The viewer is disabled because this dataset repo requires arbitrary Python code execution. Please consider
removing the
loading script
and relying on
automated data support
(you can use
convert_to_parquet
from the datasets library). If this is not possible, please
open a discussion
for direct help.
S-SYNTH
S-SYNTH is an open-source, flexible skin simulation framework to rapidly generate synthetic skin models and images using digital rendering of an anatomically inspired multi-layer, multi-component skin and growing lesion model. It allows for generation of highly-detailed 3D skin models and digitally rendered synthetic images of diverse human skin tones, with full control of underlying parameters and the image formation process.
Framework Details
S-SYNTH can be used to generate synthetic skin images with annotations (including segmentation masks) with variations of:
- Hair artifact
- Blood fraction
- Lesion shape
- Melanosome fraction
- Sample per pixel (for rendering)
- Lighting condition
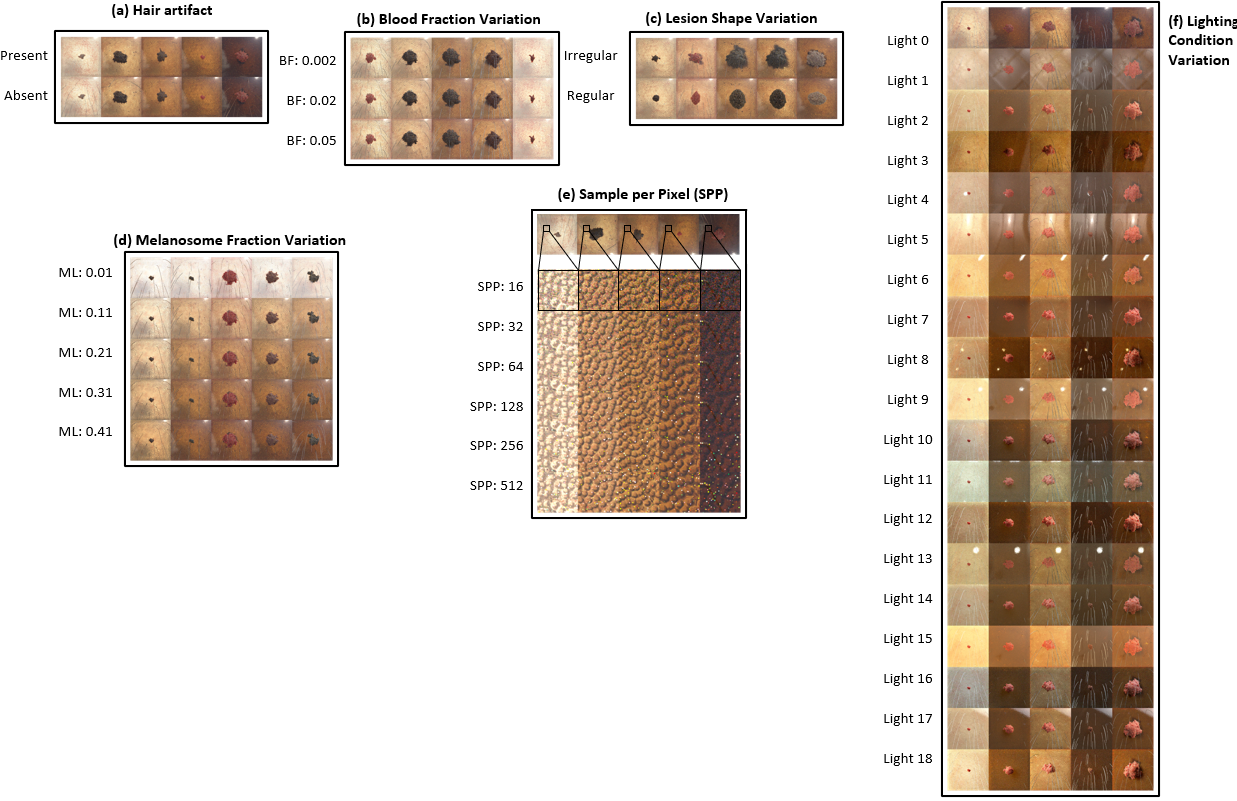
Framework Description
- Curated by: Andrea Kim, Niloufar Saharkhiz, Elena Sizikova, Miguel Lago, Berkman Sahiner, Jana Delfino, Aldo Badano
- License: Creative Commons 1.0 Universal License (CC0)
Dataset Sources
- Paper: https://arxiv.org/abs/2408.00191
- Code: https://github.com/DIDSR/ssynth-release
- Data: https://huggingface.co/datasets/didsr/ssynth_data
Uses
S-SYNTH is intended to facilitate augment limited patient datasets and identify AI performance trends by generating synthetic skin data.
