license: mit
datasets:
- adrianhenkel/lucidprots_full_data
pipeline_tag: translation
tags:
- biology
Model Card for ProstT5
ProstT5 is a protein language model (pLM) which can translate between protein sequence and structure.
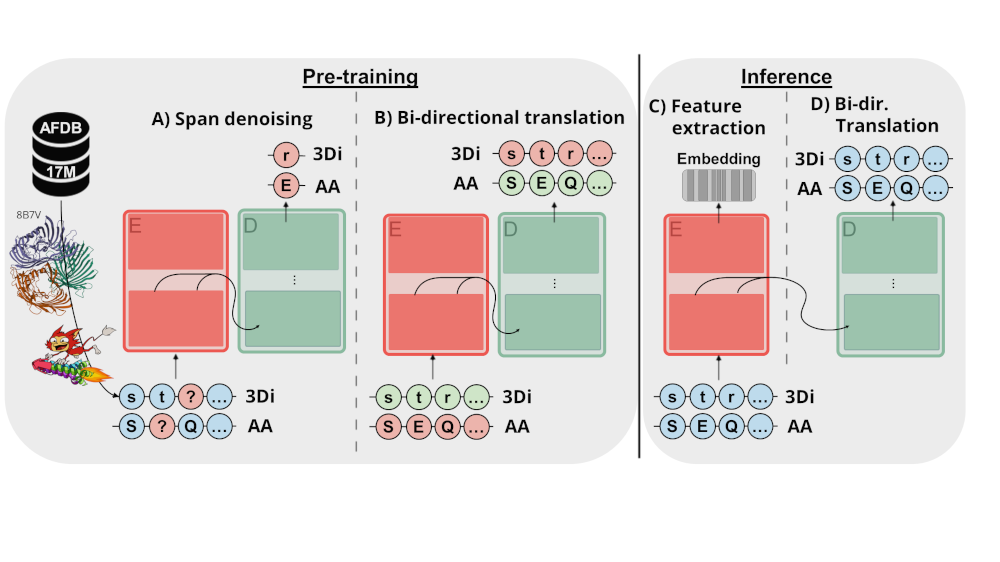
Model Details
Model Description
ProstT5 (Protein structure-sequence T5) is based on ProtT5-XL-U50, a T5 model trained on encoding protein sequences using span corruption applied on billions of protein sequences. ProstT5 finetunes ProtT5-XL-U50 on translating between protein sequence and structure using 17M proteins with high-quality 3D structure predictions from the AlphaFoldDB. Protein structure is converted from 3D to 1D using the 3Di-tokens introduced by Foldseek. In a first step, ProstT5 learnt to represent the newly introduced 3Di-tokens by continuing the original span-denoising objective applied on 3Di- and amino acid- (AA) sequences. Only in a second step, ProstT5 was trained on translating between the two modalities. The direction of the translation is indicated by two special tokens ("<fold2AA>" for translating from 3Di to AAs, “<AA2fold>” for translating from AAs to 3Di). To avoid clashes with AA tokens, 3Di-tokens were cast to lower-case (alphabets are identical otherwise).
- Developed by: Michael Heinzinger (GitHub @mheinzinger; Twitter @HeinzingerM)
- Model type: Encoder-decoder (T5)
- Language(s) (NLP): Protein sequence and structure
- License: MIT
- Finetuned from model: ProtT5-XL-U50
Uses
- The model can be used for traditional feature extraction.
For this, we recommend using only the encoder in half-precision (fp16) together with batching. Examples (currently only for original ProtT5-XL-U50 but replacing repository links and adding prefixes works): script and colab
While original ProtT5-XL-U50 could only embed AA sequences, ProstT5 can now also embed 3D structures represented by 3Di tokens. 3Di tokens can either be derived from 3D structures using Foldseek or they can be predicted from AA sequences by ProstT5. - "Folding": Translation from sequence (AAs) to structure (3Di). The resulting 3Di strings can be used together with Foldseek for remote homology detection while avoiding to compute 3D structures explicitly.
- "Inverse Folding": Translation from structure (3Di) to sequence (AA).
How to Get Started with the Model
Feature extraction:
from transformers import T5Tokenizer, T5EncoderModel
import torch
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
# Load the tokenizer
tokenizer = T5Tokenizer.from_pretrained('Rostlab/ProstT5', do_lower_case=False).to(device)
# Load the model
model = T5EncoderModel.from_pretrained("Rostlab/ProstT5").to(device)
# only GPUs support half-precision currently; if you want to run on CPU use full-precision (not recommended, much slower)
model.full() if device=='cpu' else model.half()
# prepare your protein sequences/structures as a list. Amino acid sequences are expected to be upper-case ("PRTEINO" below) while 3Di-sequences need to be lower-case ("strctr" below).
