diff --git a/.gitattributes b/.gitattributes
new file mode 100644
index 0000000000000000000000000000000000000000..c7d9f3332a950355d5a77d85000f05e6f45435ea
--- /dev/null
+++ b/.gitattributes
@@ -0,0 +1,34 @@
+*.7z filter=lfs diff=lfs merge=lfs -text
+*.arrow filter=lfs diff=lfs merge=lfs -text
+*.bin filter=lfs diff=lfs merge=lfs -text
+*.bz2 filter=lfs diff=lfs merge=lfs -text
+*.ckpt filter=lfs diff=lfs merge=lfs -text
+*.ftz filter=lfs diff=lfs merge=lfs -text
+*.gz filter=lfs diff=lfs merge=lfs -text
+*.h5 filter=lfs diff=lfs merge=lfs -text
+*.joblib filter=lfs diff=lfs merge=lfs -text
+*.lfs.* filter=lfs diff=lfs merge=lfs -text
+*.mlmodel filter=lfs diff=lfs merge=lfs -text
+*.model filter=lfs diff=lfs merge=lfs -text
+*.msgpack filter=lfs diff=lfs merge=lfs -text
+*.npy filter=lfs diff=lfs merge=lfs -text
+*.npz filter=lfs diff=lfs merge=lfs -text
+*.onnx filter=lfs diff=lfs merge=lfs -text
+*.ot filter=lfs diff=lfs merge=lfs -text
+*.parquet filter=lfs diff=lfs merge=lfs -text
+*.pb filter=lfs diff=lfs merge=lfs -text
+*.pickle filter=lfs diff=lfs merge=lfs -text
+*.pkl filter=lfs diff=lfs merge=lfs -text
+*.pt filter=lfs diff=lfs merge=lfs -text
+*.pth filter=lfs diff=lfs merge=lfs -text
+*.rar filter=lfs diff=lfs merge=lfs -text
+*.safetensors filter=lfs diff=lfs merge=lfs -text
+saved_model/**/* filter=lfs diff=lfs merge=lfs -text
+*.tar.* filter=lfs diff=lfs merge=lfs -text
+*.tflite filter=lfs diff=lfs merge=lfs -text
+*.tgz filter=lfs diff=lfs merge=lfs -text
+*.wasm filter=lfs diff=lfs merge=lfs -text
+*.xz filter=lfs diff=lfs merge=lfs -text
+*.zip filter=lfs diff=lfs merge=lfs -text
+*.zst filter=lfs diff=lfs merge=lfs -text
+*tfevents* filter=lfs diff=lfs merge=lfs -text
diff --git a/LICENSE b/LICENSE
new file mode 100644
index 0000000000000000000000000000000000000000..a584cfce171b7d0e8deb9666266c86dd15033031
--- /dev/null
+++ b/LICENSE
@@ -0,0 +1,21 @@
+MIT License
+
+Copyright (c) 2023 RosettaCommons
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
diff --git a/README.md b/README.md
new file mode 100644
index 0000000000000000000000000000000000000000..c7a1460a91dec7b3022b716406a4407e78c26966
--- /dev/null
+++ b/README.md
@@ -0,0 +1,17 @@
+---
+title: PROTEIN GENERATOR
+emoji: 🧪
+thumbnail: http://files.ipd.uw.edu/pub/sequence_diffusion/figs/diffusion_landscape.png
+colorFrom: blue
+colorTo: purple
+sdk: gradio
+sdk_version: 3.24.1
+app_file: app.py
+pinned: false
+---
+
+
+
+## Code Accessibility
+
+To download code and for more details please visit the [github](https://github.com/RosettaCommons/protein_generator)!
diff --git a/app.py b/app.py
new file mode 100644
index 0000000000000000000000000000000000000000..8555fd96dcd79380a95efa195704b2bc8dc560a3
--- /dev/null
+++ b/app.py
@@ -0,0 +1,404 @@
+import os,sys
+
+# install environment goods
+#os.system("pip -q install dgl -f https://data.dgl.ai/wheels/cu113/repo.html")
+os.system('pip install dgl==1.0.2+cu116 -f https://data.dgl.ai/wheels/cu116/repo.html')
+#os.system('pip install gradio')
+os.environ["DGLBACKEND"] = "pytorch"
+#os.system(f'pip install -r ./PROTEIN_GENERATOR/requirements.txt')
+print('Modules installed')
+
+os.system('pip install --force gradio==3.28.3')
+
+os.environ["DGLBACKEND"] = "pytorch"
+
+if not os.path.exists('./SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'):
+ print('Downloading model weights 1')
+ os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt')
+ print('Successfully Downloaded')
+
+if not os.path.exists('./SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'):
+ print('Downloading model weights 2')
+ os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt')
+ print('Successfully Downloaded')
+
+import numpy as np
+import gradio as gr
+import py3Dmol
+from io import StringIO
+import json
+import secrets
+import copy
+import matplotlib.pyplot as plt
+from utils.sampler import HuggingFace_sampler
+
+plt.rcParams.update({'font.size': 13})
+
+with open('./tmp/args.json','r') as f:
+ args = json.load(f)
+
+# manually set checkpoint to load
+args['checkpoint'] = None
+args['dump_trb'] = False
+args['dump_args'] = True
+args['save_best_plddt'] = True
+args['T'] = 25
+args['strand_bias'] = 0.0
+args['loop_bias'] = 0.0
+args['helix_bias'] = 0.0
+
+
+
+def protein_diffusion_model(sequence, seq_len, helix_bias, strand_bias, loop_bias,
+ secondary_structure, aa_bias, aa_bias_potential,
+ #target_charge, target_ph, charge_potential,
+ num_steps, noise, hydrophobic_target_score, hydrophobic_potential):
+
+ dssp_checkpoint = './SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'
+ og_checkpoint = './SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'
+
+ model_args = copy.deepcopy(args)
+
+ # make sampler
+ S = HuggingFace_sampler(args=model_args)
+
+ # get random prefix
+ S.out_prefix = './tmp/'+secrets.token_hex(nbytes=10).upper()
+
+ # set args
+ S.args['checkpoint'] = None
+ S.args['dump_trb'] = False
+ S.args['dump_args'] = True
+ S.args['save_best_plddt'] = True
+ S.args['T'] = 20
+ S.args['strand_bias'] = 0.0
+ S.args['loop_bias'] = 0.0
+ S.args['helix_bias'] = 0.0
+ S.args['potentials'] = None
+ S.args['potential_scale'] = None
+ S.args['aa_composition'] = None
+
+
+ # get sequence if entered and make sure all chars are valid
+ alt_aa_dict = {'B':['D','N'],'J':['I','L'],'U':['C'],'Z':['E','Q'],'O':['K']}
+ if sequence not in ['',None]:
+ L = len(sequence)
+ aa_seq = []
+ for aa in sequence.upper():
+ if aa in alt_aa_dict.keys():
+ aa_seq.append(np.random.choice(alt_aa_dict[aa]))
+ else:
+ aa_seq.append(aa)
+
+ S.args['sequence'] = aa_seq
+ else:
+ S.args['contigs'] = [f'{seq_len}']
+ L = int(seq_len)
+
+ if secondary_structure in ['',None]:
+ secondary_structure = None
+ else:
+ secondary_structure = ''.join(['E' if x == 'S' else x for x in secondary_structure])
+ if L < len(secondary_structure):
+ secondary_structure = secondary_structure[:len(sequence)]
+ elif L == len(secondary_structure):
+ pass
+ else:
+ dseq = L - len(secondary_structure)
+ secondary_structure += secondary_structure[-1]*dseq
+
+
+ # potentials
+ potential_list = []
+ potential_bias_list = []
+
+ if aa_bias not in ['',None]:
+ potential_list.append('aa_bias')
+ S.args['aa_composition'] = aa_bias
+ if aa_bias_potential in ['',None]:
+ aa_bias_potential = 3
+ potential_bias_list.append(str(aa_bias_potential))
+ '''
+ if target_charge not in ['',None]:
+ potential_list.append('charge')
+ if charge_potential in ['',None]:
+ charge_potential = 1
+ potential_bias_list.append(str(charge_potential))
+ S.args['target_charge'] = float(target_charge)
+ if target_ph in ['',None]:
+ target_ph = 7.4
+ S.args['target_pH'] = float(target_ph)
+ '''
+
+ if hydrophobic_target_score not in ['',None]:
+ potential_list.append('hydrophobic')
+ S.args['hydrophobic_score'] = float(hydrophobic_target_score)
+ if hydrophobic_potential in ['',None]:
+ hydrophobic_potential = 3
+ potential_bias_list.append(str(hydrophobic_potential))
+
+
+ if len(potential_list) > 0:
+ S.args['potentials'] = ','.join(potential_list)
+ S.args['potential_scale'] = ','.join(potential_bias_list)
+
+
+ # normalise secondary_structure bias from range 0-0.3
+ S.args['secondary_structure'] = secondary_structure
+ S.args['helix_bias'] = helix_bias
+ S.args['strand_bias'] = strand_bias
+ S.args['loop_bias'] = loop_bias
+
+ # set T
+ if num_steps in ['',None]:
+ S.args['T'] = 20
+ else:
+ S.args['T'] = int(num_steps)
+
+ # noise
+ if 'normal' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [0]
+ S.args['sample_distribution_gmm_variances'] = [1]
+ elif 'gmm2' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [-1,1]
+ S.args['sample_distribution_gmm_variances'] = [1,1]
+ elif 'gmm3' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [-1,0,1]
+ S.args['sample_distribution_gmm_variances'] = [1,1,1]
+
+
+
+ if secondary_structure not in ['',None] or helix_bias+strand_bias+loop_bias > 0:
+ S.args['checkpoint'] = dssp_checkpoint
+ S.args['d_t1d'] = 29
+ print('using dssp checkpoint')
+ else:
+ S.args['checkpoint'] = og_checkpoint
+ S.args['d_t1d'] = 24
+ print('using og checkpoint')
+
+
+ for k,v in S.args.items():
+ print(f"{k} --> {v}")
+
+ # init S
+ S.model_init()
+ S.diffuser_init()
+ S.setup()
+
+ # sampling loop
+ plddt_data = []
+ for j in range(S.max_t):
+ output_seq, output_pdb, plddt = S.take_step_get_outputs(j)
+ plddt_data.append(plddt)
+ yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
+
+ output_seq, output_pdb, plddt = S.get_outputs()
+
+ yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
+
+def get_plddt_plot(plddt_data, max_t):
+ x = [i+1 for i in range(len(plddt_data))]
+ fig, ax = plt.subplots(figsize=(15,6))
+ ax.plot(x,plddt_data,color='#661dbf', linewidth=3,marker='o')
+ ax.set_xticks([i+1 for i in range(max_t)])
+ ax.set_yticks([(i+1)/10 for i in range(10)])
+ ax.set_ylim([0,1])
+ ax.set_ylabel('model confidence (plddt)')
+ ax.set_xlabel('diffusion steps (t)')
+ return fig
+
+def display_pdb(path_to_pdb):
+ '''
+ #function to display pdb in py3dmol
+ '''
+ pdb = open(path_to_pdb, "r").read()
+
+ view = py3Dmol.view(width=500, height=500)
+ view.addModel(pdb, "pdb")
+ view.setStyle({'model': -1}, {"cartoon": {'colorscheme':{'prop':'b','gradient':'roygb','min':0,'max':1}}})#'linear', 'min': 0, 'max': 1, 'colors': ["#ff9ef0","#a903fc",]}}})
+ view.zoomTo()
+ output = view._make_html().replace("'", '"')
+ print(view._make_html())
+ x = f""" {output} """ # do not use ' in this input
+
+ return f""""""
+
+'''
+
+ return f""""""
+'''
+
+def toggle_seq_input(choice):
+ if choice == "protein length":
+ return gr.update(visible=True, value=None), gr.update(visible=False, value=None)
+ elif choice == "custom sequence":
+ return gr.update(visible=False, value=None), gr.update(visible=True, value=None)
+
+def toggle_secondary_structure(choice):
+ if choice == "sliders":
+ return gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=False, value=None)
+ elif choice == "explicit":
+ return gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=True, value=None)
+
+# Define the Gradio interface
+with gr.Blocks(theme='ParityError/Interstellar') as demo:
+
+ gr.Markdown(f"""# Protein Generation via Diffusion in Sequence Space""")
+
+ with gr.Row():
+ with gr.Column(min_width=500):
+ gr.Markdown(f"""
+ ## How does it work?\n
+ --- [PREPRINT](https://biorxiv.org/content/10.1101/2023.05.08.539766v1) ---
+
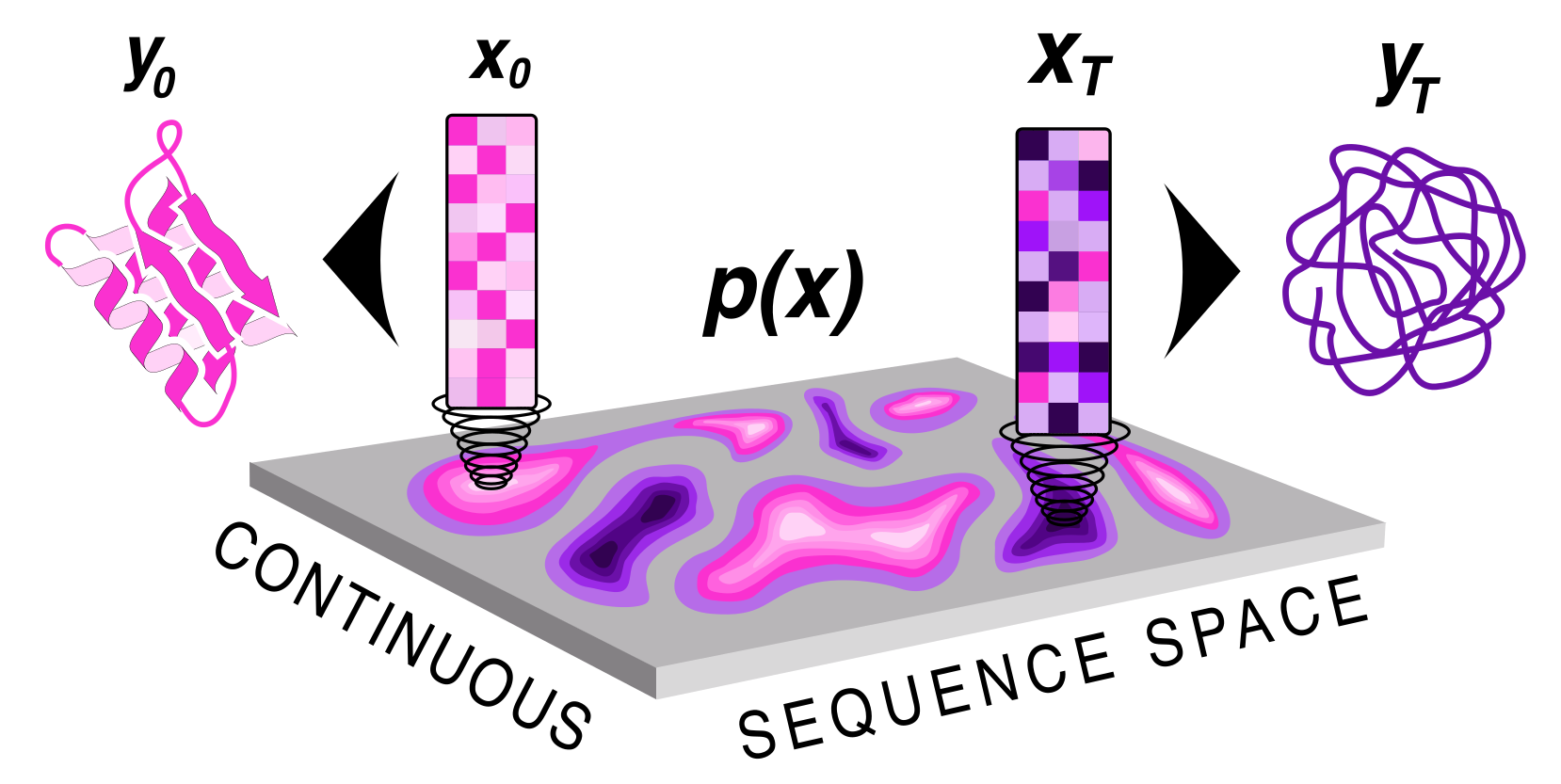
+ Protein sequence and structure co-generation is a long outstanding problem in the field of protein design. By implementing [ddpm](https://arxiv.org/abs/2006.11239) style diffusion over protein seqeuence space we generate protein sequence and structure pairs. Starting with [RoseTTAFold](https://www.science.org/doi/10.1126/science.abj8754), a protein structure prediction network, we finetuned it to predict sequence and structure given a partially noised sequence. By applying losses to both the predicted sequence and structure the model is forced to generate meaningful pairs. Diffusing in sequence space makes it easy to implement potentials to guide the diffusive process toward particular amino acid composition, net charge, and more! Furthermore, you can sample proteins from a family of sequences or even train a small sequence to function classifier to guide generation toward desired sequences.
+ 
+
+ ## How to use it?\n
+ A user can either design a custom input sequence to diffuse from or specify a length below. To scaffold a sequence use the following format where X represent residues to diffuse: XXXXXXXXSCIENCESCIENCEXXXXXXXXXXXXXXXXXXX. You can even design a protein with your name XXXXXXXXXXXXNAMEHEREXXXXXXXXXXXXX!
+
+ ### Acknowledgements\n
+ Thank you to Simon Dürr and the Hugging Face team for setting us up with a community GPU grant!
+ """)
+
+ gr.Markdown("""
+ ## Model in Action
+ 
+ """)
+
+ with gr.Row().style(equal_height=False):
+ with gr.Column():
+ gr.Markdown("""## INPUTS""")
+ gr.Markdown("""#### Start Sequence
+ Specify the protein length for complete unconditional generation, or scaffold a motif (or your name) using the custom sequence input""")
+ seq_opt = gr.Radio(["protein length","custom sequence"], label="How would you like to specify the starting sequence?", value='protein length')
+
+ sequence = gr.Textbox(label="custom sequence", lines=1, placeholder='AMINO ACIDS: A,C,D,E,F,G,H,I,K,L,M,N,P,Q,R,S,T,V,W,Y\n MASK TOKEN: X', visible=False)
+ seq_len = gr.Slider(minimum=5.0, maximum=250.0, label="protein length", value=100, visible=True)
+
+ seq_opt.change(fn=toggle_seq_input,
+ inputs=[seq_opt],
+ outputs=[seq_len, sequence],
+ queue=False)
+
+ gr.Markdown("""### Optional Parameters""")
+ with gr.Accordion(label='Secondary Structure',open=True):
+ gr.Markdown("""Try changing the sliders or inputing explicit secondary structure conditioning for each residue""")
+ sec_str_opt = gr.Radio(["sliders","explicit"], label="How would you like to specify secondary structure?", value='sliders')
+
+ secondary_structure = gr.Textbox(label="secondary structure", lines=1, placeholder='HELIX = H STRAND = S LOOP = L MASK = X(must be the same length as input sequence)', visible=False)
+
+ with gr.Column():
+ helix_bias = gr.Slider(minimum=0.0, maximum=0.05, label="helix bias", visible=True)
+ strand_bias = gr.Slider(minimum=0.0, maximum=0.05, label="strand bias", visible=True)
+ loop_bias = gr.Slider(minimum=0.0, maximum=0.20, label="loop bias", visible=True)
+
+ sec_str_opt.change(fn=toggle_secondary_structure,
+ inputs=[sec_str_opt],
+ outputs=[helix_bias,strand_bias,loop_bias,secondary_structure],
+ queue=False)
+
+ with gr.Accordion(label='Amino Acid Compositional Bias',open=False):
+ gr.Markdown("""Bias sequence composition for particular amino acids by specifying the one letter code followed by the fraction to bias. This can be input as a list for example: W0.2,E0.1""")
+ with gr.Row():
+ aa_bias = gr.Textbox(label="aa bias", lines=1, placeholder='specify one letter AA and fraction to bias, for example W0.1 or M0.1,K0.1' )
+ aa_bias_potential = gr.Textbox(label="aa bias scale", lines=1, placeholder='AA Bias potential scale (recomended range 1.0-5.0)')
+
+ '''
+ with gr.Accordion(label='Charge Bias',open=False):
+ gr.Markdown("""Bias for a specified net charge at a particular pH using the boxes below""")
+ with gr.Row():
+ target_charge = gr.Textbox(label="net charge", lines=1, placeholder='net charge to target')
+ target_ph = gr.Textbox(label="pH", lines=1, placeholder='pH at which net charge is desired')
+ charge_potential = gr.Textbox(label="charge potential scale", lines=1, placeholder='charge potential scale (recomended range 1.0-5.0)')
+ '''
+
+ with gr.Accordion(label='Hydrophobic Bias',open=False):
+ gr.Markdown("""Bias for or against hydrophobic composition, to get more soluble proteins, bias away with a negative target score (ex. -5)""")
+ with gr.Row():
+ hydrophobic_target_score = gr.Textbox(label="hydrophobic score", lines=1, placeholder='hydrophobic score to target (negative score is good for solublility)')
+ hydrophobic_potential = gr.Textbox(label="hydrophobic potential scale", lines=1, placeholder='hydrophobic potential scale (recomended range 1.0-2.0)')
+
+ with gr.Accordion(label='Diffusion Params',open=False):
+ gr.Markdown("""Increasing T to more steps can be helpful for harder design challenges, sampling from different distributions can change the sequence and structural composition""")
+ with gr.Row():
+ num_steps = gr.Textbox(label="T", lines=1, placeholder='number of diffusion steps (25 or less will speed things up)')
+ noise = gr.Dropdown(['normal','gmm2 [-1,1]','gmm3 [-1,0,1]'], label='noise type', value='normal')
+
+ btn = gr.Button("GENERATE")
+
+ #with gr.Row():
+ with gr.Column():
+ gr.Markdown("""## OUTPUTS""")
+ gr.Markdown("""#### Confidence score for generated structure at each timestep""")
+ plddt_plot = gr.Plot(label='plddt at step t')
+ gr.Markdown("""#### Output protein sequnece""")
+ output_seq = gr.Textbox(label="sequence")
+ gr.Markdown("""#### Download PDB file""")
+ output_pdb = gr.File(label="PDB file")
+ gr.Markdown("""#### Structure viewer""")
+ output_viewer = gr.HTML()
+
+ gr.Markdown("""### Don't know where to get started? Click on an example below to try it out!""")
+ gr.Examples(
+ [["","125",0.0,0.0,0.2,"","","","20","normal",'',''],
+ ["","100",0.0,0.0,0.0,"","W0.2","2","20","normal",'',''],
+ ["","100",0.0,0.0,0.0,"XXHHHHHHHHHXXXXXXXHHHHHHHHHXXXXXXXHHHHHHHHXXXXSSSSSSSSSSSXXXXXXXXSSSSSSSSSSSSXXXXXXXSSSSSSSSSXXXXXXX","","","25","normal",'',''],
+ ["XXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXXXXXPEPSEQXXXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXX","",0.0,0.0,0.0,"","","","25","normal",'','']],
+ inputs=[sequence,
+ seq_len,
+ helix_bias,
+ strand_bias,
+ loop_bias,
+ secondary_structure,
+ aa_bias,
+ aa_bias_potential,
+ #target_charge,
+ #target_ph,
+ #charge_potential,
+ num_steps,
+ noise,
+ hydrophobic_target_score,
+ hydrophobic_potential],
+ outputs=[output_seq,
+ output_pdb,
+ output_viewer,
+ plddt_plot],
+ fn=protein_diffusion_model,
+ )
+ btn.click(protein_diffusion_model,
+ [sequence,
+ seq_len,
+ helix_bias,
+ strand_bias,
+ loop_bias,
+ secondary_structure,
+ aa_bias,
+ aa_bias_potential,
+ #target_charge,
+ #target_ph,
+ #charge_potential,
+ num_steps,
+ noise,
+ hydrophobic_target_score,
+ hydrophobic_potential],
+ [output_seq,
+ output_pdb,
+ output_viewer,
+ plddt_plot])
+
+demo.queue()
+demo.launch(debug=True)
+
diff --git a/examples/aa_weights.json b/examples/aa_weights.json
new file mode 100644
index 0000000000000000000000000000000000000000..4b225021a85dbc28daa580d53b480a637bc1e0ad
--- /dev/null
+++ b/examples/aa_weights.json
@@ -0,0 +1,22 @@
+{
+ "A": 0,
+ "R": 0,
+ "N": 0,
+ "D": 0,
+ "C": 0,
+ "Q": 0,
+ "E": 0,
+ "G": 0,
+ "H": 0,
+ "I": 0,
+ "L": 0,
+ "K": 0,
+ "M": 0,
+ "F": 0,
+ "P": 0,
+ "S": 0,
+ "T": 0,
+ "W": 0,
+ "Y": 0,
+ "V": 0
+}
diff --git a/examples/binder_design.sh b/examples/binder_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..ae66f0273cb2d3fa893717d7f3102ae01bd957b7
--- /dev/null
+++ b/examples/binder_design.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/binder_design \
+ --pdb pdbs/cd86.pdb \
+ --T 25 --save_best_plddt \
+ --contigs B1-110,0 25-75 \
+ --hotspots B40,B32,B87,B96,B30
diff --git a/examples/loop_design.sh b/examples/loop_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..de6c21db8ed2bd4ab526e7c41e8fb7bd13f76754
--- /dev/null
+++ b/examples/loop_design.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --pdb pdbs/G12D_manual_mut.pdb \
+ --out out/ab_loop \
+ --contigs A2-176,0 C7-16,0 H2-95,12-15,H111-116,0 L1-45,10-12,L56-107 \
+ --T 25 --save_best_plddt --loop_design
diff --git a/examples/motif_scaffolding.sh b/examples/motif_scaffolding.sh
new file mode 100644
index 0000000000000000000000000000000000000000..ba1f0b03227cc3de8ec140c2fcc42dead2a083e2
--- /dev/null
+++ b/examples/motif_scaffolding.sh
@@ -0,0 +1,14 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --pdb pdbs/rsv5_5tpn.pdb \
+ --contigs 0-25,A163-181,25-30 --T 25 --save_best_plddt
diff --git a/examples/out/design_000000.pdb b/examples/out/design_000000.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..90f5532fa0ebdfa42f13edb342ad632b3dc18c16
--- /dev/null
+++ b/examples/out/design_000000.pdb
@@ -0,0 +1,1805 @@
+ATOM 1 N ILE A 1 -8.484 18.449 12.122 1.00 0.91
+ATOM 2 CA ILE A 1 -9.332 17.874 11.084 1.00 0.91
+ATOM 3 C ILE A 1 -9.716 16.439 11.420 1.00 0.91
+ATOM 4 O ILE A 1 -8.928 15.696 12.005 1.00 0.91
+ATOM 5 CB ILE A 1 -8.642 17.955 9.711 1.00 0.91
+ATOM 6 CG1 ILE A 1 -9.569 17.509 8.577 1.00 0.91
+ATOM 7 CG2 ILE A 1 -7.373 17.116 9.726 1.00 0.91
+ATOM 8 CD1 ILE A 1 -9.021 17.784 7.196 1.00 0.91
+ATOM 9 H ILE A 1 -8.607 18.151 13.079 1.00 0.91
+ATOM 10 HA ILE A 1 -10.245 18.464 11.018 1.00 0.91
+ATOM 11 HB ILE A 1 -8.377 18.993 9.511 1.00 0.91
+ATOM 12 1HG2 ILE A 1 -6.874 17.194 8.761 1.00 0.91
+ATOM 13 2HG2 ILE A 1 -6.707 17.478 10.509 1.00 0.91
+ATOM 14 3HG2 ILE A 1 -7.628 16.074 9.920 1.00 0.91
+ATOM 15 1HG1 ILE A 1 -9.759 16.439 8.663 1.00 0.91
+ATOM 16 2HG1 ILE A 1 -10.529 18.018 8.670 1.00 0.91
+ATOM 17 1HD1 ILE A 1 -9.734 17.440 6.446 1.00 0.91
+ATOM 18 2HD1 ILE A 1 -8.857 18.855 7.076 1.00 0.91
+ATOM 19 3HD1 ILE A 1 -8.078 17.255 7.067 1.00 0.91
+ATOM 20 N SER A 2 -11.064 16.075 11.291 1.00 0.96
+ATOM 21 CA SER A 2 -11.514 14.698 11.461 1.00 0.96
+ATOM 22 C SER A 2 -10.842 13.772 10.455 1.00 0.96
+ATOM 23 O SER A 2 -10.369 14.214 9.408 1.00 0.96
+ATOM 24 CB SER A 2 -13.020 14.614 11.309 1.00 0.96
+ATOM 25 OG SER A 2 -13.397 14.833 9.978 1.00 0.96
+ATOM 26 H SER A 2 -11.722 16.772 10.974 1.00 0.96
+ATOM 27 HG SER A 2 -12.730 14.401 9.439 1.00 0.96
+ATOM 28 HA SER A 2 -11.244 14.367 12.464 1.00 0.96
+ATOM 29 1HB SER A 2 -13.366 13.632 11.631 1.00 0.96
+ATOM 30 2HB SER A 2 -13.495 15.354 11.951 1.00 0.96
+ATOM 31 N PRO A 3 -10.783 12.424 10.716 1.00 0.97
+ATOM 32 CA PRO A 3 -10.177 11.477 9.788 1.00 0.97
+ATOM 33 C PRO A 3 -10.872 11.512 8.433 1.00 0.97
+ATOM 34 O PRO A 3 -10.224 11.433 7.390 1.00 0.97
+ATOM 35 CB PRO A 3 -10.356 10.117 10.469 1.00 0.97
+ATOM 36 CG PRO A 3 -10.316 10.398 11.932 1.00 0.97
+ATOM 37 CD PRO A 3 -10.986 11.738 12.075 1.00 0.97
+ATOM 38 HA PRO A 3 -9.107 11.710 9.680 1.00 0.97
+ATOM 39 1HB PRO A 3 -11.308 9.670 10.146 1.00 0.97
+ATOM 40 2HB PRO A 3 -9.555 9.437 10.143 1.00 0.97
+ATOM 41 1HG PRO A 3 -10.838 9.603 12.486 1.00 0.97
+ATOM 42 2HG PRO A 3 -9.276 10.404 12.290 1.00 0.97
+ATOM 43 1HD PRO A 3 -12.046 11.666 11.788 1.00 0.97
+ATOM 44 2HD PRO A 3 -10.963 12.065 13.125 1.00 0.97
+ATOM 45 N GLU A 4 -12.266 11.589 8.409 1.00 0.98
+ATOM 46 CA GLU A 4 -13.019 11.626 7.161 1.00 0.98
+ATOM 47 C GLU A 4 -12.613 12.821 6.307 1.00 0.98
+ATOM 48 O GLU A 4 -12.431 12.697 5.096 1.00 0.98
+ATOM 49 CB GLU A 4 -14.523 11.658 7.441 1.00 0.98
+ATOM 50 CG GLU A 4 -15.389 11.510 6.198 1.00 0.98
+ATOM 51 CD GLU A 4 -16.859 11.473 6.511 1.00 0.98
+ATOM 52 OE1 GLU A 4 -17.640 11.380 5.594 1.00 0.98
+ATOM 53 OE2 GLU A 4 -17.202 11.537 7.668 1.00 0.98
+ATOM 54 H GLU A 4 -12.761 11.683 9.284 1.00 0.98
+ATOM 55 HA GLU A 4 -12.797 10.719 6.597 1.00 0.98
+ATOM 56 1HB GLU A 4 -14.785 10.854 8.128 1.00 0.98
+ATOM 57 2HB GLU A 4 -14.786 12.599 7.922 1.00 0.98
+ATOM 58 1HG GLU A 4 -15.206 12.358 5.537 1.00 0.98
+ATOM 59 2HG GLU A 4 -15.128 10.580 5.693 1.00 0.98
+ATOM 60 N GLU A 5 -12.526 14.047 6.921 1.00 0.98
+ATOM 61 CA GLU A 5 -12.113 15.252 6.211 1.00 0.98
+ATOM 62 C GLU A 5 -10.711 15.100 5.636 1.00 0.98
+ATOM 63 O GLU A 5 -10.438 15.532 4.517 1.00 0.98
+ATOM 64 CB GLU A 5 -12.187 16.470 7.134 1.00 0.98
+ATOM 65 CG GLU A 5 -13.602 16.891 7.504 1.00 0.98
+ATOM 66 CD GLU A 5 -13.637 17.993 8.526 1.00 0.98
+ATOM 67 OE1 GLU A 5 -14.703 18.489 8.801 1.00 0.98
+ATOM 68 OE2 GLU A 5 -12.595 18.341 9.031 1.00 0.98
+ATOM 69 H GLU A 5 -12.708 14.099 7.913 1.00 0.98
+ATOM 70 HA GLU A 5 -12.800 15.417 5.382 1.00 0.98
+ATOM 71 1HB GLU A 5 -11.647 16.261 8.058 1.00 0.98
+ATOM 72 2HB GLU A 5 -11.700 17.320 6.656 1.00 0.98
+ATOM 73 1HG GLU A 5 -14.111 17.244 6.608 1.00 0.98
+ATOM 74 2HG GLU A 5 -14.129 16.031 7.918 1.00 0.98
+ATOM 75 N LEU A 6 -9.745 14.522 6.455 1.00 0.98
+ATOM 76 CA LEU A 6 -8.391 14.258 5.982 1.00 0.98
+ATOM 77 C LEU A 6 -8.401 13.332 4.771 1.00 0.98
+ATOM 78 O LEU A 6 -7.719 13.585 3.778 1.00 0.98
+ATOM 79 CB LEU A 6 -7.543 13.661 7.111 1.00 0.98
+ATOM 80 CG LEU A 6 -6.052 13.463 6.811 1.00 0.98
+ATOM 81 CD1 LEU A 6 -5.343 12.963 8.062 1.00 0.98
+ATOM 82 CD2 LEU A 6 -5.891 12.480 5.661 1.00 0.98
+ATOM 83 H LEU A 6 -10.007 14.229 7.386 1.00 0.98
+ATOM 84 HA LEU A 6 -7.938 15.204 5.688 1.00 0.98
+ATOM 85 1HB LEU A 6 -7.622 14.313 7.980 1.00 0.98
+ATOM 86 2HB LEU A 6 -7.960 12.688 7.374 1.00 0.98
+ATOM 87 HG LEU A 6 -5.606 14.420 6.535 1.00 0.98
+ATOM 88 1HD1 LEU A 6 -4.283 12.823 7.849 1.00 0.98
+ATOM 89 2HD1 LEU A 6 -5.457 13.695 8.862 1.00 0.98
+ATOM 90 3HD1 LEU A 6 -5.779 12.014 8.372 1.00 0.98
+ATOM 91 1HD2 LEU A 6 -4.831 12.340 5.448 1.00 0.98
+ATOM 92 2HD2 LEU A 6 -6.335 11.523 5.937 1.00 0.98
+ATOM 93 3HD2 LEU A 6 -6.392 12.870 4.776 1.00 0.98
+ATOM 94 N ILE A 7 -9.224 12.198 4.863 1.00 0.99
+ATOM 95 CA ILE A 7 -9.323 11.260 3.752 1.00 0.99
+ATOM 96 C ILE A 7 -9.830 11.950 2.492 1.00 0.99
+ATOM 97 O ILE A 7 -9.276 11.769 1.408 1.00 0.99
+ATOM 98 CB ILE A 7 -10.232 10.073 4.120 1.00 0.99
+ATOM 99 CG1 ILE A 7 -9.568 9.202 5.189 1.00 0.99
+ATOM 100 CG2 ILE A 7 -10.558 9.249 2.884 1.00 0.99
+ATOM 101 CD1 ILE A 7 -10.487 8.161 5.787 1.00 0.99
+ATOM 102 H ILE A 7 -9.745 12.015 5.709 1.00 0.99
+ATOM 103 HA ILE A 7 -8.330 10.862 3.546 1.00 0.99
+ATOM 104 HB ILE A 7 -11.161 10.447 4.551 1.00 0.99
+ATOM 105 1HG2 ILE A 7 -11.201 8.415 3.162 1.00 0.99
+ATOM 106 2HG2 ILE A 7 -11.070 9.875 2.154 1.00 0.99
+ATOM 107 3HG2 ILE A 7 -9.634 8.866 2.448 1.00 0.99
+ATOM 108 1HG1 ILE A 7 -8.707 8.691 4.758 1.00 0.99
+ATOM 109 2HG1 ILE A 7 -9.200 9.835 5.997 1.00 0.99
+ATOM 110 1HD1 ILE A 7 -9.944 7.584 6.536 1.00 0.99
+ATOM 111 2HD1 ILE A 7 -11.339 8.655 6.256 1.00 0.99
+ATOM 112 3HD1 ILE A 7 -10.840 7.494 5.002 1.00 0.99
+ATOM 113 N GLU A 8 -10.930 12.775 2.608 1.00 0.98
+ATOM 114 CA GLU A 8 -11.510 13.488 1.476 1.00 0.98
+ATOM 115 C GLU A 8 -10.498 14.437 0.847 1.00 0.98
+ATOM 116 O GLU A 8 -10.385 14.518 -0.376 1.00 0.98
+ATOM 117 CB GLU A 8 -12.764 14.252 1.907 1.00 0.98
+ATOM 118 CG GLU A 8 -13.974 13.369 2.175 1.00 0.98
+ATOM 119 CD GLU A 8 -15.164 14.142 2.672 1.00 0.98
+ATOM 120 OE1 GLU A 8 -16.179 13.538 2.923 1.00 0.98
+ATOM 121 OE2 GLU A 8 -15.058 15.339 2.800 1.00 0.98
+ATOM 122 H GLU A 8 -11.321 12.912 3.529 1.00 0.98
+ATOM 123 HA GLU A 8 -11.802 12.759 0.721 1.00 0.98
+ATOM 124 1HB GLU A 8 -12.554 14.816 2.815 1.00 0.98
+ATOM 125 2HB GLU A 8 -13.038 14.968 1.132 1.00 0.98
+ATOM 126 1HG GLU A 8 -14.259 12.869 1.249 1.00 0.98
+ATOM 127 2HG GLU A 8 -13.712 12.630 2.933 1.00 0.98
+ATOM 128 N ARG A 9 -9.729 15.202 1.689 1.00 0.98
+ATOM 129 CA ARG A 9 -8.702 16.114 1.200 1.00 0.98
+ATOM 130 C ARG A 9 -7.632 15.369 0.412 1.00 0.98
+ATOM 131 O ARG A 9 -7.235 15.796 -0.671 1.00 0.98
+ATOM 132 CB ARG A 9 -8.058 16.869 2.353 1.00 0.98
+ATOM 133 CG ARG A 9 -8.925 17.959 2.963 1.00 0.98
+ATOM 134 CD ARG A 9 -8.222 18.670 4.061 1.00 0.98
+ATOM 135 NE ARG A 9 -9.075 19.661 4.698 1.00 0.98
+ATOM 136 CZ ARG A 9 -8.706 20.431 5.740 1.00 0.98
+ATOM 137 NH1 ARG A 9 -7.500 20.314 6.249 1.00 0.98
+ATOM 138 NH2 ARG A 9 -9.557 21.305 6.250 1.00 0.98
+ATOM 139 H ARG A 9 -9.862 15.099 2.685 1.00 0.98
+ATOM 140 HA ARG A 9 -9.173 16.843 0.540 1.00 0.98
+ATOM 141 1HB ARG A 9 -7.799 16.170 3.148 1.00 0.98
+ATOM 142 2HB ARG A 9 -7.133 17.335 2.014 1.00 0.98
+ATOM 143 1HG ARG A 9 -9.188 18.687 2.195 1.00 0.98
+ATOM 144 2HG ARG A 9 -9.835 17.515 3.369 1.00 0.98
+ATOM 145 1HD ARG A 9 -7.910 17.952 4.818 1.00 0.98
+ATOM 146 2HD ARG A 9 -7.347 19.181 3.662 1.00 0.98
+ATOM 147 HE ARG A 9 -10.011 19.780 4.334 1.00 0.98
+ATOM 148 1HH1 ARG A 9 -6.849 19.646 5.860 1.00 0.98
+ATOM 149 2HH1 ARG A 9 -7.223 20.892 7.030 1.00 0.98
+ATOM 150 1HH2 ARG A 9 -10.484 21.395 5.858 1.00 0.98
+ATOM 151 2HH2 ARG A 9 -9.280 21.882 7.030 1.00 0.98
+ATOM 152 N ILE A 10 -7.122 14.206 0.993 1.00 0.99
+ATOM 153 CA ILE A 10 -6.104 13.420 0.307 1.00 0.99
+ATOM 154 C ILE A 10 -6.623 12.885 -1.021 1.00 0.99
+ATOM 155 O ILE A 10 -5.934 12.949 -2.039 1.00 0.99
+ATOM 156 CB ILE A 10 -5.601 12.271 1.199 1.00 0.99
+ATOM 157 CG1 ILE A 10 -4.820 12.822 2.395 1.00 0.99
+ATOM 158 CG2 ILE A 10 -4.740 11.311 0.393 1.00 0.99
+ATOM 159 CD1 ILE A 10 -4.511 11.787 3.452 1.00 0.99
+ATOM 160 H ILE A 10 -7.474 13.886 1.884 1.00 0.99
+ATOM 161 HA ILE A 10 -5.249 14.064 0.104 1.00 0.99
+ATOM 162 HB ILE A 10 -6.454 11.726 1.604 1.00 0.99
+ATOM 163 1HG2 ILE A 10 -4.393 10.504 1.038 1.00 0.99
+ATOM 164 2HG2 ILE A 10 -5.327 10.895 -0.424 1.00 0.99
+ATOM 165 3HG2 ILE A 10 -3.881 11.846 -0.012 1.00 0.99
+ATOM 166 1HG1 ILE A 10 -3.879 13.248 2.049 1.00 0.99
+ATOM 167 2HG1 ILE A 10 -5.389 13.626 2.862 1.00 0.99
+ATOM 168 1HD1 ILE A 10 -3.956 12.253 4.267 1.00 0.99
+ATOM 169 2HD1 ILE A 10 -5.442 11.372 3.839 1.00 0.99
+ATOM 170 3HD1 ILE A 10 -3.911 10.990 3.016 1.00 0.99
+ATOM 171 N ALA A 11 -7.864 12.279 -1.027 1.00 0.99
+ATOM 172 CA ALA A 11 -8.468 11.738 -2.238 1.00 0.99
+ATOM 173 C ALA A 11 -8.565 12.799 -3.327 1.00 0.99
+ATOM 174 O ALA A 11 -8.255 12.539 -4.490 1.00 0.99
+ATOM 175 CB ALA A 11 -9.846 11.166 -1.935 1.00 0.99
+ATOM 176 H ALA A 11 -8.384 12.247 -0.161 1.00 0.99
+ATOM 177 HA ALA A 11 -7.827 10.937 -2.608 1.00 0.99
+ATOM 178 1HB ALA A 11 -10.283 10.766 -2.850 1.00 0.99
+ATOM 179 2HB ALA A 11 -9.754 10.368 -1.198 1.00 0.99
+ATOM 180 3HB ALA A 11 -10.487 11.952 -1.540 1.00 0.99
+ATOM 181 N GLU A 12 -9.005 14.040 -2.987 1.00 0.98
+ATOM 182 CA GLU A 12 -9.129 15.146 -3.929 1.00 0.98
+ATOM 183 C GLU A 12 -7.772 15.543 -4.495 1.00 0.98
+ATOM 184 O GLU A 12 -7.643 15.823 -5.686 1.00 0.98
+ATOM 185 CB GLU A 12 -9.802 16.349 -3.264 1.00 0.98
+ATOM 186 CG GLU A 12 -11.296 16.180 -3.029 1.00 0.98
+ATOM 187 CD GLU A 12 -11.910 17.346 -2.305 1.00 0.98
+ATOM 188 OE1 GLU A 12 -13.089 17.307 -2.048 1.00 0.98
+ATOM 189 OE2 GLU A 12 -11.199 18.277 -2.010 1.00 0.98
+ATOM 190 H GLU A 12 -9.231 14.195 -2.015 1.00 0.98
+ATOM 191 HA GLU A 12 -9.758 14.824 -4.759 1.00 0.98
+ATOM 192 1HB GLU A 12 -9.332 16.543 -2.300 1.00 0.98
+ATOM 193 2HB GLU A 12 -9.659 17.235 -3.883 1.00 0.98
+ATOM 194 1HG GLU A 12 -11.796 16.079 -3.992 1.00 0.98
+ATOM 195 2HG GLU A 12 -11.460 15.285 -2.428 1.00 0.98
+ATOM 196 N ARG A 13 -6.685 15.497 -3.645 1.00 0.98
+ATOM 197 CA ARG A 13 -5.342 15.913 -4.028 1.00 0.98
+ATOM 198 C ARG A 13 -4.676 14.873 -4.920 1.00 0.98
+ATOM 199 O ARG A 13 -3.988 15.215 -5.881 1.00 0.98
+ATOM 200 CB ARG A 13 -4.488 16.174 -2.796 1.00 0.98
+ATOM 201 CG ARG A 13 -3.128 16.793 -3.074 1.00 0.98
+ATOM 202 CD ARG A 13 -2.410 17.129 -1.818 1.00 0.98
+ATOM 203 NE ARG A 13 -1.082 17.661 -2.079 1.00 0.98
+ATOM 204 CZ ARG A 13 -0.220 18.066 -1.126 1.00 0.98
+ATOM 205 NH1 ARG A 13 -0.559 17.994 0.142 1.00 0.98
+ATOM 206 NH2 ARG A 13 0.968 18.537 -1.466 1.00 0.98
+ATOM 207 H ARG A 13 -6.843 15.140 -2.714 1.00 0.98
+ATOM 208 HA ARG A 13 -5.414 16.850 -4.583 1.00 0.98
+ATOM 209 1HB ARG A 13 -5.022 16.841 -2.120 1.00 0.98
+ATOM 210 2HB ARG A 13 -4.322 15.236 -2.265 1.00 0.98
+ATOM 211 1HG ARG A 13 -2.517 16.089 -3.641 1.00 0.98
+ATOM 212 2HG ARG A 13 -3.256 17.709 -3.652 1.00 0.98
+ATOM 213 1HD ARG A 13 -2.975 17.879 -1.266 1.00 0.98
+ATOM 214 2HD ARG A 13 -2.304 16.233 -1.209 1.00 0.98
+ATOM 215 HE ARG A 13 -0.785 17.733 -3.043 1.00 0.98
+ATOM 216 1HH1 ARG A 13 -1.467 17.634 0.402 1.00 0.98
+ATOM 217 2HH1 ARG A 13 0.087 18.298 0.856 1.00 0.98
+ATOM 218 1HH2 ARG A 13 1.229 18.593 -2.441 1.00 0.98
+ATOM 219 2HH2 ARG A 13 1.613 18.840 -0.753 1.00 0.98
+ATOM 220 N TRP A 14 -4.866 13.519 -4.675 1.00 0.98
+ATOM 221 CA TRP A 14 -4.091 12.494 -5.364 1.00 0.98
+ATOM 222 C TRP A 14 -4.943 11.752 -6.386 1.00 0.98
+ATOM 223 O TRP A 14 -4.481 10.808 -7.026 1.00 0.98
+ATOM 224 CB TRP A 14 -3.479 11.514 -4.361 1.00 0.98
+ATOM 225 CG TRP A 14 -2.418 12.133 -3.503 1.00 0.98
+ATOM 226 CD1 TRP A 14 -2.563 12.583 -2.225 1.00 0.98
+ATOM 227 CD2 TRP A 14 -1.035 12.375 -3.860 1.00 0.98
+ATOM 228 NE1 TRP A 14 -1.372 13.086 -1.763 1.00 0.98
+ATOM 229 CE2 TRP A 14 -0.426 12.967 -2.750 1.00 0.98
+ATOM 230 CE3 TRP A 14 -0.278 12.141 -5.014 1.00 0.98
+ATOM 231 CZ2 TRP A 14 0.911 13.332 -2.755 1.00 0.98
+ATOM 232 CZ3 TRP A 14 1.063 12.507 -5.019 1.00 0.98
+ATOM 233 CH2 TRP A 14 1.641 13.087 -3.918 1.00 0.98
+ATOM 234 H TRP A 14 -5.587 13.237 -4.026 1.00 0.98
+ATOM 235 HA TRP A 14 -3.268 12.978 -5.890 1.00 0.98
+ATOM 236 1HB TRP A 14 -4.260 11.120 -3.710 1.00 0.98
+ATOM 237 2HB TRP A 14 -3.041 10.670 -4.894 1.00 0.98
+ATOM 238 HD1 TRP A 14 -3.488 12.548 -1.654 1.00 0.98
+ATOM 239 HE1 TRP A 14 -1.216 13.480 -0.846 1.00 0.98
+ATOM 240 HZ2 TRP A 14 1.389 13.793 -1.890 1.00 0.98
+ATOM 241 HH2 TRP A 14 2.696 13.361 -3.955 1.00 0.98
+ATOM 242 HZ3 TRP A 14 1.646 12.321 -5.921 1.00 0.98
+ATOM 243 HE3 TRP A 14 -0.732 11.681 -5.891 1.00 0.98
+ATOM 244 N GLY A 15 -6.218 11.964 -6.622 1.00 0.98
+ATOM 245 CA GLY A 15 -7.065 11.225 -7.551 1.00 0.98
+ATOM 246 C GLY A 15 -7.274 9.790 -7.082 1.00 0.98
+ATOM 247 O GLY A 15 -7.266 8.857 -7.886 1.00 0.98
+ATOM 248 H GLY A 15 -6.618 12.723 -6.090 1.00 0.98
+ATOM 249 1HA GLY A 15 -8.028 11.726 -7.643 1.00 0.98
+ATOM 250 2HA GLY A 15 -6.607 11.225 -8.539 1.00 0.98
+ATOM 251 N ILE A 16 -7.248 9.530 -5.774 1.00 0.99
+ATOM 252 CA ILE A 16 -7.457 8.250 -5.108 1.00 0.99
+ATOM 253 C ILE A 16 -8.935 8.007 -4.832 1.00 0.99
+ATOM 254 O ILE A 16 -9.692 8.945 -4.583 1.00 0.99
+ATOM 255 CB ILE A 16 -6.662 8.177 -3.791 1.00 0.99
+ATOM 256 CG1 ILE A 16 -5.154 8.253 -4.045 1.00 0.99
+ATOM 257 CG2 ILE A 16 -7.015 6.902 -3.042 1.00 0.99
+ATOM 258 CD1 ILE A 16 -4.328 8.373 -2.785 1.00 0.99
+ATOM 259 H ILE A 16 -7.082 10.354 -5.215 1.00 0.99
+ATOM 260 HA ILE A 16 -7.098 7.457 -5.762 1.00 0.99
+ATOM 261 HB ILE A 16 -6.920 9.035 -3.172 1.00 0.99
+ATOM 262 1HG2 ILE A 16 -6.460 6.865 -2.105 1.00 0.99
+ATOM 263 2HG2 ILE A 16 -8.084 6.889 -2.830 1.00 0.99
+ATOM 264 3HG2 ILE A 16 -6.755 6.037 -3.652 1.00 0.99
+ATOM 265 1HG1 ILE A 16 -4.831 7.361 -4.582 1.00 0.99
+ATOM 266 2HG1 ILE A 16 -4.934 9.112 -4.679 1.00 0.99
+ATOM 267 1HD1 ILE A 16 -3.270 8.421 -3.047 1.00 0.99
+ATOM 268 2HD1 ILE A 16 -4.611 9.279 -2.249 1.00 0.99
+ATOM 269 3HD1 ILE A 16 -4.504 7.506 -2.151 1.00 0.99
+ATOM 270 N THR A 17 -9.382 6.729 -4.692 1.00 0.99
+ATOM 271 CA THR A 17 -10.735 6.466 -4.214 1.00 0.99
+ATOM 272 C THR A 17 -10.812 6.568 -2.697 1.00 0.99
+ATOM 273 O THR A 17 -9.886 6.169 -1.990 1.00 0.99
+ATOM 274 CB THR A 17 -11.235 5.082 -4.668 1.00 0.99
+ATOM 275 OG1 THR A 17 -10.533 4.050 -3.965 1.00 0.99
+ATOM 276 CG2 THR A 17 -11.019 4.909 -6.164 1.00 0.99
+ATOM 277 H THR A 17 -8.799 5.952 -4.969 1.00 0.99
+ATOM 278 HG1 THR A 17 -9.920 4.448 -3.341 1.00 0.99
+ATOM 279 HA THR A 17 -11.402 7.218 -4.637 1.00 0.99
+ATOM 280 HB THR A 17 -12.299 4.995 -4.448 1.00 0.99
+ATOM 281 1HG2 THR A 17 -11.385 3.930 -6.473 1.00 0.99
+ATOM 282 2HG2 THR A 17 -11.562 5.686 -6.702 1.00 0.99
+ATOM 283 3HG2 THR A 17 -9.957 4.986 -6.389 1.00 0.99
+ATOM 284 N PRO A 18 -11.807 7.278 -2.112 1.00 0.99
+ATOM 285 CA PRO A 18 -11.955 7.462 -0.673 1.00 0.99
+ATOM 286 C PRO A 18 -11.897 6.129 0.062 1.00 0.99
+ATOM 287 O PRO A 18 -11.383 6.045 1.177 1.00 0.99
+ATOM 288 CB PRO A 18 -13.328 8.122 -0.519 1.00 0.99
+ATOM 289 CG PRO A 18 -13.513 8.939 -1.753 1.00 0.99
+ATOM 290 CD PRO A 18 -12.864 8.126 -2.841 1.00 0.99
+ATOM 291 HA PRO A 18 -11.165 8.137 -0.311 1.00 0.99
+ATOM 292 1HB PRO A 18 -14.097 7.344 -0.404 1.00 0.99
+ATOM 293 2HB PRO A 18 -13.343 8.727 0.399 1.00 0.99
+ATOM 294 1HG PRO A 18 -14.583 9.114 -1.936 1.00 0.99
+ATOM 295 2HG PRO A 18 -13.046 9.927 -1.627 1.00 0.99
+ATOM 296 1HD PRO A 18 -13.413 7.185 -2.990 1.00 0.99
+ATOM 297 2HD PRO A 18 -12.901 8.670 -3.795 1.00 0.99
+ATOM 298 N GLU A 19 -12.427 5.032 -0.511 1.00 0.99
+ATOM 299 CA GLU A 19 -12.398 3.698 0.076 1.00 0.99
+ATOM 300 C GLU A 19 -10.967 3.221 0.289 1.00 0.99
+ATOM 301 O GLU A 19 -10.647 2.626 1.318 1.00 0.99
+ATOM 302 CB GLU A 19 -13.159 2.705 -0.805 1.00 0.99
+ATOM 303 CG GLU A 19 -14.667 2.911 -0.820 1.00 0.99
+ATOM 304 CD GLU A 19 -15.384 1.918 -1.692 1.00 0.99
+ATOM 305 OE1 GLU A 19 -16.584 2.009 -1.800 1.00 0.99
+ATOM 306 OE2 GLU A 19 -14.732 1.068 -2.250 1.00 0.99
+ATOM 307 H GLU A 19 -12.842 5.158 -1.423 1.00 0.99
+ATOM 308 HA GLU A 19 -12.887 3.737 1.050 1.00 0.99
+ATOM 309 1HB GLU A 19 -12.801 2.780 -1.832 1.00 0.99
+ATOM 310 2HB GLU A 19 -12.963 1.689 -0.464 1.00 0.99
+ATOM 311 1HG GLU A 19 -15.048 2.805 0.196 1.00 0.99
+ATOM 312 2HG GLU A 19 -14.884 3.910 -1.198 1.00 0.99
+ATOM 313 N GLU A 20 -10.051 3.462 -0.749 1.00 0.99
+ATOM 314 CA GLU A 20 -8.648 3.090 -0.611 1.00 0.99
+ATOM 315 C GLU A 20 -8.000 3.812 0.564 1.00 0.99
+ATOM 316 O GLU A 20 -7.266 3.210 1.347 1.00 0.99
+ATOM 317 CB GLU A 20 -7.886 3.382 -1.905 1.00 0.99
+ATOM 318 CG GLU A 20 -8.295 2.514 -3.087 1.00 0.99
+ATOM 319 CD GLU A 20 -7.703 2.981 -4.388 1.00 0.99
+ATOM 320 OE1 GLU A 20 -7.531 2.170 -5.266 1.00 0.99
+ATOM 321 OE2 GLU A 20 -7.423 4.151 -4.505 1.00 0.99
+ATOM 322 H GLU A 20 -10.357 3.921 -1.596 1.00 0.99
+ATOM 323 HA GLU A 20 -8.591 2.019 -0.420 1.00 0.99
+ATOM 324 1HB GLU A 20 -8.035 4.424 -2.188 1.00 0.99
+ATOM 325 2HB GLU A 20 -6.818 3.239 -1.739 1.00 0.99
+ATOM 326 1HG GLU A 20 -7.956 1.494 -2.909 1.00 0.99
+ATOM 327 2HG GLU A 20 -9.380 2.537 -3.183 1.00 0.99
+ATOM 328 N LEU A 21 -8.202 5.171 0.663 1.00 0.99
+ATOM 329 CA LEU A 21 -7.647 5.967 1.751 1.00 0.99
+ATOM 330 C LEU A 21 -8.156 5.482 3.104 1.00 0.99
+ATOM 331 O LEU A 21 -7.395 5.388 4.067 1.00 0.99
+ATOM 332 CB LEU A 21 -7.977 7.452 1.551 1.00 0.99
+ATOM 333 CG LEU A 21 -7.270 8.160 0.389 1.00 0.99
+ATOM 334 CD1 LEU A 21 -7.878 9.542 0.190 1.00 0.99
+ATOM 335 CD2 LEU A 21 -5.780 8.256 0.683 1.00 0.99
+ATOM 336 H LEU A 21 -8.814 5.613 -0.008 1.00 0.99
+ATOM 337 HA LEU A 21 -6.562 5.865 1.735 1.00 0.99
+ATOM 338 1HB LEU A 21 -9.050 7.542 1.390 1.00 0.99
+ATOM 339 2HB LEU A 21 -7.722 7.983 2.467 1.00 0.99
+ATOM 340 HG LEU A 21 -7.422 7.594 -0.530 1.00 0.99
+ATOM 341 1HD1 LEU A 21 -7.375 10.045 -0.636 1.00 0.99
+ATOM 342 2HD1 LEU A 21 -8.939 9.443 -0.039 1.00 0.99
+ATOM 343 3HD1 LEU A 21 -7.755 10.128 1.100 1.00 0.99
+ATOM 344 1HD2 LEU A 21 -5.277 8.759 -0.143 1.00 0.99
+ATOM 345 2HD2 LEU A 21 -5.627 8.824 1.601 1.00 0.99
+ATOM 346 3HD2 LEU A 21 -5.368 7.254 0.803 1.00 0.99
+ATOM 347 N ARG A 22 -9.489 5.226 3.255 1.00 0.99
+ATOM 348 CA ARG A 22 -10.067 4.718 4.493 1.00 0.99
+ATOM 349 C ARG A 22 -9.385 3.429 4.931 1.00 0.99
+ATOM 350 O ARG A 22 -9.060 3.255 6.106 1.00 0.99
+ATOM 351 CB ARG A 22 -11.561 4.480 4.328 1.00 0.99
+ATOM 352 CG ARG A 22 -12.290 4.076 5.600 1.00 0.99
+ATOM 353 CD ARG A 22 -13.745 3.897 5.365 1.00 0.99
+ATOM 354 NE ARG A 22 -14.430 3.406 6.550 1.00 0.99
+ATOM 355 CZ ARG A 22 -15.753 3.160 6.623 1.00 0.99
+ATOM 356 NH1 ARG A 22 -16.518 3.363 5.573 1.00 0.99
+ATOM 357 NH2 ARG A 22 -16.281 2.714 7.749 1.00 0.99
+ATOM 358 H ARG A 22 -10.088 5.372 2.455 1.00 0.99
+ATOM 359 HA ARG A 22 -9.926 5.467 5.273 1.00 0.99
+ATOM 360 1HB ARG A 22 -12.033 5.387 3.952 1.00 0.99
+ATOM 361 2HB ARG A 22 -11.724 3.695 3.590 1.00 0.99
+ATOM 362 1HG ARG A 22 -11.883 3.134 5.968 1.00 0.99
+ATOM 363 2HG ARG A 22 -12.157 4.849 6.357 1.00 0.99
+ATOM 364 1HD ARG A 22 -14.189 4.852 5.086 1.00 0.99
+ATOM 365 2HD ARG A 22 -13.898 3.177 4.561 1.00 0.99
+ATOM 366 HE ARG A 22 -13.874 3.238 7.378 1.00 0.99
+ATOM 367 1HH1 ARG A 22 -16.114 3.704 4.712 1.00 0.99
+ATOM 368 2HH1 ARG A 22 -17.509 3.179 5.628 1.00 0.99
+ATOM 369 1HH2 ARG A 22 -15.692 2.558 8.556 1.00 0.99
+ATOM 370 2HH2 ARG A 22 -17.271 2.530 7.804 1.00 0.99
+ATOM 371 N GLU A 23 -9.145 2.487 3.957 1.00 0.98
+ATOM 372 CA GLU A 23 -8.451 1.240 4.254 1.00 0.98
+ATOM 373 C GLU A 23 -7.054 1.504 4.800 1.00 0.98
+ATOM 374 O GLU A 23 -6.627 0.876 5.769 1.00 0.98
+ATOM 375 CB GLU A 23 -8.380 0.356 3.008 1.00 0.98
+ATOM 376 CG GLU A 23 -9.719 -0.226 2.577 1.00 0.98
+ATOM 377 CD GLU A 23 -9.629 -1.020 1.303 1.00 0.98
+ATOM 378 OE1 GLU A 23 -10.639 -1.510 0.859 1.00 0.98
+ATOM 379 OE2 GLU A 23 -8.549 -1.136 0.774 1.00 0.98
+ATOM 380 H GLU A 23 -9.443 2.670 3.009 1.00 0.98
+ATOM 381 HA GLU A 23 -9.015 0.705 5.018 1.00 0.98
+ATOM 382 1HB GLU A 23 -7.982 0.933 2.173 1.00 0.98
+ATOM 383 2HB GLU A 23 -7.697 -0.474 3.187 1.00 0.98
+ATOM 384 1HG GLU A 23 -10.085 -0.888 3.362 1.00 0.98
+ATOM 385 2HG GLU A 23 -10.423 0.590 2.417 1.00 0.98
+ATOM 386 N ILE A 24 -6.302 2.500 4.160 1.00 0.99
+ATOM 387 CA ILE A 24 -4.959 2.841 4.614 1.00 0.99
+ATOM 388 C ILE A 24 -4.987 3.434 6.018 1.00 0.99
+ATOM 389 O ILE A 24 -4.236 3.009 6.896 1.00 0.99
+ATOM 390 CB ILE A 24 -4.273 3.815 3.639 1.00 0.99
+ATOM 391 CG1 ILE A 24 -3.913 3.109 2.329 1.00 0.99
+ATOM 392 CG2 ILE A 24 -3.033 4.417 4.282 1.00 0.99
+ATOM 393 CD1 ILE A 24 -3.443 4.046 1.240 1.00 0.99
+ATOM 394 H ILE A 24 -6.688 2.982 3.360 1.00 0.99
+ATOM 395 HA ILE A 24 -4.362 1.930 4.641 1.00 0.99
+ATOM 396 HB ILE A 24 -4.965 4.617 3.385 1.00 0.99
+ATOM 397 1HG2 ILE A 24 -2.560 5.107 3.583 1.00 0.99
+ATOM 398 2HG2 ILE A 24 -3.316 4.953 5.186 1.00 0.99
+ATOM 399 3HG2 ILE A 24 -2.332 3.621 4.535 1.00 0.99
+ATOM 400 1HG1 ILE A 24 -3.126 2.378 2.514 1.00 0.99
+ATOM 401 2HG1 ILE A 24 -4.782 2.564 1.958 1.00 0.99
+ATOM 402 1HD1 ILE A 24 -3.207 3.472 0.343 1.00 0.99
+ATOM 403 2HD1 ILE A 24 -4.232 4.765 1.013 1.00 0.99
+ATOM 404 3HD1 ILE A 24 -2.553 4.576 1.575 1.00 0.99
+ATOM 405 N TRP A 25 -5.851 4.474 6.270 1.00 0.99
+ATOM 406 CA TRP A 25 -5.936 5.168 7.550 1.00 0.99
+ATOM 407 C TRP A 25 -6.279 4.203 8.678 1.00 0.99
+ATOM 408 O TRP A 25 -5.843 4.382 9.815 1.00 0.99
+ATOM 409 CB TRP A 25 -6.970 6.293 7.486 1.00 0.99
+ATOM 410 CG TRP A 25 -6.522 7.505 6.727 1.00 0.99
+ATOM 411 CD1 TRP A 25 -7.318 8.486 6.214 1.00 0.99
+ATOM 412 CD2 TRP A 25 -5.163 7.874 6.389 1.00 0.99
+ATOM 413 NE1 TRP A 25 -6.551 9.435 5.583 1.00 0.99
+ATOM 414 CE2 TRP A 25 -5.231 9.076 5.680 1.00 0.99
+ATOM 415 CE3 TRP A 25 -3.914 7.287 6.627 1.00 0.99
+ATOM 416 CZ2 TRP A 25 -4.095 9.711 5.203 1.00 0.99
+ATOM 417 CZ3 TRP A 25 -2.775 7.925 6.149 1.00 0.99
+ATOM 418 CH2 TRP A 25 -2.864 9.105 5.455 1.00 0.99
+ATOM 419 H TRP A 25 -6.493 4.723 5.532 1.00 0.99
+ATOM 420 HA TRP A 25 -4.967 5.617 7.765 1.00 0.99
+ATOM 421 1HB TRP A 25 -7.880 5.914 7.020 1.00 0.99
+ATOM 422 2HB TRP A 25 -7.225 6.599 8.501 1.00 0.99
+ATOM 423 HD1 TRP A 25 -8.403 8.513 6.294 1.00 0.99
+ATOM 424 HE1 TRP A 25 -6.902 10.263 5.123 1.00 0.99
+ATOM 425 HZ2 TRP A 25 -4.145 10.650 4.651 1.00 0.99
+ATOM 426 HH2 TRP A 25 -1.950 9.578 5.094 1.00 0.99
+ATOM 427 HZ3 TRP A 25 -1.806 7.462 6.338 1.00 0.99
+ATOM 428 HE3 TRP A 25 -3.837 6.350 7.177 1.00 0.99
+ATOM 429 N LYS A 26 -7.066 3.121 8.429 1.00 0.98
+ATOM 430 CA LYS A 26 -7.406 2.108 9.421 1.00 0.98
+ATOM 431 C LYS A 26 -6.164 1.365 9.897 1.00 0.98
+ATOM 432 O LYS A 26 -6.146 0.806 10.994 1.00 0.98
+ATOM 433 CB LYS A 26 -8.429 1.121 8.856 1.00 0.98
+ATOM 434 CG LYS A 26 -9.840 1.680 8.733 1.00 0.98
+ATOM 435 CD LYS A 26 -10.795 0.648 8.153 1.00 0.98
+ATOM 436 CE LYS A 26 -12.198 1.215 7.999 1.00 0.98
+ATOM 437 NZ LYS A 26 -13.133 0.231 7.389 1.00 0.98
+ATOM 438 H LYS A 26 -7.422 3.028 7.489 1.00 0.98
+ATOM 439 HA LYS A 26 -7.849 2.604 10.284 1.00 0.98
+ATOM 440 1HB LYS A 26 -8.111 0.794 7.866 1.00 0.98
+ATOM 441 2HB LYS A 26 -8.473 0.238 9.494 1.00 0.98
+ATOM 442 1HG LYS A 26 -10.199 1.982 9.718 1.00 0.98
+ATOM 443 2HG LYS A 26 -9.829 2.556 8.086 1.00 0.98
+ATOM 444 1HD LYS A 26 -10.433 0.326 7.175 1.00 0.98
+ATOM 445 2HD LYS A 26 -10.834 -0.221 8.810 1.00 0.98
+ATOM 446 1HE LYS A 26 -12.669 1.450 9.006 1.00 0.98
+ATOM 447 2HE LYS A 26 -12.251 2.049 7.399 1.00 0.98
+ATOM 448 1HZ LYS A 26 -14.137 0.590 7.334 1.00 0.98
+ATOM 449 2HZ LYS A 26 -12.882 -0.085 6.503 1.00 0.98
+ATOM 450 3HZ LYS A 26 -13.274 -0.645 8.004 1.00 0.98
+ATOM 451 N TRP A 27 -5.037 1.388 9.106 1.00 0.98
+ATOM 452 CA TRP A 27 -3.835 0.612 9.388 1.00 0.98
+ATOM 453 C TRP A 27 -2.745 1.485 9.998 1.00 0.98
+ATOM 454 O TRP A 27 -1.697 0.988 10.409 1.00 0.98
+ATOM 455 CB TRP A 27 -3.313 -0.057 8.116 1.00 0.98
+ATOM 456 CG TRP A 27 -4.233 -1.102 7.559 1.00 0.98
+ATOM 457 CD1 TRP A 27 -5.075 -0.964 6.497 1.00 0.98
+ATOM 458 CD2 TRP A 27 -4.408 -2.458 8.037 1.00 0.98
+ATOM 459 NE1 TRP A 27 -5.760 -2.134 6.281 1.00 0.98
+ATOM 460 CE2 TRP A 27 -5.364 -3.059 7.214 1.00 0.98
+ATOM 461 CE3 TRP A 27 -3.839 -3.197 9.081 1.00 0.98
+ATOM 462 CZ2 TRP A 27 -5.770 -4.372 7.399 1.00 0.98
+ATOM 463 CZ3 TRP A 27 -4.246 -4.513 9.267 1.00 0.98
+ATOM 464 CH2 TRP A 27 -5.187 -5.085 8.447 1.00 0.98
+ATOM 465 H TRP A 27 -5.048 2.004 8.306 1.00 0.98
+ATOM 466 HA TRP A 27 -4.089 -0.170 10.104 1.00 0.98
+ATOM 467 1HB TRP A 27 -3.152 0.701 7.349 1.00 0.98
+ATOM 468 2HB TRP A 27 -2.350 -0.524 8.322 1.00 0.98
+ATOM 469 HD1 TRP A 27 -5.188 -0.057 5.906 1.00 0.98
+ATOM 470 HE1 TRP A 27 -6.443 -2.291 5.555 1.00 0.98
+ATOM 471 HZ2 TRP A 27 -6.515 -4.843 6.759 1.00 0.98
+ATOM 472 HH2 TRP A 27 -5.483 -6.119 8.621 1.00 0.98
+ATOM 473 HZ3 TRP A 27 -3.798 -5.082 10.082 1.00 0.98
+ATOM 474 HE3 TRP A 27 -3.092 -2.749 9.735 1.00 0.98
+ATOM 475 N ILE A 28 -2.837 2.915 9.946 1.00 0.98
+ATOM 476 CA ILE A 28 -1.792 3.854 10.335 1.00 0.98
+ATOM 477 C ILE A 28 -1.554 3.821 11.839 1.00 0.98
+ATOM 478 O ILE A 28 -0.991 4.756 12.409 1.00 0.98
+ATOM 479 CB ILE A 28 -2.140 5.276 9.860 1.00 0.98
+ATOM 480 CG1 ILE A 28 -2.051 5.358 8.334 1.00 0.98
+ATOM 481 CG2 ILE A 28 -1.216 6.298 10.505 1.00 0.98
+ATOM 482 CD1 ILE A 28 -2.581 6.652 7.760 1.00 0.98
+ATOM 483 H ILE A 28 -3.758 3.267 9.727 1.00 0.98
+ATOM 484 HA ILE A 28 -0.867 3.566 9.838 1.00 0.98
+ATOM 485 HB ILE A 28 -3.170 5.511 10.129 1.00 0.98
+ATOM 486 1HG2 ILE A 28 -1.475 7.296 10.153 1.00 0.98
+ATOM 487 2HG2 ILE A 28 -1.327 6.256 11.588 1.00 0.98
+ATOM 488 3HG2 ILE A 28 -0.183 6.075 10.238 1.00 0.98
+ATOM 489 1HG1 ILE A 28 -1.013 5.246 8.024 1.00 0.98
+ATOM 490 2HG1 ILE A 28 -2.613 4.534 7.892 1.00 0.98
+ATOM 491 1HD1 ILE A 28 -2.485 6.634 6.674 1.00 0.98
+ATOM 492 2HD1 ILE A 28 -3.632 6.766 8.029 1.00 0.98
+ATOM 493 3HD1 ILE A 28 -2.010 7.488 8.160 1.00 0.98
+ATOM 494 N ASP A 29 -1.560 2.721 12.520 1.00 0.96
+ATOM 495 CA ASP A 29 -1.134 2.519 13.899 1.00 0.96
+ATOM 496 C ASP A 29 0.255 1.896 13.960 1.00 0.96
+ATOM 497 O ASP A 29 0.641 1.127 13.079 1.00 0.96
+ATOM 498 CB ASP A 29 -2.158 1.623 14.600 1.00 0.96
+ATOM 499 CG ASP A 29 -1.859 1.335 16.065 1.00 0.96
+ATOM 500 OD1 ASP A 29 -2.254 0.296 16.539 1.00 0.96
+ATOM 501 OD2 ASP A 29 -1.239 2.157 16.696 1.00 0.96
+ATOM 502 H ASP A 29 -1.943 1.942 12.004 1.00 0.96
+ATOM 503 HA ASP A 29 -1.073 3.492 14.390 1.00 0.96
+ATOM 504 1HB ASP A 29 -3.137 2.098 14.530 1.00 0.96
+ATOM 505 2HB ASP A 29 -2.215 0.676 14.063 1.00 0.96
+ATOM 506 N PRO A 30 1.080 2.240 14.967 1.00 0.95
+ATOM 507 CA PRO A 30 2.437 1.713 15.045 1.00 0.95
+ATOM 508 C PRO A 30 2.437 0.190 15.071 1.00 0.95
+ATOM 509 O PRO A 30 3.363 -0.449 14.572 1.00 0.95
+ATOM 510 CB PRO A 30 2.992 2.294 16.349 1.00 0.95
+ATOM 511 CG PRO A 30 2.309 3.605 16.541 1.00 0.95
+ATOM 512 CD PRO A 30 0.898 3.364 16.072 1.00 0.95
+ATOM 513 HA PRO A 30 3.019 2.085 14.188 1.00 0.95
+ATOM 514 1HB PRO A 30 2.797 1.587 17.168 1.00 0.95
+ATOM 515 2HB PRO A 30 4.084 2.392 16.262 1.00 0.95
+ATOM 516 1HG PRO A 30 2.358 3.908 17.597 1.00 0.95
+ATOM 517 2HG PRO A 30 2.819 4.388 15.962 1.00 0.95
+ATOM 518 1HD PRO A 30 0.368 2.712 16.781 1.00 0.95
+ATOM 519 2HD PRO A 30 0.343 4.313 16.037 1.00 0.95
+ATOM 520 N ASP A 31 1.280 -0.461 15.574 1.00 0.94
+ATOM 521 CA ASP A 31 1.194 -1.912 15.686 1.00 0.94
+ATOM 522 C ASP A 31 0.820 -2.546 14.352 1.00 0.94
+ATOM 523 O ASP A 31 1.131 -3.710 14.099 1.00 0.94
+ATOM 524 CB ASP A 31 0.183 -2.314 16.763 1.00 0.94
+ATOM 525 CG ASP A 31 0.613 -1.902 18.164 1.00 0.94
+ATOM 526 OD1 ASP A 31 -0.151 -2.098 19.080 1.00 0.94
+ATOM 527 OD2 ASP A 31 1.699 -1.394 18.305 1.00 0.94
+ATOM 528 H ASP A 31 0.511 0.104 15.906 1.00 0.94
+ATOM 529 HA ASP A 31 2.177 -2.297 15.962 1.00 0.94
+ATOM 530 1HB ASP A 31 -0.782 -1.856 16.544 1.00 0.94
+ATOM 531 2HB ASP A 31 0.044 -3.395 16.744 1.00 0.94
+ATOM 532 N ASN A 32 -0.015 -1.818 13.479 1.00 0.97
+ATOM 533 CA ASN A 32 -0.472 -2.371 12.210 1.00 0.97
+ATOM 534 C ASN A 32 0.304 -1.781 11.040 1.00 0.97
+ATOM 535 O ASN A 32 -0.206 -1.698 9.923 1.00 0.97
+ATOM 536 CB ASN A 32 -1.966 -2.161 12.043 1.00 0.97
+ATOM 537 CG ASN A 32 -2.775 -2.911 13.064 1.00 0.97
+ATOM 538 OD1 ASN A 32 -4.011 -2.887 13.034 1.00 0.97
+ATOM 539 ND2 ASN A 32 -2.103 -3.576 13.968 1.00 0.97
+ATOM 540 H ASN A 32 -0.197 -0.847 13.686 1.00 0.97
+ATOM 541 HA ASN A 32 -0.284 -3.446 12.213 1.00 0.97
+ATOM 542 1HB ASN A 32 -2.195 -1.097 12.125 1.00 0.97
+ATOM 543 2HB ASN A 32 -2.269 -2.485 11.047 1.00 0.97
+ATOM 544 1HD2 ASN A 32 -2.589 -4.094 14.673 1.00 0.97
+ATOM 545 2HD2 ASN A 32 -1.104 -3.567 13.955 1.00 0.97
+ATOM 546 N PHE A 33 1.585 -1.332 11.271 1.00 0.98
+ATOM 547 CA PHE A 33 2.414 -0.684 10.262 1.00 0.98
+ATOM 548 C PHE A 33 2.571 -1.565 9.029 1.00 0.98
+ATOM 549 O PHE A 33 2.496 -1.085 7.898 1.00 0.98
+ATOM 550 CB PHE A 33 3.784 -0.329 10.844 1.00 0.98
+ATOM 551 CG PHE A 33 4.706 0.377 9.891 1.00 0.98
+ATOM 552 CD1 PHE A 33 4.411 1.654 9.436 1.00 0.98
+ATOM 553 CD2 PHE A 33 5.871 -0.232 9.449 1.00 0.98
+ATOM 554 CE1 PHE A 33 5.259 2.305 8.560 1.00 0.98
+ATOM 555 CE2 PHE A 33 6.720 0.416 8.574 1.00 0.98
+ATOM 556 CZ PHE A 33 6.414 1.685 8.129 1.00 0.98
+ATOM 557 H PHE A 33 1.972 -1.510 12.186 1.00 0.98
+ATOM 558 HA PHE A 33 1.923 0.237 9.950 1.00 0.98
+ATOM 559 1HB PHE A 33 3.645 0.306 11.717 1.00 0.98
+ATOM 560 2HB PHE A 33 4.276 -1.243 11.179 1.00 0.98
+ATOM 561 HD1 PHE A 33 3.499 2.143 9.777 1.00 0.98
+ATOM 562 HD2 PHE A 33 6.113 -1.236 9.801 1.00 0.98
+ATOM 563 HE1 PHE A 33 5.015 3.307 8.210 1.00 0.98
+ATOM 564 HE2 PHE A 33 7.632 -0.074 8.234 1.00 0.98
+ATOM 565 HZ PHE A 33 7.083 2.198 7.439 1.00 0.98
+ATOM 566 N ASP A 34 2.753 -2.903 9.185 1.00 0.97
+ATOM 567 CA ASP A 34 2.923 -3.814 8.059 1.00 0.97
+ATOM 568 C ASP A 34 1.705 -3.793 7.145 1.00 0.97
+ATOM 569 O ASP A 34 1.835 -3.729 5.923 1.00 0.97
+ATOM 570 CB ASP A 34 3.190 -5.238 8.554 1.00 0.97
+ATOM 571 CG ASP A 34 4.553 -5.395 9.214 1.00 0.97
+ATOM 572 OD1 ASP A 34 4.795 -6.426 9.795 1.00 0.97
+ATOM 573 OD2 ASP A 34 5.340 -4.483 9.130 1.00 0.97
+ATOM 574 H ASP A 34 2.763 -3.275 10.125 1.00 0.97
+ATOM 575 HA ASP A 34 3.777 -3.477 7.468 1.00 0.97
+ATOM 576 1HB ASP A 34 2.420 -5.522 9.271 1.00 0.97
+ATOM 577 2HB ASP A 34 3.127 -5.930 7.714 1.00 0.97
+ATOM 578 N LYS A 35 0.438 -3.945 7.743 1.00 0.99
+ATOM 579 CA LYS A 35 -0.789 -3.877 6.959 1.00 0.99
+ATOM 580 C LYS A 35 -0.889 -2.558 6.204 1.00 0.99
+ATOM 581 O LYS A 35 -1.290 -2.525 5.041 1.00 0.99
+ATOM 582 CB LYS A 35 -2.013 -4.078 7.854 1.00 0.99
+ATOM 583 CG LYS A 35 -2.163 -5.494 8.393 1.00 0.99
+ATOM 584 CD LYS A 35 -3.419 -5.633 9.241 1.00 0.99
+ATOM 585 CE LYS A 35 -3.553 -7.038 9.808 1.00 0.99
+ATOM 586 NZ LYS A 35 -4.754 -7.177 10.675 1.00 0.99
+ATOM 587 H LYS A 35 0.375 -4.028 8.748 1.00 0.99
+ATOM 588 HA LYS A 35 -0.774 -4.682 6.223 1.00 0.99
+ATOM 589 1HB LYS A 35 -1.959 -3.400 8.707 1.00 0.99
+ATOM 590 2HB LYS A 35 -2.918 -3.833 7.299 1.00 0.99
+ATOM 591 1HG LYS A 35 -2.218 -6.196 7.560 1.00 0.99
+ATOM 592 2HG LYS A 35 -1.295 -5.747 9.000 1.00 0.99
+ATOM 593 1HD LYS A 35 -3.382 -4.919 10.065 1.00 0.99
+ATOM 594 2HD LYS A 35 -4.295 -5.413 8.632 1.00 0.99
+ATOM 595 1HE LYS A 35 -3.717 -7.808 8.989 1.00 0.99
+ATOM 596 2HE LYS A 35 -2.759 -7.333 10.393 1.00 0.99
+ATOM 597 1HZ LYS A 35 -4.898 -8.174 11.029 1.00 0.99
+ATOM 598 2HZ LYS A 35 -4.780 -6.582 11.446 1.00 0.99
+ATOM 599 3HZ LYS A 35 -5.676 -7.027 10.135 1.00 0.99
+ATOM 600 N PHE A 36 -0.563 -1.409 6.919 1.00 0.99
+ATOM 601 CA PHE A 36 -0.563 -0.097 6.284 1.00 0.99
+ATOM 602 C PHE A 36 0.365 -0.069 5.076 1.00 0.99
+ATOM 603 O PHE A 36 -0.023 0.369 3.993 1.00 0.99
+ATOM 604 CB PHE A 36 -0.150 0.985 7.284 1.00 0.99
+ATOM 605 CG PHE A 36 0.072 2.338 6.671 1.00 0.99
+ATOM 606 CD1 PHE A 36 -1.003 3.130 6.295 1.00 0.99
+ATOM 607 CD2 PHE A 36 1.355 2.823 6.469 1.00 0.99
+ATOM 608 CE1 PHE A 36 -0.800 4.375 5.731 1.00 0.99
+ATOM 609 CE2 PHE A 36 1.561 4.067 5.906 1.00 0.99
+ATOM 610 CZ PHE A 36 0.483 4.843 5.536 1.00 0.99
+ATOM 611 H PHE A 36 -0.295 -1.486 7.890 1.00 0.99
+ATOM 612 HA PHE A 36 -1.571 0.116 5.931 1.00 0.99
+ATOM 613 1HB PHE A 36 -0.919 1.080 8.049 1.00 0.99
+ATOM 614 2HB PHE A 36 0.772 0.682 7.783 1.00 0.99
+ATOM 615 HD1 PHE A 36 -2.017 2.759 6.449 1.00 0.99
+ATOM 616 HD2 PHE A 36 2.208 2.209 6.761 1.00 0.99
+ATOM 617 HE1 PHE A 36 -1.654 4.986 5.440 1.00 0.99
+ATOM 618 HE2 PHE A 36 2.575 4.436 5.753 1.00 0.99
+ATOM 619 HZ PHE A 36 0.644 5.825 5.092 1.00 0.99
+ATOM 620 N LEU A 37 1.645 -0.526 5.261 1.00 0.99
+ATOM 621 CA LEU A 37 2.636 -0.527 4.191 1.00 0.99
+ATOM 622 C LEU A 37 2.161 -1.353 3.001 1.00 0.99
+ATOM 623 O LEU A 37 2.368 -0.974 1.848 1.00 0.99
+ATOM 624 CB LEU A 37 3.980 -1.057 4.708 1.00 0.99
+ATOM 625 CG LEU A 37 4.740 -0.147 5.681 1.00 0.99
+ATOM 626 CD1 LEU A 37 5.960 -0.883 6.217 1.00 0.99
+ATOM 627 CD2 LEU A 37 5.145 1.135 4.968 1.00 0.99
+ATOM 628 H LEU A 37 1.892 -0.913 6.161 1.00 0.99
+ATOM 629 HA LEU A 37 2.787 0.499 3.859 1.00 0.99
+ATOM 630 1HB LEU A 37 3.802 -2.005 5.212 1.00 0.99
+ATOM 631 2HB LEU A 37 4.627 -1.240 3.851 1.00 0.99
+ATOM 632 HG LEU A 37 4.099 0.097 6.529 1.00 0.99
+ATOM 633 1HD1 LEU A 37 6.501 -0.237 6.909 1.00 0.99
+ATOM 634 2HD1 LEU A 37 5.641 -1.785 6.740 1.00 0.99
+ATOM 635 3HD1 LEU A 37 6.614 -1.155 5.389 1.00 0.99
+ATOM 636 1HD2 LEU A 37 5.685 1.782 5.660 1.00 0.99
+ATOM 637 2HD2 LEU A 37 5.787 0.892 4.121 1.00 0.99
+ATOM 638 3HD2 LEU A 37 4.253 1.649 4.611 1.00 0.99
+ATOM 639 N GLU A 38 1.495 -2.537 3.234 1.00 0.98
+ATOM 640 CA GLU A 38 0.959 -3.360 2.156 1.00 0.98
+ATOM 641 C GLU A 38 -0.121 -2.616 1.380 1.00 0.98
+ATOM 642 O GLU A 38 -0.135 -2.629 0.150 1.00 0.98
+ATOM 643 CB GLU A 38 0.403 -4.673 2.711 1.00 0.98
+ATOM 644 CG GLU A 38 1.469 -5.661 3.163 1.00 0.98
+ATOM 645 CD GLU A 38 0.891 -6.922 3.742 1.00 0.98
+ATOM 646 OE1 GLU A 38 1.649 -7.765 4.156 1.00 0.98
+ATOM 647 OE2 GLU A 38 -0.312 -7.042 3.769 1.00 0.98
+ATOM 648 H GLU A 38 1.378 -2.840 4.190 1.00 0.98
+ATOM 649 HA GLU A 38 1.768 -3.595 1.465 1.00 0.98
+ATOM 650 1HB GLU A 38 -0.244 -4.466 3.563 1.00 0.98
+ATOM 651 2HB GLU A 38 -0.205 -5.163 1.950 1.00 0.98
+ATOM 652 1HG GLU A 38 2.084 -5.934 2.305 1.00 0.98
+ATOM 653 2HG GLU A 38 2.085 -5.190 3.929 1.00 0.98
+ATOM 654 N GLU A 39 -1.115 -1.982 2.123 1.00 0.99
+ATOM 655 CA GLU A 39 -2.167 -1.225 1.455 1.00 0.99
+ATOM 656 C GLU A 39 -1.588 -0.074 0.644 1.00 0.99
+ATOM 657 O GLU A 39 -2.021 0.188 -0.478 1.00 0.99
+ATOM 658 CB GLU A 39 -3.188 -0.710 2.472 1.00 0.99
+ATOM 659 CG GLU A 39 -4.032 -1.808 3.106 1.00 0.99
+ATOM 660 CD GLU A 39 -5.046 -1.277 4.080 1.00 0.99
+ATOM 661 OE1 GLU A 39 -5.754 -2.064 4.659 1.00 0.99
+ATOM 662 OE2 GLU A 39 -5.114 -0.081 4.244 1.00 0.99
+ATOM 663 H GLU A 39 -1.091 -2.004 3.133 1.00 0.99
+ATOM 664 HA GLU A 39 -2.689 -1.892 0.768 1.00 0.99
+ATOM 665 1HB GLU A 39 -2.675 -0.179 3.274 1.00 0.99
+ATOM 666 2HB GLU A 39 -3.864 -0.004 1.991 1.00 0.99
+ATOM 667 1HG GLU A 39 -4.566 -2.341 2.319 1.00 0.99
+ATOM 668 2HG GLU A 39 -3.375 -2.493 3.642 1.00 0.99
+ATOM 669 N LEU A 40 -0.588 0.696 1.259 1.00 0.99
+ATOM 670 CA LEU A 40 0.073 1.783 0.548 1.00 0.99
+ATOM 671 C LEU A 40 0.734 1.284 -0.732 1.00 0.99
+ATOM 672 O LEU A 40 0.612 1.905 -1.788 1.00 0.99
+ATOM 673 CB LEU A 40 1.111 2.460 1.453 1.00 0.99
+ATOM 674 CG LEU A 40 1.859 3.668 0.875 1.00 0.99
+ATOM 675 CD1 LEU A 40 0.860 4.762 0.522 1.00 0.99
+ATOM 676 CD2 LEU A 40 2.879 4.166 1.889 1.00 0.99
+ATOM 677 H LEU A 40 -0.299 0.475 2.201 1.00 0.99
+ATOM 678 HA LEU A 40 -0.676 2.528 0.281 1.00 0.99
+ATOM 679 1HB LEU A 40 0.603 2.786 2.359 1.00 0.99
+ATOM 680 2HB LEU A 40 1.854 1.711 1.728 1.00 0.99
+ATOM 681 HG LEU A 40 2.371 3.376 -0.041 1.00 0.99
+ATOM 682 1HD1 LEU A 40 1.391 5.621 0.111 1.00 0.99
+ATOM 683 2HD1 LEU A 40 0.154 4.385 -0.219 1.00 0.99
+ATOM 684 3HD1 LEU A 40 0.319 5.065 1.418 1.00 0.99
+ATOM 685 1HD2 LEU A 40 3.411 5.025 1.478 1.00 0.99
+ATOM 686 2HD2 LEU A 40 2.367 4.461 2.805 1.00 0.99
+ATOM 687 3HD2 LEU A 40 3.590 3.370 2.110 1.00 0.99
+ATOM 688 N GLU A 41 1.466 0.116 -0.654 1.00 0.99
+ATOM 689 CA GLU A 41 2.111 -0.485 -1.815 1.00 0.99
+ATOM 690 C GLU A 41 1.106 -0.746 -2.930 1.00 0.99
+ATOM 691 O GLU A 41 1.376 -0.473 -4.099 1.00 0.99
+ATOM 692 CB GLU A 41 2.812 -1.787 -1.424 1.00 0.99
+ATOM 693 CG GLU A 41 4.114 -1.589 -0.660 1.00 0.99
+ATOM 694 CD GLU A 41 4.741 -2.884 -0.226 1.00 0.99
+ATOM 695 OE1 GLU A 41 5.826 -2.850 0.303 1.00 0.99
+ATOM 696 OE2 GLU A 41 4.134 -3.911 -0.425 1.00 0.99
+ATOM 697 H GLU A 41 1.545 -0.340 0.244 1.00 0.99
+ATOM 698 HA GLU A 41 2.859 0.211 -2.194 1.00 0.99
+ATOM 699 1HB GLU A 41 2.148 -2.388 -0.802 1.00 0.99
+ATOM 700 2HB GLU A 41 3.035 -2.366 -2.320 1.00 0.99
+ATOM 701 1HG GLU A 41 4.825 -1.068 -1.303 1.00 0.99
+ATOM 702 2HG GLU A 41 3.913 -0.995 0.232 1.00 0.99
+ATOM 703 N GLU A 42 -0.102 -1.315 -2.583 1.00 0.97
+ATOM 704 CA GLU A 42 -1.126 -1.589 -3.584 1.00 0.97
+ATOM 705 C GLU A 42 -1.582 -0.308 -4.271 1.00 0.97
+ATOM 706 O GLU A 42 -1.749 -0.271 -5.490 1.00 0.97
+ATOM 707 CB GLU A 42 -2.319 -2.306 -2.948 1.00 0.97
+ATOM 708 CG GLU A 42 -2.032 -3.739 -2.520 1.00 0.97
+ATOM 709 CD GLU A 42 -3.197 -4.386 -1.825 1.00 0.97
+ATOM 710 OE1 GLU A 42 -3.071 -5.518 -1.424 1.00 0.97
+ATOM 711 OE2 GLU A 42 -4.214 -3.747 -1.694 1.00 0.97
+ATOM 712 H GLU A 42 -0.284 -1.545 -1.616 1.00 0.97
+ATOM 713 HA GLU A 42 -0.701 -2.245 -4.343 1.00 0.97
+ATOM 714 1HB GLU A 42 -2.650 -1.755 -2.068 1.00 0.97
+ATOM 715 2HB GLU A 42 -3.150 -2.328 -3.653 1.00 0.97
+ATOM 716 1HG GLU A 42 -1.795 -4.331 -3.404 1.00 0.97
+ATOM 717 2HG GLU A 42 -1.186 -3.739 -1.833 1.00 0.97
+ATOM 718 N ILE A 43 -1.803 0.809 -3.474 1.00 0.98
+ATOM 719 CA ILE A 43 -2.249 2.082 -4.027 1.00 0.98
+ATOM 720 C ILE A 43 -1.174 2.708 -4.906 1.00 0.98
+ATOM 721 O ILE A 43 -1.450 3.150 -6.021 1.00 0.98
+ATOM 722 CB ILE A 43 -2.666 3.055 -2.909 1.00 0.98
+ATOM 723 CG1 ILE A 43 -3.944 2.570 -2.220 1.00 0.98
+ATOM 724 CG2 ILE A 43 -2.859 4.455 -3.471 1.00 0.98
+ATOM 725 CD1 ILE A 43 -4.291 3.338 -0.966 1.00 0.98
+ATOM 726 H ILE A 43 -1.659 0.729 -2.477 1.00 0.98
+ATOM 727 HA ILE A 43 -3.130 1.902 -4.642 1.00 0.98
+ATOM 728 HB ILE A 43 -1.888 3.084 -2.146 1.00 0.98
+ATOM 729 1HG2 ILE A 43 -3.150 5.132 -2.668 1.00 0.98
+ATOM 730 2HG2 ILE A 43 -1.926 4.800 -3.916 1.00 0.98
+ATOM 731 3HG2 ILE A 43 -3.639 4.437 -4.232 1.00 0.98
+ATOM 732 1HG1 ILE A 43 -4.782 2.649 -2.912 1.00 0.98
+ATOM 733 2HG1 ILE A 43 -3.837 1.517 -1.956 1.00 0.98
+ATOM 734 1HD1 ILE A 43 -5.209 2.936 -0.535 1.00 0.98
+ATOM 735 2HD1 ILE A 43 -3.479 3.243 -0.244 1.00 0.98
+ATOM 736 3HD1 ILE A 43 -4.437 4.389 -1.212 1.00 0.98
+ATOM 737 N LEU A 44 0.100 2.742 -4.424 1.00 0.98
+ATOM 738 CA LEU A 44 1.215 3.357 -5.134 1.00 0.98
+ATOM 739 C LEU A 44 1.470 2.663 -6.468 1.00 0.98
+ATOM 740 O LEU A 44 1.953 3.280 -7.417 1.00 0.98
+ATOM 741 CB LEU A 44 2.481 3.330 -4.268 1.00 0.98
+ATOM 742 CG LEU A 44 2.480 4.236 -3.030 1.00 0.98
+ATOM 743 CD1 LEU A 44 3.737 3.979 -2.211 1.00 0.98
+ATOM 744 CD2 LEU A 44 2.398 5.691 -3.466 1.00 0.98
+ATOM 745 H LEU A 44 0.273 2.280 -3.543 1.00 0.98
+ATOM 746 HA LEU A 44 0.967 4.399 -5.331 1.00 0.98
+ATOM 747 1HB LEU A 44 2.641 2.307 -3.930 1.00 0.98
+ATOM 748 2HB LEU A 44 3.327 3.621 -4.891 1.00 0.98
+ATOM 749 HG LEU A 44 1.619 3.996 -2.405 1.00 0.98
+ATOM 750 1HD1 LEU A 44 3.736 4.623 -1.331 1.00 0.98
+ATOM 751 2HD1 LEU A 44 3.760 2.936 -1.896 1.00 0.98
+ATOM 752 3HD1 LEU A 44 4.616 4.195 -2.817 1.00 0.98
+ATOM 753 1HD2 LEU A 44 2.396 6.335 -2.586 1.00 0.98
+ATOM 754 2HD2 LEU A 44 3.259 5.933 -4.091 1.00 0.98
+ATOM 755 3HD2 LEU A 44 1.481 5.850 -4.034 1.00 0.98
+ATOM 756 N LYS A 45 1.172 1.283 -6.578 1.00 0.97
+ATOM 757 CA LYS A 45 1.363 0.546 -7.821 1.00 0.97
+ATOM 758 C LYS A 45 0.459 1.082 -8.923 1.00 0.97
+ATOM 759 O LYS A 45 0.740 0.908 -10.109 1.00 0.97
+ATOM 760 CB LYS A 45 1.115 -0.948 -7.607 1.00 0.97
+ATOM 761 CG LYS A 45 2.227 -1.666 -6.853 1.00 0.97
+ATOM 762 CD LYS A 45 1.893 -3.136 -6.645 1.00 0.97
+ATOM 763 CE LYS A 45 2.999 -3.853 -5.885 1.00 0.97
+ATOM 764 NZ LYS A 45 2.659 -5.277 -5.620 1.00 0.97
+ATOM 765 H LYS A 45 0.811 0.799 -5.769 1.00 0.97
+ATOM 766 HA LYS A 45 2.397 0.676 -8.145 1.00 0.97
+ATOM 767 1HB LYS A 45 0.189 -1.087 -7.049 1.00 0.97
+ATOM 768 2HB LYS A 45 0.994 -1.439 -8.572 1.00 0.97
+ATOM 769 1HG LYS A 45 3.157 -1.590 -7.417 1.00 0.97
+ATOM 770 2HG LYS A 45 2.372 -1.194 -5.882 1.00 0.97
+ATOM 771 1HD LYS A 45 0.963 -3.222 -6.083 1.00 0.97
+ATOM 772 2HD LYS A 45 1.758 -3.619 -7.613 1.00 0.97
+ATOM 773 1HE LYS A 45 3.957 -3.910 -6.493 1.00 0.97
+ATOM 774 2HE LYS A 45 3.208 -3.446 -4.964 1.00 0.97
+ATOM 775 1HZ LYS A 45 3.452 -5.812 -5.145 1.00 0.97
+ATOM 776 2HZ LYS A 45 1.850 -5.420 -5.097 1.00 0.97
+ATOM 777 3HZ LYS A 45 2.551 -5.854 -6.526 1.00 0.97
+ATOM 778 N ARG A 46 -0.724 1.754 -8.555 1.00 0.96
+ATOM 779 CA ARG A 46 -1.681 2.250 -9.537 1.00 0.96
+ATOM 780 C ARG A 46 -1.378 3.692 -9.923 1.00 0.96
+ATOM 781 O ARG A 46 -2.028 4.259 -10.801 1.00 0.96
+ATOM 782 CB ARG A 46 -3.101 2.154 -8.998 1.00 0.96
+ATOM 783 CG ARG A 46 -3.589 0.737 -8.740 1.00 0.96
+ATOM 784 CD ARG A 46 -4.976 0.722 -8.208 1.00 0.96
+ATOM 785 NE ARG A 46 -5.436 -0.629 -7.931 1.00 0.96
+ATOM 786 CZ ARG A 46 -6.639 -0.936 -7.408 1.00 0.96
+ATOM 787 NH1 ARG A 46 -7.490 0.021 -7.113 1.00 0.96
+ATOM 788 NH2 ARG A 46 -6.964 -2.199 -7.193 1.00 0.96
+ATOM 789 H ARG A 46 -0.858 1.980 -7.580 1.00 0.96
+ATOM 790 HA ARG A 46 -1.612 1.629 -10.431 1.00 0.96
+ATOM 791 1HB ARG A 46 -3.174 2.703 -8.061 1.00 0.96
+ATOM 792 2HB ARG A 46 -3.792 2.617 -9.704 1.00 0.96
+ATOM 793 1HG ARG A 46 -3.573 0.171 -9.672 1.00 0.96
+ATOM 794 2HG ARG A 46 -2.937 0.255 -8.011 1.00 0.96
+ATOM 795 1HD ARG A 46 -5.016 1.293 -7.281 1.00 0.96
+ATOM 796 2HD ARG A 46 -5.650 1.168 -8.939 1.00 0.96
+ATOM 797 HE ARG A 46 -4.808 -1.393 -8.145 1.00 0.96
+ATOM 798 1HH1 ARG A 46 -7.242 0.987 -7.277 1.00 0.96
+ATOM 799 2HH1 ARG A 46 -8.392 -0.209 -6.721 1.00 0.96
+ATOM 800 1HH2 ARG A 46 -6.309 -2.935 -7.420 1.00 0.96
+ATOM 801 2HH2 ARG A 46 -7.865 -2.428 -6.802 1.00 0.96
+ATOM 802 N MET A 47 -0.481 4.422 -9.183 1.00 0.98
+ATOM 803 CA MET A 47 -0.212 5.840 -9.393 1.00 0.98
+ATOM 804 C MET A 47 1.004 6.044 -10.287 1.00 0.98
+ATOM 805 O MET A 47 1.906 5.207 -10.325 1.00 0.98
+ATOM 806 CB MET A 47 -0.015 6.546 -8.053 1.00 0.98
+ATOM 807 CG MET A 47 -1.230 6.552 -7.136 1.00 0.98
+ATOM 808 SD MET A 47 -0.880 7.278 -5.522 1.00 0.98
+ATOM 809 CE MET A 47 -1.183 9.011 -5.858 1.00 0.98
+ATOM 810 H MET A 47 0.070 3.911 -8.508 1.00 0.98
+ATOM 811 HA MET A 47 -1.071 6.284 -9.896 1.00 0.98
+ATOM 812 1HB MET A 47 0.805 6.070 -7.517 1.00 0.98
+ATOM 813 2HB MET A 47 0.271 7.583 -8.235 1.00 0.98
+ATOM 814 1HG MET A 47 -2.035 7.119 -7.602 1.00 0.98
+ATOM 815 2HG MET A 47 -1.577 5.530 -6.985 1.00 0.98
+ATOM 816 1HE MET A 47 -0.973 9.830 -4.744 1.00 0.98
+ATOM 817 2HE MET A 47 -0.458 9.558 -6.427 1.00 0.98
+ATOM 818 3HE MET A 47 -2.177 9.413 -5.988 1.00 0.98
+ATOM 819 N SER A 48 1.106 7.168 -11.026 1.00 0.96
+ATOM 820 CA SER A 48 2.341 7.561 -11.694 1.00 0.96
+ATOM 821 C SER A 48 3.439 7.871 -10.685 1.00 0.96
+ATOM 822 O SER A 48 3.162 8.139 -9.515 1.00 0.96
+ATOM 823 CB SER A 48 2.089 8.764 -12.582 1.00 0.96
+ATOM 824 OG SER A 48 1.836 9.910 -11.818 1.00 0.96
+ATOM 825 H SER A 48 0.300 7.772 -11.089 1.00 0.96
+ATOM 826 HG SER A 48 1.755 9.610 -10.909 1.00 0.96
+ATOM 827 HA SER A 48 2.676 6.731 -12.319 1.00 0.96
+ATOM 828 1HB SER A 48 2.957 8.932 -13.219 1.00 0.96
+ATOM 829 2HB SER A 48 1.240 8.562 -13.233 1.00 0.96
+ATOM 830 N PRO A 49 4.780 7.925 -11.121 1.00 0.97
+ATOM 831 CA PRO A 49 5.846 8.051 -10.135 1.00 0.97
+ATOM 832 C PRO A 49 5.747 9.372 -9.382 1.00 0.97
+ATOM 833 O PRO A 49 5.943 9.421 -8.168 1.00 0.97
+ATOM 834 CB PRO A 49 7.125 7.978 -10.975 1.00 0.97
+ATOM 835 CG PRO A 49 6.938 6.806 -11.877 1.00 0.97
+ATOM 836 CD PRO A 49 6.252 7.373 -13.091 1.00 0.97
+ATOM 837 HA PRO A 49 5.795 7.203 -9.436 1.00 0.97
+ATOM 838 1HB PRO A 49 7.257 8.922 -11.524 1.00 0.97
+ATOM 839 2HB PRO A 49 7.994 7.869 -10.309 1.00 0.97
+ATOM 840 1HG PRO A 49 7.911 6.350 -12.115 1.00 0.97
+ATOM 841 2HG PRO A 49 6.339 6.032 -11.375 1.00 0.97
+ATOM 842 1HD PRO A 49 5.486 8.102 -12.788 1.00 0.97
+ATOM 843 2HD PRO A 49 6.976 7.915 -13.718 1.00 0.97
+ATOM 844 N GLU A 50 5.427 10.513 -10.072 1.00 0.96
+ATOM 845 CA GLU A 50 5.330 11.820 -9.434 1.00 0.96
+ATOM 846 C GLU A 50 4.205 11.851 -8.408 1.00 0.96
+ATOM 847 O GLU A 50 4.376 12.358 -7.300 1.00 0.96
+ATOM 848 CB GLU A 50 5.122 12.915 -10.483 1.00 0.96
+ATOM 849 CG GLU A 50 6.328 13.164 -11.377 1.00 0.96
+ATOM 850 CD GLU A 50 6.059 14.178 -12.454 1.00 0.96
+ATOM 851 OE1 GLU A 50 6.944 14.437 -13.234 1.00 0.96
+ATOM 852 OE2 GLU A 50 4.967 14.694 -12.497 1.00 0.96
+ATOM 853 H GLU A 50 5.240 10.433 -11.061 1.00 0.96
+ATOM 854 HA GLU A 50 6.266 12.021 -8.913 1.00 0.96
+ATOM 855 1HB GLU A 50 4.280 12.652 -11.123 1.00 0.96
+ATOM 856 2HB GLU A 50 4.875 13.853 -9.986 1.00 0.96
+ATOM 857 1HG GLU A 50 7.152 13.532 -10.765 1.00 0.96
+ATOM 858 2HG GLU A 50 6.610 12.227 -11.859 1.00 0.96
+ATOM 859 N GLN A 51 2.961 11.349 -8.788 1.00 0.98
+ATOM 860 CA GLN A 51 1.839 11.297 -7.859 1.00 0.98
+ATOM 861 C GLN A 51 2.166 10.434 -6.647 1.00 0.98
+ATOM 862 O GLN A 51 1.793 10.760 -5.521 1.00 0.98
+ATOM 863 CB GLN A 51 0.578 10.786 -8.561 1.00 0.98
+ATOM 864 CG GLN A 51 0.002 11.747 -9.587 1.00 0.98
+ATOM 865 CD GLN A 51 -1.184 11.159 -10.329 1.00 0.98
+ATOM 866 OE1 GLN A 51 -2.176 11.847 -10.586 1.00 0.98
+ATOM 867 NE2 GLN A 51 -1.088 9.881 -10.677 1.00 0.98
+ATOM 868 H GLN A 51 2.848 10.972 -9.718 1.00 0.98
+ATOM 869 HA GLN A 51 1.635 12.308 -7.506 1.00 0.98
+ATOM 870 1HB GLN A 51 0.801 9.846 -9.068 1.00 0.98
+ATOM 871 2HB GLN A 51 -0.194 10.583 -7.820 1.00 0.98
+ATOM 872 1HG GLN A 51 -0.328 12.651 -9.077 1.00 0.98
+ATOM 873 2HG GLN A 51 0.775 11.990 -10.316 1.00 0.98
+ATOM 874 1HE2 GLN A 51 -1.834 9.441 -11.165 1.00 0.98
+ATOM 875 2HE2 GLN A 51 -0.266 9.360 -10.449 1.00 0.98
+ATOM 876 N ALA A 52 2.841 9.246 -6.870 1.00 0.98
+ATOM 877 CA ALA A 52 3.271 8.378 -5.781 1.00 0.98
+ATOM 878 C ALA A 52 4.207 9.112 -4.829 1.00 0.98
+ATOM 879 O ALA A 52 4.068 9.015 -3.610 1.00 0.98
+ATOM 880 CB ALA A 52 3.934 7.123 -6.331 1.00 0.98
+ATOM 881 H ALA A 52 3.062 8.989 -7.822 1.00 0.98
+ATOM 882 HA ALA A 52 2.388 8.079 -5.217 1.00 0.98
+ATOM 883 1HB ALA A 52 4.245 6.484 -5.504 1.00 0.98
+ATOM 884 2HB ALA A 52 3.226 6.583 -6.959 1.00 0.98
+ATOM 885 3HB ALA A 52 4.805 7.401 -6.922 1.00 0.98
+ATOM 886 N ARG A 53 5.222 9.864 -5.383 1.00 0.97
+ATOM 887 CA ARG A 53 6.158 10.612 -4.553 1.00 0.97
+ATOM 888 C ARG A 53 5.434 11.647 -3.702 1.00 0.97
+ATOM 889 O ARG A 53 5.688 11.766 -2.504 1.00 0.97
+ATOM 890 CB ARG A 53 7.209 11.298 -5.414 1.00 0.97
+ATOM 891 CG ARG A 53 8.298 12.018 -4.633 1.00 0.97
+ATOM 892 CD ARG A 53 9.284 12.666 -5.536 1.00 0.97
+ATOM 893 NE ARG A 53 10.015 11.691 -6.329 1.00 0.97
+ATOM 894 CZ ARG A 53 10.957 12.000 -7.242 1.00 0.97
+ATOM 895 NH1 ARG A 53 11.268 13.257 -7.466 1.00 0.97
+ATOM 896 NH2 ARG A 53 11.566 11.038 -7.913 1.00 0.97
+ATOM 897 H ARG A 53 5.318 9.899 -6.388 1.00 0.97
+ATOM 898 HA ARG A 53 6.668 9.913 -3.890 1.00 0.97
+ATOM 899 1HB ARG A 53 7.695 10.563 -6.054 1.00 0.97
+ATOM 900 2HB ARG A 53 6.729 12.031 -6.063 1.00 0.97
+ATOM 901 1HG ARG A 53 7.847 12.789 -4.008 1.00 0.97
+ATOM 902 2HG ARG A 53 8.828 11.303 -4.003 1.00 0.97
+ATOM 903 1HD ARG A 53 8.765 13.339 -6.217 1.00 0.97
+ATOM 904 2HD ARG A 53 10.002 13.232 -4.944 1.00 0.97
+ATOM 905 HE ARG A 53 9.803 10.713 -6.184 1.00 0.97
+ATOM 906 1HH1 ARG A 53 10.802 13.992 -6.953 1.00 0.97
+ATOM 907 2HH1 ARG A 53 11.974 13.488 -8.151 1.00 0.97
+ATOM 908 1HH2 ARG A 53 11.327 10.071 -7.740 1.00 0.97
+ATOM 909 2HH2 ARG A 53 12.271 11.269 -8.597 1.00 0.97
+ATOM 910 N GLU A 54 4.496 12.452 -4.318 1.00 0.98
+ATOM 911 CA GLU A 54 3.730 13.446 -3.576 1.00 0.98
+ATOM 912 C GLU A 54 2.920 12.798 -2.460 1.00 0.98
+ATOM 913 O GLU A 54 2.844 13.323 -1.350 1.00 0.98
+ATOM 914 CB GLU A 54 2.813 14.228 -4.518 1.00 0.98
+ATOM 915 CG GLU A 54 3.548 15.164 -5.467 1.00 0.98
+ATOM 916 CD GLU A 54 2.627 15.873 -6.420 1.00 0.98
+ATOM 917 OE1 GLU A 54 3.109 16.623 -7.235 1.00 0.98
+ATOM 918 OE2 GLU A 54 1.440 15.665 -6.334 1.00 0.98
+ATOM 919 H GLU A 54 4.317 12.329 -5.304 1.00 0.98
+ATOM 920 HA GLU A 54 4.427 14.150 -3.120 1.00 0.98
+ATOM 921 1HB GLU A 54 2.227 13.534 -5.121 1.00 0.98
+ATOM 922 2HB GLU A 54 2.112 14.826 -3.935 1.00 0.98
+ATOM 923 1HG GLU A 54 4.074 15.918 -4.882 1.00 0.98
+ATOM 924 2HG GLU A 54 4.260 14.583 -6.054 1.00 0.98
+ATOM 925 N LEU A 55 2.202 11.654 -2.761 1.00 0.99
+ATOM 926 CA LEU A 55 1.443 10.937 -1.744 1.00 0.99
+ATOM 927 C LEU A 55 2.345 10.481 -0.602 1.00 0.99
+ATOM 928 O LEU A 55 2.017 10.661 0.571 1.00 0.99
+ATOM 929 CB LEU A 55 0.721 9.733 -2.362 1.00 0.99
+ATOM 930 CG LEU A 55 -0.123 8.874 -1.412 1.00 0.99
+ATOM 931 CD1 LEU A 55 -1.230 9.726 -0.806 1.00 0.99
+ATOM 932 CD2 LEU A 55 -0.699 7.690 -2.173 1.00 0.99
+ATOM 933 H LEU A 55 2.244 11.280 -3.699 1.00 0.99
+ATOM 934 HA LEU A 55 0.688 11.609 -1.338 1.00 0.99
+ATOM 935 1HB LEU A 55 0.064 10.100 -3.149 1.00 0.99
+ATOM 936 2HB LEU A 55 1.471 9.085 -2.816 1.00 0.99
+ATOM 937 HG LEU A 55 0.503 8.512 -0.596 1.00 0.99
+ATOM 938 1HD1 LEU A 55 -1.830 9.116 -0.130 1.00 0.99
+ATOM 939 2HD1 LEU A 55 -0.790 10.555 -0.251 1.00 0.99
+ATOM 940 3HD1 LEU A 55 -1.864 10.117 -1.601 1.00 0.99
+ATOM 941 1HD2 LEU A 55 -1.298 7.079 -1.497 1.00 0.99
+ATOM 942 2HD2 LEU A 55 -1.326 8.051 -2.988 1.00 0.99
+ATOM 943 3HD2 LEU A 55 0.115 7.089 -2.580 1.00 0.99
+ATOM 944 N ILE A 56 3.534 9.847 -0.932 1.00 0.98
+ATOM 945 CA ILE A 56 4.463 9.398 0.097 1.00 0.98
+ATOM 946 C ILE A 56 4.912 10.558 0.978 1.00 0.98
+ATOM 947 O ILE A 56 4.990 10.429 2.199 1.00 0.98
+ATOM 948 CB ILE A 56 5.686 8.702 -0.527 1.00 0.98
+ATOM 949 CG1 ILE A 56 5.307 7.323 -1.073 1.00 0.98
+ATOM 950 CG2 ILE A 56 6.805 8.584 0.496 1.00 0.98
+ATOM 951 CD1 ILE A 56 6.398 6.667 -1.888 1.00 0.98
+ATOM 952 H ILE A 56 3.773 9.711 -1.904 1.00 0.98
+ATOM 953 HA ILE A 56 3.956 8.669 0.728 1.00 0.98
+ATOM 954 HB ILE A 56 6.041 9.289 -1.374 1.00 0.98
+ATOM 955 1HG2 ILE A 56 7.665 8.095 0.040 1.00 0.98
+ATOM 956 2HG2 ILE A 56 7.092 9.578 0.838 1.00 0.98
+ATOM 957 3HG2 ILE A 56 6.461 7.994 1.345 1.00 0.98
+ATOM 958 1HG1 ILE A 56 5.055 6.661 -0.244 1.00 0.98
+ATOM 959 2HG1 ILE A 56 4.420 7.411 -1.701 1.00 0.98
+ATOM 960 1HD1 ILE A 56 6.055 5.694 -2.240 1.00 0.98
+ATOM 961 2HD1 ILE A 56 6.641 7.298 -2.744 1.00 0.98
+ATOM 962 3HD1 ILE A 56 7.285 6.536 -1.270 1.00 0.98
+ATOM 963 N GLU A 57 5.235 11.760 0.375 1.00 0.97
+ATOM 964 CA GLU A 57 5.627 12.941 1.135 1.00 0.97
+ATOM 965 C GLU A 57 4.530 13.361 2.105 1.00 0.97
+ATOM 966 O GLU A 57 4.800 13.673 3.265 1.00 0.97
+ATOM 967 CB GLU A 57 5.976 14.094 0.192 1.00 0.97
+ATOM 968 CG GLU A 57 7.289 13.914 -0.556 1.00 0.97
+ATOM 969 CD GLU A 57 7.548 15.006 -1.556 1.00 0.97
+ATOM 970 OE1 GLU A 57 8.581 14.980 -2.182 1.00 0.97
+ATOM 971 OE2 GLU A 57 6.712 15.867 -1.694 1.00 0.97
+ATOM 972 H GLU A 57 5.172 11.822 -0.631 1.00 0.97
+ATOM 973 HA GLU A 57 6.515 12.698 1.717 1.00 0.97
+ATOM 974 1HB GLU A 57 5.184 14.214 -0.547 1.00 0.97
+ATOM 975 2HB GLU A 57 6.039 15.023 0.758 1.00 0.97
+ATOM 976 1HG GLU A 57 8.108 13.914 0.164 1.00 0.97
+ATOM 977 2HG GLU A 57 7.263 12.965 -1.091 1.00 0.97
+ATOM 978 N TRP A 58 3.239 13.437 1.633 1.00 0.98
+ATOM 979 CA TRP A 58 2.110 13.770 2.494 1.00 0.98
+ATOM 980 C TRP A 58 1.982 12.778 3.642 1.00 0.98
+ATOM 981 O TRP A 58 1.855 13.169 4.802 1.00 0.98
+ATOM 982 CB TRP A 58 0.813 13.816 1.684 1.00 0.98
+ATOM 983 CG TRP A 58 -0.404 14.144 2.494 1.00 0.98
+ATOM 984 CD1 TRP A 58 -0.863 15.388 2.808 1.00 0.98
+ATOM 985 CD2 TRP A 58 -1.329 13.210 3.101 1.00 0.98
+ATOM 986 NE1 TRP A 58 -2.004 15.294 3.565 1.00 0.98
+ATOM 987 CE2 TRP A 58 -2.304 13.968 3.755 1.00 0.98
+ATOM 988 CE3 TRP A 58 -1.406 11.812 3.142 1.00 0.98
+ATOM 989 CZ2 TRP A 58 -3.351 13.379 4.445 1.00 0.98
+ATOM 990 CZ3 TRP A 58 -2.456 11.221 3.835 1.00 0.98
+ATOM 991 CH2 TRP A 58 -3.403 11.985 4.469 1.00 0.98
+ATOM 992 H TRP A 58 3.068 13.206 0.664 1.00 0.98
+ATOM 993 HA TRP A 58 2.277 14.762 2.914 1.00 0.98
+ATOM 994 1HB TRP A 58 0.910 14.561 0.894 1.00 0.98
+ATOM 995 2HB TRP A 58 0.656 12.849 1.205 1.00 0.98
+ATOM 996 HD1 TRP A 58 -0.392 16.321 2.503 1.00 0.98
+ATOM 997 HE1 TRP A 58 -2.537 16.072 3.925 1.00 0.98
+ATOM 998 HZ2 TRP A 58 -4.113 13.968 4.956 1.00 0.98
+ATOM 999 HH2 TRP A 58 -4.214 11.489 5.004 1.00 0.98
+ATOM 1000 HZ3 TRP A 58 -2.509 10.132 3.862 1.00 0.98
+ATOM 1001 HE3 TRP A 58 -0.657 11.201 2.640 1.00 0.98
+ATOM 1002 N TRP A 59 1.989 11.445 3.329 1.00 0.98
+ATOM 1003 CA TRP A 59 1.880 10.414 4.354 1.00 0.98
+ATOM 1004 C TRP A 59 3.028 10.506 5.352 1.00 0.98
+ATOM 1005 O TRP A 59 2.829 10.360 6.557 1.00 0.98
+ATOM 1006 CB TRP A 59 1.845 9.022 3.722 1.00 0.98
+ATOM 1007 CG TRP A 59 0.499 8.616 3.202 1.00 0.98
+ATOM 1008 CD1 TRP A 59 0.204 8.206 1.936 1.00 0.98
+ATOM 1009 CD2 TRP A 59 -0.748 8.577 3.938 1.00 0.98
+ATOM 1010 NE1 TRP A 59 -1.135 7.917 1.833 1.00 0.98
+ATOM 1011 CE2 TRP A 59 -1.732 8.138 3.048 1.00 0.98
+ATOM 1012 CE3 TRP A 59 -1.102 8.875 5.259 1.00 0.98
+ATOM 1013 CZ2 TRP A 59 -3.055 7.988 3.434 1.00 0.98
+ATOM 1014 CZ3 TRP A 59 -2.428 8.724 5.646 1.00 0.98
+ATOM 1015 CH2 TRP A 59 -3.379 8.292 4.756 1.00 0.98
+ATOM 1016 H TRP A 59 2.119 11.169 2.366 1.00 0.98
+ATOM 1017 HA TRP A 59 0.944 10.560 4.894 1.00 0.98
+ATOM 1018 1HB TRP A 59 2.556 8.987 2.896 1.00 0.98
+ATOM 1019 2HB TRP A 59 2.164 8.286 4.460 1.00 0.98
+ATOM 1020 HD1 TRP A 59 0.924 8.122 1.125 1.00 0.98
+ATOM 1021 HE1 TRP A 59 -1.604 7.594 0.999 1.00 0.98
+ATOM 1022 HZ2 TRP A 59 -3.825 7.646 2.742 1.00 0.98
+ATOM 1023 HH2 TRP A 59 -4.410 8.184 5.093 1.00 0.98
+ATOM 1024 HZ3 TRP A 59 -2.695 8.958 6.677 1.00 0.98
+ATOM 1025 HE3 TRP A 59 -0.351 9.219 5.970 1.00 0.98
+ATOM 1026 N TRP A 60 4.313 10.712 4.844 1.00 0.97
+ATOM 1027 CA TRP A 60 5.476 10.860 5.711 1.00 0.97
+ATOM 1028 C TRP A 60 5.279 11.994 6.708 1.00 0.97
+ATOM 1029 O TRP A 60 5.562 11.844 7.897 1.00 0.97
+ATOM 1030 CB TRP A 60 6.730 11.115 4.873 1.00 0.97
+ATOM 1031 CG TRP A 60 7.977 11.311 5.681 1.00 0.97
+ATOM 1032 CD1 TRP A 60 8.627 12.488 5.905 1.00 0.97
+ATOM 1033 CD2 TRP A 60 8.734 10.295 6.382 1.00 0.97
+ATOM 1034 NE1 TRP A 60 9.731 12.276 6.692 1.00 0.97
+ATOM 1035 CE2 TRP A 60 9.812 10.940 6.994 1.00 0.97
+ATOM 1036 CE3 TRP A 60 8.585 8.911 6.536 1.00 0.97
+ATOM 1037 CZ2 TRP A 60 10.745 10.250 7.753 1.00 0.97
+ATOM 1038 CZ3 TRP A 60 9.520 8.220 7.297 1.00 0.97
+ATOM 1039 CH2 TRP A 60 10.572 8.873 7.889 1.00 0.97
+ATOM 1040 H TRP A 60 4.432 10.785 3.844 1.00 0.97
+ATOM 1041 HA TRP A 60 5.613 9.933 6.267 1.00 0.97
+ATOM 1042 1HB TRP A 60 6.888 10.274 4.198 1.00 0.97
+ATOM 1043 2HB TRP A 60 6.577 12.003 4.259 1.00 0.97
+ATOM 1044 HD1 TRP A 60 8.315 13.455 5.515 1.00 0.97
+ATOM 1045 HE1 TRP A 60 10.379 12.987 7.001 1.00 0.97
+ATOM 1046 HZ2 TRP A 60 11.587 10.751 8.231 1.00 0.97
+ATOM 1047 HH2 TRP A 60 11.287 8.299 8.480 1.00 0.97
+ATOM 1048 HZ3 TRP A 60 9.397 7.142 7.413 1.00 0.97
+ATOM 1049 HE3 TRP A 60 7.752 8.388 6.068 1.00 0.97
+ATOM 1050 N LYS A 61 4.769 13.197 6.238 1.00 0.97
+ATOM 1051 CA LYS A 61 4.516 14.325 7.126 1.00 0.97
+ATOM 1052 C LYS A 61 3.503 13.961 8.205 1.00 0.97
+ATOM 1053 O LYS A 61 3.702 14.261 9.382 1.00 0.97
+ATOM 1054 CB LYS A 61 4.034 15.541 6.333 1.00 0.97
+ATOM 1055 CG LYS A 61 5.104 16.227 5.494 1.00 0.97
+ATOM 1056 CD LYS A 61 4.539 17.431 4.755 1.00 0.97
+ATOM 1057 CE LYS A 61 5.588 18.073 3.859 1.00 0.97
+ATOM 1058 NZ LYS A 61 5.044 19.242 3.115 1.00 0.97
+ATOM 1059 H LYS A 61 4.556 13.288 5.255 1.00 0.97
+ATOM 1060 HA LYS A 61 5.450 14.592 7.622 1.00 0.97
+ATOM 1061 1HB LYS A 61 3.229 15.236 5.663 1.00 0.97
+ATOM 1062 2HB LYS A 61 3.624 16.279 7.022 1.00 0.97
+ATOM 1063 1HG LYS A 61 5.917 16.558 6.142 1.00 0.97
+ATOM 1064 2HG LYS A 61 5.506 15.521 4.768 1.00 0.97
+ATOM 1065 1HD LYS A 61 3.693 17.118 4.143 1.00 0.97
+ATOM 1066 2HD LYS A 61 4.190 18.170 5.476 1.00 0.97
+ATOM 1067 1HE LYS A 61 6.447 18.508 4.462 1.00 0.97
+ATOM 1068 2HE LYS A 61 5.966 17.444 3.138 1.00 0.97
+ATOM 1069 1HZ LYS A 61 5.785 19.744 2.532 1.00 0.97
+ATOM 1070 2HZ LYS A 61 4.288 19.049 2.533 1.00 0.97
+ATOM 1071 3HZ LYS A 61 4.738 20.044 3.770 1.00 0.97
+ATOM 1072 N TRP A 62 2.345 13.323 7.810 1.00 0.97
+ATOM 1073 CA TRP A 62 1.328 12.897 8.763 1.00 0.97
+ATOM 1074 C TRP A 62 1.897 11.906 9.770 1.00 0.97
+ATOM 1075 O TRP A 62 1.706 12.054 10.977 1.00 0.97
+ATOM 1076 CB TRP A 62 0.129 12.291 8.030 1.00 0.97
+ATOM 1077 CG TRP A 62 -0.999 11.881 8.928 1.00 0.97
+ATOM 1078 CD1 TRP A 62 -1.966 12.692 9.443 1.00 0.97
+ATOM 1079 CD2 TRP A 62 -1.284 10.551 9.424 1.00 0.97
+ATOM 1080 NE1 TRP A 62 -2.830 11.962 10.221 1.00 0.97
+ATOM 1081 CE2 TRP A 62 -2.428 10.650 10.221 1.00 0.97
+ATOM 1082 CE3 TRP A 62 -0.670 9.304 9.258 1.00 0.97
+ATOM 1083 CZ2 TRP A 62 -2.976 9.547 10.856 1.00 0.97
+ATOM 1084 CZ3 TRP A 62 -1.220 8.198 9.895 1.00 0.97
+ATOM 1085 CH2 TRP A 62 -2.344 8.317 10.673 1.00 0.97
+ATOM 1086 H TRP A 62 2.234 13.089 6.833 1.00 0.97
+ATOM 1087 HA TRP A 62 0.974 13.773 9.306 1.00 0.97
+ATOM 1088 1HB TRP A 62 -0.251 13.015 7.310 1.00 0.97
+ATOM 1089 2HB TRP A 62 0.458 11.415 7.472 1.00 0.97
+ATOM 1090 HD1 TRP A 62 -2.043 13.762 9.262 1.00 0.97
+ATOM 1091 HE1 TRP A 62 -3.631 12.329 10.714 1.00 0.97
+ATOM 1092 HZ2 TRP A 62 -3.869 9.621 11.478 1.00 0.97
+ATOM 1093 HH2 TRP A 62 -2.749 7.429 11.158 1.00 0.97
+ATOM 1094 HZ3 TRP A 62 -0.736 7.230 9.760 1.00 0.97
+ATOM 1095 HE3 TRP A 62 0.223 9.203 8.641 1.00 0.97
+ATOM 1096 N ILE A 63 2.550 10.789 9.268 1.00 0.98
+ATOM 1097 CA ILE A 63 3.156 9.795 10.146 1.00 0.98
+ATOM 1098 C ILE A 63 4.151 10.439 11.103 1.00 0.98
+ATOM 1099 O ILE A 63 4.197 10.101 12.286 1.00 0.98
+ATOM 1100 CB ILE A 63 3.832 8.682 9.326 1.00 0.98
+ATOM 1101 CG1 ILE A 63 2.777 7.835 8.610 1.00 0.98
+ATOM 1102 CG2 ILE A 63 4.702 7.813 10.221 1.00 0.98
+ATOM 1103 CD1 ILE A 63 3.349 6.898 7.570 1.00 0.98
+ATOM 1104 H ILE A 63 2.667 10.699 8.269 1.00 0.98
+ATOM 1105 HA ILE A 63 2.367 9.331 10.737 1.00 0.98
+ATOM 1106 HB ILE A 63 4.456 9.128 8.552 1.00 0.98
+ATOM 1107 1HG2 ILE A 63 5.170 7.030 9.624 1.00 0.98
+ATOM 1108 2HG2 ILE A 63 5.473 8.425 10.685 1.00 0.98
+ATOM 1109 3HG2 ILE A 63 4.085 7.358 10.996 1.00 0.98
+ATOM 1110 1HG1 ILE A 63 2.231 7.240 9.342 1.00 0.98
+ATOM 1111 2HG1 ILE A 63 2.057 8.489 8.119 1.00 0.98
+ATOM 1112 1HD1 ILE A 63 2.541 6.332 7.107 1.00 0.98
+ATOM 1113 2HD1 ILE A 63 3.871 7.476 6.807 1.00 0.98
+ATOM 1114 3HD1 ILE A 63 4.047 6.210 8.045 1.00 0.98
+ATOM 1115 N GLU A 64 5.019 11.408 10.588 1.00 0.97
+ATOM 1116 CA GLU A 64 5.964 12.126 11.435 1.00 0.97
+ATOM 1117 C GLU A 64 5.248 12.851 12.568 1.00 0.97
+ATOM 1118 O GLU A 64 5.682 12.808 13.718 1.00 0.97
+ATOM 1119 CB GLU A 64 6.778 13.122 10.607 1.00 0.97
+ATOM 1120 CG GLU A 64 7.873 12.485 9.764 1.00 0.97
+ATOM 1121 CD GLU A 64 8.650 13.488 8.957 1.00 0.97
+ATOM 1122 OE1 GLU A 64 9.600 13.103 8.320 1.00 0.97
+ATOM 1123 OE2 GLU A 64 8.292 14.643 8.979 1.00 0.97
+ATOM 1124 H GLU A 64 4.973 11.631 9.604 1.00 0.97
+ATOM 1125 HA GLU A 64 6.648 11.404 11.879 1.00 0.97
+ATOM 1126 1HB GLU A 64 6.115 13.669 9.937 1.00 0.97
+ATOM 1127 2HB GLU A 64 7.247 13.850 11.269 1.00 0.97
+ATOM 1128 1HG GLU A 64 8.572 11.969 10.423 1.00 0.97
+ATOM 1129 2HG GLU A 64 7.419 11.775 9.073 1.00 0.97
+ATOM 1130 N GLU A 65 4.121 13.602 12.254 1.00 0.97
+ATOM 1131 CA GLU A 65 3.368 14.302 13.287 1.00 0.97
+ATOM 1132 C GLU A 65 2.836 13.332 14.334 1.00 0.97
+ATOM 1133 O GLU A 65 2.920 13.592 15.535 1.00 0.97
+ATOM 1134 CB GLU A 65 2.219 15.100 12.666 1.00 0.97
+ATOM 1135 CG GLU A 65 2.662 16.322 11.876 1.00 0.97
+ATOM 1136 CD GLU A 65 1.519 17.033 11.206 1.00 0.97
+ATOM 1137 OE1 GLU A 65 1.761 18.008 10.536 1.00 0.97
+ATOM 1138 OE2 GLU A 65 0.402 16.599 11.365 1.00 0.97
+ATOM 1139 H GLU A 65 3.801 13.646 11.297 1.00 0.97
+ATOM 1140 HA GLU A 65 4.036 15.003 13.787 1.00 0.97
+ATOM 1141 1HB GLU A 65 1.648 14.457 11.996 1.00 0.97
+ATOM 1142 2HB GLU A 65 1.542 15.437 13.452 1.00 0.97
+ATOM 1143 1HG GLU A 65 3.146 17.025 12.554 1.00 0.97
+ATOM 1144 2HG GLU A 65 3.363 16.007 11.102 1.00 0.97
+ATOM 1145 N GLU A 66 2.180 12.202 13.899 1.00 0.98
+ATOM 1146 CA GLU A 66 1.669 11.189 14.816 1.00 0.98
+ATOM 1147 C GLU A 66 2.784 10.619 15.683 1.00 0.98
+ATOM 1148 O GLU A 66 2.620 10.455 16.892 1.00 0.98
+ATOM 1149 CB GLU A 66 0.971 10.066 14.047 1.00 0.98
+ATOM 1150 CG GLU A 66 -0.378 10.449 13.455 1.00 0.98
+ATOM 1151 CD GLU A 66 -1.423 10.716 14.503 1.00 0.98
+ATOM 1152 OE1 GLU A 66 -2.282 11.530 14.264 1.00 0.98
+ATOM 1153 OE2 GLU A 66 -1.361 10.105 15.543 1.00 0.98
+ATOM 1154 H GLU A 66 2.121 12.039 12.904 1.00 0.98
+ATOM 1155 HA GLU A 66 0.936 11.657 15.473 1.00 0.98
+ATOM 1156 1HB GLU A 66 1.611 9.732 13.230 1.00 0.98
+ATOM 1157 2HB GLU A 66 0.815 9.215 14.710 1.00 0.98
+ATOM 1158 1HG GLU A 66 -0.258 11.355 12.860 1.00 0.98
+ATOM 1159 2HG GLU A 66 -0.732 9.633 12.825 1.00 0.98
+ATOM 1160 N TRP A 67 3.964 10.256 15.069 1.00 0.98
+ATOM 1161 CA TRP A 67 5.096 9.714 15.810 1.00 0.98
+ATOM 1162 C TRP A 67 5.617 10.717 16.831 1.00 0.98
+ATOM 1163 O TRP A 67 6.016 10.344 17.934 1.00 0.98
+ATOM 1164 CB TRP A 67 6.216 9.296 14.854 1.00 0.98
+ATOM 1165 CG TRP A 67 5.924 8.053 14.069 1.00 0.98
+ATOM 1166 CD1 TRP A 67 6.543 7.649 12.924 1.00 0.98
+ATOM 1167 CD2 TRP A 67 4.933 7.041 14.369 1.00 0.98
+ATOM 1168 NE1 TRP A 67 6.008 6.460 12.492 1.00 0.98
+ATOM 1169 CE2 TRP A 67 5.022 6.074 13.364 1.00 0.98
+ATOM 1170 CE3 TRP A 67 3.993 6.880 15.394 1.00 0.98
+ATOM 1171 CZ2 TRP A 67 4.205 4.954 13.349 1.00 0.98
+ATOM 1172 CZ3 TRP A 67 3.173 5.758 15.379 1.00 0.98
+ATOM 1173 CH2 TRP A 67 3.277 4.820 14.383 1.00 0.98
+ATOM 1174 H TRP A 67 4.059 10.424 14.077 1.00 0.98
+ATOM 1175 HA TRP A 67 4.766 8.822 16.342 1.00 0.98
+ATOM 1176 1HB TRP A 67 6.407 10.108 14.153 1.00 0.98
+ATOM 1177 2HB TRP A 67 7.130 9.134 15.425 1.00 0.98
+ATOM 1178 HD1 TRP A 67 7.344 8.190 12.425 1.00 0.98
+ATOM 1179 HE1 TRP A 67 6.293 5.952 11.667 1.00 0.98
+ATOM 1180 HZ2 TRP A 67 4.272 4.198 12.567 1.00 0.98
+ATOM 1181 HH2 TRP A 67 2.618 3.952 14.401 1.00 0.98
+ATOM 1182 HZ3 TRP A 67 2.443 5.641 16.181 1.00 0.98
+ATOM 1183 HE3 TRP A 67 3.906 7.622 16.188 1.00 0.98
+ATOM 1184 N ARG A 68 5.754 12.078 16.431 1.00 0.97
+ATOM 1185 CA ARG A 68 6.180 13.097 17.382 1.00 0.97
+ATOM 1186 C ARG A 68 5.297 13.096 18.623 1.00 0.97
+ATOM 1187 O ARG A 68 5.785 13.239 19.744 1.00 0.97
+ATOM 1188 CB ARG A 68 6.156 14.477 16.742 1.00 0.97
+ATOM 1189 CG ARG A 68 7.306 14.761 15.788 1.00 0.97
+ATOM 1190 CD ARG A 68 7.225 16.132 15.223 1.00 0.97
+ATOM 1191 NE ARG A 68 8.296 16.391 14.274 1.00 0.97
+ATOM 1192 CZ ARG A 68 8.430 17.528 13.563 1.00 0.97
+ATOM 1193 NH1 ARG A 68 7.554 18.498 13.704 1.00 0.97
+ATOM 1194 NH2 ARG A 68 9.441 17.667 12.723 1.00 0.97
+ATOM 1195 H ARG A 68 5.497 12.356 15.494 1.00 0.97
+ATOM 1196 HA ARG A 68 7.205 12.881 17.686 1.00 0.97
+ATOM 1197 1HB ARG A 68 5.228 14.603 16.185 1.00 0.97
+ATOM 1198 2HB ARG A 68 6.177 15.240 17.520 1.00 0.97
+ATOM 1199 1HG ARG A 68 8.252 14.664 16.321 1.00 0.97
+ATOM 1200 2HG ARG A 68 7.279 14.048 14.963 1.00 0.97
+ATOM 1201 1HD ARG A 68 6.274 16.258 14.706 1.00 0.97
+ATOM 1202 2HD ARG A 68 7.300 16.862 16.028 1.00 0.97
+ATOM 1203 HE ARG A 68 8.990 15.668 14.139 1.00 0.97
+ATOM 1204 1HH1 ARG A 68 6.781 18.392 14.345 1.00 0.97
+ATOM 1205 2HH1 ARG A 68 7.654 19.350 13.170 1.00 0.97
+ATOM 1206 1HH2 ARG A 68 10.114 16.921 12.615 1.00 0.97
+ATOM 1207 2HH2 ARG A 68 9.541 18.518 12.191 1.00 0.97
+ATOM 1208 N ARG A 69 3.914 13.025 18.445 1.00 0.97
+ATOM 1209 CA ARG A 69 2.997 12.950 19.576 1.00 0.97
+ATOM 1210 C ARG A 69 3.292 11.734 20.445 1.00 0.97
+ATOM 1211 O ARG A 69 3.270 11.816 21.673 1.00 0.97
+ATOM 1212 CB ARG A 69 1.553 12.904 19.097 1.00 0.97
+ATOM 1213 CG ARG A 69 1.042 14.206 18.501 1.00 0.97
+ATOM 1214 CD ARG A 69 -0.375 14.093 18.071 1.00 0.97
+ATOM 1215 NE ARG A 69 -0.853 15.319 17.451 1.00 0.97
+ATOM 1216 CZ ARG A 69 -2.087 15.486 16.937 1.00 0.97
+ATOM 1217 NH1 ARG A 69 -2.954 14.499 16.975 1.00 0.97
+ATOM 1218 NH2 ARG A 69 -2.425 16.643 16.394 1.00 0.97
+ATOM 1219 H ARG A 69 3.545 12.954 17.507 1.00 0.97
+ATOM 1220 HA ARG A 69 3.121 13.847 20.182 1.00 0.97
+ATOM 1221 1HB ARG A 69 1.444 12.129 18.339 1.00 0.97
+ATOM 1222 2HB ARG A 69 0.899 12.642 19.928 1.00 0.97
+ATOM 1223 1HG ARG A 69 1.112 14.999 19.246 1.00 0.97
+ATOM 1224 2HG ARG A 69 1.645 14.470 17.631 1.00 0.97
+ATOM 1225 1HD ARG A 69 -0.472 13.285 17.346 1.00 0.97
+ATOM 1226 2HD ARG A 69 -1.002 13.882 18.936 1.00 0.97
+ATOM 1227 HE ARG A 69 -0.213 16.102 17.404 1.00 0.97
+ATOM 1228 1HH1 ARG A 69 -2.695 13.615 17.390 1.00 0.97
+ATOM 1229 2HH1 ARG A 69 -3.879 14.624 16.590 1.00 0.97
+ATOM 1230 1HH2 ARG A 69 -1.758 17.402 16.365 1.00 0.97
+ATOM 1231 2HH2 ARG A 69 -3.350 16.768 16.009 1.00 0.97
+ATOM 1232 N TRP A 70 3.573 10.544 19.802 1.00 0.99
+ATOM 1233 CA TRP A 70 3.889 9.315 20.519 1.00 0.99
+ATOM 1234 C TRP A 70 5.167 9.466 21.335 1.00 0.99
+ATOM 1235 O TRP A 70 5.275 8.938 22.442 1.00 0.99
+ATOM 1236 CB TRP A 70 4.016 8.142 19.546 1.00 0.99
+ATOM 1237 CG TRP A 70 2.707 7.704 18.961 1.00 0.99
+ATOM 1238 CD1 TRP A 70 2.518 6.737 18.019 1.00 0.99
+ATOM 1239 CD2 TRP A 70 1.391 8.217 19.278 1.00 0.99
+ATOM 1240 NE1 TRP A 70 1.182 6.614 17.729 1.00 0.99
+ATOM 1241 CE2 TRP A 70 0.477 7.512 18.491 1.00 0.99
+ATOM 1242 CE3 TRP A 70 0.922 9.204 20.155 1.00 0.99
+ATOM 1243 CZ2 TRP A 70 -0.885 7.759 18.549 1.00 0.99
+ATOM 1244 CZ3 TRP A 70 -0.444 9.452 20.213 1.00 0.99
+ATOM 1245 CH2 TRP A 70 -1.324 8.747 19.430 1.00 0.99
+ATOM 1246 H TRP A 70 3.579 10.540 18.792 1.00 0.99
+ATOM 1247 HA TRP A 70 3.070 9.092 21.203 1.00 0.99
+ATOM 1248 1HB TRP A 70 4.681 8.418 18.728 1.00 0.99
+ATOM 1249 2HB TRP A 70 4.464 7.291 20.058 1.00 0.99
+ATOM 1250 HD1 TRP A 70 3.312 6.149 17.562 1.00 0.99
+ATOM 1251 HE1 TRP A 70 0.781 5.969 17.064 1.00 0.99
+ATOM 1252 HZ2 TRP A 70 -1.600 7.210 17.935 1.00 0.99
+ATOM 1253 HH2 TRP A 70 -2.390 8.967 19.501 1.00 0.99
+ATOM 1254 HZ3 TRP A 70 -0.801 10.221 20.898 1.00 0.99
+ATOM 1255 HE3 TRP A 70 1.616 9.766 20.778 1.00 0.99
+ATOM 1256 N ILE A 71 6.207 10.230 20.800 1.00 0.98
+ATOM 1257 CA ILE A 71 7.459 10.464 21.509 1.00 0.98
+ATOM 1258 C ILE A 71 7.229 11.266 22.784 1.00 0.98
+ATOM 1259 O ILE A 71 7.772 10.942 23.840 1.00 0.98
+ATOM 1260 CB ILE A 71 8.475 11.193 20.610 1.00 0.98
+ATOM 1261 CG1 ILE A 71 8.998 10.250 19.524 1.00 0.98
+ATOM 1262 CG2 ILE A 71 9.624 11.742 21.442 1.00 0.98
+ATOM 1263 CD1 ILE A 71 9.843 10.936 18.475 1.00 0.98
+ATOM 1264 H ILE A 71 6.093 10.602 19.868 1.00 0.98
+ATOM 1265 HA ILE A 71 7.885 9.500 21.786 1.00 0.98
+ATOM 1266 HB ILE A 71 7.981 12.019 20.099 1.00 0.98
+ATOM 1267 1HG2 ILE A 71 10.332 12.253 20.790 1.00 0.98
+ATOM 1268 2HG2 ILE A 71 9.237 12.444 22.178 1.00 0.98
+ATOM 1269 3HG2 ILE A 71 10.128 10.921 21.953 1.00 0.98
+ATOM 1270 1HG1 ILE A 71 9.595 9.463 19.982 1.00 0.98
+ATOM 1271 2HG1 ILE A 71 8.157 9.770 19.022 1.00 0.98
+ATOM 1272 1HD1 ILE A 71 10.177 10.203 17.740 1.00 0.98
+ATOM 1273 2HD1 ILE A 71 9.252 11.706 17.978 1.00 0.98
+ATOM 1274 3HD1 ILE A 71 10.710 11.394 18.949 1.00 0.98
+ATOM 1275 N GLU A 72 6.300 12.344 22.713 1.00 0.98
+ATOM 1276 CA GLU A 72 5.992 13.164 23.879 1.00 0.98
+ATOM 1277 C GLU A 72 5.427 12.319 25.013 1.00 0.98
+ATOM 1278 O GLU A 72 5.614 12.635 26.188 1.00 0.98
+ATOM 1279 CB GLU A 72 5.009 14.276 23.507 1.00 0.98
+ATOM 1280 CG GLU A 72 5.604 15.369 22.630 1.00 0.98
+ATOM 1281 CD GLU A 72 4.601 16.420 22.243 1.00 0.98
+ATOM 1282 OE1 GLU A 72 4.961 17.323 21.528 1.00 0.98
+ATOM 1283 OE2 GLU A 72 3.472 16.319 22.664 1.00 0.98
+ATOM 1284 H GLU A 72 5.843 12.546 21.835 1.00 0.98
+ATOM 1285 HA GLU A 72 6.915 13.625 24.230 1.00 0.98
+ATOM 1286 1HB GLU A 72 4.157 13.849 22.977 1.00 0.98
+ATOM 1287 2HB GLU A 72 4.627 14.745 24.414 1.00 0.98
+ATOM 1288 1HG GLU A 72 6.412 15.858 23.175 1.00 0.98
+ATOM 1289 2HG GLU A 72 5.990 14.918 21.716 1.00 0.98
+ATOM 1290 N GLU A 73 4.731 11.138 24.677 1.00 0.97
+ATOM 1291 CA GLU A 73 4.131 10.227 25.645 1.00 0.97
+ATOM 1292 C GLU A 73 5.116 9.145 26.068 1.00 0.97
+ATOM 1293 O GLU A 73 4.744 8.179 26.734 1.00 0.97
+ATOM 1294 CB GLU A 73 2.859 9.601 25.071 1.00 0.97
+ATOM 1295 CG GLU A 73 1.734 10.592 24.810 1.00 0.97
+ATOM 1296 CD GLU A 73 0.534 9.958 24.163 1.00 0.97
+ATOM 1297 OE1 GLU A 73 -0.413 10.658 23.895 1.00 0.97
+ATOM 1298 OE2 GLU A 73 0.564 8.771 23.937 1.00 0.97
+ATOM 1299 H GLU A 73 4.689 10.908 23.694 1.00 0.97
+ATOM 1300 HA GLU A 73 3.858 10.798 26.533 1.00 0.97
+ATOM 1301 1HB GLU A 73 3.091 9.103 24.130 1.00 0.97
+ATOM 1302 2HB GLU A 73 2.484 8.843 25.758 1.00 0.97
+ATOM 1303 1HG GLU A 73 1.416 11.024 25.759 1.00 0.97
+ATOM 1304 2HG GLU A 73 2.100 11.375 24.146 1.00 0.97
+ATOM 1305 N GLY A 74 6.356 9.091 25.669 1.00 0.98
+ATOM 1306 CA GLY A 74 7.406 8.121 25.955 1.00 0.98
+ATOM 1307 C GLY A 74 7.165 6.814 25.211 1.00 0.98
+ATOM 1308 O GLY A 74 7.720 5.774 25.567 1.00 0.98
+ATOM 1309 H GLY A 74 6.541 9.838 25.015 1.00 0.98
+ATOM 1310 1HA GLY A 74 8.372 8.535 25.667 1.00 0.98
+ATOM 1311 2HA GLY A 74 7.444 7.932 27.027 1.00 0.98
+ATOM 1312 N LYS A 75 6.100 6.604 24.410 1.00 0.98
+ATOM 1313 CA LYS A 75 5.845 5.426 23.590 1.00 0.98
+ATOM 1314 C LYS A 75 6.909 5.264 22.511 1.00 0.98
+ATOM 1315 O LYS A 75 7.056 4.191 21.928 1.00 0.98
+ATOM 1316 CB LYS A 75 4.454 5.498 22.958 1.00 0.98
+ATOM 1317 CG LYS A 75 3.312 5.435 23.964 1.00 0.98
+ATOM 1318 CD LYS A 75 1.960 5.444 23.266 1.00 0.98
+ATOM 1319 CE LYS A 75 0.818 5.381 24.270 1.00 0.98
+ATOM 1320 NZ LYS A 75 -0.511 5.394 23.602 1.00 0.98
+ATOM 1321 H LYS A 75 5.580 7.451 24.229 1.00 0.98
+ATOM 1322 HA LYS A 75 5.883 4.544 24.230 1.00 0.98
+ATOM 1323 1HB LYS A 75 4.354 6.427 22.396 1.00 0.98
+ATOM 1324 2HB LYS A 75 4.326 4.675 22.255 1.00 0.98
+ATOM 1325 1HG LYS A 75 3.400 4.524 24.557 1.00 0.98
+ATOM 1326 2HG LYS A 75 3.371 6.291 24.635 1.00 0.98
+ATOM 1327 1HD LYS A 75 1.862 6.355 22.674 1.00 0.98
+ATOM 1328 2HD LYS A 75 1.891 4.587 22.597 1.00 0.98
+ATOM 1329 1HE LYS A 75 0.823 4.404 24.850 1.00 0.98
+ATOM 1330 2HE LYS A 75 0.798 6.167 24.933 1.00 0.98
+ATOM 1331 1HZ LYS A 75 -1.322 5.284 24.288 1.00 0.98
+ATOM 1332 2HZ LYS A 75 -0.692 6.177 23.052 1.00 0.98
+ATOM 1333 3HZ LYS A 75 -0.669 4.530 22.975 1.00 0.98
+ATOM 1334 N ILE A 76 7.775 6.342 22.321 1.00 0.98
+ATOM 1335 CA ILE A 76 8.761 6.416 21.250 1.00 0.98
+ATOM 1336 C ILE A 76 9.914 7.338 21.626 1.00 0.98
+ATOM 1337 O ILE A 76 9.700 8.450 22.109 1.00 0.98
+ATOM 1338 CB ILE A 76 8.119 6.896 19.935 1.00 0.98
+ATOM 1339 CG1 ILE A 76 7.037 5.941 19.426 1.00 0.98
+ATOM 1340 CG2 ILE A 76 9.204 7.085 18.886 1.00 0.98
+ATOM 1341 CD1 ILE A 76 6.221 6.497 18.281 1.00 0.98
+ATOM 1342 H ILE A 76 7.604 7.158 22.890 1.00 0.98
+ATOM 1343 HA ILE A 76 9.164 5.418 21.081 1.00 0.98
+ATOM 1344 HB ILE A 76 7.633 7.854 20.118 1.00 0.98
+ATOM 1345 1HG2 ILE A 76 8.757 7.457 17.964 1.00 0.98
+ATOM 1346 2HG2 ILE A 76 9.939 7.802 19.248 1.00 0.98
+ATOM 1347 3HG2 ILE A 76 9.693 6.130 18.693 1.00 0.98
+ATOM 1348 1HG1 ILE A 76 7.498 5.011 19.097 1.00 0.98
+ATOM 1349 2HG1 ILE A 76 6.355 5.696 20.241 1.00 0.98
+ATOM 1350 1HD1 ILE A 76 5.475 5.763 17.976 1.00 0.98
+ATOM 1351 2HD1 ILE A 76 5.720 7.412 18.601 1.00 0.98
+ATOM 1352 3HD1 ILE A 76 6.877 6.717 17.441 1.00 0.98
+ATOM 1353 N THR A 77 11.224 6.994 21.705 1.00 0.98
+ATOM 1354 CA THR A 77 12.324 7.941 21.846 1.00 0.98
+ATOM 1355 C THR A 77 12.683 8.572 20.507 1.00 0.98
+ATOM 1356 O THR A 77 12.325 8.053 19.450 1.00 0.98
+ATOM 1357 CB THR A 77 13.572 7.259 22.436 1.00 0.98
+ATOM 1358 OG1 THR A 77 14.109 6.344 21.472 1.00 0.98
+ATOM 1359 CG2 THR A 77 13.223 6.502 23.708 1.00 0.98
+ATOM 1360 H THR A 77 11.432 6.012 21.592 1.00 0.98
+ATOM 1361 HG1 THR A 77 13.452 6.175 20.792 1.00 0.98
+ATOM 1362 HA THR A 77 12.009 8.738 22.520 1.00 0.98
+ATOM 1363 HB THR A 77 14.328 8.011 22.663 1.00 0.98
+ATOM 1364 1HG2 THR A 77 14.118 6.023 24.104 1.00 0.98
+ATOM 1365 2HG2 THR A 77 12.825 7.197 24.447 1.00 0.98
+ATOM 1366 3HG2 THR A 77 12.474 5.743 23.485 1.00 0.98
+ATOM 1367 N LYS A 78 13.142 9.862 20.339 1.00 0.98
+ATOM 1368 CA LYS A 78 13.972 10.378 19.257 1.00 0.98
+ATOM 1369 C LYS A 78 14.863 9.287 18.677 1.00 0.98
+ATOM 1370 O LYS A 78 14.978 9.148 17.459 1.00 0.98
+ATOM 1371 CB LYS A 78 14.817 11.555 19.746 1.00 0.98
+ATOM 1372 CG LYS A 78 15.623 12.255 18.661 1.00 0.98
+ATOM 1373 CD LYS A 78 16.383 13.449 19.220 1.00 0.98
+ATOM 1374 CE LYS A 78 17.234 14.115 18.149 1.00 0.98
+ATOM 1375 NZ LYS A 78 17.978 15.290 18.680 1.00 0.98
+ATOM 1376 H LYS A 78 13.085 10.379 21.205 1.00 0.98
+ATOM 1377 HA LYS A 78 13.318 10.734 18.459 1.00 0.98
+ATOM 1378 1HB LYS A 78 14.168 12.297 20.212 1.00 0.98
+ATOM 1379 2HB LYS A 78 15.514 11.206 20.508 1.00 0.98
+ATOM 1380 1HG LYS A 78 16.335 11.553 18.226 1.00 0.98
+ATOM 1381 2HG LYS A 78 14.952 12.599 17.874 1.00 0.98
+ATOM 1382 1HD LYS A 78 15.676 14.178 19.617 1.00 0.98
+ATOM 1383 2HD LYS A 78 17.032 13.119 20.032 1.00 0.98
+ATOM 1384 1HE LYS A 78 18.051 13.423 17.770 1.00 0.98
+ATOM 1385 2HE LYS A 78 16.696 14.473 17.348 1.00 0.98
+ATOM 1386 1HZ LYS A 78 18.630 15.730 17.957 1.00 0.98
+ATOM 1387 2HZ LYS A 78 17.424 15.999 19.050 1.00 0.98
+ATOM 1388 3HZ LYS A 78 18.690 15.019 19.444 1.00 0.98
+ATOM 1389 N GLU A 79 15.517 8.453 19.098 1.00 0.97
+ATOM 1390 CA GLU A 79 16.242 7.364 18.454 1.00 0.97
+ATOM 1391 C GLU A 79 15.284 6.335 17.868 1.00 0.97
+ATOM 1392 O GLU A 79 15.523 5.793 16.789 1.00 0.97
+ATOM 1393 CB GLU A 79 17.199 6.696 19.444 1.00 0.97
+ATOM 1394 CG GLU A 79 18.336 7.590 19.919 1.00 0.97
+ATOM 1395 CD GLU A 79 19.216 6.925 20.940 1.00 0.97
+ATOM 1396 OE1 GLU A 79 20.143 7.551 21.395 1.00 0.97
+ATOM 1397 OE2 GLU A 79 18.961 5.790 21.265 1.00 0.97
+ATOM 1398 H GLU A 79 15.522 8.518 20.106 1.00 0.97
+ATOM 1399 HA GLU A 79 16.832 7.778 17.636 1.00 0.97
+ATOM 1400 1HB GLU A 79 16.643 6.367 20.322 1.00 0.97
+ATOM 1401 2HB GLU A 79 17.639 5.811 18.985 1.00 0.97
+ATOM 1402 1HG GLU A 79 18.955 7.859 19.063 1.00 0.97
+ATOM 1403 2HG GLU A 79 17.914 8.488 20.371 1.00 0.97
+ATOM 1404 N GLU A 80 14.158 5.951 18.599 1.00 0.98
+ATOM 1405 CA GLU A 80 13.157 5.038 18.060 1.00 0.98
+ATOM 1406 C GLU A 80 12.494 5.618 16.817 1.00 0.98
+ATOM 1407 O GLU A 80 12.168 4.890 15.880 1.00 0.98
+ATOM 1408 CB GLU A 80 12.108 4.703 19.123 1.00 0.98
+ATOM 1409 CG GLU A 80 12.633 3.856 20.273 1.00 0.98
+ATOM 1410 CD GLU A 80 11.627 3.677 21.376 1.00 0.98
+ATOM 1411 OE1 GLU A 80 11.761 2.745 22.132 1.00 0.98
+ATOM 1412 OE2 GLU A 80 10.722 4.474 21.462 1.00 0.98
+ATOM 1413 H GLU A 80 14.011 6.359 19.511 1.00 0.98
+ATOM 1414 HA GLU A 80 13.653 4.110 17.775 1.00 0.98
+ATOM 1415 1HB GLU A 80 11.706 5.625 19.543 1.00 0.98
+ATOM 1416 2HB GLU A 80 11.279 4.165 18.663 1.00 0.98
+ATOM 1417 1HG GLU A 80 12.897 2.869 19.893 1.00 0.98
+ATOM 1418 2HG GLU A 80 13.514 4.341 20.695 1.00 0.98
+ATOM 1419 N TRP A 81 12.042 6.976 16.915 1.00 0.98
+ATOM 1420 CA TRP A 81 11.478 7.612 15.730 1.00 0.98
+ATOM 1421 C TRP A 81 12.379 7.414 14.518 1.00 0.98
+ATOM 1422 O TRP A 81 11.910 7.065 13.434 1.00 0.98
+ATOM 1423 CB TRP A 81 11.261 9.106 15.976 1.00 0.98
+ATOM 1424 CG TRP A 81 10.721 9.844 14.788 1.00 0.98
+ATOM 1425 CD1 TRP A 81 9.422 9.892 14.380 1.00 0.98
+ATOM 1426 CD2 TRP A 81 11.469 10.647 13.843 1.00 0.98
+ATOM 1427 NE1 TRP A 81 9.309 10.667 13.252 1.00 0.98
+ATOM 1428 CE2 TRP A 81 10.552 11.137 12.909 1.00 0.98
+ATOM 1429 CE3 TRP A 81 12.822 10.985 13.718 1.00 0.98
+ATOM 1430 CZ2 TRP A 81 10.941 11.951 11.857 1.00 0.98
+ATOM 1431 CZ3 TRP A 81 13.212 11.801 12.662 1.00 0.98
+ATOM 1432 CH2 TRP A 81 12.295 12.271 11.756 1.00 0.98
+ATOM 1433 H TRP A 81 12.211 7.535 17.739 1.00 0.98
+ATOM 1434 HA TRP A 81 10.512 7.154 15.519 1.00 0.98
+ATOM 1435 1HB TRP A 81 10.566 9.238 16.806 1.00 0.98
+ATOM 1436 2HB TRP A 81 12.206 9.565 16.264 1.00 0.98
+ATOM 1437 HD1 TRP A 81 8.594 9.389 14.876 1.00 0.98
+ATOM 1438 HE1 TRP A 81 8.452 10.860 12.754 1.00 0.98
+ATOM 1439 HZ2 TRP A 81 10.227 12.334 11.127 1.00 0.98
+ATOM 1440 HH2 TRP A 81 12.635 12.909 10.940 1.00 0.98
+ATOM 1441 HZ3 TRP A 81 14.267 12.060 12.572 1.00 0.98
+ATOM 1442 HE3 TRP A 81 13.555 10.615 14.434 1.00 0.98
+ATOM 1443 N GLU A 82 13.713 7.717 14.664 1.00 0.97
+ATOM 1444 CA GLU A 82 14.667 7.571 13.572 1.00 0.97
+ATOM 1445 C GLU A 82 14.697 6.138 13.054 1.00 0.97
+ATOM 1446 O GLU A 82 14.801 5.905 11.850 1.00 0.97
+ATOM 1447 CB GLU A 82 16.064 8.013 14.015 1.00 0.97
+ATOM 1448 CG GLU A 82 16.207 9.513 14.228 1.00 0.97
+ATOM 1449 CD GLU A 82 17.568 9.905 14.733 1.00 0.97
+ATOM 1450 OE1 GLU A 82 17.786 11.073 14.950 1.00 0.97
+ATOM 1451 OE2 GLU A 82 18.390 9.035 14.901 1.00 0.97
+ATOM 1452 H GLU A 82 14.047 7.999 15.575 1.00 0.97
+ATOM 1453 HA GLU A 82 14.356 8.218 12.752 1.00 0.97
+ATOM 1454 1HB GLU A 82 16.324 7.516 14.950 1.00 0.97
+ATOM 1455 2HB GLU A 82 16.797 7.709 13.268 1.00 0.97
+ATOM 1456 1HG GLU A 82 16.040 10.022 13.278 1.00 0.97
+ATOM 1457 2HG GLU A 82 15.468 9.838 14.961 1.00 0.97
+ATOM 1458 N ARG A 83 14.548 5.106 13.968 1.00 0.98
+ATOM 1459 CA ARG A 83 14.503 3.704 13.571 1.00 0.98
+ATOM 1460 C ARG A 83 13.245 3.399 12.768 1.00 0.98
+ATOM 1461 O ARG A 83 13.294 2.685 11.766 1.00 0.98
+ATOM 1462 CB ARG A 83 14.573 2.796 14.790 1.00 0.98
+ATOM 1463 CG ARG A 83 15.934 2.743 15.467 1.00 0.98
+ATOM 1464 CD ARG A 83 15.926 1.840 16.646 1.00 0.98
+ATOM 1465 NE ARG A 83 17.209 1.829 17.329 1.00 0.98
+ATOM 1466 CZ ARG A 83 17.472 1.134 18.454 1.00 0.98
+ATOM 1467 NH1 ARG A 83 16.532 0.401 19.007 1.00 0.98
+ATOM 1468 NH2 ARG A 83 18.674 1.191 19.000 1.00 0.98
+ATOM 1469 H ARG A 83 14.490 5.342 14.949 1.00 0.98
+ATOM 1470 HA ARG A 83 15.372 3.493 12.947 1.00 0.98
+ATOM 1471 1HB ARG A 83 13.848 3.127 15.533 1.00 0.98
+ATOM 1472 2HB ARG A 83 14.307 1.779 14.504 1.00 0.98
+ATOM 1473 1HG ARG A 83 16.678 2.377 14.759 1.00 0.98
+ATOM 1474 2HG ARG A 83 16.213 3.743 15.802 1.00 0.98
+ATOM 1475 1HD ARG A 83 15.166 2.171 17.353 1.00 0.98
+ATOM 1476 2HD ARG A 83 15.703 0.823 16.324 1.00 0.98
+ATOM 1477 HE ARG A 83 17.958 2.382 16.933 1.00 0.98
+ATOM 1478 1HH1 ARG A 83 15.613 0.358 18.590 1.00 0.98
+ATOM 1479 2HH1 ARG A 83 16.728 -0.120 19.850 1.00 0.98
+ATOM 1480 1HH2 ARG A 83 19.396 1.755 18.574 1.00 0.98
+ATOM 1481 2HH2 ARG A 83 18.870 0.670 19.842 1.00 0.98
+ATOM 1482 N LEU A 84 12.020 3.872 13.280 1.00 0.98
+ATOM 1483 CA LEU A 84 10.779 3.730 12.528 1.00 0.98
+ATOM 1484 C LEU A 84 10.902 4.344 11.138 1.00 0.98
+ATOM 1485 O LEU A 84 10.470 3.755 10.148 1.00 0.98
+ATOM 1486 CB LEU A 84 9.605 4.343 13.302 1.00 0.98
+ATOM 1487 CG LEU A 84 8.181 4.163 12.759 1.00 0.98
+ATOM 1488 CD1 LEU A 84 7.791 2.693 12.832 1.00 0.98
+ATOM 1489 CD2 LEU A 84 7.219 5.025 13.563 1.00 0.98
+ATOM 1490 H LEU A 84 12.017 4.407 14.138 1.00 0.98
+ATOM 1491 HA LEU A 84 10.563 2.668 12.416 1.00 0.98
+ATOM 1492 1HB LEU A 84 9.632 3.921 14.305 1.00 0.98
+ATOM 1493 2HB LEU A 84 9.799 5.413 13.374 1.00 0.98
+ATOM 1494 HG LEU A 84 8.150 4.465 11.712 1.00 0.98
+ATOM 1495 1HD1 LEU A 84 6.780 2.566 12.446 1.00 0.98
+ATOM 1496 2HD1 LEU A 84 8.484 2.103 12.232 1.00 0.98
+ATOM 1497 3HD1 LEU A 84 7.829 2.358 13.867 1.00 0.98
+ATOM 1498 1HD2 LEU A 84 6.207 4.898 13.177 1.00 0.98
+ATOM 1499 2HD2 LEU A 84 7.248 4.723 14.610 1.00 0.98
+ATOM 1500 3HD2 LEU A 84 7.510 6.072 13.479 1.00 0.98
+ATOM 1501 N TRP A 85 11.427 5.630 11.054 1.00 0.97
+ATOM 1502 CA TRP A 85 11.644 6.296 9.775 1.00 0.97
+ATOM 1503 C TRP A 85 12.540 5.464 8.866 1.00 0.97
+ATOM 1504 O TRP A 85 12.300 5.368 7.662 1.00 0.97
+ATOM 1505 CB TRP A 85 12.241 7.687 9.997 1.00 0.97
+ATOM 1506 CG TRP A 85 12.274 8.560 8.780 1.00 0.97
+ATOM 1507 CD1 TRP A 85 12.673 9.861 8.721 1.00 0.97
+ATOM 1508 CD2 TRP A 85 11.892 8.196 7.431 1.00 0.97
+ATOM 1509 NE1 TRP A 85 12.565 10.331 7.435 1.00 0.97
+ATOM 1510 CE2 TRP A 85 12.089 9.325 6.633 1.00 0.97
+ATOM 1511 CE3 TRP A 85 11.405 7.020 6.847 1.00 0.97
+ATOM 1512 CZ2 TRP A 85 11.817 9.319 5.274 1.00 0.97
+ATOM 1513 CZ3 TRP A 85 11.132 7.014 5.484 1.00 0.97
+ATOM 1514 CH2 TRP A 85 11.333 8.135 4.718 1.00 0.97
+ATOM 1515 H TRP A 85 11.729 6.086 11.903 1.00 0.97
+ATOM 1516 HA TRP A 85 10.680 6.424 9.284 1.00 0.97
+ATOM 1517 1HB TRP A 85 11.665 8.197 10.769 1.00 0.97
+ATOM 1518 2HB TRP A 85 13.260 7.577 10.368 1.00 0.97
+ATOM 1519 HD1 TRP A 85 13.025 10.444 9.570 1.00 0.97
+ATOM 1520 HE1 TRP A 85 12.799 11.265 7.128 1.00 0.97
+ATOM 1521 HZ2 TRP A 85 11.970 10.199 4.648 1.00 0.97
+ATOM 1522 HH2 TRP A 85 11.109 8.095 3.652 1.00 0.97
+ATOM 1523 HZ3 TRP A 85 10.754 6.094 5.037 1.00 0.97
+ATOM 1524 HE3 TRP A 85 11.243 6.127 7.450 1.00 0.97
+ATOM 1525 N GLU A 86 13.660 4.875 9.422 1.00 0.97
+ATOM 1526 CA GLU A 86 14.542 3.999 8.660 1.00 0.97
+ATOM 1527 C GLU A 86 13.788 2.789 8.124 1.00 0.97
+ATOM 1528 O GLU A 86 13.998 2.369 6.986 1.00 0.97
+ATOM 1529 CB GLU A 86 15.727 3.551 9.519 1.00 0.97
+ATOM 1530 CG GLU A 86 16.775 4.631 9.749 1.00 0.97
+ATOM 1531 CD GLU A 86 17.893 4.179 10.646 1.00 0.97
+ATOM 1532 OE1 GLU A 86 18.764 4.968 10.922 1.00 0.97
+ATOM 1533 OE2 GLU A 86 17.876 3.042 11.056 1.00 0.97
+ATOM 1534 H GLU A 86 13.854 5.040 10.399 1.00 0.97
+ATOM 1535 HA GLU A 86 14.931 4.557 7.808 1.00 0.97
+ATOM 1536 1HB GLU A 86 15.367 3.221 10.494 1.00 0.97
+ATOM 1537 2HB GLU A 86 16.220 2.702 9.047 1.00 0.97
+ATOM 1538 1HG GLU A 86 17.206 4.915 8.789 1.00 0.97
+ATOM 1539 2HG GLU A 86 16.297 5.494 10.213 1.00 0.97
+ATOM 1540 N LYS A 87 12.856 2.189 8.956 1.00 0.97
+ATOM 1541 CA LYS A 87 12.027 1.072 8.520 1.00 0.97
+ATOM 1542 C LYS A 87 11.126 1.475 7.359 1.00 0.97
+ATOM 1543 O LYS A 87 10.955 0.720 6.402 1.00 0.97
+ATOM 1544 CB LYS A 87 11.195 0.531 9.683 1.00 0.97
+ATOM 1545 CG LYS A 87 12.008 -0.205 10.740 1.00 0.97
+ATOM 1546 CD LYS A 87 11.115 -0.744 11.848 1.00 0.97
+ATOM 1547 CE LYS A 87 11.928 -1.459 12.917 1.00 0.97
+ATOM 1548 NZ LYS A 87 11.072 -1.959 14.026 1.00 0.97
+ATOM 1549 H LYS A 87 12.747 2.545 9.894 1.00 0.97
+ATOM 1550 HA LYS A 87 12.680 0.271 8.172 1.00 0.97
+ATOM 1551 1HB LYS A 87 10.674 1.354 10.175 1.00 0.97
+ATOM 1552 2HB LYS A 87 10.437 -0.155 9.305 1.00 0.97
+ATOM 1553 1HG LYS A 87 12.539 -1.037 10.276 1.00 0.97
+ATOM 1554 2HG LYS A 87 12.741 0.474 11.173 1.00 0.97
+ATOM 1555 1HD LYS A 87 10.569 0.080 12.309 1.00 0.97
+ATOM 1556 2HD LYS A 87 10.393 -1.444 11.426 1.00 0.97
+ATOM 1557 1HE LYS A 87 12.424 -2.397 12.510 1.00 0.97
+ATOM 1558 2HE LYS A 87 12.641 -0.869 13.367 1.00 0.97
+ATOM 1559 1HZ LYS A 87 11.619 -2.520 14.752 1.00 0.97
+ATOM 1560 2HZ LYS A 87 10.565 -1.276 14.499 1.00 0.97
+ATOM 1561 3HZ LYS A 87 10.363 -2.704 13.699 1.00 0.97
+ATOM 1562 N LEU A 88 10.448 2.700 7.468 1.00 0.98
+ATOM 1563 CA LEU A 88 9.626 3.203 6.375 1.00 0.98
+ATOM 1564 C LEU A 88 10.454 3.409 5.111 1.00 0.98
+ATOM 1565 O LEU A 88 10.040 3.024 4.017 1.00 0.98
+ATOM 1566 CB LEU A 88 8.935 4.510 6.785 1.00 0.98
+ATOM 1567 CG LEU A 88 7.956 5.129 5.779 1.00 0.98
+ATOM 1568 CD1 LEU A 88 6.822 4.149 5.505 1.00 0.98
+ATOM 1569 CD2 LEU A 88 7.422 6.442 6.331 1.00 0.98
+ATOM 1570 H LEU A 88 10.565 3.265 8.297 1.00 0.98
+ATOM 1571 HA LEU A 88 8.849 2.470 6.161 1.00 0.98
+ATOM 1572 1HB LEU A 88 8.388 4.324 7.708 1.00 0.98
+ATOM 1573 2HB LEU A 88 9.710 5.248 6.989 1.00 0.98
+ATOM 1574 HG LEU A 88 8.471 5.314 4.836 1.00 0.98
+ATOM 1575 1HD1 LEU A 88 6.127 4.589 4.790 1.00 0.98
+ATOM 1576 2HD1 LEU A 88 7.230 3.226 5.093 1.00 0.98
+ATOM 1577 3HD1 LEU A 88 6.298 3.931 6.435 1.00 0.98
+ATOM 1578 1HD2 LEU A 88 6.727 6.882 5.616 1.00 0.98
+ATOM 1579 2HD2 LEU A 88 6.906 6.258 7.273 1.00 0.98
+ATOM 1580 3HD2 LEU A 88 8.251 7.129 6.501 1.00 0.98
+ATOM 1581 N LYS A 89 11.666 4.088 5.230 1.00 0.97
+ATOM 1582 CA LYS A 89 12.546 4.307 4.089 1.00 0.97
+ATOM 1583 C LYS A 89 12.960 2.986 3.452 1.00 0.97
+ATOM 1584 O LYS A 89 13.018 2.867 2.228 1.00 0.97
+ATOM 1585 CB LYS A 89 13.780 5.109 4.505 1.00 0.97
+ATOM 1586 CG LYS A 89 13.508 6.586 4.759 1.00 0.97
+ATOM 1587 CD LYS A 89 14.781 7.324 5.145 1.00 0.97
+ATOM 1588 CE LYS A 89 14.509 8.798 5.408 1.00 0.97
+ATOM 1589 NZ LYS A 89 15.738 9.524 5.829 1.00 0.97
+ATOM 1590 H LYS A 89 11.960 4.398 6.145 1.00 0.97
+ATOM 1591 HA LYS A 89 12.004 4.883 3.338 1.00 0.97
+ATOM 1592 1HB LYS A 89 14.201 4.685 5.417 1.00 0.97
+ATOM 1593 2HB LYS A 89 14.542 5.038 3.729 1.00 0.97
+ATOM 1594 1HG LYS A 89 13.095 7.040 3.857 1.00 0.97
+ATOM 1595 2HG LYS A 89 12.780 6.689 5.562 1.00 0.97
+ATOM 1596 1HD LYS A 89 15.203 6.875 6.045 1.00 0.97
+ATOM 1597 2HD LYS A 89 15.511 7.237 4.341 1.00 0.97
+ATOM 1598 1HE LYS A 89 14.182 9.339 4.465 1.00 0.97
+ATOM 1599 2HE LYS A 89 13.822 8.972 6.153 1.00 0.97
+ATOM 1600 1HZ LYS A 89 15.579 10.572 5.954 1.00 0.97
+ATOM 1601 2HZ LYS A 89 16.162 9.189 6.639 1.00 0.97
+ATOM 1602 3HZ LYS A 89 16.498 9.534 5.062 1.00 0.97
+ATOM 1603 N GLU A 90 13.393 1.971 4.281 1.00 0.97
+ATOM 1604 CA GLU A 90 13.736 0.644 3.785 1.00 0.97
+ATOM 1605 C GLU A 90 12.580 0.033 3.003 1.00 0.97
+ATOM 1606 O GLU A 90 12.783 -0.572 1.950 1.00 0.97
+ATOM 1607 CB GLU A 90 14.142 -0.273 4.942 1.00 0.97
+ATOM 1608 CG GLU A 90 15.509 0.035 5.536 1.00 0.97
+ATOM 1609 CD GLU A 90 15.823 -0.802 6.745 1.00 0.97
+ATOM 1610 OE1 GLU A 90 16.877 -0.626 7.308 1.00 0.97
+ATOM 1611 OE2 GLU A 90 15.008 -1.618 7.106 1.00 0.97
+ATOM 1612 H GLU A 90 13.388 2.138 5.278 1.00 0.97
+ATOM 1613 HA GLU A 90 14.587 0.736 3.111 1.00 0.97
+ATOM 1614 1HB GLU A 90 13.405 -0.198 5.742 1.00 0.97
+ATOM 1615 2HB GLU A 90 14.151 -1.308 4.600 1.00 0.97
+ATOM 1616 1HG GLU A 90 16.273 -0.160 4.783 1.00 0.97
+ATOM 1617 2HG GLU A 90 15.537 1.083 5.834 1.00 0.97
+ATOM 1618 N TRP A 91 11.321 0.171 3.537 1.00 0.98
+ATOM 1619 CA TRP A 91 10.127 -0.284 2.835 1.00 0.98
+ATOM 1620 C TRP A 91 9.992 0.397 1.479 1.00 0.98
+ATOM 1621 O TRP A 91 9.737 -0.258 0.468 1.00 0.98
+ATOM 1622 CB TRP A 91 8.878 -0.032 3.682 1.00 0.98
+ATOM 1623 CG TRP A 91 7.598 -0.448 3.023 1.00 0.98
+ATOM 1624 CD1 TRP A 91 7.053 -1.697 3.016 1.00 0.98
+ATOM 1625 CD2 TRP A 91 6.691 0.389 2.266 1.00 0.98
+ATOM 1626 NE1 TRP A 91 5.876 -1.695 2.311 1.00 0.98
+ATOM 1627 CE2 TRP A 91 5.639 -0.427 1.844 1.00 0.98
+ATOM 1628 CE3 TRP A 91 6.689 1.746 1.917 1.00 0.98
+ATOM 1629 CZ2 TRP A 91 4.586 0.065 1.088 1.00 0.98
+ATOM 1630 CZ3 TRP A 91 5.634 2.239 1.159 1.00 0.98
+ATOM 1631 CH2 TRP A 91 4.610 1.419 0.756 1.00 0.98
+ATOM 1632 H TRP A 91 11.220 0.630 4.432 1.00 0.98
+ATOM 1633 HA TRP A 91 10.210 -1.359 2.674 1.00 0.98
+ATOM 1634 1HB TRP A 91 8.969 -0.571 4.625 1.00 0.98
+ATOM 1635 2HB TRP A 91 8.813 1.030 3.918 1.00 0.98
+ATOM 1636 HD1 TRP A 91 7.489 -2.569 3.501 1.00 0.98
+ATOM 1637 HE1 TRP A 91 5.279 -2.496 2.159 1.00 0.98
+ATOM 1638 HZ2 TRP A 91 3.763 -0.570 0.759 1.00 0.98
+ATOM 1639 HH2 TRP A 91 3.797 1.839 0.162 1.00 0.98
+ATOM 1640 HZ3 TRP A 91 5.640 3.296 0.892 1.00 0.98
+ATOM 1641 HE3 TRP A 91 7.500 2.401 2.234 1.00 0.98
+ATOM 1642 N LEU A 92 10.149 1.795 1.451 1.00 0.97
+ATOM 1643 CA LEU A 92 10.040 2.540 0.203 1.00 0.97
+ATOM 1644 C LEU A 92 11.072 2.066 -0.815 1.00 0.97
+ATOM 1645 O LEU A 92 10.757 1.874 -1.989 1.00 0.97
+ATOM 1646 CB LEU A 92 10.199 4.046 0.451 1.00 0.97
+ATOM 1647 CG LEU A 92 8.968 4.821 0.936 1.00 0.97
+ATOM 1648 CD1 LEU A 92 9.379 6.233 1.330 1.00 0.97
+ATOM 1649 CD2 LEU A 92 7.917 4.845 -0.164 1.00 0.97
+ATOM 1650 H LEU A 92 10.360 2.292 2.304 1.00 0.97
+ATOM 1651 HA LEU A 92 9.047 2.375 -0.213 1.00 0.97
+ATOM 1652 1HB LEU A 92 10.985 4.175 1.193 1.00 0.97
+ATOM 1653 2HB LEU A 92 10.531 4.500 -0.483 1.00 0.97
+ATOM 1654 HG LEU A 92 8.556 4.335 1.821 1.00 0.97
+ATOM 1655 1HD1 LEU A 92 8.504 6.784 1.674 1.00 0.97
+ATOM 1656 2HD1 LEU A 92 10.116 6.187 2.131 1.00 0.97
+ATOM 1657 3HD1 LEU A 92 9.810 6.740 0.468 1.00 0.97
+ATOM 1658 1HD2 LEU A 92 7.041 5.395 0.181 1.00 0.97
+ATOM 1659 2HD2 LEU A 92 8.327 5.332 -1.049 1.00 0.97
+ATOM 1660 3HD2 LEU A 92 7.628 3.823 -0.414 1.00 0.97
+ATOM 1661 N GLU A 93 12.374 1.887 -0.377 1.00 0.96
+ATOM 1662 CA GLU A 93 13.435 1.402 -1.252 1.00 0.96
+ATOM 1663 C GLU A 93 13.090 0.035 -1.830 1.00 0.96
+ATOM 1664 O GLU A 93 13.283 -0.213 -3.020 1.00 0.96
+ATOM 1665 CB GLU A 93 14.766 1.339 -0.500 1.00 0.96
+ATOM 1666 CG GLU A 93 15.399 2.697 -0.232 1.00 0.96
+ATOM 1667 CD GLU A 93 16.685 2.601 0.540 1.00 0.96
+ATOM 1668 OE1 GLU A 93 17.265 3.622 0.821 1.00 0.96
+ATOM 1669 OE2 GLU A 93 17.089 1.504 0.849 1.00 0.96
+ATOM 1670 H GLU A 93 12.579 2.054 0.598 1.00 0.96
+ATOM 1671 HA GLU A 93 13.544 2.100 -2.082 1.00 0.96
+ATOM 1672 1HB GLU A 93 14.619 0.843 0.459 1.00 0.96
+ATOM 1673 2HB GLU A 93 15.479 0.745 -1.070 1.00 0.96
+ATOM 1674 1HG GLU A 93 15.612 3.182 -1.185 1.00 0.96
+ATOM 1675 2HG GLU A 93 14.702 3.303 0.348 1.00 0.96
+ATOM 1676 N LYS A 94 12.606 -0.937 -0.963 1.00 0.96
+ATOM 1677 CA LYS A 94 12.175 -2.256 -1.412 1.00 0.96
+ATOM 1678 C LYS A 94 11.070 -2.148 -2.455 1.00 0.96
+ATOM 1679 O LYS A 94 11.080 -2.858 -3.460 1.00 0.96
+ATOM 1680 CB LYS A 94 11.712 -3.105 -0.228 1.00 0.96
+ATOM 1681 CG LYS A 94 12.840 -3.591 0.673 1.00 0.96
+ATOM 1682 CD LYS A 94 12.305 -4.431 1.824 1.00 0.96
+ATOM 1683 CE LYS A 94 13.425 -4.880 2.750 1.00 0.96
+ATOM 1684 NZ LYS A 94 12.912 -5.667 3.904 1.00 0.96
+ATOM 1685 H LYS A 94 12.495 -0.688 0.009 1.00 0.96
+ATOM 1686 HA LYS A 94 13.025 -2.758 -1.877 1.00 0.96
+ATOM 1687 1HB LYS A 94 11.019 -2.529 0.386 1.00 0.96
+ATOM 1688 2HB LYS A 94 11.176 -3.980 -0.594 1.00 0.96
+ATOM 1689 1HG LYS A 94 13.539 -4.192 0.090 1.00 0.96
+ATOM 1690 2HG LYS A 94 13.376 -2.734 1.079 1.00 0.96
+ATOM 1691 1HD LYS A 94 11.583 -3.847 2.396 1.00 0.96
+ATOM 1692 2HD LYS A 94 11.800 -5.312 1.427 1.00 0.96
+ATOM 1693 1HE LYS A 94 14.140 -5.593 2.229 1.00 0.96
+ATOM 1694 2HE LYS A 94 13.963 -4.107 3.165 1.00 0.96
+ATOM 1695 1HZ LYS A 94 13.691 -6.045 4.528 1.00 0.96
+ATOM 1696 2HZ LYS A 94 12.275 -5.201 4.474 1.00 0.96
+ATOM 1697 3HZ LYS A 94 12.442 -6.590 3.600 1.00 0.96
+ATOM 1698 N TRP A 95 10.070 -1.221 -2.212 1.00 0.97
+ATOM 1699 CA TRP A 95 8.965 -1.005 -3.139 1.00 0.97
+ATOM 1700 C TRP A 95 9.465 -0.482 -4.480 1.00 0.97
+ATOM 1701 O TRP A 95 9.059 -0.967 -5.536 1.00 0.97
+ATOM 1702 CB TRP A 95 7.942 -0.041 -2.535 1.00 0.97
+ATOM 1703 CG TRP A 95 6.717 0.158 -3.376 1.00 0.97
+ATOM 1704 CD1 TRP A 95 5.646 -0.679 -3.467 1.00 0.97
+ATOM 1705 CD2 TRP A 95 6.431 1.273 -4.254 1.00 0.97
+ATOM 1706 NE1 TRP A 95 4.716 -0.165 -4.337 1.00 0.97
+ATOM 1707 CE2 TRP A 95 5.180 1.029 -4.828 1.00 0.97
+ATOM 1708 CE3 TRP A 95 7.125 2.441 -4.594 1.00 0.97
+ATOM 1709 CZ2 TRP A 95 4.601 1.910 -5.728 1.00 0.97
+ATOM 1710 CZ3 TRP A 95 6.545 3.324 -5.497 1.00 0.97
+ATOM 1711 CH2 TRP A 95 5.315 3.065 -6.048 1.00 0.97
+ATOM 1712 H TRP A 95 10.120 -0.667 -1.369 1.00 0.97
+ATOM 1713 HA TRP A 95 8.463 -1.958 -3.307 1.00 0.97
+ATOM 1714 1HB TRP A 95 7.630 -0.415 -1.559 1.00 0.97
+ATOM 1715 2HB TRP A 95 8.412 0.929 -2.379 1.00 0.97
+ATOM 1716 HD1 TRP A 95 5.542 -1.619 -2.928 1.00 0.97
+ATOM 1717 HE1 TRP A 95 3.835 -0.595 -4.577 1.00 0.97
+ATOM 1718 HZ2 TRP A 95 3.626 1.722 -6.177 1.00 0.97
+ATOM 1719 HH2 TRP A 95 4.889 3.780 -6.753 1.00 0.97
+ATOM 1720 HZ3 TRP A 95 7.092 4.231 -5.756 1.00 0.97
+ATOM 1721 HE3 TRP A 95 8.101 2.651 -4.159 1.00 0.97
+ATOM 1722 N LEU A 96 10.325 0.623 -4.470 1.00 0.95
+ATOM 1723 CA LEU A 96 10.857 1.201 -5.698 1.00 0.95
+ATOM 1724 C LEU A 96 11.635 0.166 -6.503 1.00 0.95
+ATOM 1725 O LEU A 96 11.573 0.146 -7.732 1.00 0.95
+ATOM 1726 CB LEU A 96 11.742 2.413 -5.380 1.00 0.95
+ATOM 1727 CG LEU A 96 11.017 3.675 -4.896 1.00 0.95
+ATOM 1728 CD1 LEU A 96 12.042 4.704 -4.435 1.00 0.95
+ATOM 1729 CD2 LEU A 96 10.154 4.228 -6.020 1.00 0.95
+ATOM 1730 H LEU A 96 10.639 0.985 -3.580 1.00 0.95
+ATOM 1731 HA LEU A 96 10.022 1.551 -6.305 1.00 0.95
+ATOM 1732 1HB LEU A 96 12.454 2.123 -4.609 1.00 0.95
+ATOM 1733 2HB LEU A 96 12.299 2.675 -6.280 1.00 0.95
+ATOM 1734 HG LEU A 96 10.387 3.428 -4.042 1.00 0.95
+ATOM 1735 1HD1 LEU A 96 11.527 5.601 -4.091 1.00 0.95
+ATOM 1736 2HD1 LEU A 96 12.632 4.289 -3.618 1.00 0.95
+ATOM 1737 3HD1 LEU A 96 12.699 4.959 -5.266 1.00 0.95
+ATOM 1738 1HD2 LEU A 96 9.639 5.125 -5.675 1.00 0.95
+ATOM 1739 2HD2 LEU A 96 10.784 4.478 -6.874 1.00 0.95
+ATOM 1740 3HD2 LEU A 96 9.420 3.480 -6.317 1.00 0.95
+ATOM 1741 N GLU A 97 12.438 -0.737 -5.798 1.00 0.95
+ATOM 1742 CA GLU A 97 13.167 -1.824 -6.441 1.00 0.95
+ATOM 1743 C GLU A 97 12.215 -2.799 -7.122 1.00 0.95
+ATOM 1744 O GLU A 97 12.487 -3.283 -8.220 1.00 0.95
+ATOM 1745 CB GLU A 97 14.046 -2.557 -5.425 1.00 0.95
+ATOM 1746 CG GLU A 97 15.275 -1.776 -4.981 1.00 0.95
+ATOM 1747 CD GLU A 97 16.075 -2.494 -3.930 1.00 0.95
+ATOM 1748 OE1 GLU A 97 17.068 -1.959 -3.499 1.00 0.95
+ATOM 1749 OE2 GLU A 97 15.693 -3.578 -3.557 1.00 0.95
+ATOM 1750 H GLU A 97 12.479 -0.644 -4.793 1.00 0.95
+ATOM 1751 HA GLU A 97 13.817 -1.399 -7.206 1.00 0.95
+ATOM 1752 1HB GLU A 97 13.459 -2.791 -4.537 1.00 0.95
+ATOM 1753 2HB GLU A 97 14.385 -3.501 -5.851 1.00 0.95
+ATOM 1754 1HG GLU A 97 15.920 -1.614 -5.844 1.00 0.95
+ATOM 1755 2HG GLU A 97 14.955 -0.820 -4.568 1.00 0.95
+ATOM 1756 N ARG A 98 11.028 -3.129 -6.470 1.00 0.94
+ATOM 1757 CA ARG A 98 10.036 -4.036 -7.033 1.00 0.94
+ATOM 1758 C ARG A 98 9.348 -3.418 -8.244 1.00 0.94
+ATOM 1759 O ARG A 98 8.996 -4.116 -9.194 1.00 0.94
+ATOM 1760 CB ARG A 98 8.999 -4.421 -5.988 1.00 0.94
+ATOM 1761 CG ARG A 98 9.513 -5.352 -4.901 1.00 0.94
+ATOM 1762 CD ARG A 98 8.442 -5.714 -3.938 1.00 0.94
+ATOM 1763 NE ARG A 98 8.943 -6.554 -2.862 1.00 0.94
+ATOM 1764 CZ ARG A 98 8.202 -6.990 -1.824 1.00 0.94
+ATOM 1765 NH1 ARG A 98 6.933 -6.658 -1.736 1.00 0.94
+ATOM 1766 NH2 ARG A 98 8.752 -7.752 -0.894 1.00 0.94
+ATOM 1767 H ARG A 98 10.842 -2.687 -5.581 1.00 0.94
+ATOM 1768 HA ARG A 98 10.543 -4.948 -7.351 1.00 0.94
+ATOM 1769 1HB ARG A 98 8.619 -3.524 -5.502 1.00 0.94
+ATOM 1770 2HB ARG A 98 8.156 -4.913 -6.472 1.00 0.94
+ATOM 1771 1HG ARG A 98 9.891 -6.268 -5.355 1.00 0.94
+ATOM 1772 2HG ARG A 98 10.317 -4.861 -4.351 1.00 0.94
+ATOM 1773 1HD ARG A 98 8.027 -4.808 -3.498 1.00 0.94
+ATOM 1774 2HD ARG A 98 7.655 -6.259 -4.457 1.00 0.94
+ATOM 1775 HE ARG A 98 9.915 -6.831 -2.894 1.00 0.94
+ATOM 1776 1HH1 ARG A 98 6.513 -6.076 -2.447 1.00 0.94
+ATOM 1777 2HH1 ARG A 98 6.378 -6.985 -0.959 1.00 0.94
+ATOM 1778 1HH2 ARG A 98 9.727 -8.008 -0.961 1.00 0.94
+ATOM 1779 2HH2 ARG A 98 8.197 -8.079 -0.117 1.00 0.94
+ATOM 1780 N LEU A 99 9.096 -2.025 -8.224 1.00 0.92
+ATOM 1781 CA LEU A 99 8.392 -1.333 -9.297 1.00 0.92
+ATOM 1782 C LEU A 99 9.293 -1.137 -10.511 1.00 0.92
+ATOM 1783 O LEU A 99 8.814 -0.921 -11.624 1.00 0.92
+ATOM 1784 CB LEU A 99 7.858 0.018 -8.804 1.00 0.92
+ATOM 1785 CG LEU A 99 6.719 -0.032 -7.778 1.00 0.92
+ATOM 1786 CD1 LEU A 99 6.402 1.379 -7.301 1.00 0.92
+ATOM 1787 CD2 LEU A 99 5.497 -0.684 -8.407 1.00 0.92
+ATOM 1788 H LEU A 99 9.478 -1.487 -7.460 1.00 0.92
+ATOM 1789 HA LEU A 99 7.538 -1.939 -9.597 1.00 0.92
+ATOM 1790 1HB LEU A 99 8.684 0.568 -8.356 1.00 0.92
+ATOM 1791 2HB LEU A 99 7.502 0.580 -9.668 1.00 0.92
+ATOM 1792 HG LEU A 99 7.035 -0.614 -6.912 1.00 0.92
+ATOM 1793 1HD1 LEU A 99 5.592 1.343 -6.572 1.00 0.92
+ATOM 1794 2HD1 LEU A 99 7.288 1.814 -6.838 1.00 0.92
+ATOM 1795 3HD1 LEU A 99 6.098 1.990 -8.150 1.00 0.92
+ATOM 1796 1HD2 LEU A 99 4.687 -0.720 -7.678 1.00 0.92
+ATOM 1797 2HD2 LEU A 99 5.179 -0.102 -9.273 1.00 0.92
+ATOM 1798 3HD2 LEU A 99 5.746 -1.697 -8.723 1.00 0.92
+ATOM 1799 N GLY A 100 10.747 -1.181 -10.277 1.00 0.87
+ATOM 1800 CA GLY A 100 11.713 -1.143 -11.369 1.00 0.87
+ATOM 1801 C GLY A 100 11.678 -2.432 -12.180 1.00 0.87
+ATOM 1802 O GLY A 100 12.096 -2.459 -13.338 1.00 0.87
+ATOM 1803 H GLY A 100 11.073 -1.358 -9.338 1.00 0.87
+ATOM 1804 1HA GLY A 100 11.496 -0.294 -12.017 1.00 0.87
+ATOM 1805 2HA GLY A 100 12.713 -0.991 -10.964 1.00 0.87
diff --git a/examples/out/design_000000.trb b/examples/out/design_000000.trb
new file mode 100644
index 0000000000000000000000000000000000000000..90ffd20e67af718b04e90f25f604838be97dbab9
Binary files /dev/null and b/examples/out/design_000000.trb differ
diff --git a/examples/partial_diffusion.sh b/examples/partial_diffusion.sh
new file mode 100644
index 0000000000000000000000000000000000000000..80de8e8a3cc15fed0737cd3d334cf0a7f15669a7
--- /dev/null
+++ b/examples/partial_diffusion.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --pdb out/design_000.pdb \
+ --trb out/design_000.trb \
+ --out out/partial_diffusion_design \
+ --contigs 0 --sampling_temp 0.3 --T 50 --save_best_plddt
diff --git a/examples/pdbs/G12D_manual_mut.pdb b/examples/pdbs/G12D_manual_mut.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..c362fe2913fb906cb280311ed0fa9990f8489a5e
--- /dev/null
+++ b/examples/pdbs/G12D_manual_mut.pdb
@@ -0,0 +1,3257 @@
+CRYST1 270.080 270.080 270.080 90.00 90.00 90.00 P 1 0
+ATOM 1 N SER A 2 14.231 -0.987 5.628 1.00 74.84 A N
+ATOM 2 CA SER A 2 12.974 -0.625 6.266 1.00 74.84 A C
+ATOM 3 C SER A 2 11.882 -1.601 5.857 1.00 74.84 A C
+ATOM 4 O SER A 2 12.007 -2.286 4.846 1.00 74.84 A O
+ATOM 5 CB SER A 2 12.578 0.802 5.899 1.00 74.84 A C
+ATOM 6 OG SER A 2 11.201 1.023 6.128 1.00 74.84 A O
+ATOM 7 N HIS A 3 10.815 -1.667 6.650 1.00 71.81 A N
+ATOM 8 CA HIS A 3 9.689 -2.542 6.366 1.00 71.81 A C
+ATOM 9 C HIS A 3 8.421 -1.915 6.903 1.00 71.81 A C
+ATOM 10 O HIS A 3 8.457 -1.035 7.761 1.00 71.81 A O
+ATOM 11 CB HIS A 3 9.889 -3.925 6.966 1.00 71.81 A C
+ATOM 12 CG HIS A 3 11.065 -4.651 6.406 1.00 71.81 A C
+ATOM 13 CD2 HIS A 3 12.241 -5.015 6.967 1.00 71.81 A C
+ATOM 14 ND1 HIS A 3 11.123 -5.059 5.093 1.00 71.81 A N
+ATOM 15 CE1 HIS A 3 12.275 -5.666 4.874 1.00 71.81 A C
+ATOM 16 NE2 HIS A 3 12.972 -5.653 5.995 1.00 71.81 A N
+ATOM 17 N SER A 4 7.292 -2.378 6.385 1.00 65.41 A N
+ATOM 18 CA SER A 4 6.008 -1.832 6.783 1.00 65.41 A C
+ATOM 19 C SER A 4 4.990 -2.948 6.819 1.00 65.41 A C
+ATOM 20 O SER A 4 5.228 -4.053 6.335 1.00 65.41 A O
+ATOM 21 CB SER A 4 5.535 -0.747 5.826 1.00 65.41 A C
+ATOM 22 OG SER A 4 5.722 -1.156 4.487 1.00 65.41 A O
+ATOM 23 N MET A 5 3.833 -2.637 7.381 1.00 59.94 A N
+ATOM 24 CA MET A 5 2.736 -3.587 7.404 1.00 59.94 A C
+ATOM 25 C MET A 5 1.451 -2.798 7.289 1.00 59.94 A C
+ATOM 26 O MET A 5 1.131 -2.007 8.173 1.00 59.94 A O
+ATOM 27 CB MET A 5 2.784 -4.424 8.677 1.00 59.94 A C
+ATOM 28 CG MET A 5 1.594 -5.307 8.937 1.00 59.94 A C
+ATOM 29 SD MET A 5 1.749 -6.117 10.537 1.00 59.94 A S
+ATOM 30 CE MET A 5 0.532 -5.235 11.490 1.00 59.94 A C
+ATOM 31 N ARG A 6 0.725 -2.999 6.201 1.00 56.64 A N
+ATOM 32 CA ARG A 6 -0.430 -2.184 5.887 1.00 56.64 A C
+ATOM 33 C ARG A 6 -1.673 -3.049 5.941 1.00 56.64 A C
+ATOM 34 O ARG A 6 -1.616 -4.254 5.692 1.00 56.64 A O
+ATOM 35 CB ARG A 6 -0.357 -1.601 4.486 1.00 56.64 A C
+ATOM 36 CG ARG A 6 0.546 -0.420 4.255 1.00 56.64 A C
+ATOM 37 CD ARG A 6 1.942 -0.908 4.036 1.00 56.64 A C
+ATOM 38 NE ARG A 6 1.998 -1.703 2.811 1.00 56.64 A N
+ATOM 39 CZ ARG A 6 3.021 -2.478 2.479 1.00 56.64 A C
+ATOM 40 NH1 ARG A 6 3.005 -3.185 1.362 1.00 56.64 A N1+
+ATOM 41 NH2 ARG A 6 4.074 -2.527 3.270 1.00 56.64 A N
+ATOM 42 N TYR A 7 -2.806 -2.421 6.206 1.00 48.28 A N
+ATOM 43 CA TYR A 7 -4.084 -3.116 6.223 1.00 48.28 A C
+ATOM 44 C TYR A 7 -5.094 -2.278 5.456 1.00 48.28 A C
+ATOM 45 O TYR A 7 -5.602 -1.287 5.978 1.00 48.28 A O
+ATOM 46 CB TYR A 7 -4.545 -3.375 7.648 1.00 48.28 A C
+ATOM 47 CG TYR A 7 -3.875 -4.555 8.272 1.00 48.28 A C
+ATOM 48 CD1 TYR A 7 -4.119 -5.824 7.808 1.00 48.28 A C
+ATOM 49 CD2 TYR A 7 -2.988 -4.401 9.308 1.00 48.28 A C
+ATOM 50 CE1 TYR A 7 -3.498 -6.900 8.347 1.00 48.28 A C
+ATOM 51 CE2 TYR A 7 -2.366 -5.474 9.854 1.00 48.28 A C
+ATOM 52 CZ TYR A 7 -2.622 -6.723 9.371 1.00 48.28 A C
+ATOM 53 OH TYR A 7 -2.003 -7.817 9.918 1.00 48.28 A O
+ATOM 54 N PHE A 8 -5.396 -2.680 4.229 1.00 40.07 A N
+ATOM 55 CA PHE A 8 -6.361 -1.978 3.401 1.00 40.07 A C
+ATOM 56 C PHE A 8 -7.737 -2.579 3.622 1.00 40.07 A C
+ATOM 57 O PHE A 8 -7.890 -3.796 3.571 1.00 40.07 A O
+ATOM 58 CB PHE A 8 -5.966 -2.104 1.940 1.00 40.07 A C
+ATOM 59 CG PHE A 8 -4.615 -1.575 1.644 1.00 40.07 A C
+ATOM 60 CD1 PHE A 8 -3.533 -2.411 1.637 1.00 40.07 A C
+ATOM 61 CD2 PHE A 8 -4.421 -0.244 1.391 1.00 40.07 A C
+ATOM 62 CE1 PHE A 8 -2.296 -1.926 1.365 1.00 40.07 A C
+ATOM 63 CE2 PHE A 8 -3.184 0.236 1.126 1.00 40.07 A C
+ATOM 64 CZ PHE A 8 -2.123 -0.605 1.109 1.00 40.07 A C
+ATOM 65 N PHE A 9 -8.734 -1.739 3.843 1.00 40.29 A N
+ATOM 66 CA PHE A 9 -10.046 -2.223 4.261 1.00 40.29 A C
+ATOM 67 C PHE A 9 -11.120 -1.619 3.371 1.00 40.29 A C
+ATOM 68 O PHE A 9 -11.816 -0.698 3.785 1.00 40.29 A O
+ATOM 69 CB PHE A 9 -10.290 -1.859 5.699 1.00 40.29 A C
+ATOM 70 CG PHE A 9 -9.560 -2.711 6.665 1.00 40.29 A C
+ATOM 71 CD1 PHE A 9 -8.335 -2.331 7.126 1.00 40.29 A C
+ATOM 72 CD2 PHE A 9 -10.078 -3.901 7.092 1.00 40.29 A C
+ATOM 73 CE1 PHE A 9 -7.662 -3.103 8.009 1.00 40.29 A C
+ATOM 74 CE2 PHE A 9 -9.391 -4.678 7.976 1.00 40.29 A C
+ATOM 75 CZ PHE A 9 -8.180 -4.277 8.429 1.00 40.29 A C
+ATOM 76 N THR A 10 -11.317 -2.188 2.192 1.00 41.87 A N
+ATOM 77 CA THR A 10 -12.316 -1.670 1.269 1.00 41.87 A C
+ATOM 78 C THR A 10 -13.711 -2.078 1.722 1.00 41.87 A C
+ATOM 79 O THR A 10 -13.901 -3.147 2.290 1.00 41.87 A O
+ATOM 80 CB THR A 10 -12.063 -2.211 -0.129 1.00 41.87 A C
+ATOM 81 CG2 THR A 10 -12.874 -1.426 -1.110 1.00 41.87 A C
+ATOM 82 OG1 THR A 10 -10.680 -2.065 -0.462 1.00 41.87 A O
+ATOM 83 N SER A 11 -14.702 -1.234 1.457 1.00 47.15 A N
+ATOM 84 CA SER A 11 -16.055 -1.562 1.893 1.00 47.15 A C
+ATOM 85 C SER A 11 -17.085 -0.884 0.992 1.00 47.15 A C
+ATOM 86 O SER A 11 -17.527 0.222 1.300 1.00 47.15 A O
+ATOM 87 CB SER A 11 -16.246 -1.144 3.331 1.00 47.15 A C
+ATOM 88 OG SER A 11 -17.562 -1.418 3.753 1.00 47.15 A O
+ATOM 89 N VAL A 12 -17.543 -1.593 -0.039 1.00 51.01 A N
+ATOM 90 CA VAL A 12 -18.483 -1.046 -1.012 1.00 51.01 A C
+ATOM 91 C VAL A 12 -19.905 -1.246 -0.519 1.00 51.01 A C
+ATOM 92 O VAL A 12 -20.276 -2.340 -0.096 1.00 51.01 A O
+ATOM 93 CB VAL A 12 -18.289 -1.703 -2.381 1.00 51.01 A C
+ATOM 94 CG1 VAL A 12 -19.124 -0.997 -3.413 1.00 51.01 A C
+ATOM 95 CG2 VAL A 12 -16.831 -1.682 -2.758 1.00 51.01 A C
+ATOM 96 N SER A 13 -20.711 -0.201 -0.604 1.00 59.04 A N
+ATOM 97 CA SER A 13 -21.988 -0.182 0.092 1.00 59.04 A C
+ATOM 98 C SER A 13 -23.090 -0.972 -0.595 1.00 59.04 A C
+ATOM 99 O SER A 13 -24.134 -1.179 0.029 1.00 59.04 A O
+ATOM 100 CB SER A 13 -22.470 1.245 0.272 1.00 59.04 A C
+ATOM 101 OG SER A 13 -23.794 1.254 0.755 1.00 59.04 A O
+ATOM 102 N ARG A 14 -22.916 -1.352 -1.867 1.00 60.19 A N
+ATOM 103 CA ARG A 14 -23.713 -2.345 -2.599 1.00 60.19 A C
+ATOM 104 C ARG A 14 -25.171 -2.405 -2.165 1.00 60.19 A C
+ATOM 105 O ARG A 14 -25.656 -3.477 -1.799 1.00 60.19 A O
+ATOM 106 CB ARG A 14 -23.083 -3.728 -2.433 1.00 60.19 A C
+ATOM 107 CG ARG A 14 -23.498 -4.753 -3.444 1.00 60.19 A C
+ATOM 108 CD ARG A 14 -22.619 -5.971 -3.357 1.00 60.19 A C
+ATOM 109 NE ARG A 14 -22.903 -6.742 -2.160 1.00 60.19 A N
+ATOM 110 CZ ARG A 14 -22.344 -7.910 -1.881 1.00 60.19 A C
+ATOM 111 NH1 ARG A 14 -21.472 -8.442 -2.720 1.00 60.19 A N1+
+ATOM 112 NH2 ARG A 14 -22.657 -8.545 -0.763 1.00 60.19 A N
+ATOM 113 N PRO A 15 -25.900 -1.299 -2.185 1.00 65.94 A N
+ATOM 114 CA PRO A 15 -27.227 -1.283 -1.578 1.00 65.94 A C
+ATOM 115 C PRO A 15 -28.249 -1.992 -2.445 1.00 65.94 A C
+ATOM 116 O PRO A 15 -28.044 -2.233 -3.634 1.00 65.94 A O
+ATOM 117 CB PRO A 15 -27.546 0.208 -1.472 1.00 65.94 A C
+ATOM 118 CG PRO A 15 -26.853 0.786 -2.614 1.00 65.94 A C
+ATOM 119 CD PRO A 15 -25.603 -0.028 -2.851 1.00 65.94 A C
+ATOM 120 N GLY A 16 -29.370 -2.329 -1.814 1.00 70.34 A N
+ATOM 121 CA GLY A 16 -30.451 -2.998 -2.503 1.00 70.34 A C
+ATOM 122 C GLY A 16 -30.177 -4.455 -2.803 1.00 70.34 A C
+ATOM 123 O GLY A 16 -30.941 -5.330 -2.393 1.00 70.34 A O
+ATOM 124 N ARG A 17 -29.083 -4.737 -3.504 1.00 69.35 A N
+ATOM 125 CA ARG A 17 -28.819 -6.073 -4.015 1.00 69.35 A C
+ATOM 126 C ARG A 17 -27.947 -6.897 -3.078 1.00 69.35 A C
+ATOM 127 O ARG A 17 -27.160 -7.731 -3.533 1.00 69.35 A O
+ATOM 128 CB ARG A 17 -28.191 -5.997 -5.405 1.00 69.35 A C
+ATOM 129 CG ARG A 17 -26.814 -5.375 -5.456 1.00 69.35 A C
+ATOM 130 CD ARG A 17 -26.306 -5.310 -6.883 1.00 69.35 A C
+ATOM 131 NE ARG A 17 -24.873 -5.055 -6.957 1.00 69.35 A N
+ATOM 132 CZ ARG A 17 -24.322 -3.853 -6.829 1.00 69.35 A C
+ATOM 133 NH1 ARG A 17 -23.008 -3.707 -6.916 1.00 69.35 A N1+
+ATOM 134 NH2 ARG A 17 -25.087 -2.793 -6.610 1.00 69.35 A N
+ATOM 135 N GLY A 18 -28.072 -6.690 -1.785 1.00 69.58 A N
+ATOM 136 CA GLY A 18 -27.408 -7.563 -0.851 1.00 69.58 A C
+ATOM 137 C GLY A 18 -26.957 -6.806 0.390 1.00 69.58 A C
+ATOM 138 O GLY A 18 -27.703 -5.998 0.947 1.00 69.58 A O
+ATOM 139 N GLU A 19 -25.730 -7.106 0.806 1.00 68.27 A N
+ATOM 140 CA GLU A 19 -25.099 -6.617 2.015 1.00 68.27 A C
+ATOM 141 C GLU A 19 -23.742 -6.033 1.651 1.00 68.27 A C
+ATOM 142 O GLU A 19 -23.058 -6.587 0.785 1.00 68.27 A O
+ATOM 143 CB GLU A 19 -24.936 -7.768 3.015 1.00 68.27 A C
+ATOM 144 CG GLU A 19 -24.468 -7.374 4.378 1.00 68.27 A C
+ATOM 145 CD GLU A 19 -25.612 -6.982 5.266 1.00 68.27 A C
+ATOM 146 OE1 GLU A 19 -26.412 -7.862 5.630 1.00 68.27 A O
+ATOM 147 OE2 GLU A 19 -25.716 -5.793 5.596 1.00 68.27 A O1-
+ATOM 148 N PRO A 20 -23.332 -4.912 2.274 1.00 58.29 A N
+ATOM 149 CA PRO A 20 -22.075 -4.263 1.892 1.00 58.29 A C
+ATOM 150 C PRO A 20 -20.938 -5.235 1.708 1.00 58.29 A C
+ATOM 151 O PRO A 20 -20.567 -5.940 2.643 1.00 58.29 A O
+ATOM 152 CB PRO A 20 -21.798 -3.322 3.065 1.00 58.29 A C
+ATOM 153 CG PRO A 20 -23.096 -3.134 3.748 1.00 58.29 A C
+ATOM 154 CD PRO A 20 -24.127 -4.044 3.148 1.00 58.29 A C
+ATOM 155 N ARG A 21 -20.393 -5.307 0.507 1.00 54.56 A N
+ATOM 156 CA ARG A 21 -19.228 -6.136 0.332 1.00 54.56 A C
+ATOM 157 C ARG A 21 -18.134 -5.604 1.232 1.00 54.56 A C
+ATOM 158 O ARG A 21 -18.207 -4.485 1.734 1.00 54.56 A O
+ATOM 159 CB ARG A 21 -18.774 -6.158 -1.126 1.00 54.56 A C
+ATOM 160 CG ARG A 21 -17.833 -7.298 -1.398 1.00 54.56 A C
+ATOM 161 CD ARG A 21 -17.495 -7.502 -2.840 1.00 54.56 A C
+ATOM 162 NE ARG A 21 -16.524 -8.584 -2.954 1.00 54.56 A N
+ATOM 163 CZ ARG A 21 -16.074 -9.082 -4.100 1.00 54.56 A C
+ATOM 164 NH1 ARG A 21 -16.505 -8.601 -5.255 1.00 54.56 A N1+
+ATOM 165 NH2 ARG A 21 -15.191 -10.066 -4.090 1.00 54.56 A N
+ATOM 166 N PHE A 22 -17.132 -6.430 1.478 1.00 46.66 A N
+ATOM 167 CA PHE A 22 -16.050 -6.014 2.353 1.00 46.66 A C
+ATOM 168 C PHE A 22 -14.872 -6.910 2.042 1.00 46.66 A C
+ATOM 169 O PHE A 22 -14.893 -8.089 2.384 1.00 46.66 A O
+ATOM 170 CB PHE A 22 -16.478 -6.107 3.811 1.00 46.66 A C
+ATOM 171 CG PHE A 22 -15.379 -5.867 4.793 1.00 46.66 A C
+ATOM 172 CD1 PHE A 22 -14.541 -6.885 5.176 1.00 46.66 A C
+ATOM 173 CD2 PHE A 22 -15.184 -4.619 5.338 1.00 46.66 A C
+ATOM 174 CE1 PHE A 22 -13.548 -6.666 6.077 1.00 46.66 A C
+ATOM 175 CE2 PHE A 22 -14.168 -4.400 6.228 1.00 46.66 A C
+ATOM 176 CZ PHE A 22 -13.354 -5.426 6.595 1.00 46.66 A C
+ATOM 177 N ILE A 23 -13.874 -6.368 1.366 1.00 43.99 A N
+ATOM 178 CA ILE A 23 -12.610 -7.048 1.169 1.00 43.99 A C
+ATOM 179 C ILE A 23 -11.604 -6.366 2.065 1.00 43.99 A C
+ATOM 180 O ILE A 23 -11.485 -5.140 2.042 1.00 43.99 A O
+ATOM 181 CB ILE A 23 -12.162 -7.020 -0.297 1.00 43.99 A C
+ATOM 182 CG1 ILE A 23 -13.244 -7.635 -1.161 1.00 43.99 A C
+ATOM 183 CG2 ILE A 23 -10.885 -7.796 -0.477 1.00 43.99 A C
+ATOM 184 CD1 ILE A 23 -12.903 -7.688 -2.603 1.00 43.99 A C
+ATOM 185 N ALA A 24 -10.907 -7.147 2.879 1.00 40.39 A N
+ATOM 186 CA ALA A 24 -9.960 -6.612 3.846 1.00 40.39 A C
+ATOM 187 C ALA A 24 -8.632 -7.280 3.592 1.00 40.39 A C
+ATOM 188 O ALA A 24 -8.290 -8.270 4.230 1.00 40.39 A O
+ATOM 189 CB ALA A 24 -10.416 -6.835 5.255 1.00 40.39 A C
+ATOM 190 N VAL A 25 -7.882 -6.729 2.683 1.00 41.35 A N
+ATOM 191 CA VAL A 25 -6.564 -7.247 2.417 1.00 41.35 A C
+ATOM 192 C VAL A 25 -5.615 -6.781 3.512 1.00 41.35 A C
+ATOM 193 O VAL A 25 -5.845 -5.778 4.173 1.00 41.35 A O
+ATOM 194 CB VAL A 25 -6.119 -6.783 1.028 1.00 41.35 A C
+ATOM 195 CG1 VAL A 25 -4.686 -7.074 0.782 1.00 41.35 A C
+ATOM 196 CG2 VAL A 25 -6.970 -7.429 0.004 1.00 41.35 A C
+ATOM 197 N GLY A 26 -4.568 -7.543 3.734 1.00 49.17 A N
+ATOM 198 CA GLY A 26 -3.479 -7.110 4.588 1.00 49.17 A C
+ATOM 199 C GLY A 26 -2.178 -7.377 3.878 1.00 49.17 A C
+ATOM 200 O GLY A 26 -2.049 -8.360 3.159 1.00 49.17 A O
+ATOM 201 N TYR A 27 -1.215 -6.488 4.078 1.00 56.65 A N
+ATOM 202 CA TYR A 27 0.001 -6.510 3.287 1.00 56.65 A C
+ATOM 203 C TYR A 27 1.227 -6.451 4.174 1.00 56.65 A C
+ATOM 204 O TYR A 27 1.192 -5.892 5.267 1.00 56.65 A O
+ATOM 205 CB TYR A 27 0.077 -5.331 2.333 1.00 56.65 A C
+ATOM 206 CG TYR A 27 -0.521 -5.567 0.982 1.00 56.65 A C
+ATOM 207 CD1 TYR A 27 0.183 -6.236 0.008 1.00 56.65 A C
+ATOM 208 CD2 TYR A 27 -1.763 -5.068 0.662 1.00 56.65 A C
+ATOM 209 CE1 TYR A 27 -0.350 -6.442 -1.225 1.00 56.65 A C
+ATOM 210 CE2 TYR A 27 -2.296 -5.258 -0.572 1.00 56.65 A C
+ATOM 211 CZ TYR A 27 -1.588 -5.952 -1.512 1.00 56.65 A C
+ATOM 212 OH TYR A 27 -2.117 -6.152 -2.761 1.00 56.65 A O
+ATOM 213 N VAL A 28 2.326 -7.004 3.678 1.00 62.48 A N
+ATOM 214 CA VAL A 28 3.612 -6.747 4.301 1.00 62.48 A C
+ATOM 215 C VAL A 28 4.700 -6.519 3.251 1.00 62.48 A C
+ATOM 216 O VAL A 28 5.240 -7.450 2.648 1.00 62.48 A O
+ATOM 217 CB VAL A 28 3.955 -7.857 5.306 1.00 62.48 A C
+ATOM 218 CG1 VAL A 28 4.141 -9.188 4.634 1.00 62.48 A C
+ATOM 219 CG2 VAL A 28 5.177 -7.477 6.074 1.00 62.48 A C
+ATOM 220 N ASP A 29 4.986 -5.244 3.021 1.00 70.76 A N
+ATOM 221 CA ASP A 29 6.078 -4.639 2.265 1.00 70.76 A C
+ATOM 222 C ASP A 29 5.991 -4.633 0.743 1.00 70.76 A C
+ATOM 223 O ASP A 29 6.591 -3.748 0.133 1.00 70.76 A O
+ATOM 224 CB ASP A 29 7.390 -5.329 2.631 1.00 70.76 A C
+ATOM 225 CG ASP A 29 7.926 -4.880 3.955 1.00 70.76 A C
+ATOM 226 OD1 ASP A 29 7.510 -3.805 4.435 1.00 70.76 A O
+ATOM 227 OD2 ASP A 29 8.776 -5.599 4.514 1.00 70.76 A O1-
+ATOM 228 N ASP A 30 5.324 -5.592 0.104 1.00 75.51 A N
+ATOM 229 CA ASP A 30 4.374 -5.398 -0.991 1.00 75.51 A C
+ATOM 230 C ASP A 30 3.477 -6.612 -1.103 1.00 75.51 A C
+ATOM 231 O ASP A 30 2.523 -6.614 -1.881 1.00 75.51 A O
+ATOM 232 CB ASP A 30 5.026 -5.112 -2.349 1.00 75.51 A C
+ATOM 233 CG ASP A 30 5.604 -3.722 -2.448 1.00 75.51 A C
+ATOM 234 OD1 ASP A 30 5.064 -2.812 -1.787 1.00 75.51 A O
+ATOM 235 OD2 ASP A 30 6.578 -3.526 -3.194 1.00 75.51 A O1-
+ATOM 236 N THR A 31 3.789 -7.656 -0.346 1.00 64.20 A N
+ATOM 237 CA THR A 31 3.151 -8.942 -0.542 1.00 64.20 A C
+ATOM 238 C THR A 31 1.948 -9.065 0.368 1.00 64.20 A C
+ATOM 239 O THR A 31 1.884 -8.459 1.436 1.00 64.20 A O
+ATOM 240 CB THR A 31 4.110 -10.098 -0.285 1.00 64.20 A C
+ATOM 241 CG2 THR A 31 4.653 -10.030 1.099 1.00 64.20 A C
+ATOM 242 OG1 THR A 31 3.404 -11.331 -0.431 1.00 64.20 A O
+ATOM 243 N GLN A 32 0.984 -9.838 -0.087 1.00 62.19 A N
+ATOM 244 CA GLN A 32 -0.261 -10.038 0.625 1.00 62.19 A C
+ATOM 245 C GLN A 32 -0.180 -11.340 1.402 1.00 62.19 A C
+ATOM 246 O GLN A 32 0.352 -12.333 0.902 1.00 62.19 A O
+ATOM 247 CB GLN A 32 -1.420 -10.080 -0.360 1.00 62.19 A C
+ATOM 248 CG GLN A 32 -2.766 -10.011 0.268 1.00 62.19 A C
+ATOM 249 CD GLN A 32 -3.837 -10.390 -0.707 1.00 62.19 A C
+ATOM 250 NE2 GLN A 32 -5.027 -9.855 -0.521 1.00 62.19 A N
+ATOM 251 OE1 GLN A 32 -3.599 -11.173 -1.624 1.00 62.19 A O
+ATOM 252 N PHE A 33 -0.686 -11.333 2.634 1.00 57.29 A N
+ATOM 253 CA PHE A 33 -0.719 -12.556 3.413 1.00 57.29 A C
+ATOM 254 C PHE A 33 -2.091 -12.930 3.945 1.00 57.29 A C
+ATOM 255 O PHE A 33 -2.293 -14.096 4.279 1.00 57.29 A O
+ATOM 256 CB PHE A 33 0.290 -12.494 4.574 1.00 57.29 A C
+ATOM 257 CG PHE A 33 0.023 -11.415 5.575 1.00 57.29 A C
+ATOM 258 CD1 PHE A 33 -0.850 -11.610 6.611 1.00 57.29 A C
+ATOM 259 CD2 PHE A 33 0.677 -10.221 5.502 1.00 57.29 A C
+ATOM 260 CE1 PHE A 33 -1.091 -10.621 7.530 1.00 57.29 A C
+ATOM 261 CE2 PHE A 33 0.433 -9.237 6.418 1.00 57.29 A C
+ATOM 262 CZ PHE A 33 -0.452 -9.440 7.431 1.00 57.29 A C
+ATOM 263 N VAL A 34 -3.046 -12.009 3.997 1.00 52.72 A N
+ATOM 264 CA VAL A 34 -4.406 -12.354 4.370 1.00 52.72 A C
+ATOM 265 C VAL A 34 -5.378 -11.721 3.390 1.00 52.72 A C
+ATOM 266 O VAL A 34 -5.019 -10.870 2.586 1.00 52.72 A O
+ATOM 267 CB VAL A 34 -4.757 -11.938 5.807 1.00 52.72 A C
+ATOM 268 CG1 VAL A 34 -3.979 -12.739 6.790 1.00 52.72 A C
+ATOM 269 CG2 VAL A 34 -4.469 -10.493 6.009 1.00 52.72 A C
+ATOM 270 N ARG A 35 -6.625 -12.165 3.474 1.00 57.17 A N
+ATOM 271 CA ARG A 35 -7.721 -11.686 2.649 1.00 57.17 A C
+ATOM 272 C ARG A 35 -8.997 -11.917 3.430 1.00 57.17 A C
+ATOM 273 O ARG A 35 -9.015 -12.684 4.388 1.00 57.17 A O
+ATOM 274 CB ARG A 35 -7.797 -12.425 1.317 1.00 57.17 A C
+ATOM 275 CG ARG A 35 -8.577 -13.719 1.454 1.00 57.17 A C
+ATOM 276 CD ARG A 35 -8.395 -14.678 0.306 1.00 57.17 A C
+ATOM 277 NE ARG A 35 -8.988 -14.162 -0.916 1.00 57.17 A N
+ATOM 278 CZ ARG A 35 -10.288 -14.182 -1.170 1.00 57.17 A C
+ATOM 279 NH1 ARG A 35 -10.748 -13.690 -2.309 1.00 57.17 A N1+
+ATOM 280 NH2 ARG A 35 -11.132 -14.689 -0.283 1.00 57.17 A N
+ATOM 281 N PHE A 36 -10.062 -11.236 3.040 1.00 48.80 A N
+ATOM 282 CA PHE A 36 -11.349 -11.537 3.650 1.00 48.80 A C
+ATOM 283 C PHE A 36 -12.459 -10.975 2.766 1.00 48.80 A C
+ATOM 284 O PHE A 36 -12.777 -9.792 2.866 1.00 48.80 A O
+ATOM 285 CB PHE A 36 -11.405 -10.965 5.048 1.00 48.80 A C
+ATOM 286 CG PHE A 36 -12.682 -11.218 5.750 1.00 48.80 A C
+ATOM 287 CD1 PHE A 36 -12.867 -12.374 6.457 1.00 48.80 A C
+ATOM 288 CD2 PHE A 36 -13.690 -10.294 5.730 1.00 48.80 A C
+ATOM 289 CE1 PHE A 36 -14.036 -12.609 7.106 1.00 48.80 A C
+ATOM 290 CE2 PHE A 36 -14.859 -10.530 6.385 1.00 48.80 A C
+ATOM 291 CZ PHE A 36 -15.030 -11.688 7.071 1.00 48.80 A C
+ATOM 292 N ASP A 37 -13.073 -11.821 1.951 1.00 55.56 A N
+ATOM 293 CA ASP A 37 -14.253 -11.410 1.210 1.00 55.56 A C
+ATOM 294 C ASP A 37 -15.483 -11.725 2.042 1.00 55.56 A C
+ATOM 295 O ASP A 37 -15.590 -12.812 2.604 1.00 55.56 A O
+ATOM 296 CB ASP A 37 -14.355 -12.144 -0.125 1.00 55.56 A C
+ATOM 297 CG ASP A 37 -13.245 -11.793 -1.075 1.00 55.56 A C
+ATOM 298 OD1 ASP A 37 -12.707 -10.682 -0.986 1.00 55.56 A O
+ATOM 299 OD2 ASP A 37 -12.917 -12.626 -1.938 1.00 55.56 A O1-
+ATOM 300 N SER A 38 -16.419 -10.791 2.112 1.00 55.20 A N
+ATOM 301 CA SER A 38 -17.660 -11.128 2.792 1.00 55.20 A C
+ATOM 302 C SER A 38 -18.593 -11.936 1.911 1.00 55.20 A C
+ATOM 303 O SER A 38 -19.789 -12.022 2.203 1.00 55.20 A O
+ATOM 304 CB SER A 38 -18.387 -9.880 3.273 1.00 55.20 A C
+ATOM 305 OG SER A 38 -18.879 -9.148 2.179 1.00 55.20 A O
+ATOM 306 N ASP A 39 -18.082 -12.487 0.812 1.00 60.75 A N
+ATOM 307 CA ASP A 39 -18.827 -13.449 0.009 1.00 60.75 A C
+ATOM 308 C ASP A 39 -17.807 -14.358 -0.673 1.00 60.75 A C
+ATOM 309 O ASP A 39 -17.314 -14.051 -1.760 1.00 60.75 A O
+ATOM 310 CB ASP A 39 -19.736 -12.742 -0.985 1.00 60.75 A C
+ATOM 311 CG ASP A 39 -19.028 -11.657 -1.766 1.00 60.75 A C
+ATOM 312 OD1 ASP A 39 -17.805 -11.496 -1.605 1.00 60.75 A O
+ATOM 313 OD2 ASP A 39 -19.707 -10.948 -2.534 1.00 60.75 A O1-
+ATOM 314 N ALA A 40 -17.513 -15.478 -0.035 1.00 71.59 A N
+ATOM 315 CA ALA A 40 -16.539 -16.424 -0.552 1.00 71.59 A C
+ATOM 316 C ALA A 40 -16.836 -17.784 0.066 1.00 71.59 A C
+ATOM 317 O ALA A 40 -17.930 -18.018 0.592 1.00 71.59 A O
+ATOM 318 CB ALA A 40 -15.111 -15.954 -0.263 1.00 71.59 A C
+ATOM 319 N ALA A 41 -15.864 -18.695 -0.024 1.00 74.49 A N
+ATOM 320 CA ALA A 41 -15.995 -20.006 0.598 1.00 74.49 A C
+ATOM 321 C ALA A 41 -16.471 -19.889 2.038 1.00 74.49 A C
+ATOM 322 O ALA A 41 -17.567 -20.337 2.386 1.00 74.49 A O
+ATOM 323 CB ALA A 41 -14.658 -20.745 0.541 1.00 74.49 A C
+ATOM 324 N SER A 42 -15.673 -19.244 2.880 1.00 71.88 A N
+ATOM 325 CA SER A 42 -15.915 -19.189 4.310 1.00 71.88 A C
+ATOM 326 C SER A 42 -15.948 -17.743 4.773 1.00 71.88 A C
+ATOM 327 O SER A 42 -15.676 -16.817 4.008 1.00 71.88 A O
+ATOM 328 CB SER A 42 -14.834 -19.959 5.064 1.00 71.88 A C
+ATOM 329 OG SER A 42 -13.559 -19.648 4.535 1.00 71.88 A O
+ATOM 330 N GLN A 43 -16.305 -17.548 6.031 1.00 67.52 A N
+ATOM 331 CA GLN A 43 -16.146 -16.249 6.674 1.00 67.52 A C
+ATOM 332 C GLN A 43 -14.894 -16.247 7.546 1.00 67.52 A C
+ATOM 333 O GLN A 43 -14.950 -16.071 8.758 1.00 67.52 A O
+ATOM 334 CB GLN A 43 -17.383 -15.914 7.487 1.00 67.52 A C
+ATOM 335 CG GLN A 43 -18.667 -16.133 6.752 1.00 67.52 A C
+ATOM 336 CD GLN A 43 -18.895 -15.113 5.682 1.00 67.52 A C
+ATOM 337 NE2 GLN A 43 -19.274 -13.908 6.080 1.00 67.52 A N
+ATOM 338 OE1 GLN A 43 -18.739 -15.402 4.504 1.00 67.52 A O
+ATOM 339 N ARG A 44 -13.744 -16.440 6.904 1.00 69.09 A N
+ATOM 340 CA ARG A 44 -12.481 -16.598 7.605 1.00 69.09 A C
+ATOM 341 C ARG A 44 -11.414 -15.718 6.976 1.00 69.09 A C
+ATOM 342 O ARG A 44 -11.438 -15.452 5.775 1.00 69.09 A O
+ATOM 343 CB ARG A 44 -11.983 -18.056 7.560 1.00 69.09 A C
+ATOM 344 CG ARG A 44 -13.005 -19.100 7.967 1.00 69.09 A C
+ATOM 345 CD ARG A 44 -13.122 -19.244 9.455 1.00 69.09 A C
+ATOM 346 NE ARG A 44 -11.917 -19.809 10.036 1.00 69.09 A N
+ATOM 347 CZ ARG A 44 -11.649 -19.785 11.333 1.00 69.09 A C
+ATOM 348 NH1 ARG A 44 -12.507 -19.223 12.170 1.00 69.09 A N1+
+ATOM 349 NH2 ARG A 44 -10.529 -20.324 11.788 1.00 69.09 A N
+ATOM 350 N MET A 45 -10.440 -15.321 7.787 1.00 68.13 A N
+ATOM 351 CA MET A 45 -9.185 -14.804 7.256 1.00 68.13 A C
+ATOM 352 C MET A 45 -8.452 -15.937 6.568 1.00 68.13 A C
+ATOM 353 O MET A 45 -7.520 -16.537 7.102 1.00 68.13 A O
+ATOM 354 CB MET A 45 -8.310 -14.190 8.335 1.00 68.13 A C
+ATOM 355 CG MET A 45 -8.641 -12.798 8.760 1.00 68.13 A C
+ATOM 356 SD MET A 45 -7.738 -11.870 7.509 1.00 68.13 A S
+ATOM 357 CE MET A 45 -8.109 -10.181 7.887 1.00 68.13 A C
+ATOM 358 N GLU A 46 -8.884 -16.227 5.365 1.00 68.37 A N
+ATOM 359 CA GLU A 46 -8.170 -17.190 4.563 1.00 68.37 A C
+ATOM 360 C GLU A 46 -6.727 -16.732 4.433 1.00 68.37 A C
+ATOM 361 O GLU A 46 -6.482 -15.563 4.133 1.00 68.37 A O
+ATOM 362 CB GLU A 46 -8.815 -17.321 3.190 1.00 68.37 A C
+ATOM 363 CG GLU A 46 -10.301 -17.620 3.237 1.00 68.37 A C
+ATOM 364 CD GLU A 46 -10.812 -18.212 1.939 1.00 68.37 A C
+ATOM 365 OE1 GLU A 46 -10.012 -18.333 0.990 1.00 68.37 A O
+ATOM 366 OE2 GLU A 46 -12.010 -18.558 1.864 1.00 68.37 A O1-
+ATOM 367 N PRO A 47 -5.760 -17.590 4.703 1.00 66.59 A N
+ATOM 368 CA PRO A 47 -4.367 -17.202 4.525 1.00 66.59 A C
+ATOM 369 C PRO A 47 -4.107 -16.921 3.063 1.00 66.59 A C
+ATOM 370 O PRO A 47 -4.769 -17.460 2.177 1.00 66.59 A O
+ATOM 371 CB PRO A 47 -3.594 -18.437 4.982 1.00 66.59 A C
+ATOM 372 CG PRO A 47 -4.528 -19.173 5.835 1.00 66.59 A C
+ATOM 373 CD PRO A 47 -5.886 -18.930 5.283 1.00 66.59 A C
+ATOM 374 N ARG A 48 -3.124 -16.069 2.807 1.00 65.33 A N
+ATOM 375 CA ARG A 48 -2.777 -15.741 1.437 1.00 65.33 A C
+ATOM 376 C ARG A 48 -1.289 -15.873 1.175 1.00 65.33 A C
+ATOM 377 O ARG A 48 -0.832 -15.521 0.086 1.00 65.33 A O
+ATOM 378 CB ARG A 48 -3.249 -14.333 1.088 1.00 65.33 A C
+ATOM 379 CG ARG A 48 -4.697 -14.245 0.688 1.00 65.33 A C
+ATOM 380 CD ARG A 48 -4.940 -15.062 -0.555 1.00 65.33 A C
+ATOM 381 NE ARG A 48 -5.842 -14.382 -1.471 1.00 65.33 A N
+ATOM 382 CZ ARG A 48 -6.456 -14.968 -2.489 1.00 65.33 A C
+ATOM 383 NH1 ARG A 48 -7.253 -14.263 -3.278 1.00 65.33 A N1+
+ATOM 384 NH2 ARG A 48 -6.278 -16.261 -2.711 1.00 65.33 A N
+ATOM 385 N ALA A 49 -0.523 -16.371 2.137 1.00 69.50 A N
+ATOM 386 CA ALA A 49 0.883 -16.582 1.926 1.00 69.50 A C
+ATOM 387 C ALA A 49 1.300 -17.747 2.800 1.00 69.50 A C
+ATOM 388 O ALA A 49 0.797 -17.874 3.925 1.00 69.50 A O
+ATOM 389 CB ALA A 49 1.713 -15.347 2.264 1.00 69.50 A C
+ATOM 390 N PRO A 50 2.203 -18.604 2.322 1.00 73.88 A N
+ATOM 391 CA PRO A 50 2.547 -19.811 3.092 1.00 73.88 A C
+ATOM 392 C PRO A 50 3.065 -19.528 4.488 1.00 73.88 A C
+ATOM 393 O PRO A 50 2.732 -20.262 5.425 1.00 73.88 A O
+ATOM 394 CB PRO A 50 3.619 -20.476 2.221 1.00 73.88 A C
+ATOM 395 CG PRO A 50 3.373 -19.953 0.857 1.00 73.88 A C
+ATOM 396 CD PRO A 50 2.903 -18.552 1.032 1.00 73.88 A C
+ATOM 397 N TRP A 51 3.857 -18.477 4.662 1.00 74.78 A N
+ATOM 398 CA TRP A 51 4.507 -18.282 5.948 1.00 74.78 A C
+ATOM 399 C TRP A 51 3.557 -17.834 7.043 1.00 74.78 A C
+ATOM 400 O TRP A 51 4.021 -17.578 8.154 1.00 74.78 A O
+ATOM 401 CB TRP A 51 5.646 -17.276 5.812 1.00 74.78 A C
+ATOM 402 CG TRP A 51 5.254 -16.012 5.146 1.00 74.78 A C
+ATOM 403 CD1 TRP A 51 5.538 -15.645 3.872 1.00 74.78 A C
+ATOM 404 CD2 TRP A 51 4.520 -14.931 5.723 1.00 74.78 A C
+ATOM 405 CE2 TRP A 51 4.390 -13.947 4.738 1.00 74.78 A C
+ATOM 406 CE3 TRP A 51 3.960 -14.704 6.977 1.00 74.78 A C
+ATOM 407 NE1 TRP A 51 5.021 -14.406 3.614 1.00 74.78 A N
+ATOM 408 CZ2 TRP A 51 3.721 -12.758 4.964 1.00 74.78 A C
+ATOM 409 CZ3 TRP A 51 3.300 -13.528 7.197 1.00 74.78 A C
+ATOM 410 CH2 TRP A 51 3.186 -12.568 6.200 1.00 74.78 A C
+ATOM 411 N ILE A 52 2.260 -17.714 6.783 1.00 71.36 A N
+ATOM 412 CA ILE A 52 1.302 -17.459 7.847 1.00 71.36 A C
+ATOM 413 C ILE A 52 0.429 -18.668 8.128 1.00 71.36 A C
+ATOM 414 O ILE A 52 -0.380 -18.638 9.067 1.00 71.36 A O
+ATOM 415 CB ILE A 52 0.436 -16.227 7.543 1.00 71.36 A C
+ATOM 416 CG1 ILE A 52 -0.210 -15.717 8.823 1.00 71.36 A C
+ATOM 417 CG2 ILE A 52 -0.620 -16.584 6.547 1.00 71.36 A C
+ATOM 418 CD1 ILE A 52 -0.797 -14.378 8.708 1.00 71.36 A C
+ATOM 419 N GLU A 53 0.579 -19.746 7.356 1.00 76.04 A N
+ATOM 420 CA GLU A 53 -0.117 -20.986 7.676 1.00 76.04 A C
+ATOM 421 C GLU A 53 0.314 -21.529 9.028 1.00 76.04 A C
+ATOM 422 O GLU A 53 -0.482 -22.159 9.732 1.00 76.04 A O
+ATOM 423 CB GLU A 53 0.151 -22.028 6.592 1.00 76.04 A C
+ATOM 424 CG GLU A 53 -0.257 -21.625 5.189 1.00 76.04 A C
+ATOM 425 CD GLU A 53 -1.751 -21.413 5.028 1.00 76.04 A C
+ATOM 426 OE1 GLU A 53 -2.512 -21.652 5.990 1.00 76.04 A O
+ATOM 427 OE2 GLU A 53 -2.168 -21.023 3.918 1.00 76.04 A O1-
+ATOM 428 N GLN A 54 1.564 -21.281 9.406 1.00 80.33 A N
+ATOM 429 CA GLN A 54 2.129 -21.841 10.620 1.00 80.33 A C
+ATOM 430 C GLN A 54 1.368 -21.425 11.863 1.00 80.33 A C
+ATOM 431 O GLN A 54 1.459 -22.107 12.887 1.00 80.33 A O
+ATOM 432 CB GLN A 54 3.579 -21.390 10.746 1.00 80.33 A C
+ATOM 433 CG GLN A 54 4.420 -21.711 9.537 1.00 80.33 A C
+ATOM 434 CD GLN A 54 5.723 -20.948 9.527 1.00 80.33 A C
+ATOM 435 NE2 GLN A 54 6.533 -21.170 8.500 1.00 80.33 A N
+ATOM 436 OE1 GLN A 54 5.997 -20.157 10.430 1.00 80.33 A O
+ATOM 437 N GLU A 55 0.617 -20.330 11.796 1.00 77.54 A N
+ATOM 438 CA GLU A 55 0.208 -19.642 13.013 1.00 77.54 A C
+ATOM 439 C GLU A 55 -0.634 -20.530 13.918 1.00 77.54 A C
+ATOM 440 O GLU A 55 -0.402 -20.593 15.130 1.00 77.54 A O
+ATOM 441 CB GLU A 55 -0.544 -18.378 12.647 1.00 77.54 A C
+ATOM 442 CG GLU A 55 0.357 -17.190 12.458 1.00 77.54 A C
+ATOM 443 CD GLU A 55 0.857 -16.641 13.767 1.00 77.54 A C
+ATOM 444 OE1 GLU A 55 0.193 -16.877 14.797 1.00 77.54 A O
+ATOM 445 OE2 GLU A 55 1.912 -15.973 13.769 1.00 77.54 A O1-
+ATOM 446 N GLY A 56 -1.619 -21.218 13.360 1.00 75.47 A N
+ATOM 447 CA GLY A 56 -2.402 -22.135 14.148 1.00 75.47 A C
+ATOM 448 C GLY A 56 -3.773 -21.612 14.515 1.00 75.47 A C
+ATOM 449 O GLY A 56 -4.087 -20.432 14.361 1.00 75.47 A O
+ATOM 450 N PRO A 57 -4.606 -22.506 15.042 1.00 72.76 A N
+ATOM 451 CA PRO A 57 -6.041 -22.205 15.161 1.00 72.76 A C
+ATOM 452 C PRO A 57 -6.363 -20.982 15.992 1.00 72.76 A C
+ATOM 453 O PRO A 57 -7.298 -20.248 15.656 1.00 72.76 A O
+ATOM 454 CB PRO A 57 -6.603 -23.476 15.807 1.00 72.76 A C
+ATOM 455 CG PRO A 57 -5.605 -24.536 15.492 1.00 72.76 A C
+ATOM 456 CD PRO A 57 -4.279 -23.859 15.510 1.00 72.76 A C
+ATOM 457 N GLU A 58 -5.637 -20.746 17.080 1.00 75.70 A N
+ATOM 458 CA GLU A 58 -5.949 -19.587 17.907 1.00 75.70 A C
+ATOM 459 C GLU A 58 -5.746 -18.302 17.124 1.00 75.70 A C
+ATOM 460 O GLU A 58 -6.563 -17.375 17.194 1.00 75.70 A O
+ATOM 461 CB GLU A 58 -5.078 -19.588 19.159 1.00 75.70 A C
+ATOM 462 CG GLU A 58 -5.157 -18.310 19.979 1.00 75.70 A C
+ATOM 463 CD GLU A 58 -6.583 -17.911 20.309 1.00 75.70 A C
+ATOM 464 OE1 GLU A 58 -7.202 -18.590 21.153 1.00 75.70 A O
+ATOM 465 OE2 GLU A 58 -7.083 -16.915 19.745 1.00 75.70 A O1-
+ATOM 466 N TYR A 59 -4.660 -18.241 16.355 1.00 69.71 A N
+ATOM 467 CA TYR A 59 -4.354 -17.045 15.588 1.00 69.71 A C
+ATOM 468 C TYR A 59 -5.452 -16.747 14.585 1.00 69.71 A C
+ATOM 469 O TYR A 59 -5.958 -15.627 14.523 1.00 69.71 A O
+ATOM 470 CB TYR A 59 -3.024 -17.226 14.877 1.00 69.71 A C
+ATOM 471 CG TYR A 59 -2.618 -16.070 14.012 1.00 69.71 A C
+ATOM 472 CD1 TYR A 59 -2.047 -14.942 14.568 1.00 69.71 A C
+ATOM 473 CD2 TYR A 59 -2.795 -16.105 12.646 1.00 69.71 A C
+ATOM 474 CE1 TYR A 59 -1.658 -13.893 13.791 1.00 69.71 A C
+ATOM 475 CE2 TYR A 59 -2.397 -15.066 11.859 1.00 69.71 A C
+ATOM 476 CZ TYR A 59 -1.829 -13.960 12.434 1.00 69.71 A C
+ATOM 477 OH TYR A 59 -1.442 -12.904 11.645 1.00 69.71 A O
+ATOM 478 N TRP A 60 -5.823 -17.741 13.782 1.00 68.02 A N
+ATOM 479 CA TRP A 60 -6.860 -17.523 12.787 1.00 68.02 A C
+ATOM 480 C TRP A 60 -8.172 -17.173 13.451 1.00 68.02 A C
+ATOM 481 O TRP A 60 -8.913 -16.325 12.954 1.00 68.02 A O
+ATOM 482 CB TRP A 60 -7.005 -18.749 11.902 1.00 68.02 A C
+ATOM 483 CG TRP A 60 -5.816 -18.916 11.088 1.00 68.02 A C
+ATOM 484 CD1 TRP A 60 -4.950 -19.951 11.124 1.00 68.02 A C
+ATOM 485 CD2 TRP A 60 -5.308 -17.997 10.127 1.00 68.02 A C
+ATOM 486 CE2 TRP A 60 -4.130 -18.551 9.609 1.00 68.02 A C
+ATOM 487 CE3 TRP A 60 -5.740 -16.761 9.642 1.00 68.02 A C
+ATOM 488 NE1 TRP A 60 -3.931 -19.749 10.236 1.00 68.02 A N
+ATOM 489 CZ2 TRP A 60 -3.376 -17.913 8.641 1.00 68.02 A C
+ATOM 490 CZ3 TRP A 60 -4.993 -16.136 8.683 1.00 68.02 A C
+ATOM 491 CH2 TRP A 60 -3.826 -16.711 8.188 1.00 68.02 A C
+ATOM 492 N ASP A 61 -8.461 -17.783 14.596 1.00 71.99 A N
+ATOM 493 CA ASP A 61 -9.699 -17.457 15.287 1.00 71.99 A C
+ATOM 494 C ASP A 61 -9.733 -15.992 15.685 1.00 71.99 A C
+ATOM 495 O ASP A 61 -10.706 -15.283 15.409 1.00 71.99 A O
+ATOM 496 CB ASP A 61 -9.875 -18.331 16.520 1.00 71.99 A C
+ATOM 497 CG ASP A 61 -10.949 -17.801 17.437 1.00 71.99 A C
+ATOM 498 OD1 ASP A 61 -12.134 -18.081 17.177 1.00 71.99 A O
+ATOM 499 OD2 ASP A 61 -10.614 -17.085 18.403 1.00 71.99 A O1-
+ATOM 500 N GLN A 62 -8.679 -15.515 16.338 1.00 69.60 A N
+ATOM 501 CA GLN A 62 -8.753 -14.155 16.854 1.00 69.60 A C
+ATOM 502 C GLN A 62 -8.541 -13.126 15.755 1.00 69.60 A C
+ATOM 503 O GLN A 62 -9.120 -12.040 15.817 1.00 69.60 A O
+ATOM 504 CB GLN A 62 -7.793 -13.991 18.039 1.00 69.60 A C
+ATOM 505 CG GLN A 62 -6.311 -14.186 17.799 1.00 69.60 A C
+ATOM 506 CD GLN A 62 -5.593 -12.916 17.441 1.00 69.60 A C
+ATOM 507 NE2 GLN A 62 -4.504 -13.040 16.694 1.00 69.60 A N
+ATOM 508 OE1 GLN A 62 -5.984 -11.834 17.873 1.00 69.60 A O
+ATOM 509 N GLU A 63 -7.792 -13.466 14.707 1.00 64.87 A N
+ATOM 510 CA GLU A 63 -7.709 -12.590 13.546 1.00 64.87 A C
+ATOM 511 C GLU A 63 -9.047 -12.469 12.841 1.00 64.87 A C
+ATOM 512 O GLU A 63 -9.421 -11.382 12.395 1.00 64.87 A O
+ATOM 513 CB GLU A 63 -6.666 -13.108 12.579 1.00 64.87 A C
+ATOM 514 CG GLU A 63 -5.290 -12.710 12.949 1.00 64.87 A C
+ATOM 515 CD GLU A 63 -4.456 -12.504 11.731 1.00 64.87 A C
+ATOM 516 OE1 GLU A 63 -3.603 -11.598 11.732 1.00 64.87 A O
+ATOM 517 OE2 GLU A 63 -4.660 -13.253 10.759 1.00 64.87 A O1-
+ATOM 518 N THR A 64 -9.769 -13.576 12.706 1.00 63.83 A N
+ATOM 519 CA THR A 64 -11.092 -13.518 12.114 1.00 63.83 A C
+ATOM 520 C THR A 64 -12.037 -12.711 12.976 1.00 63.83 A C
+ATOM 521 O THR A 64 -12.818 -11.910 12.462 1.00 63.83 A O
+ATOM 522 CB THR A 64 -11.621 -14.922 11.904 1.00 63.83 A C
+ATOM 523 CG2 THR A 64 -12.984 -14.868 11.291 1.00 63.83 A C
+ATOM 524 OG1 THR A 64 -10.745 -15.613 11.015 1.00 63.83 A O
+ATOM 525 N ARG A 65 -11.968 -12.886 14.291 1.00 63.69 A N
+ATOM 526 CA ARG A 65 -12.802 -12.069 15.160 1.00 63.69 A C
+ATOM 527 C ARG A 65 -12.489 -10.595 14.963 1.00 63.69 A C
+ATOM 528 O ARG A 65 -13.400 -9.770 14.827 1.00 63.69 A O
+ATOM 529 CB ARG A 65 -12.599 -12.466 16.619 1.00 63.69 A C
+ATOM 530 CG ARG A 65 -12.980 -13.896 16.934 1.00 63.69 A C
+ATOM 531 CD ARG A 65 -13.232 -14.090 18.420 1.00 63.69 A C
+ATOM 532 NE ARG A 65 -12.043 -13.867 19.229 1.00 63.69 A N
+ATOM 533 CZ ARG A 65 -11.864 -12.796 19.993 1.00 63.69 A C
+ATOM 534 NH1 ARG A 65 -10.752 -12.661 20.700 1.00 63.69 A N1+
+ATOM 535 NH2 ARG A 65 -12.797 -11.857 20.043 1.00 63.69 A N
+ATOM 536 N ASN A 66 -11.203 -10.257 14.877 1.00 61.00 A N
+ATOM 537 CA ASN A 66 -10.796 -8.868 14.713 1.00 61.00 A C
+ATOM 538 C ASN A 66 -11.305 -8.287 13.402 1.00 61.00 A C
+ATOM 539 O ASN A 66 -11.932 -7.223 13.382 1.00 61.00 A O
+ATOM 540 CB ASN A 66 -9.276 -8.771 14.773 1.00 61.00 A C
+ATOM 541 CG ASN A 66 -8.726 -9.123 16.127 1.00 61.00 A C
+ATOM 542 ND2 ASN A 66 -7.428 -9.372 16.186 1.00 61.00 A N
+ATOM 543 OD1 ASN A 66 -9.457 -9.181 17.109 1.00 61.00 A O
+ATOM 544 N VAL A 67 -11.035 -8.967 12.289 1.00 55.49 A N
+ATOM 545 CA VAL A 67 -11.389 -8.392 11.001 1.00 55.49 A C
+ATOM 546 C VAL A 67 -12.892 -8.373 10.800 1.00 55.49 A C
+ATOM 547 O VAL A 67 -13.409 -7.511 10.091 1.00 55.49 A O
+ATOM 548 CB VAL A 67 -10.690 -9.120 9.856 1.00 55.49 A C
+ATOM 549 CG1 VAL A 67 -11.287 -10.457 9.661 1.00 55.49 A C
+ATOM 550 CG2 VAL A 67 -10.848 -8.325 8.615 1.00 55.49 A C
+ATOM 551 N LYS A 68 -13.633 -9.291 11.414 1.00 55.00 A N
+ATOM 552 CA LYS A 68 -15.078 -9.141 11.369 1.00 55.00 A C
+ATOM 553 C LYS A 68 -15.534 -7.986 12.244 1.00 55.00 A C
+ATOM 554 O LYS A 68 -16.554 -7.359 11.950 1.00 55.00 A O
+ATOM 555 CB LYS A 68 -15.766 -10.441 11.775 1.00 55.00 A C
+ATOM 556 CG LYS A 68 -15.378 -11.620 10.909 1.00 55.00 A C
+ATOM 557 CD LYS A 68 -16.048 -12.894 11.366 1.00 55.00 A C
+ATOM 558 CE LYS A 68 -16.977 -13.414 10.297 1.00 55.00 A C
+ATOM 559 NZ LYS A 68 -17.708 -14.616 10.745 1.00 55.00 A N1+
+ATOM 560 N ALA A 69 -14.778 -7.654 13.289 1.00 53.55 A N
+ATOM 561 CA ALA A 69 -15.082 -6.427 14.019 1.00 53.55 A C
+ATOM 562 C ALA A 69 -14.838 -5.198 13.156 1.00 53.55 A C
+ATOM 563 O ALA A 69 -15.620 -4.244 13.184 1.00 53.55 A O
+ATOM 564 CB ALA A 69 -14.251 -6.343 15.289 1.00 53.55 A C
+ATOM 565 N GLN A 70 -13.751 -5.203 12.389 1.00 51.45 A N
+ATOM 566 CA GLN A 70 -13.486 -4.108 11.460 1.00 51.45 A C
+ATOM 567 C GLN A 70 -14.590 -3.976 10.425 1.00 51.45 A C
+ATOM 568 O GLN A 70 -15.050 -2.870 10.130 1.00 51.45 A O
+ATOM 569 CB GLN A 70 -12.156 -4.322 10.757 1.00 51.45 A C
+ATOM 570 CG GLN A 70 -11.870 -3.264 9.725 1.00 51.45 A C
+ATOM 571 CD GLN A 70 -11.043 -2.119 10.239 1.00 51.45 A C
+ATOM 572 NE2 GLN A 70 -11.689 -0.990 10.499 1.00 51.45 A N
+ATOM 573 OE1 GLN A 70 -9.833 -2.242 10.391 1.00 51.45 A O
+ATOM 574 N SER A 71 -14.981 -5.095 9.821 1.00 48.62 A N
+ATOM 575 CA SER A 71 -16.087 -5.112 8.877 1.00 48.62 A C
+ATOM 576 C SER A 71 -17.343 -4.542 9.499 1.00 48.62 A C
+ATOM 577 O SER A 71 -18.073 -3.779 8.865 1.00 48.62 A O
+ATOM 578 CB SER A 71 -16.322 -6.537 8.399 1.00 48.62 A C
+ATOM 579 OG SER A 71 -17.411 -6.585 7.510 1.00 48.62 A O
+ATOM 580 N GLN A 72 -17.623 -4.918 10.738 1.00 51.34 A N
+ATOM 581 CA GLN A 72 -18.741 -4.314 11.438 1.00 51.34 A C
+ATOM 582 C GLN A 72 -18.560 -2.816 11.575 1.00 51.34 A C
+ATOM 583 O GLN A 72 -19.513 -2.056 11.412 1.00 51.34 A O
+ATOM 584 CB GLN A 72 -18.909 -4.970 12.793 1.00 51.34 A C
+ATOM 585 CG GLN A 72 -20.183 -4.623 13.489 1.00 51.34 A C
+ATOM 586 CD GLN A 72 -21.379 -4.809 12.629 1.00 51.34 A C
+ATOM 587 NE2 GLN A 72 -22.095 -3.729 12.382 1.00 51.34 A N
+ATOM 588 OE1 GLN A 72 -21.710 -5.922 12.252 1.00 51.34 A O
+ATOM 589 N THR A 73 -17.342 -2.365 11.852 1.00 53.20 A N
+ATOM 590 CA THR A 73 -17.122 -0.929 11.989 1.00 53.20 A C
+ATOM 591 C THR A 73 -17.392 -0.201 10.678 1.00 53.20 A C
+ATOM 592 O THR A 73 -18.038 0.852 10.666 1.00 53.20 A O
+ATOM 593 CB THR A 73 -15.701 -0.659 12.476 1.00 53.20 A C
+ATOM 594 CG2 THR A 73 -15.523 0.809 12.775 1.00 53.20 A C
+ATOM 595 OG1 THR A 73 -15.468 -1.393 13.680 1.00 53.20 A O
+ATOM 596 N ASP A 74 -16.920 -0.760 9.564 1.00 50.96 A N
+ATOM 597 CA ASP A 74 -17.199 -0.161 8.263 1.00 50.96 A C
+ATOM 598 C ASP A 74 -18.683 -0.163 7.948 1.00 50.96 A C
+ATOM 599 O ASP A 74 -19.200 0.806 7.399 1.00 50.96 A O
+ATOM 600 CB ASP A 74 -16.436 -0.865 7.154 1.00 50.96 A C
+ATOM 601 CG ASP A 74 -14.968 -0.609 7.231 1.00 50.96 A C
+ATOM 602 OD1 ASP A 74 -14.603 0.470 7.734 1.00 50.96 A O
+ATOM 603 OD2 ASP A 74 -14.176 -1.443 6.759 1.00 50.96 A O1-
+ATOM 604 N ARG A 75 -19.389 -1.228 8.274 1.00 55.46 A N
+ATOM 605 CA ARG A 75 -20.799 -1.203 7.938 1.00 55.46 A C
+ATOM 606 C ARG A 75 -21.577 -0.311 8.888 1.00 55.46 A C
+ATOM 607 O ARG A 75 -22.728 0.023 8.601 1.00 55.46 A O
+ATOM 608 CB ARG A 75 -21.361 -2.626 7.938 1.00 55.46 A C
+ATOM 609 CG ARG A 75 -21.824 -3.151 9.277 1.00 55.46 A C
+ATOM 610 CD ARG A 75 -22.720 -4.358 9.094 1.00 55.46 A C
+ATOM 611 NE ARG A 75 -22.137 -5.192 8.053 1.00 55.46 A N
+ATOM 612 CZ ARG A 75 -21.419 -6.283 8.294 1.00 55.46 A C
+ATOM 613 NH1 ARG A 75 -20.898 -6.968 7.296 1.00 55.46 A N1+
+ATOM 614 NH2 ARG A 75 -21.218 -6.692 9.534 1.00 55.46 A N
+ATOM 615 N VAL A 76 -20.974 0.082 10.010 1.00 55.70 A N
+ATOM 616 CA VAL A 76 -21.573 1.119 10.841 1.00 55.70 A C
+ATOM 617 C VAL A 76 -21.316 2.500 10.268 1.00 55.70 A C
+ATOM 618 O VAL A 76 -22.226 3.334 10.212 1.00 55.70 A O
+ATOM 619 CB VAL A 76 -21.072 1.030 12.291 1.00 55.70 A C
+ATOM 620 CG1 VAL A 76 -21.865 1.970 13.154 1.00 55.70 A C
+ATOM 621 CG2 VAL A 76 -21.292 -0.327 12.815 1.00 55.70 A C
+ATOM 622 N ASP A 77 -20.078 2.782 9.851 1.00 54.86 A N
+ATOM 623 CA ASP A 77 -19.745 4.140 9.440 1.00 54.86 A C
+ATOM 624 C ASP A 77 -20.042 4.411 7.976 1.00 54.86 A C
+ATOM 625 O ASP A 77 -19.961 5.563 7.552 1.00 54.86 A O
+ATOM 626 CB ASP A 77 -18.280 4.476 9.708 1.00 54.86 A C
+ATOM 627 CG ASP A 77 -17.341 3.439 9.187 1.00 54.86 A C
+ATOM 628 OD1 ASP A 77 -17.622 2.883 8.122 1.00 54.86 A O
+ATOM 629 OD2 ASP A 77 -16.297 3.204 9.821 1.00 54.86 A O1-
+ATOM 630 N LEU A 78 -20.406 3.402 7.203 1.00 51.20 A N
+ATOM 631 CA LEU A 78 -20.961 3.635 5.886 1.00 51.20 A C
+ATOM 632 C LEU A 78 -22.366 4.145 5.974 1.00 51.20 A C
+ATOM 633 O LEU A 78 -23.077 4.194 4.975 1.00 51.20 A O
+ATOM 634 CB LEU A 78 -20.929 2.341 5.077 1.00 51.20 A C
+ATOM 635 CG LEU A 78 -19.779 2.187 4.104 1.00 51.20 A C
+ATOM 636 CD1 LEU A 78 -19.875 0.896 3.349 1.00 51.20 A C
+ATOM 637 CD2 LEU A 78 -19.813 3.342 3.169 1.00 51.20 A C
+ATOM 638 N GLY A 79 -22.788 4.509 7.168 1.00 61.37 A N
+ATOM 639 CA GLY A 79 -24.112 5.021 7.394 1.00 61.37 A C
+ATOM 640 C GLY A 79 -23.961 6.317 8.136 1.00 61.37 A C
+ATOM 641 O GLY A 79 -24.940 6.997 8.435 1.00 61.37 A O
+ATOM 642 N THR A 80 -22.719 6.664 8.451 1.00 64.61 A N
+ATOM 643 CA THR A 80 -22.418 7.980 8.997 1.00 64.61 A C
+ATOM 644 C THR A 80 -21.676 8.873 8.008 1.00 64.61 A C
+ATOM 645 O THR A 80 -22.012 10.051 7.872 1.00 64.61 A O
+ATOM 646 CB THR A 80 -21.621 7.854 10.295 1.00 64.61 A C
+ATOM 647 CG2 THR A 80 -22.435 7.086 11.329 1.00 64.61 A C
+ATOM 648 OG1 THR A 80 -20.379 7.196 10.034 1.00 64.61 A O
+ATOM 649 N LEU A 81 -20.635 8.365 7.345 1.00 58.54 A N
+ATOM 650 CA LEU A 81 -20.041 9.137 6.262 1.00 58.54 A C
+ATOM 651 C LEU A 81 -21.083 9.483 5.225 1.00 58.54 A C
+ATOM 652 O LEU A 81 -21.228 10.648 4.836 1.00 58.54 A O
+ATOM 653 CB LEU A 81 -18.898 8.381 5.611 1.00 58.54 A C
+ATOM 654 CG LEU A 81 -17.718 8.178 6.534 1.00 58.54 A C
+ATOM 655 CD1 LEU A 81 -16.665 7.361 5.898 1.00 58.54 A C
+ATOM 656 CD2 LEU A 81 -17.181 9.493 6.852 1.00 58.54 A C
+ATOM 657 N ARG A 82 -21.853 8.497 4.812 1.00 65.88 A N
+ATOM 658 CA ARG A 82 -22.962 8.690 3.906 1.00 65.88 A C
+ATOM 659 C ARG A 82 -24.102 9.418 4.588 1.00 65.88 A C
+ATOM 660 O ARG A 82 -25.175 9.601 4.021 1.00 65.88 A O
+ATOM 661 CB ARG A 82 -23.389 7.320 3.351 1.00 65.88 A C
+ATOM 662 CG ARG A 82 -24.418 7.272 2.222 1.00 65.88 A C
+ATOM 663 CD ARG A 82 -25.832 7.179 2.711 1.00 65.88 A C
+ATOM 664 NE ARG A 82 -26.074 5.979 3.484 1.00 65.88 A N
+ATOM 665 CZ ARG A 82 -27.116 5.839 4.286 1.00 65.88 A C
+ATOM 666 NH1 ARG A 82 -27.951 6.851 4.443 1.00 65.88 A N1+
+ATOM 667 NH2 ARG A 82 -27.297 4.716 4.966 1.00 65.88 A N
+ATOM 668 N GLY A 83 -23.880 9.907 5.794 1.00 67.98 A N
+ATOM 669 CA GLY A 83 -24.928 10.648 6.449 1.00 67.98 A C
+ATOM 670 C GLY A 83 -24.605 12.112 6.615 1.00 67.98 A C
+ATOM 671 O GLY A 83 -25.493 12.963 6.599 1.00 67.98 A O
+ATOM 672 N TYR A 84 -23.334 12.414 6.815 1.00 75.19 A N
+ATOM 673 CA TYR A 84 -22.923 13.810 6.806 1.00 75.19 A C
+ATOM 674 C TYR A 84 -22.600 14.323 5.412 1.00 75.19 A C
+ATOM 675 O TYR A 84 -22.906 15.475 5.094 1.00 75.19 A O
+ATOM 676 CB TYR A 84 -21.725 14.058 7.711 1.00 75.19 A C
+ATOM 677 CG TYR A 84 -21.434 15.540 7.789 1.00 75.19 A C
+ATOM 678 CD1 TYR A 84 -22.460 16.469 7.915 1.00 75.19 A C
+ATOM 679 CD2 TYR A 84 -20.139 16.015 7.690 1.00 75.19 A C
+ATOM 680 CE1 TYR A 84 -22.193 17.827 7.961 1.00 75.19 A C
+ATOM 681 CE2 TYR A 84 -19.863 17.368 7.739 1.00 75.19 A C
+ATOM 682 CZ TYR A 84 -20.890 18.268 7.876 1.00 75.19 A C
+ATOM 683 OH TYR A 84 -20.600 19.610 7.924 1.00 75.19 A O
+ATOM 684 N TYR A 85 -21.993 13.506 4.561 1.00 65.60 A N
+ATOM 685 CA TYR A 85 -21.825 13.897 3.171 1.00 65.60 A C
+ATOM 686 C TYR A 85 -23.147 13.730 2.431 1.00 65.60 A C
+ATOM 687 O TYR A 85 -23.193 13.082 1.393 1.00 65.60 A O
+ATOM 688 CB TYR A 85 -20.764 13.042 2.506 1.00 65.60 A C
+ATOM 689 CG TYR A 85 -19.375 13.434 2.816 1.00 65.60 A C
+ATOM 690 CD1 TYR A 85 -18.909 14.677 2.492 1.00 65.60 A C
+ATOM 691 CD2 TYR A 85 -18.510 12.540 3.390 1.00 65.60 A C
+ATOM 692 CE1 TYR A 85 -17.629 15.031 2.758 1.00 65.60 A C
+ATOM 693 CE2 TYR A 85 -17.224 12.876 3.655 1.00 65.60 A C
+ATOM 694 CZ TYR A 85 -16.783 14.125 3.340 1.00 65.60 A C
+ATOM 695 OH TYR A 85 -15.483 14.474 3.610 1.00 65.60 A O
+ATOM 696 N ASN A 86 -24.199 14.390 2.929 1.00 71.17 A N
+ATOM 697 CA ASN A 86 -25.571 13.914 2.763 1.00 71.17 A C
+ATOM 698 C ASN A 86 -25.750 13.282 1.399 1.00 71.17 A C
+ATOM 699 O ASN A 86 -25.634 13.956 0.374 1.00 71.17 A O
+ATOM 700 CB ASN A 86 -26.543 15.083 2.902 1.00 71.17 A C
+ATOM 701 CG ASN A 86 -26.624 15.611 4.298 1.00 71.17 A C
+ATOM 702 ND2 ASN A 86 -26.650 16.930 4.427 1.00 71.17 A N
+ATOM 703 OD1 ASN A 86 -26.657 14.855 5.255 1.00 71.17 A O
+ATOM 704 N GLN A 87 -26.021 11.982 1.377 1.00 70.69 A N
+ATOM 705 CA GLN A 87 -25.689 11.223 0.185 1.00 70.69 A C
+ATOM 706 C GLN A 87 -26.745 10.164 -0.111 1.00 70.69 A C
+ATOM 707 O GLN A 87 -26.425 8.999 -0.353 1.00 70.69 A O
+ATOM 708 CB GLN A 87 -24.296 10.614 0.313 1.00 70.69 A C
+ATOM 709 CG GLN A 87 -23.794 9.884 -0.893 1.00 70.69 A C
+ATOM 710 CD GLN A 87 -22.409 9.373 -0.704 1.00 70.69 A C
+ATOM 711 NE2 GLN A 87 -21.871 8.725 -1.726 1.00 70.69 A N
+ATOM 712 OE1 GLN A 87 -21.825 9.539 0.361 1.00 70.69 A O
+ATOM 713 N SER A 88 -28.024 10.535 -0.053 1.00 80.45 A N
+ATOM 714 CA SER A 88 -28.974 9.895 -0.960 1.00 80.45 A C
+ATOM 715 C SER A 88 -29.033 8.379 -0.859 1.00 80.45 A C
+ATOM 716 O SER A 88 -28.461 7.696 -1.716 1.00 80.45 A O
+ATOM 717 CB SER A 88 -28.650 10.267 -2.406 1.00 80.45 A C
+ATOM 718 OG SER A 88 -29.338 9.414 -3.306 1.00 80.45 A O
+ATOM 719 N GLU A 89 -29.649 7.847 0.197 1.00 80.57 A N
+ATOM 720 CA GLU A 89 -29.757 6.406 0.402 1.00 80.57 A C
+ATOM 721 C GLU A 89 -30.021 5.658 -0.896 1.00 80.57 A C
+ATOM 722 O GLU A 89 -30.707 6.172 -1.785 1.00 80.57 A O
+ATOM 723 CB GLU A 89 -30.881 6.096 1.392 1.00 80.57 A C
+ATOM 724 CG GLU A 89 -30.574 6.444 2.831 1.00 80.57 A C
+ATOM 725 CD GLU A 89 -31.327 7.658 3.315 1.00 80.57 A C
+ATOM 726 OE1 GLU A 89 -31.748 7.667 4.489 1.00 80.57 A O
+ATOM 727 OE2 GLU A 89 -31.487 8.611 2.526 1.00 80.57 A O1-
+ATOM 728 N ALA A 90 -29.470 4.448 -1.006 1.00 77.02 A N
+ATOM 729 CA ALA A 90 -29.476 3.647 -2.226 1.00 77.02 A C
+ATOM 730 C ALA A 90 -28.630 4.262 -3.335 1.00 77.02 A C
+ATOM 731 O ALA A 90 -29.135 4.583 -4.413 1.00 77.02 A O
+ATOM 732 CB ALA A 90 -30.906 3.404 -2.721 1.00 77.02 A C
+ATOM 733 N GLY A 91 -27.343 4.438 -3.062 1.00 72.99 A N
+ATOM 734 CA GLY A 91 -26.339 4.660 -4.081 1.00 72.99 A C
+ATOM 735 C GLY A 91 -25.031 4.045 -3.631 1.00 72.99 A C
+ATOM 736 O GLY A 91 -24.716 4.086 -2.440 1.00 72.99 A O
+ATOM 737 N SER A 92 -24.266 3.455 -4.541 1.00 64.46 A N
+ATOM 738 CA SER A 92 -22.996 2.862 -4.151 1.00 64.46 A C
+ATOM 739 C SER A 92 -22.023 3.942 -3.711 1.00 64.46 A C
+ATOM 740 O SER A 92 -21.922 4.999 -4.336 1.00 64.46 A O
+ATOM 741 CB SER A 92 -22.396 2.065 -5.297 1.00 64.46 A C
+ATOM 742 OG SER A 92 -21.014 1.861 -5.077 1.00 64.46 A O
+ATOM 743 N HIS A 93 -21.298 3.668 -2.631 1.00 59.50 A N
+ATOM 744 CA HIS A 93 -20.414 4.653 -2.020 1.00 59.50 A C
+ATOM 745 C HIS A 93 -19.452 3.947 -1.082 1.00 59.50 A C
+ATOM 746 O HIS A 93 -19.884 3.251 -0.166 1.00 59.50 A O
+ATOM 747 CB HIS A 93 -21.237 5.717 -1.306 1.00 59.50 A C
+ATOM 748 CG HIS A 93 -22.036 5.196 -0.161 1.00 59.50 A C
+ATOM 749 CD2 HIS A 93 -21.797 5.194 1.171 1.00 59.50 A C
+ATOM 750 ND1 HIS A 93 -23.249 4.564 -0.333 1.00 59.50 A N
+ATOM 751 CE1 HIS A 93 -23.730 4.207 0.846 1.00 59.50 A C
+ATOM 752 NE2 HIS A 93 -22.864 4.571 1.775 1.00 59.50 A N
+ATOM 753 N THR A 94 -18.159 4.140 -1.305 1.00 48.70 A N
+ATOM 754 CA THR A 94 -17.092 3.320 -0.754 1.00 48.70 A C
+ATOM 755 C THR A 94 -16.489 3.954 0.511 1.00 48.70 A C
+ATOM 756 O THR A 94 -16.695 5.128 0.807 1.00 48.70 A O
+ATOM 757 CB THR A 94 -16.043 3.117 -1.844 1.00 48.70 A C
+ATOM 758 CG2 THR A 94 -14.920 2.229 -1.398 1.00 48.70 A C
+ATOM 759 OG1 THR A 94 -16.664 2.472 -2.955 1.00 48.70 A O
+ATOM 760 N ILE A 95 -15.775 3.138 1.286 1.00 42.33 A N
+ATOM 761 CA ILE A 95 -14.922 3.575 2.387 1.00 42.33 A C
+ATOM 762 C ILE A 95 -13.614 2.831 2.262 1.00 42.33 A C
+ATOM 763 O ILE A 95 -13.599 1.651 1.918 1.00 42.33 A O
+ATOM 764 CB ILE A 95 -15.527 3.294 3.768 1.00 42.33 A C
+ATOM 765 CG1 ILE A 95 -16.490 4.375 4.167 1.00 42.33 A C
+ATOM 766 CG2 ILE A 95 -14.468 3.244 4.810 1.00 42.33 A C
+ATOM 767 CD1 ILE A 95 -17.167 4.046 5.418 1.00 42.33 A C
+ATOM 768 N GLN A 96 -12.515 3.504 2.533 1.00 45.20 A N
+ATOM 769 CA GLN A 96 -11.223 2.858 2.461 1.00 45.20 A C
+ATOM 770 C GLN A 96 -10.440 3.228 3.707 1.00 45.20 A C
+ATOM 771 O GLN A 96 -10.642 4.298 4.276 1.00 45.20 A O
+ATOM 772 CB GLN A 96 -10.496 3.292 1.226 1.00 45.20 A C
+ATOM 773 CG GLN A 96 -11.171 2.843 -0.027 1.00 45.20 A C
+ATOM 774 CD GLN A 96 -10.449 3.307 -1.238 1.00 45.20 A C
+ATOM 775 NE2 GLN A 96 -10.973 2.996 -2.380 1.00 45.20 A N
+ATOM 776 OE1 GLN A 96 -9.469 4.010 -1.144 1.00 45.20 A O
+ATOM 777 N ILE A 97 -9.570 2.326 4.149 1.00 42.37 A N
+ATOM 778 CA ILE A 97 -8.834 2.518 5.391 1.00 42.37 A C
+ATOM 779 C ILE A 97 -7.461 1.889 5.277 1.00 42.37 A C
+ATOM 780 O ILE A 97 -7.331 0.721 4.914 1.00 42.37 A O
+ATOM 781 CB ILE A 97 -9.585 1.939 6.595 1.00 42.37 A C
+ATOM 782 CG1 ILE A 97 -10.758 2.821 6.972 1.00 42.37 A C
+ATOM 783 CG2 ILE A 97 -8.668 1.822 7.749 1.00 42.37 A C
+ATOM 784 CD1 ILE A 97 -11.808 2.122 7.730 1.00 42.37 A C
+ATOM 785 N MET A 98 -6.430 2.666 5.538 1.00 51.05 A N
+ATOM 786 CA MET A 98 -5.092 2.126 5.660 1.00 51.05 A C
+ATOM 787 C MET A 98 -4.691 2.081 7.138 1.00 51.05 A C
+ATOM 788 O MET A 98 -5.337 2.675 7.998 1.00 51.05 A O
+ATOM 789 CB MET A 98 -4.150 2.977 4.826 1.00 51.05 A C
+ATOM 790 CG MET A 98 -2.862 2.359 4.399 1.00 51.05 A C
+ATOM 791 SD MET A 98 -1.717 3.718 4.127 1.00 51.05 A S
+ATOM 792 CE MET A 98 -0.229 2.851 3.682 1.00 51.05 A C
+ATOM 793 N TYR A 99 -3.646 1.318 7.443 1.00 53.08 A N
+ATOM 794 CA TYR A 99 -2.969 1.363 8.731 1.00 53.08 A C
+ATOM 795 C TYR A 99 -1.496 1.318 8.429 1.00 53.08 A C
+ATOM 796 O TYR A 99 -1.087 1.351 7.268 1.00 53.08 A O
+ATOM 797 CB TYR A 99 -3.213 0.168 9.646 1.00 53.08 A C
+ATOM 798 CG TYR A 99 -4.555 0.021 10.267 1.00 53.08 A C
+ATOM 799 CD1 TYR A 99 -5.314 1.111 10.603 1.00 53.08 A C
+ATOM 800 CD2 TYR A 99 -5.029 -1.236 10.580 1.00 53.08 A C
+ATOM 801 CE1 TYR A 99 -6.538 0.946 11.178 1.00 53.08 A C
+ATOM 802 CE2 TYR A 99 -6.231 -1.407 11.146 1.00 53.08 A C
+ATOM 803 CZ TYR A 99 -6.987 -0.319 11.449 1.00 53.08 A C
+ATOM 804 OH TYR A 99 -8.208 -0.501 12.032 1.00 53.08 A O
+ATOM 805 N GLY A 100 -0.689 1.169 9.463 1.00 61.46 A N
+ATOM 806 CA GLY A 100 0.679 0.788 9.204 1.00 61.46 A C
+ATOM 807 C GLY A 100 1.554 0.995 10.407 1.00 61.46 A C
+ATOM 808 O GLY A 100 1.136 1.521 11.435 1.00 61.46 A O
+ATOM 809 N CYS A 101 2.789 0.545 10.249 1.00 71.41 A N
+ATOM 810 CA CYS A 101 3.857 0.872 11.178 1.00 71.41 A C
+ATOM 811 C CYS A 101 5.149 0.496 10.489 1.00 71.41 A C
+ATOM 812 O CYS A 101 5.328 -0.660 10.105 1.00 71.41 A O
+ATOM 813 CB CYS A 101 3.705 0.134 12.506 1.00 71.41 A C
+ATOM 814 SG CYS A 101 3.842 -1.655 12.458 1.00 71.41 A S
+ATOM 815 N ASP A 102 6.026 1.460 10.293 1.00 73.43 A N
+ATOM 816 CA ASP A 102 7.319 1.127 9.735 1.00 73.43 A C
+ATOM 817 C ASP A 102 8.251 0.648 10.832 1.00 73.43 A C
+ATOM 818 O ASP A 102 7.939 0.702 12.019 1.00 73.43 A O
+ATOM 819 CB ASP A 102 7.922 2.319 9.008 1.00 73.43 A C
+ATOM 820 CG ASP A 102 7.500 2.388 7.564 1.00 73.43 A C
+ATOM 821 OD1 ASP A 102 7.826 1.449 6.816 1.00 73.43 A O
+ATOM 822 OD2 ASP A 102 6.853 3.378 7.174 1.00 73.43 A O1-
+ATOM 823 N VAL A 103 9.403 0.148 10.417 1.00 75.80 A N
+ATOM 824 CA VAL A 103 10.473 -0.217 11.318 1.00 75.80 A C
+ATOM 825 C VAL A 103 11.788 0.188 10.667 1.00 75.80 A C
+ATOM 826 O VAL A 103 11.829 0.602 9.511 1.00 75.80 A O
+ATOM 827 CB VAL A 103 10.486 -1.722 11.638 1.00 75.80 A C
+ATOM 828 CG1 VAL A 103 9.331 -2.078 12.551 1.00 75.80 A C
+ATOM 829 CG2 VAL A 103 10.480 -2.529 10.367 1.00 75.80 A C
+ATOM 830 N GLY A 104 12.854 0.060 11.406 1.00 87.65 A N
+ATOM 831 CA GLY A 104 14.160 0.179 10.808 1.00 87.65 A C
+ATOM 832 C GLY A 104 14.643 -1.155 10.305 1.00 87.65 A C
+ATOM 833 O GLY A 104 14.046 -2.191 10.576 1.00 87.65 A O
+ATOM 834 N SER A 105 15.746 -1.116 9.559 1.00 95.51 A N
+ATOM 835 CA SER A 105 16.368 -2.361 9.124 1.00 95.51 A C
+ATOM 836 C SER A 105 16.808 -3.193 10.317 1.00 95.51 A C
+ATOM 837 O SER A 105 16.896 -4.422 10.223 1.00 95.51 A O
+ATOM 838 CB SER A 105 17.557 -2.073 8.207 1.00 95.51 A C
+ATOM 839 OG SER A 105 18.543 -1.301 8.870 1.00 95.51 A O
+ATOM 840 N ASP A 106 17.093 -2.540 11.441 1.00 89.51 A N
+ATOM 841 CA ASP A 106 17.390 -3.232 12.685 1.00 89.51 A C
+ATOM 842 C ASP A 106 16.141 -3.660 13.442 1.00 89.51 A C
+ATOM 843 O ASP A 106 16.253 -4.452 14.381 1.00 89.51 A O
+ATOM 844 CB ASP A 106 18.235 -2.334 13.581 1.00 89.51 A C
+ATOM 845 CG ASP A 106 17.474 -1.125 14.057 1.00 89.51 A C
+ATOM 846 OD1 ASP A 106 17.192 -0.242 13.226 1.00 89.51 A O
+ATOM 847 OD2 ASP A 106 17.146 -1.064 15.257 1.00 89.51 A O1-
+ATOM 848 N GLY A 107 14.967 -3.158 13.069 1.00 83.89 A N
+ATOM 849 CA GLY A 107 13.734 -3.509 13.739 1.00 83.89 A C
+ATOM 850 C GLY A 107 13.174 -2.459 14.672 1.00 83.89 A C
+ATOM 851 O GLY A 107 12.288 -2.777 15.472 1.00 83.89 A O
+ATOM 852 N ARG A 108 13.657 -1.228 14.605 1.00 84.78 A N
+ATOM 853 CA ARG A 108 13.194 -0.166 15.482 1.00 84.78 A C
+ATOM 854 C ARG A 108 12.062 0.600 14.813 1.00 84.78 A C
+ATOM 855 O ARG A 108 12.132 0.896 13.619 1.00 84.78 A O
+ATOM 856 CB ARG A 108 14.367 0.753 15.829 1.00 84.78 A C
+ATOM 857 CG ARG A 108 14.099 1.856 16.844 1.00 84.78 A C
+ATOM 858 CD ARG A 108 13.709 3.146 16.148 1.00 84.78 A C
+ATOM 859 NE ARG A 108 13.494 4.255 17.075 1.00 84.78 A N
+ATOM 860 CZ ARG A 108 12.354 4.499 17.709 1.00 84.78 A C
+ATOM 861 NH1 ARG A 108 11.304 3.710 17.529 1.00 84.78 A N1+
+ATOM 862 NH2 ARG A 108 12.264 5.539 18.527 1.00 84.78 A N
+ATOM 863 N PHE A 109 11.024 0.911 15.585 1.00 76.13 A N
+ATOM 864 CA PHE A 109 9.864 1.621 15.061 1.00 76.13 A C
+ATOM 865 C PHE A 109 10.253 2.958 14.446 1.00 76.13 A C
+ATOM 866 O PHE A 109 10.990 3.739 15.049 1.00 76.13 A O
+ATOM 867 CB PHE A 109 8.867 1.867 16.176 1.00 76.13 A C
+ATOM 868 CG PHE A 109 7.807 2.831 15.810 1.00 76.13 A C
+ATOM 869 CD1 PHE A 109 6.748 2.441 15.040 1.00 76.13 A C
+ATOM 870 CD2 PHE A 109 7.891 4.145 16.201 1.00 76.13 A C
+ATOM 871 CE1 PHE A 109 5.773 3.328 14.698 1.00 76.13 A C
+ATOM 872 CE2 PHE A 109 6.922 5.034 15.853 1.00 76.13 A C
+ATOM 873 CZ PHE A 109 5.863 4.622 15.095 1.00 76.13 A C
+ATOM 874 N LEU A 110 9.714 3.248 13.266 1.00 76.61 A N
+ATOM 875 CA LEU A 110 10.032 4.476 12.549 1.00 76.61 A C
+ATOM 876 C LEU A 110 8.878 5.458 12.500 1.00 76.61 A C
+ATOM 877 O LEU A 110 8.963 6.550 13.063 1.00 76.61 A O
+ATOM 878 CB LEU A 110 10.463 4.148 11.121 1.00 76.61 A C
+ATOM 879 CG LEU A 110 11.860 3.578 10.977 1.00 76.61 A C
+ATOM 880 CD1 LEU A 110 12.301 3.705 9.545 1.00 76.61 A C
+ATOM 881 CD2 LEU A 110 12.801 4.327 11.887 1.00 76.61 A C
+ATOM 882 N ARG A 111 7.790 5.087 11.842 1.00 71.89 A N
+ATOM 883 CA ARG A 111 6.665 5.976 11.630 1.00 71.89 A C
+ATOM 884 C ARG A 111 5.379 5.174 11.662 1.00 71.89 A C
+ATOM 885 O ARG A 111 5.359 3.986 11.337 1.00 71.89 A O
+ATOM 886 CB ARG A 111 6.794 6.710 10.302 1.00 71.89 A C
+ATOM 887 CG ARG A 111 7.240 5.805 9.186 1.00 71.89 A C
+ATOM 888 CD ARG A 111 7.613 6.566 7.939 1.00 71.89 A C
+ATOM 889 NE ARG A 111 6.700 7.675 7.692 1.00 71.89 A N
+ATOM 890 CZ ARG A 111 5.587 7.562 6.974 1.00 71.89 A C
+ATOM 891 NH1 ARG A 111 4.802 8.609 6.789 1.00 71.89 A N1+
+ATOM 892 NH2 ARG A 111 5.261 6.395 6.433 1.00 71.89 A N
+ATOM 893 N GLY A 112 4.306 5.836 12.065 1.00 68.53 A N
+ATOM 894 CA GLY A 112 3.002 5.218 12.078 1.00 68.53 A C
+ATOM 895 C GLY A 112 2.009 6.042 11.300 1.00 68.53 A C
+ATOM 896 O GLY A 112 1.943 7.261 11.458 1.00 68.53 A O
+ATOM 897 N TYR A 113 1.247 5.393 10.432 1.00 62.39 A N
+ATOM 898 CA TYR A 113 0.309 6.080 9.561 1.00 62.39 A C
+ATOM 899 C TYR A 113 -1.026 5.370 9.548 1.00 62.39 A C
+ATOM 900 O TYR A 113 -1.087 4.170 9.293 1.00 62.39 A O
+ATOM 901 CB TYR A 113 0.870 6.189 8.145 1.00 62.39 A C
+ATOM 902 CG TYR A 113 1.539 4.938 7.654 1.00 62.39 A C
+ATOM 903 CD1 TYR A 113 0.837 3.966 6.987 1.00 62.39 A C
+ATOM 904 CD2 TYR A 113 2.895 4.763 7.807 1.00 62.39 A C
+ATOM 905 CE1 TYR A 113 1.461 2.831 6.531 1.00 62.39 A C
+ATOM 906 CE2 TYR A 113 3.526 3.640 7.349 1.00 62.39 A C
+ATOM 907 CZ TYR A 113 2.803 2.674 6.709 1.00 62.39 A C
+ATOM 908 OH TYR A 113 3.415 1.540 6.240 1.00 62.39 A O
+ATOM 909 N ARG A 114 -2.085 6.115 9.835 1.00 55.70 A N
+ATOM 910 CA ARG A 114 -3.452 5.690 9.580 1.00 55.70 A C
+ATOM 911 C ARG A 114 -4.147 6.716 8.707 1.00 55.70 A C
+ATOM 912 O ARG A 114 -4.211 7.891 9.068 1.00 55.70 A O
+ATOM 913 CB ARG A 114 -4.251 5.502 10.857 1.00 55.70 A C
+ATOM 914 CG ARG A 114 -5.590 4.997 10.480 1.00 55.70 A C
+ATOM 915 CD ARG A 114 -6.485 4.675 11.607 1.00 55.70 A C
+ATOM 916 NE ARG A 114 -7.754 4.234 11.062 1.00 55.70 A N
+ATOM 917 CZ ARG A 114 -8.780 3.848 11.797 1.00 55.70 A C
+ATOM 918 NH1 ARG A 114 -9.902 3.464 11.217 1.00 55.70 A N1+
+ATOM 919 NH2 ARG A 114 -8.670 3.833 13.111 1.00 55.70 A N
+ATOM 920 N GLN A 115 -4.691 6.270 7.578 1.00 50.80 A N
+ATOM 921 CA GLN A 115 -5.281 7.144 6.574 1.00 50.80 A C
+ATOM 922 C GLN A 115 -6.660 6.624 6.197 1.00 50.80 A C
+ATOM 923 O GLN A 115 -6.804 5.445 5.877 1.00 50.80 A O
+ATOM 924 CB GLN A 115 -4.368 7.182 5.356 1.00 50.80 A C
+ATOM 925 CG GLN A 115 -3.047 7.837 5.657 1.00 50.80 A C
+ATOM 926 CD GLN A 115 -2.116 7.857 4.475 1.00 50.80 A C
+ATOM 927 NE2 GLN A 115 -0.942 8.448 4.658 1.00 50.80 A N
+ATOM 928 OE1 GLN A 115 -2.428 7.317 3.416 1.00 50.80 A O
+ATOM 929 N ASP A 116 -7.664 7.491 6.211 1.00 52.46 A N
+ATOM 930 CA ASP A 116 -9.015 7.105 5.833 1.00 52.46 A C
+ATOM 931 C ASP A 116 -9.433 7.836 4.572 1.00 52.46 A C
+ATOM 932 O ASP A 116 -9.050 8.989 4.361 1.00 52.46 A O
+ATOM 933 CB ASP A 116 -10.032 7.419 6.906 1.00 52.46 A C
+ATOM 934 CG ASP A 116 -9.860 6.572 8.114 1.00 52.46 A C
+ATOM 935 OD1 ASP A 116 -9.376 5.442 7.972 1.00 52.46 A O
+ATOM 936 OD2 ASP A 116 -10.265 7.002 9.210 1.00 52.46 A O1-
+ATOM 937 N ALA A 117 -10.212 7.163 3.731 1.00 47.91 A N
+ATOM 938 CA ALA A 117 -10.749 7.754 2.517 1.00 47.91 A C
+ATOM 939 C ALA A 117 -12.204 7.379 2.340 1.00 47.91 A C
+ATOM 940 O ALA A 117 -12.551 6.200 2.395 1.00 47.91 A O
+ATOM 941 CB ALA A 117 -9.984 7.293 1.303 1.00 47.91 A C
+ATOM 942 N TYR A 118 -13.037 8.370 2.093 1.00 52.88 A N
+ATOM 943 CA TYR A 118 -14.416 8.172 1.683 1.00 52.88 A C
+ATOM 944 C TYR A 118 -14.391 8.009 0.171 1.00 52.88 A C
+ATOM 945 O TYR A 118 -13.430 7.445 -0.356 1.00 52.88 A O
+ATOM 946 CB TYR A 118 -15.298 9.307 2.141 1.00 52.88 A C
+ATOM 947 CG TYR A 118 -16.731 8.983 1.983 1.00 52.88 A C
+ATOM 948 CD1 TYR A 118 -17.181 7.722 2.255 1.00 52.88 A C
+ATOM 949 CD2 TYR A 118 -17.644 9.938 1.646 1.00 52.88 A C
+ATOM 950 CE1 TYR A 118 -18.488 7.406 2.142 1.00 52.88 A C
+ATOM 951 CE2 TYR A 118 -18.965 9.638 1.542 1.00 52.88 A C
+ATOM 952 CZ TYR A 118 -19.385 8.367 1.788 1.00 52.88 A C
+ATOM 953 OH TYR A 118 -20.715 8.057 1.677 1.00 52.88 A O
+ATOM 954 N ASP A 119 -15.429 8.437 -0.545 1.00 58.87 A N
+ATOM 955 CA ASP A 119 -15.522 8.134 -1.970 1.00 58.87 A C
+ATOM 956 C ASP A 119 -14.331 8.678 -2.726 1.00 58.87 A C
+ATOM 957 O ASP A 119 -14.387 9.780 -3.270 1.00 58.87 A O
+ATOM 958 CB ASP A 119 -16.768 8.773 -2.582 1.00 58.87 A C
+ATOM 959 CG ASP A 119 -18.048 8.138 -2.132 1.00 58.87 A C
+ATOM 960 OD1 ASP A 119 -18.042 6.938 -1.839 1.00 58.87 A O
+ATOM 961 OD2 ASP A 119 -19.063 8.851 -2.033 1.00 58.87 A O1-
+ATOM 962 N GLY A 120 -13.241 7.922 -2.738 1.00 55.11 A N
+ATOM 963 CA GLY A 120 -12.095 8.211 -3.579 1.00 55.11 A C
+ATOM 964 C GLY A 120 -11.191 9.340 -3.132 1.00 55.11 A C
+ATOM 965 O GLY A 120 -10.007 9.355 -3.470 1.00 55.11 A O
+ATOM 966 N LYS A 121 -11.722 10.270 -2.356 1.00 61.78 A N
+ATOM 967 CA LYS A 121 -10.995 11.450 -1.912 1.00 61.78 A C
+ATOM 968 C LYS A 121 -10.262 11.113 -0.614 1.00 61.78 A C
+ATOM 969 O LYS A 121 -10.027 9.942 -0.314 1.00 61.78 A O
+ATOM 970 CB LYS A 121 -11.960 12.626 -1.767 1.00 61.78 A C
+ATOM 971 CG LYS A 121 -13.161 12.314 -0.905 1.00 61.78 A C
+ATOM 972 CD LYS A 121 -14.312 13.256 -1.190 1.00 61.78 A C
+ATOM 973 CE LYS A 121 -15.505 12.924 -0.310 1.00 61.78 A C
+ATOM 974 NZ LYS A 121 -15.086 12.709 1.103 1.00 61.78 A N1+
+ATOM 975 N ASP A 122 -9.822 12.118 0.133 1.00 58.61 A N
+ATOM 976 CA ASP A 122 -9.230 11.906 1.443 1.00 58.61 A C
+ATOM 977 C ASP A 122 -10.207 12.219 2.559 1.00 58.61 A C
+ATOM 978 O ASP A 122 -11.231 12.878 2.374 1.00 58.61 A O
+ATOM 979 CB ASP A 122 -7.971 12.742 1.651 1.00 58.61 A C
+ATOM 980 CG ASP A 122 -6.827 12.271 0.817 1.00 58.61 A C
+ATOM 981 OD1 ASP A 122 -6.775 11.064 0.528 1.00 58.61 A O
+ATOM 982 OD2 ASP A 122 -5.975 13.096 0.448 1.00 58.61 A O1-
+ATOM 983 N TYR A 123 -9.849 11.715 3.731 1.00 53.92 A N
+ATOM 984 CA TYR A 123 -10.565 11.903 4.973 1.00 53.92 A C
+ATOM 985 C TYR A 123 -9.489 12.025 6.043 1.00 53.92 A C
+ATOM 986 O TYR A 123 -8.313 12.202 5.720 1.00 53.92 A O
+ATOM 987 CB TYR A 123 -11.549 10.746 5.167 1.00 53.92 A C
+ATOM 988 CG TYR A 123 -12.705 11.053 6.057 1.00 53.92 A C
+ATOM 989 CD1 TYR A 123 -13.805 11.708 5.574 1.00 53.92 A C
+ATOM 990 CD2 TYR A 123 -12.717 10.640 7.364 1.00 53.92 A C
+ATOM 991 CE1 TYR A 123 -14.853 11.975 6.373 1.00 53.92 A C
+ATOM 992 CE2 TYR A 123 -13.759 10.902 8.164 1.00 53.92 A C
+ATOM 993 CZ TYR A 123 -14.824 11.571 7.669 1.00 53.92 A C
+ATOM 994 OH TYR A 123 -15.888 11.834 8.480 1.00 53.92 A O
+ATOM 995 N ILE A 124 -9.871 11.937 7.318 1.00 51.88 A N
+ATOM 996 CA ILE A 124 -8.901 12.046 8.403 1.00 51.88 A C
+ATOM 997 C ILE A 124 -7.673 11.174 8.171 1.00 51.88 A C
+ATOM 998 O ILE A 124 -7.756 10.074 7.619 1.00 51.88 A O
+ATOM 999 CB ILE A 124 -9.567 11.692 9.736 1.00 51.88 A C
+ATOM 1000 CG1 ILE A 124 -8.629 12.032 10.869 1.00 51.88 A C
+ATOM 1001 CG2 ILE A 124 -9.899 10.251 9.772 1.00 51.88 A C
+ATOM 1002 CD1 ILE A 124 -9.287 12.014 12.171 1.00 51.88 A C
+ATOM 1003 N ALA A 125 -6.511 11.694 8.560 1.00 57.44 A N
+ATOM 1004 CA ALA A 125 -5.252 10.964 8.493 1.00 57.44 A C
+ATOM 1005 C ALA A 125 -4.415 11.293 9.721 1.00 57.44 A C
+ATOM 1006 O ALA A 125 -4.660 12.280 10.415 1.00 57.44 A O
+ATOM 1007 CB ALA A 125 -4.471 11.285 7.219 1.00 57.44 A C
+ATOM 1008 N LEU A 126 -3.428 10.451 9.990 1.00 62.80 A N
+ATOM 1009 CA LEU A 126 -2.595 10.581 11.175 1.00 62.80 A C
+ATOM 1010 C LEU A 126 -1.256 11.213 10.823 1.00 62.80 A C
+ATOM 1011 O LEU A 126 -0.712 10.982 9.742 1.00 62.80 A O
+ATOM 1012 CB LEU A 126 -2.368 9.217 11.817 1.00 62.80 A C
+ATOM 1013 CG LEU A 126 -1.543 9.087 13.094 1.00 62.80 A C
+ATOM 1014 CD1 LEU A 126 -2.271 9.702 14.251 1.00 62.80 A C
+ATOM 1015 CD2 LEU A 126 -1.301 7.635 13.373 1.00 62.80 A C
+ATOM 1016 N ASN A 127 -0.702 11.971 11.765 1.00 70.19 A N
+ATOM 1017 CA ASN A 127 0.542 12.691 11.532 1.00 70.19 A C
+ATOM 1018 C ASN A 127 1.717 11.730 11.580 1.00 70.19 A C
+ATOM 1019 O ASN A 127 1.562 10.509 11.508 1.00 70.19 A O
+ATOM 1020 CB ASN A 127 0.721 13.803 12.550 1.00 70.19 A C
+ATOM 1021 CG ASN A 127 -0.359 14.823 12.472 1.00 70.19 A C
+ATOM 1022 ND2 ASN A 127 -0.787 15.323 13.614 1.00 70.19 A N
+ATOM 1023 OD1 ASN A 127 -0.806 15.164 11.395 1.00 70.19 A O
+ATOM 1024 N GLU A 128 2.922 12.279 11.663 1.00 76.15 A N
+ATOM 1025 CA GLU A 128 4.114 11.459 11.788 1.00 76.15 A C
+ATOM 1026 C GLU A 128 4.659 11.439 13.205 1.00 76.15 A C
+ATOM 1027 O GLU A 128 5.510 10.600 13.516 1.00 76.15 A O
+ATOM 1028 CB GLU A 128 5.189 11.956 10.825 1.00 76.15 A C
+ATOM 1029 CG GLU A 128 4.935 11.515 9.405 1.00 76.15 A C
+ATOM 1030 CD GLU A 128 4.515 10.064 9.323 1.00 76.15 A C
+ATOM 1031 OE1 GLU A 128 3.303 9.787 9.199 1.00 76.15 A O
+ATOM 1032 OE2 GLU A 128 5.403 9.196 9.372 1.00 76.15 A O1-
+ATOM 1033 N ASP A 129 4.183 12.331 14.063 1.00 77.18 A N
+ATOM 1034 CA ASP A 129 4.456 12.291 15.489 1.00 77.18 A C
+ATOM 1035 C ASP A 129 3.454 11.432 16.240 1.00 77.18 A C
+ATOM 1036 O ASP A 129 3.543 11.322 17.466 1.00 77.18 A O
+ATOM 1037 CB ASP A 129 4.458 13.711 16.048 1.00 77.18 A C
+ATOM 1038 CG ASP A 129 3.118 14.396 15.897 1.00 77.18 A C
+ATOM 1039 OD1 ASP A 129 2.194 13.792 15.324 1.00 77.18 A O
+ATOM 1040 OD2 ASP A 129 2.986 15.553 16.341 1.00 77.18 A O1-
+ATOM 1041 N LEU A 130 2.503 10.839 15.520 1.00 70.64 A N
+ATOM 1042 CA LEU A 130 1.531 9.904 16.067 1.00 70.64 A C
+ATOM 1043 C LEU A 130 0.700 10.548 17.159 1.00 70.64 A C
+ATOM 1044 O LEU A 130 0.415 9.934 18.182 1.00 70.64 A O
+ATOM 1045 CB LEU A 130 2.212 8.660 16.609 1.00 70.64 A C
+ATOM 1046 CG LEU A 130 2.904 7.834 15.553 1.00 70.64 A C
+ATOM 1047 CD1 LEU A 130 3.629 6.717 16.218 1.00 70.64 A C
+ATOM 1048 CD2 LEU A 130 1.903 7.333 14.617 1.00 70.64 A C
+ATOM 1049 N ARG A 131 0.325 11.802 16.962 1.00 75.73 A N
+ATOM 1050 CA ARG A 131 -0.308 12.501 18.069 1.00 75.73 A C
+ATOM 1051 C ARG A 131 -1.517 13.339 17.696 1.00 75.73 A C
+ATOM 1052 O ARG A 131 -2.330 13.619 18.581 1.00 75.73 A O
+ATOM 1053 CB ARG A 131 0.730 13.375 18.774 1.00 75.73 A C
+ATOM 1054 CG ARG A 131 0.334 13.889 20.135 1.00 75.73 A C
+ATOM 1055 CD ARG A 131 1.576 14.299 20.918 1.00 75.73 A C
+ATOM 1056 NE ARG A 131 2.252 13.136 21.498 1.00 75.73 A N
+ATOM 1057 CZ ARG A 131 3.217 12.443 20.901 1.00 75.73 A C
+ATOM 1058 NH1 ARG A 131 3.759 11.401 21.513 1.00 75.73 A N1+
+ATOM 1059 NH2 ARG A 131 3.652 12.791 19.700 1.00 75.73 A N
+ATOM 1060 N SER A 132 -1.696 13.744 16.445 1.00 68.39 A N
+ATOM 1061 CA SER A 132 -2.869 14.526 16.097 1.00 68.39 A C
+ATOM 1062 C SER A 132 -3.344 14.144 14.701 1.00 68.39 A C
+ATOM 1063 O SER A 132 -2.741 13.315 14.012 1.00 68.39 A O
+ATOM 1064 CB SER A 132 -2.592 16.023 16.201 1.00 68.39 A C
+ATOM 1065 OG SER A 132 -3.743 16.755 15.829 1.00 68.39 A O
+ATOM 1066 N TRP A 133 -4.436 14.777 14.284 1.00 60.92 A N
+ATOM 1067 CA TRP A 133 -5.229 14.329 13.151 1.00 60.92 A C
+ATOM 1068 C TRP A 133 -5.410 15.475 12.171 1.00 60.92 A C
+ATOM 1069 O TRP A 133 -5.940 16.525 12.541 1.00 60.92 A O
+ATOM 1070 CB TRP A 133 -6.586 13.833 13.641 1.00 60.92 A C
+ATOM 1071 CG TRP A 133 -6.496 12.634 14.535 1.00 60.92 A C
+ATOM 1072 CD1 TRP A 133 -6.814 12.577 15.857 1.00 60.92 A C
+ATOM 1073 CD2 TRP A 133 -6.112 11.305 14.160 1.00 60.92 A C
+ATOM 1074 CE2 TRP A 133 -6.198 10.507 15.308 1.00 60.92 A C
+ATOM 1075 CE3 TRP A 133 -5.701 10.717 12.967 1.00 60.92 A C
+ATOM 1076 NE1 TRP A 133 -6.621 11.307 16.336 1.00 60.92 A N
+ATOM 1077 CZ2 TRP A 133 -5.889 9.160 15.294 1.00 60.92 A C
+ATOM 1078 CZ3 TRP A 133 -5.407 9.385 12.963 1.00 60.92 A C
+ATOM 1079 CH2 TRP A 133 -5.493 8.622 14.112 1.00 60.92 A C
+ATOM 1080 N THR A 134 -5.003 15.264 10.919 1.00 61.11 A N
+ATOM 1081 CA THR A 134 -5.239 16.234 9.848 1.00 61.11 A C
+ATOM 1082 C THR A 134 -6.538 15.899 9.142 1.00 61.11 A C
+ATOM 1083 O THR A 134 -6.564 15.174 8.151 1.00 61.11 A O
+ATOM 1084 CB THR A 134 -4.102 16.247 8.841 1.00 61.11 A C
+ATOM 1085 CG2 THR A 134 -2.791 16.118 9.523 1.00 61.11 A C
+ATOM 1086 OG1 THR A 134 -4.275 15.163 7.924 1.00 61.11 A O
+ATOM 1087 N ALA A 135 -7.627 16.450 9.645 1.00 63.15 A N
+ATOM 1088 CA ALA A 135 -8.868 16.411 8.903 1.00 63.15 A C
+ATOM 1089 C ALA A 135 -8.681 17.090 7.558 1.00 63.15 A C
+ATOM 1090 O ALA A 135 -8.031 18.132 7.459 1.00 63.15 A O
+ATOM 1091 CB ALA A 135 -9.955 17.111 9.691 1.00 63.15 A C
+ATOM 1092 N ALA A 136 -9.264 16.506 6.516 1.00 67.55 A N
+ATOM 1093 CA ALA A 136 -9.098 17.003 5.157 1.00 67.55 A C
+ATOM 1094 C ALA A 136 -10.425 17.433 4.552 1.00 67.55 A C
+ATOM 1095 O ALA A 136 -10.576 17.447 3.332 1.00 67.55 A O
+ATOM 1096 CB ALA A 136 -8.430 15.955 4.272 1.00 67.55 A C
+ATOM 1097 N ASP A 137 -11.380 17.805 5.392 1.00 75.60 A N
+ATOM 1098 CA ASP A 137 -12.691 18.241 4.933 1.00 75.60 A C
+ATOM 1099 C ASP A 137 -13.481 18.687 6.152 1.00 75.60 A C
+ATOM 1100 O ASP A 137 -13.014 18.581 7.287 1.00 75.60 A O
+ATOM 1101 CB ASP A 137 -13.395 17.126 4.173 1.00 75.60 A C
+ATOM 1102 CG ASP A 137 -14.649 17.590 3.509 1.00 75.60 A C
+ATOM 1103 OD1 ASP A 137 -14.994 18.777 3.670 1.00 75.60 A O
+ATOM 1104 OD2 ASP A 137 -15.266 16.787 2.787 1.00 75.60 A O1-
+ATOM 1105 N MET A 138 -14.686 19.191 5.913 1.00 75.22 A N
+ATOM 1106 CA MET A 138 -15.578 19.596 6.991 1.00 75.22 A C
+ATOM 1107 C MET A 138 -16.330 18.433 7.610 1.00 75.22 A C
+ATOM 1108 O MET A 138 -17.158 18.651 8.498 1.00 75.22 A O
+ATOM 1109 CB MET A 138 -16.565 20.650 6.497 1.00 75.22 A C
+ATOM 1110 CG MET A 138 -15.914 21.959 6.106 1.00 75.22 A C
+ATOM 1111 SD MET A 138 -15.077 22.742 7.501 1.00 75.22 A S
+ATOM 1112 CE MET A 138 -13.361 22.350 7.165 1.00 75.22 A C
+ATOM 1113 N ALA A 139 -16.065 17.214 7.160 1.00 68.70 A N
+ATOM 1114 CA ALA A 139 -16.510 16.005 7.833 1.00 68.70 A C
+ATOM 1115 C ALA A 139 -15.405 15.361 8.643 1.00 68.70 A C
+ATOM 1116 O ALA A 139 -15.620 14.959 9.800 1.00 68.70 A O
+ATOM 1117 CB ALA A 139 -17.030 15.001 6.807 1.00 68.70 A C
+ATOM 1118 N ALA A 140 -14.211 15.277 8.068 1.00 66.90 A N
+ATOM 1119 CA ALA A 140 -13.077 14.807 8.838 1.00 66.90 A C
+ATOM 1120 C ALA A 140 -12.792 15.721 10.019 1.00 66.90 A C
+ATOM 1121 O ALA A 140 -12.118 15.297 10.953 1.00 66.90 A O
+ATOM 1122 CB ALA A 140 -11.857 14.662 7.938 1.00 66.90 A C
+ATOM 1123 N GLN A 141 -13.299 16.955 10.013 1.00 69.27 A N
+ATOM 1124 CA GLN A 141 -13.283 17.770 11.227 1.00 69.27 A C
+ATOM 1125 C GLN A 141 -14.011 17.068 12.362 1.00 69.27 A C
+ATOM 1126 O GLN A 141 -13.497 16.953 13.480 1.00 69.27 A O
+ATOM 1127 CB GLN A 141 -13.939 19.134 10.976 1.00 69.27 A C
+ATOM 1128 CG GLN A 141 -13.234 20.108 10.033 1.00 69.27 A C
+ATOM 1129 CD GLN A 141 -11.734 19.926 9.943 1.00 69.27 A C
+ATOM 1130 NE2 GLN A 141 -11.225 19.868 8.722 1.00 69.27 A N
+ATOM 1131 OE1 GLN A 141 -11.036 19.861 10.954 1.00 69.27 A O
+ATOM 1132 N ILE A 142 -15.234 16.618 12.093 1.00 65.40 A N
+ATOM 1133 CA ILE A 142 -16.036 15.954 13.110 1.00 65.40 A C
+ATOM 1134 C ILE A 142 -15.346 14.672 13.549 1.00 65.40 A C
+ATOM 1135 O ILE A 142 -15.269 14.348 14.746 1.00 65.40 A O
+ATOM 1136 CB ILE A 142 -17.440 15.681 12.560 1.00 65.40 A C
+ATOM 1137 CG1 ILE A 142 -18.110 16.978 12.149 1.00 65.40 A C
+ATOM 1138 CG2 ILE A 142 -18.281 15.168 13.653 1.00 65.40 A C
+ATOM 1139 CD1 ILE A 142 -19.586 16.831 11.888 1.00 65.40 A C
+ATOM 1140 N THR A 143 -14.821 13.928 12.581 1.00 61.39 A N
+ATOM 1141 CA THR A 143 -14.087 12.713 12.915 1.00 61.39 A C
+ATOM 1142 C THR A 143 -12.910 13.008 13.838 1.00 61.39 A C
+ATOM 1143 O THR A 143 -12.771 12.391 14.899 1.00 61.39 A O
+ATOM 1144 CB THR A 143 -13.616 12.012 11.652 1.00 61.39 A C
+ATOM 1145 CG2 THR A 143 -12.729 10.859 12.004 1.00 61.39 A C
+ATOM 1146 OG1 THR A 143 -14.752 11.521 10.947 1.00 61.39 A O
+ATOM 1147 N LYS A 144 -12.036 13.942 13.454 1.00 64.53 A N
+ATOM 1148 CA LYS A 144 -10.869 14.220 14.280 1.00 64.53 A C
+ATOM 1149 C LYS A 144 -11.257 14.757 15.641 1.00 64.53 A C
+ATOM 1150 O LYS A 144 -10.535 14.530 16.610 1.00 64.53 A O
+ATOM 1151 CB LYS A 144 -9.914 15.189 13.589 1.00 64.53 A C
+ATOM 1152 CG LYS A 144 -10.440 16.590 13.455 1.00 64.53 A C
+ATOM 1153 CD LYS A 144 -9.320 17.588 13.268 1.00 64.53 A C
+ATOM 1154 CE LYS A 144 -8.590 17.844 14.571 1.00 64.53 A C
+ATOM 1155 NZ LYS A 144 -9.449 18.582 15.534 1.00 64.53 A N1+
+ATOM 1156 N ARG A 145 -12.377 15.471 15.754 1.00 65.12 A N
+ATOM 1157 CA ARG A 145 -12.795 15.877 17.089 1.00 65.12 A C
+ATOM 1158 C ARG A 145 -13.132 14.663 17.938 1.00 65.12 A C
+ATOM 1159 O ARG A 145 -12.718 14.571 19.101 1.00 65.12 A O
+ATOM 1160 CB ARG A 145 -13.968 16.846 17.024 1.00 65.12 A C
+ATOM 1161 CG ARG A 145 -13.543 18.199 16.519 1.00 65.12 A C
+ATOM 1162 CD ARG A 145 -14.560 19.278 16.788 1.00 65.12 A C
+ATOM 1163 NE ARG A 145 -15.930 18.806 16.838 1.00 65.12 A N
+ATOM 1164 CZ ARG A 145 -16.783 18.915 15.833 1.00 65.12 A C
+ATOM 1165 NH1 ARG A 145 -18.021 18.474 15.971 1.00 65.12 A N1+
+ATOM 1166 NH2 ARG A 145 -16.400 19.473 14.695 1.00 65.12 A N
+ATOM 1167 N LYS A 146 -13.840 13.694 17.357 1.00 59.15 A N
+ATOM 1168 CA LYS A 146 -14.118 12.459 18.087 1.00 59.15 A C
+ATOM 1169 C LYS A 146 -12.870 11.649 18.392 1.00 59.15 A C
+ATOM 1170 O LYS A 146 -12.876 10.855 19.328 1.00 59.15 A O
+ATOM 1171 CB LYS A 146 -15.093 11.581 17.322 1.00 59.15 A C
+ATOM 1172 CG LYS A 146 -15.802 10.655 18.237 1.00 59.15 A C
+ATOM 1173 CD LYS A 146 -17.131 10.300 17.727 1.00 59.15 A C
+ATOM 1174 CE LYS A 146 -17.951 11.486 18.062 1.00 59.15 A C
+ATOM 1175 NZ LYS A 146 -18.035 11.555 19.522 1.00 59.15 A N1+
+ATOM 1176 N TRP A 147 -11.823 11.773 17.595 1.00 61.57 A N
+ATOM 1177 CA TRP A 147 -10.667 10.921 17.847 1.00 61.57 A C
+ATOM 1178 C TRP A 147 -9.674 11.587 18.795 1.00 61.57 A C
+ATOM 1179 O TRP A 147 -8.945 10.898 19.514 1.00 61.57 A O
+ATOM 1180 CB TRP A 147 -9.984 10.560 16.537 1.00 61.57 A C
+ATOM 1181 CG TRP A 147 -10.754 9.568 15.797 1.00 61.57 A C
+ATOM 1182 CD1 TRP A 147 -12.023 9.180 16.061 1.00 61.57 A C
+ATOM 1183 CD2 TRP A 147 -10.323 8.810 14.673 1.00 61.57 A C
+ATOM 1184 CE2 TRP A 147 -11.389 7.988 14.296 1.00 61.57 A C
+ATOM 1185 CE3 TRP A 147 -9.141 8.744 13.948 1.00 61.57 A C
+ATOM 1186 NE1 TRP A 147 -12.423 8.235 15.158 1.00 61.57 A N
+ATOM 1187 CZ2 TRP A 147 -11.309 7.114 13.231 1.00 61.57 A C
+ATOM 1188 CZ3 TRP A 147 -9.067 7.873 12.885 1.00 61.57 A C
+ATOM 1189 CH2 TRP A 147 -10.141 7.072 12.538 1.00 61.57 A C
+ATOM 1190 N GLU A 148 -9.584 12.920 18.766 1.00 66.50 A N
+ATOM 1191 CA GLU A 148 -8.836 13.635 19.792 1.00 66.50 A C
+ATOM 1192 C GLU A 148 -9.492 13.472 21.149 1.00 66.50 A C
+ATOM 1193 O GLU A 148 -8.810 13.201 22.141 1.00 66.50 A O
+ATOM 1194 CB GLU A 148 -8.703 15.118 19.446 1.00 66.50 A C
+ATOM 1195 CG GLU A 148 -7.860 15.439 18.227 1.00 66.50 A C
+ATOM 1196 CD GLU A 148 -6.391 15.101 18.414 1.00 66.50 A C
+ATOM 1197 OE1 GLU A 148 -5.894 15.190 19.556 1.00 66.50 A O
+ATOM 1198 OE2 GLU A 148 -5.717 14.791 17.411 1.00 66.50 A O1-
+ATOM 1199 N ALA A 149 -10.811 13.625 21.217 1.00 67.27 A N
+ATOM 1200 CA ALA A 149 -11.518 13.072 22.358 1.00 67.27 A C
+ATOM 1201 C ALA A 149 -11.375 11.562 22.297 1.00 67.27 A C
+ATOM 1202 O ALA A 149 -11.077 11.000 21.247 1.00 67.27 A O
+ATOM 1203 CB ALA A 149 -12.987 13.481 22.345 1.00 67.27 A C
+ATOM 1204 N ALA A 150 -11.485 10.905 23.439 1.00 70.70 A N
+ATOM 1205 CA ALA A 150 -11.430 9.450 23.537 1.00 70.70 A C
+ATOM 1206 C ALA A 150 -10.059 8.901 23.177 1.00 70.70 A C
+ATOM 1207 O ALA A 150 -9.897 7.686 23.154 1.00 70.70 A O
+ATOM 1208 CB ALA A 150 -12.469 8.738 22.661 1.00 70.70 A C
+ATOM 1209 N HIS A 151 -9.092 9.746 22.850 1.00 71.26 A N
+ATOM 1210 CA HIS A 151 -7.688 9.358 22.798 1.00 71.26 A C
+ATOM 1211 C HIS A 151 -7.478 8.148 21.906 1.00 71.26 A C
+ATOM 1212 O HIS A 151 -6.593 7.326 22.137 1.00 71.26 A O
+ATOM 1213 CB HIS A 151 -7.159 9.104 24.205 1.00 71.26 A C
+ATOM 1214 CG HIS A 151 -7.498 10.196 25.165 1.00 71.26 A C
+ATOM 1215 CD2 HIS A 151 -8.387 10.244 26.181 1.00 71.26 A C
+ATOM 1216 ND1 HIS A 151 -6.945 11.453 25.081 1.00 71.26 A N
+ATOM 1217 CE1 HIS A 151 -7.451 12.218 26.029 1.00 71.26 A C
+ATOM 1218 NE2 HIS A 151 -8.331 11.509 26.710 1.00 71.26 A N
+ATOM 1219 N GLU A 152 -8.295 8.050 20.862 1.00 70.11 A N
+ATOM 1220 CA GLU A 152 -8.085 7.000 19.876 1.00 70.11 A C
+ATOM 1221 C GLU A 152 -6.673 7.073 19.321 1.00 70.11 A C
+ATOM 1222 O GLU A 152 -6.088 6.054 18.946 1.00 70.11 A O
+ATOM 1223 CB GLU A 152 -9.117 7.121 18.760 1.00 70.11 A C
+ATOM 1224 CG GLU A 152 -9.017 6.051 17.679 1.00 70.11 A C
+ATOM 1225 CD GLU A 152 -9.091 4.635 18.218 1.00 70.11 A C
+ATOM 1226 OE1 GLU A 152 -10.211 4.168 18.497 1.00 70.11 A O
+ATOM 1227 OE2 GLU A 152 -8.033 3.985 18.354 1.00 70.11 A O1-
+ATOM 1228 N ALA A 153 -6.108 8.278 19.279 1.00 69.60 A N
+ATOM 1229 CA ALA A 153 -4.709 8.429 18.910 1.00 69.60 A C
+ATOM 1230 C ALA A 153 -3.802 7.699 19.886 1.00 69.60 A C
+ATOM 1231 O ALA A 153 -2.817 7.076 19.481 1.00 69.60 A O
+ATOM 1232 CB ALA A 153 -4.345 9.907 18.853 1.00 69.60 A C
+ATOM 1233 N GLU A 154 -4.119 7.763 21.179 1.00 77.76 A N
+ATOM 1234 CA GLU A 154 -3.274 7.122 22.181 1.00 77.76 A C
+ATOM 1235 C GLU A 154 -3.301 5.608 22.037 1.00 77.76 A C
+ATOM 1236 O GLU A 154 -2.247 4.959 21.948 1.00 77.76 A O
+ATOM 1237 CB GLU A 154 -3.751 7.520 23.575 1.00 77.76 A C
+ATOM 1238 CG GLU A 154 -3.583 8.986 23.891 1.00 77.76 A C
+ATOM 1239 CD GLU A 154 -2.140 9.397 23.972 1.00 77.76 A C
+ATOM 1240 OE1 GLU A 154 -1.311 8.557 24.377 1.00 77.76 A O
+ATOM 1241 OE2 GLU A 154 -1.832 10.559 23.632 1.00 77.76 A O1-
+ATOM 1242 N GLN A 155 -4.505 5.035 21.990 1.00 75.13 A N
+ATOM 1243 CA GLN A 155 -4.650 3.598 21.808 1.00 75.13 A C
+ATOM 1244 C GLN A 155 -3.993 3.165 20.518 1.00 75.13 A C
+ATOM 1245 O GLN A 155 -3.294 2.149 20.474 1.00 75.13 A O
+ATOM 1246 CB GLN A 155 -6.131 3.236 21.771 1.00 75.13 A C
+ATOM 1247 CG GLN A 155 -6.863 3.457 23.062 1.00 75.13 A C
+ATOM 1248 CD GLN A 155 -8.326 3.103 22.950 1.00 75.13 A C
+ATOM 1249 NE2 GLN A 155 -9.048 3.206 24.057 1.00 75.13 A N
+ATOM 1250 OE1 GLN A 155 -8.805 2.752 21.877 1.00 75.13 A O
+ATOM 1251 N LEU A 156 -4.217 3.932 19.455 1.00 70.78 A N
+ATOM 1252 CA LEU A 156 -3.650 3.592 18.165 1.00 70.78 A C
+ATOM 1253 C LEU A 156 -2.137 3.612 18.209 1.00 70.78 A C
+ATOM 1254 O LEU A 156 -1.500 2.728 17.646 1.00 70.78 A O
+ATOM 1255 CB LEU A 156 -4.173 4.540 17.091 1.00 70.78 A C
+ATOM 1256 CG LEU A 156 -3.639 4.201 15.705 1.00 70.78 A C
+ATOM 1257 CD1 LEU A 156 -4.225 2.904 15.211 1.00 70.78 A C
+ATOM 1258 CD2 LEU A 156 -3.947 5.318 14.750 1.00 70.78 A C
+ATOM 1259 N ARG A 157 -1.539 4.607 18.870 1.00 73.13 A N
+ATOM 1260 CA ARG A 157 -0.084 4.646 18.956 1.00 73.13 A C
+ATOM 1261 C ARG A 157 0.444 3.458 19.730 1.00 73.13 A C
+ATOM 1262 O ARG A 157 1.480 2.880 19.375 1.00 73.13 A O
+ATOM 1263 CB ARG A 157 0.394 5.927 19.625 1.00 73.13 A C
+ATOM 1264 CG ARG A 157 1.914 6.063 19.588 1.00 73.13 A C
+ATOM 1265 CD ARG A 157 2.412 7.288 20.330 1.00 73.13 A C
+ATOM 1266 NE ARG A 157 1.880 7.359 21.682 1.00 73.13 A N
+ATOM 1267 CZ ARG A 157 1.064 8.310 22.116 1.00 73.13 A C
+ATOM 1268 NH1 ARG A 157 0.671 9.274 21.300 1.00 73.13 A N1+
+ATOM 1269 NH2 ARG A 157 0.640 8.291 23.366 1.00 73.13 A N
+ATOM 1270 N ALA A 158 -0.238 3.108 20.822 1.00 69.79 A N
+ATOM 1271 CA ALA A 158 0.135 1.917 21.571 1.00 69.79 A C
+ATOM 1272 C ALA A 158 0.119 0.698 20.668 1.00 69.79 A C
+ATOM 1273 O ALA A 158 1.100 -0.055 20.604 1.00 69.79 A O
+ATOM 1274 CB ALA A 158 -0.812 1.727 22.751 1.00 69.79 A C
+ATOM 1275 N TYR A 159 -0.992 0.498 19.951 1.00 69.23 A N
+ATOM 1276 CA TYR A 159 -1.049 -0.554 18.945 1.00 69.23 A C
+ATOM 1277 C TYR A 159 0.159 -0.473 18.037 1.00 69.23 A C
+ATOM 1278 O TYR A 159 0.954 -1.412 17.969 1.00 69.23 A O
+ATOM 1279 CB TYR A 159 -2.313 -0.475 18.092 1.00 69.23 A C
+ATOM 1280 CG TYR A 159 -2.155 -1.350 16.856 1.00 69.23 A C
+ATOM 1281 CD1 TYR A 159 -1.711 -2.660 16.960 1.00 69.23 A C
+ATOM 1282 CD2 TYR A 159 -2.343 -0.837 15.582 1.00 69.23 A C
+ATOM 1283 CE1 TYR A 159 -1.514 -3.437 15.848 1.00 69.23 A C
+ATOM 1284 CE2 TYR A 159 -2.144 -1.618 14.464 1.00 69.23 A C
+ATOM 1285 CZ TYR A 159 -1.731 -2.912 14.610 1.00 69.23 A C
+ATOM 1286 OH TYR A 159 -1.533 -3.697 13.511 1.00 69.23 A O
+ATOM 1287 N LEU A 160 0.318 0.663 17.361 1.00 72.15 A N
+ATOM 1288 CA LEU A 160 1.307 0.838 16.315 1.00 72.15 A C
+ATOM 1289 C LEU A 160 2.647 0.380 16.840 1.00 72.15 A C
+ATOM 1290 O LEU A 160 3.049 -0.749 16.540 1.00 72.15 A O
+ATOM 1291 CB LEU A 160 1.358 2.296 15.870 1.00 72.15 A C
+ATOM 1292 CG LEU A 160 0.074 2.755 15.176 1.00 72.15 A C
+ATOM 1293 CD1 LEU A 160 0.155 4.162 14.639 1.00 72.15 A C
+ATOM 1294 CD2 LEU A 160 -0.316 1.800 14.087 1.00 72.15 A C
+ATOM 1295 N ASP A 161 3.207 1.124 17.793 1.00 73.96 A N
+ATOM 1296 CA ASP A 161 4.530 0.778 18.294 1.00 73.96 A C
+ATOM 1297 C ASP A 161 4.527 -0.641 18.837 1.00 73.96 A C
+ATOM 1298 O ASP A 161 4.997 -1.555 18.155 1.00 73.96 A O
+ATOM 1299 CB ASP A 161 4.986 1.755 19.381 1.00 73.96 A C
+ATOM 1300 CG ASP A 161 4.772 3.198 18.992 1.00 73.96 A C
+ATOM 1301 OD1 ASP A 161 4.866 3.502 17.786 1.00 73.96 A O
+ATOM 1302 OD2 ASP A 161 4.510 4.028 19.888 1.00 73.96 A O1-
+ATOM 1303 N GLY A 162 3.855 -0.872 19.962 1.00 76.55 A N
+ATOM 1304 CA GLY A 162 4.022 -2.157 20.603 1.00 76.55 A C
+ATOM 1305 C GLY A 162 3.566 -3.309 19.740 1.00 76.55 A C
+ATOM 1306 O GLY A 162 4.387 -4.068 19.215 1.00 76.55 A O
+ATOM 1307 N THR A 163 2.257 -3.415 19.542 1.00 74.09 A N
+ATOM 1308 CA THR A 163 1.724 -4.599 18.895 1.00 74.09 A C
+ATOM 1309 C THR A 163 2.142 -4.652 17.440 1.00 74.09 A C
+ATOM 1310 O THR A 163 2.582 -5.699 16.952 1.00 74.09 A O
+ATOM 1311 CB THR A 163 0.207 -4.613 19.003 1.00 74.09 A C
+ATOM 1312 CG2 THR A 163 -0.310 -5.995 18.727 1.00 74.09 A C
+ATOM 1313 OG1 THR A 163 -0.185 -4.212 20.319 1.00 74.09 A O
+ATOM 1314 N CYS A 164 1.970 -3.546 16.711 1.00 74.22 A N
+ATOM 1315 CA CYS A 164 2.216 -3.621 15.288 1.00 74.22 A C
+ATOM 1316 C CYS A 164 3.680 -3.925 15.035 1.00 74.22 A C
+ATOM 1317 O CYS A 164 4.000 -4.770 14.197 1.00 74.22 A O
+ATOM 1318 CB CYS A 164 1.793 -2.320 14.617 1.00 74.22 A C
+ATOM 1319 SG CYS A 164 1.963 -2.315 12.830 1.00 74.22 A S
+ATOM 1320 N VAL A 165 4.588 -3.294 15.791 1.00 73.77 A N
+ATOM 1321 CA VAL A 165 6.006 -3.532 15.596 1.00 73.77 A C
+ATOM 1322 C VAL A 165 6.375 -4.957 15.979 1.00 73.77 A C
+ATOM 1323 O VAL A 165 7.139 -5.619 15.270 1.00 73.77 A O
+ATOM 1324 CB VAL A 165 6.813 -2.499 16.395 1.00 73.77 A C
+ATOM 1325 CG1 VAL A 165 8.251 -2.935 16.524 1.00 73.77 A C
+ATOM 1326 CG2 VAL A 165 6.725 -1.154 15.720 1.00 73.77 A C
+ATOM 1327 N GLU A 166 5.846 -5.460 17.098 1.00 78.13 A N
+ATOM 1328 CA GLU A 166 6.257 -6.791 17.527 1.00 78.13 A C
+ATOM 1329 C GLU A 166 5.738 -7.860 16.575 1.00 78.13 A C
+ATOM 1330 O GLU A 166 6.493 -8.752 16.178 1.00 78.13 A O
+ATOM 1331 CB GLU A 166 5.808 -7.069 18.962 1.00 78.13 A C
+ATOM 1332 CG GLU A 166 4.313 -7.148 19.181 1.00 78.13 A C
+ATOM 1333 CD GLU A 166 3.945 -7.361 20.637 1.00 78.13 A C
+ATOM 1334 OE1 GLU A 166 4.863 -7.420 21.479 1.00 78.13 A O
+ATOM 1335 OE2 GLU A 166 2.739 -7.480 20.937 1.00 78.13 A O1-
+ATOM 1336 N TRP A 167 4.464 -7.786 16.174 1.00 71.28 A N
+ATOM 1337 CA TRP A 167 4.001 -8.745 15.180 1.00 71.28 A C
+ATOM 1338 C TRP A 167 4.667 -8.552 13.840 1.00 71.28 A C
+ATOM 1339 O TRP A 167 4.850 -9.529 13.116 1.00 71.28 A O
+ATOM 1340 CB TRP A 167 2.496 -8.716 14.980 1.00 71.28 A C
+ATOM 1341 CG TRP A 167 1.711 -9.229 16.090 1.00 71.28 A C
+ATOM 1342 CD1 TRP A 167 1.093 -8.526 17.060 1.00 71.28 A C
+ATOM 1343 CD2 TRP A 167 1.484 -10.606 16.374 1.00 71.28 A C
+ATOM 1344 CE2 TRP A 167 0.686 -10.666 17.520 1.00 71.28 A C
+ATOM 1345 CE3 TRP A 167 1.867 -11.794 15.753 1.00 71.28 A C
+ATOM 1346 NE1 TRP A 167 0.454 -9.380 17.924 1.00 71.28 A N
+ATOM 1347 CZ2 TRP A 167 0.267 -11.864 18.066 1.00 71.28 A C
+ATOM 1348 CZ3 TRP A 167 1.454 -12.975 16.290 1.00 71.28 A C
+ATOM 1349 CH2 TRP A 167 0.662 -13.007 17.437 1.00 71.28 A C
+ATOM 1350 N LEU A 168 5.018 -7.327 13.471 1.00 70.90 A N
+ATOM 1351 CA LEU A 168 5.722 -7.153 12.215 1.00 70.90 A C
+ATOM 1352 C LEU A 168 7.071 -7.847 12.273 1.00 70.90 A C
+ATOM 1353 O LEU A 168 7.449 -8.563 11.339 1.00 70.90 A O
+ATOM 1354 CB LEU A 168 5.848 -5.667 11.901 1.00 70.90 A C
+ATOM 1355 CG LEU A 168 6.189 -5.217 10.483 1.00 70.90 A C
+ATOM 1356 CD1 LEU A 168 5.818 -3.775 10.346 1.00 70.90 A C
+ATOM 1357 CD2 LEU A 168 7.648 -5.343 10.216 1.00 70.90 A C
+ATOM 1358 N ARG A 169 7.789 -7.696 13.386 1.00 75.02 A N
+ATOM 1359 CA ARG A 169 9.056 -8.404 13.533 1.00 75.02 A C
+ATOM 1360 C ARG A 169 8.846 -9.908 13.555 1.00 75.02 A C
+ATOM 1361 O ARG A 169 9.642 -10.659 12.984 1.00 75.02 A O
+ATOM 1362 CB ARG A 169 9.797 -7.932 14.783 1.00 75.02 A C
+ATOM 1363 CG ARG A 169 10.262 -6.502 14.673 1.00 75.02 A C
+ATOM 1364 CD ARG A 169 10.987 -6.008 15.904 1.00 75.02 A C
+ATOM 1365 NE ARG A 169 10.113 -5.889 17.059 1.00 75.02 A N
+ATOM 1366 CZ ARG A 169 10.542 -5.600 18.280 1.00 75.02 A C
+ATOM 1367 NH1 ARG A 169 9.684 -5.500 19.282 1.00 75.02 A N1+
+ATOM 1368 NH2 ARG A 169 11.831 -5.400 18.492 1.00 75.02 A N
+ATOM 1369 N ARG A 170 7.773 -10.369 14.193 1.00 74.46 A N
+ATOM 1370 CA ARG A 170 7.473 -11.794 14.168 1.00 74.46 A C
+ATOM 1371 C ARG A 170 7.252 -12.279 12.746 1.00 74.46 A C
+ATOM 1372 O ARG A 170 7.783 -13.316 12.345 1.00 74.46 A O
+ATOM 1373 CB ARG A 170 6.247 -12.102 15.015 1.00 74.46 A C
+ATOM 1374 CG ARG A 170 5.959 -13.589 15.076 1.00 74.46 A C
+ATOM 1375 CD ARG A 170 5.495 -14.031 16.440 1.00 74.46 A C
+ATOM 1376 NE ARG A 170 4.811 -15.315 16.360 1.00 74.46 A N
+ATOM 1377 CZ ARG A 170 3.831 -15.686 17.172 1.00 74.46 A C
+ATOM 1378 NH1 ARG A 170 3.264 -16.874 17.026 1.00 74.46 A N1+
+ATOM 1379 NH2 ARG A 170 3.415 -14.865 18.126 1.00 74.46 A N
+ATOM 1380 N TYR A 171 6.447 -11.549 11.977 1.00 67.55 A N
+ATOM 1381 CA TYR A 171 6.157 -11.947 10.608 1.00 67.55 A C
+ATOM 1382 C TYR A 171 7.436 -12.032 9.798 1.00 67.55 A C
+ATOM 1383 O TYR A 171 7.688 -13.022 9.105 1.00 67.55 A O
+ATOM 1384 CB TYR A 171 5.198 -10.951 9.971 1.00 67.55 A C
+ATOM 1385 CG TYR A 171 3.796 -10.996 10.498 1.00 67.55 A C
+ATOM 1386 CD1 TYR A 171 3.272 -12.142 11.041 1.00 67.55 A C
+ATOM 1387 CD2 TYR A 171 2.996 -9.883 10.443 1.00 67.55 A C
+ATOM 1388 CE1 TYR A 171 1.988 -12.171 11.515 1.00 67.55 A C
+ATOM 1389 CE2 TYR A 171 1.724 -9.900 10.917 1.00 67.55 A C
+ATOM 1390 CZ TYR A 171 1.220 -11.042 11.450 1.00 67.55 A C
+ATOM 1391 OH TYR A 171 -0.070 -11.042 11.916 1.00 67.55 A O
+ATOM 1392 N LEU A 172 8.266 -10.998 9.889 1.00 74.70 A N
+ATOM 1393 CA LEU A 172 9.543 -11.019 9.188 1.00 74.70 A C
+ATOM 1394 C LEU A 172 10.465 -12.113 9.699 1.00 74.70 A C
+ATOM 1395 O LEU A 172 11.348 -12.565 8.963 1.00 74.70 A O
+ATOM 1396 CB LEU A 172 10.190 -9.645 9.272 1.00 74.70 A C
+ATOM 1397 CG LEU A 172 9.605 -8.635 8.288 1.00 74.70 A C
+ATOM 1398 CD1 LEU A 172 8.225 -8.172 8.587 1.00 74.70 A C
+ATOM 1399 CD2 LEU A 172 10.497 -7.438 8.340 1.00 74.70 A C
+ATOM 1400 N GLU A 173 10.291 -12.554 10.943 1.00 77.94 A N
+ATOM 1401 CA GLU A 173 10.958 -13.780 11.366 1.00 77.94 A C
+ATOM 1402 C GLU A 173 10.392 -14.981 10.621 1.00 77.94 A C
+ATOM 1403 O GLU A 173 11.136 -15.823 10.107 1.00 77.94 A O
+ATOM 1404 CB GLU A 173 10.815 -13.971 12.875 1.00 77.94 A C
+ATOM 1405 CG GLU A 173 11.658 -13.029 13.705 1.00 77.94 A C
+ATOM 1406 CD GLU A 173 13.142 -13.255 13.506 1.00 77.94 A C
+ATOM 1407 OE1 GLU A 173 13.535 -14.394 13.175 1.00 77.94 A O
+ATOM 1408 OE2 GLU A 173 13.919 -12.295 13.682 1.00 77.94 A O1-
+ATOM 1409 N ASN A 174 9.068 -15.056 10.540 1.00 80.68 A N
+ATOM 1410 CA ASN A 174 8.408 -16.196 9.926 1.00 80.68 A C
+ATOM 1411 C ASN A 174 8.652 -16.218 8.427 1.00 80.68 A C
+ATOM 1412 O ASN A 174 8.991 -17.260 7.856 1.00 80.68 A O
+ATOM 1413 CB ASN A 174 6.914 -16.132 10.224 1.00 80.68 A C
+ATOM 1414 CG ASN A 174 6.604 -16.388 11.679 1.00 80.68 A C
+ATOM 1415 ND2 ASN A 174 5.685 -15.610 12.231 1.00 80.68 A N
+ATOM 1416 OD1 ASN A 174 7.185 -17.273 12.305 1.00 80.68 A O
+ATOM 1417 N GLY A 175 8.488 -15.072 7.774 1.00 82.06 A N
+ATOM 1418 CA GLY A 175 8.629 -14.983 6.338 1.00 82.06 A C
+ATOM 1419 C GLY A 175 10.034 -14.626 5.912 1.00 82.06 A C
+ATOM 1420 O GLY A 175 10.243 -14.164 4.788 1.00 82.06 A O
+ATOM 1421 N LYS A 176 11.006 -14.840 6.801 1.00 83.45 A N
+ATOM 1422 CA LYS A 176 12.359 -14.351 6.558 1.00 83.45 A C
+ATOM 1423 C LYS A 176 12.925 -14.868 5.245 1.00 83.45 A C
+ATOM 1424 O LYS A 176 13.815 -14.242 4.661 1.00 83.45 A O
+ATOM 1425 CB LYS A 176 13.278 -14.738 7.719 1.00 83.45 A C
+ATOM 1426 CG LYS A 176 13.648 -16.206 7.767 1.00 83.45 A C
+ATOM 1427 CD LYS A 176 14.757 -16.473 8.770 1.00 83.45 A C
+ATOM 1428 CE LYS A 176 14.279 -16.258 10.191 1.00 83.45 A C
+ATOM 1429 NZ LYS A 176 13.014 -16.987 10.455 1.00 83.45 A N1+
+ATOM 1430 N GLU A 177 12.426 -15.995 4.766 1.00 85.04 A N
+ATOM 1431 CA GLU A 177 12.836 -16.539 3.485 1.00 85.04 A C
+ATOM 1432 C GLU A 177 11.938 -16.077 2.349 1.00 85.04 A C
+ATOM 1433 O GLU A 177 12.143 -16.493 1.206 1.00 85.04 A O
+ATOM 1434 CB GLU A 177 12.847 -18.068 3.555 1.00 85.04 A C
+ATOM 1435 CG GLU A 177 11.478 -18.697 3.809 1.00 85.04 A C
+ATOM 1436 CD GLU A 177 11.009 -18.590 5.257 1.00 85.04 A C
+ATOM 1437 OE1 GLU A 177 11.385 -17.623 5.953 1.00 85.04 A O
+ATOM 1438 OE2 GLU A 177 10.259 -19.484 5.702 1.00 85.04 A O1-
+ATOM 1439 N THR A 178 10.959 -15.236 2.636 1.00 85.43 A N
+ATOM 1440 CA THR A 178 10.027 -14.739 1.635 1.00 85.43 A C
+ATOM 1441 C THR A 178 10.073 -13.232 1.472 1.00 85.43 A C
+ATOM 1442 O THR A 178 10.058 -12.737 0.345 1.00 85.43 A O
+ATOM 1443 CB THR A 178 8.599 -15.161 2.002 1.00 85.43 A C
+ATOM 1444 CG2 THR A 178 7.580 -14.459 1.121 1.00 85.43 A C
+ATOM 1445 OG1 THR A 178 8.463 -16.579 1.853 1.00 85.43 A O
+ATOM 1446 N LEU A 179 10.150 -12.488 2.572 1.00 83.70 A N
+ATOM 1447 CA LEU A 179 10.075 -11.035 2.529 1.00 83.70 A C
+ATOM 1448 C LEU A 179 11.441 -10.384 2.345 1.00 83.70 A C
+ATOM 1449 O LEU A 179 11.612 -9.538 1.464 1.00 83.70 A O
+ATOM 1450 CB LEU A 179 9.410 -10.520 3.803 1.00 83.70 A C
+ATOM 1451 CG LEU A 179 7.887 -10.483 3.822 1.00 83.70 A C
+ATOM 1452 CD1 LEU A 179 7.280 -11.853 3.682 1.00 83.70 A C
+ATOM 1453 CD2 LEU A 179 7.484 -9.891 5.124 1.00 83.70 A C
+ATOM 1454 N GLN A 180 12.425 -10.774 3.149 1.00 88.06 A N
+ATOM 1455 CA GLN A 180 13.760 -10.208 3.011 1.00 88.06 A C
+ATOM 1456 C GLN A 180 14.409 -10.711 1.736 1.00 88.06 A C
+ATOM 1457 O GLN A 180 15.399 -11.446 1.780 1.00 88.06 A O
+ATOM 1458 CB GLN A 180 14.638 -10.562 4.207 1.00 88.06 A C
+ATOM 1459 CG GLN A 180 14.395 -9.712 5.425 1.00 88.06 A C
+ATOM 1460 CD GLN A 180 13.175 -10.151 6.191 1.00 88.06 A C
+ATOM 1461 NE2 GLN A 180 12.693 -9.291 7.071 1.00 88.06 A N
+ATOM 1462 OE1 GLN A 180 12.673 -11.255 5.999 1.00 88.06 A O
+TER
+ATOM 1463 N VAL C 7 -1.168 -8.421 12.323 1.00 62.09 C N
+ATOM 1464 CA VAL C 7 -2.337 -8.531 13.180 1.00 62.09 C C
+ATOM 1465 C VAL C 7 -3.206 -7.297 13.123 1.00 62.09 C C
+ATOM 1466 O VAL C 7 -2.745 -6.190 13.330 1.00 62.09 C O
+ATOM 1467 CB VAL C 7 -1.927 -8.820 14.615 1.00 62.09 C C
+ATOM 1468 CG1 VAL C 7 -3.135 -8.819 15.519 1.00 62.09 C C
+ATOM 1469 CG2 VAL C 7 -1.287 -10.163 14.674 1.00 62.09 C C
+ATOM 1470 N VAL C 8 -4.486 -7.510 12.852 1.00 55.61 C N
+ATOM 1471 CA VAL C 8 -5.437 -6.418 12.840 1.00 55.61 C C
+ATOM 1472 C VAL C 8 -5.515 -5.799 14.225 1.00 55.61 C C
+ATOM 1473 O VAL C 8 -5.212 -6.441 15.231 1.00 55.61 C O
+ATOM 1474 CB VAL C 8 -6.806 -6.940 12.370 1.00 55.61 C C
+ATOM 1475 CG1 VAL C 8 -7.790 -5.812 12.158 1.00 55.61 C C
+ATOM 1476 CG2 VAL C 8 -6.627 -7.715 11.090 1.00 55.61 C C
+ATOM 1477 N VAL C 9 -5.892 -4.522 14.272 1.00 53.29 C N
+ATOM 1478 CA VAL C 9 -6.095 -3.849 15.543 1.00 53.29 C C
+ATOM 1479 C VAL C 9 -7.011 -4.679 16.416 1.00 53.29 C C
+ATOM 1480 O VAL C 9 -8.117 -5.048 16.008 1.00 53.29 C O
+ATOM 1481 CB VAL C 9 -6.669 -2.448 15.314 1.00 53.29 C C
+ATOM 1482 CG1 VAL C 9 -6.985 -1.801 16.620 1.00 53.29 C C
+ATOM 1483 CG2 VAL C 9 -5.655 -1.621 14.608 1.00 53.29 C C
+ATOM 1484 N GLY C 10 -6.549 -4.988 17.624 1.00 51.28 C N
+ATOM 1485 CA GLY C 10 -7.301 -5.884 18.476 1.00 51.28 C C
+ATOM 1486 C GLY C 10 -8.629 -5.308 18.913 1.00 51.28 C C
+ATOM 1487 O GLY C 10 -9.672 -5.938 18.742 1.00 51.28 C O
+ATOM 1488 N ALA C 11 -8.618 -4.100 19.449 1.00 51.50 C N
+ATOM 1489 CA ALA C 11 -9.825 -3.571 20.057 1.00 51.50 C C
+ATOM 1490 C ALA C 11 -10.823 -3.144 18.997 1.00 51.50 C C
+ATOM 1491 O ALA C 11 -10.464 -2.839 17.860 1.00 51.50 C O
+ATOM 1492 CB ALA C 11 -9.496 -2.393 20.964 1.00 51.50 C C
+ATOM 1493 N ASP C 12 -12.092 -3.176 19.381 1.00 0.00 C N
+ATOM 1494 CA ASP C 12 -13.065 -2.457 18.563 1.00 0.00 C C
+ATOM 1495 C ASP C 12 -12.748 -0.982 18.516 1.00 0.00 C C
+ATOM 1496 O ASP C 12 -12.796 -0.317 19.560 1.00 0.00 C O
+ATOM 1497 CB ASP C 12 -14.507 -2.699 19.092 1.00 0.00 C C
+ATOM 1498 CG ASP C 12 -15.056 -4.119 18.889 1.00 0.00 C C
+ATOM 1499 OD1 ASP C 12 -14.479 -5.078 19.442 1.00 0.00 C O
+ATOM 1500 OD2 ASP C 12 -16.040 -4.277 18.137 1.00 0.00 C O1-
+ATOM 1501 N GLY C 13 -12.361 -0.510 17.363 1.00 60.45 C N
+ATOM 1502 CA GLY C 13 -11.945 0.864 17.247 1.00 60.45 C C
+ATOM 1503 C GLY C 13 -13.109 1.816 17.362 1.00 60.45 C C
+ATOM 1504 O GLY C 13 -14.270 1.441 17.287 1.00 60.45 C O
+ATOM 1505 N VAL C 14 -12.783 3.083 17.576 1.00 62.38 C N
+ATOM 1506 CA VAL C 14 -13.815 4.098 17.543 1.00 62.38 C C
+ATOM 1507 C VAL C 14 -14.376 4.208 16.136 1.00 62.38 C C
+ATOM 1508 O VAL C 14 -13.669 4.028 15.139 1.00 62.38 C O
+ATOM 1509 CB VAL C 14 -13.270 5.431 18.084 1.00 62.38 C C
+ATOM 1510 CG1 VAL C 14 -14.305 6.538 17.980 1.00 62.38 C C
+ATOM 1511 CG2 VAL C 14 -12.890 5.260 19.542 1.00 62.38 C C
+ATOM 1512 N GLY C 15 -15.680 4.443 16.049 1.00 63.37 C N
+ATOM 1513 CA GLY C 15 -16.295 4.587 14.745 1.00 63.37 C C
+ATOM 1514 C GLY C 15 -15.786 5.827 14.034 1.00 63.37 C C
+ATOM 1515 O GLY C 15 -15.533 6.863 14.652 1.00 63.37 C O
+ATOM 1516 N LYS C 16 -15.637 5.716 12.721 1.00 59.55 C N
+ATOM 1517 CA LYS C 16 -15.237 6.848 11.898 1.00 59.55 C C
+ATOM 1518 C LYS C 16 -16.332 7.885 11.822 1.00 59.55 C C
+ATOM 1519 O LYS C 16 -16.861 8.348 12.826 1.00 59.55 C O
+ATOM 1520 CB LYS C 16 -14.869 6.391 10.490 1.00 59.55 C C
+ATOM 1521 CG LYS C 16 -13.897 7.296 9.807 1.00 59.55 C C
+ATOM 1522 CD LYS C 16 -13.608 6.839 8.422 1.00 59.55 C C
+ATOM 1523 CE LYS C 16 -13.276 5.389 8.410 1.00 59.55 C C
+ATOM 1524 NZ LYS C 16 -12.185 5.110 9.369 1.00 59.55 C N1+
+ATOM 1525 OXT LYS C 16 -16.709 8.275 10.729 1.00 59.55 C O1-
+TER
+ATOM 1526 N VAL H 2 -15.515 9.194 25.136 1.00 69.83 H N
+ATOM 1527 CA VAL H 2 -16.798 9.253 24.460 1.00 69.83 H C
+ATOM 1528 C VAL H 2 -17.886 8.603 25.306 1.00 69.83 H C
+ATOM 1529 O VAL H 2 -18.844 8.057 24.775 1.00 69.83 H O
+ATOM 1530 CB VAL H 2 -16.686 8.626 23.075 1.00 69.83 H C
+ATOM 1531 CG1 VAL H 2 -15.720 9.403 22.234 1.00 69.83 H C
+ATOM 1532 CG2 VAL H 2 -16.126 7.255 23.232 1.00 69.83 H C
+ATOM 1533 N GLN H 3 -17.746 8.692 26.627 1.00 72.01 H N
+ATOM 1534 CA GLN H 3 -18.741 8.190 27.566 1.00 72.01 H C
+ATOM 1535 C GLN H 3 -19.346 9.370 28.314 1.00 72.01 H C
+ATOM 1536 O GLN H 3 -18.630 10.263 28.770 1.00 72.01 H O
+ATOM 1537 CB GLN H 3 -18.116 7.201 28.552 1.00 72.01 H C
+ATOM 1538 CG GLN H 3 -18.081 5.764 28.049 1.00 72.01 H C
+ATOM 1539 CD GLN H 3 -17.408 5.640 26.695 1.00 72.01 H C
+ATOM 1540 NE2 GLN H 3 -18.192 5.336 25.670 1.00 72.01 H N
+ATOM 1541 OE1 GLN H 3 -16.195 5.816 26.573 1.00 72.01 H O
+ATOM 1542 N LEU H 4 -20.666 9.378 28.451 1.00 69.28 H N
+ATOM 1543 CA LEU H 4 -21.354 10.636 28.688 1.00 69.28 H C
+ATOM 1544 C LEU H 4 -22.484 10.466 29.688 1.00 69.28 H C
+ATOM 1545 O LEU H 4 -22.494 9.551 30.516 1.00 69.28 H O
+ATOM 1546 CB LEU H 4 -21.928 11.238 27.406 1.00 69.28 H C
+ATOM 1547 CG LEU H 4 -21.110 11.810 26.264 1.00 69.28 H C
+ATOM 1548 CD1 LEU H 4 -20.239 10.802 25.566 1.00 69.28 H C
+ATOM 1549 CD2 LEU H 4 -22.078 12.490 25.330 1.00 69.28 H C
+ATOM 1550 N VAL H 5 -23.414 11.419 29.621 1.00 69.85 H N
+ATOM 1551 CA VAL H 5 -24.467 11.703 30.587 1.00 69.85 H C
+ATOM 1552 C VAL H 5 -25.736 11.981 29.778 1.00 69.85 H C
+ATOM 1553 O VAL H 5 -25.829 11.598 28.610 1.00 69.85 H O
+ATOM 1554 CB VAL H 5 -24.072 12.865 31.528 1.00 69.85 H C
+ATOM 1555 CG1 VAL H 5 -25.040 13.028 32.705 1.00 69.85 H C
+ATOM 1556 CG2 VAL H 5 -22.668 12.655 32.074 1.00 69.85 H C
+ATOM 1557 N GLU H 6 -26.735 12.584 30.408 1.00 73.23 H N
+ATOM 1558 CA GLU H 6 -28.119 12.575 29.967 1.00 73.23 H C
+ATOM 1559 C GLU H 6 -28.752 13.898 30.366 1.00 73.23 H C
+ATOM 1560 O GLU H 6 -28.070 14.805 30.849 1.00 73.23 H O
+ATOM 1561 CB GLU H 6 -28.854 11.438 30.653 1.00 73.23 H C
+ATOM 1562 CG GLU H 6 -28.911 11.722 32.153 1.00 73.23 H C
+ATOM 1563 CD GLU H 6 -29.549 10.629 32.939 1.00 73.23 H C
+ATOM 1564 OE1 GLU H 6 -29.920 9.639 32.315 1.00 73.23 H O
+ATOM 1565 OE2 GLU H 6 -29.700 10.767 34.170 1.00 73.23 H O1-
+ATOM 1566 N SER H 7 -30.068 13.995 30.186 1.00 74.34 H N
+ATOM 1567 CA SER H 7 -30.872 15.093 30.728 1.00 74.34 H C
+ATOM 1568 C SER H 7 -32.342 14.809 30.445 1.00 74.34 H C
+ATOM 1569 O SER H 7 -32.711 13.709 30.023 1.00 74.34 H O
+ATOM 1570 CB SER H 7 -30.470 16.435 30.128 1.00 74.34 H C
+ATOM 1571 OG SER H 7 -30.807 16.460 28.761 1.00 74.34 H O
+ATOM 1572 N GLY H 8 -33.182 15.813 30.688 1.00 77.99 H N
+ATOM 1573 CA GLY H 8 -34.491 15.881 30.069 1.00 77.99 H C
+ATOM 1574 C GLY H 8 -35.673 16.294 30.920 1.00 77.99 H C
+ATOM 1575 O GLY H 8 -36.493 17.102 30.477 1.00 77.99 H O
+ATOM 1576 N GLY H 9 -35.772 15.788 32.134 1.00 89.06 H N
+ATOM 1577 CA GLY H 9 -36.908 16.169 32.965 1.00 89.06 H C
+ATOM 1578 C GLY H 9 -38.058 15.182 32.894 1.00 89.06 H C
+ATOM 1579 O GLY H 9 -38.299 14.523 31.884 1.00 89.06 H O
+ATOM 1580 N GLY H 10 -38.781 15.075 34.009 1.00 92.48 H N
+ATOM 1581 CA GLY H 10 -39.930 14.198 34.126 1.00 92.48 H C
+ATOM 1582 C GLY H 10 -41.194 14.912 33.707 1.00 92.48 H C
+ATOM 1583 O GLY H 10 -42.013 15.283 34.552 1.00 92.48 H O
+ATOM 1584 N LEU H 11 -41.347 15.111 32.400 1.00 99.56 H N
+ATOM 1585 CA LEU H 11 -42.351 16.014 31.852 1.00 99.56 H C
+ATOM 1586 C LEU H 11 -43.736 15.706 32.401 1.00 99.56 H C
+ATOM 1587 O LEU H 11 -44.058 14.563 32.735 1.00 99.56 H O
+ATOM 1588 CB LEU H 11 -42.359 15.909 30.329 1.00 99.56 H C
+ATOM 1589 CG LEU H 11 -43.596 16.380 29.580 1.00 99.56 H C
+ATOM 1590 CD1 LEU H 11 -43.640 17.887 29.606 1.00 99.56 H C
+ATOM 1591 CD2 LEU H 11 -43.591 15.877 28.165 1.00 99.56 H C
+ATOM 1592 N VAL H 12 -44.564 16.741 32.476 1.00106.81 H N
+ATOM 1593 CA VAL H 12 -45.911 16.625 33.009 1.00106.81 H C
+ATOM 1594 C VAL H 12 -46.875 16.824 31.854 1.00106.81 H C
+ATOM 1595 O VAL H 12 -46.447 17.021 30.715 1.00106.81 H O
+ATOM 1596 CB VAL H 12 -46.163 17.636 34.139 1.00106.81 H C
+ATOM 1597 CG1 VAL H 12 -45.075 17.519 35.199 1.00106.81 H C
+ATOM 1598 CG2 VAL H 12 -46.227 19.053 33.585 1.00106.81 H C
+ATOM 1599 N GLN H 13 -48.170 16.793 32.146 1.00116.71 H N
+ATOM 1600 CA GLN H 13 -49.230 16.558 31.172 1.00116.71 H C
+ATOM 1601 C GLN H 13 -49.105 17.390 29.900 1.00116.71 H C
+ATOM 1602 O GLN H 13 -49.266 18.616 29.922 1.00116.71 H O
+ATOM 1603 CB GLN H 13 -50.593 16.791 31.828 1.00116.71 H C
+ATOM 1604 CG GLN H 13 -51.749 16.730 30.865 1.00116.71 H C
+ATOM 1605 CD GLN H 13 -53.075 16.587 31.559 1.00116.71 H C
+ATOM 1606 NE2 GLN H 13 -53.040 16.403 32.870 1.00116.71 H N
+ATOM 1607 OE1 GLN H 13 -54.122 16.619 30.923 1.00116.71 H O
+ATOM 1608 N PRO H 14 -48.809 16.737 28.777 1.00114.35 H N
+ATOM 1609 CA PRO H 14 -48.871 17.390 27.466 1.00114.35 H C
+ATOM 1610 C PRO H 14 -50.280 17.464 26.903 1.00114.35 H C
+ATOM 1611 O PRO H 14 -50.454 17.461 25.681 1.00114.35 H O
+ATOM 1612 CB PRO H 14 -47.975 16.503 26.591 1.00114.35 H C
+ATOM 1613 CG PRO H 14 -47.994 15.199 27.263 1.00114.35 H C
+ATOM 1614 CD PRO H 14 -47.987 15.522 28.715 1.00114.35 H C
+ATOM 1615 N GLY H 15 -51.290 17.450 27.771 1.00114.55 H N
+ATOM 1616 CA GLY H 15 -52.681 17.521 27.358 1.00114.55 H C
+ATOM 1617 C GLY H 15 -52.976 18.543 26.275 1.00114.55 H C
+ATOM 1618 O GLY H 15 -54.010 18.462 25.611 1.00114.55 H O
+ATOM 1619 N GLY H 16 -52.098 19.531 26.123 1.00111.87 H N
+ATOM 1620 CA GLY H 16 -51.995 20.316 24.902 1.00111.87 H C
+ATOM 1621 C GLY H 16 -50.693 19.979 24.190 1.00111.87 H C
+ATOM 1622 O GLY H 16 -49.634 19.927 24.815 1.00111.87 H O
+ATOM 1623 N SER H 17 -50.793 19.756 22.877 1.00106.77 H N
+ATOM 1624 CA SER H 17 -49.670 19.226 22.107 1.00106.77 H C
+ATOM 1625 C SER H 17 -48.439 20.119 22.223 1.00106.77 H C
+ATOM 1626 O SER H 17 -48.526 21.341 22.074 1.00106.77 H O
+ATOM 1627 CB SER H 17 -50.079 19.068 20.645 1.00106.77 H C
+ATOM 1628 OG SER H 17 -51.105 18.100 20.512 1.00106.77 H O
+ATOM 1629 N LEU H 18 -47.288 19.503 22.477 1.00102.33 H N
+ATOM 1630 CA LEU H 18 -46.116 20.236 22.945 1.00102.33 H C
+ATOM 1631 C LEU H 18 -44.851 19.432 22.639 1.00102.33 H C
+ATOM 1632 O LEU H 18 -44.864 18.528 21.796 1.00102.33 H O
+ATOM 1633 CB LEU H 18 -46.257 20.571 24.439 1.00102.33 H C
+ATOM 1634 CG LEU H 18 -47.006 21.860 24.790 1.00102.33 H C
+ATOM 1635 CD1 LEU H 18 -47.225 21.969 26.289 1.00102.33 H C
+ATOM 1636 CD2 LEU H 18 -46.267 23.078 24.263 1.00102.33 H C
+ATOM 1637 N ARG H 19 -43.758 19.779 23.318 1.00 95.33 H N
+ATOM 1638 CA ARG H 19 -42.401 19.392 22.962 1.00 95.33 H C
+ATOM 1639 C ARG H 19 -41.654 18.899 24.197 1.00 95.33 H C
+ATOM 1640 O ARG H 19 -42.018 19.213 25.332 1.00 95.33 H O
+ATOM 1641 CB ARG H 19 -41.677 20.596 22.333 1.00 95.33 H C
+ATOM 1642 CG ARG H 19 -40.307 20.371 21.725 1.00 95.33 H C
+ATOM 1643 CD ARG H 19 -39.252 20.594 22.786 1.00 95.33 H C
+ATOM 1644 NE ARG H 19 -39.289 21.954 23.308 1.00 95.33 H N
+ATOM 1645 CZ ARG H 19 -38.706 22.326 24.443 1.00 95.33 H C
+ATOM 1646 NH1 ARG H 19 -38.058 21.434 25.179 1.00 95.33 H N1+
+ATOM 1647 NH2 ARG H 19 -38.784 23.584 24.849 1.00 95.33 H N
+ATOM 1648 N LEU H 20 -40.591 18.128 23.962 1.00 90.19 H N
+ATOM 1649 CA LEU H 20 -39.717 17.632 25.020 1.00 90.19 H C
+ATOM 1650 C LEU H 20 -38.273 17.666 24.539 1.00 90.19 H C
+ATOM 1651 O LEU H 20 -38.002 17.661 23.342 1.00 90.19 H O
+ATOM 1652 CB LEU H 20 -40.100 16.206 25.434 1.00 90.19 H C
+ATOM 1653 CG LEU H 20 -39.445 15.619 26.681 1.00 90.19 H C
+ATOM 1654 CD1 LEU H 20 -39.895 16.360 27.921 1.00 90.19 H C
+ATOM 1655 CD2 LEU H 20 -39.775 14.144 26.788 1.00 90.19 H C
+ATOM 1656 N SER H 21 -37.333 17.689 25.476 1.00 84.80 H N
+ATOM 1657 CA SER H 21 -35.945 17.843 25.075 1.00 84.80 H C
+ATOM 1658 C SER H 21 -35.032 17.112 26.039 1.00 84.80 H C
+ATOM 1659 O SER H 21 -35.352 16.948 27.216 1.00 84.80 H O
+ATOM 1660 CB SER H 21 -35.534 19.314 25.031 1.00 84.80 H C
+ATOM 1661 OG SER H 21 -35.572 19.884 26.327 1.00 84.80 H O
+ATOM 1662 N CYS H 22 -33.880 16.694 25.528 1.00 78.27 H N
+ATOM 1663 CA CYS H 22 -32.800 16.183 26.351 1.00 78.27 H C
+ATOM 1664 C CYS H 22 -31.511 16.837 25.869 1.00 78.27 H C
+ATOM 1665 O CYS H 22 -31.506 17.563 24.879 1.00 78.27 H O
+ATOM 1666 CB CYS H 22 -32.796 14.639 26.349 1.00 78.27 H C
+ATOM 1667 SG CYS H 22 -32.035 13.642 25.006 1.00 78.27 H S
+ATOM 1668 N ALA H 23 -30.417 16.638 26.594 1.00 71.14 H N
+ATOM 1669 CA ALA H 23 -29.225 17.439 26.336 1.00 71.14 H C
+ATOM 1670 C ALA H 23 -27.989 16.714 26.841 1.00 71.14 H C
+ATOM 1671 O ALA H 23 -27.977 16.220 27.966 1.00 71.14 H O
+ATOM 1672 CB ALA H 23 -29.337 18.812 27.000 1.00 71.14 H C
+ATOM 1673 N ALA H 24 -26.949 16.696 26.021 1.00 68.39 H N
+ATOM 1674 CA ALA H 24 -25.770 15.878 26.252 1.00 68.39 H C
+ATOM 1675 C ALA H 24 -24.815 16.577 27.204 1.00 68.39 H C
+ATOM 1676 O ALA H 24 -25.166 17.558 27.863 1.00 68.39 H O
+ATOM 1677 CB ALA H 24 -25.059 15.573 24.939 1.00 68.39 H C
+ATOM 1678 N SER H 25 -23.599 16.060 27.280 1.00 70.21 H N
+ATOM 1679 CA SER H 25 -22.546 16.607 28.113 1.00 70.21 H C
+ATOM 1680 C SER H 25 -21.240 15.999 27.651 1.00 70.21 H C
+ATOM 1681 O SER H 25 -21.188 15.373 26.590 1.00 70.21 H O
