
```
perf_nx
perf_sp
time_cu
time_nx
time_sp
```
___
Copyright (c) 2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
___
| github_jupyter |
##Tirmzi Analysis
n=1000 m+=1000 nm-=120 istep= 4 min=150 max=700
```
import sys
sys.path
import matplotlib.pyplot as plt
import numpy as np
import os
from scipy import signal
ls
import capsol.newanalyzecapsol as ac
ac.get_gridparameters
import glob
folders = glob.glob("FortranOutputTest/*/")
folders
all_data= dict()
for folder in folders:
params = ac.get_gridparameters(folder + 'capsol.in')
data = ac.np.loadtxt(folder + 'Z-U.dat')
process_data = ac.process_data(params, data, smoothing=False, std=5*10**-9)
all_data[folder]= (process_data)
all_params= dict()
for folder in folders:
params=ac.get_gridparameters(folder + 'capsol.in')
all_params[folder]= (params)
all_data
all_data.keys()
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 1.0}:
data=all_data[key]
thickness =all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
plt.plot(data['z'], data['c'], label= f'{rtip} nm, {er}, {thickness} nm')
plt.title('C v. Z for 1nm thick sample')
plt.ylabel("C(m)")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("C' v. Z for 1nm thick sample 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 10.0}:
data=all_data[key]
thickness =all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
plt.plot(data['z'], data['c'], label= f'{rtip} nm, {er}, {thickness} nm')
plt.title('C v. Z for 10nm thick sample')
plt.ylabel("C(m)")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("C' v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 100.0}:
data=all_data[key]
thickness =all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
plt.plot(data['z'], data['c'], label= f'{rtip} nm, {er}, {thickness} nm')
plt.title('C v. Z for 100nm sample')
plt.ylabel("C(m)")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("C' v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 500.0}:
data=all_data[key]
thickness =all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
plt.plot(data['z'], data['c'], label= f'{rtip} nm, {er}, {thickness} nm')
plt.title('C v. Z for 500nm sample')
plt.ylabel("C(m)")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("C' v. Z for varying sample thickness, 06-28-2021.png")
```
cut off last experiment because capacitance was off the scale
```
for params in all_params.values():
print(params['Thickness_sample'])
print(params['m-'])
all_params
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 1.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(4,-3)
plt.plot(data['z'][s], data['cz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Cz vs. Z for 1.0nm')
plt.ylabel("Cz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Cz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 10.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(4,-3)
plt.plot(data['z'][s], data['cz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Cz vs. Z for 10.0nm')
plt.ylabel("Cz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Cz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 100.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(4,-3)
plt.plot(data['z'][s], data['cz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Cz vs. Z for 100.0nm')
plt.ylabel("Cz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Cz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 500.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(4,-3)
plt.plot(data['z'][s], data['cz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Cz vs. Z for 500.0nm')
plt.ylabel("Cz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Cz v. Z for varying sample thickness, 06-28-2021.png")
hoepker_data= np.loadtxt("Default Dataset (2).csv" , delimiter= ",")
hoepker_data
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 1.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(5,-5)
plt.plot(data['z'][s], data['czz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Czz vs. Z for 1.0nm')
plt.ylabel("Czz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
params
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 10.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(5,-5)
plt.plot(data['z'][s], data['czz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Czz vs. Z for 10.0nm')
plt.ylabel("Czz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 100.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(5,-5)
plt.plot(data['z'][s], data['czz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Czz vs. Z for 100.0nm')
plt.ylabel("Czz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 500.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(5,-5)
plt.plot(data['z'][s], data['czz'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Czz vs. Z for 500.0 nm')
plt.ylabel("Czz")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 1.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(8,-8)
plt.plot(data['z'][s], data['alpha'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('alpha vs. Z for 1.0nm')
plt.ylabel("$\\alpha$")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Alpha v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 10.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(8,-8)
plt.plot(data['z'][s], data['alpha'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Alpha vs. Z for 10.0 nm')
plt.ylabel("$\\alpha$")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 100.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(8,-8)
plt.plot(data['z'][s], data['alpha'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Alpha vs. Z for 100.0nm')
plt.ylabel("$\\alpha$")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
for key in {key: params for key, params in all_params.items() if params['Thickness_sample'] == 500.0}:
data=all_data[key]
thickness=all_params[key]['Thickness_sample']
rtip= all_params[key]['Rtip']
er=all_params[key]['eps_r']
s=slice(8,-8)
plt.plot(data['z'][s], data['alpha'][s], label=f'{rtip} nm, {er}, {thickness} nm' )
plt.title('Alpha vs. Z for 500.0nm')
plt.ylabel("$\\alpha$")
plt.xlabel("Z(m)")
plt.legend()
plt.savefig("Czz v. Z for varying sample thickness, 06-28-2021.png")
data
from scipy.optimize import curve_fit
def Cz_model(z, a, n, b,):
return(a*z**n + b)
all_data.keys()
data= all_data['capsol-calc\\0001-capsol\\']
z= data['z'][1:-1]
cz= data['cz'][1:-1]
popt, pcov= curve_fit(Cz_model, z, cz, p0=[cz[0]*z[0], -1, 0])
a=popt[0]
n=popt[1]
b=popt[2]
std_devs= np.sqrt(pcov.diagonal())
sigma_a = std_devs[0]
sigma_n = std_devs[1]
model_output= Cz_model(z, a, n, b)
rmse= np.sqrt(np.mean((cz - model_output)**2))
f"a= {a} ± {sigma_a}"
f"n= {n}± {sigma_n}"
model_output
"Root Mean Square Error"
rmse/np.mean(-cz)
```
| github_jupyter |
```
%load_ext autoreload
%autoreload 2
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
r = np.random.randn((1000))
S0 = 1
S = np.cumsum(r) + S0
T = 2
mu = 0.
sigma = 0.01
S0 = 20
dt = 0.01
N = round(T/dt)
t = np.linspace(0, T, N)
W = np.random.standard_normal(size = N)
W = np.cumsum(W)*np.sqrt(dt) ### standard brownian motion ###
X = (mu-0.5*sigma**2)*t + sigma*W
S = S0*np.exp(X) ### geometric brownian motion ###
plt.plot(t, S)
plt.show()
from ipywidgets import interact, interactive, fixed, interact_manual
import ipywidgets as widgets
from blackscholes import geometric_brownian_motion, blackScholes
from scipy.stats import norm
geometric_brownian_motion(mu=0., sigma=0.01, s0=1, dt=0.01);
t = 2.
dt = 0.01
N = int(round(t / dt))
np.linspace(0, t, N)
tt = np.linspace(0, t, N)
W = norm((N))
@interact(mu=(-0.02, 0.05, 0.01), sigma=(0.01, 0.1, 0.005), S0=(1,100,10), dt=(0.001, 0.1, 0.001))
def plot_gbm(mu, sigma, S0, dt):
s, t = geometric_brownian_motion(mu=mu, sigma=sigma, t=2, dt=dt, s0=S0)
pd.Series(t, s).plot()
plt.show()
df.ix[0.1:,:].gamma.plot()
tau = np.clip( np.linspace(1.0, .0, 101), 0.0000001, 100)
S = 1.
K = 1.
sigma = 1
df = pd.DataFrame.from_dict(blackScholes(tau, S, K, sigma))
df.index = tau
@interact(mu=(-0.02, 0.05, 0.01), sigma=(0.01, 0.1, 0.005), S0=(1,100,10), dt=(0.001, 0.1, 0.001))
def plot_gbm(mu, sigma, S0, dt):
s, t = geometric_brownian_motion(mu=mu, sigma=sigma, t=2, dt=dt, s0=S0)
pd.Series(t, s).plot()
plt.show()
```
## Q-learning
- Initialize $V(s)$ arbitrarily
- Repeat for each episode
- Initialize s
- Repeat (for each step of episode)
- - $\alpha \leftarrow$ action given by $\pi$ for $s$
- - Take action a, observe reward r, and next state s'
- - $V(s) \leftarrow V(s) + \alpha [r = \gamma V(s') - V(s)]$
- - $s \leftarrow s'$
- until $s$ is terminal
```
import td
import scipy as sp
α = 0.05
γ = 0.1
td_learning = td.TD(α, γ)
```
## Black Scholes
$${\displaystyle d_{1}={\frac {1}{\sigma {\sqrt {T-t}}}}\left[\ln \left({\frac {S_{t}}{K}}\right)+(r-q+{\frac {1}{2}}\sigma ^{2})(T-t)\right]}$$
$${\displaystyle C(S_{t},t)=e^{-r(T-t)}[FN(d_{1})-KN(d_{2})]\,}$$
$${\displaystyle d_{2}=d_{1}-\sigma {\sqrt {T-t}}={\frac {1}{\sigma {\sqrt {T-t}}}}\left[\ln \left({\frac {S_{t}}{K}}\right)+(r-q-{\frac {1}{2}}\sigma ^{2})(T-t)\right]}$$
```
d_1 = lambda σ, T, t, S, K: 1. / σ / np.sqrt(T - t) * (np.log(S / K) + 0.5 * (σ ** 2) * (T-t))
d_2 = lambda σ, T, t, S, K: 1. / σ / np.sqrt(T - t) * (np.log(S / K) - 0.5 * (σ ** 2) * (T-t))
call = lambda σ, T, t, S, K: S * sp.stats.norm.cdf( d_1(σ, T, t, S, K) ) - K * sp.stats.norm.cdf( d_2(σ, T, t, S, K) )
plt.plot(np.linspace(0.1, 4., 100), call(1., 1., .9, np.linspace(0.1, 4., 100), 1.))
d_1(1., 1., 0., 1.9, 1)
plt.plot(d_1(1., 1., 0., np.linspace(0.1, 2.9, 10), 1))
plt.plot(np.linspace(0.01, 1.9, 100), sp.stats.norm.cdf(d_1(1., 1., 0.2, np.linspace(0.01, 1.9, 100), 1)))
plt.plot(np.linspace(0.01, 1.9, 100), sp.stats.norm.cdf(d_1(1., 1., 0.6, np.linspace(0.01, 1.9, 100), 1)))
plt.plot(np.linspace(0.01, 1.9, 100), sp.stats.norm.cdf(d_1(1., 1., 0.9, np.linspace(0.01, 1.9, 100), 1)))
plt.plot(np.linspace(0.01, 1.9, 100), sp.stats.norm.cdf(d_1(1., 1., 0.99, np.linspace(0.01, 1.9, 100), 1)))
def iterate_series(n=1000, S0 = 1):
while True:
r = np.random.randn((n))
S = np.cumsum(r) + S0
yield S, r
def iterate_world(n=1000, S0=1, N=5):
for (s, r) in take(N, iterate_series(n=n, S0=S0)):
t, t_0 = 0, 0
for t in np.linspace(0, len(s)-1, 100):
r = s[int(t)] / s[int(t_0)]
yield r, s[int(t)]
t_0 = t
from cytoolz import take
import gym
import gym_bs
from test_cem_future import *
import pandas as pd
import numpy as np
# df.iloc[3] = (0.2, 1, 3)
df
rwd, df, agent = noisy_evaluation(np.array([0.1, 0, 0]))
rwd
df
agent;
env.observation_space
```
| github_jupyter |
<a href="https://colab.research.google.com/github/mjvakili/MLcourse/blob/master/day2/nn_qso_finder.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Let's start by importing the libraries that we need for this exercise.
```
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
import matplotlib
from sklearn.model_selection import train_test_split
#matplotlib settings
matplotlib.rcParams['xtick.major.size'] = 7
matplotlib.rcParams['xtick.labelsize'] = 'x-large'
matplotlib.rcParams['ytick.major.size'] = 7
matplotlib.rcParams['ytick.labelsize'] = 'x-large'
matplotlib.rcParams['xtick.top'] = False
matplotlib.rcParams['ytick.right'] = False
matplotlib.rcParams['ytick.direction'] = 'in'
matplotlib.rcParams['xtick.direction'] = 'in'
matplotlib.rcParams['font.size'] = 15
matplotlib.rcParams['figure.figsize'] = [7,7]
#We need the astroml library to fetch the photometric datasets of sdss qsos and stars
pip install astroml
from astroML.datasets import fetch_dr7_quasar
from astroML.datasets import fetch_sdss_sspp
quasars = fetch_dr7_quasar()
stars = fetch_sdss_sspp()
# Data procesing taken from
#https://www.astroml.org/book_figures/chapter9/fig_star_quasar_ROC.html by Jake Van der Plus
# stack colors into matrix X
Nqso = len(quasars)
Nstars = len(stars)
X = np.empty((Nqso + Nstars, 4), dtype=float)
X[:Nqso, 0] = quasars['mag_u'] - quasars['mag_g']
X[:Nqso, 1] = quasars['mag_g'] - quasars['mag_r']
X[:Nqso, 2] = quasars['mag_r'] - quasars['mag_i']
X[:Nqso, 3] = quasars['mag_i'] - quasars['mag_z']
X[Nqso:, 0] = stars['upsf'] - stars['gpsf']
X[Nqso:, 1] = stars['gpsf'] - stars['rpsf']
X[Nqso:, 2] = stars['rpsf'] - stars['ipsf']
X[Nqso:, 3] = stars['ipsf'] - stars['zpsf']
y = np.zeros(Nqso + Nstars, dtype=int)
y[:Nqso] = 1
X = X/np.max(X, axis=0)
# split into training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size = 0.9)
#Now let's build a simple Sequential model in which fully connected layers come after one another
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(), #this flattens input
tf.keras.layers.Dense(128, activation = "relu"),
tf.keras.layers.Dense(64, activation = "relu"),
tf.keras.layers.Dense(32, activation = "relu"),
tf.keras.layers.Dense(32, activation = "relu"),
tf.keras.layers.Dense(1, activation="sigmoid")
])
model.compile(optimizer='adam', loss='binary_crossentropy')
history = model.fit(X_train, y_train, validation_data = (X_test, y_test), batch_size = 32, epochs=20, verbose = 1)
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(loss))
plt.plot(epochs, loss, lw = 5, label='Training loss')
plt.plot(epochs, val_loss, lw = 5, label='validation loss')
plt.title('Loss')
plt.legend(loc=0)
plt.show()
prob = model.predict_proba(X_test) #model probabilities
from sklearn.metrics import confusion_matrix
from sklearn.metrics import roc_curve
fpr, tpr, thresholds = roc_curve(y_test, prob)
plt.loglog(fpr, tpr, lw = 4)
plt.xlabel('false positive rate')
plt.ylabel('true positive rate')
plt.xlim(0.0, 0.15)
plt.ylim(0.6, 1.01)
plt.show()
plt.plot(thresholds, tpr, lw = 4)
plt.plot(thresholds, fpr, lw = 4)
plt.xlim(0,1)
plt.yscale("log")
plt.show()
#plt.xlabel('false positive rate')
#plt.ylabel('true positive rate')
##plt.xlim(0.0, 0.15)
#plt.ylim(0.6, 1.01)
#Now let's look at the confusion matrix
y_pred = model.predict(X_test)
z_pred = np.zeros(y_pred.shape[0], dtype = int)
mask = np.where(y_pred>.5)[0]
z_pred[mask] = 1
confusion_matrix(y_test, z_pred.astype(int))
import os, signal
os.kill(os.getpid(), signal.SIGKILL)
```
#Exercise1:
Try to change the number of layers, batchsize, as well as the default learning rate, one at a time. See which one can make a more significant impact on the performance of the model.
#Exercise 2:
Write a simple function for visualizing the predicted decision boundaries in the feature space. Try to identify the regions of the parameter space which contribute significantly to the false positive rates.
#Exercise 3:
This dataset is a bit imbalanced in that the QSOs are outnumbered by the stars. Can you think of a wighting scheme to pass to the loss function, such that the detection rate of QSOs increases?
| github_jupyter |
```
import numpy as np
from scipy import pi
import matplotlib.pyplot as plt
import pickle as cPickle
#Sine wave
N = 128
def get_sine_wave():
x_sin = np.array([0.0 for i in range(N)])
# print(x_sin)
for i in range(N):
# print("h")
x_sin[i] = np.sin(2.0*pi*i/16.0)
plt.plot(x_sin)
plt.title('Sine wave')
plt.show()
y_sin = np.fft.fftshift(np.fft.fft(x_sin[:16], 16))
plt.plot(abs(y_sin))
plt.title('FFT sine wave')
plt.show()
return x_sin
def get_bpsk_carrier():
x = np.fromfile('gnuradio_dumps/bpsk_carrier', dtype = 'float32')
x_bpsk_carrier = x[9000:9000+N]
plt.plot(x_bpsk_carrier)
plt.title('BPSK carrier')
plt.show()
# y_bpsk_carrier = np.fft.fft(x_bpsk_carrier, N)
# plt.plot(abs(y_bpsk_carrier))
# plt.title('FFT BPSK carrier')
# plt.show()
def get_qpsk_carrier():
x = np.fromfile('gnuradio_dumps/qpsk_carrier', dtype = 'float32')
x_qpsk_carrier = x[12000:12000+N]
plt.plot(x_qpsk_carrier)
plt.title('QPSK carrier')
plt.show()
# y_qpsk_carrier = np.fft.fft(x_qpsk_carrier, N)
# plt.plot(abs(y_qpsk_carrier))
# plt.title('FFT QPSK carrier')
# plt.show()
def get_bpsk():
x = np.fromfile('gnuradio_dumps/bpsk', dtype = 'complex64')
x_bpsk = x[9000:9000+N]
plt.plot(x_bpsk.real)
plt.plot(x_bpsk.imag)
plt.title('BPSK')
plt.show()
# y_bpsk = np.fft.fft(x_bpsk, N)
# plt.plot(abs(y_bpsk))
# plt.title('FFT BPSK')
# plt.show()
def get_qpsk():
x = np.fromfile('gnuradio_dumps/qpsk', dtype = 'complex64')
x_qpsk = x[11000:11000+N]
plt.plot(x_qpsk.real)
plt.plot(x_qpsk.imag)
plt.title('QPSK')
plt.show()
# y_qpsk = np.fft.fft(x_bpsk, N)
# plt.plot(abs(y_bqsk))
# plt.title('FFT QPSK')
# plt.show()
def load_dataset(location="../../datasets/radioml.dat"):
f = open(location, "rb")
ds = cPickle.load(f, encoding = 'latin-1')
return ds
def get_from_dataset(dataset, key):
"""Returns complex version of dataset[key][500]"""
xr = dataset[key][500][0]
xi = dataset[key][500][1]
plt.plot(xr)
plt.plot(xi)
plt.title(key)
plt.show()
return xr
x_sin = get_sine_wave()
x_bpsk_carrier = get_bpsk_carrier()
x_qpsk_carrier = get_qpsk_carrier()
x_bpsk = get_bpsk()
x_qpsk = get_qpsk()
ds = load_dataset()
x_amssb = get_from_dataset(dataset=ds, key=('AM-SSB', 16))
x_amdsb = get_from_dataset(dataset=ds, key= ('AM-DSB', 18))
x_gfsk = get_from_dataset(dataset=ds, key=('GFSK', 18))
nfft = 16
cyclo_averaging = 8
offsets = [0,1,2,3,4,5,6,7]
def compute_cyclo_fft(data, nfft):
data_reshape = np.reshape(data, (-1, nfft))
y = np.fft.fftshift(np.fft.fft(data_reshape, axis=1), axes=1)
return y.T
def compute_cyclo_ifft(data, nfft):
return np.fft.fftshift(np.fft.fft(data))
def single_fft_cyclo(fft, offset):
left = np.roll(fft, -offset)
right = np.roll(fft, offset)
spec = right * np.conj(left)
return spec
def create_sc(spec, offset):
left = np.roll(spec, -offset)
right = np.roll(spec, offset)
denom = left * right
denom_norm = np.sqrt(denom)
return np.divide(spec, denom_norm)
def cyclo_stationary(data):
# fft
cyc_fft = compute_cyclo_fft(data, nfft)
# average
num_ffts = int(cyc_fft.shape[0])
cyc_fft = cyc_fft[:num_ffts]
cyc_fft = np.mean(np.reshape(cyc_fft, (nfft, cyclo_averaging)), axis=1)
print(cyc_fft)
plt.title('cyc_fft')
plt.plot(abs(cyc_fft))
plt.show()
specs = np.zeros((len(offsets)*16), dtype=np.complex64)
scs = np.zeros((len(offsets)*16), dtype=np.complex64)
cdp = {offset: 0 for offset in offsets}
for j, offset in enumerate(offsets):
spec = single_fft_cyclo(cyc_fft, offset)
print(spec)
plt.plot(abs(spec))
plt.title(offset)
plt.show()
sc = create_sc(spec, offset)
specs[j*16:j*16+16] = spec
scs[j*16:j*16+16] = sc
cdp[offset] = max(sc)
return specs, scs, cdp
specs, scs, cdp = cyclo_stationary(x_sin)
plt.plot(np.arange(128), scs.real)
plt.plot(np.arange(128), scs.imag)
plt.show()
```
| github_jupyter |
# Exercise: Find correspondences between old and modern english
The purpose of this execise is to use two vecsigrafos, one built on UMBC and Wordnet and another one produced by directly running Swivel against a corpus of Shakespeare's complete works, to try to find corelations between old and modern English, e.g. "thou" -> "you", "dost" -> "do", "raiment" -> "clothing". For example, you can try to pick a set of 100 words in "ye olde" English corpus and see how they correlate to UMBC over WordNet.

Next, we prepare the embeddings from the Shakespeare corpus and load a UMBC vecsigrafo, which will provide the two vector spaces to correlate.
## Download a small text corpus
First, we download the corpus into our environment. We will use the Shakespeare's complete works corpus, published as part of Project Gutenberg and pbublicly available.
```
import os
%ls
#!rm -r tutorial
!git clone https://github.com/HybridNLP2018/tutorial
```
Let us see if the corpus is where we think it is:
```
%cd tutorial/lit
%ls
```
Downloading Swivel
```
!wget http://expertsystemlab.com/hybridNLP18/swivel.zip
!unzip swivel.zip
!rm swivel/*
!rm swivel.zip
```
## Learn the Swivel embeddings over the Old Shakespeare corpus
### Calculating the co-occurrence matrix
```
corpus_path = '/content/tutorial/lit/shakespeare_complete_works.txt'
coocs_path = '/content/tutorial/lit/coocs'
shard_size = 512
freq=3
!python /content/tutorial/scripts/swivel/prep.py --input={corpus_path} --output_dir={coocs_path} --shard_size={shard_size} --min_count={freq}
%ls {coocs_path} | head -n 10
```
### Learning the embeddings from the matrix
```
vec_path = '/content/tutorial/lit/vec/'
!python /content/tutorial/scripts/swivel/swivel.py --input_base_path={coocs_path} \
--output_base_path={vec_path} \
--num_epochs=20 --dim=300 \
--submatrix_rows={shard_size} --submatrix_cols={shard_size}
```
Checking the context of the 'vec' directory. Should contain checkpoints of the model plus tsv files for column and row embeddings.
```
os.listdir(vec_path)
```
Converting tsv to bin:
```
!python /content/tutorial/scripts/swivel/text2bin.py --vocab={vec_path}vocab.txt --output={vec_path}vecs.bin \
{vec_path}row_embedding.tsv \
{vec_path}col_embedding.tsv
%ls {vec_path}
```
### Read stored binary embeddings and inspect them
```
import importlib.util
spec = importlib.util.spec_from_file_location("vecs", "/content/tutorial/scripts/swivel/vecs.py")
m = importlib.util.module_from_spec(spec)
spec.loader.exec_module(m)
shakespeare_vecs = m.Vecs(vec_path + 'vocab.txt', vec_path + 'vecs.bin')
```
##Basic method to print the k nearest neighbors for a given word
```
def k_neighbors(vec, word, k=10):
res = vec.neighbors(word)
if not res:
print('%s is not in the vocabulary, try e.g. %s' % (word, vecs.random_word_in_vocab()))
else:
for word, sim in res[:10]:
print('%0.4f: %s' % (sim, word))
k_neighbors(shakespeare_vecs, 'strife')
k_neighbors(shakespeare_vecs,'youth')
```
## Load vecsigrafo from UMBC over WordNet
```
%ls
!wget https://zenodo.org/record/1446214/files/vecsigrafo_umbc_tlgs_ls_f_6e_160d_row_embedding.tar.gz
%ls
!tar -xvzf vecsigrafo_umbc_tlgs_ls_f_6e_160d_row_embedding.tar.gz
!rm vecsigrafo_umbc_tlgs_ls_f_6e_160d_row_embedding.tar.gz
umbc_wn_vec_path = '/content/tutorial/lit/vecsi_tlgs_wnscd_ls_f_6e_160d/'
```
Extracting the vocabulary from the .tsv file:
```
with open(umbc_wn_vec_path + 'vocab.txt', 'w', encoding='utf_8') as f:
with open(umbc_wn_vec_path + 'row_embedding.tsv', 'r', encoding='utf_8') as vec_lines:
vocab = [line.split('\t')[0].strip() for line in vec_lines]
for word in vocab:
print(word, file=f)
```
Converting tsv to bin:
```
!python /content/tutorial/scripts/swivel/text2bin.py --vocab={umbc_wn_vec_path}vocab.txt --output={umbc_wn_vec_path}vecs.bin \
{umbc_wn_vec_path}row_embedding.tsv
%ls
umbc_wn_vecs = m.Vecs(umbc_wn_vec_path + 'vocab.txt', umbc_wn_vec_path + 'vecs.bin')
k_neighbors(umbc_wn_vecs, 'lem_California')
```
# Add your solution to the proposed exercise here
Follow the instructions given in the prvious lesson (*Vecsigrafos for curating and interlinking knowledge graphs*) to find correlation between terms in old Enlgish extracted from the Shakespeare corpus and terms in modern English extracted from UMBC. You will need to generate a dictionary relating pairs of lemmas between the two vocabularies and use to produce a pair of translation matrices to transform vectors from one vector space to the other. Then apply the k_neighbors method to identify the correlations.
# Conclusion
This notebook proposes the use of Shakespeare's complete works and UMBC to provide the student with embeddings that can be exploited for different operations between the two vector spaces. Particularly, we propose to identify terms and their correlations over such spaces.
# Acknowledgements
In memory of Dr. Jack Brandabur, whose passion for Shakespeare and Cervantes inspired this notebook.
| github_jupyter |
<a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_04_3_regression.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# T81-558: Applications of Deep Neural Networks
**Module 4: Training for Tabular Data**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module 4 Material
* Part 4.1: Encoding a Feature Vector for Keras Deep Learning [[Video]](https://www.youtube.com/watch?v=Vxz-gfs9nMQ&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_04_1_feature_encode.ipynb)
* Part 4.2: Keras Multiclass Classification for Deep Neural Networks with ROC and AUC [[Video]](https://www.youtube.com/watch?v=-f3bg9dLMks&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_04_2_multi_class.ipynb)
* **Part 4.3: Keras Regression for Deep Neural Networks with RMSE** [[Video]](https://www.youtube.com/watch?v=wNhBUC6X5-E&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_04_3_regression.ipynb)
* Part 4.4: Backpropagation, Nesterov Momentum, and ADAM Neural Network Training [[Video]](https://www.youtube.com/watch?v=VbDg8aBgpck&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_04_4_backprop.ipynb)
* Part 4.5: Neural Network RMSE and Log Loss Error Calculation from Scratch [[Video]](https://www.youtube.com/watch?v=wmQX1t2PHJc&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_04_5_rmse_logloss.ipynb)
# Google CoLab Instructions
The following code ensures that Google CoLab is running the correct version of TensorFlow.
```
try:
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
```
# Part 4.3: Keras Regression for Deep Neural Networks with RMSE
Regression results are evaluated differently than classification. Consider the following code that trains a neural network for regression on the data set **jh-simple-dataset.csv**.
```
import pandas as pd
from scipy.stats import zscore
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
# Read the data set
df = pd.read_csv(
"https://data.heatonresearch.com/data/t81-558/jh-simple-dataset.csv",
na_values=['NA','?'])
# Generate dummies for job
df = pd.concat([df,pd.get_dummies(df['job'],prefix="job")],axis=1)
df.drop('job', axis=1, inplace=True)
# Generate dummies for area
df = pd.concat([df,pd.get_dummies(df['area'],prefix="area")],axis=1)
df.drop('area', axis=1, inplace=True)
# Generate dummies for product
df = pd.concat([df,pd.get_dummies(df['product'],prefix="product")],axis=1)
df.drop('product', axis=1, inplace=True)
# Missing values for income
med = df['income'].median()
df['income'] = df['income'].fillna(med)
# Standardize ranges
df['income'] = zscore(df['income'])
df['aspect'] = zscore(df['aspect'])
df['save_rate'] = zscore(df['save_rate'])
df['subscriptions'] = zscore(df['subscriptions'])
# Convert to numpy - Classification
x_columns = df.columns.drop('age').drop('id')
x = df[x_columns].values
y = df['age'].values
# Create train/test
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=42)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation
from tensorflow.keras.callbacks import EarlyStopping
# Build the neural network
model = Sequential()
model.add(Dense(25, input_dim=x.shape[1], activation='relu')) # Hidden 1
model.add(Dense(10, activation='relu')) # Hidden 2
model.add(Dense(1)) # Output
model.compile(loss='mean_squared_error', optimizer='adam')
monitor = EarlyStopping(monitor='val_loss', min_delta=1e-3,
patience=5, verbose=1, mode='auto', restore_best_weights=True)
model.fit(x_train,y_train,validation_data=(x_test,y_test),callbacks=[monitor],verbose=2,epochs=1000)
```
### Mean Square Error
The mean square error is the sum of the squared differences between the prediction ($\hat{y}$) and the expected ($y$). MSE values are not of a particular unit. If an MSE value has decreased for a model, that is good. However, beyond this, there is not much more you can determine. Low MSE values are desired.
$ \mbox{MSE} = \frac{1}{n} \sum_{i=1}^n \left(\hat{y}_i - y_i\right)^2 $
```
from sklearn import metrics
# Predict
pred = model.predict(x_test)
# Measure MSE error.
score = metrics.mean_squared_error(pred,y_test)
print("Final score (MSE): {}".format(score))
```
### Root Mean Square Error
The root mean square (RMSE) is essentially the square root of the MSE. Because of this, the RMSE error is in the same units as the training data outcome. Low RMSE values are desired.
$ \mbox{RMSE} = \sqrt{\frac{1}{n} \sum_{i=1}^n \left(\hat{y}_i - y_i\right)^2} $
```
import numpy as np
# Measure RMSE error. RMSE is common for regression.
score = np.sqrt(metrics.mean_squared_error(pred,y_test))
print("Final score (RMSE): {}".format(score))
```
### Lift Chart
To generate a lift chart, perform the following activities:
* Sort the data by expected output. Plot the blue line above.
* For every point on the x-axis plot the predicted value for that same data point. This is the green line above.
* The x-axis is just 0 to 100% of the dataset. The expected always starts low and ends high.
* The y-axis is ranged according to the values predicted.
Reading a lift chart:
* The expected and predict lines should be close. Notice where one is above the ot other.
* The below chart is the most accurate on lower age.
```
# Regression chart.
def chart_regression(pred, y, sort=True):
t = pd.DataFrame({'pred': pred, 'y': y.flatten()})
if sort:
t.sort_values(by=['y'], inplace=True)
plt.plot(t['y'].tolist(), label='expected')
plt.plot(t['pred'].tolist(), label='prediction')
plt.ylabel('output')
plt.legend()
plt.show()
# Plot the chart
chart_regression(pred.flatten(),y_test)
```
| github_jupyter |
```
# flake8: noqa
##########################################################
# Relative Imports
##########################################################
import sys
from os.path import isfile
from os.path import join
def find_pkg(name: str, depth: int):
if depth <= 0:
ret = None
else:
d = [".."] * depth
path_parts = d + [name, "__init__.py"]
if isfile(join(*path_parts)):
ret = d
else:
ret = find_pkg(name, depth - 1)
return ret
def find_and_ins_syspath(name: str, depth: int):
path_parts = find_pkg(name, depth)
if path_parts is None:
raise RuntimeError("Could not find {}. Try increasing depth.".format(name))
path = join(*path_parts)
if path not in sys.path:
sys.path.insert(0, path)
try:
import caldera
except ImportError:
find_and_ins_syspath("caldera", 3)
##########################################################
# Main
##########################################################
import copy
import hydra
from examples.traversals.training import TrainingModule
from examples.traversals.data import DataGenerator, DataConfig
from examples.traversals.configuration import Config
from examples.traversals.configuration.data import Uniform, DiscreteUniform
from typing import TypeVar
from pytorch_lightning import Trainer
from examples.traversals.loggers import logger
from omegaconf import DictConfig, OmegaConf
from rich.panel import Panel
from rich import print
from rich.syntax import Syntax
C = TypeVar("C")
def prime_the_model(model: TrainingModule, config: Config):
logger.info("Priming the model with data")
config_copy: DataConfig = copy.deepcopy(config.data)
config_copy.train.num_graphs = 10
config_copy.eval.num_graphs = 0
data_copy = DataGenerator(config_copy, progress_bar=False)
for a, b in data_copy.train_loader():
model.model.forward(a, 10)
break
def print_title():
print(Panel("Training Example: [red]Traversal", title="[red]caldera"))
def print_model(model: TrainingModule):
print(Panel("Network", expand=False))
print(model)
def print_yaml(cfg: Config):
print(Panel("Configuration", expand=False))
print(Syntax(OmegaConf.to_yaml(cfg), "yaml"))
# def config_override(cfg: DictConfig):
# # defaults
# cfg.hyperparameters.lr = 1e-3
# cfg.hyperparameters.train_core_processing_steps = 10
# cfg.hyperparameters.eval_core_processing_steps = 10
#
# cfg.data.train.num_graphs = 5000
# cfg.data.train.num_nodes = DiscreteUniform(10, 100)
# cfg.data.train.density = Uniform(0.01, 0.03)
# cfg.data.train.path_length = DiscreteUniform(5, 10)
# cfg.data.train.composition_density = Uniform(0.01, 0.02)
# cfg.data.train.batch_size = 512
# cfg.data.train.shuffle = False
#
# cfg.data.eval.num_graphs = 500
# cfg.data.eval.num_nodes = DiscreteUniform(10, 100)
# cfg.data.eval.density = Uniform(0.01, 0.03)
# cfg.data.eval.path_length = DiscreteUniform(5, 10)
# cfg.data.eval.composition_density = Uniform(0.01, 0.02)
# cfg.data.eval.batch_size = "${data.eval.num_graphs}"
# cfg.data.eval.shuffle = False
# @hydra.main(config_path="conf", config_name="config")
# def main(hydra_cfg: DictConfig):
# print_title()
# logger.setLevel(hydra_cfg.log_level)
# if hydra_cfg.log_level.upper() == 'DEBUG':
# verbose = True
# else:
# verbose = False
# # really unclear why hydra has so many unclear validation issues with structure configs using ConfigStore
# # this correctly assigns the correct structured config
# # and updates from the passed hydra config
# # annoying... but this resolves all these issues
# cfg = OmegaConf.structured(Config())
# cfg.update(hydra_cfg)
# # debug
# if verbose:
# print_yaml(cfg)
# from pytorch_lightning.loggers import WandbLogger
# wandb_logger = WandbLogger(project='pytorchlightning')
# # explicitly convert the DictConfig back to Config object
# # has the added benefit of performing validation upfront
# # before any expensive training or logging initiates
# config = Config.from_dict_config(cfg)
# # initialize the training module
# training_module = TrainingModule(config)
# logger.info("Priming the model with data")
# prime_the_model(training_module, config)
# logger.debug(Panel("Model", expand=False))
# if verbose:
# print_model(training_module)
# logger.info("Generating data...")
# data = DataGenerator(config.data)
# data.init()
# logger.info("Beginning training...")
# trainer = Trainer(gpus=config.gpus, logger=wandb_logger)
# trainer.fit(
# training_module,
# train_dataloader=data.train_loader(),
# val_dataloaders=data.eval_loader(),
# )
# if __name__ == "__main__":
# main()
from examples.traversals.configuration import get_config
config = get_config( as_config_class=True)
data = DataGenerator(config.data)
data.init()
training_module = TrainingModule(config)
logger.info("Priming the model with data")
prime_the_model(training_module, config)
dir(data)
from torch import optim
from tqdm.auto import tqdm
import torch
from caldera.data import GraphTuple
def mse_tuple(criterion, device, a, b):
loss = torch.tensor(0.0, dtype=torch.float32, device=device)
assert len(a) == len(b)
for i, (_a, _b) in enumerate(zip(a, b)):
assert _a.shape == _b.shape
l = criterion(_a, _b)
loss += l
return loss
def train(network, loader, cuda: bool = False):
device = 'cpu'
if cuda and torch.cuda.is_available():
device = 'cuda:' + str(torch.cuda.current_device())
network.eval()
network.to(device)
input_batch, target_batch = loader.first()
input_batch = input_batch.detach()
input_batch.to(device)
network(input_batch, 1)
optimizer = optim.AdamW(network.parameters(), lr=1e-2)
loss_func = torch.nn.MSELoss()
losses = []
for epoch in range(20):
print(epoch)
running_loss = 0.
network.train()
for input_batch, target_batch in loader:
optimizer.zero_grad()
out_batch = network(input_batch, 5)[-1]
out_tuple = GraphTuple(out_batch.e, out_batch.x, out_batch.g)
target_tuple = GraphTuple(target_batch.e, target_batch.x, target_batch.g)
loss = mse_tuple(loss_func, device, out_tuple, target_tuple)
loss.backward()
running_loss = running_loss + loss.item()
optimizer.step()
print(running_loss)
losses.append(running_loss)
return losses
# loader = DataLoaders.sigmoid_circuit(1000, 10)
train(training_module.model, data.train_loader())
inp, targ = data.eval_loader().first()
from caldera.transforms.networkx import NetworkxAttachNumpyBool
g = targ.to_networkx_list()[0]
to_bool = NetworkxAttachNumpyBool('node', 'features', 'x')
graphs = to_bool(targ.to_networkx_list())
graphs[0].nodes(data=True)
from matplotlib import cm
import matplotlib.pyplot as plt
import numpy as np
import networkx as nx
%matplotlib inline
def edge_colors(g, key, cmap):
edgecolors = list()
edgelist = list(g.edges)
edgefeat = list()
for e in edgelist:
edata = g.edges[e]
edgefeat.append(edata[key][0].item())
edgefeat = np.array(edgefeat)
edgecolors = cmap(edgefeat)
return edgecolors
nx.draw_networkx_edges(g, pos=pos, edge_color=edgecolors, arrows=False)
def node_colors(g, key, cmap):
nodecolors = list()
nodelist = list(g.nodes)
nodefeat = list()
for n in nodelist:
ndata = g.nodes[n]
nodefeat.append(ndata[key][0].item())
nodefeat = np.array(nodefeat)
nodecolors = cmap(nodefeat)
return nodecolors
nx.draw_networkx_nodes(g, pos=pos, node_size=10, node_color=nodecolors)
def plot_graph(g, ax, cmap, key='features', seed=1):
pos = nx.layout.spring_layout(g, seed=seed)
nx.draw_networkx_edges(g, ax=ax, pos=pos, edge_color=edge_colors(g, key, cmap), arrows=False);
nx.draw_networkx_nodes(g, ax=ax, pos=pos, node_size=10, node_color=node_colors(g, key, cmap))
def comparison_plot(out_g, expected_g):
fig, axes = plt.subplots(1, 2, figsize=(5, 2.5))
axes[0].axis('off')
axes[1].axis('off')
axes[0].set_title("out")
plot_graph(out_g, axes[0], cm.plasma)
axes[1].set_title("expected")
plot_graph(expected_g, axes[1], cm.plasma)
return fig, axes
def validate_compare_plot(trainer, plmodel):
eval_loader = trainer.val_dataloaders[0]
for x, y in eval_loader:
break
plmodel.eval()
y_hat = plmodel.model.forward(x, 10)[-1]
y_graphs = y.to_networkx_list()
y_hat_graphs = y_hat.to_networkx_list()
idx = 0
yg = y_graphs[idx]
yhg = y_hat_graphs[idx]
return comparison_plot(yhg, yg)
fig, axes = validate_compare_plot(trainer, training_module)
from pytorch_lightning.loggers import WandbLogger
wandb_logger = WandbLogger(project='pytorchlightning')
wandb_logger.experiment
wandb.Image?
import wandb
import io
from PIL import Image
import matplotlib.pyplot as plt
def fig_to_pil(fig):
buf = io.BytesIO()
fig.savefig(buf, format='png')
buf.seek(0)
im = Image.open(buf)
# buf.close()
return im
wandb_logger.experiment.log({'s': [wandb.Image(fig_to_pil(fig))]} )
wandb_logger.experiment.log
import io
from PIL import Image
import matplotlib.pyplot as plt
buf = io.BytesIO()
fig.savefig(buf, format='png')
buf.seek(0)
im = Image.open(buf)
im.show()
buf.close()
str(buf)
x.to_networkx_list()[0].nodes(data=True)
def comparison_plot(out_g, expected_g):
fig, axes = plt.subplots(1, 2, figsize=(5, 2.5))
axes[0].axis('off')
axes[1].axis('off')
axes[0].set_title("out")
plot_graph(out_g, axes[0], cm.plasma)
axes[1].set_title("expected")
plot_graph(expected_g, axes[1], cm.plasma)
x, y = data.eval_loader().first()
y_hat = training_module.model.forward(x, 10)[-1]
y_graphs = y.to_networkx_list()
y_hat_graphs = y_hat.to_networkx_list()
idx = 0
yg = y_graphs[idx]
yhg = y_hat_graphs[idx]
comparison_plot(yhg, yg)
g = random_graph((100, 150), d=(0.01, 0.03), e=None)
annotate_shortest_path(g)
# nx.draw(g)
pos = nx.layout.spring_layout(g)
nodelist = list(g.nodes)
node_color = []
for n in nodelist:
node_color.append(g.nodes[n]['target'][0])
edge_list = []
edge_color = []
for n1, n2, edata in g.edges(data=True):
edge_list.append((n1, n2))
edge_color.append(edata['target'][0])
print(node_color)
nx.draw_networkx_edges(g, pos=pos, width=0.5, edge_color=edge_color)
nx.draw_networkx_nodes(g, pos=pos, node_color=node_color, node_size=10)
NetworkxAttachNumpyBool?
g.nodes(data=True)
from caldera.transforms.networkx import NetworkxApplyToFeature
NetworkxApplyToFeature('features', edge_func= lambda x: list(x))(g)
import time
from rich.progress import Progress as RichProgress
from contextlib import contextmanager
from dataclasses import dataclass
@dataclass
class TaskEvent:
task_id: int
name: str
class TaskProgress(object):
DEFAULT_REFRESH_PER_SECOND = 4.
def __init__(self,
progress = None,
task_id: int = None,
refresh_rate_per_second: int = DEFAULT_REFRESH_PER_SECOND,
parent = None):
self.task_id = task_id
self.children = []
self.parent = parent
self.progress = progress or RichProgress()
self.last_updated = time.time()
self.refresh_rate_per_second = refresh_rate_per_second
def self_task(self, *args, **kwargs):
task_id = self.progress.add_task(*args, **kwargs)
self.task_id = task_id
def add_task(self, *args, **kwargs):
task_id = self.progress.add_task(*args, **kwargs)
new_task = self.__class__(self.progress, task_id, self.refresh_rate_per_second, parent=self)
self.children.append(new_task)
return new_task
@property
def _task(self):
return self.progress.tasks[self.task_id]
def listen(self, event: TaskEvent):
if event.name == 'refresh':
completed = sum(t._task.completed for t in self.children)
total = sum(t._task.total for t in self.children)
self.update(completed=completed/total, total=1., refresh=True)
elif event.name == 'finished':
self.finish()
def emit_up(self, event_name):
if self.parent:
self.parent.listen(TaskEvent(task_id=self.task_id, name=event_name))
def emit_down(self, event_name: TaskEvent):
for child in self.children:
print("sending to child")
child.listen(TaskEvent(task_id=self.task_id, name=event_name))
def update(self, *args, **kwargs):
now = time.time()
if 'refresh' not in kwargs:
if now - self.last_updated > 1. / self.refresh_rate_per_second:
kwargs['refresh'] = True
else:
kwargs['refresh'] = False
if kwargs['refresh']:
self.emit_up('refresh')
self.last_updated = now
self.progress.update(self.task_id, *args, **kwargs)
def is_complete(self):
return self.completed >= self.task.total
def finish(self):
self.progress.update(self.task_id, completed=self._task.total, refresh=True)
self.emit_down('finished')
def __enter__(self):
return self
def __exit__(self, exc_type, exc_val, exc_tb):
self.progress.__exit__(exc_type, exc_val, exc_tb)
self.finish()
with TaskProgress() as progress:
progress.self_task('main', total=10)
bar1 = progress.add_task('bar1', total=10)
bar2 = progress.add_task('bar2', total=10)
for _ in range(10):
bar1.update(advance=1)
time.sleep(0.1)
for _ in range(10):
bar2.update(advance=1)
time.sleep(0.1)
bar.progress.tasks[0].completed
import torch
target = torch.ones([1, 64], dtype=torch.float32) # 64 classes, batch size = 10
output = torch.full([1, 64], 1.5) # A prediction (logit)
print(target)
print(output)
# pos_weight = torch.ones([64]) # All weights are equal to 1
criterion = torch.nn.BCEWithLogitsLoss()
criterion(output, target) # -log(sigmoid(1.5))
from caldera.data import GraphBatch
batch = GraphBatch.random_batch(2, 5, 4, 3)
graphs = batch.to_networkx_list()
import networkx as nx
nx.draw(graphs[0])
expected = torch.randn(batch.x.shape)
x = batch.x
x = torch.nn.Softmax()(x)
print(x.sum(axis=1))
x, expected
x = torch.nn.BCELoss()(x, expected)
x
import torch
x = torch.randn(10, 10)
torch.stack([x, x]).shape
```
| github_jupyter |
# About this Notebook
In this notebook, we provide the tensor factorization implementation using an iterative Alternating Least Square (ALS), which is a good starting point for understanding tensor factorization.
```
import numpy as np
from numpy.linalg import inv as inv
```
# Part 1: Matrix Computation Concepts
## 1) Kronecker product
- **Definition**:
Given two matrices $A\in\mathbb{R}^{m_1\times n_1}$ and $B\in\mathbb{R}^{m_2\times n_2}$, then, the **Kronecker product** between these two matrices is defined as
$$A\otimes B=\left[ \begin{array}{cccc} a_{11}B & a_{12}B & \cdots & a_{1m_2}B \\ a_{21}B & a_{22}B & \cdots & a_{2m_2}B \\ \vdots & \vdots & \ddots & \vdots \\ a_{m_11}B & a_{m_12}B & \cdots & a_{m_1m_2}B \\ \end{array} \right]$$
where the symbol $\otimes$ denotes Kronecker product, and the size of resulted $A\otimes B$ is $(m_1m_2)\times (n_1n_2)$ (i.e., $m_1\times m_2$ columns and $n_1\times n_2$ rows).
- **Example**:
If $A=\left[ \begin{array}{cc} 1 & 2 \\ 3 & 4 \\ \end{array} \right]$ and $B=\left[ \begin{array}{ccc} 5 & 6 & 7\\ 8 & 9 & 10 \\ \end{array} \right]$, then, we have
$$A\otimes B=\left[ \begin{array}{cc} 1\times \left[ \begin{array}{ccc} 5 & 6 & 7\\ 8 & 9 & 10\\ \end{array} \right] & 2\times \left[ \begin{array}{ccc} 5 & 6 & 7\\ 8 & 9 & 10\\ \end{array} \right] \\ 3\times \left[ \begin{array}{ccc} 5 & 6 & 7\\ 8 & 9 & 10\\ \end{array} \right] & 4\times \left[ \begin{array}{ccc} 5 & 6 & 7\\ 8 & 9 & 10\\ \end{array} \right] \\ \end{array} \right]$$
$$=\left[ \begin{array}{cccccc} 5 & 6 & 7 & 10 & 12 & 14 \\ 8 & 9 & 10 & 16 & 18 & 20 \\ 15 & 18 & 21 & 20 & 24 & 28 \\ 24 & 27 & 30 & 32 & 36 & 40 \\ \end{array} \right]\in\mathbb{R}^{4\times 6}.$$
## 2) Khatri-Rao product (`kr_prod`)
- **Definition**:
Given two matrices $A=\left( \boldsymbol{a}_1,\boldsymbol{a}_2,...,\boldsymbol{a}_r \right)\in\mathbb{R}^{m\times r}$ and $B=\left( \boldsymbol{b}_1,\boldsymbol{b}_2,...,\boldsymbol{b}_r \right)\in\mathbb{R}^{n\times r}$ with same number of columns, then, the **Khatri-Rao product** (or **column-wise Kronecker product**) between $A$ and $B$ is given as follows,
$$A\odot B=\left( \boldsymbol{a}_1\otimes \boldsymbol{b}_1,\boldsymbol{a}_2\otimes \boldsymbol{b}_2,...,\boldsymbol{a}_r\otimes \boldsymbol{b}_r \right)\in\mathbb{R}^{(mn)\times r},$$
where the symbol $\odot$ denotes Khatri-Rao product, and $\otimes$ denotes Kronecker product.
- **Example**:
If $A=\left[ \begin{array}{cc} 1 & 2 \\ 3 & 4 \\ \end{array} \right]=\left( \boldsymbol{a}_1,\boldsymbol{a}_2 \right) $ and $B=\left[ \begin{array}{cc} 5 & 6 \\ 7 & 8 \\ 9 & 10 \\ \end{array} \right]=\left( \boldsymbol{b}_1,\boldsymbol{b}_2 \right) $, then, we have
$$A\odot B=\left( \boldsymbol{a}_1\otimes \boldsymbol{b}_1,\boldsymbol{a}_2\otimes \boldsymbol{b}_2 \right) $$
$$=\left[ \begin{array}{cc} \left[ \begin{array}{c} 1 \\ 3 \\ \end{array} \right]\otimes \left[ \begin{array}{c} 5 \\ 7 \\ 9 \\ \end{array} \right] & \left[ \begin{array}{c} 2 \\ 4 \\ \end{array} \right]\otimes \left[ \begin{array}{c} 6 \\ 8 \\ 10 \\ \end{array} \right] \\ \end{array} \right]$$
$$=\left[ \begin{array}{cc} 5 & 12 \\ 7 & 16 \\ 9 & 20 \\ 15 & 24 \\ 21 & 32 \\ 27 & 40 \\ \end{array} \right]\in\mathbb{R}^{6\times 2}.$$
```
def kr_prod(a, b):
return np.einsum('ir, jr -> ijr', a, b).reshape(a.shape[0] * b.shape[0], -1)
A = np.array([[1, 2], [3, 4]])
B = np.array([[5, 6], [7, 8], [9, 10]])
print(kr_prod(A, B))
```
## 3) CP decomposition
### CP Combination (`cp_combination`)
- **Definition**:
The CP decomposition factorizes a tensor into a sum of outer products of vectors. For example, for a third-order tensor $\mathcal{Y}\in\mathbb{R}^{m\times n\times f}$, the CP decomposition can be written as
$$\hat{\mathcal{Y}}=\sum_{s=1}^{r}\boldsymbol{u}_{s}\circ\boldsymbol{v}_{s}\circ\boldsymbol{x}_{s},$$
or element-wise,
$$\hat{y}_{ijt}=\sum_{s=1}^{r}u_{is}v_{js}x_{ts},\forall (i,j,t),$$
where vectors $\boldsymbol{u}_{s}\in\mathbb{R}^{m},\boldsymbol{v}_{s}\in\mathbb{R}^{n},\boldsymbol{x}_{s}\in\mathbb{R}^{f}$ are columns of factor matrices $U\in\mathbb{R}^{m\times r},V\in\mathbb{R}^{n\times r},X\in\mathbb{R}^{f\times r}$, respectively. The symbol $\circ$ denotes vector outer product.
- **Example**:
Given matrices $U=\left[ \begin{array}{cc} 1 & 2 \\ 3 & 4 \\ \end{array} \right]\in\mathbb{R}^{2\times 2}$, $V=\left[ \begin{array}{cc} 1 & 2 \\ 3 & 4 \\ 5 & 6 \\ \end{array} \right]\in\mathbb{R}^{3\times 2}$ and $X=\left[ \begin{array}{cc} 1 & 5 \\ 2 & 6 \\ 3 & 7 \\ 4 & 8 \\ \end{array} \right]\in\mathbb{R}^{4\times 2}$, then if $\hat{\mathcal{Y}}=\sum_{s=1}^{r}\boldsymbol{u}_{s}\circ\boldsymbol{v}_{s}\circ\boldsymbol{x}_{s}$, then, we have
$$\hat{Y}_1=\hat{\mathcal{Y}}(:,:,1)=\left[ \begin{array}{ccc} 31 & 42 & 65 \\ 63 & 86 & 135 \\ \end{array} \right],$$
$$\hat{Y}_2=\hat{\mathcal{Y}}(:,:,2)=\left[ \begin{array}{ccc} 38 & 52 & 82 \\ 78 & 108 & 174 \\ \end{array} \right],$$
$$\hat{Y}_3=\hat{\mathcal{Y}}(:,:,3)=\left[ \begin{array}{ccc} 45 & 62 & 99 \\ 93 & 130 & 213 \\ \end{array} \right],$$
$$\hat{Y}_4=\hat{\mathcal{Y}}(:,:,4)=\left[ \begin{array}{ccc} 52 & 72 & 116 \\ 108 & 152 & 252 \\ \end{array} \right].$$
```
def cp_combine(U, V, X):
return np.einsum('is, js, ts -> ijt', U, V, X)
U = np.array([[1, 2], [3, 4]])
V = np.array([[1, 3], [2, 4], [5, 6]])
X = np.array([[1, 5], [2, 6], [3, 7], [4, 8]])
print(cp_combine(U, V, X))
print()
print('tensor size:')
print(cp_combine(U, V, X).shape)
```
## 4) Tensor Unfolding (`ten2mat`)
Using numpy reshape to perform 3rd rank tensor unfold operation. [[**link**](https://stackoverflow.com/questions/49970141/using-numpy-reshape-to-perform-3rd-rank-tensor-unfold-operation)]
```
def ten2mat(tensor, mode):
return np.reshape(np.moveaxis(tensor, mode, 0), (tensor.shape[mode], -1), order = 'F')
X = np.array([[[1, 2, 3, 4], [3, 4, 5, 6]],
[[5, 6, 7, 8], [7, 8, 9, 10]],
[[9, 10, 11, 12], [11, 12, 13, 14]]])
print('tensor size:')
print(X.shape)
print('original tensor:')
print(X)
print()
print('(1) mode-1 tensor unfolding:')
print(ten2mat(X, 0))
print()
print('(2) mode-2 tensor unfolding:')
print(ten2mat(X, 1))
print()
print('(3) mode-3 tensor unfolding:')
print(ten2mat(X, 2))
```
# Part 2: Tensor CP Factorization using ALS (TF-ALS)
Regarding CP factorization as a machine learning problem, we could perform a learning task by minimizing the loss function over factor matrices, that is,
$$\min _{U, V, X} \sum_{(i, j, t) \in \Omega}\left(y_{i j t}-\sum_{r=1}^{R}u_{ir}v_{jr}x_{tr}\right)^{2}.$$
Within this optimization problem, multiplication among three factor matrices (acted as parameters) makes this problem difficult. Alternatively, we apply the ALS algorithm for CP factorization.
In particular, the optimization problem for each row $\boldsymbol{u}_{i}\in\mathbb{R}^{R},\forall i\in\left\{1,2,...,M\right\}$ of factor matrix $U\in\mathbb{R}^{M\times R}$ is given by
$$\min _{\boldsymbol{u}_{i}} \sum_{j,t:(i, j, t) \in \Omega}\left[y_{i j t}-\boldsymbol{u}_{i}^\top\left(\boldsymbol{x}_{t}\odot\boldsymbol{v}_{j}\right)\right]\left[y_{i j t}-\boldsymbol{u}_{i}^\top\left(\boldsymbol{x}_{t}\odot\boldsymbol{v}_{j}\right)\right]^\top.$$
The least square for this optimization is
$$u_{i} \Leftarrow\left(\sum_{j, t, i, j, t ) \in \Omega} \left(x_{t} \odot v_{j}\right)\left(x_{t} \odot v_{j}\right)^{\top}\right)^{-1}\left(\sum_{j, t :(i, j, t) \in \Omega} y_{i j t} \left(x_{t} \odot v_{j}\right)\right), \forall i \in\{1,2, \ldots, M\}.$$
The alternating least squares for $V\in\mathbb{R}^{N\times R}$ and $X\in\mathbb{R}^{T\times R}$ are
$$\boldsymbol{v}_{j}\Leftarrow\left(\sum_{i,t:(i,j,t)\in\Omega}\left(\boldsymbol{x}_{t}\odot\boldsymbol{u}_{i}\right)\left(\boldsymbol{x}_{t}\odot\boldsymbol{u}_{i}\right)^\top\right)^{-1}\left(\sum_{i,t:(i,j,t)\in\Omega}y_{ijt}\left(\boldsymbol{x}_{t}\odot\boldsymbol{u}_{i}\right)\right),\forall j\in\left\{1,2,...,N\right\},$$
$$\boldsymbol{x}_{t}\Leftarrow\left(\sum_{i,j:(i,j,t)\in\Omega}\left(\boldsymbol{v}_{j}\odot\boldsymbol{u}_{i}\right)\left(\boldsymbol{v}_{j}\odot\boldsymbol{u}_{i}\right)^\top\right)^{-1}\left(\sum_{i,j:(i,j,t)\in\Omega}y_{ijt}\left(\boldsymbol{v}_{j}\odot\boldsymbol{u}_{i}\right)\right),\forall t\in\left\{1,2,...,T\right\}.$$
```
def CP_ALS(sparse_tensor, rank, maxiter):
dim1, dim2, dim3 = sparse_tensor.shape
dim = np.array([dim1, dim2, dim3])
U = 0.1 * np.random.rand(dim1, rank)
V = 0.1 * np.random.rand(dim2, rank)
X = 0.1 * np.random.rand(dim3, rank)
pos = np.where(sparse_tensor != 0)
binary_tensor = np.zeros((dim1, dim2, dim3))
binary_tensor[pos] = 1
tensor_hat = np.zeros((dim1, dim2, dim3))
for iters in range(maxiter):
for order in range(dim.shape[0]):
if order == 0:
var1 = kr_prod(X, V).T
elif order == 1:
var1 = kr_prod(X, U).T
else:
var1 = kr_prod(V, U).T
var2 = kr_prod(var1, var1)
var3 = np.matmul(var2, ten2mat(binary_tensor, order).T).reshape([rank, rank, dim[order]])
var4 = np.matmul(var1, ten2mat(sparse_tensor, order).T)
for i in range(dim[order]):
var_Lambda = var3[ :, :, i]
inv_var_Lambda = inv((var_Lambda + var_Lambda.T)/2 + 10e-12 * np.eye(rank))
vec = np.matmul(inv_var_Lambda, var4[:, i])
if order == 0:
U[i, :] = vec.copy()
elif order == 1:
V[i, :] = vec.copy()
else:
X[i, :] = vec.copy()
tensor_hat = cp_combine(U, V, X)
mape = np.sum(np.abs(sparse_tensor[pos] - tensor_hat[pos])/sparse_tensor[pos])/sparse_tensor[pos].shape[0]
rmse = np.sqrt(np.sum((sparse_tensor[pos] - tensor_hat[pos]) ** 2)/sparse_tensor[pos].shape[0])
if (iters + 1) % 100 == 0:
print('Iter: {}'.format(iters + 1))
print('Training MAPE: {:.6}'.format(mape))
print('Training RMSE: {:.6}'.format(rmse))
print()
return tensor_hat, U, V, X
```
# Part 3: Data Organization
## 1) Matrix Structure
We consider a dataset of $m$ discrete time series $\boldsymbol{y}_{i}\in\mathbb{R}^{f},i\in\left\{1,2,...,m\right\}$. The time series may have missing elements. We express spatio-temporal dataset as a matrix $Y\in\mathbb{R}^{m\times f}$ with $m$ rows (e.g., locations) and $f$ columns (e.g., discrete time intervals),
$$Y=\left[ \begin{array}{cccc} y_{11} & y_{12} & \cdots & y_{1f} \\ y_{21} & y_{22} & \cdots & y_{2f} \\ \vdots & \vdots & \ddots & \vdots \\ y_{m1} & y_{m2} & \cdots & y_{mf} \\ \end{array} \right]\in\mathbb{R}^{m\times f}.$$
## 2) Tensor Structure
We consider a dataset of $m$ discrete time series $\boldsymbol{y}_{i}\in\mathbb{R}^{nf},i\in\left\{1,2,...,m\right\}$. The time series may have missing elements. We partition each time series into intervals of predifined length $f$. We express each partitioned time series as a matrix $Y_{i}$ with $n$ rows (e.g., days) and $f$ columns (e.g., discrete time intervals per day),
$$Y_{i}=\left[ \begin{array}{cccc} y_{11} & y_{12} & \cdots & y_{1f} \\ y_{21} & y_{22} & \cdots & y_{2f} \\ \vdots & \vdots & \ddots & \vdots \\ y_{n1} & y_{n2} & \cdots & y_{nf} \\ \end{array} \right]\in\mathbb{R}^{n\times f},i=1,2,...,m,$$
therefore, the resulting structure is a tensor $\mathcal{Y}\in\mathbb{R}^{m\times n\times f}$.
**How to transform a data set into something we can use for time series imputation?**
# Part 4: Experiments on Guangzhou Data Set
```
import scipy.io
tensor = scipy.io.loadmat('../datasets/Guangzhou-data-set/tensor.mat')
dense_tensor = tensor['tensor']
random_matrix = scipy.io.loadmat('../datasets/Guangzhou-data-set/random_matrix.mat')
random_matrix = random_matrix['random_matrix']
random_tensor = scipy.io.loadmat('../datasets/Guangzhou-data-set/random_tensor.mat')
random_tensor = random_tensor['random_tensor']
missing_rate = 0.2
# =============================================================================
### Random missing (RM) scenario:
binary_tensor = np.round(random_tensor + 0.5 - missing_rate)
# =============================================================================
# =============================================================================
### Non-random missing (NM) scenario:
# binary_tensor = np.zeros(dense_tensor.shape)
# for i1 in range(dense_tensor.shape[0]):
# for i2 in range(dense_tensor.shape[1]):
# binary_tensor[i1, i2, :] = np.round(random_matrix[i1, i2] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
```
**Question**: Given only the partially observed data $\mathcal{Y}\in\mathbb{R}^{m\times n\times f}$, how can we impute the unknown missing values?
The main influential factors for such imputation model are:
- `rank`.
- `maxiter`.
```
import time
start = time.time()
rank = 80
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
```
**Experiment results** of missing data imputation using TF-ALS:
| scenario |`rank`| `maxiter`| mape | rmse |
|:----------|-----:|---------:|-----------:|----------:|
|**20%, RM**| 80 | 1000 | **0.0833** | **3.5928**|
|**40%, RM**| 80 | 1000 | **0.0837** | **3.6190**|
|**20%, NM**| 10 | 1000 | **0.1027** | **4.2960**|
|**40%, NM**| 10 | 1000 | **0.1028** | **4.3274**|
# Part 5: Experiments on Birmingham Data Set
```
import scipy.io
tensor = scipy.io.loadmat('../datasets/Birmingham-data-set/tensor.mat')
dense_tensor = tensor['tensor']
random_matrix = scipy.io.loadmat('../datasets/Birmingham-data-set/random_matrix.mat')
random_matrix = random_matrix['random_matrix']
random_tensor = scipy.io.loadmat('../datasets/Birmingham-data-set/random_tensor.mat')
random_tensor = random_tensor['random_tensor']
missing_rate = 0.3
# =============================================================================
### Random missing (RM) scenario:
binary_tensor = np.round(random_tensor + 0.5 - missing_rate)
# =============================================================================
# =============================================================================
### Non-random missing (NM) scenario:
# binary_tensor = np.zeros(dense_tensor.shape)
# for i1 in range(dense_tensor.shape[0]):
# for i2 in range(dense_tensor.shape[1]):
# binary_tensor[i1, i2, :] = np.round(random_matrix[i1,i2] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 30
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
```
**Experiment results** of missing data imputation using TF-ALS:
| scenario |`rank`| `maxiter`| mape | rmse |
|:----------|-----:|---------:|-----------:|-----------:|
|**10%, RM**| 30 | 1000 | **0.0615** | **18.5005**|
|**30%, RM**| 30 | 1000 | **0.0583** | **18.9148**|
|**10%, NM**| 10 | 1000 | **0.1447** | **41.6710**|
|**30%, NM**| 10 | 1000 | **0.1765** | **63.8465**|
# Part 6: Experiments on Hangzhou Data Set
```
import scipy.io
tensor = scipy.io.loadmat('../datasets/Hangzhou-data-set/tensor.mat')
dense_tensor = tensor['tensor']
random_matrix = scipy.io.loadmat('../datasets/Hangzhou-data-set/random_matrix.mat')
random_matrix = random_matrix['random_matrix']
random_tensor = scipy.io.loadmat('../datasets/Hangzhou-data-set/random_tensor.mat')
random_tensor = random_tensor['random_tensor']
missing_rate = 0.4
# =============================================================================
### Random missing (RM) scenario:
binary_tensor = np.round(random_tensor + 0.5 - missing_rate)
# =============================================================================
# =============================================================================
### Non-random missing (NM) scenario:
# binary_tensor = np.zeros(dense_tensor.shape)
# for i1 in range(dense_tensor.shape[0]):
# for i2 in range(dense_tensor.shape[1]):
# binary_tensor[i1, i2, :] = np.round(random_matrix[i1, i2] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 50
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
```
**Experiment results** of missing data imputation using TF-ALS:
| scenario |`rank`| `maxiter`| mape | rmse |
|:----------|-----:|---------:|-----------:|----------:|
|**20%, RM**| 50 | 1000 | **0.1991** |**111.303**|
|**40%, RM**| 50 | 1000 | **0.2098** |**100.315**|
|**20%, NM**| 5 | 1000 | **0.2837** |**42.6136**|
|**40%, NM**| 5 | 1000 | **0.2811** |**38.4201**|
# Part 7: Experiments on New York Data Set
```
import scipy.io
tensor = scipy.io.loadmat('../datasets/NYC-data-set/tensor.mat')
dense_tensor = tensor['tensor']
rm_tensor = scipy.io.loadmat('../datasets/NYC-data-set/rm_tensor.mat')
rm_tensor = rm_tensor['rm_tensor']
nm_tensor = scipy.io.loadmat('../datasets/NYC-data-set/nm_tensor.mat')
nm_tensor = nm_tensor['nm_tensor']
missing_rate = 0.1
# =============================================================================
### Random missing (RM) scenario
### Set the RM scenario by:
# binary_tensor = np.round(rm_tensor + 0.5 - missing_rate)
# =============================================================================
# =============================================================================
### Non-random missing (NM) scenario
### Set the NM scenario by:
binary_tensor = np.zeros(dense_tensor.shape)
for i1 in range(dense_tensor.shape[0]):
for i2 in range(dense_tensor.shape[1]):
for i3 in range(61):
binary_tensor[i1, i2, i3 * 24 : (i3 + 1) * 24] = np.round(nm_tensor[i1, i2, i3] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 30
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
```
**Experiment results** of missing data imputation using TF-ALS:
| scenario |`rank`| `maxiter`| mape | rmse |
|:----------|-----:|---------:|-----------:|----------:|
|**10%, RM**| 30 | 1000 | **0.5262** | **6.2444**|
|**30%, RM**| 30 | 1000 | **0.5488** | **6.8968**|
|**10%, NM**| 30 | 1000 | **0.5170** | **5.9863**|
|**30%, NM**| 30 | 100 | **-** | **-**|
# Part 8: Experiments on Seattle Data Set
```
import pandas as pd
dense_mat = pd.read_csv('../datasets/Seattle-data-set/mat.csv', index_col = 0)
RM_mat = pd.read_csv('../datasets/Seattle-data-set/RM_mat.csv', index_col = 0)
dense_mat = dense_mat.values
RM_mat = RM_mat.values
dense_tensor = dense_mat.reshape([dense_mat.shape[0], 28, 288])
RM_tensor = RM_mat.reshape([RM_mat.shape[0], 28, 288])
missing_rate = 0.2
# =============================================================================
### Random missing (RM) scenario
### Set the RM scenario by:
binary_tensor = np.round(RM_tensor + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 50
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
import pandas as pd
dense_mat = pd.read_csv('../datasets/Seattle-data-set/mat.csv', index_col = 0)
RM_mat = pd.read_csv('../datasets/Seattle-data-set/RM_mat.csv', index_col = 0)
dense_mat = dense_mat.values
RM_mat = RM_mat.values
dense_tensor = dense_mat.reshape([dense_mat.shape[0], 28, 288])
RM_tensor = RM_mat.reshape([RM_mat.shape[0], 28, 288])
missing_rate = 0.4
# =============================================================================
### Random missing (RM) scenario
### Set the RM scenario by:
binary_tensor = np.round(RM_tensor + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 50
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
import pandas as pd
dense_mat = pd.read_csv('../datasets/Seattle-data-set/mat.csv', index_col = 0)
NM_mat = pd.read_csv('../datasets/Seattle-data-set/NM_mat.csv', index_col = 0)
dense_mat = dense_mat.values
NM_mat = NM_mat.values
dense_tensor = dense_mat.reshape([dense_mat.shape[0], 28, 288])
missing_rate = 0.2
# =============================================================================
### Non-random missing (NM) scenario
### Set the NM scenario by:
binary_tensor = np.zeros((dense_mat.shape[0], 28, 288))
for i1 in range(binary_tensor.shape[0]):
for i2 in range(binary_tensor.shape[1]):
binary_tensor[i1, i2, :] = np.round(NM_mat[i1, i2] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 10
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
import pandas as pd
dense_mat = pd.read_csv('../datasets/Seattle-data-set/mat.csv', index_col = 0)
NM_mat = pd.read_csv('../datasets/Seattle-data-set/NM_mat.csv', index_col = 0)
dense_mat = dense_mat.values
NM_mat = NM_mat.values
dense_tensor = dense_mat.reshape([dense_mat.shape[0], 28, 288])
missing_rate = 0.4
# =============================================================================
### Non-random missing (NM) scenario
### Set the NM scenario by:
binary_tensor = np.zeros((dense_mat.shape[0], 28, 288))
for i1 in range(binary_tensor.shape[0]):
for i2 in range(binary_tensor.shape[1]):
binary_tensor[i1, i2, :] = np.round(NM_mat[i1, i2] + 0.5 - missing_rate)
# =============================================================================
sparse_tensor = np.multiply(dense_tensor, binary_tensor)
import time
start = time.time()
rank = 10
maxiter = 1000
tensor_hat, U, V, X = CP_ALS(sparse_tensor, rank, maxiter)
pos = np.where((dense_tensor != 0) & (sparse_tensor == 0))
final_mape = np.sum(np.abs(dense_tensor[pos] - tensor_hat[pos])/dense_tensor[pos])/dense_tensor[pos].shape[0]
final_rmse = np.sqrt(np.sum((dense_tensor[pos] - tensor_hat[pos]) ** 2)/dense_tensor[pos].shape[0])
print('Final Imputation MAPE: {:.6}'.format(final_mape))
print('Final Imputation RMSE: {:.6}'.format(final_rmse))
print()
end = time.time()
print('Running time: %d seconds'%(end - start))
```
**Experiment results** of missing data imputation using TF-ALS:
| scenario |`rank`| `maxiter`| mape | rmse |
|:----------|-----:|---------:|-----------:|----------:|
|**20%, RM**| 50 | 1000 | **0.0742** |**4.4929**|
|**40%, RM**| 50 | 1000 | **0.0758** |**4.5574**|
|**20%, NM**| 10 | 1000 | **0.0995** |**5.6331**|
|**40%, NM**| 10 | 1000 | **0.1004** |**5.7034**|
| github_jupyter |
# Auditing a dataframe
In this notebook, we shall demonstrate how to use `privacypanda` to _audit_ the privacy of your data. `privacypanda` provides a simple function which prints the names of any columns which break privacy. Currently, these are:
- Addresses
- E.g. "10 Downing Street"; "221b Baker St"; "EC2R 8AH"
- Phonenumbers (UK mobile)
- E.g. "+447123456789"
- Email addresses
- Ending in ".com", ".co.uk", ".org", ".edu" (to be expanded soon)
```
%load_ext watermark
%watermark -n -p pandas,privacypanda -g
import pandas as pd
import privacypanda as pp
```
---
## Firstly, we need data
```
data = pd.DataFrame(
{
"user ID": [
1665,
1,
5287,
42,
],
"User email": [
"xxxxxxxxxxxxx",
"xxxxxxxx",
"I'm not giving you that",
"an_email@email.com",
],
"User address": [
"AB1 1AB",
"",
"XXX XXX",
"EC2R 8AH",
],
"Likes raclette": [
1,
0,
1,
1,
],
}
)
```
You will notice two things about this dataframe:
1. _Some_ of the data has already been anonymized, for example by replacing characters with "x"s. However, the person who collected this data has not been fastidious with its cleaning as there is still some raw, potentially problematic private information. As the dataset grows, it becomes easier to miss entries with private information
2. Not all columns expose privacy: "Likes raclette" is pretty benign information (but be careful, lots of benign information can be combined to form a unique fingerprint identifying an individual - let's not worry about this at the moment, though), and "user ID" is already an anonymized labelling of an individual.
---
# Auditing the data's privacy
As a data scientist, we want a simple way to tell which columns, if any break privacy. More importantly, _how_ they break privacy determines how we deal with them. For example, emails will likely be superfluous information for analysis and can therefore be removed from the data, but age may be important and so we may wish instead to apply differential privacy to the dataset.
We can use `privacypanda`'s `report_privacy` function to see which data is problematic.
```
report = pp.report_privacy(data)
print(report)
```
`report_privacy` returns a `Report` object which stores the privacy issues of each column in the data.
As `privacypanda` is in active development,
this is currently only a simple dictionary of binary "breaks"/"doesn't break" privacy for each column.
We aim to make this information _cell-level_,
i.e. removing/replacing the information in individual cells in order to protect privacy with less information loss.
| github_jupyter |
```
# Neo4J graph example
# author: Gressling, T
# license: MIT License # code: github.com/gressling/examples
# activity: single example # index: 25-2
# https://gist.github.com/korakot/328aaac51d78e589b4a176228e4bb06f
# download 3.5.8 or neo4j-enterprise-4.0.0-alpha09mr02-unix
!curl https://neo4j.com/artifact.php?name=neo4j-community-3.5.8-unix.tar.gz -v -o neo4j.tar.gz
#!curl https://s3-eu-west-1.amazonaws.com/dist.neo4j.org/neo4j-community-3.5.8-unix.tar.gz?x-amz-security-token=IQoJb3JpZ2luX2VjEBgaCXVzLWVhc3QtMSJIMEYCIQC8JQ87qLW8MutNDC7kLf%2F8lCgTEeFw6XMHe0g6JGiLwQIhALrPjMot9j4eV1EiWsysYUamjICHutsaKG%2Fa%2B05ZJKD%2BKr0DCJD%2F%2F%2F%2F%2F%2F%2F%2F%2F%2FwEQABoMMTI4OTE2Njc5MzMwIgyekSOUHQOH4V1oebkqkQMSnlGj83iqgQ1X%2Bb9lDsjjfh%2FGggoIuvkn8RO9Ur3Ws24VznIHWrxQTECnTtQfsjhruOUbGJKv%2FlKBy9VU0aLu0zdrNcxeWZedOW09We0xVS4QTBipwW4i0UubWw%2FuDp1vAKPc1wLIq3vuvgflB4sXmTgvkQ%2FcT2%2BoIrvflqmSQ%2Fr9SB9Cqj9iACjxNQZrLs3qv2WgWxUNSsVjJYGXUx1yzx0ckCtxKYZ%2BzVBvqGuG1yULIodkGo4Kfbk5bh7s69gk8N4Gli7cQvYc9ajSFGg5IHXJU7%2BvRWeekX%2F2o7JlCRQogSNlW8kvv7o9ioD6Uj1mkOnR6rMsEv4Xo%2B2buKg7LqaPobmZwyjGnMBvZdndLXq37lAT7%2BP1i%2BVNCC7rak4Aqb3HtFMDZ%2F0nmdAitcKDWG1Am1mnaXiL3s6MZQ88SoU8h5RK0k01M%2FBxU9ZPwfi%2Bm8OAC%2Bgh6QyP9f7CPqSdI%2Fy8BSthxARcwWxl2ZwhHtUu7jqFf601aTu0iof%2FP2tH9oxty4kdH%2BI64qo7JCr9%2BzDx4OT9BTrqAfGlw5dReXwyh%2BSnYxW%2BB42cGs2JDcrFohn6UGdG3BtI%2FsjAFymH0vkCtXfN3irUPPzOoFzx216v%2F4GFfGebIpWqr85hT2f%2F28fck2XPbXiv%2BXSeffQdc8UkSL7dMHbquZ%2BmdOtCNlMhOWpnov5J7aICj9uY4AR60kNUSW3N4nra3CXjNEJWy%2B8ft49e6lnR9iKlVFxdwoXb1YAEx4egpctFcffoiaIEk2GinHjShAQgApOZFgOLe%2FDC9%2BnIwhxL7rSUfP7Ox%2FbEJF%2Br6VNYJddoD6D8xF2OVo%2FxZzv4M6eyw6Squ5r6i4LM7g%3D%3D&AWSAccessKeyId=ASIAR4BAINKRGKUIBRUS&Expires=1605973931&Signature=gzC025ItqNNdXpCJkGsm%2FvQt2WU%3D -o neo4j.tar.gz
# decompress and rename
!tar -xf neo4j.tar.gz # or --strip-components=1
!mv neo4j-community-3.5.8 nj
# disable password, and start server
!sed -i '/#dbms.security.auth_enabled/s/^#//g' nj/conf/neo4j.conf
!nj/bin/neo4j start
# from neo4j import GraphDatabase
# !pip install py2neo
from py2neo import Graph
graph = Graph("bolt://localhost:7687", auth=("neo4j", "password"))
graph.delete_all()
# define the entities of the graph (nodes)
from py2neo import Node
laboratory = Node("Laboratory", name="Laboratory 1")
lab1 = Node("Person", name="Peter", employee_ID=2)
lab2 = Node("Person", name="Susan", employee_ID=4)
sample1 = Node("Sample", name="A-12213", weight=45.7)
sample2 = Node("Sample", name="B-33443", weight=48.0)
# shared sample between two experiments
sample3 = Node("Sample", name="AB-33443", weight=24.3)
experiment1 = Node("Experiment", name="Screening-45")
experiment2 = Node("Experiment", name="Screening/w/Sol")
graph.create(laboratory | lab1 | lab2 | sample1 | sample2 | experiment1 |
experiment2)
# Define the relationships of the graph (edges)
from py2neo import Relationship
graph.create(Relationship(lab1, "works in", laboratory))
graph.create(Relationship(lab2, "works in", laboratory))
graph.create(Relationship(lab1, "performs", sample1))
graph.create(Relationship(lab2, "performs", sample2))
graph.create(Relationship(lab2, "performs", sample3))
graph.create(Relationship(sample1, "partof", experiment1))
graph.create(Relationship(sample2, "partof", experiment2))
graph.create(Relationship(sample3, "partof", experiment2))
graph.create(Relationship(sample3, "partof", experiment1))
import neo4jupyter
neo4jupyter.init_notebook_mode()
neo4jupyter.draw(graph)
```
| github_jupyter |
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# tf.data: Build TensorFlow input pipelines
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/guide/data"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/data.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/guide/data.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/data.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
The `tf.data` API enables you to build complex input pipelines from simple,
reusable pieces. For example, the pipeline for an image model might aggregate
data from files in a distributed file system, apply random perturbations to each
image, and merge randomly selected images into a batch for training. The
pipeline for a text model might involve extracting symbols from raw text data,
converting them to embedding identifiers with a lookup table, and batching
together sequences of different lengths. The `tf.data` API makes it possible to
handle large amounts of data, read from different data formats, and perform
complex transformations.
The `tf.data` API introduces a `tf.data.Dataset` abstraction that represents a
sequence of elements, in which each element consists of one or more components.
For example, in an image pipeline, an element might be a single training
example, with a pair of tensor components representing the image and its label.
There are two distinct ways to create a dataset:
* A data **source** constructs a `Dataset` from data stored in memory or in
one or more files.
* A data **transformation** constructs a dataset from one or more
`tf.data.Dataset` objects.
```
import tensorflow as tf
import pathlib
import os
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
np.set_printoptions(precision=4)
```
## Basic mechanics
<a id="basic-mechanics"/>
To create an input pipeline, you must start with a data *source*. For example,
to construct a `Dataset` from data in memory, you can use
`tf.data.Dataset.from_tensors()` or `tf.data.Dataset.from_tensor_slices()`.
Alternatively, if your input data is stored in a file in the recommended
TFRecord format, you can use `tf.data.TFRecordDataset()`.
Once you have a `Dataset` object, you can *transform* it into a new `Dataset` by
chaining method calls on the `tf.data.Dataset` object. For example, you can
apply per-element transformations such as `Dataset.map()`, and multi-element
transformations such as `Dataset.batch()`. See the documentation for
`tf.data.Dataset` for a complete list of transformations.
The `Dataset` object is a Python iterable. This makes it possible to consume its
elements using a for loop:
```
dataset = tf.data.Dataset.from_tensor_slices([8, 3, 0, 8, 2, 1])
dataset
for elem in dataset:
print(elem.numpy())
```
Or by explicitly creating a Python iterator using `iter` and consuming its
elements using `next`:
```
it = iter(dataset)
print(next(it).numpy())
```
Alternatively, dataset elements can be consumed using the `reduce`
transformation, which reduces all elements to produce a single result. The
following example illustrates how to use the `reduce` transformation to compute
the sum of a dataset of integers.
```
print(dataset.reduce(0, lambda state, value: state + value).numpy())
```
<!-- TODO(jsimsa): Talk about `tf.function` support. -->
<a id="dataset_structure"></a>
### Dataset structure
A dataset contains elements that each have the same (nested) structure and the
individual components of the structure can be of any type representable by
`tf.TypeSpec`, including `tf.Tensor`, `tf.sparse.SparseTensor`, `tf.RaggedTensor`,
`tf.TensorArray`, or `tf.data.Dataset`.
The `Dataset.element_spec` property allows you to inspect the type of each
element component. The property returns a *nested structure* of `tf.TypeSpec`
objects, matching the structure of the element, which may be a single component,
a tuple of components, or a nested tuple of components. For example:
```
dataset1 = tf.data.Dataset.from_tensor_slices(tf.random.uniform([4, 10]))
dataset1.element_spec
dataset2 = tf.data.Dataset.from_tensor_slices(
(tf.random.uniform([4]),
tf.random.uniform([4, 100], maxval=100, dtype=tf.int32)))
dataset2.element_spec
dataset3 = tf.data.Dataset.zip((dataset1, dataset2))
dataset3.element_spec
# Dataset containing a sparse tensor.
dataset4 = tf.data.Dataset.from_tensors(tf.SparseTensor(indices=[[0, 0], [1, 2]], values=[1, 2], dense_shape=[3, 4]))
dataset4.element_spec
# Use value_type to see the type of value represented by the element spec
dataset4.element_spec.value_type
```
The `Dataset` transformations support datasets of any structure. When using the
`Dataset.map()`, and `Dataset.filter()` transformations,
which apply a function to each element, the element structure determines the
arguments of the function:
```
dataset1 = tf.data.Dataset.from_tensor_slices(
tf.random.uniform([4, 10], minval=1, maxval=10, dtype=tf.int32))
dataset1
for z in dataset1:
print(z.numpy())
dataset2 = tf.data.Dataset.from_tensor_slices(
(tf.random.uniform([4]),
tf.random.uniform([4, 100], maxval=100, dtype=tf.int32)))
dataset2
dataset3 = tf.data.Dataset.zip((dataset1, dataset2))
dataset3
for a, (b,c) in dataset3:
print('shapes: {a.shape}, {b.shape}, {c.shape}'.format(a=a, b=b, c=c))
```
## Reading input data
### Consuming NumPy arrays
See [Loading NumPy arrays](../tutorials/load_data/numpy.ipynb) for more examples.
If all of your input data fits in memory, the simplest way to create a `Dataset`
from them is to convert them to `tf.Tensor` objects and use
`Dataset.from_tensor_slices()`.
```
train, test = tf.keras.datasets.fashion_mnist.load_data()
images, labels = train
images = images/255
dataset = tf.data.Dataset.from_tensor_slices((images, labels))
dataset
```
Note: The above code snippet will embed the `features` and `labels` arrays
in your TensorFlow graph as `tf.constant()` operations. This works well for a
small dataset, but wastes memory---because the contents of the array will be
copied multiple times---and can run into the 2GB limit for the `tf.GraphDef`
protocol buffer.
### Consuming Python generators
Another common data source that can easily be ingested as a `tf.data.Dataset` is the python generator.
Caution: While this is a convienient approach it has limited portability and scalibility. It must run in the same python process that created the generator, and is still subject to the Python [GIL](https://en.wikipedia.org/wiki/Global_interpreter_lock).
```
def count(stop):
i = 0
while i<stop:
yield i
i += 1
for n in count(5):
print(n)
```
The `Dataset.from_generator` constructor converts the python generator to a fully functional `tf.data.Dataset`.
The constructor takes a callable as input, not an iterator. This allows it to restart the generator when it reaches the end. It takes an optional `args` argument, which is passed as the callable's arguments.
The `output_types` argument is required because `tf.data` builds a `tf.Graph` internally, and graph edges require a `tf.dtype`.
```
ds_counter = tf.data.Dataset.from_generator(count, args=[25], output_types=tf.int32, output_shapes = (), )
for count_batch in ds_counter.repeat().batch(10).take(10):
print(count_batch.numpy())
```
The `output_shapes` argument is not *required* but is highly recomended as many tensorflow operations do not support tensors with unknown rank. If the length of a particular axis is unknown or variable, set it as `None` in the `output_shapes`.
It's also important to note that the `output_shapes` and `output_types` follow the same nesting rules as other dataset methods.
Here is an example generator that demonstrates both aspects, it returns tuples of arrays, where the second array is a vector with unknown length.
```
def gen_series():
i = 0
while True:
size = np.random.randint(0, 10)
yield i, np.random.normal(size=(size,))
i += 1
for i, series in gen_series():
print(i, ":", str(series))
if i > 5:
break
```
The first output is an `int32` the second is a `float32`.
The first item is a scalar, shape `()`, and the second is a vector of unknown length, shape `(None,)`
```
ds_series = tf.data.Dataset.from_generator(
gen_series,
output_types=(tf.int32, tf.float32),
output_shapes=((), (None,)))
ds_series
```
Now it can be used like a regular `tf.data.Dataset`. Note that when batching a dataset with a variable shape, you need to use `Dataset.padded_batch`.
```
ds_series_batch = ds_series.shuffle(20).padded_batch(10)
ids, sequence_batch = next(iter(ds_series_batch))
print(ids.numpy())
print()
print(sequence_batch.numpy())
```
For a more realistic example, try wrapping `preprocessing.image.ImageDataGenerator` as a `tf.data.Dataset`.
First download the data:
```
flowers = tf.keras.utils.get_file(
'flower_photos',
'https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz',
untar=True)
```
Create the `image.ImageDataGenerator`
```
img_gen = tf.keras.preprocessing.image.ImageDataGenerator(rescale=1./255, rotation_range=20)
images, labels = next(img_gen.flow_from_directory(flowers))
print(images.dtype, images.shape)
print(labels.dtype, labels.shape)
ds = tf.data.Dataset.from_generator(
lambda: img_gen.flow_from_directory(flowers),
output_types=(tf.float32, tf.float32),
output_shapes=([32,256,256,3], [32,5])
)
ds.element_spec
for images, label in ds.take(1):
print('images.shape: ', images.shape)
print('labels.shape: ', labels.shape)
```
### Consuming TFRecord data
See [Loading TFRecords](../tutorials/load_data/tf_records.ipynb) for an end-to-end example.
The `tf.data` API supports a variety of file formats so that you can process
large datasets that do not fit in memory. For example, the TFRecord file format
is a simple record-oriented binary format that many TensorFlow applications use
for training data. The `tf.data.TFRecordDataset` class enables you to
stream over the contents of one or more TFRecord files as part of an input
pipeline.
Here is an example using the test file from the French Street Name Signs (FSNS).
```
# Creates a dataset that reads all of the examples from two files.
fsns_test_file = tf.keras.utils.get_file("fsns.tfrec", "https://storage.googleapis.com/download.tensorflow.org/data/fsns-20160927/testdata/fsns-00000-of-00001")
```
The `filenames` argument to the `TFRecordDataset` initializer can either be a
string, a list of strings, or a `tf.Tensor` of strings. Therefore if you have
two sets of files for training and validation purposes, you can create a factory
method that produces the dataset, taking filenames as an input argument:
```
dataset = tf.data.TFRecordDataset(filenames = [fsns_test_file])
dataset
```
Many TensorFlow projects use serialized `tf.train.Example` records in their TFRecord files. These need to be decoded before they can be inspected:
```
raw_example = next(iter(dataset))
parsed = tf.train.Example.FromString(raw_example.numpy())
parsed.features.feature['image/text']
```
### Consuming text data
See [Loading Text](../tutorials/load_data/text.ipynb) for an end to end example.
Many datasets are distributed as one or more text files. The
`tf.data.TextLineDataset` provides an easy way to extract lines from one or more
text files. Given one or more filenames, a `TextLineDataset` will produce one
string-valued element per line of those files.
```
directory_url = 'https://storage.googleapis.com/download.tensorflow.org/data/illiad/'
file_names = ['cowper.txt', 'derby.txt', 'butler.txt']
file_paths = [
tf.keras.utils.get_file(file_name, directory_url + file_name)
for file_name in file_names
]
dataset = tf.data.TextLineDataset(file_paths)
```
Here are the first few lines of the first file:
```
for line in dataset.take(5):
print(line.numpy())
```
To alternate lines between files use `Dataset.interleave`. This makes it easier to shuffle files together. Here are the first, second and third lines from each translation:
```
files_ds = tf.data.Dataset.from_tensor_slices(file_paths)
lines_ds = files_ds.interleave(tf.data.TextLineDataset, cycle_length=3)
for i, line in enumerate(lines_ds.take(9)):
if i % 3 == 0:
print()
print(line.numpy())
```
By default, a `TextLineDataset` yields *every* line of each file, which may
not be desirable, for example, if the file starts with a header line, or contains comments. These lines can be removed using the `Dataset.skip()` or
`Dataset.filter()` transformations. Here, you skip the first line, then filter to
find only survivors.
```
titanic_file = tf.keras.utils.get_file("train.csv", "https://storage.googleapis.com/tf-datasets/titanic/train.csv")
titanic_lines = tf.data.TextLineDataset(titanic_file)
for line in titanic_lines.take(10):
print(line.numpy())
def survived(line):
return tf.not_equal(tf.strings.substr(line, 0, 1), "0")
survivors = titanic_lines.skip(1).filter(survived)
for line in survivors.take(10):
print(line.numpy())
```
### Consuming CSV data
See [Loading CSV Files](../tutorials/load_data/csv.ipynb), and [Loading Pandas DataFrames](../tutorials/load_data/pandas.ipynb) for more examples.
The CSV file format is a popular format for storing tabular data in plain text.
For example:
```
titanic_file = tf.keras.utils.get_file("train.csv", "https://storage.googleapis.com/tf-datasets/titanic/train.csv")
df = pd.read_csv(titanic_file)
df.head()
```
If your data fits in memory the same `Dataset.from_tensor_slices` method works on dictionaries, allowing this data to be easily imported:
```
titanic_slices = tf.data.Dataset.from_tensor_slices(dict(df))
for feature_batch in titanic_slices.take(1):
for key, value in feature_batch.items():
print(" {!r:20s}: {}".format(key, value))
```
A more scalable approach is to load from disk as necessary.
The `tf.data` module provides methods to extract records from one or more CSV files that comply with [RFC 4180](https://tools.ietf.org/html/rfc4180).
The `experimental.make_csv_dataset` function is the high level interface for reading sets of csv files. It supports column type inference and many other features, like batching and shuffling, to make usage simple.
```
titanic_batches = tf.data.experimental.make_csv_dataset(
titanic_file, batch_size=4,
label_name="survived")
for feature_batch, label_batch in titanic_batches.take(1):
print("'survived': {}".format(label_batch))
print("features:")
for key, value in feature_batch.items():
print(" {!r:20s}: {}".format(key, value))
```
You can use the `select_columns` argument if you only need a subset of columns.
```
titanic_batches = tf.data.experimental.make_csv_dataset(
titanic_file, batch_size=4,
label_name="survived", select_columns=['class', 'fare', 'survived'])
for feature_batch, label_batch in titanic_batches.take(1):
print("'survived': {}".format(label_batch))
for key, value in feature_batch.items():
print(" {!r:20s}: {}".format(key, value))
```
There is also a lower-level `experimental.CsvDataset` class which provides finer grained control. It does not support column type inference. Instead you must specify the type of each column.
```
titanic_types = [tf.int32, tf.string, tf.float32, tf.int32, tf.int32, tf.float32, tf.string, tf.string, tf.string, tf.string]
dataset = tf.data.experimental.CsvDataset(titanic_file, titanic_types , header=True)
for line in dataset.take(10):
print([item.numpy() for item in line])
```
If some columns are empty, this low-level interface allows you to provide default values instead of column types.
```
%%writefile missing.csv
1,2,3,4
,2,3,4
1,,3,4
1,2,,4
1,2,3,
,,,
# Creates a dataset that reads all of the records from two CSV files, each with
# four float columns which may have missing values.
record_defaults = [999,999,999,999]
dataset = tf.data.experimental.CsvDataset("missing.csv", record_defaults)
dataset = dataset.map(lambda *items: tf.stack(items))
dataset
for line in dataset:
print(line.numpy())
```
By default, a `CsvDataset` yields *every* column of *every* line of the file,
which may not be desirable, for example if the file starts with a header line
that should be ignored, or if some columns are not required in the input.
These lines and fields can be removed with the `header` and `select_cols`
arguments respectively.
```
# Creates a dataset that reads all of the records from two CSV files with
# headers, extracting float data from columns 2 and 4.
record_defaults = [999, 999] # Only provide defaults for the selected columns
dataset = tf.data.experimental.CsvDataset("missing.csv", record_defaults, select_cols=[1, 3])
dataset = dataset.map(lambda *items: tf.stack(items))
dataset
for line in dataset:
print(line.numpy())
```
### Consuming sets of files
There are many datasets distributed as a set of files, where each file is an example.
```
flowers_root = tf.keras.utils.get_file(
'flower_photos',
'https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz',
untar=True)
flowers_root = pathlib.Path(flowers_root)
```
Note: these images are licensed CC-BY, see LICENSE.txt for details.
The root directory contains a directory for each class:
```
for item in flowers_root.glob("*"):
print(item.name)
```
The files in each class directory are examples:
```
list_ds = tf.data.Dataset.list_files(str(flowers_root/'*/*'))
for f in list_ds.take(5):
print(f.numpy())
```
Read the data using the `tf.io.read_file` function and extract the label from the path, returning `(image, label)` pairs:
```
def process_path(file_path):
label = tf.strings.split(file_path, os.sep)[-2]
return tf.io.read_file(file_path), label
labeled_ds = list_ds.map(process_path)
for image_raw, label_text in labeled_ds.take(1):
print(repr(image_raw.numpy()[:100]))
print()
print(label_text.numpy())
```
<!--
TODO(mrry): Add this section.
### Handling text data with unusual sizes
-->
## Batching dataset elements
### Simple batching
The simplest form of batching stacks `n` consecutive elements of a dataset into
a single element. The `Dataset.batch()` transformation does exactly this, with
the same constraints as the `tf.stack()` operator, applied to each component
of the elements: i.e. for each component *i*, all elements must have a tensor
of the exact same shape.
```
inc_dataset = tf.data.Dataset.range(100)
dec_dataset = tf.data.Dataset.range(0, -100, -1)
dataset = tf.data.Dataset.zip((inc_dataset, dec_dataset))
batched_dataset = dataset.batch(4)
for batch in batched_dataset.take(4):
print([arr.numpy() for arr in batch])
```
While `tf.data` tries to propagate shape information, the default settings of `Dataset.batch` result in an unknown batch size because the last batch may not be full. Note the `None`s in the shape:
```
batched_dataset
```
Use the `drop_remainder` argument to ignore that last batch, and get full shape propagation:
```
batched_dataset = dataset.batch(7, drop_remainder=True)
batched_dataset
```
### Batching tensors with padding
The above recipe works for tensors that all have the same size. However, many
models (e.g. sequence models) work with input data that can have varying size
(e.g. sequences of different lengths). To handle this case, the
`Dataset.padded_batch` transformation enables you to batch tensors of
different shape by specifying one or more dimensions in which they may be
padded.
```
dataset = tf.data.Dataset.range(100)
dataset = dataset.map(lambda x: tf.fill([tf.cast(x, tf.int32)], x))
dataset = dataset.padded_batch(4, padded_shapes=(None,))
for batch in dataset.take(2):
print(batch.numpy())
print()
```
The `Dataset.padded_batch` transformation allows you to set different padding
for each dimension of each component, and it may be variable-length (signified
by `None` in the example above) or constant-length. It is also possible to
override the padding value, which defaults to 0.
<!--
TODO(mrry): Add this section.
### Dense ragged -> tf.SparseTensor
-->
## Training workflows
### Processing multiple epochs
The `tf.data` API offers two main ways to process multiple epochs of the same
data.
The simplest way to iterate over a dataset in multiple epochs is to use the
`Dataset.repeat()` transformation. First, create a dataset of titanic data:
```
titanic_file = tf.keras.utils.get_file("train.csv", "https://storage.googleapis.com/tf-datasets/titanic/train.csv")
titanic_lines = tf.data.TextLineDataset(titanic_file)
def plot_batch_sizes(ds):
batch_sizes = [batch.shape[0] for batch in ds]
plt.bar(range(len(batch_sizes)), batch_sizes)
plt.xlabel('Batch number')
plt.ylabel('Batch size')
```
Applying the `Dataset.repeat()` transformation with no arguments will repeat
the input indefinitely.
The `Dataset.repeat` transformation concatenates its
arguments without signaling the end of one epoch and the beginning of the next
epoch. Because of this a `Dataset.batch` applied after `Dataset.repeat` will yield batches that straddle epoch boundaries:
```
titanic_batches = titanic_lines.repeat(3).batch(128)
plot_batch_sizes(titanic_batches)
```
If you need clear epoch separation, put `Dataset.batch` before the repeat:
```
titanic_batches = titanic_lines.batch(128).repeat(3)
plot_batch_sizes(titanic_batches)
```
If you would like to perform a custom computation (e.g. to collect statistics) at the end of each epoch then it's simplest to restart the dataset iteration on each epoch:
```
epochs = 3
dataset = titanic_lines.batch(128)
for epoch in range(epochs):
for batch in dataset:
print(batch.shape)
print("End of epoch: ", epoch)
```
### Randomly shuffling input data
The `Dataset.shuffle()` transformation maintains a fixed-size
buffer and chooses the next element uniformly at random from that buffer.
Note: While large buffer_sizes shuffle more thoroughly, they can take a lot of memory, and significant time to fill. Consider using `Dataset.interleave` across files if this becomes a problem.
Add an index to the dataset so you can see the effect:
```
lines = tf.data.TextLineDataset(titanic_file)
counter = tf.data.experimental.Counter()
dataset = tf.data.Dataset.zip((counter, lines))
dataset = dataset.shuffle(buffer_size=100)
dataset = dataset.batch(20)
dataset
```
Since the `buffer_size` is 100, and the batch size is 20, the first batch contains no elements with an index over 120.
```
n,line_batch = next(iter(dataset))
print(n.numpy())
```
As with `Dataset.batch` the order relative to `Dataset.repeat` matters.
`Dataset.shuffle` doesn't signal the end of an epoch until the shuffle buffer is empty. So a shuffle placed before a repeat will show every element of one epoch before moving to the next:
```
dataset = tf.data.Dataset.zip((counter, lines))
shuffled = dataset.shuffle(buffer_size=100).batch(10).repeat(2)
print("Here are the item ID's near the epoch boundary:\n")
for n, line_batch in shuffled.skip(60).take(5):
print(n.numpy())
shuffle_repeat = [n.numpy().mean() for n, line_batch in shuffled]
plt.plot(shuffle_repeat, label="shuffle().repeat()")
plt.ylabel("Mean item ID")
plt.legend()
```
But a repeat before a shuffle mixes the epoch boundaries together:
```
dataset = tf.data.Dataset.zip((counter, lines))
shuffled = dataset.repeat(2).shuffle(buffer_size=100).batch(10)
print("Here are the item ID's near the epoch boundary:\n")
for n, line_batch in shuffled.skip(55).take(15):
print(n.numpy())
repeat_shuffle = [n.numpy().mean() for n, line_batch in shuffled]
plt.plot(shuffle_repeat, label="shuffle().repeat()")
plt.plot(repeat_shuffle, label="repeat().shuffle()")
plt.ylabel("Mean item ID")
plt.legend()
```
## Preprocessing data
The `Dataset.map(f)` transformation produces a new dataset by applying a given
function `f` to each element of the input dataset. It is based on the
[`map()`](https://en.wikipedia.org/wiki/Map_\(higher-order_function\)) function
that is commonly applied to lists (and other structures) in functional
programming languages. The function `f` takes the `tf.Tensor` objects that
represent a single element in the input, and returns the `tf.Tensor` objects
that will represent a single element in the new dataset. Its implementation uses
standard TensorFlow operations to transform one element into another.
This section covers common examples of how to use `Dataset.map()`.
### Decoding image data and resizing it
<!-- TODO(markdaoust): link to image augmentation when it exists -->
When training a neural network on real-world image data, it is often necessary
to convert images of different sizes to a common size, so that they may be
batched into a fixed size.
Rebuild the flower filenames dataset:
```
list_ds = tf.data.Dataset.list_files(str(flowers_root/'*/*'))
```
Write a function that manipulates the dataset elements.
```
# Reads an image from a file, decodes it into a dense tensor, and resizes it
# to a fixed shape.
def parse_image(filename):
parts = tf.strings.split(filename, os.sep)
label = parts[-2]
image = tf.io.read_file(filename)
image = tf.image.decode_jpeg(image)
image = tf.image.convert_image_dtype(image, tf.float32)
image = tf.image.resize(image, [128, 128])
return image, label
```
Test that it works.
```
file_path = next(iter(list_ds))
image, label = parse_image(file_path)
def show(image, label):
plt.figure()
plt.imshow(image)
plt.title(label.numpy().decode('utf-8'))
plt.axis('off')
show(image, label)
```
Map it over the dataset.
```
images_ds = list_ds.map(parse_image)
for image, label in images_ds.take(2):
show(image, label)
```
### Applying arbitrary Python logic
For performance reasons, use TensorFlow operations for
preprocessing your data whenever possible. However, it is sometimes useful to
call external Python libraries when parsing your input data. You can use the `tf.py_function()` operation in a `Dataset.map()` transformation.
For example, if you want to apply a random rotation, the `tf.image` module only has `tf.image.rot90`, which is not very useful for image augmentation.
Note: `tensorflow_addons` has a TensorFlow compatible `rotate` in `tensorflow_addons.image.rotate`.
To demonstrate `tf.py_function`, try using the `scipy.ndimage.rotate` function instead:
```
import scipy.ndimage as ndimage
def random_rotate_image(image):
image = ndimage.rotate(image, np.random.uniform(-30, 30), reshape=False)
return image
image, label = next(iter(images_ds))
image = random_rotate_image(image)
show(image, label)
```
To use this function with `Dataset.map` the same caveats apply as with `Dataset.from_generator`, you need to describe the return shapes and types when you apply the function:
```
def tf_random_rotate_image(image, label):
im_shape = image.shape
[image,] = tf.py_function(random_rotate_image, [image], [tf.float32])
image.set_shape(im_shape)
return image, label
rot_ds = images_ds.map(tf_random_rotate_image)
for image, label in rot_ds.take(2):
show(image, label)
```
### Parsing `tf.Example` protocol buffer messages
Many input pipelines extract `tf.train.Example` protocol buffer messages from a
TFRecord format. Each `tf.train.Example` record contains one or more "features",
and the input pipeline typically converts these features into tensors.
```
fsns_test_file = tf.keras.utils.get_file("fsns.tfrec", "https://storage.googleapis.com/download.tensorflow.org/data/fsns-20160927/testdata/fsns-00000-of-00001")
dataset = tf.data.TFRecordDataset(filenames = [fsns_test_file])
dataset
```
You can work with `tf.train.Example` protos outside of a `tf.data.Dataset` to understand the data:
```
raw_example = next(iter(dataset))
parsed = tf.train.Example.FromString(raw_example.numpy())
feature = parsed.features.feature
raw_img = feature['image/encoded'].bytes_list.value[0]
img = tf.image.decode_png(raw_img)
plt.imshow(img)
plt.axis('off')
_ = plt.title(feature["image/text"].bytes_list.value[0])
raw_example = next(iter(dataset))
def tf_parse(eg):
example = tf.io.parse_example(
eg[tf.newaxis], {
'image/encoded': tf.io.FixedLenFeature(shape=(), dtype=tf.string),
'image/text': tf.io.FixedLenFeature(shape=(), dtype=tf.string)
})
return example['image/encoded'][0], example['image/text'][0]
img, txt = tf_parse(raw_example)
print(txt.numpy())
print(repr(img.numpy()[:20]), "...")
decoded = dataset.map(tf_parse)
decoded
image_batch, text_batch = next(iter(decoded.batch(10)))
image_batch.shape
```
<a id="time_series_windowing"></a>
### Time series windowing
For an end to end time series example see: [Time series forecasting](../../tutorials/text/time_series.ipynb).
Time series data is often organized with the time axis intact.
Use a simple `Dataset.range` to demonstrate:
```
range_ds = tf.data.Dataset.range(100000)
```
Typically, models based on this sort of data will want a contiguous time slice.
The simplest approach would be to batch the data:
#### Using `batch`
```
batches = range_ds.batch(10, drop_remainder=True)
for batch in batches.take(5):
print(batch.numpy())
```
Or to make dense predictions one step into the future, you might shift the features and labels by one step relative to each other:
```
def dense_1_step(batch):
# Shift features and labels one step relative to each other.
return batch[:-1], batch[1:]
predict_dense_1_step = batches.map(dense_1_step)
for features, label in predict_dense_1_step.take(3):
print(features.numpy(), " => ", label.numpy())
```
To predict a whole window instead of a fixed offset you can split the batches into two parts:
```
batches = range_ds.batch(15, drop_remainder=True)
def label_next_5_steps(batch):
return (batch[:-5], # Take the first 5 steps
batch[-5:]) # take the remainder
predict_5_steps = batches.map(label_next_5_steps)
for features, label in predict_5_steps.take(3):
print(features.numpy(), " => ", label.numpy())
```
To allow some overlap between the features of one batch and the labels of another, use `Dataset.zip`:
```
feature_length = 10
label_length = 3
features = range_ds.batch(feature_length, drop_remainder=True)
labels = range_ds.batch(feature_length).skip(1).map(lambda labels: labels[:label_length])
predicted_steps = tf.data.Dataset.zip((features, labels))
for features, label in predicted_steps.take(5):
print(features.numpy(), " => ", label.numpy())
```
#### Using `window`
While using `Dataset.batch` works, there are situations where you may need finer control. The `Dataset.window` method gives you complete control, but requires some care: it returns a `Dataset` of `Datasets`. See [Dataset structure](#dataset_structure) for details.
```
window_size = 5
windows = range_ds.window(window_size, shift=1)
for sub_ds in windows.take(5):
print(sub_ds)
```
The `Dataset.flat_map` method can take a dataset of datasets and flatten it into a single dataset:
```
for x in windows.flat_map(lambda x: x).take(30):
print(x.numpy(), end=' ')
```
In nearly all cases, you will want to `.batch` the dataset first:
```
def sub_to_batch(sub):
return sub.batch(window_size, drop_remainder=True)
for example in windows.flat_map(sub_to_batch).take(5):
print(example.numpy())
```
Now, you can see that the `shift` argument controls how much each window moves over.
Putting this together you might write this function:
```
def make_window_dataset(ds, window_size=5, shift=1, stride=1):
windows = ds.window(window_size, shift=shift, stride=stride)
def sub_to_batch(sub):
return sub.batch(window_size, drop_remainder=True)
windows = windows.flat_map(sub_to_batch)
return windows
ds = make_window_dataset(range_ds, window_size=10, shift = 5, stride=3)
for example in ds.take(10):
print(example.numpy())
```
Then it's easy to extract labels, as before:
```
dense_labels_ds = ds.map(dense_1_step)
for inputs,labels in dense_labels_ds.take(3):
print(inputs.numpy(), "=>", labels.numpy())
```
### Resampling
When working with a dataset that is very class-imbalanced, you may want to resample the dataset. `tf.data` provides two methods to do this. The credit card fraud dataset is a good example of this sort of problem.
Note: See [Imbalanced Data](../tutorials/keras/imbalanced_data.ipynb) for a full tutorial.
```
zip_path = tf.keras.utils.get_file(
origin='https://storage.googleapis.com/download.tensorflow.org/data/creditcard.zip',
fname='creditcard.zip',
extract=True)
csv_path = zip_path.replace('.zip', '.csv')
creditcard_ds = tf.data.experimental.make_csv_dataset(
csv_path, batch_size=1024, label_name="Class",
# Set the column types: 30 floats and an int.
column_defaults=[float()]*30+[int()])
```
Now, check the distribution of classes, it is highly skewed:
```
def count(counts, batch):
features, labels = batch
class_1 = labels == 1
class_1 = tf.cast(class_1, tf.int32)
class_0 = labels == 0
class_0 = tf.cast(class_0, tf.int32)
counts['class_0'] += tf.reduce_sum(class_0)
counts['class_1'] += tf.reduce_sum(class_1)
return counts
counts = creditcard_ds.take(10).reduce(
initial_state={'class_0': 0, 'class_1': 0},
reduce_func = count)
counts = np.array([counts['class_0'].numpy(),
counts['class_1'].numpy()]).astype(np.float32)
fractions = counts/counts.sum()
print(fractions)
```
A common approach to training with an imbalanced dataset is to balance it. `tf.data` includes a few methods which enable this workflow:
#### Datasets sampling
One approach to resampling a dataset is to use `sample_from_datasets`. This is more applicable when you have a separate `data.Dataset` for each class.
Here, just use filter to generate them from the credit card fraud data:
```
negative_ds = (
creditcard_ds
.unbatch()
.filter(lambda features, label: label==0)
.repeat())
positive_ds = (
creditcard_ds
.unbatch()
.filter(lambda features, label: label==1)
.repeat())
for features, label in positive_ds.batch(10).take(1):
print(label.numpy())
```
To use `tf.data.experimental.sample_from_datasets` pass the datasets, and the weight for each:
```
balanced_ds = tf.data.experimental.sample_from_datasets(
[negative_ds, positive_ds], [0.5, 0.5]).batch(10)
```
Now the dataset produces examples of each class with 50/50 probability:
```
for features, labels in balanced_ds.take(10):
print(labels.numpy())
```
#### Rejection resampling
One problem with the above `experimental.sample_from_datasets` approach is that
it needs a separate `tf.data.Dataset` per class. Using `Dataset.filter`
works, but results in all the data being loaded twice.
The `data.experimental.rejection_resample` function can be applied to a dataset to rebalance it, while only loading it once. Elements will be dropped from the dataset to achieve balance.
`data.experimental.rejection_resample` takes a `class_func` argument. This `class_func` is applied to each dataset element, and is used to determine which class an example belongs to for the purposes of balancing.
The elements of `creditcard_ds` are already `(features, label)` pairs. So the `class_func` just needs to return those labels:
```
def class_func(features, label):
return label
```
The resampler also needs a target distribution, and optionally an initial distribution estimate:
```
resampler = tf.data.experimental.rejection_resample(
class_func, target_dist=[0.5, 0.5], initial_dist=fractions)
```
The resampler deals with individual examples, so you must `unbatch` the dataset before applying the resampler:
```
resample_ds = creditcard_ds.unbatch().apply(resampler).batch(10)
```
The resampler returns creates `(class, example)` pairs from the output of the `class_func`. In this case, the `example` was already a `(feature, label)` pair, so use `map` to drop the extra copy of the labels:
```
balanced_ds = resample_ds.map(lambda extra_label, features_and_label: features_and_label)
```
Now the dataset produces examples of each class with 50/50 probability:
```
for features, labels in balanced_ds.take(10):
print(labels.numpy())
```
## Iterator Checkpointing
Tensorflow supports [taking checkpoints](https://www.tensorflow.org/guide/checkpoint) so that when your training process restarts it can restore the latest checkpoint to recover most of its progress. In addition to checkpointing the model variables, you can also checkpoint the progress of the dataset iterator. This could be useful if you have a large dataset and don't want to start the dataset from the beginning on each restart. Note however that iterator checkpoints may be large, since transformations such as `shuffle` and `prefetch` require buffering elements within the iterator.
To include your iterator in a checkpoint, pass the iterator to the `tf.train.Checkpoint` constructor.
```
range_ds = tf.data.Dataset.range(20)
iterator = iter(range_ds)
ckpt = tf.train.Checkpoint(step=tf.Variable(0), iterator=iterator)
manager = tf.train.CheckpointManager(ckpt, '/tmp/my_ckpt', max_to_keep=3)
print([next(iterator).numpy() for _ in range(5)])
save_path = manager.save()
print([next(iterator).numpy() for _ in range(5)])
ckpt.restore(manager.latest_checkpoint)
print([next(iterator).numpy() for _ in range(5)])
```
Note: It is not possible to checkpoint an iterator which relies on external state such as a `tf.py_function`. Attempting to do so will raise an exception complaining about the external state.
## Using tf.data with tf.keras
The `tf.keras` API simplifies many aspects of creating and executing machine
learning models. Its `.fit()` and `.evaluate()` and `.predict()` APIs support datasets as inputs. Here is a quick dataset and model setup:
```
train, test = tf.keras.datasets.fashion_mnist.load_data()
images, labels = train
images = images/255.0
labels = labels.astype(np.int32)
fmnist_train_ds = tf.data.Dataset.from_tensor_slices((images, labels))
fmnist_train_ds = fmnist_train_ds.shuffle(5000).batch(32)
model = tf.keras.Sequential([
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(10)
])
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
```
Passing a dataset of `(feature, label)` pairs is all that's needed for `Model.fit` and `Model.evaluate`:
```
model.fit(fmnist_train_ds, epochs=2)
```
If you pass an infinite dataset, for example by calling `Dataset.repeat()`, you just need to also pass the `steps_per_epoch` argument:
```
model.fit(fmnist_train_ds.repeat(), epochs=2, steps_per_epoch=20)
```
For evaluation you can pass the number of evaluation steps:
```
loss, accuracy = model.evaluate(fmnist_train_ds)
print("Loss :", loss)
print("Accuracy :", accuracy)
```
For long datasets, set the number of steps to evaluate:
```
loss, accuracy = model.evaluate(fmnist_train_ds.repeat(), steps=10)
print("Loss :", loss)
print("Accuracy :", accuracy)
```
The labels are not required in when calling `Model.predict`.
```
predict_ds = tf.data.Dataset.from_tensor_slices(images).batch(32)
result = model.predict(predict_ds, steps = 10)
print(result.shape)
```
But the labels are ignored if you do pass a dataset containing them:
```
result = model.predict(fmnist_train_ds, steps = 10)
print(result.shape)
```
| github_jupyter |
# Communication in Crisis
## Acquire
Data: [Los Angeles Parking Citations](https://www.kaggle.com/cityofLA/los-angeles-parking-citations)<br>
Load the dataset and filter for:
- Citations issued from 2017-01-01 to 2021-04-12.
- Street Sweeping violations - `Violation Description` == __"NO PARK/STREET CLEAN"__
Let's acquire the parking citations data from our file.
1. Import libraries.
1. Load the dataset.
1. Display the shape and first/last 2 rows.
1. Display general infomation about the dataset - w/ the # of unique values in each column.
1. Display the number of missing values in each column.
1. Descriptive statistics for all numeric features.
```
# Import libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import sys
import time
import folium.plugins as plugins
from IPython.display import HTML
import json
import datetime
import calplot
import folium
import math
sns.set()
from tqdm.notebook import tqdm
import src
# Filter warnings
from warnings import filterwarnings
filterwarnings('ignore')
# Load the data
df = src.get_sweep_data(prepared=False)
# Display the shape and dtypes of each column
print(df.shape)
df.info()
# Display the first two citations
df.head(2)
# Display the last two citations
df.tail(2)
# Display descriptive statistics of numeric columns
df.describe()
df.hist(figsize=(16, 8), bins=15)
plt.tight_layout();
```
__Initial findings__
- `Issue time` and `Marked Time` are quasi-normally distributed. Note: Poisson Distribution
- It's interesting to see the distribution of our activity on earth follows a normal distribution.
- Agencies 50+ write the most parking citations.
- Most fine amounts are less than $100.00
- There are a few null or invalid license plates.
# Prepare
- Remove spaces + capitalization from each column name.
- Cast `Plate Expiry Date` to datetime data type.
- Cast `Issue Date` and `Issue Time` to datetime data types.
- Drop columns missing >=74.42\% of their values.
- Drop missing values.
- Transform Latitude and Longitude columns from NAD1983StatePlaneCaliforniaVFIPS0405 feet projection to EPSG:4326 World Geodetic System 1984: used in GPS [Standard]
- Filter data for street sweeping citations only.
```
# Prepare the data using a function stored in prepare.py
df_citations = src.get_sweep_data(prepared=True)
# Display the first two rows
df_citations.head(2)
# Check the column data types and non-null counts.
df_citations.info()
```
# Exploration
## How much daily revenue is generated from street sweeper citations?
### Daily Revenue from Street Sweeper Citations
Daily street sweeper citations increased in 2020.
```
# Daily street sweeping citation revenue
daily_revenue = df_citations.groupby('issue_date').fine_amount.sum()
daily_revenue.index = pd.to_datetime(daily_revenue.index)
df_sweep = src.street_sweep(data=df_citations)
df_d = src.resample_period(data=df_sweep)
df_m = src.resample_period(data=df_sweep, period='M')
df_d.head()
sns.set_context('talk')
# Plot daily revenue from street sweeping citations
df_d.revenue.plot(figsize=(14, 7), label='Revenue', color='DodgerBlue')
plt.axhline(df_d.revenue.mean(skipna=True), color='black', label='Average Revenue')
plt.title("Daily Revenue from Street Sweeping Citations")
plt.xlabel('')
plt.ylabel("Revenue (in thousand's)")
plt.xticks(rotation=0, horizontalalignment='center', fontsize=13)
plt.yticks(range(0, 1_000_000, 200_000), ['$0', '$200', '$400', '$600', '$800',])
plt.ylim(0, 1_000_000)
plt.legend(loc=2, framealpha=.8);
```
> __Anomaly__: Between March 2020 and October 2020 a Local Emergency was Declared by the Mayor of Los Angeles in response to COVID-19. Street Sweeping was halted to help Angelenos Shelter in Place. _Street Sweeping resumed on 10/15/2020_.
### Anomaly: Declaration of Local Emergency
```
sns.set_context('talk')
# Plot daily revenue from street sweeping citations
df_d.revenue.plot(figsize=(14, 7), label='Revenue', color='DodgerBlue')
plt.axvspan('2020-03-16', '2020-10-14', color='grey', alpha=.25)
plt.text('2020-03-29', 890_000, 'Declaration of\nLocal Emergency', fontsize=11)
plt.title("Daily Revenue from Street Sweeping Citations")
plt.xlabel('')
plt.ylabel("Revenue (in thousand's)")
plt.xticks(rotation=0, horizontalalignment='center', fontsize=13)
plt.yticks(range(0, 1_000_000, 200_000), ['$0', '$200', '$400', '$600', '$800',])
plt.ylim(0, 1_000_000)
plt.legend(loc=2, framealpha=.8);
sns.set_context('talk')
# Plot daily revenue from street sweeping citations
df_d.revenue.plot(figsize=(14, 7), label='Revenue', color='DodgerBlue')
plt.axhline(df_d.revenue.mean(skipna=True), color='black', label='Average Revenue')
plt.axvline(datetime.datetime(2020, 10, 15), color='red', linestyle="--", label='October 15, 2020')
plt.title("Daily Revenue from Street Sweeping Citations")
plt.xlabel('')
plt.ylabel("Revenue (in thousand's)")
plt.xticks(rotation=0, horizontalalignment='center', fontsize=13)
plt.yticks(range(0, 1_000_000, 200_000), ['$0', '$200K', '$400K', '$600K', '$800K',])
plt.ylim(0, 1_000_000)
plt.legend(loc=2, framealpha=.8);
```
## Hypothesis Test
### General Inquiry
Is the daily citation revenue after 10/15/2020 significantly greater than average?
### Z-Score
$H_0$: The daily citation revenue after 10/15/2020 is less than or equal to the average daily revenue.
$H_a$: The daily citation revenue after 10/15/2020 is significantly greater than average.
```
confidence_interval = .997
# Directional Test
alpha = (1 - confidence_interval)/2
# Data to calculate z-scores using precovid values to calculate the mean and std
daily_revenue_precovid = df_d.loc[df_d.index < '2020-03-16']['revenue']
mean_precovid, std_precovid = daily_revenue_precovid.agg(['mean', 'std']).values
mean, std = df_d.agg(['mean', 'std']).values
# Calculating Z-Scores using precovid mean and std
z_scores_precovid = (df_d.revenue - mean_precovid)/std_precovid
z_scores_precovid.index = pd.to_datetime(z_scores_precovid.index)
sig_zscores_pre_covid = z_scores_precovid[z_scores_precovid>3]
# Calculating Z-Scores using entire data
z_scores = (df_d.revenue - mean)/std
z_scores.index = pd.to_datetime(z_scores.index)
sig_zscores = z_scores[z_scores>3]
sns.set_context('talk')
plt.figure(figsize=(12, 6))
sns.histplot(data=z_scores_precovid,
bins=50,
label='preCOVID z-scores')
sns.histplot(data=z_scores,
bins=50,
color='orange',
label='z-scores')
plt.title('Daily citation revenue after 10/15/2020 is significantly greater than average', fontsize=16)
plt.xlabel('Standard Deviations')
plt.ylabel('# of Days')
plt.axvline(3, color='Black', linestyle="--", label='3 Standard Deviations')
plt.xticks(np.linspace(-1, 9, 11))
plt.legend(fontsize=13);
a = stats.zscore(daily_revenue)
fig, ax = plt.subplots(figsize=(8, 8))
stats.probplot(a, plot=ax)
plt.xlabel("Quantile of Normal Distribution")
plt.ylabel("z-score");
```
### p-values
```
p_values_precovid = z_scores_precovid.apply(stats.norm.cdf)
p_values = z_scores_precovid.apply(stats.norm.cdf)
significant_dates_precovid = p_values_precovid[(1-p_values_precovid) < alpha]
significant_dates = p_values[(1-p_values) < alpha]
# The chance of an outcome occuring by random chance
print(f'{alpha:0.3%}')
```
### Cohen's D
```
fractions = [.1, .2, .5, .7, .9]
cohen_d = []
for percentage in fractions:
cohen_d_trial = []
for i in range(10000):
sim = daily_revenue.sample(frac=percentage)
sim_mean = sim.mean()
d = (sim_mean - mean) / (std/math.sqrt(int(len(daily_revenue)*percentage)))
cohen_d_trial.append(d)
cohen_d.append(np.mean(cohen_d_trial))
cohen_d
fractions = [.1, .2, .5, .7, .9]
cohen_d_precovid = []
for percentage in fractions:
cohen_d_trial = []
for i in range(10000):
sim = daily_revenue_precovid.sample(frac=percentage)
sim_mean = sim.mean()
d = (sim_mean - mean_precovid) / (std_precovid/math.sqrt(int(len(daily_revenue_precovid)*percentage)))
cohen_d_trial.append(d)
cohen_d_precovid.append(np.mean(cohen_d_trial))
cohen_d_precovid
```
### Significant Dates with less than a 0.15% chance of occuring
- All dates that are considered significant occur after 10/15/2020
- In the two weeks following 10/15/2020 significant events occured on __Tuesday's and Wednesday's__.
```
dates_precovid = set(list(sig_zscores_pre_covid.index))
dates = set(list(sig_zscores.index))
common_dates = list(dates.intersection(dates_precovid))
common_dates = pd.to_datetime(common_dates).sort_values()
sig_zscores
pd.Series(common_dates.day_name(),
common_dates)
np.random.seed(sum(map(ord, 'calplot')))
all_days = pd.date_range('1/1/2020', '12/22/2020', freq='D')
significant_events = pd.Series(np.ones_like(len(common_dates)), index=common_dates)
calplot.calplot(significant_events, figsize=(18, 12), cmap='coolwarm_r');
```
## Which parts of the city were impacted the most?
```
df_outliers = df_citations.loc[df_citations.issue_date.isin(list(common_dates.astype('str')))]
df_outliers.reset_index(drop=True, inplace=True)
print(df_outliers.shape)
df_outliers.head()
m = folium.Map(location=[34.0522, -118.2437],
min_zoom=8,
max_bounds=True)
mc = plugins.MarkerCluster()
for index, row in df_outliers.iterrows():
mc.add_child(
folium.Marker(location=[str(row['latitude']), str(row['longitude'])],
popup='Cited {} {} at {}'.format(row['day_of_week'],
row['issue_date'],
row['issue_time'][:-3]),
control_scale=True,
clustered_marker=True
)
)
m.add_child(mc)
```
Transfering map to Tablaeu
# Conclusions
# Appendix
## What time(s) are Street Sweeping citations issued?
Most citations are issued during the hours of 8am, 10am, and 12pm.
### Citation Times
```
# Filter street sweeping data for citations issued between
# 8 am and 2 pm, 8 and 14 respectively.
df_citation_times = df_citations.loc[(df_citations.issue_hour >= 8)&(df_citations.issue_hour < 14)]
sns.set_context('talk')
# Issue Hour Plot
df_citation_times.issue_hour.value_counts().sort_index().plot.bar(figsize=(8, 6))
# Axis labels
plt.title('Most Street Sweeper Citations are Issued at 8am')
plt.xlabel('Issue Hour (24HR)')
plt.ylabel('# of Citations (in thousands)')
# Chart Formatting
plt.xticks(rotation=0)
plt.yticks(range(100_000, 400_001,100_000), ['100', '200', '300', '400'])
plt.show()
sns.set_context('talk')
# Issue Minute Plot
df_citation_times.issue_minute.value_counts().sort_index().plot.bar(figsize=(20, 9))
# Axis labels
plt.title('Most Street Sweeper Citations are Issued in the First 30 Minutes')
plt.xlabel('Issue Minute')
plt.ylabel('# of Citations (in thousands)')
# plt.axvspan(0, 30, facecolor='grey', alpha=0.1)
# Chart Formatting
plt.xticks(rotation=0)
plt.yticks(range(5_000, 40_001, 5_000), ['5', '10', '15', '20', '25', '30', '35', '40'])
plt.tight_layout()
plt.show()
```
## Which state has the most Street Sweeping violators?
### License Plate
Over 90% of all street sweeping citations are issued to California Residents.
```
sns.set_context('talk')
fig = df_citations.rp_state_plate.value_counts(normalize=True).nlargest(3).plot.bar(figsize=(12, 6))
# Chart labels
plt.title('California residents receive the most street sweeping citations', fontsize=16)
plt.xlabel('State')
plt.ylabel('% of all Citations')
# Tick Formatting
plt.xticks(rotation=0)
plt.yticks(np.linspace(0, 1, 11), labels=[f'{i:0.0%}' for i in np.linspace(0, 1, 11)])
plt.grid(axis='x', alpha=.5)
plt.tight_layout();
```
## Which street has the most Street Sweeping citations?
The characteristics of the top 3 streets:
1. Vehicles are parked bumper to bumper leaving few parking spaces available
2. Parking spaces have a set time limit
```
df_citations['street_name'] = df_citations.location.str.replace('^[\d+]{2,}', '').str.strip()
sns.set_context('talk')
# Removing the street number and white space from the address
df_citations.street_name.value_counts().nlargest(3).plot.barh(figsize=(16, 6))
# Chart formatting
plt.title('Streets with the Most Street Sweeping Citations', fontsize=24)
plt.xlabel('# of Citations');
```
### __Abbot Kinney Blvd: "Small Boutiques, No Parking"__
> [Abbot Kinney Blvd on Google Maps](https://www.google.com/maps/@33.9923689,-118.4731719,3a,75y,112.99h,91.67t/data=!3m6!1e1!3m4!1sKD3cG40eGmdWxhwqLD1BvA!2e0!7i16384!8i8192)
<img src="./visuals/abbot.png" alt="Abbot" style="width: 450px;" align="left"/>
- Near Venice Beach
- Small businesses and name brand stores line both sides of the street
- Little to no parking in this area
- Residential area inland
- Multiplex style dwellings with available parking spaces
- Weekly Street Sweeping on Monday from 7:30 am - 9:30 am
### __Clinton Street: "Packed Street"__
> [Clinton Street on Google Maps](https://www.google.com/maps/@34.0816611,-118.3306842,3a,75y,70.72h,57.92t/data=!3m9!1e1!3m7!1sdozFgC7Ms3EvaOF4-CeNAg!2e0!7i16384!8i8192!9m2!1b1!2i37)
<img src="./visuals/clinton.png" alt="Clinton" style="width: 600px;" align="Left"/>
- All parking spaces on the street are filled
- Residential Area
- Weekly Street Sweeping on Friday from 8:00 am - 11:00 am
### __Kelton Ave: "2 Hour Time Limit"__
> [Kelton Ave on Google Maps](https://www.google.com/maps/place/Kelton+Ave,+Los+Angeles,+CA/@34.0475262,-118.437594,3a,49.9y,183.92h,85.26t/data=!3m9!1e1!3m7!1s5VICHNYMVEk9utaV5egFYg!2e0!7i16384!8i8192!9m2!1b1!2i25!4m5!3m4!1s0x80c2bb7efb3a05eb:0xe155071f3fe49df3!8m2!3d34.0542999!4d-118.4434919)
<img src="./visuals/kelton.png" width="600" height="600" align="left"/>
- Most parking spaces on this street are available. This is due to the strict 2 hour time limit for parked vehicles without the proper exception permit.
- Multiplex, Residential Area
- Weekly Street Sweeping on Thursday from 10:00 am - 1:00 pm
- Weekly Street Sweeping on Friday from 8:00 am - 10:00 am
## Which street has the most Street Sweeping citations, given the day of the week?
- __Abbot Kinney Blvd__ is the most cited street on __Monday and Tuesday__
- __4th Street East__ is the most cited street on __Saturday and Sunday__
```
# Group by the day of the week and street name
df_day_street = df_citations.groupby(by=['day_of_week', 'street_name'])\
.size()\
.sort_values()\
.groupby(level=0)\
.tail(1)\
.reset_index()\
.rename(columns={0:'count'})
# Create a new column to sort the values by the day of the
# week starting with Monday
df_day_street['order'] = [5, 6, 4, 3, 0, 2, 1]
# Display the street with the most street sweeping citations
# given the day of the week.
df_day_street.sort_values('order').set_index('order')
```
## Which Agencies issue the most street sweeping citations?
The Department of Transportation's __Western, Hollywood, and Valley__ subdivisions issue the most street sweeping citations.
```
sns.set_context('talk')
df_citations.agency.value_counts().nlargest(5).plot.barh(figsize=(12, 6));
# plt.axhspan(2.5, 5, facecolor='0.5', alpha=.8)
plt.title('Agencies With the Most Street Sweeper Citations')
plt.xlabel('# of Citations (in thousands)')
plt.xticks(np.arange(0, 400_001, 100_000), list(np.arange(0, 401, 100)))
plt.yticks([0, 1, 2, 3, 4], labels=['DOT-WESTERN',
'DOT-HOLLYWOOD',
'DOT-VALLEY',
'DOT-SOUTHERN',
'DOT-CENTRAL']);
```
When taking routes into consideration, __"Western"__ Subdivision, route 00500, has issued the most street sweeping citations.
- Is route 00500 larger than other street sweeping routes?
```
top_3_routes = df_citations.groupby(['agency', 'route'])\
.size()\
.nlargest(3)\
.sort_index()\
.rename('num_citations')\
.reset_index()\
.sort_values(by='num_citations', ascending=False)
top_3_routes.agency = ["DOT-WESTERN", "DOT-SOUTHERN", "DOT-CENTRAL"]
data = top_3_routes.set_index(['agency', 'route'])
data.plot(kind='barh', stacked=True, figsize=(12, 6), legend=None)
plt.title("Agency-Route ID's with the most Street Sweeping Citations")
plt.ylabel('')
plt.xlabel('# of Citations (in thousands)')
plt.xticks(np.arange(0, 70_001, 10_000), [str(i) for i in np.arange(0, 71, 10)]);
df_citations['issue_time_num'] = df_citations.issue_time.str.replace(":00", '')
df_citations['issue_time_num'] = df_citations.issue_time_num.str.replace(':', '').astype(np.int)
```
## What is the weekly distibution of citation times?
```
sns.set_context('talk')
plt.figure(figsize=(13, 12))
sns.boxplot(data=df_citations,
x="day_of_week",
y="issue_time_num",
order=["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"],
whis=3);
plt.title("Distribution Citation Issue Times Throughout the Week")
plt.xlabel('')
plt.ylabel('Issue Time (24HR)')
plt.yticks(np.arange(0, 2401, 200), [str(i) + ":00" for i in range(0, 25, 2)]);
```
| github_jupyter |
```
from keras.models import load_model
import pandas as pd
import keras.backend as K
from keras.callbacks import LearningRateScheduler
from keras.callbacks import Callback
import math
import numpy as np
def coeff_r2(y_true, y_pred):
from keras import backend as K
SS_res = K.sum(K.square( y_true-y_pred ))
SS_tot = K.sum(K.square( y_true - K.mean(y_true) ) )
return ( 1 - SS_res/(SS_tot + K.epsilon()) )
model = load_model('./FPV_ANN_tabulated_Standard_4Res_500n.H5')
# model = load_model('../tmp/large_next.h5',custom_objects={'coeff_r2':coeff_r2})
# model = load_model('../tmp/calc_100_3_3_cbrt.h5', custom_objects={'coeff_r2':coeff_r2})
model.summary()
import numpy as np
import pandas as pd
from sklearn.preprocessing import MinMaxScaler, StandardScaler
class data_scaler(object):
def __init__(self):
self.norm = None
self.norm_1 = None
self.std = None
self.case = None
self.scale = 1
self.bias = 1e-20
# self.bias = 1
self.switcher = {
'min_std': 'min_std',
'std2': 'std2',
'std_min':'std_min',
'min': 'min',
'no':'no',
'log': 'log',
'log_min':'log_min',
'log_std':'log_std',
'log2': 'log2',
'sqrt_std': 'sqrt_std',
'cbrt_std': 'cbrt_std',
'nrt_std':'nrt_std',
'tan': 'tan'
}
def fit_transform(self, input_data, case):
self.case = case
if self.switcher.get(self.case) == 'min_std':
self.norm = MinMaxScaler()
self.std = StandardScaler()
out = self.norm.fit_transform(input_data)
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'std2':
self.std = StandardScaler()
out = self.std.fit_transform(input_data)
if self.switcher.get(self.case) == 'std_min':
self.norm = MinMaxScaler()
self.std = StandardScaler()
out = self.std.fit_transform(input_data)
out = self.norm.fit_transform(out)
if self.switcher.get(self.case) == 'min':
self.norm = MinMaxScaler()
out = self.norm.fit_transform(input_data)
if self.switcher.get(self.case) == 'no':
self.norm = MinMaxScaler()
self.std = StandardScaler()
out = input_data
if self.switcher.get(self.case) == 'log_min':
out = - np.log(np.asarray(input_data / self.scale) + self.bias)
self.norm = MinMaxScaler()
out = self.norm.fit_transform(out)
if self.switcher.get(self.case) == 'log_std':
out = - np.log(np.asarray(input_data / self.scale) + self.bias)
self.std = StandardScaler()
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'log2':
self.norm = MinMaxScaler()
self.std = StandardScaler()
out = self.norm.fit_transform(input_data)
out = np.log(np.asarray(out) + self.bias)
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'sqrt_std':
out = np.sqrt(np.asarray(input_data / self.scale))
self.std = StandardScaler()
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'cbrt_std':
out = np.cbrt(np.asarray(input_data / self.scale))
self.std = StandardScaler()
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'nrt_std':
out = np.power(np.asarray(input_data / self.scale),1/4)
self.std = StandardScaler()
out = self.std.fit_transform(out)
if self.switcher.get(self.case) == 'tan':
self.norm = MaxAbsScaler()
self.std = StandardScaler()
out = self.std.fit_transform(input_data)
out = self.norm.fit_transform(out)
out = np.tan(out / (2 * np.pi + self.bias))
return out
def transform(self, input_data):
if self.switcher.get(self.case) == 'min_std':
out = self.norm.transform(input_data)
out = self.std.transform(out)
if self.switcher.get(self.case) == 'std2':
out = self.std.transform(input_data)
if self.switcher.get(self.case) == 'std_min':
out = self.std.transform(input_data)
out = self.norm.transform(out)
if self.switcher.get(self.case) == 'min':
out = self.norm.transform(input_data)
if self.switcher.get(self.case) == 'no':
out = input_data
if self.switcher.get(self.case) == 'log_min':
out = - np.log(np.asarray(input_data / self.scale) + self.bias)
out = self.norm.transform(out)
if self.switcher.get(self.case) == 'log_std':
out = - np.log(np.asarray(input_data / self.scale) + self.bias)
out = self.std.transform(out)
if self.switcher.get(self.case) == 'log2':
out = self.norm.transform(input_data)
out = np.log(np.asarray(out) + self.bias)
out = self.std.transform(out)
if self.switcher.get(self.case) == 'sqrt_std':
out = np.sqrt(np.asarray(input_data / self.scale))
out = self.std.transform(out)
if self.switcher.get(self.case) == 'cbrt_std':
out = np.cbrt(np.asarray(input_data / self.scale))
out = self.std.transform(out)
if self.switcher.get(self.case) == 'nrt_std':
out = np.power(np.asarray(input_data / self.scale),1/4)
out = self.std.transform(out)
if self.switcher.get(self.case) == 'tan':
out = self.std.transform(input_data)
out = self.norm.transform(out)
out = np.tan(out / (2 * np.pi + self.bias))
return out
def inverse_transform(self, input_data):
if self.switcher.get(self.case) == 'min_std':
out = self.std.inverse_transform(input_data)
out = self.norm.inverse_transform(out)
if self.switcher.get(self.case) == 'std2':
out = self.std.inverse_transform(input_data)
if self.switcher.get(self.case) == 'std_min':
out = self.norm.inverse_transform(input_data)
out = self.std.inverse_transform(out)
if self.switcher.get(self.case) == 'min':
out = self.norm.inverse_transform(input_data)
if self.switcher.get(self.case) == 'no':
out = input_data
if self.switcher.get(self.case) == 'log_min':
out = self.norm.inverse_transform(input_data)
out = (np.exp(-out) - self.bias) * self.scale
if self.switcher.get(self.case) == 'log_std':
out = self.std.inverse_transform(input_data)
out = (np.exp(-out) - self.bias) * self.scale
if self.switcher.get(self.case) == 'log2':
out = self.std.inverse_transform(input_data)
out = np.exp(out) - self.bias
out = self.norm.inverse_transform(out)
if self.switcher.get(self.case) == 'sqrt_std':
out = self.std.inverse_transform(input_data)
out = np.power(out,2) * self.scale
if self.switcher.get(self.case) == 'cbrt_std':
out = self.std.inverse_transform(input_data)
out = np.power(out,3) * self.scale
if self.switcher.get(self.case) == 'nrt_std':
out = self.std.inverse_transform(input_data)
out = np.power(out,4) * self.scale
if self.switcher.get(self.case) == 'tan':
out = (2 * np.pi + self.bias) * np.arctan(input_data)
out = self.norm.inverse_transform(out)
out = self.std.inverse_transform(out)
return out
def read_h5_data(fileName, input_features, labels):
df = pd.read_hdf(fileName)
# df = df[df['f']<0.45]
# for i in range(5):
# pv_101=df[df['pv']==1]
# pv_101['pv']=pv_101['pv']+0.002*(i+1)
# df = pd.concat([df,pv_101])
input_df=df[input_features]
in_scaler = data_scaler()
input_np = in_scaler.fit_transform(input_df.values,'std2')
label_df=df[labels].clip(0)
# if 'PVs' in labels:
# label_df['PVs']=np.log(label_df['PVs']+1)
out_scaler = data_scaler()
label_np = out_scaler.fit_transform(label_df.values,'cbrt_std')
return input_np, label_np, df, in_scaler, out_scaler
# labels = ['CH4','O2','H2O','CO','CO2','T','PVs','psi','mu','alpha']
# labels = ['T','PVs']
# labels = ['T','CH4','O2','CO2','CO','H2O','H2','OH','psi']
# labels = ['CH2OH','HNCO','CH3OH', 'CH2CHO', 'CH2O', 'C3H8', 'HNO', 'NH2', 'HCN']
# labels = np.random.choice(col_labels,20,replace=False).tolist()
# labels.append('PVs')
# labels = col_labels
# labels= ['CH4', 'CH2O', 'CH3O', 'H', 'O2', 'H2', 'O', 'OH', 'H2O', 'HO2', 'H2O2',
# 'C', 'CH', 'CH2', 'CH2(S)', 'CH3', 'CO', 'CO2', 'HCO', 'CH2OH', 'CH3OH',
# 'C2H', 'C2H2', 'C2H3', 'C2H4', 'C2H5', 'C2H6', 'HCCO', 'CH2CO', 'HCCOH',
# 'N', 'NH', 'NH2', 'NH3', 'NNH', 'NO', 'NO2', 'N2O', 'HNO', 'CN', 'HCN',
# 'H2CN', 'HCNN', 'HCNO', 'HNCO', 'NCO', 'N2', 'AR', 'C3H7', 'C3H8', 'CH2CHO', 'CH3CHO', 'T', 'PVs']
# labels.remove('AR')
# labels.remove('N2')
labels = ['H2', 'H', 'O', 'O2', 'OH', 'H2O', 'HO2', 'CH3', 'CH4', 'CO', 'CO2', 'CH2O', 'N2', 'T', 'PVs']
print(labels)
input_features=['f','zeta','pv']
# read in the data
x_input, y_label, df, in_scaler, out_scaler = read_h5_data('../data/tables_of_fgm_psi.h5',input_features=input_features, labels = labels)
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x_input,y_label, test_size=0.01)
x_test_df = pd.DataFrame(in_scaler.inverse_transform(x_test),columns=input_features)
y_test_df = pd.DataFrame(out_scaler.inverse_transform(y_test),columns=labels)
predict_val = model.predict(x_test,batch_size=1024*8)
# predict_val = model.predict(x_test,batch_size=1024*8)
predict_df = pd.DataFrame(out_scaler.inverse_transform(predict_val), columns=labels)
test_data=pd.concat([x_test_df,y_test_df],axis=1)
pred_data=pd.concat([x_test_df,predict_df],axis=1)
!rm sim_check.h5
test_data.to_hdf('sim_check.h5',key='test')
pred_data.to_hdf('sim_check.h5',key='pred')
df_test=pd.read_hdf('sim_check.h5',key='test')
df_pred=pd.read_hdf('sim_check.h5',key='pred')
zeta_level=list(set(df_test['zeta']))
zeta_level.sort()
res_sum=pd.DataFrame()
r2s=[]
r2s_i=[]
names=[]
maxs_0=[]
maxs_9=[]
for r2,name in zip(r2_score(df_test,df_pred,multioutput='raw_values'),df_test.columns):
names.append(name)
r2s.append(r2)
maxs_0.append(df_test[df_test['zeta']==zeta_level[0]][name].max())
maxs_9.append(df_test[df_test['zeta']==zeta_level[8]][name].max())
for i in zeta_level:
r2s_i.append(r2_score(df_pred[df_pred['zeta']==i][name],
df_test[df_test['zeta']==i][name]))
res_sum['name']=names
# res_sum['max_0']=maxs_0
# res_sum['max_9']=maxs_9
res_sum['z_scale']=[m_9/(m_0+1e-20) for m_9,m_0 in zip(maxs_9,maxs_0)]
# res_sum['r2']=r2s
tmp=np.asarray(r2s_i).reshape(-1,10)
for idx,z in enumerate(zeta_level):
res_sum['r2s_'+str(z)]=tmp[:,idx]
res_sum[3:]
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x_input,y_label, test_size=0.01)
x_test_df = pd.DataFrame(in_scaler.inverse_transform(x_test),columns=input_features)
y_test_df = pd.DataFrame(out_scaler.inverse_transform(y_test),columns=labels)
predict_val = student_model.predict(x_test,batch_size=1024*8)
# predict_val = model.predict(x_test,batch_size=1024*8)
predict_df = pd.DataFrame(out_scaler.inverse_transform(predict_val), columns=labels)
test_data=pd.concat([x_test_df,y_test_df],axis=1)
pred_data=pd.concat([x_test_df,predict_df],axis=1)
!rm sim_check.h5
test_data.to_hdf('sim_check.h5',key='test')
pred_data.to_hdf('sim_check.h5',key='pred')
df_test=pd.read_hdf('sim_check.h5',key='test')
df_pred=pd.read_hdf('sim_check.h5',key='pred')
zeta_level=list(set(df_test['zeta']))
zeta_level.sort()
res_sum=pd.DataFrame()
r2s=[]
r2s_i=[]
names=[]
maxs_0=[]
maxs_9=[]
for r2,name in zip(r2_score(df_test,df_pred,multioutput='raw_values'),df_test.columns):
names.append(name)
r2s.append(r2)
maxs_0.append(df_test[df_test['zeta']==zeta_level[0]][name].max())
maxs_9.append(df_test[df_test['zeta']==zeta_level[8]][name].max())
for i in zeta_level:
r2s_i.append(r2_score(df_pred[df_pred['zeta']==i][name],
df_test[df_test['zeta']==i][name]))
res_sum['name']=names
# res_sum['max_0']=maxs_0
# res_sum['max_9']=maxs_9
res_sum['z_scale']=[m_9/(m_0+1e-20) for m_9,m_0 in zip(maxs_9,maxs_0)]
# res_sum['r2']=r2s
tmp=np.asarray(r2s_i).reshape(-1,10)
for idx,z in enumerate(zeta_level):
res_sum['r2s_'+str(z)]=tmp[:,idx]
res_sum[3:]
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x_input,y_label, test_size=0.01)
x_test_df = pd.DataFrame(in_scaler.inverse_transform(x_test),columns=input_features)
y_test_df = pd.DataFrame(out_scaler.inverse_transform(y_test),columns=labels)
predict_val = model.predict(x_test,batch_size=1024*8)
# predict_val = model.predict(x_test,batch_size=1024*8)
predict_df = pd.DataFrame(out_scaler.inverse_transform(predict_val), columns=labels)
test_data=pd.concat([x_test_df,y_test_df],axis=1)
pred_data=pd.concat([x_test_df,predict_df],axis=1)
!rm sim_check.h5
test_data.to_hdf('sim_check.h5',key='test')
pred_data.to_hdf('sim_check.h5',key='pred')
df_test=pd.read_hdf('sim_check.h5',key='test')
df_pred=pd.read_hdf('sim_check.h5',key='pred')
zeta_level=list(set(df_test['zeta']))
zeta_level.sort()
res_sum=pd.DataFrame()
r2s=[]
r2s_i=[]
names=[]
maxs_0=[]
maxs_9=[]
for r2,name in zip(r2_score(df_test,df_pred,multioutput='raw_values'),df_test.columns):
names.append(name)
r2s.append(r2)
maxs_0.append(df_test[df_test['zeta']==zeta_level[0]][name].max())
maxs_9.append(df_test[df_test['zeta']==zeta_level[8]][name].max())
for i in zeta_level:
r2s_i.append(r2_score(df_pred[df_pred['zeta']==i][name],
df_test[df_test['zeta']==i][name]))
res_sum['name']=names
# res_sum['max_0']=maxs_0
# res_sum['max_9']=maxs_9
res_sum['z_scale']=[m_9/(m_0+1e-20) for m_9,m_0 in zip(maxs_9,maxs_0)]
# res_sum['r2']=r2s
tmp=np.asarray(r2s_i).reshape(-1,10)
for idx,z in enumerate(zeta_level):
res_sum['r2s_'+str(z)]=tmp[:,idx]
res_sum[3:]
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x_input,y_label, test_size=0.01)
x_test_df = pd.DataFrame(in_scaler.inverse_transform(x_test),columns=input_features)
y_test_df = pd.DataFrame(out_scaler.inverse_transform(y_test),columns=labels)
predict_val = model.predict(x_test,batch_size=1024*8)
# predict_val = model.predict(x_test,batch_size=1024*8)
predict_df = pd.DataFrame(out_scaler.inverse_transform(predict_val), columns=labels)
test_data=pd.concat([x_test_df,y_test_df],axis=1)
pred_data=pd.concat([x_test_df,predict_df],axis=1)
!rm sim_check.h5
test_data.to_hdf('sim_check.h5',key='test')
pred_data.to_hdf('sim_check.h5',key='pred')
df_test=pd.read_hdf('sim_check.h5',key='test')
df_pred=pd.read_hdf('sim_check.h5',key='pred')
zeta_level=list(set(df_test['zeta']))
zeta_level.sort()
res_sum=pd.DataFrame()
r2s=[]
r2s_i=[]
names=[]
maxs_0=[]
maxs_9=[]
for r2,name in zip(r2_score(df_test,df_pred,multioutput='raw_values'),df_test.columns):
names.append(name)
r2s.append(r2)
maxs_0.append(df_test[df_test['zeta']==zeta_level[0]][name].max())
maxs_9.append(df_test[df_test['zeta']==zeta_level[8]][name].max())
for i in zeta_level:
r2s_i.append(r2_score(df_pred[df_pred['zeta']==i][name],
df_test[df_test['zeta']==i][name]))
res_sum['name']=names
# res_sum['max_0']=maxs_0
# res_sum['max_9']=maxs_9
res_sum['z_scale']=[m_9/(m_0+1e-20) for m_9,m_0 in zip(maxs_9,maxs_0)]
# res_sum['r2']=r2s
tmp=np.asarray(r2s_i).reshape(-1,10)
for idx,z in enumerate(zeta_level):
res_sum['r2s_'+str(z)]=tmp[:,idx]
res_sum[3:]
#@title import plotly
import plotly.plotly as py
import numpy as np
from plotly.offline import init_notebook_mode, iplot
# from plotly.graph_objs import Contours, Histogram2dContour, Marker, Scatter
import plotly.graph_objs as go
def configure_plotly_browser_state():
import IPython
display(IPython.core.display.HTML('''
<script src="/static/components/requirejs/require.js"></script>
<script>
requirejs.config({
paths: {
base: '/static/base',
plotly: 'https://cdn.plot.ly/plotly-1.5.1.min.js?noext',
},
});
</script>
'''))
#@title Default title text
# species = np.random.choice(labels)
species = 'HNO' #@param {type:"string"}
z_level = 0 #@param {type:"integer"}
# configure_plotly_browser_state()
# init_notebook_mode(connected=False)
from sklearn.metrics import r2_score
df_t=df_test[df_test['zeta']==zeta_level[z_level]].sample(frac=1)
# df_p=df_pred.loc[df_pred['zeta']==zeta_level[1]].sample(frac=0.1)
df_p=df_pred.loc[df_t.index]
# error=(df_p[species]-df_t[species])
error=(df_p[species]-df_t[species])/(df_p[species]+df_t[species])
r2=round(r2_score(df_p[species],df_t[species]),4)
print(species,'r2:',r2,'max:',df_t[species].max())
fig_db = {
'data': [
{'name':'test data from table',
'x': df_t['f'],
'y': df_t['pv'],
'z': df_t[species],
'type':'scatter3d',
'mode': 'markers',
'marker':{
'size':1
}
},
{'name':'prediction from neural networks',
'x': df_p['f'],
'y': df_p['pv'],
'z': df_p[species],
'type':'scatter3d',
'mode': 'markers',
'marker':{
'size':1
},
},
{'name':'error in difference',
'x': df_p['f'],
'y': df_p['pv'],
'z': error,
'type':'scatter3d',
'mode': 'markers',
'marker':{
'size':1
},
}
],
'layout': {
'scene':{
'xaxis': {'title':'mixture fraction'},
'yaxis': {'title':'progress variable'},
'zaxis': {'title': species+'_r2:'+str(r2)}
}
}
}
# iplot(fig_db, filename='multiple-scatter')
iplot(fig_db)
%matplotlib inline
import matplotlib.pyplot as plt
z=0.22
sp='HNO'
plt.plot(df[(df.pv==1)&(df.zeta==z)]['f'],df[(df.pv==0.9)&(df.zeta==z)][sp],'rd')
from keras.models import Model
from keras.layers import Dense, Input, Dropout
n_neuron = 100
# %%
print('set up student network')
# ANN parameters
dim_input = x_train.shape[1]
dim_label = y_train.shape[1]
batch_norm = False
# This returns a tensor
inputs = Input(shape=(dim_input,),name='input_1')
# a layer instance is callable on a tensor, and returns a tensor
x = Dense(n_neuron, activation='relu')(inputs)
x = Dense(n_neuron, activation='relu')(x)
x = Dense(n_neuron, activation='relu')(x)
# x = Dropout(0.1)(x)
predictions = Dense(dim_label, activation='linear', name='output_1')(x)
student_model = Model(inputs=inputs, outputs=predictions)
student_model.summary()
import keras.backend as K
from keras.callbacks import LearningRateScheduler
import math
def cubic_loss(y_true, y_pred):
return K.mean(K.square(y_true - y_pred)*K.abs(y_true - y_pred), axis=-1)
def coeff_r2(y_true, y_pred):
from keras import backend as K
SS_res = K.sum(K.square( y_true-y_pred ))
SS_tot = K.sum(K.square( y_true - K.mean(y_true) ) )
return ( 1 - SS_res/(SS_tot + K.epsilon()) )
def step_decay(epoch):
initial_lrate = 0.002
drop = 0.5
epochs_drop = 1000.0
lrate = initial_lrate * math.pow(drop,math.floor((1+epoch)/epochs_drop))
return lrate
lrate = LearningRateScheduler(step_decay)
class SGDRScheduler(Callback):
'''Cosine annealing learning rate scheduler with periodic restarts.
# Usage
```python
schedule = SGDRScheduler(min_lr=1e-5,
max_lr=1e-2,
steps_per_epoch=np.ceil(epoch_size/batch_size),
lr_decay=0.9,
cycle_length=5,
mult_factor=1.5)
model.fit(X_train, Y_train, epochs=100, callbacks=[schedule])
```
# Arguments
min_lr: The lower bound of the learning rate range for the experiment.
max_lr: The upper bound of the learning rate range for the experiment.
steps_per_epoch: Number of mini-batches in the dataset. Calculated as `np.ceil(epoch_size/batch_size)`.
lr_decay: Reduce the max_lr after the completion of each cycle.
Ex. To reduce the max_lr by 20% after each cycle, set this value to 0.8.
cycle_length: Initial number of epochs in a cycle.
mult_factor: Scale epochs_to_restart after each full cycle completion.
# References
Blog post: jeremyjordan.me/nn-learning-rate
Original paper: http://arxiv.org/abs/1608.03983
'''
def __init__(self,
min_lr,
max_lr,
steps_per_epoch,
lr_decay=1,
cycle_length=10,
mult_factor=2):
self.min_lr = min_lr
self.max_lr = max_lr
self.lr_decay = lr_decay
self.batch_since_restart = 0
self.next_restart = cycle_length
self.steps_per_epoch = steps_per_epoch
self.cycle_length = cycle_length
self.mult_factor = mult_factor
self.history = {}
def clr(self):
'''Calculate the learning rate.'''
fraction_to_restart = self.batch_since_restart / (self.steps_per_epoch * self.cycle_length)
lr = self.min_lr + 0.5 * (self.max_lr - self.min_lr) * (1 + np.cos(fraction_to_restart * np.pi))
return lr
def on_train_begin(self, logs={}):
'''Initialize the learning rate to the minimum value at the start of training.'''
logs = logs or {}
K.set_value(self.model.optimizer.lr, self.max_lr)
def on_batch_end(self, batch, logs={}):
'''Record previous batch statistics and update the learning rate.'''
logs = logs or {}
self.history.setdefault('lr', []).append(K.get_value(self.model.optimizer.lr))
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
self.batch_since_restart += 1
K.set_value(self.model.optimizer.lr, self.clr())
def on_epoch_end(self, epoch, logs={}):
'''Check for end of current cycle, apply restarts when necessary.'''
if epoch + 1 == self.next_restart:
self.batch_since_restart = 0
self.cycle_length = np.ceil(self.cycle_length * self.mult_factor)
self.next_restart += self.cycle_length
self.max_lr *= self.lr_decay
self.best_weights = self.model.get_weights()
def on_train_end(self, logs={}):
'''Set weights to the values from the end of the most recent cycle for best performance.'''
self.model.set_weights(self.best_weights)
student_model = load_model('student.h5',custom_objects={'coeff_r2':coeff_r2})
model.summary()
gx,gy,gz=np.mgrid[0:1:600j,0:1:10j,0:1:600j]
gx=gx.reshape(-1,1)
gy=gy.reshape(-1,1)
gz=gz.reshape(-1,1)
gm=np.hstack([gx,gy,gz])
gm.shape
from keras.callbacks import ModelCheckpoint
from keras import optimizers
batch_size = 1024*16
epochs = 2000
vsplit = 0.1
loss_type='mse'
adam_op = optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999,epsilon=1e-8, decay=0.0, amsgrad=False)
student_model.compile(loss=loss_type,
# optimizer=adam_op,
optimizer='adam',
metrics=[coeff_r2])
# model.compile(loss=cubic_loss, optimizer=adam_op, metrics=['accuracy'])
# checkpoint (save the best model based validate loss)
!mkdir ./tmp
filepath = "./tmp/student_weights.best.cntk.hdf5"
checkpoint = ModelCheckpoint(filepath,
monitor='val_loss',
verbose=1,
save_best_only=True,
mode='min',
period=20)
epoch_size=x_train.shape[0]
a=0
base=2
clc=2
for i in range(5):
a+=base*clc**(i)
print(a)
epochs,c_len = a,base
schedule = SGDRScheduler(min_lr=1e-5,max_lr=1e-4,
steps_per_epoch=np.ceil(epoch_size/batch_size),
cycle_length=c_len,lr_decay=0.8,mult_factor=2)
callbacks_list = [checkpoint]
# callbacks_list = [checkpoint, schedule]
x_train_teacher = in_scaler.transform(gm)
y_train_teacher = model.predict(x_train_teacher, batch_size=1024*8)
x_train, x_test, y_train, y_test = train_test_split(x_train_teacher,y_train_teacher, test_size=0.01)
# fit the model
history = student_model.fit(
x_train, y_train,
epochs=epochs,
batch_size=batch_size,
validation_split=vsplit,
verbose=2,
callbacks=callbacks_list,
shuffle=True)
student_model.save('student_100_3.h5')
n_res = 501
pv_level = 0.996
f_1 = np.linspace(0,1,n_res)
z_1 = np.zeros(n_res)
pv_1 = np.ones(n_res)*pv_level
case_1 = np.vstack((f_1,z_1,pv_1))
# case_1 = np.vstack((pv_1,z_1,f_1))
case_1 = case_1.T
case_1.shape
out=out_scaler.inverse_transform(model.predict(case_1))
out=pd.DataFrame(out,columns=labels)
sp='PVs'
out.head()
table_val=df[(df.pv==pv_level) & (df.zeta==0)][sp]
table_val.shape
import matplotlib.pyplot as plt
plt.plot(f_1,table_val)
plt.show
plt.plot(f_1,out[sp])
plt.show
df.head()
pv_101=df[df['pv']==1][df['zeta']==0]
pv_101['pv']=pv_101['pv']+0.01
a=pd.concat([pv_101,pv_101])
pv_101.shape
a.shape
a
```
| github_jupyter |
# Training Collaborative Experts on MSR-VTT
This notebook shows how to download code that trains a Collaborative Experts model with GPT-1 + NetVLAD on the MSR-VTT Dataset.
## Setup
* Download Code and Dependencies
* Import Modules
* Download Language Model Weights
* Download Datasets
* Generate Encodings for Dataset Captions
### Code Downloading and Dependency Downloading
* Specify tensorflow version
* Clone repository from Github
* `cd` into the correct directory
* Install the requirements
```
%tensorflow_version 2.x
!git clone https://github.com/googleinterns/via-content-understanding.git
%cd via-content-understanding/videoretrieval/
!pip install -r requirements.txt
!pip install --upgrade tensorflow_addons
```
### Importing Modules
```
import tensorflow as tf
import languagemodels
import train.encoder_datasets
import train.language_model
import experts
import datasets
import datasets.msrvtt.constants
import os
import models.components
import models.encoder
import helper.precomputed_features
from tensorflow_addons.activations import mish
import tensorflow_addons as tfa
import metrics.loss
```
### Language Model Downloading
* Download GPT-1
```
gpt_model = languagemodels.OpenAIGPTModel()
```
### Dataset downloading
* Downlaod Datasets
* Download Precomputed Features
```
datasets.msrvtt_dataset.download_dataset()
```
Note: The system `curl` is more memory efficent than the download function in our codebase, so here `curl` is used rather than the download function in our codebase.
```
url = datasets.msrvtt.constants.features_tar_url
path = datasets.msrvtt.constants.features_tar_path
os.system(f"curl {url} > {path}")
helper.precomputed_features.cache_features(
datasets.msrvtt_dataset,
datasets.msrvtt.constants.expert_to_features,
datasets.msrvtt.constants.features_tar_path,)
```
### Embeddings Generation
* Generate Embeddings for MSR-VTT
* **Note: this will take 20-30 minutes on a colab, depending on the GPU**
```
train.language_model.generate_and_cache_contextual_embeddings(
gpt_model, datasets.msrvtt_dataset)
```
## Training
* Build Train Datasets
* Initialize Models
* Compile Encoders
* Fit Model
* Test Model
### Datasets Generation
```
experts_used = [
experts.i3d,
experts.r2p1d,
experts.resnext,
experts.senet,
experts.speech_expert,
experts.ocr_expert,
experts.audio_expert,
experts.densenet,
experts.face_expert]
train_ds, valid_ds, test_ds = (
train.encoder_datasets.generate_encoder_datasets(
gpt_model, datasets.msrvtt_dataset, experts_used))
```
### Model Initialization
```
class MishLayer(tf.keras.layers.Layer):
def call(self, inputs):
return mish(inputs)
mish(tf.Variable([1.0]))
text_encoder = models.components.TextEncoder(
len(experts_used),
num_netvlad_clusters=28,
ghost_clusters=1,
language_model_dimensionality=768,
encoded_expert_dimensionality=512,
residual_cls_token=False,
)
video_encoder = models.components.VideoEncoder(
num_experts=len(experts_used),
experts_use_netvlad=[False, False, False, False, True, True, True, False, False],
experts_netvlad_shape=[None, None, None, None, 19, 43, 8, None, None],
expert_aggregated_size=512,
encoded_expert_dimensionality=512,
g_mlp_layers=3,
h_mlp_layers=0,
make_activation_layer=MishLayer)
encoder = models.encoder.EncoderForFrozenLanguageModel(
video_encoder,
text_encoder,
0.0938,
[1, 5, 10, 50],
20)
```
### Encoder Compliation
```
def build_optimizer(lr=0.001):
learning_rate_scheduler = tf.keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate=lr,
decay_steps=101,
decay_rate=0.95,
staircase=True)
return tf.keras.optimizers.Adam(learning_rate_scheduler)
encoder.compile(build_optimizer(0.1), metrics.loss.bidirectional_max_margin_ranking_loss)
train_ds_prepared = (train_ds
.shuffle(1000)
.batch(64, drop_remainder=True)
.prefetch(tf.data.experimental.AUTOTUNE))
encoder.video_encoder.trainable = True
encoder.text_encoder.trainable = True
```
### Model fitting
```
encoder.fit(
train_ds_prepared,
epochs=100,
)
```
### Tests
```
captions_per_video = 20
num_videos_upper_bound = 100000
ranks = []
for caption_index in range(captions_per_video):
batch = next(iter(test_ds.shard(captions_per_video, caption_index).batch(
num_videos_upper_bound)))
video_embeddings, text_embeddings, mixture_weights = encoder.forward_pass(
batch, training=False)
similarity_matrix = metrics.loss.build_similarity_matrix(
video_embeddings,
text_embeddings,
mixture_weights,
batch[-1])
rankings = metrics.rankings.compute_ranks(similarity_matrix)
ranks += list(rankings.numpy())
def recall_at_k(ranks, k):
return len(list(filter(lambda i: i <= k, ranks))) / len(ranks)
median_rank = sorted(ranks)[len(ranks)//2]
mean_rank = sum(ranks)/len(ranks)
print(f"Median Rank: {median_rank}")
print(f"Mean Rank: {mean_rank}")
for k in [1, 5, 10, 50]:
recall = recall_at_k(ranks, k)
print(f"R@{k}: {recall}")
```
| github_jupyter |
```
import numpy as np
from scipy.spatial import Delaunay
import networkx as nx
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import pandas
import os
import graphsonchip.graphmaker
from graphsonchip.graphmaker import make_spheroids
from graphsonchip.graphmaker import graph_generation_func
from graphsonchip.graphplotter import graph_plot
```
## Generate small plot
```
cells = make_spheroids.generate_artificial_spheroid(10)['cells']
spheroid = {}
spheroid['cells'] = cells
G = graph_generation_func.generate_voronoi_graph(spheroid, dCells = 0.6)
for ind in G.nodes():
if ind % 2 == 0:
G.add_node(ind, color = 'r')
else:
G.add_node(ind, color = 'b')
graph_plot.network_plot_3D(G)
#plt.savefig('example_code.pdf')
path = r'/Users/gustaveronteix/Documents/Projets/Projets Code/3D-Segmentation-Sebastien/data'
spheroid_data = pandas.read_csv(os.path.join(path, 'spheroid_table_3.csv'))
mapper = {"centroid-0": "z", "centroid-1": "x", "centroid-2": "y"}
spheroid_data = spheroid_data.rename(columns = mapper)
spheroid = pr.single_spheroid_process(spheroid_data)
G = graph.generate_voronoi_graph(spheroid, zRatio = 1, dCells = 20)
for ind in G.nodes():
G.add_node(ind, color ='g')
pos =nx.get_node_attributes(G,'pos')
gp.network_plot_3D(G, 5)
#plt.savefig('Example_image.pdf')
path = r'/Volumes/Multicell/Sebastien/Gustave_Jeremie/spheroid_sample_Francoise.csv'
spheroid_data = pandas.read_csv(path)
spheroid = pr.single_spheroid_process(spheroid_data)
G = graph.generate_voronoi_graph(spheroid, zRatio = 2, dCells = 35)
for ind in G.nodes():
G.add_node(ind, color = 'r')
pos =nx.get_node_attributes(G,'pos')
gp.network_plot_3D(G, 20)
plt.savefig('/Volumes/Multicell/Sebastien/Gustave_Jeremie/spheroid_sample_Francoise.pdf', transparent=True)
```
## Batch analyze the data
```
spheroid_path = './utility/spheroid_sample_1.csv'
spheroid_data = pandas.read_csv(spheroid_path)
spheroid = pr.single_spheroid_process(spheroid_data[spheroid_data['area'] > 200])
G = graph.generate_voronoi_graph(spheroid, zRatio = 2, dCells = 35)
import glob
from collections import defaultdict
degree_frame_Vor = pandas.DataFrame()
i = 0
degree_frame_Geo = pandas.DataFrame()
j = 0
deg_Vor = []
deg_Geo = []
for fname in glob.glob('./utility/*.csv'):
spheroid_data = pandas.read_csv(fname)
spheroid_data['x'] *= 1.25
spheroid_data['y'] *= 1.25
spheroid_data['z'] *= 1.25
spheroid_data = spheroid_data[spheroid_data['area']>200]
spheroid = pr.single_spheroid_process(spheroid_data)
G = generate_voronoi_graph(spheroid, zRatio = 1, dCells = 55)
degree_sequence = sorted([d for n, d in G.degree()], reverse=True)
degreeCount = collections.Counter(degree_sequence)
for key in degreeCount.keys():
N_tot = 0
for k in degreeCount.keys():
N_tot += degreeCount[k]
degree_frame_Vor.loc[i, 'degree'] = key
degree_frame_Vor.loc[i, 'p'] = degreeCount[key]/N_tot
degree_frame_Vor.loc[i, 'fname'] = fname
i += 1
deg_Vor += list(degree_sequence)
G = graph.generate_geometric_graph(spheroid, zRatio = 1, dCells = 26)
degree_sequence = sorted([d for n, d in G.degree()], reverse=True)
degreeCount = collections.Counter(degree_sequence)
for key in degreeCount.keys():
N_tot = 0
for k in degreeCount.keys():
N_tot += degreeCount[k]
degree_frame_Geo.loc[j, 'degree'] = key
degree_frame_Geo.loc[j, 'p'] = degreeCount[key]/N_tot
degree_frame_Geo.loc[j, 'fname'] = fname
j += 1
deg_Geo.append(degreeCount[key])
indx = degree_frame_Vor.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).mean(axis = 1).index
mean = degree_frame_Vor.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).mean(axis = 1).values
std = degree_frame_Vor.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).std(axis = 1).values
indx_geo = degree_frame_Geo.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).mean(axis = 1).index
mean_geo = degree_frame_Geo.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).mean(axis = 1).values
std_geo = degree_frame_Geo.pivot(index = 'degree', columns = 'fname', values = 'p').fillna(0).std(axis = 1).values
import seaborn as sns
sns.set_style('white')
plt.errorbar(indx+0.3, mean, yerr=std,
marker = 's', linestyle = ' ', color = 'b',
label = 'Voronoi')
plt.errorbar(indx_geo-0.3, mean_geo, yerr=std_geo,
marker = 'o', linestyle = ' ', color = 'r',
label = 'Geometric')
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.special import factorial
from scipy.stats import poisson
# the bins should be of integer width, because poisson is an integer distribution
bins = np.arange(25)-0.5
entries, bin_edges, patches = plt.hist(deg_Vor, bins=bins, density=True, label='Data')
# calculate bin centres
bin_middles = 0.5 * (bin_edges[1:] + bin_edges[:-1])
def fit_function(k, lamb):
'''poisson function, parameter lamb is the fit parameter'''
return poisson.pmf(k, lamb)
# fit with curve_fit
parameters, cov_matrix = curve_fit(fit_function, bin_middles, entries)
# plot poisson-deviation with fitted parameter
x_plot = np.arange(0, 25)
plt.plot(
x_plot,
fit_function(x_plot, *parameters),
marker='o', linestyle='',
label='Fit result',
)
plt.legend()
plt.show()
parameters
```
| github_jupyter |
# Model Explainer Example

