The viewer is disabled because this dataset repo requires arbitrary Python code execution. Please consider
removing the
loading script
and relying on
automated data support
(you can use
convert_to_parquet
from the datasets library). If this is not possible, please
open a discussion
for direct help.
♆ Patho-Bench
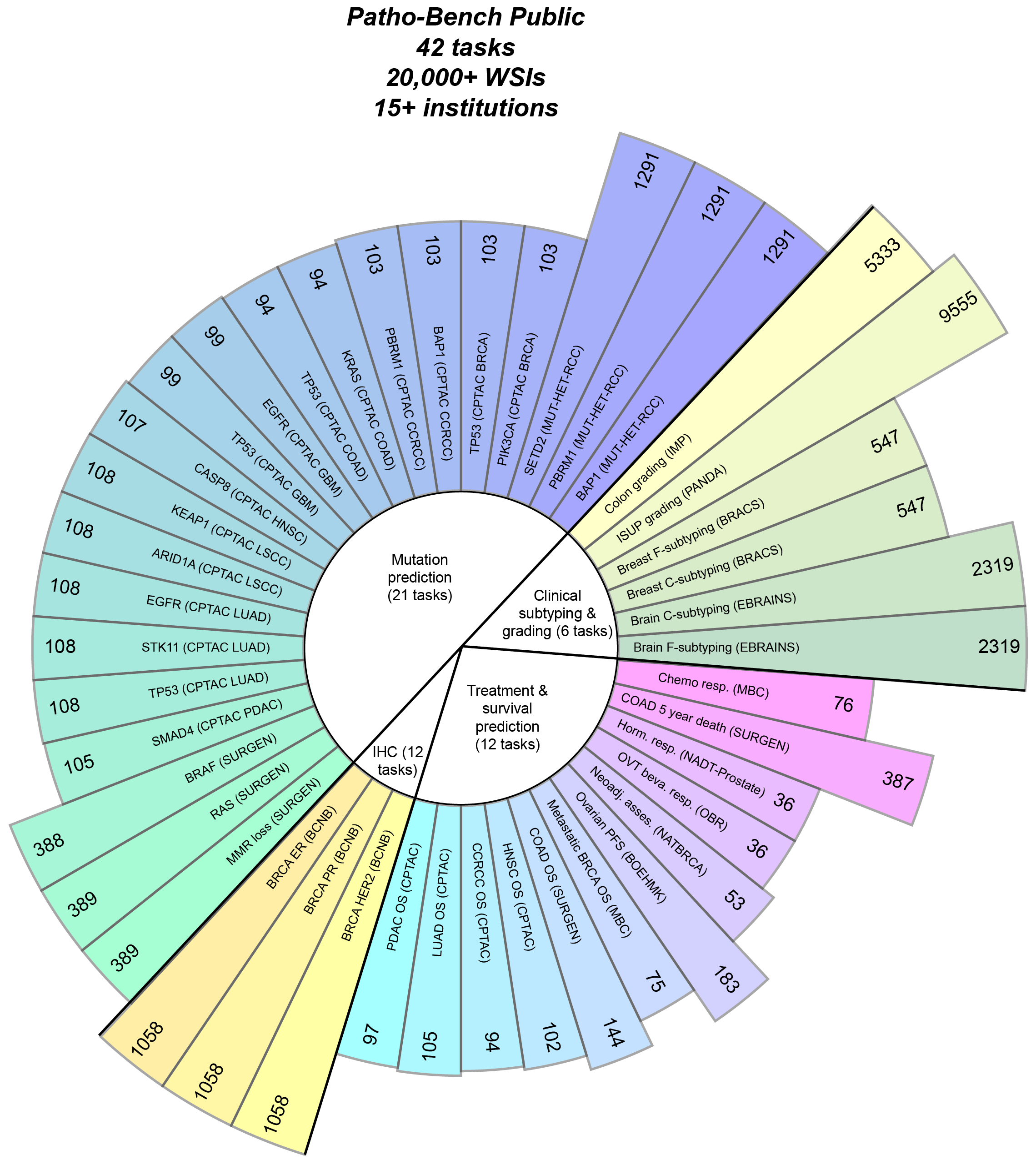
Patho-Bench is designed to evaluate patch and slide encoder foundation models for whole-slide images (WSIs).
