Upload 9 files
Browse files- README.md +64 -0
- config.json +39 -0
- index.gitignore +1 -0
- pytorch_model.bin +3 -0
- special_tokens_map.json +1 -0
- tokenizer.json +0 -0
- tokenizer_config.json +1 -0
- training_args.bin +3 -0
- vocab.txt +0 -0
README.md
ADDED
|
@@ -0,0 +1,64 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
language:
|
| 3 |
+
- en
|
| 4 |
+
tags:
|
| 5 |
+
- Named Entity Recognition
|
| 6 |
+
- SciBERT
|
| 7 |
+
- Adverse Effect
|
| 8 |
+
- Drug
|
| 9 |
+
- Medical
|
| 10 |
+
datasets:
|
| 11 |
+
- ade_corpus_v2
|
| 12 |
+
- tner/bc5cdr
|
| 13 |
+
- jnlpba
|
| 14 |
+
- bc2gm_corpus
|
| 15 |
+
- drAbreu/bc4chemd_ner
|
| 16 |
+
- linnaeus
|
| 17 |
+
- ncbi_disease
|
| 18 |
+
widget:
|
| 19 |
+
- text: "Abortion, miscarriage or uterine hemorrhage associated with misoprostol (Cytotec), a labor-inducing drug."
|
| 20 |
+
example_title: "Abortion, miscarriage, ..."
|
| 21 |
+
- text: "Addiction to many sedatives and analgesics, such as diazepam, morphine, etc."
|
| 22 |
+
example_title: "Addiction to many..."
|
| 23 |
+
- text: "Birth defects associated with thalidomide"
|
| 24 |
+
example_title: "Birth defects associated..."
|
| 25 |
+
- text: "Bleeding of the intestine associated with aspirin therapy"
|
| 26 |
+
example_title: "Bleeding of the intestine..."
|
| 27 |
+
- text: "Cardiovascular disease associated with COX-2 inhibitors (i.e. Vioxx)"
|
| 28 |
+
example_title: "Cardiovascular disease..."
|
| 29 |
+
|
| 30 |
+
---
|
| 31 |
+
|
| 32 |
+
This is a SciBERT-based model fine-tuned to perform Named Entity Recognition for drug names and adverse drug effects.
|
| 33 |
+
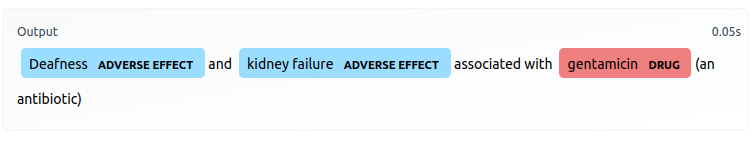
|
| 34 |
+
|
| 35 |
+
This model classifies input tokens into one of five classes:
|
| 36 |
+
|
| 37 |
+
- `B-DRUG`: beginning of a drug entity
|
| 38 |
+
- `I-DRUG`: within a drug entity
|
| 39 |
+
- `B-EFFECT`: beginning of an AE entity
|
| 40 |
+
- `I-EFFECT`: within an AE entity
|
| 41 |
+
- `O`: outside either of the above entities
|
| 42 |
+
|
| 43 |
+
To get started using this model for inference, simply set up an NER `pipeline` like below:
|
| 44 |
+
|
| 45 |
+
```python
|
| 46 |
+
from transformers import (AutoModelForTokenClassification,
|
| 47 |
+
AutoTokenizer,
|
| 48 |
+
pipeline,
|
| 49 |
+
)
|
| 50 |
+
|
| 51 |
+
model_checkpoint = "jsylee/scibert_scivocab_uncased-finetuned-ner"
|
| 52 |
+
model = AutoModelForTokenClassification.from_pretrained(model_checkpoint, num_labels=5,
|
| 53 |
+
id2label={0: 'O', 1: 'B-DRUG', 2: 'I-DRUG', 3: 'B-EFFECT', 4: 'I-EFFECT'}
|
| 54 |
+
)
|
| 55 |
+
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
|
| 56 |
+
|
| 57 |
+
model_pipeline = pipeline(task="ner", model=model, tokenizer=tokenizer)
|
| 58 |
+
|
| 59 |
+
print( model_pipeline ("Abortion, miscarriage or uterine hemorrhage associated with misoprostol (Cytotec), a labor-inducing drug."))
