Spaces:
Sleeping
Sleeping
Jacob Gershon
commited on
Commit
•
59a9ccf
0
Parent(s):
new b
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +34 -0
- LICENSE +21 -0
- README.md +17 -0
- app.py +404 -0
- examples/aa_weights.json +22 -0
- examples/binder_design.sh +16 -0
- examples/loop_design.sh +15 -0
- examples/motif_scaffolding.sh +14 -0
- examples/out/design_000000.pdb +0 -0
- examples/out/design_000000.trb +0 -0
- examples/partial_diffusion.sh +15 -0
- examples/pdbs/G12D_manual_mut.pdb +0 -0
- examples/pdbs/cd86.pdb +0 -0
- examples/pdbs/rsv5_5tpn.pdb +0 -0
- examples/secondary_structure.sh +21 -0
- examples/secondary_structure_bias.sh +15 -0
- examples/secondary_structure_from_pdb.sh +21 -0
- examples/symmetric_design.sh +16 -0
- examples/weighted_sequence.sh +15 -0
- examples/weighted_sequence_json.sh +16 -0
- model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py +140 -0
- model/Attention_module.py +411 -0
- model/AuxiliaryPredictor.py +92 -0
- model/Embeddings.py +307 -0
- model/RoseTTAFoldModel.py +140 -0
- model/SE3_network.py +83 -0
- model/Track_module.py +476 -0
- model/__pycache__/Attention_module.cpython-310.pyc +0 -0
- model/__pycache__/AuxiliaryPredictor.cpython-310.pyc +0 -0
- model/__pycache__/Embeddings.cpython-310.pyc +0 -0
- model/__pycache__/RoseTTAFoldModel.cpython-310.pyc +0 -0
- model/__pycache__/SE3_network.cpython-310.pyc +0 -0
- model/__pycache__/Track_module.cpython-310.pyc +0 -0
- model/__pycache__/ab_tools.cpython-310.pyc +0 -0
- model/__pycache__/apply_masks.cpython-310.pyc +0 -0
- model/__pycache__/arguments.cpython-310.pyc +0 -0
- model/__pycache__/chemical.cpython-310.pyc +0 -0
- model/__pycache__/data_loader.cpython-310.pyc +0 -0
- model/__pycache__/diffusion.cpython-310.pyc +0 -0
- model/__pycache__/kinematics.cpython-310.pyc +0 -0
- model/__pycache__/loss.cpython-310.pyc +0 -0
- model/__pycache__/mask_generator.cpython-310.pyc +0 -0
- model/__pycache__/parsers.cpython-310.pyc +0 -0
- model/__pycache__/scheduler.cpython-310.pyc +0 -0
- model/__pycache__/scoring.cpython-310.pyc +0 -0
- model/__pycache__/train_multi_deep.cpython-310.pyc +0 -0
- model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc +0 -0
- model/__pycache__/util.cpython-310.pyc +0 -0
- model/__pycache__/util_module.cpython-310.pyc +0 -0
- model/apply_masks.py +196 -0
.gitattributes
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
*.7z filter=lfs diff=lfs merge=lfs -text
|
| 2 |
+
*.arrow filter=lfs diff=lfs merge=lfs -text
|
| 3 |
+
*.bin filter=lfs diff=lfs merge=lfs -text
|
| 4 |
+
*.bz2 filter=lfs diff=lfs merge=lfs -text
|
| 5 |
+
*.ckpt filter=lfs diff=lfs merge=lfs -text
|
| 6 |
+
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
+
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 8 |
+
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 9 |
+
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 10 |
+
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 11 |
+
*.mlmodel filter=lfs diff=lfs merge=lfs -text
|
| 12 |
+
*.model filter=lfs diff=lfs merge=lfs -text
|
| 13 |
+
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 14 |
+
*.npy filter=lfs diff=lfs merge=lfs -text
|
| 15 |
+
*.npz filter=lfs diff=lfs merge=lfs -text
|
| 16 |
+
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 17 |
+
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 18 |
+
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 19 |
+
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 20 |
+
*.pickle filter=lfs diff=lfs merge=lfs -text
|
| 21 |
+
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 22 |
+
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 23 |
+
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 24 |
+
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 25 |
+
*.safetensors filter=lfs diff=lfs merge=lfs -text
|
| 26 |
+
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 27 |
+
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 28 |
+
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 29 |
+
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 30 |
+
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 31 |
+
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 32 |
+
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 33 |
+
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 34 |
+
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2023 RosettaCommons
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
ADDED
|
@@ -0,0 +1,17 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
title: PROTEIN GENERATOR
|
| 3 |
+
emoji: 🧪
|
| 4 |
+
thumbnail: http://files.ipd.uw.edu/pub/sequence_diffusion/figs/diffusion_landscape.png
|
| 5 |
+
colorFrom: blue
|
| 6 |
+
colorTo: purple
|
| 7 |
+
sdk: gradio
|
| 8 |
+
sdk_version: 3.24.1
|
| 9 |
+
app_file: app.py
|
| 10 |
+
pinned: false
|
| 11 |
+
---
|
| 12 |
+
|
| 13 |
+

|
| 14 |
+
|
| 15 |
+
## Code Accessibility
|
| 16 |
+
|
| 17 |
+
To download code and for more details please visit the [github](https://github.com/RosettaCommons/protein_generator)!
|
app.py
ADDED
|
@@ -0,0 +1,404 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os,sys
|
| 2 |
+
|
| 3 |
+
# install environment goods
|
| 4 |
+
#os.system("pip -q install dgl -f https://data.dgl.ai/wheels/cu113/repo.html")
|
| 5 |
+
os.system('pip install dgl==1.0.2+cu116 -f https://data.dgl.ai/wheels/cu116/repo.html')
|
| 6 |
+
#os.system('pip install gradio')
|
| 7 |
+
os.environ["DGLBACKEND"] = "pytorch"
|
| 8 |
+
#os.system(f'pip install -r ./PROTEIN_GENERATOR/requirements.txt')
|
| 9 |
+
print('Modules installed')
|
| 10 |
+
|
| 11 |
+
os.system('pip install --force gradio==3.28.3')
|
| 12 |
+
|
| 13 |
+
os.environ["DGLBACKEND"] = "pytorch"
|
| 14 |
+
|
| 15 |
+
if not os.path.exists('./SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'):
|
| 16 |
+
print('Downloading model weights 1')
|
| 17 |
+
os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt')
|
| 18 |
+
print('Successfully Downloaded')
|
| 19 |
+
|
| 20 |
+
if not os.path.exists('./SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'):
|
| 21 |
+
print('Downloading model weights 2')
|
| 22 |
+
os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt')
|
| 23 |
+
print('Successfully Downloaded')
|
| 24 |
+
|
| 25 |
+
import numpy as np
|
| 26 |
+
import gradio as gr
|
| 27 |
+
import py3Dmol
|
| 28 |
+
from io import StringIO
|
| 29 |
+
import json
|
| 30 |
+
import secrets
|
| 31 |
+
import copy
|
| 32 |
+
import matplotlib.pyplot as plt
|
| 33 |
+
from utils.sampler import HuggingFace_sampler
|
| 34 |
+
|
| 35 |
+
plt.rcParams.update({'font.size': 13})
|
| 36 |
+
|
| 37 |
+
with open('./tmp/args.json','r') as f:
|
| 38 |
+
args = json.load(f)
|
| 39 |
+
|
| 40 |
+
# manually set checkpoint to load
|
| 41 |
+
args['checkpoint'] = None
|
| 42 |
+
args['dump_trb'] = False
|
| 43 |
+
args['dump_args'] = True
|
| 44 |
+
args['save_best_plddt'] = True
|
| 45 |
+
args['T'] = 25
|
| 46 |
+
args['strand_bias'] = 0.0
|
| 47 |
+
args['loop_bias'] = 0.0
|
| 48 |
+
args['helix_bias'] = 0.0
|
| 49 |
+
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
def protein_diffusion_model(sequence, seq_len, helix_bias, strand_bias, loop_bias,
|
| 53 |
+
secondary_structure, aa_bias, aa_bias_potential,
|
| 54 |
+
#target_charge, target_ph, charge_potential,
|
| 55 |
+
num_steps, noise, hydrophobic_target_score, hydrophobic_potential):
|
| 56 |
+
|
| 57 |
+
dssp_checkpoint = './SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'
|
| 58 |
+
og_checkpoint = './SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'
|
| 59 |
+
|
| 60 |
+
model_args = copy.deepcopy(args)
|
| 61 |
+
|
| 62 |
+
# make sampler
|
| 63 |
+
S = HuggingFace_sampler(args=model_args)
|
| 64 |
+
|
| 65 |
+
# get random prefix
|
| 66 |
+
S.out_prefix = './tmp/'+secrets.token_hex(nbytes=10).upper()
|
| 67 |
+
|
| 68 |
+
# set args
|
| 69 |
+
S.args['checkpoint'] = None
|
| 70 |
+
S.args['dump_trb'] = False
|
| 71 |
+
S.args['dump_args'] = True
|
| 72 |
+
S.args['save_best_plddt'] = True
|
| 73 |
+
S.args['T'] = 20
|
| 74 |
+
S.args['strand_bias'] = 0.0
|
| 75 |
+
S.args['loop_bias'] = 0.0
|
| 76 |
+
S.args['helix_bias'] = 0.0
|
| 77 |
+
S.args['potentials'] = None
|
| 78 |
+
S.args['potential_scale'] = None
|
| 79 |
+
S.args['aa_composition'] = None
|
| 80 |
+
|
| 81 |
+
|
| 82 |
+
# get sequence if entered and make sure all chars are valid
|
| 83 |
+
alt_aa_dict = {'B':['D','N'],'J':['I','L'],'U':['C'],'Z':['E','Q'],'O':['K']}
|
| 84 |
+
if sequence not in ['',None]:
|
| 85 |
+
L = len(sequence)
|
| 86 |
+
aa_seq = []
|
| 87 |
+
for aa in sequence.upper():
|
| 88 |
+
if aa in alt_aa_dict.keys():
|
| 89 |
+
aa_seq.append(np.random.choice(alt_aa_dict[aa]))
|
| 90 |
+
else:
|
| 91 |
+
aa_seq.append(aa)
|
| 92 |
+
|
| 93 |
+
S.args['sequence'] = aa_seq
|
| 94 |
+
else:
|
| 95 |
+
S.args['contigs'] = [f'{seq_len}']
|
| 96 |
+
L = int(seq_len)
|
| 97 |
+
|
| 98 |
+
if secondary_structure in ['',None]:
|
| 99 |
+
secondary_structure = None
|
| 100 |
+
else:
|
| 101 |
+
secondary_structure = ''.join(['E' if x == 'S' else x for x in secondary_structure])
|
| 102 |
+
if L < len(secondary_structure):
|
| 103 |
+
secondary_structure = secondary_structure[:len(sequence)]
|
| 104 |
+
elif L == len(secondary_structure):
|
| 105 |
+
pass
|
| 106 |
+
else:
|
| 107 |
+
dseq = L - len(secondary_structure)
|
| 108 |
+
secondary_structure += secondary_structure[-1]*dseq
|
| 109 |
+
|
| 110 |
+
|
| 111 |
+
# potentials
|
| 112 |
+
potential_list = []
|
| 113 |
+
potential_bias_list = []
|
| 114 |
+
|
| 115 |
+
if aa_bias not in ['',None]:
|
| 116 |
+
potential_list.append('aa_bias')
|
| 117 |
+
S.args['aa_composition'] = aa_bias
|
| 118 |
+
if aa_bias_potential in ['',None]:
|
| 119 |
+
aa_bias_potential = 3
|
| 120 |
+
potential_bias_list.append(str(aa_bias_potential))
|
| 121 |
+
'''
|
| 122 |
+
if target_charge not in ['',None]:
|
| 123 |
+
potential_list.append('charge')
|
| 124 |
+
if charge_potential in ['',None]:
|
| 125 |
+
charge_potential = 1
|
| 126 |
+
potential_bias_list.append(str(charge_potential))
|
| 127 |
+
S.args['target_charge'] = float(target_charge)
|
| 128 |
+
if target_ph in ['',None]:
|
| 129 |
+
target_ph = 7.4
|
| 130 |
+
S.args['target_pH'] = float(target_ph)
|
| 131 |
+
'''
|
| 132 |
+
|
| 133 |
+
if hydrophobic_target_score not in ['',None]:
|
| 134 |
+
potential_list.append('hydrophobic')
|
| 135 |
+
S.args['hydrophobic_score'] = float(hydrophobic_target_score)
|
| 136 |
+
if hydrophobic_potential in ['',None]:
|
| 137 |
+
hydrophobic_potential = 3
|
| 138 |
+
potential_bias_list.append(str(hydrophobic_potential))
|
| 139 |
+
|
| 140 |
+
|
| 141 |
+
if len(potential_list) > 0:
|
| 142 |
+
S.args['potentials'] = ','.join(potential_list)
|
| 143 |
+
S.args['potential_scale'] = ','.join(potential_bias_list)
|
| 144 |
+
|
| 145 |
+
|
| 146 |
+
# normalise secondary_structure bias from range 0-0.3
|
| 147 |
+
S.args['secondary_structure'] = secondary_structure
|
| 148 |
+
S.args['helix_bias'] = helix_bias
|
| 149 |
+
S.args['strand_bias'] = strand_bias
|
| 150 |
+
S.args['loop_bias'] = loop_bias
|
| 151 |
+
|
| 152 |
+
# set T
|
| 153 |
+
if num_steps in ['',None]:
|
| 154 |
+
S.args['T'] = 20
|
| 155 |
+
else:
|
| 156 |
+
S.args['T'] = int(num_steps)
|
| 157 |
+
|
| 158 |
+
# noise
|
| 159 |
+
if 'normal' in noise:
|
| 160 |
+
S.args['sample_distribution'] = noise
|
| 161 |
+
S.args['sample_distribution_gmm_means'] = [0]
|
| 162 |
+
S.args['sample_distribution_gmm_variances'] = [1]
|
| 163 |
+
elif 'gmm2' in noise:
|
| 164 |
+
S.args['sample_distribution'] = noise
|
| 165 |
+
S.args['sample_distribution_gmm_means'] = [-1,1]
|
| 166 |
+
S.args['sample_distribution_gmm_variances'] = [1,1]
|
| 167 |
+
elif 'gmm3' in noise:
|
| 168 |
+
S.args['sample_distribution'] = noise
|
| 169 |
+
S.args['sample_distribution_gmm_means'] = [-1,0,1]
|
| 170 |
+
S.args['sample_distribution_gmm_variances'] = [1,1,1]
|
| 171 |
+
|
| 172 |
+
|
| 173 |
+
|
| 174 |
+
if secondary_structure not in ['',None] or helix_bias+strand_bias+loop_bias > 0:
|
| 175 |
+
S.args['checkpoint'] = dssp_checkpoint
|
| 176 |
+
S.args['d_t1d'] = 29
|
| 177 |
+
print('using dssp checkpoint')
|
| 178 |
+
else:
|
| 179 |
+
S.args['checkpoint'] = og_checkpoint
|
| 180 |
+
S.args['d_t1d'] = 24
|
| 181 |
+
print('using og checkpoint')
|
| 182 |
+
|
| 183 |
+
|
| 184 |
+
for k,v in S.args.items():
|
| 185 |
+
print(f"{k} --> {v}")
|
| 186 |
+
|
| 187 |
+
# init S
|
| 188 |
+
S.model_init()
|
| 189 |
+
S.diffuser_init()
|
| 190 |
+
S.setup()
|
| 191 |
+
|
| 192 |
+
# sampling loop
|
| 193 |
+
plddt_data = []
|
| 194 |
+
for j in range(S.max_t):
|
| 195 |
+
output_seq, output_pdb, plddt = S.take_step_get_outputs(j)
|
| 196 |
+
plddt_data.append(plddt)
|
| 197 |
+
yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
|
| 198 |
+
|
| 199 |
+
output_seq, output_pdb, plddt = S.get_outputs()
|
| 200 |
+
|
| 201 |
+
yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
|
| 202 |
+
|
| 203 |
+
def get_plddt_plot(plddt_data, max_t):
|
| 204 |
+
x = [i+1 for i in range(len(plddt_data))]
|
| 205 |
+
fig, ax = plt.subplots(figsize=(15,6))
|
| 206 |
+
ax.plot(x,plddt_data,color='#661dbf', linewidth=3,marker='o')
|
| 207 |
+
ax.set_xticks([i+1 for i in range(max_t)])
|
| 208 |
+
ax.set_yticks([(i+1)/10 for i in range(10)])
|
| 209 |
+
ax.set_ylim([0,1])
|
| 210 |
+
ax.set_ylabel('model confidence (plddt)')
|
| 211 |
+
ax.set_xlabel('diffusion steps (t)')
|
| 212 |
+
return fig
|
| 213 |
+
|
| 214 |
+
def display_pdb(path_to_pdb):
|
| 215 |
+
'''
|
| 216 |
+
#function to display pdb in py3dmol
|
| 217 |
+
'''
|
| 218 |
+
pdb = open(path_to_pdb, "r").read()
|
| 219 |
+
|
| 220 |
+
view = py3Dmol.view(width=500, height=500)
|
| 221 |
+
view.addModel(pdb, "pdb")
|
| 222 |
+
view.setStyle({'model': -1}, {"cartoon": {'colorscheme':{'prop':'b','gradient':'roygb','min':0,'max':1}}})#'linear', 'min': 0, 'max': 1, 'colors': ["#ff9ef0","#a903fc",]}}})
|
| 223 |
+
view.zoomTo()
|
| 224 |
+
output = view._make_html().replace("'", '"')
|
| 225 |
+
print(view._make_html())
|
| 226 |
+
x = f"""<!DOCTYPE html><html></center> {output} </center></html>""" # do not use ' in this input
|
| 227 |
+
|
| 228 |
+
return f"""<iframe height="500px" width="100%" name="result" allow="midi; geolocation; microphone; camera;
|
| 229 |
+
display-capture; encrypted-media;" sandbox="allow-modals allow-forms
|
| 230 |
+
allow-scripts allow-same-origin allow-popups
|
| 231 |
+
allow-top-navigation-by-user-activation allow-downloads" allowfullscreen=""
|
| 232 |
+
allowpaymentrequest="" frameborder="0" srcdoc='{x}'></iframe>"""
|
| 233 |
+
|
| 234 |
+
'''
|
| 235 |
+
|
| 236 |
+
return f"""<iframe style="width: 100%; height:700px" name="result" allow="midi; geolocation; microphone; camera;
|
| 237 |
+
display-capture; encrypted-media;" sandbox="allow-modals allow-forms
|
| 238 |
+
allow-scripts allow-same-origin allow-popups
|
| 239 |
+
allow-top-navigation-by-user-activation allow-downloads" allowfullscreen=""
|
| 240 |
+
allowpaymentrequest="" frameborder="0" srcdoc='{x}'></iframe>"""
|
| 241 |
+
'''
|
| 242 |
+
|
| 243 |
+
def toggle_seq_input(choice):
|
| 244 |
+
if choice == "protein length":
|
| 245 |
+
return gr.update(visible=True, value=None), gr.update(visible=False, value=None)
|
| 246 |
+
elif choice == "custom sequence":
|
| 247 |
+
return gr.update(visible=False, value=None), gr.update(visible=True, value=None)
|
| 248 |
+
|
| 249 |
+
def toggle_secondary_structure(choice):
|
| 250 |
+
if choice == "sliders":
|
| 251 |
+
return gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=False, value=None)
|
| 252 |
+
elif choice == "explicit":
|
| 253 |
+
return gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=True, value=None)
|
| 254 |
+
|
| 255 |
+
# Define the Gradio interface
|
| 256 |
+
with gr.Blocks(theme='ParityError/Interstellar') as demo:
|
| 257 |
+
|
| 258 |
+
gr.Markdown(f"""# Protein Generation via Diffusion in Sequence Space""")
|
| 259 |
+
|
| 260 |
+
with gr.Row():
|
| 261 |
+
with gr.Column(min_width=500):
|
| 262 |
+
gr.Markdown(f"""
|
| 263 |
+
## How does it work?\n
|
| 264 |
+
--- [PREPRINT](https://biorxiv.org/content/10.1101/2023.05.08.539766v1) ---
|
| 265 |
+
|
| 266 |
+
Protein sequence and structure co-generation is a long outstanding problem in the field of protein design. By implementing [ddpm](https://arxiv.org/abs/2006.11239) style diffusion over protein seqeuence space we generate protein sequence and structure pairs. Starting with [RoseTTAFold](https://www.science.org/doi/10.1126/science.abj8754), a protein structure prediction network, we finetuned it to predict sequence and structure given a partially noised sequence. By applying losses to both the predicted sequence and structure the model is forced to generate meaningful pairs. Diffusing in sequence space makes it easy to implement potentials to guide the diffusive process toward particular amino acid composition, net charge, and more! Furthermore, you can sample proteins from a family of sequences or even train a small sequence to function classifier to guide generation toward desired sequences.
