Datasets:
Add metadata
#2
by
egrace479
- opened
- .gitattributes +2 -0
- README.md +170 -40
- metadata/licenses.csv +0 -0
- metadata/rarespecies-catalog.csv +0 -0
- visuals/phyla_ToL_tree.html +0 -0
- visuals/phyla_ToL_tree.pdf +3 -0
- visuals/phyla_ToL_tree.png +3 -0
.gitattributes
CHANGED
|
@@ -53,3 +53,5 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 53 |
*.jpg filter=lfs diff=lfs merge=lfs -text
|
| 54 |
*.jpeg filter=lfs diff=lfs merge=lfs -text
|
| 55 |
*.webp filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
| 53 |
*.jpg filter=lfs diff=lfs merge=lfs -text
|
| 54 |
*.jpeg filter=lfs diff=lfs merge=lfs -text
|
| 55 |
*.webp filter=lfs diff=lfs merge=lfs -text
|
| 56 |
+
*.pdf filter=lfs diff=lfs merge=lfs -text
|
| 57 |
+
|
README.md
CHANGED
|
@@ -1,122 +1,252 @@
|
|
| 1 |
---
|
| 2 |
-
|
| 3 |
-
- zero-shot-classification
|
| 4 |
-
- image-classification
|
| 5 |
language:
|
| 6 |
- en
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 7 |
tags:
|
| 8 |
- biology
|
| 9 |
- image
|
| 10 |
- animals
|
| 11 |
- species
|
| 12 |
- taxonomy
|
| 13 |
-
- morphology
|
| 14 |
- rare species
|
| 15 |
- endangered species
|
| 16 |
-
-
|
| 17 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 18 |
---
|
| 19 |
|
| 20 |
# Dataset Card for Rare Species Dataset
|
| 21 |
|
| 22 |
## Dataset Description
|
| 23 |
|
| 24 |
-
- **Homepage:**
|
| 25 |
-
- **Repository:**
|
| 26 |
-
- **Paper:**
|
| 27 |
-
- **Leaderboard:**
|
| 28 |
-
- **Point of Contact:**
|
| 29 |
|
| 30 |
### Dataset Summary
|
| 31 |
|
| 32 |
-
This dataset
|
| 33 |
-
|
| 34 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 35 |
|
| 36 |
### Supported Tasks and Leaderboards
|
| 37 |
|
| 38 |
-
|
| 39 |
|
| 40 |
-
|
| 41 |
|
| 42 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 43 |
|
| 44 |
## Dataset Structure
|
| 45 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 46 |
### Data Instances
|
| 47 |
|
| 48 |
-
[
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 49 |
|
| 50 |
### Data Fields
|
| 51 |
|
| 52 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 53 |
|
| 54 |
### Data Splits
|
| 55 |
|
| 56 |
-
[
|
| 57 |
|
| 58 |
## Dataset Creation
|
| 59 |
|
| 60 |
### Curation Rationale
|
| 61 |
|
| 62 |
-
This dataset was generated with the purpose of providing a test set for the [Imageomics BioCLIP model](https://huggingface.co/imageomics/bioclip) to demonstrate robustness on data with minimal training samples available.
|
| 63 |
|
| 64 |
### Source Data
|
| 65 |
|
|
|
|
|
|
|
| 66 |
#### Initial Data Collection and Normalization
|
| 67 |
|
| 68 |
-
[
|
|
|
|
|
|
|
| 69 |
|
| 70 |
-
|
| 71 |
|
| 72 |
-
[More Information Needed]
|
| 73 |
|
| 74 |
### Annotations
|
| 75 |
|
| 76 |
#### Annotation process
|
| 77 |
|
| 78 |
-
[
|
|
|
|
|
|
|
| 79 |
|
| 80 |
#### Who are the annotators?
|
| 81 |
|
| 82 |
-
|
| 83 |
|
| 84 |
### Personal and Sensitive Information
|
| 85 |
|
| 86 |
-
[
|
| 87 |
|
| 88 |
## Considerations for Using the Data
|
| 89 |
-
|
| 90 |
### Social Impact of Dataset
|
| 91 |
|
| 92 |
-
|
| 93 |
|
| 94 |
### Discussion of Biases
|
| 95 |
|
| 96 |
-
|
| 97 |
-
|
| 98 |
-
### Other Known Limitations
|
| 99 |
|
| 100 |
-
[More Information Needed]
|
| 101 |
|
| 102 |
## Additional Information
|
| 103 |
|
| 104 |
### Dataset Curators
|
| 105 |
|
| 106 |
-
|
| 107 |
-
|
| 108 |
|
| 109 |
### Licensing Information
|
| 110 |
|
| 111 |
-
The data (images and text) contain a variety of licensing restrictions
|
| 112 |
-
|
|
|
|
|
|
|
|
|
|
| 113 |
|
| 114 |
### Citation Information
|
| 115 |
|
| 116 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 117 |
|
| 118 |
-
Please be sure to also cite the original data source(s) and all constituent parts as appropriate.
