Upload folder using huggingface_hub (#3)
Browse files- 53c5b2ec1b422daf544a0cd8c45c7e777ed646146c2787f445303436541fca1a (cbffb06d3c5974eb0d5778c3cb2945b42abc4a9a)
- c7b657bafcb5929e66fc62d2a56e7929cc838b33ac4d09d20de4684a38096019 (8c7d62d9b665cacd39a1cae351e8350ef0b8cfc9)
- b528ec290745a2609a64c92b0411895033a3cee271b45deea33ee05e84475c6e (87969cfaa17c4538e643990f2f1b9391a1ada36e)
- README.md +2 -2
- config.json +2 -2
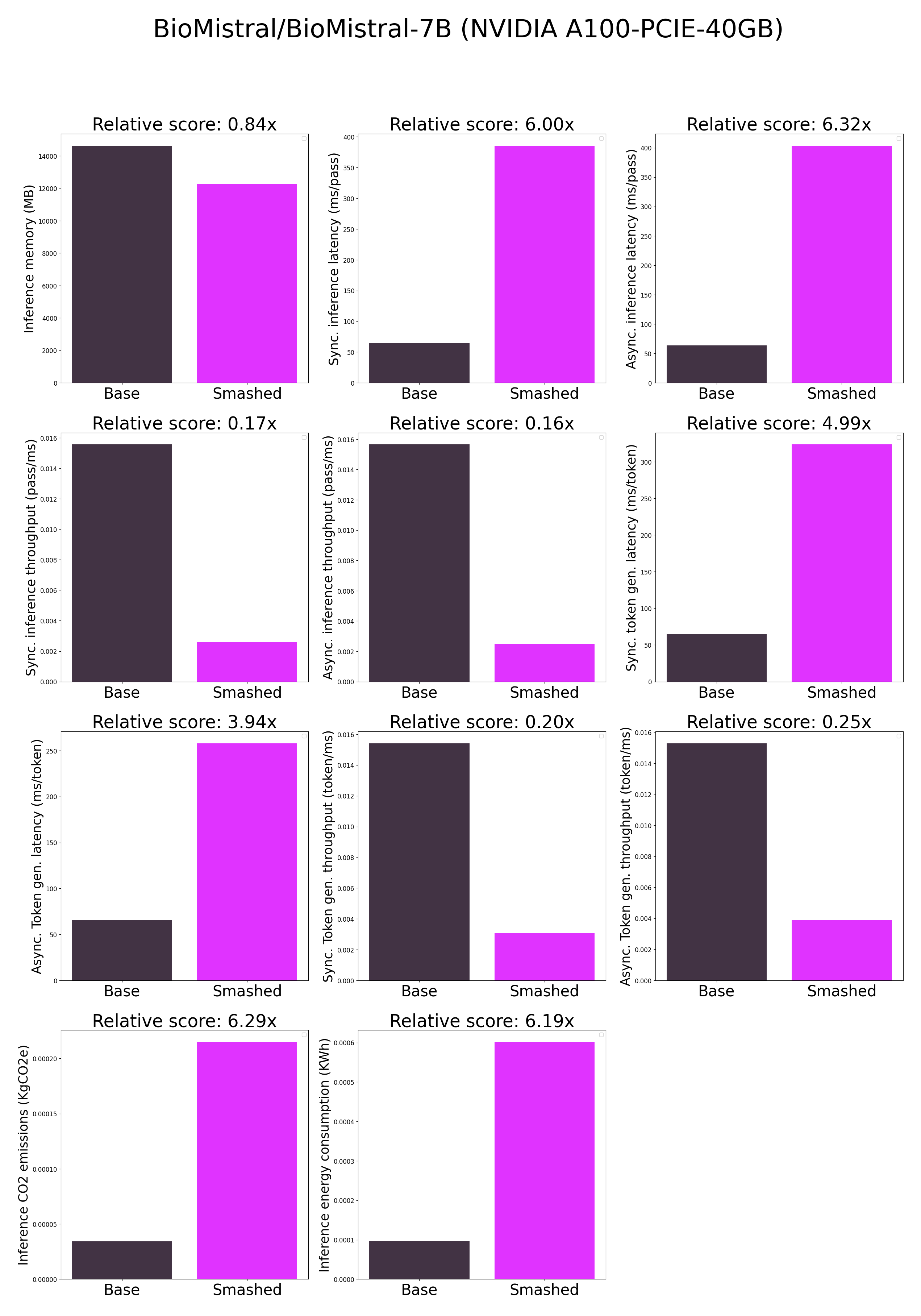
- plots.png +0 -0
- smash_config.json +1 -1
README.md
CHANGED
|
@@ -34,7 +34,7 @@ tags:
|
|
| 34 |
|
| 35 |
## Results
|
| 36 |
|
| 37 |
-
|
| 38 |
|
| 39 |
**Frequently Asked Questions**
|
| 40 |
- ***How does the compression work?*** The model is compressed with llm-int8.