sequence_examples = ["PRTEINO", "strct"]
# replace all rare/ambiguous amino acids by X (3Di sequences does not have those) and introduce white-space between all sequences (AAs and 3Di)
sequence_examples = [" ".join(list(re.sub(r"[UZOB]", "X", sequence))) for sequence in sequence_examples]
# add pre-fixes accordingly (this already expects 3Di-sequences to be lower-case)
# if you go from AAs to 3Di (or if you want to embed AAs), you need to prepend "<AA2fold>"
# if you go from 3Di to AAs (or if you want to embed 3Di), you need to prepend "<fold2AA>"
sequence_examples = [ "<AA2fold>" + " " + s if s.isupper() else "<fold2AA>" + " " + s
for s in sequence_examples
]
# tokenize sequences and pad up to the longest sequence in the batch
ids = tokenizer.batch_encode_plus(sequences_example, add_special_tokens=True, padding="longest",return_tensors='pt').to(device))
# generate embeddings
with torch.no_grad():
embedding_rpr = model(
ids.input_ids,
attention_mask=ids.attention_mask
)
# extract residue embeddings for the first ([0,:]) sequence in the batch and remove padded & special tokens, incl. prefix ([0,1:8])
emb_0 = embedding_repr.last_hidden_state[0,1:8] # shape (7 x 1024)
# same for the second ([1,:]) sequence but taking into account different sequence lengths ([1,:6])
emb_1 = embedding_repr.last_hidden_state[1,1:6] # shape (5 x 1024)
# if you want to derive a single representation (per-protein embedding) for the whole protein
emb_0_per_protein = emb_0.mean(dim=0) # shape (1024)
Translation ("folding", i.e., AA to 3Di):
from transformers import T5Tokenizer, AutoModelForSeq2SeqLM
import torch
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
# Load the tokenizer
tokenizer = T5Tokenizer.from_pretrained('Rostlab/ProstT5', do_lower_case=False).to(device)
# Load the model
model = AutoModelForSeq2SeqLM.from_pretrained("Rostlab/ProstT5").to(device)
# only GPUs support half-precision currently; if you want to run on CPU use full-precision (not recommended, much slower)
model.full() if device=='cpu' else model.half()
# prepare your protein sequences/structures as a list.
# Amino acid sequences are expected to be upper-case ("PRTEINO" below)
# while 3Di-sequences need to be lower-case.
sequence_examples = ["PRTEINO", "SEQWENCE"]
min_len = min([ len(s) for s in folding_example])
max_len = max([ len(s) for s in folding_example])
# replace all rare/ambiguous amino acids by X (3Di sequences does not have those) and introduce white-space between all sequences (AAs and 3Di)
sequence_examples = [" ".join(list(re.sub(r"[UZOB]", "X", sequence))) for sequence in sequence_examples]
# add pre-fixes accordingly. For the translation from AAs to 3Di, you need to prepend "<AA2fold>"
sequence_examples = [ "<AA2fold>" + " " + s for s in sequence_examples]
# tokenize sequences and pad up to the longest sequence in the batch
ids = tokenizer.batch_encode_plus(sequences_example,
add_special_tokens=True,
padding="longest",
return_tensors='pt').to(device))
# Generation configuration for "folding" (AA-->3Di)
gen_kwargs_aa2fold = {
"do_sample": True,
"num_beams": 3,
"top_p" : 0.95,
"temperature" : 1.2,
"top_k" : 6,
"repetition_penalty" : 1.2,
}
# translate from AA to 3Di (AA-->3Di)
with torch.no_grad():
translations = model.generate(
ids.input_ids,
attention_mask=ids.attention_mask,
max_length=max_len, # max length of generated text
min_length=min_len, # minimum length of the generated text
early_stopping=True, # stop early if end-of-text token is generated
num_return_sequences=1, # return only a single sequence
**gen_kwargs_aa2fold
)
# Decode and remove white-spaces between tokens
decoded_translations = tokenizer.batch_decode( translations, skip_special_tokens=True )
structure_sequences = [ "".join(ts.split(" ")) for ts in decoded_translations ] # predicted 3Di strings
# Now we can use the same model and invert the translation logic
# to generate an amino acid sequence from the predicted 3Di-sequence (3Di-->AA)
# add pre-fixes accordingly. For the translation from 3Di to AA (3Di-->AA), you need to prepend "<fold2AA>"
sequence_examples_backtranslation = [ "<fold2AA>" + " " + s for s in decoded_translations]
# tokenize sequences and pad up to the longest sequence in the batch
ids_backtranslation = tokenizer.batch_encode_plus(sequence_examples_backtranslation,
add_special_tokens=True,
padding="longest",
return_tensors='pt').to(device))
# Example generation configuration for "inverse folding" (3Di-->AA)
gen_kwargs_fold2AA = {
"do_sample": True,
"top_p" : 0.90,
"temperature" : 1.1,
"top_k" : 6,
"repetition_penalty" : 1.2,
}
# translate from 3Di to AA (3Di-->AA)
with torch.no_grad():
backtranslations = model.generate(
ids_backtranslation.input_ids,
attention_mask=ids_backtranslation.attention_mask,
max_length=max_len, # max length of generated text
min_length=min_len, # minimum length of the generated text
early_stopping=True, # stop early if end-of-text token is generated
num_return_sequences=1, # return only a single sequence
**gen_kwargs_fold2AA
)
# Decode and remove white-spaces between tokens
decoded_backtranslations = tokenizer.batch_decode( backtranslations, skip_special_tokens=True )
aminoAcid_sequences = [ "".join(ts.split(" ")) for ts in decoded_backtranslations ] # predicted amino acid strings
Training Details
Training Data
Pre-training data (3Di+AA sequences for 17M proteins)
Training Procedure
The first phase of the pre-training is continuing span-based denoising using 3Di- and AA-sequences using this script. For the second phase of pre-training (actual translation from 3Di- to AA-sequences and vice versa), we used this script.
Training Hyperparameters
- Training regime: we used DeepSpeed (stage-2), gradient accumulation steps (5 steps), mixed half-precision (bf16) and PyTorch2.0’s torchInductor compiler
Speed
Generating embeddings for the human proteome from the Pro(s)tT5 encoder requires around 35m (minutes) or 0.1s (seconds) per protein using batch-processing and half-precision (fp16) on a single RTX A6000 GPU with 48 GB vRAM. The translation is comparatively slow (0.6-2.5s/protein at an average length 135 and 406, respectively) due to the sequential nature of the decoding process which needs to generate left-to-right, token-by-token. We only used batch-processing with half-precision without further optimization.