|
| 60 |
+
```
|
| 61 |
+
|
| 62 |
+
SciBERT: https://huggingface.co/allenai/scibert_scivocab_uncased
|
| 63 |
+
|
| 64 |
+
Dataset: https://huggingface.co/datasets/ade_corpus_v2
|
config.json
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"_name_or_path": "allenai/scibert_scivocab_uncased",
|
| 3 |
+
"architectures": [
|
| 4 |
+
"BertForTokenClassification"
|
| 5 |
+
],
|
| 6 |
+
"attention_probs_dropout_prob": 0.1,
|
| 7 |
+
"classifier_dropout": null,
|
| 8 |
+
"hidden_act": "gelu",
|
| 9 |
+
"hidden_dropout_prob": 0.1,
|
| 10 |
+
"hidden_size": 768,
|
| 11 |
+
"id2label": {
|
| 12 |
+
"0": "LABEL_0",
|
| 13 |
+
"1": "LABEL_1",
|
| 14 |
+
"2": "LABEL_2",
|
| 15 |
+
"3": "LABEL_3",
|
| 16 |
+
"4": "LABEL_4"
|
| 17 |
+
},
|
| 18 |
+
"initializer_range": 0.02,
|
| 19 |
+
"intermediate_size": 3072,
|
| 20 |
+
"label2id": {
|
| 21 |
+
"LABEL_0": 0,
|
| 22 |
+
"LABEL_1": 1,
|
| 23 |
+
"LABEL_2": 2,
|
| 24 |
+
"LABEL_3": 3,
|
| 25 |
+
"LABEL_4": 4
|
| 26 |
+
},
|
| 27 |
+
"layer_norm_eps": 1e-12,
|
| 28 |
+
"max_position_embeddings": 512,
|
| 29 |
+
"model_type": "bert",
|
| 30 |
+
"num_attention_heads": 12,
|
| 31 |
+
"num_hidden_layers": 12,
|
| 32 |
+
"pad_token_id": 0,
|
| 33 |
+
"position_embedding_type": "absolute",
|
| 34 |
+
"torch_dtype": "float32",
|
| 35 |
+
"transformers_version": "4.11.3",
|
| 36 |
+
"type_vocab_size": 2,
|
| 37 |
+
"use_cache": true,
|
| 38 |
+
"vocab_size": 31090
|
| 39 |
+
}
|
index.gitignore
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
checkpoint-*/
|
pytorch_model.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:82f8254b4b4cbc8ad6da92d6550d2f25dd1d5b477f5f5ceb30277df548d0d482
|
| 3 |
+
size 437380979
|
special_tokens_map.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"unk_token": "[UNK]", "sep_token": "[SEP]", "pad_token": "[PAD]", "cls_token": "[CLS]", "mask_token": "[MASK]"}
|
tokenizer.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
tokenizer_config.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"do_lower_case": true, "unk_token": "[UNK]", "sep_token": "[SEP]", "pad_token": "[PAD]", "cls_token": "[CLS]", "mask_token": "[MASK]", "tokenize_chinese_chars": true, "strip_accents": null, "special_tokens_map_file": null, "name_or_path": "allenai/scibert_scivocab_uncased", "do_basic_tokenize": true, "never_split": null, "tokenizer_class": "BertTokenizer"}
|
training_args.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:222d40f49fd459c71272dc2ab29f1ef348c8ed931f0579e5856eced883daa36c
|
| 3 |
+
size 2613
|
vocab.txt
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|