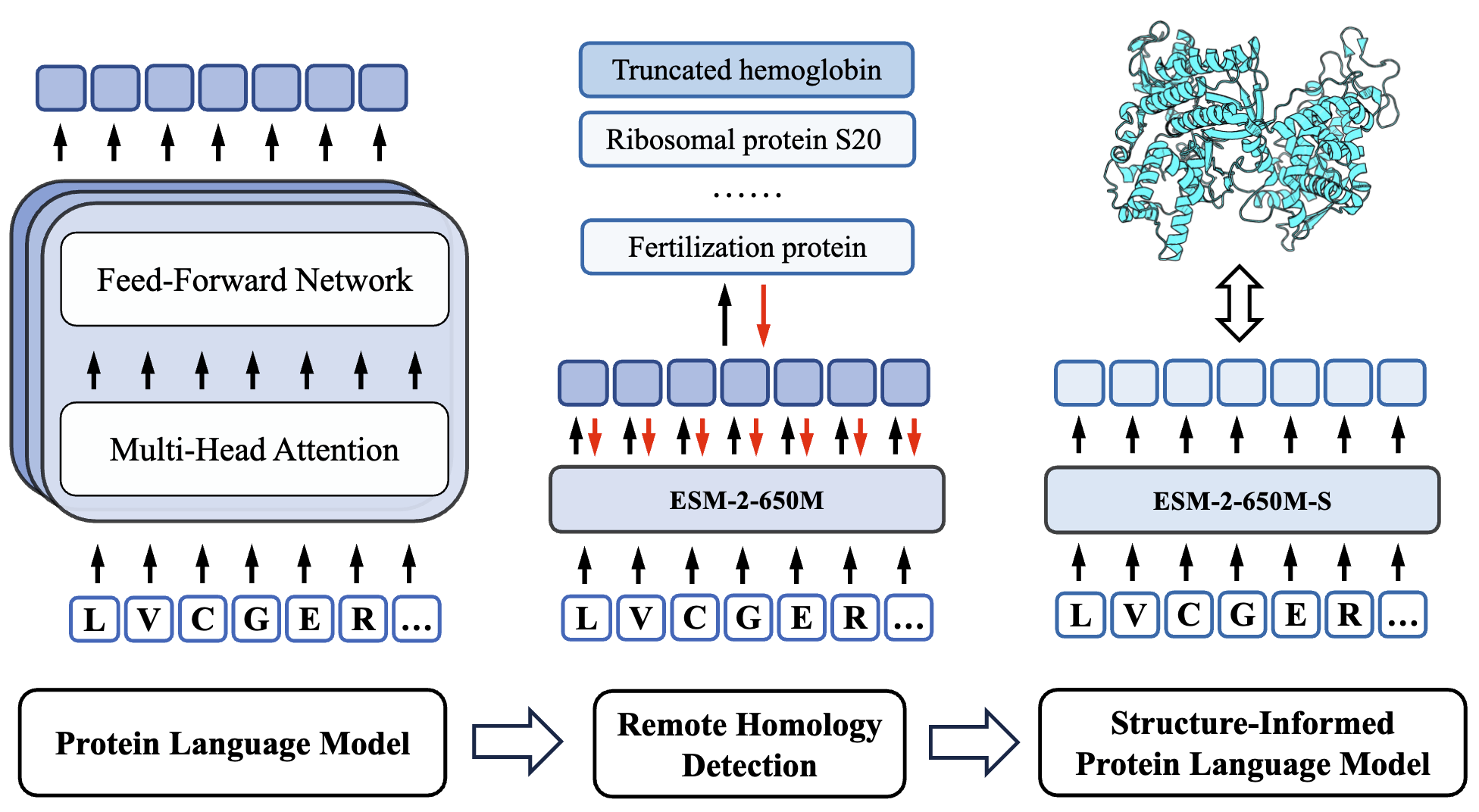
ESM-S
ESM-S (https://arxiv.org/abs/2402.05856) is a series of structure-informed protein language models, which are trained on remote homology detection tasks for distilling structural information. The corresponding datasets can be downloaded at https://huggingface.co/datasets/Oxer11/Protein-Function-Annotation. The codebase can be found at https://github.com/DeepGraphLearning/esm-s.