+ATOM 1682 CB SER H 25 -22.766 16.291 29.588 1.00 70.21 H C
+ATOM 1683 OG SER H 25 -22.402 14.952 29.855 1.00 70.21 H O
+ATOM 1684 N GLY H 26 -20.189 16.197 28.435 1.00 75.42 H N
+ATOM 1685 CA GLY H 26 -18.939 15.512 28.169 1.00 75.42 H C
+ATOM 1686 C GLY H 26 -18.476 15.808 26.767 1.00 75.42 H C
+ATOM 1687 O GLY H 26 -18.540 16.947 26.295 1.00 75.42 H O
+ATOM 1688 N PHE H 27 -18.039 14.772 26.071 1.00 78.09 H N
+ATOM 1689 CA PHE H 27 -17.557 14.966 24.718 1.00 78.09 H C
+ATOM 1690 C PHE H 27 -18.711 15.400 23.836 1.00 78.09 H C
+ATOM 1691 O PHE H 27 -19.875 15.149 24.151 1.00 78.09 H O
+ATOM 1692 CB PHE H 27 -16.923 13.686 24.191 1.00 78.09 H C
+ATOM 1693 CG PHE H 27 -15.653 13.322 24.885 1.00 78.09 H C
+ATOM 1694 CD1 PHE H 27 -14.991 14.246 25.684 1.00 78.09 H C
+ATOM 1695 CD2 PHE H 27 -15.101 12.079 24.724 1.00 78.09 H C
+ATOM 1696 CE1 PHE H 27 -13.803 13.919 26.325 1.00 78.09 H C
+ATOM 1697 CE2 PHE H 27 -13.922 11.738 25.365 1.00 78.09 H C
+ATOM 1698 CZ PHE H 27 -13.271 12.659 26.168 1.00 78.09 H C
+ATOM 1699 N ASN H 28 -18.397 16.096 22.753 1.00 77.75 H N
+ATOM 1700 CA ASN H 28 -19.386 16.364 21.721 1.00 77.75 H C
+ATOM 1701 C ASN H 28 -19.333 15.214 20.733 1.00 77.75 H C
+ATOM 1702 O ASN H 28 -18.291 14.962 20.119 1.00 77.75 H O
+ATOM 1703 CB ASN H 28 -19.124 17.679 21.004 1.00 77.75 H C
+ATOM 1704 CG ASN H 28 -20.307 18.123 20.181 1.00 77.75 H C
+ATOM 1705 ND2 ASN H 28 -20.036 18.802 19.074 1.00 77.75 H N
+ATOM 1706 OD1 ASN H 28 -21.458 17.839 20.526 1.00 77.75 H O
+ATOM 1707 N LEU H 29 -20.444 14.493 20.603 1.00 74.97 H N
+ATOM 1708 CA LEU H 29 -20.519 13.450 19.590 1.00 74.97 H C
+ATOM 1709 C LEU H 29 -20.592 14.054 18.196 1.00 74.97 H C
+ATOM 1710 O LEU H 29 -19.734 13.790 17.350 1.00 74.97 H O
+ATOM 1711 CB LEU H 29 -21.713 12.551 19.861 1.00 74.97 H C
+ATOM 1712 CG LEU H 29 -21.597 11.409 20.866 1.00 74.97 H C
+ATOM 1713 CD1 LEU H 29 -20.627 10.375 20.433 1.00 74.97 H C
+ATOM 1714 CD2 LEU H 29 -21.139 11.966 22.157 1.00 74.97 H C
+ATOM 1715 N SER H 30 -21.620 14.855 17.928 1.00 74.87 H N
+ATOM 1716 CA SER H 30 -21.714 15.686 16.738 1.00 74.87 H C
+ATOM 1717 C SER H 30 -21.653 14.897 15.454 1.00 74.87 H C
+ATOM 1718 O SER H 30 -21.679 15.497 14.388 1.00 74.87 H O
+ATOM 1719 CB SER H 30 -20.603 16.740 16.674 1.00 74.87 H C
+ATOM 1720 OG SER H 30 -20.936 17.805 15.797 1.00 74.87 H O
+ATOM 1721 N TYR H 31 -21.444 13.594 15.508 1.00 72.39 H N
+ATOM 1722 CA TYR H 31 -21.450 12.871 14.254 1.00 72.39 H C
+ATOM 1723 C TYR H 31 -22.427 11.711 14.328 1.00 72.39 H C
+ATOM 1724 O TYR H 31 -23.338 11.595 13.508 1.00 72.39 H O
+ATOM 1725 CB TYR H 31 -20.027 12.443 13.942 1.00 72.39 H C
+ATOM 1726 CG TYR H 31 -19.823 11.735 12.649 1.00 72.39 H C
+ATOM 1727 CD1 TYR H 31 -19.840 12.445 11.454 1.00 72.39 H C
+ATOM 1728 CD2 TYR H 31 -19.455 10.418 12.624 1.00 72.39 H C
+ATOM 1729 CE1 TYR H 31 -19.610 11.840 10.272 1.00 72.39 H C
+ATOM 1730 CE2 TYR H 31 -19.203 9.806 11.456 1.00 72.39 H C
+ATOM 1731 CZ TYR H 31 -19.283 10.523 10.280 1.00 72.39 H C
+ATOM 1732 OH TYR H 31 -19.044 9.891 9.089 1.00 72.39 H O
+ATOM 1733 N SER H 32 -22.234 10.853 15.321 1.00 78.73 H N
+ATOM 1734 CA SER H 32 -23.177 9.809 15.690 1.00 78.73 H C
+ATOM 1735 C SER H 32 -23.538 9.981 17.158 1.00 78.73 H C
+ATOM 1736 O SER H 32 -22.656 10.083 18.008 1.00 78.73 H O
+ATOM 1737 CB SER H 32 -22.581 8.423 15.450 1.00 78.73 H C
+ATOM 1738 OG SER H 32 -21.483 8.193 16.312 1.00 78.73 H O
+ATOM 1739 N ASP H 33 -24.828 10.068 17.445 1.00 73.64 H N
+ATOM 1740 CA ASP H 33 -25.248 10.592 18.731 1.00 73.64 H C
+ATOM 1741 C ASP H 33 -26.719 10.290 18.902 1.00 73.64 H C
+ATOM 1742 O ASP H 33 -27.181 9.240 18.469 1.00 73.64 H O
+ATOM 1743 CB ASP H 33 -24.929 12.073 18.838 1.00 73.64 H C
+ATOM 1744 CG ASP H 33 -25.205 12.658 20.205 1.00 73.64 H C
+ATOM 1745 OD1 ASP H 33 -24.445 12.414 21.155 1.00 73.64 H O
+ATOM 1746 OD2 ASP H 33 -26.212 13.372 20.317 1.00 73.64 H O1-
+ATOM 1747 N ILE H 34 -27.426 11.197 19.573 1.00 70.98 H N
+ATOM 1748 CA ILE H 34 -28.741 10.960 20.140 1.00 70.98 H C
+ATOM 1749 C ILE H 34 -29.613 10.106 19.240 1.00 70.98 H C
+ATOM 1750 O ILE H 34 -29.529 10.160 18.010 1.00 70.98 H O
+ATOM 1751 CB ILE H 34 -29.415 12.312 20.408 1.00 70.98 H C
+ATOM 1752 CG1 ILE H 34 -30.386 12.202 21.560 1.00 70.98 H C
+ATOM 1753 CG2 ILE H 34 -30.200 12.768 19.219 1.00 70.98 H C
+ATOM 1754 CD1 ILE H 34 -29.695 11.886 22.807 1.00 70.98 H C
+ATOM 1755 N HIS H 35 -30.448 9.291 19.868 1.00 69.82 H N
+ATOM 1756 CA HIS H 35 -31.241 8.297 19.176 1.00 69.82 H C
+ATOM 1757 C HIS H 35 -32.696 8.303 19.551 1.00 69.82 H C
+ATOM 1758 O HIS H 35 -33.505 7.808 18.769 1.00 69.82 H O
+ATOM 1759 CB HIS H 35 -30.700 6.902 19.462 1.00 69.82 H C
+ATOM 1760 CG HIS H 35 -29.473 6.559 18.693 1.00 69.82 H C
+ATOM 1761 CD2 HIS H 35 -28.860 7.186 17.665 1.00 69.82 H C
+ATOM 1762 ND1 HIS H 35 -28.765 5.401 18.914 1.00 69.82 H N
+ATOM 1763 CE1 HIS H 35 -27.744 5.348 18.081 1.00 69.82 H C
+ATOM 1764 NE2 HIS H 35 -27.777 6.420 17.313 1.00 69.82 H N
+ATOM 1765 N TRP H 36 -33.059 8.828 20.705 1.00 72.02 H N
+ATOM 1766 CA TRP H 36 -34.441 8.821 21.149 1.00 72.02 H C
+ATOM 1767 C TRP H 36 -35.195 7.529 20.878 1.00 72.02 H C
+ATOM 1768 O TRP H 36 -36.198 7.540 20.170 1.00 72.02 H O
+ATOM 1769 CB TRP H 36 -35.180 9.988 20.530 1.00 72.02 H C
+ATOM 1770 CG TRP H 36 -34.614 11.278 20.962 1.00 72.02 H C
+ATOM 1771 CD1 TRP H 36 -33.613 11.960 20.371 1.00 72.02 H C
+ATOM 1772 CD2 TRP H 36 -35.002 12.044 22.112 1.00 72.02 H C
+ATOM 1773 CE2 TRP H 36 -34.212 13.191 22.125 1.00 72.02 H C
+ATOM 1774 CE3 TRP H 36 -35.969 11.884 23.104 1.00 72.02 H C
+ATOM 1775 NE1 TRP H 36 -33.368 13.113 21.054 1.00 72.02 H N
+ATOM 1776 CZ2 TRP H 36 -34.346 14.173 23.091 1.00 72.02 H C
+ATOM 1777 CZ3 TRP H 36 -36.088 12.855 24.075 1.00 72.02 H C
+ATOM 1778 CH2 TRP H 36 -35.279 13.979 24.066 1.00 72.02 H C
+ATOM 1779 N VAL H 37 -34.705 6.416 21.422 1.00 61.94 H N
+ATOM 1780 CA VAL H 37 -35.480 5.184 21.476 1.00 61.94 H C
+ATOM 1781 C VAL H 37 -36.630 5.389 22.449 1.00 61.94 H C
+ATOM 1782 O VAL H 37 -36.460 6.027 23.489 1.00 61.94 H O
+ATOM 1783 CB VAL H 37 -34.581 4.021 21.910 1.00 61.94 H C
+ATOM 1784 CG1 VAL H 37 -35.376 2.791 21.973 1.00 61.94 H C
+ATOM 1785 CG2 VAL H 37 -33.409 3.845 20.970 1.00 61.94 H C
+ATOM 1786 N ARG H 38 -37.822 4.920 22.101 1.00 72.49 H N
+ATOM 1787 CA ARG H 38 -38.926 5.023 23.045 1.00 72.49 H C
+ATOM 1788 C ARG H 38 -39.425 3.652 23.484 1.00 72.49 H C
+ATOM 1789 O ARG H 38 -39.309 2.663 22.759 1.00 72.49 H O
+ATOM 1790 CB ARG H 38 -40.093 5.808 22.472 1.00 72.49 H C
+ATOM 1791 CG ARG H 38 -40.763 5.158 21.309 1.00 72.49 H C
+ATOM 1792 CD ARG H 38 -41.821 6.086 20.768 1.00 72.49 H C
+ATOM 1793 NE ARG H 38 -42.979 6.204 21.636 1.00 72.49 H N
+ATOM 1794 CZ ARG H 38 -44.028 5.403 21.556 1.00 72.49 H C
+ATOM 1795 NH1 ARG H 38 -44.053 4.456 20.638 1.00 72.49 H N1+
+ATOM 1796 NH2 ARG H 38 -45.060 5.566 22.364 1.00 72.49 H N
+ATOM 1797 N GLN H 39 -39.995 3.606 24.689 1.00 74.07 H N
+ATOM 1798 CA GLN H 39 -40.540 2.380 25.250 1.00 74.07 H C
+ATOM 1799 C GLN H 39 -41.876 2.638 25.913 1.00 74.07 H C
+ATOM 1800 O GLN H 39 -42.202 3.757 26.305 1.00 74.07 H O
+ATOM 1801 CB GLN H 39 -39.645 1.743 26.303 1.00 74.07 H C
+ATOM 1802 CG GLN H 39 -38.331 1.230 25.836 1.00 74.07 H C
+ATOM 1803 CD GLN H 39 -37.890 0.073 26.699 1.00 74.07 H C
+ATOM 1804 NE2 GLN H 39 -38.590 -0.134 27.811 1.00 74.07 H N
+ATOM 1805 OE1 GLN H 39 -36.948 -0.638 26.372 1.00 74.07 H O
+ATOM 1806 N ALA H 40 -42.625 1.561 26.052 1.00 75.03 H N
+ATOM 1807 CA ALA H 40 -43.824 1.453 26.864 1.00 75.03 H C
+ATOM 1808 C ALA H 40 -43.518 0.360 27.883 1.00 75.03 H C
+ATOM 1809 O ALA H 40 -42.455 -0.266 27.786 1.00 75.03 H O
+ATOM 1810 CB ALA H 40 -45.034 1.134 25.985 1.00 75.03 H C
+ATOM 1811 N PRO H 41 -44.384 0.080 28.865 1.00 77.29 H N
+ATOM 1812 CA PRO H 41 -43.984 -0.873 29.911 1.00 77.29 H C
+ATOM 1813 C PRO H 41 -43.819 -2.301 29.410 1.00 77.29 H C
+ATOM 1814 O PRO H 41 -44.727 -3.127 29.530 1.00 77.29 H O
+ATOM 1815 CB PRO H 41 -45.120 -0.765 30.934 1.00 77.29 H C
+ATOM 1816 CG PRO H 41 -45.695 0.572 30.709 1.00 77.29 H C
+ATOM 1817 CD PRO H 41 -45.614 0.797 29.242 1.00 77.29 H C
+ATOM 1818 N GLY H 42 -42.649 -2.589 28.837 1.00 73.24 H N
+ATOM 1819 CA GLY H 42 -42.300 -3.933 28.412 1.00 73.24 H C
+ATOM 1820 C GLY H 42 -42.069 -4.104 26.924 1.00 73.24 H C
+ATOM 1821 O GLY H 42 -42.327 -5.176 26.376 1.00 73.24 H O
+ATOM 1822 N LYS H 43 -41.561 -3.070 26.267 1.00 71.88 H N
+ATOM 1823 CA LYS H 43 -41.400 -3.037 24.816 1.00 71.88 H C
+ATOM 1824 C LYS H 43 -40.152 -2.213 24.508 1.00 71.88 H C
+ATOM 1825 O LYS H 43 -39.256 -2.095 25.347 1.00 71.88 H O
+ATOM 1826 CB LYS H 43 -42.653 -2.462 24.129 1.00 71.88 H C
+ATOM 1827 CG LYS H 43 -43.804 -3.436 23.863 1.00 71.88 H C
+ATOM 1828 CD LYS H 43 -44.666 -3.712 25.086 1.00 71.88 H C
+ATOM 1829 CE LYS H 43 -45.454 -2.486 25.496 1.00 71.88 H C
+ATOM 1830 NZ LYS H 43 -46.327 -2.753 26.670 1.00 71.88 H N1+
+ATOM 1831 N GLY H 44 -40.061 -1.696 23.281 1.00 66.60 H N
+ATOM 1832 CA GLY H 44 -39.140 -0.613 22.967 1.00 66.60 H C
+ATOM 1833 C GLY H 44 -39.057 -0.255 21.494 1.00 66.60 H C
+ATOM 1834 O GLY H 44 -38.878 -1.132 20.645 1.00 66.60 H O
+ATOM 1835 N LEU H 45 -39.124 1.040 21.184 1.00 68.51 H N
+ATOM 1836 CA LEU H 45 -39.339 1.506 19.822 1.00 68.51 H C
+ATOM 1837 C LEU H 45 -38.398 2.661 19.517 1.00 68.51 H C
+ATOM 1838 O LEU H 45 -38.134 3.506 20.375 1.00 68.51 H O
+ATOM 1839 CB LEU H 45 -40.784 1.963 19.627 1.00 68.51 H C
+ATOM 1840 CG LEU H 45 -41.865 0.892 19.731 1.00 68.51 H C
+ATOM 1841 CD1 LEU H 45 -43.232 1.522 19.671 1.00 68.51 H C
+ATOM 1842 CD2 LEU H 45 -41.713 -0.134 18.630 1.00 68.51 H C
+ATOM 1843 N GLU H 46 -37.917 2.709 18.277 1.00 65.81 H N
+ATOM 1844 CA GLU H 46 -36.969 3.727 17.825 1.00 65.81 H C
+ATOM 1845 C GLU H 46 -37.716 4.702 16.925 1.00 65.81 H C
+ATOM 1846 O GLU H 46 -37.759 4.535 15.708 1.00 65.81 H O
+ATOM 1847 CB GLU H 46 -35.816 3.100 17.095 1.00 65.81 H C
+ATOM 1848 CG GLU H 46 -34.831 4.091 16.582 1.00 65.81 H C
+ATOM 1849 CD GLU H 46 -33.710 3.422 15.844 1.00 65.81 H C
+ATOM 1850 OE1 GLU H 46 -33.987 2.443 15.141 1.00 65.81 H O
+ATOM 1851 OE2 GLU H 46 -32.549 3.856 15.962 1.00 65.81 H O1-
+ATOM 1852 N TRP H 47 -38.313 5.721 17.531 1.00 77.01 H N
+ATOM 1853 CA TRP H 47 -39.077 6.715 16.791 1.00 77.01 H C
+ATOM 1854 C TRP H 47 -38.182 7.797 16.217 1.00 77.01 H C
+ATOM 1855 O TRP H 47 -36.973 7.595 16.074 1.00 77.01 H O
+ATOM 1856 CB TRP H 47 -40.177 7.329 17.657 1.00 77.01 H C
+ATOM 1857 CG TRP H 47 -39.709 8.058 18.836 1.00 77.01 H C
+ATOM 1858 CD1 TRP H 47 -38.902 7.587 19.802 1.00 77.01 H C
+ATOM 1859 CD2 TRP H 47 -40.056 9.393 19.214 1.00 77.01 H C
+ATOM 1860 CE2 TRP H 47 -39.398 9.666 20.422 1.00 77.01 H C
+ATOM 1861 CE3 TRP H 47 -40.848 10.390 18.642 1.00 77.01 H C
+ATOM 1862 NE1 TRP H 47 -38.696 8.542 20.764 1.00 77.01 H N
+ATOM 1863 CZ2 TRP H 47 -39.510 10.893 21.075 1.00 77.01 H C
+ATOM 1864 CZ3 TRP H 47 -40.962 11.609 19.289 1.00 77.01 H C
+ATOM 1865 CH2 TRP H 47 -40.292 11.852 20.490 1.00 77.01 H C
+ATOM 1866 N VAL H 48 -38.778 8.933 15.850 1.00 80.58 H N
+ATOM 1867 CA VAL H 48 -38.069 10.029 15.197 1.00 80.58 H C
+ATOM 1868 C VAL H 48 -36.857 10.406 16.030 1.00 80.58 H C
+ATOM 1869 O VAL H 48 -36.785 10.082 17.219 1.00 80.58 H O
+ATOM 1870 CB VAL H 48 -39.013 11.234 14.981 1.00 80.58 H C
+ATOM 1871 CG1 VAL H 48 -38.304 12.450 14.420 1.00 80.58 H C
+ATOM 1872 CG2 VAL H 48 -40.046 10.858 13.976 1.00 80.58 H C
+ATOM 1873 N ALA H 49 -35.889 11.054 15.392 1.00 79.37 H N
+ATOM 1874 CA ALA H 49 -34.618 11.400 16.001 1.00 79.37 H C
+ATOM 1875 C ALA H 49 -33.727 10.178 16.160 1.00 79.37 H C
+ATOM 1876 O ALA H 49 -33.092 9.984 17.191 1.00 79.37 H O
+ATOM 1877 CB ALA H 49 -34.809 12.122 17.334 1.00 79.37 H C
+ATOM 1878 N VAL H 50 -33.672 9.357 15.124 1.00 74.57 H N
+ATOM 1879 CA VAL H 50 -32.512 8.520 14.874 1.00 74.57 H C
+ATOM 1880 C VAL H 50 -31.444 9.491 14.398 1.00 74.57 H C
+ATOM 1881 O VAL H 50 -31.755 10.665 14.188 1.00 74.57 H O
+ATOM 1882 CB VAL H 50 -32.835 7.437 13.852 1.00 74.57 H C
+ATOM 1883 CG1 VAL H 50 -31.677 6.484 13.705 1.00 74.57 H C
+ATOM 1884 CG2 VAL H 50 -34.078 6.749 14.294 1.00 74.57 H C
+ATOM 1885 N VAL H 51 -30.202 9.033 14.187 1.00 75.31 H N
+ATOM 1886 CA VAL H 51 -29.016 9.849 14.442 1.00 75.31 H C
+ATOM 1887 C VAL H 51 -29.259 11.290 14.042 1.00 75.31 H C
+ATOM 1888 O VAL H 51 -29.265 11.632 12.860 1.00 75.31 H O
+ATOM 1889 CB VAL H 51 -27.818 9.286 13.661 1.00 75.31 H C
+ATOM 1890 CG1 VAL H 51 -26.579 10.109 13.898 1.00 75.31 H C
+ATOM 1891 CG2 VAL H 51 -27.577 7.839 14.053 1.00 75.31 H C
+ATOM 1892 N MET H 52 -29.438 12.135 15.048 1.00 81.52 H N
+ATOM 1893 CA MET H 52 -29.858 13.511 14.855 1.00 81.52 H C
+ATOM 1894 C MET H 52 -28.783 14.596 14.774 1.00 81.52 H C
+ATOM 1895 O MET H 52 -28.921 15.504 13.945 1.00 81.52 H O
+ATOM 1896 CB MET H 52 -30.881 13.882 15.913 1.00 81.52 H C
+ATOM 1897 CG MET H 52 -31.691 15.054 15.458 1.00 81.52 H C
+ATOM 1898 SD MET H 52 -33.318 15.209 16.169 1.00 81.52 H S
+ATOM 1899 CE MET H 52 -34.266 14.605 14.792 1.00 81.52 H C
+ATOM 1900 N PRO H 53 -27.728 14.576 15.604 1.00 80.77 H N
+ATOM 1901 CA PRO H 53 -26.899 15.794 15.749 1.00 80.77 H C
+ATOM 1902 C PRO H 53 -26.313 16.270 14.443 1.00 80.77 H C
+ATOM 1903 O PRO H 53 -26.279 17.472 14.153 1.00 80.77 H O
+ATOM 1904 CB PRO H 53 -25.768 15.336 16.681 1.00 80.77 H C
+ATOM 1905 CG PRO H 53 -25.702 13.927 16.438 1.00 80.77 H C
+ATOM 1906 CD PRO H 53 -27.112 13.467 16.335 1.00 80.77 H C
+ATOM 1907 N ASP H 54 -25.844 15.321 13.656 1.00 81.14 H N
+ATOM 1908 CA ASP H 54 -25.174 15.500 12.380 1.00 81.14 H C
+ATOM 1909 C ASP H 54 -25.038 14.076 11.863 1.00 81.14 H C
+ATOM 1910 O ASP H 54 -25.337 13.124 12.590 1.00 81.14 H O
+ATOM 1911 CB ASP H 54 -23.849 16.241 12.565 1.00 81.14 H C
+ATOM 1912 CG ASP H 54 -23.105 16.456 11.283 1.00 81.14 H C
+ATOM 1913 OD1 ASP H 54 -22.468 15.507 10.787 1.00 81.14 H O
+ATOM 1914 OD2 ASP H 54 -23.136 17.596 10.786 1.00 81.14 H O1-
+ATOM 1915 N SER H 55 -24.589 13.927 10.620 1.00 75.11 H N
+ATOM 1916 CA SER H 55 -24.696 12.698 9.832 1.00 75.11 H C
+ATOM 1917 C SER H 55 -26.116 12.526 9.307 1.00 75.11 H C
+ATOM 1918 O SER H 55 -26.476 11.425 8.887 1.00 75.11 H O
+ATOM 1919 CB SER H 55 -24.271 11.428 10.580 1.00 75.11 H C
+ATOM 1920 OG SER H 55 -24.459 10.268 9.788 1.00 75.11 H O
+ATOM 1921 N GLY H 56 -26.943 13.564 9.373 1.00 78.40 H N
+ATOM 1922 CA GLY H 56 -28.165 13.745 8.600 1.00 78.40 H C
+ATOM 1923 C GLY H 56 -29.153 12.595 8.679 1.00 78.40 H C
+ATOM 1924 O GLY H 56 -29.053 11.699 9.523 1.00 78.40 H O
+ATOM 1925 N HIS H 57 -30.117 12.626 7.752 1.00 76.71 H N
+ATOM 1926 CA HIS H 57 -31.252 11.708 7.689 1.00 76.71 H C
+ATOM 1927 C HIS H 57 -31.716 11.353 9.089 1.00 76.71 H C
+ATOM 1928 O HIS H 57 -31.685 10.183 9.479 1.00 76.71 H O
+ATOM 1929 CB HIS H 57 -30.967 10.423 6.896 1.00 76.71 H C
+ATOM 1930 CG HIS H 57 -29.605 9.836 7.096 1.00 76.71 H C
+ATOM 1931 CD2 HIS H 57 -28.500 9.859 6.313 1.00 76.71 H C
+ATOM 1932 ND1 HIS H 57 -29.298 9.022 8.164 1.00 76.71 H N
+ATOM 1933 CE1 HIS H 57 -28.046 8.616 8.062 1.00 76.71 H C
+ATOM 1934 NE2 HIS H 57 -27.544 9.101 6.943 1.00 76.71 H N
+ATOM 1935 N THR H 58 -32.148 12.343 9.855 1.00 80.84 H N
+ATOM 1936 CA THR H 58 -32.311 12.129 11.282 1.00 80.84 H C
+ATOM 1937 C THR H 58 -33.528 11.264 11.552 1.00 80.84 H C
+ATOM 1938 O THR H 58 -34.120 11.358 12.628 1.00 80.84 H O
+ATOM 1939 CB THR H 58 -32.437 13.443 12.046 1.00 80.84 H C
+ATOM 1940 CG2 THR H 58 -31.234 14.319 11.771 1.00 80.84 H C
+ATOM 1941 OG1 THR H 58 -33.597 14.135 11.601 1.00 80.84 H O
+ATOM 1942 N ASN H 59 -33.953 10.482 10.566 1.00 81.56 H N
+ATOM 1943 CA ASN H 59 -34.915 9.408 10.780 1.00 81.56 H C
+ATOM 1944 C ASN H 59 -34.511 8.130 10.057 1.00 81.56 H C
+ATOM 1945 O ASN H 59 -35.309 7.535 9.332 1.00 81.56 H O
+ATOM 1946 CB ASN H 59 -36.315 9.807 10.345 1.00 81.56 H C
+ATOM 1947 CG ASN H 59 -36.890 10.884 11.188 1.00 81.56 H C
+ATOM 1948 ND2 ASN H 59 -37.786 11.665 10.611 1.00 81.56 H N
+ATOM 1949 OD1 ASN H 59 -36.618 10.957 12.371 1.00 81.56 H O
+ATOM 1950 N TYR H 60 -33.270 7.685 10.220 1.00 77.15 H N
+ATOM 1951 CA TYR H 60 -32.956 6.331 9.783 1.00 77.15 H C
+ATOM 1952 C TYR H 60 -33.844 5.346 10.534 1.00 77.15 H C
+ATOM 1953 O TYR H 60 -33.662 5.136 11.734 1.00 77.15 H O
+ATOM 1954 CB TYR H 60 -31.482 6.012 10.011 1.00 77.15 H C
+ATOM 1955 CG TYR H 60 -30.971 4.785 9.265 1.00 77.15 H C
+ATOM 1956 CD1 TYR H 60 -31.353 3.510 9.656 1.00 77.15 H C
+ATOM 1957 CD2 TYR H 60 -30.074 4.894 8.211 1.00 77.15 H C
+ATOM 1958 CE1 TYR H 60 -30.893 2.386 9.005 1.00 77.15 H C
+ATOM 1959 CE2 TYR H 60 -29.602 3.762 7.554 1.00 77.15 H C
+ATOM 1960 CZ TYR H 60 -30.023 2.511 7.962 1.00 77.15 H C
+ATOM 1961 OH TYR H 60 -29.576 1.371 7.343 1.00 77.15 H O
+ATOM 1962 N ALA H 61 -34.791 4.733 9.816 1.00 78.47 H N
+ATOM 1963 CA ALA H 61 -35.812 3.842 10.389 1.00 78.47 H C
+ATOM 1964 C ALA H 61 -36.842 4.609 11.221 1.00 78.47 H C
+ATOM 1965 O ALA H 61 -37.142 4.238 12.355 1.00 78.47 H O
+ATOM 1966 CB ALA H 61 -35.188 2.711 11.211 1.00 78.47 H C
+ATOM 1967 N ASP H 62 -37.392 5.681 10.648 1.00 77.61 H N
+ATOM 1968 CA ASP H 62 -38.502 6.385 11.281 1.00 77.61 H C
+ATOM 1969 C ASP H 62 -39.726 5.494 11.437 1.00 77.61 H C
+ATOM 1970 O ASP H 62 -40.111 5.127 12.552 1.00 77.61 H O
+ATOM 1971 CB ASP H 62 -38.893 7.610 10.463 1.00 77.61 H C
+ATOM 1972 CG ASP H 62 -39.432 8.713 11.321 1.00 77.61 H C
+ATOM 1973 OD1 ASP H 62 -40.110 8.364 12.299 1.00 77.61 H O
+ATOM 1974 OD2 ASP H 62 -39.328 9.899 10.963 1.00 77.61 H O1-
+ATOM 1975 N SER H 63 -40.375 5.173 10.317 1.00 76.33 H N
+ATOM 1976 CA SER H 63 -41.650 4.460 10.302 1.00 76.33 H C
+ATOM 1977 C SER H 63 -42.721 5.163 11.141 1.00 76.33 H C
+ATOM 1978 O SER H 63 -43.621 4.519 11.681 1.00 76.33 H O
+ATOM 1979 CB SER H 63 -41.473 3.012 10.753 1.00 76.33 H C
+ATOM 1980 OG SER H 63 -41.334 2.929 12.159 1.00 76.33 H O
+ATOM 1981 N VAL H 64 -42.639 6.488 11.253 1.00 79.93 H N
+ATOM 1982 CA VAL H 64 -43.746 7.296 11.759 1.00 79.93 H C
+ATOM 1983 C VAL H 64 -43.475 8.738 11.355 1.00 79.93 H C
+ATOM 1984 O VAL H 64 -42.327 9.128 11.141 1.00 79.93 H O
+ATOM 1985 CB VAL H 64 -43.916 7.134 13.290 1.00 79.93 H C
+ATOM 1986 CG1 VAL H 64 -42.654 7.542 14.005 1.00 79.93 H C
+ATOM 1987 CG2 VAL H 64 -45.122 7.906 13.787 1.00 79.93 H C
+ATOM 1988 N LYS H 65 -44.535 9.528 11.220 1.00 92.66 H N
+ATOM 1989 CA LYS H 65 -44.395 10.874 10.675 1.00 92.66 H C
+ATOM 1990 C LYS H 65 -45.609 11.698 11.096 1.00 92.66 H C
+ATOM 1991 O LYS H 65 -46.573 11.174 11.660 1.00 92.66 H O
+ATOM 1992 CB LYS H 65 -44.220 10.814 9.149 1.00 92.66 H C
+ATOM 1993 CG LYS H 65 -44.022 12.147 8.429 1.00 92.66 H C
+ATOM 1994 CD LYS H 65 -42.802 12.903 8.940 1.00 92.66 H C
+ATOM 1995 CE LYS H 65 -41.500 12.211 8.559 1.00 92.66 H C
+ATOM 1996 NZ LYS H 65 -40.303 12.980 9.008 1.00 92.66 H N1+
+ATOM 1997 N GLY H 66 -45.552 12.998 10.811 1.00 93.61 H N
+ATOM 1998 CA GLY H 66 -46.633 13.910 11.123 1.00 93.61 H C
+ATOM 1999 C GLY H 66 -46.461 14.637 12.438 1.00 93.61 H C
+ATOM 2000 O GLY H 66 -46.373 15.868 12.473 1.00 93.61 H O
+ATOM 2001 N ARG H 67 -46.428 13.885 13.532 1.00 95.28 H N
+ATOM 2002 CA ARG H 67 -46.388 14.466 14.861 1.00 95.28 H C
+ATOM 2003 C ARG H 67 -45.075 14.253 15.594 1.00 95.28 H C
+ATOM 2004 O ARG H 67 -44.739 15.066 16.461 1.00 95.28 H O
+ATOM 2005 CB ARG H 67 -47.571 13.920 15.673 1.00 95.28 H C
+ATOM 2006 CG ARG H 67 -47.365 12.569 16.323 1.00 95.28 H C
+ATOM 2007 CD ARG H 67 -48.709 12.058 16.837 1.00 95.28 H C
+ATOM 2008 NE ARG H 67 -48.622 11.202 18.016 1.00 95.28 H N
+ATOM 2009 CZ ARG H 67 -47.956 10.054 18.073 1.00 95.28 H C
+ATOM 2010 NH1 ARG H 67 -47.325 9.586 17.005 1.00 95.28 H N1+
+ATOM 2011 NH2 ARG H 67 -47.946 9.357 19.199 1.00 95.28 H N
+ATOM 2012 N PHE H 68 -44.309 13.226 15.245 1.00 89.86 H N
+ATOM 2013 CA PHE H 68 -42.973 13.030 15.795 1.00 89.86 H C
+ATOM 2014 C PHE H 68 -41.964 13.760 14.918 1.00 89.86 H C
+ATOM 2015 O PHE H 68 -41.809 13.432 13.738 1.00 89.86 H O
+ATOM 2016 CB PHE H 68 -42.638 11.545 15.894 1.00 89.86 H C
+ATOM 2017 CG PHE H 68 -43.403 10.814 16.945 1.00 89.86 H C
+ATOM 2018 CD1 PHE H 68 -44.090 11.503 17.925 1.00 89.86 H C
+ATOM 2019 CD2 PHE H 68 -43.413 9.435 16.968 1.00 89.86 H C
+ATOM 2020 CE1 PHE H 68 -44.778 10.830 18.901 1.00 89.86 H C
+ATOM 2021 CE2 PHE H 68 -44.100 8.760 17.941 1.00 89.86 H C
+ATOM 2022 CZ PHE H 68 -44.786 9.457 18.909 1.00 89.86 H C
+ATOM 2023 N THR H 69 -41.257 14.722 15.501 1.00 92.03 H N
+ATOM 2024 CA THR H 69 -40.683 15.821 14.741 1.00 92.03 H C
+ATOM 2025 C THR H 69 -39.170 15.877 14.885 1.00 92.03 H C
+ATOM 2026 O THR H 69 -38.641 15.827 15.998 1.00 92.03 H O
+ATOM 2027 CB THR H 69 -41.300 17.152 15.186 1.00 92.03 H C
+ATOM 2028 CG2 THR H 69 -40.654 18.314 14.459 1.00 92.03 H C
+ATOM 2029 OG1 THR H 69 -42.705 17.143 14.908 1.00 92.03 H O
+ATOM 2030 N ILE H 70 -38.490 16.000 13.745 1.00 92.23 H N
+ATOM 2031 CA ILE H 70 -37.070 16.321 13.717 1.00 92.23 H C
+ATOM 2032 C ILE H 70 -36.872 17.745 14.215 1.00 92.23 H C
+ATOM 2033 O ILE H 70 -37.339 18.705 13.588 1.00 92.23 H O
+ATOM 2034 CB ILE H 70 -36.532 16.174 12.288 1.00 92.23 H C
+ATOM 2035 CG1 ILE H 70 -36.633 14.724 11.812 1.00 92.23 H C
+ATOM 2036 CG2 ILE H 70 -35.125 16.765 12.173 1.00 92.23 H C
+ATOM 2037 CD1 ILE H 70 -36.369 14.567 10.330 1.00 92.23 H C
+ATOM 2038 N SER H 71 -36.165 17.900 15.338 1.00 92.31 H N
+ATOM 2039 CA SER H 71 -36.058 19.217 15.963 1.00 92.31 H C
+ATOM 2040 C SER H 71 -34.633 19.757 16.011 1.00 92.31 H C
+ATOM 2041 O SER H 71 -34.393 20.851 15.495 1.00 92.31 H O
+ATOM 2042 CB SER H 71 -36.678 19.169 17.356 1.00 92.31 H C
+ATOM 2043 OG SER H 71 -38.090 19.077 17.284 1.00 92.31 H O
+ATOM 2044 N ALA H 72 -33.676 19.046 16.605 1.00 87.74 H N
+ATOM 2045 CA ALA H 72 -32.412 19.707 16.911 1.00 87.74 H C
+ATOM 2046 C ALA H 72 -31.203 18.969 16.367 1.00 87.74 H C
+ATOM 2047 O ALA H 72 -31.348 18.030 15.585 1.00 87.74 H O
+ATOM 2048 CB ALA H 72 -32.263 19.898 18.413 1.00 87.74 H C
+ATOM 2049 N ASP H 73 -30.008 19.390 16.768 1.00 84.90 H N
+ATOM 2050 CA ASP H 73 -28.781 18.920 16.140 1.00 84.90 H C
+ATOM 2051 C ASP H 73 -27.641 19.068 17.135 1.00 84.90 H C
+ATOM 2052 O ASP H 73 -27.862 19.243 18.336 1.00 84.90 H O
+ATOM 2053 CB ASP H 73 -28.498 19.692 14.850 1.00 84.90 H C
+ATOM 2054 CG ASP H 73 -28.214 21.156 15.104 1.00 84.90 H C
+ATOM 2055 OD1 ASP H 73 -28.774 21.705 16.076 1.00 84.90 H O
+ATOM 2056 OD2 ASP H 73 -27.435 21.756 14.334 1.00 84.90 H O1-
+ATOM 2057 N THR H 74 -26.412 19.027 16.628 1.00 82.52 H N
+ATOM 2058 CA THR H 74 -25.235 19.002 17.482 1.00 82.52 H C
+ATOM 2059 C THR H 74 -24.810 20.351 18.049 1.00 82.52 H C
+ATOM 2060 O THR H 74 -23.640 20.734 17.945 1.00 82.52 H O
+ATOM 2061 CB THR H 74 -24.059 18.405 16.725 1.00 82.52 H C
+ATOM 2062 CG2 THR H 74 -23.814 19.134 15.419 1.00 82.52 H C
+ATOM 2063 OG1 THR H 74 -22.901 18.464 17.560 1.00 82.52 H O
+ATOM 2064 N SER H 75 -25.719 21.043 18.721 1.00 81.95 H N
+ATOM 2065 CA SER H 75 -25.377 22.276 19.412 1.00 81.95 H C
+ATOM 2066 C SER H 75 -24.825 22.034 20.811 1.00 81.95 H C
+ATOM 2067 O SER H 75 -24.825 22.965 21.623 1.00 81.95 H O
+ATOM 2068 CB SER H 75 -26.596 23.198 19.475 1.00 81.95 H C
+ATOM 2069 OG SER H 75 -27.615 22.643 20.275 1.00 81.95 H O
+ATOM 2070 N LYS H 76 -24.361 20.813 21.102 1.00 80.85 H N
+ATOM 2071 CA LYS H 76 -23.967 20.252 22.396 1.00 80.85 H C
+ATOM 2072 C LYS H 76 -25.182 19.852 23.216 1.00 80.85 H C
+ATOM 2073 O LYS H 76 -25.023 19.228 24.260 1.00 80.85 H O
+ATOM 2074 CB LYS H 76 -23.098 21.203 23.242 1.00 80.85 H C
+ATOM 2075 CG LYS H 76 -21.759 21.624 22.664 1.00 80.85 H C
+ATOM 2076 CD LYS H 76 -20.760 20.483 22.602 1.00 80.85 H C
+ATOM 2077 CE LYS H 76 -19.429 20.961 22.012 1.00 80.85 H C
+ATOM 2078 NZ LYS H 76 -18.716 21.912 22.909 1.00 80.85 H N1+
+ATOM 2079 N ASN H 77 -26.381 20.191 22.779 1.00 80.77 H N
+ATOM 2080 CA ASN H 77 -27.628 19.763 23.399 1.00 80.77 H C
+ATOM 2081 C ASN H 77 -28.491 19.123 22.313 1.00 80.77 H C
+ATOM 2082 O ASN H 77 -29.178 19.795 21.544 1.00 80.77 H O
+ATOM 2083 CB ASN H 77 -28.268 20.920 24.167 1.00 80.77 H C
+ATOM 2084 CG ASN H 77 -28.651 22.100 23.295 1.00 80.77 H C
+ATOM 2085 ND2 ASN H 77 -29.077 23.179 23.941 1.00 80.77 H N
+ATOM 2086 OD1 ASN H 77 -28.561 22.063 22.075 1.00 80.77 H O
+ATOM 2087 N THR H 78 -28.458 17.797 22.280 1.00 73.25 H N
+ATOM 2088 CA THR H 78 -28.430 17.109 21.002 1.00 73.25 H C
+ATOM 2089 C THR H 78 -29.815 16.791 20.452 1.00 73.25 H C
+ATOM 2090 O THR H 78 -29.910 16.222 19.362 1.00 73.25 H O
+ATOM 2091 CB THR H 78 -27.576 15.875 21.124 1.00 73.25 H C
+ATOM 2092 CG2 THR H 78 -28.112 15.044 22.206 1.00 73.25 H C
+ATOM 2093 OG1 THR H 78 -27.591 15.158 19.891 1.00 73.25 H O
+ATOM 2094 N ALA H 79 -30.894 17.081 21.184 1.00 79.84 H N
+ATOM 2095 CA ALA H 79 -32.190 17.200 20.511 1.00 79.84 H C
+ATOM 2096 C ALA H 79 -33.232 17.829 21.425 1.00 79.84 H C
+ATOM 2097 O ALA H 79 -33.057 17.890 22.640 1.00 79.84 H O
+ATOM 2098 CB ALA H 79 -32.704 15.860 19.988 1.00 79.84 H C
+ATOM 2099 N TYR H 80 -34.323 18.336 20.809 1.00 89.16 H N
+ATOM 2100 CA TYR H 80 -35.496 18.835 21.544 1.00 89.16 H C
+ATOM 2101 C TYR H 80 -36.777 18.305 20.883 1.00 89.16 H C
+ATOM 2102 O TYR H 80 -37.419 19.049 20.148 1.00 89.16 H O
+ATOM 2103 CB TYR H 80 -35.547 20.360 21.532 1.00 89.16 H C
+ATOM 2104 CG TYR H 80 -34.227 21.082 21.571 1.00 89.16 H C
+ATOM 2105 CD1 TYR H 80 -33.304 20.882 22.586 1.00 89.16 H C
+ATOM 2106 CD2 TYR H 80 -33.916 21.993 20.573 1.00 89.16 H C
+ATOM 2107 CE1 TYR H 80 -32.099 21.565 22.592 1.00 89.16 H C
+ATOM 2108 CE2 TYR H 80 -32.729 22.672 20.568 1.00 89.16 H C
+ATOM 2109 CZ TYR H 80 -31.825 22.460 21.578 1.00 89.16 H C
+ATOM 2110 OH TYR H 80 -30.644 23.161 21.545 1.00 89.16 H O
+ATOM 2111 N LEU H 81 -37.221 17.093 21.201 1.00 84.29 H N
+ATOM 2112 CA LEU H 81 -38.287 16.520 20.381 1.00 84.29 H C
+ATOM 2113 C LEU H 81 -39.681 17.011 20.751 1.00 84.29 H C
+ATOM 2114 O LEU H 81 -39.985 17.307 21.905 1.00 84.29 H O
+ATOM 2115 CB LEU H 81 -38.287 15.008 20.435 1.00 84.29 H C
+ATOM 2116 CG LEU H 81 -37.041 14.440 19.803 1.00 84.29 H C
+ATOM 2117 CD1 LEU H 81 -37.095 12.984 20.011 1.00 84.29 H C
+ATOM 2118 CD2 LEU H 81 -36.983 14.811 18.344 1.00 84.29 H C
+ATOM 2119 N GLN H 82 -40.555 17.034 19.753 1.00 95.59 H N
+ATOM 2120 CA GLN H 82 -41.889 17.598 19.881 1.00 95.59 H C
+ATOM 2121 C GLN H 82 -42.948 16.534 19.633 1.00 95.59 H C
+ATOM 2122 O GLN H 82 -42.783 15.678 18.759 1.00 95.59 H O
+ATOM 2123 CB GLN H 82 -42.089 18.745 18.910 1.00 95.59 H C
+ATOM 2124 CG GLN H 82 -43.225 19.638 19.303 1.00 95.59 H C
+ATOM 2125 CD GLN H 82 -44.489 19.189 18.641 1.00 95.59 H C
+ATOM 2126 NE2 GLN H 82 -45.628 19.632 19.156 1.00 95.59 H N
+ATOM 2127 OE1 GLN H 82 -44.439 18.442 17.669 1.00 95.59 H O
+ATOM 2128 N MET H 83 -44.037 16.598 20.402 1.00100.45 H N
+ATOM 2129 CA MET H 83 -45.038 15.534 20.382 1.00100.45 H C
+ATOM 2130 C MET H 83 -46.113 15.758 19.318 1.00100.45 H C
+ATOM 2131 O MET H 83 -46.420 14.846 18.544 1.00100.45 H O
+ATOM 2132 CB MET H 83 -45.674 15.416 21.766 1.00100.45 H C
+ATOM 2133 CG MET H 83 -46.627 14.244 21.952 1.00100.45 H C
+ATOM 2134 SD MET H 83 -45.779 12.659 21.851 1.00100.45 H S
+ATOM 2135 CE MET H 83 -47.131 11.582 21.393 1.00100.45 H C
+ATOM 2136 N ASN H 84 -46.718 16.948 19.286 1.00101.68 H N
+ATOM 2137 CA ASN H 84 -47.942 17.201 18.510 1.00101.68 H C
+ATOM 2138 C ASN H 84 -49.027 16.167 18.787 1.00101.68 H C
+ATOM 2139 O ASN H 84 -49.761 15.769 17.881 1.00101.68 H O
+ATOM 2140 CB ASN H 84 -47.676 17.259 17.005 1.00101.68 H C
+ATOM 2141 CG ASN H 84 -46.910 18.475 16.598 1.00101.68 H C
+ATOM 2142 ND2 ASN H 84 -45.935 18.289 15.723 1.00101.68 H N
+ATOM 2143 OD1 ASN H 84 -47.167 19.576 17.082 1.00101.68 H O
+ATOM 2144 N SER H 85 -49.150 15.720 20.032 1.00106.83 H N
+ATOM 2145 CA SER H 85 -50.206 14.775 20.359 1.00106.83 H C
+ATOM 2146 C SER H 85 -50.514 14.872 21.839 1.00106.83 H C
+ATOM 2147 O SER H 85 -49.614 15.070 22.657 1.00106.83 H O
+ATOM 2148 CB SER H 85 -49.817 13.341 20.000 1.00106.83 H C
+ATOM 2149 OG SER H 85 -50.775 12.417 20.482 1.00106.83 H O
+ATOM 2150 N LEU H 86 -51.791 14.731 22.171 1.00113.60 H N
+ATOM 2151 CA LEU H 86 -52.232 14.852 23.545 1.00113.60 H C
+ATOM 2152 C LEU H 86 -53.193 13.754 23.972 1.00113.60 H C
+ATOM 2153 O LEU H 86 -53.607 13.741 25.136 1.00113.60 H O
+ATOM 2154 CB LEU H 86 -52.872 16.231 23.757 1.00113.60 H C
+ATOM 2155 CG LEU H 86 -54.116 16.570 22.931 1.00113.60 H C
+ATOM 2156 CD1 LEU H 86 -55.399 16.209 23.670 1.00113.60 H C
+ATOM 2157 CD2 LEU H 86 -54.112 18.031 22.510 1.00113.60 H C
+ATOM 2158 N ARG H 87 -53.569 12.851 23.077 1.00114.12 H N
+ATOM 2159 CA ARG H 87 -54.482 11.763 23.422 1.00114.12 H C
+ATOM 2160 C ARG H 87 -54.020 10.391 22.951 1.00114.12 H C
+ATOM 2161 O ARG H 87 -54.205 9.408 23.672 1.00114.12 H O
+ATOM 2162 CB ARG H 87 -55.901 12.069 22.875 1.00114.12 H C
+ATOM 2163 CG ARG H 87 -56.084 11.929 21.363 1.00114.12 H C
+ATOM 2164 CD ARG H 87 -55.484 13.125 20.633 1.00114.12 H C
+ATOM 2165 NE ARG H 87 -55.823 13.155 19.215 1.00114.12 H N
+ATOM 2166 CZ ARG H 87 -55.244 13.963 18.332 1.00114.12 H C
+ATOM 2167 NH1 ARG H 87 -54.304 14.810 18.726 1.00114.12 H N1+
+ATOM 2168 NH2 ARG H 87 -55.610 13.929 17.058 1.00114.12 H N
+ATOM 2169 N ALA H 88 -53.410 10.293 21.778 1.00107.21 H N
+ATOM 2170 CA ALA H 88 -53.103 9.007 21.175 1.00107.21 H C
+ATOM 2171 C ALA H 88 -51.619 8.720 21.313 1.00107.21 H C
+ATOM 2172 O ALA H 88 -50.787 9.541 20.915 1.00107.21 H O
+ATOM 2173 CB ALA H 88 -53.515 8.983 19.703 1.00107.21 H C
+ATOM 2174 N GLU H 89 -51.298 7.554 21.871 1.00100.97 H N
+ATOM 2175 CA GLU H 89 -49.920 7.121 22.072 1.00100.97 H C
+ATOM 2176 C GLU H 89 -49.091 8.223 22.730 1.00100.97 H C
+ATOM 2177 O GLU H 89 -48.031 8.625 22.245 1.00100.97 H O
+ATOM 2178 CB GLU H 89 -49.299 6.671 20.753 1.00100.97 H C
+ATOM 2179 CG GLU H 89 -47.986 5.936 20.911 1.00100.97 H C
+ATOM 2180 CD GLU H 89 -47.086 6.125 19.720 1.00100.97 H C
+ATOM 2181 OE1 GLU H 89 -47.459 5.679 18.617 1.00100.97 H O
+ATOM 2182 OE2 GLU H 89 -46.014 6.740 19.882 1.00100.97 H O1-
+ATOM 2183 N ASP H 90 -49.603 8.732 23.840 1.00109.39 H N
+ATOM 2184 CA ASP H 90 -48.979 9.815 24.585 1.00109.39 H C
+ATOM 2185 C ASP H 90 -48.537 9.338 25.957 1.00109.39 H C
+ATOM 2186 O ASP H 90 -48.705 10.031 26.960 1.00109.39 H O
+ATOM 2187 CB ASP H 90 -49.933 10.998 24.706 1.00109.39 H C
+ATOM 2188 CG ASP H 90 -51.221 10.640 25.424 1.00109.39 H C
+ATOM 2189 OD1 ASP H 90 -51.437 9.445 25.714 1.00109.39 H O
+ATOM 2190 OD2 ASP H 90 -52.021 11.556 25.700 1.00109.39 H O1-
+ATOM 2191 N THR H 91 -47.966 8.139 26.014 1.00102.28 H N
+ATOM 2192 CA THR H 91 -47.808 7.478 27.300 1.00102.28 H C
+ATOM 2193 C THR H 91 -46.446 6.792 27.414 1.00102.28 H C
+ATOM 2194 O THR H 91 -46.190 6.091 28.398 1.00102.28 H O
+ATOM 2195 CB THR H 91 -48.990 6.493 27.482 1.00102.28 H C
+ATOM 2196 CG2 THR H 91 -49.025 5.858 28.870 1.00102.28 H C
+ATOM 2197 OG1 THR H 91 -50.221 7.208 27.314 1.00102.28 H O
+ATOM 2198 N ALA H 92 -45.538 7.021 26.476 1.00 82.44 H N
+ATOM 2199 CA ALA H 92 -44.283 6.296 26.492 1.00 82.44 H C
+ATOM 2200 C ALA H 92 -43.310 6.924 27.481 1.00 82.44 H C
+ATOM 2201 O ALA H 92 -43.615 7.901 28.161 1.00 82.44 H O
+ATOM 2202 CB ALA H 92 -43.674 6.259 25.096 1.00 82.44 H C
+ATOM 2203 N VAL H 93 -42.125 6.340 27.575 1.00 76.03 H N
+ATOM 2204 CA VAL H 93 -41.010 6.937 28.291 1.00 76.03 H C
+ATOM 2205 C VAL H 93 -39.880 7.071 27.284 1.00 76.03 H C
+ATOM 2206 O VAL H 93 -39.109 6.137 27.047 1.00 76.03 H O
+ATOM 2207 CB VAL H 93 -40.592 6.135 29.525 1.00 76.03 H C
+ATOM 2208 CG1 VAL H 93 -41.595 6.341 30.627 1.00 76.03 H C
+ATOM 2209 CG2 VAL H 93 -40.499 4.667 29.208 1.00 76.03 H C
+ATOM 2210 N TYR H 94 -39.785 8.247 26.688 1.00 74.79 H N
+ATOM 2211 CA TYR H 94 -38.836 8.497 25.621 1.00 74.79 H C
+ATOM 2212 C TYR H 94 -37.427 8.525 26.174 1.00 74.79 H C
+ATOM 2213 O TYR H 94 -37.207 8.896 27.325 1.00 74.79 H O
+ATOM 2214 CB TYR H 94 -39.200 9.813 24.968 1.00 74.79 H C
+ATOM 2215 CG TYR H 94 -40.618 9.781 24.489 1.00 74.79 H C
+ATOM 2216 CD1 TYR H 94 -40.976 8.997 23.416 1.00 74.79 H C
+ATOM 2217 CD2 TYR H 94 -41.617 10.440 25.182 1.00 74.79 H C
+ATOM 2218 CE1 TYR H 94 -42.271 8.936 22.987 1.00 74.79 H C
+ATOM 2219 CE2 TYR H 94 -42.921 10.380 24.768 1.00 74.79 H C
+ATOM 2220 CZ TYR H 94 -43.245 9.623 23.668 1.00 74.79 H C
+ATOM 2221 OH TYR H 94 -44.548 9.549 23.235 1.00 74.79 H O
+ATOM 2222 N TYR H 95 -36.469 8.124 25.356 1.00 70.10 H N
+ATOM 2223 CA TYR H 95 -35.113 7.979 25.856 1.00 70.10 H C
+ATOM 2224 C TYR H 95 -34.087 8.632 24.942 1.00 70.10 H C
+ATOM 2225 O TYR H 95 -34.458 9.371 24.033 1.00 70.10 H O
+ATOM 2226 CB TYR H 95 -34.834 6.508 26.077 1.00 70.10 H C
+ATOM 2227 CG TYR H 95 -35.496 6.003 27.331 1.00 70.10 H C
+ATOM 2228 CD1 TYR H 95 -35.509 6.776 28.469 1.00 70.10 H C
+ATOM 2229 CD2 TYR H 95 -36.114 4.767 27.385 1.00 70.10 H C
+ATOM 2230 CE1 TYR H 95 -36.100 6.341 29.618 1.00 70.10 H C
+ATOM 2231 CE2 TYR H 95 -36.711 4.327 28.538 1.00 70.10 H C
+ATOM 2232 CZ TYR H 95 -36.702 5.125 29.649 1.00 70.10 H C
+ATOM 2233 OH TYR H 95 -37.294 4.712 30.814 1.00 70.10 H O
+ATOM 2234 N CYS H 96 -32.801 8.415 25.202 1.00 65.85 H N
+ATOM 2235 CA CYS H 96 -31.738 9.162 24.529 1.00 65.85 H C
+ATOM 2236 C CYS H 96 -30.473 8.325 24.628 1.00 65.85 H C
+ATOM 2237 O CYS H 96 -29.940 8.171 25.722 1.00 65.85 H O
+ATOM 2238 CB CYS H 96 -31.535 10.533 25.190 1.00 65.85 H C
+ATOM 2239 SG CYS H 96 -32.897 11.780 25.098 1.00 65.85 H S
+ATOM 2240 N SER H 97 -29.990 7.775 23.521 1.00 61.70 H N
+ATOM 2241 CA SER H 97 -28.764 6.982 23.587 1.00 61.70 H C
+ATOM 2242 C SER H 97 -27.553 7.906 23.496 1.00 61.70 H C
+ATOM 2243 O SER H 97 -26.744 7.858 22.569 1.00 61.70 H O
+ATOM 2244 CB SER H 97 -28.731 5.908 22.511 1.00 61.70 H C
+ATOM 2245 OG SER H 97 -29.932 5.172 22.476 1.00 61.70 H O
+ATOM 2246 N ARG H 98 -27.394 8.706 24.542 1.00 71.86 H N
+ATOM 2247 CA ARG H 98 -26.398 9.778 24.581 1.00 71.86 H C
+ATOM 2248 C ARG H 98 -25.001 9.294 24.853 1.00 71.86 H C
+ATOM 2249 O ARG H 98 -24.284 9.881 25.664 1.00 71.86 H O
+ATOM 2250 CB ARG H 98 -26.769 10.769 25.672 1.00 71.86 H C
+ATOM 2251 CG ARG H 98 -26.205 12.120 25.455 1.00 71.86 H C
+ATOM 2252 CD ARG H 98 -27.270 12.948 24.940 1.00 71.86 H C
+ATOM 2253 NE ARG H 98 -28.140 13.101 26.079 1.00 71.86 H N
+ATOM 2254 CZ ARG H 98 -29.260 13.783 26.087 1.00 71.86 H C
+ATOM 2255 NH1 ARG H 98 -29.977 13.826 27.195 1.00 71.86 H N1+
+ATOM 2256 NH2 ARG H 98 -29.658 14.427 25.005 1.00 71.86 H N
+ATOM 2257 N ALA H 99 -24.589 8.264 24.226 1.00 69.71 H N
+ATOM 2258 CA ALA H 99 -23.167 8.000 24.176 1.00 69.71 H C
+ATOM 2259 C ALA H 99 -22.799 7.355 22.868 1.00 69.71 H C
+ATOM 2260 O ALA H 99 -21.660 6.917 22.728 1.00 69.71 H O
+ATOM 2261 CB ALA H 99 -22.719 7.129 25.344 1.00 69.71 H C
+ATOM 2262 N THR H 100 -23.730 7.243 21.928 1.00 69.76 H N
+ATOM 2263 CA THR H 100 -23.521 6.380 20.788 1.00 69.76 H C
+ATOM 2264 C THR H 100 -22.383 6.914 19.944 1.00 69.76 H C
+ATOM 2265 O THR H 100 -22.520 7.894 19.222 1.00 69.76 H O
+ATOM 2266 CB THR H 100 -24.785 6.279 19.973 1.00 69.76 H C
+ATOM 2267 CG2 THR H 100 -24.703 5.035 19.111 1.00 69.76 H C
+ATOM 2268 OG1 THR H 100 -25.889 6.160 20.872 1.00 69.76 H O
+ATOM 2269 N ASN H 101 -21.262 6.239 20.036 1.00 67.24 H N
+ATOM 2270 CA ASN H 101 -19.970 6.722 19.609 1.00 67.24 H C
+ATOM 2271 C ASN H 101 -19.098 5.677 18.951 1.00 67.24 H C
+ATOM 2272 O ASN H 101 -18.257 6.032 18.124 1.00 67.24 H O
+ATOM 2273 CB ASN H 101 -19.257 7.311 20.821 1.00 67.24 H C
+ATOM 2274 CG ASN H 101 -19.083 6.296 21.916 1.00 67.24 H C
+ATOM 2275 ND2 ASN H 101 -19.295 6.735 23.144 1.00 67.24 H N
+ATOM 2276 OD1 ASN H 101 -18.829 5.122 21.675 1.00 67.24 H O
+ATOM 2277 N ILE H 102 -19.252 4.414 19.318 1.00 63.05 H N
+ATOM 2278 CA ILE H 102 -18.543 3.301 18.700 1.00 63.05 H C
+ATOM 2279 C ILE H 102 -19.476 2.111 18.643 1.00 63.05 H C
+ATOM 2280 O ILE H 102 -20.394 1.995 19.468 1.00 63.05 H O
+ATOM 2281 CB ILE H 102 -17.268 2.958 19.481 1.00 63.05 H C
+ATOM 2282 CG1 ILE H 102 -16.265 4.123 19.451 1.00 63.05 H C
+ATOM 2283 CG2 ILE H 102 -16.657 1.633 19.055 1.00 63.05 H C
+ATOM 2284 CD1 ILE H 102 -16.293 5.199 20.487 1.00 63.05 H C
+ATOM 2285 N PRO H 103 -19.297 1.218 17.659 1.00 63.03 H N
+ATOM 2286 CA PRO H 103 -20.070 -0.030 17.695 1.00 63.03 H C
+ATOM 2287 C PRO H 103 -19.935 -0.754 19.029 1.00 63.03 H C
+ATOM 2288 O PRO H 103 -20.932 -1.275 19.549 1.00 63.03 H O
+ATOM 2289 CB PRO H 103 -19.472 -0.855 16.540 1.00 63.03 H C
+ATOM 2290 CG PRO H 103 -18.304 -0.092 16.068 1.00 63.03 H C
+ATOM 2291 CD PRO H 103 -18.552 1.323 16.403 1.00 63.03 H C
+ATOM 2292 N VAL H 104 -18.729 -0.795 19.609 1.00 61.74 H N
+ATOM 2293 CA VAL H 104 -18.526 -1.455 20.891 1.00 61.74 H C
+ATOM 2294 C VAL H 104 -17.751 -0.554 21.858 1.00 61.74 H C
+ATOM 2295 O VAL H 104 -16.529 -0.661 21.986 1.00 61.74 H O
+ATOM 2296 CB VAL H 104 -17.825 -2.808 20.645 1.00 61.74 H C
+ATOM 2297 CG1 VAL H 104 -17.683 -3.587 21.872 1.00 61.74 H C
+ATOM 2298 CG2 VAL H 104 -18.618 -3.661 19.679 1.00 61.74 H C
+ATOM 2299 N TYR H 105 -18.468 0.342 22.538 1.00 63.38 H N
+ATOM 2300 CA TYR H 105 -18.142 0.959 23.828 1.00 63.38 H C
+ATOM 2301 C TYR H 105 -19.440 1.305 24.527 1.00 63.38 H C
+ATOM 2302 O TYR H 105 -20.508 1.299 23.923 1.00 63.38 H O
+ATOM 2303 CB TYR H 105 -17.248 2.217 23.776 1.00 63.38 H C
+ATOM 2304 CG TYR H 105 -15.798 2.054 23.364 1.00 63.38 H C
+ATOM 2305 CD1 TYR H 105 -14.851 1.638 24.265 1.00 63.38 H C
+ATOM 2306 CD2 TYR H 105 -15.378 2.308 22.102 1.00 63.38 H C
+ATOM 2307 CE1 TYR H 105 -13.527 1.498 23.903 1.00 63.38 H C
+ATOM 2308 CE2 TYR H 105 -14.069 2.159 21.727 1.00 63.38 H C
+ATOM 2309 CZ TYR H 105 -13.149 1.756 22.628 1.00 63.38 H C
+ATOM 2310 OH TYR H 105 -11.838 1.616 22.241 1.00 63.38 H O
+ATOM 2311 N ALA H 106 -19.337 1.607 25.816 1.00 53.92 H N
+ATOM 2312 CA ALA H 106 -20.523 1.687 26.649 1.00 53.92 H C
+ATOM 2313 C ALA H 106 -21.502 2.684 26.066 1.00 53.92 H C
+ATOM 2314 O ALA H 106 -21.130 3.809 25.741 1.00 53.92 H O
+ATOM 2315 CB ALA H 106 -20.138 2.084 28.070 1.00 53.92 H C
+ATOM 2316 N PHE H 107 -22.750 2.279 25.915 1.00 58.60 H N
+ATOM 2317 CA PHE H 107 -23.769 3.163 25.375 1.00 58.60 H C
+ATOM 2318 C PHE H 107 -24.481 3.763 26.560 1.00 58.60 H C
+ATOM 2319 O PHE H 107 -25.578 3.329 26.909 1.00 58.60 H O
+ATOM 2320 CB PHE H 107 -24.767 2.428 24.494 1.00 58.60 H C
+ATOM 2321 CG PHE H 107 -24.188 1.924 23.233 1.00 58.60 H C
+ATOM 2322 CD1 PHE H 107 -22.971 2.374 22.790 1.00 58.60 H C
+ATOM 2323 CD2 PHE H 107 -24.852 0.987 22.491 1.00 58.60 H C
+ATOM 2324 CE1 PHE H 107 -22.427 1.894 21.620 1.00 58.60 H C
+ATOM 2325 CE2 PHE H 107 -24.317 0.501 21.321 1.00 58.60 H C
+ATOM 2326 CZ PHE H 107 -23.103 0.954 20.884 1.00 58.60 H C
+ATOM 2327 N ASP H 108 -23.895 4.794 27.137 1.00 67.51 H N
+ATOM 2328 CA ASP H 108 -24.464 5.362 28.348 1.00 67.51 H C
+ATOM 2329 C ASP H 108 -25.772 5.993 27.937 1.00 67.51 H C
+ATOM 2330 O ASP H 108 -25.842 7.176 27.634 1.00 67.51 H O
+ATOM 2331 CB ASP H 108 -23.548 6.388 28.985 1.00 67.51 H C
+ATOM 2332 CG ASP H 108 -24.083 6.868 30.311 1.00 67.51 H C
+ATOM 2333 OD1 ASP H 108 -24.002 6.111 31.301 1.00 67.51 H O
+ATOM 2334 OD2 ASP H 108 -24.659 7.970 30.349 1.00 67.51 H O1-
+ATOM 2335 N TYR H 109 -26.819 5.181 27.931 1.00 65.20 H N
+ATOM 2336 CA TYR H 109 -28.107 5.667 27.480 1.00 65.20 H C
+ATOM 2337 C TYR H 109 -28.569 6.890 28.234 1.00 65.20 H C
+ATOM 2338 O TYR H 109 -28.294 8.021 27.836 1.00 65.20 H O
+ATOM 2339 CB TYR H 109 -29.167 4.582 27.605 1.00 65.20 H C
+ATOM 2340 CG TYR H 109 -29.061 3.432 26.663 1.00 65.20 H C
+ATOM 2341 CD1 TYR H 109 -28.243 3.491 25.555 1.00 65.20 H C
+ATOM 2342 CD2 TYR H 109 -29.911 2.373 26.775 1.00 65.20 H C
+ATOM 2343 CE1 TYR H 109 -28.202 2.473 24.659 1.00 65.20 H C
+ATOM 2344 CE2 TYR H 109 -29.883 1.363 25.884 1.00 65.20 H C
+ATOM 2345 CZ TYR H 109 -29.029 1.406 24.827 1.00 65.20 H C
+ATOM 2346 OH TYR H 109 -29.007 0.366 23.926 1.00 65.20 H O
+ATOM 2347 N TRP H 110 -29.256 6.695 29.332 1.00 69.72 H N
+ATOM 2348 CA TRP H 110 -30.229 7.708 29.676 1.00 69.72 H C
+ATOM 2349 C TRP H 110 -30.770 7.421 31.055 1.00 69.72 H C
+ATOM 2350 O TRP H 110 -30.399 6.422 31.678 1.00 69.72 H O
+ATOM 2351 CB TRP H 110 -31.367 7.695 28.662 1.00 69.72 H C
+ATOM 2352 CG TRP H 110 -32.070 6.330 28.553 1.00 69.72 H C
+ATOM 2353 CD1 TRP H 110 -32.747 5.680 29.551 1.00 69.72 H C
+ATOM 2354 CD2 TRP H 110 -32.159 5.473 27.398 1.00 69.72 H C
+ATOM 2355 CE2 TRP H 110 -32.897 4.338 27.776 1.00 69.72 H C
+ATOM 2356 CE3 TRP H 110 -31.698 5.560 26.083 1.00 69.72 H C
+ATOM 2357 NE1 TRP H 110 -33.254 4.496 29.088 1.00 69.72 H N
+ATOM 2358 CZ2 TRP H 110 -33.176 3.302 26.892 1.00 69.72 H C
+ATOM 2359 CZ3 TRP H 110 -31.962 4.516 25.215 1.00 69.72 H C
+ATOM 2360 CH2 TRP H 110 -32.699 3.411 25.621 1.00 69.72 H C
+ATOM 2361 N GLY H 111 -31.644 8.287 31.543 1.00 78.03 H N
+ATOM 2362 CA GLY H 111 -32.282 7.980 32.798 1.00 78.03 H C
+ATOM 2363 C GLY H 111 -33.726 8.392 32.907 1.00 78.03 H C
+ATOM 2364 O GLY H 111 -34.293 8.269 33.994 1.00 78.03 H O
+ATOM 2365 N GLN H 112 -34.356 8.850 31.830 1.00 78.79 H N
+ATOM 2366 CA GLN H 112 -35.632 9.515 32.050 1.00 78.79 H C
+ATOM 2367 C GLN H 112 -36.286 9.803 30.705 1.00 78.79 H C
+ATOM 2368 O GLN H 112 -35.711 9.561 29.642 1.00 78.79 H O
+ATOM 2369 CB GLN H 112 -35.372 10.770 32.886 1.00 78.79 H C
+ATOM 2370 CG GLN H 112 -36.535 11.578 33.367 1.00 78.79 H C
+ATOM 2371 CD GLN H 112 -36.043 12.708 34.226 1.00 78.79 H C
+ATOM 2372 NE2 GLN H 112 -36.953 13.370 34.903 1.00 78.79 H N
+ATOM 2373 OE1 GLN H 112 -34.838 12.889 34.387 1.00 78.79 H O
+ATOM 2374 N GLY H 113 -37.504 10.325 30.771 1.00 78.11 H N
+ATOM 2375 CA GLY H 113 -38.345 10.615 29.633 1.00 78.11 H C
+ATOM 2376 C GLY H 113 -39.718 10.159 30.053 1.00 78.11 H C
+ATOM 2377 O GLY H 113 -39.831 9.098 30.667 1.00 78.11 H O
+ATOM 2378 N THR H 114 -40.767 10.923 29.776 1.00 91.77 H N
+ATOM 2379 CA THR H 114 -42.048 10.617 30.389 1.00 91.77 H C
+ATOM 2380 C THR H 114 -43.178 10.880 29.408 1.00 91.77 H C
+ATOM 2381 O THR H 114 -42.984 10.924 28.190 1.00 91.77 H O
+ATOM 2382 CB THR H 114 -42.267 11.423 31.671 1.00 91.77 H C
+ATOM 2383 CG2 THR H 114 -41.234 11.085 32.735 1.00 91.77 H C
+ATOM 2384 OG1 THR H 114 -42.238 12.821 31.363 1.00 91.77 H O
+ATOM 2385 N LEU H 115 -44.362 11.081 29.966 1.00103.43 H N
+ATOM 2386 CA LEU H 115 -45.621 10.819 29.295 1.00103.43 H C
+ATOM 2387 C LEU H 115 -46.606 11.889 29.730 1.00103.43 H C
+ATOM 2388 O LEU H 115 -46.226 12.926 30.278 1.00103.43 H O
+ATOM 2389 CB LEU H 115 -46.109 9.408 29.652 1.00103.43 H C
+ATOM 2390 CG LEU H 115 -46.649 9.200 31.081 1.00103.43 H C
+ATOM 2391 CD1 LEU H 115 -47.432 7.900 31.193 1.00103.43 H C
+ATOM 2392 CD2 LEU H 115 -45.549 9.231 32.145 1.00103.43 H C
+ATOM 2393 N VAL H 116 -47.889 11.650 29.483 1.00114.61 H N
+ATOM 2394 CA VAL H 116 -48.904 12.358 30.245 1.00114.61 H C
+ATOM 2395 C VAL H 116 -49.427 11.494 31.387 1.00114.61 H C
+ATOM 2396 O VAL H 116 -49.112 11.752 32.555 1.00114.61 H O
+ATOM 2397 CB VAL H 116 -50.029 12.870 29.317 1.00114.61 H C
+ATOM 2398 CG1 VAL H 116 -50.765 11.768 28.607 1.00114.61 H C
+ATOM 2399 CG2 VAL H 116 -51.041 13.582 30.143 1.00114.61 H C
+TER
+ATOM 2400 N ASP L 1 -39.798 -12.074 8.623 1.00 93.54 L N
+ATOM 2401 CA ASP L 1 -39.290 -10.736 8.881 1.00 93.54 L C
+ATOM 2402 C ASP L 1 -38.135 -10.777 9.871 1.00 93.54 L C
+ATOM 2403 O ASP L 1 -37.315 -11.699 9.854 1.00 93.54 L O
+ATOM 2404 CB ASP L 1 -40.407 -9.829 9.411 1.00 93.54 L C
+ATOM 2405 CG ASP L 1 -40.868 -10.221 10.802 1.00 93.54 L C
+ATOM 2406 OD1 ASP L 1 -40.836 -11.426 11.115 1.00 93.54 L O
+ATOM 2407 OD2 ASP L 1 -41.231 -9.323 11.588 1.00 93.54 L O1-
+ATOM 2408 N ILE L 2 -38.069 -9.766 10.730 1.00 89.86 L N
+ATOM 2409 CA ILE L 2 -37.156 -9.752 11.858 1.00 89.86 L C
+ATOM 2410 C ILE L 2 -38.016 -9.955 13.094 1.00 89.86 L C
+ATOM 2411 O ILE L 2 -38.915 -9.151 13.369 1.00 89.86 L O
+ATOM 2412 CB ILE L 2 -36.390 -8.431 11.954 1.00 89.86 L C
+ATOM 2413 CG1 ILE L 2 -35.801 -8.067 10.599 1.00 89.86 L C
+ATOM 2414 CG2 ILE L 2 -35.265 -8.582 12.960 1.00 89.86 L C
+ATOM 2415 CD1 ILE L 2 -34.644 -8.864 10.186 1.00 89.86 L C
+ATOM 2416 N GLN L 3 -37.762 -11.031 13.828 1.00 87.82 L N
+ATOM 2417 CA GLN L 3 -38.510 -11.321 15.045 1.00 87.82 L C
+ATOM 2418 C GLN L 3 -37.519 -11.799 16.090 1.00 87.82 L C
+ATOM 2419 O GLN L 3 -36.909 -12.856 15.920 1.00 87.82 L O
+ATOM 2420 CB GLN L 3 -39.592 -12.368 14.796 1.00 87.82 L C
+ATOM 2421 CG GLN L 3 -40.322 -12.799 16.048 1.00 87.82 L C
+ATOM 2422 CD GLN L 3 -40.843 -11.623 16.838 1.00 87.82 L C
+ATOM 2423 NE2 GLN L 3 -41.914 -11.017 16.353 1.00 87.82 L N
+ATOM 2424 OE1 GLN L 3 -40.289 -11.262 17.872 1.00 87.82 L O
+ATOM 2425 N MET L 4 -37.356 -11.028 17.158 1.00 84.13 L N
+ATOM 2426 CA MET L 4 -36.406 -11.344 18.216 1.00 84.13 L C
+ATOM 2427 C MET L 4 -37.143 -12.053 19.341 1.00 84.13 L C
+ATOM 2428 O MET L 4 -37.904 -11.427 20.081 1.00 84.13 L O
+ATOM 2429 CB MET L 4 -35.715 -10.084 18.722 1.00 84.13 L C
+ATOM 2430 CG MET L 4 -34.471 -9.752 17.952 1.00 84.13 L C
+ATOM 2431 SD MET L 4 -33.362 -11.156 17.947 1.00 84.13 L S
+ATOM 2432 CE MET L 4 -31.806 -10.314 17.782 1.00 84.13 L C
+ATOM 2433 N THR L 5 -36.911 -13.353 19.475 1.00 87.85 L N
+ATOM 2434 CA THR L 5 -37.610 -14.165 20.460 1.00 87.85 L C
+ATOM 2435 C THR L 5 -36.682 -14.452 21.627 1.00 87.85 L C
+ATOM 2436 O THR L 5 -35.506 -14.755 21.433 1.00 87.85 L O
+ATOM 2437 CB THR L 5 -38.113 -15.471 19.850 1.00 87.85 L C
+ATOM 2438 CG2 THR L 5 -39.179 -16.076 20.737 1.00 87.85 L C
+ATOM 2439 OG1 THR L 5 -38.679 -15.209 18.560 1.00 87.85 L O
+ATOM 2440 N GLN L 6 -37.232 -14.385 22.837 1.00 85.55 L N
+ATOM 2441 CA GLN L 6 -36.456 -14.093 24.031 1.00 85.55 L C
+ATOM 2442 C GLN L 6 -35.999 -15.305 24.825 1.00 85.55 L C
+ATOM 2443 O GLN L 6 -35.269 -15.116 25.799 1.00 85.55 L O
+ATOM 2444 CB GLN L 6 -37.265 -13.191 24.960 1.00 85.55 L C
+ATOM 2445 CG GLN L 6 -37.231 -11.751 24.557 1.00 85.55 L C
+ATOM 2446 CD GLN L 6 -36.083 -11.002 25.189 1.00 85.55 L C
+ATOM 2447 NE2 GLN L 6 -35.249 -10.405 24.361 1.00 85.55 L N
+ATOM 2448 OE1 GLN L 6 -35.939 -10.978 26.407 1.00 85.55 L O
+ATOM 2449 N SER L 7 -36.462 -16.516 24.491 1.00 91.62 L N
+ATOM 2450 CA SER L 7 -36.022 -17.796 25.058 1.00 91.62 L C
+ATOM 2451 C SER L 7 -35.526 -17.677 26.500 1.00 91.62 L C
+ATOM 2452 O SER L 7 -34.376 -18.028 26.789 1.00 91.62 L O
+ATOM 2453 CB SER L 7 -34.936 -18.408 24.172 1.00 91.62 L C
+ATOM 2454 OG SER L 7 -35.410 -18.654 22.860 1.00 91.62 L O
+ATOM 2455 N PRO L 8 -36.355 -17.195 27.428 1.00 96.66 L N
+ATOM 2456 CA PRO L 8 -35.842 -16.708 28.717 1.00 96.66 L C
+ATOM 2457 C PRO L 8 -35.394 -17.763 29.714 1.00 96.66 L C
+ATOM 2458 O PRO L 8 -35.247 -18.943 29.390 1.00 96.66 L O
+ATOM 2459 CB PRO L 8 -37.033 -15.922 29.281 1.00 96.66 L C
+ATOM 2460 CG PRO L 8 -37.929 -15.678 28.107 1.00 96.66 L C
+ATOM 2461 CD PRO L 8 -37.787 -16.903 27.284 1.00 96.66 L C
+ATOM 2462 N SER L 9 -35.136 -17.300 30.935 1.00100.21 L N
+ATOM 2463 CA SER L 9 -34.840 -18.098 32.118 1.00100.21 L C
+ATOM 2464 C SER L 9 -34.912 -17.159 33.312 1.00100.21 L C
+ATOM 2465 O SER L 9 -34.819 -15.943 33.149 1.00100.21 L O
+ATOM 2466 CB SER L 9 -33.476 -18.776 32.012 1.00100.21 L C
+ATOM 2467 OG SER L 9 -32.444 -17.816 31.976 1.00100.21 L O
+ATOM 2468 N SER L 10 -35.094 -17.713 34.505 1.00107.23 L N
+ATOM 2469 CA SER L 10 -35.465 -16.867 35.643 1.00107.23 L C
+ATOM 2470 C SER L 10 -35.304 -17.620 36.961 1.00107.23 L C
+ATOM 2471 O SER L 10 -34.737 -18.720 37.017 1.00107.23 L O
+ATOM 2472 CB SER L 10 -36.903 -16.370 35.508 1.00107.23 L C
+ATOM 2473 OG SER L 10 -37.278 -15.623 36.651 1.00107.23 L O
+ATOM 2474 N LEU L 11 -35.804 -16.990 38.027 1.00117.87 L N
+ATOM 2475 CA LEU L 11 -36.202 -17.517 39.329 1.00117.87 L C
+ATOM 2476 C LEU L 11 -35.071 -17.766 40.323 1.00117.87 L C
+ATOM 2477 O LEU L 11 -35.366 -18.046 41.482 1.00117.87 L O
+ATOM 2478 CB LEU L 11 -37.009 -18.827 39.217 1.00117.87 L C
+ATOM 2479 CG LEU L 11 -38.543 -18.808 39.137 1.00117.87 L C
+ATOM 2480 CD1 LEU L 11 -39.032 -18.385 37.765 1.00117.87 L C
+ATOM 2481 CD2 LEU L 11 -39.113 -20.171 39.501 1.00117.87 L C
+ATOM 2482 N SER L 12 -33.800 -17.631 39.958 1.00111.40 L N
+ATOM 2483 CA SER L 12 -32.766 -18.153 40.849 1.00111.40 L C
+ATOM 2484 C SER L 12 -31.473 -17.367 40.694 1.00111.40 L C
+ATOM 2485 O SER L 12 -30.768 -17.533 39.695 1.00111.40 L O
+ATOM 2486 CB SER L 12 -32.531 -19.626 40.559 1.00111.40 L C
+ATOM 2487 OG SER L 12 -32.181 -19.811 39.201 1.00111.40 L O
+ATOM 2488 N ALA L 13 -31.153 -16.537 41.698 1.00108.28 L N
+ATOM 2489 CA ALA L 13 -29.841 -15.897 41.744 1.00108.28 L C
+ATOM 2490 C ALA L 13 -29.251 -15.832 43.151 1.00108.28 L C
+ATOM 2491 O ALA L 13 -28.265 -15.113 43.344 1.00108.28 L O
+ATOM 2492 CB ALA L 13 -29.900 -14.462 41.173 1.00108.28 L C
+ATOM 2493 N SER L 14 -29.735 -16.642 44.095 1.00106.72 L N
+ATOM 2494 CA SER L 14 -29.699 -16.317 45.518 1.00106.72 L C
+ATOM 2495 C SER L 14 -28.464 -15.548 45.955 1.00106.72 L C
+ATOM 2496 O SER L 14 -28.580 -14.462 46.525 1.00106.72 L O
+ATOM 2497 CB SER L 14 -29.774 -17.599 46.339 1.00106.72 L C
+ATOM 2498 OG SER L 14 -28.884 -18.584 45.838 1.00106.72 L O
+ATOM 2499 N VAL L 15 -27.286 -16.111 45.719 1.00101.21 L N
+ATOM 2500 CA VAL L 15 -26.052 -15.377 45.948 1.00101.21 L C
+ATOM 2501 C VAL L 15 -25.109 -15.569 44.775 1.00101.21 L C
+ATOM 2502 O VAL L 15 -24.242 -14.728 44.519 1.00101.21 L O
+ATOM 2503 CB VAL L 15 -25.381 -15.804 47.268 1.00101.21 L C
+ATOM 2504 CG1 VAL L 15 -26.224 -15.405 48.474 1.00101.21 L C
+ATOM 2505 CG2 VAL L 15 -25.133 -17.305 47.286 1.00101.21 L C
+ATOM 2506 N GLY L 16 -25.240 -16.689 44.078 1.00 97.43 L N
+ATOM 2507 CA GLY L 16 -24.297 -17.001 43.028 1.00 97.43 L C
+ATOM 2508 C GLY L 16 -24.885 -17.796 41.888 1.00 97.43 L C
+ATOM 2509 O GLY L 16 -24.145 -18.319 41.050 1.00 97.43 L O
+ATOM 2510 N ASP L 17 -26.207 -17.913 41.852 1.00102.53 L N
+ATOM 2511 CA ASP L 17 -26.843 -18.869 40.957 1.00102.53 L C
+ATOM 2512 C ASP L 17 -26.649 -18.476 39.498 1.00102.53 L C
+ATOM 2513 O ASP L 17 -27.185 -17.465 39.036 1.00102.53 L O
+ATOM 2514 CB ASP L 17 -28.327 -18.993 41.291 1.00102.53 L C
+ATOM 2515 CG ASP L 17 -28.575 -19.177 42.783 1.00102.53 L C
+ATOM 2516 OD1 ASP L 17 -27.739 -19.812 43.455 1.00102.53 L O
+ATOM 2517 OD2 ASP L 17 -29.617 -18.709 43.283 1.00102.53 L O1-
+ATOM 2518 N ARG L 18 -25.888 -19.294 38.783 1.00 92.49 L N
+ATOM 2519 CA ARG L 18 -25.641 -19.086 37.366 1.00 92.49 L C
+ATOM 2520 C ARG L 18 -26.930 -19.249 36.571 1.00 92.49 L C
+ATOM 2521 O ARG L 18 -27.718 -20.165 36.812 1.00 92.49 L O
+ATOM 2522 CB ARG L 18 -24.580 -20.079 36.895 1.00 92.49 L C
+ATOM 2523 CG ARG L 18 -24.433 -20.293 35.400 1.00 92.49 L C
+ATOM 2524 CD ARG L 18 -23.813 -19.082 34.719 1.00 92.49 L C
+ATOM 2525 NE ARG L 18 -23.250 -19.457 33.425 1.00 92.49 L N
+ATOM 2526 CZ ARG L 18 -23.911 -19.425 32.277 1.00 92.49 L C
+ATOM 2527 NH1 ARG L 18 -25.172 -19.023 32.258 1.00 92.49 L N1+
+ATOM 2528 NH2 ARG L 18 -23.310 -19.783 31.152 1.00 92.49 L N
+ATOM 2529 N VAL L 19 -27.150 -18.338 35.625 1.00 86.12 L N
+ATOM 2530 CA VAL L 19 -28.344 -18.313 34.787 1.00 86.12 L C
+ATOM 2531 C VAL L 19 -27.896 -18.101 33.350 1.00 86.12 L C
+ATOM 2532 O VAL L 19 -26.950 -17.352 33.099 1.00 86.12 L O
+ATOM 2533 CB VAL L 19 -29.317 -17.198 35.224 1.00 86.12 L C
+ATOM 2534 CG1 VAL L 19 -30.499 -17.116 34.291 1.00 86.12 L C
+ATOM 2535 CG2 VAL L 19 -29.786 -17.418 36.641 1.00 86.12 L C
+ATOM 2536 N THR L 20 -28.541 -18.785 32.411 1.00 83.55 L N
+ATOM 2537 CA THR L 20 -28.312 -18.571 30.988 1.00 83.55 L C
+ATOM 2538 C THR L 20 -29.599 -18.126 30.321 1.00 83.55 L C
+ATOM 2539 O THR L 20 -30.627 -18.798 30.438 1.00 83.55 L O
+ATOM 2540 CB THR L 20 -27.782 -19.827 30.305 1.00 83.55 L C
+ATOM 2541 CG2 THR L 20 -27.571 -19.597 28.840 1.00 83.55 L C
+ATOM 2542 OG1 THR L 20 -26.541 -20.202 30.901 1.00 83.55 L O
+ATOM 2543 N ILE L 21 -29.535 -17.002 29.611 1.00 81.94 L N
+ATOM 2544 CA ILE L 21 -30.667 -16.450 28.880 1.00 81.94 L C
+ATOM 2545 C ILE L 21 -30.288 -16.440 27.406 1.00 81.94 L C
+ATOM 2546 O ILE L 21 -29.137 -16.161 27.063 1.00 81.94 L O
+ATOM 2547 CB ILE L 21 -31.028 -15.041 29.376 1.00 81.94 L C
+ATOM 2548 CG1 ILE L 21 -31.456 -15.088 30.831 1.00 81.94 L C
+ATOM 2549 CG2 ILE L 21 -32.187 -14.504 28.604 1.00 81.94 L C
+ATOM 2550 CD1 ILE L 21 -31.572 -13.740 31.448 1.00 81.94 L C
+ATOM 2551 N THR L 22 -31.235 -16.788 26.545 1.00 85.16 L N
+ATOM 2552 CA THR L 22 -30.988 -16.926 25.121 1.00 85.16 L C
+ATOM 2553 C THR L 22 -32.030 -16.133 24.350 1.00 85.16 L C
+ATOM 2554 O THR L 22 -33.126 -15.900 24.852 1.00 85.16 L O
+ATOM 2555 CB THR L 22 -31.042 -18.383 24.713 1.00 85.16 L C
+ATOM 2556 CG2 THR L 22 -29.842 -19.116 25.245 1.00 85.16 L C
+ATOM 2557 OG1 THR L 22 -32.209 -18.977 25.282 1.00 85.16 L O
+ATOM 2558 N CYS L 23 -31.692 -15.703 23.133 1.00 85.95 L N
+ATOM 2559 CA CYS L 23 -32.666 -15.037 22.278 1.00 85.95 L C
+ATOM 2560 C CYS L 23 -32.593 -15.552 20.846 1.00 85.95 L C
+ATOM 2561 O CYS L 23 -31.514 -15.801 20.314 1.00 85.95 L O
+ATOM 2562 CB CYS L 23 -32.487 -13.514 22.275 1.00 85.95 L C
+ATOM 2563 SG CYS L 23 -30.842 -12.916 21.847 1.00 85.95 L S
+ATOM 2564 N ARG L 24 -33.761 -15.637 20.205 1.00 89.44 L N
+ATOM 2565 CA ARG L 24 -33.936 -16.152 18.850 1.00 89.44 L C
+ATOM 2566 C ARG L 24 -34.352 -15.059 17.877 1.00 89.44 L C
+ATOM 2567 O ARG L 24 -35.024 -14.100 18.254 1.00 89.44 L O
+ATOM 2568 CB ARG L 24 -34.995 -17.267 18.830 1.00 89.44 L C
+ATOM 2569 CG ARG L 24 -35.280 -17.876 17.454 1.00 89.44 L C
+ATOM 2570 CD ARG L 24 -34.193 -18.662 16.768 1.00 89.44 L C
+ATOM 2571 NE ARG L 24 -34.744 -19.146 15.504 1.00 89.44 L N
+ATOM 2572 CZ ARG L 24 -34.042 -19.743 14.545 1.00 89.44 L C
+ATOM 2573 NH1 ARG L 24 -32.755 -19.985 14.708 1.00 89.44 L N1+
+ATOM 2574 NH2 ARG L 24 -34.629 -20.132 13.423 1.00 89.44 L N
+ATOM 2575 N ALA L 25 -33.967 -15.235 16.608 1.00 85.43 L N
+ATOM 2576 CA ALA L 25 -34.282 -14.294 15.542 1.00 85.43 L C
+ATOM 2577 C ALA L 25 -34.785 -15.016 14.294 1.00 85.43 L C
+ATOM 2578 O ALA L 25 -34.562 -16.214 14.106 1.00 85.43 L O
+ATOM 2579 CB ALA L 25 -33.064 -13.444 15.177 1.00 85.43 L C
+ATOM 2580 N SER L 26 -35.479 -14.258 13.438 1.00 84.32 L N
+ATOM 2581 CA SER L 26 -35.953 -14.703 12.131 1.00 84.32 L C
+ATOM 2582 C SER L 26 -35.066 -14.231 10.981 1.00 84.32 L C
+ATOM 2583 O SER L 26 -35.416 -14.431 9.815 1.00 84.32 L O
+ATOM 2584 CB SER L 26 -37.381 -14.217 11.912 1.00 84.32 L C
+ATOM 2585 OG SER L 26 -38.171 -14.525 13.040 1.00 84.32 L O
+ATOM 2586 N GLN L 27 -33.947 -13.592 11.297 1.00 81.18 L N
+ATOM 2587 CA GLN L 27 -32.897 -13.168 10.381 1.00 81.18 L C
+ATOM 2588 C GLN L 27 -31.585 -13.813 10.786 1.00 81.18 L C
+ATOM 2589 O GLN L 27 -31.517 -14.543 11.776 1.00 81.18 L O
+ATOM 2590 CB GLN L 27 -32.801 -11.647 10.356 1.00 81.18 L C
+ATOM 2591 CG GLN L 27 -32.455 -11.022 11.716 1.00 81.18 L C
+ATOM 2592 CD GLN L 27 -31.004 -10.978 12.084 1.00 81.18 L C
+ATOM 2593 NE2 GLN L 27 -30.161 -10.916 11.069 1.00 81.18 L N
+ATOM 2594 OE1 GLN L 27 -30.634 -11.022 13.247 1.00 81.18 L O
+ATOM 2595 N ASP L 28 -30.550 -13.603 9.989 1.00 80.68 L N
+ATOM 2596 CA ASP L 28 -29.278 -14.254 10.250 1.00 80.68 L C
+ATOM 2597 C ASP L 28 -28.225 -13.194 10.545 1.00 80.68 L C
+ATOM 2598 O ASP L 28 -28.080 -12.229 9.790 1.00 80.68 L O
+ATOM 2599 CB ASP L 28 -28.872 -15.138 9.070 1.00 80.68 L C
+ATOM 2600 CG ASP L 28 -27.673 -16.012 9.378 1.00 80.68 L C
+ATOM 2601 OD1 ASP L 28 -27.138 -15.909 10.496 1.00 80.68 L O
+ATOM 2602 OD2 ASP L 28 -27.287 -16.832 8.516 1.00 80.68 L O1-
+ATOM 2603 N VAL L 29 -27.487 -13.388 11.639 1.00 75.75 L N
+ATOM 2604 CA VAL L 29 -26.477 -12.461 12.142 1.00 75.75 L C
+ATOM 2605 C VAL L 29 -25.101 -12.940 11.712 1.00 75.75 L C
+ATOM 2606 O VAL L 29 -24.960 -14.044 11.182 1.00 75.75 L O
+ATOM 2607 CB VAL L 29 -26.550 -12.354 13.669 1.00 75.75 L C
+ATOM 2608 CG1 VAL L 29 -27.856 -11.782 14.082 1.00 75.75 L C
+ATOM 2609 CG2 VAL L 29 -26.388 -13.715 14.290 1.00 75.75 L C
+ATOM 2610 N ASN L 30 -24.075 -12.122 11.908 1.00 73.62 L N
+ATOM 2611 CA ASN L 30 -22.710 -12.612 11.680 1.00 73.62 L C
+ATOM 2612 C ASN L 30 -22.108 -13.178 12.962 1.00 73.62 L C
+ATOM 2613 O ASN L 30 -21.873 -14.386 13.060 1.00 73.62 L O
+ATOM 2614 CB ASN L 30 -21.820 -11.498 11.131 1.00 73.62 L C
+ATOM 2615 CG ASN L 30 -20.534 -12.021 10.538 1.00 73.62 L C
+ATOM 2616 ND2 ASN L 30 -20.282 -11.685 9.279 1.00 73.62 L N
+ATOM 2617 OD1 ASN L 30 -19.770 -12.711 11.204 1.00 73.62 L O
+ATOM 2618 N THR L 31 -21.847 -12.320 13.954 1.00 67.73 L N
+ATOM 2619 CA THR L 31 -21.416 -12.774 15.271 1.00 67.73 L C
+ATOM 2620 C THR L 31 -21.966 -11.897 16.382 1.00 67.73 L C
+ATOM 2621 O THR L 31 -21.218 -11.535 17.290 1.00 67.73 L O
+ATOM 2622 CB THR L 31 -19.893 -12.792 15.415 1.00 67.73 L C
+ATOM 2623 CG2 THR L 31 -19.181 -13.278 14.182 1.00 67.73 L C
+ATOM 2624 OG1 THR L 31 -19.439 -11.471 15.715 1.00 67.73 L O
+ATOM 2625 N ALA L 32 -23.238 -11.504 16.356 1.00 67.02 L N
+ATOM 2626 CA ALA L 32 -23.573 -10.537 17.399 1.00 67.02 L C
+ATOM 2627 C ALA L 32 -25.073 -10.318 17.551 1.00 67.02 L C
+ATOM 2628 O ALA L 32 -25.847 -10.547 16.621 1.00 67.02 L O
+ATOM 2629 CB ALA L 32 -22.900 -9.182 17.130 1.00 67.02 L C
+ATOM 2630 N VAL L 33 -25.461 -9.891 18.764 1.00 68.92 L N
+ATOM 2631 CA VAL L 33 -26.671 -9.123 19.074 1.00 68.92 L C
+ATOM 2632 C VAL L 33 -26.276 -8.052 20.084 1.00 68.92 L C
+ATOM 2633 O VAL L 33 -25.088 -7.818 20.305 1.00 68.92 L O
+ATOM 2634 CB VAL L 33 -27.817 -9.978 19.627 1.00 68.92 L C
+ATOM 2635 CG1 VAL L 33 -28.411 -10.830 18.532 1.00 68.92 L C
+ATOM 2636 CG2 VAL L 33 -27.343 -10.780 20.805 1.00 68.92 L C
+ATOM 2637 N ALA L 34 -27.250 -7.353 20.657 1.00 64.25 L N
+ATOM 2638 CA ALA L 34 -27.001 -6.441 21.763 1.00 64.25 L C
+ATOM 2639 C ALA L 34 -27.748 -6.887 23.010 1.00 64.25 L C
+ATOM 2640 O ALA L 34 -28.797 -7.522 22.925 1.00 64.25 L O
+ATOM 2641 CB ALA L 34 -27.427 -5.026 21.405 1.00 64.25 L C
+ATOM 2642 N TRP L 35 -27.217 -6.518 24.179 1.00 63.07 L N
+ATOM 2643 CA TRP L 35 -27.824 -6.862 25.460 1.00 63.07 L C
+ATOM 2644 C TRP L 35 -27.988 -5.622 26.308 1.00 63.07 L C
+ATOM 2645 O TRP L 35 -27.227 -4.669 26.195 1.00 63.07 L O
+ATOM 2646 CB TRP L 35 -27.015 -7.884 26.261 1.00 63.07 L C
+ATOM 2647 CG TRP L 35 -27.255 -9.275 25.853 1.00 63.07 L C
+ATOM 2648 CD1 TRP L 35 -26.437 -10.057 25.115 1.00 63.07 L C
+ATOM 2649 CD2 TRP L 35 -28.412 -10.054 26.141 1.00 63.07 L C
+ATOM 2650 CE2 TRP L 35 -28.228 -11.304 25.546 1.00 63.07 L C
+ATOM 2651 CE3 TRP L 35 -29.584 -9.815 26.854 1.00 63.07 L C
+ATOM 2652 NE1 TRP L 35 -27.009 -11.284 24.925 1.00 63.07 L N
+ATOM 2653 CZ2 TRP L 35 -29.171 -12.313 25.638 1.00 63.07 L C
+ATOM 2654 CZ3 TRP L 35 -30.520 -10.807 26.933 1.00 63.07 L C
+ATOM 2655 CH2 TRP L 35 -30.309 -12.043 26.334 1.00 63.07 L C
+ATOM 2656 N TYR L 36 -28.977 -5.662 27.185 1.00 64.02 L N
+ATOM 2657 CA TYR L 36 -29.492 -4.446 27.789 1.00 64.02 L C
+ATOM 2658 C TYR L 36 -30.252 -4.783 29.068 1.00 64.02 L C
+ATOM 2659 O TYR L 36 -31.410 -5.192 28.992 1.00 64.02 L O
+ATOM 2660 CB TYR L 36 -30.371 -3.771 26.759 1.00 64.02 L C
+ATOM 2661 CG TYR L 36 -31.170 -2.560 27.141 1.00 64.02 L C
+ATOM 2662 CD1 TYR L 36 -30.657 -1.514 27.869 1.00 64.02 L C
+ATOM 2663 CD2 TYR L 36 -32.472 -2.471 26.705 1.00 64.02 L C
+ATOM 2664 CE1 TYR L 36 -31.457 -0.429 28.184 1.00 64.02 L C
+ATOM 2665 CE2 TYR L 36 -33.254 -1.417 26.995 1.00 64.02 L C
+ATOM 2666 CZ TYR L 36 -32.762 -0.397 27.726 1.00 64.02 L C
+ATOM 2667 OH TYR L 36 -33.641 0.625 27.960 1.00 64.02 L O
+ATOM 2668 N GLN L 37 -29.633 -4.553 30.227 1.00 63.96 L N
+ATOM 2669 CA GLN L 37 -30.413 -4.673 31.448 1.00 63.96 L C
+ATOM 2670 C GLN L 37 -31.522 -3.643 31.453 1.00 63.96 L C
+ATOM 2671 O GLN L 37 -31.546 -2.709 30.664 1.00 63.96 L O
+ATOM 2672 CB GLN L 37 -29.596 -4.449 32.721 1.00 63.96 L C
+ATOM 2673 CG GLN L 37 -28.315 -5.209 32.932 1.00 63.96 L C
+ATOM 2674 CD GLN L 37 -27.921 -5.230 34.413 1.00 63.96 L C
+ATOM 2675 NE2 GLN L 37 -26.629 -5.307 34.698 1.00 63.96 L N
+ATOM 2676 OE1 GLN L 37 -28.783 -5.179 35.283 1.00 63.96 L O
+ATOM 2677 N GLN L 38 -32.445 -3.819 32.377 1.00 67.31 L N
+ATOM 2678 CA GLN L 38 -33.446 -2.800 32.631 1.00 67.31 L C
+ATOM 2679 C GLN L 38 -33.846 -2.973 34.088 1.00 67.31 L C
+ATOM 2680 O GLN L 38 -34.714 -3.791 34.398 1.00 67.31 L O
+ATOM 2681 CB GLN L 38 -34.631 -2.949 31.700 1.00 67.31 L C
+ATOM 2682 CG GLN L 38 -35.254 -1.644 31.306 1.00 67.31 L C
+ATOM 2683 CD GLN L 38 -36.342 -1.227 32.240 1.00 67.31 L C
+ATOM 2684 NE2 GLN L 38 -36.697 0.046 32.206 1.00 67.31 L N
+ATOM 2685 OE1 GLN L 38 -36.871 -2.042 32.984 1.00 67.31 L O
+ATOM 2686 N LYS L 39 -33.212 -2.209 34.966 1.00 74.72 L N
+ATOM 2687 CA LYS L 39 -33.412 -2.397 36.390 1.00 74.72 L C
+ATOM 2688 C LYS L 39 -34.875 -2.136 36.742 1.00 74.72 L C
+ATOM 2689 O LYS L 39 -35.537 -1.319 36.100 1.00 74.72 L O
+ATOM 2690 CB LYS L 39 -32.494 -1.466 37.172 1.00 74.72 L C
+ATOM 2691 CG LYS L 39 -31.070 -1.430 36.633 1.00 74.72 L C
+ATOM 2692 CD LYS L 39 -30.403 -2.801 36.693 1.00 74.72 L C
+ATOM 2693 CE LYS L 39 -28.985 -2.706 37.239 1.00 74.72 L C
+ATOM 2694 NZ LYS L 39 -28.222 -3.977 37.087 1.00 74.72 L N1+
+ATOM 2695 N PRO L 40 -35.410 -2.830 37.730 1.00 78.78 L N
+ATOM 2696 CA PRO L 40 -36.858 -2.785 37.957 1.00 78.78 L C
+ATOM 2697 C PRO L 40 -37.351 -1.413 38.366 1.00 78.78 L C
+ATOM 2698 O PRO L 40 -37.006 -0.913 39.440 1.00 78.78 L O
+ATOM 2699 CB PRO L 40 -37.070 -3.804 39.082 1.00 78.78 L C
+ATOM 2700 CG PRO L 40 -35.814 -4.597 39.137 1.00 78.78 L C
+ATOM 2701 CD PRO L 40 -34.735 -3.674 38.722 1.00 78.78 L C
+ATOM 2702 N GLY L 41 -38.162 -0.798 37.510 1.00 80.49 L N
+ATOM 2703 CA GLY L 41 -38.793 0.464 37.821 1.00 80.49 L C
+ATOM 2704 C GLY L 41 -37.943 1.693 37.606 1.00 80.49 L C
+ATOM 2705 O GLY L 41 -38.410 2.798 37.900 1.00 80.49 L O
+ATOM 2706 N LYS L 42 -36.721 1.544 37.098 1.00 79.43 L N
+ATOM 2707 CA LYS L 42 -35.851 2.689 36.880 1.00 79.43 L C
+ATOM 2708 C LYS L 42 -34.615 2.258 36.103 1.00 79.43 L C
+ATOM 2709 O LYS L 42 -34.192 1.102 36.179 1.00 79.43 L O
+ATOM 2710 CB LYS L 42 -35.429 3.326 38.210 1.00 79.43 L C
+ATOM 2711 CG LYS L 42 -34.712 2.372 39.147 1.00 79.43 L C
+ATOM 2712 CD LYS L 42 -34.634 2.918 40.571 1.00 79.43 L C
+ATOM 2713 CE LYS L 42 -35.779 2.398 41.425 1.00 79.43 L C
+ATOM 2714 NZ LYS L 42 -35.589 0.961 41.775 1.00 79.43 L N1+
+ATOM 2715 N ALA L 43 -34.040 3.210 35.365 1.00 80.95 L N
+ATOM 2716 CA ALA L 43 -32.656 3.160 34.896 1.00 80.95 L C
+ATOM 2717 C ALA L 43 -32.320 2.016 33.947 1.00 80.95 L C
+ATOM 2718 O ALA L 43 -31.724 1.016 34.366 1.00 80.95 L O
+ATOM 2719 CB ALA L 43 -31.711 3.104 36.097 1.00 80.95 L C
+ATOM 2720 N PRO L 44 -32.693 2.116 32.675 1.00 69.79 L N
+ATOM 2721 CA PRO L 44 -32.109 1.237 31.660 1.00 69.79 L C
+ATOM 2722 C PRO L 44 -30.604 1.397 31.563 1.00 69.79 L C
+ATOM 2723 O PRO L 44 -30.054 2.459 31.845 1.00 69.79 L O
+ATOM 2724 CB PRO L 44 -32.790 1.724 30.394 1.00 69.79 L C
+ATOM 2725 CG PRO L 44 -34.134 2.036 30.913 1.00 69.79 L C
+ATOM 2726 CD PRO L 44 -33.746 2.930 32.057 1.00 69.79 L C
+ATOM 2727 N LYS L 45 -29.931 0.326 31.157 1.00 60.30 L N
+ATOM 2728 CA LYS L 45 -28.483 0.280 31.257 1.00 60.30 L C
+ATOM 2729 C LYS L 45 -27.918 -0.606 30.173 1.00 60.30 L C
+ATOM 2730 O LYS L 45 -28.451 -1.683 29.919 1.00 60.30 L O
+ATOM 2731 CB LYS L 45 -28.034 -0.288 32.597 1.00 60.30 L C
+ATOM 2732 CG LYS L 45 -28.159 0.635 33.766 1.00 60.30 L C
+ATOM 2733 CD LYS L 45 -27.858 -0.111 35.048 1.00 60.30 L C
+ATOM 2734 CE LYS L 45 -26.676 -1.054 34.884 1.00 60.30 L C
+ATOM 2735 NZ LYS L 45 -25.373 -0.361 35.041 1.00 60.30 L N1+
+ATOM 2736 N LEU L 46 -26.814 -0.185 29.577 1.00 51.46 L N
+ATOM 2737 CA LEU L 46 -26.108 -1.118 28.726 1.00 51.46 L C
+ATOM 2738 C LEU L 46 -25.646 -2.259 29.619 1.00 51.46 L C
+ATOM 2739 O LEU L 46 -25.488 -2.099 30.829 1.00 51.46 L O
+ATOM 2740 CB LEU L 46 -24.884 -0.488 28.058 1.00 51.46 L C
+ATOM 2741 CG LEU L 46 -24.208 -1.440 27.067 1.00 51.46 L C
+ATOM 2742 CD1 LEU L 46 -25.141 -1.719 25.911 1.00 51.46 L C
+ATOM 2743 CD2 LEU L 46 -22.793 -1.189 26.639 1.00 51.46 L C
+ATOM 2744 N LEU L 47 -25.423 -3.413 29.013 1.00 59.03 L N
+ATOM 2745 CA LEU L 47 -24.884 -4.583 29.675 1.00 59.03 L C
+ATOM 2746 C LEU L 47 -23.883 -5.134 28.675 1.00 59.03 L C
+ATOM 2747 O LEU L 47 -23.355 -4.384 27.856 1.00 59.03 L O
+ATOM 2748 CB LEU L 47 -26.021 -5.524 30.086 1.00 59.03 L C
+ATOM 2749 CG LEU L 47 -25.659 -6.785 30.858 1.00 59.03 L C
+ATOM 2750 CD1 LEU L 47 -25.038 -6.393 32.168 1.00 59.03 L C
+ATOM 2751 CD2 LEU L 47 -26.870 -7.666 31.062 1.00 59.03 L C
+ATOM 2752 N ILE L 48 -23.560 -6.412 28.745 1.00 60.66 L N
+ATOM 2753 CA ILE L 48 -22.790 -7.047 27.683 1.00 60.66 L C
+ATOM 2754 C ILE L 48 -23.294 -6.533 26.340 1.00 60.66 L C
+ATOM 2755 O ILE L 48 -24.491 -6.292 26.183 1.00 60.66 L O
+ATOM 2756 CB ILE L 48 -22.929 -8.580 27.784 1.00 60.66 L C
+ATOM 2757 CG1 ILE L 48 -22.208 -9.120 29.023 1.00 60.66 L C
+ATOM 2758 CG2 ILE L 48 -22.485 -9.279 26.502 1.00 60.66 L C
+ATOM 2759 CD1 ILE L 48 -20.719 -9.208 28.884 1.00 60.66 L C
+ATOM 2760 N TYR L 49 -22.407 -6.274 25.379 1.00 63.32 L N
+ATOM 2761 CA TYR L 49 -23.003 -5.832 24.129 1.00 63.32 L C
+ATOM 2762 C TYR L 49 -23.217 -6.926 23.106 1.00 63.32 L C
+ATOM 2763 O TYR L 49 -24.357 -7.227 22.763 1.00 63.32 L O
+ATOM 2764 CB TYR L 49 -22.208 -4.749 23.414 1.00 63.32 L C
+ATOM 2765 CG TYR L 49 -23.166 -4.002 22.516 1.00 63.32 L C
+ATOM 2766 CD1 TYR L 49 -24.352 -3.512 23.032 1.00 63.32 L C
+ATOM 2767 CD2 TYR L 49 -22.986 -3.936 21.144 1.00 63.32 L C
+ATOM 2768 CE1 TYR L 49 -25.265 -2.865 22.244 1.00 63.32 L C
+ATOM 2769 CE2 TYR L 49 -23.921 -3.303 20.341 1.00 63.32 L C
+ATOM 2770 CZ TYR L 49 -25.060 -2.777 20.909 1.00 63.32 L C
+ATOM 2771 OH TYR L 49 -26.006 -2.140 20.159 1.00 63.32 L O
+ATOM 2772 N SER L 50 -22.151 -7.532 22.608 1.00 62.35 L N
+ATOM 2773 CA SER L 50 -22.308 -8.335 21.401 1.00 62.35 L C
+ATOM 2774 C SER L 50 -21.756 -9.721 21.602 1.00 62.35 L C
+ATOM 2775 O SER L 50 -21.692 -10.230 22.720 1.00 62.35 L O
+ATOM 2776 CB SER L 50 -21.608 -7.743 20.190 1.00 62.35 L C
+ATOM 2777 OG SER L 50 -20.211 -7.868 20.338 1.00 62.35 L O
+ATOM 2778 N ALA L 51 -21.434 -10.360 20.495 1.00 68.40 L N
+ATOM 2779 CA ALA L 51 -20.653 -11.577 20.520 1.00 68.40 L C
+ATOM 2780 C ALA L 51 -19.418 -11.410 19.644 1.00 68.40 L C
+ATOM 2781 O ALA L 51 -19.345 -10.497 18.817 1.00 68.40 L O
+ATOM 2782 CB ALA L 51 -21.493 -12.744 20.062 1.00 68.40 L C
+ATOM 2783 N SER L 52 -18.422 -12.261 19.887 1.00 67.76 L N
+ATOM 2784 CA SER L 52 -17.102 -12.328 19.269 1.00 67.76 L C
+ATOM 2785 C SER L 52 -16.252 -11.167 19.739 1.00 67.76 L C
+ATOM 2786 O SER L 52 -15.033 -11.216 19.609 1.00 67.76 L O
+ATOM 2787 CB SER L 52 -17.148 -12.334 17.729 1.00 67.76 L C
+ATOM 2788 OG SER L 52 -15.857 -12.367 17.169 1.00 67.76 L O
+ATOM 2789 N PHE L 53 -16.856 -10.151 20.331 1.00 60.75 L N
+ATOM 2790 CA PHE L 53 -16.339 -9.086 21.136 1.00 60.75 L C
+ATOM 2791 C PHE L 53 -17.473 -8.712 22.076 1.00 60.75 L C
+ATOM 2792 O PHE L 53 -18.393 -7.990 21.732 1.00 60.75 L O
+ATOM 2793 CB PHE L 53 -15.850 -7.883 20.329 1.00 60.75 L C
+ATOM 2794 CG PHE L 53 -14.639 -8.158 19.525 1.00 60.75 L C
+ATOM 2795 CD1 PHE L 53 -13.403 -8.127 20.126 1.00 60.75 L C
+ATOM 2796 CD2 PHE L 53 -14.715 -8.481 18.207 1.00 60.75 L C
+ATOM 2797 CE1 PHE L 53 -12.270 -8.381 19.416 1.00 60.75 L C
+ATOM 2798 CE2 PHE L 53 -13.584 -8.737 17.504 1.00 60.75 L C
+ATOM 2799 CZ PHE L 53 -12.363 -8.686 18.103 1.00 60.75 L C
+ATOM 2800 N LEU L 54 -17.430 -9.258 23.277 1.00 59.34 L N
+ATOM 2801 CA LEU L 54 -18.285 -8.729 24.314 1.00 59.34 L C
+ATOM 2802 C LEU L 54 -17.694 -7.414 24.774 1.00 59.34 L C
+ATOM 2803 O LEU L 54 -16.488 -7.194 24.677 1.00 59.34 L O
+ATOM 2804 CB LEU L 54 -18.412 -9.697 25.491 1.00 59.34 L C
+ATOM 2805 CG LEU L 54 -17.249 -9.913 26.471 1.00 59.34 L C
+ATOM 2806 CD1 LEU L 54 -17.727 -10.607 27.741 1.00 59.34 L C
+ATOM 2807 CD2 LEU L 54 -16.116 -10.706 25.832 1.00 59.34 L C
+ATOM 2808 N TYR L 55 -18.542 -6.516 25.211 1.00 65.14 L N
+ATOM 2809 CA TYR L 55 -18.073 -5.376 25.965 1.00 65.14 L C
+ATOM 2810 C TYR L 55 -19.066 -5.124 27.075 1.00 65.14 L C
+ATOM 2811 O TYR L 55 -20.272 -5.094 26.835 1.00 65.14 L O
+ATOM 2812 CB TYR L 55 -17.947 -4.169 25.105 1.00 65.14 L C
+ATOM 2813 CG TYR L 55 -17.420 -2.956 25.803 1.00 65.14 L C
+ATOM 2814 CD1 TYR L 55 -16.076 -2.846 26.102 1.00 65.14 L C
+ATOM 2815 CD2 TYR L 55 -18.259 -1.918 26.160 1.00 65.14 L C
+ATOM 2816 CE1 TYR L 55 -15.577 -1.734 26.737 1.00 65.14 L C
+ATOM 2817 CE2 TYR L 55 -17.769 -0.800 26.803 1.00 65.14 L C
+ATOM 2818 CZ TYR L 55 -16.429 -0.713 27.084 1.00 65.14 L C
+ATOM 2819 OH TYR L 55 -15.935 0.403 27.717 1.00 65.14 L O
+ATOM 2820 N SER L 56 -18.559 -4.905 28.275 1.00 61.13 L N
+ATOM 2821 CA SER L 56 -19.356 -5.053 29.474 1.00 61.13 L C
+ATOM 2822 C SER L 56 -19.469 -3.711 30.172 1.00 61.13 L C
+ATOM 2823 O SER L 56 -18.607 -2.846 30.034 1.00 61.13 L O
+ATOM 2824 CB SER L 56 -18.714 -6.073 30.408 1.00 61.13 L C
+ATOM 2825 OG SER L 56 -18.676 -7.358 29.823 1.00 61.13 L O
+ATOM 2826 N GLY L 57 -20.534 -3.548 30.939 1.00 60.13 L N
+ATOM 2827 CA GLY L 57 -20.643 -2.418 31.833 1.00 60.13 L C
+ATOM 2828 C GLY L 57 -19.742 -2.643 33.029 1.00 60.13 L C
+ATOM 2829 O GLY L 57 -18.866 -3.511 33.023 1.00 60.13 L O
+ATOM 2830 N VAL L 58 -19.943 -1.836 34.073 1.00 63.69 L N
+ATOM 2831 CA VAL L 58 -19.166 -2.027 35.301 1.00 63.69 L C
+ATOM 2832 C VAL L 58 -19.562 -3.297 36.040 1.00 63.69 L C
+ATOM 2833 O VAL L 58 -18.708 -3.870 36.729 1.00 63.69 L O
+ATOM 2834 CB VAL L 58 -19.212 -0.755 36.163 1.00 63.69 L C
+ATOM 2835 CG1 VAL L 58 -18.838 0.449 35.317 1.00 63.69 L C
+ATOM 2836 CG2 VAL L 58 -20.551 -0.537 36.839 1.00 63.69 L C
+ATOM 2837 N PRO L 59 -20.808 -3.815 35.948 1.00 61.94 L N
+ATOM 2838 CA PRO L 59 -20.979 -5.230 36.339 1.00 61.94 L C
+ATOM 2839 C PRO L 59 -20.389 -6.177 35.300 1.00 61.94 L C
+ATOM 2840 O PRO L 59 -21.066 -6.730 34.446 1.00 61.94 L O
+ATOM 2841 CB PRO L 59 -22.498 -5.371 36.434 1.00 61.94 L C
+ATOM 2842 CG PRO L 59 -22.985 -3.991 36.663 1.00 61.94 L C
+ATOM 2843 CD PRO L 59 -22.121 -3.168 35.796 1.00 61.94 L C
+ATOM 2844 N SER L 60 -19.080 -6.386 35.401 1.00 64.96 L N
+ATOM 2845 CA SER L 60 -18.291 -7.073 34.380 1.00 64.96 L C
+ATOM 2846 C SER L 60 -18.081 -8.544 34.688 1.00 64.96 L C
+ATOM 2847 O SER L 60 -17.032 -9.109 34.366 1.00 64.96 L O
+ATOM 2848 CB SER L 60 -16.952 -6.362 34.219 1.00 64.96 L C
+ATOM 2849 OG SER L 60 -16.306 -6.741 33.016 1.00 64.96 L O
+ATOM 2850 N ARG L 61 -19.063 -9.183 35.327 1.00 68.14 L N
+ATOM 2851 CA ARG L 61 -18.990 -10.582 35.738 1.00 68.14 L C
+ATOM 2852 C ARG L 61 -19.738 -11.526 34.804 1.00 68.14 L C
+ATOM 2853 O ARG L 61 -19.549 -12.743 34.901 1.00 68.14 L O
+ATOM 2854 CB ARG L 61 -19.575 -10.736 37.138 1.00 68.14 L C
+ATOM 2855 CG ARG L 61 -21.042 -10.430 37.133 1.00 68.14 L C
+ATOM 2856 CD ARG L 61 -21.644 -10.591 38.471 1.00 68.14 L C
+ATOM 2857 NE ARG L 61 -21.115 -9.642 39.427 1.00 68.14 L N
+ATOM 2858 CZ ARG L 61 -21.630 -8.437 39.610 1.00 68.14 L C
+ATOM 2859 NH1 ARG L 61 -22.701 -8.073 38.928 1.00 68.14 L N1+
+ATOM 2860 NH2 ARG L 61 -21.099 -7.610 40.498 1.00 68.14 L N
+ATOM 2861 N PHE L 62 -20.599 -10.995 33.934 1.00 68.05 L N
+ATOM 2862 CA PHE L 62 -21.379 -11.804 33.006 1.00 68.05 L C
+ATOM 2863 C PHE L 62 -20.540 -12.268 31.830 1.00 68.05 L C
+ATOM 2864 O PHE L 62 -19.387 -11.871 31.664 1.00 68.05 L O
+ATOM 2865 CB PHE L 62 -22.572 -11.026 32.463 1.00 68.05 L C
+ATOM 2866 CG PHE L 62 -23.545 -10.593 33.500 1.00 68.05 L C
+ATOM 2867 CD1 PHE L 62 -23.425 -10.993 34.807 1.00 68.05 L C
+ATOM 2868 CD2 PHE L 62 -24.612 -9.807 33.150 1.00 68.05 L C
+ATOM 2869 CE1 PHE L 62 -24.336 -10.592 35.743 1.00 68.05 L C
+ATOM 2870 CE2 PHE L 62 -25.526 -9.407 34.077 1.00 68.05 L C
+ATOM 2871 CZ PHE L 62 -25.390 -9.798 35.374 1.00 68.05 L C
+ATOM 2872 N SER L 63 -21.122 -13.156 31.033 1.00 74.72 L N
+ATOM 2873 CA SER L 63 -20.602 -13.498 29.720 1.00 74.72 L C
+ATOM 2874 C SER L 63 -21.745 -13.524 28.713 1.00 74.72 L C
+ATOM 2875 O SER L 63 -22.747 -14.210 28.926 1.00 74.72 L O
+ATOM 2876 CB SER L 63 -19.881 -14.844 29.749 1.00 74.72 L C
+ATOM 2877 OG SER L 63 -19.180 -15.068 28.541 1.00 74.72 L O
+ATOM 2878 N GLY L 64 -21.592 -12.778 27.622 1.00 72.04 L N
+ATOM 2879 CA GLY L 64 -22.480 -12.861 26.475 1.00 72.04 L C
+ATOM 2880 C GLY L 64 -21.737 -13.541 25.338 1.00 72.04 L C
+ATOM 2881 O GLY L 64 -20.581 -13.218 25.068 1.00 72.04 L O
+ATOM 2882 N SER L 65 -22.398 -14.497 24.698 1.00 73.22 L N
+ATOM 2883 CA SER L 65 -21.681 -15.448 23.867 1.00 73.22 L C
+ATOM 2884 C SER L 65 -22.258 -15.525 22.467 1.00 73.22 L C
+ATOM 2885 O SER L 65 -23.443 -15.275 22.240 1.00 73.22 L O
+ATOM 2886 CB SER L 65 -21.683 -16.847 24.478 1.00 73.22 L C
+ATOM 2887 OG SER L 65 -20.694 -17.658 23.865 1.00 73.22 L O
+ATOM 2888 N ARG L 66 -21.382 -15.878 21.534 1.00 81.62 L N
+ATOM 2889 CA ARG L 66 -21.762 -16.031 20.141 1.00 81.62 L C
+ATOM 2890 C ARG L 66 -22.525 -17.324 19.919 1.00 81.62 L C
+ATOM 2891 O ARG L 66 -22.335 -18.313 20.629 1.00 81.62 L O
+ATOM 2892 CB ARG L 66 -20.523 -16.006 19.246 1.00 81.62 L C
+ATOM 2893 CG ARG L 66 -19.680 -17.269 19.291 1.00 81.62 L C
+ATOM 2894 CD ARG L 66 -18.960 -17.476 17.968 1.00 81.62 L C
+ATOM 2895 NE ARG L 66 -18.063 -16.370 17.664 1.00 81.62 L N
+ATOM 2896 CZ ARG L 66 -16.785 -16.331 18.023 1.00 81.62 L C
+ATOM 2897 NH1 ARG L 66 -16.250 -17.343 18.685 1.00 81.62 L N1+
+ATOM 2898 NH2 ARG L 66 -16.035 -15.291 17.709 1.00 81.62 L N
+ATOM 2899 N SER L 67 -23.393 -17.307 18.923 1.00 85.10 L N
+ATOM 2900 CA SER L 67 -24.035 -18.517 18.449 1.00 85.10 L C
+ATOM 2901 C SER L 67 -23.653 -18.876 17.022 1.00 85.10 L C
+ATOM 2902 O SER L 67 -23.970 -19.983 16.576 1.00 85.10 L O
+ATOM 2903 CB SER L 67 -25.548 -18.364 18.559 1.00 85.10 L C
+ATOM 2904 OG SER L 67 -26.226 -19.566 18.262 1.00 85.10 L O
+ATOM 2905 N GLY L 68 -22.965 -17.994 16.308 1.00 83.54 L N
+ATOM 2906 CA GLY L 68 -22.666 -18.267 14.914 1.00 83.54 L C
+ATOM 2907 C GLY L 68 -23.970 -18.376 14.163 1.00 83.54 L C
+ATOM 2908 O GLY L 68 -24.576 -17.378 13.760 1.00 83.54 L O
+ATOM 2909 N THR L 69 -24.399 -19.618 13.965 1.00 88.69 L N
+ATOM 2910 CA THR L 69 -25.769 -19.884 13.559 1.00 88.69 L C
+ATOM 2911 C THR L 69 -26.706 -19.246 14.567 1.00 88.69 L C
+ATOM 2912 O THR L 69 -26.400 -19.165 15.757 1.00 88.69 L O
+ATOM 2913 CB THR L 69 -26.025 -21.386 13.504 1.00 88.69 L C
+ATOM 2914 CG2 THR L 69 -24.787 -22.096 13.010 1.00 88.69 L C
+ATOM 2915 OG1 THR L 69 -26.308 -21.862 14.822 1.00 88.69 L O
+ATOM 2916 N ASP L 70 -27.840 -18.753 14.094 1.00 90.92 L N
+ATOM 2917 CA ASP L 70 -28.838 -18.229 15.020 1.00 90.92 L C
+ATOM 2918 C ASP L 70 -29.725 -19.365 15.527 1.00 90.92 L C
+ATOM 2919 O ASP L 70 -30.921 -19.425 15.267 1.00 90.92 L O
+ATOM 2920 CB ASP L 70 -29.654 -17.119 14.368 1.00 90.92 L C
+ATOM 2921 CG ASP L 70 -30.786 -16.624 15.251 1.00 90.92 L C
+ATOM 2922 OD1 ASP L 70 -31.883 -16.377 14.702 1.00 90.92 L O
+ATOM 2923 OD2 ASP L 70 -30.587 -16.537 16.486 1.00 90.92 L O1-
+ATOM 2924 N PHE L 71 -29.109 -20.303 16.251 1.00 93.36 L N
+ATOM 2925 CA PHE L 71 -29.913 -21.065 17.198 1.00 93.36 L C
+ATOM 2926 C PHE L 71 -30.557 -20.125 18.208 1.00 93.36 L C
+ATOM 2927 O PHE L 71 -31.782 -20.114 18.365 1.00 93.36 L O
+ATOM 2928 CB PHE L 71 -29.070 -22.106 17.938 1.00 93.36 L C
+ATOM 2929 CG PHE L 71 -28.589 -23.241 17.084 1.00 93.36 L C
+ATOM 2930 CD1 PHE L 71 -27.269 -23.644 17.125 1.00 93.36 L C
+ATOM 2931 CD2 PHE L 71 -29.471 -23.939 16.278 1.00 93.36 L C
+ATOM 2932 CE1 PHE L 71 -26.833 -24.704 16.356 1.00 93.36 L C
+ATOM 2933 CE2 PHE L 71 -29.039 -25.000 15.507 1.00 93.36 L C
+ATOM 2934 CZ PHE L 71 -27.718 -25.380 15.546 1.00 93.36 L C
+ATOM 2935 N THR L 72 -29.743 -19.288 18.841 1.00 87.95 L N
+ATOM 2936 CA THR L 72 -30.102 -18.306 19.864 1.00 87.95 L C
+ATOM 2937 C THR L 72 -28.991 -17.261 19.866 1.00 87.95 L C
+ATOM 2938 O THR L 72 -28.098 -17.299 19.021 1.00 87.95 L O
+ATOM 2939 CB THR L 72 -30.251 -18.916 21.266 1.00 87.95 L C
+ATOM 2940 CG2 THR L 72 -31.240 -20.071 21.328 1.00 87.95 L C
+ATOM 2941 OG1 THR L 72 -28.968 -19.319 21.769 1.00 87.95 L O
+ATOM 2942 N LEU L 73 -29.069 -16.306 20.796 1.00 84.00 L N
+ATOM 2943 CA LEU L 73 -27.877 -15.685 21.374 1.00 84.00 L C
+ATOM 2944 C LEU L 73 -28.023 -15.615 22.885 1.00 84.00 L C
+ATOM 2945 O LEU L 73 -29.109 -15.315 23.387 1.00 84.00 L O
+ATOM 2946 CB LEU L 73 -27.586 -14.292 20.846 1.00 84.00 L C
+ATOM 2947 CG LEU L 73 -26.713 -14.149 19.597 1.00 84.00 L C
+ATOM 2948 CD1 LEU L 73 -25.289 -14.624 19.903 1.00 84.00 L C
+ATOM 2949 CD2 LEU L 73 -27.255 -14.682 18.274 1.00 84.00 L C
+ATOM 2950 N THR L 74 -26.913 -15.830 23.602 1.00 77.88 L N
+ATOM 2951 CA THR L 74 -26.930 -16.279 24.989 1.00 77.88 L C
+ATOM 2952 C THR L 74 -26.216 -15.290 25.903 1.00 77.88 L C
+ATOM 2953 O THR L 74 -25.286 -14.600 25.482 1.00 77.88 L O
+ATOM 2954 CB THR L 74 -26.239 -17.629 25.123 1.00 77.88 L C
+ATOM 2955 CG2 THR L 74 -26.730 -18.589 24.054 1.00 77.88 L C
+ATOM 2956 OG1 THR L 74 -24.836 -17.451 24.910 1.00 77.88 L O
+ATOM 2957 N ILE L 75 -26.657 -15.232 27.165 1.00 74.66 L N
+ATOM 2958 CA ILE L 75 -26.000 -14.471 28.228 1.00 74.66 L C
+ATOM 2959 C ILE L 75 -25.913 -15.316 29.496 1.00 74.66 L C
+ATOM 2960 O ILE L 75 -26.751 -16.191 29.725 1.00 74.66 L O
+ATOM 2961 CB ILE L 75 -26.734 -13.153 28.531 1.00 74.66 L C
+ATOM 2962 CG1 ILE L 75 -25.824 -12.204 29.300 1.00 74.66 L C
+ATOM 2963 CG2 ILE L 75 -27.968 -13.405 29.316 1.00 74.66 L C
+ATOM 2964 CD1 ILE L 75 -26.502 -10.948 29.723 1.00 74.66 L C
+ATOM 2965 N SER L 76 -24.904 -15.048 30.324 1.00 78.02 L N
+ATOM 2966 CA SER L 76 -24.711 -15.711 31.612 1.00 78.02 L C
+ATOM 2967 C SER L 76 -24.838 -14.709 32.750 1.00 78.02 L C
+ATOM 2968 O SER L 76 -24.023 -13.793 32.857 1.00 78.02 L O
+ATOM 2969 CB SER L 76 -23.337 -16.367 31.675 1.00 78.02 L C
+ATOM 2970 OG SER L 76 -23.083 -17.131 30.515 1.00 78.02 L O
+ATOM 2971 N SER L 77 -25.806 -14.916 33.631 1.00 78.66 L N
+ATOM 2972 CA SER L 77 -26.006 -14.054 34.787 1.00 78.66 L C
+ATOM 2973 C SER L 77 -25.712 -14.837 36.064 1.00 78.66 L C
+ATOM 2974 O SER L 77 -26.058 -16.013 36.168 1.00 78.66 L O
+ATOM 2975 CB SER L 77 -27.441 -13.522 34.814 1.00 78.66 L C
+ATOM 2976 OG SER L 77 -27.704 -12.704 33.697 1.00 78.66 L O
+ATOM 2977 N LEU L 78 -25.052 -14.198 37.038 1.00 87.04 L N
+ATOM 2978 CA LEU L 78 -24.603 -14.925 38.227 1.00 87.04 L C
+ATOM 2979 C LEU L 78 -25.150 -14.414 39.559 1.00 87.04 L C
+ATOM 2980 O LEU L 78 -25.756 -15.200 40.292 1.00 87.04 L O
+ATOM 2981 CB LEU L 78 -23.071 -14.979 38.258 1.00 87.04 L C
+ATOM 2982 CG LEU L 78 -22.427 -16.093 37.432 1.00 87.04 L C
+ATOM 2983 CD1 LEU L 78 -22.543 -15.887 35.949 1.00 87.04 L C
+ATOM 2984 CD2 LEU L 78 -20.980 -16.228 37.821 1.00 87.04 L C
+ATOM 2985 N GLN L 79 -24.970 -13.151 39.915 1.00 87.99 L N
+ATOM 2986 CA GLN L 79 -25.183 -12.724 41.296 1.00 87.99 L C
+ATOM 2987 C GLN L 79 -26.599 -12.210 41.508 1.00 87.99 L C
+ATOM 2988 O GLN L 79 -27.400 -12.140 40.579 1.00 87.99 L O
+ATOM 2989 CB GLN L 79 -24.142 -11.677 41.672 1.00 87.99 L C
+ATOM 2990 CG GLN L 79 -22.748 -12.050 41.270 1.00 87.99 L C
+ATOM 2991 CD GLN L 79 -22.384 -13.470 41.605 1.00 87.99 L C
+ATOM 2992 NE2 GLN L 79 -22.749 -13.908 42.799 1.00 87.99 L N
+ATOM 2993 OE1 GLN L 79 -21.739 -14.156 40.816 1.00 87.99 L O
+ATOM 2994 N PRO L 80 -26.972 -11.913 42.753 1.00 96.09 L N
+ATOM 2995 CA PRO L 80 -28.356 -11.512 43.061 1.00 96.09 L C
+ATOM 2996 C PRO L 80 -28.650 -10.025 43.222 1.00 96.09 L C
+ATOM 2997 O PRO L 80 -29.812 -9.683 43.462 1.00 96.09 L O
+ATOM 2998 CB PRO L 80 -28.583 -12.217 44.396 1.00 96.09 L C
+ATOM 2999 CG PRO L 80 -27.262 -12.091 45.071 1.00 96.09 L C
+ATOM 3000 CD PRO L 80 -26.267 -12.349 43.974 1.00 96.09 L C
+ATOM 3001 N GLU L 81 -27.674 -9.145 43.135 1.00 87.13 L N
+ATOM 3002 CA GLU L 81 -27.985 -7.725 43.135 1.00 87.13 L C
+ATOM 3003 C GLU L 81 -28.314 -7.215 41.738 1.00 87.13 L C
+ATOM 3004 O GLU L 81 -28.747 -6.067 41.591 1.00 87.13 L O
+ATOM 3005 CB GLU L 81 -26.812 -6.940 43.733 1.00 87.13 L C
+ATOM 3006 CG GLU L 81 -25.455 -7.195 43.064 1.00 87.13 L C
+ATOM 3007 CD GLU L 81 -24.836 -8.537 43.432 1.00 87.13 L C
+ATOM 3008 OE1 GLU L 81 -25.585 -9.470 43.775 1.00 87.13 L O
+ATOM 3009 OE2 GLU L 81 -23.595 -8.655 43.382 1.00 87.13 L O1-
+ATOM 3010 N ASP L 82 -28.112 -8.050 40.726 1.00 80.80 L N
+ATOM 3011 CA ASP L 82 -28.323 -7.712 39.324 1.00 80.80 L C
+ATOM 3012 C ASP L 82 -29.641 -8.285 38.811 1.00 80.80 L C
+ATOM 3013 O ASP L 82 -29.657 -9.072 37.877 1.00 80.80 L O
+ATOM 3014 CB ASP L 82 -27.161 -8.240 38.499 1.00 80.80 L C
+ATOM 3015 CG ASP L 82 -25.831 -7.825 39.053 1.00 80.80 L C
+ATOM 3016 OD1 ASP L 82 -25.756 -6.740 39.656 1.00 80.80 L O
+ATOM 3017 OD2 ASP L 82 -24.858 -8.587 38.901 1.00 80.80 L O1-
+ATOM 3018 N PHE L 83 -30.758 -7.902 39.412 1.00 87.28 L N
+ATOM 3019 CA PHE L 83 -32.047 -8.461 39.010 1.00 87.28 L C
+ATOM 3020 C PHE L 83 -32.783 -7.454 38.138 1.00 87.28 L C
+ATOM 3021 O PHE L 83 -33.347 -6.480 38.636 1.00 87.28 L O
+ATOM 3022 CB PHE L 83 -32.881 -8.903 40.212 1.00 87.28 L C
+ATOM 3023 CG PHE L 83 -33.091 -7.855 41.270 1.00 87.28 L C
+ATOM 3024 CD1 PHE L 83 -32.045 -7.429 42.078 1.00 87.28 L C
+ATOM 3025 CD2 PHE L 83 -34.357 -7.348 41.504 1.00 87.28 L C
+ATOM 3026 CE1 PHE L 83 -32.251 -6.483 43.067 1.00 87.28 L C
+ATOM 3027 CE2 PHE L 83 -34.571 -6.402 42.492 1.00 87.28 L C
+ATOM 3028 CZ PHE L 83 -33.516 -5.969 43.274 1.00 87.28 L C
+ATOM 3029 N ALA L 84 -32.789 -7.713 36.828 1.00 73.79 L N
+ATOM 3030 CA ALA L 84 -33.367 -6.818 35.837 1.00 73.79 L C
+ATOM 3031 C ALA L 84 -33.891 -7.620 34.653 1.00 73.79 L C
+ATOM 3032 O ALA L 84 -33.409 -8.712 34.363 1.00 73.79 L O
+ATOM 3033 CB ALA L 84 -32.341 -5.791 35.356 1.00 73.79 L C
+ATOM 3034 N THR L 85 -34.887 -7.057 33.973 1.00 68.25 L N
+ATOM 3035 CA THR L 85 -35.478 -7.664 32.784 1.00 68.25 L C
+ATOM 3036 C THR L 85 -34.535 -7.495 31.610 1.00 68.25 L C
+ATOM 3037 O THR L 85 -34.405 -6.393 31.083 1.00 68.25 L O
+ATOM 3038 CB THR L 85 -36.807 -6.991 32.474 1.00 68.25 L C
+ATOM 3039 CG2 THR L 85 -37.364 -7.487 31.164 1.00 68.25 L C
+ATOM 3040 OG1 THR L 85 -37.740 -7.251 33.524 1.00 68.25 L O
+ATOM 3041 N TYR L 86 -33.883 -8.552 31.166 1.00 67.53 L N
+ATOM 3042 CA TYR L 86 -32.894 -8.338 30.125 1.00 67.53 L C
+ATOM 3043 C TYR L 86 -33.537 -8.429 28.748 1.00 67.53 L C
+ATOM 3044 O TYR L 86 -34.524 -9.137 28.544 1.00 67.53 L O
+ATOM 3045 CB TYR L 86 -31.725 -9.316 30.214 1.00 67.53 L C
+ATOM 3046 CG TYR L 86 -30.975 -9.260 31.519 1.00 67.53 L C
+ATOM 3047 CD1 TYR L 86 -31.275 -8.314 32.473 1.00 67.53 L C
+ATOM 3048 CD2 TYR L 86 -29.885 -10.071 31.742 1.00 67.53 L C
+ATOM 3049 CE1 TYR L 86 -30.594 -8.251 33.662 1.00 67.53 L C
+ATOM 3050 CE2 TYR L 86 -29.181 -9.999 32.922 1.00 67.53 L C
+ATOM 3051 CZ TYR L 86 -29.545 -9.084 33.877 1.00 67.53 L C
+ATOM 3052 OH TYR L 86 -28.853 -8.990 35.056 1.00 67.53 L O
+ATOM 3053 N TYR L 87 -32.969 -7.683 27.803 1.00 64.40 L N
+ATOM 3054 CA TYR L 87 -33.490 -7.544 26.456 1.00 64.40 L C
+ATOM 3055 C TYR L 87 -32.444 -7.997 25.457 1.00 64.40 L C
+ATOM 3056 O TYR L 87 -31.248 -7.866 25.691 1.00 64.40 L O
+ATOM 3057 CB TYR L 87 -33.829 -6.104 26.157 1.00 64.40 L C
+ATOM 3058 CG TYR L 87 -35.011 -5.563 26.874 1.00 64.40 L C
+ATOM 3059 CD1 TYR L 87 -36.285 -5.907 26.501 1.00 64.40 L C
+ATOM 3060 CD2 TYR L 87 -34.853 -4.670 27.904 1.00 64.40 L C
+ATOM 3061 CE1 TYR L 87 -37.364 -5.385 27.145 1.00 64.40 L C
+ATOM 3062 CE2 TYR L 87 -35.922 -4.144 28.550 1.00 64.40 L C
+ATOM 3063 CZ TYR L 87 -37.173 -4.502 28.167 1.00 64.40 L C
+ATOM 3064 OH TYR L 87 -38.246 -3.976 28.825 1.00 64.40 L O
+ATOM 3065 N CYS L 88 -32.900 -8.473 24.307 1.00 70.48 L N
+ATOM 3066 CA CYS L 88 -32.013 -8.886 23.227 1.00 70.48 L C
+ATOM 3067 C CYS L 88 -32.295 -7.947 22.068 1.00 70.48 L C
+ATOM 3068 O CYS L 88 -33.444 -7.818 21.635 1.00 70.48 L O
+ATOM 3069 CB CYS L 88 -32.226 -10.344 22.829 1.00 70.48 L C
+ATOM 3070 SG CYS L 88 -31.083 -10.919 21.541 1.00 70.48 L S
+ATOM 3071 N GLN L 89 -31.257 -7.277 21.588 1.00 64.99 L N
+ATOM 3072 CA GLN L 89 -31.407 -6.080 20.787 1.00 64.99 L C
+ATOM 3073 C GLN L 89 -30.623 -6.329 19.501 1.00 64.99 L C
+ATOM 3074 O GLN L 89 -29.727 -7.170 19.486 1.00 64.99 L O
+ATOM 3075 CB GLN L 89 -30.828 -4.918 21.577 1.00 64.99 L C
+ATOM 3076 CG GLN L 89 -31.004 -3.621 20.998 1.00 64.99 L C
+ATOM 3077 CD GLN L 89 -32.462 -3.367 20.965 1.00 64.99 L C
+ATOM 3078 NE2 GLN L 89 -33.014 -3.135 22.153 1.00 64.99 L N
+ATOM 3079 OE1 GLN L 89 -33.111 -3.429 19.931 1.00 64.99 L O
+ATOM 3080 N GLN L 90 -30.913 -5.608 18.414 1.00 65.24 L N
+ATOM 3081 CA GLN L 90 -30.451 -6.074 17.102 1.00 65.24 L C
+ATOM 3082 C GLN L 90 -29.094 -5.514 16.643 1.00 65.24 L C
+ATOM 3083 O GLN L 90 -28.890 -4.298 16.565 1.00 65.24 L O
+ATOM 3084 CB GLN L 90 -31.496 -5.804 16.020 1.00 65.24 L C
+ATOM 3085 CG GLN L 90 -31.244 -6.682 14.785 1.00 65.24 L C
+ATOM 3086 CD GLN L 90 -31.709 -6.083 13.461 1.00 65.24 L C
+ATOM 3087 NE2 GLN L 90 -31.014 -6.429 12.384 1.00 65.24 L N
+ATOM 3088 OE1 GLN L 90 -32.687 -5.341 13.405 1.00 65.24 L O
+ATOM 3089 N SER L 91 -28.179 -6.448 16.340 1.00 64.81 L N
+ATOM 3090 CA SER L 91 -26.947 -6.418 15.535 1.00 64.81 L C
+ATOM 3091 C SER L 91 -27.196 -6.832 14.075 1.00 64.81 L C
+ATOM 3092 O SER L 91 -28.176 -6.389 13.469 1.00 64.81 L O
+ATOM 3093 CB SER L 91 -25.837 -7.240 16.176 1.00 64.81 L C
+ATOM 3094 OG SER L 91 -25.574 -6.756 17.477 1.00 64.81 L O
+ATOM 3095 N TYR L 92 -26.364 -7.713 13.506 1.00 70.09 L N
+ATOM 3096 CA TYR L 92 -25.549 -7.634 12.296 1.00 70.09 L C
+ATOM 3097 C TYR L 92 -25.704 -6.511 11.281 1.00 70.09 L C
+ATOM 3098 O TYR L 92 -24.732 -5.776 11.096 1.00 70.09 L O
+ATOM 3099 CB TYR L 92 -25.828 -8.928 11.532 1.00 70.09 L C
+ATOM 3100 CG TYR L 92 -25.039 -9.219 10.274 1.00 70.09 L C
+ATOM 3101 CD1 TYR L 92 -23.690 -8.996 10.189 1.00 70.09 L C
+ATOM 3102 CD2 TYR L 92 -25.676 -9.748 9.162 1.00 70.09 L C
+ATOM 3103 CE1 TYR L 92 -22.994 -9.293 9.029 1.00 70.09 L C
+ATOM 3104 CE2 TYR L 92 -24.988 -10.047 8.004 1.00 70.09 L C
+ATOM 3105 CZ TYR L 92 -23.647 -9.819 7.944 1.00 70.09 L C
+ATOM 3106 OH TYR L 92 -22.953 -10.107 6.798 1.00 70.09 L O
+ATOM 3107 N TYR L 93 -26.821 -6.325 10.585 1.00 77.30 L N
+ATOM 3108 CA TYR L 93 -26.875 -5.099 9.788 1.00 77.30 L C
+ATOM 3109 C TYR L 93 -27.115 -3.921 10.709 1.00 77.30 L C
+ATOM 3110 O TYR L 93 -28.158 -3.843 11.364 1.00 77.30 L O
+ATOM 3111 CB TYR L 93 -27.928 -5.132 8.681 1.00 77.30 L C
+ATOM 3112 CG TYR L 93 -27.895 -3.836 7.872 1.00 77.30 L C
+ATOM 3113 CD1 TYR L 93 -26.897 -3.573 6.942 1.00 77.30 L C
+ATOM 3114 CD2 TYR L 93 -28.859 -2.864 8.070 1.00 77.30 L C
+ATOM 3115 CE1 TYR L 93 -26.877 -2.377 6.228 1.00 77.30 L C
+ATOM 3116 CE2 TYR L 93 -28.854 -1.672 7.367 1.00 77.30 L C
+ATOM 3117 CZ TYR L 93 -27.865 -1.429 6.450 1.00 77.30 L C
+ATOM 3118 OH TYR L 93 -27.880 -0.238 5.767 1.00 77.30 L O
+ATOM 3119 N TYR L 94 -26.152 -3.004 10.767 1.00 75.02 L N
+ATOM 3120 CA TYR L 94 -26.160 -2.096 11.898 1.00 75.02 L C
+ATOM 3121 C TYR L 94 -27.361 -1.193 11.713 1.00 75.02 L C
+ATOM 3122 O TYR L 94 -27.299 -0.172 11.025 1.00 75.02 L O
+ATOM 3123 CB TYR L 94 -24.851 -1.322 12.035 1.00 75.02 L C
+ATOM 3124 CG TYR L 94 -24.704 -0.633 13.402 1.00 75.02 L C
+ATOM 3125 CD1 TYR L 94 -25.152 0.663 13.630 1.00 75.02 L C
+ATOM 3126 CD2 TYR L 94 -24.181 -1.333 14.493 1.00 75.02 L C
+ATOM 3127 CE1 TYR L 94 -25.023 1.249 14.883 1.00 75.02 L C
+ATOM 3128 CE2 TYR L 94 -24.062 -0.746 15.746 1.00 75.02 L C
+ATOM 3129 CZ TYR L 94 -24.490 0.537 15.929 1.00 75.02 L C
+ATOM 3130 OH TYR L 94 -24.378 1.108 17.163 1.00 75.02 L O
+ATOM 3131 N PHE L 95 -28.484 -1.644 12.262 1.00 75.76 L N
+ATOM 3132 CA PHE L 95 -29.575 -0.774 12.655 1.00 75.76 L C
+ATOM 3133 C PHE L 95 -29.327 -0.370 14.100 1.00 75.76 L C
+ATOM 3134 O PHE L 95 -29.254 -1.230 14.986 1.00 75.76 L O
+ATOM 3135 CB PHE L 95 -30.940 -1.447 12.554 1.00 75.76 L C
+ATOM 3136 CG PHE L 95 -31.482 -1.569 11.168 1.00 75.76 L C
+ATOM 3137 CD1 PHE L 95 -32.413 -0.653 10.707 1.00 75.76 L C
+ATOM 3138 CD2 PHE L 95 -31.198 -2.669 10.383 1.00 75.76 L C
+ATOM 3139 CE1 PHE L 95 -32.974 -0.765 9.445 1.00 75.76 L C
+ATOM 3140 CE2 PHE L 95 -31.775 -2.802 9.119 1.00 75.76 L C
+ATOM 3141 CZ PHE L 95 -32.653 -1.840 8.648 1.00 75.76 L C
+ATOM 3142 N ARG L 96 -29.172 0.924 14.325 1.00 71.74 L N
+ATOM 3143 CA ARG L 96 -29.177 1.464 15.666 1.00 71.74 L C
+ATOM 3144 C ARG L 96 -30.500 1.061 16.298 1.00 71.74 L C
+ATOM 3145 O ARG L 96 -31.550 1.334 15.705 1.00 71.74 L O
+ATOM 3146 CB ARG L 96 -29.030 2.974 15.614 1.00 71.74 L C
+ATOM 3147 CG ARG L 96 -27.607 3.405 15.366 1.00 71.74 L C
+ATOM 3148 CD ARG L 96 -27.222 3.374 13.898 1.00 71.74 L C
+ATOM 3149 NE ARG L 96 -27.953 4.346 13.092 1.00 71.74 L N
+ATOM 3150 CZ ARG L 96 -27.974 4.357 11.763 1.00 71.74 L C
+ATOM 3151 NH1 ARG L 96 -27.294 3.449 11.081 1.00 71.74 L N1+
+ATOM 3152 NH2 ARG L 96 -28.652 5.293 11.121 1.00 71.74 L N
+ATOM 3153 N PRO L 97 -30.507 0.467 17.510 1.00 70.02 L N
+ATOM 3154 CA PRO L 97 -31.545 -0.515 17.859 1.00 70.02 L C
+ATOM 3155 C PRO L 97 -32.939 -0.238 17.329 1.00 70.02 L C
+ATOM 3156 O PRO L 97 -33.617 0.713 17.726 1.00 70.02 L O
+ATOM 3157 CB PRO L 97 -31.545 -0.484 19.394 1.00 70.02 L C
+ATOM 3158 CG PRO L 97 -30.247 0.099 19.807 1.00 70.02 L C
+ATOM 3159 CD PRO L 97 -29.535 0.646 18.598 1.00 70.02 L C
+ATOM 3160 N ILE L 98 -33.388 -1.139 16.462 1.00 68.00 L N
+ATOM 3161 CA ILE L 98 -34.645 -1.001 15.753 1.00 68.00 L C
+ATOM 3162 C ILE L 98 -35.690 -1.984 16.258 1.00 68.00 L C
+ATOM 3163 O ILE L 98 -36.864 -1.632 16.385 1.00 68.00 L O
+ATOM 3164 CB ILE L 98 -34.430 -1.158 14.229 1.00 68.00 L C
+ATOM 3165 CG1 ILE L 98 -35.686 -0.741 13.461 1.00 68.00 L C
+ATOM 3166 CG2 ILE L 98 -33.966 -2.554 13.864 1.00 68.00 L C
+ATOM 3167 CD1 ILE L 98 -35.511 -0.670 11.964 1.00 68.00 L C
+ATOM 3168 N THR L 99 -35.285 -3.213 16.560 1.00 64.83 L N
+ATOM 3169 CA THR L 99 -36.194 -4.269 16.983 1.00 64.83 L C
+ATOM 3170 C THR L 99 -35.724 -4.780 18.335 1.00 64.83 L C
+ATOM 3171 O THR L 99 -34.657 -5.394 18.430 1.00 64.83 L O
+ATOM 3172 CB THR L 99 -36.230 -5.401 15.956 1.00 64.83 L C
+ATOM 3173 CG2 THR L 99 -37.087 -5.020 14.750 1.00 64.83 L C
+ATOM 3174 OG1 THR L 99 -34.896 -5.688 15.517 1.00 64.83 L O
+ATOM 3175 N PHE L 100 -36.508 -4.538 19.379 1.00 63.20 L N
+ATOM 3176 CA PHE L 100 -36.122 -5.107 20.655 1.00 63.20 L C
+ATOM 3177 C PHE L 100 -36.495 -6.579 20.696 1.00 63.20 L C
+ATOM 3178 O PHE L 100 -37.193 -7.098 19.826 1.00 63.20 L O
+ATOM 3179 CB PHE L 100 -36.799 -4.419 21.848 1.00 63.20 L C
+ATOM 3180 CG PHE L 100 -36.300 -3.037 22.141 1.00 63.20 L C
+ATOM 3181 CD1 PHE L 100 -35.597 -2.314 21.204 1.00 63.20 L C
+ATOM 3182 CD2 PHE L 100 -36.424 -2.520 23.418 1.00 63.20 L C
+ATOM 3183 CE1 PHE L 100 -35.117 -1.075 21.508 1.00 63.20 L C
+ATOM 3184 CE2 PHE L 100 -35.932 -1.288 23.720 1.00 63.20 L C
+ATOM 3185 CZ PHE L 100 -35.279 -0.566 22.771 1.00 63.20 L C
+ATOM 3186 N GLY L 101 -36.038 -7.250 21.738 1.00 72.26 L N
+ATOM 3187 CA GLY L 101 -36.613 -8.516 22.138 1.00 72.26 L C
+ATOM 3188 C GLY L 101 -37.969 -8.272 22.755 1.00 72.26 L C
+ATOM 3189 O GLY L 101 -38.731 -7.404 22.316 1.00 72.26 L O
+ATOM 3190 N GLN L 102 -38.280 -9.034 23.793 1.00 75.09 L N
+ATOM 3191 CA GLN L 102 -39.554 -8.882 24.472 1.00 75.09 L C
+ATOM 3192 C GLN L 102 -39.458 -8.896 25.989 1.00 75.09 L C
+ATOM 3193 O GLN L 102 -40.315 -8.292 26.644 1.00 75.09 L O
+ATOM 3194 CB GLN L 102 -40.523 -9.979 24.021 1.00 75.09 L C
+ATOM 3195 CG GLN L 102 -41.903 -9.817 24.590 1.00 75.09 L C
+ATOM 3196 CD GLN L 102 -42.441 -8.428 24.342 1.00 75.09 L C
+ATOM 3197 NE2 GLN L 102 -42.950 -7.796 25.388 1.00 75.09 L N
+ATOM 3198 OE1 GLN L 102 -42.374 -7.916 23.228 1.00 75.09 L O
+ATOM 3199 N GLY L 103 -38.446 -9.533 26.570 1.00 79.80 L N
+ATOM 3200 CA GLY L 103 -38.296 -9.525 28.010 1.00 79.80 L C
+ATOM 3201 C GLY L 103 -38.088 -10.881 28.651 1.00 79.80 L C
+ATOM 3202 O GLY L 103 -38.781 -11.851 28.336 1.00 79.80 L O
+ATOM 3203 N THR L 104 -37.118 -10.948 29.558 1.00 82.24 L N
+ATOM 3204 CA THR L 104 -36.903 -12.095 30.426 1.00 82.24 L C
+ATOM 3205 C THR L 104 -36.950 -11.572 31.847 1.00 82.24 L C
+ATOM 3206 O THR L 104 -37.400 -10.450 32.074 1.00 82.24 L O
+ATOM 3207 CB THR L 104 -35.565 -12.778 30.162 1.00 82.24 L C
+ATOM 3208 CG2 THR L 104 -35.352 -12.952 28.686 1.00 82.24 L C
+ATOM 3209 OG1 THR L 104 -34.507 -11.970 30.678 1.00 82.24 L O
+ATOM 3210 N LYS L 105 -36.515 -12.363 32.815 1.00 91.16 L N
+ATOM 3211 CA LYS L 105 -36.568 -11.909 34.191 1.00 91.16 L C
+ATOM 3212 C LYS L 105 -35.493 -12.634 34.981 1.00 91.16 L C
+ATOM 3213 O LYS L 105 -35.179 -13.790 34.698 1.00 91.16 L O
+ATOM 3214 CB LYS L 105 -37.946 -12.169 34.785 1.00 91.16 L C
+ATOM 3215 CG LYS L 105 -38.125 -11.637 36.162 1.00 91.16 L C
+ATOM 3216 CD LYS L 105 -38.002 -10.155 36.140 1.00 91.16 L C
+ATOM 3217 CE LYS L 105 -39.150 -9.591 35.344 1.00 91.16 L C
+ATOM 3218 NZ LYS L 105 -40.441 -9.832 36.021 1.00 91.16 L N1+
+ATOM 3219 N VAL L 106 -34.905 -11.943 35.950 1.00 94.84 L N
+ATOM 3220 CA VAL L 106 -33.930 -12.542 36.853 1.00 94.84 L C
+ATOM 3221 C VAL L 106 -34.173 -11.979 38.246 1.00 94.84 L C
+ATOM 3222 O VAL L 106 -34.430 -10.781 38.389 1.00 94.84 L O
+ATOM 3223 CB VAL L 106 -32.487 -12.273 36.396 1.00 94.84 L C
+ATOM 3224 CG1 VAL L 106 -31.514 -12.843 37.394 1.00 94.84 L C
+ATOM 3225 CG2 VAL L 106 -32.232 -12.870 35.026 1.00 94.84 L C
+ATOM 3226 N GLU L 107 -34.131 -12.841 39.272 1.00112.44 L N
+ATOM 3227 CA GLU L 107 -34.346 -12.429 40.657 1.00112.44 L C
+ATOM 3228 C GLU L 107 -33.448 -13.227 41.595 1.00112.44 L C
+ATOM 3229 O GLU L 107 -32.935 -14.284 41.230 1.00112.44 L O
+ATOM 3230 CB GLU L 107 -35.804 -12.612 41.100 1.00112.44 L C
+ATOM 3231 CG GLU L 107 -36.824 -11.910 40.237 1.00112.44 L C
+ATOM 3232 CD GLU L 107 -37.661 -12.879 39.447 1.00112.44 L C
+ATOM 3233 OE1 GLU L 107 -38.850 -12.589 39.212 1.00112.44 L O
+ATOM 3234 OE2 GLU L 107 -37.128 -13.939 39.067 1.00112.44 L O1-
+ATOM 3235 N ILE L 108 -33.258 -12.700 42.809 1.00118.45 L N
+ATOM 3236 CA ILE L 108 -32.611 -13.442 43.892 1.00118.45 L C
+ATOM 3237 C ILE L 108 -33.566 -14.475 44.477 1.00118.45 L C
+ATOM 3238 O ILE L 108 -34.752 -14.192 44.689 1.00118.45 L O
+ATOM 3239 CB ILE L 108 -32.134 -12.474 44.982 1.00118.45 L C
+ATOM 3240 CG1 ILE L 108 -31.514 -13.238 46.148 1.00118.45 L C
+ATOM 3241 CG2 ILE L 108 -33.272 -11.581 45.463 1.00118.45 L C
+ATOM 3242 CD1 ILE L 108 -30.910 -12.327 47.184 1.00118.45 L C
+ATOM 3243 N LYS L 109 -33.062 -15.682 44.761 1.00118.04 L N
+ATOM 3244 CA LYS L 109 -33.923 -16.676 45.405 1.00118.04 L C
+ATOM 3245 C LYS L 109 -33.654 -16.733 46.914 1.00118.04 L C
+ATOM 3246 O LYS L 109 -32.730 -17.372 47.423 1.00118.04 L O
+ATOM 3247 CB LYS L 109 -33.816 -18.055 44.754 1.00118.04 L C
+ATOM 3248 CG LYS L 109 -32.528 -18.827 44.890 1.00118.04 L C
+ATOM 3249 CD LYS L 109 -32.635 -20.158 44.167 1.00118.04 L C
+ATOM 3250 CE LYS L 109 -31.341 -20.937 44.263 1.00118.04 L C
+ATOM 3251 NZ LYS L 109 -31.059 -21.360 45.666 1.00118.04 L N1+
+TER
+END
diff --git a/examples/pdbs/cd86.pdb b/examples/pdbs/cd86.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..9715f48a0b03147a5f1883ab6dfc042ccbad52b5
--- /dev/null
+++ b/examples/pdbs/cd86.pdb
@@ -0,0 +1,1790 @@
+SSBOND CYS B 16 CYS B 86 2.04
+ATOM 1 N MET B 1 -11.523 -4.666 -13.325 1.00 0.00 N
+ATOM 2 CA MET B 1 -11.657 -5.250 -12.001 1.00 0.00 C
+ATOM 3 C MET B 1 -11.626 -6.767 -12.081 1.00 0.00 C
+ATOM 4 O MET B 1 -12.522 -7.374 -12.658 1.00 0.00 O
+ATOM 5 CB MET B 1 -12.980 -4.828 -11.359 1.00 0.00 C
+ATOM 6 CG MET B 1 -13.186 -5.333 -9.941 1.00 0.00 C
+ATOM 7 SD MET B 1 -14.855 -4.922 -9.277 1.00 0.00 S
+ATOM 8 CE MET B 1 -15.934 -6.117 -10.125 1.00 0.00 C
+ATOM 9 1H MET B 1 -11.545 -3.669 -13.254 1.00 0.00 H
+ATOM 10 2H MET B 1 -10.654 -4.951 -13.729 1.00 0.00 H
+ATOM 11 3H MET B 1 -12.277 -4.975 -13.904 1.00 0.00 H
+ATOM 12 HA MET B 1 -10.819 -4.926 -11.381 1.00 0.00 H
+ATOM 13 1HB MET B 1 -13.042 -3.742 -11.333 1.00 0.00 H
+ATOM 14 2HB MET B 1 -13.812 -5.189 -11.966 1.00 0.00 H
+ATOM 15 1HG MET B 1 -13.052 -6.419 -9.911 1.00 0.00 H
+ATOM 16 2HG MET B 1 -12.434 -4.885 -9.288 1.00 0.00 H
+ATOM 17 1HE MET B 1 -16.969 -5.967 -9.806 1.00 0.00 H
+ATOM 18 2HE MET B 1 -15.869 -5.976 -11.204 1.00 0.00 H
+ATOM 19 3HE MET B 1 -15.623 -7.125 -9.865 1.00 0.00 H
+ATOM 20 N LEU B 2 -10.615 -7.380 -11.494 1.00 0.00 N
+ATOM 21 CA LEU B 2 -10.549 -8.838 -11.489 1.00 0.00 C
+ATOM 22 C LEU B 2 -11.553 -9.405 -10.513 1.00 0.00 C
+ATOM 23 O LEU B 2 -11.592 -8.952 -9.382 1.00 0.00 O
+ATOM 24 CB LEU B 2 -9.151 -9.285 -11.083 1.00 0.00 C
+ATOM 25 CG LEU B 2 -8.928 -10.773 -10.966 1.00 0.00 C
+ATOM 26 CD1 LEU B 2 -9.076 -11.385 -12.335 1.00 0.00 C
+ATOM 27 CD2 LEU B 2 -7.544 -11.010 -10.356 1.00 0.00 C
+ATOM 28 H LEU B 2 -9.897 -6.832 -11.012 1.00 0.00 H
+ATOM 29 HA LEU B 2 -10.786 -9.209 -12.486 1.00 0.00 H
+ATOM 30 1HB LEU B 2 -8.476 -8.941 -11.851 1.00 0.00 H
+ATOM 31 2HB LEU B 2 -8.883 -8.815 -10.155 1.00 0.00 H
+ATOM 32 HG LEU B 2 -9.682 -11.219 -10.315 1.00 0.00 H
+ATOM 33 1HD1 LEU B 2 -8.928 -12.455 -12.287 1.00 0.00 H
+ATOM 34 2HD1 LEU B 2 -10.073 -11.183 -12.706 1.00 0.00 H
+ATOM 35 3HD1 LEU B 2 -8.340 -10.950 -13.006 1.00 0.00 H
+ATOM 36 1HD2 LEU B 2 -7.373 -12.078 -10.268 1.00 0.00 H
+ATOM 37 2HD2 LEU B 2 -6.784 -10.574 -11.000 1.00 0.00 H
+ATOM 38 3HD2 LEU B 2 -7.495 -10.554 -9.373 1.00 0.00 H
+ATOM 39 N LYS B 3 -12.368 -10.368 -10.932 1.00 0.00 N
+ATOM 40 CA LYS B 3 -13.319 -11.010 -10.018 1.00 0.00 C
+ATOM 41 C LYS B 3 -12.734 -12.328 -9.530 1.00 0.00 C
+ATOM 42 O LYS B 3 -12.499 -13.232 -10.329 1.00 0.00 O
+ATOM 43 CB LYS B 3 -14.640 -11.234 -10.741 1.00 0.00 C
+ATOM 44 CG LYS B 3 -15.351 -9.936 -11.096 1.00 0.00 C
+ATOM 45 CD LYS B 3 -16.584 -10.175 -11.952 1.00 0.00 C
+ATOM 46 CE LYS B 3 -17.703 -10.886 -11.175 1.00 0.00 C
+ATOM 47 NZ LYS B 3 -18.302 -10.032 -10.077 1.00 0.00 N
+ATOM 48 H LYS B 3 -12.333 -10.678 -11.905 1.00 0.00 H
+ATOM 49 HA LYS B 3 -13.485 -10.366 -9.154 1.00 0.00 H
+ATOM 50 1HB LYS B 3 -14.454 -11.777 -11.658 1.00 0.00 H
+ATOM 51 2HB LYS B 3 -15.299 -11.835 -10.120 1.00 0.00 H
+ATOM 52 1HG LYS B 3 -15.657 -9.447 -10.168 1.00 0.00 H
+ATOM 53 2HG LYS B 3 -14.664 -9.272 -11.625 1.00 0.00 H
+ATOM 54 1HD LYS B 3 -16.959 -9.211 -12.306 1.00 0.00 H
+ATOM 55 2HD LYS B 3 -16.309 -10.777 -12.820 1.00 0.00 H
+ATOM 56 1HE LYS B 3 -18.489 -11.148 -11.881 1.00 0.00 H
+ATOM 57 2HE LYS B 3 -17.308 -11.802 -10.733 1.00 0.00 H
+ATOM 58 1HZ LYS B 3 -19.035 -10.545 -9.619 1.00 0.00 H
+ATOM 59 2HZ LYS B 3 -17.607 -9.774 -9.363 1.00 0.00 H
+ATOM 60 3HZ LYS B 3 -18.686 -9.186 -10.468 1.00 0.00 H
+ATOM 61 N ILE B 4 -12.482 -12.429 -8.236 1.00 0.00 N
+ATOM 62 CA ILE B 4 -11.827 -13.594 -7.647 1.00 0.00 C
+ATOM 63 C ILE B 4 -12.871 -14.398 -6.909 1.00 0.00 C
+ATOM 64 O ILE B 4 -13.567 -13.869 -6.035 1.00 0.00 O
+ATOM 65 CB ILE B 4 -10.711 -13.183 -6.643 1.00 0.00 C
+ATOM 66 CG1 ILE B 4 -9.664 -12.314 -7.330 1.00 0.00 C
+ATOM 67 CG2 ILE B 4 -10.055 -14.434 -6.036 1.00 0.00 C
+ATOM 68 CD1 ILE B 4 -8.648 -11.715 -6.368 1.00 0.00 C
+ATOM 69 H ILE B 4 -12.727 -11.656 -7.633 1.00 0.00 H
+ATOM 70 HA ILE B 4 -11.404 -14.213 -8.431 1.00 0.00 H
+ATOM 71 HB ILE B 4 -11.142 -12.593 -5.855 1.00 0.00 H
+ATOM 72 1HG1 ILE B 4 -9.131 -12.906 -8.065 1.00 0.00 H
+ATOM 73 2HG1 ILE B 4 -10.173 -11.498 -7.843 1.00 0.00 H
+ATOM 74 1HG2 ILE B 4 -9.285 -14.136 -5.329 1.00 0.00 H
+ATOM 75 2HG2 ILE B 4 -10.811 -15.028 -5.521 1.00 0.00 H
+ATOM 76 3HG2 ILE B 4 -9.612 -15.029 -6.832 1.00 0.00 H
+ATOM 77 1HD1 ILE B 4 -7.934 -11.110 -6.918 1.00 0.00 H
+ATOM 78 2HD1 ILE B 4 -9.162 -11.091 -5.636 1.00 0.00 H
+ATOM 79 3HD1 ILE B 4 -8.116 -12.513 -5.857 1.00 0.00 H
+ATOM 80 N GLN B 5 -13.022 -15.650 -7.303 1.00 0.00 N
+ATOM 81 CA GLN B 5 -13.997 -16.546 -6.722 1.00 0.00 C
+ATOM 82 C GLN B 5 -13.284 -17.636 -5.930 1.00 0.00 C
+ATOM 83 O GLN B 5 -12.791 -18.615 -6.487 1.00 0.00 O
+ATOM 84 CB GLN B 5 -14.860 -17.109 -7.846 1.00 0.00 C
+ATOM 85 CG GLN B 5 -15.622 -16.001 -8.572 1.00 0.00 C
+ATOM 86 CD GLN B 5 -16.449 -16.494 -9.765 1.00 0.00 C
+ATOM 87 OE1 GLN B 5 -16.414 -17.676 -10.105 1.00 0.00 O
+ATOM 88 NE2 GLN B 5 -17.165 -15.576 -10.424 1.00 0.00 N
+ATOM 89 H GLN B 5 -12.425 -15.996 -8.048 1.00 0.00 H
+ATOM 90 HA GLN B 5 -14.629 -15.990 -6.032 1.00 0.00 H
+ATOM 91 1HB GLN B 5 -14.226 -17.624 -8.571 1.00 0.00 H
+ATOM 92 2HB GLN B 5 -15.568 -17.829 -7.443 1.00 0.00 H
+ATOM 93 1HG GLN B 5 -16.292 -15.521 -7.856 1.00 0.00 H
+ATOM 94 2HG GLN B 5 -14.898 -15.273 -8.946 1.00 0.00 H
+ATOM 95 1HE2 GLN B 5 -17.715 -15.824 -11.260 1.00 0.00 H
+ATOM 96 2HE2 GLN B 5 -17.156 -14.631 -10.117 1.00 0.00 H
+ATOM 97 N ALA B 6 -13.185 -17.420 -4.630 1.00 0.00 N
+ATOM 98 CA ALA B 6 -12.441 -18.294 -3.734 1.00 0.00 C
+ATOM 99 C ALA B 6 -13.418 -18.912 -2.748 1.00 0.00 C
+ATOM 100 O ALA B 6 -14.549 -18.455 -2.663 1.00 0.00 O
+ATOM 101 CB ALA B 6 -11.363 -17.516 -3.008 1.00 0.00 C
+ATOM 102 H ALA B 6 -13.644 -16.601 -4.226 1.00 0.00 H
+ATOM 103 HA ALA B 6 -11.985 -19.092 -4.315 1.00 0.00 H
+ATOM 104 1HB ALA B 6 -10.816 -18.187 -2.349 1.00 0.00 H
+ATOM 105 2HB ALA B 6 -10.678 -17.075 -3.728 1.00 0.00 H
+ATOM 106 3HB ALA B 6 -11.838 -16.735 -2.426 1.00 0.00 H
+ATOM 107 N TYR B 7 -12.997 -19.949 -2.047 1.00 0.00 N
+ATOM 108 CA TYR B 7 -13.860 -20.618 -1.071 1.00 0.00 C
+ATOM 109 C TYR B 7 -13.493 -20.423 0.393 1.00 0.00 C
+ATOM 110 O TYR B 7 -12.336 -20.204 0.743 1.00 0.00 O
+ATOM 111 CB TYR B 7 -13.969 -22.088 -1.430 1.00 0.00 C
+ATOM 112 CG TYR B 7 -14.950 -22.333 -2.525 1.00 0.00 C
+ATOM 113 CD1 TYR B 7 -14.671 -22.032 -3.853 1.00 0.00 C
+ATOM 114 CD2 TYR B 7 -16.146 -22.904 -2.195 1.00 0.00 C
+ATOM 115 CE1 TYR B 7 -15.628 -22.284 -4.811 1.00 0.00 C
+ATOM 116 CE2 TYR B 7 -17.075 -23.164 -3.151 1.00 0.00 C
+ATOM 117 CZ TYR B 7 -16.828 -22.860 -4.443 1.00 0.00 C
+ATOM 118 OH TYR B 7 -17.779 -23.129 -5.372 1.00 0.00 O
+ATOM 119 H TYR B 7 -12.058 -20.294 -2.182 1.00 0.00 H
+ATOM 120 HA TYR B 7 -14.858 -20.197 -1.186 1.00 0.00 H
+ATOM 121 1HB TYR B 7 -13.009 -22.461 -1.756 1.00 0.00 H
+ATOM 122 2HB TYR B 7 -14.275 -22.664 -0.550 1.00 0.00 H
+ATOM 123 HD1 TYR B 7 -13.720 -21.587 -4.134 1.00 0.00 H
+ATOM 124 HD2 TYR B 7 -16.357 -23.150 -1.157 1.00 0.00 H
+ATOM 125 HE1 TYR B 7 -15.435 -22.037 -5.852 1.00 0.00 H
+ATOM 126 HE2 TYR B 7 -18.023 -23.617 -2.882 1.00 0.00 H
+ATOM 127 HH TYR B 7 -18.571 -23.448 -4.924 1.00 0.00 H
+ATOM 128 N PHE B 8 -14.515 -20.507 1.218 1.00 0.00 N
+ATOM 129 CA PHE B 8 -14.415 -20.423 2.658 1.00 0.00 C
+ATOM 130 C PHE B 8 -13.392 -21.443 3.168 1.00 0.00 C
+ATOM 131 O PHE B 8 -13.373 -22.600 2.739 1.00 0.00 O
+ATOM 132 CB PHE B 8 -15.818 -20.644 3.248 1.00 0.00 C
+ATOM 133 CG PHE B 8 -15.920 -20.531 4.706 1.00 0.00 C
+ATOM 134 CD1 PHE B 8 -15.961 -19.290 5.312 1.00 0.00 C
+ATOM 135 CD2 PHE B 8 -16.033 -21.661 5.497 1.00 0.00 C
+ATOM 136 CE1 PHE B 8 -16.085 -19.184 6.700 1.00 0.00 C
+ATOM 137 CE2 PHE B 8 -16.159 -21.558 6.871 1.00 0.00 C
+ATOM 138 CZ PHE B 8 -16.175 -20.319 7.471 1.00 0.00 C
+ATOM 139 H PHE B 8 -15.431 -20.639 0.810 1.00 0.00 H
+ATOM 140 HA PHE B 8 -14.076 -19.425 2.933 1.00 0.00 H
+ATOM 141 1HB PHE B 8 -16.504 -19.921 2.802 1.00 0.00 H
+ATOM 142 2HB PHE B 8 -16.172 -21.632 2.962 1.00 0.00 H
+ATOM 143 HD1 PHE B 8 -15.884 -18.398 4.690 1.00 0.00 H
+ATOM 144 HD2 PHE B 8 -16.015 -22.645 5.025 1.00 0.00 H
+ATOM 145 HE1 PHE B 8 -16.108 -18.210 7.183 1.00 0.00 H
+ATOM 146 HE2 PHE B 8 -16.235 -22.459 7.478 1.00 0.00 H
+ATOM 147 HZ PHE B 8 -16.270 -20.236 8.554 1.00 0.00 H
+ATOM 148 N ASN B 9 -12.515 -20.962 4.058 1.00 0.00 N
+ATOM 149 CA ASN B 9 -11.374 -21.648 4.676 1.00 0.00 C
+ATOM 150 C ASN B 9 -10.251 -21.973 3.686 1.00 0.00 C
+ATOM 151 O ASN B 9 -9.306 -22.664 4.040 1.00 0.00 O
+ATOM 152 CB ASN B 9 -11.761 -22.943 5.369 1.00 0.00 C
+ATOM 153 CG ASN B 9 -12.696 -22.805 6.526 1.00 0.00 C
+ATOM 154 OD1 ASN B 9 -13.721 -23.490 6.528 1.00 0.00 O
+ATOM 155 ND2 ASN B 9 -12.384 -21.975 7.496 1.00 0.00 N
+ATOM 156 H ASN B 9 -12.642 -19.999 4.346 1.00 0.00 H
+ATOM 157 HA ASN B 9 -10.958 -20.968 5.418 1.00 0.00 H
+ATOM 158 1HB ASN B 9 -12.203 -23.627 4.647 1.00 0.00 H
+ATOM 159 2HB ASN B 9 -10.851 -23.423 5.735 1.00 0.00 H
+ATOM 160 1HD2 ASN B 9 -12.988 -21.872 8.286 1.00 0.00 H
+ATOM 161 2HD2 ASN B 9 -11.503 -21.465 7.468 1.00 0.00 H
+ATOM 162 N GLU B 10 -10.297 -21.413 2.487 1.00 0.00 N
+ATOM 163 CA GLU B 10 -9.240 -21.594 1.503 1.00 0.00 C
+ATOM 164 C GLU B 10 -8.424 -20.309 1.466 1.00 0.00 C
+ATOM 165 O GLU B 10 -8.542 -19.482 2.368 1.00 0.00 O
+ATOM 166 CB GLU B 10 -9.796 -21.954 0.114 1.00 0.00 C
+ATOM 167 CG GLU B 10 -10.707 -23.213 0.095 1.00 0.00 C
+ATOM 168 CD GLU B 10 -10.006 -24.522 0.419 1.00 0.00 C
+ATOM 169 OE1 GLU B 10 -8.831 -24.627 0.168 1.00 0.00 O
+ATOM 170 OE2 GLU B 10 -10.658 -25.419 0.922 1.00 0.00 O
+ATOM 171 H GLU B 10 -11.111 -20.872 2.199 1.00 0.00 H
+ATOM 172 HA GLU B 10 -8.586 -22.405 1.825 1.00 0.00 H
+ATOM 173 1HB GLU B 10 -10.340 -21.114 -0.298 1.00 0.00 H
+ATOM 174 2HB GLU B 10 -8.969 -22.157 -0.562 1.00 0.00 H
+ATOM 175 1HG GLU B 10 -11.529 -23.061 0.802 1.00 0.00 H
+ATOM 176 2HG GLU B 10 -11.133 -23.305 -0.892 1.00 0.00 H
+ATOM 177 N THR B 11 -7.509 -20.198 0.514 1.00 0.00 N
+ATOM 178 CA THR B 11 -6.663 -19.013 0.422 1.00 0.00 C
+ATOM 179 C THR B 11 -6.951 -18.212 -0.864 1.00 0.00 C
+ATOM 180 O THR B 11 -7.147 -18.801 -1.923 1.00 0.00 O
+ATOM 181 CB THR B 11 -5.176 -19.439 0.521 1.00 0.00 C
+ATOM 182 OG1 THR B 11 -4.953 -20.093 1.805 1.00 0.00 O
+ATOM 183 CG2 THR B 11 -4.243 -18.262 0.408 1.00 0.00 C
+ATOM 184 H THR B 11 -7.427 -20.925 -0.182 1.00 0.00 H
+ATOM 185 HA THR B 11 -6.886 -18.367 1.267 1.00 0.00 H
+ATOM 186 HB THR B 11 -4.948 -20.136 -0.283 1.00 0.00 H
+ATOM 187 HG1 THR B 11 -4.075 -19.817 2.197 1.00 0.00 H
+ATOM 188 1HG2 THR B 11 -3.227 -18.620 0.486 1.00 0.00 H
+ATOM 189 2HG2 THR B 11 -4.383 -17.767 -0.554 1.00 0.00 H
+ATOM 190 3HG2 THR B 11 -4.430 -17.563 1.199 1.00 0.00 H
+ATOM 191 N ALA B 12 -7.056 -16.870 -0.755 1.00 0.00 N
+ATOM 192 CA ALA B 12 -7.223 -16.036 -1.955 1.00 0.00 C
+ATOM 193 C ALA B 12 -5.857 -15.540 -2.396 1.00 0.00 C
+ATOM 194 O ALA B 12 -5.035 -15.158 -1.561 1.00 0.00 O
+ATOM 195 CB ALA B 12 -8.124 -14.816 -1.725 1.00 0.00 C
+ATOM 196 H ALA B 12 -6.937 -16.446 0.165 1.00 0.00 H
+ATOM 197 HA ALA B 12 -7.648 -16.650 -2.750 1.00 0.00 H
+ATOM 198 1HB ALA B 12 -8.186 -14.248 -2.653 1.00 0.00 H
+ATOM 199 2HB ALA B 12 -9.112 -15.113 -1.433 1.00 0.00 H
+ATOM 200 3HB ALA B 12 -7.703 -14.186 -0.955 1.00 0.00 H
+ATOM 201 N ASP B 13 -5.653 -15.443 -3.695 1.00 0.00 N
+ATOM 202 CA ASP B 13 -4.438 -14.841 -4.243 1.00 0.00 C
+ATOM 203 C ASP B 13 -4.829 -13.486 -4.795 1.00 0.00 C
+ATOM 204 O ASP B 13 -5.617 -13.405 -5.734 1.00 0.00 O
+ATOM 205 CB ASP B 13 -3.903 -15.732 -5.348 1.00 0.00 C
+ATOM 206 CG ASP B 13 -3.464 -17.075 -4.812 1.00 0.00 C
+ATOM 207 OD1 ASP B 13 -4.228 -17.973 -5.017 1.00 0.00 O
+ATOM 208 OD2 ASP B 13 -2.405 -17.222 -4.238 1.00 0.00 O
+ATOM 209 H ASP B 13 -6.337 -15.845 -4.321 1.00 0.00 H
+ATOM 210 HA ASP B 13 -3.690 -14.711 -3.459 1.00 0.00 H
+ATOM 211 1HB ASP B 13 -4.669 -15.883 -6.110 1.00 0.00 H
+ATOM 212 2HB ASP B 13 -3.071 -15.257 -5.834 1.00 0.00 H
+ATOM 213 N LEU B 14 -4.296 -12.426 -4.195 1.00 0.00 N
+ATOM 214 CA LEU B 14 -4.600 -11.049 -4.566 1.00 0.00 C
+ATOM 215 C LEU B 14 -3.399 -10.477 -5.354 1.00 0.00 C
+ATOM 216 O LEU B 14 -2.393 -10.097 -4.742 1.00 0.00 O
+ATOM 217 CB LEU B 14 -4.810 -10.229 -3.279 1.00 0.00 C
+ATOM 218 CG LEU B 14 -5.888 -10.785 -2.244 1.00 0.00 C
+ATOM 219 CD1 LEU B 14 -5.885 -9.890 -0.987 1.00 0.00 C
+ATOM 220 CD2 LEU B 14 -7.297 -10.826 -2.888 1.00 0.00 C
+ATOM 221 H LEU B 14 -3.651 -12.573 -3.423 1.00 0.00 H
+ATOM 222 HA LEU B 14 -5.507 -11.032 -5.156 1.00 0.00 H
+ATOM 223 1HB LEU B 14 -3.857 -10.172 -2.754 1.00 0.00 H
+ATOM 224 2HB LEU B 14 -5.107 -9.221 -3.557 1.00 0.00 H
+ATOM 225 HG LEU B 14 -5.603 -11.797 -1.938 1.00 0.00 H
+ATOM 226 1HD1 LEU B 14 -6.606 -10.272 -0.261 1.00 0.00 H
+ATOM 227 2HD1 LEU B 14 -4.890 -9.892 -0.545 1.00 0.00 H
+ATOM 228 3HD1 LEU B 14 -6.156 -8.878 -1.262 1.00 0.00 H
+ATOM 229 1HD2 LEU B 14 -8.010 -11.213 -2.165 1.00 0.00 H
+ATOM 230 2HD2 LEU B 14 -7.601 -9.825 -3.193 1.00 0.00 H
+ATOM 231 3HD2 LEU B 14 -7.271 -11.476 -3.756 1.00 0.00 H
+ATOM 232 N PRO B 15 -3.402 -10.516 -6.698 1.00 0.00 N
+ATOM 233 CA PRO B 15 -2.265 -10.194 -7.567 1.00 0.00 C
+ATOM 234 C PRO B 15 -2.023 -8.717 -7.776 1.00 0.00 C
+ATOM 235 O PRO B 15 -2.952 -7.911 -7.717 1.00 0.00 O
+ATOM 236 CB PRO B 15 -2.670 -10.837 -8.879 1.00 0.00 C
+ATOM 237 CG PRO B 15 -4.180 -10.681 -8.908 1.00 0.00 C
+ATOM 238 CD PRO B 15 -4.632 -10.853 -7.456 1.00 0.00 C
+ATOM 239 HA PRO B 15 -1.355 -10.670 -7.177 1.00 0.00 H
+ATOM 240 1HB PRO B 15 -2.149 -10.343 -9.687 1.00 0.00 H
+ATOM 241 2HB PRO B 15 -2.349 -11.890 -8.892 1.00 0.00 H
+ATOM 242 1HG PRO B 15 -4.455 -9.699 -9.324 1.00 0.00 H
+ATOM 243 2HG PRO B 15 -4.622 -11.445 -9.574 1.00 0.00 H
+ATOM 244 1HD PRO B 15 -5.451 -10.155 -7.231 1.00 0.00 H
+ATOM 245 2HD PRO B 15 -4.915 -11.883 -7.323 1.00 0.00 H
+ATOM 246 N CYS B 16 -0.820 -8.392 -8.202 1.00 0.00 N
+ATOM 247 CA CYS B 16 -0.532 -7.050 -8.676 1.00 0.00 C
+ATOM 248 C CYS B 16 -0.563 -7.005 -10.200 1.00 0.00 C
+ATOM 249 O CYS B 16 -0.798 -5.951 -10.788 1.00 0.00 O
+ATOM 250 CB CYS B 16 0.816 -6.574 -8.149 1.00 0.00 C
+ATOM 251 SG CYS B 16 0.852 -6.438 -6.340 1.00 0.00 S
+ATOM 252 H CYS B 16 -0.074 -9.079 -8.181 1.00 0.00 H
+ATOM 253 HA CYS B 16 -1.295 -6.380 -8.299 1.00 0.00 H
+ATOM 254 1HB CYS B 16 1.600 -7.271 -8.449 1.00 0.00 H
+ATOM 255 2HB CYS B 16 1.053 -5.601 -8.572 1.00 0.00 H
+ATOM 256 N GLN B 17 -0.284 -8.144 -10.856 1.00 0.00 N
+ATOM 257 CA GLN B 17 -0.250 -8.208 -12.330 1.00 0.00 C
+ATOM 258 C GLN B 17 0.586 -7.070 -12.933 1.00 0.00 C
+ATOM 259 O GLN B 17 0.121 -6.309 -13.786 1.00 0.00 O
+ATOM 260 CB GLN B 17 -1.666 -8.247 -12.946 1.00 0.00 C
+ATOM 261 CG GLN B 17 -2.491 -9.482 -12.515 1.00 0.00 C
+ATOM 262 CD GLN B 17 -3.769 -9.708 -13.306 1.00 0.00 C
+ATOM 263 OE1 GLN B 17 -4.234 -8.873 -14.092 1.00 0.00 O
+ATOM 264 NE2 GLN B 17 -4.353 -10.883 -13.104 1.00 0.00 N
+ATOM 265 H GLN B 17 -0.101 -8.980 -10.325 1.00 0.00 H
+ATOM 266 HA GLN B 17 0.235 -9.141 -12.613 1.00 0.00 H
+ATOM 267 1HB GLN B 17 -2.220 -7.355 -12.643 1.00 0.00 H
+ATOM 268 2HB GLN B 17 -1.596 -8.241 -14.032 1.00 0.00 H
+ATOM 269 1HG GLN B 17 -1.874 -10.381 -12.573 1.00 0.00 H
+ATOM 270 2HG GLN B 17 -2.801 -9.300 -11.498 1.00 0.00 H
+ATOM 271 1HE2 GLN B 17 -5.190 -11.111 -13.600 1.00 0.00 H
+ATOM 272 2HE2 GLN B 17 -3.950 -11.560 -12.480 1.00 0.00 H
+ATOM 273 N PHE B 18 1.809 -6.938 -12.430 1.00 0.00 N
+ATOM 274 CA PHE B 18 2.723 -5.899 -12.855 1.00 0.00 C
+ATOM 275 C PHE B 18 3.839 -6.478 -13.691 1.00 0.00 C
+ATOM 276 O PHE B 18 4.627 -7.286 -13.207 1.00 0.00 O
+ATOM 277 CB PHE B 18 3.292 -5.189 -11.628 1.00 0.00 C
+ATOM 278 CG PHE B 18 4.258 -4.018 -11.891 1.00 0.00 C
+ATOM 279 CD1 PHE B 18 3.870 -2.881 -12.574 1.00 0.00 C
+ATOM 280 CD2 PHE B 18 5.517 -4.065 -11.392 1.00 0.00 C
+ATOM 281 CE1 PHE B 18 4.742 -1.821 -12.741 1.00 0.00 C
+ATOM 282 CE2 PHE B 18 6.411 -3.011 -11.539 1.00 0.00 C
+ATOM 283 CZ PHE B 18 6.017 -1.884 -12.215 1.00 0.00 C
+ATOM 284 H PHE B 18 2.112 -7.599 -11.732 1.00 0.00 H
+ATOM 285 HA PHE B 18 2.177 -5.173 -13.458 1.00 0.00 H
+ATOM 286 1HB PHE B 18 2.469 -4.817 -11.028 1.00 0.00 H
+ATOM 287 2HB PHE B 18 3.819 -5.918 -11.015 1.00 0.00 H
+ATOM 288 HD1 PHE B 18 2.865 -2.823 -12.979 1.00 0.00 H
+ATOM 289 HD2 PHE B 18 5.809 -4.960 -10.865 1.00 0.00 H
+ATOM 290 HE1 PHE B 18 4.415 -0.933 -13.283 1.00 0.00 H
+ATOM 291 HE2 PHE B 18 7.418 -3.078 -11.122 1.00 0.00 H
+ATOM 292 HZ PHE B 18 6.708 -1.051 -12.341 1.00 0.00 H
+ATOM 293 N ALA B 19 3.890 -6.062 -14.956 1.00 0.00 N
+ATOM 294 CA ALA B 19 4.885 -6.518 -15.929 1.00 0.00 C
+ATOM 295 C ALA B 19 6.290 -6.177 -15.526 1.00 0.00 C
+ATOM 296 O ALA B 19 7.238 -6.887 -15.858 1.00 0.00 O
+ATOM 297 CB ALA B 19 4.628 -5.887 -17.274 1.00 0.00 C
+ATOM 298 H ALA B 19 3.188 -5.406 -15.263 1.00 0.00 H
+ATOM 299 HA ALA B 19 4.804 -7.602 -16.013 1.00 0.00 H
+ATOM 300 1HB ALA B 19 5.349 -6.253 -18.002 1.00 0.00 H
+ATOM 301 2HB ALA B 19 3.656 -6.158 -17.562 1.00 0.00 H
+ATOM 302 3HB ALA B 19 4.709 -4.807 -17.198 1.00 0.00 H
+ATOM 303 N ASN B 20 6.446 -5.052 -14.855 1.00 0.00 N
+ATOM 304 CA ASN B 20 7.769 -4.593 -14.504 1.00 0.00 C
+ATOM 305 C ASN B 20 8.639 -4.502 -15.748 1.00 0.00 C
+ATOM 306 O ASN B 20 9.781 -4.948 -15.775 1.00 0.00 O
+ATOM 307 CB ASN B 20 8.370 -5.487 -13.443 1.00 0.00 C
+ATOM 308 CG ASN B 20 9.541 -4.898 -12.830 1.00 0.00 C
+ATOM 309 OD1 ASN B 20 9.770 -3.684 -12.936 1.00 0.00 O
+ATOM 310 ND2 ASN B 20 10.291 -5.708 -12.126 1.00 0.00 N
+ATOM 311 H ASN B 20 5.635 -4.518 -14.575 1.00 0.00 H
+ATOM 312 HA ASN B 20 7.686 -3.589 -14.096 1.00 0.00 H
+ATOM 313 1HB ASN B 20 7.627 -5.695 -12.670 1.00 0.00 H
+ATOM 314 2HB ASN B 20 8.663 -6.438 -13.876 1.00 0.00 H
+ATOM 315 1HD2 ASN B 20 11.094 -5.351 -11.611 1.00 0.00 H
+ATOM 316 2HD2 ASN B 20 10.048 -6.676 -12.053 1.00 0.00 H
+ATOM 317 N SER B 21 8.110 -3.839 -16.757 1.00 0.00 N
+ATOM 318 CA SER B 21 8.788 -3.654 -18.032 1.00 0.00 C
+ATOM 319 C SER B 21 10.092 -2.861 -17.904 1.00 0.00 C
+ATOM 320 O SER B 21 10.938 -2.895 -18.792 1.00 0.00 O
+ATOM 321 CB SER B 21 7.838 -2.960 -18.987 1.00 0.00 C
+ATOM 322 OG SER B 21 7.558 -1.640 -18.564 1.00 0.00 O
+ATOM 323 H SER B 21 7.173 -3.488 -16.651 1.00 0.00 H
+ATOM 324 HA SER B 21 9.032 -4.636 -18.437 1.00 0.00 H
+ATOM 325 1HB SER B 21 8.269 -2.946 -19.986 1.00 0.00 H
+ATOM 326 2HB SER B 21 6.910 -3.531 -19.039 1.00 0.00 H
+ATOM 327 HG SER B 21 6.784 -1.362 -19.067 1.00 0.00 H
+ATOM 328 N GLN B 22 10.239 -2.136 -16.794 1.00 0.00 N
+ATOM 329 CA GLN B 22 11.416 -1.331 -16.530 1.00 0.00 C
+ATOM 330 C GLN B 22 12.445 -2.107 -15.701 1.00 0.00 C
+ATOM 331 O GLN B 22 13.485 -1.564 -15.342 1.00 0.00 O
+ATOM 332 CB GLN B 22 11.013 -0.021 -15.849 1.00 0.00 C
+ATOM 333 CG GLN B 22 10.014 0.794 -16.688 1.00 0.00 C
+ATOM 334 CD GLN B 22 10.531 1.112 -18.067 1.00 0.00 C
+ATOM 335 OE1 GLN B 22 11.541 1.810 -18.251 1.00 0.00 O
+ATOM 336 NE2 GLN B 22 9.846 0.553 -19.071 1.00 0.00 N
+ATOM 337 H GLN B 22 9.494 -2.148 -16.116 1.00 0.00 H
+ATOM 338 HA GLN B 22 11.881 -1.084 -17.484 1.00 0.00 H
+ATOM 339 1HB GLN B 22 10.528 -0.240 -14.906 1.00 0.00 H
+ATOM 340 2HB GLN B 22 11.894 0.589 -15.641 1.00 0.00 H
+ATOM 341 1HG GLN B 22 9.089 0.226 -16.794 1.00 0.00 H
+ATOM 342 2HG GLN B 22 9.813 1.743 -16.172 1.00 0.00 H
+ATOM 343 1HE2 GLN B 22 10.140 0.693 -20.017 1.00 0.00 H
+ATOM 344 2HE2 GLN B 22 9.031 -0.034 -18.880 1.00 0.00 H
+ATOM 345 N ASN B 23 12.151 -3.378 -15.393 1.00 0.00 N
+ATOM 346 CA ASN B 23 13.048 -4.244 -14.632 1.00 0.00 C
+ATOM 347 C ASN B 23 13.444 -3.631 -13.284 1.00 0.00 C
+ATOM 348 O ASN B 23 14.614 -3.630 -12.911 1.00 0.00 O
+ATOM 349 CB ASN B 23 14.297 -4.565 -15.438 1.00 0.00 C
+ATOM 350 CG ASN B 23 15.042 -5.775 -14.920 1.00 0.00 C
+ATOM 351 OD1 ASN B 23 14.421 -6.772 -14.520 1.00 0.00 O
+ATOM 352 ND2 ASN B 23 16.363 -5.711 -14.915 1.00 0.00 N
+ATOM 353 H ASN B 23 11.273 -3.785 -15.706 1.00 0.00 H
+ATOM 354 HA ASN B 23 12.518 -5.172 -14.421 1.00 0.00 H
+ATOM 355 1HB ASN B 23 14.011 -4.756 -16.474 1.00 0.00 H
+ATOM 356 2HB ASN B 23 14.970 -3.712 -15.437 1.00 0.00 H
+ATOM 357 1HD2 ASN B 23 16.903 -6.486 -14.583 1.00 0.00 H
+ATOM 358 2HD2 ASN B 23 16.826 -4.885 -15.242 1.00 0.00 H
+ATOM 359 N GLN B 24 12.466 -3.119 -12.544 1.00 0.00 N
+ATOM 360 CA GLN B 24 12.700 -2.487 -11.255 1.00 0.00 C
+ATOM 361 C GLN B 24 12.769 -3.528 -10.155 1.00 0.00 C
+ATOM 362 O GLN B 24 12.178 -4.605 -10.272 1.00 0.00 O
+ATOM 363 CB GLN B 24 11.577 -1.501 -10.986 1.00 0.00 C
+ATOM 364 CG GLN B 24 11.580 -0.433 -11.986 1.00 0.00 C
+ATOM 365 CD GLN B 24 10.440 0.531 -11.880 1.00 0.00 C
+ATOM 366 OE1 GLN B 24 9.381 0.358 -12.499 1.00 0.00 O
+ATOM 367 NE2 GLN B 24 10.650 1.558 -11.107 1.00 0.00 N
+ATOM 368 H GLN B 24 11.512 -3.173 -12.876 1.00 0.00 H
+ATOM 369 HA GLN B 24 13.653 -1.957 -11.287 1.00 0.00 H
+ATOM 370 1HB GLN B 24 10.619 -2.009 -11.011 1.00 0.00 H
+ATOM 371 2HB GLN B 24 11.702 -1.060 -9.990 1.00 0.00 H
+ATOM 372 1HG GLN B 24 12.511 0.130 -11.895 1.00 0.00 H
+ATOM 373 2HG GLN B 24 11.540 -0.906 -12.958 1.00 0.00 H
+ATOM 374 1HE2 GLN B 24 9.957 2.298 -11.060 1.00 0.00 H
+ATOM 375 2HE2 GLN B 24 11.527 1.665 -10.640 1.00 0.00 H
+ATOM 376 N SER B 25 13.508 -3.216 -9.094 1.00 0.00 N
+ATOM 377 CA SER B 25 13.626 -4.114 -7.948 1.00 0.00 C
+ATOM 378 C SER B 25 12.602 -3.830 -6.869 1.00 0.00 C
+ATOM 379 O SER B 25 12.289 -2.675 -6.609 1.00 0.00 O
+ATOM 380 CB SER B 25 15.000 -3.986 -7.323 1.00 0.00 C
+ATOM 381 OG SER B 25 15.079 -4.726 -6.121 1.00 0.00 O
+ATOM 382 H SER B 25 13.994 -2.308 -9.056 1.00 0.00 H
+ATOM 383 HA SER B 25 13.492 -5.140 -8.298 1.00 0.00 H
+ATOM 384 1HB SER B 25 15.748 -4.347 -8.015 1.00 0.00 H
+ATOM 385 2HB SER B 25 15.211 -2.935 -7.128 1.00 0.00 H
+ATOM 386 HG SER B 25 15.126 -5.659 -6.415 1.00 0.00 H
+ATOM 387 N LEU B 26 12.185 -4.849 -6.121 1.00 0.00 N
+ATOM 388 CA LEU B 26 11.342 -4.562 -4.959 1.00 0.00 C
+ATOM 389 C LEU B 26 12.027 -3.637 -3.976 1.00 0.00 C
+ATOM 390 O LEU B 26 11.354 -2.931 -3.241 1.00 0.00 O
+ATOM 391 CB LEU B 26 10.943 -5.825 -4.182 1.00 0.00 C
+ATOM 392 CG LEU B 26 9.914 -6.720 -4.810 1.00 0.00 C
+ATOM 393 CD1 LEU B 26 9.813 -8.007 -4.022 1.00 0.00 C
+ATOM 394 CD2 LEU B 26 8.546 -5.967 -4.782 1.00 0.00 C
+ATOM 395 H LEU B 26 12.455 -5.797 -6.357 1.00 0.00 H
+ATOM 396 HA LEU B 26 10.437 -4.072 -5.311 1.00 0.00 H
+ATOM 397 1HB LEU B 26 11.838 -6.417 -4.002 1.00 0.00 H
+ATOM 398 2HB LEU B 26 10.545 -5.510 -3.224 1.00 0.00 H
+ATOM 399 HG LEU B 26 10.200 -6.962 -5.823 1.00 0.00 H
+ATOM 400 1HD1 LEU B 26 9.058 -8.654 -4.475 1.00 0.00 H
+ATOM 401 2HD1 LEU B 26 10.775 -8.517 -4.031 1.00 0.00 H
+ATOM 402 3HD1 LEU B 26 9.533 -7.775 -2.998 1.00 0.00 H
+ATOM 403 1HD2 LEU B 26 7.773 -6.590 -5.214 1.00 0.00 H
+ATOM 404 2HD2 LEU B 26 8.280 -5.734 -3.750 1.00 0.00 H
+ATOM 405 3HD2 LEU B 26 8.624 -5.041 -5.351 1.00 0.00 H
+ATOM 406 N SER B 27 13.358 -3.626 -3.958 1.00 0.00 N
+ATOM 407 CA SER B 27 14.138 -2.805 -3.044 1.00 0.00 C
+ATOM 408 C SER B 27 14.020 -1.305 -3.304 1.00 0.00 C
+ATOM 409 O SER B 27 14.498 -0.498 -2.510 1.00 0.00 O
+ATOM 410 CB SER B 27 15.570 -3.161 -3.150 1.00 0.00 C
+ATOM 411 OG SER B 27 15.986 -2.811 -4.408 1.00 0.00 O
+ATOM 412 H SER B 27 13.865 -4.195 -4.630 1.00 0.00 H
+ATOM 413 HA SER B 27 13.815 -3.011 -2.047 1.00 0.00 H
+ATOM 414 1HB SER B 27 16.127 -2.600 -2.417 1.00 0.00 H
+ATOM 415 2HB SER B 27 15.728 -4.222 -2.968 1.00 0.00 H
+ATOM 416 HG SER B 27 15.663 -3.509 -4.993 1.00 0.00 H
+ATOM 417 N GLU B 28 13.465 -0.944 -4.465 1.00 0.00 N
+ATOM 418 CA GLU B 28 13.263 0.425 -4.898 1.00 0.00 C
+ATOM 419 C GLU B 28 11.788 0.792 -4.756 1.00 0.00 C
+ATOM 420 O GLU B 28 11.393 1.935 -5.006 1.00 0.00 O
+ATOM 421 CB GLU B 28 13.683 0.540 -6.372 1.00 0.00 C
+ATOM 422 CG GLU B 28 15.156 0.217 -6.622 1.00 0.00 C
+ATOM 423 CD GLU B 28 15.564 0.148 -8.102 1.00 0.00 C
+ATOM 424 OE1 GLU B 28 14.896 -0.523 -8.893 1.00 0.00 O
+ATOM 425 OE2 GLU B 28 16.547 0.769 -8.429 1.00 0.00 O
+ATOM 426 H GLU B 28 13.120 -1.668 -5.091 1.00 0.00 H
+ATOM 427 HA GLU B 28 13.856 1.094 -4.277 1.00 0.00 H
+ATOM 428 1HB GLU B 28 13.078 -0.140 -6.979 1.00 0.00 H
+ATOM 429 2HB GLU B 28 13.493 1.553 -6.724 1.00 0.00 H
+ATOM 430 1HG GLU B 28 15.764 0.969 -6.120 1.00 0.00 H
+ATOM 431 2HG GLU B 28 15.368 -0.740 -6.154 1.00 0.00 H
+ATOM 432 N LEU B 29 10.968 -0.205 -4.428 1.00 0.00 N
+ATOM 433 CA LEU B 29 9.529 -0.030 -4.409 1.00 0.00 C
+ATOM 434 C LEU B 29 8.915 -0.208 -3.028 1.00 0.00 C
+ATOM 435 O LEU B 29 9.491 -0.855 -2.144 1.00 0.00 O
+ATOM 436 CB LEU B 29 8.873 -1.047 -5.359 1.00 0.00 C
+ATOM 437 CG LEU B 29 9.353 -1.055 -6.776 1.00 0.00 C
+ATOM 438 CD1 LEU B 29 8.685 -2.189 -7.508 1.00 0.00 C
+ATOM 439 CD2 LEU B 29 9.080 0.280 -7.438 1.00 0.00 C
+ATOM 440 H LEU B 29 11.333 -1.117 -4.171 1.00 0.00 H
+ATOM 441 HA LEU B 29 9.306 0.973 -4.760 1.00 0.00 H
+ATOM 442 1HB LEU B 29 9.033 -2.040 -4.951 1.00 0.00 H
+ATOM 443 2HB LEU B 29 7.797 -0.853 -5.378 1.00 0.00 H
+ATOM 444 HG LEU B 29 10.411 -1.233 -6.789 1.00 0.00 H
+ATOM 445 1HD1 LEU B 29 9.051 -2.221 -8.537 1.00 0.00 H
+ATOM 446 2HD1 LEU B 29 8.933 -3.116 -7.006 1.00 0.00 H
+ATOM 447 3HD1 LEU B 29 7.619 -2.050 -7.508 1.00 0.00 H
+ATOM 448 1HD2 LEU B 29 9.446 0.264 -8.464 1.00 0.00 H
+ATOM 449 2HD2 LEU B 29 8.007 0.483 -7.432 1.00 0.00 H
+ATOM 450 3HD2 LEU B 29 9.599 1.040 -6.886 1.00 0.00 H
+ATOM 451 N VAL B 30 7.737 0.357 -2.870 1.00 0.00 N
+ATOM 452 CA VAL B 30 6.932 0.112 -1.701 1.00 0.00 C
+ATOM 453 C VAL B 30 5.608 -0.454 -2.208 1.00 0.00 C
+ATOM 454 O VAL B 30 4.954 0.162 -3.055 1.00 0.00 O
+ATOM 455 CB VAL B 30 6.732 1.384 -0.886 1.00 0.00 C
+ATOM 456 CG1 VAL B 30 5.862 1.062 0.319 1.00 0.00 C
+ATOM 457 CG2 VAL B 30 8.119 1.966 -0.481 1.00 0.00 C
+ATOM 458 H VAL B 30 7.375 0.927 -3.631 1.00 0.00 H
+ATOM 459 HA VAL B 30 7.421 -0.631 -1.077 1.00 0.00 H
+ATOM 460 HB VAL B 30 6.198 2.114 -1.477 1.00 0.00 H
+ATOM 461 1HG1 VAL B 30 5.689 1.961 0.907 1.00 0.00 H
+ATOM 462 2HG1 VAL B 30 4.911 0.666 -0.023 1.00 0.00 H
+ATOM 463 3HG1 VAL B 30 6.349 0.311 0.939 1.00 0.00 H
+ATOM 464 1HG2 VAL B 30 7.978 2.872 0.095 1.00 0.00 H
+ATOM 465 2HG2 VAL B 30 8.666 1.235 0.116 1.00 0.00 H
+ATOM 466 3HG2 VAL B 30 8.692 2.199 -1.376 1.00 0.00 H
+ATOM 467 N VAL B 31 5.243 -1.658 -1.748 1.00 0.00 N
+ATOM 468 CA VAL B 31 4.024 -2.285 -2.235 1.00 0.00 C
+ATOM 469 C VAL B 31 3.044 -2.518 -1.099 1.00 0.00 C
+ATOM 470 O VAL B 31 3.362 -3.174 -0.106 1.00 0.00 O
+ATOM 471 CB VAL B 31 4.318 -3.640 -2.915 1.00 0.00 C
+ATOM 472 CG1 VAL B 31 3.003 -4.244 -3.423 1.00 0.00 C
+ATOM 473 CG2 VAL B 31 5.345 -3.453 -4.035 1.00 0.00 C
+ATOM 474 H VAL B 31 5.836 -2.126 -1.066 1.00 0.00 H
+ATOM 475 HA VAL B 31 3.556 -1.628 -2.963 1.00 0.00 H
+ATOM 476 HB VAL B 31 4.706 -4.331 -2.185 1.00 0.00 H
+ATOM 477 1HG1 VAL B 31 3.195 -5.208 -3.886 1.00 0.00 H
+ATOM 478 2HG1 VAL B 31 2.311 -4.370 -2.596 1.00 0.00 H
+ATOM 479 3HG1 VAL B 31 2.552 -3.577 -4.154 1.00 0.00 H
+ATOM 480 1HG2 VAL B 31 5.548 -4.410 -4.512 1.00 0.00 H
+ATOM 481 2HG2 VAL B 31 4.964 -2.769 -4.750 1.00 0.00 H
+ATOM 482 3HG2 VAL B 31 6.276 -3.051 -3.620 1.00 0.00 H
+ATOM 483 N PHE B 32 1.844 -1.989 -1.253 1.00 0.00 N
+ATOM 484 CA PHE B 32 0.813 -2.161 -0.241 1.00 0.00 C
+ATOM 485 C PHE B 32 -0.354 -2.937 -0.776 1.00 0.00 C
+ATOM 486 O PHE B 32 -0.732 -2.777 -1.938 1.00 0.00 O
+ATOM 487 CB PHE B 32 0.200 -0.827 0.178 1.00 0.00 C
+ATOM 488 CG PHE B 32 -0.539 -0.043 -0.823 1.00 0.00 C
+ATOM 489 CD1 PHE B 32 -1.905 -0.236 -0.992 1.00 0.00 C
+ATOM 490 CD2 PHE B 32 0.124 0.855 -1.629 1.00 0.00 C
+ATOM 491 CE1 PHE B 32 -2.593 0.463 -1.963 1.00 0.00 C
+ATOM 492 CE2 PHE B 32 -0.560 1.567 -2.613 1.00 0.00 C
+ATOM 493 CZ PHE B 32 -1.911 1.369 -2.778 1.00 0.00 C
+ATOM 494 H PHE B 32 1.643 -1.459 -2.103 1.00 0.00 H
+ATOM 495 HA PHE B 32 1.225 -2.685 0.621 1.00 0.00 H
+ATOM 496 1HB PHE B 32 -0.475 -1.008 1.015 1.00 0.00 H
+ATOM 497 2HB PHE B 32 1.000 -0.190 0.564 1.00 0.00 H
+ATOM 498 HD1 PHE B 32 -2.427 -0.946 -0.356 1.00 0.00 H
+ATOM 499 HD2 PHE B 32 1.191 0.999 -1.490 1.00 0.00 H
+ATOM 500 HE1 PHE B 32 -3.666 0.310 -2.086 1.00 0.00 H
+ATOM 501 HE2 PHE B 32 -0.025 2.278 -3.250 1.00 0.00 H
+ATOM 502 HZ PHE B 32 -2.448 1.927 -3.545 1.00 0.00 H
+ATOM 503 N TRP B 33 -0.992 -3.702 0.099 1.00 0.00 N
+ATOM 504 CA TRP B 33 -2.300 -4.241 -0.234 1.00 0.00 C
+ATOM 505 C TRP B 33 -3.255 -3.656 0.752 1.00 0.00 C
+ATOM 506 O TRP B 33 -3.005 -3.734 1.959 1.00 0.00 O
+ATOM 507 CB TRP B 33 -2.394 -5.760 -0.170 1.00 0.00 C
+ATOM 508 CG TRP B 33 -1.658 -6.440 -1.276 1.00 0.00 C
+ATOM 509 CD1 TRP B 33 -2.151 -6.975 -2.445 1.00 0.00 C
+ATOM 510 CD2 TRP B 33 -0.264 -6.621 -1.304 1.00 0.00 C
+ATOM 511 NE1 TRP B 33 -1.124 -7.492 -3.188 1.00 0.00 N
+ATOM 512 CE2 TRP B 33 0.050 -7.270 -2.504 1.00 0.00 C
+ATOM 513 CE3 TRP B 33 0.745 -6.287 -0.416 1.00 0.00 C
+ATOM 514 CZ2 TRP B 33 1.342 -7.581 -2.823 1.00 0.00 C
+ATOM 515 CZ3 TRP B 33 2.000 -6.586 -0.728 1.00 0.00 C
+ATOM 516 CH2 TRP B 33 2.304 -7.205 -1.887 1.00 0.00 C
+ATOM 517 H TRP B 33 -0.577 -3.866 1.017 1.00 0.00 H
+ATOM 518 HA TRP B 33 -2.569 -3.926 -1.230 1.00 0.00 H
+ATOM 519 1HB TRP B 33 -1.981 -6.082 0.771 1.00 0.00 H
+ATOM 520 2HB TRP B 33 -3.441 -6.067 -0.200 1.00 0.00 H
+ATOM 521 HD1 TRP B 33 -3.202 -6.989 -2.732 1.00 0.00 H
+ATOM 522 HE1 TRP B 33 -1.225 -7.954 -4.093 1.00 0.00 H
+ATOM 523 HE3 TRP B 33 0.513 -5.780 0.520 1.00 0.00 H
+ATOM 524 HZ2 TRP B 33 1.618 -8.085 -3.753 1.00 0.00 H
+ATOM 525 HZ3 TRP B 33 2.767 -6.301 -0.023 1.00 0.00 H
+ATOM 526 HH2 TRP B 33 3.330 -7.389 -2.074 1.00 0.00 H
+ATOM 527 N GLN B 34 -4.361 -3.118 0.243 1.00 0.00 N
+ATOM 528 CA GLN B 34 -5.364 -2.535 1.117 1.00 0.00 C
+ATOM 529 C GLN B 34 -6.781 -2.962 0.744 1.00 0.00 C
+ATOM 530 O GLN B 34 -7.067 -3.359 -0.387 1.00 0.00 O
+ATOM 531 CB GLN B 34 -5.247 -0.988 1.140 1.00 0.00 C
+ATOM 532 CG GLN B 34 -5.554 -0.289 -0.177 1.00 0.00 C
+ATOM 533 CD GLN B 34 -5.350 1.258 -0.098 1.00 0.00 C
+ATOM 534 OE1 GLN B 34 -4.543 1.756 0.678 1.00 0.00 O
+ATOM 535 NE2 GLN B 34 -6.071 1.999 -0.906 1.00 0.00 N
+ATOM 536 H GLN B 34 -4.468 -3.092 -0.773 1.00 0.00 H
+ATOM 537 HA GLN B 34 -5.169 -2.908 2.112 1.00 0.00 H
+ATOM 538 1HB GLN B 34 -5.931 -0.594 1.888 1.00 0.00 H
+ATOM 539 2HB GLN B 34 -4.243 -0.703 1.451 1.00 0.00 H
+ATOM 540 1HG GLN B 34 -4.896 -0.691 -0.953 1.00 0.00 H
+ATOM 541 2HG GLN B 34 -6.602 -0.478 -0.434 1.00 0.00 H
+ATOM 542 1HE2 GLN B 34 -5.979 2.991 -0.890 1.00 0.00 H
+ATOM 543 2HE2 GLN B 34 -6.721 1.571 -1.565 1.00 0.00 H
+ATOM 544 N ASP B 35 -7.661 -2.946 1.735 1.00 0.00 N
+ATOM 545 CA ASP B 35 -9.057 -3.336 1.523 1.00 0.00 C
+ATOM 546 C ASP B 35 -9.902 -2.123 1.127 1.00 0.00 C
+ATOM 547 O ASP B 35 -9.376 -1.027 0.918 1.00 0.00 O
+ATOM 548 CB ASP B 35 -9.618 -4.085 2.754 1.00 0.00 C
+ATOM 549 CG ASP B 35 -9.816 -3.257 4.032 1.00 0.00 C
+ATOM 550 OD1 ASP B 35 -9.867 -2.036 3.971 1.00 0.00 O
+ATOM 551 OD2 ASP B 35 -9.910 -3.870 5.090 1.00 0.00 O
+ATOM 552 H ASP B 35 -7.335 -2.632 2.652 1.00 0.00 H
+ATOM 553 HA ASP B 35 -9.096 -4.046 0.698 1.00 0.00 H
+ATOM 554 1HB ASP B 35 -10.567 -4.545 2.486 1.00 0.00 H
+ATOM 555 2HB ASP B 35 -8.946 -4.896 2.993 1.00 0.00 H
+ATOM 556 N GLN B 36 -11.223 -2.303 1.052 1.00 0.00 N
+ATOM 557 CA GLN B 36 -12.142 -1.258 0.626 1.00 0.00 C
+ATOM 558 C GLN B 36 -12.298 -0.106 1.624 1.00 0.00 C
+ATOM 559 O GLN B 36 -12.903 0.910 1.301 1.00 0.00 O
+ATOM 560 CB GLN B 36 -13.485 -1.891 0.272 1.00 0.00 C
+ATOM 561 CG GLN B 36 -14.233 -2.460 1.414 1.00 0.00 C
+ATOM 562 CD GLN B 36 -15.424 -3.300 0.930 1.00 0.00 C
+ATOM 563 OE1 GLN B 36 -15.281 -4.164 0.038 1.00 0.00 O
+ATOM 564 NE2 GLN B 36 -16.598 -3.039 1.512 1.00 0.00 N
+ATOM 565 H GLN B 36 -11.594 -3.221 1.240 1.00 0.00 H
+ATOM 566 HA GLN B 36 -11.739 -0.829 -0.290 1.00 0.00 H
+ATOM 567 1HB GLN B 36 -14.115 -1.169 -0.242 1.00 0.00 H
+ATOM 568 2HB GLN B 36 -13.307 -2.713 -0.407 1.00 0.00 H
+ATOM 569 1HG GLN B 36 -13.565 -3.094 1.999 1.00 0.00 H
+ATOM 570 2HG GLN B 36 -14.606 -1.642 2.034 1.00 0.00 H
+ATOM 571 1HE2 GLN B 36 -17.455 -3.551 1.259 1.00 0.00 H
+ATOM 572 2HE2 GLN B 36 -16.656 -2.331 2.218 1.00 0.00 H
+ATOM 573 N GLU B 37 -11.785 -0.270 2.852 1.00 0.00 N
+ATOM 574 CA GLU B 37 -11.830 0.780 3.870 1.00 0.00 C
+ATOM 575 C GLU B 37 -10.461 1.462 3.936 1.00 0.00 C
+ATOM 576 O GLU B 37 -10.196 2.297 4.805 1.00 0.00 O
+ATOM 577 CB GLU B 37 -12.223 0.212 5.244 1.00 0.00 C
+ATOM 578 CG GLU B 37 -13.617 -0.381 5.318 1.00 0.00 C
+ATOM 579 CD GLU B 37 -13.991 -0.876 6.715 1.00 0.00 C
+ATOM 580 OE1 GLU B 37 -13.429 -0.403 7.695 1.00 0.00 O
+ATOM 581 OE2 GLU B 37 -14.838 -1.740 6.793 1.00 0.00 O
+ATOM 582 H GLU B 37 -11.268 -1.121 3.093 1.00 0.00 H
+ATOM 583 HA GLU B 37 -12.566 1.528 3.580 1.00 0.00 H
+ATOM 584 1HB GLU B 37 -11.539 -0.595 5.500 1.00 0.00 H
+ATOM 585 2HB GLU B 37 -12.125 0.982 6.008 1.00 0.00 H
+ATOM 586 1HG GLU B 37 -14.338 0.371 5.001 1.00 0.00 H
+ATOM 587 2HG GLU B 37 -13.674 -1.217 4.616 1.00 0.00 H
+ATOM 588 N ASN B 38 -9.605 1.080 2.981 1.00 0.00 N
+ATOM 589 CA ASN B 38 -8.211 1.484 2.829 1.00 0.00 C
+ATOM 590 C ASN B 38 -7.346 1.040 4.010 1.00 0.00 C
+ATOM 591 O ASN B 38 -6.320 1.660 4.317 1.00 0.00 O
+ATOM 592 CB ASN B 38 -8.099 2.982 2.587 1.00 0.00 C
+ATOM 593 CG ASN B 38 -8.763 3.413 1.290 1.00 0.00 C
+ATOM 594 OD1 ASN B 38 -8.467 2.895 0.205 1.00 0.00 O
+ATOM 595 ND2 ASN B 38 -9.660 4.352 1.391 1.00 0.00 N
+ATOM 596 H ASN B 38 -9.919 0.401 2.295 1.00 0.00 H
+ATOM 597 HA ASN B 38 -7.819 0.975 1.945 1.00 0.00 H
+ATOM 598 1HB ASN B 38 -8.533 3.537 3.416 1.00 0.00 H
+ATOM 599 2HB ASN B 38 -7.043 3.249 2.537 1.00 0.00 H
+ATOM 600 1HD2 ASN B 38 -10.138 4.678 0.572 1.00 0.00 H
+ATOM 601 2HD2 ASN B 38 -9.876 4.743 2.287 1.00 0.00 H
+ATOM 602 N LEU B 39 -7.710 -0.078 4.634 1.00 0.00 N
+ATOM 603 CA LEU B 39 -6.893 -0.609 5.702 1.00 0.00 C
+ATOM 604 C LEU B 39 -5.835 -1.475 5.055 1.00 0.00 C
+ATOM 605 O LEU B 39 -6.090 -2.099 4.032 1.00 0.00 O
+ATOM 606 CB LEU B 39 -7.747 -1.406 6.672 1.00 0.00 C
+ATOM 607 CG LEU B 39 -8.874 -0.623 7.333 1.00 0.00 C
+ATOM 608 CD1 LEU B 39 -9.682 -1.563 8.188 1.00 0.00 C
+ATOM 609 CD2 LEU B 39 -8.298 0.533 8.127 1.00 0.00 C
+ATOM 610 H LEU B 39 -8.564 -0.587 4.368 1.00 0.00 H
+ATOM 611 HA LEU B 39 -6.406 0.209 6.234 1.00 0.00 H
+ATOM 612 1HB LEU B 39 -8.194 -2.239 6.137 1.00 0.00 H
+ATOM 613 2HB LEU B 39 -7.109 -1.804 7.457 1.00 0.00 H
+ATOM 614 HG LEU B 39 -9.531 -0.222 6.565 1.00 0.00 H
+ATOM 615 1HD1 LEU B 39 -10.507 -1.022 8.646 1.00 0.00 H
+ATOM 616 2HD1 LEU B 39 -10.079 -2.361 7.554 1.00 0.00 H
+ATOM 617 3HD1 LEU B 39 -9.065 -1.986 8.943 1.00 0.00 H
+ATOM 618 1HD2 LEU B 39 -9.097 1.089 8.583 1.00 0.00 H
+ATOM 619 2HD2 LEU B 39 -7.635 0.155 8.894 1.00 0.00 H
+ATOM 620 3HD2 LEU B 39 -7.740 1.191 7.459 1.00 0.00 H
+ATOM 621 N VAL B 40 -4.644 -1.524 5.635 1.00 0.00 N
+ATOM 622 CA VAL B 40 -3.555 -2.305 5.065 1.00 0.00 C
+ATOM 623 C VAL B 40 -3.481 -3.779 5.524 1.00 0.00 C
+ATOM 624 O VAL B 40 -3.515 -4.092 6.719 1.00 0.00 O
+ATOM 625 CB VAL B 40 -2.233 -1.558 5.338 1.00 0.00 C
+ATOM 626 CG1 VAL B 40 -1.039 -2.354 4.877 1.00 0.00 C
+ATOM 627 CG2 VAL B 40 -2.255 -0.201 4.602 1.00 0.00 C
+ATOM 628 H VAL B 40 -4.480 -0.992 6.484 1.00 0.00 H
+ATOM 629 HA VAL B 40 -3.692 -2.307 3.982 1.00 0.00 H
+ATOM 630 HB VAL B 40 -2.140 -1.402 6.376 1.00 0.00 H
+ATOM 631 1HG1 VAL B 40 -0.119 -1.807 5.091 1.00 0.00 H
+ATOM 632 2HG1 VAL B 40 -1.025 -3.290 5.396 1.00 0.00 H
+ATOM 633 3HG1 VAL B 40 -1.116 -2.532 3.802 1.00 0.00 H
+ATOM 634 1HG2 VAL B 40 -1.329 0.335 4.804 1.00 0.00 H
+ATOM 635 2HG2 VAL B 40 -2.346 -0.378 3.529 1.00 0.00 H
+ATOM 636 3HG2 VAL B 40 -3.105 0.399 4.941 1.00 0.00 H
+ATOM 637 N LEU B 41 -3.393 -4.679 4.539 1.00 0.00 N
+ATOM 638 CA LEU B 41 -3.257 -6.125 4.773 1.00 0.00 C
+ATOM 639 C LEU B 41 -1.792 -6.481 4.951 1.00 0.00 C
+ATOM 640 O LEU B 41 -1.423 -7.334 5.753 1.00 0.00 O
+ATOM 641 CB LEU B 41 -3.731 -6.944 3.573 1.00 0.00 C
+ATOM 642 CG LEU B 41 -5.224 -7.219 3.443 1.00 0.00 C
+ATOM 643 CD1 LEU B 41 -5.930 -5.933 3.056 1.00 0.00 C
+ATOM 644 CD2 LEU B 41 -5.449 -8.331 2.425 1.00 0.00 C
+ATOM 645 H LEU B 41 -3.401 -4.336 3.587 1.00 0.00 H
+ATOM 646 HA LEU B 41 -3.800 -6.397 5.670 1.00 0.00 H
+ATOM 647 1HB LEU B 41 -3.417 -6.429 2.674 1.00 0.00 H
+ATOM 648 2HB LEU B 41 -3.234 -7.896 3.608 1.00 0.00 H
+ATOM 649 HG LEU B 41 -5.599 -7.531 4.374 1.00 0.00 H
+ATOM 650 1HD1 LEU B 41 -6.989 -6.108 2.982 1.00 0.00 H
+ATOM 651 2HD1 LEU B 41 -5.748 -5.169 3.814 1.00 0.00 H
+ATOM 652 3HD1 LEU B 41 -5.554 -5.583 2.096 1.00 0.00 H
+ATOM 653 1HD2 LEU B 41 -6.507 -8.543 2.345 1.00 0.00 H
+ATOM 654 2HD2 LEU B 41 -5.064 -8.015 1.456 1.00 0.00 H
+ATOM 655 3HD2 LEU B 41 -4.925 -9.230 2.750 1.00 0.00 H
+ATOM 656 N ASN B 42 -0.961 -5.848 4.142 1.00 0.00 N
+ATOM 657 CA ASN B 42 0.476 -6.081 4.197 1.00 0.00 C
+ATOM 658 C ASN B 42 1.186 -4.943 3.500 1.00 0.00 C
+ATOM 659 O ASN B 42 0.611 -4.259 2.647 1.00 0.00 O
+ATOM 660 CB ASN B 42 0.879 -7.430 3.574 1.00 0.00 C
+ATOM 661 CG ASN B 42 2.221 -8.023 4.175 1.00 0.00 C
+ATOM 662 OD1 ASN B 42 3.019 -7.282 4.759 1.00 0.00 O
+ATOM 663 ND2 ASN B 42 2.443 -9.314 4.000 1.00 0.00 N
+ATOM 664 H ASN B 42 -1.362 -5.170 3.485 1.00 0.00 H
+ATOM 665 HA ASN B 42 0.801 -6.082 5.235 1.00 0.00 H
+ATOM 666 1HB ASN B 42 0.085 -8.160 3.722 1.00 0.00 H
+ATOM 667 2HB ASN B 42 1.006 -7.309 2.504 1.00 0.00 H
+ATOM 668 1HD2 ASN B 42 3.312 -9.759 4.321 1.00 0.00 H
+ATOM 669 2HD2 ASN B 42 1.781 -9.867 3.512 1.00 0.00 H
+ATOM 670 N GLU B 43 2.470 -4.817 3.785 1.00 0.00 N
+ATOM 671 CA GLU B 43 3.334 -3.838 3.146 1.00 0.00 C
+ATOM 672 C GLU B 43 4.698 -4.435 2.907 1.00 0.00 C
+ATOM 673 O GLU B 43 5.303 -4.995 3.822 1.00 0.00 O
+ATOM 674 CB GLU B 43 3.502 -2.586 4.048 1.00 0.00 C
+ATOM 675 CG GLU B 43 4.454 -1.470 3.466 1.00 0.00 C
+ATOM 676 CD GLU B 43 4.662 -0.228 4.385 1.00 0.00 C
+ATOM 677 OE1 GLU B 43 4.133 -0.171 5.480 1.00 0.00 O
+ATOM 678 OE2 GLU B 43 5.350 0.670 3.956 1.00 0.00 O
+ATOM 679 H GLU B 43 2.857 -5.456 4.472 1.00 0.00 H
+ATOM 680 HA GLU B 43 2.904 -3.547 2.197 1.00 0.00 H
+ATOM 681 1HB GLU B 43 2.523 -2.134 4.214 1.00 0.00 H
+ATOM 682 2HB GLU B 43 3.888 -2.884 5.015 1.00 0.00 H
+ATOM 683 1HG GLU B 43 5.426 -1.917 3.287 1.00 0.00 H
+ATOM 684 2HG GLU B 43 4.063 -1.139 2.503 1.00 0.00 H
+ATOM 685 N VAL B 44 5.209 -4.281 1.693 1.00 0.00 N
+ATOM 686 CA VAL B 44 6.568 -4.675 1.386 1.00 0.00 C
+ATOM 687 C VAL B 44 7.333 -3.372 1.131 1.00 0.00 C
+ATOM 688 O VAL B 44 7.136 -2.701 0.116 1.00 0.00 O
+ATOM 689 CB VAL B 44 6.597 -5.622 0.175 1.00 0.00 C
+ATOM 690 CG1 VAL B 44 8.029 -6.029 -0.154 1.00 0.00 C
+ATOM 691 CG2 VAL B 44 5.749 -6.855 0.490 1.00 0.00 C
+ATOM 692 H VAL B 44 4.636 -3.852 0.973 1.00 0.00 H
+ATOM 693 HA VAL B 44 7.004 -5.186 2.242 1.00 0.00 H
+ATOM 694 HB VAL B 44 6.190 -5.109 -0.680 1.00 0.00 H
+ATOM 695 1HG1 VAL B 44 8.031 -6.692 -1.018 1.00 0.00 H
+ATOM 696 2HG1 VAL B 44 8.612 -5.137 -0.381 1.00 0.00 H
+ATOM 697 3HG1 VAL B 44 8.464 -6.545 0.705 1.00 0.00 H
+ATOM 698 1HG2 VAL B 44 5.742 -7.507 -0.360 1.00 0.00 H
+ATOM 699 2HG2 VAL B 44 6.168 -7.375 1.350 1.00 0.00 H
+ATOM 700 3HG2 VAL B 44 4.745 -6.554 0.715 1.00 0.00 H
+ATOM 701 N TYR B 45 8.153 -2.984 2.097 1.00 0.00 N
+ATOM 702 CA TYR B 45 8.833 -1.701 2.069 1.00 0.00 C
+ATOM 703 C TYR B 45 10.281 -1.856 1.676 1.00 0.00 C
+ATOM 704 O TYR B 45 11.091 -2.336 2.469 1.00 0.00 O
+ATOM 705 CB TYR B 45 8.761 -1.043 3.444 1.00 0.00 C
+ATOM 706 CG TYR B 45 9.327 0.352 3.468 1.00 0.00 C
+ATOM 707 CD1 TYR B 45 8.546 1.415 3.055 1.00 0.00 C
+ATOM 708 CD2 TYR B 45 10.637 0.574 3.886 1.00 0.00 C
+ATOM 709 CE1 TYR B 45 9.053 2.690 3.040 1.00 0.00 C
+ATOM 710 CE2 TYR B 45 11.139 1.866 3.872 1.00 0.00 C
+ATOM 711 CZ TYR B 45 10.349 2.914 3.439 1.00 0.00 C
+ATOM 712 OH TYR B 45 10.848 4.189 3.395 1.00 0.00 O
+ATOM 713 H TYR B 45 8.310 -3.609 2.883 1.00 0.00 H
+ATOM 714 HA TYR B 45 8.352 -1.059 1.334 1.00 0.00 H
+ATOM 715 1HB TYR B 45 7.719 -0.990 3.762 1.00 0.00 H
+ATOM 716 2HB TYR B 45 9.295 -1.651 4.170 1.00 0.00 H
+ATOM 717 HD1 TYR B 45 7.522 1.238 2.733 1.00 0.00 H
+ATOM 718 HD2 TYR B 45 11.260 -0.257 4.212 1.00 0.00 H
+ATOM 719 HE1 TYR B 45 8.429 3.516 2.704 1.00 0.00 H
+ATOM 720 HE2 TYR B 45 12.164 2.054 4.188 1.00 0.00 H
+ATOM 721 HH TYR B 45 11.749 4.187 3.730 1.00 0.00 H
+ATOM 722 N LEU B 46 10.613 -1.505 0.449 1.00 0.00 N
+ATOM 723 CA LEU B 46 11.970 -1.652 -0.043 1.00 0.00 C
+ATOM 724 C LEU B 46 12.431 -3.118 0.082 1.00 0.00 C
+ATOM 725 O LEU B 46 13.575 -3.437 0.431 1.00 0.00 O
+ATOM 726 CB LEU B 46 12.899 -0.666 0.659 1.00 0.00 C
+ATOM 727 CG LEU B 46 12.378 0.787 0.602 1.00 0.00 C
+ATOM 728 CD1 LEU B 46 13.393 1.733 1.236 1.00 0.00 C
+ATOM 729 CD2 LEU B 46 12.071 1.173 -0.806 1.00 0.00 C
+ATOM 730 H LEU B 46 9.920 -1.139 -0.204 1.00 0.00 H
+ATOM 731 HA LEU B 46 11.966 -1.391 -1.103 1.00 0.00 H
+ATOM 732 1HB LEU B 46 13.032 -0.956 1.696 1.00 0.00 H
+ATOM 733 2HB LEU B 46 13.868 -0.694 0.157 1.00 0.00 H
+ATOM 734 HG LEU B 46 11.467 0.846 1.168 1.00 0.00 H
+ATOM 735 1HD1 LEU B 46 13.002 2.752 1.217 1.00 0.00 H
+ATOM 736 2HD1 LEU B 46 13.577 1.435 2.266 1.00 0.00 H
+ATOM 737 3HD1 LEU B 46 14.326 1.692 0.676 1.00 0.00 H
+ATOM 738 1HD2 LEU B 46 11.682 2.172 -0.806 1.00 0.00 H
+ATOM 739 2HD2 LEU B 46 12.974 1.129 -1.407 1.00 0.00 H
+ATOM 740 3HD2 LEU B 46 11.333 0.494 -1.214 1.00 0.00 H
+ATOM 741 N GLY B 47 11.503 -4.015 -0.221 1.00 0.00 N
+ATOM 742 CA GLY B 47 11.750 -5.450 -0.231 1.00 0.00 C
+ATOM 743 C GLY B 47 11.468 -6.169 1.076 1.00 0.00 C
+ATOM 744 O GLY B 47 11.420 -7.399 1.086 1.00 0.00 O
+ATOM 745 H GLY B 47 10.591 -3.673 -0.484 1.00 0.00 H
+ATOM 746 1HA GLY B 47 11.143 -5.901 -1.019 1.00 0.00 H
+ATOM 747 2HA GLY B 47 12.786 -5.622 -0.514 1.00 0.00 H
+ATOM 748 N LYS B 48 11.232 -5.450 2.166 1.00 0.00 N
+ATOM 749 CA LYS B 48 11.003 -6.168 3.409 1.00 0.00 C
+ATOM 750 C LYS B 48 9.563 -6.069 3.885 1.00 0.00 C
+ATOM 751 O LYS B 48 8.943 -4.999 3.846 1.00 0.00 O
+ATOM 752 CB LYS B 48 11.942 -5.613 4.470 1.00 0.00 C
+ATOM 753 CG LYS B 48 13.429 -5.640 4.016 1.00 0.00 C
+ATOM 754 CD LYS B 48 13.978 -7.075 3.859 1.00 0.00 C
+ATOM 755 CE LYS B 48 15.405 -7.068 3.286 1.00 0.00 C
+ATOM 756 NZ LYS B 48 16.357 -6.323 4.155 1.00 0.00 N
+ATOM 757 H LYS B 48 11.259 -4.427 2.161 1.00 0.00 H
+ATOM 758 HA LYS B 48 11.227 -7.221 3.258 1.00 0.00 H
+ATOM 759 1HB LYS B 48 11.672 -4.578 4.685 1.00 0.00 H
+ATOM 760 2HB LYS B 48 11.837 -6.184 5.392 1.00 0.00 H
+ATOM 761 1HG LYS B 48 13.525 -5.115 3.060 1.00 0.00 H
+ATOM 762 2HG LYS B 48 14.034 -5.113 4.751 1.00 0.00 H
+ATOM 763 1HD LYS B 48 13.991 -7.581 4.826 1.00 0.00 H
+ATOM 764 2HD LYS B 48 13.339 -7.644 3.178 1.00 0.00 H
+ATOM 765 1HE LYS B 48 15.744 -8.095 3.188 1.00 0.00 H
+ATOM 766 2HE LYS B 48 15.394 -6.608 2.294 1.00 0.00 H
+ATOM 767 1HZ LYS B 48 17.283 -6.361 3.733 1.00 0.00 H
+ATOM 768 2HZ LYS B 48 16.057 -5.364 4.231 1.00 0.00 H
+ATOM 769 3HZ LYS B 48 16.390 -6.740 5.071 1.00 0.00 H
+ATOM 770 N GLU B 49 9.026 -7.184 4.372 1.00 0.00 N
+ATOM 771 CA GLU B 49 7.678 -7.150 4.918 1.00 0.00 C
+ATOM 772 C GLU B 49 7.670 -6.270 6.154 1.00 0.00 C
+ATOM 773 O GLU B 49 8.558 -6.361 7.004 1.00 0.00 O
+ATOM 774 CB GLU B 49 7.152 -8.553 5.210 1.00 0.00 C
+ATOM 775 CG GLU B 49 6.804 -9.376 3.950 1.00 0.00 C
+ATOM 776 CD GLU B 49 6.242 -10.777 4.267 1.00 0.00 C
+ATOM 777 OE1 GLU B 49 5.032 -10.936 4.382 1.00 0.00 O
+ATOM 778 OE2 GLU B 49 7.036 -11.684 4.379 1.00 0.00 O
+ATOM 779 H GLU B 49 9.553 -8.045 4.375 1.00 0.00 H
+ATOM 780 HA GLU B 49 7.019 -6.701 4.178 1.00 0.00 H
+ATOM 781 1HB GLU B 49 7.888 -9.107 5.789 1.00 0.00 H
+ATOM 782 2HB GLU B 49 6.246 -8.477 5.814 1.00 0.00 H
+ATOM 783 1HG GLU B 49 6.057 -8.823 3.373 1.00 0.00 H
+ATOM 784 2HG GLU B 49 7.696 -9.474 3.333 1.00 0.00 H
+ATOM 785 N LYS B 50 6.650 -5.437 6.254 1.00 0.00 N
+ATOM 786 CA LYS B 50 6.493 -4.485 7.337 1.00 0.00 C
+ATOM 787 C LYS B 50 5.054 -4.525 7.840 1.00 0.00 C
+ATOM 788 O LYS B 50 4.126 -4.450 7.045 1.00 0.00 O
+ATOM 789 CB LYS B 50 6.877 -3.107 6.782 1.00 0.00 C
+ATOM 790 CG LYS B 50 6.904 -1.952 7.720 1.00 0.00 C
+ATOM 791 CD LYS B 50 7.506 -0.712 6.977 1.00 0.00 C
+ATOM 792 CE LYS B 50 7.433 0.560 7.808 1.00 0.00 C
+ATOM 793 NZ LYS B 50 8.314 0.509 9.006 1.00 0.00 N
+ATOM 794 H LYS B 50 5.977 -5.420 5.490 1.00 0.00 H
+ATOM 795 HA LYS B 50 7.158 -4.758 8.155 1.00 0.00 H
+ATOM 796 1HB LYS B 50 7.869 -3.172 6.324 1.00 0.00 H
+ATOM 797 2HB LYS B 50 6.181 -2.849 5.980 1.00 0.00 H
+ATOM 798 1HG LYS B 50 5.888 -1.717 8.039 1.00 0.00 H
+ATOM 799 2HG LYS B 50 7.506 -2.185 8.601 1.00 0.00 H
+ATOM 800 1HD LYS B 50 8.544 -0.910 6.713 1.00 0.00 H
+ATOM 801 2HD LYS B 50 6.949 -0.544 6.060 1.00 0.00 H
+ATOM 802 1HE LYS B 50 7.721 1.407 7.187 1.00 0.00 H
+ATOM 803 2HE LYS B 50 6.404 0.698 8.141 1.00 0.00 H
+ATOM 804 1HZ LYS B 50 8.215 1.356 9.542 1.00 0.00 H
+ATOM 805 2HZ LYS B 50 8.042 -0.278 9.618 1.00 0.00 H
+ATOM 806 3HZ LYS B 50 9.268 0.392 8.733 1.00 0.00 H
+ATOM 807 N PHE B 51 4.871 -4.682 9.155 1.00 0.00 N
+ATOM 808 CA PHE B 51 3.523 -4.811 9.721 1.00 0.00 C
+ATOM 809 C PHE B 51 3.071 -3.623 10.553 1.00 0.00 C
+ATOM 810 O PHE B 51 2.002 -3.635 11.150 1.00 0.00 O
+ATOM 811 CB PHE B 51 3.443 -6.094 10.534 1.00 0.00 C
+ATOM 812 CG PHE B 51 3.661 -7.262 9.635 1.00 0.00 C
+ATOM 813 CD1 PHE B 51 4.920 -7.808 9.481 1.00 0.00 C
+ATOM 814 CD2 PHE B 51 2.610 -7.794 8.903 1.00 0.00 C
+ATOM 815 CE1 PHE B 51 5.132 -8.858 8.599 1.00 0.00 C
+ATOM 816 CE2 PHE B 51 2.815 -8.838 8.024 1.00 0.00 C
+ATOM 817 CZ PHE B 51 4.081 -9.368 7.867 1.00 0.00 C
+ATOM 818 H PHE B 51 5.674 -4.732 9.769 1.00 0.00 H
+ATOM 819 HA PHE B 51 2.841 -4.922 8.895 1.00 0.00 H
+ATOM 820 1HB PHE B 51 4.193 -6.099 11.325 1.00 0.00 H
+ATOM 821 2HB PHE B 51 2.467 -6.169 10.986 1.00 0.00 H
+ATOM 822 HD1 PHE B 51 5.753 -7.394 10.047 1.00 0.00 H
+ATOM 823 HD2 PHE B 51 1.616 -7.372 9.020 1.00 0.00 H
+ATOM 824 HE1 PHE B 51 6.133 -9.275 8.474 1.00 0.00 H
+ATOM 825 HE2 PHE B 51 1.981 -9.241 7.442 1.00 0.00 H
+ATOM 826 HZ PHE B 51 4.249 -10.192 7.162 1.00 0.00 H
+ATOM 827 N ASP B 52 3.852 -2.562 10.528 1.00 0.00 N
+ATOM 828 CA ASP B 52 3.590 -1.373 11.323 1.00 0.00 C
+ATOM 829 C ASP B 52 2.209 -0.744 10.997 1.00 0.00 C
+ATOM 830 O ASP B 52 1.498 -0.252 11.874 1.00 0.00 O
+ATOM 831 CB ASP B 52 4.677 -0.366 11.005 1.00 0.00 C
+ATOM 832 CG ASP B 52 6.085 -0.811 11.447 1.00 0.00 C
+ATOM 833 OD1 ASP B 52 6.206 -1.741 12.196 1.00 0.00 O
+ATOM 834 OD2 ASP B 52 7.036 -0.230 10.957 1.00 0.00 O
+ATOM 835 H ASP B 52 4.690 -2.605 9.970 1.00 0.00 H
+ATOM 836 HA ASP B 52 3.622 -1.641 12.380 1.00 0.00 H
+ATOM 837 1HB ASP B 52 4.678 -0.135 9.942 1.00 0.00 H
+ATOM 838 2HB ASP B 52 4.449 0.525 11.530 1.00 0.00 H
+ATOM 839 N SER B 53 1.845 -0.774 9.707 1.00 0.00 N
+ATOM 840 CA SER B 53 0.589 -0.230 9.179 1.00 0.00 C
+ATOM 841 C SER B 53 -0.519 -1.277 9.045 1.00 0.00 C
+ATOM 842 O SER B 53 -1.613 -0.966 8.567 1.00 0.00 O
+ATOM 843 CB SER B 53 0.818 0.372 7.795 1.00 0.00 C
+ATOM 844 OG SER B 53 1.697 1.469 7.855 1.00 0.00 O
+ATOM 845 H SER B 53 2.488 -1.201 9.054 1.00 0.00 H
+ATOM 846 HA SER B 53 0.242 0.550 9.857 1.00 0.00 H
+ATOM 847 1HB SER B 53 1.235 -0.393 7.133 1.00 0.00 H
+ATOM 848 2HB SER B 53 -0.136 0.681 7.371 1.00 0.00 H
+ATOM 849 HG SER B 53 2.503 1.122 8.253 1.00 0.00 H
+ATOM 850 N VAL B 54 -0.233 -2.523 9.401 1.00 0.00 N
+ATOM 851 CA VAL B 54 -1.188 -3.586 9.164 1.00 0.00 C
+ATOM 852 C VAL B 54 -2.259 -3.661 10.219 1.00 0.00 C
+ATOM 853 O VAL B 54 -1.980 -3.648 11.418 1.00 0.00 O
+ATOM 854 CB VAL B 54 -0.467 -4.930 9.008 1.00 0.00 C
+ATOM 855 CG1 VAL B 54 -1.464 -6.060 8.882 1.00 0.00 C
+ATOM 856 CG2 VAL B 54 0.369 -4.861 7.761 1.00 0.00 C
+ATOM 857 H VAL B 54 0.643 -2.746 9.864 1.00 0.00 H
+ATOM 858 HA VAL B 54 -1.673 -3.380 8.211 1.00 0.00 H
+ATOM 859 HB VAL B 54 0.163 -5.112 9.874 1.00 0.00 H
+ATOM 860 1HG1 VAL B 54 -0.938 -7.005 8.751 1.00 0.00 H
+ATOM 861 2HG1 VAL B 54 -2.079 -6.117 9.776 1.00 0.00 H
+ATOM 862 3HG1 VAL B 54 -2.101 -5.882 8.016 1.00 0.00 H
+ATOM 863 1HG2 VAL B 54 0.892 -5.794 7.616 1.00 0.00 H
+ATOM 864 2HG2 VAL B 54 -0.287 -4.690 6.927 1.00 0.00 H
+ATOM 865 3HG2 VAL B 54 1.084 -4.047 7.831 1.00 0.00 H
+ATOM 866 N HIS B 55 -3.497 -3.693 9.761 1.00 0.00 N
+ATOM 867 CA HIS B 55 -4.616 -3.731 10.682 1.00 0.00 C
+ATOM 868 C HIS B 55 -4.629 -5.036 11.442 1.00 0.00 C
+ATOM 869 O HIS B 55 -4.393 -6.103 10.870 1.00 0.00 O
+ATOM 870 CB HIS B 55 -5.953 -3.500 9.975 1.00 0.00 C
+ATOM 871 CG HIS B 55 -7.087 -3.209 10.924 1.00 0.00 C
+ATOM 872 ND1 HIS B 55 -7.714 -4.179 11.660 1.00 0.00 N
+ATOM 873 CD2 HIS B 55 -7.702 -2.037 11.254 1.00 0.00 C
+ATOM 874 CE1 HIS B 55 -8.654 -3.619 12.406 1.00 0.00 C
+ATOM 875 NE2 HIS B 55 -8.668 -2.329 12.168 1.00 0.00 N
+ATOM 876 H HIS B 55 -3.640 -3.700 8.747 1.00 0.00 H
+ATOM 877 HA HIS B 55 -4.497 -2.951 11.419 1.00 0.00 H
+ATOM 878 1HB HIS B 55 -5.852 -2.663 9.283 1.00 0.00 H
+ATOM 879 2HB HIS B 55 -6.213 -4.377 9.399 1.00 0.00 H
+ATOM 880 HD2 HIS B 55 -7.465 -1.051 10.863 1.00 0.00 H
+ATOM 881 HE1 HIS B 55 -9.313 -4.146 13.097 1.00 0.00 H
+ATOM 882 HE2 HIS B 55 -9.304 -1.642 12.607 1.00 0.00 H
+ATOM 883 N SER B 56 -4.975 -4.959 12.729 1.00 0.00 N
+ATOM 884 CA SER B 56 -5.056 -6.128 13.612 1.00 0.00 C
+ATOM 885 C SER B 56 -5.796 -7.290 12.978 1.00 0.00 C
+ATOM 886 O SER B 56 -5.385 -8.444 13.114 1.00 0.00 O
+ATOM 887 CB SER B 56 -5.759 -5.736 14.916 1.00 0.00 C
+ATOM 888 OG SER B 56 -4.983 -4.830 15.682 1.00 0.00 O
+ATOM 889 H SER B 56 -5.164 -4.031 13.113 1.00 0.00 H
+ATOM 890 HA SER B 56 -4.042 -6.451 13.843 1.00 0.00 H
+ATOM 891 1HB SER B 56 -6.715 -5.269 14.672 1.00 0.00 H
+ATOM 892 2HB SER B 56 -5.973 -6.623 15.503 1.00 0.00 H
+ATOM 893 HG SER B 56 -5.622 -4.445 16.330 1.00 0.00 H
+ATOM 894 N LYS B 57 -6.851 -6.977 12.227 1.00 0.00 N
+ATOM 895 CA LYS B 57 -7.720 -7.902 11.521 1.00 0.00 C
+ATOM 896 C LYS B 57 -6.989 -8.834 10.549 1.00 0.00 C
+ATOM 897 O LYS B 57 -7.483 -9.914 10.224 1.00 0.00 O
+ATOM 898 CB LYS B 57 -8.776 -7.094 10.769 1.00 0.00 C
+ATOM 899 CG LYS B 57 -9.800 -7.869 10.024 1.00 0.00 C
+ATOM 900 CD LYS B 57 -10.964 -6.932 9.592 1.00 0.00 C
+ATOM 901 CE LYS B 57 -10.566 -5.999 8.462 1.00 0.00 C
+ATOM 902 NZ LYS B 57 -11.739 -5.197 7.939 1.00 0.00 N
+ATOM 903 H LYS B 57 -7.082 -5.989 12.137 1.00 0.00 H
+ATOM 904 HA LYS B 57 -8.217 -8.527 12.263 1.00 0.00 H
+ATOM 905 1HB LYS B 57 -9.304 -6.464 11.474 1.00 0.00 H
+ATOM 906 2HB LYS B 57 -8.276 -6.442 10.050 1.00 0.00 H
+ATOM 907 1HG LYS B 57 -9.339 -8.308 9.132 1.00 0.00 H
+ATOM 908 2HG LYS B 57 -10.185 -8.670 10.650 1.00 0.00 H
+ATOM 909 1HD LYS B 57 -11.812 -7.536 9.269 1.00 0.00 H
+ATOM 910 2HD LYS B 57 -11.273 -6.329 10.447 1.00 0.00 H
+ATOM 911 1HE LYS B 57 -9.781 -5.318 8.800 1.00 0.00 H
+ATOM 912 2HE LYS B 57 -10.186 -6.604 7.665 1.00 0.00 H
+ATOM 913 1HZ LYS B 57 -11.429 -4.610 7.152 1.00 0.00 H
+ATOM 914 2HZ LYS B 57 -12.453 -5.830 7.613 1.00 0.00 H
+ATOM 915 3HZ LYS B 57 -12.115 -4.614 8.668 1.00 0.00 H
+ATOM 916 N TYR B 58 -5.860 -8.380 10.007 1.00 0.00 N
+ATOM 917 CA TYR B 58 -5.115 -9.150 9.029 1.00 0.00 C
+ATOM 918 C TYR B 58 -3.818 -9.748 9.583 1.00 0.00 C
+ATOM 919 O TYR B 58 -3.107 -10.454 8.857 1.00 0.00 O
+ATOM 920 CB TYR B 58 -4.797 -8.267 7.838 1.00 0.00 C
+ATOM 921 CG TYR B 58 -6.031 -7.773 7.140 1.00 0.00 C
+ATOM 922 CD1 TYR B 58 -6.869 -8.692 6.524 1.00 0.00 C
+ATOM 923 CD2 TYR B 58 -6.325 -6.399 7.082 1.00 0.00 C
+ATOM 924 CE1 TYR B 58 -7.995 -8.265 5.847 1.00 0.00 C
+ATOM 925 CE2 TYR B 58 -7.468 -5.980 6.394 1.00 0.00 C
+ATOM 926 CZ TYR B 58 -8.288 -6.918 5.780 1.00 0.00 C
+ATOM 927 OH TYR B 58 -9.416 -6.517 5.090 1.00 0.00 O
+ATOM 928 H TYR B 58 -5.476 -7.488 10.314 1.00 0.00 H
+ATOM 929 HA TYR B 58 -5.740 -9.974 8.696 1.00 0.00 H
+ATOM 930 1HB TYR B 58 -4.224 -7.400 8.173 1.00 0.00 H
+ATOM 931 2HB TYR B 58 -4.183 -8.813 7.120 1.00 0.00 H
+ATOM 932 HD1 TYR B 58 -6.637 -9.760 6.569 1.00 0.00 H
+ATOM 933 HD2 TYR B 58 -5.666 -5.673 7.556 1.00 0.00 H
+ATOM 934 HE1 TYR B 58 -8.648 -8.993 5.360 1.00 0.00 H
+ATOM 935 HE2 TYR B 58 -7.716 -4.915 6.327 1.00 0.00 H
+ATOM 936 HH TYR B 58 -9.553 -5.547 5.200 1.00 0.00 H
+ATOM 937 N MET B 59 -3.487 -9.469 10.844 1.00 0.00 N
+ATOM 938 CA MET B 59 -2.211 -9.943 11.353 1.00 0.00 C
+ATOM 939 C MET B 59 -2.161 -11.451 11.383 1.00 0.00 C
+ATOM 940 O MET B 59 -3.079 -12.109 11.857 1.00 0.00 O
+ATOM 941 CB MET B 59 -1.907 -9.353 12.732 1.00 0.00 C
+ATOM 942 CG MET B 59 -1.561 -7.840 12.724 1.00 0.00 C
+ATOM 943 SD MET B 59 -0.037 -7.507 11.813 1.00 0.00 S
+ATOM 944 CE MET B 59 1.202 -8.272 12.841 1.00 0.00 C
+ATOM 945 H MET B 59 -4.116 -8.950 11.455 1.00 0.00 H
+ATOM 946 HA MET B 59 -1.431 -9.603 10.669 1.00 0.00 H
+ATOM 947 1HB MET B 59 -2.780 -9.491 13.375 1.00 0.00 H
+ATOM 948 2HB MET B 59 -1.075 -9.890 13.187 1.00 0.00 H
+ATOM 949 1HG MET B 59 -2.371 -7.295 12.239 1.00 0.00 H
+ATOM 950 2HG MET B 59 -1.454 -7.459 13.733 1.00 0.00 H
+ATOM 951 1HE MET B 59 2.187 -8.144 12.394 1.00 0.00 H
+ATOM 952 2HE MET B 59 1.188 -7.811 13.804 1.00 0.00 H
+ATOM 953 3HE MET B 59 0.988 -9.335 12.943 1.00 0.00 H
+ATOM 954 N GLY B 60 -1.085 -11.991 10.807 1.00 0.00 N
+ATOM 955 CA GLY B 60 -0.814 -13.422 10.755 1.00 0.00 C
+ATOM 956 C GLY B 60 -1.587 -14.141 9.651 1.00 0.00 C
+ATOM 957 O GLY B 60 -1.417 -15.348 9.451 1.00 0.00 O
+ATOM 958 H GLY B 60 -0.400 -11.362 10.412 1.00 0.00 H
+ATOM 959 1HA GLY B 60 0.254 -13.577 10.601 1.00 0.00 H
+ATOM 960 2HA GLY B 60 -1.064 -13.868 11.713 1.00 0.00 H
+ATOM 961 N ARG B 61 -2.411 -13.409 8.903 1.00 0.00 N
+ATOM 962 CA ARG B 61 -3.256 -14.037 7.904 1.00 0.00 C
+ATOM 963 C ARG B 61 -2.775 -13.882 6.478 1.00 0.00 C
+ATOM 964 O ARG B 61 -3.493 -14.262 5.554 1.00 0.00 O
+ATOM 965 CB ARG B 61 -4.658 -13.459 7.977 1.00 0.00 C
+ATOM 966 CG ARG B 61 -5.404 -13.676 9.298 1.00 0.00 C
+ATOM 967 CD ARG B 61 -6.772 -13.141 9.164 1.00 0.00 C
+ATOM 968 NE ARG B 61 -7.421 -13.865 8.158 1.00 0.00 N
+ATOM 969 CZ ARG B 61 -8.500 -13.494 7.459 1.00 0.00 C
+ATOM 970 NH1 ARG B 61 -8.905 -14.321 6.556 1.00 0.00 N
+ATOM 971 NH2 ARG B 61 -9.129 -12.321 7.659 1.00 0.00 N
+ATOM 972 H ARG B 61 -2.503 -12.401 9.058 1.00 0.00 H
+ATOM 973 HA ARG B 61 -3.312 -15.101 8.126 1.00 0.00 H
+ATOM 974 1HB ARG B 61 -4.614 -12.384 7.802 1.00 0.00 H
+ATOM 975 2HB ARG B 61 -5.267 -13.896 7.185 1.00 0.00 H
+ATOM 976 1HG ARG B 61 -5.456 -14.741 9.535 1.00 0.00 H
+ATOM 977 2HG ARG B 61 -4.896 -13.139 10.103 1.00 0.00 H
+ATOM 978 1HD ARG B 61 -7.320 -13.270 10.095 1.00 0.00 H
+ATOM 979 2HD ARG B 61 -6.758 -12.092 8.885 1.00 0.00 H
+ATOM 980 HE ARG B 61 -7.023 -14.773 7.913 1.00 0.00 H
+ATOM 981 1HH1 ARG B 61 -8.377 -15.194 6.395 1.00 0.00 H
+ATOM 982 2HH1 ARG B 61 -9.723 -14.084 6.001 1.00 0.00 H
+ATOM 983 1HH2 ARG B 61 -8.795 -11.677 8.367 1.00 0.00 H
+ATOM 984 2HH2 ARG B 61 -9.948 -12.071 7.073 1.00 0.00 H
+ATOM 985 N THR B 62 -1.595 -13.292 6.274 1.00 0.00 N
+ATOM 986 CA THR B 62 -1.148 -13.061 4.916 1.00 0.00 C
+ATOM 987 C THR B 62 0.266 -13.540 4.676 1.00 0.00 C
+ATOM 988 O THR B 62 1.080 -13.660 5.604 1.00 0.00 O
+ATOM 989 CB THR B 62 -1.196 -11.552 4.552 1.00 0.00 C
+ATOM 990 OG1 THR B 62 -0.231 -10.823 5.353 1.00 0.00 O
+ATOM 991 CG2 THR B 62 -2.602 -10.962 4.795 1.00 0.00 C
+ATOM 992 H THR B 62 -1.035 -13.004 7.060 1.00 0.00 H
+ATOM 993 HA THR B 62 -1.798 -13.600 4.244 1.00 0.00 H
+ATOM 994 HB THR B 62 -0.952 -11.439 3.510 1.00 0.00 H
+ATOM 995 HG1 THR B 62 -0.446 -9.878 5.338 1.00 0.00 H
+ATOM 996 1HG2 THR B 62 -2.601 -9.914 4.524 1.00 0.00 H
+ATOM 997 2HG2 THR B 62 -3.325 -11.499 4.179 1.00 0.00 H
+ATOM 998 3HG2 THR B 62 -2.873 -11.058 5.845 1.00 0.00 H
+ATOM 999 N SER B 63 0.570 -13.741 3.409 1.00 0.00 N
+ATOM 1000 CA SER B 63 1.942 -14.018 3.021 1.00 0.00 C
+ATOM 1001 C SER B 63 2.196 -13.443 1.637 1.00 0.00 C
+ATOM 1002 O SER B 63 1.310 -13.417 0.776 1.00 0.00 O
+ATOM 1003 CB SER B 63 2.222 -15.521 3.044 1.00 0.00 C
+ATOM 1004 OG SER B 63 1.468 -16.194 2.078 1.00 0.00 O
+ATOM 1005 H SER B 63 -0.183 -13.678 2.727 1.00 0.00 H
+ATOM 1006 HA SER B 63 2.616 -13.526 3.726 1.00 0.00 H
+ATOM 1007 1HB SER B 63 3.282 -15.697 2.867 1.00 0.00 H
+ATOM 1008 2HB SER B 63 1.989 -15.917 4.030 1.00 0.00 H
+ATOM 1009 HG SER B 63 1.841 -15.924 1.233 1.00 0.00 H
+ATOM 1010 N PHE B 64 3.424 -13.012 1.418 1.00 0.00 N
+ATOM 1011 CA PHE B 64 3.821 -12.415 0.153 1.00 0.00 C
+ATOM 1012 C PHE B 64 4.659 -13.329 -0.708 1.00 0.00 C
+ATOM 1013 O PHE B 64 5.650 -13.894 -0.254 1.00 0.00 O
+ATOM 1014 CB PHE B 64 4.567 -11.102 0.421 1.00 0.00 C
+ATOM 1015 CG PHE B 64 5.144 -10.409 -0.807 1.00 0.00 C
+ATOM 1016 CD1 PHE B 64 4.328 -9.924 -1.803 1.00 0.00 C
+ATOM 1017 CD2 PHE B 64 6.510 -10.196 -0.922 1.00 0.00 C
+ATOM 1018 CE1 PHE B 64 4.863 -9.255 -2.910 1.00 0.00 C
+ATOM 1019 CE2 PHE B 64 7.036 -9.522 -2.017 1.00 0.00 C
+ATOM 1020 CZ PHE B 64 6.204 -9.053 -3.020 1.00 0.00 C
+ATOM 1021 H PHE B 64 4.104 -13.066 2.167 1.00 0.00 H
+ATOM 1022 HA PHE B 64 2.921 -12.172 -0.409 1.00 0.00 H
+ATOM 1023 1HB PHE B 64 3.883 -10.402 0.906 1.00 0.00 H
+ATOM 1024 2HB PHE B 64 5.384 -11.284 1.121 1.00 0.00 H
+ATOM 1025 HD1 PHE B 64 3.246 -10.072 -1.723 1.00 0.00 H
+ATOM 1026 HD2 PHE B 64 7.169 -10.557 -0.134 1.00 0.00 H
+ATOM 1027 HE1 PHE B 64 4.194 -8.882 -3.674 1.00 0.00 H
+ATOM 1028 HE2 PHE B 64 8.106 -9.365 -2.081 1.00 0.00 H
+ATOM 1029 HZ PHE B 64 6.612 -8.526 -3.881 1.00 0.00 H
+ATOM 1030 N ASP B 65 4.223 -13.464 -1.959 1.00 0.00 N
+ATOM 1031 CA ASP B 65 4.945 -14.182 -2.991 1.00 0.00 C
+ATOM 1032 C ASP B 65 5.841 -13.222 -3.763 1.00 0.00 C
+ATOM 1033 O ASP B 65 5.381 -12.529 -4.676 1.00 0.00 O
+ATOM 1034 CB ASP B 65 3.966 -14.859 -3.942 1.00 0.00 C
+ATOM 1035 CG ASP B 65 4.627 -15.586 -5.083 1.00 0.00 C
+ATOM 1036 OD1 ASP B 65 5.782 -15.323 -5.383 1.00 0.00 O
+ATOM 1037 OD2 ASP B 65 3.950 -16.317 -5.752 1.00 0.00 O
+ATOM 1038 H ASP B 65 3.356 -13.008 -2.225 1.00 0.00 H
+ATOM 1039 HA ASP B 65 5.568 -14.941 -2.523 1.00 0.00 H
+ATOM 1040 1HB ASP B 65 3.354 -15.569 -3.380 1.00 0.00 H
+ATOM 1041 2HB ASP B 65 3.293 -14.111 -4.349 1.00 0.00 H
+ATOM 1042 N SER B 66 7.128 -13.212 -3.424 1.00 0.00 N
+ATOM 1043 CA SER B 66 8.071 -12.268 -4.002 1.00 0.00 C
+ATOM 1044 C SER B 66 8.490 -12.629 -5.422 1.00 0.00 C
+ATOM 1045 O SER B 66 9.240 -11.871 -6.039 1.00 0.00 O
+ATOM 1046 CB SER B 66 9.290 -12.115 -3.110 1.00 0.00 C
+ATOM 1047 OG SER B 66 10.028 -13.298 -3.042 1.00 0.00 O
+ATOM 1048 H SER B 66 7.438 -13.843 -2.699 1.00 0.00 H
+ATOM 1049 HA SER B 66 7.583 -11.304 -4.061 1.00 0.00 H
+ATOM 1050 1HB SER B 66 9.917 -11.313 -3.503 1.00 0.00 H
+ATOM 1051 2HB SER B 66 8.976 -11.824 -2.111 1.00 0.00 H
+ATOM 1052 HG SER B 66 10.113 -13.613 -3.956 1.00 0.00 H
+ATOM 1053 N ASP B 67 8.037 -13.789 -5.929 1.00 0.00 N
+ATOM 1054 CA ASP B 67 8.365 -14.192 -7.289 1.00 0.00 C
+ATOM 1055 C ASP B 67 7.294 -13.634 -8.232 1.00 0.00 C
+ATOM 1056 O ASP B 67 7.605 -13.181 -9.332 1.00 0.00 O
+ATOM 1057 CB ASP B 67 8.396 -15.732 -7.447 1.00 0.00 C
+ATOM 1058 CG ASP B 67 9.490 -16.471 -6.614 1.00 0.00 C
+ATOM 1059 OD1 ASP B 67 10.643 -16.135 -6.739 1.00 0.00 O
+ATOM 1060 OD2 ASP B 67 9.144 -17.387 -5.890 1.00 0.00 O
+ATOM 1061 H ASP B 67 7.390 -14.377 -5.394 1.00 0.00 H
+ATOM 1062 HA ASP B 67 9.335 -13.776 -7.568 1.00 0.00 H
+ATOM 1063 1HB ASP B 67 7.416 -16.134 -7.159 1.00 0.00 H
+ATOM 1064 2HB ASP B 67 8.543 -15.978 -8.500 1.00 0.00 H
+ATOM 1065 N SER B 68 6.033 -13.630 -7.776 1.00 0.00 N
+ATOM 1066 CA SER B 68 4.944 -13.125 -8.625 1.00 0.00 C
+ATOM 1067 C SER B 68 4.388 -11.761 -8.176 1.00 0.00 C
+ATOM 1068 O SER B 68 3.590 -11.152 -8.889 1.00 0.00 O
+ATOM 1069 CB SER B 68 3.782 -14.117 -8.714 1.00 0.00 C
+ATOM 1070 OG SER B 68 3.119 -14.294 -7.489 1.00 0.00 O
+ATOM 1071 H SER B 68 5.846 -14.054 -6.858 1.00 0.00 H
+ATOM 1072 HA SER B 68 5.342 -12.999 -9.630 1.00 0.00 H
+ATOM 1073 1HB SER B 68 3.068 -13.748 -9.443 1.00 0.00 H
+ATOM 1074 2HB SER B 68 4.146 -15.079 -9.074 1.00 0.00 H
+ATOM 1075 HG SER B 68 3.578 -15.036 -7.015 1.00 0.00 H
+ATOM 1076 N TRP B 69 4.780 -11.311 -6.987 1.00 0.00 N
+ATOM 1077 CA TRP B 69 4.288 -10.085 -6.343 1.00 0.00 C
+ATOM 1078 C TRP B 69 2.792 -10.184 -6.028 1.00 0.00 C
+ATOM 1079 O TRP B 69 2.003 -9.293 -6.331 1.00 0.00 O
+ATOM 1080 CB TRP B 69 4.591 -8.794 -7.104 1.00 0.00 C
+ATOM 1081 CG TRP B 69 6.075 -8.551 -7.328 1.00 0.00 C
+ATOM 1082 CD1 TRP B 69 7.093 -9.431 -7.098 1.00 0.00 C
+ATOM 1083 CD2 TRP B 69 6.703 -7.332 -7.804 1.00 0.00 C
+ATOM 1084 NE1 TRP B 69 8.295 -8.858 -7.434 1.00 0.00 N
+ATOM 1085 CE2 TRP B 69 8.074 -7.578 -7.873 1.00 0.00 C
+ATOM 1086 CE3 TRP B 69 6.221 -6.078 -8.169 1.00 0.00 C
+ATOM 1087 CZ2 TRP B 69 8.974 -6.616 -8.318 1.00 0.00 C
+ATOM 1088 CZ3 TRP B 69 7.130 -5.119 -8.590 1.00 0.00 C
+ATOM 1089 CH2 TRP B 69 8.473 -5.397 -8.672 1.00 0.00 C
+ATOM 1090 H TRP B 69 5.428 -11.876 -6.447 1.00 0.00 H
+ATOM 1091 HA TRP B 69 4.824 -10.012 -5.417 1.00 0.00 H
+ATOM 1092 1HB TRP B 69 4.084 -8.802 -8.071 1.00 0.00 H
+ATOM 1093 2HB TRP B 69 4.189 -7.950 -6.540 1.00 0.00 H
+ATOM 1094 HD1 TRP B 69 6.972 -10.445 -6.721 1.00 0.00 H
+ATOM 1095 HE1 TRP B 69 9.199 -9.313 -7.369 1.00 0.00 H
+ATOM 1096 HE3 TRP B 69 5.157 -5.856 -8.115 1.00 0.00 H
+ATOM 1097 HZ2 TRP B 69 10.048 -6.803 -8.379 1.00 0.00 H
+ATOM 1098 HZ3 TRP B 69 6.752 -4.142 -8.861 1.00 0.00 H
+ATOM 1099 HH2 TRP B 69 9.162 -4.625 -9.019 1.00 0.00 H
+ATOM 1100 N THR B 70 2.424 -11.309 -5.455 1.00 0.00 N
+ATOM 1101 CA THR B 70 1.035 -11.613 -5.092 1.00 0.00 C
+ATOM 1102 C THR B 70 0.892 -11.751 -3.585 1.00 0.00 C
+ATOM 1103 O THR B 70 1.770 -12.325 -2.935 1.00 0.00 O
+ATOM 1104 CB THR B 70 0.570 -12.931 -5.764 1.00 0.00 C
+ATOM 1105 OG1 THR B 70 0.657 -12.811 -7.194 1.00 0.00 O
+ATOM 1106 CG2 THR B 70 -0.850 -13.297 -5.379 1.00 0.00 C
+ATOM 1107 H THR B 70 3.174 -11.969 -5.252 1.00 0.00 H
+ATOM 1108 HA THR B 70 0.391 -10.794 -5.418 1.00 0.00 H
+ATOM 1109 HB THR B 70 1.222 -13.744 -5.445 1.00 0.00 H
+ATOM 1110 HG1 THR B 70 1.529 -13.192 -7.460 1.00 0.00 H
+ATOM 1111 1HG2 THR B 70 -1.104 -14.221 -5.871 1.00 0.00 H
+ATOM 1112 2HG2 THR B 70 -0.938 -13.432 -4.305 1.00 0.00 H
+ATOM 1113 3HG2 THR B 70 -1.521 -12.523 -5.692 1.00 0.00 H
+ATOM 1114 N LEU B 71 -0.185 -11.204 -3.008 1.00 0.00 N
+ATOM 1115 CA LEU B 71 -0.396 -11.403 -1.564 1.00 0.00 C
+ATOM 1116 C LEU B 71 -1.425 -12.500 -1.408 1.00 0.00 C
+ATOM 1117 O LEU B 71 -2.426 -12.520 -2.123 1.00 0.00 O
+ATOM 1118 CB LEU B 71 -0.921 -10.158 -0.846 1.00 0.00 C
+ATOM 1119 CG LEU B 71 -0.970 -10.232 0.653 1.00 0.00 C
+ATOM 1120 CD1 LEU B 71 0.463 -10.220 1.125 1.00 0.00 C
+ATOM 1121 CD2 LEU B 71 -1.740 -9.088 1.243 1.00 0.00 C
+ATOM 1122 H LEU B 71 -0.883 -10.710 -3.568 1.00 0.00 H
+ATOM 1123 HA LEU B 71 0.532 -11.721 -1.094 1.00 0.00 H
+ATOM 1124 1HB LEU B 71 -0.246 -9.388 -1.055 1.00 0.00 H
+ATOM 1125 2HB LEU B 71 -1.904 -9.897 -1.227 1.00 0.00 H
+ATOM 1126 HG LEU B 71 -1.436 -11.168 0.960 1.00 0.00 H
+ATOM 1127 1HD1 LEU B 71 0.497 -10.297 2.196 1.00 0.00 H
+ATOM 1128 2HD1 LEU B 71 0.984 -11.037 0.685 1.00 0.00 H
+ATOM 1129 3HD1 LEU B 71 0.935 -9.288 0.812 1.00 0.00 H
+ATOM 1130 1HD2 LEU B 71 -1.736 -9.173 2.322 1.00 0.00 H
+ATOM 1131 2HD2 LEU B 71 -1.262 -8.175 0.962 1.00 0.00 H
+ATOM 1132 3HD2 LEU B 71 -2.765 -9.102 0.877 1.00 0.00 H
+ATOM 1133 N ARG B 72 -1.183 -13.442 -0.521 1.00 0.00 N
+ATOM 1134 CA ARG B 72 -2.162 -14.488 -0.304 1.00 0.00 C
+ATOM 1135 C ARG B 72 -2.885 -14.192 1.002 1.00 0.00 C
+ATOM 1136 O ARG B 72 -2.234 -13.829 1.981 1.00 0.00 O
+ATOM 1137 CB ARG B 72 -1.464 -15.832 -0.241 1.00 0.00 C
+ATOM 1138 CG ARG B 72 -0.589 -16.137 -1.474 1.00 0.00 C
+ATOM 1139 CD ARG B 72 -0.020 -17.520 -1.431 1.00 0.00 C
+ATOM 1140 NE ARG B 72 -1.042 -18.514 -1.783 1.00 0.00 N
+ATOM 1141 CZ ARG B 72 -1.096 -19.796 -1.346 1.00 0.00 C
+ATOM 1142 NH1 ARG B 72 -0.195 -20.265 -0.499 1.00 0.00 N
+ATOM 1143 NH2 ARG B 72 -2.058 -20.585 -1.777 1.00 0.00 N
+ATOM 1144 H ARG B 72 -0.323 -13.423 0.018 1.00 0.00 H
+ATOM 1145 HA ARG B 72 -2.865 -14.491 -1.123 1.00 0.00 H
+ATOM 1146 1HB ARG B 72 -0.825 -15.872 0.639 1.00 0.00 H
+ATOM 1147 2HB ARG B 72 -2.184 -16.606 -0.159 1.00 0.00 H
+ATOM 1148 1HG ARG B 72 -1.199 -16.039 -2.376 1.00 0.00 H
+ATOM 1149 2HG ARG B 72 0.241 -15.422 -1.517 1.00 0.00 H
+ATOM 1150 1HD ARG B 72 0.798 -17.599 -2.144 1.00 0.00 H
+ATOM 1151 2HD ARG B 72 0.345 -17.739 -0.424 1.00 0.00 H
+ATOM 1152 HE ARG B 72 -1.748 -18.208 -2.491 1.00 0.00 H
+ATOM 1153 1HH1 ARG B 72 0.548 -19.675 -0.160 1.00 0.00 H
+ATOM 1154 2HH1 ARG B 72 -0.231 -21.256 -0.188 1.00 0.00 H
+ATOM 1155 1HH2 ARG B 72 -2.749 -20.237 -2.435 1.00 0.00 H
+ATOM 1156 2HH2 ARG B 72 -2.098 -21.544 -1.459 1.00 0.00 H
+ATOM 1157 N LEU B 73 -4.221 -14.324 1.013 1.00 0.00 N
+ATOM 1158 CA LEU B 73 -5.058 -14.118 2.216 1.00 0.00 C
+ATOM 1159 C LEU B 73 -5.578 -15.474 2.684 1.00 0.00 C
+ATOM 1160 O LEU B 73 -6.346 -16.131 1.981 1.00 0.00 O
+ATOM 1161 CB LEU B 73 -6.218 -13.134 1.918 1.00 0.00 C
+ATOM 1162 CG LEU B 73 -7.231 -12.863 3.069 1.00 0.00 C
+ATOM 1163 CD1 LEU B 73 -6.505 -12.222 4.263 1.00 0.00 C
+ATOM 1164 CD2 LEU B 73 -8.372 -11.965 2.541 1.00 0.00 C
+ATOM 1165 H LEU B 73 -4.664 -14.607 0.139 1.00 0.00 H
+ATOM 1166 HA LEU B 73 -4.442 -13.704 3.014 1.00 0.00 H
+ATOM 1167 1HB LEU B 73 -5.788 -12.176 1.627 1.00 0.00 H
+ATOM 1168 2HB LEU B 73 -6.783 -13.522 1.082 1.00 0.00 H
+ATOM 1169 HG LEU B 73 -7.654 -13.807 3.417 1.00 0.00 H
+ATOM 1170 1HD1 LEU B 73 -7.219 -12.048 5.069 1.00 0.00 H
+ATOM 1171 2HD1 LEU B 73 -5.719 -12.892 4.619 1.00 0.00 H
+ATOM 1172 3HD1 LEU B 73 -6.064 -11.272 3.961 1.00 0.00 H
+ATOM 1173 1HD2 LEU B 73 -9.087 -11.779 3.339 1.00 0.00 H
+ATOM 1174 2HD2 LEU B 73 -7.973 -11.008 2.191 1.00 0.00 H
+ATOM 1175 3HD2 LEU B 73 -8.871 -12.462 1.714 1.00 0.00 H
+ATOM 1176 N HIS B 74 -5.113 -15.911 3.852 1.00 0.00 N
+ATOM 1177 CA HIS B 74 -5.346 -17.266 4.336 1.00 0.00 C
+ATOM 1178 C HIS B 74 -6.589 -17.471 5.195 1.00 0.00 C
+ATOM 1179 O HIS B 74 -6.987 -16.580 5.944 1.00 0.00 O
+ATOM 1180 CB HIS B 74 -4.102 -17.708 5.108 1.00 0.00 C
+ATOM 1181 CG HIS B 74 -2.874 -17.819 4.227 1.00 0.00 C
+ATOM 1182 ND1 HIS B 74 -2.559 -18.965 3.513 1.00 0.00 N
+ATOM 1183 CD2 HIS B 74 -1.902 -16.904 3.931 1.00 0.00 C
+ATOM 1184 CE1 HIS B 74 -1.426 -18.753 2.840 1.00 0.00 C
+ATOM 1185 NE2 HIS B 74 -1.014 -17.509 3.075 1.00 0.00 N
+ATOM 1186 H HIS B 74 -4.532 -15.291 4.417 1.00 0.00 H
+ATOM 1187 HA HIS B 74 -5.453 -17.928 3.478 1.00 0.00 H
+ATOM 1188 1HB HIS B 74 -3.888 -16.995 5.904 1.00 0.00 H
+ATOM 1189 2HB HIS B 74 -4.286 -18.674 5.569 1.00 0.00 H
+ATOM 1190 HD2 HIS B 74 -1.835 -15.896 4.308 1.00 0.00 H
+ATOM 1191 HE1 HIS B 74 -0.933 -19.467 2.198 1.00 0.00 H
+ATOM 1192 HE2 HIS B 74 -0.164 -17.064 2.688 1.00 0.00 H
+ATOM 1193 N ASN B 75 -7.131 -18.687 5.135 1.00 0.00 N
+ATOM 1194 CA ASN B 75 -8.275 -19.126 5.944 1.00 0.00 C
+ATOM 1195 C ASN B 75 -9.463 -18.192 5.771 1.00 0.00 C
+ATOM 1196 O ASN B 75 -9.994 -17.636 6.734 1.00 0.00 O
+ATOM 1197 CB ASN B 75 -7.869 -19.267 7.403 1.00 0.00 C
+ATOM 1198 CG ASN B 75 -8.859 -20.050 8.165 1.00 0.00 C
+ATOM 1199 OD1 ASN B 75 -9.589 -20.854 7.544 1.00 0.00 O
+ATOM 1200 ND2 ASN B 75 -8.858 -19.871 9.468 1.00 0.00 N
+ATOM 1201 H ASN B 75 -6.736 -19.352 4.479 1.00 0.00 H
+ATOM 1202 HA ASN B 75 -8.582 -20.110 5.586 1.00 0.00 H
+ATOM 1203 1HB ASN B 75 -6.904 -19.733 7.480 1.00 0.00 H
+ATOM 1204 2HB ASN B 75 -7.785 -18.282 7.857 1.00 0.00 H
+ATOM 1205 1HD2 ASN B 75 -9.450 -20.423 10.093 1.00 0.00 H
+ATOM 1206 2HD2 ASN B 75 -8.213 -19.186 9.864 1.00 0.00 H
+ATOM 1207 N LEU B 76 -9.882 -18.027 4.528 1.00 0.00 N
+ATOM 1208 CA LEU B 76 -10.920 -17.061 4.184 1.00 0.00 C
+ATOM 1209 C LEU B 76 -12.225 -17.232 4.915 1.00 0.00 C
+ATOM 1210 O LEU B 76 -12.770 -18.332 5.048 1.00 0.00 O
+ATOM 1211 CB LEU B 76 -11.242 -17.179 2.702 1.00 0.00 C
+ATOM 1212 CG LEU B 76 -10.162 -16.741 1.746 1.00 0.00 C
+ATOM 1213 CD1 LEU B 76 -10.593 -17.115 0.357 1.00 0.00 C
+ATOM 1214 CD2 LEU B 76 -9.935 -15.276 1.898 1.00 0.00 C
+ATOM 1215 H LEU B 76 -9.421 -18.555 3.784 1.00 0.00 H
+ATOM 1216 HA LEU B 76 -10.537 -16.064 4.399 1.00 0.00 H
+ATOM 1217 1HB LEU B 76 -11.469 -18.223 2.480 1.00 0.00 H
+ATOM 1218 2HB LEU B 76 -12.132 -16.583 2.500 1.00 0.00 H
+ATOM 1219 HG LEU B 76 -9.237 -17.250 1.964 1.00 0.00 H
+ATOM 1220 1HD1 LEU B 76 -9.836 -16.838 -0.339 1.00 0.00 H
+ATOM 1221 2HD1 LEU B 76 -10.736 -18.195 0.305 1.00 0.00 H
+ATOM 1222 3HD1 LEU B 76 -11.522 -16.620 0.109 1.00 0.00 H
+ATOM 1223 1HD2 LEU B 76 -9.154 -14.951 1.210 1.00 0.00 H
+ATOM 1224 2HD2 LEU B 76 -10.855 -14.746 1.690 1.00 0.00 H
+ATOM 1225 3HD2 LEU B 76 -9.626 -15.069 2.922 1.00 0.00 H
+ATOM 1226 N GLN B 77 -12.735 -16.108 5.365 1.00 0.00 N
+ATOM 1227 CA GLN B 77 -14.002 -16.025 6.048 1.00 0.00 C
+ATOM 1228 C GLN B 77 -15.010 -15.344 5.151 1.00 0.00 C
+ATOM 1229 O GLN B 77 -14.647 -14.585 4.255 1.00 0.00 O
+ATOM 1230 CB GLN B 77 -13.846 -15.277 7.358 1.00 0.00 C
+ATOM 1231 CG GLN B 77 -12.824 -15.917 8.282 1.00 0.00 C
+ATOM 1232 CD GLN B 77 -13.220 -17.354 8.705 1.00 0.00 C
+ATOM 1233 OE1 GLN B 77 -14.321 -17.575 9.254 1.00 0.00 O
+ATOM 1234 NE2 GLN B 77 -12.322 -18.321 8.448 1.00 0.00 N
+ATOM 1235 H GLN B 77 -12.197 -15.246 5.231 1.00 0.00 H
+ATOM 1236 HA GLN B 77 -14.344 -17.029 6.258 1.00 0.00 H
+ATOM 1237 1HB GLN B 77 -13.513 -14.255 7.150 1.00 0.00 H
+ATOM 1238 2HB GLN B 77 -14.802 -15.220 7.873 1.00 0.00 H
+ATOM 1239 1HG GLN B 77 -11.863 -15.967 7.758 1.00 0.00 H
+ATOM 1240 2HG GLN B 77 -12.726 -15.306 9.178 1.00 0.00 H
+ATOM 1241 1HE2 GLN B 77 -12.515 -19.269 8.692 1.00 0.00 H
+ATOM 1242 2HE2 GLN B 77 -11.434 -18.082 7.987 1.00 0.00 H
+ATOM 1243 N ILE B 78 -16.286 -15.505 5.427 1.00 0.00 N
+ATOM 1244 CA ILE B 78 -17.278 -14.843 4.585 1.00 0.00 C
+ATOM 1245 C ILE B 78 -17.069 -13.327 4.613 1.00 0.00 C
+ATOM 1246 O ILE B 78 -17.215 -12.645 3.599 1.00 0.00 O
+ATOM 1247 CB ILE B 78 -18.696 -15.219 5.011 1.00 0.00 C
+ATOM 1248 CG1 ILE B 78 -18.905 -16.757 4.805 1.00 0.00 C
+ATOM 1249 CG2 ILE B 78 -19.690 -14.413 4.211 1.00 0.00 C
+ATOM 1250 CD1 ILE B 78 -18.700 -17.245 3.367 1.00 0.00 C
+ATOM 1251 H ILE B 78 -16.576 -16.113 6.182 1.00 0.00 H
+ATOM 1252 HA ILE B 78 -17.139 -15.181 3.559 1.00 0.00 H
+ATOM 1253 HB ILE B 78 -18.830 -15.005 6.072 1.00 0.00 H
+ATOM 1254 1HG1 ILE B 78 -18.212 -17.289 5.450 1.00 0.00 H
+ATOM 1255 2HG1 ILE B 78 -19.921 -17.015 5.107 1.00 0.00 H
+ATOM 1256 1HG2 ILE B 78 -20.699 -14.676 4.516 1.00 0.00 H
+ATOM 1257 2HG2 ILE B 78 -19.517 -13.353 4.390 1.00 0.00 H
+ATOM 1258 3HG2 ILE B 78 -19.558 -14.632 3.149 1.00 0.00 H
+ATOM 1259 1HD1 ILE B 78 -18.862 -18.324 3.316 1.00 0.00 H
+ATOM 1260 2HD1 ILE B 78 -19.401 -16.742 2.700 1.00 0.00 H
+ATOM 1261 3HD1 ILE B 78 -17.685 -17.021 3.049 1.00 0.00 H
+ATOM 1262 N LYS B 79 -16.702 -12.813 5.783 1.00 0.00 N
+ATOM 1263 CA LYS B 79 -16.423 -11.398 6.031 1.00 0.00 C
+ATOM 1264 C LYS B 79 -15.258 -10.823 5.194 1.00 0.00 C
+ATOM 1265 O LYS B 79 -15.065 -9.599 5.157 1.00 0.00 O
+ATOM 1266 CB LYS B 79 -16.113 -11.156 7.510 1.00 0.00 C
+ATOM 1267 CG LYS B 79 -17.289 -11.310 8.461 1.00 0.00 C
+ATOM 1268 CD LYS B 79 -16.865 -11.026 9.902 1.00 0.00 C
+ATOM 1269 CE LYS B 79 -18.039 -11.137 10.864 1.00 0.00 C
+ATOM 1270 NZ LYS B 79 -17.626 -10.880 12.274 1.00 0.00 N
+ATOM 1271 H LYS B 79 -16.607 -13.455 6.554 1.00 0.00 H
+ATOM 1272 HA LYS B 79 -17.320 -10.835 5.772 1.00 0.00 H
+ATOM 1273 1HB LYS B 79 -15.337 -11.856 7.828 1.00 0.00 H
+ATOM 1274 2HB LYS B 79 -15.712 -10.152 7.636 1.00 0.00 H
+ATOM 1275 1HG LYS B 79 -18.078 -10.613 8.178 1.00 0.00 H
+ATOM 1276 2HG LYS B 79 -17.681 -12.325 8.396 1.00 0.00 H
+ATOM 1277 1HD LYS B 79 -16.093 -11.737 10.201 1.00 0.00 H
+ATOM 1278 2HD LYS B 79 -16.451 -10.020 9.969 1.00 0.00 H
+ATOM 1279 1HE LYS B 79 -18.804 -10.413 10.582 1.00 0.00 H
+ATOM 1280 2HE LYS B 79 -18.462 -12.140 10.797 1.00 0.00 H
+ATOM 1281 1HZ LYS B 79 -18.430 -10.964 12.881 1.00 0.00 H
+ATOM 1282 2HZ LYS B 79 -16.926 -11.556 12.548 1.00 0.00 H
+ATOM 1283 3HZ LYS B 79 -17.244 -9.949 12.349 1.00 0.00 H
+ATOM 1284 N ASP B 80 -14.451 -11.684 4.571 1.00 0.00 N
+ATOM 1285 CA ASP B 80 -13.320 -11.242 3.772 1.00 0.00 C
+ATOM 1286 C ASP B 80 -13.732 -10.834 2.364 1.00 0.00 C
+ATOM 1287 O ASP B 80 -12.917 -10.323 1.595 1.00 0.00 O
+ATOM 1288 CB ASP B 80 -12.259 -12.336 3.677 1.00 0.00 C
+ATOM 1289 CG ASP B 80 -11.514 -12.600 4.974 1.00 0.00 C
+ATOM 1290 OD1 ASP B 80 -11.206 -11.679 5.683 1.00 0.00 O
+ATOM 1291 OD2 ASP B 80 -11.235 -13.751 5.253 1.00 0.00 O
+ATOM 1292 H ASP B 80 -14.640 -12.683 4.608 1.00 0.00 H
+ATOM 1293 HA ASP B 80 -12.879 -10.371 4.254 1.00 0.00 H
+ATOM 1294 1HB ASP B 80 -12.735 -13.261 3.362 1.00 0.00 H
+ATOM 1295 2HB ASP B 80 -11.537 -12.074 2.907 1.00 0.00 H
+ATOM 1296 N LYS B 81 -14.988 -11.055 2.000 1.00 0.00 N
+ATOM 1297 CA LYS B 81 -15.398 -10.647 0.668 1.00 0.00 C
+ATOM 1298 C LYS B 81 -15.326 -9.119 0.567 1.00 0.00 C
+ATOM 1299 O LYS B 81 -15.523 -8.404 1.559 1.00 0.00 O
+ATOM 1300 CB LYS B 81 -16.773 -11.236 0.354 1.00 0.00 C
+ATOM 1301 CG LYS B 81 -17.892 -10.739 1.178 1.00 0.00 C
+ATOM 1302 CD LYS B 81 -19.126 -11.606 0.884 1.00 0.00 C
+ATOM 1303 CE LYS B 81 -20.290 -11.245 1.735 1.00 0.00 C
+ATOM 1304 NZ LYS B 81 -21.483 -12.070 1.406 1.00 0.00 N
+ATOM 1305 H LYS B 81 -15.662 -11.498 2.629 1.00 0.00 H
+ATOM 1306 HA LYS B 81 -14.692 -11.058 -0.053 1.00 0.00 H
+ATOM 1307 1HB LYS B 81 -17.018 -11.034 -0.688 1.00 0.00 H
+ATOM 1308 2HB LYS B 81 -16.732 -12.320 0.477 1.00 0.00 H
+ATOM 1309 1HG LYS B 81 -17.640 -10.761 2.224 1.00 0.00 H
+ATOM 1310 2HG LYS B 81 -18.104 -9.706 0.902 1.00 0.00 H
+ATOM 1311 1HD LYS B 81 -19.405 -11.502 -0.165 1.00 0.00 H
+ATOM 1312 2HD LYS B 81 -18.874 -12.651 1.077 1.00 0.00 H
+ATOM 1313 1HE LYS B 81 -20.026 -11.394 2.781 1.00 0.00 H
+ATOM 1314 2HE LYS B 81 -20.520 -10.220 1.581 1.00 0.00 H
+ATOM 1315 1HZ LYS B 81 -22.257 -11.801 1.997 1.00 0.00 H
+ATOM 1316 2HZ LYS B 81 -21.727 -11.914 0.440 1.00 0.00 H
+ATOM 1317 3HZ LYS B 81 -21.268 -13.045 1.551 1.00 0.00 H
+ATOM 1318 N GLY B 82 -15.025 -8.623 -0.619 1.00 0.00 N
+ATOM 1319 CA GLY B 82 -14.942 -7.181 -0.827 1.00 0.00 C
+ATOM 1320 C GLY B 82 -13.826 -6.794 -1.786 1.00 0.00 C
+ATOM 1321 O GLY B 82 -13.223 -7.651 -2.441 1.00 0.00 O
+ATOM 1322 H GLY B 82 -14.885 -9.272 -1.390 1.00 0.00 H
+ATOM 1323 1HA GLY B 82 -15.897 -6.814 -1.206 1.00 0.00 H
+ATOM 1324 2HA GLY B 82 -14.775 -6.690 0.130 1.00 0.00 H
+ATOM 1325 N LEU B 83 -13.577 -5.493 -1.902 1.00 0.00 N
+ATOM 1326 CA LEU B 83 -12.523 -5.020 -2.807 1.00 0.00 C
+ATOM 1327 C LEU B 83 -11.178 -4.965 -2.144 1.00 0.00 C
+ATOM 1328 O LEU B 83 -11.046 -4.544 -0.994 1.00 0.00 O
+ATOM 1329 CB LEU B 83 -12.811 -3.632 -3.382 1.00 0.00 C
+ATOM 1330 CG LEU B 83 -13.933 -3.558 -4.272 1.00 0.00 C
+ATOM 1331 CD1 LEU B 83 -14.250 -2.090 -4.577 1.00 0.00 C
+ATOM 1332 CD2 LEU B 83 -13.606 -4.288 -5.577 1.00 0.00 C
+ATOM 1333 H LEU B 83 -14.112 -4.845 -1.320 1.00 0.00 H
+ATOM 1334 HA LEU B 83 -12.454 -5.715 -3.639 1.00 0.00 H
+ATOM 1335 1HB LEU B 83 -12.996 -2.946 -2.581 1.00 0.00 H
+ATOM 1336 2HB LEU B 83 -11.932 -3.292 -3.926 1.00 0.00 H
+ATOM 1337 HG LEU B 83 -14.735 -4.036 -3.819 1.00 0.00 H
+ATOM 1338 1HD1 LEU B 83 -15.104 -2.034 -5.253 1.00 0.00 H
+ATOM 1339 2HD1 LEU B 83 -14.493 -1.571 -3.649 1.00 0.00 H
+ATOM 1340 3HD1 LEU B 83 -13.389 -1.619 -5.047 1.00 0.00 H
+ATOM 1341 1HD2 LEU B 83 -14.469 -4.243 -6.242 1.00 0.00 H
+ATOM 1342 2HD2 LEU B 83 -12.747 -3.818 -6.056 1.00 0.00 H
+ATOM 1343 3HD2 LEU B 83 -13.372 -5.313 -5.358 1.00 0.00 H
+ATOM 1344 N TYR B 84 -10.173 -5.348 -2.924 1.00 0.00 N
+ATOM 1345 CA TYR B 84 -8.783 -5.278 -2.523 1.00 0.00 C
+ATOM 1346 C TYR B 84 -7.959 -4.573 -3.588 1.00 0.00 C
+ATOM 1347 O TYR B 84 -8.231 -4.658 -4.795 1.00 0.00 O
+ATOM 1348 CB TYR B 84 -8.219 -6.668 -2.217 1.00 0.00 C
+ATOM 1349 CG TYR B 84 -8.907 -7.305 -1.052 1.00 0.00 C
+ATOM 1350 CD1 TYR B 84 -8.403 -7.080 0.234 1.00 0.00 C
+ATOM 1351 CD2 TYR B 84 -10.035 -8.095 -1.225 1.00 0.00 C
+ATOM 1352 CE1 TYR B 84 -9.025 -7.628 1.330 1.00 0.00 C
+ATOM 1353 CE2 TYR B 84 -10.659 -8.633 -0.091 1.00 0.00 C
+ATOM 1354 CZ TYR B 84 -10.147 -8.396 1.172 1.00 0.00 C
+ATOM 1355 OH TYR B 84 -10.758 -8.923 2.291 1.00 0.00 O
+ATOM 1356 H TYR B 84 -10.401 -5.715 -3.845 1.00 0.00 H
+ATOM 1357 HA TYR B 84 -8.718 -4.683 -1.620 1.00 0.00 H
+ATOM 1358 1HB TYR B 84 -8.333 -7.312 -3.090 1.00 0.00 H
+ATOM 1359 2HB TYR B 84 -7.158 -6.588 -1.991 1.00 0.00 H
+ATOM 1360 HD1 TYR B 84 -7.519 -6.459 0.367 1.00 0.00 H
+ATOM 1361 HD2 TYR B 84 -10.443 -8.279 -2.220 1.00 0.00 H
+ATOM 1362 HE1 TYR B 84 -8.643 -7.446 2.324 1.00 0.00 H
+ATOM 1363 HE2 TYR B 84 -11.549 -9.235 -0.194 1.00 0.00 H
+ATOM 1364 HH TYR B 84 -11.534 -9.439 2.018 1.00 0.00 H
+ATOM 1365 N GLN B 85 -6.940 -3.869 -3.147 1.00 0.00 N
+ATOM 1366 CA GLN B 85 -6.119 -3.145 -4.085 1.00 0.00 C
+ATOM 1367 C GLN B 85 -4.632 -3.381 -3.849 1.00 0.00 C
+ATOM 1368 O GLN B 85 -4.156 -3.219 -2.726 1.00 0.00 O
+ATOM 1369 CB GLN B 85 -6.423 -1.649 -3.899 1.00 0.00 C
+ATOM 1370 CG GLN B 85 -7.917 -1.278 -4.021 1.00 0.00 C
+ATOM 1371 CD GLN B 85 -8.205 0.216 -3.835 1.00 0.00 C
+ATOM 1372 OE1 GLN B 85 -7.499 0.959 -3.130 1.00 0.00 O
+ATOM 1373 NE2 GLN B 85 -9.288 0.673 -4.478 1.00 0.00 N
+ATOM 1374 H GLN B 85 -6.779 -3.794 -2.145 1.00 0.00 H
+ATOM 1375 HA GLN B 85 -6.360 -3.460 -5.101 1.00 0.00 H
+ATOM 1376 1HB GLN B 85 -6.078 -1.328 -2.923 1.00 0.00 H
+ATOM 1377 2HB GLN B 85 -5.871 -1.074 -4.650 1.00 0.00 H
+ATOM 1378 1HG GLN B 85 -8.284 -1.588 -4.992 1.00 0.00 H
+ATOM 1379 2HG GLN B 85 -8.469 -1.796 -3.237 1.00 0.00 H
+ATOM 1380 1HE2 GLN B 85 -9.548 1.637 -4.399 1.00 0.00 H
+ATOM 1381 2HE2 GLN B 85 -9.847 0.049 -5.027 1.00 0.00 H
+ATOM 1382 N CYS B 86 -3.910 -3.729 -4.916 1.00 0.00 N
+ATOM 1383 CA CYS B 86 -2.444 -3.789 -4.906 1.00 0.00 C
+ATOM 1384 C CYS B 86 -1.925 -2.480 -5.391 1.00 0.00 C
+ATOM 1385 O CYS B 86 -2.259 -2.078 -6.508 1.00 0.00 O
+ATOM 1386 CB CYS B 86 -1.846 -4.851 -5.822 1.00 0.00 C
+ATOM 1387 SG CYS B 86 -0.024 -4.638 -5.947 1.00 0.00 S
+ATOM 1388 H CYS B 86 -4.391 -3.912 -5.796 1.00 0.00 H
+ATOM 1389 HA CYS B 86 -2.090 -3.965 -3.891 1.00 0.00 H
+ATOM 1390 1HB CYS B 86 -2.060 -5.850 -5.442 1.00 0.00 H
+ATOM 1391 2HB CYS B 86 -2.278 -4.763 -6.798 1.00 0.00 H
+ATOM 1392 N ILE B 87 -1.149 -1.786 -4.572 1.00 0.00 N
+ATOM 1393 CA ILE B 87 -0.664 -0.477 -4.970 1.00 0.00 C
+ATOM 1394 C ILE B 87 0.868 -0.428 -4.891 1.00 0.00 C
+ATOM 1395 O ILE B 87 1.452 -0.676 -3.834 1.00 0.00 O
+ATOM 1396 CB ILE B 87 -1.268 0.602 -4.083 1.00 0.00 C
+ATOM 1397 CG1 ILE B 87 -2.826 0.523 -4.116 1.00 0.00 C
+ATOM 1398 CG2 ILE B 87 -0.846 1.937 -4.665 1.00 0.00 C
+ATOM 1399 CD1 ILE B 87 -3.517 1.406 -3.166 1.00 0.00 C
+ATOM 1400 H ILE B 87 -0.912 -2.170 -3.659 1.00 0.00 H
+ATOM 1401 HA ILE B 87 -0.951 -0.280 -6.002 1.00 0.00 H
+ATOM 1402 HB ILE B 87 -0.917 0.489 -3.053 1.00 0.00 H
+ATOM 1403 1HG1 ILE B 87 -3.154 0.771 -5.107 1.00 0.00 H
+ATOM 1404 2HG1 ILE B 87 -3.143 -0.492 -3.882 1.00 0.00 H
+ATOM 1405 1HG2 ILE B 87 -1.256 2.755 -4.078 1.00 0.00 H
+ATOM 1406 2HG2 ILE B 87 0.228 2.006 -4.690 1.00 0.00 H
+ATOM 1407 3HG2 ILE B 87 -1.224 1.995 -5.686 1.00 0.00 H
+ATOM 1408 1HD1 ILE B 87 -4.595 1.275 -3.273 1.00 0.00 H
+ATOM 1409 2HD1 ILE B 87 -3.211 1.152 -2.149 1.00 0.00 H
+ATOM 1410 3HD1 ILE B 87 -3.257 2.440 -3.376 1.00 0.00 H
+ATOM 1411 N ILE B 88 1.508 -0.105 -6.007 1.00 0.00 N
+ATOM 1412 CA ILE B 88 2.961 -0.099 -6.076 1.00 0.00 C
+ATOM 1413 C ILE B 88 3.489 1.313 -6.240 1.00 0.00 C
+ATOM 1414 O ILE B 88 3.037 2.054 -7.125 1.00 0.00 O
+ATOM 1415 CB ILE B 88 3.487 -0.957 -7.248 1.00 0.00 C
+ATOM 1416 CG1 ILE B 88 3.003 -2.410 -7.102 1.00 0.00 C
+ATOM 1417 CG2 ILE B 88 5.014 -0.920 -7.304 1.00 0.00 C
+ATOM 1418 CD1 ILE B 88 3.384 -3.305 -8.272 1.00 0.00 C
+ATOM 1419 H ILE B 88 0.972 0.113 -6.841 1.00 0.00 H
+ATOM 1420 HA ILE B 88 3.364 -0.502 -5.147 1.00 0.00 H
+ATOM 1421 HB ILE B 88 3.108 -0.564 -8.150 1.00 0.00 H
+ATOM 1422 1HG1 ILE B 88 3.395 -2.828 -6.213 1.00 0.00 H
+ATOM 1423 2HG1 ILE B 88 1.916 -2.404 -7.024 1.00 0.00 H
+ATOM 1424 1HG2 ILE B 88 5.355 -1.514 -8.149 1.00 0.00 H
+ATOM 1425 2HG2 ILE B 88 5.347 0.102 -7.427 1.00 0.00 H
+ATOM 1426 3HG2 ILE B 88 5.429 -1.314 -6.391 1.00 0.00 H
+ATOM 1427 1HD1 ILE B 88 2.999 -4.308 -8.098 1.00 0.00 H
+ATOM 1428 2HD1 ILE B 88 2.972 -2.919 -9.189 1.00 0.00 H
+ATOM 1429 3HD1 ILE B 88 4.464 -3.344 -8.353 1.00 0.00 H
+ATOM 1430 N HIS B 89 4.455 1.684 -5.388 1.00 0.00 N
+ATOM 1431 CA HIS B 89 5.046 3.000 -5.438 1.00 0.00 C
+ATOM 1432 C HIS B 89 6.535 2.917 -5.639 1.00 0.00 C
+ATOM 1433 O HIS B 89 7.182 1.997 -5.143 1.00 0.00 O
+ATOM 1434 CB HIS B 89 4.837 3.730 -4.118 1.00 0.00 C
+ATOM 1435 CG HIS B 89 3.455 3.860 -3.694 1.00 0.00 C
+ATOM 1436 ND1 HIS B 89 2.786 2.840 -3.080 1.00 0.00 N
+ATOM 1437 CD2 HIS B 89 2.600 4.896 -3.749 1.00 0.00 C
+ATOM 1438 CE1 HIS B 89 1.572 3.238 -2.775 1.00 0.00 C
+ATOM 1439 NE2 HIS B 89 1.429 4.487 -3.178 1.00 0.00 N
+ATOM 1440 H HIS B 89 4.747 1.041 -4.655 1.00 0.00 H
+ATOM 1441 HA HIS B 89 4.629 3.561 -6.254 1.00 0.00 H
+ATOM 1442 1HB HIS B 89 5.389 3.231 -3.337 1.00 0.00 H
+ATOM 1443 2HB HIS B 89 5.238 4.731 -4.215 1.00 0.00 H
+ATOM 1444 HD1 HIS B 89 3.165 1.927 -2.862 1.00 0.00 H
+ATOM 1445 HD2 HIS B 89 2.698 5.908 -4.136 1.00 0.00 H
+ATOM 1446 HE1 HIS B 89 0.875 2.564 -2.279 1.00 0.00 H
+ATOM 1447 N HIS B 90 7.092 3.906 -6.315 1.00 0.00 N
+ATOM 1448 CA HIS B 90 8.522 4.028 -6.489 1.00 0.00 C
+ATOM 1449 C HIS B 90 9.066 4.972 -5.457 1.00 0.00 C
+ATOM 1450 O HIS B 90 8.632 6.131 -5.378 1.00 0.00 O
+ATOM 1451 CB HIS B 90 8.879 4.578 -7.861 1.00 0.00 C
+ATOM 1452 CG HIS B 90 10.343 4.703 -8.054 1.00 0.00 C
+ATOM 1453 ND1 HIS B 90 10.910 5.747 -8.746 1.00 0.00 N
+ATOM 1454 CD2 HIS B 90 11.362 3.895 -7.675 1.00 0.00 C
+ATOM 1455 CE1 HIS B 90 12.236 5.605 -8.739 1.00 0.00 C
+ATOM 1456 NE2 HIS B 90 12.534 4.481 -8.102 1.00 0.00 N
+ATOM 1457 H HIS B 90 6.490 4.611 -6.713 1.00 0.00 H
+ATOM 1458 HA HIS B 90 9.007 3.073 -6.350 1.00 0.00 H
+ATOM 1459 1HB HIS B 90 8.496 3.913 -8.633 1.00 0.00 H
+ATOM 1460 2HB HIS B 90 8.413 5.560 -8.002 1.00 0.00 H
+ATOM 1461 HD2 HIS B 90 11.266 2.972 -7.134 1.00 0.00 H
+ATOM 1462 HE1 HIS B 90 12.950 6.287 -9.199 1.00 0.00 H
+ATOM 1463 HE2 HIS B 90 13.464 4.107 -7.951 1.00 0.00 H
+ATOM 1464 N LYS B 91 9.942 4.471 -4.590 1.00 0.00 N
+ATOM 1465 CA LYS B 91 10.485 5.298 -3.524 1.00 0.00 C
+ATOM 1466 C LYS B 91 11.316 6.449 -4.111 1.00 0.00 C
+ATOM 1467 O LYS B 91 12.154 6.223 -4.988 1.00 0.00 O
+ATOM 1468 CB LYS B 91 11.315 4.457 -2.570 1.00 0.00 C
+ATOM 1469 CG LYS B 91 11.851 5.238 -1.356 1.00 0.00 C
+ATOM 1470 CD LYS B 91 10.735 5.550 -0.386 1.00 0.00 C
+ATOM 1471 CE LYS B 91 11.199 6.444 0.734 1.00 0.00 C
+ATOM 1472 NZ LYS B 91 11.310 7.873 0.285 1.00 0.00 N
+ATOM 1473 H LYS B 91 10.278 3.512 -4.697 1.00 0.00 H
+ATOM 1474 HA LYS B 91 9.656 5.732 -2.964 1.00 0.00 H
+ATOM 1475 1HB LYS B 91 10.702 3.642 -2.210 1.00 0.00 H
+ATOM 1476 2HB LYS B 91 12.160 4.020 -3.105 1.00 0.00 H
+ATOM 1477 1HG LYS B 91 12.606 4.643 -0.848 1.00 0.00 H
+ATOM 1478 2HG LYS B 91 12.307 6.175 -1.676 1.00 0.00 H
+ATOM 1479 1HD LYS B 91 9.929 6.063 -0.915 1.00 0.00 H
+ATOM 1480 2HD LYS B 91 10.342 4.619 0.032 1.00 0.00 H
+ATOM 1481 1HE LYS B 91 10.488 6.388 1.555 1.00 0.00 H
+ATOM 1482 2HE LYS B 91 12.175 6.108 1.086 1.00 0.00 H
+ATOM 1483 1HZ LYS B 91 11.614 8.453 1.044 1.00 0.00 H
+ATOM 1484 2HZ LYS B 91 11.969 7.989 -0.493 1.00 0.00 H
+ATOM 1485 3HZ LYS B 91 10.390 8.195 -0.050 1.00 0.00 H
+ATOM 1486 N LYS B 92 11.082 7.677 -3.636 1.00 0.00 N
+ATOM 1487 CA LYS B 92 11.810 8.858 -4.115 1.00 0.00 C
+ATOM 1488 C LYS B 92 12.725 9.357 -2.997 1.00 0.00 C
+ATOM 1489 O LYS B 92 12.620 8.848 -1.882 1.00 0.00 O
+ATOM 1490 CB LYS B 92 10.805 9.935 -4.521 1.00 0.00 C
+ATOM 1491 CG LYS B 92 9.835 9.478 -5.538 1.00 0.00 C
+ATOM 1492 CD LYS B 92 10.540 8.958 -6.739 1.00 0.00 C
+ATOM 1493 CE LYS B 92 11.190 10.038 -7.523 1.00 0.00 C
+ATOM 1494 NZ LYS B 92 11.793 9.494 -8.722 1.00 0.00 N
+ATOM 1495 H LYS B 92 10.364 7.819 -2.925 1.00 0.00 H
+ATOM 1496 HA LYS B 92 12.413 8.577 -4.972 1.00 0.00 H
+ATOM 1497 1HB LYS B 92 10.232 10.230 -3.653 1.00 0.00 H
+ATOM 1498 2HB LYS B 92 11.274 10.803 -4.911 1.00 0.00 H
+ATOM 1499 1HG LYS B 92 9.210 8.707 -5.147 1.00 0.00 H
+ATOM 1500 2HG LYS B 92 9.220 10.322 -5.813 1.00 0.00 H
+ATOM 1501 1HD LYS B 92 11.309 8.258 -6.431 1.00 0.00 H
+ATOM 1502 2HD LYS B 92 9.846 8.409 -7.371 1.00 0.00 H
+ATOM 1503 1HE LYS B 92 10.462 10.789 -7.806 1.00 0.00 H
+ATOM 1504 2HE LYS B 92 11.967 10.509 -6.924 1.00 0.00 H
+ATOM 1505 1HZ LYS B 92 12.227 10.241 -9.240 1.00 0.00 H
+ATOM 1506 2HZ LYS B 92 12.484 8.811 -8.491 1.00 0.00 H
+ATOM 1507 3HZ LYS B 92 11.075 9.064 -9.308 1.00 0.00 H
+ATOM 1508 N PRO B 93 13.693 10.280 -3.233 1.00 0.00 N
+ATOM 1509 CA PRO B 93 14.510 10.874 -2.172 1.00 0.00 C
+ATOM 1510 C PRO B 93 13.620 11.315 -1.001 1.00 0.00 C
+ATOM 1511 O PRO B 93 13.945 11.061 0.161 1.00 0.00 O
+ATOM 1512 CB PRO B 93 15.189 12.031 -2.911 1.00 0.00 C
+ATOM 1513 CG PRO B 93 15.340 11.506 -4.327 1.00 0.00 C
+ATOM 1514 CD PRO B 93 14.045 10.750 -4.597 1.00 0.00 C
+ATOM 1515 HA PRO B 93 15.250 10.134 -1.830 1.00 0.00 H
+ATOM 1516 1HB PRO B 93 14.576 12.932 -2.846 1.00 0.00 H
+ATOM 1517 2HB PRO B 93 16.152 12.273 -2.440 1.00 0.00 H
+ATOM 1518 1HG PRO B 93 15.493 12.348 -5.028 1.00 0.00 H
+ATOM 1519 2HG PRO B 93 16.230 10.868 -4.403 1.00 0.00 H
+ATOM 1520 1HD PRO B 93 13.317 11.456 -4.980 1.00 0.00 H
+ATOM 1521 2HD PRO B 93 14.231 9.913 -5.277 1.00 0.00 H
+ATOM 1522 N THR B 94 12.462 11.916 -1.318 1.00 0.00 N
+ATOM 1523 CA THR B 94 11.450 12.247 -0.327 1.00 0.00 C
+ATOM 1524 C THR B 94 10.119 11.747 -0.864 1.00 0.00 C
+ATOM 1525 O THR B 94 9.795 11.990 -2.025 1.00 0.00 O
+ATOM 1526 CB THR B 94 11.384 13.755 0.012 1.00 0.00 C
+ATOM 1527 OG1 THR B 94 12.630 14.180 0.575 1.00 0.00 O
+ATOM 1528 CG2 THR B 94 10.286 14.021 1.005 1.00 0.00 C
+ATOM 1529 H THR B 94 12.263 12.120 -2.285 1.00 0.00 H
+ATOM 1530 HA THR B 94 11.666 11.711 0.598 1.00 0.00 H
+ATOM 1531 HB THR B 94 11.189 14.324 -0.897 1.00 0.00 H
+ATOM 1532 HG1 THR B 94 12.751 13.736 1.425 1.00 0.00 H
+ATOM 1533 1HG2 THR B 94 10.263 15.088 1.228 1.00 0.00 H
+ATOM 1534 2HG2 THR B 94 9.331 13.721 0.587 1.00 0.00 H
+ATOM 1535 3HG2 THR B 94 10.478 13.463 1.917 1.00 0.00 H
+ATOM 1536 N GLY B 95 9.354 11.062 -0.015 1.00 0.00 N
+ATOM 1537 CA GLY B 95 8.056 10.527 -0.403 1.00 0.00 C
+ATOM 1538 C GLY B 95 8.205 9.363 -1.370 1.00 0.00 C
+ATOM 1539 O GLY B 95 9.265 8.724 -1.435 1.00 0.00 O
+ATOM 1540 H GLY B 95 9.680 10.918 0.929 1.00 0.00 H
+ATOM 1541 1HA GLY B 95 7.513 10.202 0.485 1.00 0.00 H
+ATOM 1542 2HA GLY B 95 7.463 11.314 -0.868 1.00 0.00 H
+ATOM 1543 N MET B 96 7.115 9.057 -2.067 1.00 0.00 N
+ATOM 1544 CA MET B 96 7.059 7.962 -3.009 1.00 0.00 C
+ATOM 1545 C MET B 96 6.013 8.308 -4.059 1.00 0.00 C
+ATOM 1546 O MET B 96 5.087 9.087 -3.791 1.00 0.00 O
+ATOM 1547 CB MET B 96 6.784 6.635 -2.305 1.00 0.00 C
+ATOM 1548 CG MET B 96 5.440 6.521 -1.601 1.00 0.00 C
+ATOM 1549 SD MET B 96 5.295 4.943 -0.682 1.00 0.00 S
+ATOM 1550 CE MET B 96 6.407 5.317 0.698 1.00 0.00 C
+ATOM 1551 H MET B 96 6.289 9.628 -1.948 1.00 0.00 H
+ATOM 1552 HA MET B 96 8.020 7.873 -3.508 1.00 0.00 H
+ATOM 1553 1HB MET B 96 6.865 5.830 -3.030 1.00 0.00 H
+ATOM 1554 2HB MET B 96 7.547 6.458 -1.554 1.00 0.00 H
+ATOM 1555 1HG MET B 96 5.311 7.344 -0.905 1.00 0.00 H
+ATOM 1556 2HG MET B 96 4.642 6.569 -2.343 1.00 0.00 H
+ATOM 1557 1HE MET B 96 6.443 4.467 1.380 1.00 0.00 H
+ATOM 1558 2HE MET B 96 7.407 5.522 0.317 1.00 0.00 H
+ATOM 1559 3HE MET B 96 6.042 6.192 1.238 1.00 0.00 H
+ATOM 1560 N ILE B 97 6.169 7.744 -5.241 1.00 0.00 N
+ATOM 1561 CA ILE B 97 5.244 7.946 -6.365 1.00 0.00 C
+ATOM 1562 C ILE B 97 4.537 6.691 -6.790 1.00 0.00 C
+ATOM 1563 O ILE B 97 5.175 5.685 -7.062 1.00 0.00 O
+ATOM 1564 CB ILE B 97 6.016 8.522 -7.568 1.00 0.00 C
+ATOM 1565 CG1 ILE B 97 6.546 9.916 -7.197 1.00 0.00 C
+ATOM 1566 CG2 ILE B 97 5.167 8.514 -8.856 1.00 0.00 C
+ATOM 1567 CD1 ILE B 97 7.511 10.462 -8.169 1.00 0.00 C
+ATOM 1568 H ILE B 97 6.981 7.139 -5.371 1.00 0.00 H
+ATOM 1569 HA ILE B 97 4.488 8.668 -6.061 1.00 0.00 H
+ATOM 1570 HB ILE B 97 6.894 7.896 -7.740 1.00 0.00 H
+ATOM 1571 1HG1 ILE B 97 5.704 10.610 -7.124 1.00 0.00 H
+ATOM 1572 2HG1 ILE B 97 7.025 9.862 -6.235 1.00 0.00 H
+ATOM 1573 1HG2 ILE B 97 5.756 8.891 -9.677 1.00 0.00 H
+ATOM 1574 2HG2 ILE B 97 4.859 7.496 -9.086 1.00 0.00 H
+ATOM 1575 3HG2 ILE B 97 4.279 9.136 -8.726 1.00 0.00 H
+ATOM 1576 1HD1 ILE B 97 7.847 11.446 -7.848 1.00 0.00 H
+ATOM 1577 2HD1 ILE B 97 8.346 9.806 -8.263 1.00 0.00 H
+ATOM 1578 3HD1 ILE B 97 7.047 10.536 -9.081 1.00 0.00 H
+ATOM 1579 N ARG B 98 3.216 6.731 -6.895 1.00 0.00 N
+ATOM 1580 CA ARG B 98 2.486 5.540 -7.327 1.00 0.00 C
+ATOM 1581 C ARG B 98 2.785 5.252 -8.789 1.00 0.00 C
+ATOM 1582 O ARG B 98 2.677 6.151 -9.624 1.00 0.00 O
+ATOM 1583 CB ARG B 98 0.986 5.753 -7.100 1.00 0.00 C
+ATOM 1584 CG ARG B 98 0.090 4.560 -7.362 1.00 0.00 C
+ATOM 1585 CD ARG B 98 -1.244 4.855 -6.784 1.00 0.00 C
+ATOM 1586 NE ARG B 98 -1.942 5.945 -7.464 1.00 0.00 N
+ATOM 1587 CZ ARG B 98 -2.772 5.779 -8.496 1.00 0.00 C
+ATOM 1588 NH1 ARG B 98 -3.027 4.579 -8.917 1.00 0.00 N
+ATOM 1589 NH2 ARG B 98 -3.350 6.803 -9.079 1.00 0.00 N
+ATOM 1590 H ARG B 98 2.716 7.580 -6.669 1.00 0.00 H
+ATOM 1591 HA ARG B 98 2.800 4.689 -6.733 1.00 0.00 H
+ATOM 1592 1HB ARG B 98 0.815 6.067 -6.069 1.00 0.00 H
+ATOM 1593 2HB ARG B 98 0.639 6.560 -7.744 1.00 0.00 H
+ATOM 1594 1HG ARG B 98 -0.007 4.388 -8.436 1.00 0.00 H
+ATOM 1595 2HG ARG B 98 0.505 3.670 -6.882 1.00 0.00 H
+ATOM 1596 1HD ARG B 98 -1.868 3.962 -6.863 1.00 0.00 H
+ATOM 1597 2HD ARG B 98 -1.130 5.122 -5.733 1.00 0.00 H
+ATOM 1598 HE ARG B 98 -1.769 6.892 -7.150 1.00 0.00 H
+ATOM 1599 1HH1 ARG B 98 -2.576 3.796 -8.465 1.00 0.00 H
+ATOM 1600 2HH1 ARG B 98 -3.666 4.435 -9.686 1.00 0.00 H
+ATOM 1601 1HH2 ARG B 98 -3.151 7.769 -8.778 1.00 0.00 H
+ATOM 1602 2HH2 ARG B 98 -3.979 6.638 -9.847 1.00 0.00 H
+ATOM 1603 N ILE B 99 3.137 4.007 -9.104 1.00 0.00 N
+ATOM 1604 CA ILE B 99 3.440 3.645 -10.488 1.00 0.00 C
+ATOM 1605 C ILE B 99 2.535 2.544 -11.008 1.00 0.00 C
+ATOM 1606 O ILE B 99 2.505 2.276 -12.206 1.00 0.00 O
+ATOM 1607 CB ILE B 99 4.897 3.171 -10.655 1.00 0.00 C
+ATOM 1608 CG1 ILE B 99 5.154 1.891 -9.853 1.00 0.00 C
+ATOM 1609 CG2 ILE B 99 5.874 4.264 -10.215 1.00 0.00 C
+ATOM 1610 CD1 ILE B 99 6.492 1.246 -10.157 1.00 0.00 C
+ATOM 1611 H ILE B 99 3.217 3.316 -8.370 1.00 0.00 H
+ATOM 1612 HA ILE B 99 3.292 4.521 -11.115 1.00 0.00 H
+ATOM 1613 HB ILE B 99 5.072 2.947 -11.710 1.00 0.00 H
+ATOM 1614 1HG1 ILE B 99 5.104 2.126 -8.791 1.00 0.00 H
+ATOM 1615 2HG1 ILE B 99 4.385 1.160 -10.085 1.00 0.00 H
+ATOM 1616 1HG2 ILE B 99 6.898 3.921 -10.371 1.00 0.00 H
+ATOM 1617 2HG2 ILE B 99 5.695 5.159 -10.804 1.00 0.00 H
+ATOM 1618 3HG2 ILE B 99 5.724 4.488 -9.166 1.00 0.00 H
+ATOM 1619 1HD1 ILE B 99 6.607 0.342 -9.566 1.00 0.00 H
+ATOM 1620 2HD1 ILE B 99 6.539 0.994 -11.213 1.00 0.00 H
+ATOM 1621 3HD1 ILE B 99 7.297 1.942 -9.917 1.00 0.00 H
+ATOM 1622 N HIS B 100 1.806 1.891 -10.117 1.00 0.00 N
+ATOM 1623 CA HIS B 100 0.939 0.808 -10.559 1.00 0.00 C
+ATOM 1624 C HIS B 100 -0.142 0.464 -9.556 1.00 0.00 C
+ATOM 1625 O HIS B 100 0.065 0.505 -8.349 1.00 0.00 O
+ATOM 1626 CB HIS B 100 1.753 -0.443 -10.911 1.00 0.00 C
+ATOM 1627 CG HIS B 100 0.928 -1.589 -11.408 1.00 0.00 C
+ATOM 1628 ND1 HIS B 100 0.421 -1.658 -12.691 1.00 0.00 N
+ATOM 1629 CD2 HIS B 100 0.514 -2.715 -10.773 1.00 0.00 C
+ATOM 1630 CE1 HIS B 100 -0.276 -2.798 -12.812 1.00 0.00 C
+ATOM 1631 NE2 HIS B 100 -0.217 -3.437 -11.659 1.00 0.00 N
+ATOM 1632 H HIS B 100 1.905 2.111 -9.129 1.00 0.00 H
+ATOM 1633 HA HIS B 100 0.430 1.121 -11.469 1.00 0.00 H
+ATOM 1634 1HB HIS B 100 2.468 -0.196 -11.690 1.00 0.00 H
+ATOM 1635 2HB HIS B 100 2.304 -0.777 -10.082 1.00 0.00 H
+ATOM 1636 HD2 HIS B 100 0.720 -3.001 -9.743 1.00 0.00 H
+ATOM 1637 HE1 HIS B 100 -0.796 -3.149 -13.699 1.00 0.00 H
+ATOM 1638 HE2 HIS B 100 -0.635 -4.338 -11.430 1.00 0.00 H
+ATOM 1639 N GLN B 101 -1.315 0.130 -10.062 1.00 0.00 N
+ATOM 1640 CA GLN B 101 -2.384 -0.326 -9.208 1.00 0.00 C
+ATOM 1641 C GLN B 101 -3.214 -1.406 -9.893 1.00 0.00 C
+ATOM 1642 O GLN B 101 -3.506 -1.312 -11.090 1.00 0.00 O
+ATOM 1643 CB GLN B 101 -3.271 0.840 -8.734 1.00 0.00 C
+ATOM 1644 CG GLN B 101 -4.372 0.416 -7.762 1.00 0.00 C
+ATOM 1645 CD GLN B 101 -5.030 1.582 -7.069 1.00 0.00 C
+ATOM 1646 OE1 GLN B 101 -4.413 2.634 -6.882 1.00 0.00 O
+ATOM 1647 NE2 GLN B 101 -6.272 1.401 -6.664 1.00 0.00 N
+ATOM 1648 H GLN B 101 -1.447 0.140 -11.066 1.00 0.00 H
+ATOM 1649 HA GLN B 101 -1.931 -0.764 -8.335 1.00 0.00 H
+ATOM 1650 1HB GLN B 101 -2.655 1.581 -8.229 1.00 0.00 H
+ATOM 1651 2HB GLN B 101 -3.743 1.315 -9.586 1.00 0.00 H
+ATOM 1652 1HG GLN B 101 -5.142 -0.114 -8.324 1.00 0.00 H
+ATOM 1653 2HG GLN B 101 -3.957 -0.248 -7.013 1.00 0.00 H
+ATOM 1654 1HE2 GLN B 101 -6.761 2.130 -6.183 1.00 0.00 H
+ATOM 1655 2HE2 GLN B 101 -6.727 0.519 -6.862 1.00 0.00 H
+ATOM 1656 N MET B 102 -3.592 -2.422 -9.105 1.00 0.00 N
+ATOM 1657 CA MET B 102 -4.457 -3.512 -9.556 1.00 0.00 C
+ATOM 1658 C MET B 102 -5.566 -3.769 -8.559 1.00 0.00 C
+ATOM 1659 O MET B 102 -5.338 -4.101 -7.390 1.00 0.00 O
+ATOM 1660 CB MET B 102 -3.639 -4.774 -9.753 1.00 0.00 C
+ATOM 1661 CG MET B 102 -4.421 -6.024 -10.189 1.00 0.00 C
+ATOM 1662 SD MET B 102 -5.076 -5.913 -11.832 1.00 0.00 S
+ATOM 1663 CE MET B 102 -6.218 -7.317 -11.776 1.00 0.00 C
+ATOM 1664 H MET B 102 -3.252 -2.415 -8.142 1.00 0.00 H
+ATOM 1665 HA MET B 102 -4.917 -3.231 -10.503 1.00 0.00 H
+ATOM 1666 1HB MET B 102 -2.880 -4.583 -10.504 1.00 0.00 H
+ATOM 1667 2HB MET B 102 -3.129 -5.022 -8.841 1.00 0.00 H
+ATOM 1668 1HG MET B 102 -3.759 -6.891 -10.144 1.00 0.00 H
+ATOM 1669 2HG MET B 102 -5.248 -6.199 -9.497 1.00 0.00 H
+ATOM 1670 1HE MET B 102 -6.727 -7.409 -12.734 1.00 0.00 H
+ATOM 1671 2HE MET B 102 -5.663 -8.230 -11.569 1.00 0.00 H
+ATOM 1672 3HE MET B 102 -6.949 -7.154 -10.983 1.00 0.00 H
+ATOM 1673 N ASN B 103 -6.780 -3.633 -9.033 1.00 0.00 N
+ATOM 1674 CA ASN B 103 -7.934 -3.811 -8.182 1.00 0.00 C
+ATOM 1675 C ASN B 103 -8.567 -5.168 -8.432 1.00 0.00 C
+ATOM 1676 O ASN B 103 -8.680 -5.610 -9.588 1.00 0.00 O
+ATOM 1677 CB ASN B 103 -8.948 -2.723 -8.431 1.00 0.00 C
+ATOM 1678 CG ASN B 103 -8.400 -1.335 -8.149 1.00 0.00 C
+ATOM 1679 OD1 ASN B 103 -7.460 -1.143 -7.356 1.00 0.00 O
+ATOM 1680 ND2 ASN B 103 -8.975 -0.367 -8.803 1.00 0.00 N
+ATOM 1681 H ASN B 103 -6.906 -3.378 -10.002 1.00 0.00 H
+ATOM 1682 HA ASN B 103 -7.625 -3.780 -7.141 1.00 0.00 H
+ATOM 1683 1HB ASN B 103 -9.288 -2.771 -9.467 1.00 0.00 H
+ATOM 1684 2HB ASN B 103 -9.820 -2.892 -7.795 1.00 0.00 H
+ATOM 1685 1HD2 ASN B 103 -8.681 0.582 -8.691 1.00 0.00 H
+ATOM 1686 2HD2 ASN B 103 -9.724 -0.572 -9.434 1.00 0.00 H
+ATOM 1687 N SER B 104 -9.053 -5.770 -7.365 1.00 0.00 N
+ATOM 1688 CA SER B 104 -9.775 -7.016 -7.510 1.00 0.00 C
+ATOM 1689 C SER B 104 -10.901 -7.144 -6.495 1.00 0.00 C
+ATOM 1690 O SER B 104 -10.863 -6.595 -5.392 1.00 0.00 O
+ATOM 1691 CB SER B 104 -8.800 -8.172 -7.369 1.00 0.00 C
+ATOM 1692 OG SER B 104 -8.244 -8.214 -6.094 1.00 0.00 O
+ATOM 1693 H SER B 104 -8.894 -5.367 -6.441 1.00 0.00 H
+ATOM 1694 HA SER B 104 -10.214 -7.046 -8.505 1.00 0.00 H
+ATOM 1695 1HB SER B 104 -9.318 -9.105 -7.584 1.00 0.00 H
+ATOM 1696 2HB SER B 104 -8.001 -8.062 -8.102 1.00 0.00 H
+ATOM 1697 HG SER B 104 -8.042 -7.297 -5.865 1.00 0.00 H
+ATOM 1698 N GLU B 105 -11.905 -7.890 -6.881 1.00 0.00 N
+ATOM 1699 CA GLU B 105 -13.052 -8.182 -6.042 1.00 0.00 C
+ATOM 1700 C GLU B 105 -13.013 -9.596 -5.544 1.00 0.00 C
+ATOM 1701 O GLU B 105 -12.996 -10.529 -6.339 1.00 0.00 O
+ATOM 1702 CB GLU B 105 -14.341 -8.018 -6.828 1.00 0.00 C
+ATOM 1703 CG GLU B 105 -15.594 -8.374 -6.051 1.00 0.00 C
+ATOM 1704 CD GLU B 105 -16.784 -8.464 -6.927 1.00 0.00 C
+ATOM 1705 OE1 GLU B 105 -16.774 -9.328 -7.789 1.00 0.00 O
+ATOM 1706 OE2 GLU B 105 -17.713 -7.722 -6.745 1.00 0.00 O
+ATOM 1707 H GLU B 105 -11.866 -8.294 -7.804 1.00 0.00 H
+ATOM 1708 HA GLU B 105 -13.045 -7.526 -5.175 1.00 0.00 H
+ATOM 1709 1HB GLU B 105 -14.435 -6.985 -7.134 1.00 0.00 H
+ATOM 1710 2HB GLU B 105 -14.305 -8.624 -7.733 1.00 0.00 H
+ATOM 1711 1HG GLU B 105 -15.452 -9.333 -5.558 1.00 0.00 H
+ATOM 1712 2HG GLU B 105 -15.758 -7.617 -5.280 1.00 0.00 H
+ATOM 1713 N LEU B 106 -13.043 -9.776 -4.250 1.00 0.00 N
+ATOM 1714 CA LEU B 106 -13.037 -11.113 -3.720 1.00 0.00 C
+ATOM 1715 C LEU B 106 -14.428 -11.533 -3.292 1.00 0.00 C
+ATOM 1716 O LEU B 106 -15.097 -10.832 -2.528 1.00 0.00 O
+ATOM 1717 CB LEU B 106 -12.057 -11.213 -2.546 1.00 0.00 C
+ATOM 1718 CG LEU B 106 -11.986 -12.546 -1.810 1.00 0.00 C
+ATOM 1719 CD1 LEU B 106 -11.495 -13.602 -2.721 1.00 0.00 C
+ATOM 1720 CD2 LEU B 106 -11.022 -12.400 -0.616 1.00 0.00 C
+ATOM 1721 H LEU B 106 -13.066 -8.981 -3.621 1.00 0.00 H
+ATOM 1722 HA LEU B 106 -12.715 -11.800 -4.492 1.00 0.00 H
+ATOM 1723 1HB LEU B 106 -11.055 -10.969 -2.898 1.00 0.00 H
+ATOM 1724 2HB LEU B 106 -12.356 -10.467 -1.816 1.00 0.00 H
+ATOM 1725 HG LEU B 106 -12.978 -12.825 -1.459 1.00 0.00 H
+ATOM 1726 1HD1 LEU B 106 -11.450 -14.531 -2.175 1.00 0.00 H
+ATOM 1727 2HD1 LEU B 106 -12.181 -13.706 -3.562 1.00 0.00 H
+ATOM 1728 3HD1 LEU B 106 -10.507 -13.343 -3.089 1.00 0.00 H
+ATOM 1729 1HD2 LEU B 106 -10.970 -13.339 -0.093 1.00 0.00 H
+ATOM 1730 2HD2 LEU B 106 -10.029 -12.128 -0.972 1.00 0.00 H
+ATOM 1731 3HD2 LEU B 106 -11.382 -11.630 0.059 1.00 0.00 H
+ATOM 1732 N SER B 107 -14.856 -12.687 -3.791 1.00 0.00 N
+ATOM 1733 CA SER B 107 -16.071 -13.331 -3.345 1.00 0.00 C
+ATOM 1734 C SER B 107 -15.587 -14.545 -2.574 1.00 0.00 C
+ATOM 1735 O SER B 107 -14.638 -15.207 -3.011 1.00 0.00 O
+ATOM 1736 CB SER B 107 -16.960 -13.783 -4.509 1.00 0.00 C
+ATOM 1737 OG SER B 107 -17.453 -12.701 -5.253 1.00 0.00 O
+ATOM 1738 H SER B 107 -14.290 -13.151 -4.499 1.00 0.00 H
+ATOM 1739 HA SER B 107 -16.627 -12.672 -2.679 1.00 0.00 H
+ATOM 1740 1HB SER B 107 -16.389 -14.441 -5.162 1.00 0.00 H
+ATOM 1741 2HB SER B 107 -17.792 -14.367 -4.115 1.00 0.00 H
+ATOM 1742 HG SER B 107 -16.684 -12.282 -5.656 1.00 0.00 H
+ATOM 1743 N VAL B 108 -16.237 -14.855 -1.454 1.00 0.00 N
+ATOM 1744 CA VAL B 108 -15.893 -16.062 -0.716 1.00 0.00 C
+ATOM 1745 C VAL B 108 -17.118 -16.980 -0.732 1.00 0.00 C
+ATOM 1746 O VAL B 108 -18.163 -16.638 -0.183 1.00 0.00 O
+ATOM 1747 CB VAL B 108 -15.446 -15.734 0.730 1.00 0.00 C
+ATOM 1748 CG1 VAL B 108 -15.088 -17.035 1.463 1.00 0.00 C
+ATOM 1749 CG2 VAL B 108 -14.227 -14.783 0.698 1.00 0.00 C
+ATOM 1750 H VAL B 108 -16.989 -14.264 -1.129 1.00 0.00 H
+ATOM 1751 HA VAL B 108 -15.068 -16.568 -1.212 1.00 0.00 H
+ATOM 1752 HB VAL B 108 -16.270 -15.261 1.266 1.00 0.00 H
+ATOM 1753 1HG1 VAL B 108 -14.784 -16.812 2.487 1.00 0.00 H
+ATOM 1754 2HG1 VAL B 108 -15.951 -17.696 1.477 1.00 0.00 H
+ATOM 1755 3HG1 VAL B 108 -14.269 -17.519 0.938 1.00 0.00 H
+ATOM 1756 1HG2 VAL B 108 -13.916 -14.560 1.717 1.00 0.00 H
+ATOM 1757 2HG2 VAL B 108 -13.407 -15.256 0.164 1.00 0.00 H
+ATOM 1758 3HG2 VAL B 108 -14.495 -13.857 0.196 1.00 0.00 H
+ATOM 1759 N LEU B 109 -16.965 -18.127 -1.390 1.00 0.00 N
+ATOM 1760 CA LEU B 109 -18.009 -19.112 -1.661 1.00 0.00 C
+ATOM 1761 C LEU B 109 -17.963 -20.334 -0.722 1.00 0.00 C
+ATOM 1762 O LEU B 109 -16.983 -20.557 -0.013 1.00 0.00 O
+ATOM 1763 CB LEU B 109 -17.855 -19.579 -3.102 1.00 0.00 C
+ATOM 1764 CG LEU B 109 -17.862 -18.468 -4.153 1.00 0.00 C
+ATOM 1765 CD1 LEU B 109 -17.652 -19.087 -5.516 1.00 0.00 C
+ATOM 1766 CD2 LEU B 109 -19.164 -17.690 -4.069 1.00 0.00 C
+ATOM 1767 H LEU B 109 -16.050 -18.307 -1.782 1.00 0.00 H
+ATOM 1768 HA LEU B 109 -18.976 -18.625 -1.541 1.00 0.00 H
+ATOM 1769 1HB LEU B 109 -16.903 -20.079 -3.184 1.00 0.00 H
+ATOM 1770 2HB LEU B 109 -18.642 -20.292 -3.338 1.00 0.00 H
+ATOM 1771 HG LEU B 109 -17.023 -17.785 -3.966 1.00 0.00 H
+ATOM 1772 1HD1 LEU B 109 -17.639 -18.306 -6.276 1.00 0.00 H
+ATOM 1773 2HD1 LEU B 109 -16.703 -19.621 -5.532 1.00 0.00 H
+ATOM 1774 3HD1 LEU B 109 -18.462 -19.783 -5.733 1.00 0.00 H
+ATOM 1775 1HD2 LEU B 109 -19.164 -16.895 -4.817 1.00 0.00 H
+ATOM 1776 2HD2 LEU B 109 -20.007 -18.358 -4.260 1.00 0.00 H
+ATOM 1777 3HD2 LEU B 109 -19.266 -17.252 -3.074 1.00 0.00 H
+ATOM 1778 N ALA B 110 -19.037 -21.123 -0.686 1.00 0.00 N
+ATOM 1779 CA ALA B 110 -19.022 -22.333 0.143 1.00 0.00 C
+ATOM 1780 C ALA B 110 -19.576 -23.537 -0.623 1.00 0.00 C
+ATOM 1781 O ALA B 110 -20.298 -23.345 -1.605 1.00 0.00 O
+ATOM 1782 OXT ALA B 110 -18.943 -24.602 -0.541 1.00 0.00 O
+ATOM 1783 CB ALA B 110 -19.794 -22.095 1.413 1.00 0.00 C
+ATOM 1784 H ALA B 110 -19.851 -20.904 -1.243 1.00 0.00 H
+ATOM 1785 HA ALA B 110 -17.990 -22.558 0.410 1.00 0.00 H
+ATOM 1786 1HB ALA B 110 -19.745 -22.992 2.021 1.00 0.00 H
+ATOM 1787 2HB ALA B 110 -19.355 -21.258 1.960 1.00 0.00 H
+ATOM 1788 3HB ALA B 110 -20.834 -21.873 1.177 1.00 0.00 H
+TER
diff --git a/examples/pdbs/rsv5_5tpn.pdb b/examples/pdbs/rsv5_5tpn.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..71bb52f4a629e45e46113b7af6438ddd5eb929a2
--- /dev/null
+++ b/examples/pdbs/rsv5_5tpn.pdb
@@ -0,0 +1,7077 @@
+ATOM 1 N ASN A 27 32.120 -89.596 -31.362 1.00 72.20 A N
+ATOM 2 CA ASN A 27 31.765 -90.980 -31.028 1.00 65.67 A C
+ATOM 3 C ASN A 27 30.888 -91.024 -29.775 1.00 70.28 A C
+ATOM 4 O ASN A 27 31.021 -90.212 -28.846 1.00 62.53 A O
+ATOM 5 CB ASN A 27 33.010 -91.863 -30.812 1.00 82.41 A C
+ATOM 6 CG ASN A 27 33.747 -91.546 -29.507 1.00108.25 A C
+ATOM 7 ND2 ASN A 27 34.841 -90.800 -29.610 1.00109.08 A N
+ATOM 8 OD1 ASN A 27 33.343 -91.986 -28.426 1.00122.44 A O
+ATOM 9 N ILE A 28 30.006 -91.995 -29.741 1.00 73.73 A N
+ATOM 10 CA ILE A 28 28.960 -92.036 -28.738 1.00 66.97 A C
+ATOM 11 C ILE A 28 29.318 -93.052 -27.671 1.00 72.27 A C
+ATOM 12 O ILE A 28 29.765 -94.165 -27.983 1.00 73.82 A O
+ATOM 13 CB ILE A 28 27.604 -92.343 -29.396 1.00 70.42 A C
+ATOM 14 CG1 ILE A 28 27.236 -91.185 -30.311 1.00 72.90 A C
+ATOM 15 CG2 ILE A 28 26.519 -92.523 -28.364 1.00 61.95 A C
+ATOM 16 CD1 ILE A 28 27.324 -89.849 -29.613 1.00 71.56 A C
+ATOM 17 N THR A 29 29.133 -92.652 -26.403 1.00 74.86 A N
+ATOM 18 CA THR A 29 29.375 -93.492 -25.230 1.00 67.44 A C
+ATOM 19 C THR A 29 28.154 -93.474 -24.320 1.00 71.16 A C
+ATOM 20 O THR A 29 27.337 -92.552 -24.371 1.00 73.44 A O
+ATOM 21 CB THR A 29 30.606 -93.021 -24.453 1.00 72.54 A C
+ATOM 22 CG2 THR A 29 31.882 -93.496 -25.131 1.00 68.16 A C
+ATOM 23 OG1 THR A 29 30.605 -91.588 -24.396 1.00 82.81 A O
+ATOM 24 N GLU A 30 28.007 -94.507 -23.491 1.00 68.45 A N
+ATOM 25 CA GLU A 30 26.858 -94.526 -22.588 1.00 65.76 A C
+ATOM 26 C GLU A 30 27.246 -95.046 -21.209 1.00 75.84 A C
+ATOM 27 O GLU A 30 27.927 -96.069 -21.097 1.00 71.51 A O
+ATOM 28 CB GLU A 30 25.713 -95.385 -23.126 1.00 65.49 A C
+ATOM 29 CG GLU A 30 24.358 -94.721 -22.992 1.00 67.21 A C
+ATOM 30 CD GLU A 30 23.215 -95.635 -23.387 1.00 77.78 A C
+ATOM 31 OE1 GLU A 30 23.146 -96.756 -22.837 1.00 74.02 A O
+ATOM 32 OE2 GLU A 30 22.398 -95.246 -24.254 1.00 73.88 A O1-
+ATOM 33 N GLU A 31 26.790 -94.359 -20.159 1.00 67.96 A N
+ATOM 34 CA GLU A 31 27.011 -94.800 -18.786 1.00 64.86 A C
+ATOM 35 C GLU A 31 25.677 -95.161 -18.141 1.00 62.51 A C
+ATOM 36 O GLU A 31 24.739 -94.355 -18.164 1.00 66.40 A O
+ATOM 37 CB GLU A 31 27.706 -93.715 -17.967 1.00 70.10 A C
+ATOM 38 CG GLU A 31 29.188 -93.579 -18.225 1.00 79.99 A C
+ATOM 39 CD GLU A 31 29.819 -92.451 -17.407 1.00 91.87 A C
+ATOM 40 OE1 GLU A 31 29.078 -91.760 -16.668 1.00 95.16 A O
+ATOM 41 OE2 GLU A 31 31.055 -92.258 -17.499 1.00 99.98 A O1-
+ATOM 42 N PHE A 32 25.583 -96.362 -17.563 1.00 65.37 A N
+ATOM 43 CA PHE A 32 24.382 -96.774 -16.840 1.00 70.36 A C
+ATOM 44 C PHE A 32 24.632 -96.702 -15.339 1.00 68.57 A C
+ATOM 45 O PHE A 32 25.677 -97.151 -14.864 1.00 75.91 A O
+ATOM 46 CB PHE A 32 23.946 -98.189 -17.222 1.00 65.19 A C
+ATOM 47 CG PHE A 32 22.881 -98.756 -16.313 1.00 70.64 A C
+ATOM 48 CD1 PHE A 32 21.555 -98.394 -16.464 1.00 69.16 A C
+ATOM 49 CD2 PHE A 32 23.200 -99.656 -15.325 1.00 71.95 A C
+ATOM 50 CE1 PHE A 32 20.582 -98.906 -15.643 1.00 67.88 A C
+ATOM 51 CE2 PHE A 32 22.225-100.171 -14.515 1.00 73.03 A C
+ATOM 52 CZ PHE A 32 20.918 -99.794 -14.677 1.00 68.25 A C
+ATOM 53 N TYR A 33 23.681 -96.143 -14.595 1.00 65.03 A N
+ATOM 54 CA TYR A 33 23.818 -95.980 -13.153 1.00 69.39 A C
+ATOM 55 C TYR A 33 22.828 -96.898 -12.465 1.00 77.04 A C
+ATOM 56 O TYR A 33 21.643 -96.567 -12.361 1.00 74.22 A O
+ATOM 57 CB TYR A 33 23.591 -94.526 -12.749 1.00 69.01 A C
+ATOM 58 CG TYR A 33 24.440 -93.591 -13.559 1.00 70.92 A C
+ATOM 59 CD1 TYR A 33 25.802 -93.822 -13.697 1.00 73.18 A C
+ATOM 60 CD2 TYR A 33 23.891 -92.492 -14.188 1.00 72.11 A C
+ATOM 61 CE1 TYR A 33 26.595 -92.986 -14.449 1.00 74.60 A C
+ATOM 62 CE2 TYR A 33 24.678 -91.645 -14.942 1.00 78.79 A C
+ATOM 63 CZ TYR A 33 26.033 -91.894 -15.073 1.00 82.37 A C
+ATOM 64 OH TYR A 33 26.831 -91.053 -15.827 1.00 85.82 A O
+ATOM 65 N GLN A 34 23.346 -97.999 -11.914 1.00 78.10 A N
+ATOM 66 CA GLN A 34 22.521 -99.020 -11.276 1.00 70.04 A C
+ATOM 67 C GLN A 34 21.794 -98.478 -10.059 1.00 69.60 A C
+ATOM 68 O GLN A 34 20.754 -99.025 -9.660 1.00 71.06 A O
+ATOM 69 CB GLN A 34 23.417-100.190 -10.890 1.00 75.94 A C
+ATOM 70 CG GLN A 34 22.824-101.222 -9.964 1.00 83.32 A C
+ATOM 71 CD GLN A 34 23.884-102.203 -9.506 1.00 78.64 A C
+ATOM 72 NE2 GLN A 34 23.802-103.429 -10.013 1.00 81.34 A N
+ATOM 73 OE1 GLN A 34 24.803-101.852 -8.756 1.00 81.79 A O
+ATOM 74 N SER A 35 22.324 -97.396 -9.490 1.00 66.65 A N
+ATOM 75 CA SER A 35 21.801 -96.815 -8.263 1.00 75.38 A C
+ATOM 76 C SER A 35 20.526 -96.020 -8.506 1.00 76.60 A C
+ATOM 77 O SER A 35 19.604 -96.053 -7.689 1.00 74.50 A O
+ATOM 78 CB SER A 35 22.888 -95.926 -7.671 1.00 64.71 A C
+ATOM 79 OG SER A 35 23.643 -95.313 -8.724 1.00 80.62 A O
+ATOM 80 N THR A 36 20.495 -95.237 -9.581 1.00 75.14 A N
+ATOM 81 CA THR A 36 19.382 -94.369 -9.950 1.00 77.13 A C
+ATOM 82 C THR A 36 18.491 -94.906 -11.075 1.00 80.56 A C
+ATOM 83 O THR A 36 17.590 -94.187 -11.523 1.00 77.07 A O
+ATOM 84 CB THR A 36 19.929 -92.996 -10.329 1.00 72.51 A C
+ATOM 85 CG2 THR A 36 20.525 -92.319 -9.090 1.00 70.04 A C
+ATOM 86 OG1 THR A 36 20.934 -93.156 -11.348 1.00 76.52 A O
+ATOM 87 N CYS A 37 18.741 -96.117 -11.571 1.00 69.73 A N
+ATOM 88 CA CYS A 37 17.974 -96.715 -12.670 1.00 63.55 A C
+ATOM 89 C CYS A 37 17.804 -95.715 -13.808 1.00 76.75 A C
+ATOM 90 O CYS A 37 16.714 -95.238 -14.116 1.00 78.65 A O
+ATOM 91 CB CYS A 37 16.612 -97.215 -12.197 1.00 66.00 A C
+ATOM 92 SG CYS A 37 15.899 -98.407 -13.319 1.00 79.56 A S
+ATOM 93 N SER A 38 18.950 -95.383 -14.382 1.00 75.69 A N
+ATOM 94 CA SER A 38 19.044 -94.336 -15.381 1.00 68.83 A C
+ATOM 95 C SER A 38 20.341 -94.537 -16.143 1.00 73.41 A C
+ATOM 96 O SER A 38 21.280 -95.152 -15.645 1.00 74.11 A O
+ATOM 97 CB SER A 38 19.017 -92.953 -14.727 1.00 63.08 A C
+ATOM 98 OG SER A 38 20.128 -92.804 -13.851 1.00 72.51 A O
+ATOM 99 N ALA A 39 20.399 -93.975 -17.340 1.00 65.23 A N
+ATOM 100 CA ALA A 39 21.631 -93.981 -18.106 1.00 60.04 A C
+ATOM 101 C ALA A 39 21.795 -92.620 -18.762 1.00 67.12 A C
+ATOM 102 O ALA A 39 20.866 -91.810 -18.812 1.00 61.08 A O
+ATOM 103 CB ALA A 39 21.635 -95.103 -19.145 1.00 54.43 A C
+ATOM 104 N VAL A 40 22.994 -92.353 -19.256 1.00 60.19 A N
+ATOM 105 CA VAL A 40 23.237 -91.156 -20.041 1.00 57.31 A C
+ATOM 106 C VAL A 40 23.998 -91.551 -21.293 1.00 59.99 A C
+ATOM 107 O VAL A 40 24.945 -92.347 -21.233 1.00 62.57 A O
+ATOM 108 CB VAL A 40 23.990 -90.073 -19.250 1.00 60.94 A C
+ATOM 109 CG1 VAL A 40 24.690 -89.116 -20.201 1.00 55.69 A C
+ATOM 110 CG2 VAL A 40 23.009 -89.310 -18.378 1.00 57.25 A C
+ATOM 111 N SER A 41 23.545 -91.021 -22.427 1.00 59.58 A N
+ATOM 112 CA SER A 41 24.208 -91.171 -23.711 1.00 62.27 A C
+ATOM 113 C SER A 41 24.936 -89.865 -24.013 1.00 68.14 A C
+ATOM 114 O SER A 41 24.313 -88.801 -24.123 1.00 68.21 A O
+ATOM 115 CB SER A 41 23.188 -91.513 -24.794 1.00 58.31 A C
+ATOM 116 OG SER A 41 22.163 -92.367 -24.292 1.00 69.75 A O
+ATOM 117 N LYS A 42 26.249 -89.953 -24.120 1.00 66.80 A N
+ATOM 118 CA LYS A 42 27.129 -88.801 -24.175 1.00 68.22 A C
+ATOM 119 C LYS A 42 27.880 -88.826 -25.506 1.00 68.58 A C
+ATOM 120 O LYS A 42 28.031 -89.881 -26.145 1.00 69.14 A O
+ATOM 121 CB LYS A 42 28.108 -88.808 -22.958 1.00 69.69 A C
+ATOM 122 CG LYS A 42 28.222 -87.463 -22.154 1.00 89.89 A C
+ATOM 123 CD LYS A 42 28.975 -87.587 -20.780 1.00 97.72 A C
+ATOM 124 CE LYS A 42 29.154 -86.220 -20.034 1.00103.20 A C
+ATOM 125 NZ LYS A 42 28.051 -85.808 -19.095 1.00110.07 A N1+
+ATOM 126 N GLY A 43 28.345 -87.649 -25.921 1.00 66.72 A N
+ATOM 127 CA GLY A 43 29.162 -87.526 -27.106 1.00 62.64 A C
+ATOM 128 C GLY A 43 28.483 -86.958 -28.332 1.00 70.17 A C
+ATOM 129 O GLY A 43 29.074 -87.015 -29.417 1.00 69.36 A O
+ATOM 130 N TYR A 44 27.266 -86.424 -28.204 1.00 61.49 A N
+ATOM 131 CA TYR A 44 26.544 -85.830 -29.320 1.00 59.49 A C
+ATOM 132 C TYR A 44 26.864 -84.341 -29.440 1.00 67.11 A C
+ATOM 133 O TYR A 44 27.532 -83.743 -28.592 1.00 67.92 A O
+ATOM 134 CB TYR A 44 25.034 -86.001 -29.147 1.00 61.10 A C
+ATOM 135 CG TYR A 44 24.524 -87.423 -29.144 1.00 69.67 A C
+ATOM 136 CD1 TYR A 44 24.420 -88.152 -30.313 1.00 61.28 A C
+ATOM 137 CD2 TYR A 44 24.117 -88.018 -27.968 1.00 62.82 A C
+ATOM 138 CE1 TYR A 44 23.947 -89.439 -30.303 1.00 63.60 A C
+ATOM 139 CE2 TYR A 44 23.646 -89.295 -27.948 1.00 66.76 A C
+ATOM 140 CZ TYR A 44 23.562 -90.006 -29.113 1.00 71.77 A C
+ATOM 141 OH TYR A 44 23.088 -91.295 -29.075 1.00 70.05 A O
+ATOM 142 N LEU A 45 26.300 -83.719 -30.470 1.00 67.96 A N
+ATOM 143 CA LEU A 45 26.614 -82.347 -30.850 1.00 62.06 A C
+ATOM 144 C LEU A 45 25.347 -81.522 -31.047 1.00 65.83 A C
+ATOM 145 O LEU A 45 24.343 -82.025 -31.546 1.00 68.25 A O
+ATOM 146 CB LEU A 45 27.441 -82.327 -32.137 1.00 60.41 A C
+ATOM 147 CG LEU A 45 28.935 -82.499 -31.938 1.00 74.92 A C
+ATOM 148 CD1 LEU A 45 29.591 -82.368 -33.278 1.00 66.57 A C
+ATOM 149 CD2 LEU A 45 29.428 -81.424 -30.986 1.00 74.67 A C
+ATOM 150 N SER A 46 25.332 -80.281 -30.591 1.00 58.86 A N
+ATOM 151 CA SER A 46 24.071 -79.554 -30.650 1.00 54.84 A C
+ATOM 152 C SER A 46 23.724 -78.488 -31.678 1.00 64.79 A C
+ATOM 153 O SER A 46 24.278 -77.388 -31.665 1.00 68.34 A O
+ATOM 154 CB SER A 46 23.757 -78.992 -29.256 1.00 58.87 A C
+ATOM 155 OG SER A 46 24.620 -77.916 -28.932 1.00 61.92 A O
+ATOM 156 N ALA A 47 22.765 -78.812 -32.543 1.00 59.79 A N
+ATOM 157 CA ALA A 47 22.251 -77.818 -33.469 1.00 57.25 A C
+ATOM 158 C ALA A 47 20.972 -77.279 -32.847 1.00 58.48 A C
+ATOM 159 O ALA A 47 19.938 -77.960 -32.830 1.00 61.48 A O
+ATOM 160 CB ALA A 47 22.001 -78.426 -34.842 1.00 58.81 A C
+ATOM 161 N LEU A 48 21.030 -76.051 -32.368 1.00 58.06 A N
+ATOM 162 CA LEU A 48 19.978 -75.515 -31.523 1.00 62.27 A C
+ATOM 163 C LEU A 48 19.245 -74.428 -32.282 1.00 62.27 A C
+ATOM 164 O LEU A 48 19.853 -73.431 -32.684 1.00 66.77 A O
+ATOM 165 CB LEU A 48 20.547 -74.965 -30.218 1.00 52.61 A C
+ATOM 166 CG LEU A 48 21.219 -75.974 -29.293 1.00 54.96 A C
+ATOM 167 CD1 LEU A 48 21.319 -75.372 -27.915 1.00 54.11 A C
+ATOM 168 CD2 LEU A 48 20.426 -77.268 -29.228 1.00 61.76 A C
+ATOM 169 N ARG A 49 17.939 -74.596 -32.444 1.00 60.09 A N
+ATOM 170 CA ARG A 49 17.187 -73.557 -33.117 1.00 63.78 A C
+ATOM 171 C ARG A 49 17.030 -72.434 -32.118 1.00 62.66 A C
+ATOM 172 O ARG A 49 16.297 -72.563 -31.136 1.00 59.24 A O
+ATOM 173 CB ARG A 49 15.823 -74.051 -33.579 1.00 57.80 A C
+ATOM 174 CG ARG A 49 15.219 -73.066 -34.549 1.00 60.39 A C
+ATOM 175 CD ARG A 49 13.707 -73.025 -34.493 1.00 55.28 A C
+ATOM 176 NE ARG A 49 13.185 -72.508 -33.233 1.00 60.97 A N
+ATOM 177 CZ ARG A 49 13.104 -71.219 -32.921 1.00 61.68 A C
+ATOM 178 NH1 ARG A 49 13.525 -70.306 -33.779 1.00 61.78 A N1+
+ATOM 179 NH2 ARG A 49 12.600 -70.846 -31.746 1.00 63.68 A N
+ATOM 180 N THR A 50 17.721 -71.332 -32.366 1.00 63.82 A N
+ATOM 181 CA THR A 50 17.604 -70.194 -31.486 1.00 56.36 A C
+ATOM 182 C THR A 50 16.683 -69.107 -32.017 1.00 61.31 A C
+ATOM 183 O THR A 50 16.207 -68.286 -31.220 1.00 55.88 A O
+ATOM 184 CB THR A 50 19.000 -69.633 -31.224 1.00 61.27 A C
+ATOM 185 CG2 THR A 50 19.912 -70.779 -30.873 1.00 60.88 A C
+ATOM 186 OG1 THR A 50 19.507 -69.000 -32.408 1.00 52.98 A O
+ATOM 187 N GLY A 51 16.358 -69.118 -33.307 1.00 61.17 A N
+ATOM 188 CA GLY A 51 15.647 -67.970 -33.846 1.00 58.16 A C
+ATOM 189 C GLY A 51 15.148 -68.212 -35.253 1.00 62.49 A C
+ATOM 190 O GLY A 51 15.173 -69.333 -35.753 1.00 56.96 A O
+ATOM 191 N TRP A 52 14.666 -67.147 -35.887 1.00 58.16 A N
+ATOM 192 CA TRP A 52 14.072 -67.285 -37.208 1.00 52.76 A C
+ATOM 193 C TRP A 52 14.639 -66.256 -38.172 1.00 60.30 A C
+ATOM 194 O TRP A 52 15.134 -65.192 -37.775 1.00 63.15 A O
+ATOM 195 CB TRP A 52 12.533 -67.130 -37.210 1.00 55.52 A C
+ATOM 196 CG TRP A 52 11.761 -68.041 -36.290 1.00 59.71 A C
+ATOM 197 CD1 TRP A 52 11.125 -67.684 -35.127 1.00 53.92 A C
+ATOM 198 CD2 TRP A 52 11.541 -69.458 -36.445 1.00 55.35 A C
+ATOM 199 CE2 TRP A 52 10.768 -69.885 -35.348 1.00 56.26 A C
+ATOM 200 CE3 TRP A 52 11.922 -70.400 -37.402 1.00 52.94 A C
+ATOM 201 NE1 TRP A 52 10.523 -68.785 -34.561 1.00 60.17 A N
+ATOM 202 CZ2 TRP A 52 10.376 -71.211 -35.186 1.00 57.32 A C
+ATOM 203 CZ3 TRP A 52 11.531 -71.702 -37.230 1.00 53.84 A C
+ATOM 204 CH2 TRP A 52 10.765 -72.095 -36.134 1.00 54.29 A C
+ATOM 205 N TYR A 53 14.538 -66.607 -39.460 1.00 46.59 A N
+ATOM 206 CA TYR A 53 14.768 -65.694 -40.579 1.00 42.06 A C
+ATOM 207 C TYR A 53 13.653 -65.895 -41.589 1.00 43.34 A C
+ATOM 208 O TYR A 53 13.440 -67.013 -42.069 1.00 47.84 A O
+ATOM 209 CB TYR A 53 16.126 -65.931 -41.241 1.00 52.35 A C
+ATOM 210 CG TYR A 53 16.450 -64.947 -42.337 1.00 75.87 A C
+ATOM 211 CD1 TYR A 53 17.133 -63.785 -42.052 1.00 76.30 A C
+ATOM 212 CD2 TYR A 53 16.104 -65.193 -43.649 1.00 83.29 A C
+ATOM 213 CE1 TYR A 53 17.442 -62.883 -43.026 1.00 77.04 A C
+ATOM 214 CE2 TYR A 53 16.413 -64.292 -44.641 1.00 88.43 A C
+ATOM 215 CZ TYR A 53 17.083 -63.134 -44.319 1.00 92.01 A C
+ATOM 216 OH TYR A 53 17.406 -62.206 -45.281 1.00103.48 A O
+ATOM 217 N THR A 54 12.961 -64.819 -41.923 1.00 45.05 A N
+ATOM 218 CA THR A 54 11.783 -64.888 -42.770 1.00 51.23 A C
+ATOM 219 C THR A 54 12.155 -64.549 -44.206 1.00 58.78 A C
+ATOM 220 O THR A 54 12.765 -63.507 -44.465 1.00 61.51 A O
+ATOM 221 CB THR A 54 10.719 -63.925 -42.255 1.00 51.66 A C
+ATOM 222 CG2 THR A 54 10.295 -64.327 -40.847 1.00 50.45 A C
+ATOM 223 OG1 THR A 54 11.267 -62.601 -42.205 1.00 55.42 A O
+ATOM 224 N SER A 55 11.792 -65.426 -45.135 1.00 45.33 A N
+ATOM 225 CA SER A 55 11.998 -65.194 -46.559 1.00 51.75 A C
+ATOM 226 C SER A 55 10.643 -65.070 -47.241 1.00 48.93 A C
+ATOM 227 O SER A 55 9.639 -65.614 -46.756 1.00 48.95 A O
+ATOM 228 CB SER A 55 12.777 -66.331 -47.185 1.00 55.67 A C
+ATOM 229 OG SER A 55 12.072 -67.540 -46.968 1.00 74.54 A O
+ATOM 230 N VAL A 56 10.604 -64.349 -48.361 1.00 45.30 A N
+ATOM 231 CA VAL A 56 9.357 -64.163 -49.096 1.00 36.35 A C
+ATOM 232 C VAL A 56 9.468 -64.855 -50.455 1.00 47.50 A C
+ATOM 233 O VAL A 56 10.171 -64.389 -51.365 1.00 50.37 A O
+ATOM 234 CB VAL A 56 8.994 -62.686 -49.244 1.00 46.93 A C
+ATOM 235 CG1 VAL A 56 7.793 -62.573 -50.108 1.00 52.72 A C
+ATOM 236 CG2 VAL A 56 8.692 -62.110 -47.911 1.00 36.71 A C
+ATOM 237 N ILE A 57 8.744 -65.962 -50.581 1.00 42.71 A N
+ATOM 238 CA ILE A 57 8.582 -66.712 -51.817 1.00 38.55 A C
+ATOM 239 C ILE A 57 7.404 -66.124 -52.589 1.00 50.05 A C
+ATOM 240 O ILE A 57 6.379 -65.739 -51.995 1.00 51.40 A O
+ATOM 241 CB ILE A 57 8.360 -68.201 -51.509 1.00 41.74 A C
+ATOM 242 CG1 ILE A 57 9.472 -68.701 -50.603 1.00 43.65 A C
+ATOM 243 CG2 ILE A 57 8.318 -68.994 -52.760 1.00 39.82 A C
+ATOM 244 CD1 ILE A 57 10.802 -68.400 -51.114 1.00 35.26 A C
+ATOM 245 N THR A 58 7.551 -66.026 -53.918 1.00 49.79 A N
+ATOM 246 CA THR A 58 6.504 -65.484 -54.777 1.00 44.46 A C
+ATOM 247 C THR A 58 6.397 -66.263 -56.083 1.00 49.11 A C
+ATOM 248 O THR A 58 7.317 -66.977 -56.509 1.00 54.38 A O
+ATOM 249 CB THR A 58 6.733 -64.015 -55.124 1.00 47.27 A C
+ATOM 250 CG2 THR A 58 6.799 -63.170 -53.869 1.00 49.12 A C
+ATOM 251 OG1 THR A 58 7.947 -63.894 -55.879 1.00 50.62 A O
+ATOM 252 N ILE A 59 5.233 -66.100 -56.705 1.00 46.28 A N
+ATOM 253 CA ILE A 59 4.908 -66.654 -58.008 1.00 44.24 A C
+ATOM 254 C ILE A 59 4.122 -65.602 -58.763 1.00 54.64 A C
+ATOM 255 O ILE A 59 3.124 -65.083 -58.245 1.00 63.01 A O
+ATOM 256 CB ILE A 59 4.064 -67.924 -57.897 1.00 43.70 A C
+ATOM 257 CG1 ILE A 59 4.756 -68.965 -57.058 1.00 42.15 A C
+ATOM 258 CG2 ILE A 59 3.769 -68.455 -59.257 1.00 45.90 A C
+ATOM 259 CD1 ILE A 59 3.831 -70.104 -56.753 1.00 39.09 A C
+ATOM 260 N GLU A 60 4.550 -65.295 -59.987 1.00 51.73 A N
+ATOM 261 CA GLU A 60 3.876 -64.259 -60.747 1.00 53.53 A C
+ATOM 262 C GLU A 60 2.681 -64.873 -61.468 1.00 51.07 A C
+ATOM 263 O GLU A 60 2.786 -65.951 -62.066 1.00 55.49 A O
+ATOM 264 CB GLU A 60 4.857 -63.570 -61.699 1.00 54.11 A C
+ATOM 265 CG GLU A 60 4.825 -62.032 -61.539 1.00 73.02 A C
+ATOM 266 CD GLU A 60 5.785 -61.267 -62.456 1.00 82.60 A C
+ATOM 267 OE1 GLU A 60 5.946 -61.644 -63.640 1.00 73.00 A O
+ATOM 268 OE2 GLU A 60 6.362 -60.258 -61.991 1.00102.75 A O1-
+ATOM 269 N LEU A 61 1.528 -64.218 -61.353 1.00 58.15 A N
+ATOM 270 CA LEU A 61 0.267 -64.755 -61.853 1.00 54.09 A C
+ATOM 271 C LEU A 61 -0.429 -63.725 -62.730 1.00 59.07 A C
+ATOM 272 O LEU A 61 -0.762 -62.633 -62.259 1.00 73.30 A O
+ATOM 273 CB LEU A 61 -0.660 -65.148 -60.704 1.00 59.26 A C
+ATOM 274 CG LEU A 61 -0.081 -65.937 -59.547 1.00 56.23 A C
+ATOM 275 CD1 LEU A 61 -1.015 -65.835 -58.387 1.00 61.92 A C
+ATOM 276 CD2 LEU A 61 0.015 -67.357 -59.982 1.00 51.18 A C
+ATOM 277 N SER A 62 -0.661 -64.081 -63.992 1.00 57.63 A N
+ATOM 278 CA SER A 62 -1.526 -63.295 -64.858 1.00 59.84 A C
+ATOM 279 C SER A 62 -2.927 -63.274 -64.270 1.00 71.00 A C
+ATOM 280 O SER A 62 -3.443 -64.315 -63.861 1.00 74.20 A O
+ATOM 281 CB SER A 62 -1.545 -63.908 -66.256 1.00 66.51 A C
+ATOM 282 OG SER A 62 -1.810 -62.914 -67.223 1.00 68.86 A O
+ATOM 283 N ASN A 63 -3.544 -62.092 -64.202 1.00 65.12 A N
+ATOM 284 CA ASN A 63 -4.871 -61.967 -63.601 1.00 77.30 A C
+ATOM 285 C ASN A 63 -5.874 -62.023 -64.747 1.00 72.83 A C
+ATOM 286 O ASN A 63 -6.026 -61.043 -65.479 1.00 82.32 A O
+ATOM 287 CB ASN A 63 -4.995 -60.657 -62.816 1.00 88.19 A C
+ATOM 288 CG ASN A 63 -5.705 -60.825 -61.450 1.00 97.22 A C
+ATOM 289 ND2 ASN A 63 -6.917 -60.271 -61.355 1.00100.90 A N
+ATOM 290 OD1 ASN A 63 -5.172 -61.449 -60.503 1.00101.68 A O
+ATOM 291 N ILE A 64 -6.580 -63.149 -64.887 1.00 77.79 A N
+ATOM 292 CA ILE A 64 -7.491 -63.349 -66.020 1.00 88.97 A C
+ATOM 293 C ILE A 64 -8.862 -63.771 -65.504 1.00 96.09 A C
+ATOM 294 O ILE A 64 -9.031 -64.889 -64.997 1.00 94.17 A O
+ATOM 295 CB ILE A 64 -6.966 -64.371 -67.044 1.00 80.66 A C
+ATOM 296 CG1 ILE A 64 -6.554 -65.689 -66.403 1.00 73.33 A C
+ATOM 297 CG2 ILE A 64 -5.786 -63.812 -67.783 1.00 80.77 A C
+ATOM 298 CD1 ILE A 64 -6.150 -66.672 -67.425 1.00 76.35 A C
+ATOM 299 N LYS A 65 -9.848 -62.887 -65.654 1.00106.08 A N
+ATOM 300 CA LYS A 65 -11.205 -63.233 -65.270 1.00108.87 A C
+ATOM 301 C LYS A 65 -12.045 -63.785 -66.416 1.00120.85 A C
+ATOM 302 O LYS A 65 -13.086 -64.391 -66.147 1.00115.64 A O
+ATOM 303 CB LYS A 65 -11.911 -62.016 -64.624 1.00101.95 A C
+ATOM 304 CG LYS A 65 -11.176 -61.505 -63.369 1.00 99.91 A C
+ATOM 305 CD LYS A 65 -11.964 -60.524 -62.493 1.00101.46 A C
+ATOM 306 CE LYS A 65 -11.087 -60.087 -61.296 1.00100.70 A C
+ATOM 307 NZ LYS A 65 -11.788 -59.327 -60.208 1.00 98.81 A N1+
+ATOM 308 N LYS A 66 -11.631 -63.637 -67.675 1.00 98.06 A N
+ATOM 309 CA LYS A 66 -12.538 -63.945 -68.779 1.00100.55 A C
+ATOM 310 C LYS A 66 -11.810 -64.706 -69.878 1.00 87.83 A C
+ATOM 311 O LYS A 66 -10.920 -64.143 -70.521 1.00 91.66 A O
+ATOM 312 CB LYS A 66 -13.160 -62.655 -69.340 1.00112.65 A C
+ATOM 313 CG LYS A 66 -13.643 -61.634 -68.282 1.00115.08 A C
+ATOM 314 CD LYS A 66 -14.806 -60.770 -68.786 1.00114.28 A C
+ATOM 315 CE LYS A 66 -16.141 -61.509 -68.608 1.00107.05 A C
+ATOM 316 NZ LYS A 66 -17.338 -60.744 -69.052 1.00103.06 A N1+
+ATOM 317 N ILE A 67 -12.227 -65.950 -70.140 1.00104.67 A N
+ATOM 318 CA ILE A 67 -11.690 -66.752 -71.241 1.00100.72 A C
+ATOM 319 C ILE A 67 -12.706 -66.668 -72.365 1.00103.14 A C
+ATOM 320 O ILE A 67 -13.711 -67.381 -72.331 1.00107.08 A O
+ATOM 321 CB ILE A 67 -11.510 -68.228 -70.866 1.00 86.75 A C
+ATOM 322 CG1 ILE A 67 -11.159 -68.453 -69.394 1.00 71.87 A C
+ATOM 323 CG2 ILE A 67 -10.501 -68.882 -71.795 1.00 83.05 A C
+ATOM 324 CD1 ILE A 67 -9.794 -67.985 -68.998 1.00 67.83 A C