Direct Use
S-SYNTH can be used to evaluate comparative AI performance trends in dermatologic analysis tasks, estimating the effect skin appearance, such as skin color, presence of hair, lesion shape, and blood fraction on AI performance in lesion segmentation. S-SYNTH can be used to mitigate effects of mislabeled examples and limitations of a limited patient dataset. S-SYNTH can be used to either train or test pre-trained AI models.
Out-of-Scope Use
S-SYNTH cannot be used in lieu of real patient examples to make performance determinations.
Dataset Structure
S-SYNTH is organized into a directory structure that indicates the parameters. The folder
data/synthetic_dataset/output_10k/output/skin_[SKIN_MODEL_ID]/hairModel_[HAIR_MODEL_ID]/mel_[MELANOSOME_FRACTION]/fB_[BLOOD_FRACTION]/lesion_[LESION_ID]/T_[TIMEPOINT]/[LESION_MATERIAL]/hairAlb_[HAIR_ALBEDO]/light_[LIGHT_NAME]/ROT[LESION_ROTATION_VECTOR]_[RANDOM_ANGLE]/
contains image files and paired segmentation masks with the specified parameters.
$ tree data/synthetic_dataset/output_10k/output/skin_099/hairModel_098/mel_0.47/fB_0.05/lesion_9/T_050/HbO2x1.0Epix0.25/hairAlb_0.84-0.6328-0.44/light_hospital_room_4k/ROT0-0-1_270/
data/synthetic_dataset/output_10k/output/skin_099/hairModel_098/mel_0.47/fB_0.05/lesion_9/T_050/HbO2x1.0Epix0.25/hairAlb_0.84-0.6328-0.44/light_hospital_room_4k/ROT0-0-1_270/
├── mask.png
├── image.png
├── cropped_mask.png
├── cropped_mask.png
└── crop_size.txt
Each folder contains two versions (cropped and uncropped) of a paired skin image and lesion segmentation mask. The images were cropped to ensure a variety of lesion size. The crop size is provided within the crop_size.txt file.
The accompanying data can be either directly downloaded, or accessed using the following HF data loader:
from datasets import load_dataset
data_test = load_dataset("didsr/ssynth_data", split="output_10k")
print(data_test)
Bias, Risks, and Limitations
Simulation-based testing is constrained to the parameter variability represented in the object model and the acquisition system. There is a risk of misjudging model performance if the simulated examples do not capture the variability in real patients. Please see the paper for a full discussion of biases, risks, and limitations.
How to use it
The data presented is intended to demonstrate the types of variations that the S-SYNTH skin model can simulate. The associated pre-generated datasets can be used for augmenting limited real patient training datasets for skin segmentation tasks.
Citation
@article{kim2024ssynth,
title={Knowledge-based in silico models and dataset for the comparative evaluation of mammography AI for a range of breast characteristics, lesion conspicuities and doses},
author={Kim, Andrea and Saharkhiz, Niloufar and Sizikova, Elena and Lago, Miguel, and Sahiner, Berkman and Delfino, Jana G., and Badano, Aldo},
journal={International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI)},
volume={},
pages={},
year={2024}
}
Related Links
- Downloads last month
- 357