Evaluation Performance
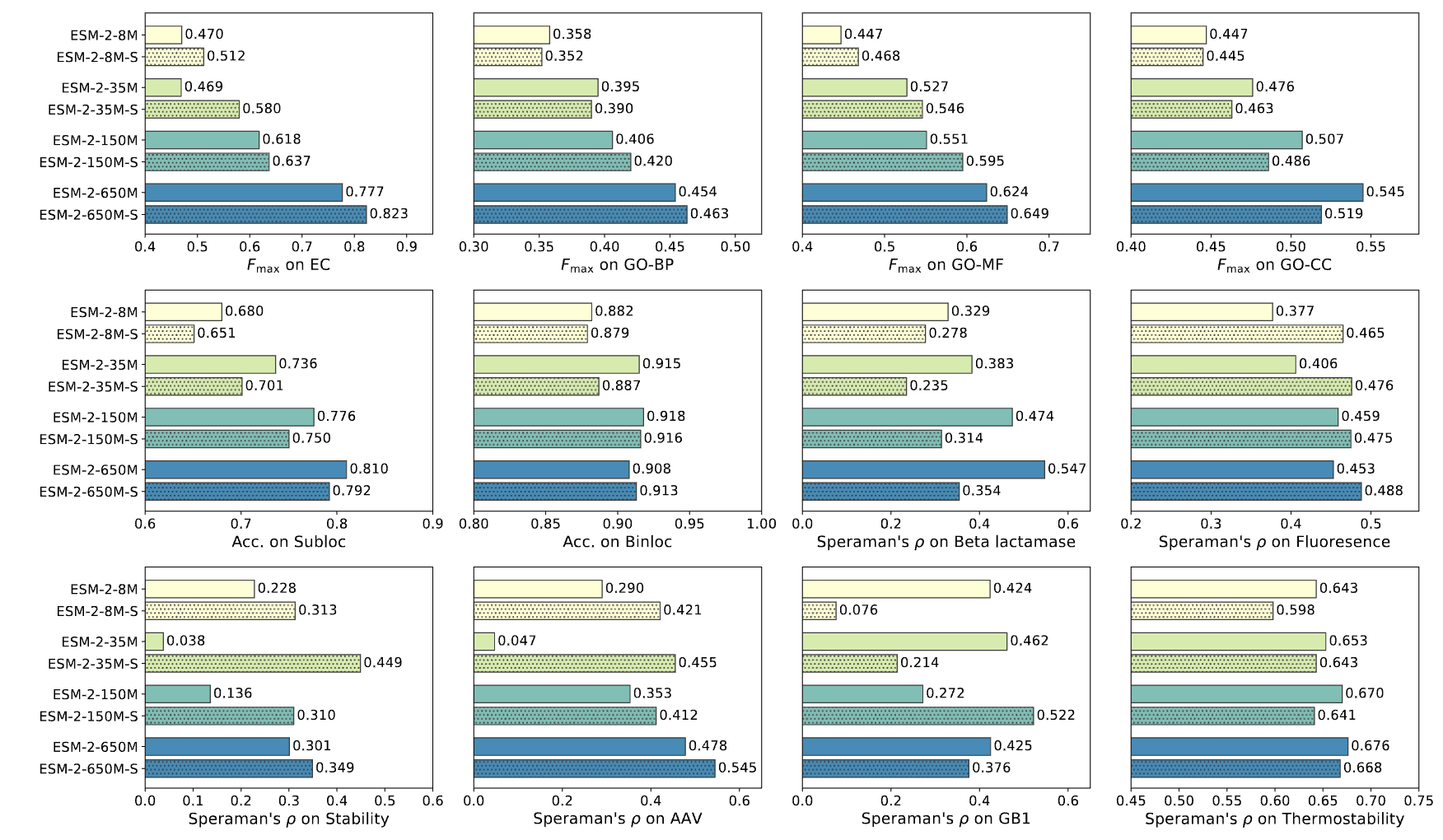
Freezing model weights and train a 2-layer MLP on downstream function prediction tasks.
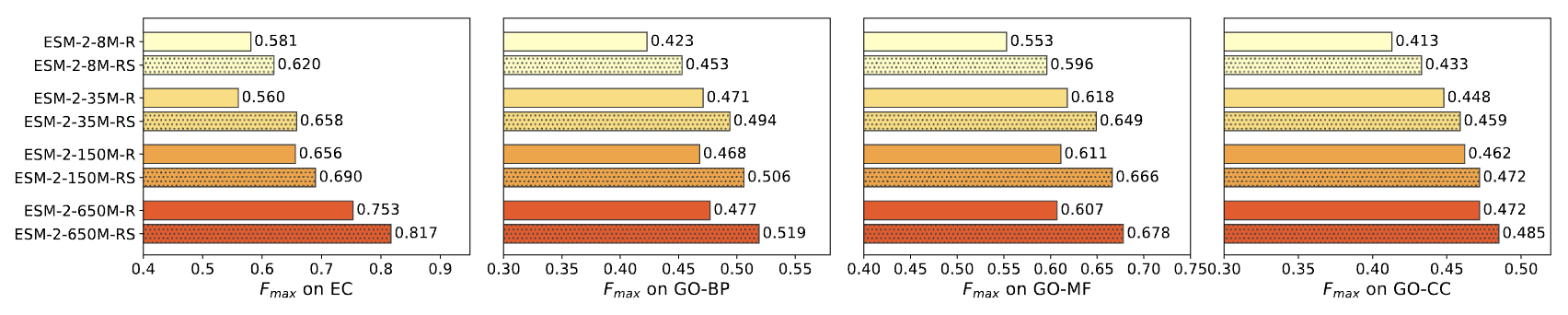
Using ESM-S representations to retrieve similar proteins for function annotation.
BibTeX
@article{zhang2024structureplm,
title={Structure-Informed Protein Language Model},
author={Zhang, Zuobai and Lu, Jiarui and Chenthamarakshan, Vijil and Lozano, Aurelie and Das, Payel and Tang, Jian},
journal={arXiv preprint arXiv:2402.05856},
year={2024}
}
- Downloads last month
- 0
Unable to determine this model's library. Check the
docs
.