|
| 119 |
|
| 120 |
### Contributions
|
| 121 |
|
| 122 |
-
The [Imageomics Institute](https://imageomics.org) is funded by the US National Science Foundation's Harnessing the Data Revolution (HDR)
|
|
|
|
| 1 |
---
|
| 2 |
+
License: cc0-1.0
|
|
|
|
|
|
|
| 3 |
language:
|
| 4 |
- en
|
| 5 |
+
- la
|
| 6 |
+
pretty_name: Rare Species Dataset
|
| 7 |
+
task_categories:
|
| 8 |
+
- image-classification
|
| 9 |
+
- zero-shot-classification
|
| 10 |
tags:
|
| 11 |
- biology
|
| 12 |
- image
|
| 13 |
- animals
|
| 14 |
- species
|
| 15 |
- taxonomy
|
|
|
|
| 16 |
- rare species
|
| 17 |
- endangered species
|
| 18 |
+
- evolutionary biology
|
| 19 |
+
- balanced
|
| 20 |
+
- CV
|
| 21 |
+
- multimodal
|
| 22 |
+
- CLIP
|
| 23 |
+
- knowledge-guided
|
| 24 |
+
size_categories: 10K<n<100K
|
| 25 |
---
|
| 26 |
|
| 27 |
# Dataset Card for Rare Species Dataset
|
| 28 |
|
| 29 |
## Dataset Description
|
| 30 |
|
| 31 |
+
<!-- - **Homepage:** -->
|
| 32 |
+
- **Repository:** [Imageomics/bioclip](https://github.com/Imageomics/bioclip)
|
| 33 |
+
- **Paper:** BioCLIP: A Vision Foundation Model for the Tree of Life ([arXiv](https://doi.org/10.48550/arXiv.2311.18803))
|
| 34 |
+
<!-- - **Leaderboard:** -->
|
|
|
|
| 35 |
|
| 36 |
### Dataset Summary
|
| 37 |
|
| 38 |
+
This dataset was generated alongside [TreeOfLife-10M](https://huggingface.co/datasets/imageomics/TreeOfLife-10M); data (images and text) were pulled from [Encyclopedia of Life (EOL)](https://eol.org) to generate a dataset consisting of rare species for zero-shot-classification and more refined image classification tasks. Here, we use "rare species" to mean species listed on [The International Union for Conservation of Nature (IUCN) Red List](https://www.iucnredlist.org/) as Near Threatened, Vulnerable, Endangered, Critically Endangered, and Extinct in the Wild.