|
| 267 |
+
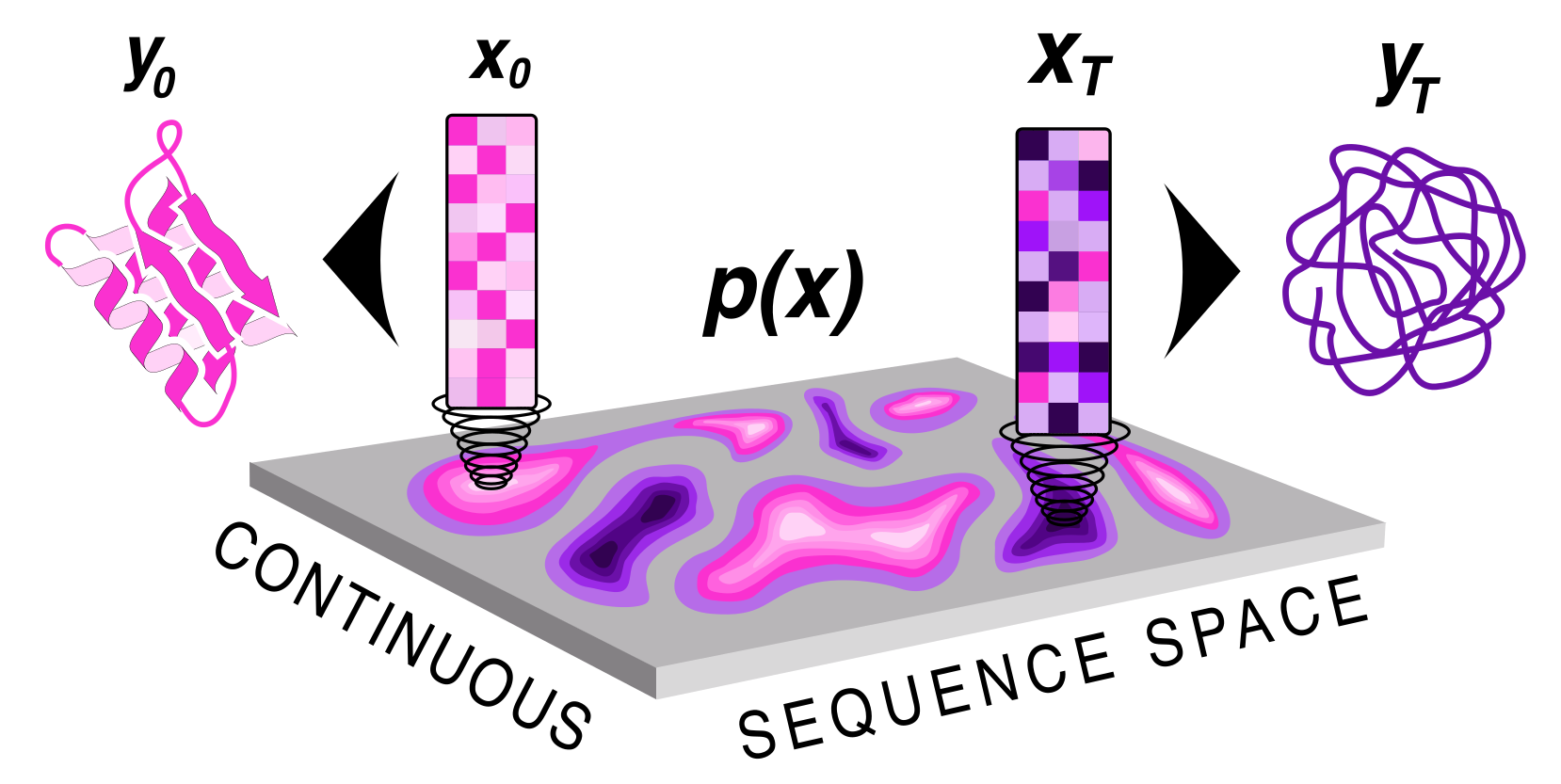
|
| 268 |
+
|
| 269 |
+
## How to use it?\n
|
| 270 |
+
A user can either design a custom input sequence to diffuse from or specify a length below. To scaffold a sequence use the following format where X represent residues to diffuse: XXXXXXXXSCIENCESCIENCEXXXXXXXXXXXXXXXXXXX. You can even design a protein with your name XXXXXXXXXXXXNAMEHEREXXXXXXXXXXXXX!
|
| 271 |
+
|
| 272 |
+
### Acknowledgements\n
|
| 273 |
+
Thank you to Simon Dürr and the Hugging Face team for setting us up with a community GPU grant!
|
| 274 |
+
""")
|
| 275 |
+
|
| 276 |
+
gr.Markdown("""
|
| 277 |
+
## Model in Action
|
| 278 |
+

|
| 279 |
+
""")
|
| 280 |
+
|
| 281 |
+
with gr.Row().style(equal_height=False):
|
| 282 |
+
with gr.Column():
|
| 283 |
+
gr.Markdown("""## INPUTS""")
|
| 284 |
+
gr.Markdown("""#### Start Sequence
|
| 285 |
+
Specify the protein length for complete unconditional generation, or scaffold a motif (or your name) using the custom sequence input""")
|
| 286 |
+
seq_opt = gr.Radio(["protein length","custom sequence"], label="How would you like to specify the starting sequence?", value='protein length')
|
| 287 |
+
|
| 288 |
+
sequence = gr.Textbox(label="custom sequence", lines=1, placeholder='AMINO ACIDS: A,C,D,E,F,G,H,I,K,L,M,N,P,Q,R,S,T,V,W,Y\n MASK TOKEN: X', visible=False)
|
| 289 |
+
seq_len = gr.Slider(minimum=5.0, maximum=250.0, label="protein length", value=100, visible=True)
|
| 290 |
+
|
| 291 |
+
seq_opt.change(fn=toggle_seq_input,
|
| 292 |
+
inputs=[seq_opt],
|
| 293 |
+
outputs=[seq_len, sequence],
|
| 294 |
+
queue=False)
|
| 295 |
+
|
| 296 |
+
gr.Markdown("""### Optional Parameters""")
|
| 297 |
+
with gr.Accordion(label='Secondary Structure',open=True):
|
| 298 |
+
gr.Markdown("""Try changing the sliders or inputing explicit secondary structure conditioning for each residue""")
|
| 299 |
+
sec_str_opt = gr.Radio(["sliders","explicit"], label="How would you like to specify secondary structure?", value='sliders')
|
| 300 |
+
|
| 301 |
+
secondary_structure = gr.Textbox(label="secondary structure", lines=1, placeholder='HELIX = H STRAND = S LOOP = L MASK = X(must be the same length as input sequence)', visible=False)
|
| 302 |
+
|
| 303 |
+
with gr.Column():
|
| 304 |
+
helix_bias = gr.Slider(minimum=0.0, maximum=0.05, label="helix bias", visible=True)
|
| 305 |
+
strand_bias = gr.Slider(minimum=0.0, maximum=0.05, label="strand bias", visible=True)
|
| 306 |
+
loop_bias = gr.Slider(minimum=0.0, maximum=0.20, label="loop bias", visible=True)
|
| 307 |
+
|
| 308 |
+
sec_str_opt.change(fn=toggle_secondary_structure,
|
| 309 |
+
inputs=[sec_str_opt],
|
| 310 |
+
outputs=[helix_bias,strand_bias,loop_bias,secondary_structure],
|
| 311 |
+
queue=False)
|
| 312 |
+
|
| 313 |
+
with gr.Accordion(label='Amino Acid Compositional Bias',open=False):
|
| 314 |
+
gr.Markdown("""Bias sequence composition for particular amino acids by specifying the one letter code followed by the fraction to bias. This can be input as a list for example: W0.2,E0.1""")
|
| 315 |
+
with gr.Row():
|
| 316 |
+
aa_bias = gr.Textbox(label="aa bias", lines=1, placeholder='specify one letter AA and fraction to bias, for example W0.1 or M0.1,K0.1' )
|
| 317 |
+
aa_bias_potential = gr.Textbox(label="aa bias scale", lines=1, placeholder='AA Bias potential scale (recomended range 1.0-5.0)')
|
| 318 |
+
|
| 319 |
+
'''
|
| 320 |
+
with gr.Accordion(label='Charge Bias',open=False):
|
| 321 |
+
gr.Markdown("""Bias for a specified net charge at a particular pH using the boxes below""")
|
| 322 |
+
with gr.Row():
|
| 323 |
+
target_charge = gr.Textbox(label="net charge", lines=1, placeholder='net charge to target')
|
| 324 |
+
target_ph = gr.Textbox(label="pH", lines=1, placeholder='pH at which net charge is desired')
|
| 325 |
+
charge_potential = gr.Textbox(label="charge potential scale", lines=1, placeholder='charge potential scale (recomended range 1.0-5.0)')
|
| 326 |
+
'''
|
| 327 |
+
|
| 328 |
+
with gr.Accordion(label='Hydrophobic Bias',open=False):
|
| 329 |
+
gr.Markdown("""Bias for or against hydrophobic composition, to get more soluble proteins, bias away with a negative target score (ex. -5)""")
|
| 330 |
+
with gr.Row():
|
| 331 |
+
hydrophobic_target_score = gr.Textbox(label="hydrophobic score", lines=1, placeholder='hydrophobic score to target (negative score is good for solublility)')
|
| 332 |
+
hydrophobic_potential = gr.Textbox(label="hydrophobic potential scale", lines=1, placeholder='hydrophobic potential scale (recomended range 1.0-2.0)')
|
| 333 |
+
|
| 334 |
+
with gr.Accordion(label='Diffusion Params',open=False):
|
| 335 |
+
gr.Markdown("""Increasing T to more steps can be helpful for harder design challenges, sampling from different distributions can change the sequence and structural composition""")
|
| 336 |
+
with gr.Row():
|
| 337 |
+
num_steps = gr.Textbox(label="T", lines=1, placeholder='number of diffusion steps (25 or less will speed things up)')
|
| 338 |
+
noise = gr.Dropdown(['normal','gmm2 [-1,1]','gmm3 [-1,0,1]'], label='noise type', value='normal')
|
| 339 |
+
|
| 340 |
+
btn = gr.Button("GENERATE")
|
| 341 |
+
|
| 342 |
+
#with gr.Row():
|
| 343 |
+
with gr.Column():
|
| 344 |
+
gr.Markdown("""## OUTPUTS""")
|
| 345 |
+
gr.Markdown("""#### Confidence score for generated structure at each timestep""")
|
| 346 |
+
plddt_plot = gr.Plot(label='plddt at step t')
|
| 347 |
+
gr.Markdown("""#### Output protein sequnece""")
|
| 348 |
+
output_seq = gr.Textbox(label="sequence")
|
| 349 |
+
gr.Markdown("""#### Download PDB file""")
|
| 350 |
+
output_pdb = gr.File(label="PDB file")
|
| 351 |
+
gr.Markdown("""#### Structure viewer""")
|
| 352 |
+
output_viewer = gr.HTML()
|
| 353 |
+
|
| 354 |
+
gr.Markdown("""### Don't know where to get started? Click on an example below to try it out!""")