In this example we will:
* [Describe the project structure](#Project-Structure)
* [Train some models](#Train-Models)
* [Create Tempo artifacts](#Create-Tempo-Artifacts)
* [Run unit tests](#Unit-Tests)
* [Save python environment for our classifier](#Save-Classifier-Environment)
* [Test Locally on Docker](#Test-Locally-on-Docker)
* [Production on Kubernetes via Tempo](#Production-Option-1-(Deploy-to-Kubernetes-with-Tempo))
* [Prodiuction on Kuebrnetes via GitOps](#Production-Option-2-(Gitops))
## Prerequisites
This notebooks needs to be run in the `tempo-examples` conda environment defined below. Create from project root folder:
```bash
conda env create --name tempo-examples --file conda/tempo-examples.yaml
```
## Project Structure
```
!tree -P "*.py" -I "__init__.py|__pycache__" -L 2
```
## Train Models
* This section is where as a data scientist you do your work of training models and creating artfacts.
* For this example we train sklearn and xgboost classification models for the iris dataset.
```
import os
import logging
import numpy as np
import json
import tempo
from tempo.utils import logger
from src.constants import ARTIFACTS_FOLDER
logger.setLevel(logging.ERROR)
logging.basicConfig(level=logging.ERROR)
from src.data import AdultData
data = AdultData()
from src.model import train_model
adult_model = train_model(ARTIFACTS_FOLDER, data)
from src.explainer import train_explainer
train_explainer(ARTIFACTS_FOLDER, data, adult_model)
```
## Create Tempo Artifacts
```
from src.tempo import create_explainer, create_adult_model
sklearn_model = create_adult_model()
Explainer = create_explainer(sklearn_model)
explainer = Explainer()
# %load src/tempo.py
import os
import dill
import numpy as np
from alibi.utils.wrappers import ArgmaxTransformer
from src.constants import ARTIFACTS_FOLDER, EXPLAINER_FOLDER, MODEL_FOLDER
from tempo.serve.metadata import ModelFramework
from tempo.serve.model import Model
from tempo.serve.pipeline import PipelineModels
from tempo.serve.utils import pipeline, predictmethod
def create_adult_model() -> Model:
sklearn_model = Model(
name="income-sklearn",
platform=ModelFramework.SKLearn,
local_folder=os.path.join(ARTIFACTS_FOLDER, MODEL_FOLDER),
uri="gs://seldon-models/test/income/model",
)
return sklearn_model
def create_explainer(model: Model):
@pipeline(
name="income-explainer",
uri="s3://tempo/explainer/pipeline",
local_folder=os.path.join(ARTIFACTS_FOLDER, EXPLAINER_FOLDER),
models=PipelineModels(sklearn=model),
)
class ExplainerPipeline(object):
def __init__(self):
pipeline = self.get_tempo()
models_folder = pipeline.details.local_folder
explainer_path = os.path.join(models_folder, "explainer.dill")
with open(explainer_path, "rb") as f:
self.explainer = dill.load(f)
def update_predict_fn(self, x):
if np.argmax(self.models.sklearn(x).shape) == 0:
self.explainer.predictor = self.models.sklearn
self.explainer.samplers[0].predictor = self.models.sklearn
else:
self.explainer.predictor = ArgmaxTransformer(self.models.sklearn)
self.explainer.samplers[0].predictor = ArgmaxTransformer(self.models.sklearn)
@predictmethod
def explain(self, payload: np.ndarray, parameters: dict) -> str:
print("Explain called with ", parameters)
self.update_predict_fn(payload)
explanation = self.explainer.explain(payload, **parameters)
return explanation.to_json()
# explainer = ExplainerPipeline()
# return sklearn_model, explainer
return ExplainerPipeline
```
## Save Explainer
```
!ls artifacts/explainer/conda.yaml
tempo.save(Explainer)
```
## Test Locally on Docker
Here we test our models using production images but running locally on Docker. This allows us to ensure the final production deployed model will behave as expected when deployed.
```
from tempo import deploy_local
remote_model = deploy_local(explainer)
r = json.loads(remote_model.predict(payload=data.X_test[0:1], parameters={"threshold":0.90}))
print(r["data"]["anchor"])
r = json.loads(remote_model.predict(payload=data.X_test[0:1], parameters={"threshold":0.99}))
print(r["data"]["anchor"])
remote_model.undeploy()
```
## Production Option 1 (Deploy to Kubernetes with Tempo)
* Here we illustrate how to run the final models in "production" on Kubernetes by using Tempo to deploy
### Prerequisites
Create a Kind Kubernetes cluster with Minio and Seldon Core installed using Ansible as described [here](https://tempo.readthedocs.io/en/latest/overview/quickstart.html#kubernetes-cluster-with-seldon-core).
```
!kubectl apply -f k8s/rbac -n production
from tempo.examples.minio import create_minio_rclone
import os
create_minio_rclone(os.getcwd()+"/rclone-minio.conf")
tempo.upload(sklearn_model)
tempo.upload(explainer)
from tempo.serve.metadata import SeldonCoreOptions
runtime_options = SeldonCoreOptions(**{
"remote_options": {
"namespace": "production",
"authSecretName": "minio-secret"
}
})
from tempo import deploy_remote
remote_model = deploy_remote(explainer, options=runtime_options)
r = json.loads(remote_model.predict(payload=data.X_test[0:1], parameters={"threshold":0.95}))
print(r["data"]["anchor"])
remote_model.undeploy()
```
## Production Option 2 (Gitops)
* We create yaml to provide to our DevOps team to deploy to a production cluster
* We add Kustomize patches to modify the base Kubernetes yaml created by Tempo
```
from tempo import manifest
from tempo.serve.metadata import SeldonCoreOptions
runtime_options = SeldonCoreOptions(**{
"remote_options": {
"namespace": "production",
"authSecretName": "minio-secret"
}
})
yaml_str = manifest(explainer, options=runtime_options)
with open(os.getcwd()+"/k8s/tempo.yaml","w") as f:
f.write(yaml_str)
!kustomize build k8s
```
| github_jupyter |
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License").
# Neural Machine Translation with Attention
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/tutorials/sequences/_nmt.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />
Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/r2/tutorials/sequences/_nmt.ipynb">
<img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />
View source on GitHub</a>
</td>
</table>
# This notebook is still under construction! Please come back later.
This notebook trains a sequence to sequence (seq2seq) model for Spanish to English translation using TF 2.0 APIs. This is an advanced example that assumes some knowledge of sequence to sequence models.
After training the model in this notebook, you will be able to input a Spanish sentence, such as *"¿todavia estan en casa?"*, and return the English translation: *"are you still at home?"*
The translation quality is reasonable for a toy example, but the generated attention plot is perhaps more interesting. This shows which parts of the input sentence has the model's attention while translating:
<img src="https://tensorflow.org/images/spanish-english.png" alt="spanish-english attention plot">
Note: This example takes approximately 10 mintues to run on a single P100 GPU.
```
import collections
import io
import itertools
import os
import random
import re
import time
import unicodedata
import numpy as np
import tensorflow as tf
assert tf.__version__.startswith('2')
import matplotlib.pyplot as plt
print(tf.__version__)
```
## Download and prepare the dataset
We'll use a language dataset provided by http://www.manythings.org/anki/. This dataset contains language translation pairs in the format:
```
May I borrow this book? ¿Puedo tomar prestado este libro?
```
There are a variety of languages available, but we'll use the English-Spanish dataset. For convenience, we've hosted a copy of this dataset on Google Cloud, but you can also download your own copy. After downloading the dataset, here are the steps we'll take to prepare the data:
1. Clean the sentences by removing special characters.
1. Add a *start* and *end* token to each sentence.
1. Create a word index and reverse word index (dictionaries mapping from word → id and id → word).
1. Pad each sentence to a maximum length.
```
# TODO(brianklee): This preprocessing should ideally be implemented in TF
# because preprocessing should be exported as part of the SavedModel.
# Converts the unicode file to ascii
# https://stackoverflow.com/a/518232/2809427
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
START_TOKEN = u'<start>'
END_TOKEN = u'<end>'
def preprocess_sentence(w):
# remove accents; lowercase everything
w = unicode_to_ascii(w.strip()).lower()
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# https://stackoverflow.com/a/3645931/3645946
w = re.sub(r'([?.!,¿])', r' \1 ', w)
# replacing everything with space except (a-z, '.', '?', '!', ',')
w = re.sub(r'[^a-z?.!,¿]+', ' ', w)
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
en_sentence = u"May I borrow this book?"
sp_sentence = u"¿Puedo tomar prestado este libro?"
print(preprocess_sentence(en_sentence))
print(preprocess_sentence(sp_sentence))
```
Training on the complete dataset of >100,000 sentences will take a long time. To train faster, we can limit the size of the dataset (of course, translation quality degrades with less data).
```
def load_anki_data(num_examples=None):
# Download the file
path_to_zip = tf.keras.utils.get_file(
'spa-eng.zip', origin='http://download.tensorflow.org/data/spa-eng.zip',
extract=True)
path_to_file = os.path.dirname(path_to_zip) + '/spa-eng/spa.txt'
with io.open(path_to_file, 'rb') as f:
lines = f.read().decode('utf8').strip().split('\n')
# Data comes as tab-separated strings; one per line.
eng_spa_pairs = [[preprocess_sentence(w) for w in line.split('\t')] for line in lines]
# The translations file is ordered from shortest to longest, so slicing from
# the front will select the shorter examples. This also speeds up training.
if num_examples is not None:
eng_spa_pairs = eng_spa_pairs[:num_examples]
eng_sentences, spa_sentences = zip(*eng_spa_pairs)
eng_tokenizer = tf.keras.preprocessing.text.Tokenizer(filters='')
spa_tokenizer = tf.keras.preprocessing.text.Tokenizer(filters='')
eng_tokenizer.fit_on_texts(eng_sentences)
spa_tokenizer.fit_on_texts(spa_sentences)
return (eng_spa_pairs, eng_tokenizer, spa_tokenizer)
NUM_EXAMPLES = 30000
sentence_pairs, english_tokenizer, spanish_tokenizer = load_anki_data(NUM_EXAMPLES)
# Turn our english/spanish pairs into TF Datasets by mapping words -> integers.
def make_dataset(eng_spa_pairs, eng_tokenizer, spa_tokenizer):
eng_sentences, spa_sentences = zip(*eng_spa_pairs)
eng_ints = eng_tokenizer.texts_to_sequences(eng_sentences)
spa_ints = spa_tokenizer.texts_to_sequences(spa_sentences)
padded_eng_ints = tf.keras.preprocessing.sequence.pad_sequences(
eng_ints, padding='post')
padded_spa_ints = tf.keras.preprocessing.sequence.pad_sequences(
spa_ints, padding='post')
dataset = tf.data.Dataset.from_tensor_slices((padded_eng_ints, padded_spa_ints))
return dataset
# Train/test split
train_size = int(len(sentence_pairs) * 0.8)
random.shuffle(sentence_pairs)
train_sentence_pairs, test_sentence_pairs = sentence_pairs[:train_size], sentence_pairs[train_size:]
# Show length
len(train_sentence_pairs), len(test_sentence_pairs)
_english, _spanish = train_sentence_pairs[0]
_eng_ints, _spa_ints = english_tokenizer.texts_to_sequences([_english])[0], spanish_tokenizer.texts_to_sequences([_spanish])[0]
print("Source language: ")
print('\n'.join('{:4d} ----> {}'.format(i, word) for i, word in zip(_eng_ints, _english.split())))
print("Target language: ")
print('\n'.join('{:4d} ----> {}'.format(i, word) for i, word in zip(_spa_ints, _spanish.split())))
# Set up datasets
BATCH_SIZE = 64
train_ds = make_dataset(train_sentence_pairs, english_tokenizer, spanish_tokenizer)
test_ds = make_dataset(test_sentence_pairs, english_tokenizer, spanish_tokenizer)
train_ds = train_ds.shuffle(len(train_sentence_pairs)).batch(BATCH_SIZE, drop_remainder=True)
test_ds = test_ds.batch(BATCH_SIZE, drop_remainder=True)
print("Dataset outputs elements with shape ({}, {})".format(
*train_ds.output_shapes))
```
## Write the encoder and decoder model
Here, we'll implement an encoder-decoder model with attention. The following diagram shows that each input word is assigned a weight by the attention mechanism which is then used by the decoder to predict the next word in the sentence.
<img src="https://www.tensorflow.org/images/seq2seq/attention_mechanism.jpg" width="500" alt="attention mechanism">
The input is put through an encoder model which gives us the encoder output of shape *(batch_size, max_length, hidden_size)* and the encoder hidden state of shape *(batch_size, hidden_size)*.
```
ENCODER_SIZE = DECODER_SIZE = 1024
EMBEDDING_DIM = 256
MAX_OUTPUT_LENGTH = train_ds.output_shapes[1][1]
def gru(units):
return tf.keras.layers.GRU(units,
return_sequences=True,
return_state=True,
recurrent_activation='sigmoid',
recurrent_initializer='glorot_uniform')
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, encoder_size):
super(Encoder, self).__init__()
self.embedding_dim = embedding_dim
self.encoder_size = encoder_size
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(encoder_size)
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state=hidden)
return output, state
def initial_hidden_state(self, batch_size):
return tf.zeros((batch_size, self.encoder_size))
```
For the decoder, we're using *Bahdanau attention*. Here are the equations that are implemented:
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_0.jpg" alt="attention equation 0" width="800">
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_1.jpg" alt="attention equation 1" width="800">
Lets decide on notation before writing the simplified form:
* FC = Fully connected (dense) layer
* EO = Encoder output
* H = hidden state
* X = input to the decoder
And the pseudo-code:
* `score = FC(tanh(FC(EO) + FC(H)))`
* `attention weights = softmax(score, axis = 1)`. Softmax by default is applied on the last axis but here we want to apply it on the *1st axis*, since the shape of score is *(batch_size, max_length, hidden_size)*. `Max_length` is the length of our input. Since we are trying to assign a weight to each input, softmax should be applied on that axis.
* `context vector = sum(attention weights * EO, axis = 1)`. Same reason as above for choosing axis as 1.
* `embedding output` = The input to the decoder X is passed through an embedding layer.
* `merged vector = concat(embedding output, context vector)`
* This merged vector is then given to the GRU
The shapes of all the vectors at each step have been specified in the comments in the code:
```
class BahdanauAttention(tf.keras.Model):
def __init__(self, units):
super(BahdanauAttention, self).__init__()
self.W1 = tf.keras.layers.Dense(units)
self.W2 = tf.keras.layers.Dense(units)
self.V = tf.keras.layers.Dense(1)
def call(self, hidden_state, enc_output):
# enc_output shape = (batch_size, max_length, hidden_size)
# (batch_size, hidden_size) -> (batch_size, 1, hidden_size)
hidden_with_time = tf.expand_dims(hidden_state, 1)
# score shape == (batch_size, max_length, 1)
score = self.V(tf.nn.tanh(self.W1(enc_output) + self.W2(hidden_with_time)))
# attention_weights shape == (batch_size, max_length, 1)
attention_weights = tf.nn.softmax(score, axis=1)
# context_vector shape after sum = (batch_size, hidden_size)
context_vector = attention_weights * enc_output
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector, attention_weights
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, decoder_size):
super(Decoder, self).__init__()
self.vocab_size = vocab_size
self.embedding_dim = embedding_dim
self.decoder_size = decoder_size
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(decoder_size)
self.fc = tf.keras.layers.Dense(vocab_size)
self.attention = BahdanauAttention(decoder_size)
def call(self, x, hidden, enc_output):
context_vector, attention_weights = self.attention(hidden, enc_output)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size, vocab)
x = self.fc(output)
return x, state, attention_weights
```
## Define a translate function
Now, let's put the encoder and decoder halves together. The encoder step is fairly straightforward; we'll just reuse Keras's dynamic unroll. For the decoder, we have to make some choices about how to feed the decoder RNN. Overall the process goes as follows:
1. Pass the *input* through the *encoder* which return *encoder output* and the *encoder hidden state*.
2. The encoder output, encoder hidden state and the <START> token is passed to the decoder.
3. The decoder returns the *predictions* and the *decoder hidden state*.
4. The encoder output, hidden state and next token is then fed back into the decoder repeatedly. This has two different behaviors under training and inference:
- during training, we use *teacher forcing*, where the correct next token is fed into the decoder, regardless of what the decoder emitted.
- during inference, we use `tf.argmax(predictions)` to select the most likely continuation and feed it back into the decoder. Another strategy that yields more robust results is called *beam search*.
5. Repeat step 4 until either the decoder emits an <END> token, indicating that it's done translating, or we run into a hardcoded length limit.
```
class NmtTranslator(tf.keras.Model):
def __init__(self, encoder, decoder, start_token_id, end_token_id):
super(NmtTranslator, self).__init__()
self.encoder = encoder
self.decoder = decoder
# (The token_id should match the decoder's language.)
# Uses start_token_id to initialize the decoder.
self.start_token_id = tf.constant(start_token_id)
# Check for sequence completion using this token_id
self.end_token_id = tf.constant(end_token_id)
@tf.function
def call(self, inp, target=None, max_output_length=MAX_OUTPUT_LENGTH):
'''Translate an input.
If target is provided, teacher forcing is used to generate the translation.
'''
batch_size = inp.shape[0]
hidden = self.encoder.initial_hidden_state(batch_size)
enc_output, enc_hidden = self.encoder(inp, hidden)
dec_hidden = enc_hidden
if target is not None:
output_length = target.shape[1]
else:
output_length = max_output_length
predictions_array = tf.TensorArray(tf.float32, size=output_length - 1)
attention_array = tf.TensorArray(tf.float32, size=output_length - 1)
# Feed <START> token to start decoder.
dec_input = tf.cast([self.start_token_id] * batch_size, tf.int32)
# Keep track of which sequences have emitted an <END> token
is_done = tf.zeros([batch_size], dtype=tf.bool)
for i in tf.range(output_length - 1):
dec_input = tf.expand_dims(dec_input, 1)
predictions, dec_hidden, attention_weights = self.decoder(dec_input, dec_hidden, enc_output)
predictions = tf.where(is_done, tf.zeros_like(predictions), predictions)
# Write predictions/attention for later visualization.
predictions_array = predictions_array.write(i, predictions)
attention_array = attention_array.write(i, attention_weights)
# Decide what to pass into the next iteration of the decoder.
if target is not None:
# if target is known, use teacher forcing
dec_input = target[:, i + 1]
else:
# Otherwise, pick the most likely continuation
dec_input = tf.argmax(predictions, axis=1, output_type=tf.int32)
# Figure out which sentences just completed.
is_done = tf.logical_or(is_done, tf.equal(dec_input, self.end_token_id))
# Exit early if all our sentences are done.
if tf.reduce_all(is_done):
break
# [time, batch, predictions] -> [batch, time, predictions]
return tf.transpose(predictions_array.stack(), [1, 0, 2]), tf.transpose(attention_array.stack(), [1, 0, 2, 3])
```
## Define the loss function
Our loss function is a word-for-word comparison between true answer and model prediction.
real = [<start>, 'This', 'is', 'the', 'correct', 'answer', '.', '<end>', '<oov>']
pred = ['This', 'is', 'what', 'the', 'model', 'emitted', '.', '<end>']
results in comparing
This/This, is/is, the/what, correct/the, answer/model, ./emitted, <end>/.
and ignoring the rest of the prediction.
```
def loss_fn(real, pred):
# The prediction doesn't include the <start> token.
real = real[:, 1:]
# Cut down the prediction to the correct shape (We ignore extra words).
pred = pred[:, :real.shape[1]]
# If real == <OOV>, then mask out the loss.
mask = 1 - np.equal(real, 0)
loss_ = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=real, logits=pred) * mask
# Sum loss over the time dimension, but average it over the batch dimension.
return tf.reduce_mean(tf.reduce_sum(loss_, axis=1))
```
## Configure model directory
We'll use one directory to save all of our relevant artifacts (summary logs, checkpoints, SavedModel exports, etc.)
```
# Where to save checkpoints, tensorboard summaries, etc.
MODEL_DIR = '/tmp/tensorflow/nmt_attention'
def apply_clean():
if tf.io.gfile.exists(MODEL_DIR):
print('Removing existing model dir: {}'.format(MODEL_DIR))
tf.io.gfile.rmtree(MODEL_DIR)
# Optional: remove existing data
apply_clean()
# Summary writers
train_summary_writer = tf.summary.create_file_writer(
os.path.join(MODEL_DIR, 'summaries', 'train'), flush_millis=10000)
test_summary_writer = tf.summary.create_file_writer(
os.path.join(MODEL_DIR, 'summaries', 'eval'), flush_millis=10000, name='test')
# Set up all stateful objects
encoder = Encoder(len(english_tokenizer.word_index) + 1, EMBEDDING_DIM, ENCODER_SIZE)
decoder = Decoder(len(spanish_tokenizer.word_index) + 1, EMBEDDING_DIM, DECODER_SIZE)
start_token_id = spanish_tokenizer.word_index[START_TOKEN]
end_token_id = spanish_tokenizer.word_index[END_TOKEN]
model = NmtTranslator(encoder, decoder, start_token_id, end_token_id)
# TODO(brianklee): Investigate whether Adam defaults have changed and whether it affects training.
optimizer = tf.keras.optimizers.Adam(epsilon=1e-8)# tf.keras.optimizers.SGD(learning_rate=0.01)#Adam()
# Checkpoints
checkpoint_dir = os.path.join(MODEL_DIR, 'checkpoints')
checkpoint_prefix = os.path.join(checkpoint_dir, 'ckpt')
checkpoint = tf.train.Checkpoint(
encoder=encoder, decoder=decoder, optimizer=optimizer)
# Restore variables on creation if a checkpoint exists.
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
# SavedModel exports
export_path = os.path.join(MODEL_DIR, 'export')
```
# Visualize the model's output
Let's visualize our model's output. (It hasn't been trained yet, so it will output gibberish.)
We'll use this visualization to check on the model's progress.
```
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence.split(), fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence.split(), fontdict=fontdict)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
plt.show()
def ints_to_words(tokenizer, ints):
return ' '.join(tokenizer.index_word[int(i)] if int(i) != 0 else '<OOV>' for i in ints)
def sentence_to_ints(tokenizer, sentence):
sentence = preprocess_sentence(sentence)
return tf.constant(tokenizer.texts_to_sequences([sentence])[0])
def translate_and_plot_ints(model, english_tokenizer, spanish_tokenizer, ints, target_ints=None):
"""Run translation on a sentence and plot an attention matrix.
Sentence should be passed in as list of integers.
"""
ints = tf.expand_dims(ints, 0)
predictions, attention = model(ints)
prediction_ids = tf.squeeze(tf.argmax(predictions, axis=-1))
attention = tf.squeeze(attention)
sentence = ints_to_words(english_tokenizer, ints[0])
predicted_sentence = ints_to_words(spanish_tokenizer, prediction_ids)
print(u'Input: {}'.format(sentence))
print(u'Predicted translation: {}'.format(predicted_sentence))
if target_ints is not None:
print(u'Correct translation: {}'.format(ints_to_words(spanish_tokenizer, target_ints)))
plot_attention(attention, sentence, predicted_sentence)
def translate_and_plot_words(model, english_tokenizer, spanish_tokenizer, sentence, target_sentence=None):
"""Same as translate_and_plot_ints, but pass in a sentence as a string."""
english_ints = sentence_to_ints(english_tokenizer, sentence)
spanish_ints = sentence_to_ints(spanish_tokenizer, target_sentence) if target_sentence is not None else None
translate_and_plot_ints(model, english_tokenizer, spanish_tokenizer, english_ints, target_ints=spanish_ints)
translate_and_plot_words(model, english_tokenizer, spanish_tokenizer, u"it's really cold here", u'hace mucho frio aqui')
```
# Train the model
```
def train(model, optimizer, dataset):
"""Trains model on `dataset` using `optimizer`."""
start = time.time()
avg_loss = tf.keras.metrics.Mean('loss', dtype=tf.float32)
for inp, target in dataset:
with tf.GradientTape() as tape:
predictions, _ = model(inp, target=target)
loss = loss_fn(target, predictions)
avg_loss(loss)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
if tf.equal(optimizer.iterations % 10, 0):
tf.summary.scalar('loss', avg_loss.result(), step=optimizer.iterations)
avg_loss.reset_states()
rate = 10 / (time.time() - start)
print('Step #%d\tLoss: %.6f (%.2f steps/sec)' % (optimizer.iterations, loss, rate))
start = time.time()
if tf.equal(optimizer.iterations % 100, 0):
# translate_and_plot_words(model, english_index, spanish_index, u"it's really cold here.", u'hace mucho frio aqui.')
translate_and_plot_ints(model, english_tokenizer, spanish_tokenizer, inp[0], target[0])
def test(model, dataset, step_num):
"""Perform an evaluation of `model` on the examples from `dataset`."""
avg_loss = tf.keras.metrics.Mean('loss', dtype=tf.float32)
for inp, target in dataset:
predictions, _ = model(inp)
loss = loss_fn(target, predictions)
avg_loss(loss)
print('Model test set loss: {:0.4f}'.format(avg_loss.result()))
tf.summary.scalar('loss', avg_loss.result(), step=step_num)
NUM_TRAIN_EPOCHS = 10
for i in range(NUM_TRAIN_EPOCHS):
start = time.time()
with train_summary_writer.as_default():
train(model, optimizer, train_ds)
end = time.time()
print('\nTrain time for epoch #{} ({} total steps): {}'.format(
i + 1, optimizer.iterations, end - start))
with test_summary_writer.as_default():
test(model, test_ds, optimizer.iterations)
checkpoint.save(checkpoint_prefix)
# TODO(brianklee): This seems to be complaining about input shapes not being set?
# tf.saved_model.save(model, export_path)
```
## Next steps
* [Download a different dataset](http://www.manythings.org/anki/) to experiment with translations, for example, English to German, or English to French.
* Experiment with training on a larger dataset, or using more epochs
```
```
| github_jupyter |
# Many to Many Classification
Simple example for Many to Many Classification (Simple pos tagger) by Recurrent Neural Networks
- Creating the **data pipeline** with `tf.data`
- Preprocessing word sequences (variable input sequence length) using `padding technique` by `user function (pad_seq)`
- Using `tf.nn.embedding_lookup` for getting vector of tokens (eg. word, character)
- Training **many to many classification** with `tf.contrib.seq2seq.sequence_loss`
- Masking unvalid token with `tf.sequence_mask`
- Creating the model as **Class**
- Reference
- https://github.com/aisolab/sample_code_of_Deep_learning_Basics/blob/master/DLEL/DLEL_12_2_RNN_(toy_example).ipynb
### Setup
```
import os, sys
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
import string
%matplotlib inline
slim = tf.contrib.slim
print(tf.__version__)
```
### Prepare example data
```
sentences = [['I', 'feel', 'hungry'],
['tensorflow', 'is', 'very', 'difficult'],
['tensorflow', 'is', 'a', 'framework', 'for', 'deep', 'learning'],
['tensorflow', 'is', 'very', 'fast', 'changing']]
pos = [['pronoun', 'verb', 'adjective'],
['noun', 'verb', 'adverb', 'adjective'],
['noun', 'verb', 'determiner', 'noun', 'preposition', 'adjective', 'noun'],
['noun', 'verb', 'adverb', 'adjective', 'verb']]
# word dic
word_list = []
for elm in sentences:
word_list += elm
word_list = list(set(word_list))
word_list.sort()
word_list = ['<pad>'] + word_list
word_dic = {word : idx for idx, word in enumerate(word_list)}
print(word_dic)
# pos dic
pos_list = []
for elm in pos:
pos_list += elm
pos_list = list(set(pos_list))
pos_list.sort()
pos_list = ['<pad>'] + pos_list
print(pos_list)
pos_dic = {pos : idx for idx, pos in enumerate(pos_list)}
pos_dic
pos_idx_to_dic = {elm[1] : elm[0] for elm in pos_dic.items()}
pos_idx_to_dic
```
### Create pad_seq function
```
def pad_seq(sequences, max_len, dic):
seq_len, seq_indices = [], []
for seq in sequences:
seq_len.append(len(seq))
seq_idx = [dic.get(char) for char in seq]
seq_idx += (max_len - len(seq_idx)) * [dic.get('<pad>')] # 0 is idx of meaningless token "<pad>"
seq_indices.append(seq_idx)
return seq_len, seq_indices
```
### Pre-process data
```
max_length = 10
X_length, X_indices = pad_seq(sequences = sentences, max_len = max_length, dic = word_dic)
print(X_length, np.shape(X_indices))
y = [elm + ['<pad>'] * (max_length - len(elm)) for elm in pos]
y = [list(map(lambda el : pos_dic.get(el), elm)) for elm in y]
print(np.shape(y))
y
```
### Define SimPosRNN
```
class SimPosRNN:
def __init__(self, X_length, X_indices, y, n_of_classes, hidden_dim, max_len, word_dic):
# Data pipeline
with tf.variable_scope('input_layer'):
# input layer를 구현해보세요
# tf.get_variable을 사용하세요
# tf.nn.embedding_lookup을 사용하세요
self._X_length = X_length
self._X_indices = X_indices
self._y = y
# RNN cell (many to many)
with tf.variable_scope('rnn_cell'):
# RNN cell을 구현해보세요
# tf.contrib.rnn.BasicRNNCell을 사용하세요
# tf.nn.dynamic_rnn을 사용하세요
# tf.contrib.rnn.OutputProjectionWrapper를 사용하세요
with tf.variable_scope('seq2seq_loss'):
# tf.sequence_mask를 사용하여 masks를 정의하세요
# tf.contrib.seq2seq.sequence_loss의 weights argument에 masks를 넣으세요
with tf.variable_scope('prediction'):
# tf.argmax를 사용하세요
def predict(self, sess, X_length, X_indices):
# predict instance method를 구현하세요
return sess.run(self._prediction, feed_dict = feed_prediction)
```
### Create a model of SimPosRNN
```
# hyper-parameter#
lr = .003
epochs = 100
batch_size = 2
total_step = int(np.shape(X_indices)[0] / batch_size)
print(total_step)
## create data pipeline with tf.data
# tf.data를 이용해서 직접 구현해보세요
# 최종적으로 model은 아래의 코드를 통해서 생성됩니다.
sim_pos_rnn = SimPosRNN(X_length = X_length_mb, X_indices = X_indices_mb, y = y_mb,
n_of_classes = 8, hidden_dim = 16, max_len = max_length, word_dic = word_dic)
```
### Creat training op and train model
```
## create training op
opt = tf.train.AdamOptimizer(learning_rate = lr)
training_op = opt.minimize(loss = sim_pos_rnn.seq2seq_loss)
sess = tf.Session()
sess.run(tf.global_variables_initializer())
tr_loss_hist = []
for epoch in range(epochs):
avg_tr_loss = 0
tr_step = 0
sess.run(tr_iterator.initializer)
try:
while True:
# 여기를 직접구현하시면 됩니다.
except tf.errors.OutOfRangeError:
pass
avg_tr_loss /= tr_step
tr_loss_hist.append(avg_tr_loss)
if (epoch + 1) % 10 == 0:
print('epoch : {:3}, tr_loss : {:.3f}'.format(epoch + 1, avg_tr_loss))
yhat = sim_pos_rnn.predict(sess = sess, X_length = X_length, X_indices = X_indices)
yhat
y
yhat = [list(map(lambda elm : pos_idx_to_dic.get(elm), row)) for row in yhat]
for elm in yhat:
print(elm)
```
| github_jupyter |
# Android的人脸识别库(NDK)
## 创建工程向导



## dlib库源代码添加到工程
* 把dlib目录下的dlib文件夹拷贝到app/src/main/
## 增加JNI接口
### 创建Java接口类
在app/src/main/java/com/wangjunjian/facerecognition下创建类FaceRecognition
```java
package com.wangjunjian.facerecognition;
import android.graphics.Rect;
public class FaceRecognition {
static {
System.loadLibrary("face-recognition");
}
public native void detect(String filename, Rect rect);
}
```
### 通过Java接口类输出C++头文件
打开Terminal窗口,输入命令(**Windows系统下要把:改为;**)
```bash
cd app/src/main/
javah -d jni -classpath /Users/wjj/Library/Android/sdk/platforms/android-21/android.jar:java com.wangjunjian.facerecognition.FaceRecognition
```
### 参考资料
* [JNI 无法确定Bitmap的签名](https://blog.csdn.net/wxxgreat/article/details/48030775)
* [在编辑JNI头文件的时候碰到无法确定Bitmap的签名问题](https://www.jianshu.com/p/b49bdcbfb5ed)
## 实现人脸检测
打开app/src/main/cpp/face-recognition.cpp
```cpp
#include <jni.h>
#include <string>
#include <dlib/image_processing/frontal_face_detector.h>
#include <dlib/image_io.h>
#include "jni/com_wangjunjian_facerecognition_FaceRecognition.h"
using namespace dlib;
using namespace std;
JNIEXPORT void JNICALL Java_com_wangjunjian_facerecognition_FaceRecognition_detect
(JNIEnv *env, jobject clazz, jstring filename, jobject rect)
{
const char* pfilename = env->GetStringUTFChars(filename, JNI_FALSE);
static frontal_face_detector detector = get_frontal_face_detector();
array2d<unsigned char> img;
load_image(img, pfilename);
env->ReleaseStringUTFChars(filename, pfilename);
std::vector<rectangle> dets = detector(img, 0);
if (dets.size() > 0)
{
rectangle faceRect = dets[0];
jclass rectClass = env->GetObjectClass(rect);
jfieldID fidLeft = env->GetFieldID(rectClass, "left", "I");
env->SetIntField(rect, fidLeft, faceRect.left());
jfieldID fidTop = env->GetFieldID(rectClass, "top", "I");
env->SetIntField(rect, fidTop, faceRect.top());
jfieldID fidRight = env->GetFieldID(rectClass, "right", "I");
env->SetIntField(rect, fidRight, faceRect.right());
jfieldID fidBottom = env->GetFieldID(rectClass, "bottom", "I");
env->SetIntField(rect, fidBottom, faceRect.bottom());
}
}
```
### 参考资料
*[Android使用JNI实现Java与C之间传递数据](https://blog.csdn.net/furongkang/article/details/6857610)
## 修改 app/CMakeLists.txt
```
# For more information about using CMake with Android Studio, read the
# documentation: https://d.android.com/studio/projects/add-native-code.html
# Sets the minimum version of CMake required to build the native library.
cmake_minimum_required(VERSION 3.4.1)
# 设置库输出路径变量
set(DISTRIBUTION_DIR ${CMAKE_SOURCE_DIR}/../distribution)
# 包含dlib的make信息
include(${CMAKE_SOURCE_DIR}/src/main/dlib/cmake)
# Creates and names a library, sets it as either STATIC
# or SHARED, and provides the relative paths to its source code.
# You can define multiple libraries, and CMake builds them for you.
# Gradle automatically packages shared libraries with your APK.
add_library( # Sets the name of the library.
face-recognition
# Sets the library as a shared library.
SHARED
# Provides a relative path to your source file(s).
src/main/cpp/face-recognition.cpp )
# 设置每个平台的ABI输出路径
set_target_properties(face-recognition PROPERTIES
LIBRARY_OUTPUT_DIRECTORY
${DISTRIBUTION_DIR}/libs/${ANDROID_ABI})
# Searches for a specified prebuilt library and stores the path as a
# variable. Because CMake includes system libraries in the search path by
# default, you only need to specify the name of the public NDK library
# you want to add. CMake verifies that the library exists before
# completing its build.
find_library( # Sets the name of the path variable.
log-lib
# Specifies the name of the NDK library that
# you want CMake to locate.
log )
# Specifies libraries CMake should link to your target library. You
# can link multiple libraries, such as libraries you define in this
# build script, prebuilt third-party libraries, or system libraries.
# 连接dlib和android
target_link_libraries( # Specifies the target library.
face-recognition
android
dlib
# Links the target library to the log library
# included in the NDK.
${log-lib} )
```
### 参考资料
* [AndroidStudio用Cmake方式编译NDK代码](https://blog.csdn.net/joe544351900/article/details/53637549)
## 修改 app/build.gradle
```
//修改为库
//apply plugin: 'com.android.application'
apply plugin: 'com.android.library'
android {
compileSdkVersion 26
defaultConfig {
//移除应用ID
//applicationId "com.wangjunjian.facerecognition"
minSdkVersion 21
targetSdkVersion 26
versionCode 1
versionName "1.0"
testInstrumentationRunner "android.support.test.runner.AndroidJUnitRunner"
externalNativeBuild {
cmake {
arguments '-DANDROID_PLATFORM=android-21',
'-DANDROID_TOOLCHAIN=clang', '-DANDROID_STL=c++_static', '-DCMAKE_BUILD_TYPE=Release ..'
cppFlags "-frtti -fexceptions -std=c++11 -O3"
}
}
//要生成的目标平台ABI
ndk {
abiFilters 'armeabi-v7a', 'arm64-v8a', 'x86', 'x86_64'
}
}
buildTypes {
release {
minifyEnabled false
proguardFiles getDefaultProguardFile('proguard-android.txt'), 'proguard-rules.pro'
}
}
externalNativeBuild {
cmake {
path "CMakeLists.txt"
}
}
//JNI库输出路径
sourceSets {
main {
jniLibs.srcDirs = ['../distribution/libs']
}
}
//消除错误 Caused by: com.android.builder.merge.DuplicateRelativeFileException: More than one file was found with OS independent path 'lib/x86/libface-recognition.so'
packagingOptions {
pickFirst 'lib/armeabi-v7a/libface-recognition.so'
pickFirst 'lib/arm64-v8a/libface-recognition.so'
pickFirst 'lib/x86/libface-recognition.so'
pickFirst 'lib/x86_64/libface-recognition.so'
}
}
//打包jar到指定路径
task makeJar(type: Copy) {
delete 'build/libs/face-recognition.jar'
from('build/intermediates/packaged-classes/release/')
into('../distribution/libs/')
include('classes.jar')
rename('classes.jar', 'face-recognition.jar')
}
makeJar.dependsOn(build)
dependencies {
implementation fileTree(dir: 'libs', include: ['*.jar'])
implementation 'com.android.support:appcompat-v7:26.1.0'
testImplementation 'junit:junit:4.12'
androidTestImplementation 'com.android.support.test:runner:1.0.2'
androidTestImplementation 'com.android.support.test.espresso:espresso-core:3.0.2'
}
```
### 参考资料
* [Android NDK samples with Android Studio](https://github.com/googlesamples/android-ndk)
* [Android studio 将 Module 打包成 Jar 包](https://www.cnblogs.com/xinaixia/p/7660173.html)
* [could not load library "libc++_shared.so" needed by "libgpg.so"](https://github.com/playgameservices/play-games-plugin-for-unity/issues/280)
* [Android NDK cannot load libc++_shared.so, gets "cannot locate symbol 'rand' reference](https://stackoverflow.com/questions/28504875/android-ndk-cannot-load-libc-shared-so-gets-cannot-locate-symbol-rand-refe)
* [记录Android-Studio遇到的各种坑](https://blog.csdn.net/u012874222/article/details/50616698)
* [Gradle flavors for android with custom source sets - what should the gradle files look like?](https://stackoverflow.com/questions/19461145/gradle-flavors-for-android-with-custom-source-sets-what-should-the-gradle-file)
* [Android Studio 2.2 gradle调用ndk-build](https://www.jianshu.com/p/0e50ae3c4d0d)
* [Android NDK: How to build for ARM64-v8a with minimumSdkVersion = 19](https://stackoverflow.com/questions/41102128/android-ndk-how-to-build-for-arm64-v8a-with-minimumsdkversion-19)
## 编译输出开发库
打开Terminal窗口,输入命令
```bash
./gradlew makeJar
```

### 参考资料
* [-bash :gradlew command not found](https://blog.csdn.net/yyh352091626/article/details/52343951)
## 查看jar文档列表
```bash
jar vtf distribution/libs/face-recognition.jar
```
### 参考资料
* [Linux环境下查看jar包的归档目录](https://blog.csdn.net/tanga842428/article/details/55101253)
## 参考资料
* [Face Landmarks In Your Android App](http://slides.com/boywang/face-landmarks-in-your-android-app/fullscreen#/)
* [dlib-android](https://github.com/tzutalin/dlib-android)
* [深入理解Android(一):Gradle详解](http://www.infoq.com/cn/articles/android-in-depth-gradle/)
* [Android NDK Gradle3.0 以上最新生成.so之旅](https://blog.csdn.net/xiaozhu0922/article/details/78835144)
* [Android Studio 手把手教你NDK打包SO库文件,并提供对应API 使用它(赋demo)](https://blog.csdn.net/u011445031/article/details/72884703)
* [Building dlib for android ndk](https://stackoverflow.com/questions/41331400/building-dlib-for-android-ndk)
* [使用 Android Studio 写出第一个 NDK 程序(超详细)](https://blog.csdn.net/young_time/article/details/80346631)
* [Android studio3.0 JNI/NDK开发流程](https://www.jianshu.com/p/a37782b56770)
* [dlib 18 android编译dlib库,运行matrix_ex demo](https://blog.csdn.net/longji/article/details/78115807)
* [Android开发——Android Studio下使用Cmake在NDK环境下移植Dlib库](https://blog.csdn.net/u012525096/article/details/78950979)
* [android编译系统makefile(Android.mk)写法](http://www.cnblogs.com/hesiming/archive/2011/03/15/1984444.html)
* [dlib-android/jni/jni_detections/jni_pedestrian_det.cpp](https://github.com/tzutalin/dlib-android/blob/master/jni/jni_detections/jni_pedestrian_det.cpp)
* [Face Detection using MTCNN and TensorFlow in android](http://androidcodehub.com/face-detection-using-mtcnn-tensorflow-android/)
| github_jupyter |
#$EXERCISE_PREAMBLE$
As always, run the setup code below before working on the questions (and if you leave this notebook and come back later, remember to run the setup code again).
```
from learntools.core import binder; binder.bind(globals())
from learntools.python.ex5 import *
print('Setup complete.')
```
# Exercises
## 1.
Have you ever felt debugging involved a bit of luck? The following program has a bug. Try to identify the bug and fix it.
```
def has_lucky_number(nums):
"""Return whether the given list of numbers is lucky. A lucky list contains
at least one number divisible by 7.
"""
for num in nums:
if num % 7 == 0:
return True
else:
return False
```
Try to identify the bug and fix it in the cell below:
```
def has_lucky_number(nums):
"""Return whether the given list of numbers is lucky. A lucky list contains
at least one number divisible by 7.
"""
for num in nums:
if num % 7 == 0:
return True
else:
return False
q1.check()
#_COMMENT_IF(PROD)_
q1.hint()
#_COMMENT_IF(PROD)_
q1.solution()
```
## 2.
### a.
Look at the Python expression below. What do you think we'll get when we run it? When you've made your prediction, uncomment the code and run the cell to see if you were right.
```
#[1, 2, 3, 4] > 2
```
### b
R and Python have some libraries (like numpy and pandas) compare each element of the list to 2 (i.e. do an 'element-wise' comparison) and give us a list of booleans like `[False, False, True, True]`.
Implement a function that reproduces this behaviour, returning a list of booleans corresponding to whether the corresponding element is greater than n.
```
def elementwise_greater_than(L, thresh):
"""Return a list with the same length as L, where the value at index i is
True if L[i] is greater than thresh, and False otherwise.
>>> elementwise_greater_than([1, 2, 3, 4], 2)
[False, False, True, True]
"""
pass
q2.check()
#_COMMENT_IF(PROD)_
q2.solution()
```
## 3.
Complete the body of the function below according to its docstring
```
def menu_is_boring(meals):
"""Given a list of meals served over some period of time, return True if the
same meal has ever been served two days in a row, and False otherwise.
"""
pass
q3.check()
#_COMMENT_IF(PROD)_
q3.hint()
#_COMMENT_IF(PROD)_
q3.solution()
```
## 4. <span title="A bit spicy" style="color: darkgreen ">🌶️</span>
Next to the Blackjack table, the Python Challenge Casino has a slot machine. You can get a result from the slot machine by calling `play_slot_machine()`. The number it returns is your winnings in dollars. Usually it returns 0. But sometimes you'll get lucky and get a big payday. Try running it below:
```
play_slot_machine()
```
By the way, did we mention that each play costs $1? Don't worry, we'll send you the bill later.
On average, how much money can you expect to gain (or lose) every time you play the machine? The casino keeps it a secret, but you can estimate the average value of each pull using a technique called the **Monte Carlo method**. To estimate the average outcome, we simulate the scenario many times, and return the average result.
Complete the following function to calculate the average value per play of the slot machine.
```
def estimate_average_slot_payout(n_runs):
"""Run the slot machine n_runs times and return the average net profit per run.
Example calls (note that return value is nondeterministic!):
>>> estimate_average_slot_payout(1)
-1
>>> estimate_average_slot_payout(1)
0.5
"""
pass
```
When you think you know the expected value per spin, uncomment the line below to see how close you were.
```
#_COMMENT_IF(PROD)_
q4.solution()
```
#$KEEP_GOING$
| github_jupyter |
# Setup
### Imports
```
import sys
sys.path.append('../')
del sys
%reload_ext autoreload
%autoreload 2
from toolbox.parsers import standard_parser, add_task_arguments, add_model_arguments
from toolbox.utils import load_task, get_pretrained_model, to_class_name
import modeling.models as models
```
### Notebook functions
```
from numpy import argmax, mean
def run_models(model_names, word2vec, bart, args, train=False):
args.word2vec = word2vec
args.bart = bart
pretrained_model = get_pretrained_model(args)
for model_name in model_names:
args.model = model_name
print(model_name)
model = getattr(models, to_class_name(args.model))(args=args, pretrained_model=pretrained_model)
model.play(task=task, args=args)
if train:
valid_scores = model.valid_scores['average_precision']
test_scores = model.test_scores['average_precision']
valid_scores = [mean(epoch_scores) for epoch_scores in valid_scores]
test_scores = [mean(epoch_scores) for epoch_scores in test_scores]
i_max = argmax(valid_scores)
print("max for epoch %i" % (i_max+1))
print("valid score: %.5f" % valid_scores[i_max])
print("test score: %.5f" % test_scores[i_max])
```
### Parameters
```
ap = standard_parser()
add_task_arguments(ap)
add_model_arguments(ap)
args = ap.parse_args(["-m", "",
"--root", ".."])
```
### Load the data
```
task = load_task(args)
```
# Basic baselines
```
run_models(model_names=["random",
"frequency"],
word2vec=False,
bart=False,
args=args)
```
# Basic baselines
```
run_models(model_names=["summaries-count",
"summaries-unique-count",
"summaries-overlap",
"activated-summaries",
"context-count",
"context-unique-count",
"summaries-context-count",
"summaries-context-unique-count",
"summaries-context-overlap"],
word2vec=False,
bart=False,
args=args)
```
# Embedding baselines
```
run_models(model_names=["summaries-average-embedding",
"summaries-overlap-average-embedding",
"context-average-embedding",
"summaries-context-average-embedding",
"summaries-context-overlap-average-embedding"],
word2vec=True,
bart=False,
args=args)
```
### Custom classifier
```
run_models(model_names=["custom-classifier"],
word2vec=True,
bart=False,
args=args,
train=True)
```
| github_jupyter |
# Implementing TF-IDF
------------------------------------
Here we implement TF-IDF, (Text Frequency - Inverse Document Frequency) for the spam-ham text data.
We will use a hybrid approach of encoding the texts with sci-kit learn's TFIDF vectorizer. Then we will use the regular TensorFlow logistic algorithm outline.
Creating the TF-IDF vectors requires us to load all the text into memory and count the occurrences of each word before we can start training our model. Because of this, it is not implemented fully in Tensorflow, so we will use Scikit-learn for creating our TF-IDF embedding, but use Tensorflow to fit the logistic model.
We start by loading the necessary libraries.
```
import tensorflow as tf
import matplotlib.pyplot as plt
import csv
import numpy as np
import os
import string
import requests
import io
import nltk
from zipfile import ZipFile
from sklearn.feature_extraction.text import TfidfVectorizer
from tensorflow.python.framework import ops
ops.reset_default_graph()
```
Start a computational graph session.
```
sess = tf.Session()
```
We set two parameters, `batch_size` and `max_features`. `batch_size` is the size of the batch we will train our logistic model on, and `max_features` is the maximum number of tf-idf textual words we will use in our logistic regression.
```
batch_size = 200
max_features = 1000
```
Check if data was downloaded, otherwise download it and save for future use
```
save_file_name = 'temp_spam_data.csv'
if os.path.isfile(save_file_name):
text_data = []
with open(save_file_name, 'r') as temp_output_file:
reader = csv.reader(temp_output_file)
for row in reader:
text_data.append(row)
else:
zip_url = 'http://archive.ics.uci.edu/ml/machine-learning-databases/00228/smsspamcollection.zip'
r = requests.get(zip_url)
z = ZipFile(io.BytesIO(r.content))
file = z.read('SMSSpamCollection')
# Format Data
text_data = file.decode()
text_data = text_data.encode('ascii',errors='ignore')
text_data = text_data.decode().split('\n')
text_data = [x.split('\t') for x in text_data if len(x)>=1]
# And write to csv
with open(save_file_name, 'w') as temp_output_file:
writer = csv.writer(temp_output_file)
writer.writerows(text_data)
```
We now clean our texts. This will decrease our vocabulary size by converting everything to lower case, removing punctuation and getting rid of numbers.
```
texts = [x[1] for x in text_data]
target = [x[0] for x in text_data]
# Relabel 'spam' as 1, 'ham' as 0
target = [1. if x=='spam' else 0. for x in target]
# Normalize text
# Lower case
texts = [x.lower() for x in texts]
# Remove punctuation
texts = [''.join(c for c in x if c not in string.punctuation) for x in texts]
# Remove numbers
texts = [''.join(c for c in x if c not in '0123456789') for x in texts]
# Trim extra whitespace
texts = [' '.join(x.split()) for x in texts]
```
Define tokenizer function and create the TF-IDF vectors with SciKit-Learn.
```
import nltk
nltk.download('punkt')
def tokenizer(text):
words = nltk.word_tokenize(text)
return words
# Create TF-IDF of texts
tfidf = TfidfVectorizer(tokenizer=tokenizer, stop_words='english', max_features=max_features)
sparse_tfidf_texts = tfidf.fit_transform(texts)
```
Split up data set into train/test.
```
train_indices = np.random.choice(sparse_tfidf_texts.shape[0], round(0.8*sparse_tfidf_texts.shape[0]), replace=False)
test_indices = np.array(list(set(range(sparse_tfidf_texts.shape[0])) - set(train_indices)))
texts_train = sparse_tfidf_texts[train_indices]
texts_test = sparse_tfidf_texts[test_indices]
target_train = np.array([x for ix, x in enumerate(target) if ix in train_indices])
target_test = np.array([x for ix, x in enumerate(target) if ix in test_indices])
```
Now we create the variables and placeholders necessary for logistic regression. After which, we declare our logistic regression operation. Remember that the sigmoid part of the logistic regression will be in the loss function.
```
# Create variables for logistic regression
A = tf.Variable(tf.random_normal(shape=[max_features,1]))
b = tf.Variable(tf.random_normal(shape=[1,1]))
# Initialize placeholders
x_data = tf.placeholder(shape=[None, max_features], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
# Declare logistic model (sigmoid in loss function)
model_output = tf.add(tf.matmul(x_data, A), b)
```
Next, we declare the loss function (which has the sigmoid in it), and the prediction function. The prediction function will have to have a sigmoid inside of it because it is not in the model output.
```
# Declare loss function (Cross Entropy loss)
loss = tf.reduce_mean(tf.nn.sigmoid_cross_entropy_with_logits(logits=model_output, labels=y_target))
# Prediction
prediction = tf.round(tf.sigmoid(model_output))
predictions_correct = tf.cast(tf.equal(prediction, y_target), tf.float32)
accuracy = tf.reduce_mean(predictions_correct)
```
Now we create the optimization function and initialize the model variables.
```
# Declare optimizer
my_opt = tf.train.GradientDescentOptimizer(0.0025)
train_step = my_opt.minimize(loss)
# Intitialize Variables
init = tf.global_variables_initializer()
sess.run(init)
```
Finally, we perform our logisitic regression on the 1000 TF-IDF features.
```
train_loss = []
test_loss = []
train_acc = []
test_acc = []
i_data = []
for i in range(10000):
rand_index = np.random.choice(texts_train.shape[0], size=batch_size)
rand_x = texts_train[rand_index].todense()
rand_y = np.transpose([target_train[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
# Only record loss and accuracy every 100 generations
if (i+1)%100==0:
i_data.append(i+1)
train_loss_temp = sess.run(loss, feed_dict={x_data: rand_x, y_target: rand_y})
train_loss.append(train_loss_temp)
test_loss_temp = sess.run(loss, feed_dict={x_data: texts_test.todense(), y_target: np.transpose([target_test])})
test_loss.append(test_loss_temp)
train_acc_temp = sess.run(accuracy, feed_dict={x_data: rand_x, y_target: rand_y})
train_acc.append(train_acc_temp)
test_acc_temp = sess.run(accuracy, feed_dict={x_data: texts_test.todense(), y_target: np.transpose([target_test])})
test_acc.append(test_acc_temp)
if (i+1)%500==0:
acc_and_loss = [i+1, train_loss_temp, test_loss_temp, train_acc_temp, test_acc_temp]
acc_and_loss = [np.round(x,2) for x in acc_and_loss]
print('Generation # {}. Train Loss (Test Loss): {:.2f} ({:.2f}). Train Acc (Test Acc): {:.2f} ({:.2f})'.format(*acc_and_loss))
```
Here is matplotlib code to plot the loss and accuracies.
```
# Plot loss over time
plt.plot(i_data, train_loss, 'k-', label='Train Loss')
plt.plot(i_data, test_loss, 'r--', label='Test Loss', linewidth=4)
plt.title('Cross Entropy Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('Cross Entropy Loss')
plt.legend(loc='upper right')
plt.show()
# Plot train and test accuracy
plt.plot(i_data, train_acc, 'k-', label='Train Set Accuracy')
plt.plot(i_data, test_acc, 'r--', label='Test Set Accuracy', linewidth=4)
plt.title('Train and Test Accuracy')
plt.xlabel('Generation')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.show()
test complete; Gopal
```
| github_jupyter |
# Table of Contents
<p><div class="lev1 toc-item"><a href="#Texte-d'oral-de-modélisation---Agrégation-Option-Informatique" data-toc-modified-id="Texte-d'oral-de-modélisation---Agrégation-Option-Informatique-1"><span class="toc-item-num">1 </span>Texte d'oral de modélisation - Agrégation Option Informatique</a></div><div class="lev2 toc-item"><a href="#Préparation-à-l'agrégation---ENS-de-Rennes,-2016-17" data-toc-modified-id="Préparation-à-l'agrégation---ENS-de-Rennes,-2016-17-11"><span class="toc-item-num">1.1 </span>Préparation à l'agrégation - ENS de Rennes, 2016-17</a></div><div class="lev2 toc-item"><a href="#À-propos-de-ce-document" data-toc-modified-id="À-propos-de-ce-document-12"><span class="toc-item-num">1.2 </span>À propos de ce document</a></div><div class="lev2 toc-item"><a href="#Implémentation" data-toc-modified-id="Implémentation-13"><span class="toc-item-num">1.3 </span>Implémentation</a></div><div class="lev3 toc-item"><a href="#Une-bonne-structure-de-donnée-pour-des-intervalles-et-des-graphes-d'intervales" data-toc-modified-id="Une-bonne-structure-de-donnée-pour-des-intervalles-et-des-graphes-d'intervales-131"><span class="toc-item-num">1.3.1 </span>Une bonne structure de donnée pour des intervalles et des graphes d'intervales</a></div><div class="lev3 toc-item"><a href="#Algorithme-de-coloriage-de-graphe-d'intervalles" data-toc-modified-id="Algorithme-de-coloriage-de-graphe-d'intervalles-132"><span class="toc-item-num">1.3.2 </span>Algorithme de coloriage de graphe d'intervalles</a></div><div class="lev3 toc-item"><a href="#Algorithme-pour-calculer-le-stable-maximum-d'un-graphe-d'intervalles" data-toc-modified-id="Algorithme-pour-calculer-le-stable-maximum-d'un-graphe-d'intervalles-133"><span class="toc-item-num">1.3.3 </span>Algorithme pour calculer le <em>stable maximum</em> d'un graphe d'intervalles</a></div><div class="lev2 toc-item"><a href="#Exemples" data-toc-modified-id="Exemples-14"><span class="toc-item-num">1.4 </span>Exemples</a></div><div class="lev3 toc-item"><a href="#Qui-a-tué-le-Duc-de-Densmore-?" data-toc-modified-id="Qui-a-tué-le-Duc-de-Densmore-?-141"><span class="toc-item-num">1.4.1 </span>Qui a tué le Duc de Densmore ?</a></div><div class="lev4 toc-item"><a href="#Comment-résoudre-ce-problème-?" data-toc-modified-id="Comment-résoudre-ce-problème-?-1411"><span class="toc-item-num">1.4.1.1 </span>Comment résoudre ce problème ?</a></div><div class="lev4 toc-item"><a href="#Solution" data-toc-modified-id="Solution-1412"><span class="toc-item-num">1.4.1.2 </span>Solution</a></div><div class="lev3 toc-item"><a href="#Le-problème-des-frigos" data-toc-modified-id="Le-problème-des-frigos-142"><span class="toc-item-num">1.4.2 </span>Le problème des frigos</a></div><div class="lev3 toc-item"><a href="#Le-problème-du-CSA" data-toc-modified-id="Le-problème-du-CSA-143"><span class="toc-item-num">1.4.3 </span>Le problème du CSA</a></div><div class="lev3 toc-item"><a href="#Le-problème-du-wagon-restaurant" data-toc-modified-id="Le-problème-du-wagon-restaurant-144"><span class="toc-item-num">1.4.4 </span>Le problème du wagon restaurant</a></div><div class="lev4 toc-item"><a href="#Solution-via-l'algorithme-de-coloriage-de-graphe-d'intervalles" data-toc-modified-id="Solution-via-l'algorithme-de-coloriage-de-graphe-d'intervalles-1441"><span class="toc-item-num">1.4.4.1 </span>Solution via l'algorithme de coloriage de graphe d'intervalles</a></div><div class="lev2 toc-item"><a href="#Bonus-?" data-toc-modified-id="Bonus-?-15"><span class="toc-item-num">1.5 </span>Bonus ?</a></div><div class="lev3 toc-item"><a href="#Visualisation-des-graphes-définis-dans-les-exemples" data-toc-modified-id="Visualisation-des-graphes-définis-dans-les-exemples-151"><span class="toc-item-num">1.5.1 </span>Visualisation des graphes définis dans les exemples</a></div><div class="lev2 toc-item"><a href="#Conclusion" data-toc-modified-id="Conclusion-16"><span class="toc-item-num">1.6 </span>Conclusion</a></div>
# Texte d'oral de modélisation - Agrégation Option Informatique
## Préparation à l'agrégation - ENS de Rennes, 2016-17
- *Date* : 3 avril 2017
- *Auteur* : [Lilian Besson](https://GitHub.com/Naereen/notebooks/)
- *Texte*: Annale 2006, "Crime Parfait"
## À propos de ce document
- Ceci est une *proposition* de correction, partielle et probablement non-optimale, pour la partie implémentation d'un [texte d'annale de l'agrégation de mathématiques, option informatique](http://Agreg.org/Textes/).
- Ce document est un [notebook Jupyter](https://www.Jupyter.org/), et [est open-source sous Licence MIT sur GitHub](https://github.com/Naereen/notebooks/tree/master/agreg/), comme les autres solutions de textes de modélisation que [j](https://GitHub.com/Naereen)'ai écrite cette année.
- L'implémentation sera faite en OCaml, version 4+ :
```
Sys.command "ocaml -version";;
```
----
## Implémentation
La question d'implémentation était la question 2) en page 7.
> « Proposer une structure de donnée adaptée pour représenter un graphe d'intervalles dont une représentation sous forme de famille d’intervalles est connue.
> Implémenter de manière efficace l’algorithme de coloriage de graphes d'intervalles et illustrer cet algorithme sur une application bien choisie citée dans le texte. »
Nous allons donc d'abord définir une structure de donnée pour une famille d'intervalles ainsi que pour un graphe d'intervalle, ainsi qu'une fonction convertissant l'un en l'autre.
Cela permettra de facilement définr les différents exemples du texte, et de les résoudre.
### Une bonne structure de donnée pour des intervalles et des graphes d'intervales
- Pour des **intervalles** à valeurs réelles, on se restreint par convénience à des valeurs entières.
```
type intervalle = (int * int);;
type intervalles = intervalle list;;
```
- Pour des **graphes d'intervalles**, on utilise une simple représentation sous forme de liste d'adjacence, plus facile à mettre en place en OCaml qu'une représentation sous forme de matrice. Ici, tous nos graphes ont pour sommets $0 \dots n - 1$.
```
type sommet = int;;
type voisins = sommet list;;
type graphe_intervalle = voisins list;;
```
> *Note:* j'ai préféré garder une structure très simple, pour les intervalles, les graphes d'intervalles et les coloriages, mais on perd un peu en lisibilité dans la fonction coloriage.
>
> Implicitement, dès qu'une liste d'intervalles est fixée, de taille $n$, ils sont numérotés de $0$ à $n-1$. Le graphe `g` aura pour sommet $0 \dots n-1$, et le coloriage sera un simple tableau de couleurs `c` (i.e., d'entiers), donnant en `c[i]` la couleur de l'intervalle numéro `i`.
>
> Une solution plus intelligente aurait été d'utiliser des tables d'association, cf. le module [Map](http://caml.inria.fr/pub/docs/manual-ocaml/libref/Map.html) de OCaml, et le code proposé par Julien durant son oral.
- On peut rapidement écrire une fonction qui va convertir une liste d'intervalle (`intervalles`) en un graphe d'intervalle. On crée les sommets du graphes, via `index_intvls` qui associe un intervalle à son indice, et ensuite on ajoute les arêtes au graphe selon les contraintes définissant un graphe d'intervalle :
$$ \forall I, I' \in V, (I,I') \in E \Leftrightarrow I \neq I' \;\text{and}\; I \cap I' \neq \emptyset $$
Donc avec des intervales $I = [x,y]$ et $I' = [a,b]$, cela donne :
$$ \forall I = [x,y], I' = [a,b] \in V, (I,I') \in E \Leftrightarrow (x,y) \neq (a,b) \;\text{and}\; \neg (b < x \;\text{or}\; y < a) $$
```
let graphe_depuis_intervalles (intvls : intervalles) : graphe_intervalle =
let n = List.length intvls in (* Nomber de sommet *)
let array_intvls = Array.of_list intvls in (* Tableau des intervalles *)
let index_intvls = Array.to_list (
Array.init n (fun i -> (
array_intvls.(i), i) (* Associe un intervalle à son indice *)
)
) in
let gr = List.map (fun (a, b) -> (* Pour chaque intervalle [a, b] *)
List.filter (fun (x, y) -> (* On ajoute [x, y] s'il intersecte [a, b] *)
(x, y) <> (a, b) (* Intervalle différent *)
&& not ( (b < x) || (y < a) ) (* pas x---y a---b ni a---b x---y *)
) intvls
) intvls in
(* On transforme la liste de liste d'intervalles en une liste de liste d'entiers *)
List.map (fun voisins ->
List.map (fun sommet -> (* Grace au tableau index_intvls *)
List.assoc sommet index_intvls
) voisins
) gr
;;
```
### Algorithme de coloriage de graphe d'intervalles
> Étant donné un graphe $G = (V, E)$, on cherche un entier $n$ minimal et une fonction $c : V \to \{1, \cdots, n\}$ telle que si $(v_1 , v_2) \in E$, alors $c(v_1) \neq c(v_2)$.
On suit les indications de l'énoncé pour implémenter facilement cet algorithme.
> Une *heuristique* simple pour résoudre ce problème consiste à appliquer l’algorithme glouton suivant :
> - tant qu'il reste reste des sommets non coloriés,
> + en choisir un
> + et le colorier avec le plus petit entier qui n’apparait pas dans les voisins déjà coloriés.
> En choisissant bien le nouveau sommet à colorier à chaque fois, cette heuristique se révelle optimale pour les graphes d’intervalles.
On peut d'abord définir un type de donnée pour un coloriage, sous la forme d'une liste de couple d'intervalle et de couleur.
Ainsi, `List.assoc` peut être utilisée pour donner le coloriage de chaque intervalle.
```
type couleur = int;;
type coloriage = (intervalle * couleur) list;;
let coloriage_depuis_couleurs (intvl : intervalles) (c : couleur array) : coloriage =
Array.to_list (Array.init (Array.length c) (fun i -> (List.nth intvl i), c.(i)));;
let quelle_couleur (intvl : intervalle) (colors : coloriage) =
List.assoc intvl colors
;;
```
Ensuite, l'ordre partiel $\prec_i$ sur les intervalles est défini comme ça :
$$ I = (a,b) \prec_i J=(x, y) \Longleftrightarrow a < x $$
```
let ordre_partiel ((a, _) : intervalle) ((x, _) : intervalle) =
a < x
;;
```
On a ensuite besoin d'une fonction qui va calculer l'inf de $\mathbb{N} \setminus \{x : x \in \mathrm{valeurs} \}$:
```
let inf_N_minus valeurs =
let res = ref 0 in (* Très important d'utiliser une référence ! *)
while List.mem !res valeurs do
incr res;
done;
!res
;;
```
On vérifie rapidement sur deux exemples :
```
inf_N_minus [0; 1; 3];; (* 2 *)
inf_N_minus [0; 1; 2; 3; 4; 5; 6; 10];; (* 7 *)
```
Enfin, on a besoin d'une fonction pour trouver l'intervalle $I \in V$, minimal pour $\prec_i$, tel que $c(I) = +\infty$.
```
let trouve_min_interval intvl (c : coloriage) (inf : couleur) =
let colorie inter = quelle_couleur inter c in
(* D'abord on extraie {I : c(I) = +oo} *)
let intvl2 = List.filter (fun i -> (colorie i) = inf) intvl in
(* Puis on parcourt la liste et on garde le plus petit pour l'ordre *)
let i0 = ref 0 in
for j = 1 to (List.length intvl2) - 1 do
if ordre_partiel (List.nth intvl2 j) (List.nth intvl2 !i0) then
i0 := j;
done;
List.nth intvl2 !i0;
;;
```
Et donc tout cela permet de finir l'algorithme, tel que décrit dans le texte :
<img style="width:65%;" alt="images/algorithme_coloriage.png" src="images/algorithme_coloriage.png">
```
let coloriage_intervalles (intvl : intervalles) : coloriage =
let n = List.length intvl in (* Nombre d'intervalles *)
let array_intvls = Array.of_list intvl in (* Tableau des intervalles *)
let index_intvls = Array.to_list (
Array.init n (fun i -> (
array_intvls.(i), i) (* Associe un intervalle à son indice *)
)
) in
let gr = graphe_depuis_intervalles intvl in
let inf = n + 10000 in (* Grande valeur, pour +oo *)
let c = Array.make n inf in (* Liste des couleurs, c(I) = +oo pour tout I *)
let maxarray = Array.fold_left max (-inf - 10000) in (* Initialisé à -oo *)
while maxarray c = inf do (* Il reste un I in V tel que c(I) = +oo *)
begin (* C'est la partie pas élégante *)
(* On récupère le coloriage depuis la liste de couleurs actuelle *)
let coloriage = (coloriage_depuis_couleurs intvl c) in
(* Puis la fonction [colorie] pour associer une couleur à un intervalle *)
let colorie inter = quelle_couleur inter coloriage in
(* On choisit un I, minimal pour ordre_partiel, tel que c(I) = +oo *)
let inter = trouve_min_interval intvl coloriage inf in
(* On trouve son indice *)
let i = List.assoc inter index_intvls in
(* On trouve les voisins de i dans le graphe *)
let adj_de_i = List.nth gr i in
(* Puis les voisins de I en tant qu'intervalles *)
let adj_de_I = List.map (fun j -> List.nth intvl j) adj_de_i in
(* Puis on récupère leurs couleurs *)
let valeurs = List.map colorie adj_de_I in
(* c(I) = inf(N - {c(J) : J adjacent a I} ) *)
c.(i) <- inf_N_minus valeurs;
end;
done;
coloriage_depuis_couleurs intvl c;
;;
```
Une fois qu'on a un coloriage, à valeurs dans $0,\dots,k$ on récupère le nombre de couleurs comme $1 + \max c$, i.e., $k+1$.
```
let max_valeurs = List.fold_left max 0;;
let nombre_chromatique (colorg : coloriage) : int =
1 + max_valeurs (List.map snd colorg)
;;
```
### Algorithme pour calculer le *stable maximum* d'un graphe d'intervalles
On répond ici à la question 7.
> « Proposer un algorithme efficace pour construire un stable maximum (i.e., un ensemble de sommets indépendants) d'un graphe d’intervalles dont on connaı̂t une représentation sous forme d'intervalles.
> On pourra chercher à quelle condition l'intervalle dont l'extrémité droite est la plus à gauche appartient à un stable maximum. »
**FIXME, je ne l'ai pas encore fait.**
----
## Exemples
On traite ici l'exemple introductif, ainsi que les trois autres exemples proposés.
### Qui a tué le Duc de Densmore ?
> On ne rappelle pas le problème, mais les données :
> - Ann dit avoir vu Betty, Cynthia, Emily, Felicia et Georgia.
- Betty dit avoir vu Ann, Cynthia et Helen.
- Cynthia dit avoir vu Ann, Betty, Diana, Emily et Helen.
- Diana dit avoir vu Cynthia et Emily.
- Emily dit avoir vu Ann, Cynthia, Diana et Felicia.
- Felicia dit avoir vu Ann et Emily.
- Georgia dit avoir vu Ann et Helen.
- Helen dit avoir vu Betty, Cynthia et Georgia.
Transcrit sous forme de graphe, cela donne :
```
(* On définit des entiers, c'est plus simple *)
let ann = 0
and betty = 1
and cynthia = 2
and diana = 3
and emily = 4
and felicia = 5
and georgia = 6
and helen = 7;;
let graphe_densmore = [
[betty; cynthia; emily; felicia; georgia]; (* Ann *)
[ann; cynthia; helen]; (* Betty *)
[ann; betty; diana; emily; helen]; (* Cynthia *)
[cynthia; emily]; (* Diana *)
[ann; cynthia; diana; felicia]; (* Emily *)
[ann; emily]; (* Felicia *)
[ann; helen]; (* Georgia *)
[betty; cynthia; georgia] (* Helen *)
];;
```

> Figure 1. Graphe d'intervalle pour le problème de l'assassinat du duc de Densmore.
Avec les prénoms plutôt que des numéros, cela donne :

> Figure 2. Graphe d'intervalle pour le problème de l'assassinat du duc de Densmore.
#### Comment résoudre ce problème ?
> Il faut utiliser la caractérisation du théorème 2 du texte, et la définition des graphes parfaits.
- Définition + Théorème 2 (point 1) :
On sait qu'un graphe d'intervalle est parfait, et donc tous ses graphes induits le sont aussi.
La caractérisation via les cordes sur les cycles de taille $\geq 4$ permet de dire qu'un quadrilatère (cycle de taille $4$) n'est pas un graphe d'intervalle.
Donc un graphe qui contient un graphe induit étant un quadrilatère ne peut être un graphe d'intervalle.
Ainsi, sur cet exemple, comme on a deux quadrilatères $A B H G$ et $A G H C$, on en déduit que $A$, $G$, ou $H$ ont menti.
- Théorème 2 (point 2) :
Ensuite, si on enlève $G$ ou $H$, le graphe ne devient pas un graphe d'intervalle, par les considérations suivantes, parce que son complémentaire n'est pas un graphe de comparaison.
En effet, par exemple si on enlève $G$, $A$ et $H$ et $D$ forment une clique dans le complémentaire $\overline{G}$ de $G$, et l'irréflexivité d'une éventuelle relation $R$ rend cela impossible. Pareil si on enlève $H$, avec $G$ et $B$ et $D$ qui formet une clique dans $\overline{G}$.
Par contre, si on enlève $A$, le graphe devient triangulé (et de comparaison, mais c'est plus dur à voir !).
Donc seule $A$ reste comme potentielle menteuse.
> « Mais... Ça semble difficile de programmer une résolution automatique de ce problème ? »
En fait, il suffit d'écrire une fonction de vérification qu'un graphe est un graphe d'intervalle, puis on essaie d'enlever chaque sommet, tant que le graphe n'est pas un graphe d'intervalle.
Si le graphe devient valide en enlevant un seul sommet, et qu'il n'y en a qu'un seul qui fonctionne, alors il y a un(e) seul(e) menteur(se) dans le graphe, et donc un(e) seul(e) coupable !
#### Solution
C'est donc $A$, i.e., Ann l'unique menteuse et donc la coupable.
> Ce n'est pas grave de ne pas avoir réussi à répondre durant l'oral !
> Au contraire, vous avez le droit de vous détacher du problème initial du texte !
> Une solution bien expliquée peut être trouvée dans [cette vidéo](https://youtu.be/ZGhSyVvOelg) :
<iframe width="640" height="360" src="https://www.youtube.com/embed/ZGhSyVvOelg" frameborder="1" allowfullscreen></iframe>
### Le problème des frigos
> Dans un grand hopital, les réductions de financement public poussent le gestionnaire du service d'immunologie à faire des économies sur le nombre de frigos à acheter pour stocker les vaccins. A peu de chose près, il lui faut stocker les vaccins suivants :
> | Numéro | Nom du vaccin | Température de conservation
| :-----: | :------------ | -------------------------: |
| 0 | Rougeole-Rubéole-Oreillons (RRO) | $4 \cdots 12$ °C
| 1 | BCG | $8 \cdots 15$ °C
| 2 | Di-Te-Per | $0 \cdots 20$ °C
| 3 | Anti-polio | $2 \cdots 3$ °C
| 4 | Anti-hépatite B | $-3 \cdots 6$ °C
| 5 | Anti-amarile | $-10 \cdots 10$ °C
| 6 | Variole | $6 \cdots 20$ °C
| 7 | Varicelle | $-5 \cdots 2$ °C
| 8 | Antihaemophilus | $-2 \cdots 8$ °C
> Combien le gestionaire doit-il acheter de frigos, et sur quelles températures doit-il les régler ?
```
let vaccins : intervalles = [
(4, 12);
(8, 15);
(0, 20);
(2, 3);
(-3, 6);
(-10, 10);
(6, 20);
(-5, 2);
(-2, 8)
]
```
Qu'on peut visualiser sous forme de graphe facilement :
```
let graphe_vaccins = graphe_depuis_intervalles vaccins;;
```

> Figure 3. Graphe d'intervalle pour le problème des frigos et des vaccins.
Avec des intervalles au lieu de numéro :

> Figure 4. Graphe d'intervalle pour le problème des frigos et des vaccins.
On peut récupérer une coloriage minimal pour ce graphe :
```
coloriage_intervalles vaccins;;
```
La couleur la plus grande est `5`, donc le nombre chromatique de ce graphe est `6`.
```
nombre_chromatique (coloriage_intervalles vaccins);;
```
Par contre, la solution au problème des frigos et des vaccins réside dans le nombre de couverture de cliques, $k(G)$, pas dans le nombre chromatique $\chi(G)$.
On peut le résoudre en répondant à la question 7, qui demandait de mettre au point un algorithme pour construire un *stable maximum* pour un graphe d'intervalle.
### Le problème du CSA
> Le Conseil Supérieur de l’Audiovisuel doit attribuer de nouvelles bandes de fréquences d’émission pour la stéréophonie numérique sous-terraine (SNS).
> Cette technologie de pointe étant encore à l'état expérimental, les appareils capables d'émettre ne peuvent utiliser que les bandes de fréquences FM suivantes :
> | Bandes de fréquence | Intervalle (kHz) |
| :-----------------: | ---------: |
| 0 | $32 \cdots 36$ |
| 1 | $24 \cdots 30$ |
| 2 | $28 \cdots 33$ |
| 3 | $22 \cdots 26$ |
| 4 | $20 \cdots 25$ |
| 5 | $30 \cdots 33$ |
| 6 | $31 \cdots 34$ |
| 7 | $27 \cdots 31$ |
> Quelles bandes de fréquences doit-on retenir pour permettre à le plus d'appareils possibles d'être utilisés, sachant que deux appareils dont les bandes de fréquences s'intersectent pleinement (pas juste sur les extrémités) sont incompatibles.
```
let csa : intervalles = [
(32, 36);
(24, 30);
(28, 33);
(22, 26);
(20, 25);
(30, 33);
(31, 34);
(27, 31)
];;
let graphe_csa = graphe_depuis_intervalles csa;;
```

> Figure 5. Graphe d'intervalle pour le problème du CSA.
Avec des intervalles au lieu de numéro :

> Figure 6. Graphe d'intervalle pour le problème du CSA.
On peut récupérer une coloriage minimal pour ce graphe :
```
coloriage_intervalles csa;;
```
La couleur la plus grande est `3`, donc le nombre chromatique de ce graphe est `4`.
```
nombre_chromatique (coloriage_intervalles csa);;
```
Par contre, la solution au problème CSA réside dans le nombre de couverture de cliques, $k(G)$, pas dans le nombre chromatique $\chi(G)$.
On peut le résoudre en répondant à la question 7, qui demandait de mettre au point un algorithme pour construire un *stable maximum* pour un graphe d'intervalle.
### Le problème du wagon restaurant
> Le chef de train de l'Orient Express doit aménager le wagon restaurant avant le départ du train. Ce wagon est assez petit et doit être le moins encombré de tables possibles, mais il faut prévoir suffisemment de tables pour accueillir toutes personnes qui ont réservé :
> | Numéro | Personnage(s) | Heures de dîner | En secondes |
| :----------------- | --------- | :---------: | :---------: |
| 0 | Le baron et la baronne Von Haussplatz | 19h30 .. 20h14 | $1170 \cdots 1214$
| 1 | Le général Cook | 20h30 .. 21h59 | $1230 \cdots 1319$
| 2 | Les époux Steinberg | 19h .. 19h59 | $1140 \cdots 1199$
| 3 | La duchesse de Colombart | 20h15 .. 20h59 | $1215 \cdots 1259$
| 4 | Le marquis de Carquamba | 21h .. 21h59 | $1260 \cdots 1319$
| 5 | La Vociafiore | 19h15 .. 20h29 | $1155 \cdots 1229$
| 6 | Le colonel Ferdinand | 20h .. 20h59 | $1200 \cdots 1259$
> Combien de tables le chef de train doit-il prévoir ?
```
let restaurant = [
(1170, 1214);
(1230, 1319);
(1140, 1199);
(1215, 1259);
(1260, 1319);
(1155, 1229);
(1200, 1259)
];;
let graphe_restaurant = graphe_depuis_intervalles restaurant;;
```

> Figure 7. Graphe d'intervalle pour le problème du wagon restaurant.
Avec des intervalles au lieu de numéro :

> Figure 8. Graphe d'intervalle pour le problème du wagon restaurant.
```
coloriage_intervalles restaurant;;
```
La couleur la plus grande est `2`, donc le nombre chromatique de ce graphe est `3`.
```
nombre_chromatique (coloriage_intervalles restaurant);;
```
#### Solution via l'algorithme de coloriage de graphe d'intervalles
Pour ce problème là, la solution est effectivement donnée par le nombre chromatique.
La couleur sera le numéro de table pour chaque passagers (ou couple de passagers), et donc le nombre minimal de table à installer dans le wagon restaurant est exactement le nombre chromatique.
Une solution peut être la suivante, avec **3 tables** :
| Numéro | Personnage(s) | Heures de dîner | Numéro de table |
| :----------------- | --------- | :---------: | :---------: |
| 0 | Le baron et la baronne Von Haussplatz | 19h30 .. 20h14 | 2
| 1 | Le général Cook | 20h30 .. 21h59 | 1
| 2 | Les époux Steinberg | 19h .. 19h59 | 0
| 3 | La duchesse de Colombart | 20h15 .. 20h59 | 2
| 4 | Le marquis de Carquamba | 21h .. 21h59 | 0
| 5 | La Vociafiore | 19h15 .. 20h29 | 1
| 6 | Le colonel Ferdinand | 20h .. 20h59 | 0
On vérifie manuellement que la solution convient.
Chaque passager devra quitter sa tableau à la minute près par contre !
On peut afficher la solution avec un graphe colorié.
La table `0` sera <span style="color:red;">rouge</span>, `1` sera <span style="color:blue;">bleu</span> et `2` sera <span style="color:yellow;">jaune</span> :

