YAML Metadata
Error:
"language[0]" must be a string
YAML Metadata
Error:
"tags[0]" must be a string
This cased model was pretrained from scratch using a custom vocabulary on the following corpora
- Pubmed
- Clinical trials corpus
- and a small subset of Bookcorpus
The pretrained model was used to do NER as is, with no fine-tuning. The approach is described in this post. Towards Data Science review
App in Spaces demonstrates this approach.
Github link to perform NER using this model in an ensemble with bert-base cased.
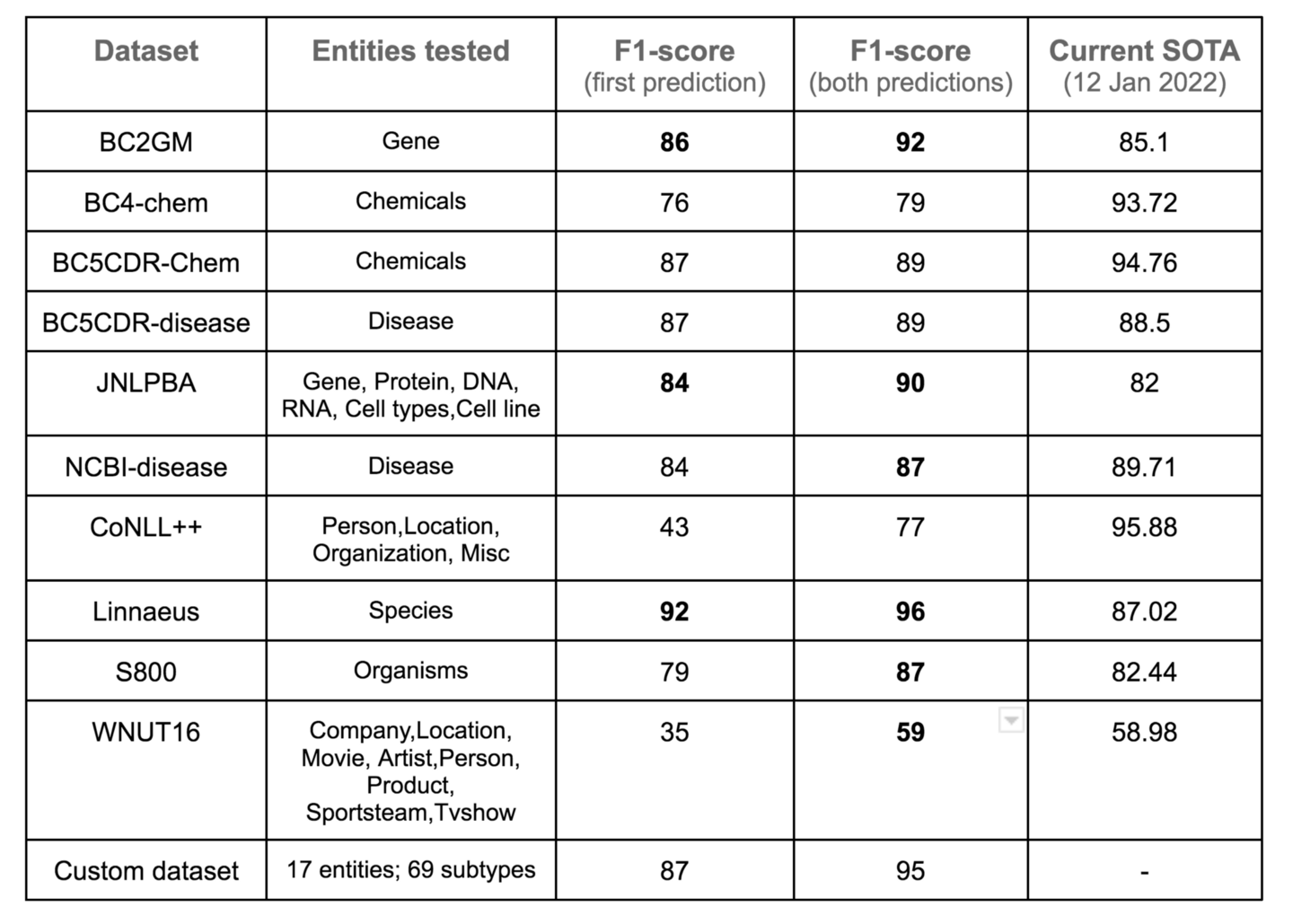
The ensemble detects 69 entity subtypes (17 broad entity groups)

Ensemble model performance
Additional notes
- The model predictions on the right do not include [CLS] predictions. Hosted inference API only returns the masked position predictions. In practice, the [CLS] predictions are just as useful as the model predictions for the masked position (if the next sentence prediction loss was low during pretraining) and are used for NER.
- Some of the top model predictions like "a", "the", punctuations, etc. while valid predictions, bear no entity information. These are filtered when harvesting descriptors for NER. The examples on the right are unfiltered results.
- Use this link to examine both fill-mask prediction and [CLS] predictions
License
MIT license

- Downloads last month
- 8
This model does not have enough activity to be deployed to Inference API (serverless) yet. Increase its social
visibility and check back later, or deploy to Inference Endpoints (dedicated)
instead.