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
<!--This dataset card aims to be a base template for new datasets. It has been generated using [this raw template](https://github.com/huggingface/huggingface_hub/blob/main/src/huggingface_hub/templates/datasetcard_template.md?plain=1). And further altered to suit Imageomics Institute needs.-->
|
| 42 |
+
|
| 43 |
+
|
| 44 |
+
|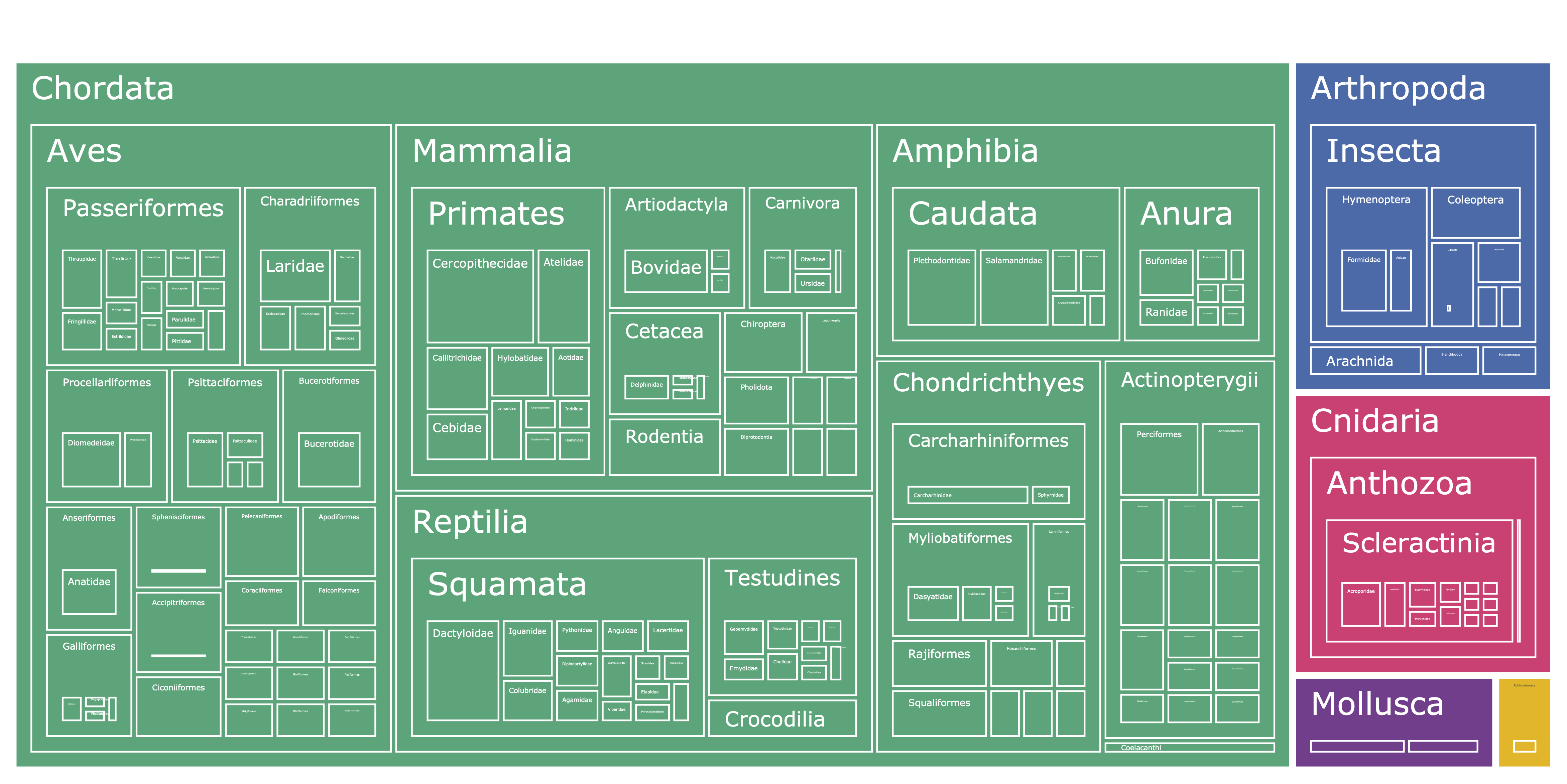|
|
| 45 |
+
|:--|
|
| 46 |
+
|**Figure 1.** Treemap from phyla down to family for Rare Species dataset. Interactive version available in [`visuals`](https://huggingface.co/imageomics/rare-species/tree/main/visuals) folder.|
|
| 47 |
+
|
| 48 |
|
| 49 |
### Supported Tasks and Leaderboards
|
| 50 |
|
| 51 |
+
Image Classification, Zero-shot and few-shot Classification.
|
| 52 |
|
| 53 |
+
Baseline for Random guessing is 0.3.