> Figure 9. Solution pour le problème du wagon restaurant.
----
## Bonus ?
### Visualisation des graphes définis dans les exemples
- J'utilise une petite fonction facile à écrire, qui convertit un graphe (`int list list`) en une chaîne de caractère au format [DOT Graph](http://www.graphviz.org/doc/info/lang.html).
- Ensuite, un appel `dot -Tpng ...` en ligne de commande convertit ce graphe en une image, que j'inclus ensuite manuellement.
```
(** Transforme un [graph] en une chaîne représentant un graphe décrit par le langage DOT,
voir http://en.wikipedia.org/wiki/DOT_language pour plus de détails sur ce langage.
@param graphname Donne le nom du graphe tel que précisé pour DOT
@param directed Vrai si le graphe doit être dirigé (c'est le cas ici) faux sinon. Change le style des arêtes ([->] ou [--])
@param verb Affiche tout dans le terminal.
@param onetoone Si on veut afficher le graphe en mode carré (échelle 1:1). Parfois bizarre, parfois génial.
*)
let graph_to_dotgraph ?(graphname = "graphname") ?(directed = false) ?(verb = false) ?(onetoone = false) (glist : int list list) =
let res = ref "" in
let log s =
if verb then print_string s; (* Si [verb] affiche dans le terminal le résultat du graphe. *)
res := !res ^ s
in
log (if directed then "digraph " else "graph ");
log graphname; log " {";
if onetoone then
log "\n size=\"1,1\";";
let g = Array.of_list (List.map Array.of_list glist) in
(* On affiche directement les arc, un à un. *)
for i = 0 to (Array.length g) - 1 do
for j = 0 to (Array.length g.(i)) - 1 do
if i < g.(i).(j) then
log ("\n \""
^ (string_of_int i) ^ "\" "
^ (if directed then "->" else "--")
^ " \"" ^ (string_of_int g.(i).(j)) ^ "\""
);
done;
done;
log "\n}\n// generated by OCaml with the function graphe_to_dotgraph.";
!res;;
(** Fonction ecrire_sortie : plus pratique que output. *)
let ecrire_sortie monoutchanel machaine =
output monoutchanel machaine 0 (String.length machaine);
flush monoutchanel;;
(** Fonction ecrire_dans_fichier : pour écrire la chaine dans le fichier à l'adresse renseignée. *)
let ecrire_dans_fichier ~chaine ~adresse =
let mon_out_channel = open_out adresse in
ecrire_sortie mon_out_channel chaine;
close_out mon_out_channel;;
let s_graphe_densmore = graph_to_dotgraph ~graphname:"densmore" ~directed:false ~verb:false graphe_densmore;;
let s_graphe_vaccins = graph_to_dotgraph ~graphname:"vaccins" ~directed:false ~verb:false graphe_vaccins;;
let s_graphe_csa = graph_to_dotgraph ~graphname:"csa" ~directed:false ~verb:false graphe_csa;;
let s_graphe_restaurant = graph_to_dotgraph ~graphname:"restaurant" ~directed:false ~verb:false graphe_restaurant;;
ecrire_dans_fichier ~chaine:s_graphe_densmore ~adresse:"/tmp/densmore.dot" ;;
(* Sys.command "fdp -Tpng /tmp/densmore.dot > images/densmore.png";; *)
ecrire_dans_fichier ~chaine:s_graphe_vaccins ~adresse:"/tmp/vaccins.dot" ;;
(* Sys.command "fdp -Tpng /tmp/vaccins.dot > images/vaccins.png";; *)
ecrire_dans_fichier ~chaine:s_graphe_csa ~adresse:"/tmp/csa.dot" ;;
(* Sys.command "fdp -Tpng /tmp/csa.dot > images/csa.png";; *)
ecrire_dans_fichier ~chaine:s_graphe_restaurant ~adresse:"/tmp/restaurant.dot" ;;
(* Sys.command "fdp -Tpng /tmp/restaurant.dot > images/restaurant.png";; *)
```
On pourrait étendre cette fonction pour qu'elle prenne les intervalles initiaux, pour afficher des bonnes étiquettes et pas des entiers, et un coloriage pour colorer directement les noeuds, mais ça prend du temps pour pas grand chose.
----
## Conclusion
Voilà pour la question obligatoire de programmation, sur l'algorithme de coloriage.
- on a décomposé le problème en sous-fonctions,
- on a fait des exemples et *on les garde* dans ce qu'on présente au jury,
- on a testé la fonction exigée sur de petits exemples et sur un exemple de taille réelle (venant du texte)
Et on a pas essayé de faire *un peu plus*.
Avec plus de temps, on aurait aussi pu écrire un algorithme pour calculer le stable maximum (ensemble de sommets indépendants de taille maximale).
> Bien-sûr, ce petit notebook ne se prétend pas être une solution optimale, ni exhaustive.
| github_jupyter |
```
import torch
from torch.autograd import grad
import torch.nn as nn
from numpy import genfromtxt
import torch.optim as optim
import matplotlib.pyplot as plt
import torch.nn.functional as F
import math
tuberculosis_data = genfromtxt('tuberculosis.csv', delimiter=',') #in the form of [t, S,L,I,T]
torch.manual_seed(1234)
%%time
PATH = 'tuberculosis'
class DINN(nn.Module):
def __init__(self, t, S_data, L_data, I_data, T_data):
super(DINN, self).__init__()
self.t = torch.tensor(t, requires_grad=True)
self.t_float = self.t.float()
self.t_batch = torch.reshape(self.t_float, (len(self.t),1)) #reshape for batch
self.S = torch.tensor(S_data)
self.L = torch.tensor(L_data)
self.I = torch.tensor(I_data)
self.T = torch.tensor(T_data)
self.N = torch.tensor(1001)
self.losses = [] #keep the losses
self.save = 2 #which file to save to
#learnable parameters
self.delta_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(500)
self.beta_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(13)
self.c_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(1)
self.mu_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(0.143)
self.k_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(0.5)
self.r_1_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(2)
self.r_2_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(1)
self.beta_prime_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(13)
self.d_tilda = torch.nn.Parameter(torch.rand(1, requires_grad=True)) #torch.tensor(0)
#matrices (x4 for S, L, I, T) for the gradients
self.m1 = torch.zeros((len(self.t), 4)); self.m1[:, 0] = 1
self.m2 = torch.zeros((len(self.t), 4)); self.m2[:, 1] = 1
self.m3 = torch.zeros((len(self.t), 4)); self.m3[:, 2] = 1
self.m4 = torch.zeros((len(self.t), 4)); self.m4[:, 3] = 1
#values for norm
self.S_max = max(self.S)
self.S_min = min(self.S)
self.L_max = max(self.L)
self.L_min = min(self.L)
self.I_max = max(self.I)
self.I_min = min(self.I)
self.T_max = max(self.T)
self.T_min = min(self.T)
#normalize
self.S_hat = (self.S - self.S_min) / (self.S_max - self.S_min)
self.L_hat = (self.L - self.L_min) / (self.L_max - self.L_min)
self.I_hat = (self.I - self.I_min) / (self.I_max - self.I_min)
self.T_hat = (self.T - self.T_min) / (self.T_max - self.T_min)
#NN
self.net_tuberculosis = self.Net_tuberculosis()
self.params = list(self.net_tuberculosis.parameters())
self.params.extend(list([self.delta_tilda ,self.beta_tilda ,self.c_tilda ,self.mu_tilda ,self.k_tilda ,self.r_1_tilda ,self.r_2_tilda ,self.beta_prime_tilda ,self.d_tilda]))
#force parameters to be in a range
@property
def delta(self):
return torch.tanh(self.delta_tilda) * 20 + 500 #self.delta_tilda
@property
def beta(self):
return torch.tanh(self.beta_tilda) * 3 + 12 #self.beta_tilda
@property
def c(self):
return torch.tanh(self.c_tilda) * 2 + 1 #self.c_tilda
@property
def mu(self):
return torch.tanh(self.mu_tilda) * 0.1 + 0.2 #self.mu_tilda
@property
def k(self):
return torch.tanh(self.k_tilda) * 0.5 + 0.5 #self.k_tilda
@property
def r_1(self):
return torch.tanh(self.r_1_tilda) + 2 #self.r_1_tilda
@property
def r_2(self):
return torch.tanh(self.r_2_tilda) * 2 + 1 #self.r_2_tilda
@property
def beta_prime(self):
return torch.tanh(self.beta_prime_tilda) * 3 + 12 #self.beta_prime_tilda
@property
def d(self):
return torch.tanh(self.d_tilda) * 0.4 #self.d_tilda
#nets
class Net_tuberculosis(nn.Module): # input = [t]
def __init__(self):
super(DINN.Net_tuberculosis, self).__init__()
self.fc1=nn.Linear(1, 20) #takes 100 t's
self.fc2=nn.Linear(20, 20)
self.fc3=nn.Linear(20, 20)
self.fc4=nn.Linear(20, 20)
self.fc5=nn.Linear(20, 20)
self.fc6=nn.Linear(20, 20)
self.fc7=nn.Linear(20, 20)
self.fc8=nn.Linear(20, 20)
self.out=nn.Linear(20, 4) #outputs S, L, I, T
def forward(self, t):
tuberculosis=F.relu(self.fc1(t))
tuberculosis=F.relu(self.fc2(tuberculosis))
tuberculosis=F.relu(self.fc3(tuberculosis))
tuberculosis=F.relu(self.fc4(tuberculosis))
tuberculosis=F.relu(self.fc5(tuberculosis))
tuberculosis=F.relu(self.fc6(tuberculosis))
tuberculosis=F.relu(self.fc7(tuberculosis))
tuberculosis=F.relu(self.fc8(tuberculosis))
tuberculosis=self.out(tuberculosis)
return tuberculosis
def net_f(self, t_batch):
tuberculosis_hat = self.net_tuberculosis(t_batch)
S_hat, L_hat, I_hat, T_hat = tuberculosis_hat[:,0], tuberculosis_hat[:,1], tuberculosis_hat[:,2], tuberculosis_hat[:,3]
#S_hat
tuberculosis_hat.backward(self.m1, retain_graph=True)
S_hat_t = self.t.grad.clone()
self.t.grad.zero_()
#L_hat
tuberculosis_hat.backward(self.m2, retain_graph=True)
L_hat_t = self.t.grad.clone()
self.t.grad.zero_()
#I_hat
tuberculosis_hat.backward(self.m3, retain_graph=True)
I_hat_t = self.t.grad.clone()
self.t.grad.zero_()
#T_hat
tuberculosis_hat.backward(self.m4, retain_graph=True)
T_hat_t = self.t.grad.clone()
self.t.grad.zero_()
#unnormalize
S = self.S_min + (self.S_max - self.S_min) * S_hat
L = self.L_min + (self.L_max - self.L_min) * L_hat
I = self.I_min + (self.I_max - self.I_min) * I_hat
T = self.T_min + (self.T_max - self.T_min) * T_hat
#equations
f1_hat = S_hat_t - (self.delta - self.beta * self.c * S * I / self.N - self.mu * S) / (self.S_max - self.S_min)
f2_hat = L_hat_t - (self.beta * self.c * S * I / self.N - (self.mu + self.k + self.r_1) * L + self.beta_prime * self.c * T * 1/self.N) / (self.L_max - self.L_min)
f3_hat = I_hat_t - (self.k*L - (self.mu + self.d) * I - self.r_2 * I) / (self.I_max - self.I_min)
f4_hat = T_hat_t - (self.r_1 * L + self.r_2 * I - self.beta_prime * self.c * T * 1/self.N - self.mu*T) / (self.T_max - self.T_min)
return f1_hat, f2_hat, f3_hat, f4_hat, S_hat, L_hat, I_hat, T_hat
def load(self):
# Load checkpoint
try:
checkpoint = torch.load(PATH + str(self.save)+'.pt')
print('\nloading pre-trained model...')
self.load_state_dict(checkpoint['model'])
self.optimizer.load_state_dict(checkpoint['optimizer_state_dict'])
self.scheduler.load_state_dict(checkpoint['scheduler'])
epoch = checkpoint['epoch']
loss = checkpoint['loss']
self.losses = checkpoint['losses']
print('loaded previous loss: ', loss)
except RuntimeError :
print('changed the architecture, ignore')
pass
except FileNotFoundError:
pass
def train(self, n_epochs):
#try loading
self.load()
#train
print('\nstarting training...\n')
for epoch in range(n_epochs):
#lists to hold the output (maintain only the final epoch)
S_pred_list = []
L_pred_list = []
I_pred_list = []
T_pred_list = []
f1_hat, f2_hat, f3_hat, f4_hat, S_hat_pred, L_hat_pred, I_hat_pred, T_hat_pred = self.net_f(self.t_batch)
self.optimizer.zero_grad()
S_pred_list.append(self.S_min + (self.S_max - self.S_min) * S_hat_pred)
L_pred_list.append(self.L_min + (self.L_max - self.L_min) * L_hat_pred)
I_pred_list.append(self.I_min + (self.I_max - self.I_min) * I_hat_pred)
T_pred_list.append(self.T_min + (self.T_max - self.T_min) * T_hat_pred)
loss = (
torch.mean(torch.square(self.S_hat - S_hat_pred)) + torch.mean(torch.square(self.L_hat - L_hat_pred)) +
torch.mean(torch.square(self.I_hat - I_hat_pred)) + torch.mean(torch.square(self.T_hat - T_hat_pred))+
torch.mean(torch.square(f1_hat)) + torch.mean(torch.square(f2_hat)) +
torch.mean(torch.square(f3_hat)) + torch.mean(torch.square(f4_hat))
)
loss.backward()
self.optimizer.step()
self.scheduler.step()
# self.scheduler.step(loss)
self.losses.append(loss.item())
if epoch % 1000 == 0:
print('\nEpoch ', epoch)
#loss + model parameters update
if epoch % 4000 == 9999:
#checkpoint save
print('\nSaving model... Loss is: ', loss)
torch.save({
'epoch': epoch,
'model': self.state_dict(),
'optimizer_state_dict': self.optimizer.state_dict(),
'scheduler': self.scheduler.state_dict(),
'loss': loss,
'losses': self.losses,
}, PATH + str(self.save)+'.pt')
if self.save % 2 > 0: #its on 3
self.save = 2 #change to 2
else: #its on 2
self.save = 3 #change to 3
print('epoch: ', epoch)
print('#################################')
#plot
plt.plot(self.losses, color = 'teal')
plt.xlabel('Epochs')
plt.ylabel('Loss')
return S_pred_list, L_pred_list, I_pred_list, T_pred_list
%%time
dinn = DINN(tuberculosis_data[0], tuberculosis_data[1], tuberculosis_data[2], tuberculosis_data[3], tuberculosis_data[4])
learning_rate = 1e-3
optimizer = optim.Adam(dinn.params, lr = learning_rate)
dinn.optimizer = optimizer
scheduler = torch.optim.lr_scheduler.CyclicLR(dinn.optimizer, base_lr=1e-7, max_lr=1e-3, step_size_up=1000, mode="exp_range", gamma=0.85, cycle_momentum=False)
dinn.scheduler = scheduler
try:
S_pred_list, L_pred_list, I_pred_list, T_pred_list = dinn.train(1) #train
except EOFError:
if dinn.save == 2:
dinn.save = 3
S_pred_list, L_pred_list, I_pred_list, T_pred_list = dinn.train(1) #train
elif dinn.save == 3:
dinn.save = 2
S_pred_list, L_pred_list, I_pred_list, T_pred_list = dinn.train(1) #train
plt.plot(dinn.losses[3000000:], color = 'teal')
plt.xlabel('Epochs')
plt.ylabel('Loss')
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111, facecolor='#dddddd', axisbelow=True)
ax.set_facecolor('xkcd:white')
ax.scatter(tuberculosis_data[0], tuberculosis_data[1], color = 'pink', alpha=0.5, lw=2, label='S Data', s=20)
ax.plot(tuberculosis_data[0], S_pred_list[0].detach().numpy(), 'navy', alpha=0.9, lw=2, label='S Prediction', linestyle='dashed')
ax.scatter(tuberculosis_data[0], tuberculosis_data[2], color = 'violet', alpha=0.5, lw=2, label='L Data', s=20)
ax.plot(tuberculosis_data[0], L_pred_list[0].detach().numpy(), 'dodgerblue', alpha=0.9, lw=2, label='L Prediction', linestyle='dashed')
ax.scatter(tuberculosis_data[0], tuberculosis_data[3], color = 'darkgreen', alpha=0.5, lw=2, label='I Data', s=20)
ax.plot(tuberculosis_data[0], I_pred_list[0].detach().numpy(), 'gold', alpha=0.9, lw=2, label='I Prediction', linestyle='dashed')
ax.scatter(tuberculosis_data[0], tuberculosis_data[4], color = 'red', alpha=0.5, lw=2, label='T Data', s=20)
ax.plot(tuberculosis_data[0], T_pred_list[0].detach().numpy(), 'blue', alpha=0.9, lw=2, label='T Prediction', linestyle='dashed')
ax.set_xlabel('Time /days',size = 20)
ax.set_ylabel('Number',size = 20)
#ax.set_ylim([-1,50])
ax.yaxis.set_tick_params(length=0)
ax.xaxis.set_tick_params(length=0)
plt.xticks(size = 20)
plt.yticks(size = 20)
# ax.grid(b=True, which='major', c='black', lw=0.2, ls='-')
legend = ax.legend(prop={'size':20})
legend.get_frame().set_alpha(0.5)
for spine in ('top', 'right', 'bottom', 'left'):
ax.spines[spine].set_visible(False)
plt.savefig('tuberculosis.pdf')
plt.show()
#vaccination!
import numpy as np
from scipy.integrate import odeint
import matplotlib.pyplot as plt
# Initial conditions
S0 = 1000
L0 = 0
I0 = 1
T0 = 0
N = 1001 #S0 + L0 + I0 + T0
# A grid of time points (in days)
t = np.linspace(0, 40, 50)
#parameters
delta = dinn.delta
print(delta)
beta = dinn.beta
print(beta)
c = dinn.c
print(c)
mu = dinn.mu
print(mu)
k = dinn.k
print(k)
r_1 = dinn.r_1
print(r_1)
r_2 = dinn.r_2
print(r_2)
beta_prime = dinn.beta_prime
print(beta_prime)
d = dinn.d
print(d)
# The SIR model differential equations.
def deriv(y, t, N, delta ,beta ,c ,mu ,k ,r_1 ,r_2 ,beta_prime,d ):
S, L, I, T= y
dSdt = delta - beta * c * S * I / N - mu * S
dLdt = beta * c * S * I / N - (mu + k + r_1) * L + beta_prime * c * T * 1/N
dIdt = k*L - (mu + d) * I - r_2 * I
dTdt = r_1 * L + r_2 * I - beta_prime * c * T * 1/N - mu*T
return dSdt, dLdt, dIdt, dTdt
# Initial conditions vector
y0 = S0, L0, I0, T0
# Integrate the SIR equations over the time grid, t.
ret = odeint(deriv, y0, t, args=(N, delta ,beta ,c ,mu ,k ,r_1 ,r_2 ,beta_prime,d ))
S, L, I, T = ret.T
# Plot the data on two separate curves for S(t), I(t)
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111, facecolor='#dddddd', axisbelow=True)
ax.set_facecolor('xkcd:white')
ax.plot(t, S, 'violet', alpha=0.5, lw=2, label='S_pred', linestyle='dashed')
ax.plot(tuberculosis_data[0], tuberculosis_data[1], 'grey', alpha=0.5, lw=2, label='S')
ax.plot(t, L, 'darkgreen', alpha=0.5, lw=2, label='L_pred', linestyle='dashed')
ax.plot(tuberculosis_data[0], tuberculosis_data[2], 'purple', alpha=0.5, lw=2, label='L')
ax.plot(t, I, 'blue', alpha=0.5, lw=2, label='I_pred', linestyle='dashed')
ax.plot(tuberculosis_data[0], tuberculosis_data[3], 'teal', alpha=0.5, lw=2, label='I')
ax.plot(t, T, 'black', alpha=0.5, lw=2, label='T_pred', linestyle='dashed')
ax.plot(tuberculosis_data[0], tuberculosis_data[4], 'red', alpha=0.5, lw=2, label='T')
ax.set_xlabel('Time /days',size = 20)
ax.set_ylabel('Number',size = 20)
#ax.set_ylim([-1,50])
ax.yaxis.set_tick_params(length=0)
ax.xaxis.set_tick_params(length=0)
plt.xticks(size = 20)
plt.yticks(size = 20)
ax.grid(b=True, which='major', c='black', lw=0.2, ls='-')
legend = ax.legend(prop={'size':20})
legend.get_frame().set_alpha(0.5)
for spine in ('top', 'right', 'bottom', 'left'):
ax.spines[spine].set_visible(False)
plt.show()
#calculate relative MSE loss
import math
S_total_loss = 0
S_den = 0
L_total_loss = 0
L_den = 0
I_total_loss = 0
I_den = 0
T_total_loss = 0
T_den = 0
for timestep in range(len(t)):
S_value = tuberculosis_data[1][timestep] - S[timestep]
S_total_loss += S_value**2
S_den += (tuberculosis_data[1][timestep])**2
L_value = tuberculosis_data[2][timestep] - L[timestep]
L_total_loss += L_value**2
L_den += (tuberculosis_data[2][timestep])**2
I_value = tuberculosis_data[3][timestep] - I[timestep]
I_total_loss += I_value**2
I_den += (tuberculosis_data[3][timestep])**2
T_value = tuberculosis_data[4][timestep] - T[timestep]
T_total_loss += T_value**2
T_den += (tuberculosis_data[4][timestep])**2
S_total_loss = math.sqrt(S_total_loss/S_den)
L_total_loss = math.sqrt(L_total_loss/L_den)
I_total_loss = math.sqrt(I_total_loss/I_den)
T_total_loss = math.sqrt(T_total_loss/T_den)
print('S_total_loss: ', S_total_loss)
print('I_total_loss: ', L_total_loss)
print('S_total_loss: ', I_total_loss)
print('I_total_loss: ', T_total_loss)
```
| github_jupyter |
```
#IMPORT SEMUA LIBARARY
#IMPORT LIBRARY PANDAS
import pandas as pd
#IMPORT LIBRARY UNTUK POSTGRE
from sqlalchemy import create_engine
import psycopg2
#IMPORT LIBRARY CHART
from matplotlib import pyplot as plt
from matplotlib import style
#IMPORT LIBRARY BASE PATH
import os
import io
#IMPORT LIBARARY PDF
from fpdf import FPDF
#IMPORT LIBARARY CHART KE BASE64
import base64
#IMPORT LIBARARY EXCEL
import xlsxwriter
#FUNGSI UNTUK MENGUPLOAD DATA DARI CSV KE POSTGRESQL
def uploadToPSQL(columns, table, filePath, engine):
#FUNGSI UNTUK MEMBACA CSV
df = pd.read_csv(
os.path.abspath(filePath),
names=columns,
keep_default_na=False
)
#APABILA ADA FIELD KOSONG DISINI DIFILTER
df.fillna('')
#MENGHAPUS COLUMN YANG TIDAK DIGUNAKAN
del df['kategori']
del df['jenis']
del df['pengiriman']
del df['satuan']
#MEMINDAHKAN DATA DARI CSV KE POSTGRESQL
df.to_sql(
table,
engine,
if_exists='replace'
)
#DIHITUNG APABILA DATA YANG DIUPLOAD BERHASIL, MAKA AKAN MENGEMBALIKAN KELUARAN TRUE(BENAR) DAN SEBALIKNYA
if len(df) == 0:
return False
else:
return True
#FUNGSI UNTUK MEMBUAT CHART, DATA YANG DIAMBIL DARI DATABASE DENGAN MENGGUNAKAN ORDER DARI TANGGAL DAN JUGA LIMIT
#DISINI JUGA MEMANGGIL FUNGSI MAKEEXCEL DAN MAKEPDF
def makeChart(host, username, password, db, port, table, judul, columns, filePath, name, subjudul, limit, negara, basePath):
#TEST KONEKSI DATABASE
try:
#KONEKSI KE DATABASE
connection = psycopg2.connect(user=username,password=password,host=host,port=port,database=db)
cursor = connection.cursor()
#MENGAMBL DATA DARI TABLE YANG DIDEFINISIKAN DIBAWAH, DAN DIORDER DARI TANGGAL TERAKHIR
#BISA DITAMBAHKAN LIMIT SUPAYA DATA YANG DIAMBIL TIDAK TERLALU BANYAK DAN BERAT
postgreSQL_select_Query = "SELECT * FROM "+table+" ORDER BY tanggal ASC LIMIT " + str(limit)
cursor.execute(postgreSQL_select_Query)
mobile_records = cursor.fetchall()
uid = []
lengthx = []
lengthy = []
#MELAKUKAN LOOPING ATAU PERULANGAN DARI DATA YANG SUDAH DIAMBIL
#KEMUDIAN DATA TERSEBUT DITEMPELKAN KE VARIABLE DIATAS INI
for row in mobile_records:
uid.append(row[0])
lengthx.append(row[1])
if row[2] == "":
lengthy.append(float(0))
else:
lengthy.append(float(row[2]))
#FUNGSI UNTUK MEMBUAT CHART
#bar
style.use('ggplot')
fig, ax = plt.subplots()
#MASUKAN DATA ID DARI DATABASE, DAN JUGA DATA TANGGAL
ax.bar(uid, lengthy, align='center')
#UNTUK JUDUL CHARTNYA
ax.set_title(judul)
ax.set_ylabel('Total')
ax.set_xlabel('Tanggal')
ax.set_xticks(uid)
#TOTAL DATA YANG DIAMBIL DARI DATABASE, DIMASUKAN DISINI
ax.set_xticklabels((lengthx))
b = io.BytesIO()
#CHART DISIMPAN KE FORMAT PNG
plt.savefig(b, format='png', bbox_inches="tight")
#CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64
barChart = base64.b64encode(b.getvalue()).decode("utf-8").replace("\n", "")
#CHART DITAMPILKAN
plt.show()
#line
#MASUKAN DATA DARI DATABASE
plt.plot(lengthx, lengthy)
plt.xlabel('Tanggal')
plt.ylabel('Total')
#UNTUK JUDUL CHARTNYA
plt.title(judul)
plt.grid(True)
l = io.BytesIO()
#CHART DISIMPAN KE FORMAT PNG
plt.savefig(l, format='png', bbox_inches="tight")
#CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64
lineChart = base64.b64encode(l.getvalue()).decode("utf-8").replace("\n", "")
#CHART DITAMPILKAN
plt.show()
#pie
#UNTUK JUDUL CHARTNYA
plt.title(judul)
#MASUKAN DATA DARI DATABASE
plt.pie(lengthy, labels=lengthx, autopct='%1.1f%%',
shadow=True, startangle=180)
plt.axis('equal')
p = io.BytesIO()
#CHART DISIMPAN KE FORMAT PNG
plt.savefig(p, format='png', bbox_inches="tight")
#CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64
pieChart = base64.b64encode(p.getvalue()).decode("utf-8").replace("\n", "")
#CHART DITAMPILKAN
plt.show()
#MENGAMBIL DATA DARI CSV YANG DIGUNAKAN SEBAGAI HEADER DARI TABLE UNTUK EXCEL DAN JUGA PDF
header = pd.read_csv(
os.path.abspath(filePath),
names=columns,
keep_default_na=False
)
#MENGHAPUS COLUMN YANG TIDAK DIGUNAKAN
header.fillna('')
del header['tanggal']
del header['total']
#MEMANGGIL FUNGSI EXCEL
makeExcel(mobile_records, header, name, limit, basePath)
#MEMANGGIL FUNGSI PDF
makePDF(mobile_records, header, judul, barChart, lineChart, pieChart, name, subjudul, limit, basePath)
#JIKA GAGAL KONEKSI KE DATABASE, MASUK KESINI UNTUK MENAMPILKAN ERRORNYA
except (Exception, psycopg2.Error) as error :
print (error)
#KONEKSI DITUTUP
finally:
if(connection):
cursor.close()
connection.close()
#FUNGSI MAKEEXCEL GUNANYA UNTUK MEMBUAT DATA YANG BERASAL DARI DATABASE DIJADIKAN FORMAT EXCEL TABLE F2
#PLUGIN YANG DIGUNAKAN ADALAH XLSXWRITER
def makeExcel(datarow, dataheader, name, limit, basePath):
#MEMBUAT FILE EXCEL
workbook = xlsxwriter.Workbook(basePath+'jupyter/BLOOMBERG/SektorHargaInflasi/excel/'+name+'.xlsx')
#MENAMBAHKAN WORKSHEET PADA FILE EXCEL TERSEBUT
worksheet = workbook.add_worksheet('sheet1')
#SETINGAN AGAR DIBERIKAN BORDER DAN FONT MENJADI BOLD
row1 = workbook.add_format({'border': 2, 'bold': 1})
row2 = workbook.add_format({'border': 2})
#MENJADIKAN DATA MENJADI ARRAY
data=list(datarow)
isihead=list(dataheader.values)
header = []
body = []
#LOOPING ATAU PERULANGAN, KEMUDIAN DATA DITAMPUNG PADA VARIABLE DIATAS
for rowhead in dataheader:
header.append(str(rowhead))
for rowhead2 in datarow:
header.append(str(rowhead2[1]))
for rowbody in isihead[1]:
body.append(str(rowbody))
for rowbody2 in data:
body.append(str(rowbody2[2]))
#MEMASUKAN DATA DARI VARIABLE DIATAS KE DALAM COLUMN DAN ROW EXCEL
for col_num, data in enumerate(header):
worksheet.write(0, col_num, data, row1)
for col_num, data in enumerate(body):
worksheet.write(1, col_num, data, row2)
#FILE EXCEL DITUTUP
workbook.close()
#FUNGSI UNTUK MEMBUAT PDF YANG DATANYA BERASAL DARI DATABASE DIJADIKAN FORMAT EXCEL TABLE F2
#PLUGIN YANG DIGUNAKAN ADALAH FPDF
def makePDF(datarow, dataheader, judul, bar, line, pie, name, subjudul, lengthPDF, basePath):
#FUNGSI UNTUK MENGATUR UKURAN KERTAS, DISINI MENGGUNAKAN UKURAN A4 DENGAN POSISI LANDSCAPE
pdf = FPDF('L', 'mm', [210,297])
#MENAMBAHKAN HALAMAN PADA PDF
pdf.add_page()
#PENGATURAN UNTUK JARAK PADDING DAN JUGA UKURAN FONT
pdf.set_font('helvetica', 'B', 20.0)
pdf.set_xy(145.0, 15.0)
#MEMASUKAN JUDUL KE DALAM PDF
pdf.cell(ln=0, h=2.0, align='C', w=10.0, txt=judul, border=0)
#PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING
pdf.set_font('arial', '', 14.0)
pdf.set_xy(145.0, 25.0)
#MEMASUKAN SUB JUDUL KE PDF
pdf.cell(ln=0, h=2.0, align='C', w=10.0, txt=subjudul, border=0)
#MEMBUAT GARIS DI BAWAH SUB JUDUL
pdf.line(10.0, 30.0, 287.0, 30.0)
pdf.set_font('times', '', 10.0)
pdf.set_xy(17.0, 37.0)
#PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING
pdf.set_font('Times','',10.0)
#MENGAMBIL DATA HEADER PDF YANG SEBELUMNYA SUDAH DIDEFINISIKAN DIATAS
datahead=list(dataheader.values)
pdf.set_font('Times','B',12.0)
pdf.ln(0.5)
th1 = pdf.font_size
#MEMBUAT TABLE PADA PDF, DAN MENAMPILKAN DATA DARI VARIABLE YANG SUDAH DIKIRIM
pdf.cell(100, 2*th1, "Kategori", border=1, align='C')
pdf.cell(177, 2*th1, datahead[0][0], border=1, align='C')
pdf.ln(2*th1)
pdf.cell(100, 2*th1, "Jenis", border=1, align='C')
pdf.cell(177, 2*th1, datahead[0][1], border=1, align='C')
pdf.ln(2*th1)
pdf.cell(100, 2*th1, "Pengiriman", border=1, align='C')
pdf.cell(177, 2*th1, datahead[0][2], border=1, align='C')
pdf.ln(2*th1)
pdf.cell(100, 2*th1, "Satuan", border=1, align='C')
pdf.cell(177, 2*th1, datahead[0][3], border=1, align='C')
pdf.ln(2*th1)
#PENGATURAN PADDING
pdf.set_xy(17.0, 75.0)
#PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING
pdf.set_font('Times','B',11.0)
data=list(datarow)
epw = pdf.w - 2*pdf.l_margin
col_width = epw/(lengthPDF+1)
#PENGATURAN UNTUK JARAK PADDING
pdf.ln(0.5)
th = pdf.font_size
#MEMASUKAN DATA HEADER YANG DIKIRIM DARI VARIABLE DIATAS KE DALAM PDF
pdf.cell(50, 2*th, str("Negara"), border=1, align='C')
for row in data:
pdf.cell(40, 2*th, str(row[1]), border=1, align='C')
pdf.ln(2*th)
#MEMASUKAN DATA ISI YANG DIKIRIM DARI VARIABLE DIATAS KE DALAM PDF
pdf.set_font('Times','B',10.0)
pdf.set_font('Arial','',9)
pdf.cell(50, 2*th, negara, border=1, align='C')
for row in data:
pdf.cell(40, 2*th, str(row[2]), border=1, align='C')
pdf.ln(2*th)
#MENGAMBIL DATA CHART, KEMUDIAN CHART TERSEBUT DIJADIKAN PNG DAN DISIMPAN PADA DIRECTORY DIBAWAH INI
#BAR CHART
bardata = base64.b64decode(bar)
barname = basePath+'jupyter/BLOOMBERG/SektorHargaInflasi/img/'+name+'-bar.png'
with open(barname, 'wb') as f:
f.write(bardata)
#LINE CHART
linedata = base64.b64decode(line)
linename = basePath+'jupyter/BLOOMBERG/SektorHargaInflasi/img/'+name+'-line.png'
with open(linename, 'wb') as f:
f.write(linedata)
#PIE CHART
piedata = base64.b64decode(pie)
piename = basePath+'jupyter/BLOOMBERG/SektorHargaInflasi/img/'+name+'-pie.png'
with open(piename, 'wb') as f:
f.write(piedata)
#PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING
pdf.set_xy(17.0, 75.0)
col = pdf.w - 2*pdf.l_margin
widthcol = col/3
#MEMANGGIL DATA GAMBAR DARI DIREKTORY DIATAS
pdf.image(barname, link='', type='',x=8, y=100, w=widthcol)
pdf.set_xy(17.0, 75.0)
col = pdf.w - 2*pdf.l_margin
pdf.image(linename, link='', type='',x=103, y=100, w=widthcol)
pdf.set_xy(17.0, 75.0)
col = pdf.w - 2*pdf.l_margin
pdf.image(piename, link='', type='',x=195, y=100, w=widthcol)
pdf.ln(2*th)
#MEMBUAT FILE PDF
pdf.output(basePath+'jupyter/BLOOMBERG/SektorHargaInflasi/pdf/'+name+'.pdf', 'F')
#DISINI TEMPAT AWAL UNTUK MENDEFINISIKAN VARIABEL VARIABEL SEBELUM NANTINYA DIKIRIM KE FUNGSI
#PERTAMA MANGGIL FUNGSI UPLOADTOPSQL DULU, KALAU SUKSES BARU MANGGIL FUNGSI MAKECHART
#DAN DI MAKECHART MANGGIL FUNGSI MAKEEXCEL DAN MAKEPDF
#DEFINISIKAN COLUMN BERDASARKAN FIELD CSV
columns = [
"kategori",
"jenis",
"tanggal",
"total",
"pengiriman",
"satuan",
]
#UNTUK NAMA FILE
name = "SektorHargaInflasi3_6"
#VARIABLE UNTUK KONEKSI KE DATABASE
host = "localhost"
username = "postgres"
password = "1234567890"
port = "5432"
database = "bloomberg_SektorHargaInflasi"
table = name.lower()
#JUDUL PADA PDF DAN EXCEL
judul = "Data Sektor Harga Inflasi"
subjudul = "Badan Perencanaan Pembangunan Nasional"
#LIMIT DATA UNTUK SELECT DI DATABASE
limitdata = int(8)
#NAMA NEGARA UNTUK DITAMPILKAN DI EXCEL DAN PDF
negara = "Indonesia"
#BASE PATH DIRECTORY
basePath = 'C:/Users/ASUS/Documents/bappenas/'
#FILE CSV
filePath = basePath+ 'data mentah/BLOOMBERG/SektorHargaInflasi/' +name+'.csv';
#KONEKSI KE DATABASE
engine = create_engine('postgresql://'+username+':'+password+'@'+host+':'+port+'/'+database)
#MEMANGGIL FUNGSI UPLOAD TO PSQL
checkUpload = uploadToPSQL(columns, table, filePath, engine)
#MENGECEK FUNGSI DARI UPLOAD PSQL, JIKA BERHASIL LANJUT MEMBUAT FUNGSI CHART, JIKA GAGAL AKAN MENAMPILKAN PESAN ERROR
if checkUpload == True:
makeChart(host, username, password, database, port, table, judul, columns, filePath, name, subjudul, limitdata, negara, basePath)
else:
print("Error When Upload CSV")
```
| github_jupyter |
```
from PIL import Image
import numpy as np
import os
import cv2
import keras
from keras.utils import np_utils
from keras.models import Sequential
from keras.layers import Conv2D,MaxPooling2D,Dense,Flatten,Dropout
import pandas as pd
import sys
import tensorflow as tf
%matplotlib inline
import matplotlib.pyplot as plt
import plotly.express as px
def readData(filepath, label):
cells = []
labels = []
file = os.listdir(filepath)
for img in file:
try:
image = cv2.imread(filepath + img)
image_from_array = Image.fromarray(image, 'RGB')
size_image = image_from_array.resize((50, 50))
cells.append(np.array(size_image))
labels.append(label)
except AttributeError as e:
print('Skipping file: ', img, e)
print(len(cells), ' Data Points Read!')
return np.array(cells), np.array(labels)
def genesis_train(file):
print('Reading Training Data')
ParasitizedCells, ParasitizedLabels = readData(file + '/Parasitized/', 1)
UninfectedCells, UninfectedLabels = readData(file + '/Uninfected/', 0)
Cells = np.concatenate((ParasitizedCells, UninfectedCells))
Labels = np.concatenate((ParasitizedLabels, UninfectedLabels))
print('Reading Testing Data')
TestParasitizedCells, TestParasitizedLabels = readData('./input/fed/test/Parasitized/', 1)
TestUninfectedCells, TestUninfectedLabels = readData('./input/fed/test/Uninfected/', 0)
TestCells = np.concatenate((TestParasitizedCells, TestUninfectedCells))
TestLabels = np.concatenate((TestParasitizedLabels, TestUninfectedLabels))
s = np.arange(Cells.shape[0])
np.random.shuffle(s)
Cells = Cells[s]
Labels = Labels[s]
sTest = np.arange(TestCells.shape[0])
np.random.shuffle(sTest)
TestCells = TestCells[sTest]
TestLabels = TestLabels[sTest]
num_classes=len(np.unique(Labels))
len_data=len(Cells)
print(len_data, ' Data Points')
(x_train,x_test)=Cells, TestCells
(y_train,y_test)=Labels, TestLabels
# Since we're working on image data, we normalize data by divinding 255.
x_train = x_train.astype('float32')/255
x_test = x_test.astype('float32')/255
train_len=len(x_train)
test_len=len(x_test)
#Doing One hot encoding as classifier has multiple classes
y_train=keras.utils.to_categorical(y_train,num_classes)
y_test=keras.utils.to_categorical(y_test,num_classes)
#creating sequential model
model=Sequential()
model.add(Conv2D(filters=16,kernel_size=2,padding="same",activation="relu",input_shape=(50,50,3)))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(500,activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(2,activation="softmax"))#2 represent output layer neurons
# model.summary()
# compile the model with loss as categorical_crossentropy and using adam optimizer
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
#Fit the model with min batch size as 50[can tune batch size to some factor of 2^power ]
model.fit(x_train, y_train, batch_size=100, epochs=5, verbose=1)
scores = model.evaluate(x_test, y_test)
print("Loss: ", scores[0]) #Loss
print("Accuracy: ", scores[1]) #Accuracy
#Saving Model
model.save("./output.h5")
return len_data, scores[1]
def update_train(file, d):
print('Reading Training Data')
ParasitizedCells, ParasitizedLabels = readData(file + '/Parasitized/', 1)
UninfectedCells, UninfectedLabels = readData(file + '/Uninfected/', 0)
Cells = np.concatenate((ParasitizedCells, UninfectedCells))
Labels = np.concatenate((ParasitizedLabels, UninfectedLabels))
print('Reading Testing Data')
TestParasitizedCells, TestParasitizedLabels = readData('./input/fed/test/Parasitized/', 1)
TestUninfectedCells, TestUninfectedLabels = readData('./input/fed/test/Uninfected/', 0)
TestCells = np.concatenate((TestParasitizedCells, TestUninfectedCells))
TestLabels = np.concatenate((TestParasitizedLabels, TestUninfectedLabels))
s = np.arange(Cells.shape[0])
np.random.shuffle(s)
Cells = Cells[s]
Labels = Labels[s]
sTest = np.arange(TestCells.shape[0])
np.random.shuffle(sTest)
TestCells = TestCells[sTest]
TestLabels = TestLabels[sTest]
num_classes=len(np.unique(Labels))
len_data=len(Cells)
print(len_data, ' Data Points')
(x_train,x_test)=Cells, TestCells
(y_train,y_test)=Labels, TestLabels
# Since we're working on image data, we normalize data by divinding 255.
x_train = x_train.astype('float32')/255
x_test = x_test.astype('float32')/255
train_len=len(x_train)
test_len=len(x_test)
#Doing One hot encoding as classifier has multiple classes
y_train=keras.utils.to_categorical(y_train,num_classes)
y_test=keras.utils.to_categorical(y_test,num_classes)
#creating sequential model
model=Sequential()
model.add(Conv2D(filters=16,kernel_size=2,padding="same",activation="relu",input_shape=(50,50,3)))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(500,activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(2,activation="softmax"))#2 represent output layer neurons
# model.summary()
model.load_weights("./output.h5")
# compile the model with loss as categorical_crossentropy and using adam optimizer
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
#Fit the model with min batch size as 50[can tune batch size to some factor of 2^power ]
model.fit(x_train, y_train, batch_size=100, epochs=5, verbose=1)
scores = model.evaluate(x_test, y_test)
print("Loss: ", scores[0]) #Loss
print("Accuracy: ", scores[1]) #Accuracy
#Saving Model
model.save("./weights/" + str(d) + ".h5")
return len_data, scores[1]
FLAccuracy = {}
# FLAccuracy['Complete Dataset'] = genesis_train('./input/cell_images')
FLAccuracy['Genesis'] = genesis_train('./input/fed/genesis')
FLAccuracy['d1'] = update_train('./input/fed/d1', 'd1')
FLAccuracy['d2'] = update_train('./input/fed/d2', 'd2')
FLAccuracy['d3'] = update_train('./input/fed/d3', 'd3')
FLAccuracy['d4'] = update_train('./input/fed/d4', 'd4')
FLAccuracy['d5'] = update_train('./input/fed/d5', 'd5')
FLAccuracy['d6'] = update_train('./input/fed/d6', 'd6')
FLAccuracy['d7'] = update_train('./input/fed/d7', 'd7')
FLAccuracy['d8'] = update_train('./input/fed/d8', 'd8')
FLAccuracy['d9'] = update_train('./input/fed/d9', 'd9')
FLAccuracy['d10'] = update_train('./input/fed/d10', 'd10')
FLAccuracy['d11'] = update_train('./input/fed/d11', 'd11')
FLAccuracy['d12'] = update_train('./input/fed/d12', 'd12')
FLAccuracy['d13'] = update_train('./input/fed/d13', 'd13')
FLAccuracy['d14'] = update_train('./input/fed/d14', 'd14')
FLAccuracy['d15'] = update_train('./input/fed/d15', 'd15')
FLAccuracy['d16'] = update_train('./input/fed/d16', 'd16')
FLAccuracy['d17'] = update_train('./input/fed/d17', 'd17')
FLAccuracy['d18'] = update_train('./input/fed/d18', 'd18')
FLAccuracy['d19'] = update_train('./input/fed/d19', 'd19')
FLAccuracy['d20'] = update_train('./input/fed/d20', 'd20')
FLAccuracy
FLAccuracyDF = pd.DataFrame.from_dict(FLAccuracy, orient='index', columns=['DataSize', 'Accuracy'])
FLAccuracyDF
FLAccuracyDF.index
n = 0
for w in FLAccuracy:
if 'Complete' in w:
continue
n += FLAccuracy[w][0]
print('Total number of data points in this round: ', n)
FLAccuracyDF['Weightage'] = FLAccuracyDF['DataSize'].apply(lambda x: x/n)
FLAccuracyDF
def scale(weight, scaler):
scaledWeights = []
for i in range(len(weight)):
scaledWeights.append(scaler * weight[i])
return scaledWeights
def getScaledWeight(d, scaler):
#creating sequential model
model=Sequential()
model.add(Conv2D(filters=16,kernel_size=2,padding="same",activation="relu",input_shape=(50,50,3)))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(500,activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(2,activation="softmax"))#2 represent output layer neurons
fpath = "./weights/"+d+".h5"
model.load_weights(fpath)
weight = model.get_weights()
scaledWeight = scale(weight, scaler)
return scaledWeight
def avgWeights(scaledWeights):
avg = list()
for weight_list_tuple in zip(*scaledWeights):
layer_mean = tf.math.reduce_sum(weight_list_tuple, axis=0)
avg.append(layer_mean)
return avg
def FedAvg(models):
scaledWeights = []
for m in models:
scaledWeights.append(getScaledWeight(m, FLAccuracyDF.loc[m]['Weightage']))
avgWeight = avgWeights(scaledWeights)
return avgWeight
models = ['d1', 'd2', 'd3', 'd4', 'd5', 'd6', 'd7', 'd8', 'd9', 'd10', 'd11', 'd12', 'd13', 'd14', 'd15', 'd16', 'd17', 'd18', 'd19', 'd20']
avgWeight = FedAvg(models)
print(avgWeight)
def testNewGlobal(weight):
print('Reading Testing Data')
TestParasitizedCells, TestParasitizedLabels = readData('./input/fed/test/Parasitized/', 1)
TestUninfectedCells, TestUninfectedLabels = readData('./input/fed/test/Uninfected/', 0)
TestCells = np.concatenate((TestParasitizedCells, TestUninfectedCells))
TestLabels = np.concatenate((TestParasitizedLabels, TestUninfectedLabels))
sTest = np.arange(TestCells.shape[0])
np.random.shuffle(sTest)
TestCells = TestCells[sTest]
TestLabels = TestLabels[sTest]
num_classes=len(np.unique(TestLabels))
(x_test) = TestCells
(y_test) = TestLabels
# Since we're working on image data, we normalize data by divinding 255.
x_test = x_test.astype('float32')/255
test_len=len(x_test)
#Doing One hot encoding as classifier has multiple classes
y_test=keras.utils.to_categorical(y_test,num_classes)
#creating sequential model
model=Sequential()
model.add(Conv2D(filters=16,kernel_size=2,padding="same",activation="relu",input_shape=(50,50,3)))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=32,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Conv2D(filters=64,kernel_size=2,padding="same",activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(500,activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(2,activation="softmax"))#2 represent output layer neurons
# model.summary()
model.set_weights(weight)
# compile the model with loss as categorical_crossentropy and using adam optimizer
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
scores = model.evaluate(x_test, y_test)
print("Loss: ", scores[0]) #Loss
print("Accuracy: ", scores[1]) #Accuracy
#Saving Model
model.save("./output.h5")
return scores[1]
testNewGlobal(avgWeight)
FLAccuracyDF
```
| github_jupyter |
# Mixture Density Networks with Edward, Keras and TensorFlow
This notebook explains how to implement Mixture Density Networks (MDN) with Edward, Keras and TensorFlow.
Keep in mind that if you want to use Keras and TensorFlow, like we do in this notebook, you need to set the backend of Keras to TensorFlow, [here](http://keras.io/backend/) it is explained how to do that.
In you are not familiar with MDNs have a look at the [following blog post](http://cbonnett.github.io/MDN.html) or at orginal [paper](http://research.microsoft.com/en-us/um/people/cmbishop/downloads/Bishop-NCRG-94-004.pdf) by Bishop.
Edward implements many probability distribution functions that are TensorFlow compatible, this makes it attractive to use Edward for MDNs.
Here are all the distributions that are currently implemented in Edward, there are more to come:
1. [Bernoulli](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L49)
2. [Beta](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L58)
3. [Binomial](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L68)
4. [Chi Squared](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L79)
5. [Dirichlet](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L89)
6. [Exponential](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L109)
7. [Gamma](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L118)
8. [Geometric](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L129)
9. [Inverse Gamma](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L138)
10. [log Normal](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L155)
11. [Multinomial](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L165)
12. [Multivariate Normal](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L194)
13. [Negative Binomial](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L283)
14. [Normal](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L294)
15. [Poisson](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L310)
16. [Student-t](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L319)
17. [Truncated Normal](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L333)
18. [Uniform](https://github.com/blei-lab/edward/blob/master/edward/stats/distributions.py#L352)
Let's start with the necessary imports.
```
# imports
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import edward as ed
import numpy as np
import tensorflow as tf
from edward.stats import norm # Normal distribution from Edward.
from keras import backend as K
from keras.layers import Dense
from sklearn.cross_validation import train_test_split
```
We will need some functions to plot the results later on, these are defined in the next code block.
```
from scipy.stats import norm as normal
def plot_normal_mix(pis, mus, sigmas, ax, label='', comp=True):
"""
Plots the mixture of Normal models to axis=ax
comp=True plots all components of mixture model
"""
x = np.linspace(-10.5, 10.5, 250)
final = np.zeros_like(x)
for i, (weight_mix, mu_mix, sigma_mix) in enumerate(zip(pis, mus, sigmas)):
temp = normal.pdf(x, mu_mix, sigma_mix) * weight_mix
final = final + temp
if comp:
ax.plot(x, temp, label='Normal ' + str(i))
ax.plot(x, final, label='Mixture of Normals ' + label)
ax.legend(fontsize=13)
def sample_from_mixture(x, pred_weights, pred_means, pred_std, amount):
"""
Draws samples from mixture model.
Returns 2 d array with input X and sample from prediction of Mixture Model
"""
samples = np.zeros((amount, 2))
n_mix = len(pred_weights[0])
to_choose_from = np.arange(n_mix)
for j,(weights, means, std_devs) in enumerate(zip(pred_weights, pred_means, pred_std)):
index = np.random.choice(to_choose_from, p=weights)
samples[j,1]= normal.rvs(means[index], std_devs[index], size=1)
samples[j,0]= x[j]
if j == amount -1:
break
return samples
```
## Making some toy-data to play with.
This is the same toy-data problem set as used in the [blog post](http://blog.otoro.net/2015/11/24/mixture-density-networks-with-tensorflow/) by Otoro where he explains MDNs. This is an inverse problem as you can see, for every ```X``` there are multiple ```y``` solutions.
```
def build_toy_dataset(nsample=40000):
y_data = np.float32(np.random.uniform(-10.5, 10.5, (1, nsample))).T
r_data = np.float32(np.random.normal(size=(nsample, 1))) # random noise
x_data = np.float32(np.sin(0.75 * y_data) * 7.0 + y_data * 0.5 + r_data * 1.0)
return train_test_split(x_data, y_data, random_state=42, train_size=0.1)
X_train, X_test, y_train, y_test = build_toy_dataset()
print("Size of features in training data: {:s}".format(X_train.shape))
print("Size of output in training data: {:s}".format(y_train.shape))
print("Size of features in test data: {:s}".format(X_test.shape))
print("Size of output in test data: {:s}".format(y_test.shape))
sns.regplot(X_train, y_train, fit_reg=False)
```
### Building a MDN using Edward, Keras and TF
We will define a class that can be used to construct MDNs. In this notebook we will be using a mixture of Normal Distributions. The advantage of defining a class is that we can easily reuse this to build other MDNs with different amount of mixture components. Furthermore, this makes it play nicely with Edward.
```
class MixtureDensityNetwork:
"""
Mixture density network for outputs y on inputs x.
p((x,y), (z,theta))
= sum_{k=1}^K pi_k(x; theta) Normal(y; mu_k(x; theta), sigma_k(x; theta))
where pi, mu, sigma are the output of a neural network taking x
as input and with parameters theta. There are no latent variables
z, which are hidden variables we aim to be Bayesian about.
"""
def __init__(self, K):
self.K = K # here K is the amount of Mixtures
def mapping(self, X):
"""pi, mu, sigma = NN(x; theta)"""
hidden1 = Dense(15, activation='relu')(X) # fully-connected layer with 15 hidden units
hidden2 = Dense(15, activation='relu')(hidden1)
self.mus = Dense(self.K)(hidden2) # the means
self.sigmas = Dense(self.K, activation=K.exp)(hidden2) # the variance
self.pi = Dense(self.K, activation=K.softmax)(hidden2) # the mixture components
def log_prob(self, xs, zs=None):
"""log p((xs,ys), (z,theta)) = sum_{n=1}^N log p((xs[n,:],ys[n]), theta)"""
# Note there are no parameters we're being Bayesian about. The
# parameters are baked into how we specify the neural networks.
X, y = xs
self.mapping(X)
result = tf.exp(norm.logpdf(y, self.mus, self.sigmas))
result = tf.mul(result, self.pi)
result = tf.reduce_sum(result, 1)
result = tf.log(result)
return tf.reduce_sum(result)
```
We can set a seed in Edward so we can reproduce all the random components. The following line:
```ed.set_seed(42)```
sets the seed in Numpy and TensorFlow under the [hood](https://github.com/blei-lab/edward/blob/master/edward/util.py#L191). We use the class we defined above to initiate the MDN with 20 mixtures, this now can be used as an Edward model.
```
ed.set_seed(42)
model = MixtureDensityNetwork(20)
```
In the following code cell we define the TensorFlow placeholders that are then used to define the Edward data model.
The following line passes the ```model``` and ```data``` to ```MAP``` from Edward which is then used to initialise the TensorFlow variables.
```inference = ed.MAP(model, data)```
MAP is a Bayesian concept and stands for Maximum A Posteriori, it tries to find the set of parameters which maximizes the posterior distribution. In the example here we don't have a prior, in a Bayesian context this means we have a flat prior. For a flat prior MAP is equivalent to Maximum Likelihood Estimation. Edward is designed to be Bayesian about its statistical inference. The cool thing about MDN's with Edward is that we could easily include priors!
```
X = tf.placeholder(tf.float32, shape=(None, 1))
y = tf.placeholder(tf.float32, shape=(None, 1))
data = ed.Data([X, y]) # Make Edward Data model
inference = ed.MAP(model, data) # Make the inference model
sess = tf.Session() # Start TF session
K.set_session(sess) # Pass session info to Keras
inference.initialize(sess=sess) # Initialize all TF variables using the Edward interface
```
Having done that we can train the MDN in TensorFlow just like we normally would, and we can get out the predictions we are interested in from ```model```, in this case:
* ```model.pi``` the mixture components,
* ```model.mus``` the means,
* ```model.sigmas``` the standard deviations.
This is done in the last line of the code cell :
```
pred_weights, pred_means, pred_std = sess.run([model.pi, model.mus, model.sigmas],
feed_dict={X: X_test})
```
The default minimisation technique used is ADAM with a decaying scale factor.
This can be seen [here](https://github.com/blei-lab/edward/blob/master/edward/inferences.py#L94) in the code base of Edward. Having a decaying scale factor is not the standard way of using ADAM, this is inspired by the Automatic Differentiation Variational Inference [(ADVI)](http://arxiv.org/abs/1603.00788) work where it was used in the RMSPROP minimizer.
The loss that is minimised in the ```MAP``` model from Edward is the negative log-likelihood, this calculation uses the ```log_prob``` method in the ```MixtureDensityNetwork``` class we defined above.
The ```build_loss``` method in the ```MAP``` class can be found [here](https://github.com/blei-lab/edward/blob/master/edward/inferences.py#L396).
However the method ```inference.loss``` used below, returns the log-likelihood, so we expect this quantity to be maximized.
```
NEPOCH = 1000
train_loss = np.zeros(NEPOCH)
test_loss = np.zeros(NEPOCH)
for i in range(NEPOCH):
_, train_loss[i] = sess.run([inference.train, inference.loss],
feed_dict={X: X_train, y: y_train})
test_loss[i] = sess.run(inference.loss, feed_dict={X: X_test, y: y_test})
pred_weights, pred_means, pred_std = sess.run([model.pi, model.mus, model.sigmas],
feed_dict={X: X_test})
```
We can plot the log-likelihood of the training and test sample as function of training epoch.
Keep in mind that ```inference.loss``` returns the total log-likelihood, so not the loss per data point, so in the plotting routine we divide by the size of the train and test data respectively.
We see that it converges after 400 training steps.
```
fig, axes = plt.subplots(nrows=1, ncols=1, figsize=(16, 3.5))
plt.plot(np.arange(NEPOCH), test_loss/len(X_test), label='Test')
plt.plot(np.arange(NEPOCH), train_loss/len(X_train), label='Train')
plt.legend(fontsize=20)
plt.xlabel('Epoch', fontsize=15)
plt.ylabel('Log-likelihood', fontsize=15)
```
Next we can have a look at how some individual examples perform. Keep in mind this is an inverse problem
so we can't get the answer correct, we can hope that the truth lies in area where the model has high probability.
In the next plot the truth is the vertical grey line while the blue line is the prediction of the mixture density network. As you can see, we didn't do too bad.
```
obj = [0, 4, 6]
fig, axes = plt.subplots(nrows=3, ncols=1, figsize=(16, 6))
plot_normal_mix(pred_weights[obj][0], pred_means[obj][0], pred_std[obj][0], axes[0], comp=False)
axes[0].axvline(x=y_test[obj][0], color='black', alpha=0.5)
plot_normal_mix(pred_weights[obj][2], pred_means[obj][2], pred_std[obj][2], axes[1], comp=False)
axes[1].axvline(x=y_test[obj][2], color='black', alpha=0.5)
plot_normal_mix(pred_weights[obj][1], pred_means[obj][1], pred_std[obj][1], axes[2], comp=False)
axes[2].axvline(x=y_test[obj][1], color='black', alpha=0.5)
```
We can check the ensemble by drawing samples of the prediction and plotting the density of those.
Seems the MDN learned what it needed too.
```
a = sample_from_mixture(X_test, pred_weights, pred_means, pred_std, amount=len(X_test))
sns.jointplot(a[:,0], a[:,1], kind="hex", color="#4CB391", ylim=(-10,10), xlim=(-14,14))
```
| github_jupyter |
```
from sklearn.model_selection import train_test_split
import pandas as pd
import numpy as np
from sklearn.preprocessing import StandardScaler
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Dense
#df = pd.read_csv(".\\Data_USD.csv", header=None,skiprows=1)
df = pd.read_csv(".\\Data_USD.csv")
df.head().to_csv(".\\test.csv")
T=df.groupby("SEX")
T.describe()
df.tail()
# X = df.drop('Y_Value',axis =1).values
# y = df['Y_Value'].values
X = df.drop('DEFAULT_PAYMENT_NEXT_MO',axis =1).values
X[2999,0]
X.shape
y = df['DEFAULT_PAYMENT_NEXT_MO'].values
#y.reshape(-1,1)
#print(X.shape)
X.shape
#print(y.shape)
y.shape
X_train, X_test, y_train, y_test = train_test_split (X,y,test_size=0.2, random_state=42)
y_test.T
X_test.shape
from sklearn.preprocessing import StandardScaler
X_scaler = StandardScaler().fit(X_train)
X_scaler
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
X_train_scaled
y_train_categorical = to_categorical(y_train)
y_test_categorical = to_categorical(y_test)
from keras.models import Sequential
#instantiate
model = Sequential()
from keras.layers import Dense
number_inputs = 10
number_hidden = 30
model.add(Dense(units = number_hidden, activation ='relu', input_dim=number_inputs))
model.add(Dense(units = 35, activation ='relu')) #second hidden layer
model.add(Dense(units = 25, activation ='relu')) #second hidden layer
model.add(Dense(units = 15, activation ='relu')) #second hidden layer
model.add(Dense(units = 5, activation ='relu')) #third hidden layer
number_classes =2 ## yes or no
model.add(Dense(units = number_classes, activation = 'softmax'))
model.summary()
#compile the model
model.compile(optimizer = 'sgd' ,
loss = 'categorical_crossentropy',
metrics =['accuracy'])
#train the model
model.fit(X_train_scaled, y_train_categorical, epochs=100,shuffle = True,verbose =2)
model.save("ccneuralnetwork.h5")
#quantify the model
model_loss, model_accuracy = model.evaluate(X_test_scaled,y_test_categorical,verbose =2)
print( model_loss )
print (model_accuracy)
```
F1, Precision Recall, and Confusion Matrix
```
from sklearn.metrics import precision_recall_fscore_support
from sklearn.metrics import recall_score
from sklearn.metrics import classification_report
y_prediction = model.predict_classes(X_test)
y_prediction.reshape(-1,1)
print("Recall score:"+ str(recall_score(y_test, y_prediction)))
print(classification_report(y_test, y_prediction,
target_names=["default", "non_default"]))
import itertools
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="red" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
# Compute confusion matrix
cnf_matrix = confusion_matrix(y_test, y_prediction)
np.set_printoptions(precision=2)
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=['Defualt', 'Non_default'],
title='Confusion matrix, without normalization')
# Plot normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=['Defualt', 'Non_default'], normalize=True,
title='Normalized confusion matrix')
plt.show()
```
| github_jupyter |
# Nonlinear recharge models
*R.A. Collenteur, University of Graz*
This notebook explains the use of the `RechargeModel` stress model to simulate the combined effect of precipitation and potential evaporation on the groundwater levels. For the computation of the groundwater recharge, three recharge models are currently available:
- `Linear` ([Berendrecht et al., 2003](#References); [von Asmuth et al., 2008](#References))
- `Berendrecht` ([Berendrecht et al., 2006](#References))
- `FlexModel` ([Collenteur et al., 2021](#References))
The first model is a simple linear function of precipitation and potential evaporation while the latter two are simulate a nonlinear response of recharge to precipitation using a soil-water balance concepts. Detailed descriptions of these models can be found in articles listed in the [References](#References) at the end of this notebook.
<div class="alert alert-info">
<b>Tip</b>
To run this notebook and the related non-linear recharge models, it is strongly recommended to install Numba (http://numba.pydata.org). This Just-In-Time (JIT) compiler compiles the computationally intensive part of the recharge calculation, making the non-linear model as fast as the Linear recharge model.
</div>
```
import pandas as pd
import pastas as ps
import matplotlib.pyplot as plt
ps.show_versions(numba=True)
ps.set_log_level("INFO")
```
## Read Input data
Input data handling is similar to other stressmodels. The only thing that is necessary to check is that the precipitation and evaporation are provided in mm/day. This is necessary because the parameters for the nonlinear recharge models are defined in mm for the length unit and days for the time unit. It is possible to use other units, but this would require manually setting the initial values and parameter boundaries for the recharge models.
```
head = pd.read_csv("../data/B32C0639001.csv", parse_dates=['date'],
index_col='date', squeeze=True)
# Make this millimeters per day
evap = ps.read_knmi("../data/etmgeg_260.txt", variables="EV24").series * 1e3
rain = ps.read_knmi("../data/etmgeg_260.txt", variables="RH").series * 1e3
fig, axes = plt.subplots(3,1, figsize=(10,6), sharex=True)
head.plot(ax=axes[0], x_compat=True, linestyle=" ", marker=".")
evap.plot(ax=axes[1], x_compat=True)
rain.plot(ax=axes[2], x_compat=True)
axes[0].set_ylabel("Head [m]")
axes[1].set_ylabel("Evap [mm/d]")
axes[2].set_ylabel("Rain [mm/d]")
plt.xlim("1985", "2005");
```
## Make a basic model
The normal workflow may be used to create and calibrate the model.
1. Create a Pastas `Model` instance
2. Choose a recharge model. All recharge models can be accessed through the recharge subpackage (`ps.rch`).
3. Create a `RechargeModel` object and add it to the model
4. Solve and visualize the model
```
ml = ps.Model(head)
# Select a recharge model
rch = ps.rch.FlexModel()
#rch = ps.rch.Berendrecht()
#rch = ps.rch.Linear()
rm = ps.RechargeModel(rain, evap, recharge=rch, rfunc=ps.Gamma, name="rch")
ml.add_stressmodel(rm)
ml.solve(noise=True, tmin="1990", report="basic")
ml.plots.results(figsize=(10,6));
```
## Analyze the estimated recharge flux
After the parameter estimation we can take a look at the recharge flux computed by the model. The flux is easy to obtain using the `get_stress` method of the model object, which automatically provides the optimal parameter values that were just estimated. After this, we can for example look at the yearly recharge flux estimated by the Pastas model.
```
recharge = ml.get_stress("rch").resample("A").sum()
ax = recharge.plot.bar(figsize=(10,3))
ax.set_xticklabels(recharge.index.year)
plt.ylabel("Recharge [mm/year]");
```
## A few things to keep in mind:
Below are a few things to keep in mind while using the (nonlinear) recharge models.
- The use of an appropriate warmup period is necessary, so make sure the precipitation and evaporation are available some time (e.g., one year) before the calibration period.
- Make sure that the units of the precipitation fluxes are in mm/day and that the DatetimeIndex matches exactly.
- It may be possible to fix or vary certain parameters, dependent on the problem. Obtaining better initial parameters may be possible by solving without a noise model first (`ml.solve(noise=False)`) and then solve it again using a noise model.
- For relatively shallow groundwater levels, it may be better to use the `Exponential` response function as the the non-linear models already cause a delayed response.
## References
- Berendrecht, W. L., Heemink, A. W., van Geer, F. C., and Gehrels, J. C. (2003) [Decoupling of modeling and measuring interval in groundwater time series analysis based on response characteristics](https://doi.org/10.1016/S0022-1694(03)00075-1), Journal of Hydrology, 278, 1–16.
- Berendrecht, W. L., Heemink, A. W., van Geer, F. C., and Gehrels, J. C. (2006) [A non-linear state space approach to model groundwater fluctuations](https://www.sciencedirect.com/science/article/abs/pii/S0309170805002113), Advances in Water Resources, 29, 959–973.
- Collenteur, R., Bakker, M., Klammler, G., and Birk, S. (2021) [Estimation of groundwater recharge from groundwater levels using nonlinear transfer function noise models and comparison to lysimeter data](https://doi.org/10.5194/hess-2020-392), Hydrol. Earth Syst. Sci., 25, 2931–2949.
- Von Asmuth, J.R., Maas, K., Bakker, M. and Petersen, J. (2008) [Modeling Time Series of Ground Water Head Fluctuations Subjected to Multiple Stresses](https://doi.org/10.1111/j.1745-6584.2007.00382.x). Groundwater, 46: 30-40.
## Data Sources
In this notebook we analysed a head time series near the town of De Bilt in the Netherlands. Data is obtained from the following resources:
- The heads (`B32C0639001.csv`) are downloaded from https://www.dinoloket.nl/
- The precipitation and potential evaporation (`etmgeg_260.txt`) are downloaded from https://knmi.nl
| github_jupyter |
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# Computing the 4-Velocity Time-Component $u^0$, the Magnetic Field Measured by a Comoving Observer $b^{\mu}$, and the Poynting Vector $S^i$
## Authors: Zach Etienne & Patrick Nelson
[comment]: <> (Abstract: TODO)
**Notebook Status:** <font color='green'><b> Validated </b></font>
**Validation Notes:** This module has been validated against a trusted code (the hand-written smallbPoynET in WVUThorns_diagnostics, which itself is based on expressions in IllinoisGRMHD... which was validated against the original GRMHD code of the Illinois NR group)
### NRPy+ Source Code for this module: [u0_smallb_Poynting__Cartesian.py](../edit/u0_smallb_Poynting__Cartesian/u0_smallb_Poynting__Cartesian.py)
[comment]: <> (Introduction: TODO)
<a id='toc'></a>
# Table of Contents
$$\label{toc}$$
This notebook is organized as follows
1. [Step 1](#u0bu): Computing $u^0$ and $b^{\mu}$
1. [Step 1.a](#4metric): Compute the 4-metric $g_{\mu\nu}$ and its inverse $g^{\mu\nu}$ from the ADM 3+1 variables, using the [`BSSN.ADMBSSN_tofrom_4metric`](../edit/BSSN/ADMBSSN_tofrom_4metric.py) ([**tutorial**](Tutorial-ADMBSSN_tofrom_4metric.ipynb)) NRPy+ module
1. [Step 1.b](#u0): Compute $u^0$ from the Valencia 3-velocity
1. [Step 1.c](#uj): Compute $u_j$ from $u^0$, the Valencia 3-velocity, and $g_{\mu\nu}$
1. [Step 1.d](#gamma): Compute $\gamma=$ `gammaDET` from the ADM 3+1 variables
1. [Step 1.e](#beta): Compute $b^\mu$
1. [Step 2](#poynting_flux): Defining the Poynting Flux Vector $S^{i}$
1. [Step 2.a](#g): Computing $g^{i\nu}$
1. [Step 2.b](#s): Computing $S^{i}$
1. [Step 3](#code_validation): Code Validation against `u0_smallb_Poynting__Cartesian` NRPy+ module
1. [Step 4](#appendix): Appendix: Proving Eqs. 53 and 56 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf)
1. [Step 5](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
<a id='u0bu'></a>
# Step 1: Computing $u^0$ and $b^{\mu}$ \[Back to [top](#toc)\]
$$\label{u0bu}$$
First some definitions. The spatial components of $b^{\mu}$ are simply the magnetic field as measured by an observer comoving with the plasma $B^{\mu}_{\rm (u)}$, divided by $\sqrt{4\pi}$. In addition, in the ideal MHD limit, $B^{\mu}_{\rm (u)}$ is orthogonal to the plasma 4-velocity $u^\mu$, which sets the $\mu=0$ component.
Note also that $B^{\mu}_{\rm (u)}$ is related to the magnetic field as measured by a *normal* observer $B^i$ via a simple projection (Eq 21 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf)), which results in the expressions (Eqs 23 and 24 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf)):
\begin{align}
\sqrt{4\pi} b^0 = B^0_{\rm (u)} &= \frac{u_j B^j}{\alpha} \\
\sqrt{4\pi} b^i = B^i_{\rm (u)} &= \frac{B^i + (u_j B^j) u^i}{\alpha u^0}\\
\end{align}
$B^i$ is related to the actual magnetic field evaluated in IllinoisGRMHD, $\tilde{B}^i$ via
$$B^i = \frac{\tilde{B}^i}{\gamma},$$
where $\gamma$ is the determinant of the spatial 3-metric.
The above expressions will require that we compute
1. the 4-metric $g_{\mu\nu}$ from the ADM 3+1 variables
1. $u^0$ from the Valencia 3-velocity
1. $u_j$ from $u^0$, the Valencia 3-velocity, and $g_{\mu\nu}$
1. $\gamma$ from the ADM 3+1 variables
<a id='4metric'></a>
## Step 1.a: Compute the 4-metric $g_{\mu\nu}$ and its inverse $g^{\mu\nu}$ from the ADM 3+1 variables, using the [`BSSN.ADMBSSN_tofrom_4metric`](../edit/BSSN/ADMBSSN_tofrom_4metric.py) ([**tutorial**](Tutorial-ADMBSSN_tofrom_4metric.ipynb)) NRPy+ module \[Back to [top](#toc)\]
$$\label{4metric}$$
We are given $\gamma_{ij}$, $\alpha$, and $\beta^i$ from ADMBase, so let's first compute
$$
g_{\mu\nu} = \begin{pmatrix}
-\alpha^2 + \beta^k \beta_k & \beta_i \\
\beta_j & \gamma_{ij}
\end{pmatrix}.
$$
```
# Step 1: Initialize needed Python/NRPy+ modules
import sympy as sp # SymPy: The Python computer algebra package upon which NRPy+ depends
import NRPy_param_funcs as par # NRPy+: Parameter interface
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm # NRPy+: Reference metric support
from outputC import * # NRPy+: Basic C code output functionality
import BSSN.ADMBSSN_tofrom_4metric as AB4m # NRPy+: ADM/BSSN <-> 4-metric conversions
# Set spatial dimension = 3
DIM=3
thismodule = "smallbPoynET"
# Step 1.a: Compute the 4-metric $g_{\mu\nu}$ and its inverse
# $g^{\mu\nu}$ from the ADM 3+1 variables, using the
# BSSN.ADMBSSN_tofrom_4metric NRPy+ module
import BSSN.ADMBSSN_tofrom_4metric as AB4m
gammaDD,betaU,alpha = AB4m.setup_ADM_quantities("ADM")
AB4m.g4DD_ito_BSSN_or_ADM("ADM",gammaDD,betaU,alpha)
g4DD = AB4m.g4DD
AB4m.g4UU_ito_BSSN_or_ADM("ADM",gammaDD,betaU,alpha)
g4UU = AB4m.g4UU
```
<a id='u0'></a>
## Step 1.b: Compute $u^0$ from the Valencia 3-velocity \[Back to [top](#toc)\]
$$\label{u0}$$
According to Eqs. 9-11 of [the IllinoisGRMHD paper](https://arxiv.org/pdf/1501.07276.pdf), the Valencia 3-velocity $v^i_{(n)}$ is related to the 4-velocity $u^\mu$ via
\begin{align}
\alpha v^i_{(n)} &= \frac{u^i}{u^0} + \beta^i \\
\implies u^i &= u^0 \left(\alpha v^i_{(n)} - \beta^i\right)
\end{align}
Defining $v^i = \frac{u^i}{u^0}$, we get
$$v^i = \alpha v^i_{(n)} - \beta^i,$$
and in terms of this variable we get
\begin{align}
g_{00} \left(u^0\right)^2 + 2 g_{0i} u^0 u^i + g_{ij} u^i u^j &= \left(u^0\right)^2 \left(g_{00} + 2 g_{0i} v^i + g_{ij} v^i v^j\right)\\
\implies u^0 &= \pm \sqrt{\frac{-1}{g_{00} + 2 g_{0i} v^i + g_{ij} v^i v^j}} \\
&= \pm \sqrt{\frac{-1}{(-\alpha^2 + \beta^2) + 2 \beta_i v^i + \gamma_{ij} v^i v^j}} \\
&= \pm \sqrt{\frac{1}{\alpha^2 - \gamma_{ij}\left(\beta^i + v^i\right)\left(\beta^j + v^j\right)}}\\
&= \pm \sqrt{\frac{1}{\alpha^2 - \alpha^2 \gamma_{ij}v^i_{(n)}v^j_{(n)}}}\\
&= \pm \frac{1}{\alpha}\sqrt{\frac{1}{1 - \gamma_{ij}v^i_{(n)}v^j_{(n)}}}
\end{align}
Generally speaking, numerical errors will occasionally drive expressions under the radical to either negative values or potentially enormous values (corresponding to enormous Lorentz factors). Thus a reliable approach for computing $u^0$ requires that we first rewrite the above expression in terms of the Lorentz factor squared: $\Gamma^2=\left(\alpha u^0\right)^2$:
\begin{align}
u^0 &= \pm \frac{1}{\alpha}\sqrt{\frac{1}{1 - \gamma_{ij}v^i_{(n)}v^j_{(n)}}}\\
\implies \left(\alpha u^0\right)^2 &= \frac{1}{1 - \gamma_{ij}v^i_{(n)}v^j_{(n)}} \\
\implies \gamma_{ij}v^i_{(n)}v^j_{(n)} &= 1 - \frac{1}{\left(\alpha u^0\right)^2} \\
&= 1 - \frac{1}{\Gamma^2}
\end{align}
In order for the bottom expression to hold true, the left-hand side must be between 0 and 1. Again, this is not guaranteed due to the appearance of numerical errors. In fact, a robust algorithm will not allow $\Gamma^2$ to become too large (which might contribute greatly to the stress-energy of a given gridpoint), so let's define $\Gamma_{\rm max}$, the largest allowed Lorentz factor.
Then our algorithm for computing $u^0$ is as follows:
If
$$R=\gamma_{ij}v^i_{(n)}v^j_{(n)}>1 - \frac{1}{\Gamma_{\rm max}^2},$$
then adjust the 3-velocity $v^i$ as follows:
$$v^i_{(n)} = \sqrt{\frac{1 - \frac{1}{\Gamma_{\rm max}^2}}{R}}v^i_{(n)}.$$
After this rescaling, we are then guaranteed that if $R$ is recomputed, it will be set to its ceiling value $R=R_{\rm max} = 1 - \frac{1}{\Gamma_{\rm max}^2}$.
Then, regardless of whether the ceiling on $R$ was applied, $u^0$ can be safely computed via
$$
u^0 = \frac{1}{\alpha \sqrt{1-R}}.
$$
```
ValenciavU = ixp.register_gridfunctions_for_single_rank1("AUX","ValenciavU",DIM=3)
# Step 1: Compute R = 1 - 1/max(Gamma)
R = sp.sympify(0)
for i in range(DIM):
for j in range(DIM):
R += gammaDD[i][j]*ValenciavU[i]*ValenciavU[j]
GAMMA_SPEED_LIMIT = par.Cparameters("REAL",thismodule,"GAMMA_SPEED_LIMIT",10.0) # Default value based on
# IllinoisGRMHD.
# GiRaFFE default = 2000.0
Rmax = 1 - 1/(GAMMA_SPEED_LIMIT*GAMMA_SPEED_LIMIT)
rescaledValenciavU = ixp.zerorank1()
for i in range(DIM):
rescaledValenciavU[i] = ValenciavU[i]*sp.sqrt(Rmax/R)
rescaledu0 = 1/(alpha*sp.sqrt(1-Rmax))
regularu0 = 1/(alpha*sp.sqrt(1-R))
computeu0_Cfunction = """
/* Function for computing u^0 from Valencia 3-velocity. */
/* Inputs: ValenciavU[], alpha, gammaDD[][], GAMMA_SPEED_LIMIT (C parameter) */
/* Output: u0=u^0 and velocity-limited ValenciavU[] */\n\n"""
computeu0_Cfunction += outputC([R,Rmax],["const double R","const double Rmax"],"returnstring",
params="includebraces=False,CSE_varprefix=tmpR,outCverbose=False")
computeu0_Cfunction += "if(R <= Rmax) "
computeu0_Cfunction += outputC(regularu0,"u0","returnstring",
params="includebraces=True,CSE_varprefix=tmpnorescale,outCverbose=False")
computeu0_Cfunction += " else "
computeu0_Cfunction += outputC([rescaledValenciavU[0],rescaledValenciavU[1],rescaledValenciavU[2],rescaledu0],
["ValenciavU0","ValenciavU1","ValenciavU2","u0"],"returnstring",
params="includebraces=True,CSE_varprefix=tmprescale,outCverbose=False")
print(computeu0_Cfunction)
```
<a id='uj'></a>
## Step 1.c: Compute $u_j$ from $u^0$, the Valencia 3-velocity, and $g_{\mu\nu}$ \[Back to [top](#toc)\]
$$\label{uj}$$
The basic equation is
\begin{align}
u_j &= g_{\mu j} u^{\mu} \\
&= g_{0j} u^0 + g_{ij} u^i \\
&= \beta_j u^0 + \gamma_{ij} u^i \\
&= \beta_j u^0 + \gamma_{ij} u^0 \left(\alpha v^i_{(n)} - \beta^i\right) \\
&= u^0 \left(\beta_j + \gamma_{ij} \left(\alpha v^i_{(n)} - \beta^i\right) \right)\\
&= \alpha u^0 \gamma_{ij} v^i_{(n)} \\
\end{align}
```
u0 = par.Cparameters("REAL",thismodule,"u0",1e300) # Will be overwritten in C code. Set to crazy value to ensure this.
uD = ixp.zerorank1()
for i in range(DIM):
for j in range(DIM):
uD[j] += alpha*u0*gammaDD[i][j]*ValenciavU[i]
```
<a id='beta'></a>
## Step 1.d: Compute $b^\mu$ \[Back to [top](#toc)\]
$$\label{beta}$$
We compute $b^\mu$ from the above expressions:
\begin{align}
\sqrt{4\pi} b^0 = B^0_{\rm (u)} &= \frac{u_j B^j}{\alpha} \\
\sqrt{4\pi} b^i = B^i_{\rm (u)} &= \frac{B^i + (u_j B^j) u^i}{\alpha u^0}\\
\end{align}
$B^i$ is exactly equal to the $B^i$ evaluated in IllinoisGRMHD/GiRaFFE.
Pulling this together, we currently have available as input:
+ $\tilde{B}^i$
+ $u_j$
+ $u^0$,
with the goal of outputting now $b^\mu$ and $b^2$:
```
M_PI = par.Cparameters("#define",thismodule,"M_PI","")
BU = ixp.register_gridfunctions_for_single_rank1("AUX","BU",DIM=3)
# uBcontraction = u_i B^i
uBcontraction = sp.sympify(0)
for i in range(DIM):
uBcontraction += uD[i]*BU[i]
# uU = 3-vector representing u^i = u^0 \left(\alpha v^i_{(n)} - \beta^i\right)
uU = ixp.zerorank1()
for i in range(DIM):
uU[i] = u0*(alpha*ValenciavU[i] - betaU[i])
smallb4U = ixp.zerorank1(DIM=4)
smallb4U[0] = uBcontraction/(alpha*sp.sqrt(4*M_PI))
for i in range(DIM):
smallb4U[1+i] = (BU[i] + uBcontraction*uU[i])/(alpha*u0*sp.sqrt(4*M_PI))
```
<a id='poynting_flux'></a>
# Step 2: Defining the Poynting Flux Vector $S^{i}$ \[Back to [top](#toc)\]
$$\label{poynting_flux}$$
The Poynting flux is defined in Eq. 11 of [Kelly *et al.*](https://arxiv.org/pdf/1710.02132.pdf) (note that we choose the minus sign convention so that the Poynting luminosity across a spherical shell is $L_{\rm EM} = \int (-\alpha T^i_{\rm EM\ 0}) \sqrt{\gamma} d\Omega = \int S^r \sqrt{\gamma} d\Omega$, as in [Farris *et al.*](https://arxiv.org/pdf/1207.3354.pdf):
$$
S^i = -\alpha T^i_{\rm EM\ 0} = -\alpha\left(b^2 u^i u_0 + \frac{1}{2} b^2 g^i{}_0 - b^i b_0\right)
$$
<a id='s'></a>
## Step 2.a: Computing $S^{i}$ \[Back to [top](#toc)\]
$$\label{s}$$
Given $g^{\mu\nu}$ computed above, we focus first on the $g^i{}_{0}$ term by computing
$$
g^\mu{}_\delta = g^{\mu\nu} g_{\nu \delta},
$$
and then the rest of the Poynting flux vector can be immediately computed from quantities defined above:
$$
S^i = -\alpha T^i_{\rm EM\ 0} = -\alpha\left(b^2 u^i u_0 + \frac{1}{2} b^2 g^i{}_0 - b^i b_0\right)
$$
```
# Step 2.a.i: compute g^\mu_\delta:
g4UD = ixp.zerorank2(DIM=4)
for mu in range(4):
for delta in range(4):
for nu in range(4):
g4UD[mu][delta] += g4UU[mu][nu]*g4DD[nu][delta]
# Step 2.a.ii: compute b_{\mu}
smallb4D = ixp.zerorank1(DIM=4)
for mu in range(4):
for nu in range(4):
smallb4D[mu] += g4DD[mu][nu]*smallb4U[nu]
# Step 2.a.iii: compute u_0 = g_{mu 0} u^{mu} = g4DD[0][0]*u0 + g4DD[i][0]*uU[i]
u_0 = g4DD[0][0]*u0
for i in range(DIM):
u_0 += g4DD[i+1][0]*uU[i]
# Step 2.a.iv: compute b^2, setting b^2 = smallb2etk, as gridfunctions with base names ending in a digit
# are forbidden in NRPy+.
smallb2etk = sp.sympify(0)
for mu in range(4):
smallb2etk += smallb4U[mu]*smallb4D[mu]
# Step 2.a.v: compute S^i
PoynSU = ixp.zerorank1()
for i in range(DIM):
PoynSU[i] = -alpha * (smallb2etk*uU[i]*u_0 + sp.Rational(1,2)*smallb2etk*g4UD[i+1][0] - smallb4U[i+1]*smallb4D[0])
```
<a id='code_validation'></a>
# Step 3: Code Validation against `u0_smallb_Poynting__Cartesian` NRPy+ module \[Back to [top](#toc)\]
$$\label{code_validation}$$
Here, as a code validation check, we verify agreement in the SymPy expressions for u0, smallbU, smallb2etk, and PoynSU between
1. this tutorial and
2. the NRPy+ [u0_smallb_Poynting__Cartesian module](../edit/u0_smallb_Poynting__Cartesian/u0_smallb_Poynting__Cartesian.py).
```
import sys
import u0_smallb_Poynting__Cartesian.u0_smallb_Poynting__Cartesian as u0etc
u0etc.compute_u0_smallb_Poynting__Cartesian(gammaDD,betaU,alpha,ValenciavU,BU)
if u0etc.computeu0_Cfunction != computeu0_Cfunction:
print("FAILURE: u0 C code has changed!")
sys.exit(1)
else:
print("PASSED: u0 C code matches!")
for i in range(4):
print("u0etc.smallb4U["+str(i)+"] - smallb4U["+str(i)+"] = "
+ str(u0etc.smallb4U[i]-smallb4U[i]))
print("u0etc.smallb2etk - smallb2etk = " + str(u0etc.smallb2etk-smallb2etk))
for i in range(DIM):
print("u0etc.PoynSU["+str(i)+"] - PoynSU["+str(i)+"] = "
+ str(u0etc.PoynSU[i]-PoynSU[i]))
```
<a id='appendix'></a>
# Step 4: Appendix: Proving Eqs. 53 and 56 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf)
$$\label{appendix}$$
$u^\mu u_\mu = -1$ implies
\begin{align}
g^{\mu\nu} u_\mu u_\nu &= g^{00} \left(u_0\right)^2 + 2 g^{0i} u_0 u_i + g^{ij} u_i u_j = -1 \\
\implies &g^{00} \left(u_0\right)^2 + 2 g^{0i} u_0 u_i + g^{ij} u_i u_j + 1 = 0\\
& a x^2 + b x + c = 0
\end{align}
Thus we have a quadratic equation for $u_0$, with solution given by
\begin{align}
u_0 &= \frac{-b \pm \sqrt{b^2 - 4 a c}}{2 a} \\
&= \frac{-2 g^{0i}u_i \pm \sqrt{\left(2 g^{0i} u_i\right)^2 - 4 g^{00} (g^{ij} u_i u_j + 1)}}{2 g^{00}}\\
&= \frac{-g^{0i}u_i \pm \sqrt{\left(g^{0i} u_i\right)^2 - g^{00} (g^{ij} u_i u_j + 1)}}{g^{00}}\\
\end{align}
Notice that (Eq. 4.49 in [Gourgoulhon](https://arxiv.org/pdf/gr-qc/0703035.pdf))
$$
g^{\mu\nu} = \begin{pmatrix}
-\frac{1}{\alpha^2} & \frac{\beta^i}{\alpha^2} \\
\frac{\beta^i}{\alpha^2} & \gamma^{ij} - \frac{\beta^i\beta^j}{\alpha^2}
\end{pmatrix},
$$
so we have
\begin{align}
u_0 &= \frac{-\beta^i u_i/\alpha^2 \pm \sqrt{\left(\beta^i u_i/\alpha^2\right)^2 + 1/\alpha^2 (g^{ij} u_i u_j + 1)}}{1/\alpha^2}\\
&= -\beta^i u_i \pm \sqrt{\left(\beta^i u_i\right)^2 + \alpha^2 (g^{ij} u_i u_j + 1)}\\
&= -\beta^i u_i \pm \sqrt{\left(\beta^i u_i\right)^2 + \alpha^2 \left(\left[\gamma^{ij} - \frac{\beta^i\beta^j}{\alpha^2}\right] u_i u_j + 1\right)}\\
&= -\beta^i u_i \pm \sqrt{\left(\beta^i u_i\right)^2 + \alpha^2 \left(\gamma^{ij}u_i u_j + 1\right) - \beta^i\beta^j u_i u_j}\\
&= -\beta^i u_i \pm \sqrt{\alpha^2 \left(\gamma^{ij}u_i u_j + 1\right)}\\
\end{align}
Now, since
$$
u^0 = g^{\alpha 0} u_\alpha = -\frac{1}{\alpha^2} u_0 + \frac{\beta^i u_i}{\alpha^2},
$$
we get
\begin{align}
u^0 &= \frac{1}{\alpha^2} \left(u_0 + \beta^i u_i\right) \\
&= \pm \frac{1}{\alpha^2} \sqrt{\alpha^2 \left(\gamma^{ij}u_i u_j + 1\right)}\\
&= \pm \frac{1}{\alpha} \sqrt{\gamma^{ij}u_i u_j + 1}\\
\end{align}
By convention, the relativistic Gamma factor is positive and given by $\alpha u^0$, so we choose the positive root. Thus we have derived Eq. 53 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf):
$$
u^0 = \frac{1}{\alpha} \sqrt{\gamma^{ij}u_i u_j + 1}.
$$
Next we evaluate
\begin{align}
u^i &= u_\mu g^{\mu i} \\
&= u_0 g^{0 i} + u_j g^{i j}\\
&= u_0 \frac{\beta^i}{\alpha^2} + u_j \left(\gamma^{ij} - \frac{\beta^i\beta^j}{\alpha^2}\right)\\
&= \gamma^{ij} u_j + u_0 \frac{\beta^i}{\alpha^2} - u_j \frac{\beta^i\beta^j}{\alpha^2}\\
&= \gamma^{ij} u_j + \frac{\beta^i}{\alpha^2} \left(u_0 - u_j \beta^j\right)\\
&= \gamma^{ij} u_j - \beta^i u^0,\\
\implies v^i &= \frac{\gamma^{ij} u_j}{u^0} - \beta^i
\end{align}
which is equivalent to Eq. 56 in [Duez *et al* (2005)](https://arxiv.org/pdf/astro-ph/0503420.pdf). Notice in the last step, we used the above definition of $u^0$.
<a id='latex_pdf_output'></a>
# Step 5: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-u0_smallb_Poynting-Cartesian.pdf](Tutorial-u0_smallb_Poynting-Cartesian.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
!jupyter nbconvert --to latex --template latex_nrpy_style.tplx --log-level='WARN' Tutorial-u0_smallb_Poynting-Cartesian.ipynb
!pdflatex -interaction=batchmode Tutorial-u0_smallb_Poynting-Cartesian.tex
!pdflatex -interaction=batchmode Tutorial-u0_smallb_Poynting-Cartesian.tex
!pdflatex -interaction=batchmode Tutorial-u0_smallb_Poynting-Cartesian.tex
!rm -f Tut*.out Tut*.aux Tut*.log
```
| github_jupyter |
```
# import re
# import tensorflow as tf
# from tensorflow.keras.preprocessing.text import text_to_word_sequence
# tokens=text_to_word_sequence("manta.com/c/mmcdqky/lily-co")
# print(tokens)
# #to map the features to a dictioanary and then convert it to a csv file.
# # Feauture extraction
# class feature_extractor(object):
# def __init__(self,url):
# self.url=url
# self.length=len(url)
# #self.domain=url.split('//')[-1].split('/')[0]
# #def entropy(self):
# #.com,.org,.net,.edu
# #has www.
# #.extension-- .htm,.html,.php,.js
# # Pattern regex = Pattern.compile(".com[,/.]")
# def domain(self):
# if re.search(".com[ .,/]",self.url):
# return 1
# elif re.search(".org[.,/]",self.url):
# return 2
# elif re.search(".net[.,/]",self.url):
# return 3
# elif re.search(".edu[.,/]",self.url):
# return 4
# else:
# return 0
# #def extension(self):
# def num_digits(self):
# return sum(n.isdigit() for n in self.url)
# def num_char(self):
# return sum(n.alpha() for n in self.url)
# def has_http(self):
# if "http" in self.url:
# return 1
# else:
# return 0
# def has_https(self):
# if "https" in self.url:
# return 1
# else:
# return 0
# #def num_special_char(self):
# #
# #def num
# def clean(input):
# tokensBySlash = str(input.encode('utf-8')).split('/')
# allTokens=[]
# for i in tokensBySlash:
# tokens = str(i).split('-')
# tokensByDot = []
# for j in range(0,len(tokens)):
# tempTokens = str(tokens[j]).split('.')
# tokentsByDot = tokensByDot + tempTokens
# allTokens = allTokens + tokens + tokensByDot
# allTokens = list(set(allTokens))
# if 'com' in allTokens:
# allTokens.remove('com')
# return allTokens
from urllib.parse import urlparse
url="http://www.pn-wuppertal.de/links/2-linkseite/5-httpwwwkrebshilfede"
def getTokens(input):
tokensBySlash = str(input.encode('utf-8')).split('/')
allTokens=[]
for i in tokensBySlash:
tokens = str(i).split('-')
tokensByDot = []
for j in range(0,len(tokens)):
tempTokens = str(tokens[j]).split('.')
tokentsByDot = tokensByDot + tempTokens
allTokens = allTokens + tokens + tokensByDot
allTokens = list(set(allTokens))
if 'com' in allTokens:
allTokens.remove('com')
return allTokens
url="http://www.pn-wuppertal.de/links/2-linkseite/5-httpwwwkrebshilfede"
x=(lambda s: sum(not((i.isalpha()) and not(i.isnumeric())) for i in s))
print((url))
from urllib.parse import urlparse
url="http://www.pn-wuppertal.de/links/2-linkseite/5-httpwwwkrebshilfede"
def fd_length(url):
urlpath= urlparse(url).path
try:
return len(urlpath.split('/')[1])
except:
return 0
print(urlparse(url))
print(fd_length(urlparse(url)))
urlparse(url).scheme
s='https://www.yandex.ru'
print(urlparse(s))
s='yourbittorrent.com/?q=anthony-hamilton-soulife'
print(urlparse(s))
print(tldextract.extract(s))
from urllib.parse import urlparse
import tldextract
s='movies.yahoo.com/shop?d=hv&cf=info&id=1800340831'
print(urlparse(s))
print(tldextract.extract(s).subdomain)
len(urlparse(s).query)
def tld_length(tld):
try:
return len(tld)
except:
return -1
import tldextract
from urllib.parse import urlparse
import tldextract
s='http://peluqueriadeautor.com/index.php?option=com_virtuemart&page=shop.browse&category_id=31&Itemid=70'
def extension(s):
domains={'com':1,'edu':2,'org':3,'net':4,'onion':5}
if s in domains.keys():
return domains[s]
else:
return 0
#s=tldextract.extract(s).suffix
#print(extension(s))
print(tldextract.extract(s))
print(urlparse(s))
from urllib.parse import urlparse
import tldextract
print(tldextract.extract("http://motthegioi.com/the-gioi-cuoi/clip-dai-gia-mac-ca-voi-co-ban-banh-my-185682.html"))
print(urlparse("http://motthegioi.vn/the-gioi-cuoi/clip-dai-gia-mac-ca-voi-co-ban-banh-my-185682.html"))
```
| github_jupyter |
**This notebook is an exercise in the [Intermediate Machine Learning](https://www.kaggle.com/learn/intermediate-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/categorical-variables).**
---
By encoding **categorical variables**, you'll obtain your best results thus far!
# Setup
The questions below will give you feedback on your work. Run the following cell to set up the feedback system.
```
# Set up code checking
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
from learntools.core import binder
binder.bind(globals())
from learntools.ml_intermediate.ex3 import *
print("Setup Complete")
```
In this exercise, you will work with data from the [Housing Prices Competition for Kaggle Learn Users](https://www.kaggle.com/c/home-data-for-ml-course).

Run the next code cell without changes to load the training and validation sets in `X_train`, `X_valid`, `y_train`, and `y_valid`. The test set is loaded in `X_test`.
```
import pandas as pd
from sklearn.model_selection import train_test_split
# Read the data
X = pd.read_csv('../input/train.csv', index_col='Id')
X_test = pd.read_csv('../input/test.csv', index_col='Id')
# Remove rows with missing target, separate target from predictors
X.dropna(axis=0, subset=['SalePrice'], inplace=True)
y = X.SalePrice
X.drop(['SalePrice'], axis=1, inplace=True)
# To keep things simple, we'll drop columns with missing values
cols_with_missing = [col for col in X.columns if X[col].isnull().any()]
X.drop(cols_with_missing, axis=1, inplace=True)
X_test.drop(cols_with_missing, axis=1, inplace=True)
# Break off validation set from training data
X_train, X_valid, y_train, y_valid = train_test_split(X, y,
train_size=0.8, test_size=0.2,
random_state=0)
```
Use the next code cell to print the first five rows of the data.
```
X_train.head()
```
Notice that the dataset contains both numerical and categorical variables. You'll need to encode the categorical data before training a model.
To compare different models, you'll use the same `score_dataset()` function from the tutorial. This function reports the [mean absolute error](https://en.wikipedia.org/wiki/Mean_absolute_error) (MAE) from a random forest model.
```
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
# function for comparing different approaches
def score_dataset(X_train, X_valid, y_train, y_valid):
model = RandomForestRegressor(n_estimators=100, random_state=0)
model.fit(X_train, y_train)
preds = model.predict(X_valid)
return mean_absolute_error(y_valid, preds)
```
# Step 1: Drop columns with categorical data
You'll get started with the most straightforward approach. Use the code cell below to preprocess the data in `X_train` and `X_valid` to remove columns with categorical data. Set the preprocessed DataFrames to `drop_X_train` and `drop_X_valid`, respectively.
```
# Fill in the lines below: drop columns in training and validation data
drop_X_train = X_train.select_dtypes(exclude=['object'])
drop_X_valid = X_valid.select_dtypes(exclude=['object'])
# Check your answers
step_1.check()
# Lines below will give you a hint or solution code
#step_1.hint()
#step_1.solution()
```
Run the next code cell to get the MAE for this approach.
```
print("MAE from Approach 1 (Drop categorical variables):")
print(score_dataset(drop_X_train, drop_X_valid, y_train, y_valid))
```
Before jumping into label encoding, we'll investigate the dataset. Specifically, we'll look at the `'Condition2'` column. The code cell below prints the unique entries in both the training and validation sets.
```
print("Unique values in 'Condition2' column in training data:", X_train['Condition2'].unique())
print("\nUnique values in 'Condition2' column in validation data:", X_valid['Condition2'].unique())
```
# Step 2: Label encoding
### Part A
If you now write code to:
- fit a label encoder to the training data, and then
- use it to transform both the training and validation data,
you'll get an error. Can you see why this is the case? (_You'll need to use the above output to answer this question._)
```
# Check your answer (Run this code cell to receive credit!)
step_2.a.check()
#step_2.a.hint()
```
This is a common problem that you'll encounter with real-world data, and there are many approaches to fixing this issue. For instance, you can write a custom label encoder to deal with new categories. The simplest approach, however, is to drop the problematic categorical columns.
Run the code cell below to save the problematic columns to a Python list `bad_label_cols`. Likewise, columns that can be safely label encoded are stored in `good_label_cols`.
```
# All categorical columns
object_cols = [col for col in X_train.columns if X_train[col].dtype == "object"]
# Columns that can be safely label encoded
good_label_cols = [col for col in object_cols if
set(X_train[col]) == set(X_valid[col])]
# Problematic columns that will be dropped from the dataset
bad_label_cols = list(set(object_cols)-set(good_label_cols))
print('Categorical columns that will be label encoded:', good_label_cols)
print('\nCategorical columns that will be dropped from the dataset:', bad_label_cols)
```
### Part B
Use the next code cell to label encode the data in `X_train` and `X_valid`. Set the preprocessed DataFrames to `label_X_train` and `label_X_valid`, respectively.
- We have provided code below to drop the categorical columns in `bad_label_cols` from the dataset.
- You should label encode the categorical columns in `good_label_cols`.
```
from sklearn.preprocessing import LabelEncoder
# Drop categorical columns that will not be encoded
label_X_train = X_train.drop(bad_label_cols, axis=1)
label_X_valid = X_valid.drop(bad_label_cols, axis=1)
# Apply label encoder
label_encoder = LabelEncoder()
for col in good_label_cols:
label_X_train[col] = label_encoder.fit_transform(label_X_train[col])
label_X_valid[col] = label_encoder.transform(label_X_valid[col])
# Check your answer
step_2.b.check()
# Lines below will give you a hint or solution code
#step_2.b.hint()
#step_2.b.solution()
```
Run the next code cell to get the MAE for this approach.
```
print("MAE from Approach 2 (Label Encoding):")
print(score_dataset(label_X_train, label_X_valid, y_train, y_valid))
```
So far, you've tried two different approaches to dealing with categorical variables. And, you've seen that encoding categorical data yields better results than removing columns from the dataset.
Soon, you'll try one-hot encoding. Before then, there's one additional topic we need to cover. Begin by running the next code cell without changes.
```
# Get number of unique entries in each column with categorical data
object_nunique = list(map(lambda col: X_train[col].nunique(), object_cols))
d = dict(zip(object_cols, object_nunique))
# Print number of unique entries by column, in ascending order
sorted(d.items(), key=lambda x: x[1])
```
# Step 3: Investigating cardinality
### Part A
The output above shows, for each column with categorical data, the number of unique values in the column. For instance, the `'Street'` column in the training data has two unique values: `'Grvl'` and `'Pave'`, corresponding to a gravel road and a paved road, respectively.
We refer to the number of unique entries of a categorical variable as the **cardinality** of that categorical variable. For instance, the `'Street'` variable has cardinality 2.
Use the output above to answer the questions below.
```
# Fill in the line below: How many categorical variables in the training data
# have cardinality greater than 10?
high_cardinality_numcols = 3
# Fill in the line below: How many columns are needed to one-hot encode the
# 'Neighborhood' variable in the training data?
num_cols_neighborhood = 25
# Check your answers
step_3.a.check()
# Lines below will give you a hint or solution code
#step_3.a.hint()
#step_3.a.solution()
```
### Part B
For large datasets with many rows, one-hot encoding can greatly expand the size of the dataset. For this reason, we typically will only one-hot encode columns with relatively low cardinality. Then, high cardinality columns can either be dropped from the dataset, or we can use label encoding.
As an example, consider a dataset with 10,000 rows, and containing one categorical column with 100 unique entries.
- If this column is replaced with the corresponding one-hot encoding, how many entries are added to the dataset?
- If we instead replace the column with the label encoding, how many entries are added?
Use your answers to fill in the lines below.
```
# Fill in the line below: How many entries are added to the dataset by
# replacing the column with a one-hot encoding?
OH_entries_added = 1e4*100 - 1e4
# Fill in the line below: How many entries are added to the dataset by
# replacing the column with a label encoding?
label_entries_added = 0
# Check your answers
step_3.b.check()
# Lines below will give you a hint or solution code
#step_3.b.hint()
#step_3.b.solution()
```
Next, you'll experiment with one-hot encoding. But, instead of encoding all of the categorical variables in the dataset, you'll only create a one-hot encoding for columns with cardinality less than 10.
Run the code cell below without changes to set `low_cardinality_cols` to a Python list containing the columns that will be one-hot encoded. Likewise, `high_cardinality_cols` contains a list of categorical columns that will be dropped from the dataset.
```
# Columns that will be one-hot encoded
low_cardinality_cols = [col for col in object_cols if X_train[col].nunique() < 10]
# Columns that will be dropped from the dataset
high_cardinality_cols = list(set(object_cols)-set(low_cardinality_cols))
print('Categorical columns that will be one-hot encoded:', low_cardinality_cols)
print('\nCategorical columns that will be dropped from the dataset:', high_cardinality_cols)
```
# Step 4: One-hot encoding
Use the next code cell to one-hot encode the data in `X_train` and `X_valid`. Set the preprocessed DataFrames to `OH_X_train` and `OH_X_valid`, respectively.
- The full list of categorical columns in the dataset can be found in the Python list `object_cols`.
- You should only one-hot encode the categorical columns in `low_cardinality_cols`. All other categorical columns should be dropped from the dataset.
```
from sklearn.preprocessing import OneHotEncoder
# Use as many lines of code as you need!
OH_encoder = OneHotEncoder(handle_unknown='ignore', sparse=False)
OH_cols_train = pd.DataFrame(OH_encoder.fit_transform(X_train[low_cardinality_cols]))
OH_cols_valid = pd.DataFrame(OH_encoder.transform(X_valid[low_cardinality_cols]))
# One-hot encoding removed index; put it back
OH_cols_train.index = X_train.index
OH_cols_valid.index = X_valid.index
# Remove categorical columns (will replace with one-hot encoding)
num_X_train = X_train.drop(object_cols, axis=1)
num_X_valid = X_valid.drop(object_cols, axis=1)
# Add one-hot encoded columns to numerical features
OH_X_train = pd.concat([num_X_train, OH_cols_train], axis=1)
OH_X_valid = pd.concat([num_X_valid, OH_cols_valid], axis=1)
# Check your answer
step_4.check()
# Lines below will give you a hint or solution code
#step_4.hint()
#step_4.solution()
```
Run the next code cell to get the MAE for this approach.
```
print("MAE from Approach 3 (One-Hot Encoding):")
print(score_dataset(OH_X_train, OH_X_valid, y_train, y_valid))
```
# Generate test predictions and submit your results
After you complete Step 4, if you'd like to use what you've learned to submit your results to the leaderboard, you'll need to preprocess the test data before generating predictions.
**This step is completely optional, and you do not need to submit results to the leaderboard to successfully complete the exercise.**
Check out the previous exercise if you need help with remembering how to [join the competition](https://www.kaggle.com/c/home-data-for-ml-course) or save your results to CSV. Once you have generated a file with your results, follow the instructions below:
1. Begin by clicking on the blue **Save Version** button in the top right corner of the window. This will generate a pop-up window.
2. Ensure that the **Save and Run All** option is selected, and then click on the blue **Save** button.
3. This generates a window in the bottom left corner of the notebook. After it has finished running, click on the number to the right of the **Save Version** button. This pulls up a list of versions on the right of the screen. Click on the ellipsis **(...)** to the right of the most recent version, and select **Open in Viewer**. This brings you into view mode of the same page. You will need to scroll down to get back to these instructions.
4. Click on the **Output** tab on the right of the screen. Then, click on the file you would like to submit, and click on the blue **Submit** button to submit your results to the leaderboard.
You have now successfully submitted to the competition!
If you want to keep working to improve your performance, select the blue **Edit** button in the top right of the screen. Then you can change your code and repeat the process. There's a lot of room to improve, and you will climb up the leaderboard as you work.
```
# (Optional) Your code here
```
# Keep going
With missing value handling and categorical encoding, your modeling process is getting complex. This complexity gets worse when you want to save your model to use in the future. The key to managing this complexity is something called **pipelines**.
**[Learn to use pipelines](https://www.kaggle.com/alexisbcook/pipelines)** to preprocess datasets with categorical variables, missing values and any other messiness your data throws at you.
---
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/161289) to chat with other Learners.*
| github_jupyter |
##### Copyright 2018 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/beta/{PATH}">
<img src="https://www.tensorflow.org/images/tf_logo_32px.png" />
View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/{PATH}.ipynb">
<img src="https://www.tensorflow.org/images/colab_logo_32px.png" />
Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/{PATH}.ipynb">
<img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />
View source on GitHub</a>
</td>
</table>
# Using TPUs
Tensor Processing Units (TPUs) are Google's specialized ASICs designed to dramatically accelerate machine learning workloads. They are available on Google Colab, the TensorFlow Research Cloud and Google Compute Engine.
In this notebook, you can try training a convolutional neural network against the Fashion MNIST dataset on Cloud TPUs using tf.keras and Distribution Strategy.
## Learning Objectives
In this Colab, you will learn how to:
* Write a standard 4-layer conv-net with drop-out and batch normalization in Keras.
* Use TPUs and Distribution Strategy to train the model.
* Run a prediction to see how well the model can predict fashion categories and output the result.
## Instructions
To use TPUs in Colab:
1. On the main menu, click Runtime and select **Change runtime type**. Set "TPU" as the hardware accelerator.
1. Click Runtime again and select **Runtime > Run All**. You can also run the cells manually with Shift-ENTER.
## Data, Model, and Training
### Download the Data
Begin by downloading the fashion MNIST dataset using `tf.keras.datasets`, as shown below. We will also need to convert the data to `float32` format, as the data types supported by TPUs are limited right now.
TPUs currently do not support Eager Execution, so we disable that with `disable_eager_execution()`.
```
from __future__ import absolute_import, division, print_function, unicode_literals
import numpy as np
from __future__ import absolute_import, division, print_function
!pip install tensorflow-gpu==2.0.0-beta1
import tensorflow as tf
tf.compat.v1.disable_eager_execution()
import numpy as np
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()
# add empty color dimension
x_train = np.expand_dims(x_train, -1)
x_test = np.expand_dims(x_test, -1)
# convert types to float32
x_train = x_train.astype(np.float32)
x_test = x_test.astype(np.float32)
y_train = y_train.astype(np.float32)
y_test = y_test.astype(np.float32)
```
### Initialize TPUStrategy
We first initialize the TPUStrategy object before creating the model, so that Keras knows that we are creating a model for TPUs.
To do this, we are first creating a TPUClusterResolver using the IP address of the TPU, and then creating a TPUStrategy object from the Cluster Resolver.
```
import os
resolver = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.tpu.experimental.initialize_tpu_system(resolver)
strategy = tf.distribute.experimental.TPUStrategy(resolver)
```
### Define the Model
The following example uses a standard conv-net that has 4 layers with drop-out and batch normalization between each layer. Note that we are creating the model within a `strategy.scope`.
```
with strategy.scope():
model = tf.keras.models.Sequential()
model.add(tf.keras.layers.BatchNormalization(input_shape=x_train.shape[1:]))
model.add(tf.keras.layers.Conv2D(64, (5, 5), padding='same', activation='elu'))
model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(128, (5, 5), padding='same', activation='elu'))
model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(256, (5, 5), padding='same', activation='elu'))
model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.Conv2D(512, (5, 5), padding='same', activation='elu'))
model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))
model.add(tf.keras.layers.Dropout(0.25))
model.add(tf.keras.layers.Flatten())
model.add(tf.keras.layers.Dense(256))
model.add(tf.keras.layers.Activation('elu'))
model.add(tf.keras.layers.Dropout(0.5))
model.add(tf.keras.layers.Dense(10))
model.add(tf.keras.layers.Activation('softmax'))
model.summary()
```
### Train on the TPU
To train on the TPU, we can simply call `model.compile` under the strategy scope, and then call `model.fit` to start training. In this case, we are training for 5 epochs with 60 steps per epoch, and running evaluation at the end of 5 epochs.
It may take a while for the training to start, as the data and model has to be transferred to the TPU and compiled before training can start.
```
with strategy.scope():
model.compile(
optimizer=tf.train.AdamOptimizer(learning_rate=1e-3),
loss=tf.keras.losses.sparse_categorical_crossentropy,
metrics=['sparse_categorical_accuracy']
)
model.fit(
(x_train, y_train),
epochs=5,
steps_per_epoch=60,
validation_data=(x_test, y_test),
validation_freq=5,
)
```
### Check our results with Inference
Now that we are done training, we can see how well the model can predict fashion categories:
```
LABEL_NAMES = ['t_shirt', 'trouser', 'pullover', 'dress', 'coat', 'sandal', 'shirt', 'sneaker', 'bag', 'ankle_boots']
from matplotlib import pyplot
%matplotlib inline
def plot_predictions(images, predictions):
n = images.shape[0]
nc = int(np.ceil(n / 4))
f, axes = pyplot.subplots(nc, 4)
for i in range(nc * 4):
y = i // 4
x = i % 4
axes[x, y].axis('off')
label = LABEL_NAMES[np.argmax(predictions[i])]
confidence = np.max(predictions[i])
if i > n:
continue
axes[x, y].imshow(images[i])
axes[x, y].text(0.5, -1.5, label + ': %.3f' % confidence, fontsize=12)
pyplot.gcf().set_size_inches(8, 8)
plot_predictions(np.squeeze(x_test[:16]),
model.predict(x_test[:16]))
```
### What's next
* Learn about [Cloud TPUs](https://cloud.google.com/tpu/docs) that Google designed and optimized specifically to speed up and scale up ML workloads for training and inference and to enable ML engineers and researchers to iterate more quickly.
* Explore the range of [Cloud TPU tutorials and Colabs](https://cloud.google.com/tpu/docs/tutorials) to find other examples that can be used when implementing your ML project.
On Google Cloud Platform, in addition to GPUs and TPUs available on pre-configured [deep learning VMs](https://cloud.google.com/deep-learning-vm/), you will find [AutoML](https://cloud.google.com/automl/)*(beta)* for training custom models without writing code and [Cloud ML Engine](https://cloud.google.com/ml-engine/docs/) which will allows you to run parallel trainings and hyperparameter tuning of your custom models on powerful distributed hardware.
| github_jupyter |
<a href="https://colab.research.google.com/github/cseveriano/spatio-temporal-forecasting/blob/master/notebooks/thesis_experiments/20200924_eMVFTS_Wind_Energy_Raw.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Forecasting experiments for GEFCOM 2012 Wind Dataset
## Install Libs
```
!pip3 install -U git+https://github.com/PYFTS/pyFTS
!pip3 install -U git+https://github.com/cseveriano/spatio-temporal-forecasting
!pip3 install -U git+https://github.com/cseveriano/evolving_clustering
!pip3 install -U git+https://github.com/cseveriano/fts2image
!pip3 install -U hyperopt
!pip3 install -U pyts
import pandas as pd
import numpy as np
from hyperopt import hp
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
from google.colab import files
import matplotlib.pyplot as plt
import pickle
import math
from pyFTS.benchmarks import Measures
from pyts.decomposition import SingularSpectrumAnalysis
from google.colab import files
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
import datetime
```
## Aux Functions
```
def normalize(df):
mindf = df.min()
maxdf = df.max()
return (df-mindf)/(maxdf-mindf)
def denormalize(norm, _min, _max):
return [(n * (_max-_min)) + _min for n in norm]
def getRollingWindow(index):
pivot = index
train_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=20)
train_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
test_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
test_end = pivot.strftime('%Y-%m-%d')
return train_start, train_end, test_start, test_end
def calculate_rolling_error(cv_name, df, forecasts, order_list):
cv_results = pd.DataFrame(columns=['Split', 'RMSE', 'SMAPE'])
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
for i in np.arange(len(forecasts)):
train_start, train_end, test_start, test_end = getRollingWindow(index)
test = df[test_start : test_end]
yhat = forecasts[i]
order = order_list[i]
rmse = Measures.rmse(test.iloc[order:], yhat[:-1])
smape = Measures.smape(test.iloc[order:], yhat[:-1])
res = {'Split' : index.strftime('%Y-%m-%d') ,'RMSE' : rmse, 'SMAPE' : smape}
cv_results = cv_results.append(res, ignore_index=True)
cv_results.to_csv(cv_name+".csv")
index = index + datetime.timedelta(days=7)
return cv_results
def get_final_forecast(norm_forecasts):
forecasts_final = []
for i in np.arange(len(norm_forecasts)):
f_raw = denormalize(norm_forecasts[i], min_raw, max_raw)
forecasts_final.append(f_raw)
return forecasts_final
from spatiotemporal.test import methods_space_oahu as ms
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
import numpy as np
from hyperopt import fmin, tpe, hp, STATUS_OK, Trials
from hyperopt import space_eval
import traceback
from . import sampling
import pickle
def calculate_error(loss_function, test_df, forecast, offset):
error = loss_function(test_df.iloc[(offset):], forecast)
print("Error : "+str(error))
return error
def method_optimize(experiment, forecast_method, train_df, test_df, space, loss_function, max_evals):
def objective(params):
print(params)
try:
_output = list(params['output'])
forecast = forecast_method(train_df, test_df, params)
_step = params.get('step', 1)
offset = params['order'] + _step - 1
error = calculate_error(loss_function, test_df[_output], forecast, offset)
except Exception:
traceback.print_exc()
error = 1000
return {'loss': error, 'status': STATUS_OK}
print("Running experiment: " + experiment)
trials = Trials()
best = fmin(objective, space, algo=tpe.suggest, max_evals=max_evals, trials=trials)
print('best parameters: ')
print(space_eval(space, best))
pickle.dump(best, open("best_" + experiment + ".pkl", "wb"))
pickle.dump(trials, open("trials_" + experiment + ".pkl", "wb"))
def run_search(methods, data, train, loss_function, max_evals=100, resample=None):
if resample:
data = sampling.resample_data(data, resample)
train_df, test_df = sampling.train_test_split(data, train)
for experiment, method, space in methods:
method_optimize(experiment, method, train_df, test_df, space, loss_function, max_evals)
```
## Load Dataset
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import math
from sklearn.metrics import mean_squared_error
#columns names
wind_farms = ['wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7']
# read raw dataset
import pandas as pd
df = pd.read_csv('https://query.data.world/s/3zx2jusk4z6zvlg2dafqgshqp3oao6', parse_dates=['date'], index_col=0)
df.index = pd.to_datetime(df.index, format="%Y%m%d%H")
interval = ((df.index >= '2009-07') & (df.index <= '2010-08'))
df = df.loc[interval]
#Normalize Data
# Save Min-Max for Denorm
min_raw = df.min()
max_raw = df.max()
# Perform Normalization
norm_df = normalize(df)
# Tuning split
tuning_df = norm_df["2009-07-01":"2009-07-31"]
norm_df = norm_df["2009-08-01":"2010-08-30"]
df = df["2009-08-01":"2010-08-30"]
```
## Forecasting Methods
### Persistence
```
def persistence_forecast(train, test, step):
predictions = []
for t in np.arange(0,len(test), step):
yhat = [test.iloc[t]] * step
predictions.extend(yhat)
return predictions
def rolling_cv_persistence(df, step):
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
yhat = persistence_forecast(train, test, step)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
forecasts_raw, order_list = rolling_cv_persistence(norm_df, 1)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_persistence", norm_df, forecasts_final, order_list)
files.download('rolling_cv_wind_raw_persistence.csv')
```
### VAR
```
from statsmodels.tsa.api import VAR, DynamicVAR
def evaluate_VAR_models(test_name, train, validation,target, maxlags_list):
var_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for lgs in maxlags_list:
model = VAR(train)
results = model.fit(maxlags=lgs, ic='aic')
order = results.k_ar
forecast = []
for i in range(len(validation)-order) :
forecast.extend(results.forecast(validation.values[i:i+order],1))
forecast_df = pd.DataFrame(columns=validation.columns, data=forecast)
rmse = Measures.rmse(validation[target].iloc[order:], forecast_df[target].values)
if rmse < best_score:
best_score, best_cfg, best_model = rmse, order, results
res = {'Order' : str(order) ,'RMSE' : rmse}
print('VAR (%s) RMSE=%.3f' % (str(order),rmse))
var_results = var_results.append(res, ignore_index=True)
var_results.to_csv(test_name+".csv")
print('Best VAR(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_model
def var_forecast(train, test, params):
order = params['order']
step = params['step']
model = VAR(train.values)
results = model.fit(maxlags=order)
lag_order = results.k_ar
print("Lag order:" + str(lag_order))
forecast = []
for i in np.arange(0,len(test)-lag_order+1,step) :
forecast.extend(results.forecast(test.values[i:i+lag_order],step))
forecast_df = pd.DataFrame(columns=test.columns, data=forecast)
return forecast_df.values, lag_order
def rolling_cv_var(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Concat train & validation for test
yhat, lag_order = var_forecast(train, test, params)
forecasts.append(yhat)
order_list.append(lag_order)
return forecasts, order_list
params_raw = {'order': 4, 'step': 1}
forecasts_raw, order_list = rolling_cv_var(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_var", df, forecasts_final, order_list)
files.download('rolling_cv_wind_raw_var.csv')
```
### e-MVFTS
```
from spatiotemporal.models.clusteredmvfts.fts import evolvingclusterfts
def evolvingfts_forecast(train_df, test_df, params, train_model=True):
_variance_limit = params['variance_limit']
_defuzzy = params['defuzzy']
_t_norm = params['t_norm']
_membership_threshold = params['membership_threshold']
_order = params['order']
_step = params['step']
model = evolvingclusterfts.EvolvingClusterFTS(variance_limit=_variance_limit, defuzzy=_defuzzy, t_norm=_t_norm,
membership_threshold=_membership_threshold)
model.fit(train_df.values, order=_order, verbose=False)
forecast = model.predict(test_df.values, steps_ahead=_step)
forecast_df = pd.DataFrame(data=forecast, columns=test_df.columns)
return forecast_df.values
def rolling_cv_evolving(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
first_time = True
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Concat train & validation for test
yhat = list(evolvingfts_forecast(train, test, params, train_model=first_time))
#yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
first_time = False
return forecasts, order_list
params_raw = {'variance_limit': 0.001, 'order': 2, 'defuzzy': 'weighted', 't_norm': 'threshold', 'membership_threshold': 0.6, 'step':1}
forecasts_raw, order_list = rolling_cv_evolving(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_emvfts", df, forecasts_final, order_list)
files.download('rolling_cv_wind_raw_emvfts.csv')
```
### MLP
```
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
from keras.constraints import maxnorm
from keras.models import Sequential
from keras.layers.core import Dense, Dropout, Activation
from keras.layers.normalization import BatchNormalization
# convert series to supervised learning
def series_to_supervised(data, n_in=1, n_out=1, dropnan=True):
n_vars = 1 if type(data) is list else data.shape[1]
df = pd.DataFrame(data)
cols, names = list(), list()
# input sequence (t-n, ... t-1)
for i in range(n_in, 0, -1):
cols.append(df.shift(i))
names += [('var%d(t-%d)' % (j+1, i)) for j in range(n_vars)]
# forecast sequence (t, t+1, ... t+n)
for i in range(0, n_out):
cols.append(df.shift(-i))
if i == 0:
names += [('var%d(t)' % (j+1)) for j in range(n_vars)]
else:
names += [('var%d(t+%d)' % (j+1, i)) for j in range(n_vars)]
# put it all together
agg = pd.concat(cols, axis=1)
agg.columns = names
# drop rows with NaN values
if dropnan:
agg.dropna(inplace=True)
return agg
```
#### MLP Parameter Tuning
```
from spatiotemporal.util import parameter_tuning, sampling
from spatiotemporal.util import experiments as ex
from sklearn.metrics import mean_squared_error
from hyperopt import hp
import numpy as np
mlp_space = {'choice':
hp.choice('num_layers',
[
{'layers': 'two',
},
{'layers': 'three',
'units3': hp.choice('units3', [8, 16, 64, 128, 256, 512]),
'dropout3': hp.choice('dropout3', [0, 0.25, 0.5, 0.75])
}
]),
'units1': hp.choice('units1', [8, 16, 64, 128, 256, 512]),
'units2': hp.choice('units2', [8, 16, 64, 128, 256, 512]),
'dropout1': hp.choice('dropout1', [0, 0.25, 0.5, 0.75]),
'dropout2': hp.choice('dropout2', [0, 0.25, 0.5, 0.75]),
'batch_size': hp.choice('batch_size', [28, 64, 128, 256, 512]),
'order': hp.choice('order', [1, 2, 3]),
'input': hp.choice('input', [wind_farms]),
'output': hp.choice('output', [wind_farms]),
'epochs': hp.choice('epochs', [100, 200, 300])}
def mlp_tuning(train_df, test_df, params):
_input = list(params['input'])
_nlags = params['order']
_epochs = params['epochs']
_batch_size = params['batch_size']
nfeat = len(train_df.columns)
nsteps = params.get('step',1)
nobs = _nlags * nfeat
output_index = -nfeat*nsteps
train_reshaped_df = series_to_supervised(train_df[_input], n_in=_nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:, :nobs].values, train_reshaped_df.iloc[:, output_index:].values
test_reshaped_df = series_to_supervised(test_df[_input], n_in=_nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:, :nobs].values, test_reshaped_df.iloc[:, output_index:].values
# design network
model = Sequential()
model.add(Dense(params['units1'], input_dim=train_X.shape[1], activation='relu'))
model.add(Dropout(params['dropout1']))
model.add(BatchNormalization())
model.add(Dense(params['units2'], activation='relu'))
model.add(Dropout(params['dropout2']))
model.add(BatchNormalization())
if params['choice']['layers'] == 'three':
model.add(Dense(params['choice']['units3'], activation='relu'))
model.add(Dropout(params['choice']['dropout3']))
model.add(BatchNormalization())
model.add(Dense(train_Y.shape[1], activation='sigmoid'))
model.compile(loss='mse', optimizer='adam')
# includes the call back object
model.fit(train_X, train_Y, epochs=_epochs, batch_size=_batch_size, verbose=False, shuffle=False)
# predict the test set
forecast = model.predict(test_X, verbose=False)
return forecast
methods = []
methods.append(("EXP_OAHU_MLP", mlp_tuning, mlp_space))
train_split = 0.6
run_search(methods, tuning_df, train_split, Measures.rmse, max_evals=30, resample=None)
```
#### MLP Forecasting
```
def mlp_multi_forecast(train_df, test_df, params):
nfeat = len(train_df.columns)
nlags = params['order']
nsteps = params.get('step',1)
nobs = nlags * nfeat
output_index = -nfeat*nsteps
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:, :nobs].values, train_reshaped_df.iloc[:, output_index:].values
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:, :nobs].values, test_reshaped_df.iloc[:, output_index:].values
# design network
model = designMLPNetwork(train_X.shape[1], train_Y.shape[1], params)
# fit network
model.fit(train_X, train_Y, epochs=500, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
# fcst = [f[0] for f in forecast]
fcst = forecast
return fcst
def designMLPNetwork(input_shape, output_shape, params):
model = Sequential()
model.add(Dense(params['units1'], input_dim=input_shape, activation='relu'))
model.add(Dropout(params['dropout1']))
model.add(BatchNormalization())
model.add(Dense(params['units2'], activation='relu'))
model.add(Dropout(params['dropout2']))
model.add(BatchNormalization())
if params['choice']['layers'] == 'three':
model.add(Dense(params['choice']['units3'], activation='relu'))
model.add(Dropout(params['choice']['dropout3']))
model.add(BatchNormalization())
model.add(Dense(output_shape, activation='sigmoid'))
model.compile(loss='mse', optimizer='adam')
return model
def rolling_cv_mlp(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Perform forecast
yhat = list(mlp_multi_forecast(train, test, params))
yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
return forecasts, order_list
# Enter best params
params_raw = {'batch_size': 64, 'choice': {'layers': 'two'}, 'dropout1': 0.25, 'dropout2': 0.5, 'epochs': 200, 'input': ('wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7'), 'order': 2, 'output': ('wp1', 'wp2', 'wp3', 'wp4', 'wp5', 'wp6', 'wp7'), 'units1': 128, 'units2': 128}
forecasts_raw, order_list = rolling_cv_mlp(norm_df, params_raw)
forecasts_final = get_final_forecast(forecasts_raw)
calculate_rolling_error("rolling_cv_wind_raw_mlp_multi", df, forecasts_final, order_list)
files.download('rolling_cv_wind_raw_mlp_multi.csv')
```
### Granular FTS
```
from pyFTS.models.multivariate import granular
from pyFTS.partitioners import Grid, Entropy
from pyFTS.models.multivariate import variable
from pyFTS.common import Membership
from pyFTS.partitioners import Grid, Entropy
```
#### Granular Parameter Tuning
```
granular_space = {
'npartitions': hp.choice('npartitions', [100, 150, 200]),
'order': hp.choice('order', [1, 2]),
'knn': hp.choice('knn', [1, 2, 3, 4, 5]),
'alpha_cut': hp.choice('alpha_cut', [0, 0.1, 0.2, 0.3]),
'input': hp.choice('input', [['wp1', 'wp2', 'wp3']]),
'output': hp.choice('output', [['wp1', 'wp2', 'wp3']])}
def granular_tuning(train_df, test_df, params):
_input = list(params['input'])
_output = list(params['output'])
_npartitions = params['npartitions']
_order = params['order']
_knn = params['knn']
_alpha_cut = params['alpha_cut']
_step = params.get('step',1)
## create explanatory variables
exp_variables = []
for vc in _input:
exp_variables.append(variable.Variable(vc, data_label=vc, alias=vc,
npart=_npartitions, func=Membership.trimf,
data=train_df, alpha_cut=_alpha_cut))
model = granular.GranularWMVFTS(explanatory_variables=exp_variables, target_variable=exp_variables[0], order=_order,
knn=_knn)
model.fit(train_df[_input], num_batches=1)
if _step > 1:
forecast = pd.DataFrame(columns=test_df.columns)
length = len(test_df.index)
for k in range(0,(length -(_order + _step - 1))):
fcst = model.predict(test_df[_input], type='multivariate', start_at=k, steps_ahead=_step)
forecast = forecast.append(fcst.tail(1))
else:
forecast = model.predict(test_df[_input], type='multivariate')
return forecast[_output].values
methods = []
methods.append(("EXP_WIND_GRANULAR", granular_tuning, granular_space))
train_split = 0.6
run_search(methods, tuning_df, train_split, Measures.rmse, max_evals=10, resample=None)
```
#### Granular Forecasting
```
def granular_forecast(train_df, test_df, params):
_input = list(params['input'])
_output = list(params['output'])
_npartitions = params['npartitions']
_knn = params['knn']
_alpha_cut = params['alpha_cut']
_order = params['order']
_step = params.get('step',1)
## create explanatory variables
exp_variables = []
for vc in _input:
exp_variables.append(variable.Variable(vc, data_label=vc, alias=vc,
npart=_npartitions, func=Membership.trimf,
data=train_df, alpha_cut=_alpha_cut))
model = granular.GranularWMVFTS(explanatory_variables=exp_variables, target_variable=exp_variables[0], order=_order,
knn=_knn)
model.fit(train_df[_input], num_batches=1)
if _step > 1:
forecast = pd.DataFrame(columns=test_df.columns)
length = len(test_df.index)
for k in range(0,(length -(_order + _step - 1))):
fcst = model.predict(test_df[_input], type='multivariate', start_at=k, steps_ahead=_step)
forecast = forecast.append(fcst.tail(1))
else:
forecast = model.predict(test_df[_input], type='multivariate')
return forecast[_output].values
def rolling_cv_granular(df, params):
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
test = df[test_start : test_end]
# Perform forecast
yhat = list(granular_forecast(train, test, params))
yhat.append(yhat[-1]) #para manter o formato do vetor de metricas
forecasts.append(yhat)
order_list.append(params['order'])
return forecasts, order_list
def granular_get_final_forecast(forecasts_raw, input):
forecasts_final = []
l_min = df[input].min()
l_max = df[input].max()
for i in np.arange(len(forecasts_raw)):
f_raw = denormalize(forecasts_raw[i], l_min, l_max)
forecasts_final.append(f_raw)
return forecasts_final
# Enter best params
params_raw = {'alpha_cut': 0.3, 'input': ('wp1', 'wp2', 'wp3'), 'knn': 5, 'npartitions': 200, 'order': 2, 'output': ('wp1', 'wp2', 'wp3')}
forecasts_raw, order_list = rolling_cv_granular(norm_df, params_raw)
forecasts_final = granular_get_final_forecast(forecasts_raw, list(params_raw['input']))
calculate_rolling_error("rolling_cv_wind_raw_granular", df[list(params_raw['input'])], forecasts_final, order_list)
files.download('rolling_cv_wind_raw_granular.csv')
```
## Result Analysis
```
import pandas as pd
from google.colab import files
files.upload()
def createBoxplot(filename, data, xticklabels, ylabel):
# Create a figure instance
fig = plt.figure(1, figsize=(9, 6))
# Create an axes instance
ax = fig.add_subplot(111)
# Create the boxplot
bp = ax.boxplot(data, patch_artist=True)
## change outline color, fill color and linewidth of the boxes
for box in bp['boxes']:
# change outline color
box.set( color='#7570b3', linewidth=2)
# change fill color
box.set( facecolor = '#AACCFF' )
## change color and linewidth of the whiskers
for whisker in bp['whiskers']:
whisker.set(color='#7570b3', linewidth=2)
## change color and linewidth of the caps
for cap in bp['caps']:
cap.set(color='#7570b3', linewidth=2)
## change color and linewidth of the medians
for median in bp['medians']:
median.set(color='#FFE680', linewidth=2)
## change the style of fliers and their fill
for flier in bp['fliers']:
flier.set(marker='o', color='#e7298a', alpha=0.5)
## Custom x-axis labels
ax.set_xticklabels(xticklabels)
ax.set_ylabel(ylabel)
plt.show()
fig.savefig(filename, bbox_inches='tight')
var_results = pd.read_csv("rolling_cv_wind_raw_var.csv")
evolving_results = pd.read_csv("rolling_cv_wind_raw_emvfts.csv")
mlp_results = pd.read_csv("rolling_cv_wind_raw_mlp_multi.csv")
granular_results = pd.read_csv("rolling_cv_wind_raw_granular.csv")
metric = 'RMSE'
results_data = [evolving_results[metric],var_results[metric], mlp_results[metric], granular_results[metric]]
xticks = ['e-MVFTS','VAR','MLP','FIG-FTS']
ylab = 'RMSE'
createBoxplot("e-mvfts_boxplot_rmse_solar", results_data, xticks, ylab)
pd.options.display.float_format = '{:.2f}'.format
metric = 'RMSE'
rmse_df = pd.DataFrame(columns=['e-MVFTS','VAR','MLP','FIG-FTS'])
rmse_df["e-MVFTS"] = evolving_results[metric]
rmse_df["VAR"] = var_results[metric]
rmse_df["MLP"] = mlp_results[metric]
rmse_df["FIG-FTS"] = granular_results[metric]
rmse_df.std()
metric = 'SMAPE'
results_data = [evolving_results[metric],var_results[metric], mlp_results[metric], granular_results[metric]]
xticks = ['e-MVFTS','VAR','MLP','FIG-FTS']
ylab = 'SMAPE'
createBoxplot("e-mvfts_boxplot_smape_solar", results_data, xticks, ylab)
metric = 'SMAPE'
smape_df = pd.DataFrame(columns=['e-MVFTS','VAR','MLP','FIG-FTS'])
smape_df["e-MVFTS"] = evolving_results[metric]
smape_df["VAR"] = var_results[metric]
smape_df["MLP"] = mlp_results[metric]
smape_df["FIG-FTS"] = granular_results[metric]
smape_df.std()
metric = "RMSE"
data = pd.DataFrame(columns=["VAR", "Evolving", "MLP", "Granular"])
data["VAR"] = var_results[metric]
data["Evolving"] = evolving_results[metric]
data["MLP"] = mlp_results[metric]
data["Granular"] = granular_results[metric]
ax = data.plot(figsize=(18,6))
ax.set(xlabel='Window', ylabel=metric)
fig = ax.get_figure()
#fig.savefig(path_images + exp_id + "_prequential.png")
x = np.arange(len(data.columns.values))
names = data.columns.values
values = data.mean().values
plt.figure(figsize=(5,6))
plt.bar(x, values, align='center', alpha=0.5, width=0.9)
plt.xticks(x, names)
#plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylabel(metric)
#plt.savefig(path_images + exp_id + "_bars.png")
metric = "SMAPE"
data = pd.DataFrame(columns=["VAR", "Evolving", "MLP", "Granular"])
data["VAR"] = var_results[metric]
data["Evolving"] = evolving_results[metric]
data["MLP"] = mlp_results[metric]
data["Granular"] = granular_results[metric]
ax = data.plot(figsize=(18,6))
ax.set(xlabel='Window', ylabel=metric)
fig = ax.get_figure()
#fig.savefig(path_images + exp_id + "_prequential.png")
x = np.arange(len(data.columns.values))
names = data.columns.values
values = data.mean().values
plt.figure(figsize=(5,6))
plt.bar(x, values, align='center', alpha=0.5, width=0.9)
plt.xticks(x, names)
#plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylabel(metric)
#plt.savefig(path_images + exp_id + "_bars.png")
```
| github_jupyter |
# Use `Lale` `AIF360` scorers to calculate and mitigate bias for credit risk AutoAI model
This notebook contains the steps and code to demonstrate support of AutoAI experiments in Watson Machine Learning service. It introduces commands for bias detecting and mitigation performed with `lale.lib.aif360` module.
Some familiarity with Python is helpful. This notebook uses Python 3.8.
## Contents
This notebook contains the following parts:
1. [Setup](#setup)
2. [Optimizer definition](#definition)
3. [Experiment Run](#run)
4. [Pipeline bias detection and mitigation](#bias)
5. [Deployment and score](#scoring)
6. [Clean up](#cleanup)
7. [Summary and next steps](#summary)
<a id="setup"></a>
## 1. Set up the environment
If you are not familiar with <a href="https://console.ng.bluemix.net/catalog/services/ibm-watson-machine-learning/" target="_blank" rel="noopener no referrer">Watson Machine Learning (WML) Service</a> and AutoAI experiments please read more about it in the sample notebook: <a href="https://github.com/IBM/watson-machine-learning-samples/blob/master/cloud/notebooks/python_sdk/experiments/autoai/Use%20AutoAI%20and%20Lale%20to%20predict%20credit%20risk.ipynb" target="_blank" rel="noopener no referrer">"Use AutoAI and Lale to predict credit risk with `ibm-watson-machine-learning`"</a>
### Install and import the `ibm-watson-machine-learning`, `lale` ,`aif360` and dependencies.
**Note:** `ibm-watson-machine-learning` documentation can be found <a href="http://ibm-wml-api-pyclient.mybluemix.net/" target="_blank" rel="noopener no referrer">here</a>.
```
!pip install -U ibm-watson-machine-learning | tail -n 1
!pip install -U scikit-learn==0.23.2 | tail -n 1
!pip install -U autoai-libs | tail -n 1
!pip install -U lale | tail -n 1
!pip install -U aif360 | tail -n 1
!pip install -U liac-arff | tail -n 1
!pip install -U cvxpy | tail -n 1
!pip install -U fairlearn | tail -n 1
```
### Connection to WML
Authenticate the Watson Machine Learning service on IBM Cloud. You need to provide Cloud `API key` and `location`.
**Tip**: Your `Cloud API key` can be generated by going to the [**Users** section of the Cloud console](https://cloud.ibm.com/iam#/users). From that page, click your name, scroll down to the **API Keys** section, and click **Create an IBM Cloud API key**. Give your key a name and click **Create**, then copy the created key and paste it below. You can also get a service specific url by going to the [**Endpoint URLs** section of the Watson Machine Learning docs](https://cloud.ibm.com/apidocs/machine-learning). You can check your instance location in your <a href="https://console.ng.bluemix.net/catalog/services/ibm-watson-machine-learning/" target="_blank" rel="noopener no referrer">Watson Machine Learning (WML) Service</a> instance details.
You can use [IBM Cloud CLI](https://cloud.ibm.com/docs/cli/index.html) to retrieve the instance `location`.
```
ibmcloud login --apikey API_KEY -a https://cloud.ibm.com
ibmcloud resource service-instance WML_INSTANCE_NAME
```
**NOTE:** You can also get a service specific apikey by going to the [**Service IDs** section of the Cloud Console](https://cloud.ibm.com/iam/serviceids). From that page, click **Create**, and then copy the created key and paste it in the following cell.
**Action**: Enter your `api_key` and `location` in the following cell.
```
api_key = 'PUT_YOUR_KEY_HERE'
location = 'us-south'
wml_credentials = {
"apikey": api_key,
"url": 'https://' + location + '.ml.cloud.ibm.com'
}
from ibm_watson_machine_learning import APIClient
client = APIClient(wml_credentials)
```
### Working with spaces
You need to create a space that will be used for your work. If you do not have a space, you can use [Deployment Spaces Dashboard](https://dataplatform.cloud.ibm.com/ml-runtime/spaces?context=cpdaas) to create one.
- Click **New Deployment Space**
- Create an empty space
- Select Cloud Object Storage
- Select Watson Machine Learning instance and press **Create**
- Copy `space_id` and paste it below
**Tip**: You can also use SDK to prepare the space for your work. More information can be found [here](https://github.com/IBM/watson-machine-learning-samples/blob/master/cloud/notebooks/python_sdk/instance-management/Space%20management.ipynb).
**Action**: assign space ID below
```
space_id = 'PASTE YOUR SPACE ID HERE'
client.spaces.list(limit=10)
client.set.default_space(space_id)
```
### Connections to COS
In next cell we read the COS credentials from the space.
```
cos_credentials = client.spaces.get_details(space_id=space_id)['entity']['storage']['properties']
```
<a id="definition"></a>
## 2. Optimizer definition
### Training data connection
Define connection information to COS bucket and training data CSV file. This example uses the [German Credit Risk dataset](https://raw.githubusercontent.com/IBM/watson-machine-learning-samples/master/cloud/data/credit_risk/credit_risk_training_light.csv).
The code in next cell uploads training data to the bucket.
```
filename = 'german_credit_data_biased_training.csv'
datasource_name = 'bluemixcloudobjectstorage'
bucketname = cos_credentials['bucket_name']
```
Download training data from git repository and split for training and test set.
```
import os, wget
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
url = 'https://raw.githubusercontent.com/IBM/watson-machine-learning-samples/master/cloud/data/credit_risk/german_credit_data_biased_training.csv'
if not os.path.isfile(filename): wget.download(url)
credit_risk_df = pd.read_csv(filename)
X = credit_risk_df.drop(['Risk'], axis=1)
y = credit_risk_df['Risk']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
credit_risk_df.head()
```
#### Create connection
```
conn_meta_props= {
client.connections.ConfigurationMetaNames.NAME: f"Connection to Database - {datasource_name} ",
client.connections.ConfigurationMetaNames.DATASOURCE_TYPE: client.connections.get_datasource_type_uid_by_name(datasource_name),
client.connections.ConfigurationMetaNames.DESCRIPTION: "Connection to external Database",
client.connections.ConfigurationMetaNames.PROPERTIES: {
'bucket': bucketname,
'access_key': cos_credentials['credentials']['editor']['access_key_id'],
'secret_key': cos_credentials['credentials']['editor']['secret_access_key'],
'iam_url': 'https://iam.cloud.ibm.com/identity/token',
'url': cos_credentials['endpoint_url']
}
}
conn_details = client.connections.create(meta_props=conn_meta_props)
```
**Note**: The above connection can be initialized alternatively with `api_key` and `resource_instance_id`.
The above cell can be replaced with:
```
conn_meta_props= {
client.connections.ConfigurationMetaNames.NAME: f"Connection to Database - {db_name} ",
client.connections.ConfigurationMetaNames.DATASOURCE_TYPE: client.connections.get_datasource_type_uid_by_name(db_name),
client.connections.ConfigurationMetaNames.DESCRIPTION: "Connection to external Database",
client.connections.ConfigurationMetaNames.PROPERTIES: {
'bucket': bucket_name,
'api_key': cos_credentials['apikey'],
'resource_instance_id': cos_credentials['resource_instance_id'],
'iam_url': 'https://iam.cloud.ibm.com/identity/token',
'url': 'https://s3.us.cloud-object-storage.appdomain.cloud'
}
}
conn_details = client.connections.create(meta_props=conn_meta_props)
```
```
connection_id = client.connections.get_uid(conn_details)
```
Define connection information to training data and upload train dataset to COS bucket.
```
from ibm_watson_machine_learning.helpers import DataConnection, S3Location
credit_risk_conn = DataConnection(
connection_asset_id=connection_id,
location=S3Location(bucket=bucketname,
path=filename))
credit_risk_conn._wml_client = client
training_data_reference=[credit_risk_conn]
credit_risk_conn.write(data=X_train.join(y_train), remote_name=filename)
```
### Optimizer configuration
Provide the input information for AutoAI optimizer:
- `name` - experiment name
- `prediction_type` - type of the problem
- `prediction_column` - target column name
- `scoring` - optimization metric
- `daub_include_only_estimators` - estimators which will be included during AutoAI training. More available estimators can be found in `experiment.ClassificationAlgorithms` enum
```
from ibm_watson_machine_learning.experiment import AutoAI
experiment = AutoAI(wml_credentials, space_id=space_id)
pipeline_optimizer = experiment.optimizer(
name='Credit Risk Bias detection in AutoAI',
prediction_type=AutoAI.PredictionType.BINARY,
prediction_column='Risk',
scoring=AutoAI.Metrics.ROC_AUC_SCORE,
include_only_estimators=[experiment.ClassificationAlgorithms.XGB]
)
```
<a id="run"></a>
## 3. Experiment run
Call the `fit()` method to trigger the AutoAI experiment. You can either use interactive mode (synchronous job) or background mode (asychronous job) by specifying `background_model=True`.
```
run_details = pipeline_optimizer.fit(
training_data_reference=training_data_reference,
background_mode=False)
pipeline_optimizer.get_run_status()
summary = pipeline_optimizer.summary()
summary
```
### Get selected pipeline model
Download pipeline model object from the AutoAI training job.
```
best_pipeline = pipeline_optimizer.get_pipeline()
```
<a id="bias"></a>
## 4. Bias detection and mitigation
The `fairness_info` dictionary contains some fairness-related metadata. The favorable and unfavorable label are values of the target class column that indicate whether the loan was granted or denied. A protected attribute is a feature that partitions the population into groups whose outcome should have parity. The credit-risk dataset has two protected attribute columns, sex and age. Each prottected attributes has privileged and unprivileged group.
Note that to use fairness metrics from lale with numpy arrays `protected_attributes.feature` need to be passed as index of the column in dataset, not as name.
```
fairness_info = {'favorable_labels': ['No Risk'],
'protected_attributes': [
{'feature': X.columns.get_loc('Sex'),'reference_group': ['male']},
{'feature': X.columns.get_loc('Age'), 'reference_group': [[26, 40]]}]}
fairness_info
```
### Calculate fairness metrics
We will calculate some model metrics. Accuracy describes how accurate is the model according to dataset.
Disparate impact is defined by comparing outcomes between a privileged group and an unprivileged group,
so it needs to check the protected attribute to determine group membership for the sample record at hand.
The third calculated metric takes the disparate impact into account along with accuracy. The best value of the score is 1.0.
```
import sklearn.metrics
from lale.lib.aif360 import disparate_impact, accuracy_and_disparate_impact
accuracy_scorer = sklearn.metrics.make_scorer(sklearn.metrics.accuracy_score)
print(f'accuracy {accuracy_scorer(best_pipeline, X_test.values, y_test.values):.1%}')
disparate_impact_scorer = disparate_impact(**fairness_info)
print(f'disparate impact {disparate_impact_scorer(best_pipeline, X_test.values, y_test.values):.2f}')
combined_scorer = accuracy_and_disparate_impact(**fairness_info)
print(f'accuracy and disparate impact metric {combined_scorer(best_pipeline, X_test.values, y_test.values):.2f}')
```
### Mitigation
`Hyperopt` minimizes (best_score - score_returned_by_the_scorer), where best_score is an argument to Hyperopt and score_returned_by_the_scorer is the value returned by the scorer for each evaluation point. We will use the `Hyperopt` to tune hyperparametres of the AutoAI pipeline to get new and more fair model.
```
from sklearn.linear_model import LogisticRegression as LR
from sklearn.tree import DecisionTreeClassifier as Tree
from sklearn.neighbors import KNeighborsClassifier as KNN
from lale.lib.lale import Hyperopt
from lale.lib.aif360 import FairStratifiedKFold
from lale import wrap_imported_operators
wrap_imported_operators()
prefix = best_pipeline.remove_last().freeze_trainable()
prefix.visualize()
new_pipeline = prefix >> (LR | Tree | KNN)
new_pipeline.visualize()
fair_cv = FairStratifiedKFold(**fairness_info, n_splits=3)
pipeline_fairer = new_pipeline.auto_configure(
X_train.values, y_train.values, optimizer=Hyperopt, cv=fair_cv,
max_evals=10, scoring=combined_scorer, best_score=1.0)
```
As with any trained model, we can evaluate and visualize the result.
```
print(f'accuracy {accuracy_scorer(pipeline_fairer, X_test.values, y_test.values):.1%}')
print(f'disparate impact {disparate_impact_scorer(pipeline_fairer, X_test.values, y_test.values):.2f}')
print(f'accuracy and disparate impact metric {combined_scorer(pipeline_fairer, X_test.values, y_test.values):.2f}')
pipeline_fairer.visualize()
```
As the result demonstrates, the best model found by AI Automation
has lower accuracy and much better disparate impact as the one we saw
before. Also, it has tuned the repair level and
has picked and tuned a classifier. These results may vary by dataset and search space.
You can get source code of the created pipeline. You just need to change the below cell type `Raw NBCovert` to `code`.
<a id="scoring"></a>
## 5. Deploy and Score
In this section you will learn how to deploy and score Lale pipeline model using WML instance.
#### Custom software_specification
Created model is AutoAI model refined with Lale. We will create new software specification based on default Python 3.7
environment extended by `autoai-libs` package.
```
base_sw_spec_uid = client.software_specifications.get_uid_by_name("default_py3.7")
print("Id of default Python 3.7 software specification is: ", base_sw_spec_uid)
url = 'https://raw.githubusercontent.com/IBM/watson-machine-learning-samples/master/cloud/configs/config.yaml'
if not os.path.isfile('config.yaml'): wget.download(url)
!cat config.yaml
```
`config.yaml` file describes details of package extention. Now you need to store new package extention with `APIClient`.
```
meta_prop_pkg_extn = {
client.package_extensions.ConfigurationMetaNames.NAME: "Scikt with autoai-libs",
client.package_extensions.ConfigurationMetaNames.DESCRIPTION: "Pkg extension for autoai-libs",
client.package_extensions.ConfigurationMetaNames.TYPE: "conda_yml"
}
pkg_extn_details = client.package_extensions.store(meta_props=meta_prop_pkg_extn, file_path="config.yaml")
pkg_extn_uid = client.package_extensions.get_uid(pkg_extn_details)
pkg_extn_url = client.package_extensions.get_href(pkg_extn_details)
```
Create new software specification and add created package extention to it.
```
meta_prop_sw_spec = {
client.software_specifications.ConfigurationMetaNames.NAME: "Mitigated AutoAI bases on scikit spec",
client.software_specifications.ConfigurationMetaNames.DESCRIPTION: "Software specification for scikt with autoai-libs",
client.software_specifications.ConfigurationMetaNames.BASE_SOFTWARE_SPECIFICATION: {"guid": base_sw_spec_uid}
}
sw_spec_details = client.software_specifications.store(meta_props=meta_prop_sw_spec)
sw_spec_uid = client.software_specifications.get_uid(sw_spec_details)
status = client.software_specifications.add_package_extension(sw_spec_uid, pkg_extn_uid)
```
You can get details of created software specification using `client.software_specifications.get_details(sw_spec_uid)`
### Store the model
```
model_props = {
client.repository.ModelMetaNames.NAME: "Fairer AutoAI model",
client.repository.ModelMetaNames.TYPE: 'scikit-learn_0.23',
client.repository.ModelMetaNames.SOFTWARE_SPEC_UID: sw_spec_uid
}
feature_vector = list(X.columns)
published_model = client.repository.store_model(
model=best_pipeline.export_to_sklearn_pipeline(),
meta_props=model_props,
training_data=X_train.values,
training_target=y_train.values,
feature_names=feature_vector,
label_column_names=['Risk']
)
published_model_uid = client.repository.get_model_id(published_model)
```
### Deployment creation
```
metadata = {
client.deployments.ConfigurationMetaNames.NAME: "Deployment of fairer model",
client.deployments.ConfigurationMetaNames.ONLINE: {}
}
created_deployment = client.deployments.create(published_model_uid, meta_props=metadata)
deployment_id = client.deployments.get_uid(created_deployment)
```
#### Deployment scoring
You need to pass scoring values as input data if the deployed model. Use `client.deployments.score()` method to get predictions from deployed model.
```
values = X_test.values
scoring_payload = {
"input_data": [{
'values': values[:5]
}]
}
predictions = client.deployments.score(deployment_id, scoring_payload)
predictions
```
<a id="cleanup"></a>
## 5. Clean up
If you want to clean up all created assets:
- experiments
- trainings
- pipelines
- model definitions
- models
- functions
- deployments
please follow up this sample [notebook](https://github.com/IBM/watson-machine-learning-samples/blob/master/cloud/notebooks/python_sdk/instance-management/Machine%20Learning%20artifacts%20management.ipynb).
<a id="summary"></a>
## 6. Summary and next steps
You successfully completed this notebook!.
Check out used packeges domuntations:
- `ibm-watson-machine-learning` [Online Documentation](https://www.ibm.com/cloud/watson-studio/autoai)
- `lale`: https://github.com/IBM/lale
- `aif360`: https://aif360.mybluemix.net/
### Authors
**Dorota Dydo-Rożniecka**, Intern in Watson Machine Learning at IBM
Copyright © 2020, 2021 IBM. This notebook and its source code are released under the terms of the MIT License.
| github_jupyter |
# Trade-off between classification accuracy and reconstruction error during dimensionality reduction
- Low-dimensional LSTM representations are excellent at dimensionality reduction, but are poor at reconstructing the original data
- On the other hand, PCs are excellent at reconstructing the original data but these high-variance components do not preserve class information
```
import numpy as np
import pandas as pd
import scipy as sp
import pickle
import os
import random
import sys
# visualizations
from _plotly_future_ import v4_subplots
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.subplots as tls
import plotly.figure_factory as ff
import plotly.io as pio
import plotly.express as px
pio.templates.default = 'plotly_white'
pio.orca.config.executable = '/home/joyneelm/fire/bin/orca'
colors = px.colors.qualitative.Plotly
class ARGS():
roi = 300
net = 7
subnet = 'wb'
train_size = 100
batch_size = 32
num_epochs = 50
zscore = 1
#gru
k_hidden = 32
k_layers = 1
dims = [3, 4, 5, 10]
args = ARGS()
def _get_results(k_dim):
RES_DIR = 'results/clip_gru_recon'
load_path = (RES_DIR +
'/roi_%d_net_%d' %(args.roi, args.net) +
'_trainsize_%d' %(args.train_size) +
'_k_hidden_%d' %(args.k_hidden) +
'_kdim_%d' %(k_dim) +
'_k_layers_%d' %(args.k_layers) +
'_batch_size_%d' %(args.batch_size) +
'_num_epochs_45' +
'_z_%d.pkl' %(args.zscore))
with open(load_path, 'rb') as f:
results = pickle.load(f)
# print(results.keys())
return results
r = {}
for k_dim in args.dims:
r[k_dim] = _get_results(k_dim)
def _plot_fig(ss):
title_text = ss
if ss=='var':
ss = 'mse'
invert = True
else:
invert = False
subplot_titles = ['train', 'test']
fig = tls.make_subplots(rows=1,
cols=2,
subplot_titles=subplot_titles,
print_grid=False)
for ii, x in enumerate(['train', 'test']):
gru_score = {'mean':[], 'ste':[]}
pca_score = {'mean':[], 'ste':[]}
for k_dim in args.dims:
a = r[k_dim]
# gru decoder
y = np.mean(a['%s_%s'%(x, ss)])
gru_score['mean'].append(y)
# pca decoder
y = np.mean(a['%s_pca_%s'%(x, ss)])
pca_score['mean'].append(y)
x = np.arange(len(args.dims))
if invert:
y = 1 - np.array(gru_score['mean'])
else:
y = gru_score['mean']
error_y = gru_score['ste']
trace = go.Bar(x=x, y=y,
name='lstm decoder',
marker_color=colors[0])
fig.add_trace(trace, 1, ii+1)
if invert:
y = 1 - np.array(pca_score['mean'])
else:
y = pca_score['mean']
error_y = pca_score['ste']
trace = go.Bar(x=x, y=y,
name='pca recon',
marker_color=colors[1])
fig.add_trace(trace, 1, ii+1)
fig.update_xaxes(tickvals=np.arange(len(args.dims)),
ticktext=args.dims)
fig.update_layout(height=350, width=700,
title_text=title_text)
return fig
```
## Mean-squared error vs number of dimensions
```
'''
mse
'''
ss = 'mse'
fig = _plot_fig(ss)
fig.show()
```
## Variance captured vs number of dimensions
```
'''
variance
'''
ss = 'var'
fig = _plot_fig(ss)
fig.show()
```
## R-squared vs number of dimensions
```
'''
r2
'''
ss = 'r2'
fig = _plot_fig(ss)
fig.show()
results = r[10]
# variance not captured by pca recon
pca_not = 1 - np.sum(results['pca_var'])
print('percent variance captured by pca components = %0.3f' %(1 - pca_not))
# this is proportional to pca mse
pca_mse = results['test_pca_mse']
# variance not captured by lstm decoder?
lstm_mse = results['test_mse']
lstm_not = lstm_mse*(pca_not/pca_mse)
print('percent variance captured by lstm recon = %0.3f' %(1 - lstm_not))
def _plot_fig_ext(ss):
title_text = ss
if ss=='var':
ss = 'mse'
invert = True
else:
invert = False
subplot_titles = ['train', 'test']
fig = go.Figure()
x = 'test'
lstm_score = {'mean':[], 'ste':[]}
pca_score = {'mean':[], 'ste':[]}
lstm_acc = {'mean':[], 'ste':[]}
pc_acc = {'mean':[], 'ste':[]}
for k_dim in args.dims:
a = r[k_dim]
# lstm encoder
k_sub = len(a['test'])
y = np.mean(a['test'])
error_y = 3/np.sqrt(k_sub)*np.std(a['test'])
lstm_acc['mean'].append(y)
lstm_acc['ste'].append(error_y)
# lstm decoder
y = np.mean(a['%s_%s'%(x, ss)])
lstm_score['mean'].append(y)
lstm_score['ste'].append(error_y)
# pca encoder
b = r_pc[k_dim]
y = np.mean(b['test'])
error_y = 3/np.sqrt(k_sub)*np.std(b['test'])
pc_acc['mean'].append(y)
pc_acc['ste'].append(error_y)
# pca decoder
y = np.mean(a['%s_pca_%s'%(x, ss)])
pca_score['mean'].append(y)
pca_score['ste'].append(error_y)
x = np.arange(len(args.dims))
y = lstm_acc['mean']
error_y = lstm_acc['ste']
trace = go.Bar(x=x, y=y,
name='GRU Accuracy',
error_y=dict(type='data',
array=error_y),
marker_color=colors[3])
fig.add_trace(trace)
y = pc_acc['mean']
error_y = pc_acc['ste']
trace = go.Bar(x=x, y=y,
name='PCA Accuracy',
error_y=dict(type='data',
array=error_y),
marker_color=colors[4])
fig.add_trace(trace)
if invert:
y = 1 - np.array(lstm_score['mean'])
else:
y = lstm_score['mean']
error_y = lstm_score['ste']
trace = go.Bar(x=x, y=y,
name='GRU Reconstruction',
error_y=dict(type='data',
array=error_y),
marker_color=colors[5])
fig.add_trace(trace)
if invert:
y = 1 - np.array(pca_score['mean'])
else:
y = pca_score['mean']
error_y = pca_score['ste']
trace = go.Bar(x=x, y=y,
name='PCA Reconstruction',
error_y=dict(type='data',
array=error_y),
marker_color=colors[2])
fig.add_trace(trace)
fig.update_yaxes(title=dict(text='Accuracy or % variance',
font_size=20),
gridwidth=1, gridcolor='#bfbfbf',
tickfont=dict(size=20))
fig.update_xaxes(title=dict(text='Number of dimensions',
font_size=20),
tickvals=np.arange(len(args.dims)),
ticktext=args.dims,
tickfont=dict(size=20))
fig.update_layout(height=470, width=570,
font_color='black',
legend_orientation='h',
legend_font_size=20,
legend_x=-0.1,
legend_y=-0.3)
return fig
def _get_pc_results(PC_DIR, k_dim):
load_path = (PC_DIR +
'/roi_%d_net_%d' %(args.roi, args.net) +
'_nw_%s' %(args.subnet) +
'_trainsize_%d' %(args.train_size) +
'_kdim_%d_batch_size_%d' %(k_dim, args.batch_size) +
'_num_epochs_%d_z_%d.pkl' %(args.num_epochs, args.zscore))
with open(load_path, 'rb') as f:
results = pickle.load(f)
print(results.keys())
return results
```
## Comparison of LSTM and PCA: classification accuracy and variance captured
```
'''
variance
'''
r_pc = {}
PC_DIR = 'results/clip_pca'
for k_dim in args.dims:
r_pc[k_dim] = _get_pc_results(PC_DIR, k_dim)
colors = px.colors.qualitative.Set3
#colors = ["#D55E00", "#009E73", "#56B4E9", "#E69F00"]
ss = 'var'
fig = _plot_fig_ext(ss)
fig.show()
fig.write_image('figures/fig3c.png')
```
| github_jupyter |
# Controlling Flow with Conditional Statements
Now that you've learned how to create conditional statements, let's learn how to use them to control the flow of our programs. This is done with `if`, `elif`, and `else` statements.
## The `if` Statement
What if we wanted to check if a number was divisible by 2 and if so then print that number out. Let's diagram that out.

- Check to see if A is even
- If yes, then print our message: "A is even"
This use case can be translated into a "if" statement. I'm going to write this out in pseudocode which looks very similar to Python.
```text
if A is even:
print "A is even"
```
```
# Let's translate this into Python code
def check_evenness(A):
if A % 2 == 0:
print(f"A ({A:02}) is even!")
for i in range(1, 11):
check_evenness(i)
# You can do multiple if statements and they're executed sequentially
A = 10
if A > 0:
print('A is positive')
if A % 2 == 0:
print('A is even!')
```
## The `else` Statement
But what if we wanted to know if the number was even OR odd? Let's diagram that out:

Again, translating this to pseudocode, we're going to use the 'else' statement:
```text
if A is even:
print "A is even"
else:
print "A is odd"
```
```
# Let's translate this into Python code
def check_evenness(A):
if A % 2 == 0:
print(f"A ({A:02}) is even!")
else:
print(f'A ({A:02}) is odd!')
for i in range(1, 11):
check_evenness(i)
```
# The 'else if' or `elif` Statement
What if we wanted to check if A is divisible by 2 or 3? Let's diagram that out:

Again, translating this into psuedocode, we're going to use the 'else if' statement.
```text
if A is divisible by 2:
print "2 divides A"
else if A is divisible by 3:
print "3 divides A"
else
print "2 and 3 don't divide A"
```
```
# Let's translate this into Python code
def check_divisible_by_2_and_3(A):
if A % 2 == 0:
print(f"2 divides A ({A:02})!")
# else if in Python is elif
elif A % 3 == 0:
print(f'3 divides A ({A:02})!')
else:
print(f'A ({A:02}) is not divisible by 2 or 3)')
for i in range(1, 11):
check_divisible_by_2_and_3(i)
```
## Order Matters
When chaining conditionals, you need to be careful how you order them. For example, what if we wanted te check if a number is divisible by 2, 3, or both:

```
# Let's translate this into Python code
def check_divisible_by_2_and_3(A):
if A % 2 == 0:
print(f"2 divides A ({A:02})!")
elif A % 3 == 0:
print(f'3 divides A ({A:02})!')
elif A % 2 == 0 and A % 3 == 0:
print(f'2 and 3 divides A ({A:02})!')
else:
print(f"2 or 3 doesn't divide A ({A:02})")
for i in range(1, 11):
check_divisible_by_2_and_3(i)
```
Wait! we would expect that 6, which is divisible by both 2 and 3 to show that! Looking back at the graphic, we can see that the flow is checking for 2 first, and since that's true we follow that path first. Let's make a correction to our diagram to fix this:

```
# Let's translate this into Python code
def check_divisible_by_2_and_3(A):
if A % 2 == 0 and A % 3 == 0:
print(f'2 and 3 divides A ({A:02})!')
elif A % 3 == 0:
print(f'3 divides A ({A:02})!')
elif A % 2 == 0:
print(f"2 divides A ({A:02})!")
else:
print(f"2 or 3 doesn't divide A ({A:02})")
for i in range(1, 11):
check_divisible_by_2_and_3(i)
```
**NOTE:** Always put your most restrictive conditional at the top of your if statements and then work your way down to the least restrictive.

## In-Class Assignments
- Create a funcition that takes two inputs variables `A` and `divisor`. Check if `divisor` divides into `A`. If it does, print `"<value of A> is divided by <value of divisor>"`. Don't forget about the `in` operator that checks if a substring is in another string.
- Create a function that takes an input variable `A` which is a string. Check if `A` has the substring `apple`, `peach`, or `blueberry` in it. Print out which of these are found within the string. Note: you could do this using just if/elif/else statements, but is there a better way using lists, for loops, and if/elif/else statements?
## Solutions
```
def is_divisible(A, divisor):
if A % divisor == 0:
print(f'{A} is divided by {divisor}')
A = 37
# this is actually a crude way to find if the number is prime
for i in range(2, int(A / 2)):
is_divisible(A, i)
# notice that nothing was printed? That's because 37 is prime
B = 27
for i in range(2, int(B / 2)):
is_divisible(B, i)
# this is ONE solution. There are more out there and probably better
# one too
def check_for_fruit(A):
found_fruit = []
if 'apple' in A:
found_fruit.append('apple')
if 'peach' in A:
found_fruit.append('peach')
if 'blueberry' in A:
found_fruit.append('blueberry')
found_fruit_str = ''
for fruit in found_fruit:
found_fruit_str += fruit
found_fruit_str += ', '
if len(found_fruit) > 0:
print(found_fruit_str + ' is found within the string')
else:
print('No fruit found in the string')
check_for_fruit('there are apples and peaches in this pie')
```
| github_jupyter |
# Geolocalizacion de dataset de escuelas argentinas
```
#Importar librerias
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
```
### Preparacion de data
```
# Vamos a cargar un padron de escuelas de Argentina
# Estos son los nombres de columna
cols = ['Jurisdicción','CUE Anexo','Nombre','Sector','Estado','Ámbito','Domicilio','CP','Teléfono','Código Localidad','Localidad','Departamento','E-mail','Ed. Común','Ed. Especial','Ed. de Jóvenes y Adultos','Ed. Artística','Ed. Hospitalaria Domiciliaria','Ed. Intercultural Bilingüe','Ed. Contexto de Encierro','Jardín maternal','Jardín de infantes','Primaria','Secundaria','Secundaria Técnica (INET)','Superior no Universitario','Superior No Universitario (INET)']
# Leer csv, remplazar las 'X' con True y los '' (NaN) con False
escuelas = pd.read_csv('../../datos/escuelas_arg.csv', names=cols).fillna(False).replace('X', True)
# Construir la columna 'dpto_link' con los codigos indetificatorios de partidos como los que teniamos
escuelas['dpto_link'] = escuelas['C\xc3\xb3digo Localidad'].astype(str).str.zfill(8).str[:5]
# Tenemos los radios censales del AMBA, que creamos en el notebook anterior. Creemos los 'dpto_link' del AMBA.
radios_censales_AMBA = pd.read_csv('../../datos/AMBA_datos', dtype=object)
dpto_links_AMBA = (radios_censales_AMBA['prov'] + radios_censales_AMBA['depto']).unique()
# Filtramos las escuelas AMBA
escuelas_AMBA = escuelas.loc[escuelas['dpto_link'].isin(dpto_links_AMBA)]
escuelas_AMBA = pd.concat([escuelas_AMBA, escuelas.loc[escuelas['Jurisdicci\xc3\xb3n'] == 'Ciudad de Buenos Aires']])
# Filtramos secundaria estatal
escuelas_AMBA_secundaria_estatal = escuelas_AMBA.loc[escuelas_AMBA['Secundaria'] & (escuelas_AMBA[u'Sector'] == 'Estatal')]
escuelas_AMBA_secundaria_estatal.reset_index(inplace=True, drop=True)
```
### Columnas de 'Address'
```
# Creamos un campo que llamamos 'Address', uniendo domicilio, localidad, departamento, jurisdiccion, y ', Argentina'
escuelas_AMBA_secundaria_estatal['Address'] = \
escuelas_AMBA_secundaria_estatal['Domicilio'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Localidad'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Departamento'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Jurisdicci\xc3\xb3n'].astype(str) +', Argentina'
pd.set_option('display.max_colwidth', -1)
import re
def filtrar_entre_calles(string):
"""
Removes substring between 'E/' and next field (delimited by ','). Case insensitive.
example:
>>> out = filtrar_entre_calles('LASCANO E/ ROMA E ISLAS MALVINAS 6213, ISIDRO CASANOVA')
>>> print out
'LASCANO 6213, ISIDRO CASANOVA'
"""
s = string.lower()
try:
m = re.search("\d", s)
start = s.index( 'e/' )
# end = s.index( last, start )
end = m.start()
return string[:start] + string[end:]
except:
return string
def filtrar_barrio(string, n = 3):
"""
Leaves only n most aggregate fields and the address.
example:
>>> out = filtrar_entre_calles('LASCANO 6213, ISIDRO CASANOVA, LA MATANZA, Buenos Aires, Argentina')
>>> print out
'LASCANO 6213, LA MATANZA, Buenos Aires, Argentina'
"""
try:
coma_partido_jurisdiccion = [m.start() for m in re.finditer(',', string)][-n]
coma_direccion = [m.start() for m in re.finditer(',', string)][0]
s = string[:coma_direccion][::-1]
if "n/s" in s.lower():
start = s.lower().index('n/s')
cut = len(s) - len('n/s') - start
else:
m = re.search("\d", s)
cut = len(s) - m.start(0)
return string[:cut] + string[coma_partido_jurisdiccion:]
except AttributeError:
return string
escuelas_AMBA_secundaria_estatal['Address_2'] = escuelas_AMBA_secundaria_estatal['Address'].apply(filtrar_entre_calles)
escuelas_AMBA_secundaria_estatal['Address_3'] = escuelas_AMBA_secundaria_estatal['Address_2'].apply(filtrar_barrio)
escuelas_AMBA_secundaria_estatal.to_csv('../../datos/escuelas_AMBA_secundaria_estatal.csv', index = False)
```
### Geolocalizacion
```
import json
import time
import urllib
import urllib2
def geolocate(inp, API_key = None, BACKOFF_TIME = 30):
# See https://developers.google.com/maps/documentation/timezone/get-api-key
# with open('googleMapsAPIkey.txt', 'r') as myfile:
# maps_key = myfile.read().replace('\n', '')
base_url = 'https://maps.googleapis.com/maps/api/geocode/json'
# This joins the parts of the URL together into one string.
url = base_url + '?' + urllib.urlencode({
'address': "%s" % (inp),
'key': API_key,
})
try:
# Get the API response.
response = str(urllib2.urlopen(url).read())
except IOError:
pass # Fall through to the retry loop.
else:
# If we didn't get an IOError then parse the result.
result = json.loads(response.replace('\\n', ''))
if result['status'] == 'OK':
return result['results'][0]
elif result['status'] != 'UNKNOWN_ERROR':
# Many API errors cannot be fixed by a retry, e.g. INVALID_REQUEST or
# ZERO_RESULTS. There is no point retrying these requests.
# raise Exception(result['error_message'])
return None
# If we're over the API limit, backoff for a while and try again later.
elif result['status'] == 'OVER_QUERY_LIMIT':
print "Hit Query Limit! Backing off for "+str(BACKOFF_TIME)+" minutes..."
time.sleep(BACKOFF_TIME * 60) # sleep for 30 minutes
geocoded = False
def set_geolocation_values(df, loc):
df.set_value(i,'lng', loc['geometry']['location']['lng'])
df.set_value(i,'lat', loc['geometry']['location']['lat'])
df.set_value(i, 'id', loc['place_id'])
dataframe = escuelas_AMBA_secundaria_estatal
col, col_2, col_3 = 'Address', 'Address_2', 'Address_3'
API_key = 'AIzaSyDjBFMZlNTyds2Sfihu2D5LTKupKDBpf6c'
for i, row in dataframe.iterrows():
loc = geolocate(row[col], API_key)
if loc:
set_geolocation_values(dataframe, loc)
else:
loc = geolocate(row[col_2], API_key)
if loc:
set_geolocation_values(dataframe, loc)
else:
loc = geolocate(row[col_3], API_key)
if loc:
set_geolocation_values(dataframe, loc)
if i%50 == 0:
print 'processed row '+str(i)
dataframe.to_csv('../../datos/esc_sec_AMBA_geoloc.csv', index = False, encoding = 'utf8')
# esc_sec_AMBA_geoloc_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_1200.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc_480_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_480_1200.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc = pd.read_csv('../../datos/esc_sec_AMBA_geoloc.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc_900_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_900_1200.csv', encoding = 'utf8')
# pd.concat([esc_sec_AMBA_geoloc[:480],esc_sec_AMBA_geoloc_480_1200[:420],esc_sec_AMBA_geoloc_900_1200, esc_sec_AMBA_geoloc_1200]).to_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', index = False, encoding = 'utf8')
print len(pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8').dropna())
print len(pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8'))
1840/2066.
import numpy as np
df = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8')
index = df['lat'].index[df['lat'].apply(np.isnan)]
plt.hist(index, 100)
# plt.xlim(900, 1300)
plt.show()
df.iloc[np.where(pd.isnull(df['lat']))][['Nombre','Address', 'Address_2', 'Address_3']].to_csv('../../datos/no_result_addresses.csv', index = False, encoding = 'utf8')
```
| github_jupyter |
# BERT finetuning on AG_news-4
## Librairy
```
# !pip install transformers==4.8.2
# !pip install datasets==1.7.0
import os
import time
import pickle
import numpy as np
import torch
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, recall_score, precision_score, f1_score
from transformers import BertTokenizer, BertTokenizerFast
from transformers import BertForSequenceClassification, AdamW
from transformers import Trainer, TrainingArguments
from transformers import EarlyStoppingCallback
from transformers.data.data_collator import DataCollatorWithPadding
from datasets import load_dataset, Dataset, concatenate_datasets
# print(torch.__version__)
# print(torch.cuda.device_count())
# print(torch.cuda.is_available())
# print(torch.cuda.get_device_name(0))
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
# if torch.cuda.is_available():
# torch.set_default_tensor_type('torch.cuda.FloatTensor')
device
```
## Global variables
```
BATCH_SIZE = 24
NB_EPOCHS = 4
RESULTS_FILE = '~/Results/BERT_finetune/ag_news-4_BERT_finetune_b'+str(BATCH_SIZE)+'_results.pkl'
RESULTS_PATH = '~/Results/BERT_finetune/ag_news-4_b'+str(BATCH_SIZE)+'/'
CACHE_DIR = '~/Data/huggignface/' # path of your folder
```
## Dataset
```
# download dataset
raw_datasets = load_dataset('ag_news', cache_dir=CACHE_DIR)
# tokenize
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
def tokenize_function(examples):
return tokenizer(examples["text"], padding=True, truncation=True)
tokenized_datasets = raw_datasets.map(tokenize_function, batched=True)
tokenized_datasets.set_format(type='torch', columns=['input_ids', 'attention_mask', 'label'])
train_dataset = tokenized_datasets["train"].shuffle(seed=42)
train_val_datasets = train_dataset.train_test_split(train_size=0.8)
train_dataset = train_val_datasets['train'].rename_column('label', 'labels')
val_dataset = train_val_datasets['test'].rename_column('label', 'labels')
test_dataset = tokenized_datasets["test"].shuffle(seed=42).rename_column('label', 'labels')
# get number of labels
num_labels = len(set(train_dataset['labels'].tolist()))
num_labels
```
## Model
#### Model
```
model = BertForSequenceClassification.from_pretrained("bert-base-uncased", num_labels=num_labels)
model.to(device)
```
#### Training
```
training_args = TrainingArguments(
# output
output_dir=RESULTS_PATH,
# params
num_train_epochs=NB_EPOCHS, # nb of epochs
per_device_train_batch_size=BATCH_SIZE, # batch size per device during training
per_device_eval_batch_size=BATCH_SIZE, # cf. paper Sun et al.
learning_rate=2e-5, # cf. paper Sun et al.
# warmup_steps=500, # number of warmup steps for learning rate scheduler
warmup_ratio=0.1, # cf. paper Sun et al.
weight_decay=0.01, # strength of weight decay
# # eval
evaluation_strategy="steps",
eval_steps=50,
# evaluation_strategy='no', # no more evaluation, takes time
# log
logging_dir=RESULTS_PATH+'logs',
logging_strategy='steps',
logging_steps=50,
# save
# save_strategy='epoch',
# save_strategy='steps',
# load_best_model_at_end=False
load_best_model_at_end=True # cf. paper Sun et al.
)
def compute_metrics(p):
pred, labels = p
pred = np.argmax(pred, axis=1)
accuracy = accuracy_score(y_true=labels, y_pred=pred)
return {"val_accuracy": accuracy}
trainer = Trainer(
model=model,
args=training_args,
tokenizer=tokenizer,
train_dataset=train_dataset,
eval_dataset=val_dataset,
# compute_metrics=compute_metrics,
# callbacks=[EarlyStoppingCallback(early_stopping_patience=5)]
)
results = trainer.train()
training_time = results.metrics["train_runtime"]
training_time_per_epoch = training_time / training_args.num_train_epochs
training_time_per_epoch
trainer.save_model(os.path.join(RESULTS_PATH, 'best_model-0'))
```
## Results
```
results_d = {}
epoch = 1
ordered_files = sorted( [f for f in os.listdir(RESULTS_PATH)
if (not f.endswith("logs")) and (f.startswith("best")) # best model eval only
],
key=lambda x: int(x.split('-')[1]) )
for filename in ordered_files:
print(filename)
# load model
model_file = os.path.join(RESULTS_PATH, filename)
finetuned_model = BertForSequenceClassification.from_pretrained(model_file, num_labels=num_labels)
finetuned_model.to(device)
finetuned_model.eval()
# compute test acc
test_trainer = Trainer(finetuned_model, data_collator=DataCollatorWithPadding(tokenizer))
raw_preds, labels, _ = test_trainer.predict(test_dataset)
preds = np.argmax(raw_preds, axis=1)
test_acc = accuracy_score(y_true=labels, y_pred=preds)
# results_d[filename] = (test_acc, training_time_per_epoch*epoch)
results_d[filename] = test_acc # best model evaluation only
print((test_acc, training_time_per_epoch*epoch))
epoch += 1
results_d['training_time'] = training_time
# save results
with open(RESULTS_FILE, 'wb') as fh:
pickle.dump(results_d, fh)
# load results
with open(RESULTS_FILE, 'rb') as fh:
results_d = pickle.load(fh)
results_d
```
| github_jupyter |
This tutorial shows how to generate an image of handwritten digits using Deep Convolutional Generative Adversarial Network (DCGAN).
Generative Adversarial Networks (GANs) are one of the most interesting fields in machine learning. The standard GAN consists of two models, a generative and a discriminator one. Two models are trained simultaneously by an adversarial process. A generative model (`the artist`) learns to generate images that look real, while the discriminator (`the art critic`) one learns to tell real images apart from the fakes.
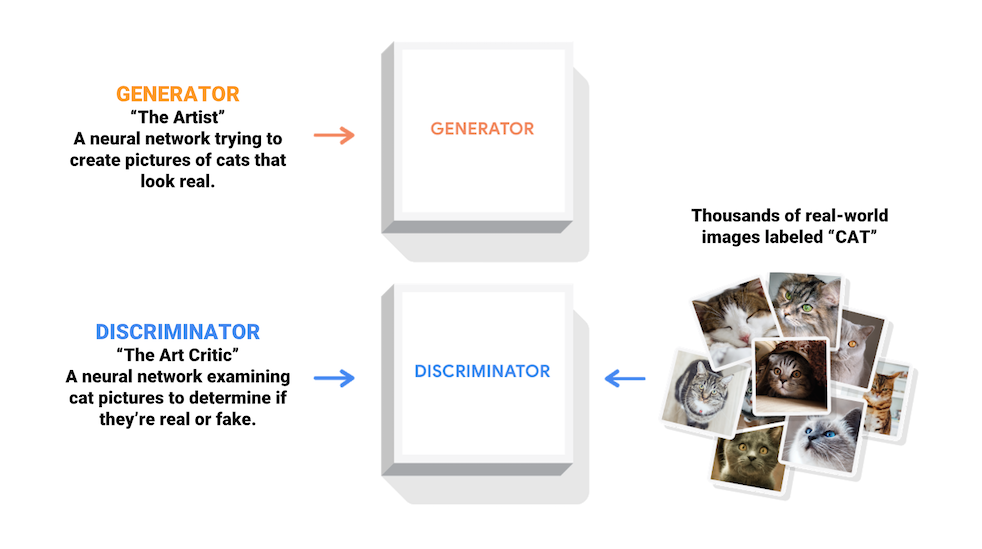
Refer to Tensorflow.org (2020).
During training, the generative model becomes progressively creating images that look real, and the discriminator model becomes progressively telling them apart. The whole process reaches equilibrium when the discriminator is no longer able to distinguish real images from fakes.
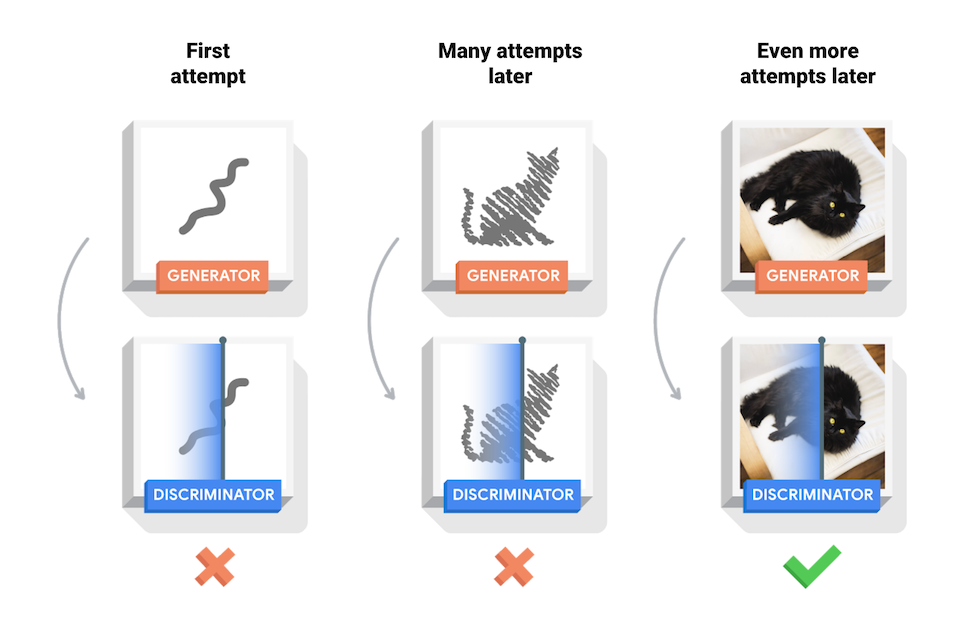
Refer to Tensorflow.org (2020).
In this demo, we show how to train a GAN model on MNIST and FASHION MNIST dataset.
```
!pip uninstall -y tensorflow
!pip install -q tf-nightly tfds-nightly
import glob
import tensorflow as tf
import tensorflow_datasets as tfds
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Conv2D, Conv2DTranspose, Dense, Flatten, BatchNormalization, ELU, LeakyReLU, Reshape, Dropout
import numpy as np
import IPython.display as display
from IPython.display import clear_output
import os
import time
import imageio
tfds.disable_progress_bar()
print("Tensorflow Version: {}".format(tf.__version__))
print("GPU {} available.".format("is" if tf.config.experimental.list_physical_devices("GPU") else "not"))
```
# Data Preprocessing
```
def normalize(image):
img = image['image']
img = (tf.cast(img, tf.float32) - 127.5) / 127.5
return img
```
## MNIST Dataset
```
raw_datasets, metadata = tfds.load(name="mnist", with_info=True)
raw_train_datasets, raw_test_datasets = raw_datasets['train'], raw_datasets['test']
raw_test_datasets, metadata
BUFFER_SIZE = 10000
BATCH_SIZE = 256
train_datasets = raw_train_datasets.map(normalize).cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE)
test_datasets = raw_test_datasets.map(normalize).batch(BATCH_SIZE)
for imgs in train_datasets.take(1):
img = imgs[0]
plt.imshow(tf.keras.preprocessing.image.array_to_img(img))
plt.axis("off")
plt.show()
```
## Fashion_MNIST Dataset
```
raw_datasets, metadata = tfds.load(name="fashion_mnist", with_info=True)
raw_train_datasets, raw_test_datasets = raw_datasets['train'], raw_datasets['test']
raw_train_datasets
for image in raw_train_datasets.take(1):
plt.imshow(tf.keras.preprocessing.image.array_to_img(image['image']))
plt.axis("off")
plt.title("Label: {}".format(image['label']))
plt.show()
BUFFER_SIZE = 10000
BATCH_SIZE = 256
train_datasets = raw_train_datasets.map(normalize).cache().prefetch(BUFFER_SIZE).batch(BATCH_SIZE)
test_datasets = raw_test_datasets.map(normalize).batch(BATCH_SIZE)
for imgs in train_datasets.take(1):
img = imgs[0]
plt.imshow(tf.keras.preprocessing.image.array_to_img(img))
plt.axis("off")
plt.show()
```
# Build the GAN Model
## The Generator
The generator uses the `tf.keras.layers.Conv2DTranspose` (upsampling) layer to produce an image from a seed input (a random noise). Start from this seed input, upsample it several times to reach the desired output (28x28x1).
```
def build_generator_model():
model = tf.keras.Sequential()
model.add(Dense(units=7 * 7 * 256, use_bias=False, input_shape=(100,)))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(Reshape(target_shape=[7,7,256]))
assert model.output_shape == (None, 7, 7, 256)
model.add(Conv2DTranspose(filters=128, kernel_size=(5,5), strides=(1,1), padding="same", use_bias=False))
model.add(BatchNormalization())
model.add(LeakyReLU())
assert model.output_shape == (None, 7, 7, 128)
model.add(Conv2DTranspose(filters=64, kernel_size=(5,5), strides=(2,2), padding='same', use_bias=False))
model.add(BatchNormalization())
model.add(LeakyReLU())
assert model.output_shape == (None, 14, 14, 64)
model.add(Conv2DTranspose(filters=1, kernel_size=(5,5), strides=(2,2), padding='same', use_bias=False,
activation="tanh"))
assert model.output_shape == (None, 28, 28, 1)
return model
generator = build_generator_model()
generator_input = tf.random.normal(shape=[1, 100])
generator_outputs = generator(generator_input, training=False)
plt.imshow(generator_outputs[0, :, :, 0], cmap='gray')
plt.show()
```
## The Discriminator
The discriminator is basically a CNN network.
```
def build_discriminator_model():
model = tf.keras.Sequential()
# [None, 28, 28, 64]
model.add(Conv2D(filters=64, kernel_size=(5,5), strides=(1,1), padding="same",
input_shape=[28,28,1]))
model.add(LeakyReLU())
model.add(Dropout(rate=0.3))
# [None, 14, 14, 128]
model.add(Conv2D(filters=128, kernel_size=(3,3), strides=(2,2), padding='same'))
model.add(LeakyReLU())
model.add(Dropout(rate=0.3))
model.add(Flatten())
model.add(Dense(units=1))
return model
```
The output of the discriminator was trained that the negative values are for the fake images and the positive values are for real ones.
```
discriminator = build_discriminator_model()
discriminator_outputs = discriminator(generator_outputs)
discriminator_outputs
```
# Define the losses and optimizers
Define the loss functions and the optimizers for both models.
```
# define the cross entropy as the helper function
cross_entropy = tf.keras.losses.BinaryCrossentropy(from_logits=True)
```
## Discriminator Loss
The discriminator's loss quantifies how well the discriminator can tell the real images from fakes. It compares the discriminator's predictions on real images to an array of 1s, and the discriminator's predictions on fake images to an array of 0s.
```
def discriminator_loss(real_output, fake_output):
real_loss = cross_entropy(tf.ones_like(real_output), real_output)
fake_loss = cross_entropy(tf.zeros_like(fake_output), fake_output)
total_loss = real_loss + fake_loss
return total_loss
```
## Generator Loss
The generator's loss quantifies how well the generator model can trick the discriminator model. If the generator performs well, the discriminator will classify the fake images as real (or 1). Here, we will compare the discriminator decisions on the generated images to an array of 1s.
```
def generator_loss(fake_output):
# the generator learns to make the discriminator predictions became real
# (or an array of 1s) on the fake images
return cross_entropy(tf.ones_like(fake_output), fake_output)
```
## Define optimizers.
```
generator_optimizer = tf.keras.optimizers.Adam(learning_rate=1e-4)
discriminator_optimizer = tf.keras.optimizers.Adam(learning_rate=1e-4)
```
## Save Checkpoints
```
ckpt_dir = "./gan_ckpt"
ckpt_prefix = os.path.join(ckpt_dir, "ckpt")
ckpt = tf.train.Checkpoint(generator_optimizer=generator_optimizer,
discriminator_optimizer=discriminator_optimizer,
generator=generator,
discriminator=discriminator)
ckpt
```
# Define the training loop
```
EPOCHS = 50
noise_dim = 100
num_generated_examples = 16
# You will reuse the seed overtime to visualize progress in the animated GIF.
seed = tf.random.normal(shape=[num_generated_examples, noise_dim])
```
In the training loop, the generator model takes the noise as the input to generate the fake images. The discriminator model takes real images and fake images to give the discriminations (or outputs) for them. Calculate the generator and discriminator losses each using the real outputs and the fake outputs. Calculate the gradients of the model trainable variables based on these losses and then apply gradients back to them.
```
@tf.function
def train_step(images):
fake_noises = tf.random.normal(shape=[BATCH_SIZE, noise_dim])
with tf.GradientTape() as disc_tape, tf.GradientTape() as gen_tape:
fake_images = generator(fake_noises, training=True)
fake_outputs = discriminator(fake_images, training=True)
real_outputs = discriminator(images, training=True)
disc_loss = discriminator_loss(real_output=real_outputs,
fake_output=fake_outputs)
gen_loss = generator_loss(fake_output=fake_outputs)
disc_gradients = disc_tape.gradient(disc_loss, discriminator.trainable_variables)
gen_gradients = gen_tape.gradient(gen_loss, generator.trainable_variables)
discriminator_optimizer.apply_gradients(zip(disc_gradients, discriminator.trainable_variables))
generator_optimizer.apply_gradients(zip(gen_gradients, generator.trainable_variables))
def generate_and_save_images(model, epoch, test_input):
"""Helps to generate the images from a fixed seed."""
predictions = model(test_input, training=False)
fig = plt.figure(figsize=(8,8))
for i in range(predictions.shape[0]):
plt.subplot(4, 4, i+1)
plt.imshow(predictions[i, :, :, 0] * 127.5 + 127.5, cmap='gray')
plt.axis("off")
plt.savefig('image_epoch_{:04d}.png'.format(epoch))
plt.show()
def train(dataset, epochs):
for epoch in range(epochs):
start = time.time()
for batch_dataset in dataset:
train_step(batch_dataset)
clear_output(wait=True)
generate_and_save_images(generator, epoch+1, seed)
if (epoch+1) % 15 == 0:
ckpt.save(file_prefix=ckpt_prefix)
print("Epoch {} in time {}.".format(epoch + 1, time.time()-start))
# after the training
clear_output(wait=True)
generate_and_save_images(generator, epoch+1, seed)
```
## Train the Model
Call the `train()` function to start the model training. Note, training GANs can be tricky. It's important that the generator and discriminator do not overpower each other (e.g. they train at a similar rate).
```
train(train_datasets, epochs=EPOCHS)
```
# Create a GIF
```
def display_image(epoch_no):
image_path = 'image_epoch_{:04d}.png'.format(epoch_no)
img = plt.imread(fname=image_path)
plt.imshow(img)
plt.margins(0)
plt.axis("off")
plt.tight_layout()
plt.show()
display_image(50)
anim_file = 'dcgan.gif'
with imageio.get_writer(anim_file, mode="I") as writer:
filenames = glob.glob('image*.png')
filenames = sorted(filenames)
for _, filename in enumerate(filenames):
image = imageio.imread(filename)
writer.append_data(image)
try:
from google.colab import files
except ImportError:
pass
else:
files.download(anim_file)
```
| github_jupyter |
## Setup
If you are running this generator locally(i.e. in a jupyter notebook in conda, just make sure you installed:
- RDKit
- DeepChem 2.5.0 & above
- Tensorflow 2.4.0 & above
Then, please skip the following part and continue from `Data Preparations`.
To increase efficiency, we recommend running this molecule generator in Colab.
Then, we'll first need to run the following lines of code, these will download conda with the deepchem environment in colab.
```
#!curl -Lo conda_installer.py https://raw.githubusercontent.com/deepchem/deepchem/master/scripts/colab_install.py
#import conda_installer
#conda_installer.install()
#!/root/miniconda/bin/conda info -e
#!pip install --pre deepchem
#import deepchem
#deepchem.__version__
```
## Data Preparations
Now we are ready to import some useful functions/packages, along with our model.
### Import Data
```
import model##our model
from rdkit import Chem
from rdkit.Chem import AllChem
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import deepchem as dc
```
Then, we are ready to import our dataset for training.
Here, for demonstration, we'll be using this dataset of in-vitro assay that detects inhibition of SARS-CoV 3CL protease via fluorescence.
The dataset is originally from [PubChem AID1706](https://pubchem.ncbi.nlm.nih.gov/bioassay/1706), previously handled by [JClinic AIcure](https://www.aicures.mit.edu/) team at MIT into this [binarized label form](https://github.com/yangkevin2/coronavirus_data/blob/master/data/AID1706_binarized_sars.csv).
```
df = pd.read_csv('AID1706_binarized_sars.csv')
```
Observe the data above, it contains a 'smiles' column, which stands for the smiles representation of the molecules. There is also an 'activity' column, in which it is the label specifying whether that molecule is considered as hit for the protein.
Here, we only need those 405 molecules considered as hits, and we'll be extracting features from them to generate new molecules that may as well be hits.
```
true = df[df['activity']==1]
```
### Set Minimum Length for molecules
Since we'll be using graphic neural network, it might be more helpful and efficient if our graph data are of the same size, thus, we'll eliminate the molecules from the training set that are shorter(i.e. lacking enough atoms) than our desired minimum size.
```
num_atoms = 6 #here the minimum length of molecules is 6
input_df = true['smiles']
df_length = []
for _ in input_df:
df_length.append(Chem.MolFromSmiles(_).GetNumAtoms() )
true['length'] = df_length #create a new column containing each molecule's length
true = true[true['length']>num_atoms] #Here we leave only the ones longer than 6
input_df = true['smiles']
input_df_smiles = input_df.apply(Chem.MolFromSmiles) #convert the smiles representations into rdkit molecules
```
Now, we are ready to apply the `featurizer` function to our molecules to convert them into graphs with nodes and edges for training.
```
#input_df = input_df.apply(Chem.MolFromSmiles)
train_set = input_df_smiles.apply( lambda x: model.featurizer(x,max_length = num_atoms))
train_set
```
We'll take one more step to make the train_set into separate nodes and edges, which fits the format later to supply to the model for training
```
nodes_train, edges_train = list(zip(*train_set) )
```
## Training
Now, we're finally ready for generating new molecules. We'll first import some necessay functions from tensorflow.
```
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
```
The network here we'll be using is Generative Adversarial Network, as mentioned in the project introduction. Here's a great [introduction](https://machinelearningmastery.com/what-are-generative-adversarial-networks-gans/).
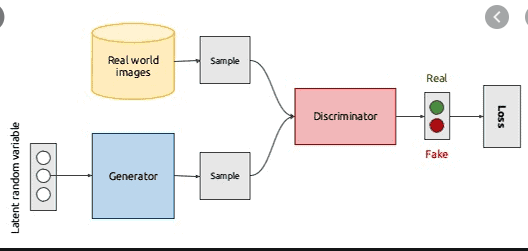
Here we'll first initiate a discriminator and a generator model with the corresponding functions in the package.
```
disc = model.make_discriminator(num_atoms)
gene = model.make_generator(num_atoms, noise_input_shape = 100)
```
Then, with the `train_batch` function, we'll supply the necessary inputs and train our network. Upon some experimentations, an epoch of around 160 would be nice for this dataset.
```
generator_trained = model.train_batch(
disc, gene,
np.array(nodes_train), np.array(edges_train),
noise_input_shape = 100, EPOCH = 160, BATCHSIZE = 2,
plot_hist = True, temp_result = False
)
```
There are two possible kind of failures regarding a GAN model: model collapse and failure of convergence. Model collapse would often mean that the generative part of the model wouldn't be able to generate diverse outcomes. Failure of convergence between the generative and the discriminative model could likely way be identified as that the loss for the discriminator has gone to zero or close to zero.
Observe the above generated plot, in the upper plot, the loss of discriminator has not gone to zero/close to zero, indicating that the model has possibily find a balance between the generator and the discriminator. In the lower plot, the accuracy is fluctuating between 1 and 0, indicating possible variability within the data generated.
Therefore, it is reasonable to conclude that within the possible range of epoch and other parameters, the model has successfully avoided the two common types of failures associated with GAN.
## Rewarding Phase
The above `train_batch` function is set to return a trained generator. Thus, we could use that function directly and observe the possible molecules we could get from that function.
```
no, ed = generator_trained(np.random.randint(0,20
, size =(1,100)))#generated nodes and edges
abs(no.numpy()).astype(int).reshape(num_atoms), abs(ed.numpy()).astype(int).reshape(num_atoms,num_atoms)
```
With the `de_featurizer`, we could convert the generated matrix into a smiles molecule and plot it out=)
```
cat, dog = model.de_featurizer(abs(no.numpy()).astype(int).reshape(num_atoms), abs(ed.numpy()).astype(int).reshape(num_atoms,num_atoms))
Chem.MolToSmiles(cat)
Chem.MolFromSmiles(Chem.MolToSmiles(cat))
```
## Brief Result Analysis
```
from rdkit import DataStructs
```
With the rdkit function of comparing similarities, here we'll demonstrate a preliminary analysis of the molecule we've generated. With "CCO" molecule as a control, we could observe that the new molecule we've generated is more similar to a random selected molecule(the fourth molecule) from the initial training set.
This may indicate that our model has indeed extracted some features from our original dataset and generated a new molecule that is relevant.
```
DataStructs.FingerprintSimilarity(Chem.RDKFingerprint(Chem.MolFromSmiles("[Li]NBBC=N")), Chem.RDKFingerprint(Chem.MolFromSmiles("CCO")))# compare with the control
#compare with one from the original data
DataStructs.FingerprintSimilarity(Chem.RDKFingerprint(Chem.MolFromSmiles("[Li]NBBC=N")), Chem.RDKFingerprint(Chem.MolFromSmiles("CCN1C2=NC(=O)N(C(=O)C2=NC(=N1)C3=CC=CC=C3)C")))
```
| github_jupyter |
# Graphs from the presentation
```
import matplotlib.pyplot as plt
%matplotlib notebook
# create a new figure
plt.figure()
# create x and y coordinates via lists
x = [99, 19, 88, 12, 95, 47, 81, 64, 83, 76]
y = [43, 18, 11, 4, 78, 47, 77, 70, 21, 24]
# scatter the points onto the figure
plt.scatter(x, y)
# create a new figure
plt.figure()
# create x and y values via lists
x = [1, 2, 3, 4, 5, 6, 7, 8]
y = [1, 4, 9, 16, 25, 36, 49, 64]
# plot the line
plt.plot(x, y)
# create a new figure
plt.figure()
# create a list of observations
observations = [5.24, 3.82, 3.73, 5.3 , 3.93, 5.32, 6.43, 4.4 , 5.79, 4.05, 5.34, 5.62, 6.02, 6.08, 6.39, 5.03, 5.34, 4.98, 3.84, 4.91, 6.62, 4.66, 5.06, 2.37, 5. , 3.7 , 5.22, 5.86, 3.88, 4.68, 4.88, 5.01, 3.09, 5.38, 4.78, 6.26, 6.29, 5.77, 4.33, 5.96, 4.74, 4.54, 7.99, 5. , 4.85, 5.68, 3.73, 4.42, 4.99, 4.47, 6.06, 5.88, 4.56, 5.37, 6.39, 4.15]
# create a histogram with 15 intervals
plt.hist(observations, bins=15)
# create a new figure
plt.figure()
# plot a red line with a transparancy of 40%. Label this 'line 1'
plt.plot(x, y, color='red', alpha=0.4, label='line 1')
# make a key appear on the plot
plt.legend()
# import pandas
import pandas as pd
# read in data from a csv
data = pd.read_csv('data/weather.csv', parse_dates=['Date'])
# create a new matplotlib figure
plt.figure()
# plot the temperature over time
plt.plot(data['Date'], data['Temp (C)'])
# add a ylabel
plt.ylabel('Temperature (C)')
plt.figure()
# create inputs
x = ['UK', 'France', 'Germany', 'Spain', 'Italy']
y = [67.5, 65.1, 83.5, 46.7, 60.6]
# plot the chart
plt.bar(x, y)
plt.ylabel('Population (M)')
plt.figure()
# create inputs
x = ['UK', 'France', 'Germany', 'Spain', 'Italy']
y = [67.5, 65.1, 83.5, 46.7, 60.6]
# create a list of colours
colour = ['red', 'green', 'blue', 'orange', 'purple']
# plot the chart with the colors and transparancy
plt.bar(x, y, color=colour, alpha=0.5)
plt.ylabel('Population (M)')
plt.figure()
x = [1, 2, 3, 4, 5, 6, 7, 8, 9]
y1 = [2, 4, 6, 8, 10, 12, 14, 16, 18]
y2 = [4, 8, 12, 16, 20, 24, 28, 32, 36]
plt.scatter(x, y1, color='cyan', s=5)
plt.scatter(x, y2, color='violet', s=15)
plt.figure()
x = [1, 2, 3, 4, 5, 6, 7, 8, 9]
y1 = [2, 4, 6, 8, 10, 12, 14, 16, 18]
y2 = [4, 8, 12, 16, 20, 24, 28, 32, 36]
size1 = [10, 20, 30, 40, 50, 60, 70, 80, 90]
size2 = [90, 80, 70, 60, 50, 40, 30, 20, 10]
plt.scatter(x, y1, color='cyan', s=size1)
plt.scatter(x, y2, color='violet', s=size2)
co2_file = '../5. Examples of Visual Analytics in Python/data/national/co2_emissions_tonnes_per_person.csv'
gdp_file = '../5. Examples of Visual Analytics in Python/data/national/gdppercapita_us_inflation_adjusted.csv'
pop_file = '../5. Examples of Visual Analytics in Python/data/national/population.csv'
co2_per_cap = pd.read_csv(co2_file, index_col=0, parse_dates=True)
gdp_per_cap = pd.read_csv(gdp_file, index_col=0, parse_dates=True)
population = pd.read_csv(pop_file, index_col=0, parse_dates=True)
plt.figure()
x = gdp_per_cap.loc['2017'] # gdp in 2017
y = co2_per_cap.loc['2017'] # co2 emmissions in 2017
# population in 2017 will give size of points (divide pop by 1M)
size = population.loc['2017'] / 1e6
# scatter points with vector size and some transparancy
plt.scatter(x, y, s=size, alpha=0.5)
# set a log-scale
plt.xscale('log')
plt.yscale('log')
plt.xlabel('GDP per capita, $US')
plt.ylabel('CO2 emissions per person per year, tonnes')
plt.figure()
# create grid of numbers
grid = [[1, 2, 3],
[4, 5, 6],
[7, 8, 9]]
# plot the grid with 'autumn' color map
plt.imshow(grid, cmap='autumn')
# add a colour key
plt.colorbar()
import pandas as pd
data = pd.read_csv("../5. Examples of Visual Analytics in Python/data/stocks/FTSE_stock_prices.csv", index_col=0)
correlation_matrix = data.pct_change().corr()
# create a new figure
plt.figure()
# imshow the grid of correlation
plt.imshow(correlation_matrix, cmap='terrain')
# add a color bar
plt.colorbar()
# remove cluttering x and y ticks
plt.xticks([])
plt.yticks([])
elevation = pd.read_csv('data/UK_elevation.csv', index_col=0)
# create figure
plt.figure()
# imshow data
plt.imshow(elevation, # grid data
vmin=-50, # minimum for colour bar
vmax=500, # maximum for colour bar
cmap='terrain', # terrain style colour map
extent=[-11, 3, 50, 60]) # [x1, x2, y1, y2] plot boundaries
# add axis labels and a title
plt.xlabel('Longitude')
plt.ylabel('Latitude')
plt.title('UK Elevation Profile')
# add a colourbar
plt.colorbar()
```
| github_jupyter |
# Introduction and Foundations: Titanic Survival Exploration
> Udacity Machine Learning Engineer Nanodegree: _Project 0_
>
> Author: _Ke Zhang_
>
> Submission Date: _2017-04-27_ (Revision 2)
## Abstract
In 1912, the ship RMS Titanic struck an iceberg on its maiden voyage and sank, resulting in the deaths of most of its passengers and crew. In this introductory project, we will explore a subset of the RMS Titanic passenger manifest to determine which features best predict whether someone survived or did not survive. To complete this project, you will need to implement several conditional predictions and answer the questions below. Your project submission will be evaluated based on the completion of the code and your responses to the questions.
## Content
- [Getting Started](#Getting-Started)
- [Making Predictions](#Making-Predictions)
- [Conclusion](#Conclusion)
- [References](#References)
- [Reproduction Environment](#Reproduction-Environment)
# Getting Started
To begin working with the RMS Titanic passenger data, we'll first need to `import` the functionality we need, and load our data into a `pandas` DataFrame.
```
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the dataset
in_file = 'titanic_data.csv'
full_data = pd.read_csv(in_file)
# Print the first few entries of the RMS Titanic data
display(full_data.head())
```
From a sample of the RMS Titanic data, we can see the various features present for each passenger on the ship:
- **Survived**: Outcome of survival (0 = No; 1 = Yes)
- **Pclass**: Socio-economic class (1 = Upper class; 2 = Middle class; 3 = Lower class)
- **Name**: Name of passenger
- **Sex**: Sex of the passenger
- **Age**: Age of the passenger (Some entries contain `NaN`)
- **SibSp**: Number of siblings and spouses of the passenger aboard
- **Parch**: Number of parents and children of the passenger aboard
- **Ticket**: Ticket number of the passenger
- **Fare**: Fare paid by the passenger
- **Cabin** Cabin number of the passenger (Some entries contain `NaN`)
- **Embarked**: Port of embarkation of the passenger (C = Cherbourg; Q = Queenstown; S = Southampton)
Since we're interested in the outcome of survival for each passenger or crew member, we can remove the **Survived** feature from this dataset and store it as its own separate variable `outcomes`. We will use these outcomes as our prediction targets.
Run the code cell below to remove **Survived** as a feature of the dataset and store it in `outcomes`.
```
# Store the 'Survived' feature in a new variable and remove it from the dataset
outcomes = full_data['Survived']
data = full_data.drop('Survived', axis = 1)
# Show the new dataset with 'Survived' removed
display(data.head())
```
The very same sample of the RMS Titanic data now shows the **Survived** feature removed from the DataFrame. Note that `data` (the passenger data) and `outcomes` (the outcomes of survival) are now *paired*. That means for any passenger `data.loc[i]`, they have the survival outcome `outcomes[i]`.
To measure the performance of our predictions, we need a metric to score our predictions against the true outcomes of survival. Since we are interested in how *accurate* our predictions are, we will calculate the proportion of passengers where our prediction of their survival is correct. Run the code cell below to create our `accuracy_score` function and test a prediction on the first five passengers.
**Think:** *Out of the first five passengers, if we predict that all of them survived, what would you expect the accuracy of our predictions to be?*
```
def accuracy_score(truth, pred):
""" Returns accuracy score for input truth and predictions. """
# Ensure that the number of predictions matches number of outcomes
if len(truth) == len(pred):
# Calculate and return the accuracy as a percent
return "Predictions have an accuracy of {:.2f}%.".format((truth == pred).mean()*100)
else:
return "Number of predictions does not match number of outcomes!"
# Test the 'accuracy_score' function
predictions = pd.Series(np.ones(5, dtype = int))
print accuracy_score(outcomes[:5], predictions)
```
> **Tip:** If you save an iPython Notebook, the output from running code blocks will also be saved. However, the state of your workspace will be reset once a new session is started. Make sure that you run all of the code blocks from your previous session to reestablish variables and functions before picking up where you last left off.
# Making Predictions
If we were asked to make a prediction about any passenger aboard the RMS Titanic whom we knew nothing about, then the best prediction we could make would be that they did not survive. This is because we can assume that a majority of the passengers (more than 50%) did not survive the ship sinking.
The `predictions_0` function below will always predict that a passenger did not survive.
```
def predictions_0(data):
""" Model with no features. Always predicts a passenger did not survive. """
predictions = []
for _, passenger in data.iterrows():
# Predict the survival of 'passenger'
predictions.append(0)
# Return our predictions
return pd.Series(predictions)
# Make the predictions
predictions = predictions_0(data)
```
### Question 1
*Using the RMS Titanic data, how accurate would a prediction be that none of the passengers survived?*
**Hint:** Run the code cell below to see the accuracy of this prediction.
```
print accuracy_score(outcomes, predictions)
```
**Answer:** The prediction accuracy is **61.62%**
***
Let's take a look at whether the feature **Sex** has any indication of survival rates among passengers using the `survival_stats` function. This function is defined in the `titanic_visualizations.py` Python script included with this project. The first two parameters passed to the function are the RMS Titanic data and passenger survival outcomes, respectively. The third parameter indicates which feature we want to plot survival statistics across.
Run the code cell below to plot the survival outcomes of passengers based on their sex.
```
vs.survival_stats(data, outcomes, 'Sex')
```
Examining the survival statistics, a large majority of males did not survive the ship sinking. However, a majority of females *did* survive the ship sinking. Let's build on our previous prediction: If a passenger was female, then we will predict that they survived. Otherwise, we will predict the passenger did not survive.
Fill in the missing code below so that the function will make this prediction.
**Hint:** You can access the values of each feature for a passenger like a dictionary. For example, `passenger['Sex']` is the sex of the passenger.
```
def predictions_1(data):
""" Model with one feature:
- Predict a passenger survived if they are female. """
predictions = []
for _, passenger in data.iterrows():
predictions.append(True if passenger['Sex'] == 'female'
else False)
# Return our predictions
return pd.Series(predictions)
# Make the predictions
predictions = predictions_1(data)
```
### Question 2
*How accurate would a prediction be that all female passengers survived and the remaining passengers did not survive?*
**Hint:** Run the code cell below to see the accuracy of this prediction.
```
print accuracy_score(outcomes, predictions)
```
**Answer**: **78.68**%
***
Using just the **Sex** feature for each passenger, we are able to increase the accuracy of our predictions by a significant margin. Now, let's consider using an additional feature to see if we can further improve our predictions. For example, consider all of the male passengers aboard the RMS Titanic: Can we find a subset of those passengers that had a higher rate of survival? Let's start by looking at the **Age** of each male, by again using the `survival_stats` function. This time, we'll use a fourth parameter to filter out the data so that only passengers with the **Sex** 'male' will be included.
Run the code cell below to plot the survival outcomes of male passengers based on their age.
```
vs.survival_stats(data, outcomes, 'Age', ["Sex == 'male'"])
```
Examining the survival statistics, the majority of males younger than 10 survived the ship sinking, whereas most males age 10 or older *did not survive* the ship sinking. Let's continue to build on our previous prediction: If a passenger was female, then we will predict they survive. If a passenger was male and younger than 10, then we will also predict they survive. Otherwise, we will predict they do not survive.
Fill in the missing code below so that the function will make this prediction.
**Hint:** You can start your implementation of this function using the prediction code you wrote earlier from `predictions_1`.
```
def predictions_2(data):
""" Model with two features:
- Predict a passenger survived if they are female.
- Predict a passenger survived if they are male and younger than 10. """
predictions = []
for _, passenger in data.iterrows():
predictions.append(True if passenger['Sex'] == 'female' or
passenger['Age'] < 10 else False)
# Return our predictions
return pd.Series(predictions)
# Make the predictions
predictions = predictions_2(data)
```
### Question 3
*How accurate would a prediction be that all female passengers and all male passengers younger than 10 survived?*
**Hint:** Run the code cell below to see the accuracy of this prediction.
```
print accuracy_score(outcomes, predictions)
```
**Answer**: **79.35**
***
Adding the feature **Age** as a condition in conjunction with **Sex** improves the accuracy by a small margin more than with simply using the feature **Sex** alone. Now it's your turn: Find a series of features and conditions to split the data on to obtain an outcome prediction accuracy of at least 80%. This may require multiple features and multiple levels of conditional statements to succeed. You can use the same feature multiple times with different conditions.
**Pclass**, **Sex**, **Age**, **SibSp**, and **Parch** are some suggested features to try.
Use the `survival_stats` function below to to examine various survival statistics.
**Hint:** To use mulitple filter conditions, put each condition in the list passed as the last argument. Example: `["Sex == 'male'", "Age < 18"]`
```
# survival by Embarked
vs.survival_stats(data, outcomes, 'Embarked')
# survival by Embarked
vs.survival_stats(data, outcomes, 'SibSp')
vs.survival_stats(data, outcomes, 'Age', ["Sex == 'male'", "Age < 18"])
```
We found out earlier that female and children had better chance to survive. In the next step we'll add another criteria 'Pclass' to further distinguish the survival rates among the different groups.
```
# female passengers in the higher pclass had great chance to survive
vs.survival_stats(data, outcomes, 'Pclass', [
"Sex == 'female'"
])
# male passengers in the higher pclass had great chance to survive
vs.survival_stats(data, outcomes, 'Pclass', [
"Sex == 'male'"
])
# more female passengers survived in all age groups
vs.survival_stats(data, outcomes, 'Age', [
"Sex == 'female'",
])
# more male passengers survived only when age < 10
vs.survival_stats(data, outcomes, 'Age', [
"Sex == 'male'",
])
```
It looks like that all female passengers under 20 survived from the accident. Let's check passengers in the lower class to complete our guess.
```
# ... but not in the lower class when they're older than 20
vs.survival_stats(data, outcomes, 'Age', [
"Sex == 'female'",
"Pclass == 3"
])
# ... actually only females under 20 had more survivers in the lower class
vs.survival_stats(data, outcomes, 'Age', [
"Sex == 'male'",
"Pclass == 3"
])
```
> We conclude that in the lower class only female under 20 had better chance to survive. In the other classes all children under 10 and female passengers had more likey survived. Let's check if we have reached our 80% target.
After exploring the survival statistics visualization, fill in the missing code below so that the function will make your prediction.
Make sure to keep track of the various features and conditions you tried before arriving at your final prediction model.
**Hint:** You can start your implementation of this function using the prediction code you wrote earlier from `predictions_2`.
```
def predictions_3(data):
"""
Model with multiple features: Sex, Age and Pclass
Makes a prediction with an accuracy of at least 80%.
"""
predictions = []
for _, passenger in data.iterrows():
if passenger['Age'] < 10:
survived = True
elif passenger['Sex'] == 'female' and not (
passenger['Pclass'] == 3 and passenger['Age'] > 20
):
survived = True
else:
survived = False
predictions.append(survived)
# Return our predictions
return pd.Series(predictions)
# Make the predictions
predictions = predictions_3(data)
```
### Question 4
*Describe the steps you took to implement the final prediction model so that it got an accuracy of at least 80%. What features did you look at? Were certain features more informative than others? Which conditions did you use to split the survival outcomes in the data? How accurate are your predictions?*
**Hint:** Run the code cell below to see the accuracy of your predictions.
```
print accuracy_score(outcomes, predictions)
```
**Answer**:
Using the features *Sex*, *Pclass* and *Age* we increased the accuracy score to **80.36%**.
We tried to plot the survival statistics with different features and chose the ones under which conditions the differences were the largest.
* some features are just not relevant like *PassengerId* or *Name*
* some features have to be decoded to be helpful like *Cabin* which could be helpful if we have more information on the location of each cabin
* some features are less informative than the others: e.g. we could use *Embarked*, *SibSp* or *Parch* to group the passengers but the resulting model would be more complicated.
* Eventually we chose *Sex*, *Pclass* and *Age* as our final features.
We derived the conditions to split the survival outcomes from the survival plots. The split conditions are:
1. All children under 10 => **survived**
2. Female passengers in the upper and middle class, or less than 20 => **survived**
3. Others => **died**
The final accuracy score was **80.36%**.
# Conclusion
After several iterations of exploring and conditioning on the data, you have built a useful algorithm for predicting the survival of each passenger aboard the RMS Titanic. The technique applied in this project is a manual implementation of a simple machine learning model, the *decision tree*. A decision tree splits a set of data into smaller and smaller groups (called *nodes*), by one feature at a time. Each time a subset of the data is split, our predictions become more accurate if each of the resulting subgroups are more homogeneous (contain similar labels) than before. The advantage of having a computer do things for us is that it will be more exhaustive and more precise than our manual exploration above. [This link](http://www.r2d3.us/visual-intro-to-machine-learning-part-1/) provides another introduction into machine learning using a decision tree.
A decision tree is just one of many models that come from *supervised learning*. In supervised learning, we attempt to use features of the data to predict or model things with objective outcome labels. That is to say, each of our data points has a known outcome value, such as a categorical, discrete label like `'Survived'`, or a numerical, continuous value like predicting the price of a house.
### Question 5
*Think of a real-world scenario where supervised learning could be applied. What would be the outcome variable that you are trying to predict? Name two features about the data used in this scenario that might be helpful for making the predictions.*
**Answer**:
A real-world scenario would be that we have a buch of animal photos labeled with the animal type on them and try to recognize new photos with supervised learning model predictions.
Useful featrues could be:
* number of legs
* size of the animal
* color of the skin or fur
* surrounding environment (tropical, water, air, iceberg etc.)
Outcome variable is the animal type.
## References
- [Udacity Website](http://www.udacity.com)
- [Pandas Documentation](http://pandas.pydata.org/pandas-docs/stable/)
## Reproduction Environment
```
import IPython
print IPython.sys_info()
!pip freeze
```
| github_jupyter |
<a href="https://colab.research.google.com/github/darshanbk/100-Days-Of-ML-Code/blob/master/Getting_started_with_BigQuery.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Before you begin
1. Use the [Cloud Resource Manager](https://console.cloud.google.com/cloud-resource-manager) to Create a Cloud Platform project if you do not already have one.
2. [Enable billing](https://support.google.com/cloud/answer/6293499#enable-billing) for the project.
3. [Enable BigQuery](https://console.cloud.google.com/flows/enableapi?apiid=bigquery) APIs for the project.
### Provide your credentials to the runtime
```
from google.colab import auth
auth.authenticate_user()
print('Authenticated')
```
## Optional: Enable data table display
Colab includes the ``google.colab.data_table`` package that can be used to display large pandas dataframes as an interactive data table.
It can be enabled with:
```
%load_ext google.colab.data_table
```
If you would prefer to return to the classic Pandas dataframe display, you can disable this by running:
```python
%unload_ext google.colab.data_table
```
# Use BigQuery via magics
The `google.cloud.bigquery` library also includes a magic command which runs a query and either displays the result or saves it to a variable as a `DataFrame`.
```
# Display query output immediately
%%bigquery --project yourprojectid
SELECT
COUNT(*) as total_rows
FROM `bigquery-public-data.samples.gsod`
# Save output in a variable `df`
%%bigquery --project yourprojectid df
SELECT
COUNT(*) as total_rows
FROM `bigquery-public-data.samples.gsod`
df
```
# Use BigQuery through google-cloud-bigquery
See [BigQuery documentation](https://cloud.google.com/bigquery/docs) and [library reference documentation](https://googlecloudplatform.github.io/google-cloud-python/latest/bigquery/usage.html).
The [GSOD sample table](https://bigquery.cloud.google.com/table/bigquery-public-data:samples.gsod) contains weather information collected by NOAA, such as precipitation amounts and wind speeds from late 1929 to early 2010.
### Declare the Cloud project ID which will be used throughout this notebook
```
project_id = '[your project ID]'
```
### Sample approximately 2000 random rows
```
from google.cloud import bigquery
client = bigquery.Client(project=project_id)
sample_count = 2000
row_count = client.query('''
SELECT
COUNT(*) as total
FROM `bigquery-public-data.samples.gsod`''').to_dataframe().total[0]
df = client.query('''
SELECT
*
FROM
`bigquery-public-data.samples.gsod`
WHERE RAND() < %d/%d
''' % (sample_count, row_count)).to_dataframe()
print('Full dataset has %d rows' % row_count)
```
### Describe the sampled data
```
df.describe()
```
### View the first 10 rows
```
df.head(10)
# 10 highest total_precipitation samples
df.sort_values('total_precipitation', ascending=False).head(10)[['station_number', 'year', 'month', 'day', 'total_precipitation']]
```
# Use BigQuery through pandas-gbq
The `pandas-gbq` library is a community led project by the pandas community. It covers basic functionality, such as writing a DataFrame to BigQuery and running a query, but as a third-party library it may not handle all BigQuery features or use cases.
[Pandas GBQ Documentation](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_gbq.html)
```
import pandas as pd
sample_count = 2000
df = pd.io.gbq.read_gbq('''
SELECT name, SUM(number) as count
FROM `bigquery-public-data.usa_names.usa_1910_2013`
WHERE state = 'TX'
GROUP BY name
ORDER BY count DESC
LIMIT 100
''', project_id=project_id, dialect='standard')
df.head()
```
# Syntax highlighting
`google.colab.syntax` can be used to add syntax highlighting to any Python string literals which are used in a query later.
```
from google.colab import syntax
query = syntax.sql('''
SELECT
COUNT(*) as total_rows
FROM
`bigquery-public-data.samples.gsod`
''')
pd.io.gbq.read_gbq(query, project_id=project_id, dialect='standard')
```
| github_jupyter |
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# 不规则张量
<table class="tfo-notebook-buttons" align="left">
<td> <a target="_blank" href="https://tensorflow.google.cn/guide/ragged_tensor"><img src="https://tensorflow.google.cn/images/tf_logo_32px.png">在 TensorFlow.org 上查看</a>
</td>
<td><a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/zh-cn/guide/ragged_tensor.ipynb"><img src="https://tensorflow.google.cn/images/colab_logo_32px.png">在 Google Colab 中运行 </a></td>
<td><a target="_blank" href="https://github.com/tensorflow/docs-l10n/blob/master/site/zh-cn/guide/ragged_tensor.ipynb"><img src="https://tensorflow.google.cn/images/GitHub-Mark-32px.png">在 Github 上查看源代码</a></td>
<td><a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/ragged_tensor.ipynb">{img}下载笔记本</a></td>
</table>
**API 文档:** [`tf.RaggedTensor`](https://tensorflow.google.cn/api_docs/python/tf/RaggedTensor) [`tf.ragged`](https://tensorflow.google.cn/api_docs/python/tf/ragged)
## 设置
```
!pip install -q tf_nightly
import math
import tensorflow as tf
```
## 概述
数据有多种形状;张量也应当有多种形状。*不规则张量*是嵌套的可变长度列表的 TensorFlow 等效项。它们使存储和处理包含非均匀形状的数据变得容易,包括:
- 可变长度特征,例如电影的演员名单。
- 成批的可变长度顺序输入,例如句子或视频剪辑。
- 分层输入,例如细分为节、段落、句子和单词的文本文档。
- 结构化输入中的各个字段,例如协议缓冲区。
### 不规则张量的功能
有一百多种 TensorFlow 运算支持不规则张量,包括数学运算(如 `tf.add` 和 `tf.reduce_mean`)、数组运算(如 `tf.concat` 和 `tf.tile`)、字符串操作运算(如 `tf.substr`)、控制流运算(如 `tf.while_loop` 和 `tf.map_fn`)等:
```
digits = tf.ragged.constant([[3, 1, 4, 1], [], [5, 9, 2], [6], []])
words = tf.ragged.constant([["So", "long"], ["thanks", "for", "all", "the", "fish"]])
print(tf.add(digits, 3))
print(tf.reduce_mean(digits, axis=1))
print(tf.concat([digits, [[5, 3]]], axis=0))
print(tf.tile(digits, [1, 2]))
print(tf.strings.substr(words, 0, 2))
print(tf.map_fn(tf.math.square, digits))
```
还有专门针对不规则张量的方法和运算,包括工厂方法、转换方法和值映射运算。有关支持的运算列表,请参阅 **`tf.ragged` 包文档**。
许多 TensorFlow API 都支持不规则张量,包括 [Keras](https://tensorflow.google.cn/guide/keras)、[Dataset](https://tensorflow.google.cn/guide/data)、[tf.function](https://tensorflow.google.cn/guide/function)、[SavedModel](https://tensorflow.google.cn/guide/saved_model) 和 [tf.Example](https://tensorflow.google.cn/tutorials/load_data/tfrecord)。有关更多信息,请参阅下面的 **TensorFlow API** 一节。
与普通张量一样,您可以使用 Python 风格的索引来访问不规则张量的特定切片。有关更多信息,请参阅下面的**索引**一节。
```
print(digits[0]) # First row
print(digits[:, :2]) # First two values in each row.
print(digits[:, -2:]) # Last two values in each row.
```
与普通张量一样,您可以使用 Python 算术和比较运算符来执行逐元素运算。有关更多信息,请参阅下面的**重载运算符**一节。
```
print(digits + 3)
print(digits + tf.ragged.constant([[1, 2, 3, 4], [], [5, 6, 7], [8], []]))
```
如果需要对 `RaggedTensor` 的值进行逐元素转换,您可以使用 `tf.ragged.map_flat_values`(它采用一个函数加上一个或多个参数的形式),并应用这个函数来转换 `RaggedTensor` 的值。
```
times_two_plus_one = lambda x: x * 2 + 1
print(tf.ragged.map_flat_values(times_two_plus_one, digits))
```
不规则张量可以转换为嵌套的 Python `list` 和 numpy `array`:
```
digits.to_list()
digits.numpy()
```
### 构造不规则张量
构造不规则张量的最简单方法是使用 `tf.ragged.constant`,它会构建与给定的嵌套 Python `list` 或 numpy `array` 相对应的 `RaggedTensor`:
```
sentences = tf.ragged.constant([
["Let's", "build", "some", "ragged", "tensors", "!"],
["We", "can", "use", "tf.ragged.constant", "."]])
print(sentences)
paragraphs = tf.ragged.constant([
[['I', 'have', 'a', 'cat'], ['His', 'name', 'is', 'Mat']],
[['Do', 'you', 'want', 'to', 'come', 'visit'], ["I'm", 'free', 'tomorrow']],
])
print(paragraphs)
```
还可以通过将扁平的*值*张量与*行分区*张量进行配对来构造不规则张量,行分区张量使用 `tf.RaggedTensor.from_value_rowids`、`tf.RaggedTensor.from_row_lengths` 和 `tf.RaggedTensor.from_row_splits` 等工厂类方法指示如何将值分成各行。
#### `tf.RaggedTensor.from_value_rowids`
如果知道每个值属于哪一行,可以使用 `value_rowids` 行分区张量构建 `RaggedTensor`:
```
print(tf.RaggedTensor.from_value_rowids(
values=[3, 1, 4, 1, 5, 9, 2],
value_rowids=[0, 0, 0, 0, 2, 2, 3]))
```
#### `tf.RaggedTensor.from_row_lengths`
如果知道每行的长度,可以使用 `row_lengths` 行分区张量:
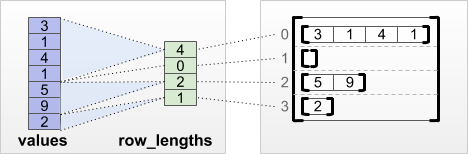
```
print(tf.RaggedTensor.from_row_lengths(
values=[3, 1, 4, 1, 5, 9, 2],
row_lengths=[4, 0, 2, 1]))
```
#### `tf.RaggedTensor.from_row_splits`
如果知道指示每行开始和结束的索引,可以使用 `row_splits` 行分区张量:
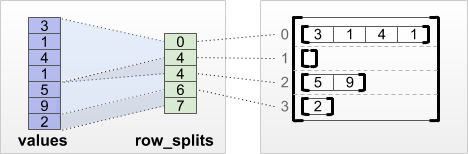
```
print(tf.RaggedTensor.from_row_splits(
values=[3, 1, 4, 1, 5, 9, 2],
row_splits=[0, 4, 4, 6, 7]))
```
有关完整的工厂方法列表,请参阅 `tf.RaggedTensor` 类文档。
注:默认情况下,这些工厂方法会添加断言,说明行分区张量结构良好且与值数量保持一致。如果您能够保证输入的结构良好且一致,可以使用 `validate=False` 参数跳过此类检查。
### 可以在不规则张量中存储什么
与普通 `Tensor` 一样,`RaggedTensor` 中的所有值必须具有相同的类型;所有值必须处于相同的嵌套深度(张量的*秩*):
```
print(tf.ragged.constant([["Hi"], ["How", "are", "you"]])) # ok: type=string, rank=2
print(tf.ragged.constant([[[1, 2], [3]], [[4, 5]]])) # ok: type=int32, rank=3
try:
tf.ragged.constant([["one", "two"], [3, 4]]) # bad: multiple types
except ValueError as exception:
print(exception)
try:
tf.ragged.constant(["A", ["B", "C"]]) # bad: multiple nesting depths
except ValueError as exception:
print(exception)
```
## 示例用例
以下示例演示了如何使用 `RaggedTensor`,通过为每个句子的开头和结尾使用特殊标记,为一批可变长度查询构造和组合一元元组与二元元组嵌入。有关本例中使用的运算的更多详细信息,请参阅 `tf.ragged` 包文档。
```
queries = tf.ragged.constant([['Who', 'is', 'Dan', 'Smith'],
['Pause'],
['Will', 'it', 'rain', 'later', 'today']])
# Create an embedding table.
num_buckets = 1024
embedding_size = 4
embedding_table = tf.Variable(
tf.random.truncated_normal([num_buckets, embedding_size],
stddev=1.0 / math.sqrt(embedding_size)))
# Look up the embedding for each word.
word_buckets = tf.strings.to_hash_bucket_fast(queries, num_buckets)
word_embeddings = tf.nn.embedding_lookup(embedding_table, word_buckets) # ①
# Add markers to the beginning and end of each sentence.
marker = tf.fill([queries.nrows(), 1], '#')
padded = tf.concat([marker, queries, marker], axis=1) # ②
# Build word bigrams & look up embeddings.
bigrams = tf.strings.join([padded[:, :-1], padded[:, 1:]], separator='+') # ③
bigram_buckets = tf.strings.to_hash_bucket_fast(bigrams, num_buckets)
bigram_embeddings = tf.nn.embedding_lookup(embedding_table, bigram_buckets) # ④
# Find the average embedding for each sentence
all_embeddings = tf.concat([word_embeddings, bigram_embeddings], axis=1) # ⑤
avg_embedding = tf.reduce_mean(all_embeddings, axis=1) # ⑥
print(avg_embedding)
```
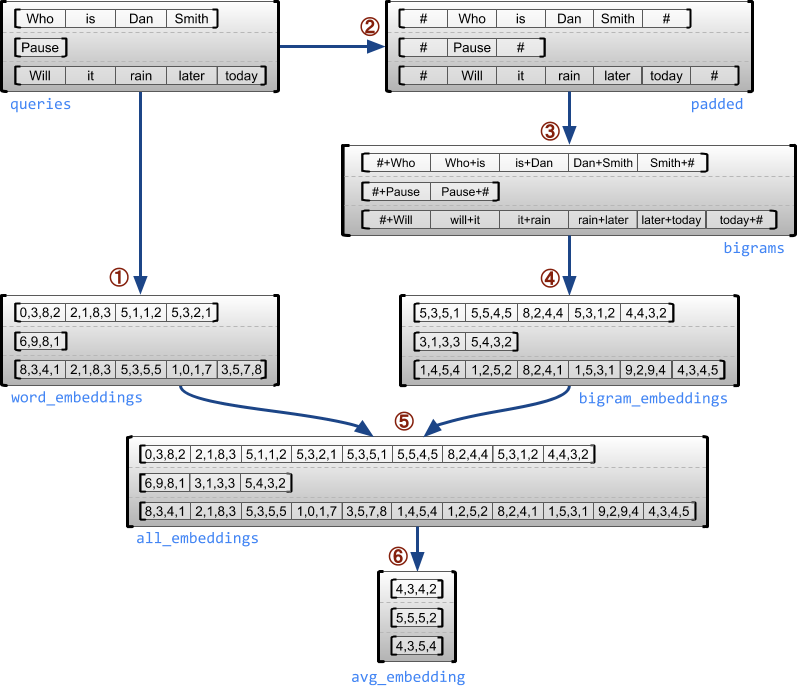
## 不规则维度和均匀维度
***不规则维度***是切片可能具有不同长度的维度。例如,`rt=[[3, 1, 4, 1], [], [5, 9, 2], [6], []]` 的内部(列)维度是不规则的,因为列切片 (`rt[0, :]`, ..., `rt[4, :]`) 具有不同的长度。切片全都具有相同长度的维度称为*均匀维度*。
不规则张量的最外层维始终是均匀维度,因为它只包含一个切片(因此不可能有不同的切片长度)。其余维度可能是不规则维度也可能是均匀维度。例如,我们可以使用形状为 `[num_sentences, (num_words), embedding_size]` 的不规则张量为一批句子中的每个单词存储单词嵌入,其中 `(num_words)` 周围的括号表示维度是不规则维度。
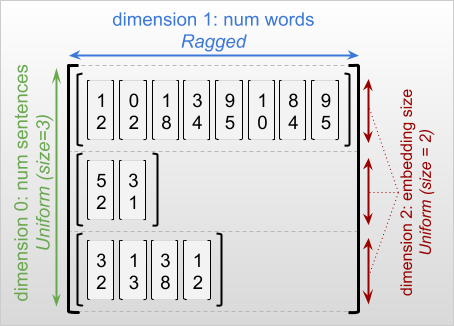
不规则张量可以有多个不规则维度。例如,我们可以使用形状为 `[num_documents, (num_paragraphs), (num_sentences), (num_words)]` 的张量存储一批结构化文本文档(其中,括号同样用于表示不规则维度)。
与 `tf.Tensor` 一样,不规则张量的***秩***是其总维数(包括不规则维度和均匀维度)。***潜在的不规则张量***是一个值,这个值可能是 `tf.Tensor` 或 `tf.RaggedTensor`。
描述 RaggedTensor 的形状时,按照惯例,不规则维度会通过括号进行指示。例如,如上面所见,存储一批句子中每个单词的单词嵌入的三维 RaggedTensor 的形状可以写为 `[num_sentences, (num_words), embedding_size]`。
`RaggedTensor.shape` 特性返回不规则张量的 `tf.TensorShape`,其中不规则维度的大小为 `None`:
```
tf.ragged.constant([["Hi"], ["How", "are", "you"]]).shape
```
可以使用方法 `tf.RaggedTensor.bounding_shape` 查找给定 `RaggedTensor` 的紧密边界形状:
```
print(tf.ragged.constant([["Hi"], ["How", "are", "you"]]).bounding_shape())
```
## 不规则张量和稀疏张量对比
不规则张量*不*应该被认为是一种稀疏张量。尤其是,稀疏张量是以紧凑的格式对相同数据建模的 *tf.Tensor 的高效编码*;而不规则张量是对扩展的数据类建模的 *tf.Tensor 的延伸*。这种区别在定义运算时至关重要:
- 对稀疏张量或密集张量应用某一运算应当始终获得相同结果。
- 对不规则张量或稀疏张量应用某一运算可能获得不同结果。
一个说明性的示例是,考虑如何为不规则张量和稀疏张量定义 `concat`、`stack` 和 `tile` 之类的数组运算。连接不规则张量时,会将每一行连在一起,形成一个具有组合长度的行:

```
ragged_x = tf.ragged.constant([["John"], ["a", "big", "dog"], ["my", "cat"]])
ragged_y = tf.ragged.constant([["fell", "asleep"], ["barked"], ["is", "fuzzy"]])
print(tf.concat([ragged_x, ragged_y], axis=1))
```
但连接稀疏张量时,相当于连接相应的密集张量,如以下示例所示(其中 Ø 表示缺失的值):
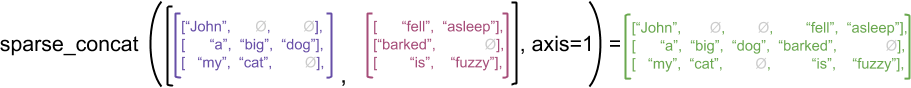
```
sparse_x = ragged_x.to_sparse()
sparse_y = ragged_y.to_sparse()
sparse_result = tf.sparse.concat(sp_inputs=[sparse_x, sparse_y], axis=1)
print(tf.sparse.to_dense(sparse_result, ''))
```
另一个说明为什么这种区别非常重要的示例是,考虑一个运算(如 `tf.reduce_mean`)的“每行平均值”的定义。对于不规则张量,一行的平均值是该行的值总和除以该行的宽度。但对于稀疏张量来说,一行的平均值是该行的值总和除以稀疏张量的总宽度(大于等于最长行的宽度)。
## TensorFlow API
### Keras
[tf.keras](https://tensorflow.google.cn/guide/keras) 是 TensorFlow 的高级 API,用于构建和训练深度学习模型。通过在 `tf.keras.Input` 或 `tf.keras.layers.InputLayer` 上设置 `ragged=True`,不规则张量可以作为输入传送到 Keras 模型。不规则张量还可以在 Keras 层之间传递,并由 Keras 模型返回。以下示例显示了一个使用不规则张量训练的小 LSTM 模型。
```
# Task: predict whether each sentence is a question or not.
sentences = tf.constant(
['What makes you think she is a witch?',
'She turned me into a newt.',
'A newt?',
'Well, I got better.'])
is_question = tf.constant([True, False, True, False])
# Preprocess the input strings.
hash_buckets = 1000
words = tf.strings.split(sentences, ' ')
hashed_words = tf.strings.to_hash_bucket_fast(words, hash_buckets)
# Build the Keras model.
keras_model = tf.keras.Sequential([
tf.keras.layers.Input(shape=[None], dtype=tf.int64, ragged=True),
tf.keras.layers.Embedding(hash_buckets, 16),
tf.keras.layers.LSTM(32, use_bias=False),
tf.keras.layers.Dense(32),
tf.keras.layers.Activation(tf.nn.relu),
tf.keras.layers.Dense(1)
])
keras_model.compile(loss='binary_crossentropy', optimizer='rmsprop')
keras_model.fit(hashed_words, is_question, epochs=5)
print(keras_model.predict(hashed_words))
```
### tf.Example
[tf.Example](https://tensorflow.google.cn/tutorials/load_data/tfrecord) 是 TensorFlow 数据的标准 [protobuf](https://developers.google.com/protocol-buffers/) 编码。使用 `tf.Example` 编码的数据往往包括可变长度特征。例如,以下代码定义了一批具有不同特征长度的四条 `tf.Example` 消息:
```
import google.protobuf.text_format as pbtext
def build_tf_example(s):
return pbtext.Merge(s, tf.train.Example()).SerializeToString()
example_batch = [
build_tf_example(r'''
features {
feature {key: "colors" value {bytes_list {value: ["red", "blue"]} } }
feature {key: "lengths" value {int64_list {value: [7]} } } }'''),
build_tf_example(r'''
features {
feature {key: "colors" value {bytes_list {value: ["orange"]} } }
feature {key: "lengths" value {int64_list {value: []} } } }'''),
build_tf_example(r'''
features {
feature {key: "colors" value {bytes_list {value: ["black", "yellow"]} } }
feature {key: "lengths" value {int64_list {value: [1, 3]} } } }'''),
build_tf_example(r'''
features {
feature {key: "colors" value {bytes_list {value: ["green"]} } }
feature {key: "lengths" value {int64_list {value: [3, 5, 2]} } } }''')]
```
我们可以使用 `tf.io.parse_example` 解析这个编码数据,它采用序列化字符串的张量和特征规范字典,并将字典映射特征名称返回给张量。要将长度可变特征读入不规则张量,我们只需在特征规范字典中使用 `tf.io.RaggedFeature` 即可:
```
feature_specification = {
'colors': tf.io.RaggedFeature(tf.string),
'lengths': tf.io.RaggedFeature(tf.int64),
}
feature_tensors = tf.io.parse_example(example_batch, feature_specification)
for name, value in feature_tensors.items():
print("{}={}".format(name, value))
```
`tf.io.RaggedFeature` 还可用于读取具有多个不规则维度的特征。有关详细信息,请参阅 [API 文档](https://tensorflow.google.cn/api_docs/python/tf/io/RaggedFeature)。
### 数据集
[tf.data](https://tensorflow.google.cn/guide/data) 是一个 API,可用于通过简单的可重用代码块构建复杂的输入流水线。它的核心数据结构是 `tf.data.Dataset`,表示一系列元素,每个元素包含一个或多个分量。
```
# Helper function used to print datasets in the examples below.
def print_dictionary_dataset(dataset):
for i, element in enumerate(dataset):
print("Element {}:".format(i))
for (feature_name, feature_value) in element.items():
print('{:>14} = {}'.format(feature_name, feature_value))
```
#### 使用不规则张量构建数据集
可以采用通过 `tf.Tensor` 或 numpy `array` 构建数据集时使用的方法,如 `Dataset.from_tensor_slices`,通过不规则张量构建数据集:
```
dataset = tf.data.Dataset.from_tensor_slices(feature_tensors)
print_dictionary_dataset(dataset)
```
注:`Dataset.from_generator` 目前还不支持不规则张量,但不久后将会支持这种张量。
#### 批处理和取消批处理具有不规则张量的数据集
可以使用 `Dataset.batch` 方法对具有不规则张量的数据集进行批处理(将 *n* 个连续元素组合成单个元素)。
```
batched_dataset = dataset.batch(2)
print_dictionary_dataset(batched_dataset)
```
相反,可以使用 `Dataset.unbatch` 将批处理后的数据集转换为扁平数据集。
```
unbatched_dataset = batched_dataset.unbatch()
print_dictionary_dataset(unbatched_dataset)
```
#### 对具有可变长度非不规则张量的数据集进行批处理
如果您有一个包含非不规则张量的数据集,而且各个元素的张量长度不同,则可以应用 `dense_to_ragged_batch` 转换,将这些非不规则张量批处理成不规则张量:
```
non_ragged_dataset = tf.data.Dataset.from_tensor_slices([1, 5, 3, 2, 8])
non_ragged_dataset = non_ragged_dataset.map(tf.range)
batched_non_ragged_dataset = non_ragged_dataset.apply(
tf.data.experimental.dense_to_ragged_batch(2))
for element in batched_non_ragged_dataset:
print(element)
```
#### 转换具有不规则张量的数据集
还可以使用 `Dataset.map` 创建或转换数据集中的不规则张量。
```
def transform_lengths(features):
return {
'mean_length': tf.math.reduce_mean(features['lengths']),
'length_ranges': tf.ragged.range(features['lengths'])}
transformed_dataset = dataset.map(transform_lengths)
print_dictionary_dataset(transformed_dataset)
```
### tf.function
[tf.function](https://tensorflow.google.cn/guide/function) 是预计算 Python 函数的 TensorFlow 计算图的装饰器,它可以大幅改善 TensorFlow 代码的性能。不规则张量能够透明地与 `@tf.function` 装饰的函数一起使用。例如,以下函数对不规则张量和非不规则张量均有效:
```
@tf.function
def make_palindrome(x, axis):
return tf.concat([x, tf.reverse(x, [axis])], axis)
make_palindrome(tf.constant([[1, 2], [3, 4], [5, 6]]), axis=1)
make_palindrome(tf.ragged.constant([[1, 2], [3], [4, 5, 6]]), axis=1)
```
如果您希望为 `tf.function` 明确指定 `input_signature`,可以使用 `tf.RaggedTensorSpec` 执行此操作。
```
@tf.function(
input_signature=[tf.RaggedTensorSpec(shape=[None, None], dtype=tf.int32)])
def max_and_min(rt):
return (tf.math.reduce_max(rt, axis=-1), tf.math.reduce_min(rt, axis=-1))
max_and_min(tf.ragged.constant([[1, 2], [3], [4, 5, 6]]))
```
#### 具体函数
[具体函数](https://tensorflow.google.cn/guide/function#obtaining_concrete_functions)封装通过 `tf.function` 构建的各个跟踪图。不规则张量可以透明地与具体函数一起使用。
```
# Preferred way to use ragged tensors with concrete functions (TF 2.3+):
try:
@tf.function
def increment(x):
return x + 1
rt = tf.ragged.constant([[1, 2], [3], [4, 5, 6]])
cf = increment.get_concrete_function(rt)
print(cf(rt))
except Exception as e:
print(f"Not supported before TF 2.3: {type(e)}: {e}")
```
### SavedModel
[SavedModel](https://tensorflow.google.cn/guide/saved_model) 是序列化 TensorFlow 程序,包括权重和计算。它可以通过 Keras 模型或自定义模型构建。在任何一种情况下,不规则张量都可以透明地与 SavedModel 定义的函数和方法一起使用。
#### 示例:保存 Keras 模型
```
import tempfile
keras_module_path = tempfile.mkdtemp()
tf.saved_model.save(keras_model, keras_module_path)
imported_model = tf.saved_model.load(keras_module_path)
imported_model(hashed_words)
```
#### 示例:保存自定义模型
```
class CustomModule(tf.Module):
def __init__(self, variable_value):
super(CustomModule, self).__init__()
self.v = tf.Variable(variable_value)
@tf.function
def grow(self, x):
return x * self.v
module = CustomModule(100.0)
# Before saving a custom model, we must ensure that concrete functions are
# built for each input signature that we will need.
module.grow.get_concrete_function(tf.RaggedTensorSpec(shape=[None, None],
dtype=tf.float32))
custom_module_path = tempfile.mkdtemp()
tf.saved_model.save(module, custom_module_path)
imported_model = tf.saved_model.load(custom_module_path)
imported_model.grow(tf.ragged.constant([[1.0, 4.0, 3.0], [2.0]]))
```
注:SavedModel [签名](https://tensorflow.google.cn/guide/saved_model#specifying_signatures_during_export)是具体函数。如上文的“具体函数”部分所述,从 TensorFlow 2.3 开始,只有具体函数才能正确处理不规则张量。如果您需要在先前版本的 TensorFlow 中使用 SavedModel 签名,建议您将不规则张量分解成其张量分量。
## 重载运算符
`RaggedTensor` 类会重载标准 Python 算术和比较运算符,使其易于执行基本的逐元素数学:
```
x = tf.ragged.constant([[1, 2], [3], [4, 5, 6]])
y = tf.ragged.constant([[1, 1], [2], [3, 3, 3]])
print(x + y)
```
由于重载运算符执行逐元素计算,因此所有二进制运算的输入必须具有相同的形状,或者可以广播至相同的形状。在最简单的广播情况下,单个标量与不规则张量中的每个值逐元素组合:
```
x = tf.ragged.constant([[1, 2], [3], [4, 5, 6]])
print(x + 3)
```
有关更高级的用例,请参阅**广播**一节。
不规则张量重载与正常 `Tensor` 相同的一组运算符:一元运算符 `-`、`~` 和 `abs()`;二元运算符 `+`、`-`、`*`、`/`、`//`、`%`、`**`、`&`、`|`、`^`、`==`、`<`、`<=`、`>` 和 `>=`。
## 索引
不规则张量支持 Python 风格的索引,包括多维索引和切片。以下示例使用二维和三维不规则张量演示了不规则张量索引。
### 索引示例:二维不规则张量
```
queries = tf.ragged.constant(
[['Who', 'is', 'George', 'Washington'],
['What', 'is', 'the', 'weather', 'tomorrow'],
['Goodnight']])
print(queries[1]) # A single query
print(queries[1, 2]) # A single word
print(queries[1:]) # Everything but the first row
print(queries[:, :3]) # The first 3 words of each query
print(queries[:, -2:]) # The last 2 words of each query
```
### 索引示例:三维不规则张量
```
rt = tf.ragged.constant([[[1, 2, 3], [4]],
[[5], [], [6]],
[[7]],
[[8, 9], [10]]])
print(rt[1]) # Second row (2-D RaggedTensor)
print(rt[3, 0]) # First element of fourth row (1-D Tensor)
print(rt[:, 1:3]) # Items 1-3 of each row (3-D RaggedTensor)
print(rt[:, -1:]) # Last item of each row (3-D RaggedTensor)
```
`RaggedTensor` 支持多维索引和切片,但有一个限制:不允许索引一个不规则维度。这种情况是有问题的,因为指示的值可能在某些行中存在,而在其他行中不存在。这种情况下,我们不知道是应该 (1) 引发 `IndexError`;(2) 使用默认值;还是 (3) 跳过该值并返回一个行数比开始时少的张量。根据 [Python 的指导原则](https://www.python.org/dev/peps/pep-0020/)(“当面对不明确的情况时,不要尝试去猜测”),我们目前不允许此运算。
## 张量类型转换
`RaggedTensor` 类定义了可用于在 `RaggedTensor` 与 `tf.Tensor` 或 `tf.SparseTensors` 之间转换的方法:
```
ragged_sentences = tf.ragged.constant([
['Hi'], ['Welcome', 'to', 'the', 'fair'], ['Have', 'fun']])
# RaggedTensor -> Tensor
print(ragged_sentences.to_tensor(default_value='', shape=[None, 10]))
# Tensor -> RaggedTensor
x = [[1, 3, -1, -1], [2, -1, -1, -1], [4, 5, 8, 9]]
print(tf.RaggedTensor.from_tensor(x, padding=-1))
#RaggedTensor -> SparseTensor
print(ragged_sentences.to_sparse())
# SparseTensor -> RaggedTensor
st = tf.SparseTensor(indices=[[0, 0], [2, 0], [2, 1]],
values=['a', 'b', 'c'],
dense_shape=[3, 3])
print(tf.RaggedTensor.from_sparse(st))
```
## 评估不规则张量
要访问不规则张量中的值,您可以:
1. 使用 `tf.RaggedTensor.to_list()` 将不规则张量转换为嵌套 Python 列表。
2. 使用 `tf.RaggedTensor.numpy()` 将不规则张量转换为 numpy 数组,数组的值是嵌套的 numpy 数组。
3. 使用 `tf.RaggedTensor.values` 和 `tf.RaggedTensor.row_splits` 属性,或 `tf.RaggedTensor.row_lengths()` 和 `tf.RaggedTensor.value_rowids()` 之类的行分区方法,将不规则张量分解成其分量。
4. 使用 Python 索引从不规则张量中选择值。
```
rt = tf.ragged.constant([[1, 2], [3, 4, 5], [6], [], [7]])
print("python list:", rt.to_list())
print("numpy array:", rt.numpy())
print("values:", rt.values.numpy())
print("splits:", rt.row_splits.numpy())
print("indexed value:", rt[1].numpy())
```
## 广播
广播是使具有不同形状的张量在进行逐元素运算时具有兼容形状的过程。有关广播的更多背景,请参阅:
- [Numpy:广播](https://docs.scipy.org/doc/numpy/user/basics.broadcasting.html)
- `tf.broadcast_dynamic_shape`
- `tf.broadcast_to`
广播两个输入 `x` 和 `y`,使其具有兼容形状的基本步骤是:
1. 如果 `x` 和 `y` 没有相同的维数,则增加外层维度(使用大小 1),直至它们具有相同的维数。
2. 对于 `x` 和 `y` 的大小不同的每一个维度:
- 如果 `x` 或 `y` 在 `d` 维中的大小为 `1`,则跨 `d` 维重复其值以匹配其他输入的大小。
- 否则,引发异常(`x` 和 `y` 非广播兼容)。
其中,均匀维度中一个张量的大小是一个数字(跨该维的切片大小);不规则维度中一个张量的大小是切片长度列表(跨该维的所有切片)。
### 广播示例
```
# x (2D ragged): 2 x (num_rows)
# y (scalar)
# result (2D ragged): 2 x (num_rows)
x = tf.ragged.constant([[1, 2], [3]])
y = 3
print(x + y)
# x (2d ragged): 3 x (num_rows)
# y (2d tensor): 3 x 1
# Result (2d ragged): 3 x (num_rows)
x = tf.ragged.constant(
[[10, 87, 12],
[19, 53],
[12, 32]])
y = [[1000], [2000], [3000]]
print(x + y)
# x (3d ragged): 2 x (r1) x 2
# y (2d ragged): 1 x 1
# Result (3d ragged): 2 x (r1) x 2
x = tf.ragged.constant(
[[[1, 2], [3, 4], [5, 6]],
[[7, 8]]],
ragged_rank=1)
y = tf.constant([[10]])
print(x + y)
# x (3d ragged): 2 x (r1) x (r2) x 1
# y (1d tensor): 3
# Result (3d ragged): 2 x (r1) x (r2) x 3
x = tf.ragged.constant(
[
[
[[1], [2]],
[],
[[3]],
[[4]],
],
[
[[5], [6]],
[[7]]
]
],
ragged_rank=2)
y = tf.constant([10, 20, 30])
print(x + y)
```
下面是一些不广播的形状示例:
```
# x (2d ragged): 3 x (r1)
# y (2d tensor): 3 x 4 # trailing dimensions do not match
x = tf.ragged.constant([[1, 2], [3, 4, 5, 6], [7]])
y = tf.constant([[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]])
try:
x + y
except tf.errors.InvalidArgumentError as exception:
print(exception)
# x (2d ragged): 3 x (r1)
# y (2d ragged): 3 x (r2) # ragged dimensions do not match.
x = tf.ragged.constant([[1, 2, 3], [4], [5, 6]])
y = tf.ragged.constant([[10, 20], [30, 40], [50]])
try:
x + y
except tf.errors.InvalidArgumentError as exception:
print(exception)
# x (3d ragged): 3 x (r1) x 2
# y (3d ragged): 3 x (r1) x 3 # trailing dimensions do not match
x = tf.ragged.constant([[[1, 2], [3, 4], [5, 6]],
[[7, 8], [9, 10]]])
y = tf.ragged.constant([[[1, 2, 0], [3, 4, 0], [5, 6, 0]],
[[7, 8, 0], [9, 10, 0]]])
try:
x + y
except tf.errors.InvalidArgumentError as exception:
print(exception)
```
## RaggedTensor 编码
不规则张量使用 `RaggedTensor` 类进行编码。在内部,每个 `RaggedTensor` 包含:
- 一个 `values` 张量,它将可变长度行连接成扁平列表。
- 一个 `row_partition`,它指示如何将这些扁平值分成各行。
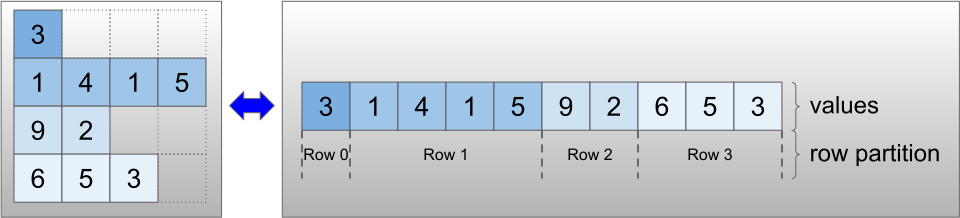
可以使用四种不同的编码存储 `row_partition`:
- `row_splits` 是一个整型向量,用于指定行之间的拆分点。
- `value_rowids` 是一个整型向量,用于指定每个值的行索引。
- `row_lengths` 是一个整型向量,用于指定每一行的长度。
- `uniform_row_length` 是一个整型标量,用于指定所有行的单个长度。
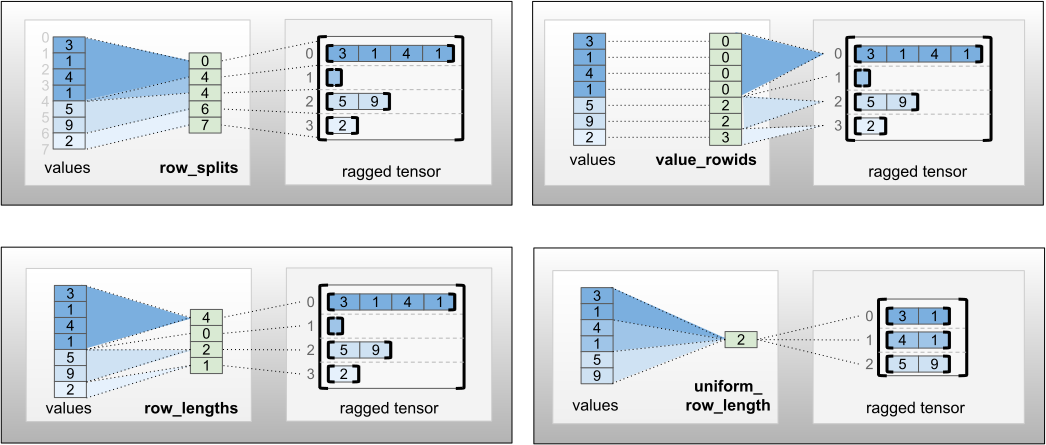
整型标量 `nrows` 还可以包含在 `row_partition` 编码中,以考虑具有 `value_rowids` 的空尾随行或具有 `uniform_row_length` 的空行。
```
rt = tf.RaggedTensor.from_row_splits(
values=[3, 1, 4, 1, 5, 9, 2],
row_splits=[0, 4, 4, 6, 7])
print(rt)
```
选择为行分区使用哪种编码由不规则张量在内部进行管理,以提高某些环境下的效率。尤其是,不同行分区方案的某些优点和缺点是:
- **高效索引**:`row_splits` 编码可以实现不规则张量的恒定时间索引和切片。
- **高效连接**:`row_lengths` 编码在连接不规则张量时更有效,因为当两个张量连接在一起时,行长度不会改变。
- **较小的编码大小**:`value_rowids` 编码在存储有大量空行的不规则张量时更有效,因为张量的大小只取决于值的总数。另一方面,`row_splits` 和 `row_lengths` 编码在存储具有较长行的不规则张量时更有效,因为它们每行只需要一个标量值。
- **兼容性**:`value_rowids` 方案与 `tf.math.segment_sum` 等运算使用的[分段](https://tensorflow.google.cn/api_docs/python/tf/math#about_segmentation)格式相匹配。`row_limits` 方案与 `tf.sequence_mask` 等运算使用的格式相匹配。
- **均匀维度**:如下文所述,`uniform_row_length` 编码用于对具有均匀维度的不规则张量进行编码。
### 多个不规则维度
具有多个不规则维度的不规则张量通过为 `values` 张量使用嵌套 `RaggedTensor` 进行编码。每个嵌套 `RaggedTensor` 都会增加一个不规则维度。
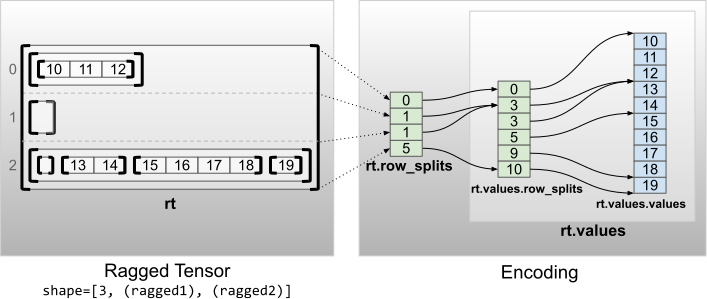
```
rt = tf.RaggedTensor.from_row_splits(
values=tf.RaggedTensor.from_row_splits(
values=[10, 11, 12, 13, 14, 15, 16, 17, 18, 19],
row_splits=[0, 3, 3, 5, 9, 10]),
row_splits=[0, 1, 1, 5])
print(rt)
print("Shape: {}".format(rt.shape))
print("Number of partitioned dimensions: {}".format(rt.ragged_rank))
```
工厂函数 `tf.RaggedTensor.from_nested_row_splits` 可用于通过提供一个 `row_splits` 张量列表,直接构造具有多个不规则维度的 RaggedTensor:
```
rt = tf.RaggedTensor.from_nested_row_splits(
flat_values=[10, 11, 12, 13, 14, 15, 16, 17, 18, 19],
nested_row_splits=([0, 1, 1, 5], [0, 3, 3, 5, 9, 10]))
print(rt)
```
### 不规则秩和扁平值
不规则张量的***不规则秩***是底层 `values` 张量的分区次数(即 `RaggedTensor` 对象的嵌套深度)。最内层的 `values` 张量称为其 ***flat_values***。在以下示例中,`conversations` 具有 ragged_rank=3,其 `flat_values` 为具有 24 个字符串的一维 `Tensor`:
```
# shape = [batch, (paragraph), (sentence), (word)]
conversations = tf.ragged.constant(
[[[["I", "like", "ragged", "tensors."]],
[["Oh", "yeah?"], ["What", "can", "you", "use", "them", "for?"]],
[["Processing", "variable", "length", "data!"]]],
[[["I", "like", "cheese."], ["Do", "you?"]],
[["Yes."], ["I", "do."]]]])
conversations.shape
assert conversations.ragged_rank == len(conversations.nested_row_splits)
conversations.ragged_rank # Number of partitioned dimensions.
conversations.flat_values.numpy()
```
### 均匀内层维度
具有均匀内层维度的不规则张量通过为 flat_values(即最内层 `values`)使用多维 `tf.Tensor` 进行编码。
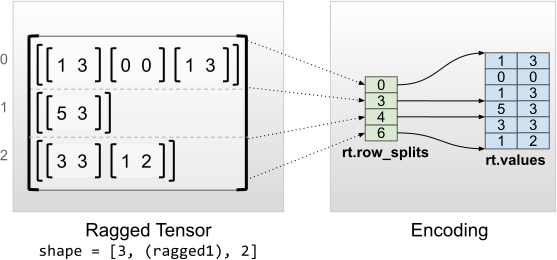
```
rt = tf.RaggedTensor.from_row_splits(
values=[[1, 3], [0, 0], [1, 3], [5, 3], [3, 3], [1, 2]],
row_splits=[0, 3, 4, 6])
print(rt)
print("Shape: {}".format(rt.shape))
print("Number of partitioned dimensions: {}".format(rt.ragged_rank))
print("Flat values shape: {}".format(rt.flat_values.shape))
print("Flat values:\n{}".format(rt.flat_values))
```
### 均匀非内层维度
具有均匀非内层维度的不规则张量通过使用 `uniform_row_length` 对行分区进行编码。
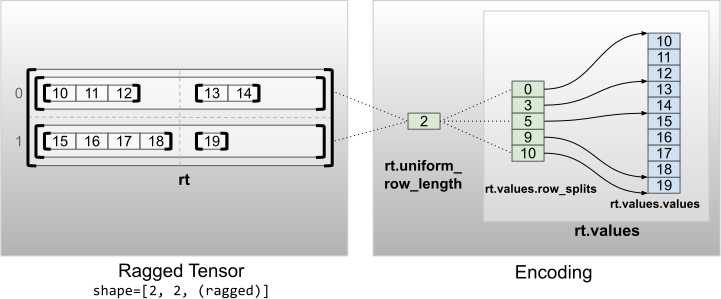
```
rt = tf.RaggedTensor.from_uniform_row_length(
values=tf.RaggedTensor.from_row_splits(
values=[10, 11, 12, 13, 14, 15, 16, 17, 18, 19],
row_splits=[0, 3, 5, 9, 10]),
uniform_row_length=2)
print(rt)
print("Shape: {}".format(rt.shape))
print("Number of partitioned dimensions: {}".format(rt.ragged_rank))
```
| github_jupyter |
# Deriving a Point-Spread Function in a Crowded Field
### following Appendix III of Peter Stetson's *User's Manual for DAOPHOT II*
### Using `pydaophot` form `astwro` python package
All *italic* text here have been taken from Stetson's manual.
The only input file for this procedure is a FITS file containing reference frame image. Here we use sample FITS form astwro package (NGC6871 I filter 20s frame). Below we get filepath for this image, as well as create instances of `Daophot` and `Allstar` classes - wrappers around `daophot` and `allstar` respectively.
One should also provide `daophot.opt`, `photo.opt` and `allstar.opt` in apropiriete constructors. Here default, build in, sample, `opt` files are used.
```
from astwro.sampledata import fits_image
frame = fits_image()
```
`Daophot` object creates temporary working directory (*runner directory*), which is passed to `Allstar` constructor to share.
```
from astwro.pydaophot import Daophot, Allstar
dp = Daophot(image=frame)
al = Allstar(dir=dp.dir)
```
Daophot got FITS file in construction, which will be automatically **ATTACH**ed.
#### *(1) Run FIND on your frame*
Daophot `FIND` parameters `Number of frames averaged, summed` are defaulted to `1,1`, below are provided for clarity.
```
res = dp.FInd(frames_av=1, frames_sum=1)
```
Check some results returned by `FIND`, every method for `daophot` command returns results object.
```
print ("{} pixels analysed, sky estimate {}, {} stars found.".format(res.pixels, res.sky, res.stars))
```
Also, take a look into *runner directory*
```
!ls -lt $dp.dir
```
We see symlinks to input image and `opt` files, and `i.coo` - result of `FIND`
#### *(2) Run PHOTOMETRY on your frame*
Below we run photometry, providing explicitly radius of aperture `A1` and `IS`, `OS` sky radiuses.
```
res = dp.PHotometry(apertures=[8], IS=35, OS=50)
```
List of stars generated by daophot commands, can be easily get as `astwro.starlist.Starlist` being essentially `pandas.DataFrame`:
```
stars = res.photometry_starlist
```
Let's check 10 stars with least A1 error (``mag_err`` column). ([pandas](https://pandas.pydata.org) style)
```
stars.sort_values('mag_err').iloc[:10]
```
#### *(3) SORT the output from PHOTOMETRY*
*in order of increasing apparent magnitude decreasing
stellar brightness with the renumbering feature. This step is optional but it can be more convenient than not.*
`SORT` command of `daophor` is not implemented (yet) in `pydaohot`. But we do sorting by ourself.
```
sorted_stars = stars.sort_values('mag')
sorted_stars.renumber()
```
Here we write sorted list back info photometry file at default name (overwriting existing one), because it's convenient to use default files in next commands.
```
dp.write_starlist(sorted_stars, 'i.ap')
!head -n20 $dp.PHotometry_result.photometry_file
dp.PHotometry_result.photometry_file
```
#### *(4) PICK to generate a set of likely PSF stars*
*How many stars you want to use is a function of the degree of variation you expect and the frequency with which stars are contaminated by cosmic rays or neighbor stars. [...]*
```
pick_res = dp.PIck(faintest_mag=20, number_of_stars_to_pick=40)
```
If no error reported, symlink to image file (renamed to `i.fits`), and all daophot output files (`i.*`) are in the working directory of runner:
```
ls $dp.dir
```
One may examine and improve `i.lst` list of PSF stars. Or use `astwro.tools.gapick.py` to obtain list of PSF stars optimised by genetic algorithm.
#### *(5) Run PSF *
*tell it the name of your complete (sorted renumbered) aperture photometry file, the name of the file with the list of PSF stars, and the name of the disk file you want the point spread function stored in (the default should be fine) [...]*
*If the frame is crowded it is probably worth your while to generate the first PSF with the "VARIABLE PSF" option set to -1 --- pure analytic PSF. That way, the companions will not generate ghosts in the model PSF that will come back to haunt you later. You should also have specified a reasonably generous fitting radius --- these stars have been preselected to be as isolated as possible and you want the best fits you can get. But remember to avoid letting neighbor stars intrude within one fitting radius of the center of any PSF star.*
For illustration we will set `VARIABLE PSF` option, before `PSf()`
```
dp.set_options('VARIABLE PSF', 2)
psf_res = dp.PSf()
```
#### *(6) Run GROUP and NSTAR or ALLSTAR on your NEI file*
*If your PSF stars have many neighbors this may take some minutes of real time. Please be patient or submit it as a batch job and perform steps on your next frame while you wait.*
We use `allstar`. (`GROUP` and `NSTAR` command are not implemented in current version of `pydaophot`). We use prepared above `Allstar` object: `al` operating on the same runner dir that `dp`.
As parameter we set input image (we haven't do that on constructor), and `nei` file produced by `PSf()`. We do not remember name `i.psf` so use `psf_res.nei_file` property.
Finally we order `allstar` to produce subtracted FITS .
```
alls_res = al.ALlstar(image_file=frame, stars=psf_res.nei_file, subtracted_image_file='is.fits')
```
All `result` objects, has `get_buffer()` method, useful to lookup unparsed `daophot` or `allstar` output:
```
print (alls_res.get_buffer())
```
#### *(8) EXIT from DAOPHOT and send this new picture to the image display *
*Examine each of the PSF stars and its environs. Have all of the PSF stars subtracted out more or less cleanly, or should some of them be rejected from further use as PSF stars? (If so use a text editor to delete these stars from the LST file.) Have the neighbors mostly disappeared, or have they left behind big zits? Have you uncovered any faint companions that FIND missed?[...]*
The absolute path to subtracted file (like for most output files) is available as result's property:
```
sub_img = alls_res.subtracted_image_file
```
We can also generate region file for psf stars:
```
from astwro.starlist.ds9 import write_ds9_regions
reg_file_path = dp.file_from_runner_dir('lst.reg')
write_ds9_regions(pick_res.picked_starlist, reg_file_path)
# One can run ds9 directly from notebook:
!ds9 $sub_img -regions $reg_file_path
```
#### *(9) Back in DAOPHOT II ATTACH the original picture and run SUBSTAR*
*specifying the file created in step (6) or in step (8f) as the stars to subtract, and the stars in the LST file as the stars to keep.*
Lookup into runner dir:
```
ls $al.dir
sub_res = dp.SUbstar(subtract=alls_res.profile_photometry_file, leave_in=pick_res.picked_stars_file)
```
*You have now created a new picture which has the PSF stars still in it but from which the known neighbors of these PSF stars have been mostly removed*
#### (10) ATTACH the new star subtracted frame and repeat step (5) to derive a new point spread function
#### (11+...) Run GROUP NSTAR or ALLSTAR
```
for i in range(3):
print ("Iteration {}: Allstar chi: {}".format(i, alls_res.als_stars.chi.mean()))
dp.image = 'is.fits'
respsf = dp.PSf()
print ("Iteration {}: PSF chi: {}".format(i, respsf.chi))
alls_res = al.ALlstar(image_file=frame, stars='i.nei')
dp.image = frame
dp.SUbstar(subtract='i.als', leave_in='i.lst')
print ("Final: Allstar chi: {}".format(alls_res.als_stars.chi.mean()))
alls_res.als_stars
```
Check last image with subtracted PSF stars neighbours.
```
!ds9 $dp.SUbstar_result.subtracted_image_file -regions $reg_file_path
```
*Once you have produced a frame in which the PSF stars and their neighbors all subtract out cleanly, one more time through PSF should produce a point-spread function you can be proud of.*
```
dp.image = 'is.fits'
psf_res = dp.PSf()
print ("PSF file: {}".format(psf_res.psf_file))
```
| github_jupyter |
```
# python standard library
import sys
import os
import operator
import itertools
import collections
import functools
import glob
import csv
import datetime
import bisect
import sqlite3
import subprocess
import random
import gc
import shutil
import shelve
import contextlib
import tempfile
import math
import pickle
# general purpose third party packages
import cython
%reload_ext Cython
import numpy as np
nnz = np.count_nonzero
import scipy
import scipy.stats
import scipy.spatial.distance
import numexpr
import h5py
import tables
import bcolz
import dask
import dask.array as da
import pandas
import IPython
from IPython.display import clear_output, display, HTML
import sklearn
import sklearn.decomposition
import sklearn.manifold
import petl as etl
etl.config.display_index_header = True
import humanize
from humanize import naturalsize, intcomma, intword
import zarr
import graphviz
import statsmodels.formula.api as sfa
# plotting setup
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from matplotlib.gridspec import GridSpec
import matplotlib_venn as venn
import seaborn as sns
sns.set_context('paper')
sns.set_style('white')
sns.set_style('ticks')
rcParams = plt.rcParams
base_font_size = 8
rcParams['font.size'] = base_font_size
rcParams['axes.titlesize'] = base_font_size
rcParams['axes.labelsize'] = base_font_size
rcParams['xtick.labelsize'] = base_font_size
rcParams['ytick.labelsize'] = base_font_size
rcParams['legend.fontsize'] = base_font_size
rcParams['axes.linewidth'] = .5
rcParams['lines.linewidth'] = .5
rcParams['patch.linewidth'] = .5
rcParams['ytick.direction'] = 'out'
rcParams['xtick.direction'] = 'out'
rcParams['savefig.jpeg_quality'] = 100
rcParams['lines.markeredgewidth'] = .5
rcParams['figure.max_open_warning'] = 1000
rcParams['figure.dpi'] = 120
rcParams['figure.facecolor'] = 'w'
# bio third party packages
import Bio
import pyfasta
# currently broken, not compatible
# import pysam
# import pysamstats
import petlx
import petlx.bio
import vcf
import anhima
import allel
sys.path.insert(0, '../agam-report-base/src/python')
from util import *
import zcache
import veff
# import hapclust
ag1k_dir = '../ngs.sanger.ac.uk/production/ag1000g'
from ag1k import phase1_ar3
phase1_ar3.init(os.path.join(ag1k_dir, 'phase1', 'AR3'))
from ag1k import phase1_ar31
phase1_ar31.init(os.path.join(ag1k_dir, 'phase1', 'AR3.1'))
from ag1k import phase2_ar1
phase2_ar1.init(os.path.join(ag1k_dir, 'phase2', 'AR1'))
region_vgsc = SeqFeature('2L', 2358158, 2431617, label='Vgsc')
```
| github_jupyter |
```
import numpy as np
from copy import deepcopy
from scipy.special import expit
from scipy.optimize import minimize
from sklearn.datasets import load_iris
from sklearn.linear_model import LogisticRegression as skLogisticRegression
from sklearn.multiclass import OneVsRestClassifier as skOneVsRestClassifier
class OneVsRestClassifier():
def __init__(self, estimator):
self.estimator = estimator
def _encode(self, y):
classes = np.unique(y)
y_train = np.zeros((y.shape[0], len(classes)))
for i, c in enumerate(classes):
y_train[y == c, i] = 1
return classes, y_train
def fit(self, X, y):
self.classes_, y_train = self._encode(y)
self.estimators_ = []
for i in range(y_train.shape[1]):
cur_y = y_train[:, i]
clf = deepcopy(self.estimator)
clf.fit(X, cur_y)
self.estimators_.append(clf)
return self
def decision_function(self, X):
scores = np.zeros((X.shape[0], len(self.classes_)))
for i, est in enumerate(self.estimators_):
scores[:, i] = est.decision_function(X)
return scores
def predict(self, X):
scores = self.decision_function(X)
indices = np.argmax(scores, axis=1)
return self.classes_[indices]
# Simplified version of LogisticRegression, only work for binary classification
class BinaryLogisticRegression():
def __init__(self, C=1.0):
self.C = C
@staticmethod
def _cost_grad(w, X, y, alpha):
def _log_logistic(x):
if x > 0:
return -np.log(1 + np.exp(-x))
else:
return x - np.log(1 + np.exp(x))
yz = y * (np.dot(X, w[:-1]) + w[-1])
cost = -np.sum(np.vectorize(_log_logistic)(yz)) + 0.5 * alpha * np.dot(w[:-1], w[:-1])
grad = np.zeros(len(w))
t = (expit(yz) - 1) * y
grad[:-1] = np.dot(X.T, t) + alpha * w[:-1]
grad[-1] = np.sum(t)
return cost, grad
def _solve_lbfgs(self, X, y):
y_train = np.full(X.shape[0], -1)
y_train[y == 1] = 1
w0 = np.zeros(X.shape[1] + 1)
res = minimize(fun=self._cost_grad, jac=True, x0=w0,
args=(X, y_train, 1 / self.C), method='L-BFGS-B')
return res.x[:-1], res.x[-1]
def fit(self, X, y):
self.coef_, self.intercept_ = self._solve_lbfgs(X, y)
return self
def decision_function(self, X):
scores = np.dot(X, self.coef_) + self.intercept_
return scores
def predict(self, X):
scores = self.decision_function(X)
indices = (scores > 0).astype(int)
return indices
for C in [0.1, 1, 10, np.inf]:
X, y = load_iris(return_X_y=True)
clf1 = OneVsRestClassifier(BinaryLogisticRegression(C=C)).fit(X, y)
clf2 = skOneVsRestClassifier(skLogisticRegression(C=C, multi_class="ovr", solver="lbfgs",
# keep consisent with scipy default
tol=1e-5, max_iter=15000)).fit(X, y)
prob1 = clf1.decision_function(X)
prob2 = clf2.decision_function(X)
pred1 = clf1.predict(X)
pred2 = clf2.predict(X)
assert np.allclose(prob1, prob2)
assert np.array_equal(pred1, pred2)
```
| github_jupyter |
# BLU15 - Model CSI
## Intro:
It often happens that your data distribution changes with time.
More than that, sometimes you don't know how a model was trained and what was the original training data.
In this learning unit we're going to try to identify whether an existing model meets our expectations and redeploy it.
## Problem statement:
As an example, we're going to use the same problem that you met in the last BLU.
You're already familiar with the problem, but just as a reminder:
> The police department has received lots of complaints about its stop and search policy. Every time a car is stopped, the police officers have to decide whether or not to search the car for contraband. According to critics, these searches have a bias against people of certain backgrounds.
You got a model from your client, and **here is the model's description:**
> It's a LightGBM model (LGBMClassifier) trained on the following features:
> - Department Name
> - InterventionLocationName
> - InterventionReasonCode
> - ReportingOfficerIdentificationID
> - ResidentIndicator
> - SearchAuthorizationCode
> - StatuteReason
> - SubjectAge
> - SubjectEthnicityCode
> - SubjectRaceCode
> - SubjectSexCode
> - TownResidentIndicator
> All the categorical feature were one-hot encoded. The only numerical feature (SubjectAge) was not changed. The rows that contain rare categorical features (the ones that appear less than N times in the dataset) were removed. Check the original_model.ipynb notebook for more details.
P.S., if you never heard about lightgbm, XGboost and other gradient boosting, I highly recommend you to read this [article](https://mlcourse.ai/articles/topic10-boosting/) or watch these videos: [part1](https://www.youtube.com/watch?v=g0ZOtzZqdqk), [part2](https://www.youtube.com/watch?v=V5158Oug4W8)
It's not essential for this BLU, so you might leave this link as a desert after you go through the learning materials and solve the exercises, but these are very good models you can use later on, so I suggest reading about them.
**Here are the requirements that the police department created:**
> - A minimum 50% success rate for searches (when a car is searched, it should be at least 50% likely that contraband is found)
> - No police sub-department should have a discrepancy bigger than 5% between the search success rate between protected classes (race, ethnicity, gender)
> - The largest possible amount of contraband found, given the constraints above.
**And here is the description of how the current model succeeds with the requirements:**
- precision score = 50%
- recall = 89.3%
- roc_auc_score for the probability predictions = 82.7%
The precision and recall above are met for probability predictions with a specified threshold equal to **0.21073452797732833**
It's not said whether the second requirement is met, and as it was not met in the previous learning unit, let's ignore it for now.
## Model diagnosing:
Let's firstly try to compare these models to the ones that we created in the previous BLU:
| Model | Baseline | Second iteration | New model | Best model |
|-------------------|---------|--------|--------|--------|
| Requirement 1 - success rate | 0.53 | 0.38 | 0.5 | 1 |
| Requirement 2 - global discrimination (race) | 0.105 | 0.11 | NaN | 1 |
| Requirement 2 - global discrimination (sex) | 0.012 | 0.014 | NaN | 1 |
| Requirement 2 - global discrimination (ethnicity) | 0.114 | 0.101 | NaN | 2 |
| Requirement 2 - # department discrimination (race) | 27 | 17 | NaN | 2 |
| Requirement 2 - # department discrimination (sex) | 19 | 23 | NaN | 1 |
| Requirement 2 - # department discrimination (ethnicity) | 24 | NaN | 23 | 2 |
| Requirement 3 - contraband found (Recall) | 0.65 | 0.76 | 0.893 | 3 |
As we can see, the last model has the exact required success rate (Requirement 1) as we need, and a very good Recall (Requirement 3).
But it might be risky to have such a specific threshold, as we might end up success rate < 0.5 really quickly. It might be a better idea to have a bigger threshold (e.g. 0.25), but let's see.
Let's imagine that the model was trained long time ago.
And now you're in the future trying to evaluate the model, because things might have changed. Data distribution is not always the same, so something that used to work even a year ago could be completely wrong today.
Especially in 2020!
<img src="media/future_2020.jpg" width=400/>
First of all, let's start the server which is running this model.
Open the shell,
```sh
python protected_server.py
```
And read a csv files with new observations from 2020:
```
import joblib
import pandas as pd
import json
import joblib
import pickle
from sklearn.metrics import precision_score, recall_score, roc_auc_score
from sklearn.metrics import confusion_matrix
import requests
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from sklearn.metrics import precision_recall_curve
%matplotlib inline
df = pd.read_csv('./data/new_observations.csv')
df.head()
```
Let's start from sending all those requests and comparing the model prediction results with the target values.
The model is already prepared to convert our observations to the format its expecting, the only thing we need to change is making department and intervention location names lowercase, and we're good to extract fields from the dataframe and put them to the post request.
```
# lowercaes departments and location names
df['Department Name'] = df['Department Name'].apply(lambda x: str(x).lower())
df['InterventionLocationName'] = df['InterventionLocationName'].apply(lambda x: str(x).lower())
url = "http://127.0.0.1:5000/predict"
headers = {'Content-Type': 'application/json'}
def send_request(index: int, obs: dict, url: str, headers: dict):
observation = {
"id": index,
"observation": {
"Department Name": obs["Department Name"],
"InterventionLocationName": obs["InterventionLocationName"],
"InterventionReasonCode": obs["InterventionReasonCode"],
"ReportingOfficerIdentificationID": obs["ReportingOfficerIdentificationID"],
"ResidentIndicator": obs["ResidentIndicator"],
"SearchAuthorizationCode": obs["SearchAuthorizationCode"],
"StatuteReason": obs["StatuteReason"],
"SubjectAge": obs["SubjectAge"],
"SubjectEthnicityCode": obs["SubjectEthnicityCode"],
"SubjectRaceCode": obs["SubjectRaceCode"],
"SubjectSexCode": obs["SubjectSexCode"],
"TownResidentIndicator": obs["TownResidentIndicator"]
}
}
r = requests.post(url, data=json.dumps(observation), headers=headers)
result = json.loads(r.text)
return result
responses = [send_request(i, obs, url, headers) for i, obs in df.iterrows()]
print(responses[0])
df['proba'] = [r['proba'] for r in responses]
threshold = 0.21073452797732833
# we're going to use the threshold we got from the client
df['prediction'] = [1 if p >= threshold else 0 for p in df['proba']]
```
**NOTE:** We could also load the model and make predictions locally (without using the api), but:
1. I wanted to show you how you might send requests in a similar situation
2. If you have a running API and some model file, you always need to understand how the API works (if it makes any kind of data preprocessing), which might sometimes be complicated, and if you're trying to analyze the model running in production, you still need to make sure that the local predictions you do are equal to the one that the production api does.
```
confusion_matrix(df['ContrabandIndicator'], df['prediction'])
```
If you're not familiar with confusion matrixes, **here is an explanation of the values:**
<img src="./media/confusion_matrix.jpg" alt="drawing" width="500"/>
These values don't seem to be good. Let's once again take a look on the client's requirements and see if we still meet them:
> A minimum 50% success rate for searches (when a car is searched, it should be at least 50% likely that contraband is found)
```
def verify_success_rate_above(y_true, y_pred, min_success_rate=0.5):
"""
Verifies the success rate on a test set is above a provided minimum
"""
precision = precision_score(y_true, y_pred, pos_label=True)
is_satisfied = (precision >= min_success_rate)
return is_satisfied, precision
verify_success_rate_above(df['ContrabandIndicator'], df['prediction'], 0.5)
```

> The largest possible amount of contraband found, given the constraints above.
As the client says, their model recall was 0.893. And what now?
```
def verify_amount_found(y_true, y_pred):
"""
Verifies the amout of contraband found in the test dataset - a.k.a the recall in our test set
"""
recall = recall_score(y_true, y_pred, pos_label=True)
return recall
verify_amount_found(df['ContrabandIndicator'], df['prediction'])
```
<img src="./media/no_please_2.jpg" alt="drawing" width="500"/>
Okay, relax, it happens. Let's start from checking different thresholds. Maybe the selected threshold was to specific and doesn't work anymore.
What about 0.25?
```
threshold = 0.25
df['prediction'] = [1 if p >= threshold else 0 for p in df['proba']]
verify_success_rate_above(df['ContrabandIndicator'], df['prediction'], 0.5)
verify_amount_found(df['ContrabandIndicator'], df['prediction'])
```
<img src="./media/poker.jpg" alt="drawing" width="200"/>
Okay, let's try the same technique to identify the best threshold as they originally did. Maybe we find something good enough.
It's not a good idea to verify such things on the test data, but we're going to use it just to confirm the model's performance, not to select the threshold.
```
precision, recall, thresholds = precision_recall_curve(df['ContrabandIndicator'], df['proba'])
precision = precision[:-1]
recall = recall[:-1]
fig=plt.figure()
ax1 = plt.subplot(211)
ax2 = plt.subplot(212)
ax1.hlines(y=0.5,xmin=0, xmax=1, colors='red')
ax1.plot(thresholds,precision)
ax2.plot(thresholds,recall)
ax1.get_shared_x_axes().join(ax1, ax2)
ax1.set_xticklabels([])
plt.xlabel('Threshold')
ax1.set_title('Precision')
ax2.set_title('Recall')
plt.show()
```
So what do we see? There is some threshold value (around 0.6) that gives us precision >= 0.5.
But the threshold is so big, that the recall at this point is really-really low.
Let's calculate the exact values:
```
min_index = [i for i, prec in enumerate(precision) if prec >= 0.5][0]
print(min_index)
thresholds[min_index]
precision[min_index]
recall[min_index]
```
<img src="./media/incredible.jpg" alt="drawing" width="400"/>
Before we move on, we need to understand why this happens, so that we can decide what kind of action to perform.
Let's try to analyze the changes in data and discuss different things we might want to do.
```
old_df = pd.read_csv('./data/train_searched.csv')
old_df.head()
```
We're going to apply the same changes to the dataset as in the original model notebook unit to understand what was the original data like and how the current dataset differs.
```
old_df = old_df[(old_df['VehicleSearchedIndicator']==True)]
# lowercaes departments and location names
old_df['Department Name'] = old_df['Department Name'].apply(lambda x: str(x).lower())
old_df['InterventionLocationName'] = old_df['InterventionLocationName'].apply(lambda x: str(x).lower())
train_features = old_df.columns.drop(['VehicleSearchedIndicator', 'ContrabandIndicator'])
categorical_features = train_features.drop(['InterventionDateTime', 'SubjectAge'])
numerical_features = ['SubjectAge']
target = 'ContrabandIndicator'
# I'm going to remove less common features.
# Let's create a dictionary with the minimum required number of appearences
min_frequency = {
"Department Name": 50,
"InterventionLocationName": 50,
"ReportingOfficerIdentificationID": 30,
"StatuteReason": 10
}
def filter_values(df: pd.DataFrame, column_name: str, threshold: int):
value_counts = df[column_name].value_counts()
to_keep = value_counts[value_counts > threshold].index
filtered = df[df[column_name].isin(to_keep)]
return filtered
for feature, threshold in min_frequency.items():
old_df = filter_values(old_df, feature, threshold)
old_df.shape
old_df.head()
old_df['ContrabandIndicator'].value_counts(normalize=True)
df['ContrabandIndicator'].value_counts(normalize=True)
```
Looks like we got a bit more contraband now, and it's already a good sign:
if the training data had a different target feature distribution than the test set, the model's predictions might have a different distribution as well. It's a good practice to have the same target feature distribution both in training and test sets.
Let's investigate further
```
new_department_names = df['Department Name'].unique()
old_department_names = old_df['Department Name'].unique()
unknown_departments = [department for department in new_department_names if department not in old_department_names]
len(unknown_departments)
df[df['Department Name'].isin(unknown_departments)].shape
```
So we have 10 departments that the original model was not trained on, but they are only 23 rows from the test set.
Let's repeat the same thing for the Intervention Location names
```
new_location_names = df['InterventionLocationName'].unique()
old_location_names = old_df['InterventionLocationName'].unique()
unknown_locations = [location for location in new_location_names if location not in old_location_names]
len(unknown_locations)
df[df['InterventionLocationName'].isin(unknown_locations)].shape[0]
print('unknown locations: ', df[df['InterventionLocationName'].isin(unknown_locations)].shape[0] * 100 / df.shape[0], '%')
```
Alright, a bit more of unknown locations.
We don't know if the feature was important for the model, so these 5.3% of unknown locations might be important or not.
But it's worth keeping it in mind.
**Here are a few ideas of what we could try to do:**
1. Reanalyze the filtered locations, e.g. filter more rare ones.
2. Create a new category for the rare locations
3. Analyze the unknown locations for containing typos
Let's go further and take a look on the relation between department names and the number of contrabands they find.
We're going to select the most common department names, and then see the percentage of contraband indicator in each one for the training and test sets
```
common_departments = df['Department Name'].value_counts().head(20).index
departments_new = df[df['Department Name'].isin(common_departments)]
departments_old = old_df[old_df['Department Name'].isin(common_departments)]
pd.crosstab(departments_new['ContrabandIndicator'], departments_new['Department Name'], normalize="columns")
pd.crosstab(departments_old['ContrabandIndicator'], departments_old['Department Name'], normalize="columns")
```
We can clearly see that some departments got a huge difference in the contraband indicator.
E.g. Bridgeport used to have 93% of False contrabands, and now has only 62%.
Similar situation with Danbury and New Haven.
Why? Hard to say. There are really a lot of variables here. Maybe the departments got instructed on how to look for contraband.
But we might need to retrain the model.
Let's just finish reviewing other columns.
```
common_location = df['InterventionLocationName'].value_counts().head(20).index
locations_new = df[df['InterventionLocationName'].isin(common_location)]
locations_old = old_df[old_df['InterventionLocationName'].isin(common_location)]
pd.crosstab(locations_new['ContrabandIndicator'], locations_new['InterventionLocationName'], normalize="columns")
pd.crosstab(locations_old['ContrabandIndicator'], locations_old['InterventionLocationName'], normalize="columns")
```
What do we see? First of all, the InterventionLocationName and the Department Name are often same.
It sounds pretty logic, as probably policeman's usually work in the area of their department. But we could try to create a feature saying whether InterventionLocationName is equal to the Department Name.
Or maybe we could just get rid of one of them, if all the values are equal.
What else?
Well, There are similar changes in the Contraband distribution as in Department Name case.
Let's move on:
```
pd.crosstab(df['ContrabandIndicator'], df['InterventionReasonCode'], normalize="columns")
pd.crosstab(old_df['ContrabandIndicator'], old_df['InterventionReasonCode'], normalize="columns")
```
There are some small changes, but they don't seem to be significant.
Especially that all the 3 values have around 33% of Contraband.
Time for officers:
```
df['ReportingOfficerIdentificationID'].value_counts()
filter_values(df, 'ReportingOfficerIdentificationID', 2)['ReportingOfficerIdentificationID'].nunique()
```
Well, looks like there are a lot of unique values for the officer id (1166 for 2000 records), and there are not so many common ones (only 206 officers have more than 2 rows in the dataset) so it doesn't make much sense to analyze it.
Let's quickly go throw the rest of the columns:
```
df.columns
rest = ['ResidentIndicator', 'SearchAuthorizationCode',
'StatuteReason', 'SubjectEthnicityCode',
'SubjectRaceCode', 'SubjectSexCode','TownResidentIndicator']
for col in rest:
display(pd.crosstab(df['ContrabandIndicator'], df[col], normalize="columns"))
display(pd.crosstab(old_df['ContrabandIndicator'], old_df[col], normalize="columns"))
```
We see that all the columns got changes, but they don't seem to be so significant as in the Departments cases.
Anyway, it seems like we need to retrain the model.
<img src="./media/retrain.jpg" alt="drawing" width="400"/>
Retraining a model is always a decision we need to think about.
Was this change in data constant, temporary or seasonal?
In other words, do we expect the data distribution to stay as it is? To change back after Covid? To change from season to season?
**Depending on that, we could retrain the model differently:**
- **If it's a seasonality**, we might want to add features like season or month and train the same model to predict differently depending on the season. We could also investigate time-series classification algorithms.
- **If it's something that is going to change back**, we might either train a new model for this particular period in case the current data distrubution changes were temporary. Otherwise, if we expect the data distribution change here and back from time to time (and we know these periods in advance), we could create a new feature that would help model understand which period it is.
> E.g. if we had a task of predicting beer consumption and had a city that has a lot of football matches, we might add a feature like **football_championship** and make the model predict differently for this occasions.
- **If the data distribution has simply changed and we know that it's never going to come back**, we can simply retrain the model.
> But in some cases we have no idea why some changes appeared (e.g. in this case of departments having more contraband).
- In this case it might be a good idea to train a new model on the new datast and create some monitoring for these features distribution, so we could react when things change.
> So, in our case we don't know what was the reason of data distribution changes, so we'd like to train a model on the new dataset.
> The only thing is the size of the dataset. Original dataset had around 50k rows, and our new set has only 2000. It's not enough to train a good model, so this time we're going to combine both the datasets and add a new feature helping model to distinguish between them. If we had more data, it would be probably better to train a completely new model.
And we're done!
<img src="./media/end.jpg" alt="drawing" width="400"/>
| github_jupyter |
```
import numpy as np
#Load the predicted 9x12 array
#1st pass
im1=np.array([[4,4,4,4,4,4,4,4,4,4,4,4],
[6,6,2,1,6,6,6,6,6,1,1,2],
[6,6,6,1,1,6,6,6,6,1,1,2],
[2,6,6,6,1,5,5,5,6,1,1,2],
[5,6,6,6,5,5,5,5,5,1,5,5],
[5,5,2,5,5,5,5,5,5,1,5,5],
[5,5,2,5,5,5,5,5,5,6,5,5],
[2,6,6,6,5,5,5,5,5,6,2,2],
[2,6,6,6,6,6,6,2,2,6,2,2]])
#zoomed into driveway
im2=np.array([[2,2,2,1,1,1,2,6,6,6,6,6],
[2,2,2,1,1,1,2,6,6,6,6,6],
[2,2,2,1,1,1,2,6,6,6,6,6],
[2,2,2,1,1,1,1,6,6,6,6,6],
[2,2,2,6,1,1,1,6,6,6,6,6],
[6,6,6,6,1,1,1,1,6,6,6,6],
[6,6,6,6,6,1,1,1,6,6,6,6],
[6,6,6,6,6,6,1,1,2,2,2,2],
[6,6,6,6,6,6,6,1,5,5,5,5]])
#%%timeit
from scipy.ndimage.measurements import label
from scipy.ndimage.measurements import center_of_mass
A=im1
#Center of the 9x12 array
img_center=np.array([4,5.5])
#Label all the driveways and roofs
driveway, num_driveway = label(A==1)
roof, num_roof = label(A==5)
#Save number of driveways into array
d=np.arange(1,num_driveway+1)
r=np.arange(1,num_roof+1)
#Find the center of the all the driveways
driveway_center=center_of_mass(A,driveway,d)
roof_center=center_of_mass(A,roof,r)
print(driveway_center)
#Function to find the closest roof/driveway
def closest(list,img_center):
closest=list[0]
for c in list:
if np.linalg.norm(c-img_center) < np.linalg.norm(closest-img_center):
closest = c
return closest
#Find the closest roof to the center of the image
closest_roof=closest(roof_center,img_center)
#Find the closest driveway to the closest roof
closest_driveway=closest(driveway_center,np.asarray(closest_roof))
print(closest_driveway)
#Look for 3x3 driveway when we have reached a certain height (maybe 5m above ground)
a=im2
#Sliding window function
def sliding_window_view(arr, shape):
n = np.array(arr.shape)
o = n - shape + 1 # output shape
strides = arr.strides
new_shape = np.concatenate((o, shape), axis=0)
new_strides = np.concatenate((strides, strides), axis=0)
return np.lib.stride_tricks.as_strided(arr ,new_shape, new_strides)
#Creates a 7x10 ndarray with all the 3x3 submatrices
sub_image=sliding_window_view(a,(3,3))
#Empty list
driveway_list=[]
#Loop through the 7x10 ndarray
for i in range(0,7):
for j in range(i,10):
#Calculate the total of the submatrices
output=sum(sum(sub_image[i,j]))
#if the output is 9, that means we have a 3x3 that is all driveway
if output==9:
#append the i(row) and j(column) to a list declared previously
#we add 1 to the i and j to find the center of the 3x3
driveway_list.append((i+1,j+1))
#Call closest function to find driveway closest to house.
closest_driveway=closest(driveway_list,np.asarray(closest_roof))
print(closest_driveway)
#Read altitude from csv & Ground Sampling
import csv
def GSD(alt):
sensor_height=4.5 #mm
sensor_width=6.17 #mm
focal_length=1.8
image_height=1080 #pixels
image_width=1920 #pixels
#GSD = (sensor height (mm) x flight height (m) x 100) / (focal lenght (mm) x image height (pixel))
GSD_x=((sensor_width*altitude*100)/(focal_length*image_width))
GSD_y=((sensor_height*altitude*100)/(focal_length*image_height))
return (GSD_x,GSD_y)
#Read alt.csv
with open('alt.csv', 'r') as csvfile:
alt_list = [line.rstrip('\n') for line in csvfile]
#chose last value in alt_list
altitude=int(alt_list[-1]) #in meters
multiplier=GSD(altitude) #cm/pixel
move_coordinates=np.asarray(closest_driveway)*np.asarray(multiplier)*40 #40 is the center of the 80x80 superpixel
print(closest_driveway)
print(multiplier)
print(move_coordinates)
# Write to CSV
import csv
with open('coordinates.csv', 'a', newline='') as csvfile:
filewriter = csv.writer(csvfile, delimiter=',')
filewriter.writerow(move_coordinates)
```
| github_jupyter |
# DB2 Jupyter Notebook Extensions
Version: 2021-08-23
This code is imported as a Jupyter notebook extension in any notebooks you create with DB2 code in it. Place the following line of code in any notebook that you want to use these commands with:
<pre>
%run db2.ipynb
</pre>
This code defines a Jupyter/Python magic command called `%sql` which allows you to execute DB2 specific calls to
the database. There are other packages available for manipulating databases, but this one has been specifically
designed for demonstrating a number of the SQL features available in DB2.
There are two ways of executing the `%sql` command. A single line SQL statement would use the
line format of the magic command:
<pre>
%sql SELECT * FROM EMPLOYEE
</pre>
If you have a large block of sql then you would place the %%sql command at the beginning of the block and then
place the SQL statements into the remainder of the block. Using this form of the `%%sql` statement means that the
notebook cell can only contain SQL and no other statements.
<pre>
%%sql
SELECT * FROM EMPLOYEE
ORDER BY LASTNAME
</pre>
You can have multiple lines in the SQL block (`%%sql`). The default SQL delimiter is the semi-column (`;`).
If you have scripts (triggers, procedures, functions) that use the semi-colon as part of the script, you
will need to use the `-d` option to change the delimiter to an at "`@`" sign.
<pre>
%%sql -d
SELECT * FROM EMPLOYEE
@
CREATE PROCEDURE ...
@
</pre>
The `%sql` command allows most DB2 commands to execute and has a special version of the CONNECT statement.
A CONNECT by itself will attempt to reconnect to the database using previously used settings. If it cannot
connect, it will prompt the user for additional information.
The CONNECT command has the following format:
<pre>
%sql CONNECT TO <database> USER <userid> USING <password | ?> HOST <ip address> PORT <port number>
</pre>
If you use a "`?`" for the password field, the system will prompt you for a password. This avoids typing the
password as clear text on the screen. If a connection is not successful, the system will print the error
message associated with the connect request.
If the connection is successful, the parameters are saved on your system and will be used the next time you
run a SQL statement, or when you issue the %sql CONNECT command with no parameters.
In addition to the -d option, there are a number different options that you can specify at the beginning of
the SQL:
- `-d, -delim` - Change SQL delimiter to "`@`" from "`;`"
- `-q, -quiet` - Quiet results - no messages returned from the function
- `-r, -array` - Return the result set as an array of values instead of a dataframe
- `-t, -time` - Time the following SQL statement and return the number of times it executes in 1 second
- `-j` - Format the first character column of the result set as a JSON record
- `-json` - Return result set as an array of json records
- `-a, -all` - Return all rows in answer set and do not limit display
- `-grid` - Display the results in a scrollable grid
- `-pb, -bar` - Plot the results as a bar chart
- `-pl, -line` - Plot the results as a line chart
- `-pp, -pie` - Plot the results as a pie chart
- `-e, -echo` - Any macro expansions are displayed in an output box
- `-sampledata` - Create and load the EMPLOYEE and DEPARTMENT tables
<p>
You can pass python variables to the `%sql` command by using the `{}` braces with the name of the
variable inbetween. Note that you will need to place proper punctuation around the variable in the event the
SQL command requires it. For instance, the following example will find employee '000010' in the EMPLOYEE table.
<pre>
empno = '000010'
%sql SELECT LASTNAME FROM EMPLOYEE WHERE EMPNO='{empno}'
</pre>
The other option is to use parameter markers. What you would need to do is use the name of the variable with a colon in front of it and the program will prepare the statement and then pass the variable to Db2 when the statement is executed. This allows you to create complex strings that might contain quote characters and other special characters and not have to worry about enclosing the string with the correct quotes. Note that you do not place the quotes around the variable even though it is a string.
<pre>
empno = '000020'
%sql SELECT LASTNAME FROM EMPLOYEE WHERE EMPNO=:empno
</pre>
## Development SQL
The previous set of `%sql` and `%%sql` commands deals with SQL statements and commands that are run in an interactive manner. There is a class of SQL commands that are more suited to a development environment where code is iterated or requires changing input. The commands that are associated with this form of SQL are:
- AUTOCOMMIT
- COMMIT/ROLLBACK
- PREPARE
- EXECUTE
Autocommit is the default manner in which SQL statements are executed. At the end of the successful completion of a statement, the results are commited to the database. There is no concept of a transaction where multiple DML/DDL statements are considered one transaction. The `AUTOCOMMIT` command allows you to turn autocommit `OFF` or `ON`. This means that the set of SQL commands run after the `AUTOCOMMIT OFF` command are executed are not commited to the database until a `COMMIT` or `ROLLBACK` command is issued.
`COMMIT` (`WORK`) will finalize all of the transactions (`COMMIT`) to the database and `ROLLBACK` will undo all of the changes. If you issue a `SELECT` statement during the execution of your block, the results will reflect all of your changes. If you `ROLLBACK` the transaction, the changes will be lost.
`PREPARE` is typically used in a situation where you want to repeatidly execute a SQL statement with different variables without incurring the SQL compilation overhead. For instance:
```
x = %sql PREPARE SELECT LASTNAME FROM EMPLOYEE WHERE EMPNO=?
for y in ['000010','000020','000030']:
%sql execute :x using :y
```
`EXECUTE` is used to execute a previously compiled statement.
To retrieve the error codes that might be associated with any SQL call, the following variables are updated after every call:
* SQLCODE
* SQLSTATE
* SQLERROR - Full error message retrieved from Db2
### Install Db2 Python Driver
If the ibm_db driver is not installed on your system, the subsequent Db2 commands will fail. In order to install the Db2 driver, issue the following command from a Jupyter notebook cell:
```
!pip install --user ibm_db
```
### Db2 Jupyter Extensions
This section of code has the import statements and global variables defined for the remainder of the functions.
```
#
# Set up Jupyter MAGIC commands "sql".
# %sql will return results from a DB2 select statement or execute a DB2 command
#
# IBM 2021: George Baklarz
# Version 2021-07-13
#
from __future__ import print_function
from IPython.display import HTML as pHTML, Image as pImage, display as pdisplay, Javascript as Javascript
from IPython.core.magic import (Magics, magics_class, line_magic,
cell_magic, line_cell_magic, needs_local_scope)
import ibm_db
import pandas
import ibm_db_dbi
import json
import matplotlib
import matplotlib.pyplot as plt
import getpass
import os
import pickle
import time
import sys
import re
import warnings
warnings.filterwarnings("ignore")
# Python Hack for Input between 2 and 3
try:
input = raw_input
except NameError:
pass
_settings = {
"maxrows" : 10,
"maxgrid" : 5,
"runtime" : 1,
"display" : "PANDAS",
"database" : "",
"hostname" : "localhost",
"port" : "50000",
"protocol" : "TCPIP",
"uid" : "DB2INST1",
"pwd" : "password",
"ssl" : ""
}
_environment = {
"jupyter" : True,
"qgrid" : True
}
_display = {
'fullWidthRows': True,
'syncColumnCellResize': True,
'forceFitColumns': False,
'defaultColumnWidth': 150,
'rowHeight': 28,
'enableColumnReorder': False,
'enableTextSelectionOnCells': True,
'editable': False,
'autoEdit': False,
'explicitInitialization': True,
'maxVisibleRows': 5,
'minVisibleRows': 5,
'sortable': True,
'filterable': False,
'highlightSelectedCell': False,
'highlightSelectedRow': True
}
# Connection settings for statements
_connected = False
_hdbc = None
_hdbi = None
_stmt = []
_stmtID = []
_stmtSQL = []
_vars = {}
_macros = {}
_flags = []
_debug = False
# Db2 Error Messages and Codes
sqlcode = 0
sqlstate = "0"
sqlerror = ""
sqlelapsed = 0
# Check to see if QGrid is installed
try:
import qgrid
qgrid.set_defaults(grid_options=_display)
except:
_environment['qgrid'] = False
# Check if we are running in iPython or Jupyter
try:
if (get_ipython().config == {}):
_environment['jupyter'] = False
_environment['qgrid'] = False
else:
_environment['jupyter'] = True
except:
_environment['jupyter'] = False
_environment['qgrid'] = False
```
## Options
There are four options that can be set with the **`%sql`** command. These options are shown below with the default value shown in parenthesis.
- **`MAXROWS n (10)`** - The maximum number of rows that will be displayed before summary information is shown. If the answer set is less than this number of rows, it will be completely shown on the screen. If the answer set is larger than this amount, only the first 5 rows and last 5 rows of the answer set will be displayed. If you want to display a very large answer set, you may want to consider using the grid option `-g` to display the results in a scrollable table. If you really want to show all results then setting MAXROWS to -1 will return all output.
- **`MAXGRID n (5)`** - The maximum size of a grid display. When displaying a result set in a grid `-g`, the default size of the display window is 5 rows. You can set this to a larger size so that more rows are shown on the screen. Note that the minimum size always remains at 5 which means that if the system is unable to display your maximum row size it will reduce the table display until it fits.
- **`DISPLAY PANDAS | GRID (PANDAS)`** - Display the results as a PANDAS dataframe (default) or as a scrollable GRID
- **`RUNTIME n (1)`** - When using the timer option on a SQL statement, the statement will execute for **`n`** number of seconds. The result that is returned is the number of times the SQL statement executed rather than the execution time of the statement. The default value for runtime is one second, so if the SQL is very complex you will need to increase the run time.
- **`LIST`** - Display the current settings
To set an option use the following syntax:
```
%sql option option_name value option_name value ....
```
The following example sets all options:
```
%sql option maxrows 100 runtime 2 display grid maxgrid 10
```
The values will **not** be saved between Jupyter notebooks sessions. If you need to retrieve the current options values, use the LIST command as the only argument:
```
%sql option list
```
```
def setOptions(inSQL):
global _settings, _display
cParms = inSQL.split()
cnt = 0
while cnt < len(cParms):
if cParms[cnt].upper() == 'MAXROWS':
if cnt+1 < len(cParms):
try:
_settings["maxrows"] = int(cParms[cnt+1])
except Exception as err:
errormsg("Invalid MAXROWS value provided.")
pass
cnt = cnt + 1
else:
errormsg("No maximum rows specified for the MAXROWS option.")
return
elif cParms[cnt].upper() == 'MAXGRID':
if cnt+1 < len(cParms):
try:
maxgrid = int(cParms[cnt+1])
if (maxgrid <= 5): # Minimum window size is 5
maxgrid = 5
_display["maxVisibleRows"] = int(cParms[cnt+1])
try:
import qgrid
qgrid.set_defaults(grid_options=_display)
except:
_environment['qgrid'] = False
except Exception as err:
errormsg("Invalid MAXGRID value provided.")
pass
cnt = cnt + 1
else:
errormsg("No maximum rows specified for the MAXROWS option.")
return
elif cParms[cnt].upper() == 'RUNTIME':
if cnt+1 < len(cParms):
try:
_settings["runtime"] = int(cParms[cnt+1])
except Exception as err:
errormsg("Invalid RUNTIME value provided.")
pass
cnt = cnt + 1
else:
errormsg("No value provided for the RUNTIME option.")
return
elif cParms[cnt].upper() == 'DISPLAY':
if cnt+1 < len(cParms):
if (cParms[cnt+1].upper() == 'GRID'):
_settings["display"] = 'GRID'
elif (cParms[cnt+1].upper() == 'PANDAS'):
_settings["display"] = 'PANDAS'
else:
errormsg("Invalid DISPLAY value provided.")
cnt = cnt + 1
else:
errormsg("No value provided for the DISPLAY option.")
return
elif (cParms[cnt].upper() == 'LIST'):
print("(MAXROWS) Maximum number of rows displayed: " + str(_settings["maxrows"]))
print("(MAXGRID) Maximum grid display size: " + str(_settings["maxgrid"]))
print("(RUNTIME) How many seconds to a run a statement for performance testing: " + str(_settings["runtime"]))
print("(DISPLAY) Use PANDAS or GRID display format for output: " + _settings["display"])
return
else:
cnt = cnt + 1
save_settings()
```
### SQL Help
The calling format of this routine is:
```
sqlhelp()
```
This code displays help related to the %sql magic command. This help is displayed when you issue a %sql or %%sql command by itself, or use the %sql -h flag.
```
def sqlhelp():
global _environment
if (_environment["jupyter"] == True):
sd = '<td style="text-align:left;">'
ed1 = '</td>'
ed2 = '</td>'
sh = '<th style="text-align:left;">'
eh1 = '</th>'
eh2 = '</th>'
sr = '<tr>'
er = '</tr>'
helpSQL = """
<h3>SQL Options</h3>
<p>The following options are available as part of a SQL statement. The options are always preceded with a
minus sign (i.e. -q).
<table>
{sr}
{sh}Option{eh1}{sh}Description{eh2}
{er}
{sr}
{sd}a, all{ed1}{sd}Return all rows in answer set and do not limit display{ed2}
{er}
{sr}
{sd}d{ed1}{sd}Change SQL delimiter to "@" from ";"{ed2}
{er}
{sr}
{sd}e, echo{ed1}{sd}Echo the SQL command that was generated after macro and variable substituion.{ed2}
{er}
{sr}
{sd}h, help{ed1}{sd}Display %sql help information.{ed2}
{er}
{sr}
{sd}j{ed1}{sd}Create a pretty JSON representation. Only the first column is formatted{ed2}
{er}
{sr}
{sd}json{ed1}{sd}Retrieve the result set as a JSON record{ed2}
{er}
{sr}
{sd}pb, bar{ed1}{sd}Plot the results as a bar chart{ed2}
{er}
{sr}
{sd}pl, line{ed1}{sd}Plot the results as a line chart{ed2}
{er}
{sr}
{sd}pp, pie{ed1}{sd}Plot Pie: Plot the results as a pie chart{ed2}
{er}
{sr}
{sd}q, quiet{ed1}{sd}Quiet results - no answer set or messages returned from the function{ed2}
{er}
{sr}
{sd}r, array{ed1}{sd}Return the result set as an array of values{ed2}
{er}
{sr}
{sd}sampledata{ed1}{sd}Create and load the EMPLOYEE and DEPARTMENT tables{ed2}
{er}
{sr}
{sd}t,time{ed1}{sd}Time the following SQL statement and return the number of times it executes in 1 second{ed2}
{er}
{sr}
{sd}grid{ed1}{sd}Display the results in a scrollable grid{ed2}
{er}
</table>
"""
else:
helpSQL = """
SQL Options
The following options are available as part of a SQL statement. Options are always
preceded with a minus sign (i.e. -q).
Option Description
a, all Return all rows in answer set and do not limit display
d Change SQL delimiter to "@" from ";"
e, echo Echo the SQL command that was generated after substitution
h, help Display %sql help information
j Create a pretty JSON representation. Only the first column is formatted
json Retrieve the result set as a JSON record
pb, bar Plot the results as a bar chart
pl, line Plot the results as a line chart
pp, pie Plot Pie: Plot the results as a pie chart
q, quiet Quiet results - no answer set or messages returned from the function
r, array Return the result set as an array of values
sampledata Create and load the EMPLOYEE and DEPARTMENT tables
t,time Time the SQL statement and return the execution count per second
grid Display the results in a scrollable grid
"""
helpSQL = helpSQL.format(**locals())
if (_environment["jupyter"] == True):
pdisplay(pHTML(helpSQL))
else:
print(helpSQL)
```
### Connection Help
The calling format of this routine is:
```
connected_help()
```
This code displays help related to the CONNECT command. This code is displayed when you issue a %sql CONNECT command with no arguments or you are running a SQL statement and there isn't any connection to a database yet.
```
def connected_help():
sd = '<td style="text-align:left;">'
ed = '</td>'
sh = '<th style="text-align:left;">'
eh = '</th>'
sr = '<tr>'
er = '</tr>'
if (_environment['jupyter'] == True):
helpConnect = """
<h3>Connecting to Db2</h3>
<p>The CONNECT command has the following format:
<p>
<pre>
%sql CONNECT TO <database> USER <userid> USING <password|?> HOST <ip address> PORT <port number> <SSL>
%sql CONNECT CREDENTIALS <varname>
%sql CONNECT CLOSE
%sql CONNECT RESET
%sql CONNECT PROMPT - use this to be prompted for values
</pre>
<p>
If you use a "?" for the password field, the system will prompt you for a password. This avoids typing the
password as clear text on the screen. If a connection is not successful, the system will print the error
message associated with the connect request.
<p>
The <b>CREDENTIALS</b> option allows you to use credentials that are supplied by Db2 on Cloud instances.
The credentials can be supplied as a variable and if successful, the variable will be saved to disk
for future use. If you create another notebook and use the identical syntax, if the variable
is not defined, the contents on disk will be used as the credentials. You should assign the
credentials to a variable that represents the database (or schema) that you are communicating with.
Using familiar names makes it easier to remember the credentials when connecting.
<p>
<b>CONNECT CLOSE</b> will close the current connection, but will not reset the database parameters. This means that
if you issue the CONNECT command again, the system should be able to reconnect you to the database.
<p>
<b>CONNECT RESET</b> will close the current connection and remove any information on the connection. You will need
to issue a new CONNECT statement with all of the connection information.
<p>
If the connection is successful, the parameters are saved on your system and will be used the next time you
run an SQL statement, or when you issue the %sql CONNECT command with no parameters.
<p>If you issue CONNECT RESET, all of the current values will be deleted and you will need to
issue a new CONNECT statement.
<p>A CONNECT command without any parameters will attempt to re-connect to the previous database you
were using. If the connection could not be established, the program to prompt you for
the values. To cancel the connection attempt, enter a blank value for any of the values. The connection
panel will request the following values in order to connect to Db2:
<table>
{sr}
{sh}Setting{eh}
{sh}Description{eh}
{er}
{sr}
{sd}Database{ed}{sd}Database name you want to connect to.{ed}
{er}
{sr}
{sd}Hostname{ed}
{sd}Use localhost if Db2 is running on your own machine, but this can be an IP address or host name.
{er}
{sr}
{sd}PORT{ed}
{sd}The port to use for connecting to Db2. This is usually 50000.{ed}
{er}
{sr}
{sd}SSL{ed}
{sd}If you are connecting to a secure port (50001) with SSL then you must include this keyword in the connect string.{ed}
{sr}
{sd}Userid{ed}
{sd}The userid to use when connecting (usually DB2INST1){ed}
{er}
{sr}
{sd}Password{ed}
{sd}No password is provided so you have to enter a value{ed}
{er}
</table>
"""
else:
helpConnect = """\
Connecting to Db2
The CONNECT command has the following format:
%sql CONNECT TO database USER userid USING password | ?
HOST ip address PORT port number SSL
%sql CONNECT CREDENTIALS varname
%sql CONNECT CLOSE
%sql CONNECT RESET
If you use a "?" for the password field, the system will prompt you for a password.
This avoids typing the password as clear text on the screen. If a connection is
not successful, the system will print the error message associated with the connect
request.
The CREDENTIALS option allows you to use credentials that are supplied by Db2 on
Cloud instances. The credentials can be supplied as a variable and if successful,
the variable will be saved to disk for future use. If you create another notebook
and use the identical syntax, if the variable is not defined, the contents on disk
will be used as the credentials. You should assign the credentials to a variable
that represents the database (or schema) that you are communicating with. Using
familiar names makes it easier to remember the credentials when connecting.
CONNECT CLOSE will close the current connection, but will not reset the database
parameters. This means that if you issue the CONNECT command again, the system
should be able to reconnect you to the database.
CONNECT RESET will close the current connection and remove any information on the
connection. You will need to issue a new CONNECT statement with all of the connection
information.
If the connection is successful, the parameters are saved on your system and will be
used the next time you run an SQL statement, or when you issue the %sql CONNECT
command with no parameters. If you issue CONNECT RESET, all of the current values
will be deleted and you will need to issue a new CONNECT statement.
A CONNECT command without any parameters will attempt to re-connect to the previous
database you were using. If the connection could not be established, the program to
prompt you for the values. To cancel the connection attempt, enter a blank value for
any of the values. The connection panel will request the following values in order
to connect to Db2:
Setting Description
Database Database name you want to connect to
Hostname Use localhost if Db2 is running on your own machine, but this can
be an IP address or host name.
PORT The port to use for connecting to Db2. This is usually 50000.
Userid The userid to use when connecting (usually DB2INST1)
Password No password is provided so you have to enter a value
SSL Include this keyword to indicate you are connecting via SSL (usually port 50001)
"""
helpConnect = helpConnect.format(**locals())
if (_environment['jupyter'] == True):
pdisplay(pHTML(helpConnect))
else:
print(helpConnect)
```
### Prompt for Connection Information
If you are running an SQL statement and have not yet connected to a database, the %sql command will prompt you for connection information. In order to connect to a database, you must supply:
- Database name
- Host name (IP address or name)
- Port number
- Userid
- Password
- Secure socket
The routine is called without any parameters:
```
connected_prompt()
```
```
# Prompt for Connection information
def connected_prompt():
global _settings
_database = ''
_hostname = ''
_port = ''
_uid = ''
_pwd = ''
_ssl = ''
print("Enter the database connection details (Any empty value will cancel the connection)")
_database = input("Enter the database name: ");
if (_database.strip() == ""): return False
_hostname = input("Enter the HOST IP address or symbolic name: ");
if (_hostname.strip() == ""): return False
_port = input("Enter the PORT number: ");
if (_port.strip() == ""): return False
_ssl = input("Is this a secure (SSL) port (y or n)");
if (_ssl.strip() == ""): return False
if (_ssl == "n"):
_ssl = ""
else:
_ssl = "Security=SSL;"
_uid = input("Enter Userid on the DB2 system: ").upper();
if (_uid.strip() == ""): return False
_pwd = getpass.getpass("Password [password]: ");
if (_pwd.strip() == ""): return False
_settings["database"] = _database.strip()
_settings["hostname"] = _hostname.strip()
_settings["port"] = _port.strip()
_settings["uid"] = _uid.strip()
_settings["pwd"] = _pwd.strip()
_settings["ssl"] = _ssl.strip()
_settings["maxrows"] = 10
_settings["maxgrid"] = 5
_settings["runtime"] = 1
return True
# Split port and IP addresses
def split_string(in_port,splitter=":"):
# Split input into an IP address and Port number
global _settings
checkports = in_port.split(splitter)
ip = checkports[0]
if (len(checkports) > 1):
port = checkports[1]
else:
port = None
return ip, port
```
### Connect Syntax Parser
The parseConnect routine is used to parse the CONNECT command that the user issued within the %sql command. The format of the command is:
```
parseConnect(inSQL)
```
The inSQL string contains the CONNECT keyword with some additional parameters. The format of the CONNECT command is one of:
```
CONNECT RESET
CONNECT CLOSE
CONNECT CREDENTIALS <variable>
CONNECT TO database USER userid USING password HOST hostname PORT portnumber <SSL>
```
If you have credentials available from Db2 on Cloud, place the contents of the credentials into a variable and then use the `CONNECT CREDENTIALS <var>` syntax to connect to the database.
In addition, supplying a question mark (?) for password will result in the program prompting you for the password rather than having it as clear text in your scripts.
When all of the information is checked in the command, the db2_doConnect function is called to actually do the connection to the database.
```
# Parse the CONNECT statement and execute if possible
def parseConnect(inSQL,local_ns):
global _settings, _connected
_connected = False
cParms = inSQL.split()
cnt = 0
_settings["ssl"] = ""
while cnt < len(cParms):
if cParms[cnt].upper() == 'TO':
if cnt+1 < len(cParms):
_settings["database"] = cParms[cnt+1].upper()
cnt = cnt + 1
else:
errormsg("No database specified in the CONNECT statement")
return
elif cParms[cnt].upper() == "SSL":
_settings["ssl"] = "Security=SSL;"
cnt = cnt + 1
elif cParms[cnt].upper() == 'CREDENTIALS':
if cnt+1 < len(cParms):
credentials = cParms[cnt+1]
tempid = eval(credentials,local_ns)
if (isinstance(tempid,dict) == False):
errormsg("The CREDENTIALS variable (" + credentials + ") does not contain a valid Python dictionary (JSON object)")
return
if (tempid == None):
fname = credentials + ".pickle"
try:
with open(fname,'rb') as f:
_id = pickle.load(f)
except:
errormsg("Unable to find credential variable or file.")
return
else:
_id = tempid
try:
_settings["database"] = _id["db"]
_settings["hostname"] = _id["hostname"]
_settings["port"] = _id["port"]
_settings["uid"] = _id["username"]
_settings["pwd"] = _id["password"]
try:
fname = credentials + ".pickle"
with open(fname,'wb') as f:
pickle.dump(_id,f)
except:
errormsg("Failed trying to write Db2 Credentials.")
return
except:
errormsg("Credentials file is missing information. db/hostname/port/username/password required.")
return
else:
errormsg("No Credentials name supplied")
return
cnt = cnt + 1
elif cParms[cnt].upper() == 'USER':
if cnt+1 < len(cParms):
_settings["uid"] = cParms[cnt+1].upper()
cnt = cnt + 1
else:
errormsg("No userid specified in the CONNECT statement")
return
elif cParms[cnt].upper() == 'USING':
if cnt+1 < len(cParms):
_settings["pwd"] = cParms[cnt+1]
if (_settings["pwd"] == '?'):
_settings["pwd"] = getpass.getpass("Password [password]: ") or "password"
cnt = cnt + 1
else:
errormsg("No password specified in the CONNECT statement")
return
elif cParms[cnt].upper() == 'HOST':
if cnt+1 < len(cParms):
hostport = cParms[cnt+1].upper()
ip, port = split_string(hostport)
if (port == None): _settings["port"] = "50000"
_settings["hostname"] = ip
cnt = cnt + 1
else:
errormsg("No hostname specified in the CONNECT statement")
return
elif cParms[cnt].upper() == 'PORT':
if cnt+1 < len(cParms):
_settings["port"] = cParms[cnt+1].upper()
cnt = cnt + 1
else:
errormsg("No port specified in the CONNECT statement")
return
elif cParms[cnt].upper() == 'PROMPT':
if (connected_prompt() == False):
print("Connection canceled.")
return
else:
cnt = cnt + 1
elif cParms[cnt].upper() in ('CLOSE','RESET') :
try:
result = ibm_db.close(_hdbc)
_hdbi.close()
except:
pass
success("Connection closed.")
if cParms[cnt].upper() == 'RESET':
_settings["database"] = ''
return
else:
cnt = cnt + 1
_ = db2_doConnect()
```
### Connect to Db2
The db2_doConnect routine is called when a connection needs to be established to a Db2 database. The command does not require any parameters since it relies on the settings variable which contains all of the information it needs to connect to a Db2 database.
```
db2_doConnect()
```
There are 4 additional variables that are used throughout the routines to stay connected with the Db2 database. These variables are:
- hdbc - The connection handle to the database
- hstmt - A statement handle used for executing SQL statements
- connected - A flag that tells the program whether or not we are currently connected to a database
- runtime - Used to tell %sql the length of time (default 1 second) to run a statement when timing it
The only database driver that is used in this program is the IBM DB2 ODBC DRIVER. This driver needs to be loaded on the system that is connecting to Db2. The Jupyter notebook that is built by this system installs the driver for you so you shouldn't have to do anything other than build the container.
If the connection is successful, the connected flag is set to True. Any subsequent %sql call will check to see if you are connected and initiate another prompted connection if you do not have a connection to a database.
```
def db2_doConnect():
global _hdbc, _hdbi, _connected, _runtime
global _settings
if _connected == False:
if len(_settings["database"]) == 0:
return False
dsn = (
"DRIVER={{IBM DB2 ODBC DRIVER}};"
"DATABASE={0};"
"HOSTNAME={1};"
"PORT={2};"
"PROTOCOL=TCPIP;"
"UID={3};"
"PWD={4};{5}").format(_settings["database"],
_settings["hostname"],
_settings["port"],
_settings["uid"],
_settings["pwd"],
_settings["ssl"])
# Get a database handle (hdbc) and a statement handle (hstmt) for subsequent access to DB2
try:
_hdbc = ibm_db.connect(dsn, "", "")
except Exception as err:
db2_error(False,True) # errormsg(str(err))
_connected = False
_settings["database"] = ''
return False
try:
_hdbi = ibm_db_dbi.Connection(_hdbc)
except Exception as err:
db2_error(False,True) # errormsg(str(err))
_connected = False
_settings["database"] = ''
return False
_connected = True
# Save the values for future use
save_settings()
success("Connection successful.")
return True
```
### Load/Save Settings
There are two routines that load and save settings between Jupyter notebooks. These routines are called without any parameters.
```
load_settings() save_settings()
```
There is a global structure called settings which contains the following fields:
```
_settings = {
"maxrows" : 10,
"maxgrid" : 5,
"runtime" : 1,
"display" : "TEXT",
"database" : "",
"hostname" : "localhost",
"port" : "50000",
"protocol" : "TCPIP",
"uid" : "DB2INST1",
"pwd" : "password"
}
```
The information in the settings structure is used for re-connecting to a database when you start up a Jupyter notebook. When the session is established for the first time, the load_settings() function is called to get the contents of the pickle file (db2connect.pickle, a Jupyter session file) that will be used for the first connection to the database. Whenever a new connection is made, the file is updated with the save_settings() function.
```
def load_settings():
# This routine will load the settings from the previous session if they exist
global _settings
fname = "db2connect.pickle"
try:
with open(fname,'rb') as f:
_settings = pickle.load(f)
# Reset runtime to 1 since it would be unexpected to keep the same value between connections
_settings["runtime"] = 1
_settings["maxgrid"] = 5
except:
pass
return
def save_settings():
# This routine will save the current settings if they exist
global _settings
fname = "db2connect.pickle"
try:
with open(fname,'wb') as f:
pickle.dump(_settings,f)
except:
errormsg("Failed trying to write Db2 Configuration Information.")
return
```
### Error and Message Functions
There are three types of messages that are thrown by the %db2 magic command. The first routine will print out a success message with no special formatting:
```
success(message)
```
The second message is used for displaying an error message that is not associated with a SQL error. This type of error message is surrounded with a red box to highlight the problem. Note that the success message has code that has been commented out that could also show a successful return code with a green box.
```
errormsg(message)
```
The final error message is based on an error occuring in the SQL code that was executed. This code will parse the message returned from the ibm_db interface and parse it to return only the error message portion (and not all of the wrapper code from the driver).
```
db2_error(quiet,connect=False)
```
The quiet flag is passed to the db2_error routine so that messages can be suppressed if the user wishes to ignore them with the -q flag. A good example of this is dropping a table that does not exist. We know that an error will be thrown so we can ignore it. The information that the db2_error routine gets is from the stmt_errormsg() function from within the ibm_db driver. The db2_error function should only be called after a SQL failure otherwise there will be no diagnostic information returned from stmt_errormsg().
If the connect flag is True, the routine will get the SQLSTATE and SQLCODE from the connection error message rather than a statement error message.
```
def db2_error(quiet,connect=False):
global sqlerror, sqlcode, sqlstate, _environment
try:
if (connect == False):
errmsg = ibm_db.stmt_errormsg().replace('\r',' ')
errmsg = errmsg[errmsg.rfind("]")+1:].strip()
else:
errmsg = ibm_db.conn_errormsg().replace('\r',' ')
errmsg = errmsg[errmsg.rfind("]")+1:].strip()
sqlerror = errmsg
msg_start = errmsg.find("SQLSTATE=")
if (msg_start != -1):
msg_end = errmsg.find(" ",msg_start)
if (msg_end == -1):
msg_end = len(errmsg)
sqlstate = errmsg[msg_start+9:msg_end]
else:
sqlstate = "0"
msg_start = errmsg.find("SQLCODE=")
if (msg_start != -1):
msg_end = errmsg.find(" ",msg_start)
if (msg_end == -1):
msg_end = len(errmsg)
sqlcode = errmsg[msg_start+8:msg_end]
try:
sqlcode = int(sqlcode)
except:
pass
else:
sqlcode = 0
except:
errmsg = "Unknown error."
sqlcode = -99999
sqlstate = "-99999"
sqlerror = errmsg
return
msg_start = errmsg.find("SQLSTATE=")
if (msg_start != -1):
msg_end = errmsg.find(" ",msg_start)
if (msg_end == -1):
msg_end = len(errmsg)
sqlstate = errmsg[msg_start+9:msg_end]
else:
sqlstate = "0"
msg_start = errmsg.find("SQLCODE=")
if (msg_start != -1):
msg_end = errmsg.find(" ",msg_start)
if (msg_end == -1):
msg_end = len(errmsg)
sqlcode = errmsg[msg_start+8:msg_end]
try:
sqlcode = int(sqlcode)
except:
pass
else:
sqlcode = 0
if quiet == True: return
if (errmsg == ""): return
html = '<p><p style="border:2px; border-style:solid; border-color:#FF0000; background-color:#ffe6e6; padding: 1em;">'
if (_environment["jupyter"] == True):
pdisplay(pHTML(html+errmsg+"</p>"))
else:
print(errmsg)
# Print out an error message
def errormsg(message):
global _environment
if (message != ""):
html = '<p><p style="border:2px; border-style:solid; border-color:#FF0000; background-color:#ffe6e6; padding: 1em;">'
if (_environment["jupyter"] == True):
pdisplay(pHTML(html + message + "</p>"))
else:
print(message)
def success(message):
if (message != ""):
print(message)
return
def debug(message,error=False):
global _environment
if (_environment["jupyter"] == True):
spacer = "<br>" + " "
else:
spacer = "\n "
if (message != ""):
lines = message.split('\n')
msg = ""
indent = 0
for line in lines:
delta = line.count("(") - line.count(")")
if (msg == ""):
msg = line
indent = indent + delta
else:
if (delta < 0): indent = indent + delta
msg = msg + spacer * (indent*2) + line
if (delta > 0): indent = indent + delta
if (indent < 0): indent = 0
if (error == True):
html = '<p><pre style="font-family: monospace; border:2px; border-style:solid; border-color:#FF0000; background-color:#ffe6e6; padding: 1em;">'
else:
html = '<p><pre style="font-family: monospace; border:2px; border-style:solid; border-color:#008000; background-color:#e6ffe6; padding: 1em;">'
if (_environment["jupyter"] == True):
pdisplay(pHTML(html + msg + "</pre></p>"))
else:
print(msg)
return
```
## Macro Processor
A macro is used to generate SQL to be executed by overriding or creating a new keyword. For instance, the base `%sql` command does not understand the `LIST TABLES` command which is usually used in conjunction with the `CLP` processor. Rather than specifically code this in the base `db2.ipynb` file, we can create a macro that can execute this code for us.
There are three routines that deal with macros.
- checkMacro is used to find the macro calls in a string. All macros are sent to parseMacro for checking.
- runMacro will evaluate the macro and return the string to the parse
- subvars is used to track the variables used as part of a macro call.
- setMacro is used to catalog a macro
### Set Macro
This code will catalog a macro call.
```
def setMacro(inSQL,parms):
global _macros
names = parms.split()
if (len(names) < 2):
errormsg("No command name supplied.")
return None
macroName = names[1].upper()
_macros[macroName] = inSQL
return
```
### Check Macro
This code will check to see if there is a macro command in the SQL. It will take the SQL that is supplied and strip out three values: the first and second keywords, and the remainder of the parameters.
For instance, consider the following statement:
```
CREATE DATABASE GEORGE options....
```
The name of the macro that we want to run is called `CREATE`. We know that there is a SQL command called `CREATE` but this code will call the macro first to see if needs to run any special code. For instance, `CREATE DATABASE` is not part of the `db2.ipynb` syntax, but we can add it in by using a macro.
The check macro logic will strip out the subcommand (`DATABASE`) and place the remainder of the string after `DATABASE` in options.
```
def checkMacro(in_sql):
global _macros
if (len(in_sql) == 0): return(in_sql) # Nothing to do
tokens = parseArgs(in_sql,None) # Take the string and reduce into tokens
macro_name = tokens[0].upper() # Uppercase the name of the token
if (macro_name not in _macros):
return(in_sql) # No macro by this name so just return the string
result = runMacro(_macros[macro_name],in_sql,tokens) # Execute the macro using the tokens we found
return(result) # Runmacro will either return the original SQL or the new one
```
### Split Assignment
This routine will return the name of a variable and it's value when the format is x=y. If y is enclosed in quotes, the quotes are removed.
```
def splitassign(arg):
var_name = "null"
var_value = "null"
arg = arg.strip()
eq = arg.find("=")
if (eq != -1):
var_name = arg[:eq].strip()
temp_value = arg[eq+1:].strip()
if (temp_value != ""):
ch = temp_value[0]
if (ch in ["'",'"']):
if (temp_value[-1:] == ch):
var_value = temp_value[1:-1]
else:
var_value = temp_value
else:
var_value = temp_value
else:
var_value = arg
return var_name, var_value
```
### Parse Args
The commands that are used in the macros need to be parsed into their separate tokens. The tokens are separated by blanks and strings that enclosed in quotes are kept together.
```
def parseArgs(argin,_vars):
quoteChar = ""
inQuote = False
inArg = True
args = []
arg = ''
for ch in argin.lstrip():
if (inQuote == True):
if (ch == quoteChar):
inQuote = False
arg = arg + ch #z
else:
arg = arg + ch
elif (ch == "\"" or ch == "\'"): # Do we have a quote
quoteChar = ch
arg = arg + ch #z
inQuote = True
elif (ch == " "):
if (arg != ""):
arg = subvars(arg,_vars)
args.append(arg)
else:
args.append("null")
arg = ""
else:
arg = arg + ch
if (arg != ""):
arg = subvars(arg,_vars)
args.append(arg)
return(args)
```
### Run Macro
This code will execute the body of the macro and return the results for that macro call.
```
def runMacro(script,in_sql,tokens):
result = ""
runIT = True
code = script.split("\n")
level = 0
runlevel = [True,False,False,False,False,False,False,False,False,False]
ifcount = 0
_vars = {}
for i in range(0,len(tokens)):
vstr = str(i)
_vars[vstr] = tokens[i]
if (len(tokens) == 0):
_vars["argc"] = "0"
else:
_vars["argc"] = str(len(tokens)-1)
for line in code:
line = line.strip()
if (line == "" or line == "\n"): continue
if (line[0] == "#"): continue # A comment line starts with a # in the first position of the line
args = parseArgs(line,_vars) # Get all of the arguments
if (args[0] == "if"):
ifcount = ifcount + 1
if (runlevel[level] == False): # You can't execute this statement
continue
level = level + 1
if (len(args) < 4):
print("Macro: Incorrect number of arguments for the if clause.")
return insql
arg1 = args[1]
arg2 = args[3]
if (len(arg2) > 2):
ch1 = arg2[0]
ch2 = arg2[-1:]
if (ch1 in ['"',"'"] and ch1 == ch2):
arg2 = arg2[1:-1].strip()
op = args[2]
if (op in ["=","=="]):
if (arg1 == arg2):
runlevel[level] = True
else:
runlevel[level] = False
elif (op in ["<=","=<"]):
if (arg1 <= arg2):
runlevel[level] = True
else:
runlevel[level] = False
elif (op in [">=","=>"]):
if (arg1 >= arg2):
runlevel[level] = True
else:
runlevel[level] = False
elif (op in ["<>","!="]):
if (arg1 != arg2):
runlevel[level] = True
else:
runlevel[level] = False
elif (op in ["<"]):
if (arg1 < arg2):
runlevel[level] = True
else:
runlevel[level] = False
elif (op in [">"]):
if (arg1 > arg2):
runlevel[level] = True
else:
runlevel[level] = False
else:
print("Macro: Unknown comparison operator in the if statement:" + op)
continue
elif (args[0] in ["exit","echo"] and runlevel[level] == True):
msg = ""
for msgline in args[1:]:
if (msg == ""):
msg = subvars(msgline,_vars)
else:
msg = msg + " " + subvars(msgline,_vars)
if (msg != ""):
if (args[0] == "echo"):
debug(msg,error=False)
else:
debug(msg,error=True)
if (args[0] == "exit"): return ''
elif (args[0] == "pass" and runlevel[level] == True):
pass
elif (args[0] == "var" and runlevel[level] == True):
value = ""
for val in args[2:]:
if (value == ""):
value = subvars(val,_vars)
else:
value = value + " " + subvars(val,_vars)
value.strip()
_vars[args[1]] = value
elif (args[0] == 'else'):
if (ifcount == level):
runlevel[level] = not runlevel[level]
elif (args[0] == 'return' and runlevel[level] == True):
return(result)
elif (args[0] == "endif"):
ifcount = ifcount - 1
if (ifcount < level):
level = level - 1
if (level < 0):
print("Macro: Unmatched if/endif pairs.")
return ''
else:
if (runlevel[level] == True):
if (result == ""):
result = subvars(line,_vars)
else:
result = result + "\n" + subvars(line,_vars)
return(result)
```
### Substitute Vars
This routine is used by the runMacro program to track variables that are used within Macros. These are kept separate from the rest of the code.
```
def subvars(script,_vars):
if (_vars == None): return script
remainder = script
result = ""
done = False
while done == False:
bv = remainder.find("{")
if (bv == -1):
done = True
continue
ev = remainder.find("}")
if (ev == -1):
done = True
continue
result = result + remainder[:bv]
vvar = remainder[bv+1:ev]
remainder = remainder[ev+1:]
upper = False
allvars = False
if (vvar[0] == "^"):
upper = True
vvar = vvar[1:]
elif (vvar[0] == "*"):
vvar = vvar[1:]
allvars = True
else:
pass
if (vvar in _vars):
if (upper == True):
items = _vars[vvar].upper()
elif (allvars == True):
try:
iVar = int(vvar)
except:
return(script)
items = ""
sVar = str(iVar)
while sVar in _vars:
if (items == ""):
items = _vars[sVar]
else:
items = items + " " + _vars[sVar]
iVar = iVar + 1
sVar = str(iVar)
else:
items = _vars[vvar]
else:
if (allvars == True):
items = ""
else:
items = "null"
result = result + items
if (remainder != ""):
result = result + remainder
return(result)
```
### SQL Timer
The calling format of this routine is:
```
count = sqlTimer(hdbc, runtime, inSQL)
```
This code runs the SQL string multiple times for one second (by default). The accuracy of the clock is not that great when you are running just one statement, so instead this routine will run the code multiple times for a second to give you an execution count. If you need to run the code for more than one second, the runtime value needs to be set to the number of seconds you want the code to run.
The return result is always the number of times that the code executed. Note, that the program will skip reading the data if it is a SELECT statement so it doesn't included fetch time for the answer set.
```
def sqlTimer(hdbc, runtime, inSQL):
count = 0
t_end = time.time() + runtime
while time.time() < t_end:
try:
stmt = ibm_db.exec_immediate(hdbc,inSQL)
if (stmt == False):
db2_error(flag(["-q","-quiet"]))
return(-1)
ibm_db.free_result(stmt)
except Exception as err:
db2_error(False)
return(-1)
count = count + 1
return(count)
```
### Split Args
This routine takes as an argument a string and then splits the arguments according to the following logic:
* If the string starts with a `(` character, it will check the last character in the string and see if it is a `)` and then remove those characters
* Every parameter is separated by a comma `,` and commas within quotes are ignored
* Each parameter returned will have three values returned - one for the value itself, an indicator which will be either True if it was quoted, or False if not, and True or False if it is numeric.
Example:
```
"abcdef",abcdef,456,"856"
```
Three values would be returned:
```
[abcdef,True,False],[abcdef,False,False],[456,False,True],[856,True,False]
```
Any quoted string will be False for numeric. The way that the parameters are handled are up to the calling program. However, in the case of Db2, the quoted strings must be in single quotes so any quoted parameter using the double quotes `"` must be wrapped with single quotes. There is always a possibility that a string contains single quotes (i.e. O'Connor) so any substituted text should use `''` so that Db2 can properly interpret the string. This routine does not adjust the strings with quotes, and depends on the variable subtitution routine to do that.
```
def splitargs(arguments):
import types
# String the string and remove the ( and ) characters if they at the beginning and end of the string
results = []
step1 = arguments.strip()
if (len(step1) == 0): return(results) # Not much to do here - no args found
if (step1[0] == '('):
if (step1[-1:] == ')'):
step2 = step1[1:-1]
step2 = step2.strip()
else:
step2 = step1
else:
step2 = step1
# Now we have a string without brackets. Start scanning for commas
quoteCH = ""
pos = 0
arg = ""
args = []
while pos < len(step2):
ch = step2[pos]
if (quoteCH == ""): # Are we in a quote?
if (ch in ('"',"'")): # Check to see if we are starting a quote
quoteCH = ch
arg = arg + ch
pos += 1
elif (ch == ","): # Are we at the end of a parameter?
arg = arg.strip()
args.append(arg)
arg = ""
inarg = False
pos += 1
else: # Continue collecting the string
arg = arg + ch
pos += 1
else:
if (ch == quoteCH): # Are we at the end of a quote?
arg = arg + ch # Add the quote to the string
pos += 1 # Increment past the quote
quoteCH = "" # Stop quote checking (maybe!)
else:
pos += 1
arg = arg + ch
if (quoteCH != ""): # So we didn't end our string
arg = arg.strip()
args.append(arg)
elif (arg != ""): # Something left over as an argument
arg = arg.strip()
args.append(arg)
else:
pass
results = []
for arg in args:
result = []
if (len(arg) > 0):
if (arg[0] in ('"',"'")):
value = arg[1:-1]
isString = True
isNumber = False
else:
isString = False
isNumber = False
try:
value = eval(arg)
if (type(value) == int):
isNumber = True
elif (isinstance(value,float) == True):
isNumber = True
else:
value = arg
except:
value = arg
else:
value = ""
isString = False
isNumber = False
result = [value,isString,isNumber]
results.append(result)
return results
```
### DataFrame Table Creation
When using dataframes, it is sometimes useful to use the definition of the dataframe to create a Db2 table. The format of the command is:
```
%sql using <df> create table <table> [with data | columns asis]
```
The value <df> is the name of the dataframe, not the contents (`:df`). The definition of the data types in the dataframe will be used to create the Db2 table using typical Db2 data types rather than generic CLOBs and FLOAT for numeric objects. The two options are used to handle how the conversion is done. If you supply `with data`, the contents of the df will be inserted into the table, otherwise the table is defined only. The column names will be uppercased and special characters (like blanks) will be replaced with underscores. If `columns asis` is specified, the column names will remain the same as in the dataframe, with each name using quotes to guarantee the same spelling as in the DF.
If the table already exists, the command will not run and an error message will be produced.
```
def createDF(hdbc,sqlin,local_ns):
import datetime
import ibm_db
global sqlcode
# Strip apart the command into tokens based on spaces
tokens = sqlin.split()
token_count = len(tokens)
if (token_count < 5): # Not enough parameters
errormsg("Insufficient arguments for USING command. %sql using df create table name [with data | columns asis]")
return
keyword_command = tokens[0].upper()
dfName = tokens[1]
keyword_create = tokens[2].upper()
keyword_table = tokens[3].upper()
table = tokens[4]
if (keyword_create not in ("CREATE","REPLACE") or keyword_table != "TABLE"):
errormsg("Incorrect syntax: %sql using <df> create table <name> [options]")
return
if (token_count % 2 != 1):
errormsg("Insufficient arguments for USING command. %sql using df create table name [with data | columns asis | keep float]")
return
flag_withdata = False
flag_asis = False
flag_float = False
flag_integer = False
limit = -1
if (keyword_create == "REPLACE"):
%sql -q DROP TABLE {table}
for token_idx in range(5,token_count,2):
option_key = tokens[token_idx].upper()
option_val = tokens[token_idx+1].upper()
if (option_key == "WITH" and option_val == "DATA"):
flag_withdata = True
elif (option_key == "COLUMNS" and option_val == "ASIS"):
flag_asis = True
elif (option_key == "KEEP" and option_val == "FLOAT64"):
flag_float = True
elif (option_key == "KEEP" and option_val == "INT64"):
flag_integer = True
elif (option_key == "LIMIT"):
if (option_val.isnumeric() == False):
errormsg("The LIMIT must be a valid number from -1 (unlimited) to the maximun number of rows to insert")
return
limit = int(option_val)
else:
errormsg("Invalid options. Must be either WITH DATA | COLUMNS ASIS | KEEP FLOAT64 | KEEP FLOAT INT64")
return
dfName = tokens[1]
if (dfName not in local_ns):
errormsg("The variable ({dfName}) does not exist in the local variable list.")
return
try:
df_value = eval(dfName,None,local_ns) # globals()[varName] # eval(varName)
except:
errormsg("The variable ({dfName}) does not contain a value.")
return
if (isinstance(df_value,pandas.DataFrame) == False): # Not a Pandas dataframe
errormsg("The variable ({dfName}) is not a Pandas dataframe.")
return
sql = []
columns = dict(df_value.dtypes)
sql.append(f'CREATE TABLE {table} (')
datatypes = []
comma = ""
for column in columns:
datatype = columns[column]
if (datatype == "object"):
datapoint = df_value[column][0]
if (isinstance(datapoint,datetime.datetime)):
type = "TIMESTAMP"
elif (isinstance(datapoint,datetime.time)):
type = "TIME"
elif (isinstance(datapoint,datetime.date)):
type = "DATE"
elif (isinstance(datapoint,float)):
if (flag_float == True):
type = "FLOAT"
else:
type = "DECFLOAT"
elif (isinstance(datapoint,int)):
if (flag_integer == True):
type = "BIGINT"
else:
type = "INTEGER"
elif (isinstance(datapoint,str)):
maxlength = df_value[column].apply(str).apply(len).max()
type = f"VARCHAR({maxlength})"
else:
type = "CLOB"
elif (datatype == "int64"):
if (flag_integer == True):
type = "BIGINT"
else:
type = "INTEGER"
elif (datatype == "float64"):
if (flag_float == True):
type = "FLOAT"
else:
type = "DECFLOAT"
elif (datatype == "datetime64"):
type = "TIMESTAMP"
elif (datatype == "bool"):
type = "BINARY"
else:
type = "CLOB"
datatypes.append(type)
if (flag_asis == False):
if (isinstance(column,str) == False):
column = str(column)
identifier = "ABCDEFGHIJKLMNOPQRSTUVWXYZ0123456789_"
column_name = column.strip().upper()
new_name = ""
for ch in column_name:
if (ch not in identifier):
new_name = new_name + "_"
else:
new_name = new_name + ch
new_name = new_name.lstrip('_').rstrip('_')
if (new_name == "" or new_name[0] not in "ABCDEFGHIJKLMNOPQRSTUVWXYZ"):
new_name = f'"{column}"'
else:
new_name = f'"{column}"'
sql.append(f" {new_name} {type}")
sql.append(")")
sqlcmd = ""
for i in range(0,len(sql)):
if (i > 0 and i < len(sql)-2):
comma = ","
else:
comma = ""
sqlcmd = "{}\n{}{}".format(sqlcmd,sql[i],comma)
print(sqlcmd)
%sql {sqlcmd}
if (sqlcode != 0):
return
if (flag_withdata == True):
autocommit = ibm_db.autocommit(hdbc)
ibm_db.autocommit(hdbc,False)
row_count = 0
insert_sql = ""
rows, cols = df_value.shape
for row in range(0,rows):
insert_row = ""
for col in range(0, cols):
value = df_value.iloc[row][col]
if (datatypes[col] == "CLOB" or "VARCHAR" in datatypes[col]):
value = str(value)
value = addquotes(value,True)
elif (datatypes[col] in ("TIME","DATE","TIMESTAMP")):
value = str(value)
value = addquotes(value,True)
elif (datatypes[col] in ("INTEGER","DECFLOAT","FLOAT","BINARY")):
strvalue = str(value)
if ("NAN" in strvalue.upper()):
value = "NULL"
else:
value = str(value)
value = addquotes(value,True)
if (insert_row == ""):
insert_row = f"{value}"
else:
insert_row = f"{insert_row},{value}"
if (insert_sql == ""):
insert_sql = f"INSERT INTO {table} VALUES ({insert_row})"
else:
insert_sql = f"{insert_sql},({insert_row})"
row_count += 1
if (row_count % 1000 == 0 or row_count == limit):
result = ibm_db.exec_immediate(hdbc, insert_sql) # Run it
if (result == False): # Error executing the code
db2_error(False)
return
ibm_db.commit(hdbc)
print(f"\r{row_count} of {rows} rows inserted.",end="")
insert_sql = ""
if (row_count == limit):
break
if (insert_sql != ""):
result = ibm_db.exec_immediate(hdbc, insert_sql) # Run it
if (result == False): # Error executing the code
db2_error(False)
ibm_db.commit(hdbc)
ibm_db.autocommit(hdbc,autocommit)
print("\nInsert completed.")
return
```
### SQL Parser
The calling format of this routine is:
```
sql_cmd, encoded_sql = sqlParser(sql_input)
```
This code will look at the SQL string that has been passed to it and parse it into two values:
- sql_cmd: First command in the list (so this may not be the actual SQL command)
- encoded_sql: SQL with the parameters removed if there are any (replaced with ? markers)
```
def sqlParser(sqlin,local_ns):
sql_cmd = ""
encoded_sql = sqlin
firstCommand = "(?:^\s*)([a-zA-Z]+)(?:\s+.*|$)"
findFirst = re.match(firstCommand,sqlin)
if (findFirst == None): # We did not find a match so we just return the empty string
return sql_cmd, encoded_sql
cmd = findFirst.group(1)
sql_cmd = cmd.upper()
#
# Scan the input string looking for variables in the format :var. If no : is found just return.
# Var must be alpha+number+_ to be valid
#
if (':' not in sqlin): # A quick check to see if parameters are in here, but not fool-proof!
return sql_cmd, encoded_sql
inVar = False
inQuote = ""
varName = ""
encoded_sql = ""
STRING = 0
NUMBER = 1
LIST = 2
RAW = 3
PANDAS = 5
for ch in sqlin:
if (inVar == True): # We are collecting the name of a variable
if (ch.upper() in "@_ABCDEFGHIJKLMNOPQRSTUVWXYZ0123456789[]"):
varName = varName + ch
continue
else:
if (varName == ""):
encode_sql = encoded_sql + ":"
elif (varName[0] in ('[',']')):
encoded_sql = encoded_sql + ":" + varName
else:
if (ch == '.'): # If the variable name is stopped by a period, assume no quotes are used
flag_quotes = False
else:
flag_quotes = True
varValue, varType = getContents(varName,flag_quotes,local_ns)
if (varType != PANDAS and varValue == None):
encoded_sql = encoded_sql + ":" + varName
else:
if (varType == STRING):
encoded_sql = encoded_sql + varValue
elif (varType == NUMBER):
encoded_sql = encoded_sql + str(varValue)
elif (varType == RAW):
encoded_sql = encoded_sql + varValue
elif (varType == PANDAS):
insertsql = ""
coltypes = varValue.dtypes
rows, cols = varValue.shape
for row in range(0,rows):
insertrow = ""
for col in range(0, cols):
value = varValue.iloc[row][col]
if (coltypes[col] == "object"):
value = str(value)
value = addquotes(value,True)
else:
strvalue = str(value)
if ("NAN" in strvalue.upper()):
value = "NULL"
if (insertrow == ""):
insertrow = f"{value}"
else:
insertrow = f"{insertrow},{value}"
if (insertsql == ""):
insertsql = f"({insertrow})"
else:
insertsql = f"{insertsql},({insertrow})"
encoded_sql = encoded_sql + insertsql
elif (varType == LIST):
start = True
for v in varValue:
if (start == False):
encoded_sql = encoded_sql + ","
if (isinstance(v,int) == True): # Integer value
encoded_sql = encoded_sql + str(v)
elif (isinstance(v,float) == True):
encoded_sql = encoded_sql + str(v)
else:
flag_quotes = True
try:
if (v.find('0x') == 0): # Just guessing this is a hex value at beginning
encoded_sql = encoded_sql + v
else:
encoded_sql = encoded_sql + addquotes(v,flag_quotes) # String
except:
encoded_sql = encoded_sql + addquotes(str(v),flag_quotes)
start = False
encoded_sql = encoded_sql + ch
varName = ""
inVar = False
elif (inQuote != ""):
encoded_sql = encoded_sql + ch
if (ch == inQuote): inQuote = ""
elif (ch in ("'",'"')):
encoded_sql = encoded_sql + ch
inQuote = ch
elif (ch == ":"): # This might be a variable
varName = ""
inVar = True
else:
encoded_sql = encoded_sql + ch
if (inVar == True):
varValue, varType = getContents(varName,True,local_ns) # We assume the end of a line is quoted
if (varType != PANDAS and varValue == None):
encoded_sql = encoded_sql + ":" + varName
else:
if (varType == STRING):
encoded_sql = encoded_sql + varValue
elif (varType == NUMBER):
encoded_sql = encoded_sql + str(varValue)
elif (varType == PANDAS):
insertsql = ""
coltypes = varValue.dtypes
rows, cols = varValue.shape
for row in range(0,rows):
insertrow = ""
for col in range(0, cols):
value = varValue.iloc[row][col]
if (coltypes[col] == "object"):
value = str(value)
value = addquotes(value,True)
else:
strvalue = str(value)
if ("NAN" in strvalue.upper()):
value = "NULL"
if (insertrow == ""):
insertrow = f"{value}"
else:
insertrow = f"{insertrow},{value}"
if (insertsql == ""):
insertsql = f"({insertrow})"
else:
insertsql = f"{insertsql},({insertrow})"
encoded_sql = encoded_sql + insertsql
elif (varType == LIST):
flag_quotes = True
start = True
for v in varValue:
if (start == False):
encoded_sql = encoded_sql + ","
if (isinstance(v,int) == True): # Integer value
encoded_sql = encoded_sql + str(v)
elif (isinstance(v,float) == True):
encoded_sql = encoded_sql + str(v)
else:
try:
if (v.find('0x') == 0): # Just guessing this is a hex value
encoded_sql = encoded_sql + v
else:
encoded_sql = encoded_sql + addquotes(v,flag_quotes) # String
except:
encoded_sql = encoded_sql + addquotes(str(v),flag_quotes)
start = False
return sql_cmd, encoded_sql
```
### Variable Contents Function
The calling format of this routine is:
```
value = getContents(varName,quote,name_space)
```
This code will take the name of a variable as input and return the contents of that variable. If the variable is not found then the program will return None which is the equivalent to empty or null. Note that this function looks at the global variable pool for Python so it is possible that the wrong version of variable is returned if it is used in different functions. For this reason, any variables used in SQL statements should use a unique namimg convention if possible.
The other thing that this function does is replace single quotes with two quotes. The reason for doing this is that Db2 will convert two single quotes into one quote when dealing with strings. This avoids problems when dealing with text that contains multiple quotes within the string. Note that this substitution is done only for single quote characters since the double quote character is used by Db2 for naming columns that are case sensitive or contain special characters.
If the quote value is True, the field will have quotes around it. The name_space is the variables currently that are registered in Python.
```
def getContents(varName,flag_quotes,local_ns):
#
# Get the contents of the variable name that is passed to the routine. Only simple
# variables are checked, i.e. arrays and lists are not parsed
#
STRING = 0
NUMBER = 1
LIST = 2
RAW = 3
DICT = 4
PANDAS = 5
try:
value = eval(varName,None,local_ns) # globals()[varName] # eval(varName)
except:
return(None,STRING)
if (isinstance(value,dict) == True): # Check to see if this is JSON dictionary
return(addquotes(value,flag_quotes),STRING)
elif(isinstance(value,list) == True): # List - tricky
return(value,LIST)
elif (isinstance(value,pandas.DataFrame) == True): # Pandas dataframe
return(value,PANDAS)
elif (isinstance(value,int) == True): # Integer value
return(value,NUMBER)
elif (isinstance(value,float) == True): # Float value
return(value,NUMBER)
else:
try:
# The pattern needs to be in the first position (0 in Python terms)
if (value.find('0x') == 0): # Just guessing this is a hex value
return(value,RAW)
else:
return(addquotes(value,flag_quotes),STRING) # String
except:
return(addquotes(str(value),flag_quotes),RAW)
```
### Add Quotes
Quotes are a challenge when dealing with dictionaries and Db2. Db2 wants strings delimited with single quotes, while Dictionaries use double quotes. That wouldn't be a problems except imbedded single quotes within these dictionaries will cause things to fail. This routine attempts to double-quote the single quotes within the dicitonary.
```
def addquotes(inString,flag_quotes):
if (isinstance(inString,dict) == True): # Check to see if this is JSON dictionary
serialized = json.dumps(inString)
else:
serialized = inString
# Replace single quotes with '' (two quotes) and wrap everything in single quotes
if (flag_quotes == False):
return(serialized)
else:
return("'"+serialized.replace("'","''")+"'") # Convert single quotes to two single quotes
```
### Create the SAMPLE Database Tables
The calling format of this routine is:
```
db2_create_sample(quiet)
```
There are a lot of examples that depend on the data within the SAMPLE database. If you are running these examples and the connection is not to the SAMPLE database, then this code will create the two (EMPLOYEE, DEPARTMENT) tables that are used by most examples. If the function finds that these tables already exist, then nothing is done. If the tables are missing then they will be created with the same data as in the SAMPLE database.
The quiet flag tells the program not to print any messages when the creation of the tables is complete.
```
def db2_create_sample(quiet):
create_department = """
BEGIN
DECLARE FOUND INTEGER;
SET FOUND = (SELECT COUNT(*) FROM SYSIBM.SYSTABLES WHERE NAME='DEPARTMENT' AND CREATOR=CURRENT USER);
IF FOUND = 0 THEN
EXECUTE IMMEDIATE('CREATE TABLE DEPARTMENT(DEPTNO CHAR(3) NOT NULL, DEPTNAME VARCHAR(36) NOT NULL,
MGRNO CHAR(6),ADMRDEPT CHAR(3) NOT NULL)');
EXECUTE IMMEDIATE('INSERT INTO DEPARTMENT VALUES
(''A00'',''SPIFFY COMPUTER SERVICE DIV.'',''000010'',''A00''),
(''B01'',''PLANNING'',''000020'',''A00''),
(''C01'',''INFORMATION CENTER'',''000030'',''A00''),
(''D01'',''DEVELOPMENT CENTER'',NULL,''A00''),
(''D11'',''MANUFACTURING SYSTEMS'',''000060'',''D01''),
(''D21'',''ADMINISTRATION SYSTEMS'',''000070'',''D01''),
(''E01'',''SUPPORT SERVICES'',''000050'',''A00''),
(''E11'',''OPERATIONS'',''000090'',''E01''),
(''E21'',''SOFTWARE SUPPORT'',''000100'',''E01''),
(''F22'',''BRANCH OFFICE F2'',NULL,''E01''),
(''G22'',''BRANCH OFFICE G2'',NULL,''E01''),
(''H22'',''BRANCH OFFICE H2'',NULL,''E01''),
(''I22'',''BRANCH OFFICE I2'',NULL,''E01''),
(''J22'',''BRANCH OFFICE J2'',NULL,''E01'')');
END IF;
END"""
%sql -d -q {create_department}
create_employee = """
BEGIN
DECLARE FOUND INTEGER;
SET FOUND = (SELECT COUNT(*) FROM SYSIBM.SYSTABLES WHERE NAME='EMPLOYEE' AND CREATOR=CURRENT USER);
IF FOUND = 0 THEN
EXECUTE IMMEDIATE('CREATE TABLE EMPLOYEE(
EMPNO CHAR(6) NOT NULL,
FIRSTNME VARCHAR(12) NOT NULL,
MIDINIT CHAR(1),
LASTNAME VARCHAR(15) NOT NULL,
WORKDEPT CHAR(3),
PHONENO CHAR(4),
HIREDATE DATE,
JOB CHAR(8),
EDLEVEL SMALLINT NOT NULL,
SEX CHAR(1),
BIRTHDATE DATE,
SALARY DECIMAL(9,2),
BONUS DECIMAL(9,2),
COMM DECIMAL(9,2)
)');
EXECUTE IMMEDIATE('INSERT INTO EMPLOYEE VALUES
(''000010'',''CHRISTINE'',''I'',''HAAS'' ,''A00'',''3978'',''1995-01-01'',''PRES '',18,''F'',''1963-08-24'',152750.00,1000.00,4220.00),
(''000020'',''MICHAEL'' ,''L'',''THOMPSON'' ,''B01'',''3476'',''2003-10-10'',''MANAGER '',18,''M'',''1978-02-02'',94250.00,800.00,3300.00),
(''000030'',''SALLY'' ,''A'',''KWAN'' ,''C01'',''4738'',''2005-04-05'',''MANAGER '',20,''F'',''1971-05-11'',98250.00,800.00,3060.00),
(''000050'',''JOHN'' ,''B'',''GEYER'' ,''E01'',''6789'',''1979-08-17'',''MANAGER '',16,''M'',''1955-09-15'',80175.00,800.00,3214.00),
(''000060'',''IRVING'' ,''F'',''STERN'' ,''D11'',''6423'',''2003-09-14'',''MANAGER '',16,''M'',''1975-07-07'',72250.00,500.00,2580.00),
(''000070'',''EVA'' ,''D'',''PULASKI'' ,''D21'',''7831'',''2005-09-30'',''MANAGER '',16,''F'',''2003-05-26'',96170.00,700.00,2893.00),
(''000090'',''EILEEN'' ,''W'',''HENDERSON'' ,''E11'',''5498'',''2000-08-15'',''MANAGER '',16,''F'',''1971-05-15'',89750.00,600.00,2380.00),
(''000100'',''THEODORE'' ,''Q'',''SPENSER'' ,''E21'',''0972'',''2000-06-19'',''MANAGER '',14,''M'',''1980-12-18'',86150.00,500.00,2092.00),
(''000110'',''VINCENZO'' ,''G'',''LUCCHESSI'' ,''A00'',''3490'',''1988-05-16'',''SALESREP'',19,''M'',''1959-11-05'',66500.00,900.00,3720.00),
(''000120'',''SEAN'' ,'' '',''O`CONNELL'' ,''A00'',''2167'',''1993-12-05'',''CLERK '',14,''M'',''1972-10-18'',49250.00,600.00,2340.00),
(''000130'',''DELORES'' ,''M'',''QUINTANA'' ,''C01'',''4578'',''2001-07-28'',''ANALYST '',16,''F'',''1955-09-15'',73800.00,500.00,1904.00),
(''000140'',''HEATHER'' ,''A'',''NICHOLLS'' ,''C01'',''1793'',''2006-12-15'',''ANALYST '',18,''F'',''1976-01-19'',68420.00,600.00,2274.00),
(''000150'',''BRUCE'' ,'' '',''ADAMSON'' ,''D11'',''4510'',''2002-02-12'',''DESIGNER'',16,''M'',''1977-05-17'',55280.00,500.00,2022.00),
(''000160'',''ELIZABETH'',''R'',''PIANKA'' ,''D11'',''3782'',''2006-10-11'',''DESIGNER'',17,''F'',''1980-04-12'',62250.00,400.00,1780.00),
(''000170'',''MASATOSHI'',''J'',''YOSHIMURA'' ,''D11'',''2890'',''1999-09-15'',''DESIGNER'',16,''M'',''1981-01-05'',44680.00,500.00,1974.00),
(''000180'',''MARILYN'' ,''S'',''SCOUTTEN'' ,''D11'',''1682'',''2003-07-07'',''DESIGNER'',17,''F'',''1979-02-21'',51340.00,500.00,1707.00),
(''000190'',''JAMES'' ,''H'',''WALKER'' ,''D11'',''2986'',''2004-07-26'',''DESIGNER'',16,''M'',''1982-06-25'',50450.00,400.00,1636.00),
(''000200'',''DAVID'' ,'' '',''BROWN'' ,''D11'',''4501'',''2002-03-03'',''DESIGNER'',16,''M'',''1971-05-29'',57740.00,600.00,2217.00),
(''000210'',''WILLIAM'' ,''T'',''JONES'' ,''D11'',''0942'',''1998-04-11'',''DESIGNER'',17,''M'',''2003-02-23'',68270.00,400.00,1462.00),
(''000220'',''JENNIFER'' ,''K'',''LUTZ'' ,''D11'',''0672'',''1998-08-29'',''DESIGNER'',18,''F'',''1978-03-19'',49840.00,600.00,2387.00),
(''000230'',''JAMES'' ,''J'',''JEFFERSON'' ,''D21'',''2094'',''1996-11-21'',''CLERK '',14,''M'',''1980-05-30'',42180.00,400.00,1774.00),
(''000240'',''SALVATORE'',''M'',''MARINO'' ,''D21'',''3780'',''2004-12-05'',''CLERK '',17,''M'',''2002-03-31'',48760.00,600.00,2301.00),
(''000250'',''DANIEL'' ,''S'',''SMITH'' ,''D21'',''0961'',''1999-10-30'',''CLERK '',15,''M'',''1969-11-12'',49180.00,400.00,1534.00),
(''000260'',''SYBIL'' ,''P'',''JOHNSON'' ,''D21'',''8953'',''2005-09-11'',''CLERK '',16,''F'',''1976-10-05'',47250.00,300.00,1380.00),
(''000270'',''MARIA'' ,''L'',''PEREZ'' ,''D21'',''9001'',''2006-09-30'',''CLERK '',15,''F'',''2003-05-26'',37380.00,500.00,2190.00),
(''000280'',''ETHEL'' ,''R'',''SCHNEIDER'' ,''E11'',''8997'',''1997-03-24'',''OPERATOR'',17,''F'',''1976-03-28'',36250.00,500.00,2100.00),
(''000290'',''JOHN'' ,''R'',''PARKER'' ,''E11'',''4502'',''2006-05-30'',''OPERATOR'',12,''M'',''1985-07-09'',35340.00,300.00,1227.00),
(''000300'',''PHILIP'' ,''X'',''SMITH'' ,''E11'',''2095'',''2002-06-19'',''OPERATOR'',14,''M'',''1976-10-27'',37750.00,400.00,1420.00),
(''000310'',''MAUDE'' ,''F'',''SETRIGHT'' ,''E11'',''3332'',''1994-09-12'',''OPERATOR'',12,''F'',''1961-04-21'',35900.00,300.00,1272.00),
(''000320'',''RAMLAL'' ,''V'',''MEHTA'' ,''E21'',''9990'',''1995-07-07'',''FIELDREP'',16,''M'',''1962-08-11'',39950.00,400.00,1596.00),
(''000330'',''WING'' ,'' '',''LEE'' ,''E21'',''2103'',''2006-02-23'',''FIELDREP'',14,''M'',''1971-07-18'',45370.00,500.00,2030.00),
(''000340'',''JASON'' ,''R'',''GOUNOT'' ,''E21'',''5698'',''1977-05-05'',''FIELDREP'',16,''M'',''1956-05-17'',43840.00,500.00,1907.00),
(''200010'',''DIAN'' ,''J'',''HEMMINGER'' ,''A00'',''3978'',''1995-01-01'',''SALESREP'',18,''F'',''1973-08-14'',46500.00,1000.00,4220.00),
(''200120'',''GREG'' ,'' '',''ORLANDO'' ,''A00'',''2167'',''2002-05-05'',''CLERK '',14,''M'',''1972-10-18'',39250.00,600.00,2340.00),
(''200140'',''KIM'' ,''N'',''NATZ'' ,''C01'',''1793'',''2006-12-15'',''ANALYST '',18,''F'',''1976-01-19'',68420.00,600.00,2274.00),
(''200170'',''KIYOSHI'' ,'' '',''YAMAMOTO'' ,''D11'',''2890'',''2005-09-15'',''DESIGNER'',16,''M'',''1981-01-05'',64680.00,500.00,1974.00),
(''200220'',''REBA'' ,''K'',''JOHN'' ,''D11'',''0672'',''2005-08-29'',''DESIGNER'',18,''F'',''1978-03-19'',69840.00,600.00,2387.00),
(''200240'',''ROBERT'' ,''M'',''MONTEVERDE'',''D21'',''3780'',''2004-12-05'',''CLERK '',17,''M'',''1984-03-31'',37760.00,600.00,2301.00),
(''200280'',''EILEEN'' ,''R'',''SCHWARTZ'' ,''E11'',''8997'',''1997-03-24'',''OPERATOR'',17,''F'',''1966-03-28'',46250.00,500.00,2100.00),
(''200310'',''MICHELLE'' ,''F'',''SPRINGER'' ,''E11'',''3332'',''1994-09-12'',''OPERATOR'',12,''F'',''1961-04-21'',35900.00,300.00,1272.00),
(''200330'',''HELENA'' ,'' '',''WONG'' ,''E21'',''2103'',''2006-02-23'',''FIELDREP'',14,''F'',''1971-07-18'',35370.00,500.00,2030.00),
(''200340'',''ROY'' ,''R'',''ALONZO'' ,''E21'',''5698'',''1997-07-05'',''FIELDREP'',16,''M'',''1956-05-17'',31840.00,500.00,1907.00)');
END IF;
END"""
%sql -d -q {create_employee}
if (quiet == False): success("Sample tables [EMPLOYEE, DEPARTMENT] created.")
```
### Check option
This function will return the original string with the option removed, and a flag or true or false of the value is found.
```
args, flag = checkOption(option_string, option, false_value, true_value)
```
Options are specified with a -x where x is the character that we are searching for. It may actually be more than one character long like -pb/-pi/etc... The false and true values are optional. By default these are the boolean values of T/F but for some options it could be a character string like ';' versus '@' for delimiters.
```
def checkOption(args_in, option, vFalse=False, vTrue=True):
args_out = args_in.strip()
found = vFalse
if (args_out != ""):
if (args_out.find(option) >= 0):
args_out = args_out.replace(option," ")
args_out = args_out.strip()
found = vTrue
return args_out, found
```
### Plot Data
This function will plot the data that is returned from the answer set. The plot value determines how we display the data. 1=Bar, 2=Pie, 3=Line, 4=Interactive.
```
plotData(flag_plot, hdbi, sql, parms)
```
The hdbi is the ibm_db_sa handle that is used by pandas dataframes to run the sql. The parms contains any of the parameters required to run the query.
```
def plotData(hdbi, sql):
try:
df = pandas.read_sql(sql,hdbi)
except Exception as err:
db2_error(False)
return
if df.empty:
errormsg("No results returned")
return
col_count = len(df.columns)
if flag(["-pb","-bar"]): # Plot 1 = bar chart
if (col_count in (1,2,3)):
if (col_count == 1):
df.index = df.index + 1
_ = df.plot(kind='bar');
_ = plt.plot();
elif (col_count == 2):
xlabel = df.columns.values[0]
ylabel = df.columns.values[1]
df.plot(kind='bar',x=xlabel,y=ylabel);
_ = plt.plot();
else:
values = df.columns.values[2]
columns = df.columns.values[0]
index = df.columns.values[1]
pivoted = pandas.pivot_table(df, values=values, columns=columns, index=index)
_ = pivoted.plot.bar();
else:
errormsg("Can't determine what columns to plot")
return
elif flag(["-pp","-pie"]): # Plot 2 = pie chart
if (col_count in (1,2)):
if (col_count == 1):
df.index = df.index + 1
yname = df.columns.values[0]
_ = df.plot(kind='pie',y=yname);
else:
xlabel = df.columns.values[0]
xname = df[xlabel].tolist()
yname = df.columns.values[1]
_ = df.plot(kind='pie',y=yname,labels=xname);
plt.show();
else:
errormsg("Can't determine what columns to plot")
return
elif flag(["-pl","-line"]): # Plot 3 = line chart
if (col_count in (1,2,3)):
if (col_count == 1):
df.index = df.index + 1
_ = df.plot(kind='line');
elif (col_count == 2):
xlabel = df.columns.values[0]
ylabel = df.columns.values[1]
_ = df.plot(kind='line',x=xlabel,y=ylabel) ;
else:
values = df.columns.values[2]
columns = df.columns.values[0]
index = df.columns.values[1]
pivoted = pandas.pivot_table(df, values=values, columns=columns, index=index)
_ = pivoted.plot();
plt.show();
else:
errormsg("Can't determine what columns to plot")
return
else:
return
```
### Find a Procedure
This routine will check to see if a procedure exists with the SCHEMA/NAME (or just NAME if no schema is supplied) and returns the number of answer sets returned. Possible values are 0, 1 (or greater) or None. If None is returned then we can't find the procedure anywhere.
```
def findProc(procname):
global _hdbc, _hdbi, _connected, _runtime
# Split the procedure name into schema.procname if appropriate
upper_procname = procname.upper()
schema, proc = split_string(upper_procname,".") # Expect schema.procname
if (proc == None):
proc = schema
# Call ibm_db.procedures to see if the procedure does exist
schema = "%"
try:
stmt = ibm_db.procedures(_hdbc, None, schema, proc)
if (stmt == False): # Error executing the code
errormsg("Procedure " + procname + " not found in the system catalog.")
return None
result = ibm_db.fetch_tuple(stmt)
resultsets = result[5]
if (resultsets >= 1): resultsets = 1
return resultsets
except Exception as err:
errormsg("Procedure " + procname + " not found in the system catalog.")
return None
```
### Parse Call Arguments
This code will parse a SQL call #name(parm1,...) and return the name and the parameters in the call.
```
def parseCallArgs(macro):
quoteChar = ""
inQuote = False
inParm = False
ignore = False
name = ""
parms = []
parm = ''
sqlin = macro.replace("\n","")
sqlin.lstrip()
for ch in sqlin:
if (inParm == False):
# We hit a blank in the name, so ignore everything after the procedure name until a ( is found
if (ch == " "):
ignore == True
elif (ch == "("): # Now we have parameters to send to the stored procedure
inParm = True
else:
if (ignore == False): name = name + ch # The name of the procedure (and no blanks)
else:
if (inQuote == True):
if (ch == quoteChar):
inQuote = False
else:
parm = parm + ch
elif (ch in ("\"","\'","[")): # Do we have a quote
if (ch == "["):
quoteChar = "]"
else:
quoteChar = ch
inQuote = True
elif (ch == ")"):
if (parm != ""):
parms.append(parm)
parm = ""
break
elif (ch == ","):
if (parm != ""):
parms.append(parm)
else:
parms.append("null")
parm = ""
else:
parm = parm + ch
if (inParm == True):
if (parm != ""):
parms.append(parm_value)
return(name,parms)
```
### Get Columns
Given a statement handle, determine what the column names are or the data types.
```
def getColumns(stmt):
columns = []
types = []
colcount = 0
try:
colname = ibm_db.field_name(stmt,colcount)
coltype = ibm_db.field_type(stmt,colcount)
while (colname != False):
columns.append(colname)
types.append(coltype)
colcount += 1
colname = ibm_db.field_name(stmt,colcount)
coltype = ibm_db.field_type(stmt,colcount)
return columns,types
except Exception as err:
db2_error(False)
return None
```
### Call a Procedure
The CALL statement is used for execution of a stored procedure. The format of the CALL statement is:
```
CALL PROC_NAME(x,y,z,...)
```
Procedures allow for the return of answer sets (cursors) as well as changing the contents of the parameters being passed to the procedure. In this implementation, the CALL function is limited to returning one answer set (or nothing). If you want to use more complex stored procedures then you will have to use the native python libraries.
```
def parseCall(hdbc, inSQL, local_ns):
global _hdbc, _hdbi, _connected, _runtime, _environment
# Check to see if we are connected first
if (_connected == False): # Check if you are connected
db2_doConnect()
if _connected == False: return None
remainder = inSQL.strip()
procName, procArgs = parseCallArgs(remainder[5:]) # Assume that CALL ... is the format
resultsets = findProc(procName)
if (resultsets == None): return None
argvalues = []
if (len(procArgs) > 0): # We have arguments to consider
for arg in procArgs:
varname = arg
if (len(varname) > 0):
if (varname[0] == ":"):
checkvar = varname[1:]
varvalue = getContents(checkvar,True,local_ns)
if (varvalue == None):
errormsg("Variable " + checkvar + " is not defined.")
return None
argvalues.append(varvalue)
else:
if (varname.upper() == "NULL"):
argvalues.append(None)
else:
argvalues.append(varname)
else:
argvalues.append(None)
try:
if (len(procArgs) > 0):
argtuple = tuple(argvalues)
result = ibm_db.callproc(_hdbc,procName,argtuple)
stmt = result[0]
else:
result = ibm_db.callproc(_hdbc,procName)
stmt = result
if (resultsets != 0 and stmt != None):
columns, types = getColumns(stmt)
if (columns == None): return None
rows = []
rowlist = ibm_db.fetch_tuple(stmt)
while ( rowlist ) :
row = []
colcount = 0
for col in rowlist:
try:
if (types[colcount] in ["int","bigint"]):
row.append(int(col))
elif (types[colcount] in ["decimal","real"]):
row.append(float(col))
elif (types[colcount] in ["date","time","timestamp"]):
row.append(str(col))
else:
row.append(col)
except:
row.append(col)
colcount += 1
rows.append(row)
rowlist = ibm_db.fetch_tuple(stmt)
if flag(["-r","-array"]):
rows.insert(0,columns)
if len(procArgs) > 0:
allresults = []
allresults.append(rows)
for x in result[1:]:
allresults.append(x)
return allresults # rows,returned_results
else:
return rows
else:
df = pandas.DataFrame.from_records(rows,columns=columns)
if flag("-grid") or _settings['display'] == 'GRID':
if (_environment['qgrid'] == False):
with pandas.option_context('display.max_rows', None, 'display.max_columns', None):
pdisplay(df)
else:
try:
pdisplay(qgrid.show_grid(df))
except:
errormsg("Grid cannot be used to display data with duplicate column names. Use option -a or %sql OPTION DISPLAY PANDAS instead.")
return
else:
if flag(["-a","-all"]) or _settings["maxrows"] == -1 : # All of the rows
with pandas.option_context('display.max_rows', None, 'display.max_columns', None):
pdisplay(df)
else:
return df
else:
if len(procArgs) > 0:
allresults = []
for x in result[1:]:
allresults.append(x)
return allresults # rows,returned_results
else:
return None
except Exception as err:
db2_error(False)
return None
```
### Parse Prepare/Execute
The PREPARE statement is used for repeated execution of a SQL statement. The PREPARE statement has the format:
```
stmt = PREPARE SELECT EMPNO FROM EMPLOYEE WHERE WORKDEPT=? AND SALARY<?
```
The SQL statement that you want executed is placed after the PREPARE statement with the location of variables marked with ? (parameter) markers. The variable stmt contains the prepared statement that need to be passed to the EXECUTE statement. The EXECUTE statement has the format:
```
EXECUTE :x USING z, y, s
```
The first variable (:x) is the name of the variable that you assigned the results of the prepare statement. The values after the USING clause are substituted into the prepare statement where the ? markers are found.
If the values in USING clause are variable names (z, y, s), a **link** is created to these variables as part of the execute statement. If you use the variable subsitution form of variable name (:z, :y, :s), the **contents** of the variable are placed into the USING clause. Normally this would not make much of a difference except when you are dealing with binary strings or JSON strings where the quote characters may cause some problems when subsituted into the statement.
```
def parsePExec(hdbc, inSQL):
import ibm_db
global _stmt, _stmtID, _stmtSQL, sqlcode
cParms = inSQL.split()
parmCount = len(cParms)
if (parmCount == 0): return(None) # Nothing to do but this shouldn't happen
keyword = cParms[0].upper() # Upper case the keyword
if (keyword == "PREPARE"): # Prepare the following SQL
uSQL = inSQL.upper()
found = uSQL.find("PREPARE")
sql = inSQL[found+7:].strip()
try:
pattern = "\?\*[0-9]+"
findparm = re.search(pattern,sql)
while findparm != None:
found = findparm.group(0)
count = int(found[2:])
markers = ('?,' * count)[:-1]
sql = sql.replace(found,markers)
findparm = re.search(pattern,sql)
stmt = ibm_db.prepare(hdbc,sql) # Check error code here
if (stmt == False):
db2_error(False)
return(False)
stmttext = str(stmt).strip()
stmtID = stmttext[33:48].strip()
if (stmtID in _stmtID) == False:
_stmt.append(stmt) # Prepare and return STMT to caller
_stmtID.append(stmtID)
else:
stmtIX = _stmtID.index(stmtID)
_stmt[stmtiX] = stmt
return(stmtID)
except Exception as err:
print(err)
db2_error(False)
return(False)
if (keyword == "EXECUTE"): # Execute the prepare statement
if (parmCount < 2): return(False) # No stmtID available
stmtID = cParms[1].strip()
if (stmtID in _stmtID) == False:
errormsg("Prepared statement not found or invalid.")
return(False)
stmtIX = _stmtID.index(stmtID)
stmt = _stmt[stmtIX]
try:
if (parmCount == 2): # Only the statement handle available
result = ibm_db.execute(stmt) # Run it
elif (parmCount == 3): # Not quite enough arguments
errormsg("Missing or invalid USING clause on EXECUTE statement.")
sqlcode = -99999
return(False)
else:
using = cParms[2].upper()
if (using != "USING"): # Bad syntax again
errormsg("Missing USING clause on EXECUTE statement.")
sqlcode = -99999
return(False)
uSQL = inSQL.upper()
found = uSQL.find("USING")
parmString = inSQL[found+5:].strip()
parmset = splitargs(parmString)
if (len(parmset) == 0):
errormsg("Missing parameters after the USING clause.")
sqlcode = -99999
return(False)
parms = []
parm_count = 0
CONSTANT = 0
VARIABLE = 1
const = [0]
const_cnt = 0
for v in parmset:
parm_count = parm_count + 1
if (v[1] == True or v[2] == True): # v[1] true if string, v[2] true if num
parm_type = CONSTANT
const_cnt = const_cnt + 1
if (v[2] == True):
if (isinstance(v[0],int) == True): # Integer value
sql_type = ibm_db.SQL_INTEGER
elif (isinstance(v[0],float) == True): # Float value
sql_type = ibm_db.SQL_DOUBLE
else:
sql_type = ibm_db.SQL_INTEGER
else:
sql_type = ibm_db.SQL_CHAR
const.append(v[0])
else:
parm_type = VARIABLE
# See if the variable has a type associated with it varname@type
varset = v[0].split("@")
parm_name = varset[0]
parm_datatype = "char"
# Does the variable exist?
if (parm_name not in globals()):
errormsg("SQL Execute parameter " + parm_name + " not found")
sqlcode = -99999
return(false)
if (len(varset) > 1): # Type provided
parm_datatype = varset[1]
if (parm_datatype == "dec" or parm_datatype == "decimal"):
sql_type = ibm_db.SQL_DOUBLE
elif (parm_datatype == "bin" or parm_datatype == "binary"):
sql_type = ibm_db.SQL_BINARY
elif (parm_datatype == "int" or parm_datatype == "integer"):
sql_type = ibm_db.SQL_INTEGER
else:
sql_type = ibm_db.SQL_CHAR
try:
if (parm_type == VARIABLE):
result = ibm_db.bind_param(stmt, parm_count, globals()[parm_name], ibm_db.SQL_PARAM_INPUT, sql_type)
else:
result = ibm_db.bind_param(stmt, parm_count, const[const_cnt], ibm_db.SQL_PARAM_INPUT, sql_type)
except:
result = False
if (result == False):
errormsg("SQL Bind on variable " + parm_name + " failed.")
sqlcode = -99999
return(false)
result = ibm_db.execute(stmt) # ,tuple(parms))
if (result == False):
errormsg("SQL Execute failed.")
return(False)
if (ibm_db.num_fields(stmt) == 0): return(True) # Command successfully completed
return(fetchResults(stmt))
except Exception as err:
db2_error(False)
return(False)
return(False)
return(False)
```
### Fetch Result Set
This code will take the stmt handle and then produce a result set of rows as either an array (`-r`,`-array`) or as an array of json records (`-json`).
```
def fetchResults(stmt):
global sqlcode
rows = []
columns, types = getColumns(stmt)
# By default we assume that the data will be an array
is_array = True
# Check what type of data we want returned - array or json
if (flag(["-r","-array"]) == False):
# See if we want it in JSON format, if not it remains as an array
if (flag("-json") == True):
is_array = False
# Set column names to lowercase for JSON records
if (is_array == False):
columns = [col.lower() for col in columns] # Convert to lowercase for each of access
# First row of an array has the column names in it
if (is_array == True):
rows.append(columns)
result = ibm_db.fetch_tuple(stmt)
rowcount = 0
while (result):
rowcount += 1
if (is_array == True):
row = []
else:
row = {}
colcount = 0
for col in result:
try:
if (types[colcount] in ["int","bigint"]):
if (is_array == True):
row.append(int(col))
else:
row[columns[colcount]] = int(col)
elif (types[colcount] in ["decimal","real"]):
if (is_array == True):
row.append(float(col))
else:
row[columns[colcount]] = float(col)
elif (types[colcount] in ["date","time","timestamp"]):
if (is_array == True):
row.append(str(col))
else:
row[columns[colcount]] = str(col)
else:
if (is_array == True):
row.append(col)
else:
row[columns[colcount]] = col
except:
if (is_array == True):
row.append(col)
else:
row[columns[colcount]] = col
colcount += 1
rows.append(row)
result = ibm_db.fetch_tuple(stmt)
if (rowcount == 0):
sqlcode = 100
else:
sqlcode = 0
return rows
```
### Parse Commit
There are three possible COMMIT verbs that can bs used:
- COMMIT [WORK] - Commit the work in progress - The WORK keyword is not checked for
- ROLLBACK - Roll back the unit of work
- AUTOCOMMIT ON/OFF - Are statements committed on or off?
The statement is passed to this routine and then checked.
```
def parseCommit(sql):
global _hdbc, _hdbi, _connected, _runtime, _stmt, _stmtID, _stmtSQL
if (_connected == False): return # Nothing to do if we are not connected
cParms = sql.split()
if (len(cParms) == 0): return # Nothing to do but this shouldn't happen
keyword = cParms[0].upper() # Upper case the keyword
if (keyword == "COMMIT"): # Commit the work that was done
try:
result = ibm_db.commit (_hdbc) # Commit the connection
if (len(cParms) > 1):
keyword = cParms[1].upper()
if (keyword == "HOLD"):
return
del _stmt[:]
del _stmtID[:]
except Exception as err:
db2_error(False)
return
if (keyword == "ROLLBACK"): # Rollback the work that was done
try:
result = ibm_db.rollback(_hdbc) # Rollback the connection
del _stmt[:]
del _stmtID[:]
except Exception as err:
db2_error(False)
return
if (keyword == "AUTOCOMMIT"): # Is autocommit on or off
if (len(cParms) > 1):
op = cParms[1].upper() # Need ON or OFF value
else:
return
try:
if (op == "OFF"):
ibm_db.autocommit(_hdbc, False)
elif (op == "ON"):
ibm_db.autocommit (_hdbc, True)
return
except Exception as err:
db2_error(False)
return
return
```
### Set Flags
This code will take the input SQL block and update the global flag list. The global flag list is just a list of options that are set at the beginning of a code block. The absence of a flag means it is false. If it exists it is true.
```
def setFlags(inSQL):
global _flags
_flags = [] # Delete all of the current flag settings
pos = 0
end = len(inSQL)-1
inFlag = False
ignore = False
outSQL = ""
flag = ""
while (pos <= end):
ch = inSQL[pos]
if (ignore == True):
outSQL = outSQL + ch
else:
if (inFlag == True):
if (ch != " "):
flag = flag + ch
else:
_flags.append(flag)
inFlag = False
else:
if (ch == "-"):
flag = "-"
inFlag = True
elif (ch == ' '):
outSQL = outSQL + ch
else:
outSQL = outSQL + ch
ignore = True
pos += 1
if (inFlag == True):
_flags.append(flag)
return outSQL
```
### Check to see if flag Exists
This function determines whether or not a flag exists in the global flag array. Absence of a value means it is false. The parameter can be a single value, or an array of values.
```
def flag(inflag):
global _flags
if isinstance(inflag,list):
for x in inflag:
if (x in _flags):
return True
return False
else:
if (inflag in _flags):
return True
else:
return False
```
### Generate a list of SQL lines based on a delimiter
Note that this function will make sure that quotes are properly maintained so that delimiters inside of quoted strings do not cause errors.
```
def splitSQL(inputString, delimiter):
pos = 0
arg = ""
results = []
quoteCH = ""
inSQL = inputString.strip()
if (len(inSQL) == 0): return(results) # Not much to do here - no args found
while pos < len(inSQL):
ch = inSQL[pos]
pos += 1
if (ch in ('"',"'")): # Is this a quote characters?
arg = arg + ch # Keep appending the characters to the current arg
if (ch == quoteCH): # Is this quote character we are in
quoteCH = ""
elif (quoteCH == ""): # Create the quote
quoteCH = ch
else:
None
elif (quoteCH != ""): # Still in a quote
arg = arg + ch
elif (ch == delimiter): # Is there a delimiter?
results.append(arg)
arg = ""
else:
arg = arg + ch
if (arg != ""):
results.append(arg)
return(results)
```
### Main %sql Magic Definition
The main %sql Magic logic is found in this section of code. This code will register the Magic command and allow Jupyter notebooks to interact with Db2 by using this extension.
```
@magics_class
class DB2(Magics):
@needs_local_scope
@line_cell_magic
def sql(self, line, cell=None, local_ns=None):
# Before we event get started, check to see if you have connected yet. Without a connection we
# can't do anything. You may have a connection request in the code, so if that is true, we run those,
# otherwise we connect immediately
# If your statement is not a connect, and you haven't connected, we need to do it for you
global _settings, _environment
global _hdbc, _hdbi, _connected, _runtime, sqlstate, sqlerror, sqlcode, sqlelapsed
# If you use %sql (line) we just run the SQL. If you use %%SQL the entire cell is run.
flag_cell = False
flag_output = False
sqlstate = "0"
sqlerror = ""
sqlcode = 0
sqlelapsed = 0
start_time = time.time()
end_time = time.time()
# Macros gets expanded before anything is done
SQL1 = setFlags(line.strip())
SQL1 = checkMacro(SQL1) # Update the SQL if any macros are in there
SQL2 = cell
if flag("-sampledata"): # Check if you only want sample data loaded
if (_connected == False):
if (db2_doConnect() == False):
errormsg('A CONNECT statement must be issued before issuing SQL statements.')
return
db2_create_sample(flag(["-q","-quiet"]))
return
if SQL1 == "?" or flag(["-h","-help"]): # Are you asking for help
sqlhelp()
return
if len(SQL1) == 0 and SQL2 == None: return # Nothing to do here
# Check for help
if SQL1.upper() == "? CONNECT": # Are you asking for help on CONNECT
connected_help()
return
sqlType,remainder = sqlParser(SQL1,local_ns) # What type of command do you have?
if (sqlType == "CONNECT"): # A connect request
parseConnect(SQL1,local_ns)
return
elif (sqlType == "USING"): # You want to use a dataframe to create a table?
createDF(_hdbc,SQL1,local_ns)
return
elif (sqlType == "DEFINE"): # Create a macro from the body
result = setMacro(SQL2,remainder)
return
elif (sqlType == "OPTION"):
setOptions(SQL1)
return
elif (sqlType == 'COMMIT' or sqlType == 'ROLLBACK' or sqlType == 'AUTOCOMMIT'):
parseCommit(remainder)
return
elif (sqlType == "PREPARE"):
pstmt = parsePExec(_hdbc, remainder)
return(pstmt)
elif (sqlType == "EXECUTE"):
result = parsePExec(_hdbc, remainder)
return(result)
elif (sqlType == "CALL"):
result = parseCall(_hdbc, remainder, local_ns)
return(result)
else:
pass
sql = SQL1
if (sql == ""): sql = SQL2
if (sql == ""): return # Nothing to do here
if (_connected == False):
if (db2_doConnect() == False):
errormsg('A CONNECT statement must be issued before issuing SQL statements.')
return
if _settings["maxrows"] == -1: # Set the return result size
pandas.reset_option('display.max_rows')
else:
pandas.options.display.max_rows = _settings["maxrows"]
runSQL = re.sub('.*?--.*$',"",sql,flags=re.M)
remainder = runSQL.replace("\n"," ")
if flag(["-d","-delim"]):
sqlLines = splitSQL(remainder,"@")
else:
sqlLines = splitSQL(remainder,";")
flag_cell = True
# For each line figure out if you run it as a command (db2) or select (sql)
for sqlin in sqlLines: # Run each command
sqlin = checkMacro(sqlin) # Update based on any macros
sqlType, sql = sqlParser(sqlin,local_ns) # Parse the SQL
if (sql.strip() == ""): continue
if flag(["-e","-echo"]): debug(sql,False)
if flag("-t"):
cnt = sqlTimer(_hdbc, _settings["runtime"], sql) # Given the sql and parameters, clock the time
if (cnt >= 0): print("Total iterations in %s second(s): %s" % (_settings["runtime"],cnt))
return(cnt)
elif flag(["-pb","-bar","-pp","-pie","-pl","-line"]): # We are plotting some results
plotData(_hdbi, sql) # Plot the data and return
return
else:
try: # See if we have an answer set
stmt = ibm_db.prepare(_hdbc,sql)
if (ibm_db.num_fields(stmt) == 0): # No, so we just execute the code
result = ibm_db.execute(stmt) # Run it
if (result == False): # Error executing the code
db2_error(flag(["-q","-quiet"]))
continue
rowcount = ibm_db.num_rows(stmt)
if (rowcount == 0 and flag(["-q","-quiet"]) == False):
errormsg("No rows found.")
continue # Continue running
elif flag(["-r","-array","-j","-json"]): # raw, json, format json
row_count = 0
resultSet = []
try:
result = ibm_db.execute(stmt) # Run it
if (result == False): # Error executing the code
db2_error(flag(["-q","-quiet"]))
return
if flag("-j"): # JSON single output
row_count = 0
json_results = []
while( ibm_db.fetch_row(stmt) ):
row_count = row_count + 1
jsonVal = ibm_db.result(stmt,0)
jsonDict = json.loads(jsonVal)
json_results.append(jsonDict)
flag_output = True
if (row_count == 0): sqlcode = 100
return(json_results)
else:
return(fetchResults(stmt))
except Exception as err:
db2_error(flag(["-q","-quiet"]))
return
else:
try:
df = pandas.read_sql(sql,_hdbi)
except Exception as err:
db2_error(False)
return
if (len(df) == 0):
sqlcode = 100
if (flag(["-q","-quiet"]) == False):
errormsg("No rows found")
continue
flag_output = True
if flag("-grid") or _settings['display'] == 'GRID': # Check to see if we can display the results
if (_environment['qgrid'] == False):
with pandas.option_context('display.max_rows', None, 'display.max_columns', None):
print(df.to_string())
else:
try:
pdisplay(qgrid.show_grid(df))
except:
errormsg("Grid cannot be used to display data with duplicate column names. Use option -a or %sql OPTION DISPLAY PANDAS instead.")
return
else:
if flag(["-a","-all"]) or _settings["maxrows"] == -1 : # All of the rows
pandas.options.display.max_rows = None
pandas.options.display.max_columns = None
return df # print(df.to_string())
else:
pandas.options.display.max_rows = _settings["maxrows"]
pandas.options.display.max_columns = None
return df # pdisplay(df) # print(df.to_string())
except:
db2_error(flag(["-q","-quiet"]))
continue # return
end_time = time.time()
sqlelapsed = end_time - start_time
if (flag_output == False and flag(["-q","-quiet"]) == False): print("Command completed.")
# Register the Magic extension in Jupyter
ip = get_ipython()
ip.register_magics(DB2)
load_settings()
success("Db2 Extensions Loaded.")
```
## Pre-defined Macros
These macros are used to simulate the LIST TABLES and DESCRIBE commands that are available from within the Db2 command line.
```
%%sql define LIST
#
# The LIST macro is used to list all of the tables in the current schema or for all schemas
#
var syntax Syntax: LIST TABLES [FOR ALL | FOR SCHEMA name]
#
# Only LIST TABLES is supported by this macro
#
if {^1} <> 'TABLES'
exit {syntax}
endif
#
# This SQL is a temporary table that contains the description of the different table types
#
WITH TYPES(TYPE,DESCRIPTION) AS (
VALUES
('A','Alias'),
('G','Created temporary table'),
('H','Hierarchy table'),
('L','Detached table'),
('N','Nickname'),
('S','Materialized query table'),
('T','Table'),
('U','Typed table'),
('V','View'),
('W','Typed view')
)
SELECT TABNAME, TABSCHEMA, T.DESCRIPTION FROM SYSCAT.TABLES S, TYPES T
WHERE T.TYPE = S.TYPE
#
# Case 1: No arguments - LIST TABLES
#
if {argc} == 1
AND OWNER = CURRENT USER
ORDER BY TABNAME, TABSCHEMA
return
endif
#
# Case 2: Need 3 arguments - LIST TABLES FOR ALL
#
if {argc} == 3
if {^2}&{^3} == 'FOR&ALL'
ORDER BY TABNAME, TABSCHEMA
return
endif
exit {syntax}
endif
#
# Case 3: Need FOR SCHEMA something here
#
if {argc} == 4
if {^2}&{^3} == 'FOR&SCHEMA'
AND TABSCHEMA = '{^4}'
ORDER BY TABNAME, TABSCHEMA
return
else
exit {syntax}
endif
endif
#
# Nothing matched - Error
#
exit {syntax}
%%sql define describe
#
# The DESCRIBE command can either use the syntax DESCRIBE TABLE <name> or DESCRIBE TABLE SELECT ...
#
var syntax Syntax: DESCRIBE [TABLE name | SELECT statement]
#
# Check to see what count of variables is... Must be at least 2 items DESCRIBE TABLE x or SELECT x
#
if {argc} < 2
exit {syntax}
endif
CALL ADMIN_CMD('{*0}');
```
Set the table formatting to left align a table in a cell. By default, tables are centered in a cell. Remove this cell if you don't want to change Jupyter notebook formatting for tables. In addition, we skip this code if you are running in a shell environment rather than a Jupyter notebook
```
#%%html
#<style>
# table {margin-left: 0 !important; text-align: left;}
#</style>
```
#### Credits: IBM 2021, George Baklarz [baklarz@ca.ibm.com]
| github_jupyter |
# Profiling TensorFlow Multi GPU Multi Node Training Job with Amazon SageMaker Debugger
This notebook will walk you through creating a TensorFlow training job with the SageMaker Debugger profiling feature enabled. It will create a multi GPU multi node training using Horovod.
### (Optional) Install SageMaker and SMDebug Python SDKs
To use the new Debugger profiling features released in December 2020, ensure that you have the latest versions of SageMaker and SMDebug SDKs installed. Use the following cell to update the libraries and restarts the Jupyter kernel to apply the updates.
```
import sys
import IPython
install_needed = False # should only be True once
if install_needed:
print("installing deps and restarting kernel")
!{sys.executable} -m pip install -U sagemaker smdebug
IPython.Application.instance().kernel.do_shutdown(True)
```
## 1. Create a Training Job with Profiling Enabled<a class="anchor" id="option-1"></a>
You will use the standard [SageMaker Estimator API for Tensorflow](https://sagemaker.readthedocs.io/en/stable/frameworks/tensorflow/sagemaker.tensorflow.html#tensorflow-estimator) to create training jobs. To enable profiling, create a `ProfilerConfig` object and pass it to the `profiler_config` parameter of the `TensorFlow` estimator.
### Define parameters for distributed training
This parameter tells SageMaker how to configure and run horovod. If you want to use more than 4 GPUs per node then change the process_per_host paramter accordingly.
```
distributions = {
"mpi": {
"enabled": True,
"processes_per_host": 4,
"custom_mpi_options": "-verbose -x HOROVOD_TIMELINE=./hvd_timeline.json -x NCCL_DEBUG=INFO -x OMPI_MCA_btl_vader_single_copy_mechanism=none",
}
}
```
### Configure rules
We specify the following rules:
- loss_not_decreasing: checks if loss is decreasing and triggers if the loss has not decreased by a certain persentage in the last few iterations
- LowGPUUtilization: checks if GPU is under-utilizated
- ProfilerReport: runs the entire set of performance rules and create a final output report with further insights and recommendations.
```
from sagemaker.debugger import Rule, ProfilerRule, rule_configs
rules = [
Rule.sagemaker(rule_configs.loss_not_decreasing()),
ProfilerRule.sagemaker(rule_configs.LowGPUUtilization()),
ProfilerRule.sagemaker(rule_configs.ProfilerReport()),
]
```
### Specify a profiler configuration
The following configuration will capture system metrics at 500 milliseconds. The system metrics include utilization per CPU, GPU, memory utilization per CPU, GPU as well I/O and network.
Debugger will capture detailed profiling information from step 5 to step 15. This information includes Horovod metrics, dataloading, preprocessing, operators running on CPU and GPU.
```
from sagemaker.debugger import ProfilerConfig, FrameworkProfile
profiler_config = ProfilerConfig(
system_monitor_interval_millis=500,
framework_profile_params=FrameworkProfile(
local_path="/opt/ml/output/profiler/", start_step=5, num_steps=10
),
)
```
### Get the image URI
The image that we will is dependent on the region that you are running this notebook in.
```
import boto3
session = boto3.session.Session()
region = session.region_name
image_uri = f"763104351884.dkr.ecr.{region}.amazonaws.com/tensorflow-training:2.3.1-gpu-py37-cu110-ubuntu18.04"
```
### Define estimator
To enable profiling, you need to pass the Debugger profiling configuration (`profiler_config`), a list of Debugger rules (`rules`), and the image URI (`image_uri`) to the estimator. Debugger enables monitoring and profiling while the SageMaker estimator requests a training job.
```
import sagemaker
from sagemaker.tensorflow import TensorFlow
estimator = TensorFlow(
role=sagemaker.get_execution_role(),
image_uri=image_uri,
instance_count=2,
instance_type="ml.p3.8xlarge",
entry_point="tf-hvd-train.py",
source_dir="entry_point",
profiler_config=profiler_config,
distribution=distributions,
rules=rules,
)
```
### Start training job
The following `estimator.fit()` with `wait=False` argument initiates the training job in the background. You can proceed to run the dashboard or analysis notebooks.
```
estimator.fit(wait=False)
```
## 2. Analyze Profiling Data
Copy outputs of the following cell (`training_job_name` and `region`) to run the analysis notebooks `profiling_generic_dashboard.ipynb`, `analyze_performance_bottlenecks.ipynb`, and `profiling_interactive_analysis.ipynb`.
```
training_job_name = estimator.latest_training_job.name
print(f"Training jobname: {training_job_name}")
print(f"Region: {region}")
```
While the training is still in progress you can visualize the performance data in SageMaker Studio or in the notebook.
Debugger provides utilities to plot system metrics in form of timeline charts or heatmaps. Checkout out the notebook
[profiling_interactive_analysis.ipynb](analysis_tools/profiling_interactive_analysis.ipynb) for more details. In the following code cell we plot the total CPU and GPU utilization as timeseries charts. To visualize other metrics such as I/O, memory, network you simply need to extend the list passed to `select_dimension` and `select_events`.
### Install the SMDebug client library to use Debugger analysis tools
```
import pip
def import_or_install(package):
try:
__import__(package)
except ImportError:
pip.main(["install", package])
import_or_install("smdebug")
```
### Access the profiling data using the SMDebug `TrainingJob` utility class
```
from smdebug.profiler.analysis.notebook_utils.training_job import TrainingJob
tj = TrainingJob(training_job_name, region)
tj.wait_for_sys_profiling_data_to_be_available()
```
### Plot time line charts
The following code shows how to use the SMDebug `TrainingJob` object, refresh the object if new event files are available, and plot time line charts of CPU and GPU usage.
```
from smdebug.profiler.analysis.notebook_utils.timeline_charts import TimelineCharts
system_metrics_reader = tj.get_systems_metrics_reader()
system_metrics_reader.refresh_event_file_list()
view_timeline_charts = TimelineCharts(
system_metrics_reader,
framework_metrics_reader=None,
select_dimensions=["CPU", "GPU"],
select_events=["total"],
)
```
## 3. Download Debugger Profiling Report
The `ProfilerReport()` rule creates an html report `profiler-report.html` with a summary of builtin rules and recommenades of next steps. You can find this report in your S3 bucket.
```
rule_output_path = estimator.output_path + estimator.latest_training_job.job_name + "/rule-output"
print(f"You will find the profiler report in {rule_output_path}")
```
For more information about how to download and open the Debugger profiling report, see [SageMaker Debugger Profiling Report](https://docs.aws.amazon.com/sagemaker/latest/dg/debugger-profiling-report.html) in the SageMaker developer guide.
| github_jupyter |
```
import sys
import os
sys.path.append(os.path.abspath("../src/"))
import extract.data_loading as data_loading
import extract.compute_predictions as compute_predictions
import extract.compute_shap as compute_shap
import extract.compute_ism as compute_ism
import model.util as model_util
import model.profile_models as profile_models
import model.binary_models as binary_models
import plot.viz_sequence as viz_sequence
import torch
import numpy as np
import scipy.stats
import matplotlib.pyplot as plt
import matplotlib.font_manager as font_manager
import json
import tqdm
tqdm.tqdm_notebook() # It is necessary to call this before the tqdm.notebook submodule is available
font_manager.fontManager.ttflist.extend(
font_manager.createFontList(
font_manager.findSystemFonts(fontpaths="/users/amtseng/modules/fonts")
)
)
plot_params = {
"axes.titlesize": 22,
"axes.labelsize": 20,
"legend.fontsize": 18,
"xtick.labelsize": 16,
"ytick.labelsize": 16,
"font.family": "Roboto",
"font.weight": "bold"
}
plt.rcParams.update(plot_params)
```
### Define paths for the model and data of interest
```
model_type = "profile"
# Shared paths/constants
reference_fasta = "/users/amtseng/genomes/hg38.fasta"
chrom_sizes = "/users/amtseng/genomes/hg38.canon.chrom.sizes"
data_base_path = "/users/amtseng/att_priors/data/processed/"
model_base_path = "/users/amtseng/att_priors/models/trained_models/%s/" % model_type
chrom_set = ["chr1"]
input_length = 1346 if model_type == "profile" else 1000
profile_length = 1000
# SPI1
condition_name = "SPI1"
files_spec_path = os.path.join(data_base_path, "ENCODE_TFChIP/%s/config/SPI1/SPI1_training_paths.json" % model_type)
num_tasks = 4
num_strands = 2
task_index = None
controls = "matched"
if model_type == "profile":
model_class = profile_models.ProfilePredictorWithMatchedControls
else:
model_class = binary_models.BinaryPredictor
noprior_model_base_path = os.path.join(model_base_path, "SPI1/")
prior_model_base_path = os.path.join(model_base_path, "SPI1_prior/")
peak_retention = "all"
# GATA2
condition_name = "GATA2"
files_spec_path = os.path.join(data_base_path, "ENCODE_TFChIP/%s/config/GATA2/GATA2_training_paths.json" % model_type)
num_tasks = 3
num_strands = 2
task_index = None
controls = "matched"
if model_type == "profile":
model_class = profile_models.ProfilePredictorWithMatchedControls
else:
model_class = binary_models.BinaryPredictor
noprior_model_base_path = os.path.join(model_base_path, "GATA2/")
prior_model_base_path = os.path.join(model_base_path, "GATA2_prior/")
peak_retention = "all"
# K562
condition_name = "K562"
files_spec_path = os.path.join(data_base_path, "ENCODE_DNase/%s/config/K562/K562_training_paths.json" % model_type)
num_tasks = 1
num_strands = 1
task_index = None
controls = "shared"
if model_type == "profile":
model_class = profile_models.ProfilePredictorWithSharedControls
else:
model_class = binary_models.BinaryPredictor
noprior_model_base_path = os.path.join(model_base_path, "K562/")
prior_model_base_path = os.path.join(model_base_path, "K562_prior/")
peak_retention = "all"
# BPNet
condition_name = "BPNet"
reference_fasta = "/users/amtseng/genomes/mm10.fasta"
chrom_sizes = "/users/amtseng/genomes/mm10.canon.chrom.sizes"
files_spec_path = os.path.join(data_base_path, "BPNet_ChIPseq/%s/config/BPNet_training_paths.json" % model_type)
num_tasks = 3
num_strands = 2
task_index = None
controls = "shared"
if model_type == "profile":
model_class = profile_models.ProfilePredictorWithSharedControls
else:
model_class = binary_models.BinaryPredictor
noprior_model_base_path = os.path.join(model_base_path, "BPNet/")
prior_model_base_path = os.path.join(model_base_path, "BPNet_prior/")
peak_retention = "all"
```
### Get all runs/epochs with random initializations
```
def import_metrics_json(model_base_path, run_num):
"""
Looks in {model_base_path}/{run_num}/metrics.json and returns the contents as a
Python dictionary. Returns None if the path does not exist.
"""
path = os.path.join(model_base_path, str(run_num), "metrics.json")
if not os.path.exists(path):
return None
with open(path, "r") as f:
return json.load(f)
def get_model_paths(
model_base_path, metric_name="val_prof_corr_losses",
reduce_func=(lambda values: np.mean(values)), compare_func=(lambda x, y: x < y),
print_found_values=True
):
"""
Looks in `model_base_path` and for each run, returns the full path to
the best epoch. By default, the best epoch in a run is determined by
the lowest validation profile loss.
"""
# Get the metrics, ignoring empty or nonexistent metrics.json files
metrics = {run_num : import_metrics_json(model_base_path, run_num) for run_num in os.listdir(model_base_path)}
metrics = {key : val for key, val in metrics.items() if val} # Remove empties
model_paths, metric_vals = [], []
for run_num in sorted(metrics.keys(), key=lambda x: int(x)):
try:
# Find the best epoch within that run
best_epoch_in_run, best_val_in_run = None, None
for i, subarr in enumerate(metrics[run_num][metric_name]["values"]):
val = reduce_func(subarr)
if best_val_in_run is None or compare_func(val, best_val_in_run):
best_epoch_in_run, best_val_in_run = i + 1, val
model_path = os.path.join(model_base_path, run_num, "model_ckpt_epoch_%d.pt" % best_epoch_in_run)
model_paths.append(model_path)
metric_vals.append(best_val_in_run)
if print_found_values:
print("\tRun %s, epoch %d: %6.2f" % (run_num, best_epoch_in_run, best_val_in_run))
except Exception:
print("Warning: Was not able to compute values for run %s" % run_num)
continue
return model_paths, metric_vals
metric_name = "val_prof_corr_losses" if model_type == "profile" else "val_corr_losses"
noprior_model_paths, noprior_metric_vals = get_model_paths(noprior_model_base_path, metric_name=metric_name)
prior_model_paths, prior_metric_vals = get_model_paths(prior_model_base_path, metric_name=metric_name)
torch.set_grad_enabled(True)
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
def restore_model(model_path):
model = model_util.restore_model(model_class, model_path)
model.eval()
model = model.to(device)
return model
```
### Data preparation
Create an input data loader, that maps coordinates or bin indices to data needed for the model
```
if model_type == "profile":
input_func = data_loading.get_profile_input_func(
files_spec_path, input_length, profile_length, reference_fasta
)
pos_examples = data_loading.get_positive_profile_coords(
files_spec_path, chrom_set=chrom_set
)
else:
input_func = data_loading.get_binary_input_func(
files_spec_path, input_length, reference_fasta
)
pos_examples = data_loading.get_positive_binary_bins(
files_spec_path, chrom_set=chrom_set
)
```
### Compute importances
```
# Pick a sample of 100 random coordinates/bins
num_samples = 100
rng = np.random.RandomState(20200318)
sample = pos_examples[rng.choice(len(pos_examples), size=num_samples, replace=False)]
# For profile models, add a random jitter to avoid center-bias
if model_type == "profile":
jitters = np.random.randint(-128, 128 + 1, size=len(sample))
sample[:, 1] = sample[:, 1] + jitters
sample[:, 2] = sample[:, 2] + jitters
def compute_gradients(model_paths, sample):
"""
Given a list of paths to M models and a list of N coordinates or bins, computes
the input gradients over all models, returning an M x N x I x 4 array of
gradient values and an N x I x 4 array of one-hot encoded sequence.
"""
num_models, num_samples = len(model_paths), len(sample)
all_input_grads = np.empty((num_models, num_samples, input_length, 4))
all_one_hot_seqs = np.empty((num_samples, input_length, 4))
for i in tqdm.notebook.trange(num_models):
model = restore_model(model_paths[i])
if model_type == "profile":
results = compute_predictions.get_profile_model_predictions(
model, sample, num_tasks, input_func, controls=controls,
return_losses=False, return_gradients=True, show_progress=False
)
else:
results = compute_predictions.get_binary_model_predictions(
model, sample, input_func,
return_losses=False, return_gradients=True, show_progress=False
)
all_input_grads[i] = results["input_grads"]
if i == 0:
all_one_hot_seqs = results["input_seqs"]
return all_input_grads, all_one_hot_seqs
def compute_shap_scores(model_paths, sample, batch_size=128):
"""
Given a list of paths to M models and a list of N coordinates or bins, computes
the SHAP scores over all models, returning an M x N x I x 4 array of
SHAP scores and an N x I x 4 array of one-hot encoded sequence.
"""
num_models, num_samples = len(model_paths), len(sample)
num_batches = int(np.ceil(num_samples / batch_size))
all_shap_scores = np.empty((num_models, num_samples, input_length, 4))
all_one_hot_seqs = np.empty((num_samples, input_length, 4))
for i in tqdm.notebook.trange(num_models):
model = restore_model(model_paths[i])
if model_type == "profile":
shap_explainer = compute_shap.create_profile_explainer(
model, input_length, profile_length, num_tasks, num_strands, controls,
task_index=task_index
)
else:
shap_explainer = compute_shap.create_binary_explainer(
model, input_length, task_index=task_index
)
for j in range(num_batches):
batch_slice = slice(j * batch_size, (j + 1) * batch_size)
batch = sample[batch_slice]
if model_type == "profile":
input_seqs, profiles = input_func(sample)
shap_scores = shap_explainer(
input_seqs, cont_profs=profiles[:, num_tasks:], hide_shap_output=True
)
else:
input_seqs, _, _ = input_func(sample)
shap_scores = shap_explainer(
input_seqs, hide_shap_output=True
)
all_shap_scores[i, batch_slice] = shap_scores
if i == 0:
all_one_hot_seqs[batch_slice] = input_seqs
return all_shap_scores, all_one_hot_seqs
# Compute the importance scores and 1-hot seqs
imp_type = ("DeepSHAP scores", "input gradients")[0]
imp_func = compute_shap_scores if imp_type == "DeepSHAP scores" else compute_gradients
noprior_scores, _ = imp_func(noprior_model_paths, sample)
prior_scores, one_hot_seqs = imp_func(prior_model_paths, sample)
```
### Compute similarity
```
def cont_jaccard(seq_1, seq_2):
"""
Takes two gradient sequences (I x 4 arrays) and computes a similarity between
them, using a continuous Jaccard metric.
"""
# L1-normalize
norm_1 = np.sum(np.abs(seq_1), axis=1, keepdims=True)
norm_2 = np.sum(np.abs(seq_2), axis=1, keepdims=True)
norm_1[norm_1 == 0] = 1
norm_2[norm_2 == 0] = 1
seq_1 = seq_1 / norm_1
seq_2 = seq_2 / norm_2
ab_1, ab_2 = np.abs(seq_1), np.abs(seq_2)
inter = np.sum(np.minimum(ab_1, ab_2) * np.sign(seq_1) * np.sign(seq_2), axis=1)
union = np.sum(np.maximum(ab_1, ab_2), axis=1)
zero_mask = union == 0
inter[zero_mask] = 0
union[zero_mask] = 1
return np.sum(inter / union)
def cosine_sim(seq_1, seq_2):
"""
Takes two gradient sequences (I x 4 arrays) and computes a similarity between
them, using a cosine similarity.
"""
seq_1, seq_2 = np.ravel(seq_1), np.ravel(seq_2)
dot = np.sum(seq_1 * seq_2)
mag_1, mag_2 = np.sqrt(np.sum(seq_1 * seq_1)), np.sqrt(np.sum(seq_2 * seq_2))
return dot / (mag_1 * mag_2) if mag_1 * mag_2 else 0
def compute_similarity_matrix(imp_scores, sim_func=cosine_sim):
"""
Given the M x N x I x 4 importance scores returned by `compute_gradients`
or `compute_shap_scores`, computes an N x M x M similarity matrix of
similarity across models (i.e. each coordinate gets a similarity matrix
across different models). By default uses cosine similarity.
"""
num_models, num_coords = imp_scores.shape[0], imp_scores.shape[1]
sim_mats = np.empty((num_coords, num_models, num_models))
for i in tqdm.notebook.trange(num_coords):
for j in range(num_models):
sim_mats[i, j, j] = 0
for k in range(j):
sim_score = sim_func(imp_scores[j][i], imp_scores[k][i])
sim_mats[i, j, k] = sim_score
sim_mats[i, k, j] = sim_score
return sim_mats
sim_type = ("Cosine", "Continuous Jaccard")[1]
sim_func = cosine_sim if sim_type == "Cosine" else cont_jaccard
noprior_sim_matrix = compute_similarity_matrix(noprior_scores, sim_func=sim_func)
prior_sim_matrix = compute_similarity_matrix(prior_scores, sim_func=sim_func)
# Plot some examples of poor consistency, particularly ones that showed an improvement
num_to_show = 100
center_view_length = 200
plot_zoom = True
midpoint = input_length // 2
start = midpoint - (center_view_length // 2)
end = start + center_view_length
center_slice = slice(550, 800)
noprior_sim_matrix_copy = noprior_sim_matrix.copy()
for i in range(len(noprior_sim_matrix_copy)):
noprior_sim_matrix_copy[i][np.diag_indices(noprior_sim_matrix.shape[1])] = np.inf # Put infinity in diagonal
diffs = np.max(prior_sim_matrix, axis=(1, 2)) - np.min(noprior_sim_matrix_copy, axis=(1, 2))
best_example_inds = np.flip(np.argsort(diffs))[:num_to_show]
best_example_inds = [7] #, 38]
for sample_index in best_example_inds:
noprior_model_ind_1, noprior_model_ind_2 = np.unravel_index(np.argmin(np.ravel(noprior_sim_matrix_copy[sample_index])), noprior_sim_matrix[sample_index].shape)
prior_model_ind_1, prior_model_ind_2 = np.unravel_index(np.argmax(np.ravel(prior_sim_matrix[sample_index])), prior_sim_matrix[sample_index].shape)
noprior_model_ind_1, noprior_model_ind_2 = 5, 17
prior_model_ind_1, prior_model_ind_2 = 13, 17
print("Sample index: %d" % sample_index)
if model_type == "binary":
bin_index = sample[sample_index]
coord = input_func(np.array([bin_index]))[2][0]
print("Coordinate: %s (bin %d)" % (str(coord), bin_index))
else:
coord = sample[sample_index]
print("Coordinate: %s" % str(coord))
print("Model indices without prior: %d vs %d" % (noprior_model_ind_1, noprior_model_ind_2))
plt.figure(figsize=(20, 2))
plt.plot(np.sum(noprior_scores[noprior_model_ind_1, sample_index] * one_hot_seqs[sample_index], axis=1), color="coral")
plt.show()
if plot_zoom:
viz_sequence.plot_weights(noprior_scores[noprior_model_ind_1, sample_index, center_slice], subticks_frequency=1000)
viz_sequence.plot_weights(noprior_scores[noprior_model_ind_1, sample_index, center_slice] * one_hot_seqs[sample_index, center_slice], subticks_frequency=1000)
plt.figure(figsize=(20, 2))
plt.plot(np.sum(noprior_scores[noprior_model_ind_2, sample_index] * one_hot_seqs[sample_index], axis=1), color="coral")
plt.show()
if plot_zoom:
viz_sequence.plot_weights(noprior_scores[noprior_model_ind_2, sample_index, center_slice], subticks_frequency=1000)
viz_sequence.plot_weights(noprior_scores[noprior_model_ind_2, sample_index, center_slice] * one_hot_seqs[sample_index, center_slice], subticks_frequency=1000)
print("Model indices with prior: %d vs %d" % (prior_model_ind_1, prior_model_ind_2))
plt.figure(figsize=(20, 2))
plt.plot(np.sum(prior_scores[prior_model_ind_1, sample_index] * one_hot_seqs[sample_index], axis=1), color="slateblue")
plt.show()
if plot_zoom:
viz_sequence.plot_weights(prior_scores[prior_model_ind_1, sample_index, center_slice], subticks_frequency=1000)
viz_sequence.plot_weights(prior_scores[prior_model_ind_1, sample_index, center_slice] * one_hot_seqs[sample_index, center_slice], subticks_frequency=1000)
plt.figure(figsize=(20, 2))
plt.plot(np.sum(prior_scores[prior_model_ind_2, sample_index] * one_hot_seqs[sample_index], axis=1), color="slateblue")
plt.show()
if plot_zoom:
viz_sequence.plot_weights(prior_scores[prior_model_ind_2, sample_index, center_slice], subticks_frequency=1000)
viz_sequence.plot_weights(prior_scores[prior_model_ind_2, sample_index, center_slice] * one_hot_seqs[sample_index, center_slice], subticks_frequency=1000)
sample_index = 7
for i in range(30):
print(i)
plt.figure(figsize=(20, 2))
plt.plot(np.sum(noprior_scores[i, sample_index] * one_hot_seqs[sample_index], axis=1), color="coral")
plt.show()
for i in range(30):
print(i)
plt.figure(figsize=(20, 2))
plt.plot(np.sum(prior_scores[i, sample_index] * one_hot_seqs[sample_index], axis=1), color="coral")
plt.show()
noprior_avg_sims, prior_avg_sims = [], []
bin_num = 30
for i in range(num_samples):
noprior_avg_sims.append(np.mean(noprior_sim_matrix[i][np.tril_indices(len(noprior_model_paths), k=-1)]))
prior_avg_sims.append(np.mean(prior_sim_matrix[i][np.tril_indices(len(prior_model_paths), k=-1)]))
noprior_avg_sims, prior_avg_sims = np.array(noprior_avg_sims), np.array(prior_avg_sims)
all_vals = np.concatenate([noprior_avg_sims, prior_avg_sims])
bins = np.linspace(np.min(all_vals), np.max(all_vals), bin_num)
fig, ax = plt.subplots(figsize=(16, 8))
ax.hist(noprior_avg_sims, bins=bins, color="coral", label="No prior", alpha=0.7)
ax.hist(prior_avg_sims, bins=bins, color="slateblue", label="With Fourier prior", alpha=0.7)
plt.legend()
plt.title(
("Mean pairwise similarities of %s between different random initializations" % imp_type) +
("\n%s %s models" % (condition_name, model_type)) +
"\nComputed over %d/%d models without/with Fourier prior on %d randomly drawn test peaks" % (len(noprior_model_paths), len(prior_model_paths), num_samples)
)
plt.xlabel("%s similarity" % sim_type)
print("Average similarity without priors: %f" % np.nanmean(noprior_avg_sims))
print("Average similarity with priors: %f" % np.nanmean(prior_avg_sims))
print("Standard error without priors: %f" % scipy.stats.sem(noprior_avg_sims, nan_policy="omit"))
print("Standard error with priors: %f" % scipy.stats.sem(prior_avg_sims, nan_policy="omit"))
w, p = scipy.stats.wilcoxon(noprior_avg_sims, prior_avg_sims, alternative="less")
print("One-sided Wilcoxon test: w = %f, p = %f" % (w, p))
avg_sim_diffs = prior_avg_sims - noprior_avg_sims
plt.figure(figsize=(16, 8))
plt.hist(avg_sim_diffs, bins=30, color="mediumorchid")
plt.title(
("Paired difference of %s similarity between different random initializations" % imp_type) +
("\n%s %s models" % (condition_name, model_type)) +
"\nComputed over %d/%d models without/with Fourier prior on %d randomly drawn test peaks" % (len(noprior_model_paths), len(prior_model_paths), num_samples)
)
plt.xlabel("Average similarity difference: with Fourier prior - no prior")
def get_bias(sim_matrix):
num_examples, num_models, _ = sim_matrix.shape
bias_vals = []
for i in range(num_models):
avg = np.sum(sim_matrix[:, i]) / (num_examples * (num_models - 1))
bias_vals.append(avg)
print("%d: %f" % (i + 1, avg))
return bias_vals
print("Model-specific bias without priors")
noprior_bias_vals = get_bias(noprior_sim_matrix)
print("Model-specific bias with priors")
prior_bias_vals = get_bias(prior_sim_matrix)
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
fig.suptitle("Model-specific average Jaccard similarity vs model performance")
ax[0].scatter(noprior_bias_vals, np.array(noprior_metric_vals)[noprior_keep_mask])
ax[0].set_title("No priors")
ax[1].scatter(prior_bias_vals, np.array(prior_metric_vals)[prior_keep_mask])
ax[1].set_title("With priors")
plt.grid(False)
fig.text(0.5, 0.04, "Average Jaccard similarity with other models over all samples", ha="center", va="center")
fig.text(0.06, 0.5, "Model profile validation loss", ha="center", va="center", rotation="vertical")
# Compute some simple bounds on the expected consistency using
# the "no-prior" scores
rng = np.random.RandomState(1234)
def shuf_none(track):
# Do nothing
return track
def shuf_bases(track):
# Shuffle the importances across each base dimension separately,
# but keep positions intact
inds = np.random.rand(*track.shape).argsort(axis=1) # Each row is 0,1,2,3 in random order
return np.take_along_axis(track, inds, axis=1)
def shuf_pos(track):
# Shuffle the importances across the positions, but keep the base
# importances at each position intact
shuf = np.copy(track)
rng.shuffle(shuf)
return shuf
def shuf_all(track):
# Shuffle the importances across positions and bases
return np.ravel(track)[rng.permutation(track.size)].reshape(track.shape)
for shuf_type, shuf_func in [
("no", shuf_none), ("base", shuf_bases), ("position", shuf_pos), ("all", shuf_all)
]:
sims = []
for i in tqdm.notebook.trange(noprior_scores.shape[0]):
for j in range(noprior_scores.shape[1]):
track = noprior_scores[i, j]
track_shuf = shuf_func(track)
sims.append(sim_func(track, track_shuf))
fig, ax = plt.subplots()
ax.hist(sims, bins=30)
ax.set_title("%s similarity with %s shuffing" % (sim_type, shuf_type))
plt.show()
print("Mean: %f" % np.mean(sims))
print("Standard deviation: %f" % np.std(sims))
```
| github_jupyter |
# 7.6 Transformerモデル(分類タスク用)の実装
- 本ファイルでは、クラス分類のTransformerモデルを実装します。
※ 本章のファイルはすべてUbuntuでの動作を前提としています。Windowsなど文字コードが違う環境での動作にはご注意下さい。
# 7.6 学習目標
1. Transformerのモジュール構成を理解する
2. LSTMやRNNを使用せずCNNベースのTransformerで自然言語処理が可能な理由を理解する
3. Transformerを実装できるようになる
# 事前準備
書籍の指示に従い、本章で使用するデータを用意します
```
import math
import numpy as np
import random
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchtext
# Setup seeds
torch.manual_seed(1234)
np.random.seed(1234)
random.seed(1234)
class Embedder(nn.Module):
'''idで示されている単語をベクトルに変換します'''
def __init__(self, text_embedding_vectors):
super(Embedder, self).__init__()
self.embeddings = nn.Embedding.from_pretrained(
embeddings=text_embedding_vectors, freeze=True)
# freeze=Trueによりバックプロパゲーションで更新されず変化しなくなります
def forward(self, x):
x_vec = self.embeddings(x)
return x_vec
# 動作確認
# 前節のDataLoaderなどを取得
from utils.dataloader import get_IMDb_DataLoaders_and_TEXT
train_dl, val_dl, test_dl, TEXT = get_IMDb_DataLoaders_and_TEXT(
max_length=256, batch_size=24)
# ミニバッチの用意
batch = next(iter(train_dl))
# モデル構築
net1 = Embedder(TEXT.vocab.vectors)
# 入出力
x = batch.Text[0]
x1 = net1(x) # 単語をベクトルに
print("入力のテンソルサイズ:", x.shape)
print("出力のテンソルサイズ:", x1.shape)
class PositionalEncoder(nn.Module):
'''入力された単語の位置を示すベクトル情報を付加する'''
def __init__(self, d_model=300, max_seq_len=256):
super().__init__()
self.d_model = d_model # 単語ベクトルの次元数
# 単語の順番(pos)と埋め込みベクトルの次元の位置(i)によって一意に定まる値の表をpeとして作成
pe = torch.zeros(max_seq_len, d_model)
# GPUが使える場合はGPUへ送る、ここでは省略。実際に学習時には使用する
# device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# pe = pe.to(device)
for pos in range(max_seq_len):
for i in range(0, d_model, 2):
pe[pos, i] = math.sin(pos / (10000 ** ((2 * i)/d_model)))
pe[pos, i + 1] = math.cos(pos /
(10000 ** ((2 * (i + 1))/d_model)))
# 表peの先頭に、ミニバッチ次元となる次元を足す
self.pe = pe.unsqueeze(0)
# 勾配を計算しないようにする
self.pe.requires_grad = False
def forward(self, x):
# 入力xとPositonal Encodingを足し算する
# xがpeよりも小さいので、大きくする
ret = math.sqrt(self.d_model)*x + self.pe
return ret
# 動作確認
# モデル構築
net1 = Embedder(TEXT.vocab.vectors)
net2 = PositionalEncoder(d_model=300, max_seq_len=256)
# 入出力
x = batch.Text[0]
x1 = net1(x) # 単語をベクトルに
x2 = net2(x1)
print("入力のテンソルサイズ:", x1.shape)
print("出力のテンソルサイズ:", x2.shape)
class Attention(nn.Module):
'''Transformerは本当はマルチヘッドAttentionですが、
分かりやすさを優先しシングルAttentionで実装します'''
def __init__(self, d_model=300):
super().__init__()
# SAGANでは1dConvを使用したが、今回は全結合層で特徴量を変換する
self.q_linear = nn.Linear(d_model, d_model)
self.v_linear = nn.Linear(d_model, d_model)
self.k_linear = nn.Linear(d_model, d_model)
# 出力時に使用する全結合層
self.out = nn.Linear(d_model, d_model)
# Attentionの大きさ調整の変数
self.d_k = d_model
def forward(self, q, k, v, mask):
# 全結合層で特徴量を変換
k = self.k_linear(k)
q = self.q_linear(q)
v = self.v_linear(v)
# Attentionの値を計算する
# 各値を足し算すると大きくなりすぎるので、root(d_k)で割って調整
weights = torch.matmul(q, k.transpose(1, 2)) / math.sqrt(self.d_k)
# ここでmaskを計算
mask = mask.unsqueeze(1)
weights = weights.masked_fill(mask == 0, -1e9)
# softmaxで規格化をする
normlized_weights = F.softmax(weights, dim=-1)
# AttentionをValueとかけ算
output = torch.matmul(normlized_weights, v)
# 全結合層で特徴量を変換
output = self.out(output)
return output, normlized_weights
class FeedForward(nn.Module):
def __init__(self, d_model, d_ff=1024, dropout=0.1):
'''Attention層から出力を単純に全結合層2つで特徴量を変換するだけのユニットです'''
super().__init__()
self.linear_1 = nn.Linear(d_model, d_ff)
self.dropout = nn.Dropout(dropout)
self.linear_2 = nn.Linear(d_ff, d_model)
def forward(self, x):
x = self.linear_1(x)
x = self.dropout(F.relu(x))
x = self.linear_2(x)
return x
class TransformerBlock(nn.Module):
def __init__(self, d_model, dropout=0.1):
super().__init__()
# LayerNormalization層
# https://pytorch.org/docs/stable/nn.html?highlight=layernorm
self.norm_1 = nn.LayerNorm(d_model)
self.norm_2 = nn.LayerNorm(d_model)
# Attention層
self.attn = Attention(d_model)
# Attentionのあとの全結合層2つ
self.ff = FeedForward(d_model)
# Dropout
self.dropout_1 = nn.Dropout(dropout)
self.dropout_2 = nn.Dropout(dropout)
def forward(self, x, mask):
# 正規化とAttention
x_normlized = self.norm_1(x)
output, normlized_weights = self.attn(
x_normlized, x_normlized, x_normlized, mask)
x2 = x + self.dropout_1(output)
# 正規化と全結合層
x_normlized2 = self.norm_2(x2)
output = x2 + self.dropout_2(self.ff(x_normlized2))
return output, normlized_weights
# 動作確認
# モデル構築
net1 = Embedder(TEXT.vocab.vectors)
net2 = PositionalEncoder(d_model=300, max_seq_len=256)
net3 = TransformerBlock(d_model=300)
# maskの作成
x = batch.Text[0]
input_pad = 1 # 単語のIDにおいて、'<pad>': 1 なので
input_mask = (x != input_pad)
print(input_mask[0])
# 入出力
x1 = net1(x) # 単語をベクトルに
x2 = net2(x1) # Positon情報を足し算
x3, normlized_weights = net3(x2, input_mask) # Self-Attentionで特徴量を変換
print("入力のテンソルサイズ:", x2.shape)
print("出力のテンソルサイズ:", x3.shape)
print("Attentionのサイズ:", normlized_weights.shape)
class ClassificationHead(nn.Module):
'''Transformer_Blockの出力を使用し、最後にクラス分類させる'''
def __init__(self, d_model=300, output_dim=2):
super().__init__()
# 全結合層
self.linear = nn.Linear(d_model, output_dim) # output_dimはポジ・ネガの2つ
# 重み初期化処理
nn.init.normal_(self.linear.weight, std=0.02)
nn.init.normal_(self.linear.bias, 0)
def forward(self, x):
x0 = x[:, 0, :] # 各ミニバッチの各文の先頭の単語の特徴量(300次元)を取り出す
out = self.linear(x0)
return out
# 動作確認
# ミニバッチの用意
batch = next(iter(train_dl))
# モデル構築
net1 = Embedder(TEXT.vocab.vectors)
net2 = PositionalEncoder(d_model=300, max_seq_len=256)
net3 = TransformerBlock(d_model=300)
net4 = ClassificationHead(output_dim=2, d_model=300)
# 入出力
x = batch.Text[0]
x1 = net1(x) # 単語をベクトルに
x2 = net2(x1) # Positon情報を足し算
x3, normlized_weights = net3(x2, input_mask) # Self-Attentionで特徴量を変換
x4 = net4(x3) # 最終出力の0単語目を使用して、分類0-1のスカラーを出力
print("入力のテンソルサイズ:", x3.shape)
print("出力のテンソルサイズ:", x4.shape)
# 最終的なTransformerモデルのクラス
class TransformerClassification(nn.Module):
'''Transformerでクラス分類させる'''
def __init__(self, text_embedding_vectors, d_model=300, max_seq_len=256, output_dim=2):
super().__init__()
# モデル構築
self.net1 = Embedder(text_embedding_vectors)
self.net2 = PositionalEncoder(d_model=d_model, max_seq_len=max_seq_len)
self.net3_1 = TransformerBlock(d_model=d_model)
self.net3_2 = TransformerBlock(d_model=d_model)
self.net4 = ClassificationHead(output_dim=output_dim, d_model=d_model)
def forward(self, x, mask):
x1 = self.net1(x) # 単語をベクトルに
x2 = self.net2(x1) # Positon情報を足し算
x3_1, normlized_weights_1 = self.net3_1(
x2, mask) # Self-Attentionで特徴量を変換
x3_2, normlized_weights_2 = self.net3_2(
x3_1, mask) # Self-Attentionで特徴量を変換
x4 = self.net4(x3_2) # 最終出力の0単語目を使用して、分類0-1のスカラーを出力
return x4, normlized_weights_1, normlized_weights_2
# 動作確認
# ミニバッチの用意
batch = next(iter(train_dl))
# モデル構築
net = TransformerClassification(
text_embedding_vectors=TEXT.vocab.vectors, d_model=300, max_seq_len=256, output_dim=2)
# 入出力
x = batch.Text[0]
input_mask = (x != input_pad)
out, normlized_weights_1, normlized_weights_2 = net(x, input_mask)
print("出力のテンソルサイズ:", out.shape)
print("出力テンソルのsigmoid:", F.softmax(out, dim=1))
```
ここまでの内容をフォルダ「utils」のtransformer.pyに別途保存しておき、次節からはこちらから読み込むようにします
以上
| github_jupyter |
# Run model module locally
```
import os
# Import os environment variables for file hyperparameters.
os.environ["TRAIN_FILE_PATTERN"] = "gs://machine-learning-1234-bucket/gan/data/cifar10/train*.tfrecord"
os.environ["EVAL_FILE_PATTERN"] = "gs://machine-learning-1234-bucket/gan/data/cifar10/test*.tfrecord"
os.environ["OUTPUT_DIR"] = "gs://machine-learning-1234-bucket/gan/cdcgan/trained_model2"
# Import os environment variables for train hyperparameters.
os.environ["TRAIN_BATCH_SIZE"] = str(100)
os.environ["TRAIN_STEPS"] = str(50000)
os.environ["SAVE_SUMMARY_STEPS"] = str(100)
os.environ["SAVE_CHECKPOINTS_STEPS"] = str(5000)
os.environ["KEEP_CHECKPOINT_MAX"] = str(10)
os.environ["INPUT_FN_AUTOTUNE"] = "False"
# Import os environment variables for eval hyperparameters.
os.environ["EVAL_BATCH_SIZE"] = str(16)
os.environ["EVAL_STEPS"] = str(10)
os.environ["START_DELAY_SECS"] = str(6000)
os.environ["THROTTLE_SECS"] = str(6000)
# Import os environment variables for image hyperparameters.
os.environ["HEIGHT"] = str(32)
os.environ["WIDTH"] = str(32)
os.environ["DEPTH"] = str(3)
# Import os environment variables for label hyperparameters.
num_classes = 10
os.environ["NUM_CLASSES"] = str(num_classes)
os.environ["LABEL_EMBEDDING_DIMENSION"] = str(10)
# Import os environment variables for generator hyperparameters.
os.environ["LATENT_SIZE"] = str(512)
os.environ["GENERATOR_PROJECTION_DIMS"] = "4,4,256"
os.environ["GENERATOR_USE_LABELS"] = "True"
os.environ["GENERATOR_EMBED_LABELS"] = "True"
os.environ["GENERATOR_CONCATENATE_LABELS"] = "True"
os.environ["GENERATOR_NUM_FILTERS"] = "128,128,128"
os.environ["GENERATOR_KERNEL_SIZES"] = "4,4,4"
os.environ["GENERATOR_STRIDES"] = "2,2,2"
os.environ["GENERATOR_FINAL_NUM_FILTERS"] = str(3)
os.environ["GENERATOR_FINAL_KERNEL_SIZE"] = str(3)
os.environ["GENERATOR_FINAL_STRIDE"] = str(1)
os.environ["GENERATOR_LEAKY_RELU_ALPHA"] = str(0.2)
os.environ["GENERATOR_FINAL_ACTIVATION"] = "tanh"
os.environ["GENERATOR_L1_REGULARIZATION_SCALE"] = str(0.)
os.environ["GENERATOR_L2_REGULARIZATION_SCALE"] = str(0.)
os.environ["GENERATOR_OPTIMIZER"] = "Adam"
os.environ["GENERATOR_LEARNING_RATE"] = str(0.0002)
os.environ["GENERATOR_ADAM_BETA1"] = str(0.5)
os.environ["GENERATOR_ADAM_BETA2"] = str(0.999)
os.environ["GENERATOR_ADAM_EPSILON"] = str(1e-8)
os.environ["GENERATOR_CLIP_GRADIENTS"] = "None"
os.environ["GENERATOR_TRAIN_STEPS"] = str(1)
# Import os environment variables for discriminator hyperparameters.
os.environ["DISCRIMINATOR_USE_LABELS"] = "True"
os.environ["DISCRIMINATOR_EMBED_LABELS"] = "True"
os.environ["DISCRIMINATOR_CONCATENATE_LABELS"] = "True"
os.environ["DISCRIMINATOR_NUM_FILTERS"] = "64,128,128,256"
os.environ["DISCRIMINATOR_KERNEL_SIZES"] = "3,3,3,3"
os.environ["DISCRIMINATOR_STRIDES"] = "1,2,2,2"
os.environ["DISCRIMINATOR_DROPOUT_RATES"] = "0.3,0.3,0.3,0.3"
os.environ["DISCRIMINATOR_LEAKY_RELU_ALPHA"] = str(0.2)
os.environ["DISCRIMINATOR_L1_REGULARIZATION_SCALE"] = str(0.)
os.environ["DISCRIMINATOR_L2_REGULARIZATION_SCALE"] = str(0.)
os.environ["DISCRIMINATOR_OPTIMIZER"] = "Adam"
os.environ["DISCRIMINATOR_LEARNING_RATE"] = str(0.0002)
os.environ["DISCRIMINATOR_ADAM_BETA1"] = str(0.5)
os.environ["DISCRIMINATOR_ADAM_BETA2"] = str(0.999)
os.environ["DISCRIMINATOR_ADAM_EPSILON"] = str(1e-8)
os.environ["DISCRIMINATOR_CLIP_GRADIENTS"] = "None"
os.environ["DISCRIMINATOR_TRAIN_STEPS"] = str(1)
os.environ["LABEL_SMOOTHING"] = str(0.9)
```
## Train cdcgan model
```
%%bash
gsutil -m rm -rf ${OUTPUT_DIR}
export PYTHONPATH=$PYTHONPATH:$PWD/cdcgan_module
python3 -m trainer.task \
--train_file_pattern=${TRAIN_FILE_PATTERN} \
--eval_file_pattern=${EVAL_FILE_PATTERN} \
--output_dir=${OUTPUT_DIR} \
--job-dir=./tmp \
\
--train_batch_size=${TRAIN_BATCH_SIZE} \
--train_steps=${TRAIN_STEPS} \
--save_summary_steps=${SAVE_SUMMARY_STEPS} \
--save_checkpoints_steps=${SAVE_CHECKPOINTS_STEPS} \
--keep_checkpoint_max=${KEEP_CHECKPOINT_MAX} \
--input_fn_autotune=${INPUT_FN_AUTOTUNE} \
\
--eval_batch_size=${EVAL_BATCH_SIZE} \
--eval_steps=${EVAL_STEPS} \
--start_delay_secs=${START_DELAY_SECS} \
--throttle_secs=${THROTTLE_SECS} \
\
--height=${HEIGHT} \
--width=${WIDTH} \
--depth=${DEPTH} \
\
--num_classes=${NUM_CLASSES} \
--label_embedding_dimension=${LABEL_EMBEDDING_DIMENSION} \
\
--latent_size=${LATENT_SIZE} \
--generator_projection_dims=${GENERATOR_PROJECTION_DIMS} \
--generator_use_labels=${GENERATOR_USE_LABELS} \
--generator_embed_labels=${GENERATOR_EMBED_LABELS} \
--generator_concatenate_labels=${GENERATOR_CONCATENATE_LABELS} \
--generator_num_filters=${GENERATOR_NUM_FILTERS} \
--generator_kernel_sizes=${GENERATOR_KERNEL_SIZES} \
--generator_strides=${GENERATOR_STRIDES} \
--generator_final_num_filters=${GENERATOR_FINAL_NUM_FILTERS} \
--generator_final_kernel_size=${GENERATOR_FINAL_KERNEL_SIZE} \
--generator_final_stride=${GENERATOR_FINAL_STRIDE} \
--generator_leaky_relu_alpha=${GENERATOR_LEAKY_RELU_ALPHA} \
--generator_final_activation=${GENERATOR_FINAL_ACTIVATION} \
--generator_l1_regularization_scale=${GENERATOR_L1_REGULARIZATION_SCALE} \
--generator_l2_regularization_scale=${GENERATOR_L2_REGULARIZATION_SCALE} \
--generator_optimizer=${GENERATOR_OPTIMIZER} \
--generator_learning_rate=${GENERATOR_LEARNING_RATE} \
--generator_adam_beta1=${GENERATOR_ADAM_BETA1} \
--generator_adam_beta2=${GENERATOR_ADAM_BETA2} \
--generator_adam_epsilon=${GENERATOR_ADAM_EPSILON} \
--generator_clip_gradients=${GENERATOR_CLIP_GRADIENTS} \
--generator_train_steps=${GENERATOR_TRAIN_STEPS} \
\
--discriminator_use_labels=${DISCRIMINATOR_USE_LABELS} \
--discriminator_embed_labels=${DISCRIMINATOR_EMBED_LABELS} \
--discriminator_concatenate_labels=${DISCRIMINATOR_CONCATENATE_LABELS} \
--discriminator_num_filters=${DISCRIMINATOR_NUM_FILTERS} \
--discriminator_kernel_sizes=${DISCRIMINATOR_KERNEL_SIZES} \
--discriminator_strides=${DISCRIMINATOR_STRIDES} \
--discriminator_dropout_rates=${DISCRIMINATOR_DROPOUT_RATES} \
--discriminator_leaky_relu_alpha=${DISCRIMINATOR_LEAKY_RELU_ALPHA} \
--discriminator_l1_regularization_scale=${DISCRIMINATOR_L1_REGULARIZATION_SCALE} \
--discriminator_l2_regularization_scale=${DISCRIMINATOR_L2_REGULARIZATION_SCALE} \
--discriminator_optimizer=${DISCRIMINATOR_OPTIMIZER} \
--discriminator_learning_rate=${DISCRIMINATOR_LEARNING_RATE} \
--discriminator_adam_beta1=${DISCRIMINATOR_ADAM_BETA1} \
--discriminator_adam_beta2=${DISCRIMINATOR_ADAM_BETA2} \
--discriminator_adam_epsilon=${DISCRIMINATOR_ADAM_EPSILON} \
--discriminator_clip_gradients=${DISCRIMINATOR_CLIP_GRADIENTS} \
--discriminator_train_steps=${DISCRIMINATOR_TRAIN_STEPS} \
--label_smoothing=${LABEL_SMOOTHING}
```
## Prediction
```
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
!gsutil ls gs://machine-learning-1234-bucket/gan/cdcgan/trained_model2/export/exporter
predict_fn = tf.contrib.predictor.from_saved_model(
"gs://machine-learning-1234-bucket/gan/cdcgan/trained_model2/export/exporter/1592859903"
)
predictions = predict_fn(
{
"Z": np.random.normal(size=(num_classes, 512)),
"label": np.arange(num_classes)
}
)
print(list(predictions.keys()))
```
Convert image back to the original scale.
```
generated_images = np.clip(
a=((predictions["generated_images"] + 1.0) * (255. / 2)).astype(np.int32),
a_min=0,
a_max=255
)
print(generated_images.shape)
def plot_images(images):
"""Plots images.
Args:
images: np.array, array of images of
[num_images, height, width, depth].
"""
num_images = len(images)
plt.figure(figsize=(20, 20))
for i in range(num_images):
image = images[i]
plt.subplot(1, num_images, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(
image,
cmap=plt.cm.binary
)
plt.show()
plot_images(generated_images)
```
| github_jupyter |
# Zircon model training notebook; (extensively) modified from Detectron2 training tutorial
This Colab Notebook will allow users to train new models to detect and segment detrital zircon from RL images using Detectron2 and the training dataset provided in the colab_zirc_dims repo. It is set up to train a Mask RCNN model (ResNet depth=101), but could be modified for other instance segmentation models provided that they are supported by Detectron2.
The training dataset should be uploaded to the user's Google Drive before running this notebook.
## Install detectron2
```
!pip install pyyaml==5.1
import torch
TORCH_VERSION = ".".join(torch.__version__.split(".")[:2])
CUDA_VERSION = torch.__version__.split("+")[-1]
print("torch: ", TORCH_VERSION, "; cuda: ", CUDA_VERSION)
# Install detectron2 that matches the above pytorch version
# See https://detectron2.readthedocs.io/tutorials/install.html for instructions
!pip install detectron2 -f https://dl.fbaipublicfiles.com/detectron2/wheels/$CUDA_VERSION/torch$TORCH_VERSION/index.html
exit(0) # Automatically restarts runtime after installation
# Some basic setup:
# Setup detectron2 logger
import detectron2
from detectron2.utils.logger import setup_logger
setup_logger()
# import some common libraries
import numpy as np
import os, json, cv2, random
from google.colab.patches import cv2_imshow
import copy
import time
import datetime
import logging
import random
import shutil
import torch
# import some common detectron2 utilities
from detectron2.engine.hooks import HookBase
from detectron2 import model_zoo
from detectron2.evaluation import inference_context, COCOEvaluator
from detectron2.engine import DefaultPredictor
from detectron2.config import get_cfg
from detectron2.utils.visualizer import Visualizer
from detectron2.utils.logger import log_every_n_seconds
from detectron2.data import MetadataCatalog, DatasetCatalog, build_detection_train_loader, DatasetMapper, build_detection_test_loader
import detectron2.utils.comm as comm
from detectron2.data import detection_utils as utils
from detectron2.config import LazyConfig
import detectron2.data.transforms as T
```
## Define Augmentations
The cell below defines augmentations used while training to ensure that models never see the same exact image twice during training. This mitigates overfitting and allows models to achieve substantially higher accuracy in their segmentations/measurements.
```
custom_transform_list = [T.ResizeShortestEdge([800,800]), #resize shortest edge of image to 800 pixels
T.RandomCrop('relative', (0.95, 0.95)), #randomly crop an area (95% size of original) from image
T.RandomLighting(100), #minor lighting randomization
T.RandomContrast(.85, 1.15), #minor contrast randomization
T.RandomFlip(prob=.5, horizontal=False, vertical=True), #random vertical flipping
T.RandomFlip(prob=.5, horizontal=True, vertical=False), #and horizontal flipping
T.RandomApply(T.RandomRotation([-30, 30], False), prob=.8), #random (80% probability) rotation up to 30 degrees; \
# more rotation does not seem to improve results
T.ResizeShortestEdge([800,800])] # resize img again for uniformity
```
## Mount Google Drive, set paths to dataset, model saving directories
```
from google.colab import drive
drive.mount('/content/drive')
#@markdown ### Add path to training dataset directory
dataset_dir = '/content/drive/MyDrive/training_dataset' #@param {type:"string"}
#@markdown ### Add path to model saving directory (automatically created if it does not yet exist)
model_save_dir = '/content/drive/MyDrive/NAME FOR MODEL SAVING FOLDER HERE' #@param {type:"string"}
os.makedirs(model_save_dir, exist_ok=True)
```
## Define dataset mapper, training, loss eval functions
```
from detectron2.engine import DefaultTrainer
from detectron2.data import DatasetMapper
from detectron2.structures import BoxMode
# a function to convert Via image annotation .json dict format to Detectron2 \
# training input dict format
def get_zircon_dicts(img_dir):
json_file = os.path.join(img_dir, "via_region_data.json")
with open(json_file) as f:
imgs_anns = json.load(f)['_via_img_metadata']
dataset_dicts = []
for idx, v in enumerate(imgs_anns.values()):
record = {}
filename = os.path.join(img_dir, v["filename"])
height, width = cv2.imread(filename).shape[:2]
record["file_name"] = filename
record["image_id"] = idx
record["height"] = height
record["width"] = width
#annos = v["regions"]
annos = {}
for n, eachitem in enumerate(v['regions']):
annos[str(n)] = eachitem
objs = []
for _, anno in annos.items():
#assert not anno["region_attributes"]
anno = anno["shape_attributes"]
px = anno["all_points_x"]
py = anno["all_points_y"]
poly = [(x + 0.5, y + 0.5) for x, y in zip(px, py)]
poly = [p for x in poly for p in x]
obj = {
"bbox": [np.min(px), np.min(py), np.max(px), np.max(py)],
"bbox_mode": BoxMode.XYXY_ABS,
"segmentation": [poly],
"category_id": 0,
}
objs.append(obj)
record["annotations"] = objs
dataset_dicts.append(record)
return dataset_dicts
# loss eval hook for getting vaidation loss, copying to metrics.json; \
# from https://gist.github.com/ortegatron/c0dad15e49c2b74de8bb09a5615d9f6b
class LossEvalHook(HookBase):
def __init__(self, eval_period, model, data_loader):
self._model = model
self._period = eval_period
self._data_loader = data_loader
def _do_loss_eval(self):
# Copying inference_on_dataset from evaluator.py
total = len(self._data_loader)
num_warmup = min(5, total - 1)
start_time = time.perf_counter()
total_compute_time = 0
losses = []
for idx, inputs in enumerate(self._data_loader):
if idx == num_warmup:
start_time = time.perf_counter()
total_compute_time = 0
start_compute_time = time.perf_counter()
if torch.cuda.is_available():
torch.cuda.synchronize()
total_compute_time += time.perf_counter() - start_compute_time
iters_after_start = idx + 1 - num_warmup * int(idx >= num_warmup)
seconds_per_img = total_compute_time / iters_after_start
if idx >= num_warmup * 2 or seconds_per_img > 5:
total_seconds_per_img = (time.perf_counter() - start_time) / iters_after_start
eta = datetime.timedelta(seconds=int(total_seconds_per_img * (total - idx - 1)))
log_every_n_seconds(
logging.INFO,
"Loss on Validation done {}/{}. {:.4f} s / img. ETA={}".format(
idx + 1, total, seconds_per_img, str(eta)
),
n=5,
)
loss_batch = self._get_loss(inputs)
losses.append(loss_batch)
mean_loss = np.mean(losses)
self.trainer.storage.put_scalar('validation_loss', mean_loss)
comm.synchronize()
return losses
def _get_loss(self, data):
# How loss is calculated on train_loop
metrics_dict = self._model(data)
metrics_dict = {
k: v.detach().cpu().item() if isinstance(v, torch.Tensor) else float(v)
for k, v in metrics_dict.items()
}
total_losses_reduced = sum(loss for loss in metrics_dict.values())
return total_losses_reduced
def after_step(self):
next_iter = self.trainer.iter + 1
is_final = next_iter == self.trainer.max_iter
if is_final or (self._period > 0 and next_iter % self._period == 0):
self._do_loss_eval()
#trainer for zircons which incorporates augmentation, hooks for eval
class ZirconTrainer(DefaultTrainer):
@classmethod
def build_train_loader(cls, cfg):
#return a custom train loader with augmentations; recompute_boxes \
# is important given cropping, rotation augs
return build_detection_train_loader(cfg, mapper=
DatasetMapper(cfg, is_train=True, recompute_boxes = True,
augmentations = custom_transform_list
),
)
@classmethod
def build_evaluator(cls, cfg, dataset_name, output_folder=None):
if output_folder is None:
output_folder = os.path.join(cfg.OUTPUT_DIR, "inference")
return COCOEvaluator(dataset_name, cfg, True, output_folder)
#set up validation loss eval hook
def build_hooks(self):
hooks = super().build_hooks()
hooks.insert(-1,LossEvalHook(
cfg.TEST.EVAL_PERIOD,
self.model,
build_detection_test_loader(
self.cfg,
self.cfg.DATASETS.TEST[0],
DatasetMapper(self.cfg,True)
)
))
return hooks
```
## Import train, val catalogs
```
#registers training, val datasets (converts annotations using get_zircon_dicts)
for d in ["train", "val"]:
DatasetCatalog.register("zircon_" + d, lambda d=d: get_zircon_dicts(dataset_dir + "/" + d))
MetadataCatalog.get("zircon_" + d).set(thing_classes=["zircon"])
zircon_metadata = MetadataCatalog.get("zircon_train")
train_cat = DatasetCatalog.get("zircon_train")
```
## Visualize train dataset
```
# visualize random sample from training dataset
dataset_dicts = get_zircon_dicts(os.path.join(dataset_dir, 'train'))
for d in random.sample(dataset_dicts, 4): #change int here to change sample size
img = cv2.imread(d["file_name"])
visualizer = Visualizer(img[:, :, ::-1], metadata=zircon_metadata, scale=0.5)
out = visualizer.draw_dataset_dict(d)
cv2_imshow(out.get_image()[:, :, ::-1])
```
# Define save to Drive function
```
# a function to save models (with iteration number in name), metrics to drive; \
# important in case training crashes or is left unattended and disconnects. \
def save_outputs_to_drive(model_name, iters):
root_output_dir = os.path.join(model_save_dir, model_name) #output_dir = save dir from user input
#creates individual model output directory if it does not already exist
os.makedirs(root_output_dir, exist_ok=True)
#creates a name for this version of model; include iteration number
curr_iters_str = str(round(iters/1000, 1)) + 'k'
curr_model_name = model_name + '_' + curr_iters_str + '.pth'
model_save_pth = os.path.join(root_output_dir, curr_model_name)
#get most recent model, current metrics, copy to drive
model_path = os.path.join(cfg.OUTPUT_DIR, "model_final.pth")
metrics_path = os.path.join(cfg.OUTPUT_DIR, 'metrics.json')
shutil.copy(model_path, model_save_pth)
shutil.copy(metrics_path, root_output_dir)
```
## Build, train model
### Set some parameters for training
```
#@markdown ### Add a base name for the model
model_save_name = 'your model name here' #@param {type:"string"}
#@markdown ### Final iteration before training stops
final_iteration = 8000 #@param {type:"slider", min:3000, max:15000, step:1000}
```
### Actually build and train model
```
#train from a pre-trained Mask RCNN model
cfg = get_cfg()
# train from base model: Default Mask RCNN
cfg.merge_from_file(model_zoo.get_config_file("COCO-InstanceSegmentation/mask_rcnn_R_101_FPN_3x.yaml"))
# Load starting weights (COCO trained) from Detectron2 model zoo.
cfg.MODEL.WEIGHTS = "https://dl.fbaipublicfiles.com/detectron2/COCO-InstanceSegmentation/mask_rcnn_R_101_FPN_3x/138205316/model_final_a3ec72.pkl"
cfg.DATASETS.TRAIN = ("zircon_train",) #load training dataset
cfg.DATASETS.TEST = ("zircon_val",) # load validation dataset
cfg.DATALOADER.NUM_WORKERS = 2
cfg.SOLVER.IMS_PER_BATCH = 2 #2 ims per batch seems to be good for model generalization
cfg.SOLVER.BASE_LR = 0.00025 # low but reasonable learning rate given pre-training; \
# by default initializes with a 1000 iteration warmup
cfg.SOLVER.MAX_ITER = 2000 #train for 2000 iterations before 1st save
cfg.SOLVER.GAMMA = 0.5
#decay learning rate by factor of GAMMA every 1000 iterations after 2000 iterations \
# and until 10000 iterations This works well for current version of training \
# dataset but should be modified (probably a longer interval) if dataset is ever\
# extended.
cfg.SOLVER.STEPS = (1999, 2999, 3999, 4999, 5999, 6999, 7999, 8999, 9999)
cfg.MODEL.ROI_HEADS.BATCH_SIZE_PER_IMAGE = 512 # use default ROI heads batch size
cfg.MODEL.ROI_HEADS.NUM_CLASSES = 1 # only class here is zircon
cfg.MODEL.RPN.NMS_THRESH = 0.1 #sets NMS threshold lower than default; should(?) eliminate overlapping regions
cfg.TEST.EVAL_PERIOD = 200 # validation eval every 200 iterations
os.makedirs(cfg.OUTPUT_DIR, exist_ok=True)
trainer = ZirconTrainer(cfg) #our zircon trainer, w/ built-in augs and val loss eval
trainer.resume_or_load(resume=False)
trainer.train() #start training
# stop training and save for the 1st time after 2000 iterations
save_outputs_to_drive(model_save_name, 2000)
# Saves, cold restarts training from saved model weights every 1000 iterations \
# until final iteration. This should probably be done via hooks without stopping \
# training but *seems* to produce faster decrease in validation loss.
for each_iters in [iter*1000 for iter in list(range(3,
int(final_iteration/1000) + 1,
1))]:
#reload model with last iteration model weights
resume_model_path = os.path.join(cfg.OUTPUT_DIR, "model_final.pth")
cfg.MODEL.WEIGHTS = resume_model_path
cfg.SOLVER.MAX_ITER = each_iters #increase max iterations
trainer = ZirconTrainer(cfg)
trainer.resume_or_load(resume=True)
trainer.train() #restart training
#save again
save_outputs_to_drive(model_save_name, each_iters)
# open tensorboard training metrics curves (metrics.json):
%load_ext tensorboard
%tensorboard --logdir output
```
## Inference & evaluation with final trained model
Initialize model from saved weights:
```
cfg.MODEL.WEIGHTS = os.path.join(cfg.OUTPUT_DIR, "model_final.pth") # final model; modify path to other non-final model to view their segmentations
cfg.MODEL.ROI_HEADS.SCORE_THRESH_TEST = 0.5 # set a custom testing threshold
cfg.MODEL.RPN.NMS_THRESH = 0.1
predictor = DefaultPredictor(cfg)
```
View model segmentations for random sample of images from zircon validation dataset:
```
from detectron2.utils.visualizer import ColorMode
dataset_dicts = get_zircon_dicts(os.path.join(dataset_dir, 'val'))
for d in random.sample(dataset_dicts, 5):
im = cv2.imread(d["file_name"])
outputs = predictor(im) # format is documented at https://detectron2.readthedocs.io/tutorials/models.html#model-output-format
v = Visualizer(im[:, :, ::-1],
metadata=zircon_metadata,
scale=1.5,
instance_mode=ColorMode.IMAGE_BW # remove the colors of unsegmented pixels. This option is only available for segmentation models
)
out = v.draw_instance_predictions(outputs["instances"].to("cpu"))
cv2_imshow(out.get_image()[:, :, ::-1])
```
Validation eval with COCO API metric:
```
from detectron2.evaluation import COCOEvaluator, inference_on_dataset
from detectron2.data import build_detection_test_loader
evaluator = COCOEvaluator("zircon_val", ("bbox", "segm"), False, output_dir="./output/")
val_loader = build_detection_test_loader(cfg, "zircon_val")
print(inference_on_dataset(trainer.model, val_loader, evaluator))
```
## Final notes:
To use newly-trained models in colab_zirc_dims:
#### Option A:
Modify the cell that initializes model(s) in colab_zirc_dims processing notebooks:
```
cfg.merge_from_file(model_zoo.get_config_file(DETECTRON2 BASE CONFIG FILE LINK FOR YOUR MODEL HERE))
cfg.MODEL.RESNETS.DEPTH = RESNET DEPTH FOR YOUR MODEL (E.G., 101) HERE
cfg.MODEL.WEIGHTS = PATH TO YOUR MODEL IN YOUR GOOGLE DRIVE HERE
```
#### Option B (more complicated but potentially useful for many models):
The dynamic model selection tool in colab_zirc_dims is populated from a .json file model library dictionary, which is by default [the current version on the GitHub repo.](https://github.com/MCSitar/colab_zirc_dims/blob/main/czd_model_library.json) The 'url' key in the dict will work with either an AWS download link for the model or the path to model in your Google Drive.
To use a custom model library dictionary:
Modify a copy of the colab_zirc_dims [.json file model library dictionary](https://github.com/MCSitar/colab_zirc_dims/blob/main/czd_model_library.json) to include download link(s)/Drive path(s) and metadata (e.g., resnet depth and config file) for your model(s). Upload this .json file to your Google Drive and change the 'model_lib_loc' variable in a processing Notebook to the .json's path for dynamic download and loading of this and other models within the Notebook.
| github_jupyter |
```
import torch
import torch.nn as nn
import numpy as np
import matplotlib.pyplot as plt
```
# Pytorch: An automatic differentiation tool
`Pytorch`를 활용하면 복잡한 함수의 미분을 손쉽게 + 효율적으로 계산할 수 있습니다!
`Pytorch`를 활용해서 복잡한 심층 신경망을 훈련할 때, 오차함수에 대한 파라미터의 편미분치를 계산을 손쉽게 수행할수 있습니다!
## Pytorch 첫만남
우리에게 아래와 같은 간단한 선형식이 주어져있다고 생각해볼까요?
$$ y = wx $$
그러면 $\frac{\partial y}{\partial w}$ 을 어떻게 계산 할 수 있을까요?
일단 직접 미분을 해보면$\frac{\partial y}{\partial w} = x$ 이 되니, 간단한
예제에서 `pytorch`로 해당 값을 계산하는 방법을 알아보도록 합시다!
```
# 랭크1 / 사이즈1 이며 값은 1*2 인 pytorch tensor를 하나 만듭니다.
x = torch.ones(1) * 2
# 랭크1 / 사이즈1 이며 값은 1 인 pytorch tensor를 하나 만듭니다.
w = torch.ones(1, requires_grad=True)
y = w * x
y
```
## 편미분 계산하기!
pytorch에서는 미분값을 계산하고 싶은 텐서에 `.backward()` 를 붙여주는 것으로, 해당 텐서 계산에 연결 되어있는 텐서 중 `gradient`를 계산해야하는 텐서(들)에 대한 편미분치들을 계산할수 있습니다. `requires_grad=True`를 통해서 어떤 텐서에 미분값을 계산할지 할당해줄 수 있습니다.
```
y.backward()
```
## 편미분값 확인하기!
`텐서.grad` 를 활용해서 특정 텐서의 gradient 값을 확인해볼 수 있습니다. 한번 `w.grad`를 활용해서 `y` 에 대한 `w`의 편미분값을 확인해볼까요?
```
w.grad
```
## 그러면 requires_grad = False 인 경우는?
```
x.grad
```
## `torch.nn`, Neural Network 패키지
`pytorch`에는 이미 다양한 neural network들의 모듈들을 구현해 놓았습니다. 그 중에 가장 간단하지만 정말 자주 쓰이는 `nn.Linear` 에 대해 알아보면서 `pytorch`의 `nn.Module`에 대해서 알아보도록 합시다.
## `nn.Linear` 돌아보기
`nn.Linear` 은 앞서 배운 선형회귀 및 다층 퍼셉트론 모델의 한 층에 해당하는 파라미터 $w$, $b$ 를 가지고 있습니다. 예시로 입력의 dimension 이 10이고 출력의 dimension 이 1인 `nn.Linear` 모듈을 만들어 봅시다!
```
lin = nn.Linear(in_features=10, out_features=1)
for p in lin.parameters():
print(p)
print(p.shape)
print('\n')
```
## `Linear` 모듈로 $y = Wx+b$ 계산하기
선형회귀식도 그랬지만, 다층 퍼셉트론 모델도 하나의 레이어는 아래의 수식을 계산했던 것을 기억하시죠?
$$y = Wx+b$$
`nn.Linear`를 활용해서 저 수식을 계산해볼까요?
검산을 쉽게 하기 위해서 W의 값은 모두 1.0 으로 b 는 5.0 으로 만들어두겠습니다.
```
lin.weight.data = torch.ones_like(lin.weight.data)
lin.bias.data = torch.ones_like(lin.bias.data) * 5.0
for p in lin.parameters():
print(p)
print(p.shape)
print('\n')
x = torch.ones(3, 10) # rank2 tensor를 만듭니다. : mini batch size = 3
y_hat = lin(x)
print(y_hat.shape)
print(y_hat)
```
## 지금 무슨일이 일어난거죠?
>Q1. 왜 Rank 2 tensor 를 입력으로 사용하나요? <br>
>A1. 파이토치의 `nn` 에 정의되어있는 클래스들은 입력의 가장 첫번째 디멘젼을 `배치 사이즈`로 해석합니다.
>Q2. lin(x) 는 도대체 무엇인가요? <br>
>A2. 파이썬에 익숙하신 분들은 `object()` 는 `object.__call__()`에 정의되어있는 함수를 실행시키신다는 것을 아실텐데요. 파이토치의 `nn.Module`은 `__call__()`을 오버라이드하는 함수인 `forward()`를 구현하는 것을 __권장__ 하고 있습니다. 일반적으로, `forward()`안에서 실제로 파라미터와 인풋을 가지고 특정 레이어의 연산과 정을 구현하게 됩니다.
여러가지 이유가 있겠지만, 파이토치가 내부적으로 foward() 의 실행의 전/후로 사용자 친화적인 환경을 제공하기위해서 추가적인 작업들을 해줍니다. 이 부분은 다음 실습에서 다층 퍼셉트론 모델을 만들면서 조금 더 자세히 설명해볼게요!
## Pytorch 로 간단히! 선형회귀 구현하기
저번 실습에서 numpy 로 구현했던 Linear regression 모델을 다시 한번 파이토치로 구현해볼까요? <br>
몇 줄이면 끝날 정도로 간단합니다 :)
```
def generate_samples(n_samples: int,
w: float = 1.0,
b: float = 0.5,
x_range=[-1.0,1.0]):
xs = np.random.uniform(low=x_range[0], high=x_range[1], size=n_samples)
ys = w * xs + b
xs = torch.tensor(xs).view(-1,1).float() # 파이토치 nn.Module 은 배치가 첫 디멘젼!
ys = torch.tensor(ys).view(-1,1).float()
return xs, ys
w = 1.0
b = 0.5
xs, ys = generate_samples(30, w=w, b=b)
lin_model = nn.Linear(in_features=1, out_features=1) # lim_model 생성
for p in lin_model.parameters():
print(p)
print(p.grad)
ys_hat = lin_model(xs) # lin_model 로 예측하기
```
## Loss 함수는? MSE!
`pytorch`에서는 자주 쓰이는 loss 함수들에 대해서도 미리 구현을 해두었습니다.
이번 실습에서는 __numpy로 선형회귀 모델 만들기__ 에서 사용됐던 MSE 를 오차함수로 사용해볼까요?
```
criteria = nn.MSELoss()
loss = criteria(ys_hat, ys)
```
## 경사하강법을 활용해서 파라미터 업데이트하기!
`pytorch`는 여러분들을 위해서 다양한 optimizer들을 구현해 두었습니다. 일단은 가장 간단한 stochastic gradient descent (SGD)를 활용해 볼까요? optimizer에 따라서 다양한 인자들을 활용하지만 기본적으로 `params` 와 `lr`을 지정해주면 나머지는 optimizer 마다 잘되는 것으로 알려진 인자들로 optimizer을 손쉽게 생성할수 있습니다.
```
opt = torch.optim.SGD(params=lin_model.parameters(), lr=0.01)
```
## 잊지마세요! opt.zero_grad()
`pytorch`로 편미분을 계산하기전에, 꼭 `opt.zero_grad()` 함수를 이용해서 편미분 계산이 필요한 텐서들의 편미분값을 초기화 해주는 것을 권장드립니다.
```
opt.zero_grad()
for p in lin_model.parameters():
print(p)
print(p.grad)
loss.backward()
opt.step()
for p in lin_model.parameters():
print(p)
print(p.grad)
```
## 경사하강법을 활용해서 최적 파라미터를 찾아봅시다!
```
def run_sgd(n_steps: int = 1000,
report_every: int = 100,
verbose=True):
lin_model = nn.Linear(in_features=1, out_features=1)
opt = torch.optim.SGD(params=lin_model.parameters(), lr=0.01)
sgd_losses = []
for i in range(n_steps):
ys_hat = lin_model(xs)
loss = criteria(ys_hat, ys)
opt.zero_grad()
loss.backward()
opt.step()
if i % report_every == 0:
if verbose:
print('\n')
print("{}th update: {}".format(i,loss))
for p in lin_model.parameters():
print(p)
sgd_losses.append(loss.log10().detach().numpy())
return sgd_losses
_ = run_sgd()
```
## 다른 Optimizer도 사용해볼까요?
수업시간에 배웠던 Adam 으로 최적화를 하면 어떤결과가 나올까요?
```
def run_adam(n_steps: int = 1000,
report_every: int = 100,
verbose=True):
lin_model = nn.Linear(in_features=1, out_features=1)
opt = torch.optim.Adam(params=lin_model.parameters(), lr=0.01)
adam_losses = []
for i in range(n_steps):
ys_hat = lin_model(xs)
loss = criteria(ys_hat, ys)
opt.zero_grad()
loss.backward()
opt.step()
if i % report_every == 0:
if verbose:
print('\n')
print("{}th update: {}".format(i,loss))
for p in lin_model.parameters():
print(p)
adam_losses.append(loss.log10().detach().numpy())
return adam_losses
_ = run_adam()
```
## 좀 더 상세하게 비교해볼까요?
`pytorch`에서 `nn.Linear`를 비롯한 많은 모듈들은 특별한 경우가 아닌이상,
모듈내에 파라미터가 임의의 값으로 __잘!__ 초기화 됩니다.
> "잘!" 에 대해서는 수업에서 다루지 않았지만, 확실히 현대 딥러닝이 잘 작동하게 하는 중요한 요소중에 하나입니다. Parameter initialization 이라고 부르는 기법들이며, 대부분의 `pytorch` 모듈들은 각각의 모듈에 따라서 일반적으로 잘 작동하는것으로 알려져있는 방식으로 파라미터들이 초기화 되게 코딩되어 있습니다.
그래서 매 번 모듈을 생성할때마다 파라미터의 초기값이 달라지게 됩니다. 이번에는 조금 공정한 비교를 위해서 위에서 했던 실험을 여러번 반복해서 평균적으로도 Adam이 좋은지 확인해볼까요?
```
sgd_losses = [run_sgd(verbose=False) for _ in range(50)]
sgd_losses = np.stack(sgd_losses)
sgd_loss_mean = np.mean(sgd_losses, axis=0)
sgd_loss_std = np.std(sgd_losses, axis=-0)
adam_losses = [run_adam(verbose=False) for _ in range(50)]
adam_losses = np.stack(adam_losses)
adam_loss_mean = np.mean(adam_losses, axis=0)
adam_loss_std = np.std(adam_losses, axis=-0)
fig, ax = plt.subplots(1,1, figsize=(10,5))
ax.grid()
ax.fill_between(x=range(sgd_loss_mean.shape[0]),
y1=sgd_loss_mean + sgd_loss_std,
y2=sgd_loss_mean - sgd_loss_std,
alpha=0.3)
ax.plot(sgd_loss_mean, label='SGD')
ax.fill_between(x=range(adam_loss_mean.shape[0]),
y1=adam_loss_mean + adam_loss_std,
y2=adam_loss_mean - adam_loss_std,
alpha=0.3)
ax.plot(adam_loss_mean, label='Adam')
ax.legend()
```
| github_jupyter |
# Callbacks and Multiple inputs
```
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from sklearn.preprocessing import scale
from keras.optimizers import SGD
from keras.layers import Dense, Input, concatenate, BatchNormalization
from keras.callbacks import EarlyStopping, TensorBoard, ModelCheckpoint
from keras.models import Model
import keras.backend as K
df = pd.read_csv("../data/titanic-train.csv")
Y = df['Survived']
df.info()
df.head()
num_features = df[['Age', 'Fare', 'SibSp', 'Parch']].fillna(0)
num_features.head()
cat_features = pd.get_dummies(df[['Pclass', 'Sex', 'Embarked']].astype('str'))
cat_features.head()
X1 = scale(num_features.values)
X2 = cat_features.values
K.clear_session()
# Numerical features branch
inputs1 = Input(shape = (X1.shape[1],))
b1 = BatchNormalization()(inputs1)
b1 = Dense(3, kernel_initializer='normal', activation = 'tanh')(b1)
b1 = BatchNormalization()(b1)
# Categorical features branch
inputs2 = Input(shape = (X2.shape[1],))
b2 = Dense(8, kernel_initializer='normal', activation = 'relu')(inputs2)
b2 = BatchNormalization()(b2)
b2 = Dense(4, kernel_initializer='normal', activation = 'relu')(b2)
b2 = BatchNormalization()(b2)
b2 = Dense(2, kernel_initializer='normal', activation = 'relu')(b2)
b2 = BatchNormalization()(b2)
merged = concatenate([b1, b2])
preds = Dense(1, activation = 'sigmoid')(merged)
# final model
model = Model([inputs1, inputs2], preds)
model.compile(loss = 'binary_crossentropy',
optimizer = 'rmsprop',
metrics = ['accuracy'])
model.summary()
outpath='/tmp/tensorflow_logs/titanic/'
early_stopper = EarlyStopping(monitor='val_acc', patience=10)
tensorboard = TensorBoard(outpath+'tensorboard/', histogram_freq=1)
checkpointer = ModelCheckpoint(outpath+'weights_epoch_{epoch:02d}_val_acc_{val_acc:.2f}.hdf5',
monitor='val_acc')
# You may have to run this a couple of times if stuck on local minimum
np.random.seed(2017)
h = model.fit([X1, X2],
Y.values,
batch_size = 32,
epochs = 40,
verbose = 1,
validation_split=0.2,
callbacks=[early_stopper,
tensorboard,
checkpointer])
import os
sorted(os.listdir(outpath))
```
Now check the tensorboard.
- If using provided aws instance, just browse to: `http://<your-ip>:6006`
- If using local, open a terminal, activate the environment and run:
```
tensorboard --logdir=/tmp/tensorflow_logs/titanic/tensorboard/
```
then open a browser at `localhost:6006`
You should see something like this:

## Exercise 1
- try modifying the parameters of the 3 callbacks provided. What are they for? What do they do?
*Copyright © 2017 CATALIT LLC. All rights reserved.*
| github_jupyter |
#Import Data
```
import numpy as np
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
# load data
import os
from google.colab import drive
drive.mount('/content/drive')
filedir = './drive/My Drive/Final/CNN_data'
with open(filedir + '/' + 'feature_extracted', 'rb') as f:
X = np.load(f)
with open(filedir + '/' + 'Y', 'rb') as f:
Y = np.load(f).astype(np.int32)
# import MFCC data
with open('./drive/My Drive/Final/mfcc_data/X', 'rb') as f:
X_mfcc = np.load(f)
with open('./drive/My Drive/Final/mfcc_data/Y', 'rb') as f:
Y_mfcc = np.load(f)
print('X_shape: {}\nY_shape: {}'.format(X_mfcc.shape, Y_mfcc.shape))
import warnings
warnings.filterwarnings("ignore")
'''
X_new = np.zeros([300,0])
for i in range(X.shape[1]):
col = X[:,i,None]
if((np.abs(col) > 1e-6).any()):
X_new = np.hstack([X_new, col])
else:
print('Yes')
print('X.shape: {}\nX_new.shape: {}\nY.shape: {}'.format(X.shape, X_new.shape, Y.shape))
print(X_new.shape)
print(np.max(X_new, axis=1) != np.max(X, axis=1))
print(np.min(X_new, axis=1))
'''
```
#CLF1 Ridge Classifier
```
'''
from sklearn.linear_model import RidgeClassifier
parameters = {'alpha':[1]}
rc = RidgeClassifier(alpha = 1)
clf = GridSearchCV(rc, parameters, cv=3)
clf.fit(X[:30], Y[:30])
clf.best_estimator_.fit(X[:30], Y[:30]).score(X, Y)
clf.best_index_
'''
from sklearn.linear_model import RidgeClassifier
def clf_RidgeClassifier(training_set, training_lable, testing_set, testing_lable):
parameters = {'alpha':[10, 1, 1e-1, 1e-2, 1e-3]}
rc = RidgeClassifier(alpha = 1)
clf = GridSearchCV(rc, parameters, cv=3, return_train_score=True, iid=False)
clf.fit(training_set, training_lable)
results = clf.cv_results_
opt_index = clf.best_index_
training_score = results['mean_train_score'][opt_index]
validation_score = results['mean_test_score'][opt_index]
testing_score = clf.best_estimator_.fit(training_set, training_lable).score(testing_set, testing_lable)
return [training_score, validation_score, testing_score], clf.best_params_
clf_RidgeClassifier(X[:240], Y[:240], X[240:], Y[240:])
```
#CLF2 SVM
```
from sklearn.svm import SVC
def clf_SVM(X_train, Y_train, X_test, Y_test):
parameters = {'C':[10, 1, 1e-1, 1e-2, 1e-3]}
svc = SVC(kernel='linear')
clf = GridSearchCV(svc, parameters, cv=3, return_train_score=True, iid=False)
clf.fit(X_train, Y_train)
results = clf.cv_results_
opt_index = clf.best_index_
training_score = results['mean_train_score'][opt_index]
validation_score = results['mean_test_score'][opt_index]
testing_score = clf.best_estimator_.fit(X_train, Y_train).score(X_test, Y_test)
return [training_score, validation_score, testing_score], clf.best_params_
clf_SVM(X[:240], Y[:240], X[240:], Y[240:])
```
#CLF3 LDA
```
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
def clf_lda(Xtrain, Ytrain, Xtest, Ytest):
"""
Input: training data, labels, testing data, labels
Output: training set mean prediciton accuracy, validation accuracy = None, testing set mean prediction accuracy
Note: LDA has no hyperparameters to tune because a model is solved in closed form
therefore there is no need for model selection via grid search cross validation
therefore there is no validation accuracy
"""
clf = LinearDiscriminantAnalysis()
clf.fit(Xtrain, Ytrain)
train_acc = clf.score(Xtrain,Ytrain)
val_acc = None
test_acc = clf.score(Xtest,Ytest)
return [train_acc,val_acc,test_acc], None
clf_lda(X[:240],Y[:240],X[240:],Y[240:])
```
#CLF4 KNN
```
from sklearn.neighbors import KNeighborsClassifier
def clf_KNN(X_train, Y_train, X_test, Y_test):
parameters = {'n_neighbors':[1,5,20]}
knn = KNeighborsClassifier(algorithm='auto', weights='uniform')
clf = GridSearchCV(knn, parameters, cv=3, return_train_score=True, iid=False)
clf.fit(X_train, Y_train)
results = clf.cv_results_
opt_index = clf.best_index_