+ATOM 325 N LYS A 68 -12.449 -65.888 -73.411 1.00 93.04 A N
+ATOM 326 CA LYS A 68 -13.386 -65.921 -74.531 1.00100.87 A C
+ATOM 327 C LYS A 68 -12.637 -66.437 -75.746 1.00107.32 A C
+ATOM 328 O LYS A 68 -12.021 -65.660 -76.475 1.00111.84 A O
+ATOM 329 CB LYS A 68 -13.991 -64.543 -74.799 1.00103.33 A C
+ATOM 330 CG LYS A 68 -15.245 -64.538 -75.716 1.00105.48 A C
+ATOM 331 CD LYS A 68 -16.498 -65.102 -74.991 1.00102.59 A C
+ATOM 332 CE LYS A 68 -17.737 -65.145 -75.898 1.00 98.19 A C
+ATOM 333 NZ LYS A 68 -18.975 -65.593 -75.177 1.00 99.02 A N1+
+ATOM 334 N CYS A 69 -12.755 -67.735 -75.988 1.00 93.39 A N
+ATOM 335 CA CYS A 69 -12.432 -68.345 -77.269 1.00 92.27 A C
+ATOM 336 C CYS A 69 -13.482 -69.412 -77.517 1.00 92.79 A C
+ATOM 337 O CYS A 69 -13.523 -70.409 -76.787 1.00 97.86 A O
+ATOM 338 CB CYS A 69 -11.031 -68.951 -77.261 1.00 86.74 A C
+ATOM 339 SG CYS A 69 -10.734 -70.142 -78.580 1.00118.72 A S
+ATOM 340 N ASN A 70 -14.213 -69.286 -78.611 1.00101.01 A N
+ATOM 341 CA ASN A 70 -15.196 -70.296 -78.942 1.00102.41 A C
+ATOM 342 C ASN A 70 -14.750 -71.467 -79.844 1.00114.52 A C
+ATOM 343 O ASN A 70 -15.631 -72.120 -80.403 1.00126.45 A O
+ATOM 344 CB ASN A 70 -16.441 -69.634 -79.545 1.00 98.13 A C
+ATOM 345 CG ASN A 70 -16.135 -68.872 -80.819 1.00 99.62 A C
+ATOM 346 ND2 ASN A 70 -16.525 -69.438 -81.956 1.00 97.85 A N
+ATOM 347 OD1 ASN A 70 -15.555 -67.787 -80.783 1.00 98.76 A O
+ATOM 348 N GLY A 71 -13.458 -71.789 -80.023 1.00113.24 A N
+ATOM 349 CA GLY A 71 -13.287 -72.923 -80.891 1.00 99.58 A C
+ATOM 350 C GLY A 71 -13.812 -74.089 -80.081 1.00 81.27 A C
+ATOM 351 O GLY A 71 -14.087 -73.960 -78.890 1.00 75.12 A O
+ATOM 352 N THR A 72 -13.975 -75.225 -80.748 1.00 94.55 A N
+ATOM 353 CA THR A 72 -14.068 -76.502 -80.046 1.00 96.02 A C
+ATOM 354 C THR A 72 -13.090 -77.438 -80.728 1.00 94.00 A C
+ATOM 355 O THR A 72 -13.378 -77.995 -81.794 1.00 77.72 A O
+ATOM 356 CB THR A 72 -15.474 -77.072 -80.078 1.00 97.19 A C
+ATOM 357 CG2 THR A 72 -16.378 -76.310 -79.106 1.00102.60 A C
+ATOM 358 OG1 THR A 72 -15.983 -76.990 -81.419 1.00100.03 A O
+ATOM 359 N ASP A 73 -11.972 -77.651 -80.061 1.00 81.63 A N
+ATOM 360 CA ASP A 73 -10.973 -78.612 -80.474 1.00 78.54 A C
+ATOM 361 C ASP A 73 -10.386 -79.153 -79.191 1.00 67.60 A C
+ATOM 362 O ASP A 73 -10.081 -78.363 -78.291 1.00 66.62 A O
+ATOM 363 CB ASP A 73 -9.899 -77.966 -81.333 1.00 70.85 A C
+ATOM 364 CG ASP A 73 -8.881 -78.950 -81.766 1.00 78.00 A C
+ATOM 365 OD1 ASP A 73 -9.248 -79.843 -82.552 1.00 78.46 A O
+ATOM 366 OD2 ASP A 73 -7.729 -78.856 -81.302 1.00 82.67 A O1-
+ATOM 367 N ALA A 74 -10.233 -80.475 -79.096 1.00 55.77 A N
+ATOM 368 CA ALA A 74 -9.920 -81.051 -77.793 1.00 63.82 A C
+ATOM 369 C ALA A 74 -8.743 -80.342 -77.146 1.00 63.12 A C
+ATOM 370 O ALA A 74 -8.763 -80.110 -75.937 1.00 67.23 A O
+ATOM 371 CB ALA A 74 -9.651 -82.551 -77.913 1.00 56.29 A C
+ATOM 372 N LYS A 75 -7.753 -79.913 -77.939 1.00 56.21 A N
+ATOM 373 CA LYS A 75 -6.595 -79.227 -77.369 1.00 57.88 A C
+ATOM 374 C LYS A 75 -7.014 -77.945 -76.663 1.00 60.40 A C
+ATOM 375 O LYS A 75 -6.751 -77.765 -75.465 1.00 60.28 A O
+ATOM 376 CB LYS A 75 -5.562 -78.969 -78.460 1.00 55.71 A C
+ATOM 377 CG LYS A 75 -5.560 -80.103 -79.457 1.00 56.52 A C
+ATOM 378 CD LYS A 75 -4.212 -80.353 -80.069 1.00 58.09 A C
+ATOM 379 CE LYS A 75 -4.274 -81.615 -80.944 1.00 62.67 A C
+ATOM 380 NZ LYS A 75 -2.918 -82.260 -81.223 1.00 70.40 A N1+
+ATOM 381 N ILE A 76 -7.713 -77.066 -77.381 1.00 55.08 A N
+ATOM 382 CA ILE A 76 -8.324 -75.893 -76.757 1.00 57.43 A C
+ATOM 383 C ILE A 76 -9.115 -76.286 -75.514 1.00 59.94 A C
+ATOM 384 O ILE A 76 -9.045 -75.617 -74.476 1.00 57.01 A O
+ATOM 385 CB ILE A 76 -9.201 -75.144 -77.780 1.00 58.15 A C
+ATOM 386 CG1 ILE A 76 -8.319 -74.338 -78.725 1.00 61.70 A C
+ATOM 387 CG2 ILE A 76 -10.184 -74.243 -77.089 1.00 52.60 A C
+ATOM 388 CD1 ILE A 76 -9.058 -73.610 -79.783 1.00 75.30 A C
+ATOM 389 N LYS A 77 -9.868 -77.382 -75.587 1.00 59.07 A N
+ATOM 390 CA LYS A 77 -10.693 -77.748 -74.444 1.00 55.83 A C
+ATOM 391 C LYS A 77 -9.835 -78.030 -73.223 1.00 57.69 A C
+ATOM 392 O LYS A 77 -10.100 -77.511 -72.136 1.00 59.86 A O
+ATOM 393 CB LYS A 77 -11.559 -78.959 -74.770 1.00 53.81 A C
+ATOM 394 CG LYS A 77 -12.388 -79.471 -73.568 1.00 67.90 A C
+ATOM 395 CD LYS A 77 -13.127 -80.777 -73.921 1.00 86.47 A C
+ATOM 396 CE LYS A 77 -14.348 -81.033 -73.043 1.00 92.39 A C
+ATOM 397 NZ LYS A 77 -15.317 -81.946 -73.723 1.00 98.83 A N1+
+ATOM 398 N LEU A 78 -8.799 -78.858 -73.385 1.00 53.26 A N
+ATOM 399 CA LEU A 78 -7.933 -79.196 -72.260 1.00 61.25 A C
+ATOM 400 C LEU A 78 -7.278 -77.947 -71.697 1.00 57.31 A C
+ATOM 401 O LEU A 78 -7.256 -77.757 -70.470 1.00 66.22 A O
+ATOM 402 CB LEU A 78 -6.869 -80.215 -72.684 1.00 48.85 A C
+ATOM 403 CG LEU A 78 -7.341 -81.476 -73.401 1.00 60.91 A C
+ATOM 404 CD1 LEU A 78 -6.168 -82.345 -73.788 1.00 58.41 A C
+ATOM 405 CD2 LEU A 78 -8.312 -82.235 -72.531 1.00 59.48 A C
+ATOM 406 N ILE A 79 -6.759 -77.074 -72.585 1.00 53.24 A N
+ATOM 407 CA ILE A 79 -6.177 -75.800 -72.147 1.00 52.56 A C
+ATOM 408 C ILE A 79 -7.180 -75.024 -71.306 1.00 55.01 A C
+ATOM 409 O ILE A 79 -6.848 -74.512 -70.234 1.00 56.74 A O
+ATOM 410 CB ILE A 79 -5.704 -74.960 -73.346 1.00 56.57 A C
+ATOM 411 CG1 ILE A 79 -4.580 -75.656 -74.092 1.00 65.94 A C
+ATOM 412 CG2 ILE A 79 -5.210 -73.626 -72.863 1.00 60.63 A C
+ATOM 413 CD1 ILE A 79 -3.373 -75.850 -73.254 1.00 67.06 A C
+ATOM 414 N LYS A 80 -8.427 -74.942 -71.774 1.00 54.62 A N
+ATOM 415 CA LYS A 80 -9.457 -74.252 -71.010 1.00 60.32 A C
+ATOM 416 C LYS A 80 -9.701 -74.935 -69.659 1.00 60.77 A C
+ATOM 417 O LYS A 80 -9.902 -74.261 -68.638 1.00 60.89 A O
+ATOM 418 CB LYS A 80 -10.743 -74.159 -71.843 1.00 52.50 A C
+ATOM 419 CG LYS A 80 -10.667 -73.172 -73.037 1.00 62.62 A C
+ATOM 420 CD LYS A 80 -11.983 -73.078 -73.866 1.00 79.98 A C
+ATOM 421 CE LYS A 80 -13.054 -72.162 -73.230 1.00 84.50 A C
+ATOM 422 NZ LYS A 80 -14.292 -72.011 -74.059 1.00 94.32 A N1+
+ATOM 423 N GLN A 81 -9.661 -76.265 -69.622 1.00 60.14 A N
+ATOM 424 CA GLN A 81 -9.854 -76.957 -68.355 1.00 59.68 A C
+ATOM 425 C GLN A 81 -8.776 -76.558 -67.358 1.00 66.16 A C
+ATOM 426 O GLN A 81 -9.075 -76.138 -66.231 1.00 61.70 A O
+ATOM 427 CB GLN A 81 -9.866 -78.466 -68.581 1.00 57.45 A C
+ATOM 428 CG GLN A 81 -11.121 -78.970 -69.289 1.00 69.03 A C
+ATOM 429 CD GLN A 81 -11.223 -80.509 -69.363 1.00 71.51 A C
+ATOM 430 NE2 GLN A 81 -12.129 -80.994 -70.229 1.00 77.15 A N
+ATOM 431 OE1 GLN A 81 -10.509 -81.250 -68.651 1.00 80.38 A O
+ATOM 432 N GLU A 82 -7.512 -76.650 -67.775 1.00 57.13 A N
+ATOM 433 CA GLU A 82 -6.417 -76.226 -66.912 1.00 55.06 A C
+ATOM 434 C GLU A 82 -6.551 -74.762 -66.509 1.00 60.54 A C
+ATOM 435 O GLU A 82 -6.237 -74.391 -65.371 1.00 61.10 A O
+ATOM 436 CB GLU A 82 -5.084 -76.452 -67.611 1.00 48.61 A C
+ATOM 437 CG GLU A 82 -3.947 -76.622 -66.639 1.00 57.90 A C
+ATOM 438 CD GLU A 82 -4.022 -77.953 -65.958 1.00 63.75 A C
+ATOM 439 OE1 GLU A 82 -4.825 -78.772 -66.456 1.00 65.18 A O
+ATOM 440 OE2 GLU A 82 -3.304 -78.186 -64.947 1.00 63.35 A O1-
+ATOM 441 N LEU A 83 -6.986 -73.909 -67.436 1.00 54.36 A N
+ATOM 442 CA LEU A 83 -7.194 -72.505 -67.106 1.00 52.22 A C
+ATOM 443 C LEU A 83 -8.174 -72.361 -65.963 1.00 56.24 A C
+ATOM 444 O LEU A 83 -7.951 -71.585 -65.035 1.00 54.40 A O
+ATOM 445 CB LEU A 83 -7.705 -71.732 -68.321 1.00 60.67 A C
+ATOM 446 CG LEU A 83 -6.611 -70.965 -69.047 1.00 72.86 A C
+ATOM 447 CD1 LEU A 83 -6.029 -70.004 -68.057 1.00 74.08 A C
+ATOM 448 CD2 LEU A 83 -5.533 -71.873 -69.587 1.00 80.54 A C
+ATOM 449 N ASP A 84 -9.282 -73.092 -66.026 1.00 61.35 A N
+ATOM 450 CA ASP A 84 -10.263 -73.007 -64.953 1.00 61.71 A C
+ATOM 451 C ASP A 84 -9.678 -73.502 -63.635 1.00 62.91 A C
+ATOM 452 O ASP A 84 -9.907 -72.896 -62.579 1.00 51.32 A O
+ATOM 453 CB ASP A 84 -11.515 -73.783 -65.353 1.00 72.84 A C
+ATOM 454 CG ASP A 84 -12.414 -72.971 -66.265 1.00 95.20 A C
+ATOM 455 OD1 ASP A 84 -12.487 -71.741 -66.045 1.00100.41 A O
+ATOM 456 OD2 ASP A 84 -13.004 -73.540 -67.215 1.00110.68 A O1-
+ATOM 457 N LYS A 85 -8.898 -74.588 -63.678 1.00 55.00 A N
+ATOM 458 CA LYS A 85 -8.173 -75.024 -62.486 1.00 54.38 A C
+ATOM 459 C LYS A 85 -7.348 -73.875 -61.889 1.00 66.00 A C
+ATOM 460 O LYS A 85 -7.415 -73.607 -60.683 1.00 63.02 A O
+ATOM 461 CB LYS A 85 -7.271 -76.214 -62.836 1.00 45.19 A C
+ATOM 462 CG LYS A 85 -7.881 -77.637 -62.838 1.00 44.23 A C
+ATOM 463 CD LYS A 85 -6.716 -78.647 -63.081 1.00 63.74 A C
+ATOM 464 CE LYS A 85 -7.045 -80.136 -62.896 1.00 78.87 A C
+ATOM 465 NZ LYS A 85 -5.797 -80.938 -62.606 1.00 85.63 A N1+
+ATOM 466 N TYR A 86 -6.576 -73.180 -62.737 1.00 56.33 A N
+ATOM 467 CA TYR A 86 -5.782 -72.020 -62.312 1.00 51.38 A C
+ATOM 468 C TYR A 86 -6.655 -70.918 -61.708 1.00 56.75 A C
+ATOM 469 O TYR A 86 -6.323 -70.353 -60.658 1.00 55.62 A O
+ATOM 470 CB TYR A 86 -4.994 -71.494 -63.518 1.00 50.36 A C
+ATOM 471 CG TYR A 86 -4.322 -70.133 -63.389 1.00 48.90 A C
+ATOM 472 CD1 TYR A 86 -2.947 -70.031 -63.273 1.00 50.17 A C
+ATOM 473 CD2 TYR A 86 -5.047 -68.959 -63.472 1.00 47.62 A C
+ATOM 474 CE1 TYR A 86 -2.323 -68.804 -63.182 1.00 51.47 A C
+ATOM 475 CE2 TYR A 86 -4.429 -67.733 -63.376 1.00 50.68 A C
+ATOM 476 CZ TYR A 86 -3.065 -67.657 -63.229 1.00 60.25 A C
+ATOM 477 OH TYR A 86 -2.435 -66.433 -63.124 1.00 58.20 A O
+ATOM 478 N LYS A 87 -7.752 -70.570 -62.387 1.00 56.70 A N
+ATOM 479 CA LYS A 87 -8.692 -69.571 -61.889 1.00 63.00 A C
+ATOM 480 C LYS A 87 -9.165 -69.907 -60.476 1.00 55.46 A C
+ATOM 481 O LYS A 87 -9.053 -69.084 -59.548 1.00 56.03 A O
+ATOM 482 CB LYS A 87 -9.883 -69.498 -62.841 1.00 64.43 A C
+ATOM 483 CG LYS A 87 -9.961 -68.277 -63.745 1.00 75.69 A C
+ATOM 484 CD LYS A 87 -11.097 -68.489 -64.749 1.00 89.93 A C
+ATOM 485 CE LYS A 87 -11.730 -67.195 -65.233 1.00 99.02 A C
+ATOM 486 NZ LYS A 87 -13.018 -67.500 -65.932 1.00101.03 A N1+
+ATOM 487 N ASN A 88 -9.693 -71.127 -60.297 1.00 54.89 A N
+ATOM 488 CA ASN A 88 -10.213 -71.545 -58.995 1.00 61.66 A C
+ATOM 489 C ASN A 88 -9.112 -71.638 -57.953 1.00 61.62 A C
+ATOM 490 O ASN A 88 -9.369 -71.490 -56.745 1.00 61.13 A O
+ATOM 491 CB ASN A 88 -10.916 -72.886 -59.127 1.00 62.31 A C
+ATOM 492 CG ASN A 88 -12.013 -72.861 -60.166 1.00 72.26 A C
+ATOM 493 ND2 ASN A 88 -12.342 -74.040 -60.697 1.00 72.00 A N
+ATOM 494 OD1 ASN A 88 -12.574 -71.797 -60.487 1.00 66.87 A O
+ATOM 495 N ALA A 89 -7.888 -71.901 -58.405 1.00 54.56 A N
+ATOM 496 CA ALA A 89 -6.741 -71.852 -57.518 1.00 53.21 A C
+ATOM 497 C ALA A 89 -6.542 -70.447 -56.988 1.00 56.09 A C
+ATOM 498 O ALA A 89 -6.424 -70.234 -55.774 1.00 62.78 A O
+ATOM 499 CB ALA A 89 -5.505 -72.322 -58.272 1.00 43.16 A C
+ATOM 500 N VAL A 90 -6.499 -69.475 -57.898 1.00 52.35 A N
+ATOM 501 CA VAL A 90 -6.356 -68.084 -57.494 1.00 55.84 A C
+ATOM 502 C VAL A 90 -7.380 -67.761 -56.424 1.00 55.87 A C
+ATOM 503 O VAL A 90 -7.038 -67.356 -55.301 1.00 56.74 A O
+ATOM 504 CB VAL A 90 -6.523 -67.152 -58.703 1.00 55.67 A C
+ATOM 505 CG1 VAL A 90 -6.586 -65.707 -58.257 1.00 47.29 A C
+ATOM 506 CG2 VAL A 90 -5.402 -67.351 -59.691 1.00 44.01 A C
+ATOM 507 N THR A 91 -8.661 -67.996 -56.752 1.00 53.68 A N
+ATOM 508 CA THR A 91 -9.745 -67.499 -55.899 1.00 51.63 A C
+ATOM 509 C THR A 91 -9.750 -68.213 -54.546 1.00 57.89 A C
+ATOM 510 O THR A 91 -9.894 -67.569 -53.491 1.00 54.28 A O
+ATOM 511 CB THR A 91 -11.074 -67.639 -56.644 1.00 55.81 A C
+ATOM 512 CG2 THR A 91 -11.044 -66.798 -57.955 1.00 53.66 A C
+ATOM 513 OG1 THR A 91 -11.281 -69.011 -56.989 1.00 66.14 A O
+ATOM 514 N GLU A 92 -9.520 -69.532 -54.561 1.00 49.94 A N
+ATOM 515 CA GLU A 92 -9.249 -70.280 -53.332 1.00 55.71 A C
+ATOM 516 C GLU A 92 -8.229 -69.536 -52.479 1.00 59.84 A C
+ATOM 517 O GLU A 92 -8.479 -69.233 -51.303 1.00 57.34 A O
+ATOM 518 CB GLU A 92 -8.722 -71.687 -53.703 1.00 58.78 A C
+ATOM 519 CG GLU A 92 -9.155 -72.913 -52.834 1.00 71.68 A C
+ATOM 520 CD GLU A 92 -8.777 -74.284 -53.476 1.00 88.76 A C
+ATOM 521 OE1 GLU A 92 -7.934 -74.995 -52.889 1.00105.43 A O
+ATOM 522 OE2 GLU A 92 -9.305 -74.648 -54.563 1.00 85.08 A O1-
+ATOM 523 N LEU A 93 -7.096 -69.170 -53.096 1.00 57.83 A N
+ATOM 524 CA LEU A 93 -6.012 -68.517 -52.374 1.00 52.66 A C
+ATOM 525 C LEU A 93 -6.427 -67.161 -51.842 1.00 54.85 A C
+ATOM 526 O LEU A 93 -5.937 -66.745 -50.790 1.00 57.29 A O
+ATOM 527 CB LEU A 93 -4.786 -68.360 -53.271 1.00 54.27 A C
+ATOM 528 CG LEU A 93 -4.001 -69.606 -53.684 1.00 56.33 A C
+ATOM 529 CD1 LEU A 93 -2.777 -69.204 -54.494 1.00 54.90 A C
+ATOM 530 CD2 LEU A 93 -3.587 -70.411 -52.453 1.00 50.90 A C
+ATOM 531 N GLN A 94 -7.310 -66.455 -52.548 1.00 49.93 A N
+ATOM 532 CA GLN A 94 -7.794 -65.171 -52.048 1.00 53.92 A C
+ATOM 533 C GLN A 94 -8.627 -65.346 -50.770 1.00 59.86 A C
+ATOM 534 O GLN A 94 -8.390 -64.665 -49.751 1.00 62.66 A O
+ATOM 535 CB GLN A 94 -8.582 -64.471 -53.158 1.00 50.17 A C
+ATOM 536 CG GLN A 94 -7.688 -63.855 -54.238 1.00 64.01 A C
+ATOM 537 CD GLN A 94 -8.369 -63.702 -55.597 1.00 75.15 A C
+ATOM 538 NE2 GLN A 94 -7.729 -62.961 -56.496 1.00 71.70 A N
+ATOM 539 OE1 GLN A 94 -9.456 -64.232 -55.832 1.00 71.18 A O
+ATOM 540 N LEU A 95 -9.598 -66.273 -50.799 1.00 57.53 A N
+ATOM 541 CA LEU A 95 -10.290 -66.670 -49.569 1.00 59.85 A C
+ATOM 542 C LEU A 95 -9.320 -67.002 -48.444 1.00 64.30 A C
+ATOM 543 O LEU A 95 -9.582 -66.681 -47.279 1.00 81.00 A O
+ATOM 544 CB LEU A 95 -11.173 -67.885 -49.819 1.00 73.91 A C
+ATOM 545 CG LEU A 95 -12.663 -67.600 -49.816 1.00 91.72 A C
+ATOM 546 CD1 LEU A 95 -13.032 -66.786 -51.061 1.00 93.63 A C
+ATOM 547 CD2 LEU A 95 -13.428 -68.914 -49.748 1.00 94.17 A C
+ATOM 548 N LEU A 96 -8.218 -67.694 -48.766 1.00 52.85 A N
+ATOM 549 CA LEU A 96 -7.191 -67.965 -47.761 1.00 58.62 A C
+ATOM 550 C LEU A 96 -6.566 -66.681 -47.236 1.00 57.35 A C
+ATOM 551 O LEU A 96 -6.270 -66.564 -46.037 1.00 68.21 A O
+ATOM 552 CB LEU A 96 -6.122 -68.878 -48.340 1.00 57.96 A C
+ATOM 553 CG LEU A 96 -6.316 -70.232 -47.698 1.00 51.77 A C
+ATOM 554 CD1 LEU A 96 -5.413 -71.236 -48.307 1.00 53.12 A C
+ATOM 555 CD2 LEU A 96 -5.994 -70.075 -46.237 1.00 53.37 A C
+ATOM 556 N MET A 97 -6.370 -65.701 -48.122 1.00 57.22 A N
+ATOM 557 CA MET A 97 -5.659 -64.481 -47.759 1.00 64.06 A C
+ATOM 558 C MET A 97 -6.455 -63.632 -46.796 1.00 77.68 A C
+ATOM 559 O MET A 97 -5.864 -62.958 -45.946 1.00 85.16 A O
+ATOM 560 CB MET A 97 -5.331 -63.671 -49.009 1.00 65.38 A C
+ATOM 561 CG MET A 97 -4.719 -62.305 -48.750 1.00 68.22 A C
+ATOM 562 SD MET A 97 -4.674 -61.333 -50.287 1.00 70.80 A S
+ATOM 563 CE MET A 97 -6.382 -61.494 -50.862 1.00 60.61 A C
+ATOM 564 N GLN A 98 -7.787 -63.645 -46.917 1.00 79.91 A N
+ATOM 565 CA GLN A 98 -8.593 -62.778 -46.055 1.00 84.50 A C
+ATOM 566 C GLN A 98 -8.407 -63.115 -44.568 1.00 86.96 A C
+ATOM 567 O GLN A 98 -8.212 -62.213 -43.743 1.00 89.87 A O
+ATOM 568 CB GLN A 98 -10.070 -62.833 -46.479 1.00 87.44 A C
+ATOM 569 CG GLN A 98 -10.840 -64.101 -46.087 1.00 93.01 A C
+ATOM 570 CD GLN A 98 -12.354 -63.904 -46.074 1.00 93.78 A C
+ATOM 571 NE2 GLN A 98 -13.076 -64.871 -45.497 1.00 85.57 A N
+ATOM 572 OE1 GLN A 98 -12.868 -62.906 -46.595 1.00 99.01 A O
+ATOM 573 N SER A 99 -8.419 -64.399 -44.210 1.00 99.34 A N
+ATOM 574 CA SER A 99 -8.332 -64.795 -42.806 1.00 98.13 A C
+ATOM 575 C SER A 99 -6.883 -64.799 -42.332 1.00 90.68 A C
+ATOM 576 O SER A 99 -5.992 -65.301 -43.028 1.00 73.63 A O
+ATOM 577 CB SER A 99 -8.961 -66.176 -42.609 1.00100.12 A C
+ATOM 578 OG SER A 99 -8.462 -67.115 -43.557 1.00105.15 A O
+ATOM 579 N THR A 100 -6.642 -64.231 -41.142 1.00 86.89 A N
+ATOM 580 CA THR A 100 -5.253 -64.104 -40.689 1.00 92.48 A C
+ATOM 581 C THR A 100 -4.767 -65.464 -40.182 1.00 90.13 A C
+ATOM 582 O THR A 100 -5.496 -66.150 -39.457 1.00 91.98 A O
+ATOM 583 CB THR A 100 -5.113 -62.983 -39.622 1.00 90.24 A C
+ATOM 584 CG2 THR A 100 -5.517 -63.444 -38.250 1.00 79.79 A C
+ATOM 585 OG1 THR A 100 -3.757 -62.530 -39.544 1.00 92.53 A O
+ATOM 586 N PRO A 101 -3.587 -65.917 -40.596 1.00 95.71 A N
+ATOM 587 CA PRO A 101 -3.237 -67.329 -40.430 1.00 80.53 A C
+ATOM 588 C PRO A 101 -2.810 -67.647 -39.009 1.00 74.48 A C
+ATOM 589 O PRO A 101 -2.197 -66.827 -38.318 1.00 68.26 A O
+ATOM 590 CB PRO A 101 -2.065 -67.511 -41.400 1.00 74.06 A C
+ATOM 591 CG PRO A 101 -1.460 -66.129 -41.527 1.00 77.69 A C
+ATOM 592 CD PRO A 101 -2.408 -65.110 -40.943 1.00 82.55 A C
+ATOM 593 N ALA A 102 -3.116 -68.870 -38.579 1.00 72.87 A N
+ATOM 594 CA ALA A 102 -2.655 -69.282 -37.263 1.00 77.74 A C
+ATOM 595 C ALA A 102 -2.498 -70.791 -37.212 1.00 73.71 A C
+ATOM 596 O ALA A 102 -3.004 -71.526 -38.066 1.00 77.89 A O
+ATOM 597 CB ALA A 102 -3.598 -68.805 -36.160 1.00 73.26 A C
+ATOM 598 N THR A 103 -1.779 -71.231 -36.182 1.00 76.63 A N
+ATOM 599 CA THR A 103 -1.452 -72.627 -35.948 1.00 80.58 A C
+ATOM 600 C THR A 103 -1.238 -72.845 -34.463 1.00 91.64 A C
+ATOM 601 O THR A 103 -1.034 -71.895 -33.704 1.00 86.59 A O
+ATOM 602 CB THR A 103 -0.188 -73.052 -36.688 1.00 74.90 A C
+ATOM 603 CG2 THR A 103 0.919 -72.065 -36.460 1.00 76.73 A C
+ATOM 604 OG1 THR A 103 0.229 -74.325 -36.187 1.00 63.67 A O
+ATOM 605 N ASN A 104 -1.274 -74.115 -34.056 1.00 90.50 A N
+ATOM 606 CA ASN A 104 -0.868 -74.521 -32.711 1.00 96.50 A C
+ATOM 607 C ASN A 104 0.292 -75.507 -32.825 1.00 91.68 A C
+ATOM 608 O ASN A 104 0.100 -76.646 -33.268 1.00 93.60 A O
+ATOM 609 CB ASN A 104 -2.035 -75.135 -31.932 1.00 97.34 A C
+ATOM 610 CG ASN A 104 -3.158 -74.145 -31.686 1.00 95.57 A C
+ATOM 611 ND2 ASN A 104 -3.308 -73.708 -30.432 1.00 87.53 A N
+ATOM 612 OD1 ASN A 104 -3.853 -73.738 -32.619 1.00 99.47 A O
+ATOM 613 N ASN A 105 1.489 -75.076 -32.414 1.00 86.80 A N
+ATOM 614 CA ASN A 105 2.669 -75.956 -32.414 1.00 81.65 A C
+ATOM 615 C ASN A 105 3.643 -75.622 -31.269 1.00 85.89 A C
+ATOM 616 O ASN A 105 3.338 -75.813 -30.081 1.00 84.49 A O
+ATOM 617 CB ASN A 105 3.410 -75.881 -33.758 1.00 78.97 A C
+ATOM 618 CG ASN A 105 2.774 -76.753 -34.840 1.00 88.86 A C
+ATOM 619 ND2 ASN A 105 1.807 -76.194 -35.551 1.00 84.27 A N
+ATOM 620 OD1 ASN A 105 3.177 -77.896 -35.056 1.00 95.34 A O
+ATOM 621 N SER A 134 3.529 -83.596 -17.837 1.00 99.06 A N
+ATOM 622 CA SER A 134 2.557 -84.576 -18.332 1.00102.75 A C
+ATOM 623 C SER A 134 2.610 -85.879 -17.517 1.00116.09 A C
+ATOM 624 O SER A 134 1.927 -86.043 -16.501 1.00139.31 A O
+ATOM 625 CB SER A 134 2.808 -84.870 -19.830 1.00 92.38 A C
+ATOM 626 OG SER A 134 2.063 -85.995 -20.294 1.00 77.34 A O
+ATOM 627 N GLY A 135 3.459 -86.784 -17.994 1.00107.84 A N
+ATOM 628 CA GLY A 135 3.731 -88.096 -17.443 1.00 97.60 A C
+ATOM 629 C GLY A 135 4.903 -88.069 -16.483 1.00 96.29 A C
+ATOM 630 O GLY A 135 4.938 -87.301 -15.514 1.00 94.12 A O
+ATOM 631 N ARG A 136 5.818 -89.007 -16.706 1.00104.86 A N
+ATOM 632 CA ARG A 136 7.122 -89.053 -16.059 1.00 92.96 A C
+ATOM 633 C ARG A 136 8.110 -88.076 -16.681 1.00 90.13 A C
+ATOM 634 O ARG A 136 9.306 -88.157 -16.388 1.00 92.59 A O
+ATOM 635 CB ARG A 136 7.697 -90.467 -16.126 1.00 85.22 A C
+ATOM 636 CG ARG A 136 7.284 -91.270 -14.939 1.00 94.41 A C
+ATOM 637 CD ARG A 136 7.982 -92.593 -14.810 1.00 92.45 A C
+ATOM 638 NE ARG A 136 7.329 -93.687 -15.526 1.00 87.28 A N
+ATOM 639 CZ ARG A 136 7.325 -94.947 -15.088 1.00 91.76 A C
+ATOM 640 NH1 ARG A 136 7.887 -95.228 -13.918 1.00 88.68 A N1+
+ATOM 641 NH2 ARG A 136 6.742 -95.919 -15.786 1.00 88.55 A N
+ATOM 642 N SER A 137 7.644 -87.211 -17.579 1.00 98.45 A N
+ATOM 643 CA SER A 137 8.438 -86.156 -18.194 1.00 89.59 A C
+ATOM 644 C SER A 137 8.169 -84.837 -17.484 1.00 80.24 A C
+ATOM 645 O SER A 137 7.109 -84.633 -16.886 1.00 82.88 A O
+ATOM 646 CB SER A 137 8.133 -85.985 -19.692 1.00 85.81 A C
+ATOM 647 OG SER A 137 6.983 -85.168 -19.934 1.00 90.41 A O
+ATOM 648 N LEU A 138 9.162 -83.959 -17.528 1.00 81.52 A N
+ATOM 649 CA LEU A 138 9.068 -82.627 -16.948 1.00 77.06 A C
+ATOM 650 C LEU A 138 8.376 -81.621 -17.858 1.00 71.83 A C
+ATOM 651 O LEU A 138 8.443 -80.419 -17.584 1.00 70.63 A O
+ATOM 652 CB LEU A 138 10.451 -82.115 -16.577 1.00 68.90 A C
+ATOM 653 CG LEU A 138 10.937 -82.736 -15.274 1.00 70.54 A C
+ATOM 654 CD1 LEU A 138 12.157 -81.990 -14.790 1.00 65.62 A C
+ATOM 655 CD2 LEU A 138 9.834 -82.752 -14.221 1.00 72.67 A C
+ATOM 656 N GLY A 139 7.768 -82.076 -18.951 1.00 73.77 A N
+ATOM 657 CA GLY A 139 7.177 -81.154 -19.910 1.00 66.17 A C
+ATOM 658 C GLY A 139 6.318 -80.056 -19.302 1.00 76.38 A C
+ATOM 659 O GLY A 139 6.438 -78.885 -19.682 1.00 84.12 A O
+ATOM 660 N PHE A 140 5.450 -80.406 -18.339 1.00 63.43 A N
+ATOM 661 CA PHE A 140 4.500 -79.421 -17.820 1.00 69.13 A C
+ATOM 662 C PHE A 140 5.197 -78.157 -17.344 1.00 75.66 A C
+ATOM 663 O PHE A 140 4.548 -77.113 -17.222 1.00 77.24 A O
+ATOM 664 CB PHE A 140 3.663 -80.003 -16.668 1.00 66.95 A C
+ATOM 665 CG PHE A 140 4.472 -80.368 -15.458 1.00 76.57 A C
+ATOM 666 CD1 PHE A 140 4.946 -81.658 -15.293 1.00 81.32 A C
+ATOM 667 CD2 PHE A 140 4.769 -79.422 -14.486 1.00 85.94 A C
+ATOM 668 CE1 PHE A 140 5.705 -81.992 -14.187 1.00 92.64 A C
+ATOM 669 CE2 PHE A 140 5.525 -79.759 -13.379 1.00 92.59 A C
+ATOM 670 CZ PHE A 140 5.992 -81.037 -13.229 1.00 93.35 A C
+ATOM 671 N LEU A 141 6.507 -78.230 -17.083 1.00 72.08 A N
+ATOM 672 CA LEU A 141 7.290 -77.078 -16.652 1.00 77.10 A C
+ATOM 673 C LEU A 141 7.603 -76.111 -17.788 1.00 74.90 A C
+ATOM 674 O LEU A 141 8.103 -75.011 -17.528 1.00 72.01 A O
+ATOM 675 CB LEU A 141 8.603 -77.541 -16.022 1.00 70.41 A C
+ATOM 676 CG LEU A 141 8.514 -78.326 -14.718 1.00 80.35 A C
+ATOM 677 CD1 LEU A 141 9.897 -78.830 -14.345 1.00 78.54 A C
+ATOM 678 CD2 LEU A 141 7.935 -77.458 -13.635 1.00 66.09 A C
+ATOM 679 N LEU A 142 7.318 -76.482 -19.028 1.00 69.72 A N
+ATOM 680 CA LEU A 142 7.776 -75.684 -20.149 1.00 72.95 A C
+ATOM 681 C LEU A 142 6.986 -74.385 -20.287 1.00 74.09 A C
+ATOM 682 O LEU A 142 5.852 -74.247 -19.813 1.00 77.23 A O
+ATOM 683 CB LEU A 142 7.682 -76.494 -21.432 1.00 64.30 A C
+ATOM 684 CG LEU A 142 8.915 -77.356 -21.603 1.00 74.87 A C
+ATOM 685 CD1 LEU A 142 8.717 -78.290 -22.746 1.00 67.74 A C
+ATOM 686 CD2 LEU A 142 10.063 -76.433 -21.884 1.00 66.01 A C
+ATOM 687 N GLY A 143 7.614 -73.422 -20.953 1.00 67.18 A N
+ATOM 688 CA GLY A 143 6.925 -72.222 -21.359 1.00 66.44 A C
+ATOM 689 C GLY A 143 6.042 -72.445 -22.576 1.00 70.48 A C
+ATOM 690 O GLY A 143 5.958 -73.530 -23.149 1.00 71.80 A O
+ATOM 691 N VAL A 144 5.381 -71.370 -22.985 1.00 69.07 A N
+ATOM 692 CA VAL A 144 4.441 -71.371 -24.097 1.00 58.78 A C
+ATOM 693 C VAL A 144 4.774 -70.223 -25.033 1.00 63.40 A C
+ATOM 694 O VAL A 144 4.916 -69.080 -24.583 1.00 68.13 A O
+ATOM 695 CB VAL A 144 3.002 -71.244 -23.587 1.00 77.99 A C
+ATOM 696 CG1 VAL A 144 2.120 -70.687 -24.655 1.00 59.68 A C
+ATOM 697 CG2 VAL A 144 2.506 -72.607 -23.228 1.00 62.67 A C
+ATOM 698 N GLY A 145 4.813 -70.502 -26.347 1.00 59.52 A N
+ATOM 699 CA GLY A 145 5.299 -69.526 -27.300 1.00 60.02 A C
+ATOM 700 C GLY A 145 4.503 -69.551 -28.587 1.00 67.01 A C
+ATOM 701 O GLY A 145 3.720 -70.463 -28.845 1.00 65.49 A O
+ATOM 702 N SER A 146 4.678 -68.503 -29.367 1.00 69.50 A N
+ATOM 703 CA SER A 146 3.980 -68.364 -30.635 1.00 56.98 A C
+ATOM 704 C SER A 146 4.955 -68.693 -31.747 1.00 60.07 A C
+ATOM 705 O SER A 146 5.792 -67.860 -32.094 1.00 61.96 A O
+ATOM 706 CB SER A 146 3.447 -66.947 -30.805 1.00 64.37 A C
+ATOM 707 OG SER A 146 2.915 -66.781 -32.099 1.00 64.41 A O
+ATOM 708 N ALA A 147 4.812 -69.872 -32.354 1.00 59.18 A N
+ATOM 709 CA ALA A 147 5.850 -70.321 -33.276 1.00 56.41 A C
+ATOM 710 C ALA A 147 5.973 -69.364 -34.468 1.00 59.84 A C
+ATOM 711 O ALA A 147 7.063 -68.850 -34.766 1.00 57.49 A O
+ATOM 712 CB ALA A 147 5.573 -71.760 -33.723 1.00 56.84 A C
+ATOM 713 N ILE A 148 4.856 -69.060 -35.121 1.00 55.88 A N
+ATOM 714 CA ILE A 148 4.931 -68.374 -36.404 1.00 56.49 A C
+ATOM 715 C ILE A 148 4.832 -66.880 -36.194 1.00 56.08 A C
+ATOM 716 O ILE A 148 4.647 -66.137 -37.164 1.00 60.01 A O
+ATOM 717 CB ILE A 148 3.850 -68.842 -37.387 1.00 54.24 A C
+ATOM 718 CG1 ILE A 148 2.472 -68.659 -36.748 1.00 58.56 A C
+ATOM 719 CG2 ILE A 148 4.143 -70.276 -37.844 1.00 53.15 A C
+ATOM 720 CD1 ILE A 148 1.303 -68.812 -37.716 1.00 57.65 A C
+ATOM 721 N ALA A 149 4.897 -66.436 -34.933 1.00 49.00 A N
+ATOM 722 CA ALA A 149 4.696 -65.016 -34.654 1.00 49.78 A C
+ATOM 723 C ALA A 149 5.534 -64.142 -35.583 1.00 55.77 A C
+ATOM 724 O ALA A 149 5.003 -63.250 -36.262 1.00 55.77 A O
+ATOM 725 CB ALA A 149 5.013 -64.713 -33.188 1.00 50.84 A C
+ATOM 726 N SER A 150 6.839 -64.421 -35.663 1.00 48.94 A N
+ATOM 727 CA SER A 150 7.759 -63.696 -36.536 1.00 48.48 A C
+ATOM 728 C SER A 150 7.236 -63.630 -37.970 1.00 50.09 A C
+ATOM 729 O SER A 150 6.852 -62.556 -38.477 1.00 53.46 A O
+ATOM 730 CB SER A 150 9.131 -64.374 -36.507 1.00 50.46 A C
+ATOM 731 OG SER A 150 8.999 -65.784 -36.387 1.00 56.77 A O
+ATOM 732 N GLY A 151 7.206 -64.801 -38.614 1.00 53.10 A N
+ATOM 733 CA GLY A 151 6.700 -64.882 -39.969 1.00 45.98 A C
+ATOM 734 C GLY A 151 5.374 -64.171 -40.153 1.00 57.53 A C
+ATOM 735 O GLY A 151 5.160 -63.496 -41.163 1.00 54.50 A O
+ATOM 736 N VAL A 152 4.471 -64.282 -39.173 1.00 58.82 A N
+ATOM 737 CA VAL A 152 3.174 -63.636 -39.345 1.00 48.15 A C
+ATOM 738 C VAL A 152 3.328 -62.124 -39.356 1.00 54.39 A C
+ATOM 739 O VAL A 152 2.646 -61.432 -40.119 1.00 63.19 A O
+ATOM 740 CB VAL A 152 2.166 -64.083 -38.276 1.00 52.20 A C
+ATOM 741 CG1 VAL A 152 0.829 -63.372 -38.506 1.00 49.31 A C
+ATOM 742 CG2 VAL A 152 1.974 -65.559 -38.338 1.00 45.44 A C
+ATOM 743 N ALA A 153 4.218 -61.581 -38.519 1.00 49.50 A N
+ATOM 744 CA ALA A 153 4.473 -60.144 -38.574 1.00 49.43 A C
+ATOM 745 C ALA A 153 4.936 -59.716 -39.968 1.00 53.98 A C
+ATOM 746 O ALA A 153 4.480 -58.690 -40.499 1.00 62.94 A O
+ATOM 747 CB ALA A 153 5.499 -59.747 -37.514 1.00 54.25 A C
+ATOM 748 N VAL A 154 5.819 -60.502 -40.591 1.00 50.21 A N
+ATOM 749 CA VAL A 154 6.226 -60.182 -41.963 1.00 54.89 A C
+ATOM 750 C VAL A 154 5.026 -60.240 -42.913 1.00 52.46 A C
+ATOM 751 O VAL A 154 4.816 -59.338 -43.742 1.00 55.68 A O
+ATOM 752 CB VAL A 154 7.355 -61.120 -42.420 1.00 50.70 A C
+ATOM 753 CG1 VAL A 154 7.821 -60.739 -43.794 1.00 42.79 A C
+ATOM 754 CG2 VAL A 154 8.501 -61.047 -41.442 1.00 45.08 A C
+ATOM 755 N SER A 155 4.227 -61.310 -42.815 1.00 43.62 A N
+ATOM 756 CA SER A 155 3.015 -61.429 -43.619 1.00 54.21 A C
+ATOM 757 C SER A 155 2.143 -60.192 -43.501 1.00 57.16 A C
+ATOM 758 O SER A 155 1.634 -59.678 -44.502 1.00 65.83 A O
+ATOM 759 CB SER A 155 2.222 -62.659 -43.195 1.00 50.03 A C
+ATOM 760 OG SER A 155 0.856 -62.515 -43.543 1.00 58.33 A O
+ATOM 761 N LYS A 156 1.941 -59.706 -42.284 1.00 51.55 A N
+ATOM 762 CA LYS A 156 1.235 -58.446 -42.143 1.00 54.59 A C
+ATOM 763 C LYS A 156 1.927 -57.375 -42.973 1.00 57.08 A C
+ATOM 764 O LYS A 156 1.314 -56.810 -43.882 1.00 61.09 A O
+ATOM 765 CB LYS A 156 1.127 -58.046 -40.669 1.00 55.63 A C
+ATOM 766 CG LYS A 156 0.505 -59.135 -39.745 1.00 79.38 A C
+ATOM 767 CD LYS A 156 -1.028 -59.260 -39.831 1.00102.05 A C
+ATOM 768 CE LYS A 156 -1.586 -60.114 -38.673 1.00111.09 A C
+ATOM 769 NZ LYS A 156 -3.082 -60.254 -38.652 1.00109.62 A N1+
+ATOM 770 N VAL A 157 3.240 -57.176 -42.760 1.00 47.86 A N
+ATOM 771 CA VAL A 157 3.970 -56.087 -43.432 1.00 52.04 A C
+ATOM 772 C VAL A 157 3.765 -56.103 -44.943 1.00 58.00 A C
+ATOM 773 O VAL A 157 3.801 -55.049 -45.593 1.00 76.55 A O
+ATOM 774 CB VAL A 157 5.479 -56.138 -43.090 1.00 58.81 A C
+ATOM 775 CG1 VAL A 157 6.280 -55.134 -43.934 1.00 63.40 A C
+ATOM 776 CG2 VAL A 157 5.711 -55.883 -41.620 1.00 64.04 A C
+ATOM 777 N LEU A 158 3.530 -57.286 -45.526 1.00 62.93 A N
+ATOM 778 CA LEU A 158 3.366 -57.368 -46.982 1.00 58.29 A C
+ATOM 779 C LEU A 158 2.054 -56.737 -47.460 1.00 70.17 A C
+ATOM 780 O LEU A 158 2.012 -56.145 -48.551 1.00 70.23 A O
+ATOM 781 CB LEU A 158 3.442 -58.819 -47.459 1.00 55.93 A C
+ATOM 782 CG LEU A 158 4.732 -59.597 -47.235 1.00 58.15 A C
+ATOM 783 CD1 LEU A 158 4.651 -60.827 -48.056 1.00 53.74 A C
+ATOM 784 CD2 LEU A 158 5.958 -58.797 -47.636 1.00 59.25 A C
+ATOM 785 N HIS A 159 0.963 -56.881 -46.683 1.00 58.12 A N
+ATOM 786 CA HIS A 159 -0.321 -56.316 -47.108 1.00 61.96 A C
+ATOM 787 C HIS A 159 -0.250 -54.805 -47.252 1.00 76.30 A C
+ATOM 788 O HIS A 159 -0.965 -54.228 -48.082 1.00 91.05 A O
+ATOM 789 CB HIS A 159 -1.429 -56.628 -46.111 1.00 59.52 A C
+ATOM 790 CG HIS A 159 -1.868 -58.053 -46.083 1.00 63.24 A C
+ATOM 791 CD2 HIS A 159 -3.037 -58.628 -46.451 1.00 63.02 A C
+ATOM 792 ND1 HIS A 159 -1.077 -59.067 -45.587 1.00 78.41 A N
+ATOM 793 CE1 HIS A 159 -1.740 -60.208 -45.651 1.00 70.68 A C
+ATOM 794 NE2 HIS A 159 -2.927 -59.971 -46.182 1.00 59.22 A N
+ATOM 795 N LEU A 160 0.599 -54.162 -46.445 1.00 60.03 A N
+ATOM 796 CA LEU A 160 0.705 -52.710 -46.402 1.00 68.46 A C
+ATOM 797 C LEU A 160 0.807 -52.148 -47.808 1.00 79.88 A C
+ATOM 798 O LEU A 160 1.555 -52.668 -48.643 1.00 89.41 A O
+ATOM 799 CB LEU A 160 1.939 -52.306 -45.602 1.00 66.72 A C
+ATOM 800 CG LEU A 160 1.978 -52.302 -44.088 1.00 65.65 A C
+ATOM 801 CD1 LEU A 160 1.370 -53.527 -43.509 1.00 60.23 A C
+ATOM 802 CD2 LEU A 160 3.441 -52.251 -43.718 1.00 59.64 A C
+ATOM 803 N GLU A 161 0.028 -51.102 -48.075 1.00 77.11 A N
+ATOM 804 CA GLU A 161 -0.134 -50.662 -49.450 1.00 87.36 A C
+ATOM 805 C GLU A 161 1.216 -50.239 -50.016 1.00 78.14 A C
+ATOM 806 O GLU A 161 1.993 -49.538 -49.351 1.00 71.49 A O
+ATOM 807 CB GLU A 161 -1.162 -49.532 -49.543 1.00107.07 A C
+ATOM 808 CG GLU A 161 -1.814 -49.469 -50.917 1.00117.39 A C
+ATOM 809 CD GLU A 161 -2.302 -50.835 -51.393 1.00128.90 A C
+ATOM 810 OE1 GLU A 161 -2.903 -51.574 -50.578 1.00133.12 A O
+ATOM 811 OE2 GLU A 161 -2.053 -51.178 -52.574 1.00132.63 A O1-
+ATOM 812 N GLY A 162 1.513 -50.740 -51.227 1.00 83.85 A N
+ATOM 813 CA GLY A 162 2.754 -50.487 -51.928 1.00 83.99 A C
+ATOM 814 C GLY A 162 3.952 -51.284 -51.454 1.00 89.60 A C
+ATOM 815 O GLY A 162 5.036 -51.136 -52.035 1.00 93.14 A O
+ATOM 816 N GLU A 163 3.814 -52.106 -50.407 1.00 81.31 A N
+ATOM 817 CA GLU A 163 4.923 -52.976 -50.032 1.00 75.19 A C
+ATOM 818 C GLU A 163 5.193 -54.009 -51.117 1.00 73.62 A C
+ATOM 819 O GLU A 163 6.351 -54.254 -51.472 1.00 76.76 A O
+ATOM 820 CB GLU A 163 4.651 -53.652 -48.687 1.00 71.33 A C
+ATOM 821 CG GLU A 163 5.407 -53.009 -47.517 1.00 84.90 A C
+ATOM 822 CD GLU A 163 6.941 -53.069 -47.666 1.00 91.50 A C
+ATOM 823 OE1 GLU A 163 7.470 -54.107 -48.124 1.00 79.35 A O
+ATOM 824 OE2 GLU A 163 7.628 -52.071 -47.346 1.00101.98 A O1-
+ATOM 825 N VAL A 164 4.137 -54.604 -51.676 1.00 73.31 A N
+ATOM 826 CA VAL A 164 4.314 -55.572 -52.756 1.00 71.78 A C
+ATOM 827 C VAL A 164 5.197 -54.985 -53.849 1.00 72.91 A C
+ATOM 828 O VAL A 164 6.177 -55.603 -54.278 1.00 71.46 A O
+ATOM 829 CB VAL A 164 2.953 -56.008 -53.325 1.00 85.03 A C
+ATOM 830 CG1 VAL A 164 3.138 -57.141 -54.288 1.00 76.45 A C
+ATOM 831 CG2 VAL A 164 2.005 -56.388 -52.221 1.00 92.72 A C
+ATOM 832 N ASN A 165 4.881 -53.766 -54.291 1.00 72.62 A N
+ATOM 833 CA ASN A 165 5.603 -53.186 -55.419 1.00 80.72 A C
+ATOM 834 C ASN A 165 7.099 -53.051 -55.166 1.00 79.55 A C
+ATOM 835 O ASN A 165 7.882 -53.202 -56.107 1.00 74.76 A O
+ATOM 836 CB ASN A 165 5.011 -51.840 -55.787 1.00 94.37 A C
+ATOM 837 CG ASN A 165 3.890 -51.982 -56.744 1.00106.62 A C
+ATOM 838 ND2 ASN A 165 4.177 -52.570 -57.897 1.00104.39 A N
+ATOM 839 OD1 ASN A 165 2.764 -51.588 -56.458 1.00115.86 A O
+ATOM 840 N LYS A 166 7.524 -52.751 -53.931 1.00 76.41 A N
+ATOM 841 CA LYS A 166 8.950 -52.831 -53.619 1.00 73.03 A C
+ATOM 842 C LYS A 166 9.521 -54.179 -54.035 1.00 71.12 A C
+ATOM 843 O LYS A 166 10.584 -54.255 -54.660 1.00 71.71 A O
+ATOM 844 CB LYS A 166 9.210 -52.598 -52.129 1.00 67.34 A C
+ATOM 845 CG LYS A 166 8.993 -51.192 -51.605 1.00 77.40 A C
+ATOM 846 CD LYS A 166 9.489 -51.131 -50.149 1.00 85.63 A C
+ATOM 847 CE LYS A 166 9.369 -49.734 -49.557 1.00 92.33 A C
+ATOM 848 NZ LYS A 166 9.843 -48.714 -50.550 1.00 96.83 A N1+
+ATOM 849 N ILE A 167 8.808 -55.256 -53.713 1.00 70.57 A N
+ATOM 850 CA ILE A 167 9.296 -56.596 -54.010 1.00 68.78 A C
+ATOM 851 C ILE A 167 9.264 -56.862 -55.511 1.00 69.41 A C
+ATOM 852 O ILE A 167 10.215 -57.417 -56.079 1.00 68.00 A O
+ATOM 853 CB ILE A 167 8.474 -57.627 -53.216 1.00 56.10 A C
+ATOM 854 CG1 ILE A 167 8.733 -57.452 -51.724 1.00 51.77 A C
+ATOM 855 CG2 ILE A 167 8.781 -59.043 -53.648 1.00 49.46 A C
+ATOM 856 CD1 ILE A 167 8.187 -58.581 -50.903 1.00 51.71 A C
+ATOM 857 N LYS A 168 8.181 -56.471 -56.183 1.00 62.94 A N
+ATOM 858 CA LYS A 168 8.133 -56.678 -57.624 1.00 63.49 A C
+ATOM 859 C LYS A 168 9.232 -55.895 -58.319 1.00 67.56 A C
+ATOM 860 O LYS A 168 9.778 -56.351 -59.329 1.00 69.95 A O
+ATOM 861 CB LYS A 168 6.748 -56.317 -58.176 1.00 67.36 A C
+ATOM 862 CG LYS A 168 5.812 -57.552 -58.291 1.00 78.06 A C
+ATOM 863 CD LYS A 168 4.333 -57.220 -58.538 1.00 79.94 A C
+ATOM 864 CE LYS A 168 4.088 -56.630 -59.905 1.00 78.97 A C
+ATOM 865 NZ LYS A 168 2.792 -55.925 -59.903 1.00 77.41 A N1+
+ATOM 866 N SER A 169 9.610 -54.751 -57.766 1.00 58.82 A N
+ATOM 867 CA SER A 169 10.664 -53.959 -58.374 1.00 66.64 A C
+ATOM 868 C SER A 169 12.048 -54.440 -57.992 1.00 63.94 A C
+ATOM 869 O SER A 169 13.018 -54.057 -58.646 1.00 69.99 A O
+ATOM 870 CB SER A 169 10.516 -52.485 -57.995 1.00 77.00 A C
+ATOM 871 OG SER A 169 9.431 -51.889 -58.693 1.00 83.83 A O
+ATOM 872 N ALA A 170 12.176 -55.251 -56.949 1.00 66.40 A N
+ATOM 873 CA ALA A 170 13.497 -55.789 -56.652 1.00 68.53 A C
+ATOM 874 C ALA A 170 13.862 -56.956 -57.553 1.00 71.84 A C
+ATOM 875 O ALA A 170 15.050 -57.259 -57.711 1.00 69.05 A O
+ATOM 876 CB ALA A 170 13.593 -56.233 -55.194 1.00 66.25 A C
+ATOM 877 N LEU A 171 12.877 -57.627 -58.131 1.00 74.73 A N
+ATOM 878 CA LEU A 171 13.130 -58.847 -58.874 1.00 69.58 A C
+ATOM 879 C LEU A 171 13.219 -58.635 -60.379 1.00 72.28 A C
+ATOM 880 O LEU A 171 13.093 -59.615 -61.128 1.00 68.31 A O
+ATOM 881 CB LEU A 171 12.090 -59.903 -58.533 1.00 55.03 A C
+ATOM 882 CG LEU A 171 12.777 -60.647 -57.390 1.00 52.13 A C
+ATOM 883 CD1 LEU A 171 12.568 -59.899 -56.067 1.00 56.70 A C
+ATOM 884 CD2 LEU A 171 12.397 -62.125 -57.268 1.00 56.44 A C
+ATOM 885 N LEU A 172 13.316 -57.382 -60.847 1.00 52.23 A N
+ATOM 886 CA LEU A 172 13.342 -57.128 -62.291 1.00 57.87 A C
+ATOM 887 C LEU A 172 14.557 -57.772 -62.960 1.00 69.54 A C
+ATOM 888 O LEU A 172 14.439 -58.424 -64.009 1.00 60.94 A O
+ATOM 889 CB LEU A 172 13.340 -55.623 -62.566 1.00 62.23 A C
+ATOM 890 CG LEU A 172 12.256 -54.724 -61.981 1.00 63.65 A C
+ATOM 891 CD1 LEU A 172 12.745 -53.322 -62.112 1.00 55.26 A C
+ATOM 892 CD2 LEU A 172 10.911 -54.868 -62.661 1.00 56.93 A C
+ATOM 893 N SER A 173 15.739 -57.552 -62.394 1.00 54.84 A N
+ATOM 894 CA SER A 173 16.993 -57.933 -63.018 1.00 59.62 A C
+ATOM 895 C SER A 173 17.563 -59.253 -62.484 1.00 49.88 A C
+ATOM 896 O SER A 173 18.711 -59.589 -62.796 1.00 50.62 A O
+ATOM 897 CB SER A 173 17.994 -56.800 -62.868 1.00 65.86 A C
+ATOM 898 OG SER A 173 18.915 -56.851 -63.933 1.00 70.17 A O
+ATOM 899 N THR A 174 16.760 -60.033 -61.757 1.00 65.92 A N
+ATOM 900 CA THR A 174 17.208 -61.340 -61.251 1.00 56.37 A C
+ATOM 901 C THR A 174 16.371 -61.860 -60.077 1.00 52.93 A C
+ATOM 902 O THR A 174 15.982 -61.076 -59.211 1.00 48.42 A O
+ATOM 903 CB THR A 174 18.690 -61.302 -60.830 1.00 52.33 A C
+ATOM 904 CG2 THR A 174 18.948 -60.131 -59.895 1.00 45.24 A C
+ATOM 905 OG1 THR A 174 19.032 -62.524 -60.164 1.00 58.69 A O
+ATOM 906 N ASN A 175 16.091 -63.167 -60.028 1.00 47.85 A N
+ATOM 907 CA ASN A 175 15.302 -63.666 -58.901 1.00 46.18 A C
+ATOM 908 C ASN A 175 16.147 -63.531 -57.611 1.00 47.66 A C
+ATOM 909 O ASN A 175 17.170 -62.830 -57.609 1.00 45.27 A O
+ATOM 910 CB ASN A 175 14.783 -65.060 -59.208 1.00 43.29 A C
+ATOM 911 CG ASN A 175 15.863 -66.003 -59.488 1.00 58.74 A C
+ATOM 912 ND2 ASN A 175 15.974 -67.018 -58.671 1.00 66.15 A N
+ATOM 913 OD1 ASN A 175 16.581 -65.855 -60.456 1.00 65.69 A O
+ATOM 914 N LYS A 176 15.703 -64.106 -56.474 1.00 43.49 A N
+ATOM 915 CA LYS A 176 16.529 -64.138 -55.251 1.00 43.59 A C
+ATOM 916 C LYS A 176 17.256 -62.817 -54.990 1.00 51.42 A C
+ATOM 917 O LYS A 176 18.458 -62.698 -55.248 1.00 74.13 A O
+ATOM 918 CB LYS A 176 17.561 -65.288 -55.286 1.00 44.61 A C
+ATOM 919 CG LYS A 176 18.136 -65.739 -53.899 1.00 48.09 A C
+ATOM 920 CD LYS A 176 19.585 -66.280 -53.963 1.00 60.58 A C
+ATOM 921 CE LYS A 176 19.815 -67.203 -55.206 1.00 61.48 A C
+ATOM 922 NZ LYS A 176 21.204 -67.815 -55.488 1.00 61.87 A N1+
+ATOM 923 N ALA A 177 16.549 -61.791 -54.542 1.00 59.61 A N
+ATOM 924 CA ALA A 177 17.250 -60.585 -54.146 1.00 56.48 A C
+ATOM 925 C ALA A 177 16.697 -60.058 -52.825 1.00 59.26 A C
+ATOM 926 O ALA A 177 15.570 -60.386 -52.424 1.00 63.92 A O
+ATOM 927 CB ALA A 177 17.155 -59.539 -55.246 1.00 49.89 A C
+ATOM 928 N VAL A 178 17.519 -59.227 -52.156 1.00 58.19 A N
+ATOM 929 CA VAL A 178 17.169 -58.590 -50.882 1.00 58.37 A C
+ATOM 930 C VAL A 178 16.230 -57.408 -51.070 1.00 64.12 A C
+ATOM 931 O VAL A 178 16.514 -56.471 -51.830 1.00 71.99 A O
+ATOM 932 CB VAL A 178 18.427 -58.159 -50.114 1.00 54.00 A C
+ATOM 933 CG1 VAL A 178 18.114 -56.989 -49.255 1.00 58.08 A C
+ATOM 934 CG2 VAL A 178 18.757 -59.217 -49.142 1.00 50.61 A C
+ATOM 935 N VAL A 179 15.136 -57.432 -50.312 1.00 62.66 A N
+ATOM 936 CA VAL A 179 14.237 -56.302 -50.155 1.00 65.68 A C
+ATOM 937 C VAL A 179 14.251 -55.890 -48.691 1.00 73.44 A C
+ATOM 938 O VAL A 179 14.188 -56.748 -47.802 1.00 78.07 A O
+ATOM 939 CB VAL A 179 12.812 -56.676 -50.595 1.00 62.19 A C
+ATOM 940 CG1 VAL A 179 11.966 -55.425 -50.773 1.00 58.77 A C
+ATOM 941 CG2 VAL A 179 12.866 -57.505 -51.849 1.00 60.78 A C
+ATOM 942 N SER A 180 14.342 -54.588 -48.434 1.00 66.05 A N
+ATOM 943 CA SER A 180 14.077 -54.054 -47.105 1.00 76.44 A C
+ATOM 944 C SER A 180 12.650 -53.532 -47.090 1.00 70.19 A C
+ATOM 945 O SER A 180 12.220 -52.858 -48.039 1.00 72.77 A O
+ATOM 946 CB SER A 180 15.065 -52.953 -46.743 1.00 91.73 A C
+ATOM 947 OG SER A 180 16.275 -53.534 -46.305 1.00 99.23 A O
+ATOM 948 N LEU A 181 11.902 -53.875 -46.046 1.00 80.56 A N
+ATOM 949 CA LEU A 181 10.484 -53.545 -46.047 1.00 85.88 A C
+ATOM 950 C LEU A 181 10.247 -52.202 -45.352 1.00 97.23 A C
+ATOM 951 O LEU A 181 11.163 -51.588 -44.796 1.00104.36 A O
+ATOM 952 CB LEU A 181 9.680 -54.676 -45.402 1.00 77.17 A C
+ATOM 953 CG LEU A 181 9.980 -56.075 -45.963 1.00 69.70 A C
+ATOM 954 CD1 LEU A 181 9.498 -57.178 -45.030 1.00 55.21 A C
+ATOM 955 CD2 LEU A 181 9.396 -56.271 -47.366 1.00 59.24 A C
+ATOM 956 N SER A 182 8.988 -51.751 -45.368 1.00 82.47 A N
+ATOM 957 CA SER A 182 8.647 -50.435 -44.834 1.00 85.73 A C
+ATOM 958 C SER A 182 9.146 -50.254 -43.412 1.00 85.42 A C
+ATOM 959 O SER A 182 9.585 -49.166 -43.034 1.00 94.88 A O
+ATOM 960 CB SER A 182 7.136 -50.234 -44.877 1.00 85.09 A C
+ATOM 961 OG SER A 182 6.650 -50.463 -46.182 1.00 88.04 A O
+ATOM 962 N ASN A 183 9.081 -51.304 -42.611 1.00 75.55 A N
+ATOM 963 CA ASN A 183 9.603 -51.281 -41.260 1.00 79.68 A C
+ATOM 964 C ASN A 183 11.083 -51.596 -41.223 1.00 91.16 A C
+ATOM 965 O ASN A 183 11.629 -51.840 -40.142 1.00 92.51 A O
+ATOM 966 CB ASN A 183 8.848 -52.266 -40.382 1.00 70.17 A C
+ATOM 967 CG ASN A 183 9.147 -53.702 -40.741 1.00 75.39 A C
+ATOM 968 ND2 ASN A 183 8.825 -54.611 -39.828 1.00 71.20 A N
+ATOM 969 OD1 ASN A 183 9.648 -53.997 -41.833 1.00 85.11 A O
+ATOM 970 N GLY A 184 11.717 -51.705 -42.383 1.00 88.83 A N
+ATOM 971 CA GLY A 184 13.155 -51.816 -42.400 1.00 87.75 A C
+ATOM 972 C GLY A 184 13.689 -53.145 -41.937 1.00 80.69 A C
+ATOM 973 O GLY A 184 14.900 -53.271 -41.735 1.00 85.83 A O
+ATOM 974 N VAL A 185 12.825 -54.133 -41.733 1.00 89.78 A N
+ATOM 975 CA VAL A 185 13.269 -55.515 -41.592 1.00 82.85 A C
+ATOM 976 C VAL A 185 13.592 -56.025 -42.989 1.00 76.18 A C
+ATOM 977 O VAL A 185 12.741 -55.980 -43.886 1.00 71.53 A O
+ATOM 978 CB VAL A 185 12.188 -56.373 -40.922 1.00 84.62 A C
+ATOM 979 CG1 VAL A 185 12.669 -57.795 -40.769 1.00 80.15 A C
+ATOM 980 CG2 VAL A 185 11.797 -55.782 -39.581 1.00 87.85 A C
+ATOM 981 N SER A 186 14.827 -56.473 -43.200 1.00 81.73 A N
+ATOM 982 CA SER A 186 15.215 -56.855 -44.552 1.00 78.98 A C
+ATOM 983 C SER A 186 15.165 -58.373 -44.686 1.00 67.77 A C
+ATOM 984 O SER A 186 15.499 -59.121 -43.759 1.00 72.58 A O
+ATOM 985 CB SER A 186 16.596 -56.312 -44.971 1.00 83.30 A C
+ATOM 986 OG SER A 186 17.649 -57.236 -44.766 1.00 85.27 A O
+ATOM 987 N VAL A 187 14.702 -58.809 -45.848 1.00 70.97 A N
+ATOM 988 CA VAL A 187 14.360 -60.194 -46.096 1.00 59.17 A C
+ATOM 989 C VAL A 187 14.789 -60.545 -47.510 1.00 66.51 A C
+ATOM 990 O VAL A 187 14.853 -59.686 -48.401 1.00 78.02 A O
+ATOM 991 CB VAL A 187 12.847 -60.442 -45.907 1.00 54.62 A C
+ATOM 992 CG1 VAL A 187 12.417 -60.101 -44.473 1.00 67.49 A C
+ATOM 993 CG2 VAL A 187 12.040 -59.630 -46.944 1.00 55.09 A C
+ATOM 994 N LEU A 188 15.123 -61.818 -47.690 1.00 55.31 A N
+ATOM 995 CA LEU A 188 15.295 -62.363 -49.021 1.00 47.69 A C
+ATOM 996 C LEU A 188 13.927 -62.523 -49.658 1.00 55.55 A C
+ATOM 997 O LEU A 188 12.951 -62.874 -48.984 1.00 52.33 A O
+ATOM 998 CB LEU A 188 16.001 -63.710 -48.952 1.00 47.72 A C
+ATOM 999 CG LEU A 188 17.444 -63.573 -48.526 1.00 71.76 A C
+ATOM 1000 CD1 LEU A 188 17.946 -64.899 -48.045 1.00 75.89 A C
+ATOM 1001 CD2 LEU A 188 18.242 -63.065 -49.692 1.00 73.84 A C
+ATOM 1002 N THR A 189 13.848 -62.227 -50.951 1.00 53.11 A N
+ATOM 1003 CA THR A 189 12.687 -62.578 -51.748 1.00 43.96 A C
+ATOM 1004 C THR A 189 13.141 -63.405 -52.929 1.00 44.61 A C
+ATOM 1005 O THR A 189 14.061 -63.003 -53.655 1.00 50.37 A O
+ATOM 1006 CB THR A 189 11.927 -61.359 -52.240 1.00 51.48 A C
+ATOM 1007 CG2 THR A 189 10.852 -61.774 -53.215 1.00 52.49 A C
+ATOM 1008 OG1 THR A 189 11.310 -60.727 -51.122 1.00 61.56 A O
+ATOM 1009 N SER A 190 12.488 -64.553 -53.116 1.00 45.31 A N
+ATOM 1010 CA SER A 190 12.760 -65.443 -54.234 1.00 47.19 A C
+ATOM 1011 C SER A 190 11.477 -65.661 -55.019 1.00 51.12 A C
+ATOM 1012 O SER A 190 10.424 -65.922 -54.422 1.00 56.62 A O
+ATOM 1013 CB SER A 190 13.320 -66.778 -53.746 1.00 47.59 A C
+ATOM 1014 OG SER A 190 13.464 -67.673 -54.837 1.00 62.41 A O
+ATOM 1015 N LYS A 191 11.561 -65.537 -56.354 1.00 55.72 A N
+ATOM 1016 CA LYS A 191 10.431 -65.843 -57.228 1.00 49.62 A C
+ATOM 1017 C LYS A 191 10.660 -67.213 -57.832 1.00 48.88 A C
+ATOM 1018 O LYS A 191 11.502 -67.368 -58.719 1.00 47.90 A O
+ATOM 1019 CB LYS A 191 10.256 -64.824 -58.347 1.00 48.28 A C
+ATOM 1020 CG LYS A 191 9.240 -65.286 -59.374 1.00 46.34 A C
+ATOM 1021 CD LYS A 191 9.432 -64.599 -60.698 1.00 53.83 A C
+ATOM 1022 CE LYS A 191 8.817 -63.217 -60.748 1.00 55.84 A C
+ATOM 1023 NZ LYS A 191 9.119 -62.580 -62.075 1.00 64.35 A N1+
+ATOM 1024 N VAL A 192 9.890 -68.208 -57.384 1.00 46.18 A N
+ATOM 1025 CA VAL A 192 10.135 -69.545 -57.900 1.00 54.34 A C
+ATOM 1026 C VAL A 192 9.272 -69.905 -59.094 1.00 55.48 A C
+ATOM 1027 O VAL A 192 9.516 -70.948 -59.713 1.00 52.68 A O
+ATOM 1028 CB VAL A 192 9.954 -70.612 -56.811 1.00 51.07 A C
+ATOM 1029 CG1 VAL A 192 10.741 -70.233 -55.620 1.00 43.88 A C
+ATOM 1030 CG2 VAL A 192 8.544 -70.687 -56.435 1.00 42.01 A C
+ATOM 1031 N LEU A 193 8.262 -69.115 -59.442 1.00 55.49 A N
+ATOM 1032 CA LEU A 193 7.518 -69.492 -60.636 1.00 52.45 A C
+ATOM 1033 C LEU A 193 6.975 -68.278 -61.394 1.00 56.59 A C
+ATOM 1034 O LEU A 193 6.317 -67.418 -60.795 1.00 58.81 A O
+ATOM 1035 CB LEU A 193 6.402 -70.433 -60.235 1.00 52.73 A C
+ATOM 1036 CG LEU A 193 5.666 -70.897 -61.457 1.00 50.89 A C
+ATOM 1037 CD1 LEU A 193 6.724 -71.458 -62.418 1.00 58.17 A C
+ATOM 1038 CD2 LEU A 193 4.625 -71.927 -61.015 1.00 45.98 A C
+ATOM 1039 N ASP A 194 7.213 -68.217 -62.711 1.00 58.41 A N
+ATOM 1040 CA ASP A 194 6.880 -67.034 -63.506 1.00 58.26 A C
+ATOM 1041 C ASP A 194 5.551 -67.135 -64.263 1.00 56.72 A C
+ATOM 1042 O ASP A 194 5.395 -66.470 -65.296 1.00 61.09 A O
+ATOM 1043 CB ASP A 194 8.001 -66.656 -64.462 1.00 59.04 A C
+ATOM 1044 CG ASP A 194 8.032 -65.139 -64.736 1.00 66.76 A C
+ATOM 1045 OD1 ASP A 194 6.985 -64.462 -64.549 1.00 73.37 A O
+ATOM 1046 OD2 ASP A 194 9.107 -64.624 -65.127 1.00 67.84 A O1-
+ATOM 1047 N LEU A 195 4.618 -67.981 -63.807 1.00 52.56 A N
+ATOM 1048 CA LEU A 195 3.436 -68.417 -64.550 1.00 53.53 A C
+ATOM 1049 C LEU A 195 2.811 -67.337 -65.420 1.00 60.64 A C
+ATOM 1050 O LEU A 195 2.392 -67.619 -66.549 1.00 58.97 A O
+ATOM 1051 CB LEU A 195 2.373 -68.896 -63.567 1.00 45.34 A C
+ATOM 1052 CG LEU A 195 2.327 -70.408 -63.443 1.00 46.72 A C
+ATOM 1053 CD1 LEU A 195 1.193 -70.900 -62.545 1.00 45.45 A C
+ATOM 1054 CD2 LEU A 195 2.191 -70.948 -64.817 1.00 47.13 A C
+ATOM 1055 N LYS A 196 2.750 -66.102 -64.914 1.00 61.74 A N
+ATOM 1056 CA LYS A 196 2.289 -64.993 -65.745 1.00 58.09 A C
+ATOM 1057 C LYS A 196 3.083 -64.930 -67.044 1.00 70.01 A C
+ATOM 1058 O LYS A 196 2.501 -64.762 -68.120 1.00 54.01 A O
+ATOM 1059 CB LYS A 196 2.375 -63.663 -64.977 1.00 61.41 A C
+ATOM 1060 CG LYS A 196 2.064 -62.397 -65.807 1.00 64.02 A C
+ATOM 1061 CD LYS A 196 3.344 -61.581 -66.021 1.00 66.30 A C
+ATOM 1062 CE LYS A 196 3.153 -60.320 -66.870 1.00 77.19 A C
+ATOM 1063 NZ LYS A 196 2.828 -59.064 -66.128 1.00 83.12 A N1+
+ATOM 1064 N ASN A 197 4.408 -65.105 -66.978 1.00 64.32 A N
+ATOM 1065 CA ASN A 197 5.187 -65.060 -68.212 1.00 58.55 A C
+ATOM 1066 C ASN A 197 4.721 -66.130 -69.187 1.00 54.17 A C
+ATOM 1067 O ASN A 197 4.440 -65.832 -70.352 1.00 54.09 A O
+ATOM 1068 CB ASN A 197 6.685 -65.210 -67.950 1.00 66.85 A C
+ATOM 1069 CG ASN A 197 7.498 -65.312 -69.249 1.00 77.66 A C
+ATOM 1070 ND2 ASN A 197 7.908 -66.533 -69.606 1.00 78.49 A N
+ATOM 1071 OD1 ASN A 197 7.746 -64.306 -69.915 1.00 79.00 A O
+ATOM 1072 N TYR A 198 4.613 -67.383 -68.737 1.00 59.82 A N
+ATOM 1073 CA TYR A 198 4.227 -68.435 -69.672 1.00 59.39 A C
+ATOM 1074 C TYR A 198 2.864 -68.155 -70.266 1.00 58.04 A C
+ATOM 1075 O TYR A 198 2.672 -68.285 -71.478 1.00 64.12 A O
+ATOM 1076 CB TYR A 198 4.234 -69.799 -69.004 1.00 44.23 A C
+ATOM 1077 CG TYR A 198 3.856 -70.893 -69.953 1.00 49.09 A C
+ATOM 1078 CD1 TYR A 198 4.815 -71.499 -70.732 1.00 49.49 A C
+ATOM 1079 CD2 TYR A 198 2.560 -71.355 -70.042 1.00 47.82 A C
+ATOM 1080 CE1 TYR A 198 4.493 -72.509 -71.609 1.00 49.44 A C
+ATOM 1081 CE2 TYR A 198 2.232 -72.374 -70.904 1.00 50.83 A C
+ATOM 1082 CZ TYR A 198 3.204 -72.941 -71.694 1.00 54.32 A C
+ATOM 1083 OH TYR A 198 2.918 -73.956 -72.580 1.00 62.15 A O
+ATOM 1084 N ILE A 199 1.910 -67.751 -69.426 1.00 55.54 A N
+ATOM 1085 CA ILE A 199 0.585 -67.376 -69.916 1.00 54.80 A C
+ATOM 1086 C ILE A 199 0.688 -66.366 -71.057 1.00 59.69 A C
+ATOM 1087 O ILE A 199 0.006 -66.502 -72.082 1.00 63.33 A O
+ATOM 1088 CB ILE A 199 -0.264 -66.826 -68.760 1.00 51.97 A C
+ATOM 1089 CG1 ILE A 199 -0.661 -67.944 -67.815 1.00 48.25 A C
+ATOM 1090 CG2 ILE A 199 -1.497 -66.165 -69.294 1.00 50.38 A C
+ATOM 1091 CD1 ILE A 199 -1.639 -67.477 -66.778 1.00 53.29 A C
+ATOM 1092 N ASP A 200 1.544 -65.344 -70.904 1.00 47.55 A N
+ATOM 1093 CA ASP A 200 1.673 -64.308 -71.932 1.00 55.72 A C
+ATOM 1094 C ASP A 200 2.352 -64.847 -73.182 1.00 54.43 A C
+ATOM 1095 O ASP A 200 1.800 -64.806 -74.287 1.00 62.23 A O
+ATOM 1096 CB ASP A 200 2.488 -63.125 -71.402 1.00 57.15 A C
+ATOM 1097 CG ASP A 200 1.890 -62.478 -70.161 1.00 61.49 A C
+ATOM 1098 OD1 ASP A 200 0.645 -62.531 -69.950 1.00 57.97 A O
+ATOM 1099 OD2 ASP A 200 2.700 -61.908 -69.395 1.00 68.84 A O1-
+ATOM 1100 N LYS A 201 3.559 -65.368 -73.009 1.00 60.34 A N
+ATOM 1101 CA LYS A 201 4.392 -65.845 -74.096 1.00 56.45 A C
+ATOM 1102 C LYS A 201 3.794 -67.074 -74.779 1.00 54.97 A C
+ATOM 1103 O LYS A 201 4.375 -67.569 -75.750 1.00 63.33 A O
+ATOM 1104 CB LYS A 201 5.807 -66.128 -73.549 1.00 72.13 A C
+ATOM 1105 CG LYS A 201 6.996 -65.461 -74.303 1.00 87.88 A C
+ATOM 1106 CD LYS A 201 8.217 -65.162 -73.366 1.00 90.29 A C
+ATOM 1107 CE LYS A 201 9.337 -66.246 -73.393 1.00 93.38 A C
+ATOM 1108 NZ LYS A 201 10.513 -65.979 -72.481 1.00 89.93 A N1+
+ATOM 1109 N GLN A 202 2.691 -67.619 -74.270 1.00 56.51 A N
+ATOM 1110 CA GLN A 202 2.215 -68.899 -74.783 1.00 57.18 A C
+ATOM 1111 C GLN A 202 0.712 -68.992 -75.028 1.00 59.96 A C
+ATOM 1112 O GLN A 202 0.277 -69.471 -76.073 1.00 62.89 A O
+ATOM 1113 CB GLN A 202 2.698 -70.022 -73.870 1.00 58.04 A C
+ATOM 1114 CG GLN A 202 4.119 -70.428 -74.178 1.00 74.55 A C
+ATOM 1115 CD GLN A 202 4.236 -71.037 -75.554 1.00 87.80 A C
+ATOM 1116 NE2 GLN A 202 4.598 -70.218 -76.539 1.00 91.58 A N
+ATOM 1117 OE1 GLN A 202 4.003 -72.235 -75.733 1.00 89.23 A O
+ATOM 1118 N LEU A 203 -0.092 -68.720 -74.023 1.00 68.98 A N
+ATOM 1119 CA LEU A 203 -1.515 -68.957 -74.149 1.00 56.78 A C
+ATOM 1120 C LEU A 203 -2.328 -67.717 -74.506 1.00 64.58 A C
+ATOM 1121 O LEU A 203 -3.560 -67.794 -74.506 1.00 70.09 A O
+ATOM 1122 CB LEU A 203 -2.056 -69.577 -72.868 1.00 50.67 A C
+ATOM 1123 CG LEU A 203 -1.780 -71.053 -72.615 1.00 58.73 A C
+ATOM 1124 CD1 LEU A 203 -0.335 -71.384 -72.630 1.00 66.52 A C
+ATOM 1125 CD2 LEU A 203 -2.340 -71.390 -71.250 1.00 52.84 A C
+ATOM 1126 N LEU A 204 -1.707 -66.570 -74.773 1.00 49.69 A N
+ATOM 1127 CA LEU A 204 -2.513 -65.354 -74.868 1.00 64.72 A C
+ATOM 1128 C LEU A 204 -3.592 -65.371 -75.951 1.00 75.34 A C
+ATOM 1129 O LEU A 204 -4.628 -64.721 -75.726 1.00 85.31 A O
+ATOM 1130 CB LEU A 204 -1.616 -64.125 -75.011 1.00 76.63 A C
+ATOM 1131 CG LEU A 204 -1.428 -63.568 -73.600 1.00 74.79 A C
+ATOM 1132 CD1 LEU A 204 -0.877 -62.162 -73.612 1.00 76.49 A C
+ATOM 1133 CD2 LEU A 204 -2.730 -63.643 -72.833 1.00 72.18 A C
+ATOM 1134 N PRO A 205 -3.429 -66.033 -77.120 1.00 69.42 A N
+ATOM 1135 CA PRO A 205 -4.589 -66.216 -78.025 1.00 78.88 A C
+ATOM 1136 C PRO A 205 -5.879 -66.695 -77.344 1.00 83.93 A C
+ATOM 1137 O PRO A 205 -6.960 -66.159 -77.634 1.00 83.80 A O
+ATOM 1138 CB PRO A 205 -4.072 -67.259 -79.031 1.00 77.47 A C
+ATOM 1139 CG PRO A 205 -2.634 -66.971 -79.131 1.00 71.50 A C
+ATOM 1140 CD PRO A 205 -2.168 -66.392 -77.797 1.00 65.26 A C
+ATOM 1141 N ILE A 206 -5.784 -67.658 -76.415 1.00 60.81 A N
+ATOM 1142 CA ILE A 206 -6.951 -68.377 -75.900 1.00 78.88 A C
+ATOM 1143 C ILE A 206 -7.775 -67.564 -74.919 1.00 94.81 A C
+ATOM 1144 O ILE A 206 -8.936 -67.907 -74.670 1.00100.72 A O
+ATOM 1145 CB ILE A 206 -6.541 -69.689 -75.216 1.00 76.07 A C
+ATOM 1146 CG1 ILE A 206 -5.369 -70.346 -75.949 1.00 87.46 A C
+ATOM 1147 CG2 ILE A 206 -7.701 -70.635 -75.154 1.00 66.12 A C
+ATOM 1148 CD1 ILE A 206 -5.624 -70.658 -77.422 1.00 95.91 A C
+ATOM 1149 N VAL A 207 -7.211 -66.531 -74.305 1.00 88.91 A N
+ATOM 1150 CA VAL A 207 -8.045 -65.671 -73.480 1.00 87.69 A C
+ATOM 1151 C VAL A 207 -8.836 -64.687 -74.343 1.00 88.23 A C
+ATOM 1152 O VAL A 207 -10.067 -64.601 -74.236 1.00 96.02 A O
+ATOM 1153 CB VAL A 207 -7.183 -64.956 -72.418 1.00 85.21 A C
+ATOM 1154 CG1 VAL A 207 -7.956 -63.819 -71.756 1.00 80.15 A C
+ATOM 1155 CG2 VAL A 207 -6.698 -65.948 -71.361 1.00 85.30 A C
+ATOM 1156 N ASN A 208 -8.150 -63.933 -75.214 1.00105.49 A N
+ATOM 1157 CA ASN A 208 -8.743 -62.755 -75.852 1.00 92.53 A C
+ATOM 1158 C ASN A 208 -9.170 -62.926 -77.318 1.00 88.83 A C
+ATOM 1159 O ASN A 208 -9.756 -61.997 -77.878 1.00 87.67 A O
+ATOM 1160 CB ASN A 208 -7.790 -61.556 -75.713 1.00 85.00 A C
+ATOM 1161 CG ASN A 208 -7.280 -61.368 -74.272 1.00 80.78 A C
+ATOM 1162 ND2 ASN A 208 -8.000 -60.579 -73.472 1.00 69.99 A N
+ATOM 1163 OD1 ASN A 208 -6.265 -61.941 -73.889 1.00 88.75 A O
+ATOM 1164 N LYS A 209 -8.926 -64.067 -77.953 1.00100.78 A N
+ATOM 1165 CA LYS A 209 -9.385 -64.297 -79.320 1.00102.44 A C
+ATOM 1166 C LYS A 209 -10.493 -65.354 -79.330 1.00101.97 A C
+ATOM 1167 O LYS A 209 -10.558 -66.212 -78.449 1.00100.26 A O
+ATOM 1168 CB LYS A 209 -8.225 -64.752 -80.230 1.00 99.70 A C
+ATOM 1169 CG LYS A 209 -7.543 -63.678 -81.101 1.00 98.60 A C
+ATOM 1170 CD LYS A 209 -6.065 -64.043 -81.471 1.00 97.69 A C
+ATOM 1171 CE LYS A 209 -5.929 -65.189 -82.500 1.00101.20 A C
+ATOM 1172 NZ LYS A 209 -4.533 -65.723 -82.676 1.00 99.59 A N1+
+ATOM 1173 N GLN A 210 -11.326 -65.269 -80.361 1.00112.88 A N
+ATOM 1174 CA GLN A 210 -12.374 -66.223 -80.658 1.00111.92 A C
+ATOM 1175 C GLN A 210 -11.616 -66.711 -81.869 1.00113.52 A C
+ATOM 1176 O GLN A 210 -10.994 -65.888 -82.541 1.00112.10 A O
+ATOM 1177 CB GLN A 210 -13.658 -65.515 -81.071 1.00117.43 A C
+ATOM 1178 CG GLN A 210 -13.442 -64.130 -81.660 1.00122.04 A C
+ATOM 1179 CD GLN A 210 -13.158 -63.084 -80.600 1.00123.60 A C
+ATOM 1180 NE2 GLN A 210 -12.486 -62.009 -80.994 1.00123.11 A N
+ATOM 1181 OE1 GLN A 210 -13.540 -63.240 -79.440 1.00125.33 A O
+ATOM 1182 N SER A 211 -11.590 -68.001 -82.178 1.00114.18 A N
+ATOM 1183 CA SER A 211 -10.740 -68.340 -83.336 1.00106.26 A C
+ATOM 1184 C SER A 211 -9.627 -67.268 -83.442 1.00109.00 A C
+ATOM 1185 O SER A 211 -9.679 -66.468 -84.375 1.00115.83 A O
+ATOM 1186 CB SER A 211 -11.573 -68.423 -84.642 1.00100.71 A C
+ATOM 1187 OG SER A 211 -10.838 -68.907 -85.765 1.00 96.47 A O
+ATOM 1188 N CYS A 212 -8.631 -67.193 -82.538 1.00114.50 A N
+ATOM 1189 CA CYS A 212 -8.142 -68.193 -81.558 1.00100.74 A C
+ATOM 1190 C CYS A 212 -7.596 -69.443 -82.218 1.00 81.30 A C
+ATOM 1191 O CYS A 212 -8.231 -70.489 -82.243 1.00 67.61 A O
+ATOM 1192 CB CYS A 212 -9.212 -68.546 -80.531 1.00104.49 A C
+ATOM 1193 SG CYS A 212 -8.849 -69.763 -79.278 1.00 98.48 A S
+ATOM 1194 N SER A 213 -6.405 -69.268 -82.788 1.00 87.31 A N
+ATOM 1195 CA SER A 213 -5.631 -70.344 -83.393 1.00 79.74 A C
+ATOM 1196 C SER A 213 -5.396 -71.494 -82.409 1.00 76.03 A C
+ATOM 1197 O SER A 213 -5.256 -71.278 -81.200 1.00 85.80 A O
+ATOM 1198 CB SER A 213 -4.306 -69.753 -83.877 1.00 85.43 A C
+ATOM 1199 OG SER A 213 -3.886 -68.708 -83.000 1.00104.05 A O
+ATOM 1200 N ILE A 214 -5.324 -72.719 -82.945 1.00 74.70 A N
+ATOM 1201 CA ILE A 214 -5.414 -73.964 -82.157 1.00 70.33 A C
+ATOM 1202 C ILE A 214 -4.041 -74.331 -81.591 1.00 67.27 A C
+ATOM 1203 O ILE A 214 -3.076 -74.471 -82.362 1.00 72.07 A O
+ATOM 1204 CB ILE A 214 -5.976 -75.110 -83.014 1.00 75.15 A C
+ATOM 1205 CG1 ILE A 214 -7.510 -75.164 -82.935 1.00 75.79 A C
+ATOM 1206 CG2 ILE A 214 -5.339 -76.461 -82.650 1.00 69.33 A C
+ATOM 1207 CD1 ILE A 214 -8.142 -76.282 -83.771 1.00 78.73 A C
+ATOM 1208 N PRO A 215 -3.924 -74.551 -80.277 1.00 61.77 A N
+ATOM 1209 CA PRO A 215 -2.631 -74.912 -79.683 1.00 62.58 A C
+ATOM 1210 C PRO A 215 -2.256 -76.363 -79.910 1.00 60.46 A C
+ATOM 1211 O PRO A 215 -3.092 -77.269 -79.911 1.00 59.32 A O
+ATOM 1212 CB PRO A 215 -2.847 -74.653 -78.192 1.00 56.60 A C
+ATOM 1213 CG PRO A 215 -4.307 -74.906 -77.989 1.00 63.72 A C
+ATOM 1214 CD PRO A 215 -4.990 -74.463 -79.263 1.00 64.35 A C
+ATOM 1215 N ASN A 216 -0.965 -76.581 -80.069 1.00 64.91 A N
+ATOM 1216 CA ASN A 216 -0.479 -77.936 -80.242 1.00 64.00 A C
+ATOM 1217 C ASN A 216 -0.443 -78.644 -78.898 1.00 61.72 A C
+ATOM 1218 O ASN A 216 -0.301 -78.017 -77.840 1.00 59.64 A O
+ATOM 1219 CB ASN A 216 0.911 -77.932 -80.875 1.00 64.83 A C
+ATOM 1220 CG ASN A 216 1.929 -77.211 -80.020 1.00 72.32 A C
+ATOM 1221 ND2 ASN A 216 2.211 -75.942 -80.356 1.00 72.73 A N
+ATOM 1222 OD1 ASN A 216 2.447 -77.784 -79.052 1.00 64.50 A O
+ATOM 1223 N ILE A 217 -0.554 -79.969 -78.954 1.00 64.80 A N
+ATOM 1224 CA ILE A 217 -0.719 -80.751 -77.735 1.00 66.09 A C
+ATOM 1225 C ILE A 217 0.488 -80.611 -76.809 1.00 63.01 A C
+ATOM 1226 O ILE A 217 0.341 -80.669 -75.573 1.00 71.79 A O
+ATOM 1227 CB ILE A 217 -1.026 -82.216 -78.104 1.00 66.97 A C
+ATOM 1228 CG1 ILE A 217 -1.110 -83.084 -76.855 1.00 65.03 A C
+ATOM 1229 CG2 ILE A 217 -0.039 -82.734 -79.126 1.00 67.05 A C
+ATOM 1230 CD1 ILE A 217 -2.095 -84.204 -76.996 1.00 70.05 A C
+ATOM 1231 N GLU A 218 1.685 -80.380 -77.372 1.00 61.84 A N
+ATOM 1232 CA GLU A 218 2.852 -80.118 -76.533 1.00 61.13 A C
+ATOM 1233 C GLU A 218 2.597 -78.970 -75.567 1.00 59.34 A C
+ATOM 1234 O GLU A 218 3.001 -79.023 -74.394 1.00 65.15 A O
+ATOM 1235 CB GLU A 218 4.068 -79.802 -77.393 1.00 62.54 A C
+ATOM 1236 CG GLU A 218 4.555 -80.959 -78.185 1.00 64.53 A C
+ATOM 1237 CD GLU A 218 3.800 -81.125 -79.467 1.00 67.23 A C
+ATOM 1238 OE1 GLU A 218 2.954 -82.041 -79.521 1.00 78.20 A O
+ATOM 1239 OE2 GLU A 218 4.056 -80.353 -80.418 1.00 72.39 A O1-
+ATOM 1240 N THR A 219 1.919 -77.921 -76.038 1.00 57.64 A N
+ATOM 1241 CA THR A 219 1.597 -76.824 -75.138 1.00 57.42 A C
+ATOM 1242 C THR A 219 0.632 -77.270 -74.054 1.00 65.68 A C
+ATOM 1243 O THR A 219 0.752 -76.840 -72.904 1.00 64.18 A O
+ATOM 1244 CB THR A 219 1.025 -75.653 -75.915 1.00 57.54 A C
+ATOM 1245 CG2 THR A 219 0.474 -74.635 -74.947 1.00 56.66 A C
+ATOM 1246 OG1 THR A 219 2.074 -75.051 -76.683 1.00 56.42 A O
+ATOM 1247 N VAL A 220 -0.313 -78.149 -74.389 1.00 57.95 A N
+ATOM 1248 CA VAL A 220 -1.228 -78.669 -73.375 1.00 59.85 A C
+ATOM 1249 C VAL A 220 -0.448 -79.307 -72.238 1.00 59.99 A C
+ATOM 1250 O VAL A 220 -0.608 -78.936 -71.062 1.00 64.11 A O
+ATOM 1251 CB VAL A 220 -2.209 -79.677 -73.986 1.00 65.52 A C
+ATOM 1252 CG1 VAL A 220 -3.083 -80.231 -72.887 1.00 57.58 A C
+ATOM 1253 CG2 VAL A 220 -3.035 -79.023 -75.059 1.00 58.89 A C
+ATOM 1254 N ILE A 221 0.409 -80.282 -72.580 1.00 61.49 A N
+ATOM 1255 CA ILE A 221 1.235 -80.945 -71.565 1.00 56.02 A C
+ATOM 1256 C ILE A 221 2.012 -79.914 -70.764 1.00 57.45 A C
+ATOM 1257 O ILE A 221 2.033 -79.931 -69.522 1.00 49.97 A O
+ATOM 1258 CB ILE A 221 2.196 -81.953 -72.215 1.00 54.72 A C
+ATOM 1259 CG1 ILE A 221 1.535 -82.644 -73.396 1.00 60.87 A C
+ATOM 1260 CG2 ILE A 221 2.651 -82.973 -71.195 1.00 52.53 A C
+ATOM 1261 CD1 ILE A 221 0.406 -83.568 -73.008 1.00 63.85 A C
+ATOM 1262 N GLU A 222 2.666 -78.998 -71.473 1.00 55.59 A N
+ATOM 1263 CA GLU A 222 3.526 -78.055 -70.789 1.00 54.40 A C
+ATOM 1264 C GLU A 222 2.734 -77.255 -69.769 1.00 49.02 A C
+ATOM 1265 O GLU A 222 3.115 -77.174 -68.599 1.00 50.98 A O
+ATOM 1266 CB GLU A 222 4.219 -77.156 -71.803 1.00 52.93 A C
+ATOM 1267 CG GLU A 222 5.407 -76.443 -71.215 1.00 60.85 A C
+ATOM 1268 CD GLU A 222 6.283 -75.876 -72.288 1.00 71.78 A C
+ATOM 1269 OE1 GLU A 222 5.787 -75.739 -73.437 1.00 72.89 A O
+ATOM 1270 OE2 GLU A 222 7.464 -75.590 -71.984 1.00 84.71 A O1-
+ATOM 1271 N PHE A 223 1.597 -76.711 -70.170 1.00 45.99 A N
+ATOM 1272 CA PHE A 223 0.859 -75.896 -69.226 1.00 48.36 A C
+ATOM 1273 C PHE A 223 0.430 -76.715 -68.016 1.00 48.57 A C
+ATOM 1274 O PHE A 223 0.510 -76.231 -66.882 1.00 47.23 A O
+ATOM 1275 CB PHE A 223 -0.345 -75.267 -69.891 1.00 43.70 A C
+ATOM 1276 CG PHE A 223 -1.001 -74.241 -69.055 1.00 47.85 A C
+ATOM 1277 CD1 PHE A 223 -0.301 -73.135 -68.645 1.00 44.61 A C
+ATOM 1278 CD2 PHE A 223 -2.305 -74.387 -68.653 1.00 51.77 A C
+ATOM 1279 CE1 PHE A 223 -0.897 -72.162 -67.850 1.00 39.41 A C
+ATOM 1280 CE2 PHE A 223 -2.915 -73.431 -67.862 1.00 53.99 A C
+ATOM 1281 CZ PHE A 223 -2.207 -72.311 -67.460 1.00 46.89 A C
+ATOM 1282 N GLN A 224 -0.001 -77.968 -68.230 1.00 47.57 A N
+ATOM 1283 CA GLN A 224 -0.393 -78.816 -67.097 1.00 46.90 A C
+ATOM 1284 C GLN A 224 0.757 -78.980 -66.112 1.00 48.09 A C
+ATOM 1285 O GLN A 224 0.579 -78.847 -64.887 1.00 51.70 A O
+ATOM 1286 CB GLN A 224 -0.857 -80.180 -67.600 1.00 48.04 A C
+ATOM 1287 CG GLN A 224 -2.138 -80.133 -68.446 1.00 78.76 A C
+ATOM 1288 CD GLN A 224 -2.634 -81.520 -68.864 1.00 86.35 A C
+ATOM 1289 NE2 GLN A 224 -3.952 -81.647 -69.039 1.00 86.61 A N
+ATOM 1290 OE1 GLN A 224 -1.845 -82.466 -69.014 1.00 86.72 A O
+ATOM 1291 N GLN A 225 1.951 -79.265 -66.649 1.00 50.70 A N
+ATOM 1292 CA GLN A 225 3.191 -79.292 -65.876 1.00 48.87 A C
+ATOM 1293 C GLN A 225 3.365 -78.021 -65.053 1.00 43.31 A C
+ATOM 1294 O GLN A 225 3.517 -78.073 -63.827 1.00 60.24 A O
+ATOM 1295 CB GLN A 225 4.386 -79.444 -66.828 1.00 58.21 A C
+ATOM 1296 CG GLN A 225 4.526 -80.793 -67.437 1.00 69.50 A C
+ATOM 1297 CD GLN A 225 4.448 -81.839 -66.383 1.00 82.80 A C
+ATOM 1298 NE2 GLN A 225 3.332 -82.557 -66.341 1.00 82.93 A N
+ATOM 1299 OE1 GLN A 225 5.363 -81.980 -65.573 1.00 90.75 A O
+ATOM 1300 N LYS A 226 3.353 -76.861 -65.722 1.00 37.79 A N
+ATOM 1301 CA LYS A 226 3.692 -75.616 -65.041 1.00 38.98 A C
+ATOM 1302 C LYS A 226 2.663 -75.284 -63.970 1.00 52.58 A C
+ATOM 1303 O LYS A 226 3.009 -75.079 -62.803 1.00 60.15 A O
+ATOM 1304 CB LYS A 226 3.812 -74.478 -66.055 1.00 35.30 A C
+ATOM 1305 CG LYS A 226 5.102 -74.451 -66.850 1.00 68.09 A C
+ATOM 1306 CD LYS A 226 5.976 -73.293 -66.416 1.00 69.06 A C
+ATOM 1307 CE LYS A 226 7.161 -73.129 -67.353 1.00 76.01 A C
+ATOM 1308 NZ LYS A 226 8.128 -72.097 -66.887 1.00 81.45 A N1+
+ATOM 1309 N ASN A 227 1.389 -75.279 -64.351 1.00 52.91 A N
+ATOM 1310 CA ASN A 227 0.296 -74.885 -63.471 1.00 51.97 A C
+ATOM 1311 C ASN A 227 0.160 -75.807 -62.260 1.00 57.03 A C
+ATOM 1312 O ASN A 227 -0.333 -75.387 -61.187 1.00 54.19 A O
+ATOM 1313 CB ASN A 227 -0.985 -74.875 -64.309 1.00 43.91 A C
+ATOM 1314 CG ASN A 227 -2.175 -74.319 -63.571 1.00 46.22 A C
+ATOM 1315 ND2 ASN A 227 -3.226 -75.139 -63.419 1.00 41.95 A N
+ATOM 1316 OD1 ASN A 227 -2.175 -73.143 -63.182 1.00 48.78 A O
+ATOM 1317 N ASN A 228 0.586 -77.070 -62.416 1.00 53.69 A N
+ATOM 1318 CA ASN A 228 0.435 -78.039 -61.333 1.00 46.65 A C
+ATOM 1319 C ASN A 228 0.994 -77.515 -60.012 1.00 50.62 A C
+ATOM 1320 O ASN A 228 0.342 -77.622 -58.969 1.00 55.88 A O
+ATOM 1321 CB ASN A 228 1.095 -79.363 -61.703 1.00 53.43 A C
+ATOM 1322 CG ASN A 228 1.031 -80.350 -60.580 1.00 61.29 A C
+ATOM 1323 ND2 ASN A 228 -0.157 -80.899 -60.320 1.00 60.40 A N
+ATOM 1324 OD1 ASN A 228 2.031 -80.592 -59.925 1.00 64.10 A O
+ATOM 1325 N ARG A 229 2.192 -76.925 -60.042 1.00 49.47 A N
+ATOM 1326 CA ARG A 229 2.789 -76.410 -58.809 1.00 51.20 A C
+ATOM 1327 C ARG A 229 1.815 -75.514 -58.050 1.00 53.93 A C
+ATOM 1328 O ARG A 229 1.555 -75.721 -56.860 1.00 58.93 A O
+ATOM 1329 CB ARG A 229 4.087 -75.657 -59.128 1.00 45.92 A C
+ATOM 1330 CG ARG A 229 4.716 -74.988 -57.925 1.00 50.33 A C
+ATOM 1331 CD ARG A 229 6.227 -74.867 -58.062 1.00 52.45 A C
+ATOM 1332 NE ARG A 229 6.799 -74.608 -56.742 1.00 51.13 A N
+ATOM 1333 CZ ARG A 229 8.004 -74.087 -56.500 1.00 53.22 A C
+ATOM 1334 NH1 ARG A 229 8.828 -73.770 -57.495 1.00 51.22 A N1+
+ATOM 1335 NH2 ARG A 229 8.392 -73.890 -55.243 1.00 59.13 A N
+ATOM 1336 N LEU A 230 1.237 -74.531 -58.744 1.00 54.22 A N
+ATOM 1337 CA LEU A 230 0.240 -73.661 -58.121 1.00 45.71 A C
+ATOM 1338 C LEU A 230 -0.907 -74.472 -57.541 1.00 49.45 A C
+ATOM 1339 O LEU A 230 -1.368 -74.220 -56.414 1.00 50.15 A O
+ATOM 1340 CB LEU A 230 -0.294 -72.659 -59.145 1.00 44.61 A C
+ATOM 1341 CG LEU A 230 -1.235 -71.600 -58.607 1.00 52.80 A C
+ATOM 1342 CD1 LEU A 230 -0.558 -70.944 -57.452 1.00 46.03 A C
+ATOM 1343 CD2 LEU A 230 -1.484 -70.614 -59.710 1.00 54.44 A C
+ATOM 1344 N LEU A 231 -1.380 -75.461 -58.296 1.00 48.46 A N
+ATOM 1345 CA LEU A 231 -2.463 -76.262 -57.755 1.00 49.25 A C
+ATOM 1346 C LEU A 231 -2.044 -76.919 -56.440 1.00 57.79 A C
+ATOM 1347 O LEU A 231 -2.771 -76.825 -55.449 1.00 61.77 A O
+ATOM 1348 CB LEU A 231 -2.922 -77.267 -58.804 1.00 46.33 A C
+ATOM 1349 CG LEU A 231 -3.302 -76.539 -60.109 1.00 59.58 A C
+ATOM 1350 CD1 LEU A 231 -3.509 -77.525 -61.234 1.00 51.66 A C
+ATOM 1351 CD2 LEU A 231 -4.539 -75.658 -59.939 1.00 50.86 A C
+ATOM 1352 N GLU A 232 -0.839 -77.499 -56.373 1.00 56.85 A N
+ATOM 1353 CA GLU A 232 -0.475 -78.219 -55.153 1.00 56.85 A C
+ATOM 1354 C GLU A 232 -0.236 -77.272 -53.994 1.00 56.85 A C
+ATOM 1355 O GLU A 232 -0.612 -77.580 -52.852 1.00 56.85 A O
+ATOM 1356 CB GLU A 232 0.763 -79.090 -55.342 1.00 56.85 A C
+ATOM 1357 CG GLU A 232 0.667 -80.109 -56.434 1.00 56.85 A C
+ATOM 1358 CD GLU A 232 -0.465 -81.132 -56.242 1.00 56.85 A C
+ATOM 1359 OE1 GLU A 232 -1.063 -81.207 -55.133 1.00 56.85 A O
+ATOM 1360 OE2 GLU A 232 -0.747 -81.877 -57.229 1.00 56.85 A O1-
+ATOM 1361 N ILE A 233 0.411 -76.133 -54.254 1.00 51.64 A N
+ATOM 1362 CA ILE A 233 0.619 -75.166 -53.185 1.00 46.86 A C
+ATOM 1363 C ILE A 233 -0.721 -74.729 -52.621 1.00 48.91 A C
+ATOM 1364 O ILE A 233 -0.896 -74.587 -51.397 1.00 51.43 A O
+ATOM 1365 CB ILE A 233 1.416 -73.962 -53.692 1.00 47.76 A C
+ATOM 1366 CG1 ILE A 233 2.662 -74.418 -54.423 1.00 51.96 A C
+ATOM 1367 CG2 ILE A 233 1.788 -73.089 -52.526 1.00 43.38 A C
+ATOM 1368 CD1 ILE A 233 3.598 -73.280 -54.796 1.00 46.09 A C
+ATOM 1369 N THR A 234 -1.701 -74.545 -53.507 1.00 46.51 A N
+ATOM 1370 CA THR A 234 -3.034 -74.176 -53.066 1.00 54.35 A C
+ATOM 1371 C THR A 234 -3.667 -75.282 -52.228 1.00 54.36 A C
+ATOM 1372 O THR A 234 -4.171 -75.018 -51.135 1.00 59.01 A O
+ATOM 1373 CB THR A 234 -3.886 -73.860 -54.275 1.00 48.68 A C
+ATOM 1374 CG2 THR A 234 -5.233 -73.386 -53.822 1.00 40.88 A C
+ATOM 1375 OG1 THR A 234 -3.261 -72.809 -55.009 1.00 45.97 A O
+ATOM 1376 N ARG A 235 -3.640 -76.531 -52.713 1.00 46.49 A N
+ATOM 1377 CA ARG A 235 -4.227 -77.643 -51.959 1.00 44.99 A C
+ATOM 1378 C ARG A 235 -3.674 -77.690 -50.528 1.00 45.66 A C
+ATOM 1379 O ARG A 235 -4.435 -77.759 -49.543 1.00 59.70 A O
+ATOM 1380 CB ARG A 235 -3.986 -78.971 -52.704 1.00 53.04 A C
+ATOM 1381 CG ARG A 235 -4.637 -79.020 -54.106 1.00 56.15 A C
+ATOM 1382 CD ARG A 235 -4.089 -80.121 -55.045 1.00 64.34 A C
+ATOM 1383 NE ARG A 235 -4.565 -79.960 -56.437 1.00 73.96 A N
+ATOM 1384 CZ ARG A 235 -4.378 -80.850 -57.429 1.00 82.91 A C
+ATOM 1385 NH1 ARG A 235 -3.739 -81.987 -57.185 1.00 78.78 A N1+
+ATOM 1386 NH2 ARG A 235 -4.835 -80.626 -58.670 1.00 87.59 A N
+ATOM 1387 N GLU A 236 -2.344 -77.604 -50.401 1.00 45.23 A N
+ATOM 1388 CA GLU A 236 -1.720 -77.652 -49.084 1.00 48.81 A C
+ATOM 1389 C GLU A 236 -2.203 -76.516 -48.210 1.00 50.19 A C
+ATOM 1390 O GLU A 236 -2.639 -76.731 -47.076 1.00 48.41 A O
+ATOM 1391 CB GLU A 236 -0.210 -77.568 -49.203 1.00 42.54 A C
+ATOM 1392 CG GLU A 236 0.415 -78.623 -50.046 1.00 49.02 A C
+ATOM 1393 CD GLU A 236 1.909 -78.608 -49.886 1.00 59.32 A C
+ATOM 1394 OE1 GLU A 236 2.361 -78.065 -48.832 1.00 63.55 A O
+ATOM 1395 OE2 GLU A 236 2.611 -79.106 -50.815 1.00 77.27 A O1-
+ATOM 1396 N PHE A 237 -2.095 -75.289 -48.705 1.00 41.57 A N
+ATOM 1397 CA PHE A 237 -2.603 -74.182 -47.915 1.00 54.24 A C
+ATOM 1398 C PHE A 237 -4.082 -74.366 -47.552 1.00 53.04 A C
+ATOM 1399 O PHE A 237 -4.508 -73.882 -46.500 1.00 57.88 A O
+ATOM 1400 CB PHE A 237 -2.384 -72.884 -48.668 1.00 52.28 A C
+ATOM 1401 CG PHE A 237 -1.019 -72.307 -48.500 1.00 46.09 A C
+ATOM 1402 CD1 PHE A 237 -0.329 -72.461 -47.328 1.00 46.78 A C
+ATOM 1403 CD2 PHE A 237 -0.418 -71.613 -49.540 1.00 48.60 A C
+ATOM 1404 CE1 PHE A 237 0.939 -71.904 -47.184 1.00 43.05 A C
+ATOM 1405 CE2 PHE A 237 0.838 -71.073 -49.413 1.00 45.70 A C
+ATOM 1406 CZ PHE A 237 1.521 -71.217 -48.240 1.00 40.54 A C
+ATOM 1407 N SER A 238 -4.876 -75.080 -48.380 1.00 46.83 A N
+ATOM 1408 CA SER A 238 -6.305 -75.260 -48.076 1.00 54.06 A C
+ATOM 1409 C SER A 238 -6.526 -76.193 -46.897 1.00 51.43 A C
+ATOM 1410 O SER A 238 -7.239 -75.844 -45.946 1.00 58.02 A O
+ATOM 1411 CB SER A 238 -7.109 -75.777 -49.279 1.00 52.93 A C
+ATOM 1412 OG SER A 238 -7.635 -74.732 -50.091 1.00 88.79 A O
+ATOM 1413 N VAL A 239 -5.963 -77.397 -46.938 1.00 45.76 A N
+ATOM 1414 CA VAL A 239 -6.320 -78.238 -45.809 1.00 51.20 A C
+ATOM 1415 C VAL A 239 -5.650 -77.747 -44.528 1.00 47.32 A C
+ATOM 1416 O VAL A 239 -6.126 -78.046 -43.424 1.00 57.92 A O
+ATOM 1417 CB VAL A 239 -5.955 -79.703 -46.048 1.00 55.02 A C
+ATOM 1418 CG1 VAL A 239 -6.355 -80.120 -47.460 1.00 58.03 A C
+ATOM 1419 CG2 VAL A 239 -4.505 -79.957 -45.718 1.00 45.79 A C
+ATOM 1420 N ASN A 240 -4.553 -76.999 -44.641 1.00 49.70 A N
+ATOM 1421 CA ASN A 240 -3.737 -76.610 -43.498 1.00 49.45 A C
+ATOM 1422 C ASN A 240 -4.055 -75.224 -42.956 1.00 53.81 A C
+ATOM 1423 O ASN A 240 -3.276 -74.700 -42.154 1.00 57.22 A O
+ATOM 1424 CB ASN A 240 -2.264 -76.723 -43.852 1.00 51.26 A C
+ATOM 1425 CG ASN A 240 -1.737 -78.111 -43.596 1.00 57.58 A C
+ATOM 1426 ND2 ASN A 240 -1.402 -78.373 -42.333 1.00 52.16 A N
+ATOM 1427 OD1 ASN A 240 -1.665 -78.957 -44.497 1.00 53.70 A O
+ATOM 1428 N ALA A 241 -5.134 -74.595 -43.429 1.00 53.29 A N
+ATOM 1429 CA ALA A 241 -5.559 -73.288 -42.934 1.00 55.28 A C
+ATOM 1430 C ALA A 241 -4.423 -72.279 -43.032 1.00 53.04 A C
+ATOM 1431 O ALA A 241 -4.220 -71.450 -42.145 1.00 60.20 A O
+ATOM 1432 CB ALA A 241 -6.086 -73.380 -41.503 1.00 46.86 A C
+ATOM 1433 N GLY A 242 -3.648 -72.386 -44.102 1.00 48.00 A N
+ATOM 1434 CA GLY A 242 -2.692 -71.370 -44.463 1.00 47.26 A C
+ATOM 1435 C GLY A 242 -1.339 -71.447 -43.792 1.00 50.60 A C
+ATOM 1436 O GLY A 242 -0.535 -70.520 -43.967 1.00 54.53 A O
+ATOM 1437 N VAL A 243 -1.055 -72.496 -43.016 1.00 53.63 A N
+ATOM 1438 CA VAL A 243 0.265 -72.656 -42.404 1.00 52.31 A C
+ATOM 1439 C VAL A 243 0.701 -74.107 -42.531 1.00 48.48 A C
+ATOM 1440 O VAL A 243 0.046 -74.998 -41.981 1.00 57.33 A O
+ATOM 1441 CB VAL A 243 0.267 -72.250 -40.923 1.00 51.16 A C
+ATOM 1442 CG1 VAL A 243 1.683 -72.201 -40.413 1.00 52.15 A C
+ATOM 1443 CG2 VAL A 243 -0.446 -70.921 -40.717 1.00 54.55 A C
+ATOM 1444 N THR A 244 1.816 -74.358 -43.203 1.00 49.32 A N
+ATOM 1445 CA THR A 244 2.272 -75.732 -43.341 1.00 49.02 A C
+ATOM 1446 C THR A 244 3.616 -75.929 -42.666 1.00 55.80 A C
+ATOM 1447 O THR A 244 4.494 -75.066 -42.740 1.00 51.30 A O
+ATOM 1448 CB THR A 244 2.369 -76.173 -44.808 1.00 48.77 A C
+ATOM 1449 CG2 THR A 244 1.148 -75.727 -45.559 1.00 40.24 A C
+ATOM 1450 OG1 THR A 244 3.545 -75.636 -45.430 1.00 49.81 A O
+ATOM 1451 N THR A 245 3.750 -77.048 -41.968 1.00 56.17 A N
+ATOM 1452 CA THR A 245 5.063 -77.586 -41.639 1.00 52.18 A C
+ATOM 1453 C THR A 245 5.058 -79.101 -41.547 1.00 53.20 A C
+ATOM 1454 O THR A 245 4.086 -79.680 -41.097 1.00 51.19 A O
+ATOM 1455 CB THR A 245 5.585 -77.029 -40.332 1.00 50.63 A C
+ATOM 1456 CG2 THR A 245 4.499 -77.058 -39.297 1.00 43.13 A C
+ATOM 1457 OG1 THR A 245 6.721 -77.804 -39.914 1.00 55.14 A O
+ATOM 1458 N PRO A 246 6.143 -79.755 -41.971 1.00 51.61 A N
+ATOM 1459 CA PRO A 246 7.322 -79.226 -42.659 1.00 52.34 A C
+ATOM 1460 C PRO A 246 7.002 -78.573 -44.014 1.00 53.80 A C
+ATOM 1461 O PRO A 246 5.847 -78.583 -44.470 1.00 54.42 A O
+ATOM 1462 CB PRO A 246 8.184 -80.477 -42.859 1.00 53.35 A C
+ATOM 1463 CG PRO A 246 7.238 -81.630 -42.705 1.00 52.58 A C
+ATOM 1464 CD PRO A 246 6.303 -81.178 -41.656 1.00 45.81 A C
+ATOM 1465 N VAL A 247 8.001 -78.000 -44.662 1.00 54.58 A N
+ATOM 1466 CA VAL A 247 7.762 -77.237 -45.878 1.00 47.21 A C
+ATOM 1467 C VAL A 247 8.088 -78.131 -47.055 1.00 52.87 A C
+ATOM 1468 O VAL A 247 9.111 -78.823 -47.060 1.00 52.15 A O
+ATOM 1469 CB VAL A 247 8.594 -75.944 -45.889 1.00 52.33 A C
+ATOM 1470 CG1 VAL A 247 7.965 -74.898 -44.981 1.00 48.23 A C
+ATOM 1471 CG2 VAL A 247 9.953 -76.252 -45.364 1.00 49.09 A C
+ATOM 1472 N SER A 248 7.236 -78.085 -48.071 1.00 49.89 A N
+ATOM 1473 CA SER A 248 7.194 -79.074 -49.137 1.00 64.63 A C
+ATOM 1474 C SER A 248 8.377 -78.943 -50.087 1.00 57.77 A C
+ATOM 1475 O SER A 248 9.150 -77.987 -50.058 1.00 56.78 A O
+ATOM 1476 CB SER A 248 5.939 -78.862 -49.965 1.00 54.52 A C
+ATOM 1477 OG SER A 248 4.842 -78.757 -49.087 1.00 59.14 A O
+ATOM 1478 N THR A 249 8.459 -79.900 -50.998 1.00 56.45 A N
+ATOM 1479 CA THR A 249 9.234 -79.676 -52.200 1.00 60.68 A C
+ATOM 1480 C THR A 249 8.479 -78.746 -53.130 1.00 57.08 A C
+ATOM 1481 O THR A 249 9.107 -78.073 -53.951 1.00 66.12 A O
+ATOM 1482 CB THR A 249 9.547 -81.014 -52.894 1.00 57.52 A C
+ATOM 1483 CG2 THR A 249 10.492 -80.851 -54.108 1.00 55.23 A C
+ATOM 1484 OG1 THR A 249 10.164 -81.911 -51.962 1.00 57.17 A O
+ATOM 1485 N TYR A 250 7.141 -78.683 -53.004 1.00 58.91 A N
+ATOM 1486 CA TYR A 250 6.345 -77.744 -53.794 1.00 58.34 A C
+ATOM 1487 C TYR A 250 6.476 -76.315 -53.288 1.00 62.18 A C
+ATOM 1488 O TYR A 250 6.395 -75.366 -54.076 1.00 65.11 A O
+ATOM 1489 CB TYR A 250 4.865 -78.106 -53.771 1.00 51.48 A C
+ATOM 1490 CG TYR A 250 4.492 -79.456 -54.322 1.00 53.73 A C
+ATOM 1491 CD1 TYR A 250 4.933 -79.883 -55.577 1.00 49.38 A C
+ATOM 1492 CD2 TYR A 250 3.617 -80.273 -53.620 1.00 53.87 A C
+ATOM 1493 CE1 TYR A 250 4.555 -81.122 -56.088 1.00 46.28 A C
+ATOM 1494 CE2 TYR A 250 3.229 -81.508 -54.127 1.00 53.13 A C
+ATOM 1495 CZ TYR A 250 3.702 -81.933 -55.355 1.00 59.37 A C
+ATOM 1496 OH TYR A 250 3.301 -83.175 -55.811 1.00 71.15 A O
+ATOM 1497 N MET A 251 6.634 -76.144 -51.978 1.00 56.68 A N
+ATOM 1498 CA MET A 251 6.810 -74.809 -51.440 1.00 54.54 A C
+ATOM 1499 C MET A 251 8.219 -74.291 -51.691 1.00 53.29 A C
+ATOM 1500 O MET A 251 8.378 -73.136 -52.083 1.00 51.04 A O
+ATOM 1501 CB MET A 251 6.484 -74.815 -49.958 1.00 45.34 A C
+ATOM 1502 CG MET A 251 5.087 -75.293 -49.618 1.00 59.15 A C
+ATOM 1503 SD MET A 251 3.944 -73.900 -49.617 1.00 41.36 A S
+ATOM 1504 CE MET A 251 2.526 -74.566 -48.721 1.00 52.33 A C
+ATOM 1505 N LEU A 252 9.251 -75.112 -51.453 1.00 50.54 A N
+ATOM 1506 CA LEU A 252 10.627 -74.803 -51.856 1.00 42.99 A C
+ATOM 1507 C LEU A 252 11.313 -76.060 -52.380 1.00 45.86 A C
+ATOM 1508 O LEU A 252 11.352 -77.091 -51.698 1.00 47.52 A O
+ATOM 1509 CB LEU A 252 11.451 -74.231 -50.706 1.00 41.63 A C
+ATOM 1510 CG LEU A 252 11.109 -72.812 -50.300 1.00 52.49 A C
+ATOM 1511 CD1 LEU A 252 12.076 -72.322 -49.259 1.00 55.47 A C
+ATOM 1512 CD2 LEU A 252 11.163 -71.953 -51.501 1.00 51.85 A C
+ATOM 1513 N THR A 253 11.848 -75.971 -53.592 1.00 55.09 A N
+ATOM 1514 CA THR A 253 12.663 -77.036 -54.149 1.00 56.99 A C
+ATOM 1515 C THR A 253 13.987 -77.087 -53.423 1.00 56.39 A C
+ATOM 1516 O THR A 253 14.482 -76.060 -52.952 1.00 54.34 A O
+ATOM 1517 CB THR A 253 12.915 -76.762 -55.621 1.00 56.94 A C
+ATOM 1518 CG2 THR A 253 13.646 -77.959 -56.341 1.00 47.22 A C
+ATOM 1519 OG1 THR A 253 11.662 -76.429 -56.238 1.00 53.73 A O
+ATOM 1520 N ASN A 254 14.579 -78.287 -53.349 1.00 45.69 A N
+ATOM 1521 CA ASN A 254 15.906 -78.381 -52.739 1.00 53.09 A C
+ATOM 1522 C ASN A 254 16.844 -77.336 -53.320 1.00 52.44 A C
+ATOM 1523 O ASN A 254 17.563 -76.656 -52.575 1.00 67.08 A O
+ATOM 1524 CB ASN A 254 16.497 -79.781 -52.898 1.00 52.67 A C
+ATOM 1525 CG ASN A 254 16.285 -80.639 -51.658 1.00 68.29 A C
+ATOM 1526 ND2 ASN A 254 17.359 -81.268 -51.209 1.00 69.84 A N
+ATOM 1527 OD1 ASN A 254 15.181 -80.715 -51.094 1.00 80.05 A O
+ATOM 1528 N SER A 255 16.783 -77.132 -54.644 1.00 50.64 A N
+ATOM 1529 CA SER A 255 17.670 -76.166 -55.286 1.00 53.31 A C
+ATOM 1530 C SER A 255 17.382 -74.751 -54.805 1.00 52.49 A C
+ATOM 1531 O SER A 255 18.307 -73.956 -54.566 1.00 68.20 A O
+ATOM 1532 CB SER A 255 17.532 -76.264 -56.801 1.00 53.34 A C
+ATOM 1533 OG SER A 255 18.673 -75.691 -57.420 1.00 65.86 A O
+ATOM 1534 N GLU A 256 16.101 -74.434 -54.641 1.00 48.80 A N
+ATOM 1535 CA GLU A 256 15.722 -73.112 -54.168 1.00 46.97 A C
+ATOM 1536 C GLU A 256 16.119 -72.927 -52.710 1.00 50.56 A C
+ATOM 1537 O GLU A 256 16.646 -71.876 -52.330 1.00 57.47 A O
+ATOM 1538 CB GLU A 256 14.214 -72.906 -54.368 1.00 42.52 A C
+ATOM 1539 CG GLU A 256 13.739 -73.030 -55.825 1.00 52.32 A C
+ATOM 1540 CD GLU A 256 12.244 -73.359 -55.953 1.00 61.81 A C
+ATOM 1541 OE1 GLU A 256 11.645 -73.783 -54.948 1.00 60.74 A O
+ATOM 1542 OE2 GLU A 256 11.664 -73.205 -57.053 1.00 54.63 A O1-
+ATOM 1543 N LEU A 257 15.891 -73.940 -51.880 1.00 49.08 A N
+ATOM 1544 CA LEU A 257 16.125 -73.744 -50.455 1.00 44.72 A C
+ATOM 1545 C LEU A 257 17.601 -73.647 -50.170 1.00 55.08 A C
+ATOM 1546 O LEU A 257 18.030 -72.816 -49.362 1.00 54.46 A O
+ATOM 1547 CB LEU A 257 15.499 -74.863 -49.626 1.00 33.69 A C
+ATOM 1548 CG LEU A 257 15.731 -74.520 -48.165 1.00 50.71 A C
+ATOM 1549 CD1 LEU A 257 14.440 -74.505 -47.439 1.00 41.92 A C
+ATOM 1550 CD2 LEU A 257 16.634 -75.544 -47.572 1.00 39.46 A C
+ATOM 1551 N LEU A 258 18.398 -74.479 -50.832 1.00 52.14 A N
+ATOM 1552 CA LEU A 258 19.836 -74.309 -50.726 1.00 52.02 A C
+ATOM 1553 C LEU A 258 20.250 -72.934 -51.236 1.00 49.06 A C
+ATOM 1554 O LEU A 258 20.997 -72.220 -50.554 1.00 62.60 A O
+ATOM 1555 CB LEU A 258 20.557 -75.430 -51.469 1.00 43.99 A C
+ATOM 1556 CG LEU A 258 20.208 -76.806 -50.907 1.00 52.63 A C
+ATOM 1557 CD1 LEU A 258 20.819 -77.955 -51.700 1.00 45.94 A C
+ATOM 1558 CD2 LEU A 258 20.663 -76.832 -49.482 1.00 43.54 A C
+ATOM 1559 N SER A 259 19.724 -72.510 -52.414 1.00 52.57 A N
+ATOM 1560 CA SER A 259 20.171 -71.218 -52.957 1.00 59.70 A C
+ATOM 1561 C SER A 259 19.617 -70.029 -52.159 1.00 53.46 A C
+ATOM 1562 O SER A 259 19.946 -68.871 -52.451 1.00 70.05 A O
+ATOM 1563 CB SER A 259 19.833 -71.079 -54.449 1.00 67.72 A C
+ATOM 1564 OG SER A 259 20.766 -70.192 -55.094 1.00 81.81 A O
+ATOM 1565 N LEU A 260 18.778 -70.300 -51.167 1.00 56.53 A N
+ATOM 1566 CA LEU A 260 18.354 -69.322 -50.179 1.00 54.78 A C
+ATOM 1567 C LEU A 260 19.330 -69.303 -49.014 1.00 62.69 A C
+ATOM 1568 O LEU A 260 19.887 -68.256 -48.666 1.00 62.51 A O
+ATOM 1569 CB LEU A 260 16.943 -69.658 -49.700 1.00 46.78 A C
+ATOM 1570 CG LEU A 260 16.050 -68.454 -49.435 1.00 51.30 A C
+ATOM 1571 CD1 LEU A 260 16.225 -67.399 -50.518 1.00 46.43 A C
+ATOM 1572 CD2 LEU A 260 14.627 -68.935 -49.412 1.00 50.28 A C
+ATOM 1573 N ILE A 261 19.500 -70.464 -48.388 1.00 55.56 A N
+ATOM 1574 CA ILE A 261 20.428 -70.702 -47.297 1.00 51.84 A C
+ATOM 1575 C ILE A 261 21.729 -70.018 -47.635 1.00 57.79 A C
+ATOM 1576 O ILE A 261 22.248 -69.236 -46.841 1.00 57.88 A O
+ATOM 1577 CB ILE A 261 20.678 -72.205 -47.148 1.00 46.59 A C
+ATOM 1578 CG1 ILE A 261 19.512 -72.909 -46.490 1.00 46.63 A C
+ATOM 1579 CG2 ILE A 261 21.976 -72.460 -46.449 1.00 42.83 A C
+ATOM 1580 CD1 ILE A 261 19.769 -74.369 -46.378 1.00 47.20 A C
+ATOM 1581 N ASN A 262 22.233 -70.282 -48.844 1.00 57.54 A N
+ATOM 1582 CA ASN A 262 23.542 -69.783 -49.242 1.00 56.35 A C
+ATOM 1583 C ASN A 262 23.648 -68.280 -49.081 1.00 63.25 A C
+ATOM 1584 O ASN A 262 24.730 -67.756 -48.792 1.00 62.53 A O
+ATOM 1585 CB ASN A 262 23.833 -70.138 -50.685 1.00 55.06 A C
+ATOM 1586 CG ASN A 262 25.299 -70.111 -50.986 1.00 70.86 A C
+ATOM 1587 ND2 ASN A 262 25.672 -70.712 -52.096 1.00 69.44 A N
+ATOM 1588 OD1 ASN A 262 26.095 -69.558 -50.233 1.00 80.05 A O
+ATOM 1589 N ASP A 263 22.567 -67.559 -49.335 1.00 50.66 A N
+ATOM 1590 CA ASP A 263 22.625 -66.111 -49.281 1.00 59.03 A C
+ATOM 1591 C ASP A 263 22.028 -65.521 -48.008 1.00 59.74 A C
+ATOM 1592 O ASP A 263 22.001 -64.298 -47.856 1.00 68.83 A O
+ATOM 1593 CB ASP A 263 21.974 -65.543 -50.536 1.00 63.06 A C
+ATOM 1594 CG ASP A 263 22.876 -65.695 -51.774 1.00 74.82 A C
+ATOM 1595 OD1 ASP A 263 24.107 -65.895 -51.594 1.00 78.90 A O
+ATOM 1596 OD2 ASP A 263 22.355 -65.613 -52.909 1.00 78.50 A O1-
+ATOM 1597 N MET A 264 21.548 -66.346 -47.090 1.00 62.09 A N
+ATOM 1598 CA MET A 264 21.194 -65.812 -45.798 1.00 60.87 A C
+ATOM 1599 C MET A 264 22.423 -65.205 -45.138 1.00 77.50 A C
+ATOM 1600 O MET A 264 23.562 -65.521 -45.501 1.00 85.95 A O
+ATOM 1601 CB MET A 264 20.623 -66.894 -44.891 1.00 44.22 A C
+ATOM 1602 CG MET A 264 19.325 -67.544 -45.332 1.00 40.80 A C
+ATOM 1603 SD MET A 264 18.887 -68.948 -44.244 1.00 51.75 A S
+ATOM 1604 CE MET A 264 17.902 -68.196 -42.962 1.00 40.42 A C
+ATOM 1605 N PRO A 265 22.199 -64.278 -44.139 1.00 52.90 A N
+ATOM 1606 CA PRO A 265 23.277 -63.734 -43.278 1.00 67.95 A C
+ATOM 1607 C PRO A 265 23.680 -64.648 -42.125 1.00 69.96 A C
+ATOM 1608 O PRO A 265 23.237 -64.491 -40.995 1.00104.67 A O
+ATOM 1609 CB PRO A 265 22.656 -62.439 -42.748 1.00 75.96 A C
+ATOM 1610 CG PRO A 265 21.192 -62.687 -42.776 1.00 70.17 A C
+ATOM 1611 CD PRO A 265 20.938 -63.530 -43.978 1.00 61.31 A C
+ATOM 1612 N ILE A 266 24.541 -65.631 -42.398 1.00 91.92 A N
+ATOM 1613 CA ILE A 266 24.989 -66.531 -41.337 1.00 65.95 A C
+ATOM 1614 C ILE A 266 26.410 -67.023 -41.582 1.00 62.66 A C
+ATOM 1615 O ILE A 266 27.001 -66.724 -42.628 1.00 62.03 A O
+ATOM 1616 CB ILE A 266 24.033 -67.727 -41.187 1.00 61.73 A C
+ATOM 1617 CG1 ILE A 266 23.606 -68.263 -42.549 1.00 52.37 A C
+ATOM 1618 CG2 ILE A 266 22.824 -67.367 -40.368 1.00 68.52 A C
+ATOM 1619 CD1 ILE A 266 22.849 -69.548 -42.429 1.00 51.18 A C
+ATOM 1620 N THR A 267 26.945 -67.803 -40.624 1.00 53.31 A N
+ATOM 1621 CA THR A 267 28.268 -68.425 -40.724 1.00 58.17 A C
+ATOM 1622 C THR A 267 28.336 -69.381 -41.897 1.00 58.71 A C
+ATOM 1623 O THR A 267 27.358 -70.052 -42.230 1.00 67.21 A O
+ATOM 1624 CB THR A 267 28.581 -69.284 -39.504 1.00 70.96 A C
+ATOM 1625 CG2 THR A 267 30.085 -69.512 -39.406 1.00 79.34 A C
+ATOM 1626 OG1 THR A 267 28.102 -68.667 -38.307 1.00 68.93 A O
+ATOM 1627 N ASN A 268 29.525 -69.534 -42.470 1.00 58.76 A N
+ATOM 1628 CA ASN A 268 29.673 -70.651 -43.394 1.00 60.07 A C
+ATOM 1629 C ASN A 268 29.509 -71.982 -42.684 1.00 62.03 A C
+ATOM 1630 O ASN A 268 29.124 -72.977 -43.309 1.00 61.17 A O
+ATOM 1631 CB ASN A 268 31.003 -70.584 -44.129 1.00 52.58 A C
+ATOM 1632 CG ASN A 268 31.009 -69.499 -45.156 1.00 69.38 A C
+ATOM 1633 ND2 ASN A 268 31.651 -69.757 -46.295 1.00 67.71 A N
+ATOM 1634 OD1 ASN A 268 30.400 -68.443 -44.950 1.00 74.14 A O
+ATOM 1635 N ASP A 269 29.769 -72.023 -41.380 1.00 69.59 A N
+ATOM 1636 CA ASP A 269 29.454 -73.239 -40.646 1.00 71.11 A C
+ATOM 1637 C ASP A 269 27.947 -73.434 -40.566 1.00 59.18 A C
+ATOM 1638 O ASP A 269 27.453 -74.559 -40.720 1.00 57.81 A O
+ATOM 1639 CB ASP A 269 30.077 -73.201 -39.250 1.00 73.77 A C
+ATOM 1640 CG ASP A 269 31.515 -72.725 -39.268 1.00 79.26 A C
+ATOM 1641 OD1 ASP A 269 32.239 -73.064 -40.229 1.00 83.10 A O
+ATOM 1642 OD2 ASP A 269 31.926 -72.034 -38.311 1.00 79.00 A O1-
+ATOM 1643 N GLN A 270 27.199 -72.346 -40.344 1.00 47.28 A N
+ATOM 1644 CA GLN A 270 25.745 -72.440 -40.350 1.00 53.49 A C
+ATOM 1645 C GLN A 270 25.222 -72.770 -41.744 1.00 50.42 A C
+ATOM 1646 O GLN A 270 24.387 -73.669 -41.908 1.00 49.32 A O
+ATOM 1647 CB GLN A 270 25.130 -71.144 -39.854 1.00 61.99 A C
+ATOM 1648 CG GLN A 270 25.580 -70.749 -38.501 1.00 67.58 A C
+ATOM 1649 CD GLN A 270 24.965 -69.446 -38.087 1.00 78.52 A C
+ATOM 1650 NE2 GLN A 270 23.767 -69.516 -37.534 1.00 71.70 A N
+ATOM 1651 OE1 GLN A 270 25.551 -68.381 -38.269 1.00 88.51 A O
+ATOM 1652 N LYS A 271 25.682 -72.043 -42.763 1.00 51.94 A N
+ATOM 1653 CA LYS A 271 25.279 -72.381 -44.123 1.00 47.38 A C
+ATOM 1654 C LYS A 271 25.522 -73.871 -44.389 1.00 46.75 A C
+ATOM 1655 O LYS A 271 24.587 -74.617 -44.721 1.00 57.75 A O
+ATOM 1656 CB LYS A 271 26.032 -71.499 -45.127 1.00 43.85 A C
+ATOM 1657 CG LYS A 271 25.646 -70.034 -45.118 1.00 59.33 A C
+ATOM 1658 CD LYS A 271 26.504 -69.267 -46.102 1.00 73.13 A C
+ATOM 1659 CE LYS A 271 26.265 -67.781 -45.979 1.00 73.69 A C
+ATOM 1660 NZ LYS A 271 27.160 -67.007 -46.867 1.00 78.48 A N1+
+ATOM 1661 N LYS A 272 26.768 -74.330 -44.167 1.00 47.78 A N
+ATOM 1662 CA LYS A 272 27.127 -75.735 -44.362 1.00 51.24 A C
+ATOM 1663 C LYS A 272 26.210 -76.656 -43.567 1.00 56.70 A C
+ATOM 1664 O LYS A 272 25.841 -77.741 -44.037 1.00 55.89 A O
+ATOM 1665 CB LYS A 272 28.598 -75.941 -43.969 1.00 67.26 A C
+ATOM 1666 CG LYS A 272 29.159 -77.374 -44.112 1.00 86.26 A C
+ATOM 1667 CD LYS A 272 30.427 -77.578 -43.228 1.00101.63 A C
+ATOM 1668 CE LYS A 272 31.016 -79.004 -43.273 1.00108.72 A C
+ATOM 1669 NZ LYS A 272 31.844 -79.295 -44.490 1.00111.53 A N1+
+ATOM 1670 N LEU A 273 25.783 -76.205 -42.393 1.00 57.88 A N
+ATOM 1671 CA LEU A 273 25.014 -77.038 -41.480 1.00 49.38 A C
+ATOM 1672 C LEU A 273 23.573 -77.222 -41.943 1.00 51.08 A C
+ATOM 1673 O LEU A 273 23.033 -78.334 -41.890 1.00 50.88 A O
+ATOM 1674 CB LEU A 273 25.048 -76.432 -40.079 1.00 47.93 A C
+ATOM 1675 CG LEU A 273 23.981 -77.088 -39.226 1.00 53.74 A C
+ATOM 1676 CD1 LEU A 273 24.318 -78.544 -38.994 1.00 50.20 A C
+ATOM 1677 CD2 LEU A 273 23.840 -76.348 -37.951 1.00 51.30 A C
+ATOM 1678 N MET A 274 22.912 -76.145 -42.361 1.00 44.81 A N
+ATOM 1679 CA MET A 274 21.580 -76.315 -42.923 1.00 47.78 A C
+ATOM 1680 C MET A 274 21.658 -77.111 -44.220 1.00 52.55 A C
+ATOM 1681 O MET A 274 21.008 -78.156 -44.371 1.00 51.61 A O
+ATOM 1682 CB MET A 274 20.927 -74.955 -43.147 1.00 51.15 A C
+ATOM 1683 CG MET A 274 21.101 -74.062 -41.961 1.00 45.36 A C
+ATOM 1684 SD MET A 274 20.052 -72.602 -41.935 1.00 59.02 A S
+ATOM 1685 CE MET A 274 20.500 -71.833 -43.440 1.00 49.54 A C
+ATOM 1686 N SER A 275 22.506 -76.665 -45.146 1.00 52.89 A N
+ATOM 1687 CA SER A 275 22.517 -77.297 -46.458 1.00 51.18 A C
+ATOM 1688 C SER A 275 23.008 -78.744 -46.423 1.00 57.08 A C
+ATOM 1689 O SER A 275 22.774 -79.477 -47.387 1.00 63.49 A O
+ATOM 1690 CB SER A 275 23.313 -76.429 -47.466 1.00 55.92 A C
+ATOM 1691 OG SER A 275 24.408 -75.721 -46.902 1.00 62.23 A O
+ATOM 1692 N ASN A 276 23.644 -79.195 -45.345 1.00 56.18 A N
+ATOM 1693 CA ASN A 276 23.826 -80.637 -45.217 1.00 56.00 A C
+ATOM 1694 C ASN A 276 22.570 -81.316 -44.709 1.00 60.58 A C
+ATOM 1695 O ASN A 276 22.362 -82.508 -44.954 1.00 74.32 A O
+ATOM 1696 CB ASN A 276 24.960 -80.957 -44.257 1.00 59.96 A C
+ATOM 1697 CG ASN A 276 26.293 -80.770 -44.872 1.00 67.79 A C
+ATOM 1698 ND2 ASN A 276 27.263 -80.356 -44.059 1.00 63.76 A N
+ATOM 1699 OD1 ASN A 276 26.463 -80.973 -46.073 1.00 65.95 A O
+ATOM 1700 N ASN A 277 21.796 -80.606 -43.908 1.00 55.64 A N
+ATOM 1701 CA ASN A 277 20.683 -81.181 -43.180 1.00 57.07 A C
+ATOM 1702 C ASN A 277 19.309 -80.804 -43.717 1.00 57.92 A C
+ATOM 1703 O ASN A 277 18.342 -80.808 -42.942 1.00 60.68 A O
+ATOM 1704 CB ASN A 277 20.830 -80.861 -41.715 1.00 49.29 A C
+ATOM 1705 CG ASN A 277 21.839 -81.728 -41.101 1.00 59.28 A C
+ATOM 1706 ND2 ASN A 277 21.460 -82.971 -40.807 1.00 51.74 A N
+ATOM 1707 OD1 ASN A 277 22.991 -81.335 -40.986 1.00 66.28 A O
+ATOM 1708 N VAL A 278 19.211 -80.386 -44.980 1.00 51.88 A N
+ATOM 1709 CA VAL A 278 17.986 -79.797 -45.518 1.00 47.62 A C
+ATOM 1710 C VAL A 278 16.738 -80.500 -44.995 1.00 45.74 A C
+ATOM 1711 O VAL A 278 15.830 -79.833 -44.493 1.00 53.40 A O
+ATOM 1712 CB VAL A 278 17.980 -79.844 -47.052 1.00 53.33 A C
+ATOM 1713 CG1 VAL A 278 16.813 -79.077 -47.556 1.00 57.62 A C
+ATOM 1714 CG2 VAL A 278 19.269 -79.307 -47.607 1.00 50.72 A C
+ATOM 1715 N GLN A 279 16.710 -81.842 -45.046 1.00 44.50 A N
+ATOM 1716 CA GLN A 279 15.560 -82.596 -44.535 1.00 45.63 A C
+ATOM 1717 C GLN A 279 15.129 -82.145 -43.127 1.00 49.69 A C
+ATOM 1718 O GLN A 279 13.923 -82.057 -42.840 1.00 52.38 A O
+ATOM 1719 CB GLN A 279 15.866 -84.101 -44.543 1.00 54.56 A C
+ATOM 1720 CG GLN A 279 17.030 -84.548 -43.653 1.00 74.65 A C
+ATOM 1721 CD GLN A 279 18.363 -84.539 -44.387 1.00 97.39 A C
+ATOM 1722 NE2 GLN A 279 19.308 -85.344 -43.911 1.00103.93 A N
+ATOM 1723 OE1 GLN A 279 18.540 -83.817 -45.370 1.00104.68 A O
+ATOM 1724 N ILE A 280 16.088 -81.858 -42.232 1.00 48.74 A N
+ATOM 1725 CA ILE A 280 15.733 -81.329 -40.914 1.00 50.08 A C
+ATOM 1726 C ILE A 280 15.235 -79.899 -41.027 1.00 47.90 A C
+ATOM 1727 O ILE A 280 14.196 -79.540 -40.456 1.00 53.07 A O
+ATOM 1728 CB ILE A 280 16.922 -81.410 -39.949 1.00 59.33 A C
+ATOM 1729 CG1 ILE A 280 17.447 -82.821 -39.929 1.00 52.05 A C
+ATOM 1730 CG2 ILE A 280 16.492 -80.994 -38.559 1.00 47.76 A C
+ATOM 1731 CD1 ILE A 280 16.391 -83.773 -39.476 1.00 52.26 A C
+ATOM 1732 N VAL A 281 15.985 -79.059 -41.751 1.00 41.63 A N
+ATOM 1733 CA VAL A 281 15.602 -77.661 -41.928 1.00 44.71 A C
+ATOM 1734 C VAL A 281 14.144 -77.556 -42.334 1.00 51.82 A C
+ATOM 1735 O VAL A 281 13.409 -76.688 -41.852 1.00 48.52 A O
+ATOM 1736 CB VAL A 281 16.513 -76.995 -42.966 1.00 50.59 A C
+ATOM 1737 CG1 VAL A 281 16.393 -75.499 -42.851 1.00 49.71 A C
+ATOM 1738 CG2 VAL A 281 17.933 -77.466 -42.778 1.00 43.64 A C
+ATOM 1739 N ARG A 282 13.696 -78.454 -43.210 1.00 48.93 A N
+ATOM 1740 CA ARG A 282 12.307 -78.428 -43.641 1.00 42.21 A C
+ATOM 1741 C ARG A 282 11.372 -78.584 -42.460 1.00 49.06 A C
+ATOM 1742 O ARG A 282 10.404 -77.832 -42.320 1.00 53.41 A O
+ATOM 1743 CB ARG A 282 12.039 -79.523 -44.668 1.00 41.63 A C
+ATOM 1744 CG ARG A 282 12.573 -79.243 -46.035 1.00 50.24 A C
+ATOM 1745 CD ARG A 282 11.887 -80.129 -47.028 1.00 48.78 A C
+ATOM 1746 NE ARG A 282 12.530 -80.051 -48.326 1.00 49.94 A N
+ATOM 1747 CZ ARG A 282 12.522 -78.966 -49.095 1.00 48.33 A C
+ATOM 1748 NH1 ARG A 282 11.890 -77.889 -48.669 1.00 42.78 A N1+
+ATOM 1749 NH2 ARG A 282 13.139 -78.956 -50.282 1.00 58.58 A N
+ATOM 1750 N GLN A 283 11.647 -79.548 -41.594 1.00 47.61 A N
+ATOM 1751 CA GLN A 283 10.689 -79.822 -40.539 1.00 49.02 A C
+ATOM 1752 C GLN A 283 10.641 -78.720 -39.490 1.00 52.67 A C
+ATOM 1753 O GLN A 283 9.641 -78.586 -38.772 1.00 51.80 A O
+ATOM 1754 CB GLN A 283 11.027 -81.151 -39.901 1.00 40.51 A C
+ATOM 1755 CG GLN A 283 11.134 -82.271 -40.899 1.00 49.23 A C
+ATOM 1756 CD GLN A 283 10.700 -83.571 -40.261 1.00 50.51 A C
+ATOM 1757 NE2 GLN A 283 9.779 -84.282 -40.921 1.00 65.94 A N
+ATOM 1758 OE1 GLN A 283 11.183 -83.935 -39.180 1.00 70.82 A O
+ATOM 1759 N GLN A 284 11.697 -77.940 -39.389 1.00 44.51 A N
+ATOM 1760 CA GLN A 284 11.760 -76.831 -38.467 1.00 50.51 A C
+ATOM 1761 C GLN A 284 11.326 -75.527 -39.116 1.00 58.12 A C
+ATOM 1762 O GLN A 284 11.520 -74.468 -38.522 1.00 64.25 A O
+ATOM 1763 CB GLN A 284 13.178 -76.710 -37.904 1.00 49.12 A C
+ATOM 1764 CG GLN A 284 13.724 -78.032 -37.353 1.00 47.47 A C
+ATOM 1765 CD GLN A 284 14.423 -77.879 -36.001 1.00 52.55 A C
+ATOM 1766 NE2 GLN A 284 15.312 -76.886 -35.911 1.00 57.46 A N
+ATOM 1767 OE1 GLN A 284 14.184 -78.661 -35.055 1.00 61.16 A O
+ATOM 1768 N SER A 285 10.789 -75.568 -40.332 1.00 51.45 A N
+ATOM 1769 CA SER A 285 10.407 -74.367 -41.059 1.00 48.07 A C
+ATOM 1770 C SER A 285 8.904 -74.298 -41.227 1.00 44.97 A C
+ATOM 1771 O SER A 285 8.193 -75.291 -41.135 1.00 48.44 A O
+ATOM 1772 CB SER A 285 11.039 -74.316 -42.446 1.00 42.21 A C
+ATOM 1773 OG SER A 285 12.440 -74.237 -42.355 1.00 50.91 A O
+ATOM 1774 N TYR A 286 8.417 -73.108 -41.470 1.00 52.09 A N
+ATOM 1775 CA TYR A 286 7.008 -72.956 -41.757 1.00 50.14 A C
+ATOM 1776 C TYR A 286 6.831 -72.252 -43.098 1.00 59.69 A C
+ATOM 1777 O TYR A 286 7.690 -71.480 -43.540 1.00 55.07 A O
+ATOM 1778 CB TYR A 286 6.308 -72.168 -40.646 1.00 51.64 A C
+ATOM 1779 CG TYR A 286 6.170 -72.871 -39.310 1.00 49.35 A C
+ATOM 1780 CD1 TYR A 286 7.165 -72.765 -38.356 1.00 43.60 A C
+ATOM 1781 CD2 TYR A 286 5.011 -73.559 -38.965 1.00 57.36 A C
+ATOM 1782 CE1 TYR A 286 7.040 -73.375 -37.094 1.00 49.91 A C
+ATOM 1783 CE2 TYR A 286 4.867 -74.159 -37.706 1.00 48.67 A C
+ATOM 1784 CZ TYR A 286 5.893 -74.070 -36.779 1.00 46.70 A C
+ATOM 1785 OH TYR A 286 5.799 -74.658 -35.541 1.00 64.43 A O
+ATOM 1786 N SER A 287 5.713 -72.539 -43.753 1.00 45.16 A N
+ATOM 1787 CA SER A 287 5.250 -71.773 -44.900 1.00 43.36 A C
+ATOM 1788 C SER A 287 3.929 -71.147 -44.504 1.00 46.69 A C
+ATOM 1789 O SER A 287 2.992 -71.854 -44.112 1.00 49.59 A O
+ATOM 1790 CB SER A 287 5.081 -72.641 -46.149 1.00 50.95 A C
+ATOM 1791 OG SER A 287 4.429 -71.916 -47.174 1.00 50.22 A O
+ATOM 1792 N ILE A 288 3.869 -69.829 -44.590 1.00 47.55 A N
+ATOM 1793 CA ILE A 288 2.703 -69.065 -44.199 1.00 45.95 A C
+ATOM 1794 C ILE A 288 2.159 -68.378 -45.438 1.00 50.63 A C
+ATOM 1795 O ILE A 288 2.827 -67.502 -46.010 1.00 51.10 A O
+ATOM 1796 CB ILE A 288 3.036 -68.031 -43.128 1.00 53.53 A C
+ATOM 1797 CG1 ILE A 288 3.910 -68.652 -42.057 1.00 49.22 A C
+ATOM 1798 CG2 ILE A 288 1.765 -67.538 -42.523 1.00 53.32 A C
+ATOM 1799 CD1 ILE A 288 4.377 -67.642 -41.041 1.00 51.29 A C
+ATOM 1800 N MET A 289 0.946 -68.761 -45.845 1.00 45.19 A N
+ATOM 1801 CA MET A 289 0.223 -67.997 -46.853 1.00 43.03 A C
+ATOM 1802 C MET A 289 0.045 -66.556 -46.408 1.00 48.24 A C
+ATOM 1803 O MET A 289 -0.446 -66.287 -45.308 1.00 55.29 A O
+ATOM 1804 CB MET A 289 -1.142 -68.607 -47.110 1.00 49.46 A C
+ATOM 1805 CG MET A 289 -2.030 -67.667 -47.882 1.00 47.86 A C
+ATOM 1806 SD MET A 289 -1.344 -67.446 -49.514 1.00 60.09 A S
+ATOM 1807 CE MET A 289 -2.552 -66.361 -50.273 1.00 58.46 A C
+ATOM 1808 N SER A 290 0.428 -65.630 -47.279 1.00 52.78 A N
+ATOM 1809 CA SER A 290 0.341 -64.217 -46.949 1.00 55.52 A C
+ATOM 1810 C SER A 290 -0.652 -63.481 -47.840 1.00 59.96 A C
+ATOM 1811 O SER A 290 -1.771 -63.185 -47.403 1.00 62.50 A O
+ATOM 1812 CB SER A 290 1.720 -63.577 -47.053 1.00 47.75 A C
+ATOM 1813 OG SER A 290 1.659 -62.204 -46.723 1.00 57.69 A O
+ATOM 1814 N ILE A 291 -0.280 -63.175 -49.084 1.00 47.30 A N
+ATOM 1815 CA ILE A 291 -1.172 -62.406 -49.942 1.00 54.18 A C
+ATOM 1816 C ILE A 291 -1.137 -62.970 -51.342 1.00 51.55 A C
+ATOM 1817 O ILE A 291 -0.225 -63.703 -51.731 1.00 60.61 A O
+ATOM 1818 CB ILE A 291 -0.845 -60.901 -50.075 1.00 62.85 A C
+ATOM 1819 CG1 ILE A 291 0.533 -60.695 -50.675 1.00 72.52 A C
+ATOM 1820 CG2 ILE A 291 -0.878 -60.207 -48.750 1.00 70.32 A C
+ATOM 1821 CD1 ILE A 291 0.914 -59.259 -50.676 1.00 81.12 A C
+ATOM 1822 N ILE A 292 -2.158 -62.597 -52.100 1.00 48.65 A N
+ATOM 1823 CA ILE A 292 -2.137 -62.657 -53.547 1.00 58.40 A C
+ATOM 1824 C ILE A 292 -2.769 -61.353 -54.019 1.00 55.03 A C
+ATOM 1825 O ILE A 292 -3.938 -61.087 -53.731 1.00 64.24 A O
+ATOM 1826 CB ILE A 292 -2.869 -63.892 -54.088 1.00 55.02 A C
+ATOM 1827 CG1 ILE A 292 -3.022 -63.814 -55.587 1.00 55.55 A C
+ATOM 1828 CG2 ILE A 292 -4.189 -64.126 -53.377 1.00 53.16 A C
+ATOM 1829 CD1 ILE A 292 -3.735 -65.011 -56.135 1.00 60.19 A C
+ATOM 1830 N LYS A 293 -1.983 -60.512 -54.678 1.00 56.95 A N
+ATOM 1831 CA LYS A 293 -2.510 -59.259 -55.203 1.00 67.51 A C
+ATOM 1832 C LYS A 293 -1.522 -58.740 -56.239 1.00 72.08 A C
+ATOM 1833 O LYS A 293 -0.418 -59.265 -56.380 1.00 71.43 A O
+ATOM 1834 CB LYS A 293 -2.721 -58.226 -54.090 1.00 72.50 A C
+ATOM 1835 CG LYS A 293 -1.418 -57.732 -53.465 1.00 82.50 A C
+ATOM 1836 CD LYS A 293 -1.658 -56.644 -52.425 1.00 87.13 A C
+ATOM 1837 CE LYS A 293 -1.915 -55.279 -53.034 1.00 98.60 A C
+ATOM 1838 NZ LYS A 293 -1.916 -54.234 -51.971 1.00101.73 A N1+
+ATOM 1839 N GLU A 294 -1.925 -57.675 -56.940 1.00 80.86 A N
+ATOM 1840 CA GLU A 294 -1.072 -57.000 -57.912 1.00 81.95 A C
+ATOM 1841 C GLU A 294 -0.300 -57.992 -58.777 1.00 66.84 A C
+ATOM 1842 O GLU A 294 0.889 -57.774 -59.035 1.00 56.09 A O
+ATOM 1843 CB GLU A 294 -0.110 -56.035 -57.206 1.00 91.99 A C
+ATOM 1844 CG GLU A 294 -0.806 -54.920 -56.411 1.00110.43 A C
+ATOM 1845 CD GLU A 294 0.166 -53.884 -55.837 1.00126.62 A C
+ATOM 1846 OE1 GLU A 294 1.323 -53.829 -56.313 1.00123.18 A O
+ATOM 1847 OE2 GLU A 294 -0.232 -53.112 -54.927 1.00136.21 A O1-
+ATOM 1848 N GLU A 295 -0.957 -59.090 -59.195 1.00 63.22 A N
+ATOM 1849 CA GLU A 295 -0.410 -60.109 -60.115 1.00 65.80 A C
+ATOM 1850 C GLU A 295 0.768 -60.906 -59.525 1.00 52.33 A C
+ATOM 1851 O GLU A 295 1.605 -61.449 -60.259 1.00 69.17 A O
+ATOM 1852 CB GLU A 295 -0.002 -59.490 -61.457 1.00 75.33 A C
+ATOM 1853 CG GLU A 295 -1.178 -59.145 -62.351 1.00 80.60 A C
+ATOM 1854 CD GLU A 295 -0.810 -59.117 -63.829 1.00 98.05 A C
+ATOM 1855 OE1 GLU A 295 0.390 -59.290 -64.149 1.00 98.69 A O
+ATOM 1856 OE2 GLU A 295 -1.719 -58.918 -64.669 1.00109.05 A O1-
+ATOM 1857 N VAL A 296 0.839 -61.018 -58.205 1.00 51.62 A N
+ATOM 1858 CA VAL A 296 1.837 -61.842 -57.546 1.00 60.33 A C
+ATOM 1859 C VAL A 296 1.203 -62.536 -56.345 1.00 53.19 A C
+ATOM 1860 O VAL A 296 0.530 -61.897 -55.526 1.00 60.62 A O
+ATOM 1861 CB VAL A 296 3.062 -61.002 -57.142 1.00 60.33 A C
+ATOM 1862 CG1 VAL A 296 2.627 -59.803 -56.388 1.00 62.98 A C
+ATOM 1863 CG2 VAL A 296 3.979 -61.818 -56.252 1.00 56.28 A C
+ATOM 1864 N LEU A 297 1.372 -63.856 -56.278 1.00 51.51 A N
+ATOM 1865 CA LEU A 297 1.147 -64.624 -55.060 1.00 51.11 A C
+ATOM 1866 C LEU A 297 2.427 -64.622 -54.243 1.00 47.28 A C
+ATOM 1867 O LEU A 297 3.512 -64.869 -54.780 1.00 50.62 A O
+ATOM 1868 CB LEU A 297 0.748 -66.063 -55.380 1.00 50.01 A C
+ATOM 1869 CG LEU A 297 1.106 -67.126 -54.337 1.00 47.86 A C
+ATOM 1870 CD1 LEU A 297 0.273 -66.954 -53.106 1.00 48.14 A C
+ATOM 1871 CD2 LEU A 297 0.919 -68.514 -54.901 1.00 57.08 A C
+ATOM 1872 N ALA A 298 2.301 -64.356 -52.947 1.00 47.52 A N
+ATOM 1873 CA ALA A 298 3.462 -64.257 -52.073 1.00 51.76 A C
+ATOM 1874 C ALA A 298 3.151 -64.881 -50.722 1.00 49.81 A C
+ATOM 1875 O ALA A 298 2.107 -64.601 -50.117 1.00 57.44 A O
+ATOM 1876 CB ALA A 298 3.892 -62.798 -51.896 1.00 42.93 A C
+ATOM 1877 N TYR A 299 4.070 -65.719 -50.255 1.00 44.12 A N
+ATOM 1878 CA TYR A 299 3.964 -66.347 -48.953 1.00 48.36 A C
+ATOM 1879 C TYR A 299 5.335 -66.328 -48.315 1.00 53.91 A C
+ATOM 1880 O TYR A 299 6.353 -66.253 -49.003 1.00 56.05 A O
+ATOM 1881 CB TYR A 299 3.429 -67.774 -49.045 1.00 43.65 A C
+ATOM 1882 CG TYR A 299 4.184 -68.716 -49.954 1.00 40.70 A C
+ATOM 1883 CD1 TYR A 299 4.935 -69.759 -49.439 1.00 46.74 A C
+ATOM 1884 CD2 TYR A 299 4.129 -68.576 -51.337 1.00 37.57 A C
+ATOM 1885 CE1 TYR A 299 5.608 -70.647 -50.285 1.00 43.48 A C
+ATOM 1886 CE2 TYR A 299 4.813 -69.448 -52.188 1.00 36.04 A C
+ATOM 1887 CZ TYR A 299 5.546 -70.478 -51.656 1.00 53.11 A C
+ATOM 1888 OH TYR A 299 6.209 -71.342 -52.493 1.00 43.83 A O
+ATOM 1889 N VAL A 300 5.352 -66.362 -46.988 1.00 54.71 A N
+ATOM 1890 CA VAL A 300 6.586 -66.261 -46.225 1.00 52.16 A C
+ATOM 1891 C VAL A 300 6.965 -67.646 -45.761 1.00 46.31 A C
+ATOM 1892 O VAL A 300 6.125 -68.380 -45.226 1.00 52.42 A O
+ATOM 1893 CB VAL A 300 6.432 -65.327 -45.021 1.00 45.81 A C
+ATOM 1894 CG1 VAL A 300 7.549 -65.569 -44.055 1.00 39.89 A C
+ATOM 1895 CG2 VAL A 300 6.456 -63.899 -45.474 1.00 49.20 A C
+ATOM 1896 N VAL A 301 8.221 -68.007 -45.967 1.00 49.31 A N
+ATOM 1897 CA VAL A 301 8.797 -69.197 -45.368 1.00 45.14 A C
+ATOM 1898 C VAL A 301 9.754 -68.716 -44.292 1.00 48.27 A C
+ATOM 1899 O VAL A 301 10.787 -68.104 -44.606 1.00 53.90 A O
+ATOM 1900 CB VAL A 301 9.500 -70.064 -46.409 1.00 53.82 A C
+ATOM 1901 CG1 VAL A 301 9.965 -71.344 -45.795 1.00 53.55 A C
+ATOM 1902 CG2 VAL A 301 8.538 -70.355 -47.478 1.00 38.66 A C
+ATOM 1903 N GLN A 302 9.394 -68.949 -43.022 1.00 54.57 A N
+ATOM 1904 CA GLN A 302 10.272 -68.616 -41.912 1.00 49.05 A C
+ATOM 1905 C GLN A 302 11.116 -69.830 -41.590 1.00 52.60 A C
+ATOM 1906 O GLN A 302 10.587 -70.923 -41.362 1.00 55.47 A O
+ATOM 1907 CB GLN A 302 9.497 -68.153 -40.684 1.00 51.38 A C
+ATOM 1908 CG GLN A 302 8.815 -69.224 -39.864 1.00 42.31 A C
+ATOM 1909 CD GLN A 302 8.197 -68.647 -38.579 1.00 48.25 A C
+ATOM 1910 NE2 GLN A 302 8.255 -69.420 -37.477 1.00 45.57 A N
+ATOM 1911 OE1 GLN A 302 7.678 -67.518 -38.582 1.00 51.53 A O
+ATOM 1912 N LEU A 303 12.421 -69.632 -41.588 1.00 48.52 A N
+ATOM 1913 CA LEU A 303 13.429 -70.671 -41.575 1.00 47.70 A C
+ATOM 1914 C LEU A 303 14.239 -70.635 -40.285 1.00 52.04 A C
+ATOM 1915 O LEU A 303 14.523 -69.552 -39.753 1.00 44.78 A O
+ATOM 1916 CB LEU A 303 14.365 -70.484 -42.763 1.00 52.27 A C
+ATOM 1917 CG LEU A 303 13.651 -70.540 -44.101 1.00 55.44 A C
+ATOM 1918 CD1 LEU A 303 14.562 -69.970 -45.143 1.00 54.89 A C
+ATOM 1919 CD2 LEU A 303 13.302 -71.968 -44.446 1.00 38.19 A C
+ATOM 1920 N PRO A 304 14.620 -71.795 -39.761 1.00 46.02 A N
+ATOM 1921 CA PRO A 304 15.288 -71.829 -38.457 1.00 46.44 A C
+ATOM 1922 C PRO A 304 16.712 -71.284 -38.517 1.00 52.76 A C
+ATOM 1923 O PRO A 304 17.541 -71.725 -39.320 1.00 64.40 A O
+ATOM 1924 CB PRO A 304 15.268 -73.318 -38.091 1.00 50.19 A C
+ATOM 1925 CG PRO A 304 15.278 -73.991 -39.384 1.00 49.21 A C
+ATOM 1926 CD PRO A 304 14.462 -73.136 -40.326 1.00 50.97 A C
+ATOM 1927 N LEU A 305 16.999 -70.355 -37.603 1.00 57.28 A N
+ATOM 1928 CA LEU A 305 18.350 -69.893 -37.313 1.00 49.94 A C
+ATOM 1929 C LEU A 305 18.909 -70.712 -36.151 1.00 49.77 A C
+ATOM 1930 O LEU A 305 18.373 -70.660 -35.023 1.00 61.84 A O
+ATOM 1931 CB LEU A 305 18.349 -68.398 -36.987 1.00 64.31 A C
+ATOM 1932 CG LEU A 305 17.958 -67.560 -38.198 1.00 79.86 A C
+ATOM 1933 CD1 LEU A 305 18.151 -66.084 -37.951 1.00 81.20 A C
+ATOM 1934 CD2 LEU A 305 18.765 -68.021 -39.400 1.00 82.84 A C
+ATOM 1935 N TYR A 306 19.976 -71.472 -36.448 1.00 59.68 A N
+ATOM 1936 CA TYR A 306 20.711 -72.266 -35.467 1.00 64.85 A C
+ATOM 1937 C TYR A 306 21.862 -71.419 -34.945 1.00 62.15 A C
+ATOM 1938 O TYR A 306 22.909 -71.301 -35.585 1.00 60.13 A O
+ATOM 1939 CB TYR A 306 21.232 -73.553 -36.104 1.00 56.68 A C
+ATOM 1940 CG TYR A 306 20.164 -74.460 -36.674 1.00 53.24 A C
+ATOM 1941 CD1 TYR A 306 19.090 -74.845 -35.891 1.00 57.61 A C
+ATOM 1942 CD2 TYR A 306 20.212 -74.917 -37.995 1.00 57.10 A C
+ATOM 1943 CE1 TYR A 306 18.090 -75.676 -36.390 1.00 53.83 A C
+ATOM 1944 CE2 TYR A 306 19.212 -75.745 -38.506 1.00 52.09 A C
+ATOM 1945 CZ TYR A 306 18.150 -76.122 -37.687 1.00 50.64 A C
+ATOM 1946 OH TYR A 306 17.121 -76.936 -38.113 1.00 56.47 A O
+ATOM 1947 N GLY A 307 21.707 -70.911 -33.734 1.00 64.36 A N
+ATOM 1948 CA GLY A 307 22.668 -70.000 -33.169 1.00 58.65 A C
+ATOM 1949 C GLY A 307 23.596 -70.648 -32.185 1.00 61.79 A C
+ATOM 1950 O GLY A 307 24.412 -69.962 -31.565 1.00 63.96 A O
+ATOM 1951 N VAL A 308 23.462 -71.946 -31.972 1.00 48.25 A N
+ATOM 1952 CA VAL A 308 24.356 -72.684 -31.106 1.00 60.64 A C
+ATOM 1953 C VAL A 308 24.674 -73.974 -31.843 1.00 62.58 A C
+ATOM 1954 O VAL A 308 23.796 -74.832 -32.003 1.00 57.52 A O
+ATOM 1955 CB VAL A 308 23.728 -72.966 -29.736 1.00 57.97 A C
+ATOM 1956 CG1 VAL A 308 24.645 -73.833 -28.899 1.00 59.17 A C
+ATOM 1957 CG2 VAL A 308 23.420 -71.658 -29.020 1.00 56.98 A C
+ATOM 1958 N ILE A 309 25.931 -74.130 -32.260 1.00 56.58 A N
+ATOM 1959 CA ILE A 309 26.373 -75.218 -33.129 1.00 53.60 A C
+ATOM 1960 C ILE A 309 27.548 -75.900 -32.462 1.00 50.71 A C
+ATOM 1961 O ILE A 309 28.480 -75.228 -32.012 1.00 55.08 A O
+ATOM 1962 CB ILE A 309 26.782 -74.718 -34.537 1.00 60.13 A C
+ATOM 1963 CG1 ILE A 309 25.569 -74.254 -35.353 1.00 60.01 A C
+ATOM 1964 CG2 ILE A 309 27.658 -75.719 -35.259 1.00 66.28 A C
+ATOM 1965 CD1 ILE A 309 24.310 -74.975 -35.024 1.00 72.90 A C
+ATOM 1966 N ASP A 310 27.505 -77.227 -32.411 1.00 65.41 A N
+ATOM 1967 CA ASP A 310 28.629 -78.046 -31.959 1.00 65.14 A C
+ATOM 1968 C ASP A 310 29.025 -77.716 -30.510 1.00 58.68 A C
+ATOM 1969 O ASP A 310 30.140 -77.258 -30.225 1.00 67.91 A O
+ATOM 1970 CB ASP A 310 29.838 -77.898 -32.909 1.00 62.93 A C
+ATOM 1971 CG ASP A 310 29.573 -78.412 -34.342 1.00 79.05 A C
+ATOM 1972 OD1 ASP A 310 28.864 -79.437 -34.543 1.00 78.11 A O
+ATOM 1973 OD2 ASP A 310 30.113 -77.780 -35.277 1.00 92.68 A O1-
+ATOM 1974 N THR A 311 28.101 -77.977 -29.587 1.00 64.55 A N
+ATOM 1975 CA THR A 311 28.418 -78.043 -28.170 1.00 66.94 A C
+ATOM 1976 C THR A 311 28.023 -79.440 -27.720 1.00 61.38 A C
+ATOM 1977 O THR A 311 27.322 -80.149 -28.452 1.00 66.76 A O
+ATOM 1978 CB THR A 311 27.657 -77.004 -27.344 1.00 60.62 A C
+ATOM 1979 CG2 THR A 311 27.849 -75.657 -27.970 1.00 57.31 A C
+ATOM 1980 OG1 THR A 311 26.264 -77.289 -27.403 1.00 54.38 A O
+ATOM 1981 N PRO A 312 28.487 -79.898 -26.558 1.00 70.98 A N
+ATOM 1982 CA PRO A 312 28.166 -81.272 -26.149 1.00 66.03 A C
+ATOM 1983 C PRO A 312 26.721 -81.415 -25.697 1.00 70.72 A C
+ATOM 1984 O PRO A 312 26.132 -80.510 -25.090 1.00 69.46 A O
+ATOM 1985 CB PRO A 312 29.131 -81.541 -24.996 1.00 62.83 A C
+ATOM 1986 CG PRO A 312 29.514 -80.192 -24.505 1.00 72.29 A C
+ATOM 1987 CD PRO A 312 29.498 -79.286 -25.682 1.00 63.94 A C
+ATOM 1988 N CYS A 313 26.160 -82.585 -26.010 1.00 65.11 A N
+ATOM 1989 CA CYS A 313 24.779 -82.924 -25.702 1.00 64.93 A C
+ATOM 1990 C CYS A 313 24.727 -84.364 -25.219 1.00 69.98 A C
+ATOM 1991 O CYS A 313 25.517 -85.207 -25.647 1.00 69.84 A O
+ATOM 1992 CB CYS A 313 23.873 -82.764 -26.925 1.00 59.28 A C
+ATOM 1993 SG CYS A 313 23.805 -81.102 -27.643 1.00 82.09 A S
+ATOM 1994 N TRP A 314 23.788 -84.644 -24.329 1.00 67.42 A N
+ATOM 1995 CA TRP A 314 23.648 -85.988 -23.802 1.00 66.76 A C
+ATOM 1996 C TRP A 314 22.167 -86.241 -23.583 1.00 69.43 A C
+ATOM 1997 O TRP A 314 21.395 -85.310 -23.333 1.00 72.52 A O
+ATOM 1998 CB TRP A 314 24.454 -86.166 -22.505 1.00 60.62 A C
+ATOM 1999 CG TRP A 314 24.187 -85.084 -21.455 1.00 70.58 A C
+ATOM 2000 CD1 TRP A 314 23.212 -85.096 -20.489 1.00 64.47 A C
+ATOM 2001 CD2 TRP A 314 24.905 -83.852 -21.272 1.00 67.71 A C
+ATOM 2002 CE2 TRP A 314 24.302 -83.170 -20.185 1.00 68.43 A C
+ATOM 2003 CE3 TRP A 314 25.995 -83.255 -21.923 1.00 72.10 A C
+ATOM 2004 NE1 TRP A 314 23.277 -83.952 -19.726 1.00 69.76 A N
+ATOM 2005 CZ2 TRP A 314 24.749 -81.927 -19.736 1.00 75.82 A C
+ATOM 2006 CZ3 TRP A 314 26.443 -82.005 -21.469 1.00 68.27 A C
+ATOM 2007 CH2 TRP A 314 25.817 -81.362 -20.386 1.00 73.35 A C
+ATOM 2008 N LYS A 315 21.770 -87.500 -23.718 1.00 64.85 A N
+ATOM 2009 CA LYS A 315 20.376 -87.900 -23.574 1.00 64.39 A C
+ATOM 2010 C LYS A 315 20.248 -88.721 -22.303 1.00 65.02 A C
+ATOM 2011 O LYS A 315 20.867 -89.784 -22.183 1.00 64.94 A O
+ATOM 2012 CB LYS A 315 19.895 -88.699 -24.785 1.00 68.39 A C
+ATOM 2013 CG LYS A 315 18.382 -88.897 -24.870 1.00 68.55 A C
+ATOM 2014 CD LYS A 315 18.030 -89.922 -25.955 1.00 69.09 A C
+ATOM 2015 CE LYS A 315 16.529 -90.035 -26.195 1.00 63.12 A C
+ATOM 2016 NZ LYS A 315 16.166 -89.715 -27.627 1.00 66.56 A N1+
+ATOM 2017 N LEU A 316 19.455 -88.223 -21.364 1.00 66.61 A N
+ATOM 2018 CA LEU A 316 19.279 -88.863 -20.072 1.00 70.06 A C
+ATOM 2019 C LEU A 316 18.076 -89.785 -20.145 1.00 73.02 A C
+ATOM 2020 O LEU A 316 16.972 -89.331 -20.476 1.00 70.06 A O
+ATOM 2021 CB LEU A 316 19.070 -87.808 -18.989 1.00 63.81 A C
+ATOM 2022 CG LEU A 316 19.203 -88.180 -17.507 1.00 65.41 A C
+ATOM 2023 CD1 LEU A 316 19.512 -86.922 -16.687 1.00 62.67 A C
+ATOM 2024 CD2 LEU A 316 17.968 -88.861 -16.982 1.00 59.85 A C
+ATOM 2025 N HIS A 317 18.281 -91.057 -19.800 1.00 70.33 A N
+ATOM 2026 CA HIS A 317 17.231 -92.072 -19.808 1.00 63.89 A C
+ATOM 2027 C HIS A 317 16.872 -92.493 -18.377 1.00 68.55 A C
+ATOM 2028 O HIS A 317 17.756 -92.656 -17.527 1.00 74.72 A O
+ATOM 2029 CB HIS A 317 17.664 -93.309 -20.613 1.00 61.72 A C
+ATOM 2030 CG HIS A 317 18.215 -93.008 -21.977 1.00 68.77 A C
+ATOM 2031 CD2 HIS A 317 19.311 -92.310 -22.364 1.00 60.79 A C
+ATOM 2032 ND1 HIS A 317 17.649 -93.502 -23.136 1.00 69.35 A N
+ATOM 2033 CE1 HIS A 317 18.356 -93.094 -24.178 1.00 67.83 A C
+ATOM 2034 NE2 HIS A 317 19.374 -92.376 -23.738 1.00 69.90 A N
+ATOM 2035 N THR A 318 15.568 -92.692 -18.119 1.00 69.89 A N
+ATOM 2036 CA THR A 318 15.049 -93.012 -16.792 1.00 64.77 A C
+ATOM 2037 C THR A 318 14.088 -94.199 -16.834 1.00 68.84 A C
+ATOM 2038 O THR A 318 13.280 -94.327 -17.761 1.00 71.33 A O
+ATOM 2039 CB THR A 318 14.288 -91.846 -16.200 1.00 62.69 A C
+ATOM 2040 CG2 THR A 318 14.991 -90.567 -16.479 1.00 68.28 A C
+ATOM 2041 OG1 THR A 318 12.993 -91.792 -16.803 1.00 80.04 A O
+ATOM 2042 N SER A 319 14.125 -95.022 -15.788 1.00 76.29 A N
+ATOM 2043 CA SER A 319 13.257 -96.187 -15.681 1.00 74.47 A C
+ATOM 2044 C SER A 319 12.895 -96.415 -14.220 1.00 75.40 A C
+ATOM 2045 O SER A 319 13.640 -95.994 -13.319 1.00 78.14 A O
+ATOM 2046 CB SER A 319 13.942 -97.419 -16.281 1.00 77.11 A C
+ATOM 2047 OG SER A 319 13.103 -98.556 -16.254 1.00 86.38 A O
+ATOM 2048 N PRO A 320 11.754 -97.072 -13.936 1.00 81.68 A N
+ATOM 2049 CA PRO A 320 11.337 -97.311 -12.546 1.00 80.35 A C
+ATOM 2050 C PRO A 320 12.380 -98.048 -11.706 1.00 83.07 A C
+ATOM 2051 O PRO A 320 12.971 -99.043 -12.144 1.00 84.35 A O
+ATOM 2052 CB PRO A 320 10.071 -98.162 -12.693 1.00 74.49 A C
+ATOM 2053 CG PRO A 320 9.560 -97.894 -14.024 1.00 80.85 A C
+ATOM 2054 CD PRO A 320 10.698 -97.463 -14.893 1.00 80.08 A C
+ATOM 2055 N LEU A 321 12.598 -97.552 -10.493 1.00 77.13 A N
+ATOM 2056 CA LEU A 321 13.507 -98.173 -9.543 1.00 76.07 A C
+ATOM 2057 C LEU A 321 12.496 -98.854 -8.646 1.00 75.77 A C
+ATOM 2058 O LEU A 321 11.302 -98.592 -8.797 1.00 80.69 A O
+ATOM 2059 CB LEU A 321 14.293 -97.120 -8.769 1.00 77.74 A C
+ATOM 2060 CG LEU A 321 15.488 -97.627 -7.959 1.00 77.69 A C
+ATOM 2061 CD1 LEU A 321 16.158 -98.796 -8.664 1.00 77.14 A C
+ATOM 2062 CD2 LEU A 321 16.484 -96.504 -7.710 1.00 77.32 A C
+ATOM 2063 N CYS A 322 12.896 -99.721 -7.722 1.00 87.18 A N
+ATOM 2064 CA CYS A 322 11.830-100.313 -6.941 1.00 86.76 A C
+ATOM 2065 C CYS A 322 12.412-101.415 -6.059 1.00 92.16 A C
+ATOM 2066 O CYS A 322 13.537-101.872 -6.281 1.00 91.55 A O
+ATOM 2067 CB CYS A 322 10.778-100.817 -7.915 1.00 88.14 A C
+ATOM 2068 SG CYS A 322 9.092-100.677 -7.479 1.00 97.37 A S
+ATOM 2069 N THR A 323 11.650-101.833 -5.044 1.00 83.17 A N
+ATOM 2070 CA THR A 323 12.186-102.860 -4.162 1.00 82.85 A C
+ATOM 2071 C THR A 323 11.756-104.256 -4.633 1.00 90.32 A C
+ATOM 2072 O THR A 323 10.896-104.415 -5.504 1.00 87.84 A O
+ATOM 2073 CB THR A 323 11.739-102.592 -2.721 1.00 90.30 A C
+ATOM 2074 CG2 THR A 323 12.287-101.243 -2.263 1.00 78.56 A C
+ATOM 2075 OG1 THR A 323 10.307-102.531 -2.662 1.00 87.58 A O
+ATOM 2076 N THR A 324 12.368-105.273 -4.029 1.00 86.72 A N
+ATOM 2077 CA THR A 324 12.156-106.676 -4.371 1.00 89.30 A C
+ATOM 2078 C THR A 324 11.255-107.415 -3.384 1.00 94.57 A C
+ATOM 2079 O THR A 324 11.125-108.641 -3.492 1.00 97.73 A O
+ATOM 2080 CB THR A 324 13.503-107.387 -4.495 1.00 82.62 A C
+ATOM 2081 CG2 THR A 324 13.413-108.609 -5.418 1.00 80.95 A C
+ATOM 2082 OG1 THR A 324 14.447-106.469 -5.050 1.00 84.38 A O
+ATOM 2083 N ASN A 325 10.662-106.712 -2.410 1.00 98.50 A N
+ATOM 2084 CA ASN A 325 9.958-107.362 -1.303 1.00 98.15 A C
+ATOM 2085 C ASN A 325 8.986-108.414 -1.822 1.00100.90 A C
+ATOM 2086 O ASN A 325 8.127-108.128 -2.664 1.00102.52 A O
+ATOM 2087 CB ASN A 325 9.200-106.318 -0.478 1.00102.34 A C
+ATOM 2088 CG ASN A 325 10.078-105.633 0.546 1.00110.75 A C
+ATOM 2089 ND2 ASN A 325 9.778-104.372 0.836 1.00111.23 A N
+ATOM 2090 OD1 ASN A 325 11.017-106.230 1.071 1.00114.96 A O
+ATOM 2091 N THR A 326 9.124-109.637 -1.294 1.00104.82 A N
+ATOM 2092 CA THR A 326 8.501-110.803 -1.921 1.00116.59 A C
+ATOM 2093 C THR A 326 6.973-110.729 -1.878 1.00122.02 A C
+ATOM 2094 O THR A 326 6.302-111.226 -2.795 1.00118.52 A O
+ATOM 2095 CB THR A 326 9.028-112.082 -1.259 1.00117.87 A C
+ATOM 2096 CG2 THR A 326 8.824-112.052 0.266 1.00113.63 A C
+ATOM 2097 OG1 THR A 326 8.373-113.220 -1.824 1.00117.01 A O
+ATOM 2098 N LYS A 327 6.409-110.091 -0.847 1.00125.96 A N
+ATOM 2099 CA LYS A 327 4.970-109.861 -0.779 1.00122.23 A C
+ATOM 2100 C LYS A 327 4.580-108.649 -1.616 1.00124.25 A C
+ATOM 2101 O LYS A 327 5.296-107.643 -1.660 1.00122.20 A O
+ATOM 2102 CB LYS A 327 4.515-109.666 0.673 1.00115.91 A C
+ATOM 2103 CG LYS A 327 3.321-110.549 1.100 1.00121.43 A C
+ATOM 2104 CD LYS A 327 3.643-112.047 1.073 1.00127.23 A C
+ATOM 2105 CE LYS A 327 2.467-112.877 1.574 1.00129.31 A C
+ATOM 2106 NZ LYS A 327 2.751-114.337 1.545 1.00128.44 A N1+
+ATOM 2107 N GLU A 328 3.424-108.756 -2.267 1.00121.86 A N
+ATOM 2108 CA GLU A 328 2.933-107.771 -3.225 1.00124.99 A C
+ATOM 2109 C GLU A 328 2.293-106.562 -2.545 1.00120.53 A C
+ATOM 2110 O GLU A 328 1.754-106.649 -1.437 1.00121.39 A O
+ATOM 2111 CB GLU A 328 1.932-108.428 -4.184 1.00135.63 A C
+ATOM 2112 CG GLU A 328 0.615-108.907 -3.541 1.00143.01 A C
+ATOM 2113 CD GLU A 328 0.798-110.052 -2.522 1.00143.75 A C
+ATOM 2114 OE1 GLU A 328 1.927-110.586 -2.392 1.00137.59 A O
+ATOM 2115 OE2 GLU A 328 -0.192-110.421 -1.848 1.00143.06 A O1-
+ATOM 2116 N GLY A 329 2.372-105.417 -3.229 1.00115.74 A N
+ATOM 2117 CA GLY A 329 1.875-104.162 -2.711 1.00106.64 A C
+ATOM 2118 C GLY A 329 2.684-103.578 -1.579 1.00 95.32 A C
+ATOM 2119 O GLY A 329 2.420-102.442 -1.174 1.00 96.43 A O
+ATOM 2120 N SER A 330 3.663-104.312 -1.059 1.00103.66 A N
+ATOM 2121 CA SER A 330 4.511-103.877 0.042 1.00101.43 A C
+ATOM 2122 C SER A 330 5.737-103.114 -0.439 1.00 93.32 A C
+ATOM 2123 O SER A 330 6.600-102.769 0.380 1.00 94.21 A O
+ATOM 2124 CB SER A 330 4.932-105.089 0.889 1.00101.88 A C
+ATOM 2125 OG SER A 330 5.754-105.997 0.164 1.00101.02 A O
+ATOM 2126 N ASN A 331 5.827-102.844 -1.742 1.00 80.57 A N
+ATOM 2127 CA ASN A 331 7.013-102.260 -2.346 1.00 85.18 A C
+ATOM 2128 C ASN A 331 6.893-100.748 -2.471 1.00 84.99 A C
+ATOM 2129 O ASN A 331 5.812-100.212 -2.732 1.00 90.37 A O
+ATOM 2130 CB ASN A 331 7.266-102.858 -3.720 1.00 79.94 A C
+ATOM 2131 CG ASN A 331 7.938-104.182 -3.642 1.00 93.78 A C
+ATOM 2132 ND2 ASN A 331 7.231-105.231 -4.053 1.00 94.30 A N
+ATOM 2133 OD1 ASN A 331 9.089-104.275 -3.220 1.00 93.96 A O
+ATOM 2134 N ILE A 332 8.019-100.074 -2.288 1.00 82.32 A N
+ATOM 2135 CA ILE A 332 8.147 -98.645 -2.521 1.00 82.88 A C
+ATOM 2136 C ILE A 332 8.942 -98.446 -3.799 1.00 81.54 A C
+ATOM 2137 O ILE A 332 9.822 -99.251 -4.136 1.00 85.12 A O
+ATOM 2138 CB ILE A 332 8.849 -97.941 -1.355 1.00 77.65 A C
+ATOM 2139 CG1 ILE A 332 10.234 -98.557 -1.163 1.00 73.27 A C
+ATOM 2140 CG2 ILE A 332 8.016 -98.067 -0.122 1.00 72.32 A C
+ATOM 2141 CD1 ILE A 332 11.131 -97.778 -0.242 1.00 79.47 A C
+ATOM 2142 N CYS A 333 8.650 -97.349 -4.504 1.00 80.85 A N
+ATOM 2143 CA CYS A 333 9.104 -97.243 -5.879 1.00 78.12 A C
+ATOM 2144 C CYS A 333 9.405 -95.776 -6.225 1.00 83.57 A C
+ATOM 2145 O CYS A 333 8.665 -94.870 -5.835 1.00 80.78 A O
+ATOM 2146 CB CYS A 333 8.022 -97.890 -6.745 1.00 69.65 A C
+ATOM 2147 SG CYS A 333 8.576 -98.829 -8.173 1.00 81.00 A S
+ATOM 2148 N LEU A 334 10.502 -95.556 -6.960 1.00 79.48 A N
+ATOM 2149 CA LEU A 334 11.079 -94.234 -7.229 1.00 77.15 A C
+ATOM 2150 C LEU A 334 11.648 -94.141 -8.647 1.00 74.14 A C
+ATOM 2151 O LEU A 334 12.478 -94.981 -9.009 1.00 74.74 A O
+ATOM 2152 CB LEU A 334 12.179 -93.977 -6.199 1.00 86.66 A C
+ATOM 2153 CG LEU A 334 13.172 -92.834 -6.266 1.00 89.98 A C
+ATOM 2154 CD1 LEU A 334 13.687 -92.627 -4.888 1.00 82.97 A C
+ATOM 2155 CD2 LEU A 334 14.339 -93.278 -7.108 1.00 76.65 A C
+ATOM 2156 N THR A 335 11.277 -93.098 -9.425 1.00 71.71 A N
+ATOM 2157 CA THR A 335 11.777 -92.938 -10.804 1.00 75.02 A C
+ATOM 2158 C THR A 335 12.170 -91.499 -11.122 1.00 74.66 A C
+ATOM 2159 O THR A 335 11.354 -90.587 -10.949 1.00 77.04 A O
+ATOM 2160 CB THR A 335 10.742 -93.369 -11.846 1.00 72.35 A C
+ATOM 2161 CG2 THR A 335 11.373 -93.360 -13.229 1.00 73.23 A C
+ATOM 2162 OG1 THR A 335 10.290 -94.694 -11.565 1.00 78.77 A O
+ATOM 2163 N ARG A 336 13.387 -91.302 -11.645 1.00 63.13 A N
+ATOM 2164 CA ARG A 336 13.853 -89.955 -11.971 1.00 70.26 A C
+ATOM 2165 C ARG A 336 13.029 -89.366 -13.106 1.00 72.94 A C
+ATOM 2166 O ARG A 336 12.734 -90.041 -14.091 1.00 61.11 A O
+ATOM 2167 CB ARG A 336 15.313 -89.955 -12.399 1.00 62.41 A C
+ATOM 2168 CG ARG A 336 16.321 -90.448 -11.434 1.00 63.44 A C
+ATOM 2169 CD ARG A 336 17.664 -90.407 -12.137 1.00 68.16 A C
+ATOM 2170 NE ARG A 336 18.016 -89.051 -12.563 1.00 68.92 A N
+ATOM 2171 CZ ARG A 336 19.092 -88.733 -13.287 1.00 60.10 A C
+ATOM 2172 NH1 ARG A 336 19.941 -89.670 -13.690 1.00 65.52 A N1+
+ATOM 2173 NH2 ARG A 336 19.322 -87.463 -13.598 1.00 67.04 A N
+ATOM 2174 N THR A 337 12.647 -88.105 -12.974 1.00 68.06 A N
+ATOM 2175 CA THR A 337 11.811 -87.473 -13.983 1.00 65.45 A C
+ATOM 2176 C THR A 337 12.560 -86.536 -14.941 1.00 74.66 A C
+ATOM 2177 O THR A 337 11.934 -85.938 -15.826 1.00 66.45 A O
+ATOM 2178 CB THR A 337 10.668 -86.764 -13.282 1.00 66.57 A C
+ATOM 2179 CG2 THR A 337 9.806 -87.812 -12.600 1.00 64.50 A C
+ATOM 2180 OG1 THR A 337 11.204 -85.893 -12.284 1.00 69.07 A O
+ATOM 2181 N ASP A 338 13.880 -86.433 -14.837 1.00 67.29 A N
+ATOM 2182 CA ASP A 338 14.688 -85.519 -15.653 1.00 62.79 A C
+ATOM 2183 C ASP A 338 14.998 -86.067 -17.081 1.00 65.64 A C
+ATOM 2184 O ASP A 338 16.016 -85.699 -17.681 1.00 74.18 A O
+ATOM 2185 CB ASP A 338 15.945 -85.045 -14.881 1.00 64.29 A C
+ATOM 2186 CG ASP A 338 16.578 -86.122 -13.969 1.00 75.12 A C
+ATOM 2187 OD1 ASP A 338 16.150 -87.295 -14.007 1.00 72.94 A O
+ATOM 2188 OD2 ASP A 338 17.529 -85.780 -13.210 1.00 71.80 A O1-
+ATOM 2189 N ARG A 339 14.139 -86.968 -17.580 1.00 66.63 A N
+ATOM 2190 CA ARG A 339 14.235 -87.556 -18.920 1.00 63.23 A C
+ATOM 2191 C ARG A 339 14.526 -86.499 -19.971 1.00 61.44 A C
+ATOM 2192 O ARG A 339 14.092 -85.347 -19.847 1.00 59.55 A O
+ATOM 2193 CB ARG A 339 12.900 -88.213 -19.314 1.00 61.15 A C
+ATOM 2194 CG ARG A 339 12.736 -89.670 -18.988 1.00 74.43 A C
+ATOM 2195 CD ARG A 339 11.260 -90.093 -19.054 1.00 89.28 A C
+ATOM 2196 NE ARG A 339 11.075 -91.459 -18.547 1.00121.85 A N
+ATOM 2197 CZ ARG A 339 9.913 -92.111 -18.504 1.00116.95 A C
+ATOM 2198 NH1 ARG A 339 8.802 -91.534 -18.948 1.00104.58 A N1+
+ATOM 2199 NH2 ARG A 339 9.865 -93.347 -18.014 1.00114.90 A N
+ATOM 2200 N GLY A 340 15.254 -86.913 -21.027 1.00 57.14 A N
+ATOM 2201 CA GLY A 340 15.383 -86.120 -22.242 1.00 56.44 A C
+ATOM 2202 C GLY A 340 16.768 -85.533 -22.462 1.00 64.00 A C
+ATOM 2203 O GLY A 340 17.763 -85.957 -21.867 1.00 69.87 A O
+ATOM 2204 N TRP A 341 16.818 -84.571 -23.393 1.00 62.26 A N
+ATOM 2205 CA TRP A 341 18.067 -84.048 -23.947 1.00 59.86 A C
+ATOM 2206 C TRP A 341 18.640 -82.870 -23.158 1.00 67.97 A C
+ATOM 2207 O TRP A 341 17.902 -82.010 -22.671 1.00 63.25 A O
+ATOM 2208 CB TRP A 341 17.846 -83.613 -25.395 1.00 60.63 A C
+ATOM 2209 CG TRP A 341 17.892 -84.745 -26.360 1.00 58.55 A C
+ATOM 2210 CD1 TRP A 341 16.831 -85.384 -26.921 1.00 53.94 A C
+ATOM 2211 CD2 TRP A 341 19.065 -85.391 -26.866 1.00 55.12 A C
+ATOM 2212 CE2 TRP A 341 18.642 -86.409 -27.730 1.00 60.32 A C
+ATOM 2213 CE3 TRP A 341 20.432 -85.203 -26.670 1.00 66.73 A C
+ATOM 2214 NE1 TRP A 341 17.273 -86.382 -27.757 1.00 65.00 A N
+ATOM 2215 CZ2 TRP A 341 19.535 -87.242 -28.393 1.00 60.31 A C
+ATOM 2216 CZ3 TRP A 341 21.319 -86.032 -27.337 1.00 71.40 A C
+ATOM 2217 CH2 TRP A 341 20.867 -87.038 -28.184 1.00 67.23 A C
+ATOM 2218 N TYR A 342 19.973 -82.824 -23.060 1.00 66.93 A N
+ATOM 2219 CA TYR A 342 20.690 -81.711 -22.443 1.00 55.35 A C
+ATOM 2220 C TYR A 342 21.827 -81.270 -23.354 1.00 71.68 A C
+ATOM 2221 O TYR A 342 22.589 -82.108 -23.850 1.00 65.66 A O
+ATOM 2222 CB TYR A 342 21.254 -82.083 -21.058 1.00 55.37 A C
+ATOM 2223 CG TYR A 342 20.212 -82.450 -20.006 1.00 65.55 A C
+ATOM 2224 CD1 TYR A 342 19.661 -83.722 -19.949 1.00 61.78 A C
+ATOM 2225 CD2 TYR A 342 19.783 -81.523 -19.072 1.00 57.18 A C
+ATOM 2226 CE1 TYR A 342 18.709 -84.052 -19.005 1.00 62.08 A C
+ATOM 2227 CE2 TYR A 342 18.832 -81.845 -18.133 1.00 60.80 A C
+ATOM 2228 CZ TYR A 342 18.304 -83.108 -18.106 1.00 66.76 A C
+ATOM 2229 OH TYR A 342 17.361 -83.429 -17.163 1.00 67.70 A O
+ATOM 2230 N CYS A 343 21.947 -79.953 -23.571 1.00 60.79 A N
+ATOM 2231 CA CYS A 343 23.069 -79.405 -24.326 1.00 57.89 A C
+ATOM 2232 C CYS A 343 23.692 -78.225 -23.602 1.00 63.76 A C
+ATOM 2233 O CYS A 343 22.992 -77.406 -22.993 1.00 73.42 A O
+ATOM 2234 CB CYS A 343 22.666 -78.940 -25.719 1.00 58.63 A C
+ATOM 2235 SG CYS A 343 22.147 -80.248 -26.795 1.00 73.67 A S
+ATOM 2236 N ASP A 344 25.014 -78.129 -23.701 1.00 66.33 A N
+ATOM 2237 CA ASP A 344 25.692 -76.936 -23.212 1.00 67.40 A C
+ATOM 2238 C ASP A 344 25.243 -75.731 -24.044 1.00 73.06 A C
+ATOM 2239 O ASP A 344 25.349 -75.743 -25.276 1.00 69.88 A O
+ATOM 2240 CB ASP A 344 27.223 -77.108 -23.300 1.00 65.32 A C
+ATOM 2241 CG ASP A 344 27.804 -78.116 -22.282 1.00 76.00 A C
+ATOM 2242 OD1 ASP A 344 27.042 -78.820 -21.581 1.00 80.85 A O
+ATOM 2243 OD2 ASP A 344 29.052 -78.189 -22.193 1.00 89.96 A O1-
+ATOM 2244 N ASN A 345 24.716 -74.700 -23.387 1.00 73.34 A N
+ATOM 2245 CA ASN A 345 24.368 -73.456 -24.073 1.00 72.89 A C
+ATOM 2246 C ASN A 345 24.821 -72.293 -23.197 1.00 71.39 A C
+ATOM 2247 O ASN A 345 24.230 -72.054 -22.137 1.00 74.39 A O
+ATOM 2248 CB ASN A 345 22.853 -73.409 -24.329 1.00 63.46 A C
+ATOM 2249 CG ASN A 345 22.420 -72.307 -25.304 1.00 72.17 A C
+ATOM 2250 ND2 ASN A 345 21.417 -72.614 -26.111 1.00 61.89 A N
+ATOM 2251 OD1 ASN A 345 22.934 -71.190 -25.292 1.00 66.49 A O
+ATOM 2252 N ALA A 346 25.832 -71.545 -23.649 1.00 76.67 A N
+ATOM 2253 CA ALA A 346 26.171 -70.228 -23.077 1.00 71.85 A C
+ATOM 2254 C ALA A 346 26.448 -70.277 -21.569 1.00 77.34 A C
+ATOM 2255 O ALA A 346 25.903 -69.488 -20.789 1.00 76.88 A O
+ATOM 2256 CB ALA A 346 25.066 -69.208 -23.379 1.00 73.36 A C
+ATOM 2257 N GLY A 347 27.306 -71.214 -21.154 1.00 77.96 A N
+ATOM 2258 CA GLY A 347 27.613 -71.396 -19.747 1.00 75.51 A C
+ATOM 2259 C GLY A 347 26.526 -72.058 -18.925 1.00 75.00 A C
+ATOM 2260 O GLY A 347 26.795 -72.481 -17.790 1.00 75.74 A O
+ATOM 2261 N SER A 348 25.315 -72.171 -19.449 1.00 70.91 A N
+ATOM 2262 CA SER A 348 24.267 -72.940 -18.810 1.00 70.17 A C
+ATOM 2263 C SER A 348 24.092 -74.260 -19.544 1.00 74.41 A C
+ATOM 2264 O SER A 348 24.785 -74.573 -20.513 1.00 73.01 A O
+ATOM 2265 CB SER A 348 22.959 -72.153 -18.787 1.00 70.18 A C
+ATOM 2266 OG SER A 348 23.028 -71.111 -17.831 1.00 75.82 A O
+ATOM 2267 N VAL A 349 23.155 -75.056 -19.062 1.00 67.57 A N
+ATOM 2268 CA VAL A 349 22.700 -76.240 -19.770 1.00 66.77 A C
+ATOM 2269 C VAL A 349 21.238 -76.016 -20.091 1.00 71.41 A C
+ATOM 2270 O VAL A 349 20.466 -75.620 -19.208 1.00 67.22 A O
+ATOM 2271 CB VAL A 349 22.916 -77.519 -18.943 1.00 70.92 A C
+ATOM 2272 CG1 VAL A 349 21.916 -78.584 -19.307 1.00 69.82 A C
+ATOM 2273 CG2 VAL A 349 24.294 -78.042 -19.219 1.00 65.79 A C
+ATOM 2274 N SER A 350 20.885 -76.249 -21.346 1.00 61.88 A N
+ATOM 2275 CA SER A 350 19.518 -76.116 -21.776 1.00 59.55 A C
+ATOM 2276 C SER A 350 18.941 -77.449 -21.364 1.00 70.07 A C
+ATOM 2277 O SER A 350 19.641 -78.276 -20.779 1.00 67.22 A O
+ATOM 2278 CB SER A 350 19.437 -75.941 -23.290 1.00 62.15 A C
+ATOM 2279 OG SER A 350 20.730 -75.844 -23.862 1.00 67.95 A O
+ATOM 2280 N PHE A 351 17.675 -77.672 -21.662 1.00 61.92 A N
+ATOM 2281 CA PHE A 351 17.053 -78.931 -21.301 1.00 62.12 A C
+ATOM 2282 C PHE A 351 15.804 -79.084 -22.145 1.00 72.03 A C
+ATOM 2283 O PHE A 351 14.905 -78.229 -22.097 1.00 78.00 A O
+ATOM 2284 CB PHE A 351 16.694 -78.934 -19.813 1.00 58.25 A C
+ATOM 2285 CG PHE A 351 15.884 -80.123 -19.365 1.00 66.63 A C
+ATOM 2286 CD1 PHE A 351 15.986 -81.346 -19.994 1.00 67.41 A C
+ATOM 2287 CD2 PHE A 351 14.990 -79.995 -18.321 1.00 66.14 A C
+ATOM 2288 CE1 PHE A 351 15.214 -82.425 -19.575 1.00 72.41 A C
+ATOM 2289 CE2 PHE A 351 14.228 -81.067 -17.901 1.00 69.33 A C
+ATOM 2290 CZ PHE A 351 14.335 -82.286 -18.527 1.00 71.60 A C
+ATOM 2291 N PHE A 352 15.755 -80.175 -22.903 1.00 62.99 A N
+ATOM 2292 CA PHE A 352 14.604 -80.532 -23.715 1.00 61.93 A C
+ATOM 2293 C PHE A 352 13.925 -81.737 -23.090 1.00 68.66 A C
+ATOM 2294 O PHE A 352 14.453 -82.862 -23.146 1.00 70.38 A O
+ATOM 2295 CB PHE A 352 15.065 -80.809 -25.139 1.00 62.07 A C
+ATOM 2296 CG PHE A 352 15.986 -79.757 -25.640 1.00 66.79 A C
+ATOM 2297 CD1 PHE A 352 15.484 -78.551 -26.087 1.00 66.49 A C
+ATOM 2298 CD2 PHE A 352 17.351 -79.937 -25.603 1.00 65.48 A C
+ATOM 2299 CE1 PHE A 352 16.323 -77.558 -26.530 1.00 66.06 A C
+ATOM 2300 CE2 PHE A 352 18.206 -78.944 -26.050 1.00 70.83 A C
+ATOM 2301 CZ PHE A 352 17.691 -77.757 -26.517 1.00 69.43 A C
+ATOM 2302 N PRO A 353 12.797 -81.525 -22.414 1.00 66.34 A N
+ATOM 2303 CA PRO A 353 12.119 -82.618 -21.721 1.00 67.45 A C
+ATOM 2304 C PRO A 353 11.352 -83.542 -22.644 1.00 68.13 A C
+ATOM 2305 O PRO A 353 10.917 -84.614 -22.201 1.00 71.99 A O
+ATOM 2306 CB PRO A 353 11.188 -81.872 -20.759 1.00 60.83 A C
+ATOM 2307 CG PRO A 353 10.823 -80.643 -21.522 1.00 58.61 A C
+ATOM 2308 CD PRO A 353 11.967 -80.311 -22.464 1.00 59.26 A C
+ATOM 2309 N GLN A 354 11.113 -83.132 -23.884 1.00 63.86 A N
+ATOM 2310 CA GLN A 354 10.196 -83.815 -24.788 1.00 69.82 A C
+ATOM 2311 C GLN A 354 10.983 -84.362 -25.972 1.00 74.43 A C
+ATOM 2312 O GLN A 354 11.630 -83.594 -26.690 1.00 74.72 A O
+ATOM 2313 CB GLN A 354 9.111 -82.839 -25.259 1.00 60.01 A C
+ATOM 2314 CG GLN A 354 8.459 -82.031 -24.144 1.00 84.61 A C
+ATOM 2315 CD GLN A 354 6.952 -82.038 -24.226 1.00120.59 A C
+ATOM 2316 NE2 GLN A 354 6.309 -82.868 -23.405 1.00136.37 A N
+ATOM 2317 OE1 GLN A 354 6.369 -81.306 -25.024 1.00125.24 A O
+ATOM 2318 N ALA A 355 10.930 -85.682 -26.188 1.00 89.59 A N
+ATOM 2319 CA ALA A 355 11.707 -86.250 -27.293 1.00 86.31 A C
+ATOM 2320 C ALA A 355 11.236 -85.741 -28.652 1.00 85.54 A C
+ATOM 2321 O ALA A 355 11.994 -85.824 -29.625 1.00 91.19 A O
+ATOM 2322 CB ALA A 355 11.669 -87.787 -27.274 1.00 72.10 A C
+ATOM 2323 N GLU A 356 10.012 -85.210 -28.737 1.00 79.04 A N
+ATOM 2324 CA GLU A 356 9.546 -84.632 -29.993 1.00 83.17 A C
+ATOM 2325 C GLU A 356 10.329 -83.370 -30.364 1.00 78.45 A C
+ATOM 2326 O GLU A 356 10.556 -83.113 -31.555 1.00 92.89 A O
+ATOM 2327 CB GLU A 356 8.045 -84.329 -29.904 1.00 94.89 A C
+ATOM 2328 CG GLU A 356 7.537 -83.382 -30.996 1.00121.49 A C
+ATOM 2329 CD GLU A 356 6.189 -82.759 -30.673 1.00134.61 A C
+ATOM 2330 OE1 GLU A 356 5.520 -83.220 -29.718 1.00134.99 A O
+ATOM 2331 OE2 GLU A 356 5.808 -81.797 -31.377 1.00136.96 A O1-
+ATOM 2332 N THR A 357 10.746 -82.571 -29.371 1.00 74.82 A N
+ATOM 2333 CA THR A 357 11.484 -81.347 -29.676 1.00 76.60 A C
+ATOM 2334 C THR A 357 12.803 -81.652 -30.386 1.00 79.68 A C
+ATOM 2335 O THR A 357 13.258 -80.859 -31.224 1.00 72.32 A O
+ATOM 2336 CB THR A 357 11.731 -80.527 -28.398 1.00 72.07 A C
+ATOM 2337 CG2 THR A 357 12.598 -79.307 -28.689 1.00 73.61 A C
+ATOM 2338 OG1 THR A 357 10.483 -80.093 -27.840 1.00 83.65 A O
+ATOM 2339 N CYS A 358 13.405 -82.805 -30.113 1.00 73.26 A N
+ATOM 2340 CA CYS A 358 14.732 -83.112 -30.616 1.00 66.18 A C
+ATOM 2341 C CYS A 358 14.681 -84.179 -31.692 1.00 71.12 A C
+ATOM 2342 O CYS A 358 13.980 -85.186 -31.556 1.00 74.15 A O
+ATOM 2343 CB CYS A 358 15.631 -83.565 -29.487 1.00 59.81 A C
+ATOM 2344 SG CYS A 358 15.952 -82.237 -28.361 1.00 68.83 A S
+ATOM 2345 N LYS A 359 15.432 -83.949 -32.760 1.00 59.86 A N
+ATOM 2346 CA LYS A 359 15.603 -84.909 -33.837 1.00 58.91 A C
+ATOM 2347 C LYS A 359 17.093 -85.182 -33.972 1.00 63.56 A C
+ATOM 2348 O LYS A 359 17.916 -84.271 -33.833 1.00 61.06 A O
+ATOM 2349 CB LYS A 359 14.998 -84.368 -35.138 1.00 75.26 A C
+ATOM 2350 CG LYS A 359 13.611 -83.785 -34.911 1.00 95.07 A C
+ATOM 2351 CD LYS A 359 13.203 -82.792 -35.973 1.00 97.02 A C
+ATOM 2352 CE LYS A 359 11.979 -82.015 -35.509 1.00 81.18 A C
+ATOM 2353 NZ LYS A 359 12.321 -81.050 -34.400 1.00 84.62 A N1+
+ATOM 2354 N VAL A 360 17.439 -86.437 -34.214 1.00 68.13 A N
+ATOM 2355 CA VAL A 360 18.817 -86.890 -34.108 1.00 59.62 A C
+ATOM 2356 C VAL A 360 19.320 -87.258 -35.497 1.00 64.38 A C
+ATOM 2357 O VAL A 360 18.997 -88.343 -35.995 1.00 64.89 A O
+ATOM 2358 CB VAL A 360 18.908 -88.105 -33.181 1.00 65.07 A C
+ATOM 2359 CG1 VAL A 360 20.309 -88.256 -32.668 1.00 64.15 A C
+ATOM 2360 CG2 VAL A 360 17.876 -88.005 -32.079 1.00 59.72 A C
+ATOM 2361 N GLN A 361 20.179 -86.438 -36.116 1.00 57.94 A N
+ATOM 2362 CA GLN A 361 20.818 -86.913 -37.344 1.00 65.10 A C
+ATOM 2363 C GLN A 361 22.276 -87.230 -37.015 1.00 70.07 A C
+ATOM 2364 O GLN A 361 23.153 -86.361 -36.995 1.00 74.35 A O
+ATOM 2365 CB GLN A 361 20.678 -85.915 -38.489 1.00 59.61 A C
+ATOM 2366 CG GLN A 361 19.235 -85.582 -38.833 1.00 71.58 A C
+ATOM 2367 CD GLN A 361 18.531 -86.610 -39.724 1.00 84.62 A C
+ATOM 2368 NE2 GLN A 361 17.583 -87.358 -39.147 1.00 80.13 A N
+ATOM 2369 OE1 GLN A 361 18.793 -86.687 -40.926 1.00 87.02 A O
+ATOM 2370 N SER A 362 22.487 -88.546 -36.970 1.00 78.63 A N
+ATOM 2371 CA SER A 362 23.776 -89.174 -36.773 1.00 79.81 A C
+ATOM 2372 C SER A 362 24.308 -88.714 -35.464 1.00 71.90 A C
+ATOM 2373 O SER A 362 23.865 -89.164 -34.407 1.00 71.84 A O
+ATOM 2374 CB SER A 362 24.738 -88.789 -37.898 1.00 84.83 A C
+ATOM 2375 OG SER A 362 25.304 -89.940 -38.502 1.00 91.81 A O
+ATOM 2376 N ASN A 363 25.259 -87.801 -35.518 1.00 76.23 A N
+ATOM 2377 CA ASN A 363 25.776 -87.315 -34.238 1.00 77.73 A C
+ATOM 2378 C ASN A 363 25.289 -85.924 -33.844 1.00 72.04 A C
+ATOM 2379 O ASN A 363 25.599 -85.480 -32.732 1.00 62.25 A O
+ATOM 2380 CB ASN A 363 27.320 -87.391 -34.172 1.00 88.22 A C
+ATOM 2381 CG ASN A 363 28.008 -87.123 -35.504 1.00 96.97 A C
+ATOM 2382 ND2 ASN A 363 29.077 -87.877 -35.763 1.00 97.18 A N
+ATOM 2383 OD1 ASN A 363 27.597 -86.262 -36.289 1.00 99.26 A O
+ATOM 2384 N ARG A 364 24.587 -85.206 -34.723 1.00 59.75 A N
+ATOM 2385 CA ARG A 364 24.064 -83.891 -34.388 1.00 56.48 A C
+ATOM 2386 C ARG A 364 22.640 -84.090 -33.860 1.00 53.27 A C
+ATOM 2387 O ARG A 364 21.917 -84.992 -34.297 1.00 65.34 A O
+ATOM 2388 CB ARG A 364 24.122 -82.927 -35.605 1.00 61.76 A C
+ATOM 2389 CG ARG A 364 25.533 -82.235 -35.990 1.00 66.75 A C
+ATOM 2390 CD ARG A 364 25.498 -81.403 -37.345 1.00 75.86 A C
+ATOM 2391 NE ARG A 364 26.635 -81.524 -38.316 1.00 73.83 A N
+ATOM 2392 CZ ARG A 364 26.561 -81.909 -39.613 1.00 68.36 A C
+ATOM 2393 NH1 ARG A 364 25.437 -82.318 -40.151 1.00 66.58 A N1+
+ATOM 2394 NH2 ARG A 364 27.633 -81.933 -40.391 1.00 66.91 A N
+ATOM 2395 N VAL A 365 22.285 -83.308 -32.850 1.00 56.17 A N
+ATOM 2396 CA VAL A 365 20.967 -83.296 -32.231 1.00 53.14 A C
+ATOM 2397 C VAL A 365 20.363 -81.931 -32.500 1.00 60.48 A C
+ATOM 2398 O VAL A 365 20.763 -80.934 -31.886 1.00 60.45 A O
+ATOM 2399 CB VAL A 365 21.055 -83.558 -30.726 1.00 58.03 A C
+ATOM 2400 CG1 VAL A 365 19.692 -83.716 -30.128 1.00 64.18 A C
+ATOM 2401 CG2 VAL A 365 21.864 -84.794 -30.489 1.00 72.48 A C
+ATOM 2402 N PHE A 366 19.395 -81.879 -33.395 1.00 58.52 A N
+ATOM 2403 CA PHE A 366 18.695 -80.645 -33.697 1.00 57.73 A C
+ATOM 2404 C PHE A 366 17.548 -80.494 -32.707 1.00 62.93 A C
+ATOM 2405 O PHE A 366 16.648 -81.343 -32.678 1.00 67.62 A O
+ATOM 2406 CB PHE A 366 18.149 -80.683 -35.121 1.00 51.33 A C
+ATOM 2407 CG PHE A 366 19.194 -80.573 -36.190 1.00 52.72 A C
+ATOM 2408 CD1 PHE A 366 19.941 -81.678 -36.568 1.00 53.22 A C
+ATOM 2409 CD2 PHE A 366 19.396 -79.374 -36.851 1.00 53.72 A C
+ATOM 2410 CE1 PHE A 366 20.886 -81.590 -37.571 1.00 58.79 A C
+ATOM 2411 CE2 PHE A 366 20.353 -79.277 -37.850 1.00 56.89 A C
+ATOM 2412 CZ PHE A 366 21.097 -80.391 -38.211 1.00 58.07 A C
+ATOM 2413 N CYS A 367 17.562 -79.422 -31.906 1.00 65.25 A N
+ATOM 2414 CA CYS A 367 16.473 -79.197 -30.954 1.00 61.61 A C
+ATOM 2415 C CYS A 367 15.944 -77.785 -31.082 1.00 70.28 A C
+ATOM 2416 O CYS A 367 16.560 -76.921 -31.704 1.00 77.54 A O
+ATOM 2417 CB CYS A 367 16.894 -79.456 -29.506 1.00 58.06 A C
+ATOM 2418 SG CYS A 367 17.468 -81.154 -29.249 1.00 68.16 A S
+ATOM 2419 N ASP A 368 14.780 -77.565 -30.492 1.00 66.07 A N
+ATOM 2420 CA ASP A 368 14.118 -76.274 -30.566 1.00 55.30 A C
+ATOM 2421 C ASP A 368 14.109 -75.643 -29.182 1.00 64.09 A C
+ATOM 2422 O ASP A 368 13.437 -76.140 -28.273 1.00 65.16 A O
+ATOM 2423 CB ASP A 368 12.707 -76.416 -31.114 1.00 52.77 A C
+ATOM 2424 CG ASP A 368 12.173 -75.106 -31.661 1.00 53.95 A C
+ATOM 2425 OD1 ASP A 368 12.854 -74.077 -31.448 1.00 66.55 A O
+ATOM 2426 OD2 ASP A 368 11.091 -75.096 -32.300 1.00 70.59 A O1-
+ATOM 2427 N THR A 369 14.838 -74.535 -29.025 1.00 55.10 A N
+ATOM 2428 CA THR A 369 14.933 -73.928 -27.707 1.00 64.77 A C
+ATOM 2429 C THR A 369 13.583 -73.449 -27.216 1.00 58.83 A C
+ATOM 2430 O THR A 369 13.406 -73.312 -25.999 1.00 64.60 A O
+ATOM 2431 CB THR A 369 15.895 -72.746 -27.707 1.00 65.27 A C
+ATOM 2432 CG2 THR A 369 17.235 -73.164 -28.307 1.00 63.76 A C
+ATOM 2433 OG1 THR A 369 15.342 -71.675 -28.480 1.00 68.44 A O
+ATOM 2434 N MET A 370 12.631 -73.191 -28.130 1.00 63.44 A N
+ATOM 2435 CA MET A 370 11.313 -72.696 -27.719 1.00 65.09 A C
+ATOM 2436 C MET A 370 10.661 -73.619 -26.692 1.00 66.96 A C
+ATOM 2437 O MET A 370 10.116 -73.145 -25.686 1.00 70.03 A O
+ATOM 2438 CB MET A 370 10.384 -72.512 -28.915 1.00 63.66 A C
+ATOM 2439 CG MET A 370 9.175 -71.674 -28.538 1.00 82.88 A C
+ATOM 2440 SD MET A 370 7.902 -71.577 -29.810 1.00 93.00 A S
+ATOM 2441 CE MET A 370 8.840 -72.014 -31.276 1.00100.38 A C
+ATOM 2442 N ASN A 371 10.711 -74.943 -26.912 1.00 65.40 A N
+ATOM 2443 CA ASN A 371 10.315 -75.861 -25.846 1.00 65.08 A C
+ATOM 2444 C ASN A 371 11.577 -76.424 -25.197 1.00 68.11 A C
+ATOM 2445 O ASN A 371 12.038 -77.520 -25.521 1.00 67.40 A O
+ATOM 2446 CB ASN A 371 9.429 -76.960 -26.410 1.00 77.00 A C
+ATOM 2447 CG ASN A 371 7.968 -76.553 -26.451 1.00 92.45 A C
+ATOM 2448 ND2 ASN A 371 7.059 -77.541 -26.417 1.00 79.91 A N
+ATOM 2449 OD1 ASN A 371 7.657 -75.359 -26.471 1.00108.09 A O
+ATOM 2450 N SER A 372 12.043 -75.714 -24.172 1.00 65.04 A N
+ATOM 2451 CA SER A 372 13.265 -76.004 -23.436 1.00 61.37 A C
+ATOM 2452 C SER A 372 13.253 -75.163 -22.177 1.00 62.69 A C
+ATOM 2453 O SER A 372 12.595 -74.120 -22.125 1.00 67.83 A O
+ATOM 2454 CB SER A 372 14.511 -75.672 -24.253 1.00 62.98 A C
+ATOM 2455 OG SER A 372 14.805 -74.289 -24.131 1.00 62.96 A O
+ATOM 2456 N LEU A 373 14.050 -75.578 -21.195 1.00 69.45 A N
+ATOM 2457 CA LEU A 373 14.382 -74.753 -20.035 1.00 60.25 A C
+ATOM 2458 C LEU A 373 15.891 -74.564 -19.990 1.00 62.43 A C
+ATOM 2459 O LEU A 373 16.639 -75.428 -20.441 1.00 67.43 A O
+ATOM 2460 CB LEU A 373 13.922 -75.400 -18.728 1.00 60.05 A C
+ATOM 2461 CG LEU A 373 12.456 -75.804 -18.681 1.00 70.84 A C
+ATOM 2462 CD1 LEU A 373 12.137 -76.434 -17.348 1.00 68.20 A C
+ATOM 2463 CD2 LEU A 373 11.544 -74.616 -18.948 1.00 65.04 A C
+ATOM 2464 N THR A 374 16.366 -73.454 -19.446 1.00 57.59 A N
+ATOM 2465 CA THR A 374 17.809 -73.314 -19.263 1.00 56.60 A C
+ATOM 2466 C THR A 374 18.110 -73.266 -17.770 1.00 62.79 A C
+ATOM 2467 O THR A 374 17.798 -72.280 -17.091 1.00 65.68 A O
+ATOM 2468 CB THR A 374 18.343 -72.096 -19.994 1.00 64.05 A C
+ATOM 2469 CG2 THR A 374 18.034 -72.233 -21.452 1.00 71.85 A C
+ATOM 2470 OG1 THR A 374 17.658 -70.945 -19.524 1.00 66.08 A O
+ATOM 2471 N LEU A 375 18.714 -74.365 -17.261 1.00 61.49 A N
+ATOM 2472 CA LEU A 375 19.175 -74.956 -16.014 1.00 65.45 A C
+ATOM 2473 C LEU A 375 20.651 -74.663 -15.776 1.00 64.43 A C
+ATOM 2474 O LEU A 375 21.470 -74.750 -16.705 1.00 64.35 A O
+ATOM 2475 CB LEU A 375 18.966 -76.458 -16.015 1.00 58.50 A C
+ATOM 2476 CG LEU A 375 17.610 -76.924 -16.514 1.00 68.21 A C
+ATOM 2477 CD1 LEU A 375 17.390 -78.366 -16.093 1.00 57.18 A C
+ATOM 2478 CD2 LEU A 375 16.499 -76.037 -16.004 1.00 61.97 A C
+ATOM 2479 N PRO A 376 20.975 -74.318 -14.534 1.00 76.23 A N
+ATOM 2480 CA PRO A 376 22.381 -74.141 -14.163 1.00 70.49 A C
+ATOM 2481 C PRO A 376 23.134 -75.459 -14.251 1.00 74.94 A C
+ATOM 2482 O PRO A 376 22.598 -76.529 -13.943 1.00 75.23 A O
+ATOM 2483 CB PRO A 376 22.302 -73.628 -12.721 1.00 71.44 A C
+ATOM 2484 CG PRO A 376 20.842 -73.121 -12.557 1.00 85.28 A C
+ATOM 2485 CD PRO A 376 20.058 -74.057 -13.406 1.00 76.77 A C
+ATOM 2486 N SER A 377 24.411 -75.355 -14.644 1.00 70.61 A N
+ATOM 2487 CA SER A 377 25.198 -76.519 -15.056 1.00 68.53 A C
+ATOM 2488 C SER A 377 25.149 -77.668 -14.052 1.00 67.60 A C
+ATOM 2489 O SER A 377 25.252 -78.840 -14.445 1.00 72.12 A O
+ATOM 2490 CB SER A 377 26.650 -76.094 -15.289 1.00 68.56 A C
+ATOM 2491 OG SER A 377 27.456 -77.215 -15.611 1.00 84.66 A O
+ATOM 2492 N GLU A 378 24.948 -77.356 -12.763 1.00 70.43 A N
+ATOM 2493 CA GLU A 378 25.018 -78.340 -11.683 1.00 74.35 A C
+ATOM 2494 C GLU A 378 23.984 -79.451 -11.822 1.00 77.12 A C
+ATOM 2495 O GLU A 378 24.027 -80.425 -11.060 1.00 85.42 A O
+ATOM 2496 CB GLU A 378 24.848 -77.659 -10.318 1.00 72.15 A C
+ATOM 2497 CG GLU A 378 26.068 -76.879 -9.833 1.00 81.54 A C
+ATOM 2498 CD GLU A 378 26.329 -75.633 -10.654 1.00 90.49 A C
+ATOM 2499 OE1 GLU A 378 25.345 -75.031 -11.131 1.00 89.02 A O
+ATOM 2500 OE2 GLU A 378 27.508 -75.264 -10.842 1.00 96.87 A O1-
+ATOM 2501 N VAL A 379 23.045 -79.327 -12.756 1.00 75.84 A N
+ATOM 2502 CA VAL A 379 22.144 -80.440 -13.009 1.00 74.33 A C
+ATOM 2503 C VAL A 379 22.906 -81.675 -13.478 1.00 76.35 A C
+ATOM 2504 O VAL A 379 22.460 -82.807 -13.243 1.00 85.41 A O
+ATOM 2505 CB VAL A 379 21.064 -80.002 -14.004 1.00 77.36 A C
+ATOM 2506 CG1 VAL A 379 21.714 -79.405 -15.215 1.00 64.19 A C
+ATOM 2507 CG2 VAL A 379 20.198 -81.186 -14.388 1.00 73.25 A C
+ATOM 2508 N ASN A 380 24.094 -81.501 -14.078 1.00 74.44 A N
+ATOM 2509 CA ASN A 380 24.853 -82.695 -14.450 1.00 77.46 A C
+ATOM 2510 C ASN A 380 25.262 -83.495 -13.228 1.00 75.75 A C
+ATOM 2511 O ASN A 380 25.825 -84.586 -13.382 1.00 76.88 A O
+ATOM 2512 CB ASN A 380 26.101 -82.370 -15.297 1.00 78.37 A C
+ATOM 2513 CG ASN A 380 26.628 -83.608 -16.104 1.00 89.96 A C
+ATOM 2514 ND2 ASN A 380 26.513 -84.797 -15.523 1.00 93.91 A N
+ATOM 2515 OD1 ASN A 380 27.129 -83.474 -17.228 1.00 91.63 A O
+ATOM 2516 N LEU A 381 24.995 -82.988 -12.022 1.00 81.16 A N
+ATOM 2517 CA LEU A 381 25.340 -83.744 -10.827 1.00 76.16 A C
+ATOM 2518 C LEU A 381 24.568 -85.061 -10.749 1.00 78.46 A C
+ATOM 2519 O LEU A 381 25.161 -86.119 -10.512 1.00 82.39 A O
+ATOM 2520 CB LEU A 381 25.126 -82.888 -9.579 1.00 73.12 A C
+ATOM 2521 CG LEU A 381 26.136 -81.739 -9.450 1.00 84.24 A C
+ATOM 2522 CD1 LEU A 381 26.186 -81.275 -8.028 1.00 76.64 A C
+ATOM 2523 CD2 LEU A 381 27.539 -82.076 -9.959 1.00 75.44 A C
+ATOM 2524 N CYS A 382 23.262 -85.034 -11.005 1.00 73.23 A N
+ATOM 2525 CA CYS A 382 22.442 -86.186 -10.632 1.00 81.63 A C
+ATOM 2526 C CYS A 382 22.746 -87.453 -11.389 1.00 82.45 A C
+ATOM 2527 O CYS A 382 22.193 -88.512 -11.062 1.00 79.78 A O
+ATOM 2528 CB CYS A 382 21.002 -85.823 -10.785 1.00 75.03 A C
+ATOM 2529 SG CYS A 382 20.843 -84.578 -9.588 1.00 81.78 A S
+ATOM 2530 N ASN A 383 23.590 -87.374 -12.387 1.00 73.44 A N
+ATOM 2531 CA ASN A 383 24.120 -88.592 -12.946 1.00 79.10 A C
+ATOM 2532 C ASN A 383 25.240 -89.172 -12.076 1.00 78.74 A C
+ATOM 2533 O ASN A 383 25.513 -90.375 -12.177 1.00 95.67 A O
+ATOM 2534 CB ASN A 383 24.507 -88.272 -14.389 1.00 73.60 A C
+ATOM 2535 CG ASN A 383 23.370 -87.524 -15.128 1.00 73.82 A C
+ATOM 2536 ND2 ASN A 383 23.710 -86.739 -16.151 1.00 71.45 A N
+ATOM 2537 OD1 ASN A 383 22.205 -87.638 -14.740 1.00 77.12 A O
+ATOM 2538 N VAL A 384 25.821 -88.366 -11.160 1.00 70.56 A N
+ATOM 2539 CA VAL A 384 26.795 -88.829 -10.160 1.00 84.52 A C
+ATOM 2540 C VAL A 384 26.230 -88.883 -8.732 1.00 85.95 A C
+ATOM 2541 O VAL A 384 25.827 -89.955 -8.258 1.00 87.44 A O
+ATOM 2542 CB VAL A 384 28.039 -87.931 -10.194 1.00100.53 A C
+ATOM 2543 CG1 VAL A 384 29.191 -88.614 -9.476 1.00107.46 A C
+ATOM 2544 CG2 VAL A 384 28.381 -87.574 -11.632 1.00102.53 A C
+ATOM 2545 N ASP A 385 26.149 -87.734 -8.039 1.00101.92 A N
+ATOM 2546 CA ASP A 385 25.519 -87.685 -6.710 1.00 94.28 A C
+ATOM 2547 C ASP A 385 24.118 -87.150 -6.889 1.00 86.66 A C
+ATOM 2548 O ASP A 385 23.912 -85.932 -6.946 1.00 90.59 A O
+ATOM 2549 CB ASP A 385 26.237 -86.780 -5.709 1.00 98.21 A C
+ATOM 2550 CG ASP A 385 27.702 -87.088 -5.566 1.00111.22 A C
+ATOM 2551 OD1 ASP A 385 28.149 -88.140 -6.080 1.00113.40 A O
+ATOM 2552 OD2 ASP A 385 28.402 -86.274 -4.913 1.00128.49 A O1-
+ATOM 2553 N ILE A 386 23.145 -88.060 -6.847 1.00 88.93 A N
+ATOM 2554 CA ILE A 386 21.757 -87.645 -6.974 1.00 81.01 A C
+ATOM 2555 C ILE A 386 21.303 -86.995 -5.679 1.00 98.06 A C
+ATOM 2556 O ILE A 386 20.319 -86.232 -5.657 1.00 89.74 A O
+ATOM 2557 CB ILE A 386 20.904 -88.858 -7.389 1.00 82.28 A C
+ATOM 2558 CG1 ILE A 386 19.425 -88.493 -7.472 1.00 85.52 A C
+ATOM 2559 CG2 ILE A 386 21.135 -90.009 -6.446 1.00 83.72 A C
+ATOM 2560 CD1 ILE A 386 18.620 -89.511 -8.232 1.00 83.31 A C
+ATOM 2561 N PHE A 387 22.055 -87.241 -4.610 1.00 77.36 A N
+ATOM 2562 CA PHE A 387 21.797 -86.694 -3.295 1.00 76.30 A C
+ATOM 2563 C PHE A 387 22.571 -85.411 -3.035 1.00 83.70 A C
+ATOM 2564 O PHE A 387 22.457 -84.846 -1.943 1.00 90.15 A O
+ATOM 2565 CB PHE A 387 22.134 -87.752 -2.255 1.00 75.39 A C
+ATOM 2566 CG PHE A 387 21.130 -88.848 -2.190 1.00 80.16 A C
+ATOM 2567 CD1 PHE A 387 19.786 -88.557 -2.051 1.00 81.56 A C
+ATOM 2568 CD2 PHE A 387 21.514 -90.168 -2.311 1.00 85.36 A C
+ATOM 2569 CE1 PHE A 387 18.832 -89.577 -1.997 1.00 82.37 A C
+ATOM 2570 CE2 PHE A 387 20.566 -91.194 -2.261 1.00 82.24 A C
+ATOM 2571 CZ PHE A 387 19.222 -90.895 -2.105 1.00 82.90 A C
+ATOM 2572 N ASN A 388 23.332 -84.928 -4.012 1.00 92.68 A N
+ATOM 2573 CA ASN A 388 24.123 -83.726 -3.799 1.00 99.33 A C
+ATOM 2574 C ASN A 388 23.237 -82.501 -3.598 1.00 97.39 A C
+ATOM 2575 O ASN A 388 22.391 -82.208 -4.447 1.00 97.01 A O
+ATOM 2576 CB ASN A 388 25.038 -83.469 -4.964 1.00103.89 A C
+ATOM 2577 CG ASN A 388 26.306 -82.836 -4.520 1.00110.07 A C
+ATOM 2578 ND2 ASN A 388 27.248 -83.666 -4.079 1.00102.30 A N
+ATOM 2579 OD1 ASN A 388 26.457 -81.609 -4.571 1.00108.86 A O
+ATOM 2580 N PRO A 389 23.430 -81.738 -2.522 1.00 99.64 A N
+ATOM 2581 CA PRO A 389 22.496 -80.642 -2.216 1.00 97.28 A C
+ATOM 2582 C PRO A 389 22.632 -79.426 -3.123 1.00 94.14 A C
+ATOM 2583 O PRO A 389 21.712 -78.597 -3.139 1.00111.27 A O
+ATOM 2584 CB PRO A 389 22.832 -80.295 -0.760 1.00 94.36 A C
+ATOM 2585 CG PRO A 389 24.310 -80.587 -0.662 1.00 94.50 A C
+ATOM 2586 CD PRO A 389 24.633 -81.694 -1.673 1.00 95.26 A C
+ATOM 2587 N LYS A 390 23.748 -79.271 -3.850 1.00 90.27 A N
+ATOM 2588 CA LYS A 390 23.922 -78.076 -4.679 1.00 93.76 A C
+ATOM 2589 C LYS A 390 22.883 -77.994 -5.789 1.00100.43 A C
+ATOM 2590 O LYS A 390 22.523 -76.890 -6.224 1.00 95.24 A O
+ATOM 2591 CB LYS A 390 25.332 -78.023 -5.277 1.00 89.84 A C
+ATOM 2592 CG LYS A 390 26.384 -77.512 -4.304 1.00 95.10 A C
+ATOM 2593 CD LYS A 390 27.557 -76.847 -5.009 1.00108.32 A C
+ATOM 2594 CE LYS A 390 28.394 -77.849 -5.760 1.00121.02 A C
+ATOM 2595 NZ LYS A 390 29.551 -77.166 -6.383 1.00140.55 A N1+
+ATOM 2596 N TYR A 391 22.403 -79.140 -6.267 1.00 96.27 A N
+ATOM 2597 CA TYR A 391 21.299 -79.201 -7.215 1.00 88.66 A C
+ATOM 2598 C TYR A 391 20.318 -80.261 -6.756 1.00 88.24 A C
+ATOM 2599 O TYR A 391 20.697 -81.424 -6.589 1.00 94.83 A O
+ATOM 2600 CB TYR A 391 21.787 -79.518 -8.625 1.00 83.71 A C
+ATOM 2601 CG TYR A 391 20.685 -79.400 -9.649 1.00 89.91 A C
+ATOM 2602 CD1 TYR A 391 20.278 -78.152 -10.109 1.00 86.71 A C
+ATOM 2603 CD2 TYR A 391 20.055 -80.531 -10.163 1.00 87.92 A C
+ATOM 2604 CE1 TYR A 391 19.266 -78.026 -11.044 1.00 89.10 A C
+ATOM 2605 CE2 TYR A 391 19.046 -80.418 -11.103 1.00 86.51 A C
+ATOM 2606 CZ TYR A 391 18.656 -79.159 -11.542 1.00 91.53 A C
+ATOM 2607 OH TYR A 391 17.652 -79.014 -12.478 1.00 94.45 A O
+ATOM 2608 N ASP A 392 19.060 -79.873 -6.587 1.00 80.46 A N
+ATOM 2609 CA ASP A 392 18.050 -80.760 -6.017 1.00 82.24 A C
+ATOM 2610 C ASP A 392 17.329 -81.481 -7.159 1.00 85.32 A C
+ATOM 2611 O ASP A 392 16.521 -80.873 -7.866 1.00 83.59 A O
+ATOM 2612 CB ASP A 392 17.092 -79.937 -5.150 1.00 87.92 A C
+ATOM 2613 CG ASP A 392 15.806 -80.677 -4.782 1.00 91.75 A C
+ATOM 2614 OD1 ASP A 392 14.753 -80.013 -4.813 1.00 93.11 A O
+ATOM 2615 OD2 ASP A 392 15.831 -81.885 -4.447 1.00 94.00 A O1-
+ATOM 2616 N CYS A 393 17.578 -82.782 -7.326 1.00 80.71 A N
+ATOM 2617 CA CYS A 393 17.017 -83.473 -8.478 1.00 77.49 A C
+ATOM 2618 C CYS A 393 15.600 -83.951 -8.329 1.00 88.37 A C
+ATOM 2619 O CYS A 393 15.173 -84.410 -7.271 1.00 83.49 A O
+ATOM 2620 CB CYS A 393 17.894 -84.622 -8.933 1.00 78.32 A C
+ATOM 2621 SG CYS A 393 19.046 -83.829 -10.033 1.00 89.64 A S
+ATOM 2622 N LYS A 394 14.912 -83.908 -9.454 1.00 69.44 A N
+ATOM 2623 CA LYS A 394 13.497 -84.142 -9.530 1.00 71.71 A C
+ATOM 2624 C LYS A 394 13.243 -85.615 -9.780 1.00 74.36 A C
+ATOM 2625 O LYS A 394 13.931 -86.246 -10.589 1.00 68.37 A O
+ATOM 2626 CB LYS A 394 12.908 -83.273 -10.633 1.00 66.33 A C
+ATOM 2627 CG LYS A 394 13.204 -81.800 -10.416 1.00 68.64 A C
+ATOM 2628 CD LYS A 394 12.074 -80.966 -10.932 1.00 90.23 A C
+ATOM 2629 CE LYS A 394 10.749 -81.497 -10.426 1.00 82.78 A C
+ATOM 2630 NZ LYS A 394 10.511 -81.190 -9.005 1.00 85.83 A N1+
+ATOM 2631 N ILE A 395 12.264 -86.155 -9.047 1.00 77.76 A N
+ATOM 2632 CA ILE A 395 11.899 -87.569 -9.070 1.00 75.92 A C
+ATOM 2633 C ILE A 395 10.417 -87.698 -8.746 1.00 75.47 A C
+ATOM 2634 O ILE A 395 9.834 -86.865 -8.042 1.00 75.79 A O
+ATOM 2635 CB ILE A 395 12.717 -88.401 -8.057 1.00 73.33 A C
+ATOM 2636 CG1 ILE A 395 12.687 -87.703 -6.696 1.00 76.95 A C
+ATOM 2637 CG2 ILE A 395 14.127 -88.645 -8.555 1.00 78.31 A C
+ATOM 2638 CD1 ILE A 395 13.198 -88.541 -5.586 1.00 77.25 A C
+ATOM 2639 N MET A 396 9.816 -88.774 -9.239 1.00 77.69 A N
+ATOM 2640 CA MET A 396 8.471 -89.155 -8.852 1.00 73.87 A C
+ATOM 2641 C MET A 396 8.585 -90.397 -7.982 1.00 71.77 A C
+ATOM 2642 O MET A 396 9.553 -91.165 -8.083 1.00 73.81 A O
+ATOM 2643 CB MET A 396 7.584 -89.469 -10.057 1.00 70.15 A C
+ATOM 2644 CG MET A 396 7.444 -90.957 -10.347 1.00 86.86 A C
+ATOM 2645 SD MET A 396 6.310 -91.196 -11.695 1.00109.63 A S
+ATOM 2646 CE MET A 396 5.015 -90.094 -11.164 1.00 82.59 A C
+ATOM 2647 N THR A 397 7.605 -90.574 -7.108 1.00 75.02 A N
+ATOM 2648 CA THR A 397 7.546 -91.716 -6.216 1.00 79.84 A C
+ATOM 2649 C THR A 397 6.170 -92.350 -6.306 1.00 79.89 A C
+ATOM 2650 O THR A 397 5.168 -91.654 -6.491 1.00 90.08 A O
+ATOM 2651 CB THR A 397 7.824 -91.279 -4.796 1.00 82.77 A C
+ATOM 2652 CG2 THR A 397 9.199 -90.647 -4.697 1.00 83.38 A C
+ATOM 2653 OG1 THR A 397 6.815 -90.336 -4.396 1.00 84.90 A O
+ATOM 2654 N SER A 398 6.120 -93.667 -6.177 1.00 80.66 A N
+ATOM 2655 CA SER A 398 4.828 -94.336 -6.128 1.00 78.44 A C
+ATOM 2656 C SER A 398 5.022 -95.769 -5.663 1.00 78.53 A C
+ATOM 2657 O SER A 398 6.144 -96.264 -5.524 1.00 75.55 A O
+ATOM 2658 CB SER A 398 4.121 -94.304 -7.483 1.00 74.74 A C
+ATOM 2659 OG SER A 398 2.917 -95.053 -7.443 1.00 83.70 A O
+ATOM 2660 N LYS A 399 3.892 -96.441 -5.490 1.00 83.69 A N
+ATOM 2661 CA LYS A 399 3.772 -97.773 -4.920 1.00 79.39 A C
+ATOM 2662 C LYS A 399 3.722 -98.869 -5.974 1.00 87.12 A C
+ATOM 2663 O LYS A 399 3.697-100.052 -5.616 1.00 95.66 A O
+ATOM 2664 CB LYS A 399 2.493 -97.830 -4.063 1.00 88.77 A C
+ATOM 2665 CG LYS A 399 2.316 -99.029 -3.142 1.00 99.22 A C
+ATOM 2666 CD LYS A 399 0.966 -98.938 -2.440 1.00111.54 A C
+ATOM 2667 CE LYS A 399 0.884 -99.865 -1.245 1.00116.69 A C
+ATOM 2668 NZ LYS A 399 -0.326 -99.626 -0.414 1.00107.13 A N1+
+ATOM 2669 N THR A 400 3.706 -98.514 -7.259 1.00 78.79 A N
+ATOM 2670 CA THR A 400 3.276 -99.443 -8.304 1.00 85.84 A C
+ATOM 2671 C THR A 400 4.517-100.005 -8.989 1.00 78.88 A C
+ATOM 2672 O THR A 400 5.133 -99.345 -9.827 1.00 77.36 A O
+ATOM 2673 CB THR A 400 2.362 -98.747 -9.312 1.00 98.44 A C
+ATOM 2674 CG2 THR A 400 1.516 -99.776 -10.021 1.00 97.63 A C
+ATOM 2675 OG1 THR A 400 1.476 -97.851 -8.632 1.00106.26 A O
+ATOM 2676 N ASP A 401 4.846-101.253 -8.684 1.00 83.28 A N
+ATOM 2677 CA ASP A 401 5.999-101.907 -9.285 1.00 80.06 A C
+ATOM 2678 C ASP A 401 5.488-102.663 -10.501 1.00 86.85 A C
+ATOM 2679 O ASP A 401 4.784-103.673 -10.363 1.00 90.00 A O
+ATOM 2680 CB ASP A 401 6.687-102.834 -8.280 1.00 85.74 A C
+ATOM 2681 CG ASP A 401 5.705-103.685 -7.470 1.00 90.66 A C
+ATOM 2682 OD1 ASP A 401 4.930-103.128 -6.665 1.00 83.73 A O
+ATOM 2683 OD2 ASP A 401 5.750-104.926 -7.585 1.00 91.67 A O1-
+ATOM 2684 N VAL A 402 5.823-102.167 -11.695 1.00 76.80 A N
+ATOM 2685 CA VAL A 402 5.358-102.746 -12.953 1.00 67.23 A C
+ATOM 2686 C VAL A 402 6.550-102.895 -13.879 1.00 64.23 A C
+ATOM 2687 O VAL A 402 7.376-101.980 -13.994 1.00 72.47 A O
+ATOM 2688 CB VAL A 402 4.276-101.885 -13.626 1.00 78.16 A C
+ATOM 2689 CG1 VAL A 402 3.525-102.710 -14.627 1.00 92.97 A C
+ATOM 2690 CG2 VAL A 402 3.352-101.291 -12.597 1.00 79.09 A C
+ATOM 2691 N SER A 403 6.630-104.032 -14.561 1.00 71.61 A N
+ATOM 2692 CA SER A 403 7.791-104.291 -15.401 1.00 68.58 A C
+ATOM 2693 C SER A 403 7.809-103.340 -16.596 1.00 69.88 A C
+ATOM 2694 O SER A 403 6.869-103.280 -17.394 1.00 73.54 A O
+ATOM 2695 CB SER A 403 7.804-105.744 -15.853 1.00 66.41 A C
+ATOM 2696 OG SER A 403 7.718-106.595 -14.729 1.00 68.69 A O
+ATOM 2697 N SER A 404 8.891-102.600 -16.708 1.00 69.86 A N
+ATOM 2698 CA SER A 404 9.068-101.615 -17.746 1.00 73.43 A C
+ATOM 2699 C SER A 404 10.417-101.883 -18.395 1.00 74.92 A C
+ATOM 2700 O SER A 404 11.232-102.657 -17.892 1.00 73.63 A O
+ATOM 2701 CB SER A 404 8.976-100.195 -17.161 1.00 69.53 A C
+ATOM 2702 OG SER A 404 9.210 -99.195 -18.144 1.00 75.34 A O
+ATOM 2703 N SER A 405 10.647-101.272 -19.542 1.00 77.53 A N
+ATOM 2704 CA SER A 405 11.977-101.313 -20.111 1.00 67.97 A C
+ATOM 2705 C SER A 405 12.154-100.081 -20.963 1.00 73.00 A C
+ATOM 2706 O SER A 405 11.180 -99.528 -21.469 1.00 68.54 A O
+ATOM 2707 CB SER A 405 12.210-102.560 -20.952 1.00 66.13 A C
+ATOM 2708 OG SER A 405 13.472-102.456 -21.585 1.00 70.62 A O
+ATOM 2709 N VAL A 406 13.395 -99.643 -21.118 1.00 64.24 A N
+ATOM 2710 CA VAL A 406 13.652 -98.447 -21.904 1.00 60.80 A C
+ATOM 2711 C VAL A 406 14.859 -98.700 -22.789 1.00 68.47 A C
+ATOM 2712 O VAL A 406 15.960 -98.941 -22.277 1.00 71.86 A O
+ATOM 2713 CB VAL A 406 13.873 -97.216 -21.014 1.00 66.58 A C
+ATOM 2714 CG1 VAL A 406 14.623 -96.150 -21.754 1.00 57.18 A C
+ATOM 2715 CG2 VAL A 406 12.538 -96.670 -20.578 1.00 56.51 A C
+ATOM 2716 N ILE A 407 14.662 -98.637 -24.111 1.00 61.64 A N
+ATOM 2717 CA ILE A 407 15.766 -98.818 -25.047 1.00 58.31 A C
+ATOM 2718 C ILE A 407 16.503 -97.496 -25.136 1.00 61.30 A C
+ATOM 2719 O ILE A 407 15.914 -96.476 -25.510 1.00 67.14 A O
+ATOM 2720 CB ILE A 407 15.287 -99.251 -26.435 1.00 64.53 A C
+ATOM 2721 CG1 ILE A 407 14.208-100.320 -26.350 1.00 58.99 A C
+ATOM 2722 CG2 ILE A 407 16.471 -99.838 -27.178 1.00 57.13 A C
+ATOM 2723 CD1 ILE A 407 14.657-101.555 -25.675 1.00 56.87 A C
+ATOM 2724 N THR A 408 17.782 -97.510 -24.790 1.00 64.86 A N
+ATOM 2725 CA THR A 408 18.607 -96.321 -24.826 1.00 60.14 A C
+ATOM 2726 C THR A 408 19.395 -96.311 -26.125 1.00 64.92 A C
+ATOM 2727 O THR A 408 19.227 -97.190 -26.976 1.00 64.60 A O
+ATOM 2728 CB THR A 408 19.537 -96.271 -23.619 1.00 60.68 A C
+ATOM 2729 CG2 THR A 408 18.771 -96.615 -22.358 1.00 59.62 A C
+ATOM 2730 OG1 THR A 408 20.612 -97.204 -23.814 1.00 67.55 A O
+ATOM 2731 N SER A 409 20.281 -95.317 -26.265 1.00 62.57 A N
+ATOM 2732 CA SER A 409 21.088 -95.175 -27.477 1.00 66.13 A C
+ATOM 2733 C SER A 409 21.991 -96.385 -27.684 1.00 68.31 A C
+ATOM 2734 O SER A 409 21.942 -97.045 -28.730 1.00 67.21 A O
+ATOM 2735 CB SER A 409 21.909 -93.887 -27.409 1.00 60.20 A C
+ATOM 2736 OG SER A 409 21.052 -92.778 -27.236 1.00 70.65 A O
+ATOM 2737 N LEU A 410 22.830 -96.687 -26.697 1.00 62.39 A N
+ATOM 2738 CA LEU A 410 23.757 -97.802 -26.789 1.00 65.02 A C
+ATOM 2739 C LEU A 410 23.283 -99.069 -26.087 1.00 73.87 A C
+ATOM 2740 O LEU A 410 24.040-100.042 -26.034 1.00 68.95 A O
+ATOM 2741 CB LEU A 410 25.121 -97.383 -26.250 1.00 65.18 A C
+ATOM 2742 CG LEU A 410 26.147 -97.073 -27.332 1.00 68.41 A C
+ATOM 2743 CD1 LEU A 410 25.558 -96.165 -28.362 1.00 69.39 A C
+ATOM 2744 CD2 LEU A 410 27.336 -96.397 -26.697 1.00 71.93 A C
+ATOM 2745 N GLY A 411 22.078 -99.098 -25.539 1.00 64.35 A N
+ATOM 2746 CA GLY A 411 21.658-100.292 -24.838 1.00 58.08 A C
+ATOM 2747 C GLY A 411 20.240-100.197 -24.338 1.00 64.67 A C
+ATOM 2748 O GLY A 411 19.432 -99.419 -24.848 1.00 66.98 A O
+ATOM 2749 N ALA A 412 19.943-101.029 -23.337 1.00 66.89 A N
+ATOM 2750 CA ALA A 412 18.597-101.140 -22.788 1.00 63.72 A C
+ATOM 2751 C ALA A 412 18.632-101.248 -21.269 1.00 61.83 A C
+ATOM 2752 O ALA A 412 19.388-102.047 -20.707 1.00 65.65 A O
+ATOM 2753 CB ALA A 412 17.855-102.348 -23.371 1.00 60.07 A C
+ATOM 2754 N ILE A 413 17.823-100.427 -20.624 1.00 59.69 A N
+ATOM 2755 CA ILE A 413 17.506-100.539 -19.213 1.00 59.06 A C
+ATOM 2756 C ILE A 413 16.298-101.445 -19.079 1.00 64.62 A C
+ATOM 2757 O ILE A 413 15.389-101.406 -19.916 1.00 60.51 A O
+ATOM 2758 CB ILE A 413 17.213 -99.156 -18.621 1.00 59.92 A C
+ATOM 2759 CG1 ILE A 413 18.379 -98.203 -18.859 1.00 59.66 A C
+ATOM 2760 CG2 ILE A 413 16.894 -99.290 -17.177 1.00 51.09 A C
+ATOM 2761 CD1 ILE A 413 18.147 -96.818 -18.302 1.00 55.32 A C
+ATOM 2762 N VAL A 414 16.267-102.259 -18.032 1.00 60.01 A N
+ATOM 2763 CA VAL A 414 15.136-103.148 -17.796 1.00 59.48 A C
+ATOM 2764 C VAL A 414 14.748-103.099 -16.319 1.00 61.90 A C
+ATOM 2765 O VAL A 414 15.597-103.242 -15.435 1.00 64.68 A O
+ATOM 2766 CB VAL A 414 15.459-104.584 -18.245 1.00 67.62 A C
+ATOM 2767 CG1 VAL A 414 14.437-105.552 -17.687 1.00 64.90 A C
+ATOM 2768 CG2 VAL A 414 15.483-104.641 -19.746 1.00 63.07 A C
+ATOM 2769 N SER A 415 13.469-102.888 -16.051 1.00 64.28 A N
+ATOM 2770 CA SER A 415 12.942-102.926 -14.696 1.00 68.71 A C
+ATOM 2771 C SER A 415 12.008-104.124 -14.593 1.00 77.19 A C
+ATOM 2772 O SER A 415 10.862-104.065 -15.059 1.00 72.68 A O
+ATOM 2773 CB SER A 415 12.214-101.625 -14.370 1.00 62.45 A C
+ATOM 2774 OG SER A 415 13.075-100.525 -14.571 1.00 74.16 A O
+ATOM 2775 N CYS A 416 12.471-105.181 -13.917 1.00 80.02 A N
+ATOM 2776 CA CYS A 416 11.753-106.450 -13.837 1.00 80.17 A C
+ATOM 2777 C CYS A 416 11.239-106.655 -12.419 1.00 82.72 A C
+ATOM 2778 O CYS A 416 12.016-106.970 -11.512 1.00 84.32 A O
+ATOM 2779 CB CYS A 416 12.665-107.601 -14.238 1.00 77.48 A C
+ATOM 2780 SG CYS A 416 11.749-108.975 -14.921 1.00 91.59 A S
+ATOM 2781 N TYR A 417 9.928-106.517 -12.241 1.00 72.19 A N
+ATOM 2782 CA TYR A 417 9.289-106.605 -10.935 1.00 77.24 A C
+ATOM 2783 C TYR A 417 8.126-107.586 -11.006 1.00 79.83 A C
+ATOM 2784 O TYR A 417 7.397-107.631 -12.004 1.00 74.85 A O
+ATOM 2785 CB TYR A 417 8.793-105.239 -10.487 1.00 77.72 A C
+ATOM 2786 CG TYR A 417 9.873-104.192 -10.514 1.00 84.18 A C
+ATOM 2787 CD1 TYR A 417 11.183-104.510 -10.170 1.00 79.46 A C
+ATOM 2788 CD2 TYR A 417 9.587-102.879 -10.869 1.00 79.48 A C
+ATOM 2789 CE1 TYR A 417 12.187-103.564 -10.202 1.00 82.90 A C
+ATOM 2790 CE2 TYR A 417 10.586-101.913 -10.905 1.00 78.21 A C
+ATOM 2791 CZ TYR A 417 11.888-102.263 -10.564 1.00 82.46 A C
+ATOM 2792 OH TYR A 417 12.890-101.313 -10.585 1.00 82.25 A O
+ATOM 2793 N GLY A 418 7.947-108.372 -9.947 1.00 75.57 A N
+ATOM 2794 CA GLY A 418 6.903-109.381 -9.966 1.00 71.99 A C
+ATOM 2795 C GLY A 418 7.342-110.634 -10.705 1.00 82.51 A C
+ATOM 2796 O GLY A 418 8.529-110.874 -10.962 1.00 79.60 A O
+ATOM 2797 N LYS A 419 6.359-111.434 -11.081 1.00 81.78 A N
+ATOM 2798 CA LYS A 419 6.701-112.679 -11.745 1.00 81.69 A C
+ATOM 2799 C LYS A 419 6.993-112.486 -13.226 1.00 79.13 A C
+ATOM 2800 O LYS A 419 7.106-113.482 -13.952 1.00 77.07 A O
+ATOM 2801 CB LYS A 419 5.610-113.742 -11.551 1.00 77.35 A C
+ATOM 2802 CG LYS A 419 5.818-114.601 -10.282 1.00 78.17 A C
+ATOM 2803 CD LYS A 419 5.282-116.038 -10.429 1.00 87.47 A C
+ATOM 2804 CE LYS A 419 3.848-116.188 -9.949 1.00 84.00 A C
+ATOM 2805 NZ LYS A 419 3.718-115.794 -8.514 1.00 84.60 A N1+
+ATOM 2806 N THR A 420 7.062-111.236 -13.691 1.00 76.14 A N
+ATOM 2807 CA THR A 420 7.272-110.957 -15.107 1.00 81.85 A C
+ATOM 2808 C THR A 420 8.609-111.497 -15.588 1.00 76.62 A C
+ATOM 2809 O THR A 420 9.623-111.383 -14.894 1.00 74.80 A O
+ATOM 2810 CB THR A 420 7.232-109.460 -15.363 1.00 79.71 A C
+ATOM 2811 CG2 THR A 420 7.149-109.202 -16.841 1.00 72.61 A C
+ATOM 2812 OG1 THR A 420 6.109-108.871 -14.693 1.00 73.57 A O
+ATOM 2813 N LYS A 421 8.612-112.064 -16.799 1.00 75.45 A N
+ATOM 2814 CA LYS A 421 9.821-112.651 -17.371 1.00 80.75 A C
+ATOM 2815 C LYS A 421 10.544-111.610 -18.224 1.00 80.40 A C
+ATOM 2816 O LYS A 421 9.963-111.068 -19.174 1.00 76.79 A O
+ATOM 2817 CB LYS A 421 9.501-113.900 -18.201 1.00 73.94 A C
+ATOM 2818 CG LYS A 421 10.715-114.848 -18.345 1.00100.87 A C
+ATOM 2819 CD LYS A 421 11.137-115.417 -16.966 1.00111.93 A C
+ATOM 2820 CE LYS A 421 12.572-115.964 -16.934 1.00120.42 A C
+ATOM 2821 NZ LYS A 421 12.754-117.328 -17.489 1.00115.98 A N1+
+ATOM 2822 N CYS A 422 11.807-111.336 -17.888 1.00 73.03 A N
+ATOM 2823 CA CYS A 422 12.624-110.363 -18.600 1.00 70.60 A C
+ATOM 2824 C CYS A 422 13.901-111.032 -19.069 1.00 80.73 A C
+ATOM 2825 O CYS A 422 14.610-111.653 -18.276 1.00 73.53 A O
+ATOM 2826 CB CYS A 422 12.961-109.159 -17.722 1.00 69.70 A C
+ATOM 2827 SG CYS A 422 11.528-108.450 -16.889 1.00 76.17 A S
+ATOM 2828 N THR A 423 14.187-110.907 -20.358 1.00 61.89 A N
+ATOM 2829 CA THR A 423 15.309-111.603 -20.960 1.00 63.03 A C
+ATOM 2830 C THR A 423 15.889-110.744 -22.070 1.00 71.76 A C
+ATOM 2831 O THR A 423 15.222-109.866 -22.619 1.00 69.63 A O
+ATOM 2832 CB THR A 423 14.897-112.968 -21.537 1.00 64.32 A C
+ATOM 2833 CG2 THR A 423 14.289-113.877 -20.470 1.00 64.69 A C
+ATOM 2834 OG1 THR A 423 13.955-112.784 -22.604 1.00 71.50 A O
+ATOM 2835 N ALA A 424 17.143-111.015 -22.399 1.00 61.15 A N
+ATOM 2836 CA ALA A 424 17.794-110.431 -23.556 1.00 61.44 A C
+ATOM 2837 C ALA A 424 18.336-111.568 -24.398 1.00 68.43 A C
+ATOM 2838 O ALA A 424 18.922-112.518 -23.861 1.00 66.55 A O
+ATOM 2839 CB ALA A 424 18.920-109.471 -23.162 1.00 58.29 A C
+ATOM 2840 N SER A 425 18.131-111.465 -25.714 1.00 60.10 A N
+ATOM 2841 CA SER A 425 18.546-112.482 -26.672 1.00 65.90 A C
+ATOM 2842 C SER A 425 19.284-111.879 -27.872 1.00 70.84 A C
+ATOM 2843 O SER A 425 19.180-110.685 -28.189 1.00 71.68 A O
+ATOM 2844 CB SER A 425 17.340-113.329 -27.137 1.00 67.14 A C
+ATOM 2845 OG SER A 425 16.148-112.570 -27.190 1.00 84.86 A O
+ATOM 2846 N ASN A 426 20.052-112.757 -28.517 1.00 66.30 A N
+ATOM 2847 CA ASN A 426 20.841-112.519 -29.718 1.00 69.09 A C
+ATOM 2848 C ASN A 426 20.197-113.252 -30.887 1.00 85.38 A C
+ATOM 2849 O ASN A 426 19.555-114.290 -30.708 1.00 93.65 A O
+ATOM 2850 CB ASN A 426 22.277-113.025 -29.531 1.00 74.55 A C
+ATOM 2851 CG ASN A 426 23.206-112.610 -30.662 1.00 92.90 A C
+ATOM 2852 ND2 ASN A 426 24.069-111.637 -30.384 1.00 95.59 A N
+ATOM 2853 OD1 ASN A 426 23.174-113.179 -31.762 1.00103.76 A O
+ATOM 2854 N LYS A 427 20.374-112.713 -32.092 1.00 79.42 A N
+ATOM 2855 CA LYS A 427 19.685-113.299 -33.234 1.00 78.66 A C
+ATOM 2856 C LYS A 427 20.134-114.729 -33.516 1.00 68.00 A C
+ATOM 2857 O LYS A 427 19.394-115.471 -34.178 1.00 70.27 A O
+ATOM 2858 CB LYS A 427 19.865-112.423 -34.485 1.00 81.32 A C
+ATOM 2859 CG LYS A 427 21.191-112.565 -35.206 1.00 95.98 A C
+ATOM 2860 CD LYS A 427 21.286-111.625 -36.398 1.00 97.49 A C
+ATOM 2861 CE LYS A 427 21.304-110.178 -35.950 1.00 96.05 A C
+ATOM 2862 NZ LYS A 427 21.662-109.268 -37.056 1.00 97.37 A N1+
+ATOM 2863 N ASN A 428 21.315-115.135 -33.015 1.00 74.69 A N
+ATOM 2864 CA ASN A 428 21.885-116.468 -33.234 1.00 80.66 A C
+ATOM 2865 C ASN A 428 21.778-117.343 -31.992 1.00 90.93 A C
+ATOM 2866 O ASN A 428 21.115-118.383 -32.005 1.00103.56 A O
+ATOM 2867 CB ASN A 428 23.363-116.375 -33.627 1.00 84.68 A C
+ATOM 2868 CG ASN A 428 23.585-115.531 -34.839 1.00 94.59 A C
+ATOM 2869 ND2 ASN A 428 23.215-116.052 -36.011 1.00 97.48 A N
+ATOM 2870 OD1 ASN A 428 24.073-114.400 -34.726 1.00 95.79 A O
+ATOM 2871 N ARG A 429 22.440-116.935 -30.913 1.00 91.11 A N
+ATOM 2872 CA ARG A 429 22.593-117.741 -29.716 1.00 95.45 A C
+ATOM 2873 C ARG A 429 21.285-117.902 -28.926 1.00 82.40 A C
+ATOM 2874 O ARG A 429 21.289-118.629 -27.924 1.00 84.68 A O
+ATOM 2875 CB ARG A 429 23.710-117.141 -28.833 1.00 99.34 A C
+ATOM 2876 CG ARG A 429 25.167-117.412 -29.317 1.00105.62 A C
+ATOM 2877 CD ARG A 429 26.201-116.368 -28.811 1.00115.76 A C
+ATOM 2878 NE ARG A 429 26.203-115.101 -29.565 1.00114.69 A N
+ATOM 2879 CZ ARG A 429 27.068-114.761 -30.532 1.00112.04 A C
+ATOM 2880 NH1 ARG A 429 28.049-115.583 -30.892 1.00 96.93 A N1+
+ATOM 2881 NH2 ARG A 429 26.956-113.583 -31.143 1.00118.07 A N
+ATOM 2882 N GLY A 430 20.175-117.267 -29.329 1.00 93.68 A N
+ATOM 2883 CA GLY A 430 18.955-117.324 -28.524 1.00 72.48 A C
+ATOM 2884 C GLY A 430 19.035-116.391 -27.318 1.00 81.65 A C
+ATOM 2885 O GLY A 430 19.808-115.420 -27.307 1.00 79.39 A O
+ATOM 2886 N ILE A 431 18.237-116.683 -26.283 1.00 69.20 A N
+ATOM 2887 CA ILE A 431 18.350-115.887 -25.063 1.00 72.67 A C
+ATOM 2888 C ILE A 431 19.759-116.028 -24.515 1.00 65.34 A C
+ATOM 2889 O ILE A 431 20.223-117.145 -24.267 1.00 70.01 A O
+ATOM 2890 CB ILE A 431 17.341-116.337 -24.015 1.00 63.89 A C
+ATOM 2891 CG1 ILE A 431 15.915-116.032 -24.449 1.00 71.11 A C
+ATOM 2892 CG2 ILE A 431 17.664-115.672 -22.686 1.00 62.63 A C
+ATOM 2893 CD1 ILE A 431 14.878-116.269 -23.334 1.00 64.70 A C
+ATOM 2894 N ILE A 432 20.470-114.911 -24.362 1.00 59.14 A N
+ATOM 2895 CA ILE A 432 21.775-114.950 -23.710 1.00 69.51 A C
+ATOM 2896 C ILE A 432 21.799-114.366 -22.303 1.00 68.97 A C
+ATOM 2897 O ILE A 432 22.850-114.444 -21.649 1.00 79.37 A O
+ATOM 2898 CB ILE A 432 22.862-114.291 -24.577 1.00 75.48 A C
+ATOM 2899 CG1 ILE A 432 22.477-112.865 -24.935 1.00 66.90 A C
+ATOM 2900 CG2 ILE A 432 23.056-115.108 -25.841 1.00 79.72 A C
+ATOM 2901 CD1 ILE A 432 23.477-112.198 -25.879 1.00 72.61 A C
+ATOM 2902 N LYS A 433 20.717-113.770 -21.812 1.00 69.95 A N
+ATOM 2903 CA LYS A 433 20.704-113.415 -20.395 1.00 69.82 A C
+ATOM 2904 C LYS A 433 19.272-113.387 -19.886 1.00 69.36 A C
+ATOM 2905 O LYS A 433 18.372-112.921 -20.593 1.00 71.24 A O
+ATOM 2906 CB LYS A 433 21.380-112.060 -20.125 1.00 68.44 A C
+ATOM 2907 CG LYS A 433 21.261-111.597 -18.670 1.00 67.11 A C
+ATOM 2908 CD LYS A 433 22.172-110.409 -18.330 1.00 81.17 A C
+ATOM 2909 CE LYS A 433 22.253-110.191 -16.797 1.00 85.57 A C
+ATOM 2910 NZ LYS A 433 23.276-109.188 -16.320 1.00 84.85 A N1+
+ATOM 2911 N THR A 434 19.066-113.863 -18.656 1.00 67.96 A N
+ATOM 2912 CA THR A 434 17.787-113.735 -17.963 1.00 66.15 A C
+ATOM 2913 C THR A 434 17.964-112.806 -16.774 1.00 69.56 A C
+ATOM 2914 O THR A 434 18.920-112.949 -16.006 1.00 74.70 A O
+ATOM 2915 CB THR A 434 17.244-115.087 -17.499 1.00 67.11 A C
+ATOM 2916 CG2 THR A 434 15.917-114.909 -16.783 1.00 66.18 A C
+ATOM 2917 OG1 THR A 434 17.059-115.945 -18.630 1.00 68.05 A O
+ATOM 2918 N PHE A 435 17.051-111.854 -16.640 1.00 69.20 A N
+ATOM 2919 CA PHE A 435 17.146-110.822 -15.623 1.00 73.90 A C
+ATOM 2920 C PHE A 435 16.410-111.246 -14.362 1.00 79.34 A C
+ATOM 2921 O PHE A 435 15.316-111.823 -14.428 1.00 82.34 A O
+ATOM 2922 CB PHE A 435 16.550-109.506 -16.118 1.00 69.65 A C
+ATOM 2923 CG PHE A 435 17.236-108.918 -17.319 1.00 78.10 A C
+ATOM 2924 CD1 PHE A 435 18.484-108.325 -17.200 1.00 77.88 A C
+ATOM 2925 CD2 PHE A 435 16.598-108.886 -18.547 1.00 77.52 A C
+ATOM 2926 CE1 PHE A 435 19.103-107.752 -18.300 1.00 80.25 A C
+ATOM 2927 CE2 PHE A 435 17.208-108.316 -19.645 1.00 69.70 A C
+ATOM 2928 CZ PHE A 435 18.460-107.746 -19.525 1.00 77.67 A C
+ATOM 2929 N SER A 436 17.012-110.934 -13.216 1.00 79.87 A N
+ATOM 2930 CA SER A 436 16.375-111.137 -11.921 1.00 87.84 A C
+ATOM 2931 C SER A 436 15.459-109.947 -11.630 1.00 85.25 A C
+ATOM 2932 O SER A 436 15.221-109.089 -12.484 1.00 84.19 A O
+ATOM 2933 CB SER A 436 17.428-111.342 -10.832 1.00 77.18 A C
+ATOM 2934 OG SER A 436 18.185-112.516 -11.070 1.00124.51 A O
+ATOM 2935 N ASN A 437 14.920-109.886 -10.417 1.00 75.25 A N
+ATOM 2936 CA ASN A 437 14.049-108.781 -10.051 1.00 76.16 A C
+ATOM 2937 C ASN A 437 14.897-107.576 -9.660 1.00 73.76 A C
+ATOM 2938 O ASN A 437 15.829-107.690 -8.859 1.00 79.84 A O
+ATOM 2939 CB ASN A 437 13.100-109.195 -8.924 1.00 73.56 A C
+ATOM 2940 CG ASN A 437 11.922-110.043 -9.427 1.00 89.27 A C
+ATOM 2941 ND2 ASN A 437 12.149-111.347 -9.596 1.00118.62 A N
+ATOM 2942 OD1 ASN A 437 10.822-109.528 -9.647 1.00 85.51 A O
+ATOM 2943 N GLY A 438 14.611-106.439 -10.272 1.00 74.49 A N
+ATOM 2944 CA GLY A 438 15.370-105.232 -10.027 1.00 73.48 A C
+ATOM 2945 C GLY A 438 15.457-104.410 -11.296 1.00 77.75 A C
+ATOM 2946 O GLY A 438 14.794-104.692 -12.287 1.00 68.46 A O
+ATOM 2947 N CYS A 439 16.302-103.387 -11.244 1.00 64.49 A N
+ATOM 2948 CA CYS A 439 16.609-102.555 -12.401 1.00 65.51 A C
+ATOM 2949 C CYS A 439 18.021-102.881 -12.888 1.00 68.04 A C
+ATOM 2950 O CYS A 439 18.999-102.651 -12.167 1.00 70.32 A O
+ATOM 2951 CB CYS A 439 16.473-101.073 -12.048 1.00 57.45 A C
+ATOM 2952 SG CYS A 439 17.184 -99.968 -13.267 1.00147.39 A S
+ATOM 2953 N ASP A 440 18.127-103.421 -14.104 1.00 65.94 A N
+ATOM 2954 CA ASP A 440 19.402-103.804 -14.699 1.00 66.78 A C
+ATOM 2955 C ASP A 440 19.565-103.109 -16.054 1.00 73.16 A C
+ATOM 2956 O ASP A 440 18.715-102.331 -16.497 1.00 67.24 A O
+ATOM 2957 CB ASP A 440 19.504-105.328 -14.878 1.00 70.41 A C
+ATOM 2958 CG ASP A 440 20.942-105.840 -14.785 1.00 73.02 A C
+ATOM 2959 OD1 ASP A 440 21.863-105.004 -14.673 1.00 78.88 A O
+ATOM 2960 OD2 ASP A 440 21.157-107.071 -14.875 1.00 86.92 A O1-
+ATOM 2961 N TYR A 441 20.668-103.422 -16.730 1.00 67.79 A N
+ATOM 2962 CA TYR A 441 21.001-102.837 -18.020 1.00 67.13 A C
+ATOM 2963 C TYR A 441 21.788-103.857 -18.836 1.00 65.92 A C
+ATOM 2964 O TYR A 441 22.421-104.767 -18.290 1.00 68.03 A O
+ATOM 2965 CB TYR A 441 21.798-101.544 -17.834 1.00 66.95 A C
+ATOM 2966 CG TYR A 441 22.409-100.975 -19.093 1.00 69.82 A C
+ATOM 2967 CD1 TYR A 441 21.807 -99.928 -19.762 1.00 72.57 A C
+ATOM 2968 CD2 TYR A 441 23.607-101.470 -19.602 1.00 71.52 A C
+ATOM 2969 CE1 TYR A 441 22.369 -99.395 -20.907 1.00 62.47 A C
+ATOM 2970 CE2 TYR A 441 24.165-100.955 -20.750 1.00 67.28 A C
+ATOM 2971 CZ TYR A 441 23.544 -99.910 -21.390 1.00 69.14 A C
+ATOM 2972 OH TYR A 441 24.098 -99.376 -22.524 1.00 72.64 A O
+ATOM 2973 N VAL A 442 21.731-103.699 -20.158 1.00 59.95 A N
+ATOM 2974 CA VAL A 442 22.526-104.513 -21.070 1.00 61.34 A C
+ATOM 2975 C VAL A 442 22.931-103.660 -22.269 1.00 68.59 A C
+ATOM 2976 O VAL A 442 22.144-102.855 -22.776 1.00 62.64 A O
+ATOM 2977 CB VAL A 442 21.770-105.799 -21.479 1.00 68.62 A C
+ATOM 2978 CG1 VAL A 442 20.365-105.503 -21.962 1.00 60.86 A C
+ATOM 2979 CG2 VAL A 442 22.529-106.501 -22.547 1.00 69.29 A C
+ATOM 2980 N SER A 443 24.179-103.821 -22.702 1.00 67.36 A N
+ATOM 2981 CA SER A 443 24.693-103.073 -23.837 1.00 79.20 A C
+ATOM 2982 C SER A 443 24.091-103.588 -25.137 1.00 73.91 A C
+ATOM 2983 O SER A 443 23.584-104.712 -25.210 1.00 75.19 A O
+ATOM 2984 CB SER A 443 26.209-103.189 -23.924 1.00 69.77 A C
+ATOM 2985 OG SER A 443 26.570-104.503 -24.322 1.00 81.54 A O
+ATOM 2986 N ASN A 444 24.168-102.747 -26.180 1.00 73.11 A N
+ATOM 2987 CA ASN A 444 23.744-103.148 -27.523 1.00 71.64 A C
+ATOM 2988 C ASN A 444 24.743-104.074 -28.202 1.00 70.88 A C
+ATOM 2989 O ASN A 444 24.388-104.709 -29.203 1.00 85.04 A O
+ATOM 2990 CB ASN A 444 23.512-101.921 -28.406 1.00 66.49 A C
+ATOM 2991 CG ASN A 444 24.808-101.301 -28.900 1.00 68.43 A C
+ATOM 2992 ND2 ASN A 444 25.551-100.676 -27.986 1.00 74.40 A N
+ATOM 2993 OD1 ASN A 444 25.137-101.373 -30.091 1.00 88.58 A O
+ATOM 2994 N LYS A 445 25.982-104.156 -27.698 1.00 65.85 A N
+ATOM 2995 CA LYS A 445 26.960-105.083 -28.263 1.00 77.34 A C
+ATOM 2996 C LYS A 445 26.516-106.511 -27.980 1.00 79.77 A C
+ATOM 2997 O LYS A 445 26.213-106.863 -26.836 1.00 81.22 A O
+ATOM 2998 CB LYS A 445 28.363-104.848 -27.687 1.00 74.63 A C
+ATOM 2999 CG LYS A 445 28.801-103.375 -27.521 1.00 90.64 A C
+ATOM 3000 CD LYS A 445 30.175-103.286 -26.805 1.00110.45 A C
+ATOM 3001 CE LYS A 445 30.622-101.851 -26.482 1.00125.19 A C
+ATOM 3002 NZ LYS A 445 29.738-101.179 -25.491 1.00130.94 A N1+
+ATOM 3003 N GLY A 446 26.446-107.325 -29.031 1.00 82.56 A N
+ATOM 3004 CA GLY A 446 26.082-108.721 -28.890 1.00 79.74 A C
+ATOM 3005 C GLY A 446 24.630-108.990 -28.563 1.00 84.67 A C
+ATOM 3006 O GLY A 446 24.234-110.163 -28.480 1.00 90.41 A O
+ATOM 3007 N VAL A 447 23.821-107.954 -28.372 1.00 80.82 A N
+ATOM 3008 CA VAL A 447 22.398-108.114 -28.116 1.00 81.00 A C
+ATOM 3009 C VAL A 447 21.632-107.530 -29.285 1.00 83.88 A C
+ATOM 3010 O VAL A 447 22.029-106.514 -29.872 1.00 87.21 A O
+ATOM 3011 CB VAL A 447 21.959-107.439 -26.806 1.00 77.64 A C
+ATOM 3012 CG1 VAL A 447 20.572-107.936 -26.390 1.00 76.04 A C
+ATOM 3013 CG2 VAL A 447 22.991-107.692 -25.751 1.00 72.58 A C
+ATOM 3014 N ASP A 448 20.534-108.187 -29.624 1.00 71.38 A N
+ATOM 3015 CA ASP A 448 19.579-107.674 -30.586 1.00 72.88 A C
+ATOM 3016 C ASP A 448 18.255-107.361 -29.897 1.00 70.25 A C
+ATOM 3017 O ASP A 448 17.793-106.213 -29.881 1.00 70.86 A O
+ATOM 3018 CB ASP A 448 19.439-108.687 -31.729 1.00 78.81 A C
+ATOM 3019 CG ASP A 448 20.751-108.877 -32.493 1.00 89.17 A C
+ATOM 3020 OD1 ASP A 448 21.460-107.878 -32.731 1.00 92.04 A O
+ATOM 3021 OD2 ASP A 448 21.090-110.027 -32.833 1.00 97.51 A O1-
+ATOM 3022 N THR A 449 17.639-108.351 -29.278 1.00 71.64 A N
+ATOM 3023 CA THR A 449 16.318-108.157 -28.721 1.00 68.52 A C
+ATOM 3024 C THR A 449 16.322-108.145 -27.201 1.00 73.88 A C
+ATOM 3025 O THR A 449 17.098-108.850 -26.554 1.00 70.23 A O
+ATOM 3026 CB THR A 449 15.418-109.254 -29.220 1.00 74.12 A C
+ATOM 3027 CG2 THR A 449 14.016-108.869 -29.012 1.00 73.18 A C
+ATOM 3028 OG1 THR A 449 15.699-109.464 -30.612 1.00 78.05 A O
+ATOM 3029 N VAL A 450 15.430-107.346 -26.634 1.00 67.35 A N
+ATOM 3030 CA VAL A 450 15.175-107.338 -25.202 1.00 62.73 A C
+ATOM 3031 C VAL A 450 13.680-107.546 -25.009 1.00 56.73 A C
+ATOM 3032 O VAL A 450 12.873-106.682 -25.374 1.00 63.82 A O
+ATOM 3033 CB VAL A 450 15.646-106.042 -24.544 1.00 67.22 A C
+ATOM 3034 CG1 VAL A 450 15.012-105.886 -23.195 1.00 75.50 A C
+ATOM 3035 CG2 VAL A 450 17.142-106.082 -24.414 1.00 70.08 A C
+ATOM 3036 N SER A 451 13.307-108.687 -24.441 1.00 62.57 A N
+ATOM 3037 CA SER A 451 11.913-109.075 -24.290 1.00 64.64 A C
+ATOM 3038 C SER A 451 11.529-108.944 -22.822 1.00 67.52 A C
+ATOM 3039 O SER A 451 12.250-109.423 -21.941 1.00 67.49 A O
+ATOM 3040 CB SER A 451 11.689-110.501 -24.813 1.00 64.74 A C
+ATOM 3041 OG SER A 451 12.491-110.763 -25.972 1.00 83.91 A O
+ATOM 3042 N VAL A 452 10.425-108.248 -22.561 1.00 62.63 A N
+ATOM 3043 CA VAL A 452 9.960-107.972 -21.202 1.00 58.83 A C
+ATOM 3044 C VAL A 452 8.458-108.219 -21.151 1.00 59.59 A C
+ATOM 3045 O VAL A 452 7.686-107.560 -21.864 1.00 74.13 A O
+ATOM 3046 CB VAL A 452 10.267-106.535 -20.751 1.00 71.20 A C
+ATOM 3047 CG1 VAL A 452 9.846-106.355 -19.329 1.00 65.54 A C
+ATOM 3048 CG2 VAL A 452 11.735-106.228 -20.903 1.00 74.49 A C
+ATOM 3049 N GLY A 453 8.038-109.146 -20.309 1.00 59.02 A N
+ATOM 3050 CA GLY A 453 6.639-109.521 -20.357 1.00 62.75 A C
+ATOM 3051 C GLY A 453 6.303-110.039 -21.735 1.00 71.86 A C
+ATOM 3052 O GLY A 453 7.004-110.896 -22.278 1.00 72.47 A O
+ATOM 3053 N ASN A 454 5.232-109.520 -22.325 1.00 68.36 A N
+ATOM 3054 CA ASN A 454 4.874-109.920 -23.676 1.00 64.65 A C
+ATOM 3055 C ASN A 454 5.355-108.953 -24.753 1.00 64.15 A C
+ATOM 3056 O ASN A 454 5.081-109.188 -25.936 1.00 70.84 A O
+ATOM 3057 CB ASN A 454 3.360-110.109 -23.800 1.00 64.54 A C
+ATOM 3058 CG ASN A 454 2.838-111.212 -22.906 1.00 76.11 A C
+ATOM 3059 ND2 ASN A 454 1.645-111.007 -22.365 1.00 71.80 A N
+ATOM 3060 OD1 ASN A 454 3.501-112.235 -22.693 1.00 77.06 A O
+ATOM 3061 N THR A 455 6.057-107.881 -24.397 1.00 52.75 A N
+ATOM 3062 CA THR A 455 6.491-106.923 -25.402 1.00 60.11 A C
+ATOM 3063 C THR A 455 7.937-107.233 -25.778 1.00 66.25 A C
+ATOM 3064 O THR A 455 8.786-107.431 -24.900 1.00 67.38 A O
+ATOM 3065 CB THR A 455 6.351-105.493 -24.885 1.00 63.33 A C
+ATOM 3066 CG2 THR A 455 5.197-105.395 -23.887 1.00 61.51 A C
+ATOM 3067 OG1 THR A 455 7.547-105.127 -24.207 1.00 63.72 A O
+ATOM 3068 N LEU A 456 8.209-107.308 -27.081 1.00 66.20 A N
+ATOM 3069 CA LEU A 456 9.545-107.598 -27.605 1.00 59.11 A C
+ATOM 3070 C LEU A 456 10.174-106.349 -28.209 1.00 63.81 A C
+ATOM 3071 O LEU A 456 9.758-105.905 -29.283 1.00 66.08 A O
+ATOM 3072 CB LEU A 456 9.454-108.716 -28.632 1.00 68.50 A C
+ATOM 3073 CG LEU A 456 10.791-109.115 -29.219 1.00 64.01 A C
+ATOM 3074 CD1 LEU A 456 10.914-110.617 -29.329 1.00 67.54 A C
+ATOM 3075 CD2 LEU A 456 10.883-108.523 -30.601 1.00 81.85 A C
+ATOM 3076 N TYR A 457 11.194-105.802 -27.542 1.00 60.67 A N
+ATOM 3077 CA TYR A 457 11.855-104.574 -27.993 1.00 59.19 A C
+ATOM 3078 C TYR A 457 13.097-104.912 -28.825 1.00 60.19 A C
+ATOM 3079 O TYR A 457 13.845-105.838 -28.491 1.00 63.36 A O
+ATOM 3080 CB TYR A 457 12.264-103.689 -26.800 1.00 54.51 A C
+ATOM 3081 CG TYR A 457 11.134-103.128 -25.939 1.00 68.18 A C
+ATOM 3082 CD1 TYR A 457 10.478-101.957 -26.290 1.00 75.41 A C
+ATOM 3083 CD2 TYR A 457 10.758-103.744 -24.759 1.00 78.10 A C
+ATOM 3084 CE1 TYR A 457 9.447-101.440 -25.517 1.00 88.58 A C
+ATOM 3085 CE2 TYR A 457 9.742-103.222 -23.970 1.00 89.16 A C
+ATOM 3086 CZ TYR A 457 9.085-102.074 -24.354 1.00 97.30 A C
+ATOM 3087 OH TYR A 457 8.065-101.562 -23.570 1.00101.83 A O
+ATOM 3088 N TYR A 458 13.318-104.179 -29.915 1.00 64.10 A N
+ATOM 3089 CA TYR A 458 14.610-104.266 -30.590 1.00 66.28 A C
+ATOM 3090 C TYR A 458 15.459-103.102 -30.102 1.00 61.30 A C
+ATOM 3091 O TYR A 458 14.931-102.024 -29.810 1.00 59.93 A O
+ATOM 3092 CB TYR A 458 14.487-104.198 -32.117 1.00 64.78 A C
+ATOM 3093 CG TYR A 458 13.992-105.445 -32.838 1.00 67.84 A C
+ATOM 3094 CD1 TYR A 458 12.671-105.842 -32.735 1.00 57.58 A C
+ATOM 3095 CD2 TYR A 458 14.809-106.156 -33.709 1.00 61.41 A C
+ATOM 3096 CE1 TYR A 458 12.193-106.961 -33.418 1.00 60.43 A C
+ATOM 3097 CE2 TYR A 458 14.326-107.282 -34.399 1.00 63.09 A C
+ATOM 3098 CZ TYR A 458 13.012-107.670 -34.242 1.00 67.86 A C
+ATOM 3099 OH TYR A 458 12.474-108.755 -34.895 1.00 70.83 A O
+ATOM 3100 N VAL A 459 16.780-103.332 -29.981 1.00 56.83 A N
+ATOM 3101 CA VAL A 459 17.711-102.295 -29.529 1.00 64.15 A C
+ATOM 3102 C VAL A 459 18.373-101.667 -30.743 1.00 66.90 A C
+ATOM 3103 O VAL A 459 18.601-102.338 -31.754 1.00 66.61 A O
+ATOM 3104 CB VAL A 459 18.779-102.833 -28.555 1.00 69.93 A C
+ATOM 3105 CG1 VAL A 459 18.129-103.406 -27.345 1.00 64.61 A C
+ATOM 3106 CG2 VAL A 459 19.650-103.872 -29.236 1.00 70.02 A C
+ATOM 3107 N ASN A 460 18.661-100.363 -30.658 1.00 63.16 A N
+ATOM 3108 CA ASN A 460 19.425 -99.706 -31.714 1.00 56.42 A C
+ATOM 3109 C ASN A 460 20.795-100.355 -31.822 1.00 64.49 A C
+ATOM 3110 O ASN A 460 21.405-100.713 -30.813 1.00 66.83 A O
+ATOM 3111 CB ASN A 460 19.582 -98.214 -31.435 1.00 70.48 A C
+ATOM 3112 CG ASN A 460 18.388 -97.643 -30.765 1.00 70.61 A C
+ATOM 3113 ND2 ASN A 460 17.475 -97.079 -31.524 1.00 63.26 A N
+ATOM 3114 OD1 ASN A 460 18.270 -97.746 -29.564 1.00 69.31 A O
+ATOM 3115 N LYS A 461 21.262-100.555 -33.049 1.00 61.51 A N
+ATOM 3116 CA LYS A 461 22.553-101.188 -33.239 1.00 69.64 A C
+ATOM 3117 C LYS A 461 23.687-100.195 -33.491 1.00 73.86 A C
+ATOM 3118 O LYS A 461 24.797-100.633 -33.795 1.00 91.95 A O
+ATOM 3119 CB LYS A 461 22.469-102.251 -34.337 1.00 68.86 A C
+ATOM 3120 CG LYS A 461 21.683-103.513 -33.882 1.00 69.19 A C
+ATOM 3121 CD LYS A 461 22.322-104.261 -32.669 1.00 82.40 A C
+ATOM 3122 CE LYS A 461 23.338-105.364 -33.093 1.00 81.52 A C
+ATOM 3123 NZ LYS A 461 24.136-106.101 -32.020 1.00 68.05 A N1+
+ATOM 3124 N GLN A 462 23.455 -98.880 -33.377 1.00 88.07 A N
+ATOM 3125 CA GLN A 462 24.549 -97.934 -33.609 1.00 87.64 A C
+ATOM 3126 C GLN A 462 25.646 -98.080 -32.555 1.00 81.42 A C
+ATOM 3127 O GLN A 462 25.390 -98.381 -31.388 1.00 85.53 A O
+ATOM 3128 CB GLN A 462 24.062 -96.478 -33.653 1.00 94.29 A C
+ATOM 3129 CG GLN A 462 25.156 -95.454 -34.188 1.00 98.48 A C
+ATOM 3130 CD GLN A 462 25.568 -95.603 -35.707 1.00113.79 A C
+ATOM 3131 NE2 GLN A 462 25.261 -94.574 -36.510 1.00116.60 A N
+ATOM 3132 OE1 GLN A 462 26.186 -96.601 -36.119 1.00125.60 A O
+ATOM 3133 N GLU A 463 26.885 -97.860 -32.999 1.00 85.35 A N
+ATOM 3134 CA GLU A 463 28.096 -98.264 -32.302 1.00 95.23 A C
+ATOM 3135 C GLU A 463 28.473 -97.287 -31.193 1.00 91.56 A C
+ATOM 3136 O GLU A 463 28.095 -96.112 -31.203 1.00 88.41 A O
+ATOM 3137 CB GLU A 463 29.259 -98.396 -33.287 1.00 95.60 A C
+ATOM 3138 CG GLU A 463 29.063 -99.443 -34.400 1.00116.35 A C
+ATOM 3139 CD GLU A 463 29.002-100.900 -33.896 1.00128.88 A C
+ATOM 3140 OE1 GLU A 463 29.342-101.160 -32.719 1.00138.11 A O
+ATOM 3141 OE2 GLU A 463 28.623-101.791 -34.695 1.00132.91 A O1-
+ATOM 3142 N GLY A 464 29.230 -97.801 -30.230 1.00 93.41 A N
+ATOM 3143 CA GLY A 464 29.799 -97.003 -29.159 1.00 83.31 A C
+ATOM 3144 C GLY A 464 30.130 -97.861 -27.958 1.00 87.26 A C
+ATOM 3145 O GLY A 464 29.743 -99.033 -27.865 1.00 87.85 A O
+ATOM 3146 N LYS A 465 30.845 -97.251 -27.008 1.00 83.34 A N
+ATOM 3147 CA LYS A 465 31.296 -97.976 -25.826 1.00 79.32 A C
+ATOM 3148 C LYS A 465 30.374 -97.697 -24.646 1.00 83.58 A C
+ATOM 3149 O LYS A 465 29.963 -96.552 -24.399 1.00 83.52 A O
+ATOM 3150 CB LYS A 465 32.749 -97.649 -25.453 1.00 83.05 A C
+ATOM 3151 CG LYS A 465 33.397 -98.741 -24.567 1.00 97.14 A C
+ATOM 3152 CD LYS A 465 34.884 -98.496 -24.300 1.00118.38 A C
+ATOM 3153 CE LYS A 465 35.450 -99.523 -23.326 1.00118.61 A C
+ATOM 3154 NZ LYS A 465 35.206-100.917 -23.794 1.00126.44 A N1+
+ATOM 3155 N SER A 466 30.063 -98.766 -23.917 1.00 81.39 A N
+ATOM 3156 CA SER A 466 29.034 -98.763 -22.892 1.00 79.95 A C
+ATOM 3157 C SER A 466 29.649 -99.210 -21.572 1.00 78.51 A C
+ATOM 3158 O SER A 466 30.272-100.276 -21.505 1.00 88.71 A O
+ATOM 3159 CB SER A 466 27.882 -99.683 -23.309 1.00 74.29 A C
+ATOM 3160 OG SER A 466 26.669 -99.297 -22.697 1.00 98.19 A O
+ATOM 3161 N LEU A 467 29.471 -98.400 -20.528 1.00 77.90 A N
+ATOM 3162 CA LEU A 467 30.048 -98.648 -19.213 1.00 74.17 A C
+ATOM 3163 C LEU A 467 28.943 -98.783 -18.171 1.00 79.08 A C
+ATOM 3164 O LEU A 467 28.133 -97.859 -17.993 1.00 71.79 A O
+ATOM 3165 CB LEU A 467 31.001 -97.522 -18.814 1.00 78.16 A C
+ATOM 3166 CG LEU A 467 31.686 -97.734 -17.457 1.00 90.99 A C
+ATOM 3167 CD1 LEU A 467 32.817 -98.754 -17.554 1.00 83.80 A C
+ATOM 3168 CD2 LEU A 467 32.174 -96.419 -16.862 1.00 78.67 A C
+ATOM 3169 N TYR A 468 28.931 -99.927 -17.476 1.00 86.53 A N
+ATOM 3170 CA TYR A 468 28.017-100.199 -16.365 1.00 79.19 A C
+ATOM 3171 C TYR A 468 28.625 -99.687 -15.066 1.00 76.53 A C
+ATOM 3172 O TYR A 468 29.743-100.074 -14.716 1.00 83.19 A O
+ATOM 3173 CB TYR A 468 27.770-101.700 -16.259 1.00 75.62 A C
+ATOM 3174 CG TYR A 468 26.557-102.129 -15.464 1.00 73.81 A C
+ATOM 3175 CD1 TYR A 468 25.575-102.926 -16.052 1.00 76.06 A C
+ATOM 3176 CD2 TYR A 468 26.411-101.784 -14.123 1.00 66.81 A C
+ATOM 3177 CE1 TYR A 468 24.474-103.358 -15.342 1.00 68.11 A C
+ATOM 3178 CE2 TYR A 468 25.306-102.211 -13.394 1.00 72.59 A C
+ATOM 3179 CZ TYR A 468 24.333-103.001 -14.014 1.00 75.00 A C
+ATOM 3180 OH TYR A 468 23.214-103.432 -13.314 1.00 77.50 A O
+ATOM 3181 N VAL A 469 27.898 -98.839 -14.342 1.00 72.66 A N
+ATOM 3182 CA VAL A 469 28.403 -98.236 -13.110 1.00 79.42 A C
+ATOM 3183 C VAL A 469 27.552 -98.724 -11.940 1.00 74.95 A C
+ATOM 3184 O VAL A 469 26.433 -98.231 -11.720 1.00 73.60 A O
+ATOM 3185 CB VAL A 469 28.421 -96.706 -13.197 1.00 81.21 A C
+ATOM 3186 CG1 VAL A 469 28.764 -96.111 -11.871 1.00 68.36 A C
+ATOM 3187 CG2 VAL A 469 29.449 -96.285 -14.202 1.00 74.21 A C
+ATOM 3188 N LYS A 470 28.121 -99.627 -11.138 1.00 73.05 A N
+ATOM 3189 CA LYS A 470 27.386-100.244 -10.044 1.00 78.53 A C
+ATOM 3190 C LYS A 470 27.030 -99.197 -8.989 1.00 74.93 A C
+ATOM 3191 O LYS A 470 27.549 -98.079 -8.973 1.00 77.36 A O
+ATOM 3192 CB LYS A 470 28.195-101.389 -9.432 1.00 77.83 A C
+ATOM 3193 CG LYS A 470 28.671-102.400 -10.463 1.00 79.45 A C
+ATOM 3194 CD LYS A 470 29.627-103.467 -9.896 1.00 96.69 A C
+ATOM 3195 CE LYS A 470 28.885-104.635 -9.218 1.00105.54 A C
+ATOM 3196 NZ LYS A 470 29.786-105.773 -8.830 1.00108.31 A N1+
+ATOM 3197 N GLY A 471 26.073 -99.544 -8.148 1.00 80.95 A N
+ATOM 3198 CA GLY A 471 25.658 -98.640 -7.096 1.00 73.33 A C
+ATOM 3199 C GLY A 471 24.641 -99.304 -6.197 1.00 77.25 A C
+ATOM 3200 O GLY A 471 24.006-100.304 -6.567 1.00 86.28 A O
+ATOM 3201 N GLU A 472 24.503 -98.717 -4.938 1.00 83.41 A N
+ATOM 3202 CA GLU A 472 23.379 -99.454 -4.389 1.00 77.61 A C
+ATOM 3203 C GLU A 472 22.082 -98.747 -4.770 1.00 74.57 A C
+ATOM 3204 O GLU A 472 22.027 -97.507 -4.763 1.00 72.95 A O
+ATOM 3205 CB GLU A 472 23.483 -99.568 -2.874 1.00 75.98 A C
+ATOM 3206 CG GLU A 472 22.627-100.687 -2.303 1.00 90.66 A C
+ATOM 3207 CD GLU A 472 22.338-100.510 -0.831 1.00104.29 A C
+ATOM 3208 OE1 GLU A 472 23.074 -99.745 -0.168 1.00107.68 A O
+ATOM 3209 OE2 GLU A 472 21.378-101.138 -0.335 1.00112.09 A O1-
+ATOM 3210 N PRO A 473 21.052 -99.511 -5.138 1.00 78.23 A N
+ATOM 3211 CA PRO A 473 19.807 -98.880 -5.591 1.00 75.68 A C
+ATOM 3212 C PRO A 473 19.252 -97.979 -4.504 1.00 76.77 A C
+ATOM 3213 O PRO A 473 18.863 -98.437 -3.432 1.00 74.95 A O
+ATOM 3214 CB PRO A 473 18.888-100.075 -5.856 1.00 73.75 A C
+ATOM 3215 CG PRO A 473 19.820-101.203 -6.166 1.00 73.68 A C
+ATOM 3216 CD PRO A 473 21.043-100.973 -5.334 1.00 76.47 A C
+ATOM 3217 N ILE A 474 19.121 -96.697 -4.833 1.00 72.91 A N
+ATOM 3218 CA ILE A 474 18.826 -95.706 -3.820 1.00 80.68 A C
+ATOM 3219 C ILE A 474 17.462 -95.927 -3.213 1.00 77.82 A C
+ATOM 3220 O ILE A 474 17.162 -95.360 -2.164 1.00 88.19 A O
+ATOM 3221 CB ILE A 474 18.930 -94.285 -4.409 1.00 78.52 A C
+ATOM 3222 CG1 ILE A 474 17.832 -94.063 -5.446 1.00 74.34 A C
+ATOM 3223 CG2 ILE A 474 20.274 -94.097 -5.072 1.00 73.93 A C
+ATOM 3224 CD1 ILE A 474 17.863 -92.702 -6.064 1.00 79.87 A C
+ATOM 3225 N ILE A 475 16.621 -96.746 -3.846 1.00 86.44 A N
+ATOM 3226 CA ILE A 475 15.371 -97.116 -3.205 1.00 82.71 A C
+ATOM 3227 C ILE A 475 15.625 -97.814 -1.883 1.00 82.41 A C
+ATOM 3228 O ILE A 475 14.739 -97.846 -1.021 1.00 87.04 A O
+ATOM 3229 CB ILE A 475 14.510 -98.011 -4.122 1.00 82.53 A C
+ATOM 3230 CG1 ILE A 475 13.048 -97.997 -3.658 1.00 80.78 A C
+ATOM 3231 CG2 ILE A 475 15.048 -99.435 -4.150 1.00 84.22 A C
+ATOM 3232 CD1 ILE A 475 12.495 -96.598 -3.499 1.00 74.11 A C
+ATOM 3233 N ASN A 476 16.823 -98.375 -1.691 1.00 77.48 A N
+ATOM 3234 CA ASN A 476 17.097 -99.123 -0.471 1.00 77.64 A C
+ATOM 3235 C ASN A 476 17.334 -98.222 0.740 1.00 83.03 A C
+ATOM 3236 O ASN A 476 17.077 -98.659 1.868 1.00 86.37 A O
+ATOM 3237 CB ASN A 476 18.280-100.066 -0.705 1.00 82.21 A C
+ATOM 3238 CG ASN A 476 17.911-101.246 -1.591 1.00 93.07 A C
+ATOM 3239 ND2 ASN A 476 18.870-102.116 -1.850 1.00 95.81 A N
+ATOM 3240 OD1 ASN A 476 16.779-101.359 -2.049 1.00 89.57 A O
+ATOM 3241 N PHE A 477 17.729 -96.961 0.535 1.00 89.53 A N
+ATOM 3242 CA PHE A 477 18.058 -96.057 1.637 1.00 83.21 A C
+ATOM 3243 C PHE A 477 16.824 -95.457 2.290 1.00 83.86 A C
+ATOM 3244 O PHE A 477 16.956 -94.594 3.159 1.00 91.18 A O
+ATOM 3245 CB PHE A 477 18.951 -94.893 1.181 1.00 84.30 A C
+ATOM 3246 CG PHE A 477 20.216 -95.300 0.460 1.00 82.45 A C
+ATOM 3247 CD1 PHE A 477 21.059 -94.317 -0.048 1.00 81.34 A C
+ATOM 3248 CD2 PHE A 477 20.575 -96.628 0.287 1.00 78.83 A C
+ATOM 3249 CE1 PHE A 477 22.231 -94.644 -0.723 1.00 88.06 A C
+ATOM 3250 CE2 PHE A 477 21.746 -96.963 -0.387 1.00 78.05 A C
+ATOM 3251 CZ PHE A 477 22.576 -95.967 -0.895 1.00 76.63 A C
+ATOM 3252 N TYR A 478 15.638 -95.839 1.858 1.00 90.36 A N
+ATOM 3253 CA TYR A 478 14.408 -95.233 2.331 1.00 83.53 A C
+ATOM 3254 C TYR A 478 13.692 -96.194 3.272 1.00 86.44 A C
+ATOM 3255 O TYR A 478 13.660 -97.407 3.034 1.00 82.42 A O
+ATOM 3256 CB TYR A 478 13.517 -94.844 1.130 1.00 73.56 A C
+ATOM 3257 CG TYR A 478 13.952 -93.554 0.439 1.00 75.81 A C
+ATOM 3258 CD1 TYR A 478 13.429 -92.336 0.844 1.00 74.36 A C
+ATOM 3259 CD2 TYR A 478 14.913 -93.552 -0.576 1.00 73.88 A C
+ATOM 3260 CE1 TYR A 478 13.826 -91.152 0.258 1.00 76.73 A C
+ATOM 3261 CE2 TYR A 478 15.327 -92.362 -1.167 1.00 71.50 A C
+ATOM 3262 CZ TYR A 478 14.773 -91.159 -0.739 1.00 76.29 A C
+ATOM 3263 OH TYR A 478 15.138 -89.939 -1.284 1.00 82.33 A O
+ATOM 3264 N ASP A 479 13.140 -95.656 4.352 1.00 80.73 A N
+ATOM 3265 CA ASP A 479 12.340 -96.471 5.260 1.00 82.71 A C
+ATOM 3266 C ASP A 479 10.922 -96.581 4.722 1.00 91.45 A C
+ATOM 3267 O ASP A 479 10.225 -95.562 4.644 1.00 93.94 A O
+ATOM 3268 CB ASP A 479 12.323 -95.866 6.668 1.00 84.13 A C
+ATOM 3269 CG ASP A 479 12.065 -96.917 7.785 1.00 98.86 A C
+ATOM 3270 OD1 ASP A 479 11.260 -97.862 7.578 1.00 99.53 A O
+ATOM 3271 OD2 ASP A 479 12.680 -96.794 8.877 1.00105.26 A O1-
+ATOM 3272 N PRO A 480 10.451 -97.784 4.366 1.00 82.33 A N
+ATOM 3273 CA PRO A 480 9.097 -97.922 3.808 1.00 82.49 A C
+ATOM 3274 C PRO A 480 8.040 -97.186 4.588 1.00 83.36 A C
+ATOM 3275 O PRO A 480 7.057 -96.720 3.998 1.00 88.45 A O
+ATOM 3276 CB PRO A 480 8.851 -99.438 3.850 1.00 78.53 A C
+ATOM 3277 CG PRO A 480 10.176-100.017 3.743 1.00 80.25 A C
+ATOM 3278 CD PRO A 480 11.147 -99.074 4.426 1.00 78.35 A C
+ATOM 3279 N LEU A 481 8.214 -97.042 5.898 1.00 82.76 A N
+ATOM 3280 CA LEU A 481 7.113 -96.560 6.721 1.00 87.87 A C
+ATOM 3281 C LEU A 481 6.937 -95.053 6.592 1.00 90.34 A C
+ATOM 3282 O LEU A 481 5.807 -94.566 6.505 1.00 93.37 A O
+ATOM 3283 CB LEU A 481 7.326 -96.995 8.177 1.00 95.15 A C
+ATOM 3284 CG LEU A 481 7.160 -98.511 8.457 1.00111.94 A C
+ATOM 3285 CD1 LEU A 481 8.408 -99.363 8.135 1.00107.82 A C
+ATOM 3286 CD2 LEU A 481 6.709 -98.764 9.885 1.00120.66 A C
+ATOM 3287 N VAL A 482 8.038 -94.303 6.539 1.00 99.57 A N
+ATOM 3288 CA VAL A 482 7.970 -92.849 6.408 1.00100.32 A C
+ATOM 3289 C VAL A 482 8.108 -92.376 4.966 1.00 99.57 A C
+ATOM 3290 O VAL A 482 8.140 -91.160 4.720 1.00106.15 A O
+ATOM 3291 CB VAL A 482 9.044 -92.159 7.270 1.00101.95 A C
+ATOM 3292 CG1 VAL A 482 8.604 -92.111 8.734 1.00100.96 A C
+ATOM 3293 CG2 VAL A 482 10.406 -92.849 7.073 1.00 96.43 A C
+ATOM 3294 N PHE A 483 8.201 -93.297 4.009 1.00 96.53 A N
+ATOM 3295 CA PHE A 483 8.496 -92.951 2.625 1.00100.03 A C
+ATOM 3296 C PHE A 483 7.196 -92.665 1.877 1.00105.17 A C
+ATOM 3297 O PHE A 483 6.377 -93.584 1.703 1.00106.91 A O
+ATOM 3298 CB PHE A 483 9.260 -94.096 1.973 1.00 91.20 A C
+ATOM 3299 CG PHE A 483 9.312 -94.028 0.491 1.00 97.52 A C
+ATOM 3300 CD1 PHE A 483 10.321 -93.324 -0.138 1.00 94.79 A C
+ATOM 3301 CD2 PHE A 483 8.357 -94.678 -0.278 1.00 86.83 A C
+ATOM 3302 CE1 PHE A 483 10.374 -93.265 -1.506 1.00 94.47 A C
+ATOM 3303 CE2 PHE A 483 8.396 -94.615 -1.640 1.00 93.51 A C
+ATOM 3304 CZ PHE A 483 9.409 -93.913 -2.261 1.00 97.98 A C
+ATOM 3305 N PRO A 484 6.937 -91.416 1.457 1.00 96.90 A N
+ATOM 3306 CA PRO A 484 5.686 -91.116 0.737 1.00 92.62 A C
+ATOM 3307 C PRO A 484 5.719 -91.626 -0.699 1.00 96.43 A C
+ATOM 3308 O PRO A 484 6.688 -91.415 -1.428 1.00 98.25 A O
+ATOM 3309 CB PRO A 484 5.603 -89.587 0.790 1.00 89.61 A C
+ATOM 3310 CG PRO A 484 7.035 -89.155 0.844 1.00 96.99 A C
+ATOM 3311 CD PRO A 484 7.835 -90.250 1.514 1.00 95.91 A C
+ATOM 3312 N SER A 485 4.654 -92.311 -1.094 1.00 88.12 A N
+ATOM 3313 CA SER A 485 4.403 -92.683 -2.477 1.00 95.86 A C
+ATOM 3314 C SER A 485 3.382 -91.779 -3.158 1.00 99.16 A C
+ATOM 3315 O SER A 485 3.038 -92.016 -4.318 1.00100.46 A O
+ATOM 3316 CB SER A 485 3.955 -94.145 -2.542 1.00 92.93 A C
+ATOM 3317 OG SER A 485 2.818 -94.366 -1.718 1.00110.52 A O
+ATOM 3318 N ASP A 486 2.875 -90.766 -2.464 1.00101.59 A N
+ATOM 3319 CA ASP A 486 1.861 -89.885 -3.023 1.00 97.65 A C
+ATOM 3320 C ASP A 486 2.467 -88.727 -3.799 1.00103.90 A C
+ATOM 3321 O ASP A 486 1.729 -87.830 -4.231 1.00113.10 A O
+ATOM 3322 CB ASP A 486 0.923 -89.344 -1.924 1.00 97.24 A C
+ATOM 3323 CG ASP A 486 1.659 -88.569 -0.813 1.00106.10 A C
+ATOM 3324 OD1 ASP A 486 2.905 -88.403 -0.892 1.00 97.68 A O
+ATOM 3325 OD2 ASP A 486 0.966 -88.087 0.123 1.00100.08 A O1-
+ATOM 3326 N GLN A 487 3.788 -88.720 -3.984 1.00 90.44 A N
+ATOM 3327 CA GLN A 487 4.458 -87.618 -4.662 1.00 92.67 A C
+ATOM 3328 C GLN A 487 4.543 -87.920 -6.156 1.00 92.72 A C
+ATOM 3329 O GLN A 487 5.326 -88.777 -6.585 1.00 96.61 A O
+ATOM 3330 CB GLN A 487 5.835 -87.384 -4.048 1.00 89.19 A C
+ATOM 3331 CG GLN A 487 5.776 -87.127 -2.555 1.00 89.19 A C
+ATOM 3332 CD GLN A 487 4.981 -85.859 -2.206 1.00 94.87 A C
+ATOM 3333 NE2 GLN A 487 4.022 -85.983 -1.283 1.00 92.43 A N
+ATOM 3334 OE1 GLN A 487 5.231 -84.784 -2.766 1.00100.86 A O
+ATOM 3335 N PHE A 488 3.755 -87.183 -6.949 1.00 83.71 A N
+ATOM 3336 CA PHE A 488 3.739 -87.357 -8.402 1.00 90.40 A C
+ATOM 3337 C PHE A 488 4.871 -86.549 -9.030 1.00 81.72 A C
+ATOM 3338 O PHE A 488 5.833 -87.113 -9.560 1.00 82.18 A O
+ATOM 3339 CB PHE A 488 2.367 -86.945 -8.949 1.00 88.42 A C
+ATOM 3340 CG PHE A 488 2.088 -87.424 -10.348 1.00 90.23 A C
+ATOM 3341 CD1 PHE A 488 1.983 -88.772 -10.625 1.00 86.03 A C
+ATOM 3342 CD2 PHE A 488 1.870 -86.518 -11.375 1.00 97.38 A C
+ATOM 3343 CE1 PHE A 488 1.713 -89.215 -11.911 1.00 94.78 A C
+ATOM 3344 CE2 PHE A 488 1.592 -86.952 -12.667 1.00 90.03 A C
+ATOM 3345 CZ PHE A 488 1.515 -88.304 -12.933 1.00 84.69 A C
+ATOM 3346 N ASP A 489 4.774 -85.218 -8.952 1.00 82.03 A N
+ATOM 3347 CA ASP A 489 5.874 -84.307 -9.267 1.00 84.27 A C
+ATOM 3348 C ASP A 489 6.584 -83.926 -7.973 1.00 84.32 A C
+ATOM 3349 O ASP A 489 5.978 -83.301 -7.089 1.00 94.67 A O
+ATOM 3350 CB ASP A 489 5.378 -83.041 -9.962 1.00 94.20 A C
+ATOM 3351 CG ASP A 489 6.379 -81.910 -9.870 1.00100.20 A C
+ATOM 3352 OD1 ASP A 489 7.552 -82.127 -10.239 1.00 94.00 A O
+ATOM 3353 OD2 ASP A 489 6.007 -80.824 -9.384 1.00112.31 A O1-
+ATOM 3354 N ALA A 490 7.867 -84.271 -7.876 1.00 83.95 A N
+ATOM 3355 CA ALA A 490 8.586 -84.141 -6.616 1.00 85.30 A C
+ATOM 3356 C ALA A 490 10.078 -84.015 -6.889 1.00 85.53 A C
+ATOM 3357 O ALA A 490 10.545 -84.220 -8.010 1.00 76.06 A O
+ATOM 3358 CB ALA A 490 8.287 -85.324 -5.695 1.00 81.71 A C
+ATOM 3359 N SER A 491 10.813 -83.629 -5.851 1.00 78.80 A N
+ATOM 3360 CA SER A 491 12.273 -83.581 -5.845 1.00 80.39 A C
+ATOM 3361 C SER A 491 12.827 -84.588 -4.837 1.00 82.80 A C
+ATOM 3362 O SER A 491 12.083 -85.321 -4.184 1.00 85.14 A O
+ATOM 3363 CB SER A 491 12.766 -82.175 -5.513 1.00 83.18 A C
+ATOM 3364 OG SER A 491 12.003 -81.617 -4.461 1.00 85.74 A O
+ATOM 3365 N ILE A 492 14.158 -84.629 -4.722 1.00 78.59 A N
+ATOM 3366 CA ILE A 492 14.777 -85.442 -3.678 1.00 75.88 A C
+ATOM 3367 C ILE A 492 14.502 -84.831 -2.315 1.00 87.26 A C
+ATOM 3368 O ILE A 492 14.086 -85.520 -1.369 1.00 84.99 A O
+ATOM 3369 CB ILE A 492 16.289 -85.592 -3.922 1.00 76.26 A C
+ATOM 3370 CG1 ILE A 492 16.564 -86.628 -5.000 1.00 82.25 A C
+ATOM 3371 CG2 ILE A 492 16.986 -86.006 -2.658 1.00 72.65 A C
+ATOM 3372 CD1 ILE A 492 16.525 -88.035 -4.492 1.00 81.82 A C
+ATOM 3373 N SER A 493 14.704 -83.517 -2.211 1.00 76.46 A N
+ATOM 3374 CA SER A 493 14.561 -82.835 -0.934 1.00 94.87 A C
+ATOM 3375 C SER A 493 13.098 -82.700 -0.506 1.00 89.58 A C
+ATOM 3376 O SER A 493 12.819 -82.635 0.694 1.00 92.61 A O
+ATOM 3377 CB SER A 493 15.240 -81.468 -1.013 1.00 77.86 A C
+ATOM 3378 OG SER A 493 14.516 -80.582 -1.853 1.00107.73 A O
+ATOM 3379 N GLN A 494 12.156 -82.639 -1.455 1.00 79.64 A N
+ATOM 3380 CA GLN A 494 10.741 -82.609 -1.084 1.00 83.34 A C
+ATOM 3381 C GLN A 494 10.305 -83.941 -0.483 1.00 83.12 A C
+ATOM 3382 O GLN A 494 9.475 -83.969 0.437 1.00 81.93 A O
+ATOM 3383 CB GLN A 494 9.879 -82.256 -2.304 1.00 85.97 A C
+ATOM 3384 CG GLN A 494 8.417 -81.873 -2.017 1.00106.44 A C
+ATOM 3385 CD GLN A 494 7.663 -81.439 -3.278 1.00128.44 A C
+ATOM 3386 NE2 GLN A 494 6.388 -81.825 -3.376 1.00140.60 A N
+ATOM 3387 OE1 GLN A 494 8.224 -80.765 -4.150 1.00134.93 A O
+ATOM 3388 N VAL A 495 10.856 -85.049 -0.986 1.00 83.18 A N
+ATOM 3389 CA VAL A 495 10.634 -86.347 -0.360 1.00 84.75 A C
+ATOM 3390 C VAL A 495 11.279 -86.384 1.018 1.00 94.32 A C
+ATOM 3391 O VAL A 495 10.659 -86.819 2.001 1.00 97.39 A O
+ATOM 3392 CB VAL A 495 11.162 -87.471 -1.261 1.00 87.30 A C
+ATOM 3393 CG1 VAL A 495 11.554 -88.660 -0.431 1.00 80.74 A C
+ATOM 3394 CG2 VAL A 495 10.090 -87.865 -2.233 1.00 78.39 A C
+ATOM 3395 N ASN A 496 12.534 -85.914 1.113 1.00 87.03 A N
+ATOM 3396 CA ASN A 496 13.166 -85.762 2.422 1.00101.65 A C
+ATOM 3397 C ASN A 496 12.302 -84.948 3.372 1.00101.37 A C
+ATOM 3398 O ASN A 496 12.321 -85.191 4.579 1.00 98.60 A O
+ATOM 3399 CB ASN A 496 14.529 -85.087 2.292 1.00 89.80 A C
+ATOM 3400 CG ASN A 496 15.621 -86.060 1.934 1.00100.02 A C
+ATOM 3401 ND2 ASN A 496 16.860 -85.555 1.821 1.00 91.41 A N
+ATOM 3402 OD1 ASN A 496 15.364 -87.265 1.773 1.00101.56 A O
+ATOM 3403 N GLU A 497 11.542 -83.985 2.841 1.00 81.00 A N
+ATOM 3404 CA GLU A 497 10.744 -83.064 3.641 1.00 83.56 A C
+ATOM 3405 C GLU A 497 9.458 -83.721 4.135 1.00 87.49 A C
+ATOM 3406 O GLU A 497 9.111 -83.606 5.318 1.00 89.74 A O
+ATOM 3407 CB GLU A 497 10.438 -81.812 2.813 1.00 85.32 A C
+ATOM 3408 CG GLU A 497 9.921 -80.610 3.594 1.00118.00 A C
+ATOM 3409 CD GLU A 497 9.762 -79.370 2.719 1.00144.91 A C
+ATOM 3410 OE1 GLU A 497 9.770 -79.504 1.474 1.00157.59 A O
+ATOM 3411 OE2 GLU A 497 9.642 -78.259 3.279 1.00146.63 A O1-
+ATOM 3412 N LYS A 498 8.731 -84.408 3.244 1.00 85.99 A N
+ATOM 3413 CA LYS A 498 7.555 -85.151 3.694 1.00 88.45 A C
+ATOM 3414 C LYS A 498 7.968 -86.256 4.673 1.00 91.19 A C
+ATOM 3415 O LYS A 498 7.217 -86.594 5.610 1.00 92.08 A O
+ATOM 3416 CB LYS A 498 6.776 -85.685 2.470 1.00 86.50 A C
+ATOM 3417 CG LYS A 498 5.324 -86.188 2.726 1.00 94.22 A C
+ATOM 3418 CD LYS A 498 4.195 -85.130 2.587 1.00 94.59 A C
+ATOM 3419 CE LYS A 498 2.812 -85.781 2.911 1.00104.46 A C
+ATOM 3420 NZ LYS A 498 1.591 -84.902 2.988 1.00105.82 A N1+
+ATOM 3421 N ILE A 499 9.193 -86.771 4.522 1.00 83.81 A N
+ATOM 3422 CA ILE A 499 9.714 -87.729 5.489 1.00 88.55 A C
+ATOM 3423 C ILE A 499 10.015 -87.049 6.816 1.00 94.00 A C
+ATOM 3424 O ILE A 499 9.629 -87.542 7.875 1.00101.03 A O
+ATOM 3425 CB ILE A 499 10.952 -88.454 4.939 1.00 82.42 A C
+ATOM 3426 CG1 ILE A 499 10.545 -89.421 3.825 1.00 74.87 A C
+ATOM 3427 CG2 ILE A 499 11.663 -89.150 6.061 1.00 78.83 A C
+ATOM 3428 CD1 ILE A 499 11.530 -90.543 3.553 1.00 78.25 A C
+ATOM 3429 N ASN A 500 10.712 -85.914 6.782 1.00 83.29 A N
+ATOM 3430 CA ASN A 500 11.054 -85.218 8.022 1.00 87.07 A C
+ATOM 3431 C ASN A 500 9.803 -84.802 8.796 1.00 90.24 A C
+ATOM 3432 O ASN A 500 9.827 -84.794 10.030 1.00 96.78 A O
+ATOM 3433 CB ASN A 500 11.945 -83.997 7.732 1.00 94.17 A C
+ATOM 3434 CG ASN A 500 13.446 -84.295 7.889 1.00107.46 A C
+ATOM 3435 ND2 ASN A 500 14.232 -83.262 8.194 1.00119.87 A N
+ATOM 3436 OD1 ASN A 500 13.887 -85.434 7.720 1.00109.37 A O
+ATOM 3437 N GLN A 501 8.698 -84.476 8.104 1.00 86.90 A N
+ATOM 3438 CA GLN A 501 7.444 -84.196 8.817 1.00 92.35 A C
+ATOM 3439 C GLN A 501 6.838 -85.477 9.415 1.00 98.27 A C
+ATOM 3440 O GLN A 501 6.393 -85.482 10.581 1.00103.05 A O
+ATOM 3441 CB GLN A 501 6.430 -83.475 7.893 1.00 94.84 A C
+ATOM 3442 CG GLN A 501 5.263 -84.356 7.306 1.00106.95 A C
+ATOM 3443 CD GLN A 501 4.225 -83.590 6.461 1.00128.48 A C
+ATOM 3444 NE2 GLN A 501 3.271 -84.326 5.886 1.00136.92 A N
+ATOM 3445 OE1 GLN A 501 4.277 -82.365 6.338 1.00132.66 A O
+ATOM 3446 N SER A 502 6.828 -86.584 8.651 1.00 91.83 A N
+ATOM 3447 CA SER A 502 6.194 -87.788 9.180 1.00 91.85 A C
+ATOM 3448 C SER A 502 7.044 -88.488 10.245 1.00 97.09 A C
+ATOM 3449 O SER A 502 6.517 -89.328 10.987 1.00104.65 A O
+ATOM 3450 CB SER A 502 5.857 -88.751 8.041 1.00 93.50 A C
+ATOM 3451 OG SER A 502 4.924 -89.725 8.477 1.00 92.32 A O
+ATOM 3452 N LEU A 503 8.346 -88.181 10.319 1.00 90.03 A N
+ATOM 3453 CA LEU A 503 9.189 -88.576 11.448 1.00 94.62 A C
+ATOM 3454 C LEU A 503 9.025 -87.610 12.611 1.00106.09 A C
+ATOM 3455 O LEU A 503 9.054 -88.022 13.780 1.00106.30 A O
+ATOM 3456 CB LEU A 503 10.664 -88.631 11.034 1.00 82.19 A C
+ATOM 3457 CG LEU A 503 11.196 -89.852 10.285 1.00 94.86 A C
+ATOM 3458 CD1 LEU A 503 12.631 -89.613 9.904 1.00 95.86 A C
+ATOM 3459 CD2 LEU A 503 11.078 -91.107 11.115 1.00 81.33 A C
+ATOM 3460 N ALA A 504 8.877 -86.319 12.298 1.00 99.08 A N
+ATOM 3461 CA ALA A 504 8.663 -85.317 13.332 1.00100.32 A C
+ATOM 3462 C ALA A 504 7.476 -85.686 14.205 1.00106.78 A C
+ATOM 3463 O ALA A 504 7.610 -85.784 15.430 1.00 98.93 A O
+ATOM 3464 CB ALA A 504 8.463 -83.939 12.703 1.00100.52 A C
+ATOM 3465 N PHE A 505 6.304 -85.931 13.598 1.00101.97 A N
+ATOM 3466 CA PHE A 505 5.148 -86.193 14.458 1.00106.89 A C
+ATOM 3467 C PHE A 505 5.309 -87.479 15.271 1.00109.70 A C
+ATOM 3468 O PHE A 505 4.676 -87.616 16.329 1.00118.00 A O
+ATOM 3469 CB PHE A 505 3.839 -86.169 13.649 1.00110.85 A C
+ATOM 3470 CG PHE A 505 3.154 -84.792 13.637 1.00116.60 A C
+ATOM 3471 CD1 PHE A 505 3.255 -83.932 14.750 1.00122.94 A C
+ATOM 3472 CD2 PHE A 505 2.394 -84.362 12.547 1.00115.52 A C
+ATOM 3473 CE1 PHE A 505 2.636 -82.665 14.769 1.00124.94 A C
+ATOM 3474 CE2 PHE A 505 1.765 -83.089 12.568 1.00111.73 A C
+ATOM 3475 CZ PHE A 505 1.896 -82.246 13.683 1.00117.10 A C
+ATOM 3476 N ILE A 506 6.206 -88.379 14.856 1.00103.40 A N
+ATOM 3477 CA ILE A 506 6.601 -89.493 15.718 1.00102.28 A C
+ATOM 3478 C ILE A 506 7.436 -88.994 16.890 1.00101.76 A C
+ATOM 3479 O ILE A 506 7.240 -89.418 18.036 1.00116.17 A O
+ATOM 3480 CB ILE A 506 7.371 -90.554 14.914 1.00 95.41 A C
+ATOM 3481 CG1 ILE A 506 6.537 -91.008 13.718 1.00 99.11 A C
+ATOM 3482 CG2 ILE A 506 7.739 -91.725 15.816 1.00 86.15 A C
+ATOM 3483 CD1 ILE A 506 7.242 -91.970 12.823 1.00101.16 A C
+ATOM 3484 N ARG A 507 8.396 -88.106 16.620 1.00101.60 A N
+ATOM 3485 CA ARG A 507 9.229 -87.579 17.696 1.00110.06 A C
+ATOM 3486 C ARG A 507 8.367 -86.876 18.744 1.00127.25 A C
+ATOM 3487 O ARG A 507 8.447 -87.191 19.938 1.00119.03 A O
+ATOM 3488 CB ARG A 507 10.297 -86.636 17.122 1.00 99.96 A C
+ATOM 3489 CG ARG A 507 11.385 -86.202 18.110 1.00102.16 A C
+ATOM 3490 CD ARG A 507 12.214 -85.028 17.578 1.00112.67 A C
+ATOM 3491 NE ARG A 507 11.355 -83.959 17.053 1.00113.23 A N
+ATOM 3492 CZ ARG A 507 10.902 -82.919 17.761 1.00124.85 A C
+ATOM 3493 NH1 ARG A 507 11.226 -82.790 19.044 1.00134.76 A N1+
+ATOM 3494 NH2 ARG A 507 10.120 -82.002 17.187 1.00124.95 A N
+ATOM 3495 N LYS A 508 7.497 -85.954 18.307 1.00109.65 A N
+ATOM 3496 CA LYS A 508 6.631 -85.250 19.250 1.00113.83 A C
+ATOM 3497 C LYS A 508 5.720 -86.208 20.004 1.00113.00 A C
+ATOM 3498 O LYS A 508 5.590 -86.105 21.227 1.00116.37 A O
+ATOM 3499 CB LYS A 508 5.794 -84.177 18.551 1.00115.16 A C
+ATOM 3500 CG LYS A 508 6.538 -82.885 18.273 1.00117.99 A C
+ATOM 3501 CD LYS A 508 5.648 -81.860 17.552 1.00119.78 A C
+ATOM 3502 CE LYS A 508 6.479 -80.781 16.839 1.00116.25 A C
+ATOM 3503 NZ LYS A 508 5.649 -79.920 15.945 1.00111.60 A N1+
+ATOM 3504 N SER A 509 5.068 -87.144 19.307 1.00115.23 A N
+ATOM 3505 CA SER A 509 4.130 -87.997 20.037 1.00109.68 A C
+ATOM 3506 C SER A 509 4.848 -88.908 21.031 1.00113.97 A C
+ATOM 3507 O SER A 509 4.331 -89.158 22.127 1.00122.37 A O
+ATOM 3508 CB SER A 509 3.268 -88.807 19.070 1.00 95.58 A C
+ATOM 3509 OG SER A 509 4.052 -89.558 18.164 1.00 92.75 A O
+ATOM 3510 N ASP A 510 6.048 -89.388 20.689 1.00111.10 A N
+ATOM 3511 CA ASP A 510 6.792 -90.236 21.622 1.00111.10 A C
+ATOM 3512 C ASP A 510 7.334 -89.445 22.804 1.00111.10 A C
+ATOM 3513 O ASP A 510 7.368 -89.952 23.935 1.00111.10 A O
+ATOM 3514 CB ASP A 510 7.937 -90.948 20.912 1.00111.10 A C
+ATOM 3515 CG ASP A 510 7.481 -92.186 20.194 1.00111.10 A C
+ATOM 3516 OD1 ASP A 510 6.351 -92.635 20.487 1.00111.10 A O
+ATOM 3517 OD2 ASP A 510 8.250 -92.713 19.355 1.00111.10 A O1-
+ATOM 3518 N GLU A 511 7.776 -88.210 22.571 1.00111.31 A N
+ATOM 3519 CA GLU A 511 8.256 -87.409 23.687 1.00111.31 A C
+ATOM 3520 C GLU A 511 7.119 -86.936 24.591 1.00111.31 A C
+ATOM 3521 O GLU A 511 7.319 -86.822 25.804 1.00111.31 A O
+ATOM 3522 CB GLU A 511 9.075 -86.240 23.165 1.00111.31 A C
+ATOM 3523 CG GLU A 511 10.362 -86.691 22.499 1.00111.31 A C
+ATOM 3524 CD GLU A 511 11.141 -85.533 21.896 1.00111.31 A C
+ATOM 3525 OE1 GLU A 511 10.557 -84.424 21.782 1.00111.31 A O
+ATOM 3526 OE2 GLU A 511 12.337 -85.727 21.553 1.00111.31 A O1-
+ATOM 3527 N LEU A 512 5.926 -86.663 24.044 1.00 98.81 A N
+ATOM 3528 CA LEU A 512 4.781 -86.372 24.907 1.00 98.81 A C
+ATOM 3529 C LEU A 512 4.388 -87.595 25.718 1.00 98.81 A C
+ATOM 3530 O LEU A 512 4.138 -87.496 26.923 1.00 98.81 A O
+ATOM 3531 CB LEU A 512 3.575 -85.888 24.105 1.00 98.81 A C
+ATOM 3532 CG LEU A 512 3.513 -84.440 23.617 1.00 98.81 A C
+ATOM 3533 CD1 LEU A 512 2.056 -84.052 23.354 1.00 98.81 A C
+ATOM 3534 CD2 LEU A 512 4.171 -83.467 24.572 1.00 98.81 A C
+ATOM 3535 N LEU A 513 4.318 -88.759 25.075 1.00101.12 A N
+ATOM 3536 CA LEU A 513 3.996 -89.963 25.830 1.00101.12 A C
+ATOM 3537 C LEU A 513 5.024 -90.247 26.922 1.00101.12 A C
+ATOM 3538 O LEU A 513 4.659 -90.741 27.993 1.00101.12 A O
+ATOM 3539 CB LEU A 513 3.869 -91.160 24.892 1.00101.12 A C
+ATOM 3540 CG LEU A 513 3.334 -92.460 25.501 1.00101.12 A C
+ATOM 3541 CD1 LEU A 513 2.275 -93.113 24.608 1.00101.12 A C
+ATOM 3542 CD2 LEU A 513 4.493 -93.423 25.767 1.00101.12 A C
+ATOM 3543 N SER A 514 6.307 -89.952 26.688 1.00105.32 A N
+ATOM 3544 CA SER A 514 7.273 -90.159 27.764 1.00105.32 A C
+ATOM 3545 C SER A 514 7.267 -89.021 28.781 1.00105.32 A C
+ATOM 3546 O SER A 514 7.740 -89.218 29.909 1.00105.32 A O
+ATOM 3547 CB SER A 514 8.692 -90.347 27.209 1.00105.32 A C
+ATOM 3548 OG SER A 514 9.419 -89.126 27.197 1.00105.32 A O
+ATOM 3549 N ALA A 515 6.731 -87.846 28.419 1.00102.87 A N
+ATOM 3550 CA ALA A 515 6.725 -86.704 29.332 1.00102.87 A C
+ATOM 3551 C ALA A 515 5.656 -86.834 30.408 1.00102.87 A C
+ATOM 3552 O ALA A 515 5.835 -86.304 31.512 1.00102.87 A O
+ATOM 3553 CB ALA A 515 6.529 -85.391 28.568 1.00102.87 A C
+ATOM 3554 N ILE A 516 4.544 -87.525 30.117 1.00 99.24 A N
+ATOM 3555 CA ILE A 516 3.556 -87.815 31.159 1.00 99.24 A C
+ATOM 3556 C ILE A 516 3.976 -88.979 32.045 1.00 99.24 A C
+ATOM 3557 O ILE A 516 3.285 -89.277 33.029 1.00 99.24 A O
+ATOM 3558 CB ILE A 516 2.155 -88.079 30.568 1.00 99.24 A C
+ATOM 3559 CG1 ILE A 516 2.175 -89.269 29.600 1.00 99.24 A C
+ATOM 3560 CG2 ILE A 516 1.614 -86.821 29.905 1.00 99.24 A C
+ATOM 3561 CD1 ILE A 516 0.819 -89.960 29.438 1.00 99.24 A C
+ATOM 3562 N GLY A 517 5.084 -89.654 31.725 1.00 97.92 A N
+ATOM 3563 CA GLY A 517 5.647 -90.602 32.670 1.00 97.92 A C
+ATOM 3564 C GLY A 517 5.950 -89.946 34.002 1.00 97.92 A C
+ATOM 3565 O GLY A 517 5.539 -90.433 35.060 1.00 97.92 A O
+ATOM 3566 N GLY A 518 6.609 -88.785 33.960 1.00 89.76 A N
+ATOM 3567 CA GLY A 518 7.027 -88.063 35.142 1.00 89.76 A C
+ATOM 3568 C GLY A 518 5.925 -87.392 35.933 1.00 89.76 A C
+ATOM 3569 O GLY A 518 6.213 -86.584 36.820 1.00 89.76 A O
+ATOM 3570 N TYR A 519 4.666 -87.683 35.638 1.00 88.53 A N
+ATOM 3571 CA TYR A 519 3.540 -87.107 36.353 1.00 88.53 A C
+ATOM 3572 C TYR A 519 2.852 -88.211 37.143 1.00 88.53 A C
+ATOM 3573 O TYR A 519 2.939 -89.392 36.787 1.00 88.53 A O
+ATOM 3574 CB TYR A 519 2.593 -86.408 35.369 1.00 88.53 A C
+ATOM 3575 CG TYR A 519 3.225 -85.144 34.831 1.00 88.53 A C
+ATOM 3576 CD1 TYR A 519 4.330 -85.199 33.982 1.00 88.53 A C
+ATOM 3577 CD2 TYR A 519 2.741 -83.895 35.186 1.00 88.53 A C
+ATOM 3578 CE1 TYR A 519 4.941 -84.039 33.505 1.00 88.53 A C
+ATOM 3579 CE2 TYR A 519 3.338 -82.726 34.710 1.00 88.53 A C
+ATOM 3580 CZ TYR A 519 4.435 -82.803 33.870 1.00 88.53 A C
+ATOM 3581 OH TYR A 519 5.015 -81.641 33.404 1.00 88.53 A O
+ATOM 3582 N ILE A 520 2.235 -87.839 38.264 1.00 75.89 A N
+ATOM 3583 CA ILE A 520 1.763 -88.870 39.190 1.00 75.89 A C
+ATOM 3584 C ILE A 520 0.296 -89.234 38.971 1.00 75.89 A C
+ATOM 3585 O ILE A 520 -0.546 -88.346 38.733 1.00 75.89 A O
+ATOM 3586 CB ILE A 520 2.018 -88.474 40.655 1.00 75.89 A C
+ATOM 3587 CG1 ILE A 520 1.333 -87.170 41.016 1.00 75.89 A C
+ATOM 3588 CG2 ILE A 520 3.496 -88.342 40.947 1.00 75.89 A C
+ATOM 3589 CD1 ILE A 520 1.557 -86.857 42.468 1.00 75.89 A C
+ATOM 3590 N PRO A 521 -0.047 -90.546 39.074 1.00 71.26 A N
+ATOM 3591 CA PRO A 521 -1.436 -90.995 38.904 1.00 71.26 A C
+ATOM 3592 C PRO A 521 -2.347 -90.546 40.023 1.00 71.26 A C
+ATOM 3593 O PRO A 521 -1.934 -89.814 40.928 1.00 71.26 A O
+ATOM 3594 CB PRO A 521 -1.312 -92.520 38.935 1.00 71.26 A C
+ATOM 3595 CG PRO A 521 -0.204 -92.731 39.886 1.00 71.26 A C
+ATOM 3596 CD PRO A 521 0.809 -91.654 39.517 1.00 71.26 A C
+ATOM 3597 N GLU A 522 -3.589 -91.011 39.986 1.00 72.58 A N
+ATOM 3598 CA GLU A 522 -4.490 -90.795 41.105 1.00 72.58 A C
+ATOM 3599 C GLU A 522 -4.199 -91.833 42.178 1.00 72.58 A C
+ATOM 3600 O GLU A 522 -3.930 -93.006 41.887 1.00 72.58 A O
+ATOM 3601 CB GLU A 522 -5.954 -90.870 40.657 1.00 72.58 A C
+ATOM 3602 CG GLU A 522 -6.974 -90.769 41.776 1.00 72.58 A C
+ATOM 3603 CD GLU A 522 -6.896 -89.441 42.503 1.00 72.58 A C
+ATOM 3604 OE1 GLU A 522 -6.668 -88.426 41.809 1.00 72.58 A O
+ATOM 3605 OE2 GLU A 522 -7.047 -89.418 43.761 1.00 72.58 A O1-
+ATOM 3606 N ALA A 523 -4.203 -91.377 43.419 1.00 66.35 A N
+ATOM 3607 CA ALA A 523 -4.130 -92.276 44.541 1.00 66.35 A C
+ATOM 3608 C ALA A 523 -5.423 -93.081 44.652 1.00 66.35 A C
+ATOM 3609 O ALA A 523 -6.449 -92.709 44.079 1.00 66.35 A O
+ATOM 3610 CB ALA A 523 -3.879 -91.474 45.812 1.00 66.35 A C
+ATOM 3611 N PRO A 524 -5.405 -94.168 45.415 1.00 61.84 A N
+ATOM 3612 CA PRO A 524 -6.623 -94.960 45.613 1.00 61.84 A C
+ATOM 3613 C PRO A 524 -7.831 -94.113 46.004 1.00 61.84 A C
+ATOM 3614 O PRO A 524 -7.706 -92.990 46.498 1.00 61.84 A O
+ATOM 3615 CB PRO A 524 -6.229 -95.916 46.743 1.00 61.84 A C
+ATOM 3616 CG PRO A 524 -4.769 -96.093 46.576 1.00 61.84 A C
+ATOM 3617 CD PRO A 524 -4.208 -94.902 45.842 1.00 61.84 A C
+ATOM 3618 N ARG A 525 -9.016 -94.636 45.693 1.00 76.81 A N
+ATOM 3619 CA ARG A 525 -10.277 -93.966 45.977 1.00 76.81 A C
+ATOM 3620 C ARG A 525 -10.927 -94.442 47.271 1.00 76.81 A C
+ATOM 3621 O ARG A 525 -12.118 -94.189 47.492 1.00 76.81 A O
+ATOM 3622 CB ARG A 525 -11.221 -94.087 44.790 1.00 76.81 A C
+ATOM 3623 CG ARG A 525 -10.794 -93.135 43.693 1.00 76.81 A C
+ATOM 3624 CD ARG A 525 -11.777 -93.049 42.542 1.00 76.81 A C
+ATOM 3625 NE ARG A 525 -11.179 -92.341 41.409 1.00 76.81 A N
+ATOM 3626 CZ ARG A 525 -11.397 -91.054 41.142 1.00 76.81 A C
+ATOM 3627 NH1 ARG A 525 -12.211 -90.343 41.926 1.00 76.81 A N1+
+ATOM 3628 NH2 ARG A 525 -10.808 -90.478 40.094 1.00 76.81 A N
+ATOM 3629 N ASP A 526 -10.180 -95.134 48.118 1.00 69.18 A N
+ATOM 3630 CA ASP A 526 -10.723 -95.991 49.161 1.00 69.18 A C
+ATOM 3631 C ASP A 526 -11.189 -95.250 50.401 1.00 69.18 A C
+ATOM 3632 O ASP A 526 -11.396 -95.900 51.431 1.00 69.18 A O
+ATOM 3633 CB ASP A 526 -9.668 -96.980 49.599 1.00 69.18 A C
+ATOM 3634 CG ASP A 526 -8.447 -96.287 50.069 1.00 69.18 A C
+ATOM 3635 OD1 ASP A 526 -8.451 -95.030 50.073 1.00 69.18 A O
+ATOM 3636 OD2 ASP A 526 -7.490 -96.987 50.435 1.00 69.18 A O1-
+ATOM 3637 N GLY A 527 -11.230 -93.926 50.399 1.00 68.06 A N
+ATOM 3638 CA GLY A 527 -11.642 -93.257 51.620 1.00 68.06 A C
+ATOM 3639 C GLY A 527 -10.543 -93.095 52.644 1.00 68.06 A C
+ATOM 3640 O GLY A 527 -10.831 -92.818 53.821 1.00 68.06 A O
+ATOM 3641 N GLN A 528 -9.297 -93.274 52.228 1.00 61.82 A N
+ATOM 3642 CA GLN A 528 -8.136 -92.943 53.026 1.00 61.82 A C
+ATOM 3643 C GLN A 528 -7.364 -91.814 52.359 1.00 61.82 A C
+ATOM 3644 O GLN A 528 -7.479 -91.584 51.151 1.00 61.82 A O
+ATOM 3645 CB GLN A 528 -7.245 -94.169 53.225 1.00 61.82 A C
+ATOM 3646 CG GLN A 528 -7.941 -95.253 54.032 1.00 61.82 A C
+ATOM 3647 CD GLN A 528 -7.148 -96.541 54.114 1.00 61.82 A C
+ATOM 3648 NE2 GLN A 528 -7.246 -97.351 53.072 1.00 61.82 A N
+ATOM 3649 OE1 GLN A 528 -6.474 -96.820 55.112 1.00 61.82 A O
+ATOM 3650 N ALA A 529 -6.615 -91.082 53.174 1.00 54.32 A N
+ATOM 3651 CA ALA A 529 -5.753 -90.019 52.689 1.00 54.32 A C
+ATOM 3652 C ALA A 529 -4.381 -90.590 52.389 1.00 54.32 A C
+ATOM 3653 O ALA A 529 -3.857 -91.414 53.147 1.00 54.32 A O
+ATOM 3654 CB ALA A 529 -5.639 -88.896 53.723 1.00 54.32 A C
+ATOM 3655 N TYR A 530 -3.793 -90.137 51.295 1.00 52.96 A N
+ATOM 3656 CA TYR A 530 -2.524 -90.673 50.838 1.00 52.96 A C
+ATOM 3657 C TYR A 530 -1.485 -89.576 50.668 1.00 52.96 A C
+ATOM 3658 O TYR A 530 -1.791 -88.474 50.222 1.00 52.96 A O
+ATOM 3659 CB TYR A 530 -2.715 -91.408 49.528 1.00 52.96 A C
+ATOM 3660 CG TYR A 530 -3.306 -92.768 49.708 1.00 52.96 A C
+ATOM 3661 CD1 TYR A 530 -2.502 -93.838 50.041 1.00 52.96 A C
+ATOM 3662 CD2 TYR A 530 -4.678 -92.983 49.581 1.00 52.96 A C
+ATOM 3663 CE1 TYR A 530 -3.039 -95.099 50.221 1.00 52.96 A C
+ATOM 3664 CE2 TYR A 530 -5.224 -94.244 49.755 1.00 52.96 A C
+ATOM 3665 CZ TYR A 530 -4.394 -95.300 50.077 1.00 52.96 A C
+ATOM 3666 OH TYR A 530 -4.885 -96.568 50.262 1.00 52.96 A O
+ATOM 3667 N VAL A 531 -0.248 -89.904 50.989 1.00 49.90 A N
+ATOM 3668 CA VAL A 531 0.872 -88.998 50.862 1.00 49.90 A C
+ATOM 3669 C VAL A 531 1.851 -89.628 49.871 1.00 49.90 A C
+ATOM 3670 O VAL A 531 1.837 -90.843 49.653 1.00 49.90 A O
+ATOM 3671 CB VAL A 531 1.501 -88.802 52.237 1.00 49.90 A C
+ATOM 3672 CG1 VAL A 531 2.396 -87.621 52.265 1.00 49.90 A C
+ATOM 3673 CG2 VAL A 531 0.422 -88.690 53.240 1.00 49.90 A C
+ATOM 3674 N ARG A 532 2.712 -88.806 49.262 1.00 57.79 A N
+ATOM 3675 CA ARG A 532 3.686 -89.298 48.290 1.00 57.79 A C
+ATOM 3676 C ARG A 532 5.005 -89.607 48.990 1.00 57.79 A C
+ATOM 3677 O ARG A 532 5.550 -88.734 49.676 1.00 57.79 A O
+ATOM 3678 CB ARG A 532 3.899 -88.261 47.197 1.00 57.79 A C
+ATOM 3679 CG ARG A 532 4.450 -88.836 45.922 1.00 57.79 A C
+ATOM 3680 CD ARG A 532 3.454 -89.841 45.360 1.00 57.79 A C
+ATOM 3681 NE ARG A 532 3.836 -90.343 44.045 1.00 57.79 A N
+ATOM 3682 CZ ARG A 532 3.227 -91.351 43.433 1.00 57.79 A C
+ATOM 3683 NH1 ARG A 532 2.218 -91.979 44.020 1.00 57.79 A N1+
+ATOM 3684 NH2 ARG A 532 3.636 -91.742 42.238 1.00 57.79 A N
+ATOM 3685 N LYS A 533 5.501 -90.848 48.848 1.00 56.42 A N
+ATOM 3686 CA LYS A 533 6.831 -91.241 49.345 1.00 56.42 A C
+ATOM 3687 C LYS A 533 7.515 -92.154 48.337 1.00 56.42 A C
+ATOM 3688 O LYS A 533 6.944 -93.192 47.970 1.00 56.42 A O
+ATOM 3689 CB LYS A 533 6.758 -91.964 50.687 1.00 56.42 A C
+ATOM 3690 CG LYS A 533 8.106 -92.349 51.267 1.00 56.42 A C
+ATOM 3691 CD LYS A 533 7.890 -93.204 52.498 1.00 56.42 A C
+ATOM 3692 CE LYS A 533 9.153 -93.422 53.298 1.00 56.42 A C
+ATOM 3693 NZ LYS A 533 8.834 -94.083 54.608 1.00 56.42 A N1+
+ATOM 3694 N ASP A 534 8.714 -91.749 47.872 1.00 66.00 A N
+ATOM 3695 CA ASP A 534 9.548 -92.439 46.867 1.00 66.00 A C
+ATOM 3696 C ASP A 534 8.715 -93.230 45.847 1.00 66.00 A C
+ATOM 3697 O ASP A 534 8.670 -94.471 45.868 1.00 66.00 A O
+ATOM 3698 CB ASP A 534 10.702 -93.318 47.438 1.00 66.00 A C
+ATOM 3699 CG ASP A 534 10.380 -94.064 48.714 1.00 66.00 A C
+ATOM 3700 OD1 ASP A 534 9.246 -94.553 48.842 1.00 66.00 A O
+ATOM 3701 OD2 ASP A 534 11.308 -94.227 49.558 1.00 66.00 A O1-
+ATOM 3702 N GLY A 535 8.011 -92.500 44.981 1.00 68.89 A N
+ATOM 3703 CA GLY A 535 7.323 -93.088 43.861 1.00 68.89 A C
+ATOM 3704 C GLY A 535 5.925 -93.588 44.134 1.00 68.89 A C
+ATOM 3705 O GLY A 535 5.183 -93.838 43.176 1.00 68.89 A O
+ATOM 3706 N GLU A 536 5.521 -93.746 45.391 1.00 72.34 A N
+ATOM 3707 CA GLU A 536 4.215 -94.337 45.625 1.00 72.34 A C
+ATOM 3708 C GLU A 536 3.343 -93.476 46.524 1.00 72.34 A C
+ATOM 3709 O GLU A 536 3.771 -92.458 47.083 1.00 72.34 A O
+ATOM 3710 CB GLU A 536 4.365 -95.722 46.229 1.00 72.34 A C
+ATOM 3711 CG GLU A 536 4.703 -96.731 45.180 1.00 72.34 A C
+ATOM 3712 CD GLU A 536 4.519 -98.147 45.684 1.00 72.34 A C
+ATOM 3713 OE1 GLU A 536 4.457 -98.331 46.944 1.00 72.34 A O
+ATOM 3714 OE2 GLU A 536 4.426 -99.061 44.812 1.00 72.34 A O1-
+ATOM 3715 N TRP A 537 2.102 -93.922 46.666 1.00 57.12 A N
+ATOM 3716 CA TRP A 537 1.234 -93.409 47.705 1.00 57.12 A C
+ATOM 3717 C TRP A 537 1.329 -94.297 48.935 1.00 57.12 A C
+ATOM 3718 O TRP A 537 1.378 -95.525 48.826 1.00 57.12 A O
+ATOM 3719 CB TRP A 537 -0.209 -93.331 47.233 1.00 57.12 A C
+ATOM 3720 CG TRP A 537 -0.422 -92.412 46.098 1.00 57.12 A C
+ATOM 3721 CD1 TRP A 537 -0.637 -92.753 44.804 1.00 57.12 A C
+ATOM 3722 CD2 TRP A 537 -0.462 -90.978 46.146 1.00 57.12 A C
+ATOM 3723 CE2 TRP A 537 -0.712 -90.521 44.840 1.00 57.12 A C
+ATOM 3724 CE3 TRP A 537 -0.314 -90.038 47.169 1.00 57.12 A C
+ATOM 3725 NE1 TRP A 537 -0.817 -91.621 44.033 1.00 57.12 A N
+ATOM 3726 CZ2 TRP A 537 -0.811 -89.174 44.533 1.00 57.12 A C
+ATOM 3727 CZ3 TRP A 537 -0.416 -88.706 46.860 1.00 57.12 A C
+ATOM 3728 CH2 TRP A 537 -0.664 -88.283 45.558 1.00 57.12 A C
+ATOM 3729 N VAL A 538 1.391 -93.666 50.099 1.00 50.16 A N
+ATOM 3730 CA VAL A 538 1.376 -94.336 51.389 1.00 50.16 A C
+ATOM 3731 C VAL A 538 0.203 -93.757 52.158 1.00 50.16 A C
+ATOM 3732 O VAL A 538 -0.250 -92.640 51.886 1.00 50.16 A O
+ATOM 3733 CB VAL A 538 2.672 -94.100 52.158 1.00 50.16 A C
+ATOM 3734 CG1 VAL A 538 3.824 -94.508 51.316 1.00 50.16 A C
+ATOM 3735 CG2 VAL A 538 2.781 -92.644 52.503 1.00 50.16 A C
+ATOM 3736 N LEU A 539 -0.301 -94.499 53.131 1.00 47.24 A N
+ATOM 3737 CA LEU A 539 -1.433 -93.947 53.859 1.00 47.24 A C
+ATOM 3738 C LEU A 539 -0.919 -92.920 54.857 1.00 47.24 A C
+ATOM 3739 O LEU A 539 0.158 -93.085 55.440 1.00 47.24 A O
+ATOM 3740 CB LEU A 539 -2.243 -95.043 54.546 1.00 47.24 A C
+ATOM 3741 CG LEU A 539 -2.432 -96.182 53.544 1.00 47.24 A C
+ATOM 3742 CD1 LEU A 539 -1.463 -97.366 53.843 1.00 47.24 A C
+ATOM 3743 CD2 LEU A 539 -3.878 -96.610 53.358 1.00 47.24 A C
+ATOM 3744 N LEU A 540 -1.652 -91.817 55.000 1.00 41.44 A N
+ATOM 3745 CA LEU A 540 -1.275 -90.875 56.041 1.00 41.44 A C
+ATOM 3746 C LEU A 540 -1.255 -91.590 57.383 1.00 41.44 A C
+ATOM 3747 O LEU A 540 -0.368 -91.364 58.223 1.00 41.44 A O
+ATOM 3748 CB LEU A 540 -2.249 -89.707 56.060 1.00 41.44 A C
+ATOM 3749 CG LEU A 540 -1.904 -88.563 56.996 1.00 41.44 A C
+ATOM 3750 CD1 LEU A 540 -0.476 -88.147 56.807 1.00 41.44 A C
+ATOM 3751 CD2 LEU A 540 -2.854 -87.441 56.717 1.00 41.44 A C
+ATOM 3752 N SER A 541 -2.200 -92.517 57.557 1.00 47.65 A N
+ATOM 3753 CA SER A 541 -2.338 -93.289 58.784 1.00 47.65 A C
+ATOM 3754 C SER A 541 -1.011 -93.860 59.222 1.00 47.65 A C
+ATOM 3755 O SER A 541 -0.714 -93.902 60.415 1.00 47.65 A O
+ATOM 3756 CB SER A 541 -3.341 -94.404 58.531 1.00 47.65 A C
+ATOM 3757 OG SER A 541 -4.339 -93.909 57.637 1.00 47.65 A O
+ATOM 3758 N THR A 542 -0.186 -94.264 58.260 1.00 48.64 A N
+ATOM 3759 CA THR A 542 1.133 -94.799 58.558 1.00 48.64 A C
+ATOM 3760 C THR A 542 1.879 -93.931 59.542 1.00 48.64 A C
+ATOM 3761 O THR A 542 2.591 -94.439 60.422 1.00 48.64 A O
+ATOM 3762 CB THR A 542 1.965 -94.874 57.293 1.00 48.64 A C
+ATOM 3763 CG2 THR A 542 3.164 -95.798 57.519 1.00 48.64 A C
+ATOM 3764 OG1 THR A 542 1.149 -95.355 56.222 1.00 48.64 A O
+ATOM 3765 N PHE A 543 1.761 -92.612 59.384 1.00 48.10 A N
+ATOM 3766 CA PHE A 543 2.598 -91.679 60.116 1.00 48.10 A C
+ATOM 3767 C PHE A 543 1.938 -91.120 61.363 1.00 48.10 A C
+ATOM 3768 O PHE A 543 2.613 -90.429 62.130 1.00 48.10 A O
+ATOM 3769 CB PHE A 543 3.043 -90.550 59.191 1.00 48.10 A C
+ATOM 3770 CG PHE A 543 3.724 -91.042 57.952 1.00 48.10 A C
+ATOM 3771 CD1 PHE A 543 2.994 -91.361 56.825 1.00 48.10 A C
+ATOM 3772 CD2 PHE A 543 5.088 -91.232 57.931 1.00 48.10 A C
+ATOM 3773 CE1 PHE A 543 3.616 -91.824 55.697 1.00 48.10 A C
+ATOM 3774 CE2 PHE A 543 5.721 -91.698 56.796 1.00 48.10 A C
+ATOM 3775 CZ PHE A 543 4.984 -91.995 55.679 1.00 48.10 A C
+ATOM 3776 N LEU A 544 0.670 -91.442 61.625 1.00 38.38 A N
+ATOM 3777 CA LEU A 544 0.005 -90.911 62.822 1.00 38.38 A C
+ATOM 3778 C LEU A 544 -0.484 -92.018 63.785 1.00 38.38 A C
+ATOM 3779 O LEU A 544 0.151 -92.327 64.815 1.00 38.38 A O
+ATOM 3780 CB LEU A 544 -1.171 -90.025 62.404 1.00 38.38 A C
+ATOM 3781 CG LEU A 544 -1.032 -89.143 61.163 1.00 38.38 A C
+ATOM 3782 CD1 LEU A 544 -2.280 -88.341 60.997 1.00 38.38 A C
+ATOM 3783 CD2 LEU A 544 0.130 -88.239 61.294 1.00 38.38 A C
+TER
+ATOM 3784 N VAL H 2 24.712 -62.728 -82.954 1.00 62.22 B N
+ATOM 3785 CA VAL H 2 24.045 -61.988 -81.866 1.00 58.35 B C
+ATOM 3786 C VAL H 2 24.512 -60.545 -81.702 1.00 59.47 B C
+ATOM 3787 O VAL H 2 25.583 -60.299 -81.141 1.00 60.10 B O
+ATOM 3788 CB VAL H 2 24.236 -62.666 -80.519 1.00 61.67 B C
+ATOM 3789 CG1 VAL H 2 23.507 -61.859 -79.436 1.00 61.67 B C
+ATOM 3790 CG2 VAL H 2 23.769 -64.098 -80.588 1.00 61.67 B C
+ATOM 3791 N GLN H 3 23.700 -59.578 -82.121 1.00 67.34 B N
+ATOM 3792 CA GLN H 3 24.203 -58.223 -82.247 1.00 69.24 B C
+ATOM 3793 C GLN H 3 23.197 -57.207 -81.729 1.00 65.06 B C
+ATOM 3794 O GLN H 3 21.980 -57.397 -81.812 1.00 64.92 B O
+ATOM 3795 CB GLN H 3 24.544 -57.927 -83.699 1.00 73.55 B C
+ATOM 3796 CG GLN H 3 25.765 -58.696 -84.144 1.00 73.55 B C
+ATOM 3797 CD GLN H 3 26.842 -57.817 -84.761 1.00 73.55 B C
+ATOM 3798 NE2 GLN H 3 28.122 -58.200 -84.573 1.00 73.55 B N
+ATOM 3799 OE1 GLN H 3 26.532 -56.807 -85.407 1.00 73.55 B O
+ATOM 3800 N LEU H 4 23.744 -56.123 -81.185 1.00 53.68 B N
+ATOM 3801 CA LEU H 4 23.022 -54.910 -80.822 1.00 55.13 B C
+ATOM 3802 C LEU H 4 23.747 -53.744 -81.481 1.00 56.69 B C
+ATOM 3803 O LEU H 4 24.961 -53.574 -81.302 1.00 57.19 B O
+ATOM 3804 CB LEU H 4 22.990 -54.725 -79.309 1.00 63.45 B C
+ATOM 3805 CG LEU H 4 21.674 -55.001 -78.614 1.00 63.45 B C
+ATOM 3806 CD1 LEU H 4 21.213 -56.345 -79.051 1.00 63.45 B C
+ATOM 3807 CD2 LEU H 4 21.887 -54.992 -77.126 1.00 63.45 B C
+ATOM 3808 N VAL H 5 23.030 -52.961 -82.271 1.00 57.51 B N
+ATOM 3809 CA VAL H 5 23.685 -51.927 -83.053 1.00 57.42 B C
+ATOM 3810 C VAL H 5 23.027 -50.599 -82.738 1.00 59.81 B C
+ATOM 3811 O VAL H 5 21.853 -50.378 -83.068 1.00 61.62 B O
+ATOM 3812 CB VAL H 5 23.653 -52.227 -84.552 1.00 67.33 B C
+ATOM 3813 CG1 VAL H 5 24.566 -53.394 -84.824 1.00 67.33 B C
+ATOM 3814 CG2 VAL H 5 22.259 -52.560 -85.017 1.00 67.33 B C
+ATOM 3815 N GLU H 6 23.779 -49.723 -82.084 1.00 57.30 B N
+ATOM 3816 CA GLU H 6 23.312 -48.384 -81.788 1.00 60.63 B C
+ATOM 3817 C GLU H 6 23.420 -47.509 -83.024 1.00 60.38 B C
+ATOM 3818 O GLU H 6 24.299 -47.698 -83.868 1.00 56.38 B O
+ATOM 3819 CB GLU H 6 24.120 -47.783 -80.647 1.00 66.68 B C