|
| 54 |
|
| 55 |
+
|
| 56 |
+
| Model | | Rare Species Classification Results | |
|
| 57 |
+
| ---- | :----: | :----: | :----: |
|
| 58 |
+
| | _Zero-Shot Classification_ | _One-Shot Classification_ | _Five-Shot Classification_ |
|
| 59 |
+
| CLIP | 31.81 | 29.26 | 47.45 |
|
| 60 |
+
| OpenCLIP | 29.85 | 28.52 | 46.07 |
|
| 61 |
+
| BioCLIP | **38.09** | **44.9** | **65.7** |
|
| 62 |
+
| --iNat21 Only | 21.33 | 36.94 | 55.65 |
|
| 63 |
+
|
| 64 |
+
| |
|
| 65 |
+
| -- |
|
| 66 |
+
| Zero-, one- and five-shot classification top-1 accuracy for different CLIP models. **Bold** indicates best accuracy. All models use the same architecture: ViT-B/16 vision encoders, 77-token text encoder. "iNat21 Only" follows the same procedure as BioCLIP but uses iNat21 instead of TreeOfLife-10M. CLIP and OpenCLIP are tested on common name, while BioCLIP and iNat21 Only were tested on full taxonomic name + common name. In this manner, we compare the optimal CLIP and OpenCLIP performance (both were primarily trained with common names). |
|
| 67 |
+
|
| 68 |
+
### Languages
|
| 69 |
+
English, Latin
|
| 70 |
|
| 71 |
## Dataset Structure
|
| 72 |
|
| 73 |
+
```
|
| 74 |
+
/dataset/
|
| 75 |
+
<kingdom-phylum-class-order-family-genus-species-1>/
|
| 76 |
+
<eol_content_id_1>_<eol_page_id>_eol_full-size-copy.jpg
|
| 77 |
+
<eol_content_id_2>_<eol_page_id>_eol_full-size-copy.jpg
|
| 78 |
+
...
|
| 79 |
+
<eol_content_id_30>_<eol_page_id>_eol_full-size-copy.jpg
|
| 80 |
+
<kingdom-phylum-class-order-family-genus-species-2>/
|
| 81 |
+
<eol_content_id_1>_<eol_page_id>_eol_full-size-copy.jpg
|
| 82 |
+
<eol_content_id_2>_<eol_page_id>_eol_full-size-copy.jpg
|
| 83 |
+
...
|
| 84 |
+
<eol_content_id_30>_<eol_page_id>_eol_full-size-copy.jpg
|
| 85 |
+
...
|
| 86 |
+
<kingdom-phylum-class-order-family-genus-species-400>/
|
| 87 |
+
<eol_content_id_1>_<eol_page_id>_eol_full-size-copy.jpg
|
| 88 |
+
<eol_content_id_2>_<eol_page_id>_eol_full-size-copy.jpg
|
| 89 |
+
...
|
| 90 |
+
<eol_content_id_30>_<eol_page_id>_eol_full-size-copy.jpg
|
| 91 |
+
metadata/
|
| 92 |
+
rarespecies-catalog.csv
|
| 93 |
+
licenses.csv
|
| 94 |
+
visuals/
|
| 95 |
+
phyla_ToL_tree.html
|
| 96 |
+
phyla_ToL_tree.pdf
|
| 97 |
+
phyla_ToL_tree.png
|
| 98 |
+
```
|
| 99 |
+
|
| 100 |
### Data Instances
|
| 101 |
|
| 102 |
+
This dataset is a collection of images with associated text. The text matched to images contains both [Linnaean taxonomy](https://www.britannica.com/science/taxonomy/The-objectives-of-biological-classification) (kingdom through species) for the particular subject of the image and its scientific name (`<genus> <species>`). All images have full 7-rank taxonomy filled, and are included in the [IUCN Red List](https://www.iucnredlist.org/) categories Near Threatened, Vulnerable, Endangered, Critically Endangered, and Extinct in the Wild. There are 30 images per species for the 400 species included.*
|
| 103 |
+
|
| 104 |
+
The images in this dataset are JPGs with filenames `<eol_content_id>_<eol_page_id>_eol_full-size-copy.jpg`. See Metadata Files below for definition of the IDs.
|
| 105 |
+
|
| 106 |
+
*It was discovered after training on TreeOfLife-10M that of the 400 species held out, 5 did not actually have 30 unique images, despite each image having unique EOL content IDs and EOL full-size image URLs. These species are as follows:
|
| 107 |
+
| Species | Number of Unique Images |
|
| 108 |
+
| --- | -- |
|
| 109 |
+
| _Pheidole elecebra_ | 21 |
|
| 110 |
+
| _Calumma ambreense_ | 27 |
|
| 111 |
+
| _Acanthochelys macrocephala_ | 27 |
|
| 112 |
+
| _Haliaeetus vociferoides_ | 29 |
|
| 113 |
+
| _Wallago attu_ | 29 |
|
| 114 |
+
|
| 115 |
|
| 116 |
### Data Fields
|
| 117 |
|
| 118 |
+
#### Metadata Files
|
| 119 |
+
|
| 120 |
+
`rarespecies-catalog.csv`: contains the following metadata associated with each image in the dataset
|
| 121 |
+
- `rarespecies_id`: unique identifier for the image in the dataset.
