tags:
- molecular language model
- SELFIES
- molecule generation
widget:
- text: '[C][=C][C][=C][C][=C][Ring1][=Branch1]'
inference: false
MolGen-large
MolGen-large was introduced in the paper "Domain-Agnostic Molecular Generation with Self-feedback" and first released in this repository. It is a pre-trained molecular generative model built using the 100% robust molecular language representation, SELFIES.
Model description
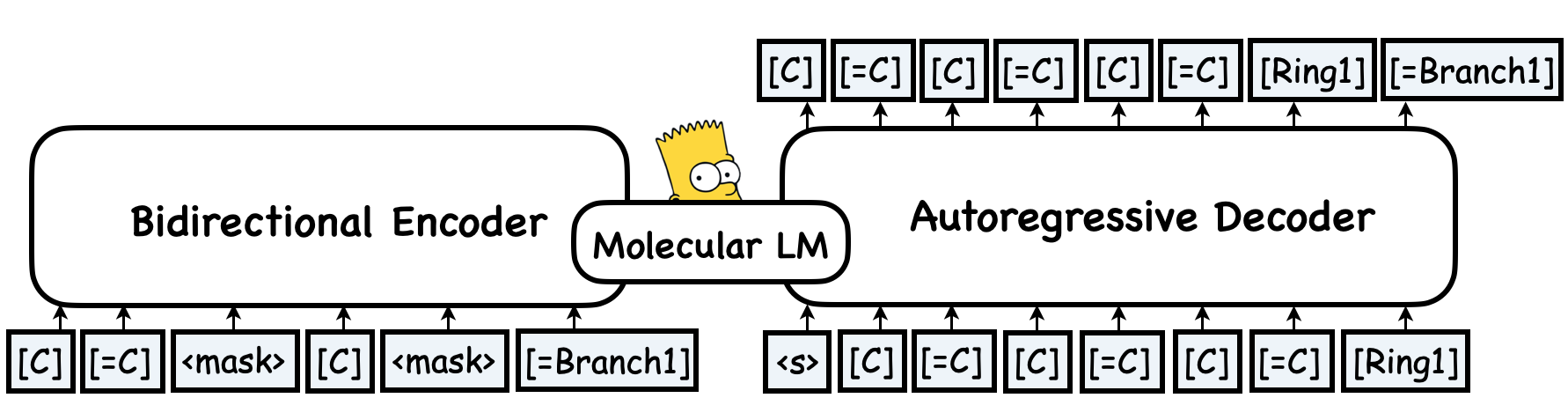
MolGen-large is the first pre-trained model that only produces chemically valid molecules. With a training corpus of over 100 million molecules in SELFIES representation, MolGen-large learns the intrinsic structural patterns of molecules by mapping corrupted SELFIES to their original forms. Specifically, MolGen-large employs a bidirectional Transformer as its encoder and an autoregressive Transformer as its decoder. Through its carefully designed multi-task molecular prefix tuning (MPT), MolGen-large can generate molecules with desired properties, making it a valuable tool for molecular optimization.
Intended uses
You can use the raw model for molecule generation or fine-tune it to a downstream task. Please take note that the following examples only demonstrate the utilization of our pre-trained model for molecule generation. See the repository to look for fine-tune details on a task that interests you.
How to use
Molecule generation example:
>>> from transformers import AutoTokenizer, AutoModelForSeq2SeqLM
>>> tokenizer = AutoTokenizer.from_pretrained("zjunlp/MolGen-large")
>>> model = AutoModelForSeq2SeqLM.from_pretrained("zjunlp/MolGen-large")
>>> sf_input = tokenizer("[C][=C][C][=C][C][=C][Ring1][=Branch1]", return_tensors="pt")
>>> # beam search
>>> molecules = model.generate(input_ids=sf_input["input_ids"],
attention_mask=sf_input["attention_mask"],
max_length=15,
min_length=5,
num_return_sequences=5,
num_beams=5)
>>> sf_output = [tokenizer.decode(g, skip_special_tokens=True, clean_up_tokenization_spaces=True).replace(" ","") for g in molecules]
['[C][=C][C][=C][C][=C][Ring1][=Branch1]',
'[C][=C][C][=C][C][=C][C][=C][Ring1][=Branch1]',
'[C][=C][C][=C][C][=C][Ring1][=Branch1][C][=C][C][=C]',
'[C][=C][C][=C][C][=C][Ring1][=Branch1][C@H1][C][=C][C]',
'[C][=C][C][=C][C][=C][Ring1][=Branch1][C@H1][=C][C][=C]']
BibTeX entry and citation info
@inproceedings{fang2023domain,
author = {Yin Fang and
Ningyu Zhang and
Zhuo Chen and
Xiaohui Fan and
Huajun Chen},
title = {Domain-Agnostic Molecular Generation with Chemical Feedback},
booktitle = {{ICLR}},
publisher = {OpenReview.net},
year = {2024},
url = {https://openreview.net/pdf?id=9rPyHyjfwP}
}