|
| 355 |
+
gr.Examples(
|
| 356 |
+
[["","125",0.0,0.0,0.2,"","","","20","normal",'',''],
|
| 357 |
+
["","100",0.0,0.0,0.0,"","W0.2","2","20","normal",'',''],
|
| 358 |
+
["","100",0.0,0.0,0.0,"XXHHHHHHHHHXXXXXXXHHHHHHHHHXXXXXXXHHHHHHHHXXXXSSSSSSSSSSSXXXXXXXXSSSSSSSSSSSSXXXXXXXSSSSSSSSSXXXXXXX","","","25","normal",'',''],
|
| 359 |
+
["XXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXXXXXPEPSEQXXXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXX","",0.0,0.0,0.0,"","","","25","normal",'','']],
|
| 360 |
+
inputs=[sequence,
|
| 361 |
+
seq_len,
|
| 362 |
+
helix_bias,
|
| 363 |
+
strand_bias,
|
| 364 |
+
loop_bias,
|
| 365 |
+
secondary_structure,
|
| 366 |
+
aa_bias,
|
| 367 |
+
aa_bias_potential,
|
| 368 |
+
#target_charge,
|
| 369 |
+
#target_ph,
|
| 370 |
+
#charge_potential,
|
| 371 |
+
num_steps,
|
| 372 |
+
noise,
|
| 373 |
+
hydrophobic_target_score,
|
| 374 |
+
hydrophobic_potential],
|
| 375 |
+
outputs=[output_seq,
|
| 376 |
+
output_pdb,
|
| 377 |
+
output_viewer,
|
| 378 |
+
plddt_plot],
|
| 379 |
+
fn=protein_diffusion_model,
|
| 380 |
+
)
|
| 381 |
+
btn.click(protein_diffusion_model,
|
| 382 |
+
[sequence,
|
| 383 |
+
seq_len,
|
| 384 |
+
helix_bias,
|
| 385 |
+
strand_bias,
|
| 386 |
+
loop_bias,
|
| 387 |
+
secondary_structure,
|
| 388 |
+
aa_bias,
|
| 389 |
+
aa_bias_potential,
|
| 390 |
+
#target_charge,
|
| 391 |
+
#target_ph,
|
| 392 |
+
#charge_potential,
|
| 393 |
+
num_steps,
|
| 394 |
+
noise,
|
| 395 |
+
hydrophobic_target_score,
|
| 396 |
+
hydrophobic_potential],
|
| 397 |
+
[output_seq,
|
| 398 |
+
output_pdb,
|
| 399 |
+
output_viewer,
|
| 400 |
+
plddt_plot])
|
| 401 |
+
|
| 402 |
+
demo.queue()
|
| 403 |
+
demo.launch(debug=True)
|
| 404 |
+
|
examples/aa_weights.json
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"A": 0,
|
| 3 |
+
"R": 0,
|
| 4 |
+
"N": 0,
|
| 5 |
+
"D": 0,
|
| 6 |
+
"C": 0,
|
| 7 |
+
"Q": 0,
|
| 8 |
+
"E": 0,
|
| 9 |
+
"G": 0,
|
| 10 |
+
"H": 0,
|
| 11 |
+
"I": 0,
|
| 12 |
+
"L": 0,
|
| 13 |
+
"K": 0,
|
| 14 |
+
"M": 0,
|
| 15 |
+
"F": 0,
|
| 16 |
+
"P": 0,
|
| 17 |
+
"S": 0,
|
| 18 |
+
"T": 0,
|
| 19 |
+
"W": 0,
|
| 20 |
+
"Y": 0,
|
| 21 |
+
"V": 0
|
| 22 |
+
}
|
examples/binder_design.sh
ADDED
|
@@ -0,0 +1,16 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/binder_design \
|
| 13 |
+
--pdb pdbs/cd86.pdb \
|
| 14 |
+
--T 25 --save_best_plddt \
|
| 15 |
+
--contigs B1-110,0 25-75 \
|
| 16 |
+
--hotspots B40,B32,B87,B96,B30
|
examples/loop_design.sh
ADDED
|
@@ -0,0 +1,15 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--pdb pdbs/G12D_manual_mut.pdb \
|
| 13 |
+
--out out/ab_loop \
|
| 14 |
+
--contigs A2-176,0 C7-16,0 H2-95,12-15,H111-116,0 L1-45,10-12,L56-107 \
|
| 15 |
+
--T 25 --save_best_plddt --loop_design
|
examples/motif_scaffolding.sh
ADDED
|
@@ -0,0 +1,14 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--pdb pdbs/rsv5_5tpn.pdb \
|
| 14 |
+
--contigs 0-25,A163-181,25-30 --T 25 --save_best_plddt
|
examples/out/design_000000.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
examples/out/design_000000.trb
ADDED
|
Binary file (3.51 kB). View file
|
|
|
examples/partial_diffusion.sh
ADDED
|
@@ -0,0 +1,15 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--pdb out/design_000.pdb \
|
| 13 |
+
--trb out/design_000.trb \
|
| 14 |
+
--out out/partial_diffusion_design \
|
| 15 |
+
--contigs 0 --sampling_temp 0.3 --T 50 --save_best_plddt
|
examples/pdbs/G12D_manual_mut.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
examples/pdbs/cd86.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
examples/pdbs/rsv5_5tpn.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
examples/secondary_structure.sh
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--contigs 100 \
|
| 14 |
+
--T 25 --save_best_plddt \
|
| 15 |
+
--secondary_structure XXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXX
|
| 16 |
+
|
| 17 |
+
# FOR SECONDARY STRUCTURE:
|
| 18 |
+
# X - mask
|
| 19 |
+
# H - helix
|
| 20 |
+
# E - strand
|
| 21 |
+
# L - loop
|
examples/secondary_structure_bias.sh
ADDED
|
@@ -0,0 +1,15 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--contigs 100 \
|
| 14 |
+
--T 25 --save_best_plddt \
|
| 15 |
+
--helix_bias 0.01 --strand_bias 0.01 --loop_bias 0.0
|
examples/secondary_structure_from_pdb.sh
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--contigs 110 \
|
| 14 |
+
--T 25 --save_best_plddt \
|
| 15 |
+
--dssp_pdb ./pdbs/cd86.pdb
|
| 16 |
+
|
| 17 |
+
# FOR SECONDARY STRUCTURE:
|
| 18 |
+
# X - mask
|
| 19 |
+
# H - helix
|
| 20 |
+
# E - strand
|
| 21 |
+
# L - loop
|
examples/symmetric_design.sh
ADDED
|
@@ -0,0 +1,16 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/symmetric_design \
|
| 13 |
+
--contigs 25,0 25,0 25,0 \
|
| 14 |
+
--T 50 \
|
| 15 |
+
--save_best_plddt \
|
| 16 |
+
--symmetry 3
|
examples/weighted_sequence.sh
ADDED
|
@@ -0,0 +1,15 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--contigs 100 \
|
| 14 |
+
--T 25 --save_best_plddt \
|
| 15 |
+
--aa_composition W0.2 --potential_scale 1.75
|
examples/weighted_sequence_json.sh
ADDED
|
@@ -0,0 +1,16 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -J seq_diff
|
| 3 |
+
#SBATCH -p gpu
|
| 4 |
+
#SBATCH --mem=8g
|
| 5 |
+
#SBATCH --gres=gpu:a6000:1
|
| 6 |
+
#SBATCH -o ./out/slurm/slurm_%j.out
|
| 7 |
+
|
| 8 |
+
source activate /software/conda/envs/SE3nv
|
| 9 |
+
|
| 10 |
+
srun python ../inference.py \
|
| 11 |
+
--num_designs 10 \
|
| 12 |
+
--out out/design \
|
| 13 |
+
--contigs 75 \
|
| 14 |
+
--aa_weights_json aa_weights.json \
|
| 15 |
+
--add_weight_every_n 5 --add_weight_every_n \
|
| 16 |
+
--T 25 --save_best_plddt
|
model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py
ADDED
|
@@ -0,0 +1,140 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
|
| 4 |
+
from Track_module import IterativeSimulator
|
| 5 |
+
from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
|
| 6 |
+
from util import INIT_CRDS
|
| 7 |
+
from opt_einsum import contract as einsum
|
| 8 |
+
from icecream import ic
|
| 9 |
+
|
| 10 |
+
class RoseTTAFoldModule(nn.Module):
|
| 11 |
+
def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
|
| 12 |
+
d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
|
| 13 |
+
n_head_msa=8, n_head_pair=4, n_head_templ=4,
|
| 14 |
+
d_hidden=32, d_hidden_templ=64,
|
| 15 |
+
p_drop=0.15, d_t1d=24, d_t2d=44,
|
| 16 |
+
SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 17 |
+
SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 18 |
+
):
|
| 19 |
+
super(RoseTTAFoldModule, self).__init__()
|
| 20 |
+
#
|
| 21 |
+
# Input Embeddings
|
| 22 |
+
d_state = SE3_param_topk['l0_out_features']
|
| 23 |
+
self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
|
| 24 |
+
self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
|
| 25 |
+
self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
|
| 26 |
+
n_head=n_head_templ,
|
| 27 |
+
d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
|
| 28 |
+
# Update inputs with outputs from previous round
|
| 29 |
+
self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
|
| 30 |
+
#
|
| 31 |
+
self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
|
| 32 |
+
n_main_block=n_main_block,
|
| 33 |
+
n_ref_block=n_ref_block,
|
| 34 |
+
d_msa=d_msa, d_msa_full=d_msa_full,
|
| 35 |
+
d_pair=d_pair, d_hidden=d_hidden,
|
| 36 |
+
n_head_msa=n_head_msa,
|
| 37 |
+
n_head_pair=n_head_pair,
|
| 38 |
+
SE3_param_full=SE3_param_full,
|
| 39 |
+
SE3_param_topk=SE3_param_topk,
|
| 40 |
+
p_drop=p_drop)
|
| 41 |
+
##
|
| 42 |
+
self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
|
| 43 |
+
self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
|
| 44 |
+
self.lddt_pred = LDDTNetwork(d_state)
|
| 45 |
+
|
| 46 |
+
self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
|
| 47 |
+
|
| 48 |
+
def forward(self, msa_latent, msa_full, seq, xyz, idx,
|
| 49 |
+
seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
|
| 50 |
+
msa_prev=None, pair_prev=None, state_prev=None,
|
| 51 |
+
return_raw=False, return_full=False,
|
| 52 |
+
use_checkpoint=False, return_infer=False):
|
| 53 |
+
B, N, L = msa_latent.shape[:3]
|
| 54 |
+
# Get embeddings
|
| 55 |
+
#ic(seq.shape)
|
| 56 |
+
#ic(msa_latent.shape)
|
| 57 |
+
#ic(seq1hot.shape)
|
| 58 |
+
#ic(idx.shape)
|
| 59 |
+
#ic(xyz.shape)
|
| 60 |
+
#ic(seq1hot.shape)
|
| 61 |
+
#ic(t1d.shape)
|
| 62 |
+
#ic(t2d.shape)
|
| 63 |
+
|
| 64 |
+
idx = idx.long()
|
| 65 |
+
msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
|
| 66 |
+
|
| 67 |
+
msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
|
| 68 |
+
#
|
| 69 |
+
# Do recycling
|
| 70 |
+
if msa_prev == None:
|
| 71 |
+
msa_prev = torch.zeros_like(msa_latent[:,0])
|
| 72 |
+
if pair_prev == None:
|
| 73 |
+
pair_prev = torch.zeros_like(pair)
|
| 74 |
+
if state_prev == None:
|
| 75 |
+
state_prev = torch.zeros_like(state)
|
| 76 |
+
|
| 77 |
+
#ic(seq.shape)
|
| 78 |
+
#ic(msa_prev.shape)
|
| 79 |
+
#ic(pair_prev.shape)
|
| 80 |
+
#ic(xyz.shape)
|
| 81 |
+
#ic(state_prev.shape)
|
| 82 |
+
|
| 83 |
+
|
| 84 |
+
msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
|
| 85 |
+
msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
|
| 86 |
+
pair = pair + pair_recycle
|
| 87 |
+
state = state + state_recycle
|
| 88 |
+
#
|
| 89 |
+
#ic(t1d.dtype)
|
| 90 |
+
#ic(t2d.dtype)
|
| 91 |
+
#ic(alpha_t.dtype)
|
| 92 |
+
#ic(xyz_t.dtype)
|
| 93 |
+
#ic(pair.dtype)
|
| 94 |
+
#ic(state.dtype)
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
#import pdb; pdb.set_trace()
|
| 98 |
+
|
| 99 |
+
# add template embedding
|
| 100 |
+
pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
|
| 101 |
+
|
| 102 |
+
#ic(seq.dtype)
|
| 103 |
+
#ic(msa_latent.dtype)
|
| 104 |
+
#ic(msa_full.dtype)
|
| 105 |
+
#ic(pair.dtype)
|
| 106 |
+
#ic(xyz.dtype)
|
| 107 |
+
#ic(state.dtype)
|
| 108 |
+
#ic(idx.dtype)
|
| 109 |
+
|
| 110 |
+
# Predict coordinates from given inputs
|
| 111 |
+
msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
|
| 112 |
+
state, idx, use_checkpoint=use_checkpoint)
|
| 113 |
+
|
| 114 |
+
if return_raw:
|
| 115 |
+
# get last structure
|
| 116 |
+
xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
|
| 117 |
+
return msa[:,0], pair, xyz, state, alpha_s[-1]
|
| 118 |
+
|
| 119 |
+
# predict masked amino acids
|
| 120 |
+
logits_aa = self.aa_pred(msa)
|
| 121 |
+
#
|
| 122 |
+
# predict distogram & orientograms
|
| 123 |
+
logits = self.c6d_pred(pair)
|
| 124 |
+
|
| 125 |
+
# Predict LDDT
|
| 126 |
+
lddt = self.lddt_pred(state)
|
| 127 |
+
|
| 128 |
+
# predict experimentally resolved or not
|
| 129 |
+
logits_exp = self.exp_pred(msa[:,0], state)
|
| 130 |
+
|
| 131 |
+
if return_infer:
|
| 132 |
+
#get last structure
|
| 133 |
+
xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
|
| 134 |
+
return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
# get all intermediate bb structures
|
| 138 |
+
xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
|
| 139 |
+
|
| 140 |
+
return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
|
model/Attention_module.py
ADDED
|
@@ -0,0 +1,411 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
import torch.nn.functional as F
|
| 4 |
+
import math
|
| 5 |
+
from opt_einsum import contract as einsum
|
| 6 |
+
from util_module import init_lecun_normal
|
| 7 |
+
from icecream import ic
|
| 8 |
+
|
| 9 |
+
class FeedForwardLayer(nn.Module):
|
| 10 |
+
def __init__(self, d_model, r_ff, p_drop=0.1):
|
| 11 |
+
super(FeedForwardLayer, self).__init__()
|
| 12 |
+
self.norm = nn.LayerNorm(d_model)
|
| 13 |
+
self.linear1 = nn.Linear(d_model, d_model*r_ff)
|
| 14 |
+
self.dropout = nn.Dropout(p_drop)
|
| 15 |
+
self.linear2 = nn.Linear(d_model*r_ff, d_model)
|
| 16 |
+
|
| 17 |
+
self.reset_parameter()
|
| 18 |
+
|
| 19 |
+
def reset_parameter(self):
|
| 20 |
+
# initialize linear layer right before ReLu: He initializer (kaiming normal)
|
| 21 |
+
nn.init.kaiming_normal_(self.linear1.weight, nonlinearity='relu')
|
| 22 |
+
nn.init.zeros_(self.linear1.bias)
|
| 23 |
+
|
| 24 |
+
# initialize linear layer right before residual connection: zero initialize
|
| 25 |
+
nn.init.zeros_(self.linear2.weight)
|
| 26 |
+
nn.init.zeros_(self.linear2.bias)
|
| 27 |
+
|
| 28 |
+
def forward(self, src):
|
| 29 |
+
src = self.norm(src)
|
| 30 |
+
src = self.linear2(self.dropout(F.relu_(self.linear1(src))))
|
| 31 |
+
return src
|
| 32 |
+
|
| 33 |
+
class Attention(nn.Module):
|
| 34 |
+
# calculate multi-head attention
|
| 35 |
+
def __init__(self, d_query, d_key, n_head, d_hidden, d_out, p_drop=0.1):
|
| 36 |
+
super(Attention, self).__init__()
|
| 37 |
+
self.h = n_head
|
| 38 |
+
self.dim = d_hidden
|
| 39 |
+
#
|
| 40 |
+
self.to_q = nn.Linear(d_query, n_head*d_hidden, bias=False)
|
| 41 |
+
self.to_k = nn.Linear(d_key, n_head*d_hidden, bias=False)
|
| 42 |
+
self.to_v = nn.Linear(d_key, n_head*d_hidden, bias=False)
|
| 43 |
+
#
|
| 44 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_out)
|
| 45 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 46 |
+
#
|
| 47 |
+
# initialize all parameters properly
|
| 48 |
+
self.reset_parameter()
|
| 49 |
+
|
| 50 |
+
def reset_parameter(self):
|
| 51 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 52 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 53 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 54 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 55 |
+
|
| 56 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 57 |
+
nn.init.zeros_(self.to_out.weight)
|
| 58 |
+
nn.init.zeros_(self.to_out.bias)
|
| 59 |
+
|
| 60 |
+
def forward(self, query, key, value):
|
| 61 |
+
B, Q = query.shape[:2]
|
| 62 |
+
B, K = key.shape[:2]
|
| 63 |
+
#
|
| 64 |
+
query = self.to_q(query).reshape(B, Q, self.h, self.dim)
|
| 65 |
+
key = self.to_k(key).reshape(B, K, self.h, self.dim)
|
| 66 |
+
value = self.to_v(value).reshape(B, K, self.h, self.dim)
|
| 67 |
+
#
|
| 68 |
+
query = query * self.scaling
|
| 69 |
+
attn = einsum('bqhd,bkhd->bhqk', query, key)
|
| 70 |
+
attn = F.softmax(attn, dim=-1)
|
| 71 |
+
#
|
| 72 |
+
out = einsum('bhqk,bkhd->bqhd', attn, value)
|
| 73 |
+
out = out.reshape(B, Q, self.h*self.dim)
|
| 74 |
+
#
|
| 75 |
+
out = self.to_out(out)
|
| 76 |
+
|
| 77 |
+
return out
|
| 78 |
+
|
| 79 |
+