|
| 122 |
+
- `eol_content_id`: unique identifier within EOL database for images sourced from [EOL](https://eol.org). Note that EOL content IDs are not stable.
|
| 123 |
+
- `eol_page_id`: identifier of page from which images from EOL are sourced. Note that an image's association to a particular page ID may change with updates to the EOL (or image provider's) hierarchy. However, EOL taxon page IDs are stable.
|
| 124 |
+
The remaining terms describe the Linnaean taxonomy of the subject of the images; application of these labels is described below in the [annotation process](#annotation-process).
|
| 125 |
+
- `kingdom`: kingdom to which the subject of the image belongs (all `Animalia`).
|
| 126 |
+
- `phylum`: phylum to which the subject of the image belongs.
|
| 127 |
+
- `class`: class to which the subject of the image belongs.
|
| 128 |
+
- `order`: order to which the subject of the image belongs.
|
| 129 |
+
- `family`: family to which the subject of the image belongs.
|
| 130 |
+
- `genus`: genus to which the subject of the image belongs.
|
| 131 |
+
- `species`: species to which the subject of the image belongs.
|
| 132 |
+
- `sciName`: scientific name associated with the subject of the image (`genus-species`).
|
| 133 |
+
- `common`: common name associated with the subject of the image. Note that there are only 398 unique common names; it is not uncommon for species of the same genera to share a common name. The two specific instances are _Acropora acuminata_ and _Acropora millepora_, which share the common name staghorn coral, and both _Tylototriton shanjing_ and _Tylototriton verrucosus_ have the common name Yunnan Newt.
|
| 134 |
+
|
| 135 |
+
`licenses.csv`: File with license, source, and copyright holder associated to each image listed in `rarespecies-catalog.csv`; `rarespecies_id` is the shared unique identifier to link the two files. Columns are
|
| 136 |
+
- `rarespecies_id`, `eol_content_id`, and `eol_page_id` are as defined above.
|
| 137 |
+
- `md5`: MD5 hash of the image.
|
| 138 |
+
- `medium_source_url`: URL pointing to source of image.
|
| 139 |
+
- `eol_full_size_copy_url`: URL to access the full-sized image; this is the URL from which the image was downloaded for this dataset (see [Initial Data Collection and Normalization](#initial-data-collection-and-normalization) for more information on this process).
|
| 140 |
+
- `license_name`: name of license attached to the image (eg., `cc-by`).
|
| 141 |
+
- `copyright_owner`: copyright holder for the image, filled with `not provided` if no copyright owner was provided.
|
| 142 |
+
- `license_link`: URL to the listed license, left null in the case that `License Name` is `No known copyright restrictions`.
|
| 143 |
+
- `title`: title provided for the image, filled with `not provided` if no title was provided.
|
| 144 |
+
|
| 145 |
+
|
| 146 |
+
|
| 147 |
+
The visuals folder has treemaps that were generated by feeding `rarespecies-catalog.csv` to the `taxa_viz` script in the [BioCLIP GitHub repository](https://github.com/Imageomics/bioclip).
|
| 148 |
+
|
| 149 |
|
| 150 |
### Data Splits
|
| 151 |
|
| 152 |
+
This entire dataset was used for testing the [BioCLIP model](https://huggingface.co/imageomics/bioclip), which was trained on [TreeOfLife-10M](https://huggingface.co/datasets/imageomics/TreeOfLife-10M).
|
| 153 |
|
| 154 |
## Dataset Creation
|
| 155 |
|
| 156 |
### Curation Rationale
|
| 157 |
|
| 158 |
+
This dataset was generated with the purpose of providing a biologically meaningful test set for the [Imageomics BioCLIP model](https://huggingface.co/imageomics/bioclip) to demonstrate robustness on data with minimal training samples available and biologically meaningful potential applications.