This HuggingFace repository contains the data splits for the public Patho-Bench tasks. Please visit our codebase on GitHub for the full codebase and benchmark implementation.
This project was developed by the Mahmood Lab at Harvard Medical School and Brigham and Women's Hospital. This work was funded by NIH NIGMS R35GM138216.
Contributions are welcome! If you'd like to submit a new dataset and/or task for inclusion in Patho-Bench, please reach out to us via the Issues tab of our Github repo.
Currently, Patho-Bench contains the following task families. We will add more tasks in the future. For further details on each task, please refer to the THREADS foundation model paper.
| Family | Description | Tasks |
|---|---|---|
| Morphological Subtyping | Classifying distinct morphological patterns associated with different disease subtypes | 4 |
| Tumor Grading | Assigning a grade based on cellular differentiation and growth patterns | 2 |
| Molecular Subtyping | Predicting antigen presence (e.g., via IHC staining) | 3 |
| Mutation Prediction | Predicting specific genetic mutations in tumors | 21 |
| Treatment Response & Assessment | Evaluating patient response to treatment | 6 |
| Survival Prediction | Predicting survival outcomes and risk stratification | 6 |
🔥 Latest updates
- February 2025: Patho-Bench is now available on HuggingFace.
⚡ Installation
Install the required packages:
pip install --upgrade datasets
pip install --upgrade huggingface_hub
🔑 Authentication
from huggingface_hub import login
login(token="YOUR_HUGGINGFACE_TOKEN")
⬇️ Usage
The Patho-Bench data splits are designed for use with the Patho-Bench software package. However, you are welcome to use the data splits in your custom pipeline. Each task is associated with a YAML file containing task metadata and a TSV file containing the sample IDs, slide IDs, and labels.
Patho-Bench only provides the data splits and labels, NOT the raw image data. You will need to download the raw image data from the respective dataset repositories (see links below).
Download an individual task
import datasets
dataset='cptac_coad'
task='KRAS_mutation'
datasets.load_dataset(
'MahmoodLab/Patho-Bench',
cache_dir='/path/to/saveto',
dataset_to_download=dataset, # Throws error if source not found
task_in_dataset=task, # Throws error if task not found in dataset
trust_remote_code=True
)
Download all tasks from a dataset
import datasets
dataset='cptac_coad'
task='*'
datasets.load_dataset(
'MahmoodLab/Patho-Bench',
cache_dir='/path/to/saveto',
dataset_to_download=dataset,
task_in_dataset=task,
trust_remote_code=True
)
Download entire Patho-Bench [4.2 MB]
import datasets
dataset='*'
datasets.load_dataset(
'MahmoodLab/Patho-Bench',
cache_dir='/path/to/saveto',
dataset_to_download=dataset,
trust_remote_code=True
)
📢 Image data access links
For each dataset in Patho-Bench, please visit the respective repository below to download the raw image data.
📇 Contact
For any questions, contact:
- Faisal Mahmood (faisalmahmood@bwh.harvard.edu)
- Anurag Vaidya (avaidya@mit.edu)
- Andrew Zhang (andrewzh@mit.edu)
- Guillaume Jaume (gjaume@bwh.harvard.edu)
📜 Data description
Developed by: Mahmood Lab AI for Pathology @ Harvard/BWH Repository: GitHub License: CC-BY-NC-4.0
🤝 Acknowledgements
Patho-Bench tasks were compiled from public image datasets and repositories (linked above). We thank the authors of these datasets for making their data publicly available.
📰 How to cite
If Patho-Bench contributes to your research, please cite:
@article{vaidya2025molecular,
title={Molecular-driven Foundation Model for Oncologic Pathology},
author={Vaidya, Anurag and Zhang, Andrew and Jaume, Guillaume and Song, Andrew H and Ding, Tong and Wagner, Sophia J and Lu, Ming Y and Doucet, Paul and Robertson, Harry and Almagro-Perez, Cristina and others},
journal={arXiv preprint arXiv:2501.16652},
year={2025}
}
@article{zhang2025standardizing,
title={Accelerating Data Processing and Benchmarking of AI Models for Pathology},
author={Zhang, Andrew and Jaume, Guillaume and Vaidya, Anurag and Ding, Tong and Mahmood, Faisal},
journal={arXiv preprint arXiv:2502.06750},
year={2025}
}
- Downloads last month
- 2,910