class AttentionWithBias(nn.Module):
|
| 80 |
+
def __init__(self, d_in=256, d_bias=128, n_head=8, d_hidden=32):
|
| 81 |
+
super(AttentionWithBias, self).__init__()
|
| 82 |
+
self.norm_in = nn.LayerNorm(d_in)
|
| 83 |
+
self.norm_bias = nn.LayerNorm(d_bias)
|
| 84 |
+
#
|
| 85 |
+
self.to_q = nn.Linear(d_in, n_head*d_hidden, bias=False)
|
| 86 |
+
self.to_k = nn.Linear(d_in, n_head*d_hidden, bias=False)
|
| 87 |
+
self.to_v = nn.Linear(d_in, n_head*d_hidden, bias=False)
|
| 88 |
+
self.to_b = nn.Linear(d_bias, n_head, bias=False)
|
| 89 |
+
self.to_g = nn.Linear(d_in, n_head*d_hidden)
|
| 90 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_in)
|
| 91 |
+
|
| 92 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 93 |
+
self.h = n_head
|
| 94 |
+
self.dim = d_hidden
|
| 95 |
+
|
| 96 |
+
self.reset_parameter()
|
| 97 |
+
|
| 98 |
+
def reset_parameter(self):
|
| 99 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 100 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 101 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 102 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 103 |
+
|
| 104 |
+
# bias: normal distribution
|
| 105 |
+
self.to_b = init_lecun_normal(self.to_b)
|
| 106 |
+
|
| 107 |
+
# gating: zero weights, one biases (mostly open gate at the begining)
|
| 108 |
+
nn.init.zeros_(self.to_g.weight)
|
| 109 |
+
nn.init.ones_(self.to_g.bias)
|
| 110 |
+
|
| 111 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 112 |
+
nn.init.zeros_(self.to_out.weight)
|
| 113 |
+
nn.init.zeros_(self.to_out.bias)
|
| 114 |
+
|
| 115 |
+
def forward(self, x, bias):
|
| 116 |
+
B, L = x.shape[:2]
|
| 117 |
+
#
|
| 118 |
+
x = self.norm_in(x)
|
| 119 |
+
bias = self.norm_bias(bias)
|
| 120 |
+
#
|
| 121 |
+
query = self.to_q(x).reshape(B, L, self.h, self.dim)
|
| 122 |
+
key = self.to_k(x).reshape(B, L, self.h, self.dim)
|
| 123 |
+
value = self.to_v(x).reshape(B, L, self.h, self.dim)
|
| 124 |
+
bias = self.to_b(bias) # (B, L, L, h)
|
| 125 |
+
gate = torch.sigmoid(self.to_g(x))
|
| 126 |
+
#
|
| 127 |
+
key = key * self.scaling
|
| 128 |
+
attn = einsum('bqhd,bkhd->bqkh', query, key)
|
| 129 |
+
attn = attn + bias
|
| 130 |
+
attn = F.softmax(attn, dim=-2)
|
| 131 |
+
#
|
| 132 |
+
out = einsum('bqkh,bkhd->bqhd', attn, value).reshape(B, L, -1)
|
| 133 |
+
out = gate * out
|
| 134 |
+
#
|
| 135 |
+
out = self.to_out(out)
|
| 136 |
+
return out
|
| 137 |
+
|
| 138 |
+
# MSA Attention (row/column) from AlphaFold architecture
|
| 139 |
+
class SequenceWeight(nn.Module):
|
| 140 |
+
def __init__(self, d_msa, n_head, d_hidden, p_drop=0.1):
|
| 141 |
+
super(SequenceWeight, self).__init__()
|
| 142 |
+
self.h = n_head
|
| 143 |
+
self.dim = d_hidden
|
| 144 |
+
self.scale = 1.0 / math.sqrt(self.dim)
|
| 145 |
+
|
| 146 |
+
self.to_query = nn.Linear(d_msa, n_head*d_hidden)
|
| 147 |
+
self.to_key = nn.Linear(d_msa, n_head*d_hidden)
|
| 148 |
+
self.dropout = nn.Dropout(p_drop)
|
| 149 |
+
|
| 150 |
+
self.reset_parameter()
|
| 151 |
+
|
| 152 |
+
def reset_parameter(self):
|
| 153 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 154 |
+
nn.init.xavier_uniform_(self.to_query.weight)
|
| 155 |
+
nn.init.xavier_uniform_(self.to_key.weight)
|
| 156 |
+
|
| 157 |
+
def forward(self, msa):
|
| 158 |
+
B, N, L = msa.shape[:3]
|
| 159 |
+
|
| 160 |
+
tar_seq = msa[:,0]
|
| 161 |
+
|
| 162 |
+
q = self.to_query(tar_seq).view(B, 1, L, self.h, self.dim)
|
| 163 |
+
k = self.to_key(msa).view(B, N, L, self.h, self.dim)
|
| 164 |
+
|
| 165 |
+
q = q * self.scale
|
| 166 |
+
attn = einsum('bqihd,bkihd->bkihq', q, k)
|
| 167 |
+
attn = F.softmax(attn, dim=1)
|
| 168 |
+
return self.dropout(attn)
|
| 169 |
+
|
| 170 |
+
class MSARowAttentionWithBias(nn.Module):
|
| 171 |
+
def __init__(self, d_msa=256, d_pair=128, n_head=8, d_hidden=32):
|
| 172 |
+
super(MSARowAttentionWithBias, self).__init__()
|
| 173 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 174 |
+
self.norm_pair = nn.LayerNorm(d_pair)
|
| 175 |
+
#
|
| 176 |
+
self.seq_weight = SequenceWeight(d_msa, n_head, d_hidden, p_drop=0.1)
|
| 177 |
+
self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 178 |
+
self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 179 |
+
self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 180 |
+
self.to_b = nn.Linear(d_pair, n_head, bias=False)
|
| 181 |
+
self.to_g = nn.Linear(d_msa, n_head*d_hidden)
|
| 182 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_msa)
|
| 183 |
+
|
| 184 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 185 |
+
self.h = n_head
|
| 186 |
+
self.dim = d_hidden
|
| 187 |
+
|
| 188 |
+
self.reset_parameter()
|
| 189 |
+
|
| 190 |
+
def reset_parameter(self):
|
| 191 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 192 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 193 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 194 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 195 |
+
|
| 196 |
+
# bias: normal distribution
|
| 197 |
+
self.to_b = init_lecun_normal(self.to_b)
|
| 198 |
+
|
| 199 |
+
# gating: zero weights, one biases (mostly open gate at the begining)
|
| 200 |
+
nn.init.zeros_(self.to_g.weight)
|
| 201 |
+
nn.init.ones_(self.to_g.bias)
|
| 202 |
+
|
| 203 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 204 |
+
nn.init.zeros_(self.to_out.weight)
|
| 205 |
+
nn.init.zeros_(self.to_out.bias)
|
| 206 |
+
|
| 207 |
+
def forward(self, msa, pair): # TODO: make this as tied-attention
|
| 208 |
+
B, N, L = msa.shape[:3]
|
| 209 |
+
#
|
| 210 |
+
msa = self.norm_msa(msa)
|
| 211 |
+
pair = self.norm_pair(pair)
|
| 212 |
+
#
|
| 213 |
+
seq_weight = self.seq_weight(msa) # (B, N, L, h, 1)
|
| 214 |
+
query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
|
| 215 |
+
key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
|
| 216 |
+
value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
|
| 217 |
+
bias = self.to_b(pair) # (B, L, L, h)
|
| 218 |
+
gate = torch.sigmoid(self.to_g(msa))
|
| 219 |
+
#
|
| 220 |
+
query = query * seq_weight.expand(-1, -1, -1, -1, self.dim)
|
| 221 |
+
key = key * self.scaling
|
| 222 |
+
attn = einsum('bsqhd,bskhd->bqkh', query, key)
|
| 223 |
+
attn = attn + bias
|
| 224 |
+
attn = F.softmax(attn, dim=-2)
|
| 225 |
+
#
|
| 226 |
+
out = einsum('bqkh,bskhd->bsqhd', attn, value).reshape(B, N, L, -1)
|
| 227 |
+
out = gate * out
|
| 228 |
+
#
|
| 229 |
+
out = self.to_out(out)
|
| 230 |
+
return out
|
| 231 |
+
|
| 232 |
+
class MSAColAttention(nn.Module):
|
| 233 |
+
def __init__(self, d_msa=256, n_head=8, d_hidden=32):
|
| 234 |
+
super(MSAColAttention, self).__init__()
|
| 235 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 236 |
+
#
|
| 237 |
+
self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 238 |
+
self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 239 |
+
self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 240 |
+
self.to_g = nn.Linear(d_msa, n_head*d_hidden)
|
| 241 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_msa)
|
| 242 |
+
|
| 243 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 244 |
+
self.h = n_head
|
| 245 |
+
self.dim = d_hidden
|
| 246 |
+
|
| 247 |
+
self.reset_parameter()
|
| 248 |
+
|
| 249 |
+
def reset_parameter(self):
|
| 250 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 251 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 252 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 253 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 254 |
+
|
| 255 |
+
# gating: zero weights, one biases (mostly open gate at the begining)
|
| 256 |
+
nn.init.zeros_(self.to_g.weight)
|
| 257 |
+
nn.init.ones_(self.to_g.bias)
|
| 258 |
+
|
| 259 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 260 |
+
nn.init.zeros_(self.to_out.weight)
|
| 261 |
+
nn.init.zeros_(self.to_out.bias)
|
| 262 |
+
|
| 263 |
+
def forward(self, msa):
|
| 264 |
+
B, N, L = msa.shape[:3]
|
| 265 |
+
#
|
| 266 |
+
msa = self.norm_msa(msa)
|
| 267 |
+
#
|
| 268 |
+
query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
|
| 269 |
+
key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
|
| 270 |
+
value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
|
| 271 |
+
gate = torch.sigmoid(self.to_g(msa))
|
| 272 |
+
#
|
| 273 |
+
query = query * self.scaling
|
| 274 |
+
attn = einsum('bqihd,bkihd->bihqk', query, key)
|
| 275 |
+
attn = F.softmax(attn, dim=-1)
|
| 276 |
+
#
|
| 277 |
+
out = einsum('bihqk,bkihd->bqihd', attn, value).reshape(B, N, L, -1)
|
| 278 |
+
out = gate * out
|
| 279 |
+
#
|
| 280 |
+
out = self.to_out(out)
|
| 281 |
+
return out
|
| 282 |
+
|
| 283 |
+
class MSAColGlobalAttention(nn.Module):
|
| 284 |
+
def __init__(self, d_msa=64, n_head=8, d_hidden=8):
|
| 285 |
+
super(MSAColGlobalAttention, self).__init__()
|
| 286 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 287 |
+
#
|
| 288 |
+
self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
|
| 289 |
+
self.to_k = nn.Linear(d_msa, d_hidden, bias=False)
|
| 290 |
+
self.to_v = nn.Linear(d_msa, d_hidden, bias=False)
|
| 291 |
+
self.to_g = nn.Linear(d_msa, n_head*d_hidden)
|
| 292 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_msa)
|
| 293 |
+
|
| 294 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 295 |
+
self.h = n_head
|
| 296 |
+
self.dim = d_hidden
|
| 297 |
+
|
| 298 |
+
self.reset_parameter()
|
| 299 |
+
|
| 300 |
+
def reset_parameter(self):
|
| 301 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 302 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 303 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 304 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 305 |
+
|
| 306 |
+
# gating: zero weights, one biases (mostly open gate at the begining)
|
| 307 |
+
nn.init.zeros_(self.to_g.weight)
|
| 308 |
+
nn.init.ones_(self.to_g.bias)
|
| 309 |
+
|
| 310 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 311 |
+
nn.init.zeros_(self.to_out.weight)
|
| 312 |
+
nn.init.zeros_(self.to_out.bias)
|
| 313 |
+
|
| 314 |
+
def forward(self, msa):
|
| 315 |
+
B, N, L = msa.shape[:3]
|
| 316 |
+
#
|
| 317 |
+
msa = self.norm_msa(msa)
|
| 318 |
+
#
|
| 319 |
+
query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
|
| 320 |
+
query = query.mean(dim=1) # (B, L, h, dim)
|
| 321 |
+
key = self.to_k(msa) # (B, N, L, dim)
|
| 322 |
+
value = self.to_v(msa) # (B, N, L, dim)
|
| 323 |
+
gate = torch.sigmoid(self.to_g(msa)) # (B, N, L, h*dim)
|
| 324 |
+
#
|
| 325 |
+
query = query * self.scaling
|
| 326 |
+
attn = einsum('bihd,bkid->bihk', query, key) # (B, L, h, N)
|
| 327 |
+
attn = F.softmax(attn, dim=-1)
|
| 328 |
+
#
|
| 329 |
+
out = einsum('bihk,bkid->bihd', attn, value).reshape(B, 1, L, -1) # (B, 1, L, h*dim)
|
| 330 |
+
out = gate * out # (B, N, L, h*dim)
|
| 331 |
+
#
|
| 332 |
+
out = self.to_out(out)
|
| 333 |
+
return out
|
| 334 |
+
|
| 335 |
+
# Instead of triangle attention, use Tied axail attention with bias from coordinates..?
|
| 336 |
+
class BiasedAxialAttention(nn.Module):
|
| 337 |
+
def __init__(self, d_pair, d_bias, n_head, d_hidden, p_drop=0.1, is_row=True):
|
| 338 |
+
super(BiasedAxialAttention, self).__init__()
|
| 339 |
+
#
|
| 340 |
+
self.is_row = is_row
|
| 341 |
+
self.norm_pair = nn.LayerNorm(d_pair)
|
| 342 |
+
self.norm_bias = nn.LayerNorm(d_bias)
|
| 343 |
+
|
| 344 |
+
self.to_q = nn.Linear(d_pair, n_head*d_hidden, bias=False)
|
| 345 |
+
self.to_k = nn.Linear(d_pair, n_head*d_hidden, bias=False)
|
| 346 |
+
self.to_v = nn.Linear(d_pair, n_head*d_hidden, bias=False)
|
| 347 |
+
self.to_b = nn.Linear(d_bias, n_head, bias=False)
|
| 348 |
+
self.to_g = nn.Linear(d_pair, n_head*d_hidden)
|
| 349 |
+
self.to_out = nn.Linear(n_head*d_hidden, d_pair)
|
| 350 |
+
|
| 351 |
+
self.scaling = 1/math.sqrt(d_hidden)
|
| 352 |
+
self.h = n_head
|
| 353 |
+
self.dim = d_hidden
|
| 354 |
+
|
| 355 |
+
# initialize all parameters properly
|
| 356 |
+
self.reset_parameter()
|
| 357 |
+
|
| 358 |
+
def reset_parameter(self):
|
| 359 |
+
# query/key/value projection: Glorot uniform / Xavier uniform
|
| 360 |
+
nn.init.xavier_uniform_(self.to_q.weight)
|
| 361 |
+
nn.init.xavier_uniform_(self.to_k.weight)
|
| 362 |
+
nn.init.xavier_uniform_(self.to_v.weight)
|
| 363 |
+
|
| 364 |
+
# bias: normal distribution
|
| 365 |
+
self.to_b = init_lecun_normal(self.to_b)
|
| 366 |
+
|
| 367 |
+
# gating: zero weights, one biases (mostly open gate at the begining)
|
| 368 |
+
nn.init.zeros_(self.to_g.weight)
|
| 369 |
+
nn.init.ones_(self.to_g.bias)
|
| 370 |
+
|
| 371 |
+
# to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
|
| 372 |
+
nn.init.zeros_(self.to_out.weight)
|
| 373 |
+
nn.init.zeros_(self.to_out.bias)
|
| 374 |
+
|
| 375 |
+
def forward(self, pair, bias, same_chain = None):
|
| 376 |
+
# pair: (B, L, L, d_pair)
|
| 377 |
+
B, L = pair.shape[:2]
|
| 378 |
+
|
| 379 |
+
if self.is_row:
|
| 380 |
+
pair = pair.permute(0,2,1,3)
|
| 381 |
+
bias = bias.permute(0,2,1,3)
|
| 382 |
+
|
| 383 |
+
pair = self.norm_pair(pair)
|
| 384 |
+
bias = self.norm_bias(bias)
|
| 385 |
+
|
| 386 |
+
query = self.to_q(pair).reshape(B, L, L, self.h, self.dim)
|
| 387 |
+
key = self.to_k(pair).reshape(B, L, L, self.h, self.dim)
|
| 388 |
+
value = self.to_v(pair).reshape(B, L, L, self.h, self.dim)
|
| 389 |
+
bias = self.to_b(bias) # (B, L, L, h)
|
| 390 |
+
gate = torch.sigmoid(self.to_g(pair)) # (B, L, L, h*dim)
|
| 391 |
+
|
| 392 |
+
query = query * self.scaling
|
| 393 |
+
key = key / math.sqrt(L) # normalize for tied attention
|
| 394 |
+
attn = einsum('bnihk,bnjhk->bijh', query, key) # tied attention
|
| 395 |
+
attn = attn + bias # apply bias
|
| 396 |
+
attn = F.softmax(attn, dim=-2) # (B, L, L, h)
|
| 397 |
+
|
| 398 |
+
if same_chain is not None:
|
| 399 |
+
ic(same_chain)
|
| 400 |
+
ic(attn)
|
| 401 |
+
ic(attn[~same_chain])
|
| 402 |
+
attn[~same_chain] *= 1.1
|
| 403 |
+
|
| 404 |
+
out = einsum('bijh,bkjhd->bikhd', attn, value).reshape(B, L, L, -1)
|
| 405 |
+
out = gate * out
|
| 406 |
+
|
| 407 |
+
out = self.to_out(out)
|
| 408 |
+
if self.is_row:
|
| 409 |
+
out = out.permute(0,2,1,3)
|
| 410 |
+
return out
|
| 411 |
+
|
model/AuxiliaryPredictor.py
ADDED
|
@@ -0,0 +1,92 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
|
| 4 |
+
class DistanceNetwork(nn.Module):
|
| 5 |
+