|
| 159 |
|
| 160 |
### Source Data
|
| 161 |
|
| 162 |
+
[EOL](https://eol.org) and [IUCN Red List](https://www.iucnredlist.org/)
|
| 163 |
+
|
| 164 |
#### Initial Data Collection and Normalization
|
| 165 |
|
| 166 |
+
The IUCN Red List of Threatened Species categorization of animals was pulled from the [IUCN website](https://www.iucnredlist.org/). There are approximately 25,000 species that fall into the categories Near Threatened, Vulnerable, Endangered, Critically Endangered, and Extinct in the Wild (as of July 13, 2023), though image availability on EOL is not consistent across species. We select 400 species from the list under the condition there are at least 30 images per species available and they are not species in [iNat21](https://kaggle.com/competitions/inaturalist-2021) or [BIOSCAN-1M](https://zenodo.org/doi/10.5281/zenodo.8030064) datasets which were also used to generate [TreeOfLife-10M](https://huggingface.co/datasets/imageomics/TreeOfLife-10M). A random subset of 30 images is then selected for each species in this collection.
|
| 167 |
+
|
| 168 |
+
This dataset was generated concurrently with [TreeOfLife-10M](https://huggingface.co/datasets/imageomics/TreeOfLife-10M), so the process is as described [there](https://huggingface.co/datasets/imageomics/TreeOfLife-10M#initial-data-collection-and-normalization), with the exception that these images were entirely sourced from EOL, and the species represented were excluded from the TreeOfLife-10M dataset.
|
| 169 |
|
| 170 |
+
The IUCN data was used for selection of the included species, and is not reproduced here. [This link](https://www.iucnredlist.org/search?permalink=ab8daad6-d564-4370-b8e6-9c5ac9f8336f) provides the search used to gather the list of species classified as Near Threatened to Extinct in the Wild. The results were downloaded on July 13, 2023, but note the results are subject to change with IUCN Red List Updates ([IUCN Update Schedule](https://www.iucnredlist.org/assessment/updates)).
|
| 171 |
|
|
|
|
| 172 |
|
| 173 |
### Annotations
|
| 174 |
|
| 175 |
#### Annotation process
|
| 176 |
|
| 177 |
+
Annotations were primarily sourced from EOL (image source provider) following the procedure described in the [TreeOfLife-10M annotation process](https://huggingface.co/datasets/imageomics/TreeOfLife-10M#annotation-process). [IUCN Red List](https://www.iucnredlist.org/) was then used for filtering these taxa out of [TreeOfLife-10M](https://huggingface.co/datasets/imageomics/TreeOfLife-10M) to create this Rare Species dataset.
|
| 178 |
+
|
| 179 |
+
The scientific name (`genus-species`, as labeled by EOL) was used to look up the higher-order taxa from EOL aggregate datasets (described below), then matched against the ITIS hierarchy for the higher-order taxa standardization. A small number of these are [homonyms](https://en.wikipedia.org/wiki/Homonym_(biology)), for which a list was generated to ensure proper matching of higher-order taxa. After these resources were exhausted, any remaining unresolved taxa were fed through the [Global Names Resolver (GNR) API](https://resolver.globalnames.org/api).
|
| 180 |
|
| 181 |
#### Who are the annotators?
|
| 182 |
|
| 183 |
+
Samuel Stevens, Jiaman Wu, Matthew J. Thompson, and Elizabeth G. Campolongo
|
| 184 |
|
| 185 |
### Personal and Sensitive Information
|
| 186 |
|
| 187 |
+
All animals included in this dataset are listed as Near Threatened, Vulnerable, Endangered, Critically Endangered, or Extinct in the Wild by the [IUCN Red List](https://www.iucnredlist.org/) as of July 13, 2023 (IUCN generally updates classifications twice each year (see the [IUCN Update Schedule](https://www.iucnredlist.org/assessment/updates) for more information)). However, the specific ranking is not tied to any individual, and there is no geographical information included.
|
| 188 |
|
| 189 |
## Considerations for Using the Data
|
|
|
|
| 190 |
### Social Impact of Dataset
|
| 191 |
|
| 192 |
+
The hope is that this dataset could be helpful in conservation efforts or biodiversity research.
|
| 193 |
|
| 194 |
### Discussion of Biases
|
| 195 |
|
| 196 |
+
Inclusion of a species in this dataset required that EOL provided at least 30 images of it, so there are only 400 of the 25,000 species in these categories included, and only 30 images per species. Additionally, all included species are in the kingdom, _Animalia_, and within 5 phyla.