training_score = results['mean_train_score'][opt_index]
validation_score = results['mean_test_score'][opt_index]
testing_score = clf.best_estimator_.fit(X_train, Y_train).score(X_test, Y_test)
return [training_score, validation_score, testing_score], clf.best_params_
clf_KNN(X[:240], Y[:240], X[240:], Y[240:])
```
#CLF5 Decision Tree
```
from sklearn.tree import DecisionTreeClassifier
def clf_DecisionTree(X_train, Y_train, X_test, Y_test):
parameters = {'max_depth':[5,10,15,20,25], 'criterion':['entropy', 'gini']}
dtc = DecisionTreeClassifier()
clf = GridSearchCV(dtc, parameters, cv=3, return_train_score=True, iid=False)
clf.fit(X_train, Y_train)
results = clf.cv_results_
opt_index = clf.best_index_
training_score = results['mean_train_score'][opt_index]
validation_score = results['mean_test_score'][opt_index]
testing_score = clf.best_estimator_.fit(X_train, Y_train).score(X_test, Y_test)
return [training_score, validation_score, testing_score], clf.best_params_
clf_DecisionTree(X[:240], Y[:240], X[240:], Y[240:])
```
#Testing On Data
```
clf_list = [clf_RidgeClassifier, clf_SVM, clf_lda, clf_KNN, clf_DecisionTree]
def test_trial(X_shuffled, Y_shuffled):
global clf_list
error = np.zeros((3,5,3)) # partition(3) * clf(5) * error(3)
# (8/2,5/5,2/8) * (clf_list) * (trn,val,tst)
opt_param = np.empty((3,5), dtype=dict) # partition(3) * clf(5)
sample_size = len(X_shuffled)
# 80/20 split
train_size = int(sample_size * 0.8)
X_train = X_shuffled[:train_size]
Y_train = Y_shuffled[:train_size]
X_test = X_shuffled[train_size:]
Y_test = Y_shuffled[train_size:]
for i in range(len(clf_list)):
clffn = clf_list[i]
error[0,i,:], opt_param[0,i] = clffn(X_train, Y_train, X_test, Y_test)
# 50/50 split
train_size = int(sample_size * 0.5)
X_train = X_shuffled[:train_size]
Y_train = Y_shuffled[:train_size]
X_test = X_shuffled[train_size:]
Y_test = Y_shuffled[train_size:]
for i in range(len(clf_list)):
clffn = clf_list[i]
error[1,i,:], opt_param[1,i] = clffn(X_train, Y_train, X_test, Y_test)
# 80/20 split
train_size = int(sample_size * 0.2)
X_train = X_shuffled[:train_size]
Y_train = Y_shuffled[:train_size]
X_test = X_shuffled[train_size:]
Y_test = Y_shuffled[train_size:]
for i in range(len(clf_list)):
clffn = clf_list[i]
error[2,i,:], opt_param[2,i] = clffn(X_train, Y_train, X_test, Y_test)
# return error array
return error, opt_param
from sklearn.utils import shuffle
def test_data(X, Y):
error = np.zeros((3,3,5,3)) # trial(3) * error_from_test_trial(3*5*3)
opt_param = np.empty((3,3,5), dtype=dict) # trial(3) * opt_param_from_test_trial(3*5)
# trial 1
X_shuffled, Y_shuffled = shuffle(X, Y)
error[0], opt_param[0] = test_trial(X_shuffled, Y_shuffled)
# trial 2
X_shuffled, Y_shuffled = shuffle(X_shuffled, Y_shuffled)
error[1], opt_param[1] = test_trial(X_shuffled, Y_shuffled)
# trial 3
X_shuffled, Y_shuffled = shuffle(X_shuffled, Y_shuffled)
error[2], opt_param[2] = test_trial(X_shuffled, Y_shuffled)
return error, opt_param
# test on CNN-extracted features
acc_CNN, opt_param_CNN = test_data(X, Y)
np.mean(acc_CNN[:,:,:,:], axis=0)
acc_clf, opt_param = test_data(X_mfcc, Y_mfcc)
avg_cnn_acc = np.mean(acc_CNN, axis=0)
avg_clf_acc = np.mean(acc_clf, axis=0)
print('cnn: {}'.format(avg_cnn_acc))
print('clf: {}'.format(avg_clf_acc))
# partition_accuracy plot
from matplotlib import rcParams
rcParams['figure.figsize'] = (8,8)
colors = ['cyan', 'green', 'red', 'orange','black']
clf = ['RidgeRegression', 'SVM', 'LDA', 'KNN', 'DecisionTree']
for clfid in range(5):
plt.plot(avg_cnn_acc[:,clfid,-1], color=colors[clfid], linestyle='solid', label='CNN '+clf[clfid])
plt.plot(avg_clf_acc[:,clfid,-1], color=colors[clfid], linestyle='dashed', label='MFCC '+clf[clfid])
plt.legend(loc='lower left')
plt.xticks((0,1,2),['80/20', '50/50', '20/80'])
plt.xlabel('partition (train/test)')
plt.ylabel('average test accuracy')
plt.savefig('./drive/My Drive/Final/graphs/partition_accuracy.png', bbox='tight')
# SVM hyperparameter error plot
parameters = {'C':[10, 1, 1e-1, 1e-2, 1e-3]}
svc = SVC(kernel='linear')
clf = GridSearchCV(svc, parameters, cv=3, return_train_score=True, iid=False)
clf.fit(X[:240], Y[:240])
results = clf.cv_results_
opt_index = clf.best_index_
training_score = results['mean_train_score']
validation_score = results['mean_test_score']
param_x = results['param_C'].data.astype(np.float32)
plt.plot(param_x, training_score, 'r-', label='training')
plt.plot(param_x, validation_score, 'b-', label='validation')
plt.legend(loc='lower left')
plt.xticks([0,2.5,5,7.5,10], ['10','1','1e-1','1e-2','1e-3'])
plt.xlabel('param_C')
plt.ylabel('accuracy')
#plt.show()
plt.savefig('./drive/My Drive/Final/graphs/SVM_hyperparameter_accuracy.png')
# avg cross-partition accuracy
cnn_cp_acc = np.mean(avg_cnn_acc[:,:,-1], axis=0)
clf_cp_acc = np.mean(avg_clf_acc[:,:,-1], axis=0)
print('cnn_cp_acc: {}'.format(cnn_cp_acc))
print('clf_cp_acc: {}'.format(clf_cp_acc))
avg_totalcp_acc = (cnn_cp_acc + clf_cp_acc) / 2
print(avg_totalcp_acc)
(avg_cnn_acc + avg_clf_acc)/2
opt_param
opt_param_CNN
max_ind_cnn = np.argpartition(np.sum(X, axis=0), -2)[-2:]
std_ind_cnn = np.argpartition(np.std(X, axis=0), -2)[-2:]
max_ind_clf = np.argpartition(np.sum(X_mfcc, axis=0), -2)[-2:]
std_ind_clf = np.argpartition(np.std(X_mfcc, axis=0), -2)[-2:]
max_cnn = X[:,max_ind_cnn]
std_cnn = X[:,std_ind_cnn]
max_clf = X_mfcc[:,max_ind_clf]
std_clf = X_mfcc[:,std_ind_clf]
def plot_features(X, Y):
return X[Y==0,:], X[Y==1,:]
# 2 max features from cnn plotted
plt.clf()
feature0, feature1 = plot_features(max_cnn, Y)
plt.plot(feature0[:,0], feature0[:,1],'ro', label='digit 0')
plt.plot(feature1[:,0], feature1[:,1],'go', label='digit 1')
plt.legend(loc='lower right')
plt.show()
#plt.savefig('./drive/My Drive/Final/graphs/2_max_sum_cnn_features.png')
# 2 var features from cnn plotted
feature0, feature1 = plot_features(std_cnn, Y)
plt.plot(feature0[:,0], feature0[:,1],'ro', label='digit 0')
plt.plot(feature1[:,0], feature1[:,1],'go', label='digit 1')
plt.legend(loc='lower right')
#plt.show()
plt.savefig('./drive/My Drive/Final/graphs/2_max_var_cnn_features.png')
# 2 max features from mfcc plotted
feature0, feature1 = plot_features(max_clf, Y)
plt.plot(feature0[:,0], feature0[:,1],'ro', label='digit 0')
plt.plot(feature1[:,0], feature1[:,1],'go', label='digit 1')
plt.legend(loc='lower right')
#plt.show()
plt.savefig('./drive/My Drive/Final/graphs/2_max_sum_mfcc_features.png')
# 2 var features from mfcc plotted
feature0, feature1 = plot_features(std_clf, Y)
plt.plot(feature0[:,0], feature0[:,1],'ro', label='digit 0')
plt.plot(feature1[:,0], feature1[:,1],'go', label='digit 1')
plt.legend(loc='lower right')
#plt.show()
plt.savefig('./drive/My Drive/Final/graphs/2_max_var_mfcc_features.png')
```
| github_jupyter |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Image/extract_value_to_points.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Image/extract_value_to_points.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Image/extract_value_to_points.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Image/extract_value_to_points.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
```
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
# Input imagery is a cloud-free Landsat 8 composite.
l8 = ee.ImageCollection('LANDSAT/LC08/C01/T1')
image = ee.Algorithms.Landsat.simpleComposite(**{
'collection': l8.filterDate('2018-01-01', '2018-12-31'),
'asFloat': True
})
# Use these bands for prediction.
bands = ['B2', 'B3', 'B4', 'B5', 'B6', 'B7', 'B10', 'B11']
# Load training points. The numeric property 'class' stores known labels.
points = ee.FeatureCollection('GOOGLE/EE/DEMOS/demo_landcover_labels')
# This property of the table stores the land cover labels.
label = 'landcover'
# Overlay the points on the imagery to get training.
training = image.select(bands).sampleRegions(**{
'collection': points,
'properties': [label],
'scale': 30
})
# Define visualization parameters in an object literal.
vizParams = {'bands': ['B5', 'B4', 'B3'],
'min': 0, 'max': 1, 'gamma': 1.3}
Map.centerObject(points, 10)
Map.addLayer(image, vizParams, 'Image')
Map.addLayer(points, {'color': "yellow"}, 'Training points')
first = training.first()
print(first.getInfo())
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
| github_jupyter |
# Fmriprep
Today, many excellent general-purpose, open-source neuroimaging software packages exist: [SPM](https://www.fil.ion.ucl.ac.uk/spm/) (Matlab-based), [FSL](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki), [AFNI](https://afni.nimh.nih.gov/), and [Freesurfer](https://surfer.nmr.mgh.harvard.edu/) (with a shell interface). We argue that there is not one single package that is always the best choice for every step in your preprocessing pipeline. Fortunately, people from the [Poldrack lab](https://poldracklab.stanford.edu/) created [fmriprep](https://fmriprep.readthedocs.io/en/stable/), a software package that offers a preprocessing pipeline which "glues together" functionality from different neuroimaging software packages (such as Freesurfer and FSL), such that each step in the pipeline is executed by the software package that (arguably) does it best.
We have been using *Fmriprep* for preprocessing of our own data and we strongly recommend it. It is relatively simple to use, requires minimal user intervention, and creates extensive visual reports for users to do visual quality control (to check whether each step in the pipeline worked as expected). The *only* requirement to use Fmriprep is that your data is formatted as specified in the Brain Imaging Data Structure (BIDS).
## The BIDS-format
[BIDS](https://bids.neuroimaging.io/) is a specification on how to format, name, and organize your MRI dataset. It specifies the file format of MRI files (i.e., compressed Nifti: `.nii.gz` files), lays out rules for how you should name your files (i.e., with "key-value" pairs, such as: `sub-01_ses-1_task-1back_run-1_bold.nii.gz`), and outlines the file/folder structure of your dataset (where each subject has its own directory with separate subdirectories for different MRI modalities, including fieldmaps, functional, diffusion, and anatomical MRI). Additionally, it specifies a way to include "metadata" about the (MRI) files in your dataset with [JSON](https://en.wikipedia.org/wiki/JSON) files: plain-text files with key-value pairs (in the form "parameter: value"). Given that your dataset is BIDS-formatted and contains the necessary metadata, you can use `fmriprep` on your dataset. (You can use the awesome [bids-validator](https://bids-standard.github.io/bids-validator/) to see whether your dataset is completely valid according to BIDS.)
There are different tools to convert your "raw" scanner data (e.g., in DICOM or PAR/REC format) to BIDS, including [heudiconv](https://heudiconv.readthedocs.io/en/latest/), [bidscoin](https://github.com/Donders-Institute/bidscoin), and [bidsify](https://github.com/NILAB-UvA/bidsify) (created by Lukas). We'll skip over this step and assume that you'll be able to convert your data to BIDS.
## Installing Fmriprep
Now, having your data in BIDS is an important step in getting started with Fmriprep. The next step is installing the package. Technically, Fmriprep is a Python package, so it can be installed as such (using `pip install fmriprep`), but we do not recommend this "bare metal" installation, because it depends on a host of neuroimaging software packages (including FSL, Freesurfer, AFNI, and ANTs). So if you'd want to directly install Fmriprep, you'd need to install those extra neuroimaging software packages as well (which is not worth your time, trust us).
Fortunately, Fmriprep also offers a "Docker container" in which Fmriprep and all the associated dependencies are already installed. [Docker](https://www.docker.com/) is software that allows you to create "containers", which are like lightweight "virtual machines" ([VM](https://en.wikipedia.org/wiki/Virtual_machine)) that are like a separate (Linux-based) operating system with a specific software configuration. You can download the Fmriprep-specific docker "image", which is like a "recipe", build the Fmriprep-specific "container" according to this "recipe" on your computer, and finally use this container to run Fmriprep on your computer as if all dependencies were actually installed on your computer! Docker is available on Linux, Mac, and Windows. To install Docker, google something like "install docker for {Windows,Mac,Linux}" to find a google walkthrough.
Note that you need administrator ("root") privilege on your computer (which is likely the case for your own computer, but not on shared analysis servers) to run Docker. If you don't have root access on your computer/server, ask you administrator/sysadmin to install [singularity](https://fmriprep.readthedocs.io/en/stable/installation.html#singularity-container), which allows you to convert Docker images to Singularity images, which you can run without administrator privileges.
Assuming you have installed Docker, you can run the "containerized" Fmriprep from your command line directly, which involves a fairly long and complicated command (i.e., `docker run -it --rm -v bids_dir /data ... etc`), or using the `fmriprep-docker` Python package. This `fmriprep-docker` package is just a simple wrapper around the appropriate Docker command to run the complicated "containerized" Fmriprep command. We strongly recommend this method.
To install `fmriprep-docker`, you can use `pip` (from your command line):
```
pip install fmriprep-docker
```
Now, you should have access to the `fmriprep-docker` command on your command line and you're ready to start preprocessing your dataset. For more detailed information about installing Fmriprep, check out their [website](https://fmriprep.readthedocs.io/en/stable/installation.html).
## Running Fmriprep
Assuming you have Docker and `fmriprep-docker` installed, you're ready to run Fmriprep. The basic format of the `fmriprep-docker` command is as follows:
```
fmriprep-docker <your bids-folder> <your output-folder>
```
This means that `fmriprep-docker` has two mandatory positional arguments: the first one being your BIDS-folder (i.e., the path to your folder with BIDS-formattefd data), and the second one being the output-folder (i.e., where you want Fmriprep to output the preprocessed data). We recommend setting your output-folder to a subfolder of your BIDS-folder named "derivatives": `<your bids-folder>/derivatives`.
Then, you can add a bunch of extra "flags" (parameters) to the command to specify the preprocessing pipeline as you like it. We highlight a couple of important ones here, but for the full list of parameters, check out the [Fmriprep](https://fmriprep.readthedocs.io/en/stable/usage.html) website.
### Freesurfer
When running Fmriprep from Docker, you don't need to have Freesurfer installed, but you *do* need a Freesurfer license. You can download this here: https://surfer.nmr.mgh.harvard.edu/fswiki/License. Then, you need to supply the `--fs-license-file <path to license file>` parameter to your `fmriprep-docker` command:
```
fmriprep-docker <your bids-folder> <your output-folder> --fs-license-file /home/lukas/license.txt
```
### Configuring what is preprocessed
If you just run Fmriprep with the mandatory BIDS-folder and output-folder arguments, it will preprocess everything it finds in the BIDS-folder. Sometimes, however, you may just want to run one (or several) specific participants, or one (or more) specific tasks (e.g., only the MRI files associated with the localizer runs, but not the working memory runs). You can do this by adding the `--participant` and `--task` flags to the command:
```
fmriprep-docker <your bids-folder> <your output-folder> --participant sub-01 --task localizer
```
You can also specify some things to be ignored during preprocessing using the `--ignore` parameters (like `fieldmaps`):
```
fmriprep-docker <your bids-folder> <your output-folder> --ignore fieldmaps
```
### Handling performance
It's very easy to parallelize the preprocessing pipeline by setting the `--nthreads` and `--omp-nthreads` parameters, which refer to the number of threads that should be used to run Fmriprep on. Note that laptops usually have 4 threads available (but analysis servers usually have more!). You can also specify the maximum of RAM that Fmriprep is allowed to use by the `--mem_mb` parameters. So, if you for example want to run Fmriprep with 3 threads and a maximum of 3GB of RAM, you can run:
```
fmriprep-docker <your bids-folder> <your output-folder> --nthreads 3 --omp-nthreads 3 --mem_mb 3000
```
In our experience, however, specifying the `--mem_mb` parameter is rarely necessary if you don't parallelize too much.
### Output spaces
Specifying your "output spaces" (with the `--output-spaces` flag) tells Fmriprep to what "space(s)" you want your preprocessed data registered to. For example, you can specify `T1w` to have your functional data registered to the participant's T1 scan. You can, instead or in addition to, also specify some standard template, like the MNI template (`MNI152NLin2009cAsym` or `MNI152NLin6Asym`). You can even specify surface templates if you want (like `fsaverage`), which will sample your volumetric functional data onto the surface (as computed by freesurfer). In addition to the specific output space(s), you can add a resolution "modifier" to the parameter to specify in what spatial resolution you want your resampled data to be. Without any resolution modifier, the native resolution of your functional files (e.g., $3\times3\times3$ mm.) will be kept intact. But if you want to upsample your resampled files to 2mm, you can add `YourTemplate:2mm`. For example, if you want to use the FSL-style MNI template (`MNI152NLin6Asym`) resampled at 2 mm, you'd use:
```
fmriprep-docker <your bids-folder> <your output-folder> --output-spaces MNI152NLin6Asym:2mm
```
You can of course specify multiple output-spaces:
```
fmriprep-docker <your bids-folder> <your output-folder> --output-spaces MNI152NLin6Asym:2mm T1w fsaverage
```
### Other parameters
There are many options that you can set when running Fmriprep. Check out the [Fmriprep website](https://fmriprep.readthedocs.io/) (under "Usage") for a list of all options!
## Issues, errors, and troubleshooting
While Fmriprep often works out-of-the-box (assuming your data are properly BIDS-formatted), it may happen that it crashes or otherwise gives unexpected results. A great place to start looking for help is [neurostars.org](https://neurostars.org). This website is dedicated to helping neuroscientists with neuroimaging/neuroscience-related questions. Make sure to check whether your question has been asked here already and, if not, pose it here!
If you encounter Fmriprep-specific bugs, you can also submit and issue at the [Github repository](https://github.com/poldracklab/fmriprep) of Fmriprep.
## Fmriprep output/reports
After Fmriprep has run, it outputs, for each participants separately, a directory with results (i.e., preprocessed files) and an HTML-file with a summary and figures of the different steps in the preprocessing pipeline.
We ran Fmriprep on a single run/task (`flocBLOCKED`) from a single subject (`sub-03`) some data with the following command:
```
fmriprep-docker /home/lsnoek1/ni-edu/bids /home/lsnoek1/ni-edu/bids/derivatives --participant-label sub-03 --output-spaces T1w MNI152NLin2009cAsym
```
We've copied the Fmriprep output for this subject (`sub-03`) in the `fmriprep` subdirectory of the `week_4` directory. Let's check its contents:
```
import os
print(os.listdir('bids/derivatives/fmriprep'))
```
As said, Fmriprep outputs a directory with results (`sub-03`) and an associated HTML-file with a summary of the (intermediate and final) results. Let's check the directory with results first:
```
from pprint import pprint # pprint stands for "pretty print",
sub_path = os.path.join('bids/derivatives/fmriprep', 'sub-03')
pprint(sorted(os.listdir(sub_path)))
```
The `figures` directory contains several figures with the result of different preprocessing stages (like functional → high-res anatomical registration), but these figures are also included in the HTML-file, so we'll leave that for now. The other two directories, `anat` and `func`, contain the preprocessed anatomical and functional files, respectively. Let's inspect the `anat` directory:
```
anat_path = os.path.join(sub_path, 'anat')
pprint(os.listdir(anat_path))
```
Here, we see a couple of different files. There are both (preprocessed) nifti images (`*.nii.gz`) and associated meta-data (plain-text files in JSON format: `*.json`).
Importantly, the nifti outputs are in two different spaces: one set of files are in the original "T1 space", so without any resampling to another space (these files have the same resolution and orientation as the original T1 anatomical scan). For example, the `sub_03_desc-preproc_T1w.nii.gz` scan is the preprocessed (i.e., bias-corrected) T1 scan. In addition, most files are also available in `MNI152NLin2009cAsym` space, a standard template. For example, the `sub-03_space-MNI152NLin2009cAsym_desc-preproc_T1w.nii.gz` is the same file as `sub_03_desc-preproc_T1w.nii.gz`, but resampled to the `MNI152NLin2009cAsym` template. In addition, there are subject-specific brain parcellations (the `*aparcaseg_dseg.nii.gz `and `*aseg_dseg.nii.gz` files), files with registration parameters (`*from- ... -to ...` files), probabilistic tissue segmentation files (`*label-{CSF,GM,WM}_probseg.nii.gz`) files, and brain masks (to outline what is brain and not skull/dura/etc; `*brain_mask.nii.gz`).
Again, on the [Fmriprep website](https://fmriprep.readthedocs.io/), you can find more information about the specific outputs.
Now, let's check out the `func` directory:
```
func_path = os.path.join(sub_path, 'func')
pprint(os.listdir(func_path))
```
Again, like the files in the `anat` folder, the functional outputs are available in two spaces: `T1w` and `MNI152NLin2009cAsym`. In terms of actual images, there are preprocessed BOLD files (ending in `preproc_bold.nii.gz`), the functional volume used for "functional → anatomical" registration (ending in `boldref.nii.gz`), brain parcellations in functional space (ending in `dseg.nii.gz`), and brain masks (ending in `brain_mask.nii.gz`). In addition, there are files with "confounds" (ending in `confounds_regressors.tsv`) which contain variables that you might want to include as nuisance regressors in your first-level analysis. These confound files are speadsheet-like files (like `csv` files, but instead of being comma-delimited, they are tab-delimited) and can be easily loaded in Python using the [pandas](https://pandas.pydata.org/) package:
```
import pandas as pd
conf_path = os.path.join(func_path, 'sub-03_task-flocBLOCKED_desc-confounds_regressors.tsv')
conf = pd.read_csv(conf_path, sep='\t')
conf.head()
```
Confound files from Fmriprep contain a large set of confounds, ranging from motion parameters (`rot_x`, `rot_y`, `rot_z`, `trans_x`, `trans_y`, and `trans_z`) and their derivatives (`*derivative1`) and squares (`*_power2`) to the average signal from the brain's white matter and cerebrospinal fluid (CSF), which should contain sources of noise such as respiratory, cardiac, or motion related signals (but not signal from neural sources, which should be largely constrained to gray matter). For a full list and explanation of Fmriprep's estimated confounds, check their website. Also, check [this thread](https://neurostars.org/t/confounds-from-fmriprep-which-one-would-you-use-for-glm/326) on Neurostars for a discussion on which confounds to include in your analyses.
In addition to the actual preprocessed outputs, Fmriprep also provides you with a nice (visual) summary of the different (major) preprocessing steps in an HTML-file, which you'd normally open in any standard browser to view. Here. we load this file for our example participants (`sub-03`) inside the notebook below. Scroll through it to see which preprocessing steps are highlighted. Note that the images from the HTML-file are not properly rendered in Jupyter notebooks, but you can right-click the image links (e.g., `sub-03/figures/sub-03_dseg.svg`) and click "Open link in new tab" to view the image.
```
from IPython.display import IFrame
IFrame(src='./bids/derivatives/fmriprep/sub-03.html', width=700, height=600)
```
| github_jupyter |
# Pattern Mining
## Library
```
source("https://raw.githubusercontent.com/eogasawara/mylibrary/master/myPreprocessing.R")
loadlibrary("arules")
loadlibrary("arulesViz")
loadlibrary("arulesSequences")
data(AdultUCI)
dim(AdultUCI)
head(AdultUCI)
```
## Removing attributes
```
AdultUCI$fnlwgt <- NULL
AdultUCI$"education-num" <- NULL
```
## Conceptual Hierarchy and Binning
```
AdultUCI$age <- ordered(cut(AdultUCI$age, c(15,25,45,65,100)),
labels = c("Young", "Middle-aged", "Senior", "Old"))
AdultUCI$"hours-per-week" <- ordered(cut(AdultUCI$"hours-per-week",
c(0,25,40,60,168)),
labels = c("Part-time", "Full-time", "Over-time", "Workaholic"))
AdultUCI$"capital-gain" <- ordered(cut(AdultUCI$"capital-gain",
c(-Inf,0,median(AdultUCI$"capital-gain"[AdultUCI$"capital-gain">0]),
Inf)), labels = c("None", "Low", "High"))
AdultUCI$"capital-loss" <- ordered(cut(AdultUCI$"capital-loss",
c(-Inf,0, median(AdultUCI$"capital-loss"[AdultUCI$"capital-loss">0]),
Inf)), labels = c("None", "Low", "High"))
head(AdultUCI)
```
## Convert to transactions
```
AdultTrans <- as(AdultUCI, "transactions")
```
## A Priori
```
rules <- apriori(AdultTrans, parameter=list(supp = 0.5, conf = 0.9, minlen=2, maxlen= 10, target = "rules"),
appearance=list(rhs = c("capital-gain=None"), default="lhs"), control=NULL)
inspect(rules)
rules_a <- as(rules, "data.frame")
head(rules_a)
```
## Analysis of Rules
```
imrules <- interestMeasure(rules, transactions = AdultTrans)
head(imrules)
```
## Removing redundant rules
```
nrules <- rules[!is.redundant(rules)]
arules::inspect(nrules)
```
## Showing the transactions that support the rules
In this example, we can see the transactions (trans) that support rules 1.
```
st <- supportingTransactions(nrules[1], AdultTrans)
trans <- unique(st@data@i)
length(trans)
print(c(length(trans)/length(AdultTrans), nrules[1]@quality$support))
```
Now we can see the transactions (trans) that support rules 1 and 2.
As can be observed, the support for both rules is not the sum of the support of each rule.
```
st <- supportingTransactions(nrules[1:2], AdultTrans)
trans <- unique(st@data@i)
length(trans)
print(c(length(trans)/length(AdultTrans), nrules[1:2]@quality$support))
```
## Rules visualization
```
options(repr.plot.width=7, repr.plot.height=4)
plot(rules)
options(repr.plot.width=7, repr.plot.height=4)
plot(rules, method="paracoord", control=list(reorder=TRUE))
```
# Sequence Mining
```
x <- read_baskets(con = system.file("misc", "zaki.txt", package = "arulesSequences"), info = c("sequenceID","eventID","SIZE"))
as(x, "data.frame")
s1 <- cspade(x, parameter = list(support = 0.4), control = list(verbose = TRUE))
as(s1, "data.frame")
```
| github_jupyter |
<a href="https://colab.research.google.com/github/yohanesnuwara/reservoir-geomechanics/blob/master/delft%20course%20dr%20weijermars/stress_tensor.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import matplotlib.pyplot as plt
```
# Introduction to vectors
Plot vector that has notation (2,4,4). Another vector has notation (1,2,3). Find the direction cosines of each vector, the angles of each vector to the three axes, and the angle between the two vectors!
```
from mpl_toolkits.mplot3d import axes3d
X = np.array((0, 0))
Y= np.array((0, 0))
Z = np.array((0, 0))
U = np.array((2, 1))
V = np.array((4, 2))
W = np.array((4, 3))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.quiver(X, Y, Z, U, V, W)
ax.set_xlim([-4, 4])
ax.set_ylim([-4, 4])
ax.set_zlim([-4, 4])
# vector A and B
A_mag = np.sqrt(((U[0] - X[0])**2) + ((V[0] - Y[0])**2) + ((W[0] - Z[0])**2))
print('Magnitude of vector A:', A_mag, 'units')
B_mag = np.sqrt(((U[1] - X[1])**2) + ((V[1] - Y[1])**2) + ((W[1] - Z[1])**2))
print('Magnitude of vector B:', B_mag, 'units')
# direction cosines
l_A = (U[0] - X[0]) / A_mag
m_A = (V[0] - Y[0]) / A_mag
n_A = (W[0] - Z[0]) / A_mag
print('Direction cosine to x axis (cos alpha):', l_A, "to y axis (cos beta):", m_A, "to z axis (cos gamma):", n_A)
print('Pythagorean Sum of direction cosines of vector A:', l_A**2 + m_A**2 + n_A**2, "and must be equals to 1")
l_B = (U[1] - X[1]) / B_mag
m_B = (V[1] - Y[1]) / B_mag
n_B = (W[1] - Z[1]) / B_mag
print('Direction cosine to x axis (cos alpha):', l_B, "to y axis (cos beta):", m_B, "to z axis (cos gamma):", n_B)
print('Pythagorean Sum of direction cosines of vector B:', l_B**2 + m_B**2 + n_B**2, "and must be equals to 1")
# angles
alpha_A = np.rad2deg(np.arccos(l_A))
beta_A = np.rad2deg(np.arccos(m_A))
gamma_A = np.rad2deg(np.arccos(n_A))
print('Angle to x axis (alpha):', alpha_A, "to y axis (beta):", beta_A, "to z axis (gamma):", gamma_A)
alpha_B = np.rad2deg(np.arccos(l_B))
beta_B= np.rad2deg(np.arccos(m_B))
gamma_B = np.rad2deg(np.arccos(n_B))
print('Angle to x axis (alpha):', alpha_B, "to y axis (beta):", beta_B, "to z axis (gamma):", gamma_B)
# angle between two vectors
cosine_angle = (l_A * l_B) + (m_A * m_B) + (n_A * n_B)
angle = np.rad2deg(np.arccos(cosine_angle))
print('Angle between vector A and B:', angle, 'degrees')
```
# Exercise 10-3. Effective, Normal, and Shear Stress on a Plane
Consider a plane that makes an angle 60 degrees with $\sigma_1$ and 60 degrees with $\sigma_3$. The principal stresses are: -600, -400, -200 MPa. Calculate:
* Total effective stress
* Normal stress
* Shear stress
```
# principle stresses
sigma_1 = -600; sigma_2 = -400; sigma_3 = -200
# calculate the angle of plane to second principal stress sigma 2
# using pythagorean
alpha = 60; gamma = 60
l = np.cos(np.deg2rad(alpha))
n = np.cos(np.deg2rad(gamma))
m = np.sqrt(1 - l**2 - n**2)
beta = np.rad2deg(np.arccos(m))
print("The second principal stress sigma 2 makes angle:", beta, "degrees to the plane")
# effective stress
sigma_eff = np.sqrt(((sigma_1**2) * (l**2)) + ((sigma_2**2) * (m**2)) + ((sigma_3**2) * (n**2)))
print("The effective stress is:", -sigma_eff, "MPa (minus because it's compressive)")
# normal stress
sigma_normal = (sigma_1 * (l**2)) + (sigma_2 * (m**2)) + (sigma_3 * (n**2))
print("The normal stress is:", sigma_normal, "MPa")
# shear stress
sigma_shear = np.sqrt((sigma_eff**2) - (sigma_normal**2))
print("The shear stress is:", sigma_shear, "MPa")
```
# Stress Tensor Components
```
stress_tensor = [[sigma_xx, sigma_xy, sigma_xz],
[sigma_yx, sigma_yy, sigma_yz],
[sigma_zx, sigma_zy, sigma_zz]]
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.pyplot as plt
# point of cube
points = np.array([[-5, -5, -5],
[5, -5, -5 ],
[5, 5, -5],
[-5, 5, -5],
[-5, -5, 5],
[5, -5, 5 ],
[5, 5, 5],
[-5, 5, 5]])
# vector
a = np.array((0, 0))
b= np.array((0, 0))
c = np.array((0, 0))
u = np.array((0, -4))
v = np.array((5, 0))
w = np.array((0, -4))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.quiver(a, b, c, u, v, w, color='black')
ax.set_xlim([-5, 5])
ax.set_ylim([-5, 5])
ax.set_zlim([-5, 5])
r = [-5,5]
X, Y = np.meshgrid(r, r)
one = np.array([5, 5, 5, 5])
one = one.reshape(2, 2)
ax.plot_wireframe(X,Y,one, alpha=0.5)
ax.plot_wireframe(X,Y,-one, alpha=0.5)
ax.plot_wireframe(X,-one,Y, alpha=0.5)
ax.plot_wireframe(X,one,Y, alpha=0.5)
ax.plot_wireframe(one,X,Y, alpha=0.5)
ax.plot_wireframe(-one,X,Y, alpha=0.5)
ax.scatter3D(points[:, 0], points[:, 1], points[:, 2])
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_zlabel('Z')
plt.show()
np.ones(4)
```
# Exercise 10-7 Total Stress, Deviatoric Stress, Effective Stress, Cauchy Summation
$$\sigma_{ij}=\tau_{ij}+P_{ij}$$
$$P_{ij}=P \cdot \delta_{ij}$$
Pressure is: $P=|\sigma_{mean}|=|\frac{\sigma_{xx}+\sigma_{yy}+\sigma_{zz}}{3}|$
Knorecker Delta is: $\delta_{ij}=\begin{bmatrix} 1 & 0 & 0 \\ 0 & 1 & 0 \\ 0 & 0 & 1 \end{bmatrix}$
Pressure tensor is: $P_{ij}=P \cdot \delta_{ij}$
So, overall the total stress is: $\sigma_{ij}=\begin{bmatrix} P+\tau_{xx} & \tau_{xy} & \tau_{zx} \\ \tau_{yx} & P+\tau_{yy} & \tau_{yz} \\ \tau_{zx} & \tau_{zy} & P+\tau_{zz} \end{bmatrix}$
Cauchy summation to calculate the components of effective stress
$$\sigma_{eff}=\begin{bmatrix} \sigma_x \\ \sigma_y \\ \sigma_z \end{bmatrix}=\begin{bmatrix} \sigma_{xx} & \sigma_{xy} & \sigma_{xz} \\ \sigma_{yx} & \sigma_{yy} & \sigma_{zy} \\ \sigma_{zx} & \sigma_{zy} & \sigma_{zz} \end{bmatrix} \cdot \begin{bmatrix} l \\ m \\ n \end{bmatrix}$$
**Known**: direction cosines of plane ABC, total stress tensor.
**Task**:
* Determine the deviatoric stress tensor
* Calculate the components of effective stress on plane ABC (use Cauchy's summation)
* Calculate total effective stress, total normal stress, total shear stress
```
# known
l, m, n = 0.7, 0.5, 0.5 # direction cosines
alpha, beta, gamma = 45, 60, 60 # angles
stress_ij = np.array([[-40, -40, -35],
[-40, 45, -50],
[-35, -50, -20]]) # total stress tensor
# calculate pressure
P = np.abs(np.mean(np.array([(stress_ij[0][0]), (stress_ij[1][1]), (stress_ij[2][2])])))
print("Pressure:", P, "MPa")
# pressure TENSOR
kronecker = np.array([[1, 0, 0],
[0, 1, 0],
[0, 0, 1]])
P_ij = P * kronecker
print('Pressure tensor:')
print(P_ij)
# deviatoric stress TENSOR
tau_ij = stress_ij - P_ij
print('Deviatoric stress tensor:')
print(tau_ij)
# direction cosines VECTOR
lmn = np.array([[l],
[m],
[n]])
# effective stress VECTOR
stress_eff = np.dot(stress_ij, lmn)
stress_eff_1 = stress_eff[0][0]
stress_eff_2 = stress_eff[1][0]
stress_eff_3 = stress_eff[2][0]
print('Effective stress vector:')
print(stress_eff)
print('X component of effective stress:', stress_eff_1, 'MPa')
print('Y component of effective stress:', stress_eff_2, 'MPa')
print('Z component of effective stress:', stress_eff_3, 'MPa')
# total / magnitude of effective stress, is SCALAR
sigma_eff = np.sqrt((stress_eff_1**2) + (stress_eff_2**2) + (stress_eff_3**2))
print("The total effective stress is:", -sigma_eff, "MPa")
# principal stresses
sigma_1 = stress_eff_1 / l
sigma_2 = stress_eff_2 / m
sigma_3 = stress_eff_3 / n
print('X component of principal stress:', sigma_1, 'MPa')
print('Y component of principal stress:', sigma_2, 'MPa')
print('Z component of principal stress:', sigma_3, 'MPa')
# total normal stress
sigma_normal = (sigma_1 * (l**2)) + (sigma_2 * (m**2)) + (sigma_3 * (n**2))
print("The normal stress is:", sigma_normal, "MPa")
print("Because normal stress", sigma_normal, "MPa nearly equals to sigma 1", sigma_1, "MPa, the plane is nearly normal to sigma 1")
# total shear stress
sigma_shear = np.sqrt((sigma_eff**2) - (sigma_normal**2))
print("The shear stress is:", sigma_shear, "MPa")
```
<div>
<img src="https://user-images.githubusercontent.com/51282928/77084625-cdfbe280-6a31-11ea-9c3f-c4e592d5cfd9.jpeg" width="500"/>
</div>
# Exercise 10-8 Transforming Stress Tensor (Containing all the 9 tensors of shear and normal) to Principal Stress Tensor using Cubic Equation
```
sigma_ij = np.array([[0, 0, 100],
[0, 0, 0],
[-100, 0, 0]]) # stress tensor
# cubic equation
coeff3 = 1
coeff2 = -((sigma_ij[0][0] + sigma_ij[1][1] + sigma_ij[2][2]))
coeff1 = (sigma_ij[0][0] * sigma_ij[1][1]) + (sigma_ij[1][1] * sigma_ij[2][2]) + (sigma_ij[2][2] * sigma_ij[0][0]) - ((sigma_ij[0][1])**2) - ((sigma_ij[1][2])**2) - ((sigma_ij[2][0])**2)
coeff0 = -((sigma_ij[0][0] * sigma_ij[1][1] * sigma_ij[2][2]) + (2 * sigma_ij[0][1] * sigma_ij[1][2] * sigma_ij[2][0]) - (sigma_ij[0][0] * (sigma_ij[1][2])**2) - (sigma_ij[1][1] * (sigma_ij[2][0])**2) - (sigma_ij[2][2]* (sigma_ij[0][1])**2))
roots = np.roots([coeff3, coeff2, coeff1, coeff0])
sigma = np.sort(roots)
sigma_1 = sigma[2]
sigma_2 = sigma[1]
sigma_3 = sigma[0]
sigma_principal = np.array([[sigma_1, 0, 0],
[0, sigma_2, 0],
[0, 0, sigma_3]])
print("The principal stresses are, sigma 1:", sigma_1, "MPa, sigma 2:", sigma_2, "MPa, and sigma 3:", sigma_3, "MPa")
print("Principal stress tensor:")
print(sigma_principal)
denominator_l = (sigma_ij[0][0] * sigma_ij[2][2]) - (sigma_ij[1][1] * sigma_1) - (sigma_ij[2][2] * sigma_1) + (sigma_1)**2 - (sigma_ij[1][2])**2
denominator_m = (sigma_2 * sigma_ij[0][1]) + (sigma_ij[2][0] * sigma_ij[1][2]) - (sigma_ij[0][1] * sigma_ij[2][2])
denominator_n = (sigma_3 * sigma_ij[2][0]) + (sigma_ij[0][1] * sigma_ij[1][2]) - (sigma_ij[2][0] * sigma_ij[1][1])
denominator_l, denominator_m, denominator_n
```
# ***
```
from mpl_toolkits.mplot3d import axes3d
X = np.array((0))
Y= np.array((0))
U = np.array((0))
V = np.array((4))
fig, ax = plt.subplots()
q = ax.quiver(X, Y, U, V,units='xy' ,scale=1)
plt.grid()
ax.set_aspect('equal')
plt.xlim(-5,5)
plt.ylim(-5,5)
from mpl_toolkits.mplot3d import axes3d
X = np.array((0))
Y= np.array((0))
Z = np.array((0))
U = np.array((1))
V = np.array((1))
W = np.array((1))
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.quiver(X, Y, Z, U, V, W)
ax.set_xlim([-1, 1])
ax.set_ylim([-1, 1])
ax.set_zlim([-1, 1])
from mpl_toolkits.mplot3d import axes3d
vx_mag = v_mag * l
vy_mag = v_mag * m
vz_mag = v_mag * n
x = 0; y = 0; z = 0
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.quiver(x, y, z, vx_mag, vy_mag, vz_mag)
ax.set_xlim(0, 10); ax.set_ylim(0, 10); ax.set_zlim(0, 5)
```
| github_jupyter |
# AutoGluon Tabular with SageMaker
[AutoGluon](https://github.com/awslabs/autogluon) automates machine learning tasks enabling you to easily achieve strong predictive performance in your applications. With just a few lines of code, you can train and deploy high-accuracy deep learning models on tabular, image, and text data.
This notebook shows how to use AutoGluon-Tabular with Amazon SageMaker by creating custom containers.
## Prerequisites
If using a SageMaker hosted notebook, select kernel `conda_mxnet_p36`.
```
# Make sure docker compose is set up properly for local mode
!./setup.sh
# Imports
import os
import boto3
import sagemaker
from time import sleep
from collections import Counter
import numpy as np
import pandas as pd
from sagemaker import get_execution_role, local, Model, utils, fw_utils, s3
from sagemaker.estimator import Estimator
from sagemaker.predictor import RealTimePredictor, csv_serializer, StringDeserializer
from sklearn.metrics import accuracy_score, classification_report
from IPython.core.display import display, HTML
from IPython.core.interactiveshell import InteractiveShell
# Print settings
InteractiveShell.ast_node_interactivity = "all"
pd.set_option('display.max_columns', 500)
pd.set_option('display.max_rows', 10)
# Account/s3 setup
session = sagemaker.Session()
local_session = local.LocalSession()
bucket = session.default_bucket()
prefix = 'sagemaker/autogluon-tabular'
region = session.boto_region_name
role = get_execution_role()
client = session.boto_session.client(
"sts", region_name=region, endpoint_url=utils.sts_regional_endpoint(region)
)
account = client.get_caller_identity()['Account']
ecr_uri_prefix = utils.get_ecr_image_uri_prefix(account, region)
registry_id = fw_utils._registry_id(region, 'mxnet', 'py3', account, '1.6.0')
registry_uri = utils.get_ecr_image_uri_prefix(registry_id, region)
```
### Build docker images
First, build autogluon package to copy into docker image.
```
if not os.path.exists('package'):
!pip install PrettyTable -t package
!pip install --upgrade boto3 -t package
!pip install bokeh -t package
!pip install --upgrade matplotlib -t package
!pip install autogluon -t package
```
Now build the training/inference image and push to ECR
```
training_algorithm_name = 'autogluon-sagemaker-training'
inference_algorithm_name = 'autogluon-sagemaker-inference'
!./container-training/build_push_training.sh {account} {region} {training_algorithm_name} {ecr_uri_prefix} {registry_id} {registry_uri}
!./container-inference/build_push_inference.sh {account} {region} {inference_algorithm_name} {ecr_uri_prefix} {registry_id} {registry_uri}
```
### Get the data
In this example we'll use the direct-marketing dataset to build a binary classification model that predicts whether customers will accept or decline a marketing offer.
First we'll download the data and split it into train and test sets. AutoGluon does not require a separate validation set (it uses bagged k-fold cross-validation).
```
# Download and unzip the data
!aws s3 cp --region {region} s3://sagemaker-sample-data-{region}/autopilot/direct_marketing/bank-additional.zip .
!unzip -qq -o bank-additional.zip
!rm bank-additional.zip
local_data_path = './bank-additional/bank-additional-full.csv'
data = pd.read_csv(local_data_path)
# Split train/test data
train = data.sample(frac=0.7, random_state=42)
test = data.drop(train.index)
# Split test X/y
label = 'y'
y_test = test[label]
X_test = test.drop(columns=[label])
```
##### Check the data
```
train.head(3)
train.shape
test.head(3)
test.shape
X_test.head(3)
X_test.shape
```
Upload the data to s3
```
train_file = 'train.csv'
train.to_csv(train_file,index=False)
train_s3_path = session.upload_data(train_file, key_prefix='{}/data'.format(prefix))
test_file = 'test.csv'
test.to_csv(test_file,index=False)
test_s3_path = session.upload_data(test_file, key_prefix='{}/data'.format(prefix))
X_test_file = 'X_test.csv'
X_test.to_csv(X_test_file,index=False)
X_test_s3_path = session.upload_data(X_test_file, key_prefix='{}/data'.format(prefix))
```
## Hyperparameter Selection
The minimum required settings for training is just a target label, `fit_args['label']`.
Additional optional hyperparameters can be passed to the `autogluon.task.TabularPrediction.fit` function via `fit_args`.
Below shows a more in depth example of AutoGluon-Tabular hyperparameters from the example [Predicting Columns in a Table - In Depth](https://autogluon.mxnet.io/tutorials/tabular_prediction/tabular-indepth.html#model-ensembling-with-stacking-bagging). Please see [fit parameters](https://autogluon.mxnet.io/api/autogluon.task.html?highlight=eval_metric#autogluon.task.TabularPrediction.fit) for further information. Note that in order for hyperparameter ranges to work in SageMaker, values passed to the `fit_args['hyperparameters']` must be represented as strings.
```python
nn_options = {
'num_epochs': "10",
'learning_rate': "ag.space.Real(1e-4, 1e-2, default=5e-4, log=True)",
'activation': "ag.space.Categorical('relu', 'softrelu', 'tanh')",
'layers': "ag.space.Categorical([100],[1000],[200,100],[300,200,100])",
'dropout_prob': "ag.space.Real(0.0, 0.5, default=0.1)"
}
gbm_options = {
'num_boost_round': "100",
'num_leaves': "ag.space.Int(lower=26, upper=66, default=36)"
}
model_hps = {'NN': nn_options, 'GBM': gbm_options}
fit_args = {
'label': 'y',
'presets': ['best_quality', 'optimize_for_deployment'],
'time_limits': 60*10,
'hyperparameters': model_hps,
'hyperparameter_tune': True,
'search_strategy': 'skopt'
}
hyperparameters = {
'fit_args': fit_args,
'feature_importance': True
}
```
**Note:** Your hyperparameter choices may affect the size of the model package, which could result in additional time taken to upload your model and complete training. Including `'optimize_for_deployment'` in the list of `fit_args['presets']` is recommended to greatly reduce upload times.
<br>
```
# Define required label and optional additional parameters
fit_args = {
'label': 'y',
# Adding 'best_quality' to presets list will result in better performance (but longer runtime)
'presets': ['optimize_for_deployment'],
}
# Pass fit_args to SageMaker estimator hyperparameters
hyperparameters = {
'fit_args': fit_args,
'feature_importance': True
}
```
## Train
For local training set `train_instance_type` to `local` .
For non-local training the recommended instance type is `ml.m5.2xlarge`.
**Note:** Depending on how many underlying models are trained, `train_volume_size` may need to be increased so that they all fit on disk.
```
%%time
instance_type = 'ml.m5.2xlarge'
#instance_type = 'local'
ecr_image = f'{ecr_uri_prefix}/{training_algorithm_name}:latest'
estimator = Estimator(image_name=ecr_image,
role=role,
train_instance_count=1,
train_instance_type=instance_type,
hyperparameters=hyperparameters,
train_volume_size=100)
# Set inputs. Test data is optional, but requires a label column.
inputs = {'training': train_s3_path, 'testing': test_s3_path}
estimator.fit(inputs)
```
### Create Model
```
# Create predictor object
class AutoGluonTabularPredictor(RealTimePredictor):
def __init__(self, *args, **kwargs):
super().__init__(*args, content_type='text/csv',
serializer=csv_serializer,
deserializer=StringDeserializer(), **kwargs)
ecr_image = f'{ecr_uri_prefix}/{inference_algorithm_name}:latest'
if instance_type == 'local':
model = estimator.create_model(image=ecr_image, role=role)
else:
model_uri = os.path.join(estimator.output_path, estimator._current_job_name, "output", "model.tar.gz")
model = Model(model_uri, ecr_image, role=role, sagemaker_session=session, predictor_cls=AutoGluonTabularPredictor)
```
### Batch Transform
For local mode, either `s3://<bucket>/<prefix>/output/` or `file:///<absolute_local_path>` can be used as outputs.
By including the label column in the test data, you can also evaluate prediction performance (In this case, passing `test_s3_path` instead of `X_test_s3_path`).
```
output_path = f's3://{bucket}/{prefix}/output/'
# output_path = f'file://{os.getcwd()}'
transformer = model.transformer(instance_count=1,
instance_type=instance_type,
strategy='MultiRecord',
max_payload=6,
max_concurrent_transforms=1,
output_path=output_path)
transformer.transform(test_s3_path, content_type='text/csv', split_type='Line')
transformer.wait()
```
### Endpoint
##### Deploy remote or local endpoint
```
instance_type = 'ml.m5.2xlarge'
#instance_type = 'local'
predictor = model.deploy(initial_instance_count=1,
instance_type=instance_type)
```
##### Attach to endpoint (or reattach if kernel was restarted)
```
# Select standard or local session based on instance_type
if instance_type == 'local':
sess = local_session
else:
sess = session
# Attach to endpoint
predictor = AutoGluonTabularPredictor(predictor.endpoint, sagemaker_session=sess)
```
##### Predict on unlabeled test data
```
results = predictor.predict(X_test.to_csv(index=False)).splitlines()
# Check output
print(Counter(results))
```
##### Predict on data that includes label column
Prediction performance metrics will be printed to endpoint logs.
```
results = predictor.predict(test.to_csv(index=False)).splitlines()
# Check output
print(Counter(results))
```
##### Check that classification performance metrics match evaluation printed to endpoint logs as expected
```
y_results = np.array(results)
print("accuracy: {}".format(accuracy_score(y_true=y_test, y_pred=y_results)))
print(classification_report(y_true=y_test, y_pred=y_results, digits=6))
```
##### Clean up endpoint
```
predictor.delete_endpoint()
```
| github_jupyter |
# Homework - Random Walks (18 pts)
## Continuous random walk in three dimensions
Write a program simulating a three-dimensional random walk in a continuous space. Let 1000 independent particles all start at random positions within a cube with corners at (0,0,0) and (1,1,1). At each time step each particle will move in a random direction by a random amount between -1 and 1 along each axis (x, y, z).
1. (3 pts) Create data structure(s) to store your simulated particle positions for each of 2000 time steps and initialize them with the particles starting positions.
```
import numpy as np
numTimeSteps = 2000
numParticles = 1000
positions = np.zeros( (numParticles, 3, numTimeSteps) )
# initialize starting positions on first time step
positions[:,:,0] = np.random.random( (numParticles, 3) )
```
2. (3 pts) Write code to run your simulation for 2000 time steps.
```
for t in range(numTimeSteps-1):
# 2 * [0 to 1] - 1 --> [-1 to 1]
jumpsForAllParticles = 2 * np.random.random((numParticles, 3)) - 1
positions[:,:,t+1] = positions[:,:,t] + jumpsForAllParticles
# just for fun, here's another way to run the simulation above without a loop
jumpsForAllParticlesAndAllTimeSteps = 2 * np.random.random((numParticles, 3, numTimeSteps-1)) - 1
positions[:,:,1:] = positions[:,:,0].reshape(numParticles, 3, 1) + np.cumsum(jumpsForAllParticlesAndAllTimeSteps, axis=2)
```
3. (3 pts) Generate a series of four 3D scatter plots at selected time points to visually convey what is going on. Arrange the plots in a single row from left to right. Make sure you indicate which time points you are showing.
```
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
lim = 70
plt.figure(figsize=(12,3))
for (i,t) in enumerate([0, 100, 1000, 1999]):
ax = plt.subplot(1, 4, i+1, projection='3d')
x = positions[:,0,t]
y = positions[:,1,t]
z = positions[:,2,t]
ax.scatter(x, y, z)
plt.xlim([-lim, lim])
plt.ylim([-lim, lim])
ax.set_zlim([-lim, lim])
plt.xlabel("x")
plt.ylabel("y")
ax.set_zlabel("z")
plt.title(f"Time {t}");
```
4. (3 pts) Draw the path of a single particle (your choice) across all time steps in a 3D plot.
```
ax = plt.subplot(1, 1, 1, projection='3d')
i = 10 # particle index
x = positions[i,0,:]
y = positions[i,1,:]
z = positions[i,2,:]
plt.plot(x, y, z)
plt.xlabel("x")
plt.ylabel("y")
ax.set_zlabel("z")
plt.title(f"Particle {i}");
```
5. (3 pts) Find the minimum, maximum, mean and variance for the jump distances of all particles throughout the entire simulation. Jump distance is the euclidean distance moved on each time step $\sqrt(dx^2+dy^2+dz^2)$. *Hint: numpy makes this very simple.*
```
jumpsXYZForAllParticlesAndAllTimeSteps = positions[:,:,1:] - positions[:,:,:-1]
jumpDistancesForAllParticlesAndAllTimeSteps = np.sqrt(np.sum(jumpsXYZForAllParticlesAndAllTimeSteps**2, axis=1))
print(f"min = {jumpDistancesForAllParticlesAndAllTimeSteps.min()}")
print(f"max = {jumpDistancesForAllParticlesAndAllTimeSteps.max()}")
print(f"mean = {jumpDistancesForAllParticlesAndAllTimeSteps.mean()}")
print(f"var = {jumpDistancesForAllParticlesAndAllTimeSteps.var()}")
```
6. (3 pts) Repeat the simulation, but this time confine the particles to a unit cell of dimension 10x10x10. Make it so that if a particle leaves one edge of the cell, it enters on the opposite edge (this is the sort of thing most molecular dynamics simulations do). Show plots as in #3 to visualize the simulation (note that most interesting stuff liekly happens in the first 100 time steps).
```
for t in range(numTimeSteps-1):
# 2 * [0 to 1] - 1 --> [-1 to 1]
jumpsForAllParticles = 2 * np.random.random((numParticles, 3)) - 1
positions[:,:,t+1] = positions[:,:,t] + jumpsForAllParticles
# check for out-of-bounds and warp to opposite bound
for i in range(numParticles):
for j in range(3):
if positions[i,j,t+1] < 0:
positions[i,j,t+1] += 10
elif positions[i,j,t+1] > 10:
positions[i,j,t+1] -= 10
plt.figure(figsize=(12,3))
for (i,t) in enumerate([0, 3, 10, 1999]):
ax = plt.subplot(1, 4, i+1, projection='3d')
x = positions[:,0,t]
y = positions[:,1,t]
z = positions[:,2,t]
ax.scatter(x, y, z)
plt.xlim([0, 10])
plt.ylim([0, 10])
ax.set_zlim([0, 10])
plt.xlabel("x")
plt.ylabel("y")
ax.set_zlabel("z")
plt.title(f"Time {t}");
```
| github_jupyter |
# Data preparation for tutorial
This notebook contains the code to convert raw downloaded external data into a cleaned or simplified version for tutorial purposes.
The raw data is expected to be in the `./raw` sub-directory (not included in the git repo).
```
%matplotlib inline
import pandas as pd
import geopandas
```
## Countries dataset
http://www.naturalearthdata.com/downloads/110m-cultural-vectors/110m-admin-0-countries/
```
countries = geopandas.read_file("zip://./raw/original_data_ne/ne_110m_admin_0_countries.zip")
countries.head()
len(countries)
countries_subset = countries[['ADM0_A3', 'NAME', 'CONTINENT', 'POP_EST', 'GDP_MD_EST', 'geometry']]
countries_subset.columns = countries_subset.columns.str.lower()
countries_subset = countries_subset.rename(columns={'adm0_a3': 'iso_a3'})
countries_subset.head()
countries_subset.to_file("ne_110m_admin_0_countries.shp")
```
## Natural Earth - Cities dataset
http://www.naturalearthdata.com/downloads/110m-cultural-vectors/110m-populated-places/ (simple, version 4.0.0, downloaded May 2018)
```
cities = geopandas.read_file("zip://./raw/original_data_ne/ne_110m_populated_places_simple.zip")
cities.head()
len(cities)
cities_subset = cities[['name', 'geometry']]
cities_subset.head()
cities_subset.to_file("ne_110m_populated_places.shp")
```
## Natural Earth - Rivers dataset
http://www.naturalearthdata.com/downloads/50m-physical-vectors/50m-rivers-lake-centerlines/ (version 4.0.0, downloaded May 2018)
```
rivers = geopandas.read_file("zip://./raw/ne_50m_rivers_lake_centerlines.zip")
rivers.head()
```
Remove rows with missing geometry:
```
len(rivers)
rivers = rivers[~rivers.geometry.isna()].reset_index(drop=True)
len(rivers)
```
Subset of the columns:
```
rivers_subset = rivers[['featurecla', 'name_en', 'geometry']].rename(columns={'name_en': 'name'})
rivers_subset.head()
rivers_subset.to_file("ne_50m_rivers_lake_centerlines.shp")
```
## Paris districts
Source: https://opendata.paris.fr/explore/dataset/quartier_paris/ (downloaded as GeoJSON file on August 20, 2018)
Administrative districts, polygon dataset
```
districts = geopandas.read_file("./raw/quartier_paris.geojson")
districts.head()
districts = districts.rename(columns={'l_qu': 'district_name', 'c_qu': 'id'}).sort_values('id').reset_index(drop=True)
```
Add population data (based on pdfs downloaded from ..):
```
population = pd.read_csv("./raw/paris-population.csv")
population['temp'] = population.district_name.str.lower()
population['temp'] = population['temp'].replace({
'javel': 'javel 15art',
'saint avoye': 'sainte avoie',
"saint germain l'auxerrois": "st germain l'auxerrois",
'plaine monceau': 'plaine de monceaux',
'la chapelle': 'la chapelle'})
districts['temp'] = (districts.district_name.str.lower().str.replace('-', ' ')
.str.replace('é', 'e').str.replace('è', 'e').str.replace('ê', 'e').str.replace('ô', 'o'))
res = pd.merge(districts, population[['population', 'temp']], on='temp', how='outer')
assert len(res) == len(districts)
districts = res[['id', 'district_name', 'population', 'geometry']]
districts.head()
districts.to_file("processed/paris_districts.geojson", driver='GeoJSON')
districts = districts.to_crs(epsg=32631)
districts.to_file("paris_districts_utm.geojson", driver='GeoJSON')
```
## Commerces de Paris
Source: https://opendata.paris.fr/explore/dataset/commercesparis/ (downloaded as csv file (`commercesparis.csv`) on October 30, 2018)
```
df = pd.read_csv("./raw/commercesparis.csv", sep=';')
df.iloc[0]
```
Take subset of the restaurants:
```
restaurants = df[df['CODE ACTIVITE'].str.startswith('CH1', na=False)].copy()
restaurants['LIBELLE ACTIVITE'].value_counts()
restaurants = restaurants.dropna(subset=['XY']).reset_index(drop=True)
```
Translate the restaurants and rename column:
```
restaurants['LIBELLE ACTIVITE'] = restaurants['LIBELLE ACTIVITE'].replace({
'Restaurant traditionnel français': 'Traditional French restaurant',
'Restaurant asiatique': 'Asian restaurant',
'Restaurant européen': 'European restuarant',
'Restaurant indien, pakistanais et Moyen Orient': 'Indian / Middle Eastern restaurant',
'Restaurant maghrébin': 'Maghrebian restaurant',
'Restaurant africain': 'African restaurant',
'Autre restaurant du monde': 'Other world restaurant',
'Restaurant central et sud américain': 'Central and South American restuarant',
'Restaurant antillais': 'Caribbean restaurant'
})
restaurants = restaurants.rename(columns={'LIBELLE ACTIVITE': 'type'})
```
Create GeoDataFrame
```
from shapely.geometry import Point
restaurants['geometry'] = restaurants['XY'].str.split(', ').map(lambda x: Point(float(x[1]), float(x[0])))
restaurants = geopandas.GeoDataFrame(restaurants[['type', 'geometry']], crs={'init': 'epsg:4326'})
restaurants.head()
restaurants.to_file("processed/paris_restaurants.gpkg", driver='GPKG')
```
| github_jupyter |
# Another attempt at MC Simulation on AHP/ANP
The ideas are the following:
1. There is a class MCAnp that has a sim() method that will simulate any Prioritizer
2. MCAnp also has a sim_fill() function that does fills in the data needed for a single simulation
## Import needed libs
```
import pandas as pd
import sys
import os
sys.path.insert(0, os.path.abspath("../"))
import numpy as np
from scipy.stats import triang
from copy import deepcopy
from pyanp.priority import pri_eigen
from pyanp.pairwise import Pairwise
from pyanp.ahptree import AHPTree, AHPTreeNode
from pyanp.direct import Direct
```
# MCAnp class
```
def ascale_mscale(val:(float,int))->float:
if val is None:
return 0
elif val < 0:
val = -val
val += 1
val = 1.0/val
return val
else:
return val+1
def mscale_ascale(val:(float,int))->float:
if val == 0:
return None
elif val >= 1:
return val - 1
else:
val = 1/val
val = val-1
return -val
DEFAULT_DISTRIB = triang(c=0.5, loc=-1.5, scale=3.0)
def avote_random(avote):
"""
Returns a random additive vote in the neighborhood of the additive vote avote
according to the default disribution DEFAULT_DISTRIB
"""
if avote is None:
return None
raw_val = DEFAULT_DISTRIB.rvs(size=1)[0]
return avote+raw_val
def mvote_random(mvote):
"""
Returns a random multiplicative vote in the neighborhhod of the multiplicative vote mvote
according to the default distribution DEFAULT_DISTRIB. This is handled by converting
the multiplicative vote to an additive vote, calling avote_random() and converting the
result back to an additive vote
"""
avote = mscale_ascale(mvote)
rval_a = avote_random(avote)
rval = ascale_mscale(rval_a)
return rval
def direct_random(direct, max_percent_chg=0.2)->float:
"""
Returns a random direct data value near the value `direct'. This function
creates a random percent change, between -max_percent_chg and +max_percent_chg, and
then changes the direct value by that factor, and returns it.
"""
pchg = np.random.uniform(low=-max_percent_chg, high=max_percent_chg)
return direct * (1 + pchg)
class MCAnp:
def __init__(self):
# Setup the random pairwise vote generator
self.pwvote_random = mvote_random
# Setup the random direct vote generator
self.directvote_random = direct_random
# Set the default user to use across the simulation
# follows the standard from Pairwise class, i.e. it can be a list
# of usernames, a single username, or None (which means total group average)
self.username = None
# What is the pairwise priority calculation?
self.pwprioritycalc = pri_eigen
def sim_fill(self, src, dest):
"""
Fills in data on a structure prior to doing the simulation calculations.
This function calls sim_NAME_fill depending on the class of the src object.
If the dest object is None, we create a dest object by calling deepcopy().
In either case, we always return the allocated dest object
"""
if dest is None:
dest = deepcopy(src)
# Which kind of src do we have
if isinstance(src, np.ndarray):
# We are simulating on a pairwise comparison matrix
return self.sim_pwmat_fill(src, dest)
elif isinstance(src, Pairwise):
# We are simulating on a multi-user pairwise comparison object
return self.sim_pw_fill(src, dest)
elif isinstance(src, AHPTree):
# We are simulating on an ahp tree object
return self.sim_ahptree_fill(src, dest)
elif isinstance(src, Direct):
# We are simulating on an ahp direct data
return self.sim_direct_fill(src, dest)
else:
raise ValueError("Src class is not handled, it is "+type(src).__name__)
def sim_pwmat_fill(self, pwsrc:np.ndarray, pwdest:np.ndarray=None)->np.ndarray:
"""
Fills in a pairwise comparison matrix with noisy votes based on pwsrc
If pwsrc is None, we create a new matrix, otherwise we fill in pwdest
with noisy values based on pwsrc and the self.pwvote_random parameter.
In either case, we return the resulting noisy matrix
"""
if pwdest is None:
pwdest = deepcopy(pwsrc)
size = len(pwsrc)
for row in range(size):
pwdest[row,row] = 1.0
for col in range(row+1, size):
val = pwsrc[row,col]
if val >= 1:
nvote = self.pwvote_random(val)
pwdest[row, col]=nvote
pwdest[col, row]=1/nvote
elif val!= 0:
nvote = self.pwvote_random(1/val)
pwdest[col, row] = nvote
pwdest[row, col] = 1/nvote
else:
pwdest[row, col] = nvote
pwdest[col, row] = nvote
return pwdest
def sim_pwmat(self, pwsrc:np.ndarray, pwdest:np.ndarray=None)->np.ndarray:
"""
creates a noisy pw comparison matrix from pwsrc, stores the matrix in pwdest (which
is created if pwdest is None) calculates the resulting priority and returns that
"""
pwdest = self.sim_pwmat_fill(pwsrc, pwdest)
rval = self.pwprioritycalc(pwdest)
return rval
def sim_pw(self, pwsrc:Pairwise, pwdest:Pairwise)->np.ndarray:
"""
Performs a simulation on a pairwise comparison matrix object and returns the
resulting priorities
"""
pwdest = self.sim_pw_fill(pwsrc, pwdest)
mat = pwdest.matrix(self.username)
rval = self.pwprioritycalc(mat)
return rval
def sim_pw_fill(self, pwsrc:Pairwise, pwdest:Pairwise=None)->Pairwise:
"""
Fills in the pairwise comparison structure of pwdest with noisy pairwise data from pwsrc.
If pwdest is None, we create one first, then fill in. In either case, we return the pwdest
object with new noisy data in it.
"""
if pwdest is None:
pwdest = deepcopy(pwsrc)
for user in pwsrc.usernames():
srcmat = pwsrc.matrix(user)
destmat = pwdest.matrix(user)
self.sim_pwmat_fill(srcmat, destmat)
return pwdest
def sim_direct_fill(self, directsrc:Direct, directdest:Direct=None)->Direct:
"""
Fills in the direct data structure of directdest with noisy data from directsrc.
If directdest is None, we create on as a deep copy of directsrc, then fill in.
In either case, we return the directdest object with new noisy data in it.
"""
if directdest is None:
directdest = deepcopy(directsrc)
for altpos in range(len(directdest)):
orig = directsrc[altpos]
newvote = self.directvote_random(orig)
directdest.data[altpos] = newvote
return directdest
def sim_direct(self, directsrc:Direct, directdest:Direct=None)->np.ndarray:
"""
Simulates for direct data
"""
directdest = self.sim_direct_fill(directsrc, directdest)
return directdest.priority()
def sim_ahptree_fill(self, ahpsrc:AHPTree, ahpdest:AHPTree)->AHPTree:
"""
Fills in the ahp tree structure of ahpdest with noisy data from ahpsrc.
If ahpdest is None, we create one as a deepcopy of ahpsrc, then fill in.
In either case, we return the ahpdest object with new noisy data in it.
"""
if ahpdest is None:
ahpdest = deepcopy(ahpsrc)
self.sim_ahptreenode_fill(ahpsrc.root, ahpdest.root)
return ahpdest
def sim_ahptreenode_fill(self, nodesrc:AHPTreeNode, nodedest:AHPTreeNode)->AHPTreeNode:
"""
Fills in data in an AHPTree
"""
#Okay, first we fill in for the alt_prioritizer
if nodesrc.alt_prioritizer is not None:
self.sim_fill(nodesrc.alt_prioritizer, nodedest.alt_prioritizer)
#Now wefill in the child prioritizer
if nodesrc.child_prioritizer is not None:
self.sim_fill(nodesrc.child_prioritizer, nodedest.child_prioritizer)
#Now for each child, fill in
for childsrc, childdest in zip(nodesrc.children, nodedest.children):
self.sim_ahptreenode_fill(childsrc, childdest)
#We are done, return the dest
return nodedest
def sim_ahptree(self, ahpsrc:AHPTree, ahpdest:AHPTree)->np.ndarray:
"""
Perform the actual simulation
"""
ahpdest = self.sim_ahptree_fill(ahpsrc, ahpdest)
return ahpdest.priority()
mc = MCAnp()
pw = np.array([
[1, 1/2, 3],
[2, 1, 5],
[1/3, 1/5, 1]
])
rpw= mc.sim_pwmat_fill(pw)
rpw
[mc.sim_pwmat(pw) for i in range(20)]
pwobj = Pairwise(alts=['alt '+str(i) for i in range(3)])
pwobj.vote_matrix(user_name='u1', val=pw)
```
## Checking that the deep copy is actually a deep copy
For some reason deepcopy was not copying the matrix, I had to overwrite
__deepcopy__ in Pairwise
```
pwobj.matrix('u1')
rpwobj = pwobj.__deepcopy__()
a=rpwobj
b=pwobj
a.df
display(a.df.loc['u1', 'Matrix'])
display(b.df.loc['u1', 'Matrix'])
display(a.matrix('u1') is b.matrix('u1'))
display(a.matrix('u1') == b.matrix('u1'))
```
## Now let's try to simulate
```
[mc.sim_pw(pwobj, rpwobj) for i in range(20)]
pwobj.matrix('u1')
```
## Try to simulate a direct data
```
dd = Direct(alt_names=['a1', 'a2', 'a3'])
dd.data[0]=0.5
dd.data[1]=0.3
dd.data[2]=0.2
rdd=mc.sim_direct_fill(dd)
rdd.data
```
## Simulate an ahptree
```
alts=['alt '+str(i) for i in range(3)]
tree = AHPTree(alt_names=alts)
kids = ['crit '+str(i) for i in range(4)]
for kid in kids:
tree.add_child(kid)
node = tree.get_node(kid)
direct = node.alt_prioritizer
s = 0
for alt in alts:
direct[alt] = np.random.uniform()
s += direct[alt]
if s != 0:
for alt in alts:
direct[alt] /= s
tree.priority()
mc.sim_ahptree(tree, None)
tree.priority()
```
| github_jupyter |
# Laboratorio 5
## Datos: _European Union lesbian, gay, bisexual and transgender survey (2012)_
Link a los datos [aquí](https://www.kaggle.com/ruslankl/european-union-lgbt-survey-2012).
### Contexto
La FRA (Agencia de Derechos Fundamentales) realizó una encuesta en línea para identificar cómo las personas lesbianas, gays, bisexuales y transgénero (LGBT) que viven en la Unión Europea y Croacia experimentan el cumplimiento de sus derechos fundamentales. La evidencia producida por la encuesta apoyará el desarrollo de leyes y políticas más efectivas para combatir la discriminación, la violencia y el acoso, mejorando la igualdad de trato en toda la sociedad. La necesidad de una encuesta de este tipo en toda la UE se hizo evidente después de la publicación en 2009 del primer informe de la FRA sobre la homofobia y la discriminación por motivos de orientación sexual o identidad de género, que destacó la ausencia de datos comparables. La Comisión Europea solicitó a FRA que recopilara datos comparables en toda la UE sobre este tema. FRA organizó la recopilación de datos en forma de una encuesta en línea que abarca todos los Estados miembros de la UE y Croacia. Los encuestados eran personas mayores de 18 años, que se identifican como lesbianas, homosexuales, bisexuales o transgénero, de forma anónima. La encuesta se hizo disponible en línea, de abril a julio de 2012, en los 23 idiomas oficiales de la UE (excepto irlandés) más catalán, croata, luxemburgués, ruso y turco. En total, 93,079 personas LGBT completaron la encuesta. Los expertos internos de FRA diseñaron la encuesta que fue implementada por Gallup, uno de los líderes del mercado en encuestas a gran escala. Además, organizaciones de la sociedad civil como ILGA-Europa (Región Europea de la Asociación Internacional de Lesbianas, Gays, Bisexuales, Trans e Intersexuales) y Transgender Europe (TGEU) brindaron asesoramiento sobre cómo acercarse mejor a las personas LGBT.
Puede encontrar más información sobre la metodología de la encuesta en el [__Informe técnico de la encuesta LGBT de la UE. Metodología, encuesta en línea, cuestionario y muestra__](https://fra.europa.eu/sites/default/files/eu-lgbt-survey-technical-report_en.pdf).
### Contenido
El conjunto de datos consta de 5 archivos .csv que representan 5 bloques de preguntas: vida cotidiana, discriminación, violencia y acoso, conciencia de los derechos, preguntas específicas de personas transgénero.
El esquema de todas las tablas es idéntico:
* `CountryCode` - name of the country
* `subset` - Lesbian, Gay, Bisexual women, Bisexual men or Transgender (for Transgender Specific Questions table the value is only Transgender)
* `question_code` - unique code ID for the question
* `question_label` - full question text
* `answer` - answer given
* `percentage`
* `notes` - [0]: small sample size; [1]: NA due to small sample size; [2]: missing value
En el laboratorio de hoy solo utilizaremos los relacionados a la vida cotidiana, disponibles en el archivo `LGBT_Survey_DailyLife.csv` dentro de la carpeta `data`.
```
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
daily_life_raw = pd.read_csv(os.path.join("..", "data", "LGBT_Survey_DailyLife.csv"))
daily_life_raw.head()
daily_life_raw.info()
daily_life_raw.describe(include="all").T
questions = (
daily_life_raw.loc[: , ["question_code", "question_label"]]
.drop_duplicates()
.set_index("question_code")
.squeeze()
)
for idx, value in questions.items():
print(f"Question code {idx}:\n\n{value}\n\n")
```
### Preprocesamiento de datos
¿Te fijaste que la columna `percentage` no es numérica? Eso es por los registros con notes `[1]`, por lo que los eliminaremos.
```
daily_life_raw.notes.unique()
daily_life = (
daily_life_raw.query("notes != ' [1] '")
.astype({"percentage": "int"})
.drop(columns=["question_label", "notes"])
.rename(columns={"CountryCode": "country"})
)
daily_life.head()
```
## Ejercicio 1
(1 pto)
¿A qué tipo de dato (nominal, ordinal, discreto, continuo) corresponde cada columna del DataFrame `daily_life`?
Recomendación, mira los valores únicos de cada columna.
```
daily_life.dtypes
# FREE STYLE #
```
__Respuesta:__
* `country`:
* `subset`:
* `question_code`:
* `answer`:
* `percentage`:
## Ejercicio 2
(1 pto)
Crea un nuevo dataframe `df1` tal que solo posea registros de Bélgica, la pregunta con código `b1_b` y que hayan respondido _Very widespread_.
Ahora, crea un gráfico de barras vertical con la función `bar` de `matplotlib` para mostrar el porcentaje de respuestas por cada grupo. La figura debe ser de tamaño 10 x 6 y el color de las barras verde.
```
print(f"Question b1_b:\n\n{questions['b1_b']}")
df1 = # FIX ME #
df1
x = # FIX ME #
y = # FIX ME #
fig = plt.figure(# FIX ME #)
plt# FIX ME #
plt.show()
```
## Ejercicio 3
(1 pto)
Respecto a la pregunta con código `g5`, ¿Cuál es el porcentaje promedio por cada valor de la respuesta (notar que la respuestas a las preguntas son numéricas)?
```
print(f"Question g5:\n\n{questions['g5']}")
```
Crea un DataFrame llamado `df2` tal que:
1. Solo sean registros con la pregunta con código `g5`
2. Cambia el tipo de la columna `answer` a `int`.
3. Agrupa por país y respuesta y calcula el promedio a la columna porcentaje (usa `agg`).
4. Resetea los índices.
```
df2 = (
# FIX ME #
)
df2
```
Crea un DataFrame llamado `df2_mean` tal que:
1. Agrupa `df2` por respuesta y calcula el promedio del porcentaje.
2. Resetea los índices.
```
df2_mean = df2.# FIX ME #
df2_mean.head()
```
Ahora, grafica lo siguiente:
1. Una figura con dos columnas, tamaño de figura 15 x 12 y que compartan eje x y eje y. Usar `plt.subplots`.
2. Para el primer _Axe_ (`ax1`), haz un _scatter plot_ tal que el eje x sea los valores de respuestas de `df2`, y el eye y corresponda a los porcentajes de `df2`. Recuerda que en este caso corresponde a promedios por país, por lo que habrán más de 10 puntos en el gráfico..
3. Para el segundo _Axe_ (`ax2`), haz un gráfico de barras horizontal tal que el eje x sea los valores de respuestas de `df2_mean`, y el eye y corresponda a los porcentajes de `df2_mean`.
```
x = # FIX ME #
y = # FIX ME #
x_mean = # FIX ME #s
y_mean = # FIX ME #
fig, (ax1, ax2) = plt.subplots(# FIX ME #)
ax1.# FIX ME #
ax1.grid(alpha=0.3)
ax2.# FIX ME #
ax2.grid(alpha=0.3)
fig.show()
```
## Ejercicio 4
(1 pto)
Respecto a la misma pregunta `g5`, cómo se distribuyen los porcentajes en promedio para cada país - grupo?
Utilizaremos el mapa de calor presentado en la clase, para ello es necesario procesar un poco los datos para conformar los elementos que se necesitan.
Crea un DataFrame llamado `df3` tal que:
1. Solo sean registros con la pregunta con código `g5`
2. Cambia el tipo de la columna `answer` a `int`.
3. Agrupa por país y subset, luego calcula el promedio a la columna porcentaje (usa `agg`).
4. Resetea los índices.
5. Pivotea tal que los índices sean los países, las columnas los grupos y los valores el promedio de porcentajes.
6. Llena los valores nulos con cero. Usa `fillna`.
```
## Code from:
# https://matplotlib.org/3.1.1/gallery/images_contours_and_fields/image_annotated_heatmap.html#sphx-glr-gallery-images-contours-and-fields-image-annotated-heatmap-py
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
def heatmap(data, row_labels, col_labels, ax=None,
cbar_kw={}, cbarlabel="", **kwargs):
"""
Create a heatmap from a numpy array and two lists of labels.
Parameters
----------
data
A 2D numpy array of shape (N, M).
row_labels
A list or array of length N with the labels for the rows.
col_labels
A list or array of length M with the labels for the columns.
ax
A `matplotlib.axes.Axes` instance to which the heatmap is plotted. If
not provided, use current axes or create a new one. Optional.
cbar_kw
A dictionary with arguments to `matplotlib.Figure.colorbar`. Optional.
cbarlabel
The label for the colorbar. Optional.
**kwargs
All other arguments are forwarded to `imshow`.
"""
if not ax:
ax = plt.gca()
# Plot the heatmap
im = ax.imshow(data, **kwargs)
# Create colorbar
cbar = ax.figure.colorbar(im, ax=ax, **cbar_kw)
cbar.ax.set_ylabel(cbarlabel, rotation=-90, va="bottom")
# We want to show all ticks...
ax.set_xticks(np.arange(data.shape[1]))
ax.set_yticks(np.arange(data.shape[0]))
# ... and label them with the respective list entries.
ax.set_xticklabels(col_labels)
ax.set_yticklabels(row_labels)
# Let the horizontal axes labeling appear on top.
ax.tick_params(top=True, bottom=False,
labeltop=True, labelbottom=False)
# Rotate the tick labels and set their alignment.
plt.setp(ax.get_xticklabels(), rotation=-30, ha="right",
rotation_mode="anchor")
# Turn spines off and create white grid.
for edge, spine in ax.spines.items():
spine.set_visible(False)
ax.set_xticks(np.arange(data.shape[1]+1)-.5, minor=True)
ax.set_yticks(np.arange(data.shape[0]+1)-.5, minor=True)
ax.grid(which="minor", color="w", linestyle='-', linewidth=3)
ax.tick_params(which="minor", bottom=False, left=False)
return im, cbar
def annotate_heatmap(im, data=None, valfmt="{x:.2f}",
textcolors=["black", "white"],
threshold=None, **textkw):
"""
A function to annotate a heatmap.
Parameters
----------
im
The AxesImage to be labeled.
data
Data used to annotate. If None, the image's data is used. Optional.
valfmt
The format of the annotations inside the heatmap. This should either
use the string format method, e.g. "$ {x:.2f}", or be a
`matplotlib.ticker.Formatter`. Optional.
textcolors
A list or array of two color specifications. The first is used for
values below a threshold, the second for those above. Optional.
threshold
Value in data units according to which the colors from textcolors are
applied. If None (the default) uses the middle of the colormap as
separation. Optional.
**kwargs
All other arguments are forwarded to each call to `text` used to create
the text labels.
"""
if not isinstance(data, (list, np.ndarray)):
data = im.get_array()
# Normalize the threshold to the images color range.
if threshold is not None:
threshold = im.norm(threshold)
else:
threshold = im.norm(data.max())/2.
# Set default alignment to center, but allow it to be
# overwritten by textkw.
kw = dict(horizontalalignment="center",
verticalalignment="center")
kw.update(textkw)
# Get the formatter in case a string is supplied
if isinstance(valfmt, str):
valfmt = matplotlib.ticker.StrMethodFormatter(valfmt)
# Loop over the data and create a `Text` for each "pixel".
# Change the text's color depending on the data.
texts = []
for i in range(data.shape[0]):
for j in range(data.shape[1]):
kw.update(color=textcolors[int(im.norm(data[i, j]) > threshold)])
text = im.axes.text(j, i, valfmt(data[i, j], None), **kw)
texts.append(text)
return texts
df3 = (
# FIX ME #
)
df3.head()
```
Finalmente, los ingredientes para el heat map son:
```
countries = df3.index.tolist()
subsets = df3.columns.tolist()
answers = df3.values
```
El mapa de calor debe ser de la siguiente manera:
* Tamaño figura: 15 x 20
* cmap = "YlGn"
* cbarlabel = "Porcentaje promedio (%)"
* Precición en las anotaciones: Flotante con dos decimales.
```
fig, ax = plt.subplots(# FIX ME #)
im, cbar = heatmap(# FIX ME #")
texts = annotate_heatmap(# FIX ME #)
fig.tight_layout()
plt.show()
```
| github_jupyter |
# Talktorial 1
# Compound data acquisition (ChEMBL)
#### Developed in the CADD seminars 2017 and 2018, AG Volkamer, Charité/FU Berlin
Paula Junge and Svetlana Leng
## Aim of this talktorial
We learn how to extract data from ChEMBL:
* Find ligands which were tested on a certain target
* Filter by available bioactivity data
* Calculate pIC50 values
* Merge dataframes and draw extracted molecules
## Learning goals
### Theory
* ChEMBL database
* ChEMBL web services
* ChEMBL webresource client
* Compound activity measures
* IC50
* pIC50
### Practical
Goal: Get list of compounds with bioactivity data for a given target
* Connect to ChEMBL database
* Get target data (EGFR kinase)
* Bioactivity data
* Download and filter bioactivities
* Clean and convert
* Compound data
* Get list of compounds
* Prepare output data
* Output
* Draw molecules with highest pIC50
* Write output file
## References
* ChEMBL bioactivity database (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5210557/)
* ChEMBL web services: <i>Nucleic Acids Res.</i> (2015), <b>43</b>, 612-620 (https://academic.oup.com/nar/article/43/W1/W612/2467881)
* ChEMBL webrescource client GitHub (https://github.com/chembl/chembl_webresource_client)
* myChEMBL webservices version 2.x (https://github.com/chembl/mychembl/blob/master/ipython_notebooks/09_myChEMBL_web_services.ipynb)
* ChEMBL web-interface (https://www.ebi.ac.uk/chembl/)
* EBI-RDF platform (https://www.ncbi.nlm.nih.gov/pubmed/24413672)
* IC50 and pIC50 (https://en.wikipedia.org/wiki/IC50)
* UniProt website (https://www.uniprot.org/)
_____________________________________________________________________________________________________________________
## Theory
### ChEMBL database
* Open large-scale bioactivity database
* **Current data content (as of 10.2018):**
* \>1.8 million distinct compound structures
* \>15 million activity values from 1 million assays
* Assays are mapped to ∼12 000 targets
* **Data sources** include scientific literature, PubChem bioassays, Drugs for Neglected Diseases Initiative (DNDi), BindingDB database, ...
* ChEMBL data can be accessed via a [web-interface](https://www.ebi.ac.uk/chembl/), the [EBI-RDF platform](https://www.ncbi.nlm.nih.gov/pubmed/24413672) and the [ChEMBL web services](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4489243/#B5)
#### ChEMBL web services
* RESTful web service
* ChEMBL web service version 2.x resource schema:
[](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4489243/figure/F2/)
*Figure 1:*
"ChEMBL web service schema diagram. The oval shapes represent ChEMBL web service resources and the line between two resources indicates that they share a common attribute. The arrow direction shows where the primary information about a resource type can be found. A dashed line indicates the relationship between two resources behaves differently. For example, the `Image` resource provides a graphical based representation of a `Molecule`."
Figure and description taken from: [<i>Nucleic Acids Res.</i> (2015), <b>43</b>, 612-620](https://academic.oup.com/nar/article/43/W1/W612/2467881).
#### ChEMBL webresource client
* Python client library for accessing ChEMBL data
* Handles interaction with the HTTPS protocol
* Lazy evaluation of results -> reduced number of network requests
### Compound activity measures
#### IC50
* [Half maximal inhibitory concentration](https://en.wikipedia.org/wiki/IC50)
* Indicates how much of a particular drug or other substance is needed to inhibit a given biological process by half
[<img src="https://upload.wikimedia.org/wikipedia/commons/8/81/Example_IC50_curve_demonstrating_visually_how_IC50_is_derived.png" width="450" align="center" >](https://commons.wikimedia.org/wiki/File:Example_IC50_curve_demonstrating_visually_how_IC50_is_derived.png)
*Figure 2:* Visual demonstration of how to derive an IC50 value: Arrange data with inhibition on vertical axis and log(concentration) on horizontal axis; then identify max and min inhibition; then the IC50 is the concentration at which the curve passes through the 50% inhibition level.
#### pIC50
* To facilitate the comparison of IC50 values, we define pIC50 values on a logarithmic scale, such that <br />
$ pIC_{50} = -log_{10}(IC_{50}) $ where $ IC_{50}$ is specified in units of M.
* Higher pIC50 values indicate exponentially greater potency of the drug
* pIC50 is given in terms of molar concentration (mol/L or M) <br />
* IC50 should be specified in M to convert to pIC50
* For nM: $pIC_{50} = -log_{10}(IC_{50}*10^{-9})= 9-log_{10}(IC_{50}) $
Besides, IC50 and pIC50, other bioactivity measures are used, such as the equilibrium constant [KI](https://en.wikipedia.org/wiki/Equilibrium_constant) and the half maximal effective concentration [EC50](https://en.wikipedia.org/wiki/EC50).
## Practical
In the following, we want to download all molecules that have been tested against our target of interest, the EGFR kinase.
### Connect to ChEMBL database
First, the ChEMBL webresource client as well as other python libraries are imported.
```
from chembl_webresource_client.new_client import new_client
import pandas as pd
import math
from rdkit.Chem import PandasTools
```
Create resource objects for API access.
```
targets = new_client.target
compounds = new_client.molecule
bioactivities = new_client.activity
```
## Target data
* Get UniProt-ID (http://www.uniprot.org/uniprot/P00533) of the target of interest (EGFR kinase) from UniProt website (https://www.uniprot.org/)
* Use UniProt-ID to get target information
* Select a different UniProt-ID if you are interested into another target
```
uniprot_id = 'P00533'
# Get target information from ChEMBL but restrict to specified values only
target_P00533 = targets.get(target_components__accession=uniprot_id) \
.only('target_chembl_id', 'organism', 'pref_name', 'target_type')
print(type(target_P00533))
pd.DataFrame.from_records(target_P00533)
```
### After checking the entries, we select the first entry as our target of interest
`CHEMBL203`: It is a single protein and represents the human Epidermal growth factor receptor (EGFR, also named erbB1)
```
target = target_P00533[0]
target
```
Save selected ChEMBL-ID.
```
chembl_id = target['target_chembl_id']
chembl_id
```
### Bioactivity data
Now, we want to query bioactivity data for the target of interest.
#### Download and filter bioactivities for the target
In this step, we download and filter the bioactivity data and only consider
* human proteins
* bioactivity type IC50
* exact measurements (relation '=')
* binding data (assay type 'B')
```
bioact = bioactivities.filter(target_chembl_id = chembl_id) \
.filter(type = 'IC50') \
.filter(relation = '=') \
.filter(assay_type = 'B') \
.only('activity_id','assay_chembl_id', 'assay_description', 'assay_type', \
'molecule_chembl_id', 'type', 'units', 'relation', 'value', \
'target_chembl_id', 'target_organism')
len(bioact), len(bioact[0]), type(bioact), type(bioact[0])
```
If you experience difficulties to query the ChEMBL database, we provide here a file containing the results for the query in the previous cell (11 April 2019). We do this using the Python package pickle which serializes Python objects so they can be saved to a file, and loaded in a program again later on.
(Learn more about object serialization on [DataCamp](https://www.datacamp.com/community/tutorials/pickle-python-tutorial))
You can load the "pickled" compounds by uncommenting and running the next cell.
```
#import pickle
#bioact = pickle.load(open("../data/T1/EGFR_compounds_from_chembl_query_20190411.p", "rb"))
```
#### Clean and convert bioactivity data
The data is stored as a list of dictionaries
```
bioact[0]
```
Convert to pandas dataframe (this might take some minutes).
```
bioact_df = pd.DataFrame.from_records(bioact)
bioact_df.head(10)
bioact_df.shape
```
Delete entries with missing values.
```
bioact_df = bioact_df.dropna(axis=0, how = 'any')
bioact_df.shape
```
Delete duplicates:
Sometimes the same molecule (`molecule_chembl_id`) has been tested more than once, in this case, we only keep the first one.
```
bioact_df = bioact_df.drop_duplicates('molecule_chembl_id', keep = 'first')
bioact_df.shape
```
We would like to only keep bioactivity data measured in molar units. The following print statements will help us to see what units are contained and to control what is kept after dropping some rows.
```
print(bioact_df.units.unique())
bioact_df = bioact_df.drop(bioact_df.index[~bioact_df.units.str.contains('M')])
print(bioact_df.units.unique())
bioact_df.shape
```
Since we deleted some rows, but we want to iterate over the index later, we reset index to be continuous.
```
bioact_df = bioact_df.reset_index(drop=True)
bioact_df.head()
```
To allow further comparison of the IC50 values, we convert all units to nM. First, we write a helper function, which can be applied to the whole dataframe in the next step.
```
def convert_to_NM(unit, bioactivity):
# c=0
# for i, unit in enumerate(bioact_df.units):
if unit != "nM":
if unit == "pM":
value = float(bioactivity)/1000
elif unit == "10'-11M":
value = float(bioactivity)/100
elif unit == "10'-10M":
value = float(bioactivity)/10
elif unit == "10'-8M":
value = float(bioactivity)*10
elif unit == "10'-1microM" or unit == "10'-7M":
value = float(bioactivity)*100
elif unit == "uM" or unit == "/uM" or unit == "10'-6M":
value = float(bioactivity)*1000
elif unit == "10'1 uM":
value = float(bioactivity)*10000
elif unit == "10'2 uM":
value = float(bioactivity)*100000
elif unit == "mM":
value = float(bioactivity)*1000000
elif unit == "M":
value = float(bioactivity)*1000000000
else:
print ('unit not recognized...', unit)
return value
else: return bioactivity
bioactivity_nM = []
for i, row in bioact_df.iterrows():
bioact_nM = convert_to_NM(row['units'], row['value'])
bioactivity_nM.append(bioact_nM)
bioact_df['value'] = bioactivity_nM
bioact_df['units'] = 'nM'
bioact_df.head()
```
### Compound data
We have a data frame containing all molecules tested (with the respective measure) against EGFR. Now, we want to get the molecules that are stored behind the respective ChEMBL IDs.
#### Get list of compounds
Let's have a look at the compounds from ChEMBL we have defined bioactivity data for. First, we retrieve ChEMBL ID and structures for the compounds with desired bioactivity data.
```
cmpd_id_list = list(bioact_df['molecule_chembl_id'])
compound_list = compounds.filter(molecule_chembl_id__in = cmpd_id_list) \
.only('molecule_chembl_id','molecule_structures')
```
Then, we convert the list to a pandas dataframe and delete duplicates (again, the pandas from_records function might take some time).
```
compound_df = pd.DataFrame.from_records(compound_list)
compound_df = compound_df.drop_duplicates('molecule_chembl_id', keep = 'first')
print(compound_df.shape)
print(bioact_df.shape)
compound_df.head()
```
So far, we have multiple different molecular structure representations. We only want to keep the canonical SMILES.
```
for i, cmpd in compound_df.iterrows():
if compound_df.loc[i]['molecule_structures'] != None:
compound_df.loc[i]['molecule_structures'] = cmpd['molecule_structures']['canonical_smiles']
print (compound_df.shape)
```
#### Prepare output data
Merge values of interest in one dataframe on ChEMBL-IDs:
* ChEMBL-IDs
* SMILES
* units
* IC50
```
output_df = pd.merge(bioact_df[['molecule_chembl_id','units','value']], compound_df, on='molecule_chembl_id')
print(output_df.shape)
output_df.head()
```
For distinct column names, we rename IC50 and SMILES columns.
```
output_df = output_df.rename(columns= {'molecule_structures':'smiles', 'value':'IC50'})
output_df.shape
```
If we do not have a SMILES representation of a compound, we can not further use it in the following talktorials. Therefore, we delete compounds without SMILES column.
```
output_df = output_df[~output_df['smiles'].isnull()]
print(output_df.shape)
output_df.head()
```
In the next cell, you see that the low IC50 values are difficult to read. Therefore, we prefer to convert the IC50 values to pIC50.
```
output_df = output_df.reset_index(drop=True)
ic50 = output_df.IC50.astype(float)
print(len(ic50))
print(ic50.head(10))
# Convert IC50 to pIC50 and add pIC50 column:
pIC50 = pd.Series()
i = 0
while i < len(output_df.IC50):
value = 9 - math.log10(ic50[i]) # pIC50=-log10(IC50 mol/l) --> for nM: -log10(IC50*10**-9)= 9-log10(IC50)
if value < 0:
print("Negative pIC50 value at index"+str(i))
pIC50.at[i] = value
i += 1
output_df['pIC50'] = pIC50
output_df.head()
```
### Collected bioactivity data for EGFR
Let's have a look at our collected data set.
#### Draw molecules
In the next steps, we add a molecule column to our datafame and look at the structures of the molecules with the highest pIC50 values.
```
PandasTools.AddMoleculeColumnToFrame(output_df, smilesCol='smiles')
```
Sort molecules by pIC50.
```
output_df.sort_values(by="pIC50", ascending=False, inplace=True)
output_df.reset_index(drop=True, inplace=True)
```
Show the most active molecules = molecules with the highest pIC50 values.
```
output_df.drop("smiles", axis=1).head()
```
#### Write output file
To use the data for the following talktorials, we save the data as csv file. Note that it is advisable to drop the molecule column (only contains an image of the molecules) when saving the data.
```
output_df.drop("ROMol", axis=1).to_csv("../data/T1/EGFR_compounds.csv")
```
## Discussion
In this tutorial, we collected all available bioactivity data for our target of interest from the ChEMBL database. We filtered the data set to only contain molecules with measured IC50 or pIC50 bioactivity values.
Be aware that ChEMBL data originates from various sources. Compound data has been generated in different labs by different people all over the world. Therefore, we have to be cautious with the predictions we make using this dataset. It is always important to consider the source of the data and consistency of data production assays when interpreting the results and determining how much confidence we have in our predictions.
In the next tutorials we will filter our acquired data by the Lipinski rule of five and by unwanted substructures. Another important step would be to clean the data and remove duplicates. As this is not shown in any of our talktorials (yet), we would like to refer to the standardiser library ([github Francis Atkinson](https://github.com/flatkinson/standardiser)) or [MolVS](https://molvs.readthedocs.io/en/latest/) as possible tools for this task.
## Quiz
* We have downloaded in this talktorial molecules and bioactivity data from ChEMBL. What else is the ChEMBL database useful for?
* What is the difference between IC50 and EC50?
* What can we use the data extracted from ChEMBL for?
| github_jupyter |
<a href="https://colab.research.google.com/github/BreakoutMentors/Data-Science-and-Machine-Learning/blob/main/machine_learning/lesson%204%20-%20ML%20Apps/Gradio/EMNIST_Gradio_Tutorial.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Making ML Applications with Gradio
[Gradio](https://www.gradio.app/) is a python library that provides web interfaces for your models. This library is very high-level with it being the easiest to learn for beginners. Here we use a dataset called [EMNIST](https://pytorch.org/vision/stable/datasets.html#emnist) which is an addition to the MNIST(dataset of images with numbers) datasest, by including images of capital and lowercase letters with a total of 62 classes.
Using Gradio, an interface is created at the bottom using the model trained in this notebook to accept our drawings of images or numbers to then predict.
## Importing libraries and Installing Gradio using PIP
Google does not have Gradio automatically installed on their Google Colab machines, so it is necessary to install it to the specific machine you are using right now. If you choose another runtime machine, it is necessary to repeat this step.
**Also, please run this code with a GPU**
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# Importing PyTorch
import torch
import torch.nn as nn
# Importing torchvision for dataset
import torchvision
import torchvision.transforms as transforms
# Installing gradio using PIP
!pip install gradio
```
## Downloading and Preparing EMNIST Dataset
**Note:** Even though the images in the EMNIST dataset are 28x28 images just like the regular MNIST dataset, there are necessary transforms needed for EMNIST dataset. If not transformed, the images are rotated 90° counter-clockwise and are flipped vertically. To undo these two issues, we first rotate it 90° counter-clockwise and then flip it horizontally
Here is the image before processing:
<img src="https://raw.githubusercontent.com/BreakoutMentors/Data-Science-and-Machine-Learning/main/machine_learning/lesson%204%20-%20ML%20Apps/images/image_before_processing.jpg" width=200>
Here is the image after processing:
<img src="https://github.com/BreakoutMentors/Data-Science-and-Machine-Learning/blob/main/machine_learning/lesson%204%20-%20ML%20Apps/images/image_after_processing.jpg?raw=true" width=200>
```
# Getting Dataset
!mkdir EMNIST
root = '/content/EMNIST'
# Creating Transforms
transforms = transforms.Compose([
# Rotating image 90 degrees counter-clockwise
transforms.RandomRotation((-90,-90)),
# Flipping images horizontally
transforms.RandomHorizontalFlip(p=1),
# Converting images to tensor
transforms.ToTensor()
])
# Getting dataset
training_dataset = torchvision.datasets.EMNIST(root,
split='byclass',
train=True,
download=True,
transform=transforms)
test_dataset = torchvision.datasets.EMNIST(root,
split='byclass',
train=False,
download=True,
transform=transforms)
# Loading Dataset into dataloaders
batch_size = 2048
training_dataloader = torch.utils.data.DataLoader(training_dataset, batch_size=batch_size, shuffle=True)
test_dataloader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size, shuffle=False)
# Getting shapes of dataset
print('Shape of the training dataset:', training_dataset.data.shape)
print('Shape of the test dataset:', test_dataset.data.shape)
# Getting reverted class_to_idx dictionary to get classes by idx
idx_to_class = {val:key for key, val in training_dataset.class_to_idx.items()}
# Plotting 5 images with classes
plt.figure(figsize=(10,2))
for i in range(5):
plt.subplot(1,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(training_dataset[i][0].squeeze().numpy(), cmap=plt.cm.binary)
plt.xlabel(idx_to_class[training_dataset[i][1]])
```
## Building the Model
```
class Neural_Network(nn.Module):
# Constructor
def __init__(self, num_classes):
super(Neural_Network, self).__init__()
# Defining Fully-Connected Layers
self.fc1 = nn.Linear(28*28, 392) # 28*28 since each image is 28*28
self.fc2 = nn.Linear(392, 196)
self.fc3 = nn.Linear(196, 98)
self.fc4 = nn.Linear(98, num_classes)
# Activation function
self.relu = nn.ReLU()
def forward(self, x):
# Need to flatten each image in the batch
x = x.flatten(start_dim=1)
# Input it into the Fully connected layers
x = self.relu(self.fc1(x))
x = self.relu(self.fc2(x))
x = self.relu(self.fc3(x))
x = self.fc4(x)
return x
# Getting number of classes
num_classes = len(idx_to_class)
model = Neural_Network(num_classes)
print(model)
```
## Defining Loss Function and Optimizer
```
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
```
## Moving model to GPU
If you have not changed the runtime type to a GPU, please do so now. This helps with the speed of training.
```
# Use GPU if available
device = "cuda" if torch.cuda.is_available() else "cpu"
# Moving model to use GPU
model.to(device)
```
## Training the Model
```
# Function that returns a torch tensor with predictions to compare with labels
def get_preds_from_logits(logits):
# Using softmax to get an array that sums to 1, and then getting the index with the highest value
return torch.nn.functional.softmax(logits, dim=1).argmax(dim=1)
epochs = 10
train_losses = []
train_accuracies = []
for epoch in range(1, epochs+1):
train_loss = 0.0
train_counts = 0
###################
# train the model #
###################
# Setting model to train mode
model.train()
for images, labels in training_dataloader:
# Moving data to GPU if available
images, labels = images.to(device), labels.to(device)
# Setting all gradients to zero
optimizer.zero_grad()
# Calculate Output
output = model(images)
# Calculate Loss
loss = criterion(output, labels)
# Calculate Gradients
loss.backward()
# Perform Gradient Descent Step
optimizer.step()
# Saving loss
train_loss += loss.item()
# Get Predictions
train_preds = get_preds_from_logits(output)
# Saving number of right predictions for accuracy
train_counts += train_preds.eq(labels).sum().item()
# Averaging and Saving Losses
train_loss/=len(training_dataset)
train_losses.append(train_loss)
# Getting accuracies and saving them
train_acc = train_counts/len(training_dataset)
train_accuracies.append(train_acc)
print('Epoch: {} \tTraining Loss: {:.6f} \tTraining Accuracy: {:.2f}%'.format(epoch, train_loss, train_acc*100))
plt.plot(train_losses)
plt.xlabel('epoch')
plt.ylabel('Mean Squared Error')
plt.title('Training Loss')
plt.show()
plt.plot(train_accuracies)
plt.xlabel('epoch')
plt.ylabel('Accuracy')
plt.title('Training Accuracy')
plt.show()
```
## Evaluating the model
Here we will display the test loss and accuracy and examples of images that were misclassified.
```
test_loss = 0.0
test_counts = 0
# Setting model to evaluation mode, no parameters will change
model.eval()
for images, labels in test_dataloader:
# Moving to GPU if available
images, labels = images.to(device), labels.to(device)
# Calculate Output
output = model(images)
# Calculate Loss
loss = criterion(output, labels)
# Saving loss
test_loss += loss.item()
# Get Predictions
test_preds = get_preds_from_logits(output)
# Saving number of right predictions for accuracy
test_counts += test_preds.eq(labels).sum().item()
# Calculating test accuracy
test_acc = test_counts/len(test_dataset)
print('Test Loss: {:.6f} \tTest Accuracy: {:.2f}%'.format(test_loss, test_acc*100))
import torchvision.transforms as transforms
# Have to another set of transforms to rotate and flip testing data
test_transforms = transforms.Compose([
# Rotating image 90 degrees counter-clockwise
transforms.RandomRotation((-90,-90)),
# Flipping images horizontally
transforms.RandomHorizontalFlip(p=1)
])
# Transforming the data and normalizing them
test_images = test_transforms(test_dataset.data).to(device)/255
# Getting Predictions
predictions = get_preds_from_logits(model(test_images))
# Getting Labels
test_labels = test_dataset.targets.to(device)
# Getting misclassified booleans
correct_bools = test_labels.eq(predictions)
misclassified_indices = []
for i in range(len(correct_bools)):
if correct_bools[i] == False:
misclassified_indices.append(i)
# Plotting 5 misclassified images
plt.figure(figsize=(10,2))
for i in range(5):
idx = misclassified_indices[i]
plt.subplot(1,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(test_images[idx].squeeze().cpu().numpy(), cmap=plt.cm.binary)
true_label = idx_to_class[test_labels[idx].item()]
pred_label = idx_to_class[predictions[idx].item()]
plt.xlabel(f'True: {true_label}, Pred: {pred_label}')
```
# How to use Gradio
There are three parts of using Gradio
1. Define a function that takes input and returns your model's output
2. Define what type of input the interface will use
3. Define what type of output the interface will give
The function `recognize_image` takes a 28x28 image that is not yet normalized and returns a dictionary with the keys being the classes and the values being the probabilities for that class.
The class [`gradio.inputs.Image`](https://www.gradio.app/docs#i_image) is used as the input that provides a window in the Gradio interface, but there are many customizations you can provide.
These are some the parameters:
1. shape - (width, height) shape to crop and resize image to; if None, matches input image size.
2. image_mode - "RGB" if color, or "L" if black and white.
3. invert_colors - whether to invert the image as a preprocessing step.
4. source - Source of image. "upload" creates a box where user can drop an image file, "webcam" allows user to take snapshot from their webcam, "canvas" defaults to a white image that can be edited and drawn upon with tools.
The class [gradio.outputs.Label](https://www.gradio.app/docs#o_label) is used as the output, which provides probabilities to the interface for the purpose of displaying them.
These are the parameters:
1. num_top_classes - number of most confident classes to show.
2. type - Type of value to be passed to component. "value" expects a single out label, "confidences" expects a dictionary mapping labels to confidence scores, "auto" detects return type.
3. label - component name in interface.
The interface class [gradio.Interface](https://www.gradio.app/docs#interface) is responsible of creating the interface that compiles the type of inputs and outputs. There is a `.launch()` method that launches the interface in this notebook after compiling.
These are the parameters used in this interface:
1. fn - the function to wrap an interface around.
2. inputs - a single Gradio input component, or list of Gradio input components. Components can either be passed as instantiated objects, or referred to by their string shortcuts. The number of input components should match the number of parameters in fn.
3. outputs - a single Gradio output component, or list of Gradio output components. Components can either be passed as instantiated objects, or referred to by their string shortcuts. The number of output components should match the number of values returned by fn.
4. title - a title for the interface; if provided, appears above the input and output components.
5. description - a description for the interface; if provided, appears above the input and output components.
6. live - whether the interface should automatically reload on change.
7. interpretation - function that provides interpretation explaining prediction output. Pass "default" to use built-in interpreter.
I will enourage you to view the [documentation](https://www.gradio.app/docs) for the interface, inputs and outputs, you can find all the information you need there. It is helpful to refer to the documentation to understand other parameters that are not used in this lesson.
```
import gradio
import gradio as gr
# Function that returns a torch tensor with predictions to compare with labels
def get_probs_from_logits(logits):
# Using softmax to get probabilities from the logits
return torch.nn.functional.softmax(logits, dim=1)
# Function that takes the img drawn in the Gradio interface, then gives probabilities
def recognize_image(img):
# Normalizes inputted image and converts it to a tensor for the model
img = torch.tensor(img/255, dtype=torch.float).unsqueeze(dim=0).to(device)
# Getting output
output = model(img)
# Getting probabilites of the image
probabilities = get_probs_from_logits(output).flatten()
# Returns a dictionary with the key being the class and val being the probability
probabilities_dict = {idx_to_class[i]:probabilities[i].item() for i in range(num_classes)}
return probabilities_dict
im = gradio.inputs.Image(shape=(28, 28),
image_mode='L',
invert_colors=True,
source="canvas")
title = "Number and Letter Classifier App"
description = """This app is able to guess the number or letter you draw below.
The ML model was trained on the EMNIST dataset, please use below!"""
iface = gr.Interface(fn=recognize_image,
inputs=im,
outputs=gradio.outputs.Label(num_top_classes=5),
title=title,
description=description,
live=True,
interpretation="default")
iface.launch()
```
# What's next?
The next challenge will cover pretrained models, which are models that are already trained for us and gives us the availability of using the model to make predictions automatically. You will create another Gradio app that uses pretrained models to classify images.
| github_jupyter |
# Backprop Core Example: Text Summarisation
Text summarisation takes a chunk of text, and extracts the key information.
```
# Set your API key to do inference on Backprop's platform
# Leave as None to run locally
api_key = None
import backprop
summarisation = backprop.Summarisation(api_key=api_key)
# Change this up.
input_text = """
Britain began its third COVID-19 lockdown on Tuesday with the government calling for one last major national effort to defeat the spread of a virus that has infected an estimated one in 50 citizens before mass vaccinations turn the tide.
Finance minister Rishi Sunak announced a new package of business grants worth 4.6 billion pounds ($6.2 billion) to help keep people in jobs and firms afloat until measures are relaxed gradually, at the earliest from mid-February but likely later.
Britain has been among the countries worst-hit by COVID-19, with the second highest death toll in Europe and an economy that suffered the sharpest contraction of any in the Group of Seven during the first wave of infections last spring.
Prime Minister Boris Johnson said the latest data showed 2% of the population were currently infected - more than a million people in England.
“When everybody looks at the position, people understand overwhelmingly that we have no choice,” he told a news conference.
More than 1.3 million people in Britain have already received their first dose of a COVID-19 vaccination, but this is not enough to have an impact on transmission yet.
Johnson announced the new lockdown late on Monday, saying the highly contagious new coronavirus variant first identified in Britain was spreading so fast the National Health Service risked being overwhelmed within 21 days.
In England alone, some 27,000 people are in hospital with COVID, 40% more than during the first peak in April, with infection numbers expected to rise further after increased socialising during the Christmas period.
Since the start of the pandemic, more than 75,000 people have died in the United Kingdom within 28 days of testing positive for coronavirus, according to official figures. The number of daily new infections passed 60,000 for the first time on Tuesday.
A Savanta-ComRes poll taken just after Johnson’s address suggested four in five adults in England supported the lockdown.
“I definitely think it was the right decision to make,” said Londoner Kaitlin Colucci, 28. “I just hope that everyone doesn’t struggle too much with having to be indoors again.”
Downing Street said Johnson had cancelled a visit to India later this month to focus on the response to the virus, and Buckingham Palace called off its traditional summer garden parties this year.
nder the new rules in England, schools are closed to most pupils, people should work from home if possible, and all hospitality and non-essential shops are closed. Semi-autonomous executives in Scotland, Wales and Northern Ireland have imposed similar measures.
As infection rates soar across Europe, other countries are also clamping down on public life. Germany is set to extend its strict lockdown until the end of the month, and Italy will keep nationwide restrictions in place this weekend while relaxing curbs on weekdays.
Sunak’s latest package of grants adds to the eye-watering 280 billion pounds in UK government support already announced for this financial year to stave off total economic collapse.
The new lockdown is likely to cause the economy to shrink again, though not as much as during the first lockdown last spring. JP Morgan economist Allan Monks said he expected the economy to shrink by 2.5% in the first quarter of 2021 -- compared with almost 20% in the second quarter of 2020.
To end the cycle of lockdowns, the government is pinning its hopes on vaccines. It aims to vaccinate all elderly care home residents and their carers, everyone over the age of 70, all frontline health and social care workers, and everyone who is clinically extremely vulnerable by mid-February.
"""
summary = summarisation(input_text)
print(summary)
```
| github_jupyter |
```
%pylab --no-import-all
%matplotlib inline
import PyDSTool as pdt
ab = np.loadtxt('birdsynth/test/ba_example_ab.dat')
#ab = np.zeros((40000, 2))
ab[:, 0] += np.random.normal(0, 0.01, len(ab))
t_mom = np.linspace(0, len(ab)/44100, len(ab))
inputs = pdt.pointset_to_traj(pdt.Pointset(coorddict={'a': ab[:, 1], 'b':ab[:, 0]}, indepvardict={'t': t_mom}))
```
# Jacobian calculation
```
x = pdt.Var('x')
y = pdt.Var('y')
gm = pdt.Par('gm')
a = pdt.Par('a')
b = pdt.Par('b')
t = pdt.Var('t')
xdot = pdt.Fun(y, [y], 'xdot')
ydot = pdt.Fun(-a*gm*gm - b*gm*gm*x -gm*gm*x*x*x -gm*x*x*y + gm*gm*x*x - gm*x*y, [x, y], 'ydot')
F = pdt.Fun([xdot(y), ydot(x, y)], [x,y], 'F')
jac = pdt.Fun(pdt.Diff(F, [x, y]), [t, x, y], 'Jacobian')
jac.simplify()
print(jac.eval(t=t, x=x, y=y))
```
# Simple model
```
icdict = {'x': 0, 'y': 0}
pardict = {
'gm': 2 # g is γ in Boari 2015
}
vardict = {
'x': xdot(y),
'y': ydot(x,y),
}
args = pdt.args()
args.name = 'birdsynth'
args.fnspecs = [jac, xdot, ydot]
args.ics = icdict
args.pars = pardict
args.inputs = inputs
args.tdata = [0, 1]
args.varspecs = vardict
ds = pdt.Generator.Vode_ODEsystem(args)
ds.haveJacobian()
traj = ds.compute('demo')
plt.plot(traj.sample(dt=1/(44100*20))['x'])
auxdict = {'Pi':(['t', 'x', 'a_'], 'if(t > 0, a_ * x - r * 1, 0)'),
'Pt':(['t', 'x', 'a_'], '(1 - r) * Pi(t - 0.5 * T, x, a_)')
}
icdict = {'x': 0, 'y': 0, 'o1':0, 'i1':0, 'i3':0}
pardict = {'g': 2400, # g is γ in Boari 2015
'T': 0.2,
'r': 0.1,
'a_p': -540e6,
'b_p': -7800,
'c_p': 1.8e8,
'd_p': 1.2e-2,
'e_p': 7.2e-1,
'f_p': -0.83e-2,
'g_p': -5e2,
'h_p': 1e-4
}
vardict = {'x': 'y',
'y': '-a*Pow(g, 2) - b * Pow(g, 2) * x - Pow(g, 2) * Pow(x, 3) - g * Pow(x, 2) * y + Pow(g, 2) * x * x'
'- g * x * y',
'i1': 'o1',
'o1': 'a_p * i1 + b_p * o1 + c_p * i3 + d_p * Pt(t, x, a) + e_p * Pt(t, x, a)',
'i3': 'f_p * o1 + g_p * i3 + h_p * Pt(t, x, a)'
}
args = pdt.args()
args.name = 'birdsynth'
args.ics = icdict
args.pars = pardict
args.fnspecs = auxdict
args.inputs = inputs
args.tdata = [0, len(ab)/44100]
args.varspecs = vardict
ds = pdt.Generator.Vode_ODEsystem(args)
traj = ds.compute('demo')
pts = traj.sample(dt=1/(44100))
plt.plot(pts['t'], pts['x'])
x = ds.variables['x']
y_0 = pdt.Var('-a*Pow(g, 2) - b * Pow(g, 2) * x - Pow(g, 2) * Pow(x, 3) - g * Pow(x, 2) * y + Pow(g, 2) * x * x'
'- g * x * y', 'y_0')
Pi(2)
```
| github_jupyter |
# Bayesian Hierarchical Modeling
This jupyter notebook accompanies the Bayesian Hierarchical Modeling lecture(s) delivered by Stephen Feeney as part of David Hogg's [Computational Data Analysis class](http://dwh.gg/FlatironCDA). As part of the lecture(s) you will be asked to complete a number of tasks, some of which will involve direct coding into the notebook; these sections are marked by task. This notebook requires numpy, matplotlib, scipy, [corner](https://github.com/sfeeney/bhm_lecture.git), [pystan](https://pystan.readthedocs.io/en/latest/getting_started.html) and pickle to run (the last two are required solely for the final task).
The model we're going to be inferring is below.
<img src="bhm_plot.png" alt="drawing" width="500"/>
We start with imports...
```
from __future__ import print_function
# make sure everything we need is installed if running on Google Colab
def is_colab():
try:
cfg = get_ipython().config
if cfg['IPKernelApp']['kernel_class'] == 'google.colab._kernel.Kernel':
return True
else:
return False
except NameError:
return False
if is_colab():
!pip install --quiet numpy matplotlib scipy corner pystan
import numpy as np
import numpy.random as npr
import matplotlib.pyplot as mp
%matplotlib inline
```
... and immediately move to...
## Task 2
In which I ask you to write a Python function to generate a simulated Cepheid sample using the period-luminosity relation $m_{ij} = \mu_i + M^* + s\,\log p_{ij} + \epsilon(\sigma_{\rm int})$. For simplicity, assume Gaussian priors on everything, Gaussian intrinsic scatter and Gaussian measurement uncertainties. Assume only the first host has a distance modulus estimate.
```
# setup
n_gal = 2
n_star = 200
n_samples = 50000
# PL relation parameters
abs_bar = -26.0 # mean of standard absolute magnitude prior
abs_sig = 4.0 # std dev of standard absolute magnitude prior
s_bar = -1.0 # mean of slope prior
s_sig = 1.0 # std dev of slope prior
mu_bar = 30.0 # mean of distance modulus prior
mu_sig = 5.0 # std dev of distance modulus prior
m_sig_int = 0.05 # intrinsic scatter, assumed known
# uncertainties
mu_hat_sig = 0.01 # distance modulus measurement uncertainty
m_hat_sig = 0.02 # apparent magnitude measurement uncertainty
def simulate(n_gal, n_star, abs_bar, abs_sig, s_bar, s_sig, mu_bar, mu_sig, mu_hat_sig, m_sig_int, m_hat_sig):
# draw CPL parameters from Gaussian prior with means abs_bar and s_bar and standard deviations
# abs_sig and s_sig
#abs_true = abs_bar
#s_true = s_bar
abs_true = abs_bar + npr.randn() * abs_sig
s_true = s_bar + npr.randn() * s_sig
# draw n_gal distance moduli from Gaussian prior with mean mu_bar and standard deviation mu_sig
# i've chosen to sort here so the closest galaxy is the one with the measured distance modulus
mu_true = np.sort(mu_bar + npr.randn(n_gal) * mu_sig)
# measure ONLY ONE galaxy's distance modulus noisily. the noise here is assumed Gaussian with
# zero mean and standard deviation mu_hat_sig
mu_hat = mu_true[0] + npr.randn() * mu_hat_sig
# draw log periods. these are assumed to be perfectly observed in this model, so they
# are simply a set of pre-specified numbers. i have chosen to generate new values with
# each simulation, drawn such that log-periods are uniformly drawn in the range 1-2 (i.e.,
# 10 to 100 days). you can have these for free!
lp_true = 1.0 + npr.rand(n_gal, n_star)
# draw true apparent magnitudes. these are distributed around the Cepheid period-luminosity
# relation with Gaussian intrinsic scatter (mean 0, standard deviation m_sig_int)
m_true = np.zeros((n_gal, n_star))
for i in range(n_gal):
m_true[i, :] = mu_true[i] + abs_true + s_true * lp_true[i, :] + npr.randn(n_star) * m_sig_int
# measure the apparent magnitudes noisily, all with the same measurement uncertainty m_hat_sig
m_hat = m_true + npr.randn(n_gal, n_star) * m_hat_sig
# return!
return (abs_true, s_true, mu_true, lp_true, m_true, mu_hat, m_hat)
```
Let's check that the simulation generates something sane. A simple test that the magnitude measurements errors are correctly generated.
```
# simulate
abs_true, s_true, mu_true, lp_true, m_true, mu_hat, m_hat = \
simulate(n_gal, n_star, abs_bar, abs_sig, s_bar, s_sig, mu_bar, mu_sig, mu_hat_sig, m_sig_int, m_hat_sig)
# plot difference between true and observed apparent magnitudes. this should be the
# noise, which is Gaussian distributed with mean zero and std dev m_hat_sig
outs = mp.hist((m_true - m_hat).flatten())
dm_grid = np.linspace(np.min(outs[1]), np.max(outs[1]))
mp.plot(dm_grid, np.exp(-0.5 * (dm_grid/m_hat_sig) ** 2) * np.max(outs[0]))
mp.xlabel(r'$m_{ij} - \hat{m}_{ij}$')
mp.ylabel(r'$N \left(m_{ij} - \hat{m}_{ij}\right)$')
```
And another test that the intrinsic scatter is added as expected.
```
# plot difference between true apparent magnitudes and expected apparent
# magnitude given a perfect (i.e., intrinsic-scatter-free) period-luminosity
# relation. this should be the intrinsic scatter, which is Gaussian-
# distributed with mean zero and std dev m_sig_int
eps = np.zeros((n_gal, n_star))
for i in range(n_gal):
eps[i, :] = mu_true[i] + abs_true + s_true * lp_true[i, :] - m_true[i, :]
outs = mp.hist(eps.flatten())
dm_grid = np.linspace(np.min(outs[1]), np.max(outs[1]))
mp.plot(dm_grid, np.exp(-0.5 * (dm_grid/m_sig_int) ** 2) * np.max(outs[0]))
mp.xlabel(r'$m_{ij} - \hat{m}_{ij}$')
mp.ylabel(r'$N \left(m_{ij} - \hat{m}_{ij}\right)$')
```
## Generalized Least Squares Demo
Coding up the [GLS estimator](https://en.wikipedia.org/wiki/Generalized_least_squares) is a little involved, so I've done it for you below. Note that, rather unhelpfully, I've done so in a different order than in the notes. When I get a chance I will re-write. For now, you can simply evaluate the cells and bask in the glory of the fastest inference you will ever do!
```
def gls_fit(n_gal, n_star, mu_hat, mu_hat_sig, m_hat, m_sig_int, m_hat_sig, \
lp_true, priors=None):
# setup
# n_obs is one anchor constraint and one magnitude per Cepheid.
# n_par is one mu per Cepheid host and 2 CPL params. if priors
# are used, we add on n_gal + 2 observations: one prior constraint
# on each host distance modulus and CPL parameter
n_obs = n_gal * n_star + 1
n_par = n_gal + 2
if priors is not None:
n_obs += n_gal + 2
data = np.zeros(n_obs)
design = np.zeros((n_obs, n_par))
cov_inv = np.zeros((n_obs, n_obs))
# anchor
data[0] = mu_hat
design[0, 0] = 1.0
cov_inv[0, 0] = 1.0 / mu_hat_sig ** 2
# Cepheids
k = 1
for i in range(0, n_gal):
for j in range(0, n_star):
data[k] = m_hat[i, j]
design[k, i] = 1.0
design[k, n_gal] = 1.0
design[k, n_gal + 1] = lp_true[i, j]
cov_inv[k, k] = 1.0 / (m_hat_sig ** 2 + m_sig_int ** 2)
k += 1
# and, finally, priors if desired
if priors is not None:
abs_bar, abs_sig, s_bar, s_sig, mu_bar, mu_sig = priors
for i in range(n_gal):
data[k] = mu_bar
design[k, i] = 1.0
cov_inv[k, k] = 1.0 / mu_sig ** 2
k += 1
data[k] = abs_bar
design[k, n_gal] = 1.0
cov_inv[k, k] = 1.0 / abs_sig ** 2
k += 1
data[k] = s_bar
design[k, n_gal + 1] = 1.0
cov_inv[k, k] = 1.0 / s_sig ** 2
k += 1
# fit and return
destci = np.dot(design.transpose(), cov_inv)
pars_cov = np.linalg.inv(np.dot(destci, design))
pars = np.dot(np.dot(pars_cov, destci), data)
res = data - np.dot(design, pars)
dof = n_obs - n_par
chisq_dof = np.dot(res.transpose(), np.dot(cov_inv, res))
return pars, pars_cov, chisq_dof
gls_pars, gls_pars_cov, gls_chisq = gls_fit(n_gal, n_star, mu_hat, mu_hat_sig, m_hat, \
m_sig_int, m_hat_sig, lp_true, \
priors=[abs_bar, abs_sig, s_bar, s_sig, mu_bar, mu_sig])
```
In order to plot the outputs of the GLS fit we could draw a large number of samples from the resulting multivariate Gaussian posterior and pass them to something like [`corner`](https://corner.readthedocs.io/en/latest/); however, as we have analytic results we might as well use those directly. I've coded up something totally hacky here in order to do so. Information on how to draw confidence ellipses can be found in [Dan Coe's note](https://arxiv.org/pdf/0906.4123.pdf).
```
# this is a hacky function designed to transform the analytic GLS outputs
# into a corner.py style triangle plot, containing 1D and 2D marginalized
# posteriors
import scipy.stats as sps
import matplotlib.patches as mpp
def schmorner(par_mean, par_cov, par_true, par_label):
# setup
par_std = np.sqrt(np.diag(par_cov))
x_min = par_mean[0] - 3.5 * par_std[0]
x_max = par_mean[0] + 3.5 * par_std[0]
y_min = par_mean[1] - 3.5 * par_std[1]
y_max = par_mean[1] + 3.5 * par_std[1]
fig, axes = mp.subplots(2, 2)
# 1D marge
x = np.linspace(x_min, x_max, 100)
axes[0, 0].plot(x, sps.norm.pdf(x, par_mean[0], par_std[0]), 'k')
axes[0, 0].axvline(par_true[0])
axes[1, 0].axvline(par_true[0])
axes[0, 0].set_xticklabels([])
axes[0, 0].set_yticklabels([])
axes[0, 0].set_xlim(x_min, x_max)
axes[0, 0].set_title(par_label[0])
axes[0, 0].set_title(par_label[0] + r'$=' + '{:6.2f}'.format(par_mean[0]) + \
r'\pm' + '{:4.2f}'.format(par_std[0]) + r'$')
y = np.linspace(y_min, y_max, 100)
axes[1, 1].plot(y, sps.norm.pdf(y, par_mean[1], par_std[1]), 'k')
axes[1, 0].axhline(par_true[1])
axes[1, 1].axvline(par_true[1])
axes[1, 1].tick_params(labelleft=False)
axes[1, 1].set_xlim(y_min, y_max)
for tick in axes[1, 1].get_xticklabels():
tick.set_rotation(45)
axes[1, 1].set_title(par_label[1] + r'$=' + '{:5.2f}'.format(par_mean[1]) + \
r'\pm' + '{:4.2f}'.format(par_std[1]) + r'$')
# 2D marge
vals, vecs = np.linalg.eig(par_cov)
theta = np.degrees(np.arctan2(*vecs[::-1, 0]))
w, h = 2 * np.sqrt(vals)
ell = mpp.Ellipse(xy=par_mean, width=w, height=h,
angle=theta, color='k')
ell.set_facecolor("none")
axes[1, 0].add_artist(ell)
ell = mpp.Ellipse(xy=par_mean, width=2*w, height=2*h,
angle=theta, color='k')
ell.set_facecolor("none")
axes[1, 0].add_artist(ell)
axes[1, 0].set_xlim(x_min, x_max)
axes[1, 0].set_ylim(y_min, y_max)
for tick in axes[1, 0].get_xticklabels():
tick.set_rotation(45)
for tick in axes[1, 0].get_yticklabels():
tick.set_rotation(45)
axes[1, 0].set_xlabel(par_label[0])
axes[1, 0].set_ylabel(par_label[1])
fig.delaxes(axes[0, 1])
fig.subplots_adjust(hspace=0, wspace=0)
test = schmorner(gls_pars[n_gal:], gls_pars_cov[n_gal:, n_gal:], \
[abs_true, s_true], [r'$M$', r'$s$'])
#
#lazy = npr.multivariate_normal(gls_pars[n_gal:], gls_pars_cov[n_gal:, n_gal:], n_samples)
#fig = corner.corner(samples.T, labels=[r"$M$", r"$s$"],
# show_titles=True, truths=[abs_bar, s_bar])
```
## Task 3B
Below I've written the majority of a Gibbs sampler to infer the hyper-parameters of the Cepheid PL relation from our simulated sample. One component is missing: drawing from the conditional distribution of the standard absolute magnitude, $M^*$. Please fill it in, using the results of whiteboard/paper Task 3A.
```
def gibbs_sample(n_samples, n_gal, n_star, abs_bar, abs_sig, \
s_bar, s_sig, mu_bar, mu_sig, mu_hat_sig, \
m_sig_int, m_hat_sig, mu_hat, lp_true, m_hat):
# storage
abs_samples = np.zeros(n_samples)
s_samples = np.zeros(n_samples)
mu_samples = np.zeros((n_gal, n_samples))
m_samples = np.zeros((n_gal, n_star, n_samples))
# initialize sampler
abs_samples[0] = abs_bar + npr.randn() * abs_sig
s_samples[0] = s_bar + npr.randn() * s_sig
mu_samples[:, 0] = mu_bar + npr.randn(n_gal) * mu_bar
for i in range(n_gal):
m_samples[i, :, 0] = mu_samples[i, 0] + abs_samples[0] + s_samples[0] * lp_true[i, :]
# sample!
for k in range(1, n_samples):
# sample abs mag
abs_sig_pl = m_sig_int / np.sqrt(n_gal * n_star)
abs_bar_pl = 0.0
for j in range(n_gal):
abs_bar_pl += np.sum(m_samples[j, :, k - 1] - mu_samples[j, k - 1] - s_samples[k - 1] * lp_true[j, :])
abs_bar_pl /= (n_gal * n_star)
abs_std = np.sqrt((abs_sig * abs_sig_pl) ** 2 / (abs_sig ** 2 + abs_sig_pl ** 2))
abs_mean = (abs_sig ** 2 * abs_bar_pl + abs_sig_pl ** 2 * abs_bar) / \
(abs_sig ** 2 + abs_sig_pl ** 2)
abs_samples[k] = abs_mean + npr.randn() * abs_std
# sample slope
s_sig_pl = m_sig_int / np.sqrt(np.sum(lp_true ** 2))
s_bar_pl = 0.0
for j in range(n_gal):
s_bar_pl += np.sum((m_samples[j, :, k - 1] - mu_samples[j, k - 1] - abs_samples[k]) * lp_true[j, :])
s_bar_pl /= np.sum(lp_true ** 2)
s_std = np.sqrt((s_sig * s_sig_pl) ** 2 / (s_sig ** 2 + s_sig_pl ** 2))
s_mean = (s_sig ** 2 * s_bar_pl + s_sig_pl ** 2 * s_bar) / \
(s_sig ** 2 + s_sig_pl ** 2)
s_samples[k] = s_mean + npr.randn() * s_std
# sample apparent magnitudes
for j in range(n_gal):
m_mean_pl = mu_samples[j, k - 1] + abs_samples[k] + s_samples[k] * lp_true[j, :]
m_std = np.sqrt(m_sig_int ** 2 * m_hat_sig ** 2 / (m_sig_int ** 2 + m_hat_sig ** 2))
m_mean = (m_sig_int ** 2 * m_hat[j, :] + m_hat_sig ** 2 * m_mean_pl) / (m_sig_int ** 2 + m_hat_sig ** 2)
m_samples[j, :, k] = m_mean + npr.randn(n_star) * m_std
# sample distance moduli
mu_sig_pl = m_sig_int / np.sqrt(n_star)
mu_bar_pl = np.mean(m_samples[0, :, k] - abs_samples[k] - s_samples[k] * lp_true[0, :])
mu_var = 1.0 / (1.0 / mu_sig ** 2 + 1.0 / mu_hat_sig ** 2 + 1.0 / mu_sig_pl ** 2)
mu_mean = (mu_bar / mu_sig ** 2 + mu_hat / mu_hat_sig ** 2 + mu_bar_pl / mu_sig_pl ** 2) * mu_var
mu_samples[0, k] = mu_mean + npr.randn() * np.sqrt(mu_var)
for j in range(1, n_gal):
mu_sig_pl = m_sig_int / np.sqrt(n_star)
mu_bar_pl = np.mean(m_samples[j, :, k] - abs_samples[k] - s_samples[k] * lp_true[j, :])
mu_std = (mu_sig * mu_sig_pl) ** 2 / (mu_sig ** 2 + mu_sig_pl ** 2)
mu_mean = (mu_sig ** 2 * mu_bar_pl + mu_sig_pl ** 2 * mu_bar) / \
(mu_sig ** 2 + mu_sig_pl ** 2)
mu_samples[j, k] = mu_mean + npr.randn() * mu_std
return (abs_samples, s_samples, mu_samples, m_samples)
```
Now let's sample, setting aside the first half of the samples as warmup.
```
all_samples = gibbs_sample(n_samples, n_gal, n_star, abs_bar, abs_sig, \
s_bar, s_sig, mu_bar, mu_sig, mu_hat_sig, \
m_sig_int, m_hat_sig, mu_hat, lp_true, m_hat)
n_warmup = int(n_samples / 2)
g_samples = [samples[n_warmup:] for samples in all_samples]
```
Let's make sure that the absolute magnitude is being inferred as expected. First, generate a trace plot of the absolute magnitude samples (the first entry in `g_samples`), overlaying the ground truth. Then print out the mean and standard deviation of the marginalized absolute magnitude posterior. Recall that marginalizing is as simple as throwing away the samples of all other parameters.
```
mp.plot(g_samples[0])
mp.axhline(abs_true)
mp.xlabel('sample')
mp.ylabel(r'$M^*$')
print('Truth {:6.2f}; inferred {:6.2f} +/- {:4.2f}'.format(abs_true, np.mean(g_samples[0]), np.std(g_samples[0])))
```
Now let's generate some marginalized parameter posteriors (by simply discarding all samples of the latent parameters) using DFM's [`corner`](https://corner.readthedocs.io/en/latest/) package. Note the near identical nature of this plot to the `schmorner` plot we generated above.
```
import corner
samples = np.stack((g_samples[0], g_samples[1]))
fig = corner.corner(samples.T, labels=[r"$M^*$", r"$s$"],
show_titles=True, truths=[abs_true, s_true])
```
## Task 4
The final task is to write a [Stan model](https://pystan.readthedocs.io/en/latest/getting_started.html) to infer the parameters of the period-luminosity relation. I've coded up the other two blocks required (`data` and `parameters`), so all that is required is for you to write the joint posterior (factorized into its individual components) in Stan's sampling-statement-based syntax. Essentially all you need are Gaussian sampling statements (`abs_true ~ normal(abs_bar, abs_sig);`) and for loops (`for(i in 1: n_gal){...}`).
When you evaluate this cell, Stan will translate your model into `c++` code and compile it. We will then pickle the compiled model so you can re-use it rapidly without recompiling. To do so, please set `recompile = False` in the notebook.
```
import sys
import pystan as ps
import pickle
stan_code = """
data {
int<lower=0> n_gal;
int<lower=0> n_star;
real mu_hat;
real mu_hat_sig;
real m_hat[n_gal, n_star];
real m_hat_sig;
real m_sig_int;
real lp_true[n_gal, n_star];
real abs_bar;
real abs_sig;
real s_bar;
real s_sig;
real mu_bar;
real mu_sig;
}
parameters {
real mu_true[n_gal];
real m_true[n_gal, n_star];
real abs_true;
real s_true;
}
model {
// priors
abs_true ~ normal(abs_bar, abs_sig);
s_true ~ normal(s_bar, s_sig);
mu_true ~ normal(mu_bar, mu_sig);
// whatevers
for(i in 1: n_gal){
for(j in 1: n_star){
m_true[i, j] ~ normal(mu_true[i] + abs_true + s_true * lp_true[i, j], m_sig_int);
}
}
// likelihoods
mu_hat ~ normal(mu_true[1], mu_hat_sig);
for(i in 1: n_gal){
for(j in 1: n_star){
m_hat[i, j] ~ normal(m_true[i, j], m_hat_sig);
}
}
}
"""
n_samples_stan = 5000
recompile = True
pkl_fname = 'bhms_stan_model_v{:d}p{:d}p{:d}.pkl'.format(sys.version_info[0], \
sys.version_info[1], \
sys.version_info[2])
if recompile:
stan_model = ps.StanModel(model_code=stan_code)
with open(pkl_fname, 'wb') as f:
pickle.dump(stan_model, f)
else:
try:
with open(pkl_fname, 'rb') as f:
stan_model = pickle.load(f)
except EnvironmentError:
print('ERROR: pickled Stan model (' + pkl_fname + ') not found. ' + \
'Please set recompile = True')
```
Now let's sample...
```
stan_data = {'n_gal': n_gal, 'n_star': n_star, 'mu_hat': mu_hat, 'mu_hat_sig': mu_hat_sig, \
'm_hat': m_hat, 'm_hat_sig': m_hat_sig, 'm_sig_int': m_sig_int, 'lp_true': lp_true, \
'abs_bar': abs_bar, 'abs_sig': abs_sig, 's_bar': s_bar, 's_sig': s_sig, \
'mu_bar': mu_bar, 'mu_sig': mu_sig}
fit = stan_model.sampling(data=stan_data, iter=n_samples_stan, chains=4)
```
... print out Stan's posterior summary (note this is for _all_ parameters)...
```
samples = fit.extract(permuted=True)
print(fit)
```
... and plot the marginalized posterior of the PL parameters, as with the Gibbs sampler.
```
c_samples = np.stack((samples['abs_true'], samples['s_true']))
fig = corner.corner(c_samples.T, labels=[r"$M^*$", r"$s$"],
show_titles=True, truths=[abs_true, s_true])
```
Our work here is done!
| github_jupyter |
# Detecting Loops in Linked Lists
In this notebook, you'll implement a function that detects if a loop exists in a linked list. The way we'll do this is by having two pointers, called "runners", moving through the list at different rates. Typically we have a "slow" runner which moves at one node per step and a "fast" runner that moves at two nodes per step.
If a loop exists in the list, the fast runner will eventually move behind the slow runner as it moves to the beginning of the loop. Eventually it will catch up to the slow runner and both runners will be pointing to the same node at the same time. If this happens then you know there is a loop in the linked list. Below is an example where we have a slow runner (the green arrow) and a fast runner (the red arrow).
<center><img src='assets/two_runners_circular.png' alt="Visual walk through of the steps described above to determine if a loop exists in a linked list." width=300px></center>
```
class Node:
def __init__(self, value):
self.value = value
self.next = None
class LinkedList:
def __init__(self, init_list=None):
self.head = None
if init_list:
for value in init_list:
self.append(value)
def append(self, value):
if self.head is None:
self.head = Node(value)
return
# Move to the tail (the last node)
node = self.head
while node.next:
node = node.next
node.next = Node(value)
return
def __iter__(self):
node = self.head
while node:
yield node.value
node = node.next
def __repr__(self):
return str([i for i in self])
list_with_loop = LinkedList([2, -1, 3, 0, 5])
# Creating a loop where the last node points back to the second node
loop_start = list_with_loop.head.next
node = list_with_loop.head
while node.next:
node = node.next
node.next = loop_start
# You will encouter the unlimited loop
# Click on stop
# Then right click on `clear outpit`
for i in list_with_loop:
print(i)
```
### Write the function definition here
**Exercise:** Given a linked list, implement a function `iscircular` that returns `True` if a loop exists in the list and `False` otherwise.
```
def iscircular(linked_list):
"""
Determine whether the Linked List is circular or not
Args:
linked_list(obj): Linked List to be checked
Returns:
bool: Return True if the linked list is circular, return False otherwise
"""
# TODO: Write function to check if linked list is circular
if linked_list is None:
return False
slow, fast = linked_list.head, linked_list.head
while fast and fast.next:
slow, fast = slow.next, fast.next.next
if slow == fast:
return True
return False
```
### Let's test your function
```
iscircular(list_with_loop)
# Test Cases
# Create another circular linked list
small_loop = LinkedList([0])
small_loop.head.next = small_loop.head
print ("Pass" if iscircular(list_with_loop) else "Fail") # Pass
print ("Pass" if iscircular(LinkedList([-4, 7, 2, 5, -1])) else "Fail") # Fail
print ("Pass" if iscircular(LinkedList([1])) else "Fail") # Fail
print ("Pass" if iscircular(small_loop) else "Fail") # Pass
print ("Pass" if iscircular(LinkedList([])) else "Fail") # Fail
```
<span class="graffiti-highlight graffiti-id_tuhz4y1-id_fy0906u"><i></i><button>Show Solution</button></span>
| github_jupyter |
# Neural Networks
In the previous part of this exercise, you implemented multi-class logistic re gression to recognize handwritten digits. However, logistic regression cannot form more complex hypotheses as it is only a linear classifier.<br><br>
In this part of the exercise, you will implement a neural network to recognize handwritten digits using the same training set as before. The <strong>neural network</strong> will be able to represent complex models that form <strong>non-linear hypotheses</strong>. For this week, you will be using parameters from <strong>a neural network that we have already trained</strong>. Your goal is to implement the <strong>feedforward propagation algorithm to use our weights for prediction</strong>. In next week’s exercise, you will write the backpropagation algorithm for learning the neural network parameters.<br><br>
The file <strong><em>ex3data1</em></strong> contains a training set.<br>
The structure of the dataset described blow:<br>
1. X array = <strong>400 columns describe the values of pixels of 20*20 images in flatten format for 5000 samples</strong>
2. y array = <strong>Value of image (number between 0-9)</strong>
<br><br>
<strong>
Our assignment has these sections:
1. Visualizing the Data
1. Converting .mat to .csv
2. Loading Dataset and Trained Neural Network Weights
3. Ploting Data
2. Model Representation
3. Feedforward Propagation and Prediction
</strong>
In each section full description provided.
## 1. Visualizing the Dataset
Before starting on any task, it is often useful to understand the data by visualizing it.<br>
### 1.A Converting .mat to .csv
In this specific assignment, the instructor added a .mat file as training set and weights of trained neural network. But we have to convert it to .csv to use in python.<br>
After all we now ready to import our new csv files to pandas dataframes and do preprocessing on it and make it ready for next steps.
```
# import libraries
import scipy.io
import numpy as np
data = scipy.io.loadmat("ex3data1")
weights = scipy.io.loadmat('ex3weights')
```
Now we extract X and y variables from the .mat file and save them into .csv file for further usage. After running the below code <strong>you should see X.csv and y.csv files</strong> in your directory.
```
for i in data:
if '__' not in i and 'readme' not in i:
np.savetxt((i+".csv"),data[i],delimiter=',')
for i in weights:
if '__' not in i and 'readme' not in i:
np.savetxt((i+".csv"),weights[i],delimiter=',')
```
### 1.B Loading Dataset and Trained Neural Network Weights
First we import .csv files into pandas dataframes then save them into numpy arrays.<br><br>
There are <strong>5000 training examples</strong> in ex3data1.mat, where each training example is a <strong>20 pixel by 20 pixel <em>grayscale</em> image of the digit</strong>. Each pixel is represented by a floating point number indicating the <strong>grayscale intensity</strong> at that location. The 20 by 20 grid of pixels is <strong>"flatten" into a 400-dimensional vector</strong>. <strong>Each of these training examples becomes a single row in our data matrix X</strong>. This gives us a 5000 by 400 matrix X where every row is a training example for a handwritten digit image.<br><br>
The second part of the training set is a <strong>5000-dimensional vector y that contains labels</strong> for the training set.<br><br>
<strong>Notice: In dataset, the digit zero mapped to the value ten. Therefore, a "0" digit is labeled as "10", while the digits "1" to "9" are labeled as "1" to "9" in their natural order.<br></strong>
But this make thing harder so we bring it back to natural order for 0!
```
# import library
import pandas as pd
# saving .csv files to pandas dataframes
x_df = pd.read_csv('X.csv',names= np.arange(0,400))
y_df = pd.read_csv('y.csv',names=['label'])
# saving .csv files to pandas dataframes
Theta1_df = pd.read_csv('Theta1.csv',names = np.arange(0,401))
Theta2_df = pd.read_csv('Theta2.csv',names = np.arange(0,26))
# saving x_df and y_df into numpy arrays
x = x_df.iloc[:,:].values
y = y_df.iloc[:,:].values
m, n = x.shape
# bring back 0 to 0 !!!
y = y.reshape(m,)
y[y==10] = 0
y = y.reshape(m,1)
print('#{} Number of training samples, #{} features per sample'.format(m,n))
# saving Theta1_df and Theta2_df into numpy arrays
theta1 = Theta1_df.iloc[:,:].values
theta2 = Theta2_df.iloc[:,:].values
```
### 1.C Plotting Data
You will begin by visualizing a subset of the training set. In first part, the code <strong>randomly selects selects 100 rows from X</strong> and passes those rows to the <strong>display_data</strong> function. This function maps each row to a 20 pixel by 20 pixel grayscale image and displays the images together.<br>
After plotting, you should see an image like this:<img src='img/plot.jpg'>
```
import numpy as np
import matplotlib.pyplot as plt
import random
amount = 100
lines = 10
columns = 10
image = np.zeros((amount, 20, 20))
number = np.zeros(amount)
for i in range(amount):
rnd = random.randint(0,4999)
image[i] = x[rnd].reshape(20, 20)
y_temp = y.reshape(m,)
number[i] = y_temp[rnd]
fig = plt.figure(figsize=(8,8))
for i in range(amount):
ax = fig.add_subplot(lines, columns, 1 + i)
# Turn off tick labels
ax.set_yticklabels([])
ax.set_xticklabels([])
plt.imshow(image[i], cmap='binary')
plt.show()
print(number)
```
# 2. Model Representation
Our neural network is shown in below figure. It has <strong>3 layers an input layer, a hidden layer and an output layer</strong>. Recall that our <strong>inputs are pixel</strong> values of digit images. Since the images are of <strong>size 20×20</strong>, this gives us <strong>400 input layer units</strong> (excluding the extra bias unit which always outputs +1).<br><br><img src='img/nn.jpg'><br>
You have been provided with a set of <strong>network parameters (Θ<sup>(1)</sup>; Θ<sup>(2)</sup>)</strong> already trained by instructor.<br><br>
<strong>Theta1 and Theta2 The parameters have dimensions that are sized for a neural network with 25 units in the second layer and 10 output units (corresponding to the 10 digit classes).</strong>
```
print('theta1 shape = {}, theta2 shape = {}'.format(theta1.shape,theta2.shape))
```
It seems our weights are transposed, so we transpose them to have them in a way our neural network is.
```
theta1 = theta1.transpose()
theta2 = theta2.transpose()
print('theta1 shape = {}, theta2 shape = {}'.format(theta1.shape,theta2.shape))
```
# 3. Feedforward Propagation and Prediction
Now you will implement feedforward propagation for the neural network.<br>
You should implement the <strong>feedforward computation</strong> that computes <strong>h<sub>θ</sub>(x<sup>(i)</sup>)</strong> for every example i and returns the associated predictions. Similar to the one-vs-all classification strategy, the prediction from the neural network will be the <strong>label</strong> that has the <strong>largest output <strong>h<sub>θ</sub>(x)<sub>k</sub></strong></strong>.
<strong>Implementation Note:</strong> The matrix X contains the examples in rows. When you complete the code, <strong>you will need to add the column of 1’s</strong> to the matrix. The matrices <strong>Theta1 and Theta2 contain the parameters for each unit in rows.</strong> Specifically, the first row of Theta1 corresponds to the first hidden unit in the second layer. <br>
You must get <strong>a<sup>(l)</sup></strong> as a column vector.<br><br>
You should see that the <strong>accuracy is about 97.5%</strong>.
```
# adding column of 1's to x
x = np.append(np.ones(shape=(m,1)),x,axis = 1)
```
<strong>h = hypothesis(x,theta)</strong> will compute <strong>sigmoid</strong> function on <strong>θ<sup>T</sup>X</strong> and return a number which <strong>0<=h<=1</strong>.<br>
You can use <a href='https://docs.scipy.org/doc/scipy-0.15.1/reference/generated/scipy.special.expit.html'>this</a> library for calculating sigmoid.
```
def sigmoid(z):
return 1/(1+np.exp(-z))
def lr_hypothesis(x,theta):
return np.dot(x,theta)
```
<strong>predict(theta1, theta2, x):</strong> outputs the predicted label of x given the trained weights of a neural network (theta1, theta2).
```
layers = 3
num_labels = 10
```
<strong>Becuase the initial dataset has changed and mapped 0 to "10", so the weights also are changed. So we just rotate columns one step to right, to predict correct values.<br>
Recall we have changed mapping 0 to "10" to 0 to "0" but we cannot detect this mapping in weights of neural netwrok. So we have to this rotation on final output of probabilities.</strong>
```
def rotate_column(array):
array_ = np.zeros(shape=(m,num_labels))
temp = np.zeros(num_labels,)
temp= array[:,9]
array_[:,1:10] = array[:,0:9]
array_[:,0] = temp
return array_
def predict(theta1,theta2,x):
z2 = np.dot(x,theta1) # hidden layer
a2 = sigmoid(z2) # hidden layer
# adding column of 1's to a2
a2 = np.append(np.ones(shape=(m,1)),a2,axis = 1)
z3 = np.dot(a2,theta2)
a3 = sigmoid(z3)
# mapping problem. Rotate left one step
y_prob = rotate_column(a3)
# prediction on activation a2
y_pred = np.argmax(y_prob, axis=1).reshape(-1,1)
return y_pred
y_pred = predict(theta1,theta2,x)
y_pred.shape
```
Now we will compare our predicted result to the true one with <a href='http://scikit-learn.org/stable/modules/generated/sklearn.metrics.confusion_matrix.html'>confusion_matrix</a> of numpy library.
```
from sklearn.metrics import confusion_matrix
# Function for accuracy
def acc(confusion_matrix):
t = 0
for i in range(num_labels):
t += confusion_matrix[i][i]
f = m-t
ac = t/(m)
return (t,f,ac)
#import library
from sklearn.metrics import confusion_matrix
cm_train = confusion_matrix(y.reshape(m,),y_pred.reshape(m,))
t,f,ac = acc(cm_train)
print('With #{} correct, #{} wrong ==========> accuracy = {}%'
.format(t,f,ac*100))
cm_train
```
| github_jupyter |
```
# This cell is added by sphinx-gallery
!pip install mrsimulator --quiet
%matplotlib inline
import mrsimulator
print(f'You are using mrsimulator v{mrsimulator.__version__}')
```
# ²⁹Si 1D MAS spinning sideband (CSA)
After acquiring an NMR spectrum, we often require a least-squares analysis to
determine site populations and nuclear spin interaction parameters. Generally, this
comprises of two steps:
- create a fitting model, and
- determine the model parameters that give the best fit to the spectrum.
Here, we will use the mrsimulator objects to create a fitting model, and use the
`LMFIT <https://lmfit.github.io/lmfit-py/>`_ library for performing the least-squares
fitting optimization.
In this example, we use a synthetic $^{29}\text{Si}$ NMR spectrum of cuspidine,
generated from the tensor parameters reported by Hansen `et al.` [#f1]_, to
demonstrate a simple fitting procedure.
We will begin by importing relevant modules and establishing figure size.
```
import csdmpy as cp
import matplotlib.pyplot as plt
from lmfit import Minimizer, Parameters
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator.methods import BlochDecaySpectrum
from mrsimulator import signal_processing as sp
from mrsimulator.utils import spectral_fitting as sf
```
## Import the dataset
Use the `csdmpy <https://csdmpy.readthedocs.io/en/stable/index.html>`_
module to load the synthetic dataset as a CSDM object.
```
file_ = "https://sandbox.zenodo.org/record/835664/files/synthetic_cuspidine_test.csdf?"
synthetic_experiment = cp.load(file_).real
# standard deviation of noise from the dataset
sigma = 0.03383338
# convert the dimension coordinates from Hz to ppm
synthetic_experiment.x[0].to("ppm", "nmr_frequency_ratio")
# Plot of the synthetic dataset.
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(synthetic_experiment, "k", alpha=0.5)
ax.set_xlim(50, -200)
plt.grid()
plt.tight_layout()
plt.show()
```
## Create a fitting model
Before you can fit a simulation to an experiment, in this case, the synthetic dataset,
you will first need to create a fitting model. We will use the ``mrsimulator`` objects
as tools in creating a model for the least-squares fitting.
**Step 1:** Create initial guess sites and spin systems.
The initial guess is often based on some prior knowledge about the system under
investigation. For the current example, we know that Cuspidine is a crystalline silica
polymorph with one crystallographic Si site. Therefore, our initial guess model is a
single $^{29}\text{Si}$ site spin system. For non-linear fitting algorithms, as
a general recommendation, the initial guess model parameters should be a good starting
point for the algorithms to converge.
```
# the guess model comprising of a single site spin system
site = Site(
isotope="29Si",
isotropic_chemical_shift=-82.0, # in ppm,
shielding_symmetric={"zeta": -63, "eta": 0.4}, # zeta in ppm
)
spin_system = SpinSystem(
name="Si Site",
description="A 29Si site in cuspidine",
sites=[site], # from the above code
abundance=100,
)
```
**Step 2:** Create the method object.
The method should be the same as the one used
in the measurement. In this example, we use the `BlochDecaySpectrum` method. Note,
when creating the method object, the value of the method parameters must match the
respective values used in the experiment.
```
MAS = BlochDecaySpectrum(
channels=["29Si"],
magnetic_flux_density=7.1, # in T
rotor_frequency=780, # in Hz
spectral_dimensions=[
{
"count": 2048,
"spectral_width": 25000, # in Hz
"reference_offset": -5000, # in Hz
}
],
experiment=synthetic_experiment, # add the measurement to the method.
)
```
**Step 3:** Create the Simulator object, add the method and spin system objects, and
run the simulation.
```
sim = Simulator(spin_systems=[spin_system], methods=[MAS])
sim.run()
```
**Step 4:** Create a SignalProcessor class and apply post simulation processing.
```
processor = sp.SignalProcessor(
operations=[
sp.IFFT(), # inverse FFT to convert frequency based spectrum to time domain.
sp.apodization.Exponential(FWHM="200 Hz"), # apodization of time domain signal.
sp.FFT(), # forward FFT to convert time domain signal to frequency spectrum.
sp.Scale(factor=3), # scale the frequency spectrum.
]
)
processed_data = processor.apply_operations(data=sim.methods[0].simulation).real
```
**Step 5:** The plot the spectrum. We also plot the synthetic dataset for comparison.
```
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(synthetic_experiment, "k", linewidth=1, label="Experiment")
ax.plot(processed_data, "r", alpha=0.75, linewidth=1, label="guess spectrum")
ax.set_xlim(50, -200)
plt.legend()
plt.grid()
plt.tight_layout()
plt.show()
```
## Setup a Least-squares minimization
Now that our model is ready, the next step is to set up a least-squares minimization.
You may use any optimization package of choice, here we show an application using
LMFIT. You may read more on the LMFIT
`documentation page <https://lmfit.github.io/lmfit-py/index.html>`_.
### Create fitting parameters
Next, you will need a list of parameters that will be used in the fit. The *LMFIT*
library provides a `Parameters <https://lmfit.github.io/lmfit-py/parameters.html>`_
class to create a list of parameters.
```
site1 = spin_system.sites[0]
params = Parameters()
params.add(name="iso", value=site1.isotropic_chemical_shift)
params.add(name="eta", value=site1.shielding_symmetric.eta, min=0, max=1)
params.add(name="zeta", value=site1.shielding_symmetric.zeta)
params.add(name="FWHM", value=processor.operations[1].FWHM)
params.add(name="factor", value=processor.operations[3].factor)
```
### Create a minimization function
Note, the above set of parameters does not know about the model. You will need to
set up a function that will
- update the parameters of the `Simulator` and `SignalProcessor` object based on the
LMFIT parameter updates,
- re-simulate the spectrum based on the updated values, and
- return the difference between the experiment and simulation.
```
def minimization_function(params, sim, processor, sigma=1):
values = params.valuesdict()
# the experiment data as a Numpy array
intensity = sim.methods[0].experiment.y[0].components[0].real
# Here, we update simulation parameters iso, eta, and zeta for the site object
site = sim.spin_systems[0].sites[0]
site.isotropic_chemical_shift = values["iso"]
site.shielding_symmetric.eta = values["eta"]
site.shielding_symmetric.zeta = values["zeta"]
# run the simulation
sim.run()
# update the SignalProcessor parameter and apply line broadening.
# update the scaling factor parameter at index 3 of operations list.
processor.operations[3].factor = values["factor"]
# update the exponential apodization FWHM parameter at index 1 of operations list.
processor.operations[1].FWHM = values["FWHM"]
# apply signal processing
processed_data = processor.apply_operations(sim.methods[0].simulation)
# return the difference vector.
diff = intensity - processed_data.y[0].components[0].real
return diff / sigma
```
<div class="alert alert-info"><h4>Note</h4><p>To automate the fitting process, we provide a function to parse the
``Simulator`` and ``SignalProcessor`` objects for parameters and construct an
*LMFIT* ``Parameters`` object. Similarly, a minimization function, analogous to
the above `minimization_function`, is also included in the *mrsimulator*
library. See the next example for usage instructions.</p></div>
### Perform the least-squares minimization
With the synthetic dataset, simulation, and the initial guess parameters, we are ready
to perform the fit. To fit, we use the *LMFIT*
`Minimizer <https://lmfit.github.io/lmfit-py/fitting.html>`_ class.
```
minner = Minimizer(minimization_function, params, fcn_args=(sim, processor, sigma))
result = minner.minimize()
result
```
The plot of the fit, measurement and the residuals is shown below.
```
best_fit = sf.bestfit(sim, processor)[0]
residuals = sf.residuals(sim, processor)[0]
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(synthetic_experiment, "k", linewidth=1, label="Experiment")
ax.plot(best_fit, "r", alpha=0.75, linewidth=1, label="Best Fit")
ax.plot(residuals, alpha=0.75, linewidth=1, label="Residuals")
ax.set_xlabel("Frequency / Hz")
ax.set_xlim(50, -200)
plt.legend()
plt.grid()
plt.tight_layout()
plt.show()
```
.. [#f1] Hansen, M. R., Jakobsen, H. J., Skibsted, J., $^{29}\text{Si}$
Chemical Shift Anisotropies in Calcium Silicates from High-Field
$^{29}\text{Si}$ MAS NMR Spectroscopy, Inorg. Chem. 2003,
**42**, *7*, 2368-2377.
`DOI: 10.1021/ic020647f <https://doi.org/10.1021/ic020647f>`_
| github_jupyter |
# Data Science Boot Camp
## Introduction to Pandas Part 1
* __Pandas__ is a Python package providing fast, flexible, and expressive data structures designed to work with *relational* or *labeled* data both.<br>
<br>
* It is a fundamental high-level building block for doing practical, real world data analysis in Python.<br>
<br>
* Python has always been great for prepping and munging data, but it's never been great for analysis - you'd usually end up using R or loading it into a database and using SQL. Pandas makes Python great for analysis.<br>
* Pandas is well suited for:<br>
<br>
* Tabular data with heterogeneously-typed columns, as in an SQL table or Excel spreadsheet<br>
<br>
* Ordered and unordered (not necessarily fixed-frequency) time series data.<br>
<br>
* Arbitrary matrix data (homogeneously typed or heterogeneous) with row and column labels<br>
<br>
* Any other form of observational / statistical data sets. The data actually need not be labeled at all to be placed into a pandas data structure<br>
* Key features of Pandas:<br>
<br>
* Easy handling of __missing data__<br>
<br>
* __Size mutability__: columns can be inserted and deleted from DataFrame and higher dimensional objects.<br>
<br>
* Automatic and explicit __data alignment__: objects can be explicitly aligned to a set of labels, or the data can be aligned automatically.<br>
<br>
* __Fast__ and __efficient__ DataFrame object with default and customized indexing.<br>
<br>
* __Reshaping__ and __pivoting__ of data sets.<br>
* Key features of Pandas (Continued):<br>
<br>
* Label-based __slicing__, __indexing__, __fancy indexing__ and __subsetting__ of large data sets.<br>
<br>
* __Group by__ data for aggregation and transformations.<br>
<br>
* High performance __merging__ and __joining__ of data.<br>
<br>
* __IO Tools__ for loading data into in-memory data objects from different file formats.<br>
<br>
* __Time Series__ functionality.<br>
* First thing we have to import pandas and numpy library under the aliases pd and np.<br>
<br>
* Then check our pandas version.<br>
```
%matplotlib inline
import pandas as pd
import numpy as np
print(pd.__version__)
```
* Let's set some options for `Pandas`
```
pd.set_option('display.notebook_repr_html', False)
pd.set_option('max_columns', 10)
pd.set_option('max_rows', 10)
```
## Pandas Objects
* At the very basic level, Pandas objects can be thought of as enhanced versions of NumPy structured arrays in which the rows and columns are identified with labels rather than simple integer indices.<br>
<br>
* There are three fundamental Pandas data structures: the Series, DataFrame, and Index.
### Series
* A __Series__ is a single vector of data (like a NumPy array) with an *index* that labels each element in the vector.<br><br>
* It can be created from a list or array as follows:
```
counts = pd.Series([15029231, 7529491, 7499740, 5445026, 2702492, 2742534, 4279677, 2133548, 2146129])
counts
```
* If an index is not specified, a default sequence of integers is assigned as the index. A NumPy array comprises the values of the `Series`, while the index is a pandas `Index` object.
```
counts.values
counts.index
```
* We can assign meaningful labels to the index, if they are available:
```
population = pd.Series([15029231, 7529491, 7499740, 5445026, 2702492, 2742534, 4279677, 2133548, 2146129],
index=['Istanbul Total', 'Istanbul Males', 'Istanbul Females', 'Ankara Total', 'Ankara Males', 'Ankara Females', 'Izmir Total', 'Izmir Males', 'Izmir Females'])
population
```
* These labels can be used to refer to the values in the `Series`.
```
population['Istanbul Total']
mask = [city.endswith('Females') for city in population.index]
mask
population[mask]
```
* As you noticed that we can masking in series.<br>
<br>
* Also we can still use positional indexing even we assign meaningful labels to the index, if we wish.<br>
```
population[0]
```
* We can give both the array of values and the index meaningful labels themselves:<br>
```
population.name = 'population'
population.index.name = 'city'
population
```
* Also, NumPy's math functions and other operations can be applied to Series without losing the data structure.<br>
```
np.ceil(population / 1000000) * 1000000
```
* We can also filter according to the values in the `Series` like in the Numpy's:
```
population[population>3000000]
```
* A `Series` can be thought of as an ordered key-value store. In fact, we can create one from a `dict`:
```
populationDict = {'Istanbul Total': 15029231, 'Ankara Total': 5445026, 'Izmir Total': 4279677}
pd.Series(populationDict)
```
* Notice that the `Series` is created in key-sorted order.<br>
<br>
* If we pass a custom index to `Series`, it will select the corresponding values from the dict, and treat indices without corrsponding values as missing. Pandas uses the `NaN` (not a number) type for missing values.<br>
```
population2 = pd.Series(populationDict, index=['Istanbul Total','Ankara Total','Izmir Total','Bursa Total', 'Antalya Total'])
population2
population2.isnull()
```
* Critically, the labels are used to **align data** when used in operations with other Series objects:
```
population + population2
```
* Contrast this with NumPy arrays, where arrays of the same length will combine values element-wise; adding Series combined values with the same label in the resulting series. Notice also that the missing values were propogated by addition.
### DataFrame
* A `DataFrame` represents a tabular, spreadsheet-like data structure containing an or- dered collection of columns, each of which can be a different value type (numeric, string, boolean, etc.).<br>
<br>
* `DataFrame` has both a row and column index; it can be thought of as a dict of Series (one for all sharing the same index).
```
areaDict = {'Istanbul': 5461, 'Ankara': 25632, 'Izmir': 11891,
'Bursa': 10813, 'Antalya': 20177}
area = pd.Series(areaDict)
area
populationDict = {'Istanbul': 15029231, 'Ankara': 5445026, 'Izmir': 4279677, 'Bursa': 2936803, 'Antalya': 2364396}
population3 = pd.Series(populationDict)
population3
```
* Now that we have 2 Series population by cities and areas by cities, we can use a dictionary to construct a single two-dimensional object containing this information:
```
cities = pd.DataFrame({'population': population3, 'area': area})
cities
```
* Or we can create our cities `DataFrame` with lists and indexes.
```
cities = pd.DataFrame({
'population':[15029231, 5445026, 4279677, 2936803, 2364396],
'area':[5461, 25632, 11891, 10813, 20177],
'city':['Istanbul', 'Ankara', 'Izmir', 'Bursa', 'Antalya']
})
cities
```
Notice the `DataFrame` is sorted by column name. We can change the order by indexing them in the order we desire:
```
cities[['city','area', 'population']]
```
* A `DataFrame` has a second index, representing the columns:
```
cities.columns
```
* If we wish to access columns, we can do so either by dictionary like indexing or by attribute:
```
cities['area']
cities.area
type(cities.area)
type(cities[['area']])
```
* Notice this is different than with `Series`, where dictionary like indexing retrieved a particular element (row). If we want access to a row in a `DataFrame`, we index its `iloc` attribute.
```
cities.iloc[2]
cities.iloc[0:2]
```
Alternatively, we can create a `DataFrame` with a dict of dicts:
```
cities = pd.DataFrame({
0: {'city': 'Istanbul', 'area': 5461, 'population': 15029231},
1: {'city': 'Ankara', 'area': 25632, 'population': 5445026},
2: {'city': 'Izmir', 'area': 11891, 'population': 4279677},
3: {'city': 'Bursa', 'area': 10813, 'population': 2936803},
4: {'city': 'Antalya', 'area': 20177, 'population': 2364396},
})
cities
```
* We probably want this transposed:
```
cities = cities.T
cities
```
* It's important to note that the Series returned when a DataFrame is indexted is merely a **view** on the DataFrame, and not a copy of the data itself. <br>
<br>
* So you must be cautious when manipulating this data just like in the Numpy.<br>
```
areas = cities.area
areas
areas[3] = 0
areas
cities
```
* It's a usefull behavior for large data sets but for preventing this you can use copy method.<br>
```
areas = cities.area.copy()
areas[3] = 10813
areas
cities
```
* We can create or modify columns by assignment:<br>
```
cities.area[3] = 10813
cities
cities['year'] = 2017
cities
```
* But note that, we can not use the attribute indexing method to add a new column:<br>
```
cities.projection2020 = 20000000
cities
```
* It creates another variable.<br>
```
cities.projection2020
```
* Specifying a `Series` as a new columns cause its values to be added according to the `DataFrame`'s index:
```
populationIn2000 = pd.Series([11076840, 3889199, 3431204, 2150571, 1430539])
populationIn2000
cities['population_2000'] = populationIn2000
cities
```
* Other Python data structures (ones without an index) need to be the same length as the `DataFrame`:
```
populationIn2007 = [12573836, 4466756, 3739353, 2439876]
cities['population_2007'] = populationIn2007
```
* We can use `del` to remove columns, in the same way `dict` entries can be removed:
```
cities
del cities['population_2000']
cities
```
* We can extract the underlying data as a simple `ndarray` by accessing the `values` attribute:<br>
```
cities.values
```
* Notice that because of the mix of string and integer (and could be`NaN`) values, the dtype of the array is `object`.
* The dtype will automatically be chosen to be as general as needed to accomodate all the columns.
```
df = pd.DataFrame({'integers': [1,2,3], 'floatNumbers':[0.5, -1.25, 2.5]})
df
print(df.values.dtype)
df.values
```
* Pandas uses a custom data structure to represent the indices of Series and DataFrames.
```
cities.index
```
* Index objects are immutable:
```
cities.index[0] = 15
```
* This is so that Index objects can be shared between data structures without fear that they will be changed.
* That means you can move, copy your meaningful labels to other `DataFrames`
```
cities
cities.index = population2.index
cities
```
## Importing data
* A key, but often under appreciated, step in data analysis is importing the data that we wish to analyze.<br>
<br>
* Though it is easy to load basic data structures into Python using built-in tools or those provided by packages like NumPy, it is non-trivial to import structured data well, and to easily convert this input into a robust data structure.<br>
<br>
* Pandas provides a convenient set of functions for importing tabular data in a number of formats directly into a `DataFrame` object.
* Let's start with some more population data, stored in csv format.
```
!cat data/population.csv
```
* This table can be read into a DataFrame using `read_csv`:
```
populationDF = pd.read_csv("data/population.csv")
populationDF
```
* Notice that `read_csv` automatically considered the first row in the file to be a header row.<br>
<br>
* We can override default behavior by customizing some the arguments, like `header`, `names` or `index_col`.<br>
* `read_csv` is just a convenience function for `read_table`, since csv is such a common format:<br>
```
pd.set_option('max_columns', 5)
populationDF = pd.read_table("data/population_missing.csv", sep=';')
populationDF
```
* The `sep` argument can be customized as needed to accomodate arbitrary separators.<br>
* If we have sections of data that we do not wish to import (for example, in this example empty rows), we can populate the `skiprows` argument:
```
populationDF = pd.read_csv("data/population_missing.csv", sep=';', skiprows=[1,2])
populationDF
```
* For a more useful index, we can specify the first column, which provide a unique index to the data.
```
populationDF = pd.read_csv("data/population.csv", sep=';', index_col='Provinces')
populationDF.index
```
Conversely, if we only want to import a small number of rows from, say, a very large data file we can use `nrows`:
```
pd.read_csv("data/population.csv", sep=';', nrows=4)
```
* Most real-world data is incomplete, with values missing due to incomplete observation, data entry or transcription error, or other reasons. Pandas will automatically recognize and parse common missing data indicators, including `NA`, `NaN`, `NULL`.
```
pd.read_csv("data/population_missing.csv", sep=';').head(10)
```
Above, Pandas recognized `NaN` and an empty field as missing data.
```
pd.isnull(pd.read_csv("data/population_missing.csv", sep=';')).head(10)
```
### Microsoft Excel
* Since so much financial and scientific data ends up in Excel spreadsheets, Pandas' ability to directly import Excel spreadsheets is valuable. <br>
<br>
* This support is contingent on having one or two dependencies (depending on what version of Excel file is being imported) installed: `xlrd` and `openpyxl`.<br>
<br>
* Importing Excel data to Pandas is a two-step process. First, we create an `ExcelFile` object using the path of the file:
```
excel_file = pd.ExcelFile('data/population.xlsx')
excel_file
```
* Then, since modern spreadsheets consist of one or more "sheets", we parse the sheet with the data of interest:
```
excelDf = excel_file.parse("Sheet 1 ")
excelDf
```
* Also, there is a `read_excel` conveneince function in Pandas that combines these steps into a single call:
```
excelDf2 = pd.read_excel('data/population.xlsx', sheet_name='Sheet 1 ')
excelDf2.head(10)
```
* In, the first day we learned how to read and write `JSON` Files, with that way you can also import JSON files to `DataFrames`.
* Also, you can connect to databases and import your data into `DataFrames` by help of 3rd party libraries.
## Pandas Fundamentals
* This section introduces the new user to the key functionality of Pandas that is required to use the software effectively.<br>
<br>
* For some variety, we will leave our population data behind and employ some `Superhero` data.<br>
* The data comes from Marvel Wikia.<br>
<br>
* The file has the following variables:<br>
<table>
<table>
<thead>
<tr>
<th style="text-align:left;">Variable</th>
<th style="text-align:left;">Definition</th>
</tr>
</thead>
<tbody>
<tr>
<td style="text-align:left;">page_id</td>
<td style="text-align:left;">The unique identifier for that characters page within the wikia</td>
</tr>
<tr>
<td style="text-align:left;">name</td>
<td style="text-align:left;">The name of the character</td>
</tr>
<tr>
<td style="text-align:left;">urlslug</td>
<td style="text-align:left;">The unique url within the wikia that takes you to the character</td>
</tr>
<tr>
<td style="text-align:left;">ID</td>
<td style="text-align:left;">The identity status of the character (Secret Identity, Public identity No Dual Identity)</td>
</tr>
<tr>
<td style="text-align:left;">ALIGN</td>
<td style="text-align:left;">If the character is Good, Bad or Neutral</td>
</tr>
<tr>
<td style="text-align:left;">EYE</td>
<td style="text-align:left;">Eye color of the character</td>
</tr>
<tr>
<td style="text-align:left;">HAIR</td>
<td style="text-align:left;">Hair color of the character</td>
</tr>
<tr>
<td style="text-align:left;">SEX</td>
<td style="text-align:left;">Sex of the character (e.g. Male, Female, etc.)</td>
</tr>
<tr>
<td style="text-align:left;">GSM</td>
<td style="text-align:left;">If the character is a gender or sexual minority (e.g. Homosexual characters, bisexual characters)</td>
</tr>
<tr>
<td style="text-align:left;">ALIVE</td>
<td style="text-align:left;">If the character is alive or deceased</td>
</tr>
<tr>
<td style="text-align:left;">APPEARANCES</td>
<td style="text-align:left;">The number of appareances of the character in comic books (as of Sep. 2, 2014. Number will become increasingly out of date as time goes on.)</td>
</tr>
<tr>
<td style="text-align:left;">FIRST APPEARANCE</td>
<td style="text-align:left;">The month and year of the character's first appearance in a comic book, if available</td>
</tr>
<tr>
<td style="text-align:left;">YEAR</td>
<td style="text-align:left;">The year of the character's first appearance in a comic book, if available</td>
</tr>
</tbody>
</table>
```
pd.set_option('max_columns', 12)
pd.set_option('display.notebook_repr_html', True)
marvelDF = pd.read_csv("data/marvel-wikia-data.csv", index_col='page_id')
marvelDF.head(5)
```
* Notice that we specified the `page_id` column as the index, since it appears to be a unique identifier. We could try to create a unique index ourselves by trimming `name`:
* First, import the regex module of python.<br>
<br>
* Then, trim the name column with regex.<br>
```
import re
pattern = re.compile('([a-zA-Z]|-|\s|\.|\')*([a-zA-Z])')
heroName = []
for name in marvelDF.name:
match = re.search(pattern, name)
if match:
heroName.append(match.group())
else:
heroName.append(name)
heroName
```
* This looks okay, let's copy '__marvelDF__' to '__marvelDF_newID__' and assign new indexes.<br>
```
marvelDF_newID = marvelDF.copy()
marvelDF_newID.index = heroName
marvelDF_newID.head(5)
```
* Let's check the uniqueness of ID's:
```
marvelDF_newID.index.is_unique
```
* So, indices need not be unique. Our choice is not unique because some of superheros have some differenet variations.
```
pd.Series(marvelDF_newID.index).value_counts()
```
* The most important consequence of a non-unique index is that indexing by label will return multiple values for some labels:
```
marvelDF_newID.loc['Peter Parker']
```
* Let's give a truly unique index by not triming `name` column:
```
hero_id = marvelDF.name
marvelDF_newID = marvelDF.copy()
marvelDF_newID.index = hero_id
marvelDF_newID.head()
marvelDF_newID.index.is_unique
```
* We can create meaningful indices more easily using a hierarchical index.<br>
<br>
* For now, we will stick with the numeric IDs as our index for '__NewID__' DataFrame.<br>
```
marvelDF_newID.index = range(16376)
marvelDF.index = marvelDF['name']
marvelDF_newID.head(5)
```
### Manipulating indices
* __Reindexing__ allows users to manipulate the data labels in a DataFrame. <br>
<br>
* It forces a DataFrame to conform to the new index, and optionally, fill in missing data if requested.<br>
<br>
* A simple use of `reindex` is reverse the order of the rows:
```
marvelDF_newID.reindex(marvelDF_newID.index[::-1]).head()
```
* Keep in mind that `reindex` does not work if we pass a non-unique index series.
* We can remove rows or columns via the `drop` method:
```
marvelDF_newID.shape
marvelDF_dropped = marvelDF_newID.drop([16375, 16374])
print(marvelDF_newID.shape)
print(marvelDF_dropped.shape)
marvelDF_dropped = marvelDF_newID.drop(['EYE','HAIR'], axis=1)
print(marvelDF_newID.shape)
print(marvelDF_dropped.shape)
```
## Indexing and Selection
* Indexing works like indexing in NumPy arrays, except we can use the labels in the `Index` object to extract values in addition to arrays of integers.<br>
```
heroAppearances = marvelDF.APPEARANCES
heroAppearances
```
* Let's start with Numpy style indexing:
```
heroAppearances[:3]
```
* Indexing by Label:
```
heroAppearances[['Spider-Man (Peter Parker)','Hulk (Robert Bruce Banner)']]
```
* We can also slice with data labels, since they have an intrinsic order within the Index:
```
heroAppearances['Spider-Man (Peter Parker)':'Matthew Murdock (Earth-616)']
```
* You can change sliced array, and if you get warning it's ok.<br>
```
heroAppearances['Minister of Castile D\'or (Earth-616)':'Yologarch (Earth-616)'] = 0
heroAppearances
```
* In a `DataFrame` we can slice along either or both axes:
```
marvelDF[['SEX','ALIGN']]
mask = marvelDF.APPEARANCES>50
marvelDF[mask]
```
* The indexing field `loc` allows us to select subsets of rows and columns in an intuitive way:
```
marvelDF.loc['Spider-Man (Peter Parker)', ['ID', 'EYE', 'HAIR']]
marvelDF.loc[['Spider-Man (Peter Parker)','Thor (Thor Odinson)'],['ID', 'EYE', 'HAIR']]
```
## Operations
* `DataFrame` and `Series` objects allow for several operations to take place either on a single object, or between two or more objects.<br>
<br>
* For example, we can perform arithmetic on the elements of two objects, such as change in population across years:
```
populationDF
pop2000 = populationDF['2000']
pop2017 = populationDF['2017']
pop2000DF = pd.Series(pop2000.values, index=populationDF.index)
pop2017DF = pd.Series(pop2017.values, index=populationDF.index)
popDiff = pop2017DF - pop2000DF
popDiff
```
* Let's assume our '__pop2000DF__' DataFrame has not row which index is "Yalova"
```
pop2000DF["Yalova"] = np.nan
pop2000DF
popDiff = pop2017DF - pop2000DF
popDiff
```
* For accessing not null elements, we can use Pandas'notnull function.
```
popDiff[popDiff.notnull()]
```
* We can add `fill_value` argument to insert a zero for home `NaN` values.
```
pop2017DF.subtract(pop2000DF, fill_value=0)
```
* We can also use functions to each column or row of a `DataFrame`
```
minPop = pop2017DF.values.min()
indexOfMinPop = pop2017DF.index[pop2017DF.values.argmin()]
print(indexOfMinPop + " -> " + str(minPop))
populationDF['2000'] = np.ceil(populationDF['2000'] / 10000) * 10000
populationDF
```
## Sorting and Ranking
* Pandas objects include methods for re-ordering data.
```
populationDF.sort_index(ascending=True).head()
populationDF.sort_index().head()
populationDF.sort_index(axis=1, ascending=False).head()
```
* We can also use `order` to sort a `Series` by value, rather than by label.
* For a `DataFrame`, we can sort according to the values of one or more columns using the `by` argument of `sort_values`:
```
populationDF[['2017','2001']].sort_values(by=['2017', '2001'],ascending=[False,True]).head(10)
```
* __Ranking__ does not re-arrange data, but instead returns an index that ranks each value relative to others in the Series.
```
populationDF['2010'].rank(ascending=False)
populationDF[['2017','2001']].sort_values(by=['2017', '2001'],ascending=[False,True]).rank(ascending=False)
```
* Ties are assigned the mean value of the tied ranks, which may result in decimal values.
```
pd.Series([50,60,50]).rank()
```
* Alternatively, you can break ties via one of several methods, such as by the order in which they occur in the dataset:
```
pd.Series([100,50,100]).rank(method='first')
```
* Calling the `DataFrame`'s `rank` method results in the ranks of all columns:
```
populationDF.rank(ascending=False)
```
## Hierarchical indexing
* Hierarchical indexing is an important feature of pandas enabling you to have multiple (two or more) index levels on an axis.<br>
<br>
* Somewhat abstractly, it provides a way for you to work with higher dimensional data in a lower dimensional form.<br>
* Let’s create a Series with a list of lists or arrays as the index:
```
data = pd.Series(np.random.randn(10),
index=[['a', 'a', 'a', 'b', 'b', 'b', 'c', 'c', 'd', 'd'],
[1, 2, 3, 1, 2, 3, 1, 2, 2, 3]])
data
data.index
```
* With a hierarchically-indexed object, so-called partial indexing is possible, enabling you to concisely select subsets of the data:
```
data['b']
data['a':'c']
```
* Selection is even possible in some cases from an “inner” level:
```
data[:, 1]
```
* Hierarchical indexing plays a critical role in reshaping data and group-based operations like forming a pivot table. For example, this data could be rearranged into a DataFrame using its unstack method:
```
dataDF = data.unstack()
dataDF
```
* The inverse operation of unstack is stack:
```
dataDF.stack()
```
## Missing data
* The occurence of missing data is so prevalent that it pays to use tools like Pandas, which seamlessly integrates missing data handling so that it can be dealt with easily, and in the manner required by the analysis at hand.
* Missing data are represented in `Series` and `DataFrame` objects by the `NaN` floating point value. However, `None` is also treated as missing, since it is commonly used as such in other contexts (NumPy).
```
weirdSeries = pd.Series([np.nan, None, 'string', 1])
weirdSeries
weirdSeries.isnull()
```
* Missing values may be dropped or indexed out:
```
population2
population2.dropna()
population2[population2.notnull()]
dataDF
```
* By default, `dropna` drops entire rows in which one or more values are missing.
```
dataDF.dropna()
```
* This can be overridden by passing the `how='all'` argument, which only drops a row when every field is a missing value.
```
dataDF.dropna(how='all')
```
* This can be customized further by specifying how many values need to be present before a row is dropped via the `thresh` argument.
```
dataDF[2]['c'] = np.nan
dataDF
dataDF.dropna(thresh=2)
```
* If we want to drop missing values column-wise instead of row-wise, we use `axis=1`.
```
dataDF[1]['d'] = np.random.randn(1)
dataDF
dataDF.dropna(axis=1)
```
* Rather than omitting missing data from an analysis, in some cases it may be suitable to fill the missing value in, either with a default value (such as zero) or a value that is either imputed or carried forward/backward from similar data points. <br>
<br>
* We can do this programmatically in Pandas with the `fillna` argument.<br>
```
dataDF
dataDF.fillna(0)
dataDF.fillna({2: 1.5, 3:0.50})
```
* Notice that `fillna` by default returns a new object with the desired filling behavior, rather than changing the `Series` or `DataFrame` in place.
```
dataDF
```
* If you don't like this behaviour you can alter values in-place using `inplace=True`.
```
dataDF.fillna({2: 1.5, 3:0.50}, inplace=True)
dataDF
```
* Missing values can also be interpolated, using any one of a variety of methods:
```
dataDF[2]['c'] = np.nan
dataDF[3]['d'] = np.nan
dataDF
```
* We can also propagate non-null values forward or backward.
```
dataDF.fillna(method='ffill')
dataDF.fillna(dataDF.mean())
```
## Data summarization
* We often wish to summarize data in `Series` or `DataFrame` objects, so that they can more easily be understood or compared with similar data.<br>
<br>
* The NumPy package contains several functions that are useful here, but several summarization or reduction methods are built into Pandas data structures.<br>
```
marvelDF.sum()
```
* Clearly, `sum` is more meaningful for some columns than others.(Total Appearances)<br>
* For methods like `mean` for which application to string variables is not just meaningless, but impossible, these columns are automatically exculded:
```
marvelDF.mean()
```
* The important difference between NumPy's functions and Pandas' methods is that Numpy have different functions for handling missing data like 'nansum' but Pandas use same functions.
```
dataDF
dataDF.mean()
```
* Sometimes we may not want to ignore missing values, and allow the `nan` to propagate.
```
dataDF.mean(skipna=False)
```
* A useful summarization that gives a quick snapshot of multiple statistics for a `Series` or `DataFrame` is `describe`:
```
dataDF.describe()
```
* `describe` can detect non-numeric data and sometimes yield useful information about it.
## Writing Data to Files
* Pandas can also export data to a variety of storage formats.<br>
<br>
* We will bring your attention to just a couple of these.
```
myDF = populationDF['2000']
myDF.to_csv("data/roundedPopulation2000.csv")
```
* The `to_csv` method writes a `DataFrame` to a comma-separated values (csv) file.<br>
<br>
* You can specify custom delimiters (via `sep` argument), how missing values are written (via `na_rep` argument), whether the index is writen (via `index` argument), whether the header is included (via `header` argument), among other options.
| github_jupyter |
```
# coding=utf-8
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from keras.utils import np_utils
from keras.models import Sequential,load_model,save_model
from keras.layers import Dense, Dropout, Activation,LeakyReLU
from keras.optimizers import SGD, Adam
from keras.callbacks import EarlyStopping,ModelCheckpoint
from keras import backend as K
from sklearn import preprocessing
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score,accuracy_score
from scipy import sparse
import gc
from time import strftime, localtime
# 打印当前时间
def printTime():
print(strftime("%Y-%m-%d %H:%M:%S", localtime()))
return
printTime()
csr_trainData0 = sparse.load_npz(r'../trainTestData/trainData13100.npz')
csr_trainData0.shape
csr_trainData1 = sparse.load_npz(r'../trainTestData/trainData15112.npz')
csr_trainData1.shape
csr_trainData = sparse.hstack((csr_trainData0,csr_trainData1),format='csr')
del csr_trainData0,csr_trainData1
gc.collect()
age_train = pd.read_csv(r'../data/age_train.csv',header=None)
label = age_train[1].values
print(label.shape)
import time
seed = 7
np.random.seed(seed)
kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=seed)
model_filePath = r'../model/model28212_NN_'
currK = 0
val_index_list, score = [], []
val_probability = np.zeros((2010000,7))
printTime()
for train_index, val_index in kfold.split(csr_trainData,label):
K.clear_session()
trainData, trainLabel, valData, valLabel = csr_trainData[train_index,:], label[train_index], csr_trainData[val_index,:] , label[val_index]
trainLabel,valLabel = np_utils.to_categorical(trainLabel,num_classes=7),np_utils.to_categorical(valLabel,num_classes=7)
print('----------------------------------------------------------------------------------------------------------------------------------')
print(currK,'split Done!\n')
# 全连接模型
model = Sequential()
model.add(Dense(4000, activation='tanh', input_shape=(csr_trainData.shape[1],)))
model.add(Dense(2000, activation='relu'))
model.add(Dense(1000, activation='sigmoid'))
model.add(Dense(7, activation='softmax'))
#损失函数使用交叉熵
adam = Adam(lr=0.0003)
model.compile(loss='categorical_crossentropy',
optimizer = adam,
metrics=['accuracy'])
#模型训练
batch_size = 1024
epochs = 100
early_stopping = EarlyStopping(monitor='val_loss', patience=5, verbose=2)
bestModel = ModelCheckpoint(model_filePath + str(currK) + r'.h5', monitor='val_loss', verbose=0, save_best_only=True, save_weights_only=False, mode='auto', period=1)
hist = model.fit(trainData, trainLabel,
batch_size=batch_size,
epochs=epochs,
verbose=1,
shuffle=True,
validation_data=(valData,valLabel),
callbacks=[early_stopping,bestModel],
)
print('\n',currK,'train Done!')
printTime()
K.clear_session()
model = load_model(model_filePath + str(currK) + r'.h5')
probability = model.predict(valData,batch_size=1024)
val_probability[val_index,:] = probability
score.append(np.max(hist.history['val_acc']))
y_label = label[val_index]
val_label = np.argmax(probability,axis=1)
print(currK,'val_acc:',accuracy_score(val_label,y_label),'\n\n')
currK += 1
K.clear_session()
del trainData, valData, trainLabel,valLabel,model
print('----------------------------------------------------------------------------------------------------------------------------------')
print('mean val_acc:', np.mean(score))
printTime()
accuracy_score(np.argmax(val_probability,axis=1) ,label)
del csr_trainData
import gc
gc.collect()
```
# 验证集
```
val_probability = pd.DataFrame(val_probability)
print(val_probability.shape)
print(val_probability.head())
val_probability.drop(labels=[0],axis=1,inplace=True)
val_probability.to_csv(r'../processed/val_probability_28212.csv',header=None,index=False)
```
# 测试集
```
import os
model_file = r'../model/model28212_NN_'
csr_testData0 = sparse.load_npz(r'../trainTestData/trainData13100.npz')
csr_testData0.shape
csr_testData1 = sparse.load_npz(r'../trainTestData/trainData15112.npz')
csr_testData1.shape
csr_testData = sparse.hstack((csr_testData0, csr_testData1),format='csr')
del csr_trainData0,csr_trainData1
gc.collect()
age_test = pd.read_csv(r'../data/age_test.csv',header=None,usecols=[0])
printTime()
proflag = True
model_Num = 0
for i in list(range(10)):
model = load_model(model_file + str(i) + '.h5')
if proflag==True:
probability = model.predict(csr_testData,batch_size=1024,verbose=1)
proflag = False
else:
probability += model.predict(csr_testData,batch_size=1024,verbose=1)
model_Num += 1
print(model_Num)
K.clear_session()
del model
printTime()
model_Num
probability /= model_Num
age = np.argmax(probability,axis=1)
age_test = pd.read_csv(r'../data/age_test.csv',header=None,usecols=[0])
age_test = age_test.values
type(age_test)
print(probability.shape)
pro = np.column_stack((age_test,probability))
pro = pd.DataFrame(pro)
pro.drop(labels=[0,1],axis=1,inplace=True)
print(pro.shape)
pro.to_csv(r'../processed/test_probability_28212.csv',index=False,header=False)
```
| github_jupyter |
```
import torch
import numpy as np
import pandas as pd
from sklearn.cluster import KMeans
from statsmodels.discrete.discrete_model import Probit
import patsy
import matplotlib.pylab as plt
import tqdm
import itertools
ax = np.newaxis
```
Make sure you have installed the pygfe package. You can simply call `pip install pygrpfe` in the terminal or call the magic command `!pip install pygrpfe` from within the notebook. If you are using the binder link, then `pygrpfe` is already installed. You can import the package directly.
```
import pygrpfe as gfe
```
# A simple model of wage and participation
\begin{align*}
Y^*_{it} & = \alpha_i + \epsilon_{it} \\
D_{it} &= 1\big[ u(\alpha_i) \geq c(D_{it-1}) + V_{it} \big] \\
Y_{it} &= D_{it} Y^*_{it} \\
\end{align*}
where we use
$$u(\alpha) = \frac{e^{(1-\gamma) \alpha } -1}{1-\gamma}$$
and use as initial conditions $D_{i1} = 1\big[ u(\alpha_i) \geq c(1) + V_{i1} \big]$.
```
def dgp_simulate(ni,nt,gamma=2.0,eps_sd=1.0):
""" simulates according to the model """
alpha = np.random.normal(size=(ni))
eps = np.random.normal(size=(ni,nt))
v = np.random.normal(size=(ni,nt))
# non-censored outcome
W = alpha[:,ax] + eps*eps_sd
# utility
U = (np.exp( alpha * (1-gamma)) - 1)/(1-gamma)
U = U - U.mean()
# costs
C1 = -1; C0=0;
# binary decision
Y = np.ones((ni,nt))
Y[:,0] = U.squeeze() > C1 + v[:,0]
for t in range(1,nt):
Y[:,t] = U > C1*Y[:,t-1] + C0*(1-Y[:,t-1]) + v[:,t]
W = W * Y
return(W,Y)
```
# Estimating the model
We show the steps to estimating the model. Later on, we will run a Monte-Carlo Simulation.
We simulate from the DGP we have defined.
```
ni = 1000
nt = 50
Y,D = dgp_simulate(ni,nt,2.0)
```
## Step 1: grouping observations
We group individuals based on their outcomes. We consider as moments the average value of $Y$ and the average value of $D$. We give our gfe function the $t$ sepcific values so that it can compute the within individual variation. This is a measure used to pick the nubmer of groups.
The `group` function chooses the number of groups based on the rule described in the paper.
```
# we create the moments
# this has dimension ni x nt x nm
M_itm = np.stack([Y,D],axis=2)
# we use our sugar function to get the groups
G_i,_ = gfe.group(M_itm)
print("Number of groups = {:d}".format(G_i.max()))
```
We can plot the grouping:
```
dd = pd.DataFrame({'Y':Y.mean(1),'G':G_i,'D':D.mean(1)})
plt.scatter(dd.Y,dd.D,c=dd.G*1.0)
plt.show()
```
## Step 2: Estimate the likelihood model with group specific parameters
In the model we proposed, this second step is a probit. We can then directly use python probit routine with group dummies.
```
ni,nt = D.shape
# next we minimize using groups as FE
dd = pd.DataFrame({
'd': D[:,range(1,nt)].flatten(),
'dl':D[:,range(nt-1)].flatten(),
'gi':np.broadcast_to(G_i[:,ax], (ni,nt-1)).flatten()})
yv,Xv = patsy.dmatrices("d ~ 0 + dl + C(gi)", dd, return_type='matrix')
mod = Probit(dd['d'], Xv)
res = mod.fit(maxiter=2000,method='bfgs')
print("Estimated cost parameters = {:.3f}".format(res.params[-1]))
```
## Step 2 (alternative implementation): Pytorch and auto-diff
We next write down a likelihood that we want to optimize. Instead of using the Python routine for the Probit, we make use of automatic differentiation from PyTorch. This makes it easy to modify the estimating model to accomodate for less standard likelihoods!
We create a class which initializes the parameters in the `__init__` method and computes the loss in the `loss` method. We will see later how we can use this to define a fixed effect estimator.
```
class GrpProbit:
# initialize parameters and data
def __init__(self,D,G_i):
# define parameters and tell PyTorch to keep track of gradients
self.alpha = torch.tensor( np.ones(G_i.max()+1), requires_grad=True)
self.cost = torch.tensor( np.random.normal(1), requires_grad=True)
self.params = [self.alpha,self.cost]
# predefine some components
ni,nt = D.shape
self.ni = ni
self.G_i = G_i
self.Dlag = torch.tensor(D[:,range(0,nt-1)])
self.Dout = torch.tensor(D[:,range(1,nt)])
self.N = torch.distributions.normal.Normal(0,1)
# define our loss function
def loss(self):
Id = self.alpha[self.G_i].reshape(self.ni,1) + self.cost * self.Dlag
lik_it = self.Dout * torch.log( torch.clamp( self.N.cdf( Id ), min=1e-7)) + \
(1-self.Dout)*torch.log( torch.clamp( self.N.cdf( -Id ), min=1e-7) )
return(- lik_it.mean())
# initialize the model with groups and estimate it
model = GrpProbit(D,G_i)
gfe.train(model)
print("Estimated cost parameters = {:.3f}".format(model.params[1]))
```
## Use PyTorch to estimate Fixed Effect version
Since Pytorch makes use of efficient automatic differentiation, we can use it with many variables. This allows us to give each individual their own group, effectivily estimating a fixed-effect model.
```
model_fe = GrpProbit(D,np.arange(ni))
gfe.train(model_fe)
print("Estimated cost parameters FE = {:.3f}".format(model_fe.params[1]))
```
# Monte-Carlo
We finish with running a short Monte-Carlo exercise.
```
all = []
import itertools
ll = list(itertools.product(range(50), [10,20,30,40]))
for r, nt in tqdm.tqdm(ll):
ni = 1000
gamma =2.0
Y,D = dgp_simulate(ni,nt,gamma)
M_itm = np.stack([Y,D],axis=2)
G_i,_ = blm2.group(M_itm,scale=True)
model_fe = GrpProbit(D,np.arange(ni))
gfe.train(model_fe)
model_gfe = GrpProbit(D,G_i)
gfe.train(model_gfe)
all.append({
'c_fe' : model_fe.params[1].item(),
'c_gfe': model_gfe.params[1].item(),
'ni':ni,
'nt':nt,
'gamma':gamma,
'ng':G_i.max()+1})
df = pd.DataFrame(all)
df2 = df.groupby(['ni','nt','gamma']).mean().reset_index()
plt.plot(df2['nt'],df2['c_gfe'],label="gfe",color="orange")
plt.plot(df2['nt'],df2['c_fe'],label="fe",color="red")
plt.axhline(1.0,label="true",color="black",linestyle=":")
plt.xlabel("T")
plt.legend()
plt.show()
df.groupby(['ni','nt','gamma']).mean()
```
| github_jupyter |
# GDP and life expectancy
Richer countries can afford to invest more on healthcare, on work and road safety, and other measures that reduce mortality. On the other hand, richer countries may have less healthy lifestyles. Is there any relation between the wealth of a country and the life expectancy of its inhabitants?
The following analysis checks whether there is any correlation between the total gross domestic product (GDP) of a country in 2013 and the life expectancy of people born in that country in 2013.
Getting the data
Two datasets of the World Bank are considered. One dataset, available at http://data.worldbank.org/indicator/NY.GDP.MKTP.CD, lists the GDP of the world's countries in current US dollars, for various years. The use of a common currency allows us to compare GDP values across countries. The other dataset, available at http://data.worldbank.org/indicator/SP.DYN.LE00.IN, lists the life expectancy of the world's countries. The datasets were downloaded as CSV files in March 2016.
```
import warnings
warnings.simplefilter('ignore', FutureWarning)
import pandas as pd
YEAR = 2018
GDP_INDICATOR = 'NY.GDP.MKTP.CD'
gdpReset = pd.read_csv('WB 2018 GDP.csv')
LIFE_INDICATOR = 'SP.DYN.LE00.IN_'
lifeReset = pd.read_csv('WB 2018 LE.csv')
lifeReset.head()
```
## Cleaning the data
Inspecting the data with `head()` and `tail()` shows that:
1. the first 34 rows are aggregated data, for the Arab World, the Caribbean small states, and other country groups used by the World Bank;
- GDP and life expectancy values are missing for some countries.
The data is therefore cleaned by:
1. removing the first 34 rows;
- removing rows with unavailable values.
```
gdpCountries = gdpReset.dropna()
lifeCountries = lifeReset.dropna()
```
## Transforming the data
The World Bank reports GDP in US dollars and cents. To make the data easier to read, the GDP is converted to millions of British pounds (the author's local currency) with the following auxiliary functions, using the average 2013 dollar-to-pound conversion rate provided by <http://www.ukforex.co.uk/forex-tools/historical-rate-tools/yearly-average-rates>.
```
def roundToMillions (value):
return round(value / 1000000)
def usdToGBP (usd):
return usd / 1.334801
GDP = 'GDP (£m)'
gdpCountries[GDP] = gdpCountries[GDP_INDICATOR].apply(usdToGBP).apply(roundToMillions)
gdpCountries.head()
COUNTRY = 'Country Name'
headings = [COUNTRY, GDP]
gdpClean = gdpCountries[headings]
gdpClean.head()
LIFE = 'Life expectancy (years)'
lifeCountries[LIFE] = lifeCountries[LIFE_INDICATOR].apply(round)
headings = [COUNTRY, LIFE]
lifeClean = lifeCountries[headings]
lifeClean.head()
gdpVsLife = pd.merge(gdpClean, lifeClean, on=COUNTRY, how='inner')
gdpVsLife.head()
```
## Calculating the correlation
To measure if the life expectancy and the GDP grow together, the Spearman rank correlation coefficient is used. It is a number from -1 (perfect inverse rank correlation: if one indicator increases, the other decreases) to 1 (perfect direct rank correlation: if one indicator increases, so does the other), with 0 meaning there is no rank correlation. A perfect correlation doesn't imply any cause-effect relation between the two indicators. A p-value below 0.05 means the correlation is statistically significant.
```
from scipy.stats import spearmanr
gdpColumn = gdpVsLife[GDP]
lifeColumn = gdpVsLife[LIFE]
(correlation, pValue) = spearmanr(gdpColumn, lifeColumn)
print('The correlation is', correlation)
if pValue < 0.05:
print('It is statistically significant.')
else:
print('It is not statistically significant.')
```
The value shows a direct correlation, i.e. richer countries tend to have longer life expectancy.
## Showing the data
Measures of correlation can be misleading, so it is best to see the overall picture with a scatterplot. The GDP axis uses a logarithmic scale to better display the vast range of GDP values, from a few million to several billion (million of million) pounds.
```
%matplotlib inline
gdpVsLife.plot(x=GDP, y=LIFE, kind='scatter', grid=True, logx=True, figsize=(10, 4))
```
The plot shows there is no clear correlation: there are rich countries with low life expectancy, poor countries with high expectancy, and countries with around 10 thousand (104) million pounds GDP have almost the full range of values, from below 50 to over 80 years. Towards the lower and higher end of GDP, the variation diminishes. Above 40 thousand million pounds of GDP (3rd tick mark to the right of 104), most countries have an expectancy of 70 years or more, whilst below that threshold most countries' life expectancy is below 70 years.
Comparing the 10 poorest countries and the 10 countries with the lowest life expectancy shows that total GDP is a rather crude measure. The population size should be taken into account for a more precise definiton of what 'poor' and 'rich' means. Furthermore, looking at the countries below, droughts and internal conflicts may also play a role in life expectancy.
```
# the 10 countries with lowest GDP
gdpVsLife.sort_values(GDP).head(10)
# the 10 countries with lowest life expectancy
gdpVsLife.sort_values(LIFE).head(10)
```
## Conclusions
To sum up, there is no strong correlation between a country's wealth and the life expectancy of its inhabitants: there is often a wide variation of life expectancy for countries with similar GDP, countries with the lowest life expectancy are not the poorest countries, and countries with the highest expectancy are not the richest countries. Nevertheless there is some relationship, because the vast majority of countries with a life expectancy below 70 years is on the left half of the scatterplot.
| github_jupyter |
# American Gut Project example
This notebook was created from a question we recieved from a user of MGnify.
The question was:
```
I am attempting to retrieve some of the MGnify results from samples that are part of the American Gut Project based on sample location.
However latitude and longitude do not appear to be searchable fields.
Is it possible to query these fields myself or to work with someone to retrieve a list of samples from a specific geographic range? I am interested in samples from people in Hawaii, so 20.5 - 20.7 and -154.0 - -161.2.
```
Let's decompose the question:
- project "American Gut Project"
- Metadata filtration using the geographic location of a sample.
- Get samples for Hawai: 20.5 - 20.7 ; -154.0 - -161.2
Each sample if MGnify it's obtained from [ENA](https://www.ebi.ac.uk/ena).
## Get samples
The first step is to obtain the samples using [ENA advanced search API](https://www.ebi.ac.uk/ena/browser/advanced-search).
```
from pandas import DataFrame
import requests
base_url = 'https://www.ebi.ac.uk/ena/portal/api/search'
# parameters
params = {
'result': 'sample',
'query': ' AND '.join([
'geo_box1(16.9175,-158.4687,21.6593,-152.7969)',
'description="*American Gut Project*"'
]),
'fields': ','.join(['secondary_sample_accession', 'lat', 'lon']),
'format': 'json',
}
response = requests.post(base_url, data=params)
agp_samples = response.json()
df = DataFrame(columns=('secondary_sample_accession', 'lat', 'lon'))
df.index.name = 'accession'
for s in agp_samples:
df.loc[s.get('accession')] = [
s.get('secondary_sample_accession'),
s.get('lat'),
s.get('lon')
]
df
```
Now we can use EMG API to get the information.
```
#!/bin/usr/env python
import requests
import sys
def get_links(data):
return data["links"]["related"]
if __name__ == "__main__":
samples_url = "https://www.ebi.ac.uk/metagenomics/api/v1/samples/"
tsv = sys.argv[1] if len(sys.argv) == 2 else None
if not tsv:
print("The first arg is the tsv file")
exit(1)
tsv_fh = open(tsv, "r")
# header
next(tsv_fh)
for record in tsv_fh:
# get the runs first
# mgnify references the secondary accession
_, sec_acc, *_ = record.split("\t")
samples_res = requests.get(samples_url + sec_acc)
if samples_res.status_code == 404:
print(sec_acc + " not found in MGnify")
continue
# then the analysis for that run
runs_url = get_links(samples_res.json()["data"]["relationships"]["runs"])
if not runs_url:
print("No runs for sample " + sec_acc)
continue
print("Getting the runs: " + runs_url)
run_res = requests.get(runs_url)
if run_res.status_code != 200:
print(run_url + " failed", file=sys.stderr)
continue
# iterate over the sample runs
run_data = run_res.json()
# this script doesn't consider pagination, it's just an example
# there could be more that one page of runs
# use links -> next to get the next page
for run in run_data["data"]:
analyses_url = get_links(run["relationships"]["analyses"])
if not analyses_url:
print("No analyses for run " + run)
continue
analyses_res = requests.get(analyses_url)
if analyses_res.status_code != 200:
print(analyses_url + " failed", file=sys.stderr)
continue
# dump
print("Raw analyses data")
print(analyses_res.json())
print("=" * 30)
tsv_fh.close()
```
| github_jupyter |
# Employee Attrition Prediction
There is a class of problems that predict that some event happens after N years. Examples are employee attrition, hard drive failure, life expectancy, etc.
Usually these kind of problems are considered simple problems and are the models have vairous degree of performance. Usually it is treated as a classification problem, predicting if after exactly N years the event happens. The problem with this approach is that people care not so much about the likelihood that the event happens exactly after N years, but the probability that the event happens today. While you can infer this using Bayes theorem, doing it during prediction will not give you good accuracy because the Bayesian inference will be based on one piece of data. It is better to do this kind of inference during training time, and learn the probability than the likelihood function.
Thus, the problem is learning a conditional probability of the person quitting, given he has not quit yet, and is similar to the Hazard function in survival analysis problem
```
#Import
import numpy as np
import pandas as pd
import numpy.random
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from sklearn.preprocessing import MinMaxScaler
import math
%matplotlib inline
numpy.random.seed(1239)
# Read the data
# Source: https://www.ibm.com/communities/analytics/watson-analytics-blog/hr-employee-attrition/
raw_data = pd.read_csv('data/WA_Fn-UseC_-HR-Employee-Attrition.csv')
#Check if any is nan. If no nans, we don't need to worry about dealing with them
raw_data.isna().sum().sum()
def prepare_data(raw_data):
'''
Prepare the data
1. Set EmployeeNumber as the index
2. Drop redundant columns
3. Reorder columns to make YearsAtCompany first
4. Change OverTime to the boolean type
5. Do 1-hot encoding
'''
labels = raw_data.Attrition == 'Yes'
employee_data = raw_data.set_index('EmployeeNumber').drop(columns=['Attrition', 'EmployeeCount', 'Over18'])
employee_data.loc[:, 'OverTime'] = (employee_data.OverTime == 'Yes').astype('float')
employee_data = pd.get_dummies(employee_data)
employee_data = pd.concat([employee_data.YearsAtCompany, employee_data.drop(columns='YearsAtCompany')], axis=1)
return employee_data, labels
#Split to features and labels
employee_data, labels = prepare_data(raw_data)
```
First we will work on the synthetic set of data, for this reason we will not split the dataset to train/test yet
```
#Now scale the entire dataset, but not the first column (YearsAtCompany). Instead scale the dataset to be similar range
#to the first column
max_year = employee_data.YearsAtCompany.max()
scaler = MinMaxScaler(feature_range=(0, max_year))
scaled_data = pd.DataFrame(scaler.fit_transform(employee_data.values.astype('float')),
columns=employee_data.columns,
index=employee_data.index)
```
Based on the chart it seems like a realistic data set.
Now we need to construct our loss function. It will have an additional parameter: number of years
We define probability $p(x, t)$ that the person quits this very day, where t is the number of years and x is the remaining features. Then the likelihood that the person has quit after the year $t$ is
$$P(x,t) = (\prod_{l=0}^{t-1} (1-p(x,l))) p(x,t) $$ whereas the likelihood that the person will remain after the year $t$ is
$$P(x,t) = \prod_{l=0}^{t} (1-p(x,l)) $$
Strictly speaking x is also dependent on t, but we don't have the historical data for this, so we assume that x is independent of t.
Using the principle of maximum likelihood, we derive the loss function taking negative log of the likelihood function:
$$\mathscr{L}(y,p) = -\sum_{l=0}^{t-1} \log(1-p(x,l)) - y \log{p} - (1-y) \log(1-p) $$
Where y is an indicator if the person has quit after working exactly t years or not.
Notice that the last two terms is the cross-entropy loss function, and the first term is a hitorical term.
We will use a modified Cox Hazard function mechanism and model the conditional probability $p(x,l)$ a sigmoid function (for simplicity we include bias in the list of weights, and so the weight for the t parameter): $$p=\frac{1}{1 + e^{-\bf{w}\bf{x}}}$$
To create a synthetic set we assume that p does not depend on anything. Then the maximum likelihood gives us this simple formula: $$Pos=M p \bar{t}$$
Here Pos is the number of positive example (people who quit) and M is the total number of examples and $\bar{t}$ is the mean time (number of years)
```
#pick a p
p = 0.01
#Get the maximum years. We need it to make sure that the product of p YearsAtCompany never exceeds 1.
#In reality that is not a problem, but we will use it to correctly create synthetic labels
scaled_data.YearsAtCompany.max()
#Create the synthetic labels.
synthetic_labels = numpy.random.rand(employee_data.shape[0]) < p * employee_data.YearsAtCompany
#Plot the data with the synthetic labels
sns.swarmplot(y='years', x='quit', data=pd.DataFrame({"quit":synthetic_labels, 'years':employee_data.YearsAtCompany}));
#We expect the probability based on the synthesized data (but we are getting something else....) to be close to p
synthetic_labels.sum()/len(synthetic_labels)/employee_data.YearsAtCompany.mean()
```
Indeed pretty close to the value of p we set beforehand
## Logistic Regression with the synthetic labels
In this version of the POC we will use TensorFlow
We need to add ones to the dataframe.
But since we scaled everything to be between `0` and `40`, the convergence will be faster if we add `40.0` instead of `1`
```
#Add 1 to the employee data.
#But to make convergence fa
scaled_data['Ones'] = 40.0
scaled_data
def reset_graph(seed=1239):
tf.reset_default_graph()
tf.set_random_seed(seed)
np.random.seed(seed)
def create_year_column(X, w, year):
year_term = tf.reshape(X[:,0]-year, (-1,1)) * w[0]
year_column = tf.reshape(X @ w - year_term,(-1,))
return year_column * tf.cast(tf.greater(X[:,0],year), dtype=tf.float32)
def logit(X, w):
'''
IMPORTANT: This assumes that the weight for the temporal variable is w[0]
TODO: Remove this assumption and allow to specify the index of the temporal variable
'''
max_year_tf = tf.reduce_max(X[:,0])
tensors = tf.map_fn(lambda year: create_year_column(X, w, year), tf.range(max_year_tf))
return tf.transpose(tensors)
logit_result = logit(X,weights)
init = tf.global_variables_initializer()
with tf.Session() as sess:
sess.run(init)
result = logit_result.eval()
result[1]
def get_loss(X, y, w):
'''
The loss function
'''
#The first term
logit_ = logit(X, w)
temp_tensor = tf.sigmoid(logit_) * tf.cast(tf.greater(logit_, 0), tf.float32)
sum_loss = tf.reduce_sum(tf.log(1-temp_tensor),1)
sum_loss = tf.reshape(sum_loss, (-1,1))
logistic_prob = tf.sigmoid(X @ w)
return -sum_loss - y * tf.log(logistic_prob) - (1-y) * tf.log(1-logistic_prob)
loss_result = get_loss(X, y, weights/100)
init = tf.global_variables_initializer()
with tf.Session() as sess:
sess.run(init)
result = loss_result.eval()
result
reset_graph()
learning_rate = 0.0005
l2 = 2.0
X = tf.constant(scaled_data.values, dtype=tf.float32, name="X")
y = tf.constant(synthetic_labels.values.reshape(-1, 1), dtype=tf.float32, name="y")
weights = tf.Variable(tf.random_uniform([scaled_data.values.shape[1], 1], -0.01, 0.01, seed=1239), name="weights")
loss = get_loss(X, y, weights)
l2_regularizer = tf.nn.l2_loss(weights) - 0.5 * weights[-1] ** 2
cost = tf.reduce_mean(loss) + l2 * l2_regularizer
optimizer = tf.train.GradientDescentOptimizer(learning_rate)
training_op = optimizer.minimize(cost)
init = tf.global_variables_initializer()
n_epochs = 20000
with tf.Session() as sess:
sess.run(init)
for epoch in range(n_epochs):
if epoch % 1000 == 0:
print("Epoch", epoch, "Cost =", cost.eval())
print(f'w: {weights[-1].eval()}')
sess.run(training_op)
best_theta = weights.eval()
```
The cost will never go down to zero, because of the additional term in the loss function.
```
#We will print the learned weights.
learned_weights = [(column_name,float(best_theta[column_num])) \
for column_num, column_name in enumerate(scaled_data.columns)]
#We print the weights sorted by the absolute value of the value
sorted(learned_weights, key=lambda x: abs(x[1]), reverse=True)
```
To compare with the other result we need to multiplty the last weight by 40
```
print(f'The predicted probability is: {float(1/(1+np.exp(-best_theta[-1]*40)))}')
```
This is very close indeed to the value `0.01` we created for the synthetic dataset of
| github_jupyter |
```
# Configuration --- Change to your setup and preferences!
CAFFE_ROOT = "~/caffe2"
# What image do you want to test? Can be local or URL.
# IMAGE_LOCATION = "images/cat.jpg"
# IMAGE_LOCATION = "https://upload.wikimedia.org/wikipedia/commons/thumb/f/f8/Whole-Lemon.jpg/1235px-Whole-Lemon.jpg"
# IMAGE_LOCATION = "https://upload.wikimedia.org/wikipedia/commons/7/7b/Orange-Whole-%26-Split.jpg"
# IMAGE_LOCATION = "https://upload.wikimedia.org/wikipedia/commons/7/7c/Zucchini-Whole.jpg"
# IMAGE_LOCATION = "https://upload.wikimedia.org/wikipedia/commons/a/ac/Pretzel.jpg"
IMAGE_LOCATION = "https://cdn.pixabay.com/photo/2015/02/10/21/28/flower-631765_1280.jpg"
# What model are we using? You should have already converted or downloaded one.
# format below is the model's:
# folder, init_net, predict_net, mean, input image size
# you can switch the comments on MODEL to try out different model conversions
MODEL = 'squeezenet', 'init_net.pb', 'run_net.pb', 'ilsvrc_2012_mean.npy', 227
# googlenet will fail with "enforce fail at fully_connected_op.h:25"
# MODEL = 'bvlc_googlenet', 'init_net.pb', 'predict_net.pb', 'ilsvrc_2012_mean.npy', 224
# these will run out of memory and fail... waiting for C++ version of predictor
# MODEL = 'bvlc_alexnet', 'init_net.pb', 'predict_net.pb', 'ilsvrc_2012_mean.npy', 224
# MODEL = 'finetune_flickr_style', 'init_net.pb', 'predict_net.pb', 'ilsvrc_2012_mean.npy', 224
# The list of output codes for the AlexNet models (squeezenet)
codes = "https://gist.githubusercontent.com/maraoz/388eddec39d60c6d52d4/raw/791d5b370e4e31a4e9058d49005be4888ca98472/gistfile1.txt"
print "Config set!"
%matplotlib inline
from caffe2.proto import caffe2_pb2
import numpy as np
import skimage.io
import skimage.transform
from matplotlib import pyplot
import os
from caffe2.python import core, workspace
import urllib2
print("Required modules imported.")
def crop_center(img,cropx,cropy):
y,x,c = img.shape
startx = x//2-(cropx//2)
starty = y//2-(cropy//2)
return img[starty:starty+cropy,startx:startx+cropx]
def rescale(img, input_height, input_width):
print("Original image shape:" + str(img.shape) + " and remember it should be in H, W, C!")
print("Model's input shape is %dx%d") % (input_height, input_width)
aspect = img.shape[1]/float(img.shape[0])
print("Orginal aspect ratio: " + str(aspect))
if(aspect>1):
# landscape orientation - wide image
res = int(aspect * input_height)
imgScaled = skimage.transform.resize(img, (input_width, res))
if(aspect<1):
# portrait orientation - tall image
res = int(input_width/aspect)
imgScaled = skimage.transform.resize(img, (res, input_height))
if(aspect == 1):
imgScaled = skimage.transform.resize(img, (input_width, input_height))
pyplot.figure()
pyplot.imshow(imgScaled)
pyplot.axis('on')
pyplot.title('Rescaled image')
print("New image shape:" + str(imgScaled.shape) + " in HWC")
return imgScaled
print "Functions set."
# set paths and variables from model choice
CAFFE_ROOT = os.path.expanduser(CAFFE_ROOT)
CAFFE_MODELS = os.path.join(CAFFE_ROOT, 'models')
MEAN_FILE = os.path.join(CAFFE_MODELS, MODEL[0], MODEL[3])
if not os.path.exists(MEAN_FILE):
mean = 128
else:
mean = np.load(MEAN_FILE).mean(1).mean(1)
mean = mean[:, np.newaxis, np.newaxis]
print "mean was set to: ", mean
INPUT_IMAGE_SIZE = MODEL[4]
if not os.path.exists(CAFFE_ROOT):
print("Houston, you may have a problem.")
INIT_NET = os.path.join(CAFFE_MODELS, MODEL[0], MODEL[1])
PREDICT_NET = os.path.join(CAFFE_MODELS, MODEL[0], MODEL[2])
if not os.path.exists(INIT_NET):
print(INIT_NET + " not found!")
else:
print "Found ", INIT_NET, "...Now looking for", PREDICT_NET
if not os.path.exists(PREDICT_NET):
print "Caffe model file, " + PREDICT_NET + " was not found!"
else:
print "All needed files found! Loading the model in the next block."
# initialize the neural net
p = workspace.Predictor(INIT_NET, PREDICT_NET)
# load and transform image
img = skimage.img_as_float(skimage.io.imread(IMAGE_LOCATION)).astype(np.float32)
img = rescale(img, INPUT_IMAGE_SIZE, INPUT_IMAGE_SIZE)
img = crop_center(img, INPUT_IMAGE_SIZE, INPUT_IMAGE_SIZE)
print "After crop: " , img.shape
pyplot.figure()
pyplot.imshow(img)
pyplot.axis('on')
pyplot.title('Cropped')
# switch to CHW
img = img.swapaxes(1, 2).swapaxes(0, 1)
pyplot.figure()
for i in range(3):
# For some reason, pyplot subplot follows Matlab's indexing
# convention (starting with 1). Well, we'll just follow it...
pyplot.subplot(1, 3, i+1)
pyplot.imshow(img[i])
pyplot.axis('off')
pyplot.title('RGB channel %d' % (i+1))
# switch to BGR
img = img[(2, 1, 0), :, :]
# remove mean for better results
img = img * 255 - mean
# add batch size
img = img[np.newaxis, :, :, :].astype(np.float32)
print "NCHW: ", img.shape
# run the net and return prediction
results = p.run([img])
results = np.asarray(results)
results = np.delete(results, 1)
index = 0
highest = 0
arr = np.empty((0,2), dtype=object)
arr[:,0] = int(10)
arr[:,1:] = float(10)
for i, r in enumerate(results):
# imagenet index begins with 1!# imagenet index begins with 1!
i=i+1
arr = np.append(arr, np.array([[i,r]]), axis=0)
if (r > highest):
highest = r
index = i
print index, " :: ", highest
# top 3
# sorted(arr, key=lambda x: x[1], reverse=True)[:3]
response = urllib2.urlopen(codes)
for line in response:
code, result = line.partition(":")[::2]
if (code.strip() == str(index)):
print result.strip()[1:-2]
```
Check [this list](https://gist.github.com/maraoz/388eddec39d60c6d52d4) to verify the results.
| github_jupyter |
# LassoLars Regression with Robust Scaler
This Code template is for the regression analysis using a simple LassoLars Regression. It is a lasso model implemented using the LARS algorithm and feature scaling using Robust Scaler in a Pipeline
### Required Packages
```
import warnings
import numpy as np
import pandas as pd
import seaborn as se
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import RobustScaler
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.linear_model import LassoLars
warnings.filterwarnings('ignore')
```
### Initialization
Filepath of CSV file
```
#filepath
file_path= ""
```
List of features which are required for model training .
```
#x_values
features=[]
```
Target feature for prediction.
```
#y_value
target=''
```
### Data Fetching
Pandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.
We will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry.
```
df=pd.read_csv(file_path)
df.head()
```
### Feature Selections
It is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.
We will assign all the required input features to X and target/outcome to Y.
```
X=df[features]
Y=df[target]
```
### Data Preprocessing
Since the majority of the machine learning models in the Sklearn library doesn't handle string category data and Null value, we have to explicitly remove or replace null values. The below snippet have functions, which removes the null value if any exists. And convert the string classes data in the datasets by encoding them to integer classes.
```
def NullClearner(df):
if(isinstance(df, pd.Series) and (df.dtype in ["float64","int64"])):
df.fillna(df.mean(),inplace=True)
return df
elif(isinstance(df, pd.Series)):
df.fillna(df.mode()[0],inplace=True)
return df
else:return df
def EncodeX(df):
return pd.get_dummies(df)
```
Calling preprocessing functions on the feature and target set.
```
x=X.columns.to_list()
for i in x:
X[i]=NullClearner(X[i])
X=EncodeX(X)
Y=NullClearner(Y)
X.head()
```
#### Correlation Map
In order to check the correlation between the features, we will plot a correlation matrix. It is effective in summarizing a large amount of data where the goal is to see patterns.
```
f,ax = plt.subplots(figsize=(18, 18))
matrix = np.triu(X.corr())
se.heatmap(X.corr(), annot=True, linewidths=.5, fmt= '.1f',ax=ax, mask=matrix)
plt.show()
```
### Data Splitting
The train-test split is a procedure for evaluating the performance of an algorithm. The procedure involves taking a dataset and dividing it into two subsets. The first subset is utilized to fit/train the model. The second subset is used for prediction. The main motive is to estimate the performance of the model on new data.
```
x_train,x_test,y_train,y_test=train_test_split(X,Y,test_size=0.2,random_state=123)
```
### Model
LassoLars is a lasso model implemented using the LARS algorithm, and unlike the implementation based on coordinate descent, this yields the exact solution, which is piecewise linear as a function of the norm of its coefficients.
### Tuning parameters
> **fit_intercept** -> whether to calculate the intercept for this model. If set to false, no intercept will be used in calculations
> **alpha** -> Constant that multiplies the penalty term. Defaults to 1.0. alpha = 0 is equivalent to an ordinary least square, solved by LinearRegression. For numerical reasons, using alpha = 0 with the LassoLars object is not advised and you should prefer the LinearRegression object.
> **eps** -> The machine-precision regularization in the computation of the Cholesky diagonal factors. Increase this for very ill-conditioned systems. Unlike the tol parameter in some iterative optimization-based algorithms, this parameter does not control the tolerance of the optimization.
> **max_iter** -> Maximum number of iterations to perform.
> **positive** -> Restrict coefficients to be >= 0. Be aware that you might want to remove fit_intercept which is set True by default. Under the positive restriction the model coefficients will not converge to the ordinary-least-squares solution for small values of alpha. Only coefficients up to the smallest alpha value (alphas_[alphas_ > 0.].min() when fit_path=True) reached by the stepwise Lars-Lasso algorithm are typically in congruence with the solution of the coordinate descent Lasso estimator.
> **precompute** -> Whether to use a precomputed Gram matrix to speed up calculations.
### Feature Scaling
Robust Scaler scale features using statistics that are robust to outliers.
This Scaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range). The IQR is the range between the 1st quartile (25th quantile) and the 3rd quartile (75th quantile).<br>
For more information... [click here](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.RobustScaler.html)
```
model=make_pipeline(RobustScaler(),LassoLars())
model.fit(x_train,y_train)
```
#### Model Accuracy
We will use the trained model to make a prediction on the test set.Then use the predicted value for measuring the accuracy of our model.
score: The score function returns the coefficient of determination R2 of the prediction.
```
print("Accuracy score {:.2f} %\n".format(model.score(x_test,y_test)*100))
```
> **r2_score**: The **r2_score** function computes the percentage variablility explained by our model, either the fraction or the count of correct predictions.
> **mae**: The **mean abosolute error** function calculates the amount of total error(absolute average distance between the real data and the predicted data) by our model.
> **mse**: The **mean squared error** function squares the error(penalizes the model for large errors) by our model.
```
y_pred=model.predict(x_test)
print("R2 Score: {:.2f} %".format(r2_score(y_test,y_pred)*100))
print("Mean Absolute Error {:.2f}".format(mean_absolute_error(y_test,y_pred)))
print("Mean Squared Error {:.2f}".format(mean_squared_error(y_test,y_pred)))
```
#### Prediction Plot
First, we make use of a plot to plot the actual observations, with x_train on the x-axis and y_train on the y-axis.
For the regression line, we will use x_train on the x-axis and then the predictions of the x_train observations on the y-axis.
```
plt.figure(figsize=(14,10))
plt.plot(range(20),y_test[0:20], color = "green")
plt.plot(range(20),model.predict(x_test[0:20]), color = "red")
plt.legend(["Actual","prediction"])
plt.title("Predicted vs True Value")
plt.xlabel("Record number")
plt.ylabel(target)
plt.show()
```
#### Creator: Anu Rithiga , Github: [Profile](https://github.com/iamgrootsh7)
| github_jupyter |
# SLAM算法介绍
## 1. 名词解释:
### 1.1 什么是SLAM?
SLAM,即Simultaneous localization and mapping,中文可译作“同时定位与地图构建”。它描述的是这样一类过程:机器人在陌生环境中运动,通过处理各类传感器收集的机器人自身及环境信息,精确地获取对机器人自身位置的估计(即“定位”),再通过机器人自身位置确定周围环境特征的位置(即“建图”)
在SLAM过程中,机器人不断地在收集各类传感器信息,如激光雷达的点云、相机的图像、imu的信息、里程计的信息等,通过对这些不断变化的传感器的一系列分析计算,机器人会实时地得出自身行进的轨迹(比如一系列时刻的位姿),但得到的轨迹往往包含很大误差,因此需要进行修正优化,修正的过程很可能不再是实时进行的。实时得出自身行进轨迹的过程一般称作“前端”,修正优化的过程一般称作“后端”。
实现后端优化的处理方法可以分为滤波和优化两类。
### 1.2 什么是滤波?
滤波在一般工程领域指的是根据一定规则对信号进行筛选,保留需要的内容,如各种高通滤波、低通滤波、带通滤波等。但在SLAM算法的语境下,滤波指的是“贝叶斯滤波”概念下的一系列“滤波器”,它们通过概率分析,使用传感器读数、传感器参数、机器人上一时刻位姿等信息,对机器人的下一时刻位姿作出修正:机器人不够准确的粗略轨迹经过”过滤“,变得更准确了。
SLAM中常见滤波有:EKF扩展卡尔曼滤波、UKF无迹卡尔曼滤波、particle filter粒子滤波等。
### 1.3 什么是优化问题?什么是非线性最小二乘优化问题?
各种滤波手段在SLAM问题中曾经占据主导地位,但随着地图规模的扩大(如机器人行进的面积范围增大、引入视觉算法后地图更“精细”),滤波方法所需要的计算量会不断增大。因此现阶段各种优化算法成为了SLAM问题后端处理方法的主流。
什么是优化问题呢?假设有一个函数f,以x为输入,以y为输出,那么一个优化问题就是通过某种手段找到一个x,使y的值最大/最小。而一个SLAM问题的优化中,x通常指的是各种待确定的状态量,比如机器人在各个时刻的位姿、地图中特征点的空间位置等,y通常指的是各种误差,比如传感器测量的量与状态量的差。SLAM问题待优化的函数f通常是非线性的,而且是以二次方项加和的形式存在的,因此属于非线性最小二乘优化问题。
解决非线性优化的开源库如google的Ceres,应用于cartographer、VINS等算法中。
### 1.4 什么是图优化?
图优化指的是把一个优化问题以一个“图”(graph)的形式表示出来(注:这里的”图“可以看做是指一种特殊的数据结构),可以用到图论相关的性质和算法,本质上还是一个优化问题。可以简单理解:待优化的状态量,即机器人在各个时刻的位姿、地图中特征点的空间位置,可以表示为graph的各个顶点,相关的顶点间以边连接,各个边代表的就是误差项,所以图优化问题就是通过优化各个顶点的”位置“,使所有的边加起来的和最小。
解决图优化的开源库如g2o,应用于ORB SLAM等算法中。
### 1.5 什么是约束?
在图优化问题中,顶点与顶点间连接的边就称为一个“约束”(constraint),这个约束可以表示如激光测量量与位置状态量之间的差值、imu测量量与位置状态量之间的差值等。
### 1.6 什么是回环检测
回环检测,也可以称为闭环检测等。简单理解就是,机器人“看到”了看到过的场景,就叫做回环检测成功。回环检测在SLAM问题中,对后端优化具有重要作用。
### 1.7 一个最简单的例子:
[graph slam tutorial : 从推导到应用1](https://heyijia.blog.csdn.net/article/details/47686523)
## 2. 举例分析
主武器与辅助武器:
对于一辆坦克来说,炮塔中央的主炮显然就是主武器,其他辅助武器可以有:机枪、反坦克导弹等。
相似地,对于激光slam算法,激光雷达是主武器,imu、里程计等属于辅助武器;对于视觉slam算法,相机就是主武器,imu、里程计等属于辅助武器。
### 2.1 激光slam举例:
cartographer