+ATOM 3820 CG GLU H 6 24.026 -48.580 -79.375 1.00 66.68 B C
+ATOM 3821 CD GLU H 6 25.124 -49.593 -79.260 1.00 66.68 B C
+ATOM 3822 OE1 GLU H 6 25.723 -49.919 -80.312 1.00 66.68 B O
+ATOM 3823 OE2 GLU H 6 25.422 -50.007 -78.112 1.00 66.68 B O1-
+ATOM 3824 N SER H 7 22.515 -46.545 -83.120 1.00 62.25 B N
+ATOM 3825 CA SER H 7 22.409 -45.671 -84.273 1.00 58.61 B C
+ATOM 3826 C SER H 7 21.862 -44.327 -83.815 1.00 58.51 B C
+ATOM 3827 O SER H 7 21.043 -44.268 -82.893 1.00 57.45 B O
+ATOM 3828 CB SER H 7 21.496 -46.295 -85.326 1.00 67.75 B C
+ATOM 3829 OG SER H 7 21.106 -45.315 -86.259 1.00 67.75 B O
+ATOM 3830 N GLY H 8 22.309 -43.246 -84.447 1.00 63.57 B N
+ATOM 3831 CA GLY H 8 21.642 -41.968 -84.259 1.00 62.26 B C
+ATOM 3832 C GLY H 8 22.295 -40.981 -83.312 1.00 67.14 B C
+ATOM 3833 O GLY H 8 21.774 -39.868 -83.140 1.00 71.88 B O
+ATOM 3834 N GLY H 9 23.397 -41.351 -82.679 1.00 69.25 B N
+ATOM 3835 CA GLY H 9 24.230 -40.353 -82.044 1.00 69.79 B C
+ATOM 3836 C GLY H 9 24.788 -39.369 -83.053 1.00 78.64 B C
+ATOM 3837 O GLY H 9 24.822 -39.607 -84.264 1.00 86.22 B O
+ATOM 3838 N GLY H 11 25.246 -38.244 -82.533 1.00 68.38 B N
+ATOM 3839 CA GLY H 11 25.710 -37.189 -83.397 1.00 64.09 B C
+ATOM 3840 C GLY H 11 26.208 -36.030 -82.573 1.00 64.71 B C
+ATOM 3841 O GLY H 11 26.596 -36.193 -81.415 1.00 65.19 B O
+ATOM 3842 N LEU H 12 26.210 -34.860 -83.205 1.00 53.71 B N
+ATOM 3843 CA LEU H 12 26.625 -33.599 -82.606 1.00 52.35 B C
+ATOM 3844 C LEU H 12 25.398 -32.712 -82.457 1.00 53.60 B C
+ATOM 3845 O LEU H 12 24.776 -32.315 -83.451 1.00 57.55 B O
+ATOM 3846 CB LEU H 12 27.677 -32.931 -83.475 1.00 62.49 B C
+ATOM 3847 CG LEU H 12 28.716 -32.124 -82.759 1.00 62.49 B C
+ATOM 3848 CD1 LEU H 12 29.595 -31.585 -83.837 1.00 62.49 B C
+ATOM 3849 CD2 LEU H 12 28.049 -31.019 -81.996 1.00 62.49 B C
+ATOM 3850 N VAL H 13 25.053 -32.403 -81.227 1.00 55.40 B N
+ATOM 3851 CA VAL H 13 23.791 -31.753 -80.929 1.00 57.90 B C
+ATOM 3852 C VAL H 13 24.072 -30.566 -80.030 1.00 62.89 B C
+ATOM 3853 O VAL H 13 24.869 -30.661 -79.089 1.00 67.15 B O
+ATOM 3854 CB VAL H 13 22.794 -32.729 -80.268 1.00 64.70 B C
+ATOM 3855 CG1 VAL H 13 21.407 -32.099 -80.172 1.00 64.70 B C
+ATOM 3856 CG2 VAL H 13 22.730 -34.031 -81.054 1.00 64.70 B C
+ATOM 3857 N GLN H 14 23.421 -29.445 -80.325 1.00 71.77 B N
+ATOM 3858 CA GLN H 14 23.582 -28.263 -79.499 1.00 72.52 B C
+ATOM 3859 C GLN H 14 22.869 -28.437 -78.159 1.00 71.06 B C
+ATOM 3860 O GLN H 14 21.866 -29.152 -78.064 1.00 68.94 B O
+ATOM 3861 CB GLN H 14 23.042 -27.029 -80.214 1.00 74.20 B C
+ATOM 3862 CG GLN H 14 23.899 -26.573 -81.388 1.00 74.20 B C
+ATOM 3863 CD GLN H 14 24.629 -25.260 -81.119 1.00 74.20 B C
+ATOM 3864 NE2 GLN H 14 25.692 -25.006 -81.890 1.00 74.20 B N
+ATOM 3865 OE1 GLN H 14 24.237 -24.479 -80.235 1.00 74.20 B O
+ATOM 3866 N PRO H 15 23.339 -27.754 -77.130 1.00 67.74 B N
+ATOM 3867 CA PRO H 15 22.711 -27.851 -75.803 1.00 66.40 B C
+ATOM 3868 C PRO H 15 21.220 -27.556 -75.816 1.00 62.57 B C
+ATOM 3869 O PRO H 15 20.706 -27.028 -76.799 1.00 63.65 B O
+ATOM 3870 CB PRO H 15 23.465 -26.795 -74.989 1.00 68.28 B C
+ATOM 3871 CG PRO H 15 24.805 -26.721 -75.649 1.00 68.28 B C
+ATOM 3872 CD PRO H 15 24.628 -27.047 -77.099 1.00 68.28 B C
+ATOM 3873 N ARG H 16 20.494 -27.977 -74.785 1.00 86.46 B N
+ATOM 3874 CA ARG H 16 19.096 -27.620 -74.570 1.00 86.52 B C
+ATOM 3875 C ARG H 16 18.160 -28.188 -75.656 1.00 89.49 B C
+ATOM 3876 O ARG H 16 16.945 -28.036 -75.525 1.00 93.16 B O
+ATOM 3877 CB ARG H 16 18.934 -26.093 -74.441 1.00 92.39 B C
+ATOM 3878 CG ARG H 16 17.743 -25.578 -73.623 1.00 92.39 B C
+ATOM 3879 CD ARG H 16 17.515 -24.030 -73.843 1.00 92.39 B C
+ATOM 3880 NE ARG H 16 17.547 -23.625 -75.272 1.00 92.39 B N
+ATOM 3881 CZ ARG H 16 16.944 -22.552 -75.806 1.00 92.39 B C
+ATOM 3882 NH1 ARG H 16 16.205 -21.744 -75.050 1.00 92.39 B N1+
+ATOM 3883 NH2 ARG H 16 17.062 -22.305 -77.113 1.00 92.39 B N
+ATOM 3884 N ARG H 17 18.676 -28.875 -76.698 1.00 87.08 B N
+ATOM 3885 CA ARG H 17 17.880 -29.413 -77.804 1.00 86.03 B C
+ATOM 3886 C ARG H 17 17.823 -30.953 -77.759 1.00 85.49 B C
+ATOM 3887 O ARG H 17 18.249 -31.581 -76.790 1.00 81.59 B O
+ATOM 3888 CB ARG H 17 18.408 -28.891 -79.147 1.00 88.79 B C
+ATOM 3889 CG ARG H 17 17.313 -28.825 -80.239 1.00 88.79 B C
+ATOM 3890 CD ARG H 17 17.926 -28.674 -81.622 1.00 88.79 B C
+ATOM 3891 NE ARG H 17 17.293 -29.514 -82.641 1.00 88.79 B N
+ATOM 3892 CZ ARG H 17 17.785 -29.687 -83.868 1.00 88.79 B C
+ATOM 3893 NH1 ARG H 17 18.891 -29.058 -84.235 1.00 88.79 B N1+
+ATOM 3894 NH2 ARG H 17 17.166 -30.476 -84.732 1.00 88.79 B N
+ATOM 3895 N SER H 18 17.237 -31.567 -78.792 1.00 78.28 B N
+ATOM 3896 CA SER H 18 16.760 -32.944 -78.724 1.00 78.79 B C
+ATOM 3897 C SER H 18 17.432 -33.860 -79.741 1.00 77.87 B C
+ATOM 3898 O SER H 18 17.766 -33.437 -80.852 1.00 78.69 B O
+ATOM 3899 CB SER H 18 15.253 -32.988 -78.961 1.00 84.10 B C
+ATOM 3900 OG SER H 18 14.805 -34.321 -79.159 1.00 84.10 B O
+ATOM 3901 N LEU H 19 17.558 -35.141 -79.368 1.00 64.34 B N
+ATOM 3902 CA LEU H 19 18.151 -36.184 -80.211 1.00 68.35 B C
+ATOM 3903 C LEU H 19 17.522 -37.548 -79.904 1.00 68.76 B C
+ATOM 3904 O LEU H 19 17.132 -37.812 -78.764 1.00 76.13 B O
+ATOM 3905 CB LEU H 19 19.671 -36.244 -80.006 1.00 74.62 B C
+ATOM 3906 CG LEU H 19 20.359 -37.291 -80.861 1.00 74.62 B C
+ATOM 3907 CD1 LEU H 19 20.082 -36.987 -82.307 1.00 74.62 B C
+ATOM 3908 CD2 LEU H 19 21.833 -37.299 -80.580 1.00 74.62 B C
+ATOM 3909 N ARG H 20 17.425 -38.425 -80.917 1.00 69.11 B N
+ATOM 3910 CA ARG H 20 16.907 -39.785 -80.730 1.00 70.46 B C
+ATOM 3911 C ARG H 20 17.957 -40.821 -81.093 1.00 68.38 B C
+ATOM 3912 O ARG H 20 18.570 -40.740 -82.166 1.00 68.64 B O
+ATOM 3913 CB ARG H 20 15.673 -40.083 -81.576 1.00 76.94 B C
+ATOM 3914 CG ARG H 20 15.669 -41.541 -82.074 1.00 76.94 B C
+ATOM 3915 CD ARG H 20 14.405 -41.960 -82.789 1.00 76.94 B C
+ATOM 3916 NE ARG H 20 13.324 -42.203 -81.843 1.00 76.94 B N
+ATOM 3917 CZ ARG H 20 12.148 -42.735 -82.169 1.00 76.94 B C
+ATOM 3918 NH1 ARG H 20 11.905 -43.110 -83.437 1.00 76.94 B N1+
+ATOM 3919 NH2 ARG H 20 11.226 -42.914 -81.214 1.00 76.94 B N
+ATOM 3920 N LEU H 21 18.121 -41.810 -80.209 1.00 60.07 B N
+ATOM 3921 CA LEU H 21 19.002 -42.957 -80.390 1.00 59.75 B C
+ATOM 3922 C LEU H 21 18.158 -44.202 -80.598 1.00 64.51 B C
+ATOM 3923 O LEU H 21 17.150 -44.397 -79.911 1.00 63.61 B O
+ATOM 3924 CB LEU H 21 19.900 -43.157 -79.164 1.00 69.03 B C
+ATOM 3925 CG LEU H 21 21.166 -42.339 -78.897 1.00 69.03 B C
+ATOM 3926 CD1 LEU H 21 20.973 -40.860 -78.983 1.00 69.03 B C
+ATOM 3927 CD2 LEU H 21 21.549 -42.647 -77.484 1.00 69.03 B C
+ATOM 3928 N SER H 22 18.576 -45.054 -81.529 1.00 62.43 B N
+ATOM 3929 CA SER H 22 17.894 -46.315 -81.777 1.00 58.77 B C
+ATOM 3930 C SER H 22 18.885 -47.477 -81.698 1.00 60.00 B C
+ATOM 3931 O SER H 22 20.092 -47.316 -81.891 1.00 57.65 B O
+ATOM 3932 CB SER H 22 17.197 -46.296 -83.135 1.00 67.92 B C
+ATOM 3933 OG SER H 22 18.165 -46.323 -84.166 1.00 67.92 B O
+ATOM 3934 N CYS H 23 18.359 -48.664 -81.402 1.00 66.01 B N
+ATOM 3935 CA CYS H 23 19.154 -49.879 -81.294 1.00 66.64 B C
+ATOM 3936 C CYS H 23 18.460 -50.977 -82.078 1.00 65.91 B C
+ATOM 3937 O CYS H 23 17.303 -51.288 -81.807 1.00 62.18 B O
+ATOM 3938 CB CYS H 23 19.317 -50.286 -79.832 1.00 73.36 B C
+ATOM 3939 SG CYS H 23 20.273 -51.816 -79.543 1.00 73.36 B S
+ATOM 3940 N ALA H 24 19.142 -51.550 -83.053 1.00 63.59 B N
+ATOM 3941 CA ALA H 24 18.568 -52.660 -83.795 1.00 65.16 B C
+ATOM 3942 C ALA H 24 19.175 -53.949 -83.286 1.00 63.56 B C
+ATOM 3943 O ALA H 24 20.377 -54.007 -83.003 1.00 65.11 B O
+ATOM 3944 CB ALA H 24 18.813 -52.546 -85.296 1.00 63.07 B C
+ATOM 3945 N ALA H 25 18.338 -54.983 -83.182 1.00 65.59 B N
+ATOM 3946 CA ALA H 25 18.696 -56.245 -82.554 1.00 64.49 B C
+ATOM 3947 C ALA H 25 18.815 -57.341 -83.594 1.00 67.14 B C
+ATOM 3948 O ALA H 25 18.097 -57.354 -84.596 1.00 69.41 B O
+ATOM 3949 CB ALA H 25 17.659 -56.650 -81.518 1.00 64.17 B C
+ATOM 3950 N SER H 26 19.702 -58.279 -83.330 1.00 60.73 B N
+ATOM 3951 CA SER H 26 20.034 -59.274 -84.325 1.00 61.26 B C
+ATOM 3952 C SER H 26 20.400 -60.580 -83.650 1.00 60.93 B C
+ATOM 3953 O SER H 26 21.006 -60.569 -82.577 1.00 58.54 B O
+ATOM 3954 CB SER H 26 21.198 -58.783 -85.170 1.00 65.71 B C
+ATOM 3955 OG SER H 26 21.913 -59.889 -85.673 1.00 65.71 B O
+ATOM 3956 N GLY H 27 20.026 -61.699 -84.273 1.00 69.02 B N
+ATOM 3957 CA GLY H 27 20.596 -62.999 -83.926 1.00 64.25 B C
+ATOM 3958 C GLY H 27 20.195 -63.609 -82.593 1.00 66.78 B C
+ATOM 3959 O GLY H 27 20.830 -64.579 -82.149 1.00 70.51 B O
+ATOM 3960 N PHE H 28 19.186 -63.065 -81.923 1.00 63.94 B N
+ATOM 3961 CA PHE H 28 18.634 -63.679 -80.729 1.00 63.94 B C
+ATOM 3962 C PHE H 28 17.188 -63.221 -80.653 1.00 63.94 B C
+ATOM 3963 O PHE H 28 16.845 -62.152 -81.166 1.00 63.94 B O
+ATOM 3964 CB PHE H 28 19.401 -63.285 -79.457 1.00 63.94 B C
+ATOM 3965 CG PHE H 28 19.179 -61.856 -79.046 1.00 63.94 B C
+ATOM 3966 CD1 PHE H 28 18.108 -61.510 -78.223 1.00 63.94 B C
+ATOM 3967 CD2 PHE H 28 20.013 -60.860 -79.493 1.00 63.94 B C
+ATOM 3968 CE1 PHE H 28 17.875 -60.202 -77.850 1.00 63.94 B C
+ATOM 3969 CE2 PHE H 28 19.779 -59.547 -79.121 1.00 63.94 B C
+ATOM 3970 CZ PHE H 28 18.704 -59.221 -78.293 1.00 63.94 B C
+ATOM 3971 N THR H 29 16.340 -64.006 -79.991 1.00 71.07 B N
+ATOM 3972 CA THR H 29 14.916 -63.693 -80.051 1.00 71.07 B C
+ATOM 3973 C THR H 29 14.680 -62.502 -79.129 1.00 71.07 B C
+ATOM 3974 O THR H 29 14.806 -62.605 -77.904 1.00 71.07 B O
+ATOM 3975 CB THR H 29 14.062 -64.914 -79.691 1.00 71.07 B C
+ATOM 3976 CG2 THR H 29 14.325 -66.072 -80.688 1.00 71.07 B C
+ATOM 3977 OG1 THR H 29 14.359 -65.361 -78.363 1.00 71.07 B O
+ATOM 3978 N PHE H 30 14.295 -61.376 -79.728 1.00 69.79 B N
+ATOM 3979 CA PHE H 30 14.451 -60.097 -79.053 1.00 69.79 B C
+ATOM 3980 C PHE H 30 13.492 -59.960 -77.883 1.00 69.79 B C
+ATOM 3981 O PHE H 30 13.894 -59.504 -76.810 1.00 69.79 B O
+ATOM 3982 CB PHE H 30 14.230 -58.961 -80.039 1.00 69.79 B C
+ATOM 3983 CG PHE H 30 14.374 -57.589 -79.441 1.00 69.79 B C
+ATOM 3984 CD1 PHE H 30 15.607 -57.117 -79.055 1.00 69.79 B C
+ATOM 3985 CD2 PHE H 30 13.276 -56.767 -79.283 1.00 69.79 B C
+ATOM 3986 CE1 PHE H 30 15.742 -55.844 -78.542 1.00 69.79 B C
+ATOM 3987 CE2 PHE H 30 13.404 -55.499 -78.766 1.00 69.79 B C
+ATOM 3988 CZ PHE H 30 14.630 -55.037 -78.395 1.00 69.79 B C
+ATOM 3989 N ASP H 35 12.226 -60.355 -78.070 1.00 78.95 B N
+ATOM 3990 CA ASP H 35 11.199 -60.065 -77.072 1.00 78.95 B C
+ATOM 3991 C ASP H 35 11.585 -60.609 -75.710 1.00 78.95 B C
+ATOM 3992 O ASP H 35 11.384 -59.944 -74.688 1.00 78.95 B O
+ATOM 3993 CB ASP H 35 9.854 -60.648 -77.493 1.00 78.95 B C
+ATOM 3994 CG ASP H 35 9.498 -60.317 -78.928 1.00 78.95 B C
+ATOM 3995 OD1 ASP H 35 10.201 -59.478 -79.543 1.00 78.95 B O
+ATOM 3996 OD2 ASP H 35 8.502 -60.886 -79.441 1.00 78.95 B O1-
+ATOM 3997 N ASP H 36 12.197 -61.786 -75.677 1.00 75.30 B N
+ATOM 3998 CA ASP H 36 12.418 -62.463 -74.410 1.00 75.30 B C
+ATOM 3999 C ASP H 36 13.306 -61.696 -73.430 1.00 75.30 B C
+ATOM 4000 O ASP H 36 13.212 -61.940 -72.220 1.00 75.30 B O
+ATOM 4001 CB ASP H 36 12.976 -63.842 -74.706 1.00 75.30 B C
+ATOM 4002 CG ASP H 36 11.977 -64.689 -75.437 1.00 75.30 B C
+ATOM 4003 OD1 ASP H 36 10.779 -64.296 -75.400 1.00 75.30 B O
+ATOM 4004 OD2 ASP H 36 12.374 -65.718 -76.037 1.00 75.30 B O1-
+ATOM 4005 N TYR H 37 14.140 -60.766 -73.891 1.00 67.24 B N
+ATOM 4006 CA TYR H 37 15.091 -60.104 -73.006 1.00 66.21 B C
+ATOM 4007 C TYR H 37 14.668 -58.686 -72.690 1.00 66.21 B C
+ATOM 4008 O TYR H 37 14.009 -58.010 -73.484 1.00 66.65 B O
+ATOM 4009 CB TYR H 37 16.498 -60.079 -73.596 1.00 57.33 B C
+ATOM 4010 CG TYR H 37 16.979 -61.455 -73.806 1.00 57.33 B C
+ATOM 4011 CD1 TYR H 37 17.513 -62.179 -72.767 1.00 57.33 B C
+ATOM 4012 CD2 TYR H 37 16.787 -62.077 -75.006 1.00 57.33 B C
+ATOM 4013 CE1 TYR H 37 17.917 -63.485 -72.949 1.00 57.33 B C
+ATOM 4014 CE2 TYR H 37 17.170 -63.378 -75.197 1.00 57.33 B C
+ATOM 4015 CZ TYR H 37 17.739 -64.080 -74.174 1.00 57.33 B C
+ATOM 4016 OH TYR H 37 18.123 -65.380 -74.400 1.00 57.33 B O
+ATOM 4017 N THR H 38 15.060 -58.252 -71.511 1.00 59.89 B N
+ATOM 4018 CA THR H 38 14.884 -56.859 -71.169 1.00 59.11 B C
+ATOM 4019 C THR H 38 16.177 -56.119 -71.503 1.00 59.11 B C
+ATOM 4020 O THR H 38 17.272 -56.691 -71.456 1.00 59.11 B O
+ATOM 4021 CB THR H 38 14.487 -56.729 -69.697 1.00 54.91 B C
+ATOM 4022 CG2 THR H 38 13.687 -57.995 -69.303 1.00 54.91 B C
+ATOM 4023 OG1 THR H 38 15.653 -56.623 -68.871 1.00 54.91 B O
+ATOM 4024 N ILE H 39 16.041 -54.866 -71.933 1.00 48.59 B N
+ATOM 4025 CA ILE H 39 17.170 -54.147 -72.511 1.00 48.59 B C
+ATOM 4026 C ILE H 39 17.375 -52.828 -71.789 1.00 48.59 B C
+ATOM 4027 O ILE H 39 16.404 -52.167 -71.396 1.00 48.59 B O
+ATOM 4028 CB ILE H 39 16.983 -53.894 -74.011 1.00 51.77 B C
+ATOM 4029 CG1 ILE H 39 16.385 -55.123 -74.689 1.00 51.77 B C
+ATOM 4030 CG2 ILE H 39 18.300 -53.565 -74.609 1.00 51.77 B C
+ATOM 4031 CD1 ILE H 39 17.352 -56.250 -74.893 1.00 51.77 B C
+ATOM 4032 N HIS H 40 18.650 -52.446 -71.624 1.00 47.82 B N
+ATOM 4033 CA HIS H 40 19.064 -51.229 -70.938 1.00 47.82 B C
+ATOM 4034 C HIS H 40 19.874 -50.344 -71.872 1.00 47.82 B C
+ATOM 4035 O HIS H 40 20.586 -50.833 -72.755 1.00 47.82 B O
+ATOM 4036 CB HIS H 40 19.934 -51.501 -69.693 1.00 54.40 B C
+ATOM 4037 CG HIS H 40 19.433 -52.598 -68.806 1.00 54.40 B C
+ATOM 4038 CD2 HIS H 40 20.043 -53.715 -68.340 1.00 54.40 B C
+ATOM 4039 ND1 HIS H 40 18.168 -52.603 -68.265 1.00 54.40 B N
+ATOM 4040 CE1 HIS H 40 18.012 -53.687 -67.523 1.00 54.40 B C
+ATOM 4041 NE2 HIS H 40 19.135 -54.379 -67.552 1.00 54.40 B N
+ATOM 4042 N TRP H 41 19.749 -49.036 -71.668 1.00 54.06 B N
+ATOM 4043 CA TRP H 41 20.686 -48.057 -72.186 1.00 48.76 B C
+ATOM 4044 C TRP H 41 21.646 -47.720 -71.075 1.00 48.76 B C
+ATOM 4045 O TRP H 41 21.207 -47.310 -69.980 1.00 48.76 B O
+ATOM 4046 CB TRP H 41 19.991 -46.788 -72.641 1.00 57.76 B C
+ATOM 4047 CG TRP H 41 19.383 -46.943 -73.937 1.00 57.76 B C
+ATOM 4048 CD1 TRP H 41 18.076 -47.204 -74.203 1.00 57.76 B C
+ATOM 4049 CD2 TRP H 41 20.047 -46.869 -75.188 1.00 57.76 B C
+ATOM 4050 CE2 TRP H 41 19.082 -47.086 -76.185 1.00 57.76 B C
+ATOM 4051 CE3 TRP H 41 21.363 -46.621 -75.568 1.00 57.76 B C
+ATOM 4052 NE1 TRP H 41 17.882 -47.290 -75.556 1.00 57.76 B N
+ATOM 4053 CZ2 TRP H 41 19.393 -47.072 -77.548 1.00 57.76 B C
+ATOM 4054 CZ3 TRP H 41 21.672 -46.608 -76.931 1.00 57.76 B C
+ATOM 4055 CH2 TRP H 41 20.689 -46.837 -77.900 1.00 57.76 B C
+ATOM 4056 N VAL H 42 22.941 -47.905 -71.355 1.00 43.76 B N
+ATOM 4057 CA VAL H 42 24.021 -47.578 -70.427 1.00 43.76 B C
+ATOM 4058 C VAL H 42 25.001 -46.658 -71.137 1.00 46.28 B C
+ATOM 4059 O VAL H 42 25.464 -46.965 -72.240 1.00 45.10 B O
+ATOM 4060 CB VAL H 42 24.749 -48.829 -69.908 1.00 54.19 B C
+ATOM 4061 CG1 VAL H 42 25.972 -48.422 -69.157 1.00 54.19 B C
+ATOM 4062 CG2 VAL H 42 23.870 -49.570 -68.983 1.00 54.19 B C
+ATOM 4063 N ARG H 43 25.335 -45.543 -70.501 1.00 47.65 B N
+ATOM 4064 CA ARG H 43 26.223 -44.574 -71.114 1.00 47.65 B C
+ATOM 4065 C ARG H 43 27.542 -44.556 -70.373 1.00 47.65 B C
+ATOM 4066 O ARG H 43 27.634 -44.969 -69.211 1.00 47.65 B O
+ATOM 4067 CB ARG H 43 25.618 -43.168 -71.129 1.00 58.22 B C
+ATOM 4068 CG ARG H 43 25.498 -42.521 -69.773 1.00 58.22 B C
+ATOM 4069 CD ARG H 43 25.830 -41.062 -69.909 1.00 58.22 B C
+ATOM 4070 NE ARG H 43 25.347 -40.231 -68.816 1.00 58.22 B N
+ATOM 4071 CZ ARG H 43 24.141 -39.676 -68.813 1.00 58.22 B C
+ATOM 4072 NH1 ARG H 43 23.350 -39.866 -69.859 1.00 58.22 B N1+
+ATOM 4073 NH2 ARG H 43 23.736 -38.919 -67.794 1.00 58.22 B N
+ATOM 4074 N GLN H 44 28.575 -44.130 -71.095 1.00 50.16 B N
+ATOM 4075 CA GLN H 44 29.919 -43.975 -70.539 1.00 50.16 B C
+ATOM 4076 C GLN H 44 30.472 -42.643 -71.011 1.00 50.16 B C
+ATOM 4077 O GLN H 44 30.731 -42.483 -72.210 1.00 51.20 B O
+ATOM 4078 CB GLN H 44 30.829 -45.117 -70.990 1.00 60.51 B C
+ATOM 4079 CG GLN H 44 32.216 -45.134 -70.396 1.00 60.51 B C
+ATOM 4080 CD GLN H 44 32.875 -46.491 -70.576 1.00 60.51 B C
+ATOM 4081 NE2 GLN H 44 33.592 -46.930 -69.550 1.00 60.51 B N
+ATOM 4082 OE1 GLN H 44 32.736 -47.144 -71.627 1.00 60.51 B O
+ATOM 4083 N ALA H 45 30.687 -41.704 -70.080 1.00 56.14 B N
+ATOM 4084 CA ALA H 45 31.370 -40.470 -70.431 1.00 61.17 B C
+ATOM 4085 C ALA H 45 32.786 -40.807 -70.909 1.00 63.10 B C
+ATOM 4086 O ALA H 45 33.278 -41.918 -70.668 1.00 66.58 B O
+ATOM 4087 CB ALA H 45 31.399 -39.523 -69.235 1.00 58.01 B C
+ATOM 4088 N PRO H 46 33.445 -39.898 -71.643 1.00 59.76 B N
+ATOM 4089 CA PRO H 46 34.795 -40.211 -72.136 1.00 60.94 B C
+ATOM 4090 C PRO H 46 35.807 -40.208 -70.991 1.00 59.31 B C
+ATOM 4091 O PRO H 46 35.782 -39.334 -70.114 1.00 62.54 B O
+ATOM 4092 CB PRO H 46 35.063 -39.098 -73.146 1.00 69.43 B C
+ATOM 4093 CG PRO H 46 34.301 -37.922 -72.571 1.00 69.43 B C
+ATOM 4094 CD PRO H 46 33.055 -38.509 -71.968 1.00 69.43 B C
+ATOM 4095 N GLY H 47 36.673 -41.228 -70.989 1.00 64.00 B N
+ATOM 4096 CA GLY H 47 37.606 -41.457 -69.901 1.00 62.74 B C
+ATOM 4097 C GLY H 47 36.970 -41.385 -68.522 1.00 65.98 B C
+ATOM 4098 O GLY H 47 37.484 -40.699 -67.630 1.00 66.57 B O
+ATOM 4099 N LYS H 48 35.846 -42.078 -68.343 1.00 55.15 B N
+ATOM 4100 CA LYS H 48 35.039 -42.038 -67.119 1.00 58.10 B C
+ATOM 4101 C LYS H 48 34.378 -43.407 -66.963 1.00 57.43 B C
+ATOM 4102 O LYS H 48 34.730 -44.364 -67.660 1.00 56.07 B O
+ATOM 4103 CB LYS H 48 34.019 -40.888 -67.176 1.00 66.13 B C
+ATOM 4104 CG LYS H 48 34.553 -39.479 -66.905 1.00 66.13 B C
+ATOM 4105 CD LYS H 48 34.540 -39.178 -65.411 1.00 66.13 B C
+ATOM 4106 CE LYS H 48 35.140 -37.811 -65.078 1.00 66.13 B C
+ATOM 4107 NZ LYS H 48 35.192 -37.555 -63.594 1.00 66.13 B N1+
+ATOM 4108 N GLY H 49 33.405 -43.502 -66.055 1.00 57.89 B N
+ATOM 4109 CA GLY H 49 32.783 -44.769 -65.719 1.00 56.80 B C
+ATOM 4110 C GLY H 49 31.520 -45.090 -66.511 1.00 58.45 B C
+ATOM 4111 O GLY H 49 31.200 -44.473 -67.531 1.00 57.95 B O
+ATOM 4112 N LEU H 50 30.789 -46.087 -66.010 1.00 43.33 B N
+ATOM 4113 CA LEU H 50 29.563 -46.559 -66.636 1.00 43.33 B C
+ATOM 4114 C LEU H 50 28.371 -46.050 -65.843 1.00 43.33 B C
+ATOM 4115 O LEU H 50 28.312 -46.220 -64.622 1.00 43.33 B O
+ATOM 4116 CB LEU H 50 29.536 -48.084 -66.680 1.00 54.06 B C
+ATOM 4117 CG LEU H 50 30.600 -48.796 -67.505 1.00 54.06 B C
+ATOM 4118 CD1 LEU H 50 30.850 -50.175 -66.902 1.00 54.06 B C
+ATOM 4119 CD2 LEU H 50 30.172 -48.901 -68.946 1.00 54.06 B C
+ATOM 4120 N GLU H 51 27.423 -45.439 -66.529 1.00 45.00 B N
+ATOM 4121 CA GLU H 51 26.207 -44.965 -65.889 1.00 45.90 B C
+ATOM 4122 C GLU H 51 25.013 -45.634 -66.547 1.00 45.00 B C
+ATOM 4123 O GLU H 51 24.862 -45.589 -67.781 1.00 45.00 B O
+ATOM 4124 CB GLU H 51 26.079 -43.451 -65.986 1.00 55.61 B C
+ATOM 4125 CG GLU H 51 24.843 -42.896 -65.324 1.00 55.61 B C
+ATOM 4126 CD GLU H 51 24.853 -41.382 -65.298 1.00 55.61 B C
+ATOM 4127 OE1 GLU H 51 25.387 -40.784 -66.256 1.00 55.61 B O
+ATOM 4128 OE2 GLU H 51 24.359 -40.790 -64.313 1.00 55.61 B O1-
+ATOM 4129 N TRP H 52 24.184 -46.276 -65.728 1.00 47.28 B N
+ATOM 4130 CA TRP H 52 22.947 -46.847 -66.235 1.00 47.28 B C
+ATOM 4131 C TRP H 52 21.987 -45.729 -66.577 1.00 47.66 B C
+ATOM 4132 O TRP H 52 21.699 -44.867 -65.740 1.00 48.28 B O
+ATOM 4133 CB TRP H 52 22.336 -47.766 -65.196 1.00 57.57 B C
+ATOM 4134 CG TRP H 52 21.065 -48.438 -65.601 1.00 57.57 B C
+ATOM 4135 CD1 TRP H 52 20.922 -49.505 -66.434 1.00 57.57 B C
+ATOM 4136 CD2 TRP H 52 19.752 -48.109 -65.141 1.00 57.57 B C
+ATOM 4137 CE2 TRP H 52 18.860 -49.011 -65.748 1.00 57.57 B C
+ATOM 4138 CE3 TRP H 52 19.245 -47.133 -64.279 1.00 57.57 B C
+ATOM 4139 NE1 TRP H 52 19.600 -49.858 -66.532 1.00 57.57 B N
+ATOM 4140 CZ2 TRP H 52 17.487 -48.962 -65.521 1.00 57.57 B C
+ATOM 4141 CZ3 TRP H 52 17.896 -47.090 -64.049 1.00 57.57 B C
+ATOM 4142 CH2 TRP H 52 17.028 -47.996 -64.667 1.00 57.57 B C
+ATOM 4143 N VAL H 53 21.507 -45.726 -67.810 1.00 43.55 B N
+ATOM 4144 CA VAL H 53 20.660 -44.625 -68.234 1.00 43.69 B C
+ATOM 4145 C VAL H 53 19.215 -44.982 -67.985 1.00 49.67 B C
+ATOM 4146 O VAL H 53 18.511 -44.258 -67.277 1.00 52.72 B O
+ATOM 4147 CB VAL H 53 20.869 -44.249 -69.696 1.00 53.17 B C
+ATOM 4148 CG1 VAL H 53 20.006 -43.079 -69.983 1.00 53.17 B C
+ATOM 4149 CG2 VAL H 53 22.315 -43.904 -69.918 1.00 53.17 B C
+ATOM 4150 N SER H 54 18.749 -46.060 -68.604 1.00 56.97 B N
+ATOM 4151 CA SER H 54 17.390 -46.531 -68.318 1.00 53.84 B C
+ATOM 4152 C SER H 54 17.234 -47.950 -68.853 1.00 54.60 B C
+ATOM 4153 O SER H 54 18.193 -48.555 -69.347 1.00 57.62 B O
+ATOM 4154 CB SER H 54 16.353 -45.610 -68.923 1.00 58.75 B C
+ATOM 4155 OG SER H 54 16.860 -45.128 -70.124 1.00 58.75 B O
+ATOM 4156 N GLY H 55 16.016 -48.488 -68.735 1.00 56.69 B N
+ATOM 4157 CA GLY H 55 15.758 -49.807 -69.286 1.00 56.26 B C
+ATOM 4158 C GLY H 55 14.300 -50.207 -69.307 1.00 56.99 B C
+ATOM 4159 O GLY H 55 13.520 -49.746 -68.461 1.00 58.69 B O
+ATOM 4160 N ILE H 56 13.932 -51.096 -70.237 1.00 66.00 B N
+ATOM 4161 CA ILE H 56 12.568 -51.596 -70.379 1.00 65.76 B C
+ATOM 4162 C ILE H 56 12.599 -53.116 -70.330 1.00 67.25 B C
+ATOM 4163 O ILE H 56 13.651 -53.744 -70.491 1.00 69.80 B O
+ATOM 4164 CB ILE H 56 11.903 -51.134 -71.690 1.00 57.59 B C
+ATOM 4165 CG1 ILE H 56 12.912 -51.207 -72.809 1.00 57.59 B C
+ATOM 4166 CG2 ILE H 56 11.489 -49.710 -71.665 1.00 57.59 B C
+ATOM 4167 CD1 ILE H 56 13.055 -52.576 -73.380 1.00 57.59 B C
+ATOM 4168 N THR H 57 11.454 -53.760 -70.107 1.00 74.89 B N
+ATOM 4169 CA THR H 57 11.533 -55.219 -69.992 1.00 73.95 B C
+ATOM 4170 C THR H 57 10.576 -56.259 -70.612 1.00 73.95 B C
+ATOM 4171 O THR H 57 9.780 -56.872 -69.899 1.00 75.03 B O
+ATOM 4172 CB THR H 57 11.723 -55.613 -68.506 1.00 55.92 B C
+ATOM 4173 CG2 THR H 57 10.494 -55.238 -67.693 1.00 55.92 B C
+ATOM 4174 OG1 THR H 57 11.944 -57.025 -68.408 1.00 55.92 B O
+ATOM 4175 N TRP H 58 10.683 -56.495 -71.918 1.00 71.27 B N
+ATOM 4176 CA TRP H 58 9.931 -57.590 -72.545 1.00 71.27 B C
+ATOM 4177 C TRP H 58 8.533 -57.210 -73.048 1.00 71.27 B C
+ATOM 4178 O TRP H 58 8.035 -57.794 -74.006 1.00 71.27 B O
+ATOM 4179 CB TRP H 58 9.807 -58.766 -71.562 1.00 71.27 B C
+ATOM 4180 CG TRP H 58 8.518 -59.481 -71.736 1.00 71.27 B C
+ATOM 4181 CD1 TRP H 58 7.444 -59.397 -70.931 1.00 71.27 B C
+ATOM 4182 CD2 TRP H 58 8.165 -60.390 -72.781 1.00 71.27 B C
+ATOM 4183 CE2 TRP H 58 6.836 -60.786 -72.555 1.00 71.27 B C
+ATOM 4184 CE3 TRP H 58 8.842 -60.905 -73.899 1.00 71.27 B C
+ATOM 4185 NE1 TRP H 58 6.419 -60.165 -71.414 1.00 71.27 B N
+ATOM 4186 CZ2 TRP H 58 6.168 -61.685 -73.390 1.00 71.27 B C
+ATOM 4187 CZ3 TRP H 58 8.180 -61.802 -74.738 1.00 71.27 B C
+ATOM 4188 CH2 TRP H 58 6.854 -62.178 -74.476 1.00 71.27 B C
+ATOM 4189 N ASN H 59 7.971 -56.180 -72.446 1.00 70.45 B N
+ATOM 4190 CA ASN H 59 6.698 -55.679 -72.871 1.00 70.45 B C
+ATOM 4191 C ASN H 59 6.572 -54.252 -72.368 1.00 70.45 B C
+ATOM 4192 O ASN H 59 5.517 -53.870 -71.861 1.00 70.45 B O
+ATOM 4193 CB ASN H 59 5.544 -56.513 -72.310 1.00 70.45 B C
+ATOM 4194 CG ASN H 59 5.470 -56.462 -70.797 1.00 70.45 B C
+ATOM 4195 ND2 ASN H 59 4.357 -56.929 -70.243 1.00 70.45 B N
+ATOM 4196 OD1 ASN H 59 6.402 -56.008 -70.133 1.00 70.45 B O
+ATOM 4197 N SER H 62 7.627 -53.438 -72.477 1.00 69.75 B N
+ATOM 4198 CA SER H 62 7.378 -52.119 -71.942 1.00 69.75 B C
+ATOM 4199 C SER H 62 6.407 -52.246 -70.776 1.00 69.75 B C
+ATOM 4200 O SER H 62 5.547 -51.405 -70.572 1.00 69.75 B O
+ATOM 4201 CB SER H 62 6.846 -51.166 -73.016 1.00 69.75 B C
+ATOM 4202 OG SER H 62 7.907 -50.544 -73.753 1.00 69.75 B O
+ATOM 4203 N GLY H 63 6.565 -53.305 -69.994 1.00 70.01 B N
+ATOM 4204 CA GLY H 63 5.781 -53.477 -68.791 1.00 70.01 B C
+ATOM 4205 C GLY H 63 6.273 -52.641 -67.627 1.00 70.01 B C
+ATOM 4206 O GLY H 63 5.503 -51.941 -66.960 1.00 70.01 B O
+ATOM 4207 N TYR H 64 7.566 -52.709 -67.367 1.00 75.94 B N
+ATOM 4208 CA TYR H 64 8.195 -51.868 -66.372 1.00 75.94 B C
+ATOM 4209 C TYR H 64 9.327 -51.148 -67.082 1.00 75.94 B C
+ATOM 4210 O TYR H 64 10.082 -51.776 -67.825 1.00 77.82 B O
+ATOM 4211 CB TYR H 64 8.702 -52.709 -65.205 1.00 59.63 B C
+ATOM 4212 CG TYR H 64 9.190 -51.902 -64.020 1.00 59.63 B C
+ATOM 4213 CD1 TYR H 64 8.526 -51.941 -62.783 1.00 59.63 B C
+ATOM 4214 CD2 TYR H 64 10.325 -51.078 -64.130 1.00 59.63 B C
+ATOM 4215 CE1 TYR H 64 8.997 -51.167 -61.671 1.00 59.63 B C
+ATOM 4216 CE2 TYR H 64 10.800 -50.300 -63.038 1.00 59.63 B C
+ATOM 4217 CZ TYR H 64 10.139 -50.346 -61.815 1.00 59.63 B C
+ATOM 4218 OH TYR H 64 10.639 -49.576 -60.772 1.00 59.63 B O
+ATOM 4219 N ILE H 65 9.429 -49.834 -66.910 1.00 63.81 B N
+ATOM 4220 CA ILE H 65 10.583 -49.133 -67.452 1.00 65.70 B C
+ATOM 4221 C ILE H 65 11.073 -48.224 -66.346 1.00 64.48 B C
+ATOM 4222 O ILE H 65 10.289 -47.765 -65.512 1.00 64.71 B O
+ATOM 4223 CB ILE H 65 10.289 -48.285 -68.705 1.00 58.43 B C
+ATOM 4224 CG1 ILE H 65 9.654 -46.956 -68.333 1.00 58.43 B C
+ATOM 4225 CG2 ILE H 65 9.203 -48.956 -69.552 1.00 58.43 B C
+ATOM 4226 CD1 ILE H 65 9.907 -45.870 -69.385 1.00 58.43 B C
+ATOM 4227 N GLY H 66 12.359 -47.930 -66.357 1.00 61.69 B N
+ATOM 4228 CA GLY H 66 12.931 -47.135 -65.285 1.00 61.69 B C
+ATOM 4229 C GLY H 66 14.125 -46.338 -65.746 1.00 61.69 B C
+ATOM 4230 O GLY H 66 14.815 -46.705 -66.701 1.00 61.69 B O
+ATOM 4231 N TYR H 67 14.384 -45.250 -65.040 1.00 71.67 B N
+ATOM 4232 CA TYR H 67 15.326 -44.241 -65.475 1.00 70.38 B C
+ATOM 4233 C TYR H 67 16.328 -43.985 -64.364 1.00 71.35 B C
+ATOM 4234 O TYR H 67 16.089 -44.318 -63.199 1.00 72.26 B O
+ATOM 4235 CB TYR H 67 14.599 -42.944 -65.819 1.00 62.53 B C
+ATOM 4236 CG TYR H 67 13.712 -43.031 -67.036 1.00 62.53 B C
+ATOM 4237 CD1 TYR H 67 12.822 -42.013 -67.358 1.00 62.53 B C
+ATOM 4238 CD2 TYR H 67 13.783 -44.110 -67.879 1.00 62.53 B C
+ATOM 4239 CE1 TYR H 67 12.016 -42.094 -68.489 1.00 62.53 B C
+ATOM 4240 CE2 TYR H 67 12.998 -44.203 -69.006 1.00 62.53 B C
+ATOM 4241 CZ TYR H 67 12.115 -43.197 -69.310 1.00 62.53 B C
+ATOM 4242 OH TYR H 67 11.339 -43.329 -70.437 1.00 62.53 B O
+ATOM 4243 N ALA H 68 17.456 -43.389 -64.728 1.00 67.00 B N
+ATOM 4244 CA ALA H 68 18.338 -42.836 -63.717 1.00 66.55 B C
+ATOM 4245 C ALA H 68 17.763 -41.519 -63.222 1.00 69.80 B C
+ATOM 4246 O ALA H 68 16.988 -40.855 -63.914 1.00 72.28 B O
+ATOM 4247 CB ALA H 68 19.751 -42.633 -64.265 1.00 66.55 B C
+ATOM 4248 N ASP H 69 18.118 -41.155 -61.994 1.00 90.44 B N
+ATOM 4249 CA ASP H 69 17.687 -39.862 -61.482 1.00 97.36 B C
+ATOM 4250 C ASP H 69 18.196 -38.735 -62.365 1.00 96.24 B C
+ATOM 4251 O ASP H 69 17.520 -37.716 -62.520 1.00 92.96 B O
+ATOM 4252 CB ASP H 69 18.167 -39.693 -60.039 1.00 72.01 B C
+ATOM 4253 CG ASP H 69 17.470 -40.670 -59.068 1.00 72.01 B C
+ATOM 4254 OD1 ASP H 69 16.261 -40.953 -59.306 1.00 72.01 B O
+ATOM 4255 OD2 ASP H 69 18.120 -41.158 -58.088 1.00 72.01 B O1-
+ATOM 4256 N SER H 70 19.358 -38.935 -62.989 1.00 81.83 B N
+ATOM 4257 CA SER H 70 19.988 -37.914 -63.819 1.00 82.18 B C
+ATOM 4258 C SER H 70 19.099 -37.502 -64.973 1.00 84.14 B C
+ATOM 4259 O SER H 70 18.877 -36.313 -65.207 1.00 84.02 B O
+ATOM 4260 CB SER H 70 21.301 -38.448 -64.368 1.00 64.37 B C
+ATOM 4261 OG SER H 70 21.926 -39.280 -63.408 1.00 64.37 B O
+ATOM 4262 N VAL H 71 18.631 -38.464 -65.743 1.00 87.73 B N
+ATOM 4263 CA VAL H 71 17.744 -38.195 -66.863 1.00 86.23 B C
+ATOM 4264 C VAL H 71 16.353 -38.617 -66.430 1.00 91.44 B C
+ATOM 4265 O VAL H 71 16.060 -39.811 -66.341 1.00 95.78 B O
+ATOM 4266 CB VAL H 71 18.204 -38.928 -68.139 1.00 65.73 B C
+ATOM 4267 CG1 VAL H 71 19.547 -38.389 -68.575 1.00 65.73 B C
+ATOM 4268 CG2 VAL H 71 18.379 -40.428 -67.928 1.00 65.73 B C
+ATOM 4269 N LYS H 72 15.474 -37.679 -66.129 1.00 98.13 B N
+ATOM 4270 CA LYS H 72 14.139 -38.107 -65.741 1.00106.19 B C
+ATOM 4271 C LYS H 72 13.167 -37.137 -66.371 1.00110.40 B C
+ATOM 4272 O LYS H 72 13.324 -35.923 -66.226 1.00115.16 B O
+ATOM 4273 CB LYS H 72 13.985 -38.185 -64.219 1.00 72.38 B C
+ATOM 4274 CG LYS H 72 12.581 -38.559 -63.752 1.00 72.38 B C
+ATOM 4275 CD LYS H 72 12.507 -39.968 -63.125 1.00 72.38 B C
+ATOM 4276 CE LYS H 72 11.138 -40.201 -62.392 1.00 72.38 B C
+ATOM 4277 NZ LYS H 72 10.932 -41.540 -61.690 1.00 72.38 B N1+
+ATOM 4278 N GLY H 74 12.242 -37.663 -67.165 1.00 99.57 B N
+ATOM 4279 CA GLY H 74 11.403 -36.812 -67.974 1.00 95.47 B C
+ATOM 4280 C GLY H 74 12.152 -36.077 -69.062 1.00 98.76 B C
+ATOM 4281 O GLY H 74 11.521 -35.471 -69.935 1.00100.38 B O
+ATOM 4282 N ARG H 75 13.479 -36.099 -69.030 1.00106.92 B N
+ATOM 4283 CA ARG H 75 14.273 -35.662 -70.161 1.00100.32 B C
+ATOM 4284 C ARG H 75 14.374 -36.761 -71.203 1.00 94.99 B C
+ATOM 4285 O ARG H 75 14.326 -36.491 -72.408 1.00 99.23 B O
+ATOM 4286 CB ARG H 75 15.656 -35.277 -69.676 1.00 68.51 B C
+ATOM 4287 CG ARG H 75 15.795 -33.877 -69.245 1.00 68.51 B C
+ATOM 4288 CD ARG H 75 17.232 -33.488 -69.426 1.00 68.51 B C
+ATOM 4289 NE ARG H 75 18.068 -34.115 -68.412 1.00 68.51 B N
+ATOM 4290 CZ ARG H 75 19.371 -34.322 -68.556 1.00 68.51 B C
+ATOM 4291 NH1 ARG H 75 19.964 -33.972 -69.689 1.00 68.51 B N1+
+ATOM 4292 NH2 ARG H 75 20.073 -34.905 -67.588 1.00 68.51 B N
+ATOM 4293 N PHE H 76 14.512 -37.999 -70.744 1.00 86.73 B N
+ATOM 4294 CA PHE H 76 14.737 -39.166 -71.577 1.00 88.91 B C
+ATOM 4295 C PHE H 76 13.495 -40.043 -71.591 1.00 91.84 B C
+ATOM 4296 O PHE H 76 12.743 -40.089 -70.614 1.00 95.35 B O
+ATOM 4297 CB PHE H 76 15.923 -39.996 -71.058 1.00 62.97 B C
+ATOM 4298 CG PHE H 76 17.293 -39.456 -71.431 1.00 62.97 B C
+ATOM 4299 CD1 PHE H 76 17.463 -38.165 -71.890 1.00 62.97 B C
+ATOM 4300 CD2 PHE H 76 18.404 -40.266 -71.333 1.00 62.97 B C
+ATOM 4301 CE1 PHE H 76 18.720 -37.698 -72.227 1.00 62.97 B C
+ATOM 4302 CE2 PHE H 76 19.642 -39.799 -71.663 1.00 62.97 B C
+ATOM 4303 CZ PHE H 76 19.805 -38.510 -72.099 1.00 62.97 B C
+ATOM 4304 N THR H 77 13.276 -40.730 -72.709 1.00 84.50 B N
+ATOM 4305 CA THR H 77 12.208 -41.720 -72.784 1.00 80.33 B C
+ATOM 4306 C THR H 77 12.556 -43.021 -73.504 1.00 81.75 B C
+ATOM 4307 O THR H 77 12.915 -43.007 -74.686 1.00 78.67 B O
+ATOM 4308 CB THR H 77 10.928 -41.077 -73.326 1.00 57.29 B C
+ATOM 4309 CG2 THR H 77 10.087 -40.442 -72.222 1.00 57.29 B C
+ATOM 4310 OG1 THR H 77 11.279 -40.104 -74.315 1.00 57.29 B O
+ATOM 4311 N ILE H 78 12.430 -44.171 -72.857 1.00 74.60 B N
+ATOM 4312 CA ILE H 78 12.715 -45.406 -73.601 1.00 77.51 B C
+ATOM 4313 C ILE H 78 11.693 -46.465 -74.040 1.00 80.73 B C
+ATOM 4314 O ILE H 78 11.170 -47.210 -73.211 1.00 78.96 B O
+ATOM 4315 CB ILE H 78 13.736 -46.203 -72.763 1.00 56.16 B C
+ATOM 4316 CG1 ILE H 78 14.257 -47.402 -73.557 1.00 56.16 B C
+ATOM 4317 CG2 ILE H 78 13.111 -46.656 -71.452 1.00 56.16 B C
+ATOM 4318 CD1 ILE H 78 15.477 -48.056 -72.946 1.00 56.16 B C
+ATOM 4319 N SER H 79 11.375 -46.505 -75.338 1.00 82.66 B N
+ATOM 4320 CA SER H 79 10.400 -47.484 -75.805 1.00 80.23 B C
+ATOM 4321 C SER H 79 10.932 -48.440 -76.837 1.00 83.60 B C
+ATOM 4322 O SER H 79 11.939 -48.194 -77.493 1.00 82.52 B O
+ATOM 4323 CB SER H 79 9.079 -46.921 -76.304 1.00 59.76 B C
+ATOM 4324 OG SER H 79 8.221 -46.705 -75.188 1.00 59.76 B O
+ATOM 4325 N ARG H 80 10.225 -49.545 -76.967 1.00 81.05 B N
+ATOM 4326 CA ARG H 80 10.618 -50.606 -77.864 1.00 81.38 B C
+ATOM 4327 C ARG H 80 9.481 -50.914 -78.819 1.00 84.96 B C
+ATOM 4328 O ARG H 80 8.325 -50.571 -78.573 1.00 87.16 B O
+ATOM 4329 CB ARG H 80 10.990 -51.848 -77.081 1.00 60.67 B C
+ATOM 4330 CG ARG H 80 9.900 -52.199 -76.099 1.00 60.67 B C
+ATOM 4331 CD ARG H 80 10.281 -53.363 -75.227 1.00 60.67 B C
+ATOM 4332 NE ARG H 80 11.320 -54.171 -75.849 1.00 60.67 B N
+ATOM 4333 CZ ARG H 80 12.000 -55.106 -75.203 1.00 60.67 B C
+ATOM 4334 NH1 ARG H 80 11.749 -55.334 -73.930 1.00 60.67 B N1+
+ATOM 4335 NH2 ARG H 80 12.930 -55.806 -75.825 1.00 60.67 B N
+ATOM 4336 N ASP H 81 9.844 -51.564 -79.920 1.00102.17 B N
+ATOM 4337 CA ASP H 81 8.903 -52.030 -80.926 1.00102.34 B C
+ATOM 4338 C ASP H 81 8.611 -53.513 -80.742 1.00101.33 B C
+ATOM 4339 O ASP H 81 7.447 -53.919 -80.658 1.00101.11 B O
+ATOM 4340 CB ASP H 81 9.443 -51.746 -82.334 1.00 72.20 B C
+ATOM 4341 CG ASP H 81 8.321 -51.528 -83.374 1.00 72.20 B C
+ATOM 4342 OD1 ASP H 81 7.134 -51.394 -82.967 1.00 72.20 B O
+ATOM 4343 OD2 ASP H 81 8.626 -51.456 -84.598 1.00 72.20 B O1-
+ATOM 4344 N ASN H 82 9.661 -54.334 -80.775 1.00 97.20 B N
+ATOM 4345 CA ASN H 82 9.617 -55.792 -80.721 1.00 96.28 B C
+ATOM 4346 C ASN H 82 9.166 -56.282 -82.071 1.00 97.45 B C
+ATOM 4347 O ASN H 82 9.371 -57.438 -82.438 1.00 98.87 B O
+ATOM 4348 CB ASN H 82 8.677 -56.337 -79.645 1.00 69.39 B C
+ATOM 4349 CG ASN H 82 9.214 -56.156 -78.249 1.00 69.39 B C
+ATOM 4350 ND2 ASN H 82 8.310 -56.033 -77.266 1.00 69.39 B N
+ATOM 4351 OD1 ASN H 82 10.429 -56.151 -78.042 1.00 69.39 B O
+ATOM 4352 N ALA H 83 8.567 -55.387 -82.826 1.00111.84 B N
+ATOM 4353 CA ALA H 83 8.275 -55.696 -84.203 1.00113.30 B C
+ATOM 4354 C ALA H 83 9.510 -55.434 -85.041 1.00115.20 B C
+ATOM 4355 O ALA H 83 10.128 -56.361 -85.571 1.00115.16 B O
+ATOM 4356 CB ALA H 83 7.086 -54.860 -84.657 1.00 33.06 B C
+ATOM 4357 N ARG H 84 9.925 -54.173 -85.098 1.00118.66 B N
+ATOM 4358 CA ARG H 84 11.135 -53.778 -85.801 1.00118.44 B C
+ATOM 4359 C ARG H 84 12.349 -54.407 -85.112 1.00116.03 B C
+ATOM 4360 O ARG H 84 13.481 -54.270 -85.592 1.00111.79 B O
+ATOM 4361 CB ARG H 84 11.250 -52.236 -85.875 1.00 80.38 B C
+ATOM 4362 CG ARG H 84 10.524 -51.523 -87.086 1.00 80.38 B C
+ATOM 4363 CD ARG H 84 10.838 -49.966 -87.238 1.00 80.38 B C
+ATOM 4364 NE ARG H 84 12.279 -49.651 -87.417 1.00 80.38 B N
+ATOM 4365 CZ ARG H 84 12.846 -48.434 -87.349 1.00 80.38 B C
+ATOM 4366 NH1 ARG H 84 12.111 -47.343 -87.125 1.00 80.38 B N1+
+ATOM 4367 NH2 ARG H 84 14.168 -48.310 -87.506 1.00 80.38 B N
+ATOM 4368 N ASN H 85 12.130 -55.095 -83.989 1.00107.39 B N
+ATOM 4369 CA ASN H 85 13.218 -55.598 -83.149 1.00100.85 B C
+ATOM 4370 C ASN H 85 14.161 -54.455 -82.809 1.00101.08 B C
+ATOM 4371 O ASN H 85 15.353 -54.473 -83.113 1.00101.32 B O
+ATOM 4372 CB ASN H 85 13.978 -56.755 -83.808 1.00 69.23 B C
+ATOM 4373 CG ASN H 85 13.444 -58.132 -83.396 1.00 69.23 B C
+ATOM 4374 ND2 ASN H 85 14.205 -59.185 -83.713 1.00 69.23 B N
+ATOM 4375 OD1 ASN H 85 12.367 -58.246 -82.797 1.00 69.23 B O
+ATOM 4376 N SER H 86 13.575 -53.423 -82.221 1.00 95.76 B N
+ATOM 4377 CA SER H 86 14.256 -52.155 -82.086 1.00 91.67 B C
+ATOM 4378 C SER H 86 13.993 -51.558 -80.717 1.00 94.66 B C
+ATOM 4379 O SER H 86 13.005 -51.860 -80.048 1.00 97.56 B O
+ATOM 4380 CB SER H 86 13.825 -51.176 -83.179 1.00 64.08 B C
+ATOM 4381 OG SER H 86 14.609 -51.343 -84.358 1.00 64.08 B O
+ATOM 4382 N LEU H 87 14.907 -50.697 -80.323 1.00 88.64 B N
+ATOM 4383 CA LEU H 87 14.871 -50.001 -79.061 1.00 88.64 B C
+ATOM 4384 C LEU H 87 15.117 -48.536 -79.347 1.00 88.64 B C
+ATOM 4385 O LEU H 87 15.839 -48.197 -80.279 1.00 89.43 B O
+ATOM 4386 CB LEU H 87 15.926 -50.556 -78.121 1.00 49.73 B C
+ATOM 4387 CG LEU H 87 15.868 -49.962 -76.717 1.00 49.73 B C
+ATOM 4388 CD1 LEU H 87 14.427 -49.890 -76.256 1.00 49.73 B C
+ATOM 4389 CD2 LEU H 87 16.683 -50.783 -75.755 1.00 49.73 B C
+ATOM 4390 N TYR H 88 14.504 -47.667 -78.560 1.00 88.92 B N
+ATOM 4391 CA TYR H 88 14.565 -46.236 -78.812 1.00 89.47 B C
+ATOM 4392 C TYR H 88 14.734 -45.510 -77.489 1.00 89.00 B C
+ATOM 4393 O TYR H 88 14.025 -45.804 -76.522 1.00 92.98 B O
+ATOM 4394 CB TYR H 88 13.296 -45.752 -79.543 1.00 67.14 B C
+ATOM 4395 CG TYR H 88 12.972 -46.514 -80.827 1.00 67.14 B C
+ATOM 4396 CD1 TYR H 88 13.353 -46.028 -82.078 1.00 67.14 B C
+ATOM 4397 CD2 TYR H 88 12.321 -47.744 -80.786 1.00 67.14 B C
+ATOM 4398 CE1 TYR H 88 13.065 -46.746 -83.255 1.00 67.14 B C
+ATOM 4399 CE2 TYR H 88 12.034 -48.467 -81.948 1.00 67.14 B C
+ATOM 4400 CZ TYR H 88 12.404 -47.969 -83.175 1.00 67.14 B C
+ATOM 4401 OH TYR H 88 12.109 -48.704 -84.308 1.00 67.14 B O
+ATOM 4402 N LEU H 89 15.677 -44.574 -77.447 1.00 79.27 B N
+ATOM 4403 CA LEU H 89 15.835 -43.675 -76.311 1.00 78.57 B C
+ATOM 4404 C LEU H 89 15.818 -42.244 -76.825 1.00 80.71 B C
+ATOM 4405 O LEU H 89 16.531 -41.914 -77.777 1.00 80.73 B O
+ATOM 4406 CB LEU H 89 17.136 -43.951 -75.546 1.00 57.10 B C
+ATOM 4407 CG LEU H 89 17.512 -42.922 -74.474 1.00 57.10 B C
+ATOM 4408 CD1 LEU H 89 16.644 -43.105 -73.288 1.00 57.10 B C
+ATOM 4409 CD2 LEU H 89 18.950 -43.018 -74.066 1.00 57.10 B C
+ATOM 4410 N GLN H 90 15.013 -41.397 -76.198 1.00 94.70 B N
+ATOM 4411 CA GLN H 90 14.836 -40.025 -76.642 1.00 91.07 B C
+ATOM 4412 C GLN H 90 15.365 -39.029 -75.625 1.00 95.01 B C
+ATOM 4413 O GLN H 90 15.105 -39.151 -74.424 1.00 99.63 B O
+ATOM 4414 CB GLN H 90 13.366 -39.735 -76.924 1.00 65.19 B C
+ATOM 4415 CG GLN H 90 13.125 -38.300 -77.334 1.00 65.19 B C
+ATOM 4416 CD GLN H 90 13.027 -38.110 -78.837 1.00 65.19 B C
+ATOM 4417 NE2 GLN H 90 13.358 -36.889 -79.276 1.00 65.19 B N
+ATOM 4418 OE1 GLN H 90 12.646 -39.032 -79.604 1.00 65.19 B O
+ATOM 4419 N MET H 91 16.042 -38.009 -76.140 1.00 97.01 B N
+ATOM 4420 CA MET H 91 16.898 -37.106 -75.382 1.00 98.12 B C
+ATOM 4421 C MET H 91 16.368 -35.690 -75.553 1.00 99.96 B C
+ATOM 4422 O MET H 91 16.477 -35.119 -76.646 1.00100.69 B O
+ATOM 4423 CB MET H 91 18.315 -37.151 -75.937 1.00 67.01 B C
+ATOM 4424 CG MET H 91 19.051 -38.481 -75.878 1.00 67.01 B C
+ATOM 4425 SD MET H 91 20.064 -38.758 -74.437 1.00 67.01 B S
+ATOM 4426 CE MET H 91 21.585 -39.240 -75.251 1.00 67.01 B C
+ATOM 4427 N ASN H 92 15.860 -35.094 -74.479 1.00 98.65 B N
+ATOM 4428 CA ASN H 92 15.269 -33.767 -74.560 1.00101.51 B C
+ATOM 4429 C ASN H 92 15.822 -32.898 -73.447 1.00104.58 B C
+ATOM 4430 O ASN H 92 16.306 -33.402 -72.432 1.00105.96 B O
+ATOM 4431 CB ASN H 92 13.748 -33.837 -74.466 1.00 70.39 B C
+ATOM 4432 CG ASN H 92 13.140 -34.626 -75.612 1.00 70.39 B C
+ATOM 4433 ND2 ASN H 92 13.084 -34.034 -76.805 1.00 70.39 B N
+ATOM 4434 OD1 ASN H 92 12.746 -35.775 -75.425 1.00 70.39 B O
+ATOM 4435 N SER H 93 15.740 -31.580 -73.646 1.00106.04 B N
+ATOM 4436 CA SER H 93 16.436 -30.631 -72.778 1.00107.13 B C
+ATOM 4437 C SER H 93 17.850 -31.126 -72.546 1.00102.60 B C
+ATOM 4438 O SER H 93 18.391 -31.050 -71.446 1.00100.37 B O
+ATOM 4439 CB SER H 93 15.700 -30.415 -71.444 1.00 64.54 B C
+ATOM 4440 OG SER H 93 16.310 -29.408 -70.608 1.00 64.54 B O
+ATOM 4441 N LEU H 94 18.430 -31.680 -73.591 1.00 94.69 B N
+ATOM 4442 CA LEU H 94 19.668 -32.408 -73.441 1.00 94.97 B C
+ATOM 4443 C LEU H 94 20.702 -31.503 -72.809 1.00100.87 B C
+ATOM 4444 O LEU H 94 21.054 -30.470 -73.373 1.00102.56 B O
+ATOM 4445 CB LEU H 94 20.101 -32.865 -74.823 1.00 57.97 B C
+ATOM 4446 CG LEU H 94 21.220 -33.835 -75.157 1.00 57.97 B C
+ATOM 4447 CD1 LEU H 94 21.135 -35.082 -74.298 1.00 57.97 B C
+ATOM 4448 CD2 LEU H 94 21.160 -34.198 -76.648 1.00 57.97 B C
+ATOM 4449 N ARG H 95 21.200 -31.881 -71.643 1.00119.44 B N
+ATOM 4450 CA ARG H 95 22.159 -30.999 -71.011 1.00118.59 B C
+ATOM 4451 C ARG H 95 23.533 -31.373 -71.553 1.00111.02 B C
+ATOM 4452 O ARG H 95 23.683 -32.298 -72.354 1.00108.46 B O
+ATOM 4453 CB ARG H 95 22.054 -31.043 -69.474 1.00 76.27 B C
+ATOM 4454 CG ARG H 95 20.796 -30.290 -68.866 1.00 76.27 B C
+ATOM 4455 CD ARG H 95 20.608 -28.845 -69.510 1.00 76.27 B C
+ATOM 4456 NE ARG H 95 19.265 -28.212 -69.390 1.00 76.27 B N
+ATOM 4457 CZ ARG H 95 19.018 -26.960 -68.953 1.00 76.27 B C
+ATOM 4458 NH1 ARG H 95 20.014 -26.140 -68.592 1.00 76.27 B N1+
+ATOM 4459 NH2 ARG H 95 17.757 -26.512 -68.896 1.00 76.27 B N
+ATOM 4460 N ALA H 96 24.546 -30.632 -71.131 1.00 77.72 B N
+ATOM 4461 CA ALA H 96 25.858 -30.723 -71.753 1.00 81.14 B C
+ATOM 4462 C ALA H 96 26.719 -31.838 -71.185 1.00 80.37 B C
+ATOM 4463 O ALA H 96 27.764 -32.150 -71.759 1.00 83.73 B O
+ATOM 4464 CB ALA H 96 26.575 -29.385 -71.593 1.00 78.76 B C
+ATOM 4465 N GLU H 97 26.329 -32.413 -70.061 1.00 79.55 B N
+ATOM 4466 CA GLU H 97 27.055 -33.515 -69.460 1.00 83.82 B C
+ATOM 4467 C GLU H 97 26.505 -34.854 -69.890 1.00 84.47 B C
+ATOM 4468 O GLU H 97 27.023 -35.887 -69.462 1.00 88.20 B O
+ATOM 4469 CB GLU H 97 27.017 -33.413 -67.937 1.00 89.53 B C
+ATOM 4470 CG GLU H 97 27.604 -32.113 -67.391 1.00 89.53 B C
+ATOM 4471 CD GLU H 97 26.626 -30.943 -67.426 1.00 89.53 B C
+ATOM 4472 OE1 GLU H 97 25.411 -31.173 -67.641 1.00 89.53 B O
+ATOM 4473 OE2 GLU H 97 27.073 -29.790 -67.237 1.00 89.53 B O1-
+ATOM 4474 N ASP H 98 25.459 -34.861 -70.711 1.00 75.56 B N
+ATOM 4475 CA ASP H 98 24.964 -36.122 -71.235 1.00 72.34 B C
+ATOM 4476 C ASP H 98 25.880 -36.695 -72.291 1.00 71.16 B C
+ATOM 4477 O ASP H 98 25.669 -37.838 -72.704 1.00 69.73 B O
+ATOM 4478 CB ASP H 98 23.562 -35.963 -71.825 1.00 79.58 B C
+ATOM 4479 CG ASP H 98 22.481 -35.886 -70.760 1.00 79.58 B C
+ATOM 4480 OD1 ASP H 98 22.694 -36.393 -69.625 1.00 79.58 B O
+ATOM 4481 OD2 ASP H 98 21.407 -35.322 -71.063 1.00 79.58 B O1-
+ATOM 4482 N THR H 99 26.872 -35.934 -72.741 1.00 64.99 B N
+ATOM 4483 CA THR H 99 27.751 -36.429 -73.785 1.00 63.29 B C
+ATOM 4484 C THR H 99 28.507 -37.644 -73.276 1.00 62.88 B C
+ATOM 4485 O THR H 99 29.008 -37.654 -72.148 1.00 62.63 B O
+ATOM 4486 CB THR H 99 28.734 -35.357 -74.198 1.00 71.04 B C
+ATOM 4487 CG2 THR H 99 28.140 -34.037 -73.937 1.00 71.04 B C
+ATOM 4488 OG1 THR H 99 29.876 -35.456 -73.360 1.00 71.04 B O
+ATOM 4489 N ALA H 100 28.576 -38.674 -74.111 1.00 62.73 B N
+ATOM 4490 CA ALA H 100 29.062 -39.983 -73.695 1.00 61.26 B C
+ATOM 4491 C ALA H 100 28.912 -40.915 -74.883 1.00 57.14 B C
+ATOM 4492 O ALA H 100 28.229 -40.603 -75.867 1.00 59.38 B O
+ATOM 4493 CB ALA H 100 28.293 -40.558 -72.506 1.00 57.14 B C
+ATOM 4494 N LEU H 101 29.541 -42.075 -74.773 1.00 47.91 B N
+ATOM 4495 CA LEU H 101 29.265 -43.144 -75.703 1.00 47.79 B C
+ATOM 4496 C LEU H 101 28.080 -43.898 -75.132 1.00 47.79 B C
+ATOM 4497 O LEU H 101 28.089 -44.250 -73.946 1.00 50.20 B O
+ATOM 4498 CB LEU H 101 30.476 -44.041 -75.866 1.00 58.00 B C
+ATOM 4499 CG LEU H 101 30.294 -44.754 -77.181 1.00 58.00 B C
+ATOM 4500 CD1 LEU H 101 31.614 -44.749 -77.896 1.00 58.00 B C
+ATOM 4501 CD2 LEU H 101 29.850 -46.128 -76.913 1.00 58.00 B C
+ATOM 4502 N TYR H 102 27.034 -44.075 -75.944 1.00 46.71 B N
+ATOM 4503 CA TYR H 102 25.773 -44.671 -75.495 1.00 46.71 B C
+ATOM 4504 C TYR H 102 25.671 -46.108 -75.992 1.00 46.71 B C
+ATOM 4505 O TYR H 102 25.774 -46.372 -77.206 1.00 46.71 B O
+ATOM 4506 CB TYR H 102 24.566 -43.860 -75.963 1.00 57.38 B C
+ATOM 4507 CG TYR H 102 24.270 -42.691 -75.066 1.00 57.38 B C
+ATOM 4508 CD1 TYR H 102 23.290 -42.761 -74.102 1.00 57.38 B C
+ATOM 4509 CD2 TYR H 102 25.014 -41.528 -75.162 1.00 57.38 B C
+ATOM 4510 CE1 TYR H 102 23.041 -41.701 -73.283 1.00 57.38 B C
+ATOM 4511 CE2 TYR H 102 24.773 -40.469 -74.354 1.00 57.38 B C
+ATOM 4512 CZ TYR H 102 23.786 -40.559 -73.413 1.00 57.38 B C
+ATOM 4513 OH TYR H 102 23.549 -39.482 -72.598 1.00 57.38 B O
+ATOM 4514 N TYR H 103 25.450 -47.014 -75.036 1.00 48.49 B N
+ATOM 4515 CA TYR H 103 25.559 -48.461 -75.172 1.00 48.49 B C
+ATOM 4516 C TYR H 103 24.183 -49.094 -75.077 1.00 48.49 B C
+ATOM 4517 O TYR H 103 23.433 -48.833 -74.119 1.00 51.52 B O
+ATOM 4518 CB TYR H 103 26.416 -49.048 -74.051 1.00 58.68 B C
+ATOM 4519 CG TYR H 103 27.894 -48.986 -74.238 1.00 58.68 B C
+ATOM 4520 CD1 TYR H 103 28.473 -49.410 -75.422 1.00 58.68 B C
+ATOM 4521 CD2 TYR H 103 28.716 -48.509 -73.224 1.00 58.68 B C
+ATOM 4522 CE1 TYR H 103 29.838 -49.365 -75.598 1.00 58.68 B C
+ATOM 4523 CE2 TYR H 103 30.085 -48.449 -73.385 1.00 58.68 B C
+ATOM 4524 CZ TYR H 103 30.643 -48.876 -74.580 1.00 58.68 B C
+ATOM 4525 OH TYR H 103 32.010 -48.828 -74.771 1.00 58.68 B O
+ATOM 4526 N CYS H 104 23.873 -49.940 -76.047 1.00 57.06 B N
+ATOM 4527 CA CYS H 104 22.735 -50.842 -75.977 1.00 56.66 B C
+ATOM 4528 C CYS H 104 23.176 -52.125 -75.270 1.00 58.26 B C
+ATOM 4529 O CYS H 104 24.161 -52.751 -75.688 1.00 59.14 B O
+ATOM 4530 CB CYS H 104 22.256 -51.124 -77.392 1.00 67.66 B C
+ATOM 4531 SG CYS H 104 20.607 -51.721 -77.518 1.00 67.66 B S
+ATOM 4532 N VAL H 105 22.485 -52.506 -74.187 1.00 49.90 B N
+ATOM 4533 CA VAL H 105 22.970 -53.574 -73.307 1.00 49.90 B C
+ATOM 4534 C VAL H 105 21.842 -54.509 -72.926 1.00 51.74 B C
+ATOM 4535 O VAL H 105 20.905 -54.088 -72.240 1.00 50.00 B O
+ATOM 4536 CB VAL H 105 23.583 -53.039 -72.014 1.00 58.06 B C
+ATOM 4537 CG1 VAL H 105 24.272 -54.170 -71.334 1.00 58.06 B C
+ATOM 4538 CG2 VAL H 105 24.540 -51.909 -72.303 1.00 58.06 B C
+ATOM 4539 N ARG H 106 21.987 -55.794 -73.257 1.00 54.17 B N
+ATOM 4540 CA ARG H 106 20.950 -56.785 -72.992 1.00 54.92 B C
+ATOM 4541 C ARG H 106 21.116 -57.412 -71.614 1.00 51.72 B C
+ATOM 4542 O ARG H 106 22.223 -57.765 -71.202 1.00 51.82 B O
+ATOM 4543 CB ARG H 106 20.983 -57.868 -74.056 1.00 55.79 B C
+ATOM 4544 CG ARG H 106 20.251 -59.125 -73.704 1.00 55.79 B C
+ATOM 4545 CD ARG H 106 21.072 -60.234 -74.234 1.00 55.79 B C
+ATOM 4546 NE ARG H 106 20.316 -61.345 -74.785 1.00 55.79 B N
+ATOM 4547 CZ ARG H 106 20.915 -62.455 -75.196 1.00 55.79 B C
+ATOM 4548 NH1 ARG H 106 22.229 -62.531 -75.102 1.00 55.79 B N1+
+ATOM 4549 NH2 ARG H 106 20.242 -63.474 -75.716 1.00 55.79 B N
+ATOM 4550 N ASP H 107 20.014 -57.545 -70.898 1.00 59.29 B N
+ATOM 4551 CA ASP H 107 20.059 -58.096 -69.554 1.00 59.29 B C
+ATOM 4552 C ASP H 107 19.818 -59.603 -69.620 1.00 59.29 B C
+ATOM 4553 O ASP H 107 18.724 -60.046 -69.993 1.00 59.29 B O
+ATOM 4554 CB ASP H 107 19.031 -57.385 -68.679 1.00 59.29 B C
+ATOM 4555 CG ASP H 107 19.243 -57.644 -67.198 1.00 59.29 B C
+ATOM 4556 OD1 ASP H 107 20.262 -58.282 -66.836 1.00 59.29 B O
+ATOM 4557 OD2 ASP H 107 18.405 -57.192 -66.378 1.00 59.29 B O1-
+ATOM 4558 N ALA H 108 20.836 -60.388 -69.243 1.00 54.40 B N
+ATOM 4559 CA ALA H 108 20.834 -61.842 -69.413 1.00 54.40 B C
+ATOM 4560 C ALA H 108 19.820 -62.569 -68.554 1.00 54.40 B C
+ATOM 4561 O ALA H 108 19.671 -63.780 -68.723 1.00 54.40 B O
+ATOM 4562 CB ALA H 108 22.201 -62.443 -69.095 1.00 54.40 B C
+ATOM 4563 N TYR H 109 19.137 -61.894 -67.639 1.00 64.04 B N
+ATOM 4564 CA TYR H 109 18.170 -62.562 -66.775 1.00 64.04 B C
+ATOM 4565 C TYR H 109 16.814 -62.561 -67.456 1.00 64.04 B C
+ATOM 4566 O TYR H 109 16.161 -61.513 -67.561 1.00 64.04 B O
+ATOM 4567 CB TYR H 109 18.086 -61.884 -65.415 1.00 64.04 B C
+ATOM 4568 CG TYR H 109 16.958 -62.438 -64.597 1.00 64.04 B C
+ATOM 4569 CD1 TYR H 109 17.061 -63.664 -63.976 1.00 64.04 B C
+ATOM 4570 CD2 TYR H 109 15.770 -61.745 -64.470 1.00 64.04 B C
+ATOM 4571 CE1 TYR H 109 16.012 -64.190 -63.214 1.00 64.04 B C
+ATOM 4572 CE2 TYR H 109 14.713 -62.258 -63.712 1.00 64.04 B C
+ATOM 4573 CZ TYR H 109 14.840 -63.487 -63.079 1.00 64.04 B C
+ATOM 4574 OH TYR H 109 13.792 -63.996 -62.322 1.00 64.04 B O
+ATOM 4575 N VAL H 110 16.381 -63.744 -67.894 1.00 68.66 B N
+ATOM 4576 CA VAL H 110 15.201 -63.842 -68.743 1.00 68.66 B C
+ATOM 4577 C VAL H 110 13.891 -63.907 -67.960 1.00 68.66 B C
+ATOM 4578 O VAL H 110 12.837 -63.554 -68.509 1.00 68.66 B O
+ATOM 4579 CB VAL H 110 15.311 -65.078 -69.641 1.00 68.66 B C
+ATOM 4580 CG1 VAL H 110 15.086 -66.334 -68.785 1.00 68.66 B C
+ATOM 4581 CG2 VAL H 110 14.362 -64.964 -70.820 1.00 68.66 B C
+ATOM 4582 N SER H 111 13.929 -64.326 -66.694 1.00 69.05 B N
+ATOM 4583 CA SER H 111 12.722 -64.531 -65.895 1.00 69.05 B C
+ATOM 4584 C SER H 111 11.750 -65.529 -66.493 1.00 69.05 B C
+ATOM 4585 O SER H 111 10.825 -65.161 -67.219 1.00 69.05 B O
+ATOM 4586 CB SER H 111 11.954 -63.249 -65.612 1.00 69.05 B C
+ATOM 4587 OG SER H 111 11.027 -63.536 -64.561 1.00 69.05 B O
+ATOM 4588 N GLY H 111A 11.974 -66.796 -66.227 1.00 67.69 B N
+ATOM 4589 CA GLY H 111A 10.990 -67.775 -66.573 1.00 67.69 B C
+ATOM 4590 C GLY H 111A 11.429 -68.737 -67.614 1.00 67.69 B C
+ATOM 4591 O GLY H 111A 10.670 -69.661 -67.906 1.00 67.69 B O
+ATOM 4592 N SER H 111B 12.606 -68.563 -68.206 1.00 78.31 B N
+ATOM 4593 CA SER H 111B 13.186 -69.729 -68.863 1.00 78.31 B C
+ATOM 4594 C SER H 111B 13.778 -70.673 -67.814 1.00 78.31 B C
+ATOM 4595 O SER H 111B 13.954 -70.318 -66.646 1.00 78.31 B O
+ATOM 4596 CB SER H 111B 14.224 -69.344 -69.929 1.00 78.31 B C
+ATOM 4597 OG SER H 111B 15.553 -69.374 -69.431 1.00 78.31 B O
+ATOM 4598 N ASP H 112C 14.033 -71.910 -68.242 1.00 82.12 B N
+ATOM 4599 CA ASP H 112C 14.343 -72.967 -67.288 1.00 82.12 B C
+ATOM 4600 C ASP H 112C 15.725 -72.794 -66.671 1.00 82.12 B C
+ATOM 4601 O ASP H 112C 15.921 -73.115 -65.493 1.00 82.12 B O
+ATOM 4602 CB ASP H 112C 14.225 -74.329 -67.959 1.00 82.12 B C
+ATOM 4603 CG ASP H 112C 12.790 -74.684 -68.296 1.00 82.12 B C
+ATOM 4604 OD1 ASP H 112C 11.888 -73.883 -67.951 1.00 82.12 B O
+ATOM 4605 OD2 ASP H 112C 12.562 -75.761 -68.897 1.00 82.12 B O1-
+ATOM 4606 N TYR H 112B 16.699 -72.317 -67.438 1.00 85.22 B N
+ATOM 4607 CA TYR H 112B 18.031 -72.021 -66.917 1.00 85.22 B C
+ATOM 4608 C TYR H 112B 18.256 -70.513 -66.965 1.00 85.22 B C
+ATOM 4609 O TYR H 112B 18.200 -69.906 -68.043 1.00 85.22 B O
+ATOM 4610 CB TYR H 112B 19.104 -72.774 -67.705 1.00 85.22 B C
+ATOM 4611 CG TYR H 112B 20.485 -72.928 -67.034 1.00 85.22 B C
+ATOM 4612 CD1 TYR H 112B 21.293 -74.047 -67.304 1.00 85.22 B C
+ATOM 4613 CD2 TYR H 112B 21.021 -71.932 -66.196 1.00 85.22 B C
+ATOM 4614 CE1 TYR H 112B 22.576 -74.191 -66.722 1.00 85.22 B C
+ATOM 4615 CE2 TYR H 112B 22.307 -72.068 -65.611 1.00 85.22 B C
+ATOM 4616 CZ TYR H 112B 23.074 -73.200 -65.882 1.00 85.22 B C
+ATOM 4617 OH TYR H 112B 24.331 -73.349 -65.321 1.00 85.22 B O
+ATOM 4618 N TYR H 112A 18.519 -69.921 -65.791 1.00 61.55 B N
+ATOM 4619 CA TYR H 112A 18.706 -68.483 -65.621 1.00 61.55 B C
+ATOM 4620 C TYR H 112A 20.150 -68.062 -65.881 1.00 61.55 B C
+ATOM 4621 O TYR H 112A 21.095 -68.762 -65.515 1.00 61.55 B O
+ATOM 4622 CB TYR H 112A 18.306 -68.069 -64.205 1.00 61.55 B C
+ATOM 4623 CG TYR H 112A 16.823 -68.080 -63.945 1.00 61.55 B C
+ATOM 4624 CD1 TYR H 112A 15.979 -67.207 -64.598 1.00 61.55 B C
+ATOM 4625 CD2 TYR H 112A 16.269 -68.958 -63.038 1.00 61.55 B C
+ATOM 4626 CE1 TYR H 112A 14.623 -67.203 -64.358 1.00 61.55 B C
+ATOM 4627 CE2 TYR H 112A 14.904 -68.976 -62.789 1.00 61.55 B C
+ATOM 4628 CZ TYR H 112A 14.083 -68.092 -63.454 1.00 61.55 B C
+ATOM 4629 OH TYR H 112A 12.721 -68.094 -63.211 1.00 61.55 B O
+ATOM 4630 N TYR H 112 20.317 -66.916 -66.518 1.00 62.31 B N
+ATOM 4631 CA TYR H 112 21.613 -66.266 -66.589 1.00 62.31 B C
+ATOM 4632 C TYR H 112 21.442 -64.857 -66.049 1.00 62.31 B C
+ATOM 4633 O TYR H 112 20.326 -64.345 -65.990 1.00 62.31 B O
+ATOM 4634 CB TYR H 112 22.171 -66.241 -68.024 1.00 62.31 B C
+ATOM 4635 CG TYR H 112 22.365 -67.627 -68.589 1.00 62.31 B C
+ATOM 4636 CD1 TYR H 112 23.540 -68.331 -68.380 1.00 62.31 B C
+ATOM 4637 CD2 TYR H 112 21.371 -68.234 -69.335 1.00 62.31 B C
+ATOM 4638 CE1 TYR H 112 23.709 -69.619 -68.886 1.00 62.31 B C
+ATOM 4639 CE2 TYR H 112 21.534 -69.518 -69.851 1.00 62.31 B C
+ATOM 4640 CZ TYR H 112 22.702 -70.207 -69.623 1.00 62.31 B C
+ATOM 4641 OH TYR H 112 22.848 -71.482 -70.137 1.00 62.31 B O
+ATOM 4642 N TYR H 113 22.544 -64.227 -65.637 1.00 52.74 B N
+ATOM 4643 CA TYR H 113 22.481 -62.894 -65.036 1.00 52.74 B C
+ATOM 4644 C TYR H 113 23.574 -62.000 -65.618 1.00 52.74 B C
+ATOM 4645 O TYR H 113 24.385 -62.424 -66.447 1.00 52.74 B O
+ATOM 4646 CB TYR H 113 22.604 -62.969 -63.511 1.00 52.74 B C
+ATOM 4647 CG TYR H 113 21.710 -63.995 -62.887 1.00 52.74 B C
+ATOM 4648 CD1 TYR H 113 20.491 -63.650 -62.371 1.00 52.74 B C
+ATOM 4649 CD2 TYR H 113 22.086 -65.316 -62.818 1.00 52.74 B C
+ATOM 4650 CE1 TYR H 113 19.666 -64.608 -61.784 1.00 52.74 B C
+ATOM 4651 CE2 TYR H 113 21.266 -66.278 -62.248 1.00 52.74 B C
+ATOM 4652 CZ TYR H 113 20.057 -65.927 -61.728 1.00 52.74 B C
+ATOM 4653 OH TYR H 113 19.240 -66.890 -61.155 1.00 52.74 B O
+ATOM 4654 N GLY H 114 23.598 -60.747 -65.166 1.00 54.79 B N
+ATOM 4655 CA GLY H 114 24.577 -59.797 -65.648 1.00 54.79 B C
+ATOM 4656 C GLY H 114 24.123 -59.172 -66.951 1.00 54.79 B C
+ATOM 4657 O GLY H 114 22.996 -59.384 -67.416 1.00 54.79 B O
+ATOM 4658 N LEU H 115 25.025 -58.393 -67.558 1.00 46.31 B N
+ATOM 4659 CA LEU H 115 24.774 -57.769 -68.856 1.00 47.65 B C
+ATOM 4660 C LEU H 115 25.683 -58.456 -69.867 1.00 52.78 B C
+ATOM 4661 O LEU H 115 26.890 -58.205 -69.899 1.00 54.63 B O
+ATOM 4662 CB LEU H 115 25.052 -56.279 -68.759 1.00 44.41 B C
+ATOM 4663 CG LEU H 115 24.267 -55.691 -67.600 1.00 44.41 B C
+ATOM 4664 CD1 LEU H 115 24.627 -54.264 -67.395 1.00 44.41 B C
+ATOM 4665 CD2 LEU H 115 22.803 -55.834 -67.881 1.00 44.41 B C
+ATOM 4666 N ASP H 116 25.111 -59.344 -70.672 1.00 54.96 B N
+ATOM 4667 CA ASP H 116 25.940 -60.176 -71.531 1.00 54.96 B C
+ATOM 4668 C ASP H 116 26.238 -59.565 -72.889 1.00 54.96 B C
+ATOM 4669 O ASP H 116 27.245 -59.933 -73.497 1.00 54.96 B O
+ATOM 4670 CB ASP H 116 25.309 -61.549 -71.713 1.00 59.63 B C
+ATOM 4671 CG ASP H 116 23.885 -61.451 -72.097 1.00 59.63 B C
+ATOM 4672 OD1 ASP H 116 23.149 -60.766 -71.364 1.00 59.63 B O
+ATOM 4673 OD2 ASP H 116 23.484 -62.043 -73.115 1.00 59.63 B O1-
+ATOM 4674 N VAL H 117 25.395 -58.682 -73.424 1.00 55.04 B N
+ATOM 4675 CA VAL H 117 25.556 -58.237 -74.813 1.00 55.04 B C
+ATOM 4676 C VAL H 117 25.550 -56.710 -74.910 1.00 55.04 B C
+ATOM 4677 O VAL H 117 24.579 -56.061 -74.488 1.00 55.04 B O
+ATOM 4678 CB VAL H 117 24.475 -58.833 -75.710 1.00 62.31 B C
+ATOM 4679 CG1 VAL H 117 24.483 -58.118 -77.057 1.00 62.31 B C
+ATOM 4680 CG2 VAL H 117 24.727 -60.323 -75.847 1.00 62.31 B C
+ATOM 4681 N TRP H 118 26.621 -56.144 -75.492 1.00 45.98 B N
+ATOM 4682 CA TRP H 118 26.830 -54.701 -75.586 1.00 41.15 B C
+ATOM 4683 C TRP H 118 27.150 -54.318 -77.017 1.00 42.63 B C
+ATOM 4684 O TRP H 118 28.039 -54.913 -77.633 1.00 48.64 B O
+ATOM 4685 CB TRP H 118 28.001 -54.234 -74.717 1.00 51.78 B C
+ATOM 4686 CG TRP H 118 27.863 -54.503 -73.277 1.00 51.78 B C
+ATOM 4687 CD1 TRP H 118 27.728 -55.711 -72.695 1.00 51.78 B C
+ATOM 4688 CD2 TRP H 118 27.877 -53.538 -72.212 1.00 51.78 B C
+ATOM 4689 CE2 TRP H 118 27.724 -54.236 -71.019 1.00 51.78 B C
+ATOM 4690 CE3 TRP H 118 27.999 -52.157 -72.163 1.00 51.78 B C
+ATOM 4691 NE1 TRP H 118 27.627 -55.563 -71.336 1.00 51.78 B N
+ATOM 4692 CZ2 TRP H 118 27.680 -53.609 -69.797 1.00 51.78 B C
+ATOM 4693 CZ3 TRP H 118 27.958 -51.539 -70.951 1.00 51.78 B C
+ATOM 4694 CH2 TRP H 118 27.803 -52.260 -69.786 1.00 51.78 B C
+ATOM 4695 N GLY H 119 26.486 -53.300 -77.530 1.00 57.81 B N
+ATOM 4696 CA GLY H 119 26.886 -52.780 -78.820 1.00 57.81 B C
+ATOM 4697 C GLY H 119 28.246 -52.096 -78.762 1.00 57.81 B C
+ATOM 4698 O GLY H 119 28.824 -51.864 -77.694 1.00 57.81 B O
+ATOM 4699 N ARG H 120 28.780 -51.794 -79.957 1.00 66.11 B N
+ATOM 4700 CA ARG H 120 29.911 -50.875 -80.048 1.00 70.13 B C
+ATOM 4701 C ARG H 120 29.572 -49.534 -79.424 1.00 68.50 B C
+ATOM 4702 O ARG H 120 30.450 -48.852 -78.890 1.00 71.08 B O
+ATOM 4703 CB ARG H 120 30.323 -50.660 -81.505 1.00 77.75 B C
+ATOM 4704 CG ARG H 120 30.706 -51.889 -82.306 1.00 77.75 B C
+ATOM 4705 CD ARG H 120 30.776 -51.550 -83.828 1.00 77.75 B C
+ATOM 4706 NE ARG H 120 32.095 -51.086 -84.318 1.00 77.75 B N
+ATOM 4707 CZ ARG H 120 32.373 -49.860 -84.802 1.00 77.75 B C
+ATOM 4708 NH1 ARG H 120 31.419 -48.925 -84.917 1.00 77.75 B N1+
+ATOM 4709 NH2 ARG H 120 33.617 -49.578 -85.209 1.00 77.75 B N
+ATOM 4710 N GLY H 121 28.320 -49.132 -79.521 1.00 61.49 B N
+ATOM 4711 CA GLY H 121 27.770 -47.914 -78.956 1.00 63.37 B C
+ATOM 4712 C GLY H 121 27.973 -46.728 -79.867 1.00 64.20 B C
+ATOM 4713 O GLY H 121 28.975 -46.615 -80.568 1.00 67.54 B O
+ATOM 4714 N THR H 122 27.007 -45.814 -79.851 1.00 62.63 B N
+ATOM 4715 CA THR H 122 27.089 -44.604 -80.659 1.00 62.63 B C
+ATOM 4716 C THR H 122 27.941 -43.600 -79.894 1.00 63.25 B C
+ATOM 4717 O THR H 122 28.634 -43.976 -78.948 1.00 67.22 B O
+ATOM 4718 CB THR H 122 25.698 -44.000 -80.923 1.00 73.01 B C
+ATOM 4719 CG2 THR H 122 25.099 -43.460 -79.633 1.00 73.01 B C
+ATOM 4720 OG1 THR H 122 25.810 -42.936 -81.875 1.00 73.01 B O
+ATOM 4721 N LEU H 123 27.901 -42.330 -80.284 1.00 60.34 B N
+ATOM 4722 CA LEU H 123 28.696 -41.335 -79.572 1.00 60.34 B C
+ATOM 4723 C LEU H 123 28.029 -39.958 -79.640 1.00 61.59 B C
+ATOM 4724 O LEU H 123 28.052 -39.291 -80.677 1.00 65.66 B O
+ATOM 4725 CB LEU H 123 30.103 -41.382 -80.171 1.00 70.34 B C
+ATOM 4726 CG LEU H 123 30.156 -41.251 -81.720 1.00 70.34 B C
+ATOM 4727 CD1 LEU H 123 31.420 -40.586 -82.194 1.00 70.34 B C
+ATOM 4728 CD2 LEU H 123 29.888 -42.530 -82.582 1.00 70.34 B C
+ATOM 4729 N VAL H 124 27.459 -39.524 -78.516 1.00 58.80 B N
+ATOM 4730 CA VAL H 124 26.602 -38.340 -78.434 1.00 55.96 B C
+ATOM 4731 C VAL H 124 27.359 -37.219 -77.746 1.00 62.40 B C
+ATOM 4732 O VAL H 124 27.766 -37.374 -76.587 1.00 63.92 B O
+ATOM 4733 CB VAL H 124 25.328 -38.636 -77.643 1.00 66.55 B C
+ATOM 4734 CG1 VAL H 124 24.480 -37.392 -77.580 1.00 66.55 B C
+ATOM 4735 CG2 VAL H 124 24.595 -39.780 -78.270 1.00 66.55 B C
+ATOM 4736 N THR H 125 27.516 -36.072 -78.412 1.00 57.12 B N
+ATOM 4737 CA THR H 125 28.274 -34.976 -77.812 1.00 53.18 B C
+ATOM 4738 C THR H 125 27.442 -33.702 -77.829 1.00 54.85 B C
+ATOM 4739 O THR H 125 27.104 -33.189 -78.895 1.00 56.08 B O
+ATOM 4740 CB THR H 125 29.607 -34.738 -78.526 1.00 62.04 B C
+ATOM 4741 CG2 THR H 125 30.197 -36.040 -79.027 1.00 62.04 B C
+ATOM 4742 OG1 THR H 125 29.376 -33.919 -79.662 1.00 62.04 B O
+ATOM 4743 N VAL H 126 27.157 -33.174 -76.651 1.00 60.27 B N
+ATOM 4744 CA VAL H 126 26.325 -31.994 -76.479 1.00 57.07 B C
+ATOM 4745 C VAL H 126 27.246 -30.817 -76.217 1.00 61.62 B C
+ATOM 4746 O VAL H 126 27.703 -30.615 -75.092 1.00 62.07 B O
+ATOM 4747 CB VAL H 126 25.336 -32.194 -75.337 1.00 61.95 B C
+ATOM 4748 CG1 VAL H 126 24.451 -31.003 -75.177 1.00 61.95 B C
+ATOM 4749 CG2 VAL H 126 24.533 -33.424 -75.651 1.00 61.95 B C
+ATOM 4750 N SER H 127 27.425 -29.990 -77.240 1.00 65.99 B N
+ATOM 4751 CA SER H 127 28.364 -28.880 -77.218 1.00 64.73 B C
+ATOM 4752 C SER H 127 27.852 -27.802 -78.160 1.00 69.92 B C
+ATOM 4753 O SER H 127 27.191 -28.090 -79.162 1.00 73.21 B O
+ATOM 4754 CB SER H 127 29.780 -29.330 -77.629 1.00 60.38 B C
+ATOM 4755 OG SER H 127 30.732 -28.268 -77.607 1.00 60.38 B O
+ATOM 4756 N SER H 128 28.152 -26.559 -77.824 1.00 78.66 B N
+ATOM 4757 CA SER H 128 27.788 -25.443 -78.681 1.00 80.49 B C
+ATOM 4758 C SER H 128 28.900 -25.030 -79.639 1.00 79.87 B C
+ATOM 4759 O SER H 128 28.665 -24.215 -80.536 1.00 83.23 B O
+ATOM 4760 CB SER H 128 27.351 -24.264 -77.806 1.00 53.40 B C
+ATOM 4761 OG SER H 128 28.305 -24.031 -76.775 1.00 53.40 B O
+ATOM 4762 N ALA H 129 30.091 -25.579 -79.493 1.00 63.84 B N
+ATOM 4763 CA ALA H 129 31.135 -25.257 -80.443 1.00 65.44 B C
+ATOM 4764 C ALA H 129 30.770 -25.776 -81.827 1.00 69.35 B C
+ATOM 4765 O ALA H 129 30.000 -26.726 -81.980 1.00 71.52 B O
+ATOM 4766 CB ALA H 129 32.464 -25.863 -80.002 1.00 62.31 B C
+ATOM 4767 N SER H 130 31.351 -25.153 -82.842 1.00 56.39 B N
+ATOM 4768 CA SER H 130 31.091 -25.544 -84.218 1.00 62.50 B C
+ATOM 4769 C SER H 130 32.235 -26.388 -84.752 1.00 63.20 B C
+ATOM 4770 O SER H 130 33.391 -26.177 -84.395 1.00 66.74 B O
+ATOM 4771 CB SER H 130 30.889 -24.317 -85.108 1.00 65.60 B C
+ATOM 4772 OG SER H 130 29.853 -23.485 -84.602 1.00 70.32 B O
+ATOM 4773 N THR H 131 31.902 -27.340 -85.619 1.00 65.02 B N
+ATOM 4774 CA THR H 131 32.921 -28.179 -86.242 1.00 60.59 B C
+ATOM 4775 C THR H 131 34.025 -27.304 -86.808 1.00 60.41 B C
+ATOM 4776 O THR H 131 33.748 -26.293 -87.449 1.00 63.37 B O
+ATOM 4777 CB THR H 131 32.302 -29.024 -87.365 1.00 58.81 B C
+ATOM 4778 CG2 THR H 131 33.375 -29.828 -88.133 1.00 54.78 B C
+ATOM 4779 OG1 THR H 131 31.308 -29.909 -86.821 1.00 59.71 B O
+ATOM 4780 N LYS H 132 35.276 -27.660 -86.532 1.00 64.47 B N
+ATOM 4781 CA LYS H 132 36.396 -26.853 -86.996 1.00 62.71 B C
+ATOM 4782 C LYS H 132 37.571 -27.732 -87.384 1.00 58.67 B C
+ATOM 4783 O LYS H 132 37.960 -28.623 -86.625 1.00 57.26 B O
+ATOM 4784 CB LYS H 132 36.848 -25.860 -85.924 1.00 64.12 B C
+ATOM 4785 CG LYS H 132 37.876 -24.886 -86.444 1.00 66.54 B C
+ATOM 4786 CD LYS H 132 38.216 -23.820 -85.427 1.00 70.23 B C
+ATOM 4787 CE LYS H 132 39.350 -22.922 -85.934 1.00 71.47 B C
+ATOM 4788 NZ LYS H 132 40.608 -23.666 -86.270 1.00 73.18 B N1+
+ATOM 4789 N GLY H 133 38.166 -27.457 -88.538 1.00 65.97 B N
+ATOM 4790 CA GLY H 133 39.433 -28.063 -88.864 1.00 64.64 B C
+ATOM 4791 C GLY H 133 40.493 -27.691 -87.840 1.00 65.66 B C
+ATOM 4792 O GLY H 133 40.424 -26.640 -87.194 1.00 66.29 B O
+ATOM 4793 N PRO H 134 41.479 -28.565 -87.650 1.00 59.03 B N
+ATOM 4794 CA PRO H 134 42.598 -28.244 -86.757 1.00 57.88 B C
+ATOM 4795 C PRO H 134 43.594 -27.291 -87.391 1.00 62.36 B C
+ATOM 4796 O PRO H 134 43.909 -27.401 -88.577 1.00 65.21 B O
+ATOM 4797 CB PRO H 134 43.259 -29.604 -86.536 1.00 56.67 B C
+ATOM 4798 CG PRO H 134 42.971 -30.345 -87.767 1.00 59.97 B C
+ATOM 4799 CD PRO H 134 41.619 -29.897 -88.250 1.00 61.39 B C
+ATOM 4800 N SER H 135 44.123 -26.376 -86.580 1.00 61.83 B N
+ATOM 4801 CA SER H 135 45.355 -25.669 -86.921 1.00 62.14 B C
+ATOM 4802 C SER H 135 46.522 -26.512 -86.421 1.00 64.53 B C
+ATOM 4803 O SER H 135 46.516 -26.944 -85.271 1.00 69.21 B O
+ATOM 4804 CB SER H 135 45.398 -24.270 -86.292 1.00 60.14 B C
+ATOM 4805 OG SER H 135 44.348 -23.429 -86.750 1.00 62.09 B O
+ATOM 4806 N VAL H 136 47.512 -26.755 -87.278 1.00 65.66 B N
+ATOM 4807 CA VAL H 136 48.575 -27.719 -86.990 1.00 64.24 B C
+ATOM 4808 C VAL H 136 49.919 -27.007 -87.027 1.00 70.64 B C
+ATOM 4809 O VAL H 136 50.385 -26.606 -88.101 1.00 77.68 B O
+ATOM 4810 CB VAL H 136 48.559 -28.888 -87.978 1.00 60.36 B C
+ATOM 4811 CG1 VAL H 136 49.853 -29.680 -87.883 1.00 58.24 B C
+ATOM 4812 CG2 VAL H 136 47.333 -29.753 -87.739 1.00 58.66 B C
+ATOM 4813 N PHE H 137 50.575 -26.905 -85.874 1.00 80.01 B N
+ATOM 4814 CA PHE H 137 51.816 -26.158 -85.791 1.00 75.10 B C
+ATOM 4815 C PHE H 137 52.960 -27.092 -85.450 1.00 76.23 B C
+ATOM 4816 O PHE H 137 52.760 -28.050 -84.705 1.00 76.45 B O
+ATOM 4817 CB PHE H 137 51.718 -25.096 -84.709 1.00 70.79 B C
+ATOM 4818 CG PHE H 137 50.477 -24.330 -84.776 1.00 70.97 B C
+ATOM 4819 CD1 PHE H 137 50.272 -23.426 -85.790 1.00 71.56 B C
+ATOM 4820 CD2 PHE H 137 49.496 -24.512 -83.829 1.00 71.78 B C
+ATOM 4821 CE1 PHE H 137 49.097 -22.708 -85.868 1.00 71.82 B C
+ATOM 4822 CE2 PHE H 137 48.313 -23.801 -83.890 1.00 72.15 B C
+ATOM 4823 CZ PHE H 137 48.113 -22.891 -84.909 1.00 71.90 B C
+ATOM 4824 N PRO H 138 54.166 -26.847 -85.965 1.00 81.08 B N
+ATOM 4825 CA PRO H 138 55.266 -27.803 -85.757 1.00 81.77 B C
+ATOM 4826 C PRO H 138 55.916 -27.685 -84.380 1.00 79.81 B C
+ATOM 4827 O PRO H 138 56.091 -26.589 -83.840 1.00 80.25 B O
+ATOM 4828 CB PRO H 138 56.259 -27.438 -86.865 1.00 82.19 B C
+ATOM 4829 CG PRO H 138 56.034 -25.981 -87.066 1.00 82.69 B C
+ATOM 4830 CD PRO H 138 54.574 -25.703 -86.792 1.00 81.71 B C
+ATOM 4831 N LEU H 139 56.306 -28.845 -83.827 1.00 78.61 B N
+ATOM 4832 CA LEU H 139 57.067 -28.947 -82.581 1.00 75.70 B C
+ATOM 4833 C LEU H 139 58.511 -29.310 -82.934 1.00 81.07 B C
+ATOM 4834 O LEU H 139 58.801 -30.448 -83.332 1.00 85.99 B O
+ATOM 4835 CB LEU H 139 56.435 -29.970 -81.646 1.00 76.00 B C
+ATOM 4836 CG LEU H 139 55.067 -29.491 -81.175 1.00 75.16 B C
+ATOM 4837 CD1 LEU H 139 54.447 -30.518 -80.266 1.00 67.93 B C
+ATOM 4838 CD2 LEU H 139 55.164 -28.135 -80.488 1.00 71.46 B C
+ATOM 4839 N ALA H 140 59.423 -28.329 -82.756 1.00 81.43 B N
+ATOM 4840 CA ALA H 140 60.730 -28.221 -83.392 1.00 81.43 B C
+ATOM 4841 C ALA H 140 61.851 -28.710 -82.487 1.00 81.43 B C
+ATOM 4842 O ALA H 140 61.898 -28.320 -81.306 1.00 81.43 B O
+ATOM 4843 CB ALA H 140 60.977 -26.770 -83.782 1.00 81.43 B C
+ATOM 4844 N PRO H 141 62.772 -29.555 -83.006 1.00 72.68 B N
+ATOM 4845 CA PRO H 141 64.018 -29.990 -82.331 1.00 76.72 B C
+ATOM 4846 C PRO H 141 64.984 -28.846 -81.865 1.00 82.91 B C
+ATOM 4847 O PRO H 141 65.647 -28.860 -80.795 1.00 82.58 B O
+ATOM 4848 CB PRO H 141 64.696 -30.888 -83.397 1.00 75.85 B C
+ATOM 4849 CG PRO H 141 63.872 -30.775 -84.650 1.00 74.63 B C
+ATOM 4850 CD PRO H 141 62.518 -30.310 -84.248 1.00 71.60 B C
+ATOM 4851 N GLY H 149 70.926 -37.392 -76.822 1.00109.78 B N
+ATOM 4852 CA GLY H 149 71.668 -38.016 -77.907 1.00103.08 B C
+ATOM 4853 C GLY H 149 70.772 -38.397 -79.069 1.00 92.70 B C
+ATOM 4854 O GLY H 149 71.222 -38.597 -80.202 1.00 92.48 B O
+ATOM 4855 N THR H 150 69.483 -38.513 -78.767 1.00119.37 B N
+ATOM 4856 CA THR H 150 68.435 -38.645 -79.766 1.00106.19 B C
+ATOM 4857 C THR H 150 67.416 -37.527 -79.561 1.00 92.35 B C
+ATOM 4858 O THR H 150 67.213 -37.046 -78.436 1.00 91.24 B O
+ATOM 4859 CB THR H 150 67.764 -40.009 -79.682 1.00105.69 B C
+ATOM 4860 CG2 THR H 150 68.741 -41.103 -80.125 1.00106.85 B C
+ATOM 4861 OG1 THR H 150 67.343 -40.242 -78.332 1.00101.91 B O
+ATOM 4862 N ALA H 151 66.785 -37.101 -80.659 1.00102.35 B N
+ATOM 4863 CA ALA H 151 66.041 -35.847 -80.692 1.00 92.41 B C
+ATOM 4864 C ALA H 151 64.541 -36.084 -80.819 1.00 90.57 B C
+ATOM 4865 O ALA H 151 64.091 -36.946 -81.587 1.00 90.88 B O
+ATOM 4866 CB ALA H 151 66.529 -34.959 -81.840 1.00 82.20 B C
+ATOM 4867 N ALA H 152 63.773 -35.297 -80.066 1.00 83.29 B N
+ATOM 4868 CA ALA H 152 62.325 -35.415 -80.024 1.00 78.15 B C
+ATOM 4869 C ALA H 152 61.691 -34.224 -80.725 1.00 78.87 B C
+ATOM 4870 O ALA H 152 62.039 -33.065 -80.444 1.00 78.14 B O
+ATOM 4871 CB ALA H 152 61.820 -35.510 -78.586 1.00 79.12 B C
+ATOM 4872 N LEU H 153 60.766 -34.528 -81.641 1.00 75.92 B N
+ATOM 4873 CA LEU H 153 60.034 -33.541 -82.426 1.00 76.60 B C
+ATOM 4874 C LEU H 153 58.618 -34.050 -82.626 1.00 78.77 B C
+ATOM 4875 O LEU H 153 58.318 -35.215 -82.379 1.00 79.77 B O
+ATOM 4876 CB LEU H 153 60.703 -33.266 -83.779 1.00 77.15 B C
+ATOM 4877 CG LEU H 153 60.910 -34.451 -84.730 1.00 81.93 B C
+ATOM 4878 CD1 LEU H 153 59.654 -34.863 -85.504 1.00 83.31 B C
+ATOM 4879 CD2 LEU H 153 62.080 -34.148 -85.670 1.00 82.68 B C
+ATOM 4880 N GLY H 154 57.749 -33.172 -83.092 1.00 78.87 B N
+ATOM 4881 CA GLY H 154 56.355 -33.531 -83.198 1.00 78.35 B C
+ATOM 4882 C GLY H 154 55.582 -32.438 -83.880 1.00 80.24 B C
+ATOM 4883 O GLY H 154 56.151 -31.615 -84.595 1.00 82.86 B O
+ATOM 4884 N CYS H 155 54.269 -32.466 -83.687 1.00 79.99 B N
+ATOM 4885 CA CYS H 155 53.404 -31.403 -84.168 1.00 80.51 B C
+ATOM 4886 C CYS H 155 52.177 -31.355 -83.276 1.00 79.30 B C
+ATOM 4887 O CYS H 155 51.779 -32.354 -82.670 1.00 79.36 B O
+ATOM 4888 CB CYS H 155 53.026 -31.556 -85.661 1.00 70.11 B C
+ATOM 4889 SG CYS H 155 52.387 -33.195 -86.136 1.00 77.29 B S
+ATOM 4890 N LEU H 156 51.616 -30.160 -83.186 1.00 68.08 B N
+ATOM 4891 CA LEU H 156 50.587 -29.784 -82.232 1.00 65.67 B C
+ATOM 4892 C LEU H 156 49.330 -29.573 -83.041 1.00 67.35 B C
+ATOM 4893 O LEU H 156 49.283 -28.666 -83.873 1.00 68.98 B O
+ATOM 4894 CB LEU H 156 50.976 -28.494 -81.515 1.00 63.54 B C
+ATOM 4895 CG LEU H 156 50.266 -27.992 -80.278 1.00 61.13 B C
+ATOM 4896 CD1 LEU H 156 50.914 -26.685 -79.879 1.00 61.81 B C
+ATOM 4897 CD2 LEU H 156 48.827 -27.771 -80.535 1.00 62.89 B C
+ATOM 4898 N VAL H 157 48.309 -30.356 -82.769 1.00 58.58 B N
+ATOM 4899 CA VAL H 157 47.038 -30.255 -83.467 1.00 57.54 B C
+ATOM 4900 C VAL H 157 46.092 -29.477 -82.558 1.00 58.89 B C
+ATOM 4901 O VAL H 157 45.584 -30.015 -81.566 1.00 58.60 B O
+ATOM 4902 CB VAL H 157 46.498 -31.646 -83.799 1.00 59.57 B C
+ATOM 4903 CG1 VAL H 157 45.224 -31.552 -84.585 1.00 61.64 B C
+ATOM 4904 CG2 VAL H 157 47.561 -32.435 -84.553 1.00 57.02 B C
+ATOM 4905 N LYS H 158 45.823 -28.211 -82.890 1.00 59.02 B N
+ATOM 4906 CA LYS H 158 45.200 -27.283 -81.956 1.00 58.21 B C
+ATOM 4907 C LYS H 158 43.885 -26.767 -82.502 1.00 58.35 B C
+ATOM 4908 O LYS H 158 43.721 -26.601 -83.718 1.00 58.93 B O
+ATOM 4909 CB LYS H 158 46.099 -26.093 -81.657 1.00 63.15 B C
+ATOM 4910 CG LYS H 158 45.687 -25.335 -80.425 1.00 67.45 B C
+ATOM 4911 CD LYS H 158 46.612 -24.165 -80.226 1.00 67.02 B C
+ATOM 4912 CE LYS H 158 46.368 -23.461 -78.895 1.00 68.15 B C
+ATOM 4913 NZ LYS H 158 44.943 -23.149 -78.610 1.00 69.26 B N1+
+ATOM 4914 N ASP H 159 42.956 -26.514 -81.579 1.00 65.08 B N
+ATOM 4915 CA ASP H 159 41.676 -25.881 -81.893 1.00 64.52 B C
+ATOM 4916 C ASP H 159 40.993 -26.589 -83.065 1.00 68.57 B C
+ATOM 4917 O ASP H 159 40.992 -26.126 -84.209 1.00 70.44 B O
+ATOM 4918 CB ASP H 159 41.866 -24.386 -82.162 1.00 63.95 B C
+ATOM 4919 CG ASP H 159 42.289 -23.629 -80.918 1.00 69.25 B C
+ATOM 4920 OD1 ASP H 159 41.792 -23.959 -79.809 1.00 66.50 B O
+ATOM 4921 OD2 ASP H 159 43.127 -22.713 -81.054 1.00 69.97 B O1-
+ATOM 4922 N TYR H 160 40.482 -27.779 -82.742 1.00 65.85 B N
+ATOM 4923 CA TYR H 160 39.686 -28.586 -83.653 1.00 66.07 B C
+ATOM 4924 C TYR H 160 38.513 -29.160 -82.870 1.00 64.57 B C
+ATOM 4925 O TYR H 160 38.482 -29.140 -81.633 1.00 63.17 B O
+ATOM 4926 CB TYR H 160 40.515 -29.708 -84.295 1.00 69.59 B C
+ATOM 4927 CG TYR H 160 40.766 -30.860 -83.355 1.00 68.33 B C
+ATOM 4928 CD1 TYR H 160 39.959 -31.988 -83.392 1.00 67.67 B C
+ATOM 4929 CD2 TYR H 160 41.769 -30.796 -82.389 1.00 64.37 B C
+ATOM 4930 CE1 TYR H 160 40.160 -33.032 -82.522 1.00 64.79 B C
+ATOM 4931 CE2 TYR H 160 41.974 -31.838 -81.510 1.00 66.70 B C
+ATOM 4932 CZ TYR H 160 41.159 -32.953 -81.580 1.00 66.86 B C
+ATOM 4933 OH TYR H 160 41.335 -34.012 -80.722 1.00 69.17 B O
+ATOM 4934 N PHE H 161 37.557 -29.701 -83.609 1.00 59.92 B N
+ATOM 4935 CA PHE H 161 36.274 -30.094 -83.050 1.00 60.09 B C
+ATOM 4936 C PHE H 161 35.426 -30.732 -84.143 1.00 62.29 B C
+ATOM 4937 O PHE H 161 35.524 -30.352 -85.296 1.00 65.88 B O
+ATOM 4938 CB PHE H 161 35.556 -28.881 -82.469 1.00 61.46 B C
+ATOM 4939 CG PHE H 161 34.299 -29.217 -81.759 1.00 64.04 B C
+ATOM 4940 CD1 PHE H 161 34.296 -29.396 -80.391 1.00 61.37 B C
+ATOM 4941 CD2 PHE H 161 33.118 -29.368 -82.453 1.00 65.56 B C
+ATOM 4942 CE1 PHE H 161 33.122 -29.706 -79.725 1.00 61.02 B C
+ATOM 4943 CE2 PHE H 161 31.962 -29.685 -81.802 1.00 65.29 B C
+ATOM 4944 CZ PHE H 161 31.959 -29.854 -80.440 1.00 63.11 B C
+ATOM 4945 N PRO H 162 34.626 -31.739 -83.796 1.00 63.84 B N
+ATOM 4946 CA PRO H 162 34.733 -32.431 -82.520 1.00 62.16 B C
+ATOM 4947 C PRO H 162 35.706 -33.594 -82.718 1.00 62.88 B C
+ATOM 4948 O PRO H 162 36.155 -33.797 -83.869 1.00 64.91 B O
+ATOM 4949 CB PRO H 162 33.307 -32.889 -82.268 1.00 64.42 B C
+ATOM 4950 CG PRO H 162 32.807 -33.195 -83.667 1.00 59.66 B C
+ATOM 4951 CD PRO H 162 33.532 -32.281 -84.618 1.00 64.20 B C
+ATOM 4952 N GLU H 163 36.028 -34.339 -81.660 1.00 60.86 B N
+ATOM 4953 CA GLU H 163 36.986 -35.419 -81.820 1.00 61.42 B C
+ATOM 4954 C GLU H 163 36.313 -36.444 -82.682 1.00 64.14 B C
+ATOM 4955 O GLU H 163 35.105 -36.458 -82.754 1.00 65.11 B O
+ATOM 4956 CB GLU H 163 37.416 -35.978 -80.474 1.00 58.64 B C
+ATOM 4957 CG GLU H 163 38.915 -35.861 -80.283 1.00 62.88 B C
+ATOM 4958 CD GLU H 163 39.422 -36.562 -79.045 1.00 64.52 B C
+ATOM 4959 OE1 GLU H 163 38.686 -36.545 -78.021 1.00 64.85 B O
+ATOM 4960 OE2 GLU H 163 40.557 -37.119 -79.100 1.00 66.16 B O1-
+ATOM 4961 N PRO H 164 37.064 -37.337 -83.319 1.00 64.90 B N
+ATOM 4962 CA PRO H 164 38.476 -37.706 -83.323 1.00 63.13 B C
+ATOM 4963 C PRO H 164 39.354 -37.069 -84.367 1.00 65.37 B C
+ATOM 4964 O PRO H 164 38.890 -36.523 -85.359 1.00 64.88 B O
+ATOM 4965 CB PRO H 164 38.413 -39.175 -83.667 1.00 57.21 B C
+ATOM 4966 CG PRO H 164 37.330 -39.185 -84.710 1.00 58.63 B C
+ATOM 4967 CD PRO H 164 36.305 -38.181 -84.248 1.00 60.52 B C
+ATOM 4968 N VAL H 165 40.644 -37.343 -84.222 1.00 62.38 B N
+ATOM 4969 CA VAL H 165 41.653 -36.936 -85.184 1.00 62.54 B C
+ATOM 4970 C VAL H 165 42.726 -38.007 -85.164 1.00 63.05 B C
+ATOM 4971 O VAL H 165 42.961 -38.647 -84.138 1.00 59.05 B O
+ATOM 4972 CB VAL H 165 42.196 -35.524 -84.848 1.00 62.36 B C
+ATOM 4973 CG1 VAL H 165 42.506 -35.429 -83.386 1.00 62.69 B C
+ATOM 4974 CG2 VAL H 165 43.457 -35.228 -85.643 1.00 64.27 B C
+ATOM 4975 N THR H 166 43.334 -38.252 -86.318 1.00 62.46 B N
+ATOM 4976 CA THR H 166 44.372 -39.269 -86.423 1.00 61.34 B C
+ATOM 4977 C THR H 166 45.628 -38.623 -86.983 1.00 61.54 B C
+ATOM 4978 O THR H 166 45.566 -37.933 -88.004 1.00 65.42 B O
+ATOM 4979 CB THR H 166 43.932 -40.449 -87.314 1.00 62.62 B C
+ATOM 4980 CG2 THR H 166 42.629 -41.055 -86.803 1.00 60.91 B C
+ATOM 4981 OG1 THR H 166 43.748 -40.013 -88.671 1.00 67.16 B O
+ATOM 4982 N VAL H 167 46.759 -38.824 -86.311 1.00 59.69 B N
+ATOM 4983 CA VAL H 167 48.059 -38.417 -86.827 1.00 61.82 B C
+ATOM 4984 C VAL H 167 48.822 -39.680 -87.171 1.00 66.72 B C
+ATOM 4985 O VAL H 167 48.865 -40.621 -86.365 1.00 70.63 B O
+ATOM 4986 CB VAL H 167 48.853 -37.561 -85.826 1.00 61.32 B C
+ATOM 4987 CG1 VAL H 167 50.207 -37.247 -86.393 1.00 59.80 B C
+ATOM 4988 CG2 VAL H 167 48.122 -36.269 -85.513 1.00 57.19 B C
+ATOM 4989 N SER H 168 49.370 -39.721 -88.385 1.00 74.26 B N
+ATOM 4990 CA SER H 168 50.345 -40.718 -88.800 1.00 74.98 B C
+ATOM 4991 C SER H 168 51.484 -39.978 -89.467 1.00 76.59 B C
+ATOM 4992 O SER H 168 51.261 -38.958 -90.130 1.00 75.21 B O
+ATOM 4993 CB SER H 168 49.754 -41.760 -89.752 1.00 76.10 B C
+ATOM 4994 OG SER H 168 49.006 -42.736 -89.033 1.00 79.37 B O
+ATOM 4995 N TRP H 169 52.701 -40.476 -89.270 1.00 78.76 B N
+ATOM 4996 CA TRP H 169 53.906 -39.725 -89.599 1.00 78.81 B C
+ATOM 4997 C TRP H 169 54.538 -40.216 -90.894 1.00 84.92 B C
+ATOM 4998 O TRP H 169 54.526 -41.416 -91.189 1.00 88.37 B O
+ATOM 4999 CB TRP H 169 54.920 -39.811 -88.462 1.00 73.06 B C
+ATOM 5000 CG TRP H 169 54.620 -38.853 -87.387 1.00 73.73 B C
+ATOM 5001 CD1 TRP H 169 53.817 -39.061 -86.315 1.00 73.84 B C
+ATOM 5002 CD2 TRP H 169 55.105 -37.518 -87.274 1.00 74.31 B C
+ATOM 5003 CE2 TRP H 169 54.555 -36.973 -86.101 1.00 73.21 B C
+ATOM 5004 CE3 TRP H 169 55.955 -36.732 -88.045 1.00 73.33 B C
+ATOM 5005 NE1 TRP H 169 53.768 -37.938 -85.534 1.00 72.65 B N
+ATOM 5006 CZ2 TRP H 169 54.832 -35.677 -85.675 1.00 71.32 B C
+ATOM 5007 CZ3 TRP H 169 56.231 -35.441 -87.618 1.00 73.20 B C
+ATOM 5008 CH2 TRP H 169 55.670 -34.928 -86.443 1.00 72.93 B C
+ATOM 5009 N ASN H 170 55.109 -39.273 -91.653 1.00 85.74 B N
+ATOM 5010 CA ASN H 170 55.732 -39.573 -92.937 1.00 88.17 B C
+ATOM 5011 C ASN H 170 54.812 -40.459 -93.777 1.00 88.64 B C
+ATOM 5012 O ASN H 170 55.218 -41.483 -94.331 1.00 92.14 B O
+ATOM 5013 CB ASN H 170 57.110 -40.212 -92.742 1.00 87.29 B C
+ATOM 5014 CG ASN H 170 58.204 -39.191 -92.449 1.00 86.17 B C
+ATOM 5015 ND2 ASN H 170 59.460 -39.596 -92.654 1.00 81.81 B N
+ATOM 5016 OD1 ASN H 170 57.929 -38.064 -92.025 1.00 89.68 B O
+ATOM 5017 N SER H 171 53.534 -40.062 -93.833 1.00 95.11 B N
+ATOM 5018 CA SER H 171 52.515 -40.743 -94.642 1.00 94.67 B C
+ATOM 5019 C SER H 171 52.434 -42.224 -94.293 1.00 95.22 B C
+ATOM 5020 O SER H 171 52.147 -43.064 -95.148 1.00 99.89 B O
+ATOM 5021 CB SER H 171 52.772 -40.547 -96.143 1.00 95.01 B C
+ATOM 5022 OG SER H 171 52.884 -39.166 -96.487 1.00 97.47 B O
+ATOM 5023 N GLY H 172 52.693 -42.545 -93.030 1.00 86.75 B N
+ATOM 5024 CA GLY H 172 52.718 -43.913 -92.566 1.00 86.87 B C
+ATOM 5025 C GLY H 172 54.087 -44.553 -92.479 1.00 91.01 B C
+ATOM 5026 O GLY H 172 54.178 -45.721 -92.069 1.00 95.01 B O
+ATOM 5027 N ALA H 173 55.158 -43.826 -92.816 1.00 91.27 B N
+ATOM 5028 CA ALA H 173 56.492 -44.427 -92.807 1.00 93.52 B C
+ATOM 5029 C ALA H 173 56.957 -44.676 -91.382 1.00 95.32 B C
+ATOM 5030 O ALA H 173 57.136 -45.829 -90.965 1.00 98.48 B O
+ATOM 5031 CB ALA H 173 57.502 -43.539 -93.542 1.00 95.26 B C
+ATOM 5032 N LEU H 174 57.086 -43.602 -90.601 1.00 86.83 B N
+ATOM 5033 CA LEU H 174 57.727 -43.638 -89.290 1.00 84.86 B C
+ATOM 5034 C LEU H 174 56.663 -44.064 -88.292 1.00 80.68 B C
+ATOM 5035 O LEU H 174 55.790 -43.278 -87.921 1.00 81.02 B O
+ATOM 5036 CB LEU H 174 58.293 -42.254 -88.988 1.00 85.62 B C
+ATOM 5037 CG LEU H 174 58.601 -41.717 -87.600 1.00 85.33 B C
+ATOM 5038 CD1 LEU H 174 59.643 -42.546 -86.866 1.00 86.68 B C
+ATOM 5039 CD2 LEU H 174 59.070 -40.294 -87.769 1.00 81.02 B C
+ATOM 5040 N THR H 175 56.721 -45.331 -87.879 1.00 82.11 B N
+ATOM 5041 CA THR H 175 55.716 -45.900 -86.994 1.00 82.56 B C
+ATOM 5042 C THR H 175 56.179 -46.061 -85.550 1.00 85.35 B C
+ATOM 5043 O THR H 175 55.353 -46.375 -84.685 1.00 89.01 B O
+ATOM 5044 CB THR H 175 55.261 -47.256 -87.542 1.00 84.23 B C
+ATOM 5045 CG2 THR H 175 54.658 -47.069 -88.919 1.00 87.11 B C
+ATOM 5046 OG1 THR H 175 56.388 -48.131 -87.652 1.00 84.69 B O
+ATOM 5047 N SER H 176 57.451 -45.812 -85.258 1.00 89.05 B N
+ATOM 5048 CA SER H 176 58.060 -46.227 -84.003 1.00 91.47 B C
+ATOM 5049 C SER H 176 58.356 -45.033 -83.102 1.00 91.48 B C
+ATOM 5050 O SER H 176 58.766 -43.961 -83.568 1.00 92.84 B O
+ATOM 5051 CB SER H 176 59.359 -46.979 -84.261 1.00 94.99 B C
+ATOM 5052 OG SER H 176 60.411 -46.035 -84.393 1.00 99.63 B O
+ATOM 5053 N GLY H 177 58.196 -45.252 -81.800 1.00 88.93 B N
+ATOM 5054 CA GLY H 177 58.414 -44.179 -80.846 1.00 84.09 B C
+ATOM 5055 C GLY H 177 57.556 -42.982 -81.166 1.00 83.02 B C
+ATOM 5056 O GLY H 177 58.038 -41.845 -81.137 1.00 83.11 B O
+ATOM 5057 N VAL H 178 56.299 -43.224 -81.516 1.00 76.19 B N
+ATOM 5058 CA VAL H 178 55.354 -42.183 -81.877 1.00 73.31 B C
+ATOM 5059 C VAL H 178 54.277 -42.168 -80.805 1.00 74.36 B C
+ATOM 5060 O VAL H 178 53.435 -43.070 -80.741 1.00 78.67 B O
+ATOM 5061 CB VAL H 178 54.746 -42.438 -83.258 1.00 71.87 B C
+ATOM 5062 CG1 VAL H 178 53.970 -41.224 -83.719 1.00 70.10 B C
+ATOM 5063 CG2 VAL H 178 55.818 -42.833 -84.247 1.00 72.22 B C
+ATOM 5064 N HIS H 179 54.279 -41.150 -79.971 1.00 66.99 B N
+ATOM 5065 CA HIS H 179 53.277 -41.004 -78.926 1.00 66.66 B C
+ATOM 5066 C HIS H 179 52.365 -39.850 -79.323 1.00 67.84 B C
+ATOM 5067 O HIS H 179 52.811 -38.698 -79.362 1.00 68.98 B O
+ATOM 5068 CB HIS H 179 53.917 -40.705 -77.571 1.00 70.79 B C
+ATOM 5069 CG HIS H 179 54.938 -41.703 -77.116 1.00 74.55 B C
+ATOM 5070 CD2 HIS H 179 56.271 -41.775 -77.341 1.00 75.97 B C
+ATOM 5071 ND1 HIS H 179 54.636 -42.737 -76.255 1.00 77.18 B N
+ATOM 5072 CE1 HIS H 179 55.735 -43.420 -75.994 1.00 75.46 B C
+ATOM 5073 NE2 HIS H 179 56.741 -42.856 -76.638 1.00 73.22 B N
+ATOM 5074 N THR H 180 51.092 -40.136 -79.583 1.00 64.14 B N
+ATOM 5075 CA THR H 180 50.123 -39.088 -79.882 1.00 62.06 B C
+ATOM 5076 C THR H 180 49.089 -39.094 -78.770 1.00 58.18 B C
+ATOM 5077 O THR H 180 48.446 -40.112 -78.514 1.00 60.44 B O
+ATOM 5078 CB THR H 180 49.458 -39.320 -81.229 1.00 61.12 B C
+ATOM 5079 CG2 THR H 180 50.497 -39.722 -82.275 1.00 60.12 B C
+ATOM 5080 OG1 THR H 180 48.579 -40.433 -81.092 1.00 62.34 B O
+ATOM 5081 N PHE H 181 48.928 -37.978 -78.131 1.00 59.10 B N
+ATOM 5082 CA PHE H 181 48.331 -37.943 -76.811 1.00 58.77 B C
+ATOM 5083 C PHE H 181 46.814 -37.927 -76.859 1.00 57.50 B C
+ATOM 5084 O PHE H 181 46.217 -37.732 -77.916 1.00 59.96 B O
+ATOM 5085 CB PHE H 181 48.900 -36.743 -76.074 1.00 53.42 B C
+ATOM 5086 CG PHE H 181 50.313 -36.976 -75.661 1.00 56.14 B C
+ATOM 5087 CD1 PHE H 181 51.331 -36.926 -76.594 1.00 56.99 B C
+ATOM 5088 CD2 PHE H 181 50.635 -37.216 -74.348 1.00 55.63 B C
+ATOM 5089 CE1 PHE H 181 52.628 -37.177 -76.240 1.00 57.36 B C
+ATOM 5090 CE2 PHE H 181 51.948 -37.437 -73.990 1.00 57.37 B C
+ATOM 5091 CZ PHE H 181 52.941 -37.416 -74.945 1.00 57.84 B C
+ATOM 5092 N PRO H 182 46.166 -38.231 -75.740 1.00 51.82 B N
+ATOM 5093 CA PRO H 182 44.748 -37.916 -75.616 1.00 52.31 B C
+ATOM 5094 C PRO H 182 44.529 -36.451 -75.892 1.00 56.63 B C
+ATOM 5095 O PRO H 182 45.297 -35.594 -75.442 1.00 63.29 B O
+ATOM 5096 CB PRO H 182 44.434 -38.259 -74.163 1.00 53.63 B C
+ATOM 5097 CG PRO H 182 45.312 -39.388 -73.896 1.00 56.47 B C
+ATOM 5098 CD PRO H 182 46.593 -39.164 -74.693 1.00 54.79 B C
+ATOM 5099 N ALA H 183 43.494 -36.172 -76.673 1.00 57.26 B N
+ATOM 5100 CA ALA H 183 43.046 -34.799 -76.776 1.00 53.28 B C
+ATOM 5101 C ALA H 183 42.704 -34.286 -75.391 1.00 57.82 B C
+ATOM 5102 O ALA H 183 42.301 -35.035 -74.503 1.00 61.59 B O
+ATOM 5103 CB ALA H 183 41.833 -34.687 -77.691 1.00 56.89 B C
+ATOM 5104 N VAL H 184 42.912 -33.017 -75.195 1.00 59.11 B N
+ATOM 5105 CA VAL H 184 42.407 -32.373 -74.006 1.00 57.60 B C
+ATOM 5106 C VAL H 184 41.394 -31.341 -74.468 1.00 54.06 B C
+ATOM 5107 O VAL H 184 41.431 -30.899 -75.619 1.00 54.82 B O
+ATOM 5108 CB VAL H 184 43.554 -31.761 -73.189 1.00 59.76 B C
+ATOM 5109 CG1 VAL H 184 44.240 -30.669 -73.966 1.00 65.00 B C
+ATOM 5110 CG2 VAL H 184 43.016 -31.231 -71.910 1.00 53.00 B C
+ATOM 5111 N LEU H 185 40.447 -31.001 -73.606 1.00 58.17 B N
+ATOM 5112 CA LEU H 185 39.352 -30.119 -73.994 1.00 60.36 B C
+ATOM 5113 C LEU H 185 39.638 -28.730 -73.459 1.00 64.65 B C
+ATOM 5114 O LEU H 185 39.588 -28.488 -72.248 1.00 65.63 B O
+ATOM 5115 CB LEU H 185 38.004 -30.635 -73.499 1.00 59.83 B C
+ATOM 5116 CG LEU H 185 36.776 -29.730 -73.632 1.00 61.51 B C
+ATOM 5117 CD1 LEU H 185 36.769 -28.937 -74.906 1.00 60.88 B C
+ATOM 5118 CD2 LEU H 185 35.572 -30.606 -73.612 1.00 65.58 B C
+ATOM 5119 N GLN H 186 39.897 -27.818 -74.373 1.00 72.48 B N
+ATOM 5120 CA GLN H 186 40.328 -26.503 -73.978 1.00 73.64 B C
+ATOM 5121 C GLN H 186 39.133 -25.756 -73.396 1.00 78.38 B C
+ATOM 5122 O GLN H 186 37.982 -26.167 -73.567 1.00 81.19 B O
+ATOM 5123 CB GLN H 186 40.949 -25.847 -75.198 1.00 70.25 B C
+ATOM 5124 CG GLN H 186 42.083 -24.929 -74.951 1.00 70.81 B C
+ATOM 5125 CD GLN H 186 42.977 -24.922 -76.190 1.00 72.18 B C
+ATOM 5126 NE2 GLN H 186 42.596 -25.740 -77.198 1.00 68.43 B N
+ATOM 5127 OE1 GLN H 186 44.033 -24.260 -76.221 1.00 73.89 B O
+ATOM 5128 N SER H 187 39.410 -24.694 -72.632 1.00 77.52 B N
+ATOM 5129 CA SER H 187 38.333 -23.904 -72.025 1.00 73.65 B C
+ATOM 5130 C SER H 187 37.403 -23.288 -73.065 1.00 72.96 B C
+ATOM 5131 O SER H 187 36.301 -22.860 -72.721 1.00 76.07 B O
+ATOM 5132 CB SER H 187 38.941 -22.820 -71.138 1.00 74.17 B C
+ATOM 5133 OG SER H 187 40.158 -22.348 -71.701 1.00 75.29 B O
+ATOM 5134 N SER H 188 37.810 -23.305 -74.328 1.00 71.55 B N
+ATOM 5135 CA SER H 188 37.140 -22.728 -75.480 1.00 69.94 B C
+ATOM 5136 C SER H 188 36.132 -23.667 -76.122 1.00 74.49 B C
+ATOM 5137 O SER H 188 35.718 -23.416 -77.256 1.00 76.87 B O
+ATOM 5138 CB SER H 188 38.183 -22.354 -76.532 1.00 65.98 B C
+ATOM 5139 OG SER H 188 38.777 -23.528 -77.097 1.00 68.37 B O
+ATOM 5140 N GLY H 189 35.824 -24.797 -75.494 1.00 68.36 B N
+ATOM 5141 CA GLY H 189 35.017 -25.803 -76.159 1.00 66.75 B C
+ATOM 5142 C GLY H 189 35.712 -26.518 -77.290 1.00 68.49 B C
+ATOM 5143 O GLY H 189 35.069 -27.305 -77.989 1.00 66.50 B O
+ATOM 5144 N LEU H 190 37.012 -26.286 -77.472 1.00 76.30 B N
+ATOM 5145 CA LEU H 190 37.785 -26.806 -78.595 1.00 71.34 B C
+ATOM 5146 C LEU H 190 38.869 -27.772 -78.123 1.00 69.37 B C
+ATOM 5147 O LEU H 190 39.551 -27.524 -77.119 1.00 71.08 B O
+ATOM 5148 CB LEU H 190 38.434 -25.659 -79.366 1.00 69.72 B C
+ATOM 5149 CG LEU H 190 37.643 -25.203 -80.579 1.00 70.43 B C
+ATOM 5150 CD1 LEU H 190 37.856 -26.174 -81.698 1.00 65.65 B C
+ATOM 5151 CD2 LEU H 190 36.150 -25.091 -80.255 1.00 66.83 B C
+ATOM 5152 N TYR H 191 39.050 -28.863 -78.859 1.00 61.72 B N
+ATOM 5153 CA TYR H 191 40.106 -29.782 -78.475 1.00 64.58 B C
+ATOM 5154 C TYR H 191 41.474 -29.296 -78.936 1.00 65.30 B C
+ATOM 5155 O TYR H 191 41.613 -28.501 -79.866 1.00 65.49 B O
+ATOM 5156 CB TYR H 191 39.852 -31.175 -79.031 1.00 62.69 B C
+ATOM 5157 CG TYR H 191 38.647 -31.807 -78.408 1.00 65.84 B C
+ATOM 5158 CD1 TYR H 191 38.216 -31.399 -77.154 1.00 63.53 B C
+ATOM 5159 CD2 TYR H 191 37.946 -32.821 -79.052 1.00 66.17 B C
+ATOM 5160 CE1 TYR H 191 37.115 -31.957 -76.572 1.00 60.69 B C
+ATOM 5161 CE2 TYR H 191 36.837 -33.392 -78.471 1.00 64.45 B C
+ATOM 5162 CZ TYR H 191 36.431 -32.950 -77.230 1.00 60.48 B C
+ATOM 5163 OH TYR H 191 35.335 -33.500 -76.621 1.00 62.60 B O
+ATOM 5164 N SER H 192 42.499 -29.786 -78.257 1.00 56.62 B N
+ATOM 5165 CA SER H 192 43.853 -29.665 -78.759 1.00 55.53 B C
+ATOM 5166 C SER H 192 44.664 -30.849 -78.250 1.00 55.25 B C
+ATOM 5167 O SER H 192 44.440 -31.333 -77.138 1.00 60.27 B O
+ATOM 5168 CB SER H 192 44.471 -28.348 -78.318 1.00 62.29 B C
+ATOM 5169 OG SER H 192 45.868 -28.505 -78.269 1.00 68.00 B O
+ATOM 5170 N LEU H 193 45.587 -31.340 -79.072 1.00 53.08 B N
+ATOM 5171 CA LEU H 193 46.439 -32.444 -78.642 1.00 52.00 B C
+ATOM 5172 C LEU H 193 47.749 -32.400 -79.406 1.00 52.98 B C
+ATOM 5173 O LEU H 193 47.805 -31.939 -80.545 1.00 55.52 B O
+ATOM 5174 CB LEU H 193 45.803 -33.816 -78.871 1.00 56.12 B C
+ATOM 5175 CG LEU H 193 45.582 -34.258 -80.319 1.00 57.55 B C
+ATOM 5176 CD1 LEU H 193 46.790 -34.985 -80.915 1.00 53.83 B C
+ATOM 5177 CD2 LEU H 193 44.375 -35.141 -80.358 1.00 57.32 B C
+ATOM 5178 N SER H 194 48.786 -32.969 -78.802 1.00 55.97 B N
+ATOM 5179 CA SER H 194 50.110 -33.014 -79.396 1.00 55.45 B C
+ATOM 5180 C SER H 194 50.428 -34.438 -79.818 1.00 54.04 B C
+ATOM 5181 O SER H 194 49.892 -35.399 -79.261 1.00 56.48 B O
+ATOM 5182 CB SER H 194 51.140 -32.509 -78.404 1.00 62.07 B C
+ATOM 5183 OG SER H 194 50.727 -32.897 -77.107 1.00 76.59 B O
+ATOM 5184 N SER H 195 51.255 -34.569 -80.850 1.00 60.69 B N
+ATOM 5185 CA SER H 195 51.711 -35.877 -81.320 1.00 61.38 B C
+ATOM 5186 C SER H 195 53.198 -35.766 -81.584 1.00 63.91 B C
+ATOM 5187 O SER H 195 53.615 -34.934 -82.397 1.00 64.66 B O
+ATOM 5188 CB SER H 195 50.985 -36.329 -82.586 1.00 62.29 B C
+ATOM 5189 OG SER H 195 51.777 -37.270 -83.278 1.00 68.66 B O
+ATOM 5190 N VAL H 196 53.992 -36.590 -80.899 1.00 62.49 B N
+ATOM 5191 CA VAL H 196 55.441 -36.469 -80.928 1.00 64.11 B C
+ATOM 5192 C VAL H 196 56.067 -37.787 -81.342 1.00 69.44 B C
+ATOM 5193 O VAL H 196 55.464 -38.858 -81.262 1.00 73.82 B O
+ATOM 5194 CB VAL H 196 55.996 -36.038 -79.573 1.00 60.12 B C
+ATOM 5195 CG1 VAL H 196 55.390 -34.720 -79.188 1.00 55.88 B C
+ATOM 5196 CG2 VAL H 196 55.670 -37.085 -78.555 1.00 58.26 B C
+ATOM 5197 N VAL H 197 57.300 -37.684 -81.791 1.00 74.25 B N
+ATOM 5198 CA VAL H 197 58.103 -38.831 -82.158 1.00 73.63 B C
+ATOM 5199 C VAL H 197 59.537 -38.554 -81.727 1.00 77.75 B C
+ATOM 5200 O VAL H 197 60.036 -37.422 -81.818 1.00 80.53 B O
+ATOM 5201 CB VAL H 197 58.010 -39.150 -83.664 1.00 74.30 B C
+ATOM 5202 CG1 VAL H 197 58.760 -38.110 -84.483 1.00 74.86 B C
+ATOM 5203 CG2 VAL H 197 58.546 -40.543 -83.947 1.00 73.57 B C
+ATOM 5204 N THR H 198 60.160 -39.590 -81.187 1.00 84.18 B N
+ATOM 5205 CA THR H 198 61.575 -39.610 -80.876 1.00 86.54 B C
+ATOM 5206 C THR H 198 62.304 -40.204 -82.068 1.00 92.12 B C
+ATOM 5207 O THR H 198 61.880 -41.230 -82.610 1.00 92.77 B O
+ATOM 5208 CB THR H 198 61.809 -40.444 -79.617 1.00 84.58 B C
+ATOM 5209 CG2 THR H 198 61.287 -39.722 -78.403 1.00 81.62 B C
+ATOM 5210 OG1 THR H 198 61.133 -41.707 -79.749 1.00 88.54 B O
+ATOM 5211 N VAL H 199 63.375 -39.550 -82.498 1.00101.70 B N
+ATOM 5212 CA VAL H 199 64.054 -40.006 -83.705 1.00102.48 B C
+ATOM 5213 C VAL H 199 65.556 -39.795 -83.546 1.00103.83 B C
+ATOM 5214 O VAL H 199 65.968 -38.858 -82.838 1.00107.37 B O
+ATOM 5215 CB VAL H 199 63.487 -39.298 -84.947 1.00102.50 B C
+ATOM 5216 CG1 VAL H 199 64.166 -37.940 -85.200 1.00101.38 B C
+ATOM 5217 CG2 VAL H 199 63.554 -40.217 -86.154 1.00 99.30 B C
+ATOM 5218 N PRO H 200 66.400 -40.656 -84.134 1.00103.56 B N
+ATOM 5219 CA PRO H 200 67.857 -40.476 -84.000 1.00106.66 B C
+ATOM 5220 C PRO H 200 68.335 -39.139 -84.562 1.00107.40 B C
+ATOM 5221 O PRO H 200 68.008 -38.764 -85.695 1.00109.66 B O
+ATOM 5222 CB PRO H 200 68.424 -41.664 -84.784 1.00109.22 B C
+ATOM 5223 CG PRO H 200 67.403 -42.733 -84.567 1.00106.95 B C
+ATOM 5224 CD PRO H 200 66.070 -42.017 -84.606 1.00101.81 B C
+ATOM 5225 N SER H 201 69.130 -38.424 -83.749 1.00 96.53 B N
+ATOM 5226 CA SER H 201 69.491 -37.042 -84.045 1.00101.23 B C
+ATOM 5227 C SER H 201 70.499 -36.915 -85.179 1.00101.06 B C
+ATOM 5228 O SER H 201 70.690 -35.804 -85.692 1.00104.47 B O
+ATOM 5229 CB SER H 201 70.036 -36.351 -82.794 1.00107.54 B C
+ATOM 5230 OG SER H 201 70.330 -34.983 -83.057 1.00113.70 B O
+ATOM 5231 N SER H 202 71.151 -38.010 -85.577 1.00111.85 B N
+ATOM 5232 CA SER H 202 72.003 -37.964 -86.760 1.00115.33 B C
+ATOM 5233 C SER H 202 71.185 -37.930 -88.052 1.00117.18 B C
+ATOM 5234 O SER H 202 71.732 -37.609 -89.113 1.00120.51 B O
+ATOM 5235 CB SER H 202 72.985 -39.150 -86.757 1.00117.50 B C
+ATOM 5236 OG SER H 202 72.347 -40.392 -86.497 1.00118.52 B O
+ATOM 5237 N SER H 203 69.893 -38.246 -87.993 1.00112.78 B N
+ATOM 5238 CA SER H 203 69.028 -38.166 -89.158 1.00115.57 B C
+ATOM 5239 C SER H 203 68.232 -36.869 -89.228 1.00112.54 B C
+ATOM 5240 O SER H 203 67.377 -36.733 -90.108 1.00111.51 B O
+ATOM 5241 CB SER H 203 68.094 -39.369 -89.205 1.00120.80 B C
+ATOM 5242 OG SER H 203 68.736 -40.431 -89.885 1.00122.25 B O
+ATOM 5243 N LEU H 204 68.478 -35.921 -88.321 1.00109.00 B N
+ATOM 5244 CA LEU H 204 67.765 -34.645 -88.355 1.00109.73 B C
+ATOM 5245 C LEU H 204 67.897 -33.957 -89.712 1.00115.46 B C
+ATOM 5246 O LEU H 204 66.893 -33.656 -90.367 1.00117.90 B O
+ATOM 5247 CB LEU H 204 68.262 -33.747 -87.221 1.00105.63 B C
+ATOM 5248 CG LEU H 204 67.316 -33.670 -86.011 1.00101.62 B C
+ATOM 5249 CD1 LEU H 204 66.963 -35.066 -85.507 1.00 98.61 B C
+ATOM 5250 CD2 LEU H 204 67.932 -32.820 -84.875 1.00100.32 B C
+ATOM 5251 N GLY H 205 69.130 -33.716 -90.159 1.00 96.74 B N
+ATOM 5252 CA GLY H 205 69.350 -33.141 -91.476 1.00102.97 B C
+ATOM 5253 C GLY H 205 69.336 -34.131 -92.625 1.00107.20 B C
+ATOM 5254 O GLY H 205 69.346 -33.712 -93.785 1.00108.83 B O
+ATOM 5255 N THR H 206 69.314 -35.435 -92.329 1.00112.64 B N
+ATOM 5256 CA THR H 206 69.235 -36.461 -93.373 1.00115.03 B C
+ATOM 5257 C THR H 206 67.790 -36.639 -93.829 1.00112.25 B C
+ATOM 5258 O THR H 206 67.410 -36.202 -94.922 1.00113.55 B O
+ATOM 5259 CB THR H 206 69.797 -37.799 -92.860 1.00119.28 B C
+ATOM 5260 CG2 THR H 206 69.478 -38.948 -93.836 1.00123.48 B C
+ATOM 5261 OG1 THR H 206 71.218 -37.710 -92.703 1.00122.11 B O
+ATOM 5262 N GLN H 207 66.968 -37.267 -92.986 1.00114.92 B N
+ATOM 5263 CA GLN H 207 65.579 -37.504 -93.338 1.00107.19 B C
+ATOM 5264 C GLN H 207 64.718 -36.280 -93.015 1.00101.71 B C
+ATOM 5265 O GLN H 207 65.007 -35.500 -92.095 1.00 99.19 B O
+ATOM 5266 CB GLN H 207 65.040 -38.764 -92.649 1.00106.87 B C
+ATOM 5267 CG GLN H 207 63.607 -39.045 -93.058 1.00110.32 B C
+ATOM 5268 CD GLN H 207 63.330 -40.497 -93.270 1.00114.75 B C
+ATOM 5269 NE2 GLN H 207 62.336 -40.792 -94.107 1.00116.98 B N
+ATOM 5270 OE1 GLN H 207 64.019 -41.356 -92.724 1.00117.29 B O
+ATOM 5271 N THR H 208 63.682 -36.094 -93.836 1.00102.83 B N
+ATOM 5272 CA THR H 208 62.698 -35.030 -93.694 1.00103.62 B C
+ATOM 5273 C THR H 208 61.430 -35.602 -93.066 1.00100.32 B C
+ATOM 5274 O THR H 208 60.872 -36.582 -93.575 1.00 99.12 B O
+ATOM 5275 CB THR H 208 62.396 -34.432 -95.062 1.00107.86 B C
+ATOM 5276 CG2 THR H 208 63.634 -33.711 -95.595 1.00110.07 B C
+ATOM 5277 OG1 THR H 208 62.065 -35.500 -95.956 1.00108.00 B O
+ATOM 5278 N TYR H 209 60.982 -34.993 -91.956 1.00100.46 B N
+ATOM 5279 CA TYR H 209 59.895 -35.519 -91.128 1.00 93.84 B C
+ATOM 5280 C TYR H 209 58.620 -34.703 -91.343 1.00 90.95 B C
+ATOM 5281 O TYR H 209 58.574 -33.509 -91.009 1.00 90.63 B O
+ATOM 5282 CB TYR H 209 60.307 -35.528 -89.655 1.00 91.64 B C
+ATOM 5283 CG TYR H 209 61.430 -36.499 -89.391 1.00 92.11 B C
+ATOM 5284 CD1 TYR H 209 61.235 -37.866 -89.562 1.00 91.31 B C
+ATOM 5285 CD2 TYR H 209 62.684 -36.063 -88.986 1.00 92.77 B C
+ATOM 5286 CE1 TYR H 209 62.256 -38.780 -89.347 1.00 93.58 B C
+ATOM 5287 CE2 TYR H 209 63.717 -36.974 -88.763 1.00 93.43 B C
+ATOM 5288 CZ TYR H 209 63.495 -38.335 -88.948 1.00 95.05 B C
+ATOM 5289 OH TYR H 209 64.508 -39.254 -88.740 1.00 95.87 B O
+ATOM 5290 N ILE H 210 57.581 -35.360 -91.876 1.00 87.54 B N
+ATOM 5291 CA ILE H 210 56.293 -34.729 -92.177 1.00 87.28 B C
+ATOM 5292 C ILE H 210 55.189 -35.364 -91.328 1.00 85.61 B C
+ATOM 5293 O ILE H 210 54.911 -36.573 -91.408 1.00 87.70 B O
+ATOM 5294 CB ILE H 210 55.966 -34.750 -93.685 1.00 88.54 B C
+ATOM 5295 CG1 ILE H 210 55.884 -36.189 -94.243 1.00 92.11 B C
+ATOM 5296 CG2 ILE H 210 56.949 -33.840 -94.462 1.00 85.42 B C
+ATOM 5297 CD1 ILE H 210 55.257 -36.336 -95.656 1.00 94.37 B C
+ATOM 5298 N CYS H 211 54.636 -34.528 -90.461 1.00 83.39 B N
+ATOM 5299 CA CYS H 211 53.411 -34.729 -89.709 1.00 81.99 B C
+ATOM 5300 C CYS H 211 52.217 -34.944 -90.633 1.00 81.29 B C
+ATOM 5301 O CYS H 211 52.026 -34.181 -91.587 1.00 81.11 B O
+ATOM 5302 CB CYS H 211 53.256 -33.470 -88.856 1.00 76.90 B C
+ATOM 5303 SG CYS H 211 51.766 -33.173 -88.053 1.00 83.11 B S
+ATOM 5304 N ASN H 212 51.385 -35.942 -90.339 1.00 76.50 B N
+ATOM 5305 CA ASN H 212 50.274 -36.240 -91.233 1.00 80.33 B C
+ATOM 5306 C ASN H 212 49.002 -36.253 -90.399 1.00 81.29 B C
+ATOM 5307 O ASN H 212 48.764 -37.216 -89.661 1.00 84.61 B O
+ATOM 5308 CB ASN H 212 50.508 -37.596 -91.891 1.00 81.53 B C
+ATOM 5309 CG ASN H 212 50.251 -37.583 -93.352 1.00 82.03 B C
+ATOM 5310 ND2 ASN H 212 51.326 -37.687 -94.138 1.00 81.35 B N
+ATOM 5311 OD1 ASN H 212 49.108 -37.479 -93.793 1.00 81.58 B O
+ATOM 5312 N VAL H 213 48.146 -35.234 -90.546 1.00 69.94 B N
+ATOM 5313 CA VAL H 213 46.934 -35.149 -89.722 1.00 68.91 B C
+ATOM 5314 C VAL H 213 45.746 -35.647 -90.534 1.00 71.22 B C
+ATOM 5315 O VAL H 213 45.788 -35.735 -91.764 1.00 75.21 B O
+ATOM 5316 CB VAL H 213 46.679 -33.702 -89.217 1.00 64.42 B C
+ATOM 5317 CG1 VAL H 213 45.628 -33.660 -88.101 1.00 66.61 B C
+ATOM 5318 CG2 VAL H 213 47.955 -33.082 -88.747 1.00 58.44 B C
+ATOM 5319 N ASN H 214 44.646 -35.937 -89.842 1.00 65.95 B N
+ATOM 5320 CA ASN H 214 43.377 -36.143 -90.513 1.00 68.48 B C
+ATOM 5321 C ASN H 214 42.254 -35.916 -89.525 1.00 69.43 B C
+ATOM 5322 O ASN H 214 42.299 -36.427 -88.405 1.00 70.52 B O
+ATOM 5323 CB ASN H 214 43.287 -37.542 -91.109 1.00 71.01 B C
+ATOM 5324 CG ASN H 214 42.434 -37.568 -92.339 1.00 76.90 B C
+ATOM 5325 ND2 ASN H 214 42.942 -38.189 -93.406 1.00 82.35 B N
+ATOM 5326 OD1 ASN H 214 41.339 -36.991 -92.354 1.00 77.99 B O
+ATOM 5327 N HIS H 215 41.222 -35.212 -89.968 1.00 72.38 B N
+ATOM 5328 CA HIS H 215 40.032 -34.958 -89.161 1.00 68.48 B C
+ATOM 5329 C HIS H 215 38.860 -35.334 -90.050 1.00 70.78 B C
+ATOM 5330 O HIS H 215 38.552 -34.608 -91.000 1.00 71.11 B O
+ATOM 5331 CB HIS H 215 39.962 -33.479 -88.752 1.00 59.05 B C
+ATOM 5332 CG HIS H 215 38.942 -33.152 -87.700 1.00 63.87 B C
+ATOM 5333 CD2 HIS H 215 38.409 -33.890 -86.699 1.00 63.13 B C
+ATOM 5334 ND1 HIS H 215 38.426 -31.882 -87.547 1.00 64.99 B N
+ATOM 5335 CE1 HIS H 215 37.587 -31.864 -86.526 1.00 65.35 B C
+ATOM 5336 NE2 HIS H 215 37.559 -33.068 -85.990 1.00 64.70 B N
+ATOM 5337 N LYS H 216 38.184 -36.440 -89.755 1.00 60.18 B N
+ATOM 5338 CA LYS H 216 37.046 -36.772 -90.604 1.00 65.81 B C
+ATOM 5339 C LYS H 216 35.927 -35.758 -90.425 1.00 66.32 B C
+ATOM 5340 O LYS H 216 35.431 -35.240 -91.441 1.00 70.73 B O
+ATOM 5341 CB LYS H 216 36.597 -38.210 -90.357 1.00 67.62 B C
+ATOM 5342 CG LYS H 216 36.912 -39.152 -91.521 1.00 72.03 B C
+ATOM 5343 CD LYS H 216 38.284 -39.834 -91.377 1.00 77.11 B C
+ATOM 5344 CE LYS H 216 38.427 -41.084 -92.263 1.00 80.72 B C
+ATOM 5345 NZ LYS H 216 39.794 -41.677 -92.158 1.00 82.88 B N1+
+ATOM 5346 N PRO H 217 35.523 -35.378 -89.205 1.00 68.49 B N
+ATOM 5347 CA PRO H 217 34.373 -34.458 -89.069 1.00 64.67 B C
+ATOM 5348 C PRO H 217 34.486 -33.150 -89.832 1.00 68.98 B C
+ATOM 5349 O PRO H 217 33.448 -32.532 -90.085 1.00 71.64 B O
+ATOM 5350 CB PRO H 217 34.297 -34.212 -87.558 1.00 63.41 B C
+ATOM 5351 CG PRO H 217 34.770 -35.480 -86.976 1.00 63.24 B C
+ATOM 5352 CD PRO H 217 35.869 -35.986 -87.908 1.00 64.81 B C
+ATOM 5353 N SER H 218 35.682 -32.661 -90.147 1.00 65.04 B N
+ATOM 5354 CA SER H 218 35.789 -31.480 -90.998 1.00 66.81 B C
+ATOM 5355 C SER H 218 36.171 -31.794 -92.440 1.00 71.57 B C
+ATOM 5356 O SER H 218 36.259 -30.868 -93.248 1.00 71.01 B O
+ATOM 5357 CB SER H 218 36.777 -30.461 -90.398 1.00 65.01 B C
+ATOM 5358 OG SER H 218 38.074 -31.004 -90.218 1.00 66.13 B O
+ATOM 5359 N ASN H 219 36.390 -33.064 -92.780 1.00 58.50 B N
+ATOM 5360 CA ASN H 219 37.005 -33.464 -94.055 1.00 62.40 B C
+ATOM 5361 C ASN H 219 38.324 -32.744 -94.310 1.00 61.31 B C
+ATOM 5362 O ASN H 219 38.674 -32.420 -95.447 1.00 65.79 B O
+ATOM 5363 CB ASN H 219 36.068 -33.266 -95.241 1.00 67.20 B C
+ATOM 5364 CG ASN H 219 36.432 -34.166 -96.407 1.00 71.76 B C
+ATOM 5365 ND2 ASN H 219 36.451 -33.598 -97.611 1.00 73.58 B N
+ATOM 5366 OD1 ASN H 219 36.725 -35.356 -96.224 1.00 74.85 B O
+ATOM 5367 N THR H 220 39.082 -32.519 -93.250 1.00 68.52 B N
+ATOM 5368 CA THR H 220 40.388 -31.899 -93.363 1.00 65.23 B C
+ATOM 5369 C THR H 220 41.486 -32.947 -93.368 1.00 62.54 B C
+ATOM 5370 O THR H 220 41.450 -33.904 -92.593 1.00 62.78 B O
+ATOM 5371 CB THR H 220 40.625 -30.937 -92.211 1.00 64.97 B C
+ATOM 5372 CG2 THR H 220 41.760 -29.974 -92.563 1.00 58.15 B C
+ATOM 5373 OG1 THR H 220 39.416 -30.213 -91.958 1.00 67.91 B O
+ATOM 5374 N LYS H 221 42.460 -32.762 -94.250 1.00 63.35 B N
+ATOM 5375 CA LYS H 221 43.732 -33.467 -94.172 1.00 66.50 B C
+ATOM 5376 C LYS H 221 44.829 -32.419 -94.275 1.00 69.30 B C
+ATOM 5377 O LYS H 221 44.953 -31.750 -95.304 1.00 72.29 B O
+ATOM 5378 CB LYS H 221 43.864 -34.529 -95.263 1.00 70.35 B C
+ATOM 5379 CG LYS H 221 45.141 -35.359 -95.156 1.00 74.80 B C
+ATOM 5380 CD LYS H 221 44.970 -36.780 -95.737 1.00 79.54 B C
+ATOM 5381 CE LYS H 221 46.250 -37.633 -95.540 1.00 82.48 B C
+ATOM 5382 NZ LYS H 221 46.102 -39.079 -95.927 1.00 84.24 B N1+
+ATOM 5383 N VAL H 222 45.600 -32.266 -93.199 1.00 66.71 B N
+ATOM 5384 CA VAL H 222 46.728 -31.344 -93.121 1.00 66.07 B C
+ATOM 5385 C VAL H 222 48.009 -32.164 -93.071 1.00 66.28 B C
+ATOM 5386 O VAL H 222 48.098 -33.135 -92.308 1.00 67.38 B O
+ATOM 5387 CB VAL H 222 46.629 -30.440 -91.884 1.00 65.77 B C
+ATOM 5388 CG1 VAL H 222 47.845 -29.563 -91.782 1.00 65.98 B C
+ATOM 5389 CG2 VAL H 222 45.383 -29.610 -91.952 1.00 67.87 B C
+ATOM 5390 N ASP H 223 48.990 -31.795 -93.893 1.00 69.93 B N
+ATOM 5391 CA ASP H 223 50.350 -32.305 -93.781 1.00 72.29 B C
+ATOM 5392 C ASP H 223 51.235 -31.145 -93.365 1.00 76.99 B C
+ATOM 5393 O ASP H 223 51.216 -30.092 -94.011 1.00 81.24 B O
+ATOM 5394 CB ASP H 223 50.851 -32.897 -95.099 1.00 74.34 B C
+ATOM 5395 CG ASP H 223 50.208 -34.231 -95.427 1.00 77.66 B C
+ATOM 5396 OD1 ASP H 223 49.084 -34.490 -94.938 1.00 77.77 B O
+ATOM 5397 OD2 ASP H 223 50.819 -35.016 -96.197 1.00 80.69 B O1-
+ATOM 5398 N LYS H 224 51.984 -31.323 -92.279 1.00 85.12 B N
+ATOM 5399 CA LYS H 224 52.875 -30.282 -91.788 1.00 81.58 B C
+ATOM 5400 C LYS H 224 54.320 -30.762 -91.886 1.00 75.42 B C
+ATOM 5401 O LYS H 224 54.624 -31.931 -91.618 1.00 74.24 B O
+ATOM 5402 CB LYS H 224 52.531 -29.893 -90.356 1.00 84.88 B C
+ATOM 5403 CG LYS H 224 53.141 -28.584 -89.912 1.00 89.53 B C
+ATOM 5404 CD LYS H 224 52.737 -27.451 -90.833 1.00 95.72 B C
+ATOM 5405 CE LYS H 224 53.511 -26.171 -90.519 1.00100.38 B C
+ATOM 5406 NZ LYS H 224 53.131 -25.041 -91.422 1.00103.64 B N1+
+ATOM 5407 N LYS H 225 55.205 -29.871 -92.317 1.00 83.71 B N
+ATOM 5408 CA LYS H 225 56.619 -30.188 -92.390 1.00 84.81 B C
+ATOM 5409 C LYS H 225 57.267 -29.783 -91.075 1.00 82.44 B C
+ATOM 5410 O LYS H 225 56.939 -28.733 -90.508 1.00 85.19 B O
+ATOM 5411 CB LYS H 225 57.269 -29.471 -93.573 1.00 90.18 B C
+ATOM 5412 CG LYS H 225 58.275 -30.331 -94.345 1.00 96.96 B C
+ATOM 5413 CD LYS H 225 58.799 -29.641 -95.619 1.00103.49 B C
+ATOM 5414 CE LYS H 225 60.239 -30.044 -95.902 1.00109.87 B C
+ATOM 5415 NZ LYS H 225 60.401 -31.520 -95.984 1.00113.63 B N1+
+ATOM 5416 N VAL H 226 58.158 -30.640 -90.568 1.00 86.22 B N
+ATOM 5417 CA VAL H 226 58.788 -30.428 -89.265 1.00 86.59 B C
+ATOM 5418 C VAL H 226 60.306 -30.432 -89.430 1.00 90.11 B C
+ATOM 5419 O VAL H 226 60.899 -31.460 -89.792 1.00 90.77 B O
+ATOM 5420 CB VAL H 226 58.341 -31.473 -88.233 1.00 84.08 B C
+ATOM 5421 CG1 VAL H 226 58.831 -31.071 -86.839 1.00 81.80 B C
+ATOM 5422 CG2 VAL H 226 56.810 -31.634 -88.262 1.00 82.68 B C
+ATOM 5423 N GLU H 227 60.918 -29.284 -89.147 1.00 78.62 B N
+ATOM 5424 CA GLU H 227 62.331 -28.976 -89.305 1.00 85.24 B C
+ATOM 5425 C GLU H 227 62.786 -28.164 -88.101 1.00 81.46 B C
+ATOM 5426 O GLU H 227 61.968 -27.507 -87.445 1.00 82.90 B O
+ATOM 5427 CB GLU H 227 62.600 -28.162 -90.581 1.00 94.96 B C
+ATOM 5428 CG GLU H 227 61.806 -28.588 -91.805 1.00104.99 B C
+ATOM 5429 CD GLU H 227 61.648 -27.465 -92.822 1.00112.53 B C
+ATOM 5430 OE1 GLU H 227 61.925 -26.294 -92.467 1.00113.70 B O
+ATOM 5431 OE2 GLU H 227 61.253 -27.758 -93.974 1.00116.02 B O1-
+ATOM 5432 N PRO H 228 64.097 -28.175 -87.792 1.00 78.37 B N
+ATOM 5433 CA PRO H 228 64.690 -27.390 -86.684 1.00 84.15 B C
+ATOM 5434 C PRO H 228 64.552 -25.861 -86.793 1.00 92.08 B C
+ATOM 5435 O PRO H 228 64.914 -25.156 -85.843 1.00 93.07 B O
+ATOM 5436 CB PRO H 228 66.176 -27.784 -86.730 1.00 81.53 B C
+ATOM 5437 CG PRO H 228 66.363 -28.513 -88.060 1.00 80.67 B C
+ATOM 5438 CD PRO H 228 65.053 -29.146 -88.355 1.00 78.16 B C
+TER
+ATOM 5439 N GLU L 1 19.563 -45.257 -54.751 1.00122.44 C N
+ATOM 5440 CA GLU L 1 20.329 -46.343 -55.361 1.00110.26 C C
+ATOM 5441 C GLU L 1 21.521 -46.709 -54.507 1.00 98.29 C C
+ATOM 5442 O GLU L 1 22.020 -45.880 -53.746 1.00 98.59 C O
+ATOM 5443 CB GLU L 1 20.810 -45.960 -56.767 1.00 90.62 C C
+ATOM 5444 CG GLU L 1 22.245 -45.424 -56.897 1.00 90.62 C C
+ATOM 5445 CD GLU L 1 22.393 -44.034 -56.326 1.00 90.62 C C
+ATOM 5446 OE1 GLU L 1 21.344 -43.415 -56.004 1.00 90.62 C O
+ATOM 5447 OE2 GLU L 1 23.549 -43.553 -56.234 1.00 90.62 C O1-
+ATOM 5448 N ILE L 2 21.994 -47.942 -54.633 1.00 80.45 C N
+ATOM 5449 CA ILE L 2 23.112 -48.387 -53.815 1.00 72.28 C C
+ATOM 5450 C ILE L 2 24.372 -48.150 -54.607 1.00 72.52 C C
+ATOM 5451 O ILE L 2 24.387 -48.321 -55.826 1.00 74.39 C O
+ATOM 5452 CB ILE L 2 22.976 -49.864 -53.415 1.00 66.75 C C
+ATOM 5453 CG1 ILE L 2 22.773 -50.723 -54.629 1.00 66.75 C C
+ATOM 5454 CG2 ILE L 2 21.746 -50.068 -52.531 1.00 66.75 C C
+ATOM 5455 CD1 ILE L 2 22.403 -52.090 -54.251 1.00 66.75 C C
+ATOM 5456 N VAL L 3 25.428 -47.754 -53.918 1.00 63.33 C N
+ATOM 5457 CA VAL L 3 26.638 -47.250 -54.548 1.00 64.77 C C
+ATOM 5458 C VAL L 3 27.697 -48.330 -54.527 1.00 63.76 C C
+ATOM 5459 O VAL L 3 28.182 -48.698 -53.454 1.00 65.06 C O
+ATOM 5460 CB VAL L 3 27.153 -46.001 -53.826 1.00 50.28 C C
+ATOM 5461 CG1 VAL L 3 28.485 -45.591 -54.402 1.00 50.28 C C
+ATOM 5462 CG2 VAL L 3 26.135 -44.879 -53.920 1.00 50.28 C C
+ATOM 5463 N MET L 4 28.098 -48.815 -55.689 1.00 59.21 C N
+ATOM 5464 CA MET L 4 29.291 -49.637 -55.705 1.00 59.21 C C
+ATOM 5465 C MET L 4 30.511 -48.767 -55.513 1.00 63.13 C C
+ATOM 5466 O MET L 4 30.631 -47.712 -56.132 1.00 66.98 C O
+ATOM 5467 CB MET L 4 29.412 -50.405 -56.998 1.00 50.04 C C
+ATOM 5468 CG MET L 4 28.241 -51.292 -57.259 1.00 50.04 C C
+ATOM 5469 SD MET L 4 27.877 -52.313 -55.845 1.00 50.04 C S
+ATOM 5470 CE MET L 4 26.430 -51.454 -55.213 1.00 50.04 C C
+ATOM 5471 N THR L 5 31.407 -49.203 -54.646 1.00 57.01 C N
+ATOM 5472 CA THR L 5 32.651 -48.503 -54.380 1.00 53.42 C C
+ATOM 5473 C THR L 5 33.791 -49.398 -54.842 1.00 53.68 C C
+ATOM 5474 O THR L 5 34.162 -50.365 -54.162 1.00 53.36 C O
+ATOM 5475 CB THR L 5 32.757 -48.193 -52.911 1.00 48.57 C C
+ATOM 5476 CG2 THR L 5 33.761 -47.104 -52.715 1.00 48.57 C C
+ATOM 5477 OG1 THR L 5 31.478 -47.753 -52.438 1.00 48.57 C O
+ATOM 5478 N SER L 6 34.365 -49.045 -55.981 1.00 53.67 C N
+ATOM 5479 CA SER L 6 35.446 -49.825 -56.540 1.00 53.67 C C
+ATOM 5480 C SER L 6 36.692 -49.563 -55.705 1.00 55.07 C C
+ATOM 5481 O SER L 6 36.787 -48.537 -55.037 1.00 63.61 C O
+ATOM 5482 CB SER L 6 35.628 -49.441 -58.014 1.00 51.39 C C
+ATOM 5483 OG SER L 6 36.191 -50.494 -58.780 1.00 51.39 C O
+ATOM 5484 N SER L 7 37.638 -50.501 -55.712 1.00 61.71 C N
+ATOM 5485 CA SER L 7 38.904 -50.278 -54.978 1.00 62.11 C C
+ATOM 5486 C SER L 7 39.907 -51.404 -55.110 1.00 61.27 C C
+ATOM 5487 O SER L 7 39.545 -52.563 -55.079 1.00 61.11 C O
+ATOM 5488 CB SER L 7 38.626 -50.050 -53.490 1.00 52.39 C C
+ATOM 5489 OG SER L 7 38.198 -51.255 -52.865 1.00 52.39 C O
+ATOM 5490 N PRO L 8 41.186 -51.082 -55.259 1.00 58.15 C N
+ATOM 5491 CA PRO L 8 41.908 -49.821 -55.334 1.00 57.03 C C
+ATOM 5492 C PRO L 8 41.604 -48.981 -56.576 1.00 57.15 C C
+ATOM 5493 O PRO L 8 41.081 -49.481 -57.569 1.00 56.72 C O
+ATOM 5494 CB PRO L 8 43.351 -50.285 -55.406 1.00 50.72 C C
+ATOM 5495 CG PRO L 8 43.260 -51.527 -56.228 1.00 50.72 C C
+ATOM 5496 CD PRO L 8 42.054 -52.204 -55.652 1.00 50.72 C C
+ATOM 5497 N ALA L 9 41.971 -47.705 -56.518 1.00 63.77 C N
+ATOM 5498 CA ALA L 9 41.970 -46.848 -57.696 1.00 65.47 C C
+ATOM 5499 C ALA L 9 42.644 -47.551 -58.868 1.00 62.93 C C
+ATOM 5500 O ALA L 9 41.996 -47.929 -59.851 1.00 67.49 C O
+ATOM 5501 CB ALA L 9 42.701 -45.537 -57.375 1.00 59.88 C C
+ATOM 5502 N THR L 10 43.958 -47.721 -58.753 1.00 62.28 C N
+ATOM 5503 CA THR L 10 44.787 -48.467 -59.682 1.00 65.20 C C
+ATOM 5504 C THR L 10 45.408 -49.614 -58.920 1.00 63.97 C C
+ATOM 5505 O THR L 10 45.488 -49.591 -57.691 1.00 66.57 C O
+ATOM 5506 CB THR L 10 45.901 -47.606 -60.274 1.00 63.44 C C
+ATOM 5507 CG2 THR L 10 46.420 -48.204 -61.560 1.00 63.44 C C
+ATOM 5508 OG1 THR L 10 45.395 -46.293 -60.534 1.00 63.44 C O
+ATOM 5509 N LEU L 11 45.830 -50.627 -59.655 1.00 61.57 C N
+ATOM 5510 CA LEU L 11 46.546 -51.738 -59.058 1.00 60.71 C C
+ATOM 5511 C LEU L 11 47.626 -52.165 -60.029 1.00 60.71 C C
+ATOM 5512 O LEU L 11 47.357 -52.368 -61.216 1.00 65.86 C O
+ATOM 5513 CB LEU L 11 45.605 -52.898 -58.740 1.00 55.74 C C
+ATOM 5514 CG LEU L 11 46.271 -54.105 -58.112 1.00 55.74 C C
+ATOM 5515 CD1 LEU L 11 45.447 -54.603 -56.938 1.00 55.74 C C
+ATOM 5516 CD2 LEU L 11 46.388 -55.166 -59.176 1.00 55.74 C C
+ATOM 5517 N SER L 12 48.845 -52.295 -59.535 1.00 64.93 C N
+ATOM 5518 CA SER L 12 49.979 -52.623 -60.384 1.00 64.93 C C
+ATOM 5519 C SER L 12 50.526 -53.962 -59.928 1.00 69.01 C C
+ATOM 5520 O SER L 12 50.906 -54.117 -58.764 1.00 73.52 C O
+ATOM 5521 CB SER L 12 51.027 -51.493 -60.355 1.00 57.27 C C
+ATOM 5522 OG SER L 12 51.094 -50.842 -59.087 1.00 57.27 C O
+ATOM 5523 N VAL L 13 50.506 -54.944 -60.818 1.00 66.97 C N
+ATOM 5524 CA VAL L 13 50.995 -56.273 -60.484 1.00 70.16 C C
+ATOM 5525 C VAL L 13 51.740 -56.828 -61.677 1.00 66.40 C C
+ATOM 5526 O VAL L 13 51.301 -56.674 -62.819 1.00 65.70 C O
+ATOM 5527 CB VAL L 13 49.863 -57.232 -60.063 1.00 55.85 C C
+ATOM 5528 CG1 VAL L 13 49.421 -56.962 -58.596 1.00 55.85 C C
+ATOM 5529 CG2 VAL L 13 48.722 -57.114 -60.974 1.00 55.85 C C
+ATOM 5530 N SER L 14 52.857 -57.491 -61.402 1.00 72.38 C N
+ATOM 5531 CA SER L 14 53.761 -57.917 -62.454 1.00 71.84 C C
+ATOM 5532 C SER L 14 53.188 -59.117 -63.196 1.00 74.14 C C
+ATOM 5533 O SER L 14 52.470 -59.924 -62.608 1.00 72.08 C O
+ATOM 5534 CB SER L 14 55.108 -58.283 -61.856 1.00 57.00 C C
+ATOM 5535 OG SER L 14 55.679 -57.145 -61.247 1.00 57.00 C O
+ATOM 5536 N PRO L 15 53.505 -59.270 -64.480 1.00 73.36 C N
+ATOM 5537 CA PRO L 15 52.907 -60.364 -65.255 1.00 73.42 C C
+ATOM 5538 C PRO L 15 53.091 -61.705 -64.556 1.00 75.20 C C
+ATOM 5539 O PRO L 15 54.111 -61.964 -63.915 1.00 75.84 C O
+ATOM 5540 CB PRO L 15 53.645 -60.301 -66.598 1.00 55.96 C C
+ATOM 5541 CG PRO L 15 54.913 -59.573 -66.291 1.00 55.96 C C
+ATOM 5542 CD PRO L 15 54.545 -58.566 -65.238 1.00 55.96 C C
+ATOM 5543 N GLY L 16 52.055 -62.531 -64.622 1.00 77.05 C N
+ATOM 5544 CA GLY L 16 52.031 -63.766 -63.871 1.00 77.05 C C
+ATOM 5545 C GLY L 16 52.004 -63.581 -62.369 1.00 77.05 C C
+ATOM 5546 O GLY L 16 52.926 -64.029 -61.692 1.00 77.24 C O
+ATOM 5547 N GLU L 17 51.007 -62.896 -61.825 1.00 91.57 C N
+ATOM 5548 CA GLU L 17 50.899 -62.783 -60.379 1.00 91.57 C C
+ATOM 5549 C GLU L 17 49.436 -62.629 -60.027 1.00 91.57 C C
+ATOM 5550 O GLU L 17 48.664 -62.058 -60.800 1.00 91.57 C O
+ATOM 5551 CB GLU L 17 51.660 -61.584 -59.823 1.00 73.14 C C
+ATOM 5552 CG GLU L 17 53.139 -61.576 -60.130 1.00 73.14 C C
+ATOM 5553 CD GLU L 17 53.975 -62.368 -59.140 1.00 73.14 C C
+ATOM 5554 OE1 GLU L 17 53.532 -62.539 -57.966 1.00 73.14 C O
+ATOM 5555 OE2 GLU L 17 55.075 -62.829 -59.562 1.00 73.14 C O1-
+ATOM 5556 N ARG L 18 49.056 -63.126 -58.854 1.00 97.70 C N
+ATOM 5557 CA ARG L 18 47.674 -62.958 -58.437 1.00 96.86 C C
+ATOM 5558 C ARG L 18 47.399 -61.481 -58.206 1.00101.41 C C
+ATOM 5559 O ARG L 18 48.255 -60.729 -57.729 1.00103.63 C O
+ATOM 5560 CB ARG L 18 47.336 -63.791 -57.183 1.00 74.93 C C
+ATOM 5561 CG ARG L 18 46.544 -65.108 -57.471 1.00 74.93 C C
+ATOM 5562 CD ARG L 18 46.067 -65.859 -56.221 1.00 74.93 C C
+ATOM 5563 NE ARG L 18 47.043 -65.784 -55.133 1.00 74.93 C N
+ATOM 5564 CZ ARG L 18 48.200 -66.473 -55.085 1.00 74.93 C C
+ATOM 5565 NH1 ARG L 18 48.548 -67.301 -56.089 1.00 74.93 C N1+
+ATOM 5566 NH2 ARG L 18 49.036 -66.321 -54.039 1.00 74.93 C N
+ATOM 5567 N VAL L 19 46.214 -61.063 -58.617 1.00 90.22 C N
+ATOM 5568 CA VAL L 19 45.743 -59.713 -58.409 1.00 90.06 C C
+ATOM 5569 C VAL L 19 44.352 -59.800 -57.830 1.00 87.46 C C
+ATOM 5570 O VAL L 19 43.565 -60.673 -58.209 1.00 87.12 C O
+ATOM 5571 CB VAL L 19 45.727 -58.932 -59.705 1.00 32.96 C C
+ATOM 5572 N THR L 20 44.060 -58.910 -56.905 1.00 70.03 C N
+ATOM 5573 CA THR L 20 42.768 -58.932 -56.248 1.00 71.42 C C
+ATOM 5574 C THR L 20 42.181 -57.536 -56.232 1.00 74.19 C C
+ATOM 5575 O THR L 20 42.858 -56.565 -55.880 1.00 76.66 C O
+ATOM 5576 CB THR L 20 42.847 -59.446 -54.813 1.00 58.82 C C
+ATOM 5577 CG2 THR L 20 43.088 -60.955 -54.798 1.00 58.82 C C
+ATOM 5578 OG1 THR L 20 43.896 -58.757 -54.122 1.00 58.82 C O
+ATOM 5579 N LEU L 21 40.908 -57.464 -56.595 1.00 58.22 C N
+ATOM 5580 CA LEU L 21 40.161 -56.237 -56.752 1.00 57.32 C C
+ATOM 5581 C LEU L 21 38.977 -56.326 -55.814 1.00 58.21 C C
+ATOM 5582 O LEU L 21 38.480 -57.418 -55.542 1.00 58.38 C O
+ATOM 5583 CB LEU L 21 39.694 -56.080 -58.206 1.00 49.87 C C
+ATOM 5584 CG LEU L 21 40.729 -55.611 -59.249 1.00 49.87 C C
+ATOM 5585 CD1 LEU L 21 42.049 -56.397 -59.257 1.00 49.87 C C
+ATOM 5586 CD2 LEU L 21 40.132 -55.646 -60.612 1.00 49.87 C C
+ATOM 5587 N PHE L 22 38.525 -55.193 -55.322 1.00 66.94 C N
+ATOM 5588 CA PHE L 22 37.528 -55.170 -54.263 1.00 63.76 C C
+ATOM 5589 C PHE L 22 36.334 -54.287 -54.624 1.00 65.33 C C
+ATOM 5590 O PHE L 22 36.495 -53.226 -55.242 1.00 68.86 C O
+ATOM 5591 CB PHE L 22 38.159 -54.672 -52.956 1.00 55.08 C C
+ATOM 5592 CG PHE L 22 39.418 -55.434 -52.519 1.00 55.08 C C
+ATOM 5593 CD1 PHE L 22 39.585 -56.787 -52.796 1.00 55.08 C C
+ATOM 5594 CD2 PHE L 22 40.422 -54.780 -51.797 1.00 55.08 C C
+ATOM 5595 CE1 PHE L 22 40.723 -57.463 -52.385 1.00 55.08 C C
+ATOM 5596 CE2 PHE L 22 41.555 -55.462 -51.379 1.00 55.08 C C
+ATOM 5597 CZ PHE L 22 41.705 -56.804 -51.679 1.00 55.08 C C
+ATOM 5598 N CYS L 23 35.135 -54.714 -54.213 1.00 61.19 C N
+ATOM 5599 CA CYS L 23 33.904 -53.956 -54.455 1.00 65.85 C C
+ATOM 5600 C CYS L 23 33.089 -53.885 -53.162 1.00 63.24 C C
+ATOM 5601 O CYS L 23 32.600 -54.907 -52.684 1.00 62.02 C O
+ATOM 5602 CB CYS L 23 33.105 -54.616 -55.585 1.00 53.02 C C
+ATOM 5603 SG CYS L 23 31.573 -53.796 -56.217 1.00 53.02 C S
+ATOM 5604 N ARG L 24 32.934 -52.691 -52.589 1.00 63.26 C N
+ATOM 5605 CA ARG L 24 32.074 -52.488 -51.423 1.00 63.26 C C
+ATOM 5606 C ARG L 24 30.729 -52.007 -51.946 1.00 63.26 C C
+ATOM 5607 O ARG L 24 30.661 -51.366 -52.988 1.00 63.26 C O
+ATOM 5608 CB ARG L 24 32.684 -51.467 -50.436 1.00 56.88 C C
+ATOM 5609 CG ARG L 24 31.766 -50.870 -49.320 1.00 56.88 C C
+ATOM 5610 CD ARG L 24 32.367 -49.593 -48.549 1.00 56.88 C C
+ATOM 5611 NE ARG L 24 31.873 -48.258 -49.019 1.00 56.88 C N
+ATOM 5612 CZ ARG L 24 30.889 -47.494 -48.467 1.00 56.88 C C
+ATOM 5613 NH1 ARG L 24 30.220 -47.868 -47.363 1.00 56.88 C N1+
+ATOM 5614 NH2 ARG L 24 30.554 -46.323 -49.036 1.00 56.88 C N
+ATOM 5615 N ALA L 25 29.648 -52.371 -51.279 1.00 62.91 C N
+ATOM 5616 CA ALA L 25 28.327 -51.943 -51.723 1.00 62.91 C C
+ATOM 5617 C ALA L 25 27.618 -51.155 -50.624 1.00 63.97 C C
+ATOM 5618 O ALA L 25 27.748 -51.459 -49.438 1.00 65.10 C O
+ATOM 5619 CB ALA L 25 27.486 -53.139 -52.166 1.00 62.91 C C
+ATOM 5620 N SER L 26 26.881 -50.128 -51.023 1.00 70.46 C N
+ATOM 5621 CA SER L 26 26.226 -49.249 -50.067 1.00 70.46 C C
+ATOM 5622 C SER L 26 25.362 -50.048 -49.098 1.00 72.18 C C
+ATOM 5623 O SER L 26 25.721 -50.193 -47.931 1.00 75.25 C O
+ATOM 5624 CB SER L 26 25.411 -48.198 -50.834 1.00 69.00 C C
+ATOM 5625 OG SER L 26 24.335 -47.635 -50.080 1.00 69.00 C O
+ATOM 5626 N GLN L 27 24.240 -50.584 -49.559 1.00 80.90 C N
+ATOM 5627 CA GLN L 27 23.516 -51.572 -48.783 1.00 81.29 C C
+ATOM 5628 C GLN L 27 23.795 -52.953 -49.371 1.00 84.03 C C
+ATOM 5629 O GLN L 27 24.443 -53.090 -50.409 1.00 82.95 C O
+ATOM 5630 CB GLN L 27 22.008 -51.243 -48.734 1.00 77.85 C C
+ATOM 5631 CG GLN L 27 21.135 -52.178 -47.769 1.00 77.85 C C
+ATOM 5632 CD GLN L 27 21.605 -52.277 -46.240 1.00 77.85 C C
+ATOM 5633 NE2 GLN L 27 21.222 -51.267 -45.432 1.00 77.85 C N
+ATOM 5634 OE1 GLN L 27 22.271 -53.267 -45.816 1.00 77.85 C O
+ATOM 5635 N SER L 28 23.298 -53.982 -48.685 1.00 67.73 C N
+ATOM 5636 CA SER L 28 23.635 -55.351 -49.028 1.00 67.73 C C
+ATOM 5637 C SER L 28 23.107 -55.714 -50.385 1.00 67.73 C C
+ATOM 5638 O SER L 28 21.928 -55.533 -50.679 1.00 67.73 C O
+ATOM 5639 CB SER L 28 23.042 -56.336 -48.042 1.00 67.73 C C
+ATOM 5640 OG SER L 28 22.885 -57.573 -48.730 1.00 67.73 C O
+ATOM 5641 N VAL L 29 23.989 -56.247 -51.224 1.00 61.03 C N
+ATOM 5642 CA VAL L 29 23.617 -56.662 -52.570 1.00 62.01 C C
+ATOM 5643 C VAL L 29 23.483 -58.180 -52.626 1.00 61.40 C C
+ATOM 5644 O VAL L 29 23.149 -58.748 -53.666 1.00 63.45 C O
+ATOM 5645 CB VAL L 29 24.655 -56.206 -53.611 1.00 62.12 C C
+ATOM 5646 CG1 VAL L 29 24.033 -56.165 -54.998 1.00 62.12 C C
+ATOM 5647 CG2 VAL L 29 25.222 -54.846 -53.233 1.00 62.12 C C
+ATOM 5648 N ILE L 36 23.747 -58.829 -51.497 1.00 61.88 C N
+ATOM 5649 CA ILE L 36 23.660 -60.283 -51.399 1.00 60.82 C C
+ATOM 5650 C ILE L 36 24.726 -60.965 -52.257 1.00 59.08 C C
+ATOM 5651 O ILE L 36 25.886 -61.060 -51.857 1.00 67.25 C O
+ATOM 5652 CB ILE L 36 22.257 -60.794 -51.785 1.00 67.10 C C
+ATOM 5653 CG1 ILE L 36 21.188 -60.123 -50.921 1.00 67.10 C C
+ATOM 5654 CG2 ILE L 36 22.186 -62.308 -51.651 1.00 67.10 C C
+ATOM 5655 CD1 ILE L 36 21.030 -60.747 -49.552 1.00 67.10 C C
+ATOM 5656 N SER L 37 24.330 -61.435 -53.436 1.00 58.75 C N
+ATOM 5657 CA SER L 37 25.250 -62.103 -54.347 1.00 59.92 C C
+ATOM 5658 C SER L 37 25.087 -61.548 -55.756 1.00 59.07 C C
+ATOM 5659 O SER L 37 26.037 -61.513 -56.538 1.00 58.75 C O
+ATOM 5660 CB SER L 37 25.010 -63.613 -54.344 1.00 57.65 C C
+ATOM 5661 OG SER L 37 23.832 -63.944 -55.059 1.00 57.65 C O
+ATOM 5662 N ASN L 38 23.871 -61.113 -56.070 1.00 58.34 C N
+ATOM 5663 CA ASN L 38 23.565 -60.549 -57.383 1.00 58.34 C C
+ATOM 5664 C ASN L 38 24.545 -59.419 -57.670 1.00 58.34 C C
+ATOM 5665 O ASN L 38 24.242 -58.246 -57.502 1.00 58.93 C O
+ATOM 5666 CB ASN L 38 22.152 -59.973 -57.384 1.00 54.26 C C
+ATOM 5667 CG ASN L 38 21.097 -61.019 -57.306 1.00 54.26 C C
+ATOM 5668 ND2 ASN L 38 20.021 -60.727 -56.564 1.00 54.26 C N
+ATOM 5669 OD1 ASN L 38 21.236 -62.097 -57.910 1.00 54.26 C O
+ATOM 5670 N LEU L 39 25.645 -59.768 -58.322 1.00 53.74 C N
+ATOM 5671 CA LEU L 39 26.718 -58.834 -58.607 1.00 53.74 C C
+ATOM 5672 C LEU L 39 27.513 -59.452 -59.731 1.00 53.74 C C
+ATOM 5673 O LEU L 39 27.512 -60.669 -59.895 1.00 53.74 C O
+ATOM 5674 CB LEU L 39 27.590 -58.600 -57.383 1.00 47.75 C C
+ATOM 5675 CG LEU L 39 28.798 -57.691 -57.568 1.00 47.75 C C
+ATOM 5676 CD1 LEU L 39 28.906 -56.657 -56.427 1.00 47.75 C C
+ATOM 5677 CD2 LEU L 39 30.077 -58.526 -57.731 1.00 47.75 C C
+ATOM 5678 N ALA L 40 28.212 -58.618 -60.487 1.00 45.49 C N
+ATOM 5679 CA ALA L 40 28.880 -59.081 -61.687 1.00 45.49 C C
+ATOM 5680 C ALA L 40 30.164 -58.303 -61.816 1.00 45.49 C C
+ATOM 5681 O ALA L 40 30.299 -57.214 -61.257 1.00 45.49 C O
+ATOM 5682 CB ALA L 40 28.015 -58.899 -62.933 1.00 45.49 C C
+ATOM 5683 N TRP L 41 31.110 -58.882 -62.541 1.00 47.38 C N
+ATOM 5684 CA TRP L 41 32.396 -58.264 -62.803 1.00 47.38 C C
+ATOM 5685 C TRP L 41 32.582 -58.192 -64.300 1.00 47.38 C C
+ATOM 5686 O TRP L 41 32.364 -59.197 -64.988 1.00 50.08 C O
+ATOM 5687 CB TRP L 41 33.550 -59.065 -62.197 1.00 45.54 C C
+ATOM 5688 CG TRP L 41 33.604 -58.987 -60.751 1.00 45.54 C C
+ATOM 5689 CD1 TRP L 41 33.027 -59.839 -59.857 1.00 45.54 C C
+ATOM 5690 CD2 TRP L 41 34.227 -57.968 -59.995 1.00 45.54 C C
+ATOM 5691 CE2 TRP L 41 34.006 -58.255 -58.641 1.00 45.54 C C
+ATOM 5692 CE3 TRP L 41 34.944 -56.832 -60.328 1.00 45.54 C C
+ATOM 5693 NE1 TRP L 41 33.275 -59.413 -58.576 1.00 45.54 C N
+ATOM 5694 CZ2 TRP L 41 34.480 -57.439 -57.624 1.00 45.54 C C
+ATOM 5695 CZ3 TRP L 41 35.420 -56.025 -59.315 1.00 45.54 C C
+ATOM 5696 CH2 TRP L 41 35.187 -56.328 -57.982 1.00 45.54 C C
+ATOM 5697 N TYR L 42 33.028 -57.032 -64.792 1.00 44.77 C N
+ATOM 5698 CA TYR L 42 33.263 -56.770 -66.206 1.00 44.77 C C
+ATOM 5699 C TYR L 42 34.713 -56.393 -66.445 1.00 44.77 C C
+ATOM 5700 O TYR L 42 35.336 -55.723 -65.618 1.00 44.77 C O
+ATOM 5701 CB TYR L 42 32.374 -55.652 -66.721 1.00 45.15 C C
+ATOM 5702 CG TYR L 42 30.935 -55.965 -66.549 1.00 45.15 C C
+ATOM 5703 CD1 TYR L 42 30.163 -56.313 -67.632 1.00 45.15 C C
+ATOM 5704 CD2 TYR L 42 30.345 -55.954 -65.300 1.00 45.15 C C
+ATOM 5705 CE1 TYR L 42 28.814 -56.624 -67.482 1.00 45.15 C C
+ATOM 5706 CE2 TYR L 42 29.008 -56.261 -65.133 1.00 45.15 C C
+ATOM 5707 CZ TYR L 42 28.241 -56.592 -66.227 1.00 45.15 C C
+ATOM 5708 OH TYR L 42 26.904 -56.902 -66.088 1.00 45.15 C O
+ATOM 5709 N GLN L 43 35.241 -56.826 -67.579 1.00 53.14 C N
+ATOM 5710 CA GLN L 43 36.555 -56.420 -68.050 1.00 53.14 C C
+ATOM 5711 C GLN L 43 36.334 -55.499 -69.242 1.00 53.98 C C
+ATOM 5712 O GLN L 43 35.461 -55.755 -70.078 1.00 58.30 C O
+ATOM 5713 CB GLN L 43 37.402 -57.650 -68.434 1.00 51.34 C C
+ATOM 5714 CG GLN L 43 38.800 -57.412 -69.003 1.00 51.34 C C
+ATOM 5715 CD GLN L 43 39.152 -58.408 -70.112 1.00 51.34 C C
+ATOM 5716 NE2 GLN L 43 40.005 -59.392 -69.799 1.00 51.34 C N
+ATOM 5717 OE1 GLN L 43 38.658 -58.290 -71.240 1.00 51.34 C O
+ATOM 5718 N GLN L 44 37.061 -54.388 -69.278 1.00 58.73 C N
+ATOM 5719 CA GLN L 44 36.992 -53.453 -70.393 1.00 58.73 C C
+ATOM 5720 C GLN L 44 38.409 -53.142 -70.833 1.00 58.89 C C
+ATOM 5721 O GLN L 44 39.139 -52.442 -70.119 1.00 60.66 C O
+ATOM 5722 CB GLN L 44 36.259 -52.176 -69.996 1.00 52.75 C C
+ATOM 5723 CG GLN L 44 35.796 -51.337 -71.166 1.00 52.75 C C
+ATOM 5724 CD GLN L 44 35.118 -50.039 -70.732 1.00 52.75 C C
+ATOM 5725 NE2 GLN L 44 34.035 -49.669 -71.436 1.00 52.75 C N
+ATOM 5726 OE1 GLN L 44 35.575 -49.362 -69.795 1.00 52.75 C O
+ATOM 5727 N LYS L 45 38.781 -53.647 -72.012 1.00 73.48 C N
+ATOM 5728 CA LYS L 45 40.059 -53.315 -72.622 1.00 73.48 C C
+ATOM 5729 C LYS L 45 39.878 -51.984 -73.332 1.00 74.86 C C
+ATOM 5730 O LYS L 45 38.785 -51.672 -73.813 1.00 77.04 C O
+ATOM 5731 CB LYS L 45 40.511 -54.397 -73.617 1.00 66.99 C C
+ATOM 5732 CG LYS L 45 40.161 -55.856 -73.234 1.00 66.99 C C
+ATOM 5733 CD LYS L 45 41.037 -56.917 -73.959 1.00 66.99 C C
+ATOM 5734 CE LYS L 45 40.404 -58.354 -73.940 1.00 66.99 C C
+ATOM 5735 NZ LYS L 45 39.197 -58.560 -74.871 1.00 66.99 C N1+
+ATOM 5736 N SER L 46 40.941 -51.181 -73.367 1.00 73.55 C N
+ATOM 5737 CA SER L 46 40.787 -49.799 -73.813 1.00 75.05 C C
+ATOM 5738 C SER L 46 40.198 -49.735 -75.218 1.00 81.10 C C
+ATOM 5739 O SER L 46 40.624 -50.451 -76.130 1.00 81.93 C O
+ATOM 5740 CB SER L 46 42.123 -49.056 -73.748 1.00 67.40 C C
+ATOM 5741 OG SER L 46 43.078 -49.622 -74.627 1.00 67.40 C O
+ATOM 5742 N GLY L 47 39.179 -48.901 -75.365 1.00 70.02 C N
+ATOM 5743 CA GLY L 47 38.567 -48.696 -76.652 1.00 68.29 C C
+ATOM 5744 C GLY L 47 37.695 -49.832 -77.120 1.00 69.65 C C
+ATOM 5745 O GLY L 47 37.701 -50.153 -78.313 1.00 70.72 C O
+ATOM 5746 N GLN L 48 36.945 -50.460 -76.222 1.00 73.92 C N
+ATOM 5747 CA GLN L 48 35.901 -51.389 -76.644 1.00 75.92 C C
+ATOM 5748 C GLN L 48 34.901 -51.520 -75.508 1.00 75.33 C C
+ATOM 5749 O GLN L 48 35.040 -50.892 -74.456 1.00 75.82 C O
+ATOM 5750 CB GLN L 48 36.486 -52.736 -77.087 1.00 66.17 C C
+ATOM 5751 CG GLN L 48 37.532 -53.331 -76.163 1.00 66.17 C C
+ATOM 5752 CD GLN L 48 38.342 -54.440 -76.846 1.00 66.17 C C
+ATOM 5753 NE2 GLN L 48 39.049 -54.081 -77.920 1.00 66.17 C N
+ATOM 5754 OE1 GLN L 48 38.363 -55.592 -76.392 1.00 66.17 C O
+ATOM 5755 N ALA L 49 33.868 -52.309 -75.741 1.00 63.85 C N
+ATOM 5756 CA ALA L 49 32.789 -52.428 -74.780 1.00 61.21 C C
+ATOM 5757 C ALA L 49 33.230 -53.292 -73.608 1.00 59.70 C C
+ATOM 5758 O ALA L 49 34.321 -53.868 -73.606 1.00 59.70 C O
+ATOM 5759 CB ALA L 49 31.548 -52.998 -75.460 1.00 59.70 C C
+ATOM 5760 N PRO L 50 32.428 -53.350 -72.555 1.00 53.48 C N
+ATOM 5761 CA PRO L 50 32.659 -54.363 -71.519 1.00 53.48 C C
+ATOM 5762 C PRO L 50 32.514 -55.790 -72.045 1.00 53.48 C C
+ATOM 5763 O PRO L 50 31.663 -56.085 -72.884 1.00 57.15 C O
+ATOM 5764 CB PRO L 50 31.584 -54.038 -70.479 1.00 47.84 C C
+ATOM 5765 CG PRO L 50 31.337 -52.555 -70.662 1.00 47.84 C C
+ATOM 5766 CD PRO L 50 31.510 -52.285 -72.118 1.00 47.84 C C
+ATOM 5767 N ARG L 51 33.387 -56.669 -71.568 1.00 67.46 C N
+ATOM 5768 CA ARG L 51 33.163 -58.110 -71.578 1.00 67.22 C C
+ATOM 5769 C ARG L 51 32.639 -58.458 -70.194 1.00 66.83 C C
+ATOM 5770 O ARG L 51 33.083 -57.873 -69.199 1.00 67.22 C O
+ATOM 5771 CB ARG L 51 34.461 -58.884 -71.882 1.00 61.05 C C
+ATOM 5772 CG ARG L 51 34.359 -60.336 -72.452 1.00 61.05 C C
+ATOM 5773 CD ARG L 51 35.709 -60.835 -73.146 1.00 61.05 C C
+ATOM 5774 NE ARG L 51 36.722 -61.398 -72.198 1.00 61.05 C N
+ATOM 5775 CZ ARG L 51 38.008 -61.716 -72.497 1.00 61.05 C C
+ATOM 5776 NH1 ARG L 51 38.457 -61.534 -73.750 1.00 61.05 C N1+
+ATOM 5777 NH2 ARG L 51 38.860 -62.212 -71.558 1.00 61.05 C N
+ATOM 5778 N LEU L 52 31.655 -59.358 -70.127 1.00 53.55 C N
+ATOM 5779 CA LEU L 52 31.109 -59.781 -68.838 1.00 53.55 C C
+ATOM 5780 C LEU L 52 31.943 -60.941 -68.316 1.00 53.55 C C
+ATOM 5781 O LEU L 52 32.045 -61.981 -68.972 1.00 53.55 C O
+ATOM 5782 CB LEU L 52 29.638 -60.184 -68.933 1.00 47.72 C C
+ATOM 5783 CG LEU L 52 29.187 -61.005 -67.715 1.00 47.72 C C
+ATOM 5784 CD1 LEU L 52 29.103 -60.159 -66.483 1.00 47.72 C C
+ATOM 5785 CD2 LEU L 52 27.860 -61.668 -67.944 1.00 47.72 C C
+ATOM 5786 N LEU L 53 32.538 -60.760 -67.142 1.00 52.17 C N
+ATOM 5787 CA LEU L 53 33.435 -61.757 -66.568 1.00 52.17 C C
+ATOM 5788 C LEU L 53 32.715 -62.717 -65.630 1.00 52.58 C C
+ATOM 5789 O LEU L 53 32.687 -63.928 -65.869 1.00 52.17 C O
+ATOM 5790 CB LEU L 53 34.570 -61.066 -65.821 1.00 45.82 C C
+ATOM 5791 CG LEU L 53 35.852 -61.060 -66.608 1.00 45.82 C C
+ATOM 5792 CD1 LEU L 53 35.568 -60.593 -68.018 1.00 45.82 C C
+ATOM 5793 CD2 LEU L 53 36.755 -60.119 -65.881 1.00 45.82 C C
+ATOM 5794 N ILE L 54 32.176 -62.203 -64.533 1.00 51.09 C N
+ATOM 5795 CA ILE L 54 31.509 -63.056 -63.569 1.00 51.09 C C
+ATOM 5796 C ILE L 54 30.113 -62.511 -63.314 1.00 51.09 C C
+ATOM 5797 O ILE L 54 29.838 -61.326 -63.508 1.00 51.09 C O
+ATOM 5798 CB ILE L 54 32.286 -63.117 -62.260 1.00 43.43 C C
+ATOM 5799 CG1 ILE L 54 33.778 -63.148 -62.543 1.00 43.43 C C
+ATOM 5800 CG2 ILE L 54 31.846 -64.306 -61.438 1.00 43.43 C C
+ATOM 5801 CD1 ILE L 54 34.197 -64.393 -63.155 1.00 43.43 C C
+ATOM 5802 N TYR L 55 29.231 -63.386 -62.843 1.00 52.24 C N
+ATOM 5803 CA TYR L 55 27.926 -62.950 -62.378 1.00 52.24 C C
+ATOM 5804 C TYR L 55 27.437 -63.896 -61.294 1.00 52.24 C C
+ATOM 5805 O TYR L 55 27.923 -65.019 -61.165 1.00 53.95 C O
+ATOM 5806 CB TYR L 55 26.942 -62.860 -63.529 1.00 50.27 C C
+ATOM 5807 CG TYR L 55 26.589 -64.191 -64.114 1.00 50.27 C C
+ATOM 5808 CD1 TYR L 55 25.528 -64.934 -63.625 1.00 50.27 C C
+ATOM 5809 CD2 TYR L 55 27.303 -64.695 -65.190 1.00 50.27 C C
+ATOM 5810 CE1 TYR L 55 25.199 -66.154 -64.185 1.00 50.27 C C
+ATOM 5811 CE2 TYR L 55 26.981 -65.913 -65.766 1.00 50.27 C C
+ATOM 5812 CZ TYR L 55 25.929 -66.639 -65.261 1.00 50.27 C C
+ATOM 5813 OH TYR L 55 25.610 -67.856 -65.822 1.00 50.27 C O
+ATOM 5814 N GLY L 56 26.472 -63.427 -60.502 1.00 51.20 C N
+ATOM 5815 CA GLY L 56 26.164 -64.111 -59.260 1.00 51.20 C C
+ATOM 5816 C GLY L 56 27.320 -64.087 -58.298 1.00 51.20 C C
+ATOM 5817 O GLY L 56 27.375 -64.892 -57.364 1.00 51.20 C O
+ATOM 5818 N ALA L 57 28.290 -63.223 -58.573 1.00 57.17 C N
+ATOM 5819 CA ALA L 57 29.514 -62.961 -57.829 1.00 57.17 C C
+ATOM 5820 C ALA L 57 30.498 -64.114 -57.948 1.00 57.35 C C
+ATOM 5821 O ALA L 57 31.716 -63.922 -57.875 1.00 57.20 C O
+ATOM 5822 CB ALA L 57 29.200 -62.702 -56.355 1.00 57.17 C C
+ATOM 5823 N SER L 65 29.975 -65.309 -58.150 1.00 61.10 C N
+ATOM 5824 CA SER L 65 30.787 -66.499 -58.067 1.00 60.00 C C
+ATOM 5825 C SER L 65 30.965 -67.281 -59.361 1.00 59.91 C C
+ATOM 5826 O SER L 65 31.771 -68.210 -59.380 1.00 62.40 C O
+ATOM 5827 CB SER L 65 30.179 -67.407 -56.998 1.00 57.75 C C
+ATOM 5828 OG SER L 65 28.759 -67.254 -57.028 1.00 57.75 C O
+ATOM 5829 N THR L 66 30.242 -66.985 -60.428 1.00 56.21 C N
+ATOM 5830 CA THR L 66 30.110 -67.966 -61.499 1.00 56.21 C C
+ATOM 5831 C THR L 66 30.587 -67.329 -62.790 1.00 57.54 C C
+ATOM 5832 O THR L 66 29.998 -66.347 -63.248 1.00 56.32 C O
+ATOM 5833 CB THR L 66 28.658 -68.432 -61.647 1.00 50.35 C C
+ATOM 5834 CG2 THR L 66 27.891 -68.338 -60.312 1.00 50.35 C C
+ATOM 5835 OG1 THR L 66 27.989 -67.566 -62.559 1.00 50.35 C O
+ATOM 5836 N ARG L 67 31.645 -67.856 -63.381 1.00 68.08 C N
+ATOM 5837 CA ARG L 67 32.187 -67.162 -64.538 1.00 66.16 C C
+ATOM 5838 C ARG L 67 31.343 -67.438 -65.768 1.00 67.24 C C
+ATOM 5839 O ARG L 67 30.789 -68.523 -65.926 1.00 71.18 C O
+ATOM 5840 CB ARG L 67 33.654 -67.517 -64.778 1.00 58.71 C C
+ATOM 5841 CG ARG L 67 34.058 -68.910 -64.454 1.00 58.71 C C
+ATOM 5842 CD ARG L 67 35.570 -69.070 -64.510 1.00 58.71 C C
+ATOM 5843 NE ARG L 67 35.820 -70.452 -64.889 1.00 58.71 C N
+ATOM 5844 CZ ARG L 67 35.762 -71.482 -64.040 1.00 58.71 C C
+ATOM 5845 NH1 ARG L 67 35.521 -71.252 -62.729 1.00 58.71 C N1+
+ATOM 5846 NH2 ARG L 67 35.968 -72.738 -64.501 1.00 58.71 C N
+ATOM 5847 N ALA L 68 31.194 -66.412 -66.598 1.00 67.82 C N
+ATOM 5848 CA ALA L 68 30.370 -66.508 -67.790 1.00 68.33 C C
+ATOM 5849 C ALA L 68 31.085 -67.349 -68.825 1.00 69.01 C C
+ATOM 5850 O ALA L 68 32.302 -67.538 -68.765 1.00 68.23 C O
+ATOM 5851 CB ALA L 68 30.065 -65.125 -68.371 1.00 46.80 C C
+ATOM 5852 N THR L 69 30.320 -67.865 -69.781 1.00 76.89 C N
+ATOM 5853 CA THR L 69 30.920 -68.772 -70.747 1.00 80.55 C C
+ATOM 5854 C THR L 69 31.946 -68.018 -71.584 1.00 79.65 C C
+ATOM 5855 O THR L 69 31.698 -66.909 -72.066 1.00 76.54 C O
+ATOM 5856 CB THR L 69 29.860 -69.441 -71.631 1.00 56.08 C C
+ATOM 5857 CG2 THR L 69 29.005 -70.467 -70.815 1.00 56.08 C C
+ATOM 5858 OG1 THR L 69 29.011 -68.447 -72.218 1.00 56.08 C O
+ATOM 5859 N GLY L 70 33.115 -68.628 -71.717 1.00 65.00 C N
+ATOM 5860 CA GLY L 70 34.280 -68.054 -72.318 1.00 63.75 C C
+ATOM 5861 C GLY L 70 35.329 -67.592 -71.325 1.00 67.32 C C
+ATOM 5862 O GLY L 70 36.522 -67.637 -71.637 1.00 69.93 C O
+ATOM 5863 N ILE L 71 34.935 -67.238 -70.118 1.00 62.89 C N
+ATOM 5864 CA ILE L 71 35.930 -66.656 -69.220 1.00 63.18 C C
+ATOM 5865 C ILE L 71 36.852 -67.760 -68.726 1.00 64.86 C C
+ATOM 5866 O ILE L 71 36.363 -68.808 -68.279 1.00 69.24 C O
+ATOM 5867 CB ILE L 71 35.247 -65.952 -68.055 1.00 51.06 C C
+ATOM 5868 CG1 ILE L 71 34.801 -64.540 -68.444 1.00 51.06 C C
+ATOM 5869 CG2 ILE L 71 36.189 -65.880 -66.870 1.00 51.06 C C
+ATOM 5870 CD1 ILE L 71 33.658 -64.507 -69.380 1.00 51.06 C C
+ATOM 5871 N PRO L 72 38.169 -67.590 -68.791 1.00 62.53 C N
+ATOM 5872 CA PRO L 72 39.083 -68.630 -68.294 1.00 62.53 C C
+ATOM 5873 C PRO L 72 39.043 -68.732 -66.775 1.00 62.53 C C
+ATOM 5874 O PRO L 72 38.443 -67.916 -66.074 1.00 62.88 C O
+ATOM 5875 CB PRO L 72 40.459 -68.181 -68.798 1.00 58.36 C C
+ATOM 5876 CG PRO L 72 40.157 -67.202 -69.912 1.00 58.36 C C
+ATOM 5877 CD PRO L 72 38.895 -66.511 -69.468 1.00 58.36 C C
+ATOM 5878 N SER L 74 39.662 -69.787 -66.260 1.00 69.94 C N
+ATOM 5879 CA SER L 74 39.529 -70.031 -64.831 1.00 69.94 C C
+ATOM 5880 C SER L 74 40.496 -69.209 -63.987 1.00 69.94 C C
+ATOM 5881 O SER L 74 40.537 -69.382 -62.761 1.00 73.70 C O
+ATOM 5882 CB SER L 74 39.697 -71.521 -64.530 1.00 27.51 C C
+ATOM 5883 N ARG L 75 41.289 -68.332 -64.595 1.00 69.94 C N
+ATOM 5884 CA ARG L 75 41.983 -67.353 -63.773 1.00 69.94 C C
+ATOM 5885 C ARG L 75 40.986 -66.476 -63.028 1.00 71.35 C C
+ATOM 5886 O ARG L 75 41.155 -66.201 -61.837 1.00 73.40 C O
+ATOM 5887 CB ARG L 75 42.947 -66.501 -64.610 1.00 59.35 C C
+ATOM 5888 CG ARG L 75 43.622 -67.195 -65.783 1.00 59.35 C C
+ATOM 5889 CD ARG L 75 44.402 -66.190 -66.613 1.00 59.35 C C
+ATOM 5890 NE ARG L 75 44.340 -66.394 -68.054 1.00 59.35 C N
+ATOM 5891 CZ ARG L 75 43.604 -65.672 -68.882 1.00 59.35 C C
+ATOM 5892 NH1 ARG L 75 42.898 -64.679 -68.392 1.00 59.35 C N1+
+ATOM 5893 NH2 ARG L 75 43.602 -65.921 -70.194 1.00 59.35 C N
+ATOM 5894 N PHE L 76 39.915 -66.072 -63.697 1.00 67.88 C N
+ATOM 5895 CA PHE L 76 38.990 -65.106 -63.125 1.00 67.05 C C
+ATOM 5896 C PHE L 76 38.002 -65.775 -62.174 1.00 68.58 C C
+ATOM 5897 O PHE L 76 37.332 -66.755 -62.529 1.00 70.54 C O
+ATOM 5898 CB PHE L 76 38.251 -64.366 -64.235 1.00 55.56 C C
+ATOM 5899 CG PHE L 76 39.159 -63.701 -65.208 1.00 55.56 C C
+ATOM 5900 CD1 PHE L 76 39.636 -62.426 -64.962 1.00 55.56 C C
+ATOM 5901 CD2 PHE L 76 39.528 -64.341 -66.370 1.00 55.56 C C
+ATOM 5902 CE1 PHE L 76 40.482 -61.811 -65.847 1.00 55.56 C C
+ATOM 5903 CE2 PHE L 76 40.361 -63.730 -67.260 1.00 55.56 C C
+ATOM 5904 CZ PHE L 76 40.846 -62.462 -67.000 1.00 55.56 C C
+ATOM 5905 N SER L 77 37.916 -65.236 -60.963 1.00 64.58 C N
+ATOM 5906 CA SER L 77 37.131 -65.870 -59.921 1.00 64.46 C C
+ATOM 5907 C SER L 77 36.681 -64.819 -58.922 1.00 64.46 C C
+ATOM 5908 O SER L 77 37.426 -63.888 -58.613 1.00 64.46 C O
+ATOM 5909 CB SER L 77 37.972 -66.932 -59.233 1.00 59.92 C C
+ATOM 5910 OG SER L 77 39.314 -66.465 -59.193 1.00 59.92 C O
+ATOM 5911 N GLY L 78 35.502 -65.023 -58.338 1.00 65.01 C N
+ATOM 5912 CA GLY L 78 34.909 -63.995 -57.511 1.00 65.01 C C
+ATOM 5913 C GLY L 78 34.185 -64.572 -56.313 1.00 65.01 C C
+ATOM 5914 O GLY L 78 33.865 -65.766 -56.253 1.00 68.74 C O
+ATOM 5915 N SER L 79 34.003 -63.749 -55.287 1.00 69.63 C N
+ATOM 5916 CA SER L 79 33.350 -64.189 -54.056 1.00 69.63 C C
+ATOM 5917 C SER L 79 32.188 -63.252 -53.862 1.00 69.83 C C
+ATOM 5918 O SER L 79 31.815 -62.561 -54.810 1.00 69.63 C O
+ATOM 5919 CB SER L 79 34.311 -64.101 -52.871 1.00 60.57 C C
+ATOM 5920 OG SER L 79 35.626 -63.796 -53.302 1.00 60.57 C O
+ATOM 5921 N GLY L 80 31.589 -63.181 -52.676 1.00 59.39 C N
+ATOM 5922 CA GLY L 80 30.496 -62.228 -52.640 1.00 59.39 C C
+ATOM 5923 C GLY L 80 29.341 -62.450 -51.688 1.00 59.39 C C
+ATOM 5924 O GLY L 80 29.514 -63.020 -50.609 1.00 59.39 C O
+ATOM 5925 N SER L 83 28.162 -61.927 -52.055 1.00 70.19 C N
+ATOM 5926 CA SER L 83 26.931 -62.023 -51.255 1.00 70.19 C C
+ATOM 5927 C SER L 83 27.093 -61.294 -49.919 1.00 70.19 C C
+ATOM 5928 O SER L 83 27.302 -61.880 -48.863 1.00 70.19 C O
+ATOM 5929 CB SER L 83 26.506 -63.488 -51.040 1.00 70.19 C C
+ATOM 5930 OG SER L 83 25.478 -63.611 -50.060 1.00 70.19 C O
+ATOM 5931 N GLY L 84 27.017 -59.976 -50.015 1.00 65.17 C N
+ATOM 5932 CA GLY L 84 27.199 -59.150 -48.837 1.00 65.17 C C
+ATOM 5933 C GLY L 84 27.265 -57.683 -49.202 1.00 65.17 C C
+ATOM 5934 O GLY L 84 26.554 -57.213 -50.093 1.00 65.17 C O
+ATOM 5935 N THR L 85 27.991 -56.947 -48.386 1.00 67.74 C N
+ATOM 5936 CA THR L 85 28.504 -55.659 -48.798 1.00 67.74 C C
+ATOM 5937 C THR L 85 29.850 -55.740 -49.509 1.00 67.74 C C
+ATOM 5938 O THR L 85 30.081 -54.995 -50.458 1.00 67.74 C O
+ATOM 5939 CB THR L 85 28.549 -54.763 -47.576 1.00 67.74 C C
+ATOM 5940 CG2 THR L 85 29.391 -53.504 -47.801 1.00 67.74 C C
+ATOM 5941 OG1 THR L 85 27.198 -54.425 -47.241 1.00 67.74 C O
+ATOM 5942 N GLU L 86 30.765 -56.601 -49.084 1.00 75.19 C N
+ATOM 5943 CA GLU L 86 32.101 -56.595 -49.659 1.00 75.19 C C
+ATOM 5944 C GLU L 86 32.296 -57.793 -50.583 1.00 75.19 C C
+ATOM 5945 O GLU L 86 31.734 -58.868 -50.360 1.00 78.73 C O
+ATOM 5946 CB GLU L 86 33.172 -56.579 -48.570 1.00 61.05 C C
+ATOM 5947 CG GLU L 86 34.406 -55.737 -48.940 1.00 61.05 C C
+ATOM 5948 CD GLU L 86 34.278 -54.222 -48.575 1.00 61.05 C C
+ATOM 5949 OE1 GLU L 86 33.270 -53.848 -47.894 1.00 61.05 C O
+ATOM 5950 OE2 GLU L 86 35.198 -53.419 -48.963 1.00 61.05 C O1-
+ATOM 5951 N PHE L 87 33.089 -57.586 -51.636 1.00 61.05 C N
+ATOM 5952 CA PHE L 87 33.194 -58.520 -52.752 1.00 61.05 C C
+ATOM 5953 C PHE L 87 34.607 -58.505 -53.301 1.00 63.26 C C
+ATOM 5954 O PHE L 87 35.340 -57.523 -53.155 1.00 67.91 C O
+ATOM 5955 CB PHE L 87 32.267 -58.164 -53.916 1.00 49.75 C C
+ATOM 5956 CG PHE L 87 30.809 -58.250 -53.609 1.00 49.75 C C
+ATOM 5957 CD1 PHE L 87 30.059 -59.292 -54.085 1.00 49.75 C C
+ATOM 5958 CD2 PHE L 87 30.171 -57.260 -52.897 1.00 49.75 C C
+ATOM 5959 CE1 PHE L 87 28.701 -59.369 -53.823 1.00 49.75 C C
+ATOM 5960 CE2 PHE L 87 28.817 -57.327 -52.634 1.00 49.75 C C
+ATOM 5961 CZ PHE L 87 28.083 -58.380 -53.101 1.00 49.75 C C
+ATOM 5962 N THR L 88 34.945 -59.571 -54.022 1.00 57.55 C N
+ATOM 5963 CA THR L 88 36.302 -59.771 -54.512 1.00 57.55 C C
+ATOM 5964 C THR L 88 36.347 -60.356 -55.906 1.00 60.92 C C
+ATOM 5965 O THR L 88 35.702 -61.380 -56.167 1.00 65.60 C O
+ATOM 5966 CB THR L 88 37.067 -60.715 -53.612 1.00 49.83 C C
+ATOM 5967 CG2 THR L 88 38.343 -61.151 -54.291 1.00 49.83 C C
+ATOM 5968 OG1 THR L 88 37.358 -60.047 -52.388 1.00 49.83 C O
+ATOM 5969 N LEU L 89 37.157 -59.741 -56.769 1.00 57.05 C N
+ATOM 5970 CA LEU L 89 37.557 -60.326 -58.037 1.00 54.34 C C
+ATOM 5971 C LEU L 89 39.022 -60.697 -57.949 1.00 54.51 C C
+ATOM 5972 O LEU L 89 39.824 -59.932 -57.419 1.00 56.50 C O
+ATOM 5973 CB LEU L 89 37.330 -59.371 -59.215 1.00 51.13 C C
+ATOM 5974 CG LEU L 89 37.894 -59.856 -60.565 1.00 51.13 C C
+ATOM 5975 CD1 LEU L 89 37.365 -61.215 -60.989 1.00 51.13 C C
+ATOM 5976 CD2 LEU L 89 37.576 -58.869 -61.651 1.00 51.13 C C
+ATOM 5977 N THR L 90 39.362 -61.873 -58.462 1.00 62.73 C N
+ATOM 5978 CA THR L 90 40.701 -62.438 -58.355 1.00 62.73 C C
+ATOM 5979 C THR L 90 41.139 -62.964 -59.699 1.00 65.41 C C
+ATOM 5980 O THR L 90 40.475 -63.840 -60.270 1.00 70.60 C O
+ATOM 5981 CB THR L 90 40.769 -63.580 -57.351 1.00 59.58 C C
+ATOM 5982 CG2 THR L 90 42.108 -64.251 -57.428 1.00 59.58 C C
+ATOM 5983 OG1 THR L 90 40.526 -63.088 -56.033 1.00 59.58 C O
+ATOM 5984 N ILE L 91 42.274 -62.460 -60.173 1.00 75.82 C N
+ATOM 5985 CA ILE L 91 42.901 -62.940 -61.392 1.00 74.55 C C
+ATOM 5986 C ILE L 91 44.146 -63.701 -60.984 1.00 75.41 C C
+ATOM 5987 O ILE L 91 45.021 -63.138 -60.316 1.00 80.16 C O
+ATOM 5988 CB ILE L 91 43.260 -61.795 -62.344 1.00 62.15 C C
+ATOM 5989 CG1 ILE L 91 42.069 -60.874 -62.539 1.00 62.15 C C
+ATOM 5990 CG2 ILE L 91 43.551 -62.371 -63.682 1.00 62.15 C C
+ATOM 5991 CD1 ILE L 91 42.301 -59.774 -63.565 1.00 62.15 C C
+ATOM 5992 N SER L 92 44.249 -64.952 -61.443 1.00 94.39 C N
+ATOM 5993 CA SER L 92 45.185 -65.919 -60.872 1.00 91.50 C C
+ATOM 5994 C SER L 92 46.619 -65.609 -61.255 1.00 91.50 C C
+ATOM 5995 O SER L 92 47.479 -65.426 -60.392 1.00 94.47 C O
+ATOM 5996 CB SER L 92 44.828 -67.324 -61.334 1.00 67.47 C C
+ATOM 5997 OG SER L 92 43.474 -67.600 -61.030 1.00 67.47 C O
+ATOM 5998 N SER L 93 46.921 -65.637 -62.540 1.00 83.18 C N
+ATOM 5999 CA SER L 93 48.213 -65.171 -63.013 1.00 83.18 C C
+ATOM 6000 C SER L 93 47.925 -64.163 -64.106 1.00 83.18 C C
+ATOM 6001 O SER L 93 47.414 -64.524 -65.172 1.00 83.18 C O
+ATOM 6002 CB SER L 93 49.067 -66.323 -63.527 1.00 60.32 C C
+ATOM 6003 OG SER L 93 48.727 -66.604 -64.878 1.00 60.32 C O
+ATOM 6004 N LEU L 94 48.246 -62.906 -63.843 1.00 84.53 C N
+ATOM 6005 CA LEU L 94 47.906 -61.869 -64.797 1.00 84.53 C C
+ATOM 6006 C LEU L 94 48.563 -62.156 -66.131 1.00 84.53 C C
+ATOM 6007 O LEU L 94 49.755 -62.440 -66.203 1.00 86.87 C O
+ATOM 6008 CB LEU L 94 48.340 -60.502 -64.284 1.00 62.54 C C
+ATOM 6009 CG LEU L 94 47.182 -59.758 -63.628 1.00 62.54 C C
+ATOM 6010 CD1 LEU L 94 47.406 -58.272 -63.613 1.00 62.54 C C
+ATOM 6011 CD2 LEU L 94 45.929 -60.080 -64.387 1.00 62.54 C C
+ATOM 6012 N GLN L 95 47.792 -62.088 -67.190 1.00100.29 C N
+ATOM 6013 CA GLN L 95 48.355 -62.353 -68.492 1.00100.29 C C
+ATOM 6014 C GLN L 95 48.121 -61.136 -69.383 1.00101.20 C C
+ATOM 6015 O GLN L 95 47.313 -60.261 -69.069 1.00100.29 C O
+ATOM 6016 CB GLN L 95 47.747 -63.615 -69.091 1.00 69.87 C C
+ATOM 6017 CG GLN L 95 48.560 -64.154 -70.246 1.00 69.87 C C
+ATOM 6018 CD GLN L 95 47.675 -64.760 -71.330 1.00 69.87 C C
+ATOM 6019 NE2 GLN L 95 48.082 -64.597 -72.613 1.00 69.87 C N
+ATOM 6020 OE1 GLN L 95 46.629 -65.365 -71.022 1.00 69.87 C O
+ATOM 6021 N SER L 96 48.829 -61.103 -70.516 1.00 96.11 C N
+ATOM 6022 CA SER L 96 48.963 -59.893 -71.326 1.00 96.11 C C
+ATOM 6023 C SER L 96 47.639 -59.180 -71.520 1.00101.08 C C
+ATOM 6024 O SER L 96 47.531 -57.968 -71.324 1.00107.79 C O
+ATOM 6025 CB SER L 96 49.531 -60.259 -72.693 1.00 67.77 C C
+ATOM 6026 OG SER L 96 50.483 -61.314 -72.611 1.00 67.77 C O
+ATOM 6027 N GLU L 97 46.617 -59.922 -71.885 1.00103.20 C N
+ATOM 6028 CA GLU L 97 45.334 -59.347 -72.230 1.00103.19 C C
+ATOM 6029 C GLU L 97 44.420 -59.187 -71.027 1.00100.63 C C
+ATOM 6030 O GLU L 97 43.227 -58.918 -71.201 1.00102.77 C O
+ATOM 6031 CB GLU L 97 44.675 -60.202 -73.307 1.00 74.91 C C
+ATOM 6032 CG GLU L 97 44.235 -61.591 -72.819 1.00 74.91 C C
+ATOM 6033 CD GLU L 97 45.298 -62.693 -73.034 1.00 74.91 C C
+ATOM 6034 OE1 GLU L 97 46.483 -62.356 -73.320 1.00 74.91 C O
+ATOM 6035 OE2 GLU L 97 44.922 -63.895 -72.943 1.00 74.91 C O1-
+ATOM 6036 N ASP L 98 44.928 -59.419 -69.822 1.00 83.07 C N
+ATOM 6037 CA ASP L 98 44.156 -59.157 -68.619 1.00 83.06 C C
+ATOM 6038 C ASP L 98 44.409 -57.783 -68.038 1.00 83.93 C C
+ATOM 6039 O ASP L 98 43.700 -57.383 -67.115 1.00 83.81 C O
+ATOM 6040 CB ASP L 98 44.452 -60.193 -67.543 1.00 65.01 C C
+ATOM 6041 CG ASP L 98 44.331 -61.594 -68.061 1.00 65.01 C C
+ATOM 6042 OD1 ASP L 98 43.707 -61.752 -69.141 1.00 65.01 C O
+ATOM 6043 OD2 ASP L 98 44.835 -62.530 -67.394 1.00 65.01 C O1-
+ATOM 6044 N PHE L 99 45.381 -57.045 -68.551 1.00 80.82 C N
+ATOM 6045 CA PHE L 99 45.604 -55.703 -68.040 1.00 80.35 C C
+ATOM 6046 C PHE L 99 44.609 -54.801 -68.740 1.00 80.35 C C
+ATOM 6047 O PHE L 99 44.728 -54.543 -69.939 1.00 80.35 C O
+ATOM 6048 CB PHE L 99 47.037 -55.248 -68.304 1.00 60.07 C C
+ATOM 6049 CG PHE L 99 48.087 -56.065 -67.589 1.00 60.07 C C
+ATOM 6050 CD1 PHE L 99 48.493 -55.731 -66.314 1.00 60.07 C C
+ATOM 6051 CD2 PHE L 99 48.669 -57.153 -68.196 1.00 60.07 C C
+ATOM 6052 CE1 PHE L 99 49.445 -56.479 -65.664 1.00 60.07 C C
+ATOM 6053 CE2 PHE L 99 49.625 -57.894 -67.542 1.00 60.07 C C
+ATOM 6054 CZ PHE L 99 50.011 -57.556 -66.278 1.00 60.07 C C
+ATOM 6055 N ALA L 100 43.655 -54.297 -67.988 1.00 61.28 C N
+ATOM 6056 CA ALA L 100 42.508 -53.574 -68.525 1.00 61.28 C C
+ATOM 6057 C ALA L 100 41.777 -52.981 -67.333 1.00 61.28 C C
+ATOM 6058 O ALA L 100 42.256 -53.076 -66.199 1.00 61.28 C O
+ATOM 6059 CB ALA L 100 41.621 -54.491 -69.357 1.00 62.23 C C
+ATOM 6060 N VAL L 101 40.637 -52.368 -67.545 1.00 50.32 C N
+ATOM 6061 CA VAL L 101 39.910 -51.846 -66.396 1.00 50.32 C C
+ATOM 6062 C VAL L 101 38.798 -52.802 -66.006 1.00 51.87 C C
+ATOM 6063 O VAL L 101 38.150 -53.398 -66.869 1.00 52.47 C O
+ATOM 6064 CB VAL L 101 39.369 -50.447 -66.679 1.00 51.95 C C
+ATOM 6065 CG1 VAL L 101 38.487 -49.989 -65.513 1.00 51.95 C C
+ATOM 6066 CG2 VAL L 101 40.554 -49.535 -66.876 1.00 51.95 C C
+ATOM 6067 N TYR L 102 38.578 -52.966 -64.707 1.00 49.04 C N
+ATOM 6068 CA TYR L 102 37.560 -53.884 -64.225 1.00 49.04 C C
+ATOM 6069 C TYR L 102 36.482 -53.103 -63.502 1.00 49.04 C C
+ATOM 6070 O TYR L 102 36.779 -52.128 -62.813 1.00 49.15 C O
+ATOM 6071 CB TYR L 102 38.169 -54.953 -63.325 1.00 50.47 C C
+ATOM 6072 CG TYR L 102 39.026 -55.899 -64.125 1.00 50.47 C C
+ATOM 6073 CD1 TYR L 102 38.522 -57.101 -64.606 1.00 50.47 C C
+ATOM 6074 CD2 TYR L 102 40.319 -55.561 -64.474 1.00 50.47 C C
+ATOM 6075 CE1 TYR L 102 39.310 -57.960 -65.374 1.00 50.47 C C
+ATOM 6076 CE2 TYR L 102 41.100 -56.411 -65.246 1.00 50.47 C C
+ATOM 6077 CZ TYR L 102 40.592 -57.603 -65.688 1.00 50.47 C C
+ATOM 6078 OH TYR L 102 41.376 -58.433 -66.443 1.00 50.47 C O
+ATOM 6079 N PHE L 103 35.230 -53.511 -63.706 1.00 45.86 C N
+ATOM 6080 CA PHE L 103 34.069 -52.839 -63.148 1.00 45.86 C C
+ATOM 6081 C PHE L 103 33.208 -53.867 -62.454 1.00 46.09 C C
+ATOM 6082 O PHE L 103 33.048 -54.972 -62.964 1.00 47.99 C O
+ATOM 6083 CB PHE L 103 33.214 -52.170 -64.211 1.00 49.42 C C
+ATOM 6084 CG PHE L 103 33.850 -50.985 -64.831 1.00 49.42 C C
+ATOM 6085 CD1 PHE L 103 33.901 -49.771 -64.146 1.00 49.42 C C
+ATOM 6086 CD2 PHE L 103 34.364 -51.062 -66.118 1.00 49.42 C C
+ATOM 6087 CE1 PHE L 103 34.493 -48.659 -64.725 1.00 49.42 C C
+ATOM 6088 CE2 PHE L 103 34.951 -49.969 -66.716 1.00 49.42 C C
+ATOM 6089 CZ PHE L 103 35.021 -48.761 -66.022 1.00 49.42 C C
+ATOM 6090 N CYS L 104 32.635 -53.504 -61.310 1.00 49.22 C N
+ATOM 6091 CA CYS L 104 31.645 -54.340 -60.654 1.00 49.22 C C
+ATOM 6092 C CYS L 104 30.279 -53.691 -60.751 1.00 49.22 C C
+ATOM 6093 O CYS L 104 30.161 -52.470 -60.818 1.00 49.22 C O
+ATOM 6094 CB CYS L 104 31.999 -54.586 -59.199 1.00 49.62 C C
+ATOM 6095 SG CYS L 104 31.989 -53.115 -58.170 1.00 49.62 C S
+ATOM 6096 N GLN L 105 29.251 -54.522 -60.789 1.00 53.52 C N
+ATOM 6097 CA GLN L 105 27.882 -54.038 -60.888 1.00 53.52 C C
+ATOM 6098 C GLN L 105 27.002 -54.868 -59.982 1.00 53.52 C C
+ATOM 6099 O GLN L 105 27.227 -56.070 -59.874 1.00 53.93 C O
+ATOM 6100 CB GLN L 105 27.363 -54.150 -62.315 1.00 51.60 C C
+ATOM 6101 CG GLN L 105 25.878 -53.987 -62.405 1.00 51.60 C C
+ATOM 6102 CD GLN L 105 25.271 -54.928 -63.395 1.00 51.60 C C
+ATOM 6103 NE2 GLN L 105 23.983 -55.249 -63.221 1.00 51.60 C N
+ATOM 6104 OE1 GLN L 105 25.952 -55.360 -64.324 1.00 51.60 C O
+ATOM 6105 N GLN L 106 26.006 -54.276 -59.331 1.00 60.80 C N
+ATOM 6106 CA GLN L 106 25.018 -55.170 -58.754 1.00 60.80 C C
+ATOM 6107 C GLN L 106 23.741 -55.066 -59.576 1.00 60.80 C C
+ATOM 6108 O GLN L 106 23.480 -54.050 -60.213 1.00 60.80 C O
+ATOM 6109 CB GLN L 106 24.680 -54.904 -57.279 1.00 60.80 C C
+ATOM 6110 CG GLN L 106 23.456 -54.023 -57.022 1.00 60.80 C C
+ATOM 6111 CD GLN L 106 23.460 -52.716 -57.696 1.00 60.80 C C
+ATOM 6112 NE2 GLN L 106 22.333 -52.025 -57.581 1.00 60.80 C N
+ATOM 6113 OE1 GLN L 106 24.371 -52.389 -58.466 1.00 60.80 C O
+ATOM 6114 N TYR L 107 23.011 -56.164 -59.669 1.00 59.53 C N
+ATOM 6115 CA TYR L 107 21.665 -56.148 -60.225 1.00 59.53 C C
+ATOM 6116 C TYR L 107 20.564 -56.311 -59.177 1.00 59.53 C C
+ATOM 6117 O TYR L 107 19.422 -56.618 -59.543 1.00 59.53 C O
+ATOM 6118 CB TYR L 107 21.535 -57.158 -61.374 1.00 53.20 C C
+ATOM 6119 CG TYR L 107 22.271 -58.416 -61.156 1.00 53.20 C C
+ATOM 6120 CD1 TYR L 107 21.601 -59.549 -60.803 1.00 53.20 C C
+ATOM 6121 CD2 TYR L 107 23.633 -58.476 -61.321 1.00 53.20 C C
+ATOM 6122 CE1 TYR L 107 22.258 -60.711 -60.591 1.00 53.20 C C
+ATOM 6123 CE2 TYR L 107 24.308 -59.625 -61.109 1.00 53.20 C C
+ATOM 6124 CZ TYR L 107 23.615 -60.752 -60.741 1.00 53.20 C C
+ATOM 6125 OH TYR L 107 24.260 -61.950 -60.517 1.00 53.20 C O
+ATOM 6126 N ASN L 108 20.882 -56.214 -57.888 1.00 68.38 C N
+ATOM 6127 CA ASN L 108 19.869 -56.507 -56.876 1.00 67.95 C C
+ATOM 6128 C ASN L 108 18.667 -55.590 -56.992 1.00 68.84 C C
+ATOM 6129 O ASN L 108 17.522 -56.038 -56.863 1.00 77.93 C O
+ATOM 6130 CB ASN L 108 20.454 -56.385 -55.480 1.00 61.24 C C
+ATOM 6131 CG ASN L 108 19.688 -57.193 -54.460 1.00 61.24 C C
+ATOM 6132 ND2 ASN L 108 18.697 -56.553 -53.805 1.00 61.24 C N
+ATOM 6133 OD1 ASN L 108 20.000 -58.381 -54.236 1.00 61.24 C O
+ATOM 6134 N ASN L 109 18.906 -54.298 -57.188 1.00 75.43 C N
+ATOM 6135 CA ASN L 109 17.836 -53.350 -57.461 1.00 78.14 C C
+ATOM 6136 C ASN L 109 18.117 -52.579 -58.731 1.00 79.52 C C
+ATOM 6137 O ASN L 109 19.262 -52.459 -59.180 1.00 78.63 C O
+ATOM 6138 CB ASN L 109 17.639 -52.343 -56.341 1.00 69.45 C C
+ATOM 6139 CG ASN L 109 16.725 -52.853 -55.285 1.00 69.45 C C
+ATOM 6140 ND2 ASN L 109 15.426 -52.550 -55.434 1.00 69.45 C N
+ATOM 6141 OD1 ASN L 109 17.157 -53.553 -54.352 1.00 69.45 C O
+ATOM 6142 N TRP L 110 17.051 -52.055 -59.306 1.00 63.52 C N
+ATOM 6143 CA TRP L 110 17.238 -50.991 -60.251 1.00 64.14 C C
+ATOM 6144 C TRP L 110 17.339 -49.745 -59.378 1.00 62.01 C C
+ATOM 6145 O TRP L 110 16.697 -49.708 -58.333 1.00 65.77 C O
+ATOM 6146 CB TRP L 110 16.085 -50.924 -61.239 1.00 54.69 C C
+ATOM 6147 CG TRP L 110 16.140 -51.984 -62.308 1.00 54.69 C C
+ATOM 6148 CD1 TRP L 110 16.896 -53.127 -62.310 1.00 54.69 C C
+ATOM 6149 CD2 TRP L 110 15.424 -51.979 -63.545 1.00 54.69 C C
+ATOM 6150 CE2 TRP L 110 15.775 -53.149 -64.240 1.00 54.69 C C
+ATOM 6151 CE3 TRP L 110 14.513 -51.096 -64.130 1.00 54.69 C C
+ATOM 6152 NE1 TRP L 110 16.677 -53.837 -63.469 1.00 54.69 C N
+ATOM 6153 CZ2 TRP L 110 15.251 -53.451 -65.488 1.00 54.69 C C
+ATOM 6154 CZ3 TRP L 110 13.998 -51.400 -65.362 1.00 54.69 C C
+ATOM 6155 CH2 TRP L 110 14.364 -52.566 -66.029 1.00 54.69 C C
+ATOM 6156 N PRO L 111 18.147 -48.735 -59.770 1.00 58.63 C N
+ATOM 6157 CA PRO L 111 18.969 -48.643 -60.963 1.00 58.31 C C
+ATOM 6158 C PRO L 111 20.257 -49.448 -60.826 1.00 61.47 C C
+ATOM 6159 O PRO L 111 20.856 -49.514 -59.750 1.00 62.71 C O
+ATOM 6160 CB PRO L 111 19.254 -47.153 -61.046 1.00 49.03 C C
+ATOM 6161 CG PRO L 111 19.364 -46.754 -59.689 1.00 49.03 C C
+ATOM 6162 CD PRO L 111 18.424 -47.598 -58.884 1.00 49.03 C C
+ATOM 6163 N LEU L 112 20.667 -50.056 -61.933 1.00 29.48 C N
+ATOM 6164 CA LEU L 112 21.923 -50.761 -61.975 1.00169.14 C C
+ATOM 6165 C LEU L 112 23.025 -49.753 -61.834 1.00169.72 C C
+ATOM 6166 O LEU L 112 23.045 -48.720 -62.501 1.00168.67 C O
+ATOM 6167 CB LEU L 112 22.077 -51.515 -63.276 1.00 46.33 C C
+ATOM 6168 CG LEU L 112 20.972 -52.521 -63.509 1.00 46.33 C C
+ATOM 6169 CD1 LEU L 112 21.168 -53.181 -64.847 1.00 46.33 C C
+ATOM 6170 CD2 LEU L 112 20.934 -53.515 -62.399 1.00 46.33 C C
+ATOM 6171 N THR L 113 23.911 -50.031 -60.936 1.00 74.08 C N
+ATOM 6172 CA THR L 113 25.004 -49.147 -60.704 1.00 75.36 C C
+ATOM 6173 C THR L 113 26.298 -49.885 -60.973 1.00 74.90 C C
+ATOM 6174 O THR L 113 26.322 -51.091 -61.214 1.00 76.29 C O
+ATOM 6175 CB THR L 113 24.967 -48.667 -59.273 1.00 50.85 C C
+ATOM 6176 CG2 THR L 113 23.554 -48.406 -58.885 1.00 50.85 C C
+ATOM 6177 OG1 THR L 113 25.454 -49.725 -58.448 1.00 50.85 C O
+ATOM 6178 N PHE L 114 27.389 -49.154 -60.871 1.00 56.89 C N
+ATOM 6179 CA PHE L 114 28.696 -49.713 -61.095 1.00 56.89 C C
+ATOM 6180 C PHE L 114 29.651 -49.055 -60.133 1.00 59.09 C C
+ATOM 6181 O PHE L 114 29.340 -48.049 -59.501 1.00 56.89 C O
+ATOM 6182 CB PHE L 114 29.178 -49.469 -62.515 1.00 49.74 C C
+ATOM 6183 CG PHE L 114 28.299 -50.060 -63.598 1.00 49.74 C C
+ATOM 6184 CD1 PHE L 114 28.459 -51.372 -64.006 1.00 49.74 C C
+ATOM 6185 CD2 PHE L 114 27.367 -49.277 -64.258 1.00 49.74 C C
+ATOM 6186 CE1 PHE L 114 27.688 -51.894 -65.022 1.00 49.74 C C
+ATOM 6187 CE2 PHE L 114 26.594 -49.803 -65.286 1.00 49.74 C C
+ATOM 6188 CZ PHE L 114 26.755 -51.105 -65.661 1.00 49.74 C C
+ATOM 6189 N GLY L 115 30.817 -49.643 -60.018 1.00 49.54 C N
+ATOM 6190 CA GLY L 115 31.921 -48.970 -59.389 1.00 48.13 C C
+ATOM 6191 C GLY L 115 32.587 -48.014 -60.357 1.00 48.63 C C
+ATOM 6192 O GLY L 115 32.203 -47.866 -61.523 1.00 50.71 C O
+ATOM 6193 N GLY L 116 33.615 -47.341 -59.836 1.00 52.58 C N
+ATOM 6194 CA GLY L 116 34.415 -46.477 -60.678 1.00 56.71 C C
+ATOM 6195 C GLY L 116 35.379 -47.263 -61.533 1.00 56.69 C C
+ATOM 6196 O GLY L 116 35.738 -46.839 -62.640 1.00 61.94 C O
+ATOM 6197 N GLY L 117 35.784 -48.421 -61.057 1.00 54.22 C N
+ATOM 6198 CA GLY L 117 36.707 -49.229 -61.816 1.00 52.34 C C
+ATOM 6199 C GLY L 117 38.077 -49.262 -61.175 1.00 54.54 C C
+ATOM 6200 O GLY L 117 38.454 -48.394 -60.388 1.00 60.61 C O
+ATOM 6201 N THR L 118 38.820 -50.301 -61.522 1.00 56.81 C N
+ATOM 6202 CA THR L 118 40.172 -50.511 -61.036 1.00 53.10 C C
+ATOM 6203 C THR L 118 41.043 -50.719 -62.266 1.00 54.65 C C
+ATOM 6204 O THR L 118 40.827 -51.674 -63.015 1.00 54.05 C O
+ATOM 6205 CB THR L 118 40.218 -51.729 -60.095 1.00 49.89 C C
+ATOM 6206 CG2 THR L 118 41.593 -51.925 -59.492 1.00 49.89 C C
+ATOM 6207 OG1 THR L 118 39.250 -51.581 -59.044 1.00 49.89 C O
+ATOM 6208 N GLN L 119 41.981 -49.813 -62.520 1.00 67.42 C N
+ATOM 6209 CA GLN L 119 42.937 -50.069 -63.588 1.00 68.58 C C
+ATOM 6210 C GLN L 119 43.964 -51.046 -63.040 1.00 67.82 C C
+ATOM 6211 O GLN L 119 44.469 -50.841 -61.934 1.00 73.88 C O
+ATOM 6212 CB GLN L 119 43.627 -48.779 -64.043 1.00 62.55 C C
+ATOM 6213 CG GLN L 119 44.549 -48.919 -65.298 1.00 62.55 C C
+ATOM 6214 CD GLN L 119 45.369 -47.626 -65.658 1.00 62.55 C C
+ATOM 6215 NE2 GLN L 119 45.807 -47.520 -66.929 1.00 62.55 C N
+ATOM 6216 OE1 GLN L 119 45.583 -46.742 -64.808 1.00 62.55 C O
+ATOM 6217 N VAL L 120 44.261 -52.116 -63.773 1.00 60.98 C N
+ATOM 6218 CA VAL L 120 45.419 -52.947 -63.447 1.00 60.98 C C
+ATOM 6219 C VAL L 120 46.519 -52.608 -64.439 1.00 60.98 C C
+ATOM 6220 O VAL L 120 46.412 -52.915 -65.631 1.00 64.63 C O
+ATOM 6221 CB VAL L 120 45.107 -54.449 -63.460 1.00 54.33 C C
+ATOM 6222 CG1 VAL L 120 44.382 -54.845 -62.211 1.00 54.33 C C
+ATOM 6223 CG2 VAL L 120 44.321 -54.816 -64.654 1.00 54.33 C C
+ATOM 6224 N ASN L 121 47.581 -51.964 -63.978 1.00 73.12 C N
+ATOM 6225 CA ASN L 121 48.631 -51.661 -64.928 1.00 73.12 C C
+ATOM 6226 C ASN L 121 49.766 -52.627 -64.668 1.00 73.41 C C
+ATOM 6227 O ASN L 121 49.943 -53.124 -63.552 1.00 73.12 C O
+ATOM 6228 CB ASN L 121 49.111 -50.185 -64.900 1.00 59.98 C C
+ATOM 6229 CG ASN L 121 50.015 -49.795 -63.688 1.00 59.98 C C
+ATOM 6230 ND2 ASN L 121 51.235 -49.312 -63.997 1.00 59.98 C N
+ATOM 6231 OD1 ASN L 121 49.596 -49.828 -62.525 1.00 59.98 C O
+ATOM 6232 N VAL L 122 50.472 -52.957 -65.745 1.00 65.88 C N
+ATOM 6233 CA VAL L 122 51.603 -53.865 -65.659 1.00 68.94 C C
+ATOM 6234 C VAL L 122 52.752 -53.159 -64.937 1.00 73.04 C C
+ATOM 6235 O VAL L 122 53.070 -52.001 -65.236 1.00 76.16 C O
+ATOM 6236 CB VAL L 122 51.954 -54.384 -67.070 1.00 49.22 C C
+ATOM 6237 CG1 VAL L 122 52.113 -53.256 -68.069 1.00 49.22 C C
+ATOM 6238 CG2 VAL L 122 53.196 -55.262 -67.041 1.00 49.22 C C
+ATOM 6239 N GLN L 123 53.326 -53.829 -63.921 1.00 65.32 C N
+ATOM 6240 CA GLN L 123 54.294 -53.205 -63.017 1.00 65.32 C C
+ATOM 6241 C GLN L 123 55.723 -53.434 -63.495 1.00 65.32 C C
+ATOM 6242 O GLN L 123 56.099 -54.542 -63.883 1.00 65.32 C O
+ATOM 6243 CB GLN L 123 54.154 -53.709 -61.578 1.00 65.32 C C
+ATOM 6244 CG GLN L 123 54.931 -52.847 -60.575 1.00 65.32 C C
+ATOM 6245 CD GLN L 123 54.864 -53.334 -59.103 1.00 65.32 C C
+ATOM 6246 NE2 GLN L 123 55.033 -52.402 -58.165 1.00 65.32 C N
+ATOM 6247 OE1 GLN L 123 54.670 -54.521 -58.819 1.00 65.32 C O
+ATOM 6248 N ARG L 124 56.516 -52.364 -63.426 1.00 63.16 C N
+ATOM 6249 CA ARG L 124 57.797 -52.186 -64.093 1.00 60.56 C C
+ATOM 6250 C ARG L 124 58.800 -51.687 -63.066 1.00 59.12 C C
+ATOM 6251 O ARG L 124 58.425 -51.246 -61.978 1.00 58.05 C O
+ATOM 6252 CB ARG L 124 57.678 -51.170 -65.240 1.00 60.53 C C
+ATOM 6253 CG ARG L 124 58.861 -51.136 -66.145 1.00 61.35 C C
+ATOM 6254 CD ARG L 124 59.039 -49.788 -66.755 1.00 62.56 C C
+ATOM 6255 NE ARG L 124 59.976 -49.857 -67.873 1.00 66.82 C N
+ATOM 6256 CZ ARG L 124 61.306 -49.778 -67.751 1.00 68.50 C C
+ATOM 6257 NH1 ARG L 124 61.850 -49.620 -66.541 1.00 67.78 C N1+
+ATOM 6258 NH2 ARG L 124 62.101 -49.857 -68.833 1.00 71.80 C N
+ATOM 6259 N THR L 125 60.085 -51.753 -63.415 1.00 57.81 C N
+ATOM 6260 CA THR L 125 61.113 -51.078 -62.628 1.00 59.77 C C
+ATOM 6261 C THR L 125 61.004 -49.572 -62.848 1.00 65.47 C C
+ATOM 6262 O THR L 125 60.730 -49.123 -63.964 1.00 67.25 C O
+ATOM 6263 CB THR L 125 62.494 -51.566 -63.046 1.00 62.62 C C
+ATOM 6264 CG2 THR L 125 62.392 -53.007 -63.555 1.00 62.40 C C
+ATOM 6265 OG1 THR L 125 62.976 -50.761 -64.126 1.00 64.91 C O
+ATOM 6266 N VAL L 126 61.208 -48.785 -61.788 1.00 56.60 C N
+ATOM 6267 CA VAL L 126 61.042 -47.341 -61.921 1.00 50.72 C C
+ATOM 6268 C VAL L 126 61.899 -46.834 -63.066 1.00 52.02 C C
+ATOM 6269 O VAL L 126 63.056 -47.235 -63.220 1.00 58.98 C O
+ATOM 6270 CB VAL L 126 61.416 -46.625 -60.614 1.00 50.52 C C
+ATOM 6271 CG1 VAL L 126 61.091 -45.156 -60.717 1.00 45.87 C C
+ATOM 6272 CG2 VAL L 126 60.696 -47.235 -59.462 1.00 48.98 C C
+ATOM 6273 N ALA L 127 61.332 -45.964 -63.895 1.00 49.16 C N
+ATOM 6274 CA ALA L 127 62.046 -45.432 -65.051 1.00 49.70 C C
+ATOM 6275 C ALA L 127 61.845 -43.928 -65.117 1.00 54.24 C C
+ATOM 6276 O ALA L 127 60.707 -43.459 -65.212 1.00 54.36 C O
+ATOM 6277 CB ALA L 127 61.563 -46.089 -66.336 1.00 47.87 C C
+ATOM 6278 N ALA L 128 62.941 -43.173 -65.073 1.00 49.94 C N
+ATOM 6279 CA ALA L 128 62.814 -41.726 -64.941 1.00 47.10 C C
+ATOM 6280 C ALA L 128 62.477 -41.113 -66.294 1.00 44.86 C C
+ATOM 6281 O ALA L 128 62.926 -41.617 -67.317 1.00 45.25 C O
+ATOM 6282 CB ALA L 128 64.101 -41.126 -64.397 1.00 46.93 C C
+ATOM 6283 N PRO L 129 61.676 -40.054 -66.338 1.00 45.99 C N
+ATOM 6284 CA PRO L 129 61.202 -39.532 -67.623 1.00 46.47 C C
+ATOM 6285 C PRO L 129 62.240 -38.663 -68.309 1.00 47.05 C C
+ATOM 6286 O PRO L 129 63.047 -38.001 -67.656 1.00 46.76 C O
+ATOM 6287 CB PRO L 129 59.991 -38.692 -67.211 1.00 45.22 C C
+ATOM 6288 CG PRO L 129 60.419 -38.119 -65.887 1.00 44.96 C C
+ATOM 6289 CD PRO L 129 61.306 -39.163 -65.228 1.00 46.98 C C
+ATOM 6290 N SER L 130 62.196 -38.628 -69.642 1.00 52.98 C N
+ATOM 6291 CA SER L 130 63.075 -37.720 -70.389 1.00 49.80 C C
+ATOM 6292 C SER L 130 62.328 -36.446 -70.761 1.00 53.40 C C
+ATOM 6293 O SER L 130 61.275 -36.506 -71.398 1.00 54.87 C O
+ATOM 6294 CB SER L 130 63.643 -38.376 -71.651 1.00 52.12 C C
+ATOM 6295 OG SER L 130 64.286 -39.606 -71.353 1.00 63.19 C O
+ATOM 6296 N VAL L 131 62.881 -35.298 -70.393 1.00 57.41 C N
+ATOM 6297 CA VAL L 131 62.173 -34.030 -70.510 1.00 53.50 C C
+ATOM 6298 C VAL L 131 62.611 -33.280 -71.760 1.00 58.37 C C
+ATOM 6299 O VAL L 131 63.805 -33.104 -72.015 1.00 60.24 C O
+ATOM 6300 CB VAL L 131 62.383 -33.170 -69.263 1.00 52.09 C C
+ATOM 6301 CG1 VAL L 131 61.690 -31.836 -69.438 1.00 49.82 C C
+ATOM 6302 CG2 VAL L 131 61.857 -33.909 -68.059 1.00 53.16 C C
+ATOM 6303 N PHE L 132 61.646 -32.824 -72.532 1.00 69.23 C N
+ATOM 6304 CA PHE L 132 61.909 -31.961 -73.664 1.00 65.58 C C
+ATOM 6305 C PHE L 132 60.945 -30.808 -73.607 1.00 66.20 C C
+ATOM 6306 O PHE L 132 59.760 -31.006 -73.344 1.00 65.48 C O
+ATOM 6307 CB PHE L 132 61.688 -32.639 -74.961 1.00 64.75 C C
+ATOM 6308 CG PHE L 132 62.506 -33.825 -75.151 1.00 64.68 C C
+ATOM 6309 CD1 PHE L 132 62.232 -34.984 -74.454 1.00 62.17 C C
+ATOM 6310 CD2 PHE L 132 63.533 -33.803 -76.077 1.00 63.99 C C
+ATOM 6311 CE1 PHE L 132 62.988 -36.109 -74.640 1.00 61.44 C C
+ATOM 6312 CE2 PHE L 132 64.300 -34.922 -76.292 1.00 67.40 C C
+ATOM 6313 CZ PHE L 132 64.027 -36.091 -75.560 1.00 65.54 C C
+ATOM 6314 N ILE L 133 61.428 -29.616 -73.887 1.00 67.39 C N
+ATOM 6315 CA ILE L 133 60.553 -28.465 -73.982 1.00 61.57 C C
+ATOM 6316 C ILE L 133 60.505 -28.037 -75.437 1.00 61.76 C C
+ATOM 6317 O ILE L 133 61.488 -28.174 -76.176 1.00 61.02 C O
+ATOM 6318 CB ILE L 133 61.000 -27.329 -73.048 1.00 60.98 C C
+ATOM 6319 CG1 ILE L 133 59.894 -26.305 -72.934 1.00 60.25 C C
+ATOM 6320 CG2 ILE L 133 62.265 -26.714 -73.522 1.00 58.39 C C
+ATOM 6321 CD1 ILE L 133 59.725 -25.797 -71.541 1.00 60.05 C C
+ATOM 6322 N PHE L 134 59.336 -27.589 -75.864 1.00 72.22 C N
+ATOM 6323 CA PHE L 134 59.164 -26.993 -77.164 1.00 73.11 C C
+ATOM 6324 C PHE L 134 58.576 -25.600 -76.983 1.00 75.13 C C
+ATOM 6325 O PHE L 134 57.665 -25.409 -76.153 1.00 76.78 C O
+ATOM 6326 CB PHE L 134 58.229 -27.836 -78.033 1.00 68.51 C C
+ATOM 6327 CG PHE L 134 58.685 -29.256 -78.214 1.00 70.82 C C
+ATOM 6328 CD1 PHE L 134 59.646 -29.579 -79.163 1.00 73.43 C C
+ATOM 6329 CD2 PHE L 134 58.145 -30.271 -77.449 1.00 72.32 C C
+ATOM 6330 CE1 PHE L 134 60.057 -30.895 -79.344 1.00 76.10 C C
+ATOM 6331 CE2 PHE L 134 58.555 -31.582 -77.621 1.00 75.69 C C
+ATOM 6332 CZ PHE L 134 59.509 -31.894 -78.567 1.00 75.54 C C
+ATOM 6333 N PRO L 135 59.077 -24.612 -77.723 1.00 73.15 C N
+ATOM 6334 CA PRO L 135 58.525 -23.267 -77.629 1.00 77.08 C C
+ATOM 6335 C PRO L 135 57.470 -23.080 -78.701 1.00 78.43 C C
+ATOM 6336 O PRO L 135 57.395 -23.893 -79.634 1.00 78.87 C O
+ATOM 6337 CB PRO L 135 59.752 -22.380 -77.883 1.00 78.92 C C
+ATOM 6338 CG PRO L 135 60.856 -23.357 -78.430 1.00 78.18 C C
+ATOM 6339 CD PRO L 135 60.185 -24.671 -78.688 1.00 74.57 C C
+ATOM 6340 N PRO L 136 56.701 -21.995 -78.669 1.00 79.85 C N
+ATOM 6341 CA PRO L 136 55.579 -21.881 -79.609 1.00 78.75 C C
+ATOM 6342 C PRO L 136 56.070 -21.828 -81.050 1.00 81.62 C C
+ATOM 6343 O PRO L 136 57.261 -21.672 -81.326 1.00 83.39 C O
+ATOM 6344 CB PRO L 136 54.863 -20.589 -79.183 1.00 76.77 C C
+ATOM 6345 CG PRO L 136 55.788 -19.907 -78.253 1.00 78.94 C C
+ATOM 6346 CD PRO L 136 56.622 -20.977 -77.612 1.00 78.98 C C
+ATOM 6347 N SER L 137 55.136 -22.035 -81.980 1.00 88.79 C N
+ATOM 6348 CA SER L 137 55.481 -22.097 -83.393 1.00 88.46 C C
+ATOM 6349 C SER L 137 55.623 -20.706 -83.979 1.00 89.15 C C
+ATOM 6350 O SER L 137 54.859 -19.792 -83.643 1.00 87.98 C O
+ATOM 6351 CB SER L 137 54.429 -22.867 -84.182 1.00 89.05 C C
+ATOM 6352 OG SER L 137 54.461 -24.235 -83.837 1.00 89.34 C O
+ATOM 6353 N ASP L 138 56.617 -20.548 -84.859 1.00 87.77 C N
+ATOM 6354 CA ASP L 138 56.780 -19.274 -85.543 1.00 93.67 C C
+ATOM 6355 C ASP L 138 55.497 -18.908 -86.265 1.00 93.69 C C
+ATOM 6356 O ASP L 138 54.988 -17.789 -86.120 1.00 91.43 C O
+ATOM 6357 CB ASP L 138 57.966 -19.338 -86.499 1.00 98.34 C C
+ATOM 6358 CG ASP L 138 59.265 -18.997 -85.809 1.00102.52 C C
+ATOM 6359 OD1 ASP L 138 59.254 -18.121 -84.909 1.00101.74 C O
+ATOM 6360 OD2 ASP L 138 60.292 -19.612 -86.156 1.00104.94 C O1-
+ATOM 6361 N GLU L 139 54.918 -19.874 -86.984 1.00 88.00 C N
+ATOM 6362 CA GLU L 139 53.601 -19.676 -87.584 1.00 87.61 C C
+ATOM 6363 C GLU L 139 52.556 -19.337 -86.525 1.00 82.77 C C
+ATOM 6364 O GLU L 139 51.937 -18.267 -86.564 1.00 83.77 C O
+ATOM 6365 CB GLU L 139 53.189 -20.927 -88.359 1.00 95.07 C C
+ATOM 6366 CG GLU L 139 51.714 -20.927 -88.734 1.00102.94 C C
+ATOM 6367 CD GLU L 139 51.303 -22.134 -89.556 1.00110.49 C C
+ATOM 6368 OE1 GLU L 139 52.186 -22.957 -89.907 1.00113.87 C O
+ATOM 6369 OE2 GLU L 139 50.088 -22.253 -89.842 1.00113.32 C O1-
+ATOM 6370 N GLN L 140 52.371 -20.232 -85.548 1.00 88.65 C N
+ATOM 6371 CA GLN L 140 51.340 -20.040 -84.531 1.00 87.44 C C
+ATOM 6372 C GLN L 140 51.443 -18.677 -83.869 1.00 87.59 C C
+ATOM 6373 O GLN L 140 50.442 -18.133 -83.380 1.00 88.77 C O
+ATOM 6374 CB GLN L 140 51.442 -21.125 -83.452 1.00 83.26 C C
+ATOM 6375 CG GLN L 140 50.464 -20.924 -82.294 1.00 80.48 C C
+ATOM 6376 CD GLN L 140 50.840 -21.704 -81.062 1.00 80.66 C C
+ATOM 6377 NE2 GLN L 140 51.644 -22.752 -81.243 1.00 78.45 C N
+ATOM 6378 OE1 GLN L 140 50.421 -21.366 -79.953 1.00 80.90 C O
+ATOM 6379 N LEU L 141 52.644 -18.113 -83.830 1.00 81.84 C N
+ATOM 6380 CA LEU L 141 52.798 -16.850 -83.139 1.00 85.35 C C
+ATOM 6381 C LEU L 141 52.036 -15.744 -83.848 1.00 91.16 C C
+ATOM 6382 O LEU L 141 51.581 -14.792 -83.199 1.00 94.38 C O
+ATOM 6383 CB LEU L 141 54.277 -16.533 -82.994 1.00 82.67 C C
+ATOM 6384 CG LEU L 141 54.717 -17.241 -81.716 1.00 82.93 C C
+ATOM 6385 CD1 LEU L 141 56.202 -17.133 -81.545 1.00 83.32 C C
+ATOM 6386 CD2 LEU L 141 53.981 -16.682 -80.500 1.00 83.99 C C
+ATOM 6387 N LYS L 142 51.837 -15.878 -85.164 1.00 80.28 C N
+ATOM 6388 CA LYS L 142 50.997 -14.916 -85.873 1.00 88.59 C C
+ATOM 6389 C LYS L 142 49.574 -14.915 -85.315 1.00 91.80 C C
+ATOM 6390 O LYS L 142 48.947 -13.855 -85.197 1.00 96.76 C O
+ATOM 6391 CB LYS L 142 51.008 -15.210 -87.380 1.00 95.37 C C
+ATOM 6392 CG LYS L 142 52.404 -15.161 -88.005 1.00102.36 C C
+ATOM 6393 CD LYS L 142 52.366 -15.231 -89.523 1.00106.93 C C
+ATOM 6394 CE LYS L 142 52.733 -13.885 -90.143 1.00107.84 C C
+ATOM 6395 NZ LYS L 142 52.893 -13.957 -91.621 1.00109.88 C N1+
+ATOM 6396 N SER L 143 49.079 -16.085 -84.898 1.00 89.68 C N
+ATOM 6397 CA SER L 143 47.685 -16.266 -84.503 1.00 89.28 C C
+ATOM 6398 C SER L 143 47.307 -15.549 -83.207 1.00 83.83 C C
+ATOM 6399 O SER L 143 46.114 -15.491 -82.876 1.00 87.87 C O
+ATOM 6400 CB SER L 143 47.389 -17.759 -84.363 1.00 94.84 C C
+ATOM 6401 OG SER L 143 47.809 -18.457 -85.517 1.00100.25 C O
+ATOM 6402 N GLY L 144 48.267 -15.014 -82.462 1.00 91.93 C N
+ATOM 6403 CA GLY L 144 47.928 -14.349 -81.225 1.00 89.80 C C
+ATOM 6404 C GLY L 144 47.829 -15.246 -80.010 1.00 87.82 C C
+ATOM 6405 O GLY L 144 47.457 -14.757 -78.932 1.00 88.28 C O
+ATOM 6406 N THR L 145 48.137 -16.545 -80.147 1.00 91.12 C N
+ATOM 6407 CA THR L 145 48.304 -17.450 -79.011 1.00 89.06 C C
+ATOM 6408 C THR L 145 49.634 -18.180 -79.134 1.00 86.43 C C
+ATOM 6409 O THR L 145 50.047 -18.559 -80.240 1.00 85.39 C O
+ATOM 6410 CB THR L 145 47.184 -18.512 -78.890 1.00 89.73 C C
+ATOM 6411 CG2 THR L 145 45.876 -17.891 -78.389 1.00 86.80 C C
+ATOM 6412 OG1 THR L 145 46.998 -19.189 -80.145 1.00 92.25 C O
+ATOM 6413 N ALA L 146 50.284 -18.378 -77.983 1.00 84.67 C N
+ATOM 6414 CA ALA L 146 51.526 -19.136 -77.864 1.00 80.83 C C
+ATOM 6415 C ALA L 146 51.272 -20.383 -77.024 1.00 79.09 C C
+ATOM 6416 O ALA L 146 50.799 -20.289 -75.881 1.00 80.78 C O
+ATOM 6417 CB ALA L 146 52.636 -18.283 -77.245 1.00 80.34 C C
+ATOM 6418 N SER L 147 51.574 -21.546 -77.596 1.00 75.37 C N
+ATOM 6419 CA SER L 147 51.498 -22.820 -76.895 1.00 70.83 C C
+ATOM 6420 C SER L 147 52.910 -23.307 -76.638 1.00 70.35 C C
+ATOM 6421 O SER L 147 53.655 -23.573 -77.586 1.00 74.97 C O
+ATOM 6422 CB SER L 147 50.724 -23.864 -77.702 1.00 74.20 C C
+ATOM 6423 OG SER L 147 49.326 -23.635 -77.666 1.00 75.21 C O
+ATOM 6424 N VAL L 148 53.275 -23.422 -75.369 1.00 69.31 C N
+ATOM 6425 CA VAL L 148 54.533 -24.039 -74.973 1.00 67.27 C C
+ATOM 6426 C VAL L 148 54.245 -25.467 -74.555 1.00 69.08 C C
+ATOM 6427 O VAL L 148 53.286 -25.722 -73.812 1.00 69.50 C O
+ATOM 6428 CB VAL L 148 55.190 -23.275 -73.824 1.00 67.19 C C
+ATOM 6429 CG1 VAL L 148 56.614 -23.756 -73.641 1.00 64.45 C C
+ATOM 6430 CG2 VAL L 148 55.121 -21.822 -74.117 1.00 65.75 C C
+ATOM 6431 N VAL L 149 55.072 -26.405 -75.002 1.00 64.98 C N
+ATOM 6432 CA VAL L 149 54.762 -27.811 -74.785 1.00 63.76 C C
+ATOM 6433 C VAL L 149 55.935 -28.496 -74.100 1.00 66.57 C C
+ATOM 6434 O VAL L 149 57.012 -28.629 -74.686 1.00 66.50 C O
+ATOM 6435 CB VAL L 149 54.383 -28.519 -76.090 1.00 65.78 C C
+ATOM 6436 CG1 VAL L 149 54.540 -30.021 -75.946 1.00 62.38 C C
+ATOM 6437 CG2 VAL L 149 52.929 -28.188 -76.433 1.00 61.66 C C
+ATOM 6438 N CYS L 150 55.719 -28.946 -72.868 1.00 54.24 C N
+ATOM 6439 CA CYS L 150 56.676 -29.779 -72.157 1.00 56.83 C C
+ATOM 6440 C CYS L 150 56.287 -31.250 -72.273 1.00 60.70 C C
+ATOM 6441 O CYS L 150 55.128 -31.616 -72.043 1.00 60.50 C O
+ATOM 6442 CB CYS L 150 56.761 -29.375 -70.691 1.00 55.53 C C
+ATOM 6443 SG CYS L 150 58.188 -30.087 -69.890 1.00 67.63 C S
+ATOM 6444 N LEU L 151 57.269 -32.081 -72.608 1.00 56.81 C N
+ATOM 6445 CA LEU L 151 57.107 -33.506 -72.860 1.00 53.94 C C
+ATOM 6446 C LEU L 151 57.935 -34.307 -71.867 1.00 53.38 C C
+ATOM 6447 O LEU L 151 59.100 -33.982 -71.624 1.00 57.50 C O
+ATOM 6448 CB LEU L 151 57.536 -33.846 -74.281 1.00 56.15 C C
+ATOM 6449 CG LEU L 151 57.966 -35.282 -74.552 1.00 54.85 C C
+ATOM 6450 CD1 LEU L 151 56.813 -36.207 -74.334 1.00 53.32 C C
+ATOM 6451 CD2 LEU L 151 58.441 -35.418 -75.992 1.00 53.45 C C
+ATOM 6452 N LEU L 152 57.328 -35.344 -71.292 1.00 40.29 C N
+ATOM 6453 CA LEU L 152 57.994 -36.294 -70.413 1.00 44.04 C C
+ATOM 6454 C LEU L 152 57.858 -37.654 -71.068 1.00 49.15 C C
+ATOM 6455 O LEU L 152 56.740 -38.144 -71.259 1.00 49.05 C O
+ATOM 6456 CB LEU L 152 57.379 -36.306 -69.015 1.00 43.88 C C
+ATOM 6457 CG LEU L 152 57.364 -35.028 -68.165 1.00 49.60 C C
+ATOM 6458 CD1 LEU L 152 56.577 -33.888 -68.776 1.00 51.51 C C
+ATOM 6459 CD2 LEU L 152 56.861 -35.326 -66.770 1.00 48.38 C C
+ATOM 6460 N ASN L 153 58.979 -38.257 -71.438 1.00 54.58 C N
+ATOM 6461 CA ASN L 153 58.939 -39.435 -72.289 1.00 52.84 C C
+ATOM 6462 C ASN L 153 59.379 -40.697 -71.562 1.00 56.44 C C
+ATOM 6463 O ASN L 153 60.464 -40.741 -70.950 1.00 58.11 C O
+ATOM 6464 CB ASN L 153 59.774 -39.247 -73.544 1.00 54.47 C C
+ATOM 6465 CG ASN L 153 59.082 -39.806 -74.756 1.00 57.51 C C
+ATOM 6466 ND2 ASN L 153 59.433 -39.295 -75.926 1.00 56.95 C N
+ATOM 6467 OD1 ASN L 153 58.200 -40.665 -74.639 1.00 60.68 C O
+ATOM 6468 N ASN L 154 58.486 -41.695 -71.622 1.00 62.60 C N
+ATOM 6469 CA ASN L 154 58.740 -43.104 -71.336 1.00 61.26 C C
+ATOM 6470 C ASN L 154 59.209 -43.301 -69.893 1.00 61.31 C C
+ATOM 6471 O ASN L 154 60.355 -43.634 -69.611 1.00 65.47 C O
+ATOM 6472 CB ASN L 154 59.728 -43.656 -72.373 1.00 60.48 C C
+ATOM 6473 CG ASN L 154 59.068 -43.913 -73.729 1.00 64.43 C C
+ATOM 6474 ND2 ASN L 154 59.876 -44.244 -74.728 1.00 72.38 C N
+ATOM 6475 OD1 ASN L 154 57.854 -43.778 -73.878 1.00 62.49 C O
+ATOM 6476 N PHE L 155 58.280 -43.078 -68.976 1.00 59.63 C N
+ATOM 6477 CA PHE L 155 58.606 -43.156 -67.565 1.00 55.45 C C
+ATOM 6478 C PHE L 155 57.670 -44.145 -66.894 1.00 56.91 C C
+ATOM 6479 O PHE L 155 56.778 -44.709 -67.534 1.00 56.56 C O
+ATOM 6480 CB PHE L 155 58.520 -41.782 -66.901 1.00 53.65 C C
+ATOM 6481 CG PHE L 155 57.171 -41.139 -67.005 1.00 53.58 C C
+ATOM 6482 CD1 PHE L 155 56.891 -40.245 -68.025 1.00 52.65 C C
+ATOM 6483 CD2 PHE L 155 56.189 -41.394 -66.054 1.00 51.06 C C
+ATOM 6484 CE1 PHE L 155 55.649 -39.637 -68.108 1.00 49.77 C C
+ATOM 6485 CE2 PHE L 155 54.945 -40.791 -66.134 1.00 51.29 C C
+ATOM 6486 CZ PHE L 155 54.675 -39.911 -67.163 1.00 50.47 C C
+ATOM 6487 N TYR L 156 57.850 -44.302 -65.580 1.00 55.88 C N
+ATOM 6488 CA TYR L 156 57.087 -45.221 -64.744 1.00 51.50 C C
+ATOM 6489 C TYR L 156 57.524 -44.947 -63.332 1.00 55.18 C C
+ATOM 6490 O TYR L 156 58.712 -44.818 -63.077 1.00 58.87 C O
+ATOM 6491 CB TYR L 156 57.352 -46.698 -65.084 1.00 53.64 C C
+ATOM 6492 CG TYR L 156 56.417 -47.608 -64.350 1.00 51.07 C C
+ATOM 6493 CD1 TYR L 156 56.587 -47.846 -63.010 1.00 51.51 C C
+ATOM 6494 CD2 TYR L 156 55.356 -48.210 -64.987 1.00 47.95 C C
+ATOM 6495 CE1 TYR L 156 55.721 -48.618 -62.289 1.00 55.22 C C
+ATOM 6496 CE2 TYR L 156 54.469 -49.017 -64.280 1.00 47.04 C C
+ATOM 6497 CZ TYR L 156 54.660 -49.221 -62.906 1.00 53.11 C C
+ATOM 6498 OH TYR L 156 53.811 -50.017 -62.119 1.00 59.24 C O
+ATOM 6499 N PRO L 157 56.573 -44.886 -62.394 1.00 57.81 C N
+ATOM 6500 CA PRO L 157 55.114 -44.965 -62.579 1.00 56.71 C C
+ATOM 6501 C PRO L 157 54.452 -43.663 -63.090 1.00 55.01 C C
+ATOM 6502 O PRO L 157 55.156 -42.660 -63.256 1.00 54.11 C O
+ATOM 6503 CB PRO L 157 54.612 -45.314 -61.179 1.00 58.08 C C
+ATOM 6504 CG PRO L 157 55.670 -44.806 -60.261 1.00 58.63 C C
+ATOM 6505 CD PRO L 157 56.965 -44.937 -60.976 1.00 56.96 C C
+ATOM 6506 N ARG L 158 53.127 -43.688 -63.323 1.00 55.40 C N
+ATOM 6507 CA ARG L 158 52.440 -42.572 -63.983 1.00 59.54 C C
+ATOM 6508 C ARG L 158 52.581 -41.263 -63.194 1.00 64.01 C C
+ATOM 6509 O ARG L 158 52.741 -40.189 -63.789 1.00 65.29 C O
+ATOM 6510 CB ARG L 158 50.965 -42.945 -64.212 1.00 61.82 C C
+ATOM 6511 CG ARG L 158 50.007 -41.799 -64.589 1.00 69.97 C C
+ATOM 6512 CD ARG L 158 48.575 -42.303 -64.923 1.00 79.15 C C
+ATOM 6513 NE ARG L 158 47.614 -41.215 -65.184 1.00 85.09 C N
+ATOM 6514 CZ ARG L 158 46.332 -41.385 -65.532 1.00 89.37 C C
+ATOM 6515 NH1 ARG L 158 45.841 -42.614 -65.682 1.00 91.79 C N1+
+ATOM 6516 NH2 ARG L 158 45.537 -40.327 -65.733 1.00 91.49 C N
+ATOM 6517 N GLU L 159 52.574 -41.331 -61.861 1.00 60.26 C N
+ATOM 6518 CA GLU L 159 52.538 -40.121 -61.044 1.00 61.22 C C
+ATOM 6519 C GLU L 159 53.781 -39.279 -61.282 1.00 62.42 C C
+ATOM 6520 O GLU L 159 54.902 -39.754 -61.093 1.00 61.31 C O
+ATOM 6521 CB GLU L 159 52.429 -40.501 -59.569 1.00 67.91 C C
+ATOM 6522 CG GLU L 159 51.181 -41.305 -59.246 1.00 76.15 C C
+ATOM 6523 CD GLU L 159 51.244 -42.774 -59.695 1.00 77.54 C C
+ATOM 6524 OE1 GLU L 159 51.895 -43.072 -60.727 1.00 78.79 C O
+ATOM 6525 OE2 GLU L 159 50.593 -43.631 -59.044 1.00 76.49 C O1-
+ATOM 6526 N ALA L 160 53.586 -38.030 -61.705 1.00 58.15 C N
+ATOM 6527 CA ALA L 160 54.685 -37.084 -61.861 1.00 54.78 C C
+ATOM 6528 C ALA L 160 54.135 -35.675 -61.736 1.00 56.70 C C
+ATOM 6529 O ALA L 160 52.999 -35.412 -62.132 1.00 57.72 C O
+ATOM 6530 CB ALA L 160 55.391 -37.257 -63.203 1.00 51.38 C C
+ATOM 6531 N LYS L 161 54.936 -34.762 -61.192 1.00 58.93 C N
+ATOM 6532 CA LYS L 161 54.513 -33.369 -61.074 1.00 62.59 C C
+ATOM 6533 C LYS L 161 55.238 -32.548 -62.131 1.00 57.75 C C
+ATOM 6534 O LYS L 161 56.472 -32.434 -62.098 1.00 53.73 C O
+ATOM 6535 CB LYS L 161 54.755 -32.801 -59.669 1.00 71.17 C C
+ATOM 6536 CG LYS L 161 53.439 -32.646 -58.862 1.00 79.91 C C
+ATOM 6537 CD LYS L 161 53.560 -32.053 -57.430 1.00 88.90 C C
+ATOM 6538 CE LYS L 161 52.209 -32.220 -56.679 1.00 95.67 C C
+ATOM 6539 NZ LYS L 161 52.206 -31.795 -55.247 1.00100.21 C N1+
+ATOM 6540 N VAL L 162 54.475 -32.005 -63.083 1.00 55.15 C N
+ATOM 6541 CA VAL L 162 54.986 -31.019 -64.028 1.00 57.76 C C
+ATOM 6542 C VAL L 162 54.622 -29.639 -63.511 1.00 56.99 C C
+ATOM 6543 O VAL L 162 53.471 -29.395 -63.127 1.00 64.47 C O
+ATOM 6544 CB VAL L 162 54.431 -31.239 -65.440 1.00 55.82 C C
+ATOM 6545 CG1 VAL L 162 54.788 -30.071 -66.294 1.00 55.38 C C
+ATOM 6546 CG2 VAL L 162 55.039 -32.474 -66.039 1.00 57.51 C C
+ATOM 6547 N GLN L 163 55.599 -28.743 -63.497 1.00 65.52 C N
+ATOM 6548 CA GLN L 163 55.456 -27.402 -62.961 1.00 65.82 C C
+ATOM 6549 C GLN L 163 55.869 -26.397 -64.030 1.00 61.33 C C
+ATOM 6550 O GLN L 163 56.763 -26.676 -64.838 1.00 57.85 C O
+ATOM 6551 CB GLN L 163 56.321 -27.250 -61.734 1.00 61.48 C C
+ATOM 6552 CG GLN L 163 55.892 -26.171 -60.822 1.00 58.82 C C
+ATOM 6553 CD GLN L 163 55.129 -26.688 -59.618 1.00 68.00 C C
+ATOM 6554 NE2 GLN L 163 53.791 -26.655 -59.688 1.00 70.59 C N
+ATOM 6555 OE1 GLN L 163 55.740 -27.069 -58.610 1.00 74.20 C O
+ATOM 6556 N TRP L 164 55.206 -25.233 -64.064 1.00 63.57 C N
+ATOM 6557 CA TRP L 164 55.552 -24.191 -65.027 1.00 62.00 C C
+ATOM 6558 C TRP L 164 55.978 -22.936 -64.292 1.00 65.09 C C
+ATOM 6559 O TRP L 164 55.327 -22.520 -63.327 1.00 68.66 C O
+ATOM 6560 CB TRP L 164 54.396 -23.839 -65.948 1.00 57.49 C C
+ATOM 6561 CG TRP L 164 54.143 -24.814 -67.010 1.00 57.19 C C
+ATOM 6562 CD1 TRP L 164 53.206 -25.791 -67.000 1.00 57.38 C C
+ATOM 6563 CD2 TRP L 164 54.812 -24.908 -68.263 1.00 56.45 C C
+ATOM 6564 CE2 TRP L 164 54.229 -25.979 -68.967 1.00 60.17 C C
+ATOM 6565 CE3 TRP L 164 55.844 -24.195 -68.861 1.00 56.80 C C
+ATOM 6566 NE1 TRP L 164 53.249 -26.501 -68.170 1.00 59.87 C N
+ATOM 6567 CZ2 TRP L 164 54.650 -26.363 -70.245 1.00 62.64 C C
+ATOM 6568 CZ3 TRP L 164 56.264 -24.573 -70.143 1.00 58.70 C C
+ATOM 6569 CH2 TRP L 164 55.666 -25.649 -70.816 1.00 60.49 C C
+ATOM 6570 N LYS L 165 57.072 -22.333 -64.752 1.00 72.93 C N
+ATOM 6571 CA LYS L 165 57.493 -21.027 -64.256 1.00 71.37 C C
+ATOM 6572 C LYS L 165 57.913 -20.158 -65.431 1.00 73.57 C C
+ATOM 6573 O LYS L 165 58.833 -20.512 -66.181 1.00 72.15 C O
+ATOM 6574 CB LYS L 165 58.627 -21.156 -63.235 1.00 70.05 C C
+ATOM 6575 CG LYS L 165 58.225 -21.915 -61.970 1.00 71.20 C C
+ATOM 6576 CD LYS L 165 59.355 -21.965 -60.957 1.00 77.05 C C
+ATOM 6577 CE LYS L 165 59.804 -23.384 -60.721 1.00 88.31 C C
+ATOM 6578 NZ LYS L 165 60.492 -23.500 -59.420 1.00 92.93 C N1+
+ATOM 6579 N VAL L 166 57.218 -19.039 -65.610 1.00 84.56 C N
+ATOM 6580 CA VAL L 166 57.632 -18.028 -66.574 1.00 81.90 C C
+ATOM 6581 C VAL L 166 58.434 -16.974 -65.819 1.00 84.43 C C
+ATOM 6582 O VAL L 166 58.011 -16.502 -64.748 1.00 91.03 C O
+ATOM 6583 CB VAL L 166 56.425 -17.417 -67.324 1.00 81.87 C C
+ATOM 6584 CG1 VAL L 166 55.536 -18.499 -67.902 1.00 83.98 C C
+ATOM 6585 CG2 VAL L 166 55.612 -16.476 -66.448 1.00 75.78 C C
+ATOM 6586 N ASP L 167 59.625 -16.648 -66.347 1.00 84.82 C N
+ATOM 6587 CA ASP L 167 60.571 -15.765 -65.663 1.00 85.31 C C
+ATOM 6588 C ASP L 167 60.661 -16.166 -64.204 1.00 84.99 C C
+ATOM 6589 O ASP L 167 60.681 -15.331 -63.297 1.00 88.22 C O
+ATOM 6590 CB ASP L 167 60.197 -14.290 -65.815 1.00 85.01 C C
+ATOM 6591 CG ASP L 167 60.560 -13.739 -67.183 1.00 88.74 C C
+ATOM 6592 OD1 ASP L 167 61.425 -14.341 -67.868 1.00 87.84 C O
+ATOM 6593 OD2 ASP L 167 59.966 -12.709 -67.577 1.00 90.62 C O1-
+ATOM 6594 N ASN L 168 60.628 -17.478 -63.996 1.00 83.48 C N
+ATOM 6595 CA ASN L 168 60.882 -18.121 -62.725 1.00 87.61 C C
+ATOM 6596 C ASN L 168 59.819 -17.793 -61.670 1.00 90.86 C C
+ATOM 6597 O ASN L 168 60.075 -17.937 -60.471 1.00 91.57 C O
+ATOM 6598 CB ASN L 168 62.291 -17.730 -62.257 1.00 95.78 C C
+ATOM 6599 CG ASN L 168 63.120 -18.915 -61.837 1.00101.13 C C
+ATOM 6600 ND2 ASN L 168 64.406 -18.897 -62.212 1.00100.16 C N
+ATOM 6601 OD1 ASN L 168 62.613 -19.856 -61.211 1.00105.13 C O
+ATOM 6602 N ALA L 169 58.615 -17.383 -62.089 1.00 80.25 C N
+ATOM 6603 CA ALA L 169 57.456 -17.248 -61.206 1.00 89.58 C C
+ATOM 6604 C ALA L 169 56.416 -18.309 -61.563 1.00 86.20 C C
+ATOM 6605 O ALA L 169 56.210 -18.622 -62.744 1.00 85.08 C O
+ATOM 6606 CB ALA L 169 56.846 -15.849 -61.298 1.00 90.42 C C
+ATOM 6607 N LEU L 170 55.757 -18.858 -60.541 1.00 78.33 C N
+ATOM 6608 CA LEU L 170 55.029 -20.117 -60.689 1.00 75.96 C C
+ATOM 6609 C LEU L 170 53.658 -19.907 -61.336 1.00 79.19 C C
+ATOM 6610 O LEU L 170 52.789 -19.235 -60.767 1.00 80.68 C O
+ATOM 6611 CB LEU L 170 54.865 -20.783 -59.327 1.00 77.16 C C
+ATOM 6612 CG LEU L 170 54.175 -22.127 -59.486 1.00 75.23 C C
+ATOM 6613 CD1 LEU L 170 55.075 -22.971 -60.315 1.00 74.75 C C
+ATOM 6614 CD2 LEU L 170 53.877 -22.775 -58.154 1.00 73.52 C C
+ATOM 6615 N GLN L 171 53.439 -20.533 -62.492 1.00 81.00 C N
+ATOM 6616 CA GLN L 171 52.193 -20.372 -63.230 1.00 80.76 C C
+ATOM 6617 C GLN L 171 51.147 -21.343 -62.713 1.00 81.75 C C
+ATOM 6618 O GLN L 171 51.459 -22.495 -62.398 1.00 81.42 C O
+ATOM 6619 CB GLN L 171 52.418 -20.621 -64.716 1.00 77.44 C C
+ATOM 6620 CG GLN L 171 53.477 -19.744 -65.292 1.00 81.09 C C
+ATOM 6621 CD GLN L 171 53.030 -18.316 -65.365 1.00 83.95 C C
+ATOM 6622 NE2 GLN L 171 52.174 -18.014 -66.336 1.00 79.29 C N
+ATOM 6623 OE1 GLN L 171 53.447 -17.481 -64.561 1.00 89.03 C O
+ATOM 6624 N SER L 172 49.898 -20.882 -62.644 1.00 83.45 C N
+ATOM 6625 CA SER L 172 48.804 -21.705 -62.133 1.00 85.46 C C
+ATOM 6626 C SER L 172 47.555 -21.517 -62.989 1.00 85.69 C C
+ATOM 6627 O SER L 172 47.046 -20.400 -63.119 1.00 82.50 C O
+ATOM 6628 CB SER L 172 48.509 -21.366 -60.664 1.00 83.63 C C
+ATOM 6629 OG SER L 172 47.715 -22.365 -60.049 1.00 83.79 C O
+ATOM 6630 N GLY L 173 47.050 -22.609 -63.554 1.00 80.76 C N
+ATOM 6631 CA GLY L 173 45.739 -22.625 -64.161 1.00 69.41 C C
+ATOM 6632 C GLY L 173 45.719 -22.504 -65.664 1.00 73.80 C C
+ATOM 6633 O GLY L 173 44.655 -22.689 -66.262 1.00 78.98 C O
+ATOM 6634 N ASN L 174 46.840 -22.165 -66.285 1.00 73.00 C N
+ATOM 6635 CA ASN L 174 46.935 -22.007 -67.731 1.00 73.41 C C
+ATOM 6636 C ASN L 174 47.511 -23.226 -68.465 1.00 71.25 C C
+ATOM 6637 O ASN L 174 47.785 -23.145 -69.672 1.00 78.44 C O
+ATOM 6638 CB ASN L 174 47.732 -20.744 -68.045 1.00 65.27 C C
+ATOM 6639 CG ASN L 174 48.893 -20.556 -67.099 1.00 73.90 C C
+ATOM 6640 ND2 ASN L 174 49.840 -19.684 -67.475 1.00 70.92 C N
+ATOM 6641 OD1 ASN L 174 48.966 -21.231 -66.057 1.00 76.90 C O
+ATOM 6642 N SER L 175 47.721 -24.338 -67.767 1.00 65.87 C N
+ATOM 6643 CA SER L 175 48.299 -25.542 -68.346 1.00 66.75 C C
+ATOM 6644 C SER L 175 47.301 -26.696 -68.361 1.00 67.96 C C
+ATOM 6645 O SER L 175 46.433 -26.801 -67.486 1.00 68.44 C O
+ATOM 6646 CB SER L 175 49.523 -25.973 -67.545 1.00 65.79 C C
+ATOM 6647 OG SER L 175 49.093 -26.664 -66.380 1.00 68.62 C O
+ATOM 6648 N GLN L 176 47.459 -27.591 -69.344 1.00 57.40 C N
+ATOM 6649 CA GLN L 176 46.699 -28.840 -69.403 1.00 59.71 C C
+ATOM 6650 C GLN L 176 47.620 -30.032 -69.635 1.00 58.95 C C
+ATOM 6651 O GLN L 176 48.604 -29.939 -70.378 1.00 59.30 C O
+ATOM 6652 CB GLN L 176 45.651 -28.811 -70.502 1.00 62.72 C C
+ATOM 6653 CG GLN L 176 44.266 -28.468 -70.024 1.00 62.19 C C
+ATOM 6654 CD GLN L 176 43.424 -27.921 -71.153 1.00 66.07 C C
+ATOM 6655 NE2 GLN L 176 42.199 -27.502 -70.840 1.00 67.37 C N
+ATOM 6656 OE1 GLN L 176 43.872 -27.880 -72.302 1.00 71.31 C O
+ATOM 6657 N GLU L 177 47.279 -31.158 -69.007 1.00 57.54 C N
+ATOM 6658 CA GLU L 177 48.076 -32.379 -69.070 1.00 55.42 C C
+ATOM 6659 C GLU L 177 47.263 -33.489 -69.718 1.00 56.57 C C
+ATOM 6660 O GLU L 177 46.116 -33.735 -69.329 1.00 57.04 C O
+ATOM 6661 CB GLU L 177 48.534 -32.822 -67.666 1.00 56.24 C C
+ATOM 6662 CG GLU L 177 49.709 -32.033 -67.074 1.00 66.12 C C
+ATOM 6663 CD GLU L 177 50.239 -32.634 -65.765 1.00 70.38 C C
+ATOM 6664 OE1 GLU L 177 49.999 -33.842 -65.512 1.00 73.83 C O
+ATOM 6665 OE2 GLU L 177 50.899 -31.891 -64.990 1.00 69.86 C O1-
+ATOM 6666 N SER L 178 47.861 -34.160 -70.687 1.00 57.18 C N
+ATOM 6667 CA SER L 178 47.337 -35.406 -71.220 1.00 53.57 C C
+ATOM 6668 C SER L 178 48.407 -36.478 -71.048 1.00 55.52 C C
+ATOM 6669 O SER L 178 49.594 -36.156 -70.986 1.00 57.67 C O
+ATOM 6670 CB SER L 178 46.954 -35.233 -72.691 1.00 54.89 C C
+ATOM 6671 OG SER L 178 46.741 -36.468 -73.354 1.00 60.85 C O
+ATOM 6672 N VAL L 179 48.015 -37.754 -70.948 1.00 50.81 C N
+ATOM 6673 CA VAL L 179 48.983 -38.799 -70.616 1.00 47.83 C C
+ATOM 6674 C VAL L 179 48.703 -40.063 -71.428 1.00 52.96 C C
+ATOM 6675 O VAL L 179 47.548 -40.463 -71.607 1.00 55.58 C O
+ATOM 6676 CB VAL L 179 48.979 -39.084 -69.097 1.00 52.82 C C
+ATOM 6677 CG1 VAL L 179 47.767 -39.916 -68.676 1.00 59.38 C C
+ATOM 6678 CG2 VAL L 179 50.261 -39.723 -68.681 1.00 53.60 C C
+ATOM 6679 N THR L 180 49.775 -40.712 -71.888 1.00 59.86 C N
+ATOM 6680 CA THR L 180 49.681 -41.941 -72.671 1.00 60.14 C C
+ATOM 6681 C THR L 180 48.992 -43.027 -71.871 1.00 58.81 C C
+ATOM 6682 O THR L 180 48.967 -43.009 -70.640 1.00 58.53 C O
+ATOM 6683 CB THR L 180 51.070 -42.485 -73.031 1.00 59.73 C C
+ATOM 6684 CG2 THR L 180 51.022 -43.399 -74.245 1.00 58.56 C C
+ATOM 6685 OG1 THR L 180 51.962 -41.408 -73.305 1.00 61.76 C O
+ATOM 6686 N GLU L 181 48.444 -43.994 -72.577 1.00 72.48 C N
+ATOM 6687 CA GLU L 181 48.127 -45.242 -71.915 1.00 74.40 C C
+ATOM 6688 C GLU L 181 49.369 -46.120 -71.941 1.00 73.16 C C
+ATOM 6689 O GLU L 181 50.162 -46.066 -72.886 1.00 72.90 C O
+ATOM 6690 CB GLU L 181 46.932 -45.926 -72.579 1.00 73.33 C C
+ATOM 6691 CG GLU L 181 45.646 -45.073 -72.563 1.00 76.81 C C
+ATOM 6692 CD GLU L 181 45.251 -44.523 -71.143 1.00 83.32 C C
+ATOM 6693 OE1 GLU L 181 45.618 -45.151 -70.091 1.00 86.28 C O
+ATOM 6694 OE2 GLU L 181 44.567 -43.447 -71.099 1.00 87.66 C O1-
+ATOM 6695 N GLN L 182 49.557 -46.900 -70.876 1.00 63.48 C N
+ATOM 6696 CA GLN L 182 50.826 -47.593 -70.660 1.00 65.25 C C
+ATOM 6697 C GLN L 182 51.217 -48.428 -71.865 1.00 69.06 C C
+ATOM 6698 O GLN L 182 50.447 -49.268 -72.326 1.00 72.85 C O
+ATOM 6699 CB GLN L 182 50.716 -48.471 -69.418 1.00 64.08 C C
+ATOM 6700 CG GLN L 182 52.019 -49.127 -69.004 1.00 69.45 C C
+ATOM 6701 CD GLN L 182 51.861 -49.986 -67.753 1.00 68.38 C C
+ATOM 6702 NE2 GLN L 182 52.964 -50.215 -67.043 1.00 70.49 C N
+ATOM 6703 OE1 GLN L 182 50.759 -50.421 -67.422 1.00 67.83 C O
+ATOM 6704 N ASP L 183 52.414 -48.196 -72.381 1.00 67.64 C N
+ATOM 6705 CA ASP L 183 52.788 -48.811 -73.647 1.00 73.20 C C
+ATOM 6706 C ASP L 183 52.917 -50.325 -73.523 1.00 81.44 C C
+ATOM 6707 O ASP L 183 53.289 -50.859 -72.478 1.00 82.56 C O
+ATOM 6708 CB ASP L 183 54.098 -48.224 -74.165 1.00 72.53 C C
+ATOM 6709 CG ASP L 183 54.544 -48.865 -75.467 1.00 78.54 C C
+ATOM 6710 OD1 ASP L 183 53.698 -49.504 -76.137 1.00 78.98 C O
+ATOM 6711 OD2 ASP L 183 55.727 -48.724 -75.840 1.00 83.34 C O1-
+ATOM 6712 N SER L 184 52.618 -51.015 -74.626 1.00 78.05 C N
+ATOM 6713 CA SER L 184 52.684 -52.476 -74.651 1.00 81.42 C C
+ATOM 6714 C SER L 184 54.114 -52.989 -74.457 1.00 82.87 C C
+ATOM 6715 O SER L 184 54.361 -53.849 -73.600 1.00 88.74 C O
+ATOM 6716 CB SER L 184 52.114 -52.987 -75.969 1.00 81.52 C C
+ATOM 6717 OG SER L 184 52.891 -52.500 -77.050 1.00 83.64 C O
+ATOM 6718 N LYS L 185 55.065 -52.488 -75.258 1.00 82.92 C N
+ATOM 6719 CA LYS L 185 56.428 -53.019 -75.236 1.00 85.93 C C
+ATOM 6720 C LYS L 185 57.134 -52.741 -73.911 1.00 79.41 C C
+ATOM 6721 O LYS L 185 57.562 -53.674 -73.223 1.00 82.24 C O
+ATOM 6722 CB LYS L 185 57.243 -52.444 -76.404 1.00 94.41 C C
+ATOM 6723 CG LYS L 185 57.151 -53.185 -77.763 1.00104.03 C C
+ATOM 6724 CD LYS L 185 57.929 -54.521 -77.700 1.00111.84 C C
+ATOM 6725 CE LYS L 185 57.918 -55.309 -79.015 1.00116.41 C C
+ATOM 6726 NZ LYS L 185 57.976 -56.799 -78.807 1.00120.21 C N1+
+ATOM 6727 N ASP L 186 57.290 -51.463 -73.545 1.00 74.79 C N
+ATOM 6728 CA ASP L 186 58.119 -51.075 -72.397 1.00 72.91 C C
+ATOM 6729 C ASP L 186 57.343 -50.731 -71.132 1.00 73.07 C C
+ATOM 6730 O ASP L 186 57.961 -50.366 -70.135 1.00 75.39 C O
+ATOM 6731 CB ASP L 186 59.015 -49.890 -72.763 1.00 73.30 C C
+ATOM 6732 CG ASP L 186 58.232 -48.715 -73.277 1.00 76.16 C C
+ATOM 6733 OD1 ASP L 186 57.028 -48.902 -73.549 1.00 72.17 C O
+ATOM 6734 OD2 ASP L 186 58.826 -47.623 -73.442 1.00 82.31 C O1-
+ATOM 6735 N SER L 187 56.019 -50.790 -71.160 1.00 68.17 C N
+ATOM 6736 CA SER L 187 55.184 -50.492 -69.997 1.00 66.19 C C
+ATOM 6737 C SER L 187 55.515 -49.131 -69.393 1.00 68.82 C C
+ATOM 6738 O SER L 187 55.657 -48.983 -68.175 1.00 71.69 C O
+ATOM 6739 CB SER L 187 55.300 -51.592 -68.947 1.00 67.09 C C
+ATOM 6740 OG SER L 187 54.913 -52.833 -69.495 1.00 75.41 C O
+ATOM 6741 N THR L 188 55.620 -48.117 -70.254 1.00 66.91 C N
+ATOM 6742 CA THR L 188 55.916 -46.759 -69.815 1.00 65.15 C C
+ATOM 6743 C THR L 188 54.830 -45.780 -70.234 1.00 67.36 C C
+ATOM 6744 O THR L 188 54.465 -45.694 -71.417 1.00 69.35 C O
+ATOM 6745 CB THR L 188 57.244 -46.262 -70.369 1.00 67.46 C C
+ATOM 6746 CG2 THR L 188 58.315 -47.324 -70.197 1.00 72.49 C C
+ATOM 6747 OG1 THR L 188 57.077 -45.937 -71.759 1.00 63.36 C O
+ATOM 6748 N TYR L 189 54.362 -45.011 -69.264 1.00 61.68 C N
+ATOM 6749 CA TYR L 189 53.599 -43.812 -69.551 1.00 58.86 C C
+ATOM 6750 C TYR L 189 54.502 -42.732 -70.163 1.00 58.34 C C
+ATOM 6751 O TYR L 189 55.722 -42.720 -69.953 1.00 64.10 C O
+ATOM 6752 CB TYR L 189 52.955 -43.315 -68.260 1.00 55.85 C C
+ATOM 6753 CG TYR L 189 52.132 -44.380 -67.593 1.00 55.81 C C
+ATOM 6754 CD1 TYR L 189 50.780 -44.493 -67.846 1.00 58.57 C C
+ATOM 6755 CD2 TYR L 189 52.716 -45.301 -66.748 1.00 57.76 C C
+ATOM 6756 CE1 TYR L 189 50.021 -45.484 -67.249 1.00 59.40 C C
+ATOM 6757 CE2 TYR L 189 51.977 -46.295 -66.149 1.00 57.51 C C
+ATOM 6758 CZ TYR L 189 50.629 -46.384 -66.399 1.00 64.66 C C
+ATOM 6759 OH TYR L 189 49.881 -47.373 -65.804 1.00 74.23 C O
+ATOM 6760 N SER L 190 53.892 -41.840 -70.964 1.00 55.97 C N
+ATOM 6761 CA SER L 190 54.480 -40.554 -71.347 1.00 57.42 C C
+ATOM 6762 C SER L 190 53.413 -39.468 -71.214 1.00 54.95 C C
+ATOM 6763 O SER L 190 52.211 -39.742 -71.307 1.00 58.50 C O
+ATOM 6764 CB SER L 190 55.039 -40.570 -72.767 1.00 56.85 C C
+ATOM 6765 OG SER L 190 55.585 -41.842 -73.057 1.00 57.96 C O
+ATOM 6766 N LEU L 191 53.848 -38.231 -70.978 1.00 51.38 C N
+ATOM 6767 CA LEU L 191 52.933 -37.173 -70.560 1.00 51.76 C C
+ATOM 6768 C LEU L 191 53.238 -35.894 -71.321 1.00 52.11 C C
+ATOM 6769 O LEU L 191 54.402 -35.547 -71.525 1.00 52.75 C O
+ATOM 6770 CB LEU L 191 53.028 -36.912 -69.047 1.00 51.94 C C
+ATOM 6771 CG LEU L 191 52.212 -35.798 -68.393 1.00 50.40 C C
+ATOM 6772 CD1 LEU L 191 52.132 -36.044 -66.900 1.00 42.45 C C
+ATOM 6773 CD2 LEU L 191 52.802 -34.440 -68.617 1.00 50.23 C C
+ATOM 6774 N SER L 192 52.181 -35.185 -71.706 1.00 52.96 C N
+ATOM 6775 CA SER L 192 52.263 -33.877 -72.338 1.00 56.20 C C
+ATOM 6776 C SER L 192 51.635 -32.824 -71.433 1.00 60.09 C C
+ATOM 6777 O SER L 192 50.549 -33.036 -70.871 1.00 60.82 C O
+ATOM 6778 CB SER L 192 51.558 -33.877 -73.691 1.00 55.50 C C
+ATOM 6779 OG SER L 192 51.107 -32.564 -73.993 1.00 64.21 C O
+ATOM 6780 N SER L 193 52.322 -31.697 -71.289 1.00 65.75 C N
+ATOM 6781 CA SER L 193 51.760 -30.540 -70.608 1.00 62.75 C C
+ATOM 6782 C SER L 193 51.886 -29.344 -71.532 1.00 62.41 C C
+ATOM 6783 O SER L 193 52.940 -29.129 -72.143 1.00 64.01 C O
+ATOM 6784 CB SER L 193 52.453 -30.246 -69.279 1.00 62.87 C C
+ATOM 6785 OG SER L 193 51.727 -29.266 -68.558 1.00 65.19 C O
+ATOM 6786 N THR L 194 50.815 -28.569 -71.634 1.00 62.14 C N
+ATOM 6787 CA THR L 194 50.768 -27.439 -72.543 1.00 59.68 C C
+ATOM 6788 C THR L 194 50.419 -26.192 -71.739 1.00 62.33 C C
+ATOM 6789 O THR L 194 49.412 -26.165 -71.013 1.00 65.86 C O
+ATOM 6790 CB THR L 194 49.766 -27.706 -73.662 1.00 65.03 C C
+ATOM 6791 CG2 THR L 194 49.678 -26.523 -74.541 1.00 62.60 C C
+ATOM 6792 OG1 THR L 194 50.207 -28.835 -74.432 1.00 68.56 C O
+ATOM 6793 N LEU L 195 51.289 -25.188 -71.825 1.00 67.38 C N
+ATOM 6794 CA LEU L 195 51.044 -23.872 -71.257 1.00 65.13 C C
+ATOM 6795 C LEU L 195 50.601 -22.975 -72.397 1.00 69.28 C C
+ATOM 6796 O LEU L 195 51.403 -22.628 -73.274 1.00 72.22 C O
+ATOM 6797 CB LEU L 195 52.291 -23.309 -70.584 1.00 67.86 C C
+ATOM 6798 CG LEU L 195 51.954 -22.049 -69.788 1.00 70.27 C C
+ATOM 6799 CD1 LEU L 195 50.820 -22.397 -68.867 1.00 68.07 C C
+ATOM 6800 CD2 LEU L 195 53.145 -21.524 -68.992 1.00 73.32 C C
+ATOM 6801 N THR L 196 49.322 -22.616 -72.389 1.00 70.74 C N
+ATOM 6802 CA THR L 196 48.715 -21.814 -73.445 1.00 76.23 C C
+ATOM 6803 C THR L 196 48.583 -20.394 -72.926 1.00 78.99 C C
+ATOM 6804 O THR L 196 47.817 -20.148 -71.988 1.00 82.49 C O
+ATOM 6805 CB THR L 196 47.353 -22.380 -73.846 1.00 80.39 C C
+ATOM 6806 CG2 THR L 196 46.548 -21.363 -74.618 1.00 78.72 C C
+ATOM 6807 OG1 THR L 196 47.518 -23.563 -74.645 1.00 84.97 C O
+ATOM 6808 N LEU L 197 49.337 -19.462 -73.517 1.00 80.42 C N
+ATOM 6809 CA LEU L 197 49.277 -18.072 -73.081 1.00 80.47 C C
+ATOM 6810 C LEU L 197 49.194 -17.106 -74.255 1.00 78.74 C C
+ATOM 6811 O LEU L 197 49.715 -17.358 -75.348 1.00 81.58 C O
+ATOM 6812 CB LEU L 197 50.458 -17.687 -72.176 1.00 83.22 C C
+ATOM 6813 CG LEU L 197 49.992 -17.602 -70.711 1.00 86.73 C C
+ATOM 6814 CD1 LEU L 197 51.014 -16.898 -69.831 1.00 85.59 C C
+ATOM 6815 CD2 LEU L 197 48.620 -16.915 -70.587 1.00 91.40 C C
+ATOM 6816 N SER L 198 48.501 -15.998 -73.991 1.00 84.31 C N
+ATOM 6817 CA SER L 198 48.355 -14.904 -74.936 1.00 87.88 C C
+ATOM 6818 C SER L 198 49.712 -14.461 -75.461 1.00 87.91 C C
+ATOM 6819 O SER L 198 50.656 -14.281 -74.685 1.00 86.23 C O
+ATOM 6820 CB SER L 198 47.655 -13.745 -74.224 1.00 94.62 C C
+ATOM 6821 OG SER L 198 48.061 -12.490 -74.734 1.00 99.09 C O
+ATOM 6822 N LYS L 199 49.808 -14.291 -76.785 1.00 77.12 C N
+ATOM 6823 CA LYS L 199 51.042 -13.783 -77.389 1.00 84.05 C C
+ATOM 6824 C LYS L 199 51.521 -12.511 -76.685 1.00 84.29 C C
+ATOM 6825 O LYS L 199 52.728 -12.337 -76.435 1.00 84.01 C O
+ATOM 6826 CB LYS L 199 50.812 -13.557 -78.882 1.00 88.38 C C
+ATOM 6827 CG LYS L 199 51.959 -12.980 -79.682 1.00 90.00 C C
+ATOM 6828 CD LYS L 199 51.455 -12.674 -81.096 1.00 98.26 C C
+ATOM 6829 CE LYS L 199 52.489 -12.017 -81.995 1.00107.82 C C
+ATOM 6830 NZ LYS L 199 52.657 -10.570 -81.707 1.00112.96 C N1+
+ATOM 6831 N ALA L 200 50.574 -11.643 -76.302 1.00 90.60 C N
+ATOM 6832 CA ALA L 200 50.875 -10.470 -75.482 1.00 88.79 C C
+ATOM 6833 C ALA L 200 51.672 -10.849 -74.237 1.00 91.37 C C
+ATOM 6834 O ALA L 200 52.783 -10.348 -74.014 1.00 91.96 C O
+ATOM 6835 CB ALA L 200 49.574 -9.765 -75.084 1.00 84.64 C C
+ATOM 6836 N ASP L 201 51.121 -11.755 -73.420 1.00 83.64 C N
+ATOM 6837 CA ASP L 201 51.831 -12.192 -72.223 1.00 85.37 C C
+ATOM 6838 C ASP L 201 53.139 -12.878 -72.578 1.00 83.68 C C
+ATOM 6839 O ASP L 201 54.102 -12.801 -71.809 1.00 84.67 C O
+ATOM 6840 CB ASP L 201 50.956 -13.140 -71.409 1.00 86.79 C C
+ATOM 6841 CG ASP L 201 49.742 -12.455 -70.829 1.00 89.71 C C
+ATOM 6842 OD1 ASP L 201 49.907 -11.470 -70.069 1.00 94.19 C O
+ATOM 6843 OD2 ASP L 201 48.619 -12.901 -71.159 1.00 89.12 C O1-
+ATOM 6844 N TYR L 202 53.195 -13.545 -73.736 1.00 80.51 C N
+ATOM 6845 CA TYR L 202 54.370 -14.342 -74.067 1.00 80.81 C C
+ATOM 6846 C TYR L 202 55.585 -13.462 -74.325 1.00 83.36 C C
+ATOM 6847 O TYR L 202 56.712 -13.833 -73.971 1.00 85.51 C O
+ATOM 6848 CB TYR L 202 54.096 -15.253 -75.268 1.00 81.10 C C
+ATOM 6849 CG TYR L 202 55.325 -16.020 -75.760 1.00 79.31 C C
+ATOM 6850 CD1 TYR L 202 55.618 -17.292 -75.297 1.00 76.92 C C
+ATOM 6851 CD2 TYR L 202 56.179 -15.471 -76.712 1.00 81.58 C C
+ATOM 6852 CE1 TYR L 202 56.734 -17.982 -75.756 1.00 77.06 C C
+ATOM 6853 CE2 TYR L 202 57.293 -16.156 -77.166 1.00 79.71 C C
+ATOM 6854 CZ TYR L 202 57.566 -17.406 -76.685 1.00 77.96 C C
+ATOM 6855 OH TYR L 202 58.673 -18.072 -77.150 1.00 77.26 C O
+ATOM 6856 N GLU L 203 55.395 -12.302 -74.947 1.00 85.03 C N
+ATOM 6857 CA GLU L 203 56.553 -11.425 -75.097 1.00 89.15 C C
+ATOM 6858 C GLU L 203 56.908 -10.696 -73.798 1.00 90.80 C C
+ATOM 6859 O GLU L 203 58.080 -10.342 -73.598 1.00 90.06 C O
+ATOM 6860 CB GLU L 203 56.297 -10.447 -76.239 1.00 92.54 C C
+ATOM 6861 CG GLU L 203 56.034 -11.143 -77.565 1.00 99.37 C C
+ATOM 6862 CD GLU L 203 54.845 -10.539 -78.309 1.00106.89 C C
+ATOM 6863 OE1 GLU L 203 54.175 -9.648 -77.736 1.00108.77 C O
+ATOM 6864 OE2 GLU L 203 54.573 -10.952 -79.459 1.00109.81 C O1-
+ATOM 6865 N LYS L 204 55.931 -10.507 -72.897 1.00 90.88 C N
+ATOM 6866 CA LYS L 204 56.183 -9.910 -71.584 1.00 90.53 C C
+ATOM 6867 C LYS L 204 57.259 -10.649 -70.792 1.00 88.94 C C
+ATOM 6868 O LYS L 204 57.788 -10.095 -69.819 1.00 94.83 C O
+ATOM 6869 CB LYS L 204 54.887 -9.876 -70.760 1.00 96.00 C C
+ATOM 6870 CG LYS L 204 53.922 -8.754 -71.096 1.00102.59 C C
+ATOM 6871 CD LYS L 204 52.666 -8.827 -70.217 1.00108.90 C C
+ATOM 6872 CE LYS L 204 52.925 -8.547 -68.739 1.00116.09 C C
+ATOM 6873 NZ LYS L 204 51.663 -8.706 -67.937 1.00119.52 C N1+
+ATOM 6874 N HIS L 205 57.582 -11.888 -71.166 1.00 89.25 C N
+ATOM 6875 CA HIS L 205 58.463 -12.736 -70.385 1.00 85.84 C C
+ATOM 6876 C HIS L 205 59.541 -13.324 -71.282 1.00 83.49 C C
+ATOM 6877 O HIS L 205 59.426 -13.331 -72.514 1.00 85.66 C O
+ATOM 6878 CB HIS L 205 57.654 -13.825 -69.682 1.00 85.88 C C
+ATOM 6879 CG HIS L 205 56.543 -13.282 -68.835 1.00 91.76 C C
+ATOM 6880 CD2 HIS L 205 55.254 -13.674 -68.690 1.00 94.05 C C
+ATOM 6881 ND1 HIS L 205 56.692 -12.160 -68.046 1.00 93.74 C N
+ATOM 6882 CE1 HIS L 205 55.550 -11.900 -67.433 1.00 94.66 C C
+ATOM 6883 NE2 HIS L 205 54.661 -12.804 -67.806 1.00 97.45 C N
+ATOM 6884 N LYS L 206 60.608 -13.805 -70.640 1.00 85.17 C N
+ATOM 6885 CA LYS L 206 61.829 -14.213 -71.334 1.00 86.96 C C
+ATOM 6886 C LYS L 206 62.175 -15.676 -71.079 1.00 85.75 C C
+ATOM 6887 O LYS L 206 62.330 -16.435 -72.043 1.00 89.56 C O
+ATOM 6888 CB LYS L 206 62.992 -13.287 -70.939 1.00 85.38 C C
+ATOM 6889 CG LYS L 206 64.154 -13.210 -71.961 1.00 92.04 C C
+ATOM 6890 CD LYS L 206 65.263 -12.245 -71.469 1.00103.57 C C
+ATOM 6891 CE LYS L 206 66.623 -12.470 -72.145 1.00109.92 C C
+ATOM 6892 NZ LYS L 206 66.645 -12.116 -73.594 1.00113.31 C N1+
+ATOM 6893 N VAL L 207 62.363 -16.082 -69.818 1.00 83.78 C N
+ATOM 6894 CA VAL L 207 62.753 -17.451 -69.471 1.00 84.69 C C
+ATOM 6895 C VAL L 207 61.512 -18.289 -69.183 1.00 89.50 C C
+ATOM 6896 O VAL L 207 60.764 -18.011 -68.235 1.00 95.12 C O
+ATOM 6897 CB VAL L 207 63.704 -17.478 -68.265 1.00 84.02 C C
+ATOM 6898 CG1 VAL L 207 63.836 -18.906 -67.727 1.00 83.00 C C
+ATOM 6899 CG2 VAL L 207 65.071 -16.918 -68.635 1.00 81.72 C C
+ATOM 6900 N TYR L 208 61.326 -19.345 -69.976 1.00 82.70 C N
+ATOM 6901 CA TYR L 208 60.217 -20.282 -69.841 1.00 79.34 C C
+ATOM 6902 C TYR L 208 60.744 -21.624 -69.359 1.00 80.74 C C
+ATOM 6903 O TYR L 208 61.714 -22.155 -69.918 1.00 80.46 C O
+ATOM 6904 CB TYR L 208 59.474 -20.437 -71.171 1.00 78.70 C C
+ATOM 6905 CG TYR L 208 58.654 -19.218 -71.488 1.00 80.65 C C
+ATOM 6906 CD1 TYR L 208 58.692 -18.638 -72.742 1.00 77.60 C C
+ATOM 6907 CD2 TYR L 208 57.867 -18.617 -70.502 1.00 79.49 C C
+ATOM 6908 CE1 TYR L 208 57.969 -17.509 -73.007 1.00 79.00 C C
+ATOM 6909 CE2 TYR L 208 57.134 -17.489 -70.765 1.00 81.88 C C
+ATOM 6910 CZ TYR L 208 57.186 -16.946 -72.024 1.00 84.04 C C
+ATOM 6911 OH TYR L 208 56.469 -15.818 -72.306 1.00 88.50 C O
+ATOM 6912 N ALA L 209 60.111 -22.166 -68.319 1.00 70.28 C N
+ATOM 6913 CA ALA L 209 60.673 -23.337 -67.664 1.00 71.32 C C
+ATOM 6914 C ALA L 209 59.602 -24.331 -67.236 1.00 71.12 C C
+ATOM 6915 O ALA L 209 58.609 -23.973 -66.585 1.00 70.12 C O
+ATOM 6916 CB ALA L 209 61.515 -22.924 -66.454 1.00 69.57 C C
+ATOM 6917 N CYS L 210 59.853 -25.586 -67.591 1.00 57.61 C N
+ATOM 6918 CA CYS L 210 59.052 -26.740 -67.218 1.00 61.04 C C
+ATOM 6919 C CYS L 210 59.864 -27.575 -66.232 1.00 58.67 C C
+ATOM 6920 O CYS L 210 60.951 -28.055 -66.580 1.00 55.81 C O
+ATOM 6921 CB CYS L 210 58.702 -27.547 -68.465 1.00 69.29 C C
+ATOM 6922 SG CYS L 210 58.164 -29.164 -68.103 1.00 84.38 C S
+ATOM 6923 N GLU L 211 59.352 -27.732 -65.006 1.00 56.22 C N
+ATOM 6924 CA GLU L 211 60.070 -28.381 -63.914 1.00 55.41 C C
+ATOM 6925 C GLU L 211 59.346 -29.664 -63.524 1.00 52.92 C C
+ATOM 6926 O GLU L 211 58.253 -29.618 -62.955 1.00 51.95 C O
+ATOM 6927 CB GLU L 211 60.185 -27.435 -62.727 1.00 61.07 C C
+ATOM 6928 CG GLU L 211 60.971 -27.977 -61.553 1.00 73.83 C C
+ATOM 6929 CD GLU L 211 60.909 -27.045 -60.353 1.00 77.54 C C
+ATOM 6930 OE1 GLU L 211 59.780 -26.624 -60.010 1.00 72.34 C O
+ATOM 6931 OE2 GLU L 211 61.978 -26.714 -59.777 1.00 87.61 C O1-
+ATOM 6932 N VAL L 212 59.970 -30.802 -63.814 1.00 51.24 C N
+ATOM 6933 CA VAL L 212 59.419 -32.131 -63.583 1.00 52.00 C C
+ATOM 6934 C VAL L 212 59.990 -32.692 -62.297 1.00 55.09 C C
+ATOM 6935 O VAL L 212 61.199 -32.594 -62.053 1.00 52.97 C O
+ATOM 6936 CB VAL L 212 59.770 -33.075 -64.741 1.00 50.38 C C
+ATOM 6937 CG1 VAL L 212 59.355 -34.479 -64.391 1.00 50.16 C C
+ATOM 6938 CG2 VAL L 212 59.134 -32.621 -66.019 1.00 51.15 C C
+ATOM 6939 N THR L 213 59.147 -33.317 -61.488 1.00 55.05 C N
+ATOM 6940 CA THR L 213 59.634 -34.119 -60.375 1.00 58.21 C C
+ATOM 6941 C THR L 213 58.971 -35.494 -60.449 1.00 54.84 C C
+ATOM 6942 O THR L 213 57.761 -35.598 -60.687 1.00 55.45 C O
+ATOM 6943 CB THR L 213 59.337 -33.459 -59.034 1.00 61.05 C C
+ATOM 6944 CG2 THR L 213 59.276 -31.944 -59.187 1.00 59.10 C C
+ATOM 6945 OG1 THR L 213 58.056 -33.890 -58.592 1.00 67.78 C O
+ATOM 6946 N HIS L 214 59.762 -36.548 -60.256 1.00 48.82 C N
+ATOM 6947 CA HIS L 214 59.274 -37.906 -60.451 1.00 51.30 C C
+ATOM 6948 C HIS L 214 60.114 -38.850 -59.613 1.00 56.22 C C
+ATOM 6949 O HIS L 214 61.231 -38.517 -59.202 1.00 55.73 C O
+ATOM 6950 CB HIS L 214 59.323 -38.313 -61.919 1.00 47.34 C C
+ATOM 6951 CG HIS L 214 58.852 -39.710 -62.172 1.00 48.37 C C
+ATOM 6952 CD2 HIS L 214 57.845 -40.427 -61.621 1.00 51.07 C C
+ATOM 6953 ND1 HIS L 214 59.442 -40.539 -63.105 1.00 46.46 C N
+ATOM 6954 CE1 HIS L 214 58.822 -41.704 -63.118 1.00 51.42 C C
+ATOM 6955 NE2 HIS L 214 57.851 -41.664 -62.224 1.00 50.12 C N
+ATOM 6956 N GLN L 215 59.557 -40.047 -59.380 1.00 50.83 C N
+ATOM 6957 CA GLN L 215 60.151 -40.988 -58.434 1.00 49.42 C C
+ATOM 6958 C GLN L 215 61.538 -41.445 -58.879 1.00 47.37 C C
+ATOM 6959 O GLN L 215 62.383 -41.776 -58.027 1.00 53.14 C O
+ATOM 6960 CB GLN L 215 59.218 -42.187 -58.229 1.00 57.07 C C
+ATOM 6961 CG GLN L 215 59.763 -43.196 -57.225 1.00 67.22 C C
+ATOM 6962 CD GLN L 215 58.832 -44.368 -56.961 1.00 75.19 C C
+ATOM 6963 NE2 GLN L 215 59.343 -45.387 -56.272 1.00 79.18 C N
+ATOM 6964 OE1 GLN L 215 57.668 -44.353 -57.353 1.00 76.59 C O
+ATOM 6965 N GLY L 216 61.801 -41.441 -60.198 1.00 40.93 C N
+ATOM 6966 CA GLY L 216 63.093 -41.798 -60.753 1.00 45.85 C C
+ATOM 6967 C GLY L 216 64.119 -40.681 -60.869 1.00 52.80 C C
+ATOM 6968 O GLY L 216 65.268 -40.938 -61.260 1.00 51.90 C O
+ATOM 6969 N LEU L 217 63.743 -39.444 -60.551 1.00 48.23 C N
+ATOM 6970 CA LEU L 217 64.665 -38.320 -60.582 1.00 48.64 C C
+ATOM 6971 C LEU L 217 65.167 -38.033 -59.168 1.00 53.33 C C
+ATOM 6972 O LEU L 217 64.370 -37.863 -58.233 1.00 46.74 C O
+ATOM 6973 CB LEU L 217 63.998 -37.088 -61.189 1.00 44.55 C C
+ATOM 6974 CG LEU L 217 63.224 -37.311 -62.494 1.00 43.21 C C
+ATOM 6975 CD1 LEU L 217 62.492 -36.061 -62.962 1.00 39.90 C C
+ATOM 6976 CD2 LEU L 217 64.118 -37.781 -63.584 1.00 40.40 C C
+ATOM 6977 N SER L 218 66.499 -37.988 -59.019 1.00 49.97 C N
+ATOM 6978 CA SER L 218 67.153 -37.784 -57.730 1.00 59.61 C C
+ATOM 6979 C SER L 218 66.964 -36.379 -57.228 1.00 52.70 C C
+ATOM 6980 O SER L 218 67.385 -36.076 -56.102 1.00 53.74 C O
+ATOM 6981 CB SER L 218 68.645 -38.050 -57.842 1.00 70.07 C C
+ATOM 6982 OG SER L 218 69.091 -37.562 -59.090 1.00 76.07 C O
+ATOM 6983 N SER L 219 66.405 -35.525 -58.075 1.00 65.47 C N
+ATOM 6984 CA SER L 219 65.916 -34.195 -57.769 1.00 60.20 C C
+ATOM 6985 C SER L 219 65.319 -33.600 -59.043 1.00 51.75 C C
+ATOM 6986 O SER L 219 65.707 -33.981 -60.155 1.00 43.43 C O
+ATOM 6987 CB SER L 219 67.037 -33.340 -57.185 1.00 64.91 C C
+ATOM 6988 OG SER L 219 67.640 -32.484 -58.116 1.00 69.18 C O
+ATOM 6989 N PRO L 220 64.365 -32.688 -58.920 1.00 54.84 C N
+ATOM 6990 CA PRO L 220 63.557 -32.294 -60.077 1.00 50.91 C C
+ATOM 6991 C PRO L 220 64.386 -31.831 -61.263 1.00 49.01 C C
+ATOM 6992 O PRO L 220 65.216 -30.931 -61.152 1.00 50.40 C O
+ATOM 6993 CB PRO L 220 62.708 -31.150 -59.525 1.00 53.34 C C
+ATOM 6994 CG PRO L 220 62.678 -31.370 -58.059 1.00 56.90 C C
+ATOM 6995 CD PRO L 220 63.959 -32.000 -57.688 1.00 59.03 C C
+ATOM 6996 N VAL L 221 64.115 -32.423 -62.416 1.00 53.88 C N
+ATOM 6997 CA VAL L 221 64.729 -31.995 -63.667 1.00 50.49 C C
+ATOM 6998 C VAL L 221 63.979 -30.793 -64.205 1.00 50.92 C C
+ATOM 6999 O VAL L 221 62.747 -30.812 -64.291 1.00 48.14 C O
+ATOM 7000 CB VAL L 221 64.697 -33.127 -64.691 1.00 48.86 C C
+ATOM 7001 CG1 VAL L 221 65.312 -32.663 -65.970 1.00 48.54 C C
+ATOM 7002 CG2 VAL L 221 65.396 -34.324 -64.133 1.00 53.74 C C
+ATOM 7003 N THR L 222 64.708 -29.763 -64.621 1.00 61.50 C N
+ATOM 7004 CA THR L 222 64.082 -28.566 -65.171 1.00 56.84 C C
+ATOM 7005 C THR L 222 64.622 -28.272 -66.564 1.00 57.48 C C
+ATOM 7006 O THR L 222 65.839 -28.280 -66.787 1.00 61.73 C O
+ATOM 7007 CB THR L 222 64.293 -27.365 -64.257 1.00 53.02 C C
+ATOM 7008 CG2 THR L 222 63.723 -26.125 -64.914 1.00 53.87 C C
+ATOM 7009 OG1 THR L 222 63.643 -27.603 -62.997 1.00 58.05 C O
+ATOM 7010 N LYS L 223 63.712 -28.027 -67.495 1.00 63.76 C N
+ATOM 7011 CA LYS L 223 64.045 -27.705 -68.870 1.00 60.37 C C
+ATOM 7012 C LYS L 223 63.541 -26.300 -69.150 1.00 60.24 C C
+ATOM 7013 O LYS L 223 62.506 -25.901 -68.608 1.00 60.82 C O
+ATOM 7014 CB LYS L 223 63.419 -28.705 -69.810 1.00 64.98 C C
+ATOM 7015 CG LYS L 223 64.413 -29.352 -70.726 1.00 69.95 C C
+ATOM 7016 CD LYS L 223 65.260 -30.428 -70.048 1.00 72.65 C C
+ATOM 7017 CE LYS L 223 66.213 -31.062 -71.097 1.00 76.45 C C
+ATOM 7018 NZ LYS L 223 66.829 -32.383 -70.709 1.00 76.60 C N1+
+ATOM 7019 N SER L 224 64.281 -25.530 -69.956 1.00 71.97 C N
+ATOM 7020 CA SER L 224 63.955 -24.112 -70.114 1.00 69.10 C C
+ATOM 7021 C SER L 224 64.493 -23.569 -71.434 1.00 71.83 C C
+ATOM 7022 O SER L 224 65.321 -24.191 -72.108 1.00 74.75 C O
+ATOM 7023 CB SER L 224 64.490 -23.273 -68.946 1.00 74.95 C C
+ATOM 7024 OG SER L 224 65.838 -23.598 -68.667 1.00 80.77 C O
+ATOM 7025 N PHE L 225 63.983 -22.389 -71.796 1.00 82.30 C N
+ATOM 7026 CA PHE L 225 64.436 -21.654 -72.972 1.00 84.83 C C
+ATOM 7027 C PHE L 225 64.195 -20.162 -72.753 1.00 88.78 C C
+ATOM 7028 O PHE L 225 63.402 -19.765 -71.890 1.00 87.65 C O
+ATOM 7029 CB PHE L 225 63.736 -22.145 -74.254 1.00 82.25 C C
+ATOM 7030 CG PHE L 225 62.260 -21.785 -74.351 1.00 83.51 C C
+ATOM 7031 CD1 PHE L 225 61.845 -20.649 -75.018 1.00 82.48 C C
+ATOM 7032 CD2 PHE L 225 61.290 -22.609 -73.819 1.00 84.84 C C
+ATOM 7033 CE1 PHE L 225 60.500 -20.335 -75.117 1.00 86.99 C C
+ATOM 7034 CE2 PHE L 225 59.945 -22.297 -73.933 1.00 87.54 C C
+ATOM 7035 CZ PHE L 225 59.557 -21.166 -74.577 1.00 88.46 C C
+ATOM 7036 N ASN L 226 64.886 -19.329 -73.541 1.00 93.89 C N
+ATOM 7037 CA ASN L 226 64.684 -17.884 -73.492 1.00 93.65 C C
+ATOM 7038 C ASN L 226 63.970 -17.382 -74.742 1.00 92.39 C C
+ATOM 7039 O ASN L 226 64.301 -17.794 -75.862 1.00 94.64 C O
+ATOM 7040 CB ASN L 226 66.005 -17.139 -73.297 1.00 95.67 C C
+ATOM 7041 CG ASN L 226 66.395 -17.037 -71.830 1.00102.99 C C
+ATOM 7042 ND2 ASN L 226 66.773 -15.836 -71.391 1.00107.52 C N
+ATOM 7043 OD1 ASN L 226 66.315 -18.021 -71.091 1.00105.09 C O
+ATOM 7044 N ARG L 227 63.032 -16.440 -74.530 1.00 88.77 C N
+ATOM 7045 CA ARG L 227 62.014 -16.104 -75.529 1.00 97.54 C C
+ATOM 7046 C ARG L 227 62.617 -15.911 -76.911 1.00111.28 C C
+ATOM 7047 O ARG L 227 62.193 -16.549 -77.881 1.00120.87 C O
+ATOM 7048 CB ARG L 227 61.258 -14.833 -75.107 1.00 91.09 C C
+ATOM 7049 CG ARG L 227 60.443 -14.181 -76.230 1.00 89.35 C C
+ATOM 7050 CD ARG L 227 59.767 -12.877 -75.790 1.00 91.59 C C
+ATOM 7051 NE ARG L 227 60.727 -11.814 -75.411 1.00 93.21 C N
+ATOM 7052 CZ ARG L 227 61.174 -10.821 -76.212 1.00 97.39 C C
+ATOM 7053 NH1 ARG L 227 60.777 -10.755 -77.483 1.00 99.05 C N1+
+ATOM 7054 NH2 ARG L 227 62.046 -9.896 -75.760 1.00103.01 C N
+ATOM 7055 N GLY L 228 63.613 -15.037 -77.014 1.00 98.05 C N
+ATOM 7056 CA GLY L 228 64.191 -14.702 -78.298 1.00109.87 C C
+ATOM 7057 C GLY L 228 65.318 -15.598 -78.764 1.00117.73 C C
+ATOM 7058 O GLY L 228 65.477 -15.820 -79.970 1.00121.38 C O
+ATOM 7059 N GLU L 229 66.092 -16.142 -77.818 1.00107.10 C N
+ATOM 7060 CA GLU L 229 67.339 -16.842 -78.131 1.00109.83 C C
+ATOM 7061 C GLU L 229 67.124 -18.230 -78.736 1.00118.76 C C
+ATOM 7062 O GLU L 229 68.119 -18.870 -79.126 1.00119.81 C O
+ATOM 7063 CB GLU L 229 68.204 -16.907 -76.872 1.00105.02 C C
+ATOM 7064 CG GLU L 229 68.631 -15.512 -76.418 1.00103.20 C C
+ATOM 7065 CD GLU L 229 68.935 -15.415 -74.933 1.00102.82 C C
+ATOM 7066 OE1 GLU L 229 69.771 -16.199 -74.433 1.00105.21 C O
+ATOM 7067 OE2 GLU L 229 68.341 -14.535 -74.265 1.00100.61 C O1-
+ATOM 7068 N CYS L 230 65.866 -18.689 -78.818 1.00109.79 C N
+ATOM 7069 CA CYS L 230 65.472 -19.909 -79.544 1.00120.44 C C
+ATOM 7070 C CYS L 230 63.992 -19.849 -79.942 1.00119.61 C C
+ATOM 7071 O CYS L 230 63.288 -18.889 -79.622 1.00121.00 C O
+ATOM 7072 CB CYS L 230 65.735 -21.168 -78.702 1.00124.82 C C
+ATOM 7073 SG CYS L 230 65.858 -22.707 -79.653 1.00205.08 C S
+TER
+END
diff --git a/examples/secondary_structure.sh b/examples/secondary_structure.sh
new file mode 100644
index 0000000000000000000000000000000000000000..bab74c9f80d34c962d7379c431f2ae63750c9559
--- /dev/null
+++ b/examples/secondary_structure.sh
@@ -0,0 +1,21 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --secondary_structure XXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXX
+
+# FOR SECONDARY STRUCTURE:
+# X - mask
+# H - helix
+# E - strand
+# L - loop
diff --git a/examples/secondary_structure_bias.sh b/examples/secondary_structure_bias.sh
new file mode 100644
index 0000000000000000000000000000000000000000..c76d6747e1508af285efc18331e266e456d842b6
--- /dev/null
+++ b/examples/secondary_structure_bias.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --helix_bias 0.01 --strand_bias 0.01 --loop_bias 0.0
diff --git a/examples/secondary_structure_from_pdb.sh b/examples/secondary_structure_from_pdb.sh
new file mode 100644
index 0000000000000000000000000000000000000000..b548870e8eade138a4c59ebada89fb4ab2a9ec68
--- /dev/null
+++ b/examples/secondary_structure_from_pdb.sh
@@ -0,0 +1,21 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 110 \
+ --T 25 --save_best_plddt \
+ --dssp_pdb ./pdbs/cd86.pdb
+
+# FOR SECONDARY STRUCTURE:
+# X - mask
+# H - helix
+# E - strand
+# L - loop
diff --git a/examples/symmetric_design.sh b/examples/symmetric_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..cca1065f1aa383c16f7f5338de0a6a44c6109479
--- /dev/null
+++ b/examples/symmetric_design.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/symmetric_design \
+ --contigs 25,0 25,0 25,0 \
+ --T 50 \
+ --save_best_plddt \
+ --symmetry 3
diff --git a/examples/weighted_sequence.sh b/examples/weighted_sequence.sh
new file mode 100644
index 0000000000000000000000000000000000000000..779abda1d21255ed8d68198f72ac55729839be26
--- /dev/null
+++ b/examples/weighted_sequence.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --aa_composition W0.2 --potential_scale 1.75
diff --git a/examples/weighted_sequence_json.sh b/examples/weighted_sequence_json.sh
new file mode 100644
index 0000000000000000000000000000000000000000..701d7d8d8c2352e2cc95255732d7de341979083a
--- /dev/null
+++ b/examples/weighted_sequence_json.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 75 \
+ --aa_weights_json aa_weights.json \
+ --add_weight_every_n 5 --add_weight_every_n \
+ --T 25 --save_best_plddt
diff --git a/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py b/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..ec42a3cd8a5a5bd8f59cff12963908760eff59b7
--- /dev/null
+++ b/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py
@@ -0,0 +1,140 @@
+import torch
+import torch.nn as nn
+from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
+from Track_module import IterativeSimulator
+from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
+from util import INIT_CRDS
+from opt_einsum import contract as einsum
+from icecream import ic
+
+class RoseTTAFoldModule(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
+ d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
+ n_head_msa=8, n_head_pair=4, n_head_templ=4,
+ d_hidden=32, d_hidden_templ=64,
+ p_drop=0.15, d_t1d=24, d_t2d=44,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ ):
+ super(RoseTTAFoldModule, self).__init__()
+ #
+ # Input Embeddings
+ d_state = SE3_param_topk['l0_out_features']
+ self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
+ self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
+ self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
+ n_head=n_head_templ,
+ d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
+ # Update inputs with outputs from previous round
+ self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
+ #
+ self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
+ n_main_block=n_main_block,
+ n_ref_block=n_ref_block,
+ d_msa=d_msa, d_msa_full=d_msa_full,
+ d_pair=d_pair, d_hidden=d_hidden,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ SE3_param_full=SE3_param_full,
+ SE3_param_topk=SE3_param_topk,
+ p_drop=p_drop)
+ ##
+ self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
+ self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
+ self.lddt_pred = LDDTNetwork(d_state)
+
+ self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
+
+ def forward(self, msa_latent, msa_full, seq, xyz, idx,
+ seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
+ msa_prev=None, pair_prev=None, state_prev=None,
+ return_raw=False, return_full=False,
+ use_checkpoint=False, return_infer=False):
+ B, N, L = msa_latent.shape[:3]
+ # Get embeddings
+ #ic(seq.shape)
+ #ic(msa_latent.shape)
+ #ic(seq1hot.shape)
+ #ic(idx.shape)
+ #ic(xyz.shape)
+ #ic(seq1hot.shape)
+ #ic(t1d.shape)
+ #ic(t2d.shape)
+
+ idx = idx.long()
+ msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
+
+ msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
+ #
+ # Do recycling
+ if msa_prev == None:
+ msa_prev = torch.zeros_like(msa_latent[:,0])
+ if pair_prev == None:
+ pair_prev = torch.zeros_like(pair)
+ if state_prev == None:
+ state_prev = torch.zeros_like(state)
+
+ #ic(seq.shape)
+ #ic(msa_prev.shape)
+ #ic(pair_prev.shape)
+ #ic(xyz.shape)
+ #ic(state_prev.shape)
+
+
+ msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
+ msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
+ pair = pair + pair_recycle
+ state = state + state_recycle
+ #
+ #ic(t1d.dtype)
+ #ic(t2d.dtype)
+ #ic(alpha_t.dtype)
+ #ic(xyz_t.dtype)
+ #ic(pair.dtype)
+ #ic(state.dtype)
+
+
+ #import pdb; pdb.set_trace()
+
+ # add template embedding
+ pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
+
+ #ic(seq.dtype)
+ #ic(msa_latent.dtype)
+ #ic(msa_full.dtype)
+ #ic(pair.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+ # Predict coordinates from given inputs
+ msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
+ state, idx, use_checkpoint=use_checkpoint)
+
+ if return_raw:
+ # get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return msa[:,0], pair, xyz, state, alpha_s[-1]
+
+ # predict masked amino acids
+ logits_aa = self.aa_pred(msa)
+ #
+ # predict distogram & orientograms
+ logits = self.c6d_pred(pair)
+
+ # Predict LDDT
+ lddt = self.lddt_pred(state)
+
+ # predict experimentally resolved or not
+ logits_exp = self.exp_pred(msa[:,0], state)
+
+ if return_infer:
+ #get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
+
+
+ # get all intermediate bb structures
+ xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
+
+ return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
diff --git a/model/Attention_module.py b/model/Attention_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..44310ebc94cc40f7cca9eda31463e172188a0084
--- /dev/null
+++ b/model/Attention_module.py
@@ -0,0 +1,411 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+import math
+from opt_einsum import contract as einsum
+from util_module import init_lecun_normal
+from icecream import ic
+
+class FeedForwardLayer(nn.Module):
+ def __init__(self, d_model, r_ff, p_drop=0.1):
+ super(FeedForwardLayer, self).__init__()
+ self.norm = nn.LayerNorm(d_model)
+ self.linear1 = nn.Linear(d_model, d_model*r_ff)
+ self.dropout = nn.Dropout(p_drop)
+ self.linear2 = nn.Linear(d_model*r_ff, d_model)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize linear layer right before ReLu: He initializer (kaiming normal)
+ nn.init.kaiming_normal_(self.linear1.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear1.bias)
+
+ # initialize linear layer right before residual connection: zero initialize
+ nn.init.zeros_(self.linear2.weight)
+ nn.init.zeros_(self.linear2.bias)
+
+ def forward(self, src):
+ src = self.norm(src)
+ src = self.linear2(self.dropout(F.relu_(self.linear1(src))))
+ return src
+
+class Attention(nn.Module):
+ # calculate multi-head attention
+ def __init__(self, d_query, d_key, n_head, d_hidden, d_out, p_drop=0.1):
+ super(Attention, self).__init__()
+ self.h = n_head
+ self.dim = d_hidden
+ #
+ self.to_q = nn.Linear(d_query, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_key, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_key, n_head*d_hidden, bias=False)
+ #
+ self.to_out = nn.Linear(n_head*d_hidden, d_out)
+ self.scaling = 1/math.sqrt(d_hidden)
+ #
+ # initialize all parameters properly
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, query, key, value):
+ B, Q = query.shape[:2]
+ B, K = key.shape[:2]
+ #
+ query = self.to_q(query).reshape(B, Q, self.h, self.dim)
+ key = self.to_k(key).reshape(B, K, self.h, self.dim)
+ value = self.to_v(value).reshape(B, K, self.h, self.dim)
+ #
+ query = query * self.scaling
+ attn = einsum('bqhd,bkhd->bhqk', query, key)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bhqk,bkhd->bqhd', attn, value)
+ out = out.reshape(B, Q, self.h*self.dim)
+ #
+ out = self.to_out(out)
+
+ return out
+
+class AttentionWithBias(nn.Module):
+ def __init__(self, d_in=256, d_bias=128, n_head=8, d_hidden=32):
+ super(AttentionWithBias, self).__init__()
+ self.norm_in = nn.LayerNorm(d_in)
+ self.norm_bias = nn.LayerNorm(d_bias)
+ #
+ self.to_q = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_bias, n_head, bias=False)
+ self.to_g = nn.Linear(d_in, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_in)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, x, bias):
+ B, L = x.shape[:2]
+ #
+ x = self.norm_in(x)
+ bias = self.norm_bias(bias)
+ #
+ query = self.to_q(x).reshape(B, L, self.h, self.dim)
+ key = self.to_k(x).reshape(B, L, self.h, self.dim)
+ value = self.to_v(x).reshape(B, L, self.h, self.dim)
+ bias = self.to_b(bias) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(x))
+ #
+ key = key * self.scaling
+ attn = einsum('bqhd,bkhd->bqkh', query, key)
+ attn = attn + bias
+ attn = F.softmax(attn, dim=-2)
+ #
+ out = einsum('bqkh,bkhd->bqhd', attn, value).reshape(B, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+# MSA Attention (row/column) from AlphaFold architecture
+class SequenceWeight(nn.Module):
+ def __init__(self, d_msa, n_head, d_hidden, p_drop=0.1):
+ super(SequenceWeight, self).__init__()
+ self.h = n_head
+ self.dim = d_hidden
+ self.scale = 1.0 / math.sqrt(self.dim)
+
+ self.to_query = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_key = nn.Linear(d_msa, n_head*d_hidden)
+ self.dropout = nn.Dropout(p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_query.weight)
+ nn.init.xavier_uniform_(self.to_key.weight)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+
+ tar_seq = msa[:,0]
+
+ q = self.to_query(tar_seq).view(B, 1, L, self.h, self.dim)
+ k = self.to_key(msa).view(B, N, L, self.h, self.dim)
+
+ q = q * self.scale
+ attn = einsum('bqihd,bkihd->bkihq', q, k)
+ attn = F.softmax(attn, dim=1)
+ return self.dropout(attn)
+
+class MSARowAttentionWithBias(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, n_head=8, d_hidden=32):
+ super(MSARowAttentionWithBias, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ #
+ self.seq_weight = SequenceWeight(d_msa, n_head, d_hidden, p_drop=0.1)
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_pair, n_head, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa, pair): # TODO: make this as tied-attention
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ pair = self.norm_pair(pair)
+ #
+ seq_weight = self.seq_weight(msa) # (B, N, L, h, 1)
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
+ value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
+ bias = self.to_b(pair) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(msa))
+ #
+ query = query * seq_weight.expand(-1, -1, -1, -1, self.dim)
+ key = key * self.scaling
+ attn = einsum('bsqhd,bskhd->bqkh', query, key)
+ attn = attn + bias
+ attn = F.softmax(attn, dim=-2)
+ #
+ out = einsum('bqkh,bskhd->bsqhd', attn, value).reshape(B, N, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+class MSAColAttention(nn.Module):
+ def __init__(self, d_msa=256, n_head=8, d_hidden=32):
+ super(MSAColAttention, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ #
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ #
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
+ value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
+ gate = torch.sigmoid(self.to_g(msa))
+ #
+ query = query * self.scaling
+ attn = einsum('bqihd,bkihd->bihqk', query, key)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bihqk,bkihd->bqihd', attn, value).reshape(B, N, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+class MSAColGlobalAttention(nn.Module):
+ def __init__(self, d_msa=64, n_head=8, d_hidden=8):
+ super(MSAColGlobalAttention, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ #
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, d_hidden, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ #
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ query = query.mean(dim=1) # (B, L, h, dim)
+ key = self.to_k(msa) # (B, N, L, dim)
+ value = self.to_v(msa) # (B, N, L, dim)
+ gate = torch.sigmoid(self.to_g(msa)) # (B, N, L, h*dim)
+ #
+ query = query * self.scaling
+ attn = einsum('bihd,bkid->bihk', query, key) # (B, L, h, N)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bihk,bkid->bihd', attn, value).reshape(B, 1, L, -1) # (B, 1, L, h*dim)
+ out = gate * out # (B, N, L, h*dim)
+ #
+ out = self.to_out(out)
+ return out
+
+# Instead of triangle attention, use Tied axail attention with bias from coordinates..?
