Datasets:
The dataset viewer is not available because its heuristics could not detect any supported data files. You can try uploading some data files, or configuring the data files location manually.
Otter UBC Dataset Card
UBC is a dataset comprising entities (Proteins/Drugs) from Uniprot (U), BindingDB (B) and. ChemBL (C). It contains 6,207,654 triples.
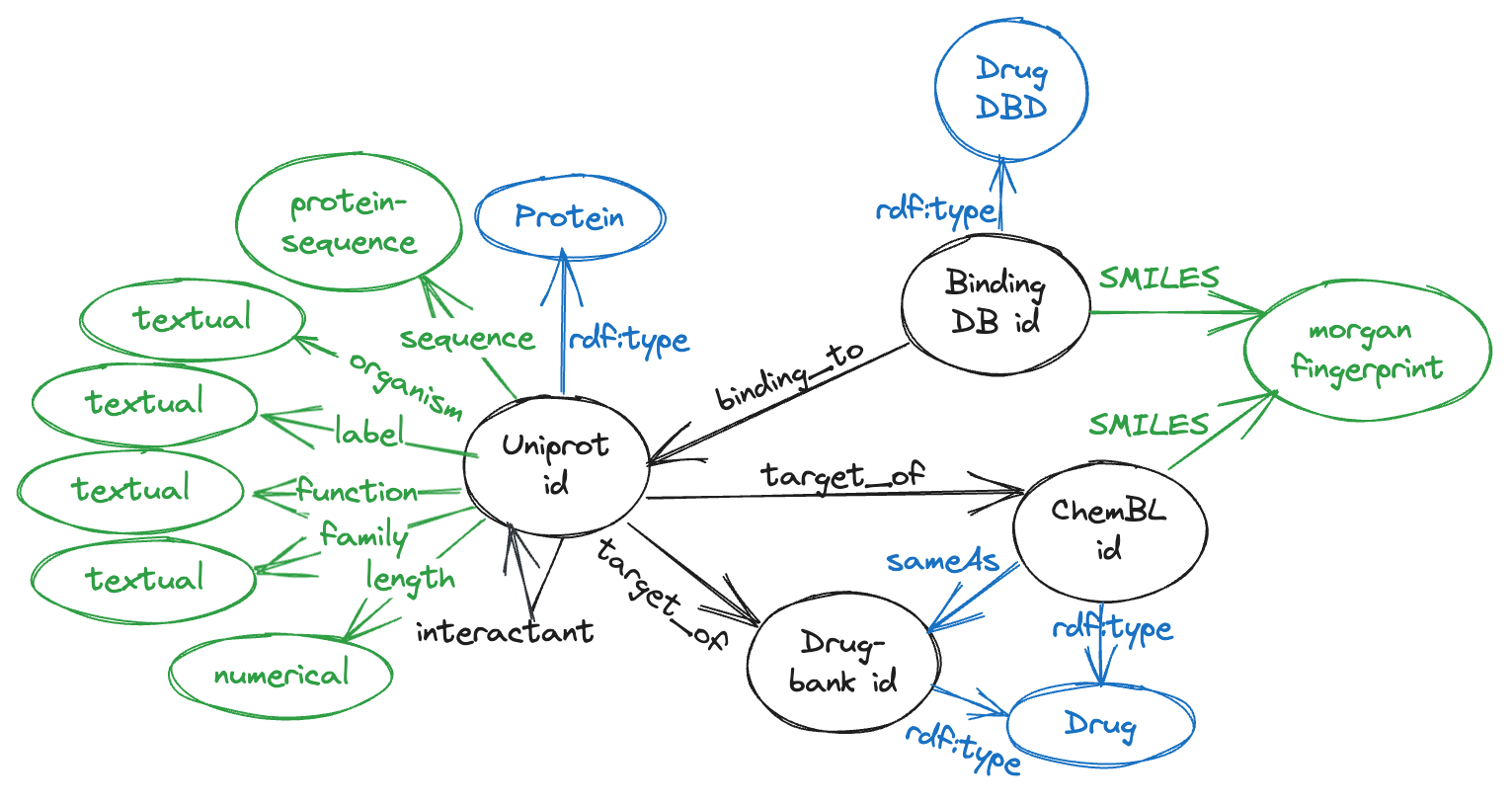
Dataset details
Uniprot
Uniprot comprises of 573,227 proteins from SwissProt, which is the subset of manually curated entries within UniProt, including attributes with different modalities like the sequence (567,483 of them), full name, organism, protein family, description of its function, catalytics activity, pathways and its length. The number of edges are 38,665 of type target_of from Uniprot ids to both ChEMBL and Drugbank ids, and 196,133 interactants between Uniprot protein ids.