在SLAM问题的工程实践中,所谓的非线性优化,其实不止出现在后端的全局优化阶段。以google的cartographer为例:
算法前端接收一帧接一帧的激光扫描数据scans,插入到一个小范围的子图(submap)中(比如规定90帧scans组成一个子图),通过调用非线性优化解算库Ceres解决scan在submap中的插入位置问题,在这个优化过程中,imu和里程计负责提供初始值;后端负责进行“回环检测”,寻找新建立的子图submap和之前的scan间的约束,调用非线性优化解算库Ceres计算这个约束,使用一种叫”分支定界“的方法提供这类优化的初始值;最终,后端还要根据约束对所有已有的scan和submap进行全局优化,再次调用非线性优化解算库Ceres解决这个问题。
所以可以粗略地认为,在cartographer中有三处都运用了非线性优化。
### 2.2 视觉slam举例:
VINS-mono

港科大的VINS是视觉融合imu信息处理SLAM问题的典范。以单目视觉算法为主的VINS-mono为例:
首先进行”初始化“步骤,在此步骤中,视觉图像和imu信息互相辅助,imu解决了单目图像无法测量深度的问题,并提供了重力方向,视觉图像标定了imu的某些内部参数;
通过”滑窗“方法,使用图像、imu信息建立非线性优化问题,解算每帧图像的优化后位姿,以上内容组成了VIO,即所谓”视觉imu里程计“,可以算是前端的内容,但实际上这个前端也是在使用非线性优化在一直优化每帧的位姿的。
如果回环检测成功检测到了闭环,那么通过非线性优化进行”重定位“,调整滑窗内的位姿;最终通过全局优化,使用非线性优化方法修正所有帧的位姿。
以下是论文中对于重定位及全局优化的配图:

为便于理解,总结一下imu在不同slam算法中的作用:
1. imu在cartographer中的主要作用:通过scan match插入一帧激光建立submap前,预估机器人新位姿,给非线性优化提供初始值。
2. imu在VINS中的主要作用:在“初始化”阶段,获取图像深度尺度等参数;参与VIO优化约束建立。
| github_jupyter |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import gc
plt.style.use('ggplot')
dtypes = {
'ip' : 'uint32',
'app' : 'uint16',
'device' : 'uint16',
'os' : 'uint16',
'channel' : 'uint16',
'is_attributed' : 'uint8',
}
random = pd.read_csv('train_random_10_percent.csv', dtype=dtypes)
df = random.sample(3000000)
# prepare test data
test = pd.read_csv("test.csv", dtype=dtypes)
df = df.sort_values(['ip','click_time'])
test = test.sort_values(['ip','click_time'])
df.shape
gc.collect()
df['click_time'] = pd.to_datetime(df.click_time)
df['attributed_time'] = pd.to_datetime(df.attributed_time)
test['click_time'] = pd.to_datetime(test.click_time)
did_download = df[df.is_attributed==1].ip.values
did_download
df[df.is_attributed==1]
#ip of people that downloaded an application at some point
did_download = df[df.ip.apply(lambda x: x in did_download)]
did_download
did_download.shape
ip_ad_exposure = did_download.ip.value_counts()
ip_ad_exposure
app_or_channel = did_download[did_download.is_attributed == 1]
app_or_channel.shape
downloaded = did_download.dropna()
#lets explore more just the adds that led to download
time_of_exposure = did_download.attributed_time.dropna().groupby(did_download.attributed_time.dt.hour).count()
time_of_exposure
t = downloaded.attributed_time - downloaded.click_time
channel_success = did_download.groupby(['channel']).is_attributed.mean()
channel_success.head(10)
app_success = did_download.groupby(['app']).is_attributed.mean()
channel_success = channel_success.to_dict()
app_success = app_success.to_dict()
df['channel_success'] = df.channel.map(channel_success)
df['app_success'] = df.channel.map(app_success)
df.channel_success.fillna(0,inplace=True)
df.app_success.fillna(0,inplace=True)
df.head(10)
s = df.groupby(['ip']).os.value_counts().to_frame().rename(columns={'os':'ip_os_count'}).reset_index()
u = test.groupby(['ip']).os.value_counts().to_frame().rename(columns={'os':'ip_os_count'}).reset_index()
s.head(10)
gc.collect()
df = pd.merge(df,s,on=['ip','os'])
df['ip_os_count'] = df.ip_os_count.astype('float')
test = pd.merge(test,u,on=['ip','os'])
test['ip_os_count'] = test.ip_os_count.astype('float')
df.head(10)
n_chans = df.groupby(['ip','app']).channel.count().reset_index().rename(columns={'channel':'ip_app_count'})
df = df.merge(n_chans,on=['ip','app'],how='left')
x_chans = test.groupby(['ip','app']).channel.count().reset_index().rename(columns={'channel':'ip_app_count'})
test = test.merge(x_chans,on=['ip','app'],how='left')
test.head(10)
df['clicked'] = np.ones(df.shape[0],dtype= np.float64)
df['app_exposure'] = df.groupby(['ip','app',]).clicked.cumsum()
df['channel_exposure'] = df.groupby(['ip','channel',]).clicked.cumsum()
test['clicked'] = np.ones(test.shape[0],dtype= np.float64)
test['app_exposure'] = test.groupby(['ip','app',]).clicked.cumsum()
test['channel_exposure'] = test.groupby(['ip','channel',]).clicked.cumsum()
df.head(10)
df['daily_usage'] = df.groupby(['ip',df.click_time.dt.day]).clicked.cumsum()
df.head(10)
df['hour'] = df.click_time.dt.hour
df['hour_cumative_clicks'] = df.groupby(['ip',df.click_time.dt.hour]).clicked.cumsum()
df.head(10)
gc.collect()
test['daily_usage'] = test.groupby(['ip', test.click_time.dt.day]).clicked.cumsum()
test['hour'] = test.click_time.dt.hour
test['hour_cumative_clicks'] = test.groupby(['ip', test.click_time.dt.hour]).clicked.cumsum()
gc.collect()
from sklearn.model_selection import train_test_split
X = df[['app','device','os','channel','app_exposure','daily_usage','hour','hour_cumative_clicks','ip_os_count']]
y = df.is_attributed
X_test = test[['app','device','os','channel','app_exposure','daily_usage','hour','hour_cumative_clicks','ip_os_count']]
gc.collect()
from catboost import CatBoostClassifier
categorical_features_indices = np.where(X.dtypes != np.float)[0]
categorical_features_indices = np.where(X_test.dtypes != np.float)[0]
cat = CatBoostClassifier()
model = cat.fit(X, y,cat_features=categorical_features_indices,plot=False,verbose=True)
y_pred_prob = model.predict_proba(X_test)
gc.collect()
output = pd.DataFrame(test['click_id'])
output['is_attributed'] = y_pred_prob[:,1]
output = output.set_index('click_id')
output.to_csv("submission_stackF.csv")
```
| github_jupyter |
# 2章 微分積分
## 2.1 関数
```
# 必要ライブラリの宣言
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# PDF出力用
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('png', 'pdf')
def f(x):
return x**2 +1
f(1)
f(2)
```
### 図2-2 点(x, f(x))のプロットとy=f(x)のグラフ
```
x = np.linspace(-3, 3, 601)
y = f(x)
x1 = np.linspace(-3, 3, 7)
y1 = f(x1)
plt.figure(figsize=(6,6))
plt.ylim(-2,10)
plt.plot([-3,3],[0,0],c='k')
plt.plot([0,0],[-2,10],c='k')
plt.scatter(x1,y1,c='k',s=50)
plt.grid()
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.show()
x2 = np.linspace(-3, 3, 31)
y2 = f(x2)
plt.figure(figsize=(6,6))
plt.ylim(-2,10)
plt.plot([-3,3],[0,0],c='k')
plt.plot([0,0],[-2,10],c='k')
plt.scatter(x2,y2,c='k',s=50)
plt.grid()
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.show()
plt.figure(figsize=(6,6))
plt.plot(x,y,c='k')
plt.ylim(-2,10)
plt.plot([-3,3],[0,0],c='k')
plt.plot([0,0],[-2,10],c='k')
plt.scatter([1,2],[2,5],c='k',s=50)
plt.grid()
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.show()
```
## 2.2 合成関数・逆関数
### 図2.6 逆関数のグラフ
```
def f(x):
return(x**2 + 1)
def g(x):
return(np.sqrt(x - 1))
xx1 = np.linspace(0.0, 4.0, 200)
xx2 = np.linspace(1.0, 4.0, 200)
yy1 = f(xx1)
yy2 = g(xx2)
plt.figure(figsize=(6,6))
plt.xlabel('$x$',fontsize=14)
plt.ylabel('$y$',fontsize=14)
plt.ylim(-2.0, 4.0)
plt.xlim(-2.0, 4.0)
plt.grid()
plt.plot(xx1,yy1, linestyle='-', c='k', label='$y=x^2+1$')
plt.plot(xx2,yy2, linestyle='-.', c='k', label='$y=\sqrt{x-1}$')
plt.plot([-2,4],[-2,4], color='black')
plt.plot([-2,4],[0,0], color='black')
plt.plot([0,0],[-2,4],color='black')
plt.legend(fontsize=14)
plt.show()
```
## 2.3 微分と極限
### 図2-7 関数のグラフを拡大したときの様子
```
from matplotlib import pyplot as plt
import numpy as np
def f(x):
return(x**3 - x)
delta = 2.0
x = np.linspace(0.5-delta, 0.5+delta, 200)
y = f(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-3.0/8.0-delta, -3.0/8.0+delta)
plt.xlim(0.5-delta, 0.5+delta)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.scatter([0.5], [-3.0/8.0])
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.grid()
plt.title('delta = %.4f' % delta, fontsize=14)
plt.show()
delta = 0.2
x = np.linspace(0.5-delta, 0.5+delta, 200)
y = f(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-3.0/8.0-delta, -3.0/8.0+delta)
plt.xlim(0.5-delta, 0.5+delta)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.scatter([0.5], [-3.0/8.0])
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.grid()
plt.title('delta = %.4f' % delta, fontsize=14)
plt.show()
delta = 0.01
x = np.linspace(0.5-delta, 0.5+delta, 200)
y = f(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-3.0/8.0-delta, -3.0/8.0+delta)
plt.xlim(0.5-delta, 0.5+delta)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.scatter(0.5, -3.0/8.0)
plt.xlabel('x',fontsize=14)
plt.ylabel('y',fontsize=14)
plt.grid()
plt.title('delta = %.4f' % delta, fontsize=14)
plt.show()
```
### 図2-8 関数のグラフ上の2点を結んだ直線の傾き
```
delta = 2.0
x = np.linspace(0.5-delta, 0.5+delta, 200)
x1 = 0.6
x2 = 1.0
y = f(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-1, 0.5)
plt.xlim(0, 1.5)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.scatter([x1, x2], [f(x1), f(x2)], c='k', lw=1)
plt.plot([x1, x2], [f(x1), f(x2)], c='k', lw=1)
plt.plot([x1, x2, x2], [f(x1), f(x1), f(x2)], c='k', lw=1)
plt.tick_params(labelbottom=False, labelleft=False, labelright=False, labeltop=False)
plt.tick_params(color='white')
plt.show()
```
### 図2-10 接線の方程式
```
def f(x):
return(x**2 - 4*x)
def g(x):
return(-2*x -1)
x = np.linspace(-2, 6, 500)
fig = plt.figure(figsize=(6,6))
plt.scatter([1],[-3],c='k')
plt.plot(x, f(x), 'b-', lw=1, c='k')
plt.plot(x, g(x), 'b-', lw=1, c='b')
plt.plot([x.min(), x.max()], [0, 0], lw=2, c='k')
plt.plot([0, 0], [g(x).min(), f(x).max()], lw=2, c='k')
plt.grid(lw=2)
plt.tick_params(labelbottom=False, labelleft=False, labelright=False, labeltop=False)
plt.tick_params(color='white')
plt.xlabel('X')
plt.show()
```
## 2.4 極大・極小
### 図2-11 y= x3-3xのグラフと極大・極小
```
def f1(x):
return(x**3 - 3*x)
x = np.linspace(-3, 3, 500)
y = f1(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-4, 4)
plt.xlim(-3, 3)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.plot([0,0],[-4,4],c='k')
plt.plot([-3,3],[0,0],c='k')
plt.grid()
plt.show()
```
### 図2-12 極大でも極小でもない例 (y=x3のグラフ)
```
def f2(x):
return(x**3)
x = np.linspace(-3, 3, 500)
y = f2(x)
fig = plt.figure(figsize=(6,6))
plt.ylim(-4, 4)
plt.xlim(-3, 3)
plt.plot(x, y, 'b-', lw=1, c='k')
plt.plot([0,0],[-4,4],c='k')
plt.plot([-3,3],[0,0],c='k')
plt.grid()
plt.show()
```
## 2.7 合成関数の微分
### 図2-14 逆関数の微分
```
#逆関数の微分
def f(x):
return(x**2 + 1)
def g(x):
return(np.sqrt(x - 1))
xx1 = np.linspace(0.0, 4.0, 200)
xx2 = np.linspace(1.0, 4.0, 200)
yy1 = f(xx1)
yy2 = g(xx2)
plt.figure(figsize=(6,6))
plt.xlabel('$x$',fontsize=14)
plt.ylabel('$y$',fontsize=14)
plt.ylim(-2.0, 4.0)
plt.xlim(-2.0, 4.0)
plt.grid()
plt.plot(xx1,yy1, linestyle='-', color='blue')
plt.plot(xx2,yy2, linestyle='-', color='blue')
plt.plot([-2,4],[-2,4], color='black')
plt.plot([-2,4],[0,0], color='black')
plt.plot([0,0],[-2,4],color='black')
plt.show()
```
## 2.9 積分
### 図2-15 面積を表す関数S(x)とf(x)の関係
```
def f(x) :
return x**2 + 1
xx = np.linspace(-4.0, 4.0, 200)
yy = f(xx)
plt.figure(figsize=(6,6))
plt.xlim(-2,2)
plt.ylim(-1,4)
plt.plot(xx, yy)
plt.plot([-2,2],[0,0],c='k',lw=1)
plt.plot([0,0],[-1,4],c='k',lw=1)
plt.plot([0,0],[0,f(0)],c='b')
plt.plot([1,1],[0,f(1)],c='b')
plt.plot([1.5,1.5],[0,f(1.5)],c='b')
plt.plot([1,1.5],[f(1),f(1)],c='b')
plt.tick_params(labelbottom=False, labelleft=False, labelright=False, labeltop=False)
plt.tick_params(color='white')
plt.show()
```
### 図2-16 グラフの面積と定積分
```
plt.figure(figsize=(6,6))
plt.xlim(-2,2)
plt.ylim(-1,4)
plt.plot(xx, yy)
plt.plot([-2,2],[0,0],c='k',lw=1)
plt.plot([0,0],[-1,4],c='k',lw=1)
plt.plot([0,0],[0,f(0)],c='b')
plt.plot([1,1],[0,f(1)],c='b')
plt.plot([1.5,1.5],[0,f(1.5)],c='b')
plt.tick_params(labelbottom=False, labelleft=False, labelright=False, labeltop=False)
plt.tick_params(color='white')
plt.show()
```
### 図2-17 積分と面積の関係
```
def f(x) :
return x**2 + 1
x = np.linspace(-1.0, 2.0, 200)
y = f(x)
N = 10
xx = np.linspace(0.5, 1.5, N+1)
yy = f(xx)
print(xx)
plt.figure(figsize=(6,6))
plt.xlim(-1,2)
plt.ylim(-1,4)
plt.plot(x, y)
plt.plot([-1,2],[0,0],c='k',lw=2)
plt.plot([0,0],[-1,4],c='k',lw=2)
plt.plot([0.5,0.5],[0,f(0.5)],c='b')
plt.plot([1.5,1.5],[0,f(1.5)],c='b')
plt.bar(xx[:-1], yy[:-1], align='edge', width=1/N*0.9)
plt.tick_params(labelbottom=False, labelleft=False, labelright=False, labeltop=False)
plt.tick_params(color='white')
plt.grid()
plt.show()
```
| github_jupyter |
## 1. Meet Dr. Ignaz Semmelweis
<p><img style="float: left;margin:5px 20px 5px 1px" src="https://assets.datacamp.com/production/project_20/img/ignaz_semmelweis_1860.jpeg"></p>
<!--
<img style="float: left;margin:5px 20px 5px 1px" src="https://assets.datacamp.com/production/project_20/datasets/ignaz_semmelweis_1860.jpeg">
-->
<p>This is Dr. Ignaz Semmelweis, a Hungarian physician born in 1818 and active at the Vienna General Hospital. If Dr. Semmelweis looks troubled it's probably because he's thinking about <em>childbed fever</em>: A deadly disease affecting women that just have given birth. He is thinking about it because in the early 1840s at the Vienna General Hospital as many as 10% of the women giving birth die from it. He is thinking about it because he knows the cause of childbed fever: It's the contaminated hands of the doctors delivering the babies. And they won't listen to him and <em>wash their hands</em>!</p>
<p>In this notebook, we're going to reanalyze the data that made Semmelweis discover the importance of <em>handwashing</em>. Let's start by looking at the data that made Semmelweis realize that something was wrong with the procedures at Vienna General Hospital.</p>
```
# Importing modules
import pandas as pd
# Read datasets/yearly_deaths_by_clinic.csv into yearly
yearly = pd.read_csv("datasets/yearly_deaths_by_clinic.csv")
# Print out yearly
yearly
```
## 2. The alarming number of deaths
<p>The table above shows the number of women giving birth at the two clinics at the Vienna General Hospital for the years 1841 to 1846. You'll notice that giving birth was very dangerous; an <em>alarming</em> number of women died as the result of childbirth, most of them from childbed fever.</p>
<p>We see this more clearly if we look at the <em>proportion of deaths</em> out of the number of women giving birth. Let's zoom in on the proportion of deaths at Clinic 1.</p>
```
# Calculate proportion of deaths per no. births
yearly["proportion_deaths"] = yearly["deaths"] / yearly["births"]
# Extract Clinic 1 data into clinic_1 and Clinic 2 data into clinic_2
clinic_1 = yearly[yearly["clinic"] == "clinic 1"]
clinic_2 = yearly[yearly["clinic"] == "clinic 2"]
# Print out clinic_1
clinic_1
```
## 3. Death at the clinics
<p>If we now plot the proportion of deaths at both Clinic 1 and Clinic 2 we'll see a curious pattern…</p>
```
# This makes plots appear in the notebook
%matplotlib inline
# Plot yearly proportion of deaths at the two clinics
ax = clinic_1.plot(x="year", y="proportion_deaths", label="Clinic 1")
clinic_2.plot(x="year", y="proportion_deaths", label="Clinic 2", ax=ax, ylabel="Proportion deaths")
```
## 4. The handwashing begins
<p>Why is the proportion of deaths consistently so much higher in Clinic 1? Semmelweis saw the same pattern and was puzzled and distressed. The only difference between the clinics was that many medical students served at Clinic 1, while mostly midwife students served at Clinic 2. While the midwives only tended to the women giving birth, the medical students also spent time in the autopsy rooms examining corpses. </p>
<p>Semmelweis started to suspect that something on the corpses spread from the hands of the medical students, caused childbed fever. So in a desperate attempt to stop the high mortality rates, he decreed: <em>Wash your hands!</em> This was an unorthodox and controversial request, nobody in Vienna knew about bacteria at this point in time. </p>
<p>Let's load in monthly data from Clinic 1 to see if the handwashing had any effect.</p>
```
# Read datasets/monthly_deaths.csv into monthly
monthly = pd.read_csv("datasets/monthly_deaths.csv", parse_dates=["date"])
# Calculate proportion of deaths per no. births
monthly["proportion_deaths"] = monthly["deaths"] / monthly["births"]
# Print out the first rows in monthly
monthly.head()
```
## 5. The effect of handwashing
<p>With the data loaded we can now look at the proportion of deaths over time. In the plot below we haven't marked where obligatory handwashing started, but it reduced the proportion of deaths to such a degree that you should be able to spot it!</p>
```
# Plot monthly proportion of deaths
ax = monthly.plot(x="date", y="proportion_deaths", ylabel="Proportion deaths")
```
## 6. The effect of handwashing highlighted
<p>Starting from the summer of 1847 the proportion of deaths is drastically reduced and, yes, this was when Semmelweis made handwashing obligatory. </p>
<p>The effect of handwashing is made even more clear if we highlight this in the graph.</p>
```
# Date when handwashing was made mandatory
handwashing_start = pd.to_datetime('1847-06-01')
# Split monthly into before and after handwashing_start
before_washing = monthly[monthly["date"] < handwashing_start]
after_washing = monthly[monthly["date"] >= handwashing_start]
# Plot monthly proportion of deaths before and after handwashing
ax = before_washing.plot(x="date", y="proportion_deaths",
label="Before handwashing")
after_washing.plot(x="date", y="proportion_deaths",
label="After handwashing", ax=ax, ylabel="Proportion deaths")
```
## 7. More handwashing, fewer deaths?
<p>Again, the graph shows that handwashing had a huge effect. How much did it reduce the monthly proportion of deaths on average?</p>
```
# Difference in mean monthly proportion of deaths due to handwashing
before_proportion = before_washing["proportion_deaths"]
after_proportion = after_washing["proportion_deaths"]
mean_diff = after_proportion.mean() - before_proportion.mean()
mean_diff
```
## 8. A Bootstrap analysis of Semmelweis handwashing data
<p>It reduced the proportion of deaths by around 8 percentage points! From 10% on average to just 2% (which is still a high number by modern standards). </p>
<p>To get a feeling for the uncertainty around how much handwashing reduces mortalities we could look at a confidence interval (here calculated using the bootstrap method).</p>
```
# A bootstrap analysis of the reduction of deaths due to handwashing
boot_mean_diff = []
for i in range(3000):
boot_before = before_proportion.sample(frac=1, replace=True)
boot_after = after_proportion.sample(frac=1, replace=True)
boot_mean_diff.append( boot_after.mean() - boot_before.mean() )
# Calculating a 95% confidence interval from boot_mean_diff
confidence_interval = pd.Series(boot_mean_diff).quantile([0.025, 0.975])
confidence_interval
```
## 9. The fate of Dr. Semmelweis
<p>So handwashing reduced the proportion of deaths by between 6.7 and 10 percentage points, according to a 95% confidence interval. All in all, it would seem that Semmelweis had solid evidence that handwashing was a simple but highly effective procedure that could save many lives.</p>
<p>The tragedy is that, despite the evidence, Semmelweis' theory — that childbed fever was caused by some "substance" (what we today know as <em>bacteria</em>) from autopsy room corpses — was ridiculed by contemporary scientists. The medical community largely rejected his discovery and in 1849 he was forced to leave the Vienna General Hospital for good.</p>
<p>One reason for this was that statistics and statistical arguments were uncommon in medical science in the 1800s. Semmelweis only published his data as long tables of raw data, but he didn't show any graphs nor confidence intervals. If he would have had access to the analysis we've just put together he might have been more successful in getting the Viennese doctors to wash their hands.</p>
```
# The data Semmelweis collected points to that:
doctors_should_wash_their_hands = True
```
| github_jupyter |
```
import os
from glob import glob
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
```
## Cleaning Up (& Stats About It)
- For each annotator:
- How many annotation files?
- How many txt files?
- Number of empty .ann files
- How many non-empty .ann files have a `TranscriptionError_Document`/`DuplicatePage` tag?
- How many .ann files have ONLY one of those two tags and are empty o/w? -> remove if so
=> remove corresponding .txt files
=> create new corpus
```
def get_all_files(annotator):
""" collapsing folder structure per annotator"""
data_dir = "../Data/"
ann_dir = data_dir+annotator+"/"
for cur_dir in glob(ann_dir+"/6*"):
txt_files = sorted(glob(cur_dir+"/*.txt"))
ann_files = sorted(glob(cur_dir+"/*.ann"))
yield from zip(txt_files, ann_files)
def has_error_tag(any_string):
"""Return strings with error tags"""
return "TranscriptionError_Document" in any_string or\
"DuplicatePage" in any_string
def remove_error_tag_lines(ann_file_content):
return [line for line in ann_file_content.strip().split("\n")
if not has_error_tag(line)]
annotators = "A B C Silja Yolien".split()
results = {}
print("Total Annotation Files Per Annotator\n")
for anno in annotators:
empty = []
cur_keep = []
error_tag = []
error_tag_but_non_empty = []
ann_files = list(get_all_files(anno))
print(anno, len(ann_files))
for txt, ann in ann_files:
with open(ann) as handle:
contents = handle.read()
if not contents.strip():
empty.append((txt, ann))
elif has_error_tag(contents):
error_tags_removed = remove_error_tag_lines(
contents
)
if error_tags_removed == []:
error_tag.append((txt, ann))
else:
error_tag_but_non_empty.append((txt, ann))
else:
cur_keep.append((txt, ann))
results[anno] = [cur_keep, empty, error_tag, error_tag_but_non_empty]
from tabulate import tabulate
stats = pd.DataFrame([
[k, sum(map(len, v))]+
[len(v[0])+len(v[-1])]+
list(map(len, v)) for k, v in results.items()
],
columns=["Annotator", "Total", "Keep",
"Non-empty-No error", "Empty", "Error", "Err.&Non-Empty"]).set_index("Annotator")
print(stats)
stats_T = pd.melt(stats[["Total", "Empty", "Keep", "Error"]].reset_index(),
id_vars=["Annotator"], value_name="Number")
plt.figure(figsize=(10, 7))
sns.barplot(data=stats_T, x='Annotator', y="Number", hue="variable")
keep = {anno: v[0]+v[-1] for anno, v in results.items()}
{k: len(v) for k, v in keep.items()}
# keep
```
### Make New Corpus
by copying files
```
from shutil import copy2
already_copied = True
if not already_copied:
from tqdm import tqdm
os.makedirs('Keep')
for anno, ls in tqdm(keep.items()):
cur_dir = f"Keep/{anno}"
os.makedirs(cur_dir)
for txt, ann in ls:
copy2(txt, cur_dir)
copy2(ann, cur_dir)
else:
print("Already copied, doing nothing!")
```
# Pairwise Intersections of Annotation Files
```
def only_names(file_list):
"returns only names of files in a particular list"
return [ann.split("/")[-1] for txt, ann in file_list]
ls = []
for a1, fs1 in keep.items():
for a2, fs2 in keep.items():
if not a1 == a2:
names1, names2 = only_names(fs1), only_names(fs2)
inter = set(names1) & set(names2) #names of files are identical
val = len(inter)/len(names1)
total_names1 = only_names(tup for ls in results[a1] for tup in ls)
total_names2 = only_names(tup for ls in results[a2] for tup in ls)
total_inter = set(total_names1) & set(total_names2)
total_val = len(total_inter)/len(total_names1)
jacc_val = len(set(names1).intersection(set(names2)))/len(set(names1).union(set(names2)))
jacc_val_2 = len(set(total_names1).intersection(set(total_names2)))/len(set(total_names1).union(set(total_names2)))
ls.append([a1, a2, len(inter), val,
len(total_inter), total_val, jacc_val, jacc_val_2])
inter_stats = pd.DataFrame(ls,
columns=["Anno1", "Anno2",
"Intersection", "normed_Intersection",
"total_Intersection", "total_normed_Intersection", "Jaccard_distance", "Jaccard_Distance_2"])
# inter_stats
```
#### Jaccard Distance to Understand Overlap Pages between Annotators
```
inter_stats_T = inter_stats.pivot_table(
values="Jaccard_distance",
index="Anno1", columns="Anno2"
)
sns.heatmap(inter_stats_T*100, annot=True, cmap="YlGnBu")
_ = plt.title("Before Clean Up: Jaccard Distance (percentage)")
plt.show()
inter_stats_T = inter_stats.pivot_table(
values="Jaccard_Distance_2",
index="Anno1", columns="Anno2"
)
sns.heatmap(inter_stats_T*100, annot=True, cmap="YlGnBu")
_ = plt.title("After Clean Up: Jaccard Distance (percentage)")
plt.show()
# inter_stats_T = inter_stats.pivot_table(
# values="Intersection",
# index="Anno1", columns="Anno2"
# )
# sns.heatmap(inter_stats_T,
# annot=True, cmap="YlGnBu")
# _ = plt.title("Before Clean Up: Raw Counts")
```
**Conclusion**: Each pair of annotators annotated on average have 6% overlap (over the total documents they annotated together).
## Check Tag Distributions
```
def get_lines(ann_file):
with open(ann_file) as handle:
for l in handle:
if not l.strip(): continue
yield l.strip().split("\t")
def get_entities(ann_file):
for line in get_lines(ann_file):
if line[0].startswith("T") and len(line) >= 2:
tag_type, tag, string = line
yield tag.split()[0]
ents = {a: [e for txt, ann in files for e in get_entities(ann)]
for a, files in keep.items()}
from collections import Counter
entity_stats = pd.DataFrame(
[[a, e, c] for a in ents for e, c in Counter(ents[a]).items() if not e in ["DuplicatePage", "Noteworthy", "TranscriptionError_Document"]],
columns=["Annotator", "EntityType", "Count"]
)
plt.figure(figsize=(10, 7))
_ = sns.barplot(data=entity_stats, x='Annotator', y="Count", hue="EntityType")
```
**Conclusion**:
Here we see that most annotators follow a similar trend in entities annotated, only annotator who stands out is '3'.
| github_jupyter |
# ORF recognition by CNN
Compare to ORF_CNN_101.
Use 2-layer CNN.
Run on Mac.
```
PC_SEQUENCES=20000 # how many protein-coding sequences
NC_SEQUENCES=20000 # how many non-coding sequences
PC_TESTS=1000
NC_TESTS=1000
BASES=1000 # how long is each sequence
ALPHABET=4 # how many different letters are possible
INPUT_SHAPE_2D = (BASES,ALPHABET,1) # Conv2D needs 3D inputs
INPUT_SHAPE = (BASES,ALPHABET) # Conv1D needs 2D inputs
FILTERS = 32 # how many different patterns the model looks for
NEURONS = 16
WIDTH = 3 # how wide each pattern is, in bases
STRIDE_2D = (1,1) # For Conv2D how far in each direction
STRIDE = 1 # For Conv1D, how far between pattern matches, in bases
EPOCHS=10 # how many times to train on all the data
SPLITS=5 # SPLITS=3 means train on 2/3 and validate on 1/3
FOLDS=5 # train the model this many times (range 1 to SPLITS)
import sys
try:
from google.colab import drive
IN_COLAB = True
print("On Google CoLab, mount cloud-local file, get our code from GitHub.")
PATH='/content/drive/'
#drive.mount(PATH,force_remount=True) # hardly ever need this
#drive.mount(PATH) # Google will require login credentials
DATAPATH=PATH+'My Drive/data/' # must end in "/"
import requests
r = requests.get('https://raw.githubusercontent.com/ShepherdCode/Soars2021/master/SimTools/RNA_gen.py')
with open('RNA_gen.py', 'w') as f:
f.write(r.text)
from RNA_gen import *
r = requests.get('https://raw.githubusercontent.com/ShepherdCode/Soars2021/master/SimTools/RNA_describe.py')
with open('RNA_describe.py', 'w') as f:
f.write(r.text)
from RNA_describe import *
r = requests.get('https://raw.githubusercontent.com/ShepherdCode/Soars2021/master/SimTools/RNA_prep.py')
with open('RNA_prep.py', 'w') as f:
f.write(r.text)
from RNA_prep import *
except:
print("CoLab not working. On my PC, use relative paths.")
IN_COLAB = False
DATAPATH='data/' # must end in "/"
sys.path.append("..") # append parent dir in order to use sibling dirs
from SimTools.RNA_gen import *
from SimTools.RNA_describe import *
from SimTools.RNA_prep import *
MODELPATH="BestModel" # saved on cloud instance and lost after logout
#MODELPATH=DATAPATH+MODELPATH # saved on Google Drive but requires login
if not assert_imported_RNA_gen():
print("ERROR: Cannot use RNA_gen.")
if not assert_imported_RNA_prep():
print("ERROR: Cannot use RNA_prep.")
from os import listdir
import time # datetime
import csv
from zipfile import ZipFile
import numpy as np
import pandas as pd
from scipy import stats # mode
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from keras.models import Sequential
from keras.layers import Dense,Embedding
from keras.layers import Conv1D,Conv2D
from keras.layers import Flatten,MaxPooling1D,MaxPooling2D
from keras.losses import BinaryCrossentropy
# tf.keras.losses.BinaryCrossentropy
import matplotlib.pyplot as plt
from matplotlib import colors
mycmap = colors.ListedColormap(['red','blue']) # list color for label 0 then 1
np.set_printoptions(precision=2)
t = time.time()
time.strftime('%Y-%m-%d %H:%M:%S %Z', time.localtime(t))
# Use code from our SimTools library.
def make_generators(seq_len):
pcgen = Collection_Generator()
pcgen.get_len_oracle().set_mean(seq_len)
pcgen.set_seq_oracle(Transcript_Oracle())
ncgen = Collection_Generator()
ncgen.get_len_oracle().set_mean(seq_len)
return pcgen,ncgen
pc_sim,nc_sim = make_generators(BASES)
pc_train = pc_sim.get_sequences(PC_SEQUENCES)
nc_train = nc_sim.get_sequences(NC_SEQUENCES)
print("Train on",len(pc_train),"PC seqs")
print("Train on",len(nc_train),"NC seqs")
# Use code from our LearnTools library.
X,y = prepare_inputs_len_x_alphabet(pc_train,nc_train,ALPHABET) # shuffles
print("Data ready.")
def make_DNN():
print("make_DNN")
print("input shape:",INPUT_SHAPE)
dnn = Sequential()
#dnn.add(Embedding(input_dim=INPUT_SHAPE,output_dim=INPUT_SHAPE))
dnn.add(Conv1D(filters=FILTERS,kernel_size=WIDTH,strides=STRIDE,padding="same",
input_shape=INPUT_SHAPE))
dnn.add(Conv1D(filters=FILTERS,kernel_size=WIDTH,strides=STRIDE,padding="same"))
dnn.add(MaxPooling1D())
dnn.add(Conv1D(filters=FILTERS,kernel_size=WIDTH,strides=STRIDE,padding="same"))
dnn.add(Conv1D(filters=FILTERS,kernel_size=WIDTH,strides=STRIDE,padding="same"))
dnn.add(MaxPooling1D())
dnn.add(Flatten())
dnn.add(Dense(NEURONS,activation="sigmoid",dtype=np.float32))
dnn.add(Dense(1,activation="sigmoid",dtype=np.float32))
dnn.compile(optimizer='adam',
loss=BinaryCrossentropy(from_logits=False),
metrics=['accuracy']) # add to default metrics=loss
dnn.build(input_shape=INPUT_SHAPE)
#ln_rate = tf.keras.optimizers.Adam(learning_rate = LN_RATE)
#bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)
#model.compile(loss=bc, optimizer=ln_rate, metrics=["accuracy"])
return dnn
model = make_DNN()
print(model.summary())
from keras.callbacks import ModelCheckpoint
def do_cross_validation(X,y):
cv_scores = []
fold=0
mycallbacks = [ModelCheckpoint(
filepath=MODELPATH, save_best_only=True,
monitor='val_accuracy', mode='max')]
splitter = KFold(n_splits=SPLITS) # this does not shuffle
for train_index,valid_index in splitter.split(X):
if fold < FOLDS:
fold += 1
X_train=X[train_index] # inputs for training
y_train=y[train_index] # labels for training
X_valid=X[valid_index] # inputs for validation
y_valid=y[valid_index] # labels for validation
print("MODEL")
# Call constructor on each CV. Else, continually improves the same model.
model = model = make_DNN()
print("FIT") # model.fit() implements learning
start_time=time.time()
history=model.fit(X_train, y_train,
epochs=EPOCHS,
verbose=1, # ascii art while learning
callbacks=mycallbacks, # called at end of each epoch
validation_data=(X_valid,y_valid))
end_time=time.time()
elapsed_time=(end_time-start_time)
print("Fold %d, %d epochs, %d sec"%(fold,EPOCHS,elapsed_time))
# print(history.history.keys()) # all these keys will be shown in figure
pd.DataFrame(history.history).plot(figsize=(8,5))
plt.grid(True)
plt.gca().set_ylim(0,1) # any losses > 1 will be off the scale
plt.show()
do_cross_validation(X,y)
from keras.models import load_model
pc_test = pc_sim.get_sequences(PC_TESTS)
nc_test = nc_sim.get_sequences(NC_TESTS)
X,y = prepare_inputs_len_x_alphabet(pc_test,nc_test,ALPHABET)
best_model=load_model(MODELPATH)
scores = best_model.evaluate(X, y, verbose=0)
print("The best model parameters were saved during cross-validation.")
print("Best was defined as maximum validation accuracy at end of any epoch.")
print("Now re-load the best model and test it on previously unseen data.")
print("Test on",len(pc_test),"PC seqs")
print("Test on",len(nc_test),"NC seqs")
print("%s: %.2f%%" % (best_model.metrics_names[1], scores[1]*100))
from sklearn.metrics import roc_curve
from sklearn.metrics import roc_auc_score
ns_probs = [0 for _ in range(len(y))]
bm_probs = best_model.predict(X)
ns_auc = roc_auc_score(y, ns_probs)
bm_auc = roc_auc_score(y, bm_probs)
ns_fpr, ns_tpr, _ = roc_curve(y, ns_probs)
bm_fpr, bm_tpr, _ = roc_curve(y, bm_probs)
plt.plot(ns_fpr, ns_tpr, linestyle='--', label='Guess, auc=%.4f'%ns_auc)
plt.plot(bm_fpr, bm_tpr, marker='.', label='Model, auc=%.4f'%bm_auc)
plt.title('ROC')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.legend()
plt.show()
print("%s: %.2f%%" %('AUC',bm_auc))
```
| github_jupyter |
```
"""
You can run either this notebook locally (if you have all the dependencies and a GPU) or on Google Colab.
Instructions for setting up Colab are as follows:
1. Open a new Python 3 notebook.
2. Import this notebook from GitHub (File -> Upload Notebook -> "GITHUB" tab -> copy/paste GitHub URL)
3. Connect to an instance with a GPU (Runtime -> Change runtime type -> select "GPU" for hardware accelerator)
4. Run this cell to set up dependencies.
"""
# If you're using Google Colab and not running locally, run this cell.
## Install dependencies
!pip install wget
!apt-get install sox libsndfile1 ffmpeg
!pip install unidecode
# ## Install NeMo
!python -m pip install --upgrade git+https://github.com/NVIDIA/NeMo.git#egg=nemo_toolkit[all]
## Install TorchAudio
!pip install torchaudio>=0.6.0 -f https://download.pytorch.org/whl/torch_stable.html
## Grab the config we'll use in this example
!mkdir configs
```
# minGPT License
*This notebook port's the [minGPT codebase](https://github.com/karpathy/minGPT) into equivalent NeMo code. The license for minGPT has therefore been attached here.*
```
The MIT License (MIT) Copyright (c) 2020 Andrej Karpathy
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
```
# torch-rnn License
*This notebook utilizes the `tiny-shakespeare` dataset from the [torch-rnn](https://github.com/jcjohnson/torch-rnn) codebase. The license for torch-rnn has therefore been attached here.*
```
The MIT License (MIT)
Copyright (c) 2016 Justin Johnson
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
```
-------
***Note: This notebook will intentionally introduce some errors to show the power of Neural Types or model development concepts, inside the cells marked with `[ERROR CELL]`. The explanation of and resolution of such errors can be found in the subsequent cells.***
-----
# The NeMo Model
NeMo comes with many state of the art pre-trained Conversational AI models for users to quickly be able to start training and fine-tuning on their own datasets.
In the previous [NeMo Primer](https://colab.research.google.com/github/NVIDIA/NeMo/blob/main/tutorials/00_NeMo_Primer.ipynb) notebook, we learned how to download pretrained checkpoints with NeMo and we also discussed the fundamental concepts of the NeMo Model. The previous tutorial showed us how to use, modify, save, and restore NeMo Models.
In this tutorial we will learn how to develop a non-trivial NeMo model from scratch. This helps us to understand the underlying components and how they interact with the overall PyTorch ecosystem.
-------
At the heart of NeMo lies the concept of the "Model". For NeMo developers, a "Model" is the neural network(s) as well as all the infrastructure supporting those network(s), wrapped into a singular, cohesive unit. As such, most NeMo models are constructed to contain the following out of the box (note: some NeMo models support additional functionality specific to the domain/use case!) -
- Neural Network architecture - all of the modules that are required for the model.
- Dataset + Data Loaders - all of the components that prepare the data for consumption during training or evaluation.
- Preprocessing + Postprocessing - any of the components that process the datasets so the modules can easily consume them.
- Optimizer + Schedulers - basic defaults that work out of the box and allow further experimentation with ease.
- Any other supporting infrastructure - tokenizers, language model configuration, data augmentation, etc.
# Constructing a NeMo Model
NeMo "Models" are comprised of a few key components, so let's tackle them one by one. We will attempt to go in the order that's stated above.
To make this slightly challenging, let's port a model from the NLP domain this time. Transformers are all the rage, with BERT and his friends from Sesame Street forming the core infrastructure for many NLP tasks.
An excellent (yet simple) implementation of one such model - GPT - can be found in the `minGPT` repository - https://github.com/karpathy/minGPT. While the script is short, it explains and succinctly explores all of the core components we expect in a NeMo model, so it's a prime candidate for NeMo! Sidenote: NeMo supports GPT in its NLP collection, and as such, this notebook aims to be an in-depth development walkthrough for such models.
In the following notebook, we will attempt to port minGPT to NeMo, and along the way, discuss some core concepts of NeMo itself.
# Constructing the Neural Network Architecture
First, on the list - the neural network that forms the backbone of the NeMo Model.
So how do we create such a model? Using PyTorch! As you'll see below, NeMo components are compatible with all of PyTorch, so you can augment your workflow without ever losing the flexibility of PyTorch itself!
Let's start with a couple of imports -
```
import torch
import nemo
from nemo.core import NeuralModule
from nemo.core import typecheck
```
## Neural Module
Wait, what's `NeuralModule`? Where is the wonderful `torch.nn.Module`?
`NeuralModule` is a subclass of `torch.nn.Module`, and it brings with it a few additional functionalities.
In addition to being a `torch.nn.Module`, thereby being entirely compatible with the PyTorch ecosystem, it has the following capabilities -
1) `Typing` - It adds support for `Neural Type Checking` to the model. `Typing` is optional but quite useful, as we will discuss below!
2) `Serialization` - Remember the `OmegaConf` config dict and YAML config files? Well, all `NeuralModules` inherently supports serialization/deserialization from such config dictionaries!
3) `FileIO` - This is another entirely optional file serialization system. Does your `NeuralModule` require some way to preserve data that can't be saved into a PyTorch checkpoint? Write your serialization and deserialization logic in two handy methods! **Note**: When you create the final NeMo Model, this will be implemented for you! Automatic serialization and deserialization support of NeMo models!
```
class MyEmptyModule(NeuralModule):
def forward(self):
print("Neural Module ~ hello world!")
x = MyEmptyModule()
x()
```
## Neural Types
Neural Types? You might be wondering what that term refers to.
Almost all NeMo components inherit the class `Typing`. `Typing` is a simple class that adds two properties to the class that inherits it - `input_types` and `output_types`. A NeuralType, by its shortest definition, is simply a semantic tensor. It contains information regarding the semantic shape the tensor should hold, as well as the semantic information of what that tensor represents. That's it.
So what semantic information does such a typed tensor contain? Let's take an example below.
------
Across the Deep Learning domain, we often encounter cases where tensor shapes may match, but the semantics don't match at all. For example take a look at the following rank 3 tensors -
```
# Case 1:
embedding = torch.nn.Embedding(num_embeddings=10, embedding_dim=30)
x = torch.randint(high=10, size=(1, 5))
print("x :", x)
print("embedding(x) :", embedding(x).shape)
# Case 2
lstm = torch.nn.LSTM(1, 30, batch_first=True)
x = torch.randn(1, 5, 1)
print("x :", x)
print("lstm(x) :", lstm(x)[0].shape) # Let's take all timestep outputs of the LSTM
```
-------
As you can see, the output of Case 1 is an embedding of shape [1, 5, 30], and the output of Case 2 is an LSTM output (state `h` over all time steps), also of the same shape [1, 5, 30].
Do they have the same shape? **Yes**. <br>If we do a Case 1 .shape == Case 2 .shape, will we get True as an output? **Yes**. <br>
Do they represent the same concept? **No**. <br>
The ability to recognize that the two tensors do not represent the same semantic information is precisely why we utilize Neural Types. It contains the information of both the shape and the semantic concept of what that tensor represents. If we performed a neural type check between the two outputs of those tensors, it would raise an error saying semantically they were different things (more technically, it would say that they are `INCOMPATIBLE` with each other)!
--------
You may have read of concepts such as [Named Tensors](https://pytorch.org/docs/stable/named_tensor.html). While conceptually similar, Neural Types attached by NeMo are not as tightly bound to the PyTorch ecosystem - practically any object of a class can be attached with a neural type!
## Neural Types - Usage
Neural Types sound interesting, so how do we go about adding them? Let's take a few cases below.
Neural Types are one of the core foundations of NeMo - you will find them in a vast majority of Neural Modules, and every NeMo Model will have its Neural Types defined. While they are entirely optional and unintrusive, NeMo takes great care to support it so that there is no semantic incompatibility between components being used by users.
Let's start with a basic example of a type checked module.
```
from nemo.core.neural_types import NeuralType
from nemo.core.neural_types import *
class EmbeddingModule(NeuralModule):
def __init__(self):
super().__init__()
self.embedding = torch.nn.Embedding(num_embeddings=10, embedding_dim=30)
@typecheck()
def forward(self, x):
return self.embedding(x)
@property
def input_types(self):
return {
'x': NeuralType(axes=('B', 'T'), elements_type=Index())
}
@property
def output_types(self):
return {
'y': NeuralType(axes=('B', 'T', 'C'), elements_type=EmbeddedTextType())
}
```
To show the benefit of Neural Types, we are going to replicate the above cases inside NeuralModules.
Let's discuss how we added type checking support to the above class.
1) `forward` has a decorator `@typecheck()` on it.
2) `input_types` and `output_types` properties are defined.
That's it!
-------
Let's expand on each of the above steps.
- `@typecheck()` is a simple decorator that takes any class that inherits `Typing` (NeuralModule does this for us) and adds the two default properties of `input_types` and `output_types`, which by default returns None.
The `@typecheck()` decorator's explicit use ensures that, by default, neural type checking is **disabled**. NeMo does not wish to intrude on the development process of models. So users can "opt-in" to type checking by overriding the two properties. Therefore, the decorator ensures that users are not burdened with type checking before they wish to have it.
So what is `@typecheck()`? Simply put, you can wrap **any** function of a class that inherits `Typing` with this decorator, and it will look up the definition of the types of that class and enforce them. Typically, `torch.nn.Module` subclasses only implement `forward()` so it is most common to wrap that method, but `@typecheck()` is a very flexible decorator. Inside NeMo, we will show some advanced use cases (which are quite crucial to particular domains such as TTS).
------
As we see above, `@typecheck()` enforces the types. How then, do we provide this type of information to NeMo?
By overriding `input_types` and `output_types` properties of the class, we can return a dictionary mapping a string name to a `NeuralType`.
In the above case, we define a `NeuralType` as two components -
- `axes`: This is the semantic information of the carried by the axes themselves. The most common axes information is from single character notation.
> `B` = Batch <br>
> `C` / `D` - Channel / Dimension (treated the same) <br>
> `T` - Time <br>
> `H` / `W` - Height / Width <br>
- `elements_type`: This is the semantic information of "what the tensor represents". All such types are derived from the basic `ElementType`, and merely subclassing `ElementType` allows us to build a hierarchy of custom semantic types that can be used by NeMo!
Here, we declare that the input is an element_type of `Index` (index of the character in the vocabulary) and that the output is an element_type of `EmbeddedTextType` (the text embedding)
```
embedding_module = EmbeddingModule()
```
Now let's construct the equivalent of the Case 2 above, but as a `NeuralModule`.
```
class LSTMModule(NeuralModule):
def __init__(self):
super().__init__()
self.lstm = torch.nn.LSTM(1, 30, batch_first=True)
@typecheck()
def forward(self, x):
return self.lstm(x)
@property
def input_types(self):
return {
'x': NeuralType(axes=('B', 'T', 'C'), elements_type=SpectrogramType())
}
@property
def output_types(self):
return {
'y': NeuralType(axes=('B', 'T', 'C'), elements_type=EncodedRepresentation())
}
```
------
Here, we define the LSTM module from the Case 2 above.
We changed the input to be a rank three tensor, now representing a "SpectrogramType". We intentionally keep it generic - it can be a `MelSpectrogramType` or a `MFCCSpectrogramType` as it's input!
The output of an LSTM is now an `EncodedRepresentation`. Practically, this can be the output of a CNN layer, a Transformer block, or in this case, an LSTM layer. We can, of course, specialize by subclassing EncodedRepresentation and then using that!
```
lstm_module = LSTMModule()
```
------
Now for the test !
```
# Case 1 [ERROR CELL]
x1 = torch.randint(high=10, size=(1, 5))
print("x :", x1)
print("embedding(x) :", embedding_module(x1).shape)
```
-----
You might be wondering why we get a `TypeError` right off the bat. This `TypeError` is raised by design.
Positional arguments can cause significant issues during model development, mostly when the model/module design is not finalized. To reduce the potential for mistakes caused by wrong positional arguments and enforce the name of arguments provided to the function, `Typing` requires you to **call all of your type-checked functions by kwargs only**.
```
# Case 1
print("x :", x1)
print("embedding(x) :", embedding_module(x=x1).shape)
```
Now let's try the same for the `LSTMModule` in Case 2
```
# Case 2 [ERROR CELL]
x2 = torch.randn(1, 5, 1)
print("x :", x2)
print("lstm(x) :", lstm_module(x=x2)[0].shape) # Let's take all timestep outputs of the LSTM
```
-----
Now we get a type error stating that the number of output arguments provided does not match what is expected.
What exactly is going on here? Well, inside our `LSTMModule` class, we declare the output types to be a single NeuralType - an `EncodedRepresentation` of shape [B, T, C].
But the output of an LSTM layer is a tuple of two state values - the hidden state `h` and the cell state `c`!
So the neural type system raises an error saying that the number of output arguments does not match what is expected.
Let's fix the above.
```
class CorrectLSTMModule(LSTMModule): # Let's inherit the wrong class to make it easy to override
@property
def output_types(self):
return {
'h': NeuralType(axes=('B', 'T', 'C'), elements_type=EncodedRepresentation()),
'c': NeuralType(axes=('B', 'T', 'C'), elements_type=EncodedRepresentation()),
}
lstm_module = CorrectLSTMModule()
# Case 2
x2 = torch.randn(1, 5, 1)
print("x :", x2)
print("lstm(x) :", lstm_module(x=x2)[0].shape) # Let's take all timestep outputs of the LSTM `h` gate
```
------
Great! So now, the type checking system is happy.
If you looked closely, the outputs were ordinary Torch Tensors (this is good news; we don't want to be incompatible with torch Tensors after all!). So, where exactly is the type of information stored?
When the `output_types` is overridden, and valid torch tensors are returned as a result, these tensors are attached with the attribute `neural_type`. Let's inspect this -
```
emb_out = embedding_module(x=x1)
lstm_out = lstm_module(x=x2)[0]
assert hasattr(emb_out, 'neural_type')
assert hasattr(lstm_out, 'neural_type')
print("Embedding tensor :", emb_out.neural_type)
print("LSTM tensor :", lstm_out.neural_type)
```
-------
So we see that these tensors now have this attribute called `neural_type` and are the same shape.
This exercise's entire goal was to assert that the two outputs are semantically **not** the same object, even if they are the same shape.
Let's test this!
```
emb_out.neural_type.compare(lstm_out.neural_type)
emb_out.neural_type == lstm_out.neural_type
```
## Neural Types - Limitations
You might have noticed one interesting fact - our inputs were just `torch.Tensor` to both typed function calls, and they had no `neural_type` assigned to them.
So why did the type check system not raise any error?
This is to maintain compatibility - type checking is meant to work on a chain of function calls - and each of these functions should themselves be wrapped with the `@typecheck()` decorator. This is also done because we don't want to overtax the forward call with dozens of checks, and therefore we only type modules that perform some higher-order logical computation.
------
As an example, it is mostly unnecessary (but still possible) to type the input and output of every residual block of a ResNet model. However, it is practically important to type the encoder (no matter how many layers is inside it) and the decoder (the classification head) separately so that when one does fine-tuning, there is no semantic mismatch of the tensors input to the encoder and bound to the decoder.
-------
For this case, since it would be impractical to extend a class to attach a type to the input tensor, we can take a shortcut and directly attach the neural type to the input!
```
embedding_module = EmbeddingModule()
x1 = torch.randint(high=10, size=(1, 5))
# Attach correct neural type
x1.neural_type = NeuralType(('B', 'T'), Index())
print("embedding(x) :", embedding_module(x=x1).shape)
# Attach wrong neural type [ERROR CELL]
x1.neural_type = NeuralType(('B', 'T'), LabelsType())
print("embedding(x) :", embedding_module(x=x1).shape)
```
## Let's create the minGPT components
Now that we have a somewhat firm grasp of neural type checking, let's begin porting the minGPT example code. Once again, most of the code will be a direct port from the [minGPT repository](https://github.com/karpathy/minGPT).
Here, you will notice one thing. By just changing class imports, one `@typecheck()` on forward, and adding `input_types` and `output_types` (which are also entirely optional!), we are almost entirely done with the PyTorch Lightning port!
```
import math
from typing import List, Set, Dict, Tuple, Optional
import torch
import torch.nn as nn
from torch.nn import functional as F
```
## Creating Element Types
Till now, we have used the Neural Types provided by the NeMo core. But we need not be restricted to the pre-defined element types !
Users have total flexibility in defining any hierarchy of element types as they please!
```
class AttentionType(EncodedRepresentation):
"""Basic Attention Element Type"""
class SelfAttentionType(AttentionType):
"""Self Attention Element Type"""
class CausalSelfAttentionType(SelfAttentionType):
"""Causal Self Attention Element Type"""
```
## Creating the modules
Neural Modules are generally top-level modules but can be used at any level of the module hierarchy.
For demonstration, we will treat an encoder comprising a block of Causal Self Attention modules as a typed Neural Module. Of course, we can also treat each Causal Self Attention layer itself as a neural module if we require it, but top-level modules are generally preferred.
```
class CausalSelfAttention(nn.Module):
"""
A vanilla multi-head masked self-attention layer with a projection at the end.
It is possible to use torch.nn.MultiheadAttention here but I am including an
explicit implementation here to show that there is nothing too scary here.
"""
def __init__(self, n_embd, block_size, n_head, attn_pdrop, resid_pdrop):
super().__init__()
assert n_embd % n_head == 0
self.n_head = n_head
# key, query, value projections for all heads
self.key = nn.Linear(n_embd, n_embd)
self.query = nn.Linear(n_embd, n_embd)
self.value = nn.Linear(n_embd, n_embd)
# regularization
self.attn_drop = nn.Dropout(attn_pdrop)
self.resid_drop = nn.Dropout(resid_pdrop)
# output projection
self.proj = nn.Linear(n_embd, n_embd)
# causal mask to ensure that attention is only applied to the left in the input sequence
self.register_buffer("mask", torch.tril(torch.ones(block_size, block_size))
.view(1, 1, block_size, block_size))
def forward(self, x, layer_past=None):
B, T, C = x.size()
# calculate query, key, values for all heads in batch and move head forward to be the batch dim
k = self.key(x).view(B, T, self.n_head, C // self.n_head).transpose(1, 2) # (B, nh, T, hs)
q = self.query(x).view(B, T, self.n_head, C // self.n_head).transpose(1, 2) # (B, nh, T, hs)
v = self.value(x).view(B, T, self.n_head, C // self.n_head).transpose(1, 2) # (B, nh, T, hs)
# causal self-attention; Self-attend: (B, nh, T, hs) x (B, nh, hs, T) -> (B, nh, T, T)
att = (q @ k.transpose(-2, -1)) * (1.0 / math.sqrt(k.size(-1)))
att = att.masked_fill(self.mask[:,:,:T,:T] == 0, float('-inf'))
att = F.softmax(att, dim=-1)
att = self.attn_drop(att)
y = att @ v # (B, nh, T, T) x (B, nh, T, hs) -> (B, nh, T, hs)
y = y.transpose(1, 2).contiguous().view(B, T, C) # re-assemble all head outputs side by side
# output projection
y = self.resid_drop(self.proj(y))
return y
class Block(nn.Module):
""" an unassuming Transformer block """
def __init__(self, n_embd, block_size, n_head, attn_pdrop, resid_pdrop):
super().__init__()
self.ln1 = nn.LayerNorm(n_embd)
self.ln2 = nn.LayerNorm(n_embd)
self.attn = CausalSelfAttention(n_embd, block_size, n_head, attn_pdrop, resid_pdrop)
self.mlp = nn.Sequential(
nn.Linear(n_embd, 4 * n_embd),
nn.GELU(),
nn.Linear(4 * n_embd, n_embd),
nn.Dropout(resid_pdrop),
)
def forward(self, x):
x = x + self.attn(self.ln1(x))
x = x + self.mlp(self.ln2(x))
return x
```
## Building the NeMo Model
Since a NeMo Model is comprised of various parts, we are going to iterate on the model step by step inside this notebook. As such, we will have multiple intermediate NeMo "Models", which will be partial implementations, and they will inherit each other iteratively.
In a complete implementation of a NeMo Model (as found in the NeMo collections), all of these components will generally be found in a single class.
Let's start by inheriting `ModelPT` - the core class of a PyTorch NeMo Model, which inherits the PyTorch Lightning Module.
-------
**Remember**:
- The NeMo equivalent of `torch.nn.Module` is the `NeuralModule.
- The NeMo equivalent of the `LightningModule` is `ModelPT`.
```
import pytorch_lightning as ptl
from nemo.core import ModelPT
from omegaconf import OmegaConf
```
------
Next, let's construct the bare minimum implementation of the NeMo Model - just the constructor, the initializer of weights, and the forward method.
Initially, we will follow the steps followed by the minGPT implementation, and progressively refactor for NeMo
```
class PTLGPT(ptl.LightningModule):
def __init__(self,
# model definition args
vocab_size: int, # size of the vocabulary (number of possible tokens)
block_size: int, # length of the model's context window in time
n_layer: int, # depth of the model; number of Transformer blocks in sequence
n_embd: int, # the "width" of the model, number of channels in each Transformer
n_head: int, # number of heads in each multi-head attention inside each Transformer block
# model optimization args
learning_rate: float = 3e-4, # the base learning rate of the model
weight_decay: float = 0.1, # amount of regularizing L2 weight decay on MatMul ops
betas: Tuple[float, float] = (0.9, 0.95), # momentum terms (betas) for the Adam optimizer
embd_pdrop: float = 0.1, # \in [0,1]: amount of dropout on input embeddings
resid_pdrop: float = 0.1, # \in [0,1]: amount of dropout in each residual connection
attn_pdrop: float = 0.1, # \in [0,1]: amount of dropout on the attention matrix
):
super().__init__()
# save these for optimizer init later
self.learning_rate = learning_rate
self.weight_decay = weight_decay
self.betas = betas
# input embedding stem: drop(content + position)
self.tok_emb = nn.Embedding(vocab_size, n_embd)
self.pos_emb = nn.Parameter(torch.zeros(1, block_size, n_embd))
self.drop = nn.Dropout(embd_pdrop)
# deep transformer: just a sequence of transformer blocks
self.blocks = nn.Sequential(*[Block(n_embd, block_size, n_head, attn_pdrop, resid_pdrop) for _ in range(n_layer)])
# decoder: at the end one more layernorm and decode the answers
self.ln_f = nn.LayerNorm(n_embd)
self.head = nn.Linear(n_embd, vocab_size, bias=False) # no need for extra bias due to one in ln_f
self.block_size = block_size
self.apply(self._init_weights)
print("number of parameters: %e" % sum(p.numel() for p in self.parameters()))
def forward(self, idx):
b, t = idx.size()
assert t <= self.block_size, "Cannot forward, model block size is exhausted."
# forward the GPT model
token_embeddings = self.tok_emb(idx) # each index maps to a (learnable) vector
position_embeddings = self.pos_emb[:, :t, :] # each position maps to a (learnable) vector
x = self.drop(token_embeddings + position_embeddings)
x = self.blocks(x)
x = self.ln_f(x)
logits = self.head(x)
return logits
def get_block_size(self):
return self.block_size
def _init_weights(self, module):
"""
Vanilla model initialization:
- all MatMul weights \in N(0, 0.02) and biases to zero
- all LayerNorm post-normalization scaling set to identity, so weight=1, bias=0
"""
if isinstance(module, (nn.Linear, nn.Embedding)):
module.weight.data.normal_(mean=0.0, std=0.02)
if isinstance(module, nn.Linear) and module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
```
------
Let's create a PyTorch Lightning Model above, just to make sure it works !
```
m = PTLGPT(vocab_size=100, block_size=32, n_layer=1, n_embd=32, n_head=4)
```
------
Now, let's convert the above easily into a NeMo Model.
A NeMo Model constructor generally accepts only two things -
1) `cfg`: An OmegaConf DictConfig object that defines precisely the components required by the model to define its neural network architecture, data loader setup, optimizer setup, and any additional components needed for the model itself.
2) `trainer`: An optional Trainer from PyTorch Lightning if the NeMo model will be used for training. It can be set after construction (if required) using the `set_trainer` method. For this notebook, we will not be constructing the config for the Trainer object.
## Refactoring Neural Modules
As we discussed above, Neural Modules are generally higher-level components of the Model and can potentially be replaced by equivalent Neural Modules.
As we see above, the embedding modules, deep transformer network, and final decoder layer have all been combined inside the PyTorch Lightning implementation constructor.
------
However, the decoder could have been an RNN instead of a simple Linear layer, or it could have been a 1D-CNN instead.
Likewise, the deep encoder could potentially have a different implementation of Self Attention modules.
These changes cannot be easily implemented any more inside the above implementation. However, if we refactor these components into their respective NeuralModules, then we can easily replace them with equivalent modules we construct in the future!
### Refactoring the Embedding module
Let's first refactor out the embedding module from the above implementation
```
class GPTEmbedding(NeuralModule):
def __init__(self, vocab_size: int, n_embd: int, block_size: int, embd_pdrop: float = 0.0):
super().__init__()
# input embedding stem: drop(content + position)
self.tok_emb = nn.Embedding(vocab_size, n_embd)
self.pos_emb = nn.Parameter(torch.zeros(1, block_size, n_embd))
self.drop = nn.Dropout(embd_pdrop)
@typecheck()
def forward(self, idx):
b, t = idx.size()
# forward the GPT model
token_embeddings = self.tok_emb(idx) # each index maps to a (learnable) vector
position_embeddings = self.pos_emb[:, :t, :] # each position maps to a (learnable) vector
x = self.drop(token_embeddings + position_embeddings)
return x
@property
def input_types(self):
return {
'idx': NeuralType(('B', 'T'), Index())
}
@property
def output_types(self):
return {
'embeddings': NeuralType(('B', 'T', 'C'), EmbeddedTextType())
}
```
### Refactoring the Encoder
Next, let's refactor the Encoder - the multi layer Transformer Encoder
```
class GPTTransformerEncoder(NeuralModule):
def __init__(self, n_embd: int, block_size: int, n_head: int, n_layer: int, attn_pdrop: float = 0.0, resid_pdrop: float = 0.0):
super().__init__()
self.blocks = nn.Sequential(*[Block(n_embd, block_size, n_head, attn_pdrop, resid_pdrop)
for _ in range(n_layer)])
@typecheck()
def forward(self, embed):
return self.blocks(embed)
@property
def input_types(self):
return {
'embed': NeuralType(('B', 'T', 'C'), EmbeddedTextType())
}
@property
def output_types(self):
return {
'encoding': NeuralType(('B', 'T', 'C'), CausalSelfAttentionType())
}
```
### Refactoring the Decoder
Finally, let's refactor the Decoder - the small one-layer feed-forward network to decode the answer.
-------
Note an interesting detail - The `input_types` of the Decoder accepts the generic `EncoderRepresentation()`, where as the `neural_type` of the `GPTTransformerEncoder` has the `output_type` of `CausalSelfAttentionType`.
This is semantically *not* a mismatch! As you can see above in the inheritance chart, we declare `EncodedRepresentation` -> `AttentionType` -> `SelfAttentionType` -> `CausalSelfAttentionType`.
Such an inheritance hierarchy for the `element_type` allows future encoders (which also have a neural output type of at least `EncodedRepresentation`) to be swapped in place of the current GPT Causal Self Attention Encoder while keeping the rest of the NeMo model working just fine!
```
class GPTDecoder(NeuralModule):
def __init__(self, n_embd: int, vocab_size: int):
super().__init__()
self.ln_f = nn.LayerNorm(n_embd)
self.head = nn.Linear(n_embd, vocab_size, bias=False) # no need for extra bias due to one in ln_f
@typecheck()
def forward(self, encoding):
x = self.ln_f(encoding)
logits = self.head(x)
return logits
@property
def input_types(self):
return {
'encoding': NeuralType(('B', 'T', 'C'), EncodedRepresentation())
}
@property
def output_types(self):
return {
'logits': NeuralType(('B', 'T', 'C'), LogitsType())
}
```
### Refactoring the NeMo GPT Model
Now that we have 3 NeuralModules for the embedding, the encoder, and the decoder, let's refactor the NeMo model to take advantage of this refactor!
This time, we inherit from `ModelPT` instead of the general `LightningModule`.
```
class AbstractNeMoGPT(ModelPT):
def __init__(self, cfg: OmegaConf, trainer: ptl.Trainer = None):
super().__init__(cfg=cfg, trainer=trainer)
# input embedding stem: drop(content + position)
self.embedding = self.from_config_dict(self.cfg.embedding)
# deep transformer: just a sequence of transformer blocks
self.encoder = self.from_config_dict(self.cfg.encoder)
# decoder: at the end one more layernorm and decode the answers
self.decoder = self.from_config_dict(self.cfg.decoder)
self.block_size = self.cfg.embedding.block_size
self.apply(self._init_weights)
print("number of parameters: %e" % self.num_weights)
@typecheck()
def forward(self, idx):
b, t = idx.size()
assert t <= self.block_size, "Cannot forward, model block size is exhausted."
# forward the GPT model
# Remember: Only kwargs are allowed !
e = self.embedding(idx=idx)
x = self.encoder(embed=e)
logits = self.decoder(encoding=x)
return logits
def get_block_size(self):
return self.block_size
def _init_weights(self, module):
"""
Vanilla model initialization:
- all MatMul weights \in N(0, 0.02) and biases to zero
- all LayerNorm post-normalization scaling set to identity, so weight=1, bias=0
"""
if isinstance(module, (nn.Linear, nn.Embedding)):
module.weight.data.normal_(mean=0.0, std=0.02)
if isinstance(module, nn.Linear) and module.bias is not None:
module.bias.data.zero_()
elif isinstance(module, nn.LayerNorm):
module.bias.data.zero_()
module.weight.data.fill_(1.0)
@property
def input_types(self):
return {
'idx': NeuralType(('B', 'T'), Index())
}
@property
def output_types(self):
return {
'logits': NeuralType(('B', 'T', 'C'), LogitsType())
}
```
## Creating a config for a Model
At first glance, not much changed compared to the PyTorch Lightning implementation above. Other than the constructor, which now accepts a config, nothing changed at all!
NeMo operates on the concept of a NeMo Model being accompanied by a corresponding config dict (instantiated as an OmegaConf object). This enables us to prototype the model by utilizing Hydra rapidly. This includes various other benefits - such as hyperparameter optimization and serialization/deserialization of NeMo models.
Let's look at how actually to construct such config objects!
```
# model definition args (required)
# ================================
# vocab_size: int # size of the vocabulary (number of possible tokens)
# block_size: int # length of the model's context window in time
# n_layer: int # depth of the model; number of Transformer blocks in sequence
# n_embd: int # the "width" of the model, number of channels in each Transformer
# n_head: int # number of heads in each multi-head attention inside each Transformer block
# model definition args (optional)
# ================================
# embd_pdrop: float = 0.1, # \in [0,1]: amount of dropout on input embeddings
# resid_pdrop: float = 0.1, # \in [0,1]: amount of dropout in each residual connection
# attn_pdrop: float = 0.1, # \in [0,1]: amount of dropout on the attention matrix
```
------
As we look at the required parameters above, we need a way to tell OmegaConf that these values are currently not set, but the user should set them before we use them.
OmegaConf supports such behavior using the `MISSING` value. A similar effect can be achieved in YAML configs by using `???` as a placeholder.
```
from omegaconf import MISSING
# Let's create a utility for building the class path
def get_class_path(cls):
return f'{cls.__module__}.{cls.__name__}'
```
### Structure of a Model config
Let's first create a config for the common components of the model level config -
```
common_config = OmegaConf.create({
'vocab_size': MISSING,
'block_size': MISSING,
'n_layer': MISSING,
'n_embd': MISSING,
'n_head': MISSING,
})
```
-----
The model config right now is still being built - it needs to contain a lot more details!
A complete Model Config should have the sub-configs of all of its top-level modules as well. This means the configs of the `embedding`, `encoder`, and the `decoder`.
### Structure of sub-module config
For top-level models, we generally don't change the actual module very often, and instead, primarily change the hyperparameters of that model.
So we will make use of `Hydra`'s Class instantiation method - which can easily be accessed via the class method `ModelPT.from_config_dict()`.
Let's take a few examples below -
```
embedding_config = OmegaConf.create({
'_target_': get_class_path(GPTEmbedding),
'vocab_size': '${model.vocab_size}',
'n_embd': '${model.n_embd}',
'block_size': '${model.block_size}',
'embd_pdrop': 0.1
})
encoder_config = OmegaConf.create({
'_target_': get_class_path(GPTTransformerEncoder),
'n_embd': '${model.n_embd}',
'block_size': '${model.block_size}',
'n_head': '${model.n_head}',
'n_layer': '${model.n_layer}',
'attn_pdrop': 0.1,
'resid_pdrop': 0.1
})
decoder_config = OmegaConf.create({
'_target_': get_class_path(GPTDecoder),
# n_embd: int, vocab_size: int
'n_embd': '${model.n_embd}',
'vocab_size': '${model.vocab_size}'
})
```
##### What is `_target_`?
--------
In the above config, we see a `_target_` in the config. `_target_` is usually a full classpath to the actual class in the python package/user local directory. It is required for Hydra to locate and instantiate the model from its path correctly.
So why do we want to set a classpath?
In general, when developing models, we don't often change the encoder or the decoder, but we do change the hyperparameters of the encoder and decoder.
This notation helps us keep the Model level declaration of the forward step neat and precise. It also logically helps us demark which parts of the model can be easily replaced - in the future, we can easily replace the encoder with some other type of self-attention block or the decoder with an RNN or 1D-CNN neural module (as long as they have the same Neural Type definition as the current blocks).
##### What is the `${}` syntax?
-------
OmegaConf, and by extension, Hydra, supports Variable Interpolation. As you can see in the `__init__` of embedding, encoder, and decoder neural modules, they often share many parameters between each other.
It would become tedious and error-prone to set each of these constructors' values separately in each of the embedding, encoder, and decoder configs.
So instead, we define standard keys inside of the `model` level config and then interpolate these values inside of the respective configs!
### Attaching the model and module-level configs
So now, we have a Model level and per-module level configs for the core components. Sub-module configs generally fall under the "model" namespace, but you have the flexibility to define the structure as you require.
Let's attach them!
```
model_config = OmegaConf.create({
'model': common_config
})
# Then let's attach the sub-module configs
model_config.model.embedding = embedding_config
model_config.model.encoder = encoder_config
model_config.model.decoder = decoder_config
```
-----
Let's print this config!
```
print(OmegaConf.to_yaml(model_config))
```
-----
Wait, why did OmegaConf not fill in the value of the variable interpolation for the configs yet?
This is because OmegaConf takes a deferred approach to variable interpolation. To force it ahead of time, we can use the following snippet -
```
temp_config = OmegaConf.create(OmegaConf.to_container(model_config, resolve=True))
print(OmegaConf.to_yaml(temp_config))
```
-----
Now that we have a config, let's try to create an object of the NeMo Model !
```
import copy
# Let's work on a copy of the model config and update it before we send it into the Model.
cfg = copy.deepcopy(model_config)
# Let's set the values of the config (for some plausible small model)
cfg.model.vocab_size = 100
cfg.model.block_size = 128
cfg.model.n_layer = 1
cfg.model.n_embd = 32
cfg.model.n_head = 4
print(OmegaConf.to_yaml(cfg))
# Try to create a model with this config [ERROR CELL]
m = AbstractNeMoGPT(cfg.model)
```
-----
You will note that we added the `Abstract` tag for a reason to this NeMo Model and that when we try to instantiate it - it raises an error that we need to implement specific methods.
1) `setup_training_data` & `setup_validation_data` - All NeMo models should implement two data loaders - the training data loader and the validation data loader. Optionally, they can go one step further and also implement the `setup_test_data` method to add support for evaluating the Model on its own.
Why do we enforce this? NeMo Models are meant to be a unified, cohesive object containing the details about the neural network underlying that Model and the data loaders to train, validate, and optionally test those models.
In doing so, once the Model is created/deserialized, it would take just a few more steps to train the Model from scratch / fine-tune/evaluate the Model on any data that the user provides, as long as this user-provided dataset is in a format supported by the Dataset / DataLoader that is used by this Model!
2) `list_available_models` - This is a utility method to provide a list of pre-trained NeMo models to the user from the cloud.
Typically, NeMo models can be easily packaged into a tar file (which we call a .nemo file in the earlier primer notebook). These tar files contain the model config + the pre-trained checkpoint weights of the Model, and can easily be downloaded from some cloud service.
For this notebook, we will not be implementing this method.
--------
Finally, let's create a concrete implementation of the above NeMo Model!
```
from nemo.core.classes.common import PretrainedModelInfo
class BasicNeMoGPT(AbstractNeMoGPT):
@classmethod
def list_available_models(cls) -> PretrainedModelInfo:
return None
def setup_training_data(self, train_data_config: OmegaConf):
self._train_dl = None
def setup_validation_data(self, val_data_config: OmegaConf):
self._validation_dl = None
def setup_test_data(self, test_data_config: OmegaConf):
self._test_dl = None
```
------
Now let's try to create an object of the `BasicNeMoGPT` model
```
m = BasicNeMoGPT(cfg.model)
```
## Setting up train-val-test steps
The above `BasicNeMoGPT` Model is a basic PyTorch Lightning Module, with some added functionality -
1) Neural Type checks support - as defined in the Model as well as the internal modules.
2) Save and restore of the Model (in the trivial case) to a tarfile.
But as the Model is right now, it crucially does not support PyTorch Lightning's `Trainer`. As such, while this Model can be called manually, it cannot be easily trained or evaluated by using the PyTorch Lightning framework.
------
Let's begin adding support for this then -
```
class BasicNeMoGPTWithSteps(BasicNeMoGPT):
def step_(self, split, batch, batch_idx=None):
idx, targets = batch
logits = self(idx=idx)
loss = F.cross_entropy(logits.view(-1, logits.size(-1)), targets.view(-1))
key = 'loss' if split == 'train' else f"{split}_loss"
return {key: loss}
def training_step(self, *args, **kwargs):
return self.step_('train', *args, **kwargs)
def validation_step(self, *args, **kwargs):
return self.step_('val', *args, **kwargs)
def test_step(self, *args, **kwargs):
return self.step_('test', *args, **kwargs)
# This is useful for multiple validation data loader setup
def multi_validation_epoch_end(self, outputs, dataloader_idx: int = 0):
val_loss_mean = torch.stack([x['val_loss'] for x in outputs]).mean()
return {'val_loss': val_loss_mean}
# This is useful for multiple test data loader setup
def multi_test_epoch_end(self, outputs, dataloader_idx: int = 0):
test_loss_mean = torch.stack([x['test_loss'] for x in outputs]).mean()
return {'test_loss': test_loss_mean}
m = BasicNeMoGPTWithSteps(cfg=cfg.model)
```
### Setup for Multi Validation and Multi Test data loaders
As discussed in the NeMo Primer, NeMo has in-built support for multiple data loaders for validation and test steps. Therefore, as an example of how easy it is to add such support, we include the `multi_validation_epoch_end` and `multi_test_epoch_end` overrides.
It is also practically essential to collate results from more than one distributed GPUs, and then aggregate results properly at the end of the epoch. NeMo strictly enforces the correct collation of results, even if you will work on only one device! Future-proofing is baked into the model design for this case!
Therefore NeMo provides the above two generic methods to support aggregation and simultaneously support multiple datasets!
**Please note, you can prepend your already existing `validation_epoch_end` and `test_epoch_end` implementations with the `multi_` in the name, and that alone is sufficient to enable multi-dataset and multi-GPU support!**
------
**Note: To disable multi-dataset support, simply override `validation_epoch_end` and `test_epoch_end` instead of `multi_validation_epoch_end` and `multi_test_epoch_end`!**
## Setting up the optimizer / scheduler
We are relatively close to reaching feature parity with the MinGPT Model! But we are missing a crucial piece - the optimizer.
All NeMo Model's come with a default implementation of `setup_optimization()`, which will parse the provided model config to obtain the `optim` and `sched` sub-configs, and automatically configure the optimizer and scheduler.
If training GPT was as simple as plugging in an Adam optimizer over all the parameters with a cosine weight decay schedule, we could do that from the config alone.
-------
But GPT is not such a trivial model - more specifically, it requires weight decay to be applied to the weight matrices but not to the biases, the embedding matrix, or the LayerNorm layers.
We can drop the support that Nemo provides for such special cases and instead utilize the PyTorch Lightning method `configure_optimizers` to perform the same task.
-------
Note, for NeMo Models; the `configure_optimizers` is implemented as a trivial call to `setup_optimization()` followed by returning the generated optimizer and scheduler! So we can override the `configure_optimizer` method and manage the optimizer creation manually!
NeMo's goal is to provide usable defaults for the general case and simply back off to either PyTorch Lightning or PyTorch nn.Module itself in cases which the additional flexibility becomes necessary!
```
class BasicNeMoGPTWithOptim(BasicNeMoGPTWithSteps):
def configure_optimizers(self):
"""
This long function is unfortunately doing something very simple and is being very defensive:
We are separating out all parameters of the model into two buckets: those that will experience
weight decay for regularization and those that won't (biases, and layernorm/embedding weights).
We are then returning the PyTorch optimizer object.
"""
# separate out all parameters to those that will and won't experience weight decay
decay = set()
no_decay = set()
whitelist_weight_modules = (torch.nn.Linear, )
blacklist_weight_modules = (torch.nn.LayerNorm, torch.nn.Embedding)
for mn, m in self.named_modules():
for pn, p in m.named_parameters():
fpn = '%s.%s' % (mn, pn) if mn else pn # full param name
if pn.endswith('bias'):
# all biases will not be decayed
no_decay.add(fpn)
elif pn.endswith('weight') and isinstance(m, whitelist_weight_modules):
# weights of whitelist modules will be weight decayed
decay.add(fpn)
elif pn.endswith('weight') and isinstance(m, blacklist_weight_modules):
# weights of blacklist modules will NOT be weight decayed
no_decay.add(fpn)
# special case the position embedding parameter in the root GPT module as not decayed
no_decay.add('embedding.pos_emb')
# validate that we considered every parameter
param_dict = {pn: p for pn, p in self.named_parameters()}
inter_params = decay & no_decay
union_params = decay | no_decay
assert len(inter_params) == 0, "parameters %s made it into both decay/no_decay sets!" % (str(inter_params), )
assert len(param_dict.keys() - union_params) == 0, "parameters %s were not separated into either decay/no_decay set!" \
% (str(param_dict.keys() - union_params), )
# create the pytorch optimizer object
optim_groups = [
{"params": [param_dict[pn] for pn in sorted(list(decay))], "weight_decay": self.cfg.optim.weight_decay},
{"params": [param_dict[pn] for pn in sorted(list(no_decay))], "weight_decay": 0.0},
]
optimizer = torch.optim.AdamW(optim_groups, lr=self.cfg.optim.lr, betas=self.cfg.optim.betas)
return optimizer
m = BasicNeMoGPTWithOptim(cfg=cfg.model)
```
-----
Now let's setup the config for the optimizer !
```
OmegaConf.set_struct(cfg.model, False)
optim_config = OmegaConf.create({
'lr': 3e-4,
'weight_decay': 0.1,
'betas': [0.9, 0.95]
})
cfg.model.optim = optim_config
OmegaConf.set_struct(cfg.model, True)
```
## Setting up the dataset / data loaders
So we were able almost entirely to replicate the MinGPT implementation.
Remember, NeMo models should contain all of the logic to load the Dataset and DataLoader for at least the train and validation step.
We temporarily provided empty implementations to get around it till now, but let's fill that in now!
-------
**Note for datasets**: Below, we will show an example using a very small dataset called `tiny_shakespeare`, found at the original [char-rnn repository](https://github.com/karpathy/char-rnn), but practically you could use any text corpus. The one suggested in minGPT is available at http://mattmahoney.net/dc/textdata.html
### Creating the Dataset
NeMo has Neural Type checking support, even for Datasets! It's just a minor change of the import in most cases and one difference in how we handle `collate_fn`.
We could paste the dataset info from minGPT, and you'd only need to make 2 changes!
-----
In this example, we will be writing a thin subclass over the datasets provided by `nlp` from HuggingFace!
```
from nemo.core import Dataset
from torch.utils import data
from torch.utils.data.dataloader import DataLoader
class TinyShakespeareDataset(Dataset):
def __init__(self, data_path, block_size, crop=None, override_vocab=None):
# load the data and crop it appropriately
with open(data_path, 'r') as f:
if crop is None:
data = f.read()
else:
f.seek(crop[0])
data = f.read(crop[1])
# build a vocabulary from data or inherit it
vocab = sorted(list(set(data))) if override_vocab is None else override_vocab
# Add UNK
special_tokens = ['<PAD>', '<UNK>'] # We use just <UNK> and <PAD> in the call, but can add others.
if not override_vocab:
vocab = [*special_tokens, *vocab] # Update train vocab with special tokens
data_size, vocab_size = len(data), len(vocab)
print('data of crop %s has %d characters, vocab of size %d.' % (str(crop), data_size, vocab_size))
print('Num samples in dataset : %d' % (data_size // block_size))
self.stoi = { ch:i for i,ch in enumerate(vocab) }
self.itos = { i:ch for i,ch in enumerate(vocab) }
self.block_size = block_size
self.vocab_size = vocab_size
self.data = data
self.vocab = vocab
self.special_tokens = special_tokens
def __len__(self):
return len(self.data) // self.block_size
def __getitem__(self, idx):
# attempt to fetch a chunk of (block_size + 1) items, but (block_size) will work too
chunk = self.data[idx*self.block_size : min(len(self.data), (idx+1)*self.block_size + 1)]
# map the string into a sequence of integers
ixes = [self.stoi[s] if s in self.stoi else self.stoi['<UNK>'] for s in chunk ]
# if stars align (last idx and len(self.data) % self.block_size == 0), pad with <PAD>
if len(ixes) < self.block_size + 1:
assert len(ixes) == self.block_size # i believe this is the only way this could happen, make sure
ixes.append(self.stoi['<PAD>'])
dix = torch.tensor(ixes, dtype=torch.long)
return dix[:-1], dix[1:]
@property
def output_types(self):
return {
'input': NeuralType(('B', 'T'), Index()),
'target': NeuralType(('B', 'T'), LabelsType())
}
```
------
We didn't have to change anything until here. How then is type-checking done?
NeMo does type-checking inside of the collate function implementation itself! In this case, it is not necessary to override the `collate_fn` inside the Dataset, but if we did need to override it, **NeMo requires that the private method `_collate_fn` be overridden instead**.
We can then use data loaders with minor modifications!
**Also, there is no need to implement the `input_types` for Dataset, as they are the ones generating the input for the model!**
-----
Let's prepare the dataset that we are going to use - Tiny Shakespeare from the following codebase [char-rnn](https://github.com/karpathy/char-rnn).
```
import os
if not os.path.exists('tiny-shakespeare.txt'):
!wget https://raw.githubusercontent.com/jcjohnson/torch-rnn/master/data/tiny-shakespeare.txt
!head -n 5 tiny-shakespeare.txt
train_dataset = TinyShakespeareDataset('tiny-shakespeare.txt', cfg.model.block_size, crop=(0, int(1e6)))
val_dataset = TinyShakespeareDataset('tiny-shakespeare.txt', cfg.model.block_size, crop=(int(1e6), int(50e3)), override_vocab=train_dataset.vocab)
test_dataset = TinyShakespeareDataset('tiny-shakespeare.txt', cfg.model.block_size, crop=(int(1.05e6), int(100e3)), override_vocab=train_dataset.vocab)
```
### Setting up dataset/data loader support in the Model
So we now know our data loader works. Let's integrate it as part of the Model itself!
To do this, we use the three special attributes of the NeMo Model - `self._train_dl`, `self._validation_dl` and `self._test_dl`. Once you construct your DataLoader, place your data loader to these three variables.
For multi-data loader support, the same applies! NeMo will automatically handle the management of multiple data loaders for you!
```
class NeMoGPT(BasicNeMoGPTWithOptim):
def _setup_data_loader(self, cfg):
if self.vocab is None:
override_vocab = None
else:
override_vocab = self.vocab
dataset = TinyShakespeareDataset(
data_path=cfg.data_path,
block_size=cfg.block_size,
crop=tuple(cfg.crop) if 'crop' in cfg else None,
override_vocab=override_vocab
)
if self.vocab is None:
self.vocab = dataset.vocab
return DataLoader(
dataset=dataset,
batch_size=cfg.batch_size,
shuffle=cfg.shuffle,
collate_fn=dataset.collate_fn, # <-- this is necessary for type checking
pin_memory=cfg.pin_memory if 'pin_memory' in cfg else False,
num_workers=cfg.num_workers if 'num_workers' in cfg else 0
)
def setup_training_data(self, train_data_config: OmegaConf):
self.vocab = None
self._train_dl = self._setup_data_loader(train_data_config)
def setup_validation_data(self, val_data_config: OmegaConf):
self._validation_dl = self._setup_data_loader(val_data_config)
def setup_test_data(self, test_data_config: OmegaConf):
self._test_dl = self._setup_data_loader(test_data_config)
```
### Creating the dataset / dataloader config
The final step to setup this model is to add the `train_ds`, `validation_ds` and `test_ds` configs inside the model config!
```
OmegaConf.set_struct(cfg.model, False)
# Set the data path and update vocabular size
cfg.model.data_path = 'tiny-shakespeare.txt'
cfg.model.vocab_size = train_dataset.vocab_size
OmegaConf.set_struct(cfg.model, True)
train_ds = OmegaConf.create({
'data_path': '${model.data_path}',
'block_size': '${model.block_size}',
'crop': [0, int(1e6)],
'batch_size': 64,
'shuffle': True,
})
validation_ds = OmegaConf.create({
'data_path': '${model.data_path}',
'block_size': '${model.block_size}',
'crop': [int(1e6), int(50e3)],
'batch_size': 4,
'shuffle': False,
})
test_ds = OmegaConf.create({
'data_path': '${model.data_path}',
'block_size': '${model.block_size}',
'crop': [int(1.05e6), int(100e3)],
'batch_size': 4,
'shuffle': False,
})
# Attach to the model config
OmegaConf.set_struct(cfg.model, False)
cfg.model.train_ds = train_ds
cfg.model.validation_ds = validation_ds
cfg.model.test_ds = test_ds
OmegaConf.set_struct(cfg.model, True)
# Let's see the config now !
print(OmegaConf.to_yaml(cfg))
# Let's try creating a model now !
model = NeMoGPT(cfg=cfg.model)
```
-----
All the data loaders load properly ! Yay!
# Evaluate the model - end to end!
Now that the data loaders have been set up, all that's left is to train and test the model! We have most of the components required by this model - the train, val and test data loaders, the optimizer, and the type-checked forward step to perform the train-validation-test steps!
But training a GPT model from scratch is not the goal of this primer, so instead, let's do a sanity check by merely testing the model for a few steps using random initial weights.
The above will ensure that -
1) Our data loaders work as intended
2) The type checking system assures us that our Neural Modules are performing their forward step correctly.
3) The loss is calculated, and therefore the model runs end to end, ultimately supporting PyTorch Lightning.
```
if torch.cuda.is_available():
cuda = 1
else:
cuda = 0
trainer = ptl.Trainer(gpus=cuda, test_percent_check=1.0)
trainer.test(model)
```
# Saving and restoring models
NeMo internally keeps track of the model configuration, as well as the model checkpoints and parameters.
As long as your NeMo follows the above general guidelines, you can call the `save_to` and `restore_from` methods to save and restore your models!
```
model.save_to('gpt_model.nemo')
!ls -d -- *.nemo
temp_model = NeMoGPT.restore_from('gpt_model.nemo')
# [ERROR CELL]
temp_model.setup_test_data(temp_model.cfg.test_ds)
```
-----
Hmm, it seems it wasn't so easy in this case. Non-trivial models have non-trivial issues!
Remember, our NeMoGPT model sets its self.vocab inside the `setup_train_data` step. But that depends on the vocabulary generated by the train set... which is **not** restored during model restoration (unless you call `setup_train_data` explicitly!).
We can quickly resolve this issue by constructing an external data file to enable save and restore support, and NeMo supports that too! We will use the `register_artifact` API in NeMo to support external files being attached to the .nemo checkpoint.
```
class NeMoGPTv2(NeMoGPT):
def setup_training_data(self, train_data_config: OmegaConf):
self.vocab = None
self._train_dl = self._setup_data_loader(train_data_config)
# Save the vocab into a text file for now
with open('vocab.txt', 'w') as f:
for token in self.vocab:
f.write(f"{token}<SEP>")
# This is going to register the file into .nemo!
# When you later use .save_to(), it will copy this file into the tar file.
self.register_artifact(None, 'vocab.txt')
def setup_validation_data(self, val_data_config: OmegaConf):
# This is going to try to find the same file, and if it fails,
# it will use the copy in .nemo
vocab_file = self.register_artifact(None, 'vocab.txt')
with open(vocab_file, 'r') as f:
vocab = []
vocab = f.read().split('<SEP>')[:-1] # the -1 here is for the dangling <SEP> token in the file
self.vocab = vocab
self._validation_dl = self._setup_data_loader(val_data_config)
def setup_test_data(self, test_data_config: OmegaConf):
# This is going to try to find the same file, and if it fails,
# it will use the copy in .nemo
vocab_file = self.register_artifact(None, 'vocab.txt')
with open(vocab_file, 'r') as f:
vocab = []
vocab = f.read().split('<SEP>')[:-1] # the -1 here is for the dangling <SEP> token in the file
self.vocab = vocab
self._test_dl = self._setup_data_loader(test_data_config)
# Let's try creating a model now !
model = NeMoGPTv2(cfg=cfg.model)
# Now let's try to save and restore !
model.save_to('gpt_model.nemo')
temp_model = NeMoGPTv2.restore_from('gpt_model.nemo')
temp_model.setup_multiple_test_data(temp_model.cfg.test_ds)
if torch.cuda.is_available():
cuda = 1
else:
cuda = 0
trainer = ptl.Trainer(gpus=cuda, test_percent_check=1.0)
trainer.test(model)
```
------
There we go ! Now our model's can be serialized and de-serialized without any issue, even with an external vocab file !
| github_jupyter |
```
import pandas as pd
import numpy as np
#upload the csv file or
#!git clone
#and locate the csv and change location
df=pd.read_csv("/content/T1.csv", engine='python')
df.head()
lst=df["Wind Speed (m/s)"]
lst
max(lst)
min(lst)
lst=list(df["Wind Speed (m/s)"])
# Python program to get average of a list
def Average(lst):
return sum(lst) / len(lst)
# Driver Code
average = Average(lst)
# Printing average of the list
print("Average of the list =", round(average, 2))
for i in range(len(lst)):
lst[i]=round(lst[i],0)
lst
# Python program to count the frequency of
# elements in a list using a dictionary
def CountFrequency(my_list):
# Creating an empty dictionary
freq = {}
for item in my_list:
if (item in freq):
freq[item] += 1
else:
freq[item] = 1
for key, value in freq.items():
print ("% d : % d"%(key, value))
return freq
f=CountFrequency(lst)
dictionary_items = f.items()
sorted_items = sorted(dictionary_items)
sorted_items
#x wind speed
#y frequency
x=[]
y=[]
for each in sorted_items:
print(each)
x.append(each[0])
y.append(each[1])
x
y
ybar=np.array(y)/5
ybar=ybar/10
xbar=np.array(x)
from scipy import stats
import matplotlib.pyplot as plt
plt.figure(figsize=(20,8))
plt.style.use('dark_background')
#plt.rcParams["font.family"] = "Times New Roman"
plt.rcParams["font.size"] = "16"
plt.title('Actual Distribution of Wind Speed in a Practical Scenario', fontsize=30)
plt.grid(False)
from scipy.interpolate import make_interp_spline, BSpline
T,power=xbar,ybar
# 300 represents number of points to make between T.min and T.max
xnew = np.linspace(T.min(), T.max(), 300)
spl = make_interp_spline(T, power, k=3) # type: BSpline
power_smooth = spl(xnew)
plt.plot(xnew, power_smooth,color="w")
#plt.show()
#plt.plot(xbar, ybar)
#plt.hist(x, bins=np.linspace(0, 16, 33), normed=True, alpha=0.5);
width=0.8
bar1=plt.bar(xbar, ybar, width,color="y")
for rect,val in zip(bar1,ybar):
height = rect.get_height()
#print(val)
if(val==0):
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.01, str("-"), ha='center', va='bottom',fontsize=20)
else:
plt.text(rect.get_x() + rect.get_width()/2.0, height+2, str(int(round(val,0))), ha='center', va='bottom',fontsize=12)
#plt.xticks(np.arange(25) + width , list(range(25)))
plt.rcParams['xtick.labelsize']=16
plt.rcParams['ytick.labelsize']=16
plt.xlabel('Wind Speed(m/s)', fontsize=18)
plt.ylabel('Frequency[%]', fontsize=18)
plt.show()
def percentage(y):
#print(y)
tot=y.sum()
#print(tot)
y=y/tot
return y*100
ybar=percentage(np.array(y))
#print(ybar)
xbar=np.array(x)
from scipy import stats
import matplotlib.pyplot as plt
plt.figure(figsize=(20,8))
plt.style.use('dark_background')
#plt.rcParams["font.family"] = "Times New Roman"
plt.rcParams["font.size"] = "16"
plt.title('Actual Distribution of Wind Speed in a Practical Scenario', fontsize=30)
plt.grid(False)
from scipy.interpolate import make_interp_spline, BSpline
T,power=xbar,ybar
# 300 represents number of points to make between T.min and T.max
xnew = np.linspace(T.min(), T.max(), 300)
spl = make_interp_spline(T, power, k=3) # type: BSpline
power_smooth = spl(xnew)
plt.plot(xnew, power_smooth,color="w")
#plt.show()
#plt.plot(xbar, ybar)
#plt.hist(x, bins=np.linspace(0, 16, 33), normed=True, alpha=0.5);
width=0.8
bar1=plt.bar(xbar, ybar, width,color="y")
for rect,val in zip(bar1,ybar):
height = rect.get_height()
#print(val)
if(val==0):
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.01, str("-"), ha='center', va='bottom',fontsize=20)
else:
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.2, str(round(val,1)), ha='center', va='bottom',fontsize=12)
#plt.xticks(np.arange(25) + width , list(range(25)))
plt.rcParams['xtick.labelsize']=16
plt.rcParams['ytick.labelsize']=16
plt.xlabel('Wind Speed(m/s)', fontsize=18)
plt.ylabel('Frequency[%]', fontsize=18)
plt.savefig("actual_distribution.png" ,dpi=100)
plt.show()
from scipy import stats
import matplotlib.pyplot as plt
#input for pseudo data
N = 100
Kappa_in = 2.08
Lambda_in = 8.97
a_in = 1
loc_in = 0
#Generate data from given input
data = stats.exponweib.rvs(a=a_in,c=Kappa_in, loc=loc_in, scale=Lambda_in, size = N)
#The a and loc are fixed in the fit since it is standard to assume they are known
a_out, Kappa_out, loc_out, Lambda_out = stats.exponweib.fit(data, f0=a_in,floc=loc_in)
#Plot
bins = range(25)
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
y=stats.exponweib.pdf(bins, a=a_out,c=Kappa_out,loc=loc_out,scale = Lambda_out)
ax.plot(bins,y*1000)
#ax.hist(data, bins = bins , alpha=0.5)
#ax.annotate("Shape: $k = %.2f$ \n Scale: $\lambda = %.2f$"%(Kappa_out,Lambda_out), xy=(0.7, 0.85), xycoords=ax.transAxes)
plt.show()
def percentage(y):
#print(y)
tot=y.sum()
#print(tot)
y=y/tot
return y*100
ybar=percentage(np.array(y))
from scipy import stats
import matplotlib.pyplot as plt
#input for pseudo data
N = 100
Kappa_in = 2.08
Lambda_in = 8.97
a_in = 1
loc_in = 0
#Generate data from given input
data = stats.exponweib.rvs(a=a_in,c=Kappa_in, loc=loc_in, scale=Lambda_in, size = N)
#The a and loc are fixed in the fit since it is standard to assume they are known
a_out, Kappa_out, loc_out, Lambda_out = stats.exponweib.fit(data, f0=a_in,floc=loc_in)
#Plot
#print(ybar)
xbar=np.array(x)
from scipy import stats
import matplotlib.pyplot as plt
plt.figure(figsize=(20,8))
plt.style.use('dark_background')
bins = range(25)
#fig = plt.figure()
#ax = fig.add_subplot(1, 1, 1)
yhat=stats.exponweib.pdf(bins, a=a_out,c=Kappa_out,loc=loc_out,scale = Lambda_out)
plt.plot(bins,yhat*100, linewidth=4,markersize=12,marker='o',color='green')
#ax.hist(data, bins = bins , alpha=0.5)
#ax.annotate("Shape: $k = %.2f$ \n Scale: $\lambda = %.2f$"%(Kappa_out,Lambda_out), xy=(0.7, 0.85), xycoords=ax.transAxes)
#plt.show()
#plt.rcParams["font.family"] = "Times New Roman"
plt.rcParams["font.size"] = "16"
plt.title('Comparitive Distribution of Wind Speed', fontsize=30)
plt.grid(False)
from scipy.interpolate import make_interp_spline, BSpline
T,power=xbar[:-1],ybar
print(xbar.shape,ybar.shape)
# 300 represents number of points to make between T.min and T.max
xnew = np.linspace(T.min(), T.max(), 300)
spl = make_interp_spline(T, power, k=3) # type: BSpline
power_smooth = spl(xnew)
plt.plot(xnew, power_smooth,color="red" ,linewidth=4,markersize=12,marker='+')
#plt.show()
#plt.plot(xbar, ybar)
#plt.hist(x, bins=np.linspace(0, 16, 33), normed=True, alpha=0.5);
width=0.8
#bar1=plt.bar(xbar, ybar, width,color="y")
"""
for rect,val in zip(bar1,ybar):
height = rect.get_height()
#print(val)
if(val==0):
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.01, str("-"), ha='center', va='bottom',fontsize=20)
else:
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.2, str(round(val,1)), ha='center', va='bottom',fontsize=12)
"""
#plt.xticks(np.arange(25) + width , list(range(25)))
plt.rcParams['xtick.labelsize']=16
plt.rcParams['ytick.labelsize']=16
plt.xlabel('Wind Speed(m/s)', fontsize=18)
plt.ylabel('Frequency[%]', fontsize=18)
plt.savefig("new_distribution.png" ,dpi=100)
plt.show()
def percentage(y):
#print(y)
tot=y.sum()
#print(tot)
y=y/tot
return y*100
ybar=percentage(np.array(y))
from scipy import stats
import matplotlib.pyplot as plt
#input for pseudo data
N = 100
Kappa_in = 2.08
Lambda_in = 8.97
a_in = 1
loc_in = 0
#Generate data from given input
data = stats.exponweib.rvs(a=a_in,c=Kappa_in, loc=loc_in, scale=Lambda_in, size = N)
#The a and loc are fixed in the fit since it is standard to assume they are known
a_out, Kappa_out, loc_out, Lambda_out = stats.exponweib.fit(data, f0=a_in,floc=loc_in)
#Plot
#print(ybar)
xbar=np.array(x)
from scipy import stats
import matplotlib.pyplot as plt
plt.figure(figsize=(20,8))
plt.style.use('dark_background')
bins = range(25)
#fig = plt.figure()
#ax = fig.add_subplot(1, 1, 1)
yhat=stats.exponweib.pdf(bins, a=a_out,c=Kappa_out,loc=loc_out,scale = Lambda_out)
plt.plot(bins,yhat*100, linewidth=4,color='chartreuse',label="Theoretical Weibull Distribution")
#ax.hist(data, bins = bins , alpha=0.5)
#ax.annotate("Shape: $k = %.2f$ \n Scale: $\lambda = %.2f$"%(Kappa_out,Lambda_out), xy=(0.7, 0.85), xycoords=ax.transAxes)
#plt.show()
#plt.rcParams["font.family"] = "Times New Roman"
plt.rcParams["font.size"] = "16"
plt.title('Comparative Distribution of Wind Speed', fontsize=30)
plt.grid(False)
from scipy.interpolate import make_interp_spline, BSpline
T,power=xbar[:-1],ybar
# 300 represents number of points to make between T.min and T.max
xnew = np.linspace(T.min(), T.max(), 300)
spl = make_interp_spline(T, power, k=3) # type: BSpline
power_smooth = spl(xnew)
plt.plot(xnew, power_smooth,color="red" ,linewidth=4,label=" Practical Distribution")
#plt.show()
#plt.plot(xbar, ybar)
#plt.hist(x, bins=np.linspace(0, 16, 33), normed=True, alpha=0.5);
width=0.8
#bar1=plt.bar(xbar, ybar, width,color="y")
"""
for rect,val in zip(bar1,ybar):
height = rect.get_height()
#print(val)
if(val==0):
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.01, str("-"), ha='center', va='bottom',fontsize=20)
else:
plt.text(rect.get_x() + rect.get_width()/2.0, height+0.2, str(round(val,1)), ha='center', va='bottom',fontsize=12)
"""
#plt.xticks(np.arange(25) + width , list(range(25)))
plt.rcParams['xtick.labelsize']=16
plt.rcParams['ytick.labelsize']=16
lg=plt.legend(loc='best',title='Distribution Type', prop={'size': 20})
lg.get_title().set_fontsize(20)
lg._legend_box.align = "center"
plt.xlabel('Wind Speed(m/s)', fontsize=18)
plt.ylabel('Frequency[%]', fontsize=18)
plt.savefig("new_distribution.png" ,dpi=100)
plt.show()
1. Sort data in ascending order
2. Assign them a rank, such that the lowest data point is 1, second lowest is 2, etc.
3. Assign each data point a probability. For beginners, i recommend (i-0.5)/n, where i and n are rank and sample size, respectively.
4. Take natural log of data.
5. Calculate ln (-ln (1-P)) for every data, where P is probabiliyy calculated in step 3.
6. Linear regression with results of Step 5 as Y and results of Step 4 as X. Altrrnatively, you can fit a trendline in Excel.
7. Slope of the regression line is the shape parameter, aka Weibull modulus. The intercept is the negative of the product of shape parameter and natural log of scale parameter.
from scipy.interpolate import make_interp_spline, BSpline
T,power=xbar,ybar
# 300 represents number of points to make between T.min and T.max
xnew = np.linspace(T.min(), T.max(), 300)
spl = make_interp_spline(T, power, k=3) # type: BSpline
power_smooth = spl(xnew)
plt.plot(xnew, power_smooth)
plt.show()
#x = np.random.normal(size=100)
import seaborn as sns
sns.distplot(x);
sns.jointplot(x=x, y=y);
```
| github_jupyter |
```
%matplotlib inline
import matplotlib.pyplot as plt
import sys,os
sys.path.insert(0,'../')
from ml_tools.descriptors import RawSoapInternal
from ml_tools.models.KRR import KRR,TrainerCholesky,KRRFastCV
from ml_tools.kernels import KernelPower,KernelSum
from ml_tools.utils import get_mae,get_rmse,get_sup,get_spearman,get_score,load_pck,tqdm_cs
from ml_tools.split import KFold,LCSplit,ShuffleSplit
from ml_tools.compressor import FPSFilter
import numpy as np
from ase.io import read,write
from ase.visualize import view
```
# Build a kernel Matrix
```
# load the structures
frames = read('data/dft-smiles_500.xyz',':')
global_species = []
for frame in frames:
global_species.extend(frame.get_atomic_numbers())
global_species = np.unique(global_species)
# split the structures in 2 sets
frames_train = frames[:300]
frames_test = frames[300:]
# set up the soap parameters
soap_params = dict(rc=3.5, nmax=6, lmax=6, awidth=0.4,
global_species=global_species,nocenters=[])
representation = RawSoapInternal(**soap_params)
# set up the kernel parameters
kernel = KernelSum(KernelPower(zeta = 2),chunk_shape=[100,100])
# compute the soap vectors
rawsoaps = representation.transform(frames_train)
X_train = dict(feature_matrix=rawsoaps,strides=representation.strides)
# compute the soap vectors
rawsoaps = representation.transform(frames_test)
X_test = dict(feature_matrix=rawsoaps,strides=representation.strides)
# compute the square kernel matrix
Kmat = kernel.transform(X_train)
# compute a rectangular kernel matrix
Kmat_rect = kernel.transform(X_test,X_train)
```
# FPS selection of the samples
```
# load the structures
frames = read('data/dft-smiles_500.xyz',':300')
global_species = []
for frame in frames:
global_species.extend(frame.get_atomic_numbers())
global_species = np.unique(global_species)
# set up the soap parameters
soap_params = dict(rc=3.5, nmax=6, lmax=6, awidth=0.4,
global_species=global_species,nocenters=[])
representation = RawSoapInternal(**soap_params)
# set up the kernel parameters
kernel = KernelSum(KernelPower(zeta = 2),chunk_shape=[100,100])
# compute the soap vectors
rawsoaps = representation.transform(frames)
X = dict(feature_matrix=rawsoaps,strides=representation.strides)
# run the fps selection on the set and plot the minmax distance
Nselect = 250
compressor = FPSFilter(Nselect,kernel,act_on='sample',precompute_kernel=True,disable_pbar=True)
compressor.fit(X,dry_run=True)
compressor.plot()
# select the appropriate number of samples to select
compressor.Nselect = 250
# and compress
X_compressed = compressor.transform(X)
compressor.selected_ids[:compressor.Nselect]
X['feature_matrix'].shape
X_compressed['feature_matrix'].shape
X_compressed['strides'].shape
```
# FPS selection of the features
```
# load the structures
frames = read('data/dft-smiles_500.xyz',':300')
global_species = []
for frame in frames:
global_species.extend(frame.get_atomic_numbers())
global_species = np.unique(global_species)
# set up the soap parameters
soap_params = dict(rc=3.5, nmax=6, lmax=6, awidth=0.4,
global_species=global_species,nocenters=[])
representation = RawSoapInternal(**soap_params)
# set up the kernel parameters
kernel = KernelPower(zeta = 2)
# compute the soap vectors
X = representation.transform(frames)
# run the fps selection on the set and plot the minmax distance
Nselect = 250
compressor = FPSFilter(Nselect,kernel,act_on='feature',precompute_kernel=True,disable_pbar=True)
compressor.fit(X,dry_run=True)
compressor.plot()
# select the appropriate number of samples to select
compressor.Nselect = 500
# and compress
X_compressed = compressor.transform(X)
compressor.selected_ids[:compressor.Nselect]
```
# get a cross validation score
```
# load the structures
frames = read('data/dft-smiles_500.xyz',':')
global_species = []
y = []
for frame in frames:
global_species.extend(frame.get_atomic_numbers())
y.append(frame.info['dft_formation_energy_per_atom_in_eV'])
y = np.array(y)
global_species = np.unique(global_species)
# set up the soap parameters
soap_params = dict(rc=3.5, nmax=6, lmax=6, awidth=0.4,
global_species=global_species,nocenters=[])
representation = RawSoapInternal(**soap_params)
# set up the kernel parameters
kernel = KernelSum(KernelPower(zeta = 2),chunk_shape=[100,100])
# set the splitting rational
cv = KFold(n_splits=6,random_state=10,shuffle=True)
# set up the regression model
jitter = 1e-8
krr = KRRFastCV(jitter, 1.,cv)
# compute the soap vectors
rawsoaps = representation.transform(frames)
X = dict(feature_matrix=rawsoaps,strides=representation.strides)
rawsoaps.shape
# compute the kernel matrix for the dataset
Kmat = kernel.transform(X)
# fit the model
krr.fit(Kmat,y)
# get the predictions for each folds
y_pred = krr.predict()
# compute the CV score for the dataset
get_score(y_pred,y)
```
# LC
```
# load the structures
frames = read('data/dft-smiles_500.xyz',':')
global_species = []
y = []
for frame in frames:
global_species.extend(frame.get_atomic_numbers())
y.append(frame.info['dft_formation_energy_per_atom_in_eV'])
y = np.array(y)
global_species = np.unique(global_species)
# set up the soap parameters
soap_params = dict(rc=3.5, nmax=6, lmax=6, awidth=0.4,
global_species=global_species,nocenters=[])
representation = RawSoapInternal(**soap_params)
# set up the kernel parameters
kernel = KernelSum(KernelPower(zeta = 2),chunk_shape=[100,100])
# set the splitting rational
trainer = TrainerCholesky(memory_efficient=True)
# set up the regression model
jitter = 1e-8
krr = KRR(jitter,1.,trainer)
train_sizes=[20,50,100]
lc = LCSplit(ShuffleSplit, n_repeats=[20,20,20],train_sizes=train_sizes,test_size=100, random_state=10)
rawsoaps = representation.transform(frames)
X = dict(feature_matrix=rawsoaps,strides=representation.strides)
K = kernel.transform(X)
scores = {size:[] for size in train_sizes}
for train,test in tqdm_cs(lc.split(y),total=lc.n_splits):
Ntrain = len(train)
k_train = K[np.ix_(train,train)]
y_train = y[train]
k_test = K[np.ix_(test,train)]
krr.fit(k_train,y_train)
y_pred = krr.predict(k_test)
scores[Ntrain].append(get_score(y_pred,y[test]))
sc_name = 'RMSE'
Ntrains = []
avg_scores = []
for Ntrain, score in scores.items():
avg = 0
for sc in score:
avg += sc[sc_name]
avg /= len(score)
avg_scores.append(avg)
Ntrains.append(Ntrain)
plt.plot(Ntrains,avg_scores,'--o')
plt.xlabel('Number of training samples')
plt.ylabel('Test {}'.format(sc_name))
plt.xscale('log')
plt.yscale('log')
```
| github_jupyter |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.