+class BiasedAxialAttention(nn.Module):
+ def __init__(self, d_pair, d_bias, n_head, d_hidden, p_drop=0.1, is_row=True):
+ super(BiasedAxialAttention, self).__init__()
+ #
+ self.is_row = is_row
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_bias = nn.LayerNorm(d_bias)
+
+ self.to_q = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_bias, n_head, bias=False)
+ self.to_g = nn.Linear(d_pair, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_pair)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ # initialize all parameters properly
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, pair, bias, same_chain = None):
+ # pair: (B, L, L, d_pair)
+ B, L = pair.shape[:2]
+
+ if self.is_row:
+ pair = pair.permute(0,2,1,3)
+ bias = bias.permute(0,2,1,3)
+
+ pair = self.norm_pair(pair)
+ bias = self.norm_bias(bias)
+
+ query = self.to_q(pair).reshape(B, L, L, self.h, self.dim)
+ key = self.to_k(pair).reshape(B, L, L, self.h, self.dim)
+ value = self.to_v(pair).reshape(B, L, L, self.h, self.dim)
+ bias = self.to_b(bias) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(pair)) # (B, L, L, h*dim)
+
+ query = query * self.scaling
+ key = key / math.sqrt(L) # normalize for tied attention
+ attn = einsum('bnihk,bnjhk->bijh', query, key) # tied attention
+ attn = attn + bias # apply bias
+ attn = F.softmax(attn, dim=-2) # (B, L, L, h)
+
+ if same_chain is not None:
+ ic(same_chain)
+ ic(attn)
+ ic(attn[~same_chain])
+ attn[~same_chain] *= 1.1
+
+ out = einsum('bijh,bkjhd->bikhd', attn, value).reshape(B, L, L, -1)
+ out = gate * out
+
+ out = self.to_out(out)
+ if self.is_row:
+ out = out.permute(0,2,1,3)
+ return out
+
diff --git a/model/AuxiliaryPredictor.py b/model/AuxiliaryPredictor.py
new file mode 100644
index 0000000000000000000000000000000000000000..af392f9a9e8a00775c690fdaec2c3db6b39afc11
--- /dev/null
+++ b/model/AuxiliaryPredictor.py
@@ -0,0 +1,92 @@
+import torch
+import torch.nn as nn
+
+class DistanceNetwork(nn.Module):
+ def __init__(self, n_feat, p_drop=0.1):
+ super(DistanceNetwork, self).__init__()
+ #
+ self.proj_symm = nn.Linear(n_feat, 37*2)
+ self.proj_asymm = nn.Linear(n_feat, 37+19)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize linear layer for final logit prediction
+ nn.init.zeros_(self.proj_symm.weight)
+ nn.init.zeros_(self.proj_asymm.weight)
+ nn.init.zeros_(self.proj_symm.bias)
+ nn.init.zeros_(self.proj_asymm.bias)
+
+ def forward(self, x):
+ # input: pair info (B, L, L, C)
+
+ # predict theta, phi (non-symmetric)
+ logits_asymm = self.proj_asymm(x)
+ logits_theta = logits_asymm[:,:,:,:37].permute(0,3,1,2)
+ logits_phi = logits_asymm[:,:,:,37:].permute(0,3,1,2)
+
+ # predict dist, omega
+ logits_symm = self.proj_symm(x)
+ logits_symm = logits_symm + logits_symm.permute(0,2,1,3)
+ logits_dist = logits_symm[:,:,:,:37].permute(0,3,1,2)
+ logits_omega = logits_symm[:,:,:,37:].permute(0,3,1,2)
+
+ return logits_dist, logits_omega, logits_theta, logits_phi
+
+class MaskedTokenNetwork(nn.Module):
+ def __init__(self, n_feat, p_drop=0.1):
+ super(MaskedTokenNetwork, self).__init__()
+ self.proj = nn.Linear(n_feat, 21)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, x):
+ B, N, L = x.shape[:3]
+ logits = self.proj(x).permute(0,3,1,2).reshape(B, -1, N*L)
+
+ return logits
+
+class LDDTNetwork(nn.Module):
+ def __init__(self, n_feat, n_bin_lddt=50):
+ super(LDDTNetwork, self).__init__()
+ self.proj = nn.Linear(n_feat, n_bin_lddt)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, x):
+ logits = self.proj(x) # (B, L, 50)
+
+ return logits.permute(0,2,1)
+
+class ExpResolvedNetwork(nn.Module):
+ def __init__(self, d_msa, d_state, p_drop=0.1):
+ super(ExpResolvedNetwork, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.proj = nn.Linear(d_msa+d_state, 1)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, seq, state):
+ B, L = seq.shape[:2]
+
+ seq = self.norm_msa(seq)
+ state = self.norm_state(state)
+ feat = torch.cat((seq, state), dim=-1)
+ logits = self.proj(feat)
+ return logits.reshape(B, L)
+
+
+
diff --git a/model/Embeddings.py b/model/Embeddings.py
new file mode 100644
index 0000000000000000000000000000000000000000..d4558dd4eb9646070d333011d0d8783249c85264
--- /dev/null
+++ b/model/Embeddings.py
@@ -0,0 +1,307 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import torch.utils.checkpoint as checkpoint
+from util import get_tips
+from util_module import Dropout, create_custom_forward, rbf, init_lecun_normal
+from Attention_module import Attention, FeedForwardLayer, AttentionWithBias
+from Track_module import PairStr2Pair
+from icecream import ic
+
+# Module contains classes and functions to generate initial embeddings
+
+class PositionalEncoding2D(nn.Module):
+ # Add relative positional encoding to pair features
+ def __init__(self, d_model, minpos=-32, maxpos=32, p_drop=0.1):
+ super(PositionalEncoding2D, self).__init__()
+ self.minpos = minpos
+ self.maxpos = maxpos
+ self.nbin = abs(minpos)+maxpos+1
+ self.emb = nn.Embedding(self.nbin, d_model)
+ self.drop = nn.Dropout(p_drop)
+
+ def forward(self, x, idx):
+ bins = torch.arange(self.minpos, self.maxpos, device=x.device)
+ seqsep = idx[:,None,:] - idx[:,:,None] # (B, L, L)
+ #
+ ib = torch.bucketize(seqsep, bins).long() # (B, L, L)
+ emb = self.emb(ib) #(B, L, L, d_model)
+ x = x + emb # add relative positional encoding
+ return self.drop(x)
+
+class MSA_emb(nn.Module):
+ # Get initial seed MSA embedding
+ def __init__(self, d_msa=256, d_pair=128, d_state=32, d_init=22+22+2+2,
+ minpos=-32, maxpos=32, p_drop=0.1):
+ super(MSA_emb, self).__init__()
+ self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
+ self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence -- used for MSA embedding
+ self.emb_left = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
+ self.emb_right = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
+ self.emb_state = nn.Embedding(22, d_state)
+ self.drop = nn.Dropout(p_drop)
+ self.pos = PositionalEncoding2D(d_pair, minpos=minpos, maxpos=maxpos, p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ self.emb_q = init_lecun_normal(self.emb_q)
+ self.emb_left = init_lecun_normal(self.emb_left)
+ self.emb_right = init_lecun_normal(self.emb_right)
+ self.emb_state = init_lecun_normal(self.emb_state)
+
+ nn.init.zeros_(self.emb.bias)
+
+ def forward(self, msa, seq, idx, seq1hot=None):
+ # Inputs:
+ # - msa: Input MSA (B, N, L, d_init)
+ # - seq: Input Sequence (B, L)
+ # - idx: Residue index
+ # Outputs:
+ # - msa: Initial MSA embedding (B, N, L, d_msa)
+ # - pair: Initial Pair embedding (B, L, L, d_pair)
+
+ N = msa.shape[1] # number of sequenes in MSA
+
+ # msa embedding
+ msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
+ seq = seq.long()
+ tmp = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ msa = msa + tmp.expand(-1, N, -1, -1) # adding query embedding to MSA
+ msa = self.drop(msa)
+
+ # pair embedding
+ if seq1hot is not None:
+ left = (seq1hot @ self.emb_left.weight)[:,None] # (B, 1, L, d_pair)
+ right = (seq1hot @ self.emb_right.weight)[:,:,None] # (B, L, 1, d_pair)
+ else:
+ left = self.emb_left(seq)[:,None] # (B, 1, L, d_pair)
+ right = self.emb_right(seq)[:,:,None] # (B, L, 1, d_pair)
+ #ic(torch.norm(self.emb_left.weight, dim=1))
+ #ic(torch.norm(self.emb_right.weight, dim=1))
+ pair = left + right # (B, L, L, d_pair)
+ pair = self.pos(pair, idx) # add relative position
+
+ # state embedding
+ state = self.drop(self.emb_state(seq))
+
+ return msa, pair, state
+
+class Extra_emb(nn.Module):
+ # Get initial seed MSA embedding
+ def __init__(self, d_msa=256, d_init=22+1+2, p_drop=0.1):
+ super(Extra_emb, self).__init__()
+ self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
+ self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence
+ self.drop = nn.Dropout(p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ nn.init.zeros_(self.emb.bias)
+
+ def forward(self, msa, seq, idx, seq1hot=None):
+ # Inputs:
+ # - msa: Input MSA (B, N, L, d_init)
+ # - seq: Input Sequence (B, L)
+ # - idx: Residue index
+ # Outputs:
+ # - msa: Initial MSA embedding (B, N, L, d_msa)
+ N = msa.shape[1] # number of sequenes in MSA
+ msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
+ if seq1hot is not None:
+ seq = (seq1hot @ self.emb_q.weight).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ else:
+ seq = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ #ic(torch.norm(self.emb_q.weight, dim=1))
+ msa = msa + seq.expand(-1, N, -1, -1) # adding query embedding to MSA
+ return self.drop(msa)
+
+class TemplatePairStack(nn.Module):
+ # process template pairwise features
+ # use structure-biased attention
+ def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.25):
+ super(TemplatePairStack, self).__init__()
+ self.n_block = n_block
+ proc_s = [PairStr2Pair(d_pair=d_templ, n_head=n_head, d_hidden=d_hidden, p_drop=p_drop) for i in range(n_block)]
+ self.block = nn.ModuleList(proc_s)
+ self.norm = nn.LayerNorm(d_templ)
+ def forward(self, templ, rbf_feat, use_checkpoint=False):
+ B, T, L = templ.shape[:3]
+ templ = templ.reshape(B*T, L, L, -1)
+
+ for i_block in range(self.n_block):
+ if use_checkpoint:
+ templ = checkpoint.checkpoint(create_custom_forward(self.block[i_block]), templ, rbf_feat)
+ else:
+ templ = self.block[i_block](templ, rbf_feat)
+ return self.norm(templ).reshape(B, T, L, L, -1)
+
+class TemplateTorsionStack(nn.Module):
+ def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.15):
+ super(TemplateTorsionStack, self).__init__()
+ self.n_block=n_block
+ self.proj_pair = nn.Linear(d_templ+36, d_templ)
+ proc_s = [AttentionWithBias(d_in=d_templ, d_bias=d_templ,
+ n_head=n_head, d_hidden=d_hidden) for i in range(n_block)]
+ self.row_attn = nn.ModuleList(proc_s)
+ proc_s = [FeedForwardLayer(d_templ, 4, p_drop=p_drop) for i in range(n_block)]
+ self.ff = nn.ModuleList(proc_s)
+ self.norm = nn.LayerNorm(d_templ)
+
+ def reset_parameter(self):
+ self.proj_pair = init_lecun_normal(self.proj_pair)
+ nn.init.zeros_(self.proj_pair.bias)
+
+ def forward(self, tors, pair, rbf_feat, use_checkpoint=False):
+ B, T, L = tors.shape[:3]
+ tors = tors.reshape(B*T, L, -1)
+ pair = pair.reshape(B*T, L, L, -1)
+ pair = torch.cat((pair, rbf_feat), dim=-1)
+ pair = self.proj_pair(pair)
+
+ for i_block in range(self.n_block):
+ if use_checkpoint:
+ tors = tors + checkpoint.checkpoint(create_custom_forward(self.row_attn[i_block]), tors, pair)
+ else:
+ tors = tors + self.row_attn[i_block](tors, pair)
+ tors = tors + self.ff[i_block](tors)
+ return self.norm(tors).reshape(B, T, L, -1)
+
+class Templ_emb(nn.Module):
+ # Get template embedding
+ # Features are
+ # t2d:
+ # - 37 distogram bins + 6 orientations (43)
+ # - Mask (missing/unaligned) (1)
+ # t1d:
+ # - tiled AA sequence (20 standard aa + gap)
+ # - seq confidence (1)
+ # - global time step (1)
+ # - struc confidence (1)
+ #
+ def __init__(self, d_t1d=21+1+1+1, d_t2d=43+1, d_tor=30, d_pair=128, d_state=32,
+ n_block=2, d_templ=64,
+ n_head=4, d_hidden=16, p_drop=0.25):
+ super(Templ_emb, self).__init__()
+ # process 2D features
+ self.emb = nn.Linear(d_t1d*2+d_t2d, d_templ)
+ self.templ_stack = TemplatePairStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
+ d_hidden=d_hidden, p_drop=p_drop)
+
+ self.attn = Attention(d_pair, d_templ, n_head, d_hidden, d_pair, p_drop=p_drop)
+
+ # process torsion angles
+ self.emb_t1d = nn.Linear(d_t1d+d_tor, d_templ)
+ self.proj_t1d = nn.Linear(d_templ, d_templ)
+ #self.tor_stack = TemplateTorsionStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
+ # d_hidden=d_hidden, p_drop=p_drop)
+ self.attn_tor = Attention(d_state, d_templ, n_head, d_hidden, d_state, p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ #nn.init.zeros_(self.emb.weight) #init weights to zero
+ nn.init.zeros_(self.emb.bias)
+
+ nn.init.kaiming_normal_(self.emb_t1d.weight, nonlinearity='relu')
+ #nn.init.zeros_(self.emb_t1d.weight)
+ nn.init.zeros_(self.emb_t1d.bias)
+
+ self.proj_t1d = init_lecun_normal(self.proj_t1d)
+ nn.init.zeros_(self.proj_t1d.bias)
+
+ def forward(self, t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=False):
+ # Input
+ # - t1d: 1D template info (B, T, L, 23) 24 SL
+ # - t2d: 2D template info (B, T, L, L, 44)
+ B, T, L, _ = t1d.shape
+
+ # Prepare 2D template features
+ left = t1d.unsqueeze(3).expand(-1,-1,-1,L,-1)
+ right = t1d.unsqueeze(2).expand(-1,-1,L,-1,-1)
+ #
+ templ = torch.cat((t2d, left, right), -1) # (B, T, L, L, 88)
+
+ #ic(templ.shape)
+ #ic(templ.dtype)
+ #ic(self.emb.weight.dtype)
+ templ = self.emb(templ) # Template templures (B, T, L, L, d_templ)
+ # process each template features
+ xyz_t = xyz_t.reshape(B*T, L, -1, 3)
+ rbf_feat = rbf(torch.cdist(xyz_t[:,:,1], xyz_t[:,:,1]))
+ templ = self.templ_stack(templ, rbf_feat, use_checkpoint=use_checkpoint) # (B, T, L,L, d_templ)
+
+ # Prepare 1D template torsion angle features
+ t1d = torch.cat((t1d, alpha_t), dim=-1) # (B, T, L, 22+30)
+ # process each template features
+ t1d = self.proj_t1d(F.relu_(self.emb_t1d(t1d)))
+
+ # mixing query state features to template state features
+ state = state.reshape(B*L, 1, -1)
+ t1d = t1d.permute(0,2,1,3).reshape(B*L, T, -1)
+ if use_checkpoint:
+ out = checkpoint.checkpoint(create_custom_forward(self.attn_tor), state, t1d, t1d)
+ out = out.reshape(B, L, -1)
+ else:
+ out = self.attn_tor(state, t1d, t1d).reshape(B, L, -1)
+ state = state.reshape(B, L, -1)
+ state = state + out
+
+ # mixing query pair features to template information (Template pointwise attention)
+ pair = pair.reshape(B*L*L, 1, -1)
+ templ = templ.permute(0, 2, 3, 1, 4).reshape(B*L*L, T, -1)
+ if use_checkpoint:
+ out = checkpoint.checkpoint(create_custom_forward(self.attn), pair, templ, templ)
+ out = out.reshape(B, L, L, -1)
+ else:
+ out = self.attn(pair, templ, templ).reshape(B, L, L, -1)
+ #
+ pair = pair.reshape(B, L, L, -1)
+ pair = pair + out
+
+ return pair, state
+
+class Recycling(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_state=32):
+ super(Recycling, self).__init__()
+ self.proj_dist = nn.Linear(36+d_state*2, d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_msa = nn.LayerNorm(d_msa)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.proj_dist = init_lecun_normal(self.proj_dist)
+ nn.init.zeros_(self.proj_dist.bias)
+
+ def forward(self, seq, msa, pair, xyz, state):
+ B, L = pair.shape[:2]
+ state = self.norm_state(state)
+ #
+ left = state.unsqueeze(2).expand(-1,-1,L,-1)
+ right = state.unsqueeze(1).expand(-1,L,-1,-1)
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ dist = rbf(torch.cdist(Cb, Cb))
+ dist = torch.cat((dist, left, right), dim=-1)
+ dist = self.proj_dist(dist)
+ pair = dist + self.norm_pair(pair)
+ msa = self.norm_msa(msa)
+ return msa, pair, state
+
diff --git a/model/RoseTTAFoldModel.py b/model/RoseTTAFoldModel.py
new file mode 100644
index 0000000000000000000000000000000000000000..ec42a3cd8a5a5bd8f59cff12963908760eff59b7
--- /dev/null
+++ b/model/RoseTTAFoldModel.py
@@ -0,0 +1,140 @@
+import torch
+import torch.nn as nn
+from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
+from Track_module import IterativeSimulator
+from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
+from util import INIT_CRDS
+from opt_einsum import contract as einsum
+from icecream import ic
+
+class RoseTTAFoldModule(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
+ d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
+ n_head_msa=8, n_head_pair=4, n_head_templ=4,
+ d_hidden=32, d_hidden_templ=64,
+ p_drop=0.15, d_t1d=24, d_t2d=44,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ ):
+ super(RoseTTAFoldModule, self).__init__()
+ #
+ # Input Embeddings
+ d_state = SE3_param_topk['l0_out_features']
+ self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
+ self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
+ self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
+ n_head=n_head_templ,
+ d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
+ # Update inputs with outputs from previous round
+ self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
+ #
+ self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
+ n_main_block=n_main_block,
+ n_ref_block=n_ref_block,
+ d_msa=d_msa, d_msa_full=d_msa_full,
+ d_pair=d_pair, d_hidden=d_hidden,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ SE3_param_full=SE3_param_full,
+ SE3_param_topk=SE3_param_topk,
+ p_drop=p_drop)
+ ##
+ self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
+ self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
+ self.lddt_pred = LDDTNetwork(d_state)
+
+ self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
+
+ def forward(self, msa_latent, msa_full, seq, xyz, idx,
+ seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
+ msa_prev=None, pair_prev=None, state_prev=None,
+ return_raw=False, return_full=False,
+ use_checkpoint=False, return_infer=False):
+ B, N, L = msa_latent.shape[:3]
+ # Get embeddings
+ #ic(seq.shape)
+ #ic(msa_latent.shape)
+ #ic(seq1hot.shape)
+ #ic(idx.shape)
+ #ic(xyz.shape)
+ #ic(seq1hot.shape)
+ #ic(t1d.shape)
+ #ic(t2d.shape)
+
+ idx = idx.long()
+ msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
+
+ msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
+ #
+ # Do recycling
+ if msa_prev == None:
+ msa_prev = torch.zeros_like(msa_latent[:,0])
+ if pair_prev == None:
+ pair_prev = torch.zeros_like(pair)
+ if state_prev == None:
+ state_prev = torch.zeros_like(state)
+
+ #ic(seq.shape)
+ #ic(msa_prev.shape)
+ #ic(pair_prev.shape)
+ #ic(xyz.shape)
+ #ic(state_prev.shape)
+
+
+ msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
+ msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
+ pair = pair + pair_recycle
+ state = state + state_recycle
+ #
+ #ic(t1d.dtype)
+ #ic(t2d.dtype)
+ #ic(alpha_t.dtype)
+ #ic(xyz_t.dtype)
+ #ic(pair.dtype)
+ #ic(state.dtype)
+
+
+ #import pdb; pdb.set_trace()
+
+ # add template embedding
+ pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
+
+ #ic(seq.dtype)
+ #ic(msa_latent.dtype)
+ #ic(msa_full.dtype)
+ #ic(pair.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+ # Predict coordinates from given inputs
+ msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
+ state, idx, use_checkpoint=use_checkpoint)
+
+ if return_raw:
+ # get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return msa[:,0], pair, xyz, state, alpha_s[-1]
+
+ # predict masked amino acids
+ logits_aa = self.aa_pred(msa)
+ #
+ # predict distogram & orientograms
+ logits = self.c6d_pred(pair)
+
+ # Predict LDDT
+ lddt = self.lddt_pred(state)
+
+ # predict experimentally resolved or not
+ logits_exp = self.exp_pred(msa[:,0], state)
+
+ if return_infer:
+ #get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
+
+
+ # get all intermediate bb structures
+ xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
+
+ return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
diff --git a/model/SE3_network.py b/model/SE3_network.py
new file mode 100644
index 0000000000000000000000000000000000000000..a1f01d116af573f19d84a91432498a68d5b47950
--- /dev/null
+++ b/model/SE3_network.py
@@ -0,0 +1,83 @@
+import torch
+import torch.nn as nn
+
+#from equivariant_attention.modules import get_basis_and_r, GSE3Res, GNormBias
+#from equivariant_attention.modules import GConvSE3, GNormSE3
+#from equivariant_attention.fibers import Fiber
+
+from util_module import init_lecun_normal_param
+from se3_transformer.model import SE3Transformer
+from se3_transformer.model.fiber import Fiber
+
+class SE3TransformerWrapper(nn.Module):
+ """SE(3) equivariant GCN with attention"""
+ def __init__(self, num_layers=2, num_channels=32, num_degrees=3, n_heads=4, div=4,
+ l0_in_features=32, l0_out_features=32,
+ l1_in_features=3, l1_out_features=2,
+ num_edge_features=32):
+ super().__init__()
+ # Build the network
+ self.l1_in = l1_in_features
+ #
+ fiber_edge = Fiber({0: num_edge_features})
+ if l1_out_features > 0:
+ if l1_in_features > 0:
+ fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
+ else:
+ fiber_in = Fiber({0: l0_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
+ else:
+ if l1_in_features > 0:
+ fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features})
+ else:
+ fiber_in = Fiber({0: l0_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features})
+
+ self.se3 = SE3Transformer(num_layers=num_layers,
+ fiber_in=fiber_in,
+ fiber_hidden=fiber_hidden,
+ fiber_out = fiber_out,
+ num_heads=n_heads,
+ channels_div=div,
+ fiber_edge=fiber_edge,
+ use_layer_norm=True)
+ #use_layer_norm=False)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+
+ # make sure linear layer before ReLu are initialized with kaiming_normal_
+ for n, p in self.se3.named_parameters():
+ if "bias" in n:
+ nn.init.zeros_(p)
+ elif len(p.shape) == 1:
+ continue
+ else:
+ if "radial_func" not in n:
+ p = init_lecun_normal_param(p)
+ else:
+ if "net.6" in n:
+ nn.init.zeros_(p)
+ else:
+ nn.init.kaiming_normal_(p, nonlinearity='relu')
+
+ # make last layers to be zero-initialized
+ #self.se3.graph_modules[-1].to_kernel_self['0'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['0'])
+ #self.se3.graph_modules[-1].to_kernel_self['1'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['1'])
+ nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['0'])
+ nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['1'])
+
+ def forward(self, G, type_0_features, type_1_features=None, edge_features=None):
+ if self.l1_in > 0:
+ node_features = {'0': type_0_features, '1': type_1_features}
+ else:
+ node_features = {'0': type_0_features}
+ edge_features = {'0': edge_features}
+ return self.se3(G, node_features, edge_features)
diff --git a/model/Track_module.py b/model/Track_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..e710dd3763b9eb6475fc609a1d26ee19063bc13a
--- /dev/null
+++ b/model/Track_module.py
@@ -0,0 +1,476 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import torch.utils.checkpoint as checkpoint
+from util import cross_product_matrix
+from util_module import *
+from Attention_module import *
+from SE3_network import SE3TransformerWrapper
+from icecream import ic
+
+# Components for three-track blocks
+# 1. MSA -> MSA update (biased attention. bias from pair & structure)
+# 2. Pair -> Pair update (biased attention. bias from structure)
+# 3. MSA -> Pair update (extract coevolution signal)
+# 4. Str -> Str update (node from MSA, edge from Pair)
+
+# Update MSA with biased self-attention. bias from Pair & Str
+class MSAPairStr2MSA(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, n_head=8, d_state=16,
+ d_hidden=32, p_drop=0.15, use_global_attn=False):
+ super(MSAPairStr2MSA, self).__init__()
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.proj_pair = nn.Linear(d_pair+36, d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.proj_state = nn.Linear(d_state, d_msa)
+ self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
+ self.row_attn = MSARowAttentionWithBias(d_msa=d_msa, d_pair=d_pair,
+ n_head=n_head, d_hidden=d_hidden)
+ if use_global_attn:
+ self.col_attn = MSAColGlobalAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
+ else:
+ self.col_attn = MSAColAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
+ self.ff = FeedForwardLayer(d_msa, 4, p_drop=p_drop)
+
+ # Do proper initialization
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize weights to normal distrib
+ self.proj_pair = init_lecun_normal(self.proj_pair)
+ self.proj_state = init_lecun_normal(self.proj_state)
+
+ # initialize bias to zeros
+ nn.init.zeros_(self.proj_pair.bias)
+ nn.init.zeros_(self.proj_state.bias)
+
+ def forward(self, msa, pair, rbf_feat, state):
+ '''
+ Inputs:
+ - msa: MSA feature (B, N, L, d_msa)
+ - pair: Pair feature (B, L, L, d_pair)
+ - rbf_feat: Ca-Ca distance feature calculated from xyz coordinates (B, L, L, 36)
+ - xyz: xyz coordinates (B, L, n_atom, 3)
+ - state: updated node features after SE(3)-Transformer layer (B, L, d_state)
+ Output:
+ - msa: Updated MSA feature (B, N, L, d_msa)
+ '''
+ B, N, L = msa.shape[:3]
+
+ # prepare input bias feature by combining pair & coordinate info
+ pair = self.norm_pair(pair)
+ pair = torch.cat((pair, rbf_feat), dim=-1)
+ pair = self.proj_pair(pair) # (B, L, L, d_pair)
+ #
+ # update query sequence feature (first sequence in the MSA) with feedbacks (state) from SE3
+ state = self.norm_state(state)
+ state = self.proj_state(state).reshape(B, 1, L, -1)
+
+ msa = msa.index_add(1, torch.tensor([0,], device=state.device), state.type(torch.float32))
+ #
+ # Apply row/column attention to msa & transform
+ msa = msa + self.drop_row(self.row_attn(msa, pair))
+ msa = msa + self.col_attn(msa)
+ msa = msa + self.ff(msa)
+
+ return msa
+
+class PairStr2Pair(nn.Module):
+ def __init__(self, d_pair=128, n_head=4, d_hidden=32, d_rbf=36, p_drop=0.15):
+ super(PairStr2Pair, self).__init__()
+
+ self.emb_rbf = nn.Linear(d_rbf, d_hidden)
+ self.proj_rbf = nn.Linear(d_hidden, d_pair)
+
+ self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
+ self.drop_col = Dropout(broadcast_dim=2, p_drop=p_drop)
+
+ self.row_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=True)
+ self.col_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=False)
+
+ self.ff = FeedForwardLayer(d_pair, 2)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.kaiming_normal_(self.emb_rbf.weight, nonlinearity='relu')
+ nn.init.zeros_(self.emb_rbf.bias)
+
+ self.proj_rbf = init_lecun_normal(self.proj_rbf)
+ nn.init.zeros_(self.proj_rbf.bias)
+
+ def forward(self, pair, rbf_feat):
+ B, L = pair.shape[:2]
+
+ rbf_feat = self.proj_rbf(F.relu_(self.emb_rbf(rbf_feat)))
+
+ pair = pair + self.drop_row(self.row_attn(pair, rbf_feat))
+ pair = pair + self.drop_col(self.col_attn(pair, rbf_feat))
+ pair = pair + self.ff(pair)
+ return pair
+
+class MSA2Pair(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_hidden=32, p_drop=0.15):
+ super(MSA2Pair, self).__init__()
+ self.norm = nn.LayerNorm(d_msa)
+ self.proj_left = nn.Linear(d_msa, d_hidden)
+ self.proj_right = nn.Linear(d_msa, d_hidden)
+ self.proj_out = nn.Linear(d_hidden*d_hidden, d_pair)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # normal initialization
+ self.proj_left = init_lecun_normal(self.proj_left)
+ self.proj_right = init_lecun_normal(self.proj_right)
+ nn.init.zeros_(self.proj_left.bias)
+ nn.init.zeros_(self.proj_right.bias)
+
+ # zero initialize output
+ nn.init.zeros_(self.proj_out.weight)
+ nn.init.zeros_(self.proj_out.bias)
+
+ def forward(self, msa, pair):
+ B, N, L = msa.shape[:3]
+ msa = self.norm(msa)
+ left = self.proj_left(msa)
+ right = self.proj_right(msa)
+ right = right / float(N)
+ out = einsum('bsli,bsmj->blmij', left, right).reshape(B, L, L, -1)
+ out = self.proj_out(out)
+
+ pair = pair + out
+
+ return pair
+
+class SCPred(nn.Module):
+ def __init__(self, d_msa=256, d_state=32, d_hidden=128, p_drop=0.15):
+ super(SCPred, self).__init__()
+ self.norm_s0 = nn.LayerNorm(d_msa)
+ self.norm_si = nn.LayerNorm(d_state)
+ self.linear_s0 = nn.Linear(d_msa, d_hidden)
+ self.linear_si = nn.Linear(d_state, d_hidden)
+
+ # ResNet layers
+ self.linear_1 = nn.Linear(d_hidden, d_hidden)
+ self.linear_2 = nn.Linear(d_hidden, d_hidden)
+ self.linear_3 = nn.Linear(d_hidden, d_hidden)
+ self.linear_4 = nn.Linear(d_hidden, d_hidden)
+
+ # Final outputs
+ self.linear_out = nn.Linear(d_hidden, 20)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # normal initialization
+ self.linear_s0 = init_lecun_normal(self.linear_s0)
+ self.linear_si = init_lecun_normal(self.linear_si)
+ self.linear_out = init_lecun_normal(self.linear_out)
+ nn.init.zeros_(self.linear_s0.bias)
+ nn.init.zeros_(self.linear_si.bias)
+ nn.init.zeros_(self.linear_out.bias)
+
+ # right before relu activation: He initializer (kaiming normal)
+ nn.init.kaiming_normal_(self.linear_1.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear_1.bias)
+ nn.init.kaiming_normal_(self.linear_3.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear_3.bias)
+
+ # right before residual connection: zero initialize
+ nn.init.zeros_(self.linear_2.weight)
+ nn.init.zeros_(self.linear_2.bias)
+ nn.init.zeros_(self.linear_4.weight)
+ nn.init.zeros_(self.linear_4.bias)
+
+ def forward(self, seq, state):
+ '''
+ Predict side-chain torsion angles along with backbone torsions
+ Inputs:
+ - seq: hidden embeddings corresponding to query sequence (B, L, d_msa)
+ - state: state feature (output l0 feature) from previous SE3 layer (B, L, d_state)
+ Outputs:
+ - si: predicted torsion angles (phi, psi, omega, chi1~4 with cos/sin, Cb bend, Cb twist, CG) (B, L, 10, 2)
+ '''
+ B, L = seq.shape[:2]
+ seq = self.norm_s0(seq)
+ state = self.norm_si(state)
+ si = self.linear_s0(seq) + self.linear_si(state)
+
+ si = si + self.linear_2(F.relu_(self.linear_1(F.relu_(si))))
+ si = si + self.linear_4(F.relu_(self.linear_3(F.relu_(si))))
+
+ si = self.linear_out(F.relu_(si))
+ return si.view(B, L, 10, 2)
+
+
+class Str2Str(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_state=16,
+ SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}, p_drop=0.1):
+ super(Str2Str, self).__init__()
+
+ # initial node & pair feature process
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+
+ self.embed_x = nn.Linear(d_msa+d_state, SE3_param['l0_in_features'])
+ self.embed_e1 = nn.Linear(d_pair, SE3_param['num_edge_features'])
+ self.embed_e2 = nn.Linear(SE3_param['num_edge_features']+36+1, SE3_param['num_edge_features'])
+
+ self.norm_node = nn.LayerNorm(SE3_param['l0_in_features'])
+ self.norm_edge1 = nn.LayerNorm(SE3_param['num_edge_features'])
+ self.norm_edge2 = nn.LayerNorm(SE3_param['num_edge_features'])
+
+ self.se3 = SE3TransformerWrapper(**SE3_param)
+ self.sc_predictor = SCPred(d_msa=d_msa, d_state=SE3_param['l0_out_features'],
+ p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize weights to normal distribution
+ self.embed_x = init_lecun_normal(self.embed_x)
+ self.embed_e1 = init_lecun_normal(self.embed_e1)
+ self.embed_e2 = init_lecun_normal(self.embed_e2)
+
+ # initialize bias to zeros
+ nn.init.zeros_(self.embed_x.bias)
+ nn.init.zeros_(self.embed_e1.bias)
+ nn.init.zeros_(self.embed_e2.bias)
+
+ @torch.cuda.amp.autocast(enabled=False)
+ def forward(self, msa, pair, R_in, T_in, xyz, state, idx, top_k=64, eps=1e-5):
+ B, N, L = msa.shape[:3]
+
+ state = state.type(torch.float32)
+ mas = msa.type(torch.float32)
+ pair = pair.type(torch.float32)
+ R_in = R_in.type(torch.float32)
+ T_in = T_in.type(torch.float32)
+ xyz = xyz.type(torch.float32)
+
+ #ic(msa.dtype)
+ #ic(pair.dtype)
+ #ic(R_in.dtype)
+ #ic(T_in.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+
+ # process msa & pair features
+ node = self.norm_msa(msa[:,0])
+ pair = self.norm_pair(pair)
+ state = self.norm_state(state)
+
+ node = torch.cat((node, state), dim=-1)
+ node = self.norm_node(self.embed_x(node))
+ pair = self.norm_edge1(self.embed_e1(pair))
+
+ neighbor = get_seqsep(idx)
+ rbf_feat = rbf(torch.cdist(xyz[:,:,1], xyz[:,:,1]))
+ pair = torch.cat((pair, rbf_feat, neighbor), dim=-1)
+ pair = self.norm_edge2(self.embed_e2(pair))
+
+ # define graph
+ if top_k != 0:
+ G, edge_feats = make_topk_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
+ else:
+ G, edge_feats = make_full_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
+ l1_feats = xyz - xyz[:,:,1,:].unsqueeze(2)
+ l1_feats = l1_feats.reshape(B*L, -1, 3)
+
+ # apply SE(3) Transformer & update coordinates
+ shift = self.se3(G, node.reshape(B*L, -1, 1), l1_feats, edge_feats)
+
+ state = shift['0'].reshape(B, L, -1) # (B, L, C)
+
+ offset = shift['1'].reshape(B, L, 2, 3)
+ delTi = offset[:,:,0,:] / 10.0 # translation
+ R = offset[:,:,1,:] / 100.0 # rotation
+
+ Qnorm = torch.sqrt( 1 + torch.sum(R*R, dim=-1) )
+ qA, qB, qC, qD = 1/Qnorm, R[:,:,0]/Qnorm, R[:,:,1]/Qnorm, R[:,:,2]/Qnorm
+
+ delRi = torch.zeros((B,L,3,3), device=xyz.device)
+ delRi[:,:,0,0] = qA*qA+qB*qB-qC*qC-qD*qD
+ delRi[:,:,0,1] = 2*qB*qC - 2*qA*qD
+ delRi[:,:,0,2] = 2*qB*qD + 2*qA*qC
+ delRi[:,:,1,0] = 2*qB*qC + 2*qA*qD
+ delRi[:,:,1,1] = qA*qA-qB*qB+qC*qC-qD*qD
+ delRi[:,:,1,2] = 2*qC*qD - 2*qA*qB
+ delRi[:,:,2,0] = 2*qB*qD - 2*qA*qC
+ delRi[:,:,2,1] = 2*qC*qD + 2*qA*qB
+ delRi[:,:,2,2] = qA*qA-qB*qB-qC*qC+qD*qD
+ #
+ ## convert vector to rotation matrix
+ #R_angle = torch.norm(R, dim=-1, keepdim=True) # (B, L, 1)
+ #cos_angle = torch.cos(R_angle).unsqueeze(2) # (B, L, 1, 1)
+ #sin_angle = torch.sin(R_angle).unsqueeze(2) # (B, L, 1, 1)
+ #R_vector = R / (R_angle+eps) # (B, L, 3)
+
+ #delRi = cos_angle*torch.eye(3, device=R.device).reshape(1,1,3,3) \
+ # + sin_angle*cross_product_matrix(R_vector) \
+ # + (1.0-cos_angle)*einsum('bni,bnj->bnij', R_vector, R_vector)
+
+ Ri = einsum('bnij,bnjk->bnik', delRi, R_in)
+ Ti = delTi + T_in #einsum('bnij,bnj->bni', delRi, T_in) + delTi
+
+ alpha = self.sc_predictor(msa[:,0], state)
+ return Ri, Ti, state, alpha
+
+class IterBlock(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128,
+ n_head_msa=8, n_head_pair=4,
+ use_global_attn=False,
+ d_hidden=32, d_hidden_msa=None, p_drop=0.15,
+ SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}):
+ super(IterBlock, self).__init__()
+ if d_hidden_msa == None:
+ d_hidden_msa = d_hidden
+
+ self.msa2msa = MSAPairStr2MSA(d_msa=d_msa, d_pair=d_pair,
+ n_head=n_head_msa,
+ d_state=SE3_param['l0_out_features'],
+ use_global_attn=use_global_attn,
+ d_hidden=d_hidden_msa, p_drop=p_drop)
+ self.msa2pair = MSA2Pair(d_msa=d_msa, d_pair=d_pair,
+ d_hidden=d_hidden//2, p_drop=p_drop)
+ #d_hidden=d_hidden, p_drop=p_drop)
+ self.pair2pair = PairStr2Pair(d_pair=d_pair, n_head=n_head_pair,
+ d_hidden=d_hidden, p_drop=p_drop)
+ self.str2str = Str2Str(d_msa=d_msa, d_pair=d_pair,
+ d_state=SE3_param['l0_out_features'],
+ SE3_param=SE3_param,
+ p_drop=p_drop)
+
+ def forward(self, msa, pair, R_in, T_in, xyz, state, idx, use_checkpoint=False):
+ rbf_feat = rbf(torch.cdist(xyz[:,:,1,:], xyz[:,:,1,:]))
+ if use_checkpoint:
+ msa = checkpoint.checkpoint(create_custom_forward(self.msa2msa), msa, pair, rbf_feat, state)
+ pair = checkpoint.checkpoint(create_custom_forward(self.msa2pair), msa, pair)
+ pair = checkpoint.checkpoint(create_custom_forward(self.pair2pair), pair, rbf_feat)
+ R, T, state, alpha = checkpoint.checkpoint(create_custom_forward(self.str2str, top_k=0), msa, pair, R_in, T_in, xyz, state, idx)
+ else:
+ msa = self.msa2msa(msa, pair, rbf_feat, state)
+ pair = self.msa2pair(msa, pair)
+ pair = self.pair2pair(pair, rbf_feat)
+ R, T, state, alpha = self.str2str(msa, pair, R_in, T_in, xyz, state, idx, top_k=0)
+
+ return msa, pair, R, T, state, alpha
+
+class IterativeSimulator(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=12, n_ref_block=4,
+ d_msa=256, d_msa_full=64, d_pair=128, d_hidden=32,
+ n_head_msa=8, n_head_pair=4,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ p_drop=0.15):
+ super(IterativeSimulator, self).__init__()
+ self.n_extra_block = n_extra_block
+ self.n_main_block = n_main_block
+ self.n_ref_block = n_ref_block
+
+ self.proj_state = nn.Linear(SE3_param_topk['l0_out_features'], SE3_param_full['l0_out_features'])
+ # Update with extra sequences
+ if n_extra_block > 0:
+ self.extra_block = nn.ModuleList([IterBlock(d_msa=d_msa_full, d_pair=d_pair,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ d_hidden_msa=8,
+ d_hidden=d_hidden,
+ p_drop=p_drop,
+ use_global_attn=True,
+ SE3_param=SE3_param_full)
+ for i in range(n_extra_block)])
+
+ # Update with seed sequences
+ if n_main_block > 0:
+ self.main_block = nn.ModuleList([IterBlock(d_msa=d_msa, d_pair=d_pair,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ d_hidden=d_hidden,
+ p_drop=p_drop,
+ use_global_attn=False,
+ SE3_param=SE3_param_full)
+ for i in range(n_main_block)])
+
+ self.proj_state2 = nn.Linear(SE3_param_full['l0_out_features'], SE3_param_topk['l0_out_features'])
+ # Final SE(3) refinement
+ if n_ref_block > 0:
+ self.str_refiner = Str2Str(d_msa=d_msa, d_pair=d_pair,
+ d_state=SE3_param_topk['l0_out_features'],
+ SE3_param=SE3_param_topk,
+ p_drop=p_drop)
+
+ self.reset_parameter()
+ def reset_parameter(self):
+ self.proj_state = init_lecun_normal(self.proj_state)
+ nn.init.zeros_(self.proj_state.bias)
+ self.proj_state2 = init_lecun_normal(self.proj_state2)
+ nn.init.zeros_(self.proj_state2.bias)
+
+ def forward(self, seq, msa, msa_full, pair, xyz_in, state, idx, use_checkpoint=False):
+ # input:
+ # seq: query sequence (B, L)
+ # msa: seed MSA embeddings (B, N, L, d_msa)
+ # msa_full: extra MSA embeddings (B, N, L, d_msa_full)
+ # pair: initial residue pair embeddings (B, L, L, d_pair)
+ # xyz_in: initial BB coordinates (B, L, n_atom, 3)
+ # state: initial state features containing mixture of query seq, sidechain, accuracy info (B, L, d_state)
+ # idx: residue index
+
+ B, L = pair.shape[:2]
+
+ R_in = torch.eye(3, device=xyz_in.device).reshape(1,1,3,3).expand(B, L, -1, -1)
+ T_in = xyz_in[:,:,1].clone()
+ xyz_in = xyz_in - T_in.unsqueeze(-2)
+
+ state = self.proj_state(state)
+
+ R_s = list()
+ T_s = list()
+ alpha_s = list()
+ for i_m in range(self.n_extra_block):
+ R_in = R_in.detach() # detach rotation (for stability)
+ T_in = T_in.detach()
+ # Get current BB structure
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+
+ msa_full, pair, R_in, T_in, state, alpha = self.extra_block[i_m](msa_full, pair,
+ R_in, T_in, xyz, state, idx,
+ use_checkpoint=use_checkpoint)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ for i_m in range(self.n_main_block):
+ R_in = R_in.detach()
+ T_in = T_in.detach()
+ # Get current BB structure
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+
+ msa, pair, R_in, T_in, state, alpha = self.main_block[i_m](msa, pair,
+ R_in, T_in, xyz, state, idx,
+ use_checkpoint=use_checkpoint)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ state = self.proj_state2(state)
+ for i_m in range(self.n_ref_block):
+ R_in = R_in.detach()
+ T_in = T_in.detach()
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+ R_in, T_in, state, alpha = self.str_refiner(msa, pair, R_in, T_in, xyz, state, idx, top_k=64)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ R_s = torch.stack(R_s, dim=0)
+ T_s = torch.stack(T_s, dim=0)
+ alpha_s = torch.stack(alpha_s, dim=0)
+
+ return msa, pair, R_s, T_s, alpha_s, state
diff --git a/model/__pycache__/Attention_module.cpython-310.pyc b/model/__pycache__/Attention_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b3322289b0411833d51fa3ec56561e167490a97c
Binary files /dev/null and b/model/__pycache__/Attention_module.cpython-310.pyc differ
diff --git a/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc b/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0640806f046ee39a07f43474a92e7863eac08249
Binary files /dev/null and b/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc differ
diff --git a/model/__pycache__/Embeddings.cpython-310.pyc b/model/__pycache__/Embeddings.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0e1de9e9bc00e4678d998dca23d1c4937120641a
Binary files /dev/null and b/model/__pycache__/Embeddings.cpython-310.pyc differ
diff --git a/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc b/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5d1316d2c7827ae620c416d0c6dce0ad4384589b
Binary files /dev/null and b/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc differ
diff --git a/model/__pycache__/SE3_network.cpython-310.pyc b/model/__pycache__/SE3_network.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9bd3c3173207da5f95e9fca5350bfefd7b00d763
Binary files /dev/null and b/model/__pycache__/SE3_network.cpython-310.pyc differ
diff --git a/model/__pycache__/Track_module.cpython-310.pyc b/model/__pycache__/Track_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b8bc6b2787f3fbde9891f4aa45c3e78949b10b4b
Binary files /dev/null and b/model/__pycache__/Track_module.cpython-310.pyc differ
diff --git a/model/__pycache__/ab_tools.cpython-310.pyc b/model/__pycache__/ab_tools.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9865c1ff58c72107bf69cfcf86336b8880d0b70e
Binary files /dev/null and b/model/__pycache__/ab_tools.cpython-310.pyc differ
diff --git a/model/__pycache__/apply_masks.cpython-310.pyc b/model/__pycache__/apply_masks.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..fe3081116d10b9f1d83620ff7bc99f231ed5632a
Binary files /dev/null and b/model/__pycache__/apply_masks.cpython-310.pyc differ
diff --git a/model/__pycache__/arguments.cpython-310.pyc b/model/__pycache__/arguments.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..fb972ac10b616e96ee62c25f471b52cfee4fe07a
Binary files /dev/null and b/model/__pycache__/arguments.cpython-310.pyc differ
diff --git a/model/__pycache__/chemical.cpython-310.pyc b/model/__pycache__/chemical.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5150406fe03402a121d1ba5c5057166aca9942dd
Binary files /dev/null and b/model/__pycache__/chemical.cpython-310.pyc differ
diff --git a/model/__pycache__/data_loader.cpython-310.pyc b/model/__pycache__/data_loader.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..8c14615387c282c92028e4ccb224c5e25f3a1141
Binary files /dev/null and b/model/__pycache__/data_loader.cpython-310.pyc differ
diff --git a/model/__pycache__/diffusion.cpython-310.pyc b/model/__pycache__/diffusion.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..184f706857ce375fce3d08ea762e3265d18231d0
Binary files /dev/null and b/model/__pycache__/diffusion.cpython-310.pyc differ
diff --git a/model/__pycache__/kinematics.cpython-310.pyc b/model/__pycache__/kinematics.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..75973ac48c41c032972159fba097a8f07e3e886d
Binary files /dev/null and b/model/__pycache__/kinematics.cpython-310.pyc differ
diff --git a/model/__pycache__/loss.cpython-310.pyc b/model/__pycache__/loss.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..63a1aee0f9ff190da7197bd870322242fb4b0716
Binary files /dev/null and b/model/__pycache__/loss.cpython-310.pyc differ
diff --git a/model/__pycache__/mask_generator.cpython-310.pyc b/model/__pycache__/mask_generator.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0d07980de4880838df2a2a548d2145b800a09b09
Binary files /dev/null and b/model/__pycache__/mask_generator.cpython-310.pyc differ
diff --git a/model/__pycache__/parsers.cpython-310.pyc b/model/__pycache__/parsers.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..3188471a2c59eb6a67aa63b841e9aef1548a1e4c
Binary files /dev/null and b/model/__pycache__/parsers.cpython-310.pyc differ
diff --git a/model/__pycache__/scheduler.cpython-310.pyc b/model/__pycache__/scheduler.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5a8f61d5257f9f26cb71049fdc8c68d9352d9aa3
Binary files /dev/null and b/model/__pycache__/scheduler.cpython-310.pyc differ
diff --git a/model/__pycache__/scoring.cpython-310.pyc b/model/__pycache__/scoring.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..dfd273eea8119c39592a933e7dd72b3f642a1371
Binary files /dev/null and b/model/__pycache__/scoring.cpython-310.pyc differ
diff --git a/model/__pycache__/train_multi_deep.cpython-310.pyc b/model/__pycache__/train_multi_deep.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..302fe224c624840c1200a68e22db9990b8e070f9
Binary files /dev/null and b/model/__pycache__/train_multi_deep.cpython-310.pyc differ
diff --git a/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc b/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..d3e9bafc9b0645924d26dfdc4a2dafa9e774705c
Binary files /dev/null and b/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc differ
diff --git a/model/__pycache__/util.cpython-310.pyc b/model/__pycache__/util.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b3d1ff3bb6e5fb456825033bb2e9c9be41a3a6ce
Binary files /dev/null and b/model/__pycache__/util.cpython-310.pyc differ
diff --git a/model/__pycache__/util_module.cpython-310.pyc b/model/__pycache__/util_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..bc0740e035a77ef694f6060b13d4683f99e5b4f6
Binary files /dev/null and b/model/__pycache__/util_module.cpython-310.pyc differ
diff --git a/model/apply_masks.py b/model/apply_masks.py
new file mode 100755
index 0000000000000000000000000000000000000000..ecb0fd70f8fd973aa4abb992188cd2335db0cc57
--- /dev/null
+++ b/model/apply_masks.py
@@ -0,0 +1,196 @@
+import sys, os
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+sys.path.append('../')
+from utils.calc_dssp import annotate_sse
+
+ic.configureOutput(includeContext=True)
+
+def mask_inputs(seq,
+ msa_masked,
+ msa_full,
+ xyz_t,
+ t1d,
+ mask_msa,
+ input_seq_mask=None,
+ input_str_mask=None,
+ input_floating_mask=None,
+ input_t1dconf_mask=None,
+ loss_seq_mask=None,
+ loss_str_mask=None,
+ loss_str_mask_2d=None,
+ dssp=False,
+ hotspots=False,
+ diffuser=None,
+ t=None,
+ freeze_seq_emb=False,
+ mutate_seq=False,
+ no_clamp_seq=False,
+ norm_input=False,
+ contacts=None,
+ frac_provide_dssp=0.5,
+ dssp_mask_percentage=[0,100],
+ frac_provide_contacts=0.5,
+ struc_cond=False):
+ """
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+
+
+
+ #ic(input_seq_mask.shape)
+ #ic(seq.shape)
+ #ic(msa_masked.shape)
+ #ic(msa_full.shape)
+ #ic(t1d.shape)
+ #ic(xyz_t.shape)
+ #ic(input_str_mask.shape)
+ #ic(mask_msa.shape)
+
+ ###########
+ seq_mask = input_seq_mask
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+
+ str_mask = input_str_mask
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+ seq_diffused = diffuser.q_sample(x_0,t,mask=seq_mask)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t]),mask=msa_seq_mask)
+
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #make msa_full same size as msa_masked
+ #ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ #to do add structure confidence metric; need to expand dimensions of chkpt b4
+ #if t1d_24: JG - changed to be default
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ if dssp:
+ print(f'adding dssp {frac_provide_dssp} of time')
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],4)).float()), -1).to(seq.device)
+ #dssp info
+ #mask some percentage of dssp info in range dssp_mask_percentage[0],dssp_mask_percentage[1]
+ percentage_mask=random.randint(dssp_mask_percentage[0], dssp_mask_percentage[1])
+ dssp=annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=percentage_mask)
+ #dssp_unmasked = annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=0)
+ if np.random.rand()>frac_provide_dssp:
+ print('masking dssp')
+ dssp[...]=0 #replace with mask token
+ dssp[:,-1]=1
+ t1d[...,24:]=dssp
+
+ if hotspots:
+ print(f"adding hotspots {frac_provide_contacts} of time")
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ #mask all contacts some fraction of the time
+ if np.random.rand()>frac_provide_contacts:
+ print('masking contacts')
+ contacts = torch.zeros(L)
+ t1d[...,-1] = contacts
+
+ ### xyz_t ###
+ #############
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+ #Sequence masking
+ xyz_t[:,:,3:,:] = float('nan')
+ # Structure masking
+ if struc_cond:
+ print("non-autoregressive structure conditioning")
+ r = diffuser.alphas_cumprod[t]
+ xyz_mask = (torch.rand(xyz_t.shape[1]) > r).to(torch.bool).to(seq.device)
+ xyz_mask = torch.logical_and(xyz_mask,~str_mask)
+ xyz_t[:,xyz_mask,:,:] = float('nan')
+ else:
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ ### mask_msa ###
+ ################
+ # NOTE: this is for loss scoring
+ mask_msa[:,:,~loss_seq_mask] = False
+
+ out=dict(
+ seq= seq,
+ msa_masked= msa_masked,
+ msa_full= msa_full,
+ xyz_t= xyz_t,
+ t1d= t1d,
+ mask_msa= mask_msa,
+ seq_diffused= seq_diffused
+ )
+
+ return out
diff --git a/model/chemical.py b/model/chemical.py
new file mode 100644
index 0000000000000000000000000000000000000000..206adc88ee09010b80e265c2d16b2c6fd447fa43
--- /dev/null
+++ b/model/chemical.py
@@ -0,0 +1,570 @@
+import torch
+import numpy as np
+
+num2aa=[
+ 'ALA','ARG','ASN','ASP','CYS',
+ 'GLN','GLU','GLY','HIS','ILE',
+ 'LEU','LYS','MET','PHE','PRO',
+ 'SER','THR','TRP','TYR','VAL',
+ 'UNK','MAS',
+ ]
+
+aa2num= {x:i for i,x in enumerate(num2aa)}
+
+# full sc atom representation (Nx14)
+aa2long=[
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # ala
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," NE "," CZ "," NH1"," NH2", None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD "," HE ","1HH1","2HH1","1HH2","2HH2"), # arg
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," ND2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HD2","2HD2", None, None, None, None, None, None, None), # asn
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," OD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None, None), # asp
+ (" N "," CA "," C "," O "," CB "," SG ", None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ", None, None, None, None, None, None, None, None), # cys
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," NE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE2","2HE2", None, None, None, None, None), # gln
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," OE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ", None, None, None, None, None, None, None), # glu
+ (" N "," CA "," C "," O ", None, None, None, None, None, None, None, None, None, None," H ","1HA ","2HA ", None, None, None, None, None, None, None, None, None, None), # gly
+ (" N "," CA "," C "," O "," CB "," CG "," ND1"," CD2"," CE1"," NE2", None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1"," HE2", None, None, None, None, None, None), # his
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2"," CD1", None, None, None, None, None, None," H "," HA "," HB ","1HG2","2HG2","3HG2","1HG1","2HG1","1HD1","2HD1","3HD1", None, None), # ile
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ","1HD1","2HD1","3HD1","1HD2","2HD2","3HD2", None, None), # leu
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," CE "," NZ ", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ","1HE ","2HE ","1HZ ","2HZ ","3HZ "), # lys
+ (" N "," CA "," C "," O "," CB "," CG "," SD "," CE ", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE ","2HE ","3HE ", None, None, None, None), # met
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," CE1"," CE2"," CZ ", None, None, None," H "," HA ","1HB ","2HB "," HD1"," HD2"," HE1"," HE2"," HZ ", None, None, None, None), # phe
+ (" N "," CA "," C "," O "," CB "," CG "," CD ", None, None, None, None, None, None, None," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ", None, None, None, None, None, None), # pro
+ (" N "," CA "," C "," O "," CB "," OG ", None, None, None, None, None, None, None, None," H "," HG "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None), # ser
+ (" N "," CA "," C "," O "," CB "," OG1"," CG2", None, None, None, None, None, None, None," H "," HG1"," HA "," HB ","1HG2","2HG2","3HG2", None, None, None, None, None, None), # thr
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," NE1"," CE2"," CE3"," CZ2"," CZ3"," CH2"," H "," HA ","1HB ","2HB "," HD1"," HE1"," HZ2"," HH2"," HZ3"," HE3", None, None, None), # trp
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," CE1"," CE2"," CZ "," OH ", None, None," H "," HA ","1HB ","2HB "," HD1"," HE1"," HE2"," HD2"," HH ", None, None, None, None), # tyr
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2", None, None, None, None, None, None, None," H "," HA "," HB ","1HG1","2HG1","3HG1","1HG2","2HG2","3HG2", None, None, None, None), # val
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # unk
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # mask
+]
+
+# build the "alternate" sc mapping
+aa2longalt=[
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # ala
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," NE "," CZ "," NH1"," NH2", None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD "," HE ","1HH1","2HH1","1HH2","2HH2"), # arg
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," ND2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HD2","2HD2", None, None, None, None, None, None, None), # asn
+ (" N "," CA "," C "," O "," CB "," CG "," OD2"," OD1", None, None, None, None, None, None," H "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None, None), # asp
+ (" N "," CA "," C "," O "," CB "," SG ", None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ", None, None, None, None, None, None, None, None), # cys
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," NE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE2","2HE2", None, None, None, None, None), # gln
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE2"," OE1", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ", None, None, None, None, None, None, None), # glu
+ (" N "," CA "," C "," O ", None, None, None, None, None, None, None, None, None, None," H ","1HA ","2HA ", None, None, None, None, None, None, None, None, None, None), # gly
+ (" N "," CA "," C "," O "," CB "," CG "," NE2"," CD2"," CE1"," ND1", None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1"," HE2", None, None, None, None, None, None), # his
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2"," CD1", None, None, None, None, None, None," H "," HA "," HB ","1HG2","2HG2","3HG2","1HG1","2HG1","1HD1","2HD1","3HD1", None, None), # ile
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ","1HD1","2HD1","3HD1","1HD2","2HD2","3HD2", None, None), # leu
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," CE "," NZ ", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ","1HE ","2HE ","1HZ ","2HZ ","3HZ "), # lys
+ (" N "," CA "," C "," O "," CB "," CG "," SD "," CE ", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE ","2HE ","3HE ", None, None, None, None), # met
+ (" N "," CA "," C "," O "," CB "," CG "," CD2"," CD1"," CE2"," CE1"," CZ ", None, None, None," H "," HD2"," HE2"," HZ "," HE1"," HD1"," HA ","1HB ","2HB ", None, None, None, None), # phe
+ (" N "," CA "," C "," O "," CB "," CG "," CD ", None, None, None, None, None, None, None," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ", None, None, None, None, None, None), # pro
+ (" N "," CA "," C "," O "," CB "," OG ", None, None, None, None, None, None, None, None," H "," HG "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None), # ser
+ (" N "," CA "," C "," O "," CB "," OG1"," CG2", None, None, None, None, None, None, None," H "," HG1"," HA "," HB ","1HG2","2HG2","3HG2", None, None, None, None, None, None), # thr
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," NE1"," CE2"," CE3"," CZ2"," CZ3"," CH2"," H "," HA ","1HB ","2HB "," HD1"," HE1"," HZ2"," HH2"," HZ3"," HE3", None, None, None), # trp
+ (" N "," CA "," C "," O "," CB "," CG "," CD2"," CD1"," CE2"," CE1"," CZ "," OH ", None, None," H "," HA ","1HB ","2HB "," HD2"," HE2"," HE1"," HD1"," HH ", None, None, None, None), # tyr
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2", None, None, None, None, None, None, None," H "," HA "," HB ","1HG1","2HG1","3HG1","1HG2","2HG2","3HG2", None, None, None, None), # val
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # unk
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # mask
+]
+
+aabonds=[
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # ala
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," NE "),(" CD ","1HD "),(" CD ","2HD "),(" NE "," CZ "),(" NE "," HE "),(" CZ "," NH1"),(" CZ "," NH2"),(" NH1","1HH1"),(" NH1","2HH1"),(" NH2","1HH2"),(" NH2","2HH2")) , # arg
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," OD1"),(" CG "," ND2"),(" ND2","1HD2"),(" ND2","2HD2")) , # asn
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," OD1"),(" CG "," OD2")) , # asp
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," SG "),(" CB ","1HB "),(" CB ","2HB "),(" SG "," HG ")) , # cys
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," OE1"),(" CD "," NE2"),(" NE2","1HE2"),(" NE2","2HE2")) , # gln
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," OE1"),(" CD "," OE2")) , # glu
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA ","1HA "),(" CA ","2HA "),(" C "," O ")) , # gly
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," ND1"),(" CG "," CD2"),(" ND1"," CE1"),(" CD2"," NE2"),(" CD2"," HD2"),(" CE1"," NE2"),(" CE1"," HE1"),(" NE2"," HE2")) , # his
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG1"),(" CB "," CG2"),(" CB "," HB "),(" CG1"," CD1"),(" CG1","1HG1"),(" CG1","2HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2"),(" CD1","1HD1"),(" CD1","2HD1"),(" CD1","3HD1")) , # ile
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CG "," HG "),(" CD1","1HD1"),(" CD1","2HD1"),(" CD1","3HD1"),(" CD2","1HD2"),(" CD2","2HD2"),(" CD2","3HD2")) , # leu
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," CE "),(" CD ","1HD "),(" CD ","2HD "),(" CE "," NZ "),(" CE ","1HE "),(" CE ","2HE "),(" NZ ","1HZ "),(" NZ ","2HZ "),(" NZ ","3HZ ")) , # lys
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," SD "),(" CG ","1HG "),(" CG ","2HG "),(" SD "," CE "),(" CE ","1HE "),(" CE ","2HE "),(" CE ","3HE ")) , # met
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," CE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," HD2"),(" CE1"," CZ "),(" CE1"," HE1"),(" CE2"," CZ "),(" CE2"," HE2"),(" CZ "," HZ ")) , # phe
+ ((" N "," CA "),(" N "," CD "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD ","1HD "),(" CD ","2HD ")) , # pro
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," OG "),(" CB ","1HB "),(" CB ","2HB "),(" OG "," HG ")) , # ser
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," OG1"),(" CB "," CG2"),(" CB "," HB "),(" OG1"," HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2")) , # thr
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," NE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," CE3"),(" NE1"," CE2"),(" NE1"," HE1"),(" CE2"," CZ2"),(" CE3"," CZ3"),(" CE3"," HE3"),(" CZ2"," CH2"),(" CZ2"," HZ2"),(" CZ3"," CH2"),(" CZ3"," HZ3"),(" CH2"," HH2")) , # trp
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," CE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," HD2"),(" CE1"," CZ "),(" CE1"," HE1"),(" CE2"," CZ "),(" CE2"," HE2"),(" CZ "," OH "),(" OH "," HH ")) , # tyr
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG1"),(" CB "," CG2"),(" CB "," HB "),(" CG1","1HG1"),(" CG1","2HG1"),(" CG1","3HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2")), # val
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # unk
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # mask
+]
+
+aa2type = [
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # ala
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CH2", "NtrR","aroC","Narg","Narg", None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol","Hpol","Hpol","Hpol"), # arg
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CNH2","ONH2","NH2O", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hpol","Hpol", None, None, None, None, None, None, None), # asn
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "COO", "OOC", "OOC", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None, None), # asp
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "SH1", None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","HS", None, None, None, None, None, None, None, None), # cys
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CNH2","ONH2","NH2O", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol", None, None, None, None, None), # gln
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "COO", "OOC", "OOC", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None), # glu
+ ("Nbb", "CAbb","CObb","OCbb", None, None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo", None, None, None, None, None, None, None, None, None, None), # gly
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "Nhis","aroC","aroC","Ntrp", None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hpol","Hapo","Hapo", None, None, None, None, None, None), # his
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "CH2", "CH3", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None), # ile
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH1", "CH3", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None), # leu
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CH2", "CH2", "Nlys", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol","Hpol"), # lys
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "S", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None), # met
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","aroC","aroC","aroC","aroC", None, None, None,"HNbb","Hapo","Hapo","Hapo","Haro","Haro","Haro","Haro","Haro", None, None, None, None), # phe
+ ("Npro","CAbb","CObb","OCbb","CH2", "CH2", "CH2", None, None, None, None, None, None, None,"Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None), # pro
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "OH", None, None, None, None, None, None, None, None,"HNbb","Hpol","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # ser
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "OH", "CH3", None, None, None, None, None, None, None,"HNbb","Hpol","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None), # thr
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","CH0", "Ntrp","CH0", "aroC","aroC","aroC","aroC","HNbb","Haro","Hapo","Hapo","Hapo","Hpol","Haro","Haro","Haro","Haro", None, None, None), # trp
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","aroC","aroC","aroC","CH0", "OHY", None, None,"HNbb","Haro","Haro","Haro","Haro","Hapo","Hapo","Hapo","Hpol", None, None, None, None), # tyr
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "CH3", "CH3", None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None), # val
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # unk
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # mask
+]
+
+# tip atom
+aa2tip = [
+ " CB ", # ala
+ " CZ ", # arg
+ " ND2", # asn
+ " CG ", # asp
+ " SG ", # cys
+ " NE2", # gln
+ " CD ", # glu
+ " CA ", # gly
+ " NE2", # his
+ " CD1", # ile
+ " CG ", # leu
+ " NZ ", # lys
+ " SD ", # met
+ " CZ ", # phe
+ " CG ", # pro
+ " OG ", # ser
+ " OG1", # thr
+ " CH2", # trp
+ " OH ", # tyr
+ " CB ", # val
+ " CB ", # unknown (gap etc)
+ " CB " # masked
+ ]
+
+
+torsions=[
+ [ None, None, None, None ], # ala
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," NE "], [" CG "," CD "," NE "," CZ "] ], # arg
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," OD1"], None, None ], # asn
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," OD1"], None, None ], # asp
+ [ [" N "," CA "," CB "," SG "], [" CA "," CB "," SG "," HG "], None, None ], # cys
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," OE1"], None ], # gln
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," OE1"], None ], # glu
+ [ None, None, None, None ], # gly
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," ND1"], [" CD2"," CE1"," HE1"," NE2"], None ], # his (protonation handled as a pseudo-torsion)
+ [ [" N "," CA "," CB "," CG1"], [" CA "," CB "," CG1"," CD1"], None, None ], # ile
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # leu
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," CE "], [" CG "," CD "," CE "," NZ "] ], # lys
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," SD "], [" CB "," CG "," SD "," CE "], None ], # met
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # phe
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD ","1HD "], None ], # pro
+ [ [" N "," CA "," CB "," OG "], [" CA "," CB "," OG "," HG "], None, None ], # ser
+ [ [" N "," CA "," CB "," OG1"], [" CA "," CB "," OG1"," HG1"], None, None ], # thr
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # trp
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], [" CE1"," CZ "," OH "," HH "], None ], # tyr
+ [ [" N "," CA "," CB "," CG1"], None, None, None ], # val
+ [ None, None, None, None ], # unk
+ [ None, None, None, None ], # mask
+]
+
+# ideal N, CA, C initial coordinates
+init_N = torch.tensor([-0.5272, 1.3593, 0.000]).float()
+init_CA = torch.zeros_like(init_N)
+init_C = torch.tensor([1.5233, 0.000, 0.000]).float()
+INIT_CRDS = torch.full((27, 3), np.nan)
+INIT_CRDS[:3] = torch.stack((init_N, init_CA, init_C), dim=0) # (3,3)
+
+norm_N = init_N / (torch.norm(init_N, dim=-1, keepdim=True) + 1e-5)
+norm_C = init_C / (torch.norm(init_C, dim=-1, keepdim=True) + 1e-5)
+cos_ideal_NCAC = torch.sum(norm_N*norm_C, dim=-1) # cosine of ideal N-CA-C bond angle
+
+#fd Rosetta ideal coords
+#fd - uses same "frame-building" as AF2
+ideal_coords = [
+ [ # 0 ala
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+ [ # 1 arg
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3467, -0.5055, 0.9018)],
+ [' CB ', 8, (-0.5042,-0.7698,-1.2118)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3639, -0.5323, -0.8789)],
+ [' CG ', 4, (0.6396,1.3794, 0.000)],
+ ['1HG ', 5, (0.3639, -0.5139, 0.8900)],
+ ['2HG ', 5, (0.3641, -0.5140, -0.8903)],
+ [' CD ', 5, (0.5492,1.3801, 0.000)],
+ ['1HD ', 6, (0.3637, -0.5135, 0.8895)],
+ ['2HD ', 6, (0.3636, -0.5134, -0.8893)],
+ [' NE ', 6, (0.5423,1.3491, 0.000)],
+ [' NH1', 7, (0.2012,2.2965, 0.000)],
+ [' NH2', 7, (2.0824,1.0030, 0.000)],
+ [' CZ ', 7, (0.7650,1.1090, 0.000)],
+ [' HE ', 7, (0.4701,-0.8955, 0.000)],
+ ['1HH1', 7, (-0.8059,2.3776, 0.000)],
+ ['1HH2', 7, (2.5160,0.0898, 0.000)],
+ ['2HH1', 7, (0.7745,3.1277, 0.000)],
+ ['2HH2', 7, (2.6554,1.8336, 0.000)],
+ ],
+ [ # 2 asn
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3233, -0.4967, 0.9162)],
+ [' CB ', 8, (-0.5341,-0.7799,-1.1874)],
+ ['1HB ', 4, ( 0.3641, -0.5327, 0.8795)],
+ ['2HB ', 4, ( 0.3639, -0.5323, -0.8789)],
+ [' CG ', 4, (0.5778,1.3881, 0.000)],
+ [' ND2', 5, (0.5839,-1.1711, 0.000)],
+ [' OD1', 5, (0.6331,1.0620, 0.000)],
+ ['1HD2', 5, (1.5825, -1.2322, 0.000)],
+ ['2HD2', 5, (0.0323, -2.0046, 0.000)],
+ ],
+ [ # 3 asp
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3233, -0.4967, 0.9162)],
+ [' CB ', 8, (-0.5162,-0.7757,-1.2144)],
+ ['1HB ', 4, ( 0.3639, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3640, -0.5325, -0.8792)],
+ [' CG ', 4, (0.5926,1.4028, 0.000)],
+ [' OD1', 5, (0.5746,1.0629, 0.000)],
+ [' OD2', 5, (0.5738,-1.0627, 0.000)],
+ ],
+ [ # 4 cys
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3481, -0.5059, 0.9006)],
+ [' CB ', 8, (-0.5046,-0.7727,-1.2189)],
+ ['1HB ', 4, ( 0.3639, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8787)],
+ [' SG ', 4, (0.7386,1.6511, 0.000)],
+ [' HG ', 5, (0.1387,1.3221, 0.000)],
+ ],
+ [ # 5 gln
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3363, -0.5013, 0.9074)],
+ [' CB ', 8, (-0.5226,-0.7776,-1.2109)],
+ ['1HB ', 4, ( 0.3638, -0.5323, 0.8789)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8788)],
+ [' CG ', 4, (0.6225,1.3857, 0.000)],
+ ['1HG ', 5, ( 0.3531, -0.5156, 0.8931)],
+ ['2HG ', 5, ( 0.3531, -0.5156, -0.8931)],
+ [' CD ', 5, (0.5788,1.4021, 0.000)],
+ [' NE2', 6, (0.5908,-1.1895, 0.000)],
+ [' OE1', 6, (0.6347,1.0584, 0.000)],
+ ['1HE2', 6, (1.5825, -1.2525, 0.000)],
+ ['2HE2', 6, (0.0380, -2.0229, 0.000)],
+ ],
+ [ # 6 glu
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3363, -0.5013, 0.9074)],
+ [' CB ', 8, (-0.5197,-0.7737,-1.2137)],
+ ['1HB ', 4, ( 0.3638, -0.5323, 0.8789)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8788)],
+ [' CG ', 4, (0.6287,1.3862, 0.000)],
+ ['1HG ', 5, ( 0.3531, -0.5156, 0.8931)],
+ ['2HG ', 5, ( 0.3531, -0.5156, -0.8931)],
+ [' CD ', 5, (0.5850,1.3849, 0.000)],
+ [' OE1', 6, (0.5752,1.0618, 0.000)],
+ [' OE2', 6, (0.5741,-1.0635, 0.000)],
+ ],
+ [ # 7 gly
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ ['1HA ', 0, ( -0.3676, -0.5329, 0.8771)],
+ ['2HA ', 0, ( -0.3674, -0.5325, -0.8765)],
+ ],
+ [ # 8 his
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3299, -0.5180, 0.9001)],
+ [' CB ', 8, (-0.5163,-0.7809,-1.2129)],
+ ['1HB ', 4, ( 0.3640, -0.5325, 0.8793)],
+ ['2HB ', 4, ( 0.3637, -0.5321, -0.8786)],
+ [' CG ', 4, (0.6016,1.3710, 0.000)],
+ [' CD2', 5, (0.8918,-1.0184, 0.000)],
+ [' CE1', 5, (2.0299,0.8564, 0.000)],
+ [' HE1', 5, (2.8542, 1.5693, 0.000)],
+ [' HD2', 5, ( 0.6584, -2.0835, 0.000) ],
+ [' ND1', 6, (-1.8631, -1.0722, 0.000)],
+ [' NE2', 6, (-1.8625, 1.0707, 0.000)],
+ [' HE2', 6, (-1.5439, 2.0292, 0.000)],
+ ],
+ [ # 9 ile
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3405, -0.5028, 0.9044)],
+ [' CB ', 8, (-0.5140,-0.7885,-1.2184)],
+ [' HB ', 4, (0.3637, -0.4714, 0.9125)],
+ [' CG1', 4, (0.5339,1.4348,0.000)],
+ [' CG2', 4, (0.5319,-0.7693,-1.1994)],
+ ['1HG2', 4, (1.6215, -0.7588, -1.1842)],
+ ['2HG2', 4, (0.1785, -1.7986, -1.1569)],
+ ['3HG2', 4, (0.1773, -0.3016, -2.1180)],
+ [' CD1', 5, (0.6106,1.3829, 0.000)],
+ ['1HG1', 5, (0.3637, -0.5338, 0.8774)],
+ ['2HG1', 5, (0.3640, -0.5322, -0.8793)],
+ ['1HD1', 5, (1.6978, 1.3006, 0.000)],
+ ['2HD1', 5, (0.2873, 1.9236, -0.8902)],
+ ['3HD1', 5, (0.2888, 1.9224, 0.8896)],
+ ],
+ [ # 10 leu
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.525, -0.000, -0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3435, -0.5040, 0.9027)],
+ [' CB ', 8, (-0.5175,-0.7692,-1.2220)],
+ ['1HB ', 4, ( 0.3473, -0.5346, 0.8827)],
+ ['2HB ', 4, ( 0.3476, -0.5351, -0.8836)],
+ [' CG ', 4, (0.6652,1.3823, 0.000)],
+ [' CD1', 5, (0.5083,1.4353, 0.000)],
+ [' CD2', 5, (0.5079,-0.7600,1.2163)],
+ [' HG ', 5, (0.3640, -0.4825, -0.9075)],
+ ['1HD1', 5, (1.5984, 1.4353, 0.000)],
+ ['2HD1', 5, (0.1462, 1.9496, -0.8903)],
+ ['3HD1', 5, (0.1459, 1.9494, 0.8895)],
+ ['1HD2', 5, (1.5983, -0.7606, 1.2158)],
+ ['2HD2', 5, (0.1456, -0.2774, 2.1243)],
+ ['3HD2', 5, (0.1444, -1.7871, 1.1815)],
+ ],
+ [ # 11 lys
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3335, -0.5005, 0.9097)],
+ ['1HB ', 4, ( 0.3640, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3639, -0.5324, -0.8790)],
+ [' CB ', 8, (-0.5259,-0.7785,-1.2069)],
+ ['1HG ', 5, (0.3641, -0.5229, 0.8852)],
+ ['2HG ', 5, (0.3637, -0.5227, -0.8841)],
+ [' CG ', 4, (0.6291,1.3869, 0.000)],
+ [' CD ', 5, (0.5526,1.4174, 0.000)],
+ ['1HD ', 6, (0.3641, -0.5239, 0.8848)],
+ ['2HD ', 6, (0.3638, -0.5219, -0.8850)],
+ [' CE ', 6, (0.5544,1.4170, 0.000)],
+ [' NZ ', 7, (0.5566,1.3801, 0.000)],
+ ['1HE ', 7, (0.4199, -0.4638, 0.9482)],
+ ['2HE ', 7, (0.4202, -0.4631, -0.8172)],
+ ['1HZ ', 7, (1.6223, 1.3980, 0.0658)],
+ ['2HZ ', 7, (0.2970, 1.9326, -0.7584)],
+ ['3HZ ', 7, (0.2981, 1.9319, 0.8909)],
+ ],
+ [ # 12 met
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3303, -0.4990, 0.9108)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3641, -0.5326, -0.8795)],
+ [' CB ', 8, (-0.5331,-0.7727,-1.2048)],
+ ['1HG ', 5, (0.3637, -0.5256, 0.8823)],
+ ['2HG ', 5, (0.3638, -0.5249, -0.8831)],
+ [' CG ', 4, (0.6298,1.3858,0.000)],
+ [' SD ', 5, (0.6953,1.6645,0.000)],
+ [' CE ', 6, (0.3383,1.7581,0.000)],
+ ['1HE ', 6, (1.7054, 2.0532, -0.0063)],
+ ['2HE ', 6, (0.1906, 2.3099, -0.9072)],
+ ['3HE ', 6, (0.1917, 2.3792, 0.8720)],
+ ],
+ [ # 13 phe
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3303, -0.4990, 0.9108)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3641, -0.5326, -0.8795)],
+ [' CB ', 8, (-0.5150,-0.7729,-1.2156)],
+ [' CG ', 4, (0.6060,1.3746, 0.000)],
+ [' CD1', 5, (0.7078,1.1928, 0.000)],
+ [' CD2', 5, (0.7084,-1.1920, 0.000)],
+ [' CE1', 5, (2.0900,1.1940, 0.000)],
+ [' CE2', 5, (2.0897,-1.1939, 0.000)],
+ [' CZ ', 5, (2.7809, 0.000, 0.000)],
+ [' HD1', 5, (0.1613, 2.1362, 0.000)],
+ [' HD2', 5, (0.1621, -2.1360, 0.000)],
+ [' HE1', 5, (2.6335, 2.1384, 0.000)],
+ [' HE2', 5, (2.6344, -2.1378, 0.000)],
+ [' HZ ', 5, (3.8700, 0.000, 0.000)],
+ ],
+ [ # 14 pro
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' HA ', 0, (-0.3868, -0.5380, 0.8781)],
+ ['1HB ', 4, ( 0.3762, -0.5355, 0.8842)],
+ ['2HB ', 4, ( 0.3762, -0.5355, -0.8842)],
+ [' CB ', 8, (-0.5649,-0.5888,-1.2966)],
+ [' CG ', 4, (0.3657,1.4451,0.0000)],
+ [' CD ', 5, (0.3744,1.4582, 0.0)],
+ ['1HG ', 5, (0.3798, -0.5348, 0.8830)],
+ ['2HG ', 5, (0.3798, -0.5348, -0.8830)],
+ ['1HD ', 6, (0.3798, -0.5348, 0.8830)],
+ ['2HD ', 6, (0.3798, -0.5348, -0.8830)],
+ ],
+ [ # 15 ser
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3425, -0.5041, 0.9048)],
+ ['1HB ', 4, ( 0.3637, -0.5321, 0.8786)],
+ ['2HB ', 4, ( 0.3636, -0.5319, -0.8782)],
+ [' CB ', 8, (-0.5146,-0.7595,-1.2073)],
+ [' OG ', 4, (0.5021,1.3081, 0.000)],
+ [' HG ', 5, (0.2647, 0.9230, 0.000)],
+ ],
+ [ # 16 thr
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3364, -0.5015, 0.9078)],
+ [' HB ', 4, ( 0.3638, -0.5006, 0.8971)],
+ ['1HG2', 4, ( 1.6231, -0.7142, -1.2097)],
+ ['2HG2', 4, ( 0.1792, -1.7546, -1.2237)],
+ ['3HG2', 4, ( 0.1808, -0.2222, -2.1269)],
+ [' CB ', 8, (-0.5172,-0.7952,-1.2130)],
+ [' CG2', 4, (0.5334,-0.7239,-1.2267)],
+ [' OG1', 4, (0.4804,1.3506,0.000)],
+ [' HG1', 5, (0.3194, 0.9056, 0.000)],
+ ],
+ [ # 17 trp
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3436, -0.5042, 0.9031)],
+ ['1HB ', 4, ( 0.3639, -0.5323, 0.8790)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8787)],
+ [' CB ', 8, (-0.5136,-0.7712,-1.2173)],
+ [' CG ', 4, (0.5984,1.3741, 0.000)],
+ [' CD1', 5, (0.8151,1.0921, 0.000)],
+ [' CD2', 5, (0.8753,-1.1538, 0.000)],
+ [' CE2', 5, (2.1865,-0.6707, 0.000)],
+ [' CE3', 5, (0.6541,-2.5366, 0.000)],
+ [' NE1', 5, (2.1309,0.7003, 0.000)],
+ [' CH2', 5, (3.0315,-2.8930, 0.000)],
+ [' CZ2', 5, (3.2813,-1.5205, 0.000)],
+ [' CZ3', 5, (1.7521,-3.3888, 0.000)],
+ [' HD1', 5, (0.4722, 2.1252, 0.000)],
+ [' HE1', 5, ( 2.9291, 1.3191, 0.000)],
+ [' HE3', 5, (-0.3597, -2.9356, 0.000)],
+ [' HZ2', 5, (4.3053, -1.1462, 0.000)],
+ [' HZ3', 5, ( 1.5712, -4.4640, 0.000)],
+ [' HH2', 5, ( 3.8700, -3.5898, 0.000)],
+ ],
+ [ # 18 tyr
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3305, -0.4992, 0.9112)],
+ ['1HB ', 4, ( 0.3642, -0.5327, 0.8797)],
+ ['2HB ', 4, ( 0.3637, -0.5321, -0.8785)],
+ [' CB ', 8, (-0.5305,-0.7799,-1.2051)],
+ [' CG ', 4, (0.6104,1.3840, 0.000)],
+ [' CD1', 5, (0.6936,1.2013, 0.000)],
+ [' CD2', 5, (0.6934,-1.2011, 0.000)],
+ [' CE1', 5, (2.0751,1.2013, 0.000)],
+ [' CE2', 5, (2.0748,-1.2011, 0.000)],
+ [' OH ', 5, (4.1408, 0.000, 0.000)],
+ [' CZ ', 5, (2.7648, 0.000, 0.000)],
+ [' HD1', 5, (0.1485, 2.1455, 0.000)],
+ [' HD2', 5, (0.1484, -2.1451, 0.000)],
+ [' HE1', 5, (2.6200, 2.1450, 0.000)],
+ [' HE2', 5, (2.6199, -2.1453, 0.000)],
+ [' HH ', 6, (0.3190, 0.9057, 0.000)],
+ ],
+ [ # 19 val
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3497, -0.5068, 0.9002)],
+ [' CB ', 8, (-0.5105,-0.7712,-1.2317)],
+ [' CG1', 4, (0.5326,1.4252, 0.000)],
+ [' CG2', 4, (0.5177,-0.7693,1.2057)],
+ [' HB ', 4, (0.3541, -0.4754, -0.9148)],
+ ['1HG1', 4, (1.6228, 1.4063, 0.000)],
+ ['2HG1', 4, (0.1790, 1.9457, -0.8898)],
+ ['3HG1', 4, (0.1798, 1.9453, 0.8903)],
+ ['1HG2', 4, (1.6073, -0.7659, 1.1989)],
+ ['2HG2', 4, (0.1586, -0.2971, 2.1203)],
+ ['3HG2', 4, (0.1582, -1.7976, 1.1631)],
+ ],
+ [ # 20 unk
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+ [ # 21 mask
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+]
diff --git a/model/coords6d.py b/model/coords6d.py
new file mode 100644
index 0000000000000000000000000000000000000000..9f10d593851674d083b9a906c84ee2762839f1d2
--- /dev/null
+++ b/model/coords6d.py
@@ -0,0 +1,80 @@
+import numpy as np
+import scipy
+import scipy.spatial
+
+# calculate dihedral angles defined by 4 sets of points
+def get_dihedrals(a, b, c, d):
+
+ b0 = -1.0*(b - a)
+ b1 = c - b
+ b2 = d - c
+
+ b1 /= np.linalg.norm(b1, axis=-1)[:,None]
+
+ v = b0 - np.sum(b0*b1, axis=-1)[:,None]*b1
+ w = b2 - np.sum(b2*b1, axis=-1)[:,None]*b1
+
+ x = np.sum(v*w, axis=-1)
+ y = np.sum(np.cross(b1, v)*w, axis=-1)
+
+ return np.arctan2(y, x)
+
+# calculate planar angles defined by 3 sets of points
+def get_angles(a, b, c):
+
+ v = a - b
+ v /= np.linalg.norm(v, axis=-1)[:,None]
+
+ w = c - b
+ w /= np.linalg.norm(w, axis=-1)[:,None]
+
+ x = np.sum(v*w, axis=1)
+
+ #return np.arccos(x)
+ return np.arccos(np.clip(x, -1.0, 1.0))
+
+# get 6d coordinates from x,y,z coords of N,Ca,C atoms
+def get_coords6d(xyz, dmax):
+
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[0]
+ Ca = xyz[1]
+ C = xyz[2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = np.cross(b, c)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # fast neighbors search to collect all
+ # Cb-Cb pairs within dmax
+ kdCb = scipy.spatial.cKDTree(Cb)
+ indices = kdCb.query_ball_tree(kdCb, dmax)
+
+ # indices of contacting residues
+ idx = np.array([[i,j] for i in range(len(indices)) for j in indices[i] if i != j]).T
+ idx0 = idx[0]
+ idx1 = idx[1]
+
+ # Cb-Cb distance matrix
+ dist6d = np.full((nres, nres),999.9, dtype=np.float32)
+ dist6d[idx0,idx1] = np.linalg.norm(Cb[idx1]-Cb[idx0], axis=-1)
+
+ # matrix of Ca-Cb-Cb-Ca dihedrals
+ omega6d = np.zeros((nres, nres), dtype=np.float32)
+ omega6d[idx0,idx1] = get_dihedrals(Ca[idx0], Cb[idx0], Cb[idx1], Ca[idx1])
+
+ # matrix of polar coord theta
+ theta6d = np.zeros((nres, nres), dtype=np.float32)
+ theta6d[idx0,idx1] = get_dihedrals(N[idx0], Ca[idx0], Cb[idx0], Cb[idx1])
+
+ # matrix of polar coord phi
+ phi6d = np.zeros((nres, nres), dtype=np.float32)
+ phi6d[idx0,idx1] = get_angles(Ca[idx0], Cb[idx0], Cb[idx1])
+
+ mask = np.zeros((nres, nres), dtype=np.float32)
+ mask[idx0, idx1] = 1.0
+ return dist6d, omega6d, theta6d, phi6d, mask
diff --git a/model/diffusion.py b/model/diffusion.py
new file mode 100644
index 0000000000000000000000000000000000000000..8699f7d8acb3c30d0fed7991f29ab0f27b370861
--- /dev/null
+++ b/model/diffusion.py
@@ -0,0 +1,217 @@
+import enum
+import math
+
+import numpy as np
+import torch as th
+
+
+##########################################################################################
+
+# DIFFUSION CODE BASE FOR PROTEIN SEQUENCE DIFFUSION WAS ADAPTED FROM LM-DIFFUSION #
+
+ # (https://github.com/XiangLi1999/Diffusion-LM) #
+
+##########################################################################################
+
+class GaussianDiffusion_SEQDIFF:
+ """
+ T = number of timesteps to set up diffuser with
+
+ schedule = type of noise schedule to use linear, cosine, gaussian
+
+ noise = type of ditribution to sample from; DEFAULT - normal_gaussian
+
+ """
+
+ def __init__(self,
+ T=1000,
+ schedule='sqrt',
+ sample_distribution='normal',
+ sample_distribution_gmm_means=[-1.0, 1.0],
+ sample_distribution_gmm_variances=[1.0, 1.0],
+ F=1,
+ ):
+
+ # Use float64 for accuracy.
+ betas = np.array(get_named_beta_schedule(schedule, T), dtype=np.float64)
+ self.betas = betas
+ assert len(betas.shape) == 1, "betas must be 1-D"
+ assert (betas > 0).all() and (betas <= 1).all()
+
+ self.num_timesteps = int(betas.shape[0])
+ self.F = F
+
+ alphas = 1.0 - betas
+ self.alphas_cumprod = np.cumprod(alphas, axis=0)
+ self.alphas_cumprod_prev = np.append(1.0, self.alphas_cumprod[:-1])
+ self.alphas_cumprod_next = np.append(self.alphas_cumprod[1:], 0.0)
+ assert self.alphas_cumprod_prev.shape == (self.num_timesteps,)
+
+ # calculations for posterior q(x_{t-1} | x_t, x_0)
+ self.posterior_variance = (betas * (1.0 - self.alphas_cumprod_prev) / (1.0 - self.alphas_cumprod))
+ # log calculation clipped because the posterior variance is 0 at the
+ # beginning of the diffusion chain.
+ self.posterior_log_variance_clipped = np.log(np.append(self.posterior_variance[1], self.posterior_variance[1:]))
+ self.posterior_mean_coef1 = (betas * np.sqrt(self.alphas_cumprod_prev) / (1.0 - self.alphas_cumprod))
+ self.posterior_mean_coef2 = ((1.0 - self.alphas_cumprod_prev) * np.sqrt(alphas) / (1.0 - self.alphas_cumprod))
+
+ # calculations for diffusion q(x_t | x_{t-1}) and others
+ self.sqrt_alphas_cumprod = np.sqrt(self.alphas_cumprod)
+ self.sqrt_one_minus_alphas_cumprod = np.sqrt(1.0 - self.alphas_cumprod)
+ self.log_one_minus_alphas_cumprod = np.log(1.0 - self.alphas_cumprod)
+ self.sqrt_recip_alphas_cumprod = np.sqrt(1.0 / self.alphas_cumprod)
+
+ # sample_distribution_params
+ self.sample_distribution = sample_distribution
+ self.sample_distribution_gmm_means = [float(mean) for mean in sample_distribution_gmm_means]
+ self.sample_distribution_gmm_variances = [float(variance) for variance in sample_distribution_gmm_variances]
+
+ if self.sample_distribution == 'normal':
+ self.noise_function = th.randn_like
+ else:
+ self.noise_function = self.randnmixture_like
+
+
+ def q_mean_variance(self, x_start, t):
+ """
+ Get the distribution q(x_t | x_0).
+ :param x_start: the [N x C x ...] tensor of noiseless inputs.
+ :param t: the number of diffusion steps (minus 1). Here, 0 means one step.
+ :return: A tuple (mean, variance, log_variance), all of x_start's shape.
+ """
+ mean = (
+ _extract(self.sqrt_alphas_cumprod, t, x_start.shape) * x_start
+ )
+ variance = _extract(1.0 - self.alphas_cumprod, t, x_start.shape)
+ log_variance = _extract(
+ self.log_one_minus_alphas_cumprod, t, x_start.shape
+ )
+ return mean, variance, log_variance
+
+ def q_sample(self, x_start, t, mask=None, DEVICE=None):
+ """
+ Diffuse the data for a given number of diffusion steps.
+ In other words, sample from q(x_t | x_0).
+ :param x_start: the initial data batch.
+ :param t: the number of diffusion steps (minus 1). Here, 0 means one step.
+ :param noise: if specified, the split-out normal noise.
+ :return: A noisy version of x_start.
+ """
+
+ # noise_function is determined in init depending on type of noise specified
+ noise = self.noise_function(x_start)*(self.F**2)
+ if DEVICE != None:
+ noise = noise.to(DEVICE)
+
+ assert noise.shape == x_start.shape
+ x_sample = (
+ _extract(self.sqrt_alphas_cumprod, t, x_start.shape) * x_start
+ + _extract(self.sqrt_one_minus_alphas_cumprod, t, x_start.shape)
+ * noise)
+
+ if mask is not None:
+ x_sample[mask]=x_start[mask]
+
+ return x_sample
+
+
+ def q_posterior_mean_variance(self, x_start, x_t, t):
+ """
+ Compute the mean and variance of the diffusion posterior:
+ q(x_{t-1} | x_t, x_0)
+ """
+ assert x_start.shape == x_t.shape
+
+ posterior_mean = (_extract(self.posterior_mean_coef1, t, x_t.shape) * x_start
+ + _extract(self.posterior_mean_coef2, t, x_t.shape) * x_t)
+
+ posterior_variance = _extract(self.posterior_variance, t, x_t.shape)
+
+ posterior_log_variance_clipped = _extract(self.posterior_log_variance_clipped, t, x_t.shape)
+
+ assert (
+ posterior_mean.shape[0]
+ == posterior_variance.shape[0]
+ == posterior_log_variance_clipped.shape[0]
+ == x_start.shape[0]
+ )
+ return posterior_mean, posterior_variance, posterior_log_variance_clipped
+
+
+ def randnmixture_like(self, tensor_like, number_normal=3, weights_normal=None):
+
+ if self.sample_distribution_gmm_means and self.sample_distribution_gmm_variances:
+ assert len(self.sample_distribution_gmm_means) == len(self.sample_distribution_gmm_variances)
+
+ if not weights_normal:
+ mix = th.distributions.Categorical(th.ones(len(self.sample_distribution_gmm_means))) #number_normal
+ else:
+ assert len(weights_normal) == number_normal
+ mix = th.distributions.Categorical(weights_normal)
+ #comp = torch.distributions.Normal(torch.randn(number_normal), torch.rand(number_normal))
+ comp = th.distributions.Normal(th.tensor(self.sample_distribution_gmm_means), th.tensor(self.sample_distribution_gmm_variances))
+ #comp = torch.distributions.Normal([-3, 3], [1, 1])
+ #comp = torch.distributions.Normal([-3, 0, 3], [1, 1, 1])
+ #comp = torch.distributions.Normal([-3, 0, 3], [1, 1, 1])
+ gmm = th.distributions.mixture_same_family.MixtureSameFamily(mix, comp)
+ return th.tensor([gmm.sample() for _ in range(np.prod(tensor_like.shape))]).reshape(tensor_like.shape)
+
+
+
+def get_named_beta_schedule(schedule_name, num_diffusion_timesteps):
+ """
+ Get a pre-defined beta schedule for the given name.
+ The beta schedule library consists of beta schedules which remain similar
+ in the limit of num_diffusion_timesteps.
+ Beta schedules may be added, but should not be removed or changed once
+ they are committed to maintain backwards compatibility.
+ """
+ if schedule_name == "linear":
+ # Linear schedule from Ho et al, extended to work for any number of
+ # diffusion steps.
+ scale = 1000 / num_diffusion_timesteps
+ beta_start = scale * 0.0001
+ beta_end = scale * 0.02
+ return np.linspace(beta_start, beta_end, num_diffusion_timesteps, dtype=np.float64)
+
+ elif schedule_name == "cosine":
+ return betas_for_alpha_bar(num_diffusion_timesteps, lambda t: math.cos((t + 0.008) / 1.008 * math.pi / 2) ** 2,)
+
+ elif schedule_name == 'sqrt':
+ return betas_for_alpha_bar(num_diffusion_timesteps, lambda t: 1-np.sqrt(t + 0.0001),)
+
+ else:
+ raise NotImplementedError(f"unknown beta schedule: {schedule_name}")
+
+def betas_for_alpha_bar(num_diffusion_timesteps, alpha_bar, max_beta=0.999):
+ """
+ Create a beta schedule that discretizes the given alpha_t_bar function,
+ which defines the cumulative product of (1-beta) over time from t = [0,1].
+ :param num_diffusion_timesteps: the number of betas to produce.
+ :param alpha_bar: a lambda that takes an argument t from 0 to 1 and
+ produces the cumulative product of (1-beta) up to that
+ part of the diffusion process.
+ :param max_beta: the maximum beta to use; use values lower than 1 to
+ prevent singularities.
+ """
+ betas = []
+ for i in range(num_diffusion_timesteps):
+ t1 = i / num_diffusion_timesteps
+ t2 = (i + 1) / num_diffusion_timesteps
+ betas.append(min(1 - alpha_bar(t2) / alpha_bar(t1), max_beta))
+ return np.array(betas)
+
+
+def _extract(arr, timesteps, broadcast_shape):
+ """
+ Extract values from a 1-D numpy array for a batch of indices.
+ :param arr: the 1-D numpy array.
+ :param timesteps: a tensor of indices into the array to extract.
+ :param broadcast_shape: a larger shape of K dimensions with the batch
+ dimension equal to the length of timesteps.
+ :return: a tensor of shape [batch_size, 1, ...] where the shape has K dims.
+ """
+ res = th.from_numpy(arr).to(device=timesteps.device)[timesteps].float()
+ while len(res.shape) < len(broadcast_shape):
+ res = res[..., None]
+ return res.expand(broadcast_shape)
diff --git a/model/kinematics.py b/model/kinematics.py
new file mode 100644
index 0000000000000000000000000000000000000000..392dd8ccc124a6c20ed2b4a3d654db331a920d88
--- /dev/null
+++ b/model/kinematics.py
@@ -0,0 +1,307 @@
+import numpy as np
+import torch
+from chemical import INIT_CRDS
+
+PARAMS = {
+ "DMIN" : 2.0,
+ "DMAX" : 20.0,
+ "DBINS" : 36,
+ "ABINS" : 36,
+}
+
+# ============================================================
+def get_pair_dist(a, b):
+ """calculate pair distances between two sets of points
+
+ Parameters
+ ----------
+ a,b : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of two sets of atoms
+ Returns
+ -------
+ dist : pytorch tensor of shape [batch,nres,nres]
+ stores paitwise distances between atoms in a and b
+ """
+
+ dist = torch.cdist(a, b, p=2)
+ return dist
+
+# ============================================================
+def get_ang(a, b, c):
+ """calculate planar angles for all consecutive triples (a[i],b[i],c[i])
+ from Cartesian coordinates of three sets of atoms a,b,c
+
+ Parameters
+ ----------
+ a,b,c : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of three sets of atoms
+ Returns
+ -------
+ ang : pytorch tensor of shape [batch,nres]
+ stores resulting planar angles
+ """
+ v = a - b
+ w = c - b
+ v /= torch.norm(v, dim=-1, keepdim=True)
+ w /= torch.norm(w, dim=-1, keepdim=True)
+ vw = torch.sum(v*w, dim=-1)
+
+ return torch.acos(vw)
+
+# ============================================================
+def get_dih(a, b, c, d):
+ """calculate dihedral angles for all consecutive quadruples (a[i],b[i],c[i],d[i])
+ given Cartesian coordinates of four sets of atoms a,b,c,d
+
+ Parameters
+ ----------
+ a,b,c,d : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of four sets of atoms
+ Returns
+ -------
+ dih : pytorch tensor of shape [batch,nres]
+ stores resulting dihedrals
+ """
+ b0 = a - b
+ b1 = c - b
+ b2 = d - c
+
+ b1 /= torch.norm(b1, dim=-1, keepdim=True)
+
+ v = b0 - torch.sum(b0*b1, dim=-1, keepdim=True)*b1
+ w = b2 - torch.sum(b2*b1, dim=-1, keepdim=True)*b1
+
+ x = torch.sum(v*w, dim=-1)
+ y = torch.sum(torch.cross(b1,v,dim=-1)*w, dim=-1)
+
+ return torch.atan2(y, x)
+
+
+# ============================================================
+def xyz_to_c6d(xyz, params=PARAMS):
+ """convert cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz : pytorch tensor of shape [batch,nres,3,3]
+ stores Cartesian coordinates of backbone N,Ca,C atoms
+ Returns
+ -------
+ c6d : pytorch tensor of shape [batch,nres,nres,4]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+
+ batch = xyz.shape[0]
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # 6d coordinates order: (dist,omega,theta,phi)
+ c6d = torch.zeros([batch,nres,nres,4],dtype=xyz.dtype,device=xyz.device)
+
+ dist = get_pair_dist(Cb,Cb)
+ dist[torch.isnan(dist)] = 999.9
+ c6d[...,0] = dist + 999.9*torch.eye(nres,device=xyz.device)[None,...]
+ b,i,j = torch.where(c6d[...,0]=params['DMAX']] = 999.9
+
+ mask = torch.zeros((batch, nres,nres), dtype=xyz.dtype, device=xyz.device)
+ mask[b,i,j] = 1.0
+ return c6d, mask
+
+def xyz_to_t2d(xyz_t, params=PARAMS):
+ """convert template cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz_t : pytorch tensor of shape [batch,templ,nres,3,3]
+ stores Cartesian coordinates of template backbone N,Ca,C atoms
+
+ Returns
+ -------
+ t2d : pytorch tensor of shape [batch,nres,nres,37+6+3]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+ B, T, L = xyz_t.shape[:3]
+ c6d, mask = xyz_to_c6d(xyz_t[:,:,:,:3].view(B*T,L,3,3), params=params)
+ c6d = c6d.view(B, T, L, L, 4)
+ mask = mask.view(B, T, L, L, 1)
+ #
+ # dist to one-hot encoded
+ dist = dist_to_onehot(c6d[...,0], params)
+ orien = torch.cat((torch.sin(c6d[...,1:]), torch.cos(c6d[...,1:])), dim=-1)*mask # (B, T, L, L, 6)
+ #
+ mask = ~torch.isnan(c6d[:,:,:,:,0]) # (B, T, L, L)
+ t2d = torch.cat((dist, orien, mask.unsqueeze(-1)), dim=-1)
+ t2d[torch.isnan(t2d)] = 0.0
+ return t2d
+
+def xyz_to_chi1(xyz_t):
+ '''convert template cartesian coordinates into chi1 angles
+
+ Parameters
+ ----------
+ xyz_t: pytorch tensor of shape [batch, templ, nres, 14, 3]
+ stores Cartesian coordinates of template atoms. For missing atoms, it should be NaN
+
+ Returns
+ -------
+ chi1 : pytorch tensor of shape [batch, templ, nres, 2]
+ stores cos and sin chi1 angle
+ '''
+ B, T, L = xyz_t.shape[:3]
+ xyz_t = xyz_t.reshape(B*T, L, 14, 3)
+
+ # chi1 angle: N, CA, CB, CG
+ chi1 = get_dih(xyz_t[:,:,0], xyz_t[:,:,1], xyz_t[:,:,4], xyz_t[:,:,5]) # (B*T, L)
+ cos_chi1 = torch.cos(chi1)
+ sin_chi1 = torch.sin(chi1)
+ mask_chi1 = ~torch.isnan(chi1)
+ chi1 = torch.stack((cos_chi1, sin_chi1, mask_chi1), dim=-1) # (B*T, L, 3)
+ chi1[torch.isnan(chi1)] = 0.0
+ chi1 = chi1.reshape(B, T, L, 3)
+ return chi1
+
+def xyz_to_bbtor(xyz, params=PARAMS):
+ batch = xyz.shape[0]
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ next_N = torch.roll(N, -1, dims=1)
+ prev_C = torch.roll(C, 1, dims=1)
+ phi = get_dih(prev_C, N, Ca, C)
+ psi = get_dih(N, Ca, C, next_N)
+ #
+ phi[:,0] = 0.0
+ psi[:,-1] = 0.0
+ #
+ astep = 2.0*np.pi / params['ABINS']
+ phi_bin = torch.round((phi+np.pi-astep/2)/astep)
+ psi_bin = torch.round((psi+np.pi-astep/2)/astep)
+ return torch.stack([phi_bin, psi_bin], axis=-1).long()
+
+# ============================================================
+def dist_to_onehot(dist, params=PARAMS):
+ dist[torch.isnan(dist)] = 999.9
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=dist.dtype,device=dist.device)
+ db = torch.bucketize(dist.contiguous(),dbins).long()
+ dist = torch.nn.functional.one_hot(db, num_classes=params['DBINS']+1).float()
+ return dist
+
+def c6d_to_bins(c6d,params=PARAMS):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=c6d.dtype,device=c6d.device)
+ ab360 = torch.linspace(-np.pi+astep, np.pi, params['ABINS'],dtype=c6d.dtype,device=c6d.device)
+ ab180 = torch.linspace(astep, np.pi, params['ABINS']//2,dtype=c6d.dtype,device=c6d.device)
+
+ db = torch.bucketize(c6d[...,0].contiguous(),dbins)
+ ob = torch.bucketize(c6d[...,1].contiguous(),ab360)
+ tb = torch.bucketize(c6d[...,2].contiguous(),ab360)
+ pb = torch.bucketize(c6d[...,3].contiguous(),ab180)
+
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).to(torch.uint8)
+
+
+# ============================================================
+def dist_to_bins(dist,params=PARAMS):
+ """bin 2d distance maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ db = torch.round((dist-params['DMIN']-dstep/2)/dstep)
+
+ db[db<0] = 0
+ db[db>params['DBINS']] = params['DBINS']
+
+ return db.long()
+
+
+# ============================================================
+def c6d_to_bins2(c6d, same_chain, negative=False, params=PARAMS):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ db = torch.round((c6d[...,0]-params['DMIN']-dstep/2)/dstep)
+ ob = torch.round((c6d[...,1]+np.pi-astep/2)/astep)
+ tb = torch.round((c6d[...,2]+np.pi-astep/2)/astep)
+ pb = torch.round((c6d[...,3]-astep/2)/astep)
+
+ # put all dparams['DBINS']] = params['DBINS']
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ if negative:
+ db = torch.where(same_chain.bool(), db.long(), params['DBINS'])
+ ob = torch.where(same_chain.bool(), ob.long(), params['ABINS'])
+ tb = torch.where(same_chain.bool(), tb.long(), params['ABINS'])
+ pb = torch.where(same_chain.bool(), pb.long(), params['ABINS']//2)
+
+ return torch.stack([db,ob,tb,pb],axis=-1).long()
+
+def get_init_xyz(xyz_t):
+ # input: xyz_t (B, T, L, 14, 3)
+ # ouput: xyz (B, T, L, 14, 3)
+ B, T, L = xyz_t.shape[:3]
+ init = INIT_CRDS.to(xyz_t.device).reshape(1,1,1,27,3).repeat(B,T,L,1,1)
+ if torch.isnan(xyz_t).all():
+ return init
+
+ mask = torch.isnan(xyz_t[:,:,:,:3]).any(dim=-1).any(dim=-1) # (B, T, L)
+ #
+ center_CA = ((~mask[:,:,:,None]) * torch.nan_to_num(xyz_t[:,:,:,1,:])).sum(dim=2) / ((~mask[:,:,:,None]).sum(dim=2)+1e-4) # (B, T, 3)
+ xyz_t = xyz_t - center_CA.view(B,T,1,1,3)
+ #
+ idx_s = list()
+ for i_b in range(B):
+ for i_T in range(T):
+ if mask[i_b, i_T].all():
+ continue
+ exist_in_templ = torch.where(~mask[i_b, i_T])[0] # (L_sub)
+ seqmap = (torch.arange(L, device=xyz_t.device)[:,None] - exist_in_templ[None,:]).abs() # (L, L_sub)
+ seqmap = torch.argmin(seqmap, dim=-1) # (L)
+ idx = torch.gather(exist_in_templ, -1, seqmap) # (L)
+ offset_CA = torch.gather(xyz_t[i_b, i_T, :, 1, :], 0, idx.reshape(L,1).expand(-1,3))
+ init[i_b,i_T] += offset_CA.reshape(L,1,3)
+ #
+ xyz = torch.where(mask.view(B, T, L, 1, 1), init, xyz_t)
+ return xyz
diff --git a/model/parsers.py b/model/parsers.py
new file mode 100644
index 0000000000000000000000000000000000000000..001a086b66cb60df21085a0295874ecf3606a1f2
--- /dev/null
+++ b/model/parsers.py
@@ -0,0 +1,219 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+import gzip
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+# read A3M and convert letters into
+# integers in the 0..20 range,
+# also keep track of insertions
+def parse_a3m(filename):
+
+ msa = []
+ ins = []
+
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+
+ #print(filename)
+
+ if filename.split('.')[-1] == 'gz':
+ fp = gzip.open(filename, 'rt')
+ else:
+ fp = open(filename, 'r')
+
+ # read file line by line
+ for line in fp:
+
+ # skip labels
+ if line[0] == '>':
+ continue
+
+ # remove right whitespaces
+ line = line.rstrip()
+
+ if len(line) == 0:
+ continue
+
+ # remove lowercase letters and append to MSA
+ msa.append(line.translate(table))
+
+ # sequence length
+ L = len(msa[-1])
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in line])
+ i = np.zeros((L))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+
+ # append to the matrix of insetions
+ i[pos] = num
+
+ ins.append(i)
+ if len(msa) == 10000:
+ break
+
+ # convert letters into numbers
+ alphabet = np.array(list("ARNDCQEGHILKMFPSTWYV-"), dtype='|S1').view(np.uint8)
+ msa = np.array([list(s) for s in msa], dtype='|S1').view(np.uint8)
+ for i in range(alphabet.shape[0]):
+ msa[msa == alphabet[i]] = i
+
+ # treat all unknown characters as gaps
+ msa[msa > 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return msa,ins
+
+
+# read and extract xyz coords of N,Ca,C atoms
+# from a PDB file
+def parse_pdb(filename):
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines)
+
+#'''
+def parse_pdb_lines(lines):
+
+ # indices of residues observed in the structure
+ idx_s = [int(l[22:26]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(idx_s), 14, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ resNo, atom, aa = int(l[22:26]), l[12:16], l[17:20]
+ idx = idx_s.index(resNo)
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm == atom:
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ return xyz,mask,np.array(idx_s)
+#'''
+
+'''
+def parse_pdb_lines(lines):
+
+ # indices of residues observed in the structure
+ #idx_s = [int(l[22:26]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ idx_s = [int(r[0]) for r in res]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(idx_s), 14, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ resNo, atom, aa = int(l[22:26]), l[12:16], l[17:20]
+ idx = idx_s.index(resNo)
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm == atom:
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ return xyz,mask,np.array(idx_s), np.array(seq)
+'''
+
+
+def parse_templates(item, params):
+
+ # init FFindexDB of templates
+ ### and extract template IDs
+ ### present in the DB
+ ffdb = FFindexDB(read_index(params['FFDB']+'_pdb.ffindex'),
+ read_data(params['FFDB']+'_pdb.ffdata'))
+ #ffids = set([i.name for i in ffdb.index])
+
+ # process tabulated hhsearch output to get
+ # matched positions and positional scores
+ infile = params['DIR']+'/hhr/'+item[-2:]+'/'+item+'.atab'
+ hits = []
+ for l in open(infile, "r").readlines():
+ if l[0]=='>':
+ key = l[1:].split()[0]
+ hits.append([key,[],[]])
+ elif "score" in l or "dssp" in l:
+ continue
+ else:
+ hi = l.split()[:5]+[0.0,0.0,0.0]
+ hits[-1][1].append([int(hi[0]),int(hi[1])])
+ hits[-1][2].append([float(hi[2]),float(hi[3]),float(hi[4])])
+
+ # get per-hit statistics from an .hhr file
+ # (!!! assume that .hhr and .atab have the same hits !!!)
+ # [Probab, E-value, Score, Aligned_cols,
+ # Identities, Similarity, Sum_probs, Template_Neff]
+ lines = open(infile[:-4]+'hhr', "r").readlines()
+ pos = [i+1 for i,l in enumerate(lines) if l[0]=='>']
+ for i,posi in enumerate(pos):
+ hits[i].append([float(s) for s in re.sub('[=%]',' ',lines[posi]).split()[1::2]])
+
+ # parse templates from FFDB
+ for hi in hits:
+ #if hi[0] not in ffids:
+ # continue
+ entry = get_entry_by_name(hi[0], ffdb.index)
+ if entry == None:
+ continue
+ data = read_entry_lines(entry, ffdb.data)
+ hi += list(parse_pdb_lines(data))
+
+ # process hits
+ counter = 0
+ xyz,qmap,mask,f0d,f1d,ids = [],[],[],[],[],[]
+ for data in hits:
+ if len(data)<7:
+ continue
+
+ qi,ti = np.array(data[1]).T
+ _,sel1,sel2 = np.intersect1d(ti, data[6], return_indices=True)
+ ncol = sel1.shape[0]
+ if ncol < 10:
+ continue
+
+ ids.append(data[0])
+ f0d.append(data[3])
+ f1d.append(np.array(data[2])[sel1])
+ xyz.append(data[4][sel2])
+ mask.append(data[5][sel2])
+ qmap.append(np.stack([qi[sel1]-1,[counter]*ncol],axis=-1))
+ counter += 1
+
+ xyz = np.vstack(xyz).astype(np.float32)
+ mask = np.vstack(mask).astype(np.bool)
+ qmap = np.vstack(qmap).astype(np.long)
+ f0d = np.vstack(f0d).astype(np.float32)
+ f1d = np.vstack(f1d).astype(np.float32)
+ ids = ids
+
+ return xyz,mask,qmap,f0d,f1d,ids
diff --git a/model/scoring.py b/model/scoring.py
new file mode 100644
index 0000000000000000000000000000000000000000..21377f66cec15b7b01c23031f9b5b5357cf38e38
--- /dev/null
+++ b/model/scoring.py
@@ -0,0 +1,300 @@
+
+##
+## lk and lk term
+#(LJ_RADIUS LJ_WDEPTH LK_DGFREE LK_LAMBDA LK_VOLUME)
+type2ljlk = {
+ "CNH2":(1.968297,0.094638,3.077030,3.5000,13.500000),
+ "COO":(1.916661,0.141799,-3.332648,3.5000,14.653000),
+ "CH0":(2.011760,0.062642,1.409284,3.5000,8.998000),
+ "CH1":(2.011760,0.062642,-3.538387,3.5000,10.686000),
+ "CH2":(2.011760,0.062642,-1.854658,3.5000,18.331000),
+ "CH3":(2.011760,0.062642,7.292929,3.5000,25.855000),
+ "aroC":(2.016441,0.068775,1.797950,3.5000,16.704000),
+ "Ntrp":(1.802452,0.161725,-8.413116,3.5000,9.522100),
+ "Nhis":(1.802452,0.161725,-9.739606,3.5000,9.317700),
+ "NtrR":(1.802452,0.161725,-5.158080,3.5000,9.779200),
+ "NH2O":(1.802452,0.161725,-8.101638,3.5000,15.689000),
+ "Nlys":(1.802452,0.161725,-20.864641,3.5000,16.514000),
+ "Narg":(1.802452,0.161725,-8.968351,3.5000,15.717000),
+ "Npro":(1.802452,0.161725,-0.984585,3.5000,3.718100),
+ "OH":(1.542743,0.161947,-8.133520,3.5000,10.722000),
+ "OHY":(1.542743,0.161947,-8.133520,3.5000,10.722000),
+ "ONH2":(1.548662,0.182924,-6.591644,3.5000,10.102000),
+ "OOC":(1.492871,0.099873,-9.239832,3.5000,9.995600),
+ "S":(1.975967,0.455970,-1.707229,3.5000,17.640000),
+ "SH1":(1.975967,0.455970,3.291643,3.5000,23.240000),
+ "Nbb":(1.802452,0.161725,-9.969494,3.5000,15.992000),
+ "CAbb":(2.011760,0.062642,2.533791,3.5000,12.137000),
+ "CObb":(1.916661,0.141799,3.104248,3.5000,13.221000),
+ "OCbb":(1.540580,0.142417,-8.006829,3.5000,12.196000),
+ "HNbb":(0.901681,0.005000,0.0000,3.5000,0.0000),
+ "Hapo":(1.421272,0.021808,0.0000,3.5000,0.0000),
+ "Haro":(1.374914,0.015909,0.0000,3.5000,0.0000),
+ "Hpol":(0.901681,0.005000,0.0000,3.5000,0.0000),
+ "HS":(0.363887,0.050836,0.0000,3.5000,0.0000),
+}
+
+# hbond donor/acceptors
+class HbAtom:
+ NO = 0
+ DO = 1 # donor
+ AC = 2 # acceptor
+ DA = 3 # donor & acceptor
+ HP = 4 # polar H
+
+type2hb = {
+ "CNH2":HbAtom.NO, "COO":HbAtom.NO, "CH0":HbAtom.NO, "CH1":HbAtom.NO,
+ "CH2":HbAtom.NO, "CH3":HbAtom.NO, "aroC":HbAtom.NO, "Ntrp":HbAtom.DO,
+ "Nhis":HbAtom.AC, "NtrR":HbAtom.DO, "NH2O":HbAtom.DO, "Nlys":HbAtom.DO,
+ "Narg":HbAtom.DO, "Npro":HbAtom.NO, "OH":HbAtom.DA, "OHY":HbAtom.DA,
+ "ONH2":HbAtom.AC, "OOC":HbAtom.AC, "S":HbAtom.NO, "SH1":HbAtom.NO,
+ "Nbb":HbAtom.DO, "CAbb":HbAtom.NO, "CObb":HbAtom.NO, "OCbb":HbAtom.AC,
+ "HNbb":HbAtom.HP, "Hapo":HbAtom.NO, "Haro":HbAtom.NO, "Hpol":HbAtom.HP,
+ "HS":HbAtom.HP, # HP in rosetta(?)
+}
+
+##
+## hbond term
+class HbDonType:
+ PBA = 0
+ IND = 1
+ IME = 2
+ GDE = 3
+ CXA = 4
+ AMO = 5
+ HXL = 6
+ AHX = 7
+ NTYPES = 8
+
+class HbAccType:
+ PBA = 0
+ CXA = 1
+ CXL = 2
+ HXL = 3
+ AHX = 4
+ IME = 5
+ NTYPES = 6
+
+class HbHybType:
+ SP2 = 0
+ SP3 = 1
+ RING = 2
+ NTYPES = 3
+
+type2dontype = {
+ "Nbb": HbDonType.PBA,
+ "Ntrp": HbDonType.IND,
+ "NtrR": HbDonType.GDE,
+ "Narg": HbDonType.GDE,
+ "NH2O": HbDonType.CXA,
+ "Nlys": HbDonType.AMO,
+ "OH": HbDonType.HXL,
+ "OHY": HbDonType.AHX,
+}
+
+type2acctype = {
+ "OCbb": HbAccType.PBA,
+ "ONH2": HbAccType.CXA,
+ "OOC": HbAccType.CXL,
+ "OH": HbAccType.HXL,
+ "OHY": HbAccType.AHX,
+ "Nhis": HbAccType.IME,
+}
+
+type2hybtype = {
+ "OCbb": HbHybType.SP2,
+ "ONH2": HbHybType.SP2,
+ "OOC": HbHybType.SP2,
+ "OHY": HbHybType.SP3,
+ "OH": HbHybType.SP3,
+ "Nhis": HbHybType.RING,
+}
+
+dontype2wt = {
+ HbDonType.PBA: 1.45,
+ HbDonType.IND: 1.15,
+ HbDonType.IME: 1.42,
+ HbDonType.GDE: 1.11,
+ HbDonType.CXA: 1.29,
+ HbDonType.AMO: 1.17,
+ HbDonType.HXL: 0.99,
+ HbDonType.AHX: 1.00,
+}
+
+acctype2wt = {
+ HbAccType.PBA: 1.19,
+ HbAccType.CXA: 1.21,
+ HbAccType.CXL: 1.10,
+ HbAccType.HXL: 1.15,
+ HbAccType.AHX: 1.15,
+ HbAccType.IME: 1.17,
+}
+
+class HbPolyType:
+ ahdist_aASN_dARG = 0
+ ahdist_aASN_dASN = 1
+ ahdist_aASN_dGLY = 2
+ ahdist_aASN_dHIS = 3
+ ahdist_aASN_dLYS = 4
+ ahdist_aASN_dSER = 5
+ ahdist_aASN_dTRP = 6
+ ahdist_aASN_dTYR = 7
+ ahdist_aASP_dARG = 8
+ ahdist_aASP_dASN = 9
+ ahdist_aASP_dGLY = 10
+ ahdist_aASP_dHIS = 11
+ ahdist_aASP_dLYS = 12
+ ahdist_aASP_dSER = 13
+ ahdist_aASP_dTRP = 14
+ ahdist_aASP_dTYR = 15
+ ahdist_aGLY_dARG = 16
+ ahdist_aGLY_dASN = 17
+ ahdist_aGLY_dGLY = 18
+ ahdist_aGLY_dHIS = 19
+ ahdist_aGLY_dLYS = 20
+ ahdist_aGLY_dSER = 21
+ ahdist_aGLY_dTRP = 22
+ ahdist_aGLY_dTYR = 23
+ ahdist_aHIS_dARG = 24
+ ahdist_aHIS_dASN = 25
+ ahdist_aHIS_dGLY = 26
+ ahdist_aHIS_dHIS = 27
+ ahdist_aHIS_dLYS = 28
+ ahdist_aHIS_dSER = 29
+ ahdist_aHIS_dTRP = 30
+ ahdist_aHIS_dTYR = 31
+ ahdist_aSER_dARG = 32
+ ahdist_aSER_dASN = 33
+ ahdist_aSER_dGLY = 34
+ ahdist_aSER_dHIS = 35
+ ahdist_aSER_dLYS = 36
+ ahdist_aSER_dSER = 37
+ ahdist_aSER_dTRP = 38
+ ahdist_aSER_dTYR = 39
+ ahdist_aTYR_dARG = 40
+ ahdist_aTYR_dASN = 41
+ ahdist_aTYR_dGLY = 42
+ ahdist_aTYR_dHIS = 43
+ ahdist_aTYR_dLYS = 44
+ ahdist_aTYR_dSER = 45
+ ahdist_aTYR_dTRP = 46
+ ahdist_aTYR_dTYR = 47
+ cosBAH_off = 48
+ cosBAH_7 = 49
+ cosBAH_6i = 50
+ AHD_1h = 51
+ AHD_1i = 52
+ AHD_1j = 53
+ AHD_1k = 54
+
+# map donor:acceptor pairs to polynomials
+hbtypepair2poly = {
+ (HbDonType.PBA,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.CXA,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.IME,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.IND,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.AMO,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.AHX,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.CXA): (HbPolyType.ahdist_aASN_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.CXA,HbAccType.CXA): (HbPolyType.ahdist_aASN_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IME,HbAccType.CXA): (HbPolyType.ahdist_aASN_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IND,HbAccType.CXA): (HbPolyType.ahdist_aASN_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AMO,HbAccType.CXA): (HbPolyType.ahdist_aASN_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.CXA): (HbPolyType.ahdist_aASN_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AHX,HbAccType.CXA): (HbPolyType.ahdist_aASN_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.CXA): (HbPolyType.ahdist_aASN_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.CXL): (HbPolyType.ahdist_aASP_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.CXA,HbAccType.CXL): (HbPolyType.ahdist_aASP_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IME,HbAccType.CXL): (HbPolyType.ahdist_aASP_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IND,HbAccType.CXL): (HbPolyType.ahdist_aASP_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AMO,HbAccType.CXL): (HbPolyType.ahdist_aASP_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.CXL): (HbPolyType.ahdist_aASP_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AHX,HbAccType.CXL): (HbPolyType.ahdist_aASP_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.CXL): (HbPolyType.ahdist_aASP_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.IME): (HbPolyType.ahdist_aHIS_dGLY,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.IME): (HbPolyType.ahdist_aHIS_dASN,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.IME): (HbPolyType.ahdist_aHIS_dHIS,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.IND,HbAccType.IME): (HbPolyType.ahdist_aHIS_dTRP,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.IME): (HbPolyType.ahdist_aHIS_dLYS,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.IME): (HbPolyType.ahdist_aHIS_dARG,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.AHX,HbAccType.IME): (HbPolyType.ahdist_aHIS_dTYR,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.IME): (HbPolyType.ahdist_aHIS_dSER,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.PBA,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dGLY,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dASN,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dHIS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IND,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dTRP,HbPolyType.cosBAH_6i,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dLYS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dARG,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.AHX,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dTYR,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dSER,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.PBA,HbAccType.HXL): (HbPolyType.ahdist_aSER_dGLY,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.HXL): (HbPolyType.ahdist_aSER_dASN,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.HXL): (HbPolyType.ahdist_aSER_dHIS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IND,HbAccType.HXL): (HbPolyType.ahdist_aSER_dTRP,HbPolyType.cosBAH_6i,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.HXL): (HbPolyType.ahdist_aSER_dLYS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.HXL): (HbPolyType.ahdist_aSER_dARG,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.AHX,HbAccType.HXL): (HbPolyType.ahdist_aSER_dTYR,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.HXL): (HbPolyType.ahdist_aSER_dSER,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+}
+
+
+# polynomials are triplets, (x_min, x_max), (y[xx_max]), (c_9,...,c_0)
+hbpolytype2coeffs = { # Parameters imported from rosetta sp2_elec_params @v2017.48-dev59886
+ HbPolyType.ahdist_aASN_dARG: ((0.7019094761929999, 2.86820307153,),(1.1, 1.1,),( 0.58376113, -9.29345473, 64.86270904, -260.3946711, 661.43138077, -1098.01378958, 1183.58371466, -790.82929582, 291.33125475, -43.01629727,)),
+ HbPolyType.ahdist_aASN_dASN: ((0.625841094801, 2.75107708444,),(1.1, 1.1,),( -1.31243015, 18.6745072, -112.63858313, 373.32878091, -734.99145504, 861.38324861, -556.21026097, 143.5626977, 20.03238394, -11.52167705,)),
+ HbPolyType.ahdist_aASN_dGLY: ((0.7477341047139999, 2.6796350782799996,),(1.1, 1.1,),( -1.61294554, 23.3150793, -144.11313069, 496.13575, -1037.83809166, 1348.76826073, -1065.14368678, 473.89008925, -100.41142701, 7.44453515,)),
+ HbPolyType.ahdist_aASN_dHIS: ((0.344789524346, 2.8303582266000005,),(1.1, 1.1,),( -0.2657122, 4.1073775, -26.9099632, 97.10486507, -209.96002602, 277.33057268, -218.74766996, 97.42852213, -24.07382402, 3.73962807,)),
+ HbPolyType.ahdist_aASN_dLYS: ((0.542905671869, 2.45259389314,),(1.1, 1.1,),( 1.38531754, -18.48733797, 106.14444613, -344.70585054, 698.91577956, -917.0879402, 775.32787908, -403.09588787, 113.65054778, -11.66516403,)),
+ HbPolyType.ahdist_aASN_dSER: ((1.0812774602500002, 2.6832123582599996,),(1.1, 1.1,),( -3.51524353, 47.54032873, -254.40168577, 617.84606386, -255.49935027, -2361.56230539, 6426.85797934, -7760.4403891, 4694.08106855, -1149.83549068,)),
+ HbPolyType.ahdist_aASN_dTRP: ((0.6689984999999999, 3.0704254,),(1.1, 1.1,),( -0.5284840422, 8.3510150838, -56.4100479414, 212.4884326254, -488.3178610608, 703.7762350506, -628.9936994633999, 331.4294356146, -93.265817571, 11.9691623698,)),
+ HbPolyType.ahdist_aASN_dTYR: ((1.08950268805, 2.6887046709400004,),(1.1, 1.1,),( -4.4488705, 63.27696281, -371.44187037, 1121.71921621, -1638.11394306, 142.99988401, 3436.65879147, -5496.07011787, 3709.30505237, -962.79669688,)),
+ HbPolyType.ahdist_aASP_dARG: ((0.8100404642229999, 2.9851230124799994,),(1.1, 1.1,),( -0.66430344, 10.41343145, -70.12656205, 265.12578414, -617.05849171, 911.39378582, -847.25013928, 472.09090981, -141.71513167, 18.57721132,)),
+ HbPolyType.ahdist_aASP_dASN: ((1.05401125073, 3.11129675908,),(1.1, 1.1,),( 0.02090728, -0.24144928, -0.19578075, 16.80904547, -117.70216251, 407.18551288, -809.95195924, 939.83137947, -593.94527692, 159.57610528,)),
+ HbPolyType.ahdist_aASP_dGLY: ((0.886260952629, 2.66843608743,),(1.1, 1.1,),( -7.00699267, 107.33021779, -713.45752385, 2694.43092298, -6353.05100287, 9667.94098394, -9461.9261027, 5721.0086877, -1933.97818198, 279.47763789,)),
+ HbPolyType.ahdist_aASP_dHIS: ((1.03597611139, 2.78208509117,),(1.1, 1.1,),( -1.34823406, 17.08925926, -78.75087193, 106.32795459, 400.18459698, -2041.04320193, 4033.83557387, -4239.60530204, 2324.00877252, -519.38410941,)),
+ HbPolyType.ahdist_aASP_dLYS: ((0.97789485082, 2.50496946108,),(1.1, 1.1,),( -0.41300315, 6.59243438, -44.44525308, 163.11796012, -351.2307798, 443.2463146, -297.84582856, 62.38600547, 33.77496227, -14.11652182,)),
+ HbPolyType.ahdist_aASP_dSER: ((0.542905671869, 2.45259389314,),(1.1, 1.1,),( 1.38531754, -18.48733797, 106.14444613, -344.70585054, 698.91577956, -917.0879402, 775.32787908, -403.09588787, 113.65054778, -11.66516403,)),
+ HbPolyType.ahdist_aASP_dTRP: ((0.419155746414, 3.0486938610500003,),(1.1, 1.1,),( -0.24563471, 3.85598551, -25.75176874, 95.36525025, -214.13175785, 299.76133553, -259.0691378, 132.06975835, -37.15612683, 5.60445773,)),
+ HbPolyType.ahdist_aASP_dTYR: ((1.01057521468, 2.7207545786900003,),(1.1, 1.1,),( -0.15808672, -10.21398871, 178.80080949, -1238.0583801, 4736.25248274, -11071.96777725, 16239.07550047, -14593.21092621, 7335.66765017, -1575.08145078,)),
+ HbPolyType.ahdist_aGLY_dARG: ((0.499016667857, 2.9377031027599996,),(1.1, 1.1,),( -0.15923533, 2.5526639, -17.38788803, 65.71046957, -151.13491186, 218.78048387, -199.15882919, 110.56568974, -35.95143745, 6.47580213,)),
+ HbPolyType.ahdist_aGLY_dASN: ((0.7194388032060001, 2.9303772333599998,),(1.1, 1.1,),( -1.40718342, 23.65929694, -172.97144348, 720.64417348, -1882.85420815, 3194.87197776, -3515.52467458, 2415.75238278, -941.47705161, 159.84784277,)),
+ HbPolyType.ahdist_aGLY_dGLY: ((1.38403812683, 2.9981039433,),(1.1, 1.1,),( -0.5307601, 6.47949946, -22.39522814, -55.14303544, 708.30945242, -2619.49318162, 5227.8805795, -6043.31211632, 3806.04676175, -1007.66024144,)),
+ HbPolyType.ahdist_aGLY_dHIS: ((0.47406840932899996, 2.9234200830400003,),(1.1, 1.1,),( -0.12881679, 1.933838, -12.03134888, 39.92691227, -75.41519959, 78.87968016, -37.82769801, -0.13178679, 4.50193019, 0.45408359,)),
+ HbPolyType.ahdist_aGLY_dLYS: ((0.545347533475, 2.42624380351,),(1.1, 1.1,),( -0.22921901, 2.07015714, -6.2947417, 0.66645697, 45.21805416, -130.26668981, 176.32401031, -126.68226346, 43.96744431, -4.40105281,)),
+ HbPolyType.ahdist_aGLY_dSER: ((1.2803349239700001, 2.2465996077400003,),(1.1, 1.1,),( 6.72508613, -86.98495585, 454.18518444, -1119.89141452, 715.624663, 3172.36852982, -9455.49113097, 11797.38766934, -7363.28302948, 1885.50119665,)),
+ HbPolyType.ahdist_aGLY_dTRP: ((0.686512740494, 3.02901351815,),(1.1, 1.1,),( -0.1051487, 1.41597708, -7.42149173, 17.31830704, -6.98293652, -54.76605063, 130.95272289, -132.77575305, 62.75460448, -9.89110842,)),
+ HbPolyType.ahdist_aGLY_dTYR: ((1.28894687639, 2.26335316892,),(1.1, 1.1,),( 13.84536925, -169.40579865, 893.79467505, -2670.60617561, 5016.46234701, -6293.79378818, 5585.1049063, -3683.50722701, 1709.48661405, -399.5712153,)),
+ HbPolyType.ahdist_aHIS_dARG: ((0.8967400957230001, 2.96809434226,),(1.1, 1.1,),( 0.43460495, -10.52727665, 103.16979807, -551.42887412, 1793.25378923, -3701.08304991, 4861.05155388, -3922.4285529, 1763.82137881, -335.43441944,)),
+ HbPolyType.ahdist_aHIS_dASN: ((0.887120931718, 2.59166903153,),(1.1, 1.1,),( -3.50289894, 54.42813924, -368.14395507, 1418.90186454, -3425.60485859, 5360.92334837, -5428.54462336, 3424.68800187, -1221.49631986, 189.27122436,)),
+ HbPolyType.ahdist_aHIS_dGLY: ((1.01629363411, 2.58523052904,),(1.1, 1.1,),( -1.68095217, 21.31894078, -107.72203494, 251.81021758, -134.07465831, -707.64527046, 1894.6282743, -2156.85951846, 1216.83585872, -275.48078944,)),
+ HbPolyType.ahdist_aHIS_dHIS: ((0.9773010778919999, 2.72533796329,),(1.1, 1.1,),( -2.33350626, 35.66072412, -233.98966111, 859.13714961, -1925.30958567, 2685.35293578, -2257.48067507, 1021.49796136, -169.36082523, -12.1348055,)),
+ HbPolyType.ahdist_aHIS_dLYS: ((0.7080936539849999, 2.47191718632,),(1.1, 1.1,),( -1.88479369, 28.38084382, -185.74039957, 690.81875917, -1605.11404391, 2414.83545623, -2355.9723201, 1442.24496229, -506.45880637, 79.47512505,)),
+ HbPolyType.ahdist_aHIS_dSER: ((0.90846809159, 2.5477956147,),(1.1, 1.1,),( -0.92004641, 15.91841533, -117.83979251, 488.22211296, -1244.13047376, 2017.43704053, -2076.04468019, 1302.42621488, -451.29138643, 67.15812575,)),
+ HbPolyType.ahdist_aHIS_dTRP: ((0.991999676806, 2.81296584506,),(1.1, 1.1,),( -1.29358587, 19.97152857, -131.89796017, 485.29199356, -1084.0466445, 1497.3352889, -1234.58042682, 535.8048197, -75.58951691, -9.91148332,)),
+ HbPolyType.ahdist_aHIS_dTYR: ((0.882661836357, 2.5469016429900004,),(1.1, 1.1,),( -6.94700143, 109.07997256, -747.64035726, 2929.83959536, -7220.15788571, 11583.34170519, -12078.443492, 7881.85479715, -2918.19482068, 468.23988622,)),
+ HbPolyType.ahdist_aSER_dARG: ((1.0204658147399999, 2.8899566041900004,),(1.1, 1.1,),( 0.33887327, -7.54511361, 70.87316645, -371.88263665, 1206.67454443, -2516.82084076, 3379.45432693, -2819.73384601, 1325.33307517, -265.54533008,)),
+ HbPolyType.ahdist_aSER_dASN: ((1.01393052233, 3.0024434159299997,),(1.1, 1.1,),( 0.37012361, -7.46486204, 64.85775924, -318.6047209, 974.66322243, -1924.37334018, 2451.63840629, -1943.1915675, 867.07870559, -163.83771761,)),
+ HbPolyType.ahdist_aSER_dGLY: ((1.3856562156299999, 2.74160605537,),(1.1, 1.1,),( -1.32847415, 22.67528654, -172.53450064, 770.79034865, -2233.48829652, 4354.38807288, -5697.35144236, 4803.38686157, -2361.48028857, 518.28202382,)),
+ HbPolyType.ahdist_aSER_dHIS: ((0.550992321207, 2.68549261999,),(1.1, 1.1,),( -1.98041793, 29.59668639, -190.36751773, 688.43324385, -1534.68894765, 2175.66568976, -1952.07622113, 1066.28943929, -324.23381388, 43.41006168,)),
+ HbPolyType.ahdist_aSER_dLYS: ((0.8603189393170001, 2.77729502744,),(1.1, 1.1,),( 0.90884741, -17.24690746, 141.78469099, -661.85989315, 1929.7674992, -3636.43392779, 4419.00727923, -3332.43482061, 1410.78913266, -253.53829424,)),
+ HbPolyType.ahdist_aSER_dSER: ((1.10866545921, 2.61727781204,),(1.1, 1.1,),( -0.38264308, 4.41779675, -10.7016645, -81.91314845, 668.91174735, -2187.50684758, 3983.56103269, -4213.32320546, 2418.41531442, -580.28918569,)),
+ HbPolyType.ahdist_aSER_dTRP: ((1.4092077245899999, 2.8066121197099996,),(1.1, 1.1,),( 0.73762477, -11.70741276, 73.05154232, -205.00144794, 89.58794368, 1082.94541375, -3343.98293188, 4601.70815729, -3178.53568678, 896.59487831,)),
+ HbPolyType.ahdist_aSER_dTYR: ((1.10773547919, 2.60403567341,),(1.1, 1.1,),( -1.13249925, 14.66643161, -69.01708791, 93.96846742, 380.56063898, -1984.56675689, 4074.08891127, -4492.76927139, 2613.13168054, -627.71933508,)),
+ HbPolyType.ahdist_aTYR_dARG: ((1.05581400627, 2.85499888099,),(1.1, 1.1,),( -0.30396592, 5.30288548, -39.75788579, 167.5416547, -435.15958911, 716.52357586, -735.95195083, 439.76284677, -130.00400085, 13.23827556,)),
+ HbPolyType.ahdist_aTYR_dASN: ((1.0994919065200002, 2.8400869077900004,),(1.1, 1.1,),( 0.33548259, -3.5890451, 8.97769025, 48.1492734, -400.5983616, 1269.89613211, -2238.03101675, 2298.33009115, -1290.42961162, 308.43185147,)),
+ HbPolyType.ahdist_aTYR_dGLY: ((1.36546155066, 2.7303075916400004,),(1.1, 1.1,),( -1.55312915, 18.62092487, -70.91365499, -41.83066505, 1248.88835245, -4719.81948329, 9186.09528168, -10266.11434548, 6266.21959533, -1622.19652457,)),
+ HbPolyType.ahdist_aTYR_dHIS: ((0.5955982461899999, 2.6643551317500003,),(1.1, 1.1,),( -0.47442788, 7.16629863, -46.71287553, 171.46128947, -388.17484011, 558.45202337, -506.35587481, 276.46237273, -83.52554392, 12.05709329,)),
+ HbPolyType.ahdist_aTYR_dLYS: ((0.7978598238760001, 2.7620933782,),(1.1, 1.1,),( -0.20201464, 1.69684984, 0.27677515, -55.05786347, 286.29918332, -725.92372531, 1054.771746, -889.33602341, 401.11342256, -73.02221189,)),
+ HbPolyType.ahdist_aTYR_dSER: ((0.7083554962559999, 2.7032011990599996,),(1.1, 1.1,),( -0.70764192, 11.67978065, -82.80447482, 329.83401367, -810.58976486, 1269.57613941, -1261.04047117, 761.72890446, -254.37526011, 37.24301861,)),
+ HbPolyType.ahdist_aTYR_dTRP: ((1.10934023051, 2.8819112108,),(1.1, 1.1,),( -11.58453967, 204.88308091, -1589.77384548, 7100.84791905, -20113.61354433, 37457.83646055, -45850.02969172, 35559.8805122, -15854.78726237, 3098.04931146,)),
+ HbPolyType.ahdist_aTYR_dTYR: ((1.1105954899400001, 2.60081798685,),(1.1, 1.1,),( -1.63120628, 19.48493187, -81.0332905, 56.80517706, 687.42717782, -2842.77799908, 5385.52231471, -5656.74159307, 3178.83470588, -744.70042777,)),
+ HbPolyType.AHD_1h: ((1.76555274367, 3.1416,),(1.1, 1.1,),( 0.62725838, -9.98558225, 59.39060071, -120.82930213, -333.26536028, 2603.13082592, -6895.51207142, 9651.25238056, -7127.13394872, 2194.77244026,)),
+ HbPolyType.AHD_1i: ((1.59914724347, 3.1416,),(1.1, 1.1,),( -0.18888801, 3.48241679, -25.65508662, 89.57085435, -95.91708218, -367.93452341, 1589.6904702, -2662.3582135, 2184.40194483, -723.28383545,)),
+ HbPolyType.AHD_1j: ((1.1435646388, 3.1416,),(1.1, 1.1,),( 0.47683259, -9.54524724, 83.62557693, -420.55867774, 1337.19354878, -2786.26265686, 3803.178227, -3278.62879901, 1619.04116204, -347.50157909,)),
+ HbPolyType.AHD_1k: ((1.15651981164, 3.1416,),(1.1, 1.1,),( -0.10757999, 2.0276542, -16.51949978, 75.83866839, -214.18025678, 380.55117567, -415.47847283, 255.66998474, -69.94662165, 3.21313428,)),
+ HbPolyType.cosBAH_off: ((-1234.0, 1.1,),(1.1, 1.1,),( 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,)),
+ HbPolyType.cosBAH_6i: ((-0.23538144897100002, 1.1,),(1.1, 1.1,),( -0.822093, -3.75364636, 46.88852157, -129.5440564, 146.69151428, -67.60598792, 2.91683129, 9.26673173, -3.84488178, 0.05706659,)),
+ HbPolyType.cosBAH_7: ((-0.019373850666900002, 1.1,),(1.1, 1.1,),( 0.0, -27.942923450028, 136.039920253368, -268.06959056747, 275.400462507919, -153.502076215949, 39.741591385461, 0.693861510121, -3.885952320499, 1.024765090788892)),
+}
\ No newline at end of file
diff --git a/model/se3_transformer/__init__.py b/model/se3_transformer/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..e69de29bb2d1d6434b8b29ae775ad8c2e48c5391
diff --git a/model/se3_transformer/__pycache__/__init__.cpython-310.pyc b/model/se3_transformer/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..01f8e2d61996d7fe7617616463cf01fb6b19dee2
Binary files /dev/null and b/model/se3_transformer/__pycache__/__init__.cpython-310.pyc differ
diff --git a/model/se3_transformer/data_loading/__init__.py b/model/se3_transformer/data_loading/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..67a9e0ed7b9c787830f1b99cc0132a14bbcaee43
--- /dev/null
+++ b/model/se3_transformer/data_loading/__init__.py
@@ -0,0 +1 @@
+from .qm9 import QM9DataModule
diff --git a/model/se3_transformer/data_loading/data_module.py b/model/se3_transformer/data_loading/data_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..1047d41d16b81c05e2f14130311b42ab5b36eb36
--- /dev/null
+++ b/model/se3_transformer/data_loading/data_module.py
@@ -0,0 +1,63 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import torch.distributed as dist
+from abc import ABC
+from torch.utils.data import DataLoader, DistributedSampler, Dataset
+
+from se3_transformer.runtime.utils import get_local_rank
+
+
+def _get_dataloader(dataset: Dataset, shuffle: bool, **kwargs) -> DataLoader:
+ # Classic or distributed dataloader depending on the context
+ sampler = DistributedSampler(dataset, shuffle=shuffle) if dist.is_initialized() else None
+ return DataLoader(dataset, shuffle=(shuffle and sampler is None), sampler=sampler, **kwargs)
+
+
+class DataModule(ABC):
+ """ Abstract DataModule. Children must define self.ds_{train | val | test}. """
+
+ def __init__(self, **dataloader_kwargs):
+ super().__init__()
+ if get_local_rank() == 0:
+ self.prepare_data()
+
+ # Wait until rank zero has prepared the data (download, preprocessing, ...)
+ if dist.is_initialized():
+ dist.barrier(device_ids=[get_local_rank()])
+
+ self.dataloader_kwargs = {'pin_memory': True, 'persistent_workers': True, **dataloader_kwargs}
+ self.ds_train, self.ds_val, self.ds_test = None, None, None
+
+ def prepare_data(self):
+ """ Method called only once per node. Put here any downloading or preprocessing """
+ pass
+
+ def train_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_train, shuffle=True, **self.dataloader_kwargs)
+
+ def val_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_val, shuffle=False, **self.dataloader_kwargs)
+
+ def test_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_test, shuffle=False, **self.dataloader_kwargs)
diff --git a/model/se3_transformer/data_loading/qm9.py b/model/se3_transformer/data_loading/qm9.py
new file mode 100644
index 0000000000000000000000000000000000000000..b45839868626f56d3a42ce859b2033ce1526373e
--- /dev/null
+++ b/model/se3_transformer/data_loading/qm9.py
@@ -0,0 +1,171 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+from typing import Tuple
+
+import dgl
+import pathlib
+import torch
+from dgl.data import QM9EdgeDataset
+from dgl import DGLGraph
+from torch import Tensor
+from torch.utils.data import random_split, DataLoader, Dataset
+from tqdm import tqdm
+
+from se3_transformer.data_loading.data_module import DataModule
+from se3_transformer.model.basis import get_basis
+from se3_transformer.runtime.utils import get_local_rank, str2bool, using_tensor_cores
+
+
+def _get_relative_pos(qm9_graph: DGLGraph) -> Tensor:
+ x = qm9_graph.ndata['pos']
+ src, dst = qm9_graph.edges()
+ rel_pos = x[dst] - x[src]
+ return rel_pos
+
+
+def _get_split_sizes(full_dataset: Dataset) -> Tuple[int, int, int]:
+ len_full = len(full_dataset)
+ len_train = 100_000
+ len_test = int(0.1 * len_full)
+ len_val = len_full - len_train - len_test
+ return len_train, len_val, len_test
+
+
+class QM9DataModule(DataModule):
+ """
+ Datamodule wrapping https://docs.dgl.ai/en/latest/api/python/dgl.data.html#qm9edge-dataset
+ Training set is 100k molecules. Test set is 10% of the dataset. Validation set is the rest.
+ This includes all the molecules from QM9 except the ones that are uncharacterized.
+ """
+
+ NODE_FEATURE_DIM = 6
+ EDGE_FEATURE_DIM = 4
+
+ def __init__(self,
+ data_dir: pathlib.Path,
+ task: str = 'homo',
+ batch_size: int = 240,
+ num_workers: int = 8,
+ num_degrees: int = 4,
+ amp: bool = False,
+ precompute_bases: bool = False,
+ **kwargs):
+ self.data_dir = data_dir # This needs to be before __init__ so that prepare_data has access to it
+ super().__init__(batch_size=batch_size, num_workers=num_workers, collate_fn=self._collate)
+ self.amp = amp
+ self.task = task
+ self.batch_size = batch_size
+ self.num_degrees = num_degrees
+
+ qm9_kwargs = dict(label_keys=[self.task], verbose=False, raw_dir=str(data_dir))
+ if precompute_bases:
+ bases_kwargs = dict(max_degree=num_degrees - 1, use_pad_trick=using_tensor_cores(amp), amp=amp)
+ full_dataset = CachedBasesQM9EdgeDataset(bases_kwargs=bases_kwargs, batch_size=batch_size, **qm9_kwargs)
+ else:
+ full_dataset = QM9EdgeDataset(**qm9_kwargs)
+
+ self.ds_train, self.ds_val, self.ds_test = random_split(full_dataset, _get_split_sizes(full_dataset),
+ generator=torch.Generator().manual_seed(0))
+
+ train_targets = full_dataset.targets[self.ds_train.indices, full_dataset.label_keys[0]]
+ self.targets_mean = train_targets.mean()
+ self.targets_std = train_targets.std()
+
+ def prepare_data(self):
+ # Download the QM9 preprocessed data
+ QM9EdgeDataset(verbose=True, raw_dir=str(self.data_dir))
+
+ def _collate(self, samples):
+ graphs, y, *bases = map(list, zip(*samples))
+ batched_graph = dgl.batch(graphs)
+ edge_feats = {'0': batched_graph.edata['edge_attr'][..., None]}
+ batched_graph.edata['rel_pos'] = _get_relative_pos(batched_graph)
+ # get node features
+ node_feats = {'0': batched_graph.ndata['attr'][:, :6, None]}
+ targets = (torch.cat(y) - self.targets_mean) / self.targets_std
+
+ if bases:
+ # collate bases
+ all_bases = {
+ key: torch.cat([b[key] for b in bases[0]], dim=0)
+ for key in bases[0][0].keys()
+ }
+
+ return batched_graph, node_feats, edge_feats, all_bases, targets
+ else:
+ return batched_graph, node_feats, edge_feats, targets
+
+ @staticmethod
+ def add_argparse_args(parent_parser):
+ parser = parent_parser.add_argument_group("QM9 dataset")
+ parser.add_argument('--task', type=str, default='homo', const='homo', nargs='?',
+ choices=['mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'U0', 'U', 'H', 'G', 'Cv',
+ 'U0_atom', 'U_atom', 'H_atom', 'G_atom', 'A', 'B', 'C'],
+ help='Regression task to train on')
+ parser.add_argument('--precompute_bases', type=str2bool, nargs='?', const=True, default=False,
+ help='Precompute bases at the beginning of the script during dataset initialization,'
+ ' instead of computing them at the beginning of each forward pass.')
+ return parent_parser
+
+ def __repr__(self):
+ return f'QM9({self.task})'
+
+
+class CachedBasesQM9EdgeDataset(QM9EdgeDataset):
+ """ Dataset extending the QM9 dataset from DGL with precomputed (cached in RAM) pairwise bases """
+
+ def __init__(self, bases_kwargs: dict, batch_size: int, *args, **kwargs):
+ """
+ :param bases_kwargs: Arguments to feed the bases computation function
+ :param batch_size: Batch size to use when iterating over the dataset for computing bases
+ """
+ self.bases_kwargs = bases_kwargs
+ self.batch_size = batch_size
+ self.bases = None
+ super().__init__(*args, **kwargs)
+
+ def load(self):
+ super().load()
+ # Iterate through the dataset and compute bases (pairwise only)
+ # Potential improvement: use multi-GPU and reduction
+ dataloader = DataLoader(self, shuffle=False, batch_size=self.batch_size,
+ collate_fn=lambda samples: dgl.batch([sample[0] for sample in samples]))
+ bases = []
+ for i, graph in tqdm(enumerate(dataloader), total=len(dataloader), desc='Precomputing QM9 bases',
+ disable=get_local_rank() != 0):
+ rel_pos = _get_relative_pos(graph)
+ # Compute the bases with the GPU but convert the result to CPU to store in RAM
+ bases.append({k: v.cpu() for k, v in get_basis(rel_pos.cuda(), **self.bases_kwargs).items()})
+ self.bases = bases # Assign at the end so that __getitem__ isn't confused
+
+ def __getitem__(self, idx: int):
+ graph, label = super().__getitem__(idx)
+
+ if self.bases:
+ bases_idx = idx // self.batch_size
+ bases_cumsum_idx = self.ne_cumsum[idx] - self.ne_cumsum[bases_idx * self.batch_size]
+ bases_cumsum_next_idx = self.ne_cumsum[idx + 1] - self.ne_cumsum[bases_idx * self.batch_size]
+ return graph, label, {key: basis[bases_cumsum_idx:bases_cumsum_next_idx] for key, basis in
+ self.bases[bases_idx].items()}
+ else:
+ return graph, label
diff --git a/model/se3_transformer/model/__init__.py b/model/se3_transformer/model/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..628d01e98d45d360187fb7fcce63f33ebf05d3b7
--- /dev/null
+++ b/model/se3_transformer/model/__init__.py
@@ -0,0 +1,2 @@
+from .transformer import SE3Transformer, SE3TransformerPooled
+from .fiber import Fiber
diff --git a/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc b/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b86f94e8876762ea1a611443581478df39a4dbc9
Binary files /dev/null and b/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc b/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..86e5265ee87961d2d03093c0756a829b408edde9
Binary files /dev/null and b/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc b/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..f40874b4b024c1305a9a92ea21435108db7b1271
Binary files /dev/null and b/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc b/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..3472eec408c815c3f5ea1dd15437135eaa4c7f5e
Binary files /dev/null and b/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/basis.py b/model/se3_transformer/model/basis.py
new file mode 100644
index 0000000000000000000000000000000000000000..74f04a0fada04ad63f9d9fc17f4fe7bb038d806b
--- /dev/null
+++ b/model/se3_transformer/model/basis.py
@@ -0,0 +1,178 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from functools import lru_cache
+from typing import Dict, List
+
+import e3nn.o3 as o3
+import torch
+import torch.nn.functional as F
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.runtime.utils import degree_to_dim
+
+
+@lru_cache(maxsize=None)
+def get_clebsch_gordon(J: int, d_in: int, d_out: int, device) -> Tensor:
+ """ Get the (cached) Q^{d_out,d_in}_J matrices from equation (8) """
+ return o3.wigner_3j(J, d_in, d_out, dtype=torch.float64, device=device).permute(2, 1, 0)
+
+
+@lru_cache(maxsize=None)
+def get_all_clebsch_gordon(max_degree: int, device) -> List[List[Tensor]]:
+ all_cb = []
+ for d_in in range(max_degree + 1):
+ for d_out in range(max_degree + 1):
+ K_Js = []
+ for J in range(abs(d_in - d_out), d_in + d_out + 1):
+ K_Js.append(get_clebsch_gordon(J, d_in, d_out, device))
+ all_cb.append(K_Js)
+ return all_cb
+
+
+def get_spherical_harmonics(relative_pos: Tensor, max_degree: int) -> List[Tensor]:
+ all_degrees = list(range(2 * max_degree + 1))
+ with nvtx_range('spherical harmonics'):
+ sh = o3.spherical_harmonics(all_degrees, relative_pos, normalize=True)
+ return torch.split(sh, [degree_to_dim(d) for d in all_degrees], dim=1)
+
+
+@torch.jit.script
+def get_basis_script(max_degree: int,
+ use_pad_trick: bool,
+ spherical_harmonics: List[Tensor],
+ clebsch_gordon: List[List[Tensor]],
+ amp: bool) -> Dict[str, Tensor]:
+ """
+ Compute pairwise bases matrices for degrees up to max_degree
+ :param max_degree: Maximum input or output degree
+ :param use_pad_trick: Pad some of the odd dimensions for a better use of Tensor Cores
+ :param spherical_harmonics: List of computed spherical harmonics
+ :param clebsch_gordon: List of computed CB-coefficients
+ :param amp: When true, return bases in FP16 precision
+ """
+ basis = {}
+ idx = 0
+ # Double for loop instead of product() because of JIT script
+ for d_in in range(max_degree + 1):
+ for d_out in range(max_degree + 1):
+ key = f'{d_in},{d_out}'
+ K_Js = []
+ for freq_idx, J in enumerate(range(abs(d_in - d_out), d_in + d_out + 1)):
+ Q_J = clebsch_gordon[idx][freq_idx]
+ K_Js.append(torch.einsum('n f, k l f -> n l k', spherical_harmonics[J].float(), Q_J.float()))
+
+ basis[key] = torch.stack(K_Js, 2) # Stack on second dim so order is n l f k
+ if amp:
+ basis[key] = basis[key].half()
+ if use_pad_trick:
+ basis[key] = F.pad(basis[key], (0, 1)) # Pad the k dimension, that can be sliced later
+
+ idx += 1
+
+ return basis
+
+
+@torch.jit.script
+def update_basis_with_fused(basis: Dict[str, Tensor],
+ max_degree: int,
+ use_pad_trick: bool,
+ fully_fused: bool) -> Dict[str, Tensor]:
+ """ Update the basis dict with partially and optionally fully fused bases """
+ num_edges = basis['0,0'].shape[0]
+ device = basis['0,0'].device
+ dtype = basis['0,0'].dtype
+ sum_dim = sum([degree_to_dim(d) for d in range(max_degree + 1)])
+
+ # Fused per output degree
+ for d_out in range(max_degree + 1):
+ sum_freq = sum([degree_to_dim(min(d, d_out)) for d in range(max_degree + 1)])
+ basis_fused = torch.zeros(num_edges, sum_dim, sum_freq, degree_to_dim(d_out) + int(use_pad_trick),
+ device=device, dtype=dtype)
+ acc_d, acc_f = 0, 0
+ for d_in in range(max_degree + 1):
+ basis_fused[:, acc_d:acc_d + degree_to_dim(d_in), acc_f:acc_f + degree_to_dim(min(d_out, d_in)),
+ :degree_to_dim(d_out)] = basis[f'{d_in},{d_out}'][:, :, :, :degree_to_dim(d_out)]
+
+ acc_d += degree_to_dim(d_in)
+ acc_f += degree_to_dim(min(d_out, d_in))
+
+ basis[f'out{d_out}_fused'] = basis_fused
+
+ # Fused per input degree
+ for d_in in range(max_degree + 1):
+ sum_freq = sum([degree_to_dim(min(d, d_in)) for d in range(max_degree + 1)])
+ basis_fused = torch.zeros(num_edges, degree_to_dim(d_in), sum_freq, sum_dim,
+ device=device, dtype=dtype)
+ acc_d, acc_f = 0, 0
+ for d_out in range(max_degree + 1):
+ basis_fused[:, :, acc_f:acc_f + degree_to_dim(min(d_out, d_in)), acc_d:acc_d + degree_to_dim(d_out)] \
+ = basis[f'{d_in},{d_out}'][:, :, :, :degree_to_dim(d_out)]
+
+ acc_d += degree_to_dim(d_out)
+ acc_f += degree_to_dim(min(d_out, d_in))
+
+ basis[f'in{d_in}_fused'] = basis_fused
+
+ if fully_fused:
+ # Fully fused
+ # Double sum this way because of JIT script
+ sum_freq = sum([
+ sum([degree_to_dim(min(d_in, d_out)) for d_in in range(max_degree + 1)]) for d_out in range(max_degree + 1)
+ ])
+ basis_fused = torch.zeros(num_edges, sum_dim, sum_freq, sum_dim, device=device, dtype=dtype)
+
+ acc_d, acc_f = 0, 0
+ for d_out in range(max_degree + 1):
+ b = basis[f'out{d_out}_fused']
+ basis_fused[:, :, acc_f:acc_f + b.shape[2], acc_d:acc_d + degree_to_dim(d_out)] = b[:, :, :,
+ :degree_to_dim(d_out)]
+ acc_f += b.shape[2]
+ acc_d += degree_to_dim(d_out)
+
+ basis['fully_fused'] = basis_fused
+
+ del basis['0,0'] # We know that the basis for l = k = 0 is filled with a constant
+ return basis
+
+
+def get_basis(relative_pos: Tensor,
+ max_degree: int = 4,
+ compute_gradients: bool = False,
+ use_pad_trick: bool = False,
+ amp: bool = False) -> Dict[str, Tensor]:
+ with nvtx_range('spherical harmonics'):
+ spherical_harmonics = get_spherical_harmonics(relative_pos, max_degree)
+ with nvtx_range('CB coefficients'):
+ clebsch_gordon = get_all_clebsch_gordon(max_degree, relative_pos.device)
+
+ with torch.autograd.set_grad_enabled(compute_gradients):
+ with nvtx_range('bases'):
+ basis = get_basis_script(max_degree=max_degree,
+ use_pad_trick=use_pad_trick,
+ spherical_harmonics=spherical_harmonics,
+ clebsch_gordon=clebsch_gordon,
+ amp=amp)
+ return basis
diff --git a/model/se3_transformer/model/fiber.py b/model/se3_transformer/model/fiber.py
new file mode 100644
index 0000000000000000000000000000000000000000..38db33b0d27d70116c92650176170e9b3cf9a9c7
--- /dev/null
+++ b/model/se3_transformer/model/fiber.py
@@ -0,0 +1,144 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from collections import namedtuple
+from itertools import product
+from typing import Dict
+
+import torch
+from torch import Tensor
+
+from se3_transformer.runtime.utils import degree_to_dim
+
+FiberEl = namedtuple('FiberEl', ['degree', 'channels'])
+
+
+class Fiber(dict):
+ """
+ Describes the structure of some set of features.
+ Features are split into types (0, 1, 2, 3, ...). A feature of type k has a dimension of 2k+1.
+ Type-0 features: invariant scalars
+ Type-1 features: equivariant 3D vectors
+ Type-2 features: equivariant symmetric traceless matrices
+ ...
+
+ As inputs to a SE3 layer, there can be many features of the same types, and many features of different types.
+ The 'multiplicity' or 'number of channels' is the number of features of a given type.
+ This class puts together all the degrees and their multiplicities in order to describe
+ the inputs, outputs or hidden features of SE3 layers.
+ """
+
+ def __init__(self, structure):
+ if isinstance(structure, dict):
+ structure = [FiberEl(int(d), int(m)) for d, m in sorted(structure.items(), key=lambda x: x[1])]
+ elif not isinstance(structure[0], FiberEl):
+ structure = list(map(lambda t: FiberEl(*t), sorted(structure, key=lambda x: x[1])))
+ self.structure = structure
+ super().__init__({d: m for d, m in self.structure})
+
+ @property
+ def degrees(self):
+ return sorted([t.degree for t in self.structure])
+
+ @property
+ def channels(self):
+ return [self[d] for d in self.degrees]
+
+ @property
+ def num_features(self):
+ """ Size of the resulting tensor if all features were concatenated together """
+ return sum(t.channels * degree_to_dim(t.degree) for t in self.structure)
+
+ @staticmethod
+ def create(num_degrees: int, num_channels: int):
+ """ Create a Fiber with degrees 0..num_degrees-1, all with the same multiplicity """
+ return Fiber([(degree, num_channels) for degree in range(num_degrees)])
+
+ @staticmethod
+ def from_features(feats: Dict[str, Tensor]):
+ """ Infer the Fiber structure from a feature dict """
+ structure = {}
+ for k, v in feats.items():
+ degree = int(k)
+ assert len(v.shape) == 3, 'Feature shape should be (N, C, 2D+1)'
+ assert v.shape[-1] == degree_to_dim(degree)
+ structure[degree] = v.shape[-2]
+ return Fiber(structure)
+
+ def __getitem__(self, degree: int):
+ """ fiber[degree] returns the multiplicity for this degree """
+ return dict(self.structure).get(degree, 0)
+
+ def __iter__(self):
+ """ Iterate over namedtuples (degree, channels) """
+ return iter(self.structure)
+
+ def __mul__(self, other):
+ """
+ If other in an int, multiplies all the multiplicities by other.
+ If other is a fiber, returns the cartesian product.
+ """
+ if isinstance(other, Fiber):
+ return product(self.structure, other.structure)
+ elif isinstance(other, int):
+ return Fiber({t.degree: t.channels * other for t in self.structure})
+
+ def __add__(self, other):
+ """
+ If other in an int, add other to all the multiplicities.
+ If other is a fiber, add the multiplicities of the fibers together.
+ """
+ if isinstance(other, Fiber):
+ return Fiber({t.degree: t.channels + other[t.degree] for t in self.structure})
+ elif isinstance(other, int):
+ return Fiber({t.degree: t.channels + other for t in self.structure})
+
+ def __repr__(self):
+ return str(self.structure)
+
+ @staticmethod
+ def combine_max(f1, f2):
+ """ Combine two fiber by taking the maximum multiplicity for each degree in both fibers """
+ new_dict = dict(f1.structure)
+ for k, m in f2.structure:
+ new_dict[k] = max(new_dict.get(k, 0), m)
+
+ return Fiber(list(new_dict.items()))
+
+ @staticmethod
+ def combine_selectively(f1, f2):
+ """ Combine two fiber by taking the sum of multiplicities for each degree in the first fiber """
+ # only use orders which occur in fiber f1
+ new_dict = dict(f1.structure)
+ for k in f1.degrees:
+ if k in f2.degrees:
+ new_dict[k] += f2[k]
+ return Fiber(list(new_dict.items()))
+
+ def to_attention_heads(self, tensors: Dict[str, Tensor], num_heads: int):
+ # dict(N, num_channels, 2d+1) -> (N, num_heads, -1)
+ fibers = [tensors[str(degree)].reshape(*tensors[str(degree)].shape[:-2], num_heads, -1) for degree in
+ self.degrees]
+ fibers = torch.cat(fibers, -1)
+ return fibers
diff --git a/model/se3_transformer/model/layers/._attention.py b/model/se3_transformer/model/layers/._attention.py
new file mode 100644
index 0000000000000000000000000000000000000000..e104c6317b4c2bd3aed2efb7c5db8556cd8d985e
Binary files /dev/null and b/model/se3_transformer/model/layers/._attention.py differ
diff --git a/model/se3_transformer/model/layers/__init__.py b/model/se3_transformer/model/layers/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..9eb9e3ced5ef94e9a5c5be5883cc14ebaabe31f1
--- /dev/null
+++ b/model/se3_transformer/model/layers/__init__.py
@@ -0,0 +1,5 @@
+from .linear import LinearSE3
+from .norm import NormSE3
+from .pooling import GPooling
+from .convolution import ConvSE3
+from .attention import AttentionBlockSE3
\ No newline at end of file
diff --git a/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b290d94285440c9f97f61ca3c6ea68c7ebcf1644
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9da62209e5b13745f7e89a936519b085fd9a4e05
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..e42942454c78b14df9935261fe61eb959245849d
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..1c09095ac7efe0bda53fef2fd11ccf8475d0e99f
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..e4288c55b9b01dc2b17e125898fc76928bc729ae
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc b/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..90f9fc414b3be6c849f369d2ea9a0fd4053f201e
Binary files /dev/null and b/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc differ
diff --git a/model/se3_transformer/model/layers/attention.py b/model/se3_transformer/model/layers/attention.py
new file mode 100644
index 0000000000000000000000000000000000000000..1dc68fe95faaee99f362678399fa7e47010e1af1
--- /dev/null
+++ b/model/se3_transformer/model/layers/attention.py
@@ -0,0 +1,180 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import dgl
+import numpy as np
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from dgl.ops import edge_softmax
+from torch import Tensor
+from typing import Dict, Optional, Union
+
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.model.layers.convolution import ConvSE3, ConvSE3FuseLevel
+from se3_transformer.model.layers.linear import LinearSE3
+from se3_transformer.runtime.utils import degree_to_dim, aggregate_residual, unfuse_features
+from torch.cuda.nvtx import range as nvtx_range
+
+
+class AttentionSE3(nn.Module):
+ """ Multi-headed sparse graph self-attention (SE(3)-equivariant) """
+
+ def __init__(
+ self,
+ num_heads: int,
+ key_fiber: Fiber,
+ value_fiber: Fiber
+ ):
+ """
+ :param num_heads: Number of attention heads
+ :param key_fiber: Fiber for the keys (and also for the queries)
+ :param value_fiber: Fiber for the values
+ """
+ super().__init__()
+ self.num_heads = num_heads
+ self.key_fiber = key_fiber
+ self.value_fiber = value_fiber
+
+ def forward(
+ self,
+ value: Union[Tensor, Dict[str, Tensor]], # edge features (may be fused)
+ key: Union[Tensor, Dict[str, Tensor]], # edge features (may be fused)
+ query: Dict[str, Tensor], # node features
+ graph: DGLGraph
+ ):
+ with nvtx_range('AttentionSE3'):
+ with nvtx_range('reshape keys and queries'):
+ if isinstance(key, Tensor):
+ # case where features of all types are fused
+ key = key.reshape(key.shape[0], self.num_heads, -1)
+ # need to reshape queries that way to keep the same layout as keys
+ out = torch.cat([query[str(d)] for d in self.key_fiber.degrees], dim=-1)
+ query = out.reshape(list(query.values())[0].shape[0], self.num_heads, -1)
+ else:
+ # features are not fused, need to fuse and reshape them
+ key = self.key_fiber.to_attention_heads(key, self.num_heads)
+ query = self.key_fiber.to_attention_heads(query, self.num_heads)
+
+ with nvtx_range('attention dot product + softmax'):
+ # Compute attention weights (softmax of inner product between key and query)
+ edge_weights = dgl.ops.e_dot_v(graph, key, query).squeeze(-1)
+ edge_weights /= np.sqrt(self.key_fiber.num_features)
+ edge_weights = edge_softmax(graph, edge_weights)
+ edge_weights = edge_weights[..., None, None]
+
+ with nvtx_range('weighted sum'):
+ if isinstance(value, Tensor):
+ # features of all types are fused
+ v = value.view(value.shape[0], self.num_heads, -1, value.shape[-1])
+ weights = edge_weights * v
+ feat_out = dgl.ops.copy_e_sum(graph, weights)
+ feat_out = feat_out.view(feat_out.shape[0], -1, feat_out.shape[-1]) # merge heads
+ out = unfuse_features(feat_out, self.value_fiber.degrees)
+ else:
+ out = {}
+ for degree, channels in self.value_fiber:
+ v = value[str(degree)].view(-1, self.num_heads, channels // self.num_heads,
+ degree_to_dim(degree))
+ weights = edge_weights * v
+ res = dgl.ops.copy_e_sum(graph, weights)
+ out[str(degree)] = res.view(-1, channels, degree_to_dim(degree)) # merge heads
+
+ return out
+
+
+class AttentionBlockSE3(nn.Module):
+ """ Multi-headed sparse graph self-attention block with skip connection, linear projection (SE(3)-equivariant) """
+
+ def __init__(
+ self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Optional[Fiber] = None,
+ num_heads: int = 4,
+ channels_div: int = 2,
+ use_layer_norm: bool = False,
+ max_degree: bool = 4,
+ fuse_level: ConvSE3FuseLevel = ConvSE3FuseLevel.FULL,
+ **kwargs
+ ):
+ """
+ :param fiber_in: Fiber describing the input features
+ :param fiber_out: Fiber describing the output features
+ :param fiber_edge: Fiber describing the edge features (node distances excluded)
+ :param num_heads: Number of attention heads
+ :param channels_div: Divide the channels by this integer for computing values
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param max_degree: Maximum degree used in the bases computation
+ :param fuse_level: Maximum fuse level to use in TFN convolutions
+ """
+ super().__init__()
+ if fiber_edge is None:
+ fiber_edge = Fiber({})
+ self.fiber_in = fiber_in
+ # value_fiber has same structure as fiber_out but #channels divided by 'channels_div'
+ value_fiber = Fiber([(degree, channels // channels_div) for degree, channels in fiber_out])
+ # key_query_fiber has the same structure as fiber_out, but only degrees which are in in_fiber
+ # (queries are merely projected, hence degrees have to match input)
+ key_query_fiber = Fiber([(fe.degree, fe.channels) for fe in value_fiber if fe.degree in fiber_in.degrees])
+
+ self.to_key_value = ConvSE3(fiber_in, value_fiber + key_query_fiber, pool=False, fiber_edge=fiber_edge,
+ use_layer_norm=use_layer_norm, max_degree=max_degree, fuse_level=fuse_level,
+ allow_fused_output=True)
+ self.to_query = LinearSE3(fiber_in, key_query_fiber)
+ self.attention = AttentionSE3(num_heads, key_query_fiber, value_fiber)
+ self.project = LinearSE3(value_fiber + fiber_in, fiber_out)
+
+ def forward(
+ self,
+ node_features: Dict[str, Tensor],
+ edge_features: Dict[str, Tensor],
+ graph: DGLGraph,
+ basis: Dict[str, Tensor]
+ ):
+ with nvtx_range('AttentionBlockSE3'):
+ with nvtx_range('keys / values'):
+ fused_key_value = self.to_key_value(node_features, edge_features, graph, basis)
+ key, value = self._get_key_value_from_fused(fused_key_value)
+
+ with nvtx_range('queries'):
+ query = self.to_query(node_features)
+
+ z = self.attention(value, key, query, graph)
+ z_concat = aggregate_residual(node_features, z, 'cat')
+ return self.project(z_concat)
+
+ def _get_key_value_from_fused(self, fused_key_value):
+ # Extract keys and queries features from fused features
+ if isinstance(fused_key_value, Tensor):
+ # Previous layer was a fully fused convolution
+ value, key = torch.chunk(fused_key_value, chunks=2, dim=-2)
+ else:
+ key, value = {}, {}
+ for degree, feat in fused_key_value.items():
+ if int(degree) in self.fiber_in.degrees:
+ value[degree], key[degree] = torch.chunk(feat, chunks=2, dim=-2)
+ else:
+ value[degree] = feat
+
+ return key, value
diff --git a/model/se3_transformer/model/layers/convolution.py b/model/se3_transformer/model/layers/convolution.py
new file mode 100644
index 0000000000000000000000000000000000000000..fcbb1c22dd8e7b14ab61e717ddbe6e0a19164137
--- /dev/null
+++ b/model/se3_transformer/model/layers/convolution.py
@@ -0,0 +1,335 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from enum import Enum
+from itertools import product
+from typing import Dict
+
+import dgl
+import numpy as np
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.runtime.utils import degree_to_dim, unfuse_features
+
+
+class ConvSE3FuseLevel(Enum):
+ """
+ Enum to select a maximum level of fusing optimizations that will be applied when certain conditions are met.
+ If a desired level L is picked and the level L cannot be applied to a level, other fused ops < L are considered.
+ A higher level means faster training, but also more memory usage.
+ If you are tight on memory and want to feed large inputs to the network, choose a low value.
+ If you want to train fast, choose a high value.
+ Recommended value is FULL with AMP.
+
+ Fully fused TFN convolutions requirements:
+ - all input channels are the same
+ - all output channels are the same
+ - input degrees span the range [0, ..., max_degree]
+ - output degrees span the range [0, ..., max_degree]
+
+ Partially fused TFN convolutions requirements:
+ * For fusing by output degree:
+ - all input channels are the same
+ - input degrees span the range [0, ..., max_degree]
+ * For fusing by input degree:
+ - all output channels are the same
+ - output degrees span the range [0, ..., max_degree]
+
+ Original TFN pairwise convolutions: no requirements
+ """
+
+ FULL = 2
+ PARTIAL = 1
+ NONE = 0
+
+
+class RadialProfile(nn.Module):
+ """
+ Radial profile function.
+ Outputs weights used to weigh basis matrices in order to get convolution kernels.
+ In TFN notation: $R^{l,k}$
+ In SE(3)-Transformer notation: $\phi^{l,k}$
+
+ Note:
+ In the original papers, this function only depends on relative node distances ||x||.
+ Here, we allow this function to also take as input additional invariant edge features.
+ This does not break equivariance and adds expressive power to the model.
+
+ Diagram:
+ invariant edge features (node distances included) ───> MLP layer (shared across edges) ───> radial weights
+ """
+
+ def __init__(
+ self,
+ num_freq: int,
+ channels_in: int,
+ channels_out: int,
+ edge_dim: int = 1,
+ mid_dim: int = 32,
+ use_layer_norm: bool = False
+ ):
+ """
+ :param num_freq: Number of frequencies
+ :param channels_in: Number of input channels
+ :param channels_out: Number of output channels
+ :param edge_dim: Number of invariant edge features (input to the radial function)
+ :param mid_dim: Size of the hidden MLP layers
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ """
+ super().__init__()
+ modules = [
+ nn.Linear(edge_dim, mid_dim),
+ nn.LayerNorm(mid_dim) if use_layer_norm else None,
+ nn.ReLU(),
+ nn.Linear(mid_dim, mid_dim),
+ nn.LayerNorm(mid_dim) if use_layer_norm else None,
+ nn.ReLU(),
+ nn.Linear(mid_dim, num_freq * channels_in * channels_out, bias=False)
+ ]
+
+ self.net = nn.Sequential(*[m for m in modules if m is not None])
+
+ def forward(self, features: Tensor) -> Tensor:
+ return self.net(features)
+
+
+class VersatileConvSE3(nn.Module):
+ """
+ Building block for TFN convolutions.
+ This single module can be used for fully fused convolutions, partially fused convolutions, or pairwise convolutions.
+ """
+
+ def __init__(self,
+ freq_sum: int,
+ channels_in: int,
+ channels_out: int,
+ edge_dim: int,
+ use_layer_norm: bool,
+ fuse_level: ConvSE3FuseLevel):
+ super().__init__()
+ self.freq_sum = freq_sum
+ self.channels_out = channels_out
+ self.channels_in = channels_in
+ self.fuse_level = fuse_level
+ self.radial_func = RadialProfile(num_freq=freq_sum,
+ channels_in=channels_in,
+ channels_out=channels_out,
+ edge_dim=edge_dim,
+ use_layer_norm=use_layer_norm)
+
+ def forward(self, features: Tensor, invariant_edge_feats: Tensor, basis: Tensor):
+ with nvtx_range(f'VersatileConvSE3'):
+ num_edges = features.shape[0]
+ in_dim = features.shape[2]
+ with nvtx_range(f'RadialProfile'):
+ radial_weights = self.radial_func(invariant_edge_feats) \
+ .view(-1, self.channels_out, self.channels_in * self.freq_sum)
+
+ if basis is not None:
+ # This block performs the einsum n i l, n o i f, n l f k -> n o k
+ out_dim = basis.shape[-1]
+ if self.fuse_level != ConvSE3FuseLevel.FULL:
+ out_dim += out_dim % 2 - 1 # Account for padded basis
+ basis_view = basis.view(num_edges, in_dim, -1)
+ tmp = (features @ basis_view).view(num_edges, -1, basis.shape[-1])
+ return (radial_weights @ tmp)[:, :, :out_dim]
+ else:
+ # k = l = 0 non-fused case
+ return radial_weights @ features
+
+
+class ConvSE3(nn.Module):
+ """
+ SE(3)-equivariant graph convolution (Tensor Field Network convolution).
+ This convolution can map an arbitrary input Fiber to an arbitrary output Fiber, while preserving equivariance.
+ Features of different degrees interact together to produce output features.
+
+ Note 1:
+ The option is given to not pool the output. This means that the convolution sum over neighbors will not be
+ done, and the returned features will be edge features instead of node features.
+
+ Note 2:
+ Unlike the original paper and implementation, this convolution can handle edge feature of degree greater than 0.
+ Input edge features are concatenated with input source node features before the kernel is applied.
+ """
+
+ def __init__(
+ self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Fiber,
+ pool: bool = True,
+ use_layer_norm: bool = False,
+ self_interaction: bool = False,
+ max_degree: int = 4,
+ fuse_level: ConvSE3FuseLevel = ConvSE3FuseLevel.FULL,
+ allow_fused_output: bool = False
+ ):
+ """
+ :param fiber_in: Fiber describing the input features
+ :param fiber_out: Fiber describing the output features
+ :param fiber_edge: Fiber describing the edge features (node distances excluded)
+ :param pool: If True, compute final node features by averaging incoming edge features
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param self_interaction: Apply self-interaction of nodes
+ :param max_degree: Maximum degree used in the bases computation
+ :param fuse_level: Maximum fuse level to use in TFN convolutions
+ :param allow_fused_output: Allow the module to output a fused representation of features
+ """
+ super().__init__()
+ self.pool = pool
+ self.fiber_in = fiber_in
+ self.fiber_out = fiber_out
+ self.self_interaction = self_interaction
+ self.max_degree = max_degree
+ self.allow_fused_output = allow_fused_output
+
+ # channels_in: account for the concatenation of edge features
+ channels_in_set = set([f.channels + fiber_edge[f.degree] * (f.degree > 0) for f in self.fiber_in])
+ channels_out_set = set([f.channels for f in self.fiber_out])
+ unique_channels_in = (len(channels_in_set) == 1)
+ unique_channels_out = (len(channels_out_set) == 1)
+ degrees_up_to_max = list(range(max_degree + 1))
+ common_args = dict(edge_dim=fiber_edge[0] + 1, use_layer_norm=use_layer_norm)
+
+ if fuse_level.value >= ConvSE3FuseLevel.FULL.value and \
+ unique_channels_in and fiber_in.degrees == degrees_up_to_max and \
+ unique_channels_out and fiber_out.degrees == degrees_up_to_max:
+ # Single fused convolution
+ self.used_fuse_level = ConvSE3FuseLevel.FULL
+
+ sum_freq = sum([
+ degree_to_dim(min(d_in, d_out))
+ for d_in, d_out in product(degrees_up_to_max, degrees_up_to_max)
+ ])
+
+ self.conv = VersatileConvSE3(sum_freq, list(channels_in_set)[0], list(channels_out_set)[0],
+ fuse_level=self.used_fuse_level, **common_args)
+
+ elif fuse_level.value >= ConvSE3FuseLevel.PARTIAL.value and \
+ unique_channels_in and fiber_in.degrees == degrees_up_to_max:
+ # Convolutions fused per output degree
+ self.used_fuse_level = ConvSE3FuseLevel.PARTIAL
+ self.conv_out = nn.ModuleDict()
+ for d_out, c_out in fiber_out:
+ sum_freq = sum([degree_to_dim(min(d_out, d)) for d in fiber_in.degrees])
+ self.conv_out[str(d_out)] = VersatileConvSE3(sum_freq, list(channels_in_set)[0], c_out,
+ fuse_level=self.used_fuse_level, **common_args)
+
+ elif fuse_level.value >= ConvSE3FuseLevel.PARTIAL.value and \
+ unique_channels_out and fiber_out.degrees == degrees_up_to_max:
+ # Convolutions fused per input degree
+ self.used_fuse_level = ConvSE3FuseLevel.PARTIAL
+ self.conv_in = nn.ModuleDict()
+ for d_in, c_in in fiber_in:
+ sum_freq = sum([degree_to_dim(min(d_in, d)) for d in fiber_out.degrees])
+ self.conv_in[str(d_in)] = VersatileConvSE3(sum_freq, c_in, list(channels_out_set)[0],
+ fuse_level=ConvSE3FuseLevel.FULL, **common_args)
+ else:
+ # Use pairwise TFN convolutions
+ self.used_fuse_level = ConvSE3FuseLevel.NONE
+ self.conv = nn.ModuleDict()
+ for (degree_in, channels_in), (degree_out, channels_out) in (self.fiber_in * self.fiber_out):
+ dict_key = f'{degree_in},{degree_out}'
+ channels_in_new = channels_in + fiber_edge[degree_in] * (degree_in > 0)
+ sum_freq = degree_to_dim(min(degree_in, degree_out))
+ self.conv[dict_key] = VersatileConvSE3(sum_freq, channels_in_new, channels_out,
+ fuse_level=self.used_fuse_level, **common_args)
+
+ if self_interaction:
+ self.to_kernel_self = nn.ParameterDict()
+ for degree_out, channels_out in fiber_out:
+ if fiber_in[degree_out]:
+ self.to_kernel_self[str(degree_out)] = nn.Parameter(
+ torch.randn(channels_out, fiber_in[degree_out]) / np.sqrt(fiber_in[degree_out]))
+
+ def forward(
+ self,
+ node_feats: Dict[str, Tensor],
+ edge_feats: Dict[str, Tensor],
+ graph: DGLGraph,
+ basis: Dict[str, Tensor]
+ ):
+ with nvtx_range(f'ConvSE3'):
+ invariant_edge_feats = edge_feats['0'].squeeze(-1)
+ src, dst = graph.edges()
+ out = {}
+ in_features = []
+
+ # Fetch all input features from edge and node features
+ for degree_in in self.fiber_in.degrees:
+ src_node_features = node_feats[str(degree_in)][src]
+ if degree_in > 0 and str(degree_in) in edge_feats:
+ # Handle edge features of any type by concatenating them to node features
+ src_node_features = torch.cat([src_node_features, edge_feats[str(degree_in)]], dim=1)
+ in_features.append(src_node_features)
+
+ if self.used_fuse_level == ConvSE3FuseLevel.FULL:
+ in_features_fused = torch.cat(in_features, dim=-1)
+ out = self.conv(in_features_fused, invariant_edge_feats, basis['fully_fused'])
+
+ if not self.allow_fused_output or self.self_interaction or self.pool:
+ out = unfuse_features(out, self.fiber_out.degrees)
+
+ elif self.used_fuse_level == ConvSE3FuseLevel.PARTIAL and hasattr(self, 'conv_out'):
+ in_features_fused = torch.cat(in_features, dim=-1)
+ for degree_out in self.fiber_out.degrees:
+ out[str(degree_out)] = self.conv_out[str(degree_out)](in_features_fused, invariant_edge_feats,
+ basis[f'out{degree_out}_fused'])
+
+ elif self.used_fuse_level == ConvSE3FuseLevel.PARTIAL and hasattr(self, 'conv_in'):
+ out = 0
+ for degree_in, feature in zip(self.fiber_in.degrees, in_features):
+ out += self.conv_in[str(degree_in)](feature, invariant_edge_feats,
+ basis[f'in{degree_in}_fused'])
+ if not self.allow_fused_output or self.self_interaction or self.pool:
+ out = unfuse_features(out, self.fiber_out.degrees)
+ else:
+ # Fallback to pairwise TFN convolutions
+ for degree_out in self.fiber_out.degrees:
+ out_feature = 0
+ for degree_in, feature in zip(self.fiber_in.degrees, in_features):
+ dict_key = f'{degree_in},{degree_out}'
+ out_feature = out_feature + self.conv[dict_key](feature, invariant_edge_feats,
+ basis.get(dict_key, None))
+ out[str(degree_out)] = out_feature
+
+ for degree_out in self.fiber_out.degrees:
+ if self.self_interaction and str(degree_out) in self.to_kernel_self:
+ with nvtx_range(f'self interaction'):
+ dst_features = node_feats[str(degree_out)][dst]
+ kernel_self = self.to_kernel_self[str(degree_out)]
+ out[str(degree_out)] += kernel_self @ dst_features
+
+ if self.pool:
+ with nvtx_range(f'pooling'):
+ if isinstance(out, dict):
+ out[str(degree_out)] = dgl.ops.copy_e_sum(graph, out[str(degree_out)])
+ else:
+ out = dgl.ops.copy_e_sum(graph, out)
+ return out
diff --git a/model/se3_transformer/model/layers/linear.py b/model/se3_transformer/model/layers/linear.py
new file mode 100644
index 0000000000000000000000000000000000000000..f720d77ecc540423a6a6545f9e50c117ad1c08db
--- /dev/null
+++ b/model/se3_transformer/model/layers/linear.py
@@ -0,0 +1,59 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from typing import Dict
+
+import numpy as np
+import torch
+import torch.nn as nn
+from torch import Tensor
+
+from se3_transformer.model.fiber import Fiber
+
+
+class LinearSE3(nn.Module):
+ """
+ Graph Linear SE(3)-equivariant layer, equivalent to a 1x1 convolution.
+ Maps a fiber to a fiber with the same degrees (channels may be different).
+ No interaction between degrees, but interaction between channels.
+
+ type-0 features (C_0 channels) ────> Linear(bias=False) ────> type-0 features (C'_0 channels)
+ type-1 features (C_1 channels) ────> Linear(bias=False) ────> type-1 features (C'_1 channels)
+ :
+ type-k features (C_k channels) ────> Linear(bias=False) ────> type-k features (C'_k channels)
+ """
+
+ def __init__(self, fiber_in: Fiber, fiber_out: Fiber):
+ super().__init__()
+ self.weights = nn.ParameterDict({
+ str(degree_out): nn.Parameter(
+ torch.randn(channels_out, fiber_in[degree_out]) / np.sqrt(fiber_in[degree_out]))
+ for degree_out, channels_out in fiber_out
+ })
+
+ def forward(self, features: Dict[str, Tensor], *args, **kwargs) -> Dict[str, Tensor]:
+ return {
+ degree: self.weights[degree] @ features[degree]
+ for degree, weight in self.weights.items()
+ }
diff --git a/model/se3_transformer/model/layers/norm.py b/model/se3_transformer/model/layers/norm.py
new file mode 100644
index 0000000000000000000000000000000000000000..acbe23d774d50c26e22a60336a3d05b66a164eb3
--- /dev/null
+++ b/model/se3_transformer/model/layers/norm.py
@@ -0,0 +1,83 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from typing import Dict
+
+import torch
+import torch.nn as nn
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.model.fiber import Fiber
+
+
+class NormSE3(nn.Module):
+ """
+ Norm-based SE(3)-equivariant nonlinearity.
+
+ ┌──> feature_norm ──> LayerNorm() ──> ReLU() ──┐
+ feature_in ──┤ * ──> feature_out
+ └──> feature_phase ────────────────────────────┘
+ """
+
+ NORM_CLAMP = 2 ** -24 # Minimum positive subnormal for FP16
+
+ def __init__(self, fiber: Fiber, nonlinearity: nn.Module = nn.ReLU()):
+ super().__init__()
+ self.fiber = fiber
+ self.nonlinearity = nonlinearity
+
+ if len(set(fiber.channels)) == 1:
+ # Fuse all the layer normalizations into a group normalization
+ self.group_norm = nn.GroupNorm(num_groups=len(fiber.degrees), num_channels=sum(fiber.channels))
+ else:
+ # Use multiple layer normalizations
+ self.layer_norms = nn.ModuleDict({
+ str(degree): nn.LayerNorm(channels)
+ for degree, channels in fiber
+ })
+
+ def forward(self, features: Dict[str, Tensor], *args, **kwargs) -> Dict[str, Tensor]:
+ with nvtx_range('NormSE3'):
+ output = {}
+ if hasattr(self, 'group_norm'):
+ # Compute per-degree norms of features
+ norms = [features[str(d)].norm(dim=-1, keepdim=True).clamp(min=self.NORM_CLAMP)
+ for d in self.fiber.degrees]
+ fused_norms = torch.cat(norms, dim=-2)
+
+ # Transform the norms only
+ new_norms = self.nonlinearity(self.group_norm(fused_norms.squeeze(-1))).unsqueeze(-1)
+ new_norms = torch.chunk(new_norms, chunks=len(self.fiber.degrees), dim=-2)
+
+ # Scale features to the new norms
+ for norm, new_norm, d in zip(norms, new_norms, self.fiber.degrees):
+ output[str(d)] = features[str(d)] / norm * new_norm
+ else:
+ for degree, feat in features.items():
+ norm = feat.norm(dim=-1, keepdim=True).clamp(min=self.NORM_CLAMP)
+ new_norm = self.nonlinearity(self.layer_norms[degree](norm.squeeze(-1)).unsqueeze(-1))
+ output[degree] = new_norm * feat / norm
+
+ return output
diff --git a/model/se3_transformer/model/layers/pooling.py b/model/se3_transformer/model/layers/pooling.py
new file mode 100644
index 0000000000000000000000000000000000000000..e42c5383ba3239e3d93c928fa83a61a9e19b9437
--- /dev/null
+++ b/model/se3_transformer/model/layers/pooling.py
@@ -0,0 +1,53 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from typing import Dict, Literal
+
+import torch.nn as nn
+from dgl import DGLGraph
+from dgl.nn.pytorch import AvgPooling, MaxPooling
+from torch import Tensor
+
+
+class GPooling(nn.Module):
+ """
+ Graph max/average pooling on a given feature type.
+ The average can be taken for any feature type, and equivariance will be maintained.
+ The maximum can only be taken for invariant features (type 0).
+ If you want max-pooling for type > 0 features, look into Vector Neurons.
+ """
+
+ def __init__(self, feat_type: int = 0, pool: Literal['max', 'avg'] = 'max'):
+ """
+ :param feat_type: Feature type to pool
+ :param pool: Type of pooling: max or avg
+ """
+ super().__init__()
+ assert pool in ['max', 'avg'], f'Unknown pooling: {pool}'
+ assert feat_type == 0 or pool == 'avg', 'Max pooling on type > 0 features will break equivariance'
+ self.feat_type = feat_type
+ self.pool = MaxPooling() if pool == 'max' else AvgPooling()
+
+ def forward(self, features: Dict[str, Tensor], graph: DGLGraph, **kwargs) -> Tensor:
+ pooled = self.pool(graph, features[str(self.feat_type)])
+ return pooled.squeeze(dim=-1)
diff --git a/model/se3_transformer/model/transformer.py b/model/se3_transformer/model/transformer.py
new file mode 100644
index 0000000000000000000000000000000000000000..ab89f77a12b8b40b16c402647059260606c98c60
--- /dev/null
+++ b/model/se3_transformer/model/transformer.py
@@ -0,0 +1,222 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+from typing import Optional, Literal, Dict
+
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from torch import Tensor
+
+from se3_transformer.model.basis import get_basis, update_basis_with_fused
+from se3_transformer.model.layers.attention import AttentionBlockSE3
+from se3_transformer.model.layers.convolution import ConvSE3, ConvSE3FuseLevel
+from se3_transformer.model.layers.norm import NormSE3
+from se3_transformer.model.layers.pooling import GPooling
+from se3_transformer.runtime.utils import str2bool
+from se3_transformer.model.fiber import Fiber
+
+
+class Sequential(nn.Sequential):
+ """ Sequential module with arbitrary forward args and kwargs. Used to pass graph, basis and edge features. """
+
+ def forward(self, input, *args, **kwargs):
+ for module in self:
+ input = module(input, *args, **kwargs)
+ return input
+
+
+def get_populated_edge_features(relative_pos: Tensor, edge_features: Optional[Dict[str, Tensor]] = None):
+ """ Add relative positions to existing edge features """
+ edge_features = edge_features.copy() if edge_features else {}
+ r = relative_pos.norm(dim=-1, keepdim=True)
+ if '0' in edge_features:
+ edge_features['0'] = torch.cat([edge_features['0'], r[..., None]], dim=1)
+ else:
+ edge_features['0'] = r[..., None]
+
+ return edge_features
+
+
+class SE3Transformer(nn.Module):
+ def __init__(self,
+ num_layers: int,
+ fiber_in: Fiber,
+ fiber_hidden: Fiber,
+ fiber_out: Fiber,
+ num_heads: int,
+ channels_div: int,
+ fiber_edge: Fiber = Fiber({}),
+ return_type: Optional[int] = None,
+ pooling: Optional[Literal['avg', 'max']] = None,
+ norm: bool = True,
+ use_layer_norm: bool = True,
+ tensor_cores: bool = False,
+ low_memory: bool = False,
+ **kwargs):
+ """
+ :param num_layers: Number of attention layers
+ :param fiber_in: Input fiber description
+ :param fiber_hidden: Hidden fiber description
+ :param fiber_out: Output fiber description
+ :param fiber_edge: Input edge fiber description
+ :param num_heads: Number of attention heads
+ :param channels_div: Channels division before feeding to attention layer
+ :param return_type: Return only features of this type
+ :param pooling: 'avg' or 'max' graph pooling before MLP layers
+ :param norm: Apply a normalization layer after each attention block
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param tensor_cores: True if using Tensor Cores (affects the use of fully fused convs, and padded bases)
+ :param low_memory: If True, will use slower ops that use less memory
+ """
+ super().__init__()
+ self.num_layers = num_layers
+ self.fiber_edge = fiber_edge
+ self.num_heads = num_heads
+ self.channels_div = channels_div
+ self.return_type = return_type
+ self.pooling = pooling
+ self.max_degree = max(*fiber_in.degrees, *fiber_hidden.degrees, *fiber_out.degrees)
+ self.tensor_cores = tensor_cores
+ self.low_memory = low_memory
+
+ if low_memory and not tensor_cores:
+ logging.warning('Low memory mode will have no effect with no Tensor Cores')
+
+ # Fully fused convolutions when using Tensor Cores (and not low memory mode)
+ fuse_level = ConvSE3FuseLevel.FULL if tensor_cores and not low_memory else ConvSE3FuseLevel.PARTIAL
+
+ graph_modules = []
+ for i in range(num_layers):
+ graph_modules.append(AttentionBlockSE3(fiber_in=fiber_in,
+ fiber_out=fiber_hidden,
+ fiber_edge=fiber_edge,
+ num_heads=num_heads,
+ channels_div=channels_div,
+ use_layer_norm=use_layer_norm,
+ max_degree=self.max_degree,
+ fuse_level=fuse_level))
+ if norm:
+ graph_modules.append(NormSE3(fiber_hidden))
+ fiber_in = fiber_hidden
+
+ graph_modules.append(ConvSE3(fiber_in=fiber_in,
+ fiber_out=fiber_out,
+ fiber_edge=fiber_edge,
+ self_interaction=True,
+ use_layer_norm=use_layer_norm,
+ max_degree=self.max_degree))
+ self.graph_modules = Sequential(*graph_modules)
+
+ if pooling is not None:
+ assert return_type is not None, 'return_type must be specified when pooling'
+ self.pooling_module = GPooling(pool=pooling, feat_type=return_type)
+
+ def forward(self, graph: DGLGraph, node_feats: Dict[str, Tensor],
+ edge_feats: Optional[Dict[str, Tensor]] = None,
+ basis: Optional[Dict[str, Tensor]] = None):
+ # Compute bases in case they weren't precomputed as part of the data loading
+ basis = basis or get_basis(graph.edata['rel_pos'], max_degree=self.max_degree, compute_gradients=False,
+ use_pad_trick=self.tensor_cores and not self.low_memory,
+ amp=torch.is_autocast_enabled())
+
+ # Add fused bases (per output degree, per input degree, and fully fused) to the dict
+ basis = update_basis_with_fused(basis, self.max_degree, use_pad_trick=self.tensor_cores and not self.low_memory,
+ fully_fused=self.tensor_cores and not self.low_memory)
+
+ edge_feats = get_populated_edge_features(graph.edata['rel_pos'], edge_feats)
+
+ node_feats = self.graph_modules(node_feats, edge_feats, graph=graph, basis=basis)
+
+ if self.pooling is not None:
+ return self.pooling_module(node_feats, graph=graph)
+
+ if self.return_type is not None:
+ return node_feats[str(self.return_type)]
+
+ return node_feats
+
+ @staticmethod
+ def add_argparse_args(parser):
+ parser.add_argument('--num_layers', type=int, default=7,
+ help='Number of stacked Transformer layers')
+ parser.add_argument('--num_heads', type=int, default=8,
+ help='Number of heads in self-attention')
+ parser.add_argument('--channels_div', type=int, default=2,
+ help='Channels division before feeding to attention layer')
+ parser.add_argument('--pooling', type=str, default=None, const=None, nargs='?', choices=['max', 'avg'],
+ help='Type of graph pooling')
+ parser.add_argument('--norm', type=str2bool, nargs='?', const=True, default=False,
+ help='Apply a normalization layer after each attention block')
+ parser.add_argument('--use_layer_norm', type=str2bool, nargs='?', const=True, default=False,
+ help='Apply layer normalization between MLP layers')
+ parser.add_argument('--low_memory', type=str2bool, nargs='?', const=True, default=False,
+ help='If true, will use fused ops that are slower but that use less memory '
+ '(expect 25 percent less memory). '
+ 'Only has an effect if AMP is enabled on Volta GPUs, or if running on Ampere GPUs')
+
+ return parser
+
+
+class SE3TransformerPooled(nn.Module):
+ def __init__(self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Fiber,
+ num_degrees: int,
+ num_channels: int,
+ output_dim: int,
+ **kwargs):
+ super().__init__()
+ kwargs['pooling'] = kwargs['pooling'] or 'max'
+ self.transformer = SE3Transformer(
+ fiber_in=fiber_in,
+ fiber_hidden=Fiber.create(num_degrees, num_channels),
+ fiber_out=fiber_out,
+ fiber_edge=fiber_edge,
+ return_type=0,
+ **kwargs
+ )
+
+ n_out_features = fiber_out.num_features
+ self.mlp = nn.Sequential(
+ nn.Linear(n_out_features, n_out_features),
+ nn.ReLU(),
+ nn.Linear(n_out_features, output_dim)
+ )
+
+ def forward(self, graph, node_feats, edge_feats, basis=None):
+ feats = self.transformer(graph, node_feats, edge_feats, basis).squeeze(-1)
+ y = self.mlp(feats).squeeze(-1)
+ return y
+
+ @staticmethod
+ def add_argparse_args(parent_parser):
+ parser = parent_parser.add_argument_group("Model architecture")
+ SE3Transformer.add_argparse_args(parser)
+ parser.add_argument('--num_degrees',
+ help='Number of degrees to use. Hidden features will have types [0, ..., num_degrees - 1]',
+ type=int, default=4)
+ parser.add_argument('--num_channels', help='Number of channels for the hidden features', type=int, default=32)
+ return parent_parser
diff --git a/model/se3_transformer/runtime/__init__.py b/model/se3_transformer/runtime/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..e69de29bb2d1d6434b8b29ae775ad8c2e48c5391
diff --git a/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc b/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..2df9df1f20c3e458717d3abe34924e313d57d1b3
Binary files /dev/null and b/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc differ
diff --git a/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc b/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..c77046360b569d12450ee583b5b0338545860ba1
Binary files /dev/null and b/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc differ
diff --git a/model/se3_transformer/runtime/arguments.py b/model/se3_transformer/runtime/arguments.py
new file mode 100644
index 0000000000000000000000000000000000000000..d35d5ee1d05a150ea40c411ec47c633213ca9d9f
--- /dev/null
+++ b/model/se3_transformer/runtime/arguments.py
@@ -0,0 +1,70 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import argparse
+import pathlib
+
+from se3_transformer.data_loading import QM9DataModule
+from se3_transformer.model import SE3TransformerPooled
+from se3_transformer.runtime.utils import str2bool
+
+PARSER = argparse.ArgumentParser(description='SE(3)-Transformer')
+
+paths = PARSER.add_argument_group('Paths')
+paths.add_argument('--data_dir', type=pathlib.Path, default=pathlib.Path('./data'),
+ help='Directory where the data is located or should be downloaded')
+paths.add_argument('--log_dir', type=pathlib.Path, default=pathlib.Path('/results'),
+ help='Directory where the results logs should be saved')
+paths.add_argument('--dllogger_name', type=str, default='dllogger_results.json',
+ help='Name for the resulting DLLogger JSON file')
+paths.add_argument('--save_ckpt_path', type=pathlib.Path, default=None,
+ help='File where the checkpoint should be saved')
+paths.add_argument('--load_ckpt_path', type=pathlib.Path, default=None,
+ help='File of the checkpoint to be loaded')
+
+optimizer = PARSER.add_argument_group('Optimizer')
+optimizer.add_argument('--optimizer', choices=['adam', 'sgd', 'lamb'], default='adam')
+optimizer.add_argument('--learning_rate', '--lr', dest='learning_rate', type=float, default=0.002)
+optimizer.add_argument('--min_learning_rate', '--min_lr', dest='min_learning_rate', type=float, default=None)
+optimizer.add_argument('--momentum', type=float, default=0.9)
+optimizer.add_argument('--weight_decay', type=float, default=0.1)
+
+PARSER.add_argument('--epochs', type=int, default=100, help='Number of training epochs')
+PARSER.add_argument('--batch_size', type=int, default=240, help='Batch size')
+PARSER.add_argument('--seed', type=int, default=None, help='Set a seed globally')
+PARSER.add_argument('--num_workers', type=int, default=8, help='Number of dataloading workers')
+
+PARSER.add_argument('--amp', type=str2bool, nargs='?', const=True, default=False, help='Use Automatic Mixed Precision')
+PARSER.add_argument('--gradient_clip', type=float, default=None, help='Clipping of the gradient norms')
+PARSER.add_argument('--accumulate_grad_batches', type=int, default=1, help='Gradient accumulation')
+PARSER.add_argument('--ckpt_interval', type=int, default=-1, help='Save a checkpoint every N epochs')
+PARSER.add_argument('--eval_interval', dest='eval_interval', type=int, default=1,
+ help='Do an evaluation round every N epochs')
+PARSER.add_argument('--silent', type=str2bool, nargs='?', const=True, default=False,
+ help='Minimize stdout output')
+
+PARSER.add_argument('--benchmark', type=str2bool, nargs='?', const=True, default=False,
+ help='Benchmark mode')
+
+QM9DataModule.add_argparse_args(PARSER)
+SE3TransformerPooled.add_argparse_args(PARSER)
diff --git a/model/se3_transformer/runtime/callbacks.py b/model/se3_transformer/runtime/callbacks.py
new file mode 100644
index 0000000000000000000000000000000000000000..b49d670c8305b3c4dea53ebf5c428cac628f2cf7
--- /dev/null
+++ b/model/se3_transformer/runtime/callbacks.py
@@ -0,0 +1,160 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+import time
+from abc import ABC, abstractmethod
+from typing import Optional
+
+import numpy as np
+import torch
+
+from se3_transformer.runtime.loggers import Logger
+from se3_transformer.runtime.metrics import MeanAbsoluteError
+
+
+class BaseCallback(ABC):
+ def on_fit_start(self, optimizer, args):
+ pass
+
+ def on_fit_end(self):
+ pass
+
+ def on_epoch_end(self):
+ pass
+
+ def on_batch_start(self):
+ pass
+
+ def on_validation_step(self, input, target, pred):
+ pass
+
+ def on_validation_end(self, epoch=None):
+ pass
+
+ def on_checkpoint_load(self, checkpoint):
+ pass
+
+ def on_checkpoint_save(self, checkpoint):
+ pass
+
+
+class LRSchedulerCallback(BaseCallback):
+ def __init__(self, logger: Optional[Logger] = None):
+ self.logger = logger
+ self.scheduler = None
+
+ @abstractmethod
+ def get_scheduler(self, optimizer, args):
+ pass
+
+ def on_fit_start(self, optimizer, args):
+ self.scheduler = self.get_scheduler(optimizer, args)
+
+ def on_checkpoint_load(self, checkpoint):
+ self.scheduler.load_state_dict(checkpoint['scheduler_state_dict'])
+
+ def on_checkpoint_save(self, checkpoint):
+ checkpoint['scheduler_state_dict'] = self.scheduler.state_dict()
+
+ def on_epoch_end(self):
+ if self.logger is not None:
+ self.logger.log_metrics({'learning rate': self.scheduler.get_last_lr()[0]}, step=self.scheduler.last_epoch)
+ self.scheduler.step()
+
+
+class QM9MetricCallback(BaseCallback):
+ """ Logs the rescaled mean absolute error for QM9 regression tasks """
+
+ def __init__(self, logger, targets_std, prefix=''):
+ self.mae = MeanAbsoluteError()
+ self.logger = logger
+ self.targets_std = targets_std
+ self.prefix = prefix
+ self.best_mae = float('inf')
+
+ def on_validation_step(self, input, target, pred):
+ self.mae(pred.detach(), target.detach())
+
+ def on_validation_end(self, epoch=None):
+ mae = self.mae.compute() * self.targets_std
+ logging.info(f'{self.prefix} MAE: {mae}')
+ self.logger.log_metrics({f'{self.prefix} MAE': mae}, epoch)
+ self.best_mae = min(self.best_mae, mae)
+
+ def on_fit_end(self):
+ if self.best_mae != float('inf'):
+ self.logger.log_metrics({f'{self.prefix} best MAE': self.best_mae})
+
+
+class QM9LRSchedulerCallback(LRSchedulerCallback):
+ def __init__(self, logger, epochs):
+ super().__init__(logger)
+ self.epochs = epochs
+
+ def get_scheduler(self, optimizer, args):
+ min_lr = args.min_learning_rate if args.min_learning_rate else args.learning_rate / 10.0
+ return torch.optim.lr_scheduler.CosineAnnealingWarmRestarts(optimizer, self.epochs, eta_min=min_lr)
+
+
+class PerformanceCallback(BaseCallback):
+ def __init__(self, logger, batch_size: int, warmup_epochs: int = 1, mode: str = 'train'):
+ self.batch_size = batch_size
+ self.warmup_epochs = warmup_epochs
+ self.epoch = 0
+ self.timestamps = []
+ self.mode = mode
+ self.logger = logger
+
+ def on_batch_start(self):
+ if self.epoch >= self.warmup_epochs:
+ self.timestamps.append(time.time() * 1000.0)
+
+ def _log_perf(self):
+ stats = self.process_performance_stats()
+ for k, v in stats.items():
+ logging.info(f'performance {k}: {v}')
+
+ self.logger.log_metrics(stats)
+
+ def on_epoch_end(self):
+ self.epoch += 1
+
+ def on_fit_end(self):
+ if self.epoch > self.warmup_epochs:
+ self._log_perf()
+ self.timestamps = []
+
+ def process_performance_stats(self):
+ timestamps = np.asarray(self.timestamps)
+ deltas = np.diff(timestamps)
+ throughput = (self.batch_size / deltas).mean()
+ stats = {
+ f"throughput_{self.mode}": throughput,
+ f"latency_{self.mode}_mean": deltas.mean(),
+ f"total_time_{self.mode}": timestamps[-1] - timestamps[0],
+ }
+ for level in [90, 95, 99]:
+ stats.update({f"latency_{self.mode}_{level}": np.percentile(deltas, level)})
+
+ return stats
diff --git a/model/se3_transformer/runtime/gpu_affinity.py b/model/se3_transformer/runtime/gpu_affinity.py
new file mode 100644
index 0000000000000000000000000000000000000000..9d46215bbbcb04f87e4c8250e08b575f8e529693
--- /dev/null
+++ b/model/se3_transformer/runtime/gpu_affinity.py
@@ -0,0 +1,325 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import collections
+import itertools
+import math
+import os
+import pathlib
+import re
+
+import pynvml
+
+
+class Device:
+ # assumes nvml returns list of 64 bit ints
+ _nvml_affinity_elements = math.ceil(os.cpu_count() / 64)
+
+ def __init__(self, device_idx):
+ super().__init__()
+ self.handle = pynvml.nvmlDeviceGetHandleByIndex(device_idx)
+
+ def get_name(self):
+ return pynvml.nvmlDeviceGetName(self.handle)
+
+ def get_uuid(self):
+ return pynvml.nvmlDeviceGetUUID(self.handle)
+
+ def get_cpu_affinity(self):
+ affinity_string = ""
+ for j in pynvml.nvmlDeviceGetCpuAffinity(self.handle, Device._nvml_affinity_elements):
+ # assume nvml returns list of 64 bit ints
+ affinity_string = "{:064b}".format(j) + affinity_string
+
+ affinity_list = [int(x) for x in affinity_string]
+ affinity_list.reverse() # so core 0 is in 0th element of list
+
+ ret = [i for i, e in enumerate(affinity_list) if e != 0]
+ return ret
+
+
+def get_thread_siblings_list():
+ """
+ Returns a list of 2-element integer tuples representing pairs of
+ hyperthreading cores.
+ """
+ path = "/sys/devices/system/cpu/cpu*/topology/thread_siblings_list"
+ thread_siblings_list = []
+ pattern = re.compile(r"(\d+)\D(\d+)")
+ for fname in pathlib.Path(path[0]).glob(path[1:]):
+ with open(fname) as f:
+ content = f.read().strip()
+ res = pattern.findall(content)
+ if res:
+ pair = tuple(map(int, res[0]))
+ thread_siblings_list.append(pair)
+ return thread_siblings_list
+
+
+def check_socket_affinities(socket_affinities):
+ # sets of cores should be either identical or disjoint
+ for i, j in itertools.product(socket_affinities, socket_affinities):
+ if not set(i) == set(j) and not set(i).isdisjoint(set(j)):
+ raise RuntimeError(f"Sets of cores should be either identical or disjoint, " f"but got {i} and {j}.")
+
+
+def get_socket_affinities(nproc_per_node, exclude_unavailable_cores=True):
+ devices = [Device(i) for i in range(nproc_per_node)]
+ socket_affinities = [dev.get_cpu_affinity() for dev in devices]
+
+ if exclude_unavailable_cores:
+ available_cores = os.sched_getaffinity(0)
+ socket_affinities = [list(set(affinity) & available_cores) for affinity in socket_affinities]
+
+ check_socket_affinities(socket_affinities)
+
+ return socket_affinities
+
+
+def set_socket_affinity(gpu_id):
+ """
+ The process is assigned with all available logical CPU cores from the CPU
+ socket connected to the GPU with a given id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ dev = Device(gpu_id)
+ affinity = dev.get_cpu_affinity()
+ os.sched_setaffinity(0, affinity)
+
+
+def set_single_affinity(gpu_id):
+ """
+ The process is assigned with the first available logical CPU core from the
+ list of all CPU cores from the CPU socket connected to the GPU with a given
+ id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ dev = Device(gpu_id)
+ affinity = dev.get_cpu_affinity()
+
+ # exclude unavailable cores
+ available_cores = os.sched_getaffinity(0)
+ affinity = list(set(affinity) & available_cores)
+ os.sched_setaffinity(0, affinity[:1])
+
+
+def set_single_unique_affinity(gpu_id, nproc_per_node):
+ """
+ The process is assigned with a single unique available physical CPU core
+ from the list of all CPU cores from the CPU socket connected to the GPU with
+ a given id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ socket_affinities = get_socket_affinities(nproc_per_node)
+
+ siblings_list = get_thread_siblings_list()
+ siblings_dict = dict(siblings_list)
+
+ # remove siblings
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities[idx] = list(set(socket_affinity) - set(siblings_dict.values()))
+
+ affinities = []
+ assigned = []
+
+ for socket_affinity in socket_affinities:
+ for core in socket_affinity:
+ if core not in assigned:
+ affinities.append([core])
+ assigned.append(core)
+ break
+ os.sched_setaffinity(0, affinities[gpu_id])
+
+
+def set_socket_unique_affinity(gpu_id, nproc_per_node, mode, balanced=True):
+ """
+ The process is assigned with an unique subset of available physical CPU
+ cores from the CPU socket connected to a GPU with a given id.
+ Assignment automatically includes hyperthreading siblings (if siblings are
+ available).
+
+ Args:
+ gpu_id: index of a GPU
+ nproc_per_node: total number of processes per node
+ mode: mode
+ balanced: assign an equal number of physical cores to each process
+ """
+ socket_affinities = get_socket_affinities(nproc_per_node)
+
+ siblings_list = get_thread_siblings_list()
+ siblings_dict = dict(siblings_list)
+
+ # remove hyperthreading siblings
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities[idx] = list(set(socket_affinity) - set(siblings_dict.values()))
+
+ socket_affinities_to_device_ids = collections.defaultdict(list)
+
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities_to_device_ids[tuple(socket_affinity)].append(idx)
+
+ # compute minimal number of physical cores per GPU across all GPUs and
+ # sockets, code assigns this number of cores per GPU if balanced == True
+ min_physical_cores_per_gpu = min(
+ [len(cores) // len(gpus) for cores, gpus in socket_affinities_to_device_ids.items()]
+ )
+
+ for socket_affinity, device_ids in socket_affinities_to_device_ids.items():
+ devices_per_group = len(device_ids)
+ if balanced:
+ cores_per_device = min_physical_cores_per_gpu
+ socket_affinity = socket_affinity[: devices_per_group * min_physical_cores_per_gpu]
+ else:
+ cores_per_device = len(socket_affinity) // devices_per_group
+
+ for group_id, device_id in enumerate(device_ids):
+ if device_id == gpu_id:
+
+ # In theory there should be no difference in performance between
+ # 'interleaved' and 'continuous' pattern on Intel-based DGX-1,
+ # but 'continuous' should be better for DGX A100 because on AMD
+ # Rome 4 consecutive cores are sharing L3 cache.
+ # TODO: code doesn't attempt to automatically detect layout of
+ # L3 cache, also external environment may already exclude some
+ # cores, this code makes no attempt to detect it and to align
+ # mapping to multiples of 4.
+
+ if mode == "interleaved":
+ affinity = list(socket_affinity[group_id::devices_per_group])
+ elif mode == "continuous":
+ affinity = list(socket_affinity[group_id * cores_per_device: (group_id + 1) * cores_per_device])
+ else:
+ raise RuntimeError("Unknown set_socket_unique_affinity mode")
+
+ # unconditionally reintroduce hyperthreading siblings, this step
+ # may result in a different numbers of logical cores assigned to
+ # each GPU even if balanced == True (if hyperthreading siblings
+ # aren't available for a subset of cores due to some external
+ # constraints, siblings are re-added unconditionally, in the
+ # worst case unavailable logical core will be ignored by
+ # os.sched_setaffinity().
+ affinity += [siblings_dict[aff] for aff in affinity if aff in siblings_dict]
+ os.sched_setaffinity(0, affinity)
+
+
+def set_affinity(gpu_id, nproc_per_node, mode="socket_unique_continuous", balanced=True):
+ """
+ The process is assigned with a proper CPU affinity which matches hardware
+ architecture on a given platform. Usually it improves and stabilizes
+ performance of deep learning training workloads.
+
+ This function assumes that the workload is running in multi-process
+ single-device mode (there are multiple training processes and each process
+ is running on a single GPU), which is typical for multi-GPU training
+ workloads using `torch.nn.parallel.DistributedDataParallel`.
+
+ Available affinity modes:
+ * 'socket' - the process is assigned with all available logical CPU cores
+ from the CPU socket connected to the GPU with a given id.
+ * 'single' - the process is assigned with the first available logical CPU
+ core from the list of all CPU cores from the CPU socket connected to the GPU
+ with a given id (multiple GPUs could be assigned with the same CPU core).
+ * 'single_unique' - the process is assigned with a single unique available
+ physical CPU core from the list of all CPU cores from the CPU socket
+ connected to the GPU with a given id.
+ * 'socket_unique_interleaved' - the process is assigned with an unique
+ subset of available physical CPU cores from the CPU socket connected to a
+ GPU with a given id, hyperthreading siblings are included automatically,
+ cores are assigned with interleaved indexing pattern
+ * 'socket_unique_continuous' - (the default) the process is assigned with an
+ unique subset of available physical CPU cores from the CPU socket connected
+ to a GPU with a given id, hyperthreading siblings are included
+ automatically, cores are assigned with continuous indexing pattern
+
+ 'socket_unique_continuous' is the recommended mode for deep learning
+ training workloads on NVIDIA DGX machines.
+
+ Args:
+ gpu_id: integer index of a GPU
+ nproc_per_node: number of processes per node
+ mode: affinity mode
+ balanced: assign an equal number of physical cores to each process,
+ affects only 'socket_unique_interleaved' and
+ 'socket_unique_continuous' affinity modes
+
+ Returns a set of logical CPU cores on which the process is eligible to run.
+
+ Example:
+
+ import argparse
+ import os
+
+ import gpu_affinity
+ import torch
+
+
+ def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument(
+ '--local_rank',
+ type=int,
+ default=os.getenv('LOCAL_RANK', 0),
+ )
+ args = parser.parse_args()
+
+ nproc_per_node = torch.cuda.device_count()
+
+ affinity = gpu_affinity.set_affinity(args.local_rank, nproc_per_node)
+ print(f'{args.local_rank}: core affinity: {affinity}')
+
+
+ if __name__ == "__main__":
+ main()
+
+ Launch the example with:
+ python -m torch.distributed.launch --nproc_per_node <#GPUs> example.py
+
+
+ WARNING: On DGX A100 only a half of CPU cores have direct access to GPUs.
+ This function restricts execution only to the CPU cores directly connected
+ to GPUs, so on DGX A100 it will limit the code to half of CPU cores and half
+ of CPU memory bandwidth (which may be fine for many DL models).
+ """
+ pynvml.nvmlInit()
+
+ if mode == "socket":
+ set_socket_affinity(gpu_id)
+ elif mode == "single":
+ set_single_affinity(gpu_id)
+ elif mode == "single_unique":
+ set_single_unique_affinity(gpu_id, nproc_per_node)
+ elif mode == "socket_unique_interleaved":
+ set_socket_unique_affinity(gpu_id, nproc_per_node, "interleaved", balanced)
+ elif mode == "socket_unique_continuous":
+ set_socket_unique_affinity(gpu_id, nproc_per_node, "continuous", balanced)
+ else:
+ raise RuntimeError("Unknown affinity mode")
+
+ affinity = os.sched_getaffinity(0)
+ return affinity
diff --git a/model/se3_transformer/runtime/inference.py b/model/se3_transformer/runtime/inference.py
new file mode 100644
index 0000000000000000000000000000000000000000..21e9125b24f9865b89cff29063ad997e77297d21
--- /dev/null
+++ b/model/se3_transformer/runtime/inference.py
@@ -0,0 +1,131 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from typing import List
+
+import torch
+import torch.nn as nn
+from torch.nn.parallel import DistributedDataParallel
+from torch.utils.data import DataLoader
+from tqdm import tqdm
+
+from se3_transformer.runtime import gpu_affinity
+from se3_transformer.runtime.arguments import PARSER
+from se3_transformer.runtime.callbacks import BaseCallback
+from se3_transformer.runtime.loggers import DLLogger
+from se3_transformer.runtime.utils import to_cuda, get_local_rank
+
+
+@torch.inference_mode()
+def evaluate(model: nn.Module,
+ dataloader: DataLoader,
+ callbacks: List[BaseCallback],
+ args):
+ model.eval()
+ for i, batch in tqdm(enumerate(dataloader), total=len(dataloader), unit='batch', desc=f'Evaluation',
+ leave=False, disable=(args.silent or get_local_rank() != 0)):
+ *input, target = to_cuda(batch)
+
+ for callback in callbacks:
+ callback.on_batch_start()
+
+ with torch.cuda.amp.autocast(enabled=args.amp):
+ pred = model(*input)
+
+ for callback in callbacks:
+ callback.on_validation_step(input, target, pred)
+
+
+if __name__ == '__main__':
+ from se3_transformer.runtime.callbacks import QM9MetricCallback, PerformanceCallback
+ from se3_transformer.runtime.utils import init_distributed, seed_everything
+ from se3_transformer.model import SE3TransformerPooled, Fiber
+ from se3_transformer.data_loading import QM9DataModule
+ import torch.distributed as dist
+ import logging
+ import sys
+
+ is_distributed = init_distributed()
+ local_rank = get_local_rank()
+ args = PARSER.parse_args()
+
+ logging.getLogger().setLevel(logging.CRITICAL if local_rank != 0 or args.silent else logging.INFO)
+
+ logging.info('====== SE(3)-Transformer ======')
+ logging.info('| Inference on the test set |')
+ logging.info('===============================')
+
+ if not args.benchmark and args.load_ckpt_path is None:
+ logging.error('No load_ckpt_path provided, you need to provide a saved model to evaluate')
+ sys.exit(1)
+
+ if args.benchmark:
+ logging.info('Running benchmark mode with one warmup pass')
+
+ if args.seed is not None:
+ seed_everything(args.seed)
+
+ major_cc, minor_cc = torch.cuda.get_device_capability()
+
+ logger = DLLogger(args.log_dir, filename=args.dllogger_name)
+ datamodule = QM9DataModule(**vars(args))
+ model = SE3TransformerPooled(
+ fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}),
+ fiber_out=Fiber({0: args.num_degrees * args.num_channels}),
+ fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}),
+ output_dim=1,
+ tensor_cores=(args.amp and major_cc >= 7) or major_cc >= 8, # use Tensor Cores more effectively
+ **vars(args)
+ )
+ callbacks = [QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix='test')]
+
+ model.to(device=torch.cuda.current_device())
+ if args.load_ckpt_path is not None:
+ checkpoint = torch.load(str(args.load_ckpt_path), map_location={'cuda:0': f'cuda:{local_rank}'})
+ model.load_state_dict(checkpoint['state_dict'])
+
+ if is_distributed:
+ nproc_per_node = torch.cuda.device_count()
+ affinity = gpu_affinity.set_affinity(local_rank, nproc_per_node)
+ model = DistributedDataParallel(model, device_ids=[local_rank], output_device=local_rank)
+
+ test_dataloader = datamodule.test_dataloader() if not args.benchmark else datamodule.train_dataloader()
+ evaluate(model,
+ test_dataloader,
+ callbacks,
+ args)
+
+ for callback in callbacks:
+ callback.on_validation_end()
+
+ if args.benchmark:
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+ callbacks = [PerformanceCallback(logger, args.batch_size * world_size, warmup_epochs=1, mode='inference')]
+ for _ in range(6):
+ evaluate(model,
+ test_dataloader,
+ callbacks,
+ args)
+ callbacks[0].on_epoch_end()
+
+ callbacks[0].on_fit_end()
diff --git a/model/se3_transformer/runtime/loggers.py b/model/se3_transformer/runtime/loggers.py
new file mode 100644
index 0000000000000000000000000000000000000000..591486d8814c5dff43b8652823900e05add06c83
--- /dev/null
+++ b/model/se3_transformer/runtime/loggers.py
@@ -0,0 +1,134 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import pathlib
+from abc import ABC, abstractmethod
+from enum import Enum
+from typing import Dict, Any, Callable, Optional
+
+import dllogger
+import torch.distributed as dist
+import wandb
+from dllogger import Verbosity
+
+from se3_transformer.runtime.utils import rank_zero_only
+
+
+class Logger(ABC):
+ @rank_zero_only
+ @abstractmethod
+ def log_hyperparams(self, params):
+ pass
+
+ @rank_zero_only
+ @abstractmethod
+ def log_metrics(self, metrics, step=None):
+ pass
+
+ @staticmethod
+ def _sanitize_params(params):
+ def _sanitize(val):
+ if isinstance(val, Callable):
+ try:
+ _val = val()
+ if isinstance(_val, Callable):
+ return val.__name__
+ return _val
+ except Exception:
+ return getattr(val, "__name__", None)
+ elif isinstance(val, pathlib.Path) or isinstance(val, Enum):
+ return str(val)
+ return val
+
+ return {key: _sanitize(val) for key, val in params.items()}
+
+
+class LoggerCollection(Logger):
+ def __init__(self, loggers):
+ super().__init__()
+ self.loggers = loggers
+
+ def __getitem__(self, index):
+ return [logger for logger in self.loggers][index]
+
+ @rank_zero_only
+ def log_metrics(self, metrics, step=None):
+ for logger in self.loggers:
+ logger.log_metrics(metrics, step)
+
+ @rank_zero_only
+ def log_hyperparams(self, params):
+ for logger in self.loggers:
+ logger.log_hyperparams(params)
+
+
+class DLLogger(Logger):
+ def __init__(self, save_dir: pathlib.Path, filename: str):
+ super().__init__()
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ save_dir.mkdir(parents=True, exist_ok=True)
+ dllogger.init(
+ backends=[dllogger.JSONStreamBackend(Verbosity.DEFAULT, str(save_dir / filename))])
+
+ @rank_zero_only
+ def log_hyperparams(self, params):
+ params = self._sanitize_params(params)
+ dllogger.log(step="PARAMETER", data=params)
+
+ @rank_zero_only
+ def log_metrics(self, metrics, step=None):
+ if step is None:
+ step = tuple()
+
+ dllogger.log(step=step, data=metrics)
+
+
+class WandbLogger(Logger):
+ def __init__(
+ self,
+ name: str,
+ save_dir: pathlib.Path,
+ id: Optional[str] = None,
+ project: Optional[str] = None
+ ):
+ super().__init__()
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ save_dir.mkdir(parents=True, exist_ok=True)
+ self.experiment = wandb.init(name=name,
+ project=project,
+ id=id,
+ dir=str(save_dir),
+ resume='allow',
+ anonymous='must')
+
+ @rank_zero_only
+ def log_hyperparams(self, params: Dict[str, Any]) -> None:
+ params = self._sanitize_params(params)
+ self.experiment.config.update(params, allow_val_change=True)
+
+ @rank_zero_only
+ def log_metrics(self, metrics: Dict[str, float], step: Optional[int] = None) -> None:
+ if step is not None:
+ self.experiment.log({**metrics, 'epoch': step})
+ else:
+ self.experiment.log(metrics)
diff --git a/model/se3_transformer/runtime/metrics.py b/model/se3_transformer/runtime/metrics.py
new file mode 100644
index 0000000000000000000000000000000000000000..63026a71989441603df6abd447555524f2fd1e85
--- /dev/null
+++ b/model/se3_transformer/runtime/metrics.py
@@ -0,0 +1,83 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from abc import ABC, abstractmethod
+
+import torch
+import torch.distributed as dist
+from torch import Tensor
+
+
+class Metric(ABC):
+ """ Metric class with synchronization capabilities similar to TorchMetrics """
+
+ def __init__(self):
+ self.states = {}
+
+ def add_state(self, name: str, default: Tensor):
+ assert name not in self.states
+ self.states[name] = default.clone()
+ setattr(self, name, default)
+
+ def synchronize(self):
+ if dist.is_initialized():
+ for state in self.states:
+ dist.all_reduce(getattr(self, state), op=dist.ReduceOp.SUM, group=dist.group.WORLD)
+
+ def __call__(self, *args, **kwargs):
+ self.update(*args, **kwargs)
+
+ def reset(self):
+ for name, default in self.states.items():
+ setattr(self, name, default.clone())
+
+ def compute(self):
+ self.synchronize()
+ value = self._compute().item()
+ self.reset()
+ return value
+
+ @abstractmethod
+ def _compute(self):
+ pass
+
+ @abstractmethod
+ def update(self, preds: Tensor, targets: Tensor):
+ pass
+
+
+class MeanAbsoluteError(Metric):
+ def __init__(self):
+ super().__init__()
+ self.add_state('error', torch.tensor(0, dtype=torch.float32, device='cuda'))
+ self.add_state('total', torch.tensor(0, dtype=torch.int32, device='cuda'))
+
+ def update(self, preds: Tensor, targets: Tensor):
+ preds = preds.detach()
+ n = preds.shape[0]
+ error = torch.abs(preds.view(n, -1) - targets.view(n, -1)).sum()
+ self.total += n
+ self.error += error
+
+ def _compute(self):
+ return self.error / self.total
diff --git a/model/se3_transformer/runtime/training.py b/model/se3_transformer/runtime/training.py
new file mode 100644
index 0000000000000000000000000000000000000000..2d66f08f45ce7bc753ac463489fc9a8d14a6b8b6
--- /dev/null
+++ b/model/se3_transformer/runtime/training.py
@@ -0,0 +1,238 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+import pathlib
+from typing import List
+
+import numpy as np
+import torch
+import torch.distributed as dist
+import torch.nn as nn
+from apex.optimizers import FusedAdam, FusedLAMB
+from torch.nn.modules.loss import _Loss
+from torch.nn.parallel import DistributedDataParallel
+from torch.optim import Optimizer
+from torch.utils.data import DataLoader, DistributedSampler
+from tqdm import tqdm
+
+from se3_transformer.data_loading import QM9DataModule
+from se3_transformer.model import SE3TransformerPooled
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.runtime import gpu_affinity
+from se3_transformer.runtime.arguments import PARSER
+from se3_transformer.runtime.callbacks import QM9MetricCallback, QM9LRSchedulerCallback, BaseCallback, \
+ PerformanceCallback
+from se3_transformer.runtime.inference import evaluate
+from se3_transformer.runtime.loggers import LoggerCollection, DLLogger, WandbLogger, Logger
+from se3_transformer.runtime.utils import to_cuda, get_local_rank, init_distributed, seed_everything, \
+ using_tensor_cores, increase_l2_fetch_granularity
+
+
+def save_state(model: nn.Module, optimizer: Optimizer, epoch: int, path: pathlib.Path, callbacks: List[BaseCallback]):
+ """ Saves model, optimizer and epoch states to path (only once per node) """
+ if get_local_rank() == 0:
+ state_dict = model.module.state_dict() if isinstance(model, DistributedDataParallel) else model.state_dict()
+ checkpoint = {
+ 'state_dict': state_dict,
+ 'optimizer_state_dict': optimizer.state_dict(),
+ 'epoch': epoch
+ }
+ for callback in callbacks:
+ callback.on_checkpoint_save(checkpoint)
+
+ torch.save(checkpoint, str(path))
+ logging.info(f'Saved checkpoint to {str(path)}')
+
+
+def load_state(model: nn.Module, optimizer: Optimizer, path: pathlib.Path, callbacks: List[BaseCallback]):
+ """ Loads model, optimizer and epoch states from path """
+ checkpoint = torch.load(str(path), map_location={'cuda:0': f'cuda:{get_local_rank()}'})
+ if isinstance(model, DistributedDataParallel):
+ model.module.load_state_dict(checkpoint['state_dict'])
+ else:
+ model.load_state_dict(checkpoint['state_dict'])
+ optimizer.load_state_dict(checkpoint['optimizer_state_dict'])
+
+ for callback in callbacks:
+ callback.on_checkpoint_load(checkpoint)
+
+ logging.info(f'Loaded checkpoint from {str(path)}')
+ return checkpoint['epoch']
+
+
+def train_epoch(train_dataloader, loss_fn, epoch_idx, grad_scaler, optimizer, local_rank, callbacks, args):
+ losses = []
+ for i, batch in tqdm(enumerate(train_dataloader), total=len(train_dataloader), unit='batch',
+ desc=f'Epoch {epoch_idx}', disable=(args.silent or local_rank != 0)):
+ *inputs, target = to_cuda(batch)
+
+ for callback in callbacks:
+ callback.on_batch_start()
+
+ with torch.cuda.amp.autocast(enabled=args.amp):
+ pred = model(*inputs)
+ loss = loss_fn(pred, target) / args.accumulate_grad_batches
+
+ grad_scaler.scale(loss).backward()
+
+ # gradient accumulation
+ if (i + 1) % args.accumulate_grad_batches == 0 or (i + 1) == len(train_dataloader):
+ if args.gradient_clip:
+ grad_scaler.unscale_(optimizer)
+ torch.nn.utils.clip_grad_norm_(model.parameters(), args.gradient_clip)
+
+ grad_scaler.step(optimizer)
+ grad_scaler.update()
+ optimizer.zero_grad()
+
+ losses.append(loss.item())
+
+ return np.mean(losses)
+
+
+def train(model: nn.Module,
+ loss_fn: _Loss,
+ train_dataloader: DataLoader,
+ val_dataloader: DataLoader,
+ callbacks: List[BaseCallback],
+ logger: Logger,
+ args):
+ device = torch.cuda.current_device()
+ model.to(device=device)
+ local_rank = get_local_rank()
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+
+ if dist.is_initialized():
+ model = DistributedDataParallel(model, device_ids=[local_rank], output_device=local_rank)
+
+ model.train()
+ grad_scaler = torch.cuda.amp.GradScaler(enabled=args.amp)
+ if args.optimizer == 'adam':
+ optimizer = FusedAdam(model.parameters(), lr=args.learning_rate, betas=(args.momentum, 0.999),
+ weight_decay=args.weight_decay)
+ elif args.optimizer == 'lamb':
+ optimizer = FusedLAMB(model.parameters(), lr=args.learning_rate, betas=(args.momentum, 0.999),
+ weight_decay=args.weight_decay)
+ else:
+ optimizer = torch.optim.SGD(model.parameters(), lr=args.learning_rate, momentum=args.momentum,
+ weight_decay=args.weight_decay)
+
+ epoch_start = load_state(model, optimizer, args.load_ckpt_path, callbacks) if args.load_ckpt_path else 0
+
+ for callback in callbacks:
+ callback.on_fit_start(optimizer, args)
+
+ for epoch_idx in range(epoch_start, args.epochs):
+ if isinstance(train_dataloader.sampler, DistributedSampler):
+ train_dataloader.sampler.set_epoch(epoch_idx)
+
+ loss = train_epoch(train_dataloader, loss_fn, epoch_idx, grad_scaler, optimizer, local_rank, callbacks, args)
+ if dist.is_initialized():
+ loss = torch.tensor(loss, dtype=torch.float, device=device)
+ torch.distributed.all_reduce(loss)
+ loss = (loss / world_size).item()
+
+ logging.info(f'Train loss: {loss}')
+ logger.log_metrics({'train loss': loss}, epoch_idx)
+
+ for callback in callbacks:
+ callback.on_epoch_end()
+
+ if not args.benchmark and args.save_ckpt_path is not None and args.ckpt_interval > 0 \
+ and (epoch_idx + 1) % args.ckpt_interval == 0:
+ save_state(model, optimizer, epoch_idx, args.save_ckpt_path, callbacks)
+
+ if not args.benchmark and args.eval_interval > 0 and (epoch_idx + 1) % args.eval_interval == 0:
+ evaluate(model, val_dataloader, callbacks, args)
+ model.train()
+
+ for callback in callbacks:
+ callback.on_validation_end(epoch_idx)
+
+ if args.save_ckpt_path is not None and not args.benchmark:
+ save_state(model, optimizer, args.epochs, args.save_ckpt_path, callbacks)
+
+ for callback in callbacks:
+ callback.on_fit_end()
+
+
+def print_parameters_count(model):
+ num_params_trainable = sum(p.numel() for p in model.parameters() if p.requires_grad)
+ logging.info(f'Number of trainable parameters: {num_params_trainable}')
+
+
+if __name__ == '__main__':
+ is_distributed = init_distributed()
+ local_rank = get_local_rank()
+ args = PARSER.parse_args()
+
+ logging.getLogger().setLevel(logging.CRITICAL if local_rank != 0 or args.silent else logging.INFO)
+
+ logging.info('====== SE(3)-Transformer ======')
+ logging.info('| Training procedure |')
+ logging.info('===============================')
+
+ if args.seed is not None:
+ logging.info(f'Using seed {args.seed}')
+ seed_everything(args.seed)
+
+ logger = LoggerCollection([
+ DLLogger(save_dir=args.log_dir, filename=args.dllogger_name),
+ WandbLogger(name=f'QM9({args.task})', save_dir=args.log_dir, project='se3-transformer')
+ ])
+
+ datamodule = QM9DataModule(**vars(args))
+ model = SE3TransformerPooled(
+ fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}),
+ fiber_out=Fiber({0: args.num_degrees * args.num_channels}),
+ fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}),
+ output_dim=1,
+ tensor_cores=using_tensor_cores(args.amp), # use Tensor Cores more effectively
+ **vars(args)
+ )
+ loss_fn = nn.L1Loss()
+
+ if args.benchmark:
+ logging.info('Running benchmark mode')
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+ callbacks = [PerformanceCallback(logger, args.batch_size * world_size)]
+ else:
+ callbacks = [QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix='validation'),
+ QM9LRSchedulerCallback(logger, epochs=args.epochs)]
+
+ if is_distributed:
+ gpu_affinity.set_affinity(gpu_id=get_local_rank(), nproc_per_node=torch.cuda.device_count())
+
+ print_parameters_count(model)
+ logger.log_hyperparams(vars(args))
+ increase_l2_fetch_granularity()
+ train(model,
+ loss_fn,
+ datamodule.train_dataloader(),
+ datamodule.val_dataloader(),
+ callbacks,
+ logger,
+ args)
+
+ logging.info('Training finished successfully')
diff --git a/model/se3_transformer/runtime/utils.py b/model/se3_transformer/runtime/utils.py
new file mode 100644
index 0000000000000000000000000000000000000000..b6248fa6e5d94a14df6726eb73274a4727281a63
--- /dev/null
+++ b/model/se3_transformer/runtime/utils.py
@@ -0,0 +1,130 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import argparse
+import ctypes
+import logging
+import os
+import random
+from functools import wraps
+from typing import Union, List, Dict
+
+import numpy as np
+import torch
+import torch.distributed as dist
+from torch import Tensor
+
+
+def aggregate_residual(feats1, feats2, method: str):
+ """ Add or concatenate two fiber features together. If degrees don't match, will use the ones of feats2. """
+ if method in ['add', 'sum']:
+ return {k: (v + feats1[k]) if k in feats1 else v for k, v in feats2.items()}
+ elif method in ['cat', 'concat']:
+ return {k: torch.cat([v, feats1[k]], dim=1) if k in feats1 else v for k, v in feats2.items()}
+ else:
+ raise ValueError('Method must be add/sum or cat/concat')
+
+
+def degree_to_dim(degree: int) -> int:
+ return 2 * degree + 1
+
+
+def unfuse_features(features: Tensor, degrees: List[int]) -> Dict[str, Tensor]:
+ return dict(zip(map(str, degrees), features.split([degree_to_dim(deg) for deg in degrees], dim=-1)))
+
+
+def str2bool(v: Union[bool, str]) -> bool:
+ if isinstance(v, bool):
+ return v
+ if v.lower() in ('yes', 'true', 't', 'y', '1'):
+ return True
+ elif v.lower() in ('no', 'false', 'f', 'n', '0'):
+ return False
+ else:
+ raise argparse.ArgumentTypeError('Boolean value expected.')
+
+
+def to_cuda(x):
+ """ Try to convert a Tensor, a collection of Tensors or a DGLGraph to CUDA """
+ if isinstance(x, Tensor):
+ return x.cuda(non_blocking=True)
+ elif isinstance(x, tuple):
+ return (to_cuda(v) for v in x)
+ elif isinstance(x, list):
+ return [to_cuda(v) for v in x]
+ elif isinstance(x, dict):
+ return {k: to_cuda(v) for k, v in x.items()}
+ else:
+ # DGLGraph or other objects
+ return x.to(device=torch.cuda.current_device())
+
+
+def get_local_rank() -> int:
+ return int(os.environ.get('LOCAL_RANK', 0))
+
+
+def init_distributed() -> bool:
+ world_size = int(os.environ.get('WORLD_SIZE', 1))
+ distributed = world_size > 1
+ if distributed:
+ backend = 'nccl' if torch.cuda.is_available() else 'gloo'
+ dist.init_process_group(backend=backend, init_method='env://')
+ if backend == 'nccl':
+ torch.cuda.set_device(get_local_rank())
+ else:
+ logging.warning('Running on CPU only!')
+ assert torch.distributed.is_initialized()
+ return distributed
+
+
+def increase_l2_fetch_granularity():
+ # maximum fetch granularity of L2: 128 bytes
+ _libcudart = ctypes.CDLL('libcudart.so')
+ # set device limit on the current device
+ # cudaLimitMaxL2FetchGranularity = 0x05
+ pValue = ctypes.cast((ctypes.c_int * 1)(), ctypes.POINTER(ctypes.c_int))
+ _libcudart.cudaDeviceSetLimit(ctypes.c_int(0x05), ctypes.c_int(128))
+ _libcudart.cudaDeviceGetLimit(pValue, ctypes.c_int(0x05))
+ assert pValue.contents.value == 128
+
+
+def seed_everything(seed):
+ seed = int(seed)
+ random.seed(seed)
+ np.random.seed(seed)
+ torch.manual_seed(seed)
+ torch.cuda.manual_seed_all(seed)
+
+
+def rank_zero_only(fn):
+ @wraps(fn)
+ def wrapped_fn(*args, **kwargs):
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ return fn(*args, **kwargs)
+
+ return wrapped_fn
+
+
+def using_tensor_cores(amp: bool) -> bool:
+ major_cc, minor_cc = torch.cuda.get_device_capability()
+ return (amp and major_cc >= 7) or major_cc >= 8
diff --git a/model/util.py b/model/util.py
new file mode 100644
index 0000000000000000000000000000000000000000..017f7028636ce8bf5597294913273fc257c2c844
--- /dev/null
+++ b/model/util.py
@@ -0,0 +1,522 @@
+import sys
+
+import numpy as np
+import torch
+
+import scipy.sparse
+
+from chemical import *
+from scoring import *
+
+def th_ang_v(ab,bc,eps:float=1e-8):
+ def th_norm(x,eps:float=1e-8):
+ return x.square().sum(-1,keepdim=True).add(eps).sqrt()
+ def th_N(x,alpha:float=0):
+ return x/th_norm(x).add(alpha)
+ ab, bc = th_N(ab),th_N(bc)
+ cos_angle = torch.clamp( (ab*bc).sum(-1), -1, 1)
+ sin_angle = torch.sqrt(1-cos_angle.square() + eps)
+ dih = torch.stack((cos_angle,sin_angle),-1)
+ return dih
+
+def th_dih_v(ab,bc,cd):
+ def th_cross(a,b):
+ a,b = torch.broadcast_tensors(a,b)
+ return torch.cross(a,b, dim=-1)
+ def th_norm(x,eps:float=1e-8):
+ return x.square().sum(-1,keepdim=True).add(eps).sqrt()
+ def th_N(x,alpha:float=0):
+ return x/th_norm(x).add(alpha)
+
+ ab, bc, cd = th_N(ab),th_N(bc),th_N(cd)
+ n1 = th_N( th_cross(ab,bc) )
+ n2 = th_N( th_cross(bc,cd) )
+ sin_angle = (th_cross(n1,bc)*n2).sum(-1)
+ cos_angle = (n1*n2).sum(-1)
+ dih = torch.stack((cos_angle,sin_angle),-1)
+ return dih
+
+def th_dih(a,b,c,d):
+ return th_dih_v(a-b,b-c,c-d)
+
+# More complicated version splits error in CA-N and CA-C (giving more accurate CB position)
+# It returns the rigid transformation from local frame to global frame
+def rigid_from_3_points(N, Ca, C, non_ideal=False, eps=1e-8):
+ #N, Ca, C - [B,L, 3]
+ #R - [B,L, 3, 3], det(R)=1, inv(R) = R.T, R is a rotation matrix
+ B,L = N.shape[:2]
+
+ v1 = C-Ca
+ v2 = N-Ca
+ e1 = v1/(torch.norm(v1, dim=-1, keepdim=True)+eps)
+ u2 = v2-(torch.einsum('bli, bli -> bl', e1, v2)[...,None]*e1)
+ e2 = u2/(torch.norm(u2, dim=-1, keepdim=True)+eps)
+ e3 = torch.cross(e1, e2, dim=-1)
+ R = torch.cat([e1[...,None], e2[...,None], e3[...,None]], axis=-1) #[B,L,3,3] - rotation matrix
+
+ if non_ideal:
+ v2 = v2/(torch.norm(v2, dim=-1, keepdim=True)+eps)
+ cosref = torch.sum(e1*v2, dim=-1) # cosine of current N-CA-C bond angle
+ costgt = cos_ideal_NCAC.item()
+ cos2del = torch.clamp( cosref*costgt + torch.sqrt((1-cosref*cosref)*(1-costgt*costgt)+eps), min=-1.0, max=1.0 )
+ cosdel = torch.sqrt(0.5*(1+cos2del)+eps)
+ sindel = torch.sign(costgt-cosref) * torch.sqrt(1-0.5*(1+cos2del)+eps)
+ Rp = torch.eye(3, device=N.device).repeat(B,L,1,1)
+ Rp[:,:,0,0] = cosdel
+ Rp[:,:,0,1] = -sindel
+ Rp[:,:,1,0] = sindel
+ Rp[:,:,1,1] = cosdel
+
+ R = torch.einsum('blij,bljk->blik', R,Rp)
+
+ return R, Ca
+
+def get_tor_mask(seq, torsion_indices, mask_in=None):
+ B,L = seq.shape[:2]
+ tors_mask = torch.ones((B,L,10), dtype=torch.bool, device=seq.device)
+ tors_mask[...,3:7] = torsion_indices[seq,:,-1] > 0
+ tors_mask[:,0,1] = False
+ tors_mask[:,-1,0] = False
+
+ # mask for additional angles
+ tors_mask[:,:,7] = seq!=aa2num['GLY']
+ tors_mask[:,:,8] = seq!=aa2num['GLY']
+ tors_mask[:,:,9] = torch.logical_and( seq!=aa2num['GLY'], seq!=aa2num['ALA'] )
+ tors_mask[:,:,9] = torch.logical_and( tors_mask[:,:,9], seq!=aa2num['UNK'] )
+ tors_mask[:,:,9] = torch.logical_and( tors_mask[:,:,9], seq!=aa2num['MAS'] )
+
+ if mask_in != None:
+ # mask for missing atoms
+ # chis
+ ti0 = torch.gather(mask_in,2,torsion_indices[seq,:,0])
+ ti1 = torch.gather(mask_in,2,torsion_indices[seq,:,1])
+ ti2 = torch.gather(mask_in,2,torsion_indices[seq,:,2])
+ ti3 = torch.gather(mask_in,2,torsion_indices[seq,:,3])
+ is_valid = torch.stack((ti0, ti1, ti2, ti3), dim=-2).all(dim=-1)
+ tors_mask[...,3:7] = torch.logical_and(tors_mask[...,3:7], is_valid)
+ tors_mask[:,:,7] = torch.logical_and(tors_mask[:,:,7], mask_in[:,:,4]) # CB exist?
+ tors_mask[:,:,8] = torch.logical_and(tors_mask[:,:,8], mask_in[:,:,4]) # CB exist?
+ tors_mask[:,:,9] = torch.logical_and(tors_mask[:,:,9], mask_in[:,:,5]) # XG exist?
+
+ return tors_mask
+
+def get_torsions(xyz_in, seq, torsion_indices, torsion_can_flip, ref_angles, mask_in=None):
+ B,L = xyz_in.shape[:2]
+
+ tors_mask = get_tor_mask(seq, torsion_indices, mask_in)
+
+ # torsions to restrain to 0 or 180degree
+ tors_planar = torch.zeros((B, L, 10), dtype=torch.bool, device=xyz_in.device)
+ tors_planar[:,:,5] = seq == aa2num['TYR'] # TYR chi 3 should be planar
+
+ # idealize given xyz coordinates before computing torsion angles
+ xyz = xyz_in.clone()
+ Rs, Ts = rigid_from_3_points(xyz[...,0,:],xyz[...,1,:],xyz[...,2,:])
+ Nideal = torch.tensor([-0.5272, 1.3593, 0.000], device=xyz_in.device)
+ Cideal = torch.tensor([1.5233, 0.000, 0.000], device=xyz_in.device)
+ xyz[...,0,:] = torch.einsum('brij,j->bri', Rs, Nideal) + Ts
+ xyz[...,2,:] = torch.einsum('brij,j->bri', Rs, Cideal) + Ts
+
+ torsions = torch.zeros( (B,L,10,2), device=xyz.device )
+ # avoid undefined angles for H generation
+ torsions[:,0,1,0] = 1.0
+ torsions[:,-1,0,0] = 1.0
+
+ # omega
+ torsions[:,:-1,0,:] = th_dih(xyz[:,:-1,1,:],xyz[:,:-1,2,:],xyz[:,1:,0,:],xyz[:,1:,1,:])
+ # phi
+ torsions[:,1:,1,:] = th_dih(xyz[:,:-1,2,:],xyz[:,1:,0,:],xyz[:,1:,1,:],xyz[:,1:,2,:])
+ # psi
+ torsions[:,:,2,:] = -1 * th_dih(xyz[:,:,0,:],xyz[:,:,1,:],xyz[:,:,2,:],xyz[:,:,3,:])
+
+ # chis
+ ti0 = torch.gather(xyz,2,torsion_indices[seq,:,0,None].repeat(1,1,1,3))
+ ti1 = torch.gather(xyz,2,torsion_indices[seq,:,1,None].repeat(1,1,1,3))
+ ti2 = torch.gather(xyz,2,torsion_indices[seq,:,2,None].repeat(1,1,1,3))
+ ti3 = torch.gather(xyz,2,torsion_indices[seq,:,3,None].repeat(1,1,1,3))
+ torsions[:,:,3:7,:] = th_dih(ti0,ti1,ti2,ti3)
+
+ # CB bend
+ NC = 0.5*( xyz[:,:,0,:3] + xyz[:,:,2,:3] )
+ CA = xyz[:,:,1,:3]
+ CB = xyz[:,:,4,:3]
+ t = th_ang_v(CB-CA,NC-CA)
+ t0 = ref_angles[seq][...,0,:]
+ torsions[:,:,7,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ # CB twist
+ NCCA = NC-CA
+ NCp = xyz[:,:,2,:3] - xyz[:,:,0,:3]
+ NCpp = NCp - torch.sum(NCp*NCCA, dim=-1, keepdim=True)/ torch.sum(NCCA*NCCA, dim=-1, keepdim=True) * NCCA
+ t = th_ang_v(CB-CA,NCpp)
+ t0 = ref_angles[seq][...,1,:]
+ torsions[:,:,8,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ # CG bend
+ CG = xyz[:,:,5,:3]
+ t = th_ang_v(CG-CB,CA-CB)
+ t0 = ref_angles[seq][...,2,:]
+ torsions[:,:,9,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ mask0 = torch.isnan(torsions[...,0]).nonzero()
+ mask1 = torch.isnan(torsions[...,1]).nonzero()
+ torsions[mask0[:,0],mask0[:,1],mask0[:,2],0] = 1.0
+ torsions[mask1[:,0],mask1[:,1],mask1[:,2],1] = 0.0
+
+ # alt chis
+ torsions_alt = torsions.clone()
+ torsions_alt[torsion_can_flip[seq,:]] *= -1
+
+ return torsions, torsions_alt, tors_mask, tors_planar
+
+def get_tips(xyz, seq):
+ B,L = xyz.shape[:2]
+
+ xyz_tips = torch.gather(xyz, 2, tip_indices.to(xyz.device)[seq][:,:,None,None].expand(-1,-1,-1,3)).reshape(B, L, 3)
+ mask = ~(torch.isnan(xyz_tips[:,:,0]))
+ if torch.isnan(xyz_tips).any(): # replace NaN tip atom with virtual Cb atom
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ xyz_tips = torch.where(torch.isnan(xyz_tips), Cb, xyz_tips)
+ return xyz_tips, mask
+
+# process ideal frames
+def make_frame(X, Y):
+ Xn = X / torch.linalg.norm(X)
+ Y = Y - torch.dot(Y, Xn) * Xn
+ Yn = Y / torch.linalg.norm(Y)
+ Z = torch.cross(Xn,Yn)
+ Zn = Z / torch.linalg.norm(Z)
+
+ return torch.stack((Xn,Yn,Zn), dim=-1)
+
+def cross_product_matrix(u):
+ B, L = u.shape[:2]
+ matrix = torch.zeros((B, L, 3, 3), device=u.device)
+ matrix[:,:,0,1] = -u[...,2]
+ matrix[:,:,0,2] = u[...,1]
+ matrix[:,:,1,0] = u[...,2]
+ matrix[:,:,1,2] = -u[...,0]
+ matrix[:,:,2,0] = -u[...,1]
+ matrix[:,:,2,1] = u[...,0]
+ return matrix
+
+# writepdb
+def writepdb(filename, atoms, seq, idx_pdb=None, bfacts=None):
+ f = open(filename,"w")
+ ctr = 1
+ scpu = seq.cpu().squeeze()
+ atomscpu = atoms.cpu().squeeze()
+ if bfacts is None:
+ bfacts = torch.zeros(atomscpu.shape[0])
+ if idx_pdb is None:
+ idx_pdb = 1 + torch.arange(atomscpu.shape[0])
+
+ Bfacts = torch.clamp( bfacts.cpu(), 0, 1)
+ for i,s in enumerate(scpu):
+ if (len(atomscpu.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", num2aa[s],
+ "A", idx_pdb[i], atomscpu[i,0], atomscpu[i,1], atomscpu[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ elif atomscpu.shape[1]==3:
+ for j,atm_j in enumerate([" N "," CA "," C "]):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ "A", idx_pdb[i], atomscpu[i,j,0], atomscpu[i,j,1], atomscpu[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ natoms = atomscpu.shape[1]
+ if (natoms!=14 and natoms!=27):
+ print ('bad size!', atoms.shape)
+ assert(False)
+ atms = aa2long[s]
+ # his prot hack
+ if (s==8 and torch.linalg.norm( atomscpu[i,9,:]-atomscpu[i,5,:] ) < 1.7):
+ atms = (
+ " N "," CA "," C "," O "," CB "," CG "," NE2"," CD2"," CE1"," ND1",
+ None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1",
+ " HD1", None, None, None, None, None, None) # his_d
+
+ for j,atm_j in enumerate(atms):
+ if (j=4] = 4
+ num_bonds[i,...] = torch.tensor(num_bonds_i)
+
+
+# LJ/LK scoring parameters
+ljlk_parameters = torch.zeros((22,27,5), dtype=torch.float)
+lj_correction_parameters = torch.zeros((22,27,4), dtype=bool) # donor/acceptor/hpol/disulf
+for i in range(22):
+ for j,a in enumerate(aa2type[i]):
+ if (a is not None):
+ ljlk_parameters[i,j,:] = torch.tensor( type2ljlk[a] )
+ lj_correction_parameters[i,j,0] = (type2hb[a]==HbAtom.DO)+(type2hb[a]==HbAtom.DA)
+ lj_correction_parameters[i,j,1] = (type2hb[a]==HbAtom.AC)+(type2hb[a]==HbAtom.DA)
+ lj_correction_parameters[i,j,2] = (type2hb[a]==HbAtom.HP)
+ lj_correction_parameters[i,j,3] = (a=="SH1" or a=="HS")
+
+# hbond scoring parameters
+def donorHs(D,bonds,atoms):
+ dHs = []
+ for (i,j) in bonds:
+ if (i==D):
+ idx_j = atoms.index(j)
+ if (idx_j>=14): # if atom j is a hydrogen
+ dHs.append(idx_j)
+ if (j==D):
+ idx_i = atoms.index(i)
+ if (idx_i>=14): # if atom j is a hydrogen
+ dHs.append(idx_i)
+ assert (len(dHs)>0)
+ return dHs
+
+def acceptorBB0(A,hyb,bonds,atoms):
+ if (hyb == HbHybType.SP2):
+ for (i,j) in bonds:
+ if (i==A):
+ B = atoms.index(j)
+ if (B<14):
+ break
+ if (j==A):
+ B = atoms.index(i)
+ if (B<14):
+ break
+ for (i,j) in bonds:
+ if (i==atoms[B]):
+ B0 = atoms.index(j)
+ if (B0<14):
+ break
+ if (j==atoms[B]):
+ B0 = atoms.index(i)
+ if (B0<14):
+ break
+ elif (hyb == HbHybType.SP3 or hyb == HbHybType.RING):
+ for (i,j) in bonds:
+ if (i==A):
+ B = atoms.index(j)
+ if (B<14):
+ break
+ if (j==A):
+ B = atoms.index(i)
+ if (B<14):
+ break
+ for (i,j) in bonds:
+ if (i==A and j!=atoms[B]):
+ B0 = atoms.index(j)
+ break
+ if (j==A and i!=atoms[B]):
+ B0 = atoms.index(i)
+ break
+
+ return B,B0
+
+
+hbtypes = torch.full((22,27,3),-1, dtype=torch.long) # (donortype, acceptortype, acchybtype)
+hbbaseatoms = torch.full((22,27,2),-1, dtype=torch.long) # (B,B0) for acc; (D,-1) for don
+hbpolys = torch.zeros((HbDonType.NTYPES,HbAccType.NTYPES,3,15)) # weight,xmin,xmax,ymin,ymax,c9,...,c0
+
+for i in range(22):
+ for j,a in enumerate(aa2type[i]):
+ if (a in type2dontype):
+ j_hs = donorHs(aa2long[i][j],aabonds[i],aa2long[i])
+ for j_h in j_hs:
+ hbtypes[i,j_h,0] = type2dontype[a]
+ hbbaseatoms[i,j_h,0] = j
+ if (a in type2acctype):
+ j_b, j_b0 = acceptorBB0(aa2long[i][j],type2hybtype[a],aabonds[i],aa2long[i])
+ hbtypes[i,j,1] = type2acctype[a]
+ hbtypes[i,j,2] = type2hybtype[a]
+ hbbaseatoms[i,j,0] = j_b
+ hbbaseatoms[i,j,1] = j_b0
+
+for i in range(HbDonType.NTYPES):
+ for j in range(HbAccType.NTYPES):
+ weight = dontype2wt[i]*acctype2wt[j]
+
+ pdist,pbah,pahd = hbtypepair2poly[(i,j)]
+ xrange,yrange,coeffs = hbpolytype2coeffs[pdist]
+ hbpolys[i,j,0,0] = weight
+ hbpolys[i,j,0,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,0,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,0,5:] = torch.tensor(coeffs)
+ xrange,yrange,coeffs = hbpolytype2coeffs[pahd]
+ hbpolys[i,j,1,0] = weight
+ hbpolys[i,j,1,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,1,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,1,5:] = torch.tensor(coeffs)
+ xrange,yrange,coeffs = hbpolytype2coeffs[pbah]
+ hbpolys[i,j,2,0] = weight
+ hbpolys[i,j,2,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,2,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,2,5:] = torch.tensor(coeffs)
+
+# kinematic parameters
+base_indices = torch.full((22,27),0, dtype=torch.long)
+xyzs_in_base_frame = torch.ones((22,27,4))
+RTs_by_torsion = torch.eye(4).repeat(22,7,1,1)
+reference_angles = torch.ones((22,3,2))
+
+for i in range(22):
+ i_l = aa2long[i]
+ for name, base, coords in ideal_coords[i]:
+ idx = i_l.index(name)
+ base_indices[i,idx] = base
+ xyzs_in_base_frame[i,idx,:3] = torch.tensor(coords)
+
+ # omega frame
+ RTs_by_torsion[i,0,:3,:3] = torch.eye(3)
+ RTs_by_torsion[i,0,:3,3] = torch.zeros(3)
+
+ # phi frame
+ RTs_by_torsion[i,1,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,0,:3] - xyzs_in_base_frame[i,1,:3],
+ torch.tensor([1.,0.,0.])
+ )
+ RTs_by_torsion[i,1,:3,3] = xyzs_in_base_frame[i,0,:3]
+
+ # psi frame
+ RTs_by_torsion[i,2,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,2,:3] - xyzs_in_base_frame[i,1,:3],
+ xyzs_in_base_frame[i,1,:3] - xyzs_in_base_frame[i,0,:3]
+ )
+ RTs_by_torsion[i,2,:3,3] = xyzs_in_base_frame[i,2,:3]
+
+ # chi1 frame
+ if torsions[i][0] is not None:
+ a0,a1,a2 = torsion_indices[i,0,0:3]
+ RTs_by_torsion[i,3,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3]-xyzs_in_base_frame[i,a1,:3],
+ xyzs_in_base_frame[i,a0,:3]-xyzs_in_base_frame[i,a1,:3],
+ )
+ RTs_by_torsion[i,3,:3,3] = xyzs_in_base_frame[i,a2,:3]
+
+ # chi2~4 frame
+ for j in range(1,4):
+ if torsions[i][j] is not None:
+ a2 = torsion_indices[i,j,2]
+ if ((i==18 and j==2) or (i==8 and j==2)): # TYR CZ-OH & HIS CE1-HE1 a special case
+ a0,a1 = torsion_indices[i,j,0:2]
+ RTs_by_torsion[i,3+j,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3]-xyzs_in_base_frame[i,a1,:3],
+ xyzs_in_base_frame[i,a0,:3]-xyzs_in_base_frame[i,a1,:3] )
+ else:
+ RTs_by_torsion[i,3+j,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3],
+ torch.tensor([-1.,0.,0.]), )
+ RTs_by_torsion[i,3+j,:3,3] = xyzs_in_base_frame[i,a2,:3]
+
+
+ # CB/CG angles
+ NCr = 0.5*(xyzs_in_base_frame[i,0,:3]+xyzs_in_base_frame[i,2,:3])
+ CAr = xyzs_in_base_frame[i,1,:3]
+ CBr = xyzs_in_base_frame[i,4,:3]
+ CGr = xyzs_in_base_frame[i,5,:3]
+ reference_angles[i,0,:]=th_ang_v(CBr-CAr,NCr-CAr)
+ NCp = xyzs_in_base_frame[i,2,:3]-xyzs_in_base_frame[i,0,:3]
+ NCpp = NCp - torch.dot(NCp,NCr)/ torch.dot(NCr,NCr) * NCr
+ reference_angles[i,1,:]=th_ang_v(CBr-CAr,NCpp)
+ reference_angles[i,2,:]=th_ang_v(CGr,torch.tensor([-1.,0.,0.]))
+
+def get_rmsd(a, b, eps=1e-6):
+ '''
+ align crds b to a : always use all alphas
+ expexted tensor of shape (L,3)
+ jake's torch adapted version
+ '''
+ assert a.shape == b.shape, 'make sure tensors are the same size'
+ L = a.shape[0]
+ assert a.shape == torch.Size([L,3]), 'make sure tensors are in format [L,3]'
+
+ # center to CA centroid
+ a = a - a.mean(dim=0)
+ b = b - b.mean(dim=0)
+
+ # Computation of the covariance matrix
+ C = torch.einsum('kj,ji->ki', torch.transpose(b.type(torch.float32),0,1), a.type(torch.float32))
+
+ # Compute optimal rotation matrix using SVD
+ V, S, W = torch.linalg.svd(C)
+
+ # get sign to ensure right-handedness
+ d = torch.ones([3,3])
+ d[:,-1] = torch.sign(torch.linalg.det(V)*torch.linalg.det(W))
+
+ # Rotation matrix U
+ U = torch.einsum('kj,ji->ki',(d*V),W)
+
+ # Rotate xyz_hal
+ rP = torch.einsum('kj,ji->ki',b.type(torch.float32),U.type(torch.float32))
+
+ L = rP.shape[0]
+ rmsd = torch.sqrt(torch.sum((rP-a)*(rP-a), axis=(0,1)) / L + eps)
+
+ return rmsd, U
+
diff --git a/model/util_module.py b/model/util_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..4ba2b5723434b195468c0d11473a8a33f295f4cf
--- /dev/null
+++ b/model/util_module.py
@@ -0,0 +1,310 @@
+import numpy as np
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import copy
+import dgl
+from util import base_indices, RTs_by_torsion, xyzs_in_base_frame, rigid_from_3_points
+
+def init_lecun_normal(module, scale=1.0):
+ def truncated_normal(uniform, mu=0.0, sigma=1.0, a=-2, b=2):
+ normal = torch.distributions.normal.Normal(0, 1)
+
+ alpha = (a - mu) / sigma
+ beta = (b - mu) / sigma
+
+ alpha_normal_cdf = normal.cdf(torch.tensor(alpha))
+ p = alpha_normal_cdf + (normal.cdf(torch.tensor(beta)) - alpha_normal_cdf) * uniform
+
+ v = torch.clamp(2 * p - 1, -1 + 1e-8, 1 - 1e-8)
+ x = mu + sigma * np.sqrt(2) * torch.erfinv(v)
+ x = torch.clamp(x, a, b)
+
+ return x
+
+ def sample_truncated_normal(shape, scale=1.0):
+ stddev = np.sqrt(scale/shape[-1])/.87962566103423978 # shape[-1] = fan_in
+ return stddev * truncated_normal(torch.rand(shape))
+
+ module.weight = torch.nn.Parameter( (sample_truncated_normal(module.weight.shape)) )
+ return module
+
+def init_lecun_normal_param(weight, scale=1.0):
+ def truncated_normal(uniform, mu=0.0, sigma=1.0, a=-2, b=2):
+ normal = torch.distributions.normal.Normal(0, 1)
+
+ alpha = (a - mu) / sigma
+ beta = (b - mu) / sigma
+
+ alpha_normal_cdf = normal.cdf(torch.tensor(alpha))
+ p = alpha_normal_cdf + (normal.cdf(torch.tensor(beta)) - alpha_normal_cdf) * uniform
+
+ v = torch.clamp(2 * p - 1, -1 + 1e-8, 1 - 1e-8)
+ x = mu + sigma * np.sqrt(2) * torch.erfinv(v)
+ x = torch.clamp(x, a, b)
+
+ return x
+
+ def sample_truncated_normal(shape, scale=1.0):
+ stddev = np.sqrt(scale/shape[-1])/.87962566103423978 # shape[-1] = fan_in
+ return stddev * truncated_normal(torch.rand(shape))
+
+ weight = torch.nn.Parameter( (sample_truncated_normal(weight.shape)) )
+ return weight
+
+# for gradient checkpointing
+def create_custom_forward(module, **kwargs):
+ def custom_forward(*inputs):
+ return module(*inputs, **kwargs)
+ return custom_forward
+
+def get_clones(module, N):
+ return nn.ModuleList([copy.deepcopy(module) for i in range(N)])
+
+class Dropout(nn.Module):
+ # Dropout entire row or column
+ def __init__(self, broadcast_dim=None, p_drop=0.15):
+ super(Dropout, self).__init__()
+ # give ones with probability of 1-p_drop / zeros with p_drop
+ self.sampler = torch.distributions.bernoulli.Bernoulli(torch.tensor([1-p_drop]))
+ self.broadcast_dim=broadcast_dim
+ self.p_drop=p_drop
+ def forward(self, x):
+ if not self.training: # no drophead during evaluation mode
+ return x
+ shape = list(x.shape)
+ if not self.broadcast_dim == None:
+ shape[self.broadcast_dim] = 1
+ mask = self.sampler.sample(shape).to(x.device).view(shape)
+
+ x = mask * x / (1.0 - self.p_drop)
+ return x
+
+def rbf(D):
+ # Distance radial basis function
+ D_min, D_max, D_count = 0., 20., 36
+ D_mu = torch.linspace(D_min, D_max, D_count).to(D.device)
+ D_mu = D_mu[None,:]
+ D_sigma = (D_max - D_min) / D_count
+ D_expand = torch.unsqueeze(D, -1)
+ RBF = torch.exp(-((D_expand - D_mu) / D_sigma)**2)
+ return RBF
+
+def get_seqsep(idx):
+ '''
+ Input:
+ - idx: residue indices of given sequence (B,L)
+ Output:
+ - seqsep: sequence separation feature with sign (B, L, L, 1)
+ Sergey found that having sign in seqsep features helps a little
+ '''
+ seqsep = idx[:,None,:] - idx[:,:,None]
+ sign = torch.sign(seqsep)
+ neigh = torch.abs(seqsep)
+ neigh[neigh > 1] = 0.0 # if bonded -- 1.0 / else 0.0
+ neigh = sign * neigh
+ return neigh.unsqueeze(-1)
+
+def make_full_graph(xyz, pair, idx, top_k=64, kmin=9):
+ '''
+ Input:
+ - xyz: current backbone cooordinates (B, L, 3, 3)
+ - pair: pair features from Trunk (B, L, L, E)
+ - idx: residue index from ground truth pdb
+ Output:
+ - G: defined graph
+ '''
+
+ B, L = xyz.shape[:2]
+ device = xyz.device
+
+ # seq sep
+ sep = idx[:,None,:] - idx[:,:,None]
+ b,i,j = torch.where(sep.abs() > 0)
+
+ src = b*L+i
+ tgt = b*L+j
+ G = dgl.graph((src, tgt), num_nodes=B*L).to(device)
+ G.edata['rel_pos'] = (xyz[b,j,:] - xyz[b,i,:]).detach() # no gradient through basis function
+
+ return G, pair[b,i,j][...,None]
+
+def make_topk_graph(xyz, pair, idx, top_k=64, kmin=32, eps=1e-6):
+ '''
+ Input:
+ - xyz: current backbone cooordinates (B, L, 3, 3)
+ - pair: pair features from Trunk (B, L, L, E)
+ - idx: residue index from ground truth pdb
+ Output:
+ - G: defined graph
+ '''
+
+ B, L = xyz.shape[:2]
+ device = xyz.device
+
+ # distance map from current CA coordinates
+ D = torch.cdist(xyz, xyz) + torch.eye(L, device=device).unsqueeze(0)*999.9 # (B, L, L)
+ # seq sep
+ sep = idx[:,None,:] - idx[:,:,None]
+ sep = sep.abs() + torch.eye(L, device=device).unsqueeze(0)*999.9
+ D = D + sep*eps
+
+ # get top_k neighbors
+ D_neigh, E_idx = torch.topk(D, min(top_k, L), largest=False) # shape of E_idx: (B, L, top_k)
+ topk_matrix = torch.zeros((B, L, L), device=device)
+ topk_matrix.scatter_(2, E_idx, 1.0)
+
+ # put an edge if any of the 3 conditions are met:
+ # 1) |i-j| <= kmin (connect sequentially adjacent residues)
+ # 2) top_k neighbors
+ cond = torch.logical_or(topk_matrix > 0.0, sep < kmin)
+ b,i,j = torch.where(cond)
+
+ src = b*L+i
+ tgt = b*L+j
+ G = dgl.graph((src, tgt), num_nodes=B*L).to(device)
+ G.edata['rel_pos'] = (xyz[b,j,:] - xyz[b,i,:]).detach() # no gradient through basis function
+
+ return G, pair[b,i,j][...,None]
+
+def make_rotX(angs, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ RTs[:,:,1,1] = angs[:,:,0]/NORM
+ RTs[:,:,1,2] = -angs[:,:,1]/NORM
+ RTs[:,:,2,1] = angs[:,:,1]/NORM
+ RTs[:,:,2,2] = angs[:,:,0]/NORM
+ return RTs
+
+# rotate about the z axis
+def make_rotZ(angs, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ RTs[:,:,0,0] = angs[:,:,0]/NORM
+ RTs[:,:,0,1] = -angs[:,:,1]/NORM
+ RTs[:,:,1,0] = angs[:,:,1]/NORM
+ RTs[:,:,1,1] = angs[:,:,0]/NORM
+ return RTs
+
+# rotate about an arbitrary axis
+def make_rot_axis(angs, u, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ ct = angs[:,:,0]/NORM
+ st = angs[:,:,1]/NORM
+ u0 = u[:,:,0]
+ u1 = u[:,:,1]
+ u2 = u[:,:,2]
+
+ RTs[:,:,0,0] = ct+u0*u0*(1-ct)
+ RTs[:,:,0,1] = u0*u1*(1-ct)-u2*st
+ RTs[:,:,0,2] = u0*u2*(1-ct)+u1*st
+ RTs[:,:,1,0] = u0*u1*(1-ct)+u2*st
+ RTs[:,:,1,1] = ct+u1*u1*(1-ct)
+ RTs[:,:,1,2] = u1*u2*(1-ct)-u0*st
+ RTs[:,:,2,0] = u0*u2*(1-ct)-u1*st
+ RTs[:,:,2,1] = u1*u2*(1-ct)+u0*st
+ RTs[:,:,2,2] = ct+u2*u2*(1-ct)
+ return RTs
+
+class ComputeAllAtomCoords(nn.Module):
+ def __init__(self):
+ super(ComputeAllAtomCoords, self).__init__()
+
+ self.base_indices = nn.Parameter(base_indices, requires_grad=False)
+ self.RTs_in_base_frame = nn.Parameter(RTs_by_torsion, requires_grad=False)
+ self.xyzs_in_base_frame = nn.Parameter(xyzs_in_base_frame, requires_grad=False)
+
+ def forward(self, seq, xyz, alphas, non_ideal=False, use_H=True):
+ B,L = xyz.shape[:2]
+
+ Rs, Ts = rigid_from_3_points(xyz[...,0,:],xyz[...,1,:],xyz[...,2,:], non_ideal=non_ideal)
+
+ RTF0 = torch.eye(4).repeat(B,L,1,1).to(device=Rs.device)
+
+ # bb
+ RTF0[:,:,:3,:3] = Rs
+ RTF0[:,:,:3,3] = Ts
+
+ # omega
+ RTF1 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,0,:], make_rotX(alphas[:,:,0,:]))
+
+ # phi
+ RTF2 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,1,:], make_rotX(alphas[:,:,1,:]))
+
+ # psi
+ RTF3 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,2,:], make_rotX(alphas[:,:,2,:]))
+
+ # CB bend
+ basexyzs = self.xyzs_in_base_frame[seq]
+ NCr = 0.5*(basexyzs[:,:,2,:3]+basexyzs[:,:,0,:3])
+ CAr = (basexyzs[:,:,1,:3])
+ CBr = (basexyzs[:,:,4,:3])
+ CBrotaxis1 = (CBr-CAr).cross(NCr-CAr)
+ CBrotaxis1 /= torch.linalg.norm(CBrotaxis1, dim=-1, keepdim=True)+1e-8
+
+ # CB twist
+ NCp = basexyzs[:,:,2,:3] - basexyzs[:,:,0,:3]
+ NCpp = NCp - torch.sum(NCp*NCr, dim=-1, keepdim=True)/ torch.sum(NCr*NCr, dim=-1, keepdim=True) * NCr
+ CBrotaxis2 = (CBr-CAr).cross(NCpp)
+ CBrotaxis2 /= torch.linalg.norm(CBrotaxis2, dim=-1, keepdim=True)+1e-8
+
+ CBrot1 = make_rot_axis(alphas[:,:,7,:], CBrotaxis1 )
+ CBrot2 = make_rot_axis(alphas[:,:,8,:], CBrotaxis2 )
+
+ RTF8 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, CBrot1,CBrot2)
+
+ # chi1 + CG bend
+ RTF4 = torch.einsum(
+ 'brij,brjk,brkl,brlm->brim',
+ RTF8,
+ self.RTs_in_base_frame[seq,3,:],
+ make_rotX(alphas[:,:,3,:]),
+ make_rotZ(alphas[:,:,9,:]))
+
+ # chi2
+ RTF5 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF4, self.RTs_in_base_frame[seq,4,:],make_rotX(alphas[:,:,4,:]))
+
+ # chi3
+ RTF6 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF5,self.RTs_in_base_frame[seq,5,:],make_rotX(alphas[:,:,5,:]))
+
+ # chi4
+ RTF7 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF6,self.RTs_in_base_frame[seq,6,:],make_rotX(alphas[:,:,6,:]))
+
+ RTframes = torch.stack((
+ RTF0,RTF1,RTF2,RTF3,RTF4,RTF5,RTF6,RTF7,RTF8
+ ),dim=2)
+
+ xyzs = torch.einsum(
+ 'brtij,brtj->brti',
+ RTframes.gather(2,self.base_indices[seq][...,None,None].repeat(1,1,1,4,4)), basexyzs
+ )
+
+ if use_H:
+ return RTframes, xyzs[...,:3]
+ else:
+ return RTframes, xyzs[...,:14,:3]
diff --git a/model/utils/.inf_methods.py.swp b/model/utils/.inf_methods.py.swp
new file mode 100644
index 0000000000000000000000000000000000000000..1638aa8c4baf5a8ded77e8d71bda84a9f34fce3c
Binary files /dev/null and b/model/utils/.inf_methods.py.swp differ
diff --git a/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py b/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..fe2b975316f2de89d021d4dff442182192d5b7f8
--- /dev/null
+++ b/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py
@@ -0,0 +1,234 @@
+#@title get secondary structure (SSE) from given PDB file
+#@markdown So far it seems the best solution is to steal code from biotite
+#@markdown which calculates the SSE of a peptide chain based on the P-SEA algorithm (Labesse 1997)
+# CODE FROM BIOKITE
+# From Krypton
+import numpy as np
+import random
+import torch
+
+def vector_dot(v1,v2):
+ return (v1*v2).sum(axis=-1)
+
+def norm_vector(v):
+ factor = np.linalg.norm(v, axis=-1)
+ if isinstance(factor, np.ndarray):
+ v /= factor[..., np.newaxis]
+ else:
+ v /= factor
+ return v
+
+def coord(x):
+ return np.asarray(x)
+def displacement(atoms1, atoms2):
+ v1 = coord(atoms1)
+ v2 = coord(atoms2)
+ if len(v1.shape) <= len(v2.shape):
+ diff = v2 - v1
+ else:
+ diff = -(v1 - v2)
+ return diff
+def distance(atoms1, atoms2):
+ diff = displacement(atoms1, atoms2)
+ return np.sqrt(vector_dot(diff, diff))
+
+def angle(atoms1, atoms2, atoms3):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms3, atoms2)
+ norm_vector(v1)
+ norm_vector(v2)
+ return np.arccos(vector_dot(v1,v2))
+
+def dihedral(atoms1, atoms2, atoms3, atoms4):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms2, atoms3)
+ v3 = displacement(atoms3, atoms4)
+ norm_vector(v1)
+ norm_vector(v2)
+ norm_vector(v3)
+
+ n1 = np.cross(v1, v2)
+ n2 = np.cross(v2, v3)
+
+ # Calculation using atan2, to ensure the correct sign of the angle
+ x = vector_dot(n1,n2)
+ y = vector_dot(np.cross(n1,n2), v2)
+ return np.arctan2(y,x)
+
+def replace_letters(arr):
+ # Create a dictionary that maps the letters 'a', 'b', and 'c' to the corresponding numbers
+ letter_to_number = {'a': 0, 'b': 1, 'c': 2}
+
+ # Create a new array that will hold the numbers
+ nums = []
+
+ # Loop through the input array and replace the letters with the corresponding numbers
+ for letter in arr:
+ if letter in letter_to_number:
+ nums.append(letter_to_number[letter])
+ else:
+ nums.append(letter)
+
+ return np.array(nums)
+
+def replace_with_mask(arr, percentage, replace_loops=False):
+ # Make sure the percentage is between 0 and 100
+ percentage = min(max(percentage, 0), 100)
+
+ # Calculate the number of values to replace
+ num_to_replace = int(len(arr) * percentage / 100)
+
+ # Choose a random subset of the array to replace
+ replace_indices = random.sample(range(len(arr)), num_to_replace)
+
+ # Replace the values at the chosen indices with the number 3
+ for i in replace_indices:
+ arr[i] = 3
+
+ if replace_loops:
+ for i in arr:
+ if arr[i] == 2:
+ arr[i] = 3
+
+ return arr
+
+def annotate_sse(ca_coord, percentage_mask=0, replace_loops=False):
+ _radians_to_angle = 2*np.pi/360
+
+ _r_helix = ((89-12)*_radians_to_angle, (89+12)*_radians_to_angle)
+ _a_helix = ((50-20)*_radians_to_angle, (50+20)*_radians_to_angle)
+ _d2_helix = ((5.5-0.5), (5.5+0.5))
+ _d3_helix = ((5.3-0.5), (5.3+0.5))
+ _d4_helix = ((6.4-0.6), (6.4+0.6))
+
+ _r_strand = ((124-14)*_radians_to_angle, (124+14)*_radians_to_angle)
+ _a_strand = ((-180)*_radians_to_angle, (-125)*_radians_to_angle,
+ (145)*_radians_to_angle, (180)*_radians_to_angle)
+ _d2_strand = ((6.7-0.6), (6.7+0.6))
+ _d3_strand = ((9.9-0.9), (9.9+0.9))
+ _d4_strand = ((12.4-1.1), (12.4+1.1))
+
+ # Filter all CA atoms in the relevant chain.
+
+ d2i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d3i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d4i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ ri_coord = np.full(( len(ca_coord), 3, 3 ), np.nan)
+ ai_coord = np.full(( len(ca_coord), 4, 3 ), np.nan)
+
+ # The distances and angles are not defined for the entire interval,
+ # therefore the indices do not have the full range
+ # Values that are not defined are NaN
+ for i in range(1, len(ca_coord)-1):
+ d2i_coord[i] = (ca_coord[i-1], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ d3i_coord[i] = (ca_coord[i-1], ca_coord[i+2])
+ for i in range(1, len(ca_coord)-3):
+ d4i_coord[i] = (ca_coord[i-1], ca_coord[i+3])
+ for i in range(1, len(ca_coord)-1):
+ ri_coord[i] = (ca_coord[i-1], ca_coord[i], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ ai_coord[i] = (ca_coord[i-1], ca_coord[i],
+ ca_coord[i+1], ca_coord[i+2])
+
+ d2i = distance(d2i_coord[:,0], d2i_coord[:,1])
+ d3i = distance(d3i_coord[:,0], d3i_coord[:,1])
+ d4i = distance(d4i_coord[:,0], d4i_coord[:,1])
+ ri = angle(ri_coord[:,0], ri_coord[:,1], ri_coord[:,2])
+ ai = dihedral(ai_coord[:,0], ai_coord[:,1],
+ ai_coord[:,2], ai_coord[:,3])
+
+ sse = np.full(len(ca_coord), "c", dtype="U1")
+
+ # Annotate helices
+ # Find CA that meet criteria for potential helices
+ is_pot_helix = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if (
+ d3i[i] >= _d3_helix[0] and d3i[i] <= _d3_helix[1]
+ and d4i[i] >= _d4_helix[0] and d4i[i] <= _d4_helix[1]
+ ) or (
+ ri[i] >= _r_helix[0] and ri[i] <= _r_helix[1]
+ and ai[i] >= _a_helix[0] and ai[i] <= _a_helix[1]
+ ):
+ is_pot_helix[i] = True
+ # Real helices are 5 consecutive helix elements
+ is_helix = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ for i in range(len(sse)):
+ if is_pot_helix[i]:
+ counter += 1
+ else:
+ if counter >= 5:
+ is_helix[i-counter : i] = True
+ counter = 0
+ # Extend the helices by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_helix[i]:
+ sse[i] = "a"
+ if (
+ d3i[i-1] >= _d3_helix[0] and d3i[i-1] <= _d3_helix[1]
+ ) or (
+ ri[i-1] >= _r_helix[0] and ri[i-1] <= _r_helix[1]
+ ):
+ sse[i-1] = "a"
+ sse[i] = "a"
+ if (
+ d3i[i+1] >= _d3_helix[0] and d3i[i+1] <= _d3_helix[1]
+ ) or (
+ ri[i+1] >= _r_helix[0] and ri[i+1] <= _r_helix[1]
+ ):
+ sse[i+1] = "a"
+ i += 1
+
+ # Annotate sheets
+ # Find CA that meet criteria for potential strands
+ is_pot_strand = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if ( d2i[i] >= _d2_strand[0] and d2i[i] <= _d2_strand[1]
+ and d3i[i] >= _d3_strand[0] and d3i[i] <= _d3_strand[1]
+ and d4i[i] >= _d4_strand[0] and d4i[i] <= _d4_strand[1]
+ ) or (
+ ri[i] >= _r_strand[0] and ri[i] <= _r_strand[1]
+ and ( (ai[i] >= _a_strand[0] and ai[i] <= _a_strand[1])
+ or (ai[i] >= _a_strand[2] and ai[i] <= _a_strand[3]))
+ ):
+ is_pot_strand[i] = True
+ # Real strands are 5 consecutive strand elements,
+ # or shorter fragments of at least 3 consecutive strand residues,
+ # if they are in hydrogen bond proximity to 5 other residues
+ pot_strand_coord = ca_coord[is_pot_strand]
+ is_strand = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ contacts = 0
+ for i in range(len(sse)):
+ if is_pot_strand[i]:
+ counter += 1
+ coord = ca_coord[i]
+ for strand_coord in ca_coord:
+ dist = distance(coord, strand_coord)
+ if dist >= 4.2 and dist <= 5.2:
+ contacts += 1
+ else:
+ if counter >= 4:
+ is_strand[i-counter : i] = True
+ elif counter == 3 and contacts >= 5:
+ is_strand[i-counter : i] = True
+ counter = 0
+ contacts = 0
+ # Extend the strands by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_strand[i]:
+ sse[i] = "b"
+ if d3i[i-1] >= _d3_strand[0] and d3i[i-1] <= _d3_strand[1]:
+ sse[i-1] = "b"
+ sse[i] = "b"
+ if d3i[i+1] >= _d3_strand[0] and d3i[i+1] <= _d3_strand[1]:
+ sse[i+1] = "b"
+ i += 1
+ sse=replace_letters(sse)
+ sse=replace_with_mask(sse, percentage_mask, replace_loops=replace_loops)
+ sse=torch.nn.functional.one_hot(torch.tensor(sse), num_classes=4)
+ return sse
diff --git a/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py b/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..2fde867439dc3a604f03a9af67f88992148db71a
--- /dev/null
+++ b/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
@@ -0,0 +1,292 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None):
+ """
+ RFnar inference
+ """
+ seq_mask = ~input_seq_mask
+ str_mask = input_str_mask
+ nar = Nonautoregressive()
+ r = t
+ print(f'USING THIS R: {r}')
+
+ #mask sequence
+ if mask_seq_token:
+ print("MASK SEQ TOKEN")
+ seq_corrupt[seq_mask] = self.masked_token
+ elif mask_seq_random:
+ print("MASK SEQ RANDOM")
+
+
+ ic(seq_corrupt)
+
+ seq=seq_corrupt.repeat(seq.shape[0], 1)
+ seq_corrupt_onehot=torch.nn.functional.one_hot(seq_corrupt,num_classes=22).float()
+
+ ### msa_masked ###
+ ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_corrupt_onehot
+
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ #msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_corrupt_onehot
+ #msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ # msa_full #
+ #make msa_full same size as msa_masked
+ ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_corrupt_onehot
+ #msa_full[:,1:,:,:22] = msa_diffused
+
+ ###########
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_corrupt_onehot[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,seq_mask,21] = 0.0
+ t1d[:,~seq_mask,21] = 1.0
+
+ #to do add structure confidence metric; need to expand dimensions of chkpt b4
+ if t1d_24:
+ print("expanding t1d dim to 24")
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,str_mask,23] = 0.0
+ t1d[:,~str_mask,23] = 1.0
+
+ t1d[:1,:,22] = r
+
+ ################
+ #mask structure#
+ ################
+ if mask_xyz_hole:
+ print("MASK XYZ BLACK HOLE")
+ ic(xyz_t.shape)
+ xyz_corrupt, xyz_mask = nar.xyz_mask_0(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True, cos=True)
+ ic(xyz_corrupt.shape)
+ elif mask_xyz_random:
+ print("MASK XYZ RANDOM")
+ xyz_corrupt, xyz_mask = nar.xyz_mask_random(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True, cos=True)
+ #only corrupt first template
+ xyz_t[0]=xyz_corrupt
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+ mask_msa[:,:,~loss_seq_mask] = False
+
+ seq_diffused = seq_corrupt_onehot
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, RFnar = False):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = None #seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+#def take_step_nar():
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+
diff --git a/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py b/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..0fc7f7ef6e3b7619fd8902acae4e5d8f69ad993b
--- /dev/null
+++ b/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py
@@ -0,0 +1,376 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+def mask_inputs_RFnar(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, nar= None, t=None, T= None, mask_seq_token=True, mask_seq_random=False, mask_xyz_hole=True, mask_xyz_random=False):
+ """
+ RFnar inference
+ """
+ ic(seq.shape)
+ seq = seq[0,:1]
+ ic(seq.shape)
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = ~input_seq_mask[0]
+ ic(seq_mask.shape)
+
+ str_mask = ~input_str_mask[0]
+ #nar = Nonautoregressive()
+ r = (T-t)/T
+ print(f'USING THIS R: {r}')
+
+ #mask sequence
+ if mask_seq_token:
+ print("MASK SEQ TOKEN")
+ seq_corrupt = seq.clone()
+ seq_corrupt[:,seq_mask] = 21
+ ic(seq)
+ ic(seq_corrupt)
+ elif mask_seq_random:
+ print("MASK SEQ RANDOM")
+
+
+ ic(seq_corrupt)
+
+ seq=seq_corrupt.repeat(seq.shape[0], 1)
+ ic(seq.shape)
+ seq_corrupt_onehot=torch.nn.functional.one_hot(seq_corrupt,num_classes=22).float()[0]
+ ic(seq_corrupt_onehot.shape)
+
+ ### msa_masked ###
+ ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_corrupt_onehot
+
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ #msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_corrupt_onehot
+ #msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ # msa_full #
+ #make msa_full same size as msa_masked
+ ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_corrupt_onehot
+ #msa_full[:,1:,:,:22] = msa_diffused
+
+ ###########
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_corrupt_onehot[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,seq_mask,21] = 0.0
+ t1d[:,~seq_mask,21] = 1.0
+
+ t1d[:,str_mask,23] = 0.0
+ t1d[:,~str_mask,23] = 1.0
+
+ t1d[:1,:,22] = r
+
+ ################
+ #mask structure#
+ ################
+ if mask_xyz_hole:
+ print("MASK XYZ BLACK HOLE")
+ ic(xyz_t.shape)
+ xyz_corrupt, xyz_mask = nar.xyz_mask_0(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True)
+ ic(xyz_corrupt.shape)
+ elif mask_xyz_random:
+ print("MASK XYZ RANDOM")
+ xyz_corrupt, xyz_mask = nar.xyz_mask_random(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True)
+ #only corrupt first template
+ xyz_t[0]=xyz_corrupt
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+ seq_diffused = seq_corrupt_onehot
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, T=None, RFnar = False):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ if RFnar:
+
+ return mask_inputs_RFnar(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=input_seq_mask,
+ input_str_mask=input_str_mask, input_t1dconf_mask=input_t1dconf_mask, nar=diffuser, t=t, T=T)
+
+
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape )
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(xyz_prev.shape)
+ #xyz_prev = xyz_prev[-1]
+ #ic(xyz_prev.shape)
+
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ #ic(t1d.shape)
+ t1d[:,:,:,:21] = logit_aa_s[0,:21,:].permute(1,0)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
diff --git a/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py b/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..4350df63bd2ad5f5397d0d032c6cf2f200378c99
--- /dev/null
+++ b/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'wt') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py b/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..1a8bc9aca6de872a4e2b4388937c4d85e6b8d6b1
--- /dev/null
+++ b/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
@@ -0,0 +1,151 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ i[pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/model/utils/__pycache__/calc_dssp.cpython-310.pyc b/model/utils/__pycache__/calc_dssp.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..a8099164d1068e06d85c4b12051a8f548eaaddfc
Binary files /dev/null and b/model/utils/__pycache__/calc_dssp.cpython-310.pyc differ
diff --git a/model/utils/calc_dssp.py b/model/utils/calc_dssp.py
new file mode 100644
index 0000000000000000000000000000000000000000..fe2b975316f2de89d021d4dff442182192d5b7f8
--- /dev/null
+++ b/model/utils/calc_dssp.py
@@ -0,0 +1,234 @@
+#@title get secondary structure (SSE) from given PDB file
+#@markdown So far it seems the best solution is to steal code from biotite
+#@markdown which calculates the SSE of a peptide chain based on the P-SEA algorithm (Labesse 1997)
+# CODE FROM BIOKITE
+# From Krypton
+import numpy as np
+import random
+import torch
+
+def vector_dot(v1,v2):
+ return (v1*v2).sum(axis=-1)
+
+def norm_vector(v):
+ factor = np.linalg.norm(v, axis=-1)
+ if isinstance(factor, np.ndarray):
+ v /= factor[..., np.newaxis]
+ else:
+ v /= factor
+ return v
+
+def coord(x):
+ return np.asarray(x)
+def displacement(atoms1, atoms2):
+ v1 = coord(atoms1)
+ v2 = coord(atoms2)
+ if len(v1.shape) <= len(v2.shape):
+ diff = v2 - v1
+ else:
+ diff = -(v1 - v2)
+ return diff
+def distance(atoms1, atoms2):
+ diff = displacement(atoms1, atoms2)
+ return np.sqrt(vector_dot(diff, diff))
+
+def angle(atoms1, atoms2, atoms3):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms3, atoms2)
+ norm_vector(v1)
+ norm_vector(v2)
+ return np.arccos(vector_dot(v1,v2))
+
+def dihedral(atoms1, atoms2, atoms3, atoms4):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms2, atoms3)
+ v3 = displacement(atoms3, atoms4)
+ norm_vector(v1)
+ norm_vector(v2)
+ norm_vector(v3)
+
+ n1 = np.cross(v1, v2)
+ n2 = np.cross(v2, v3)
+
+ # Calculation using atan2, to ensure the correct sign of the angle
+ x = vector_dot(n1,n2)
+ y = vector_dot(np.cross(n1,n2), v2)
+ return np.arctan2(y,x)
+
+def replace_letters(arr):
+ # Create a dictionary that maps the letters 'a', 'b', and 'c' to the corresponding numbers
+ letter_to_number = {'a': 0, 'b': 1, 'c': 2}
+
+ # Create a new array that will hold the numbers
+ nums = []
+
+ # Loop through the input array and replace the letters with the corresponding numbers
+ for letter in arr:
+ if letter in letter_to_number:
+ nums.append(letter_to_number[letter])
+ else:
+ nums.append(letter)
+
+ return np.array(nums)
+
+def replace_with_mask(arr, percentage, replace_loops=False):
+ # Make sure the percentage is between 0 and 100
+ percentage = min(max(percentage, 0), 100)
+
+ # Calculate the number of values to replace
+ num_to_replace = int(len(arr) * percentage / 100)
+
+ # Choose a random subset of the array to replace
+ replace_indices = random.sample(range(len(arr)), num_to_replace)
+
+ # Replace the values at the chosen indices with the number 3
+ for i in replace_indices:
+ arr[i] = 3
+
+ if replace_loops:
+ for i in arr:
+ if arr[i] == 2:
+ arr[i] = 3
+
+ return arr
+
+def annotate_sse(ca_coord, percentage_mask=0, replace_loops=False):
+ _radians_to_angle = 2*np.pi/360
+
+ _r_helix = ((89-12)*_radians_to_angle, (89+12)*_radians_to_angle)
+ _a_helix = ((50-20)*_radians_to_angle, (50+20)*_radians_to_angle)
+ _d2_helix = ((5.5-0.5), (5.5+0.5))
+ _d3_helix = ((5.3-0.5), (5.3+0.5))
+ _d4_helix = ((6.4-0.6), (6.4+0.6))
+
+ _r_strand = ((124-14)*_radians_to_angle, (124+14)*_radians_to_angle)
+ _a_strand = ((-180)*_radians_to_angle, (-125)*_radians_to_angle,
+ (145)*_radians_to_angle, (180)*_radians_to_angle)
+ _d2_strand = ((6.7-0.6), (6.7+0.6))
+ _d3_strand = ((9.9-0.9), (9.9+0.9))
+ _d4_strand = ((12.4-1.1), (12.4+1.1))
+
+ # Filter all CA atoms in the relevant chain.