def __init__(self, n_feat, p_drop=0.1):
|
| 6 |
+
super(DistanceNetwork, self).__init__()
|
| 7 |
+
#
|
| 8 |
+
self.proj_symm = nn.Linear(n_feat, 37*2)
|
| 9 |
+
self.proj_asymm = nn.Linear(n_feat, 37+19)
|
| 10 |
+
|
| 11 |
+
self.reset_parameter()
|
| 12 |
+
|
| 13 |
+
def reset_parameter(self):
|
| 14 |
+
# initialize linear layer for final logit prediction
|
| 15 |
+
nn.init.zeros_(self.proj_symm.weight)
|
| 16 |
+
nn.init.zeros_(self.proj_asymm.weight)
|
| 17 |
+
nn.init.zeros_(self.proj_symm.bias)
|
| 18 |
+
nn.init.zeros_(self.proj_asymm.bias)
|
| 19 |
+
|
| 20 |
+
def forward(self, x):
|
| 21 |
+
# input: pair info (B, L, L, C)
|
| 22 |
+
|
| 23 |
+
# predict theta, phi (non-symmetric)
|
| 24 |
+
logits_asymm = self.proj_asymm(x)
|
| 25 |
+
logits_theta = logits_asymm[:,:,:,:37].permute(0,3,1,2)
|
| 26 |
+
logits_phi = logits_asymm[:,:,:,37:].permute(0,3,1,2)
|
| 27 |
+
|
| 28 |
+
# predict dist, omega
|
| 29 |
+
logits_symm = self.proj_symm(x)
|
| 30 |
+
logits_symm = logits_symm + logits_symm.permute(0,2,1,3)
|
| 31 |
+
logits_dist = logits_symm[:,:,:,:37].permute(0,3,1,2)
|
| 32 |
+
logits_omega = logits_symm[:,:,:,37:].permute(0,3,1,2)
|
| 33 |
+
|
| 34 |
+
return logits_dist, logits_omega, logits_theta, logits_phi
|
| 35 |
+
|
| 36 |
+
class MaskedTokenNetwork(nn.Module):
|
| 37 |
+
def __init__(self, n_feat, p_drop=0.1):
|
| 38 |
+
super(MaskedTokenNetwork, self).__init__()
|
| 39 |
+
self.proj = nn.Linear(n_feat, 21)
|
| 40 |
+
|
| 41 |
+
self.reset_parameter()
|
| 42 |
+
|
| 43 |
+
def reset_parameter(self):
|
| 44 |
+
nn.init.zeros_(self.proj.weight)
|
| 45 |
+
nn.init.zeros_(self.proj.bias)
|
| 46 |
+
|
| 47 |
+
def forward(self, x):
|
| 48 |
+
B, N, L = x.shape[:3]
|
| 49 |
+
logits = self.proj(x).permute(0,3,1,2).reshape(B, -1, N*L)
|
| 50 |
+
|
| 51 |
+
return logits
|
| 52 |
+
|
| 53 |
+
class LDDTNetwork(nn.Module):
|
| 54 |
+
def __init__(self, n_feat, n_bin_lddt=50):
|
| 55 |
+
super(LDDTNetwork, self).__init__()
|
| 56 |
+
self.proj = nn.Linear(n_feat, n_bin_lddt)
|
| 57 |
+
|
| 58 |
+
self.reset_parameter()
|
| 59 |
+
|
| 60 |
+
def reset_parameter(self):
|
| 61 |
+
nn.init.zeros_(self.proj.weight)
|
| 62 |
+
nn.init.zeros_(self.proj.bias)
|
| 63 |
+
|
| 64 |
+
def forward(self, x):
|
| 65 |
+
logits = self.proj(x) # (B, L, 50)
|
| 66 |
+
|
| 67 |
+
return logits.permute(0,2,1)
|
| 68 |
+
|
| 69 |
+
class ExpResolvedNetwork(nn.Module):
|
| 70 |
+
def __init__(self, d_msa, d_state, p_drop=0.1):
|
| 71 |
+
super(ExpResolvedNetwork, self).__init__()
|
| 72 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 73 |
+
self.norm_state = nn.LayerNorm(d_state)
|
| 74 |
+
self.proj = nn.Linear(d_msa+d_state, 1)
|
| 75 |
+
|
| 76 |
+
self.reset_parameter()
|
| 77 |
+
|
| 78 |
+
def reset_parameter(self):
|
| 79 |
+
nn.init.zeros_(self.proj.weight)
|
| 80 |
+
nn.init.zeros_(self.proj.bias)
|
| 81 |
+
|
| 82 |
+
def forward(self, seq, state):
|
| 83 |
+
B, L = seq.shape[:2]
|
| 84 |
+
|
| 85 |
+
seq = self.norm_msa(seq)
|
| 86 |
+
state = self.norm_state(state)
|
| 87 |
+
feat = torch.cat((seq, state), dim=-1)
|
| 88 |
+
logits = self.proj(feat)
|
| 89 |
+
return logits.reshape(B, L)
|
| 90 |
+
|
| 91 |
+
|
| 92 |
+
|
model/Embeddings.py
ADDED
|
@@ -0,0 +1,307 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
import torch.nn.functional as F
|
| 4 |
+
from opt_einsum import contract as einsum
|
| 5 |
+
import torch.utils.checkpoint as checkpoint
|
| 6 |
+
from util import get_tips
|
| 7 |
+
from util_module import Dropout, create_custom_forward, rbf, init_lecun_normal
|
| 8 |
+
from Attention_module import Attention, FeedForwardLayer, AttentionWithBias
|
| 9 |
+
from Track_module import PairStr2Pair
|
| 10 |
+
from icecream import ic
|
| 11 |
+
|
| 12 |
+
# Module contains classes and functions to generate initial embeddings
|
| 13 |
+
|
| 14 |
+
class PositionalEncoding2D(nn.Module):
|
| 15 |
+
# Add relative positional encoding to pair features
|
| 16 |
+
def __init__(self, d_model, minpos=-32, maxpos=32, p_drop=0.1):
|
| 17 |
+
super(PositionalEncoding2D, self).__init__()
|
| 18 |
+
self.minpos = minpos
|
| 19 |
+
self.maxpos = maxpos
|
| 20 |
+
self.nbin = abs(minpos)+maxpos+1
|
| 21 |
+
self.emb = nn.Embedding(self.nbin, d_model)
|
| 22 |
+
self.drop = nn.Dropout(p_drop)
|
| 23 |
+
|
| 24 |
+
def forward(self, x, idx):
|
| 25 |
+
bins = torch.arange(self.minpos, self.maxpos, device=x.device)
|
| 26 |
+
seqsep = idx[:,None,:] - idx[:,:,None] # (B, L, L)
|
| 27 |
+
#
|
| 28 |
+
ib = torch.bucketize(seqsep, bins).long() # (B, L, L)
|
| 29 |
+
emb = self.emb(ib) #(B, L, L, d_model)
|
| 30 |
+
x = x + emb # add relative positional encoding
|
| 31 |
+
return self.drop(x)
|
| 32 |
+
|
| 33 |
+
class MSA_emb(nn.Module):
|
| 34 |
+
# Get initial seed MSA embedding
|
| 35 |
+
def __init__(self, d_msa=256, d_pair=128, d_state=32, d_init=22+22+2+2,
|
| 36 |
+
minpos=-32, maxpos=32, p_drop=0.1):
|
| 37 |
+
super(MSA_emb, self).__init__()
|
| 38 |
+
self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
|
| 39 |
+
self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence -- used for MSA embedding
|
| 40 |
+
self.emb_left = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
|
| 41 |
+
self.emb_right = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
|
| 42 |
+
self.emb_state = nn.Embedding(22, d_state)
|
| 43 |
+
self.drop = nn.Dropout(p_drop)
|
| 44 |
+
self.pos = PositionalEncoding2D(d_pair, minpos=minpos, maxpos=maxpos, p_drop=p_drop)
|
| 45 |
+
|
| 46 |
+
self.reset_parameter()
|
| 47 |
+
|
| 48 |
+
def reset_parameter(self):
|
| 49 |
+
self.emb = init_lecun_normal(self.emb)
|
| 50 |
+
self.emb_q = init_lecun_normal(self.emb_q)
|
| 51 |
+
self.emb_left = init_lecun_normal(self.emb_left)
|
| 52 |
+
self.emb_right = init_lecun_normal(self.emb_right)
|
| 53 |
+
self.emb_state = init_lecun_normal(self.emb_state)
|
| 54 |
+
|
| 55 |
+
nn.init.zeros_(self.emb.bias)
|
| 56 |
+
|
| 57 |
+
def forward(self, msa, seq, idx, seq1hot=None):
|
| 58 |
+
# Inputs:
|
| 59 |
+
# - msa: Input MSA (B, N, L, d_init)
|
| 60 |
+
# - seq: Input Sequence (B, L)
|
| 61 |
+
# - idx: Residue index
|
| 62 |
+
# Outputs:
|
| 63 |
+
# - msa: Initial MSA embedding (B, N, L, d_msa)
|
| 64 |
+
# - pair: Initial Pair embedding (B, L, L, d_pair)
|
| 65 |
+
|
| 66 |
+
N = msa.shape[1] # number of sequenes in MSA
|
| 67 |
+
|
| 68 |
+
# msa embedding
|
| 69 |
+
msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
|
| 70 |
+
seq = seq.long()
|
| 71 |
+
tmp = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
|
| 72 |
+
msa = msa + tmp.expand(-1, N, -1, -1) # adding query embedding to MSA
|
| 73 |
+
msa = self.drop(msa)
|
| 74 |
+
|
| 75 |
+
# pair embedding
|
| 76 |
+
if seq1hot is not None:
|
| 77 |
+
left = (seq1hot @ self.emb_left.weight)[:,None] # (B, 1, L, d_pair)
|
| 78 |
+
right = (seq1hot @ self.emb_right.weight)[:,:,None] # (B, L, 1, d_pair)
|
| 79 |
+
else:
|
| 80 |
+
left = self.emb_left(seq)[:,None] # (B, 1, L, d_pair)
|
| 81 |
+
right = self.emb_right(seq)[:,:,None] # (B, L, 1, d_pair)
|
| 82 |
+
#ic(torch.norm(self.emb_left.weight, dim=1))
|
| 83 |
+
#ic(torch.norm(self.emb_right.weight, dim=1))
|
| 84 |
+
pair = left + right # (B, L, L, d_pair)
|
| 85 |
+
pair = self.pos(pair, idx) # add relative position
|
| 86 |
+
|
| 87 |
+
# state embedding
|
| 88 |
+
state = self.drop(self.emb_state(seq))
|
| 89 |
+
|
| 90 |
+
return msa, pair, state
|
| 91 |
+
|
| 92 |
+
class Extra_emb(nn.Module):
|
| 93 |
+
# Get initial seed MSA embedding
|
| 94 |
+
def __init__(self, d_msa=256, d_init=22+1+2, p_drop=0.1):
|
| 95 |
+
super(Extra_emb, self).__init__()
|
| 96 |
+
self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
|
| 97 |
+
self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence
|
| 98 |
+
self.drop = nn.Dropout(p_drop)
|
| 99 |
+
|
| 100 |
+
self.reset_parameter()
|
| 101 |
+
|
| 102 |
+
def reset_parameter(self):
|
| 103 |
+
self.emb = init_lecun_normal(self.emb)
|
| 104 |
+
nn.init.zeros_(self.emb.bias)
|
| 105 |
+
|
| 106 |
+
def forward(self, msa, seq, idx, seq1hot=None):
|
| 107 |
+
# Inputs:
|
| 108 |
+
# - msa: Input MSA (B, N, L, d_init)
|
| 109 |
+
# - seq: Input Sequence (B, L)
|
| 110 |
+
# - idx: Residue index
|
| 111 |
+
# Outputs:
|
| 112 |
+
# - msa: Initial MSA embedding (B, N, L, d_msa)
|
| 113 |
+
N = msa.shape[1] # number of sequenes in MSA
|
| 114 |
+
msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
|
| 115 |
+
if seq1hot is not None:
|
| 116 |
+
seq = (seq1hot @ self.emb_q.weight).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
|
| 117 |
+
else:
|
| 118 |
+
seq = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
|
| 119 |
+
#ic(torch.norm(self.emb_q.weight, dim=1))
|
| 120 |
+
msa = msa + seq.expand(-1, N, -1, -1) # adding query embedding to MSA
|
| 121 |
+
return self.drop(msa)
|
| 122 |
+
|
| 123 |
+
class TemplatePairStack(nn.Module):
|
| 124 |
+
# process template pairwise features
|
| 125 |
+
# use structure-biased attention
|
| 126 |
+
def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.25):
|
| 127 |
+
super(TemplatePairStack, self).__init__()
|
| 128 |
+
self.n_block = n_block
|
| 129 |
+
proc_s = [PairStr2Pair(d_pair=d_templ, n_head=n_head, d_hidden=d_hidden, p_drop=p_drop) for i in range(n_block)]
|
| 130 |
+
self.block = nn.ModuleList(proc_s)
|
| 131 |
+
self.norm = nn.LayerNorm(d_templ)
|
| 132 |
+
def forward(self, templ, rbf_feat, use_checkpoint=False):
|
| 133 |
+
B, T, L = templ.shape[:3]
|
| 134 |
+
templ = templ.reshape(B*T, L, L, -1)
|
| 135 |
+
|
| 136 |
+
for i_block in range(self.n_block):
|
| 137 |
+
if use_checkpoint:
|
| 138 |
+
templ = checkpoint.checkpoint(create_custom_forward(self.block[i_block]), templ, rbf_feat)
|
| 139 |
+
else:
|
| 140 |
+
templ = self.block[i_block](templ, rbf_feat)
|
| 141 |
+
return self.norm(templ).reshape(B, T, L, L, -1)
|
| 142 |
+
|
| 143 |
+
class TemplateTorsionStack(nn.Module):
|
| 144 |
+
def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.15):
|
| 145 |
+
super(TemplateTorsionStack, self).__init__()
|
| 146 |
+
self.n_block=n_block
|
| 147 |
+
self.proj_pair = nn.Linear(d_templ+36, d_templ)
|
| 148 |
+
proc_s = [AttentionWithBias(d_in=d_templ, d_bias=d_templ,
|
| 149 |
+
n_head=n_head, d_hidden=d_hidden) for i in range(n_block)]
|
| 150 |
+
self.row_attn = nn.ModuleList(proc_s)
|
| 151 |
+
proc_s = [FeedForwardLayer(d_templ, 4, p_drop=p_drop) for i in range(n_block)]
|
| 152 |
+
self.ff = nn.ModuleList(proc_s)
|
| 153 |
+
self.norm = nn.LayerNorm(d_templ)
|
| 154 |
+
|
| 155 |
+
def reset_parameter(self):
|
| 156 |
+
self.proj_pair = init_lecun_normal(self.proj_pair)
|
| 157 |
+
nn.init.zeros_(self.proj_pair.bias)
|
| 158 |
+
|
| 159 |
+
def forward(self, tors, pair, rbf_feat, use_checkpoint=False):
|
| 160 |
+
B, T, L = tors.shape[:3]
|
| 161 |
+
tors = tors.reshape(B*T, L, -1)
|
| 162 |
+
pair = pair.reshape(B*T, L, L, -1)
|
| 163 |
+
pair = torch.cat((pair, rbf_feat), dim=-1)
|
| 164 |
+
pair = self.proj_pair(pair)
|
| 165 |
+
|
| 166 |
+
for i_block in range(self.n_block):
|
| 167 |
+
if use_checkpoint:
|
| 168 |
+
tors = tors + checkpoint.checkpoint(create_custom_forward(self.row_attn[i_block]), tors, pair)
|
| 169 |
+
else:
|
| 170 |
+
tors = tors + self.row_attn[i_block](tors, pair)
|
| 171 |
+
tors = tors + self.ff[i_block](tors)
|
| 172 |
+
return self.norm(tors).reshape(B, T, L, -1)
|
| 173 |
+
|
| 174 |
+
class Templ_emb(nn.Module):
|
| 175 |
+
# Get template embedding
|
| 176 |
+
# Features are
|
| 177 |
+
# t2d:
|
| 178 |
+
# - 37 distogram bins + 6 orientations (43)
|
| 179 |
+
# - Mask (missing/unaligned) (1)
|
| 180 |
+
# t1d:
|
| 181 |
+
# - tiled AA sequence (20 standard aa + gap)
|
| 182 |
+
# - seq confidence (1)
|
| 183 |
+
# - global time step (1)
|
| 184 |
+
# - struc confidence (1)
|
| 185 |
+
#
|
| 186 |
+
def __init__(self, d_t1d=21+1+1+1, d_t2d=43+1, d_tor=30, d_pair=128, d_state=32,
|
| 187 |
+
n_block=2, d_templ=64,
|
| 188 |
+
n_head=4, d_hidden=16, p_drop=0.25):
|
| 189 |
+
super(Templ_emb, self).__init__()
|
| 190 |
+
# process 2D features
|
| 191 |
+
self.emb = nn.Linear(d_t1d*2+d_t2d, d_templ)
|
| 192 |
+
self.templ_stack = TemplatePairStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
|
| 193 |
+
d_hidden=d_hidden, p_drop=p_drop)
|
| 194 |
+
|
| 195 |
+
self.attn = Attention(d_pair, d_templ, n_head, d_hidden, d_pair, p_drop=p_drop)
|
| 196 |
+
|
| 197 |
+
# process torsion angles
|
| 198 |
+
self.emb_t1d = nn.Linear(d_t1d+d_tor, d_templ)
|
| 199 |
+
self.proj_t1d = nn.Linear(d_templ, d_templ)
|
| 200 |
+
#self.tor_stack = TemplateTorsionStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
|
| 201 |
+
# d_hidden=d_hidden, p_drop=p_drop)
|
| 202 |
+
self.attn_tor = Attention(d_state, d_templ, n_head, d_hidden, d_state, p_drop=p_drop)
|
| 203 |
+
|
| 204 |
+
self.reset_parameter()
|
| 205 |
+
|
| 206 |
+
def reset_parameter(self):
|
| 207 |
+
self.emb = init_lecun_normal(self.emb)
|
| 208 |
+
#nn.init.zeros_(self.emb.weight) #init weights to zero
|
| 209 |
+
nn.init.zeros_(self.emb.bias)
|
| 210 |
+
|
| 211 |
+
nn.init.kaiming_normal_(self.emb_t1d.weight, nonlinearity='relu')
|
| 212 |
+
#nn.init.zeros_(self.emb_t1d.weight)
|
| 213 |
+
nn.init.zeros_(self.emb_t1d.bias)
|
| 214 |
+
|
| 215 |
+
self.proj_t1d = init_lecun_normal(self.proj_t1d)
|
| 216 |
+
nn.init.zeros_(self.proj_t1d.bias)
|
| 217 |
+
|
| 218 |
+
def forward(self, t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=False):
|
| 219 |
+
# Input
|
| 220 |
+
# - t1d: 1D template info (B, T, L, 23) 24 SL
|
| 221 |
+
# - t2d: 2D template info (B, T, L, L, 44)
|
| 222 |
+
B, T, L, _ = t1d.shape
|
| 223 |
+
|
| 224 |
+
# Prepare 2D template features
|
| 225 |
+
left = t1d.unsqueeze(3).expand(-1,-1,-1,L,-1)
|
| 226 |
+
right = t1d.unsqueeze(2).expand(-1,-1,L,-1,-1)
|
| 227 |
+
#
|
| 228 |
+
templ = torch.cat((t2d, left, right), -1) # (B, T, L, L, 88)
|
| 229 |
+
|
| 230 |
+
#ic(templ.shape)
|
| 231 |
+
#ic(templ.dtype)
|
| 232 |
+
#ic(self.emb.weight.dtype)
|
| 233 |
+
templ = self.emb(templ) # Template templures (B, T, L, L, d_templ)
|
| 234 |
+
# process each template features
|
| 235 |
+
xyz_t = xyz_t.reshape(B*T, L, -1, 3)
|
| 236 |
+
rbf_feat = rbf(torch.cdist(xyz_t[:,:,1], xyz_t[:,:,1]))
|
| 237 |
+