|
|
|
|
|
|
|
| 197 |
|
|
|
|
| 198 |
|
| 199 |
## Additional Information
|
| 200 |
|
| 201 |
### Dataset Curators
|
| 202 |
|
| 203 |
+
Samuel Stevens, Jiaman Wu, Matthew J. Thompson, and Elizabeth G. Campolongo
|
|
|
|
| 204 |
|
| 205 |
### Licensing Information
|
| 206 |
|
| 207 |
+
The data (images and text) contain a variety of licensing restrictions ranging from [CC0](https://creativecommons.org/publicdomain/zero/1.0/) to [CC BY-NC-SA](https://creativecommons.org/licenses/by-nc-sa/4.0/). Each image and text in this dataset is provided under the least restrictive terms allowed by its licensing requirements as provided to us (i.e, we impose no additional restrictions past those specified by licenses in the license file).
|
| 208 |
+
|
| 209 |
+
This dataset (the compilation) has been marked as dedicated to the public domain by applying the [CC0 Public Domain Waiver](https://creativecommons.org/publicdomain/zero/1.0/). However, images may be licensed under different terms (as noted above).
|
| 210 |
+
For license and citation information by image, see our [license file](https://huggingface.co/datasets/imageomics/rare-species/blob/main/metadata/licenses.csv).
|
| 211 |
+
|
| 212 |
|
| 213 |
### Citation Information
|
| 214 |
|
| 215 |
+
```
|
| 216 |
+
@dataset{rare_species_2023,
|
| 217 |
+
author = {Samuel Stevens and Jiaman Wu and Matthew J Thompson and Elizabeth G Campolongo and Chan Hee Song and David Edward Carlyn and Li Dong and Wasila M Dahdul and Charles Stewart and Tanya Berger-Wolf and Wei-Lun Chao and Yu Su},
|
| 218 |
+
title = {Rare Species},
|
| 219 |
+
year = {2023},
|
| 220 |
+
url = {https://huggingface.co/datasets/imageomics/rare-species},
|
| 221 |
+
doi = {<doi once generated>},
|
| 222 |
+
publisher = {Hugging Face}
|
| 223 |
+
}
|
| 224 |
+
```
|
| 225 |
+
Please also cite our paper:
|
| 226 |
+
```
|
| 227 |
+
@article{stevens2023bioclip,
|
| 228 |
+
title = {BIOCLIP: A Vision Foundation Model for the Tree of Life},
|
| 229 |
+
author = {Samuel Stevens and Jiaman Wu and Matthew J Thompson and Elizabeth G Campolongo and Chan Hee Song and David Edward Carlyn and Li Dong and Wasila M Dahdul and Charles Stewart and Tanya Berger-Wolf and Wei-Lun Chao and Yu Su},
|
| 230 |
+
year = {2023},
|
| 231 |
+
eprint = {2311.18803},
|
| 232 |
+
archivePrefix = {arXiv},
|
| 233 |
+
primaryClass = {cs.CV}}
|
| 234 |
+
```
|
| 235 |
+
|
| 236 |
+
|
| 237 |
+
Please be sure to also cite the original data sources and all constituent parts as appropriate.
|
| 238 |
+
|
| 239 |
+
|
| 240 |
+
**EOL and IUCN classification data:**
|
| 241 |
+
|
| 242 |
+
IUCN. 2022. The IUCN Red List of Threatened Species. Version 2022-2. https://www.iucnredlist.org. Accessed on 5 July 2023. https://www.iucnredlist.org/search?permalink=ab8daad6-d564-4370-b8e6-9c5ac9f8336f.
|
| 243 |
+
|
| 244 |
+
Encyclopedia of Life. Available from http://eol.org. Accessed 29 July 2023.
|
| 245 |
+
|
| 246 |
+
|
| 247 |
+
For license and citation information by image, see our [license file](https://huggingface.co/datasets/imageomics/rare-species/blob/main/metadata/licenses.csv).
|
| 248 |
|
|
|
|
| 249 |
|
| 250 |
### Contributions
|
| 251 |
|
| 252 |
+
The [Imageomics Institute](https://imageomics.org) is funded by the US National Science Foundation's Harnessing the Data Revolution (HDR) program under [Award #2118240](https://www.nsf.gov/awardsearch/showAward?AWD_ID=2118240) (Imageomics: A New Frontier of Biological Information Powered by Knowledge-Guided Machine Learning). Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
|
metadata/licenses.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
metadata/rarespecies-catalog.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
visuals/phyla_ToL_tree.html
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
visuals/phyla_ToL_tree.pdf
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0cb51b358b978b256077586eb940aecc2b53da2566f95ff758b284e70cd69b2e
|
| 3 |
+
size 1048892
|
visuals/phyla_ToL_tree.png
ADDED

|
Git LFS Details
|