+
+ d2i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d3i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d4i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ ri_coord = np.full(( len(ca_coord), 3, 3 ), np.nan)
+ ai_coord = np.full(( len(ca_coord), 4, 3 ), np.nan)
+
+ # The distances and angles are not defined for the entire interval,
+ # therefore the indices do not have the full range
+ # Values that are not defined are NaN
+ for i in range(1, len(ca_coord)-1):
+ d2i_coord[i] = (ca_coord[i-1], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ d3i_coord[i] = (ca_coord[i-1], ca_coord[i+2])
+ for i in range(1, len(ca_coord)-3):
+ d4i_coord[i] = (ca_coord[i-1], ca_coord[i+3])
+ for i in range(1, len(ca_coord)-1):
+ ri_coord[i] = (ca_coord[i-1], ca_coord[i], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ ai_coord[i] = (ca_coord[i-1], ca_coord[i],
+ ca_coord[i+1], ca_coord[i+2])
+
+ d2i = distance(d2i_coord[:,0], d2i_coord[:,1])
+ d3i = distance(d3i_coord[:,0], d3i_coord[:,1])
+ d4i = distance(d4i_coord[:,0], d4i_coord[:,1])
+ ri = angle(ri_coord[:,0], ri_coord[:,1], ri_coord[:,2])
+ ai = dihedral(ai_coord[:,0], ai_coord[:,1],
+ ai_coord[:,2], ai_coord[:,3])
+
+ sse = np.full(len(ca_coord), "c", dtype="U1")
+
+ # Annotate helices
+ # Find CA that meet criteria for potential helices
+ is_pot_helix = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if (
+ d3i[i] >= _d3_helix[0] and d3i[i] <= _d3_helix[1]
+ and d4i[i] >= _d4_helix[0] and d4i[i] <= _d4_helix[1]
+ ) or (
+ ri[i] >= _r_helix[0] and ri[i] <= _r_helix[1]
+ and ai[i] >= _a_helix[0] and ai[i] <= _a_helix[1]
+ ):
+ is_pot_helix[i] = True
+ # Real helices are 5 consecutive helix elements
+ is_helix = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ for i in range(len(sse)):
+ if is_pot_helix[i]:
+ counter += 1
+ else:
+ if counter >= 5:
+ is_helix[i-counter : i] = True
+ counter = 0
+ # Extend the helices by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_helix[i]:
+ sse[i] = "a"
+ if (
+ d3i[i-1] >= _d3_helix[0] and d3i[i-1] <= _d3_helix[1]
+ ) or (
+ ri[i-1] >= _r_helix[0] and ri[i-1] <= _r_helix[1]
+ ):
+ sse[i-1] = "a"
+ sse[i] = "a"
+ if (
+ d3i[i+1] >= _d3_helix[0] and d3i[i+1] <= _d3_helix[1]
+ ) or (
+ ri[i+1] >= _r_helix[0] and ri[i+1] <= _r_helix[1]
+ ):
+ sse[i+1] = "a"
+ i += 1
+
+ # Annotate sheets
+ # Find CA that meet criteria for potential strands
+ is_pot_strand = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if ( d2i[i] >= _d2_strand[0] and d2i[i] <= _d2_strand[1]
+ and d3i[i] >= _d3_strand[0] and d3i[i] <= _d3_strand[1]
+ and d4i[i] >= _d4_strand[0] and d4i[i] <= _d4_strand[1]
+ ) or (
+ ri[i] >= _r_strand[0] and ri[i] <= _r_strand[1]
+ and ( (ai[i] >= _a_strand[0] and ai[i] <= _a_strand[1])
+ or (ai[i] >= _a_strand[2] and ai[i] <= _a_strand[3]))
+ ):
+ is_pot_strand[i] = True
+ # Real strands are 5 consecutive strand elements,
+ # or shorter fragments of at least 3 consecutive strand residues,
+ # if they are in hydrogen bond proximity to 5 other residues
+ pot_strand_coord = ca_coord[is_pot_strand]
+ is_strand = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ contacts = 0
+ for i in range(len(sse)):
+ if is_pot_strand[i]:
+ counter += 1
+ coord = ca_coord[i]
+ for strand_coord in ca_coord:
+ dist = distance(coord, strand_coord)
+ if dist >= 4.2 and dist <= 5.2:
+ contacts += 1
+ else:
+ if counter >= 4:
+ is_strand[i-counter : i] = True
+ elif counter == 3 and contacts >= 5:
+ is_strand[i-counter : i] = True
+ counter = 0
+ contacts = 0
+ # Extend the strands by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_strand[i]:
+ sse[i] = "b"
+ if d3i[i-1] >= _d3_strand[0] and d3i[i-1] <= _d3_strand[1]:
+ sse[i-1] = "b"
+ sse[i] = "b"
+ if d3i[i+1] >= _d3_strand[0] and d3i[i+1] <= _d3_strand[1]:
+ sse[i+1] = "b"
+ i += 1
+ sse=replace_letters(sse)
+ sse=replace_with_mask(sse, percentage_mask, replace_loops=replace_loops)
+ sse=torch.nn.functional.one_hot(torch.tensor(sse), num_classes=4)
+ return sse
diff --git a/model/utils/contigs.py b/model/utils/contigs.py
new file mode 100644
index 0000000000000000000000000000000000000000..f0057e729528392d3b297d49aec8a7db901b12f2
--- /dev/null
+++ b/model/utils/contigs.py
@@ -0,0 +1,1415 @@
+# utility functions for dealing with contigs during hallucination
+import numpy as np
+import random, copy, torch, geometry, os, sys
+from kinematics import xyz_to_t2d
+
+def parse_range_string(el):
+ ''' Splits string with integer or integer range into start and end ints. '''
+ if '-' in el:
+ s,e = el.split('-')
+ s,e = int(s), int(e)
+ else:
+ s,e = int(el), int(el)
+ return s,e
+
+def ranges_to_indexes(range_string):
+ '''Converts a string containig comma-separated numeric ranges to a list of integers'''
+ idx = []
+ for x in range_string.split(','):
+ start, end = parse_range_string(x)
+ idx.extend(np.arange(start, end+1))
+ return np.array(idx)
+
+def parse_contigs(contig_input, pdb_id):
+ '''
+ Input: contig start/end by pdb chain and residue number as in the pdb file
+ ex - B12-17
+ Output: corresponding start/end indices of the "features" numpy array (idx0)
+ '''
+ contigs = []
+ for con in contig_input.split(','):
+ pdb_ch = con[0]
+ pdb_s, pdb_e = parse_range_string(con[1:])
+
+ np_s = pdb_id.index((pdb_ch, pdb_s))
+ np_e = pdb_id.index((pdb_ch, pdb_e))
+
+ contigs.append([np_s, np_e])
+ return contigs
+
+
+def mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out):
+ #####################################
+ # rearrange ref features according to hal_2_ref_idx0
+ #####################################
+ #1. find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ #2. rearrange the 6D features
+ hal_len = len(hal_2_ref_idx0)
+ if 'feat' in pdb_out:
+ d_feat = pdb_out['feat'].shape[3:]
+
+ feat_hal = np.zeros((1, hal_len, hal_len) + d_feat)
+ feat_ref = pdb_out['feat'] # (B,L,L,...)
+
+ feat_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ else:
+ feat_hal = None
+
+ #3. make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+
+ #####################################
+ # mappings between hal and ref
+ #####################################
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_out['pdb_idx'][i] for i in ref_idx0],
+ 'mask_1d': mask_1d,
+ }
+
+ return feat_hal, mappings
+
+def scatter_feats(template_mask, feat_1d_ref=None, feat_2d_ref=None, pdb_idx=None):
+ '''
+ Scatters 1D and/or 2D reference features according to mappings in hal_2_ref_idx0
+
+ Inputs
+ ----------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ feat_1d_ref: (np.array; (batch, L_ref, ...))
+ 1D refence features to scatter
+ feat_1d_ref: (np.array; (batch, L_ref, L_ref, ...))
+ pdb_idx: (list)
+ List of pdb chain and residue numbers, in the order that pdb features were read/parsed.
+
+ Outputs
+ ----------
+ feat_1d_hal: (np.array, (batch, L_hal, ...))
+ Scattered 1d reference features. "None" mappings are 0.
+ feat_2d_hal: (np.array, (batch, L_hal, L_hal, ...))
+ Scattered 2d reference features. "None" mappings are 0.
+ mappings: (dict)
+ Keeps track of corresponding possitions in ref and hal proteins.
+ '''
+ hal_2_ref_idx0, _ = contigs.sample_mask(template_mask, pdb_idx)
+ out = {}
+
+ # Find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+ hal_len = len(hal_2_ref_idx0)
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ # Make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+ # scatter 2D features
+ if feat_2d_ref is not None:
+ B = feat_2d_ref.shape[0]
+ d_feat = feat_2d_ref.shape[3:]
+ feat_2d_hal = np.zeros((B, hal_len, hal_len)+d_feat)
+ feat_2d_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_2d_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ out['feat_2d_hal'] = feat_2d_hal
+
+ # scatter 1D features
+ if feat_1d_ref is not None:
+ B = feat_1d_ref.shape[0]
+ d_feat = feat_1d_ref.shape[2:]
+ feat_1d_hal = np.zeros((B, hal_len)+d_feat)
+ feat_1d_hal[:, hal_idx0] = feat_1d_ref[:, ref_idx0]
+ out['feat_1d_hal'] = feat_1d_hal
+
+ # Mappings between hal and ref
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'mask_1d': mask_1d,
+ }
+
+ if pdb_idx is not None:
+ mappings.update({
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_idx[i] for i in ref_idx0],
+ })
+
+ out['mappings'] = mappings
+
+ return out
+
+def scatter_contigs(contigs, pdb_out, L_range, keep_order=False, min_gap=0):
+ '''
+ Randomly places contigs in a protein within the length range.
+
+ Inputs
+ Contig: A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number. Comma separated
+ ex: B6-11,A12-19
+ pdb_out: dictionary from the prep_input function
+ L_range: String range of possible lengths.
+ ex: 90-110
+ ex: 70
+ keep_order: keep contigs in the provided order or randomly permute
+ min_gap: minimum number of amino acids separating contigs
+
+ Outputs
+ feat_hal: target pdb features to hallucinate
+ mappings: dictionary of ways to convert from the hallucinated protein
+ to the reference protein
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #####################################
+ # make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ #####################################
+ #1. Permute contig order
+ contigs = contigs.split(',')
+
+ if not keep_order:
+ random.shuffle(contigs)
+
+ #2. convert to ref_idx0
+ contigs_ref_idx0 = []
+ for con in contigs:
+ chain = con[0]
+ s, e = parse_range_string(con[1:])
+ contigs_ref_idx0.append( [ref_pdb_2_idx0[(chain, i)] for i in range(s, e+1)] )
+
+ #3. Add minimum gap size
+ for i in range(len(contigs_ref_idx0) - 1):
+ contigs_ref_idx0[i] += [None] * min_gap
+
+ #4. Sample protein length
+ L_low, L_high = parse_range_string(L_range)
+ L_hal = np.random.randint(L_low, L_high+1)
+
+ L_con = 0
+ for con in contigs_ref_idx0:
+ L_con += len(con)
+
+ L_gaps = L_hal - L_con
+
+ if L_gaps <= 1:
+ print("Error: The protein isn't long enough to incorporate all the contigs."
+ "Consider reduce the min_gap or increasing L_range")
+ return
+
+ #5. Randomly insert contigs into gaps
+ hal_2_ref_idx0 = np.array([None] * L_gaps, dtype=float) # inserting contigs into this
+ n_contigs = len(contigs_ref_idx0)
+ insertion_idxs = np.random.randint(L_gaps + 1, size=n_contigs)
+ insertion_idxs.sort()
+
+ for idx, con in zip(insertion_idxs[::-1], contigs_ref_idx0[::-1]):
+ hal_2_ref_idx0 = np.insert(hal_2_ref_idx0, idx, con)
+
+ #6. Convert mask to feat_hal and mappings
+ hal_2_ref_idx0 = [int(el) if ~np.isnan(el) else None for el in hal_2_ref_idx0] # convert nan to None
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #7. Generate str of the sampled mask
+ contig_positive = np.array(hal_2_ref_idx0) != None
+ boundaries = np.where(np.diff(contig_positive))[0]
+ start_idx0 = np.concatenate([np.array([0]), boundaries+1])
+ end_idx0 = np.concatenate([boundaries, np.array([contig_positive.shape[0]])-1])
+ lengths = end_idx0 - start_idx0 + 1
+ is_contig = contig_positive[start_idx0]
+
+ sampled_mask = []
+ con_counter = 0
+
+ for i, is_con in enumerate(is_contig):
+ if is_con:
+ sampled_mask.append(contigs[con_counter])
+ con_counter += 1
+ else:
+ len_gap = lengths[i]
+ sampled_mask.append(f'{len_gap}-{len_gap}')
+
+ sampled_mask = ','.join(sampled_mask)
+ mappings['sampled_mask'] = sampled_mask
+
+ return feat_hal, mappings
+
+def get_receptor_contig(ref_pdb_idx):
+ rec_pdb_idx = [idx for idx in ref_pdb_idx if idx[0]=='R']
+ return SampledMask.contract(rec_pdb_idx)
+
+def mk_con_to_set(mask, set_id=None, args=None, ref_pdb_idx=None):
+ '''
+ Maps a mask or list of contigs to a set_id. If no set_id is provided, it treats
+ everything as set 0.
+
+ Input
+ -----------
+ mask (str): Mask or list of contigs. Ex: 3,B6-11,12,A12-19,9 or Ex: B6-11,A12-19
+ ref_pdb_idx (List(ch, res)): pdb idxs of the reference pdb. Ex: [(A, 2), (A, 3), ...]
+ args: Arguments object. Must have args.receptor
+ set_id (list): List of integers. Length must match contigs in mask. Ex: [0,1]
+
+ Output
+ -----------
+ con_to_set (dict): Maps str of contig to integer
+ '''
+
+ # Extract contigs
+ cons = [l for l in mask.split(',') if l[0].isalpha()]
+
+ # Assign all contigs to set 0 if set_id is not passed
+ if set_id is None:
+ set_id = [0] * len(cons)
+
+ con_to_set = dict(zip(cons, set_id))
+
+ # Assign receptor to set 0
+ if args.receptor:
+ receptor_contig = get_receptor_contig(ref_pdb_idx)
+ con_to_set.update({receptor_contig: 0})
+
+ return con_to_set
+
+def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = parse_range(contig)
+
+ return ch, s, e
+
+def mask_as_list(sampled_mask):
+ '''
+ Make a length L_hal list, with each position pointing to a ref_pdb_idx (or None)
+ '''
+ mask_list = []
+ for l in sampled_mask.split(','):
+ ch, s, e = parse_contig(l)
+ # contig
+ if ch is not None:
+ mask_list += [(ch, idx) for idx in range(s, e+1)]
+ # gap
+ else:
+ mask_list += [None for _ in range(s, e+1)]
+
+ return mask_list
+
+def mask_subset(sampled_mask, subset):
+ '''
+ Returns a 1D boolean array of where a subset of the contig is in the hallucinated protein
+
+ Input
+ ---------
+ subset (str): Some chain and residue subset of the contigs. Ex: A10-15
+ Can also just pass chain. All contig residues from that chain are selected. Ex: R
+
+ Ouput
+ ---------
+ m_1d (np.array): Boolean array where subset appears in the hallucinated protein
+
+ '''
+ mask_list = mask_as_list(sampled_mask)
+ m_1d = []
+
+ ch_subset, s, e = parse_contig(subset)
+ assert ch_subset.isalpha(), '"Subset" must include a chain reference'
+
+ if (s is None) or (e is None):
+ s = -np.inf
+ e = np.inf
+
+ for l in mask_list:
+ if l is None:
+ continue
+
+ ch, idx = l
+ if (ch == ch_subset) and (idx >= s) and (idx <= e):
+ m_1d.append(True)
+ else:
+ m_1d.append(False)
+
+ return np.array(m_1d)
+
+def mk_cce_and_hal_mask_2d(sampled_mask, con_to_set=None):
+ '''
+ Makes masks for ij pixels where the cce and hallucination loss should be applied.
+
+ Inputs
+ ---------------
+ sampled_mask (str): String of where contigs should be applied. Ex: 3,B6-11,12,A12-19,9
+ cce_cutoff (float): Apply cce loss to cb-cb distances less than this value. Angstroms.
+ con_to_set (dict): Dictionary mapping the string of a contig (ex: 'B6-11') to an integer.
+ L_rec (int): Length of the receptor, if hallucinating in the context of the receptor.
+
+ Outputs
+ ---------------
+ mask_cce (np.array, (L_hal, L_hal)): Boolean array. True where cce loss should be applied.
+ mask_hal (np.array, (L_hal, L_hal)): Boolean array. True where hallucination loss should be applied.
+ '''
+ if con_to_set is None:
+ con_to_set = mk_con_to_set(sampled_mask)
+
+ # Length of hallucinated protein
+ L_hal, L_max = mask_len(sampled_mask)
+ assert L_hal == L_max, 'A sampled mask must have gaps of a single length.'
+
+ # Map each contig to a 1D boolean mask
+ m_con = dict()
+ start_idx = 0
+ for l in sampled_mask.split(','):
+ if l[0].isalpha():
+ s, e = parse_range_string(l[1:])
+ L_con = e - s + 1
+ m = np.zeros(L_hal, dtype=bool)
+ m[start_idx:start_idx+L_con] = True
+
+ m_con[l] = m
+ start_idx += L_con
+ else:
+ L_gap, _ = parse_range_string(l)
+ start_idx += L_gap
+
+ # Combine contigs masks from each set to make 2D mask
+ mask_cce = np.zeros((L_hal, L_hal), dtype=bool)
+ for set_id in set(con_to_set.values()):
+ # gather all masks from contigs in the same set
+ masks = [m_con[k] for k,v in con_to_set.items() if v == set_id]
+ mask_1D = np.any(masks, axis=0)
+ update = mask_1D[:,None] * mask_1D[None,:]
+ mask_cce = np.any([mask_cce, update], axis=0)
+
+ # Make mask_hal
+ mask_hal = ~mask_cce
+
+ # Don't apply ANY losses on diagonal
+ mask_cce[np.arange(L_hal), np.arange(L_hal)] = False
+ mask_hal[np.arange(L_hal), np.arange(L_hal)] = False
+
+ # Don't apply ANY losses to receptor
+ m_1d_rec = mask_subset(sampled_mask, 'R')
+ m_2d_rec = m_1d_rec[:, None] * m_1d_rec[None, :]
+ mask_cce *= ~m_2d_rec
+ mask_hal *= ~m_2d_rec
+
+ return mask_cce, mask_hal
+
+
+def apply_mask(mask, pdb_out):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ pdb_out: dictionary from the prep_input function
+
+
+ Outputs
+ -------------
+ feat_hal: features from pdb_out scattered according to the sampled mask
+ mappings: dict keeping track of corresponding positions in the ref and hal features
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ #2. Convert mask to feat_hal and mappings
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #3. Record the mask that was sampled
+ mappings['sampled_mask'] = ','.join(sampled_mask)
+
+ return feat_hal, mappings
+
+
+def sample_mask(mask, pdb_idx):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ Outputs
+ -------------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ sampled_mask: (str)
+ string of the sampled mask, so the transformations can be reapplied
+ ex - '3-3,B6-11,9-9,A36-42,20-20,A12-24,5-5'
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_i:i for i, pdb_i in enumerate(pdb_idx)}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ return hal_2_ref_idx0, sampled_mask
+
+
+class GapResampler():
+ def __init__(self, use_bkg=True):
+ '''
+
+ '''
+
+ self.counts_passed = {} # dictionary for tallying counts of gap lengths for designs passing some threshold
+ self.counts_bkg = {}
+ self.use_bkg = use_bkg
+
+
+ def clean_mask(self, mask):
+ '''
+ Makes mask into a cononical form.
+ Ensures masks always alternate gap, contig and that
+ masks begin and end with a gap (even of length 0)
+
+ Input
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+
+ Output
+ -----------
+ A canonicalized mask. Ex: N,9,A12-19,15,B45-52,0,C
+ '''
+ mask = mask.split(',')
+ mask_out = []
+ was_contig = True
+ was_gap = False
+
+ for i, el in enumerate(mask):
+ is_contig = el[0].isalpha()
+ is_gap = not is_contig
+ is_last = i == len(mask) - 1
+
+ # accepting gaps as either x-x or just x
+ if is_gap:
+ if '-' in el:
+ x1, x2 = el.split('-')
+ if x1 != x2:
+ print(f"Error: Gap must not be a range: {mask}")
+ return None
+ gap = x1
+ else:
+ gap = el
+
+ if is_contig:
+ contig = el
+
+ # gap -> contig: just append new contig
+ if (was_gap and is_contig):
+ mask_out.append(contig)
+
+ # contig -> gap: just append gap
+ elif (was_contig and is_gap):
+ mask_out.append(gap)
+
+ # contig -> contig: insert gap of 0, then add contig
+ elif (was_contig and is_contig):
+ mask_out.append('0')
+ mask_out.append(contig)
+
+ # gap -> gap: add them
+ elif (was_gap and is_gap):
+ combined_len = int(mask_out[-1]) + int(gap)
+ mask_out[-1] = str(combined_len)
+
+ # ensure last mask element is a gap
+ if (is_last and is_contig):
+ mask_out.append('0')
+
+ # update what previous element was
+ was_contig = el[0].isalpha()
+ was_gap = ~is_contig
+
+ # add 'N' and 'C' contigs
+ mask_out.insert(0, 'N')
+ mask_out.append('C')
+
+ return ','.join(mask_out)
+
+
+ def add_mask(self, mask, counting_dict):
+ '''
+ Adds counts of gap lengths to counting_dict
+
+ Inputs
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+ '''
+ mask = self.clean_mask(mask)
+ mask = mask.split(',')
+ n_gaps = len(mask) // 2
+
+ # count occurances of contig,gap,contig triples
+ for i in range(n_gaps):
+ con1, gap, con2 = mask[2*i : 2*i+3]
+
+ # count gap length
+ if con1 in counting_dict:
+ if (gap, con2) in counting_dict[con1]:
+ counting_dict[con1][(gap, con2)] += 1
+ else:
+ counting_dict[con1][(gap, con2)] = 1
+ else:
+ counting_dict[con1] = {(gap, con2): 1}
+
+
+ def add_mask_pass(self, mask):
+ '''
+ Add a mask that passed to self.counts_passed
+ '''
+ self.add_mask(mask, self.counts_passed)
+
+
+ def add_mask_bkg(self, mask):
+ '''
+ Add a mask that passed to self.counts_bkg
+ '''
+ self.add_mask(mask, self.counts_bkg)
+
+
+ def get_enrichment(self):
+ '''
+ Calculate the ratio of counts_passed / count_bkg
+ Also notes all contigs
+ '''
+ if self.use_bkg is False:
+ print('Please pass in background masks and set self.use_bkg=True')
+ return
+
+ self.counts_enrich = copy.copy(self.counts_passed)
+ self.con_all = set()
+
+ for con1 in self.counts_enrich.keys():
+ self.con_all |= set([con1])
+
+ for gap, con2 in self.counts_enrich[con1].keys():
+ self.con_all |= set([con2])
+ bkg = self.counts_bkg[con1][(gap, con2)]
+ cnt = self.counts_passed[con1][(gap, con2)]
+ self.counts_enrich[con1][(gap, con2)] = cnt / bkg
+
+ def sample_mask(self):
+ '''
+ Sample a mask
+ '''
+ searching = True
+ while searching:
+ n_gaps = len(self.con_all) - 1
+ mask = ['N']
+
+ if self.use_bkg:
+ counts = self.counts_enrich
+ else:
+ counts = self.counts_passed
+
+ for i in range(n_gaps):
+ con_last = mask[-1]
+
+ # only allow jump to C as last option
+ if i == n_gaps - 1:
+ con_used = set(mask[::2])
+ else:
+ con_used = set(mask[::2]+['C'])
+
+ con_free = self.con_all - con_used
+
+ # get available "jumps" (con -> gap, con) you can make
+ jumps_all = counts[con_last]
+ jumps_free = {k:v for k,v in jumps_all.items() if k[1] in con_free}
+
+ if len(jumps_free) == 0:
+ print('No available jumps to continue the mask. Sampling again...')
+ else:
+ # normalize counts and sample move
+ mvs, cnt = zip(*jumps_free.items())
+ cnt = np.array(cnt)
+ prob = cnt / cnt.sum()
+ idx = np.random.choice(len(prob), p=prob)
+ mv = mvs[idx]
+
+ # add to the mask
+ mask.append(mv[0])
+ mask.append(mv[1])
+
+ # check that mask has the right number of elements
+ if len(mask) == 2*n_gaps + 1:
+ searching = False
+ else:
+ searching = True
+
+ return ','.join(mask[1:-1])
+
+
+ def gaps_as_ranges(self, mask):
+ '''
+ Convert gaps of a single int to ranges, for
+ backwards compatibility reasons
+ '''
+
+ mask_out = []
+ for el in mask.split(','):
+ if el[0].isalpha():
+ mask_out.append(el)
+ else:
+ mask_out.append(f'{el}-{el}')
+
+ return ','.join(mask_out)
+
+
+def recover_mask(trb):
+ '''
+ Recover the string of the sampled mask given the trb file
+ '''
+
+ L_hal = trb['mask_contig'].shape[0]
+ mask = []
+
+ for idx0 in range(L_hal):
+ # what is the current idx
+ if idx0 in trb['con_hal_idx0']:
+ is_con = True
+ is_gap = False
+ else:
+ is_con = False
+ is_gap = True
+
+ # dealing with the first entry
+ if idx0 == 0:
+ if is_gap:
+ L_gap = 1
+ elif is_con:
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+
+ # take action based on what happend last time
+ else:
+ if (was_gap) and (is_gap):
+ L_gap +=1
+ #elif (was_con) and (is_con):
+ # continue
+ elif (was_gap) and (is_con):
+ # end gap
+ mask.append(f'{L_gap}-{L_gap}')
+ # start con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+ elif (was_con) and (is_gap):
+ # end con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ mask.append(f'{con_start}-{idx}')
+ # start gap
+ L_gap = 1
+
+ # dealing with last entry
+ if idx0 == L_hal-1:
+ if is_gap:
+ mask.append(f'{L_gap}-{L_gap}')
+ elif is_con: # (edge case not handled: con starts and ends on last idx)
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0-1) ]
+ mask.append(f'{con_start}-{idx}')
+
+ # update what last position was
+ was_con = copy.copy(is_con)
+ was_gap = copy.copy(is_gap)
+
+ return ','.join(mask)
+
+
+def mask_len(mask):
+ '''
+ Calculate the min and max possible length that can
+ be sampled given a mask
+ '''
+ L_min = 0
+ L_max = 0
+
+ for el in mask.split(','):
+ if el[0].isalpha(): # is con
+ con_s, con_e = el[1:].split('-')
+ con_s, con_e = int(con_s), int(con_e)
+ L_con = con_e - con_s + 1
+ L_min += L_con
+ L_max += L_con
+
+ else: # is gap
+ if '-' in el:
+ gap_min, gap_max = el.split('-')
+ gap_min, gap_max = int(gap_min), int(gap_max)
+ L_min += gap_min
+ L_max += gap_max
+ else:
+ L_min += int(el)
+ L_max += int(el)
+
+ return L_min, L_max
+
+class SampledMask():
+ def __init__(self, mask_str, ref_pdb_idx, con_to_set=None):
+ self.str = mask_str
+ self.L_hal = len(self)
+ self.L_ref = len(ref_pdb_idx)
+
+ #################
+ # con indices in hal and ref
+ #################
+ self.ref_pdb_idx = ref_pdb_idx
+ self.hal_pdb_idx = [('A', i) for i in range(1, len(self)+1)]
+
+ hal_idx0 = 0
+ con_ref_pdb_idx = []
+ con_hal_pdb_idx = []
+ con_ref_idx0 = []
+ con_hal_idx0 = []
+
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ for res in range(s, e+1):
+ con_ref_pdb_idx.append((ch, res))
+ con_hal_pdb_idx.append(('A', hal_idx0+1))
+ con_ref_idx0.append(self.ref_pdb_idx.index((ch, res)))
+ con_hal_idx0.append(hal_idx0)
+ hal_idx0 += 1
+ # gap
+ else:
+ for _ in range(s):
+ hal_idx0 += 1
+
+ self.con_mappings = {
+ 'ref_pdb_idx': con_ref_pdb_idx,
+ 'hal_pdb_idx': con_hal_pdb_idx,
+ 'ref_idx0': con_ref_idx0,
+ 'hal_idx0': con_hal_idx0,
+ }
+
+ #################
+ # con_to_set mapping
+ #################
+ if con_to_set:
+ self.con_to_set = con_to_set
+ else:
+ contigs = self.get_contigs()
+ self.con_to_set = dict(zip(contigs, len(contigs)*[0]))
+
+ # set_to_con mapping
+ set_to_con = {}
+ for k, v in self.con_to_set.items():
+ set_to_con[v] = set_to_con.get(v, []) + [k] # invert a dictionary with non-unique values
+ self.set_to_con = set_to_con
+
+ def __len__(self,):
+ _, L_max = self.mask_len(self.str)
+ return L_max
+
+ def map(self, sel, src, dst):
+ '''
+ Convert the contig selection in one indexing scheme to another.
+ Will return None if selection is not in a contig.
+
+ Input
+ ----------
+ sel (str): selection of a contig range or idx0 range. Can take multiple comma separated values of same type. Ex: A5-10,B2-8 or 3-8,14-21
+ src (str): <'ref', 'hal'>
+ dst (str): <'ref_pdb_idx', 'hal_pdb_idx', 'ref_idx0', 'hal_idx0>
+ '''
+ out = []
+ for con in sel.split(','):
+
+ ch, s, e = SampledMask.parse_contig(con)
+
+ # selection type is pdb_idx
+ if ch:
+ src_long = f'{src}_pdb_idx'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get((ch, res)) for res in range(s, e+1)]
+
+ # selection type is idx0
+ else:
+ src_long = f'{src}_idx0'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get(i) for i in range(s, e+1)]
+
+ return out
+
+ @staticmethod
+ def expand(mask_str):
+ '''
+ Ex: '2,A3-5,3' --> [None, None, (A,3), (A,4), (A,5), None, None, None]
+ '''
+ expanded = []
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ expanded += [(ch, res) for res in range(s, e+1)]
+ # gap
+ else:
+ expanded += [None for _ in range(s)]
+
+ return expanded
+
+ @staticmethod
+ def contract(pdb_idx):
+ '''
+ Inverse of expand
+ Ex: [None, None, (A,3), (A,4), (A,5), None, None, None] --> '2,A3-5,3'
+ '''
+
+ contracted = []
+ l_prev = (None, -200)
+ first_el_written = False
+
+ for l_curr in pdb_idx:
+ if l_curr is None:
+ l_curr = (None, -100)
+
+ # extend gap
+ if l_curr == l_prev:
+ L_gap += 1
+
+ # extend con
+ elif l_curr == (l_prev[0], l_prev[1]+1):
+ con_e = l_curr[1]
+
+ # new gap
+ elif (l_curr != l_prev) and (l_curr[0] is None):
+ # write prev con
+ if 'con_ch' in locals():
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+
+ L_gap = 1
+
+ # new con
+ elif (l_curr != l_prev) and isinstance(l_curr[0], str):
+ # write prev con
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ # write prev gap
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ con_ch = l_curr[0]
+ con_s = l_curr[1]
+ con_e = l_curr[1]
+
+ # update l_prev
+ l_prev = l_curr
+
+ # write last element
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ return ','.join(contracted)
+
+ def subset(self, sub):
+ '''
+ Make a mask_str that is a subset of the original mask_str
+ Ex: self.mask_str = '2,A5-20,4', sub='A5-10' --> '2,A5-10,14'
+ '''
+
+ # map from hal_idx0 to ref_pdb_idx
+ hal_idx0 = self.map(sub, 'ref', 'hal_idx0')
+ ref_pdb_idx = SampledMask.expand(sub)
+ mapping = dict(zip(hal_idx0, ref_pdb_idx))
+
+ expanded = [mapping.get(idx0) for idx0 in range(len(self))]
+
+ return self.contract(expanded)
+
+ def mask_len(self, mask):
+ '''
+ Technically, can take both sampled and unsampled mask
+ '''
+ L_min = 0
+ L_max = 0
+ for l in self.str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ L_min += e - s + 1
+ L_max += e - s + 1
+ # gap
+ else:
+ L_min += s
+ L_max += e
+
+ return L_min, L_max
+
+ def get_contigs(self, include_receptor=True):
+ '''
+ Get a list of all contigs in the mask
+ '''
+ [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0]]
+
+ contigs = []
+ for con in self.str.split(','):
+ ch = SampledMask.parse_contig(con)[0]
+ if ch == 'R' and include_receptor == False:
+ continue
+ if ch:
+ contigs.append(con)
+
+ return contigs
+
+ def get_gaps(self,):
+ '''
+ Get a list of all gaps in the mask
+ '''
+ return [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0] is None]
+
+ @staticmethod
+ def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+ @staticmethod
+ def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = SampledMask.parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = SampledMask.parse_range(contig)
+
+ return ch, s, e
+
+ def remove_diag(self, m_2d):
+ '''
+ Set the diagonal of a 2D boolean array to False
+ '''
+ L = m_2d.shape[0]
+ m_2d[np.arange(L), np.arange(L)] = False
+
+ return m_2d
+
+ def get_receptor_contig(self,):
+ '''
+ Returns None if there is no chain R in the mask_str
+ '''
+ receptor_contig = [l for l in self.get_contigs() if 'R' in l]
+
+ if len(receptor_contig) == 0:
+ receptor_contig = None
+ else:
+ receptor_contig = ','.join(receptor_contig)
+
+ return receptor_contig
+
+ def remove_receptor(self, m_2d):
+ '''
+ Remove intra-receptor contacts (chain R) from a mask
+ '''
+ receptor_contig = self.get_receptor_contig()
+
+ if receptor_contig: # has chain R
+ m_1d = np.zeros(self.L_hal, dtype=bool)
+ idx = np.array(self.map(receptor_contig, 'ref', 'hal_idx0'))
+ m_1d[idx] = True
+ update = m_1d[:, None] * m_1d[None, :]
+ m_2d = m_2d * ~update
+
+ return m_2d
+
+ def get_mask_con(self, include_receptor=False):
+ # Make a 2D boolean mask for each contig set
+ L = self.L_hal
+ mask_con = np.zeros([L, L], dtype=bool)
+
+ for set_id, contigs in self.set_to_con.items():
+ m_1d = np.zeros(L, dtype=bool)
+ for con in contigs:
+ idx = self.map(con, 'ref', 'hal_idx0')
+ idx = [l for l in idx if l != None]
+ idx = np.array(idx, dtype=int)
+ m_1d[idx] = True
+
+ update = m_1d[:, None] * m_1d[None, :]
+ mask_con = np.any([mask_con, update], axis=0)
+
+ # clean up
+ mask_con = self.remove_diag(mask_con)
+
+ if not include_receptor:
+ mask_con = self.remove_receptor(mask_con)
+
+ return mask_con
+
+ def get_mask_hal(self,):
+ mask_hal = ~self.get_mask_con()
+ mask_hal = self.remove_diag(mask_hal)
+ mask_hal = self.remove_receptor(mask_hal)
+
+ return mask_hal
+
+ def get_mask_cce(self, pdb, cce_cutoff=20., include_receptor=False):
+ '''
+ Remove ij pixels where contig distances are greater than cce_cutoff.
+ '''
+ # start with mask_con
+ mask_con = self.get_mask_con(include_receptor=include_receptor)
+
+ # get ref dists
+ xyz_ref = torch.tensor(pdb['xyz'][:,:3,:]).float()
+ c6d_ref = geometry.xyz_to_c6d(xyz_ref[None].permute(0,2,1,3),{'DMAX':20.0}).numpy()
+ dist = c6d_ref[0,:,:,0] # (L_ref, L_ref)
+
+ # scatter
+ dist_scattered = self.scatter_2d(dist)
+
+ # apply cce cuttoff
+ update = dist_scattered < cce_cutoff
+ mask_cce = np.all([mask_con, update], axis=0)
+
+ return mask_cce
+
+ def scatter_2d(self, ref_feat_2d):
+ '''
+ Inputs
+ ---------
+ ref_feat_2d (np.array; (L_ref, L_ref, ...)): Features to be scattered. The first two leading dimensions must be equal to L_ref.
+ '''
+ assert ref_feat_2d.shape[:2] == (self.L_ref, self.L_ref), 'ERROR: feat_2d must have leading dimensions of (L_ref, L_ref)'
+
+ trailing_dims = ref_feat_2d.shape[2:]
+ dtype = ref_feat_2d.dtype
+ hal_feat_2d = np.zeros((self.L_hal, self.L_hal)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_2d[con_hal_idx0[:, None], con_hal_idx0[None, :]] = ref_feat_2d[ref_hal_idx0[:, None], ref_hal_idx0[None, :]]
+
+ return hal_feat_2d
+
+ def scatter_1d(self, ref_feat_1d):
+ '''
+ Inputs
+ ---------
+ ref_feat_1d (np.array; (L_ref, ...)): Features to be scattered. The first leading dimension must be equal to L_ref.
+ '''
+ assert ref_feat_1d.shape[0] == self.L_ref, 'ERROR: feat_1d must have leading dimensions of (L_ref,)'
+
+ trailing_dims = ref_feat_1d.shape[1:]
+ dtype = ref_feat_1d.dtype
+ hal_feat_1d = np.zeros((self.L_hal,)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_1d[con_hal_idx0] = ref_feat_1d[ref_hal_idx0]
+
+ return hal_feat_1d
+
+ def idx_for_template(self, gap=200):
+ '''
+ Essentially return hal_idx0, except have a large jump for chain B,
+ to simulate a chain break. If B contains internal jumps in residue
+ numbering, these are preserved.
+ '''
+
+ is_rec = self.m1d_receptor()
+ resi_rec = np.array([idx[1] for idx in SampledMask.expand(self.str)
+ if idx is not None and idx[0]=='R'])
+ L_binder = sum(~is_rec)
+
+
+ if len(resi_rec)>0:
+ if is_rec[0]:
+ # receptor first
+ idx_tmpl = np.arange(resi_rec[-1]+gap+1, resi_rec[-1]+gap+1+L_binder)
+ idx_tmpl = np.concatenate([resi_rec, idx_tmpl])
+ else:
+ # receptor second
+ idx_tmpl = np.arange(L_binder)
+ if resi_rec[0] <= idx_tmpl[-1]+gap:
+ resi_rec += idx_tmpl[-1] - resi_rec[0] + gap + 1
+ idx_tmpl = np.concatenate([idx_tmpl, resi_rec])
+ else:
+ #when no receptor
+ idx_tmpl = np.arange(L_binder)
+ return idx_tmpl
+
+ def m1d_receptor(self,):
+ '''
+ Get a boolean array, True if the position corresponds to the receptor
+ '''
+ m1d = [(l is not None) and (l[0] == 'R') for l in SampledMask.expand(self.str)]
+ return np.array(m1d)
+
+ def erode(self, N_term=True, C_term=True):
+ '''
+ Reduce non-receptor contigs by 1 residue from the N and/or C terminus.
+ '''
+ x = SampledMask.expand(self.str)
+
+ if N_term:
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ if C_term:
+ x = x[::-1]
+
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ x = x[::-1]
+
+ self.str = self.contract(x)
+
+ return
+
+ def len_contigs(self, include_receptor=False):
+ con_str = ','.join(self.get_contigs(include_receptor))
+ return len(SampledMask.expand(con_str))
+
+
+def make_template_features(pdb, args, device, hal_2_ref_idx0=None, sm_loss=None):
+ '''
+ Inputs
+ ----------
+ sm_loss: Instance of a contig.SampledMask object used for making the loss masks.
+ '''
+ PARAMS = {
+ "DMIN" : 2.0,
+ "DMAX" : 20.0,
+ "DBINS" : 36,
+ "ABINS" : 36,
+ }
+ if args.use_template:
+ B,T = 1,1 # batch, templates
+
+ # spoof reference features
+ xyz_t = torch.tensor(pdb['xyz'][:, :3][None, None]) # (batch,templ,nres,3,3)
+ t0d = torch.ones((1,1,3)) # (batch, templ, 3)
+
+ t2d_ref = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS) # (B,T,L,L,...)
+ L_ref = t2d_ref.shape[2]
+ #t1d_ref = torch.ones(size=(B,T,L_ref,3), dtype=torch.float32, device=device)
+ a = 2 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ b = 0 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ c = 1 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+
+ t1d_ref = torch.stack([a,b,c], axis=-1)
+
+ # Get the mask_str for scattering template features
+ #1. Template mask = sampled mask
+ if (args.use_template.lower() == 't') or (args.use_template.lower() == 'true'):
+ sm_tmpl = sm_loss
+ #2. Template mask is a subset of the sampled mask
+ else:
+ subset_contigs = args.use_template
+
+ if args.receptor:
+ receptor_contig = sm_loss.get_receptor_contig()
+ subset_contigs = ','.join([subset_contigs, receptor_contig])
+
+ mask_str_tmpl = sm_loss.subset(subset_contigs)
+ sm_tmpl = SampledMask(mask_str=mask_str_tmpl, ref_pdb_idx=pdb['pdb_idx'])
+
+ # scatter template features
+ # make leading dims (L,(L),...)
+ t1d_ref = t1d_ref.permute(2,3,0,1) # (L, ..., B, T)
+ t2d_ref = t2d_ref.permute(2,3,4,0,1) # (L, L, ..., B, T)
+
+ t1d_tmpl = sm_tmpl.scatter_1d(t1d_ref.cpu().numpy())
+ t2d_tmpl = sm_tmpl.scatter_2d(t2d_ref.cpu().numpy())
+
+ # update t2d_tmpl with mask_con (could update with mask_cce instead?)
+ mask_con = sm_tmpl.get_mask_con(include_receptor=True)
+ t2d_tmpl = (t2d_tmpl.T * mask_con.T).T # trick to broadcast arrays if leading dimensions match
+
+ t1d_tmpl = torch.tensor(t1d_tmpl, device=device)
+ t2d_tmpl = torch.tensor(t2d_tmpl, device=device)
+
+ # Permute B and T dims back to front
+ t1d_tmpl = t1d_tmpl.permute(2,3,0,1)
+ t2d_tmpl = t2d_tmpl.permute(3,4,0,1,2)
+
+ # Make last 3 idx of last dim all 1 to mimick Ivan's template feature
+ t2d_tmpl[..., -3:] = 1.
+
+ idx = torch.tensor(sm_tmpl.idx_for_template(gap=200), device=device)[None]
+
+ net_kwargs = {
+ 'idx': idx,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ elif args.template_pdbs is not None:
+ B,T = 1, len(args.template_pdbs) # batch, templates
+
+ # get xyz features of all templates
+ xyz_t = [torch.tensor(parse_pdb(f_pdb)['xyz'][:, :3]) for f_pdb in args.template_pdbs]
+ xyz_t = torch.stack(xyz_t, axis=0)[None] # (batch, template, nres, 3, 3)
+ t0d = torch.ones(B,T,3)
+
+ t2d_tmpl = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS).to(device) # (B,T,L,L,...)
+ L_tmpl = t2d_tmpl.shape[2]
+ t1d_tmpl = torch.ones(size=(B,T,L_tmpl,3), dtype=torch.float32, device=device)
+
+ # spoof pdb idx
+ idx_tmpl = torch.range(0, L_tmpl-1, dtype=torch.long, device=device)[None]
+
+ # Net() kwargs
+ net_kwargs = {
+ 'idx': idx_tmpl,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ else:
+ net_kwargs = {}
+
+ return net_kwargs
diff --git a/model/utils/diff_utils.py b/model/utils/diff_utils.py
new file mode 100644
index 0000000000000000000000000000000000000000..9a6b71a8cc0af83e027e1d8e7b5ad21ab5ccbbe9
--- /dev/null
+++ b/model/utils/diff_utils.py
@@ -0,0 +1,283 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, MODEL_PARAM=None, hotspots=None, dssp=None):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ # EXPAND t1d to match model params
+ if MODEL_PARAM['d_t1d'] == 29:
+ ## added t1d features ##
+ # 24 -- dssp helix
+ # 25 -- dssp sheet
+ # 26 -- dssp loop
+ # 27 -- dssp mask
+ # 28 -- hotspot resi on target
+ t1d = torch.cat((t1d,torch.zeros(t1d.shape[0],t1d.shape[1],5)),dim=-1)
+ t1d[:,:,24:28] = dssp
+ t1d[:,:,28] = hotspots
+ t1d[:,str_mask,24:27] = 0.0
+ t1d[:,str_mask,27] = 1.0
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape )
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(xyz_prev.shape)
+ #xyz_prev = xyz_prev[-1]
+ #ic(xyz_prev.shape)
+
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ #ic(t1d.shape)
+ t1d[:,:,:,:21] = logit_aa_s[0,:21,:].permute(1,0)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
diff --git a/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg b/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..a514787d2c75eeede8bfab24f78440746debd28e
Binary files /dev/null and b/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg differ
diff --git a/model/utils/figs/fig1.jpg b/model/utils/figs/fig1.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..a514787d2c75eeede8bfab24f78440746debd28e
Binary files /dev/null and b/model/utils/figs/fig1.jpg differ
diff --git a/model/utils/figs/fig2.jpg b/model/utils/figs/fig2.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..053bafd7c530d636e94fb6e3c58ee0c1231b45c9
Binary files /dev/null and b/model/utils/figs/fig2.jpg differ
diff --git a/model/utils/figs/fig3.jpg b/model/utils/figs/fig3.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..d4bce6dfae5115b075ef199b592d3b32e4e3ca4a
Binary files /dev/null and b/model/utils/figs/fig3.jpg differ
diff --git a/model/utils/geometry.py b/model/utils/geometry.py
new file mode 100644
index 0000000000000000000000000000000000000000..58edab102102bf5650d11c72a7d5a76bb1abfb33
--- /dev/null
+++ b/model/utils/geometry.py
@@ -0,0 +1,200 @@
+import numpy as np
+import torch
+
+# ============================================================
+def get_pair_dist(a, b):
+ """calculate pair distances between two sets of points
+
+ Parameters
+ ----------
+ a,b : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of two sets of atoms
+ Returns
+ -------
+ dist : pytorch tensor of shape [batch,nres,nres]
+ stores paitwise distances between atoms in a and b
+ """
+
+ dist = torch.cdist(a, b, p=2)
+ return dist
+
+# ============================================================
+def get_ang(a, b, c):
+ """calculate planar angles for all consecutive triples (a[i],b[i],c[i])
+ from Cartesian coordinates of three sets of atoms a,b,c
+
+ Parameters
+ ----------
+ a,b,c : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of three sets of atoms
+ Returns
+ -------
+ ang : pytorch tensor of shape [batch,nres]
+ stores resulting planar angles
+ """
+ v = a - b
+ w = c - b
+ v = v / torch.norm(v, dim=-1, keepdim=True)
+ w = w / torch.norm(w, dim=-1, keepdim=True)
+
+ # this is not stable at the poles
+ #vw = torch.sum(v*w, dim=-1)
+ #ang = torch.acos(vw)
+
+ # this is better
+ # https://math.stackexchange.com/questions/1143354/numerically-stable-method-for-angle-between-3d-vectors/1782769
+ y = torch.norm(v-w,dim=-1)
+ x = torch.norm(v+w,dim=-1)
+ ang = 2*torch.atan2(y, x)
+
+ return ang
+
+# ============================================================
+def get_dih(a, b, c, d):
+ """calculate dihedral angles for all consecutive quadruples (a[i],b[i],c[i],d[i])
+ given Cartesian coordinates of four sets of atoms a,b,c,d
+
+ Parameters
+ ----------
+ a,b,c,d : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of four sets of atoms
+ Returns
+ -------
+ dih : pytorch tensor of shape [batch,nres]
+ stores resulting dihedrals
+ """
+ b0 = a - b
+ b1r = c - b
+ b2 = d - c
+
+ b1 = b1r/torch.norm(b1r, dim=-1, keepdim=True)
+
+ v = b0 - torch.sum(b0*b1, dim=-1, keepdim=True)*b1
+ w = b2 - torch.sum(b2*b1, dim=-1, keepdim=True)*b1
+
+ x = torch.sum(v*w, dim=-1)
+ y = torch.sum(torch.cross(b1,v,dim=-1)*w, dim=-1)
+ ang = torch.atan2(y, x)
+
+ return ang
+
+
+# ============================================================
+def xyz_to_c6d(xyz, params):
+ """convert cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz : pytorch tensor of shape [batch,3,nres,3]
+ stores Cartesian coordinates of backbone N,Ca,C atoms
+ Returns
+ -------
+ c6d : pytorch tensor of shape [batch,nres,nres,4]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+
+ batch = xyz.shape[0]
+ nres = xyz.shape[2]
+
+ # three anchor atoms
+ N = xyz[:,0]
+ Ca = xyz[:,1]
+ C = xyz[:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # 6d coordinates order: (dist,omega,theta,phi)
+ c6d = torch.zeros([batch,nres,nres,4],dtype=xyz.dtype,device=xyz.device)
+
+ dist = get_pair_dist(Cb,Cb)
+ dist[torch.isnan(dist)] = 999.9
+ c6d[...,0] = dist + 999.9*torch.eye(nres,device=xyz.device)[None,...]
+ b,i,j = torch.where(c6d[...,0]=params['DMAX']] = 999.9
+
+ return c6d
+
+
+# ============================================================
+def c6d_to_bins(c6d,params):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=c6d.dtype,device=c6d.device)
+ ab360 = torch.linspace(-np.pi+astep, np.pi, params['ABINS'],dtype=c6d.dtype,device=c6d.device)
+ ab180 = torch.linspace(astep, np.pi, params['ABINS']//2,dtype=c6d.dtype,device=c6d.device)
+
+ db = torch.bucketize(c6d[...,0].contiguous(),dbins)
+ ob = torch.bucketize(c6d[...,1].contiguous(),ab360)
+ tb = torch.bucketize(c6d[...,2].contiguous(),ab360)
+ pb = torch.bucketize(c6d[...,3].contiguous(),ab180)
+
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).to(torch.uint8)
+
+
+# ============================================================
+def dist_to_bins(dist,params):
+ """bin 2d distance maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ db = torch.round((dist-params['DMIN']-dstep/2)/dstep)
+
+ db[db<0] = 0
+ db[db>params['DBINS']] = params['DBINS']
+
+ return db.long()
+
+
+# ============================================================
+def c6d_to_bins2(c6d,params):
+ """bin 2d distance and orientation maps
+ (alternative slightly simpler version)
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ db = torch.round((c6d[...,0]-params['DMIN']-dstep/2)/dstep)
+ ob = torch.round((c6d[...,1]+np.pi-astep/2)/astep)
+ tb = torch.round((c6d[...,2]+np.pi-astep/2)/astep)
+ pb = torch.round((c6d[...,3]-astep/2)/astep)
+
+ # put all dparams['DBINS']] = params['DBINS']
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).long()
+
+
+# ============================================================
+def get_cb(N,Ca,C):
+ """recreate Cb given N,Ca,C"""
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+ return Cb
diff --git a/model/utils/inpainting_util.py b/model/utils/inpainting_util.py
new file mode 100644
index 0000000000000000000000000000000000000000..4350df63bd2ad5f5397d0d032c6cf2f200378c99
--- /dev/null
+++ b/model/utils/inpainting_util.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'wt') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/model/utils/parsers_inference.py b/model/utils/parsers_inference.py
new file mode 100644
index 0000000000000000000000000000000000000000..1a8bc9aca6de872a4e2b4388937c4d85e6b8d6b1
--- /dev/null
+++ b/model/utils/parsers_inference.py
@@ -0,0 +1,151 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ i[pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/requirements.txt b/requirements.txt
new file mode 100644
index 0000000000000000000000000000000000000000..4381612685cf0b7d085d42b23b6afb294096a74a
--- /dev/null
+++ b/requirements.txt
@@ -0,0 +1,7 @@
+torch==1.9
+e3nn==0.3.3
+pynvml==11.0.0
+decorator==5.1.0
+icecream
+biopython
+py3Dmol
diff --git a/tmp/args.json b/tmp/args.json
new file mode 100644
index 0000000000000000000000000000000000000000..fe157435b9a5d55dff83f0afb4fbfe34be96f777
--- /dev/null
+++ b/tmp/args.json
@@ -0,0 +1 @@
+{"F": 1, "T": 25, "aa_composition": "", "aa_spec": null, "aa_weight": null, "aa_weights_json": null, "add_weight_every_n": 1, "argmax_seq": false, "cautious": false, "checkpoint": "./SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt", "clamp_seqout": false, "contigs": ["0"], "d_t1d": 29, "dssp_pdb": null, "dump_all": false, "dump_npz": false, "dump_pdb": true, "dump_trb": true, "frac_seq_to_weight": 0.0, "hal_idx": null, "helix_bias": 0.0, "hotspots": null, "idx_rf": null, "inpaint_seq": null, "inpaint_seq_tensor": null, "inpaint_str": null, "inpaint_str_tensor": null, "input_json": null, "length": null, "loop_bias": 0.0, "loop_design": false, "min_decoding_distance": 15, "multi_templates": null, "multi_tmpl_conf": null, "n_cycle": 4, "noise_schedule": "sqrt", "num_designs": 500, "one_weight_per_position": false, "out": "./tmp/", "pdb": null, "potential_scale": null, "ref_idx": null, "sampling_temp": 1.0, "save_all_steps": false, "save_best_plddt": true, "save_seqs": false, "scheduled_str_cond": false, "secondary_structure": null, "softmax_seqout": false, "start_num": 0, "strand_bias": 0.01, "struc_cond_sc": false, "symmetry": 1, "symmetry_cap": 0, "temperature": 1.0, "tmpl_conf": "1", "trb": null, "sequence": "", "save_args": true, "potentials": null, "target_charge": -10, "charge_loss_type": "complex", "target_pH": 7.4, "hydrophobic_score": -10, "hydrophobic_loss_type": "complex", "sample_distribution": "normal", "sample_distribution_gmm_means": [-1, 1], "sample_distribution_gmm_variances": [1, 1], "gradio": true}
diff --git a/utils/.inf_methods.py.swp b/utils/.inf_methods.py.swp
new file mode 100644
index 0000000000000000000000000000000000000000..1638aa8c4baf5a8ded77e8d71bda84a9f34fce3c
Binary files /dev/null and b/utils/.inf_methods.py.swp differ
diff --git a/utils/.ipynb_checkpoints/diff_utils-checkpoint.py b/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..9a6b71a8cc0af83e027e1d8e7b5ad21ab5ccbbe9
--- /dev/null
+++ b/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
@@ -0,0 +1,283 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, MODEL_PARAM=None, hotspots=None, dssp=None):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ # EXPAND t1d to match model params
+ if MODEL_PARAM['d_t1d'] == 29:
+ ## added t1d features ##
+ # 24 -- dssp helix
+ # 25 -- dssp sheet
+ # 26 -- dssp loop
+ # 27 -- dssp mask
+ # 28 -- hotspot resi on target
+ t1d = torch.cat((t1d,torch.zeros(t1d.shape[0],t1d.shape[1],5)),dim=-1)
+ t1d[:,:,24:28] = dssp
+ t1d[:,:,28] = hotspots
+ t1d[:,str_mask,24:27] = 0.0
+ t1d[:,str_mask,27] = 1.0
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape )
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(xyz_prev.shape)
+ #xyz_prev = xyz_prev[-1]
+ #ic(xyz_prev.shape)
+
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ #ic(t1d.shape)
+ t1d[:,:,:,:21] = logit_aa_s[0,:21,:].permute(1,0)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
diff --git a/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py b/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..9acb5356ac24fba71f65eb09bb777c62ccb97a45
--- /dev/null
+++ b/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'w+') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py b/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..1a8bc9aca6de872a4e2b4388937c4d85e6b8d6b1
--- /dev/null
+++ b/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
@@ -0,0 +1,151 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ i[pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/utils/LISCENCE b/utils/LISCENCE
new file mode 100644
index 0000000000000000000000000000000000000000..a584cfce171b7d0e8deb9666266c86dd15033031
--- /dev/null
+++ b/utils/LISCENCE
@@ -0,0 +1,21 @@
+MIT License
+
+Copyright (c) 2023 RosettaCommons
+
+Permission is hereby granted, free of charge, to any person obtaining a copy
+of this software and associated documentation files (the "Software"), to deal
+in the Software without restriction, including without limitation the rights
+to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
+copies of the Software, and to permit persons to whom the Software is
+furnished to do so, subject to the following conditions:
+
+The above copyright notice and this permission notice shall be included in all
+copies or substantial portions of the Software.
+
+THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
+AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
+OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
+SOFTWARE.
diff --git a/utils/README.md b/utils/README.md
new file mode 100644
index 0000000000000000000000000000000000000000..06b2c403cbf5ef306c6400960bb39b2d9fa2ee63
--- /dev/null
+++ b/utils/README.md
@@ -0,0 +1,132 @@
+---
+title: PROTEIN GENERATOR
+emoji: 🐨
+thumbnail: http://files.ipd.uw.edu/pub/sequence_diffusion/figs/diffusion_landscape.png
+colorFrom: blue
+colorTo: purple
+sdk: gradio
+sdk_version: 3.24.1
+app_file: app.py
+pinned: false
+---
+
+
+
+## TLDR but I know how to use inpainting
+Submit jobs with all the same args as [inpainting](https://git.ipd.uw.edu/jwatson3/proteininpainting) plus some new ones
+- --T specify number of timesteps to use, good choices are 5,25,50,100 (try lower T first)
+- --save_best_plddt recommended arg to save best plddt str/seq in trajectory
+- --loop_design use when generating loops for binder design (or Ab loop design), will load finetuned checkpoint
+- --symmetry integer to divide the input up with for symmetric update (input of X will divide the sequence into X symmetric motifs)
+- --trb specify trb when partially diffusing to use same mask as the design, must match pdb input (contigs will not get used)
+- --sampling_temp fraction of diffusion trajectory to use for partial diffusion (default is 1.0 for fully diffused) values around 0.3 seem to give good diversity
+
+**environment to use**
+```
+source activate /software/conda/envs/SE3nv
+```
+**example command**
+```
+python inference.py --pdb examples/pdbs/rsv5_5tpn.pdb --out examples/out/design \
+ --contigs 0-25,A163-181,25-30 --T 25 --save_best_plddt
+```
+For jobs with inputs <75 residues it is feesible to run on CPUs. It's helpful to redesign output backbones with [MPNN](https://git.ipd.uw.edu/justas/proteinmpnn) (not sure if useful yet when using --loop_design). Check back for more updates.
+
+## Getting started
+
+Check out the templates in the example folder to see how you can set up jobs for the various design strategies
+
+- [ ] [Motif or active-site scaffolding](examples/motif_scaffolding.sh)
+- [ ] [Partial diffusion (design diversification)](examples/partial_diffusion.sh)
+- [ ] [Anitbody / Loop design](examples/loop_design.sh)
+- [ ] [Symmetric design](examples/symmetric_design.sh)
+
+## Weighted Sequence design
+Biasing the sequence by weighting certain amino acid types is a nice way to control and guide the diffusion process, generate interesting folds, and repeat units. It is possible to combine this technique with motif scaffolding as well. here are a few different ways to set up sequence potentials:
+
+The --aa_spec argument used in combination with the --aa_weight allows you to specify the complete amino acid weighting pattern for a sequence. The pattern specified in aa_spec will be repeated along the entire length of the design.
+ - --aa_spec base repeat unit to weight sequence with, X is used as a mask token, for example --aa_spec XXAXXLXX will generate solenoid folds like the one below
+ - --aa_weight weights to assign for non masked residues in aa_spec, for example --aa_weight 2,2 will weight alanine to 2 and leucine to 2
+
+**Make solenoids with a little bias!**
+
+  +
+
+
+**example job set up for sequene weighting**
+```
+python inference.py \
+ --num_designs 10 \
+ --out examples/out/seq_bias \
+ --contigs 100 --symmetry 5 \
+ --T 25 --save_best_plddt \
+ --aa_spec XXAXXLXX --aa_weight 2,2
+```
+
+In addition to the contigs above users can also use a disctionary to specify sequence weighting with [aa_weights](examples/aa_weights.json) for more generic uses. These weights can be specified with the --aa_weights_json arg and used in combination with the --add_weight_every_n arg or --frac_seq_to_weight arg. Each of these args defines where weights in the aa_weights dictionary will be applied to the sequence (you cannot specify both simultaneously). To add the weight every 5 residues use --add_weight_every_n 5. To add weight to a randomly sampled 40% of the sequence use --frac_seq_to_weight 0.4. If you add weight to multiple amino acid types in aa_weights, use the --one_weight_per_position flag to specify that a randomly sampled amino acid from aa_weight with a positive value should be chosen where the sequence bias is added. This allows the user to specify multiple amino acid types you want to upweight while ensuring to only bias for one type at each position, this usually is more effective.
+
+
+## Motif and active site scaffolding
+An example for motif scaffolding submission is written below, if you are inputing an active site with single residue inputs this can be specified in the contigs like 10-20,A10-10,20-30,A50-50,5-15 to scaffold just the 10th and 50th residues of chain A. Setting the model at higher T usually results in higher success rates, but it can still be useful to try problems out with just a few steps (T = 5, 15, or 25), before increasing the number of steps further. It is recommended to use [MPNN](https://git.ipd.uw.edu/justas/proteinmpnn) on the output backbones before alphafolding for validation.
+
+```
+python inference.py \
+ --num_designs 10 \
+ --out examples/out/design \
+ --pdb examples/pdbs/rsv5_5tpn.pdb \
+ --contigs 0-25,A163-181,25-30 --T 25 --save_best_plddt
+```
+
+## Partial diffusion
+To sample diverse and highquality desing fast, it can be useful to run many designs with T=5, and then after MPNN and alphafold filtering partially diffuse the successful designs to generate more diversity around designs that seem to be working. By using the --trb flag the script will enter partial diffusion mode. With the --T flag you can specify the total number of steps inthe trajectory and with the --sampling_temp flag you can determine how far into the trajectory the inputs will be diffused. Setting the sampling temp to 1.0 would be full diffused. In this mode the contigs will be ignored, and the mask used from the original design will be used.
+
+```
+python inference.py \
+ --num_designs 10 \
+ --pdb examples/out/design_000.pdb \
+ --trb examples/out/design_000.trb \
+ --out examples/out/partial_diffusion_design \
+ --contigs 0 --sampling_temp 0.3 --T 50 --save_best_plddt
+```
+
+
+## Symmetric design
+In symmetric design mode, the --symmetry flag is used to specify the number of partitions to make from the input sequence length. Each partition will be updated symmetric according to the first in the sequence. This requires that your sequence length (L) is divisible by the symmetry input. Symmetric motif scaffolding should be possible with the right contigs, but has not been experimented with yet.
+
+```
+python inference.py \
+ --num_designs 10 \
+ --pdb examples/pdbs/rsv5_5tpn.pdb \
+ --out examples/out/symmetric_design \
+ --contigs 25,0 25,0 25,0 \
+ --T 50 --save_best_plddt --symmetry 3
+```
+
+
+## Antibody and loop design
+Using the --loop_desing flag will load a version of the model finetuned on antibody CDR loops. This is useful if you are looking to design new CDR loops or are strcutred loops for binder design. It is helpful to run the designs with a target input too.
+
+```
+python inference.py \
+ --num_designs 10 \
+ --pdb examples/pdbs/G12D_manual_mut.pdb \
+ --out examples/out/ab_loop \
+ --contigs A2-176,0 C7-16,0 H2-95,12-15,H111-116,0 L1-45,10-12,L56-107 \
+ --T 25 --save_best_plddt --loop_design
+```
+
+
+
+## About the model
+Sequence diffusion is trained on the same dataset and uses the same architecture as RoseTTAFold. To train the model, a ground truth sequence is transformed into an Lx20 continuous space and gaussian noise is added to diffuse the sequence to the sampled timestep. To condition on structure and sequence, the structre for a motif is given and then corresponding sequence is denoised in the input. The rest of the structure is blackhole initialized. For each example the model is trained to predict Xo and losses are applied on the structure and sequence respectively. During training big T is set to 1000 steps, and a square root schedule is used to add noise.
+
+
+
+
+## Looking ahead
+We are interested in problems where diffusing in sequence space is useful, if you would like to chat more or join in our effort for sequence diffusion come talk to Sidney or Jake!
+
+
+## Acknowledgements
+A project by Sidney Lisanza and Jake Gershon. Thanks to Sam Tipps for implementing symmetric sequence diffusion. Thank you to Minkyung Baek and Frank Dimaio for developing RoseTTAFold, Joe Watson and David Juergens for the developing inpainting inference script which the inference code is built on top of.
+
diff --git a/utils/__pycache__/calc_dssp.cpython-310.pyc b/utils/__pycache__/calc_dssp.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..71b35c2ced09a7d59783b56b939e28908edb3915
Binary files /dev/null and b/utils/__pycache__/calc_dssp.cpython-310.pyc differ
diff --git a/utils/__pycache__/contigs.cpython-310.pyc b/utils/__pycache__/contigs.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..697beeced76697149d5e4947bf4ab8b664cb1e39
Binary files /dev/null and b/utils/__pycache__/contigs.cpython-310.pyc differ
diff --git a/utils/__pycache__/diff_utils.cpython-310.pyc b/utils/__pycache__/diff_utils.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..f389761c5c1bddb8c7f0a7bfeb1aa74c844684e3
Binary files /dev/null and b/utils/__pycache__/diff_utils.cpython-310.pyc differ
diff --git a/utils/__pycache__/geometry.cpython-310.pyc b/utils/__pycache__/geometry.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5bff0e27eb9ed8b37e5080ecfc5f440e936df375
Binary files /dev/null and b/utils/__pycache__/geometry.cpython-310.pyc differ
diff --git a/utils/__pycache__/inpainting_util.cpython-310.pyc b/utils/__pycache__/inpainting_util.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..178d30ef7a5c3d53a2ff71326e787f6f352b0f9a
Binary files /dev/null and b/utils/__pycache__/inpainting_util.cpython-310.pyc differ
diff --git a/utils/__pycache__/parsers_inference.cpython-310.pyc b/utils/__pycache__/parsers_inference.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..29e7013b9ad9508b4392917d8df65ced65ade5c3
Binary files /dev/null and b/utils/__pycache__/parsers_inference.cpython-310.pyc differ
diff --git a/utils/__pycache__/potentials.cpython-310.pyc b/utils/__pycache__/potentials.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..6b1290254defb69e37e77689b6c74082aa98bcb1
Binary files /dev/null and b/utils/__pycache__/potentials.cpython-310.pyc differ
diff --git a/utils/__pycache__/sampler.cpython-310.pyc b/utils/__pycache__/sampler.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..28d84845305b468bf62527bea260995818b16934
Binary files /dev/null and b/utils/__pycache__/sampler.cpython-310.pyc differ
diff --git a/utils/app.py b/utils/app.py
new file mode 100644
index 0000000000000000000000000000000000000000..8efb331948e8d2e41d8d82dafcedc0c573d2c9ba
--- /dev/null
+++ b/utils/app.py
@@ -0,0 +1,383 @@
+import os,sys
+
+# install environment goods
+#os.system("pip -q install dgl -f https://data.dgl.ai/wheels/cu113/repo.html")
+os.system('pip install dgl==1.0.2+cu116 -f https://data.dgl.ai/wheels/cu116/repo.html')
+#os.system('pip install gradio')
+os.environ["DGLBACKEND"] = "pytorch"
+#os.system(f'pip install -r ./PROTEIN_GENERATOR/requirements.txt')
+print('Modules installed')
+
+os.environ["DGLBACKEND"] = "pytorch"
+
+if not os.path.exists('./SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'):
+ print('Downloading model weights 1')
+ os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt')
+ print('Successfully Downloaded')
+
+if not os.path.exists('./SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'):
+ print('Downloading model weights 2')
+ os.system('wget http://files.ipd.uw.edu/pub/sequence_diffusion/checkpoints/SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt')
+ print('Successfully Downloaded')
+
+import numpy as np
+import gradio as gr
+import py3Dmol
+from io import StringIO
+import json
+import secrets
+import copy
+import matplotlib.pyplot as plt
+from utils.sampler import HuggingFace_sampler
+
+plt.rcParams.update({'font.size': 13})
+
+with open('./tmp/args.json','r') as f:
+ args = json.load(f)
+
+# manually set checkpoint to load
+args['checkpoint'] = None
+args['dump_trb'] = False
+args['dump_args'] = True
+args['save_best_plddt'] = True
+args['T'] = 25
+args['strand_bias'] = 0.0
+args['loop_bias'] = 0.0
+args['helix_bias'] = 0.0
+
+
+
+def protein_diffusion_model(sequence, seq_len, helix_bias, strand_bias, loop_bias,
+ secondary_structure, aa_bias, aa_bias_potential, target_charge, target_ph, charge_potential,
+ num_steps, noise, hydrophobic_target_score, hydrophobic_potential):
+
+ dssp_checkpoint = './SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'
+ og_checkpoint = './SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt'
+
+ model_args = copy.deepcopy(args)
+
+ # make sampler
+ S = HuggingFace_sampler(args=model_args)
+
+ # get random prefix
+ S.out_prefix = './tmp/'+secrets.token_hex(nbytes=10).upper()
+
+ # set args
+ S.args['checkpoint'] = None
+ S.args['dump_trb'] = False
+ S.args['dump_args'] = True
+ S.args['save_best_plddt'] = True
+ S.args['T'] = 20
+ S.args['strand_bias'] = 0.0
+ S.args['loop_bias'] = 0.0
+ S.args['helix_bias'] = 0.0
+ S.args['potentials'] = None
+ S.args['potential_scale'] = None
+ S.args['aa_composition'] = None
+
+
+ # get sequence if entered and make sure all chars are valid
+ alt_aa_dict = {'B':['D','N'],'J':['I','L'],'U':['C'],'Z':['E','Q'],'O':['K']}
+ if sequence not in ['',None]:
+ aa_seq = []
+ for aa in sequence.upper():
+ if aa in alt_aa_dict.keys():
+ aa_seq.append(np.random.choice(alt_aa_dict[aa]))
+ else:
+ aa_seq.append(aa)
+
+ S.args['sequence'] = aa_seq
+ else:
+ S.args['contigs'] = [f'{seq_len}']
+
+ if secondary_structure in ['',None]:
+ secondary_structure = None
+ else:
+ secondary_structure = ''.join(['E' if x == 'S' else x for x in secondary_structure])
+ if len(sequence) < len(secondary_structure):
+ secondary_structure = secondary_structure[:len(sequence)]
+ elif len(sequence) == len(secondary_structure):
+ pass
+ else:
+ dseq = len(sequence) - len(secondary_structure)
+ secondary_structure += secondary_structure[-1]*dseq
+
+
+ # potentials
+ potential_list = []
+ potential_bias_list = []
+
+ if aa_bias not in ['',None]:
+ potential_list.append('aa_bias')
+ S.args['aa_composition'] = aa_bias
+ if aa_bias_potential in ['',None]:
+ aa_bias_potential = 3
+ potential_bias_list.append(str(aa_bias_potential))
+
+ if target_charge not in ['',None]:
+ potential_list.append('charge')
+ if charge_potential in ['',None]:
+ charge_potential = 1
+ potential_bias_list.append(str(charge_potential))
+ S.args['target_charge'] = int(target_charge)
+ if target_ph in ['',None]:
+ target_ph = 7.4
+ S.args['target_pH'] = float(target_ph)
+
+
+ if hydrophobic_target_score not in ['',None]:
+ potential_list.append('hydrophobic')
+ S.args['hydrophobic_score'] = float(hydrophobic_target_score)
+ if hydrophobic_potential in ['',None]:
+ hydrophobic_potential = 3
+ potential_bias_list.append(str(hydrophobic_potential))
+
+
+ if len(potential_list) > 0:
+ S.args['potentials'] = ','.join(potential_list)
+ S.args['potential_scale'] = ','.join(potential_bias_list)
+
+
+ # normalise secondary_structure bias from range 0-0.3
+ S.args['secondary_structure'] = secondary_structure
+ S.args['helix_bias'] = helix_bias
+ S.args['strand_bias'] = strand_bias
+ S.args['loop_bias'] = loop_bias
+
+ # set T
+ if num_steps in ['',None]:
+ S.args['T'] = 20
+ else:
+ S.args['T'] = int(num_steps)
+
+ # noise
+ if 'normal' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [0]
+ S.args['sample_distribution_gmm_variances'] = [1]
+ elif 'gmm2' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [-1,1]
+ S.args['sample_distribution_gmm_variances'] = [1,1]
+ elif 'gmm3' in noise:
+ S.args['sample_distribution'] = noise
+ S.args['sample_distribution_gmm_means'] = [-1,0,1]
+ S.args['sample_distribution_gmm_variances'] = [1,1,1]
+
+
+
+ if secondary_structure not in ['',None] or helix_bias+strand_bias+loop_bias > 0:
+ S.args['checkpoint'] = dssp_checkpoint
+ S.args['d_t1d'] = 29
+ print('using dssp checkpoint')
+ else:
+ S.args['checkpoint'] = og_checkpoint
+ S.args['d_t1d'] = 24
+ print('using og checkpoint')
+
+
+ for k,v in S.args.items():
+ print(f"{k} --> {v}")
+
+ # init S
+ S.model_init()
+ S.diffuser_init()
+ S.setup()
+
+ # sampling loop
+ plddt_data = []
+ for j in range(S.max_t):
+ output_seq, output_pdb, plddt = S.take_step_get_outputs(j)
+ plddt_data.append(plddt)
+ yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
+
+ output_seq, output_pdb, plddt = S.get_outputs()
+
+ yield output_seq, output_pdb, display_pdb(output_pdb), get_plddt_plot(plddt_data, S.max_t)
+
+def get_plddt_plot(plddt_data, max_t):
+ x = [i+1 for i in range(len(plddt_data))]
+ fig, ax = plt.subplots(figsize=(15,6))
+ ax.plot(x,plddt_data,color='#661dbf', linewidth=3,marker='o')
+ ax.set_xticks([i+1 for i in range(max_t)])
+ ax.set_yticks([(i+1)/10 for i in range(10)])
+ ax.set_ylim([0,1])
+ ax.set_ylabel('model confidence (plddt)')
+ ax.set_xlabel('diffusion steps (t)')
+ return fig
+
+def display_pdb(path_to_pdb):
+ '''
+ #function to display pdb in py3dmol
+ '''
+ pdb = open(path_to_pdb, "r").read()
+
+ view = py3Dmol.view(width=500, height=500)
+ view.addModel(pdb, "pdb")
+ view.setStyle({'model': -1}, {"cartoon": {'colorscheme':{'prop':'b','gradient':'roygb','min':0,'max':1}}})#'linear', 'min': 0, 'max': 1, 'colors': ["#ff9ef0","#a903fc",]}}})
+ view.zoomTo()
+ output = view._make_html().replace("'", '"')
+ print(view._make_html())
+ x = f""" {output} """ # do not use ' in this input
+
+ return f""""""
+
+'''
+
+ return f""""""
+'''
+
+def toggle_seq_input(choice):
+ if choice == "protein length":
+ return gr.update(visible=True, value=None), gr.update(visible=False, value=None)
+ elif choice == "custom sequence":
+ return gr.update(visible=False, value=None), gr.update(visible=True, value=None)
+
+def toggle_secondary_structure(choice):
+ if choice == "sliders":
+ return gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=True, value=None),gr.update(visible=False, value=None)
+ elif choice == "explicit":
+ return gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=False, value=None),gr.update(visible=True, value=None)
+
+# Define the Gradio interface
+with gr.Blocks(theme='ParityError/Interstellar') as demo:
+
+ gr.Markdown(f"""# Protein Generation via Diffusion in Sequence Space""")
+
+ with gr.Row():
+ with gr.Column(min_width=500):
+ gr.Markdown(f"""
+ ## How does it work?\n
+ Protein sequence and structure co-generation is a long outstanding problem in the field of protein design. By implementing [ddpm](https://arxiv.org/abs/2006.11239) style diffusion over protein seqeuence space we generate protein sequence and structure pairs. Starting with [RoseTTAFold](https://www.science.org/doi/10.1126/science.abj8754), a protein structure prediction network, we finetuned it to predict sequence and structure given a partially noised sequence. By applying losses to both the predicted sequence and structure the model is forced to generate meaningful pairs. Diffusing in sequence space makes it easy to implement potentials to guide the diffusive process toward particular amino acid composition, net charge, and more! Furthermore, you can sample proteins from a family of sequences or even train a small sequence to function classifier to guide generation toward desired sequences.
+ 
+
+ ## How to use it?\n
+ A user can either design a custom input sequence to diffuse from or specify a length below. To scaffold a sequence use the following format where X represent residues to diffuse: XXXXXXXXSCIENCESCIENCEXXXXXXXXXXXXXXXXXXX. You can even design a protein with your name XXXXXXXXXXXXNAMEHEREXXXXXXXXXXXXX!
+
+ ### Acknowledgements\n
+ Thank you to Simon Dürr and the Hugging Face team for setting us up with a community GPU grant!
+ """)
+
+ gr.Markdown("""
+ ## Model in Action
+ 
+ """)
+
+ with gr.Row().style(equal_height=False):
+ with gr.Column():
+ gr.Markdown("""## INPUTS""")
+ gr.Markdown("""#### Start Sequence
+ Specify the protein length for complete unconditional generation, or scaffold a motif (or your name) using the custom sequence input""")
+ seq_opt = gr.Radio(["protein length","custom sequence"], label="How would you like to specify the starting sequence?", value='protein length')
+
+ sequence = gr.Textbox(label="custom sequence", lines=1, placeholder='AMINO ACIDS: A,C,D,E,F,G,H,I,K,L,M,N,P,Q,R,S,T,V,W,Y\n MASK TOKEN: X', visible=False)
+ seq_len = gr.Slider(minimum=5.0, maximum=250.0, label="protein length", value=100, visible=True)
+
+ seq_opt.change(fn=toggle_seq_input,
+ inputs=[seq_opt],
+ outputs=[seq_len, sequence],
+ queue=False)
+
+ gr.Markdown("""### Optional Parameters""")
+ with gr.Accordion(label='Secondary Structure',open=True):
+ gr.Markdown("""Try changing the sliders or inputing explicit secondary structure conditioning for each residue""")
+ sec_str_opt = gr.Radio(["sliders","explicit"], label="How would you like to specify secondary structure?", value='sliders')
+
+ secondary_structure = gr.Textbox(label="secondary structure", lines=1, placeholder='HELIX = H STRAND = S LOOP = L MASK = X(must be the same length as input sequence)', visible=False)
+
+ with gr.Column():
+ helix_bias = gr.Slider(minimum=0.0, maximum=0.05, label="helix bias", visible=True)
+ strand_bias = gr.Slider(minimum=0.0, maximum=0.05, label="strand bias", visible=True)
+ loop_bias = gr.Slider(minimum=0.0, maximum=0.20, label="loop bias", visible=True)
+
+ sec_str_opt.change(fn=toggle_secondary_structure,
+ inputs=[sec_str_opt],
+ outputs=[helix_bias,strand_bias,loop_bias,secondary_structure],
+ queue=False)
+
+ with gr.Accordion(label='Amino Acid Compositional Bias',open=False):
+ gr.Markdown("""Bias sequence composition for particular amino acids by specifying the one letter code followed by the fraction to bias. This can be input as a list for example: W0.2,E0.1""")
+ with gr.Row():
+ aa_bias = gr.Textbox(label="aa bias", lines=1, placeholder='specify one letter AA and fraction to bias, for example W0.1 or M0.1,K0.1' )
+ aa_bias_potential = gr.Textbox(label="aa bias scale", lines=1, placeholder='AA Bias potential scale (recomended range 1.0-5.0)')
+
+ with gr.Accordion(label='Charge Bias',open=False):
+ gr.Markdown("""Bias for a specified net charge at a particular pH using the boxes below""")
+ with gr.Row():
+ target_charge = gr.Textbox(label="net charge", lines=1, placeholder='net charge to target')
+ target_ph = gr.Textbox(label="pH", lines=1, placeholder='pH at which net charge is desired')
+ charge_potential = gr.Textbox(label="charge potential scale", lines=1, placeholder='charge potential scale (recomended range 1.0-5.0)')
+
+ with gr.Accordion(label='Hydrophobic Bias',open=False):
+ gr.Markdown("""Bias for or against hydrophobic composition, to get more soluble proteins, bias away with a negative target score (ex. -5)""")
+ with gr.Row():
+ hydrophobic_target_score = gr.Textbox(label="hydrophobic score", lines=1, placeholder='hydrophobic score to target (negative score is good for solublility)')
+ hydrophobic_potential = gr.Textbox(label="hydrophobic potential scale", lines=1, placeholder='hydrophobic potential scale (recomended range 1.0-2.0)')
+
+ with gr.Accordion(label='Diffusion Params',open=False):
+ gr.Markdown("""Increasing T to more steps can be helpful for harder design challenges, sampling from different distributions can change the sequence and structural composition""")
+ with gr.Row():
+ num_steps = gr.Textbox(label="T", lines=1, placeholder='number of diffusion steps (25 or less will speed things up)')
+ noise = gr.Dropdown(['normal','gmm2 [-1,1]','gmm3 [-1,0,1]'], label='noise type', value='normal')
+
+ btn = gr.Button("GENERATE")
+
+ #with gr.Row():
+ with gr.Column():
+ gr.Markdown("""## OUTPUTS""")
+ gr.Markdown("""#### Confidence score for generated structure at each timestep""")
+ plddt_plot = gr.Plot(label='plddt at step t')
+ gr.Markdown("""#### Output protein sequnece""")
+ output_seq = gr.Textbox(label="sequence")
+ gr.Markdown("""#### Download PDB file""")
+ output_pdb = gr.File(label="PDB file")
+ gr.Markdown("""#### Structure viewer""")
+ output_viewer = gr.HTML()
+
+ gr.Markdown("""### Don't know where to get started? Click on an example below to try it out!""")
+ gr.Examples(
+ [["","125",0.0,0.0,0.2,"","","","","","","20","normal"],
+ ["","100",0.0,0.0,0.0,"","W0.2","2","","","","20","normal"],
+ ["","100",0.0,0.0,0.0,"XXHHHHHHHHHXXXXXXXHHHHHHHHHXXXXXXXHHHHHHHHXXXXSSSSSSSSSSSXXXXXXXXSSSSSSSSSSSSXXXXXXXSSSSSSSSSXXXXXXX","","","","","","25","normal"],
+ ["XXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXXXXXPEPSEQXXXXXXXXXXXXXXXXXXXXXXXXXXIPDXXXXXXXXXXXXXXXXXXX","",0.0,0.0,0.0,"","","","","","","25","normal"]],
+ inputs=[sequence, seq_len, helix_bias,
+ strand_bias, loop_bias,
+ secondary_structure, aa_bias,
+ aa_bias_potential, target_charge,
+ target_ph, charge_potential,
+ num_steps, noise],
+ outputs=[output_seq,output_pdb,output_viewer,plddt_plot],
+ fn=protein_diffusion_model,
+ )
+ btn.click(protein_diffusion_model,
+ [sequence,
+ seq_len,
+ helix_bias,
+ strand_bias,
+ loop_bias,
+ secondary_structure,
+ aa_bias,
+ aa_bias_potential,
+ target_charge,
+ target_ph,
+ charge_potential,
+ num_steps,
+ noise,
+ hydrophobic_target_score,
+ hydrophobic_potential],
+ [output_seq,
+ output_pdb,
+ output_viewer,
+ plddt_plot])
+
+demo.queue()
+demo.launch(debug=True)
+
diff --git a/utils/calc_dssp.py b/utils/calc_dssp.py
new file mode 100644
index 0000000000000000000000000000000000000000..fe2b975316f2de89d021d4dff442182192d5b7f8
--- /dev/null
+++ b/utils/calc_dssp.py
@@ -0,0 +1,234 @@
+#@title get secondary structure (SSE) from given PDB file
+#@markdown So far it seems the best solution is to steal code from biotite
+#@markdown which calculates the SSE of a peptide chain based on the P-SEA algorithm (Labesse 1997)
+# CODE FROM BIOKITE
+# From Krypton
+import numpy as np
+import random
+import torch
+
+def vector_dot(v1,v2):
+ return (v1*v2).sum(axis=-1)
+
+def norm_vector(v):
+ factor = np.linalg.norm(v, axis=-1)
+ if isinstance(factor, np.ndarray):
+ v /= factor[..., np.newaxis]
+ else:
+ v /= factor
+ return v
+
+def coord(x):
+ return np.asarray(x)
+def displacement(atoms1, atoms2):
+ v1 = coord(atoms1)
+ v2 = coord(atoms2)
+ if len(v1.shape) <= len(v2.shape):
+ diff = v2 - v1
+ else:
+ diff = -(v1 - v2)
+ return diff
+def distance(atoms1, atoms2):
+ diff = displacement(atoms1, atoms2)
+ return np.sqrt(vector_dot(diff, diff))
+
+def angle(atoms1, atoms2, atoms3):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms3, atoms2)
+ norm_vector(v1)
+ norm_vector(v2)
+ return np.arccos(vector_dot(v1,v2))
+
+def dihedral(atoms1, atoms2, atoms3, atoms4):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms2, atoms3)
+ v3 = displacement(atoms3, atoms4)
+ norm_vector(v1)
+ norm_vector(v2)
+ norm_vector(v3)
+
+ n1 = np.cross(v1, v2)
+ n2 = np.cross(v2, v3)
+
+ # Calculation using atan2, to ensure the correct sign of the angle
+ x = vector_dot(n1,n2)
+ y = vector_dot(np.cross(n1,n2), v2)
+ return np.arctan2(y,x)
+
+def replace_letters(arr):
+ # Create a dictionary that maps the letters 'a', 'b', and 'c' to the corresponding numbers
+ letter_to_number = {'a': 0, 'b': 1, 'c': 2}
+
+ # Create a new array that will hold the numbers
+ nums = []
+
+ # Loop through the input array and replace the letters with the corresponding numbers
+ for letter in arr:
+ if letter in letter_to_number:
+ nums.append(letter_to_number[letter])
+ else:
+ nums.append(letter)
+
+ return np.array(nums)
+
+def replace_with_mask(arr, percentage, replace_loops=False):
+ # Make sure the percentage is between 0 and 100
+ percentage = min(max(percentage, 0), 100)
+
+ # Calculate the number of values to replace
+ num_to_replace = int(len(arr) * percentage / 100)
+
+ # Choose a random subset of the array to replace
+ replace_indices = random.sample(range(len(arr)), num_to_replace)
+
+ # Replace the values at the chosen indices with the number 3
+ for i in replace_indices:
+ arr[i] = 3
+
+ if replace_loops:
+ for i in arr:
+ if arr[i] == 2:
+ arr[i] = 3
+
+ return arr
+
+def annotate_sse(ca_coord, percentage_mask=0, replace_loops=False):
+ _radians_to_angle = 2*np.pi/360
+
+ _r_helix = ((89-12)*_radians_to_angle, (89+12)*_radians_to_angle)
+ _a_helix = ((50-20)*_radians_to_angle, (50+20)*_radians_to_angle)
+ _d2_helix = ((5.5-0.5), (5.5+0.5))
+ _d3_helix = ((5.3-0.5), (5.3+0.5))
+ _d4_helix = ((6.4-0.6), (6.4+0.6))
+
+ _r_strand = ((124-14)*_radians_to_angle, (124+14)*_radians_to_angle)
+ _a_strand = ((-180)*_radians_to_angle, (-125)*_radians_to_angle,
+ (145)*_radians_to_angle, (180)*_radians_to_angle)
+ _d2_strand = ((6.7-0.6), (6.7+0.6))
+ _d3_strand = ((9.9-0.9), (9.9+0.9))
+ _d4_strand = ((12.4-1.1), (12.4+1.1))
+
+ # Filter all CA atoms in the relevant chain.
+
+ d2i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d3i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d4i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ ri_coord = np.full(( len(ca_coord), 3, 3 ), np.nan)
+ ai_coord = np.full(( len(ca_coord), 4, 3 ), np.nan)
+
+ # The distances and angles are not defined for the entire interval,
+ # therefore the indices do not have the full range
+ # Values that are not defined are NaN
+ for i in range(1, len(ca_coord)-1):
+ d2i_coord[i] = (ca_coord[i-1], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ d3i_coord[i] = (ca_coord[i-1], ca_coord[i+2])
+ for i in range(1, len(ca_coord)-3):
+ d4i_coord[i] = (ca_coord[i-1], ca_coord[i+3])
+ for i in range(1, len(ca_coord)-1):
+ ri_coord[i] = (ca_coord[i-1], ca_coord[i], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ ai_coord[i] = (ca_coord[i-1], ca_coord[i],
+ ca_coord[i+1], ca_coord[i+2])
+
+ d2i = distance(d2i_coord[:,0], d2i_coord[:,1])
+ d3i = distance(d3i_coord[:,0], d3i_coord[:,1])
+ d4i = distance(d4i_coord[:,0], d4i_coord[:,1])
+ ri = angle(ri_coord[:,0], ri_coord[:,1], ri_coord[:,2])
+ ai = dihedral(ai_coord[:,0], ai_coord[:,1],
+ ai_coord[:,2], ai_coord[:,3])
+
+ sse = np.full(len(ca_coord), "c", dtype="U1")
+
+ # Annotate helices
+ # Find CA that meet criteria for potential helices
+ is_pot_helix = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if (
+ d3i[i] >= _d3_helix[0] and d3i[i] <= _d3_helix[1]
+ and d4i[i] >= _d4_helix[0] and d4i[i] <= _d4_helix[1]
+ ) or (
+ ri[i] >= _r_helix[0] and ri[i] <= _r_helix[1]
+ and ai[i] >= _a_helix[0] and ai[i] <= _a_helix[1]
+ ):
+ is_pot_helix[i] = True
+ # Real helices are 5 consecutive helix elements
+ is_helix = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ for i in range(len(sse)):
+ if is_pot_helix[i]:
+ counter += 1
+ else:
+ if counter >= 5:
+ is_helix[i-counter : i] = True
+ counter = 0
+ # Extend the helices by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_helix[i]:
+ sse[i] = "a"
+ if (
+ d3i[i-1] >= _d3_helix[0] and d3i[i-1] <= _d3_helix[1]
+ ) or (
+ ri[i-1] >= _r_helix[0] and ri[i-1] <= _r_helix[1]
+ ):
+ sse[i-1] = "a"
+ sse[i] = "a"
+ if (
+ d3i[i+1] >= _d3_helix[0] and d3i[i+1] <= _d3_helix[1]
+ ) or (
+ ri[i+1] >= _r_helix[0] and ri[i+1] <= _r_helix[1]
+ ):
+ sse[i+1] = "a"
+ i += 1
+
+ # Annotate sheets
+ # Find CA that meet criteria for potential strands
+ is_pot_strand = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if ( d2i[i] >= _d2_strand[0] and d2i[i] <= _d2_strand[1]
+ and d3i[i] >= _d3_strand[0] and d3i[i] <= _d3_strand[1]
+ and d4i[i] >= _d4_strand[0] and d4i[i] <= _d4_strand[1]
+ ) or (
+ ri[i] >= _r_strand[0] and ri[i] <= _r_strand[1]
+ and ( (ai[i] >= _a_strand[0] and ai[i] <= _a_strand[1])
+ or (ai[i] >= _a_strand[2] and ai[i] <= _a_strand[3]))
+ ):
+ is_pot_strand[i] = True
+ # Real strands are 5 consecutive strand elements,
+ # or shorter fragments of at least 3 consecutive strand residues,
+ # if they are in hydrogen bond proximity to 5 other residues
+ pot_strand_coord = ca_coord[is_pot_strand]
+ is_strand = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ contacts = 0
+ for i in range(len(sse)):
+ if is_pot_strand[i]:
+ counter += 1
+ coord = ca_coord[i]
+ for strand_coord in ca_coord:
+ dist = distance(coord, strand_coord)
+ if dist >= 4.2 and dist <= 5.2:
+ contacts += 1
+ else:
+ if counter >= 4:
+ is_strand[i-counter : i] = True
+ elif counter == 3 and contacts >= 5:
+ is_strand[i-counter : i] = True
+ counter = 0
+ contacts = 0
+ # Extend the strands by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_strand[i]:
+ sse[i] = "b"
+ if d3i[i-1] >= _d3_strand[0] and d3i[i-1] <= _d3_strand[1]:
+ sse[i-1] = "b"
+ sse[i] = "b"
+ if d3i[i+1] >= _d3_strand[0] and d3i[i+1] <= _d3_strand[1]:
+ sse[i+1] = "b"
+ i += 1
+ sse=replace_letters(sse)
+ sse=replace_with_mask(sse, percentage_mask, replace_loops=replace_loops)
+ sse=torch.nn.functional.one_hot(torch.tensor(sse), num_classes=4)
+ return sse
diff --git a/utils/contigs.py b/utils/contigs.py
new file mode 100644
index 0000000000000000000000000000000000000000..f0057e729528392d3b297d49aec8a7db901b12f2
--- /dev/null
+++ b/utils/contigs.py
@@ -0,0 +1,1415 @@
+# utility functions for dealing with contigs during hallucination
+import numpy as np
+import random, copy, torch, geometry, os, sys
+from kinematics import xyz_to_t2d
+
+def parse_range_string(el):
+ ''' Splits string with integer or integer range into start and end ints. '''
+ if '-' in el:
+ s,e = el.split('-')
+ s,e = int(s), int(e)
+ else:
+ s,e = int(el), int(el)
+ return s,e
+
+def ranges_to_indexes(range_string):
+ '''Converts a string containig comma-separated numeric ranges to a list of integers'''
+ idx = []
+ for x in range_string.split(','):
+ start, end = parse_range_string(x)
+ idx.extend(np.arange(start, end+1))
+ return np.array(idx)
+
+def parse_contigs(contig_input, pdb_id):
+ '''
+ Input: contig start/end by pdb chain and residue number as in the pdb file
+ ex - B12-17
+ Output: corresponding start/end indices of the "features" numpy array (idx0)
+ '''
+ contigs = []
+ for con in contig_input.split(','):
+ pdb_ch = con[0]
+ pdb_s, pdb_e = parse_range_string(con[1:])
+
+ np_s = pdb_id.index((pdb_ch, pdb_s))
+ np_e = pdb_id.index((pdb_ch, pdb_e))
+
+ contigs.append([np_s, np_e])
+ return contigs
+
+
+def mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out):
+ #####################################
+ # rearrange ref features according to hal_2_ref_idx0
+ #####################################
+ #1. find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ #2. rearrange the 6D features
+ hal_len = len(hal_2_ref_idx0)
+ if 'feat' in pdb_out:
+ d_feat = pdb_out['feat'].shape[3:]
+
+ feat_hal = np.zeros((1, hal_len, hal_len) + d_feat)
+ feat_ref = pdb_out['feat'] # (B,L,L,...)
+
+ feat_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ else:
+ feat_hal = None
+
+ #3. make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+
+ #####################################
+ # mappings between hal and ref
+ #####################################
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_out['pdb_idx'][i] for i in ref_idx0],
+ 'mask_1d': mask_1d,
+ }
+
+ return feat_hal, mappings
+
+def scatter_feats(template_mask, feat_1d_ref=None, feat_2d_ref=None, pdb_idx=None):
+ '''
+ Scatters 1D and/or 2D reference features according to mappings in hal_2_ref_idx0
+
+ Inputs
+ ----------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ feat_1d_ref: (np.array; (batch, L_ref, ...))
+ 1D refence features to scatter
+ feat_1d_ref: (np.array; (batch, L_ref, L_ref, ...))
+ pdb_idx: (list)
+ List of pdb chain and residue numbers, in the order that pdb features were read/parsed.
+
+ Outputs
+ ----------
+ feat_1d_hal: (np.array, (batch, L_hal, ...))
+ Scattered 1d reference features. "None" mappings are 0.
+ feat_2d_hal: (np.array, (batch, L_hal, L_hal, ...))
+ Scattered 2d reference features. "None" mappings are 0.
+ mappings: (dict)
+ Keeps track of corresponding possitions in ref and hal proteins.
+ '''
+ hal_2_ref_idx0, _ = contigs.sample_mask(template_mask, pdb_idx)
+ out = {}
+
+ # Find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+ hal_len = len(hal_2_ref_idx0)
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ # Make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+ # scatter 2D features
+ if feat_2d_ref is not None:
+ B = feat_2d_ref.shape[0]
+ d_feat = feat_2d_ref.shape[3:]
+ feat_2d_hal = np.zeros((B, hal_len, hal_len)+d_feat)
+ feat_2d_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_2d_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ out['feat_2d_hal'] = feat_2d_hal
+
+ # scatter 1D features
+ if feat_1d_ref is not None:
+ B = feat_1d_ref.shape[0]
+ d_feat = feat_1d_ref.shape[2:]
+ feat_1d_hal = np.zeros((B, hal_len)+d_feat)
+ feat_1d_hal[:, hal_idx0] = feat_1d_ref[:, ref_idx0]
+ out['feat_1d_hal'] = feat_1d_hal
+
+ # Mappings between hal and ref
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'mask_1d': mask_1d,
+ }
+
+ if pdb_idx is not None:
+ mappings.update({
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_idx[i] for i in ref_idx0],
+ })
+
+ out['mappings'] = mappings
+
+ return out
+
+def scatter_contigs(contigs, pdb_out, L_range, keep_order=False, min_gap=0):
+ '''
+ Randomly places contigs in a protein within the length range.
+
+ Inputs
+ Contig: A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number. Comma separated
+ ex: B6-11,A12-19
+ pdb_out: dictionary from the prep_input function
+ L_range: String range of possible lengths.
+ ex: 90-110
+ ex: 70
+ keep_order: keep contigs in the provided order or randomly permute
+ min_gap: minimum number of amino acids separating contigs
+
+ Outputs
+ feat_hal: target pdb features to hallucinate
+ mappings: dictionary of ways to convert from the hallucinated protein
+ to the reference protein
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #####################################
+ # make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ #####################################
+ #1. Permute contig order
+ contigs = contigs.split(',')
+
+ if not keep_order:
+ random.shuffle(contigs)
+
+ #2. convert to ref_idx0
+ contigs_ref_idx0 = []
+ for con in contigs:
+ chain = con[0]
+ s, e = parse_range_string(con[1:])
+ contigs_ref_idx0.append( [ref_pdb_2_idx0[(chain, i)] for i in range(s, e+1)] )
+
+ #3. Add minimum gap size
+ for i in range(len(contigs_ref_idx0) - 1):
+ contigs_ref_idx0[i] += [None] * min_gap
+
+ #4. Sample protein length
+ L_low, L_high = parse_range_string(L_range)
+ L_hal = np.random.randint(L_low, L_high+1)
+
+ L_con = 0
+ for con in contigs_ref_idx0:
+ L_con += len(con)
+
+ L_gaps = L_hal - L_con
+
+ if L_gaps <= 1:
+ print("Error: The protein isn't long enough to incorporate all the contigs."
+ "Consider reduce the min_gap or increasing L_range")
+ return
+
+ #5. Randomly insert contigs into gaps
+ hal_2_ref_idx0 = np.array([None] * L_gaps, dtype=float) # inserting contigs into this
+ n_contigs = len(contigs_ref_idx0)
+ insertion_idxs = np.random.randint(L_gaps + 1, size=n_contigs)
+ insertion_idxs.sort()
+
+ for idx, con in zip(insertion_idxs[::-1], contigs_ref_idx0[::-1]):
+ hal_2_ref_idx0 = np.insert(hal_2_ref_idx0, idx, con)
+
+ #6. Convert mask to feat_hal and mappings
+ hal_2_ref_idx0 = [int(el) if ~np.isnan(el) else None for el in hal_2_ref_idx0] # convert nan to None
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #7. Generate str of the sampled mask
+ contig_positive = np.array(hal_2_ref_idx0) != None
+ boundaries = np.where(np.diff(contig_positive))[0]
+ start_idx0 = np.concatenate([np.array([0]), boundaries+1])
+ end_idx0 = np.concatenate([boundaries, np.array([contig_positive.shape[0]])-1])
+ lengths = end_idx0 - start_idx0 + 1
+ is_contig = contig_positive[start_idx0]
+
+ sampled_mask = []
+ con_counter = 0
+
+ for i, is_con in enumerate(is_contig):
+ if is_con:
+ sampled_mask.append(contigs[con_counter])
+ con_counter += 1
+ else:
+ len_gap = lengths[i]
+ sampled_mask.append(f'{len_gap}-{len_gap}')
+
+ sampled_mask = ','.join(sampled_mask)
+ mappings['sampled_mask'] = sampled_mask
+
+ return feat_hal, mappings
+
+def get_receptor_contig(ref_pdb_idx):
+ rec_pdb_idx = [idx for idx in ref_pdb_idx if idx[0]=='R']
+ return SampledMask.contract(rec_pdb_idx)
+
+def mk_con_to_set(mask, set_id=None, args=None, ref_pdb_idx=None):
+ '''
+ Maps a mask or list of contigs to a set_id. If no set_id is provided, it treats
+ everything as set 0.
+
+ Input
+ -----------
+ mask (str): Mask or list of contigs. Ex: 3,B6-11,12,A12-19,9 or Ex: B6-11,A12-19
+ ref_pdb_idx (List(ch, res)): pdb idxs of the reference pdb. Ex: [(A, 2), (A, 3), ...]
+ args: Arguments object. Must have args.receptor
+ set_id (list): List of integers. Length must match contigs in mask. Ex: [0,1]
+
+ Output
+ -----------
+ con_to_set (dict): Maps str of contig to integer
+ '''
+
+ # Extract contigs
+ cons = [l for l in mask.split(',') if l[0].isalpha()]
+
+ # Assign all contigs to set 0 if set_id is not passed
+ if set_id is None:
+ set_id = [0] * len(cons)
+
+ con_to_set = dict(zip(cons, set_id))
+
+ # Assign receptor to set 0
+ if args.receptor:
+ receptor_contig = get_receptor_contig(ref_pdb_idx)
+ con_to_set.update({receptor_contig: 0})
+
+ return con_to_set
+
+def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = parse_range(contig)
+
+ return ch, s, e
+
+def mask_as_list(sampled_mask):
+ '''
+ Make a length L_hal list, with each position pointing to a ref_pdb_idx (or None)
+ '''
+ mask_list = []
+ for l in sampled_mask.split(','):
+ ch, s, e = parse_contig(l)
+ # contig
+ if ch is not None:
+ mask_list += [(ch, idx) for idx in range(s, e+1)]
+ # gap
+ else:
+ mask_list += [None for _ in range(s, e+1)]
+
+ return mask_list
+
+def mask_subset(sampled_mask, subset):
+ '''
+ Returns a 1D boolean array of where a subset of the contig is in the hallucinated protein
+
+ Input
+ ---------
+ subset (str): Some chain and residue subset of the contigs. Ex: A10-15
+ Can also just pass chain. All contig residues from that chain are selected. Ex: R
+
+ Ouput
+ ---------
+ m_1d (np.array): Boolean array where subset appears in the hallucinated protein
+
+ '''
+ mask_list = mask_as_list(sampled_mask)
+ m_1d = []
+
+ ch_subset, s, e = parse_contig(subset)
+ assert ch_subset.isalpha(), '"Subset" must include a chain reference'
+
+ if (s is None) or (e is None):
+ s = -np.inf
+ e = np.inf
+
+ for l in mask_list:
+ if l is None:
+ continue
+
+ ch, idx = l
+ if (ch == ch_subset) and (idx >= s) and (idx <= e):
+ m_1d.append(True)
+ else:
+ m_1d.append(False)
+
+ return np.array(m_1d)
+
+def mk_cce_and_hal_mask_2d(sampled_mask, con_to_set=None):
+ '''
+ Makes masks for ij pixels where the cce and hallucination loss should be applied.
+
+ Inputs
+ ---------------
+ sampled_mask (str): String of where contigs should be applied. Ex: 3,B6-11,12,A12-19,9
+ cce_cutoff (float): Apply cce loss to cb-cb distances less than this value. Angstroms.
+ con_to_set (dict): Dictionary mapping the string of a contig (ex: 'B6-11') to an integer.
+ L_rec (int): Length of the receptor, if hallucinating in the context of the receptor.
+
+ Outputs
+ ---------------
+ mask_cce (np.array, (L_hal, L_hal)): Boolean array. True where cce loss should be applied.
+ mask_hal (np.array, (L_hal, L_hal)): Boolean array. True where hallucination loss should be applied.
+ '''
+ if con_to_set is None:
+ con_to_set = mk_con_to_set(sampled_mask)
+
+ # Length of hallucinated protein
+ L_hal, L_max = mask_len(sampled_mask)
+ assert L_hal == L_max, 'A sampled mask must have gaps of a single length.'
+
+ # Map each contig to a 1D boolean mask
+ m_con = dict()
+ start_idx = 0
+ for l in sampled_mask.split(','):
+ if l[0].isalpha():
+ s, e = parse_range_string(l[1:])
+ L_con = e - s + 1
+ m = np.zeros(L_hal, dtype=bool)
+ m[start_idx:start_idx+L_con] = True
+
+ m_con[l] = m
+ start_idx += L_con
+ else:
+ L_gap, _ = parse_range_string(l)
+ start_idx += L_gap
+
+ # Combine contigs masks from each set to make 2D mask
+ mask_cce = np.zeros((L_hal, L_hal), dtype=bool)
+ for set_id in set(con_to_set.values()):
+ # gather all masks from contigs in the same set
+ masks = [m_con[k] for k,v in con_to_set.items() if v == set_id]
+ mask_1D = np.any(masks, axis=0)
+ update = mask_1D[:,None] * mask_1D[None,:]
+ mask_cce = np.any([mask_cce, update], axis=0)
+
+ # Make mask_hal
+ mask_hal = ~mask_cce
+
+ # Don't apply ANY losses on diagonal
+ mask_cce[np.arange(L_hal), np.arange(L_hal)] = False
+ mask_hal[np.arange(L_hal), np.arange(L_hal)] = False
+
+ # Don't apply ANY losses to receptor
+ m_1d_rec = mask_subset(sampled_mask, 'R')
+ m_2d_rec = m_1d_rec[:, None] * m_1d_rec[None, :]
+ mask_cce *= ~m_2d_rec
+ mask_hal *= ~m_2d_rec
+
+ return mask_cce, mask_hal
+
+
+def apply_mask(mask, pdb_out):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ pdb_out: dictionary from the prep_input function
+
+
+ Outputs
+ -------------
+ feat_hal: features from pdb_out scattered according to the sampled mask
+ mappings: dict keeping track of corresponding positions in the ref and hal features
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ #2. Convert mask to feat_hal and mappings
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #3. Record the mask that was sampled
+ mappings['sampled_mask'] = ','.join(sampled_mask)
+
+ return feat_hal, mappings
+
+
+def sample_mask(mask, pdb_idx):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ Outputs
+ -------------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ sampled_mask: (str)
+ string of the sampled mask, so the transformations can be reapplied
+ ex - '3-3,B6-11,9-9,A36-42,20-20,A12-24,5-5'
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_i:i for i, pdb_i in enumerate(pdb_idx)}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ return hal_2_ref_idx0, sampled_mask
+
+
+class GapResampler():
+ def __init__(self, use_bkg=True):
+ '''
+
+ '''
+
+ self.counts_passed = {} # dictionary for tallying counts of gap lengths for designs passing some threshold
+ self.counts_bkg = {}
+ self.use_bkg = use_bkg
+
+
+ def clean_mask(self, mask):
+ '''
+ Makes mask into a cononical form.
+ Ensures masks always alternate gap, contig and that
+ masks begin and end with a gap (even of length 0)
+
+ Input
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+
+ Output
+ -----------
+ A canonicalized mask. Ex: N,9,A12-19,15,B45-52,0,C
+ '''
+ mask = mask.split(',')
+ mask_out = []
+ was_contig = True
+ was_gap = False
+
+ for i, el in enumerate(mask):
+ is_contig = el[0].isalpha()
+ is_gap = not is_contig
+ is_last = i == len(mask) - 1
+
+ # accepting gaps as either x-x or just x
+ if is_gap:
+ if '-' in el:
+ x1, x2 = el.split('-')
+ if x1 != x2:
+ print(f"Error: Gap must not be a range: {mask}")
+ return None
+ gap = x1
+ else:
+ gap = el
+
+ if is_contig:
+ contig = el
+
+ # gap -> contig: just append new contig
+ if (was_gap and is_contig):
+ mask_out.append(contig)
+
+ # contig -> gap: just append gap
+ elif (was_contig and is_gap):
+ mask_out.append(gap)
+
+ # contig -> contig: insert gap of 0, then add contig
+ elif (was_contig and is_contig):
+ mask_out.append('0')
+ mask_out.append(contig)
+
+ # gap -> gap: add them
+ elif (was_gap and is_gap):
+ combined_len = int(mask_out[-1]) + int(gap)
+ mask_out[-1] = str(combined_len)
+
+ # ensure last mask element is a gap
+ if (is_last and is_contig):
+ mask_out.append('0')
+
+ # update what previous element was
+ was_contig = el[0].isalpha()
+ was_gap = ~is_contig
+
+ # add 'N' and 'C' contigs
+ mask_out.insert(0, 'N')
+ mask_out.append('C')
+
+ return ','.join(mask_out)
+
+
+ def add_mask(self, mask, counting_dict):
+ '''
+ Adds counts of gap lengths to counting_dict
+
+ Inputs
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+ '''
+ mask = self.clean_mask(mask)
+ mask = mask.split(',')
+ n_gaps = len(mask) // 2
+
+ # count occurances of contig,gap,contig triples
+ for i in range(n_gaps):
+ con1, gap, con2 = mask[2*i : 2*i+3]
+
+ # count gap length
+ if con1 in counting_dict:
+ if (gap, con2) in counting_dict[con1]:
+ counting_dict[con1][(gap, con2)] += 1
+ else:
+ counting_dict[con1][(gap, con2)] = 1
+ else:
+ counting_dict[con1] = {(gap, con2): 1}
+
+
+ def add_mask_pass(self, mask):
+ '''
+ Add a mask that passed to self.counts_passed
+ '''
+ self.add_mask(mask, self.counts_passed)
+
+
+ def add_mask_bkg(self, mask):
+ '''
+ Add a mask that passed to self.counts_bkg
+ '''
+ self.add_mask(mask, self.counts_bkg)
+
+
+ def get_enrichment(self):
+ '''
+ Calculate the ratio of counts_passed / count_bkg
+ Also notes all contigs
+ '''
+ if self.use_bkg is False:
+ print('Please pass in background masks and set self.use_bkg=True')
+ return
+
+ self.counts_enrich = copy.copy(self.counts_passed)
+ self.con_all = set()
+
+ for con1 in self.counts_enrich.keys():
+ self.con_all |= set([con1])
+
+ for gap, con2 in self.counts_enrich[con1].keys():
+ self.con_all |= set([con2])
+ bkg = self.counts_bkg[con1][(gap, con2)]
+ cnt = self.counts_passed[con1][(gap, con2)]
+ self.counts_enrich[con1][(gap, con2)] = cnt / bkg
+
+ def sample_mask(self):
+ '''
+ Sample a mask
+ '''
+ searching = True
+ while searching:
+ n_gaps = len(self.con_all) - 1
+ mask = ['N']
+
+ if self.use_bkg:
+ counts = self.counts_enrich
+ else:
+ counts = self.counts_passed
+
+ for i in range(n_gaps):
+ con_last = mask[-1]
+
+ # only allow jump to C as last option
+ if i == n_gaps - 1:
+ con_used = set(mask[::2])
+ else:
+ con_used = set(mask[::2]+['C'])
+
+ con_free = self.con_all - con_used
+
+ # get available "jumps" (con -> gap, con) you can make
+ jumps_all = counts[con_last]
+ jumps_free = {k:v for k,v in jumps_all.items() if k[1] in con_free}
+
+ if len(jumps_free) == 0:
+ print('No available jumps to continue the mask. Sampling again...')
+ else:
+ # normalize counts and sample move
+ mvs, cnt = zip(*jumps_free.items())
+ cnt = np.array(cnt)
+ prob = cnt / cnt.sum()
+ idx = np.random.choice(len(prob), p=prob)
+ mv = mvs[idx]
+
+ # add to the mask
+ mask.append(mv[0])
+ mask.append(mv[1])
+
+ # check that mask has the right number of elements
+ if len(mask) == 2*n_gaps + 1:
+ searching = False
+ else:
+ searching = True
+
+ return ','.join(mask[1:-1])
+
+
+ def gaps_as_ranges(self, mask):
+ '''
+ Convert gaps of a single int to ranges, for
+ backwards compatibility reasons
+ '''
+
+ mask_out = []
+ for el in mask.split(','):
+ if el[0].isalpha():
+ mask_out.append(el)
+ else:
+ mask_out.append(f'{el}-{el}')
+
+ return ','.join(mask_out)
+
+
+def recover_mask(trb):
+ '''
+ Recover the string of the sampled mask given the trb file
+ '''
+
+ L_hal = trb['mask_contig'].shape[0]
+ mask = []
+
+ for idx0 in range(L_hal):
+ # what is the current idx
+ if idx0 in trb['con_hal_idx0']:
+ is_con = True
+ is_gap = False
+ else:
+ is_con = False
+ is_gap = True
+
+ # dealing with the first entry
+ if idx0 == 0:
+ if is_gap:
+ L_gap = 1
+ elif is_con:
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+
+ # take action based on what happend last time
+ else:
+ if (was_gap) and (is_gap):
+ L_gap +=1
+ #elif (was_con) and (is_con):
+ # continue
+ elif (was_gap) and (is_con):
+ # end gap
+ mask.append(f'{L_gap}-{L_gap}')
+ # start con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+ elif (was_con) and (is_gap):
+ # end con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ mask.append(f'{con_start}-{idx}')
+ # start gap
+ L_gap = 1
+
+ # dealing with last entry
+ if idx0 == L_hal-1:
+ if is_gap:
+ mask.append(f'{L_gap}-{L_gap}')
+ elif is_con: # (edge case not handled: con starts and ends on last idx)
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0-1) ]
+ mask.append(f'{con_start}-{idx}')
+
+ # update what last position was
+ was_con = copy.copy(is_con)
+ was_gap = copy.copy(is_gap)
+
+ return ','.join(mask)
+
+
+def mask_len(mask):
+ '''
+ Calculate the min and max possible length that can
+ be sampled given a mask
+ '''
+ L_min = 0
+ L_max = 0
+
+ for el in mask.split(','):
+ if el[0].isalpha(): # is con
+ con_s, con_e = el[1:].split('-')
+ con_s, con_e = int(con_s), int(con_e)
+ L_con = con_e - con_s + 1
+ L_min += L_con
+ L_max += L_con
+
+ else: # is gap
+ if '-' in el:
+ gap_min, gap_max = el.split('-')
+ gap_min, gap_max = int(gap_min), int(gap_max)
+ L_min += gap_min
+ L_max += gap_max
+ else:
+ L_min += int(el)
+ L_max += int(el)
+
+ return L_min, L_max
+
+class SampledMask():
+ def __init__(self, mask_str, ref_pdb_idx, con_to_set=None):
+ self.str = mask_str
+ self.L_hal = len(self)
+ self.L_ref = len(ref_pdb_idx)
+
+ #################
+ # con indices in hal and ref
+ #################
+ self.ref_pdb_idx = ref_pdb_idx
+ self.hal_pdb_idx = [('A', i) for i in range(1, len(self)+1)]
+
+ hal_idx0 = 0
+ con_ref_pdb_idx = []
+ con_hal_pdb_idx = []
+ con_ref_idx0 = []
+ con_hal_idx0 = []
+
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ for res in range(s, e+1):
+ con_ref_pdb_idx.append((ch, res))
+ con_hal_pdb_idx.append(('A', hal_idx0+1))
+ con_ref_idx0.append(self.ref_pdb_idx.index((ch, res)))
+ con_hal_idx0.append(hal_idx0)
+ hal_idx0 += 1
+ # gap
+ else:
+ for _ in range(s):
+ hal_idx0 += 1
+
+ self.con_mappings = {
+ 'ref_pdb_idx': con_ref_pdb_idx,
+ 'hal_pdb_idx': con_hal_pdb_idx,
+ 'ref_idx0': con_ref_idx0,
+ 'hal_idx0': con_hal_idx0,
+ }
+
+ #################
+ # con_to_set mapping
+ #################
+ if con_to_set:
+ self.con_to_set = con_to_set
+ else:
+ contigs = self.get_contigs()
+ self.con_to_set = dict(zip(contigs, len(contigs)*[0]))
+
+ # set_to_con mapping
+ set_to_con = {}
+ for k, v in self.con_to_set.items():
+ set_to_con[v] = set_to_con.get(v, []) + [k] # invert a dictionary with non-unique values
+ self.set_to_con = set_to_con
+
+ def __len__(self,):
+ _, L_max = self.mask_len(self.str)
+ return L_max
+
+ def map(self, sel, src, dst):
+ '''
+ Convert the contig selection in one indexing scheme to another.
+ Will return None if selection is not in a contig.
+
+ Input
+ ----------
+ sel (str): selection of a contig range or idx0 range. Can take multiple comma separated values of same type. Ex: A5-10,B2-8 or 3-8,14-21
+ src (str): <'ref', 'hal'>
+ dst (str): <'ref_pdb_idx', 'hal_pdb_idx', 'ref_idx0', 'hal_idx0>
+ '''
+ out = []
+ for con in sel.split(','):
+
+ ch, s, e = SampledMask.parse_contig(con)
+
+ # selection type is pdb_idx
+ if ch:
+ src_long = f'{src}_pdb_idx'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get((ch, res)) for res in range(s, e+1)]
+
+ # selection type is idx0
+ else:
+ src_long = f'{src}_idx0'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get(i) for i in range(s, e+1)]
+
+ return out
+
+ @staticmethod
+ def expand(mask_str):
+ '''
+ Ex: '2,A3-5,3' --> [None, None, (A,3), (A,4), (A,5), None, None, None]
+ '''
+ expanded = []
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ expanded += [(ch, res) for res in range(s, e+1)]
+ # gap
+ else:
+ expanded += [None for _ in range(s)]
+
+ return expanded
+
+ @staticmethod
+ def contract(pdb_idx):
+ '''
+ Inverse of expand
+ Ex: [None, None, (A,3), (A,4), (A,5), None, None, None] --> '2,A3-5,3'
+ '''
+
+ contracted = []
+ l_prev = (None, -200)
+ first_el_written = False
+
+ for l_curr in pdb_idx:
+ if l_curr is None:
+ l_curr = (None, -100)
+
+ # extend gap
+ if l_curr == l_prev:
+ L_gap += 1
+
+ # extend con
+ elif l_curr == (l_prev[0], l_prev[1]+1):
+ con_e = l_curr[1]
+
+ # new gap
+ elif (l_curr != l_prev) and (l_curr[0] is None):
+ # write prev con
+ if 'con_ch' in locals():
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+
+ L_gap = 1
+
+ # new con
+ elif (l_curr != l_prev) and isinstance(l_curr[0], str):
+ # write prev con
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ # write prev gap
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ con_ch = l_curr[0]
+ con_s = l_curr[1]
+ con_e = l_curr[1]
+
+ # update l_prev
+ l_prev = l_curr
+
+ # write last element
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ return ','.join(contracted)
+
+ def subset(self, sub):
+ '''
+ Make a mask_str that is a subset of the original mask_str
+ Ex: self.mask_str = '2,A5-20,4', sub='A5-10' --> '2,A5-10,14'
+ '''
+
+ # map from hal_idx0 to ref_pdb_idx
+ hal_idx0 = self.map(sub, 'ref', 'hal_idx0')
+ ref_pdb_idx = SampledMask.expand(sub)
+ mapping = dict(zip(hal_idx0, ref_pdb_idx))
+
+ expanded = [mapping.get(idx0) for idx0 in range(len(self))]
+
+ return self.contract(expanded)
+
+ def mask_len(self, mask):
+ '''
+ Technically, can take both sampled and unsampled mask
+ '''
+ L_min = 0
+ L_max = 0
+ for l in self.str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ L_min += e - s + 1
+ L_max += e - s + 1
+ # gap
+ else:
+ L_min += s
+ L_max += e
+
+ return L_min, L_max
+
+ def get_contigs(self, include_receptor=True):
+ '''
+ Get a list of all contigs in the mask
+ '''
+ [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0]]
+
+ contigs = []
+ for con in self.str.split(','):
+ ch = SampledMask.parse_contig(con)[0]
+ if ch == 'R' and include_receptor == False:
+ continue
+ if ch:
+ contigs.append(con)
+
+ return contigs
+
+ def get_gaps(self,):
+ '''
+ Get a list of all gaps in the mask
+ '''
+ return [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0] is None]
+
+ @staticmethod
+ def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+ @staticmethod
+ def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = SampledMask.parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = SampledMask.parse_range(contig)
+
+ return ch, s, e
+
+ def remove_diag(self, m_2d):
+ '''
+ Set the diagonal of a 2D boolean array to False
+ '''
+ L = m_2d.shape[0]
+ m_2d[np.arange(L), np.arange(L)] = False
+
+ return m_2d
+
+ def get_receptor_contig(self,):
+ '''
+ Returns None if there is no chain R in the mask_str
+ '''
+ receptor_contig = [l for l in self.get_contigs() if 'R' in l]
+
+ if len(receptor_contig) == 0:
+ receptor_contig = None
+ else:
+ receptor_contig = ','.join(receptor_contig)
+
+ return receptor_contig
+
+ def remove_receptor(self, m_2d):
+ '''
+ Remove intra-receptor contacts (chain R) from a mask
+ '''
+ receptor_contig = self.get_receptor_contig()
+
+ if receptor_contig: # has chain R
+ m_1d = np.zeros(self.L_hal, dtype=bool)
+ idx = np.array(self.map(receptor_contig, 'ref', 'hal_idx0'))
+ m_1d[idx] = True
+ update = m_1d[:, None] * m_1d[None, :]
+ m_2d = m_2d * ~update
+
+ return m_2d
+
+ def get_mask_con(self, include_receptor=False):
+ # Make a 2D boolean mask for each contig set
+ L = self.L_hal
+ mask_con = np.zeros([L, L], dtype=bool)
+
+ for set_id, contigs in self.set_to_con.items():
+ m_1d = np.zeros(L, dtype=bool)
+ for con in contigs:
+ idx = self.map(con, 'ref', 'hal_idx0')
+ idx = [l for l in idx if l != None]
+ idx = np.array(idx, dtype=int)
+ m_1d[idx] = True
+
+ update = m_1d[:, None] * m_1d[None, :]
+ mask_con = np.any([mask_con, update], axis=0)
+
+ # clean up
+ mask_con = self.remove_diag(mask_con)
+
+ if not include_receptor:
+ mask_con = self.remove_receptor(mask_con)
+
+ return mask_con
+
+ def get_mask_hal(self,):
+ mask_hal = ~self.get_mask_con()
+ mask_hal = self.remove_diag(mask_hal)
+ mask_hal = self.remove_receptor(mask_hal)
+
+ return mask_hal
+
+ def get_mask_cce(self, pdb, cce_cutoff=20., include_receptor=False):
+ '''
+ Remove ij pixels where contig distances are greater than cce_cutoff.
+ '''
+ # start with mask_con
+ mask_con = self.get_mask_con(include_receptor=include_receptor)
+
+ # get ref dists
+ xyz_ref = torch.tensor(pdb['xyz'][:,:3,:]).float()
+ c6d_ref = geometry.xyz_to_c6d(xyz_ref[None].permute(0,2,1,3),{'DMAX':20.0}).numpy()
+ dist = c6d_ref[0,:,:,0] # (L_ref, L_ref)
+
+ # scatter
+ dist_scattered = self.scatter_2d(dist)
+
+ # apply cce cuttoff
+ update = dist_scattered < cce_cutoff
+ mask_cce = np.all([mask_con, update], axis=0)
+
+ return mask_cce
+
+ def scatter_2d(self, ref_feat_2d):
+ '''
+ Inputs
+ ---------
+ ref_feat_2d (np.array; (L_ref, L_ref, ...)): Features to be scattered. The first two leading dimensions must be equal to L_ref.
+ '''
+ assert ref_feat_2d.shape[:2] == (self.L_ref, self.L_ref), 'ERROR: feat_2d must have leading dimensions of (L_ref, L_ref)'
+
+ trailing_dims = ref_feat_2d.shape[2:]
+ dtype = ref_feat_2d.dtype
+ hal_feat_2d = np.zeros((self.L_hal, self.L_hal)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_2d[con_hal_idx0[:, None], con_hal_idx0[None, :]] = ref_feat_2d[ref_hal_idx0[:, None], ref_hal_idx0[None, :]]
+
+ return hal_feat_2d
+
+ def scatter_1d(self, ref_feat_1d):
+ '''
+ Inputs
+ ---------
+ ref_feat_1d (np.array; (L_ref, ...)): Features to be scattered. The first leading dimension must be equal to L_ref.
+ '''
+ assert ref_feat_1d.shape[0] == self.L_ref, 'ERROR: feat_1d must have leading dimensions of (L_ref,)'
+
+ trailing_dims = ref_feat_1d.shape[1:]
+ dtype = ref_feat_1d.dtype
+ hal_feat_1d = np.zeros((self.L_hal,)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_1d[con_hal_idx0] = ref_feat_1d[ref_hal_idx0]
+
+ return hal_feat_1d
+
+ def idx_for_template(self, gap=200):
+ '''
+ Essentially return hal_idx0, except have a large jump for chain B,
+ to simulate a chain break. If B contains internal jumps in residue
+ numbering, these are preserved.
+ '''
+
+ is_rec = self.m1d_receptor()
+ resi_rec = np.array([idx[1] for idx in SampledMask.expand(self.str)
+ if idx is not None and idx[0]=='R'])
+ L_binder = sum(~is_rec)
+
+
+ if len(resi_rec)>0:
+ if is_rec[0]:
+ # receptor first
+ idx_tmpl = np.arange(resi_rec[-1]+gap+1, resi_rec[-1]+gap+1+L_binder)
+ idx_tmpl = np.concatenate([resi_rec, idx_tmpl])
+ else:
+ # receptor second
+ idx_tmpl = np.arange(L_binder)
+ if resi_rec[0] <= idx_tmpl[-1]+gap:
+ resi_rec += idx_tmpl[-1] - resi_rec[0] + gap + 1
+ idx_tmpl = np.concatenate([idx_tmpl, resi_rec])
+ else:
+ #when no receptor
+ idx_tmpl = np.arange(L_binder)
+ return idx_tmpl
+
+ def m1d_receptor(self,):
+ '''
+ Get a boolean array, True if the position corresponds to the receptor
+ '''
+ m1d = [(l is not None) and (l[0] == 'R') for l in SampledMask.expand(self.str)]
+ return np.array(m1d)
+
+ def erode(self, N_term=True, C_term=True):
+ '''
+ Reduce non-receptor contigs by 1 residue from the N and/or C terminus.
+ '''
+ x = SampledMask.expand(self.str)
+
+ if N_term:
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ if C_term:
+ x = x[::-1]
+
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ x = x[::-1]
+
+ self.str = self.contract(x)
+
+ return
+
+ def len_contigs(self, include_receptor=False):
+ con_str = ','.join(self.get_contigs(include_receptor))
+ return len(SampledMask.expand(con_str))
+
+
+def make_template_features(pdb, args, device, hal_2_ref_idx0=None, sm_loss=None):
+ '''
+ Inputs
+ ----------
+ sm_loss: Instance of a contig.SampledMask object used for making the loss masks.
+ '''
+ PARAMS = {
+ "DMIN" : 2.0,
+ "DMAX" : 20.0,
+ "DBINS" : 36,
+ "ABINS" : 36,
+ }
+ if args.use_template:
+ B,T = 1,1 # batch, templates
+
+ # spoof reference features
+ xyz_t = torch.tensor(pdb['xyz'][:, :3][None, None]) # (batch,templ,nres,3,3)
+ t0d = torch.ones((1,1,3)) # (batch, templ, 3)
+
+ t2d_ref = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS) # (B,T,L,L,...)
+ L_ref = t2d_ref.shape[2]
+ #t1d_ref = torch.ones(size=(B,T,L_ref,3), dtype=torch.float32, device=device)
+ a = 2 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ b = 0 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ c = 1 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+
+ t1d_ref = torch.stack([a,b,c], axis=-1)
+
+ # Get the mask_str for scattering template features
+ #1. Template mask = sampled mask
+ if (args.use_template.lower() == 't') or (args.use_template.lower() == 'true'):
+ sm_tmpl = sm_loss
+ #2. Template mask is a subset of the sampled mask
+ else:
+ subset_contigs = args.use_template
+
+ if args.receptor:
+ receptor_contig = sm_loss.get_receptor_contig()
+ subset_contigs = ','.join([subset_contigs, receptor_contig])
+
+ mask_str_tmpl = sm_loss.subset(subset_contigs)
+ sm_tmpl = SampledMask(mask_str=mask_str_tmpl, ref_pdb_idx=pdb['pdb_idx'])
+
+ # scatter template features
+ # make leading dims (L,(L),...)
+ t1d_ref = t1d_ref.permute(2,3,0,1) # (L, ..., B, T)
+ t2d_ref = t2d_ref.permute(2,3,4,0,1) # (L, L, ..., B, T)
+
+ t1d_tmpl = sm_tmpl.scatter_1d(t1d_ref.cpu().numpy())
+ t2d_tmpl = sm_tmpl.scatter_2d(t2d_ref.cpu().numpy())
+
+ # update t2d_tmpl with mask_con (could update with mask_cce instead?)
+ mask_con = sm_tmpl.get_mask_con(include_receptor=True)
+ t2d_tmpl = (t2d_tmpl.T * mask_con.T).T # trick to broadcast arrays if leading dimensions match
+
+ t1d_tmpl = torch.tensor(t1d_tmpl, device=device)
+ t2d_tmpl = torch.tensor(t2d_tmpl, device=device)
+
+ # Permute B and T dims back to front
+ t1d_tmpl = t1d_tmpl.permute(2,3,0,1)
+ t2d_tmpl = t2d_tmpl.permute(3,4,0,1,2)
+
+ # Make last 3 idx of last dim all 1 to mimick Ivan's template feature
+ t2d_tmpl[..., -3:] = 1.
+
+ idx = torch.tensor(sm_tmpl.idx_for_template(gap=200), device=device)[None]
+
+ net_kwargs = {
+ 'idx': idx,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ elif args.template_pdbs is not None:
+ B,T = 1, len(args.template_pdbs) # batch, templates
+
+ # get xyz features of all templates
+ xyz_t = [torch.tensor(parse_pdb(f_pdb)['xyz'][:, :3]) for f_pdb in args.template_pdbs]
+ xyz_t = torch.stack(xyz_t, axis=0)[None] # (batch, template, nres, 3, 3)
+ t0d = torch.ones(B,T,3)
+
+ t2d_tmpl = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS).to(device) # (B,T,L,L,...)
+ L_tmpl = t2d_tmpl.shape[2]
+ t1d_tmpl = torch.ones(size=(B,T,L_tmpl,3), dtype=torch.float32, device=device)
+
+ # spoof pdb idx
+ idx_tmpl = torch.range(0, L_tmpl-1, dtype=torch.long, device=device)[None]
+
+ # Net() kwargs
+ net_kwargs = {
+ 'idx': idx_tmpl,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ else:
+ net_kwargs = {}
+
+ return net_kwargs
diff --git a/utils/diff_utils.py b/utils/diff_utils.py
new file mode 100644
index 0000000000000000000000000000000000000000..0efce5b9e934f3eae6eec8b3523312e492050f2e
--- /dev/null
+++ b/utils/diff_utils.py
@@ -0,0 +1,276 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None,
+ MODEL_PARAM=None, hotspots=None, dssp=None, v2_mode=False):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ # EXPAND t1d to match model params
+ if MODEL_PARAM['d_t1d'] == 29:
+ ## added t1d features ##
+ # 24 -- dssp helix
+ # 25 -- dssp sheet
+ # 26 -- dssp loop
+ # 27 -- dssp mask
+ # 28 -- hotspot resi on target
+ t1d = torch.cat((t1d,torch.zeros(t1d.shape[0],t1d.shape[1],5)),dim=-1)
+ t1d[:,:,24:28] = dssp
+ t1d[:,:,28] = hotspots
+ t1d[:,str_mask,24:27] = 0.0
+ t1d[:,str_mask,27] = 1.0
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ if not v2_mode:
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def get_alphas(t1d, xyz_t, B, L, ti_dev, ti_flip, ang_ref):
+ # get torsion angles from templates
+ seq_tmp = t1d[...,:21].argmax(dim=-1).reshape(-1,L)
+ alpha, _, alpha_mask, _ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B, -1, L, 30)
+ return alpha, alpha_t
diff --git a/utils/examples/aa_weights.json b/utils/examples/aa_weights.json
new file mode 100644
index 0000000000000000000000000000000000000000..4b225021a85dbc28daa580d53b480a637bc1e0ad
--- /dev/null
+++ b/utils/examples/aa_weights.json
@@ -0,0 +1,22 @@
+{
+ "A": 0,
+ "R": 0,
+ "N": 0,
+ "D": 0,
+ "C": 0,
+ "Q": 0,
+ "E": 0,
+ "G": 0,
+ "H": 0,
+ "I": 0,
+ "L": 0,
+ "K": 0,
+ "M": 0,
+ "F": 0,
+ "P": 0,
+ "S": 0,
+ "T": 0,
+ "W": 0,
+ "Y": 0,
+ "V": 0
+}
diff --git a/utils/examples/binder_design.sh b/utils/examples/binder_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..ae66f0273cb2d3fa893717d7f3102ae01bd957b7
--- /dev/null
+++ b/utils/examples/binder_design.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/binder_design \
+ --pdb pdbs/cd86.pdb \
+ --T 25 --save_best_plddt \
+ --contigs B1-110,0 25-75 \
+ --hotspots B40,B32,B87,B96,B30
diff --git a/utils/examples/loop_design.sh b/utils/examples/loop_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..de6c21db8ed2bd4ab526e7c41e8fb7bd13f76754
--- /dev/null
+++ b/utils/examples/loop_design.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --pdb pdbs/G12D_manual_mut.pdb \
+ --out out/ab_loop \
+ --contigs A2-176,0 C7-16,0 H2-95,12-15,H111-116,0 L1-45,10-12,L56-107 \
+ --T 25 --save_best_plddt --loop_design
diff --git a/utils/examples/motif_scaffolding.sh b/utils/examples/motif_scaffolding.sh
new file mode 100644
index 0000000000000000000000000000000000000000..ba1f0b03227cc3de8ec140c2fcc42dead2a083e2
--- /dev/null
+++ b/utils/examples/motif_scaffolding.sh
@@ -0,0 +1,14 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --pdb pdbs/rsv5_5tpn.pdb \
+ --contigs 0-25,A163-181,25-30 --T 25 --save_best_plddt
diff --git a/utils/examples/out/design_000000.pdb b/utils/examples/out/design_000000.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..90f5532fa0ebdfa42f13edb342ad632b3dc18c16
--- /dev/null
+++ b/utils/examples/out/design_000000.pdb
@@ -0,0 +1,1805 @@
+ATOM 1 N ILE A 1 -8.484 18.449 12.122 1.00 0.91
+ATOM 2 CA ILE A 1 -9.332 17.874 11.084 1.00 0.91
+ATOM 3 C ILE A 1 -9.716 16.439 11.420 1.00 0.91
+ATOM 4 O ILE A 1 -8.928 15.696 12.005 1.00 0.91
+ATOM 5 CB ILE A 1 -8.642 17.955 9.711 1.00 0.91
+ATOM 6 CG1 ILE A 1 -9.569 17.509 8.577 1.00 0.91
+ATOM 7 CG2 ILE A 1 -7.373 17.116 9.726 1.00 0.91
+ATOM 8 CD1 ILE A 1 -9.021 17.784 7.196 1.00 0.91
+ATOM 9 H ILE A 1 -8.607 18.151 13.079 1.00 0.91
+ATOM 10 HA ILE A 1 -10.245 18.464 11.018 1.00 0.91
+ATOM 11 HB ILE A 1 -8.377 18.993 9.511 1.00 0.91
+ATOM 12 1HG2 ILE A 1 -6.874 17.194 8.761 1.00 0.91
+ATOM 13 2HG2 ILE A 1 -6.707 17.478 10.509 1.00 0.91
+ATOM 14 3HG2 ILE A 1 -7.628 16.074 9.920 1.00 0.91
+ATOM 15 1HG1 ILE A 1 -9.759 16.439 8.663 1.00 0.91
+ATOM 16 2HG1 ILE A 1 -10.529 18.018 8.670 1.00 0.91
+ATOM 17 1HD1 ILE A 1 -9.734 17.440 6.446 1.00 0.91
+ATOM 18 2HD1 ILE A 1 -8.857 18.855 7.076 1.00 0.91
+ATOM 19 3HD1 ILE A 1 -8.078 17.255 7.067 1.00 0.91
+ATOM 20 N SER A 2 -11.064 16.075 11.291 1.00 0.96
+ATOM 21 CA SER A 2 -11.514 14.698 11.461 1.00 0.96
+ATOM 22 C SER A 2 -10.842 13.772 10.455 1.00 0.96
+ATOM 23 O SER A 2 -10.369 14.214 9.408 1.00 0.96
+ATOM 24 CB SER A 2 -13.020 14.614 11.309 1.00 0.96
+ATOM 25 OG SER A 2 -13.397 14.833 9.978 1.00 0.96
+ATOM 26 H SER A 2 -11.722 16.772 10.974 1.00 0.96
+ATOM 27 HG SER A 2 -12.730 14.401 9.439 1.00 0.96
+ATOM 28 HA SER A 2 -11.244 14.367 12.464 1.00 0.96
+ATOM 29 1HB SER A 2 -13.366 13.632 11.631 1.00 0.96
+ATOM 30 2HB SER A 2 -13.495 15.354 11.951 1.00 0.96
+ATOM 31 N PRO A 3 -10.783 12.424 10.716 1.00 0.97
+ATOM 32 CA PRO A 3 -10.177 11.477 9.788 1.00 0.97
+ATOM 33 C PRO A 3 -10.872 11.512 8.433 1.00 0.97
+ATOM 34 O PRO A 3 -10.224 11.433 7.390 1.00 0.97
+ATOM 35 CB PRO A 3 -10.356 10.117 10.469 1.00 0.97
+ATOM 36 CG PRO A 3 -10.316 10.398 11.932 1.00 0.97
+ATOM 37 CD PRO A 3 -10.986 11.738 12.075 1.00 0.97
+ATOM 38 HA PRO A 3 -9.107 11.710 9.680 1.00 0.97
+ATOM 39 1HB PRO A 3 -11.308 9.670 10.146 1.00 0.97
+ATOM 40 2HB PRO A 3 -9.555 9.437 10.143 1.00 0.97
+ATOM 41 1HG PRO A 3 -10.838 9.603 12.486 1.00 0.97
+ATOM 42 2HG PRO A 3 -9.276 10.404 12.290 1.00 0.97
+ATOM 43 1HD PRO A 3 -12.046 11.666 11.788 1.00 0.97
+ATOM 44 2HD PRO A 3 -10.963 12.065 13.125 1.00 0.97
+ATOM 45 N GLU A 4 -12.266 11.589 8.409 1.00 0.98
+ATOM 46 CA GLU A 4 -13.019 11.626 7.161 1.00 0.98
+ATOM 47 C GLU A 4 -12.613 12.821 6.307 1.00 0.98
+ATOM 48 O GLU A 4 -12.431 12.697 5.096 1.00 0.98
+ATOM 49 CB GLU A 4 -14.523 11.658 7.441 1.00 0.98
+ATOM 50 CG GLU A 4 -15.389 11.510 6.198 1.00 0.98
+ATOM 51 CD GLU A 4 -16.859 11.473 6.511 1.00 0.98
+ATOM 52 OE1 GLU A 4 -17.640 11.380 5.594 1.00 0.98
+ATOM 53 OE2 GLU A 4 -17.202 11.537 7.668 1.00 0.98
+ATOM 54 H GLU A 4 -12.761 11.683 9.284 1.00 0.98
+ATOM 55 HA GLU A 4 -12.797 10.719 6.597 1.00 0.98
+ATOM 56 1HB GLU A 4 -14.785 10.854 8.128 1.00 0.98
+ATOM 57 2HB GLU A 4 -14.786 12.599 7.922 1.00 0.98
+ATOM 58 1HG GLU A 4 -15.206 12.358 5.537 1.00 0.98
+ATOM 59 2HG GLU A 4 -15.128 10.580 5.693 1.00 0.98
+ATOM 60 N GLU A 5 -12.526 14.047 6.921 1.00 0.98
+ATOM 61 CA GLU A 5 -12.113 15.252 6.211 1.00 0.98
+ATOM 62 C GLU A 5 -10.711 15.100 5.636 1.00 0.98
+ATOM 63 O GLU A 5 -10.438 15.532 4.517 1.00 0.98
+ATOM 64 CB GLU A 5 -12.187 16.470 7.134 1.00 0.98
+ATOM 65 CG GLU A 5 -13.602 16.891 7.504 1.00 0.98
+ATOM 66 CD GLU A 5 -13.637 17.993 8.526 1.00 0.98
+ATOM 67 OE1 GLU A 5 -14.703 18.489 8.801 1.00 0.98
+ATOM 68 OE2 GLU A 5 -12.595 18.341 9.031 1.00 0.98
+ATOM 69 H GLU A 5 -12.708 14.099 7.913 1.00 0.98
+ATOM 70 HA GLU A 5 -12.800 15.417 5.382 1.00 0.98
+ATOM 71 1HB GLU A 5 -11.647 16.261 8.058 1.00 0.98
+ATOM 72 2HB GLU A 5 -11.700 17.320 6.656 1.00 0.98
+ATOM 73 1HG GLU A 5 -14.111 17.244 6.608 1.00 0.98
+ATOM 74 2HG GLU A 5 -14.129 16.031 7.918 1.00 0.98
+ATOM 75 N LEU A 6 -9.745 14.522 6.455 1.00 0.98
+ATOM 76 CA LEU A 6 -8.391 14.258 5.982 1.00 0.98
+ATOM 77 C LEU A 6 -8.401 13.332 4.771 1.00 0.98
+ATOM 78 O LEU A 6 -7.719 13.585 3.778 1.00 0.98
+ATOM 79 CB LEU A 6 -7.543 13.661 7.111 1.00 0.98
+ATOM 80 CG LEU A 6 -6.052 13.463 6.811 1.00 0.98
+ATOM 81 CD1 LEU A 6 -5.343 12.963 8.062 1.00 0.98
+ATOM 82 CD2 LEU A 6 -5.891 12.480 5.661 1.00 0.98
+ATOM 83 H LEU A 6 -10.007 14.229 7.386 1.00 0.98
+ATOM 84 HA LEU A 6 -7.938 15.204 5.688 1.00 0.98
+ATOM 85 1HB LEU A 6 -7.622 14.313 7.980 1.00 0.98
+ATOM 86 2HB LEU A 6 -7.960 12.688 7.374 1.00 0.98
+ATOM 87 HG LEU A 6 -5.606 14.420 6.535 1.00 0.98
+ATOM 88 1HD1 LEU A 6 -4.283 12.823 7.849 1.00 0.98
+ATOM 89 2HD1 LEU A 6 -5.457 13.695 8.862 1.00 0.98
+ATOM 90 3HD1 LEU A 6 -5.779 12.014 8.372 1.00 0.98
+ATOM 91 1HD2 LEU A 6 -4.831 12.340 5.448 1.00 0.98
+ATOM 92 2HD2 LEU A 6 -6.335 11.523 5.937 1.00 0.98
+ATOM 93 3HD2 LEU A 6 -6.392 12.870 4.776 1.00 0.98
+ATOM 94 N ILE A 7 -9.224 12.198 4.863 1.00 0.99
+ATOM 95 CA ILE A 7 -9.323 11.260 3.752 1.00 0.99
+ATOM 96 C ILE A 7 -9.830 11.950 2.492 1.00 0.99
+ATOM 97 O ILE A 7 -9.276 11.769 1.408 1.00 0.99
+ATOM 98 CB ILE A 7 -10.232 10.073 4.120 1.00 0.99
+ATOM 99 CG1 ILE A 7 -9.568 9.202 5.189 1.00 0.99
+ATOM 100 CG2 ILE A 7 -10.558 9.249 2.884 1.00 0.99
+ATOM 101 CD1 ILE A 7 -10.487 8.161 5.787 1.00 0.99
+ATOM 102 H ILE A 7 -9.745 12.015 5.709 1.00 0.99
+ATOM 103 HA ILE A 7 -8.330 10.862 3.546 1.00 0.99
+ATOM 104 HB ILE A 7 -11.161 10.447 4.551 1.00 0.99
+ATOM 105 1HG2 ILE A 7 -11.201 8.415 3.162 1.00 0.99
+ATOM 106 2HG2 ILE A 7 -11.070 9.875 2.154 1.00 0.99
+ATOM 107 3HG2 ILE A 7 -9.634 8.866 2.448 1.00 0.99
+ATOM 108 1HG1 ILE A 7 -8.707 8.691 4.758 1.00 0.99
+ATOM 109 2HG1 ILE A 7 -9.200 9.835 5.997 1.00 0.99
+ATOM 110 1HD1 ILE A 7 -9.944 7.584 6.536 1.00 0.99
+ATOM 111 2HD1 ILE A 7 -11.339 8.655 6.256 1.00 0.99
+ATOM 112 3HD1 ILE A 7 -10.840 7.494 5.002 1.00 0.99
+ATOM 113 N GLU A 8 -10.930 12.775 2.608 1.00 0.98
+ATOM 114 CA GLU A 8 -11.510 13.488 1.476 1.00 0.98
+ATOM 115 C GLU A 8 -10.498 14.437 0.847 1.00 0.98
+ATOM 116 O GLU A 8 -10.385 14.518 -0.376 1.00 0.98
+ATOM 117 CB GLU A 8 -12.764 14.252 1.907 1.00 0.98
+ATOM 118 CG GLU A 8 -13.974 13.369 2.175 1.00 0.98
+ATOM 119 CD GLU A 8 -15.164 14.142 2.672 1.00 0.98
+ATOM 120 OE1 GLU A 8 -16.179 13.538 2.923 1.00 0.98
+ATOM 121 OE2 GLU A 8 -15.058 15.339 2.800 1.00 0.98
+ATOM 122 H GLU A 8 -11.321 12.912 3.529 1.00 0.98
+ATOM 123 HA GLU A 8 -11.802 12.759 0.721 1.00 0.98
+ATOM 124 1HB GLU A 8 -12.554 14.816 2.815 1.00 0.98
+ATOM 125 2HB GLU A 8 -13.038 14.968 1.132 1.00 0.98
+ATOM 126 1HG GLU A 8 -14.259 12.869 1.249 1.00 0.98
+ATOM 127 2HG GLU A 8 -13.712 12.630 2.933 1.00 0.98
+ATOM 128 N ARG A 9 -9.729 15.202 1.689 1.00 0.98
+ATOM 129 CA ARG A 9 -8.702 16.114 1.200 1.00 0.98
+ATOM 130 C ARG A 9 -7.632 15.369 0.412 1.00 0.98
+ATOM 131 O ARG A 9 -7.235 15.796 -0.671 1.00 0.98
+ATOM 132 CB ARG A 9 -8.058 16.869 2.353 1.00 0.98
+ATOM 133 CG ARG A 9 -8.925 17.959 2.963 1.00 0.98
+ATOM 134 CD ARG A 9 -8.222 18.670 4.061 1.00 0.98
+ATOM 135 NE ARG A 9 -9.075 19.661 4.698 1.00 0.98
+ATOM 136 CZ ARG A 9 -8.706 20.431 5.740 1.00 0.98
+ATOM 137 NH1 ARG A 9 -7.500 20.314 6.249 1.00 0.98
+ATOM 138 NH2 ARG A 9 -9.557 21.305 6.250 1.00 0.98
+ATOM 139 H ARG A 9 -9.862 15.099 2.685 1.00 0.98
+ATOM 140 HA ARG A 9 -9.173 16.843 0.540 1.00 0.98
+ATOM 141 1HB ARG A 9 -7.799 16.170 3.148 1.00 0.98
+ATOM 142 2HB ARG A 9 -7.133 17.335 2.014 1.00 0.98
+ATOM 143 1HG ARG A 9 -9.188 18.687 2.195 1.00 0.98
+ATOM 144 2HG ARG A 9 -9.835 17.515 3.369 1.00 0.98
+ATOM 145 1HD ARG A 9 -7.910 17.952 4.818 1.00 0.98
+ATOM 146 2HD ARG A 9 -7.347 19.181 3.662 1.00 0.98
+ATOM 147 HE ARG A 9 -10.011 19.780 4.334 1.00 0.98
+ATOM 148 1HH1 ARG A 9 -6.849 19.646 5.860 1.00 0.98
+ATOM 149 2HH1 ARG A 9 -7.223 20.892 7.030 1.00 0.98
+ATOM 150 1HH2 ARG A 9 -10.484 21.395 5.858 1.00 0.98
+ATOM 151 2HH2 ARG A 9 -9.280 21.882 7.030 1.00 0.98
+ATOM 152 N ILE A 10 -7.122 14.206 0.993 1.00 0.99
+ATOM 153 CA ILE A 10 -6.104 13.420 0.307 1.00 0.99
+ATOM 154 C ILE A 10 -6.623 12.885 -1.021 1.00 0.99
+ATOM 155 O ILE A 10 -5.934 12.949 -2.039 1.00 0.99
+ATOM 156 CB ILE A 10 -5.601 12.271 1.199 1.00 0.99
+ATOM 157 CG1 ILE A 10 -4.820 12.822 2.395 1.00 0.99
+ATOM 158 CG2 ILE A 10 -4.740 11.311 0.393 1.00 0.99
+ATOM 159 CD1 ILE A 10 -4.511 11.787 3.452 1.00 0.99
+ATOM 160 H ILE A 10 -7.474 13.886 1.884 1.00 0.99
+ATOM 161 HA ILE A 10 -5.249 14.064 0.104 1.00 0.99
+ATOM 162 HB ILE A 10 -6.454 11.726 1.604 1.00 0.99
+ATOM 163 1HG2 ILE A 10 -4.393 10.504 1.038 1.00 0.99
+ATOM 164 2HG2 ILE A 10 -5.327 10.895 -0.424 1.00 0.99
+ATOM 165 3HG2 ILE A 10 -3.881 11.846 -0.012 1.00 0.99
+ATOM 166 1HG1 ILE A 10 -3.879 13.248 2.049 1.00 0.99
+ATOM 167 2HG1 ILE A 10 -5.389 13.626 2.862 1.00 0.99
+ATOM 168 1HD1 ILE A 10 -3.956 12.253 4.267 1.00 0.99
+ATOM 169 2HD1 ILE A 10 -5.442 11.372 3.839 1.00 0.99
+ATOM 170 3HD1 ILE A 10 -3.911 10.990 3.016 1.00 0.99
+ATOM 171 N ALA A 11 -7.864 12.279 -1.027 1.00 0.99
+ATOM 172 CA ALA A 11 -8.468 11.738 -2.238 1.00 0.99
+ATOM 173 C ALA A 11 -8.565 12.799 -3.327 1.00 0.99
+ATOM 174 O ALA A 11 -8.255 12.539 -4.490 1.00 0.99
+ATOM 175 CB ALA A 11 -9.846 11.166 -1.935 1.00 0.99
+ATOM 176 H ALA A 11 -8.384 12.247 -0.161 1.00 0.99
+ATOM 177 HA ALA A 11 -7.827 10.937 -2.608 1.00 0.99
+ATOM 178 1HB ALA A 11 -10.283 10.766 -2.850 1.00 0.99
+ATOM 179 2HB ALA A 11 -9.754 10.368 -1.198 1.00 0.99
+ATOM 180 3HB ALA A 11 -10.487 11.952 -1.540 1.00 0.99
+ATOM 181 N GLU A 12 -9.005 14.040 -2.987 1.00 0.98
+ATOM 182 CA GLU A 12 -9.129 15.146 -3.929 1.00 0.98
+ATOM 183 C GLU A 12 -7.772 15.543 -4.495 1.00 0.98
+ATOM 184 O GLU A 12 -7.643 15.823 -5.686 1.00 0.98
+ATOM 185 CB GLU A 12 -9.802 16.349 -3.264 1.00 0.98
+ATOM 186 CG GLU A 12 -11.296 16.180 -3.029 1.00 0.98
+ATOM 187 CD GLU A 12 -11.910 17.346 -2.305 1.00 0.98
+ATOM 188 OE1 GLU A 12 -13.089 17.307 -2.048 1.00 0.98
+ATOM 189 OE2 GLU A 12 -11.199 18.277 -2.010 1.00 0.98
+ATOM 190 H GLU A 12 -9.231 14.195 -2.015 1.00 0.98
+ATOM 191 HA GLU A 12 -9.758 14.824 -4.759 1.00 0.98
+ATOM 192 1HB GLU A 12 -9.332 16.543 -2.300 1.00 0.98
+ATOM 193 2HB GLU A 12 -9.659 17.235 -3.883 1.00 0.98
+ATOM 194 1HG GLU A 12 -11.796 16.079 -3.992 1.00 0.98
+ATOM 195 2HG GLU A 12 -11.460 15.285 -2.428 1.00 0.98
+ATOM 196 N ARG A 13 -6.685 15.497 -3.645 1.00 0.98
+ATOM 197 CA ARG A 13 -5.342 15.913 -4.028 1.00 0.98
+ATOM 198 C ARG A 13 -4.676 14.873 -4.920 1.00 0.98
+ATOM 199 O ARG A 13 -3.988 15.215 -5.881 1.00 0.98
+ATOM 200 CB ARG A 13 -4.488 16.174 -2.796 1.00 0.98
+ATOM 201 CG ARG A 13 -3.128 16.793 -3.074 1.00 0.98
+ATOM 202 CD ARG A 13 -2.410 17.129 -1.818 1.00 0.98
+ATOM 203 NE ARG A 13 -1.082 17.661 -2.079 1.00 0.98
+ATOM 204 CZ ARG A 13 -0.220 18.066 -1.126 1.00 0.98
+ATOM 205 NH1 ARG A 13 -0.559 17.994 0.142 1.00 0.98
+ATOM 206 NH2 ARG A 13 0.968 18.537 -1.466 1.00 0.98
+ATOM 207 H ARG A 13 -6.843 15.140 -2.714 1.00 0.98
+ATOM 208 HA ARG A 13 -5.414 16.850 -4.583 1.00 0.98
+ATOM 209 1HB ARG A 13 -5.022 16.841 -2.120 1.00 0.98
+ATOM 210 2HB ARG A 13 -4.322 15.236 -2.265 1.00 0.98
+ATOM 211 1HG ARG A 13 -2.517 16.089 -3.641 1.00 0.98
+ATOM 212 2HG ARG A 13 -3.256 17.709 -3.652 1.00 0.98
+ATOM 213 1HD ARG A 13 -2.975 17.879 -1.266 1.00 0.98
+ATOM 214 2HD ARG A 13 -2.304 16.233 -1.209 1.00 0.98
+ATOM 215 HE ARG A 13 -0.785 17.733 -3.043 1.00 0.98
+ATOM 216 1HH1 ARG A 13 -1.467 17.634 0.402 1.00 0.98
+ATOM 217 2HH1 ARG A 13 0.087 18.298 0.856 1.00 0.98
+ATOM 218 1HH2 ARG A 13 1.229 18.593 -2.441 1.00 0.98
+ATOM 219 2HH2 ARG A 13 1.613 18.840 -0.753 1.00 0.98
+ATOM 220 N TRP A 14 -4.866 13.519 -4.675 1.00 0.98
+ATOM 221 CA TRP A 14 -4.091 12.494 -5.364 1.00 0.98
+ATOM 222 C TRP A 14 -4.943 11.752 -6.386 1.00 0.98
+ATOM 223 O TRP A 14 -4.481 10.808 -7.026 1.00 0.98
+ATOM 224 CB TRP A 14 -3.479 11.514 -4.361 1.00 0.98
+ATOM 225 CG TRP A 14 -2.418 12.133 -3.503 1.00 0.98
+ATOM 226 CD1 TRP A 14 -2.563 12.583 -2.225 1.00 0.98
+ATOM 227 CD2 TRP A 14 -1.035 12.375 -3.860 1.00 0.98
+ATOM 228 NE1 TRP A 14 -1.372 13.086 -1.763 1.00 0.98
+ATOM 229 CE2 TRP A 14 -0.426 12.967 -2.750 1.00 0.98
+ATOM 230 CE3 TRP A 14 -0.278 12.141 -5.014 1.00 0.98
+ATOM 231 CZ2 TRP A 14 0.911 13.332 -2.755 1.00 0.98
+ATOM 232 CZ3 TRP A 14 1.063 12.507 -5.019 1.00 0.98
+ATOM 233 CH2 TRP A 14 1.641 13.087 -3.918 1.00 0.98
+ATOM 234 H TRP A 14 -5.587 13.237 -4.026 1.00 0.98
+ATOM 235 HA TRP A 14 -3.268 12.978 -5.890 1.00 0.98
+ATOM 236 1HB TRP A 14 -4.260 11.120 -3.710 1.00 0.98
+ATOM 237 2HB TRP A 14 -3.041 10.670 -4.894 1.00 0.98
+ATOM 238 HD1 TRP A 14 -3.488 12.548 -1.654 1.00 0.98
+ATOM 239 HE1 TRP A 14 -1.216 13.480 -0.846 1.00 0.98
+ATOM 240 HZ2 TRP A 14 1.389 13.793 -1.890 1.00 0.98
+ATOM 241 HH2 TRP A 14 2.696 13.361 -3.955 1.00 0.98
+ATOM 242 HZ3 TRP A 14 1.646 12.321 -5.921 1.00 0.98
+ATOM 243 HE3 TRP A 14 -0.732 11.681 -5.891 1.00 0.98
+ATOM 244 N GLY A 15 -6.218 11.964 -6.622 1.00 0.98
+ATOM 245 CA GLY A 15 -7.065 11.225 -7.551 1.00 0.98
+ATOM 246 C GLY A 15 -7.274 9.790 -7.082 1.00 0.98
+ATOM 247 O GLY A 15 -7.266 8.857 -7.886 1.00 0.98
+ATOM 248 H GLY A 15 -6.618 12.723 -6.090 1.00 0.98
+ATOM 249 1HA GLY A 15 -8.028 11.726 -7.643 1.00 0.98
+ATOM 250 2HA GLY A 15 -6.607 11.225 -8.539 1.00 0.98
+ATOM 251 N ILE A 16 -7.248 9.530 -5.774 1.00 0.99
+ATOM 252 CA ILE A 16 -7.457 8.250 -5.108 1.00 0.99
+ATOM 253 C ILE A 16 -8.935 8.007 -4.832 1.00 0.99
+ATOM 254 O ILE A 16 -9.692 8.945 -4.583 1.00 0.99
+ATOM 255 CB ILE A 16 -6.662 8.177 -3.791 1.00 0.99
+ATOM 256 CG1 ILE A 16 -5.154 8.253 -4.045 1.00 0.99
+ATOM 257 CG2 ILE A 16 -7.015 6.902 -3.042 1.00 0.99
+ATOM 258 CD1 ILE A 16 -4.328 8.373 -2.785 1.00 0.99
+ATOM 259 H ILE A 16 -7.082 10.354 -5.215 1.00 0.99
+ATOM 260 HA ILE A 16 -7.098 7.457 -5.762 1.00 0.99
+ATOM 261 HB ILE A 16 -6.920 9.035 -3.172 1.00 0.99
+ATOM 262 1HG2 ILE A 16 -6.460 6.865 -2.105 1.00 0.99
+ATOM 263 2HG2 ILE A 16 -8.084 6.889 -2.830 1.00 0.99
+ATOM 264 3HG2 ILE A 16 -6.755 6.037 -3.652 1.00 0.99
+ATOM 265 1HG1 ILE A 16 -4.831 7.361 -4.582 1.00 0.99
+ATOM 266 2HG1 ILE A 16 -4.934 9.112 -4.679 1.00 0.99
+ATOM 267 1HD1 ILE A 16 -3.270 8.421 -3.047 1.00 0.99
+ATOM 268 2HD1 ILE A 16 -4.611 9.279 -2.249 1.00 0.99
+ATOM 269 3HD1 ILE A 16 -4.504 7.506 -2.151 1.00 0.99
+ATOM 270 N THR A 17 -9.382 6.729 -4.692 1.00 0.99
+ATOM 271 CA THR A 17 -10.735 6.466 -4.214 1.00 0.99
+ATOM 272 C THR A 17 -10.812 6.568 -2.697 1.00 0.99
+ATOM 273 O THR A 17 -9.886 6.169 -1.990 1.00 0.99
+ATOM 274 CB THR A 17 -11.235 5.082 -4.668 1.00 0.99
+ATOM 275 OG1 THR A 17 -10.533 4.050 -3.965 1.00 0.99
+ATOM 276 CG2 THR A 17 -11.019 4.909 -6.164 1.00 0.99
+ATOM 277 H THR A 17 -8.799 5.952 -4.969 1.00 0.99
+ATOM 278 HG1 THR A 17 -9.920 4.448 -3.341 1.00 0.99
+ATOM 279 HA THR A 17 -11.402 7.218 -4.637 1.00 0.99
+ATOM 280 HB THR A 17 -12.299 4.995 -4.448 1.00 0.99
+ATOM 281 1HG2 THR A 17 -11.385 3.930 -6.473 1.00 0.99
+ATOM 282 2HG2 THR A 17 -11.562 5.686 -6.702 1.00 0.99
+ATOM 283 3HG2 THR A 17 -9.957 4.986 -6.389 1.00 0.99
+ATOM 284 N PRO A 18 -11.807 7.278 -2.112 1.00 0.99
+ATOM 285 CA PRO A 18 -11.955 7.462 -0.673 1.00 0.99
+ATOM 286 C PRO A 18 -11.897 6.129 0.062 1.00 0.99
+ATOM 287 O PRO A 18 -11.383 6.045 1.177 1.00 0.99
+ATOM 288 CB PRO A 18 -13.328 8.122 -0.519 1.00 0.99
+ATOM 289 CG PRO A 18 -13.513 8.939 -1.753 1.00 0.99
+ATOM 290 CD PRO A 18 -12.864 8.126 -2.841 1.00 0.99
+ATOM 291 HA PRO A 18 -11.165 8.137 -0.311 1.00 0.99
+ATOM 292 1HB PRO A 18 -14.097 7.344 -0.404 1.00 0.99
+ATOM 293 2HB PRO A 18 -13.343 8.727 0.399 1.00 0.99
+ATOM 294 1HG PRO A 18 -14.583 9.114 -1.936 1.00 0.99
+ATOM 295 2HG PRO A 18 -13.046 9.927 -1.627 1.00 0.99
+ATOM 296 1HD PRO A 18 -13.413 7.185 -2.990 1.00 0.99
+ATOM 297 2HD PRO A 18 -12.901 8.670 -3.795 1.00 0.99
+ATOM 298 N GLU A 19 -12.427 5.032 -0.511 1.00 0.99
+ATOM 299 CA GLU A 19 -12.398 3.698 0.076 1.00 0.99
+ATOM 300 C GLU A 19 -10.967 3.221 0.289 1.00 0.99
+ATOM 301 O GLU A 19 -10.647 2.626 1.318 1.00 0.99
+ATOM 302 CB GLU A 19 -13.159 2.705 -0.805 1.00 0.99
+ATOM 303 CG GLU A 19 -14.667 2.911 -0.820 1.00 0.99
+ATOM 304 CD GLU A 19 -15.384 1.918 -1.692 1.00 0.99
+ATOM 305 OE1 GLU A 19 -16.584 2.009 -1.800 1.00 0.99
+ATOM 306 OE2 GLU A 19 -14.732 1.068 -2.250 1.00 0.99
+ATOM 307 H GLU A 19 -12.842 5.158 -1.423 1.00 0.99
+ATOM 308 HA GLU A 19 -12.887 3.737 1.050 1.00 0.99
+ATOM 309 1HB GLU A 19 -12.801 2.780 -1.832 1.00 0.99
+ATOM 310 2HB GLU A 19 -12.963 1.689 -0.464 1.00 0.99
+ATOM 311 1HG GLU A 19 -15.048 2.805 0.196 1.00 0.99
+ATOM 312 2HG GLU A 19 -14.884 3.910 -1.198 1.00 0.99
+ATOM 313 N GLU A 20 -10.051 3.462 -0.749 1.00 0.99
+ATOM 314 CA GLU A 20 -8.648 3.090 -0.611 1.00 0.99
+ATOM 315 C GLU A 20 -8.000 3.812 0.564 1.00 0.99
+ATOM 316 O GLU A 20 -7.266 3.210 1.347 1.00 0.99
+ATOM 317 CB GLU A 20 -7.886 3.382 -1.905 1.00 0.99
+ATOM 318 CG GLU A 20 -8.295 2.514 -3.087 1.00 0.99
+ATOM 319 CD GLU A 20 -7.703 2.981 -4.388 1.00 0.99
+ATOM 320 OE1 GLU A 20 -7.531 2.170 -5.266 1.00 0.99
+ATOM 321 OE2 GLU A 20 -7.423 4.151 -4.505 1.00 0.99
+ATOM 322 H GLU A 20 -10.357 3.921 -1.596 1.00 0.99
+ATOM 323 HA GLU A 20 -8.591 2.019 -0.420 1.00 0.99
+ATOM 324 1HB GLU A 20 -8.035 4.424 -2.188 1.00 0.99
+ATOM 325 2HB GLU A 20 -6.818 3.239 -1.739 1.00 0.99
+ATOM 326 1HG GLU A 20 -7.956 1.494 -2.909 1.00 0.99
+ATOM 327 2HG GLU A 20 -9.380 2.537 -3.183 1.00 0.99
+ATOM 328 N LEU A 21 -8.202 5.171 0.663 1.00 0.99
+ATOM 329 CA LEU A 21 -7.647 5.967 1.751 1.00 0.99
+ATOM 330 C LEU A 21 -8.156 5.482 3.104 1.00 0.99
+ATOM 331 O LEU A 21 -7.395 5.388 4.067 1.00 0.99
+ATOM 332 CB LEU A 21 -7.977 7.452 1.551 1.00 0.99
+ATOM 333 CG LEU A 21 -7.270 8.160 0.389 1.00 0.99
+ATOM 334 CD1 LEU A 21 -7.878 9.542 0.190 1.00 0.99
+ATOM 335 CD2 LEU A 21 -5.780 8.256 0.683 1.00 0.99
+ATOM 336 H LEU A 21 -8.814 5.613 -0.008 1.00 0.99
+ATOM 337 HA LEU A 21 -6.562 5.865 1.735 1.00 0.99
+ATOM 338 1HB LEU A 21 -9.050 7.542 1.390 1.00 0.99
+ATOM 339 2HB LEU A 21 -7.722 7.983 2.467 1.00 0.99
+ATOM 340 HG LEU A 21 -7.422 7.594 -0.530 1.00 0.99
+ATOM 341 1HD1 LEU A 21 -7.375 10.045 -0.636 1.00 0.99
+ATOM 342 2HD1 LEU A 21 -8.939 9.443 -0.039 1.00 0.99
+ATOM 343 3HD1 LEU A 21 -7.755 10.128 1.100 1.00 0.99
+ATOM 344 1HD2 LEU A 21 -5.277 8.759 -0.143 1.00 0.99
+ATOM 345 2HD2 LEU A 21 -5.627 8.824 1.601 1.00 0.99
+ATOM 346 3HD2 LEU A 21 -5.368 7.254 0.803 1.00 0.99
+ATOM 347 N ARG A 22 -9.489 5.226 3.255 1.00 0.99
+ATOM 348 CA ARG A 22 -10.067 4.718 4.493 1.00 0.99
+ATOM 349 C ARG A 22 -9.385 3.429 4.931 1.00 0.99
+ATOM 350 O ARG A 22 -9.060 3.255 6.106 1.00 0.99
+ATOM 351 CB ARG A 22 -11.561 4.480 4.328 1.00 0.99
+ATOM 352 CG ARG A 22 -12.290 4.076 5.600 1.00 0.99
+ATOM 353 CD ARG A 22 -13.745 3.897 5.365 1.00 0.99
+ATOM 354 NE ARG A 22 -14.430 3.406 6.550 1.00 0.99
+ATOM 355 CZ ARG A 22 -15.753 3.160 6.623 1.00 0.99
+ATOM 356 NH1 ARG A 22 -16.518 3.363 5.573 1.00 0.99
+ATOM 357 NH2 ARG A 22 -16.281 2.714 7.749 1.00 0.99
+ATOM 358 H ARG A 22 -10.088 5.372 2.455 1.00 0.99
+ATOM 359 HA ARG A 22 -9.926 5.467 5.273 1.00 0.99
+ATOM 360 1HB ARG A 22 -12.033 5.387 3.952 1.00 0.99
+ATOM 361 2HB ARG A 22 -11.724 3.695 3.590 1.00 0.99
+ATOM 362 1HG ARG A 22 -11.883 3.134 5.968 1.00 0.99
+ATOM 363 2HG ARG A 22 -12.157 4.849 6.357 1.00 0.99
+ATOM 364 1HD ARG A 22 -14.189 4.852 5.086 1.00 0.99
+ATOM 365 2HD ARG A 22 -13.898 3.177 4.561 1.00 0.99
+ATOM 366 HE ARG A 22 -13.874 3.238 7.378 1.00 0.99
+ATOM 367 1HH1 ARG A 22 -16.114 3.704 4.712 1.00 0.99
+ATOM 368 2HH1 ARG A 22 -17.509 3.179 5.628 1.00 0.99
+ATOM 369 1HH2 ARG A 22 -15.692 2.558 8.556 1.00 0.99
+ATOM 370 2HH2 ARG A 22 -17.271 2.530 7.804 1.00 0.99
+ATOM 371 N GLU A 23 -9.145 2.487 3.957 1.00 0.98
+ATOM 372 CA GLU A 23 -8.451 1.240 4.254 1.00 0.98
+ATOM 373 C GLU A 23 -7.054 1.504 4.800 1.00 0.98
+ATOM 374 O GLU A 23 -6.627 0.876 5.769 1.00 0.98
+ATOM 375 CB GLU A 23 -8.380 0.356 3.008 1.00 0.98
+ATOM 376 CG GLU A 23 -9.719 -0.226 2.577 1.00 0.98
+ATOM 377 CD GLU A 23 -9.629 -1.020 1.303 1.00 0.98
+ATOM 378 OE1 GLU A 23 -10.639 -1.510 0.859 1.00 0.98
+ATOM 379 OE2 GLU A 23 -8.549 -1.136 0.774 1.00 0.98
+ATOM 380 H GLU A 23 -9.443 2.670 3.009 1.00 0.98
+ATOM 381 HA GLU A 23 -9.015 0.705 5.018 1.00 0.98
+ATOM 382 1HB GLU A 23 -7.982 0.933 2.173 1.00 0.98
+ATOM 383 2HB GLU A 23 -7.697 -0.474 3.187 1.00 0.98
+ATOM 384 1HG GLU A 23 -10.085 -0.888 3.362 1.00 0.98
+ATOM 385 2HG GLU A 23 -10.423 0.590 2.417 1.00 0.98
+ATOM 386 N ILE A 24 -6.302 2.500 4.160 1.00 0.99
+ATOM 387 CA ILE A 24 -4.959 2.841 4.614 1.00 0.99
+ATOM 388 C ILE A 24 -4.987 3.434 6.018 1.00 0.99
+ATOM 389 O ILE A 24 -4.236 3.009 6.896 1.00 0.99
+ATOM 390 CB ILE A 24 -4.273 3.815 3.639 1.00 0.99
+ATOM 391 CG1 ILE A 24 -3.913 3.109 2.329 1.00 0.99
+ATOM 392 CG2 ILE A 24 -3.033 4.417 4.282 1.00 0.99
+ATOM 393 CD1 ILE A 24 -3.443 4.046 1.240 1.00 0.99
+ATOM 394 H ILE A 24 -6.688 2.982 3.360 1.00 0.99
+ATOM 395 HA ILE A 24 -4.362 1.930 4.641 1.00 0.99
+ATOM 396 HB ILE A 24 -4.965 4.617 3.385 1.00 0.99
+ATOM 397 1HG2 ILE A 24 -2.560 5.107 3.583 1.00 0.99
+ATOM 398 2HG2 ILE A 24 -3.316 4.953 5.186 1.00 0.99
+ATOM 399 3HG2 ILE A 24 -2.332 3.621 4.535 1.00 0.99
+ATOM 400 1HG1 ILE A 24 -3.126 2.378 2.514 1.00 0.99
+ATOM 401 2HG1 ILE A 24 -4.782 2.564 1.958 1.00 0.99
+ATOM 402 1HD1 ILE A 24 -3.207 3.472 0.343 1.00 0.99
+ATOM 403 2HD1 ILE A 24 -4.232 4.765 1.013 1.00 0.99
+ATOM 404 3HD1 ILE A 24 -2.553 4.576 1.575 1.00 0.99
+ATOM 405 N TRP A 25 -5.851 4.474 6.270 1.00 0.99
+ATOM 406 CA TRP A 25 -5.936 5.168 7.550 1.00 0.99
+ATOM 407 C TRP A 25 -6.279 4.203 8.678 1.00 0.99
+ATOM 408 O TRP A 25 -5.843 4.382 9.815 1.00 0.99
+ATOM 409 CB TRP A 25 -6.970 6.293 7.486 1.00 0.99
+ATOM 410 CG TRP A 25 -6.522 7.505 6.727 1.00 0.99
+ATOM 411 CD1 TRP A 25 -7.318 8.486 6.214 1.00 0.99
+ATOM 412 CD2 TRP A 25 -5.163 7.874 6.389 1.00 0.99
+ATOM 413 NE1 TRP A 25 -6.551 9.435 5.583 1.00 0.99
+ATOM 414 CE2 TRP A 25 -5.231 9.076 5.680 1.00 0.99
+ATOM 415 CE3 TRP A 25 -3.914 7.287 6.627 1.00 0.99
+ATOM 416 CZ2 TRP A 25 -4.095 9.711 5.203 1.00 0.99
+ATOM 417 CZ3 TRP A 25 -2.775 7.925 6.149 1.00 0.99
+ATOM 418 CH2 TRP A 25 -2.864 9.105 5.455 1.00 0.99
+ATOM 419 H TRP A 25 -6.493 4.723 5.532 1.00 0.99
+ATOM 420 HA TRP A 25 -4.967 5.617 7.765 1.00 0.99
+ATOM 421 1HB TRP A 25 -7.880 5.914 7.020 1.00 0.99
+ATOM 422 2HB TRP A 25 -7.225 6.599 8.501 1.00 0.99
+ATOM 423 HD1 TRP A 25 -8.403 8.513 6.294 1.00 0.99
+ATOM 424 HE1 TRP A 25 -6.902 10.263 5.123 1.00 0.99
+ATOM 425 HZ2 TRP A 25 -4.145 10.650 4.651 1.00 0.99
+ATOM 426 HH2 TRP A 25 -1.950 9.578 5.094 1.00 0.99
+ATOM 427 HZ3 TRP A 25 -1.806 7.462 6.338 1.00 0.99
+ATOM 428 HE3 TRP A 25 -3.837 6.350 7.177 1.00 0.99
+ATOM 429 N LYS A 26 -7.066 3.121 8.429 1.00 0.98
+ATOM 430 CA LYS A 26 -7.406 2.108 9.421 1.00 0.98
+ATOM 431 C LYS A 26 -6.164 1.365 9.897 1.00 0.98
+ATOM 432 O LYS A 26 -6.146 0.806 10.994 1.00 0.98
+ATOM 433 CB LYS A 26 -8.429 1.121 8.856 1.00 0.98
+ATOM 434 CG LYS A 26 -9.840 1.680 8.733 1.00 0.98
+ATOM 435 CD LYS A 26 -10.795 0.648 8.153 1.00 0.98
+ATOM 436 CE LYS A 26 -12.198 1.215 7.999 1.00 0.98
+ATOM 437 NZ LYS A 26 -13.133 0.231 7.389 1.00 0.98
+ATOM 438 H LYS A 26 -7.422 3.028 7.489 1.00 0.98
+ATOM 439 HA LYS A 26 -7.849 2.604 10.284 1.00 0.98
+ATOM 440 1HB LYS A 26 -8.111 0.794 7.866 1.00 0.98
+ATOM 441 2HB LYS A 26 -8.473 0.238 9.494 1.00 0.98
+ATOM 442 1HG LYS A 26 -10.199 1.982 9.718 1.00 0.98
+ATOM 443 2HG LYS A 26 -9.829 2.556 8.086 1.00 0.98
+ATOM 444 1HD LYS A 26 -10.433 0.326 7.175 1.00 0.98
+ATOM 445 2HD LYS A 26 -10.834 -0.221 8.810 1.00 0.98
+ATOM 446 1HE LYS A 26 -12.669 1.450 9.006 1.00 0.98
+ATOM 447 2HE LYS A 26 -12.251 2.049 7.399 1.00 0.98
+ATOM 448 1HZ LYS A 26 -14.137 0.590 7.334 1.00 0.98
+ATOM 449 2HZ LYS A 26 -12.882 -0.085 6.503 1.00 0.98
+ATOM 450 3HZ LYS A 26 -13.274 -0.645 8.004 1.00 0.98
+ATOM 451 N TRP A 27 -5.037 1.388 9.106 1.00 0.98
+ATOM 452 CA TRP A 27 -3.835 0.612 9.388 1.00 0.98
+ATOM 453 C TRP A 27 -2.745 1.485 9.998 1.00 0.98
+ATOM 454 O TRP A 27 -1.697 0.988 10.409 1.00 0.98
+ATOM 455 CB TRP A 27 -3.313 -0.057 8.116 1.00 0.98
+ATOM 456 CG TRP A 27 -4.233 -1.102 7.559 1.00 0.98
+ATOM 457 CD1 TRP A 27 -5.075 -0.964 6.497 1.00 0.98
+ATOM 458 CD2 TRP A 27 -4.408 -2.458 8.037 1.00 0.98
+ATOM 459 NE1 TRP A 27 -5.760 -2.134 6.281 1.00 0.98
+ATOM 460 CE2 TRP A 27 -5.364 -3.059 7.214 1.00 0.98
+ATOM 461 CE3 TRP A 27 -3.839 -3.197 9.081 1.00 0.98
+ATOM 462 CZ2 TRP A 27 -5.770 -4.372 7.399 1.00 0.98
+ATOM 463 CZ3 TRP A 27 -4.246 -4.513 9.267 1.00 0.98
+ATOM 464 CH2 TRP A 27 -5.187 -5.085 8.447 1.00 0.98
+ATOM 465 H TRP A 27 -5.048 2.004 8.306 1.00 0.98
+ATOM 466 HA TRP A 27 -4.089 -0.170 10.104 1.00 0.98
+ATOM 467 1HB TRP A 27 -3.152 0.701 7.349 1.00 0.98
+ATOM 468 2HB TRP A 27 -2.350 -0.524 8.322 1.00 0.98
+ATOM 469 HD1 TRP A 27 -5.188 -0.057 5.906 1.00 0.98
+ATOM 470 HE1 TRP A 27 -6.443 -2.291 5.555 1.00 0.98
+ATOM 471 HZ2 TRP A 27 -6.515 -4.843 6.759 1.00 0.98
+ATOM 472 HH2 TRP A 27 -5.483 -6.119 8.621 1.00 0.98
+ATOM 473 HZ3 TRP A 27 -3.798 -5.082 10.082 1.00 0.98
+ATOM 474 HE3 TRP A 27 -3.092 -2.749 9.735 1.00 0.98
+ATOM 475 N ILE A 28 -2.837 2.915 9.946 1.00 0.98
+ATOM 476 CA ILE A 28 -1.792 3.854 10.335 1.00 0.98
+ATOM 477 C ILE A 28 -1.554 3.821 11.839 1.00 0.98
+ATOM 478 O ILE A 28 -0.991 4.756 12.409 1.00 0.98
+ATOM 479 CB ILE A 28 -2.140 5.276 9.860 1.00 0.98
+ATOM 480 CG1 ILE A 28 -2.051 5.358 8.334 1.00 0.98
+ATOM 481 CG2 ILE A 28 -1.216 6.298 10.505 1.00 0.98
+ATOM 482 CD1 ILE A 28 -2.581 6.652 7.760 1.00 0.98
+ATOM 483 H ILE A 28 -3.758 3.267 9.727 1.00 0.98
+ATOM 484 HA ILE A 28 -0.867 3.566 9.838 1.00 0.98
+ATOM 485 HB ILE A 28 -3.170 5.511 10.129 1.00 0.98
+ATOM 486 1HG2 ILE A 28 -1.475 7.296 10.153 1.00 0.98
+ATOM 487 2HG2 ILE A 28 -1.327 6.256 11.588 1.00 0.98
+ATOM 488 3HG2 ILE A 28 -0.183 6.075 10.238 1.00 0.98
+ATOM 489 1HG1 ILE A 28 -1.013 5.246 8.024 1.00 0.98
+ATOM 490 2HG1 ILE A 28 -2.613 4.534 7.892 1.00 0.98
+ATOM 491 1HD1 ILE A 28 -2.485 6.634 6.674 1.00 0.98
+ATOM 492 2HD1 ILE A 28 -3.632 6.766 8.029 1.00 0.98
+ATOM 493 3HD1 ILE A 28 -2.010 7.488 8.160 1.00 0.98
+ATOM 494 N ASP A 29 -1.560 2.721 12.520 1.00 0.96
+ATOM 495 CA ASP A 29 -1.134 2.519 13.899 1.00 0.96
+ATOM 496 C ASP A 29 0.255 1.896 13.960 1.00 0.96
+ATOM 497 O ASP A 29 0.641 1.127 13.079 1.00 0.96
+ATOM 498 CB ASP A 29 -2.158 1.623 14.600 1.00 0.96
+ATOM 499 CG ASP A 29 -1.859 1.335 16.065 1.00 0.96
+ATOM 500 OD1 ASP A 29 -2.254 0.296 16.539 1.00 0.96
+ATOM 501 OD2 ASP A 29 -1.239 2.157 16.696 1.00 0.96
+ATOM 502 H ASP A 29 -1.943 1.942 12.004 1.00 0.96
+ATOM 503 HA ASP A 29 -1.073 3.492 14.390 1.00 0.96
+ATOM 504 1HB ASP A 29 -3.137 2.098 14.530 1.00 0.96
+ATOM 505 2HB ASP A 29 -2.215 0.676 14.063 1.00 0.96
+ATOM 506 N PRO A 30 1.080 2.240 14.967 1.00 0.95
+ATOM 507 CA PRO A 30 2.437 1.713 15.045 1.00 0.95
+ATOM 508 C PRO A 30 2.437 0.190 15.071 1.00 0.95
+ATOM 509 O PRO A 30 3.363 -0.449 14.572 1.00 0.95
+ATOM 510 CB PRO A 30 2.992 2.294 16.349 1.00 0.95
+ATOM 511 CG PRO A 30 2.309 3.605 16.541 1.00 0.95
+ATOM 512 CD PRO A 30 0.898 3.364 16.072 1.00 0.95
+ATOM 513 HA PRO A 30 3.019 2.085 14.188 1.00 0.95
+ATOM 514 1HB PRO A 30 2.797 1.587 17.168 1.00 0.95
+ATOM 515 2HB PRO A 30 4.084 2.392 16.262 1.00 0.95
+ATOM 516 1HG PRO A 30 2.358 3.908 17.597 1.00 0.95
+ATOM 517 2HG PRO A 30 2.819 4.388 15.962 1.00 0.95
+ATOM 518 1HD PRO A 30 0.368 2.712 16.781 1.00 0.95
+ATOM 519 2HD PRO A 30 0.343 4.313 16.037 1.00 0.95
+ATOM 520 N ASP A 31 1.280 -0.461 15.574 1.00 0.94
+ATOM 521 CA ASP A 31 1.194 -1.912 15.686 1.00 0.94
+ATOM 522 C ASP A 31 0.820 -2.546 14.352 1.00 0.94
+ATOM 523 O ASP A 31 1.131 -3.710 14.099 1.00 0.94
+ATOM 524 CB ASP A 31 0.183 -2.314 16.763 1.00 0.94
+ATOM 525 CG ASP A 31 0.613 -1.902 18.164 1.00 0.94
+ATOM 526 OD1 ASP A 31 -0.151 -2.098 19.080 1.00 0.94
+ATOM 527 OD2 ASP A 31 1.699 -1.394 18.305 1.00 0.94
+ATOM 528 H ASP A 31 0.511 0.104 15.906 1.00 0.94
+ATOM 529 HA ASP A 31 2.177 -2.297 15.962 1.00 0.94
+ATOM 530 1HB ASP A 31 -0.782 -1.856 16.544 1.00 0.94
+ATOM 531 2HB ASP A 31 0.044 -3.395 16.744 1.00 0.94
+ATOM 532 N ASN A 32 -0.015 -1.818 13.479 1.00 0.97
+ATOM 533 CA ASN A 32 -0.472 -2.371 12.210 1.00 0.97
+ATOM 534 C ASN A 32 0.304 -1.781 11.040 1.00 0.97
+ATOM 535 O ASN A 32 -0.206 -1.698 9.923 1.00 0.97
+ATOM 536 CB ASN A 32 -1.966 -2.161 12.043 1.00 0.97
+ATOM 537 CG ASN A 32 -2.775 -2.911 13.064 1.00 0.97
+ATOM 538 OD1 ASN A 32 -4.011 -2.887 13.034 1.00 0.97
+ATOM 539 ND2 ASN A 32 -2.103 -3.576 13.968 1.00 0.97
+ATOM 540 H ASN A 32 -0.197 -0.847 13.686 1.00 0.97
+ATOM 541 HA ASN A 32 -0.284 -3.446 12.213 1.00 0.97
+ATOM 542 1HB ASN A 32 -2.195 -1.097 12.125 1.00 0.97
+ATOM 543 2HB ASN A 32 -2.269 -2.485 11.047 1.00 0.97
+ATOM 544 1HD2 ASN A 32 -2.589 -4.094 14.673 1.00 0.97
+ATOM 545 2HD2 ASN A 32 -1.104 -3.567 13.955 1.00 0.97
+ATOM 546 N PHE A 33 1.585 -1.332 11.271 1.00 0.98
+ATOM 547 CA PHE A 33 2.414 -0.684 10.262 1.00 0.98
+ATOM 548 C PHE A 33 2.571 -1.565 9.029 1.00 0.98
+ATOM 549 O PHE A 33 2.496 -1.085 7.898 1.00 0.98
+ATOM 550 CB PHE A 33 3.784 -0.329 10.844 1.00 0.98
+ATOM 551 CG PHE A 33 4.706 0.377 9.891 1.00 0.98
+ATOM 552 CD1 PHE A 33 4.411 1.654 9.436 1.00 0.98
+ATOM 553 CD2 PHE A 33 5.871 -0.232 9.449 1.00 0.98
+ATOM 554 CE1 PHE A 33 5.259 2.305 8.560 1.00 0.98
+ATOM 555 CE2 PHE A 33 6.720 0.416 8.574 1.00 0.98
+ATOM 556 CZ PHE A 33 6.414 1.685 8.129 1.00 0.98
+ATOM 557 H PHE A 33 1.972 -1.510 12.186 1.00 0.98
+ATOM 558 HA PHE A 33 1.923 0.237 9.950 1.00 0.98
+ATOM 559 1HB PHE A 33 3.645 0.306 11.717 1.00 0.98
+ATOM 560 2HB PHE A 33 4.276 -1.243 11.179 1.00 0.98
+ATOM 561 HD1 PHE A 33 3.499 2.143 9.777 1.00 0.98
+ATOM 562 HD2 PHE A 33 6.113 -1.236 9.801 1.00 0.98
+ATOM 563 HE1 PHE A 33 5.015 3.307 8.210 1.00 0.98
+ATOM 564 HE2 PHE A 33 7.632 -0.074 8.234 1.00 0.98
+ATOM 565 HZ PHE A 33 7.083 2.198 7.439 1.00 0.98
+ATOM 566 N ASP A 34 2.753 -2.903 9.185 1.00 0.97
+ATOM 567 CA ASP A 34 2.923 -3.814 8.059 1.00 0.97
+ATOM 568 C ASP A 34 1.705 -3.793 7.145 1.00 0.97
+ATOM 569 O ASP A 34 1.835 -3.729 5.923 1.00 0.97
+ATOM 570 CB ASP A 34 3.190 -5.238 8.554 1.00 0.97
+ATOM 571 CG ASP A 34 4.553 -5.395 9.214 1.00 0.97
+ATOM 572 OD1 ASP A 34 4.795 -6.426 9.795 1.00 0.97
+ATOM 573 OD2 ASP A 34 5.340 -4.483 9.130 1.00 0.97
+ATOM 574 H ASP A 34 2.763 -3.275 10.125 1.00 0.97
+ATOM 575 HA ASP A 34 3.777 -3.477 7.468 1.00 0.97
+ATOM 576 1HB ASP A 34 2.420 -5.522 9.271 1.00 0.97
+ATOM 577 2HB ASP A 34 3.127 -5.930 7.714 1.00 0.97
+ATOM 578 N LYS A 35 0.438 -3.945 7.743 1.00 0.99
+ATOM 579 CA LYS A 35 -0.789 -3.877 6.959 1.00 0.99
+ATOM 580 C LYS A 35 -0.889 -2.558 6.204 1.00 0.99
+ATOM 581 O LYS A 35 -1.290 -2.525 5.041 1.00 0.99
+ATOM 582 CB LYS A 35 -2.013 -4.078 7.854 1.00 0.99
+ATOM 583 CG LYS A 35 -2.163 -5.494 8.393 1.00 0.99
+ATOM 584 CD LYS A 35 -3.419 -5.633 9.241 1.00 0.99
+ATOM 585 CE LYS A 35 -3.553 -7.038 9.808 1.00 0.99
+ATOM 586 NZ LYS A 35 -4.754 -7.177 10.675 1.00 0.99
+ATOM 587 H LYS A 35 0.375 -4.028 8.748 1.00 0.99
+ATOM 588 HA LYS A 35 -0.774 -4.682 6.223 1.00 0.99
+ATOM 589 1HB LYS A 35 -1.959 -3.400 8.707 1.00 0.99
+ATOM 590 2HB LYS A 35 -2.918 -3.833 7.299 1.00 0.99
+ATOM 591 1HG LYS A 35 -2.218 -6.196 7.560 1.00 0.99
+ATOM 592 2HG LYS A 35 -1.295 -5.747 9.000 1.00 0.99
+ATOM 593 1HD LYS A 35 -3.382 -4.919 10.065 1.00 0.99
+ATOM 594 2HD LYS A 35 -4.295 -5.413 8.632 1.00 0.99
+ATOM 595 1HE LYS A 35 -3.717 -7.808 8.989 1.00 0.99
+ATOM 596 2HE LYS A 35 -2.759 -7.333 10.393 1.00 0.99
+ATOM 597 1HZ LYS A 35 -4.898 -8.174 11.029 1.00 0.99
+ATOM 598 2HZ LYS A 35 -4.780 -6.582 11.446 1.00 0.99
+ATOM 599 3HZ LYS A 35 -5.676 -7.027 10.135 1.00 0.99
+ATOM 600 N PHE A 36 -0.563 -1.409 6.919 1.00 0.99
+ATOM 601 CA PHE A 36 -0.563 -0.097 6.284 1.00 0.99
+ATOM 602 C PHE A 36 0.365 -0.069 5.076 1.00 0.99
+ATOM 603 O PHE A 36 -0.023 0.369 3.993 1.00 0.99
+ATOM 604 CB PHE A 36 -0.150 0.985 7.284 1.00 0.99
+ATOM 605 CG PHE A 36 0.072 2.338 6.671 1.00 0.99
+ATOM 606 CD1 PHE A 36 -1.003 3.130 6.295 1.00 0.99
+ATOM 607 CD2 PHE A 36 1.355 2.823 6.469 1.00 0.99
+ATOM 608 CE1 PHE A 36 -0.800 4.375 5.731 1.00 0.99
+ATOM 609 CE2 PHE A 36 1.561 4.067 5.906 1.00 0.99
+ATOM 610 CZ PHE A 36 0.483 4.843 5.536 1.00 0.99
+ATOM 611 H PHE A 36 -0.295 -1.486 7.890 1.00 0.99
+ATOM 612 HA PHE A 36 -1.571 0.116 5.931 1.00 0.99
+ATOM 613 1HB PHE A 36 -0.919 1.080 8.049 1.00 0.99
+ATOM 614 2HB PHE A 36 0.772 0.682 7.783 1.00 0.99
+ATOM 615 HD1 PHE A 36 -2.017 2.759 6.449 1.00 0.99
+ATOM 616 HD2 PHE A 36 2.208 2.209 6.761 1.00 0.99
+ATOM 617 HE1 PHE A 36 -1.654 4.986 5.440 1.00 0.99
+ATOM 618 HE2 PHE A 36 2.575 4.436 5.753 1.00 0.99
+ATOM 619 HZ PHE A 36 0.644 5.825 5.092 1.00 0.99
+ATOM 620 N LEU A 37 1.645 -0.526 5.261 1.00 0.99
+ATOM 621 CA LEU A 37 2.636 -0.527 4.191 1.00 0.99
+ATOM 622 C LEU A 37 2.161 -1.353 3.001 1.00 0.99
+ATOM 623 O LEU A 37 2.368 -0.974 1.848 1.00 0.99
+ATOM 624 CB LEU A 37 3.980 -1.057 4.708 1.00 0.99
+ATOM 625 CG LEU A 37 4.740 -0.147 5.681 1.00 0.99
+ATOM 626 CD1 LEU A 37 5.960 -0.883 6.217 1.00 0.99
+ATOM 627 CD2 LEU A 37 5.145 1.135 4.968 1.00 0.99
+ATOM 628 H LEU A 37 1.892 -0.913 6.161 1.00 0.99
+ATOM 629 HA LEU A 37 2.787 0.499 3.859 1.00 0.99
+ATOM 630 1HB LEU A 37 3.802 -2.005 5.212 1.00 0.99
+ATOM 631 2HB LEU A 37 4.627 -1.240 3.851 1.00 0.99
+ATOM 632 HG LEU A 37 4.099 0.097 6.529 1.00 0.99
+ATOM 633 1HD1 LEU A 37 6.501 -0.237 6.909 1.00 0.99
+ATOM 634 2HD1 LEU A 37 5.641 -1.785 6.740 1.00 0.99
+ATOM 635 3HD1 LEU A 37 6.614 -1.155 5.389 1.00 0.99
+ATOM 636 1HD2 LEU A 37 5.685 1.782 5.660 1.00 0.99
+ATOM 637 2HD2 LEU A 37 5.787 0.892 4.121 1.00 0.99
+ATOM 638 3HD2 LEU A 37 4.253 1.649 4.611 1.00 0.99
+ATOM 639 N GLU A 38 1.495 -2.537 3.234 1.00 0.98
+ATOM 640 CA GLU A 38 0.959 -3.360 2.156 1.00 0.98
+ATOM 641 C GLU A 38 -0.121 -2.616 1.380 1.00 0.98
+ATOM 642 O GLU A 38 -0.135 -2.629 0.150 1.00 0.98
+ATOM 643 CB GLU A 38 0.403 -4.673 2.711 1.00 0.98
+ATOM 644 CG GLU A 38 1.469 -5.661 3.163 1.00 0.98
+ATOM 645 CD GLU A 38 0.891 -6.922 3.742 1.00 0.98
+ATOM 646 OE1 GLU A 38 1.649 -7.765 4.156 1.00 0.98
+ATOM 647 OE2 GLU A 38 -0.312 -7.042 3.769 1.00 0.98
+ATOM 648 H GLU A 38 1.378 -2.840 4.190 1.00 0.98
+ATOM 649 HA GLU A 38 1.768 -3.595 1.465 1.00 0.98
+ATOM 650 1HB GLU A 38 -0.244 -4.466 3.563 1.00 0.98
+ATOM 651 2HB GLU A 38 -0.205 -5.163 1.950 1.00 0.98
+ATOM 652 1HG GLU A 38 2.084 -5.934 2.305 1.00 0.98
+ATOM 653 2HG GLU A 38 2.085 -5.190 3.929 1.00 0.98
+ATOM 654 N GLU A 39 -1.115 -1.982 2.123 1.00 0.99
+ATOM 655 CA GLU A 39 -2.167 -1.225 1.455 1.00 0.99
+ATOM 656 C GLU A 39 -1.588 -0.074 0.644 1.00 0.99
+ATOM 657 O GLU A 39 -2.021 0.188 -0.478 1.00 0.99
+ATOM 658 CB GLU A 39 -3.188 -0.710 2.472 1.00 0.99
+ATOM 659 CG GLU A 39 -4.032 -1.808 3.106 1.00 0.99
+ATOM 660 CD GLU A 39 -5.046 -1.277 4.080 1.00 0.99
+ATOM 661 OE1 GLU A 39 -5.754 -2.064 4.659 1.00 0.99
+ATOM 662 OE2 GLU A 39 -5.114 -0.081 4.244 1.00 0.99
+ATOM 663 H GLU A 39 -1.091 -2.004 3.133 1.00 0.99
+ATOM 664 HA GLU A 39 -2.689 -1.892 0.768 1.00 0.99
+ATOM 665 1HB GLU A 39 -2.675 -0.179 3.274 1.00 0.99
+ATOM 666 2HB GLU A 39 -3.864 -0.004 1.991 1.00 0.99
+ATOM 667 1HG GLU A 39 -4.566 -2.341 2.319 1.00 0.99
+ATOM 668 2HG GLU A 39 -3.375 -2.493 3.642 1.00 0.99
+ATOM 669 N LEU A 40 -0.588 0.696 1.259 1.00 0.99
+ATOM 670 CA LEU A 40 0.073 1.783 0.548 1.00 0.99
+ATOM 671 C LEU A 40 0.734 1.284 -0.732 1.00 0.99
+ATOM 672 O LEU A 40 0.612 1.905 -1.788 1.00 0.99
+ATOM 673 CB LEU A 40 1.111 2.460 1.453 1.00 0.99
+ATOM 674 CG LEU A 40 1.859 3.668 0.875 1.00 0.99
+ATOM 675 CD1 LEU A 40 0.860 4.762 0.522 1.00 0.99
+ATOM 676 CD2 LEU A 40 2.879 4.166 1.889 1.00 0.99
+ATOM 677 H LEU A 40 -0.299 0.475 2.201 1.00 0.99
+ATOM 678 HA LEU A 40 -0.676 2.528 0.281 1.00 0.99
+ATOM 679 1HB LEU A 40 0.603 2.786 2.359 1.00 0.99
+ATOM 680 2HB LEU A 40 1.854 1.711 1.728 1.00 0.99
+ATOM 681 HG LEU A 40 2.371 3.376 -0.041 1.00 0.99
+ATOM 682 1HD1 LEU A 40 1.391 5.621 0.111 1.00 0.99
+ATOM 683 2HD1 LEU A 40 0.154 4.385 -0.219 1.00 0.99
+ATOM 684 3HD1 LEU A 40 0.319 5.065 1.418 1.00 0.99
+ATOM 685 1HD2 LEU A 40 3.411 5.025 1.478 1.00 0.99
+ATOM 686 2HD2 LEU A 40 2.367 4.461 2.805 1.00 0.99
+ATOM 687 3HD2 LEU A 40 3.590 3.370 2.110 1.00 0.99
+ATOM 688 N GLU A 41 1.466 0.116 -0.654 1.00 0.99
+ATOM 689 CA GLU A 41 2.111 -0.485 -1.815 1.00 0.99
+ATOM 690 C GLU A 41 1.106 -0.746 -2.930 1.00 0.99
+ATOM 691 O GLU A 41 1.376 -0.473 -4.099 1.00 0.99
+ATOM 692 CB GLU A 41 2.812 -1.787 -1.424 1.00 0.99
+ATOM 693 CG GLU A 41 4.114 -1.589 -0.660 1.00 0.99
+ATOM 694 CD GLU A 41 4.741 -2.884 -0.226 1.00 0.99
+ATOM 695 OE1 GLU A 41 5.826 -2.850 0.303 1.00 0.99
+ATOM 696 OE2 GLU A 41 4.134 -3.911 -0.425 1.00 0.99
+ATOM 697 H GLU A 41 1.545 -0.340 0.244 1.00 0.99
+ATOM 698 HA GLU A 41 2.859 0.211 -2.194 1.00 0.99
+ATOM 699 1HB GLU A 41 2.148 -2.388 -0.802 1.00 0.99
+ATOM 700 2HB GLU A 41 3.035 -2.366 -2.320 1.00 0.99
+ATOM 701 1HG GLU A 41 4.825 -1.068 -1.303 1.00 0.99
+ATOM 702 2HG GLU A 41 3.913 -0.995 0.232 1.00 0.99
+ATOM 703 N GLU A 42 -0.102 -1.315 -2.583 1.00 0.97
+ATOM 704 CA GLU A 42 -1.126 -1.589 -3.584 1.00 0.97
+ATOM 705 C GLU A 42 -1.582 -0.308 -4.271 1.00 0.97
+ATOM 706 O GLU A 42 -1.749 -0.271 -5.490 1.00 0.97
+ATOM 707 CB GLU A 42 -2.319 -2.306 -2.948 1.00 0.97
+ATOM 708 CG GLU A 42 -2.032 -3.739 -2.520 1.00 0.97
+ATOM 709 CD GLU A 42 -3.197 -4.386 -1.825 1.00 0.97
+ATOM 710 OE1 GLU A 42 -3.071 -5.518 -1.424 1.00 0.97
+ATOM 711 OE2 GLU A 42 -4.214 -3.747 -1.694 1.00 0.97
+ATOM 712 H GLU A 42 -0.284 -1.545 -1.616 1.00 0.97
+ATOM 713 HA GLU A 42 -0.701 -2.245 -4.343 1.00 0.97
+ATOM 714 1HB GLU A 42 -2.650 -1.755 -2.068 1.00 0.97
+ATOM 715 2HB GLU A 42 -3.150 -2.328 -3.653 1.00 0.97
+ATOM 716 1HG GLU A 42 -1.795 -4.331 -3.404 1.00 0.97
+ATOM 717 2HG GLU A 42 -1.186 -3.739 -1.833 1.00 0.97
+ATOM 718 N ILE A 43 -1.803 0.809 -3.474 1.00 0.98
+ATOM 719 CA ILE A 43 -2.249 2.082 -4.027 1.00 0.98
+ATOM 720 C ILE A 43 -1.174 2.708 -4.906 1.00 0.98
+ATOM 721 O ILE A 43 -1.450 3.150 -6.021 1.00 0.98
+ATOM 722 CB ILE A 43 -2.666 3.055 -2.909 1.00 0.98
+ATOM 723 CG1 ILE A 43 -3.944 2.570 -2.220 1.00 0.98
+ATOM 724 CG2 ILE A 43 -2.859 4.455 -3.471 1.00 0.98
+ATOM 725 CD1 ILE A 43 -4.291 3.338 -0.966 1.00 0.98
+ATOM 726 H ILE A 43 -1.659 0.729 -2.477 1.00 0.98
+ATOM 727 HA ILE A 43 -3.130 1.902 -4.642 1.00 0.98
+ATOM 728 HB ILE A 43 -1.888 3.084 -2.146 1.00 0.98
+ATOM 729 1HG2 ILE A 43 -3.150 5.132 -2.668 1.00 0.98
+ATOM 730 2HG2 ILE A 43 -1.926 4.800 -3.916 1.00 0.98
+ATOM 731 3HG2 ILE A 43 -3.639 4.437 -4.232 1.00 0.98
+ATOM 732 1HG1 ILE A 43 -4.782 2.649 -2.912 1.00 0.98
+ATOM 733 2HG1 ILE A 43 -3.837 1.517 -1.956 1.00 0.98
+ATOM 734 1HD1 ILE A 43 -5.209 2.936 -0.535 1.00 0.98
+ATOM 735 2HD1 ILE A 43 -3.479 3.243 -0.244 1.00 0.98
+ATOM 736 3HD1 ILE A 43 -4.437 4.389 -1.212 1.00 0.98
+ATOM 737 N LEU A 44 0.100 2.742 -4.424 1.00 0.98
+ATOM 738 CA LEU A 44 1.215 3.357 -5.134 1.00 0.98
+ATOM 739 C LEU A 44 1.470 2.663 -6.468 1.00 0.98
+ATOM 740 O LEU A 44 1.953 3.280 -7.417 1.00 0.98
+ATOM 741 CB LEU A 44 2.481 3.330 -4.268 1.00 0.98
+ATOM 742 CG LEU A 44 2.480 4.236 -3.030 1.00 0.98
+ATOM 743 CD1 LEU A 44 3.737 3.979 -2.211 1.00 0.98
+ATOM 744 CD2 LEU A 44 2.398 5.691 -3.466 1.00 0.98
+ATOM 745 H LEU A 44 0.273 2.280 -3.543 1.00 0.98
+ATOM 746 HA LEU A 44 0.967 4.399 -5.331 1.00 0.98
+ATOM 747 1HB LEU A 44 2.641 2.307 -3.930 1.00 0.98
+ATOM 748 2HB LEU A 44 3.327 3.621 -4.891 1.00 0.98
+ATOM 749 HG LEU A 44 1.619 3.996 -2.405 1.00 0.98
+ATOM 750 1HD1 LEU A 44 3.736 4.623 -1.331 1.00 0.98
+ATOM 751 2HD1 LEU A 44 3.760 2.936 -1.896 1.00 0.98
+ATOM 752 3HD1 LEU A 44 4.616 4.195 -2.817 1.00 0.98
+ATOM 753 1HD2 LEU A 44 2.396 6.335 -2.586 1.00 0.98
+ATOM 754 2HD2 LEU A 44 3.259 5.933 -4.091 1.00 0.98
+ATOM 755 3HD2 LEU A 44 1.481 5.850 -4.034 1.00 0.98
+ATOM 756 N LYS A 45 1.172 1.283 -6.578 1.00 0.97
+ATOM 757 CA LYS A 45 1.363 0.546 -7.821 1.00 0.97
+ATOM 758 C LYS A 45 0.459 1.082 -8.923 1.00 0.97
+ATOM 759 O LYS A 45 0.740 0.908 -10.109 1.00 0.97
+ATOM 760 CB LYS A 45 1.115 -0.948 -7.607 1.00 0.97
+ATOM 761 CG LYS A 45 2.227 -1.666 -6.853 1.00 0.97
+ATOM 762 CD LYS A 45 1.893 -3.136 -6.645 1.00 0.97
+ATOM 763 CE LYS A 45 2.999 -3.853 -5.885 1.00 0.97
+ATOM 764 NZ LYS A 45 2.659 -5.277 -5.620 1.00 0.97
+ATOM 765 H LYS A 45 0.811 0.799 -5.769 1.00 0.97
+ATOM 766 HA LYS A 45 2.397 0.676 -8.145 1.00 0.97
+ATOM 767 1HB LYS A 45 0.189 -1.087 -7.049 1.00 0.97
+ATOM 768 2HB LYS A 45 0.994 -1.439 -8.572 1.00 0.97
+ATOM 769 1HG LYS A 45 3.157 -1.590 -7.417 1.00 0.97
+ATOM 770 2HG LYS A 45 2.372 -1.194 -5.882 1.00 0.97
+ATOM 771 1HD LYS A 45 0.963 -3.222 -6.083 1.00 0.97
+ATOM 772 2HD LYS A 45 1.758 -3.619 -7.613 1.00 0.97
+ATOM 773 1HE LYS A 45 3.957 -3.910 -6.493 1.00 0.97
+ATOM 774 2HE LYS A 45 3.208 -3.446 -4.964 1.00 0.97
+ATOM 775 1HZ LYS A 45 3.452 -5.812 -5.145 1.00 0.97
+ATOM 776 2HZ LYS A 45 1.850 -5.420 -5.097 1.00 0.97
+ATOM 777 3HZ LYS A 45 2.551 -5.854 -6.526 1.00 0.97
+ATOM 778 N ARG A 46 -0.724 1.754 -8.555 1.00 0.96
+ATOM 779 CA ARG A 46 -1.681 2.250 -9.537 1.00 0.96
+ATOM 780 C ARG A 46 -1.378 3.692 -9.923 1.00 0.96
+ATOM 781 O ARG A 46 -2.028 4.259 -10.801 1.00 0.96
+ATOM 782 CB ARG A 46 -3.101 2.154 -8.998 1.00 0.96
+ATOM 783 CG ARG A 46 -3.589 0.737 -8.740 1.00 0.96
+ATOM 784 CD ARG A 46 -4.976 0.722 -8.208 1.00 0.96
+ATOM 785 NE ARG A 46 -5.436 -0.629 -7.931 1.00 0.96
+ATOM 786 CZ ARG A 46 -6.639 -0.936 -7.408 1.00 0.96
+ATOM 787 NH1 ARG A 46 -7.490 0.021 -7.113 1.00 0.96
+ATOM 788 NH2 ARG A 46 -6.964 -2.199 -7.193 1.00 0.96
+ATOM 789 H ARG A 46 -0.858 1.980 -7.580 1.00 0.96
+ATOM 790 HA ARG A 46 -1.612 1.629 -10.431 1.00 0.96
+ATOM 791 1HB ARG A 46 -3.174 2.703 -8.061 1.00 0.96
+ATOM 792 2HB ARG A 46 -3.792 2.617 -9.704 1.00 0.96
+ATOM 793 1HG ARG A 46 -3.573 0.171 -9.672 1.00 0.96
+ATOM 794 2HG ARG A 46 -2.937 0.255 -8.011 1.00 0.96
+ATOM 795 1HD ARG A 46 -5.016 1.293 -7.281 1.00 0.96
+ATOM 796 2HD ARG A 46 -5.650 1.168 -8.939 1.00 0.96
+ATOM 797 HE ARG A 46 -4.808 -1.393 -8.145 1.00 0.96
+ATOM 798 1HH1 ARG A 46 -7.242 0.987 -7.277 1.00 0.96
+ATOM 799 2HH1 ARG A 46 -8.392 -0.209 -6.721 1.00 0.96
+ATOM 800 1HH2 ARG A 46 -6.309 -2.935 -7.420 1.00 0.96
+ATOM 801 2HH2 ARG A 46 -7.865 -2.428 -6.802 1.00 0.96
+ATOM 802 N MET A 47 -0.481 4.422 -9.183 1.00 0.98
+ATOM 803 CA MET A 47 -0.212 5.840 -9.393 1.00 0.98
+ATOM 804 C MET A 47 1.004 6.044 -10.287 1.00 0.98
+ATOM 805 O MET A 47 1.906 5.207 -10.325 1.00 0.98
+ATOM 806 CB MET A 47 -0.015 6.546 -8.053 1.00 0.98
+ATOM 807 CG MET A 47 -1.230 6.552 -7.136 1.00 0.98
+ATOM 808 SD MET A 47 -0.880 7.278 -5.522 1.00 0.98
+ATOM 809 CE MET A 47 -1.183 9.011 -5.858 1.00 0.98
+ATOM 810 H MET A 47 0.070 3.911 -8.508 1.00 0.98
+ATOM 811 HA MET A 47 -1.071 6.284 -9.896 1.00 0.98
+ATOM 812 1HB MET A 47 0.805 6.070 -7.517 1.00 0.98
+ATOM 813 2HB MET A 47 0.271 7.583 -8.235 1.00 0.98
+ATOM 814 1HG MET A 47 -2.035 7.119 -7.602 1.00 0.98
+ATOM 815 2HG MET A 47 -1.577 5.530 -6.985 1.00 0.98
+ATOM 816 1HE MET A 47 -0.973 9.830 -4.744 1.00 0.98
+ATOM 817 2HE MET A 47 -0.458 9.558 -6.427 1.00 0.98
+ATOM 818 3HE MET A 47 -2.177 9.413 -5.988 1.00 0.98
+ATOM 819 N SER A 48 1.106 7.168 -11.026 1.00 0.96
+ATOM 820 CA SER A 48 2.341 7.561 -11.694 1.00 0.96
+ATOM 821 C SER A 48 3.439 7.871 -10.685 1.00 0.96
+ATOM 822 O SER A 48 3.162 8.139 -9.515 1.00 0.96
+ATOM 823 CB SER A 48 2.089 8.764 -12.582 1.00 0.96
+ATOM 824 OG SER A 48 1.836 9.910 -11.818 1.00 0.96
+ATOM 825 H SER A 48 0.300 7.772 -11.089 1.00 0.96
+ATOM 826 HG SER A 48 1.755 9.610 -10.909 1.00 0.96
+ATOM 827 HA SER A 48 2.676 6.731 -12.319 1.00 0.96
+ATOM 828 1HB SER A 48 2.957 8.932 -13.219 1.00 0.96
+ATOM 829 2HB SER A 48 1.240 8.562 -13.233 1.00 0.96
+ATOM 830 N PRO A 49 4.780 7.925 -11.121 1.00 0.97
+ATOM 831 CA PRO A 49 5.846 8.051 -10.135 1.00 0.97
+ATOM 832 C PRO A 49 5.747 9.372 -9.382 1.00 0.97
+ATOM 833 O PRO A 49 5.943 9.421 -8.168 1.00 0.97
+ATOM 834 CB PRO A 49 7.125 7.978 -10.975 1.00 0.97
+ATOM 835 CG PRO A 49 6.938 6.806 -11.877 1.00 0.97
+ATOM 836 CD PRO A 49 6.252 7.373 -13.091 1.00 0.97
+ATOM 837 HA PRO A 49 5.795 7.203 -9.436 1.00 0.97
+ATOM 838 1HB PRO A 49 7.257 8.922 -11.524 1.00 0.97
+ATOM 839 2HB PRO A 49 7.994 7.869 -10.309 1.00 0.97
+ATOM 840 1HG PRO A 49 7.911 6.350 -12.115 1.00 0.97
+ATOM 841 2HG PRO A 49 6.339 6.032 -11.375 1.00 0.97
+ATOM 842 1HD PRO A 49 5.486 8.102 -12.788 1.00 0.97
+ATOM 843 2HD PRO A 49 6.976 7.915 -13.718 1.00 0.97
+ATOM 844 N GLU A 50 5.427 10.513 -10.072 1.00 0.96
+ATOM 845 CA GLU A 50 5.330 11.820 -9.434 1.00 0.96
+ATOM 846 C GLU A 50 4.205 11.851 -8.408 1.00 0.96
+ATOM 847 O GLU A 50 4.376 12.358 -7.300 1.00 0.96
+ATOM 848 CB GLU A 50 5.122 12.915 -10.483 1.00 0.96
+ATOM 849 CG GLU A 50 6.328 13.164 -11.377 1.00 0.96
+ATOM 850 CD GLU A 50 6.059 14.178 -12.454 1.00 0.96
+ATOM 851 OE1 GLU A 50 6.944 14.437 -13.234 1.00 0.96
+ATOM 852 OE2 GLU A 50 4.967 14.694 -12.497 1.00 0.96
+ATOM 853 H GLU A 50 5.240 10.433 -11.061 1.00 0.96
+ATOM 854 HA GLU A 50 6.266 12.021 -8.913 1.00 0.96
+ATOM 855 1HB GLU A 50 4.280 12.652 -11.123 1.00 0.96
+ATOM 856 2HB GLU A 50 4.875 13.853 -9.986 1.00 0.96
+ATOM 857 1HG GLU A 50 7.152 13.532 -10.765 1.00 0.96
+ATOM 858 2HG GLU A 50 6.610 12.227 -11.859 1.00 0.96
+ATOM 859 N GLN A 51 2.961 11.349 -8.788 1.00 0.98
+ATOM 860 CA GLN A 51 1.839 11.297 -7.859 1.00 0.98
+ATOM 861 C GLN A 51 2.166 10.434 -6.647 1.00 0.98
+ATOM 862 O GLN A 51 1.793 10.760 -5.521 1.00 0.98
+ATOM 863 CB GLN A 51 0.578 10.786 -8.561 1.00 0.98
+ATOM 864 CG GLN A 51 0.002 11.747 -9.587 1.00 0.98
+ATOM 865 CD GLN A 51 -1.184 11.159 -10.329 1.00 0.98
+ATOM 866 OE1 GLN A 51 -2.176 11.847 -10.586 1.00 0.98
+ATOM 867 NE2 GLN A 51 -1.088 9.881 -10.677 1.00 0.98
+ATOM 868 H GLN A 51 2.848 10.972 -9.718 1.00 0.98
+ATOM 869 HA GLN A 51 1.635 12.308 -7.506 1.00 0.98
+ATOM 870 1HB GLN A 51 0.801 9.846 -9.068 1.00 0.98
+ATOM 871 2HB GLN A 51 -0.194 10.583 -7.820 1.00 0.98
+ATOM 872 1HG GLN A 51 -0.328 12.651 -9.077 1.00 0.98
+ATOM 873 2HG GLN A 51 0.775 11.990 -10.316 1.00 0.98
+ATOM 874 1HE2 GLN A 51 -1.834 9.441 -11.165 1.00 0.98
+ATOM 875 2HE2 GLN A 51 -0.266 9.360 -10.449 1.00 0.98
+ATOM 876 N ALA A 52 2.841 9.246 -6.870 1.00 0.98
+ATOM 877 CA ALA A 52 3.271 8.378 -5.781 1.00 0.98
+ATOM 878 C ALA A 52 4.207 9.112 -4.829 1.00 0.98
+ATOM 879 O ALA A 52 4.068 9.015 -3.610 1.00 0.98
+ATOM 880 CB ALA A 52 3.934 7.123 -6.331 1.00 0.98
+ATOM 881 H ALA A 52 3.062 8.989 -7.822 1.00 0.98
+ATOM 882 HA ALA A 52 2.388 8.079 -5.217 1.00 0.98
+ATOM 883 1HB ALA A 52 4.245 6.484 -5.504 1.00 0.98
+ATOM 884 2HB ALA A 52 3.226 6.583 -6.959 1.00 0.98
+ATOM 885 3HB ALA A 52 4.805 7.401 -6.922 1.00 0.98
+ATOM 886 N ARG A 53 5.222 9.864 -5.383 1.00 0.97
+ATOM 887 CA ARG A 53 6.158 10.612 -4.553 1.00 0.97
+ATOM 888 C ARG A 53 5.434 11.647 -3.702 1.00 0.97
+ATOM 889 O ARG A 53 5.688 11.766 -2.504 1.00 0.97
+ATOM 890 CB ARG A 53 7.209 11.298 -5.414 1.00 0.97
+ATOM 891 CG ARG A 53 8.298 12.018 -4.633 1.00 0.97
+ATOM 892 CD ARG A 53 9.284 12.666 -5.536 1.00 0.97
+ATOM 893 NE ARG A 53 10.015 11.691 -6.329 1.00 0.97
+ATOM 894 CZ ARG A 53 10.957 12.000 -7.242 1.00 0.97
+ATOM 895 NH1 ARG A 53 11.268 13.257 -7.466 1.00 0.97
+ATOM 896 NH2 ARG A 53 11.566 11.038 -7.913 1.00 0.97
+ATOM 897 H ARG A 53 5.318 9.899 -6.388 1.00 0.97
+ATOM 898 HA ARG A 53 6.668 9.913 -3.890 1.00 0.97
+ATOM 899 1HB ARG A 53 7.695 10.563 -6.054 1.00 0.97
+ATOM 900 2HB ARG A 53 6.729 12.031 -6.063 1.00 0.97
+ATOM 901 1HG ARG A 53 7.847 12.789 -4.008 1.00 0.97
+ATOM 902 2HG ARG A 53 8.828 11.303 -4.003 1.00 0.97
+ATOM 903 1HD ARG A 53 8.765 13.339 -6.217 1.00 0.97
+ATOM 904 2HD ARG A 53 10.002 13.232 -4.944 1.00 0.97
+ATOM 905 HE ARG A 53 9.803 10.713 -6.184 1.00 0.97
+ATOM 906 1HH1 ARG A 53 10.802 13.992 -6.953 1.00 0.97
+ATOM 907 2HH1 ARG A 53 11.974 13.488 -8.151 1.00 0.97
+ATOM 908 1HH2 ARG A 53 11.327 10.071 -7.740 1.00 0.97
+ATOM 909 2HH2 ARG A 53 12.271 11.269 -8.597 1.00 0.97
+ATOM 910 N GLU A 54 4.496 12.452 -4.318 1.00 0.98
+ATOM 911 CA GLU A 54 3.730 13.446 -3.576 1.00 0.98
+ATOM 912 C GLU A 54 2.920 12.798 -2.460 1.00 0.98
+ATOM 913 O GLU A 54 2.844 13.323 -1.350 1.00 0.98
+ATOM 914 CB GLU A 54 2.813 14.228 -4.518 1.00 0.98
+ATOM 915 CG GLU A 54 3.548 15.164 -5.467 1.00 0.98
+ATOM 916 CD GLU A 54 2.627 15.873 -6.420 1.00 0.98
+ATOM 917 OE1 GLU A 54 3.109 16.623 -7.235 1.00 0.98
+ATOM 918 OE2 GLU A 54 1.440 15.665 -6.334 1.00 0.98
+ATOM 919 H GLU A 54 4.317 12.329 -5.304 1.00 0.98
+ATOM 920 HA GLU A 54 4.427 14.150 -3.120 1.00 0.98
+ATOM 921 1HB GLU A 54 2.227 13.534 -5.121 1.00 0.98
+ATOM 922 2HB GLU A 54 2.112 14.826 -3.935 1.00 0.98
+ATOM 923 1HG GLU A 54 4.074 15.918 -4.882 1.00 0.98
+ATOM 924 2HG GLU A 54 4.260 14.583 -6.054 1.00 0.98
+ATOM 925 N LEU A 55 2.202 11.654 -2.761 1.00 0.99
+ATOM 926 CA LEU A 55 1.443 10.937 -1.744 1.00 0.99
+ATOM 927 C LEU A 55 2.345 10.481 -0.602 1.00 0.99
+ATOM 928 O LEU A 55 2.017 10.661 0.571 1.00 0.99
+ATOM 929 CB LEU A 55 0.721 9.733 -2.362 1.00 0.99
+ATOM 930 CG LEU A 55 -0.123 8.874 -1.412 1.00 0.99
+ATOM 931 CD1 LEU A 55 -1.230 9.726 -0.806 1.00 0.99
+ATOM 932 CD2 LEU A 55 -0.699 7.690 -2.173 1.00 0.99
+ATOM 933 H LEU A 55 2.244 11.280 -3.699 1.00 0.99
+ATOM 934 HA LEU A 55 0.688 11.609 -1.338 1.00 0.99
+ATOM 935 1HB LEU A 55 0.064 10.100 -3.149 1.00 0.99
+ATOM 936 2HB LEU A 55 1.471 9.085 -2.816 1.00 0.99
+ATOM 937 HG LEU A 55 0.503 8.512 -0.596 1.00 0.99
+ATOM 938 1HD1 LEU A 55 -1.830 9.116 -0.130 1.00 0.99
+ATOM 939 2HD1 LEU A 55 -0.790 10.555 -0.251 1.00 0.99
+ATOM 940 3HD1 LEU A 55 -1.864 10.117 -1.601 1.00 0.99
+ATOM 941 1HD2 LEU A 55 -1.298 7.079 -1.497 1.00 0.99
+ATOM 942 2HD2 LEU A 55 -1.326 8.051 -2.988 1.00 0.99
+ATOM 943 3HD2 LEU A 55 0.115 7.089 -2.580 1.00 0.99
+ATOM 944 N ILE A 56 3.534 9.847 -0.932 1.00 0.98
+ATOM 945 CA ILE A 56 4.463 9.398 0.097 1.00 0.98
+ATOM 946 C ILE A 56 4.912 10.558 0.978 1.00 0.98
+ATOM 947 O ILE A 56 4.990 10.429 2.199 1.00 0.98
+ATOM 948 CB ILE A 56 5.686 8.702 -0.527 1.00 0.98
+ATOM 949 CG1 ILE A 56 5.307 7.323 -1.073 1.00 0.98
+ATOM 950 CG2 ILE A 56 6.805 8.584 0.496 1.00 0.98
+ATOM 951 CD1 ILE A 56 6.398 6.667 -1.888 1.00 0.98
+ATOM 952 H ILE A 56 3.773 9.711 -1.904 1.00 0.98
+ATOM 953 HA ILE A 56 3.956 8.669 0.728 1.00 0.98
+ATOM 954 HB ILE A 56 6.041 9.289 -1.374 1.00 0.98
+ATOM 955 1HG2 ILE A 56 7.665 8.095 0.040 1.00 0.98
+ATOM 956 2HG2 ILE A 56 7.092 9.578 0.838 1.00 0.98
+ATOM 957 3HG2 ILE A 56 6.461 7.994 1.345 1.00 0.98
+ATOM 958 1HG1 ILE A 56 5.055 6.661 -0.244 1.00 0.98
+ATOM 959 2HG1 ILE A 56 4.420 7.411 -1.701 1.00 0.98
+ATOM 960 1HD1 ILE A 56 6.055 5.694 -2.240 1.00 0.98
+ATOM 961 2HD1 ILE A 56 6.641 7.298 -2.744 1.00 0.98
+ATOM 962 3HD1 ILE A 56 7.285 6.536 -1.270 1.00 0.98
+ATOM 963 N GLU A 57 5.235 11.760 0.375 1.00 0.97
+ATOM 964 CA GLU A 57 5.627 12.941 1.135 1.00 0.97
+ATOM 965 C GLU A 57 4.530 13.361 2.105 1.00 0.97
+ATOM 966 O GLU A 57 4.800 13.673 3.265 1.00 0.97
+ATOM 967 CB GLU A 57 5.976 14.094 0.192 1.00 0.97
+ATOM 968 CG GLU A 57 7.289 13.914 -0.556 1.00 0.97
+ATOM 969 CD GLU A 57 7.548 15.006 -1.556 1.00 0.97
+ATOM 970 OE1 GLU A 57 8.581 14.980 -2.182 1.00 0.97
+ATOM 971 OE2 GLU A 57 6.712 15.867 -1.694 1.00 0.97
+ATOM 972 H GLU A 57 5.172 11.822 -0.631 1.00 0.97
+ATOM 973 HA GLU A 57 6.515 12.698 1.717 1.00 0.97
+ATOM 974 1HB GLU A 57 5.184 14.214 -0.547 1.00 0.97
+ATOM 975 2HB GLU A 57 6.039 15.023 0.758 1.00 0.97
+ATOM 976 1HG GLU A 57 8.108 13.914 0.164 1.00 0.97
+ATOM 977 2HG GLU A 57 7.263 12.965 -1.091 1.00 0.97
+ATOM 978 N TRP A 58 3.239 13.437 1.633 1.00 0.98
+ATOM 979 CA TRP A 58 2.110 13.770 2.494 1.00 0.98
+ATOM 980 C TRP A 58 1.982 12.778 3.642 1.00 0.98
+ATOM 981 O TRP A 58 1.855 13.169 4.802 1.00 0.98
+ATOM 982 CB TRP A 58 0.813 13.816 1.684 1.00 0.98
+ATOM 983 CG TRP A 58 -0.404 14.144 2.494 1.00 0.98
+ATOM 984 CD1 TRP A 58 -0.863 15.388 2.808 1.00 0.98
+ATOM 985 CD2 TRP A 58 -1.329 13.210 3.101 1.00 0.98
+ATOM 986 NE1 TRP A 58 -2.004 15.294 3.565 1.00 0.98
+ATOM 987 CE2 TRP A 58 -2.304 13.968 3.755 1.00 0.98
+ATOM 988 CE3 TRP A 58 -1.406 11.812 3.142 1.00 0.98
+ATOM 989 CZ2 TRP A 58 -3.351 13.379 4.445 1.00 0.98
+ATOM 990 CZ3 TRP A 58 -2.456 11.221 3.835 1.00 0.98
+ATOM 991 CH2 TRP A 58 -3.403 11.985 4.469 1.00 0.98
+ATOM 992 H TRP A 58 3.068 13.206 0.664 1.00 0.98
+ATOM 993 HA TRP A 58 2.277 14.762 2.914 1.00 0.98
+ATOM 994 1HB TRP A 58 0.910 14.561 0.894 1.00 0.98
+ATOM 995 2HB TRP A 58 0.656 12.849 1.205 1.00 0.98
+ATOM 996 HD1 TRP A 58 -0.392 16.321 2.503 1.00 0.98
+ATOM 997 HE1 TRP A 58 -2.537 16.072 3.925 1.00 0.98
+ATOM 998 HZ2 TRP A 58 -4.113 13.968 4.956 1.00 0.98
+ATOM 999 HH2 TRP A 58 -4.214 11.489 5.004 1.00 0.98
+ATOM 1000 HZ3 TRP A 58 -2.509 10.132 3.862 1.00 0.98
+ATOM 1001 HE3 TRP A 58 -0.657 11.201 2.640 1.00 0.98
+ATOM 1002 N TRP A 59 1.989 11.445 3.329 1.00 0.98
+ATOM 1003 CA TRP A 59 1.880 10.414 4.354 1.00 0.98
+ATOM 1004 C TRP A 59 3.028 10.506 5.352 1.00 0.98
+ATOM 1005 O TRP A 59 2.829 10.360 6.557 1.00 0.98
+ATOM 1006 CB TRP A 59 1.845 9.022 3.722 1.00 0.98
+ATOM 1007 CG TRP A 59 0.499 8.616 3.202 1.00 0.98
+ATOM 1008 CD1 TRP A 59 0.204 8.206 1.936 1.00 0.98
+ATOM 1009 CD2 TRP A 59 -0.748 8.577 3.938 1.00 0.98
+ATOM 1010 NE1 TRP A 59 -1.135 7.917 1.833 1.00 0.98
+ATOM 1011 CE2 TRP A 59 -1.732 8.138 3.048 1.00 0.98
+ATOM 1012 CE3 TRP A 59 -1.102 8.875 5.259 1.00 0.98
+ATOM 1013 CZ2 TRP A 59 -3.055 7.988 3.434 1.00 0.98
+ATOM 1014 CZ3 TRP A 59 -2.428 8.724 5.646 1.00 0.98
+ATOM 1015 CH2 TRP A 59 -3.379 8.292 4.756 1.00 0.98
+ATOM 1016 H TRP A 59 2.119 11.169 2.366 1.00 0.98
+ATOM 1017 HA TRP A 59 0.944 10.560 4.894 1.00 0.98
+ATOM 1018 1HB TRP A 59 2.556 8.987 2.896 1.00 0.98
+ATOM 1019 2HB TRP A 59 2.164 8.286 4.460 1.00 0.98
+ATOM 1020 HD1 TRP A 59 0.924 8.122 1.125 1.00 0.98
+ATOM 1021 HE1 TRP A 59 -1.604 7.594 0.999 1.00 0.98
+ATOM 1022 HZ2 TRP A 59 -3.825 7.646 2.742 1.00 0.98
+ATOM 1023 HH2 TRP A 59 -4.410 8.184 5.093 1.00 0.98
+ATOM 1024 HZ3 TRP A 59 -2.695 8.958 6.677 1.00 0.98
+ATOM 1025 HE3 TRP A 59 -0.351 9.219 5.970 1.00 0.98
+ATOM 1026 N TRP A 60 4.313 10.712 4.844 1.00 0.97
+ATOM 1027 CA TRP A 60 5.476 10.860 5.711 1.00 0.97
+ATOM 1028 C TRP A 60 5.279 11.994 6.708 1.00 0.97
+ATOM 1029 O TRP A 60 5.562 11.844 7.897 1.00 0.97
+ATOM 1030 CB TRP A 60 6.730 11.115 4.873 1.00 0.97
+ATOM 1031 CG TRP A 60 7.977 11.311 5.681 1.00 0.97
+ATOM 1032 CD1 TRP A 60 8.627 12.488 5.905 1.00 0.97
+ATOM 1033 CD2 TRP A 60 8.734 10.295 6.382 1.00 0.97
+ATOM 1034 NE1 TRP A 60 9.731 12.276 6.692 1.00 0.97
+ATOM 1035 CE2 TRP A 60 9.812 10.940 6.994 1.00 0.97
+ATOM 1036 CE3 TRP A 60 8.585 8.911 6.536 1.00 0.97
+ATOM 1037 CZ2 TRP A 60 10.745 10.250 7.753 1.00 0.97
+ATOM 1038 CZ3 TRP A 60 9.520 8.220 7.297 1.00 0.97
+ATOM 1039 CH2 TRP A 60 10.572 8.873 7.889 1.00 0.97
+ATOM 1040 H TRP A 60 4.432 10.785 3.844 1.00 0.97
+ATOM 1041 HA TRP A 60 5.613 9.933 6.267 1.00 0.97
+ATOM 1042 1HB TRP A 60 6.888 10.274 4.198 1.00 0.97
+ATOM 1043 2HB TRP A 60 6.577 12.003 4.259 1.00 0.97
+ATOM 1044 HD1 TRP A 60 8.315 13.455 5.515 1.00 0.97
+ATOM 1045 HE1 TRP A 60 10.379 12.987 7.001 1.00 0.97
+ATOM 1046 HZ2 TRP A 60 11.587 10.751 8.231 1.00 0.97
+ATOM 1047 HH2 TRP A 60 11.287 8.299 8.480 1.00 0.97
+ATOM 1048 HZ3 TRP A 60 9.397 7.142 7.413 1.00 0.97
+ATOM 1049 HE3 TRP A 60 7.752 8.388 6.068 1.00 0.97
+ATOM 1050 N LYS A 61 4.769 13.197 6.238 1.00 0.97
+ATOM 1051 CA LYS A 61 4.516 14.325 7.126 1.00 0.97
+ATOM 1052 C LYS A 61 3.503 13.961 8.205 1.00 0.97
+ATOM 1053 O LYS A 61 3.702 14.261 9.382 1.00 0.97
+ATOM 1054 CB LYS A 61 4.034 15.541 6.333 1.00 0.97
+ATOM 1055 CG LYS A 61 5.104 16.227 5.494 1.00 0.97
+ATOM 1056 CD LYS A 61 4.539 17.431 4.755 1.00 0.97
+ATOM 1057 CE LYS A 61 5.588 18.073 3.859 1.00 0.97
+ATOM 1058 NZ LYS A 61 5.044 19.242 3.115 1.00 0.97
+ATOM 1059 H LYS A 61 4.556 13.288 5.255 1.00 0.97
+ATOM 1060 HA LYS A 61 5.450 14.592 7.622 1.00 0.97
+ATOM 1061 1HB LYS A 61 3.229 15.236 5.663 1.00 0.97
+ATOM 1062 2HB LYS A 61 3.624 16.279 7.022 1.00 0.97
+ATOM 1063 1HG LYS A 61 5.917 16.558 6.142 1.00 0.97
+ATOM 1064 2HG LYS A 61 5.506 15.521 4.768 1.00 0.97
+ATOM 1065 1HD LYS A 61 3.693 17.118 4.143 1.00 0.97
+ATOM 1066 2HD LYS A 61 4.190 18.170 5.476 1.00 0.97
+ATOM 1067 1HE LYS A 61 6.447 18.508 4.462 1.00 0.97
+ATOM 1068 2HE LYS A 61 5.966 17.444 3.138 1.00 0.97
+ATOM 1069 1HZ LYS A 61 5.785 19.744 2.532 1.00 0.97
+ATOM 1070 2HZ LYS A 61 4.288 19.049 2.533 1.00 0.97
+ATOM 1071 3HZ LYS A 61 4.738 20.044 3.770 1.00 0.97
+ATOM 1072 N TRP A 62 2.345 13.323 7.810 1.00 0.97
+ATOM 1073 CA TRP A 62 1.328 12.897 8.763 1.00 0.97
+ATOM 1074 C TRP A 62 1.897 11.906 9.770 1.00 0.97
+ATOM 1075 O TRP A 62 1.706 12.054 10.977 1.00 0.97
+ATOM 1076 CB TRP A 62 0.129 12.291 8.030 1.00 0.97
+ATOM 1077 CG TRP A 62 -0.999 11.881 8.928 1.00 0.97
+ATOM 1078 CD1 TRP A 62 -1.966 12.692 9.443 1.00 0.97
+ATOM 1079 CD2 TRP A 62 -1.284 10.551 9.424 1.00 0.97
+ATOM 1080 NE1 TRP A 62 -2.830 11.962 10.221 1.00 0.97
+ATOM 1081 CE2 TRP A 62 -2.428 10.650 10.221 1.00 0.97
+ATOM 1082 CE3 TRP A 62 -0.670 9.304 9.258 1.00 0.97
+ATOM 1083 CZ2 TRP A 62 -2.976 9.547 10.856 1.00 0.97
+ATOM 1084 CZ3 TRP A 62 -1.220 8.198 9.895 1.00 0.97
+ATOM 1085 CH2 TRP A 62 -2.344 8.317 10.673 1.00 0.97
+ATOM 1086 H TRP A 62 2.234 13.089 6.833 1.00 0.97
+ATOM 1087 HA TRP A 62 0.974 13.773 9.306 1.00 0.97
+ATOM 1088 1HB TRP A 62 -0.251 13.015 7.310 1.00 0.97
+ATOM 1089 2HB TRP A 62 0.458 11.415 7.472 1.00 0.97
+ATOM 1090 HD1 TRP A 62 -2.043 13.762 9.262 1.00 0.97
+ATOM 1091 HE1 TRP A 62 -3.631 12.329 10.714 1.00 0.97
+ATOM 1092 HZ2 TRP A 62 -3.869 9.621 11.478 1.00 0.97
+ATOM 1093 HH2 TRP A 62 -2.749 7.429 11.158 1.00 0.97
+ATOM 1094 HZ3 TRP A 62 -0.736 7.230 9.760 1.00 0.97
+ATOM 1095 HE3 TRP A 62 0.223 9.203 8.641 1.00 0.97
+ATOM 1096 N ILE A 63 2.550 10.789 9.268 1.00 0.98
+ATOM 1097 CA ILE A 63 3.156 9.795 10.146 1.00 0.98
+ATOM 1098 C ILE A 63 4.151 10.439 11.103 1.00 0.98
+ATOM 1099 O ILE A 63 4.197 10.101 12.286 1.00 0.98
+ATOM 1100 CB ILE A 63 3.832 8.682 9.326 1.00 0.98
+ATOM 1101 CG1 ILE A 63 2.777 7.835 8.610 1.00 0.98
+ATOM 1102 CG2 ILE A 63 4.702 7.813 10.221 1.00 0.98
+ATOM 1103 CD1 ILE A 63 3.349 6.898 7.570 1.00 0.98
+ATOM 1104 H ILE A 63 2.667 10.699 8.269 1.00 0.98
+ATOM 1105 HA ILE A 63 2.367 9.331 10.737 1.00 0.98
+ATOM 1106 HB ILE A 63 4.456 9.128 8.552 1.00 0.98
+ATOM 1107 1HG2 ILE A 63 5.170 7.030 9.624 1.00 0.98
+ATOM 1108 2HG2 ILE A 63 5.473 8.425 10.685 1.00 0.98
+ATOM 1109 3HG2 ILE A 63 4.085 7.358 10.996 1.00 0.98
+ATOM 1110 1HG1 ILE A 63 2.231 7.240 9.342 1.00 0.98
+ATOM 1111 2HG1 ILE A 63 2.057 8.489 8.119 1.00 0.98
+ATOM 1112 1HD1 ILE A 63 2.541 6.332 7.107 1.00 0.98
+ATOM 1113 2HD1 ILE A 63 3.871 7.476 6.807 1.00 0.98
+ATOM 1114 3HD1 ILE A 63 4.047 6.210 8.045 1.00 0.98
+ATOM 1115 N GLU A 64 5.019 11.408 10.588 1.00 0.97
+ATOM 1116 CA GLU A 64 5.964 12.126 11.435 1.00 0.97
+ATOM 1117 C GLU A 64 5.248 12.851 12.568 1.00 0.97
+ATOM 1118 O GLU A 64 5.682 12.808 13.718 1.00 0.97
+ATOM 1119 CB GLU A 64 6.778 13.122 10.607 1.00 0.97
+ATOM 1120 CG GLU A 64 7.873 12.485 9.764 1.00 0.97
+ATOM 1121 CD GLU A 64 8.650 13.488 8.957 1.00 0.97
+ATOM 1122 OE1 GLU A 64 9.600 13.103 8.320 1.00 0.97
+ATOM 1123 OE2 GLU A 64 8.292 14.643 8.979 1.00 0.97
+ATOM 1124 H GLU A 64 4.973 11.631 9.604 1.00 0.97
+ATOM 1125 HA GLU A 64 6.648 11.404 11.879 1.00 0.97
+ATOM 1126 1HB GLU A 64 6.115 13.669 9.937 1.00 0.97
+ATOM 1127 2HB GLU A 64 7.247 13.850 11.269 1.00 0.97
+ATOM 1128 1HG GLU A 64 8.572 11.969 10.423 1.00 0.97
+ATOM 1129 2HG GLU A 64 7.419 11.775 9.073 1.00 0.97
+ATOM 1130 N GLU A 65 4.121 13.602 12.254 1.00 0.97
+ATOM 1131 CA GLU A 65 3.368 14.302 13.287 1.00 0.97
+ATOM 1132 C GLU A 65 2.836 13.332 14.334 1.00 0.97
+ATOM 1133 O GLU A 65 2.920 13.592 15.535 1.00 0.97
+ATOM 1134 CB GLU A 65 2.219 15.100 12.666 1.00 0.97
+ATOM 1135 CG GLU A 65 2.662 16.322 11.876 1.00 0.97
+ATOM 1136 CD GLU A 65 1.519 17.033 11.206 1.00 0.97
+ATOM 1137 OE1 GLU A 65 1.761 18.008 10.536 1.00 0.97
+ATOM 1138 OE2 GLU A 65 0.402 16.599 11.365 1.00 0.97
+ATOM 1139 H GLU A 65 3.801 13.646 11.297 1.00 0.97
+ATOM 1140 HA GLU A 65 4.036 15.003 13.787 1.00 0.97
+ATOM 1141 1HB GLU A 65 1.648 14.457 11.996 1.00 0.97
+ATOM 1142 2HB GLU A 65 1.542 15.437 13.452 1.00 0.97
+ATOM 1143 1HG GLU A 65 3.146 17.025 12.554 1.00 0.97
+ATOM 1144 2HG GLU A 65 3.363 16.007 11.102 1.00 0.97
+ATOM 1145 N GLU A 66 2.180 12.202 13.899 1.00 0.98
+ATOM 1146 CA GLU A 66 1.669 11.189 14.816 1.00 0.98
+ATOM 1147 C GLU A 66 2.784 10.619 15.683 1.00 0.98
+ATOM 1148 O GLU A 66 2.620 10.455 16.892 1.00 0.98
+ATOM 1149 CB GLU A 66 0.971 10.066 14.047 1.00 0.98
+ATOM 1150 CG GLU A 66 -0.378 10.449 13.455 1.00 0.98
+ATOM 1151 CD GLU A 66 -1.423 10.716 14.503 1.00 0.98
+ATOM 1152 OE1 GLU A 66 -2.282 11.530 14.264 1.00 0.98
+ATOM 1153 OE2 GLU A 66 -1.361 10.105 15.543 1.00 0.98
+ATOM 1154 H GLU A 66 2.121 12.039 12.904 1.00 0.98
+ATOM 1155 HA GLU A 66 0.936 11.657 15.473 1.00 0.98
+ATOM 1156 1HB GLU A 66 1.611 9.732 13.230 1.00 0.98
+ATOM 1157 2HB GLU A 66 0.815 9.215 14.710 1.00 0.98
+ATOM 1158 1HG GLU A 66 -0.258 11.355 12.860 1.00 0.98
+ATOM 1159 2HG GLU A 66 -0.732 9.633 12.825 1.00 0.98
+ATOM 1160 N TRP A 67 3.964 10.256 15.069 1.00 0.98
+ATOM 1161 CA TRP A 67 5.096 9.714 15.810 1.00 0.98
+ATOM 1162 C TRP A 67 5.617 10.717 16.831 1.00 0.98
+ATOM 1163 O TRP A 67 6.016 10.344 17.934 1.00 0.98
+ATOM 1164 CB TRP A 67 6.216 9.296 14.854 1.00 0.98
+ATOM 1165 CG TRP A 67 5.924 8.053 14.069 1.00 0.98
+ATOM 1166 CD1 TRP A 67 6.543 7.649 12.924 1.00 0.98
+ATOM 1167 CD2 TRP A 67 4.933 7.041 14.369 1.00 0.98
+ATOM 1168 NE1 TRP A 67 6.008 6.460 12.492 1.00 0.98
+ATOM 1169 CE2 TRP A 67 5.022 6.074 13.364 1.00 0.98
+ATOM 1170 CE3 TRP A 67 3.993 6.880 15.394 1.00 0.98
+ATOM 1171 CZ2 TRP A 67 4.205 4.954 13.349 1.00 0.98
+ATOM 1172 CZ3 TRP A 67 3.173 5.758 15.379 1.00 0.98
+ATOM 1173 CH2 TRP A 67 3.277 4.820 14.383 1.00 0.98
+ATOM 1174 H TRP A 67 4.059 10.424 14.077 1.00 0.98
+ATOM 1175 HA TRP A 67 4.766 8.822 16.342 1.00 0.98
+ATOM 1176 1HB TRP A 67 6.407 10.108 14.153 1.00 0.98
+ATOM 1177 2HB TRP A 67 7.130 9.134 15.425 1.00 0.98
+ATOM 1178 HD1 TRP A 67 7.344 8.190 12.425 1.00 0.98
+ATOM 1179 HE1 TRP A 67 6.293 5.952 11.667 1.00 0.98
+ATOM 1180 HZ2 TRP A 67 4.272 4.198 12.567 1.00 0.98
+ATOM 1181 HH2 TRP A 67 2.618 3.952 14.401 1.00 0.98
+ATOM 1182 HZ3 TRP A 67 2.443 5.641 16.181 1.00 0.98
+ATOM 1183 HE3 TRP A 67 3.906 7.622 16.188 1.00 0.98
+ATOM 1184 N ARG A 68 5.754 12.078 16.431 1.00 0.97
+ATOM 1185 CA ARG A 68 6.180 13.097 17.382 1.00 0.97
+ATOM 1186 C ARG A 68 5.297 13.096 18.623 1.00 0.97
+ATOM 1187 O ARG A 68 5.785 13.239 19.744 1.00 0.97
+ATOM 1188 CB ARG A 68 6.156 14.477 16.742 1.00 0.97
+ATOM 1189 CG ARG A 68 7.306 14.761 15.788 1.00 0.97
+ATOM 1190 CD ARG A 68 7.225 16.132 15.223 1.00 0.97
+ATOM 1191 NE ARG A 68 8.296 16.391 14.274 1.00 0.97
+ATOM 1192 CZ ARG A 68 8.430 17.528 13.563 1.00 0.97
+ATOM 1193 NH1 ARG A 68 7.554 18.498 13.704 1.00 0.97
+ATOM 1194 NH2 ARG A 68 9.441 17.667 12.723 1.00 0.97
+ATOM 1195 H ARG A 68 5.497 12.356 15.494 1.00 0.97
+ATOM 1196 HA ARG A 68 7.205 12.881 17.686 1.00 0.97
+ATOM 1197 1HB ARG A 68 5.228 14.603 16.185 1.00 0.97
+ATOM 1198 2HB ARG A 68 6.177 15.240 17.520 1.00 0.97
+ATOM 1199 1HG ARG A 68 8.252 14.664 16.321 1.00 0.97
+ATOM 1200 2HG ARG A 68 7.279 14.048 14.963 1.00 0.97
+ATOM 1201 1HD ARG A 68 6.274 16.258 14.706 1.00 0.97
+ATOM 1202 2HD ARG A 68 7.300 16.862 16.028 1.00 0.97
+ATOM 1203 HE ARG A 68 8.990 15.668 14.139 1.00 0.97
+ATOM 1204 1HH1 ARG A 68 6.781 18.392 14.345 1.00 0.97
+ATOM 1205 2HH1 ARG A 68 7.654 19.350 13.170 1.00 0.97
+ATOM 1206 1HH2 ARG A 68 10.114 16.921 12.615 1.00 0.97
+ATOM 1207 2HH2 ARG A 68 9.541 18.518 12.191 1.00 0.97
+ATOM 1208 N ARG A 69 3.914 13.025 18.445 1.00 0.97
+ATOM 1209 CA ARG A 69 2.997 12.950 19.576 1.00 0.97
+ATOM 1210 C ARG A 69 3.292 11.734 20.445 1.00 0.97
+ATOM 1211 O ARG A 69 3.270 11.816 21.673 1.00 0.97
+ATOM 1212 CB ARG A 69 1.553 12.904 19.097 1.00 0.97
+ATOM 1213 CG ARG A 69 1.042 14.206 18.501 1.00 0.97
+ATOM 1214 CD ARG A 69 -0.375 14.093 18.071 1.00 0.97
+ATOM 1215 NE ARG A 69 -0.853 15.319 17.451 1.00 0.97
+ATOM 1216 CZ ARG A 69 -2.087 15.486 16.937 1.00 0.97
+ATOM 1217 NH1 ARG A 69 -2.954 14.499 16.975 1.00 0.97
+ATOM 1218 NH2 ARG A 69 -2.425 16.643 16.394 1.00 0.97
+ATOM 1219 H ARG A 69 3.545 12.954 17.507 1.00 0.97
+ATOM 1220 HA ARG A 69 3.121 13.847 20.182 1.00 0.97
+ATOM 1221 1HB ARG A 69 1.444 12.129 18.339 1.00 0.97
+ATOM 1222 2HB ARG A 69 0.899 12.642 19.928 1.00 0.97
+ATOM 1223 1HG ARG A 69 1.112 14.999 19.246 1.00 0.97
+ATOM 1224 2HG ARG A 69 1.645 14.470 17.631 1.00 0.97
+ATOM 1225 1HD ARG A 69 -0.472 13.285 17.346 1.00 0.97
+ATOM 1226 2HD ARG A 69 -1.002 13.882 18.936 1.00 0.97
+ATOM 1227 HE ARG A 69 -0.213 16.102 17.404 1.00 0.97
+ATOM 1228 1HH1 ARG A 69 -2.695 13.615 17.390 1.00 0.97
+ATOM 1229 2HH1 ARG A 69 -3.879 14.624 16.590 1.00 0.97
+ATOM 1230 1HH2 ARG A 69 -1.758 17.402 16.365 1.00 0.97
+ATOM 1231 2HH2 ARG A 69 -3.350 16.768 16.009 1.00 0.97
+ATOM 1232 N TRP A 70 3.573 10.544 19.802 1.00 0.99
+ATOM 1233 CA TRP A 70 3.889 9.315 20.519 1.00 0.99
+ATOM 1234 C TRP A 70 5.167 9.466 21.335 1.00 0.99
+ATOM 1235 O TRP A 70 5.275 8.938 22.442 1.00 0.99
+ATOM 1236 CB TRP A 70 4.016 8.142 19.546 1.00 0.99
+ATOM 1237 CG TRP A 70 2.707 7.704 18.961 1.00 0.99
+ATOM 1238 CD1 TRP A 70 2.518 6.737 18.019 1.00 0.99
+ATOM 1239 CD2 TRP A 70 1.391 8.217 19.278 1.00 0.99
+ATOM 1240 NE1 TRP A 70 1.182 6.614 17.729 1.00 0.99
+ATOM 1241 CE2 TRP A 70 0.477 7.512 18.491 1.00 0.99
+ATOM 1242 CE3 TRP A 70 0.922 9.204 20.155 1.00 0.99
+ATOM 1243 CZ2 TRP A 70 -0.885 7.759 18.549 1.00 0.99
+ATOM 1244 CZ3 TRP A 70 -0.444 9.452 20.213 1.00 0.99
+ATOM 1245 CH2 TRP A 70 -1.324 8.747 19.430 1.00 0.99
+ATOM 1246 H TRP A 70 3.579 10.540 18.792 1.00 0.99
+ATOM 1247 HA TRP A 70 3.070 9.092 21.203 1.00 0.99
+ATOM 1248 1HB TRP A 70 4.681 8.418 18.728 1.00 0.99
+ATOM 1249 2HB TRP A 70 4.464 7.291 20.058 1.00 0.99
+ATOM 1250 HD1 TRP A 70 3.312 6.149 17.562 1.00 0.99
+ATOM 1251 HE1 TRP A 70 0.781 5.969 17.064 1.00 0.99
+ATOM 1252 HZ2 TRP A 70 -1.600 7.210 17.935 1.00 0.99
+ATOM 1253 HH2 TRP A 70 -2.390 8.967 19.501 1.00 0.99
+ATOM 1254 HZ3 TRP A 70 -0.801 10.221 20.898 1.00 0.99
+ATOM 1255 HE3 TRP A 70 1.616 9.766 20.778 1.00 0.99
+ATOM 1256 N ILE A 71 6.207 10.230 20.800 1.00 0.98
+ATOM 1257 CA ILE A 71 7.459 10.464 21.509 1.00 0.98
+ATOM 1258 C ILE A 71 7.229 11.266 22.784 1.00 0.98
+ATOM 1259 O ILE A 71 7.772 10.942 23.840 1.00 0.98
+ATOM 1260 CB ILE A 71 8.475 11.193 20.610 1.00 0.98
+ATOM 1261 CG1 ILE A 71 8.998 10.250 19.524 1.00 0.98
+ATOM 1262 CG2 ILE A 71 9.624 11.742 21.442 1.00 0.98
+ATOM 1263 CD1 ILE A 71 9.843 10.936 18.475 1.00 0.98
+ATOM 1264 H ILE A 71 6.093 10.602 19.868 1.00 0.98
+ATOM 1265 HA ILE A 71 7.885 9.500 21.786 1.00 0.98
+ATOM 1266 HB ILE A 71 7.981 12.019 20.099 1.00 0.98
+ATOM 1267 1HG2 ILE A 71 10.332 12.253 20.790 1.00 0.98
+ATOM 1268 2HG2 ILE A 71 9.237 12.444 22.178 1.00 0.98
+ATOM 1269 3HG2 ILE A 71 10.128 10.921 21.953 1.00 0.98
+ATOM 1270 1HG1 ILE A 71 9.595 9.463 19.982 1.00 0.98
+ATOM 1271 2HG1 ILE A 71 8.157 9.770 19.022 1.00 0.98
+ATOM 1272 1HD1 ILE A 71 10.177 10.203 17.740 1.00 0.98
+ATOM 1273 2HD1 ILE A 71 9.252 11.706 17.978 1.00 0.98
+ATOM 1274 3HD1 ILE A 71 10.710 11.394 18.949 1.00 0.98
+ATOM 1275 N GLU A 72 6.300 12.344 22.713 1.00 0.98
+ATOM 1276 CA GLU A 72 5.992 13.164 23.879 1.00 0.98
+ATOM 1277 C GLU A 72 5.427 12.319 25.013 1.00 0.98
+ATOM 1278 O GLU A 72 5.614 12.635 26.188 1.00 0.98
+ATOM 1279 CB GLU A 72 5.009 14.276 23.507 1.00 0.98
+ATOM 1280 CG GLU A 72 5.604 15.369 22.630 1.00 0.98
+ATOM 1281 CD GLU A 72 4.601 16.420 22.243 1.00 0.98
+ATOM 1282 OE1 GLU A 72 4.961 17.323 21.528 1.00 0.98
+ATOM 1283 OE2 GLU A 72 3.472 16.319 22.664 1.00 0.98
+ATOM 1284 H GLU A 72 5.843 12.546 21.835 1.00 0.98
+ATOM 1285 HA GLU A 72 6.915 13.625 24.230 1.00 0.98
+ATOM 1286 1HB GLU A 72 4.157 13.849 22.977 1.00 0.98
+ATOM 1287 2HB GLU A 72 4.627 14.745 24.414 1.00 0.98
+ATOM 1288 1HG GLU A 72 6.412 15.858 23.175 1.00 0.98
+ATOM 1289 2HG GLU A 72 5.990 14.918 21.716 1.00 0.98
+ATOM 1290 N GLU A 73 4.731 11.138 24.677 1.00 0.97
+ATOM 1291 CA GLU A 73 4.131 10.227 25.645 1.00 0.97
+ATOM 1292 C GLU A 73 5.116 9.145 26.068 1.00 0.97
+ATOM 1293 O GLU A 73 4.744 8.179 26.734 1.00 0.97
+ATOM 1294 CB GLU A 73 2.859 9.601 25.071 1.00 0.97
+ATOM 1295 CG GLU A 73 1.734 10.592 24.810 1.00 0.97
+ATOM 1296 CD GLU A 73 0.534 9.958 24.163 1.00 0.97
+ATOM 1297 OE1 GLU A 73 -0.413 10.658 23.895 1.00 0.97
+ATOM 1298 OE2 GLU A 73 0.564 8.771 23.937 1.00 0.97
+ATOM 1299 H GLU A 73 4.689 10.908 23.694 1.00 0.97
+ATOM 1300 HA GLU A 73 3.858 10.798 26.533 1.00 0.97
+ATOM 1301 1HB GLU A 73 3.091 9.103 24.130 1.00 0.97
+ATOM 1302 2HB GLU A 73 2.484 8.843 25.758 1.00 0.97
+ATOM 1303 1HG GLU A 73 1.416 11.024 25.759 1.00 0.97
+ATOM 1304 2HG GLU A 73 2.100 11.375 24.146 1.00 0.97
+ATOM 1305 N GLY A 74 6.356 9.091 25.669 1.00 0.98
+ATOM 1306 CA GLY A 74 7.406 8.121 25.955 1.00 0.98
+ATOM 1307 C GLY A 74 7.165 6.814 25.211 1.00 0.98
+ATOM 1308 O GLY A 74 7.720 5.774 25.567 1.00 0.98
+ATOM 1309 H GLY A 74 6.541 9.838 25.015 1.00 0.98
+ATOM 1310 1HA GLY A 74 8.372 8.535 25.667 1.00 0.98
+ATOM 1311 2HA GLY A 74 7.444 7.932 27.027 1.00 0.98
+ATOM 1312 N LYS A 75 6.100 6.604 24.410 1.00 0.98
+ATOM 1313 CA LYS A 75 5.845 5.426 23.590 1.00 0.98
+ATOM 1314 C LYS A 75 6.909 5.264 22.511 1.00 0.98
+ATOM 1315 O LYS A 75 7.056 4.191 21.928 1.00 0.98
+ATOM 1316 CB LYS A 75 4.454 5.498 22.958 1.00 0.98
+ATOM 1317 CG LYS A 75 3.312 5.435 23.964 1.00 0.98
+ATOM 1318 CD LYS A 75 1.960 5.444 23.266 1.00 0.98
+ATOM 1319 CE LYS A 75 0.818 5.381 24.270 1.00 0.98
+ATOM 1320 NZ LYS A 75 -0.511 5.394 23.602 1.00 0.98
+ATOM 1321 H LYS A 75 5.580 7.451 24.229 1.00 0.98
+ATOM 1322 HA LYS A 75 5.883 4.544 24.230 1.00 0.98
+ATOM 1323 1HB LYS A 75 4.354 6.427 22.396 1.00 0.98
+ATOM 1324 2HB LYS A 75 4.326 4.675 22.255 1.00 0.98
+ATOM 1325 1HG LYS A 75 3.400 4.524 24.557 1.00 0.98
+ATOM 1326 2HG LYS A 75 3.371 6.291 24.635 1.00 0.98
+ATOM 1327 1HD LYS A 75 1.862 6.355 22.674 1.00 0.98
+ATOM 1328 2HD LYS A 75 1.891 4.587 22.597 1.00 0.98
+ATOM 1329 1HE LYS A 75 0.823 4.404 24.850 1.00 0.98
+ATOM 1330 2HE LYS A 75 0.798 6.167 24.933 1.00 0.98
+ATOM 1331 1HZ LYS A 75 -1.322 5.284 24.288 1.00 0.98
+ATOM 1332 2HZ LYS A 75 -0.692 6.177 23.052 1.00 0.98
+ATOM 1333 3HZ LYS A 75 -0.669 4.530 22.975 1.00 0.98
+ATOM 1334 N ILE A 76 7.775 6.342 22.321 1.00 0.98
+ATOM 1335 CA ILE A 76 8.761 6.416 21.250 1.00 0.98
+ATOM 1336 C ILE A 76 9.914 7.338 21.626 1.00 0.98
+ATOM 1337 O ILE A 76 9.700 8.450 22.109 1.00 0.98
+ATOM 1338 CB ILE A 76 8.119 6.896 19.935 1.00 0.98
+ATOM 1339 CG1 ILE A 76 7.037 5.941 19.426 1.00 0.98
+ATOM 1340 CG2 ILE A 76 9.204 7.085 18.886 1.00 0.98
+ATOM 1341 CD1 ILE A 76 6.221 6.497 18.281 1.00 0.98
+ATOM 1342 H ILE A 76 7.604 7.158 22.890 1.00 0.98
+ATOM 1343 HA ILE A 76 9.164 5.418 21.081 1.00 0.98
+ATOM 1344 HB ILE A 76 7.633 7.854 20.118 1.00 0.98
+ATOM 1345 1HG2 ILE A 76 8.757 7.457 17.964 1.00 0.98
+ATOM 1346 2HG2 ILE A 76 9.939 7.802 19.248 1.00 0.98
+ATOM 1347 3HG2 ILE A 76 9.693 6.130 18.693 1.00 0.98
+ATOM 1348 1HG1 ILE A 76 7.498 5.011 19.097 1.00 0.98
+ATOM 1349 2HG1 ILE A 76 6.355 5.696 20.241 1.00 0.98
+ATOM 1350 1HD1 ILE A 76 5.475 5.763 17.976 1.00 0.98
+ATOM 1351 2HD1 ILE A 76 5.720 7.412 18.601 1.00 0.98
+ATOM 1352 3HD1 ILE A 76 6.877 6.717 17.441 1.00 0.98
+ATOM 1353 N THR A 77 11.224 6.994 21.705 1.00 0.98
+ATOM 1354 CA THR A 77 12.324 7.941 21.846 1.00 0.98
+ATOM 1355 C THR A 77 12.683 8.572 20.507 1.00 0.98
+ATOM 1356 O THR A 77 12.325 8.053 19.450 1.00 0.98
+ATOM 1357 CB THR A 77 13.572 7.259 22.436 1.00 0.98
+ATOM 1358 OG1 THR A 77 14.109 6.344 21.472 1.00 0.98
+ATOM 1359 CG2 THR A 77 13.223 6.502 23.708 1.00 0.98
+ATOM 1360 H THR A 77 11.432 6.012 21.592 1.00 0.98
+ATOM 1361 HG1 THR A 77 13.452 6.175 20.792 1.00 0.98
+ATOM 1362 HA THR A 77 12.009 8.738 22.520 1.00 0.98
+ATOM 1363 HB THR A 77 14.328 8.011 22.663 1.00 0.98
+ATOM 1364 1HG2 THR A 77 14.118 6.023 24.104 1.00 0.98
+ATOM 1365 2HG2 THR A 77 12.825 7.197 24.447 1.00 0.98
+ATOM 1366 3HG2 THR A 77 12.474 5.743 23.485 1.00 0.98
+ATOM 1367 N LYS A 78 13.142 9.862 20.339 1.00 0.98
+ATOM 1368 CA LYS A 78 13.972 10.378 19.257 1.00 0.98
+ATOM 1369 C LYS A 78 14.863 9.287 18.677 1.00 0.98
+ATOM 1370 O LYS A 78 14.978 9.148 17.459 1.00 0.98
+ATOM 1371 CB LYS A 78 14.817 11.555 19.746 1.00 0.98
+ATOM 1372 CG LYS A 78 15.623 12.255 18.661 1.00 0.98
+ATOM 1373 CD LYS A 78 16.383 13.449 19.220 1.00 0.98
+ATOM 1374 CE LYS A 78 17.234 14.115 18.149 1.00 0.98
+ATOM 1375 NZ LYS A 78 17.978 15.290 18.680 1.00 0.98
+ATOM 1376 H LYS A 78 13.085 10.379 21.205 1.00 0.98
+ATOM 1377 HA LYS A 78 13.318 10.734 18.459 1.00 0.98
+ATOM 1378 1HB LYS A 78 14.168 12.297 20.212 1.00 0.98
+ATOM 1379 2HB LYS A 78 15.514 11.206 20.508 1.00 0.98
+ATOM 1380 1HG LYS A 78 16.335 11.553 18.226 1.00 0.98
+ATOM 1381 2HG LYS A 78 14.952 12.599 17.874 1.00 0.98
+ATOM 1382 1HD LYS A 78 15.676 14.178 19.617 1.00 0.98
+ATOM 1383 2HD LYS A 78 17.032 13.119 20.032 1.00 0.98
+ATOM 1384 1HE LYS A 78 18.051 13.423 17.770 1.00 0.98
+ATOM 1385 2HE LYS A 78 16.696 14.473 17.348 1.00 0.98
+ATOM 1386 1HZ LYS A 78 18.630 15.730 17.957 1.00 0.98
+ATOM 1387 2HZ LYS A 78 17.424 15.999 19.050 1.00 0.98
+ATOM 1388 3HZ LYS A 78 18.690 15.019 19.444 1.00 0.98
+ATOM 1389 N GLU A 79 15.517 8.453 19.098 1.00 0.97
+ATOM 1390 CA GLU A 79 16.242 7.364 18.454 1.00 0.97
+ATOM 1391 C GLU A 79 15.284 6.335 17.868 1.00 0.97
+ATOM 1392 O GLU A 79 15.523 5.793 16.789 1.00 0.97
+ATOM 1393 CB GLU A 79 17.199 6.696 19.444 1.00 0.97
+ATOM 1394 CG GLU A 79 18.336 7.590 19.919 1.00 0.97
+ATOM 1395 CD GLU A 79 19.216 6.925 20.940 1.00 0.97
+ATOM 1396 OE1 GLU A 79 20.143 7.551 21.395 1.00 0.97
+ATOM 1397 OE2 GLU A 79 18.961 5.790 21.265 1.00 0.97
+ATOM 1398 H GLU A 79 15.522 8.518 20.106 1.00 0.97
+ATOM 1399 HA GLU A 79 16.832 7.778 17.636 1.00 0.97
+ATOM 1400 1HB GLU A 79 16.643 6.367 20.322 1.00 0.97
+ATOM 1401 2HB GLU A 79 17.639 5.811 18.985 1.00 0.97
+ATOM 1402 1HG GLU A 79 18.955 7.859 19.063 1.00 0.97
+ATOM 1403 2HG GLU A 79 17.914 8.488 20.371 1.00 0.97
+ATOM 1404 N GLU A 80 14.158 5.951 18.599 1.00 0.98
+ATOM 1405 CA GLU A 80 13.157 5.038 18.060 1.00 0.98
+ATOM 1406 C GLU A 80 12.494 5.618 16.817 1.00 0.98
+ATOM 1407 O GLU A 80 12.168 4.890 15.880 1.00 0.98
+ATOM 1408 CB GLU A 80 12.108 4.703 19.123 1.00 0.98
+ATOM 1409 CG GLU A 80 12.633 3.856 20.273 1.00 0.98
+ATOM 1410 CD GLU A 80 11.627 3.677 21.376 1.00 0.98
+ATOM 1411 OE1 GLU A 80 11.761 2.745 22.132 1.00 0.98
+ATOM 1412 OE2 GLU A 80 10.722 4.474 21.462 1.00 0.98
+ATOM 1413 H GLU A 80 14.011 6.359 19.511 1.00 0.98
+ATOM 1414 HA GLU A 80 13.653 4.110 17.775 1.00 0.98
+ATOM 1415 1HB GLU A 80 11.706 5.625 19.543 1.00 0.98
+ATOM 1416 2HB GLU A 80 11.279 4.165 18.663 1.00 0.98
+ATOM 1417 1HG GLU A 80 12.897 2.869 19.893 1.00 0.98
+ATOM 1418 2HG GLU A 80 13.514 4.341 20.695 1.00 0.98
+ATOM 1419 N TRP A 81 12.042 6.976 16.915 1.00 0.98
+ATOM 1420 CA TRP A 81 11.478 7.612 15.730 1.00 0.98
+ATOM 1421 C TRP A 81 12.379 7.414 14.518 1.00 0.98
+ATOM 1422 O TRP A 81 11.910 7.065 13.434 1.00 0.98
+ATOM 1423 CB TRP A 81 11.261 9.106 15.976 1.00 0.98
+ATOM 1424 CG TRP A 81 10.721 9.844 14.788 1.00 0.98
+ATOM 1425 CD1 TRP A 81 9.422 9.892 14.380 1.00 0.98
+ATOM 1426 CD2 TRP A 81 11.469 10.647 13.843 1.00 0.98
+ATOM 1427 NE1 TRP A 81 9.309 10.667 13.252 1.00 0.98
+ATOM 1428 CE2 TRP A 81 10.552 11.137 12.909 1.00 0.98
+ATOM 1429 CE3 TRP A 81 12.822 10.985 13.718 1.00 0.98
+ATOM 1430 CZ2 TRP A 81 10.941 11.951 11.857 1.00 0.98
+ATOM 1431 CZ3 TRP A 81 13.212 11.801 12.662 1.00 0.98
+ATOM 1432 CH2 TRP A 81 12.295 12.271 11.756 1.00 0.98
+ATOM 1433 H TRP A 81 12.211 7.535 17.739 1.00 0.98
+ATOM 1434 HA TRP A 81 10.512 7.154 15.519 1.00 0.98
+ATOM 1435 1HB TRP A 81 10.566 9.238 16.806 1.00 0.98
+ATOM 1436 2HB TRP A 81 12.206 9.565 16.264 1.00 0.98
+ATOM 1437 HD1 TRP A 81 8.594 9.389 14.876 1.00 0.98
+ATOM 1438 HE1 TRP A 81 8.452 10.860 12.754 1.00 0.98
+ATOM 1439 HZ2 TRP A 81 10.227 12.334 11.127 1.00 0.98
+ATOM 1440 HH2 TRP A 81 12.635 12.909 10.940 1.00 0.98
+ATOM 1441 HZ3 TRP A 81 14.267 12.060 12.572 1.00 0.98
+ATOM 1442 HE3 TRP A 81 13.555 10.615 14.434 1.00 0.98
+ATOM 1443 N GLU A 82 13.713 7.717 14.664 1.00 0.97
+ATOM 1444 CA GLU A 82 14.667 7.571 13.572 1.00 0.97
+ATOM 1445 C GLU A 82 14.697 6.138 13.054 1.00 0.97
+ATOM 1446 O GLU A 82 14.801 5.905 11.850 1.00 0.97
+ATOM 1447 CB GLU A 82 16.064 8.013 14.015 1.00 0.97
+ATOM 1448 CG GLU A 82 16.207 9.513 14.228 1.00 0.97
+ATOM 1449 CD GLU A 82 17.568 9.905 14.733 1.00 0.97
+ATOM 1450 OE1 GLU A 82 17.786 11.073 14.950 1.00 0.97
+ATOM 1451 OE2 GLU A 82 18.390 9.035 14.901 1.00 0.97
+ATOM 1452 H GLU A 82 14.047 7.999 15.575 1.00 0.97
+ATOM 1453 HA GLU A 82 14.356 8.218 12.752 1.00 0.97
+ATOM 1454 1HB GLU A 82 16.324 7.516 14.950 1.00 0.97
+ATOM 1455 2HB GLU A 82 16.797 7.709 13.268 1.00 0.97
+ATOM 1456 1HG GLU A 82 16.040 10.022 13.278 1.00 0.97
+ATOM 1457 2HG GLU A 82 15.468 9.838 14.961 1.00 0.97
+ATOM 1458 N ARG A 83 14.548 5.106 13.968 1.00 0.98
+ATOM 1459 CA ARG A 83 14.503 3.704 13.571 1.00 0.98
+ATOM 1460 C ARG A 83 13.245 3.399 12.768 1.00 0.98
+ATOM 1461 O ARG A 83 13.294 2.685 11.766 1.00 0.98
+ATOM 1462 CB ARG A 83 14.573 2.796 14.790 1.00 0.98
+ATOM 1463 CG ARG A 83 15.934 2.743 15.467 1.00 0.98
+ATOM 1464 CD ARG A 83 15.926 1.840 16.646 1.00 0.98
+ATOM 1465 NE ARG A 83 17.209 1.829 17.329 1.00 0.98
+ATOM 1466 CZ ARG A 83 17.472 1.134 18.454 1.00 0.98
+ATOM 1467 NH1 ARG A 83 16.532 0.401 19.007 1.00 0.98
+ATOM 1468 NH2 ARG A 83 18.674 1.191 19.000 1.00 0.98
+ATOM 1469 H ARG A 83 14.490 5.342 14.949 1.00 0.98
+ATOM 1470 HA ARG A 83 15.372 3.493 12.947 1.00 0.98
+ATOM 1471 1HB ARG A 83 13.848 3.127 15.533 1.00 0.98
+ATOM 1472 2HB ARG A 83 14.307 1.779 14.504 1.00 0.98
+ATOM 1473 1HG ARG A 83 16.678 2.377 14.759 1.00 0.98
+ATOM 1474 2HG ARG A 83 16.213 3.743 15.802 1.00 0.98
+ATOM 1475 1HD ARG A 83 15.166 2.171 17.353 1.00 0.98
+ATOM 1476 2HD ARG A 83 15.703 0.823 16.324 1.00 0.98
+ATOM 1477 HE ARG A 83 17.958 2.382 16.933 1.00 0.98
+ATOM 1478 1HH1 ARG A 83 15.613 0.358 18.590 1.00 0.98
+ATOM 1479 2HH1 ARG A 83 16.728 -0.120 19.850 1.00 0.98
+ATOM 1480 1HH2 ARG A 83 19.396 1.755 18.574 1.00 0.98
+ATOM 1481 2HH2 ARG A 83 18.870 0.670 19.842 1.00 0.98
+ATOM 1482 N LEU A 84 12.020 3.872 13.280 1.00 0.98
+ATOM 1483 CA LEU A 84 10.779 3.730 12.528 1.00 0.98
+ATOM 1484 C LEU A 84 10.902 4.344 11.138 1.00 0.98
+ATOM 1485 O LEU A 84 10.470 3.755 10.148 1.00 0.98
+ATOM 1486 CB LEU A 84 9.605 4.343 13.302 1.00 0.98
+ATOM 1487 CG LEU A 84 8.181 4.163 12.759 1.00 0.98
+ATOM 1488 CD1 LEU A 84 7.791 2.693 12.832 1.00 0.98
+ATOM 1489 CD2 LEU A 84 7.219 5.025 13.563 1.00 0.98
+ATOM 1490 H LEU A 84 12.017 4.407 14.138 1.00 0.98
+ATOM 1491 HA LEU A 84 10.563 2.668 12.416 1.00 0.98
+ATOM 1492 1HB LEU A 84 9.632 3.921 14.305 1.00 0.98
+ATOM 1493 2HB LEU A 84 9.799 5.413 13.374 1.00 0.98
+ATOM 1494 HG LEU A 84 8.150 4.465 11.712 1.00 0.98
+ATOM 1495 1HD1 LEU A 84 6.780 2.566 12.446 1.00 0.98
+ATOM 1496 2HD1 LEU A 84 8.484 2.103 12.232 1.00 0.98
+ATOM 1497 3HD1 LEU A 84 7.829 2.358 13.867 1.00 0.98
+ATOM 1498 1HD2 LEU A 84 6.207 4.898 13.177 1.00 0.98
+ATOM 1499 2HD2 LEU A 84 7.248 4.723 14.610 1.00 0.98
+ATOM 1500 3HD2 LEU A 84 7.510 6.072 13.479 1.00 0.98
+ATOM 1501 N TRP A 85 11.427 5.630 11.054 1.00 0.97
+ATOM 1502 CA TRP A 85 11.644 6.296 9.775 1.00 0.97
+ATOM 1503 C TRP A 85 12.540 5.464 8.866 1.00 0.97
+ATOM 1504 O TRP A 85 12.300 5.368 7.662 1.00 0.97
+ATOM 1505 CB TRP A 85 12.241 7.687 9.997 1.00 0.97
+ATOM 1506 CG TRP A 85 12.274 8.560 8.780 1.00 0.97
+ATOM 1507 CD1 TRP A 85 12.673 9.861 8.721 1.00 0.97
+ATOM 1508 CD2 TRP A 85 11.892 8.196 7.431 1.00 0.97
+ATOM 1509 NE1 TRP A 85 12.565 10.331 7.435 1.00 0.97
+ATOM 1510 CE2 TRP A 85 12.089 9.325 6.633 1.00 0.97
+ATOM 1511 CE3 TRP A 85 11.405 7.020 6.847 1.00 0.97
+ATOM 1512 CZ2 TRP A 85 11.817 9.319 5.274 1.00 0.97
+ATOM 1513 CZ3 TRP A 85 11.132 7.014 5.484 1.00 0.97
+ATOM 1514 CH2 TRP A 85 11.333 8.135 4.718 1.00 0.97
+ATOM 1515 H TRP A 85 11.729 6.086 11.903 1.00 0.97
+ATOM 1516 HA TRP A 85 10.680 6.424 9.284 1.00 0.97
+ATOM 1517 1HB TRP A 85 11.665 8.197 10.769 1.00 0.97
+ATOM 1518 2HB TRP A 85 13.260 7.577 10.368 1.00 0.97
+ATOM 1519 HD1 TRP A 85 13.025 10.444 9.570 1.00 0.97
+ATOM 1520 HE1 TRP A 85 12.799 11.265 7.128 1.00 0.97
+ATOM 1521 HZ2 TRP A 85 11.970 10.199 4.648 1.00 0.97
+ATOM 1522 HH2 TRP A 85 11.109 8.095 3.652 1.00 0.97
+ATOM 1523 HZ3 TRP A 85 10.754 6.094 5.037 1.00 0.97
+ATOM 1524 HE3 TRP A 85 11.243 6.127 7.450 1.00 0.97
+ATOM 1525 N GLU A 86 13.660 4.875 9.422 1.00 0.97
+ATOM 1526 CA GLU A 86 14.542 3.999 8.660 1.00 0.97
+ATOM 1527 C GLU A 86 13.788 2.789 8.124 1.00 0.97
+ATOM 1528 O GLU A 86 13.998 2.369 6.986 1.00 0.97
+ATOM 1529 CB GLU A 86 15.727 3.551 9.519 1.00 0.97
+ATOM 1530 CG GLU A 86 16.775 4.631 9.749 1.00 0.97
+ATOM 1531 CD GLU A 86 17.893 4.179 10.646 1.00 0.97
+ATOM 1532 OE1 GLU A 86 18.764 4.968 10.922 1.00 0.97
+ATOM 1533 OE2 GLU A 86 17.876 3.042 11.056 1.00 0.97
+ATOM 1534 H GLU A 86 13.854 5.040 10.399 1.00 0.97
+ATOM 1535 HA GLU A 86 14.931 4.557 7.808 1.00 0.97
+ATOM 1536 1HB GLU A 86 15.367 3.221 10.494 1.00 0.97
+ATOM 1537 2HB GLU A 86 16.220 2.702 9.047 1.00 0.97
+ATOM 1538 1HG GLU A 86 17.206 4.915 8.789 1.00 0.97
+ATOM 1539 2HG GLU A 86 16.297 5.494 10.213 1.00 0.97
+ATOM 1540 N LYS A 87 12.856 2.189 8.956 1.00 0.97
+ATOM 1541 CA LYS A 87 12.027 1.072 8.520 1.00 0.97
+ATOM 1542 C LYS A 87 11.126 1.475 7.359 1.00 0.97
+ATOM 1543 O LYS A 87 10.955 0.720 6.402 1.00 0.97
+ATOM 1544 CB LYS A 87 11.195 0.531 9.683 1.00 0.97
+ATOM 1545 CG LYS A 87 12.008 -0.205 10.740 1.00 0.97
+ATOM 1546 CD LYS A 87 11.115 -0.744 11.848 1.00 0.97
+ATOM 1547 CE LYS A 87 11.928 -1.459 12.917 1.00 0.97
+ATOM 1548 NZ LYS A 87 11.072 -1.959 14.026 1.00 0.97
+ATOM 1549 H LYS A 87 12.747 2.545 9.894 1.00 0.97
+ATOM 1550 HA LYS A 87 12.680 0.271 8.172 1.00 0.97
+ATOM 1551 1HB LYS A 87 10.674 1.354 10.175 1.00 0.97
+ATOM 1552 2HB LYS A 87 10.437 -0.155 9.305 1.00 0.97
+ATOM 1553 1HG LYS A 87 12.539 -1.037 10.276 1.00 0.97
+ATOM 1554 2HG LYS A 87 12.741 0.474 11.173 1.00 0.97
+ATOM 1555 1HD LYS A 87 10.569 0.080 12.309 1.00 0.97
+ATOM 1556 2HD LYS A 87 10.393 -1.444 11.426 1.00 0.97
+ATOM 1557 1HE LYS A 87 12.424 -2.397 12.510 1.00 0.97
+ATOM 1558 2HE LYS A 87 12.641 -0.869 13.367 1.00 0.97
+ATOM 1559 1HZ LYS A 87 11.619 -2.520 14.752 1.00 0.97
+ATOM 1560 2HZ LYS A 87 10.565 -1.276 14.499 1.00 0.97
+ATOM 1561 3HZ LYS A 87 10.363 -2.704 13.699 1.00 0.97
+ATOM 1562 N LEU A 88 10.448 2.700 7.468 1.00 0.98
+ATOM 1563 CA LEU A 88 9.626 3.203 6.375 1.00 0.98
+ATOM 1564 C LEU A 88 10.454 3.409 5.111 1.00 0.98
+ATOM 1565 O LEU A 88 10.040 3.024 4.017 1.00 0.98
+ATOM 1566 CB LEU A 88 8.935 4.510 6.785 1.00 0.98
+ATOM 1567 CG LEU A 88 7.956 5.129 5.779 1.00 0.98
+ATOM 1568 CD1 LEU A 88 6.822 4.149 5.505 1.00 0.98
+ATOM 1569 CD2 LEU A 88 7.422 6.442 6.331 1.00 0.98
+ATOM 1570 H LEU A 88 10.565 3.265 8.297 1.00 0.98
+ATOM 1571 HA LEU A 88 8.849 2.470 6.161 1.00 0.98
+ATOM 1572 1HB LEU A 88 8.388 4.324 7.708 1.00 0.98
+ATOM 1573 2HB LEU A 88 9.710 5.248 6.989 1.00 0.98
+ATOM 1574 HG LEU A 88 8.471 5.314 4.836 1.00 0.98
+ATOM 1575 1HD1 LEU A 88 6.127 4.589 4.790 1.00 0.98
+ATOM 1576 2HD1 LEU A 88 7.230 3.226 5.093 1.00 0.98
+ATOM 1577 3HD1 LEU A 88 6.298 3.931 6.435 1.00 0.98
+ATOM 1578 1HD2 LEU A 88 6.727 6.882 5.616 1.00 0.98
+ATOM 1579 2HD2 LEU A 88 6.906 6.258 7.273 1.00 0.98
+ATOM 1580 3HD2 LEU A 88 8.251 7.129 6.501 1.00 0.98
+ATOM 1581 N LYS A 89 11.666 4.088 5.230 1.00 0.97
+ATOM 1582 CA LYS A 89 12.546 4.307 4.089 1.00 0.97
+ATOM 1583 C LYS A 89 12.960 2.986 3.452 1.00 0.97
+ATOM 1584 O LYS A 89 13.018 2.867 2.228 1.00 0.97
+ATOM 1585 CB LYS A 89 13.780 5.109 4.505 1.00 0.97
+ATOM 1586 CG LYS A 89 13.508 6.586 4.759 1.00 0.97
+ATOM 1587 CD LYS A 89 14.781 7.324 5.145 1.00 0.97
+ATOM 1588 CE LYS A 89 14.509 8.798 5.408 1.00 0.97
+ATOM 1589 NZ LYS A 89 15.738 9.524 5.829 1.00 0.97
+ATOM 1590 H LYS A 89 11.960 4.398 6.145 1.00 0.97
+ATOM 1591 HA LYS A 89 12.004 4.883 3.338 1.00 0.97
+ATOM 1592 1HB LYS A 89 14.201 4.685 5.417 1.00 0.97
+ATOM 1593 2HB LYS A 89 14.542 5.038 3.729 1.00 0.97
+ATOM 1594 1HG LYS A 89 13.095 7.040 3.857 1.00 0.97
+ATOM 1595 2HG LYS A 89 12.780 6.689 5.562 1.00 0.97
+ATOM 1596 1HD LYS A 89 15.203 6.875 6.045 1.00 0.97
+ATOM 1597 2HD LYS A 89 15.511 7.237 4.341 1.00 0.97
+ATOM 1598 1HE LYS A 89 14.182 9.339 4.465 1.00 0.97
+ATOM 1599 2HE LYS A 89 13.822 8.972 6.153 1.00 0.97
+ATOM 1600 1HZ LYS A 89 15.579 10.572 5.954 1.00 0.97
+ATOM 1601 2HZ LYS A 89 16.162 9.189 6.639 1.00 0.97
+ATOM 1602 3HZ LYS A 89 16.498 9.534 5.062 1.00 0.97
+ATOM 1603 N GLU A 90 13.393 1.971 4.281 1.00 0.97
+ATOM 1604 CA GLU A 90 13.736 0.644 3.785 1.00 0.97
+ATOM 1605 C GLU A 90 12.580 0.033 3.003 1.00 0.97
+ATOM 1606 O GLU A 90 12.783 -0.572 1.950 1.00 0.97
+ATOM 1607 CB GLU A 90 14.142 -0.273 4.942 1.00 0.97
+ATOM 1608 CG GLU A 90 15.509 0.035 5.536 1.00 0.97
+ATOM 1609 CD GLU A 90 15.823 -0.802 6.745 1.00 0.97
+ATOM 1610 OE1 GLU A 90 16.877 -0.626 7.308 1.00 0.97
+ATOM 1611 OE2 GLU A 90 15.008 -1.618 7.106 1.00 0.97
+ATOM 1612 H GLU A 90 13.388 2.138 5.278 1.00 0.97
+ATOM 1613 HA GLU A 90 14.587 0.736 3.111 1.00 0.97
+ATOM 1614 1HB GLU A 90 13.405 -0.198 5.742 1.00 0.97
+ATOM 1615 2HB GLU A 90 14.151 -1.308 4.600 1.00 0.97
+ATOM 1616 1HG GLU A 90 16.273 -0.160 4.783 1.00 0.97
+ATOM 1617 2HG GLU A 90 15.537 1.083 5.834 1.00 0.97
+ATOM 1618 N TRP A 91 11.321 0.171 3.537 1.00 0.98
+ATOM 1619 CA TRP A 91 10.127 -0.284 2.835 1.00 0.98
+ATOM 1620 C TRP A 91 9.992 0.397 1.479 1.00 0.98
+ATOM 1621 O TRP A 91 9.737 -0.258 0.468 1.00 0.98
+ATOM 1622 CB TRP A 91 8.878 -0.032 3.682 1.00 0.98
+ATOM 1623 CG TRP A 91 7.598 -0.448 3.023 1.00 0.98
+ATOM 1624 CD1 TRP A 91 7.053 -1.697 3.016 1.00 0.98
+ATOM 1625 CD2 TRP A 91 6.691 0.389 2.266 1.00 0.98
+ATOM 1626 NE1 TRP A 91 5.876 -1.695 2.311 1.00 0.98
+ATOM 1627 CE2 TRP A 91 5.639 -0.427 1.844 1.00 0.98
+ATOM 1628 CE3 TRP A 91 6.689 1.746 1.917 1.00 0.98
+ATOM 1629 CZ2 TRP A 91 4.586 0.065 1.088 1.00 0.98
+ATOM 1630 CZ3 TRP A 91 5.634 2.239 1.159 1.00 0.98
+ATOM 1631 CH2 TRP A 91 4.610 1.419 0.756 1.00 0.98
+ATOM 1632 H TRP A 91 11.220 0.630 4.432 1.00 0.98
+ATOM 1633 HA TRP A 91 10.210 -1.359 2.674 1.00 0.98
+ATOM 1634 1HB TRP A 91 8.969 -0.571 4.625 1.00 0.98
+ATOM 1635 2HB TRP A 91 8.813 1.030 3.918 1.00 0.98
+ATOM 1636 HD1 TRP A 91 7.489 -2.569 3.501 1.00 0.98
+ATOM 1637 HE1 TRP A 91 5.279 -2.496 2.159 1.00 0.98
+ATOM 1638 HZ2 TRP A 91 3.763 -0.570 0.759 1.00 0.98
+ATOM 1639 HH2 TRP A 91 3.797 1.839 0.162 1.00 0.98
+ATOM 1640 HZ3 TRP A 91 5.640 3.296 0.892 1.00 0.98
+ATOM 1641 HE3 TRP A 91 7.500 2.401 2.234 1.00 0.98
+ATOM 1642 N LEU A 92 10.149 1.795 1.451 1.00 0.97
+ATOM 1643 CA LEU A 92 10.040 2.540 0.203 1.00 0.97
+ATOM 1644 C LEU A 92 11.072 2.066 -0.815 1.00 0.97
+ATOM 1645 O LEU A 92 10.757 1.874 -1.989 1.00 0.97
+ATOM 1646 CB LEU A 92 10.199 4.046 0.451 1.00 0.97
+ATOM 1647 CG LEU A 92 8.968 4.821 0.936 1.00 0.97
+ATOM 1648 CD1 LEU A 92 9.379 6.233 1.330 1.00 0.97
+ATOM 1649 CD2 LEU A 92 7.917 4.845 -0.164 1.00 0.97
+ATOM 1650 H LEU A 92 10.360 2.292 2.304 1.00 0.97
+ATOM 1651 HA LEU A 92 9.047 2.375 -0.213 1.00 0.97
+ATOM 1652 1HB LEU A 92 10.985 4.175 1.193 1.00 0.97
+ATOM 1653 2HB LEU A 92 10.531 4.500 -0.483 1.00 0.97
+ATOM 1654 HG LEU A 92 8.556 4.335 1.821 1.00 0.97
+ATOM 1655 1HD1 LEU A 92 8.504 6.784 1.674 1.00 0.97
+ATOM 1656 2HD1 LEU A 92 10.116 6.187 2.131 1.00 0.97
+ATOM 1657 3HD1 LEU A 92 9.810 6.740 0.468 1.00 0.97
+ATOM 1658 1HD2 LEU A 92 7.041 5.395 0.181 1.00 0.97
+ATOM 1659 2HD2 LEU A 92 8.327 5.332 -1.049 1.00 0.97
+ATOM 1660 3HD2 LEU A 92 7.628 3.823 -0.414 1.00 0.97
+ATOM 1661 N GLU A 93 12.374 1.887 -0.377 1.00 0.96
+ATOM 1662 CA GLU A 93 13.435 1.402 -1.252 1.00 0.96
+ATOM 1663 C GLU A 93 13.090 0.035 -1.830 1.00 0.96
+ATOM 1664 O GLU A 93 13.283 -0.213 -3.020 1.00 0.96
+ATOM 1665 CB GLU A 93 14.766 1.339 -0.500 1.00 0.96
+ATOM 1666 CG GLU A 93 15.399 2.697 -0.232 1.00 0.96
+ATOM 1667 CD GLU A 93 16.685 2.601 0.540 1.00 0.96
+ATOM 1668 OE1 GLU A 93 17.265 3.622 0.821 1.00 0.96
+ATOM 1669 OE2 GLU A 93 17.089 1.504 0.849 1.00 0.96
+ATOM 1670 H GLU A 93 12.579 2.054 0.598 1.00 0.96
+ATOM 1671 HA GLU A 93 13.544 2.100 -2.082 1.00 0.96
+ATOM 1672 1HB GLU A 93 14.619 0.843 0.459 1.00 0.96
+ATOM 1673 2HB GLU A 93 15.479 0.745 -1.070 1.00 0.96
+ATOM 1674 1HG GLU A 93 15.612 3.182 -1.185 1.00 0.96
+ATOM 1675 2HG GLU A 93 14.702 3.303 0.348 1.00 0.96
+ATOM 1676 N LYS A 94 12.606 -0.937 -0.963 1.00 0.96
+ATOM 1677 CA LYS A 94 12.175 -2.256 -1.412 1.00 0.96
+ATOM 1678 C LYS A 94 11.070 -2.148 -2.455 1.00 0.96
+ATOM 1679 O LYS A 94 11.080 -2.858 -3.460 1.00 0.96
+ATOM 1680 CB LYS A 94 11.712 -3.105 -0.228 1.00 0.96
+ATOM 1681 CG LYS A 94 12.840 -3.591 0.673 1.00 0.96
+ATOM 1682 CD LYS A 94 12.305 -4.431 1.824 1.00 0.96
+ATOM 1683 CE LYS A 94 13.425 -4.880 2.750 1.00 0.96
+ATOM 1684 NZ LYS A 94 12.912 -5.667 3.904 1.00 0.96
+ATOM 1685 H LYS A 94 12.495 -0.688 0.009 1.00 0.96
+ATOM 1686 HA LYS A 94 13.025 -2.758 -1.877 1.00 0.96
+ATOM 1687 1HB LYS A 94 11.019 -2.529 0.386 1.00 0.96
+ATOM 1688 2HB LYS A 94 11.176 -3.980 -0.594 1.00 0.96
+ATOM 1689 1HG LYS A 94 13.539 -4.192 0.090 1.00 0.96
+ATOM 1690 2HG LYS A 94 13.376 -2.734 1.079 1.00 0.96
+ATOM 1691 1HD LYS A 94 11.583 -3.847 2.396 1.00 0.96
+ATOM 1692 2HD LYS A 94 11.800 -5.312 1.427 1.00 0.96
+ATOM 1693 1HE LYS A 94 14.140 -5.593 2.229 1.00 0.96
+ATOM 1694 2HE LYS A 94 13.963 -4.107 3.165 1.00 0.96
+ATOM 1695 1HZ LYS A 94 13.691 -6.045 4.528 1.00 0.96
+ATOM 1696 2HZ LYS A 94 12.275 -5.201 4.474 1.00 0.96
+ATOM 1697 3HZ LYS A 94 12.442 -6.590 3.600 1.00 0.96
+ATOM 1698 N TRP A 95 10.070 -1.221 -2.212 1.00 0.97
+ATOM 1699 CA TRP A 95 8.965 -1.005 -3.139 1.00 0.97
+ATOM 1700 C TRP A 95 9.465 -0.482 -4.480 1.00 0.97
+ATOM 1701 O TRP A 95 9.059 -0.967 -5.536 1.00 0.97
+ATOM 1702 CB TRP A 95 7.942 -0.041 -2.535 1.00 0.97
+ATOM 1703 CG TRP A 95 6.717 0.158 -3.376 1.00 0.97
+ATOM 1704 CD1 TRP A 95 5.646 -0.679 -3.467 1.00 0.97
+ATOM 1705 CD2 TRP A 95 6.431 1.273 -4.254 1.00 0.97
+ATOM 1706 NE1 TRP A 95 4.716 -0.165 -4.337 1.00 0.97
+ATOM 1707 CE2 TRP A 95 5.180 1.029 -4.828 1.00 0.97
+ATOM 1708 CE3 TRP A 95 7.125 2.441 -4.594 1.00 0.97
+ATOM 1709 CZ2 TRP A 95 4.601 1.910 -5.728 1.00 0.97
+ATOM 1710 CZ3 TRP A 95 6.545 3.324 -5.497 1.00 0.97
+ATOM 1711 CH2 TRP A 95 5.315 3.065 -6.048 1.00 0.97
+ATOM 1712 H TRP A 95 10.120 -0.667 -1.369 1.00 0.97
+ATOM 1713 HA TRP A 95 8.463 -1.958 -3.307 1.00 0.97
+ATOM 1714 1HB TRP A 95 7.630 -0.415 -1.559 1.00 0.97
+ATOM 1715 2HB TRP A 95 8.412 0.929 -2.379 1.00 0.97
+ATOM 1716 HD1 TRP A 95 5.542 -1.619 -2.928 1.00 0.97
+ATOM 1717 HE1 TRP A 95 3.835 -0.595 -4.577 1.00 0.97
+ATOM 1718 HZ2 TRP A 95 3.626 1.722 -6.177 1.00 0.97
+ATOM 1719 HH2 TRP A 95 4.889 3.780 -6.753 1.00 0.97
+ATOM 1720 HZ3 TRP A 95 7.092 4.231 -5.756 1.00 0.97
+ATOM 1721 HE3 TRP A 95 8.101 2.651 -4.159 1.00 0.97
+ATOM 1722 N LEU A 96 10.325 0.623 -4.470 1.00 0.95
+ATOM 1723 CA LEU A 96 10.857 1.201 -5.698 1.00 0.95
+ATOM 1724 C LEU A 96 11.635 0.166 -6.503 1.00 0.95
+ATOM 1725 O LEU A 96 11.573 0.146 -7.732 1.00 0.95
+ATOM 1726 CB LEU A 96 11.742 2.413 -5.380 1.00 0.95
+ATOM 1727 CG LEU A 96 11.017 3.675 -4.896 1.00 0.95
+ATOM 1728 CD1 LEU A 96 12.042 4.704 -4.435 1.00 0.95
+ATOM 1729 CD2 LEU A 96 10.154 4.228 -6.020 1.00 0.95
+ATOM 1730 H LEU A 96 10.639 0.985 -3.580 1.00 0.95
+ATOM 1731 HA LEU A 96 10.022 1.551 -6.305 1.00 0.95
+ATOM 1732 1HB LEU A 96 12.454 2.123 -4.609 1.00 0.95
+ATOM 1733 2HB LEU A 96 12.299 2.675 -6.280 1.00 0.95
+ATOM 1734 HG LEU A 96 10.387 3.428 -4.042 1.00 0.95
+ATOM 1735 1HD1 LEU A 96 11.527 5.601 -4.091 1.00 0.95
+ATOM 1736 2HD1 LEU A 96 12.632 4.289 -3.618 1.00 0.95
+ATOM 1737 3HD1 LEU A 96 12.699 4.959 -5.266 1.00 0.95
+ATOM 1738 1HD2 LEU A 96 9.639 5.125 -5.675 1.00 0.95
+ATOM 1739 2HD2 LEU A 96 10.784 4.478 -6.874 1.00 0.95
+ATOM 1740 3HD2 LEU A 96 9.420 3.480 -6.317 1.00 0.95
+ATOM 1741 N GLU A 97 12.438 -0.737 -5.798 1.00 0.95
+ATOM 1742 CA GLU A 97 13.167 -1.824 -6.441 1.00 0.95
+ATOM 1743 C GLU A 97 12.215 -2.799 -7.122 1.00 0.95
+ATOM 1744 O GLU A 97 12.487 -3.283 -8.220 1.00 0.95
+ATOM 1745 CB GLU A 97 14.046 -2.557 -5.425 1.00 0.95
+ATOM 1746 CG GLU A 97 15.275 -1.776 -4.981 1.00 0.95
+ATOM 1747 CD GLU A 97 16.075 -2.494 -3.930 1.00 0.95
+ATOM 1748 OE1 GLU A 97 17.068 -1.959 -3.499 1.00 0.95
+ATOM 1749 OE2 GLU A 97 15.693 -3.578 -3.557 1.00 0.95
+ATOM 1750 H GLU A 97 12.479 -0.644 -4.793 1.00 0.95
+ATOM 1751 HA GLU A 97 13.817 -1.399 -7.206 1.00 0.95
+ATOM 1752 1HB GLU A 97 13.459 -2.791 -4.537 1.00 0.95
+ATOM 1753 2HB GLU A 97 14.385 -3.501 -5.851 1.00 0.95
+ATOM 1754 1HG GLU A 97 15.920 -1.614 -5.844 1.00 0.95
+ATOM 1755 2HG GLU A 97 14.955 -0.820 -4.568 1.00 0.95
+ATOM 1756 N ARG A 98 11.028 -3.129 -6.470 1.00 0.94
+ATOM 1757 CA ARG A 98 10.036 -4.036 -7.033 1.00 0.94
+ATOM 1758 C ARG A 98 9.348 -3.418 -8.244 1.00 0.94
+ATOM 1759 O ARG A 98 8.996 -4.116 -9.194 1.00 0.94
+ATOM 1760 CB ARG A 98 8.999 -4.421 -5.988 1.00 0.94
+ATOM 1761 CG ARG A 98 9.513 -5.352 -4.901 1.00 0.94
+ATOM 1762 CD ARG A 98 8.442 -5.714 -3.938 1.00 0.94
+ATOM 1763 NE ARG A 98 8.943 -6.554 -2.862 1.00 0.94
+ATOM 1764 CZ ARG A 98 8.202 -6.990 -1.824 1.00 0.94
+ATOM 1765 NH1 ARG A 98 6.933 -6.658 -1.736 1.00 0.94
+ATOM 1766 NH2 ARG A 98 8.752 -7.752 -0.894 1.00 0.94
+ATOM 1767 H ARG A 98 10.842 -2.687 -5.581 1.00 0.94
+ATOM 1768 HA ARG A 98 10.543 -4.948 -7.351 1.00 0.94
+ATOM 1769 1HB ARG A 98 8.619 -3.524 -5.502 1.00 0.94
+ATOM 1770 2HB ARG A 98 8.156 -4.913 -6.472 1.00 0.94
+ATOM 1771 1HG ARG A 98 9.891 -6.268 -5.355 1.00 0.94
+ATOM 1772 2HG ARG A 98 10.317 -4.861 -4.351 1.00 0.94
+ATOM 1773 1HD ARG A 98 8.027 -4.808 -3.498 1.00 0.94
+ATOM 1774 2HD ARG A 98 7.655 -6.259 -4.457 1.00 0.94
+ATOM 1775 HE ARG A 98 9.915 -6.831 -2.894 1.00 0.94
+ATOM 1776 1HH1 ARG A 98 6.513 -6.076 -2.447 1.00 0.94
+ATOM 1777 2HH1 ARG A 98 6.378 -6.985 -0.959 1.00 0.94
+ATOM 1778 1HH2 ARG A 98 9.727 -8.008 -0.961 1.00 0.94
+ATOM 1779 2HH2 ARG A 98 8.197 -8.079 -0.117 1.00 0.94
+ATOM 1780 N LEU A 99 9.096 -2.025 -8.224 1.00 0.92
+ATOM 1781 CA LEU A 99 8.392 -1.333 -9.297 1.00 0.92
+ATOM 1782 C LEU A 99 9.293 -1.137 -10.511 1.00 0.92
+ATOM 1783 O LEU A 99 8.814 -0.921 -11.624 1.00 0.92
+ATOM 1784 CB LEU A 99 7.858 0.018 -8.804 1.00 0.92
+ATOM 1785 CG LEU A 99 6.719 -0.032 -7.778 1.00 0.92
+ATOM 1786 CD1 LEU A 99 6.402 1.379 -7.301 1.00 0.92
+ATOM 1787 CD2 LEU A 99 5.497 -0.684 -8.407 1.00 0.92
+ATOM 1788 H LEU A 99 9.478 -1.487 -7.460 1.00 0.92
+ATOM 1789 HA LEU A 99 7.538 -1.939 -9.597 1.00 0.92
+ATOM 1790 1HB LEU A 99 8.684 0.568 -8.356 1.00 0.92
+ATOM 1791 2HB LEU A 99 7.502 0.580 -9.668 1.00 0.92
+ATOM 1792 HG LEU A 99 7.035 -0.614 -6.912 1.00 0.92
+ATOM 1793 1HD1 LEU A 99 5.592 1.343 -6.572 1.00 0.92
+ATOM 1794 2HD1 LEU A 99 7.288 1.814 -6.838 1.00 0.92