templ = self.templ_stack(templ, rbf_feat, use_checkpoint=use_checkpoint) # (B, T, L,L, d_templ)
|
| 238 |
+
|
| 239 |
+
# Prepare 1D template torsion angle features
|
| 240 |
+
t1d = torch.cat((t1d, alpha_t), dim=-1) # (B, T, L, 22+30)
|
| 241 |
+
# process each template features
|
| 242 |
+
t1d = self.proj_t1d(F.relu_(self.emb_t1d(t1d)))
|
| 243 |
+
|
| 244 |
+
# mixing query state features to template state features
|
| 245 |
+
state = state.reshape(B*L, 1, -1)
|
| 246 |
+
t1d = t1d.permute(0,2,1,3).reshape(B*L, T, -1)
|
| 247 |
+
if use_checkpoint:
|
| 248 |
+
out = checkpoint.checkpoint(create_custom_forward(self.attn_tor), state, t1d, t1d)
|
| 249 |
+
out = out.reshape(B, L, -1)
|
| 250 |
+
else:
|
| 251 |
+
out = self.attn_tor(state, t1d, t1d).reshape(B, L, -1)
|
| 252 |
+
state = state.reshape(B, L, -1)
|
| 253 |
+
state = state + out
|
| 254 |
+
|
| 255 |
+
# mixing query pair features to template information (Template pointwise attention)
|
| 256 |
+
pair = pair.reshape(B*L*L, 1, -1)
|
| 257 |
+
templ = templ.permute(0, 2, 3, 1, 4).reshape(B*L*L, T, -1)
|
| 258 |
+
if use_checkpoint:
|
| 259 |
+
out = checkpoint.checkpoint(create_custom_forward(self.attn), pair, templ, templ)
|
| 260 |
+
out = out.reshape(B, L, L, -1)
|
| 261 |
+
else:
|
| 262 |
+
out = self.attn(pair, templ, templ).reshape(B, L, L, -1)
|
| 263 |
+
#
|
| 264 |
+
pair = pair.reshape(B, L, L, -1)
|
| 265 |
+
pair = pair + out
|
| 266 |
+
|
| 267 |
+
return pair, state
|
| 268 |
+
|
| 269 |
+
class Recycling(nn.Module):
|
| 270 |
+
def __init__(self, d_msa=256, d_pair=128, d_state=32):
|
| 271 |
+
super(Recycling, self).__init__()
|
| 272 |
+
self.proj_dist = nn.Linear(36+d_state*2, d_pair)
|
| 273 |
+
self.norm_state = nn.LayerNorm(d_state)
|
| 274 |
+
self.norm_pair = nn.LayerNorm(d_pair)
|
| 275 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 276 |
+
|
| 277 |
+
self.reset_parameter()
|
| 278 |
+
|
| 279 |
+
def reset_parameter(self):
|
| 280 |
+
self.proj_dist = init_lecun_normal(self.proj_dist)
|
| 281 |
+
nn.init.zeros_(self.proj_dist.bias)
|
| 282 |
+
|
| 283 |
+
def forward(self, seq, msa, pair, xyz, state):
|
| 284 |
+
B, L = pair.shape[:2]
|
| 285 |
+
state = self.norm_state(state)
|
| 286 |
+
#
|
| 287 |
+
left = state.unsqueeze(2).expand(-1,-1,L,-1)
|
| 288 |
+
right = state.unsqueeze(1).expand(-1,L,-1,-1)
|
| 289 |
+
|
| 290 |
+
# three anchor atoms
|
| 291 |
+
N = xyz[:,:,0]
|
| 292 |
+
Ca = xyz[:,:,1]
|
| 293 |
+
C = xyz[:,:,2]
|
| 294 |
+
|
| 295 |
+
# recreate Cb given N,Ca,C
|
| 296 |
+
b = Ca - N
|
| 297 |
+
c = C - Ca
|
| 298 |
+
a = torch.cross(b, c, dim=-1)
|
| 299 |
+
Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
|
| 300 |
+
|
| 301 |
+
dist = rbf(torch.cdist(Cb, Cb))
|
| 302 |
+
dist = torch.cat((dist, left, right), dim=-1)
|
| 303 |
+
dist = self.proj_dist(dist)
|
| 304 |
+
pair = dist + self.norm_pair(pair)
|
| 305 |
+
msa = self.norm_msa(msa)
|
| 306 |
+
return msa, pair, state
|
| 307 |
+
|
model/RoseTTAFoldModel.py
ADDED
|
@@ -0,0 +1,140 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
|
| 4 |
+
from Track_module import IterativeSimulator
|
| 5 |
+
from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
|
| 6 |
+
from util import INIT_CRDS
|
| 7 |
+
from opt_einsum import contract as einsum
|
| 8 |
+
from icecream import ic
|
| 9 |
+
|
| 10 |
+
class RoseTTAFoldModule(nn.Module):
|
| 11 |
+
def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
|
| 12 |
+
d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
|
| 13 |
+
n_head_msa=8, n_head_pair=4, n_head_templ=4,
|
| 14 |
+
d_hidden=32, d_hidden_templ=64,
|
| 15 |
+
p_drop=0.15, d_t1d=24, d_t2d=44,
|
| 16 |
+
SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 17 |
+
SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 18 |
+
):
|
| 19 |
+
super(RoseTTAFoldModule, self).__init__()
|
| 20 |
+
#
|
| 21 |
+
# Input Embeddings
|
| 22 |
+
d_state = SE3_param_topk['l0_out_features']
|
| 23 |
+
self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
|
| 24 |
+
self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
|
| 25 |
+
self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
|
| 26 |
+
n_head=n_head_templ,
|
| 27 |
+
d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
|
| 28 |
+
# Update inputs with outputs from previous round
|
| 29 |
+
self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
|
| 30 |
+
#
|
| 31 |
+
self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
|
| 32 |
+
n_main_block=n_main_block,
|
| 33 |
+
n_ref_block=n_ref_block,
|
| 34 |
+
d_msa=d_msa, d_msa_full=d_msa_full,
|
| 35 |
+
d_pair=d_pair, d_hidden=d_hidden,
|
| 36 |
+
n_head_msa=n_head_msa,
|
| 37 |
+
n_head_pair=n_head_pair,
|
| 38 |
+
SE3_param_full=SE3_param_full,
|
| 39 |
+
SE3_param_topk=SE3_param_topk,
|
| 40 |
+
p_drop=p_drop)
|
| 41 |
+
##
|
| 42 |
+
self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
|
| 43 |
+
self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
|
| 44 |
+
self.lddt_pred = LDDTNetwork(d_state)
|
| 45 |
+
|
| 46 |
+
self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
|
| 47 |
+
|
| 48 |
+
def forward(self, msa_latent, msa_full, seq, xyz, idx,
|
| 49 |
+
seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
|
| 50 |
+
msa_prev=None, pair_prev=None, state_prev=None,
|
| 51 |
+
return_raw=False, return_full=False,
|
| 52 |
+
use_checkpoint=False, return_infer=False):
|
| 53 |
+
B, N, L = msa_latent.shape[:3]
|
| 54 |
+
# Get embeddings
|
| 55 |
+
#ic(seq.shape)
|
| 56 |
+
#ic(msa_latent.shape)
|
| 57 |
+
#ic(seq1hot.shape)
|
| 58 |
+
#ic(idx.shape)
|
| 59 |
+
#ic(xyz.shape)
|
| 60 |
+
#ic(seq1hot.shape)
|
| 61 |
+
#ic(t1d.shape)
|
| 62 |
+
#ic(t2d.shape)
|
| 63 |
+
|
| 64 |
+
idx = idx.long()
|
| 65 |
+
msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
|
| 66 |
+
|
| 67 |
+
msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
|
| 68 |
+
#
|
| 69 |
+
# Do recycling
|
| 70 |
+
if msa_prev == None:
|
| 71 |
+
msa_prev = torch.zeros_like(msa_latent[:,0])
|
| 72 |
+
if pair_prev == None:
|
| 73 |
+
pair_prev = torch.zeros_like(pair)
|
| 74 |
+
if state_prev == None:
|
| 75 |
+
state_prev = torch.zeros_like(state)
|
| 76 |
+
|
| 77 |
+
#ic(seq.shape)
|
| 78 |
+
#ic(msa_prev.shape)
|
| 79 |
+
#ic(pair_prev.shape)
|
| 80 |
+
#ic(xyz.shape)
|
| 81 |
+
#ic(state_prev.shape)
|
| 82 |
+
|
| 83 |
+
|
| 84 |
+
msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
|
| 85 |
+
msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
|
| 86 |
+
pair = pair + pair_recycle
|
| 87 |
+
state = state + state_recycle
|
| 88 |
+
#
|
| 89 |
+
#ic(t1d.dtype)
|
| 90 |
+
#ic(t2d.dtype)
|
| 91 |
+
#ic(alpha_t.dtype)
|
| 92 |
+
#ic(xyz_t.dtype)
|
| 93 |
+
#ic(pair.dtype)
|
| 94 |
+
#ic(state.dtype)
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
#import pdb; pdb.set_trace()
|
| 98 |
+
|
| 99 |
+
# add template embedding
|
| 100 |
+
pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
|
| 101 |
+
|
| 102 |
+
#ic(seq.dtype)
|
| 103 |
+
#ic(msa_latent.dtype)
|
| 104 |
+
#ic(msa_full.dtype)
|
| 105 |
+
#ic(pair.dtype)
|
| 106 |
+
#ic(xyz.dtype)
|
| 107 |
+
#ic(state.dtype)
|
| 108 |
+
#ic(idx.dtype)
|
| 109 |
+
|
| 110 |
+
# Predict coordinates from given inputs
|
| 111 |
+
msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
|
| 112 |
+
state, idx, use_checkpoint=use_checkpoint)
|
| 113 |
+
|
| 114 |
+
if return_raw:
|
| 115 |
+
# get last structure
|
| 116 |
+
xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
|
| 117 |
+
return msa[:,0], pair, xyz, state, alpha_s[-1]
|
| 118 |
+
|
| 119 |
+
# predict masked amino acids
|
| 120 |
+
logits_aa = self.aa_pred(msa)
|
| 121 |
+
#
|
| 122 |
+
# predict distogram & orientograms
|
| 123 |
+
logits = self.c6d_pred(pair)
|
| 124 |
+
|
| 125 |
+
# Predict LDDT
|
| 126 |
+
lddt = self.lddt_pred(state)
|
| 127 |
+
|
| 128 |
+
# predict experimentally resolved or not
|
| 129 |
+
logits_exp = self.exp_pred(msa[:,0], state)
|
| 130 |
+
|
| 131 |
+
if return_infer:
|
| 132 |
+
#get last structure
|
| 133 |
+
xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
|
| 134 |
+
return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
# get all intermediate bb structures
|
| 138 |
+
xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
|
| 139 |
+
|
| 140 |
+
return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
|
model/SE3_network.py
ADDED
|
@@ -0,0 +1,83 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
|
| 4 |
+
#from equivariant_attention.modules import get_basis_and_r, GSE3Res, GNormBias
|
| 5 |
+
#from equivariant_attention.modules import GConvSE3, GNormSE3
|
| 6 |
+
#from equivariant_attention.fibers import Fiber
|
| 7 |
+
|
| 8 |
+
from util_module import init_lecun_normal_param
|
| 9 |
+
from se3_transformer.model import SE3Transformer
|
| 10 |
+
from se3_transformer.model.fiber import Fiber
|
| 11 |
+
|
| 12 |
+
class SE3TransformerWrapper(nn.Module):
|
| 13 |
+
"""SE(3) equivariant GCN with attention"""
|
| 14 |
+
def __init__(self, num_layers=2, num_channels=32, num_degrees=3, n_heads=4, div=4,
|
| 15 |
+
l0_in_features=32, l0_out_features=32,
|
| 16 |
+
l1_in_features=3, l1_out_features=2,
|
| 17 |
+
num_edge_features=32):
|
| 18 |
+
super().__init__()
|
| 19 |
+
# Build the network
|
| 20 |
+
self.l1_in = l1_in_features
|
| 21 |
+
#
|
| 22 |
+
fiber_edge = Fiber({0: num_edge_features})
|
| 23 |
+
if l1_out_features > 0:
|
| 24 |
+
if l1_in_features > 0:
|
| 25 |
+
fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
|
| 26 |
+
fiber_hidden = Fiber.create(num_degrees, num_channels)
|
| 27 |
+
fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
|
| 28 |
+
else:
|
| 29 |
+
fiber_in = Fiber({0: l0_in_features})
|
| 30 |
+
fiber_hidden = Fiber.create(num_degrees, num_channels)
|
| 31 |
+
fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
|
| 32 |
+
else:
|
| 33 |
+
if l1_in_features > 0:
|
| 34 |
+
fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
|
| 35 |
+
fiber_hidden = Fiber.create(num_degrees, num_channels)
|
| 36 |
+
fiber_out = Fiber({0: l0_out_features})
|
| 37 |
+
else:
|
| 38 |
+
fiber_in = Fiber({0: l0_in_features})
|
| 39 |
+
fiber_hidden = Fiber.create(num_degrees, num_channels)
|
| 40 |
+
fiber_out = Fiber({0: l0_out_features})
|
| 41 |
+
|
| 42 |
+
self.se3 = SE3Transformer(num_layers=num_layers,
|
| 43 |
+
fiber_in=fiber_in,
|
| 44 |
+
fiber_hidden=fiber_hidden,
|
| 45 |
+
fiber_out = fiber_out,
|
| 46 |
+
num_heads=n_heads,
|
| 47 |
+
channels_div=div,
|
| 48 |
+
fiber_edge=fiber_edge,
|
| 49 |
+
use_layer_norm=True)
|
| 50 |
+
#use_layer_norm=False)
|
| 51 |
+
|
| 52 |
+
self.reset_parameter()
|
| 53 |
+
|
| 54 |
+
def reset_parameter(self):
|
| 55 |
+
|
| 56 |
+
# make sure linear layer before ReLu are initialized with kaiming_normal_
|
| 57 |
+
for n, p in self.se3.named_parameters():
|
| 58 |
+
if "bias" in n:
|
| 59 |
+
nn.init.zeros_(p)
|
| 60 |
+
elif len(p.shape) == 1:
|
| 61 |
+
continue
|
| 62 |
+
else:
|
| 63 |
+
if "radial_func" not in n:
|
| 64 |
+
p = init_lecun_normal_param(p)
|
| 65 |
+
else:
|
| 66 |
+
if "net.6" in n:
|
| 67 |
+
nn.init.zeros_(p)
|
| 68 |
+
else:
|
| 69 |
+
nn.init.kaiming_normal_(p, nonlinearity='relu')
|
| 70 |
+
|
| 71 |
+
# make last layers to be zero-initialized
|
| 72 |
+
#self.se3.graph_modules[-1].to_kernel_self['0'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['0'])
|
| 73 |
+
#self.se3.graph_modules[-1].to_kernel_self['1'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['1'])
|
| 74 |
+
nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['0'])
|
| 75 |
+
nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['1'])
|
| 76 |
+
|
| 77 |
+
def forward(self, G, type_0_features, type_1_features=None, edge_features=None):
|
| 78 |
+
if self.l1_in > 0:
|
| 79 |
+
node_features = {'0': type_0_features, '1': type_1_features}
|
| 80 |
+
else:
|
| 81 |
+
node_features = {'0': type_0_features}
|
| 82 |
+
edge_features = {'0': edge_features}
|
| 83 |
+
return self.se3(G, node_features, edge_features)
|
model/Track_module.py
ADDED
|
@@ -0,0 +1,476 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
import torch.nn.functional as F
|
| 4 |
+
from opt_einsum import contract as einsum
|
| 5 |
+
import torch.utils.checkpoint as checkpoint
|
| 6 |
+
from util import cross_product_matrix
|
| 7 |
+
from util_module import *
|
| 8 |
+
from Attention_module import *
|
| 9 |
+
from SE3_network import SE3TransformerWrapper
|
| 10 |
+
from icecream import ic
|
| 11 |
+
|
| 12 |
+
# Components for three-track blocks
|
| 13 |
+
# 1. MSA -> MSA update (biased attention. bias from pair & structure)
|
| 14 |
+
# 2. Pair -> Pair update (biased attention. bias from structure)
|
| 15 |
+
# 3. MSA -> Pair update (extract coevolution signal)
|
| 16 |
+
# 4. Str -> Str update (node from MSA, edge from Pair)
|
| 17 |
+
|
| 18 |
+
# Update MSA with biased self-attention. bias from Pair & Str
|
| 19 |
+
class MSAPairStr2MSA(nn.Module):
|
| 20 |
+
def __init__(self, d_msa=256, d_pair=128, n_head=8, d_state=16,
|
| 21 |
+
d_hidden=32, p_drop=0.15, use_global_attn=False):
|
| 22 |
+
super(MSAPairStr2MSA, self).__init__()
|
| 23 |
+
self.norm_pair = nn.LayerNorm(d_pair)
|
| 24 |
+
self.proj_pair = nn.Linear(d_pair+36, d_pair)
|
| 25 |
+
self.norm_state = nn.LayerNorm(d_state)
|
| 26 |
+
self.proj_state = nn.Linear(d_state, d_msa)
|
| 27 |
+
self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
|
| 28 |
+
self.row_attn = MSARowAttentionWithBias(d_msa=d_msa, d_pair=d_pair,
|
| 29 |
+
n_head=n_head, d_hidden=d_hidden)
|
| 30 |
+
if use_global_attn:
|
| 31 |
+
self.col_attn = MSAColGlobalAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
|
| 32 |
+
else:
|
| 33 |
+
self.col_attn = MSAColAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
|
| 34 |
+
self.ff = FeedForwardLayer(d_msa, 4, p_drop=p_drop)
|
| 35 |
+
|
| 36 |
+
# Do proper initialization
|
| 37 |
+
self.reset_parameter()
|
| 38 |
+
|
| 39 |
+
def reset_parameter(self):
|
| 40 |
+
# initialize weights to normal distrib
|
| 41 |
+
self.proj_pair = init_lecun_normal(self.proj_pair)
|
| 42 |
+
self.proj_state = init_lecun_normal(self.proj_state)
|
| 43 |
+
|
| 44 |
+
# initialize bias to zeros
|
| 45 |
+
nn.init.zeros_(self.proj_pair.bias)
|
| 46 |
+
nn.init.zeros_(self.proj_state.bias)
|
| 47 |
+
|
| 48 |