BindingDB
BindingDB consists of 2,656,221 data points, involving 1.2 million compounds and 9,000 targets. Instead of utilizing the affinity score, we generate a triple for each combination of drugs and proteins. In order to prevent any data leakage, we eliminate overlapping triples with the TDC DTI dataset. As a result, the dataset concludes with a total of 2,232,392 triples.
ChEMBL
ChemBL comprises of drug-like bioactive molecules, 10,261 ChEMBL ids with their corresponding SMILES were downloaded from OpenTargets, from which 7,610 have a sameAs link to drugbank id molecules.
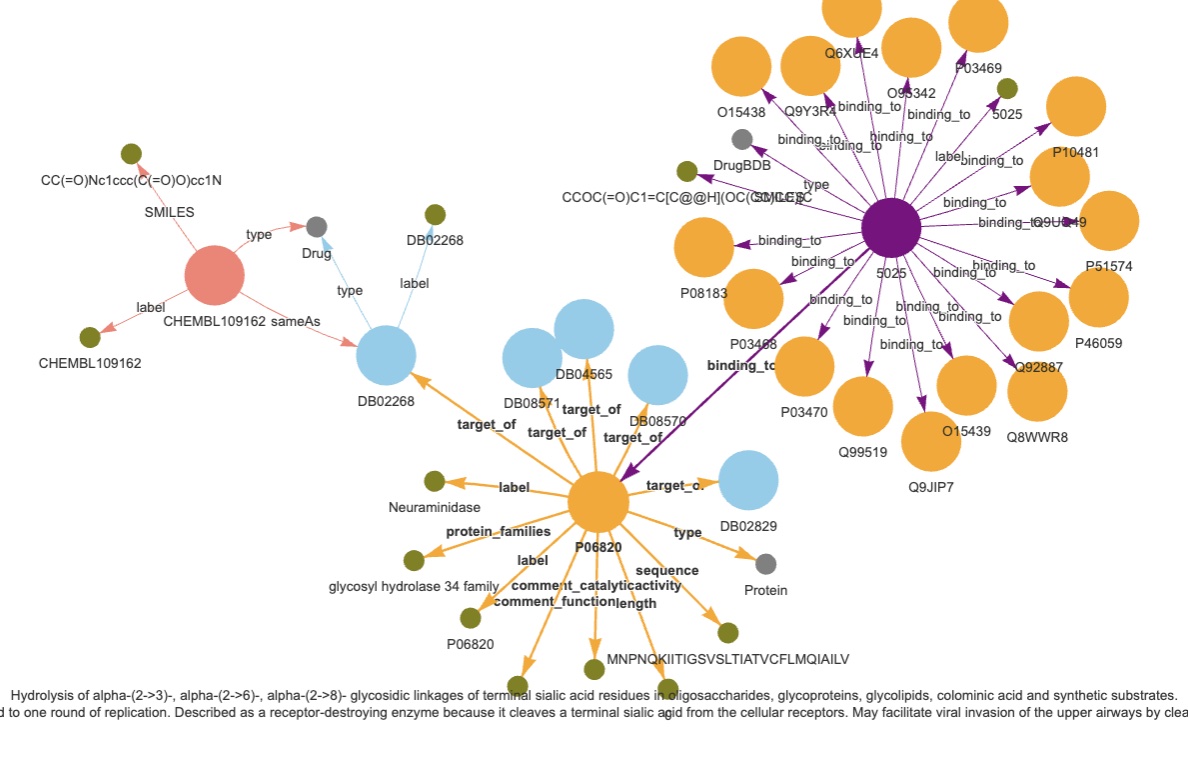
Original datasets:
- Uniprot: The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Research, 51(D1):D523–D531, 11 2022. ISSN 0305-1048. doi: 10.1093/nar/gkac1052. URL https://doi.org/10.1093/nar/gkac1052
- BindingDB: Tiqing Liu, Yuhmei Lin, Xin Wen, Robert N Jorissen, and Michael K Gilson. Bindingdb: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic acids research, 35(suppl_1):D198–D201, 2007.
- ChemBL: Anna Gaulton, Louisa J. Bellis, A. Patricia Bento, Jon Chambers, Mark Davies, Anne Hersey, Yvonne Light, Shaun McGlinchey, David Michalovich, Bissan Al-Lazikani, and John P. Overington. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Research, 40(D1):D1100–D1107, 09 2011. ISSN 0305-1048. doi: 10.1093/nar/gkr777. URL https://doi.org/10.1093/nar/gkr777
Paper or resources for more information:
License:
MIT
Where to send questions or comments about the dataset:
Models trained on Otter UBC
- Downloads last month
- 48