+ATOM 1795 3HD1 LEU A 99 6.098 1.990 -8.150 1.00 0.92
+ATOM 1796 1HD2 LEU A 99 4.687 -0.720 -7.678 1.00 0.92
+ATOM 1797 2HD2 LEU A 99 5.179 -0.102 -9.273 1.00 0.92
+ATOM 1798 3HD2 LEU A 99 5.746 -1.697 -8.723 1.00 0.92
+ATOM 1799 N GLY A 100 10.747 -1.181 -10.277 1.00 0.87
+ATOM 1800 CA GLY A 100 11.713 -1.143 -11.369 1.00 0.87
+ATOM 1801 C GLY A 100 11.678 -2.432 -12.180 1.00 0.87
+ATOM 1802 O GLY A 100 12.096 -2.459 -13.338 1.00 0.87
+ATOM 1803 H GLY A 100 11.073 -1.358 -9.338 1.00 0.87
+ATOM 1804 1HA GLY A 100 11.496 -0.294 -12.017 1.00 0.87
+ATOM 1805 2HA GLY A 100 12.713 -0.991 -10.964 1.00 0.87
diff --git a/utils/examples/out/design_000000.trb b/utils/examples/out/design_000000.trb
new file mode 100644
index 0000000000000000000000000000000000000000..90ffd20e67af718b04e90f25f604838be97dbab9
Binary files /dev/null and b/utils/examples/out/design_000000.trb differ
diff --git a/utils/examples/partial_diffusion.sh b/utils/examples/partial_diffusion.sh
new file mode 100644
index 0000000000000000000000000000000000000000..80de8e8a3cc15fed0737cd3d334cf0a7f15669a7
--- /dev/null
+++ b/utils/examples/partial_diffusion.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --pdb out/design_000.pdb \
+ --trb out/design_000.trb \
+ --out out/partial_diffusion_design \
+ --contigs 0 --sampling_temp 0.3 --T 50 --save_best_plddt
diff --git a/utils/examples/pdbs/G12D_manual_mut.pdb b/utils/examples/pdbs/G12D_manual_mut.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..c362fe2913fb906cb280311ed0fa9990f8489a5e
--- /dev/null
+++ b/utils/examples/pdbs/G12D_manual_mut.pdb
@@ -0,0 +1,3257 @@
+CRYST1 270.080 270.080 270.080 90.00 90.00 90.00 P 1 0
+ATOM 1 N SER A 2 14.231 -0.987 5.628 1.00 74.84 A N
+ATOM 2 CA SER A 2 12.974 -0.625 6.266 1.00 74.84 A C
+ATOM 3 C SER A 2 11.882 -1.601 5.857 1.00 74.84 A C
+ATOM 4 O SER A 2 12.007 -2.286 4.846 1.00 74.84 A O
+ATOM 5 CB SER A 2 12.578 0.802 5.899 1.00 74.84 A C
+ATOM 6 OG SER A 2 11.201 1.023 6.128 1.00 74.84 A O
+ATOM 7 N HIS A 3 10.815 -1.667 6.650 1.00 71.81 A N
+ATOM 8 CA HIS A 3 9.689 -2.542 6.366 1.00 71.81 A C
+ATOM 9 C HIS A 3 8.421 -1.915 6.903 1.00 71.81 A C
+ATOM 10 O HIS A 3 8.457 -1.035 7.761 1.00 71.81 A O
+ATOM 11 CB HIS A 3 9.889 -3.925 6.966 1.00 71.81 A C
+ATOM 12 CG HIS A 3 11.065 -4.651 6.406 1.00 71.81 A C
+ATOM 13 CD2 HIS A 3 12.241 -5.015 6.967 1.00 71.81 A C
+ATOM 14 ND1 HIS A 3 11.123 -5.059 5.093 1.00 71.81 A N
+ATOM 15 CE1 HIS A 3 12.275 -5.666 4.874 1.00 71.81 A C
+ATOM 16 NE2 HIS A 3 12.972 -5.653 5.995 1.00 71.81 A N
+ATOM 17 N SER A 4 7.292 -2.378 6.385 1.00 65.41 A N
+ATOM 18 CA SER A 4 6.008 -1.832 6.783 1.00 65.41 A C
+ATOM 19 C SER A 4 4.990 -2.948 6.819 1.00 65.41 A C
+ATOM 20 O SER A 4 5.228 -4.053 6.335 1.00 65.41 A O
+ATOM 21 CB SER A 4 5.535 -0.747 5.826 1.00 65.41 A C
+ATOM 22 OG SER A 4 5.722 -1.156 4.487 1.00 65.41 A O
+ATOM 23 N MET A 5 3.833 -2.637 7.381 1.00 59.94 A N
+ATOM 24 CA MET A 5 2.736 -3.587 7.404 1.00 59.94 A C
+ATOM 25 C MET A 5 1.451 -2.798 7.289 1.00 59.94 A C
+ATOM 26 O MET A 5 1.131 -2.007 8.173 1.00 59.94 A O
+ATOM 27 CB MET A 5 2.784 -4.424 8.677 1.00 59.94 A C
+ATOM 28 CG MET A 5 1.594 -5.307 8.937 1.00 59.94 A C
+ATOM 29 SD MET A 5 1.749 -6.117 10.537 1.00 59.94 A S
+ATOM 30 CE MET A 5 0.532 -5.235 11.490 1.00 59.94 A C
+ATOM 31 N ARG A 6 0.725 -2.999 6.201 1.00 56.64 A N
+ATOM 32 CA ARG A 6 -0.430 -2.184 5.887 1.00 56.64 A C
+ATOM 33 C ARG A 6 -1.673 -3.049 5.941 1.00 56.64 A C
+ATOM 34 O ARG A 6 -1.616 -4.254 5.692 1.00 56.64 A O
+ATOM 35 CB ARG A 6 -0.357 -1.601 4.486 1.00 56.64 A C
+ATOM 36 CG ARG A 6 0.546 -0.420 4.255 1.00 56.64 A C
+ATOM 37 CD ARG A 6 1.942 -0.908 4.036 1.00 56.64 A C
+ATOM 38 NE ARG A 6 1.998 -1.703 2.811 1.00 56.64 A N
+ATOM 39 CZ ARG A 6 3.021 -2.478 2.479 1.00 56.64 A C
+ATOM 40 NH1 ARG A 6 3.005 -3.185 1.362 1.00 56.64 A N1+
+ATOM 41 NH2 ARG A 6 4.074 -2.527 3.270 1.00 56.64 A N
+ATOM 42 N TYR A 7 -2.806 -2.421 6.206 1.00 48.28 A N
+ATOM 43 CA TYR A 7 -4.084 -3.116 6.223 1.00 48.28 A C
+ATOM 44 C TYR A 7 -5.094 -2.278 5.456 1.00 48.28 A C
+ATOM 45 O TYR A 7 -5.602 -1.287 5.978 1.00 48.28 A O
+ATOM 46 CB TYR A 7 -4.545 -3.375 7.648 1.00 48.28 A C
+ATOM 47 CG TYR A 7 -3.875 -4.555 8.272 1.00 48.28 A C
+ATOM 48 CD1 TYR A 7 -4.119 -5.824 7.808 1.00 48.28 A C
+ATOM 49 CD2 TYR A 7 -2.988 -4.401 9.308 1.00 48.28 A C
+ATOM 50 CE1 TYR A 7 -3.498 -6.900 8.347 1.00 48.28 A C
+ATOM 51 CE2 TYR A 7 -2.366 -5.474 9.854 1.00 48.28 A C
+ATOM 52 CZ TYR A 7 -2.622 -6.723 9.371 1.00 48.28 A C
+ATOM 53 OH TYR A 7 -2.003 -7.817 9.918 1.00 48.28 A O
+ATOM 54 N PHE A 8 -5.396 -2.680 4.229 1.00 40.07 A N
+ATOM 55 CA PHE A 8 -6.361 -1.978 3.401 1.00 40.07 A C
+ATOM 56 C PHE A 8 -7.737 -2.579 3.622 1.00 40.07 A C
+ATOM 57 O PHE A 8 -7.890 -3.796 3.571 1.00 40.07 A O
+ATOM 58 CB PHE A 8 -5.966 -2.104 1.940 1.00 40.07 A C
+ATOM 59 CG PHE A 8 -4.615 -1.575 1.644 1.00 40.07 A C
+ATOM 60 CD1 PHE A 8 -3.533 -2.411 1.637 1.00 40.07 A C
+ATOM 61 CD2 PHE A 8 -4.421 -0.244 1.391 1.00 40.07 A C
+ATOM 62 CE1 PHE A 8 -2.296 -1.926 1.365 1.00 40.07 A C
+ATOM 63 CE2 PHE A 8 -3.184 0.236 1.126 1.00 40.07 A C
+ATOM 64 CZ PHE A 8 -2.123 -0.605 1.109 1.00 40.07 A C
+ATOM 65 N PHE A 9 -8.734 -1.739 3.843 1.00 40.29 A N
+ATOM 66 CA PHE A 9 -10.046 -2.223 4.261 1.00 40.29 A C
+ATOM 67 C PHE A 9 -11.120 -1.619 3.371 1.00 40.29 A C
+ATOM 68 O PHE A 9 -11.816 -0.698 3.785 1.00 40.29 A O
+ATOM 69 CB PHE A 9 -10.290 -1.859 5.699 1.00 40.29 A C
+ATOM 70 CG PHE A 9 -9.560 -2.711 6.665 1.00 40.29 A C
+ATOM 71 CD1 PHE A 9 -8.335 -2.331 7.126 1.00 40.29 A C
+ATOM 72 CD2 PHE A 9 -10.078 -3.901 7.092 1.00 40.29 A C
+ATOM 73 CE1 PHE A 9 -7.662 -3.103 8.009 1.00 40.29 A C
+ATOM 74 CE2 PHE A 9 -9.391 -4.678 7.976 1.00 40.29 A C
+ATOM 75 CZ PHE A 9 -8.180 -4.277 8.429 1.00 40.29 A C
+ATOM 76 N THR A 10 -11.317 -2.188 2.192 1.00 41.87 A N
+ATOM 77 CA THR A 10 -12.316 -1.670 1.269 1.00 41.87 A C
+ATOM 78 C THR A 10 -13.711 -2.078 1.722 1.00 41.87 A C
+ATOM 79 O THR A 10 -13.901 -3.147 2.290 1.00 41.87 A O
+ATOM 80 CB THR A 10 -12.063 -2.211 -0.129 1.00 41.87 A C
+ATOM 81 CG2 THR A 10 -12.874 -1.426 -1.110 1.00 41.87 A C
+ATOM 82 OG1 THR A 10 -10.680 -2.065 -0.462 1.00 41.87 A O
+ATOM 83 N SER A 11 -14.702 -1.234 1.457 1.00 47.15 A N
+ATOM 84 CA SER A 11 -16.055 -1.562 1.893 1.00 47.15 A C
+ATOM 85 C SER A 11 -17.085 -0.884 0.992 1.00 47.15 A C
+ATOM 86 O SER A 11 -17.527 0.222 1.300 1.00 47.15 A O
+ATOM 87 CB SER A 11 -16.246 -1.144 3.331 1.00 47.15 A C
+ATOM 88 OG SER A 11 -17.562 -1.418 3.753 1.00 47.15 A O
+ATOM 89 N VAL A 12 -17.543 -1.593 -0.039 1.00 51.01 A N
+ATOM 90 CA VAL A 12 -18.483 -1.046 -1.012 1.00 51.01 A C
+ATOM 91 C VAL A 12 -19.905 -1.246 -0.519 1.00 51.01 A C
+ATOM 92 O VAL A 12 -20.276 -2.340 -0.096 1.00 51.01 A O
+ATOM 93 CB VAL A 12 -18.289 -1.703 -2.381 1.00 51.01 A C
+ATOM 94 CG1 VAL A 12 -19.124 -0.997 -3.413 1.00 51.01 A C
+ATOM 95 CG2 VAL A 12 -16.831 -1.682 -2.758 1.00 51.01 A C
+ATOM 96 N SER A 13 -20.711 -0.201 -0.604 1.00 59.04 A N
+ATOM 97 CA SER A 13 -21.988 -0.182 0.092 1.00 59.04 A C
+ATOM 98 C SER A 13 -23.090 -0.972 -0.595 1.00 59.04 A C
+ATOM 99 O SER A 13 -24.134 -1.179 0.029 1.00 59.04 A O
+ATOM 100 CB SER A 13 -22.470 1.245 0.272 1.00 59.04 A C
+ATOM 101 OG SER A 13 -23.794 1.254 0.755 1.00 59.04 A O
+ATOM 102 N ARG A 14 -22.916 -1.352 -1.867 1.00 60.19 A N
+ATOM 103 CA ARG A 14 -23.713 -2.345 -2.599 1.00 60.19 A C
+ATOM 104 C ARG A 14 -25.171 -2.405 -2.165 1.00 60.19 A C
+ATOM 105 O ARG A 14 -25.656 -3.477 -1.799 1.00 60.19 A O
+ATOM 106 CB ARG A 14 -23.083 -3.728 -2.433 1.00 60.19 A C
+ATOM 107 CG ARG A 14 -23.498 -4.753 -3.444 1.00 60.19 A C
+ATOM 108 CD ARG A 14 -22.619 -5.971 -3.357 1.00 60.19 A C
+ATOM 109 NE ARG A 14 -22.903 -6.742 -2.160 1.00 60.19 A N
+ATOM 110 CZ ARG A 14 -22.344 -7.910 -1.881 1.00 60.19 A C
+ATOM 111 NH1 ARG A 14 -21.472 -8.442 -2.720 1.00 60.19 A N1+
+ATOM 112 NH2 ARG A 14 -22.657 -8.545 -0.763 1.00 60.19 A N
+ATOM 113 N PRO A 15 -25.900 -1.299 -2.185 1.00 65.94 A N
+ATOM 114 CA PRO A 15 -27.227 -1.283 -1.578 1.00 65.94 A C
+ATOM 115 C PRO A 15 -28.249 -1.992 -2.445 1.00 65.94 A C
+ATOM 116 O PRO A 15 -28.044 -2.233 -3.634 1.00 65.94 A O
+ATOM 117 CB PRO A 15 -27.546 0.208 -1.472 1.00 65.94 A C
+ATOM 118 CG PRO A 15 -26.853 0.786 -2.614 1.00 65.94 A C
+ATOM 119 CD PRO A 15 -25.603 -0.028 -2.851 1.00 65.94 A C
+ATOM 120 N GLY A 16 -29.370 -2.329 -1.814 1.00 70.34 A N
+ATOM 121 CA GLY A 16 -30.451 -2.998 -2.503 1.00 70.34 A C
+ATOM 122 C GLY A 16 -30.177 -4.455 -2.803 1.00 70.34 A C
+ATOM 123 O GLY A 16 -30.941 -5.330 -2.393 1.00 70.34 A O
+ATOM 124 N ARG A 17 -29.083 -4.737 -3.504 1.00 69.35 A N
+ATOM 125 CA ARG A 17 -28.819 -6.073 -4.015 1.00 69.35 A C
+ATOM 126 C ARG A 17 -27.947 -6.897 -3.078 1.00 69.35 A C
+ATOM 127 O ARG A 17 -27.160 -7.731 -3.533 1.00 69.35 A O
+ATOM 128 CB ARG A 17 -28.191 -5.997 -5.405 1.00 69.35 A C
+ATOM 129 CG ARG A 17 -26.814 -5.375 -5.456 1.00 69.35 A C
+ATOM 130 CD ARG A 17 -26.306 -5.310 -6.883 1.00 69.35 A C
+ATOM 131 NE ARG A 17 -24.873 -5.055 -6.957 1.00 69.35 A N
+ATOM 132 CZ ARG A 17 -24.322 -3.853 -6.829 1.00 69.35 A C
+ATOM 133 NH1 ARG A 17 -23.008 -3.707 -6.916 1.00 69.35 A N1+
+ATOM 134 NH2 ARG A 17 -25.087 -2.793 -6.610 1.00 69.35 A N
+ATOM 135 N GLY A 18 -28.072 -6.690 -1.785 1.00 69.58 A N
+ATOM 136 CA GLY A 18 -27.408 -7.563 -0.851 1.00 69.58 A C
+ATOM 137 C GLY A 18 -26.957 -6.806 0.390 1.00 69.58 A C
+ATOM 138 O GLY A 18 -27.703 -5.998 0.947 1.00 69.58 A O
+ATOM 139 N GLU A 19 -25.730 -7.106 0.806 1.00 68.27 A N
+ATOM 140 CA GLU A 19 -25.099 -6.617 2.015 1.00 68.27 A C
+ATOM 141 C GLU A 19 -23.742 -6.033 1.651 1.00 68.27 A C
+ATOM 142 O GLU A 19 -23.058 -6.587 0.785 1.00 68.27 A O
+ATOM 143 CB GLU A 19 -24.936 -7.768 3.015 1.00 68.27 A C
+ATOM 144 CG GLU A 19 -24.468 -7.374 4.378 1.00 68.27 A C
+ATOM 145 CD GLU A 19 -25.612 -6.982 5.266 1.00 68.27 A C
+ATOM 146 OE1 GLU A 19 -26.412 -7.862 5.630 1.00 68.27 A O
+ATOM 147 OE2 GLU A 19 -25.716 -5.793 5.596 1.00 68.27 A O1-
+ATOM 148 N PRO A 20 -23.332 -4.912 2.274 1.00 58.29 A N
+ATOM 149 CA PRO A 20 -22.075 -4.263 1.892 1.00 58.29 A C
+ATOM 150 C PRO A 20 -20.938 -5.235 1.708 1.00 58.29 A C
+ATOM 151 O PRO A 20 -20.567 -5.940 2.643 1.00 58.29 A O
+ATOM 152 CB PRO A 20 -21.798 -3.322 3.065 1.00 58.29 A C
+ATOM 153 CG PRO A 20 -23.096 -3.134 3.748 1.00 58.29 A C
+ATOM 154 CD PRO A 20 -24.127 -4.044 3.148 1.00 58.29 A C
+ATOM 155 N ARG A 21 -20.393 -5.307 0.507 1.00 54.56 A N
+ATOM 156 CA ARG A 21 -19.228 -6.136 0.332 1.00 54.56 A C
+ATOM 157 C ARG A 21 -18.134 -5.604 1.232 1.00 54.56 A C
+ATOM 158 O ARG A 21 -18.207 -4.485 1.734 1.00 54.56 A O
+ATOM 159 CB ARG A 21 -18.774 -6.158 -1.126 1.00 54.56 A C
+ATOM 160 CG ARG A 21 -17.833 -7.298 -1.398 1.00 54.56 A C
+ATOM 161 CD ARG A 21 -17.495 -7.502 -2.840 1.00 54.56 A C
+ATOM 162 NE ARG A 21 -16.524 -8.584 -2.954 1.00 54.56 A N
+ATOM 163 CZ ARG A 21 -16.074 -9.082 -4.100 1.00 54.56 A C
+ATOM 164 NH1 ARG A 21 -16.505 -8.601 -5.255 1.00 54.56 A N1+
+ATOM 165 NH2 ARG A 21 -15.191 -10.066 -4.090 1.00 54.56 A N
+ATOM 166 N PHE A 22 -17.132 -6.430 1.478 1.00 46.66 A N
+ATOM 167 CA PHE A 22 -16.050 -6.014 2.353 1.00 46.66 A C
+ATOM 168 C PHE A 22 -14.872 -6.910 2.042 1.00 46.66 A C
+ATOM 169 O PHE A 22 -14.893 -8.089 2.384 1.00 46.66 A O
+ATOM 170 CB PHE A 22 -16.478 -6.107 3.811 1.00 46.66 A C
+ATOM 171 CG PHE A 22 -15.379 -5.867 4.793 1.00 46.66 A C
+ATOM 172 CD1 PHE A 22 -14.541 -6.885 5.176 1.00 46.66 A C
+ATOM 173 CD2 PHE A 22 -15.184 -4.619 5.338 1.00 46.66 A C
+ATOM 174 CE1 PHE A 22 -13.548 -6.666 6.077 1.00 46.66 A C
+ATOM 175 CE2 PHE A 22 -14.168 -4.400 6.228 1.00 46.66 A C
+ATOM 176 CZ PHE A 22 -13.354 -5.426 6.595 1.00 46.66 A C
+ATOM 177 N ILE A 23 -13.874 -6.368 1.366 1.00 43.99 A N
+ATOM 178 CA ILE A 23 -12.610 -7.048 1.169 1.00 43.99 A C
+ATOM 179 C ILE A 23 -11.604 -6.366 2.065 1.00 43.99 A C
+ATOM 180 O ILE A 23 -11.485 -5.140 2.042 1.00 43.99 A O
+ATOM 181 CB ILE A 23 -12.162 -7.020 -0.297 1.00 43.99 A C
+ATOM 182 CG1 ILE A 23 -13.244 -7.635 -1.161 1.00 43.99 A C
+ATOM 183 CG2 ILE A 23 -10.885 -7.796 -0.477 1.00 43.99 A C
+ATOM 184 CD1 ILE A 23 -12.903 -7.688 -2.603 1.00 43.99 A C
+ATOM 185 N ALA A 24 -10.907 -7.147 2.879 1.00 40.39 A N
+ATOM 186 CA ALA A 24 -9.960 -6.612 3.846 1.00 40.39 A C
+ATOM 187 C ALA A 24 -8.632 -7.280 3.592 1.00 40.39 A C
+ATOM 188 O ALA A 24 -8.290 -8.270 4.230 1.00 40.39 A O
+ATOM 189 CB ALA A 24 -10.416 -6.835 5.255 1.00 40.39 A C
+ATOM 190 N VAL A 25 -7.882 -6.729 2.683 1.00 41.35 A N
+ATOM 191 CA VAL A 25 -6.564 -7.247 2.417 1.00 41.35 A C
+ATOM 192 C VAL A 25 -5.615 -6.781 3.512 1.00 41.35 A C
+ATOM 193 O VAL A 25 -5.845 -5.778 4.173 1.00 41.35 A O
+ATOM 194 CB VAL A 25 -6.119 -6.783 1.028 1.00 41.35 A C
+ATOM 195 CG1 VAL A 25 -4.686 -7.074 0.782 1.00 41.35 A C
+ATOM 196 CG2 VAL A 25 -6.970 -7.429 0.004 1.00 41.35 A C
+ATOM 197 N GLY A 26 -4.568 -7.543 3.734 1.00 49.17 A N
+ATOM 198 CA GLY A 26 -3.479 -7.110 4.588 1.00 49.17 A C
+ATOM 199 C GLY A 26 -2.178 -7.377 3.878 1.00 49.17 A C
+ATOM 200 O GLY A 26 -2.049 -8.360 3.159 1.00 49.17 A O
+ATOM 201 N TYR A 27 -1.215 -6.488 4.078 1.00 56.65 A N
+ATOM 202 CA TYR A 27 0.001 -6.510 3.287 1.00 56.65 A C
+ATOM 203 C TYR A 27 1.227 -6.451 4.174 1.00 56.65 A C
+ATOM 204 O TYR A 27 1.192 -5.892 5.267 1.00 56.65 A O
+ATOM 205 CB TYR A 27 0.077 -5.331 2.333 1.00 56.65 A C
+ATOM 206 CG TYR A 27 -0.521 -5.567 0.982 1.00 56.65 A C
+ATOM 207 CD1 TYR A 27 0.183 -6.236 0.008 1.00 56.65 A C
+ATOM 208 CD2 TYR A 27 -1.763 -5.068 0.662 1.00 56.65 A C
+ATOM 209 CE1 TYR A 27 -0.350 -6.442 -1.225 1.00 56.65 A C
+ATOM 210 CE2 TYR A 27 -2.296 -5.258 -0.572 1.00 56.65 A C
+ATOM 211 CZ TYR A 27 -1.588 -5.952 -1.512 1.00 56.65 A C
+ATOM 212 OH TYR A 27 -2.117 -6.152 -2.761 1.00 56.65 A O
+ATOM 213 N VAL A 28 2.326 -7.004 3.678 1.00 62.48 A N
+ATOM 214 CA VAL A 28 3.612 -6.747 4.301 1.00 62.48 A C
+ATOM 215 C VAL A 28 4.700 -6.519 3.251 1.00 62.48 A C
+ATOM 216 O VAL A 28 5.240 -7.450 2.648 1.00 62.48 A O
+ATOM 217 CB VAL A 28 3.955 -7.857 5.306 1.00 62.48 A C
+ATOM 218 CG1 VAL A 28 4.141 -9.188 4.634 1.00 62.48 A C
+ATOM 219 CG2 VAL A 28 5.177 -7.477 6.074 1.00 62.48 A C
+ATOM 220 N ASP A 29 4.986 -5.244 3.021 1.00 70.76 A N
+ATOM 221 CA ASP A 29 6.078 -4.639 2.265 1.00 70.76 A C
+ATOM 222 C ASP A 29 5.991 -4.633 0.743 1.00 70.76 A C
+ATOM 223 O ASP A 29 6.591 -3.748 0.133 1.00 70.76 A O
+ATOM 224 CB ASP A 29 7.390 -5.329 2.631 1.00 70.76 A C
+ATOM 225 CG ASP A 29 7.926 -4.880 3.955 1.00 70.76 A C
+ATOM 226 OD1 ASP A 29 7.510 -3.805 4.435 1.00 70.76 A O
+ATOM 227 OD2 ASP A 29 8.776 -5.599 4.514 1.00 70.76 A O1-
+ATOM 228 N ASP A 30 5.324 -5.592 0.104 1.00 75.51 A N
+ATOM 229 CA ASP A 30 4.374 -5.398 -0.991 1.00 75.51 A C
+ATOM 230 C ASP A 30 3.477 -6.612 -1.103 1.00 75.51 A C
+ATOM 231 O ASP A 30 2.523 -6.614 -1.881 1.00 75.51 A O
+ATOM 232 CB ASP A 30 5.026 -5.112 -2.349 1.00 75.51 A C
+ATOM 233 CG ASP A 30 5.604 -3.722 -2.448 1.00 75.51 A C
+ATOM 234 OD1 ASP A 30 5.064 -2.812 -1.787 1.00 75.51 A O
+ATOM 235 OD2 ASP A 30 6.578 -3.526 -3.194 1.00 75.51 A O1-
+ATOM 236 N THR A 31 3.789 -7.656 -0.346 1.00 64.20 A N
+ATOM 237 CA THR A 31 3.151 -8.942 -0.542 1.00 64.20 A C
+ATOM 238 C THR A 31 1.948 -9.065 0.368 1.00 64.20 A C
+ATOM 239 O THR A 31 1.884 -8.459 1.436 1.00 64.20 A O
+ATOM 240 CB THR A 31 4.110 -10.098 -0.285 1.00 64.20 A C
+ATOM 241 CG2 THR A 31 4.653 -10.030 1.099 1.00 64.20 A C
+ATOM 242 OG1 THR A 31 3.404 -11.331 -0.431 1.00 64.20 A O
+ATOM 243 N GLN A 32 0.984 -9.838 -0.087 1.00 62.19 A N
+ATOM 244 CA GLN A 32 -0.261 -10.038 0.625 1.00 62.19 A C
+ATOM 245 C GLN A 32 -0.180 -11.340 1.402 1.00 62.19 A C
+ATOM 246 O GLN A 32 0.352 -12.333 0.902 1.00 62.19 A O
+ATOM 247 CB GLN A 32 -1.420 -10.080 -0.360 1.00 62.19 A C
+ATOM 248 CG GLN A 32 -2.766 -10.011 0.268 1.00 62.19 A C
+ATOM 249 CD GLN A 32 -3.837 -10.390 -0.707 1.00 62.19 A C
+ATOM 250 NE2 GLN A 32 -5.027 -9.855 -0.521 1.00 62.19 A N
+ATOM 251 OE1 GLN A 32 -3.599 -11.173 -1.624 1.00 62.19 A O
+ATOM 252 N PHE A 33 -0.686 -11.333 2.634 1.00 57.29 A N
+ATOM 253 CA PHE A 33 -0.719 -12.556 3.413 1.00 57.29 A C
+ATOM 254 C PHE A 33 -2.091 -12.930 3.945 1.00 57.29 A C
+ATOM 255 O PHE A 33 -2.293 -14.096 4.279 1.00 57.29 A O
+ATOM 256 CB PHE A 33 0.290 -12.494 4.574 1.00 57.29 A C
+ATOM 257 CG PHE A 33 0.023 -11.415 5.575 1.00 57.29 A C
+ATOM 258 CD1 PHE A 33 -0.850 -11.610 6.611 1.00 57.29 A C
+ATOM 259 CD2 PHE A 33 0.677 -10.221 5.502 1.00 57.29 A C
+ATOM 260 CE1 PHE A 33 -1.091 -10.621 7.530 1.00 57.29 A C
+ATOM 261 CE2 PHE A 33 0.433 -9.237 6.418 1.00 57.29 A C
+ATOM 262 CZ PHE A 33 -0.452 -9.440 7.431 1.00 57.29 A C
+ATOM 263 N VAL A 34 -3.046 -12.009 3.997 1.00 52.72 A N
+ATOM 264 CA VAL A 34 -4.406 -12.354 4.370 1.00 52.72 A C
+ATOM 265 C VAL A 34 -5.378 -11.721 3.390 1.00 52.72 A C
+ATOM 266 O VAL A 34 -5.019 -10.870 2.586 1.00 52.72 A O
+ATOM 267 CB VAL A 34 -4.757 -11.938 5.807 1.00 52.72 A C
+ATOM 268 CG1 VAL A 34 -3.979 -12.739 6.790 1.00 52.72 A C
+ATOM 269 CG2 VAL A 34 -4.469 -10.493 6.009 1.00 52.72 A C
+ATOM 270 N ARG A 35 -6.625 -12.165 3.474 1.00 57.17 A N
+ATOM 271 CA ARG A 35 -7.721 -11.686 2.649 1.00 57.17 A C
+ATOM 272 C ARG A 35 -8.997 -11.917 3.430 1.00 57.17 A C
+ATOM 273 O ARG A 35 -9.015 -12.684 4.388 1.00 57.17 A O
+ATOM 274 CB ARG A 35 -7.797 -12.425 1.317 1.00 57.17 A C
+ATOM 275 CG ARG A 35 -8.577 -13.719 1.454 1.00 57.17 A C
+ATOM 276 CD ARG A 35 -8.395 -14.678 0.306 1.00 57.17 A C
+ATOM 277 NE ARG A 35 -8.988 -14.162 -0.916 1.00 57.17 A N
+ATOM 278 CZ ARG A 35 -10.288 -14.182 -1.170 1.00 57.17 A C
+ATOM 279 NH1 ARG A 35 -10.748 -13.690 -2.309 1.00 57.17 A N1+
+ATOM 280 NH2 ARG A 35 -11.132 -14.689 -0.283 1.00 57.17 A N
+ATOM 281 N PHE A 36 -10.062 -11.236 3.040 1.00 48.80 A N
+ATOM 282 CA PHE A 36 -11.349 -11.537 3.650 1.00 48.80 A C
+ATOM 283 C PHE A 36 -12.459 -10.975 2.766 1.00 48.80 A C
+ATOM 284 O PHE A 36 -12.777 -9.792 2.866 1.00 48.80 A O
+ATOM 285 CB PHE A 36 -11.405 -10.965 5.048 1.00 48.80 A C
+ATOM 286 CG PHE A 36 -12.682 -11.218 5.750 1.00 48.80 A C
+ATOM 287 CD1 PHE A 36 -12.867 -12.374 6.457 1.00 48.80 A C
+ATOM 288 CD2 PHE A 36 -13.690 -10.294 5.730 1.00 48.80 A C
+ATOM 289 CE1 PHE A 36 -14.036 -12.609 7.106 1.00 48.80 A C
+ATOM 290 CE2 PHE A 36 -14.859 -10.530 6.385 1.00 48.80 A C
+ATOM 291 CZ PHE A 36 -15.030 -11.688 7.071 1.00 48.80 A C
+ATOM 292 N ASP A 37 -13.073 -11.821 1.951 1.00 55.56 A N
+ATOM 293 CA ASP A 37 -14.253 -11.410 1.210 1.00 55.56 A C
+ATOM 294 C ASP A 37 -15.483 -11.725 2.042 1.00 55.56 A C
+ATOM 295 O ASP A 37 -15.590 -12.812 2.604 1.00 55.56 A O
+ATOM 296 CB ASP A 37 -14.355 -12.144 -0.125 1.00 55.56 A C
+ATOM 297 CG ASP A 37 -13.245 -11.793 -1.075 1.00 55.56 A C
+ATOM 298 OD1 ASP A 37 -12.707 -10.682 -0.986 1.00 55.56 A O
+ATOM 299 OD2 ASP A 37 -12.917 -12.626 -1.938 1.00 55.56 A O1-
+ATOM 300 N SER A 38 -16.419 -10.791 2.112 1.00 55.20 A N
+ATOM 301 CA SER A 38 -17.660 -11.128 2.792 1.00 55.20 A C
+ATOM 302 C SER A 38 -18.593 -11.936 1.911 1.00 55.20 A C
+ATOM 303 O SER A 38 -19.789 -12.022 2.203 1.00 55.20 A O
+ATOM 304 CB SER A 38 -18.387 -9.880 3.273 1.00 55.20 A C
+ATOM 305 OG SER A 38 -18.879 -9.148 2.179 1.00 55.20 A O
+ATOM 306 N ASP A 39 -18.082 -12.487 0.812 1.00 60.75 A N
+ATOM 307 CA ASP A 39 -18.827 -13.449 0.009 1.00 60.75 A C
+ATOM 308 C ASP A 39 -17.807 -14.358 -0.673 1.00 60.75 A C
+ATOM 309 O ASP A 39 -17.314 -14.051 -1.760 1.00 60.75 A O
+ATOM 310 CB ASP A 39 -19.736 -12.742 -0.985 1.00 60.75 A C
+ATOM 311 CG ASP A 39 -19.028 -11.657 -1.766 1.00 60.75 A C
+ATOM 312 OD1 ASP A 39 -17.805 -11.496 -1.605 1.00 60.75 A O
+ATOM 313 OD2 ASP A 39 -19.707 -10.948 -2.534 1.00 60.75 A O1-
+ATOM 314 N ALA A 40 -17.513 -15.478 -0.035 1.00 71.59 A N
+ATOM 315 CA ALA A 40 -16.539 -16.424 -0.552 1.00 71.59 A C
+ATOM 316 C ALA A 40 -16.836 -17.784 0.066 1.00 71.59 A C
+ATOM 317 O ALA A 40 -17.930 -18.018 0.592 1.00 71.59 A O
+ATOM 318 CB ALA A 40 -15.111 -15.954 -0.263 1.00 71.59 A C
+ATOM 319 N ALA A 41 -15.864 -18.695 -0.024 1.00 74.49 A N
+ATOM 320 CA ALA A 41 -15.995 -20.006 0.598 1.00 74.49 A C
+ATOM 321 C ALA A 41 -16.471 -19.889 2.038 1.00 74.49 A C
+ATOM 322 O ALA A 41 -17.567 -20.337 2.386 1.00 74.49 A O
+ATOM 323 CB ALA A 41 -14.658 -20.745 0.541 1.00 74.49 A C
+ATOM 324 N SER A 42 -15.673 -19.244 2.880 1.00 71.88 A N
+ATOM 325 CA SER A 42 -15.915 -19.189 4.310 1.00 71.88 A C
+ATOM 326 C SER A 42 -15.948 -17.743 4.773 1.00 71.88 A C
+ATOM 327 O SER A 42 -15.676 -16.817 4.008 1.00 71.88 A O
+ATOM 328 CB SER A 42 -14.834 -19.959 5.064 1.00 71.88 A C
+ATOM 329 OG SER A 42 -13.559 -19.648 4.535 1.00 71.88 A O
+ATOM 330 N GLN A 43 -16.305 -17.548 6.031 1.00 67.52 A N
+ATOM 331 CA GLN A 43 -16.146 -16.249 6.674 1.00 67.52 A C
+ATOM 332 C GLN A 43 -14.894 -16.247 7.546 1.00 67.52 A C
+ATOM 333 O GLN A 43 -14.950 -16.071 8.758 1.00 67.52 A O
+ATOM 334 CB GLN A 43 -17.383 -15.914 7.487 1.00 67.52 A C
+ATOM 335 CG GLN A 43 -18.667 -16.133 6.752 1.00 67.52 A C
+ATOM 336 CD GLN A 43 -18.895 -15.113 5.682 1.00 67.52 A C
+ATOM 337 NE2 GLN A 43 -19.274 -13.908 6.080 1.00 67.52 A N
+ATOM 338 OE1 GLN A 43 -18.739 -15.402 4.504 1.00 67.52 A O
+ATOM 339 N ARG A 44 -13.744 -16.440 6.904 1.00 69.09 A N
+ATOM 340 CA ARG A 44 -12.481 -16.598 7.605 1.00 69.09 A C
+ATOM 341 C ARG A 44 -11.414 -15.718 6.976 1.00 69.09 A C
+ATOM 342 O ARG A 44 -11.438 -15.452 5.775 1.00 69.09 A O
+ATOM 343 CB ARG A 44 -11.983 -18.056 7.560 1.00 69.09 A C
+ATOM 344 CG ARG A 44 -13.005 -19.100 7.967 1.00 69.09 A C
+ATOM 345 CD ARG A 44 -13.122 -19.244 9.455 1.00 69.09 A C
+ATOM 346 NE ARG A 44 -11.917 -19.809 10.036 1.00 69.09 A N
+ATOM 347 CZ ARG A 44 -11.649 -19.785 11.333 1.00 69.09 A C
+ATOM 348 NH1 ARG A 44 -12.507 -19.223 12.170 1.00 69.09 A N1+
+ATOM 349 NH2 ARG A 44 -10.529 -20.324 11.788 1.00 69.09 A N
+ATOM 350 N MET A 45 -10.440 -15.321 7.787 1.00 68.13 A N
+ATOM 351 CA MET A 45 -9.185 -14.804 7.256 1.00 68.13 A C
+ATOM 352 C MET A 45 -8.452 -15.937 6.568 1.00 68.13 A C
+ATOM 353 O MET A 45 -7.520 -16.537 7.102 1.00 68.13 A O
+ATOM 354 CB MET A 45 -8.310 -14.190 8.335 1.00 68.13 A C
+ATOM 355 CG MET A 45 -8.641 -12.798 8.760 1.00 68.13 A C
+ATOM 356 SD MET A 45 -7.738 -11.870 7.509 1.00 68.13 A S
+ATOM 357 CE MET A 45 -8.109 -10.181 7.887 1.00 68.13 A C
+ATOM 358 N GLU A 46 -8.884 -16.227 5.365 1.00 68.37 A N
+ATOM 359 CA GLU A 46 -8.170 -17.190 4.563 1.00 68.37 A C
+ATOM 360 C GLU A 46 -6.727 -16.732 4.433 1.00 68.37 A C
+ATOM 361 O GLU A 46 -6.482 -15.563 4.133 1.00 68.37 A O
+ATOM 362 CB GLU A 46 -8.815 -17.321 3.190 1.00 68.37 A C
+ATOM 363 CG GLU A 46 -10.301 -17.620 3.237 1.00 68.37 A C
+ATOM 364 CD GLU A 46 -10.812 -18.212 1.939 1.00 68.37 A C
+ATOM 365 OE1 GLU A 46 -10.012 -18.333 0.990 1.00 68.37 A O
+ATOM 366 OE2 GLU A 46 -12.010 -18.558 1.864 1.00 68.37 A O1-
+ATOM 367 N PRO A 47 -5.760 -17.590 4.703 1.00 66.59 A N
+ATOM 368 CA PRO A 47 -4.367 -17.202 4.525 1.00 66.59 A C
+ATOM 369 C PRO A 47 -4.107 -16.921 3.063 1.00 66.59 A C
+ATOM 370 O PRO A 47 -4.769 -17.460 2.177 1.00 66.59 A O
+ATOM 371 CB PRO A 47 -3.594 -18.437 4.982 1.00 66.59 A C
+ATOM 372 CG PRO A 47 -4.528 -19.173 5.835 1.00 66.59 A C
+ATOM 373 CD PRO A 47 -5.886 -18.930 5.283 1.00 66.59 A C
+ATOM 374 N ARG A 48 -3.124 -16.069 2.807 1.00 65.33 A N
+ATOM 375 CA ARG A 48 -2.777 -15.741 1.437 1.00 65.33 A C
+ATOM 376 C ARG A 48 -1.289 -15.873 1.175 1.00 65.33 A C
+ATOM 377 O ARG A 48 -0.832 -15.521 0.086 1.00 65.33 A O
+ATOM 378 CB ARG A 48 -3.249 -14.333 1.088 1.00 65.33 A C
+ATOM 379 CG ARG A 48 -4.697 -14.245 0.688 1.00 65.33 A C
+ATOM 380 CD ARG A 48 -4.940 -15.062 -0.555 1.00 65.33 A C
+ATOM 381 NE ARG A 48 -5.842 -14.382 -1.471 1.00 65.33 A N
+ATOM 382 CZ ARG A 48 -6.456 -14.968 -2.489 1.00 65.33 A C
+ATOM 383 NH1 ARG A 48 -7.253 -14.263 -3.278 1.00 65.33 A N1+
+ATOM 384 NH2 ARG A 48 -6.278 -16.261 -2.711 1.00 65.33 A N
+ATOM 385 N ALA A 49 -0.523 -16.371 2.137 1.00 69.50 A N
+ATOM 386 CA ALA A 49 0.883 -16.582 1.926 1.00 69.50 A C
+ATOM 387 C ALA A 49 1.300 -17.747 2.800 1.00 69.50 A C
+ATOM 388 O ALA A 49 0.797 -17.874 3.925 1.00 69.50 A O
+ATOM 389 CB ALA A 49 1.713 -15.347 2.264 1.00 69.50 A C
+ATOM 390 N PRO A 50 2.203 -18.604 2.322 1.00 73.88 A N
+ATOM 391 CA PRO A 50 2.547 -19.811 3.092 1.00 73.88 A C
+ATOM 392 C PRO A 50 3.065 -19.528 4.488 1.00 73.88 A C
+ATOM 393 O PRO A 50 2.732 -20.262 5.425 1.00 73.88 A O
+ATOM 394 CB PRO A 50 3.619 -20.476 2.221 1.00 73.88 A C
+ATOM 395 CG PRO A 50 3.373 -19.953 0.857 1.00 73.88 A C
+ATOM 396 CD PRO A 50 2.903 -18.552 1.032 1.00 73.88 A C
+ATOM 397 N TRP A 51 3.857 -18.477 4.662 1.00 74.78 A N
+ATOM 398 CA TRP A 51 4.507 -18.282 5.948 1.00 74.78 A C
+ATOM 399 C TRP A 51 3.557 -17.834 7.043 1.00 74.78 A C
+ATOM 400 O TRP A 51 4.021 -17.578 8.154 1.00 74.78 A O
+ATOM 401 CB TRP A 51 5.646 -17.276 5.812 1.00 74.78 A C
+ATOM 402 CG TRP A 51 5.254 -16.012 5.146 1.00 74.78 A C
+ATOM 403 CD1 TRP A 51 5.538 -15.645 3.872 1.00 74.78 A C
+ATOM 404 CD2 TRP A 51 4.520 -14.931 5.723 1.00 74.78 A C
+ATOM 405 CE2 TRP A 51 4.390 -13.947 4.738 1.00 74.78 A C
+ATOM 406 CE3 TRP A 51 3.960 -14.704 6.977 1.00 74.78 A C
+ATOM 407 NE1 TRP A 51 5.021 -14.406 3.614 1.00 74.78 A N
+ATOM 408 CZ2 TRP A 51 3.721 -12.758 4.964 1.00 74.78 A C
+ATOM 409 CZ3 TRP A 51 3.300 -13.528 7.197 1.00 74.78 A C
+ATOM 410 CH2 TRP A 51 3.186 -12.568 6.200 1.00 74.78 A C
+ATOM 411 N ILE A 52 2.260 -17.714 6.783 1.00 71.36 A N
+ATOM 412 CA ILE A 52 1.302 -17.459 7.847 1.00 71.36 A C
+ATOM 413 C ILE A 52 0.429 -18.668 8.128 1.00 71.36 A C
+ATOM 414 O ILE A 52 -0.380 -18.638 9.067 1.00 71.36 A O
+ATOM 415 CB ILE A 52 0.436 -16.227 7.543 1.00 71.36 A C
+ATOM 416 CG1 ILE A 52 -0.210 -15.717 8.823 1.00 71.36 A C
+ATOM 417 CG2 ILE A 52 -0.620 -16.584 6.547 1.00 71.36 A C
+ATOM 418 CD1 ILE A 52 -0.797 -14.378 8.708 1.00 71.36 A C
+ATOM 419 N GLU A 53 0.579 -19.746 7.356 1.00 76.04 A N
+ATOM 420 CA GLU A 53 -0.117 -20.986 7.676 1.00 76.04 A C
+ATOM 421 C GLU A 53 0.314 -21.529 9.028 1.00 76.04 A C
+ATOM 422 O GLU A 53 -0.482 -22.159 9.732 1.00 76.04 A O
+ATOM 423 CB GLU A 53 0.151 -22.028 6.592 1.00 76.04 A C
+ATOM 424 CG GLU A 53 -0.257 -21.625 5.189 1.00 76.04 A C
+ATOM 425 CD GLU A 53 -1.751 -21.413 5.028 1.00 76.04 A C
+ATOM 426 OE1 GLU A 53 -2.512 -21.652 5.990 1.00 76.04 A O
+ATOM 427 OE2 GLU A 53 -2.168 -21.023 3.918 1.00 76.04 A O1-
+ATOM 428 N GLN A 54 1.564 -21.281 9.406 1.00 80.33 A N
+ATOM 429 CA GLN A 54 2.129 -21.841 10.620 1.00 80.33 A C
+ATOM 430 C GLN A 54 1.368 -21.425 11.863 1.00 80.33 A C
+ATOM 431 O GLN A 54 1.459 -22.107 12.887 1.00 80.33 A O
+ATOM 432 CB GLN A 54 3.579 -21.390 10.746 1.00 80.33 A C
+ATOM 433 CG GLN A 54 4.420 -21.711 9.537 1.00 80.33 A C
+ATOM 434 CD GLN A 54 5.723 -20.948 9.527 1.00 80.33 A C
+ATOM 435 NE2 GLN A 54 6.533 -21.170 8.500 1.00 80.33 A N
+ATOM 436 OE1 GLN A 54 5.997 -20.157 10.430 1.00 80.33 A O
+ATOM 437 N GLU A 55 0.617 -20.330 11.796 1.00 77.54 A N
+ATOM 438 CA GLU A 55 0.208 -19.642 13.013 1.00 77.54 A C
+ATOM 439 C GLU A 55 -0.634 -20.530 13.918 1.00 77.54 A C
+ATOM 440 O GLU A 55 -0.402 -20.593 15.130 1.00 77.54 A O
+ATOM 441 CB GLU A 55 -0.544 -18.378 12.647 1.00 77.54 A C
+ATOM 442 CG GLU A 55 0.357 -17.190 12.458 1.00 77.54 A C
+ATOM 443 CD GLU A 55 0.857 -16.641 13.767 1.00 77.54 A C
+ATOM 444 OE1 GLU A 55 0.193 -16.877 14.797 1.00 77.54 A O
+ATOM 445 OE2 GLU A 55 1.912 -15.973 13.769 1.00 77.54 A O1-
+ATOM 446 N GLY A 56 -1.619 -21.218 13.360 1.00 75.47 A N
+ATOM 447 CA GLY A 56 -2.402 -22.135 14.148 1.00 75.47 A C
+ATOM 448 C GLY A 56 -3.773 -21.612 14.515 1.00 75.47 A C
+ATOM 449 O GLY A 56 -4.087 -20.432 14.361 1.00 75.47 A O
+ATOM 450 N PRO A 57 -4.606 -22.506 15.042 1.00 72.76 A N
+ATOM 451 CA PRO A 57 -6.041 -22.205 15.161 1.00 72.76 A C
+ATOM 452 C PRO A 57 -6.363 -20.982 15.992 1.00 72.76 A C
+ATOM 453 O PRO A 57 -7.298 -20.248 15.656 1.00 72.76 A O
+ATOM 454 CB PRO A 57 -6.603 -23.476 15.807 1.00 72.76 A C
+ATOM 455 CG PRO A 57 -5.605 -24.536 15.492 1.00 72.76 A C
+ATOM 456 CD PRO A 57 -4.279 -23.859 15.510 1.00 72.76 A C
+ATOM 457 N GLU A 58 -5.637 -20.746 17.080 1.00 75.70 A N
+ATOM 458 CA GLU A 58 -5.949 -19.587 17.907 1.00 75.70 A C
+ATOM 459 C GLU A 58 -5.746 -18.302 17.124 1.00 75.70 A C
+ATOM 460 O GLU A 58 -6.563 -17.375 17.194 1.00 75.70 A O
+ATOM 461 CB GLU A 58 -5.078 -19.588 19.159 1.00 75.70 A C
+ATOM 462 CG GLU A 58 -5.157 -18.310 19.979 1.00 75.70 A C
+ATOM 463 CD GLU A 58 -6.583 -17.911 20.309 1.00 75.70 A C
+ATOM 464 OE1 GLU A 58 -7.202 -18.590 21.153 1.00 75.70 A O
+ATOM 465 OE2 GLU A 58 -7.083 -16.915 19.745 1.00 75.70 A O1-
+ATOM 466 N TYR A 59 -4.660 -18.241 16.355 1.00 69.71 A N
+ATOM 467 CA TYR A 59 -4.354 -17.045 15.588 1.00 69.71 A C
+ATOM 468 C TYR A 59 -5.452 -16.747 14.585 1.00 69.71 A C
+ATOM 469 O TYR A 59 -5.958 -15.627 14.523 1.00 69.71 A O
+ATOM 470 CB TYR A 59 -3.024 -17.226 14.877 1.00 69.71 A C
+ATOM 471 CG TYR A 59 -2.618 -16.070 14.012 1.00 69.71 A C
+ATOM 472 CD1 TYR A 59 -2.047 -14.942 14.568 1.00 69.71 A C
+ATOM 473 CD2 TYR A 59 -2.795 -16.105 12.646 1.00 69.71 A C
+ATOM 474 CE1 TYR A 59 -1.658 -13.893 13.791 1.00 69.71 A C
+ATOM 475 CE2 TYR A 59 -2.397 -15.066 11.859 1.00 69.71 A C
+ATOM 476 CZ TYR A 59 -1.829 -13.960 12.434 1.00 69.71 A C
+ATOM 477 OH TYR A 59 -1.442 -12.904 11.645 1.00 69.71 A O
+ATOM 478 N TRP A 60 -5.823 -17.741 13.782 1.00 68.02 A N
+ATOM 479 CA TRP A 60 -6.860 -17.523 12.787 1.00 68.02 A C
+ATOM 480 C TRP A 60 -8.172 -17.173 13.451 1.00 68.02 A C
+ATOM 481 O TRP A 60 -8.913 -16.325 12.954 1.00 68.02 A O
+ATOM 482 CB TRP A 60 -7.005 -18.749 11.902 1.00 68.02 A C
+ATOM 483 CG TRP A 60 -5.816 -18.916 11.088 1.00 68.02 A C
+ATOM 484 CD1 TRP A 60 -4.950 -19.951 11.124 1.00 68.02 A C
+ATOM 485 CD2 TRP A 60 -5.308 -17.997 10.127 1.00 68.02 A C
+ATOM 486 CE2 TRP A 60 -4.130 -18.551 9.609 1.00 68.02 A C
+ATOM 487 CE3 TRP A 60 -5.740 -16.761 9.642 1.00 68.02 A C
+ATOM 488 NE1 TRP A 60 -3.931 -19.749 10.236 1.00 68.02 A N
+ATOM 489 CZ2 TRP A 60 -3.376 -17.913 8.641 1.00 68.02 A C
+ATOM 490 CZ3 TRP A 60 -4.993 -16.136 8.683 1.00 68.02 A C
+ATOM 491 CH2 TRP A 60 -3.826 -16.711 8.188 1.00 68.02 A C
+ATOM 492 N ASP A 61 -8.461 -17.783 14.596 1.00 71.99 A N
+ATOM 493 CA ASP A 61 -9.699 -17.457 15.287 1.00 71.99 A C
+ATOM 494 C ASP A 61 -9.733 -15.992 15.685 1.00 71.99 A C
+ATOM 495 O ASP A 61 -10.706 -15.283 15.409 1.00 71.99 A O
+ATOM 496 CB ASP A 61 -9.875 -18.331 16.520 1.00 71.99 A C
+ATOM 497 CG ASP A 61 -10.949 -17.801 17.437 1.00 71.99 A C
+ATOM 498 OD1 ASP A 61 -12.134 -18.081 17.177 1.00 71.99 A O
+ATOM 499 OD2 ASP A 61 -10.614 -17.085 18.403 1.00 71.99 A O1-
+ATOM 500 N GLN A 62 -8.679 -15.515 16.338 1.00 69.60 A N
+ATOM 501 CA GLN A 62 -8.753 -14.155 16.854 1.00 69.60 A C
+ATOM 502 C GLN A 62 -8.541 -13.126 15.755 1.00 69.60 A C
+ATOM 503 O GLN A 62 -9.120 -12.040 15.817 1.00 69.60 A O
+ATOM 504 CB GLN A 62 -7.793 -13.991 18.039 1.00 69.60 A C
+ATOM 505 CG GLN A 62 -6.311 -14.186 17.799 1.00 69.60 A C
+ATOM 506 CD GLN A 62 -5.593 -12.916 17.441 1.00 69.60 A C
+ATOM 507 NE2 GLN A 62 -4.504 -13.040 16.694 1.00 69.60 A N
+ATOM 508 OE1 GLN A 62 -5.984 -11.834 17.873 1.00 69.60 A O
+ATOM 509 N GLU A 63 -7.792 -13.466 14.707 1.00 64.87 A N
+ATOM 510 CA GLU A 63 -7.709 -12.590 13.546 1.00 64.87 A C
+ATOM 511 C GLU A 63 -9.047 -12.469 12.841 1.00 64.87 A C
+ATOM 512 O GLU A 63 -9.421 -11.382 12.395 1.00 64.87 A O
+ATOM 513 CB GLU A 63 -6.666 -13.108 12.579 1.00 64.87 A C
+ATOM 514 CG GLU A 63 -5.290 -12.710 12.949 1.00 64.87 A C
+ATOM 515 CD GLU A 63 -4.456 -12.504 11.731 1.00 64.87 A C
+ATOM 516 OE1 GLU A 63 -3.603 -11.598 11.732 1.00 64.87 A O
+ATOM 517 OE2 GLU A 63 -4.660 -13.253 10.759 1.00 64.87 A O1-
+ATOM 518 N THR A 64 -9.769 -13.576 12.706 1.00 63.83 A N
+ATOM 519 CA THR A 64 -11.092 -13.518 12.114 1.00 63.83 A C
+ATOM 520 C THR A 64 -12.037 -12.711 12.976 1.00 63.83 A C
+ATOM 521 O THR A 64 -12.818 -11.910 12.462 1.00 63.83 A O
+ATOM 522 CB THR A 64 -11.621 -14.922 11.904 1.00 63.83 A C
+ATOM 523 CG2 THR A 64 -12.984 -14.868 11.291 1.00 63.83 A C
+ATOM 524 OG1 THR A 64 -10.745 -15.613 11.015 1.00 63.83 A O
+ATOM 525 N ARG A 65 -11.968 -12.886 14.291 1.00 63.69 A N
+ATOM 526 CA ARG A 65 -12.802 -12.069 15.160 1.00 63.69 A C
+ATOM 527 C ARG A 65 -12.489 -10.595 14.963 1.00 63.69 A C
+ATOM 528 O ARG A 65 -13.400 -9.770 14.827 1.00 63.69 A O
+ATOM 529 CB ARG A 65 -12.599 -12.466 16.619 1.00 63.69 A C
+ATOM 530 CG ARG A 65 -12.980 -13.896 16.934 1.00 63.69 A C
+ATOM 531 CD ARG A 65 -13.232 -14.090 18.420 1.00 63.69 A C
+ATOM 532 NE ARG A 65 -12.043 -13.867 19.229 1.00 63.69 A N
+ATOM 533 CZ ARG A 65 -11.864 -12.796 19.993 1.00 63.69 A C
+ATOM 534 NH1 ARG A 65 -10.752 -12.661 20.700 1.00 63.69 A N1+
+ATOM 535 NH2 ARG A 65 -12.797 -11.857 20.043 1.00 63.69 A N
+ATOM 536 N ASN A 66 -11.203 -10.257 14.877 1.00 61.00 A N
+ATOM 537 CA ASN A 66 -10.796 -8.868 14.713 1.00 61.00 A C
+ATOM 538 C ASN A 66 -11.305 -8.287 13.402 1.00 61.00 A C
+ATOM 539 O ASN A 66 -11.932 -7.223 13.382 1.00 61.00 A O
+ATOM 540 CB ASN A 66 -9.276 -8.771 14.773 1.00 61.00 A C
+ATOM 541 CG ASN A 66 -8.726 -9.123 16.127 1.00 61.00 A C
+ATOM 542 ND2 ASN A 66 -7.428 -9.372 16.186 1.00 61.00 A N
+ATOM 543 OD1 ASN A 66 -9.457 -9.181 17.109 1.00 61.00 A O
+ATOM 544 N VAL A 67 -11.035 -8.967 12.289 1.00 55.49 A N
+ATOM 545 CA VAL A 67 -11.389 -8.392 11.001 1.00 55.49 A C
+ATOM 546 C VAL A 67 -12.892 -8.373 10.800 1.00 55.49 A C
+ATOM 547 O VAL A 67 -13.409 -7.511 10.091 1.00 55.49 A O
+ATOM 548 CB VAL A 67 -10.690 -9.120 9.856 1.00 55.49 A C
+ATOM 549 CG1 VAL A 67 -11.287 -10.457 9.661 1.00 55.49 A C
+ATOM 550 CG2 VAL A 67 -10.848 -8.325 8.615 1.00 55.49 A C
+ATOM 551 N LYS A 68 -13.633 -9.291 11.414 1.00 55.00 A N
+ATOM 552 CA LYS A 68 -15.078 -9.141 11.369 1.00 55.00 A C
+ATOM 553 C LYS A 68 -15.534 -7.986 12.244 1.00 55.00 A C
+ATOM 554 O LYS A 68 -16.554 -7.359 11.950 1.00 55.00 A O
+ATOM 555 CB LYS A 68 -15.766 -10.441 11.775 1.00 55.00 A C
+ATOM 556 CG LYS A 68 -15.378 -11.620 10.909 1.00 55.00 A C
+ATOM 557 CD LYS A 68 -16.048 -12.894 11.366 1.00 55.00 A C
+ATOM 558 CE LYS A 68 -16.977 -13.414 10.297 1.00 55.00 A C
+ATOM 559 NZ LYS A 68 -17.708 -14.616 10.745 1.00 55.00 A N1+
+ATOM 560 N ALA A 69 -14.778 -7.654 13.289 1.00 53.55 A N
+ATOM 561 CA ALA A 69 -15.082 -6.427 14.019 1.00 53.55 A C
+ATOM 562 C ALA A 69 -14.838 -5.198 13.156 1.00 53.55 A C
+ATOM 563 O ALA A 69 -15.620 -4.244 13.184 1.00 53.55 A O
+ATOM 564 CB ALA A 69 -14.251 -6.343 15.289 1.00 53.55 A C
+ATOM 565 N GLN A 70 -13.751 -5.203 12.389 1.00 51.45 A N
+ATOM 566 CA GLN A 70 -13.486 -4.108 11.460 1.00 51.45 A C
+ATOM 567 C GLN A 70 -14.590 -3.976 10.425 1.00 51.45 A C
+ATOM 568 O GLN A 70 -15.050 -2.870 10.130 1.00 51.45 A O
+ATOM 569 CB GLN A 70 -12.156 -4.322 10.757 1.00 51.45 A C
+ATOM 570 CG GLN A 70 -11.870 -3.264 9.725 1.00 51.45 A C
+ATOM 571 CD GLN A 70 -11.043 -2.119 10.239 1.00 51.45 A C
+ATOM 572 NE2 GLN A 70 -11.689 -0.990 10.499 1.00 51.45 A N
+ATOM 573 OE1 GLN A 70 -9.833 -2.242 10.391 1.00 51.45 A O
+ATOM 574 N SER A 71 -14.981 -5.095 9.821 1.00 48.62 A N
+ATOM 575 CA SER A 71 -16.087 -5.112 8.877 1.00 48.62 A C
+ATOM 576 C SER A 71 -17.343 -4.542 9.499 1.00 48.62 A C
+ATOM 577 O SER A 71 -18.073 -3.779 8.865 1.00 48.62 A O
+ATOM 578 CB SER A 71 -16.322 -6.537 8.399 1.00 48.62 A C
+ATOM 579 OG SER A 71 -17.411 -6.585 7.510 1.00 48.62 A O
+ATOM 580 N GLN A 72 -17.623 -4.918 10.738 1.00 51.34 A N
+ATOM 581 CA GLN A 72 -18.741 -4.314 11.438 1.00 51.34 A C
+ATOM 582 C GLN A 72 -18.560 -2.816 11.575 1.00 51.34 A C
+ATOM 583 O GLN A 72 -19.513 -2.056 11.412 1.00 51.34 A O
+ATOM 584 CB GLN A 72 -18.909 -4.970 12.793 1.00 51.34 A C
+ATOM 585 CG GLN A 72 -20.183 -4.623 13.489 1.00 51.34 A C
+ATOM 586 CD GLN A 72 -21.379 -4.809 12.629 1.00 51.34 A C
+ATOM 587 NE2 GLN A 72 -22.095 -3.729 12.382 1.00 51.34 A N
+ATOM 588 OE1 GLN A 72 -21.710 -5.922 12.252 1.00 51.34 A O
+ATOM 589 N THR A 73 -17.342 -2.365 11.852 1.00 53.20 A N
+ATOM 590 CA THR A 73 -17.122 -0.929 11.989 1.00 53.20 A C
+ATOM 591 C THR A 73 -17.392 -0.201 10.678 1.00 53.20 A C
+ATOM 592 O THR A 73 -18.038 0.852 10.666 1.00 53.20 A O
+ATOM 593 CB THR A 73 -15.701 -0.659 12.476 1.00 53.20 A C
+ATOM 594 CG2 THR A 73 -15.523 0.809 12.775 1.00 53.20 A C
+ATOM 595 OG1 THR A 73 -15.468 -1.393 13.680 1.00 53.20 A O
+ATOM 596 N ASP A 74 -16.920 -0.760 9.564 1.00 50.96 A N
+ATOM 597 CA ASP A 74 -17.199 -0.161 8.263 1.00 50.96 A C
+ATOM 598 C ASP A 74 -18.683 -0.163 7.948 1.00 50.96 A C
+ATOM 599 O ASP A 74 -19.200 0.806 7.399 1.00 50.96 A O
+ATOM 600 CB ASP A 74 -16.436 -0.865 7.154 1.00 50.96 A C
+ATOM 601 CG ASP A 74 -14.968 -0.609 7.231 1.00 50.96 A C
+ATOM 602 OD1 ASP A 74 -14.603 0.470 7.734 1.00 50.96 A O
+ATOM 603 OD2 ASP A 74 -14.176 -1.443 6.759 1.00 50.96 A O1-
+ATOM 604 N ARG A 75 -19.389 -1.228 8.274 1.00 55.46 A N
+ATOM 605 CA ARG A 75 -20.799 -1.203 7.938 1.00 55.46 A C
+ATOM 606 C ARG A 75 -21.577 -0.311 8.888 1.00 55.46 A C
+ATOM 607 O ARG A 75 -22.728 0.023 8.601 1.00 55.46 A O
+ATOM 608 CB ARG A 75 -21.361 -2.626 7.938 1.00 55.46 A C
+ATOM 609 CG ARG A 75 -21.824 -3.151 9.277 1.00 55.46 A C
+ATOM 610 CD ARG A 75 -22.720 -4.358 9.094 1.00 55.46 A C
+ATOM 611 NE ARG A 75 -22.137 -5.192 8.053 1.00 55.46 A N
+ATOM 612 CZ ARG A 75 -21.419 -6.283 8.294 1.00 55.46 A C
+ATOM 613 NH1 ARG A 75 -20.898 -6.968 7.296 1.00 55.46 A N1+
+ATOM 614 NH2 ARG A 75 -21.218 -6.692 9.534 1.00 55.46 A N
+ATOM 615 N VAL A 76 -20.974 0.082 10.010 1.00 55.70 A N
+ATOM 616 CA VAL A 76 -21.573 1.119 10.841 1.00 55.70 A C
+ATOM 617 C VAL A 76 -21.316 2.500 10.268 1.00 55.70 A C
+ATOM 618 O VAL A 76 -22.226 3.334 10.212 1.00 55.70 A O
+ATOM 619 CB VAL A 76 -21.072 1.030 12.291 1.00 55.70 A C
+ATOM 620 CG1 VAL A 76 -21.865 1.970 13.154 1.00 55.70 A C
+ATOM 621 CG2 VAL A 76 -21.292 -0.327 12.815 1.00 55.70 A C
+ATOM 622 N ASP A 77 -20.078 2.782 9.851 1.00 54.86 A N
+ATOM 623 CA ASP A 77 -19.745 4.140 9.440 1.00 54.86 A C
+ATOM 624 C ASP A 77 -20.042 4.411 7.976 1.00 54.86 A C
+ATOM 625 O ASP A 77 -19.961 5.563 7.552 1.00 54.86 A O
+ATOM 626 CB ASP A 77 -18.280 4.476 9.708 1.00 54.86 A C
+ATOM 627 CG ASP A 77 -17.341 3.439 9.187 1.00 54.86 A C
+ATOM 628 OD1 ASP A 77 -17.622 2.883 8.122 1.00 54.86 A O
+ATOM 629 OD2 ASP A 77 -16.297 3.204 9.821 1.00 54.86 A O1-
+ATOM 630 N LEU A 78 -20.406 3.402 7.203 1.00 51.20 A N
+ATOM 631 CA LEU A 78 -20.961 3.635 5.886 1.00 51.20 A C
+ATOM 632 C LEU A 78 -22.366 4.145 5.974 1.00 51.20 A C
+ATOM 633 O LEU A 78 -23.077 4.194 4.975 1.00 51.20 A O
+ATOM 634 CB LEU A 78 -20.929 2.341 5.077 1.00 51.20 A C
+ATOM 635 CG LEU A 78 -19.779 2.187 4.104 1.00 51.20 A C
+ATOM 636 CD1 LEU A 78 -19.875 0.896 3.349 1.00 51.20 A C
+ATOM 637 CD2 LEU A 78 -19.813 3.342 3.169 1.00 51.20 A C
+ATOM 638 N GLY A 79 -22.788 4.509 7.168 1.00 61.37 A N
+ATOM 639 CA GLY A 79 -24.112 5.021 7.394 1.00 61.37 A C
+ATOM 640 C GLY A 79 -23.961 6.317 8.136 1.00 61.37 A C
+ATOM 641 O GLY A 79 -24.940 6.997 8.435 1.00 61.37 A O
+ATOM 642 N THR A 80 -22.719 6.664 8.451 1.00 64.61 A N
+ATOM 643 CA THR A 80 -22.418 7.980 8.997 1.00 64.61 A C
+ATOM 644 C THR A 80 -21.676 8.873 8.008 1.00 64.61 A C
+ATOM 645 O THR A 80 -22.012 10.051 7.872 1.00 64.61 A O
+ATOM 646 CB THR A 80 -21.621 7.854 10.295 1.00 64.61 A C
+ATOM 647 CG2 THR A 80 -22.435 7.086 11.329 1.00 64.61 A C
+ATOM 648 OG1 THR A 80 -20.379 7.196 10.034 1.00 64.61 A O
+ATOM 649 N LEU A 81 -20.635 8.365 7.345 1.00 58.54 A N
+ATOM 650 CA LEU A 81 -20.041 9.137 6.262 1.00 58.54 A C
+ATOM 651 C LEU A 81 -21.083 9.483 5.225 1.00 58.54 A C
+ATOM 652 O LEU A 81 -21.228 10.648 4.836 1.00 58.54 A O
+ATOM 653 CB LEU A 81 -18.898 8.381 5.611 1.00 58.54 A C
+ATOM 654 CG LEU A 81 -17.718 8.178 6.534 1.00 58.54 A C
+ATOM 655 CD1 LEU A 81 -16.665 7.361 5.898 1.00 58.54 A C
+ATOM 656 CD2 LEU A 81 -17.181 9.493 6.852 1.00 58.54 A C
+ATOM 657 N ARG A 82 -21.853 8.497 4.812 1.00 65.88 A N
+ATOM 658 CA ARG A 82 -22.962 8.690 3.906 1.00 65.88 A C
+ATOM 659 C ARG A 82 -24.102 9.418 4.588 1.00 65.88 A C
+ATOM 660 O ARG A 82 -25.175 9.601 4.021 1.00 65.88 A O
+ATOM 661 CB ARG A 82 -23.389 7.320 3.351 1.00 65.88 A C
+ATOM 662 CG ARG A 82 -24.418 7.272 2.222 1.00 65.88 A C
+ATOM 663 CD ARG A 82 -25.832 7.179 2.711 1.00 65.88 A C
+ATOM 664 NE ARG A 82 -26.074 5.979 3.484 1.00 65.88 A N
+ATOM 665 CZ ARG A 82 -27.116 5.839 4.286 1.00 65.88 A C
+ATOM 666 NH1 ARG A 82 -27.951 6.851 4.443 1.00 65.88 A N1+
+ATOM 667 NH2 ARG A 82 -27.297 4.716 4.966 1.00 65.88 A N
+ATOM 668 N GLY A 83 -23.880 9.907 5.794 1.00 67.98 A N
+ATOM 669 CA GLY A 83 -24.928 10.648 6.449 1.00 67.98 A C
+ATOM 670 C GLY A 83 -24.605 12.112 6.615 1.00 67.98 A C
+ATOM 671 O GLY A 83 -25.493 12.963 6.599 1.00 67.98 A O
+ATOM 672 N TYR A 84 -23.334 12.414 6.815 1.00 75.19 A N
+ATOM 673 CA TYR A 84 -22.923 13.810 6.806 1.00 75.19 A C
+ATOM 674 C TYR A 84 -22.600 14.323 5.412 1.00 75.19 A C
+ATOM 675 O TYR A 84 -22.906 15.475 5.094 1.00 75.19 A O
+ATOM 676 CB TYR A 84 -21.725 14.058 7.711 1.00 75.19 A C
+ATOM 677 CG TYR A 84 -21.434 15.540 7.789 1.00 75.19 A C
+ATOM 678 CD1 TYR A 84 -22.460 16.469 7.915 1.00 75.19 A C
+ATOM 679 CD2 TYR A 84 -20.139 16.015 7.690 1.00 75.19 A C
+ATOM 680 CE1 TYR A 84 -22.193 17.827 7.961 1.00 75.19 A C
+ATOM 681 CE2 TYR A 84 -19.863 17.368 7.739 1.00 75.19 A C
+ATOM 682 CZ TYR A 84 -20.890 18.268 7.876 1.00 75.19 A C
+ATOM 683 OH TYR A 84 -20.600 19.610 7.924 1.00 75.19 A O
+ATOM 684 N TYR A 85 -21.993 13.506 4.561 1.00 65.60 A N
+ATOM 685 CA TYR A 85 -21.825 13.897 3.171 1.00 65.60 A C
+ATOM 686 C TYR A 85 -23.147 13.730 2.431 1.00 65.60 A C
+ATOM 687 O TYR A 85 -23.193 13.082 1.393 1.00 65.60 A O
+ATOM 688 CB TYR A 85 -20.764 13.042 2.506 1.00 65.60 A C
+ATOM 689 CG TYR A 85 -19.375 13.434 2.816 1.00 65.60 A C
+ATOM 690 CD1 TYR A 85 -18.909 14.677 2.492 1.00 65.60 A C
+ATOM 691 CD2 TYR A 85 -18.510 12.540 3.390 1.00 65.60 A C
+ATOM 692 CE1 TYR A 85 -17.629 15.031 2.758 1.00 65.60 A C
+ATOM 693 CE2 TYR A 85 -17.224 12.876 3.655 1.00 65.60 A C
+ATOM 694 CZ TYR A 85 -16.783 14.125 3.340 1.00 65.60 A C
+ATOM 695 OH TYR A 85 -15.483 14.474 3.610 1.00 65.60 A O
+ATOM 696 N ASN A 86 -24.199 14.390 2.929 1.00 71.17 A N
+ATOM 697 CA ASN A 86 -25.571 13.914 2.763 1.00 71.17 A C
+ATOM 698 C ASN A 86 -25.750 13.282 1.399 1.00 71.17 A C
+ATOM 699 O ASN A 86 -25.634 13.956 0.374 1.00 71.17 A O
+ATOM 700 CB ASN A 86 -26.543 15.083 2.902 1.00 71.17 A C
+ATOM 701 CG ASN A 86 -26.624 15.611 4.298 1.00 71.17 A C
+ATOM 702 ND2 ASN A 86 -26.650 16.930 4.427 1.00 71.17 A N
+ATOM 703 OD1 ASN A 86 -26.657 14.855 5.255 1.00 71.17 A O
+ATOM 704 N GLN A 87 -26.021 11.982 1.377 1.00 70.69 A N
+ATOM 705 CA GLN A 87 -25.689 11.223 0.185 1.00 70.69 A C
+ATOM 706 C GLN A 87 -26.745 10.164 -0.111 1.00 70.69 A C
+ATOM 707 O GLN A 87 -26.425 8.999 -0.353 1.00 70.69 A O
+ATOM 708 CB GLN A 87 -24.296 10.614 0.313 1.00 70.69 A C
+ATOM 709 CG GLN A 87 -23.794 9.884 -0.893 1.00 70.69 A C
+ATOM 710 CD GLN A 87 -22.409 9.373 -0.704 1.00 70.69 A C
+ATOM 711 NE2 GLN A 87 -21.871 8.725 -1.726 1.00 70.69 A N
+ATOM 712 OE1 GLN A 87 -21.825 9.539 0.361 1.00 70.69 A O
+ATOM 713 N SER A 88 -28.024 10.535 -0.053 1.00 80.45 A N
+ATOM 714 CA SER A 88 -28.974 9.895 -0.960 1.00 80.45 A C
+ATOM 715 C SER A 88 -29.033 8.379 -0.859 1.00 80.45 A C
+ATOM 716 O SER A 88 -28.461 7.696 -1.716 1.00 80.45 A O
+ATOM 717 CB SER A 88 -28.650 10.267 -2.406 1.00 80.45 A C
+ATOM 718 OG SER A 88 -29.338 9.414 -3.306 1.00 80.45 A O
+ATOM 719 N GLU A 89 -29.649 7.847 0.197 1.00 80.57 A N
+ATOM 720 CA GLU A 89 -29.757 6.406 0.402 1.00 80.57 A C
+ATOM 721 C GLU A 89 -30.021 5.658 -0.896 1.00 80.57 A C
+ATOM 722 O GLU A 89 -30.707 6.172 -1.785 1.00 80.57 A O
+ATOM 723 CB GLU A 89 -30.881 6.096 1.392 1.00 80.57 A C
+ATOM 724 CG GLU A 89 -30.574 6.444 2.831 1.00 80.57 A C
+ATOM 725 CD GLU A 89 -31.327 7.658 3.315 1.00 80.57 A C
+ATOM 726 OE1 GLU A 89 -31.748 7.667 4.489 1.00 80.57 A O
+ATOM 727 OE2 GLU A 89 -31.487 8.611 2.526 1.00 80.57 A O1-
+ATOM 728 N ALA A 90 -29.470 4.448 -1.006 1.00 77.02 A N
+ATOM 729 CA ALA A 90 -29.476 3.647 -2.226 1.00 77.02 A C
+ATOM 730 C ALA A 90 -28.630 4.262 -3.335 1.00 77.02 A C
+ATOM 731 O ALA A 90 -29.135 4.583 -4.413 1.00 77.02 A O
+ATOM 732 CB ALA A 90 -30.906 3.404 -2.721 1.00 77.02 A C
+ATOM 733 N GLY A 91 -27.343 4.438 -3.062 1.00 72.99 A N
+ATOM 734 CA GLY A 91 -26.339 4.660 -4.081 1.00 72.99 A C
+ATOM 735 C GLY A 91 -25.031 4.045 -3.631 1.00 72.99 A C
+ATOM 736 O GLY A 91 -24.716 4.086 -2.440 1.00 72.99 A O
+ATOM 737 N SER A 92 -24.266 3.455 -4.541 1.00 64.46 A N
+ATOM 738 CA SER A 92 -22.996 2.862 -4.151 1.00 64.46 A C
+ATOM 739 C SER A 92 -22.023 3.942 -3.711 1.00 64.46 A C
+ATOM 740 O SER A 92 -21.922 4.999 -4.336 1.00 64.46 A O
+ATOM 741 CB SER A 92 -22.396 2.065 -5.297 1.00 64.46 A C
+ATOM 742 OG SER A 92 -21.014 1.861 -5.077 1.00 64.46 A O
+ATOM 743 N HIS A 93 -21.298 3.668 -2.631 1.00 59.50 A N
+ATOM 744 CA HIS A 93 -20.414 4.653 -2.020 1.00 59.50 A C
+ATOM 745 C HIS A 93 -19.452 3.947 -1.082 1.00 59.50 A C
+ATOM 746 O HIS A 93 -19.884 3.251 -0.166 1.00 59.50 A O
+ATOM 747 CB HIS A 93 -21.237 5.717 -1.306 1.00 59.50 A C
+ATOM 748 CG HIS A 93 -22.036 5.196 -0.161 1.00 59.50 A C
+ATOM 749 CD2 HIS A 93 -21.797 5.194 1.171 1.00 59.50 A C
+ATOM 750 ND1 HIS A 93 -23.249 4.564 -0.333 1.00 59.50 A N
+ATOM 751 CE1 HIS A 93 -23.730 4.207 0.846 1.00 59.50 A C
+ATOM 752 NE2 HIS A 93 -22.864 4.571 1.775 1.00 59.50 A N
+ATOM 753 N THR A 94 -18.159 4.140 -1.305 1.00 48.70 A N
+ATOM 754 CA THR A 94 -17.092 3.320 -0.754 1.00 48.70 A C
+ATOM 755 C THR A 94 -16.489 3.954 0.511 1.00 48.70 A C
+ATOM 756 O THR A 94 -16.695 5.128 0.807 1.00 48.70 A O
+ATOM 757 CB THR A 94 -16.043 3.117 -1.844 1.00 48.70 A C
+ATOM 758 CG2 THR A 94 -14.920 2.229 -1.398 1.00 48.70 A C
+ATOM 759 OG1 THR A 94 -16.664 2.472 -2.955 1.00 48.70 A O
+ATOM 760 N ILE A 95 -15.775 3.138 1.286 1.00 42.33 A N
+ATOM 761 CA ILE A 95 -14.922 3.575 2.387 1.00 42.33 A C
+ATOM 762 C ILE A 95 -13.614 2.831 2.262 1.00 42.33 A C
+ATOM 763 O ILE A 95 -13.599 1.651 1.918 1.00 42.33 A O
+ATOM 764 CB ILE A 95 -15.527 3.294 3.768 1.00 42.33 A C
+ATOM 765 CG1 ILE A 95 -16.490 4.375 4.167 1.00 42.33 A C
+ATOM 766 CG2 ILE A 95 -14.468 3.244 4.810 1.00 42.33 A C
+ATOM 767 CD1 ILE A 95 -17.167 4.046 5.418 1.00 42.33 A C
+ATOM 768 N GLN A 96 -12.515 3.504 2.533 1.00 45.20 A N
+ATOM 769 CA GLN A 96 -11.223 2.858 2.461 1.00 45.20 A C
+ATOM 770 C GLN A 96 -10.440 3.228 3.707 1.00 45.20 A C
+ATOM 771 O GLN A 96 -10.642 4.298 4.276 1.00 45.20 A O
+ATOM 772 CB GLN A 96 -10.496 3.292 1.226 1.00 45.20 A C
+ATOM 773 CG GLN A 96 -11.171 2.843 -0.027 1.00 45.20 A C
+ATOM 774 CD GLN A 96 -10.449 3.307 -1.238 1.00 45.20 A C
+ATOM 775 NE2 GLN A 96 -10.973 2.996 -2.380 1.00 45.20 A N
+ATOM 776 OE1 GLN A 96 -9.469 4.010 -1.144 1.00 45.20 A O
+ATOM 777 N ILE A 97 -9.570 2.326 4.149 1.00 42.37 A N
+ATOM 778 CA ILE A 97 -8.834 2.518 5.391 1.00 42.37 A C
+ATOM 779 C ILE A 97 -7.461 1.889 5.277 1.00 42.37 A C
+ATOM 780 O ILE A 97 -7.331 0.721 4.914 1.00 42.37 A O
+ATOM 781 CB ILE A 97 -9.585 1.939 6.595 1.00 42.37 A C
+ATOM 782 CG1 ILE A 97 -10.758 2.821 6.972 1.00 42.37 A C
+ATOM 783 CG2 ILE A 97 -8.668 1.822 7.749 1.00 42.37 A C
+ATOM 784 CD1 ILE A 97 -11.808 2.122 7.730 1.00 42.37 A C
+ATOM 785 N MET A 98 -6.430 2.666 5.538 1.00 51.05 A N
+ATOM 786 CA MET A 98 -5.092 2.126 5.660 1.00 51.05 A C
+ATOM 787 C MET A 98 -4.691 2.081 7.138 1.00 51.05 A C
+ATOM 788 O MET A 98 -5.337 2.675 7.998 1.00 51.05 A O
+ATOM 789 CB MET A 98 -4.150 2.977 4.826 1.00 51.05 A C
+ATOM 790 CG MET A 98 -2.862 2.359 4.399 1.00 51.05 A C
+ATOM 791 SD MET A 98 -1.717 3.718 4.127 1.00 51.05 A S
+ATOM 792 CE MET A 98 -0.229 2.851 3.682 1.00 51.05 A C
+ATOM 793 N TYR A 99 -3.646 1.318 7.443 1.00 53.08 A N
+ATOM 794 CA TYR A 99 -2.969 1.363 8.731 1.00 53.08 A C
+ATOM 795 C TYR A 99 -1.496 1.318 8.429 1.00 53.08 A C
+ATOM 796 O TYR A 99 -1.087 1.351 7.268 1.00 53.08 A O
+ATOM 797 CB TYR A 99 -3.213 0.168 9.646 1.00 53.08 A C
+ATOM 798 CG TYR A 99 -4.555 0.021 10.267 1.00 53.08 A C
+ATOM 799 CD1 TYR A 99 -5.314 1.111 10.603 1.00 53.08 A C
+ATOM 800 CD2 TYR A 99 -5.029 -1.236 10.580 1.00 53.08 A C
+ATOM 801 CE1 TYR A 99 -6.538 0.946 11.178 1.00 53.08 A C
+ATOM 802 CE2 TYR A 99 -6.231 -1.407 11.146 1.00 53.08 A C
+ATOM 803 CZ TYR A 99 -6.987 -0.319 11.449 1.00 53.08 A C
+ATOM 804 OH TYR A 99 -8.208 -0.501 12.032 1.00 53.08 A O
+ATOM 805 N GLY A 100 -0.689 1.169 9.463 1.00 61.46 A N
+ATOM 806 CA GLY A 100 0.679 0.788 9.204 1.00 61.46 A C
+ATOM 807 C GLY A 100 1.554 0.995 10.407 1.00 61.46 A C
+ATOM 808 O GLY A 100 1.136 1.521 11.435 1.00 61.46 A O
+ATOM 809 N CYS A 101 2.789 0.545 10.249 1.00 71.41 A N
+ATOM 810 CA CYS A 101 3.857 0.872 11.178 1.00 71.41 A C
+ATOM 811 C CYS A 101 5.149 0.496 10.489 1.00 71.41 A C
+ATOM 812 O CYS A 101 5.328 -0.660 10.105 1.00 71.41 A O
+ATOM 813 CB CYS A 101 3.705 0.134 12.506 1.00 71.41 A C
+ATOM 814 SG CYS A 101 3.842 -1.655 12.458 1.00 71.41 A S
+ATOM 815 N ASP A 102 6.026 1.460 10.293 1.00 73.43 A N
+ATOM 816 CA ASP A 102 7.319 1.127 9.735 1.00 73.43 A C
+ATOM 817 C ASP A 102 8.251 0.648 10.832 1.00 73.43 A C
+ATOM 818 O ASP A 102 7.939 0.702 12.019 1.00 73.43 A O
+ATOM 819 CB ASP A 102 7.922 2.319 9.008 1.00 73.43 A C
+ATOM 820 CG ASP A 102 7.500 2.388 7.564 1.00 73.43 A C
+ATOM 821 OD1 ASP A 102 7.826 1.449 6.816 1.00 73.43 A O
+ATOM 822 OD2 ASP A 102 6.853 3.378 7.174 1.00 73.43 A O1-
+ATOM 823 N VAL A 103 9.403 0.148 10.417 1.00 75.80 A N
+ATOM 824 CA VAL A 103 10.473 -0.217 11.318 1.00 75.80 A C
+ATOM 825 C VAL A 103 11.788 0.188 10.667 1.00 75.80 A C
+ATOM 826 O VAL A 103 11.829 0.602 9.511 1.00 75.80 A O
+ATOM 827 CB VAL A 103 10.486 -1.722 11.638 1.00 75.80 A C
+ATOM 828 CG1 VAL A 103 9.331 -2.078 12.551 1.00 75.80 A C
+ATOM 829 CG2 VAL A 103 10.480 -2.529 10.367 1.00 75.80 A C
+ATOM 830 N GLY A 104 12.854 0.060 11.406 1.00 87.65 A N
+ATOM 831 CA GLY A 104 14.160 0.179 10.808 1.00 87.65 A C
+ATOM 832 C GLY A 104 14.643 -1.155 10.305 1.00 87.65 A C
+ATOM 833 O GLY A 104 14.046 -2.191 10.576 1.00 87.65 A O
+ATOM 834 N SER A 105 15.746 -1.116 9.559 1.00 95.51 A N
+ATOM 835 CA SER A 105 16.368 -2.361 9.124 1.00 95.51 A C
+ATOM 836 C SER A 105 16.808 -3.193 10.317 1.00 95.51 A C
+ATOM 837 O SER A 105 16.896 -4.422 10.223 1.00 95.51 A O
+ATOM 838 CB SER A 105 17.557 -2.073 8.207 1.00 95.51 A C
+ATOM 839 OG SER A 105 18.543 -1.301 8.870 1.00 95.51 A O
+ATOM 840 N ASP A 106 17.093 -2.540 11.441 1.00 89.51 A N
+ATOM 841 CA ASP A 106 17.390 -3.232 12.685 1.00 89.51 A C
+ATOM 842 C ASP A 106 16.141 -3.660 13.442 1.00 89.51 A C
+ATOM 843 O ASP A 106 16.253 -4.452 14.381 1.00 89.51 A O
+ATOM 844 CB ASP A 106 18.235 -2.334 13.581 1.00 89.51 A C
+ATOM 845 CG ASP A 106 17.474 -1.125 14.057 1.00 89.51 A C
+ATOM 846 OD1 ASP A 106 17.192 -0.242 13.226 1.00 89.51 A O
+ATOM 847 OD2 ASP A 106 17.146 -1.064 15.257 1.00 89.51 A O1-
+ATOM 848 N GLY A 107 14.967 -3.158 13.069 1.00 83.89 A N
+ATOM 849 CA GLY A 107 13.734 -3.509 13.739 1.00 83.89 A C
+ATOM 850 C GLY A 107 13.174 -2.459 14.672 1.00 83.89 A C
+ATOM 851 O GLY A 107 12.288 -2.777 15.472 1.00 83.89 A O
+ATOM 852 N ARG A 108 13.657 -1.228 14.605 1.00 84.78 A N
+ATOM 853 CA ARG A 108 13.194 -0.166 15.482 1.00 84.78 A C
+ATOM 854 C ARG A 108 12.062 0.600 14.813 1.00 84.78 A C
+ATOM 855 O ARG A 108 12.132 0.896 13.619 1.00 84.78 A O
+ATOM 856 CB ARG A 108 14.367 0.753 15.829 1.00 84.78 A C
+ATOM 857 CG ARG A 108 14.099 1.856 16.844 1.00 84.78 A C
+ATOM 858 CD ARG A 108 13.709 3.146 16.148 1.00 84.78 A C
+ATOM 859 NE ARG A 108 13.494 4.255 17.075 1.00 84.78 A N
+ATOM 860 CZ ARG A 108 12.354 4.499 17.709 1.00 84.78 A C
+ATOM 861 NH1 ARG A 108 11.304 3.710 17.529 1.00 84.78 A N1+
+ATOM 862 NH2 ARG A 108 12.264 5.539 18.527 1.00 84.78 A N
+ATOM 863 N PHE A 109 11.024 0.911 15.585 1.00 76.13 A N
+ATOM 864 CA PHE A 109 9.864 1.621 15.061 1.00 76.13 A C
+ATOM 865 C PHE A 109 10.253 2.958 14.446 1.00 76.13 A C
+ATOM 866 O PHE A 109 10.990 3.739 15.049 1.00 76.13 A O
+ATOM 867 CB PHE A 109 8.867 1.867 16.176 1.00 76.13 A C
+ATOM 868 CG PHE A 109 7.807 2.831 15.810 1.00 76.13 A C
+ATOM 869 CD1 PHE A 109 6.748 2.441 15.040 1.00 76.13 A C
+ATOM 870 CD2 PHE A 109 7.891 4.145 16.201 1.00 76.13 A C
+ATOM 871 CE1 PHE A 109 5.773 3.328 14.698 1.00 76.13 A C
+ATOM 872 CE2 PHE A 109 6.922 5.034 15.853 1.00 76.13 A C
+ATOM 873 CZ PHE A 109 5.863 4.622 15.095 1.00 76.13 A C
+ATOM 874 N LEU A 110 9.714 3.248 13.266 1.00 76.61 A N
+ATOM 875 CA LEU A 110 10.032 4.476 12.549 1.00 76.61 A C
+ATOM 876 C LEU A 110 8.878 5.458 12.500 1.00 76.61 A C
+ATOM 877 O LEU A 110 8.963 6.550 13.063 1.00 76.61 A O
+ATOM 878 CB LEU A 110 10.463 4.148 11.121 1.00 76.61 A C
+ATOM 879 CG LEU A 110 11.860 3.578 10.977 1.00 76.61 A C
+ATOM 880 CD1 LEU A 110 12.301 3.705 9.545 1.00 76.61 A C
+ATOM 881 CD2 LEU A 110 12.801 4.327 11.887 1.00 76.61 A C
+ATOM 882 N ARG A 111 7.790 5.087 11.842 1.00 71.89 A N
+ATOM 883 CA ARG A 111 6.665 5.976 11.630 1.00 71.89 A C
+ATOM 884 C ARG A 111 5.379 5.174 11.662 1.00 71.89 A C
+ATOM 885 O ARG A 111 5.359 3.986 11.337 1.00 71.89 A O
+ATOM 886 CB ARG A 111 6.794 6.710 10.302 1.00 71.89 A C
+ATOM 887 CG ARG A 111 7.240 5.805 9.186 1.00 71.89 A C
+ATOM 888 CD ARG A 111 7.613 6.566 7.939 1.00 71.89 A C
+ATOM 889 NE ARG A 111 6.700 7.675 7.692 1.00 71.89 A N
+ATOM 890 CZ ARG A 111 5.587 7.562 6.974 1.00 71.89 A C
+ATOM 891 NH1 ARG A 111 4.802 8.609 6.789 1.00 71.89 A N1+
+ATOM 892 NH2 ARG A 111 5.261 6.395 6.433 1.00 71.89 A N
+ATOM 893 N GLY A 112 4.306 5.836 12.065 1.00 68.53 A N
+ATOM 894 CA GLY A 112 3.002 5.218 12.078 1.00 68.53 A C
+ATOM 895 C GLY A 112 2.009 6.042 11.300 1.00 68.53 A C
+ATOM 896 O GLY A 112 1.943 7.261 11.458 1.00 68.53 A O
+ATOM 897 N TYR A 113 1.247 5.393 10.432 1.00 62.39 A N
+ATOM 898 CA TYR A 113 0.309 6.080 9.561 1.00 62.39 A C
+ATOM 899 C TYR A 113 -1.026 5.370 9.548 1.00 62.39 A C
+ATOM 900 O TYR A 113 -1.087 4.170 9.293 1.00 62.39 A O
+ATOM 901 CB TYR A 113 0.870 6.189 8.145 1.00 62.39 A C
+ATOM 902 CG TYR A 113 1.539 4.938 7.654 1.00 62.39 A C
+ATOM 903 CD1 TYR A 113 0.837 3.966 6.987 1.00 62.39 A C
+ATOM 904 CD2 TYR A 113 2.895 4.763 7.807 1.00 62.39 A C
+ATOM 905 CE1 TYR A 113 1.461 2.831 6.531 1.00 62.39 A C
+ATOM 906 CE2 TYR A 113 3.526 3.640 7.349 1.00 62.39 A C
+ATOM 907 CZ TYR A 113 2.803 2.674 6.709 1.00 62.39 A C
+ATOM 908 OH TYR A 113 3.415 1.540 6.240 1.00 62.39 A O
+ATOM 909 N ARG A 114 -2.085 6.115 9.835 1.00 55.70 A N
+ATOM 910 CA ARG A 114 -3.452 5.690 9.580 1.00 55.70 A C
+ATOM 911 C ARG A 114 -4.147 6.716 8.707 1.00 55.70 A C
+ATOM 912 O ARG A 114 -4.211 7.891 9.068 1.00 55.70 A O
+ATOM 913 CB ARG A 114 -4.251 5.502 10.857 1.00 55.70 A C
+ATOM 914 CG ARG A 114 -5.590 4.997 10.480 1.00 55.70 A C
+ATOM 915 CD ARG A 114 -6.485 4.675 11.607 1.00 55.70 A C
+ATOM 916 NE ARG A 114 -7.754 4.234 11.062 1.00 55.70 A N
+ATOM 917 CZ ARG A 114 -8.780 3.848 11.797 1.00 55.70 A C
+ATOM 918 NH1 ARG A 114 -9.902 3.464 11.217 1.00 55.70 A N1+
+ATOM 919 NH2 ARG A 114 -8.670 3.833 13.111 1.00 55.70 A N
+ATOM 920 N GLN A 115 -4.691 6.270 7.578 1.00 50.80 A N
+ATOM 921 CA GLN A 115 -5.281 7.144 6.574 1.00 50.80 A C
+ATOM 922 C GLN A 115 -6.660 6.624 6.197 1.00 50.80 A C
+ATOM 923 O GLN A 115 -6.804 5.445 5.877 1.00 50.80 A O
+ATOM 924 CB GLN A 115 -4.368 7.182 5.356 1.00 50.80 A C
+ATOM 925 CG GLN A 115 -3.047 7.837 5.657 1.00 50.80 A C
+ATOM 926 CD GLN A 115 -2.116 7.857 4.475 1.00 50.80 A C
+ATOM 927 NE2 GLN A 115 -0.942 8.448 4.658 1.00 50.80 A N
+ATOM 928 OE1 GLN A 115 -2.428 7.317 3.416 1.00 50.80 A O
+ATOM 929 N ASP A 116 -7.664 7.491 6.211 1.00 52.46 A N
+ATOM 930 CA ASP A 116 -9.015 7.105 5.833 1.00 52.46 A C
+ATOM 931 C ASP A 116 -9.433 7.836 4.572 1.00 52.46 A C
+ATOM 932 O ASP A 116 -9.050 8.989 4.361 1.00 52.46 A O
+ATOM 933 CB ASP A 116 -10.032 7.419 6.906 1.00 52.46 A C
+ATOM 934 CG ASP A 116 -9.860 6.572 8.114 1.00 52.46 A C
+ATOM 935 OD1 ASP A 116 -9.376 5.442 7.972 1.00 52.46 A O
+ATOM 936 OD2 ASP A 116 -10.265 7.002 9.210 1.00 52.46 A O1-
+ATOM 937 N ALA A 117 -10.212 7.163 3.731 1.00 47.91 A N
+ATOM 938 CA ALA A 117 -10.749 7.754 2.517 1.00 47.91 A C
+ATOM 939 C ALA A 117 -12.204 7.379 2.340 1.00 47.91 A C
+ATOM 940 O ALA A 117 -12.551 6.200 2.395 1.00 47.91 A O
+ATOM 941 CB ALA A 117 -9.984 7.293 1.303 1.00 47.91 A C
+ATOM 942 N TYR A 118 -13.037 8.370 2.093 1.00 52.88 A N
+ATOM 943 CA TYR A 118 -14.416 8.172 1.683 1.00 52.88 A C
+ATOM 944 C TYR A 118 -14.391 8.009 0.171 1.00 52.88 A C
+ATOM 945 O TYR A 118 -13.430 7.445 -0.356 1.00 52.88 A O
+ATOM 946 CB TYR A 118 -15.298 9.307 2.141 1.00 52.88 A C
+ATOM 947 CG TYR A 118 -16.731 8.983 1.983 1.00 52.88 A C
+ATOM 948 CD1 TYR A 118 -17.181 7.722 2.255 1.00 52.88 A C
+ATOM 949 CD2 TYR A 118 -17.644 9.938 1.646 1.00 52.88 A C
+ATOM 950 CE1 TYR A 118 -18.488 7.406 2.142 1.00 52.88 A C
+ATOM 951 CE2 TYR A 118 -18.965 9.638 1.542 1.00 52.88 A C
+ATOM 952 CZ TYR A 118 -19.385 8.367 1.788 1.00 52.88 A C
+ATOM 953 OH TYR A 118 -20.715 8.057 1.677 1.00 52.88 A O
+ATOM 954 N ASP A 119 -15.429 8.437 -0.545 1.00 58.87 A N
+ATOM 955 CA ASP A 119 -15.522 8.134 -1.970 1.00 58.87 A C
+ATOM 956 C ASP A 119 -14.331 8.678 -2.726 1.00 58.87 A C
+ATOM 957 O ASP A 119 -14.387 9.780 -3.270 1.00 58.87 A O
+ATOM 958 CB ASP A 119 -16.768 8.773 -2.582 1.00 58.87 A C
+ATOM 959 CG ASP A 119 -18.048 8.138 -2.132 1.00 58.87 A C
+ATOM 960 OD1 ASP A 119 -18.042 6.938 -1.839 1.00 58.87 A O
+ATOM 961 OD2 ASP A 119 -19.063 8.851 -2.033 1.00 58.87 A O1-
+ATOM 962 N GLY A 120 -13.241 7.922 -2.738 1.00 55.11 A N
+ATOM 963 CA GLY A 120 -12.095 8.211 -3.579 1.00 55.11 A C
+ATOM 964 C GLY A 120 -11.191 9.340 -3.132 1.00 55.11 A C
+ATOM 965 O GLY A 120 -10.007 9.355 -3.470 1.00 55.11 A O
+ATOM 966 N LYS A 121 -11.722 10.270 -2.356 1.00 61.78 A N
+ATOM 967 CA LYS A 121 -10.995 11.450 -1.912 1.00 61.78 A C
+ATOM 968 C LYS A 121 -10.262 11.113 -0.614 1.00 61.78 A C
+ATOM 969 O LYS A 121 -10.027 9.942 -0.314 1.00 61.78 A O
+ATOM 970 CB LYS A 121 -11.960 12.626 -1.767 1.00 61.78 A C
+ATOM 971 CG LYS A 121 -13.161 12.314 -0.905 1.00 61.78 A C
+ATOM 972 CD LYS A 121 -14.312 13.256 -1.190 1.00 61.78 A C
+ATOM 973 CE LYS A 121 -15.505 12.924 -0.310 1.00 61.78 A C
+ATOM 974 NZ LYS A 121 -15.086 12.709 1.103 1.00 61.78 A N1+
+ATOM 975 N ASP A 122 -9.822 12.118 0.133 1.00 58.61 A N
+ATOM 976 CA ASP A 122 -9.230 11.906 1.443 1.00 58.61 A C
+ATOM 977 C ASP A 122 -10.207 12.219 2.559 1.00 58.61 A C
+ATOM 978 O ASP A 122 -11.231 12.878 2.374 1.00 58.61 A O
+ATOM 979 CB ASP A 122 -7.971 12.742 1.651 1.00 58.61 A C
+ATOM 980 CG ASP A 122 -6.827 12.271 0.817 1.00 58.61 A C
+ATOM 981 OD1 ASP A 122 -6.775 11.064 0.528 1.00 58.61 A O
+ATOM 982 OD2 ASP A 122 -5.975 13.096 0.448 1.00 58.61 A O1-
+ATOM 983 N TYR A 123 -9.849 11.715 3.731 1.00 53.92 A N
+ATOM 984 CA TYR A 123 -10.565 11.903 4.973 1.00 53.92 A C
+ATOM 985 C TYR A 123 -9.489 12.025 6.043 1.00 53.92 A C
+ATOM 986 O TYR A 123 -8.313 12.202 5.720 1.00 53.92 A O
+ATOM 987 CB TYR A 123 -11.549 10.746 5.167 1.00 53.92 A C
+ATOM 988 CG TYR A 123 -12.705 11.053 6.057 1.00 53.92 A C
+ATOM 989 CD1 TYR A 123 -13.805 11.708 5.574 1.00 53.92 A C
+ATOM 990 CD2 TYR A 123 -12.717 10.640 7.364 1.00 53.92 A C
+ATOM 991 CE1 TYR A 123 -14.853 11.975 6.373 1.00 53.92 A C
+ATOM 992 CE2 TYR A 123 -13.759 10.902 8.164 1.00 53.92 A C
+ATOM 993 CZ TYR A 123 -14.824 11.571 7.669 1.00 53.92 A C
+ATOM 994 OH TYR A 123 -15.888 11.834 8.480 1.00 53.92 A O
+ATOM 995 N ILE A 124 -9.871 11.937 7.318 1.00 51.88 A N
+ATOM 996 CA ILE A 124 -8.901 12.046 8.403 1.00 51.88 A C
+ATOM 997 C ILE A 124 -7.673 11.174 8.171 1.00 51.88 A C
+ATOM 998 O ILE A 124 -7.756 10.074 7.619 1.00 51.88 A O
+ATOM 999 CB ILE A 124 -9.567 11.692 9.736 1.00 51.88 A C
+ATOM 1000 CG1 ILE A 124 -8.629 12.032 10.869 1.00 51.88 A C
+ATOM 1001 CG2 ILE A 124 -9.899 10.251 9.772 1.00 51.88 A C
+ATOM 1002 CD1 ILE A 124 -9.287 12.014 12.171 1.00 51.88 A C
+ATOM 1003 N ALA A 125 -6.511 11.694 8.560 1.00 57.44 A N
+ATOM 1004 CA ALA A 125 -5.252 10.964 8.493 1.00 57.44 A C
+ATOM 1005 C ALA A 125 -4.415 11.293 9.721 1.00 57.44 A C
+ATOM 1006 O ALA A 125 -4.660 12.280 10.415 1.00 57.44 A O
+ATOM 1007 CB ALA A 125 -4.471 11.285 7.219 1.00 57.44 A C
+ATOM 1008 N LEU A 126 -3.428 10.451 9.990 1.00 62.80 A N
+ATOM 1009 CA LEU A 126 -2.595 10.581 11.175 1.00 62.80 A C
+ATOM 1010 C LEU A 126 -1.256 11.213 10.823 1.00 62.80 A C
+ATOM 1011 O LEU A 126 -0.712 10.982 9.742 1.00 62.80 A O
+ATOM 1012 CB LEU A 126 -2.368 9.217 11.817 1.00 62.80 A C
+ATOM 1013 CG LEU A 126 -1.543 9.087 13.094 1.00 62.80 A C
+ATOM 1014 CD1 LEU A 126 -2.271 9.702 14.251 1.00 62.80 A C
+ATOM 1015 CD2 LEU A 126 -1.301 7.635 13.373 1.00 62.80 A C
+ATOM 1016 N ASN A 127 -0.702 11.971 11.765 1.00 70.19 A N
+ATOM 1017 CA ASN A 127 0.542 12.691 11.532 1.00 70.19 A C
+ATOM 1018 C ASN A 127 1.717 11.730 11.580 1.00 70.19 A C
+ATOM 1019 O ASN A 127 1.562 10.509 11.508 1.00 70.19 A O
+ATOM 1020 CB ASN A 127 0.721 13.803 12.550 1.00 70.19 A C
+ATOM 1021 CG ASN A 127 -0.359 14.823 12.472 1.00 70.19 A C
+ATOM 1022 ND2 ASN A 127 -0.787 15.323 13.614 1.00 70.19 A N
+ATOM 1023 OD1 ASN A 127 -0.806 15.164 11.395 1.00 70.19 A O
+ATOM 1024 N GLU A 128 2.922 12.279 11.663 1.00 76.15 A N
+ATOM 1025 CA GLU A 128 4.114 11.459 11.788 1.00 76.15 A C
+ATOM 1026 C GLU A 128 4.659 11.439 13.205 1.00 76.15 A C
+ATOM 1027 O GLU A 128 5.510 10.600 13.516 1.00 76.15 A O
+ATOM 1028 CB GLU A 128 5.189 11.956 10.825 1.00 76.15 A C
+ATOM 1029 CG GLU A 128 4.935 11.515 9.405 1.00 76.15 A C
+ATOM 1030 CD GLU A 128 4.515 10.064 9.323 1.00 76.15 A C
+ATOM 1031 OE1 GLU A 128 3.303 9.787 9.199 1.00 76.15 A O
+ATOM 1032 OE2 GLU A 128 5.403 9.196 9.372 1.00 76.15 A O1-
+ATOM 1033 N ASP A 129 4.183 12.331 14.063 1.00 77.18 A N
+ATOM 1034 CA ASP A 129 4.456 12.291 15.489 1.00 77.18 A C
+ATOM 1035 C ASP A 129 3.454 11.432 16.240 1.00 77.18 A C
+ATOM 1036 O ASP A 129 3.543 11.322 17.466 1.00 77.18 A O
+ATOM 1037 CB ASP A 129 4.458 13.711 16.048 1.00 77.18 A C
+ATOM 1038 CG ASP A 129 3.118 14.396 15.897 1.00 77.18 A C
+ATOM 1039 OD1 ASP A 129 2.194 13.792 15.324 1.00 77.18 A O
+ATOM 1040 OD2 ASP A 129 2.986 15.553 16.341 1.00 77.18 A O1-
+ATOM 1041 N LEU A 130 2.503 10.839 15.520 1.00 70.64 A N
+ATOM 1042 CA LEU A 130 1.531 9.904 16.067 1.00 70.64 A C
+ATOM 1043 C LEU A 130 0.700 10.548 17.159 1.00 70.64 A C
+ATOM 1044 O LEU A 130 0.415 9.934 18.182 1.00 70.64 A O
+ATOM 1045 CB LEU A 130 2.212 8.660 16.609 1.00 70.64 A C
+ATOM 1046 CG LEU A 130 2.904 7.834 15.553 1.00 70.64 A C
+ATOM 1047 CD1 LEU A 130 3.629 6.717 16.218 1.00 70.64 A C
+ATOM 1048 CD2 LEU A 130 1.903 7.333 14.617 1.00 70.64 A C
+ATOM 1049 N ARG A 131 0.325 11.802 16.962 1.00 75.73 A N
+ATOM 1050 CA ARG A 131 -0.308 12.501 18.069 1.00 75.73 A C
+ATOM 1051 C ARG A 131 -1.517 13.339 17.696 1.00 75.73 A C
+ATOM 1052 O ARG A 131 -2.330 13.619 18.581 1.00 75.73 A O
+ATOM 1053 CB ARG A 131 0.730 13.375 18.774 1.00 75.73 A C
+ATOM 1054 CG ARG A 131 0.334 13.889 20.135 1.00 75.73 A C
+ATOM 1055 CD ARG A 131 1.576 14.299 20.918 1.00 75.73 A C
+ATOM 1056 NE ARG A 131 2.252 13.136 21.498 1.00 75.73 A N
+ATOM 1057 CZ ARG A 131 3.217 12.443 20.901 1.00 75.73 A C
+ATOM 1058 NH1 ARG A 131 3.759 11.401 21.513 1.00 75.73 A N1+
+ATOM 1059 NH2 ARG A 131 3.652 12.791 19.700 1.00 75.73 A N
+ATOM 1060 N SER A 132 -1.696 13.744 16.445 1.00 68.39 A N
+ATOM 1061 CA SER A 132 -2.869 14.526 16.097 1.00 68.39 A C
+ATOM 1062 C SER A 132 -3.344 14.144 14.701 1.00 68.39 A C
+ATOM 1063 O SER A 132 -2.741 13.315 14.012 1.00 68.39 A O
+ATOM 1064 CB SER A 132 -2.592 16.023 16.201 1.00 68.39 A C
+ATOM 1065 OG SER A 132 -3.743 16.755 15.829 1.00 68.39 A O
+ATOM 1066 N TRP A 133 -4.436 14.777 14.284 1.00 60.92 A N
+ATOM 1067 CA TRP A 133 -5.229 14.329 13.151 1.00 60.92 A C
+ATOM 1068 C TRP A 133 -5.410 15.475 12.171 1.00 60.92 A C
+ATOM 1069 O TRP A 133 -5.940 16.525 12.541 1.00 60.92 A O
+ATOM 1070 CB TRP A 133 -6.586 13.833 13.641 1.00 60.92 A C
+ATOM 1071 CG TRP A 133 -6.496 12.634 14.535 1.00 60.92 A C
+ATOM 1072 CD1 TRP A 133 -6.814 12.577 15.857 1.00 60.92 A C
+ATOM 1073 CD2 TRP A 133 -6.112 11.305 14.160 1.00 60.92 A C
+ATOM 1074 CE2 TRP A 133 -6.198 10.507 15.308 1.00 60.92 A C
+ATOM 1075 CE3 TRP A 133 -5.701 10.717 12.967 1.00 60.92 A C
+ATOM 1076 NE1 TRP A 133 -6.621 11.307 16.336 1.00 60.92 A N
+ATOM 1077 CZ2 TRP A 133 -5.889 9.160 15.294 1.00 60.92 A C
+ATOM 1078 CZ3 TRP A 133 -5.407 9.385 12.963 1.00 60.92 A C
+ATOM 1079 CH2 TRP A 133 -5.493 8.622 14.112 1.00 60.92 A C
+ATOM 1080 N THR A 134 -5.003 15.264 10.919 1.00 61.11 A N
+ATOM 1081 CA THR A 134 -5.239 16.234 9.848 1.00 61.11 A C
+ATOM 1082 C THR A 134 -6.538 15.899 9.142 1.00 61.11 A C
+ATOM 1083 O THR A 134 -6.564 15.174 8.151 1.00 61.11 A O
+ATOM 1084 CB THR A 134 -4.102 16.247 8.841 1.00 61.11 A C
+ATOM 1085 CG2 THR A 134 -2.791 16.118 9.523 1.00 61.11 A C
+ATOM 1086 OG1 THR A 134 -4.275 15.163 7.924 1.00 61.11 A O
+ATOM 1087 N ALA A 135 -7.627 16.450 9.645 1.00 63.15 A N
+ATOM 1088 CA ALA A 135 -8.868 16.411 8.903 1.00 63.15 A C
+ATOM 1089 C ALA A 135 -8.681 17.090 7.558 1.00 63.15 A C
+ATOM 1090 O ALA A 135 -8.031 18.132 7.459 1.00 63.15 A O
+ATOM 1091 CB ALA A 135 -9.955 17.111 9.691 1.00 63.15 A C
+ATOM 1092 N ALA A 136 -9.264 16.506 6.516 1.00 67.55 A N
+ATOM 1093 CA ALA A 136 -9.098 17.003 5.157 1.00 67.55 A C
+ATOM 1094 C ALA A 136 -10.425 17.433 4.552 1.00 67.55 A C
+ATOM 1095 O ALA A 136 -10.576 17.447 3.332 1.00 67.55 A O
+ATOM 1096 CB ALA A 136 -8.430 15.955 4.272 1.00 67.55 A C
+ATOM 1097 N ASP A 137 -11.380 17.805 5.392 1.00 75.60 A N
+ATOM 1098 CA ASP A 137 -12.691 18.241 4.933 1.00 75.60 A C
+ATOM 1099 C ASP A 137 -13.481 18.687 6.152 1.00 75.60 A C
+ATOM 1100 O ASP A 137 -13.014 18.581 7.287 1.00 75.60 A O
+ATOM 1101 CB ASP A 137 -13.395 17.126 4.173 1.00 75.60 A C
+ATOM 1102 CG ASP A 137 -14.649 17.590 3.509 1.00 75.60 A C
+ATOM 1103 OD1 ASP A 137 -14.994 18.777 3.670 1.00 75.60 A O
+ATOM 1104 OD2 ASP A 137 -15.266 16.787 2.787 1.00 75.60 A O1-
+ATOM 1105 N MET A 138 -14.686 19.191 5.913 1.00 75.22 A N
+ATOM 1106 CA MET A 138 -15.578 19.596 6.991 1.00 75.22 A C
+ATOM 1107 C MET A 138 -16.330 18.433 7.610 1.00 75.22 A C
+ATOM 1108 O MET A 138 -17.158 18.651 8.498 1.00 75.22 A O
+ATOM 1109 CB MET A 138 -16.565 20.650 6.497 1.00 75.22 A C
+ATOM 1110 CG MET A 138 -15.914 21.959 6.106 1.00 75.22 A C
+ATOM 1111 SD MET A 138 -15.077 22.742 7.501 1.00 75.22 A S
+ATOM 1112 CE MET A 138 -13.361 22.350 7.165 1.00 75.22 A C
+ATOM 1113 N ALA A 139 -16.065 17.214 7.160 1.00 68.70 A N
+ATOM 1114 CA ALA A 139 -16.510 16.005 7.833 1.00 68.70 A C
+ATOM 1115 C ALA A 139 -15.405 15.361 8.643 1.00 68.70 A C
+ATOM 1116 O ALA A 139 -15.620 14.959 9.800 1.00 68.70 A O
+ATOM 1117 CB ALA A 139 -17.030 15.001 6.807 1.00 68.70 A C
+ATOM 1118 N ALA A 140 -14.211 15.277 8.068 1.00 66.90 A N
+ATOM 1119 CA ALA A 140 -13.077 14.807 8.838 1.00 66.90 A C
+ATOM 1120 C ALA A 140 -12.792 15.721 10.019 1.00 66.90 A C
+ATOM 1121 O ALA A 140 -12.118 15.297 10.953 1.00 66.90 A O
+ATOM 1122 CB ALA A 140 -11.857 14.662 7.938 1.00 66.90 A C
+ATOM 1123 N GLN A 141 -13.299 16.955 10.013 1.00 69.27 A N
+ATOM 1124 CA GLN A 141 -13.283 17.770 11.227 1.00 69.27 A C
+ATOM 1125 C GLN A 141 -14.011 17.068 12.362 1.00 69.27 A C
+ATOM 1126 O GLN A 141 -13.497 16.953 13.480 1.00 69.27 A O
+ATOM 1127 CB GLN A 141 -13.939 19.134 10.976 1.00 69.27 A C
+ATOM 1128 CG GLN A 141 -13.234 20.108 10.033 1.00 69.27 A C
+ATOM 1129 CD GLN A 141 -11.734 19.926 9.943 1.00 69.27 A C
+ATOM 1130 NE2 GLN A 141 -11.225 19.868 8.722 1.00 69.27 A N
+ATOM 1131 OE1 GLN A 141 -11.036 19.861 10.954 1.00 69.27 A O
+ATOM 1132 N ILE A 142 -15.234 16.618 12.093 1.00 65.40 A N
+ATOM 1133 CA ILE A 142 -16.036 15.954 13.110 1.00 65.40 A C
+ATOM 1134 C ILE A 142 -15.346 14.672 13.549 1.00 65.40 A C
+ATOM 1135 O ILE A 142 -15.269 14.348 14.746 1.00 65.40 A O
+ATOM 1136 CB ILE A 142 -17.440 15.681 12.560 1.00 65.40 A C
+ATOM 1137 CG1 ILE A 142 -18.110 16.978 12.149 1.00 65.40 A C
+ATOM 1138 CG2 ILE A 142 -18.281 15.168 13.653 1.00 65.40 A C
+ATOM 1139 CD1 ILE A 142 -19.586 16.831 11.888 1.00 65.40 A C
+ATOM 1140 N THR A 143 -14.821 13.928 12.581 1.00 61.39 A N
+ATOM 1141 CA THR A 143 -14.087 12.713 12.915 1.00 61.39 A C
+ATOM 1142 C THR A 143 -12.910 13.008 13.838 1.00 61.39 A C
+ATOM 1143 O THR A 143 -12.771 12.391 14.899 1.00 61.39 A O
+ATOM 1144 CB THR A 143 -13.616 12.012 11.652 1.00 61.39 A C
+ATOM 1145 CG2 THR A 143 -12.729 10.859 12.004 1.00 61.39 A C
+ATOM 1146 OG1 THR A 143 -14.752 11.521 10.947 1.00 61.39 A O
+ATOM 1147 N LYS A 144 -12.036 13.942 13.454 1.00 64.53 A N
+ATOM 1148 CA LYS A 144 -10.869 14.220 14.280 1.00 64.53 A C
+ATOM 1149 C LYS A 144 -11.257 14.757 15.641 1.00 64.53 A C
+ATOM 1150 O LYS A 144 -10.535 14.530 16.610 1.00 64.53 A O
+ATOM 1151 CB LYS A 144 -9.914 15.189 13.589 1.00 64.53 A C
+ATOM 1152 CG LYS A 144 -10.440 16.590 13.455 1.00 64.53 A C
+ATOM 1153 CD LYS A 144 -9.320 17.588 13.268 1.00 64.53 A C
+ATOM 1154 CE LYS A 144 -8.590 17.844 14.571 1.00 64.53 A C
+ATOM 1155 NZ LYS A 144 -9.449 18.582 15.534 1.00 64.53 A N1+
+ATOM 1156 N ARG A 145 -12.377 15.471 15.754 1.00 65.12 A N
+ATOM 1157 CA ARG A 145 -12.795 15.877 17.089 1.00 65.12 A C
+ATOM 1158 C ARG A 145 -13.132 14.663 17.938 1.00 65.12 A C
+ATOM 1159 O ARG A 145 -12.718 14.571 19.101 1.00 65.12 A O
+ATOM 1160 CB ARG A 145 -13.968 16.846 17.024 1.00 65.12 A C
+ATOM 1161 CG ARG A 145 -13.543 18.199 16.519 1.00 65.12 A C
+ATOM 1162 CD ARG A 145 -14.560 19.278 16.788 1.00 65.12 A C
+ATOM 1163 NE ARG A 145 -15.930 18.806 16.838 1.00 65.12 A N
+ATOM 1164 CZ ARG A 145 -16.783 18.915 15.833 1.00 65.12 A C
+ATOM 1165 NH1 ARG A 145 -18.021 18.474 15.971 1.00 65.12 A N1+
+ATOM 1166 NH2 ARG A 145 -16.400 19.473 14.695 1.00 65.12 A N
+ATOM 1167 N LYS A 146 -13.840 13.694 17.357 1.00 59.15 A N
+ATOM 1168 CA LYS A 146 -14.118 12.459 18.087 1.00 59.15 A C
+ATOM 1169 C LYS A 146 -12.870 11.649 18.392 1.00 59.15 A C
+ATOM 1170 O LYS A 146 -12.876 10.855 19.328 1.00 59.15 A O
+ATOM 1171 CB LYS A 146 -15.093 11.581 17.322 1.00 59.15 A C
+ATOM 1172 CG LYS A 146 -15.802 10.655 18.237 1.00 59.15 A C
+ATOM 1173 CD LYS A 146 -17.131 10.300 17.727 1.00 59.15 A C
+ATOM 1174 CE LYS A 146 -17.951 11.486 18.062 1.00 59.15 A C
+ATOM 1175 NZ LYS A 146 -18.035 11.555 19.522 1.00 59.15 A N1+
+ATOM 1176 N TRP A 147 -11.823 11.773 17.595 1.00 61.57 A N
+ATOM 1177 CA TRP A 147 -10.667 10.921 17.847 1.00 61.57 A C
+ATOM 1178 C TRP A 147 -9.674 11.587 18.795 1.00 61.57 A C
+ATOM 1179 O TRP A 147 -8.945 10.898 19.514 1.00 61.57 A O
+ATOM 1180 CB TRP A 147 -9.984 10.560 16.537 1.00 61.57 A C
+ATOM 1181 CG TRP A 147 -10.754 9.568 15.797 1.00 61.57 A C
+ATOM 1182 CD1 TRP A 147 -12.023 9.180 16.061 1.00 61.57 A C
+ATOM 1183 CD2 TRP A 147 -10.323 8.810 14.673 1.00 61.57 A C
+ATOM 1184 CE2 TRP A 147 -11.389 7.988 14.296 1.00 61.57 A C
+ATOM 1185 CE3 TRP A 147 -9.141 8.744 13.948 1.00 61.57 A C
+ATOM 1186 NE1 TRP A 147 -12.423 8.235 15.158 1.00 61.57 A N
+ATOM 1187 CZ2 TRP A 147 -11.309 7.114 13.231 1.00 61.57 A C
+ATOM 1188 CZ3 TRP A 147 -9.067 7.873 12.885 1.00 61.57 A C
+ATOM 1189 CH2 TRP A 147 -10.141 7.072 12.538 1.00 61.57 A C
+ATOM 1190 N GLU A 148 -9.584 12.920 18.766 1.00 66.50 A N
+ATOM 1191 CA GLU A 148 -8.836 13.635 19.792 1.00 66.50 A C
+ATOM 1192 C GLU A 148 -9.492 13.472 21.149 1.00 66.50 A C
+ATOM 1193 O GLU A 148 -8.810 13.201 22.141 1.00 66.50 A O
+ATOM 1194 CB GLU A 148 -8.703 15.118 19.446 1.00 66.50 A C
+ATOM 1195 CG GLU A 148 -7.860 15.439 18.227 1.00 66.50 A C
+ATOM 1196 CD GLU A 148 -6.391 15.101 18.414 1.00 66.50 A C
+ATOM 1197 OE1 GLU A 148 -5.894 15.190 19.556 1.00 66.50 A O
+ATOM 1198 OE2 GLU A 148 -5.717 14.791 17.411 1.00 66.50 A O1-
+ATOM 1199 N ALA A 149 -10.811 13.625 21.217 1.00 67.27 A N
+ATOM 1200 CA ALA A 149 -11.518 13.072 22.358 1.00 67.27 A C
+ATOM 1201 C ALA A 149 -11.375 11.562 22.297 1.00 67.27 A C
+ATOM 1202 O ALA A 149 -11.077 11.000 21.247 1.00 67.27 A O
+ATOM 1203 CB ALA A 149 -12.987 13.481 22.345 1.00 67.27 A C
+ATOM 1204 N ALA A 150 -11.485 10.905 23.439 1.00 70.70 A N
+ATOM 1205 CA ALA A 150 -11.430 9.450 23.537 1.00 70.70 A C
+ATOM 1206 C ALA A 150 -10.059 8.901 23.177 1.00 70.70 A C
+ATOM 1207 O ALA A 150 -9.897 7.686 23.154 1.00 70.70 A O
+ATOM 1208 CB ALA A 150 -12.469 8.738 22.661 1.00 70.70 A C
+ATOM 1209 N HIS A 151 -9.092 9.746 22.850 1.00 71.26 A N
+ATOM 1210 CA HIS A 151 -7.688 9.358 22.798 1.00 71.26 A C
+ATOM 1211 C HIS A 151 -7.478 8.148 21.906 1.00 71.26 A C
+ATOM 1212 O HIS A 151 -6.593 7.326 22.137 1.00 71.26 A O
+ATOM 1213 CB HIS A 151 -7.159 9.104 24.205 1.00 71.26 A C
+ATOM 1214 CG HIS A 151 -7.498 10.196 25.165 1.00 71.26 A C
+ATOM 1215 CD2 HIS A 151 -8.387 10.244 26.181 1.00 71.26 A C
+ATOM 1216 ND1 HIS A 151 -6.945 11.453 25.081 1.00 71.26 A N
+ATOM 1217 CE1 HIS A 151 -7.451 12.218 26.029 1.00 71.26 A C
+ATOM 1218 NE2 HIS A 151 -8.331 11.509 26.710 1.00 71.26 A N
+ATOM 1219 N GLU A 152 -8.295 8.050 20.862 1.00 70.11 A N
+ATOM 1220 CA GLU A 152 -8.085 7.000 19.876 1.00 70.11 A C
+ATOM 1221 C GLU A 152 -6.673 7.073 19.321 1.00 70.11 A C
+ATOM 1222 O GLU A 152 -6.088 6.054 18.946 1.00 70.11 A O
+ATOM 1223 CB GLU A 152 -9.117 7.121 18.760 1.00 70.11 A C
+ATOM 1224 CG GLU A 152 -9.017 6.051 17.679 1.00 70.11 A C
+ATOM 1225 CD GLU A 152 -9.091 4.635 18.218 1.00 70.11 A C
+ATOM 1226 OE1 GLU A 152 -10.211 4.168 18.497 1.00 70.11 A O
+ATOM 1227 OE2 GLU A 152 -8.033 3.985 18.354 1.00 70.11 A O1-
+ATOM 1228 N ALA A 153 -6.108 8.278 19.279 1.00 69.60 A N
+ATOM 1229 CA ALA A 153 -4.709 8.429 18.910 1.00 69.60 A C
+ATOM 1230 C ALA A 153 -3.802 7.699 19.886 1.00 69.60 A C
+ATOM 1231 O ALA A 153 -2.817 7.076 19.481 1.00 69.60 A O
+ATOM 1232 CB ALA A 153 -4.345 9.907 18.853 1.00 69.60 A C
+ATOM 1233 N GLU A 154 -4.119 7.763 21.179 1.00 77.76 A N
+ATOM 1234 CA GLU A 154 -3.274 7.122 22.181 1.00 77.76 A C
+ATOM 1235 C GLU A 154 -3.301 5.608 22.037 1.00 77.76 A C
+ATOM 1236 O GLU A 154 -2.247 4.959 21.948 1.00 77.76 A O
+ATOM 1237 CB GLU A 154 -3.751 7.520 23.575 1.00 77.76 A C
+ATOM 1238 CG GLU A 154 -3.583 8.986 23.891 1.00 77.76 A C
+ATOM 1239 CD GLU A 154 -2.140 9.397 23.972 1.00 77.76 A C
+ATOM 1240 OE1 GLU A 154 -1.311 8.557 24.377 1.00 77.76 A O
+ATOM 1241 OE2 GLU A 154 -1.832 10.559 23.632 1.00 77.76 A O1-
+ATOM 1242 N GLN A 155 -4.505 5.035 21.990 1.00 75.13 A N
+ATOM 1243 CA GLN A 155 -4.650 3.598 21.808 1.00 75.13 A C
+ATOM 1244 C GLN A 155 -3.993 3.165 20.518 1.00 75.13 A C
+ATOM 1245 O GLN A 155 -3.294 2.149 20.474 1.00 75.13 A O
+ATOM 1246 CB GLN A 155 -6.131 3.236 21.771 1.00 75.13 A C
+ATOM 1247 CG GLN A 155 -6.863 3.457 23.062 1.00 75.13 A C
+ATOM 1248 CD GLN A 155 -8.326 3.103 22.950 1.00 75.13 A C
+ATOM 1249 NE2 GLN A 155 -9.048 3.206 24.057 1.00 75.13 A N
+ATOM 1250 OE1 GLN A 155 -8.805 2.752 21.877 1.00 75.13 A O
+ATOM 1251 N LEU A 156 -4.217 3.932 19.455 1.00 70.78 A N
+ATOM 1252 CA LEU A 156 -3.650 3.592 18.165 1.00 70.78 A C
+ATOM 1253 C LEU A 156 -2.137 3.612 18.209 1.00 70.78 A C
+ATOM 1254 O LEU A 156 -1.500 2.728 17.646 1.00 70.78 A O
+ATOM 1255 CB LEU A 156 -4.173 4.540 17.091 1.00 70.78 A C
+ATOM 1256 CG LEU A 156 -3.639 4.201 15.705 1.00 70.78 A C
+ATOM 1257 CD1 LEU A 156 -4.225 2.904 15.211 1.00 70.78 A C
+ATOM 1258 CD2 LEU A 156 -3.947 5.318 14.750 1.00 70.78 A C
+ATOM 1259 N ARG A 157 -1.539 4.607 18.870 1.00 73.13 A N
+ATOM 1260 CA ARG A 157 -0.084 4.646 18.956 1.00 73.13 A C
+ATOM 1261 C ARG A 157 0.444 3.458 19.730 1.00 73.13 A C
+ATOM 1262 O ARG A 157 1.480 2.880 19.375 1.00 73.13 A O
+ATOM 1263 CB ARG A 157 0.394 5.927 19.625 1.00 73.13 A C
+ATOM 1264 CG ARG A 157 1.914 6.063 19.588 1.00 73.13 A C
+ATOM 1265 CD ARG A 157 2.412 7.288 20.330 1.00 73.13 A C
+ATOM 1266 NE ARG A 157 1.880 7.359 21.682 1.00 73.13 A N
+ATOM 1267 CZ ARG A 157 1.064 8.310 22.116 1.00 73.13 A C
+ATOM 1268 NH1 ARG A 157 0.671 9.274 21.300 1.00 73.13 A N1+
+ATOM 1269 NH2 ARG A 157 0.640 8.291 23.366 1.00 73.13 A N
+ATOM 1270 N ALA A 158 -0.238 3.108 20.822 1.00 69.79 A N
+ATOM 1271 CA ALA A 158 0.135 1.917 21.571 1.00 69.79 A C
+ATOM 1272 C ALA A 158 0.119 0.698 20.668 1.00 69.79 A C
+ATOM 1273 O ALA A 158 1.100 -0.055 20.604 1.00 69.79 A O
+ATOM 1274 CB ALA A 158 -0.812 1.727 22.751 1.00 69.79 A C
+ATOM 1275 N TYR A 159 -0.992 0.498 19.951 1.00 69.23 A N
+ATOM 1276 CA TYR A 159 -1.049 -0.554 18.945 1.00 69.23 A C
+ATOM 1277 C TYR A 159 0.159 -0.473 18.037 1.00 69.23 A C
+ATOM 1278 O TYR A 159 0.954 -1.412 17.969 1.00 69.23 A O
+ATOM 1279 CB TYR A 159 -2.313 -0.475 18.092 1.00 69.23 A C
+ATOM 1280 CG TYR A 159 -2.155 -1.350 16.856 1.00 69.23 A C
+ATOM 1281 CD1 TYR A 159 -1.711 -2.660 16.960 1.00 69.23 A C
+ATOM 1282 CD2 TYR A 159 -2.343 -0.837 15.582 1.00 69.23 A C
+ATOM 1283 CE1 TYR A 159 -1.514 -3.437 15.848 1.00 69.23 A C
+ATOM 1284 CE2 TYR A 159 -2.144 -1.618 14.464 1.00 69.23 A C
+ATOM 1285 CZ TYR A 159 -1.731 -2.912 14.610 1.00 69.23 A C
+ATOM 1286 OH TYR A 159 -1.533 -3.697 13.511 1.00 69.23 A O
+ATOM 1287 N LEU A 160 0.318 0.663 17.361 1.00 72.15 A N
+ATOM 1288 CA LEU A 160 1.307 0.838 16.315 1.00 72.15 A C
+ATOM 1289 C LEU A 160 2.647 0.380 16.840 1.00 72.15 A C
+ATOM 1290 O LEU A 160 3.049 -0.749 16.540 1.00 72.15 A O
+ATOM 1291 CB LEU A 160 1.358 2.296 15.870 1.00 72.15 A C
+ATOM 1292 CG LEU A 160 0.074 2.755 15.176 1.00 72.15 A C
+ATOM 1293 CD1 LEU A 160 0.155 4.162 14.639 1.00 72.15 A C
+ATOM 1294 CD2 LEU A 160 -0.316 1.800 14.087 1.00 72.15 A C
+ATOM 1295 N ASP A 161 3.207 1.124 17.793 1.00 73.96 A N
+ATOM 1296 CA ASP A 161 4.530 0.778 18.294 1.00 73.96 A C
+ATOM 1297 C ASP A 161 4.527 -0.641 18.837 1.00 73.96 A C
+ATOM 1298 O ASP A 161 4.997 -1.555 18.155 1.00 73.96 A O
+ATOM 1299 CB ASP A 161 4.986 1.755 19.381 1.00 73.96 A C
+ATOM 1300 CG ASP A 161 4.772 3.198 18.992 1.00 73.96 A C
+ATOM 1301 OD1 ASP A 161 4.866 3.502 17.786 1.00 73.96 A O
+ATOM 1302 OD2 ASP A 161 4.510 4.028 19.888 1.00 73.96 A O1-
+ATOM 1303 N GLY A 162 3.855 -0.872 19.962 1.00 76.55 A N
+ATOM 1304 CA GLY A 162 4.022 -2.157 20.603 1.00 76.55 A C
+ATOM 1305 C GLY A 162 3.566 -3.309 19.740 1.00 76.55 A C
+ATOM 1306 O GLY A 162 4.387 -4.068 19.215 1.00 76.55 A O
+ATOM 1307 N THR A 163 2.257 -3.415 19.542 1.00 74.09 A N
+ATOM 1308 CA THR A 163 1.724 -4.599 18.895 1.00 74.09 A C
+ATOM 1309 C THR A 163 2.142 -4.652 17.440 1.00 74.09 A C
+ATOM 1310 O THR A 163 2.582 -5.699 16.952 1.00 74.09 A O
+ATOM 1311 CB THR A 163 0.207 -4.613 19.003 1.00 74.09 A C
+ATOM 1312 CG2 THR A 163 -0.310 -5.995 18.727 1.00 74.09 A C
+ATOM 1313 OG1 THR A 163 -0.185 -4.212 20.319 1.00 74.09 A O
+ATOM 1314 N CYS A 164 1.970 -3.546 16.711 1.00 74.22 A N
+ATOM 1315 CA CYS A 164 2.216 -3.621 15.288 1.00 74.22 A C
+ATOM 1316 C CYS A 164 3.680 -3.925 15.035 1.00 74.22 A C
+ATOM 1317 O CYS A 164 4.000 -4.770 14.197 1.00 74.22 A O
+ATOM 1318 CB CYS A 164 1.793 -2.320 14.617 1.00 74.22 A C
+ATOM 1319 SG CYS A 164 1.963 -2.315 12.830 1.00 74.22 A S
+ATOM 1320 N VAL A 165 4.588 -3.294 15.791 1.00 73.77 A N
+ATOM 1321 CA VAL A 165 6.006 -3.532 15.596 1.00 73.77 A C
+ATOM 1322 C VAL A 165 6.375 -4.957 15.979 1.00 73.77 A C
+ATOM 1323 O VAL A 165 7.139 -5.619 15.270 1.00 73.77 A O
+ATOM 1324 CB VAL A 165 6.813 -2.499 16.395 1.00 73.77 A C
+ATOM 1325 CG1 VAL A 165 8.251 -2.935 16.524 1.00 73.77 A C
+ATOM 1326 CG2 VAL A 165 6.725 -1.154 15.720 1.00 73.77 A C
+ATOM 1327 N GLU A 166 5.846 -5.460 17.098 1.00 78.13 A N
+ATOM 1328 CA GLU A 166 6.257 -6.791 17.527 1.00 78.13 A C
+ATOM 1329 C GLU A 166 5.738 -7.860 16.575 1.00 78.13 A C
+ATOM 1330 O GLU A 166 6.493 -8.752 16.178 1.00 78.13 A O
+ATOM 1331 CB GLU A 166 5.808 -7.069 18.962 1.00 78.13 A C
+ATOM 1332 CG GLU A 166 4.313 -7.148 19.181 1.00 78.13 A C
+ATOM 1333 CD GLU A 166 3.945 -7.361 20.637 1.00 78.13 A C
+ATOM 1334 OE1 GLU A 166 4.863 -7.420 21.479 1.00 78.13 A O
+ATOM 1335 OE2 GLU A 166 2.739 -7.480 20.937 1.00 78.13 A O1-
+ATOM 1336 N TRP A 167 4.464 -7.786 16.174 1.00 71.28 A N
+ATOM 1337 CA TRP A 167 4.001 -8.745 15.180 1.00 71.28 A C
+ATOM 1338 C TRP A 167 4.667 -8.552 13.840 1.00 71.28 A C
+ATOM 1339 O TRP A 167 4.850 -9.529 13.116 1.00 71.28 A O
+ATOM 1340 CB TRP A 167 2.496 -8.716 14.980 1.00 71.28 A C
+ATOM 1341 CG TRP A 167 1.711 -9.229 16.090 1.00 71.28 A C
+ATOM 1342 CD1 TRP A 167 1.093 -8.526 17.060 1.00 71.28 A C
+ATOM 1343 CD2 TRP A 167 1.484 -10.606 16.374 1.00 71.28 A C
+ATOM 1344 CE2 TRP A 167 0.686 -10.666 17.520 1.00 71.28 A C
+ATOM 1345 CE3 TRP A 167 1.867 -11.794 15.753 1.00 71.28 A C
+ATOM 1346 NE1 TRP A 167 0.454 -9.380 17.924 1.00 71.28 A N
+ATOM 1347 CZ2 TRP A 167 0.267 -11.864 18.066 1.00 71.28 A C
+ATOM 1348 CZ3 TRP A 167 1.454 -12.975 16.290 1.00 71.28 A C
+ATOM 1349 CH2 TRP A 167 0.662 -13.007 17.437 1.00 71.28 A C
+ATOM 1350 N LEU A 168 5.018 -7.327 13.471 1.00 70.90 A N
+ATOM 1351 CA LEU A 168 5.722 -7.153 12.215 1.00 70.90 A C
+ATOM 1352 C LEU A 168 7.071 -7.847 12.273 1.00 70.90 A C
+ATOM 1353 O LEU A 168 7.449 -8.563 11.339 1.00 70.90 A O
+ATOM 1354 CB LEU A 168 5.848 -5.667 11.901 1.00 70.90 A C
+ATOM 1355 CG LEU A 168 6.189 -5.217 10.483 1.00 70.90 A C
+ATOM 1356 CD1 LEU A 168 5.818 -3.775 10.346 1.00 70.90 A C
+ATOM 1357 CD2 LEU A 168 7.648 -5.343 10.216 1.00 70.90 A C
+ATOM 1358 N ARG A 169 7.789 -7.696 13.386 1.00 75.02 A N
+ATOM 1359 CA ARG A 169 9.056 -8.404 13.533 1.00 75.02 A C
+ATOM 1360 C ARG A 169 8.846 -9.908 13.555 1.00 75.02 A C
+ATOM 1361 O ARG A 169 9.642 -10.659 12.984 1.00 75.02 A O
+ATOM 1362 CB ARG A 169 9.797 -7.932 14.783 1.00 75.02 A C
+ATOM 1363 CG ARG A 169 10.262 -6.502 14.673 1.00 75.02 A C
+ATOM 1364 CD ARG A 169 10.987 -6.008 15.904 1.00 75.02 A C
+ATOM 1365 NE ARG A 169 10.113 -5.889 17.059 1.00 75.02 A N
+ATOM 1366 CZ ARG A 169 10.542 -5.600 18.280 1.00 75.02 A C
+ATOM 1367 NH1 ARG A 169 9.684 -5.500 19.282 1.00 75.02 A N1+
+ATOM 1368 NH2 ARG A 169 11.831 -5.400 18.492 1.00 75.02 A N
+ATOM 1369 N ARG A 170 7.773 -10.369 14.193 1.00 74.46 A N
+ATOM 1370 CA ARG A 170 7.473 -11.794 14.168 1.00 74.46 A C
+ATOM 1371 C ARG A 170 7.252 -12.279 12.746 1.00 74.46 A C
+ATOM 1372 O ARG A 170 7.783 -13.316 12.345 1.00 74.46 A O
+ATOM 1373 CB ARG A 170 6.247 -12.102 15.015 1.00 74.46 A C
+ATOM 1374 CG ARG A 170 5.959 -13.589 15.076 1.00 74.46 A C
+ATOM 1375 CD ARG A 170 5.495 -14.031 16.440 1.00 74.46 A C
+ATOM 1376 NE ARG A 170 4.811 -15.315 16.360 1.00 74.46 A N
+ATOM 1377 CZ ARG A 170 3.831 -15.686 17.172 1.00 74.46 A C
+ATOM 1378 NH1 ARG A 170 3.264 -16.874 17.026 1.00 74.46 A N1+
+ATOM 1379 NH2 ARG A 170 3.415 -14.865 18.126 1.00 74.46 A N
+ATOM 1380 N TYR A 171 6.447 -11.549 11.977 1.00 67.55 A N
+ATOM 1381 CA TYR A 171 6.157 -11.947 10.608 1.00 67.55 A C
+ATOM 1382 C TYR A 171 7.436 -12.032 9.798 1.00 67.55 A C
+ATOM 1383 O TYR A 171 7.688 -13.022 9.105 1.00 67.55 A O
+ATOM 1384 CB TYR A 171 5.198 -10.951 9.971 1.00 67.55 A C
+ATOM 1385 CG TYR A 171 3.796 -10.996 10.498 1.00 67.55 A C
+ATOM 1386 CD1 TYR A 171 3.272 -12.142 11.041 1.00 67.55 A C
+ATOM 1387 CD2 TYR A 171 2.996 -9.883 10.443 1.00 67.55 A C
+ATOM 1388 CE1 TYR A 171 1.988 -12.171 11.515 1.00 67.55 A C
+ATOM 1389 CE2 TYR A 171 1.724 -9.900 10.917 1.00 67.55 A C
+ATOM 1390 CZ TYR A 171 1.220 -11.042 11.450 1.00 67.55 A C
+ATOM 1391 OH TYR A 171 -0.070 -11.042 11.916 1.00 67.55 A O
+ATOM 1392 N LEU A 172 8.266 -10.998 9.889 1.00 74.70 A N
+ATOM 1393 CA LEU A 172 9.543 -11.019 9.188 1.00 74.70 A C
+ATOM 1394 C LEU A 172 10.465 -12.113 9.699 1.00 74.70 A C
+ATOM 1395 O LEU A 172 11.348 -12.565 8.963 1.00 74.70 A O
+ATOM 1396 CB LEU A 172 10.190 -9.645 9.272 1.00 74.70 A C
+ATOM 1397 CG LEU A 172 9.605 -8.635 8.288 1.00 74.70 A C
+ATOM 1398 CD1 LEU A 172 8.225 -8.172 8.587 1.00 74.70 A C
+ATOM 1399 CD2 LEU A 172 10.497 -7.438 8.340 1.00 74.70 A C
+ATOM 1400 N GLU A 173 10.291 -12.554 10.943 1.00 77.94 A N
+ATOM 1401 CA GLU A 173 10.958 -13.780 11.366 1.00 77.94 A C
+ATOM 1402 C GLU A 173 10.392 -14.981 10.621 1.00 77.94 A C
+ATOM 1403 O GLU A 173 11.136 -15.823 10.107 1.00 77.94 A O
+ATOM 1404 CB GLU A 173 10.815 -13.971 12.875 1.00 77.94 A C
+ATOM 1405 CG GLU A 173 11.658 -13.029 13.705 1.00 77.94 A C
+ATOM 1406 CD GLU A 173 13.142 -13.255 13.506 1.00 77.94 A C
+ATOM 1407 OE1 GLU A 173 13.535 -14.394 13.175 1.00 77.94 A O
+ATOM 1408 OE2 GLU A 173 13.919 -12.295 13.682 1.00 77.94 A O1-
+ATOM 1409 N ASN A 174 9.068 -15.056 10.540 1.00 80.68 A N
+ATOM 1410 CA ASN A 174 8.408 -16.196 9.926 1.00 80.68 A C
+ATOM 1411 C ASN A 174 8.652 -16.218 8.427 1.00 80.68 A C
+ATOM 1412 O ASN A 174 8.991 -17.260 7.856 1.00 80.68 A O
+ATOM 1413 CB ASN A 174 6.914 -16.132 10.224 1.00 80.68 A C
+ATOM 1414 CG ASN A 174 6.604 -16.388 11.679 1.00 80.68 A C
+ATOM 1415 ND2 ASN A 174 5.685 -15.610 12.231 1.00 80.68 A N
+ATOM 1416 OD1 ASN A 174 7.185 -17.273 12.305 1.00 80.68 A O
+ATOM 1417 N GLY A 175 8.488 -15.072 7.774 1.00 82.06 A N
+ATOM 1418 CA GLY A 175 8.629 -14.983 6.338 1.00 82.06 A C
+ATOM 1419 C GLY A 175 10.034 -14.626 5.912 1.00 82.06 A C
+ATOM 1420 O GLY A 175 10.243 -14.164 4.788 1.00 82.06 A O
+ATOM 1421 N LYS A 176 11.006 -14.840 6.801 1.00 83.45 A N
+ATOM 1422 CA LYS A 176 12.359 -14.351 6.558 1.00 83.45 A C
+ATOM 1423 C LYS A 176 12.925 -14.868 5.245 1.00 83.45 A C
+ATOM 1424 O LYS A 176 13.815 -14.242 4.661 1.00 83.45 A O
+ATOM 1425 CB LYS A 176 13.278 -14.738 7.719 1.00 83.45 A C
+ATOM 1426 CG LYS A 176 13.648 -16.206 7.767 1.00 83.45 A C
+ATOM 1427 CD LYS A 176 14.757 -16.473 8.770 1.00 83.45 A C
+ATOM 1428 CE LYS A 176 14.279 -16.258 10.191 1.00 83.45 A C
+ATOM 1429 NZ LYS A 176 13.014 -16.987 10.455 1.00 83.45 A N1+
+ATOM 1430 N GLU A 177 12.426 -15.995 4.766 1.00 85.04 A N
+ATOM 1431 CA GLU A 177 12.836 -16.539 3.485 1.00 85.04 A C
+ATOM 1432 C GLU A 177 11.938 -16.077 2.349 1.00 85.04 A C
+ATOM 1433 O GLU A 177 12.143 -16.493 1.206 1.00 85.04 A O
+ATOM 1434 CB GLU A 177 12.847 -18.068 3.555 1.00 85.04 A C
+ATOM 1435 CG GLU A 177 11.478 -18.697 3.809 1.00 85.04 A C
+ATOM 1436 CD GLU A 177 11.009 -18.590 5.257 1.00 85.04 A C
+ATOM 1437 OE1 GLU A 177 11.385 -17.623 5.953 1.00 85.04 A O
+ATOM 1438 OE2 GLU A 177 10.259 -19.484 5.702 1.00 85.04 A O1-
+ATOM 1439 N THR A 178 10.959 -15.236 2.636 1.00 85.43 A N
+ATOM 1440 CA THR A 178 10.027 -14.739 1.635 1.00 85.43 A C
+ATOM 1441 C THR A 178 10.073 -13.232 1.472 1.00 85.43 A C
+ATOM 1442 O THR A 178 10.058 -12.737 0.345 1.00 85.43 A O
+ATOM 1443 CB THR A 178 8.599 -15.161 2.002 1.00 85.43 A C
+ATOM 1444 CG2 THR A 178 7.580 -14.459 1.121 1.00 85.43 A C
+ATOM 1445 OG1 THR A 178 8.463 -16.579 1.853 1.00 85.43 A O
+ATOM 1446 N LEU A 179 10.150 -12.488 2.572 1.00 83.70 A N
+ATOM 1447 CA LEU A 179 10.075 -11.035 2.529 1.00 83.70 A C
+ATOM 1448 C LEU A 179 11.441 -10.384 2.345 1.00 83.70 A C
+ATOM 1449 O LEU A 179 11.612 -9.538 1.464 1.00 83.70 A O
+ATOM 1450 CB LEU A 179 9.410 -10.520 3.803 1.00 83.70 A C
+ATOM 1451 CG LEU A 179 7.887 -10.483 3.822 1.00 83.70 A C
+ATOM 1452 CD1 LEU A 179 7.280 -11.853 3.682 1.00 83.70 A C
+ATOM 1453 CD2 LEU A 179 7.484 -9.891 5.124 1.00 83.70 A C
+ATOM 1454 N GLN A 180 12.425 -10.774 3.149 1.00 88.06 A N
+ATOM 1455 CA GLN A 180 13.760 -10.208 3.011 1.00 88.06 A C
+ATOM 1456 C GLN A 180 14.409 -10.711 1.736 1.00 88.06 A C
+ATOM 1457 O GLN A 180 15.399 -11.446 1.780 1.00 88.06 A O
+ATOM 1458 CB GLN A 180 14.638 -10.562 4.207 1.00 88.06 A C
+ATOM 1459 CG GLN A 180 14.395 -9.712 5.425 1.00 88.06 A C
+ATOM 1460 CD GLN A 180 13.175 -10.151 6.191 1.00 88.06 A C
+ATOM 1461 NE2 GLN A 180 12.693 -9.291 7.071 1.00 88.06 A N
+ATOM 1462 OE1 GLN A 180 12.673 -11.255 5.999 1.00 88.06 A O
+TER
+ATOM 1463 N VAL C 7 -1.168 -8.421 12.323 1.00 62.09 C N
+ATOM 1464 CA VAL C 7 -2.337 -8.531 13.180 1.00 62.09 C C
+ATOM 1465 C VAL C 7 -3.206 -7.297 13.123 1.00 62.09 C C
+ATOM 1466 O VAL C 7 -2.745 -6.190 13.330 1.00 62.09 C O
+ATOM 1467 CB VAL C 7 -1.927 -8.820 14.615 1.00 62.09 C C
+ATOM 1468 CG1 VAL C 7 -3.135 -8.819 15.519 1.00 62.09 C C
+ATOM 1469 CG2 VAL C 7 -1.287 -10.163 14.674 1.00 62.09 C C
+ATOM 1470 N VAL C 8 -4.486 -7.510 12.852 1.00 55.61 C N
+ATOM 1471 CA VAL C 8 -5.437 -6.418 12.840 1.00 55.61 C C
+ATOM 1472 C VAL C 8 -5.515 -5.799 14.225 1.00 55.61 C C
+ATOM 1473 O VAL C 8 -5.212 -6.441 15.231 1.00 55.61 C O
+ATOM 1474 CB VAL C 8 -6.806 -6.940 12.370 1.00 55.61 C C
+ATOM 1475 CG1 VAL C 8 -7.790 -5.812 12.158 1.00 55.61 C C
+ATOM 1476 CG2 VAL C 8 -6.627 -7.715 11.090 1.00 55.61 C C
+ATOM 1477 N VAL C 9 -5.892 -4.522 14.272 1.00 53.29 C N
+ATOM 1478 CA VAL C 9 -6.095 -3.849 15.543 1.00 53.29 C C
+ATOM 1479 C VAL C 9 -7.011 -4.679 16.416 1.00 53.29 C C
+ATOM 1480 O VAL C 9 -8.117 -5.048 16.008 1.00 53.29 C O
+ATOM 1481 CB VAL C 9 -6.669 -2.448 15.314 1.00 53.29 C C
+ATOM 1482 CG1 VAL C 9 -6.985 -1.801 16.620 1.00 53.29 C C
+ATOM 1483 CG2 VAL C 9 -5.655 -1.621 14.608 1.00 53.29 C C
+ATOM 1484 N GLY C 10 -6.549 -4.988 17.624 1.00 51.28 C N
+ATOM 1485 CA GLY C 10 -7.301 -5.884 18.476 1.00 51.28 C C
+ATOM 1486 C GLY C 10 -8.629 -5.308 18.913 1.00 51.28 C C
+ATOM 1487 O GLY C 10 -9.672 -5.938 18.742 1.00 51.28 C O
+ATOM 1488 N ALA C 11 -8.618 -4.100 19.449 1.00 51.50 C N
+ATOM 1489 CA ALA C 11 -9.825 -3.571 20.057 1.00 51.50 C C
+ATOM 1490 C ALA C 11 -10.823 -3.144 18.997 1.00 51.50 C C
+ATOM 1491 O ALA C 11 -10.464 -2.839 17.860 1.00 51.50 C O
+ATOM 1492 CB ALA C 11 -9.496 -2.393 20.964 1.00 51.50 C C
+ATOM 1493 N ASP C 12 -12.092 -3.176 19.381 1.00 0.00 C N
+ATOM 1494 CA ASP C 12 -13.065 -2.457 18.563 1.00 0.00 C C
+ATOM 1495 C ASP C 12 -12.748 -0.982 18.516 1.00 0.00 C C
+ATOM 1496 O ASP C 12 -12.796 -0.317 19.560 1.00 0.00 C O
+ATOM 1497 CB ASP C 12 -14.507 -2.699 19.092 1.00 0.00 C C
+ATOM 1498 CG ASP C 12 -15.056 -4.119 18.889 1.00 0.00 C C
+ATOM 1499 OD1 ASP C 12 -14.479 -5.078 19.442 1.00 0.00 C O
+ATOM 1500 OD2 ASP C 12 -16.040 -4.277 18.137 1.00 0.00 C O1-
+ATOM 1501 N GLY C 13 -12.361 -0.510 17.363 1.00 60.45 C N
+ATOM 1502 CA GLY C 13 -11.945 0.864 17.247 1.00 60.45 C C
+ATOM 1503 C GLY C 13 -13.109 1.816 17.362 1.00 60.45 C C
+ATOM 1504 O GLY C 13 -14.270 1.441 17.287 1.00 60.45 C O
+ATOM 1505 N VAL C 14 -12.783 3.083 17.576 1.00 62.38 C N
+ATOM 1506 CA VAL C 14 -13.815 4.098 17.543 1.00 62.38 C C
+ATOM 1507 C VAL C 14 -14.376 4.208 16.136 1.00 62.38 C C
+ATOM 1508 O VAL C 14 -13.669 4.028 15.139 1.00 62.38 C O
+ATOM 1509 CB VAL C 14 -13.270 5.431 18.084 1.00 62.38 C C
+ATOM 1510 CG1 VAL C 14 -14.305 6.538 17.980 1.00 62.38 C C
+ATOM 1511 CG2 VAL C 14 -12.890 5.260 19.542 1.00 62.38 C C
+ATOM 1512 N GLY C 15 -15.680 4.443 16.049 1.00 63.37 C N
+ATOM 1513 CA GLY C 15 -16.295 4.587 14.745 1.00 63.37 C C
+ATOM 1514 C GLY C 15 -15.786 5.827 14.034 1.00 63.37 C C
+ATOM 1515 O GLY C 15 -15.533 6.863 14.652 1.00 63.37 C O
+ATOM 1516 N LYS C 16 -15.637 5.716 12.721 1.00 59.55 C N
+ATOM 1517 CA LYS C 16 -15.237 6.848 11.898 1.00 59.55 C C
+ATOM 1518 C LYS C 16 -16.332 7.885 11.822 1.00 59.55 C C
+ATOM 1519 O LYS C 16 -16.861 8.348 12.826 1.00 59.55 C O
+ATOM 1520 CB LYS C 16 -14.869 6.391 10.490 1.00 59.55 C C
+ATOM 1521 CG LYS C 16 -13.897 7.296 9.807 1.00 59.55 C C
+ATOM 1522 CD LYS C 16 -13.608 6.839 8.422 1.00 59.55 C C
+ATOM 1523 CE LYS C 16 -13.276 5.389 8.410 1.00 59.55 C C
+ATOM 1524 NZ LYS C 16 -12.185 5.110 9.369 1.00 59.55 C N1+
+ATOM 1525 OXT LYS C 16 -16.709 8.275 10.729 1.00 59.55 C O1-
+TER
+ATOM 1526 N VAL H 2 -15.515 9.194 25.136 1.00 69.83 H N
+ATOM 1527 CA VAL H 2 -16.798 9.253 24.460 1.00 69.83 H C
+ATOM 1528 C VAL H 2 -17.886 8.603 25.306 1.00 69.83 H C
+ATOM 1529 O VAL H 2 -18.844 8.057 24.775 1.00 69.83 H O
+ATOM 1530 CB VAL H 2 -16.686 8.626 23.075 1.00 69.83 H C
+ATOM 1531 CG1 VAL H 2 -15.720 9.403 22.234 1.00 69.83 H C
+ATOM 1532 CG2 VAL H 2 -16.126 7.255 23.232 1.00 69.83 H C
+ATOM 1533 N GLN H 3 -17.746 8.692 26.627 1.00 72.01 H N
+ATOM 1534 CA GLN H 3 -18.741 8.190 27.566 1.00 72.01 H C
+ATOM 1535 C GLN H 3 -19.346 9.370 28.314 1.00 72.01 H C
+ATOM 1536 O GLN H 3 -18.630 10.263 28.770 1.00 72.01 H O
+ATOM 1537 CB GLN H 3 -18.116 7.201 28.552 1.00 72.01 H C
+ATOM 1538 CG GLN H 3 -18.081 5.764 28.049 1.00 72.01 H C
+ATOM 1539 CD GLN H 3 -17.408 5.640 26.695 1.00 72.01 H C
+ATOM 1540 NE2 GLN H 3 -18.192 5.336 25.670 1.00 72.01 H N
+ATOM 1541 OE1 GLN H 3 -16.195 5.816 26.573 1.00 72.01 H O
+ATOM 1542 N LEU H 4 -20.666 9.378 28.451 1.00 69.28 H N
+ATOM 1543 CA LEU H 4 -21.354 10.636 28.688 1.00 69.28 H C
+ATOM 1544 C LEU H 4 -22.484 10.466 29.688 1.00 69.28 H C
+ATOM 1545 O LEU H 4 -22.494 9.551 30.516 1.00 69.28 H O
+ATOM 1546 CB LEU H 4 -21.928 11.238 27.406 1.00 69.28 H C
+ATOM 1547 CG LEU H 4 -21.110 11.810 26.264 1.00 69.28 H C
+ATOM 1548 CD1 LEU H 4 -20.239 10.802 25.566 1.00 69.28 H C
+ATOM 1549 CD2 LEU H 4 -22.078 12.490 25.330 1.00 69.28 H C
+ATOM 1550 N VAL H 5 -23.414 11.419 29.621 1.00 69.85 H N
+ATOM 1551 CA VAL H 5 -24.467 11.703 30.587 1.00 69.85 H C
+ATOM 1552 C VAL H 5 -25.736 11.981 29.778 1.00 69.85 H C
+ATOM 1553 O VAL H 5 -25.829 11.598 28.610 1.00 69.85 H O
+ATOM 1554 CB VAL H 5 -24.072 12.865 31.528 1.00 69.85 H C
+ATOM 1555 CG1 VAL H 5 -25.040 13.028 32.705 1.00 69.85 H C
+ATOM 1556 CG2 VAL H 5 -22.668 12.655 32.074 1.00 69.85 H C
+ATOM 1557 N GLU H 6 -26.735 12.584 30.408 1.00 73.23 H N
+ATOM 1558 CA GLU H 6 -28.119 12.575 29.967 1.00 73.23 H C
+ATOM 1559 C GLU H 6 -28.752 13.898 30.366 1.00 73.23 H C
+ATOM 1560 O GLU H 6 -28.070 14.805 30.849 1.00 73.23 H O
+ATOM 1561 CB GLU H 6 -28.854 11.438 30.653 1.00 73.23 H C
+ATOM 1562 CG GLU H 6 -28.911 11.722 32.153 1.00 73.23 H C
+ATOM 1563 CD GLU H 6 -29.549 10.629 32.939 1.00 73.23 H C
+ATOM 1564 OE1 GLU H 6 -29.920 9.639 32.315 1.00 73.23 H O
+ATOM 1565 OE2 GLU H 6 -29.700 10.767 34.170 1.00 73.23 H O1-
+ATOM 1566 N SER H 7 -30.068 13.995 30.186 1.00 74.34 H N
+ATOM 1567 CA SER H 7 -30.872 15.093 30.728 1.00 74.34 H C
+ATOM 1568 C SER H 7 -32.342 14.809 30.445 1.00 74.34 H C
+ATOM 1569 O SER H 7 -32.711 13.709 30.023 1.00 74.34 H O
+ATOM 1570 CB SER H 7 -30.470 16.435 30.128 1.00 74.34 H C
+ATOM 1571 OG SER H 7 -30.807 16.460 28.761 1.00 74.34 H O
+ATOM 1572 N GLY H 8 -33.182 15.813 30.688 1.00 77.99 H N
+ATOM 1573 CA GLY H 8 -34.491 15.881 30.069 1.00 77.99 H C
+ATOM 1574 C GLY H 8 -35.673 16.294 30.920 1.00 77.99 H C
+ATOM 1575 O GLY H 8 -36.493 17.102 30.477 1.00 77.99 H O
+ATOM 1576 N GLY H 9 -35.772 15.788 32.134 1.00 89.06 H N
+ATOM 1577 CA GLY H 9 -36.908 16.169 32.965 1.00 89.06 H C
+ATOM 1578 C GLY H 9 -38.058 15.182 32.894 1.00 89.06 H C
+ATOM 1579 O GLY H 9 -38.299 14.523 31.884 1.00 89.06 H O
+ATOM 1580 N GLY H 10 -38.781 15.075 34.009 1.00 92.48 H N
+ATOM 1581 CA GLY H 10 -39.930 14.198 34.126 1.00 92.48 H C
+ATOM 1582 C GLY H 10 -41.194 14.912 33.707 1.00 92.48 H C
+ATOM 1583 O GLY H 10 -42.013 15.283 34.552 1.00 92.48 H O
+ATOM 1584 N LEU H 11 -41.347 15.111 32.400 1.00 99.56 H N
+ATOM 1585 CA LEU H 11 -42.351 16.014 31.852 1.00 99.56 H C
+ATOM 1586 C LEU H 11 -43.736 15.706 32.401 1.00 99.56 H C
+ATOM 1587 O LEU H 11 -44.058 14.563 32.735 1.00 99.56 H O
+ATOM 1588 CB LEU H 11 -42.359 15.909 30.329 1.00 99.56 H C
+ATOM 1589 CG LEU H 11 -43.596 16.380 29.580 1.00 99.56 H C
+ATOM 1590 CD1 LEU H 11 -43.640 17.887 29.606 1.00 99.56 H C
+ATOM 1591 CD2 LEU H 11 -43.591 15.877 28.165 1.00 99.56 H C
+ATOM 1592 N VAL H 12 -44.564 16.741 32.476 1.00106.81 H N
+ATOM 1593 CA VAL H 12 -45.911 16.625 33.009 1.00106.81 H C
+ATOM 1594 C VAL H 12 -46.875 16.824 31.854 1.00106.81 H C
+ATOM 1595 O VAL H 12 -46.447 17.021 30.715 1.00106.81 H O
+ATOM 1596 CB VAL H 12 -46.163 17.636 34.139 1.00106.81 H C
+ATOM 1597 CG1 VAL H 12 -45.075 17.519 35.199 1.00106.81 H C
+ATOM 1598 CG2 VAL H 12 -46.227 19.053 33.585 1.00106.81 H C
+ATOM 1599 N GLN H 13 -48.170 16.793 32.146 1.00116.71 H N
+ATOM 1600 CA GLN H 13 -49.230 16.558 31.172 1.00116.71 H C
+ATOM 1601 C GLN H 13 -49.105 17.390 29.900 1.00116.71 H C
+ATOM 1602 O GLN H 13 -49.266 18.616 29.922 1.00116.71 H O
+ATOM 1603 CB GLN H 13 -50.593 16.791 31.828 1.00116.71 H C
+ATOM 1604 CG GLN H 13 -51.749 16.730 30.865 1.00116.71 H C
+ATOM 1605 CD GLN H 13 -53.075 16.587 31.559 1.00116.71 H C
+ATOM 1606 NE2 GLN H 13 -53.040 16.403 32.870 1.00116.71 H N
+ATOM 1607 OE1 GLN H 13 -54.122 16.619 30.923 1.00116.71 H O
+ATOM 1608 N PRO H 14 -48.809 16.737 28.777 1.00114.35 H N
+ATOM 1609 CA PRO H 14 -48.871 17.390 27.466 1.00114.35 H C
+ATOM 1610 C PRO H 14 -50.280 17.464 26.903 1.00114.35 H C
+ATOM 1611 O PRO H 14 -50.454 17.461 25.681 1.00114.35 H O
+ATOM 1612 CB PRO H 14 -47.975 16.503 26.591 1.00114.35 H C
+ATOM 1613 CG PRO H 14 -47.994 15.199 27.263 1.00114.35 H C
+ATOM 1614 CD PRO H 14 -47.987 15.522 28.715 1.00114.35 H C
+ATOM 1615 N GLY H 15 -51.290 17.450 27.771 1.00114.55 H N
+ATOM 1616 CA GLY H 15 -52.681 17.521 27.358 1.00114.55 H C
+ATOM 1617 C GLY H 15 -52.976 18.543 26.275 1.00114.55 H C
+ATOM 1618 O GLY H 15 -54.010 18.462 25.611 1.00114.55 H O
+ATOM 1619 N GLY H 16 -52.098 19.531 26.123 1.00111.87 H N
+ATOM 1620 CA GLY H 16 -51.995 20.316 24.902 1.00111.87 H C
+ATOM 1621 C GLY H 16 -50.693 19.979 24.190 1.00111.87 H C
+ATOM 1622 O GLY H 16 -49.634 19.927 24.815 1.00111.87 H O
+ATOM 1623 N SER H 17 -50.793 19.756 22.877 1.00106.77 H N
+ATOM 1624 CA SER H 17 -49.670 19.226 22.107 1.00106.77 H C
+ATOM 1625 C SER H 17 -48.439 20.119 22.223 1.00106.77 H C
+ATOM 1626 O SER H 17 -48.526 21.341 22.074 1.00106.77 H O
+ATOM 1627 CB SER H 17 -50.079 19.068 20.645 1.00106.77 H C
+ATOM 1628 OG SER H 17 -51.105 18.100 20.512 1.00106.77 H O
+ATOM 1629 N LEU H 18 -47.288 19.503 22.477 1.00102.33 H N
+ATOM 1630 CA LEU H 18 -46.116 20.236 22.945 1.00102.33 H C
+ATOM 1631 C LEU H 18 -44.851 19.432 22.639 1.00102.33 H C
+ATOM 1632 O LEU H 18 -44.864 18.528 21.796 1.00102.33 H O
+ATOM 1633 CB LEU H 18 -46.257 20.571 24.439 1.00102.33 H C
+ATOM 1634 CG LEU H 18 -47.006 21.860 24.790 1.00102.33 H C
+ATOM 1635 CD1 LEU H 18 -47.225 21.969 26.289 1.00102.33 H C
+ATOM 1636 CD2 LEU H 18 -46.267 23.078 24.263 1.00102.33 H C
+ATOM 1637 N ARG H 19 -43.758 19.779 23.318 1.00 95.33 H N
+ATOM 1638 CA ARG H 19 -42.401 19.392 22.962 1.00 95.33 H C
+ATOM 1639 C ARG H 19 -41.654 18.899 24.197 1.00 95.33 H C
+ATOM 1640 O ARG H 19 -42.018 19.213 25.332 1.00 95.33 H O
+ATOM 1641 CB ARG H 19 -41.677 20.596 22.333 1.00 95.33 H C
+ATOM 1642 CG ARG H 19 -40.307 20.371 21.725 1.00 95.33 H C
+ATOM 1643 CD ARG H 19 -39.252 20.594 22.786 1.00 95.33 H C
+ATOM 1644 NE ARG H 19 -39.289 21.954 23.308 1.00 95.33 H N
+ATOM 1645 CZ ARG H 19 -38.706 22.326 24.443 1.00 95.33 H C
+ATOM 1646 NH1 ARG H 19 -38.058 21.434 25.179 1.00 95.33 H N1+
+ATOM 1647 NH2 ARG H 19 -38.784 23.584 24.849 1.00 95.33 H N
+ATOM 1648 N LEU H 20 -40.591 18.128 23.962 1.00 90.19 H N
+ATOM 1649 CA LEU H 20 -39.717 17.632 25.020 1.00 90.19 H C
+ATOM 1650 C LEU H 20 -38.273 17.666 24.539 1.00 90.19 H C
+ATOM 1651 O LEU H 20 -38.002 17.661 23.342 1.00 90.19 H O
+ATOM 1652 CB LEU H 20 -40.100 16.206 25.434 1.00 90.19 H C
+ATOM 1653 CG LEU H 20 -39.445 15.619 26.681 1.00 90.19 H C
+ATOM 1654 CD1 LEU H 20 -39.895 16.360 27.921 1.00 90.19 H C
+ATOM 1655 CD2 LEU H 20 -39.775 14.144 26.788 1.00 90.19 H C
+ATOM 1656 N SER H 21 -37.333 17.689 25.476 1.00 84.80 H N
+ATOM 1657 CA SER H 21 -35.945 17.843 25.075 1.00 84.80 H C
+ATOM 1658 C SER H 21 -35.032 17.112 26.039 1.00 84.80 H C
+ATOM 1659 O SER H 21 -35.352 16.948 27.216 1.00 84.80 H O
+ATOM 1660 CB SER H 21 -35.534 19.314 25.031 1.00 84.80 H C
+ATOM 1661 OG SER H 21 -35.572 19.884 26.327 1.00 84.80 H O
+ATOM 1662 N CYS H 22 -33.880 16.694 25.528 1.00 78.27 H N
+ATOM 1663 CA CYS H 22 -32.800 16.183 26.351 1.00 78.27 H C
+ATOM 1664 C CYS H 22 -31.511 16.837 25.869 1.00 78.27 H C
+ATOM 1665 O CYS H 22 -31.506 17.563 24.879 1.00 78.27 H O
+ATOM 1666 CB CYS H 22 -32.796 14.639 26.349 1.00 78.27 H C
+ATOM 1667 SG CYS H 22 -32.035 13.642 25.006 1.00 78.27 H S
+ATOM 1668 N ALA H 23 -30.417 16.638 26.594 1.00 71.14 H N
+ATOM 1669 CA ALA H 23 -29.225 17.439 26.336 1.00 71.14 H C
+ATOM 1670 C ALA H 23 -27.989 16.714 26.841 1.00 71.14 H C
+ATOM 1671 O ALA H 23 -27.977 16.220 27.966 1.00 71.14 H O
+ATOM 1672 CB ALA H 23 -29.337 18.812 27.000 1.00 71.14 H C
+ATOM 1673 N ALA H 24 -26.949 16.696 26.021 1.00 68.39 H N
+ATOM 1674 CA ALA H 24 -25.770 15.878 26.252 1.00 68.39 H C
+ATOM 1675 C ALA H 24 -24.815 16.577 27.204 1.00 68.39 H C
+ATOM 1676 O ALA H 24 -25.166 17.558 27.863 1.00 68.39 H O
+ATOM 1677 CB ALA H 24 -25.059 15.573 24.939 1.00 68.39 H C
+ATOM 1678 N SER H 25 -23.599 16.060 27.280 1.00 70.21 H N
+ATOM 1679 CA SER H 25 -22.546 16.607 28.113 1.00 70.21 H C
+ATOM 1680 C SER H 25 -21.240 15.999 27.651 1.00 70.21 H C
+ATOM 1681 O SER H 25 -21.188 15.373 26.590 1.00 70.21 H O
+ATOM 1682 CB SER H 25 -22.766 16.291 29.588 1.00 70.21 H C
+ATOM 1683 OG SER H 25 -22.402 14.952 29.855 1.00 70.21 H O
+ATOM 1684 N GLY H 26 -20.189 16.197 28.435 1.00 75.42 H N
+ATOM 1685 CA GLY H 26 -18.939 15.512 28.169 1.00 75.42 H C
+ATOM 1686 C GLY H 26 -18.476 15.808 26.767 1.00 75.42 H C
+ATOM 1687 O GLY H 26 -18.540 16.947 26.295 1.00 75.42 H O
+ATOM 1688 N PHE H 27 -18.039 14.772 26.071 1.00 78.09 H N
+ATOM 1689 CA PHE H 27 -17.557 14.966 24.718 1.00 78.09 H C
+ATOM 1690 C PHE H 27 -18.711 15.400 23.836 1.00 78.09 H C
+ATOM 1691 O PHE H 27 -19.875 15.149 24.151 1.00 78.09 H O
+ATOM 1692 CB PHE H 27 -16.923 13.686 24.191 1.00 78.09 H C
+ATOM 1693 CG PHE H 27 -15.653 13.322 24.885 1.00 78.09 H C
+ATOM 1694 CD1 PHE H 27 -14.991 14.246 25.684 1.00 78.09 H C
+ATOM 1695 CD2 PHE H 27 -15.101 12.079 24.724 1.00 78.09 H C
+ATOM 1696 CE1 PHE H 27 -13.803 13.919 26.325 1.00 78.09 H C
+ATOM 1697 CE2 PHE H 27 -13.922 11.738 25.365 1.00 78.09 H C
+ATOM 1698 CZ PHE H 27 -13.271 12.659 26.168 1.00 78.09 H C
+ATOM 1699 N ASN H 28 -18.397 16.096 22.753 1.00 77.75 H N
+ATOM 1700 CA ASN H 28 -19.386 16.364 21.721 1.00 77.75 H C
+ATOM 1701 C ASN H 28 -19.333 15.214 20.733 1.00 77.75 H C
+ATOM 1702 O ASN H 28 -18.291 14.962 20.119 1.00 77.75 H O
+ATOM 1703 CB ASN H 28 -19.124 17.679 21.004 1.00 77.75 H C
+ATOM 1704 CG ASN H 28 -20.307 18.123 20.181 1.00 77.75 H C
+ATOM 1705 ND2 ASN H 28 -20.036 18.802 19.074 1.00 77.75 H N
+ATOM 1706 OD1 ASN H 28 -21.458 17.839 20.526 1.00 77.75 H O
+ATOM 1707 N LEU H 29 -20.444 14.493 20.603 1.00 74.97 H N
+ATOM 1708 CA LEU H 29 -20.519 13.450 19.590 1.00 74.97 H C
+ATOM 1709 C LEU H 29 -20.592 14.054 18.196 1.00 74.97 H C
+ATOM 1710 O LEU H 29 -19.734 13.790 17.350 1.00 74.97 H O
+ATOM 1711 CB LEU H 29 -21.713 12.551 19.861 1.00 74.97 H C
+ATOM 1712 CG LEU H 29 -21.597 11.409 20.866 1.00 74.97 H C
+ATOM 1713 CD1 LEU H 29 -20.627 10.375 20.433 1.00 74.97 H C
+ATOM 1714 CD2 LEU H 29 -21.139 11.966 22.157 1.00 74.97 H C
+ATOM 1715 N SER H 30 -21.620 14.855 17.928 1.00 74.87 H N
+ATOM 1716 CA SER H 30 -21.714 15.686 16.738 1.00 74.87 H C
+ATOM 1717 C SER H 30 -21.653 14.897 15.454 1.00 74.87 H C
+ATOM 1718 O SER H 30 -21.679 15.497 14.388 1.00 74.87 H O
+ATOM 1719 CB SER H 30 -20.603 16.740 16.674 1.00 74.87 H C
+ATOM 1720 OG SER H 30 -20.936 17.805 15.797 1.00 74.87 H O
+ATOM 1721 N TYR H 31 -21.444 13.594 15.508 1.00 72.39 H N
+ATOM 1722 CA TYR H 31 -21.450 12.871 14.254 1.00 72.39 H C
+ATOM 1723 C TYR H 31 -22.427 11.711 14.328 1.00 72.39 H C
+ATOM 1724 O TYR H 31 -23.338 11.595 13.508 1.00 72.39 H O
+ATOM 1725 CB TYR H 31 -20.027 12.443 13.942 1.00 72.39 H C
+ATOM 1726 CG TYR H 31 -19.823 11.735 12.649 1.00 72.39 H C
+ATOM 1727 CD1 TYR H 31 -19.840 12.445 11.454 1.00 72.39 H C
+ATOM 1728 CD2 TYR H 31 -19.455 10.418 12.624 1.00 72.39 H C
+ATOM 1729 CE1 TYR H 31 -19.610 11.840 10.272 1.00 72.39 H C
+ATOM 1730 CE2 TYR H 31 -19.203 9.806 11.456 1.00 72.39 H C
+ATOM 1731 CZ TYR H 31 -19.283 10.523 10.280 1.00 72.39 H C
+ATOM 1732 OH TYR H 31 -19.044 9.891 9.089 1.00 72.39 H O
+ATOM 1733 N SER H 32 -22.234 10.853 15.321 1.00 78.73 H N
+ATOM 1734 CA SER H 32 -23.177 9.809 15.690 1.00 78.73 H C
+ATOM 1735 C SER H 32 -23.538 9.981 17.158 1.00 78.73 H C
+ATOM 1736 O SER H 32 -22.656 10.083 18.008 1.00 78.73 H O
+ATOM 1737 CB SER H 32 -22.581 8.423 15.450 1.00 78.73 H C
+ATOM 1738 OG SER H 32 -21.483 8.193 16.312 1.00 78.73 H O
+ATOM 1739 N ASP H 33 -24.828 10.068 17.445 1.00 73.64 H N
+ATOM 1740 CA ASP H 33 -25.248 10.592 18.731 1.00 73.64 H C
+ATOM 1741 C ASP H 33 -26.719 10.290 18.902 1.00 73.64 H C
+ATOM 1742 O ASP H 33 -27.181 9.240 18.469 1.00 73.64 H O
+ATOM 1743 CB ASP H 33 -24.929 12.073 18.838 1.00 73.64 H C
+ATOM 1744 CG ASP H 33 -25.205 12.658 20.205 1.00 73.64 H C
+ATOM 1745 OD1 ASP H 33 -24.445 12.414 21.155 1.00 73.64 H O
+ATOM 1746 OD2 ASP H 33 -26.212 13.372 20.317 1.00 73.64 H O1-
+ATOM 1747 N ILE H 34 -27.426 11.197 19.573 1.00 70.98 H N
+ATOM 1748 CA ILE H 34 -28.741 10.960 20.140 1.00 70.98 H C
+ATOM 1749 C ILE H 34 -29.613 10.106 19.240 1.00 70.98 H C
+ATOM 1750 O ILE H 34 -29.529 10.160 18.010 1.00 70.98 H O
+ATOM 1751 CB ILE H 34 -29.415 12.312 20.408 1.00 70.98 H C
+ATOM 1752 CG1 ILE H 34 -30.386 12.202 21.560 1.00 70.98 H C
+ATOM 1753 CG2 ILE H 34 -30.200 12.768 19.219 1.00 70.98 H C
+ATOM 1754 CD1 ILE H 34 -29.695 11.886 22.807 1.00 70.98 H C
+ATOM 1755 N HIS H 35 -30.448 9.291 19.868 1.00 69.82 H N
+ATOM 1756 CA HIS H 35 -31.241 8.297 19.176 1.00 69.82 H C
+ATOM 1757 C HIS H 35 -32.696 8.303 19.551 1.00 69.82 H C
+ATOM 1758 O HIS H 35 -33.505 7.808 18.769 1.00 69.82 H O
+ATOM 1759 CB HIS H 35 -30.700 6.902 19.462 1.00 69.82 H C
+ATOM 1760 CG HIS H 35 -29.473 6.559 18.693 1.00 69.82 H C
+ATOM 1761 CD2 HIS H 35 -28.860 7.186 17.665 1.00 69.82 H C
+ATOM 1762 ND1 HIS H 35 -28.765 5.401 18.914 1.00 69.82 H N
+ATOM 1763 CE1 HIS H 35 -27.744 5.348 18.081 1.00 69.82 H C
+ATOM 1764 NE2 HIS H 35 -27.777 6.420 17.313 1.00 69.82 H N
+ATOM 1765 N TRP H 36 -33.059 8.828 20.705 1.00 72.02 H N
+ATOM 1766 CA TRP H 36 -34.441 8.821 21.149 1.00 72.02 H C
+ATOM 1767 C TRP H 36 -35.195 7.529 20.878 1.00 72.02 H C
+ATOM 1768 O TRP H 36 -36.198 7.540 20.170 1.00 72.02 H O
+ATOM 1769 CB TRP H 36 -35.180 9.988 20.530 1.00 72.02 H C
+ATOM 1770 CG TRP H 36 -34.614 11.278 20.962 1.00 72.02 H C
+ATOM 1771 CD1 TRP H 36 -33.613 11.960 20.371 1.00 72.02 H C
+ATOM 1772 CD2 TRP H 36 -35.002 12.044 22.112 1.00 72.02 H C
+ATOM 1773 CE2 TRP H 36 -34.212 13.191 22.125 1.00 72.02 H C
+ATOM 1774 CE3 TRP H 36 -35.969 11.884 23.104 1.00 72.02 H C
+ATOM 1775 NE1 TRP H 36 -33.368 13.113 21.054 1.00 72.02 H N
+ATOM 1776 CZ2 TRP H 36 -34.346 14.173 23.091 1.00 72.02 H C
+ATOM 1777 CZ3 TRP H 36 -36.088 12.855 24.075 1.00 72.02 H C
+ATOM 1778 CH2 TRP H 36 -35.279 13.979 24.066 1.00 72.02 H C
+ATOM 1779 N VAL H 37 -34.705 6.416 21.422 1.00 61.94 H N
+ATOM 1780 CA VAL H 37 -35.480 5.184 21.476 1.00 61.94 H C
+ATOM 1781 C VAL H 37 -36.630 5.389 22.449 1.00 61.94 H C
+ATOM 1782 O VAL H 37 -36.460 6.027 23.489 1.00 61.94 H O
+ATOM 1783 CB VAL H 37 -34.581 4.021 21.910 1.00 61.94 H C
+ATOM 1784 CG1 VAL H 37 -35.376 2.791 21.973 1.00 61.94 H C
+ATOM 1785 CG2 VAL H 37 -33.409 3.845 20.970 1.00 61.94 H C
+ATOM 1786 N ARG H 38 -37.822 4.920 22.101 1.00 72.49 H N
+ATOM 1787 CA ARG H 38 -38.926 5.023 23.045 1.00 72.49 H C
+ATOM 1788 C ARG H 38 -39.425 3.652 23.484 1.00 72.49 H C
+ATOM 1789 O ARG H 38 -39.309 2.663 22.759 1.00 72.49 H O
+ATOM 1790 CB ARG H 38 -40.093 5.808 22.472 1.00 72.49 H C
+ATOM 1791 CG ARG H 38 -40.763 5.158 21.309 1.00 72.49 H C
+ATOM 1792 CD ARG H 38 -41.821 6.086 20.768 1.00 72.49 H C
+ATOM 1793 NE ARG H 38 -42.979 6.204 21.636 1.00 72.49 H N
+ATOM 1794 CZ ARG H 38 -44.028 5.403 21.556 1.00 72.49 H C
+ATOM 1795 NH1 ARG H 38 -44.053 4.456 20.638 1.00 72.49 H N1+
+ATOM 1796 NH2 ARG H 38 -45.060 5.566 22.364 1.00 72.49 H N
+ATOM 1797 N GLN H 39 -39.995 3.606 24.689 1.00 74.07 H N
+ATOM 1798 CA GLN H 39 -40.540 2.380 25.250 1.00 74.07 H C
+ATOM 1799 C GLN H 39 -41.876 2.638 25.913 1.00 74.07 H C
+ATOM 1800 O GLN H 39 -42.202 3.757 26.305 1.00 74.07 H O
+ATOM 1801 CB GLN H 39 -39.645 1.743 26.303 1.00 74.07 H C
+ATOM 1802 CG GLN H 39 -38.331 1.230 25.836 1.00 74.07 H C
+ATOM 1803 CD GLN H 39 -37.890 0.073 26.699 1.00 74.07 H C
+ATOM 1804 NE2 GLN H 39 -38.590 -0.134 27.811 1.00 74.07 H N
+ATOM 1805 OE1 GLN H 39 -36.948 -0.638 26.372 1.00 74.07 H O
+ATOM 1806 N ALA H 40 -42.625 1.561 26.052 1.00 75.03 H N
+ATOM 1807 CA ALA H 40 -43.824 1.453 26.864 1.00 75.03 H C
+ATOM 1808 C ALA H 40 -43.518 0.360 27.883 1.00 75.03 H C
+ATOM 1809 O ALA H 40 -42.455 -0.266 27.786 1.00 75.03 H O
+ATOM 1810 CB ALA H 40 -45.034 1.134 25.985 1.00 75.03 H C
+ATOM 1811 N PRO H 41 -44.384 0.080 28.865 1.00 77.29 H N
+ATOM 1812 CA PRO H 41 -43.984 -0.873 29.911 1.00 77.29 H C
+ATOM 1813 C PRO H 41 -43.819 -2.301 29.410 1.00 77.29 H C
+ATOM 1814 O PRO H 41 -44.727 -3.127 29.530 1.00 77.29 H O
+ATOM 1815 CB PRO H 41 -45.120 -0.765 30.934 1.00 77.29 H C
+ATOM 1816 CG PRO H 41 -45.695 0.572 30.709 1.00 77.29 H C
+ATOM 1817 CD PRO H 41 -45.614 0.797 29.242 1.00 77.29 H C
+ATOM 1818 N GLY H 42 -42.649 -2.589 28.837 1.00 73.24 H N
+ATOM 1819 CA GLY H 42 -42.300 -3.933 28.412 1.00 73.24 H C
+ATOM 1820 C GLY H 42 -42.069 -4.104 26.924 1.00 73.24 H C
+ATOM 1821 O GLY H 42 -42.327 -5.176 26.376 1.00 73.24 H O
+ATOM 1822 N LYS H 43 -41.561 -3.070 26.267 1.00 71.88 H N
+ATOM 1823 CA LYS H 43 -41.400 -3.037 24.816 1.00 71.88 H C
+ATOM 1824 C LYS H 43 -40.152 -2.213 24.508 1.00 71.88 H C
+ATOM 1825 O LYS H 43 -39.256 -2.095 25.347 1.00 71.88 H O
+ATOM 1826 CB LYS H 43 -42.653 -2.462 24.129 1.00 71.88 H C
+ATOM 1827 CG LYS H 43 -43.804 -3.436 23.863 1.00 71.88 H C
+ATOM 1828 CD LYS H 43 -44.666 -3.712 25.086 1.00 71.88 H C
+ATOM 1829 CE LYS H 43 -45.454 -2.486 25.496 1.00 71.88 H C
+ATOM 1830 NZ LYS H 43 -46.327 -2.753 26.670 1.00 71.88 H N1+
+ATOM 1831 N GLY H 44 -40.061 -1.696 23.281 1.00 66.60 H N
+ATOM 1832 CA GLY H 44 -39.140 -0.613 22.967 1.00 66.60 H C
+ATOM 1833 C GLY H 44 -39.057 -0.255 21.494 1.00 66.60 H C
+ATOM 1834 O GLY H 44 -38.878 -1.132 20.645 1.00 66.60 H O
+ATOM 1835 N LEU H 45 -39.124 1.040 21.184 1.00 68.51 H N
+ATOM 1836 CA LEU H 45 -39.339 1.506 19.822 1.00 68.51 H C
+ATOM 1837 C LEU H 45 -38.398 2.661 19.517 1.00 68.51 H C
+ATOM 1838 O LEU H 45 -38.134 3.506 20.375 1.00 68.51 H O
+ATOM 1839 CB LEU H 45 -40.784 1.963 19.627 1.00 68.51 H C
+ATOM 1840 CG LEU H 45 -41.865 0.892 19.731 1.00 68.51 H C
+ATOM 1841 CD1 LEU H 45 -43.232 1.522 19.671 1.00 68.51 H C
+ATOM 1842 CD2 LEU H 45 -41.713 -0.134 18.630 1.00 68.51 H C
+ATOM 1843 N GLU H 46 -37.917 2.709 18.277 1.00 65.81 H N
+ATOM 1844 CA GLU H 46 -36.969 3.727 17.825 1.00 65.81 H C
+ATOM 1845 C GLU H 46 -37.716 4.702 16.925 1.00 65.81 H C
+ATOM 1846 O GLU H 46 -37.759 4.535 15.708 1.00 65.81 H O
+ATOM 1847 CB GLU H 46 -35.816 3.100 17.095 1.00 65.81 H C
+ATOM 1848 CG GLU H 46 -34.831 4.091 16.582 1.00 65.81 H C
+ATOM 1849 CD GLU H 46 -33.710 3.422 15.844 1.00 65.81 H C
+ATOM 1850 OE1 GLU H 46 -33.987 2.443 15.141 1.00 65.81 H O
+ATOM 1851 OE2 GLU H 46 -32.549 3.856 15.962 1.00 65.81 H O1-
+ATOM 1852 N TRP H 47 -38.313 5.721 17.531 1.00 77.01 H N
+ATOM 1853 CA TRP H 47 -39.077 6.715 16.791 1.00 77.01 H C
+ATOM 1854 C TRP H 47 -38.182 7.797 16.217 1.00 77.01 H C
+ATOM 1855 O TRP H 47 -36.973 7.595 16.074 1.00 77.01 H O
+ATOM 1856 CB TRP H 47 -40.177 7.329 17.657 1.00 77.01 H C
+ATOM 1857 CG TRP H 47 -39.709 8.058 18.836 1.00 77.01 H C
+ATOM 1858 CD1 TRP H 47 -38.902 7.587 19.802 1.00 77.01 H C
+ATOM 1859 CD2 TRP H 47 -40.056 9.393 19.214 1.00 77.01 H C
+ATOM 1860 CE2 TRP H 47 -39.398 9.666 20.422 1.00 77.01 H C
+ATOM 1861 CE3 TRP H 47 -40.848 10.390 18.642 1.00 77.01 H C
+ATOM 1862 NE1 TRP H 47 -38.696 8.542 20.764 1.00 77.01 H N
+ATOM 1863 CZ2 TRP H 47 -39.510 10.893 21.075 1.00 77.01 H C
+ATOM 1864 CZ3 TRP H 47 -40.962 11.609 19.289 1.00 77.01 H C
+ATOM 1865 CH2 TRP H 47 -40.292 11.852 20.490 1.00 77.01 H C
+ATOM 1866 N VAL H 48 -38.778 8.933 15.850 1.00 80.58 H N
+ATOM 1867 CA VAL H 48 -38.069 10.029 15.197 1.00 80.58 H C
+ATOM 1868 C VAL H 48 -36.857 10.406 16.030 1.00 80.58 H C
+ATOM 1869 O VAL H 48 -36.785 10.082 17.219 1.00 80.58 H O
+ATOM 1870 CB VAL H 48 -39.013 11.234 14.981 1.00 80.58 H C
+ATOM 1871 CG1 VAL H 48 -38.304 12.450 14.420 1.00 80.58 H C
+ATOM 1872 CG2 VAL H 48 -40.046 10.858 13.976 1.00 80.58 H C
+ATOM 1873 N ALA H 49 -35.889 11.054 15.392 1.00 79.37 H N
+ATOM 1874 CA ALA H 49 -34.618 11.400 16.001 1.00 79.37 H C
+ATOM 1875 C ALA H 49 -33.727 10.178 16.160 1.00 79.37 H C
+ATOM 1876 O ALA H 49 -33.092 9.984 17.191 1.00 79.37 H O
+ATOM 1877 CB ALA H 49 -34.809 12.122 17.334 1.00 79.37 H C
+ATOM 1878 N VAL H 50 -33.672 9.357 15.124 1.00 74.57 H N
+ATOM 1879 CA VAL H 50 -32.512 8.520 14.874 1.00 74.57 H C
+ATOM 1880 C VAL H 50 -31.444 9.491 14.398 1.00 74.57 H C
+ATOM 1881 O VAL H 50 -31.755 10.665 14.188 1.00 74.57 H O
+ATOM 1882 CB VAL H 50 -32.835 7.437 13.852 1.00 74.57 H C
+ATOM 1883 CG1 VAL H 50 -31.677 6.484 13.705 1.00 74.57 H C
+ATOM 1884 CG2 VAL H 50 -34.078 6.749 14.294 1.00 74.57 H C
+ATOM 1885 N VAL H 51 -30.202 9.033 14.187 1.00 75.31 H N
+ATOM 1886 CA VAL H 51 -29.016 9.849 14.442 1.00 75.31 H C
+ATOM 1887 C VAL H 51 -29.259 11.290 14.042 1.00 75.31 H C
+ATOM 1888 O VAL H 51 -29.265 11.632 12.860 1.00 75.31 H O
+ATOM 1889 CB VAL H 51 -27.818 9.286 13.661 1.00 75.31 H C
+ATOM 1890 CG1 VAL H 51 -26.579 10.109 13.898 1.00 75.31 H C
+ATOM 1891 CG2 VAL H 51 -27.577 7.839 14.053 1.00 75.31 H C
+ATOM 1892 N MET H 52 -29.438 12.135 15.048 1.00 81.52 H N
+ATOM 1893 CA MET H 52 -29.858 13.511 14.855 1.00 81.52 H C
+ATOM 1894 C MET H 52 -28.783 14.596 14.774 1.00 81.52 H C
+ATOM 1895 O MET H 52 -28.921 15.504 13.945 1.00 81.52 H O
+ATOM 1896 CB MET H 52 -30.881 13.882 15.913 1.00 81.52 H C
+ATOM 1897 CG MET H 52 -31.691 15.054 15.458 1.00 81.52 H C
+ATOM 1898 SD MET H 52 -33.318 15.209 16.169 1.00 81.52 H S
+ATOM 1899 CE MET H 52 -34.266 14.605 14.792 1.00 81.52 H C
+ATOM 1900 N PRO H 53 -27.728 14.576 15.604 1.00 80.77 H N
+ATOM 1901 CA PRO H 53 -26.899 15.794 15.749 1.00 80.77 H C
+ATOM 1902 C PRO H 53 -26.313 16.270 14.443 1.00 80.77 H C
+ATOM 1903 O PRO H 53 -26.279 17.472 14.153 1.00 80.77 H O
+ATOM 1904 CB PRO H 53 -25.768 15.336 16.681 1.00 80.77 H C
+ATOM 1905 CG PRO H 53 -25.702 13.927 16.438 1.00 80.77 H C
+ATOM 1906 CD PRO H 53 -27.112 13.467 16.335 1.00 80.77 H C
+ATOM 1907 N ASP H 54 -25.844 15.321 13.656 1.00 81.14 H N
+ATOM 1908 CA ASP H 54 -25.174 15.500 12.380 1.00 81.14 H C
+ATOM 1909 C ASP H 54 -25.038 14.076 11.863 1.00 81.14 H C
+ATOM 1910 O ASP H 54 -25.337 13.124 12.590 1.00 81.14 H O
+ATOM 1911 CB ASP H 54 -23.849 16.241 12.565 1.00 81.14 H C
+ATOM 1912 CG ASP H 54 -23.105 16.456 11.283 1.00 81.14 H C
+ATOM 1913 OD1 ASP H 54 -22.468 15.507 10.787 1.00 81.14 H O
+ATOM 1914 OD2 ASP H 54 -23.136 17.596 10.786 1.00 81.14 H O1-
+ATOM 1915 N SER H 55 -24.589 13.927 10.620 1.00 75.11 H N
+ATOM 1916 CA SER H 55 -24.696 12.698 9.832 1.00 75.11 H C
+ATOM 1917 C SER H 55 -26.116 12.526 9.307 1.00 75.11 H C
+ATOM 1918 O SER H 55 -26.476 11.425 8.887 1.00 75.11 H O
+ATOM 1919 CB SER H 55 -24.271 11.428 10.580 1.00 75.11 H C
+ATOM 1920 OG SER H 55 -24.459 10.268 9.788 1.00 75.11 H O
+ATOM 1921 N GLY H 56 -26.943 13.564 9.373 1.00 78.40 H N
+ATOM 1922 CA GLY H 56 -28.165 13.745 8.600 1.00 78.40 H C
+ATOM 1923 C GLY H 56 -29.153 12.595 8.679 1.00 78.40 H C
+ATOM 1924 O GLY H 56 -29.053 11.699 9.523 1.00 78.40 H O
+ATOM 1925 N HIS H 57 -30.117 12.626 7.752 1.00 76.71 H N
+ATOM 1926 CA HIS H 57 -31.252 11.708 7.689 1.00 76.71 H C
+ATOM 1927 C HIS H 57 -31.716 11.353 9.089 1.00 76.71 H C
+ATOM 1928 O HIS H 57 -31.685 10.183 9.479 1.00 76.71 H O
+ATOM 1929 CB HIS H 57 -30.967 10.423 6.896 1.00 76.71 H C
+ATOM 1930 CG HIS H 57 -29.605 9.836 7.096 1.00 76.71 H C
+ATOM 1931 CD2 HIS H 57 -28.500 9.859 6.313 1.00 76.71 H C
+ATOM 1932 ND1 HIS H 57 -29.298 9.022 8.164 1.00 76.71 H N
+ATOM 1933 CE1 HIS H 57 -28.046 8.616 8.062 1.00 76.71 H C
+ATOM 1934 NE2 HIS H 57 -27.544 9.101 6.943 1.00 76.71 H N
+ATOM 1935 N THR H 58 -32.148 12.343 9.855 1.00 80.84 H N
+ATOM 1936 CA THR H 58 -32.311 12.129 11.282 1.00 80.84 H C
+ATOM 1937 C THR H 58 -33.528 11.264 11.552 1.00 80.84 H C
+ATOM 1938 O THR H 58 -34.120 11.358 12.628 1.00 80.84 H O
+ATOM 1939 CB THR H 58 -32.437 13.443 12.046 1.00 80.84 H C
+ATOM 1940 CG2 THR H 58 -31.234 14.319 11.771 1.00 80.84 H C
+ATOM 1941 OG1 THR H 58 -33.597 14.135 11.601 1.00 80.84 H O
+ATOM 1942 N ASN H 59 -33.953 10.482 10.566 1.00 81.56 H N
+ATOM 1943 CA ASN H 59 -34.915 9.408 10.780 1.00 81.56 H C
+ATOM 1944 C ASN H 59 -34.511 8.130 10.057 1.00 81.56 H C
+ATOM 1945 O ASN H 59 -35.309 7.535 9.332 1.00 81.56 H O
+ATOM 1946 CB ASN H 59 -36.315 9.807 10.345 1.00 81.56 H C
+ATOM 1947 CG ASN H 59 -36.890 10.884 11.188 1.00 81.56 H C
+ATOM 1948 ND2 ASN H 59 -37.786 11.665 10.611 1.00 81.56 H N
+ATOM 1949 OD1 ASN H 59 -36.618 10.957 12.371 1.00 81.56 H O
+ATOM 1950 N TYR H 60 -33.270 7.685 10.220 1.00 77.15 H N
+ATOM 1951 CA TYR H 60 -32.956 6.331 9.783 1.00 77.15 H C
+ATOM 1952 C TYR H 60 -33.844 5.346 10.534 1.00 77.15 H C
+ATOM 1953 O TYR H 60 -33.662 5.136 11.734 1.00 77.15 H O
+ATOM 1954 CB TYR H 60 -31.482 6.012 10.011 1.00 77.15 H C
+ATOM 1955 CG TYR H 60 -30.971 4.785 9.265 1.00 77.15 H C
+ATOM 1956 CD1 TYR H 60 -31.353 3.510 9.656 1.00 77.15 H C
+ATOM 1957 CD2 TYR H 60 -30.074 4.894 8.211 1.00 77.15 H C
+ATOM 1958 CE1 TYR H 60 -30.893 2.386 9.005 1.00 77.15 H C
+ATOM 1959 CE2 TYR H 60 -29.602 3.762 7.554 1.00 77.15 H C
+ATOM 1960 CZ TYR H 60 -30.023 2.511 7.962 1.00 77.15 H C
+ATOM 1961 OH TYR H 60 -29.576 1.371 7.343 1.00 77.15 H O
+ATOM 1962 N ALA H 61 -34.791 4.733 9.816 1.00 78.47 H N
+ATOM 1963 CA ALA H 61 -35.812 3.842 10.389 1.00 78.47 H C
+ATOM 1964 C ALA H 61 -36.842 4.609 11.221 1.00 78.47 H C
+ATOM 1965 O ALA H 61 -37.142 4.238 12.355 1.00 78.47 H O
+ATOM 1966 CB ALA H 61 -35.188 2.711 11.211 1.00 78.47 H C
+ATOM 1967 N ASP H 62 -37.392 5.681 10.648 1.00 77.61 H N
+ATOM 1968 CA ASP H 62 -38.502 6.385 11.281 1.00 77.61 H C
+ATOM 1969 C ASP H 62 -39.726 5.494 11.437 1.00 77.61 H C
+ATOM 1970 O ASP H 62 -40.111 5.127 12.552 1.00 77.61 H O
+ATOM 1971 CB ASP H 62 -38.893 7.610 10.463 1.00 77.61 H C
+ATOM 1972 CG ASP H 62 -39.432 8.713 11.321 1.00 77.61 H C
+ATOM 1973 OD1 ASP H 62 -40.110 8.364 12.299 1.00 77.61 H O
+ATOM 1974 OD2 ASP H 62 -39.328 9.899 10.963 1.00 77.61 H O1-
+ATOM 1975 N SER H 63 -40.375 5.173 10.317 1.00 76.33 H N
+ATOM 1976 CA SER H 63 -41.650 4.460 10.302 1.00 76.33 H C
+ATOM 1977 C SER H 63 -42.721 5.163 11.141 1.00 76.33 H C
+ATOM 1978 O SER H 63 -43.621 4.519 11.681 1.00 76.33 H O
+ATOM 1979 CB SER H 63 -41.473 3.012 10.753 1.00 76.33 H C
+ATOM 1980 OG SER H 63 -41.334 2.929 12.159 1.00 76.33 H O
+ATOM 1981 N VAL H 64 -42.639 6.488 11.253 1.00 79.93 H N
+ATOM 1982 CA VAL H 64 -43.746 7.296 11.759 1.00 79.93 H C
+ATOM 1983 C VAL H 64 -43.475 8.738 11.355 1.00 79.93 H C
+ATOM 1984 O VAL H 64 -42.327 9.128 11.141 1.00 79.93 H O
+ATOM 1985 CB VAL H 64 -43.916 7.134 13.290 1.00 79.93 H C
+ATOM 1986 CG1 VAL H 64 -42.654 7.542 14.005 1.00 79.93 H C
+ATOM 1987 CG2 VAL H 64 -45.122 7.906 13.787 1.00 79.93 H C
+ATOM 1988 N LYS H 65 -44.535 9.528 11.220 1.00 92.66 H N
+ATOM 1989 CA LYS H 65 -44.395 10.874 10.675 1.00 92.66 H C
+ATOM 1990 C LYS H 65 -45.609 11.698 11.096 1.00 92.66 H C
+ATOM 1991 O LYS H 65 -46.573 11.174 11.660 1.00 92.66 H O
+ATOM 1992 CB LYS H 65 -44.220 10.814 9.149 1.00 92.66 H C
+ATOM 1993 CG LYS H 65 -44.022 12.147 8.429 1.00 92.66 H C
+ATOM 1994 CD LYS H 65 -42.802 12.903 8.940 1.00 92.66 H C
+ATOM 1995 CE LYS H 65 -41.500 12.211 8.559 1.00 92.66 H C
+ATOM 1996 NZ LYS H 65 -40.303 12.980 9.008 1.00 92.66 H N1+
+ATOM 1997 N GLY H 66 -45.552 12.998 10.811 1.00 93.61 H N
+ATOM 1998 CA GLY H 66 -46.633 13.910 11.123 1.00 93.61 H C
+ATOM 1999 C GLY H 66 -46.461 14.637 12.438 1.00 93.61 H C
+ATOM 2000 O GLY H 66 -46.373 15.868 12.473 1.00 93.61 H O
+ATOM 2001 N ARG H 67 -46.428 13.885 13.532 1.00 95.28 H N
+ATOM 2002 CA ARG H 67 -46.388 14.466 14.861 1.00 95.28 H C
+ATOM 2003 C ARG H 67 -45.075 14.253 15.594 1.00 95.28 H C
+ATOM 2004 O ARG H 67 -44.739 15.066 16.461 1.00 95.28 H O
+ATOM 2005 CB ARG H 67 -47.571 13.920 15.673 1.00 95.28 H C
+ATOM 2006 CG ARG H 67 -47.365 12.569 16.323 1.00 95.28 H C
+ATOM 2007 CD ARG H 67 -48.709 12.058 16.837 1.00 95.28 H C
+ATOM 2008 NE ARG H 67 -48.622 11.202 18.016 1.00 95.28 H N
+ATOM 2009 CZ ARG H 67 -47.956 10.054 18.073 1.00 95.28 H C
+ATOM 2010 NH1 ARG H 67 -47.325 9.586 17.005 1.00 95.28 H N1+
+ATOM 2011 NH2 ARG H 67 -47.946 9.357 19.199 1.00 95.28 H N
+ATOM 2012 N PHE H 68 -44.309 13.226 15.245 1.00 89.86 H N
+ATOM 2013 CA PHE H 68 -42.973 13.030 15.795 1.00 89.86 H C
+ATOM 2014 C PHE H 68 -41.964 13.760 14.918 1.00 89.86 H C
+ATOM 2015 O PHE H 68 -41.809 13.432 13.738 1.00 89.86 H O
+ATOM 2016 CB PHE H 68 -42.638 11.545 15.894 1.00 89.86 H C
+ATOM 2017 CG PHE H 68 -43.403 10.814 16.945 1.00 89.86 H C
+ATOM 2018 CD1 PHE H 68 -44.090 11.503 17.925 1.00 89.86 H C
+ATOM 2019 CD2 PHE H 68 -43.413 9.435 16.968 1.00 89.86 H C
+ATOM 2020 CE1 PHE H 68 -44.778 10.830 18.901 1.00 89.86 H C
+ATOM 2021 CE2 PHE H 68 -44.100 8.760 17.941 1.00 89.86 H C
+ATOM 2022 CZ PHE H 68 -44.786 9.457 18.909 1.00 89.86 H C
+ATOM 2023 N THR H 69 -41.257 14.722 15.501 1.00 92.03 H N
+ATOM 2024 CA THR H 69 -40.683 15.821 14.741 1.00 92.03 H C
+ATOM 2025 C THR H 69 -39.170 15.877 14.885 1.00 92.03 H C
+ATOM 2026 O THR H 69 -38.641 15.827 15.998 1.00 92.03 H O
+ATOM 2027 CB THR H 69 -41.300 17.152 15.186 1.00 92.03 H C
+ATOM 2028 CG2 THR H 69 -40.654 18.314 14.459 1.00 92.03 H C
+ATOM 2029 OG1 THR H 69 -42.705 17.143 14.908 1.00 92.03 H O
+ATOM 2030 N ILE H 70 -38.490 16.000 13.745 1.00 92.23 H N
+ATOM 2031 CA ILE H 70 -37.070 16.321 13.717 1.00 92.23 H C
+ATOM 2032 C ILE H 70 -36.872 17.745 14.215 1.00 92.23 H C
+ATOM 2033 O ILE H 70 -37.339 18.705 13.588 1.00 92.23 H O
+ATOM 2034 CB ILE H 70 -36.532 16.174 12.288 1.00 92.23 H C
+ATOM 2035 CG1 ILE H 70 -36.633 14.724 11.812 1.00 92.23 H C
+ATOM 2036 CG2 ILE H 70 -35.125 16.765 12.173 1.00 92.23 H C
+ATOM 2037 CD1 ILE H 70 -36.369 14.567 10.330 1.00 92.23 H C
+ATOM 2038 N SER H 71 -36.165 17.900 15.338 1.00 92.31 H N
+ATOM 2039 CA SER H 71 -36.058 19.217 15.963 1.00 92.31 H C
+ATOM 2040 C SER H 71 -34.633 19.757 16.011 1.00 92.31 H C
+ATOM 2041 O SER H 71 -34.393 20.851 15.495 1.00 92.31 H O
+ATOM 2042 CB SER H 71 -36.678 19.169 17.356 1.00 92.31 H C
+ATOM 2043 OG SER H 71 -38.090 19.077 17.284 1.00 92.31 H O
+ATOM 2044 N ALA H 72 -33.676 19.046 16.605 1.00 87.74 H N
+ATOM 2045 CA ALA H 72 -32.412 19.707 16.911 1.00 87.74 H C
+ATOM 2046 C ALA H 72 -31.203 18.969 16.367 1.00 87.74 H C
+ATOM 2047 O ALA H 72 -31.348 18.030 15.585 1.00 87.74 H O
+ATOM 2048 CB ALA H 72 -32.263 19.898 18.413 1.00 87.74 H C
+ATOM 2049 N ASP H 73 -30.008 19.390 16.768 1.00 84.90 H N
+ATOM 2050 CA ASP H 73 -28.781 18.920 16.140 1.00 84.90 H C
+ATOM 2051 C ASP H 73 -27.641 19.068 17.135 1.00 84.90 H C
+ATOM 2052 O ASP H 73 -27.862 19.243 18.336 1.00 84.90 H O
+ATOM 2053 CB ASP H 73 -28.498 19.692 14.850 1.00 84.90 H C
+ATOM 2054 CG ASP H 73 -28.214 21.156 15.104 1.00 84.90 H C
+ATOM 2055 OD1 ASP H 73 -28.774 21.705 16.076 1.00 84.90 H O
+ATOM 2056 OD2 ASP H 73 -27.435 21.756 14.334 1.00 84.90 H O1-
+ATOM 2057 N THR H 74 -26.412 19.027 16.628 1.00 82.52 H N
+ATOM 2058 CA THR H 74 -25.235 19.002 17.482 1.00 82.52 H C
+ATOM 2059 C THR H 74 -24.810 20.351 18.049 1.00 82.52 H C
+ATOM 2060 O THR H 74 -23.640 20.734 17.945 1.00 82.52 H O
+ATOM 2061 CB THR H 74 -24.059 18.405 16.725 1.00 82.52 H C
+ATOM 2062 CG2 THR H 74 -23.814 19.134 15.419 1.00 82.52 H C
+ATOM 2063 OG1 THR H 74 -22.901 18.464 17.560 1.00 82.52 H O
+ATOM 2064 N SER H 75 -25.719 21.043 18.721 1.00 81.95 H N
+ATOM 2065 CA SER H 75 -25.377 22.276 19.412 1.00 81.95 H C
+ATOM 2066 C SER H 75 -24.825 22.034 20.811 1.00 81.95 H C
+ATOM 2067 O SER H 75 -24.825 22.965 21.623 1.00 81.95 H O
+ATOM 2068 CB SER H 75 -26.596 23.198 19.475 1.00 81.95 H C
+ATOM 2069 OG SER H 75 -27.615 22.643 20.275 1.00 81.95 H O
+ATOM 2070 N LYS H 76 -24.361 20.813 21.102 1.00 80.85 H N
+ATOM 2071 CA LYS H 76 -23.967 20.252 22.396 1.00 80.85 H C
+ATOM 2072 C LYS H 76 -25.182 19.852 23.216 1.00 80.85 H C
+ATOM 2073 O LYS H 76 -25.023 19.228 24.260 1.00 80.85 H O
+ATOM 2074 CB LYS H 76 -23.098 21.203 23.242 1.00 80.85 H C
+ATOM 2075 CG LYS H 76 -21.759 21.624 22.664 1.00 80.85 H C
+ATOM 2076 CD LYS H 76 -20.760 20.483 22.602 1.00 80.85 H C
+ATOM 2077 CE LYS H 76 -19.429 20.961 22.012 1.00 80.85 H C
+ATOM 2078 NZ LYS H 76 -18.716 21.912 22.909 1.00 80.85 H N1+
+ATOM 2079 N ASN H 77 -26.381 20.191 22.779 1.00 80.77 H N
+ATOM 2080 CA ASN H 77 -27.628 19.763 23.399 1.00 80.77 H C
+ATOM 2081 C ASN H 77 -28.491 19.123 22.313 1.00 80.77 H C
+ATOM 2082 O ASN H 77 -29.178 19.795 21.544 1.00 80.77 H O
+ATOM 2083 CB ASN H 77 -28.268 20.920 24.167 1.00 80.77 H C
+ATOM 2084 CG ASN H 77 -28.651 22.100 23.295 1.00 80.77 H C
+ATOM 2085 ND2 ASN H 77 -29.077 23.179 23.941 1.00 80.77 H N
+ATOM 2086 OD1 ASN H 77 -28.561 22.063 22.075 1.00 80.77 H O
+ATOM 2087 N THR H 78 -28.458 17.797 22.280 1.00 73.25 H N
+ATOM 2088 CA THR H 78 -28.430 17.109 21.002 1.00 73.25 H C
+ATOM 2089 C THR H 78 -29.815 16.791 20.452 1.00 73.25 H C
+ATOM 2090 O THR H 78 -29.910 16.222 19.362 1.00 73.25 H O
+ATOM 2091 CB THR H 78 -27.576 15.875 21.124 1.00 73.25 H C
+ATOM 2092 CG2 THR H 78 -28.112 15.044 22.206 1.00 73.25 H C
+ATOM 2093 OG1 THR H 78 -27.591 15.158 19.891 1.00 73.25 H O
+ATOM 2094 N ALA H 79 -30.894 17.081 21.184 1.00 79.84 H N
+ATOM 2095 CA ALA H 79 -32.190 17.200 20.511 1.00 79.84 H C
+ATOM 2096 C ALA H 79 -33.232 17.829 21.425 1.00 79.84 H C
+ATOM 2097 O ALA H 79 -33.057 17.890 22.640 1.00 79.84 H O
+ATOM 2098 CB ALA H 79 -32.704 15.860 19.988 1.00 79.84 H C
+ATOM 2099 N TYR H 80 -34.323 18.336 20.809 1.00 89.16 H N
+ATOM 2100 CA TYR H 80 -35.496 18.835 21.544 1.00 89.16 H C
+ATOM 2101 C TYR H 80 -36.777 18.305 20.883 1.00 89.16 H C
+ATOM 2102 O TYR H 80 -37.419 19.049 20.148 1.00 89.16 H O
+ATOM 2103 CB TYR H 80 -35.547 20.360 21.532 1.00 89.16 H C
+ATOM 2104 CG TYR H 80 -34.227 21.082 21.571 1.00 89.16 H C
+ATOM 2105 CD1 TYR H 80 -33.304 20.882 22.586 1.00 89.16 H C
+ATOM 2106 CD2 TYR H 80 -33.916 21.993 20.573 1.00 89.16 H C
+ATOM 2107 CE1 TYR H 80 -32.099 21.565 22.592 1.00 89.16 H C
+ATOM 2108 CE2 TYR H 80 -32.729 22.672 20.568 1.00 89.16 H C
+ATOM 2109 CZ TYR H 80 -31.825 22.460 21.578 1.00 89.16 H C
+ATOM 2110 OH TYR H 80 -30.644 23.161 21.545 1.00 89.16 H O
+ATOM 2111 N LEU H 81 -37.221 17.093 21.201 1.00 84.29 H N
+ATOM 2112 CA LEU H 81 -38.287 16.520 20.381 1.00 84.29 H C
+ATOM 2113 C LEU H 81 -39.681 17.011 20.751 1.00 84.29 H C
+ATOM 2114 O LEU H 81 -39.985 17.307 21.905 1.00 84.29 H O
+ATOM 2115 CB LEU H 81 -38.287 15.008 20.435 1.00 84.29 H C
+ATOM 2116 CG LEU H 81 -37.041 14.440 19.803 1.00 84.29 H C
+ATOM 2117 CD1 LEU H 81 -37.095 12.984 20.011 1.00 84.29 H C
+ATOM 2118 CD2 LEU H 81 -36.983 14.811 18.344 1.00 84.29 H C
+ATOM 2119 N GLN H 82 -40.555 17.034 19.753 1.00 95.59 H N
+ATOM 2120 CA GLN H 82 -41.889 17.598 19.881 1.00 95.59 H C
+ATOM 2121 C GLN H 82 -42.948 16.534 19.633 1.00 95.59 H C
+ATOM 2122 O GLN H 82 -42.783 15.678 18.759 1.00 95.59 H O
+ATOM 2123 CB GLN H 82 -42.089 18.745 18.910 1.00 95.59 H C
+ATOM 2124 CG GLN H 82 -43.225 19.638 19.303 1.00 95.59 H C
+ATOM 2125 CD GLN H 82 -44.489 19.189 18.641 1.00 95.59 H C
+ATOM 2126 NE2 GLN H 82 -45.628 19.632 19.156 1.00 95.59 H N
+ATOM 2127 OE1 GLN H 82 -44.439 18.442 17.669 1.00 95.59 H O
+ATOM 2128 N MET H 83 -44.037 16.598 20.402 1.00100.45 H N
+ATOM 2129 CA MET H 83 -45.038 15.534 20.382 1.00100.45 H C
+ATOM 2130 C MET H 83 -46.113 15.758 19.318 1.00100.45 H C
+ATOM 2131 O MET H 83 -46.420 14.846 18.544 1.00100.45 H O
+ATOM 2132 CB MET H 83 -45.674 15.416 21.766 1.00100.45 H C
+ATOM 2133 CG MET H 83 -46.627 14.244 21.952 1.00100.45 H C
+ATOM 2134 SD MET H 83 -45.779 12.659 21.851 1.00100.45 H S
+ATOM 2135 CE MET H 83 -47.131 11.582 21.393 1.00100.45 H C
+ATOM 2136 N ASN H 84 -46.718 16.948 19.286 1.00101.68 H N
+ATOM 2137 CA ASN H 84 -47.942 17.201 18.510 1.00101.68 H C
+ATOM 2138 C ASN H 84 -49.027 16.167 18.787 1.00101.68 H C
+ATOM 2139 O ASN H 84 -49.761 15.769 17.881 1.00101.68 H O
+ATOM 2140 CB ASN H 84 -47.676 17.259 17.005 1.00101.68 H C
+ATOM 2141 CG ASN H 84 -46.910 18.475 16.598 1.00101.68 H C
+ATOM 2142 ND2 ASN H 84 -45.935 18.289 15.723 1.00101.68 H N
+ATOM 2143 OD1 ASN H 84 -47.167 19.576 17.082 1.00101.68 H O
+ATOM 2144 N SER H 85 -49.150 15.720 20.032 1.00106.83 H N
+ATOM 2145 CA SER H 85 -50.206 14.775 20.359 1.00106.83 H C
+ATOM 2146 C SER H 85 -50.514 14.872 21.839 1.00106.83 H C
+ATOM 2147 O SER H 85 -49.614 15.070 22.657 1.00106.83 H O
+ATOM 2148 CB SER H 85 -49.817 13.341 20.000 1.00106.83 H C
+ATOM 2149 OG SER H 85 -50.775 12.417 20.482 1.00106.83 H O
+ATOM 2150 N LEU H 86 -51.791 14.731 22.171 1.00113.60 H N
+ATOM 2151 CA LEU H 86 -52.232 14.852 23.545 1.00113.60 H C
+ATOM 2152 C LEU H 86 -53.193 13.754 23.972 1.00113.60 H C
+ATOM 2153 O LEU H 86 -53.607 13.741 25.136 1.00113.60 H O
+ATOM 2154 CB LEU H 86 -52.872 16.231 23.757 1.00113.60 H C
+ATOM 2155 CG LEU H 86 -54.116 16.570 22.931 1.00113.60 H C
+ATOM 2156 CD1 LEU H 86 -55.399 16.209 23.670 1.00113.60 H C
+ATOM 2157 CD2 LEU H 86 -54.112 18.031 22.510 1.00113.60 H C
+ATOM 2158 N ARG H 87 -53.569 12.851 23.077 1.00114.12 H N
+ATOM 2159 CA ARG H 87 -54.482 11.763 23.422 1.00114.12 H C
+ATOM 2160 C ARG H 87 -54.020 10.391 22.951 1.00114.12 H C
+ATOM 2161 O ARG H 87 -54.205 9.408 23.672 1.00114.12 H O
+ATOM 2162 CB ARG H 87 -55.901 12.069 22.875 1.00114.12 H C
+ATOM 2163 CG ARG H 87 -56.084 11.929 21.363 1.00114.12 H C
+ATOM 2164 CD ARG H 87 -55.484 13.125 20.633 1.00114.12 H C
+ATOM 2165 NE ARG H 87 -55.823 13.155 19.215 1.00114.12 H N
+ATOM 2166 CZ ARG H 87 -55.244 13.963 18.332 1.00114.12 H C
+ATOM 2167 NH1 ARG H 87 -54.304 14.810 18.726 1.00114.12 H N1+
+ATOM 2168 NH2 ARG H 87 -55.610 13.929 17.058 1.00114.12 H N
+ATOM 2169 N ALA H 88 -53.410 10.293 21.778 1.00107.21 H N
+ATOM 2170 CA ALA H 88 -53.103 9.007 21.175 1.00107.21 H C
+ATOM 2171 C ALA H 88 -51.619 8.720 21.313 1.00107.21 H C
+ATOM 2172 O ALA H 88 -50.787 9.541 20.915 1.00107.21 H O
+ATOM 2173 CB ALA H 88 -53.515 8.983 19.703 1.00107.21 H C
+ATOM 2174 N GLU H 89 -51.298 7.554 21.871 1.00100.97 H N
+ATOM 2175 CA GLU H 89 -49.920 7.121 22.072 1.00100.97 H C
+ATOM 2176 C GLU H 89 -49.091 8.223 22.730 1.00100.97 H C
+ATOM 2177 O GLU H 89 -48.031 8.625 22.245 1.00100.97 H O
+ATOM 2178 CB GLU H 89 -49.299 6.671 20.753 1.00100.97 H C
+ATOM 2179 CG GLU H 89 -47.986 5.936 20.911 1.00100.97 H C
+ATOM 2180 CD GLU H 89 -47.086 6.125 19.720 1.00100.97 H C
+ATOM 2181 OE1 GLU H 89 -47.459 5.679 18.617 1.00100.97 H O
+ATOM 2182 OE2 GLU H 89 -46.014 6.740 19.882 1.00100.97 H O1-
+ATOM 2183 N ASP H 90 -49.603 8.732 23.840 1.00109.39 H N
+ATOM 2184 CA ASP H 90 -48.979 9.815 24.585 1.00109.39 H C
+ATOM 2185 C ASP H 90 -48.537 9.338 25.957 1.00109.39 H C
+ATOM 2186 O ASP H 90 -48.705 10.031 26.960 1.00109.39 H O
+ATOM 2187 CB ASP H 90 -49.933 10.998 24.706 1.00109.39 H C
+ATOM 2188 CG ASP H 90 -51.221 10.640 25.424 1.00109.39 H C
+ATOM 2189 OD1 ASP H 90 -51.437 9.445 25.714 1.00109.39 H O
+ATOM 2190 OD2 ASP H 90 -52.021 11.556 25.700 1.00109.39 H O1-
+ATOM 2191 N THR H 91 -47.966 8.139 26.014 1.00102.28 H N
+ATOM 2192 CA THR H 91 -47.808 7.478 27.300 1.00102.28 H C
+ATOM 2193 C THR H 91 -46.446 6.792 27.414 1.00102.28 H C
+ATOM 2194 O THR H 91 -46.190 6.091 28.398 1.00102.28 H O
+ATOM 2195 CB THR H 91 -48.990 6.493 27.482 1.00102.28 H C
+ATOM 2196 CG2 THR H 91 -49.025 5.858 28.870 1.00102.28 H C
+ATOM 2197 OG1 THR H 91 -50.221 7.208 27.314 1.00102.28 H O
+ATOM 2198 N ALA H 92 -45.538 7.021 26.476 1.00 82.44 H N
+ATOM 2199 CA ALA H 92 -44.283 6.296 26.492 1.00 82.44 H C
+ATOM 2200 C ALA H 92 -43.310 6.924 27.481 1.00 82.44 H C
+ATOM 2201 O ALA H 92 -43.615 7.901 28.161 1.00 82.44 H O
+ATOM 2202 CB ALA H 92 -43.674 6.259 25.096 1.00 82.44 H C
+ATOM 2203 N VAL H 93 -42.125 6.340 27.575 1.00 76.03 H N
+ATOM 2204 CA VAL H 93 -41.010 6.937 28.291 1.00 76.03 H C
+ATOM 2205 C VAL H 93 -39.880 7.071 27.284 1.00 76.03 H C
+ATOM 2206 O VAL H 93 -39.109 6.137 27.047 1.00 76.03 H O
+ATOM 2207 CB VAL H 93 -40.592 6.135 29.525 1.00 76.03 H C
+ATOM 2208 CG1 VAL H 93 -41.595 6.341 30.627 1.00 76.03 H C
+ATOM 2209 CG2 VAL H 93 -40.499 4.667 29.208 1.00 76.03 H C
+ATOM 2210 N TYR H 94 -39.785 8.247 26.688 1.00 74.79 H N
+ATOM 2211 CA TYR H 94 -38.836 8.497 25.621 1.00 74.79 H C
+ATOM 2212 C TYR H 94 -37.427 8.525 26.174 1.00 74.79 H C
+ATOM 2213 O TYR H 94 -37.207 8.896 27.325 1.00 74.79 H O
+ATOM 2214 CB TYR H 94 -39.200 9.813 24.968 1.00 74.79 H C
+ATOM 2215 CG TYR H 94 -40.618 9.781 24.489 1.00 74.79 H C
+ATOM 2216 CD1 TYR H 94 -40.976 8.997 23.416 1.00 74.79 H C
+ATOM 2217 CD2 TYR H 94 -41.617 10.440 25.182 1.00 74.79 H C
+ATOM 2218 CE1 TYR H 94 -42.271 8.936 22.987 1.00 74.79 H C
+ATOM 2219 CE2 TYR H 94 -42.921 10.380 24.768 1.00 74.79 H C
+ATOM 2220 CZ TYR H 94 -43.245 9.623 23.668 1.00 74.79 H C
+ATOM 2221 OH TYR H 94 -44.548 9.549 23.235 1.00 74.79 H O
+ATOM 2222 N TYR H 95 -36.469 8.124 25.356 1.00 70.10 H N
+ATOM 2223 CA TYR H 95 -35.113 7.979 25.856 1.00 70.10 H C
+ATOM 2224 C TYR H 95 -34.087 8.632 24.942 1.00 70.10 H C
+ATOM 2225 O TYR H 95 -34.458 9.371 24.033 1.00 70.10 H O
+ATOM 2226 CB TYR H 95 -34.834 6.508 26.077 1.00 70.10 H C
+ATOM 2227 CG TYR H 95 -35.496 6.003 27.331 1.00 70.10 H C
+ATOM 2228 CD1 TYR H 95 -35.509 6.776 28.469 1.00 70.10 H C
+ATOM 2229 CD2 TYR H 95 -36.114 4.767 27.385 1.00 70.10 H C
+ATOM 2230 CE1 TYR H 95 -36.100 6.341 29.618 1.00 70.10 H C
+ATOM 2231 CE2 TYR H 95 -36.711 4.327 28.538 1.00 70.10 H C
+ATOM 2232 CZ TYR H 95 -36.702 5.125 29.649 1.00 70.10 H C
+ATOM 2233 OH TYR H 95 -37.294 4.712 30.814 1.00 70.10 H O
+ATOM 2234 N CYS H 96 -32.801 8.415 25.202 1.00 65.85 H N
+ATOM 2235 CA CYS H 96 -31.738 9.162 24.529 1.00 65.85 H C
+ATOM 2236 C CYS H 96 -30.473 8.325 24.628 1.00 65.85 H C
+ATOM 2237 O CYS H 96 -29.940 8.171 25.722 1.00 65.85 H O
+ATOM 2238 CB CYS H 96 -31.535 10.533 25.190 1.00 65.85 H C
+ATOM 2239 SG CYS H 96 -32.897 11.780 25.098 1.00 65.85 H S
+ATOM 2240 N SER H 97 -29.990 7.775 23.521 1.00 61.70 H N
+ATOM 2241 CA SER H 97 -28.764 6.982 23.587 1.00 61.70 H C
+ATOM 2242 C SER H 97 -27.553 7.906 23.496 1.00 61.70 H C
+ATOM 2243 O SER H 97 -26.744 7.858 22.569 1.00 61.70 H O
+ATOM 2244 CB SER H 97 -28.731 5.908 22.511 1.00 61.70 H C
+ATOM 2245 OG SER H 97 -29.932 5.172 22.476 1.00 61.70 H O
+ATOM 2246 N ARG H 98 -27.394 8.706 24.542 1.00 71.86 H N
+ATOM 2247 CA ARG H 98 -26.398 9.778 24.581 1.00 71.86 H C
+ATOM 2248 C ARG H 98 -25.001 9.294 24.853 1.00 71.86 H C
+ATOM 2249 O ARG H 98 -24.284 9.881 25.664 1.00 71.86 H O
+ATOM 2250 CB ARG H 98 -26.769 10.769 25.672 1.00 71.86 H C
+ATOM 2251 CG ARG H 98 -26.205 12.120 25.455 1.00 71.86 H C
+ATOM 2252 CD ARG H 98 -27.270 12.948 24.940 1.00 71.86 H C
+ATOM 2253 NE ARG H 98 -28.140 13.101 26.079 1.00 71.86 H N
+ATOM 2254 CZ ARG H 98 -29.260 13.783 26.087 1.00 71.86 H C
+ATOM 2255 NH1 ARG H 98 -29.977 13.826 27.195 1.00 71.86 H N1+
+ATOM 2256 NH2 ARG H 98 -29.658 14.427 25.005 1.00 71.86 H N
+ATOM 2257 N ALA H 99 -24.589 8.264 24.226 1.00 69.71 H N
+ATOM 2258 CA ALA H 99 -23.167 8.000 24.176 1.00 69.71 H C
+ATOM 2259 C ALA H 99 -22.799 7.355 22.868 1.00 69.71 H C
+ATOM 2260 O ALA H 99 -21.660 6.917 22.728 1.00 69.71 H O
+ATOM 2261 CB ALA H 99 -22.719 7.129 25.344 1.00 69.71 H C
+ATOM 2262 N THR H 100 -23.730 7.243 21.928 1.00 69.76 H N
+ATOM 2263 CA THR H 100 -23.521 6.380 20.788 1.00 69.76 H C
+ATOM 2264 C THR H 100 -22.383 6.914 19.944 1.00 69.76 H C
+ATOM 2265 O THR H 100 -22.520 7.894 19.222 1.00 69.76 H O
+ATOM 2266 CB THR H 100 -24.785 6.279 19.973 1.00 69.76 H C
+ATOM 2267 CG2 THR H 100 -24.703 5.035 19.111 1.00 69.76 H C
+ATOM 2268 OG1 THR H 100 -25.889 6.160 20.872 1.00 69.76 H O
+ATOM 2269 N ASN H 101 -21.262 6.239 20.036 1.00 67.24 H N
+ATOM 2270 CA ASN H 101 -19.970 6.722 19.609 1.00 67.24 H C
+ATOM 2271 C ASN H 101 -19.098 5.677 18.951 1.00 67.24 H C
+ATOM 2272 O ASN H 101 -18.257 6.032 18.124 1.00 67.24 H O
+ATOM 2273 CB ASN H 101 -19.257 7.311 20.821 1.00 67.24 H C
+ATOM 2274 CG ASN H 101 -19.083 6.296 21.916 1.00 67.24 H C
+ATOM 2275 ND2 ASN H 101 -19.295 6.735 23.144 1.00 67.24 H N
+ATOM 2276 OD1 ASN H 101 -18.829 5.122 21.675 1.00 67.24 H O
+ATOM 2277 N ILE H 102 -19.252 4.414 19.318 1.00 63.05 H N
+ATOM 2278 CA ILE H 102 -18.543 3.301 18.700 1.00 63.05 H C
+ATOM 2279 C ILE H 102 -19.476 2.111 18.643 1.00 63.05 H C
+ATOM 2280 O ILE H 102 -20.394 1.995 19.468 1.00 63.05 H O
+ATOM 2281 CB ILE H 102 -17.268 2.958 19.481 1.00 63.05 H C
+ATOM 2282 CG1 ILE H 102 -16.265 4.123 19.451 1.00 63.05 H C
+ATOM 2283 CG2 ILE H 102 -16.657 1.633 19.055 1.00 63.05 H C
+ATOM 2284 CD1 ILE H 102 -16.293 5.199 20.487 1.00 63.05 H C
+ATOM 2285 N PRO H 103 -19.297 1.218 17.659 1.00 63.03 H N
+ATOM 2286 CA PRO H 103 -20.070 -0.030 17.695 1.00 63.03 H C
+ATOM 2287 C PRO H 103 -19.935 -0.754 19.029 1.00 63.03 H C
+ATOM 2288 O PRO H 103 -20.932 -1.275 19.549 1.00 63.03 H O
+ATOM 2289 CB PRO H 103 -19.472 -0.855 16.540 1.00 63.03 H C
+ATOM 2290 CG PRO H 103 -18.304 -0.092 16.068 1.00 63.03 H C
+ATOM 2291 CD PRO H 103 -18.552 1.323 16.403 1.00 63.03 H C
+ATOM 2292 N VAL H 104 -18.729 -0.795 19.609 1.00 61.74 H N
+ATOM 2293 CA VAL H 104 -18.526 -1.455 20.891 1.00 61.74 H C
+ATOM 2294 C VAL H 104 -17.751 -0.554 21.858 1.00 61.74 H C
+ATOM 2295 O VAL H 104 -16.529 -0.661 21.986 1.00 61.74 H O
+ATOM 2296 CB VAL H 104 -17.825 -2.808 20.645 1.00 61.74 H C
+ATOM 2297 CG1 VAL H 104 -17.683 -3.587 21.872 1.00 61.74 H C
+ATOM 2298 CG2 VAL H 104 -18.618 -3.661 19.679 1.00 61.74 H C
+ATOM 2299 N TYR H 105 -18.468 0.342 22.538 1.00 63.38 H N
+ATOM 2300 CA TYR H 105 -18.142 0.959 23.828 1.00 63.38 H C
+ATOM 2301 C TYR H 105 -19.440 1.305 24.527 1.00 63.38 H C
+ATOM 2302 O TYR H 105 -20.508 1.299 23.923 1.00 63.38 H O
+ATOM 2303 CB TYR H 105 -17.248 2.217 23.776 1.00 63.38 H C
+ATOM 2304 CG TYR H 105 -15.798 2.054 23.364 1.00 63.38 H C
+ATOM 2305 CD1 TYR H 105 -14.851 1.638 24.265 1.00 63.38 H C
+ATOM 2306 CD2 TYR H 105 -15.378 2.308 22.102 1.00 63.38 H C
+ATOM 2307 CE1 TYR H 105 -13.527 1.498 23.903 1.00 63.38 H C
+ATOM 2308 CE2 TYR H 105 -14.069 2.159 21.727 1.00 63.38 H C
+ATOM 2309 CZ TYR H 105 -13.149 1.756 22.628 1.00 63.38 H C
+ATOM 2310 OH TYR H 105 -11.838 1.616 22.241 1.00 63.38 H O
+ATOM 2311 N ALA H 106 -19.337 1.607 25.816 1.00 53.92 H N
+ATOM 2312 CA ALA H 106 -20.523 1.687 26.649 1.00 53.92 H C
+ATOM 2313 C ALA H 106 -21.502 2.684 26.066 1.00 53.92 H C
+ATOM 2314 O ALA H 106 -21.130 3.809 25.741 1.00 53.92 H O
+ATOM 2315 CB ALA H 106 -20.138 2.084 28.070 1.00 53.92 H C
+ATOM 2316 N PHE H 107 -22.750 2.279 25.915 1.00 58.60 H N
+ATOM 2317 CA PHE H 107 -23.769 3.163 25.375 1.00 58.60 H C
+ATOM 2318 C PHE H 107 -24.481 3.763 26.560 1.00 58.60 H C
+ATOM 2319 O PHE H 107 -25.578 3.329 26.909 1.00 58.60 H O
+ATOM 2320 CB PHE H 107 -24.767 2.428 24.494 1.00 58.60 H C
+ATOM 2321 CG PHE H 107 -24.188 1.924 23.233 1.00 58.60 H C
+ATOM 2322 CD1 PHE H 107 -22.971 2.374 22.790 1.00 58.60 H C
+ATOM 2323 CD2 PHE H 107 -24.852 0.987 22.491 1.00 58.60 H C
+ATOM 2324 CE1 PHE H 107 -22.427 1.894 21.620 1.00 58.60 H C
+ATOM 2325 CE2 PHE H 107 -24.317 0.501 21.321 1.00 58.60 H C
+ATOM 2326 CZ PHE H 107 -23.103 0.954 20.884 1.00 58.60 H C
+ATOM 2327 N ASP H 108 -23.895 4.794 27.137 1.00 67.51 H N
+ATOM 2328 CA ASP H 108 -24.464 5.362 28.348 1.00 67.51 H C
+ATOM 2329 C ASP H 108 -25.772 5.993 27.937 1.00 67.51 H C
+ATOM 2330 O ASP H 108 -25.842 7.176 27.634 1.00 67.51 H O
+ATOM 2331 CB ASP H 108 -23.548 6.388 28.985 1.00 67.51 H C
+ATOM 2332 CG ASP H 108 -24.083 6.868 30.311 1.00 67.51 H C
+ATOM 2333 OD1 ASP H 108 -24.002 6.111 31.301 1.00 67.51 H O
+ATOM 2334 OD2 ASP H 108 -24.659 7.970 30.349 1.00 67.51 H O1-
+ATOM 2335 N TYR H 109 -26.819 5.181 27.931 1.00 65.20 H N
+ATOM 2336 CA TYR H 109 -28.107 5.667 27.480 1.00 65.20 H C
+ATOM 2337 C TYR H 109 -28.569 6.890 28.234 1.00 65.20 H C
+ATOM 2338 O TYR H 109 -28.294 8.021 27.836 1.00 65.20 H O
+ATOM 2339 CB TYR H 109 -29.167 4.582 27.605 1.00 65.20 H C
+ATOM 2340 CG TYR H 109 -29.061 3.432 26.663 1.00 65.20 H C
+ATOM 2341 CD1 TYR H 109 -28.243 3.491 25.555 1.00 65.20 H C
+ATOM 2342 CD2 TYR H 109 -29.911 2.373 26.775 1.00 65.20 H C
+ATOM 2343 CE1 TYR H 109 -28.202 2.473 24.659 1.00 65.20 H C
+ATOM 2344 CE2 TYR H 109 -29.883 1.363 25.884 1.00 65.20 H C
+ATOM 2345 CZ TYR H 109 -29.029 1.406 24.827 1.00 65.20 H C
+ATOM 2346 OH TYR H 109 -29.007 0.366 23.926 1.00 65.20 H O
+ATOM 2347 N TRP H 110 -29.256 6.695 29.332 1.00 69.72 H N
+ATOM 2348 CA TRP H 110 -30.229 7.708 29.676 1.00 69.72 H C
+ATOM 2349 C TRP H 110 -30.770 7.421 31.055 1.00 69.72 H C
+ATOM 2350 O TRP H 110 -30.399 6.422 31.678 1.00 69.72 H O
+ATOM 2351 CB TRP H 110 -31.367 7.695 28.662 1.00 69.72 H C
+ATOM 2352 CG TRP H 110 -32.070 6.330 28.553 1.00 69.72 H C
+ATOM 2353 CD1 TRP H 110 -32.747 5.680 29.551 1.00 69.72 H C
+ATOM 2354 CD2 TRP H 110 -32.159 5.473 27.398 1.00 69.72 H C
+ATOM 2355 CE2 TRP H 110 -32.897 4.338 27.776 1.00 69.72 H C
+ATOM 2356 CE3 TRP H 110 -31.698 5.560 26.083 1.00 69.72 H C
+ATOM 2357 NE1 TRP H 110 -33.254 4.496 29.088 1.00 69.72 H N
+ATOM 2358 CZ2 TRP H 110 -33.176 3.302 26.892 1.00 69.72 H C
+ATOM 2359 CZ3 TRP H 110 -31.962 4.516 25.215 1.00 69.72 H C
+ATOM 2360 CH2 TRP H 110 -32.699 3.411 25.621 1.00 69.72 H C
+ATOM 2361 N GLY H 111 -31.644 8.287 31.543 1.00 78.03 H N
+ATOM 2362 CA GLY H 111 -32.282 7.980 32.798 1.00 78.03 H C
+ATOM 2363 C GLY H 111 -33.726 8.392 32.907 1.00 78.03 H C
+ATOM 2364 O GLY H 111 -34.293 8.269 33.994 1.00 78.03 H O
+ATOM 2365 N GLN H 112 -34.356 8.850 31.830 1.00 78.79 H N
+ATOM 2366 CA GLN H 112 -35.632 9.515 32.050 1.00 78.79 H C
+ATOM 2367 C GLN H 112 -36.286 9.803 30.705 1.00 78.79 H C
+ATOM 2368 O GLN H 112 -35.711 9.561 29.642 1.00 78.79 H O
+ATOM 2369 CB GLN H 112 -35.372 10.770 32.886 1.00 78.79 H C
+ATOM 2370 CG GLN H 112 -36.535 11.578 33.367 1.00 78.79 H C
+ATOM 2371 CD GLN H 112 -36.043 12.708 34.226 1.00 78.79 H C
+ATOM 2372 NE2 GLN H 112 -36.953 13.370 34.903 1.00 78.79 H N
+ATOM 2373 OE1 GLN H 112 -34.838 12.889 34.387 1.00 78.79 H O
+ATOM 2374 N GLY H 113 -37.504 10.325 30.771 1.00 78.11 H N
+ATOM 2375 CA GLY H 113 -38.345 10.615 29.633 1.00 78.11 H C
+ATOM 2376 C GLY H 113 -39.718 10.159 30.053 1.00 78.11 H C
+ATOM 2377 O GLY H 113 -39.831 9.098 30.667 1.00 78.11 H O
+ATOM 2378 N THR H 114 -40.767 10.923 29.776 1.00 91.77 H N
+ATOM 2379 CA THR H 114 -42.048 10.617 30.389 1.00 91.77 H C
+ATOM 2380 C THR H 114 -43.178 10.880 29.408 1.00 91.77 H C
+ATOM 2381 O THR H 114 -42.984 10.924 28.190 1.00 91.77 H O
+ATOM 2382 CB THR H 114 -42.267 11.423 31.671 1.00 91.77 H C
+ATOM 2383 CG2 THR H 114 -41.234 11.085 32.735 1.00 91.77 H C
+ATOM 2384 OG1 THR H 114 -42.238 12.821 31.363 1.00 91.77 H O
+ATOM 2385 N LEU H 115 -44.362 11.081 29.966 1.00103.43 H N
+ATOM 2386 CA LEU H 115 -45.621 10.819 29.295 1.00103.43 H C
+ATOM 2387 C LEU H 115 -46.606 11.889 29.730 1.00103.43 H C
+ATOM 2388 O LEU H 115 -46.226 12.926 30.278 1.00103.43 H O
+ATOM 2389 CB LEU H 115 -46.109 9.408 29.652 1.00103.43 H C
+ATOM 2390 CG LEU H 115 -46.649 9.200 31.081 1.00103.43 H C
+ATOM 2391 CD1 LEU H 115 -47.432 7.900 31.193 1.00103.43 H C
+ATOM 2392 CD2 LEU H 115 -45.549 9.231 32.145 1.00103.43 H C
+ATOM 2393 N VAL H 116 -47.889 11.650 29.483 1.00114.61 H N
+ATOM 2394 CA VAL H 116 -48.904 12.358 30.245 1.00114.61 H C
+ATOM 2395 C VAL H 116 -49.427 11.494 31.387 1.00114.61 H C
+ATOM 2396 O VAL H 116 -49.112 11.752 32.555 1.00114.61 H O
+ATOM 2397 CB VAL H 116 -50.029 12.870 29.317 1.00114.61 H C
+ATOM 2398 CG1 VAL H 116 -50.765 11.768 28.607 1.00114.61 H C
+ATOM 2399 CG2 VAL H 116 -51.041 13.582 30.143 1.00114.61 H C
+TER
+ATOM 2400 N ASP L 1 -39.798 -12.074 8.623 1.00 93.54 L N
+ATOM 2401 CA ASP L 1 -39.290 -10.736 8.881 1.00 93.54 L C
+ATOM 2402 C ASP L 1 -38.135 -10.777 9.871 1.00 93.54 L C
+ATOM 2403 O ASP L 1 -37.315 -11.699 9.854 1.00 93.54 L O
+ATOM 2404 CB ASP L 1 -40.407 -9.829 9.411 1.00 93.54 L C
+ATOM 2405 CG ASP L 1 -40.868 -10.221 10.802 1.00 93.54 L C
+ATOM 2406 OD1 ASP L 1 -40.836 -11.426 11.115 1.00 93.54 L O
+ATOM 2407 OD2 ASP L 1 -41.231 -9.323 11.588 1.00 93.54 L O1-
+ATOM 2408 N ILE L 2 -38.069 -9.766 10.730 1.00 89.86 L N
+ATOM 2409 CA ILE L 2 -37.156 -9.752 11.858 1.00 89.86 L C
+ATOM 2410 C ILE L 2 -38.016 -9.955 13.094 1.00 89.86 L C
+ATOM 2411 O ILE L 2 -38.915 -9.151 13.369 1.00 89.86 L O
+ATOM 2412 CB ILE L 2 -36.390 -8.431 11.954 1.00 89.86 L C
+ATOM 2413 CG1 ILE L 2 -35.801 -8.067 10.599 1.00 89.86 L C
+ATOM 2414 CG2 ILE L 2 -35.265 -8.582 12.960 1.00 89.86 L C
+ATOM 2415 CD1 ILE L 2 -34.644 -8.864 10.186 1.00 89.86 L C
+ATOM 2416 N GLN L 3 -37.762 -11.031 13.828 1.00 87.82 L N
+ATOM 2417 CA GLN L 3 -38.510 -11.321 15.045 1.00 87.82 L C
+ATOM 2418 C GLN L 3 -37.519 -11.799 16.090 1.00 87.82 L C
+ATOM 2419 O GLN L 3 -36.909 -12.856 15.920 1.00 87.82 L O
+ATOM 2420 CB GLN L 3 -39.592 -12.368 14.796 1.00 87.82 L C
+ATOM 2421 CG GLN L 3 -40.322 -12.799 16.048 1.00 87.82 L C
+ATOM 2422 CD GLN L 3 -40.843 -11.623 16.838 1.00 87.82 L C
+ATOM 2423 NE2 GLN L 3 -41.914 -11.017 16.353 1.00 87.82 L N
+ATOM 2424 OE1 GLN L 3 -40.289 -11.262 17.872 1.00 87.82 L O
+ATOM 2425 N MET L 4 -37.356 -11.028 17.158 1.00 84.13 L N
+ATOM 2426 CA MET L 4 -36.406 -11.344 18.216 1.00 84.13 L C
+ATOM 2427 C MET L 4 -37.143 -12.053 19.341 1.00 84.13 L C
+ATOM 2428 O MET L 4 -37.904 -11.427 20.081 1.00 84.13 L O
+ATOM 2429 CB MET L 4 -35.715 -10.084 18.722 1.00 84.13 L C
+ATOM 2430 CG MET L 4 -34.471 -9.752 17.952 1.00 84.13 L C
+ATOM 2431 SD MET L 4 -33.362 -11.156 17.947 1.00 84.13 L S
+ATOM 2432 CE MET L 4 -31.806 -10.314 17.782 1.00 84.13 L C
+ATOM 2433 N THR L 5 -36.911 -13.353 19.475 1.00 87.85 L N
+ATOM 2434 CA THR L 5 -37.610 -14.165 20.460 1.00 87.85 L C
+ATOM 2435 C THR L 5 -36.682 -14.452 21.627 1.00 87.85 L C
+ATOM 2436 O THR L 5 -35.506 -14.755 21.433 1.00 87.85 L O
+ATOM 2437 CB THR L 5 -38.113 -15.471 19.850 1.00 87.85 L C
+ATOM 2438 CG2 THR L 5 -39.179 -16.076 20.737 1.00 87.85 L C
+ATOM 2439 OG1 THR L 5 -38.679 -15.209 18.560 1.00 87.85 L O
+ATOM 2440 N GLN L 6 -37.232 -14.385 22.837 1.00 85.55 L N
+ATOM 2441 CA GLN L 6 -36.456 -14.093 24.031 1.00 85.55 L C
+ATOM 2442 C GLN L 6 -35.999 -15.305 24.825 1.00 85.55 L C
+ATOM 2443 O GLN L 6 -35.269 -15.116 25.799 1.00 85.55 L O
+ATOM 2444 CB GLN L 6 -37.265 -13.191 24.960 1.00 85.55 L C
+ATOM 2445 CG GLN L 6 -37.231 -11.751 24.557 1.00 85.55 L C
+ATOM 2446 CD GLN L 6 -36.083 -11.002 25.189 1.00 85.55 L C
+ATOM 2447 NE2 GLN L 6 -35.249 -10.405 24.361 1.00 85.55 L N
+ATOM 2448 OE1 GLN L 6 -35.939 -10.978 26.407 1.00 85.55 L O
+ATOM 2449 N SER L 7 -36.462 -16.516 24.491 1.00 91.62 L N
+ATOM 2450 CA SER L 7 -36.022 -17.796 25.058 1.00 91.62 L C
+ATOM 2451 C SER L 7 -35.526 -17.677 26.500 1.00 91.62 L C
+ATOM 2452 O SER L 7 -34.376 -18.028 26.789 1.00 91.62 L O
+ATOM 2453 CB SER L 7 -34.936 -18.408 24.172 1.00 91.62 L C
+ATOM 2454 OG SER L 7 -35.410 -18.654 22.860 1.00 91.62 L O
+ATOM 2455 N PRO L 8 -36.355 -17.195 27.428 1.00 96.66 L N
+ATOM 2456 CA PRO L 8 -35.842 -16.708 28.717 1.00 96.66 L C
+ATOM 2457 C PRO L 8 -35.394 -17.763 29.714 1.00 96.66 L C
+ATOM 2458 O PRO L 8 -35.247 -18.943 29.390 1.00 96.66 L O
+ATOM 2459 CB PRO L 8 -37.033 -15.922 29.281 1.00 96.66 L C
+ATOM 2460 CG PRO L 8 -37.929 -15.678 28.107 1.00 96.66 L C
+ATOM 2461 CD PRO L 8 -37.787 -16.903 27.284 1.00 96.66 L C
+ATOM 2462 N SER L 9 -35.136 -17.300 30.935 1.00100.21 L N
+ATOM 2463 CA SER L 9 -34.840 -18.098 32.118 1.00100.21 L C
+ATOM 2464 C SER L 9 -34.912 -17.159 33.312 1.00100.21 L C
+ATOM 2465 O SER L 9 -34.819 -15.943 33.149 1.00100.21 L O
+ATOM 2466 CB SER L 9 -33.476 -18.776 32.012 1.00100.21 L C
+ATOM 2467 OG SER L 9 -32.444 -17.816 31.976 1.00100.21 L O
+ATOM 2468 N SER L 10 -35.094 -17.713 34.505 1.00107.23 L N
+ATOM 2469 CA SER L 10 -35.465 -16.867 35.643 1.00107.23 L C
+ATOM 2470 C SER L 10 -35.304 -17.620 36.961 1.00107.23 L C
+ATOM 2471 O SER L 10 -34.737 -18.720 37.017 1.00107.23 L O
+ATOM 2472 CB SER L 10 -36.903 -16.370 35.508 1.00107.23 L C
+ATOM 2473 OG SER L 10 -37.278 -15.623 36.651 1.00107.23 L O
+ATOM 2474 N LEU L 11 -35.804 -16.990 38.027 1.00117.87 L N
+ATOM 2475 CA LEU L 11 -36.202 -17.517 39.329 1.00117.87 L C
+ATOM 2476 C LEU L 11 -35.071 -17.766 40.323 1.00117.87 L C
+ATOM 2477 O LEU L 11 -35.366 -18.046 41.482 1.00117.87 L O
+ATOM 2478 CB LEU L 11 -37.009 -18.827 39.217 1.00117.87 L C
+ATOM 2479 CG LEU L 11 -38.543 -18.808 39.137 1.00117.87 L C
+ATOM 2480 CD1 LEU L 11 -39.032 -18.385 37.765 1.00117.87 L C
+ATOM 2481 CD2 LEU L 11 -39.113 -20.171 39.501 1.00117.87 L C
+ATOM 2482 N SER L 12 -33.800 -17.631 39.958 1.00111.40 L N
+ATOM 2483 CA SER L 12 -32.766 -18.153 40.849 1.00111.40 L C
+ATOM 2484 C SER L 12 -31.473 -17.367 40.694 1.00111.40 L C
+ATOM 2485 O SER L 12 -30.768 -17.533 39.695 1.00111.40 L O
+ATOM 2486 CB SER L 12 -32.531 -19.626 40.559 1.00111.40 L C
+ATOM 2487 OG SER L 12 -32.181 -19.811 39.201 1.00111.40 L O
+ATOM 2488 N ALA L 13 -31.153 -16.537 41.698 1.00108.28 L N
+ATOM 2489 CA ALA L 13 -29.841 -15.897 41.744 1.00108.28 L C
+ATOM 2490 C ALA L 13 -29.251 -15.832 43.151 1.00108.28 L C
+ATOM 2491 O ALA L 13 -28.265 -15.113 43.344 1.00108.28 L O
+ATOM 2492 CB ALA L 13 -29.900 -14.462 41.173 1.00108.28 L C
+ATOM 2493 N SER L 14 -29.735 -16.642 44.095 1.00106.72 L N
+ATOM 2494 CA SER L 14 -29.699 -16.317 45.518 1.00106.72 L C
+ATOM 2495 C SER L 14 -28.464 -15.548 45.955 1.00106.72 L C
+ATOM 2496 O SER L 14 -28.580 -14.462 46.525 1.00106.72 L O
+ATOM 2497 CB SER L 14 -29.774 -17.599 46.339 1.00106.72 L C
+ATOM 2498 OG SER L 14 -28.884 -18.584 45.838 1.00106.72 L O
+ATOM 2499 N VAL L 15 -27.286 -16.111 45.719 1.00101.21 L N
+ATOM 2500 CA VAL L 15 -26.052 -15.377 45.948 1.00101.21 L C
+ATOM 2501 C VAL L 15 -25.109 -15.569 44.775 1.00101.21 L C
+ATOM 2502 O VAL L 15 -24.242 -14.728 44.519 1.00101.21 L O
+ATOM 2503 CB VAL L 15 -25.381 -15.804 47.268 1.00101.21 L C
+ATOM 2504 CG1 VAL L 15 -26.224 -15.405 48.474 1.00101.21 L C
+ATOM 2505 CG2 VAL L 15 -25.133 -17.305 47.286 1.00101.21 L C
+ATOM 2506 N GLY L 16 -25.240 -16.689 44.078 1.00 97.43 L N
+ATOM 2507 CA GLY L 16 -24.297 -17.001 43.028 1.00 97.43 L C
+ATOM 2508 C GLY L 16 -24.885 -17.796 41.888 1.00 97.43 L C
+ATOM 2509 O GLY L 16 -24.145 -18.319 41.050 1.00 97.43 L O
+ATOM 2510 N ASP L 17 -26.207 -17.913 41.852 1.00102.53 L N
+ATOM 2511 CA ASP L 17 -26.843 -18.869 40.957 1.00102.53 L C
+ATOM 2512 C ASP L 17 -26.649 -18.476 39.498 1.00102.53 L C
+ATOM 2513 O ASP L 17 -27.185 -17.465 39.036 1.00102.53 L O
+ATOM 2514 CB ASP L 17 -28.327 -18.993 41.291 1.00102.53 L C
+ATOM 2515 CG ASP L 17 -28.575 -19.177 42.783 1.00102.53 L C
+ATOM 2516 OD1 ASP L 17 -27.739 -19.812 43.455 1.00102.53 L O
+ATOM 2517 OD2 ASP L 17 -29.617 -18.709 43.283 1.00102.53 L O1-
+ATOM 2518 N ARG L 18 -25.888 -19.294 38.783 1.00 92.49 L N
+ATOM 2519 CA ARG L 18 -25.641 -19.086 37.366 1.00 92.49 L C
+ATOM 2520 C ARG L 18 -26.930 -19.249 36.571 1.00 92.49 L C
+ATOM 2521 O ARG L 18 -27.718 -20.165 36.812 1.00 92.49 L O
+ATOM 2522 CB ARG L 18 -24.580 -20.079 36.895 1.00 92.49 L C
+ATOM 2523 CG ARG L 18 -24.433 -20.293 35.400 1.00 92.49 L C
+ATOM 2524 CD ARG L 18 -23.813 -19.082 34.719 1.00 92.49 L C
+ATOM 2525 NE ARG L 18 -23.250 -19.457 33.425 1.00 92.49 L N
+ATOM 2526 CZ ARG L 18 -23.911 -19.425 32.277 1.00 92.49 L C
+ATOM 2527 NH1 ARG L 18 -25.172 -19.023 32.258 1.00 92.49 L N1+
+ATOM 2528 NH2 ARG L 18 -23.310 -19.783 31.152 1.00 92.49 L N
+ATOM 2529 N VAL L 19 -27.150 -18.338 35.625 1.00 86.12 L N
+ATOM 2530 CA VAL L 19 -28.344 -18.313 34.787 1.00 86.12 L C
+ATOM 2531 C VAL L 19 -27.896 -18.101 33.350 1.00 86.12 L C
+ATOM 2532 O VAL L 19 -26.950 -17.352 33.099 1.00 86.12 L O
+ATOM 2533 CB VAL L 19 -29.317 -17.198 35.224 1.00 86.12 L C
+ATOM 2534 CG1 VAL L 19 -30.499 -17.116 34.291 1.00 86.12 L C
+ATOM 2535 CG2 VAL L 19 -29.786 -17.418 36.641 1.00 86.12 L C
+ATOM 2536 N THR L 20 -28.541 -18.785 32.411 1.00 83.55 L N
+ATOM 2537 CA THR L 20 -28.312 -18.571 30.988 1.00 83.55 L C
+ATOM 2538 C THR L 20 -29.599 -18.126 30.321 1.00 83.55 L C
+ATOM 2539 O THR L 20 -30.627 -18.798 30.438 1.00 83.55 L O
+ATOM 2540 CB THR L 20 -27.782 -19.827 30.305 1.00 83.55 L C
+ATOM 2541 CG2 THR L 20 -27.571 -19.597 28.840 1.00 83.55 L C
+ATOM 2542 OG1 THR L 20 -26.541 -20.202 30.901 1.00 83.55 L O
+ATOM 2543 N ILE L 21 -29.535 -17.002 29.611 1.00 81.94 L N
+ATOM 2544 CA ILE L 21 -30.667 -16.450 28.880 1.00 81.94 L C
+ATOM 2545 C ILE L 21 -30.288 -16.440 27.406 1.00 81.94 L C
+ATOM 2546 O ILE L 21 -29.137 -16.161 27.063 1.00 81.94 L O
+ATOM 2547 CB ILE L 21 -31.028 -15.041 29.376 1.00 81.94 L C
+ATOM 2548 CG1 ILE L 21 -31.456 -15.088 30.831 1.00 81.94 L C
+ATOM 2549 CG2 ILE L 21 -32.187 -14.504 28.604 1.00 81.94 L C
+ATOM 2550 CD1 ILE L 21 -31.572 -13.740 31.448 1.00 81.94 L C
+ATOM 2551 N THR L 22 -31.235 -16.788 26.545 1.00 85.16 L N
+ATOM 2552 CA THR L 22 -30.988 -16.926 25.121 1.00 85.16 L C
+ATOM 2553 C THR L 22 -32.030 -16.133 24.350 1.00 85.16 L C
+ATOM 2554 O THR L 22 -33.126 -15.900 24.852 1.00 85.16 L O
+ATOM 2555 CB THR L 22 -31.042 -18.383 24.713 1.00 85.16 L C
+ATOM 2556 CG2 THR L 22 -29.842 -19.116 25.245 1.00 85.16 L C
+ATOM 2557 OG1 THR L 22 -32.209 -18.977 25.282 1.00 85.16 L O
+ATOM 2558 N CYS L 23 -31.692 -15.703 23.133 1.00 85.95 L N
+ATOM 2559 CA CYS L 23 -32.666 -15.037 22.278 1.00 85.95 L C
+ATOM 2560 C CYS L 23 -32.593 -15.552 20.846 1.00 85.95 L C
+ATOM 2561 O CYS L 23 -31.514 -15.801 20.314 1.00 85.95 L O
+ATOM 2562 CB CYS L 23 -32.487 -13.514 22.275 1.00 85.95 L C
+ATOM 2563 SG CYS L 23 -30.842 -12.916 21.847 1.00 85.95 L S
+ATOM 2564 N ARG L 24 -33.761 -15.637 20.205 1.00 89.44 L N
+ATOM 2565 CA ARG L 24 -33.936 -16.152 18.850 1.00 89.44 L C
+ATOM 2566 C ARG L 24 -34.352 -15.059 17.877 1.00 89.44 L C
+ATOM 2567 O ARG L 24 -35.024 -14.100 18.254 1.00 89.44 L O
+ATOM 2568 CB ARG L 24 -34.995 -17.267 18.830 1.00 89.44 L C
+ATOM 2569 CG ARG L 24 -35.280 -17.876 17.454 1.00 89.44 L C
+ATOM 2570 CD ARG L 24 -34.193 -18.662 16.768 1.00 89.44 L C
+ATOM 2571 NE ARG L 24 -34.744 -19.146 15.504 1.00 89.44 L N
+ATOM 2572 CZ ARG L 24 -34.042 -19.743 14.545 1.00 89.44 L C
+ATOM 2573 NH1 ARG L 24 -32.755 -19.985 14.708 1.00 89.44 L N1+
+ATOM 2574 NH2 ARG L 24 -34.629 -20.132 13.423 1.00 89.44 L N
+ATOM 2575 N ALA L 25 -33.967 -15.235 16.608 1.00 85.43 L N
+ATOM 2576 CA ALA L 25 -34.282 -14.294 15.542 1.00 85.43 L C
+ATOM 2577 C ALA L 25 -34.785 -15.016 14.294 1.00 85.43 L C
+ATOM 2578 O ALA L 25 -34.562 -16.214 14.106 1.00 85.43 L O
+ATOM 2579 CB ALA L 25 -33.064 -13.444 15.177 1.00 85.43 L C
+ATOM 2580 N SER L 26 -35.479 -14.258 13.438 1.00 84.32 L N
+ATOM 2581 CA SER L 26 -35.953 -14.703 12.131 1.00 84.32 L C
+ATOM 2582 C SER L 26 -35.066 -14.231 10.981 1.00 84.32 L C
+ATOM 2583 O SER L 26 -35.416 -14.431 9.815 1.00 84.32 L O
+ATOM 2584 CB SER L 26 -37.381 -14.217 11.912 1.00 84.32 L C
+ATOM 2585 OG SER L 26 -38.171 -14.525 13.040 1.00 84.32 L O
+ATOM 2586 N GLN L 27 -33.947 -13.592 11.297 1.00 81.18 L N
+ATOM 2587 CA GLN L 27 -32.897 -13.168 10.381 1.00 81.18 L C
+ATOM 2588 C GLN L 27 -31.585 -13.813 10.786 1.00 81.18 L C
+ATOM 2589 O GLN L 27 -31.517 -14.543 11.776 1.00 81.18 L O
+ATOM 2590 CB GLN L 27 -32.801 -11.647 10.356 1.00 81.18 L C
+ATOM 2591 CG GLN L 27 -32.455 -11.022 11.716 1.00 81.18 L C
+ATOM 2592 CD GLN L 27 -31.004 -10.978 12.084 1.00 81.18 L C
+ATOM 2593 NE2 GLN L 27 -30.161 -10.916 11.069 1.00 81.18 L N
+ATOM 2594 OE1 GLN L 27 -30.634 -11.022 13.247 1.00 81.18 L O
+ATOM 2595 N ASP L 28 -30.550 -13.603 9.989 1.00 80.68 L N
+ATOM 2596 CA ASP L 28 -29.278 -14.254 10.250 1.00 80.68 L C
+ATOM 2597 C ASP L 28 -28.225 -13.194 10.545 1.00 80.68 L C
+ATOM 2598 O ASP L 28 -28.080 -12.229 9.790 1.00 80.68 L O
+ATOM 2599 CB ASP L 28 -28.872 -15.138 9.070 1.00 80.68 L C
+ATOM 2600 CG ASP L 28 -27.673 -16.012 9.378 1.00 80.68 L C
+ATOM 2601 OD1 ASP L 28 -27.138 -15.909 10.496 1.00 80.68 L O
+ATOM 2602 OD2 ASP L 28 -27.287 -16.832 8.516 1.00 80.68 L O1-
+ATOM 2603 N VAL L 29 -27.487 -13.388 11.639 1.00 75.75 L N
+ATOM 2604 CA VAL L 29 -26.477 -12.461 12.142 1.00 75.75 L C
+ATOM 2605 C VAL L 29 -25.101 -12.940 11.712 1.00 75.75 L C
+ATOM 2606 O VAL L 29 -24.960 -14.044 11.182 1.00 75.75 L O
+ATOM 2607 CB VAL L 29 -26.550 -12.354 13.669 1.00 75.75 L C
+ATOM 2608 CG1 VAL L 29 -27.856 -11.782 14.082 1.00 75.75 L C
+ATOM 2609 CG2 VAL L 29 -26.388 -13.715 14.290 1.00 75.75 L C
+ATOM 2610 N ASN L 30 -24.075 -12.122 11.908 1.00 73.62 L N
+ATOM 2611 CA ASN L 30 -22.710 -12.612 11.680 1.00 73.62 L C
+ATOM 2612 C ASN L 30 -22.108 -13.178 12.962 1.00 73.62 L C
+ATOM 2613 O ASN L 30 -21.873 -14.386 13.060 1.00 73.62 L O
+ATOM 2614 CB ASN L 30 -21.820 -11.498 11.131 1.00 73.62 L C
+ATOM 2615 CG ASN L 30 -20.534 -12.021 10.538 1.00 73.62 L C
+ATOM 2616 ND2 ASN L 30 -20.282 -11.685 9.279 1.00 73.62 L N
+ATOM 2617 OD1 ASN L 30 -19.770 -12.711 11.204 1.00 73.62 L O
+ATOM 2618 N THR L 31 -21.847 -12.320 13.954 1.00 67.73 L N
+ATOM 2619 CA THR L 31 -21.416 -12.774 15.271 1.00 67.73 L C
+ATOM 2620 C THR L 31 -21.966 -11.897 16.382 1.00 67.73 L C
+ATOM 2621 O THR L 31 -21.218 -11.535 17.290 1.00 67.73 L O
+ATOM 2622 CB THR L 31 -19.893 -12.792 15.415 1.00 67.73 L C
+ATOM 2623 CG2 THR L 31 -19.181 -13.278 14.182 1.00 67.73 L C
+ATOM 2624 OG1 THR L 31 -19.439 -11.471 15.715 1.00 67.73 L O
+ATOM 2625 N ALA L 32 -23.238 -11.504 16.356 1.00 67.02 L N
+ATOM 2626 CA ALA L 32 -23.573 -10.537 17.399 1.00 67.02 L C
+ATOM 2627 C ALA L 32 -25.073 -10.318 17.551 1.00 67.02 L C
+ATOM 2628 O ALA L 32 -25.847 -10.547 16.621 1.00 67.02 L O
+ATOM 2629 CB ALA L 32 -22.900 -9.182 17.130 1.00 67.02 L C
+ATOM 2630 N VAL L 33 -25.461 -9.891 18.764 1.00 68.92 L N
+ATOM 2631 CA VAL L 33 -26.671 -9.123 19.074 1.00 68.92 L C
+ATOM 2632 C VAL L 33 -26.276 -8.052 20.084 1.00 68.92 L C
+ATOM 2633 O VAL L 33 -25.088 -7.818 20.305 1.00 68.92 L O
+ATOM 2634 CB VAL L 33 -27.817 -9.978 19.627 1.00 68.92 L C
+ATOM 2635 CG1 VAL L 33 -28.411 -10.830 18.532 1.00 68.92 L C
+ATOM 2636 CG2 VAL L 33 -27.343 -10.780 20.805 1.00 68.92 L C
+ATOM 2637 N ALA L 34 -27.250 -7.353 20.657 1.00 64.25 L N
+ATOM 2638 CA ALA L 34 -27.001 -6.441 21.763 1.00 64.25 L C
+ATOM 2639 C ALA L 34 -27.748 -6.887 23.010 1.00 64.25 L C
+ATOM 2640 O ALA L 34 -28.797 -7.522 22.925 1.00 64.25 L O
+ATOM 2641 CB ALA L 34 -27.427 -5.026 21.405 1.00 64.25 L C
+ATOM 2642 N TRP L 35 -27.217 -6.518 24.179 1.00 63.07 L N
+ATOM 2643 CA TRP L 35 -27.824 -6.862 25.460 1.00 63.07 L C
+ATOM 2644 C TRP L 35 -27.988 -5.622 26.308 1.00 63.07 L C
+ATOM 2645 O TRP L 35 -27.227 -4.669 26.195 1.00 63.07 L O
+ATOM 2646 CB TRP L 35 -27.015 -7.884 26.261 1.00 63.07 L C
+ATOM 2647 CG TRP L 35 -27.255 -9.275 25.853 1.00 63.07 L C
+ATOM 2648 CD1 TRP L 35 -26.437 -10.057 25.115 1.00 63.07 L C
+ATOM 2649 CD2 TRP L 35 -28.412 -10.054 26.141 1.00 63.07 L C
+ATOM 2650 CE2 TRP L 35 -28.228 -11.304 25.546 1.00 63.07 L C
+ATOM 2651 CE3 TRP L 35 -29.584 -9.815 26.854 1.00 63.07 L C
+ATOM 2652 NE1 TRP L 35 -27.009 -11.284 24.925 1.00 63.07 L N
+ATOM 2653 CZ2 TRP L 35 -29.171 -12.313 25.638 1.00 63.07 L C
+ATOM 2654 CZ3 TRP L 35 -30.520 -10.807 26.933 1.00 63.07 L C
+ATOM 2655 CH2 TRP L 35 -30.309 -12.043 26.334 1.00 63.07 L C
+ATOM 2656 N TYR L 36 -28.977 -5.662 27.185 1.00 64.02 L N
+ATOM 2657 CA TYR L 36 -29.492 -4.446 27.789 1.00 64.02 L C
+ATOM 2658 C TYR L 36 -30.252 -4.783 29.068 1.00 64.02 L C
+ATOM 2659 O TYR L 36 -31.410 -5.192 28.992 1.00 64.02 L O
+ATOM 2660 CB TYR L 36 -30.371 -3.771 26.759 1.00 64.02 L C
+ATOM 2661 CG TYR L 36 -31.170 -2.560 27.141 1.00 64.02 L C
+ATOM 2662 CD1 TYR L 36 -30.657 -1.514 27.869 1.00 64.02 L C
+ATOM 2663 CD2 TYR L 36 -32.472 -2.471 26.705 1.00 64.02 L C
+ATOM 2664 CE1 TYR L 36 -31.457 -0.429 28.184 1.00 64.02 L C
+ATOM 2665 CE2 TYR L 36 -33.254 -1.417 26.995 1.00 64.02 L C
+ATOM 2666 CZ TYR L 36 -32.762 -0.397 27.726 1.00 64.02 L C
+ATOM 2667 OH TYR L 36 -33.641 0.625 27.960 1.00 64.02 L O
+ATOM 2668 N GLN L 37 -29.633 -4.553 30.227 1.00 63.96 L N
+ATOM 2669 CA GLN L 37 -30.413 -4.673 31.448 1.00 63.96 L C
+ATOM 2670 C GLN L 37 -31.522 -3.643 31.453 1.00 63.96 L C
+ATOM 2671 O GLN L 37 -31.546 -2.709 30.664 1.00 63.96 L O
+ATOM 2672 CB GLN L 37 -29.596 -4.449 32.721 1.00 63.96 L C
+ATOM 2673 CG GLN L 37 -28.315 -5.209 32.932 1.00 63.96 L C
+ATOM 2674 CD GLN L 37 -27.921 -5.230 34.413 1.00 63.96 L C
+ATOM 2675 NE2 GLN L 37 -26.629 -5.307 34.698 1.00 63.96 L N
+ATOM 2676 OE1 GLN L 37 -28.783 -5.179 35.283 1.00 63.96 L O
+ATOM 2677 N GLN L 38 -32.445 -3.819 32.377 1.00 67.31 L N
+ATOM 2678 CA GLN L 38 -33.446 -2.800 32.631 1.00 67.31 L C
+ATOM 2679 C GLN L 38 -33.846 -2.973 34.088 1.00 67.31 L C
+ATOM 2680 O GLN L 38 -34.714 -3.791 34.398 1.00 67.31 L O
+ATOM 2681 CB GLN L 38 -34.631 -2.949 31.700 1.00 67.31 L C
+ATOM 2682 CG GLN L 38 -35.254 -1.644 31.306 1.00 67.31 L C
+ATOM 2683 CD GLN L 38 -36.342 -1.227 32.240 1.00 67.31 L C
+ATOM 2684 NE2 GLN L 38 -36.697 0.046 32.206 1.00 67.31 L N
+ATOM 2685 OE1 GLN L 38 -36.871 -2.042 32.984 1.00 67.31 L O
+ATOM 2686 N LYS L 39 -33.212 -2.209 34.966 1.00 74.72 L N
+ATOM 2687 CA LYS L 39 -33.412 -2.397 36.390 1.00 74.72 L C
+ATOM 2688 C LYS L 39 -34.875 -2.136 36.742 1.00 74.72 L C
+ATOM 2689 O LYS L 39 -35.537 -1.319 36.100 1.00 74.72 L O
+ATOM 2690 CB LYS L 39 -32.494 -1.466 37.172 1.00 74.72 L C
+ATOM 2691 CG LYS L 39 -31.070 -1.430 36.633 1.00 74.72 L C
+ATOM 2692 CD LYS L 39 -30.403 -2.801 36.693 1.00 74.72 L C
+ATOM 2693 CE LYS L 39 -28.985 -2.706 37.239 1.00 74.72 L C
+ATOM 2694 NZ LYS L 39 -28.222 -3.977 37.087 1.00 74.72 L N1+
+ATOM 2695 N PRO L 40 -35.410 -2.830 37.730 1.00 78.78 L N
+ATOM 2696 CA PRO L 40 -36.858 -2.785 37.957 1.00 78.78 L C
+ATOM 2697 C PRO L 40 -37.351 -1.413 38.366 1.00 78.78 L C
+ATOM 2698 O PRO L 40 -37.006 -0.913 39.440 1.00 78.78 L O
+ATOM 2699 CB PRO L 40 -37.070 -3.804 39.082 1.00 78.78 L C
+ATOM 2700 CG PRO L 40 -35.814 -4.597 39.137 1.00 78.78 L C
+ATOM 2701 CD PRO L 40 -34.735 -3.674 38.722 1.00 78.78 L C
+ATOM 2702 N GLY L 41 -38.162 -0.798 37.510 1.00 80.49 L N
+ATOM 2703 CA GLY L 41 -38.793 0.464 37.821 1.00 80.49 L C
+ATOM 2704 C GLY L 41 -37.943 1.693 37.606 1.00 80.49 L C
+ATOM 2705 O GLY L 41 -38.410 2.798 37.900 1.00 80.49 L O
+ATOM 2706 N LYS L 42 -36.721 1.544 37.098 1.00 79.43 L N
+ATOM 2707 CA LYS L 42 -35.851 2.689 36.880 1.00 79.43 L C
+ATOM 2708 C LYS L 42 -34.615 2.258 36.103 1.00 79.43 L C
+ATOM 2709 O LYS L 42 -34.192 1.102 36.179 1.00 79.43 L O
+ATOM 2710 CB LYS L 42 -35.429 3.326 38.210 1.00 79.43 L C
+ATOM 2711 CG LYS L 42 -34.712 2.372 39.147 1.00 79.43 L C
+ATOM 2712 CD LYS L 42 -34.634 2.918 40.571 1.00 79.43 L C
+ATOM 2713 CE LYS L 42 -35.779 2.398 41.425 1.00 79.43 L C
+ATOM 2714 NZ LYS L 42 -35.589 0.961 41.775 1.00 79.43 L N1+
+ATOM 2715 N ALA L 43 -34.040 3.210 35.365 1.00 80.95 L N
+ATOM 2716 CA ALA L 43 -32.656 3.160 34.896 1.00 80.95 L C
+ATOM 2717 C ALA L 43 -32.320 2.016 33.947 1.00 80.95 L C
+ATOM 2718 O ALA L 43 -31.724 1.016 34.366 1.00 80.95 L O
+ATOM 2719 CB ALA L 43 -31.711 3.104 36.097 1.00 80.95 L C
+ATOM 2720 N PRO L 44 -32.693 2.116 32.675 1.00 69.79 L N
+ATOM 2721 CA PRO L 44 -32.109 1.237 31.660 1.00 69.79 L C
+ATOM 2722 C PRO L 44 -30.604 1.397 31.563 1.00 69.79 L C
+ATOM 2723 O PRO L 44 -30.054 2.459 31.845 1.00 69.79 L O
+ATOM 2724 CB PRO L 44 -32.790 1.724 30.394 1.00 69.79 L C
+ATOM 2725 CG PRO L 44 -34.134 2.036 30.913 1.00 69.79 L C
+ATOM 2726 CD PRO L 44 -33.746 2.930 32.057 1.00 69.79 L C
+ATOM 2727 N LYS L 45 -29.931 0.326 31.157 1.00 60.30 L N
+ATOM 2728 CA LYS L 45 -28.483 0.280 31.257 1.00 60.30 L C
+ATOM 2729 C LYS L 45 -27.918 -0.606 30.173 1.00 60.30 L C
+ATOM 2730 O LYS L 45 -28.451 -1.683 29.919 1.00 60.30 L O
+ATOM 2731 CB LYS L 45 -28.034 -0.288 32.597 1.00 60.30 L C
+ATOM 2732 CG LYS L 45 -28.159 0.635 33.766 1.00 60.30 L C
+ATOM 2733 CD LYS L 45 -27.858 -0.111 35.048 1.00 60.30 L C
+ATOM 2734 CE LYS L 45 -26.676 -1.054 34.884 1.00 60.30 L C
+ATOM 2735 NZ LYS L 45 -25.373 -0.361 35.041 1.00 60.30 L N1+
+ATOM 2736 N LEU L 46 -26.814 -0.185 29.577 1.00 51.46 L N
+ATOM 2737 CA LEU L 46 -26.108 -1.118 28.726 1.00 51.46 L C
+ATOM 2738 C LEU L 46 -25.646 -2.259 29.619 1.00 51.46 L C
+ATOM 2739 O LEU L 46 -25.488 -2.099 30.829 1.00 51.46 L O
+ATOM 2740 CB LEU L 46 -24.884 -0.488 28.058 1.00 51.46 L C
+ATOM 2741 CG LEU L 46 -24.208 -1.440 27.067 1.00 51.46 L C
+ATOM 2742 CD1 LEU L 46 -25.141 -1.719 25.911 1.00 51.46 L C
+ATOM 2743 CD2 LEU L 46 -22.793 -1.189 26.639 1.00 51.46 L C
+ATOM 2744 N LEU L 47 -25.423 -3.413 29.013 1.00 59.03 L N
+ATOM 2745 CA LEU L 47 -24.884 -4.583 29.675 1.00 59.03 L C
+ATOM 2746 C LEU L 47 -23.883 -5.134 28.675 1.00 59.03 L C
+ATOM 2747 O LEU L 47 -23.355 -4.384 27.856 1.00 59.03 L O
+ATOM 2748 CB LEU L 47 -26.021 -5.524 30.086 1.00 59.03 L C
+ATOM 2749 CG LEU L 47 -25.659 -6.785 30.858 1.00 59.03 L C
+ATOM 2750 CD1 LEU L 47 -25.038 -6.393 32.168 1.00 59.03 L C
+ATOM 2751 CD2 LEU L 47 -26.870 -7.666 31.062 1.00 59.03 L C
+ATOM 2752 N ILE L 48 -23.560 -6.412 28.745 1.00 60.66 L N
+ATOM 2753 CA ILE L 48 -22.790 -7.047 27.683 1.00 60.66 L C
+ATOM 2754 C ILE L 48 -23.294 -6.533 26.340 1.00 60.66 L C
+ATOM 2755 O ILE L 48 -24.491 -6.292 26.183 1.00 60.66 L O
+ATOM 2756 CB ILE L 48 -22.929 -8.580 27.784 1.00 60.66 L C
+ATOM 2757 CG1 ILE L 48 -22.208 -9.120 29.023 1.00 60.66 L C
+ATOM 2758 CG2 ILE L 48 -22.485 -9.279 26.502 1.00 60.66 L C
+ATOM 2759 CD1 ILE L 48 -20.719 -9.208 28.884 1.00 60.66 L C
+ATOM 2760 N TYR L 49 -22.407 -6.274 25.379 1.00 63.32 L N
+ATOM 2761 CA TYR L 49 -23.003 -5.832 24.129 1.00 63.32 L C
+ATOM 2762 C TYR L 49 -23.217 -6.926 23.106 1.00 63.32 L C
+ATOM 2763 O TYR L 49 -24.357 -7.227 22.763 1.00 63.32 L O
+ATOM 2764 CB TYR L 49 -22.208 -4.749 23.414 1.00 63.32 L C
+ATOM 2765 CG TYR L 49 -23.166 -4.002 22.516 1.00 63.32 L C
+ATOM 2766 CD1 TYR L 49 -24.352 -3.512 23.032 1.00 63.32 L C
+ATOM 2767 CD2 TYR L 49 -22.986 -3.936 21.144 1.00 63.32 L C
+ATOM 2768 CE1 TYR L 49 -25.265 -2.865 22.244 1.00 63.32 L C
+ATOM 2769 CE2 TYR L 49 -23.921 -3.303 20.341 1.00 63.32 L C
+ATOM 2770 CZ TYR L 49 -25.060 -2.777 20.909 1.00 63.32 L C
+ATOM 2771 OH TYR L 49 -26.006 -2.140 20.159 1.00 63.32 L O
+ATOM 2772 N SER L 50 -22.151 -7.532 22.608 1.00 62.35 L N
+ATOM 2773 CA SER L 50 -22.308 -8.335 21.401 1.00 62.35 L C
+ATOM 2774 C SER L 50 -21.756 -9.721 21.602 1.00 62.35 L C
+ATOM 2775 O SER L 50 -21.692 -10.230 22.720 1.00 62.35 L O
+ATOM 2776 CB SER L 50 -21.608 -7.743 20.190 1.00 62.35 L C
+ATOM 2777 OG SER L 50 -20.211 -7.868 20.338 1.00 62.35 L O
+ATOM 2778 N ALA L 51 -21.434 -10.360 20.495 1.00 68.40 L N
+ATOM 2779 CA ALA L 51 -20.653 -11.577 20.520 1.00 68.40 L C
+ATOM 2780 C ALA L 51 -19.418 -11.410 19.644 1.00 68.40 L C
+ATOM 2781 O ALA L 51 -19.345 -10.497 18.817 1.00 68.40 L O
+ATOM 2782 CB ALA L 51 -21.493 -12.744 20.062 1.00 68.40 L C
+ATOM 2783 N SER L 52 -18.422 -12.261 19.887 1.00 67.76 L N
+ATOM 2784 CA SER L 52 -17.102 -12.328 19.269 1.00 67.76 L C
+ATOM 2785 C SER L 52 -16.252 -11.167 19.739 1.00 67.76 L C
+ATOM 2786 O SER L 52 -15.033 -11.216 19.609 1.00 67.76 L O
+ATOM 2787 CB SER L 52 -17.148 -12.334 17.729 1.00 67.76 L C
+ATOM 2788 OG SER L 52 -15.857 -12.367 17.169 1.00 67.76 L O
+ATOM 2789 N PHE L 53 -16.856 -10.151 20.331 1.00 60.75 L N
+ATOM 2790 CA PHE L 53 -16.339 -9.086 21.136 1.00 60.75 L C
+ATOM 2791 C PHE L 53 -17.473 -8.712 22.076 1.00 60.75 L C
+ATOM 2792 O PHE L 53 -18.393 -7.990 21.732 1.00 60.75 L O
+ATOM 2793 CB PHE L 53 -15.850 -7.883 20.329 1.00 60.75 L C
+ATOM 2794 CG PHE L 53 -14.639 -8.158 19.525 1.00 60.75 L C
+ATOM 2795 CD1 PHE L 53 -13.403 -8.127 20.126 1.00 60.75 L C
+ATOM 2796 CD2 PHE L 53 -14.715 -8.481 18.207 1.00 60.75 L C
+ATOM 2797 CE1 PHE L 53 -12.270 -8.381 19.416 1.00 60.75 L C
+ATOM 2798 CE2 PHE L 53 -13.584 -8.737 17.504 1.00 60.75 L C
+ATOM 2799 CZ PHE L 53 -12.363 -8.686 18.103 1.00 60.75 L C
+ATOM 2800 N LEU L 54 -17.430 -9.258 23.277 1.00 59.34 L N
+ATOM 2801 CA LEU L 54 -18.285 -8.729 24.314 1.00 59.34 L C
+ATOM 2802 C LEU L 54 -17.694 -7.414 24.774 1.00 59.34 L C
+ATOM 2803 O LEU L 54 -16.488 -7.194 24.677 1.00 59.34 L O
+ATOM 2804 CB LEU L 54 -18.412 -9.697 25.491 1.00 59.34 L C
+ATOM 2805 CG LEU L 54 -17.249 -9.913 26.471 1.00 59.34 L C
+ATOM 2806 CD1 LEU L 54 -17.727 -10.607 27.741 1.00 59.34 L C
+ATOM 2807 CD2 LEU L 54 -16.116 -10.706 25.832 1.00 59.34 L C
+ATOM 2808 N TYR L 55 -18.542 -6.516 25.211 1.00 65.14 L N
+ATOM 2809 CA TYR L 55 -18.073 -5.376 25.965 1.00 65.14 L C
+ATOM 2810 C TYR L 55 -19.066 -5.124 27.075 1.00 65.14 L C
+ATOM 2811 O TYR L 55 -20.272 -5.094 26.835 1.00 65.14 L O
+ATOM 2812 CB TYR L 55 -17.947 -4.169 25.105 1.00 65.14 L C
+ATOM 2813 CG TYR L 55 -17.420 -2.956 25.803 1.00 65.14 L C
+ATOM 2814 CD1 TYR L 55 -16.076 -2.846 26.102 1.00 65.14 L C
+ATOM 2815 CD2 TYR L 55 -18.259 -1.918 26.160 1.00 65.14 L C
+ATOM 2816 CE1 TYR L 55 -15.577 -1.734 26.737 1.00 65.14 L C
+ATOM 2817 CE2 TYR L 55 -17.769 -0.800 26.803 1.00 65.14 L C
+ATOM 2818 CZ TYR L 55 -16.429 -0.713 27.084 1.00 65.14 L C
+ATOM 2819 OH TYR L 55 -15.935 0.403 27.717 1.00 65.14 L O
+ATOM 2820 N SER L 56 -18.559 -4.905 28.275 1.00 61.13 L N
+ATOM 2821 CA SER L 56 -19.356 -5.053 29.474 1.00 61.13 L C
+ATOM 2822 C SER L 56 -19.469 -3.711 30.172 1.00 61.13 L C
+ATOM 2823 O SER L 56 -18.607 -2.846 30.034 1.00 61.13 L O
+ATOM 2824 CB SER L 56 -18.714 -6.073 30.408 1.00 61.13 L C
+ATOM 2825 OG SER L 56 -18.676 -7.358 29.823 1.00 61.13 L O
+ATOM 2826 N GLY L 57 -20.534 -3.548 30.939 1.00 60.13 L N
+ATOM 2827 CA GLY L 57 -20.643 -2.418 31.833 1.00 60.13 L C
+ATOM 2828 C GLY L 57 -19.742 -2.643 33.029 1.00 60.13 L C
+ATOM 2829 O GLY L 57 -18.866 -3.511 33.023 1.00 60.13 L O
+ATOM 2830 N VAL L 58 -19.943 -1.836 34.073 1.00 63.69 L N
+ATOM 2831 CA VAL L 58 -19.166 -2.027 35.301 1.00 63.69 L C
+ATOM 2832 C VAL L 58 -19.562 -3.297 36.040 1.00 63.69 L C
+ATOM 2833 O VAL L 58 -18.708 -3.870 36.729 1.00 63.69 L O
+ATOM 2834 CB VAL L 58 -19.212 -0.755 36.163 1.00 63.69 L C
+ATOM 2835 CG1 VAL L 58 -18.838 0.449 35.317 1.00 63.69 L C
+ATOM 2836 CG2 VAL L 58 -20.551 -0.537 36.839 1.00 63.69 L C
+ATOM 2837 N PRO L 59 -20.808 -3.815 35.948 1.00 61.94 L N
+ATOM 2838 CA PRO L 59 -20.979 -5.230 36.339 1.00 61.94 L C
+ATOM 2839 C PRO L 59 -20.389 -6.177 35.300 1.00 61.94 L C
+ATOM 2840 O PRO L 59 -21.066 -6.730 34.446 1.00 61.94 L O
+ATOM 2841 CB PRO L 59 -22.498 -5.371 36.434 1.00 61.94 L C
+ATOM 2842 CG PRO L 59 -22.985 -3.991 36.663 1.00 61.94 L C
+ATOM 2843 CD PRO L 59 -22.121 -3.168 35.796 1.00 61.94 L C
+ATOM 2844 N SER L 60 -19.080 -6.386 35.401 1.00 64.96 L N
+ATOM 2845 CA SER L 60 -18.291 -7.073 34.380 1.00 64.96 L C
+ATOM 2846 C SER L 60 -18.081 -8.544 34.688 1.00 64.96 L C
+ATOM 2847 O SER L 60 -17.032 -9.109 34.366 1.00 64.96 L O
+ATOM 2848 CB SER L 60 -16.952 -6.362 34.219 1.00 64.96 L C
+ATOM 2849 OG SER L 60 -16.306 -6.741 33.016 1.00 64.96 L O
+ATOM 2850 N ARG L 61 -19.063 -9.183 35.327 1.00 68.14 L N
+ATOM 2851 CA ARG L 61 -18.990 -10.582 35.738 1.00 68.14 L C
+ATOM 2852 C ARG L 61 -19.738 -11.526 34.804 1.00 68.14 L C
+ATOM 2853 O ARG L 61 -19.549 -12.743 34.901 1.00 68.14 L O
+ATOM 2854 CB ARG L 61 -19.575 -10.736 37.138 1.00 68.14 L C
+ATOM 2855 CG ARG L 61 -21.042 -10.430 37.133 1.00 68.14 L C
+ATOM 2856 CD ARG L 61 -21.644 -10.591 38.471 1.00 68.14 L C
+ATOM 2857 NE ARG L 61 -21.115 -9.642 39.427 1.00 68.14 L N
+ATOM 2858 CZ ARG L 61 -21.630 -8.437 39.610 1.00 68.14 L C
+ATOM 2859 NH1 ARG L 61 -22.701 -8.073 38.928 1.00 68.14 L N1+
+ATOM 2860 NH2 ARG L 61 -21.099 -7.610 40.498 1.00 68.14 L N
+ATOM 2861 N PHE L 62 -20.599 -10.995 33.934 1.00 68.05 L N
+ATOM 2862 CA PHE L 62 -21.379 -11.804 33.006 1.00 68.05 L C
+ATOM 2863 C PHE L 62 -20.540 -12.268 31.830 1.00 68.05 L C
+ATOM 2864 O PHE L 62 -19.387 -11.871 31.664 1.00 68.05 L O
+ATOM 2865 CB PHE L 62 -22.572 -11.026 32.463 1.00 68.05 L C
+ATOM 2866 CG PHE L 62 -23.545 -10.593 33.500 1.00 68.05 L C
+ATOM 2867 CD1 PHE L 62 -23.425 -10.993 34.807 1.00 68.05 L C
+ATOM 2868 CD2 PHE L 62 -24.612 -9.807 33.150 1.00 68.05 L C
+ATOM 2869 CE1 PHE L 62 -24.336 -10.592 35.743 1.00 68.05 L C
+ATOM 2870 CE2 PHE L 62 -25.526 -9.407 34.077 1.00 68.05 L C
+ATOM 2871 CZ PHE L 62 -25.390 -9.798 35.374 1.00 68.05 L C
+ATOM 2872 N SER L 63 -21.122 -13.156 31.033 1.00 74.72 L N
+ATOM 2873 CA SER L 63 -20.602 -13.498 29.720 1.00 74.72 L C
+ATOM 2874 C SER L 63 -21.745 -13.524 28.713 1.00 74.72 L C
+ATOM 2875 O SER L 63 -22.747 -14.210 28.926 1.00 74.72 L O
+ATOM 2876 CB SER L 63 -19.881 -14.844 29.749 1.00 74.72 L C
+ATOM 2877 OG SER L 63 -19.180 -15.068 28.541 1.00 74.72 L O
+ATOM 2878 N GLY L 64 -21.592 -12.778 27.622 1.00 72.04 L N
+ATOM 2879 CA GLY L 64 -22.480 -12.861 26.475 1.00 72.04 L C
+ATOM 2880 C GLY L 64 -21.737 -13.541 25.338 1.00 72.04 L C
+ATOM 2881 O GLY L 64 -20.581 -13.218 25.068 1.00 72.04 L O
+ATOM 2882 N SER L 65 -22.398 -14.497 24.698 1.00 73.22 L N
+ATOM 2883 CA SER L 65 -21.681 -15.448 23.867 1.00 73.22 L C
+ATOM 2884 C SER L 65 -22.258 -15.525 22.467 1.00 73.22 L C
+ATOM 2885 O SER L 65 -23.443 -15.275 22.240 1.00 73.22 L O
+ATOM 2886 CB SER L 65 -21.683 -16.847 24.478 1.00 73.22 L C
+ATOM 2887 OG SER L 65 -20.694 -17.658 23.865 1.00 73.22 L O
+ATOM 2888 N ARG L 66 -21.382 -15.878 21.534 1.00 81.62 L N
+ATOM 2889 CA ARG L 66 -21.762 -16.031 20.141 1.00 81.62 L C
+ATOM 2890 C ARG L 66 -22.525 -17.324 19.919 1.00 81.62 L C
+ATOM 2891 O ARG L 66 -22.335 -18.313 20.629 1.00 81.62 L O
+ATOM 2892 CB ARG L 66 -20.523 -16.006 19.246 1.00 81.62 L C
+ATOM 2893 CG ARG L 66 -19.680 -17.269 19.291 1.00 81.62 L C
+ATOM 2894 CD ARG L 66 -18.960 -17.476 17.968 1.00 81.62 L C
+ATOM 2895 NE ARG L 66 -18.063 -16.370 17.664 1.00 81.62 L N
+ATOM 2896 CZ ARG L 66 -16.785 -16.331 18.023 1.00 81.62 L C
+ATOM 2897 NH1 ARG L 66 -16.250 -17.343 18.685 1.00 81.62 L N1+
+ATOM 2898 NH2 ARG L 66 -16.035 -15.291 17.709 1.00 81.62 L N
+ATOM 2899 N SER L 67 -23.393 -17.307 18.923 1.00 85.10 L N
+ATOM 2900 CA SER L 67 -24.035 -18.517 18.449 1.00 85.10 L C
+ATOM 2901 C SER L 67 -23.653 -18.876 17.022 1.00 85.10 L C
+ATOM 2902 O SER L 67 -23.970 -19.983 16.576 1.00 85.10 L O
+ATOM 2903 CB SER L 67 -25.548 -18.364 18.559 1.00 85.10 L C
+ATOM 2904 OG SER L 67 -26.226 -19.566 18.262 1.00 85.10 L O
+ATOM 2905 N GLY L 68 -22.965 -17.994 16.308 1.00 83.54 L N
+ATOM 2906 CA GLY L 68 -22.666 -18.267 14.914 1.00 83.54 L C
+ATOM 2907 C GLY L 68 -23.970 -18.376 14.163 1.00 83.54 L C
+ATOM 2908 O GLY L 68 -24.576 -17.378 13.760 1.00 83.54 L O
+ATOM 2909 N THR L 69 -24.399 -19.618 13.965 1.00 88.69 L N
+ATOM 2910 CA THR L 69 -25.769 -19.884 13.559 1.00 88.69 L C
+ATOM 2911 C THR L 69 -26.706 -19.246 14.567 1.00 88.69 L C
+ATOM 2912 O THR L 69 -26.400 -19.165 15.757 1.00 88.69 L O
+ATOM 2913 CB THR L 69 -26.025 -21.386 13.504 1.00 88.69 L C
+ATOM 2914 CG2 THR L 69 -24.787 -22.096 13.010 1.00 88.69 L C
+ATOM 2915 OG1 THR L 69 -26.308 -21.862 14.822 1.00 88.69 L O
+ATOM 2916 N ASP L 70 -27.840 -18.753 14.094 1.00 90.92 L N
+ATOM 2917 CA ASP L 70 -28.838 -18.229 15.020 1.00 90.92 L C
+ATOM 2918 C ASP L 70 -29.725 -19.365 15.527 1.00 90.92 L C
+ATOM 2919 O ASP L 70 -30.921 -19.425 15.267 1.00 90.92 L O
+ATOM 2920 CB ASP L 70 -29.654 -17.119 14.368 1.00 90.92 L C
+ATOM 2921 CG ASP L 70 -30.786 -16.624 15.251 1.00 90.92 L C
+ATOM 2922 OD1 ASP L 70 -31.883 -16.377 14.702 1.00 90.92 L O
+ATOM 2923 OD2 ASP L 70 -30.587 -16.537 16.486 1.00 90.92 L O1-
+ATOM 2924 N PHE L 71 -29.109 -20.303 16.251 1.00 93.36 L N
+ATOM 2925 CA PHE L 71 -29.913 -21.065 17.198 1.00 93.36 L C
+ATOM 2926 C PHE L 71 -30.557 -20.125 18.208 1.00 93.36 L C
+ATOM 2927 O PHE L 71 -31.782 -20.114 18.365 1.00 93.36 L O
+ATOM 2928 CB PHE L 71 -29.070 -22.106 17.938 1.00 93.36 L C
+ATOM 2929 CG PHE L 71 -28.589 -23.241 17.084 1.00 93.36 L C
+ATOM 2930 CD1 PHE L 71 -27.269 -23.644 17.125 1.00 93.36 L C
+ATOM 2931 CD2 PHE L 71 -29.471 -23.939 16.278 1.00 93.36 L C
+ATOM 2932 CE1 PHE L 71 -26.833 -24.704 16.356 1.00 93.36 L C
+ATOM 2933 CE2 PHE L 71 -29.039 -25.000 15.507 1.00 93.36 L C
+ATOM 2934 CZ PHE L 71 -27.718 -25.380 15.546 1.00 93.36 L C
+ATOM 2935 N THR L 72 -29.743 -19.288 18.841 1.00 87.95 L N
+ATOM 2936 CA THR L 72 -30.102 -18.306 19.864 1.00 87.95 L C
+ATOM 2937 C THR L 72 -28.991 -17.261 19.866 1.00 87.95 L C
+ATOM 2938 O THR L 72 -28.098 -17.299 19.021 1.00 87.95 L O
+ATOM 2939 CB THR L 72 -30.251 -18.916 21.266 1.00 87.95 L C
+ATOM 2940 CG2 THR L 72 -31.240 -20.071 21.328 1.00 87.95 L C
+ATOM 2941 OG1 THR L 72 -28.968 -19.319 21.769 1.00 87.95 L O
+ATOM 2942 N LEU L 73 -29.069 -16.306 20.796 1.00 84.00 L N
+ATOM 2943 CA LEU L 73 -27.877 -15.685 21.374 1.00 84.00 L C
+ATOM 2944 C LEU L 73 -28.023 -15.615 22.885 1.00 84.00 L C
+ATOM 2945 O LEU L 73 -29.109 -15.315 23.387 1.00 84.00 L O
+ATOM 2946 CB LEU L 73 -27.586 -14.292 20.846 1.00 84.00 L C
+ATOM 2947 CG LEU L 73 -26.713 -14.149 19.597 1.00 84.00 L C
+ATOM 2948 CD1 LEU L 73 -25.289 -14.624 19.903 1.00 84.00 L C
+ATOM 2949 CD2 LEU L 73 -27.255 -14.682 18.274 1.00 84.00 L C
+ATOM 2950 N THR L 74 -26.913 -15.830 23.602 1.00 77.88 L N
+ATOM 2951 CA THR L 74 -26.930 -16.279 24.989 1.00 77.88 L C
+ATOM 2952 C THR L 74 -26.216 -15.290 25.903 1.00 77.88 L C
+ATOM 2953 O THR L 74 -25.286 -14.600 25.482 1.00 77.88 L O
+ATOM 2954 CB THR L 74 -26.239 -17.629 25.123 1.00 77.88 L C
+ATOM 2955 CG2 THR L 74 -26.730 -18.589 24.054 1.00 77.88 L C
+ATOM 2956 OG1 THR L 74 -24.836 -17.451 24.910 1.00 77.88 L O
+ATOM 2957 N ILE L 75 -26.657 -15.232 27.165 1.00 74.66 L N
+ATOM 2958 CA ILE L 75 -26.000 -14.471 28.228 1.00 74.66 L C
+ATOM 2959 C ILE L 75 -25.913 -15.316 29.496 1.00 74.66 L C
+ATOM 2960 O ILE L 75 -26.751 -16.191 29.725 1.00 74.66 L O
+ATOM 2961 CB ILE L 75 -26.734 -13.153 28.531 1.00 74.66 L C
+ATOM 2962 CG1 ILE L 75 -25.824 -12.204 29.300 1.00 74.66 L C
+ATOM 2963 CG2 ILE L 75 -27.968 -13.405 29.316 1.00 74.66 L C
+ATOM 2964 CD1 ILE L 75 -26.502 -10.948 29.723 1.00 74.66 L C
+ATOM 2965 N SER L 76 -24.904 -15.048 30.324 1.00 78.02 L N
+ATOM 2966 CA SER L 76 -24.711 -15.711 31.612 1.00 78.02 L C
+ATOM 2967 C SER L 76 -24.838 -14.709 32.750 1.00 78.02 L C
+ATOM 2968 O SER L 76 -24.023 -13.793 32.857 1.00 78.02 L O
+ATOM 2969 CB SER L 76 -23.337 -16.367 31.675 1.00 78.02 L C
+ATOM 2970 OG SER L 76 -23.083 -17.131 30.515 1.00 78.02 L O
+ATOM 2971 N SER L 77 -25.806 -14.916 33.631 1.00 78.66 L N
+ATOM 2972 CA SER L 77 -26.006 -14.054 34.787 1.00 78.66 L C
+ATOM 2973 C SER L 77 -25.712 -14.837 36.064 1.00 78.66 L C
+ATOM 2974 O SER L 77 -26.058 -16.013 36.168 1.00 78.66 L O
+ATOM 2975 CB SER L 77 -27.441 -13.522 34.814 1.00 78.66 L C
+ATOM 2976 OG SER L 77 -27.704 -12.704 33.697 1.00 78.66 L O
+ATOM 2977 N LEU L 78 -25.052 -14.198 37.038 1.00 87.04 L N
+ATOM 2978 CA LEU L 78 -24.603 -14.925 38.227 1.00 87.04 L C
+ATOM 2979 C LEU L 78 -25.150 -14.414 39.559 1.00 87.04 L C
+ATOM 2980 O LEU L 78 -25.756 -15.200 40.292 1.00 87.04 L O
+ATOM 2981 CB LEU L 78 -23.071 -14.979 38.258 1.00 87.04 L C
+ATOM 2982 CG LEU L 78 -22.427 -16.093 37.432 1.00 87.04 L C
+ATOM 2983 CD1 LEU L 78 -22.543 -15.887 35.949 1.00 87.04 L C
+ATOM 2984 CD2 LEU L 78 -20.980 -16.228 37.821 1.00 87.04 L C
+ATOM 2985 N GLN L 79 -24.970 -13.151 39.915 1.00 87.99 L N
+ATOM 2986 CA GLN L 79 -25.183 -12.724 41.296 1.00 87.99 L C
+ATOM 2987 C GLN L 79 -26.599 -12.210 41.508 1.00 87.99 L C
+ATOM 2988 O GLN L 79 -27.400 -12.140 40.579 1.00 87.99 L O
+ATOM 2989 CB GLN L 79 -24.142 -11.677 41.672 1.00 87.99 L C
+ATOM 2990 CG GLN L 79 -22.748 -12.050 41.270 1.00 87.99 L C
+ATOM 2991 CD GLN L 79 -22.384 -13.470 41.605 1.00 87.99 L C
+ATOM 2992 NE2 GLN L 79 -22.749 -13.908 42.799 1.00 87.99 L N
+ATOM 2993 OE1 GLN L 79 -21.739 -14.156 40.816 1.00 87.99 L O
+ATOM 2994 N PRO L 80 -26.972 -11.913 42.753 1.00 96.09 L N
+ATOM 2995 CA PRO L 80 -28.356 -11.512 43.061 1.00 96.09 L C
+ATOM 2996 C PRO L 80 -28.650 -10.025 43.222 1.00 96.09 L C
+ATOM 2997 O PRO L 80 -29.812 -9.683 43.462 1.00 96.09 L O
+ATOM 2998 CB PRO L 80 -28.583 -12.217 44.396 1.00 96.09 L C
+ATOM 2999 CG PRO L 80 -27.262 -12.091 45.071 1.00 96.09 L C
+ATOM 3000 CD PRO L 80 -26.267 -12.349 43.974 1.00 96.09 L C
+ATOM 3001 N GLU L 81 -27.674 -9.145 43.135 1.00 87.13 L N
+ATOM 3002 CA GLU L 81 -27.985 -7.725 43.135 1.00 87.13 L C
+ATOM 3003 C GLU L 81 -28.314 -7.215 41.738 1.00 87.13 L C
+ATOM 3004 O GLU L 81 -28.747 -6.067 41.591 1.00 87.13 L O
+ATOM 3005 CB GLU L 81 -26.812 -6.940 43.733 1.00 87.13 L C
+ATOM 3006 CG GLU L 81 -25.455 -7.195 43.064 1.00 87.13 L C
+ATOM 3007 CD GLU L 81 -24.836 -8.537 43.432 1.00 87.13 L C
+ATOM 3008 OE1 GLU L 81 -25.585 -9.470 43.775 1.00 87.13 L O
+ATOM 3009 OE2 GLU L 81 -23.595 -8.655 43.382 1.00 87.13 L O1-
+ATOM 3010 N ASP L 82 -28.112 -8.050 40.726 1.00 80.80 L N
+ATOM 3011 CA ASP L 82 -28.323 -7.712 39.324 1.00 80.80 L C
+ATOM 3012 C ASP L 82 -29.641 -8.285 38.811 1.00 80.80 L C
+ATOM 3013 O ASP L 82 -29.657 -9.072 37.877 1.00 80.80 L O
+ATOM 3014 CB ASP L 82 -27.161 -8.240 38.499 1.00 80.80 L C
+ATOM 3015 CG ASP L 82 -25.831 -7.825 39.053 1.00 80.80 L C
+ATOM 3016 OD1 ASP L 82 -25.756 -6.740 39.656 1.00 80.80 L O
+ATOM 3017 OD2 ASP L 82 -24.858 -8.587 38.901 1.00 80.80 L O1-
+ATOM 3018 N PHE L 83 -30.758 -7.902 39.412 1.00 87.28 L N
+ATOM 3019 CA PHE L 83 -32.047 -8.461 39.010 1.00 87.28 L C
+ATOM 3020 C PHE L 83 -32.783 -7.454 38.138 1.00 87.28 L C
+ATOM 3021 O PHE L 83 -33.347 -6.480 38.636 1.00 87.28 L O
+ATOM 3022 CB PHE L 83 -32.881 -8.903 40.212 1.00 87.28 L C
+ATOM 3023 CG PHE L 83 -33.091 -7.855 41.270 1.00 87.28 L C
+ATOM 3024 CD1 PHE L 83 -32.045 -7.429 42.078 1.00 87.28 L C
+ATOM 3025 CD2 PHE L 83 -34.357 -7.348 41.504 1.00 87.28 L C
+ATOM 3026 CE1 PHE L 83 -32.251 -6.483 43.067 1.00 87.28 L C
+ATOM 3027 CE2 PHE L 83 -34.571 -6.402 42.492 1.00 87.28 L C
+ATOM 3028 CZ PHE L 83 -33.516 -5.969 43.274 1.00 87.28 L C
+ATOM 3029 N ALA L 84 -32.789 -7.713 36.828 1.00 73.79 L N
+ATOM 3030 CA ALA L 84 -33.367 -6.818 35.837 1.00 73.79 L C
+ATOM 3031 C ALA L 84 -33.891 -7.620 34.653 1.00 73.79 L C
+ATOM 3032 O ALA L 84 -33.409 -8.712 34.363 1.00 73.79 L O
+ATOM 3033 CB ALA L 84 -32.341 -5.791 35.356 1.00 73.79 L C
+ATOM 3034 N THR L 85 -34.887 -7.057 33.973 1.00 68.25 L N
+ATOM 3035 CA THR L 85 -35.478 -7.664 32.784 1.00 68.25 L C
+ATOM 3036 C THR L 85 -34.535 -7.495 31.610 1.00 68.25 L C
+ATOM 3037 O THR L 85 -34.405 -6.393 31.083 1.00 68.25 L O
+ATOM 3038 CB THR L 85 -36.807 -6.991 32.474 1.00 68.25 L C
+ATOM 3039 CG2 THR L 85 -37.364 -7.487 31.164 1.00 68.25 L C
+ATOM 3040 OG1 THR L 85 -37.740 -7.251 33.524 1.00 68.25 L O
+ATOM 3041 N TYR L 86 -33.883 -8.552 31.166 1.00 67.53 L N
+ATOM 3042 CA TYR L 86 -32.894 -8.338 30.125 1.00 67.53 L C
+ATOM 3043 C TYR L 86 -33.537 -8.429 28.748 1.00 67.53 L C
+ATOM 3044 O TYR L 86 -34.524 -9.137 28.544 1.00 67.53 L O
+ATOM 3045 CB TYR L 86 -31.725 -9.316 30.214 1.00 67.53 L C
+ATOM 3046 CG TYR L 86 -30.975 -9.260 31.519 1.00 67.53 L C
+ATOM 3047 CD1 TYR L 86 -31.275 -8.314 32.473 1.00 67.53 L C
+ATOM 3048 CD2 TYR L 86 -29.885 -10.071 31.742 1.00 67.53 L C
+ATOM 3049 CE1 TYR L 86 -30.594 -8.251 33.662 1.00 67.53 L C
+ATOM 3050 CE2 TYR L 86 -29.181 -9.999 32.922 1.00 67.53 L C
+ATOM 3051 CZ TYR L 86 -29.545 -9.084 33.877 1.00 67.53 L C
+ATOM 3052 OH TYR L 86 -28.853 -8.990 35.056 1.00 67.53 L O
+ATOM 3053 N TYR L 87 -32.969 -7.683 27.803 1.00 64.40 L N
+ATOM 3054 CA TYR L 87 -33.490 -7.544 26.456 1.00 64.40 L C
+ATOM 3055 C TYR L 87 -32.444 -7.997 25.457 1.00 64.40 L C
+ATOM 3056 O TYR L 87 -31.248 -7.866 25.691 1.00 64.40 L O
+ATOM 3057 CB TYR L 87 -33.829 -6.104 26.157 1.00 64.40 L C
+ATOM 3058 CG TYR L 87 -35.011 -5.563 26.874 1.00 64.40 L C
+ATOM 3059 CD1 TYR L 87 -36.285 -5.907 26.501 1.00 64.40 L C
+ATOM 3060 CD2 TYR L 87 -34.853 -4.670 27.904 1.00 64.40 L C
+ATOM 3061 CE1 TYR L 87 -37.364 -5.385 27.145 1.00 64.40 L C
+ATOM 3062 CE2 TYR L 87 -35.922 -4.144 28.550 1.00 64.40 L C
+ATOM 3063 CZ TYR L 87 -37.173 -4.502 28.167 1.00 64.40 L C
+ATOM 3064 OH TYR L 87 -38.246 -3.976 28.825 1.00 64.40 L O
+ATOM 3065 N CYS L 88 -32.900 -8.473 24.307 1.00 70.48 L N
+ATOM 3066 CA CYS L 88 -32.013 -8.886 23.227 1.00 70.48 L C
+ATOM 3067 C CYS L 88 -32.295 -7.947 22.068 1.00 70.48 L C
+ATOM 3068 O CYS L 88 -33.444 -7.818 21.635 1.00 70.48 L O
+ATOM 3069 CB CYS L 88 -32.226 -10.344 22.829 1.00 70.48 L C
+ATOM 3070 SG CYS L 88 -31.083 -10.919 21.541 1.00 70.48 L S
+ATOM 3071 N GLN L 89 -31.257 -7.277 21.588 1.00 64.99 L N
+ATOM 3072 CA GLN L 89 -31.407 -6.080 20.787 1.00 64.99 L C
+ATOM 3073 C GLN L 89 -30.623 -6.329 19.501 1.00 64.99 L C
+ATOM 3074 O GLN L 89 -29.727 -7.170 19.486 1.00 64.99 L O
+ATOM 3075 CB GLN L 89 -30.828 -4.918 21.577 1.00 64.99 L C
+ATOM 3076 CG GLN L 89 -31.004 -3.621 20.998 1.00 64.99 L C
+ATOM 3077 CD GLN L 89 -32.462 -3.367 20.965 1.00 64.99 L C
+ATOM 3078 NE2 GLN L 89 -33.014 -3.135 22.153 1.00 64.99 L N
+ATOM 3079 OE1 GLN L 89 -33.111 -3.429 19.931 1.00 64.99 L O
+ATOM 3080 N GLN L 90 -30.913 -5.608 18.414 1.00 65.24 L N
+ATOM 3081 CA GLN L 90 -30.451 -6.074 17.102 1.00 65.24 L C
+ATOM 3082 C GLN L 90 -29.094 -5.514 16.643 1.00 65.24 L C
+ATOM 3083 O GLN L 90 -28.890 -4.298 16.565 1.00 65.24 L O
+ATOM 3084 CB GLN L 90 -31.496 -5.804 16.020 1.00 65.24 L C
+ATOM 3085 CG GLN L 90 -31.244 -6.682 14.785 1.00 65.24 L C
+ATOM 3086 CD GLN L 90 -31.709 -6.083 13.461 1.00 65.24 L C
+ATOM 3087 NE2 GLN L 90 -31.014 -6.429 12.384 1.00 65.24 L N
+ATOM 3088 OE1 GLN L 90 -32.687 -5.341 13.405 1.00 65.24 L O
+ATOM 3089 N SER L 91 -28.179 -6.448 16.340 1.00 64.81 L N
+ATOM 3090 CA SER L 91 -26.947 -6.418 15.535 1.00 64.81 L C
+ATOM 3091 C SER L 91 -27.196 -6.832 14.075 1.00 64.81 L C
+ATOM 3092 O SER L 91 -28.176 -6.389 13.469 1.00 64.81 L O
+ATOM 3093 CB SER L 91 -25.837 -7.240 16.176 1.00 64.81 L C
+ATOM 3094 OG SER L 91 -25.574 -6.756 17.477 1.00 64.81 L O
+ATOM 3095 N TYR L 92 -26.364 -7.713 13.506 1.00 70.09 L N
+ATOM 3096 CA TYR L 92 -25.549 -7.634 12.296 1.00 70.09 L C
+ATOM 3097 C TYR L 92 -25.704 -6.511 11.281 1.00 70.09 L C
+ATOM 3098 O TYR L 92 -24.732 -5.776 11.096 1.00 70.09 L O
+ATOM 3099 CB TYR L 92 -25.828 -8.928 11.532 1.00 70.09 L C
+ATOM 3100 CG TYR L 92 -25.039 -9.219 10.274 1.00 70.09 L C
+ATOM 3101 CD1 TYR L 92 -23.690 -8.996 10.189 1.00 70.09 L C
+ATOM 3102 CD2 TYR L 92 -25.676 -9.748 9.162 1.00 70.09 L C
+ATOM 3103 CE1 TYR L 92 -22.994 -9.293 9.029 1.00 70.09 L C
+ATOM 3104 CE2 TYR L 92 -24.988 -10.047 8.004 1.00 70.09 L C
+ATOM 3105 CZ TYR L 92 -23.647 -9.819 7.944 1.00 70.09 L C
+ATOM 3106 OH TYR L 92 -22.953 -10.107 6.798 1.00 70.09 L O
+ATOM 3107 N TYR L 93 -26.821 -6.325 10.585 1.00 77.30 L N
+ATOM 3108 CA TYR L 93 -26.875 -5.099 9.788 1.00 77.30 L C
+ATOM 3109 C TYR L 93 -27.115 -3.921 10.709 1.00 77.30 L C
+ATOM 3110 O TYR L 93 -28.158 -3.843 11.364 1.00 77.30 L O
+ATOM 3111 CB TYR L 93 -27.928 -5.132 8.681 1.00 77.30 L C
+ATOM 3112 CG TYR L 93 -27.895 -3.836 7.872 1.00 77.30 L C
+ATOM 3113 CD1 TYR L 93 -26.897 -3.573 6.942 1.00 77.30 L C
+ATOM 3114 CD2 TYR L 93 -28.859 -2.864 8.070 1.00 77.30 L C
+ATOM 3115 CE1 TYR L 93 -26.877 -2.377 6.228 1.00 77.30 L C
+ATOM 3116 CE2 TYR L 93 -28.854 -1.672 7.367 1.00 77.30 L C
+ATOM 3117 CZ TYR L 93 -27.865 -1.429 6.450 1.00 77.30 L C
+ATOM 3118 OH TYR L 93 -27.880 -0.238 5.767 1.00 77.30 L O
+ATOM 3119 N TYR L 94 -26.152 -3.004 10.767 1.00 75.02 L N
+ATOM 3120 CA TYR L 94 -26.160 -2.096 11.898 1.00 75.02 L C
+ATOM 3121 C TYR L 94 -27.361 -1.193 11.713 1.00 75.02 L C
+ATOM 3122 O TYR L 94 -27.299 -0.172 11.025 1.00 75.02 L O
+ATOM 3123 CB TYR L 94 -24.851 -1.322 12.035 1.00 75.02 L C
+ATOM 3124 CG TYR L 94 -24.704 -0.633 13.402 1.00 75.02 L C
+ATOM 3125 CD1 TYR L 94 -25.152 0.663 13.630 1.00 75.02 L C
+ATOM 3126 CD2 TYR L 94 -24.181 -1.333 14.493 1.00 75.02 L C
+ATOM 3127 CE1 TYR L 94 -25.023 1.249 14.883 1.00 75.02 L C
+ATOM 3128 CE2 TYR L 94 -24.062 -0.746 15.746 1.00 75.02 L C
+ATOM 3129 CZ TYR L 94 -24.490 0.537 15.929 1.00 75.02 L C
+ATOM 3130 OH TYR L 94 -24.378 1.108 17.163 1.00 75.02 L O
+ATOM 3131 N PHE L 95 -28.484 -1.644 12.262 1.00 75.76 L N
+ATOM 3132 CA PHE L 95 -29.575 -0.774 12.655 1.00 75.76 L C
+ATOM 3133 C PHE L 95 -29.327 -0.370 14.100 1.00 75.76 L C
+ATOM 3134 O PHE L 95 -29.254 -1.230 14.986 1.00 75.76 L O
+ATOM 3135 CB PHE L 95 -30.940 -1.447 12.554 1.00 75.76 L C
+ATOM 3136 CG PHE L 95 -31.482 -1.569 11.168 1.00 75.76 L C
+ATOM 3137 CD1 PHE L 95 -32.413 -0.653 10.707 1.00 75.76 L C
+ATOM 3138 CD2 PHE L 95 -31.198 -2.669 10.383 1.00 75.76 L C
+ATOM 3139 CE1 PHE L 95 -32.974 -0.765 9.445 1.00 75.76 L C
+ATOM 3140 CE2 PHE L 95 -31.775 -2.802 9.119 1.00 75.76 L C
+ATOM 3141 CZ PHE L 95 -32.653 -1.840 8.648 1.00 75.76 L C
+ATOM 3142 N ARG L 96 -29.172 0.924 14.325 1.00 71.74 L N
+ATOM 3143 CA ARG L 96 -29.177 1.464 15.666 1.00 71.74 L C
+ATOM 3144 C ARG L 96 -30.500 1.061 16.298 1.00 71.74 L C
+ATOM 3145 O ARG L 96 -31.550 1.334 15.705 1.00 71.74 L O
+ATOM 3146 CB ARG L 96 -29.030 2.974 15.614 1.00 71.74 L C
+ATOM 3147 CG ARG L 96 -27.607 3.405 15.366 1.00 71.74 L C
+ATOM 3148 CD ARG L 96 -27.222 3.374 13.898 1.00 71.74 L C
+ATOM 3149 NE ARG L 96 -27.953 4.346 13.092 1.00 71.74 L N
+ATOM 3150 CZ ARG L 96 -27.974 4.357 11.763 1.00 71.74 L C
+ATOM 3151 NH1 ARG L 96 -27.294 3.449 11.081 1.00 71.74 L N1+
+ATOM 3152 NH2 ARG L 96 -28.652 5.293 11.121 1.00 71.74 L N
+ATOM 3153 N PRO L 97 -30.507 0.467 17.510 1.00 70.02 L N
+ATOM 3154 CA PRO L 97 -31.545 -0.515 17.859 1.00 70.02 L C
+ATOM 3155 C PRO L 97 -32.939 -0.238 17.329 1.00 70.02 L C
+ATOM 3156 O PRO L 97 -33.617 0.713 17.726 1.00 70.02 L O
+ATOM 3157 CB PRO L 97 -31.545 -0.484 19.394 1.00 70.02 L C
+ATOM 3158 CG PRO L 97 -30.247 0.099 19.807 1.00 70.02 L C
+ATOM 3159 CD PRO L 97 -29.535 0.646 18.598 1.00 70.02 L C
+ATOM 3160 N ILE L 98 -33.388 -1.139 16.462 1.00 68.00 L N
+ATOM 3161 CA ILE L 98 -34.645 -1.001 15.753 1.00 68.00 L C
+ATOM 3162 C ILE L 98 -35.690 -1.984 16.258 1.00 68.00 L C
+ATOM 3163 O ILE L 98 -36.864 -1.632 16.385 1.00 68.00 L O
+ATOM 3164 CB ILE L 98 -34.430 -1.158 14.229 1.00 68.00 L C
+ATOM 3165 CG1 ILE L 98 -35.686 -0.741 13.461 1.00 68.00 L C
+ATOM 3166 CG2 ILE L 98 -33.966 -2.554 13.864 1.00 68.00 L C
+ATOM 3167 CD1 ILE L 98 -35.511 -0.670 11.964 1.00 68.00 L C
+ATOM 3168 N THR L 99 -35.285 -3.213 16.560 1.00 64.83 L N
+ATOM 3169 CA THR L 99 -36.194 -4.269 16.983 1.00 64.83 L C
+ATOM 3170 C THR L 99 -35.724 -4.780 18.335 1.00 64.83 L C
+ATOM 3171 O THR L 99 -34.657 -5.394 18.430 1.00 64.83 L O
+ATOM 3172 CB THR L 99 -36.230 -5.401 15.956 1.00 64.83 L C
+ATOM 3173 CG2 THR L 99 -37.087 -5.020 14.750 1.00 64.83 L C
+ATOM 3174 OG1 THR L 99 -34.896 -5.688 15.517 1.00 64.83 L O
+ATOM 3175 N PHE L 100 -36.508 -4.538 19.379 1.00 63.20 L N
+ATOM 3176 CA PHE L 100 -36.122 -5.107 20.655 1.00 63.20 L C
+ATOM 3177 C PHE L 100 -36.495 -6.579 20.696 1.00 63.20 L C
+ATOM 3178 O PHE L 100 -37.193 -7.098 19.826 1.00 63.20 L O
+ATOM 3179 CB PHE L 100 -36.799 -4.419 21.848 1.00 63.20 L C
+ATOM 3180 CG PHE L 100 -36.300 -3.037 22.141 1.00 63.20 L C
+ATOM 3181 CD1 PHE L 100 -35.597 -2.314 21.204 1.00 63.20 L C
+ATOM 3182 CD2 PHE L 100 -36.424 -2.520 23.418 1.00 63.20 L C
+ATOM 3183 CE1 PHE L 100 -35.117 -1.075 21.508 1.00 63.20 L C
+ATOM 3184 CE2 PHE L 100 -35.932 -1.288 23.720 1.00 63.20 L C
+ATOM 3185 CZ PHE L 100 -35.279 -0.566 22.771 1.00 63.20 L C
+ATOM 3186 N GLY L 101 -36.038 -7.250 21.738 1.00 72.26 L N
+ATOM 3187 CA GLY L 101 -36.613 -8.516 22.138 1.00 72.26 L C
+ATOM 3188 C GLY L 101 -37.969 -8.272 22.755 1.00 72.26 L C
+ATOM 3189 O GLY L 101 -38.731 -7.404 22.316 1.00 72.26 L O
+ATOM 3190 N GLN L 102 -38.280 -9.034 23.793 1.00 75.09 L N
+ATOM 3191 CA GLN L 102 -39.554 -8.882 24.472 1.00 75.09 L C
+ATOM 3192 C GLN L 102 -39.458 -8.896 25.989 1.00 75.09 L C
+ATOM 3193 O GLN L 102 -40.315 -8.292 26.644 1.00 75.09 L O
+ATOM 3194 CB GLN L 102 -40.523 -9.979 24.021 1.00 75.09 L C
+ATOM 3195 CG GLN L 102 -41.903 -9.817 24.590 1.00 75.09 L C
+ATOM 3196 CD GLN L 102 -42.441 -8.428 24.342 1.00 75.09 L C
+ATOM 3197 NE2 GLN L 102 -42.950 -7.796 25.388 1.00 75.09 L N
+ATOM 3198 OE1 GLN L 102 -42.374 -7.916 23.228 1.00 75.09 L O
+ATOM 3199 N GLY L 103 -38.446 -9.533 26.570 1.00 79.80 L N
+ATOM 3200 CA GLY L 103 -38.296 -9.525 28.010 1.00 79.80 L C
+ATOM 3201 C GLY L 103 -38.088 -10.881 28.651 1.00 79.80 L C
+ATOM 3202 O GLY L 103 -38.781 -11.851 28.336 1.00 79.80 L O
+ATOM 3203 N THR L 104 -37.118 -10.948 29.558 1.00 82.24 L N
+ATOM 3204 CA THR L 104 -36.903 -12.095 30.426 1.00 82.24 L C
+ATOM 3205 C THR L 104 -36.950 -11.572 31.847 1.00 82.24 L C
+ATOM 3206 O THR L 104 -37.400 -10.450 32.074 1.00 82.24 L O
+ATOM 3207 CB THR L 104 -35.565 -12.778 30.162 1.00 82.24 L C
+ATOM 3208 CG2 THR L 104 -35.352 -12.952 28.686 1.00 82.24 L C
+ATOM 3209 OG1 THR L 104 -34.507 -11.970 30.678 1.00 82.24 L O
+ATOM 3210 N LYS L 105 -36.515 -12.363 32.815 1.00 91.16 L N
+ATOM 3211 CA LYS L 105 -36.568 -11.909 34.191 1.00 91.16 L C
+ATOM 3212 C LYS L 105 -35.493 -12.634 34.981 1.00 91.16 L C
+ATOM 3213 O LYS L 105 -35.179 -13.790 34.698 1.00 91.16 L O
+ATOM 3214 CB LYS L 105 -37.946 -12.169 34.785 1.00 91.16 L C
+ATOM 3215 CG LYS L 105 -38.125 -11.637 36.162 1.00 91.16 L C
+ATOM 3216 CD LYS L 105 -38.002 -10.155 36.140 1.00 91.16 L C
+ATOM 3217 CE LYS L 105 -39.150 -9.591 35.344 1.00 91.16 L C
+ATOM 3218 NZ LYS L 105 -40.441 -9.832 36.021 1.00 91.16 L N1+
+ATOM 3219 N VAL L 106 -34.905 -11.943 35.950 1.00 94.84 L N
+ATOM 3220 CA VAL L 106 -33.930 -12.542 36.853 1.00 94.84 L C
+ATOM 3221 C VAL L 106 -34.173 -11.979 38.246 1.00 94.84 L C
+ATOM 3222 O VAL L 106 -34.430 -10.781 38.389 1.00 94.84 L O
+ATOM 3223 CB VAL L 106 -32.487 -12.273 36.396 1.00 94.84 L C
+ATOM 3224 CG1 VAL L 106 -31.514 -12.843 37.394 1.00 94.84 L C
+ATOM 3225 CG2 VAL L 106 -32.232 -12.870 35.026 1.00 94.84 L C
+ATOM 3226 N GLU L 107 -34.131 -12.841 39.272 1.00112.44 L N
+ATOM 3227 CA GLU L 107 -34.346 -12.429 40.657 1.00112.44 L C
+ATOM 3228 C GLU L 107 -33.448 -13.227 41.595 1.00112.44 L C
+ATOM 3229 O GLU L 107 -32.935 -14.284 41.230 1.00112.44 L O
+ATOM 3230 CB GLU L 107 -35.804 -12.612 41.100 1.00112.44 L C
+ATOM 3231 CG GLU L 107 -36.824 -11.910 40.237 1.00112.44 L C
+ATOM 3232 CD GLU L 107 -37.661 -12.879 39.447 1.00112.44 L C
+ATOM 3233 OE1 GLU L 107 -38.850 -12.589 39.212 1.00112.44 L O
+ATOM 3234 OE2 GLU L 107 -37.128 -13.939 39.067 1.00112.44 L O1-
+ATOM 3235 N ILE L 108 -33.258 -12.700 42.809 1.00118.45 L N
+ATOM 3236 CA ILE L 108 -32.611 -13.442 43.892 1.00118.45 L C
+ATOM 3237 C ILE L 108 -33.566 -14.475 44.477 1.00118.45 L C
+ATOM 3238 O ILE L 108 -34.752 -14.192 44.689 1.00118.45 L O
+ATOM 3239 CB ILE L 108 -32.134 -12.474 44.982 1.00118.45 L C
+ATOM 3240 CG1 ILE L 108 -31.514 -13.238 46.148 1.00118.45 L C
+ATOM 3241 CG2 ILE L 108 -33.272 -11.581 45.463 1.00118.45 L C
+ATOM 3242 CD1 ILE L 108 -30.910 -12.327 47.184 1.00118.45 L C
+ATOM 3243 N LYS L 109 -33.062 -15.682 44.761 1.00118.04 L N
+ATOM 3244 CA LYS L 109 -33.923 -16.676 45.405 1.00118.04 L C
+ATOM 3245 C LYS L 109 -33.654 -16.733 46.914 1.00118.04 L C
+ATOM 3246 O LYS L 109 -32.730 -17.372 47.423 1.00118.04 L O
+ATOM 3247 CB LYS L 109 -33.816 -18.055 44.754 1.00118.04 L C
+ATOM 3248 CG LYS L 109 -32.528 -18.827 44.890 1.00118.04 L C
+ATOM 3249 CD LYS L 109 -32.635 -20.158 44.167 1.00118.04 L C
+ATOM 3250 CE LYS L 109 -31.341 -20.937 44.263 1.00118.04 L C
+ATOM 3251 NZ LYS L 109 -31.059 -21.360 45.666 1.00118.04 L N1+
+TER
+END
diff --git a/utils/examples/pdbs/cd86.pdb b/utils/examples/pdbs/cd86.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..9715f48a0b03147a5f1883ab6dfc042ccbad52b5
--- /dev/null
+++ b/utils/examples/pdbs/cd86.pdb
@@ -0,0 +1,1790 @@
+SSBOND CYS B 16 CYS B 86 2.04
+ATOM 1 N MET B 1 -11.523 -4.666 -13.325 1.00 0.00 N
+ATOM 2 CA MET B 1 -11.657 -5.250 -12.001 1.00 0.00 C
+ATOM 3 C MET B 1 -11.626 -6.767 -12.081 1.00 0.00 C
+ATOM 4 O MET B 1 -12.522 -7.374 -12.658 1.00 0.00 O
+ATOM 5 CB MET B 1 -12.980 -4.828 -11.359 1.00 0.00 C
+ATOM 6 CG MET B 1 -13.186 -5.333 -9.941 1.00 0.00 C
+ATOM 7 SD MET B 1 -14.855 -4.922 -9.277 1.00 0.00 S
+ATOM 8 CE MET B 1 -15.934 -6.117 -10.125 1.00 0.00 C
+ATOM 9 1H MET B 1 -11.545 -3.669 -13.254 1.00 0.00 H
+ATOM 10 2H MET B 1 -10.654 -4.951 -13.729 1.00 0.00 H
+ATOM 11 3H MET B 1 -12.277 -4.975 -13.904 1.00 0.00 H
+ATOM 12 HA MET B 1 -10.819 -4.926 -11.381 1.00 0.00 H
+ATOM 13 1HB MET B 1 -13.042 -3.742 -11.333 1.00 0.00 H
+ATOM 14 2HB MET B 1 -13.812 -5.189 -11.966 1.00 0.00 H
+ATOM 15 1HG MET B 1 -13.052 -6.419 -9.911 1.00 0.00 H
+ATOM 16 2HG MET B 1 -12.434 -4.885 -9.288 1.00 0.00 H
+ATOM 17 1HE MET B 1 -16.969 -5.967 -9.806 1.00 0.00 H
+ATOM 18 2HE MET B 1 -15.869 -5.976 -11.204 1.00 0.00 H
+ATOM 19 3HE MET B 1 -15.623 -7.125 -9.865 1.00 0.00 H
+ATOM 20 N LEU B 2 -10.615 -7.380 -11.494 1.00 0.00 N
+ATOM 21 CA LEU B 2 -10.549 -8.838 -11.489 1.00 0.00 C
+ATOM 22 C LEU B 2 -11.553 -9.405 -10.513 1.00 0.00 C
+ATOM 23 O LEU B 2 -11.592 -8.952 -9.382 1.00 0.00 O
+ATOM 24 CB LEU B 2 -9.151 -9.285 -11.083 1.00 0.00 C
+ATOM 25 CG LEU B 2 -8.928 -10.773 -10.966 1.00 0.00 C
+ATOM 26 CD1 LEU B 2 -9.076 -11.385 -12.335 1.00 0.00 C
+ATOM 27 CD2 LEU B 2 -7.544 -11.010 -10.356 1.00 0.00 C
+ATOM 28 H LEU B 2 -9.897 -6.832 -11.012 1.00 0.00 H
+ATOM 29 HA LEU B 2 -10.786 -9.209 -12.486 1.00 0.00 H
+ATOM 30 1HB LEU B 2 -8.476 -8.941 -11.851 1.00 0.00 H
+ATOM 31 2HB LEU B 2 -8.883 -8.815 -10.155 1.00 0.00 H
+ATOM 32 HG LEU B 2 -9.682 -11.219 -10.315 1.00 0.00 H
+ATOM 33 1HD1 LEU B 2 -8.928 -12.455 -12.287 1.00 0.00 H
+ATOM 34 2HD1 LEU B 2 -10.073 -11.183 -12.706 1.00 0.00 H
+ATOM 35 3HD1 LEU B 2 -8.340 -10.950 -13.006 1.00 0.00 H
+ATOM 36 1HD2 LEU B 2 -7.373 -12.078 -10.268 1.00 0.00 H
+ATOM 37 2HD2 LEU B 2 -6.784 -10.574 -11.000 1.00 0.00 H
+ATOM 38 3HD2 LEU B 2 -7.495 -10.554 -9.373 1.00 0.00 H
+ATOM 39 N LYS B 3 -12.368 -10.368 -10.932 1.00 0.00 N
+ATOM 40 CA LYS B 3 -13.319 -11.010 -10.018 1.00 0.00 C
+ATOM 41 C LYS B 3 -12.734 -12.328 -9.530 1.00 0.00 C
+ATOM 42 O LYS B 3 -12.499 -13.232 -10.329 1.00 0.00 O
+ATOM 43 CB LYS B 3 -14.640 -11.234 -10.741 1.00 0.00 C
+ATOM 44 CG LYS B 3 -15.351 -9.936 -11.096 1.00 0.00 C
+ATOM 45 CD LYS B 3 -16.584 -10.175 -11.952 1.00 0.00 C
+ATOM 46 CE LYS B 3 -17.703 -10.886 -11.175 1.00 0.00 C
+ATOM 47 NZ LYS B 3 -18.302 -10.032 -10.077 1.00 0.00 N
+ATOM 48 H LYS B 3 -12.333 -10.678 -11.905 1.00 0.00 H
+ATOM 49 HA LYS B 3 -13.485 -10.366 -9.154 1.00 0.00 H
+ATOM 50 1HB LYS B 3 -14.454 -11.777 -11.658 1.00 0.00 H
+ATOM 51 2HB LYS B 3 -15.299 -11.835 -10.120 1.00 0.00 H
+ATOM 52 1HG LYS B 3 -15.657 -9.447 -10.168 1.00 0.00 H
+ATOM 53 2HG LYS B 3 -14.664 -9.272 -11.625 1.00 0.00 H
+ATOM 54 1HD LYS B 3 -16.959 -9.211 -12.306 1.00 0.00 H
+ATOM 55 2HD LYS B 3 -16.309 -10.777 -12.820 1.00 0.00 H
+ATOM 56 1HE LYS B 3 -18.489 -11.148 -11.881 1.00 0.00 H
+ATOM 57 2HE LYS B 3 -17.308 -11.802 -10.733 1.00 0.00 H
+ATOM 58 1HZ LYS B 3 -19.035 -10.545 -9.619 1.00 0.00 H
+ATOM 59 2HZ LYS B 3 -17.607 -9.774 -9.363 1.00 0.00 H
+ATOM 60 3HZ LYS B 3 -18.686 -9.186 -10.468 1.00 0.00 H
+ATOM 61 N ILE B 4 -12.482 -12.429 -8.236 1.00 0.00 N
+ATOM 62 CA ILE B 4 -11.827 -13.594 -7.647 1.00 0.00 C
+ATOM 63 C ILE B 4 -12.871 -14.398 -6.909 1.00 0.00 C
+ATOM 64 O ILE B 4 -13.567 -13.869 -6.035 1.00 0.00 O
+ATOM 65 CB ILE B 4 -10.711 -13.183 -6.643 1.00 0.00 C
+ATOM 66 CG1 ILE B 4 -9.664 -12.314 -7.330 1.00 0.00 C
+ATOM 67 CG2 ILE B 4 -10.055 -14.434 -6.036 1.00 0.00 C
+ATOM 68 CD1 ILE B 4 -8.648 -11.715 -6.368 1.00 0.00 C
+ATOM 69 H ILE B 4 -12.727 -11.656 -7.633 1.00 0.00 H
+ATOM 70 HA ILE B 4 -11.404 -14.213 -8.431 1.00 0.00 H
+ATOM 71 HB ILE B 4 -11.142 -12.593 -5.855 1.00 0.00 H
+ATOM 72 1HG1 ILE B 4 -9.131 -12.906 -8.065 1.00 0.00 H
+ATOM 73 2HG1 ILE B 4 -10.173 -11.498 -7.843 1.00 0.00 H
+ATOM 74 1HG2 ILE B 4 -9.285 -14.136 -5.329 1.00 0.00 H
+ATOM 75 2HG2 ILE B 4 -10.811 -15.028 -5.521 1.00 0.00 H
+ATOM 76 3HG2 ILE B 4 -9.612 -15.029 -6.832 1.00 0.00 H
+ATOM 77 1HD1 ILE B 4 -7.934 -11.110 -6.918 1.00 0.00 H
+ATOM 78 2HD1 ILE B 4 -9.162 -11.091 -5.636 1.00 0.00 H
+ATOM 79 3HD1 ILE B 4 -8.116 -12.513 -5.857 1.00 0.00 H
+ATOM 80 N GLN B 5 -13.022 -15.650 -7.303 1.00 0.00 N
+ATOM 81 CA GLN B 5 -13.997 -16.546 -6.722 1.00 0.00 C
+ATOM 82 C GLN B 5 -13.284 -17.636 -5.930 1.00 0.00 C
+ATOM 83 O GLN B 5 -12.791 -18.615 -6.487 1.00 0.00 O
+ATOM 84 CB GLN B 5 -14.860 -17.109 -7.846 1.00 0.00 C
+ATOM 85 CG GLN B 5 -15.622 -16.001 -8.572 1.00 0.00 C
+ATOM 86 CD GLN B 5 -16.449 -16.494 -9.765 1.00 0.00 C
+ATOM 87 OE1 GLN B 5 -16.414 -17.676 -10.105 1.00 0.00 O
+ATOM 88 NE2 GLN B 5 -17.165 -15.576 -10.424 1.00 0.00 N
+ATOM 89 H GLN B 5 -12.425 -15.996 -8.048 1.00 0.00 H
+ATOM 90 HA GLN B 5 -14.629 -15.990 -6.032 1.00 0.00 H
+ATOM 91 1HB GLN B 5 -14.226 -17.624 -8.571 1.00 0.00 H
+ATOM 92 2HB GLN B 5 -15.568 -17.829 -7.443 1.00 0.00 H
+ATOM 93 1HG GLN B 5 -16.292 -15.521 -7.856 1.00 0.00 H
+ATOM 94 2HG GLN B 5 -14.898 -15.273 -8.946 1.00 0.00 H
+ATOM 95 1HE2 GLN B 5 -17.715 -15.824 -11.260 1.00 0.00 H
+ATOM 96 2HE2 GLN B 5 -17.156 -14.631 -10.117 1.00 0.00 H
+ATOM 97 N ALA B 6 -13.185 -17.420 -4.630 1.00 0.00 N
+ATOM 98 CA ALA B 6 -12.441 -18.294 -3.734 1.00 0.00 C
+ATOM 99 C ALA B 6 -13.418 -18.912 -2.748 1.00 0.00 C
+ATOM 100 O ALA B 6 -14.549 -18.455 -2.663 1.00 0.00 O
+ATOM 101 CB ALA B 6 -11.363 -17.516 -3.008 1.00 0.00 C
+ATOM 102 H ALA B 6 -13.644 -16.601 -4.226 1.00 0.00 H
+ATOM 103 HA ALA B 6 -11.985 -19.092 -4.315 1.00 0.00 H
+ATOM 104 1HB ALA B 6 -10.816 -18.187 -2.349 1.00 0.00 H
+ATOM 105 2HB ALA B 6 -10.678 -17.075 -3.728 1.00 0.00 H
+ATOM 106 3HB ALA B 6 -11.838 -16.735 -2.426 1.00 0.00 H
+ATOM 107 N TYR B 7 -12.997 -19.949 -2.047 1.00 0.00 N
+ATOM 108 CA TYR B 7 -13.860 -20.618 -1.071 1.00 0.00 C
+ATOM 109 C TYR B 7 -13.493 -20.423 0.393 1.00 0.00 C
+ATOM 110 O TYR B 7 -12.336 -20.204 0.743 1.00 0.00 O
+ATOM 111 CB TYR B 7 -13.969 -22.088 -1.430 1.00 0.00 C
+ATOM 112 CG TYR B 7 -14.950 -22.333 -2.525 1.00 0.00 C
+ATOM 113 CD1 TYR B 7 -14.671 -22.032 -3.853 1.00 0.00 C
+ATOM 114 CD2 TYR B 7 -16.146 -22.904 -2.195 1.00 0.00 C
+ATOM 115 CE1 TYR B 7 -15.628 -22.284 -4.811 1.00 0.00 C
+ATOM 116 CE2 TYR B 7 -17.075 -23.164 -3.151 1.00 0.00 C
+ATOM 117 CZ TYR B 7 -16.828 -22.860 -4.443 1.00 0.00 C
+ATOM 118 OH TYR B 7 -17.779 -23.129 -5.372 1.00 0.00 O
+ATOM 119 H TYR B 7 -12.058 -20.294 -2.182 1.00 0.00 H
+ATOM 120 HA TYR B 7 -14.858 -20.197 -1.186 1.00 0.00 H
+ATOM 121 1HB TYR B 7 -13.009 -22.461 -1.756 1.00 0.00 H
+ATOM 122 2HB TYR B 7 -14.275 -22.664 -0.550 1.00 0.00 H
+ATOM 123 HD1 TYR B 7 -13.720 -21.587 -4.134 1.00 0.00 H
+ATOM 124 HD2 TYR B 7 -16.357 -23.150 -1.157 1.00 0.00 H
+ATOM 125 HE1 TYR B 7 -15.435 -22.037 -5.852 1.00 0.00 H
+ATOM 126 HE2 TYR B 7 -18.023 -23.617 -2.882 1.00 0.00 H
+ATOM 127 HH TYR B 7 -18.571 -23.448 -4.924 1.00 0.00 H
+ATOM 128 N PHE B 8 -14.515 -20.507 1.218 1.00 0.00 N
+ATOM 129 CA PHE B 8 -14.415 -20.423 2.658 1.00 0.00 C
+ATOM 130 C PHE B 8 -13.392 -21.443 3.168 1.00 0.00 C
+ATOM 131 O PHE B 8 -13.373 -22.600 2.739 1.00 0.00 O
+ATOM 132 CB PHE B 8 -15.818 -20.644 3.248 1.00 0.00 C
+ATOM 133 CG PHE B 8 -15.920 -20.531 4.706 1.00 0.00 C
+ATOM 134 CD1 PHE B 8 -15.961 -19.290 5.312 1.00 0.00 C
+ATOM 135 CD2 PHE B 8 -16.033 -21.661 5.497 1.00 0.00 C
+ATOM 136 CE1 PHE B 8 -16.085 -19.184 6.700 1.00 0.00 C
+ATOM 137 CE2 PHE B 8 -16.159 -21.558 6.871 1.00 0.00 C
+ATOM 138 CZ PHE B 8 -16.175 -20.319 7.471 1.00 0.00 C
+ATOM 139 H PHE B 8 -15.431 -20.639 0.810 1.00 0.00 H
+ATOM 140 HA PHE B 8 -14.076 -19.425 2.933 1.00 0.00 H
+ATOM 141 1HB PHE B 8 -16.504 -19.921 2.802 1.00 0.00 H
+ATOM 142 2HB PHE B 8 -16.172 -21.632 2.962 1.00 0.00 H
+ATOM 143 HD1 PHE B 8 -15.884 -18.398 4.690 1.00 0.00 H
+ATOM 144 HD2 PHE B 8 -16.015 -22.645 5.025 1.00 0.00 H
+ATOM 145 HE1 PHE B 8 -16.108 -18.210 7.183 1.00 0.00 H
+ATOM 146 HE2 PHE B 8 -16.235 -22.459 7.478 1.00 0.00 H
+ATOM 147 HZ PHE B 8 -16.270 -20.236 8.554 1.00 0.00 H
+ATOM 148 N ASN B 9 -12.515 -20.962 4.058 1.00 0.00 N
+ATOM 149 CA ASN B 9 -11.374 -21.648 4.676 1.00 0.00 C
+ATOM 150 C ASN B 9 -10.251 -21.973 3.686 1.00 0.00 C
+ATOM 151 O ASN B 9 -9.306 -22.664 4.040 1.00 0.00 O
+ATOM 152 CB ASN B 9 -11.761 -22.943 5.369 1.00 0.00 C
+ATOM 153 CG ASN B 9 -12.696 -22.805 6.526 1.00 0.00 C
+ATOM 154 OD1 ASN B 9 -13.721 -23.490 6.528 1.00 0.00 O
+ATOM 155 ND2 ASN B 9 -12.384 -21.975 7.496 1.00 0.00 N
+ATOM 156 H ASN B 9 -12.642 -19.999 4.346 1.00 0.00 H
+ATOM 157 HA ASN B 9 -10.958 -20.968 5.418 1.00 0.00 H
+ATOM 158 1HB ASN B 9 -12.203 -23.627 4.647 1.00 0.00 H
+ATOM 159 2HB ASN B 9 -10.851 -23.423 5.735 1.00 0.00 H
+ATOM 160 1HD2 ASN B 9 -12.988 -21.872 8.286 1.00 0.00 H
+ATOM 161 2HD2 ASN B 9 -11.503 -21.465 7.468 1.00 0.00 H
+ATOM 162 N GLU B 10 -10.297 -21.413 2.487 1.00 0.00 N
+ATOM 163 CA GLU B 10 -9.240 -21.594 1.503 1.00 0.00 C
+ATOM 164 C GLU B 10 -8.424 -20.309 1.466 1.00 0.00 C
+ATOM 165 O GLU B 10 -8.542 -19.482 2.368 1.00 0.00 O
+ATOM 166 CB GLU B 10 -9.796 -21.954 0.114 1.00 0.00 C
+ATOM 167 CG GLU B 10 -10.707 -23.213 0.095 1.00 0.00 C
+ATOM 168 CD GLU B 10 -10.006 -24.522 0.419 1.00 0.00 C
+ATOM 169 OE1 GLU B 10 -8.831 -24.627 0.168 1.00 0.00 O
+ATOM 170 OE2 GLU B 10 -10.658 -25.419 0.922 1.00 0.00 O
+ATOM 171 H GLU B 10 -11.111 -20.872 2.199 1.00 0.00 H
+ATOM 172 HA GLU B 10 -8.586 -22.405 1.825 1.00 0.00 H
+ATOM 173 1HB GLU B 10 -10.340 -21.114 -0.298 1.00 0.00 H
+ATOM 174 2HB GLU B 10 -8.969 -22.157 -0.562 1.00 0.00 H
+ATOM 175 1HG GLU B 10 -11.529 -23.061 0.802 1.00 0.00 H
+ATOM 176 2HG GLU B 10 -11.133 -23.305 -0.892 1.00 0.00 H
+ATOM 177 N THR B 11 -7.509 -20.198 0.514 1.00 0.00 N
+ATOM 178 CA THR B 11 -6.663 -19.013 0.422 1.00 0.00 C
+ATOM 179 C THR B 11 -6.951 -18.212 -0.864 1.00 0.00 C
+ATOM 180 O THR B 11 -7.147 -18.801 -1.923 1.00 0.00 O
+ATOM 181 CB THR B 11 -5.176 -19.439 0.521 1.00 0.00 C
+ATOM 182 OG1 THR B 11 -4.953 -20.093 1.805 1.00 0.00 O
+ATOM 183 CG2 THR B 11 -4.243 -18.262 0.408 1.00 0.00 C
+ATOM 184 H THR B 11 -7.427 -20.925 -0.182 1.00 0.00 H
+ATOM 185 HA THR B 11 -6.886 -18.367 1.267 1.00 0.00 H
+ATOM 186 HB THR B 11 -4.948 -20.136 -0.283 1.00 0.00 H
+ATOM 187 HG1 THR B 11 -4.075 -19.817 2.197 1.00 0.00 H
+ATOM 188 1HG2 THR B 11 -3.227 -18.620 0.486 1.00 0.00 H
+ATOM 189 2HG2 THR B 11 -4.383 -17.767 -0.554 1.00 0.00 H
+ATOM 190 3HG2 THR B 11 -4.430 -17.563 1.199 1.00 0.00 H
+ATOM 191 N ALA B 12 -7.056 -16.870 -0.755 1.00 0.00 N
+ATOM 192 CA ALA B 12 -7.223 -16.036 -1.955 1.00 0.00 C
+ATOM 193 C ALA B 12 -5.857 -15.540 -2.396 1.00 0.00 C
+ATOM 194 O ALA B 12 -5.035 -15.158 -1.561 1.00 0.00 O
+ATOM 195 CB ALA B 12 -8.124 -14.816 -1.725 1.00 0.00 C
+ATOM 196 H ALA B 12 -6.937 -16.446 0.165 1.00 0.00 H
+ATOM 197 HA ALA B 12 -7.648 -16.650 -2.750 1.00 0.00 H
+ATOM 198 1HB ALA B 12 -8.186 -14.248 -2.653 1.00 0.00 H
+ATOM 199 2HB ALA B 12 -9.112 -15.113 -1.433 1.00 0.00 H
+ATOM 200 3HB ALA B 12 -7.703 -14.186 -0.955 1.00 0.00 H
+ATOM 201 N ASP B 13 -5.653 -15.443 -3.695 1.00 0.00 N
+ATOM 202 CA ASP B 13 -4.438 -14.841 -4.243 1.00 0.00 C
+ATOM 203 C ASP B 13 -4.829 -13.486 -4.795 1.00 0.00 C
+ATOM 204 O ASP B 13 -5.617 -13.405 -5.734 1.00 0.00 O
+ATOM 205 CB ASP B 13 -3.903 -15.732 -5.348 1.00 0.00 C
+ATOM 206 CG ASP B 13 -3.464 -17.075 -4.812 1.00 0.00 C
+ATOM 207 OD1 ASP B 13 -4.228 -17.973 -5.017 1.00 0.00 O
+ATOM 208 OD2 ASP B 13 -2.405 -17.222 -4.238 1.00 0.00 O
+ATOM 209 H ASP B 13 -6.337 -15.845 -4.321 1.00 0.00 H
+ATOM 210 HA ASP B 13 -3.690 -14.711 -3.459 1.00 0.00 H
+ATOM 211 1HB ASP B 13 -4.669 -15.883 -6.110 1.00 0.00 H
+ATOM 212 2HB ASP B 13 -3.071 -15.257 -5.834 1.00 0.00 H
+ATOM 213 N LEU B 14 -4.296 -12.426 -4.195 1.00 0.00 N
+ATOM 214 CA LEU B 14 -4.600 -11.049 -4.566 1.00 0.00 C
+ATOM 215 C LEU B 14 -3.399 -10.477 -5.354 1.00 0.00 C
+ATOM 216 O LEU B 14 -2.393 -10.097 -4.742 1.00 0.00 O
+ATOM 217 CB LEU B 14 -4.810 -10.229 -3.279 1.00 0.00 C
+ATOM 218 CG LEU B 14 -5.888 -10.785 -2.244 1.00 0.00 C
+ATOM 219 CD1 LEU B 14 -5.885 -9.890 -0.987 1.00 0.00 C
+ATOM 220 CD2 LEU B 14 -7.297 -10.826 -2.888 1.00 0.00 C
+ATOM 221 H LEU B 14 -3.651 -12.573 -3.423 1.00 0.00 H
+ATOM 222 HA LEU B 14 -5.507 -11.032 -5.156 1.00 0.00 H
+ATOM 223 1HB LEU B 14 -3.857 -10.172 -2.754 1.00 0.00 H
+ATOM 224 2HB LEU B 14 -5.107 -9.221 -3.557 1.00 0.00 H
+ATOM 225 HG LEU B 14 -5.603 -11.797 -1.938 1.00 0.00 H
+ATOM 226 1HD1 LEU B 14 -6.606 -10.272 -0.261 1.00 0.00 H
+ATOM 227 2HD1 LEU B 14 -4.890 -9.892 -0.545 1.00 0.00 H
+ATOM 228 3HD1 LEU B 14 -6.156 -8.878 -1.262 1.00 0.00 H
+ATOM 229 1HD2 LEU B 14 -8.010 -11.213 -2.165 1.00 0.00 H
+ATOM 230 2HD2 LEU B 14 -7.601 -9.825 -3.193 1.00 0.00 H
+ATOM 231 3HD2 LEU B 14 -7.271 -11.476 -3.756 1.00 0.00 H
+ATOM 232 N PRO B 15 -3.402 -10.516 -6.698 1.00 0.00 N
+ATOM 233 CA PRO B 15 -2.265 -10.194 -7.567 1.00 0.00 C
+ATOM 234 C PRO B 15 -2.023 -8.717 -7.776 1.00 0.00 C
+ATOM 235 O PRO B 15 -2.952 -7.911 -7.717 1.00 0.00 O
+ATOM 236 CB PRO B 15 -2.670 -10.837 -8.879 1.00 0.00 C
+ATOM 237 CG PRO B 15 -4.180 -10.681 -8.908 1.00 0.00 C
+ATOM 238 CD PRO B 15 -4.632 -10.853 -7.456 1.00 0.00 C
+ATOM 239 HA PRO B 15 -1.355 -10.670 -7.177 1.00 0.00 H
+ATOM 240 1HB PRO B 15 -2.149 -10.343 -9.687 1.00 0.00 H
+ATOM 241 2HB PRO B 15 -2.349 -11.890 -8.892 1.00 0.00 H
+ATOM 242 1HG PRO B 15 -4.455 -9.699 -9.324 1.00 0.00 H
+ATOM 243 2HG PRO B 15 -4.622 -11.445 -9.574 1.00 0.00 H
+ATOM 244 1HD PRO B 15 -5.451 -10.155 -7.231 1.00 0.00 H
+ATOM 245 2HD PRO B 15 -4.915 -11.883 -7.323 1.00 0.00 H
+ATOM 246 N CYS B 16 -0.820 -8.392 -8.202 1.00 0.00 N
+ATOM 247 CA CYS B 16 -0.532 -7.050 -8.676 1.00 0.00 C
+ATOM 248 C CYS B 16 -0.563 -7.005 -10.200 1.00 0.00 C
+ATOM 249 O CYS B 16 -0.798 -5.951 -10.788 1.00 0.00 O
+ATOM 250 CB CYS B 16 0.816 -6.574 -8.149 1.00 0.00 C
+ATOM 251 SG CYS B 16 0.852 -6.438 -6.340 1.00 0.00 S
+ATOM 252 H CYS B 16 -0.074 -9.079 -8.181 1.00 0.00 H
+ATOM 253 HA CYS B 16 -1.295 -6.380 -8.299 1.00 0.00 H
+ATOM 254 1HB CYS B 16 1.600 -7.271 -8.449 1.00 0.00 H
+ATOM 255 2HB CYS B 16 1.053 -5.601 -8.572 1.00 0.00 H
+ATOM 256 N GLN B 17 -0.284 -8.144 -10.856 1.00 0.00 N
+ATOM 257 CA GLN B 17 -0.250 -8.208 -12.330 1.00 0.00 C
+ATOM 258 C GLN B 17 0.586 -7.070 -12.933 1.00 0.00 C
+ATOM 259 O GLN B 17 0.121 -6.309 -13.786 1.00 0.00 O
+ATOM 260 CB GLN B 17 -1.666 -8.247 -12.946 1.00 0.00 C
+ATOM 261 CG GLN B 17 -2.491 -9.482 -12.515 1.00 0.00 C
+ATOM 262 CD GLN B 17 -3.769 -9.708 -13.306 1.00 0.00 C
+ATOM 263 OE1 GLN B 17 -4.234 -8.873 -14.092 1.00 0.00 O
+ATOM 264 NE2 GLN B 17 -4.353 -10.883 -13.104 1.00 0.00 N
+ATOM 265 H GLN B 17 -0.101 -8.980 -10.325 1.00 0.00 H
+ATOM 266 HA GLN B 17 0.235 -9.141 -12.613 1.00 0.00 H
+ATOM 267 1HB GLN B 17 -2.220 -7.355 -12.643 1.00 0.00 H
+ATOM 268 2HB GLN B 17 -1.596 -8.241 -14.032 1.00 0.00 H
+ATOM 269 1HG GLN B 17 -1.874 -10.381 -12.573 1.00 0.00 H
+ATOM 270 2HG GLN B 17 -2.801 -9.300 -11.498 1.00 0.00 H
+ATOM 271 1HE2 GLN B 17 -5.190 -11.111 -13.600 1.00 0.00 H
+ATOM 272 2HE2 GLN B 17 -3.950 -11.560 -12.480 1.00 0.00 H
+ATOM 273 N PHE B 18 1.809 -6.938 -12.430 1.00 0.00 N
+ATOM 274 CA PHE B 18 2.723 -5.899 -12.855 1.00 0.00 C
+ATOM 275 C PHE B 18 3.839 -6.478 -13.691 1.00 0.00 C
+ATOM 276 O PHE B 18 4.627 -7.286 -13.207 1.00 0.00 O
+ATOM 277 CB PHE B 18 3.292 -5.189 -11.628 1.00 0.00 C
+ATOM 278 CG PHE B 18 4.258 -4.018 -11.891 1.00 0.00 C
+ATOM 279 CD1 PHE B 18 3.870 -2.881 -12.574 1.00 0.00 C
+ATOM 280 CD2 PHE B 18 5.517 -4.065 -11.392 1.00 0.00 C
+ATOM 281 CE1 PHE B 18 4.742 -1.821 -12.741 1.00 0.00 C
+ATOM 282 CE2 PHE B 18 6.411 -3.011 -11.539 1.00 0.00 C
+ATOM 283 CZ PHE B 18 6.017 -1.884 -12.215 1.00 0.00 C
+ATOM 284 H PHE B 18 2.112 -7.599 -11.732 1.00 0.00 H
+ATOM 285 HA PHE B 18 2.177 -5.173 -13.458 1.00 0.00 H
+ATOM 286 1HB PHE B 18 2.469 -4.817 -11.028 1.00 0.00 H
+ATOM 287 2HB PHE B 18 3.819 -5.918 -11.015 1.00 0.00 H
+ATOM 288 HD1 PHE B 18 2.865 -2.823 -12.979 1.00 0.00 H
+ATOM 289 HD2 PHE B 18 5.809 -4.960 -10.865 1.00 0.00 H
+ATOM 290 HE1 PHE B 18 4.415 -0.933 -13.283 1.00 0.00 H
+ATOM 291 HE2 PHE B 18 7.418 -3.078 -11.122 1.00 0.00 H
+ATOM 292 HZ PHE B 18 6.708 -1.051 -12.341 1.00 0.00 H
+ATOM 293 N ALA B 19 3.890 -6.062 -14.956 1.00 0.00 N
+ATOM 294 CA ALA B 19 4.885 -6.518 -15.929 1.00 0.00 C
+ATOM 295 C ALA B 19 6.290 -6.177 -15.526 1.00 0.00 C
+ATOM 296 O ALA B 19 7.238 -6.887 -15.858 1.00 0.00 O
+ATOM 297 CB ALA B 19 4.628 -5.887 -17.274 1.00 0.00 C
+ATOM 298 H ALA B 19 3.188 -5.406 -15.263 1.00 0.00 H
+ATOM 299 HA ALA B 19 4.804 -7.602 -16.013 1.00 0.00 H
+ATOM 300 1HB ALA B 19 5.349 -6.253 -18.002 1.00 0.00 H
+ATOM 301 2HB ALA B 19 3.656 -6.158 -17.562 1.00 0.00 H
+ATOM 302 3HB ALA B 19 4.709 -4.807 -17.198 1.00 0.00 H
+ATOM 303 N ASN B 20 6.446 -5.052 -14.855 1.00 0.00 N
+ATOM 304 CA ASN B 20 7.769 -4.593 -14.504 1.00 0.00 C
+ATOM 305 C ASN B 20 8.639 -4.502 -15.748 1.00 0.00 C
+ATOM 306 O ASN B 20 9.781 -4.948 -15.775 1.00 0.00 O
+ATOM 307 CB ASN B 20 8.370 -5.487 -13.443 1.00 0.00 C
+ATOM 308 CG ASN B 20 9.541 -4.898 -12.830 1.00 0.00 C
+ATOM 309 OD1 ASN B 20 9.770 -3.684 -12.936 1.00 0.00 O
+ATOM 310 ND2 ASN B 20 10.291 -5.708 -12.126 1.00 0.00 N
+ATOM 311 H ASN B 20 5.635 -4.518 -14.575 1.00 0.00 H
+ATOM 312 HA ASN B 20 7.686 -3.589 -14.096 1.00 0.00 H
+ATOM 313 1HB ASN B 20 7.627 -5.695 -12.670 1.00 0.00 H
+ATOM 314 2HB ASN B 20 8.663 -6.438 -13.876 1.00 0.00 H
+ATOM 315 1HD2 ASN B 20 11.094 -5.351 -11.611 1.00 0.00 H
+ATOM 316 2HD2 ASN B 20 10.048 -6.676 -12.053 1.00 0.00 H
+ATOM 317 N SER B 21 8.110 -3.839 -16.757 1.00 0.00 N
+ATOM 318 CA SER B 21 8.788 -3.654 -18.032 1.00 0.00 C
+ATOM 319 C SER B 21 10.092 -2.861 -17.904 1.00 0.00 C
+ATOM 320 O SER B 21 10.938 -2.895 -18.792 1.00 0.00 O
+ATOM 321 CB SER B 21 7.838 -2.960 -18.987 1.00 0.00 C
+ATOM 322 OG SER B 21 7.558 -1.640 -18.564 1.00 0.00 O
+ATOM 323 H SER B 21 7.173 -3.488 -16.651 1.00 0.00 H
+ATOM 324 HA SER B 21 9.032 -4.636 -18.437 1.00 0.00 H
+ATOM 325 1HB SER B 21 8.269 -2.946 -19.986 1.00 0.00 H
+ATOM 326 2HB SER B 21 6.910 -3.531 -19.039 1.00 0.00 H
+ATOM 327 HG SER B 21 6.784 -1.362 -19.067 1.00 0.00 H
+ATOM 328 N GLN B 22 10.239 -2.136 -16.794 1.00 0.00 N
+ATOM 329 CA GLN B 22 11.416 -1.331 -16.530 1.00 0.00 C
+ATOM 330 C GLN B 22 12.445 -2.107 -15.701 1.00 0.00 C
+ATOM 331 O GLN B 22 13.485 -1.564 -15.342 1.00 0.00 O