+
def forward(self, msa, pair, rbf_feat, state):
|
| 49 |
+
'''
|
| 50 |
+
Inputs:
|
| 51 |
+
- msa: MSA feature (B, N, L, d_msa)
|
| 52 |
+
- pair: Pair feature (B, L, L, d_pair)
|
| 53 |
+
- rbf_feat: Ca-Ca distance feature calculated from xyz coordinates (B, L, L, 36)
|
| 54 |
+
- xyz: xyz coordinates (B, L, n_atom, 3)
|
| 55 |
+
- state: updated node features after SE(3)-Transformer layer (B, L, d_state)
|
| 56 |
+
Output:
|
| 57 |
+
- msa: Updated MSA feature (B, N, L, d_msa)
|
| 58 |
+
'''
|
| 59 |
+
B, N, L = msa.shape[:3]
|
| 60 |
+
|
| 61 |
+
# prepare input bias feature by combining pair & coordinate info
|
| 62 |
+
pair = self.norm_pair(pair)
|
| 63 |
+
pair = torch.cat((pair, rbf_feat), dim=-1)
|
| 64 |
+
pair = self.proj_pair(pair) # (B, L, L, d_pair)
|
| 65 |
+
#
|
| 66 |
+
# update query sequence feature (first sequence in the MSA) with feedbacks (state) from SE3
|
| 67 |
+
state = self.norm_state(state)
|
| 68 |
+
state = self.proj_state(state).reshape(B, 1, L, -1)
|
| 69 |
+
|
| 70 |
+
msa = msa.index_add(1, torch.tensor([0,], device=state.device), state.type(torch.float32))
|
| 71 |
+
#
|
| 72 |
+
# Apply row/column attention to msa & transform
|
| 73 |
+
msa = msa + self.drop_row(self.row_attn(msa, pair))
|
| 74 |
+
msa = msa + self.col_attn(msa)
|
| 75 |
+
msa = msa + self.ff(msa)
|
| 76 |
+
|
| 77 |
+
return msa
|
| 78 |
+
|
| 79 |
+
class PairStr2Pair(nn.Module):
|
| 80 |
+
def __init__(self, d_pair=128, n_head=4, d_hidden=32, d_rbf=36, p_drop=0.15):
|
| 81 |
+
super(PairStr2Pair, self).__init__()
|
| 82 |
+
|
| 83 |
+
self.emb_rbf = nn.Linear(d_rbf, d_hidden)
|
| 84 |
+
self.proj_rbf = nn.Linear(d_hidden, d_pair)
|
| 85 |
+
|
| 86 |
+
self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
|
| 87 |
+
self.drop_col = Dropout(broadcast_dim=2, p_drop=p_drop)
|
| 88 |
+
|
| 89 |
+
self.row_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=True)
|
| 90 |
+
self.col_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=False)
|
| 91 |
+
|
| 92 |
+
self.ff = FeedForwardLayer(d_pair, 2)
|
| 93 |
+
|
| 94 |
+
self.reset_parameter()
|
| 95 |
+
|
| 96 |
+
def reset_parameter(self):
|
| 97 |
+
nn.init.kaiming_normal_(self.emb_rbf.weight, nonlinearity='relu')
|
| 98 |
+
nn.init.zeros_(self.emb_rbf.bias)
|
| 99 |
+
|
| 100 |
+
self.proj_rbf = init_lecun_normal(self.proj_rbf)
|
| 101 |
+
nn.init.zeros_(self.proj_rbf.bias)
|
| 102 |
+
|
| 103 |
+
def forward(self, pair, rbf_feat):
|
| 104 |
+
B, L = pair.shape[:2]
|
| 105 |
+
|
| 106 |
+
rbf_feat = self.proj_rbf(F.relu_(self.emb_rbf(rbf_feat)))
|
| 107 |
+
|
| 108 |
+
pair = pair + self.drop_row(self.row_attn(pair, rbf_feat))
|
| 109 |
+
pair = pair + self.drop_col(self.col_attn(pair, rbf_feat))
|
| 110 |
+
pair = pair + self.ff(pair)
|
| 111 |
+
return pair
|
| 112 |
+
|
| 113 |
+
class MSA2Pair(nn.Module):
|
| 114 |
+
def __init__(self, d_msa=256, d_pair=128, d_hidden=32, p_drop=0.15):
|
| 115 |
+
super(MSA2Pair, self).__init__()
|
| 116 |
+
self.norm = nn.LayerNorm(d_msa)
|
| 117 |
+
self.proj_left = nn.Linear(d_msa, d_hidden)
|
| 118 |
+
self.proj_right = nn.Linear(d_msa, d_hidden)
|
| 119 |
+
self.proj_out = nn.Linear(d_hidden*d_hidden, d_pair)
|
| 120 |
+
|
| 121 |
+
self.reset_parameter()
|
| 122 |
+
|
| 123 |
+
def reset_parameter(self):
|
| 124 |
+
# normal initialization
|
| 125 |
+
self.proj_left = init_lecun_normal(self.proj_left)
|
| 126 |
+
self.proj_right = init_lecun_normal(self.proj_right)
|
| 127 |
+
nn.init.zeros_(self.proj_left.bias)
|
| 128 |
+
nn.init.zeros_(self.proj_right.bias)
|
| 129 |
+
|
| 130 |
+
# zero initialize output
|
| 131 |
+
nn.init.zeros_(self.proj_out.weight)
|
| 132 |
+
nn.init.zeros_(self.proj_out.bias)
|
| 133 |
+
|
| 134 |
+
def forward(self, msa, pair):
|
| 135 |
+
B, N, L = msa.shape[:3]
|
| 136 |
+
msa = self.norm(msa)
|
| 137 |
+
left = self.proj_left(msa)
|
| 138 |
+
right = self.proj_right(msa)
|
| 139 |
+
right = right / float(N)
|
| 140 |
+
out = einsum('bsli,bsmj->blmij', left, right).reshape(B, L, L, -1)
|
| 141 |
+
out = self.proj_out(out)
|
| 142 |
+
|
| 143 |
+
pair = pair + out
|
| 144 |
+
|
| 145 |
+
return pair
|
| 146 |
+
|
| 147 |
+
class SCPred(nn.Module):
|
| 148 |
+
def __init__(self, d_msa=256, d_state=32, d_hidden=128, p_drop=0.15):
|
| 149 |
+
super(SCPred, self).__init__()
|
| 150 |
+
self.norm_s0 = nn.LayerNorm(d_msa)
|
| 151 |
+
self.norm_si = nn.LayerNorm(d_state)
|
| 152 |
+
self.linear_s0 = nn.Linear(d_msa, d_hidden)
|
| 153 |
+
self.linear_si = nn.Linear(d_state, d_hidden)
|
| 154 |
+
|
| 155 |
+
# ResNet layers
|
| 156 |
+
self.linear_1 = nn.Linear(d_hidden, d_hidden)
|
| 157 |
+
self.linear_2 = nn.Linear(d_hidden, d_hidden)
|
| 158 |
+
self.linear_3 = nn.Linear(d_hidden, d_hidden)
|
| 159 |
+
self.linear_4 = nn.Linear(d_hidden, d_hidden)
|
| 160 |
+
|
| 161 |
+
# Final outputs
|
| 162 |
+
self.linear_out = nn.Linear(d_hidden, 20)
|
| 163 |
+
|
| 164 |
+
self.reset_parameter()
|
| 165 |
+
|
| 166 |
+
def reset_parameter(self):
|
| 167 |
+
# normal initialization
|
| 168 |
+
self.linear_s0 = init_lecun_normal(self.linear_s0)
|
| 169 |
+
self.linear_si = init_lecun_normal(self.linear_si)
|
| 170 |
+
self.linear_out = init_lecun_normal(self.linear_out)
|
| 171 |
+
nn.init.zeros_(self.linear_s0.bias)
|
| 172 |
+
nn.init.zeros_(self.linear_si.bias)
|
| 173 |
+
nn.init.zeros_(self.linear_out.bias)
|
| 174 |
+
|
| 175 |
+
# right before relu activation: He initializer (kaiming normal)
|
| 176 |
+
nn.init.kaiming_normal_(self.linear_1.weight, nonlinearity='relu')
|
| 177 |
+
nn.init.zeros_(self.linear_1.bias)
|
| 178 |
+
nn.init.kaiming_normal_(self.linear_3.weight, nonlinearity='relu')
|
| 179 |
+
nn.init.zeros_(self.linear_3.bias)
|
| 180 |
+
|
| 181 |
+
# right before residual connection: zero initialize
|
| 182 |
+
nn.init.zeros_(self.linear_2.weight)
|
| 183 |
+
nn.init.zeros_(self.linear_2.bias)
|
| 184 |
+
nn.init.zeros_(self.linear_4.weight)
|
| 185 |
+
nn.init.zeros_(self.linear_4.bias)
|
| 186 |
+
|
| 187 |
+
def forward(self, seq, state):
|
| 188 |
+
'''
|
| 189 |
+
Predict side-chain torsion angles along with backbone torsions
|
| 190 |
+
Inputs:
|
| 191 |
+
- seq: hidden embeddings corresponding to query sequence (B, L, d_msa)
|
| 192 |
+
- state: state feature (output l0 feature) from previous SE3 layer (B, L, d_state)
|
| 193 |
+
Outputs:
|
| 194 |
+
- si: predicted torsion angles (phi, psi, omega, chi1~4 with cos/sin, Cb bend, Cb twist, CG) (B, L, 10, 2)
|
| 195 |
+
'''
|
| 196 |
+
B, L = seq.shape[:2]
|
| 197 |
+
seq = self.norm_s0(seq)
|
| 198 |
+
state = self.norm_si(state)
|
| 199 |
+
si = self.linear_s0(seq) + self.linear_si(state)
|
| 200 |
+
|
| 201 |
+
si = si + self.linear_2(F.relu_(self.linear_1(F.relu_(si))))
|
| 202 |
+
si = si + self.linear_4(F.relu_(self.linear_3(F.relu_(si))))
|
| 203 |
+
|
| 204 |
+
si = self.linear_out(F.relu_(si))
|
| 205 |
+
return si.view(B, L, 10, 2)
|
| 206 |
+
|
| 207 |
+
|
| 208 |
+
class Str2Str(nn.Module):
|
| 209 |
+
def __init__(self, d_msa=256, d_pair=128, d_state=16,
|
| 210 |
+
SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}, p_drop=0.1):
|
| 211 |
+
super(Str2Str, self).__init__()
|
| 212 |
+
|
| 213 |
+
# initial node & pair feature process
|
| 214 |
+
self.norm_msa = nn.LayerNorm(d_msa)
|
| 215 |
+
self.norm_pair = nn.LayerNorm(d_pair)
|
| 216 |
+
self.norm_state = nn.LayerNorm(d_state)
|
| 217 |
+
|
| 218 |
+
self.embed_x = nn.Linear(d_msa+d_state, SE3_param['l0_in_features'])
|
| 219 |
+
self.embed_e1 = nn.Linear(d_pair, SE3_param['num_edge_features'])
|
| 220 |
+
self.embed_e2 = nn.Linear(SE3_param['num_edge_features']+36+1, SE3_param['num_edge_features'])
|
| 221 |
+
|
| 222 |
+
self.norm_node = nn.LayerNorm(SE3_param['l0_in_features'])
|
| 223 |
+
self.norm_edge1 = nn.LayerNorm(SE3_param['num_edge_features'])
|
| 224 |
+
self.norm_edge2 = nn.LayerNorm(SE3_param['num_edge_features'])
|
| 225 |
+
|
| 226 |
+
self.se3 = SE3TransformerWrapper(**SE3_param)
|
| 227 |
+
self.sc_predictor = SCPred(d_msa=d_msa, d_state=SE3_param['l0_out_features'],
|
| 228 |
+
p_drop=p_drop)
|
| 229 |
+
|
| 230 |
+
self.reset_parameter()
|
| 231 |
+
|
| 232 |
+
def reset_parameter(self):
|
| 233 |
+
# initialize weights to normal distribution
|
| 234 |
+
self.embed_x = init_lecun_normal(self.embed_x)
|
| 235 |
+
self.embed_e1 = init_lecun_normal(self.embed_e1)
|
| 236 |
+
self.embed_e2 = init_lecun_normal(self.embed_e2)
|
| 237 |
+
|
| 238 |
+
# initialize bias to zeros
|
| 239 |
+
nn.init.zeros_(self.embed_x.bias)
|
| 240 |
+
nn.init.zeros_(self.embed_e1.bias)
|
| 241 |
+
nn.init.zeros_(self.embed_e2.bias)
|
| 242 |
+
|
| 243 |
+
@torch.cuda.amp.autocast(enabled=False)
|
| 244 |
+
def forward(self, msa, pair, R_in, T_in, xyz, state, idx, top_k=64, eps=1e-5):
|
| 245 |
+
B, N, L = msa.shape[:3]
|
| 246 |
+
|
| 247 |
+
state = state.type(torch.float32)
|
| 248 |
+
mas = msa.type(torch.float32)
|
| 249 |
+
pair = pair.type(torch.float32)
|
| 250 |
+
R_in = R_in.type(torch.float32)
|
| 251 |
+
T_in = T_in.type(torch.float32)
|
| 252 |
+
xyz = xyz.type(torch.float32)
|
| 253 |
+
|
| 254 |
+
#ic(msa.dtype)
|
| 255 |
+
#ic(pair.dtype)
|
| 256 |
+
#ic(R_in.dtype)
|
| 257 |
+
#ic(T_in.dtype)
|
| 258 |
+
#ic(xyz.dtype)
|
| 259 |
+
#ic(state.dtype)
|
| 260 |
+
#ic(idx.dtype)
|
| 261 |
+
|
| 262 |
+
|
| 263 |
+
# process msa & pair features
|
| 264 |
+
node = self.norm_msa(msa[:,0])
|
| 265 |
+
pair = self.norm_pair(pair)
|
| 266 |
+
state = self.norm_state(state)
|
| 267 |
+
|
| 268 |
+
node = torch.cat((node, state), dim=-1)
|
| 269 |
+
node = self.norm_node(self.embed_x(node))
|
| 270 |
+
pair = self.norm_edge1(self.embed_e1(pair))
|
| 271 |
+
|
| 272 |
+
neighbor = get_seqsep(idx)
|
| 273 |
+
rbf_feat = rbf(torch.cdist(xyz[:,:,1], xyz[:,:,1]))
|
| 274 |
+
pair = torch.cat((pair, rbf_feat, neighbor), dim=-1)
|
| 275 |
+
pair = self.norm_edge2(self.embed_e2(pair))
|
| 276 |
+
|
| 277 |
+
# define graph
|
| 278 |
+
if top_k != 0:
|
| 279 |
+
G, edge_feats = make_topk_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
|
| 280 |
+
else:
|
| 281 |
+
G, edge_feats = make_full_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
|
| 282 |
+
l1_feats = xyz - xyz[:,:,1,:].unsqueeze(2)
|
| 283 |
+
l1_feats = l1_feats.reshape(B*L, -1, 3)
|
| 284 |
+
|
| 285 |
+
# apply SE(3) Transformer & update coordinates
|
| 286 |
+
shift = self.se3(G, node.reshape(B*L, -1, 1), l1_feats, edge_feats)
|
| 287 |
+
|
| 288 |
+
state = shift['0'].reshape(B, L, -1) # (B, L, C)
|
| 289 |
+
|
| 290 |
+
offset = shift['1'].reshape(B, L, 2, 3)
|
| 291 |
+
delTi = offset[:,:,0,:] / 10.0 # translation
|
| 292 |
+
R = offset[:,:,1,:] / 100.0 # rotation
|
| 293 |
+
|
| 294 |
+
Qnorm = torch.sqrt( 1 + torch.sum(R*R, dim=-1) )
|
| 295 |
+
qA, qB, qC, qD = 1/Qnorm, R[:,:,0]/Qnorm, R[:,:,1]/Qnorm, R[:,:,2]/Qnorm
|
| 296 |
+
|
| 297 |
+
delRi = torch.zeros((B,L,3,3), device=xyz.device)
|
| 298 |
+
delRi[:,:,0,0] = qA*qA+qB*qB-qC*qC-qD*qD
|
| 299 |
+
delRi[:,:,0,1] = 2*qB*qC - 2*qA*qD
|
| 300 |
+
delRi[:,:,0,2] = 2*qB*qD + 2*qA*qC
|
| 301 |
+
delRi[:,:,1,0] = 2*qB*qC + 2*qA*qD
|
| 302 |
+
delRi[:,:,1,1] = qA*qA-qB*qB+qC*qC-qD*qD
|
| 303 |
+
delRi[:,:,1,2] = 2*qC*qD - 2*qA*qB
|
| 304 |
+
delRi[:,:,2,0] = 2*qB*qD - 2*qA*qC
|
| 305 |
+
delRi[:,:,2,1] = 2*qC*qD + 2*qA*qB
|
| 306 |
+
delRi[:,:,2,2] = qA*qA-qB*qB-qC*qC+qD*qD
|
| 307 |
+
#
|
| 308 |
+
## convert vector to rotation matrix
|
| 309 |
+
#R_angle = torch.norm(R, dim=-1, keepdim=True) # (B, L, 1)
|
| 310 |
+
#cos_angle = torch.cos(R_angle).unsqueeze(2) # (B, L, 1, 1)
|
| 311 |
+
#sin_angle = torch.sin(R_angle).unsqueeze(2) # (B, L, 1, 1)
|
| 312 |
+
#R_vector = R / (R_angle+eps) # (B, L, 3)
|
| 313 |
+
|
| 314 |
+
#delRi = cos_angle*torch.eye(3, device=R.device).reshape(1,1,3,3) \
|
| 315 |
+
# + sin_angle*cross_product_matrix(R_vector) \
|
| 316 |
+
# + (1.0-cos_angle)*einsum('bni,bnj->bnij', R_vector, R_vector)
|
| 317 |
+
|
| 318 |
+
Ri = einsum('bnij,bnjk->bnik', delRi, R_in)
|
| 319 |
+
Ti = delTi + T_in #einsum('bnij,bnj->bni', delRi, T_in) + delTi
|
| 320 |
+
|
| 321 |
+
alpha = self.sc_predictor(msa[:,0], state)
|
| 322 |
+
return Ri, Ti, state, alpha
|
| 323 |
+
|
| 324 |
+
class IterBlock(nn.Module):
|
| 325 |
+
def __init__(self, d_msa=256, d_pair=128,
|
| 326 |
+
n_head_msa=8, n_head_pair=4,
|
| 327 |
+
use_global_attn=False,
|
| 328 |
+
d_hidden=32, d_hidden_msa=None, p_drop=0.15,
|
| 329 |
+
SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}):
|
| 330 |
+
super(IterBlock, self).__init__()
|
| 331 |
+
if d_hidden_msa == None:
|
| 332 |
+
d_hidden_msa = d_hidden
|
| 333 |
+
|
| 334 |
+
self.msa2msa = MSAPairStr2MSA(d_msa=d_msa, d_pair=d_pair,
|
| 335 |
+
n_head=n_head_msa,
|
| 336 |
+
d_state=SE3_param['l0_out_features'],
|
| 337 |
+
use_global_attn=use_global_attn,
|
| 338 |
+
d_hidden=d_hidden_msa, p_drop=p_drop)
|
| 339 |
+
self.msa2pair = MSA2Pair(d_msa=d_msa, d_pair=d_pair,
|
| 340 |
+
d_hidden=d_hidden//2, p_drop=p_drop)
|
| 341 |
+
#d_hidden=d_hidden, p_drop=p_drop)
|
| 342 |
+
self.pair2pair = PairStr2Pair(d_pair=d_pair, n_head=n_head_pair,
|
| 343 |
+
d_hidden=d_hidden, p_drop=p_drop)
|
| 344 |
+
self.str2str = Str2Str(d_msa=d_msa, d_pair=d_pair,
|
| 345 |
+
d_state=SE3_param['l0_out_features'],
|
| 346 |
+
SE3_param=SE3_param,
|
| 347 |
+
p_drop=p_drop)
|
| 348 |
+
|
| 349 |
+
def forward(self, msa, pair, R_in, T_in, xyz, state, idx, use_checkpoint=False):
|
| 350 |
+
rbf_feat = rbf(torch.cdist(xyz[:,:,1,:], xyz[:,:,1,:]))
|
| 351 |
+
if use_checkpoint:
|
| 352 |
+
msa = checkpoint.checkpoint(create_custom_forward(self.msa2msa), msa, pair, rbf_feat, state)
|
| 353 |
+
pair = checkpoint.checkpoint(create_custom_forward(self.msa2pair), msa, pair)
|
| 354 |
+
pair = checkpoint.checkpoint(create_custom_forward(self.pair2pair), pair, rbf_feat)
|
| 355 |
+
R, T, state, alpha = checkpoint.checkpoint(create_custom_forward(self.str2str, top_k=0), msa, pair, R_in, T_in, xyz, state, idx)
|
| 356 |
+
else:
|
| 357 |
+
msa = self.msa2msa(msa, pair, rbf_feat, state)
|
| 358 |
+
pair = self.msa2pair(msa, pair)
|
| 359 |
+
pair = self.pair2pair(pair, rbf_feat)
|
| 360 |
+
R, T, state, alpha = self.str2str(msa, pair, R_in, T_in, xyz, state, idx, top_k=0)
|
| 361 |
+
|
| 362 |
+
return msa, pair, R, T, state, alpha
|
| 363 |
+
|
| 364 |
+