|
|
@@ -61,7 +61,7 @@ You can run the smashed model with these steps:
|
|
| 61 |
from transformers import AutoModelForCausalLM, AutoTokenizer
|
| 62 |
|
| 63 |
model = AutoModelForCausalLM.from_pretrained("PrunaAI/BioMistral-BioMistral-7B-bnb-8bit-smashed",
|
| 64 |
-
trust_remote_code=True)
|
| 65 |
tokenizer = AutoTokenizer.from_pretrained("BioMistral/BioMistral-7B")
|
| 66 |
|
| 67 |
input_ids = tokenizer("What is the color of prunes?,", return_tensors='pt').to(model.device)["input_ids"]
|
|
|
|
| 34 |
|
| 35 |
## Results
|
| 36 |
|
| 37 |
+

|
| 38 |
|
| 39 |
**Frequently Asked Questions**
|
| 40 |
- ***How does the compression work?*** The model is compressed with llm-int8.
|
|
|
|
| 61 |
from transformers import AutoModelForCausalLM, AutoTokenizer
|
| 62 |
|
| 63 |
model = AutoModelForCausalLM.from_pretrained("PrunaAI/BioMistral-BioMistral-7B-bnb-8bit-smashed",
|
| 64 |
+
trust_remote_code=True, device_map='auto')
|
| 65 |
tokenizer = AutoTokenizer.from_pretrained("BioMistral/BioMistral-7B")
|
| 66 |
|
| 67 |
input_ids = tokenizer("What is the color of prunes?,", return_tensors='pt').to(model.device)["input_ids"]
|
config.json
CHANGED
|
@@ -1,5 +1,5 @@
|
|
| 1 |
{
|
| 2 |
-
"_name_or_path": "/tmp/
|
| 3 |
"architectures": [
|
| 4 |
"MistralForCausalLM"
|
| 5 |
],
|
|
@@ -18,7 +18,7 @@
|
|
| 18 |
"quantization_config": {
|
| 19 |
"bnb_4bit_compute_dtype": "bfloat16",
|
| 20 |
"bnb_4bit_quant_type": "fp4",
|
| 21 |
-
"bnb_4bit_use_double_quant":
|
| 22 |
"llm_int8_enable_fp32_cpu_offload": false,
|
| 23 |
"llm_int8_has_fp16_weight": false,
|
| 24 |
"llm_int8_skip_modules": [
|
|
|
|
| 1 |
{
|
| 2 |
+
"_name_or_path": "/tmp/tmpodq0q40z",
|
| 3 |
"architectures": [
|
| 4 |
"MistralForCausalLM"
|
| 5 |
],
|
|
|
|
| 18 |
"quantization_config": {
|
| 19 |
"bnb_4bit_compute_dtype": "bfloat16",
|
| 20 |
"bnb_4bit_quant_type": "fp4",
|
| 21 |
+
"bnb_4bit_use_double_quant": false,
|
| 22 |
"llm_int8_enable_fp32_cpu_offload": false,
|
| 23 |
"llm_int8_has_fp16_weight": false,
|
| 24 |
"llm_int8_skip_modules": [
|
plots.png
ADDED
|
smash_config.json
CHANGED
|
@@ -8,7 +8,7 @@
|
|
| 8 |
"compilers": "None",
|
| 9 |
"task": "text_text_generation",
|
| 10 |
"device": "cuda",
|
| 11 |
-
"cache_dir": "/ceph/hdd/staff/charpent/.cache/
|
| 12 |
"batch_size": 1,
|
| 13 |
"model_name": "BioMistral/BioMistral-7B",
|
| 14 |
"pruning_ratio": 0.0,
|
|
|
|
| 8 |
"compilers": "None",
|
| 9 |
"task": "text_text_generation",
|
| 10 |
"device": "cuda",
|
| 11 |
+
"cache_dir": "/ceph/hdd/staff/charpent/.cache/modelsnghe_ab5",
|
| 12 |
"batch_size": 1,
|
| 13 |
"model_name": "BioMistral/BioMistral-7B",
|
| 14 |
"pruning_ratio": 0.0,
|