class IterativeSimulator(nn.Module):
|
| 365 |
+
def __init__(self, n_extra_block=4, n_main_block=12, n_ref_block=4,
|
| 366 |
+
d_msa=256, d_msa_full=64, d_pair=128, d_hidden=32,
|
| 367 |
+
n_head_msa=8, n_head_pair=4,
|
| 368 |
+
SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 369 |
+
SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
|
| 370 |
+
p_drop=0.15):
|
| 371 |
+
super(IterativeSimulator, self).__init__()
|
| 372 |
+
self.n_extra_block = n_extra_block
|
| 373 |
+
self.n_main_block = n_main_block
|
| 374 |
+
self.n_ref_block = n_ref_block
|
| 375 |
+
|
| 376 |
+
self.proj_state = nn.Linear(SE3_param_topk['l0_out_features'], SE3_param_full['l0_out_features'])
|
| 377 |
+
# Update with extra sequences
|
| 378 |
+
if n_extra_block > 0:
|
| 379 |
+
self.extra_block = nn.ModuleList([IterBlock(d_msa=d_msa_full, d_pair=d_pair,
|
| 380 |
+
n_head_msa=n_head_msa,
|
| 381 |
+
n_head_pair=n_head_pair,
|
| 382 |
+
d_hidden_msa=8,
|
| 383 |
+
d_hidden=d_hidden,
|
| 384 |
+
p_drop=p_drop,
|
| 385 |
+
use_global_attn=True,
|
| 386 |
+
SE3_param=SE3_param_full)
|
| 387 |
+
for i in range(n_extra_block)])
|
| 388 |
+
|
| 389 |
+
# Update with seed sequences
|
| 390 |
+
if n_main_block > 0:
|
| 391 |
+
self.main_block = nn.ModuleList([IterBlock(d_msa=d_msa, d_pair=d_pair,
|
| 392 |
+
n_head_msa=n_head_msa,
|
| 393 |
+
n_head_pair=n_head_pair,
|
| 394 |
+
d_hidden=d_hidden,
|
| 395 |
+
p_drop=p_drop,
|
| 396 |
+
use_global_attn=False,
|
| 397 |
+
SE3_param=SE3_param_full)
|
| 398 |
+
for i in range(n_main_block)])
|
| 399 |
+
|
| 400 |
+
self.proj_state2 = nn.Linear(SE3_param_full['l0_out_features'], SE3_param_topk['l0_out_features'])
|
| 401 |
+
# Final SE(3) refinement
|
| 402 |
+
if n_ref_block > 0:
|
| 403 |
+
self.str_refiner = Str2Str(d_msa=d_msa, d_pair=d_pair,
|
| 404 |
+
d_state=SE3_param_topk['l0_out_features'],
|
| 405 |
+
SE3_param=SE3_param_topk,
|
| 406 |
+
p_drop=p_drop)
|
| 407 |
+
|
| 408 |
+
self.reset_parameter()
|
| 409 |
+
def reset_parameter(self):
|
| 410 |
+
self.proj_state = init_lecun_normal(self.proj_state)
|
| 411 |
+
nn.init.zeros_(self.proj_state.bias)
|
| 412 |
+
self.proj_state2 = init_lecun_normal(self.proj_state2)
|
| 413 |
+
nn.init.zeros_(self.proj_state2.bias)
|
| 414 |
+
|
| 415 |
+
def forward(self, seq, msa, msa_full, pair, xyz_in, state, idx, use_checkpoint=False):
|
| 416 |
+
# input:
|
| 417 |
+
# seq: query sequence (B, L)
|
| 418 |
+
# msa: seed MSA embeddings (B, N, L, d_msa)
|
| 419 |
+
# msa_full: extra MSA embeddings (B, N, L, d_msa_full)
|
| 420 |
+
# pair: initial residue pair embeddings (B, L, L, d_pair)
|
| 421 |
+
# xyz_in: initial BB coordinates (B, L, n_atom, 3)
|
| 422 |
+
# state: initial state features containing mixture of query seq, sidechain, accuracy info (B, L, d_state)
|
| 423 |
+
# idx: residue index
|
| 424 |
+
|
| 425 |
+
B, L = pair.shape[:2]
|
| 426 |
+
|
| 427 |
+
R_in = torch.eye(3, device=xyz_in.device).reshape(1,1,3,3).expand(B, L, -1, -1)
|
| 428 |
+
T_in = xyz_in[:,:,1].clone()
|
| 429 |
+
xyz_in = xyz_in - T_in.unsqueeze(-2)
|
| 430 |
+
|
| 431 |
+
state = self.proj_state(state)
|
| 432 |
+
|
| 433 |
+
R_s = list()
|
| 434 |
+
T_s = list()
|
| 435 |
+
alpha_s = list()
|
| 436 |
+
for i_m in range(self.n_extra_block):
|
| 437 |
+
R_in = R_in.detach() # detach rotation (for stability)
|
| 438 |
+
T_in = T_in.detach()
|
| 439 |
+
# Get current BB structure
|
| 440 |
+
xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
|
| 441 |
+
|
| 442 |
+
msa_full, pair, R_in, T_in, state, alpha = self.extra_block[i_m](msa_full, pair,
|
| 443 |
+
R_in, T_in, xyz, state, idx,
|
| 444 |
+
use_checkpoint=use_checkpoint)
|
| 445 |
+
R_s.append(R_in)
|
| 446 |
+
T_s.append(T_in)
|
| 447 |
+
alpha_s.append(alpha)
|
| 448 |
+
|
| 449 |
+
for i_m in range(self.n_main_block):
|
| 450 |
+
R_in = R_in.detach()
|
| 451 |
+
T_in = T_in.detach()
|
| 452 |
+
# Get current BB structure
|
| 453 |
+
xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
|
| 454 |
+
|
| 455 |
+
msa, pair, R_in, T_in, state, alpha = self.main_block[i_m](msa, pair,
|
| 456 |
+
R_in, T_in, xyz, state, idx,
|
| 457 |
+
use_checkpoint=use_checkpoint)
|
| 458 |
+
R_s.append(R_in)
|
| 459 |
+
T_s.append(T_in)
|
| 460 |
+
alpha_s.append(alpha)
|
| 461 |
+
|
| 462 |
+
state = self.proj_state2(state)
|
| 463 |
+
for i_m in range(self.n_ref_block):
|
| 464 |
+
R_in = R_in.detach()
|
| 465 |
+
T_in = T_in.detach()
|
| 466 |
+
xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
|
| 467 |
+
R_in, T_in, state, alpha = self.str_refiner(msa, pair, R_in, T_in, xyz, state, idx, top_k=64)
|
| 468 |
+
R_s.append(R_in)
|
| 469 |
+
T_s.append(T_in)
|
| 470 |
+
alpha_s.append(alpha)
|
| 471 |
+
|
| 472 |
+
R_s = torch.stack(R_s, dim=0)
|
| 473 |
+
T_s = torch.stack(T_s, dim=0)
|
| 474 |
+
alpha_s = torch.stack(alpha_s, dim=0)
|
| 475 |
+
|
| 476 |
+
return msa, pair, R_s, T_s, alpha_s, state
|
model/__pycache__/Attention_module.cpython-310.pyc
ADDED
|
Binary file (10.8 kB). View file
|
|
|
model/__pycache__/AuxiliaryPredictor.cpython-310.pyc
ADDED
|
Binary file (3.53 kB). View file
|
|
|
model/__pycache__/Embeddings.cpython-310.pyc
ADDED
|
Binary file (9.57 kB). View file
|
|
|
model/__pycache__/RoseTTAFoldModel.cpython-310.pyc
ADDED
|
Binary file (3.29 kB). View file
|
|
|
model/__pycache__/SE3_network.cpython-310.pyc
ADDED
|
Binary file (2.27 kB). View file
|
|
|
model/__pycache__/Track_module.cpython-310.pyc
ADDED
|
Binary file (13.7 kB). View file
|
|
|
model/__pycache__/ab_tools.cpython-310.pyc
ADDED
|
Binary file (17.5 kB). View file
|
|
|
model/__pycache__/apply_masks.cpython-310.pyc
ADDED
|
Binary file (4.18 kB). View file
|
|
|
model/__pycache__/arguments.cpython-310.pyc
ADDED
|
Binary file (12.1 kB). View file
|
|
|
model/__pycache__/chemical.cpython-310.pyc
ADDED
|
Binary file (19.8 kB). View file
|
|
|
model/__pycache__/data_loader.cpython-310.pyc
ADDED
|
Binary file (47.8 kB). View file
|
|
|
model/__pycache__/diffusion.cpython-310.pyc
ADDED
|
Binary file (7.34 kB). View file
|
|
|
model/__pycache__/kinematics.cpython-310.pyc
ADDED
|
Binary file (8.97 kB). View file
|
|
|
model/__pycache__/loss.cpython-310.pyc
ADDED
|
Binary file (17.4 kB). View file
|
|
|
model/__pycache__/mask_generator.cpython-310.pyc
ADDED
|
Binary file (3.01 kB). View file
|
|
|
model/__pycache__/parsers.cpython-310.pyc
ADDED
|
Binary file (4.74 kB). View file
|
|
|
model/__pycache__/scheduler.cpython-310.pyc
ADDED
|
Binary file (4.89 kB). View file
|
|
|
model/__pycache__/scoring.cpython-310.pyc
ADDED
|
Binary file (13.9 kB). View file
|
|
|
model/__pycache__/train_multi_deep.cpython-310.pyc
ADDED
|
Binary file (25.7 kB). View file
|
|
|
model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc
ADDED
|
Binary file (37.1 kB). View file
|
|
|
model/__pycache__/util.cpython-310.pyc
ADDED
|
Binary file (14.7 kB). View file
|
|
|
model/__pycache__/util_module.cpython-310.pyc
ADDED
|
Binary file (9.66 kB). View file
|
|
|
model/apply_masks.py
ADDED
|
@@ -0,0 +1,196 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import sys, os
|
| 2 |
+
import torch
|
| 3 |
+
from icecream import ic
|
| 4 |
+
import random
|
| 5 |
+
import numpy as np
|
| 6 |
+
from kinematics import get_init_xyz
|
| 7 |
+
sys.path.append('../')
|
| 8 |
+
from utils.calc_dssp import annotate_sse
|
| 9 |
+
|
| 10 |
+
ic.configureOutput(includeContext=True)
|
| 11 |
+
|
| 12 |
+
def mask_inputs(seq,
|
| 13 |
+
msa_masked,
|
| 14 |
+
msa_full,
|
| 15 |
+
xyz_t,
|
| 16 |
+
t1d,
|
| 17 |
+
mask_msa,
|
| 18 |
+
input_seq_mask=None,
|
| 19 |
+
input_str_mask=None,
|
| 20 |
+
input_floating_mask=None,
|
| 21 |
+
input_t1dconf_mask=None,
|
| 22 |
+
loss_seq_mask=None,
|
| 23 |
+
loss_str_mask=None,
|
| 24 |
+
loss_str_mask_2d=None,
|
| 25 |
+
dssp=False,
|
| 26 |
+
hotspots=False,
|
| 27 |
+
diffuser=None,
|
| 28 |
+
t=None,
|
| 29 |
+
freeze_seq_emb=False,
|
| 30 |
+
mutate_seq=False,
|
| 31 |
+
no_clamp_seq=False,
|
| 32 |
+
norm_input=False,
|
| 33 |
+
contacts=None,
|
| 34 |
+
frac_provide_dssp=0.5,
|
| 35 |
+
dssp_mask_percentage=[0,100],
|
| 36 |
+
frac_provide_contacts=0.5,
|
| 37 |
+
struc_cond=False):
|
| 38 |
+
"""
|
| 39 |
+
Parameters:
|
| 40 |
+
seq (torch.tensor, required): (I,L) integer sequence
|
| 41 |
+
|
| 42 |
+
msa_masked (torch.tensor, required): (I,N_short,L,48)
|
| 43 |
+
|
| 44 |
+
msa_full (torch,.tensor, required): (I,N_long,L,25)
|
| 45 |
+
|
| 46 |
+
xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
|
| 47 |
+
|
| 48 |
+
t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
|
| 49 |
+
|
| 50 |
+
str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
|
| 51 |
+
|
| 52 |
+
seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
|
| 53 |
+
t1d_24: is there an extra dimension to input structure confidence?
|
| 54 |
+
|
| 55 |
+
diffuser: diffuser class
|
| 56 |
+
|
| 57 |
+
t: time step
|
| 58 |
+
|
| 59 |
+
NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
|
| 60 |
+
For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
|
| 61 |
+
This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
|
| 62 |
+
"""
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
|
| 66 |
+
#ic(input_seq_mask.shape)
|
| 67 |
+
#ic(seq.shape)
|
| 68 |
+
#ic(msa_masked.shape)
|
| 69 |
+
#ic(msa_full.shape)
|
| 70 |
+
#ic(t1d.shape)
|
| 71 |
+
#ic(xyz_t.shape)
|
| 72 |
+
#ic(input_str_mask.shape)
|
| 73 |
+
#ic(mask_msa.shape)
|
| 74 |
+
|
| 75 |
+
###########
|
| 76 |
+
seq_mask = input_seq_mask
|
| 77 |
+
|
| 78 |
+
|
| 79 |
+
######################
|
| 80 |
+
###sequence diffusion###
|
| 81 |
+
######################
|
| 82 |
+
|
| 83 |
+
str_mask = input_str_mask
|
| 84 |
+
|
| 85 |
+
x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
|
| 86 |
+
seq_diffused = diffuser.q_sample(x_0,t,mask=seq_mask)
|
| 87 |
+
|
| 88 |
+
seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
|
| 89 |
+
seq=seq_tmp.repeat(seq.shape[0], 1)
|
| 90 |
+
|
| 91 |
+
###################
|
| 92 |
+
###msa diffusion###
|
| 93 |
+
###################
|
| 94 |
+
|
| 95 |
+
### msa_masked ###
|
| 96 |
+
#ic(msa_masked.shape)
|
| 97 |
+
B,N,L,_=msa_masked.shape
|
| 98 |
+
msa_masked[:,0,:,:22] = seq_diffused
|
| 99 |
+
|
| 100 |
+
x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
|
| 101 |
+
msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
|
| 102 |
+
msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t]),mask=msa_seq_mask)
|
| 103 |
+
|
| 104 |
+
msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
|
| 105 |
+
|
| 106 |
+
# index 44/45 is insertion/deletion
|
| 107 |
+
# index 43 is the masked token NOTE check this
|
| 108 |
+
# index 42 is the unknown token
|
| 109 |
+
msa_masked[:,0,:,22:44] = seq_diffused
|
| 110 |
+
msa_masked[:,1:,:,22:44] = msa_diffused
|
| 111 |
+
|
| 112 |
+
# insertion/deletion stuff
|
| 113 |
+
msa_masked[:,0,~seq_mask,44:46] = 0
|
| 114 |
+
|
| 115 |
+
### msa_full ###
|
| 116 |
+
################
|
| 117 |
+
#make msa_full same size as msa_masked
|
| 118 |
+
#ic(msa_full.shape)
|
| 119 |
+
msa_full = msa_full[:,:msa_masked.shape[1],:,:]
|
| 120 |
+
msa_full[:,0,:,:22] = seq_diffused
|
| 121 |
+
msa_full[:,1:,:,:22] = msa_diffused
|
| 122 |
+
|
| 123 |
+
### t1d ###
|
| 124 |
+
###########
|
| 125 |
+
# NOTE: adjusting t1d last dim (confidence) from sequence mask
|
| 126 |
+
t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
|
| 127 |
+
t1d[:,:,:21] = seq_diffused[...,:21]
|
| 128 |
+
|
| 129 |
+
#t1d[:,:,21] *= input_t1dconf_mask
|
| 130 |
+
#set diffused conf to 0 and everything else to 1
|
| 131 |
+
t1d[:,~seq_mask,21] = 0.0
|
| 132 |
+
t1d[:,seq_mask,21] = 1.0
|
| 133 |
+
|
| 134 |
+
t1d[:1,:,22] = 1-t/diffuser.num_timesteps
|
| 135 |
+
|
| 136 |
+
#to do add structure confidence metric; need to expand dimensions of chkpt b4
|
| 137 |
+
#if t1d_24: JG - changed to be default
|
| 138 |
+
t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
|
| 139 |
+
t1d[:,~str_mask,23] = 0.0
|
| 140 |
+
t1d[:,str_mask,23] = 1.0
|
| 141 |
+
|
| 142 |
+
if dssp:
|
| 143 |
+
print(f'adding dssp {frac_provide_dssp} of time')
|
| 144 |
+
t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],4)).float()), -1).to(seq.device)
|
| 145 |
+
#dssp info
|
| 146 |
+
#mask some percentage of dssp info in range dssp_mask_percentage[0],dssp_mask_percentage[1]
|
| 147 |
+
percentage_mask=random.randint(dssp_mask_percentage[0], dssp_mask_percentage[1])
|
| 148 |
+
dssp=annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=percentage_mask)
|
| 149 |
+
#dssp_unmasked = annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=0)
|
| 150 |
+
if np.random.rand()>frac_provide_dssp:
|
| 151 |
+
print('masking dssp')
|
| 152 |
+
dssp[...]=0 #replace with mask token
|
| 153 |
+
dssp[:,-1]=1
|
| 154 |
+
t1d[...,24:]=dssp
|
| 155 |
+
|
| 156 |
+
if hotspots:
|
| 157 |
+
print(f"adding hotspots {frac_provide_contacts} of time")
|
| 158 |
+
t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
|
| 159 |
+
#mask all contacts some fraction of the time
|
| 160 |
+
if np.random.rand()>frac_provide_contacts:
|
| 161 |
+
print('masking contacts')
|
| 162 |
+
contacts = torch.zeros(L)
|
| 163 |
+
t1d[...,-1] = contacts
|
| 164 |
+
|
| 165 |
+
### xyz_t ###
|
| 166 |
+
#############
|
| 167 |
+
xyz_t = get_init_xyz(xyz_t[None])
|
| 168 |
+
xyz_t = xyz_t[0]
|
| 169 |
+
#Sequence masking
|
| 170 |
+
xyz_t[:,:,3:,:] = float('nan')
|
| 171 |
+
# Structure masking
|
| 172 |
+
if struc_cond:
|
| 173 |
+
print("non-autoregressive structure conditioning")
|
| 174 |
+
r = diffuser.alphas_cumprod[t]
|
| 175 |
+
xyz_mask = (torch.rand(xyz_t.shape[1]) > r).to(torch.bool).to(seq.device)
|
| 176 |
+
xyz_mask = torch.logical_and(xyz_mask,~str_mask)
|
| 177 |
+
xyz_t[:,xyz_mask,:,:] = float('nan')
|
| 178 |
+
else:
|
| 179 |
+
xyz_t[:,~str_mask,:,:] = float('nan')
|
| 180 |
+
|
| 181 |
+
### mask_msa ###
|
| 182 |
+
################
|
| 183 |
+
# NOTE: this is for loss scoring
|
| 184 |
+
mask_msa[:,:,~loss_seq_mask] = False
|
| 185 |
+
|
| 186 |
+
out=dict(
|
| 187 |
+
seq= seq,
|
| 188 |
+
msa_masked= msa_masked,
|
| 189 |
+
msa_full= msa_full,
|
| 190 |
+
xyz_t= xyz_t,
|
| 191 |
+
t1d= t1d,
|
| 192 |
+
mask_msa= mask_msa,
|
| 193 |
+
seq_diffused= seq_diffused
|
| 194 |
+
)
|
| 195 |
+
|
| 196 |
+
return out
|