+ATOM 332 CB GLN B 22 11.013 -0.021 -15.849 1.00 0.00 C
+ATOM 333 CG GLN B 22 10.014 0.794 -16.688 1.00 0.00 C
+ATOM 334 CD GLN B 22 10.531 1.112 -18.067 1.00 0.00 C
+ATOM 335 OE1 GLN B 22 11.541 1.810 -18.251 1.00 0.00 O
+ATOM 336 NE2 GLN B 22 9.846 0.553 -19.071 1.00 0.00 N
+ATOM 337 H GLN B 22 9.494 -2.148 -16.116 1.00 0.00 H
+ATOM 338 HA GLN B 22 11.881 -1.084 -17.484 1.00 0.00 H
+ATOM 339 1HB GLN B 22 10.528 -0.240 -14.906 1.00 0.00 H
+ATOM 340 2HB GLN B 22 11.894 0.589 -15.641 1.00 0.00 H
+ATOM 341 1HG GLN B 22 9.089 0.226 -16.794 1.00 0.00 H
+ATOM 342 2HG GLN B 22 9.813 1.743 -16.172 1.00 0.00 H
+ATOM 343 1HE2 GLN B 22 10.140 0.693 -20.017 1.00 0.00 H
+ATOM 344 2HE2 GLN B 22 9.031 -0.034 -18.880 1.00 0.00 H
+ATOM 345 N ASN B 23 12.151 -3.378 -15.393 1.00 0.00 N
+ATOM 346 CA ASN B 23 13.048 -4.244 -14.632 1.00 0.00 C
+ATOM 347 C ASN B 23 13.444 -3.631 -13.284 1.00 0.00 C
+ATOM 348 O ASN B 23 14.614 -3.630 -12.911 1.00 0.00 O
+ATOM 349 CB ASN B 23 14.297 -4.565 -15.438 1.00 0.00 C
+ATOM 350 CG ASN B 23 15.042 -5.775 -14.920 1.00 0.00 C
+ATOM 351 OD1 ASN B 23 14.421 -6.772 -14.520 1.00 0.00 O
+ATOM 352 ND2 ASN B 23 16.363 -5.711 -14.915 1.00 0.00 N
+ATOM 353 H ASN B 23 11.273 -3.785 -15.706 1.00 0.00 H
+ATOM 354 HA ASN B 23 12.518 -5.172 -14.421 1.00 0.00 H
+ATOM 355 1HB ASN B 23 14.011 -4.756 -16.474 1.00 0.00 H
+ATOM 356 2HB ASN B 23 14.970 -3.712 -15.437 1.00 0.00 H
+ATOM 357 1HD2 ASN B 23 16.903 -6.486 -14.583 1.00 0.00 H
+ATOM 358 2HD2 ASN B 23 16.826 -4.885 -15.242 1.00 0.00 H
+ATOM 359 N GLN B 24 12.466 -3.119 -12.544 1.00 0.00 N
+ATOM 360 CA GLN B 24 12.700 -2.487 -11.255 1.00 0.00 C
+ATOM 361 C GLN B 24 12.769 -3.528 -10.155 1.00 0.00 C
+ATOM 362 O GLN B 24 12.178 -4.605 -10.272 1.00 0.00 O
+ATOM 363 CB GLN B 24 11.577 -1.501 -10.986 1.00 0.00 C
+ATOM 364 CG GLN B 24 11.580 -0.433 -11.986 1.00 0.00 C
+ATOM 365 CD GLN B 24 10.440 0.531 -11.880 1.00 0.00 C
+ATOM 366 OE1 GLN B 24 9.381 0.358 -12.499 1.00 0.00 O
+ATOM 367 NE2 GLN B 24 10.650 1.558 -11.107 1.00 0.00 N
+ATOM 368 H GLN B 24 11.512 -3.173 -12.876 1.00 0.00 H
+ATOM 369 HA GLN B 24 13.653 -1.957 -11.287 1.00 0.00 H
+ATOM 370 1HB GLN B 24 10.619 -2.009 -11.011 1.00 0.00 H
+ATOM 371 2HB GLN B 24 11.702 -1.060 -9.990 1.00 0.00 H
+ATOM 372 1HG GLN B 24 12.511 0.130 -11.895 1.00 0.00 H
+ATOM 373 2HG GLN B 24 11.540 -0.906 -12.958 1.00 0.00 H
+ATOM 374 1HE2 GLN B 24 9.957 2.298 -11.060 1.00 0.00 H
+ATOM 375 2HE2 GLN B 24 11.527 1.665 -10.640 1.00 0.00 H
+ATOM 376 N SER B 25 13.508 -3.216 -9.094 1.00 0.00 N
+ATOM 377 CA SER B 25 13.626 -4.114 -7.948 1.00 0.00 C
+ATOM 378 C SER B 25 12.602 -3.830 -6.869 1.00 0.00 C
+ATOM 379 O SER B 25 12.289 -2.675 -6.609 1.00 0.00 O
+ATOM 380 CB SER B 25 15.000 -3.986 -7.323 1.00 0.00 C
+ATOM 381 OG SER B 25 15.079 -4.726 -6.121 1.00 0.00 O
+ATOM 382 H SER B 25 13.994 -2.308 -9.056 1.00 0.00 H
+ATOM 383 HA SER B 25 13.492 -5.140 -8.298 1.00 0.00 H
+ATOM 384 1HB SER B 25 15.748 -4.347 -8.015 1.00 0.00 H
+ATOM 385 2HB SER B 25 15.211 -2.935 -7.128 1.00 0.00 H
+ATOM 386 HG SER B 25 15.126 -5.659 -6.415 1.00 0.00 H
+ATOM 387 N LEU B 26 12.185 -4.849 -6.121 1.00 0.00 N
+ATOM 388 CA LEU B 26 11.342 -4.562 -4.959 1.00 0.00 C
+ATOM 389 C LEU B 26 12.027 -3.637 -3.976 1.00 0.00 C
+ATOM 390 O LEU B 26 11.354 -2.931 -3.241 1.00 0.00 O
+ATOM 391 CB LEU B 26 10.943 -5.825 -4.182 1.00 0.00 C
+ATOM 392 CG LEU B 26 9.914 -6.720 -4.810 1.00 0.00 C
+ATOM 393 CD1 LEU B 26 9.813 -8.007 -4.022 1.00 0.00 C
+ATOM 394 CD2 LEU B 26 8.546 -5.967 -4.782 1.00 0.00 C
+ATOM 395 H LEU B 26 12.455 -5.797 -6.357 1.00 0.00 H
+ATOM 396 HA LEU B 26 10.437 -4.072 -5.311 1.00 0.00 H
+ATOM 397 1HB LEU B 26 11.838 -6.417 -4.002 1.00 0.00 H
+ATOM 398 2HB LEU B 26 10.545 -5.510 -3.224 1.00 0.00 H
+ATOM 399 HG LEU B 26 10.200 -6.962 -5.823 1.00 0.00 H
+ATOM 400 1HD1 LEU B 26 9.058 -8.654 -4.475 1.00 0.00 H
+ATOM 401 2HD1 LEU B 26 10.775 -8.517 -4.031 1.00 0.00 H
+ATOM 402 3HD1 LEU B 26 9.533 -7.775 -2.998 1.00 0.00 H
+ATOM 403 1HD2 LEU B 26 7.773 -6.590 -5.214 1.00 0.00 H
+ATOM 404 2HD2 LEU B 26 8.280 -5.734 -3.750 1.00 0.00 H
+ATOM 405 3HD2 LEU B 26 8.624 -5.041 -5.351 1.00 0.00 H
+ATOM 406 N SER B 27 13.358 -3.626 -3.958 1.00 0.00 N
+ATOM 407 CA SER B 27 14.138 -2.805 -3.044 1.00 0.00 C
+ATOM 408 C SER B 27 14.020 -1.305 -3.304 1.00 0.00 C
+ATOM 409 O SER B 27 14.498 -0.498 -2.510 1.00 0.00 O
+ATOM 410 CB SER B 27 15.570 -3.161 -3.150 1.00 0.00 C
+ATOM 411 OG SER B 27 15.986 -2.811 -4.408 1.00 0.00 O
+ATOM 412 H SER B 27 13.865 -4.195 -4.630 1.00 0.00 H
+ATOM 413 HA SER B 27 13.815 -3.011 -2.047 1.00 0.00 H
+ATOM 414 1HB SER B 27 16.127 -2.600 -2.417 1.00 0.00 H
+ATOM 415 2HB SER B 27 15.728 -4.222 -2.968 1.00 0.00 H
+ATOM 416 HG SER B 27 15.663 -3.509 -4.993 1.00 0.00 H
+ATOM 417 N GLU B 28 13.465 -0.944 -4.465 1.00 0.00 N
+ATOM 418 CA GLU B 28 13.263 0.425 -4.898 1.00 0.00 C
+ATOM 419 C GLU B 28 11.788 0.792 -4.756 1.00 0.00 C
+ATOM 420 O GLU B 28 11.393 1.935 -5.006 1.00 0.00 O
+ATOM 421 CB GLU B 28 13.683 0.540 -6.372 1.00 0.00 C
+ATOM 422 CG GLU B 28 15.156 0.217 -6.622 1.00 0.00 C
+ATOM 423 CD GLU B 28 15.564 0.148 -8.102 1.00 0.00 C
+ATOM 424 OE1 GLU B 28 14.896 -0.523 -8.893 1.00 0.00 O
+ATOM 425 OE2 GLU B 28 16.547 0.769 -8.429 1.00 0.00 O
+ATOM 426 H GLU B 28 13.120 -1.668 -5.091 1.00 0.00 H
+ATOM 427 HA GLU B 28 13.856 1.094 -4.277 1.00 0.00 H
+ATOM 428 1HB GLU B 28 13.078 -0.140 -6.979 1.00 0.00 H
+ATOM 429 2HB GLU B 28 13.493 1.553 -6.724 1.00 0.00 H
+ATOM 430 1HG GLU B 28 15.764 0.969 -6.120 1.00 0.00 H
+ATOM 431 2HG GLU B 28 15.368 -0.740 -6.154 1.00 0.00 H
+ATOM 432 N LEU B 29 10.968 -0.205 -4.428 1.00 0.00 N
+ATOM 433 CA LEU B 29 9.529 -0.030 -4.409 1.00 0.00 C
+ATOM 434 C LEU B 29 8.915 -0.208 -3.028 1.00 0.00 C
+ATOM 435 O LEU B 29 9.491 -0.855 -2.144 1.00 0.00 O
+ATOM 436 CB LEU B 29 8.873 -1.047 -5.359 1.00 0.00 C
+ATOM 437 CG LEU B 29 9.353 -1.055 -6.776 1.00 0.00 C
+ATOM 438 CD1 LEU B 29 8.685 -2.189 -7.508 1.00 0.00 C
+ATOM 439 CD2 LEU B 29 9.080 0.280 -7.438 1.00 0.00 C
+ATOM 440 H LEU B 29 11.333 -1.117 -4.171 1.00 0.00 H
+ATOM 441 HA LEU B 29 9.306 0.973 -4.760 1.00 0.00 H
+ATOM 442 1HB LEU B 29 9.033 -2.040 -4.951 1.00 0.00 H
+ATOM 443 2HB LEU B 29 7.797 -0.853 -5.378 1.00 0.00 H
+ATOM 444 HG LEU B 29 10.411 -1.233 -6.789 1.00 0.00 H
+ATOM 445 1HD1 LEU B 29 9.051 -2.221 -8.537 1.00 0.00 H
+ATOM 446 2HD1 LEU B 29 8.933 -3.116 -7.006 1.00 0.00 H
+ATOM 447 3HD1 LEU B 29 7.619 -2.050 -7.508 1.00 0.00 H
+ATOM 448 1HD2 LEU B 29 9.446 0.264 -8.464 1.00 0.00 H
+ATOM 449 2HD2 LEU B 29 8.007 0.483 -7.432 1.00 0.00 H
+ATOM 450 3HD2 LEU B 29 9.599 1.040 -6.886 1.00 0.00 H
+ATOM 451 N VAL B 30 7.737 0.357 -2.870 1.00 0.00 N
+ATOM 452 CA VAL B 30 6.932 0.112 -1.701 1.00 0.00 C
+ATOM 453 C VAL B 30 5.608 -0.454 -2.208 1.00 0.00 C
+ATOM 454 O VAL B 30 4.954 0.162 -3.055 1.00 0.00 O
+ATOM 455 CB VAL B 30 6.732 1.384 -0.886 1.00 0.00 C
+ATOM 456 CG1 VAL B 30 5.862 1.062 0.319 1.00 0.00 C
+ATOM 457 CG2 VAL B 30 8.119 1.966 -0.481 1.00 0.00 C
+ATOM 458 H VAL B 30 7.375 0.927 -3.631 1.00 0.00 H
+ATOM 459 HA VAL B 30 7.421 -0.631 -1.077 1.00 0.00 H
+ATOM 460 HB VAL B 30 6.198 2.114 -1.477 1.00 0.00 H
+ATOM 461 1HG1 VAL B 30 5.689 1.961 0.907 1.00 0.00 H
+ATOM 462 2HG1 VAL B 30 4.911 0.666 -0.023 1.00 0.00 H
+ATOM 463 3HG1 VAL B 30 6.349 0.311 0.939 1.00 0.00 H
+ATOM 464 1HG2 VAL B 30 7.978 2.872 0.095 1.00 0.00 H
+ATOM 465 2HG2 VAL B 30 8.666 1.235 0.116 1.00 0.00 H
+ATOM 466 3HG2 VAL B 30 8.692 2.199 -1.376 1.00 0.00 H
+ATOM 467 N VAL B 31 5.243 -1.658 -1.748 1.00 0.00 N
+ATOM 468 CA VAL B 31 4.024 -2.285 -2.235 1.00 0.00 C
+ATOM 469 C VAL B 31 3.044 -2.518 -1.099 1.00 0.00 C
+ATOM 470 O VAL B 31 3.362 -3.174 -0.106 1.00 0.00 O
+ATOM 471 CB VAL B 31 4.318 -3.640 -2.915 1.00 0.00 C
+ATOM 472 CG1 VAL B 31 3.003 -4.244 -3.423 1.00 0.00 C
+ATOM 473 CG2 VAL B 31 5.345 -3.453 -4.035 1.00 0.00 C
+ATOM 474 H VAL B 31 5.836 -2.126 -1.066 1.00 0.00 H
+ATOM 475 HA VAL B 31 3.556 -1.628 -2.963 1.00 0.00 H
+ATOM 476 HB VAL B 31 4.706 -4.331 -2.185 1.00 0.00 H
+ATOM 477 1HG1 VAL B 31 3.195 -5.208 -3.886 1.00 0.00 H
+ATOM 478 2HG1 VAL B 31 2.311 -4.370 -2.596 1.00 0.00 H
+ATOM 479 3HG1 VAL B 31 2.552 -3.577 -4.154 1.00 0.00 H
+ATOM 480 1HG2 VAL B 31 5.548 -4.410 -4.512 1.00 0.00 H
+ATOM 481 2HG2 VAL B 31 4.964 -2.769 -4.750 1.00 0.00 H
+ATOM 482 3HG2 VAL B 31 6.276 -3.051 -3.620 1.00 0.00 H
+ATOM 483 N PHE B 32 1.844 -1.989 -1.253 1.00 0.00 N
+ATOM 484 CA PHE B 32 0.813 -2.161 -0.241 1.00 0.00 C
+ATOM 485 C PHE B 32 -0.354 -2.937 -0.776 1.00 0.00 C
+ATOM 486 O PHE B 32 -0.732 -2.777 -1.938 1.00 0.00 O
+ATOM 487 CB PHE B 32 0.200 -0.827 0.178 1.00 0.00 C
+ATOM 488 CG PHE B 32 -0.539 -0.043 -0.823 1.00 0.00 C
+ATOM 489 CD1 PHE B 32 -1.905 -0.236 -0.992 1.00 0.00 C
+ATOM 490 CD2 PHE B 32 0.124 0.855 -1.629 1.00 0.00 C
+ATOM 491 CE1 PHE B 32 -2.593 0.463 -1.963 1.00 0.00 C
+ATOM 492 CE2 PHE B 32 -0.560 1.567 -2.613 1.00 0.00 C
+ATOM 493 CZ PHE B 32 -1.911 1.369 -2.778 1.00 0.00 C
+ATOM 494 H PHE B 32 1.643 -1.459 -2.103 1.00 0.00 H
+ATOM 495 HA PHE B 32 1.225 -2.685 0.621 1.00 0.00 H
+ATOM 496 1HB PHE B 32 -0.475 -1.008 1.015 1.00 0.00 H
+ATOM 497 2HB PHE B 32 1.000 -0.190 0.564 1.00 0.00 H
+ATOM 498 HD1 PHE B 32 -2.427 -0.946 -0.356 1.00 0.00 H
+ATOM 499 HD2 PHE B 32 1.191 0.999 -1.490 1.00 0.00 H
+ATOM 500 HE1 PHE B 32 -3.666 0.310 -2.086 1.00 0.00 H
+ATOM 501 HE2 PHE B 32 -0.025 2.278 -3.250 1.00 0.00 H
+ATOM 502 HZ PHE B 32 -2.448 1.927 -3.545 1.00 0.00 H
+ATOM 503 N TRP B 33 -0.992 -3.702 0.099 1.00 0.00 N
+ATOM 504 CA TRP B 33 -2.300 -4.241 -0.234 1.00 0.00 C
+ATOM 505 C TRP B 33 -3.255 -3.656 0.752 1.00 0.00 C
+ATOM 506 O TRP B 33 -3.005 -3.734 1.959 1.00 0.00 O
+ATOM 507 CB TRP B 33 -2.394 -5.760 -0.170 1.00 0.00 C
+ATOM 508 CG TRP B 33 -1.658 -6.440 -1.276 1.00 0.00 C
+ATOM 509 CD1 TRP B 33 -2.151 -6.975 -2.445 1.00 0.00 C
+ATOM 510 CD2 TRP B 33 -0.264 -6.621 -1.304 1.00 0.00 C
+ATOM 511 NE1 TRP B 33 -1.124 -7.492 -3.188 1.00 0.00 N
+ATOM 512 CE2 TRP B 33 0.050 -7.270 -2.504 1.00 0.00 C
+ATOM 513 CE3 TRP B 33 0.745 -6.287 -0.416 1.00 0.00 C
+ATOM 514 CZ2 TRP B 33 1.342 -7.581 -2.823 1.00 0.00 C
+ATOM 515 CZ3 TRP B 33 2.000 -6.586 -0.728 1.00 0.00 C
+ATOM 516 CH2 TRP B 33 2.304 -7.205 -1.887 1.00 0.00 C
+ATOM 517 H TRP B 33 -0.577 -3.866 1.017 1.00 0.00 H
+ATOM 518 HA TRP B 33 -2.569 -3.926 -1.230 1.00 0.00 H
+ATOM 519 1HB TRP B 33 -1.981 -6.082 0.771 1.00 0.00 H
+ATOM 520 2HB TRP B 33 -3.441 -6.067 -0.200 1.00 0.00 H
+ATOM 521 HD1 TRP B 33 -3.202 -6.989 -2.732 1.00 0.00 H
+ATOM 522 HE1 TRP B 33 -1.225 -7.954 -4.093 1.00 0.00 H
+ATOM 523 HE3 TRP B 33 0.513 -5.780 0.520 1.00 0.00 H
+ATOM 524 HZ2 TRP B 33 1.618 -8.085 -3.753 1.00 0.00 H
+ATOM 525 HZ3 TRP B 33 2.767 -6.301 -0.023 1.00 0.00 H
+ATOM 526 HH2 TRP B 33 3.330 -7.389 -2.074 1.00 0.00 H
+ATOM 527 N GLN B 34 -4.361 -3.118 0.243 1.00 0.00 N
+ATOM 528 CA GLN B 34 -5.364 -2.535 1.117 1.00 0.00 C
+ATOM 529 C GLN B 34 -6.781 -2.962 0.744 1.00 0.00 C
+ATOM 530 O GLN B 34 -7.067 -3.359 -0.387 1.00 0.00 O
+ATOM 531 CB GLN B 34 -5.247 -0.988 1.140 1.00 0.00 C
+ATOM 532 CG GLN B 34 -5.554 -0.289 -0.177 1.00 0.00 C
+ATOM 533 CD GLN B 34 -5.350 1.258 -0.098 1.00 0.00 C
+ATOM 534 OE1 GLN B 34 -4.543 1.756 0.678 1.00 0.00 O
+ATOM 535 NE2 GLN B 34 -6.071 1.999 -0.906 1.00 0.00 N
+ATOM 536 H GLN B 34 -4.468 -3.092 -0.773 1.00 0.00 H
+ATOM 537 HA GLN B 34 -5.169 -2.908 2.112 1.00 0.00 H
+ATOM 538 1HB GLN B 34 -5.931 -0.594 1.888 1.00 0.00 H
+ATOM 539 2HB GLN B 34 -4.243 -0.703 1.451 1.00 0.00 H
+ATOM 540 1HG GLN B 34 -4.896 -0.691 -0.953 1.00 0.00 H
+ATOM 541 2HG GLN B 34 -6.602 -0.478 -0.434 1.00 0.00 H
+ATOM 542 1HE2 GLN B 34 -5.979 2.991 -0.890 1.00 0.00 H
+ATOM 543 2HE2 GLN B 34 -6.721 1.571 -1.565 1.00 0.00 H
+ATOM 544 N ASP B 35 -7.661 -2.946 1.735 1.00 0.00 N
+ATOM 545 CA ASP B 35 -9.057 -3.336 1.523 1.00 0.00 C
+ATOM 546 C ASP B 35 -9.902 -2.123 1.127 1.00 0.00 C
+ATOM 547 O ASP B 35 -9.376 -1.027 0.918 1.00 0.00 O
+ATOM 548 CB ASP B 35 -9.618 -4.085 2.754 1.00 0.00 C
+ATOM 549 CG ASP B 35 -9.816 -3.257 4.032 1.00 0.00 C
+ATOM 550 OD1 ASP B 35 -9.867 -2.036 3.971 1.00 0.00 O
+ATOM 551 OD2 ASP B 35 -9.910 -3.870 5.090 1.00 0.00 O
+ATOM 552 H ASP B 35 -7.335 -2.632 2.652 1.00 0.00 H
+ATOM 553 HA ASP B 35 -9.096 -4.046 0.698 1.00 0.00 H
+ATOM 554 1HB ASP B 35 -10.567 -4.545 2.486 1.00 0.00 H
+ATOM 555 2HB ASP B 35 -8.946 -4.896 2.993 1.00 0.00 H
+ATOM 556 N GLN B 36 -11.223 -2.303 1.052 1.00 0.00 N
+ATOM 557 CA GLN B 36 -12.142 -1.258 0.626 1.00 0.00 C
+ATOM 558 C GLN B 36 -12.298 -0.106 1.624 1.00 0.00 C
+ATOM 559 O GLN B 36 -12.903 0.910 1.301 1.00 0.00 O
+ATOM 560 CB GLN B 36 -13.485 -1.891 0.272 1.00 0.00 C
+ATOM 561 CG GLN B 36 -14.233 -2.460 1.414 1.00 0.00 C
+ATOM 562 CD GLN B 36 -15.424 -3.300 0.930 1.00 0.00 C
+ATOM 563 OE1 GLN B 36 -15.281 -4.164 0.038 1.00 0.00 O
+ATOM 564 NE2 GLN B 36 -16.598 -3.039 1.512 1.00 0.00 N
+ATOM 565 H GLN B 36 -11.594 -3.221 1.240 1.00 0.00 H
+ATOM 566 HA GLN B 36 -11.739 -0.829 -0.290 1.00 0.00 H
+ATOM 567 1HB GLN B 36 -14.115 -1.169 -0.242 1.00 0.00 H
+ATOM 568 2HB GLN B 36 -13.307 -2.713 -0.407 1.00 0.00 H
+ATOM 569 1HG GLN B 36 -13.565 -3.094 1.999 1.00 0.00 H
+ATOM 570 2HG GLN B 36 -14.606 -1.642 2.034 1.00 0.00 H
+ATOM 571 1HE2 GLN B 36 -17.455 -3.551 1.259 1.00 0.00 H
+ATOM 572 2HE2 GLN B 36 -16.656 -2.331 2.218 1.00 0.00 H
+ATOM 573 N GLU B 37 -11.785 -0.270 2.852 1.00 0.00 N
+ATOM 574 CA GLU B 37 -11.830 0.780 3.870 1.00 0.00 C
+ATOM 575 C GLU B 37 -10.461 1.462 3.936 1.00 0.00 C
+ATOM 576 O GLU B 37 -10.196 2.297 4.805 1.00 0.00 O
+ATOM 577 CB GLU B 37 -12.223 0.212 5.244 1.00 0.00 C
+ATOM 578 CG GLU B 37 -13.617 -0.381 5.318 1.00 0.00 C
+ATOM 579 CD GLU B 37 -13.991 -0.876 6.715 1.00 0.00 C
+ATOM 580 OE1 GLU B 37 -13.429 -0.403 7.695 1.00 0.00 O
+ATOM 581 OE2 GLU B 37 -14.838 -1.740 6.793 1.00 0.00 O
+ATOM 582 H GLU B 37 -11.268 -1.121 3.093 1.00 0.00 H
+ATOM 583 HA GLU B 37 -12.566 1.528 3.580 1.00 0.00 H
+ATOM 584 1HB GLU B 37 -11.539 -0.595 5.500 1.00 0.00 H
+ATOM 585 2HB GLU B 37 -12.125 0.982 6.008 1.00 0.00 H
+ATOM 586 1HG GLU B 37 -14.338 0.371 5.001 1.00 0.00 H
+ATOM 587 2HG GLU B 37 -13.674 -1.217 4.616 1.00 0.00 H
+ATOM 588 N ASN B 38 -9.605 1.080 2.981 1.00 0.00 N
+ATOM 589 CA ASN B 38 -8.211 1.484 2.829 1.00 0.00 C
+ATOM 590 C ASN B 38 -7.346 1.040 4.010 1.00 0.00 C
+ATOM 591 O ASN B 38 -6.320 1.660 4.317 1.00 0.00 O
+ATOM 592 CB ASN B 38 -8.099 2.982 2.587 1.00 0.00 C
+ATOM 593 CG ASN B 38 -8.763 3.413 1.290 1.00 0.00 C
+ATOM 594 OD1 ASN B 38 -8.467 2.895 0.205 1.00 0.00 O
+ATOM 595 ND2 ASN B 38 -9.660 4.352 1.391 1.00 0.00 N
+ATOM 596 H ASN B 38 -9.919 0.401 2.295 1.00 0.00 H
+ATOM 597 HA ASN B 38 -7.819 0.975 1.945 1.00 0.00 H
+ATOM 598 1HB ASN B 38 -8.533 3.537 3.416 1.00 0.00 H
+ATOM 599 2HB ASN B 38 -7.043 3.249 2.537 1.00 0.00 H
+ATOM 600 1HD2 ASN B 38 -10.138 4.678 0.572 1.00 0.00 H
+ATOM 601 2HD2 ASN B 38 -9.876 4.743 2.287 1.00 0.00 H
+ATOM 602 N LEU B 39 -7.710 -0.078 4.634 1.00 0.00 N
+ATOM 603 CA LEU B 39 -6.893 -0.609 5.702 1.00 0.00 C
+ATOM 604 C LEU B 39 -5.835 -1.475 5.055 1.00 0.00 C
+ATOM 605 O LEU B 39 -6.090 -2.099 4.032 1.00 0.00 O
+ATOM 606 CB LEU B 39 -7.747 -1.406 6.672 1.00 0.00 C
+ATOM 607 CG LEU B 39 -8.874 -0.623 7.333 1.00 0.00 C
+ATOM 608 CD1 LEU B 39 -9.682 -1.563 8.188 1.00 0.00 C
+ATOM 609 CD2 LEU B 39 -8.298 0.533 8.127 1.00 0.00 C
+ATOM 610 H LEU B 39 -8.564 -0.587 4.368 1.00 0.00 H
+ATOM 611 HA LEU B 39 -6.406 0.209 6.234 1.00 0.00 H
+ATOM 612 1HB LEU B 39 -8.194 -2.239 6.137 1.00 0.00 H
+ATOM 613 2HB LEU B 39 -7.109 -1.804 7.457 1.00 0.00 H
+ATOM 614 HG LEU B 39 -9.531 -0.222 6.565 1.00 0.00 H
+ATOM 615 1HD1 LEU B 39 -10.507 -1.022 8.646 1.00 0.00 H
+ATOM 616 2HD1 LEU B 39 -10.079 -2.361 7.554 1.00 0.00 H
+ATOM 617 3HD1 LEU B 39 -9.065 -1.986 8.943 1.00 0.00 H
+ATOM 618 1HD2 LEU B 39 -9.097 1.089 8.583 1.00 0.00 H
+ATOM 619 2HD2 LEU B 39 -7.635 0.155 8.894 1.00 0.00 H
+ATOM 620 3HD2 LEU B 39 -7.740 1.191 7.459 1.00 0.00 H
+ATOM 621 N VAL B 40 -4.644 -1.524 5.635 1.00 0.00 N
+ATOM 622 CA VAL B 40 -3.555 -2.305 5.065 1.00 0.00 C
+ATOM 623 C VAL B 40 -3.481 -3.779 5.524 1.00 0.00 C
+ATOM 624 O VAL B 40 -3.515 -4.092 6.719 1.00 0.00 O
+ATOM 625 CB VAL B 40 -2.233 -1.558 5.338 1.00 0.00 C
+ATOM 626 CG1 VAL B 40 -1.039 -2.354 4.877 1.00 0.00 C
+ATOM 627 CG2 VAL B 40 -2.255 -0.201 4.602 1.00 0.00 C
+ATOM 628 H VAL B 40 -4.480 -0.992 6.484 1.00 0.00 H
+ATOM 629 HA VAL B 40 -3.692 -2.307 3.982 1.00 0.00 H
+ATOM 630 HB VAL B 40 -2.140 -1.402 6.376 1.00 0.00 H
+ATOM 631 1HG1 VAL B 40 -0.119 -1.807 5.091 1.00 0.00 H
+ATOM 632 2HG1 VAL B 40 -1.025 -3.290 5.396 1.00 0.00 H
+ATOM 633 3HG1 VAL B 40 -1.116 -2.532 3.802 1.00 0.00 H
+ATOM 634 1HG2 VAL B 40 -1.329 0.335 4.804 1.00 0.00 H
+ATOM 635 2HG2 VAL B 40 -2.346 -0.378 3.529 1.00 0.00 H
+ATOM 636 3HG2 VAL B 40 -3.105 0.399 4.941 1.00 0.00 H
+ATOM 637 N LEU B 41 -3.393 -4.679 4.539 1.00 0.00 N
+ATOM 638 CA LEU B 41 -3.257 -6.125 4.773 1.00 0.00 C
+ATOM 639 C LEU B 41 -1.792 -6.481 4.951 1.00 0.00 C
+ATOM 640 O LEU B 41 -1.423 -7.334 5.753 1.00 0.00 O
+ATOM 641 CB LEU B 41 -3.731 -6.944 3.573 1.00 0.00 C
+ATOM 642 CG LEU B 41 -5.224 -7.219 3.443 1.00 0.00 C
+ATOM 643 CD1 LEU B 41 -5.930 -5.933 3.056 1.00 0.00 C
+ATOM 644 CD2 LEU B 41 -5.449 -8.331 2.425 1.00 0.00 C
+ATOM 645 H LEU B 41 -3.401 -4.336 3.587 1.00 0.00 H
+ATOM 646 HA LEU B 41 -3.800 -6.397 5.670 1.00 0.00 H
+ATOM 647 1HB LEU B 41 -3.417 -6.429 2.674 1.00 0.00 H
+ATOM 648 2HB LEU B 41 -3.234 -7.896 3.608 1.00 0.00 H
+ATOM 649 HG LEU B 41 -5.599 -7.531 4.374 1.00 0.00 H
+ATOM 650 1HD1 LEU B 41 -6.989 -6.108 2.982 1.00 0.00 H
+ATOM 651 2HD1 LEU B 41 -5.748 -5.169 3.814 1.00 0.00 H
+ATOM 652 3HD1 LEU B 41 -5.554 -5.583 2.096 1.00 0.00 H
+ATOM 653 1HD2 LEU B 41 -6.507 -8.543 2.345 1.00 0.00 H
+ATOM 654 2HD2 LEU B 41 -5.064 -8.015 1.456 1.00 0.00 H
+ATOM 655 3HD2 LEU B 41 -4.925 -9.230 2.750 1.00 0.00 H
+ATOM 656 N ASN B 42 -0.961 -5.848 4.142 1.00 0.00 N
+ATOM 657 CA ASN B 42 0.476 -6.081 4.197 1.00 0.00 C
+ATOM 658 C ASN B 42 1.186 -4.943 3.500 1.00 0.00 C
+ATOM 659 O ASN B 42 0.611 -4.259 2.647 1.00 0.00 O
+ATOM 660 CB ASN B 42 0.879 -7.430 3.574 1.00 0.00 C
+ATOM 661 CG ASN B 42 2.221 -8.023 4.175 1.00 0.00 C
+ATOM 662 OD1 ASN B 42 3.019 -7.282 4.759 1.00 0.00 O
+ATOM 663 ND2 ASN B 42 2.443 -9.314 4.000 1.00 0.00 N
+ATOM 664 H ASN B 42 -1.362 -5.170 3.485 1.00 0.00 H
+ATOM 665 HA ASN B 42 0.801 -6.082 5.235 1.00 0.00 H
+ATOM 666 1HB ASN B 42 0.085 -8.160 3.722 1.00 0.00 H
+ATOM 667 2HB ASN B 42 1.006 -7.309 2.504 1.00 0.00 H
+ATOM 668 1HD2 ASN B 42 3.312 -9.759 4.321 1.00 0.00 H
+ATOM 669 2HD2 ASN B 42 1.781 -9.867 3.512 1.00 0.00 H
+ATOM 670 N GLU B 43 2.470 -4.817 3.785 1.00 0.00 N
+ATOM 671 CA GLU B 43 3.334 -3.838 3.146 1.00 0.00 C
+ATOM 672 C GLU B 43 4.698 -4.435 2.907 1.00 0.00 C
+ATOM 673 O GLU B 43 5.303 -4.995 3.822 1.00 0.00 O
+ATOM 674 CB GLU B 43 3.502 -2.586 4.048 1.00 0.00 C
+ATOM 675 CG GLU B 43 4.454 -1.470 3.466 1.00 0.00 C
+ATOM 676 CD GLU B 43 4.662 -0.228 4.385 1.00 0.00 C
+ATOM 677 OE1 GLU B 43 4.133 -0.171 5.480 1.00 0.00 O
+ATOM 678 OE2 GLU B 43 5.350 0.670 3.956 1.00 0.00 O
+ATOM 679 H GLU B 43 2.857 -5.456 4.472 1.00 0.00 H
+ATOM 680 HA GLU B 43 2.904 -3.547 2.197 1.00 0.00 H
+ATOM 681 1HB GLU B 43 2.523 -2.134 4.214 1.00 0.00 H
+ATOM 682 2HB GLU B 43 3.888 -2.884 5.015 1.00 0.00 H
+ATOM 683 1HG GLU B 43 5.426 -1.917 3.287 1.00 0.00 H
+ATOM 684 2HG GLU B 43 4.063 -1.139 2.503 1.00 0.00 H
+ATOM 685 N VAL B 44 5.209 -4.281 1.693 1.00 0.00 N
+ATOM 686 CA VAL B 44 6.568 -4.675 1.386 1.00 0.00 C
+ATOM 687 C VAL B 44 7.333 -3.372 1.131 1.00 0.00 C
+ATOM 688 O VAL B 44 7.136 -2.701 0.116 1.00 0.00 O
+ATOM 689 CB VAL B 44 6.597 -5.622 0.175 1.00 0.00 C
+ATOM 690 CG1 VAL B 44 8.029 -6.029 -0.154 1.00 0.00 C
+ATOM 691 CG2 VAL B 44 5.749 -6.855 0.490 1.00 0.00 C
+ATOM 692 H VAL B 44 4.636 -3.852 0.973 1.00 0.00 H
+ATOM 693 HA VAL B 44 7.004 -5.186 2.242 1.00 0.00 H
+ATOM 694 HB VAL B 44 6.190 -5.109 -0.680 1.00 0.00 H
+ATOM 695 1HG1 VAL B 44 8.031 -6.692 -1.018 1.00 0.00 H
+ATOM 696 2HG1 VAL B 44 8.612 -5.137 -0.381 1.00 0.00 H
+ATOM 697 3HG1 VAL B 44 8.464 -6.545 0.705 1.00 0.00 H
+ATOM 698 1HG2 VAL B 44 5.742 -7.507 -0.360 1.00 0.00 H
+ATOM 699 2HG2 VAL B 44 6.168 -7.375 1.350 1.00 0.00 H
+ATOM 700 3HG2 VAL B 44 4.745 -6.554 0.715 1.00 0.00 H
+ATOM 701 N TYR B 45 8.153 -2.984 2.097 1.00 0.00 N
+ATOM 702 CA TYR B 45 8.833 -1.701 2.069 1.00 0.00 C
+ATOM 703 C TYR B 45 10.281 -1.856 1.676 1.00 0.00 C
+ATOM 704 O TYR B 45 11.091 -2.336 2.469 1.00 0.00 O
+ATOM 705 CB TYR B 45 8.761 -1.043 3.444 1.00 0.00 C
+ATOM 706 CG TYR B 45 9.327 0.352 3.468 1.00 0.00 C
+ATOM 707 CD1 TYR B 45 8.546 1.415 3.055 1.00 0.00 C
+ATOM 708 CD2 TYR B 45 10.637 0.574 3.886 1.00 0.00 C
+ATOM 709 CE1 TYR B 45 9.053 2.690 3.040 1.00 0.00 C
+ATOM 710 CE2 TYR B 45 11.139 1.866 3.872 1.00 0.00 C
+ATOM 711 CZ TYR B 45 10.349 2.914 3.439 1.00 0.00 C
+ATOM 712 OH TYR B 45 10.848 4.189 3.395 1.00 0.00 O
+ATOM 713 H TYR B 45 8.310 -3.609 2.883 1.00 0.00 H
+ATOM 714 HA TYR B 45 8.352 -1.059 1.334 1.00 0.00 H
+ATOM 715 1HB TYR B 45 7.719 -0.990 3.762 1.00 0.00 H
+ATOM 716 2HB TYR B 45 9.295 -1.651 4.170 1.00 0.00 H
+ATOM 717 HD1 TYR B 45 7.522 1.238 2.733 1.00 0.00 H
+ATOM 718 HD2 TYR B 45 11.260 -0.257 4.212 1.00 0.00 H
+ATOM 719 HE1 TYR B 45 8.429 3.516 2.704 1.00 0.00 H
+ATOM 720 HE2 TYR B 45 12.164 2.054 4.188 1.00 0.00 H
+ATOM 721 HH TYR B 45 11.749 4.187 3.730 1.00 0.00 H
+ATOM 722 N LEU B 46 10.613 -1.505 0.449 1.00 0.00 N
+ATOM 723 CA LEU B 46 11.970 -1.652 -0.043 1.00 0.00 C
+ATOM 724 C LEU B 46 12.431 -3.118 0.082 1.00 0.00 C
+ATOM 725 O LEU B 46 13.575 -3.437 0.431 1.00 0.00 O
+ATOM 726 CB LEU B 46 12.899 -0.666 0.659 1.00 0.00 C
+ATOM 727 CG LEU B 46 12.378 0.787 0.602 1.00 0.00 C
+ATOM 728 CD1 LEU B 46 13.393 1.733 1.236 1.00 0.00 C
+ATOM 729 CD2 LEU B 46 12.071 1.173 -0.806 1.00 0.00 C
+ATOM 730 H LEU B 46 9.920 -1.139 -0.204 1.00 0.00 H
+ATOM 731 HA LEU B 46 11.966 -1.391 -1.103 1.00 0.00 H
+ATOM 732 1HB LEU B 46 13.032 -0.956 1.696 1.00 0.00 H
+ATOM 733 2HB LEU B 46 13.868 -0.694 0.157 1.00 0.00 H
+ATOM 734 HG LEU B 46 11.467 0.846 1.168 1.00 0.00 H
+ATOM 735 1HD1 LEU B 46 13.002 2.752 1.217 1.00 0.00 H
+ATOM 736 2HD1 LEU B 46 13.577 1.435 2.266 1.00 0.00 H
+ATOM 737 3HD1 LEU B 46 14.326 1.692 0.676 1.00 0.00 H
+ATOM 738 1HD2 LEU B 46 11.682 2.172 -0.806 1.00 0.00 H
+ATOM 739 2HD2 LEU B 46 12.974 1.129 -1.407 1.00 0.00 H
+ATOM 740 3HD2 LEU B 46 11.333 0.494 -1.214 1.00 0.00 H
+ATOM 741 N GLY B 47 11.503 -4.015 -0.221 1.00 0.00 N
+ATOM 742 CA GLY B 47 11.750 -5.450 -0.231 1.00 0.00 C
+ATOM 743 C GLY B 47 11.468 -6.169 1.076 1.00 0.00 C
+ATOM 744 O GLY B 47 11.420 -7.399 1.086 1.00 0.00 O
+ATOM 745 H GLY B 47 10.591 -3.673 -0.484 1.00 0.00 H
+ATOM 746 1HA GLY B 47 11.143 -5.901 -1.019 1.00 0.00 H
+ATOM 747 2HA GLY B 47 12.786 -5.622 -0.514 1.00 0.00 H
+ATOM 748 N LYS B 48 11.232 -5.450 2.166 1.00 0.00 N
+ATOM 749 CA LYS B 48 11.003 -6.168 3.409 1.00 0.00 C
+ATOM 750 C LYS B 48 9.563 -6.069 3.885 1.00 0.00 C
+ATOM 751 O LYS B 48 8.943 -4.999 3.846 1.00 0.00 O
+ATOM 752 CB LYS B 48 11.942 -5.613 4.470 1.00 0.00 C
+ATOM 753 CG LYS B 48 13.429 -5.640 4.016 1.00 0.00 C
+ATOM 754 CD LYS B 48 13.978 -7.075 3.859 1.00 0.00 C
+ATOM 755 CE LYS B 48 15.405 -7.068 3.286 1.00 0.00 C
+ATOM 756 NZ LYS B 48 16.357 -6.323 4.155 1.00 0.00 N
+ATOM 757 H LYS B 48 11.259 -4.427 2.161 1.00 0.00 H
+ATOM 758 HA LYS B 48 11.227 -7.221 3.258 1.00 0.00 H
+ATOM 759 1HB LYS B 48 11.672 -4.578 4.685 1.00 0.00 H
+ATOM 760 2HB LYS B 48 11.837 -6.184 5.392 1.00 0.00 H
+ATOM 761 1HG LYS B 48 13.525 -5.115 3.060 1.00 0.00 H
+ATOM 762 2HG LYS B 48 14.034 -5.113 4.751 1.00 0.00 H
+ATOM 763 1HD LYS B 48 13.991 -7.581 4.826 1.00 0.00 H
+ATOM 764 2HD LYS B 48 13.339 -7.644 3.178 1.00 0.00 H
+ATOM 765 1HE LYS B 48 15.744 -8.095 3.188 1.00 0.00 H
+ATOM 766 2HE LYS B 48 15.394 -6.608 2.294 1.00 0.00 H
+ATOM 767 1HZ LYS B 48 17.283 -6.361 3.733 1.00 0.00 H
+ATOM 768 2HZ LYS B 48 16.057 -5.364 4.231 1.00 0.00 H
+ATOM 769 3HZ LYS B 48 16.390 -6.740 5.071 1.00 0.00 H
+ATOM 770 N GLU B 49 9.026 -7.184 4.372 1.00 0.00 N
+ATOM 771 CA GLU B 49 7.678 -7.150 4.918 1.00 0.00 C
+ATOM 772 C GLU B 49 7.670 -6.270 6.154 1.00 0.00 C
+ATOM 773 O GLU B 49 8.558 -6.361 7.004 1.00 0.00 O
+ATOM 774 CB GLU B 49 7.152 -8.553 5.210 1.00 0.00 C
+ATOM 775 CG GLU B 49 6.804 -9.376 3.950 1.00 0.00 C
+ATOM 776 CD GLU B 49 6.242 -10.777 4.267 1.00 0.00 C
+ATOM 777 OE1 GLU B 49 5.032 -10.936 4.382 1.00 0.00 O
+ATOM 778 OE2 GLU B 49 7.036 -11.684 4.379 1.00 0.00 O
+ATOM 779 H GLU B 49 9.553 -8.045 4.375 1.00 0.00 H
+ATOM 780 HA GLU B 49 7.019 -6.701 4.178 1.00 0.00 H
+ATOM 781 1HB GLU B 49 7.888 -9.107 5.789 1.00 0.00 H
+ATOM 782 2HB GLU B 49 6.246 -8.477 5.814 1.00 0.00 H
+ATOM 783 1HG GLU B 49 6.057 -8.823 3.373 1.00 0.00 H
+ATOM 784 2HG GLU B 49 7.696 -9.474 3.333 1.00 0.00 H
+ATOM 785 N LYS B 50 6.650 -5.437 6.254 1.00 0.00 N
+ATOM 786 CA LYS B 50 6.493 -4.485 7.337 1.00 0.00 C
+ATOM 787 C LYS B 50 5.054 -4.525 7.840 1.00 0.00 C
+ATOM 788 O LYS B 50 4.126 -4.450 7.045 1.00 0.00 O
+ATOM 789 CB LYS B 50 6.877 -3.107 6.782 1.00 0.00 C
+ATOM 790 CG LYS B 50 6.904 -1.952 7.720 1.00 0.00 C
+ATOM 791 CD LYS B 50 7.506 -0.712 6.977 1.00 0.00 C
+ATOM 792 CE LYS B 50 7.433 0.560 7.808 1.00 0.00 C
+ATOM 793 NZ LYS B 50 8.314 0.509 9.006 1.00 0.00 N
+ATOM 794 H LYS B 50 5.977 -5.420 5.490 1.00 0.00 H
+ATOM 795 HA LYS B 50 7.158 -4.758 8.155 1.00 0.00 H
+ATOM 796 1HB LYS B 50 7.869 -3.172 6.324 1.00 0.00 H
+ATOM 797 2HB LYS B 50 6.181 -2.849 5.980 1.00 0.00 H
+ATOM 798 1HG LYS B 50 5.888 -1.717 8.039 1.00 0.00 H
+ATOM 799 2HG LYS B 50 7.506 -2.185 8.601 1.00 0.00 H
+ATOM 800 1HD LYS B 50 8.544 -0.910 6.713 1.00 0.00 H
+ATOM 801 2HD LYS B 50 6.949 -0.544 6.060 1.00 0.00 H
+ATOM 802 1HE LYS B 50 7.721 1.407 7.187 1.00 0.00 H
+ATOM 803 2HE LYS B 50 6.404 0.698 8.141 1.00 0.00 H
+ATOM 804 1HZ LYS B 50 8.215 1.356 9.542 1.00 0.00 H
+ATOM 805 2HZ LYS B 50 8.042 -0.278 9.618 1.00 0.00 H
+ATOM 806 3HZ LYS B 50 9.268 0.392 8.733 1.00 0.00 H
+ATOM 807 N PHE B 51 4.871 -4.682 9.155 1.00 0.00 N
+ATOM 808 CA PHE B 51 3.523 -4.811 9.721 1.00 0.00 C
+ATOM 809 C PHE B 51 3.071 -3.623 10.553 1.00 0.00 C
+ATOM 810 O PHE B 51 2.002 -3.635 11.150 1.00 0.00 O
+ATOM 811 CB PHE B 51 3.443 -6.094 10.534 1.00 0.00 C
+ATOM 812 CG PHE B 51 3.661 -7.262 9.635 1.00 0.00 C
+ATOM 813 CD1 PHE B 51 4.920 -7.808 9.481 1.00 0.00 C
+ATOM 814 CD2 PHE B 51 2.610 -7.794 8.903 1.00 0.00 C
+ATOM 815 CE1 PHE B 51 5.132 -8.858 8.599 1.00 0.00 C
+ATOM 816 CE2 PHE B 51 2.815 -8.838 8.024 1.00 0.00 C
+ATOM 817 CZ PHE B 51 4.081 -9.368 7.867 1.00 0.00 C
+ATOM 818 H PHE B 51 5.674 -4.732 9.769 1.00 0.00 H
+ATOM 819 HA PHE B 51 2.841 -4.922 8.895 1.00 0.00 H
+ATOM 820 1HB PHE B 51 4.193 -6.099 11.325 1.00 0.00 H
+ATOM 821 2HB PHE B 51 2.467 -6.169 10.986 1.00 0.00 H
+ATOM 822 HD1 PHE B 51 5.753 -7.394 10.047 1.00 0.00 H
+ATOM 823 HD2 PHE B 51 1.616 -7.372 9.020 1.00 0.00 H
+ATOM 824 HE1 PHE B 51 6.133 -9.275 8.474 1.00 0.00 H
+ATOM 825 HE2 PHE B 51 1.981 -9.241 7.442 1.00 0.00 H
+ATOM 826 HZ PHE B 51 4.249 -10.192 7.162 1.00 0.00 H
+ATOM 827 N ASP B 52 3.852 -2.562 10.528 1.00 0.00 N
+ATOM 828 CA ASP B 52 3.590 -1.373 11.323 1.00 0.00 C
+ATOM 829 C ASP B 52 2.209 -0.744 10.997 1.00 0.00 C
+ATOM 830 O ASP B 52 1.498 -0.252 11.874 1.00 0.00 O
+ATOM 831 CB ASP B 52 4.677 -0.366 11.005 1.00 0.00 C
+ATOM 832 CG ASP B 52 6.085 -0.811 11.447 1.00 0.00 C
+ATOM 833 OD1 ASP B 52 6.206 -1.741 12.196 1.00 0.00 O
+ATOM 834 OD2 ASP B 52 7.036 -0.230 10.957 1.00 0.00 O
+ATOM 835 H ASP B 52 4.690 -2.605 9.970 1.00 0.00 H
+ATOM 836 HA ASP B 52 3.622 -1.641 12.380 1.00 0.00 H
+ATOM 837 1HB ASP B 52 4.678 -0.135 9.942 1.00 0.00 H
+ATOM 838 2HB ASP B 52 4.449 0.525 11.530 1.00 0.00 H
+ATOM 839 N SER B 53 1.845 -0.774 9.707 1.00 0.00 N
+ATOM 840 CA SER B 53 0.589 -0.230 9.179 1.00 0.00 C
+ATOM 841 C SER B 53 -0.519 -1.277 9.045 1.00 0.00 C
+ATOM 842 O SER B 53 -1.613 -0.966 8.567 1.00 0.00 O
+ATOM 843 CB SER B 53 0.818 0.372 7.795 1.00 0.00 C
+ATOM 844 OG SER B 53 1.697 1.469 7.855 1.00 0.00 O
+ATOM 845 H SER B 53 2.488 -1.201 9.054 1.00 0.00 H
+ATOM 846 HA SER B 53 0.242 0.550 9.857 1.00 0.00 H
+ATOM 847 1HB SER B 53 1.235 -0.393 7.133 1.00 0.00 H
+ATOM 848 2HB SER B 53 -0.136 0.681 7.371 1.00 0.00 H
+ATOM 849 HG SER B 53 2.503 1.122 8.253 1.00 0.00 H
+ATOM 850 N VAL B 54 -0.233 -2.523 9.401 1.00 0.00 N
+ATOM 851 CA VAL B 54 -1.188 -3.586 9.164 1.00 0.00 C
+ATOM 852 C VAL B 54 -2.259 -3.661 10.219 1.00 0.00 C
+ATOM 853 O VAL B 54 -1.980 -3.648 11.418 1.00 0.00 O
+ATOM 854 CB VAL B 54 -0.467 -4.930 9.008 1.00 0.00 C
+ATOM 855 CG1 VAL B 54 -1.464 -6.060 8.882 1.00 0.00 C
+ATOM 856 CG2 VAL B 54 0.369 -4.861 7.761 1.00 0.00 C
+ATOM 857 H VAL B 54 0.643 -2.746 9.864 1.00 0.00 H
+ATOM 858 HA VAL B 54 -1.673 -3.380 8.211 1.00 0.00 H
+ATOM 859 HB VAL B 54 0.163 -5.112 9.874 1.00 0.00 H
+ATOM 860 1HG1 VAL B 54 -0.938 -7.005 8.751 1.00 0.00 H
+ATOM 861 2HG1 VAL B 54 -2.079 -6.117 9.776 1.00 0.00 H
+ATOM 862 3HG1 VAL B 54 -2.101 -5.882 8.016 1.00 0.00 H
+ATOM 863 1HG2 VAL B 54 0.892 -5.794 7.616 1.00 0.00 H
+ATOM 864 2HG2 VAL B 54 -0.287 -4.690 6.927 1.00 0.00 H
+ATOM 865 3HG2 VAL B 54 1.084 -4.047 7.831 1.00 0.00 H
+ATOM 866 N HIS B 55 -3.497 -3.693 9.761 1.00 0.00 N
+ATOM 867 CA HIS B 55 -4.616 -3.731 10.682 1.00 0.00 C
+ATOM 868 C HIS B 55 -4.629 -5.036 11.442 1.00 0.00 C
+ATOM 869 O HIS B 55 -4.393 -6.103 10.870 1.00 0.00 O
+ATOM 870 CB HIS B 55 -5.953 -3.500 9.975 1.00 0.00 C
+ATOM 871 CG HIS B 55 -7.087 -3.209 10.924 1.00 0.00 C
+ATOM 872 ND1 HIS B 55 -7.714 -4.179 11.660 1.00 0.00 N
+ATOM 873 CD2 HIS B 55 -7.702 -2.037 11.254 1.00 0.00 C
+ATOM 874 CE1 HIS B 55 -8.654 -3.619 12.406 1.00 0.00 C
+ATOM 875 NE2 HIS B 55 -8.668 -2.329 12.168 1.00 0.00 N
+ATOM 876 H HIS B 55 -3.640 -3.700 8.747 1.00 0.00 H
+ATOM 877 HA HIS B 55 -4.497 -2.951 11.419 1.00 0.00 H
+ATOM 878 1HB HIS B 55 -5.852 -2.663 9.283 1.00 0.00 H
+ATOM 879 2HB HIS B 55 -6.213 -4.377 9.399 1.00 0.00 H
+ATOM 880 HD2 HIS B 55 -7.465 -1.051 10.863 1.00 0.00 H
+ATOM 881 HE1 HIS B 55 -9.313 -4.146 13.097 1.00 0.00 H
+ATOM 882 HE2 HIS B 55 -9.304 -1.642 12.607 1.00 0.00 H
+ATOM 883 N SER B 56 -4.975 -4.959 12.729 1.00 0.00 N
+ATOM 884 CA SER B 56 -5.056 -6.128 13.612 1.00 0.00 C
+ATOM 885 C SER B 56 -5.796 -7.290 12.978 1.00 0.00 C
+ATOM 886 O SER B 56 -5.385 -8.444 13.114 1.00 0.00 O
+ATOM 887 CB SER B 56 -5.759 -5.736 14.916 1.00 0.00 C
+ATOM 888 OG SER B 56 -4.983 -4.830 15.682 1.00 0.00 O
+ATOM 889 H SER B 56 -5.164 -4.031 13.113 1.00 0.00 H
+ATOM 890 HA SER B 56 -4.042 -6.451 13.843 1.00 0.00 H
+ATOM 891 1HB SER B 56 -6.715 -5.269 14.672 1.00 0.00 H
+ATOM 892 2HB SER B 56 -5.973 -6.623 15.503 1.00 0.00 H
+ATOM 893 HG SER B 56 -5.622 -4.445 16.330 1.00 0.00 H
+ATOM 894 N LYS B 57 -6.851 -6.977 12.227 1.00 0.00 N
+ATOM 895 CA LYS B 57 -7.720 -7.902 11.521 1.00 0.00 C
+ATOM 896 C LYS B 57 -6.989 -8.834 10.549 1.00 0.00 C
+ATOM 897 O LYS B 57 -7.483 -9.914 10.224 1.00 0.00 O
+ATOM 898 CB LYS B 57 -8.776 -7.094 10.769 1.00 0.00 C
+ATOM 899 CG LYS B 57 -9.800 -7.869 10.024 1.00 0.00 C
+ATOM 900 CD LYS B 57 -10.964 -6.932 9.592 1.00 0.00 C
+ATOM 901 CE LYS B 57 -10.566 -5.999 8.462 1.00 0.00 C
+ATOM 902 NZ LYS B 57 -11.739 -5.197 7.939 1.00 0.00 N
+ATOM 903 H LYS B 57 -7.082 -5.989 12.137 1.00 0.00 H
+ATOM 904 HA LYS B 57 -8.217 -8.527 12.263 1.00 0.00 H
+ATOM 905 1HB LYS B 57 -9.304 -6.464 11.474 1.00 0.00 H
+ATOM 906 2HB LYS B 57 -8.276 -6.442 10.050 1.00 0.00 H
+ATOM 907 1HG LYS B 57 -9.339 -8.308 9.132 1.00 0.00 H
+ATOM 908 2HG LYS B 57 -10.185 -8.670 10.650 1.00 0.00 H
+ATOM 909 1HD LYS B 57 -11.812 -7.536 9.269 1.00 0.00 H
+ATOM 910 2HD LYS B 57 -11.273 -6.329 10.447 1.00 0.00 H
+ATOM 911 1HE LYS B 57 -9.781 -5.318 8.800 1.00 0.00 H
+ATOM 912 2HE LYS B 57 -10.186 -6.604 7.665 1.00 0.00 H
+ATOM 913 1HZ LYS B 57 -11.429 -4.610 7.152 1.00 0.00 H
+ATOM 914 2HZ LYS B 57 -12.453 -5.830 7.613 1.00 0.00 H
+ATOM 915 3HZ LYS B 57 -12.115 -4.614 8.668 1.00 0.00 H
+ATOM 916 N TYR B 58 -5.860 -8.380 10.007 1.00 0.00 N
+ATOM 917 CA TYR B 58 -5.115 -9.150 9.029 1.00 0.00 C
+ATOM 918 C TYR B 58 -3.818 -9.748 9.583 1.00 0.00 C
+ATOM 919 O TYR B 58 -3.107 -10.454 8.857 1.00 0.00 O
+ATOM 920 CB TYR B 58 -4.797 -8.267 7.838 1.00 0.00 C
+ATOM 921 CG TYR B 58 -6.031 -7.773 7.140 1.00 0.00 C
+ATOM 922 CD1 TYR B 58 -6.869 -8.692 6.524 1.00 0.00 C
+ATOM 923 CD2 TYR B 58 -6.325 -6.399 7.082 1.00 0.00 C
+ATOM 924 CE1 TYR B 58 -7.995 -8.265 5.847 1.00 0.00 C
+ATOM 925 CE2 TYR B 58 -7.468 -5.980 6.394 1.00 0.00 C
+ATOM 926 CZ TYR B 58 -8.288 -6.918 5.780 1.00 0.00 C
+ATOM 927 OH TYR B 58 -9.416 -6.517 5.090 1.00 0.00 O
+ATOM 928 H TYR B 58 -5.476 -7.488 10.314 1.00 0.00 H
+ATOM 929 HA TYR B 58 -5.740 -9.974 8.696 1.00 0.00 H
+ATOM 930 1HB TYR B 58 -4.224 -7.400 8.173 1.00 0.00 H
+ATOM 931 2HB TYR B 58 -4.183 -8.813 7.120 1.00 0.00 H
+ATOM 932 HD1 TYR B 58 -6.637 -9.760 6.569 1.00 0.00 H
+ATOM 933 HD2 TYR B 58 -5.666 -5.673 7.556 1.00 0.00 H
+ATOM 934 HE1 TYR B 58 -8.648 -8.993 5.360 1.00 0.00 H
+ATOM 935 HE2 TYR B 58 -7.716 -4.915 6.327 1.00 0.00 H
+ATOM 936 HH TYR B 58 -9.553 -5.547 5.200 1.00 0.00 H
+ATOM 937 N MET B 59 -3.487 -9.469 10.844 1.00 0.00 N
+ATOM 938 CA MET B 59 -2.211 -9.943 11.353 1.00 0.00 C
+ATOM 939 C MET B 59 -2.161 -11.451 11.383 1.00 0.00 C
+ATOM 940 O MET B 59 -3.079 -12.109 11.857 1.00 0.00 O
+ATOM 941 CB MET B 59 -1.907 -9.353 12.732 1.00 0.00 C
+ATOM 942 CG MET B 59 -1.561 -7.840 12.724 1.00 0.00 C
+ATOM 943 SD MET B 59 -0.037 -7.507 11.813 1.00 0.00 S
+ATOM 944 CE MET B 59 1.202 -8.272 12.841 1.00 0.00 C
+ATOM 945 H MET B 59 -4.116 -8.950 11.455 1.00 0.00 H
+ATOM 946 HA MET B 59 -1.431 -9.603 10.669 1.00 0.00 H
+ATOM 947 1HB MET B 59 -2.780 -9.491 13.375 1.00 0.00 H
+ATOM 948 2HB MET B 59 -1.075 -9.890 13.187 1.00 0.00 H
+ATOM 949 1HG MET B 59 -2.371 -7.295 12.239 1.00 0.00 H
+ATOM 950 2HG MET B 59 -1.454 -7.459 13.733 1.00 0.00 H
+ATOM 951 1HE MET B 59 2.187 -8.144 12.394 1.00 0.00 H
+ATOM 952 2HE MET B 59 1.188 -7.811 13.804 1.00 0.00 H
+ATOM 953 3HE MET B 59 0.988 -9.335 12.943 1.00 0.00 H
+ATOM 954 N GLY B 60 -1.085 -11.991 10.807 1.00 0.00 N
+ATOM 955 CA GLY B 60 -0.814 -13.422 10.755 1.00 0.00 C
+ATOM 956 C GLY B 60 -1.587 -14.141 9.651 1.00 0.00 C
+ATOM 957 O GLY B 60 -1.417 -15.348 9.451 1.00 0.00 O
+ATOM 958 H GLY B 60 -0.400 -11.362 10.412 1.00 0.00 H
+ATOM 959 1HA GLY B 60 0.254 -13.577 10.601 1.00 0.00 H
+ATOM 960 2HA GLY B 60 -1.064 -13.868 11.713 1.00 0.00 H
+ATOM 961 N ARG B 61 -2.411 -13.409 8.903 1.00 0.00 N
+ATOM 962 CA ARG B 61 -3.256 -14.037 7.904 1.00 0.00 C
+ATOM 963 C ARG B 61 -2.775 -13.882 6.478 1.00 0.00 C
+ATOM 964 O ARG B 61 -3.493 -14.262 5.554 1.00 0.00 O
+ATOM 965 CB ARG B 61 -4.658 -13.459 7.977 1.00 0.00 C
+ATOM 966 CG ARG B 61 -5.404 -13.676 9.298 1.00 0.00 C
+ATOM 967 CD ARG B 61 -6.772 -13.141 9.164 1.00 0.00 C
+ATOM 968 NE ARG B 61 -7.421 -13.865 8.158 1.00 0.00 N
+ATOM 969 CZ ARG B 61 -8.500 -13.494 7.459 1.00 0.00 C
+ATOM 970 NH1 ARG B 61 -8.905 -14.321 6.556 1.00 0.00 N
+ATOM 971 NH2 ARG B 61 -9.129 -12.321 7.659 1.00 0.00 N
+ATOM 972 H ARG B 61 -2.503 -12.401 9.058 1.00 0.00 H
+ATOM 973 HA ARG B 61 -3.312 -15.101 8.126 1.00 0.00 H
+ATOM 974 1HB ARG B 61 -4.614 -12.384 7.802 1.00 0.00 H
+ATOM 975 2HB ARG B 61 -5.267 -13.896 7.185 1.00 0.00 H
+ATOM 976 1HG ARG B 61 -5.456 -14.741 9.535 1.00 0.00 H
+ATOM 977 2HG ARG B 61 -4.896 -13.139 10.103 1.00 0.00 H
+ATOM 978 1HD ARG B 61 -7.320 -13.270 10.095 1.00 0.00 H
+ATOM 979 2HD ARG B 61 -6.758 -12.092 8.885 1.00 0.00 H
+ATOM 980 HE ARG B 61 -7.023 -14.773 7.913 1.00 0.00 H
+ATOM 981 1HH1 ARG B 61 -8.377 -15.194 6.395 1.00 0.00 H
+ATOM 982 2HH1 ARG B 61 -9.723 -14.084 6.001 1.00 0.00 H
+ATOM 983 1HH2 ARG B 61 -8.795 -11.677 8.367 1.00 0.00 H
+ATOM 984 2HH2 ARG B 61 -9.948 -12.071 7.073 1.00 0.00 H
+ATOM 985 N THR B 62 -1.595 -13.292 6.274 1.00 0.00 N
+ATOM 986 CA THR B 62 -1.148 -13.061 4.916 1.00 0.00 C
+ATOM 987 C THR B 62 0.266 -13.540 4.676 1.00 0.00 C
+ATOM 988 O THR B 62 1.080 -13.660 5.604 1.00 0.00 O
+ATOM 989 CB THR B 62 -1.196 -11.552 4.552 1.00 0.00 C
+ATOM 990 OG1 THR B 62 -0.231 -10.823 5.353 1.00 0.00 O
+ATOM 991 CG2 THR B 62 -2.602 -10.962 4.795 1.00 0.00 C
+ATOM 992 H THR B 62 -1.035 -13.004 7.060 1.00 0.00 H
+ATOM 993 HA THR B 62 -1.798 -13.600 4.244 1.00 0.00 H
+ATOM 994 HB THR B 62 -0.952 -11.439 3.510 1.00 0.00 H
+ATOM 995 HG1 THR B 62 -0.446 -9.878 5.338 1.00 0.00 H
+ATOM 996 1HG2 THR B 62 -2.601 -9.914 4.524 1.00 0.00 H
+ATOM 997 2HG2 THR B 62 -3.325 -11.499 4.179 1.00 0.00 H
+ATOM 998 3HG2 THR B 62 -2.873 -11.058 5.845 1.00 0.00 H
+ATOM 999 N SER B 63 0.570 -13.741 3.409 1.00 0.00 N
+ATOM 1000 CA SER B 63 1.942 -14.018 3.021 1.00 0.00 C
+ATOM 1001 C SER B 63 2.196 -13.443 1.637 1.00 0.00 C
+ATOM 1002 O SER B 63 1.310 -13.417 0.776 1.00 0.00 O
+ATOM 1003 CB SER B 63 2.222 -15.521 3.044 1.00 0.00 C
+ATOM 1004 OG SER B 63 1.468 -16.194 2.078 1.00 0.00 O
+ATOM 1005 H SER B 63 -0.183 -13.678 2.727 1.00 0.00 H
+ATOM 1006 HA SER B 63 2.616 -13.526 3.726 1.00 0.00 H
+ATOM 1007 1HB SER B 63 3.282 -15.697 2.867 1.00 0.00 H
+ATOM 1008 2HB SER B 63 1.989 -15.917 4.030 1.00 0.00 H
+ATOM 1009 HG SER B 63 1.841 -15.924 1.233 1.00 0.00 H
+ATOM 1010 N PHE B 64 3.424 -13.012 1.418 1.00 0.00 N
+ATOM 1011 CA PHE B 64 3.821 -12.415 0.153 1.00 0.00 C
+ATOM 1012 C PHE B 64 4.659 -13.329 -0.708 1.00 0.00 C
+ATOM 1013 O PHE B 64 5.650 -13.894 -0.254 1.00 0.00 O
+ATOM 1014 CB PHE B 64 4.567 -11.102 0.421 1.00 0.00 C
+ATOM 1015 CG PHE B 64 5.144 -10.409 -0.807 1.00 0.00 C
+ATOM 1016 CD1 PHE B 64 4.328 -9.924 -1.803 1.00 0.00 C
+ATOM 1017 CD2 PHE B 64 6.510 -10.196 -0.922 1.00 0.00 C
+ATOM 1018 CE1 PHE B 64 4.863 -9.255 -2.910 1.00 0.00 C
+ATOM 1019 CE2 PHE B 64 7.036 -9.522 -2.017 1.00 0.00 C
+ATOM 1020 CZ PHE B 64 6.204 -9.053 -3.020 1.00 0.00 C
+ATOM 1021 H PHE B 64 4.104 -13.066 2.167 1.00 0.00 H
+ATOM 1022 HA PHE B 64 2.921 -12.172 -0.409 1.00 0.00 H
+ATOM 1023 1HB PHE B 64 3.883 -10.402 0.906 1.00 0.00 H
+ATOM 1024 2HB PHE B 64 5.384 -11.284 1.121 1.00 0.00 H
+ATOM 1025 HD1 PHE B 64 3.246 -10.072 -1.723 1.00 0.00 H
+ATOM 1026 HD2 PHE B 64 7.169 -10.557 -0.134 1.00 0.00 H
+ATOM 1027 HE1 PHE B 64 4.194 -8.882 -3.674 1.00 0.00 H
+ATOM 1028 HE2 PHE B 64 8.106 -9.365 -2.081 1.00 0.00 H
+ATOM 1029 HZ PHE B 64 6.612 -8.526 -3.881 1.00 0.00 H
+ATOM 1030 N ASP B 65 4.223 -13.464 -1.959 1.00 0.00 N
+ATOM 1031 CA ASP B 65 4.945 -14.182 -2.991 1.00 0.00 C
+ATOM 1032 C ASP B 65 5.841 -13.222 -3.763 1.00 0.00 C
+ATOM 1033 O ASP B 65 5.381 -12.529 -4.676 1.00 0.00 O
+ATOM 1034 CB ASP B 65 3.966 -14.859 -3.942 1.00 0.00 C
+ATOM 1035 CG ASP B 65 4.627 -15.586 -5.083 1.00 0.00 C
+ATOM 1036 OD1 ASP B 65 5.782 -15.323 -5.383 1.00 0.00 O
+ATOM 1037 OD2 ASP B 65 3.950 -16.317 -5.752 1.00 0.00 O
+ATOM 1038 H ASP B 65 3.356 -13.008 -2.225 1.00 0.00 H
+ATOM 1039 HA ASP B 65 5.568 -14.941 -2.523 1.00 0.00 H
+ATOM 1040 1HB ASP B 65 3.354 -15.569 -3.380 1.00 0.00 H
+ATOM 1041 2HB ASP B 65 3.293 -14.111 -4.349 1.00 0.00 H
+ATOM 1042 N SER B 66 7.128 -13.212 -3.424 1.00 0.00 N
+ATOM 1043 CA SER B 66 8.071 -12.268 -4.002 1.00 0.00 C
+ATOM 1044 C SER B 66 8.490 -12.629 -5.422 1.00 0.00 C
+ATOM 1045 O SER B 66 9.240 -11.871 -6.039 1.00 0.00 O
+ATOM 1046 CB SER B 66 9.290 -12.115 -3.110 1.00 0.00 C
+ATOM 1047 OG SER B 66 10.028 -13.298 -3.042 1.00 0.00 O
+ATOM 1048 H SER B 66 7.438 -13.843 -2.699 1.00 0.00 H
+ATOM 1049 HA SER B 66 7.583 -11.304 -4.061 1.00 0.00 H
+ATOM 1050 1HB SER B 66 9.917 -11.313 -3.503 1.00 0.00 H
+ATOM 1051 2HB SER B 66 8.976 -11.824 -2.111 1.00 0.00 H
+ATOM 1052 HG SER B 66 10.113 -13.613 -3.956 1.00 0.00 H
+ATOM 1053 N ASP B 67 8.037 -13.789 -5.929 1.00 0.00 N
+ATOM 1054 CA ASP B 67 8.365 -14.192 -7.289 1.00 0.00 C
+ATOM 1055 C ASP B 67 7.294 -13.634 -8.232 1.00 0.00 C
+ATOM 1056 O ASP B 67 7.605 -13.181 -9.332 1.00 0.00 O
+ATOM 1057 CB ASP B 67 8.396 -15.732 -7.447 1.00 0.00 C
+ATOM 1058 CG ASP B 67 9.490 -16.471 -6.614 1.00 0.00 C
+ATOM 1059 OD1 ASP B 67 10.643 -16.135 -6.739 1.00 0.00 O
+ATOM 1060 OD2 ASP B 67 9.144 -17.387 -5.890 1.00 0.00 O
+ATOM 1061 H ASP B 67 7.390 -14.377 -5.394 1.00 0.00 H
+ATOM 1062 HA ASP B 67 9.335 -13.776 -7.568 1.00 0.00 H
+ATOM 1063 1HB ASP B 67 7.416 -16.134 -7.159 1.00 0.00 H
+ATOM 1064 2HB ASP B 67 8.543 -15.978 -8.500 1.00 0.00 H
+ATOM 1065 N SER B 68 6.033 -13.630 -7.776 1.00 0.00 N
+ATOM 1066 CA SER B 68 4.944 -13.125 -8.625 1.00 0.00 C
+ATOM 1067 C SER B 68 4.388 -11.761 -8.176 1.00 0.00 C
+ATOM 1068 O SER B 68 3.590 -11.152 -8.889 1.00 0.00 O
+ATOM 1069 CB SER B 68 3.782 -14.117 -8.714 1.00 0.00 C
+ATOM 1070 OG SER B 68 3.119 -14.294 -7.489 1.00 0.00 O
+ATOM 1071 H SER B 68 5.846 -14.054 -6.858 1.00 0.00 H
+ATOM 1072 HA SER B 68 5.342 -12.999 -9.630 1.00 0.00 H
+ATOM 1073 1HB SER B 68 3.068 -13.748 -9.443 1.00 0.00 H
+ATOM 1074 2HB SER B 68 4.146 -15.079 -9.074 1.00 0.00 H
+ATOM 1075 HG SER B 68 3.578 -15.036 -7.015 1.00 0.00 H
+ATOM 1076 N TRP B 69 4.780 -11.311 -6.987 1.00 0.00 N
+ATOM 1077 CA TRP B 69 4.288 -10.085 -6.343 1.00 0.00 C
+ATOM 1078 C TRP B 69 2.792 -10.184 -6.028 1.00 0.00 C
+ATOM 1079 O TRP B 69 2.003 -9.293 -6.331 1.00 0.00 O
+ATOM 1080 CB TRP B 69 4.591 -8.794 -7.104 1.00 0.00 C
+ATOM 1081 CG TRP B 69 6.075 -8.551 -7.328 1.00 0.00 C
+ATOM 1082 CD1 TRP B 69 7.093 -9.431 -7.098 1.00 0.00 C
+ATOM 1083 CD2 TRP B 69 6.703 -7.332 -7.804 1.00 0.00 C
+ATOM 1084 NE1 TRP B 69 8.295 -8.858 -7.434 1.00 0.00 N
+ATOM 1085 CE2 TRP B 69 8.074 -7.578 -7.873 1.00 0.00 C
+ATOM 1086 CE3 TRP B 69 6.221 -6.078 -8.169 1.00 0.00 C
+ATOM 1087 CZ2 TRP B 69 8.974 -6.616 -8.318 1.00 0.00 C
+ATOM 1088 CZ3 TRP B 69 7.130 -5.119 -8.590 1.00 0.00 C
+ATOM 1089 CH2 TRP B 69 8.473 -5.397 -8.672 1.00 0.00 C
+ATOM 1090 H TRP B 69 5.428 -11.876 -6.447 1.00 0.00 H
+ATOM 1091 HA TRP B 69 4.824 -10.012 -5.417 1.00 0.00 H
+ATOM 1092 1HB TRP B 69 4.084 -8.802 -8.071 1.00 0.00 H
+ATOM 1093 2HB TRP B 69 4.189 -7.950 -6.540 1.00 0.00 H
+ATOM 1094 HD1 TRP B 69 6.972 -10.445 -6.721 1.00 0.00 H
+ATOM 1095 HE1 TRP B 69 9.199 -9.313 -7.369 1.00 0.00 H
+ATOM 1096 HE3 TRP B 69 5.157 -5.856 -8.115 1.00 0.00 H
+ATOM 1097 HZ2 TRP B 69 10.048 -6.803 -8.379 1.00 0.00 H
+ATOM 1098 HZ3 TRP B 69 6.752 -4.142 -8.861 1.00 0.00 H
+ATOM 1099 HH2 TRP B 69 9.162 -4.625 -9.019 1.00 0.00 H
+ATOM 1100 N THR B 70 2.424 -11.309 -5.455 1.00 0.00 N
+ATOM 1101 CA THR B 70 1.035 -11.613 -5.092 1.00 0.00 C
+ATOM 1102 C THR B 70 0.892 -11.751 -3.585 1.00 0.00 C
+ATOM 1103 O THR B 70 1.770 -12.325 -2.935 1.00 0.00 O
+ATOM 1104 CB THR B 70 0.570 -12.931 -5.764 1.00 0.00 C
+ATOM 1105 OG1 THR B 70 0.657 -12.811 -7.194 1.00 0.00 O
+ATOM 1106 CG2 THR B 70 -0.850 -13.297 -5.379 1.00 0.00 C
+ATOM 1107 H THR B 70 3.174 -11.969 -5.252 1.00 0.00 H
+ATOM 1108 HA THR B 70 0.391 -10.794 -5.418 1.00 0.00 H
+ATOM 1109 HB THR B 70 1.222 -13.744 -5.445 1.00 0.00 H
+ATOM 1110 HG1 THR B 70 1.529 -13.192 -7.460 1.00 0.00 H
+ATOM 1111 1HG2 THR B 70 -1.104 -14.221 -5.871 1.00 0.00 H
+ATOM 1112 2HG2 THR B 70 -0.938 -13.432 -4.305 1.00 0.00 H
+ATOM 1113 3HG2 THR B 70 -1.521 -12.523 -5.692 1.00 0.00 H
+ATOM 1114 N LEU B 71 -0.185 -11.204 -3.008 1.00 0.00 N
+ATOM 1115 CA LEU B 71 -0.396 -11.403 -1.564 1.00 0.00 C
+ATOM 1116 C LEU B 71 -1.425 -12.500 -1.408 1.00 0.00 C
+ATOM 1117 O LEU B 71 -2.426 -12.520 -2.123 1.00 0.00 O
+ATOM 1118 CB LEU B 71 -0.921 -10.158 -0.846 1.00 0.00 C
+ATOM 1119 CG LEU B 71 -0.970 -10.232 0.653 1.00 0.00 C
+ATOM 1120 CD1 LEU B 71 0.463 -10.220 1.125 1.00 0.00 C
+ATOM 1121 CD2 LEU B 71 -1.740 -9.088 1.243 1.00 0.00 C
+ATOM 1122 H LEU B 71 -0.883 -10.710 -3.568 1.00 0.00 H
+ATOM 1123 HA LEU B 71 0.532 -11.721 -1.094 1.00 0.00 H
+ATOM 1124 1HB LEU B 71 -0.246 -9.388 -1.055 1.00 0.00 H
+ATOM 1125 2HB LEU B 71 -1.904 -9.897 -1.227 1.00 0.00 H
+ATOM 1126 HG LEU B 71 -1.436 -11.168 0.960 1.00 0.00 H
+ATOM 1127 1HD1 LEU B 71 0.497 -10.297 2.196 1.00 0.00 H
+ATOM 1128 2HD1 LEU B 71 0.984 -11.037 0.685 1.00 0.00 H
+ATOM 1129 3HD1 LEU B 71 0.935 -9.288 0.812 1.00 0.00 H
+ATOM 1130 1HD2 LEU B 71 -1.736 -9.173 2.322 1.00 0.00 H
+ATOM 1131 2HD2 LEU B 71 -1.262 -8.175 0.962 1.00 0.00 H
+ATOM 1132 3HD2 LEU B 71 -2.765 -9.102 0.877 1.00 0.00 H
+ATOM 1133 N ARG B 72 -1.183 -13.442 -0.521 1.00 0.00 N
+ATOM 1134 CA ARG B 72 -2.162 -14.488 -0.304 1.00 0.00 C
+ATOM 1135 C ARG B 72 -2.885 -14.192 1.002 1.00 0.00 C
+ATOM 1136 O ARG B 72 -2.234 -13.829 1.981 1.00 0.00 O
+ATOM 1137 CB ARG B 72 -1.464 -15.832 -0.241 1.00 0.00 C
+ATOM 1138 CG ARG B 72 -0.589 -16.137 -1.474 1.00 0.00 C
+ATOM 1139 CD ARG B 72 -0.020 -17.520 -1.431 1.00 0.00 C
+ATOM 1140 NE ARG B 72 -1.042 -18.514 -1.783 1.00 0.00 N
+ATOM 1141 CZ ARG B 72 -1.096 -19.796 -1.346 1.00 0.00 C
+ATOM 1142 NH1 ARG B 72 -0.195 -20.265 -0.499 1.00 0.00 N
+ATOM 1143 NH2 ARG B 72 -2.058 -20.585 -1.777 1.00 0.00 N
+ATOM 1144 H ARG B 72 -0.323 -13.423 0.018 1.00 0.00 H
+ATOM 1145 HA ARG B 72 -2.865 -14.491 -1.123 1.00 0.00 H
+ATOM 1146 1HB ARG B 72 -0.825 -15.872 0.639 1.00 0.00 H
+ATOM 1147 2HB ARG B 72 -2.184 -16.606 -0.159 1.00 0.00 H
+ATOM 1148 1HG ARG B 72 -1.199 -16.039 -2.376 1.00 0.00 H
+ATOM 1149 2HG ARG B 72 0.241 -15.422 -1.517 1.00 0.00 H
+ATOM 1150 1HD ARG B 72 0.798 -17.599 -2.144 1.00 0.00 H
+ATOM 1151 2HD ARG B 72 0.345 -17.739 -0.424 1.00 0.00 H
+ATOM 1152 HE ARG B 72 -1.748 -18.208 -2.491 1.00 0.00 H
+ATOM 1153 1HH1 ARG B 72 0.548 -19.675 -0.160 1.00 0.00 H
+ATOM 1154 2HH1 ARG B 72 -0.231 -21.256 -0.188 1.00 0.00 H
+ATOM 1155 1HH2 ARG B 72 -2.749 -20.237 -2.435 1.00 0.00 H
+ATOM 1156 2HH2 ARG B 72 -2.098 -21.544 -1.459 1.00 0.00 H
+ATOM 1157 N LEU B 73 -4.221 -14.324 1.013 1.00 0.00 N
+ATOM 1158 CA LEU B 73 -5.058 -14.118 2.216 1.00 0.00 C
+ATOM 1159 C LEU B 73 -5.578 -15.474 2.684 1.00 0.00 C
+ATOM 1160 O LEU B 73 -6.346 -16.131 1.981 1.00 0.00 O
+ATOM 1161 CB LEU B 73 -6.218 -13.134 1.918 1.00 0.00 C
+ATOM 1162 CG LEU B 73 -7.231 -12.863 3.069 1.00 0.00 C
+ATOM 1163 CD1 LEU B 73 -6.505 -12.222 4.263 1.00 0.00 C
+ATOM 1164 CD2 LEU B 73 -8.372 -11.965 2.541 1.00 0.00 C
+ATOM 1165 H LEU B 73 -4.664 -14.607 0.139 1.00 0.00 H
+ATOM 1166 HA LEU B 73 -4.442 -13.704 3.014 1.00 0.00 H
+ATOM 1167 1HB LEU B 73 -5.788 -12.176 1.627 1.00 0.00 H
+ATOM 1168 2HB LEU B 73 -6.783 -13.522 1.082 1.00 0.00 H
+ATOM 1169 HG LEU B 73 -7.654 -13.807 3.417 1.00 0.00 H
+ATOM 1170 1HD1 LEU B 73 -7.219 -12.048 5.069 1.00 0.00 H
+ATOM 1171 2HD1 LEU B 73 -5.719 -12.892 4.619 1.00 0.00 H
+ATOM 1172 3HD1 LEU B 73 -6.064 -11.272 3.961 1.00 0.00 H
+ATOM 1173 1HD2 LEU B 73 -9.087 -11.779 3.339 1.00 0.00 H
+ATOM 1174 2HD2 LEU B 73 -7.973 -11.008 2.191 1.00 0.00 H
+ATOM 1175 3HD2 LEU B 73 -8.871 -12.462 1.714 1.00 0.00 H
+ATOM 1176 N HIS B 74 -5.113 -15.911 3.852 1.00 0.00 N
+ATOM 1177 CA HIS B 74 -5.346 -17.266 4.336 1.00 0.00 C
+ATOM 1178 C HIS B 74 -6.589 -17.471 5.195 1.00 0.00 C
+ATOM 1179 O HIS B 74 -6.987 -16.580 5.944 1.00 0.00 O
+ATOM 1180 CB HIS B 74 -4.102 -17.708 5.108 1.00 0.00 C
+ATOM 1181 CG HIS B 74 -2.874 -17.819 4.227 1.00 0.00 C
+ATOM 1182 ND1 HIS B 74 -2.559 -18.965 3.513 1.00 0.00 N
+ATOM 1183 CD2 HIS B 74 -1.902 -16.904 3.931 1.00 0.00 C
+ATOM 1184 CE1 HIS B 74 -1.426 -18.753 2.840 1.00 0.00 C
+ATOM 1185 NE2 HIS B 74 -1.014 -17.509 3.075 1.00 0.00 N
+ATOM 1186 H HIS B 74 -4.532 -15.291 4.417 1.00 0.00 H
+ATOM 1187 HA HIS B 74 -5.453 -17.928 3.478 1.00 0.00 H
+ATOM 1188 1HB HIS B 74 -3.888 -16.995 5.904 1.00 0.00 H
+ATOM 1189 2HB HIS B 74 -4.286 -18.674 5.569 1.00 0.00 H
+ATOM 1190 HD2 HIS B 74 -1.835 -15.896 4.308 1.00 0.00 H
+ATOM 1191 HE1 HIS B 74 -0.933 -19.467 2.198 1.00 0.00 H
+ATOM 1192 HE2 HIS B 74 -0.164 -17.064 2.688 1.00 0.00 H
+ATOM 1193 N ASN B 75 -7.131 -18.687 5.135 1.00 0.00 N
+ATOM 1194 CA ASN B 75 -8.275 -19.126 5.944 1.00 0.00 C
+ATOM 1195 C ASN B 75 -9.463 -18.192 5.771 1.00 0.00 C
+ATOM 1196 O ASN B 75 -9.994 -17.636 6.734 1.00 0.00 O
+ATOM 1197 CB ASN B 75 -7.869 -19.267 7.403 1.00 0.00 C
+ATOM 1198 CG ASN B 75 -8.859 -20.050 8.165 1.00 0.00 C
+ATOM 1199 OD1 ASN B 75 -9.589 -20.854 7.544 1.00 0.00 O
+ATOM 1200 ND2 ASN B 75 -8.858 -19.871 9.468 1.00 0.00 N
+ATOM 1201 H ASN B 75 -6.736 -19.352 4.479 1.00 0.00 H
+ATOM 1202 HA ASN B 75 -8.582 -20.110 5.586 1.00 0.00 H
+ATOM 1203 1HB ASN B 75 -6.904 -19.733 7.480 1.00 0.00 H
+ATOM 1204 2HB ASN B 75 -7.785 -18.282 7.857 1.00 0.00 H
+ATOM 1205 1HD2 ASN B 75 -9.450 -20.423 10.093 1.00 0.00 H
+ATOM 1206 2HD2 ASN B 75 -8.213 -19.186 9.864 1.00 0.00 H
+ATOM 1207 N LEU B 76 -9.882 -18.027 4.528 1.00 0.00 N
+ATOM 1208 CA LEU B 76 -10.920 -17.061 4.184 1.00 0.00 C
+ATOM 1209 C LEU B 76 -12.225 -17.232 4.915 1.00 0.00 C
+ATOM 1210 O LEU B 76 -12.770 -18.332 5.048 1.00 0.00 O
+ATOM 1211 CB LEU B 76 -11.242 -17.179 2.702 1.00 0.00 C
+ATOM 1212 CG LEU B 76 -10.162 -16.741 1.746 1.00 0.00 C
+ATOM 1213 CD1 LEU B 76 -10.593 -17.115 0.357 1.00 0.00 C
+ATOM 1214 CD2 LEU B 76 -9.935 -15.276 1.898 1.00 0.00 C
+ATOM 1215 H LEU B 76 -9.421 -18.555 3.784 1.00 0.00 H
+ATOM 1216 HA LEU B 76 -10.537 -16.064 4.399 1.00 0.00 H
+ATOM 1217 1HB LEU B 76 -11.469 -18.223 2.480 1.00 0.00 H
+ATOM 1218 2HB LEU B 76 -12.132 -16.583 2.500 1.00 0.00 H
+ATOM 1219 HG LEU B 76 -9.237 -17.250 1.964 1.00 0.00 H
+ATOM 1220 1HD1 LEU B 76 -9.836 -16.838 -0.339 1.00 0.00 H
+ATOM 1221 2HD1 LEU B 76 -10.736 -18.195 0.305 1.00 0.00 H
+ATOM 1222 3HD1 LEU B 76 -11.522 -16.620 0.109 1.00 0.00 H
+ATOM 1223 1HD2 LEU B 76 -9.154 -14.951 1.210 1.00 0.00 H
+ATOM 1224 2HD2 LEU B 76 -10.855 -14.746 1.690 1.00 0.00 H
+ATOM 1225 3HD2 LEU B 76 -9.626 -15.069 2.922 1.00 0.00 H
+ATOM 1226 N GLN B 77 -12.735 -16.108 5.365 1.00 0.00 N
+ATOM 1227 CA GLN B 77 -14.002 -16.025 6.048 1.00 0.00 C
+ATOM 1228 C GLN B 77 -15.010 -15.344 5.151 1.00 0.00 C
+ATOM 1229 O GLN B 77 -14.647 -14.585 4.255 1.00 0.00 O
+ATOM 1230 CB GLN B 77 -13.846 -15.277 7.358 1.00 0.00 C
+ATOM 1231 CG GLN B 77 -12.824 -15.917 8.282 1.00 0.00 C
+ATOM 1232 CD GLN B 77 -13.220 -17.354 8.705 1.00 0.00 C
+ATOM 1233 OE1 GLN B 77 -14.321 -17.575 9.254 1.00 0.00 O
+ATOM 1234 NE2 GLN B 77 -12.322 -18.321 8.448 1.00 0.00 N
+ATOM 1235 H GLN B 77 -12.197 -15.246 5.231 1.00 0.00 H
+ATOM 1236 HA GLN B 77 -14.344 -17.029 6.258 1.00 0.00 H
+ATOM 1237 1HB GLN B 77 -13.513 -14.255 7.150 1.00 0.00 H
+ATOM 1238 2HB GLN B 77 -14.802 -15.220 7.873 1.00 0.00 H
+ATOM 1239 1HG GLN B 77 -11.863 -15.967 7.758 1.00 0.00 H
+ATOM 1240 2HG GLN B 77 -12.726 -15.306 9.178 1.00 0.00 H
+ATOM 1241 1HE2 GLN B 77 -12.515 -19.269 8.692 1.00 0.00 H
+ATOM 1242 2HE2 GLN B 77 -11.434 -18.082 7.987 1.00 0.00 H
+ATOM 1243 N ILE B 78 -16.286 -15.505 5.427 1.00 0.00 N
+ATOM 1244 CA ILE B 78 -17.278 -14.843 4.585 1.00 0.00 C
+ATOM 1245 C ILE B 78 -17.069 -13.327 4.613 1.00 0.00 C
+ATOM 1246 O ILE B 78 -17.215 -12.645 3.599 1.00 0.00 O
+ATOM 1247 CB ILE B 78 -18.696 -15.219 5.011 1.00 0.00 C
+ATOM 1248 CG1 ILE B 78 -18.905 -16.757 4.805 1.00 0.00 C
+ATOM 1249 CG2 ILE B 78 -19.690 -14.413 4.211 1.00 0.00 C
+ATOM 1250 CD1 ILE B 78 -18.700 -17.245 3.367 1.00 0.00 C
+ATOM 1251 H ILE B 78 -16.576 -16.113 6.182 1.00 0.00 H
+ATOM 1252 HA ILE B 78 -17.139 -15.181 3.559 1.00 0.00 H
+ATOM 1253 HB ILE B 78 -18.830 -15.005 6.072 1.00 0.00 H
+ATOM 1254 1HG1 ILE B 78 -18.212 -17.289 5.450 1.00 0.00 H
+ATOM 1255 2HG1 ILE B 78 -19.921 -17.015 5.107 1.00 0.00 H
+ATOM 1256 1HG2 ILE B 78 -20.699 -14.676 4.516 1.00 0.00 H
+ATOM 1257 2HG2 ILE B 78 -19.517 -13.353 4.390 1.00 0.00 H
+ATOM 1258 3HG2 ILE B 78 -19.558 -14.632 3.149 1.00 0.00 H
+ATOM 1259 1HD1 ILE B 78 -18.862 -18.324 3.316 1.00 0.00 H
+ATOM 1260 2HD1 ILE B 78 -19.401 -16.742 2.700 1.00 0.00 H
+ATOM 1261 3HD1 ILE B 78 -17.685 -17.021 3.049 1.00 0.00 H
+ATOM 1262 N LYS B 79 -16.702 -12.813 5.783 1.00 0.00 N
+ATOM 1263 CA LYS B 79 -16.423 -11.398 6.031 1.00 0.00 C
+ATOM 1264 C LYS B 79 -15.258 -10.823 5.194 1.00 0.00 C
+ATOM 1265 O LYS B 79 -15.065 -9.599 5.157 1.00 0.00 O
+ATOM 1266 CB LYS B 79 -16.113 -11.156 7.510 1.00 0.00 C
+ATOM 1267 CG LYS B 79 -17.289 -11.310 8.461 1.00 0.00 C
+ATOM 1268 CD LYS B 79 -16.865 -11.026 9.902 1.00 0.00 C
+ATOM 1269 CE LYS B 79 -18.039 -11.137 10.864 1.00 0.00 C
+ATOM 1270 NZ LYS B 79 -17.626 -10.880 12.274 1.00 0.00 N
+ATOM 1271 H LYS B 79 -16.607 -13.455 6.554 1.00 0.00 H
+ATOM 1272 HA LYS B 79 -17.320 -10.835 5.772 1.00 0.00 H
+ATOM 1273 1HB LYS B 79 -15.337 -11.856 7.828 1.00 0.00 H
+ATOM 1274 2HB LYS B 79 -15.712 -10.152 7.636 1.00 0.00 H
+ATOM 1275 1HG LYS B 79 -18.078 -10.613 8.178 1.00 0.00 H
+ATOM 1276 2HG LYS B 79 -17.681 -12.325 8.396 1.00 0.00 H
+ATOM 1277 1HD LYS B 79 -16.093 -11.737 10.201 1.00 0.00 H
+ATOM 1278 2HD LYS B 79 -16.451 -10.020 9.969 1.00 0.00 H
+ATOM 1279 1HE LYS B 79 -18.804 -10.413 10.582 1.00 0.00 H
+ATOM 1280 2HE LYS B 79 -18.462 -12.140 10.797 1.00 0.00 H
+ATOM 1281 1HZ LYS B 79 -18.430 -10.964 12.881 1.00 0.00 H
+ATOM 1282 2HZ LYS B 79 -16.926 -11.556 12.548 1.00 0.00 H
+ATOM 1283 3HZ LYS B 79 -17.244 -9.949 12.349 1.00 0.00 H
+ATOM 1284 N ASP B 80 -14.451 -11.684 4.571 1.00 0.00 N
+ATOM 1285 CA ASP B 80 -13.320 -11.242 3.772 1.00 0.00 C
+ATOM 1286 C ASP B 80 -13.732 -10.834 2.364 1.00 0.00 C
+ATOM 1287 O ASP B 80 -12.917 -10.323 1.595 1.00 0.00 O
+ATOM 1288 CB ASP B 80 -12.259 -12.336 3.677 1.00 0.00 C
+ATOM 1289 CG ASP B 80 -11.514 -12.600 4.974 1.00 0.00 C
+ATOM 1290 OD1 ASP B 80 -11.206 -11.679 5.683 1.00 0.00 O
+ATOM 1291 OD2 ASP B 80 -11.235 -13.751 5.253 1.00 0.00 O
+ATOM 1292 H ASP B 80 -14.640 -12.683 4.608 1.00 0.00 H
+ATOM 1293 HA ASP B 80 -12.879 -10.371 4.254 1.00 0.00 H
+ATOM 1294 1HB ASP B 80 -12.735 -13.261 3.362 1.00 0.00 H
+ATOM 1295 2HB ASP B 80 -11.537 -12.074 2.907 1.00 0.00 H
+ATOM 1296 N LYS B 81 -14.988 -11.055 2.000 1.00 0.00 N
+ATOM 1297 CA LYS B 81 -15.398 -10.647 0.668 1.00 0.00 C
+ATOM 1298 C LYS B 81 -15.326 -9.119 0.567 1.00 0.00 C
+ATOM 1299 O LYS B 81 -15.523 -8.404 1.559 1.00 0.00 O
+ATOM 1300 CB LYS B 81 -16.773 -11.236 0.354 1.00 0.00 C
+ATOM 1301 CG LYS B 81 -17.892 -10.739 1.178 1.00 0.00 C
+ATOM 1302 CD LYS B 81 -19.126 -11.606 0.884 1.00 0.00 C
+ATOM 1303 CE LYS B 81 -20.290 -11.245 1.735 1.00 0.00 C
+ATOM 1304 NZ LYS B 81 -21.483 -12.070 1.406 1.00 0.00 N
+ATOM 1305 H LYS B 81 -15.662 -11.498 2.629 1.00 0.00 H
+ATOM 1306 HA LYS B 81 -14.692 -11.058 -0.053 1.00 0.00 H
+ATOM 1307 1HB LYS B 81 -17.018 -11.034 -0.688 1.00 0.00 H
+ATOM 1308 2HB LYS B 81 -16.732 -12.320 0.477 1.00 0.00 H
+ATOM 1309 1HG LYS B 81 -17.640 -10.761 2.224 1.00 0.00 H
+ATOM 1310 2HG LYS B 81 -18.104 -9.706 0.902 1.00 0.00 H
+ATOM 1311 1HD LYS B 81 -19.405 -11.502 -0.165 1.00 0.00 H
+ATOM 1312 2HD LYS B 81 -18.874 -12.651 1.077 1.00 0.00 H
+ATOM 1313 1HE LYS B 81 -20.026 -11.394 2.781 1.00 0.00 H
+ATOM 1314 2HE LYS B 81 -20.520 -10.220 1.581 1.00 0.00 H
+ATOM 1315 1HZ LYS B 81 -22.257 -11.801 1.997 1.00 0.00 H
+ATOM 1316 2HZ LYS B 81 -21.727 -11.914 0.440 1.00 0.00 H
+ATOM 1317 3HZ LYS B 81 -21.268 -13.045 1.551 1.00 0.00 H
+ATOM 1318 N GLY B 82 -15.025 -8.623 -0.619 1.00 0.00 N
+ATOM 1319 CA GLY B 82 -14.942 -7.181 -0.827 1.00 0.00 C
+ATOM 1320 C GLY B 82 -13.826 -6.794 -1.786 1.00 0.00 C
+ATOM 1321 O GLY B 82 -13.223 -7.651 -2.441 1.00 0.00 O
+ATOM 1322 H GLY B 82 -14.885 -9.272 -1.390 1.00 0.00 H
+ATOM 1323 1HA GLY B 82 -15.897 -6.814 -1.206 1.00 0.00 H
+ATOM 1324 2HA GLY B 82 -14.775 -6.690 0.130 1.00 0.00 H
+ATOM 1325 N LEU B 83 -13.577 -5.493 -1.902 1.00 0.00 N
+ATOM 1326 CA LEU B 83 -12.523 -5.020 -2.807 1.00 0.00 C
+ATOM 1327 C LEU B 83 -11.178 -4.965 -2.144 1.00 0.00 C
+ATOM 1328 O LEU B 83 -11.046 -4.544 -0.994 1.00 0.00 O
+ATOM 1329 CB LEU B 83 -12.811 -3.632 -3.382 1.00 0.00 C
+ATOM 1330 CG LEU B 83 -13.933 -3.558 -4.272 1.00 0.00 C
+ATOM 1331 CD1 LEU B 83 -14.250 -2.090 -4.577 1.00 0.00 C
+ATOM 1332 CD2 LEU B 83 -13.606 -4.288 -5.577 1.00 0.00 C
+ATOM 1333 H LEU B 83 -14.112 -4.845 -1.320 1.00 0.00 H
+ATOM 1334 HA LEU B 83 -12.454 -5.715 -3.639 1.00 0.00 H
+ATOM 1335 1HB LEU B 83 -12.996 -2.946 -2.581 1.00 0.00 H
+ATOM 1336 2HB LEU B 83 -11.932 -3.292 -3.926 1.00 0.00 H
+ATOM 1337 HG LEU B 83 -14.735 -4.036 -3.819 1.00 0.00 H
+ATOM 1338 1HD1 LEU B 83 -15.104 -2.034 -5.253 1.00 0.00 H
+ATOM 1339 2HD1 LEU B 83 -14.493 -1.571 -3.649 1.00 0.00 H
+ATOM 1340 3HD1 LEU B 83 -13.389 -1.619 -5.047 1.00 0.00 H
+ATOM 1341 1HD2 LEU B 83 -14.469 -4.243 -6.242 1.00 0.00 H
+ATOM 1342 2HD2 LEU B 83 -12.747 -3.818 -6.056 1.00 0.00 H
+ATOM 1343 3HD2 LEU B 83 -13.372 -5.313 -5.358 1.00 0.00 H
+ATOM 1344 N TYR B 84 -10.173 -5.348 -2.924 1.00 0.00 N
+ATOM 1345 CA TYR B 84 -8.783 -5.278 -2.523 1.00 0.00 C
+ATOM 1346 C TYR B 84 -7.959 -4.573 -3.588 1.00 0.00 C
+ATOM 1347 O TYR B 84 -8.231 -4.658 -4.795 1.00 0.00 O
+ATOM 1348 CB TYR B 84 -8.219 -6.668 -2.217 1.00 0.00 C
+ATOM 1349 CG TYR B 84 -8.907 -7.305 -1.052 1.00 0.00 C
+ATOM 1350 CD1 TYR B 84 -8.403 -7.080 0.234 1.00 0.00 C
+ATOM 1351 CD2 TYR B 84 -10.035 -8.095 -1.225 1.00 0.00 C
+ATOM 1352 CE1 TYR B 84 -9.025 -7.628 1.330 1.00 0.00 C
+ATOM 1353 CE2 TYR B 84 -10.659 -8.633 -0.091 1.00 0.00 C
+ATOM 1354 CZ TYR B 84 -10.147 -8.396 1.172 1.00 0.00 C
+ATOM 1355 OH TYR B 84 -10.758 -8.923 2.291 1.00 0.00 O
+ATOM 1356 H TYR B 84 -10.401 -5.715 -3.845 1.00 0.00 H
+ATOM 1357 HA TYR B 84 -8.718 -4.683 -1.620 1.00 0.00 H
+ATOM 1358 1HB TYR B 84 -8.333 -7.312 -3.090 1.00 0.00 H
+ATOM 1359 2HB TYR B 84 -7.158 -6.588 -1.991 1.00 0.00 H
+ATOM 1360 HD1 TYR B 84 -7.519 -6.459 0.367 1.00 0.00 H
+ATOM 1361 HD2 TYR B 84 -10.443 -8.279 -2.220 1.00 0.00 H
+ATOM 1362 HE1 TYR B 84 -8.643 -7.446 2.324 1.00 0.00 H
+ATOM 1363 HE2 TYR B 84 -11.549 -9.235 -0.194 1.00 0.00 H
+ATOM 1364 HH TYR B 84 -11.534 -9.439 2.018 1.00 0.00 H
+ATOM 1365 N GLN B 85 -6.940 -3.869 -3.147 1.00 0.00 N
+ATOM 1366 CA GLN B 85 -6.119 -3.145 -4.085 1.00 0.00 C
+ATOM 1367 C GLN B 85 -4.632 -3.381 -3.849 1.00 0.00 C
+ATOM 1368 O GLN B 85 -4.156 -3.219 -2.726 1.00 0.00 O
+ATOM 1369 CB GLN B 85 -6.423 -1.649 -3.899 1.00 0.00 C
+ATOM 1370 CG GLN B 85 -7.917 -1.278 -4.021 1.00 0.00 C
+ATOM 1371 CD GLN B 85 -8.205 0.216 -3.835 1.00 0.00 C
+ATOM 1372 OE1 GLN B 85 -7.499 0.959 -3.130 1.00 0.00 O
+ATOM 1373 NE2 GLN B 85 -9.288 0.673 -4.478 1.00 0.00 N
+ATOM 1374 H GLN B 85 -6.779 -3.794 -2.145 1.00 0.00 H
+ATOM 1375 HA GLN B 85 -6.360 -3.460 -5.101 1.00 0.00 H
+ATOM 1376 1HB GLN B 85 -6.078 -1.328 -2.923 1.00 0.00 H
+ATOM 1377 2HB GLN B 85 -5.871 -1.074 -4.650 1.00 0.00 H
+ATOM 1378 1HG GLN B 85 -8.284 -1.588 -4.992 1.00 0.00 H
+ATOM 1379 2HG GLN B 85 -8.469 -1.796 -3.237 1.00 0.00 H
+ATOM 1380 1HE2 GLN B 85 -9.548 1.637 -4.399 1.00 0.00 H
+ATOM 1381 2HE2 GLN B 85 -9.847 0.049 -5.027 1.00 0.00 H
+ATOM 1382 N CYS B 86 -3.910 -3.729 -4.916 1.00 0.00 N
+ATOM 1383 CA CYS B 86 -2.444 -3.789 -4.906 1.00 0.00 C
+ATOM 1384 C CYS B 86 -1.925 -2.480 -5.391 1.00 0.00 C
+ATOM 1385 O CYS B 86 -2.259 -2.078 -6.508 1.00 0.00 O
+ATOM 1386 CB CYS B 86 -1.846 -4.851 -5.822 1.00 0.00 C
+ATOM 1387 SG CYS B 86 -0.024 -4.638 -5.947 1.00 0.00 S
+ATOM 1388 H CYS B 86 -4.391 -3.912 -5.796 1.00 0.00 H
+ATOM 1389 HA CYS B 86 -2.090 -3.965 -3.891 1.00 0.00 H
+ATOM 1390 1HB CYS B 86 -2.060 -5.850 -5.442 1.00 0.00 H
+ATOM 1391 2HB CYS B 86 -2.278 -4.763 -6.798 1.00 0.00 H
+ATOM 1392 N ILE B 87 -1.149 -1.786 -4.572 1.00 0.00 N
+ATOM 1393 CA ILE B 87 -0.664 -0.477 -4.970 1.00 0.00 C
+ATOM 1394 C ILE B 87 0.868 -0.428 -4.891 1.00 0.00 C
+ATOM 1395 O ILE B 87 1.452 -0.676 -3.834 1.00 0.00 O
+ATOM 1396 CB ILE B 87 -1.268 0.602 -4.083 1.00 0.00 C
+ATOM 1397 CG1 ILE B 87 -2.826 0.523 -4.116 1.00 0.00 C
+ATOM 1398 CG2 ILE B 87 -0.846 1.937 -4.665 1.00 0.00 C
+ATOM 1399 CD1 ILE B 87 -3.517 1.406 -3.166 1.00 0.00 C
+ATOM 1400 H ILE B 87 -0.912 -2.170 -3.659 1.00 0.00 H
+ATOM 1401 HA ILE B 87 -0.951 -0.280 -6.002 1.00 0.00 H
+ATOM 1402 HB ILE B 87 -0.917 0.489 -3.053 1.00 0.00 H
+ATOM 1403 1HG1 ILE B 87 -3.154 0.771 -5.107 1.00 0.00 H
+ATOM 1404 2HG1 ILE B 87 -3.143 -0.492 -3.882 1.00 0.00 H
+ATOM 1405 1HG2 ILE B 87 -1.256 2.755 -4.078 1.00 0.00 H
+ATOM 1406 2HG2 ILE B 87 0.228 2.006 -4.690 1.00 0.00 H
+ATOM 1407 3HG2 ILE B 87 -1.224 1.995 -5.686 1.00 0.00 H
+ATOM 1408 1HD1 ILE B 87 -4.595 1.275 -3.273 1.00 0.00 H
+ATOM 1409 2HD1 ILE B 87 -3.211 1.152 -2.149 1.00 0.00 H
+ATOM 1410 3HD1 ILE B 87 -3.257 2.440 -3.376 1.00 0.00 H
+ATOM 1411 N ILE B 88 1.508 -0.105 -6.007 1.00 0.00 N
+ATOM 1412 CA ILE B 88 2.961 -0.099 -6.076 1.00 0.00 C
+ATOM 1413 C ILE B 88 3.489 1.313 -6.240 1.00 0.00 C
+ATOM 1414 O ILE B 88 3.037 2.054 -7.125 1.00 0.00 O
+ATOM 1415 CB ILE B 88 3.487 -0.957 -7.248 1.00 0.00 C
+ATOM 1416 CG1 ILE B 88 3.003 -2.410 -7.102 1.00 0.00 C
+ATOM 1417 CG2 ILE B 88 5.014 -0.920 -7.304 1.00 0.00 C
+ATOM 1418 CD1 ILE B 88 3.384 -3.305 -8.272 1.00 0.00 C
+ATOM 1419 H ILE B 88 0.972 0.113 -6.841 1.00 0.00 H
+ATOM 1420 HA ILE B 88 3.364 -0.502 -5.147 1.00 0.00 H
+ATOM 1421 HB ILE B 88 3.108 -0.564 -8.150 1.00 0.00 H
+ATOM 1422 1HG1 ILE B 88 3.395 -2.828 -6.213 1.00 0.00 H
+ATOM 1423 2HG1 ILE B 88 1.916 -2.404 -7.024 1.00 0.00 H
+ATOM 1424 1HG2 ILE B 88 5.355 -1.514 -8.149 1.00 0.00 H
+ATOM 1425 2HG2 ILE B 88 5.347 0.102 -7.427 1.00 0.00 H
+ATOM 1426 3HG2 ILE B 88 5.429 -1.314 -6.391 1.00 0.00 H
+ATOM 1427 1HD1 ILE B 88 2.999 -4.308 -8.098 1.00 0.00 H
+ATOM 1428 2HD1 ILE B 88 2.972 -2.919 -9.189 1.00 0.00 H
+ATOM 1429 3HD1 ILE B 88 4.464 -3.344 -8.353 1.00 0.00 H
+ATOM 1430 N HIS B 89 4.455 1.684 -5.388 1.00 0.00 N
+ATOM 1431 CA HIS B 89 5.046 3.000 -5.438 1.00 0.00 C
+ATOM 1432 C HIS B 89 6.535 2.917 -5.639 1.00 0.00 C
+ATOM 1433 O HIS B 89 7.182 1.997 -5.143 1.00 0.00 O
+ATOM 1434 CB HIS B 89 4.837 3.730 -4.118 1.00 0.00 C
+ATOM 1435 CG HIS B 89 3.455 3.860 -3.694 1.00 0.00 C
+ATOM 1436 ND1 HIS B 89 2.786 2.840 -3.080 1.00 0.00 N
+ATOM 1437 CD2 HIS B 89 2.600 4.896 -3.749 1.00 0.00 C
+ATOM 1438 CE1 HIS B 89 1.572 3.238 -2.775 1.00 0.00 C
+ATOM 1439 NE2 HIS B 89 1.429 4.487 -3.178 1.00 0.00 N
+ATOM 1440 H HIS B 89 4.747 1.041 -4.655 1.00 0.00 H
+ATOM 1441 HA HIS B 89 4.629 3.561 -6.254 1.00 0.00 H
+ATOM 1442 1HB HIS B 89 5.389 3.231 -3.337 1.00 0.00 H
+ATOM 1443 2HB HIS B 89 5.238 4.731 -4.215 1.00 0.00 H
+ATOM 1444 HD1 HIS B 89 3.165 1.927 -2.862 1.00 0.00 H
+ATOM 1445 HD2 HIS B 89 2.698 5.908 -4.136 1.00 0.00 H
+ATOM 1446 HE1 HIS B 89 0.875 2.564 -2.279 1.00 0.00 H
+ATOM 1447 N HIS B 90 7.092 3.906 -6.315 1.00 0.00 N
+ATOM 1448 CA HIS B 90 8.522 4.028 -6.489 1.00 0.00 C
+ATOM 1449 C HIS B 90 9.066 4.972 -5.457 1.00 0.00 C
+ATOM 1450 O HIS B 90 8.632 6.131 -5.378 1.00 0.00 O
+ATOM 1451 CB HIS B 90 8.879 4.578 -7.861 1.00 0.00 C
+ATOM 1452 CG HIS B 90 10.343 4.703 -8.054 1.00 0.00 C
+ATOM 1453 ND1 HIS B 90 10.910 5.747 -8.746 1.00 0.00 N
+ATOM 1454 CD2 HIS B 90 11.362 3.895 -7.675 1.00 0.00 C
+ATOM 1455 CE1 HIS B 90 12.236 5.605 -8.739 1.00 0.00 C
+ATOM 1456 NE2 HIS B 90 12.534 4.481 -8.102 1.00 0.00 N
+ATOM 1457 H HIS B 90 6.490 4.611 -6.713 1.00 0.00 H
+ATOM 1458 HA HIS B 90 9.007 3.073 -6.350 1.00 0.00 H
+ATOM 1459 1HB HIS B 90 8.496 3.913 -8.633 1.00 0.00 H
+ATOM 1460 2HB HIS B 90 8.413 5.560 -8.002 1.00 0.00 H
+ATOM 1461 HD2 HIS B 90 11.266 2.972 -7.134 1.00 0.00 H
+ATOM 1462 HE1 HIS B 90 12.950 6.287 -9.199 1.00 0.00 H
+ATOM 1463 HE2 HIS B 90 13.464 4.107 -7.951 1.00 0.00 H
+ATOM 1464 N LYS B 91 9.942 4.471 -4.590 1.00 0.00 N
+ATOM 1465 CA LYS B 91 10.485 5.298 -3.524 1.00 0.00 C
+ATOM 1466 C LYS B 91 11.316 6.449 -4.111 1.00 0.00 C
+ATOM 1467 O LYS B 91 12.154 6.223 -4.988 1.00 0.00 O
+ATOM 1468 CB LYS B 91 11.315 4.457 -2.570 1.00 0.00 C
+ATOM 1469 CG LYS B 91 11.851 5.238 -1.356 1.00 0.00 C
+ATOM 1470 CD LYS B 91 10.735 5.550 -0.386 1.00 0.00 C
+ATOM 1471 CE LYS B 91 11.199 6.444 0.734 1.00 0.00 C
+ATOM 1472 NZ LYS B 91 11.310 7.873 0.285 1.00 0.00 N
+ATOM 1473 H LYS B 91 10.278 3.512 -4.697 1.00 0.00 H
+ATOM 1474 HA LYS B 91 9.656 5.732 -2.964 1.00 0.00 H
+ATOM 1475 1HB LYS B 91 10.702 3.642 -2.210 1.00 0.00 H
+ATOM 1476 2HB LYS B 91 12.160 4.020 -3.105 1.00 0.00 H
+ATOM 1477 1HG LYS B 91 12.606 4.643 -0.848 1.00 0.00 H
+ATOM 1478 2HG LYS B 91 12.307 6.175 -1.676 1.00 0.00 H
+ATOM 1479 1HD LYS B 91 9.929 6.063 -0.915 1.00 0.00 H
+ATOM 1480 2HD LYS B 91 10.342 4.619 0.032 1.00 0.00 H
+ATOM 1481 1HE LYS B 91 10.488 6.388 1.555 1.00 0.00 H
+ATOM 1482 2HE LYS B 91 12.175 6.108 1.086 1.00 0.00 H
+ATOM 1483 1HZ LYS B 91 11.614 8.453 1.044 1.00 0.00 H
+ATOM 1484 2HZ LYS B 91 11.969 7.989 -0.493 1.00 0.00 H
+ATOM 1485 3HZ LYS B 91 10.390 8.195 -0.050 1.00 0.00 H
+ATOM 1486 N LYS B 92 11.082 7.677 -3.636 1.00 0.00 N
+ATOM 1487 CA LYS B 92 11.810 8.858 -4.115 1.00 0.00 C
+ATOM 1488 C LYS B 92 12.725 9.357 -2.997 1.00 0.00 C
+ATOM 1489 O LYS B 92 12.620 8.848 -1.882 1.00 0.00 O
+ATOM 1490 CB LYS B 92 10.805 9.935 -4.521 1.00 0.00 C
+ATOM 1491 CG LYS B 92 9.835 9.478 -5.538 1.00 0.00 C
+ATOM 1492 CD LYS B 92 10.540 8.958 -6.739 1.00 0.00 C
+ATOM 1493 CE LYS B 92 11.190 10.038 -7.523 1.00 0.00 C
+ATOM 1494 NZ LYS B 92 11.793 9.494 -8.722 1.00 0.00 N
+ATOM 1495 H LYS B 92 10.364 7.819 -2.925 1.00 0.00 H
+ATOM 1496 HA LYS B 92 12.413 8.577 -4.972 1.00 0.00 H
+ATOM 1497 1HB LYS B 92 10.232 10.230 -3.653 1.00 0.00 H
+ATOM 1498 2HB LYS B 92 11.274 10.803 -4.911 1.00 0.00 H
+ATOM 1499 1HG LYS B 92 9.210 8.707 -5.147 1.00 0.00 H
+ATOM 1500 2HG LYS B 92 9.220 10.322 -5.813 1.00 0.00 H
+ATOM 1501 1HD LYS B 92 11.309 8.258 -6.431 1.00 0.00 H
+ATOM 1502 2HD LYS B 92 9.846 8.409 -7.371 1.00 0.00 H
+ATOM 1503 1HE LYS B 92 10.462 10.789 -7.806 1.00 0.00 H
+ATOM 1504 2HE LYS B 92 11.967 10.509 -6.924 1.00 0.00 H
+ATOM 1505 1HZ LYS B 92 12.227 10.241 -9.240 1.00 0.00 H
+ATOM 1506 2HZ LYS B 92 12.484 8.811 -8.491 1.00 0.00 H
+ATOM 1507 3HZ LYS B 92 11.075 9.064 -9.308 1.00 0.00 H
+ATOM 1508 N PRO B 93 13.693 10.280 -3.233 1.00 0.00 N
+ATOM 1509 CA PRO B 93 14.510 10.874 -2.172 1.00 0.00 C
+ATOM 1510 C PRO B 93 13.620 11.315 -1.001 1.00 0.00 C
+ATOM 1511 O PRO B 93 13.945 11.061 0.161 1.00 0.00 O
+ATOM 1512 CB PRO B 93 15.189 12.031 -2.911 1.00 0.00 C
+ATOM 1513 CG PRO B 93 15.340 11.506 -4.327 1.00 0.00 C
+ATOM 1514 CD PRO B 93 14.045 10.750 -4.597 1.00 0.00 C
+ATOM 1515 HA PRO B 93 15.250 10.134 -1.830 1.00 0.00 H
+ATOM 1516 1HB PRO B 93 14.576 12.932 -2.846 1.00 0.00 H
+ATOM 1517 2HB PRO B 93 16.152 12.273 -2.440 1.00 0.00 H
+ATOM 1518 1HG PRO B 93 15.493 12.348 -5.028 1.00 0.00 H
+ATOM 1519 2HG PRO B 93 16.230 10.868 -4.403 1.00 0.00 H
+ATOM 1520 1HD PRO B 93 13.317 11.456 -4.980 1.00 0.00 H
+ATOM 1521 2HD PRO B 93 14.231 9.913 -5.277 1.00 0.00 H
+ATOM 1522 N THR B 94 12.462 11.916 -1.318 1.00 0.00 N
+ATOM 1523 CA THR B 94 11.450 12.247 -0.327 1.00 0.00 C
+ATOM 1524 C THR B 94 10.119 11.747 -0.864 1.00 0.00 C
+ATOM 1525 O THR B 94 9.795 11.990 -2.025 1.00 0.00 O
+ATOM 1526 CB THR B 94 11.384 13.755 0.012 1.00 0.00 C
+ATOM 1527 OG1 THR B 94 12.630 14.180 0.575 1.00 0.00 O
+ATOM 1528 CG2 THR B 94 10.286 14.021 1.005 1.00 0.00 C
+ATOM 1529 H THR B 94 12.263 12.120 -2.285 1.00 0.00 H
+ATOM 1530 HA THR B 94 11.666 11.711 0.598 1.00 0.00 H
+ATOM 1531 HB THR B 94 11.189 14.324 -0.897 1.00 0.00 H
+ATOM 1532 HG1 THR B 94 12.751 13.736 1.425 1.00 0.00 H
+ATOM 1533 1HG2 THR B 94 10.263 15.088 1.228 1.00 0.00 H
+ATOM 1534 2HG2 THR B 94 9.331 13.721 0.587 1.00 0.00 H
+ATOM 1535 3HG2 THR B 94 10.478 13.463 1.917 1.00 0.00 H
+ATOM 1536 N GLY B 95 9.354 11.062 -0.015 1.00 0.00 N
+ATOM 1537 CA GLY B 95 8.056 10.527 -0.403 1.00 0.00 C
+ATOM 1538 C GLY B 95 8.205 9.363 -1.370 1.00 0.00 C
+ATOM 1539 O GLY B 95 9.265 8.724 -1.435 1.00 0.00 O
+ATOM 1540 H GLY B 95 9.680 10.918 0.929 1.00 0.00 H
+ATOM 1541 1HA GLY B 95 7.513 10.202 0.485 1.00 0.00 H
+ATOM 1542 2HA GLY B 95 7.463 11.314 -0.868 1.00 0.00 H
+ATOM 1543 N MET B 96 7.115 9.057 -2.067 1.00 0.00 N
+ATOM 1544 CA MET B 96 7.059 7.962 -3.009 1.00 0.00 C
+ATOM 1545 C MET B 96 6.013 8.308 -4.059 1.00 0.00 C
+ATOM 1546 O MET B 96 5.087 9.087 -3.791 1.00 0.00 O
+ATOM 1547 CB MET B 96 6.784 6.635 -2.305 1.00 0.00 C
+ATOM 1548 CG MET B 96 5.440 6.521 -1.601 1.00 0.00 C
+ATOM 1549 SD MET B 96 5.295 4.943 -0.682 1.00 0.00 S
+ATOM 1550 CE MET B 96 6.407 5.317 0.698 1.00 0.00 C
+ATOM 1551 H MET B 96 6.289 9.628 -1.948 1.00 0.00 H
+ATOM 1552 HA MET B 96 8.020 7.873 -3.508 1.00 0.00 H
+ATOM 1553 1HB MET B 96 6.865 5.830 -3.030 1.00 0.00 H
+ATOM 1554 2HB MET B 96 7.547 6.458 -1.554 1.00 0.00 H
+ATOM 1555 1HG MET B 96 5.311 7.344 -0.905 1.00 0.00 H
+ATOM 1556 2HG MET B 96 4.642 6.569 -2.343 1.00 0.00 H
+ATOM 1557 1HE MET B 96 6.443 4.467 1.380 1.00 0.00 H
+ATOM 1558 2HE MET B 96 7.407 5.522 0.317 1.00 0.00 H
+ATOM 1559 3HE MET B 96 6.042 6.192 1.238 1.00 0.00 H
+ATOM 1560 N ILE B 97 6.169 7.744 -5.241 1.00 0.00 N
+ATOM 1561 CA ILE B 97 5.244 7.946 -6.365 1.00 0.00 C
+ATOM 1562 C ILE B 97 4.537 6.691 -6.790 1.00 0.00 C
+ATOM 1563 O ILE B 97 5.175 5.685 -7.062 1.00 0.00 O
+ATOM 1564 CB ILE B 97 6.016 8.522 -7.568 1.00 0.00 C
+ATOM 1565 CG1 ILE B 97 6.546 9.916 -7.197 1.00 0.00 C
+ATOM 1566 CG2 ILE B 97 5.167 8.514 -8.856 1.00 0.00 C
+ATOM 1567 CD1 ILE B 97 7.511 10.462 -8.169 1.00 0.00 C
+ATOM 1568 H ILE B 97 6.981 7.139 -5.371 1.00 0.00 H
+ATOM 1569 HA ILE B 97 4.488 8.668 -6.061 1.00 0.00 H
+ATOM 1570 HB ILE B 97 6.894 7.896 -7.740 1.00 0.00 H
+ATOM 1571 1HG1 ILE B 97 5.704 10.610 -7.124 1.00 0.00 H
+ATOM 1572 2HG1 ILE B 97 7.025 9.862 -6.235 1.00 0.00 H
+ATOM 1573 1HG2 ILE B 97 5.756 8.891 -9.677 1.00 0.00 H
+ATOM 1574 2HG2 ILE B 97 4.859 7.496 -9.086 1.00 0.00 H
+ATOM 1575 3HG2 ILE B 97 4.279 9.136 -8.726 1.00 0.00 H
+ATOM 1576 1HD1 ILE B 97 7.847 11.446 -7.848 1.00 0.00 H
+ATOM 1577 2HD1 ILE B 97 8.346 9.806 -8.263 1.00 0.00 H
+ATOM 1578 3HD1 ILE B 97 7.047 10.536 -9.081 1.00 0.00 H
+ATOM 1579 N ARG B 98 3.216 6.731 -6.895 1.00 0.00 N
+ATOM 1580 CA ARG B 98 2.486 5.540 -7.327 1.00 0.00 C
+ATOM 1581 C ARG B 98 2.785 5.252 -8.789 1.00 0.00 C
+ATOM 1582 O ARG B 98 2.677 6.151 -9.624 1.00 0.00 O
+ATOM 1583 CB ARG B 98 0.986 5.753 -7.100 1.00 0.00 C
+ATOM 1584 CG ARG B 98 0.090 4.560 -7.362 1.00 0.00 C
+ATOM 1585 CD ARG B 98 -1.244 4.855 -6.784 1.00 0.00 C
+ATOM 1586 NE ARG B 98 -1.942 5.945 -7.464 1.00 0.00 N
+ATOM 1587 CZ ARG B 98 -2.772 5.779 -8.496 1.00 0.00 C
+ATOM 1588 NH1 ARG B 98 -3.027 4.579 -8.917 1.00 0.00 N
+ATOM 1589 NH2 ARG B 98 -3.350 6.803 -9.079 1.00 0.00 N
+ATOM 1590 H ARG B 98 2.716 7.580 -6.669 1.00 0.00 H
+ATOM 1591 HA ARG B 98 2.800 4.689 -6.733 1.00 0.00 H
+ATOM 1592 1HB ARG B 98 0.815 6.067 -6.069 1.00 0.00 H
+ATOM 1593 2HB ARG B 98 0.639 6.560 -7.744 1.00 0.00 H
+ATOM 1594 1HG ARG B 98 -0.007 4.388 -8.436 1.00 0.00 H
+ATOM 1595 2HG ARG B 98 0.505 3.670 -6.882 1.00 0.00 H
+ATOM 1596 1HD ARG B 98 -1.868 3.962 -6.863 1.00 0.00 H
+ATOM 1597 2HD ARG B 98 -1.130 5.122 -5.733 1.00 0.00 H
+ATOM 1598 HE ARG B 98 -1.769 6.892 -7.150 1.00 0.00 H
+ATOM 1599 1HH1 ARG B 98 -2.576 3.796 -8.465 1.00 0.00 H
+ATOM 1600 2HH1 ARG B 98 -3.666 4.435 -9.686 1.00 0.00 H
+ATOM 1601 1HH2 ARG B 98 -3.151 7.769 -8.778 1.00 0.00 H
+ATOM 1602 2HH2 ARG B 98 -3.979 6.638 -9.847 1.00 0.00 H
+ATOM 1603 N ILE B 99 3.137 4.007 -9.104 1.00 0.00 N
+ATOM 1604 CA ILE B 99 3.440 3.645 -10.488 1.00 0.00 C
+ATOM 1605 C ILE B 99 2.535 2.544 -11.008 1.00 0.00 C
+ATOM 1606 O ILE B 99 2.505 2.276 -12.206 1.00 0.00 O
+ATOM 1607 CB ILE B 99 4.897 3.171 -10.655 1.00 0.00 C
+ATOM 1608 CG1 ILE B 99 5.154 1.891 -9.853 1.00 0.00 C
+ATOM 1609 CG2 ILE B 99 5.874 4.264 -10.215 1.00 0.00 C
+ATOM 1610 CD1 ILE B 99 6.492 1.246 -10.157 1.00 0.00 C
+ATOM 1611 H ILE B 99 3.217 3.316 -8.370 1.00 0.00 H
+ATOM 1612 HA ILE B 99 3.292 4.521 -11.115 1.00 0.00 H
+ATOM 1613 HB ILE B 99 5.072 2.947 -11.710 1.00 0.00 H
+ATOM 1614 1HG1 ILE B 99 5.104 2.126 -8.791 1.00 0.00 H
+ATOM 1615 2HG1 ILE B 99 4.385 1.160 -10.085 1.00 0.00 H
+ATOM 1616 1HG2 ILE B 99 6.898 3.921 -10.371 1.00 0.00 H
+ATOM 1617 2HG2 ILE B 99 5.695 5.159 -10.804 1.00 0.00 H
+ATOM 1618 3HG2 ILE B 99 5.724 4.488 -9.166 1.00 0.00 H
+ATOM 1619 1HD1 ILE B 99 6.607 0.342 -9.566 1.00 0.00 H
+ATOM 1620 2HD1 ILE B 99 6.539 0.994 -11.213 1.00 0.00 H
+ATOM 1621 3HD1 ILE B 99 7.297 1.942 -9.917 1.00 0.00 H
+ATOM 1622 N HIS B 100 1.806 1.891 -10.117 1.00 0.00 N
+ATOM 1623 CA HIS B 100 0.939 0.808 -10.559 1.00 0.00 C
+ATOM 1624 C HIS B 100 -0.142 0.464 -9.556 1.00 0.00 C
+ATOM 1625 O HIS B 100 0.065 0.505 -8.349 1.00 0.00 O
+ATOM 1626 CB HIS B 100 1.753 -0.443 -10.911 1.00 0.00 C
+ATOM 1627 CG HIS B 100 0.928 -1.589 -11.408 1.00 0.00 C
+ATOM 1628 ND1 HIS B 100 0.421 -1.658 -12.691 1.00 0.00 N
+ATOM 1629 CD2 HIS B 100 0.514 -2.715 -10.773 1.00 0.00 C
+ATOM 1630 CE1 HIS B 100 -0.276 -2.798 -12.812 1.00 0.00 C
+ATOM 1631 NE2 HIS B 100 -0.217 -3.437 -11.659 1.00 0.00 N
+ATOM 1632 H HIS B 100 1.905 2.111 -9.129 1.00 0.00 H
+ATOM 1633 HA HIS B 100 0.430 1.121 -11.469 1.00 0.00 H
+ATOM 1634 1HB HIS B 100 2.468 -0.196 -11.690 1.00 0.00 H
+ATOM 1635 2HB HIS B 100 2.304 -0.777 -10.082 1.00 0.00 H
+ATOM 1636 HD2 HIS B 100 0.720 -3.001 -9.743 1.00 0.00 H
+ATOM 1637 HE1 HIS B 100 -0.796 -3.149 -13.699 1.00 0.00 H
+ATOM 1638 HE2 HIS B 100 -0.635 -4.338 -11.430 1.00 0.00 H
+ATOM 1639 N GLN B 101 -1.315 0.130 -10.062 1.00 0.00 N
+ATOM 1640 CA GLN B 101 -2.384 -0.326 -9.208 1.00 0.00 C
+ATOM 1641 C GLN B 101 -3.214 -1.406 -9.893 1.00 0.00 C
+ATOM 1642 O GLN B 101 -3.506 -1.312 -11.090 1.00 0.00 O
+ATOM 1643 CB GLN B 101 -3.271 0.840 -8.734 1.00 0.00 C
+ATOM 1644 CG GLN B 101 -4.372 0.416 -7.762 1.00 0.00 C
+ATOM 1645 CD GLN B 101 -5.030 1.582 -7.069 1.00 0.00 C
+ATOM 1646 OE1 GLN B 101 -4.413 2.634 -6.882 1.00 0.00 O
+ATOM 1647 NE2 GLN B 101 -6.272 1.401 -6.664 1.00 0.00 N
+ATOM 1648 H GLN B 101 -1.447 0.140 -11.066 1.00 0.00 H
+ATOM 1649 HA GLN B 101 -1.931 -0.764 -8.335 1.00 0.00 H
+ATOM 1650 1HB GLN B 101 -2.655 1.581 -8.229 1.00 0.00 H
+ATOM 1651 2HB GLN B 101 -3.743 1.315 -9.586 1.00 0.00 H
+ATOM 1652 1HG GLN B 101 -5.142 -0.114 -8.324 1.00 0.00 H
+ATOM 1653 2HG GLN B 101 -3.957 -0.248 -7.013 1.00 0.00 H
+ATOM 1654 1HE2 GLN B 101 -6.761 2.130 -6.183 1.00 0.00 H
+ATOM 1655 2HE2 GLN B 101 -6.727 0.519 -6.862 1.00 0.00 H
+ATOM 1656 N MET B 102 -3.592 -2.422 -9.105 1.00 0.00 N
+ATOM 1657 CA MET B 102 -4.457 -3.512 -9.556 1.00 0.00 C
+ATOM 1658 C MET B 102 -5.566 -3.769 -8.559 1.00 0.00 C
+ATOM 1659 O MET B 102 -5.338 -4.101 -7.390 1.00 0.00 O
+ATOM 1660 CB MET B 102 -3.639 -4.774 -9.753 1.00 0.00 C
+ATOM 1661 CG MET B 102 -4.421 -6.024 -10.189 1.00 0.00 C
+ATOM 1662 SD MET B 102 -5.076 -5.913 -11.832 1.00 0.00 S
+ATOM 1663 CE MET B 102 -6.218 -7.317 -11.776 1.00 0.00 C
+ATOM 1664 H MET B 102 -3.252 -2.415 -8.142 1.00 0.00 H
+ATOM 1665 HA MET B 102 -4.917 -3.231 -10.503 1.00 0.00 H
+ATOM 1666 1HB MET B 102 -2.880 -4.583 -10.504 1.00 0.00 H
+ATOM 1667 2HB MET B 102 -3.129 -5.022 -8.841 1.00 0.00 H
+ATOM 1668 1HG MET B 102 -3.759 -6.891 -10.144 1.00 0.00 H
+ATOM 1669 2HG MET B 102 -5.248 -6.199 -9.497 1.00 0.00 H
+ATOM 1670 1HE MET B 102 -6.727 -7.409 -12.734 1.00 0.00 H
+ATOM 1671 2HE MET B 102 -5.663 -8.230 -11.569 1.00 0.00 H
+ATOM 1672 3HE MET B 102 -6.949 -7.154 -10.983 1.00 0.00 H
+ATOM 1673 N ASN B 103 -6.780 -3.633 -9.033 1.00 0.00 N
+ATOM 1674 CA ASN B 103 -7.934 -3.811 -8.182 1.00 0.00 C
+ATOM 1675 C ASN B 103 -8.567 -5.168 -8.432 1.00 0.00 C
+ATOM 1676 O ASN B 103 -8.680 -5.610 -9.588 1.00 0.00 O
+ATOM 1677 CB ASN B 103 -8.948 -2.723 -8.431 1.00 0.00 C
+ATOM 1678 CG ASN B 103 -8.400 -1.335 -8.149 1.00 0.00 C
+ATOM 1679 OD1 ASN B 103 -7.460 -1.143 -7.356 1.00 0.00 O
+ATOM 1680 ND2 ASN B 103 -8.975 -0.367 -8.803 1.00 0.00 N
+ATOM 1681 H ASN B 103 -6.906 -3.378 -10.002 1.00 0.00 H
+ATOM 1682 HA ASN B 103 -7.625 -3.780 -7.141 1.00 0.00 H
+ATOM 1683 1HB ASN B 103 -9.288 -2.771 -9.467 1.00 0.00 H
+ATOM 1684 2HB ASN B 103 -9.820 -2.892 -7.795 1.00 0.00 H
+ATOM 1685 1HD2 ASN B 103 -8.681 0.582 -8.691 1.00 0.00 H
+ATOM 1686 2HD2 ASN B 103 -9.724 -0.572 -9.434 1.00 0.00 H
+ATOM 1687 N SER B 104 -9.053 -5.770 -7.365 1.00 0.00 N
+ATOM 1688 CA SER B 104 -9.775 -7.016 -7.510 1.00 0.00 C
+ATOM 1689 C SER B 104 -10.901 -7.144 -6.495 1.00 0.00 C
+ATOM 1690 O SER B 104 -10.863 -6.595 -5.392 1.00 0.00 O
+ATOM 1691 CB SER B 104 -8.800 -8.172 -7.369 1.00 0.00 C
+ATOM 1692 OG SER B 104 -8.244 -8.214 -6.094 1.00 0.00 O
+ATOM 1693 H SER B 104 -8.894 -5.367 -6.441 1.00 0.00 H
+ATOM 1694 HA SER B 104 -10.214 -7.046 -8.505 1.00 0.00 H
+ATOM 1695 1HB SER B 104 -9.318 -9.105 -7.584 1.00 0.00 H
+ATOM 1696 2HB SER B 104 -8.001 -8.062 -8.102 1.00 0.00 H
+ATOM 1697 HG SER B 104 -8.042 -7.297 -5.865 1.00 0.00 H
+ATOM 1698 N GLU B 105 -11.905 -7.890 -6.881 1.00 0.00 N
+ATOM 1699 CA GLU B 105 -13.052 -8.182 -6.042 1.00 0.00 C
+ATOM 1700 C GLU B 105 -13.013 -9.596 -5.544 1.00 0.00 C
+ATOM 1701 O GLU B 105 -12.996 -10.529 -6.339 1.00 0.00 O
+ATOM 1702 CB GLU B 105 -14.341 -8.018 -6.828 1.00 0.00 C
+ATOM 1703 CG GLU B 105 -15.594 -8.374 -6.051 1.00 0.00 C
+ATOM 1704 CD GLU B 105 -16.784 -8.464 -6.927 1.00 0.00 C
+ATOM 1705 OE1 GLU B 105 -16.774 -9.328 -7.789 1.00 0.00 O
+ATOM 1706 OE2 GLU B 105 -17.713 -7.722 -6.745 1.00 0.00 O
+ATOM 1707 H GLU B 105 -11.866 -8.294 -7.804 1.00 0.00 H
+ATOM 1708 HA GLU B 105 -13.045 -7.526 -5.175 1.00 0.00 H
+ATOM 1709 1HB GLU B 105 -14.435 -6.985 -7.134 1.00 0.00 H
+ATOM 1710 2HB GLU B 105 -14.305 -8.624 -7.733 1.00 0.00 H
+ATOM 1711 1HG GLU B 105 -15.452 -9.333 -5.558 1.00 0.00 H
+ATOM 1712 2HG GLU B 105 -15.758 -7.617 -5.280 1.00 0.00 H
+ATOM 1713 N LEU B 106 -13.043 -9.776 -4.250 1.00 0.00 N
+ATOM 1714 CA LEU B 106 -13.037 -11.113 -3.720 1.00 0.00 C
+ATOM 1715 C LEU B 106 -14.428 -11.533 -3.292 1.00 0.00 C
+ATOM 1716 O LEU B 106 -15.097 -10.832 -2.528 1.00 0.00 O
+ATOM 1717 CB LEU B 106 -12.057 -11.213 -2.546 1.00 0.00 C
+ATOM 1718 CG LEU B 106 -11.986 -12.546 -1.810 1.00 0.00 C
+ATOM 1719 CD1 LEU B 106 -11.495 -13.602 -2.721 1.00 0.00 C
+ATOM 1720 CD2 LEU B 106 -11.022 -12.400 -0.616 1.00 0.00 C
+ATOM 1721 H LEU B 106 -13.066 -8.981 -3.621 1.00 0.00 H
+ATOM 1722 HA LEU B 106 -12.715 -11.800 -4.492 1.00 0.00 H
+ATOM 1723 1HB LEU B 106 -11.055 -10.969 -2.898 1.00 0.00 H
+ATOM 1724 2HB LEU B 106 -12.356 -10.467 -1.816 1.00 0.00 H
+ATOM 1725 HG LEU B 106 -12.978 -12.825 -1.459 1.00 0.00 H
+ATOM 1726 1HD1 LEU B 106 -11.450 -14.531 -2.175 1.00 0.00 H
+ATOM 1727 2HD1 LEU B 106 -12.181 -13.706 -3.562 1.00 0.00 H
+ATOM 1728 3HD1 LEU B 106 -10.507 -13.343 -3.089 1.00 0.00 H
+ATOM 1729 1HD2 LEU B 106 -10.970 -13.339 -0.093 1.00 0.00 H
+ATOM 1730 2HD2 LEU B 106 -10.029 -12.128 -0.972 1.00 0.00 H
+ATOM 1731 3HD2 LEU B 106 -11.382 -11.630 0.059 1.00 0.00 H
+ATOM 1732 N SER B 107 -14.856 -12.687 -3.791 1.00 0.00 N
+ATOM 1733 CA SER B 107 -16.071 -13.331 -3.345 1.00 0.00 C
+ATOM 1734 C SER B 107 -15.587 -14.545 -2.574 1.00 0.00 C
+ATOM 1735 O SER B 107 -14.638 -15.207 -3.011 1.00 0.00 O
+ATOM 1736 CB SER B 107 -16.960 -13.783 -4.509 1.00 0.00 C
+ATOM 1737 OG SER B 107 -17.453 -12.701 -5.253 1.00 0.00 O
+ATOM 1738 H SER B 107 -14.290 -13.151 -4.499 1.00 0.00 H
+ATOM 1739 HA SER B 107 -16.627 -12.672 -2.679 1.00 0.00 H
+ATOM 1740 1HB SER B 107 -16.389 -14.441 -5.162 1.00 0.00 H
+ATOM 1741 2HB SER B 107 -17.792 -14.367 -4.115 1.00 0.00 H
+ATOM 1742 HG SER B 107 -16.684 -12.282 -5.656 1.00 0.00 H
+ATOM 1743 N VAL B 108 -16.237 -14.855 -1.454 1.00 0.00 N
+ATOM 1744 CA VAL B 108 -15.893 -16.062 -0.716 1.00 0.00 C
+ATOM 1745 C VAL B 108 -17.118 -16.980 -0.732 1.00 0.00 C
+ATOM 1746 O VAL B 108 -18.163 -16.638 -0.183 1.00 0.00 O
+ATOM 1747 CB VAL B 108 -15.446 -15.734 0.730 1.00 0.00 C
+ATOM 1748 CG1 VAL B 108 -15.088 -17.035 1.463 1.00 0.00 C
+ATOM 1749 CG2 VAL B 108 -14.227 -14.783 0.698 1.00 0.00 C
+ATOM 1750 H VAL B 108 -16.989 -14.264 -1.129 1.00 0.00 H
+ATOM 1751 HA VAL B 108 -15.068 -16.568 -1.212 1.00 0.00 H
+ATOM 1752 HB VAL B 108 -16.270 -15.261 1.266 1.00 0.00 H
+ATOM 1753 1HG1 VAL B 108 -14.784 -16.812 2.487 1.00 0.00 H
+ATOM 1754 2HG1 VAL B 108 -15.951 -17.696 1.477 1.00 0.00 H
+ATOM 1755 3HG1 VAL B 108 -14.269 -17.519 0.938 1.00 0.00 H
+ATOM 1756 1HG2 VAL B 108 -13.916 -14.560 1.717 1.00 0.00 H
+ATOM 1757 2HG2 VAL B 108 -13.407 -15.256 0.164 1.00 0.00 H
+ATOM 1758 3HG2 VAL B 108 -14.495 -13.857 0.196 1.00 0.00 H
+ATOM 1759 N LEU B 109 -16.965 -18.127 -1.390 1.00 0.00 N
+ATOM 1760 CA LEU B 109 -18.009 -19.112 -1.661 1.00 0.00 C
+ATOM 1761 C LEU B 109 -17.963 -20.334 -0.722 1.00 0.00 C
+ATOM 1762 O LEU B 109 -16.983 -20.557 -0.013 1.00 0.00 O
+ATOM 1763 CB LEU B 109 -17.855 -19.579 -3.102 1.00 0.00 C
+ATOM 1764 CG LEU B 109 -17.862 -18.468 -4.153 1.00 0.00 C
+ATOM 1765 CD1 LEU B 109 -17.652 -19.087 -5.516 1.00 0.00 C
+ATOM 1766 CD2 LEU B 109 -19.164 -17.690 -4.069 1.00 0.00 C
+ATOM 1767 H LEU B 109 -16.050 -18.307 -1.782 1.00 0.00 H
+ATOM 1768 HA LEU B 109 -18.976 -18.625 -1.541 1.00 0.00 H
+ATOM 1769 1HB LEU B 109 -16.903 -20.079 -3.184 1.00 0.00 H
+ATOM 1770 2HB LEU B 109 -18.642 -20.292 -3.338 1.00 0.00 H
+ATOM 1771 HG LEU B 109 -17.023 -17.785 -3.966 1.00 0.00 H
+ATOM 1772 1HD1 LEU B 109 -17.639 -18.306 -6.276 1.00 0.00 H
+ATOM 1773 2HD1 LEU B 109 -16.703 -19.621 -5.532 1.00 0.00 H
+ATOM 1774 3HD1 LEU B 109 -18.462 -19.783 -5.733 1.00 0.00 H
+ATOM 1775 1HD2 LEU B 109 -19.164 -16.895 -4.817 1.00 0.00 H
+ATOM 1776 2HD2 LEU B 109 -20.007 -18.358 -4.260 1.00 0.00 H
+ATOM 1777 3HD2 LEU B 109 -19.266 -17.252 -3.074 1.00 0.00 H
+ATOM 1778 N ALA B 110 -19.037 -21.123 -0.686 1.00 0.00 N
+ATOM 1779 CA ALA B 110 -19.022 -22.333 0.143 1.00 0.00 C
+ATOM 1780 C ALA B 110 -19.576 -23.537 -0.623 1.00 0.00 C
+ATOM 1781 O ALA B 110 -20.298 -23.345 -1.605 1.00 0.00 O
+ATOM 1782 OXT ALA B 110 -18.943 -24.602 -0.541 1.00 0.00 O
+ATOM 1783 CB ALA B 110 -19.794 -22.095 1.413 1.00 0.00 C
+ATOM 1784 H ALA B 110 -19.851 -20.904 -1.243 1.00 0.00 H
+ATOM 1785 HA ALA B 110 -17.990 -22.558 0.410 1.00 0.00 H
+ATOM 1786 1HB ALA B 110 -19.745 -22.992 2.021 1.00 0.00 H
+ATOM 1787 2HB ALA B 110 -19.355 -21.258 1.960 1.00 0.00 H
+ATOM 1788 3HB ALA B 110 -20.834 -21.873 1.177 1.00 0.00 H
+TER
diff --git a/utils/examples/pdbs/rsv5_5tpn.pdb b/utils/examples/pdbs/rsv5_5tpn.pdb
new file mode 100644
index 0000000000000000000000000000000000000000..71bb52f4a629e45e46113b7af6438ddd5eb929a2
--- /dev/null
+++ b/utils/examples/pdbs/rsv5_5tpn.pdb
@@ -0,0 +1,7077 @@
+ATOM 1 N ASN A 27 32.120 -89.596 -31.362 1.00 72.20 A N
+ATOM 2 CA ASN A 27 31.765 -90.980 -31.028 1.00 65.67 A C
+ATOM 3 C ASN A 27 30.888 -91.024 -29.775 1.00 70.28 A C
+ATOM 4 O ASN A 27 31.021 -90.212 -28.846 1.00 62.53 A O
+ATOM 5 CB ASN A 27 33.010 -91.863 -30.812 1.00 82.41 A C
+ATOM 6 CG ASN A 27 33.747 -91.546 -29.507 1.00108.25 A C
+ATOM 7 ND2 ASN A 27 34.841 -90.800 -29.610 1.00109.08 A N
+ATOM 8 OD1 ASN A 27 33.343 -91.986 -28.426 1.00122.44 A O
+ATOM 9 N ILE A 28 30.006 -91.995 -29.741 1.00 73.73 A N
+ATOM 10 CA ILE A 28 28.960 -92.036 -28.738 1.00 66.97 A C
+ATOM 11 C ILE A 28 29.318 -93.052 -27.671 1.00 72.27 A C
+ATOM 12 O ILE A 28 29.765 -94.165 -27.983 1.00 73.82 A O
+ATOM 13 CB ILE A 28 27.604 -92.343 -29.396 1.00 70.42 A C
+ATOM 14 CG1 ILE A 28 27.236 -91.185 -30.311 1.00 72.90 A C
+ATOM 15 CG2 ILE A 28 26.519 -92.523 -28.364 1.00 61.95 A C
+ATOM 16 CD1 ILE A 28 27.324 -89.849 -29.613 1.00 71.56 A C
+ATOM 17 N THR A 29 29.133 -92.652 -26.403 1.00 74.86 A N
+ATOM 18 CA THR A 29 29.375 -93.492 -25.230 1.00 67.44 A C
+ATOM 19 C THR A 29 28.154 -93.474 -24.320 1.00 71.16 A C
+ATOM 20 O THR A 29 27.337 -92.552 -24.371 1.00 73.44 A O
+ATOM 21 CB THR A 29 30.606 -93.021 -24.453 1.00 72.54 A C
+ATOM 22 CG2 THR A 29 31.882 -93.496 -25.131 1.00 68.16 A C
+ATOM 23 OG1 THR A 29 30.605 -91.588 -24.396 1.00 82.81 A O
+ATOM 24 N GLU A 30 28.007 -94.507 -23.491 1.00 68.45 A N
+ATOM 25 CA GLU A 30 26.858 -94.526 -22.588 1.00 65.76 A C
+ATOM 26 C GLU A 30 27.246 -95.046 -21.209 1.00 75.84 A C
+ATOM 27 O GLU A 30 27.927 -96.069 -21.097 1.00 71.51 A O
+ATOM 28 CB GLU A 30 25.713 -95.385 -23.126 1.00 65.49 A C
+ATOM 29 CG GLU A 30 24.358 -94.721 -22.992 1.00 67.21 A C
+ATOM 30 CD GLU A 30 23.215 -95.635 -23.387 1.00 77.78 A C
+ATOM 31 OE1 GLU A 30 23.146 -96.756 -22.837 1.00 74.02 A O
+ATOM 32 OE2 GLU A 30 22.398 -95.246 -24.254 1.00 73.88 A O1-
+ATOM 33 N GLU A 31 26.790 -94.359 -20.159 1.00 67.96 A N
+ATOM 34 CA GLU A 31 27.011 -94.800 -18.786 1.00 64.86 A C
+ATOM 35 C GLU A 31 25.677 -95.161 -18.141 1.00 62.51 A C
+ATOM 36 O GLU A 31 24.739 -94.355 -18.164 1.00 66.40 A O
+ATOM 37 CB GLU A 31 27.706 -93.715 -17.967 1.00 70.10 A C
+ATOM 38 CG GLU A 31 29.188 -93.579 -18.225 1.00 79.99 A C
+ATOM 39 CD GLU A 31 29.819 -92.451 -17.407 1.00 91.87 A C
+ATOM 40 OE1 GLU A 31 29.078 -91.760 -16.668 1.00 95.16 A O
+ATOM 41 OE2 GLU A 31 31.055 -92.258 -17.499 1.00 99.98 A O1-
+ATOM 42 N PHE A 32 25.583 -96.362 -17.563 1.00 65.37 A N
+ATOM 43 CA PHE A 32 24.382 -96.774 -16.840 1.00 70.36 A C
+ATOM 44 C PHE A 32 24.632 -96.702 -15.339 1.00 68.57 A C
+ATOM 45 O PHE A 32 25.677 -97.151 -14.864 1.00 75.91 A O
+ATOM 46 CB PHE A 32 23.946 -98.189 -17.222 1.00 65.19 A C
+ATOM 47 CG PHE A 32 22.881 -98.756 -16.313 1.00 70.64 A C
+ATOM 48 CD1 PHE A 32 21.555 -98.394 -16.464 1.00 69.16 A C
+ATOM 49 CD2 PHE A 32 23.200 -99.656 -15.325 1.00 71.95 A C
+ATOM 50 CE1 PHE A 32 20.582 -98.906 -15.643 1.00 67.88 A C
+ATOM 51 CE2 PHE A 32 22.225-100.171 -14.515 1.00 73.03 A C
+ATOM 52 CZ PHE A 32 20.918 -99.794 -14.677 1.00 68.25 A C
+ATOM 53 N TYR A 33 23.681 -96.143 -14.595 1.00 65.03 A N
+ATOM 54 CA TYR A 33 23.818 -95.980 -13.153 1.00 69.39 A C
+ATOM 55 C TYR A 33 22.828 -96.898 -12.465 1.00 77.04 A C
+ATOM 56 O TYR A 33 21.643 -96.567 -12.361 1.00 74.22 A O
+ATOM 57 CB TYR A 33 23.591 -94.526 -12.749 1.00 69.01 A C
+ATOM 58 CG TYR A 33 24.440 -93.591 -13.559 1.00 70.92 A C
+ATOM 59 CD1 TYR A 33 25.802 -93.822 -13.697 1.00 73.18 A C
+ATOM 60 CD2 TYR A 33 23.891 -92.492 -14.188 1.00 72.11 A C
+ATOM 61 CE1 TYR A 33 26.595 -92.986 -14.449 1.00 74.60 A C
+ATOM 62 CE2 TYR A 33 24.678 -91.645 -14.942 1.00 78.79 A C
+ATOM 63 CZ TYR A 33 26.033 -91.894 -15.073 1.00 82.37 A C
+ATOM 64 OH TYR A 33 26.831 -91.053 -15.827 1.00 85.82 A O
+ATOM 65 N GLN A 34 23.346 -97.999 -11.914 1.00 78.10 A N
+ATOM 66 CA GLN A 34 22.521 -99.020 -11.276 1.00 70.04 A C
+ATOM 67 C GLN A 34 21.794 -98.478 -10.059 1.00 69.60 A C
+ATOM 68 O GLN A 34 20.754 -99.025 -9.660 1.00 71.06 A O
+ATOM 69 CB GLN A 34 23.417-100.190 -10.890 1.00 75.94 A C
+ATOM 70 CG GLN A 34 22.824-101.222 -9.964 1.00 83.32 A C
+ATOM 71 CD GLN A 34 23.884-102.203 -9.506 1.00 78.64 A C
+ATOM 72 NE2 GLN A 34 23.802-103.429 -10.013 1.00 81.34 A N
+ATOM 73 OE1 GLN A 34 24.803-101.852 -8.756 1.00 81.79 A O
+ATOM 74 N SER A 35 22.324 -97.396 -9.490 1.00 66.65 A N
+ATOM 75 CA SER A 35 21.801 -96.815 -8.263 1.00 75.38 A C
+ATOM 76 C SER A 35 20.526 -96.020 -8.506 1.00 76.60 A C
+ATOM 77 O SER A 35 19.604 -96.053 -7.689 1.00 74.50 A O
+ATOM 78 CB SER A 35 22.888 -95.926 -7.671 1.00 64.71 A C
+ATOM 79 OG SER A 35 23.643 -95.313 -8.724 1.00 80.62 A O
+ATOM 80 N THR A 36 20.495 -95.237 -9.581 1.00 75.14 A N
+ATOM 81 CA THR A 36 19.382 -94.369 -9.950 1.00 77.13 A C
+ATOM 82 C THR A 36 18.491 -94.906 -11.075 1.00 80.56 A C
+ATOM 83 O THR A 36 17.590 -94.187 -11.523 1.00 77.07 A O
+ATOM 84 CB THR A 36 19.929 -92.996 -10.329 1.00 72.51 A C
+ATOM 85 CG2 THR A 36 20.525 -92.319 -9.090 1.00 70.04 A C
+ATOM 86 OG1 THR A 36 20.934 -93.156 -11.348 1.00 76.52 A O
+ATOM 87 N CYS A 37 18.741 -96.117 -11.571 1.00 69.73 A N
+ATOM 88 CA CYS A 37 17.974 -96.715 -12.670 1.00 63.55 A C
+ATOM 89 C CYS A 37 17.804 -95.715 -13.808 1.00 76.75 A C
+ATOM 90 O CYS A 37 16.714 -95.238 -14.116 1.00 78.65 A O
+ATOM 91 CB CYS A 37 16.612 -97.215 -12.197 1.00 66.00 A C
+ATOM 92 SG CYS A 37 15.899 -98.407 -13.319 1.00 79.56 A S
+ATOM 93 N SER A 38 18.950 -95.383 -14.382 1.00 75.69 A N
+ATOM 94 CA SER A 38 19.044 -94.336 -15.381 1.00 68.83 A C
+ATOM 95 C SER A 38 20.341 -94.537 -16.143 1.00 73.41 A C
+ATOM 96 O SER A 38 21.280 -95.152 -15.645 1.00 74.11 A O
+ATOM 97 CB SER A 38 19.017 -92.953 -14.727 1.00 63.08 A C
+ATOM 98 OG SER A 38 20.128 -92.804 -13.851 1.00 72.51 A O
+ATOM 99 N ALA A 39 20.399 -93.975 -17.340 1.00 65.23 A N
+ATOM 100 CA ALA A 39 21.631 -93.981 -18.106 1.00 60.04 A C
+ATOM 101 C ALA A 39 21.795 -92.620 -18.762 1.00 67.12 A C
+ATOM 102 O ALA A 39 20.866 -91.810 -18.812 1.00 61.08 A O
+ATOM 103 CB ALA A 39 21.635 -95.103 -19.145 1.00 54.43 A C
+ATOM 104 N VAL A 40 22.994 -92.353 -19.256 1.00 60.19 A N
+ATOM 105 CA VAL A 40 23.237 -91.156 -20.041 1.00 57.31 A C
+ATOM 106 C VAL A 40 23.998 -91.551 -21.293 1.00 59.99 A C
+ATOM 107 O VAL A 40 24.945 -92.347 -21.233 1.00 62.57 A O
+ATOM 108 CB VAL A 40 23.990 -90.073 -19.250 1.00 60.94 A C
+ATOM 109 CG1 VAL A 40 24.690 -89.116 -20.201 1.00 55.69 A C
+ATOM 110 CG2 VAL A 40 23.009 -89.310 -18.378 1.00 57.25 A C
+ATOM 111 N SER A 41 23.545 -91.021 -22.427 1.00 59.58 A N
+ATOM 112 CA SER A 41 24.208 -91.171 -23.711 1.00 62.27 A C
+ATOM 113 C SER A 41 24.936 -89.865 -24.013 1.00 68.14 A C
+ATOM 114 O SER A 41 24.313 -88.801 -24.123 1.00 68.21 A O
+ATOM 115 CB SER A 41 23.188 -91.513 -24.794 1.00 58.31 A C
+ATOM 116 OG SER A 41 22.163 -92.367 -24.292 1.00 69.75 A O
+ATOM 117 N LYS A 42 26.249 -89.953 -24.120 1.00 66.80 A N
+ATOM 118 CA LYS A 42 27.129 -88.801 -24.175 1.00 68.22 A C
+ATOM 119 C LYS A 42 27.880 -88.826 -25.506 1.00 68.58 A C
+ATOM 120 O LYS A 42 28.031 -89.881 -26.145 1.00 69.14 A O
+ATOM 121 CB LYS A 42 28.108 -88.808 -22.958 1.00 69.69 A C
+ATOM 122 CG LYS A 42 28.222 -87.463 -22.154 1.00 89.89 A C
+ATOM 123 CD LYS A 42 28.975 -87.587 -20.780 1.00 97.72 A C
+ATOM 124 CE LYS A 42 29.154 -86.220 -20.034 1.00103.20 A C
+ATOM 125 NZ LYS A 42 28.051 -85.808 -19.095 1.00110.07 A N1+
+ATOM 126 N GLY A 43 28.345 -87.649 -25.921 1.00 66.72 A N
+ATOM 127 CA GLY A 43 29.162 -87.526 -27.106 1.00 62.64 A C
+ATOM 128 C GLY A 43 28.483 -86.958 -28.332 1.00 70.17 A C
+ATOM 129 O GLY A 43 29.074 -87.015 -29.417 1.00 69.36 A O
+ATOM 130 N TYR A 44 27.266 -86.424 -28.204 1.00 61.49 A N
+ATOM 131 CA TYR A 44 26.544 -85.830 -29.320 1.00 59.49 A C
+ATOM 132 C TYR A 44 26.864 -84.341 -29.440 1.00 67.11 A C
+ATOM 133 O TYR A 44 27.532 -83.743 -28.592 1.00 67.92 A O
+ATOM 134 CB TYR A 44 25.034 -86.001 -29.147 1.00 61.10 A C
+ATOM 135 CG TYR A 44 24.524 -87.423 -29.144 1.00 69.67 A C
+ATOM 136 CD1 TYR A 44 24.420 -88.152 -30.313 1.00 61.28 A C
+ATOM 137 CD2 TYR A 44 24.117 -88.018 -27.968 1.00 62.82 A C
+ATOM 138 CE1 TYR A 44 23.947 -89.439 -30.303 1.00 63.60 A C
+ATOM 139 CE2 TYR A 44 23.646 -89.295 -27.948 1.00 66.76 A C
+ATOM 140 CZ TYR A 44 23.562 -90.006 -29.113 1.00 71.77 A C
+ATOM 141 OH TYR A 44 23.088 -91.295 -29.075 1.00 70.05 A O
+ATOM 142 N LEU A 45 26.300 -83.719 -30.470 1.00 67.96 A N
+ATOM 143 CA LEU A 45 26.614 -82.347 -30.850 1.00 62.06 A C
+ATOM 144 C LEU A 45 25.347 -81.522 -31.047 1.00 65.83 A C
+ATOM 145 O LEU A 45 24.343 -82.025 -31.546 1.00 68.25 A O
+ATOM 146 CB LEU A 45 27.441 -82.327 -32.137 1.00 60.41 A C
+ATOM 147 CG LEU A 45 28.935 -82.499 -31.938 1.00 74.92 A C
+ATOM 148 CD1 LEU A 45 29.591 -82.368 -33.278 1.00 66.57 A C
+ATOM 149 CD2 LEU A 45 29.428 -81.424 -30.986 1.00 74.67 A C
+ATOM 150 N SER A 46 25.332 -80.281 -30.591 1.00 58.86 A N
+ATOM 151 CA SER A 46 24.071 -79.554 -30.650 1.00 54.84 A C
+ATOM 152 C SER A 46 23.724 -78.488 -31.678 1.00 64.79 A C
+ATOM 153 O SER A 46 24.278 -77.388 -31.665 1.00 68.34 A O
+ATOM 154 CB SER A 46 23.757 -78.992 -29.256 1.00 58.87 A C
+ATOM 155 OG SER A 46 24.620 -77.916 -28.932 1.00 61.92 A O
+ATOM 156 N ALA A 47 22.765 -78.812 -32.543 1.00 59.79 A N
+ATOM 157 CA ALA A 47 22.251 -77.818 -33.469 1.00 57.25 A C
+ATOM 158 C ALA A 47 20.972 -77.279 -32.847 1.00 58.48 A C
+ATOM 159 O ALA A 47 19.938 -77.960 -32.830 1.00 61.48 A O
+ATOM 160 CB ALA A 47 22.001 -78.426 -34.842 1.00 58.81 A C
+ATOM 161 N LEU A 48 21.030 -76.051 -32.368 1.00 58.06 A N
+ATOM 162 CA LEU A 48 19.978 -75.515 -31.523 1.00 62.27 A C
+ATOM 163 C LEU A 48 19.245 -74.428 -32.282 1.00 62.27 A C
+ATOM 164 O LEU A 48 19.853 -73.431 -32.684 1.00 66.77 A O
+ATOM 165 CB LEU A 48 20.547 -74.965 -30.218 1.00 52.61 A C
+ATOM 166 CG LEU A 48 21.219 -75.974 -29.293 1.00 54.96 A C
+ATOM 167 CD1 LEU A 48 21.319 -75.372 -27.915 1.00 54.11 A C
+ATOM 168 CD2 LEU A 48 20.426 -77.268 -29.228 1.00 61.76 A C
+ATOM 169 N ARG A 49 17.939 -74.596 -32.444 1.00 60.09 A N
+ATOM 170 CA ARG A 49 17.187 -73.557 -33.117 1.00 63.78 A C
+ATOM 171 C ARG A 49 17.030 -72.434 -32.118 1.00 62.66 A C
+ATOM 172 O ARG A 49 16.297 -72.563 -31.136 1.00 59.24 A O
+ATOM 173 CB ARG A 49 15.823 -74.051 -33.579 1.00 57.80 A C
+ATOM 174 CG ARG A 49 15.219 -73.066 -34.549 1.00 60.39 A C
+ATOM 175 CD ARG A 49 13.707 -73.025 -34.493 1.00 55.28 A C
+ATOM 176 NE ARG A 49 13.185 -72.508 -33.233 1.00 60.97 A N
+ATOM 177 CZ ARG A 49 13.104 -71.219 -32.921 1.00 61.68 A C
+ATOM 178 NH1 ARG A 49 13.525 -70.306 -33.779 1.00 61.78 A N1+
+ATOM 179 NH2 ARG A 49 12.600 -70.846 -31.746 1.00 63.68 A N
+ATOM 180 N THR A 50 17.721 -71.332 -32.366 1.00 63.82 A N
+ATOM 181 CA THR A 50 17.604 -70.194 -31.486 1.00 56.36 A C
+ATOM 182 C THR A 50 16.683 -69.107 -32.017 1.00 61.31 A C
+ATOM 183 O THR A 50 16.207 -68.286 -31.220 1.00 55.88 A O
+ATOM 184 CB THR A 50 19.000 -69.633 -31.224 1.00 61.27 A C
+ATOM 185 CG2 THR A 50 19.912 -70.779 -30.873 1.00 60.88 A C
+ATOM 186 OG1 THR A 50 19.507 -69.000 -32.408 1.00 52.98 A O
+ATOM 187 N GLY A 51 16.358 -69.118 -33.307 1.00 61.17 A N
+ATOM 188 CA GLY A 51 15.647 -67.970 -33.846 1.00 58.16 A C
+ATOM 189 C GLY A 51 15.148 -68.212 -35.253 1.00 62.49 A C
+ATOM 190 O GLY A 51 15.173 -69.333 -35.753 1.00 56.96 A O
+ATOM 191 N TRP A 52 14.666 -67.147 -35.887 1.00 58.16 A N
+ATOM 192 CA TRP A 52 14.072 -67.285 -37.208 1.00 52.76 A C
+ATOM 193 C TRP A 52 14.639 -66.256 -38.172 1.00 60.30 A C
+ATOM 194 O TRP A 52 15.134 -65.192 -37.775 1.00 63.15 A O
+ATOM 195 CB TRP A 52 12.533 -67.130 -37.210 1.00 55.52 A C
+ATOM 196 CG TRP A 52 11.761 -68.041 -36.290 1.00 59.71 A C
+ATOM 197 CD1 TRP A 52 11.125 -67.684 -35.127 1.00 53.92 A C
+ATOM 198 CD2 TRP A 52 11.541 -69.458 -36.445 1.00 55.35 A C
+ATOM 199 CE2 TRP A 52 10.768 -69.885 -35.348 1.00 56.26 A C
+ATOM 200 CE3 TRP A 52 11.922 -70.400 -37.402 1.00 52.94 A C
+ATOM 201 NE1 TRP A 52 10.523 -68.785 -34.561 1.00 60.17 A N
+ATOM 202 CZ2 TRP A 52 10.376 -71.211 -35.186 1.00 57.32 A C
+ATOM 203 CZ3 TRP A 52 11.531 -71.702 -37.230 1.00 53.84 A C
+ATOM 204 CH2 TRP A 52 10.765 -72.095 -36.134 1.00 54.29 A C
+ATOM 205 N TYR A 53 14.538 -66.607 -39.460 1.00 46.59 A N
+ATOM 206 CA TYR A 53 14.768 -65.694 -40.579 1.00 42.06 A C
+ATOM 207 C TYR A 53 13.653 -65.895 -41.589 1.00 43.34 A C
+ATOM 208 O TYR A 53 13.440 -67.013 -42.069 1.00 47.84 A O
+ATOM 209 CB TYR A 53 16.126 -65.931 -41.241 1.00 52.35 A C
+ATOM 210 CG TYR A 53 16.450 -64.947 -42.337 1.00 75.87 A C
+ATOM 211 CD1 TYR A 53 17.133 -63.785 -42.052 1.00 76.30 A C
+ATOM 212 CD2 TYR A 53 16.104 -65.193 -43.649 1.00 83.29 A C
+ATOM 213 CE1 TYR A 53 17.442 -62.883 -43.026 1.00 77.04 A C
+ATOM 214 CE2 TYR A 53 16.413 -64.292 -44.641 1.00 88.43 A C
+ATOM 215 CZ TYR A 53 17.083 -63.134 -44.319 1.00 92.01 A C
+ATOM 216 OH TYR A 53 17.406 -62.206 -45.281 1.00103.48 A O
+ATOM 217 N THR A 54 12.961 -64.819 -41.923 1.00 45.05 A N
+ATOM 218 CA THR A 54 11.783 -64.888 -42.770 1.00 51.23 A C
+ATOM 219 C THR A 54 12.155 -64.549 -44.206 1.00 58.78 A C
+ATOM 220 O THR A 54 12.765 -63.507 -44.465 1.00 61.51 A O
+ATOM 221 CB THR A 54 10.719 -63.925 -42.255 1.00 51.66 A C
+ATOM 222 CG2 THR A 54 10.295 -64.327 -40.847 1.00 50.45 A C
+ATOM 223 OG1 THR A 54 11.267 -62.601 -42.205 1.00 55.42 A O
+ATOM 224 N SER A 55 11.792 -65.426 -45.135 1.00 45.33 A N
+ATOM 225 CA SER A 55 11.998 -65.194 -46.559 1.00 51.75 A C
+ATOM 226 C SER A 55 10.643 -65.070 -47.241 1.00 48.93 A C
+ATOM 227 O SER A 55 9.639 -65.614 -46.756 1.00 48.95 A O
+ATOM 228 CB SER A 55 12.777 -66.331 -47.185 1.00 55.67 A C
+ATOM 229 OG SER A 55 12.072 -67.540 -46.968 1.00 74.54 A O
+ATOM 230 N VAL A 56 10.604 -64.349 -48.361 1.00 45.30 A N
+ATOM 231 CA VAL A 56 9.357 -64.163 -49.096 1.00 36.35 A C
+ATOM 232 C VAL A 56 9.468 -64.855 -50.455 1.00 47.50 A C
+ATOM 233 O VAL A 56 10.171 -64.389 -51.365 1.00 50.37 A O
+ATOM 234 CB VAL A 56 8.994 -62.686 -49.244 1.00 46.93 A C
+ATOM 235 CG1 VAL A 56 7.793 -62.573 -50.108 1.00 52.72 A C
+ATOM 236 CG2 VAL A 56 8.692 -62.110 -47.911 1.00 36.71 A C
+ATOM 237 N ILE A 57 8.744 -65.962 -50.581 1.00 42.71 A N
+ATOM 238 CA ILE A 57 8.582 -66.712 -51.817 1.00 38.55 A C
+ATOM 239 C ILE A 57 7.404 -66.124 -52.589 1.00 50.05 A C
+ATOM 240 O ILE A 57 6.379 -65.739 -51.995 1.00 51.40 A O
+ATOM 241 CB ILE A 57 8.360 -68.201 -51.509 1.00 41.74 A C
+ATOM 242 CG1 ILE A 57 9.472 -68.701 -50.603 1.00 43.65 A C
+ATOM 243 CG2 ILE A 57 8.318 -68.994 -52.760 1.00 39.82 A C
+ATOM 244 CD1 ILE A 57 10.802 -68.400 -51.114 1.00 35.26 A C
+ATOM 245 N THR A 58 7.551 -66.026 -53.918 1.00 49.79 A N
+ATOM 246 CA THR A 58 6.504 -65.484 -54.777 1.00 44.46 A C
+ATOM 247 C THR A 58 6.397 -66.263 -56.083 1.00 49.11 A C
+ATOM 248 O THR A 58 7.317 -66.977 -56.509 1.00 54.38 A O
+ATOM 249 CB THR A 58 6.733 -64.015 -55.124 1.00 47.27 A C
+ATOM 250 CG2 THR A 58 6.799 -63.170 -53.869 1.00 49.12 A C
+ATOM 251 OG1 THR A 58 7.947 -63.894 -55.879 1.00 50.62 A O
+ATOM 252 N ILE A 59 5.233 -66.100 -56.705 1.00 46.28 A N
+ATOM 253 CA ILE A 59 4.908 -66.654 -58.008 1.00 44.24 A C
+ATOM 254 C ILE A 59 4.122 -65.602 -58.763 1.00 54.64 A C
+ATOM 255 O ILE A 59 3.124 -65.083 -58.245 1.00 63.01 A O
+ATOM 256 CB ILE A 59 4.064 -67.924 -57.897 1.00 43.70 A C
+ATOM 257 CG1 ILE A 59 4.756 -68.965 -57.058 1.00 42.15 A C
+ATOM 258 CG2 ILE A 59 3.769 -68.455 -59.257 1.00 45.90 A C
+ATOM 259 CD1 ILE A 59 3.831 -70.104 -56.753 1.00 39.09 A C
+ATOM 260 N GLU A 60 4.550 -65.295 -59.987 1.00 51.73 A N
+ATOM 261 CA GLU A 60 3.876 -64.259 -60.747 1.00 53.53 A C
+ATOM 262 C GLU A 60 2.681 -64.873 -61.468 1.00 51.07 A C
+ATOM 263 O GLU A 60 2.786 -65.951 -62.066 1.00 55.49 A O
+ATOM 264 CB GLU A 60 4.857 -63.570 -61.699 1.00 54.11 A C
+ATOM 265 CG GLU A 60 4.825 -62.032 -61.539 1.00 73.02 A C
+ATOM 266 CD GLU A 60 5.785 -61.267 -62.456 1.00 82.60 A C
+ATOM 267 OE1 GLU A 60 5.946 -61.644 -63.640 1.00 73.00 A O
+ATOM 268 OE2 GLU A 60 6.362 -60.258 -61.991 1.00102.75 A O1-
+ATOM 269 N LEU A 61 1.528 -64.218 -61.353 1.00 58.15 A N
+ATOM 270 CA LEU A 61 0.267 -64.755 -61.853 1.00 54.09 A C
+ATOM 271 C LEU A 61 -0.429 -63.725 -62.730 1.00 59.07 A C
+ATOM 272 O LEU A 61 -0.762 -62.633 -62.259 1.00 73.30 A O
+ATOM 273 CB LEU A 61 -0.660 -65.148 -60.704 1.00 59.26 A C
+ATOM 274 CG LEU A 61 -0.081 -65.937 -59.547 1.00 56.23 A C
+ATOM 275 CD1 LEU A 61 -1.015 -65.835 -58.387 1.00 61.92 A C
+ATOM 276 CD2 LEU A 61 0.015 -67.357 -59.982 1.00 51.18 A C
+ATOM 277 N SER A 62 -0.661 -64.081 -63.992 1.00 57.63 A N
+ATOM 278 CA SER A 62 -1.526 -63.295 -64.858 1.00 59.84 A C
+ATOM 279 C SER A 62 -2.927 -63.274 -64.270 1.00 71.00 A C
+ATOM 280 O SER A 62 -3.443 -64.315 -63.861 1.00 74.20 A O
+ATOM 281 CB SER A 62 -1.545 -63.908 -66.256 1.00 66.51 A C
+ATOM 282 OG SER A 62 -1.810 -62.914 -67.223 1.00 68.86 A O
+ATOM 283 N ASN A 63 -3.544 -62.092 -64.202 1.00 65.12 A N
+ATOM 284 CA ASN A 63 -4.871 -61.967 -63.601 1.00 77.30 A C
+ATOM 285 C ASN A 63 -5.874 -62.023 -64.747 1.00 72.83 A C
+ATOM 286 O ASN A 63 -6.026 -61.043 -65.479 1.00 82.32 A O
+ATOM 287 CB ASN A 63 -4.995 -60.657 -62.816 1.00 88.19 A C
+ATOM 288 CG ASN A 63 -5.705 -60.825 -61.450 1.00 97.22 A C
+ATOM 289 ND2 ASN A 63 -6.917 -60.271 -61.355 1.00100.90 A N
+ATOM 290 OD1 ASN A 63 -5.172 -61.449 -60.503 1.00101.68 A O
+ATOM 291 N ILE A 64 -6.580 -63.149 -64.887 1.00 77.79 A N
+ATOM 292 CA ILE A 64 -7.491 -63.349 -66.020 1.00 88.97 A C
+ATOM 293 C ILE A 64 -8.862 -63.771 -65.504 1.00 96.09 A C
+ATOM 294 O ILE A 64 -9.031 -64.889 -64.997 1.00 94.17 A O
+ATOM 295 CB ILE A 64 -6.966 -64.371 -67.044 1.00 80.66 A C
+ATOM 296 CG1 ILE A 64 -6.554 -65.689 -66.403 1.00 73.33 A C
+ATOM 297 CG2 ILE A 64 -5.786 -63.812 -67.783 1.00 80.77 A C
+ATOM 298 CD1 ILE A 64 -6.150 -66.672 -67.425 1.00 76.35 A C
+ATOM 299 N LYS A 65 -9.848 -62.887 -65.654 1.00106.08 A N
+ATOM 300 CA LYS A 65 -11.205 -63.233 -65.270 1.00108.87 A C
+ATOM 301 C LYS A 65 -12.045 -63.785 -66.416 1.00120.85 A C
+ATOM 302 O LYS A 65 -13.086 -64.391 -66.147 1.00115.64 A O
+ATOM 303 CB LYS A 65 -11.911 -62.016 -64.624 1.00101.95 A C
+ATOM 304 CG LYS A 65 -11.176 -61.505 -63.369 1.00 99.91 A C
+ATOM 305 CD LYS A 65 -11.964 -60.524 -62.493 1.00101.46 A C
+ATOM 306 CE LYS A 65 -11.087 -60.087 -61.296 1.00100.70 A C
+ATOM 307 NZ LYS A 65 -11.788 -59.327 -60.208 1.00 98.81 A N1+
+ATOM 308 N LYS A 66 -11.631 -63.637 -67.675 1.00 98.06 A N
+ATOM 309 CA LYS A 66 -12.538 -63.945 -68.779 1.00100.55 A C
+ATOM 310 C LYS A 66 -11.810 -64.706 -69.878 1.00 87.83 A C
+ATOM 311 O LYS A 66 -10.920 -64.143 -70.521 1.00 91.66 A O
+ATOM 312 CB LYS A 66 -13.160 -62.655 -69.340 1.00112.65 A C
+ATOM 313 CG LYS A 66 -13.643 -61.634 -68.282 1.00115.08 A C
+ATOM 314 CD LYS A 66 -14.806 -60.770 -68.786 1.00114.28 A C
+ATOM 315 CE LYS A 66 -16.141 -61.509 -68.608 1.00107.05 A C
+ATOM 316 NZ LYS A 66 -17.338 -60.744 -69.052 1.00103.06 A N1+
+ATOM 317 N ILE A 67 -12.227 -65.950 -70.140 1.00104.67 A N
+ATOM 318 CA ILE A 67 -11.690 -66.752 -71.241 1.00100.72 A C
+ATOM 319 C ILE A 67 -12.706 -66.668 -72.365 1.00103.14 A C
+ATOM 320 O ILE A 67 -13.711 -67.381 -72.331 1.00107.08 A O
+ATOM 321 CB ILE A 67 -11.510 -68.228 -70.866 1.00 86.75 A C
+ATOM 322 CG1 ILE A 67 -11.159 -68.453 -69.394 1.00 71.87 A C
+ATOM 323 CG2 ILE A 67 -10.501 -68.882 -71.795 1.00 83.05 A C
+ATOM 324 CD1 ILE A 67 -9.794 -67.985 -68.998 1.00 67.83 A C
+ATOM 325 N LYS A 68 -12.449 -65.888 -73.411 1.00 93.04 A N
+ATOM 326 CA LYS A 68 -13.386 -65.921 -74.531 1.00100.87 A C
+ATOM 327 C LYS A 68 -12.637 -66.437 -75.746 1.00107.32 A C
+ATOM 328 O LYS A 68 -12.021 -65.660 -76.475 1.00111.84 A O
+ATOM 329 CB LYS A 68 -13.991 -64.543 -74.799 1.00103.33 A C
+ATOM 330 CG LYS A 68 -15.245 -64.538 -75.716 1.00105.48 A C
+ATOM 331 CD LYS A 68 -16.498 -65.102 -74.991 1.00102.59 A C
+ATOM 332 CE LYS A 68 -17.737 -65.145 -75.898 1.00 98.19 A C
+ATOM 333 NZ LYS A 68 -18.975 -65.593 -75.177 1.00 99.02 A N1+
+ATOM 334 N CYS A 69 -12.755 -67.735 -75.988 1.00 93.39 A N
+ATOM 335 CA CYS A 69 -12.432 -68.345 -77.269 1.00 92.27 A C
+ATOM 336 C CYS A 69 -13.482 -69.412 -77.517 1.00 92.79 A C
+ATOM 337 O CYS A 69 -13.523 -70.409 -76.787 1.00 97.86 A O
+ATOM 338 CB CYS A 69 -11.031 -68.951 -77.261 1.00 86.74 A C
+ATOM 339 SG CYS A 69 -10.734 -70.142 -78.580 1.00118.72 A S
+ATOM 340 N ASN A 70 -14.213 -69.286 -78.611 1.00101.01 A N
+ATOM 341 CA ASN A 70 -15.196 -70.296 -78.942 1.00102.41 A C
+ATOM 342 C ASN A 70 -14.750 -71.467 -79.844 1.00114.52 A C
+ATOM 343 O ASN A 70 -15.631 -72.120 -80.403 1.00126.45 A O
+ATOM 344 CB ASN A 70 -16.441 -69.634 -79.545 1.00 98.13 A C
+ATOM 345 CG ASN A 70 -16.135 -68.872 -80.819 1.00 99.62 A C
+ATOM 346 ND2 ASN A 70 -16.525 -69.438 -81.956 1.00 97.85 A N
+ATOM 347 OD1 ASN A 70 -15.555 -67.787 -80.783 1.00 98.76 A O
+ATOM 348 N GLY A 71 -13.458 -71.789 -80.023 1.00113.24 A N
+ATOM 349 CA GLY A 71 -13.287 -72.923 -80.891 1.00 99.58 A C
+ATOM 350 C GLY A 71 -13.812 -74.089 -80.081 1.00 81.27 A C
+ATOM 351 O GLY A 71 -14.087 -73.960 -78.890 1.00 75.12 A O
+ATOM 352 N THR A 72 -13.975 -75.225 -80.748 1.00 94.55 A N
+ATOM 353 CA THR A 72 -14.068 -76.502 -80.046 1.00 96.02 A C
+ATOM 354 C THR A 72 -13.090 -77.438 -80.728 1.00 94.00 A C
+ATOM 355 O THR A 72 -13.378 -77.995 -81.794 1.00 77.72 A O
+ATOM 356 CB THR A 72 -15.474 -77.072 -80.078 1.00 97.19 A C
+ATOM 357 CG2 THR A 72 -16.378 -76.310 -79.106 1.00102.60 A C
+ATOM 358 OG1 THR A 72 -15.983 -76.990 -81.419 1.00100.03 A O
+ATOM 359 N ASP A 73 -11.972 -77.651 -80.061 1.00 81.63 A N
+ATOM 360 CA ASP A 73 -10.973 -78.612 -80.474 1.00 78.54 A C
+ATOM 361 C ASP A 73 -10.386 -79.153 -79.191 1.00 67.60 A C
+ATOM 362 O ASP A 73 -10.081 -78.363 -78.291 1.00 66.62 A O
+ATOM 363 CB ASP A 73 -9.899 -77.966 -81.333 1.00 70.85 A C
+ATOM 364 CG ASP A 73 -8.881 -78.950 -81.766 1.00 78.00 A C
+ATOM 365 OD1 ASP A 73 -9.248 -79.843 -82.552 1.00 78.46 A O
+ATOM 366 OD2 ASP A 73 -7.729 -78.856 -81.302 1.00 82.67 A O1-
+ATOM 367 N ALA A 74 -10.233 -80.475 -79.096 1.00 55.77 A N
+ATOM 368 CA ALA A 74 -9.920 -81.051 -77.793 1.00 63.82 A C
+ATOM 369 C ALA A 74 -8.743 -80.342 -77.146 1.00 63.12 A C
+ATOM 370 O ALA A 74 -8.763 -80.110 -75.937 1.00 67.23 A O
+ATOM 371 CB ALA A 74 -9.651 -82.551 -77.913 1.00 56.29 A C
+ATOM 372 N LYS A 75 -7.753 -79.913 -77.939 1.00 56.21 A N
+ATOM 373 CA LYS A 75 -6.595 -79.227 -77.369 1.00 57.88 A C
+ATOM 374 C LYS A 75 -7.014 -77.945 -76.663 1.00 60.40 A C
+ATOM 375 O LYS A 75 -6.751 -77.765 -75.465 1.00 60.28 A O
+ATOM 376 CB LYS A 75 -5.562 -78.969 -78.460 1.00 55.71 A C
+ATOM 377 CG LYS A 75 -5.560 -80.103 -79.457 1.00 56.52 A C
+ATOM 378 CD LYS A 75 -4.212 -80.353 -80.069 1.00 58.09 A C
+ATOM 379 CE LYS A 75 -4.274 -81.615 -80.944 1.00 62.67 A C
+ATOM 380 NZ LYS A 75 -2.918 -82.260 -81.223 1.00 70.40 A N1+
+ATOM 381 N ILE A 76 -7.713 -77.066 -77.381 1.00 55.08 A N
+ATOM 382 CA ILE A 76 -8.324 -75.893 -76.757 1.00 57.43 A C
+ATOM 383 C ILE A 76 -9.115 -76.286 -75.514 1.00 59.94 A C
+ATOM 384 O ILE A 76 -9.045 -75.617 -74.476 1.00 57.01 A O
+ATOM 385 CB ILE A 76 -9.201 -75.144 -77.780 1.00 58.15 A C
+ATOM 386 CG1 ILE A 76 -8.319 -74.338 -78.725 1.00 61.70 A C
+ATOM 387 CG2 ILE A 76 -10.184 -74.243 -77.089 1.00 52.60 A C
+ATOM 388 CD1 ILE A 76 -9.058 -73.610 -79.783 1.00 75.30 A C
+ATOM 389 N LYS A 77 -9.868 -77.382 -75.587 1.00 59.07 A N
+ATOM 390 CA LYS A 77 -10.693 -77.748 -74.444 1.00 55.83 A C
+ATOM 391 C LYS A 77 -9.835 -78.030 -73.223 1.00 57.69 A C
+ATOM 392 O LYS A 77 -10.100 -77.511 -72.136 1.00 59.86 A O
+ATOM 393 CB LYS A 77 -11.559 -78.959 -74.770 1.00 53.81 A C
+ATOM 394 CG LYS A 77 -12.388 -79.471 -73.568 1.00 67.90 A C
+ATOM 395 CD LYS A 77 -13.127 -80.777 -73.921 1.00 86.47 A C
+ATOM 396 CE LYS A 77 -14.348 -81.033 -73.043 1.00 92.39 A C
+ATOM 397 NZ LYS A 77 -15.317 -81.946 -73.723 1.00 98.83 A N1+
+ATOM 398 N LEU A 78 -8.799 -78.858 -73.385 1.00 53.26 A N
+ATOM 399 CA LEU A 78 -7.933 -79.196 -72.260 1.00 61.25 A C
+ATOM 400 C LEU A 78 -7.278 -77.947 -71.697 1.00 57.31 A C
+ATOM 401 O LEU A 78 -7.256 -77.757 -70.470 1.00 66.22 A O
+ATOM 402 CB LEU A 78 -6.869 -80.215 -72.684 1.00 48.85 A C
+ATOM 403 CG LEU A 78 -7.341 -81.476 -73.401 1.00 60.91 A C
+ATOM 404 CD1 LEU A 78 -6.168 -82.345 -73.788 1.00 58.41 A C
+ATOM 405 CD2 LEU A 78 -8.312 -82.235 -72.531 1.00 59.48 A C
+ATOM 406 N ILE A 79 -6.759 -77.074 -72.585 1.00 53.24 A N
+ATOM 407 CA ILE A 79 -6.177 -75.800 -72.147 1.00 52.56 A C
+ATOM 408 C ILE A 79 -7.180 -75.024 -71.306 1.00 55.01 A C
+ATOM 409 O ILE A 79 -6.848 -74.512 -70.234 1.00 56.74 A O
+ATOM 410 CB ILE A 79 -5.704 -74.960 -73.346 1.00 56.57 A C
+ATOM 411 CG1 ILE A 79 -4.580 -75.656 -74.092 1.00 65.94 A C
+ATOM 412 CG2 ILE A 79 -5.210 -73.626 -72.863 1.00 60.63 A C
+ATOM 413 CD1 ILE A 79 -3.373 -75.850 -73.254 1.00 67.06 A C
+ATOM 414 N LYS A 80 -8.427 -74.942 -71.774 1.00 54.62 A N
+ATOM 415 CA LYS A 80 -9.457 -74.252 -71.010 1.00 60.32 A C
+ATOM 416 C LYS A 80 -9.701 -74.935 -69.659 1.00 60.77 A C
+ATOM 417 O LYS A 80 -9.902 -74.261 -68.638 1.00 60.89 A O
+ATOM 418 CB LYS A 80 -10.743 -74.159 -71.843 1.00 52.50 A C
+ATOM 419 CG LYS A 80 -10.667 -73.172 -73.037 1.00 62.62 A C
+ATOM 420 CD LYS A 80 -11.983 -73.078 -73.866 1.00 79.98 A C
+ATOM 421 CE LYS A 80 -13.054 -72.162 -73.230 1.00 84.50 A C
+ATOM 422 NZ LYS A 80 -14.292 -72.011 -74.059 1.00 94.32 A N1+
+ATOM 423 N GLN A 81 -9.661 -76.265 -69.622 1.00 60.14 A N
+ATOM 424 CA GLN A 81 -9.854 -76.957 -68.355 1.00 59.68 A C
+ATOM 425 C GLN A 81 -8.776 -76.558 -67.358 1.00 66.16 A C
+ATOM 426 O GLN A 81 -9.075 -76.138 -66.231 1.00 61.70 A O
+ATOM 427 CB GLN A 81 -9.866 -78.466 -68.581 1.00 57.45 A C
+ATOM 428 CG GLN A 81 -11.121 -78.970 -69.289 1.00 69.03 A C
+ATOM 429 CD GLN A 81 -11.223 -80.509 -69.363 1.00 71.51 A C
+ATOM 430 NE2 GLN A 81 -12.129 -80.994 -70.229 1.00 77.15 A N
+ATOM 431 OE1 GLN A 81 -10.509 -81.250 -68.651 1.00 80.38 A O
+ATOM 432 N GLU A 82 -7.512 -76.650 -67.775 1.00 57.13 A N
+ATOM 433 CA GLU A 82 -6.417 -76.226 -66.912 1.00 55.06 A C
+ATOM 434 C GLU A 82 -6.551 -74.762 -66.509 1.00 60.54 A C
+ATOM 435 O GLU A 82 -6.237 -74.391 -65.371 1.00 61.10 A O
+ATOM 436 CB GLU A 82 -5.084 -76.452 -67.611 1.00 48.61 A C
+ATOM 437 CG GLU A 82 -3.947 -76.622 -66.639 1.00 57.90 A C
+ATOM 438 CD GLU A 82 -4.022 -77.953 -65.958 1.00 63.75 A C
+ATOM 439 OE1 GLU A 82 -4.825 -78.772 -66.456 1.00 65.18 A O
+ATOM 440 OE2 GLU A 82 -3.304 -78.186 -64.947 1.00 63.35 A O1-
+ATOM 441 N LEU A 83 -6.986 -73.909 -67.436 1.00 54.36 A N
+ATOM 442 CA LEU A 83 -7.194 -72.505 -67.106 1.00 52.22 A C
+ATOM 443 C LEU A 83 -8.174 -72.361 -65.963 1.00 56.24 A C
+ATOM 444 O LEU A 83 -7.951 -71.585 -65.035 1.00 54.40 A O
+ATOM 445 CB LEU A 83 -7.705 -71.732 -68.321 1.00 60.67 A C
+ATOM 446 CG LEU A 83 -6.611 -70.965 -69.047 1.00 72.86 A C
+ATOM 447 CD1 LEU A 83 -6.029 -70.004 -68.057 1.00 74.08 A C
+ATOM 448 CD2 LEU A 83 -5.533 -71.873 -69.587 1.00 80.54 A C
+ATOM 449 N ASP A 84 -9.282 -73.092 -66.026 1.00 61.35 A N
+ATOM 450 CA ASP A 84 -10.263 -73.007 -64.953 1.00 61.71 A C
+ATOM 451 C ASP A 84 -9.678 -73.502 -63.635 1.00 62.91 A C
+ATOM 452 O ASP A 84 -9.907 -72.896 -62.579 1.00 51.32 A O
+ATOM 453 CB ASP A 84 -11.515 -73.783 -65.353 1.00 72.84 A C
+ATOM 454 CG ASP A 84 -12.414 -72.971 -66.265 1.00 95.20 A C
+ATOM 455 OD1 ASP A 84 -12.487 -71.741 -66.045 1.00100.41 A O
+ATOM 456 OD2 ASP A 84 -13.004 -73.540 -67.215 1.00110.68 A O1-
+ATOM 457 N LYS A 85 -8.898 -74.588 -63.678 1.00 55.00 A N
+ATOM 458 CA LYS A 85 -8.173 -75.024 -62.486 1.00 54.38 A C
+ATOM 459 C LYS A 85 -7.348 -73.875 -61.889 1.00 66.00 A C
+ATOM 460 O LYS A 85 -7.415 -73.607 -60.683 1.00 63.02 A O
+ATOM 461 CB LYS A 85 -7.271 -76.214 -62.836 1.00 45.19 A C
+ATOM 462 CG LYS A 85 -7.881 -77.637 -62.838 1.00 44.23 A C
+ATOM 463 CD LYS A 85 -6.716 -78.647 -63.081 1.00 63.74 A C
+ATOM 464 CE LYS A 85 -7.045 -80.136 -62.896 1.00 78.87 A C
+ATOM 465 NZ LYS A 85 -5.797 -80.938 -62.606 1.00 85.63 A N1+
+ATOM 466 N TYR A 86 -6.576 -73.180 -62.737 1.00 56.33 A N
+ATOM 467 CA TYR A 86 -5.782 -72.020 -62.312 1.00 51.38 A C
+ATOM 468 C TYR A 86 -6.655 -70.918 -61.708 1.00 56.75 A C
+ATOM 469 O TYR A 86 -6.323 -70.353 -60.658 1.00 55.62 A O
+ATOM 470 CB TYR A 86 -4.994 -71.494 -63.518 1.00 50.36 A C
+ATOM 471 CG TYR A 86 -4.322 -70.133 -63.389 1.00 48.90 A C
+ATOM 472 CD1 TYR A 86 -2.947 -70.031 -63.273 1.00 50.17 A C
+ATOM 473 CD2 TYR A 86 -5.047 -68.959 -63.472 1.00 47.62 A C
+ATOM 474 CE1 TYR A 86 -2.323 -68.804 -63.182 1.00 51.47 A C
+ATOM 475 CE2 TYR A 86 -4.429 -67.733 -63.376 1.00 50.68 A C
+ATOM 476 CZ TYR A 86 -3.065 -67.657 -63.229 1.00 60.25 A C
+ATOM 477 OH TYR A 86 -2.435 -66.433 -63.124 1.00 58.20 A O
+ATOM 478 N LYS A 87 -7.752 -70.570 -62.387 1.00 56.70 A N
+ATOM 479 CA LYS A 87 -8.692 -69.571 -61.889 1.00 63.00 A C
+ATOM 480 C LYS A 87 -9.165 -69.907 -60.476 1.00 55.46 A C
+ATOM 481 O LYS A 87 -9.053 -69.084 -59.548 1.00 56.03 A O
+ATOM 482 CB LYS A 87 -9.883 -69.498 -62.841 1.00 64.43 A C
+ATOM 483 CG LYS A 87 -9.961 -68.277 -63.745 1.00 75.69 A C
+ATOM 484 CD LYS A 87 -11.097 -68.489 -64.749 1.00 89.93 A C
+ATOM 485 CE LYS A 87 -11.730 -67.195 -65.233 1.00 99.02 A C
+ATOM 486 NZ LYS A 87 -13.018 -67.500 -65.932 1.00101.03 A N1+
+ATOM 487 N ASN A 88 -9.693 -71.127 -60.297 1.00 54.89 A N
+ATOM 488 CA ASN A 88 -10.213 -71.545 -58.995 1.00 61.66 A C
+ATOM 489 C ASN A 88 -9.112 -71.638 -57.953 1.00 61.62 A C
+ATOM 490 O ASN A 88 -9.369 -71.490 -56.745 1.00 61.13 A O
+ATOM 491 CB ASN A 88 -10.916 -72.886 -59.127 1.00 62.31 A C
+ATOM 492 CG ASN A 88 -12.013 -72.861 -60.166 1.00 72.26 A C
+ATOM 493 ND2 ASN A 88 -12.342 -74.040 -60.697 1.00 72.00 A N
+ATOM 494 OD1 ASN A 88 -12.574 -71.797 -60.487 1.00 66.87 A O
+ATOM 495 N ALA A 89 -7.888 -71.901 -58.405 1.00 54.56 A N
+ATOM 496 CA ALA A 89 -6.741 -71.852 -57.518 1.00 53.21 A C
+ATOM 497 C ALA A 89 -6.542 -70.447 -56.988 1.00 56.09 A C
+ATOM 498 O ALA A 89 -6.424 -70.234 -55.774 1.00 62.78 A O
+ATOM 499 CB ALA A 89 -5.505 -72.322 -58.272 1.00 43.16 A C
+ATOM 500 N VAL A 90 -6.499 -69.475 -57.898 1.00 52.35 A N
+ATOM 501 CA VAL A 90 -6.356 -68.084 -57.494 1.00 55.84 A C
+ATOM 502 C VAL A 90 -7.380 -67.761 -56.424 1.00 55.87 A C
+ATOM 503 O VAL A 90 -7.038 -67.356 -55.301 1.00 56.74 A O
+ATOM 504 CB VAL A 90 -6.523 -67.152 -58.703 1.00 55.67 A C
+ATOM 505 CG1 VAL A 90 -6.586 -65.707 -58.257 1.00 47.29 A C
+ATOM 506 CG2 VAL A 90 -5.402 -67.351 -59.691 1.00 44.01 A C
+ATOM 507 N THR A 91 -8.661 -67.996 -56.752 1.00 53.68 A N
+ATOM 508 CA THR A 91 -9.745 -67.499 -55.899 1.00 51.63 A C
+ATOM 509 C THR A 91 -9.750 -68.213 -54.546 1.00 57.89 A C
+ATOM 510 O THR A 91 -9.894 -67.569 -53.491 1.00 54.28 A O
+ATOM 511 CB THR A 91 -11.074 -67.639 -56.644 1.00 55.81 A C
+ATOM 512 CG2 THR A 91 -11.044 -66.798 -57.955 1.00 53.66 A C
+ATOM 513 OG1 THR A 91 -11.281 -69.011 -56.989 1.00 66.14 A O
+ATOM 514 N GLU A 92 -9.520 -69.532 -54.561 1.00 49.94 A N
+ATOM 515 CA GLU A 92 -9.249 -70.280 -53.332 1.00 55.71 A C
+ATOM 516 C GLU A 92 -8.229 -69.536 -52.479 1.00 59.84 A C
+ATOM 517 O GLU A 92 -8.479 -69.233 -51.303 1.00 57.34 A O
+ATOM 518 CB GLU A 92 -8.722 -71.687 -53.703 1.00 58.78 A C
+ATOM 519 CG GLU A 92 -9.155 -72.913 -52.834 1.00 71.68 A C
+ATOM 520 CD GLU A 92 -8.777 -74.284 -53.476 1.00 88.76 A C
+ATOM 521 OE1 GLU A 92 -7.934 -74.995 -52.889 1.00105.43 A O
+ATOM 522 OE2 GLU A 92 -9.305 -74.648 -54.563 1.00 85.08 A O1-
+ATOM 523 N LEU A 93 -7.096 -69.170 -53.096 1.00 57.83 A N
+ATOM 524 CA LEU A 93 -6.012 -68.517 -52.374 1.00 52.66 A C
+ATOM 525 C LEU A 93 -6.427 -67.161 -51.842 1.00 54.85 A C
+ATOM 526 O LEU A 93 -5.937 -66.745 -50.790 1.00 57.29 A O
+ATOM 527 CB LEU A 93 -4.786 -68.360 -53.271 1.00 54.27 A C
+ATOM 528 CG LEU A 93 -4.001 -69.606 -53.684 1.00 56.33 A C
+ATOM 529 CD1 LEU A 93 -2.777 -69.204 -54.494 1.00 54.90 A C
+ATOM 530 CD2 LEU A 93 -3.587 -70.411 -52.453 1.00 50.90 A C
+ATOM 531 N GLN A 94 -7.310 -66.455 -52.548 1.00 49.93 A N
+ATOM 532 CA GLN A 94 -7.794 -65.171 -52.048 1.00 53.92 A C
+ATOM 533 C GLN A 94 -8.627 -65.346 -50.770 1.00 59.86 A C
+ATOM 534 O GLN A 94 -8.390 -64.665 -49.751 1.00 62.66 A O
+ATOM 535 CB GLN A 94 -8.582 -64.471 -53.158 1.00 50.17 A C
+ATOM 536 CG GLN A 94 -7.688 -63.855 -54.238 1.00 64.01 A C
+ATOM 537 CD GLN A 94 -8.369 -63.702 -55.597 1.00 75.15 A C
+ATOM 538 NE2 GLN A 94 -7.729 -62.961 -56.496 1.00 71.70 A N
+ATOM 539 OE1 GLN A 94 -9.456 -64.232 -55.832 1.00 71.18 A O
+ATOM 540 N LEU A 95 -9.598 -66.273 -50.799 1.00 57.53 A N
+ATOM 541 CA LEU A 95 -10.290 -66.670 -49.569 1.00 59.85 A C
+ATOM 542 C LEU A 95 -9.320 -67.002 -48.444 1.00 64.30 A C
+ATOM 543 O LEU A 95 -9.582 -66.681 -47.279 1.00 81.00 A O
+ATOM 544 CB LEU A 95 -11.173 -67.885 -49.819 1.00 73.91 A C
+ATOM 545 CG LEU A 95 -12.663 -67.600 -49.816 1.00 91.72 A C
+ATOM 546 CD1 LEU A 95 -13.032 -66.786 -51.061 1.00 93.63 A C
+ATOM 547 CD2 LEU A 95 -13.428 -68.914 -49.748 1.00 94.17 A C
+ATOM 548 N LEU A 96 -8.218 -67.694 -48.766 1.00 52.85 A N
+ATOM 549 CA LEU A 96 -7.191 -67.965 -47.761 1.00 58.62 A C
+ATOM 550 C LEU A 96 -6.566 -66.681 -47.236 1.00 57.35 A C
+ATOM 551 O LEU A 96 -6.270 -66.564 -46.037 1.00 68.21 A O
+ATOM 552 CB LEU A 96 -6.122 -68.878 -48.340 1.00 57.96 A C
+ATOM 553 CG LEU A 96 -6.316 -70.232 -47.698 1.00 51.77 A C
+ATOM 554 CD1 LEU A 96 -5.413 -71.236 -48.307 1.00 53.12 A C
+ATOM 555 CD2 LEU A 96 -5.994 -70.075 -46.237 1.00 53.37 A C
+ATOM 556 N MET A 97 -6.370 -65.701 -48.122 1.00 57.22 A N
+ATOM 557 CA MET A 97 -5.659 -64.481 -47.759 1.00 64.06 A C
+ATOM 558 C MET A 97 -6.455 -63.632 -46.796 1.00 77.68 A C
+ATOM 559 O MET A 97 -5.864 -62.958 -45.946 1.00 85.16 A O
+ATOM 560 CB MET A 97 -5.331 -63.671 -49.009 1.00 65.38 A C
+ATOM 561 CG MET A 97 -4.719 -62.305 -48.750 1.00 68.22 A C
+ATOM 562 SD MET A 97 -4.674 -61.333 -50.287 1.00 70.80 A S
+ATOM 563 CE MET A 97 -6.382 -61.494 -50.862 1.00 60.61 A C
+ATOM 564 N GLN A 98 -7.787 -63.645 -46.917 1.00 79.91 A N
+ATOM 565 CA GLN A 98 -8.593 -62.778 -46.055 1.00 84.50 A C
+ATOM 566 C GLN A 98 -8.407 -63.115 -44.568 1.00 86.96 A C
+ATOM 567 O GLN A 98 -8.212 -62.213 -43.743 1.00 89.87 A O
+ATOM 568 CB GLN A 98 -10.070 -62.833 -46.479 1.00 87.44 A C
+ATOM 569 CG GLN A 98 -10.840 -64.101 -46.087 1.00 93.01 A C
+ATOM 570 CD GLN A 98 -12.354 -63.904 -46.074 1.00 93.78 A C
+ATOM 571 NE2 GLN A 98 -13.076 -64.871 -45.497 1.00 85.57 A N
+ATOM 572 OE1 GLN A 98 -12.868 -62.906 -46.595 1.00 99.01 A O
+ATOM 573 N SER A 99 -8.419 -64.399 -44.210 1.00 99.34 A N
+ATOM 574 CA SER A 99 -8.332 -64.795 -42.806 1.00 98.13 A C
+ATOM 575 C SER A 99 -6.883 -64.799 -42.332 1.00 90.68 A C
+ATOM 576 O SER A 99 -5.992 -65.301 -43.028 1.00 73.63 A O
+ATOM 577 CB SER A 99 -8.961 -66.176 -42.609 1.00100.12 A C
+ATOM 578 OG SER A 99 -8.462 -67.115 -43.557 1.00105.15 A O
+ATOM 579 N THR A 100 -6.642 -64.231 -41.142 1.00 86.89 A N
+ATOM 580 CA THR A 100 -5.253 -64.104 -40.689 1.00 92.48 A C
+ATOM 581 C THR A 100 -4.767 -65.464 -40.182 1.00 90.13 A C
+ATOM 582 O THR A 100 -5.496 -66.150 -39.457 1.00 91.98 A O
+ATOM 583 CB THR A 100 -5.113 -62.983 -39.622 1.00 90.24 A C
+ATOM 584 CG2 THR A 100 -5.517 -63.444 -38.250 1.00 79.79 A C
+ATOM 585 OG1 THR A 100 -3.757 -62.530 -39.544 1.00 92.53 A O
+ATOM 586 N PRO A 101 -3.587 -65.917 -40.596 1.00 95.71 A N
+ATOM 587 CA PRO A 101 -3.237 -67.329 -40.430 1.00 80.53 A C
+ATOM 588 C PRO A 101 -2.810 -67.647 -39.009 1.00 74.48 A C
+ATOM 589 O PRO A 101 -2.197 -66.827 -38.318 1.00 68.26 A O
+ATOM 590 CB PRO A 101 -2.065 -67.511 -41.400 1.00 74.06 A C
+ATOM 591 CG PRO A 101 -1.460 -66.129 -41.527 1.00 77.69 A C
+ATOM 592 CD PRO A 101 -2.408 -65.110 -40.943 1.00 82.55 A C
+ATOM 593 N ALA A 102 -3.116 -68.870 -38.579 1.00 72.87 A N
+ATOM 594 CA ALA A 102 -2.655 -69.282 -37.263 1.00 77.74 A C
+ATOM 595 C ALA A 102 -2.498 -70.791 -37.212 1.00 73.71 A C
+ATOM 596 O ALA A 102 -3.004 -71.526 -38.066 1.00 77.89 A O
+ATOM 597 CB ALA A 102 -3.598 -68.805 -36.160 1.00 73.26 A C
+ATOM 598 N THR A 103 -1.779 -71.231 -36.182 1.00 76.63 A N
+ATOM 599 CA THR A 103 -1.452 -72.627 -35.948 1.00 80.58 A C
+ATOM 600 C THR A 103 -1.238 -72.845 -34.463 1.00 91.64 A C
+ATOM 601 O THR A 103 -1.034 -71.895 -33.704 1.00 86.59 A O
+ATOM 602 CB THR A 103 -0.188 -73.052 -36.688 1.00 74.90 A C
+ATOM 603 CG2 THR A 103 0.919 -72.065 -36.460 1.00 76.73 A C
+ATOM 604 OG1 THR A 103 0.229 -74.325 -36.187 1.00 63.67 A O
+ATOM 605 N ASN A 104 -1.274 -74.115 -34.056 1.00 90.50 A N
+ATOM 606 CA ASN A 104 -0.868 -74.521 -32.711 1.00 96.50 A C
+ATOM 607 C ASN A 104 0.292 -75.507 -32.825 1.00 91.68 A C
+ATOM 608 O ASN A 104 0.100 -76.646 -33.268 1.00 93.60 A O
+ATOM 609 CB ASN A 104 -2.035 -75.135 -31.932 1.00 97.34 A C
+ATOM 610 CG ASN A 104 -3.158 -74.145 -31.686 1.00 95.57 A C
+ATOM 611 ND2 ASN A 104 -3.308 -73.708 -30.432 1.00 87.53 A N
+ATOM 612 OD1 ASN A 104 -3.853 -73.738 -32.619 1.00 99.47 A O
+ATOM 613 N ASN A 105 1.489 -75.076 -32.414 1.00 86.80 A N
+ATOM 614 CA ASN A 105 2.669 -75.956 -32.414 1.00 81.65 A C
+ATOM 615 C ASN A 105 3.643 -75.622 -31.269 1.00 85.89 A C
+ATOM 616 O ASN A 105 3.338 -75.813 -30.081 1.00 84.49 A O
+ATOM 617 CB ASN A 105 3.410 -75.881 -33.758 1.00 78.97 A C
+ATOM 618 CG ASN A 105 2.774 -76.753 -34.840 1.00 88.86 A C
+ATOM 619 ND2 ASN A 105 1.807 -76.194 -35.551 1.00 84.27 A N
+ATOM 620 OD1 ASN A 105 3.177 -77.896 -35.056 1.00 95.34 A O
+ATOM 621 N SER A 134 3.529 -83.596 -17.837 1.00 99.06 A N
+ATOM 622 CA SER A 134 2.557 -84.576 -18.332 1.00102.75 A C
+ATOM 623 C SER A 134 2.610 -85.879 -17.517 1.00116.09 A C
+ATOM 624 O SER A 134 1.927 -86.043 -16.501 1.00139.31 A O
+ATOM 625 CB SER A 134 2.808 -84.870 -19.830 1.00 92.38 A C
+ATOM 626 OG SER A 134 2.063 -85.995 -20.294 1.00 77.34 A O
+ATOM 627 N GLY A 135 3.459 -86.784 -17.994 1.00107.84 A N
+ATOM 628 CA GLY A 135 3.731 -88.096 -17.443 1.00 97.60 A C
+ATOM 629 C GLY A 135 4.903 -88.069 -16.483 1.00 96.29 A C
+ATOM 630 O GLY A 135 4.938 -87.301 -15.514 1.00 94.12 A O
+ATOM 631 N ARG A 136 5.818 -89.007 -16.706 1.00104.86 A N
+ATOM 632 CA ARG A 136 7.122 -89.053 -16.059 1.00 92.96 A C
+ATOM 633 C ARG A 136 8.110 -88.076 -16.681 1.00 90.13 A C
+ATOM 634 O ARG A 136 9.306 -88.157 -16.388 1.00 92.59 A O
+ATOM 635 CB ARG A 136 7.697 -90.467 -16.126 1.00 85.22 A C
+ATOM 636 CG ARG A 136 7.284 -91.270 -14.939 1.00 94.41 A C
+ATOM 637 CD ARG A 136 7.982 -92.593 -14.810 1.00 92.45 A C
+ATOM 638 NE ARG A 136 7.329 -93.687 -15.526 1.00 87.28 A N
+ATOM 639 CZ ARG A 136 7.325 -94.947 -15.088 1.00 91.76 A C
+ATOM 640 NH1 ARG A 136 7.887 -95.228 -13.918 1.00 88.68 A N1+
+ATOM 641 NH2 ARG A 136 6.742 -95.919 -15.786 1.00 88.55 A N
+ATOM 642 N SER A 137 7.644 -87.211 -17.579 1.00 98.45 A N
+ATOM 643 CA SER A 137 8.438 -86.156 -18.194 1.00 89.59 A C
+ATOM 644 C SER A 137 8.169 -84.837 -17.484 1.00 80.24 A C
+ATOM 645 O SER A 137 7.109 -84.633 -16.886 1.00 82.88 A O
+ATOM 646 CB SER A 137 8.133 -85.985 -19.692 1.00 85.81 A C
+ATOM 647 OG SER A 137 6.983 -85.168 -19.934 1.00 90.41 A O
+ATOM 648 N LEU A 138 9.162 -83.959 -17.528 1.00 81.52 A N
+ATOM 649 CA LEU A 138 9.068 -82.627 -16.948 1.00 77.06 A C
+ATOM 650 C LEU A 138 8.376 -81.621 -17.858 1.00 71.83 A C
+ATOM 651 O LEU A 138 8.443 -80.419 -17.584 1.00 70.63 A O
+ATOM 652 CB LEU A 138 10.451 -82.115 -16.577 1.00 68.90 A C
+ATOM 653 CG LEU A 138 10.937 -82.736 -15.274 1.00 70.54 A C
+ATOM 654 CD1 LEU A 138 12.157 -81.990 -14.790 1.00 65.62 A C
+ATOM 655 CD2 LEU A 138 9.834 -82.752 -14.221 1.00 72.67 A C
+ATOM 656 N GLY A 139 7.768 -82.076 -18.951 1.00 73.77 A N
+ATOM 657 CA GLY A 139 7.177 -81.154 -19.910 1.00 66.17 A C
+ATOM 658 C GLY A 139 6.318 -80.056 -19.302 1.00 76.38 A C
+ATOM 659 O GLY A 139 6.438 -78.885 -19.682 1.00 84.12 A O
+ATOM 660 N PHE A 140 5.450 -80.406 -18.339 1.00 63.43 A N
+ATOM 661 CA PHE A 140 4.500 -79.421 -17.820 1.00 69.13 A C
+ATOM 662 C PHE A 140 5.197 -78.157 -17.344 1.00 75.66 A C
+ATOM 663 O PHE A 140 4.548 -77.113 -17.222 1.00 77.24 A O
+ATOM 664 CB PHE A 140 3.663 -80.003 -16.668 1.00 66.95 A C
+ATOM 665 CG PHE A 140 4.472 -80.368 -15.458 1.00 76.57 A C
+ATOM 666 CD1 PHE A 140 4.946 -81.658 -15.293 1.00 81.32 A C
+ATOM 667 CD2 PHE A 140 4.769 -79.422 -14.486 1.00 85.94 A C
+ATOM 668 CE1 PHE A 140 5.705 -81.992 -14.187 1.00 92.64 A C
+ATOM 669 CE2 PHE A 140 5.525 -79.759 -13.379 1.00 92.59 A C
+ATOM 670 CZ PHE A 140 5.992 -81.037 -13.229 1.00 93.35 A C
+ATOM 671 N LEU A 141 6.507 -78.230 -17.083 1.00 72.08 A N
+ATOM 672 CA LEU A 141 7.290 -77.078 -16.652 1.00 77.10 A C
+ATOM 673 C LEU A 141 7.603 -76.111 -17.788 1.00 74.90 A C
+ATOM 674 O LEU A 141 8.103 -75.011 -17.528 1.00 72.01 A O
+ATOM 675 CB LEU A 141 8.603 -77.541 -16.022 1.00 70.41 A C
+ATOM 676 CG LEU A 141 8.514 -78.326 -14.718 1.00 80.35 A C
+ATOM 677 CD1 LEU A 141 9.897 -78.830 -14.345 1.00 78.54 A C
+ATOM 678 CD2 LEU A 141 7.935 -77.458 -13.635 1.00 66.09 A C
+ATOM 679 N LEU A 142 7.318 -76.482 -19.028 1.00 69.72 A N
+ATOM 680 CA LEU A 142 7.776 -75.684 -20.149 1.00 72.95 A C
+ATOM 681 C LEU A 142 6.986 -74.385 -20.287 1.00 74.09 A C
+ATOM 682 O LEU A 142 5.852 -74.247 -19.813 1.00 77.23 A O
+ATOM 683 CB LEU A 142 7.682 -76.494 -21.432 1.00 64.30 A C
+ATOM 684 CG LEU A 142 8.915 -77.356 -21.603 1.00 74.87 A C
+ATOM 685 CD1 LEU A 142 8.717 -78.290 -22.746 1.00 67.74 A C
+ATOM 686 CD2 LEU A 142 10.063 -76.433 -21.884 1.00 66.01 A C
+ATOM 687 N GLY A 143 7.614 -73.422 -20.953 1.00 67.18 A N
+ATOM 688 CA GLY A 143 6.925 -72.222 -21.359 1.00 66.44 A C
+ATOM 689 C GLY A 143 6.042 -72.445 -22.576 1.00 70.48 A C
+ATOM 690 O GLY A 143 5.958 -73.530 -23.149 1.00 71.80 A O
+ATOM 691 N VAL A 144 5.381 -71.370 -22.985 1.00 69.07 A N
+ATOM 692 CA VAL A 144 4.441 -71.371 -24.097 1.00 58.78 A C
+ATOM 693 C VAL A 144 4.774 -70.223 -25.033 1.00 63.40 A C
+ATOM 694 O VAL A 144 4.916 -69.080 -24.583 1.00 68.13 A O
+ATOM 695 CB VAL A 144 3.002 -71.244 -23.587 1.00 77.99 A C
+ATOM 696 CG1 VAL A 144 2.120 -70.687 -24.655 1.00 59.68 A C
+ATOM 697 CG2 VAL A 144 2.506 -72.607 -23.228 1.00 62.67 A C
+ATOM 698 N GLY A 145 4.813 -70.502 -26.347 1.00 59.52 A N
+ATOM 699 CA GLY A 145 5.299 -69.526 -27.300 1.00 60.02 A C
+ATOM 700 C GLY A 145 4.503 -69.551 -28.587 1.00 67.01 A C
+ATOM 701 O GLY A 145 3.720 -70.463 -28.845 1.00 65.49 A O
+ATOM 702 N SER A 146 4.678 -68.503 -29.367 1.00 69.50 A N
+ATOM 703 CA SER A 146 3.980 -68.364 -30.635 1.00 56.98 A C
+ATOM 704 C SER A 146 4.955 -68.693 -31.747 1.00 60.07 A C
+ATOM 705 O SER A 146 5.792 -67.860 -32.094 1.00 61.96 A O
+ATOM 706 CB SER A 146 3.447 -66.947 -30.805 1.00 64.37 A C
+ATOM 707 OG SER A 146 2.915 -66.781 -32.099 1.00 64.41 A O
+ATOM 708 N ALA A 147 4.812 -69.872 -32.354 1.00 59.18 A N
+ATOM 709 CA ALA A 147 5.850 -70.321 -33.276 1.00 56.41 A C
+ATOM 710 C ALA A 147 5.973 -69.364 -34.468 1.00 59.84 A C
+ATOM 711 O ALA A 147 7.063 -68.850 -34.766 1.00 57.49 A O
+ATOM 712 CB ALA A 147 5.573 -71.760 -33.723 1.00 56.84 A C
+ATOM 713 N ILE A 148 4.856 -69.060 -35.121 1.00 55.88 A N
+ATOM 714 CA ILE A 148 4.931 -68.374 -36.404 1.00 56.49 A C
+ATOM 715 C ILE A 148 4.832 -66.880 -36.194 1.00 56.08 A C
+ATOM 716 O ILE A 148 4.647 -66.137 -37.164 1.00 60.01 A O
+ATOM 717 CB ILE A 148 3.850 -68.842 -37.387 1.00 54.24 A C
+ATOM 718 CG1 ILE A 148 2.472 -68.659 -36.748 1.00 58.56 A C
+ATOM 719 CG2 ILE A 148 4.143 -70.276 -37.844 1.00 53.15 A C
+ATOM 720 CD1 ILE A 148 1.303 -68.812 -37.716 1.00 57.65 A C
+ATOM 721 N ALA A 149 4.897 -66.436 -34.933 1.00 49.00 A N
+ATOM 722 CA ALA A 149 4.696 -65.016 -34.654 1.00 49.78 A C
+ATOM 723 C ALA A 149 5.534 -64.142 -35.583 1.00 55.77 A C
+ATOM 724 O ALA A 149 5.003 -63.250 -36.262 1.00 55.77 A O
+ATOM 725 CB ALA A 149 5.013 -64.713 -33.188 1.00 50.84 A C
+ATOM 726 N SER A 150 6.839 -64.421 -35.663 1.00 48.94 A N
+ATOM 727 CA SER A 150 7.759 -63.696 -36.536 1.00 48.48 A C
+ATOM 728 C SER A 150 7.236 -63.630 -37.970 1.00 50.09 A C
+ATOM 729 O SER A 150 6.852 -62.556 -38.477 1.00 53.46 A O
+ATOM 730 CB SER A 150 9.131 -64.374 -36.507 1.00 50.46 A C
+ATOM 731 OG SER A 150 8.999 -65.784 -36.387 1.00 56.77 A O
+ATOM 732 N GLY A 151 7.206 -64.801 -38.614 1.00 53.10 A N
+ATOM 733 CA GLY A 151 6.700 -64.882 -39.969 1.00 45.98 A C
+ATOM 734 C GLY A 151 5.374 -64.171 -40.153 1.00 57.53 A C
+ATOM 735 O GLY A 151 5.160 -63.496 -41.163 1.00 54.50 A O
+ATOM 736 N VAL A 152 4.471 -64.282 -39.173 1.00 58.82 A N
+ATOM 737 CA VAL A 152 3.174 -63.636 -39.345 1.00 48.15 A C
+ATOM 738 C VAL A 152 3.328 -62.124 -39.356 1.00 54.39 A C
+ATOM 739 O VAL A 152 2.646 -61.432 -40.119 1.00 63.19 A O
+ATOM 740 CB VAL A 152 2.166 -64.083 -38.276 1.00 52.20 A C
+ATOM 741 CG1 VAL A 152 0.829 -63.372 -38.506 1.00 49.31 A C
+ATOM 742 CG2 VAL A 152 1.974 -65.559 -38.338 1.00 45.44 A C
+ATOM 743 N ALA A 153 4.218 -61.581 -38.519 1.00 49.50 A N
+ATOM 744 CA ALA A 153 4.473 -60.144 -38.574 1.00 49.43 A C
+ATOM 745 C ALA A 153 4.936 -59.716 -39.968 1.00 53.98 A C
+ATOM 746 O ALA A 153 4.480 -58.690 -40.499 1.00 62.94 A O
+ATOM 747 CB ALA A 153 5.499 -59.747 -37.514 1.00 54.25 A C
+ATOM 748 N VAL A 154 5.819 -60.502 -40.591 1.00 50.21 A N
+ATOM 749 CA VAL A 154 6.226 -60.182 -41.963 1.00 54.89 A C
+ATOM 750 C VAL A 154 5.026 -60.240 -42.913 1.00 52.46 A C
+ATOM 751 O VAL A 154 4.816 -59.338 -43.742 1.00 55.68 A O
+ATOM 752 CB VAL A 154 7.355 -61.120 -42.420 1.00 50.70 A C
+ATOM 753 CG1 VAL A 154 7.821 -60.739 -43.794 1.00 42.79 A C
+ATOM 754 CG2 VAL A 154 8.501 -61.047 -41.442 1.00 45.08 A C
+ATOM 755 N SER A 155 4.227 -61.310 -42.815 1.00 43.62 A N
+ATOM 756 CA SER A 155 3.015 -61.429 -43.619 1.00 54.21 A C
+ATOM 757 C SER A 155 2.143 -60.192 -43.501 1.00 57.16 A C
+ATOM 758 O SER A 155 1.634 -59.678 -44.502 1.00 65.83 A O
+ATOM 759 CB SER A 155 2.222 -62.659 -43.195 1.00 50.03 A C
+ATOM 760 OG SER A 155 0.856 -62.515 -43.543 1.00 58.33 A O
+ATOM 761 N LYS A 156 1.941 -59.706 -42.284 1.00 51.55 A N
+ATOM 762 CA LYS A 156 1.235 -58.446 -42.143 1.00 54.59 A C
+ATOM 763 C LYS A 156 1.927 -57.375 -42.973 1.00 57.08 A C
+ATOM 764 O LYS A 156 1.314 -56.810 -43.882 1.00 61.09 A O
+ATOM 765 CB LYS A 156 1.127 -58.046 -40.669 1.00 55.63 A C
+ATOM 766 CG LYS A 156 0.505 -59.135 -39.745 1.00 79.38 A C
+ATOM 767 CD LYS A 156 -1.028 -59.260 -39.831 1.00102.05 A C
+ATOM 768 CE LYS A 156 -1.586 -60.114 -38.673 1.00111.09 A C
+ATOM 769 NZ LYS A 156 -3.082 -60.254 -38.652 1.00109.62 A N1+
+ATOM 770 N VAL A 157 3.240 -57.176 -42.760 1.00 47.86 A N
+ATOM 771 CA VAL A 157 3.970 -56.087 -43.432 1.00 52.04 A C
+ATOM 772 C VAL A 157 3.765 -56.103 -44.943 1.00 58.00 A C
+ATOM 773 O VAL A 157 3.801 -55.049 -45.593 1.00 76.55 A O
+ATOM 774 CB VAL A 157 5.479 -56.138 -43.090 1.00 58.81 A C
+ATOM 775 CG1 VAL A 157 6.280 -55.134 -43.934 1.00 63.40 A C
+ATOM 776 CG2 VAL A 157 5.711 -55.883 -41.620 1.00 64.04 A C
+ATOM 777 N LEU A 158 3.530 -57.286 -45.526 1.00 62.93 A N
+ATOM 778 CA LEU A 158 3.366 -57.368 -46.982 1.00 58.29 A C
+ATOM 779 C LEU A 158 2.054 -56.737 -47.460 1.00 70.17 A C
+ATOM 780 O LEU A 158 2.012 -56.145 -48.551 1.00 70.23 A O
+ATOM 781 CB LEU A 158 3.442 -58.819 -47.459 1.00 55.93 A C
+ATOM 782 CG LEU A 158 4.732 -59.597 -47.235 1.00 58.15 A C
+ATOM 783 CD1 LEU A 158 4.651 -60.827 -48.056 1.00 53.74 A C
+ATOM 784 CD2 LEU A 158 5.958 -58.797 -47.636 1.00 59.25 A C
+ATOM 785 N HIS A 159 0.963 -56.881 -46.683 1.00 58.12 A N
+ATOM 786 CA HIS A 159 -0.321 -56.316 -47.108 1.00 61.96 A C
+ATOM 787 C HIS A 159 -0.250 -54.805 -47.252 1.00 76.30 A C
+ATOM 788 O HIS A 159 -0.965 -54.228 -48.082 1.00 91.05 A O
+ATOM 789 CB HIS A 159 -1.429 -56.628 -46.111 1.00 59.52 A C
+ATOM 790 CG HIS A 159 -1.868 -58.053 -46.083 1.00 63.24 A C
+ATOM 791 CD2 HIS A 159 -3.037 -58.628 -46.451 1.00 63.02 A C
+ATOM 792 ND1 HIS A 159 -1.077 -59.067 -45.587 1.00 78.41 A N
+ATOM 793 CE1 HIS A 159 -1.740 -60.208 -45.651 1.00 70.68 A C
+ATOM 794 NE2 HIS A 159 -2.927 -59.971 -46.182 1.00 59.22 A N
+ATOM 795 N LEU A 160 0.599 -54.162 -46.445 1.00 60.03 A N
+ATOM 796 CA LEU A 160 0.705 -52.710 -46.402 1.00 68.46 A C
+ATOM 797 C LEU A 160 0.807 -52.148 -47.808 1.00 79.88 A C
+ATOM 798 O LEU A 160 1.555 -52.668 -48.643 1.00 89.41 A O
+ATOM 799 CB LEU A 160 1.939 -52.306 -45.602 1.00 66.72 A C
+ATOM 800 CG LEU A 160 1.978 -52.302 -44.088 1.00 65.65 A C
+ATOM 801 CD1 LEU A 160 1.370 -53.527 -43.509 1.00 60.23 A C
+ATOM 802 CD2 LEU A 160 3.441 -52.251 -43.718 1.00 59.64 A C
+ATOM 803 N GLU A 161 0.028 -51.102 -48.075 1.00 77.11 A N
+ATOM 804 CA GLU A 161 -0.134 -50.662 -49.450 1.00 87.36 A C
+ATOM 805 C GLU A 161 1.216 -50.239 -50.016 1.00 78.14 A C
+ATOM 806 O GLU A 161 1.993 -49.538 -49.351 1.00 71.49 A O
+ATOM 807 CB GLU A 161 -1.162 -49.532 -49.543 1.00107.07 A C
+ATOM 808 CG GLU A 161 -1.814 -49.469 -50.917 1.00117.39 A C
+ATOM 809 CD GLU A 161 -2.302 -50.835 -51.393 1.00128.90 A C
+ATOM 810 OE1 GLU A 161 -2.903 -51.574 -50.578 1.00133.12 A O
+ATOM 811 OE2 GLU A 161 -2.053 -51.178 -52.574 1.00132.63 A O1-
+ATOM 812 N GLY A 162 1.513 -50.740 -51.227 1.00 83.85 A N
+ATOM 813 CA GLY A 162 2.754 -50.487 -51.928 1.00 83.99 A C
+ATOM 814 C GLY A 162 3.952 -51.284 -51.454 1.00 89.60 A C
+ATOM 815 O GLY A 162 5.036 -51.136 -52.035 1.00 93.14 A O
+ATOM 816 N GLU A 163 3.814 -52.106 -50.407 1.00 81.31 A N
+ATOM 817 CA GLU A 163 4.923 -52.976 -50.032 1.00 75.19 A C
+ATOM 818 C GLU A 163 5.193 -54.009 -51.117 1.00 73.62 A C
+ATOM 819 O GLU A 163 6.351 -54.254 -51.472 1.00 76.76 A O
+ATOM 820 CB GLU A 163 4.651 -53.652 -48.687 1.00 71.33 A C
+ATOM 821 CG GLU A 163 5.407 -53.009 -47.517 1.00 84.90 A C
+ATOM 822 CD GLU A 163 6.941 -53.069 -47.666 1.00 91.50 A C
+ATOM 823 OE1 GLU A 163 7.470 -54.107 -48.124 1.00 79.35 A O
+ATOM 824 OE2 GLU A 163 7.628 -52.071 -47.346 1.00101.98 A O1-
+ATOM 825 N VAL A 164 4.137 -54.604 -51.676 1.00 73.31 A N
+ATOM 826 CA VAL A 164 4.314 -55.572 -52.756 1.00 71.78 A C
+ATOM 827 C VAL A 164 5.197 -54.985 -53.849 1.00 72.91 A C
+ATOM 828 O VAL A 164 6.177 -55.603 -54.278 1.00 71.46 A O
+ATOM 829 CB VAL A 164 2.953 -56.008 -53.325 1.00 85.03 A C
+ATOM 830 CG1 VAL A 164 3.138 -57.141 -54.288 1.00 76.45 A C
+ATOM 831 CG2 VAL A 164 2.005 -56.388 -52.221 1.00 92.72 A C
+ATOM 832 N ASN A 165 4.881 -53.766 -54.291 1.00 72.62 A N
+ATOM 833 CA ASN A 165 5.603 -53.186 -55.419 1.00 80.72 A C
+ATOM 834 C ASN A 165 7.099 -53.051 -55.166 1.00 79.55 A C
+ATOM 835 O ASN A 165 7.882 -53.202 -56.107 1.00 74.76 A O
+ATOM 836 CB ASN A 165 5.011 -51.840 -55.787 1.00 94.37 A C
+ATOM 837 CG ASN A 165 3.890 -51.982 -56.744 1.00106.62 A C
+ATOM 838 ND2 ASN A 165 4.177 -52.570 -57.897 1.00104.39 A N
+ATOM 839 OD1 ASN A 165 2.764 -51.588 -56.458 1.00115.86 A O
+ATOM 840 N LYS A 166 7.524 -52.751 -53.931 1.00 76.41 A N
+ATOM 841 CA LYS A 166 8.950 -52.831 -53.619 1.00 73.03 A C
+ATOM 842 C LYS A 166 9.521 -54.179 -54.035 1.00 71.12 A C
+ATOM 843 O LYS A 166 10.584 -54.255 -54.660 1.00 71.71 A O
+ATOM 844 CB LYS A 166 9.210 -52.598 -52.129 1.00 67.34 A C
+ATOM 845 CG LYS A 166 8.993 -51.192 -51.605 1.00 77.40 A C
+ATOM 846 CD LYS A 166 9.489 -51.131 -50.149 1.00 85.63 A C
+ATOM 847 CE LYS A 166 9.369 -49.734 -49.557 1.00 92.33 A C
+ATOM 848 NZ LYS A 166 9.843 -48.714 -50.550 1.00 96.83 A N1+
+ATOM 849 N ILE A 167 8.808 -55.256 -53.713 1.00 70.57 A N
+ATOM 850 CA ILE A 167 9.296 -56.596 -54.010 1.00 68.78 A C
+ATOM 851 C ILE A 167 9.264 -56.862 -55.511 1.00 69.41 A C
+ATOM 852 O ILE A 167 10.215 -57.417 -56.079 1.00 68.00 A O
+ATOM 853 CB ILE A 167 8.474 -57.627 -53.216 1.00 56.10 A C
+ATOM 854 CG1 ILE A 167 8.733 -57.452 -51.724 1.00 51.77 A C
+ATOM 855 CG2 ILE A 167 8.781 -59.043 -53.648 1.00 49.46 A C
+ATOM 856 CD1 ILE A 167 8.187 -58.581 -50.903 1.00 51.71 A C
+ATOM 857 N LYS A 168 8.181 -56.471 -56.183 1.00 62.94 A N
+ATOM 858 CA LYS A 168 8.133 -56.678 -57.624 1.00 63.49 A C
+ATOM 859 C LYS A 168 9.232 -55.895 -58.319 1.00 67.56 A C
+ATOM 860 O LYS A 168 9.778 -56.351 -59.329 1.00 69.95 A O
+ATOM 861 CB LYS A 168 6.748 -56.317 -58.176 1.00 67.36 A C
+ATOM 862 CG LYS A 168 5.812 -57.552 -58.291 1.00 78.06 A C
+ATOM 863 CD LYS A 168 4.333 -57.220 -58.538 1.00 79.94 A C
+ATOM 864 CE LYS A 168 4.088 -56.630 -59.905 1.00 78.97 A C
+ATOM 865 NZ LYS A 168 2.792 -55.925 -59.903 1.00 77.41 A N1+
+ATOM 866 N SER A 169 9.610 -54.751 -57.766 1.00 58.82 A N
+ATOM 867 CA SER A 169 10.664 -53.959 -58.374 1.00 66.64 A C
+ATOM 868 C SER A 169 12.048 -54.440 -57.992 1.00 63.94 A C
+ATOM 869 O SER A 169 13.018 -54.057 -58.646 1.00 69.99 A O
+ATOM 870 CB SER A 169 10.516 -52.485 -57.995 1.00 77.00 A C
+ATOM 871 OG SER A 169 9.431 -51.889 -58.693 1.00 83.83 A O
+ATOM 872 N ALA A 170 12.176 -55.251 -56.949 1.00 66.40 A N
+ATOM 873 CA ALA A 170 13.497 -55.789 -56.652 1.00 68.53 A C
+ATOM 874 C ALA A 170 13.862 -56.956 -57.553 1.00 71.84 A C
+ATOM 875 O ALA A 170 15.050 -57.259 -57.711 1.00 69.05 A O
+ATOM 876 CB ALA A 170 13.593 -56.233 -55.194 1.00 66.25 A C
+ATOM 877 N LEU A 171 12.877 -57.627 -58.131 1.00 74.73 A N
+ATOM 878 CA LEU A 171 13.130 -58.847 -58.874 1.00 69.58 A C
+ATOM 879 C LEU A 171 13.219 -58.635 -60.379 1.00 72.28 A C
+ATOM 880 O LEU A 171 13.093 -59.615 -61.128 1.00 68.31 A O
+ATOM 881 CB LEU A 171 12.090 -59.903 -58.533 1.00 55.03 A C
+ATOM 882 CG LEU A 171 12.777 -60.647 -57.390 1.00 52.13 A C
+ATOM 883 CD1 LEU A 171 12.568 -59.899 -56.067 1.00 56.70 A C
+ATOM 884 CD2 LEU A 171 12.397 -62.125 -57.268 1.00 56.44 A C
+ATOM 885 N LEU A 172 13.316 -57.382 -60.847 1.00 52.23 A N
+ATOM 886 CA LEU A 172 13.342 -57.128 -62.291 1.00 57.87 A C
+ATOM 887 C LEU A 172 14.557 -57.772 -62.960 1.00 69.54 A C
+ATOM 888 O LEU A 172 14.439 -58.424 -64.009 1.00 60.94 A O
+ATOM 889 CB LEU A 172 13.340 -55.623 -62.566 1.00 62.23 A C
+ATOM 890 CG LEU A 172 12.256 -54.724 -61.981 1.00 63.65 A C
+ATOM 891 CD1 LEU A 172 12.745 -53.322 -62.112 1.00 55.26 A C
+ATOM 892 CD2 LEU A 172 10.911 -54.868 -62.661 1.00 56.93 A C
+ATOM 893 N SER A 173 15.739 -57.552 -62.394 1.00 54.84 A N
+ATOM 894 CA SER A 173 16.993 -57.933 -63.018 1.00 59.62 A C
+ATOM 895 C SER A 173 17.563 -59.253 -62.484 1.00 49.88 A C
+ATOM 896 O SER A 173 18.711 -59.589 -62.796 1.00 50.62 A O
+ATOM 897 CB SER A 173 17.994 -56.800 -62.868 1.00 65.86 A C
+ATOM 898 OG SER A 173 18.915 -56.851 -63.933 1.00 70.17 A O
+ATOM 899 N THR A 174 16.760 -60.033 -61.757 1.00 65.92 A N
+ATOM 900 CA THR A 174 17.208 -61.340 -61.251 1.00 56.37 A C
+ATOM 901 C THR A 174 16.371 -61.860 -60.077 1.00 52.93 A C
+ATOM 902 O THR A 174 15.982 -61.076 -59.211 1.00 48.42 A O
+ATOM 903 CB THR A 174 18.690 -61.302 -60.830 1.00 52.33 A C
+ATOM 904 CG2 THR A 174 18.948 -60.131 -59.895 1.00 45.24 A C
+ATOM 905 OG1 THR A 174 19.032 -62.524 -60.164 1.00 58.69 A O
+ATOM 906 N ASN A 175 16.091 -63.167 -60.028 1.00 47.85 A N
+ATOM 907 CA ASN A 175 15.302 -63.666 -58.901 1.00 46.18 A C
+ATOM 908 C ASN A 175 16.147 -63.531 -57.611 1.00 47.66 A C
+ATOM 909 O ASN A 175 17.170 -62.830 -57.609 1.00 45.27 A O
+ATOM 910 CB ASN A 175 14.783 -65.060 -59.208 1.00 43.29 A C
+ATOM 911 CG ASN A 175 15.863 -66.003 -59.488 1.00 58.74 A C
+ATOM 912 ND2 ASN A 175 15.974 -67.018 -58.671 1.00 66.15 A N
+ATOM 913 OD1 ASN A 175 16.581 -65.855 -60.456 1.00 65.69 A O
+ATOM 914 N LYS A 176 15.703 -64.106 -56.474 1.00 43.49 A N
+ATOM 915 CA LYS A 176 16.529 -64.138 -55.251 1.00 43.59 A C
+ATOM 916 C LYS A 176 17.256 -62.817 -54.990 1.00 51.42 A C
+ATOM 917 O LYS A 176 18.458 -62.698 -55.248 1.00 74.13 A O
+ATOM 918 CB LYS A 176 17.561 -65.288 -55.286 1.00 44.61 A C
+ATOM 919 CG LYS A 176 18.136 -65.739 -53.899 1.00 48.09 A C
+ATOM 920 CD LYS A 176 19.585 -66.280 -53.963 1.00 60.58 A C
+ATOM 921 CE LYS A 176 19.815 -67.203 -55.206 1.00 61.48 A C
+ATOM 922 NZ LYS A 176 21.204 -67.815 -55.488 1.00 61.87 A N1+
+ATOM 923 N ALA A 177 16.549 -61.791 -54.542 1.00 59.61 A N
+ATOM 924 CA ALA A 177 17.250 -60.585 -54.146 1.00 56.48 A C
+ATOM 925 C ALA A 177 16.697 -60.058 -52.825 1.00 59.26 A C
+ATOM 926 O ALA A 177 15.570 -60.386 -52.424 1.00 63.92 A O
+ATOM 927 CB ALA A 177 17.155 -59.539 -55.246 1.00 49.89 A C
+ATOM 928 N VAL A 178 17.519 -59.227 -52.156 1.00 58.19 A N
+ATOM 929 CA VAL A 178 17.169 -58.590 -50.882 1.00 58.37 A C
+ATOM 930 C VAL A 178 16.230 -57.408 -51.070 1.00 64.12 A C
+ATOM 931 O VAL A 178 16.514 -56.471 -51.830 1.00 71.99 A O
+ATOM 932 CB VAL A 178 18.427 -58.159 -50.114 1.00 54.00 A C
+ATOM 933 CG1 VAL A 178 18.114 -56.989 -49.255 1.00 58.08 A C
+ATOM 934 CG2 VAL A 178 18.757 -59.217 -49.142 1.00 50.61 A C
+ATOM 935 N VAL A 179 15.136 -57.432 -50.312 1.00 62.66 A N
+ATOM 936 CA VAL A 179 14.237 -56.302 -50.155 1.00 65.68 A C
+ATOM 937 C VAL A 179 14.251 -55.890 -48.691 1.00 73.44 A C
+ATOM 938 O VAL A 179 14.188 -56.748 -47.802 1.00 78.07 A O
+ATOM 939 CB VAL A 179 12.812 -56.676 -50.595 1.00 62.19 A C
+ATOM 940 CG1 VAL A 179 11.966 -55.425 -50.773 1.00 58.77 A C
+ATOM 941 CG2 VAL A 179 12.866 -57.505 -51.849 1.00 60.78 A C
+ATOM 942 N SER A 180 14.342 -54.588 -48.434 1.00 66.05 A N
+ATOM 943 CA SER A 180 14.077 -54.054 -47.105 1.00 76.44 A C
+ATOM 944 C SER A 180 12.650 -53.532 -47.090 1.00 70.19 A C
+ATOM 945 O SER A 180 12.220 -52.858 -48.039 1.00 72.77 A O
+ATOM 946 CB SER A 180 15.065 -52.953 -46.743 1.00 91.73 A C
+ATOM 947 OG SER A 180 16.275 -53.534 -46.305 1.00 99.23 A O
+ATOM 948 N LEU A 181 11.902 -53.875 -46.046 1.00 80.56 A N
+ATOM 949 CA LEU A 181 10.484 -53.545 -46.047 1.00 85.88 A C
+ATOM 950 C LEU A 181 10.247 -52.202 -45.352 1.00 97.23 A C
+ATOM 951 O LEU A 181 11.163 -51.588 -44.796 1.00104.36 A O
+ATOM 952 CB LEU A 181 9.680 -54.676 -45.402 1.00 77.17 A C
+ATOM 953 CG LEU A 181 9.980 -56.075 -45.963 1.00 69.70 A C
+ATOM 954 CD1 LEU A 181 9.498 -57.178 -45.030 1.00 55.21 A C
+ATOM 955 CD2 LEU A 181 9.396 -56.271 -47.366 1.00 59.24 A C
+ATOM 956 N SER A 182 8.988 -51.751 -45.368 1.00 82.47 A N
+ATOM 957 CA SER A 182 8.647 -50.435 -44.834 1.00 85.73 A C
+ATOM 958 C SER A 182 9.146 -50.254 -43.412 1.00 85.42 A C
+ATOM 959 O SER A 182 9.585 -49.166 -43.034 1.00 94.88 A O
+ATOM 960 CB SER A 182 7.136 -50.234 -44.877 1.00 85.09 A C
+ATOM 961 OG SER A 182 6.650 -50.463 -46.182 1.00 88.04 A O
+ATOM 962 N ASN A 183 9.081 -51.304 -42.611 1.00 75.55 A N
+ATOM 963 CA ASN A 183 9.603 -51.281 -41.260 1.00 79.68 A C
+ATOM 964 C ASN A 183 11.083 -51.596 -41.223 1.00 91.16 A C
+ATOM 965 O ASN A 183 11.629 -51.840 -40.142 1.00 92.51 A O
+ATOM 966 CB ASN A 183 8.848 -52.266 -40.382 1.00 70.17 A C
+ATOM 967 CG ASN A 183 9.147 -53.702 -40.741 1.00 75.39 A C
+ATOM 968 ND2 ASN A 183 8.825 -54.611 -39.828 1.00 71.20 A N
+ATOM 969 OD1 ASN A 183 9.648 -53.997 -41.833 1.00 85.11 A O
+ATOM 970 N GLY A 184 11.717 -51.705 -42.383 1.00 88.83 A N
+ATOM 971 CA GLY A 184 13.155 -51.816 -42.400 1.00 87.75 A C
+ATOM 972 C GLY A 184 13.689 -53.145 -41.937 1.00 80.69 A C
+ATOM 973 O GLY A 184 14.900 -53.271 -41.735 1.00 85.83 A O
+ATOM 974 N VAL A 185 12.825 -54.133 -41.733 1.00 89.78 A N
+ATOM 975 CA VAL A 185 13.269 -55.515 -41.592 1.00 82.85 A C
+ATOM 976 C VAL A 185 13.592 -56.025 -42.989 1.00 76.18 A C
+ATOM 977 O VAL A 185 12.741 -55.980 -43.886 1.00 71.53 A O
+ATOM 978 CB VAL A 185 12.188 -56.373 -40.922 1.00 84.62 A C
+ATOM 979 CG1 VAL A 185 12.669 -57.795 -40.769 1.00 80.15 A C
+ATOM 980 CG2 VAL A 185 11.797 -55.782 -39.581 1.00 87.85 A C
+ATOM 981 N SER A 186 14.827 -56.473 -43.200 1.00 81.73 A N
+ATOM 982 CA SER A 186 15.215 -56.855 -44.552 1.00 78.98 A C
+ATOM 983 C SER A 186 15.165 -58.373 -44.686 1.00 67.77 A C
+ATOM 984 O SER A 186 15.499 -59.121 -43.759 1.00 72.58 A O
+ATOM 985 CB SER A 186 16.596 -56.312 -44.971 1.00 83.30 A C
+ATOM 986 OG SER A 186 17.649 -57.236 -44.766 1.00 85.27 A O
+ATOM 987 N VAL A 187 14.702 -58.809 -45.848 1.00 70.97 A N
+ATOM 988 CA VAL A 187 14.360 -60.194 -46.096 1.00 59.17 A C
+ATOM 989 C VAL A 187 14.789 -60.545 -47.510 1.00 66.51 A C
+ATOM 990 O VAL A 187 14.853 -59.686 -48.401 1.00 78.02 A O
+ATOM 991 CB VAL A 187 12.847 -60.442 -45.907 1.00 54.62 A C
+ATOM 992 CG1 VAL A 187 12.417 -60.101 -44.473 1.00 67.49 A C
+ATOM 993 CG2 VAL A 187 12.040 -59.630 -46.944 1.00 55.09 A C
+ATOM 994 N LEU A 188 15.123 -61.818 -47.690 1.00 55.31 A N
+ATOM 995 CA LEU A 188 15.295 -62.363 -49.021 1.00 47.69 A C
+ATOM 996 C LEU A 188 13.927 -62.523 -49.658 1.00 55.55 A C
+ATOM 997 O LEU A 188 12.951 -62.874 -48.984 1.00 52.33 A O
+ATOM 998 CB LEU A 188 16.001 -63.710 -48.952 1.00 47.72 A C
+ATOM 999 CG LEU A 188 17.444 -63.573 -48.526 1.00 71.76 A C
+ATOM 1000 CD1 LEU A 188 17.946 -64.899 -48.045 1.00 75.89 A C
+ATOM 1001 CD2 LEU A 188 18.242 -63.065 -49.692 1.00 73.84 A C
+ATOM 1002 N THR A 189 13.848 -62.227 -50.951 1.00 53.11 A N
+ATOM 1003 CA THR A 189 12.687 -62.578 -51.748 1.00 43.96 A C
+ATOM 1004 C THR A 189 13.141 -63.405 -52.929 1.00 44.61 A C
+ATOM 1005 O THR A 189 14.061 -63.003 -53.655 1.00 50.37 A O
+ATOM 1006 CB THR A 189 11.927 -61.359 -52.240 1.00 51.48 A C
+ATOM 1007 CG2 THR A 189 10.852 -61.774 -53.215 1.00 52.49 A C
+ATOM 1008 OG1 THR A 189 11.310 -60.727 -51.122 1.00 61.56 A O
+ATOM 1009 N SER A 190 12.488 -64.553 -53.116 1.00 45.31 A N
+ATOM 1010 CA SER A 190 12.760 -65.443 -54.234 1.00 47.19 A C
+ATOM 1011 C SER A 190 11.477 -65.661 -55.019 1.00 51.12 A C
+ATOM 1012 O SER A 190 10.424 -65.922 -54.422 1.00 56.62 A O
+ATOM 1013 CB SER A 190 13.320 -66.778 -53.746 1.00 47.59 A C
+ATOM 1014 OG SER A 190 13.464 -67.673 -54.837 1.00 62.41 A O
+ATOM 1015 N LYS A 191 11.561 -65.537 -56.354 1.00 55.72 A N
+ATOM 1016 CA LYS A 191 10.431 -65.843 -57.228 1.00 49.62 A C
+ATOM 1017 C LYS A 191 10.660 -67.213 -57.832 1.00 48.88 A C
+ATOM 1018 O LYS A 191 11.502 -67.368 -58.719 1.00 47.90 A O
+ATOM 1019 CB LYS A 191 10.256 -64.824 -58.347 1.00 48.28 A C
+ATOM 1020 CG LYS A 191 9.240 -65.286 -59.374 1.00 46.34 A C
+ATOM 1021 CD LYS A 191 9.432 -64.599 -60.698 1.00 53.83 A C
+ATOM 1022 CE LYS A 191 8.817 -63.217 -60.748 1.00 55.84 A C
+ATOM 1023 NZ LYS A 191 9.119 -62.580 -62.075 1.00 64.35 A N1+
+ATOM 1024 N VAL A 192 9.890 -68.208 -57.384 1.00 46.18 A N
+ATOM 1025 CA VAL A 192 10.135 -69.545 -57.900 1.00 54.34 A C
+ATOM 1026 C VAL A 192 9.272 -69.905 -59.094 1.00 55.48 A C
+ATOM 1027 O VAL A 192 9.516 -70.948 -59.713 1.00 52.68 A O
+ATOM 1028 CB VAL A 192 9.954 -70.612 -56.811 1.00 51.07 A C
+ATOM 1029 CG1 VAL A 192 10.741 -70.233 -55.620 1.00 43.88 A C
+ATOM 1030 CG2 VAL A 192 8.544 -70.687 -56.435 1.00 42.01 A C
+ATOM 1031 N LEU A 193 8.262 -69.115 -59.442 1.00 55.49 A N
+ATOM 1032 CA LEU A 193 7.518 -69.492 -60.636 1.00 52.45 A C
+ATOM 1033 C LEU A 193 6.975 -68.278 -61.394 1.00 56.59 A C
+ATOM 1034 O LEU A 193 6.317 -67.418 -60.795 1.00 58.81 A O
+ATOM 1035 CB LEU A 193 6.402 -70.433 -60.235 1.00 52.73 A C
+ATOM 1036 CG LEU A 193 5.666 -70.897 -61.457 1.00 50.89 A C
+ATOM 1037 CD1 LEU A 193 6.724 -71.458 -62.418 1.00 58.17 A C
+ATOM 1038 CD2 LEU A 193 4.625 -71.927 -61.015 1.00 45.98 A C
+ATOM 1039 N ASP A 194 7.213 -68.217 -62.711 1.00 58.41 A N
+ATOM 1040 CA ASP A 194 6.880 -67.034 -63.506 1.00 58.26 A C
+ATOM 1041 C ASP A 194 5.551 -67.135 -64.263 1.00 56.72 A C
+ATOM 1042 O ASP A 194 5.395 -66.470 -65.296 1.00 61.09 A O
+ATOM 1043 CB ASP A 194 8.001 -66.656 -64.462 1.00 59.04 A C
+ATOM 1044 CG ASP A 194 8.032 -65.139 -64.736 1.00 66.76 A C
+ATOM 1045 OD1 ASP A 194 6.985 -64.462 -64.549 1.00 73.37 A O
+ATOM 1046 OD2 ASP A 194 9.107 -64.624 -65.127 1.00 67.84 A O1-
+ATOM 1047 N LEU A 195 4.618 -67.981 -63.807 1.00 52.56 A N
+ATOM 1048 CA LEU A 195 3.436 -68.417 -64.550 1.00 53.53 A C
+ATOM 1049 C LEU A 195 2.811 -67.337 -65.420 1.00 60.64 A C
+ATOM 1050 O LEU A 195 2.392 -67.619 -66.549 1.00 58.97 A O
+ATOM 1051 CB LEU A 195 2.373 -68.896 -63.567 1.00 45.34 A C
+ATOM 1052 CG LEU A 195 2.327 -70.408 -63.443 1.00 46.72 A C
+ATOM 1053 CD1 LEU A 195 1.193 -70.900 -62.545 1.00 45.45 A C
+ATOM 1054 CD2 LEU A 195 2.191 -70.948 -64.817 1.00 47.13 A C
+ATOM 1055 N LYS A 196 2.750 -66.102 -64.914 1.00 61.74 A N
+ATOM 1056 CA LYS A 196 2.289 -64.993 -65.745 1.00 58.09 A C
+ATOM 1057 C LYS A 196 3.083 -64.930 -67.044 1.00 70.01 A C
+ATOM 1058 O LYS A 196 2.501 -64.762 -68.120 1.00 54.01 A O
+ATOM 1059 CB LYS A 196 2.375 -63.663 -64.977 1.00 61.41 A C
+ATOM 1060 CG LYS A 196 2.064 -62.397 -65.807 1.00 64.02 A C
+ATOM 1061 CD LYS A 196 3.344 -61.581 -66.021 1.00 66.30 A C
+ATOM 1062 CE LYS A 196 3.153 -60.320 -66.870 1.00 77.19 A C
+ATOM 1063 NZ LYS A 196 2.828 -59.064 -66.128 1.00 83.12 A N1+
+ATOM 1064 N ASN A 197 4.408 -65.105 -66.978 1.00 64.32 A N
+ATOM 1065 CA ASN A 197 5.187 -65.060 -68.212 1.00 58.55 A C
+ATOM 1066 C ASN A 197 4.721 -66.130 -69.187 1.00 54.17 A C
+ATOM 1067 O ASN A 197 4.440 -65.832 -70.352 1.00 54.09 A O
+ATOM 1068 CB ASN A 197 6.685 -65.210 -67.950 1.00 66.85 A C
+ATOM 1069 CG ASN A 197 7.498 -65.312 -69.249 1.00 77.66 A C
+ATOM 1070 ND2 ASN A 197 7.908 -66.533 -69.606 1.00 78.49 A N
+ATOM 1071 OD1 ASN A 197 7.746 -64.306 -69.915 1.00 79.00 A O
+ATOM 1072 N TYR A 198 4.613 -67.383 -68.737 1.00 59.82 A N
+ATOM 1073 CA TYR A 198 4.227 -68.435 -69.672 1.00 59.39 A C
+ATOM 1074 C TYR A 198 2.864 -68.155 -70.266 1.00 58.04 A C
+ATOM 1075 O TYR A 198 2.672 -68.285 -71.478 1.00 64.12 A O
+ATOM 1076 CB TYR A 198 4.234 -69.799 -69.004 1.00 44.23 A C
+ATOM 1077 CG TYR A 198 3.856 -70.893 -69.953 1.00 49.09 A C
+ATOM 1078 CD1 TYR A 198 4.815 -71.499 -70.732 1.00 49.49 A C
+ATOM 1079 CD2 TYR A 198 2.560 -71.355 -70.042 1.00 47.82 A C
+ATOM 1080 CE1 TYR A 198 4.493 -72.509 -71.609 1.00 49.44 A C
+ATOM 1081 CE2 TYR A 198 2.232 -72.374 -70.904 1.00 50.83 A C
+ATOM 1082 CZ TYR A 198 3.204 -72.941 -71.694 1.00 54.32 A C
+ATOM 1083 OH TYR A 198 2.918 -73.956 -72.580 1.00 62.15 A O
+ATOM 1084 N ILE A 199 1.910 -67.751 -69.426 1.00 55.54 A N
+ATOM 1085 CA ILE A 199 0.585 -67.376 -69.916 1.00 54.80 A C
+ATOM 1086 C ILE A 199 0.688 -66.366 -71.057 1.00 59.69 A C
+ATOM 1087 O ILE A 199 0.006 -66.502 -72.082 1.00 63.33 A O
+ATOM 1088 CB ILE A 199 -0.264 -66.826 -68.760 1.00 51.97 A C
+ATOM 1089 CG1 ILE A 199 -0.661 -67.944 -67.815 1.00 48.25 A C
+ATOM 1090 CG2 ILE A 199 -1.497 -66.165 -69.294 1.00 50.38 A C
+ATOM 1091 CD1 ILE A 199 -1.639 -67.477 -66.778 1.00 53.29 A C
+ATOM 1092 N ASP A 200 1.544 -65.344 -70.904 1.00 47.55 A N
+ATOM 1093 CA ASP A 200 1.673 -64.308 -71.932 1.00 55.72 A C
+ATOM 1094 C ASP A 200 2.352 -64.847 -73.182 1.00 54.43 A C
+ATOM 1095 O ASP A 200 1.800 -64.806 -74.287 1.00 62.23 A O
+ATOM 1096 CB ASP A 200 2.488 -63.125 -71.402 1.00 57.15 A C
+ATOM 1097 CG ASP A 200 1.890 -62.478 -70.161 1.00 61.49 A C
+ATOM 1098 OD1 ASP A 200 0.645 -62.531 -69.950 1.00 57.97 A O
+ATOM 1099 OD2 ASP A 200 2.700 -61.908 -69.395 1.00 68.84 A O1-
+ATOM 1100 N LYS A 201 3.559 -65.368 -73.009 1.00 60.34 A N
+ATOM 1101 CA LYS A 201 4.392 -65.845 -74.096 1.00 56.45 A C
+ATOM 1102 C LYS A 201 3.794 -67.074 -74.779 1.00 54.97 A C
+ATOM 1103 O LYS A 201 4.375 -67.569 -75.750 1.00 63.33 A O
+ATOM 1104 CB LYS A 201 5.807 -66.128 -73.549 1.00 72.13 A C
+ATOM 1105 CG LYS A 201 6.996 -65.461 -74.303 1.00 87.88 A C
+ATOM 1106 CD LYS A 201 8.217 -65.162 -73.366 1.00 90.29 A C
+ATOM 1107 CE LYS A 201 9.337 -66.246 -73.393 1.00 93.38 A C
+ATOM 1108 NZ LYS A 201 10.513 -65.979 -72.481 1.00 89.93 A N1+
+ATOM 1109 N GLN A 202 2.691 -67.619 -74.270 1.00 56.51 A N
+ATOM 1110 CA GLN A 202 2.215 -68.899 -74.783 1.00 57.18 A C
+ATOM 1111 C GLN A 202 0.712 -68.992 -75.028 1.00 59.96 A C
+ATOM 1112 O GLN A 202 0.277 -69.471 -76.073 1.00 62.89 A O
+ATOM 1113 CB GLN A 202 2.698 -70.022 -73.870 1.00 58.04 A C
+ATOM 1114 CG GLN A 202 4.119 -70.428 -74.178 1.00 74.55 A C
+ATOM 1115 CD GLN A 202 4.236 -71.037 -75.554 1.00 87.80 A C
+ATOM 1116 NE2 GLN A 202 4.598 -70.218 -76.539 1.00 91.58 A N
+ATOM 1117 OE1 GLN A 202 4.003 -72.235 -75.733 1.00 89.23 A O
+ATOM 1118 N LEU A 203 -0.092 -68.720 -74.023 1.00 68.98 A N
+ATOM 1119 CA LEU A 203 -1.515 -68.957 -74.149 1.00 56.78 A C
+ATOM 1120 C LEU A 203 -2.328 -67.717 -74.506 1.00 64.58 A C
+ATOM 1121 O LEU A 203 -3.560 -67.794 -74.506 1.00 70.09 A O
+ATOM 1122 CB LEU A 203 -2.056 -69.577 -72.868 1.00 50.67 A C
+ATOM 1123 CG LEU A 203 -1.780 -71.053 -72.615 1.00 58.73 A C
+ATOM 1124 CD1 LEU A 203 -0.335 -71.384 -72.630 1.00 66.52 A C
+ATOM 1125 CD2 LEU A 203 -2.340 -71.390 -71.250 1.00 52.84 A C
+ATOM 1126 N LEU A 204 -1.707 -66.570 -74.773 1.00 49.69 A N
+ATOM 1127 CA LEU A 204 -2.513 -65.354 -74.868 1.00 64.72 A C
+ATOM 1128 C LEU A 204 -3.592 -65.371 -75.951 1.00 75.34 A C
+ATOM 1129 O LEU A 204 -4.628 -64.721 -75.726 1.00 85.31 A O
+ATOM 1130 CB LEU A 204 -1.616 -64.125 -75.011 1.00 76.63 A C
+ATOM 1131 CG LEU A 204 -1.428 -63.568 -73.600 1.00 74.79 A C
+ATOM 1132 CD1 LEU A 204 -0.877 -62.162 -73.612 1.00 76.49 A C
+ATOM 1133 CD2 LEU A 204 -2.730 -63.643 -72.833 1.00 72.18 A C
+ATOM 1134 N PRO A 205 -3.429 -66.033 -77.120 1.00 69.42 A N
+ATOM 1135 CA PRO A 205 -4.589 -66.216 -78.025 1.00 78.88 A C
+ATOM 1136 C PRO A 205 -5.879 -66.695 -77.344 1.00 83.93 A C
+ATOM 1137 O PRO A 205 -6.960 -66.159 -77.634 1.00 83.80 A O
+ATOM 1138 CB PRO A 205 -4.072 -67.259 -79.031 1.00 77.47 A C
+ATOM 1139 CG PRO A 205 -2.634 -66.971 -79.131 1.00 71.50 A C
+ATOM 1140 CD PRO A 205 -2.168 -66.392 -77.797 1.00 65.26 A C
+ATOM 1141 N ILE A 206 -5.784 -67.658 -76.415 1.00 60.81 A N
+ATOM 1142 CA ILE A 206 -6.951 -68.377 -75.900 1.00 78.88 A C
+ATOM 1143 C ILE A 206 -7.775 -67.564 -74.919 1.00 94.81 A C
+ATOM 1144 O ILE A 206 -8.936 -67.907 -74.670 1.00100.72 A O
+ATOM 1145 CB ILE A 206 -6.541 -69.689 -75.216 1.00 76.07 A C
+ATOM 1146 CG1 ILE A 206 -5.369 -70.346 -75.949 1.00 87.46 A C
+ATOM 1147 CG2 ILE A 206 -7.701 -70.635 -75.154 1.00 66.12 A C
+ATOM 1148 CD1 ILE A 206 -5.624 -70.658 -77.422 1.00 95.91 A C
+ATOM 1149 N VAL A 207 -7.211 -66.531 -74.305 1.00 88.91 A N
+ATOM 1150 CA VAL A 207 -8.045 -65.671 -73.480 1.00 87.69 A C
+ATOM 1151 C VAL A 207 -8.836 -64.687 -74.343 1.00 88.23 A C
+ATOM 1152 O VAL A 207 -10.067 -64.601 -74.236 1.00 96.02 A O
+ATOM 1153 CB VAL A 207 -7.183 -64.956 -72.418 1.00 85.21 A C
+ATOM 1154 CG1 VAL A 207 -7.956 -63.819 -71.756 1.00 80.15 A C
+ATOM 1155 CG2 VAL A 207 -6.698 -65.948 -71.361 1.00 85.30 A C
+ATOM 1156 N ASN A 208 -8.150 -63.933 -75.214 1.00105.49 A N
+ATOM 1157 CA ASN A 208 -8.743 -62.755 -75.852 1.00 92.53 A C
+ATOM 1158 C ASN A 208 -9.170 -62.926 -77.318 1.00 88.83 A C
+ATOM 1159 O ASN A 208 -9.756 -61.997 -77.878 1.00 87.67 A O
+ATOM 1160 CB ASN A 208 -7.790 -61.556 -75.713 1.00 85.00 A C
+ATOM 1161 CG ASN A 208 -7.280 -61.368 -74.272 1.00 80.78 A C
+ATOM 1162 ND2 ASN A 208 -8.000 -60.579 -73.472 1.00 69.99 A N
+ATOM 1163 OD1 ASN A 208 -6.265 -61.941 -73.889 1.00 88.75 A O
+ATOM 1164 N LYS A 209 -8.926 -64.067 -77.953 1.00100.78 A N
+ATOM 1165 CA LYS A 209 -9.385 -64.297 -79.320 1.00102.44 A C
+ATOM 1166 C LYS A 209 -10.493 -65.354 -79.330 1.00101.97 A C
+ATOM 1167 O LYS A 209 -10.558 -66.212 -78.449 1.00100.26 A O
+ATOM 1168 CB LYS A 209 -8.225 -64.752 -80.230 1.00 99.70 A C
+ATOM 1169 CG LYS A 209 -7.543 -63.678 -81.101 1.00 98.60 A C
+ATOM 1170 CD LYS A 209 -6.065 -64.043 -81.471 1.00 97.69 A C
+ATOM 1171 CE LYS A 209 -5.929 -65.189 -82.500 1.00101.20 A C
+ATOM 1172 NZ LYS A 209 -4.533 -65.723 -82.676 1.00 99.59 A N1+
+ATOM 1173 N GLN A 210 -11.326 -65.269 -80.361 1.00112.88 A N
+ATOM 1174 CA GLN A 210 -12.374 -66.223 -80.658 1.00111.92 A C
+ATOM 1175 C GLN A 210 -11.616 -66.711 -81.869 1.00113.52 A C
+ATOM 1176 O GLN A 210 -10.994 -65.888 -82.541 1.00112.10 A O
+ATOM 1177 CB GLN A 210 -13.658 -65.515 -81.071 1.00117.43 A C
+ATOM 1178 CG GLN A 210 -13.442 -64.130 -81.660 1.00122.04 A C
+ATOM 1179 CD GLN A 210 -13.158 -63.084 -80.600 1.00123.60 A C
+ATOM 1180 NE2 GLN A 210 -12.486 -62.009 -80.994 1.00123.11 A N
+ATOM 1181 OE1 GLN A 210 -13.540 -63.240 -79.440 1.00125.33 A O
+ATOM 1182 N SER A 211 -11.590 -68.001 -82.178 1.00114.18 A N
+ATOM 1183 CA SER A 211 -10.740 -68.340 -83.336 1.00106.26 A C
+ATOM 1184 C SER A 211 -9.627 -67.268 -83.442 1.00109.00 A C
+ATOM 1185 O SER A 211 -9.679 -66.468 -84.375 1.00115.83 A O
+ATOM 1186 CB SER A 211 -11.573 -68.423 -84.642 1.00100.71 A C
+ATOM 1187 OG SER A 211 -10.838 -68.907 -85.765 1.00 96.47 A O
+ATOM 1188 N CYS A 212 -8.631 -67.193 -82.538 1.00114.50 A N
+ATOM 1189 CA CYS A 212 -8.142 -68.193 -81.558 1.00100.74 A C
+ATOM 1190 C CYS A 212 -7.596 -69.443 -82.218 1.00 81.30 A C
+ATOM 1191 O CYS A 212 -8.231 -70.489 -82.243 1.00 67.61 A O
+ATOM 1192 CB CYS A 212 -9.212 -68.546 -80.531 1.00104.49 A C
+ATOM 1193 SG CYS A 212 -8.849 -69.763 -79.278 1.00 98.48 A S
+ATOM 1194 N SER A 213 -6.405 -69.268 -82.788 1.00 87.31 A N
+ATOM 1195 CA SER A 213 -5.631 -70.344 -83.393 1.00 79.74 A C
+ATOM 1196 C SER A 213 -5.396 -71.494 -82.409 1.00 76.03 A C
+ATOM 1197 O SER A 213 -5.256 -71.278 -81.200 1.00 85.80 A O
+ATOM 1198 CB SER A 213 -4.306 -69.753 -83.877 1.00 85.43 A C
+ATOM 1199 OG SER A 213 -3.886 -68.708 -83.000 1.00104.05 A O
+ATOM 1200 N ILE A 214 -5.324 -72.719 -82.945 1.00 74.70 A N
+ATOM 1201 CA ILE A 214 -5.414 -73.964 -82.157 1.00 70.33 A C
+ATOM 1202 C ILE A 214 -4.041 -74.331 -81.591 1.00 67.27 A C
+ATOM 1203 O ILE A 214 -3.076 -74.471 -82.362 1.00 72.07 A O
+ATOM 1204 CB ILE A 214 -5.976 -75.110 -83.014 1.00 75.15 A C
+ATOM 1205 CG1 ILE A 214 -7.510 -75.164 -82.935 1.00 75.79 A C
+ATOM 1206 CG2 ILE A 214 -5.339 -76.461 -82.650 1.00 69.33 A C
+ATOM 1207 CD1 ILE A 214 -8.142 -76.282 -83.771 1.00 78.73 A C
+ATOM 1208 N PRO A 215 -3.924 -74.551 -80.277 1.00 61.77 A N
+ATOM 1209 CA PRO A 215 -2.631 -74.912 -79.683 1.00 62.58 A C
+ATOM 1210 C PRO A 215 -2.256 -76.363 -79.910 1.00 60.46 A C
+ATOM 1211 O PRO A 215 -3.092 -77.269 -79.911 1.00 59.32 A O
+ATOM 1212 CB PRO A 215 -2.847 -74.653 -78.192 1.00 56.60 A C
+ATOM 1213 CG PRO A 215 -4.307 -74.906 -77.989 1.00 63.72 A C
+ATOM 1214 CD PRO A 215 -4.990 -74.463 -79.263 1.00 64.35 A C
+ATOM 1215 N ASN A 216 -0.965 -76.581 -80.069 1.00 64.91 A N
+ATOM 1216 CA ASN A 216 -0.479 -77.936 -80.242 1.00 64.00 A C
+ATOM 1217 C ASN A 216 -0.443 -78.644 -78.898 1.00 61.72 A C
+ATOM 1218 O ASN A 216 -0.301 -78.017 -77.840 1.00 59.64 A O
+ATOM 1219 CB ASN A 216 0.911 -77.932 -80.875 1.00 64.83 A C
+ATOM 1220 CG ASN A 216 1.929 -77.211 -80.020 1.00 72.32 A C
+ATOM 1221 ND2 ASN A 216 2.211 -75.942 -80.356 1.00 72.73 A N
+ATOM 1222 OD1 ASN A 216 2.447 -77.784 -79.052 1.00 64.50 A O
+ATOM 1223 N ILE A 217 -0.554 -79.969 -78.954 1.00 64.80 A N
+ATOM 1224 CA ILE A 217 -0.719 -80.751 -77.735 1.00 66.09 A C
+ATOM 1225 C ILE A 217 0.488 -80.611 -76.809 1.00 63.01 A C
+ATOM 1226 O ILE A 217 0.341 -80.669 -75.573 1.00 71.79 A O
+ATOM 1227 CB ILE A 217 -1.026 -82.216 -78.104 1.00 66.97 A C
+ATOM 1228 CG1 ILE A 217 -1.110 -83.084 -76.855 1.00 65.03 A C
+ATOM 1229 CG2 ILE A 217 -0.039 -82.734 -79.126 1.00 67.05 A C
+ATOM 1230 CD1 ILE A 217 -2.095 -84.204 -76.996 1.00 70.05 A C
+ATOM 1231 N GLU A 218 1.685 -80.380 -77.372 1.00 61.84 A N
+ATOM 1232 CA GLU A 218 2.852 -80.118 -76.533 1.00 61.13 A C
+ATOM 1233 C GLU A 218 2.597 -78.970 -75.567 1.00 59.34 A C
+ATOM 1234 O GLU A 218 3.001 -79.023 -74.394 1.00 65.15 A O
+ATOM 1235 CB GLU A 218 4.068 -79.802 -77.393 1.00 62.54 A C
+ATOM 1236 CG GLU A 218 4.555 -80.959 -78.185 1.00 64.53 A C
+ATOM 1237 CD GLU A 218 3.800 -81.125 -79.467 1.00 67.23 A C
+ATOM 1238 OE1 GLU A 218 2.954 -82.041 -79.521 1.00 78.20 A O
+ATOM 1239 OE2 GLU A 218 4.056 -80.353 -80.418 1.00 72.39 A O1-
+ATOM 1240 N THR A 219 1.919 -77.921 -76.038 1.00 57.64 A N
+ATOM 1241 CA THR A 219 1.597 -76.824 -75.138 1.00 57.42 A C
+ATOM 1242 C THR A 219 0.632 -77.270 -74.054 1.00 65.68 A C
+ATOM 1243 O THR A 219 0.752 -76.840 -72.904 1.00 64.18 A O
+ATOM 1244 CB THR A 219 1.025 -75.653 -75.915 1.00 57.54 A C
+ATOM 1245 CG2 THR A 219 0.474 -74.635 -74.947 1.00 56.66 A C
+ATOM 1246 OG1 THR A 219 2.074 -75.051 -76.683 1.00 56.42 A O
+ATOM 1247 N VAL A 220 -0.313 -78.149 -74.389 1.00 57.95 A N
+ATOM 1248 CA VAL A 220 -1.228 -78.669 -73.375 1.00 59.85 A C
+ATOM 1249 C VAL A 220 -0.448 -79.307 -72.238 1.00 59.99 A C
+ATOM 1250 O VAL A 220 -0.608 -78.936 -71.062 1.00 64.11 A O
+ATOM 1251 CB VAL A 220 -2.209 -79.677 -73.986 1.00 65.52 A C
+ATOM 1252 CG1 VAL A 220 -3.083 -80.231 -72.887 1.00 57.58 A C
+ATOM 1253 CG2 VAL A 220 -3.035 -79.023 -75.059 1.00 58.89 A C
+ATOM 1254 N ILE A 221 0.409 -80.282 -72.580 1.00 61.49 A N
+ATOM 1255 CA ILE A 221 1.235 -80.945 -71.565 1.00 56.02 A C
+ATOM 1256 C ILE A 221 2.012 -79.914 -70.764 1.00 57.45 A C
+ATOM 1257 O ILE A 221 2.033 -79.931 -69.522 1.00 49.97 A O
+ATOM 1258 CB ILE A 221 2.196 -81.953 -72.215 1.00 54.72 A C
+ATOM 1259 CG1 ILE A 221 1.535 -82.644 -73.396 1.00 60.87 A C
+ATOM 1260 CG2 ILE A 221 2.651 -82.973 -71.195 1.00 52.53 A C
+ATOM 1261 CD1 ILE A 221 0.406 -83.568 -73.008 1.00 63.85 A C
+ATOM 1262 N GLU A 222 2.666 -78.998 -71.473 1.00 55.59 A N
+ATOM 1263 CA GLU A 222 3.526 -78.055 -70.789 1.00 54.40 A C
+ATOM 1264 C GLU A 222 2.734 -77.255 -69.769 1.00 49.02 A C
+ATOM 1265 O GLU A 222 3.115 -77.174 -68.599 1.00 50.98 A O
+ATOM 1266 CB GLU A 222 4.219 -77.156 -71.803 1.00 52.93 A C
+ATOM 1267 CG GLU A 222 5.407 -76.443 -71.215 1.00 60.85 A C
+ATOM 1268 CD GLU A 222 6.283 -75.876 -72.288 1.00 71.78 A C
+ATOM 1269 OE1 GLU A 222 5.787 -75.739 -73.437 1.00 72.89 A O
+ATOM 1270 OE2 GLU A 222 7.464 -75.590 -71.984 1.00 84.71 A O1-
+ATOM 1271 N PHE A 223 1.597 -76.711 -70.170 1.00 45.99 A N
+ATOM 1272 CA PHE A 223 0.859 -75.896 -69.226 1.00 48.36 A C
+ATOM 1273 C PHE A 223 0.430 -76.715 -68.016 1.00 48.57 A C
+ATOM 1274 O PHE A 223 0.510 -76.231 -66.882 1.00 47.23 A O
+ATOM 1275 CB PHE A 223 -0.345 -75.267 -69.891 1.00 43.70 A C
+ATOM 1276 CG PHE A 223 -1.001 -74.241 -69.055 1.00 47.85 A C
+ATOM 1277 CD1 PHE A 223 -0.301 -73.135 -68.645 1.00 44.61 A C
+ATOM 1278 CD2 PHE A 223 -2.305 -74.387 -68.653 1.00 51.77 A C
+ATOM 1279 CE1 PHE A 223 -0.897 -72.162 -67.850 1.00 39.41 A C
+ATOM 1280 CE2 PHE A 223 -2.915 -73.431 -67.862 1.00 53.99 A C
+ATOM 1281 CZ PHE A 223 -2.207 -72.311 -67.460 1.00 46.89 A C
+ATOM 1282 N GLN A 224 -0.001 -77.968 -68.230 1.00 47.57 A N
+ATOM 1283 CA GLN A 224 -0.393 -78.816 -67.097 1.00 46.90 A C
+ATOM 1284 C GLN A 224 0.757 -78.980 -66.112 1.00 48.09 A C
+ATOM 1285 O GLN A 224 0.579 -78.847 -64.887 1.00 51.70 A O
+ATOM 1286 CB GLN A 224 -0.857 -80.180 -67.600 1.00 48.04 A C
+ATOM 1287 CG GLN A 224 -2.138 -80.133 -68.446 1.00 78.76 A C
+ATOM 1288 CD GLN A 224 -2.634 -81.520 -68.864 1.00 86.35 A C
+ATOM 1289 NE2 GLN A 224 -3.952 -81.647 -69.039 1.00 86.61 A N
+ATOM 1290 OE1 GLN A 224 -1.845 -82.466 -69.014 1.00 86.72 A O
+ATOM 1291 N GLN A 225 1.951 -79.265 -66.649 1.00 50.70 A N
+ATOM 1292 CA GLN A 225 3.191 -79.292 -65.876 1.00 48.87 A C
+ATOM 1293 C GLN A 225 3.365 -78.021 -65.053 1.00 43.31 A C
+ATOM 1294 O GLN A 225 3.517 -78.073 -63.827 1.00 60.24 A O
+ATOM 1295 CB GLN A 225 4.386 -79.444 -66.828 1.00 58.21 A C
+ATOM 1296 CG GLN A 225 4.526 -80.793 -67.437 1.00 69.50 A C
+ATOM 1297 CD GLN A 225 4.448 -81.839 -66.383 1.00 82.80 A C
+ATOM 1298 NE2 GLN A 225 3.332 -82.557 -66.341 1.00 82.93 A N
+ATOM 1299 OE1 GLN A 225 5.363 -81.980 -65.573 1.00 90.75 A O
+ATOM 1300 N LYS A 226 3.353 -76.861 -65.722 1.00 37.79 A N
+ATOM 1301 CA LYS A 226 3.692 -75.616 -65.041 1.00 38.98 A C
+ATOM 1302 C LYS A 226 2.663 -75.284 -63.970 1.00 52.58 A C
+ATOM 1303 O LYS A 226 3.009 -75.079 -62.803 1.00 60.15 A O
+ATOM 1304 CB LYS A 226 3.812 -74.478 -66.055 1.00 35.30 A C
+ATOM 1305 CG LYS A 226 5.102 -74.451 -66.850 1.00 68.09 A C
+ATOM 1306 CD LYS A 226 5.976 -73.293 -66.416 1.00 69.06 A C
+ATOM 1307 CE LYS A 226 7.161 -73.129 -67.353 1.00 76.01 A C
+ATOM 1308 NZ LYS A 226 8.128 -72.097 -66.887 1.00 81.45 A N1+
+ATOM 1309 N ASN A 227 1.389 -75.279 -64.351 1.00 52.91 A N
+ATOM 1310 CA ASN A 227 0.296 -74.885 -63.471 1.00 51.97 A C
+ATOM 1311 C ASN A 227 0.160 -75.807 -62.260 1.00 57.03 A C
+ATOM 1312 O ASN A 227 -0.333 -75.387 -61.187 1.00 54.19 A O
+ATOM 1313 CB ASN A 227 -0.985 -74.875 -64.309 1.00 43.91 A C
+ATOM 1314 CG ASN A 227 -2.175 -74.319 -63.571 1.00 46.22 A C
+ATOM 1315 ND2 ASN A 227 -3.226 -75.139 -63.419 1.00 41.95 A N
+ATOM 1316 OD1 ASN A 227 -2.175 -73.143 -63.182 1.00 48.78 A O
+ATOM 1317 N ASN A 228 0.586 -77.070 -62.416 1.00 53.69 A N
+ATOM 1318 CA ASN A 228 0.435 -78.039 -61.333 1.00 46.65 A C
+ATOM 1319 C ASN A 228 0.994 -77.515 -60.012 1.00 50.62 A C
+ATOM 1320 O ASN A 228 0.342 -77.622 -58.969 1.00 55.88 A O
+ATOM 1321 CB ASN A 228 1.095 -79.363 -61.703 1.00 53.43 A C
+ATOM 1322 CG ASN A 228 1.031 -80.350 -60.580 1.00 61.29 A C
+ATOM 1323 ND2 ASN A 228 -0.157 -80.899 -60.320 1.00 60.40 A N
+ATOM 1324 OD1 ASN A 228 2.031 -80.592 -59.925 1.00 64.10 A O
+ATOM 1325 N ARG A 229 2.192 -76.925 -60.042 1.00 49.47 A N
+ATOM 1326 CA ARG A 229 2.789 -76.410 -58.809 1.00 51.20 A C
+ATOM 1327 C ARG A 229 1.815 -75.514 -58.050 1.00 53.93 A C
+ATOM 1328 O ARG A 229 1.555 -75.721 -56.860 1.00 58.93 A O
+ATOM 1329 CB ARG A 229 4.087 -75.657 -59.128 1.00 45.92 A C
+ATOM 1330 CG ARG A 229 4.716 -74.988 -57.925 1.00 50.33 A C
+ATOM 1331 CD ARG A 229 6.227 -74.867 -58.062 1.00 52.45 A C
+ATOM 1332 NE ARG A 229 6.799 -74.608 -56.742 1.00 51.13 A N
+ATOM 1333 CZ ARG A 229 8.004 -74.087 -56.500 1.00 53.22 A C
+ATOM 1334 NH1 ARG A 229 8.828 -73.770 -57.495 1.00 51.22 A N1+
+ATOM 1335 NH2 ARG A 229 8.392 -73.890 -55.243 1.00 59.13 A N
+ATOM 1336 N LEU A 230 1.237 -74.531 -58.744 1.00 54.22 A N
+ATOM 1337 CA LEU A 230 0.240 -73.661 -58.121 1.00 45.71 A C
+ATOM 1338 C LEU A 230 -0.907 -74.472 -57.541 1.00 49.45 A C
+ATOM 1339 O LEU A 230 -1.368 -74.220 -56.414 1.00 50.15 A O
+ATOM 1340 CB LEU A 230 -0.294 -72.659 -59.145 1.00 44.61 A C
+ATOM 1341 CG LEU A 230 -1.235 -71.600 -58.607 1.00 52.80 A C
+ATOM 1342 CD1 LEU A 230 -0.558 -70.944 -57.452 1.00 46.03 A C
+ATOM 1343 CD2 LEU A 230 -1.484 -70.614 -59.710 1.00 54.44 A C
+ATOM 1344 N LEU A 231 -1.380 -75.461 -58.296 1.00 48.46 A N
+ATOM 1345 CA LEU A 231 -2.463 -76.262 -57.755 1.00 49.25 A C
+ATOM 1346 C LEU A 231 -2.044 -76.919 -56.440 1.00 57.79 A C
+ATOM 1347 O LEU A 231 -2.771 -76.825 -55.449 1.00 61.77 A O
+ATOM 1348 CB LEU A 231 -2.922 -77.267 -58.804 1.00 46.33 A C
+ATOM 1349 CG LEU A 231 -3.302 -76.539 -60.109 1.00 59.58 A C
+ATOM 1350 CD1 LEU A 231 -3.509 -77.525 -61.234 1.00 51.66 A C
+ATOM 1351 CD2 LEU A 231 -4.539 -75.658 -59.939 1.00 50.86 A C
+ATOM 1352 N GLU A 232 -0.839 -77.499 -56.373 1.00 56.85 A N
+ATOM 1353 CA GLU A 232 -0.475 -78.219 -55.153 1.00 56.85 A C
+ATOM 1354 C GLU A 232 -0.236 -77.272 -53.994 1.00 56.85 A C
+ATOM 1355 O GLU A 232 -0.612 -77.580 -52.852 1.00 56.85 A O
+ATOM 1356 CB GLU A 232 0.763 -79.090 -55.342 1.00 56.85 A C
+ATOM 1357 CG GLU A 232 0.667 -80.109 -56.434 1.00 56.85 A C
+ATOM 1358 CD GLU A 232 -0.465 -81.132 -56.242 1.00 56.85 A C
+ATOM 1359 OE1 GLU A 232 -1.063 -81.207 -55.133 1.00 56.85 A O
+ATOM 1360 OE2 GLU A 232 -0.747 -81.877 -57.229 1.00 56.85 A O1-
+ATOM 1361 N ILE A 233 0.411 -76.133 -54.254 1.00 51.64 A N
+ATOM 1362 CA ILE A 233 0.619 -75.166 -53.185 1.00 46.86 A C
+ATOM 1363 C ILE A 233 -0.721 -74.729 -52.621 1.00 48.91 A C
+ATOM 1364 O ILE A 233 -0.896 -74.587 -51.397 1.00 51.43 A O
+ATOM 1365 CB ILE A 233 1.416 -73.962 -53.692 1.00 47.76 A C
+ATOM 1366 CG1 ILE A 233 2.662 -74.418 -54.423 1.00 51.96 A C
+ATOM 1367 CG2 ILE A 233 1.788 -73.089 -52.526 1.00 43.38 A C
+ATOM 1368 CD1 ILE A 233 3.598 -73.280 -54.796 1.00 46.09 A C
+ATOM 1369 N THR A 234 -1.701 -74.545 -53.507 1.00 46.51 A N
+ATOM 1370 CA THR A 234 -3.034 -74.176 -53.066 1.00 54.35 A C
+ATOM 1371 C THR A 234 -3.667 -75.282 -52.228 1.00 54.36 A C
+ATOM 1372 O THR A 234 -4.171 -75.018 -51.135 1.00 59.01 A O
+ATOM 1373 CB THR A 234 -3.886 -73.860 -54.275 1.00 48.68 A C
+ATOM 1374 CG2 THR A 234 -5.233 -73.386 -53.822 1.00 40.88 A C
+ATOM 1375 OG1 THR A 234 -3.261 -72.809 -55.009 1.00 45.97 A O
+ATOM 1376 N ARG A 235 -3.640 -76.531 -52.713 1.00 46.49 A N
+ATOM 1377 CA ARG A 235 -4.227 -77.643 -51.959 1.00 44.99 A C
+ATOM 1378 C ARG A 235 -3.674 -77.690 -50.528 1.00 45.66 A C
+ATOM 1379 O ARG A 235 -4.435 -77.759 -49.543 1.00 59.70 A O
+ATOM 1380 CB ARG A 235 -3.986 -78.971 -52.704 1.00 53.04 A C
+ATOM 1381 CG ARG A 235 -4.637 -79.020 -54.106 1.00 56.15 A C
+ATOM 1382 CD ARG A 235 -4.089 -80.121 -55.045 1.00 64.34 A C
+ATOM 1383 NE ARG A 235 -4.565 -79.960 -56.437 1.00 73.96 A N
+ATOM 1384 CZ ARG A 235 -4.378 -80.850 -57.429 1.00 82.91 A C
+ATOM 1385 NH1 ARG A 235 -3.739 -81.987 -57.185 1.00 78.78 A N1+
+ATOM 1386 NH2 ARG A 235 -4.835 -80.626 -58.670 1.00 87.59 A N
+ATOM 1387 N GLU A 236 -2.344 -77.604 -50.401 1.00 45.23 A N
+ATOM 1388 CA GLU A 236 -1.720 -77.652 -49.084 1.00 48.81 A C
+ATOM 1389 C GLU A 236 -2.203 -76.516 -48.210 1.00 50.19 A C
+ATOM 1390 O GLU A 236 -2.639 -76.731 -47.076 1.00 48.41 A O
+ATOM 1391 CB GLU A 236 -0.210 -77.568 -49.203 1.00 42.54 A C
+ATOM 1392 CG GLU A 236 0.415 -78.623 -50.046 1.00 49.02 A C
+ATOM 1393 CD GLU A 236 1.909 -78.608 -49.886 1.00 59.32 A C
+ATOM 1394 OE1 GLU A 236 2.361 -78.065 -48.832 1.00 63.55 A O
+ATOM 1395 OE2 GLU A 236 2.611 -79.106 -50.815 1.00 77.27 A O1-
+ATOM 1396 N PHE A 237 -2.095 -75.289 -48.705 1.00 41.57 A N
+ATOM 1397 CA PHE A 237 -2.603 -74.182 -47.915 1.00 54.24 A C
+ATOM 1398 C PHE A 237 -4.082 -74.366 -47.552 1.00 53.04 A C
+ATOM 1399 O PHE A 237 -4.508 -73.882 -46.500 1.00 57.88 A O
+ATOM 1400 CB PHE A 237 -2.384 -72.884 -48.668 1.00 52.28 A C
+ATOM 1401 CG PHE A 237 -1.019 -72.307 -48.500 1.00 46.09 A C
+ATOM 1402 CD1 PHE A 237 -0.329 -72.461 -47.328 1.00 46.78 A C
+ATOM 1403 CD2 PHE A 237 -0.418 -71.613 -49.540 1.00 48.60 A C
+ATOM 1404 CE1 PHE A 237 0.939 -71.904 -47.184 1.00 43.05 A C
+ATOM 1405 CE2 PHE A 237 0.838 -71.073 -49.413 1.00 45.70 A C
+ATOM 1406 CZ PHE A 237 1.521 -71.217 -48.240 1.00 40.54 A C
+ATOM 1407 N SER A 238 -4.876 -75.080 -48.380 1.00 46.83 A N
+ATOM 1408 CA SER A 238 -6.305 -75.260 -48.076 1.00 54.06 A C
+ATOM 1409 C SER A 238 -6.526 -76.193 -46.897 1.00 51.43 A C
+ATOM 1410 O SER A 238 -7.239 -75.844 -45.946 1.00 58.02 A O
+ATOM 1411 CB SER A 238 -7.109 -75.777 -49.279 1.00 52.93 A C
+ATOM 1412 OG SER A 238 -7.635 -74.732 -50.091 1.00 88.79 A O
+ATOM 1413 N VAL A 239 -5.963 -77.397 -46.938 1.00 45.76 A N
+ATOM 1414 CA VAL A 239 -6.320 -78.238 -45.809 1.00 51.20 A C
+ATOM 1415 C VAL A 239 -5.650 -77.747 -44.528 1.00 47.32 A C
+ATOM 1416 O VAL A 239 -6.126 -78.046 -43.424 1.00 57.92 A O
+ATOM 1417 CB VAL A 239 -5.955 -79.703 -46.048 1.00 55.02 A C
+ATOM 1418 CG1 VAL A 239 -6.355 -80.120 -47.460 1.00 58.03 A C
+ATOM 1419 CG2 VAL A 239 -4.505 -79.957 -45.718 1.00 45.79 A C
+ATOM 1420 N ASN A 240 -4.553 -76.999 -44.641 1.00 49.70 A N
+ATOM 1421 CA ASN A 240 -3.737 -76.610 -43.498 1.00 49.45 A C
+ATOM 1422 C ASN A 240 -4.055 -75.224 -42.956 1.00 53.81 A C
+ATOM 1423 O ASN A 240 -3.276 -74.700 -42.154 1.00 57.22 A O
+ATOM 1424 CB ASN A 240 -2.264 -76.723 -43.852 1.00 51.26 A C
+ATOM 1425 CG ASN A 240 -1.737 -78.111 -43.596 1.00 57.58 A C
+ATOM 1426 ND2 ASN A 240 -1.402 -78.373 -42.333 1.00 52.16 A N
+ATOM 1427 OD1 ASN A 240 -1.665 -78.957 -44.497 1.00 53.70 A O
+ATOM 1428 N ALA A 241 -5.134 -74.595 -43.429 1.00 53.29 A N
+ATOM 1429 CA ALA A 241 -5.559 -73.288 -42.934 1.00 55.28 A C
+ATOM 1430 C ALA A 241 -4.423 -72.279 -43.032 1.00 53.04 A C
+ATOM 1431 O ALA A 241 -4.220 -71.450 -42.145 1.00 60.20 A O
+ATOM 1432 CB ALA A 241 -6.086 -73.380 -41.503 1.00 46.86 A C
+ATOM 1433 N GLY A 242 -3.648 -72.386 -44.102 1.00 48.00 A N
+ATOM 1434 CA GLY A 242 -2.692 -71.370 -44.463 1.00 47.26 A C
+ATOM 1435 C GLY A 242 -1.339 -71.447 -43.792 1.00 50.60 A C
+ATOM 1436 O GLY A 242 -0.535 -70.520 -43.967 1.00 54.53 A O
+ATOM 1437 N VAL A 243 -1.055 -72.496 -43.016 1.00 53.63 A N
+ATOM 1438 CA VAL A 243 0.265 -72.656 -42.404 1.00 52.31 A C
+ATOM 1439 C VAL A 243 0.701 -74.107 -42.531 1.00 48.48 A C
+ATOM 1440 O VAL A 243 0.046 -74.998 -41.981 1.00 57.33 A O
+ATOM 1441 CB VAL A 243 0.267 -72.250 -40.923 1.00 51.16 A C
+ATOM 1442 CG1 VAL A 243 1.683 -72.201 -40.413 1.00 52.15 A C
+ATOM 1443 CG2 VAL A 243 -0.446 -70.921 -40.717 1.00 54.55 A C
+ATOM 1444 N THR A 244 1.816 -74.358 -43.203 1.00 49.32 A N
+ATOM 1445 CA THR A 244 2.272 -75.732 -43.341 1.00 49.02 A C
+ATOM 1446 C THR A 244 3.616 -75.929 -42.666 1.00 55.80 A C
+ATOM 1447 O THR A 244 4.494 -75.066 -42.740 1.00 51.30 A O
+ATOM 1448 CB THR A 244 2.369 -76.173 -44.808 1.00 48.77 A C
+ATOM 1449 CG2 THR A 244 1.148 -75.727 -45.559 1.00 40.24 A C
+ATOM 1450 OG1 THR A 244 3.545 -75.636 -45.430 1.00 49.81 A O
+ATOM 1451 N THR A 245 3.750 -77.048 -41.968 1.00 56.17 A N
+ATOM 1452 CA THR A 245 5.063 -77.586 -41.639 1.00 52.18 A C
+ATOM 1453 C THR A 245 5.058 -79.101 -41.547 1.00 53.20 A C
+ATOM 1454 O THR A 245 4.086 -79.680 -41.097 1.00 51.19 A O
+ATOM 1455 CB THR A 245 5.585 -77.029 -40.332 1.00 50.63 A C
+ATOM 1456 CG2 THR A 245 4.499 -77.058 -39.297 1.00 43.13 A C
+ATOM 1457 OG1 THR A 245 6.721 -77.804 -39.914 1.00 55.14 A O
+ATOM 1458 N PRO A 246 6.143 -79.755 -41.971 1.00 51.61 A N
+ATOM 1459 CA PRO A 246 7.322 -79.226 -42.659 1.00 52.34 A C
+ATOM 1460 C PRO A 246 7.002 -78.573 -44.014 1.00 53.80 A C
+ATOM 1461 O PRO A 246 5.847 -78.583 -44.470 1.00 54.42 A O
+ATOM 1462 CB PRO A 246 8.184 -80.477 -42.859 1.00 53.35 A C
+ATOM 1463 CG PRO A 246 7.238 -81.630 -42.705 1.00 52.58 A C
+ATOM 1464 CD PRO A 246 6.303 -81.178 -41.656 1.00 45.81 A C
+ATOM 1465 N VAL A 247 8.001 -78.000 -44.662 1.00 54.58 A N
+ATOM 1466 CA VAL A 247 7.762 -77.237 -45.878 1.00 47.21 A C
+ATOM 1467 C VAL A 247 8.088 -78.131 -47.055 1.00 52.87 A C
+ATOM 1468 O VAL A 247 9.111 -78.823 -47.060 1.00 52.15 A O
+ATOM 1469 CB VAL A 247 8.594 -75.944 -45.889 1.00 52.33 A C
+ATOM 1470 CG1 VAL A 247 7.965 -74.898 -44.981 1.00 48.23 A C
+ATOM 1471 CG2 VAL A 247 9.953 -76.252 -45.364 1.00 49.09 A C
+ATOM 1472 N SER A 248 7.236 -78.085 -48.071 1.00 49.89 A N
+ATOM 1473 CA SER A 248 7.194 -79.074 -49.137 1.00 64.63 A C
+ATOM 1474 C SER A 248 8.377 -78.943 -50.087 1.00 57.77 A C
+ATOM 1475 O SER A 248 9.150 -77.987 -50.058 1.00 56.78 A O
+ATOM 1476 CB SER A 248 5.939 -78.862 -49.965 1.00 54.52 A C
+ATOM 1477 OG SER A 248 4.842 -78.757 -49.087 1.00 59.14 A O
+ATOM 1478 N THR A 249 8.459 -79.900 -50.998 1.00 56.45 A N
+ATOM 1479 CA THR A 249 9.234 -79.676 -52.200 1.00 60.68 A C
+ATOM 1480 C THR A 249 8.479 -78.746 -53.130 1.00 57.08 A C
+ATOM 1481 O THR A 249 9.107 -78.073 -53.951 1.00 66.12 A O
+ATOM 1482 CB THR A 249 9.547 -81.014 -52.894 1.00 57.52 A C
+ATOM 1483 CG2 THR A 249 10.492 -80.851 -54.108 1.00 55.23 A C
+ATOM 1484 OG1 THR A 249 10.164 -81.911 -51.962 1.00 57.17 A O
+ATOM 1485 N TYR A 250 7.141 -78.683 -53.004 1.00 58.91 A N
+ATOM 1486 CA TYR A 250 6.345 -77.744 -53.794 1.00 58.34 A C
+ATOM 1487 C TYR A 250 6.476 -76.315 -53.288 1.00 62.18 A C
+ATOM 1488 O TYR A 250 6.395 -75.366 -54.076 1.00 65.11 A O
+ATOM 1489 CB TYR A 250 4.865 -78.106 -53.771 1.00 51.48 A C
+ATOM 1490 CG TYR A 250 4.492 -79.456 -54.322 1.00 53.73 A C
+ATOM 1491 CD1 TYR A 250 4.933 -79.883 -55.577 1.00 49.38 A C
+ATOM 1492 CD2 TYR A 250 3.617 -80.273 -53.620 1.00 53.87 A C
+ATOM 1493 CE1 TYR A 250 4.555 -81.122 -56.088 1.00 46.28 A C
+ATOM 1494 CE2 TYR A 250 3.229 -81.508 -54.127 1.00 53.13 A C
+ATOM 1495 CZ TYR A 250 3.702 -81.933 -55.355 1.00 59.37 A C
+ATOM 1496 OH TYR A 250 3.301 -83.175 -55.811 1.00 71.15 A O
+ATOM 1497 N MET A 251 6.634 -76.144 -51.978 1.00 56.68 A N
+ATOM 1498 CA MET A 251 6.810 -74.809 -51.440 1.00 54.54 A C
+ATOM 1499 C MET A 251 8.219 -74.291 -51.691 1.00 53.29 A C
+ATOM 1500 O MET A 251 8.378 -73.136 -52.083 1.00 51.04 A O
+ATOM 1501 CB MET A 251 6.484 -74.815 -49.958 1.00 45.34 A C
+ATOM 1502 CG MET A 251 5.087 -75.293 -49.618 1.00 59.15 A C
+ATOM 1503 SD MET A 251 3.944 -73.900 -49.617 1.00 41.36 A S
+ATOM 1504 CE MET A 251 2.526 -74.566 -48.721 1.00 52.33 A C
+ATOM 1505 N LEU A 252 9.251 -75.112 -51.453 1.00 50.54 A N
+ATOM 1506 CA LEU A 252 10.627 -74.803 -51.856 1.00 42.99 A C
+ATOM 1507 C LEU A 252 11.313 -76.060 -52.380 1.00 45.86 A C
+ATOM 1508 O LEU A 252 11.352 -77.091 -51.698 1.00 47.52 A O
+ATOM 1509 CB LEU A 252 11.451 -74.231 -50.706 1.00 41.63 A C
+ATOM 1510 CG LEU A 252 11.109 -72.812 -50.300 1.00 52.49 A C
+ATOM 1511 CD1 LEU A 252 12.076 -72.322 -49.259 1.00 55.47 A C
+ATOM 1512 CD2 LEU A 252 11.163 -71.953 -51.501 1.00 51.85 A C
+ATOM 1513 N THR A 253 11.848 -75.971 -53.592 1.00 55.09 A N
+ATOM 1514 CA THR A 253 12.663 -77.036 -54.149 1.00 56.99 A C
+ATOM 1515 C THR A 253 13.987 -77.087 -53.423 1.00 56.39 A C
+ATOM 1516 O THR A 253 14.482 -76.060 -52.952 1.00 54.34 A O
+ATOM 1517 CB THR A 253 12.915 -76.762 -55.621 1.00 56.94 A C
+ATOM 1518 CG2 THR A 253 13.646 -77.959 -56.341 1.00 47.22 A C
+ATOM 1519 OG1 THR A 253 11.662 -76.429 -56.238 1.00 53.73 A O
+ATOM 1520 N ASN A 254 14.579 -78.287 -53.349 1.00 45.69 A N
+ATOM 1521 CA ASN A 254 15.906 -78.381 -52.739 1.00 53.09 A C
+ATOM 1522 C ASN A 254 16.844 -77.336 -53.320 1.00 52.44 A C
+ATOM 1523 O ASN A 254 17.563 -76.656 -52.575 1.00 67.08 A O
+ATOM 1524 CB ASN A 254 16.497 -79.781 -52.898 1.00 52.67 A C
+ATOM 1525 CG ASN A 254 16.285 -80.639 -51.658 1.00 68.29 A C
+ATOM 1526 ND2 ASN A 254 17.359 -81.268 -51.209 1.00 69.84 A N
+ATOM 1527 OD1 ASN A 254 15.181 -80.715 -51.094 1.00 80.05 A O
+ATOM 1528 N SER A 255 16.783 -77.132 -54.644 1.00 50.64 A N
+ATOM 1529 CA SER A 255 17.670 -76.166 -55.286 1.00 53.31 A C
+ATOM 1530 C SER A 255 17.382 -74.751 -54.805 1.00 52.49 A C
+ATOM 1531 O SER A 255 18.307 -73.956 -54.566 1.00 68.20 A O
+ATOM 1532 CB SER A 255 17.532 -76.264 -56.801 1.00 53.34 A C
+ATOM 1533 OG SER A 255 18.673 -75.691 -57.420 1.00 65.86 A O
+ATOM 1534 N GLU A 256 16.101 -74.434 -54.641 1.00 48.80 A N
+ATOM 1535 CA GLU A 256 15.722 -73.112 -54.168 1.00 46.97 A C
+ATOM 1536 C GLU A 256 16.119 -72.927 -52.710 1.00 50.56 A C
+ATOM 1537 O GLU A 256 16.646 -71.876 -52.330 1.00 57.47 A O
+ATOM 1538 CB GLU A 256 14.214 -72.906 -54.368 1.00 42.52 A C
+ATOM 1539 CG GLU A 256 13.739 -73.030 -55.825 1.00 52.32 A C
+ATOM 1540 CD GLU A 256 12.244 -73.359 -55.953 1.00 61.81 A C
+ATOM 1541 OE1 GLU A 256 11.645 -73.783 -54.948 1.00 60.74 A O
+ATOM 1542 OE2 GLU A 256 11.664 -73.205 -57.053 1.00 54.63 A O1-
+ATOM 1543 N LEU A 257 15.891 -73.940 -51.880 1.00 49.08 A N
+ATOM 1544 CA LEU A 257 16.125 -73.744 -50.455 1.00 44.72 A C
+ATOM 1545 C LEU A 257 17.601 -73.647 -50.170 1.00 55.08 A C
+ATOM 1546 O LEU A 257 18.030 -72.816 -49.362 1.00 54.46 A O
+ATOM 1547 CB LEU A 257 15.499 -74.863 -49.626 1.00 33.69 A C
+ATOM 1548 CG LEU A 257 15.731 -74.520 -48.165 1.00 50.71 A C
+ATOM 1549 CD1 LEU A 257 14.440 -74.505 -47.439 1.00 41.92 A C
+ATOM 1550 CD2 LEU A 257 16.634 -75.544 -47.572 1.00 39.46 A C
+ATOM 1551 N LEU A 258 18.398 -74.479 -50.832 1.00 52.14 A N
+ATOM 1552 CA LEU A 258 19.836 -74.309 -50.726 1.00 52.02 A C
+ATOM 1553 C LEU A 258 20.250 -72.934 -51.236 1.00 49.06 A C
+ATOM 1554 O LEU A 258 20.997 -72.220 -50.554 1.00 62.60 A O
+ATOM 1555 CB LEU A 258 20.557 -75.430 -51.469 1.00 43.99 A C
+ATOM 1556 CG LEU A 258 20.208 -76.806 -50.907 1.00 52.63 A C
+ATOM 1557 CD1 LEU A 258 20.819 -77.955 -51.700 1.00 45.94 A C
+ATOM 1558 CD2 LEU A 258 20.663 -76.832 -49.482 1.00 43.54 A C
+ATOM 1559 N SER A 259 19.724 -72.510 -52.414 1.00 52.57 A N
+ATOM 1560 CA SER A 259 20.171 -71.218 -52.957 1.00 59.70 A C
+ATOM 1561 C SER A 259 19.617 -70.029 -52.159 1.00 53.46 A C
+ATOM 1562 O SER A 259 19.946 -68.871 -52.451 1.00 70.05 A O
+ATOM 1563 CB SER A 259 19.833 -71.079 -54.449 1.00 67.72 A C
+ATOM 1564 OG SER A 259 20.766 -70.192 -55.094 1.00 81.81 A O
+ATOM 1565 N LEU A 260 18.778 -70.300 -51.167 1.00 56.53 A N
+ATOM 1566 CA LEU A 260 18.354 -69.322 -50.179 1.00 54.78 A C
+ATOM 1567 C LEU A 260 19.330 -69.303 -49.014 1.00 62.69 A C
+ATOM 1568 O LEU A 260 19.887 -68.256 -48.666 1.00 62.51 A O
+ATOM 1569 CB LEU A 260 16.943 -69.658 -49.700 1.00 46.78 A C
+ATOM 1570 CG LEU A 260 16.050 -68.454 -49.435 1.00 51.30 A C
+ATOM 1571 CD1 LEU A 260 16.225 -67.399 -50.518 1.00 46.43 A C
+ATOM 1572 CD2 LEU A 260 14.627 -68.935 -49.412 1.00 50.28 A C
+ATOM 1573 N ILE A 261 19.500 -70.464 -48.388 1.00 55.56 A N
+ATOM 1574 CA ILE A 261 20.428 -70.702 -47.297 1.00 51.84 A C
+ATOM 1575 C ILE A 261 21.729 -70.018 -47.635 1.00 57.79 A C
+ATOM 1576 O ILE A 261 22.248 -69.236 -46.841 1.00 57.88 A O
+ATOM 1577 CB ILE A 261 20.678 -72.205 -47.148 1.00 46.59 A C
+ATOM 1578 CG1 ILE A 261 19.512 -72.909 -46.490 1.00 46.63 A C
+ATOM 1579 CG2 ILE A 261 21.976 -72.460 -46.449 1.00 42.83 A C
+ATOM 1580 CD1 ILE A 261 19.769 -74.369 -46.378 1.00 47.20 A C
+ATOM 1581 N ASN A 262 22.233 -70.282 -48.844 1.00 57.54 A N
+ATOM 1582 CA ASN A 262 23.542 -69.783 -49.242 1.00 56.35 A C
+ATOM 1583 C ASN A 262 23.648 -68.280 -49.081 1.00 63.25 A C
+ATOM 1584 O ASN A 262 24.730 -67.756 -48.792 1.00 62.53 A O
+ATOM 1585 CB ASN A 262 23.833 -70.138 -50.685 1.00 55.06 A C
+ATOM 1586 CG ASN A 262 25.299 -70.111 -50.986 1.00 70.86 A C
+ATOM 1587 ND2 ASN A 262 25.672 -70.712 -52.096 1.00 69.44 A N
+ATOM 1588 OD1 ASN A 262 26.095 -69.558 -50.233 1.00 80.05 A O
+ATOM 1589 N ASP A 263 22.567 -67.559 -49.335 1.00 50.66 A N
+ATOM 1590 CA ASP A 263 22.625 -66.111 -49.281 1.00 59.03 A C
+ATOM 1591 C ASP A 263 22.028 -65.521 -48.008 1.00 59.74 A C
+ATOM 1592 O ASP A 263 22.001 -64.298 -47.856 1.00 68.83 A O
+ATOM 1593 CB ASP A 263 21.974 -65.543 -50.536 1.00 63.06 A C
+ATOM 1594 CG ASP A 263 22.876 -65.695 -51.774 1.00 74.82 A C
+ATOM 1595 OD1 ASP A 263 24.107 -65.895 -51.594 1.00 78.90 A O
+ATOM 1596 OD2 ASP A 263 22.355 -65.613 -52.909 1.00 78.50 A O1-
+ATOM 1597 N MET A 264 21.548 -66.346 -47.090 1.00 62.09 A N
+ATOM 1598 CA MET A 264 21.194 -65.812 -45.798 1.00 60.87 A C
+ATOM 1599 C MET A 264 22.423 -65.205 -45.138 1.00 77.50 A C
+ATOM 1600 O MET A 264 23.562 -65.521 -45.501 1.00 85.95 A O
+ATOM 1601 CB MET A 264 20.623 -66.894 -44.891 1.00 44.22 A C
+ATOM 1602 CG MET A 264 19.325 -67.544 -45.332 1.00 40.80 A C
+ATOM 1603 SD MET A 264 18.887 -68.948 -44.244 1.00 51.75 A S
+ATOM 1604 CE MET A 264 17.902 -68.196 -42.962 1.00 40.42 A C
+ATOM 1605 N PRO A 265 22.199 -64.278 -44.139 1.00 52.90 A N
+ATOM 1606 CA PRO A 265 23.277 -63.734 -43.278 1.00 67.95 A C
+ATOM 1607 C PRO A 265 23.680 -64.648 -42.125 1.00 69.96 A C
+ATOM 1608 O PRO A 265 23.237 -64.491 -40.995 1.00104.67 A O
+ATOM 1609 CB PRO A 265 22.656 -62.439 -42.748 1.00 75.96 A C
+ATOM 1610 CG PRO A 265 21.192 -62.687 -42.776 1.00 70.17 A C
+ATOM 1611 CD PRO A 265 20.938 -63.530 -43.978 1.00 61.31 A C
+ATOM 1612 N ILE A 266 24.541 -65.631 -42.398 1.00 91.92 A N
+ATOM 1613 CA ILE A 266 24.989 -66.531 -41.337 1.00 65.95 A C
+ATOM 1614 C ILE A 266 26.410 -67.023 -41.582 1.00 62.66 A C
+ATOM 1615 O ILE A 266 27.001 -66.724 -42.628 1.00 62.03 A O
+ATOM 1616 CB ILE A 266 24.033 -67.727 -41.187 1.00 61.73 A C
+ATOM 1617 CG1 ILE A 266 23.606 -68.263 -42.549 1.00 52.37 A C
+ATOM 1618 CG2 ILE A 266 22.824 -67.367 -40.368 1.00 68.52 A C
+ATOM 1619 CD1 ILE A 266 22.849 -69.548 -42.429 1.00 51.18 A C
+ATOM 1620 N THR A 267 26.945 -67.803 -40.624 1.00 53.31 A N
+ATOM 1621 CA THR A 267 28.268 -68.425 -40.724 1.00 58.17 A C
+ATOM 1622 C THR A 267 28.336 -69.381 -41.897 1.00 58.71 A C
+ATOM 1623 O THR A 267 27.358 -70.052 -42.230 1.00 67.21 A O
+ATOM 1624 CB THR A 267 28.581 -69.284 -39.504 1.00 70.96 A C
+ATOM 1625 CG2 THR A 267 30.085 -69.512 -39.406 1.00 79.34 A C
+ATOM 1626 OG1 THR A 267 28.102 -68.667 -38.307 1.00 68.93 A O
+ATOM 1627 N ASN A 268 29.525 -69.534 -42.470 1.00 58.76 A N
+ATOM 1628 CA ASN A 268 29.673 -70.651 -43.394 1.00 60.07 A C
+ATOM 1629 C ASN A 268 29.509 -71.982 -42.684 1.00 62.03 A C
+ATOM 1630 O ASN A 268 29.124 -72.977 -43.309 1.00 61.17 A O
+ATOM 1631 CB ASN A 268 31.003 -70.584 -44.129 1.00 52.58 A C
+ATOM 1632 CG ASN A 268 31.009 -69.499 -45.156 1.00 69.38 A C
+ATOM 1633 ND2 ASN A 268 31.651 -69.757 -46.295 1.00 67.71 A N
+ATOM 1634 OD1 ASN A 268 30.400 -68.443 -44.950 1.00 74.14 A O
+ATOM 1635 N ASP A 269 29.769 -72.023 -41.380 1.00 69.59 A N
+ATOM 1636 CA ASP A 269 29.454 -73.239 -40.646 1.00 71.11 A C
+ATOM 1637 C ASP A 269 27.947 -73.434 -40.566 1.00 59.18 A C
+ATOM 1638 O ASP A 269 27.453 -74.559 -40.720 1.00 57.81 A O
+ATOM 1639 CB ASP A 269 30.077 -73.201 -39.250 1.00 73.77 A C
+ATOM 1640 CG ASP A 269 31.515 -72.725 -39.268 1.00 79.26 A C
+ATOM 1641 OD1 ASP A 269 32.239 -73.064 -40.229 1.00 83.10 A O
+ATOM 1642 OD2 ASP A 269 31.926 -72.034 -38.311 1.00 79.00 A O1-
+ATOM 1643 N GLN A 270 27.199 -72.346 -40.344 1.00 47.28 A N
+ATOM 1644 CA GLN A 270 25.745 -72.440 -40.350 1.00 53.49 A C
+ATOM 1645 C GLN A 270 25.222 -72.770 -41.744 1.00 50.42 A C
+ATOM 1646 O GLN A 270 24.387 -73.669 -41.908 1.00 49.32 A O
+ATOM 1647 CB GLN A 270 25.130 -71.144 -39.854 1.00 61.99 A C
+ATOM 1648 CG GLN A 270 25.580 -70.749 -38.501 1.00 67.58 A C
+ATOM 1649 CD GLN A 270 24.965 -69.446 -38.087 1.00 78.52 A C
+ATOM 1650 NE2 GLN A 270 23.767 -69.516 -37.534 1.00 71.70 A N
+ATOM 1651 OE1 GLN A 270 25.551 -68.381 -38.269 1.00 88.51 A O
+ATOM 1652 N LYS A 271 25.682 -72.043 -42.763 1.00 51.94 A N
+ATOM 1653 CA LYS A 271 25.279 -72.381 -44.123 1.00 47.38 A C
+ATOM 1654 C LYS A 271 25.522 -73.871 -44.389 1.00 46.75 A C
+ATOM 1655 O LYS A 271 24.587 -74.617 -44.721 1.00 57.75 A O
+ATOM 1656 CB LYS A 271 26.032 -71.499 -45.127 1.00 43.85 A C
+ATOM 1657 CG LYS A 271 25.646 -70.034 -45.118 1.00 59.33 A C
+ATOM 1658 CD LYS A 271 26.504 -69.267 -46.102 1.00 73.13 A C
+ATOM 1659 CE LYS A 271 26.265 -67.781 -45.979 1.00 73.69 A C
+ATOM 1660 NZ LYS A 271 27.160 -67.007 -46.867 1.00 78.48 A N1+
+ATOM 1661 N LYS A 272 26.768 -74.330 -44.167 1.00 47.78 A N
+ATOM 1662 CA LYS A 272 27.127 -75.735 -44.362 1.00 51.24 A C
+ATOM 1663 C LYS A 272 26.210 -76.656 -43.567 1.00 56.70 A C
+ATOM 1664 O LYS A 272 25.841 -77.741 -44.037 1.00 55.89 A O
+ATOM 1665 CB LYS A 272 28.598 -75.941 -43.969 1.00 67.26 A C
+ATOM 1666 CG LYS A 272 29.159 -77.374 -44.112 1.00 86.26 A C
+ATOM 1667 CD LYS A 272 30.427 -77.578 -43.228 1.00101.63 A C
+ATOM 1668 CE LYS A 272 31.016 -79.004 -43.273 1.00108.72 A C
+ATOM 1669 NZ LYS A 272 31.844 -79.295 -44.490 1.00111.53 A N1+
+ATOM 1670 N LEU A 273 25.783 -76.205 -42.393 1.00 57.88 A N
+ATOM 1671 CA LEU A 273 25.014 -77.038 -41.480 1.00 49.38 A C
+ATOM 1672 C LEU A 273 23.573 -77.222 -41.943 1.00 51.08 A C
+ATOM 1673 O LEU A 273 23.033 -78.334 -41.890 1.00 50.88 A O
+ATOM 1674 CB LEU A 273 25.048 -76.432 -40.079 1.00 47.93 A C
+ATOM 1675 CG LEU A 273 23.981 -77.088 -39.226 1.00 53.74 A C
+ATOM 1676 CD1 LEU A 273 24.318 -78.544 -38.994 1.00 50.20 A C
+ATOM 1677 CD2 LEU A 273 23.840 -76.348 -37.951 1.00 51.30 A C
+ATOM 1678 N MET A 274 22.912 -76.145 -42.361 1.00 44.81 A N
+ATOM 1679 CA MET A 274 21.580 -76.315 -42.923 1.00 47.78 A C
+ATOM 1680 C MET A 274 21.658 -77.111 -44.220 1.00 52.55 A C
+ATOM 1681 O MET A 274 21.008 -78.156 -44.371 1.00 51.61 A O
+ATOM 1682 CB MET A 274 20.927 -74.955 -43.147 1.00 51.15 A C
+ATOM 1683 CG MET A 274 21.101 -74.062 -41.961 1.00 45.36 A C
+ATOM 1684 SD MET A 274 20.052 -72.602 -41.935 1.00 59.02 A S
+ATOM 1685 CE MET A 274 20.500 -71.833 -43.440 1.00 49.54 A C
+ATOM 1686 N SER A 275 22.506 -76.665 -45.146 1.00 52.89 A N
+ATOM 1687 CA SER A 275 22.517 -77.297 -46.458 1.00 51.18 A C
+ATOM 1688 C SER A 275 23.008 -78.744 -46.423 1.00 57.08 A C
+ATOM 1689 O SER A 275 22.774 -79.477 -47.387 1.00 63.49 A O
+ATOM 1690 CB SER A 275 23.313 -76.429 -47.466 1.00 55.92 A C
+ATOM 1691 OG SER A 275 24.408 -75.721 -46.902 1.00 62.23 A O
+ATOM 1692 N ASN A 276 23.644 -79.195 -45.345 1.00 56.18 A N
+ATOM 1693 CA ASN A 276 23.826 -80.637 -45.217 1.00 56.00 A C
+ATOM 1694 C ASN A 276 22.570 -81.316 -44.709 1.00 60.58 A C
+ATOM 1695 O ASN A 276 22.362 -82.508 -44.954 1.00 74.32 A O
+ATOM 1696 CB ASN A 276 24.960 -80.957 -44.257 1.00 59.96 A C
+ATOM 1697 CG ASN A 276 26.293 -80.770 -44.872 1.00 67.79 A C
+ATOM 1698 ND2 ASN A 276 27.263 -80.356 -44.059 1.00 63.76 A N
+ATOM 1699 OD1 ASN A 276 26.463 -80.973 -46.073 1.00 65.95 A O
+ATOM 1700 N ASN A 277 21.796 -80.606 -43.908 1.00 55.64 A N
+ATOM 1701 CA ASN A 277 20.683 -81.181 -43.180 1.00 57.07 A C
+ATOM 1702 C ASN A 277 19.309 -80.804 -43.717 1.00 57.92 A C
+ATOM 1703 O ASN A 277 18.342 -80.808 -42.942 1.00 60.68 A O
+ATOM 1704 CB ASN A 277 20.830 -80.861 -41.715 1.00 49.29 A C
+ATOM 1705 CG ASN A 277 21.839 -81.728 -41.101 1.00 59.28 A C
+ATOM 1706 ND2 ASN A 277 21.460 -82.971 -40.807 1.00 51.74 A N
+ATOM 1707 OD1 ASN A 277 22.991 -81.335 -40.986 1.00 66.28 A O
+ATOM 1708 N VAL A 278 19.211 -80.386 -44.980 1.00 51.88 A N
+ATOM 1709 CA VAL A 278 17.986 -79.797 -45.518 1.00 47.62 A C
+ATOM 1710 C VAL A 278 16.738 -80.500 -44.995 1.00 45.74 A C
+ATOM 1711 O VAL A 278 15.830 -79.833 -44.493 1.00 53.40 A O
+ATOM 1712 CB VAL A 278 17.980 -79.844 -47.052 1.00 53.33 A C
+ATOM 1713 CG1 VAL A 278 16.813 -79.077 -47.556 1.00 57.62 A C
+ATOM 1714 CG2 VAL A 278 19.269 -79.307 -47.607 1.00 50.72 A C
+ATOM 1715 N GLN A 279 16.710 -81.842 -45.046 1.00 44.50 A N
+ATOM 1716 CA GLN A 279 15.560 -82.596 -44.535 1.00 45.63 A C
+ATOM 1717 C GLN A 279 15.129 -82.145 -43.127 1.00 49.69 A C
+ATOM 1718 O GLN A 279 13.923 -82.057 -42.840 1.00 52.38 A O
+ATOM 1719 CB GLN A 279 15.866 -84.101 -44.543 1.00 54.56 A C
+ATOM 1720 CG GLN A 279 17.030 -84.548 -43.653 1.00 74.65 A C
+ATOM 1721 CD GLN A 279 18.363 -84.539 -44.387 1.00 97.39 A C
+ATOM 1722 NE2 GLN A 279 19.308 -85.344 -43.911 1.00103.93 A N
+ATOM 1723 OE1 GLN A 279 18.540 -83.817 -45.370 1.00104.68 A O
+ATOM 1724 N ILE A 280 16.088 -81.858 -42.232 1.00 48.74 A N
+ATOM 1725 CA ILE A 280 15.733 -81.329 -40.914 1.00 50.08 A C
+ATOM 1726 C ILE A 280 15.235 -79.899 -41.027 1.00 47.90 A C
+ATOM 1727 O ILE A 280 14.196 -79.540 -40.456 1.00 53.07 A O
+ATOM 1728 CB ILE A 280 16.922 -81.410 -39.949 1.00 59.33 A C
+ATOM 1729 CG1 ILE A 280 17.447 -82.821 -39.929 1.00 52.05 A C
+ATOM 1730 CG2 ILE A 280 16.492 -80.994 -38.559 1.00 47.76 A C
+ATOM 1731 CD1 ILE A 280 16.391 -83.773 -39.476 1.00 52.26 A C
+ATOM 1732 N VAL A 281 15.985 -79.059 -41.751 1.00 41.63 A N
+ATOM 1733 CA VAL A 281 15.602 -77.661 -41.928 1.00 44.71 A C
+ATOM 1734 C VAL A 281 14.144 -77.556 -42.334 1.00 51.82 A C
+ATOM 1735 O VAL A 281 13.409 -76.688 -41.852 1.00 48.52 A O
+ATOM 1736 CB VAL A 281 16.513 -76.995 -42.966 1.00 50.59 A C
+ATOM 1737 CG1 VAL A 281 16.393 -75.499 -42.851 1.00 49.71 A C
+ATOM 1738 CG2 VAL A 281 17.933 -77.466 -42.778 1.00 43.64 A C
+ATOM 1739 N ARG A 282 13.696 -78.454 -43.210 1.00 48.93 A N
+ATOM 1740 CA ARG A 282 12.307 -78.428 -43.641 1.00 42.21 A C
+ATOM 1741 C ARG A 282 11.372 -78.584 -42.460 1.00 49.06 A C
+ATOM 1742 O ARG A 282 10.404 -77.832 -42.320 1.00 53.41 A O
+ATOM 1743 CB ARG A 282 12.039 -79.523 -44.668 1.00 41.63 A C
+ATOM 1744 CG ARG A 282 12.573 -79.243 -46.035 1.00 50.24 A C
+ATOM 1745 CD ARG A 282 11.887 -80.129 -47.028 1.00 48.78 A C
+ATOM 1746 NE ARG A 282 12.530 -80.051 -48.326 1.00 49.94 A N
+ATOM 1747 CZ ARG A 282 12.522 -78.966 -49.095 1.00 48.33 A C
+ATOM 1748 NH1 ARG A 282 11.890 -77.889 -48.669 1.00 42.78 A N1+
+ATOM 1749 NH2 ARG A 282 13.139 -78.956 -50.282 1.00 58.58 A N
+ATOM 1750 N GLN A 283 11.647 -79.548 -41.594 1.00 47.61 A N
+ATOM 1751 CA GLN A 283 10.689 -79.822 -40.539 1.00 49.02 A C
+ATOM 1752 C GLN A 283 10.641 -78.720 -39.490 1.00 52.67 A C
+ATOM 1753 O GLN A 283 9.641 -78.586 -38.772 1.00 51.80 A O
+ATOM 1754 CB GLN A 283 11.027 -81.151 -39.901 1.00 40.51 A C
+ATOM 1755 CG GLN A 283 11.134 -82.271 -40.899 1.00 49.23 A C
+ATOM 1756 CD GLN A 283 10.700 -83.571 -40.261 1.00 50.51 A C
+ATOM 1757 NE2 GLN A 283 9.779 -84.282 -40.921 1.00 65.94 A N
+ATOM 1758 OE1 GLN A 283 11.183 -83.935 -39.180 1.00 70.82 A O
+ATOM 1759 N GLN A 284 11.697 -77.940 -39.389 1.00 44.51 A N
+ATOM 1760 CA GLN A 284 11.760 -76.831 -38.467 1.00 50.51 A C
+ATOM 1761 C GLN A 284 11.326 -75.527 -39.116 1.00 58.12 A C
+ATOM 1762 O GLN A 284 11.520 -74.468 -38.522 1.00 64.25 A O
+ATOM 1763 CB GLN A 284 13.178 -76.710 -37.904 1.00 49.12 A C
+ATOM 1764 CG GLN A 284 13.724 -78.032 -37.353 1.00 47.47 A C
+ATOM 1765 CD GLN A 284 14.423 -77.879 -36.001 1.00 52.55 A C
+ATOM 1766 NE2 GLN A 284 15.312 -76.886 -35.911 1.00 57.46 A N
+ATOM 1767 OE1 GLN A 284 14.184 -78.661 -35.055 1.00 61.16 A O
+ATOM 1768 N SER A 285 10.789 -75.568 -40.332 1.00 51.45 A N
+ATOM 1769 CA SER A 285 10.407 -74.367 -41.059 1.00 48.07 A C
+ATOM 1770 C SER A 285 8.904 -74.298 -41.227 1.00 44.97 A C
+ATOM 1771 O SER A 285 8.193 -75.291 -41.135 1.00 48.44 A O
+ATOM 1772 CB SER A 285 11.039 -74.316 -42.446 1.00 42.21 A C
+ATOM 1773 OG SER A 285 12.440 -74.237 -42.355 1.00 50.91 A O
+ATOM 1774 N TYR A 286 8.417 -73.108 -41.470 1.00 52.09 A N
+ATOM 1775 CA TYR A 286 7.008 -72.956 -41.757 1.00 50.14 A C
+ATOM 1776 C TYR A 286 6.831 -72.252 -43.098 1.00 59.69 A C
+ATOM 1777 O TYR A 286 7.690 -71.480 -43.540 1.00 55.07 A O
+ATOM 1778 CB TYR A 286 6.308 -72.168 -40.646 1.00 51.64 A C
+ATOM 1779 CG TYR A 286 6.170 -72.871 -39.310 1.00 49.35 A C
+ATOM 1780 CD1 TYR A 286 7.165 -72.765 -38.356 1.00 43.60 A C
+ATOM 1781 CD2 TYR A 286 5.011 -73.559 -38.965 1.00 57.36 A C
+ATOM 1782 CE1 TYR A 286 7.040 -73.375 -37.094 1.00 49.91 A C
+ATOM 1783 CE2 TYR A 286 4.867 -74.159 -37.706 1.00 48.67 A C
+ATOM 1784 CZ TYR A 286 5.893 -74.070 -36.779 1.00 46.70 A C
+ATOM 1785 OH TYR A 286 5.799 -74.658 -35.541 1.00 64.43 A O
+ATOM 1786 N SER A 287 5.713 -72.539 -43.753 1.00 45.16 A N
+ATOM 1787 CA SER A 287 5.250 -71.773 -44.900 1.00 43.36 A C
+ATOM 1788 C SER A 287 3.929 -71.147 -44.504 1.00 46.69 A C
+ATOM 1789 O SER A 287 2.992 -71.854 -44.112 1.00 49.59 A O
+ATOM 1790 CB SER A 287 5.081 -72.641 -46.149 1.00 50.95 A C
+ATOM 1791 OG SER A 287 4.429 -71.916 -47.174 1.00 50.22 A O
+ATOM 1792 N ILE A 288 3.869 -69.829 -44.590 1.00 47.55 A N
+ATOM 1793 CA ILE A 288 2.703 -69.065 -44.199 1.00 45.95 A C
+ATOM 1794 C ILE A 288 2.159 -68.378 -45.438 1.00 50.63 A C
+ATOM 1795 O ILE A 288 2.827 -67.502 -46.010 1.00 51.10 A O
+ATOM 1796 CB ILE A 288 3.036 -68.031 -43.128 1.00 53.53 A C
+ATOM 1797 CG1 ILE A 288 3.910 -68.652 -42.057 1.00 49.22 A C
+ATOM 1798 CG2 ILE A 288 1.765 -67.538 -42.523 1.00 53.32 A C
+ATOM 1799 CD1 ILE A 288 4.377 -67.642 -41.041 1.00 51.29 A C
+ATOM 1800 N MET A 289 0.946 -68.761 -45.845 1.00 45.19 A N
+ATOM 1801 CA MET A 289 0.223 -67.997 -46.853 1.00 43.03 A C
+ATOM 1802 C MET A 289 0.045 -66.556 -46.408 1.00 48.24 A C
+ATOM 1803 O MET A 289 -0.446 -66.287 -45.308 1.00 55.29 A O
+ATOM 1804 CB MET A 289 -1.142 -68.607 -47.110 1.00 49.46 A C
+ATOM 1805 CG MET A 289 -2.030 -67.667 -47.882 1.00 47.86 A C
+ATOM 1806 SD MET A 289 -1.344 -67.446 -49.514 1.00 60.09 A S
+ATOM 1807 CE MET A 289 -2.552 -66.361 -50.273 1.00 58.46 A C
+ATOM 1808 N SER A 290 0.428 -65.630 -47.279 1.00 52.78 A N
+ATOM 1809 CA SER A 290 0.341 -64.217 -46.949 1.00 55.52 A C
+ATOM 1810 C SER A 290 -0.652 -63.481 -47.840 1.00 59.96 A C
+ATOM 1811 O SER A 290 -1.771 -63.185 -47.403 1.00 62.50 A O
+ATOM 1812 CB SER A 290 1.720 -63.577 -47.053 1.00 47.75 A C
+ATOM 1813 OG SER A 290 1.659 -62.204 -46.723 1.00 57.69 A O
+ATOM 1814 N ILE A 291 -0.280 -63.175 -49.084 1.00 47.30 A N
+ATOM 1815 CA ILE A 291 -1.172 -62.406 -49.942 1.00 54.18 A C
+ATOM 1816 C ILE A 291 -1.137 -62.970 -51.342 1.00 51.55 A C
+ATOM 1817 O ILE A 291 -0.225 -63.703 -51.731 1.00 60.61 A O
+ATOM 1818 CB ILE A 291 -0.845 -60.901 -50.075 1.00 62.85 A C
+ATOM 1819 CG1 ILE A 291 0.533 -60.695 -50.675 1.00 72.52 A C
+ATOM 1820 CG2 ILE A 291 -0.878 -60.207 -48.750 1.00 70.32 A C
+ATOM 1821 CD1 ILE A 291 0.914 -59.259 -50.676 1.00 81.12 A C
+ATOM 1822 N ILE A 292 -2.158 -62.597 -52.100 1.00 48.65 A N
+ATOM 1823 CA ILE A 292 -2.137 -62.657 -53.547 1.00 58.40 A C
+ATOM 1824 C ILE A 292 -2.769 -61.353 -54.019 1.00 55.03 A C
+ATOM 1825 O ILE A 292 -3.938 -61.087 -53.731 1.00 64.24 A O
+ATOM 1826 CB ILE A 292 -2.869 -63.892 -54.088 1.00 55.02 A C
+ATOM 1827 CG1 ILE A 292 -3.022 -63.814 -55.587 1.00 55.55 A C
+ATOM 1828 CG2 ILE A 292 -4.189 -64.126 -53.377 1.00 53.16 A C
+ATOM 1829 CD1 ILE A 292 -3.735 -65.011 -56.135 1.00 60.19 A C
+ATOM 1830 N LYS A 293 -1.983 -60.512 -54.678 1.00 56.95 A N
+ATOM 1831 CA LYS A 293 -2.510 -59.259 -55.203 1.00 67.51 A C
+ATOM 1832 C LYS A 293 -1.522 -58.740 -56.239 1.00 72.08 A C
+ATOM 1833 O LYS A 293 -0.418 -59.265 -56.380 1.00 71.43 A O
+ATOM 1834 CB LYS A 293 -2.721 -58.226 -54.090 1.00 72.50 A C
+ATOM 1835 CG LYS A 293 -1.418 -57.732 -53.465 1.00 82.50 A C
+ATOM 1836 CD LYS A 293 -1.658 -56.644 -52.425 1.00 87.13 A C
+ATOM 1837 CE LYS A 293 -1.915 -55.279 -53.034 1.00 98.60 A C
+ATOM 1838 NZ LYS A 293 -1.916 -54.234 -51.971 1.00101.73 A N1+
+ATOM 1839 N GLU A 294 -1.925 -57.675 -56.940 1.00 80.86 A N
+ATOM 1840 CA GLU A 294 -1.072 -57.000 -57.912 1.00 81.95 A C
+ATOM 1841 C GLU A 294 -0.300 -57.992 -58.777 1.00 66.84 A C
+ATOM 1842 O GLU A 294 0.889 -57.774 -59.035 1.00 56.09 A O
+ATOM 1843 CB GLU A 294 -0.110 -56.035 -57.206 1.00 91.99 A C
+ATOM 1844 CG GLU A 294 -0.806 -54.920 -56.411 1.00110.43 A C
+ATOM 1845 CD GLU A 294 0.166 -53.884 -55.837 1.00126.62 A C
+ATOM 1846 OE1 GLU A 294 1.323 -53.829 -56.313 1.00123.18 A O
+ATOM 1847 OE2 GLU A 294 -0.232 -53.112 -54.927 1.00136.21 A O1-
+ATOM 1848 N GLU A 295 -0.957 -59.090 -59.195 1.00 63.22 A N
+ATOM 1849 CA GLU A 295 -0.410 -60.109 -60.115 1.00 65.80 A C
+ATOM 1850 C GLU A 295 0.768 -60.906 -59.525 1.00 52.33 A C
+ATOM 1851 O GLU A 295 1.605 -61.449 -60.259 1.00 69.17 A O
+ATOM 1852 CB GLU A 295 -0.002 -59.490 -61.457 1.00 75.33 A C
+ATOM 1853 CG GLU A 295 -1.178 -59.145 -62.351 1.00 80.60 A C
+ATOM 1854 CD GLU A 295 -0.810 -59.117 -63.829 1.00 98.05 A C
+ATOM 1855 OE1 GLU A 295 0.390 -59.290 -64.149 1.00 98.69 A O
+ATOM 1856 OE2 GLU A 295 -1.719 -58.918 -64.669 1.00109.05 A O1-
+ATOM 1857 N VAL A 296 0.839 -61.018 -58.205 1.00 51.62 A N
+ATOM 1858 CA VAL A 296 1.837 -61.842 -57.546 1.00 60.33 A C
+ATOM 1859 C VAL A 296 1.203 -62.536 -56.345 1.00 53.19 A C
+ATOM 1860 O VAL A 296 0.530 -61.897 -55.526 1.00 60.62 A O
+ATOM 1861 CB VAL A 296 3.062 -61.002 -57.142 1.00 60.33 A C
+ATOM 1862 CG1 VAL A 296 2.627 -59.803 -56.388 1.00 62.98 A C
+ATOM 1863 CG2 VAL A 296 3.979 -61.818 -56.252 1.00 56.28 A C
+ATOM 1864 N LEU A 297 1.372 -63.856 -56.278 1.00 51.51 A N
+ATOM 1865 CA LEU A 297 1.147 -64.624 -55.060 1.00 51.11 A C
+ATOM 1866 C LEU A 297 2.427 -64.622 -54.243 1.00 47.28 A C
+ATOM 1867 O LEU A 297 3.512 -64.869 -54.780 1.00 50.62 A O
+ATOM 1868 CB LEU A 297 0.748 -66.063 -55.380 1.00 50.01 A C
+ATOM 1869 CG LEU A 297 1.106 -67.126 -54.337 1.00 47.86 A C
+ATOM 1870 CD1 LEU A 297 0.273 -66.954 -53.106 1.00 48.14 A C
+ATOM 1871 CD2 LEU A 297 0.919 -68.514 -54.901 1.00 57.08 A C
+ATOM 1872 N ALA A 298 2.301 -64.356 -52.947 1.00 47.52 A N
+ATOM 1873 CA ALA A 298 3.462 -64.257 -52.073 1.00 51.76 A C
+ATOM 1874 C ALA A 298 3.151 -64.881 -50.722 1.00 49.81 A C
+ATOM 1875 O ALA A 298 2.107 -64.601 -50.117 1.00 57.44 A O
+ATOM 1876 CB ALA A 298 3.892 -62.798 -51.896 1.00 42.93 A C
+ATOM 1877 N TYR A 299 4.070 -65.719 -50.255 1.00 44.12 A N
+ATOM 1878 CA TYR A 299 3.964 -66.347 -48.953 1.00 48.36 A C
+ATOM 1879 C TYR A 299 5.335 -66.328 -48.315 1.00 53.91 A C
+ATOM 1880 O TYR A 299 6.353 -66.253 -49.003 1.00 56.05 A O
+ATOM 1881 CB TYR A 299 3.429 -67.774 -49.045 1.00 43.65 A C
+ATOM 1882 CG TYR A 299 4.184 -68.716 -49.954 1.00 40.70 A C
+ATOM 1883 CD1 TYR A 299 4.935 -69.759 -49.439 1.00 46.74 A C
+ATOM 1884 CD2 TYR A 299 4.129 -68.576 -51.337 1.00 37.57 A C
+ATOM 1885 CE1 TYR A 299 5.608 -70.647 -50.285 1.00 43.48 A C
+ATOM 1886 CE2 TYR A 299 4.813 -69.448 -52.188 1.00 36.04 A C
+ATOM 1887 CZ TYR A 299 5.546 -70.478 -51.656 1.00 53.11 A C
+ATOM 1888 OH TYR A 299 6.209 -71.342 -52.493 1.00 43.83 A O
+ATOM 1889 N VAL A 300 5.352 -66.362 -46.988 1.00 54.71 A N
+ATOM 1890 CA VAL A 300 6.586 -66.261 -46.225 1.00 52.16 A C
+ATOM 1891 C VAL A 300 6.965 -67.646 -45.761 1.00 46.31 A C
+ATOM 1892 O VAL A 300 6.125 -68.380 -45.226 1.00 52.42 A O
+ATOM 1893 CB VAL A 300 6.432 -65.327 -45.021 1.00 45.81 A C
+ATOM 1894 CG1 VAL A 300 7.549 -65.569 -44.055 1.00 39.89 A C
+ATOM 1895 CG2 VAL A 300 6.456 -63.899 -45.474 1.00 49.20 A C
+ATOM 1896 N VAL A 301 8.221 -68.007 -45.967 1.00 49.31 A N
+ATOM 1897 CA VAL A 301 8.797 -69.197 -45.368 1.00 45.14 A C
+ATOM 1898 C VAL A 301 9.754 -68.716 -44.292 1.00 48.27 A C
+ATOM 1899 O VAL A 301 10.787 -68.104 -44.606 1.00 53.90 A O
+ATOM 1900 CB VAL A 301 9.500 -70.064 -46.409 1.00 53.82 A C
+ATOM 1901 CG1 VAL A 301 9.965 -71.344 -45.795 1.00 53.55 A C
+ATOM 1902 CG2 VAL A 301 8.538 -70.355 -47.478 1.00 38.66 A C
+ATOM 1903 N GLN A 302 9.394 -68.949 -43.022 1.00 54.57 A N
+ATOM 1904 CA GLN A 302 10.272 -68.616 -41.912 1.00 49.05 A C
+ATOM 1905 C GLN A 302 11.116 -69.830 -41.590 1.00 52.60 A C
+ATOM 1906 O GLN A 302 10.587 -70.923 -41.362 1.00 55.47 A O
+ATOM 1907 CB GLN A 302 9.497 -68.153 -40.684 1.00 51.38 A C
+ATOM 1908 CG GLN A 302 8.815 -69.224 -39.864 1.00 42.31 A C
+ATOM 1909 CD GLN A 302 8.197 -68.647 -38.579 1.00 48.25 A C
+ATOM 1910 NE2 GLN A 302 8.255 -69.420 -37.477 1.00 45.57 A N
+ATOM 1911 OE1 GLN A 302 7.678 -67.518 -38.582 1.00 51.53 A O
+ATOM 1912 N LEU A 303 12.421 -69.632 -41.588 1.00 48.52 A N
+ATOM 1913 CA LEU A 303 13.429 -70.671 -41.575 1.00 47.70 A C
+ATOM 1914 C LEU A 303 14.239 -70.635 -40.285 1.00 52.04 A C
+ATOM 1915 O LEU A 303 14.523 -69.552 -39.753 1.00 44.78 A O
+ATOM 1916 CB LEU A 303 14.365 -70.484 -42.763 1.00 52.27 A C
+ATOM 1917 CG LEU A 303 13.651 -70.540 -44.101 1.00 55.44 A C
+ATOM 1918 CD1 LEU A 303 14.562 -69.970 -45.143 1.00 54.89 A C
+ATOM 1919 CD2 LEU A 303 13.302 -71.968 -44.446 1.00 38.19 A C
+ATOM 1920 N PRO A 304 14.620 -71.795 -39.761 1.00 46.02 A N
+ATOM 1921 CA PRO A 304 15.288 -71.829 -38.457 1.00 46.44 A C
+ATOM 1922 C PRO A 304 16.712 -71.284 -38.517 1.00 52.76 A C
+ATOM 1923 O PRO A 304 17.541 -71.725 -39.320 1.00 64.40 A O
+ATOM 1924 CB PRO A 304 15.268 -73.318 -38.091 1.00 50.19 A C
+ATOM 1925 CG PRO A 304 15.278 -73.991 -39.384 1.00 49.21 A C
+ATOM 1926 CD PRO A 304 14.462 -73.136 -40.326 1.00 50.97 A C
+ATOM 1927 N LEU A 305 16.999 -70.355 -37.603 1.00 57.28 A N
+ATOM 1928 CA LEU A 305 18.350 -69.893 -37.313 1.00 49.94 A C
+ATOM 1929 C LEU A 305 18.909 -70.712 -36.151 1.00 49.77 A C
+ATOM 1930 O LEU A 305 18.373 -70.660 -35.023 1.00 61.84 A O
+ATOM 1931 CB LEU A 305 18.349 -68.398 -36.987 1.00 64.31 A C
+ATOM 1932 CG LEU A 305 17.958 -67.560 -38.198 1.00 79.86 A C
+ATOM 1933 CD1 LEU A 305 18.151 -66.084 -37.951 1.00 81.20 A C
+ATOM 1934 CD2 LEU A 305 18.765 -68.021 -39.400 1.00 82.84 A C
+ATOM 1935 N TYR A 306 19.976 -71.472 -36.448 1.00 59.68 A N
+ATOM 1936 CA TYR A 306 20.711 -72.266 -35.467 1.00 64.85 A C
+ATOM 1937 C TYR A 306 21.862 -71.419 -34.945 1.00 62.15 A C
+ATOM 1938 O TYR A 306 22.909 -71.301 -35.585 1.00 60.13 A O
+ATOM 1939 CB TYR A 306 21.232 -73.553 -36.104 1.00 56.68 A C
+ATOM 1940 CG TYR A 306 20.164 -74.460 -36.674 1.00 53.24 A C
+ATOM 1941 CD1 TYR A 306 19.090 -74.845 -35.891 1.00 57.61 A C
+ATOM 1942 CD2 TYR A 306 20.212 -74.917 -37.995 1.00 57.10 A C
+ATOM 1943 CE1 TYR A 306 18.090 -75.676 -36.390 1.00 53.83 A C
+ATOM 1944 CE2 TYR A 306 19.212 -75.745 -38.506 1.00 52.09 A C
+ATOM 1945 CZ TYR A 306 18.150 -76.122 -37.687 1.00 50.64 A C
+ATOM 1946 OH TYR A 306 17.121 -76.936 -38.113 1.00 56.47 A O
+ATOM 1947 N GLY A 307 21.707 -70.911 -33.734 1.00 64.36 A N
+ATOM 1948 CA GLY A 307 22.668 -70.000 -33.169 1.00 58.65 A C
+ATOM 1949 C GLY A 307 23.596 -70.648 -32.185 1.00 61.79 A C
+ATOM 1950 O GLY A 307 24.412 -69.962 -31.565 1.00 63.96 A O
+ATOM 1951 N VAL A 308 23.462 -71.946 -31.972 1.00 48.25 A N
+ATOM 1952 CA VAL A 308 24.356 -72.684 -31.106 1.00 60.64 A C
+ATOM 1953 C VAL A 308 24.674 -73.974 -31.843 1.00 62.58 A C
+ATOM 1954 O VAL A 308 23.796 -74.832 -32.003 1.00 57.52 A O
+ATOM 1955 CB VAL A 308 23.728 -72.966 -29.736 1.00 57.97 A C
+ATOM 1956 CG1 VAL A 308 24.645 -73.833 -28.899 1.00 59.17 A C
+ATOM 1957 CG2 VAL A 308 23.420 -71.658 -29.020 1.00 56.98 A C
+ATOM 1958 N ILE A 309 25.931 -74.130 -32.260 1.00 56.58 A N
+ATOM 1959 CA ILE A 309 26.373 -75.218 -33.129 1.00 53.60 A C
+ATOM 1960 C ILE A 309 27.548 -75.900 -32.462 1.00 50.71 A C
+ATOM 1961 O ILE A 309 28.480 -75.228 -32.012 1.00 55.08 A O
+ATOM 1962 CB ILE A 309 26.782 -74.718 -34.537 1.00 60.13 A C
+ATOM 1963 CG1 ILE A 309 25.569 -74.254 -35.353 1.00 60.01 A C
+ATOM 1964 CG2 ILE A 309 27.658 -75.719 -35.259 1.00 66.28 A C
+ATOM 1965 CD1 ILE A 309 24.310 -74.975 -35.024 1.00 72.90 A C
+ATOM 1966 N ASP A 310 27.505 -77.227 -32.411 1.00 65.41 A N
+ATOM 1967 CA ASP A 310 28.629 -78.046 -31.959 1.00 65.14 A C
+ATOM 1968 C ASP A 310 29.025 -77.716 -30.510 1.00 58.68 A C
+ATOM 1969 O ASP A 310 30.140 -77.258 -30.225 1.00 67.91 A O
+ATOM 1970 CB ASP A 310 29.838 -77.898 -32.909 1.00 62.93 A C
+ATOM 1971 CG ASP A 310 29.573 -78.412 -34.342 1.00 79.05 A C
+ATOM 1972 OD1 ASP A 310 28.864 -79.437 -34.543 1.00 78.11 A O
+ATOM 1973 OD2 ASP A 310 30.113 -77.780 -35.277 1.00 92.68 A O1-
+ATOM 1974 N THR A 311 28.101 -77.977 -29.587 1.00 64.55 A N
+ATOM 1975 CA THR A 311 28.418 -78.043 -28.170 1.00 66.94 A C
+ATOM 1976 C THR A 311 28.023 -79.440 -27.720 1.00 61.38 A C
+ATOM 1977 O THR A 311 27.322 -80.149 -28.452 1.00 66.76 A O
+ATOM 1978 CB THR A 311 27.657 -77.004 -27.344 1.00 60.62 A C
+ATOM 1979 CG2 THR A 311 27.849 -75.657 -27.970 1.00 57.31 A C
+ATOM 1980 OG1 THR A 311 26.264 -77.289 -27.403 1.00 54.38 A O
+ATOM 1981 N PRO A 312 28.487 -79.898 -26.558 1.00 70.98 A N
+ATOM 1982 CA PRO A 312 28.166 -81.272 -26.149 1.00 66.03 A C
+ATOM 1983 C PRO A 312 26.721 -81.415 -25.697 1.00 70.72 A C
+ATOM 1984 O PRO A 312 26.132 -80.510 -25.090 1.00 69.46 A O
+ATOM 1985 CB PRO A 312 29.131 -81.541 -24.996 1.00 62.83 A C
+ATOM 1986 CG PRO A 312 29.514 -80.192 -24.505 1.00 72.29 A C
+ATOM 1987 CD PRO A 312 29.498 -79.286 -25.682 1.00 63.94 A C
+ATOM 1988 N CYS A 313 26.160 -82.585 -26.010 1.00 65.11 A N
+ATOM 1989 CA CYS A 313 24.779 -82.924 -25.702 1.00 64.93 A C
+ATOM 1990 C CYS A 313 24.727 -84.364 -25.219 1.00 69.98 A C
+ATOM 1991 O CYS A 313 25.517 -85.207 -25.647 1.00 69.84 A O
+ATOM 1992 CB CYS A 313 23.873 -82.764 -26.925 1.00 59.28 A C
+ATOM 1993 SG CYS A 313 23.805 -81.102 -27.643 1.00 82.09 A S
+ATOM 1994 N TRP A 314 23.788 -84.644 -24.329 1.00 67.42 A N
+ATOM 1995 CA TRP A 314 23.648 -85.988 -23.802 1.00 66.76 A C
+ATOM 1996 C TRP A 314 22.167 -86.241 -23.583 1.00 69.43 A C
+ATOM 1997 O TRP A 314 21.395 -85.310 -23.333 1.00 72.52 A O
+ATOM 1998 CB TRP A 314 24.454 -86.166 -22.505 1.00 60.62 A C
+ATOM 1999 CG TRP A 314 24.187 -85.084 -21.455 1.00 70.58 A C
+ATOM 2000 CD1 TRP A 314 23.212 -85.096 -20.489 1.00 64.47 A C
+ATOM 2001 CD2 TRP A 314 24.905 -83.852 -21.272 1.00 67.71 A C
+ATOM 2002 CE2 TRP A 314 24.302 -83.170 -20.185 1.00 68.43 A C
+ATOM 2003 CE3 TRP A 314 25.995 -83.255 -21.923 1.00 72.10 A C
+ATOM 2004 NE1 TRP A 314 23.277 -83.952 -19.726 1.00 69.76 A N
+ATOM 2005 CZ2 TRP A 314 24.749 -81.927 -19.736 1.00 75.82 A C
+ATOM 2006 CZ3 TRP A 314 26.443 -82.005 -21.469 1.00 68.27 A C
+ATOM 2007 CH2 TRP A 314 25.817 -81.362 -20.386 1.00 73.35 A C
+ATOM 2008 N LYS A 315 21.770 -87.500 -23.718 1.00 64.85 A N
+ATOM 2009 CA LYS A 315 20.376 -87.900 -23.574 1.00 64.39 A C
+ATOM 2010 C LYS A 315 20.248 -88.721 -22.303 1.00 65.02 A C
+ATOM 2011 O LYS A 315 20.867 -89.784 -22.183 1.00 64.94 A O
+ATOM 2012 CB LYS A 315 19.895 -88.699 -24.785 1.00 68.39 A C
+ATOM 2013 CG LYS A 315 18.382 -88.897 -24.870 1.00 68.55 A C
+ATOM 2014 CD LYS A 315 18.030 -89.922 -25.955 1.00 69.09 A C
+ATOM 2015 CE LYS A 315 16.529 -90.035 -26.195 1.00 63.12 A C
+ATOM 2016 NZ LYS A 315 16.166 -89.715 -27.627 1.00 66.56 A N1+
+ATOM 2017 N LEU A 316 19.455 -88.223 -21.364 1.00 66.61 A N
+ATOM 2018 CA LEU A 316 19.279 -88.863 -20.072 1.00 70.06 A C
+ATOM 2019 C LEU A 316 18.076 -89.785 -20.145 1.00 73.02 A C
+ATOM 2020 O LEU A 316 16.972 -89.331 -20.476 1.00 70.06 A O
+ATOM 2021 CB LEU A 316 19.070 -87.808 -18.989 1.00 63.81 A C
+ATOM 2022 CG LEU A 316 19.203 -88.180 -17.507 1.00 65.41 A C
+ATOM 2023 CD1 LEU A 316 19.512 -86.922 -16.687 1.00 62.67 A C
+ATOM 2024 CD2 LEU A 316 17.968 -88.861 -16.982 1.00 59.85 A C
+ATOM 2025 N HIS A 317 18.281 -91.057 -19.800 1.00 70.33 A N
+ATOM 2026 CA HIS A 317 17.231 -92.072 -19.808 1.00 63.89 A C
+ATOM 2027 C HIS A 317 16.872 -92.493 -18.377 1.00 68.55 A C
+ATOM 2028 O HIS A 317 17.756 -92.656 -17.527 1.00 74.72 A O
+ATOM 2029 CB HIS A 317 17.664 -93.309 -20.613 1.00 61.72 A C
+ATOM 2030 CG HIS A 317 18.215 -93.008 -21.977 1.00 68.77 A C
+ATOM 2031 CD2 HIS A 317 19.311 -92.310 -22.364 1.00 60.79 A C
+ATOM 2032 ND1 HIS A 317 17.649 -93.502 -23.136 1.00 69.35 A N
+ATOM 2033 CE1 HIS A 317 18.356 -93.094 -24.178 1.00 67.83 A C
+ATOM 2034 NE2 HIS A 317 19.374 -92.376 -23.738 1.00 69.90 A N
+ATOM 2035 N THR A 318 15.568 -92.692 -18.119 1.00 69.89 A N
+ATOM 2036 CA THR A 318 15.049 -93.012 -16.792 1.00 64.77 A C
+ATOM 2037 C THR A 318 14.088 -94.199 -16.834 1.00 68.84 A C
+ATOM 2038 O THR A 318 13.280 -94.327 -17.761 1.00 71.33 A O
+ATOM 2039 CB THR A 318 14.288 -91.846 -16.200 1.00 62.69 A C
+ATOM 2040 CG2 THR A 318 14.991 -90.567 -16.479 1.00 68.28 A C
+ATOM 2041 OG1 THR A 318 12.993 -91.792 -16.803 1.00 80.04 A O
+ATOM 2042 N SER A 319 14.125 -95.022 -15.788 1.00 76.29 A N
+ATOM 2043 CA SER A 319 13.257 -96.187 -15.681 1.00 74.47 A C
+ATOM 2044 C SER A 319 12.895 -96.415 -14.220 1.00 75.40 A C
+ATOM 2045 O SER A 319 13.640 -95.994 -13.319 1.00 78.14 A O
+ATOM 2046 CB SER A 319 13.942 -97.419 -16.281 1.00 77.11 A C
+ATOM 2047 OG SER A 319 13.103 -98.556 -16.254 1.00 86.38 A O
+ATOM 2048 N PRO A 320 11.754 -97.072 -13.936 1.00 81.68 A N
+ATOM 2049 CA PRO A 320 11.337 -97.311 -12.546 1.00 80.35 A C
+ATOM 2050 C PRO A 320 12.380 -98.048 -11.706 1.00 83.07 A C
+ATOM 2051 O PRO A 320 12.971 -99.043 -12.144 1.00 84.35 A O
+ATOM 2052 CB PRO A 320 10.071 -98.162 -12.693 1.00 74.49 A C
+ATOM 2053 CG PRO A 320 9.560 -97.894 -14.024 1.00 80.85 A C
+ATOM 2054 CD PRO A 320 10.698 -97.463 -14.893 1.00 80.08 A C
+ATOM 2055 N LEU A 321 12.598 -97.552 -10.493 1.00 77.13 A N
+ATOM 2056 CA LEU A 321 13.507 -98.173 -9.543 1.00 76.07 A C
+ATOM 2057 C LEU A 321 12.496 -98.854 -8.646 1.00 75.77 A C
+ATOM 2058 O LEU A 321 11.302 -98.592 -8.797 1.00 80.69 A O
+ATOM 2059 CB LEU A 321 14.293 -97.120 -8.769 1.00 77.74 A C
+ATOM 2060 CG LEU A 321 15.488 -97.627 -7.959 1.00 77.69 A C
+ATOM 2061 CD1 LEU A 321 16.158 -98.796 -8.664 1.00 77.14 A C
+ATOM 2062 CD2 LEU A 321 16.484 -96.504 -7.710 1.00 77.32 A C
+ATOM 2063 N CYS A 322 12.896 -99.721 -7.722 1.00 87.18 A N
+ATOM 2064 CA CYS A 322 11.830-100.313 -6.941 1.00 86.76 A C
+ATOM 2065 C CYS A 322 12.412-101.415 -6.059 1.00 92.16 A C
+ATOM 2066 O CYS A 322 13.537-101.872 -6.281 1.00 91.55 A O
+ATOM 2067 CB CYS A 322 10.778-100.817 -7.915 1.00 88.14 A C
+ATOM 2068 SG CYS A 322 9.092-100.677 -7.479 1.00 97.37 A S
+ATOM 2069 N THR A 323 11.650-101.833 -5.044 1.00 83.17 A N
+ATOM 2070 CA THR A 323 12.186-102.860 -4.162 1.00 82.85 A C
+ATOM 2071 C THR A 323 11.756-104.256 -4.633 1.00 90.32 A C
+ATOM 2072 O THR A 323 10.896-104.415 -5.504 1.00 87.84 A O
+ATOM 2073 CB THR A 323 11.739-102.592 -2.721 1.00 90.30 A C
+ATOM 2074 CG2 THR A 323 12.287-101.243 -2.263 1.00 78.56 A C
+ATOM 2075 OG1 THR A 323 10.307-102.531 -2.662 1.00 87.58 A O
+ATOM 2076 N THR A 324 12.368-105.273 -4.029 1.00 86.72 A N
+ATOM 2077 CA THR A 324 12.156-106.676 -4.371 1.00 89.30 A C
+ATOM 2078 C THR A 324 11.255-107.415 -3.384 1.00 94.57 A C
+ATOM 2079 O THR A 324 11.125-108.641 -3.492 1.00 97.73 A O
+ATOM 2080 CB THR A 324 13.503-107.387 -4.495 1.00 82.62 A C
+ATOM 2081 CG2 THR A 324 13.413-108.609 -5.418 1.00 80.95 A C
+ATOM 2082 OG1 THR A 324 14.447-106.469 -5.050 1.00 84.38 A O
+ATOM 2083 N ASN A 325 10.662-106.712 -2.410 1.00 98.50 A N
+ATOM 2084 CA ASN A 325 9.958-107.362 -1.303 1.00 98.15 A C
+ATOM 2085 C ASN A 325 8.986-108.414 -1.822 1.00100.90 A C
+ATOM 2086 O ASN A 325 8.127-108.128 -2.664 1.00102.52 A O
+ATOM 2087 CB ASN A 325 9.200-106.318 -0.478 1.00102.34 A C
+ATOM 2088 CG ASN A 325 10.078-105.633 0.546 1.00110.75 A C
+ATOM 2089 ND2 ASN A 325 9.778-104.372 0.836 1.00111.23 A N
+ATOM 2090 OD1 ASN A 325 11.017-106.230 1.071 1.00114.96 A O
+ATOM 2091 N THR A 326 9.124-109.637 -1.294 1.00104.82 A N
+ATOM 2092 CA THR A 326 8.501-110.803 -1.921 1.00116.59 A C
+ATOM 2093 C THR A 326 6.973-110.729 -1.878 1.00122.02 A C
+ATOM 2094 O THR A 326 6.302-111.226 -2.795 1.00118.52 A O
+ATOM 2095 CB THR A 326 9.028-112.082 -1.259 1.00117.87 A C
+ATOM 2096 CG2 THR A 326 8.824-112.052 0.266 1.00113.63 A C
+ATOM 2097 OG1 THR A 326 8.373-113.220 -1.824 1.00117.01 A O
+ATOM 2098 N LYS A 327 6.409-110.091 -0.847 1.00125.96 A N
+ATOM 2099 CA LYS A 327 4.970-109.861 -0.779 1.00122.23 A C
+ATOM 2100 C LYS A 327 4.580-108.649 -1.616 1.00124.25 A C
+ATOM 2101 O LYS A 327 5.296-107.643 -1.660 1.00122.20 A O
+ATOM 2102 CB LYS A 327 4.515-109.666 0.673 1.00115.91 A C
+ATOM 2103 CG LYS A 327 3.321-110.549 1.100 1.00121.43 A C
+ATOM 2104 CD LYS A 327 3.643-112.047 1.073 1.00127.23 A C
+ATOM 2105 CE LYS A 327 2.467-112.877 1.574 1.00129.31 A C
+ATOM 2106 NZ LYS A 327 2.751-114.337 1.545 1.00128.44 A N1+
+ATOM 2107 N GLU A 328 3.424-108.756 -2.267 1.00121.86 A N
+ATOM 2108 CA GLU A 328 2.933-107.771 -3.225 1.00124.99 A C
+ATOM 2109 C GLU A 328 2.293-106.562 -2.545 1.00120.53 A C
+ATOM 2110 O GLU A 328 1.754-106.649 -1.437 1.00121.39 A O
+ATOM 2111 CB GLU A 328 1.932-108.428 -4.184 1.00135.63 A C
+ATOM 2112 CG GLU A 328 0.615-108.907 -3.541 1.00143.01 A C
+ATOM 2113 CD GLU A 328 0.798-110.052 -2.522 1.00143.75 A C
+ATOM 2114 OE1 GLU A 328 1.927-110.586 -2.392 1.00137.59 A O
+ATOM 2115 OE2 GLU A 328 -0.192-110.421 -1.848 1.00143.06 A O1-
+ATOM 2116 N GLY A 329 2.372-105.417 -3.229 1.00115.74 A N
+ATOM 2117 CA GLY A 329 1.875-104.162 -2.711 1.00106.64 A C
+ATOM 2118 C GLY A 329 2.684-103.578 -1.579 1.00 95.32 A C
+ATOM 2119 O GLY A 329 2.420-102.442 -1.174 1.00 96.43 A O
+ATOM 2120 N SER A 330 3.663-104.312 -1.059 1.00103.66 A N
+ATOM 2121 CA SER A 330 4.511-103.877 0.042 1.00101.43 A C
+ATOM 2122 C SER A 330 5.737-103.114 -0.439 1.00 93.32 A C
+ATOM 2123 O SER A 330 6.600-102.769 0.380 1.00 94.21 A O
+ATOM 2124 CB SER A 330 4.932-105.089 0.889 1.00101.88 A C
+ATOM 2125 OG SER A 330 5.754-105.997 0.164 1.00101.02 A O
+ATOM 2126 N ASN A 331 5.827-102.844 -1.742 1.00 80.57 A N
+ATOM 2127 CA ASN A 331 7.013-102.260 -2.346 1.00 85.18 A C
+ATOM 2128 C ASN A 331 6.893-100.748 -2.471 1.00 84.99 A C
+ATOM 2129 O ASN A 331 5.812-100.212 -2.732 1.00 90.37 A O
+ATOM 2130 CB ASN A 331 7.266-102.858 -3.720 1.00 79.94 A C
+ATOM 2131 CG ASN A 331 7.938-104.182 -3.642 1.00 93.78 A C
+ATOM 2132 ND2 ASN A 331 7.231-105.231 -4.053 1.00 94.30 A N
+ATOM 2133 OD1 ASN A 331 9.089-104.275 -3.220 1.00 93.96 A O
+ATOM 2134 N ILE A 332 8.019-100.074 -2.288 1.00 82.32 A N
+ATOM 2135 CA ILE A 332 8.147 -98.645 -2.521 1.00 82.88 A C
+ATOM 2136 C ILE A 332 8.942 -98.446 -3.799 1.00 81.54 A C
+ATOM 2137 O ILE A 332 9.822 -99.251 -4.136 1.00 85.12 A O
+ATOM 2138 CB ILE A 332 8.849 -97.941 -1.355 1.00 77.65 A C
+ATOM 2139 CG1 ILE A 332 10.234 -98.557 -1.163 1.00 73.27 A C
+ATOM 2140 CG2 ILE A 332 8.016 -98.067 -0.122 1.00 72.32 A C
+ATOM 2141 CD1 ILE A 332 11.131 -97.778 -0.242 1.00 79.47 A C
+ATOM 2142 N CYS A 333 8.650 -97.349 -4.504 1.00 80.85 A N
+ATOM 2143 CA CYS A 333 9.104 -97.243 -5.879 1.00 78.12 A C
+ATOM 2144 C CYS A 333 9.405 -95.776 -6.225 1.00 83.57 A C
+ATOM 2145 O CYS A 333 8.665 -94.870 -5.835 1.00 80.78 A O
+ATOM 2146 CB CYS A 333 8.022 -97.890 -6.745 1.00 69.65 A C
+ATOM 2147 SG CYS A 333 8.576 -98.829 -8.173 1.00 81.00 A S
+ATOM 2148 N LEU A 334 10.502 -95.556 -6.960 1.00 79.48 A N
+ATOM 2149 CA LEU A 334 11.079 -94.234 -7.229 1.00 77.15 A C
+ATOM 2150 C LEU A 334 11.648 -94.141 -8.647 1.00 74.14 A C
+ATOM 2151 O LEU A 334 12.478 -94.981 -9.009 1.00 74.74 A O
+ATOM 2152 CB LEU A 334 12.179 -93.977 -6.199 1.00 86.66 A C
+ATOM 2153 CG LEU A 334 13.172 -92.834 -6.266 1.00 89.98 A C
+ATOM 2154 CD1 LEU A 334 13.687 -92.627 -4.888 1.00 82.97 A C
+ATOM 2155 CD2 LEU A 334 14.339 -93.278 -7.108 1.00 76.65 A C
+ATOM 2156 N THR A 335 11.277 -93.098 -9.425 1.00 71.71 A N
+ATOM 2157 CA THR A 335 11.777 -92.938 -10.804 1.00 75.02 A C
+ATOM 2158 C THR A 335 12.170 -91.499 -11.122 1.00 74.66 A C
+ATOM 2159 O THR A 335 11.354 -90.587 -10.949 1.00 77.04 A O
+ATOM 2160 CB THR A 335 10.742 -93.369 -11.846 1.00 72.35 A C
+ATOM 2161 CG2 THR A 335 11.373 -93.360 -13.229 1.00 73.23 A C
+ATOM 2162 OG1 THR A 335 10.290 -94.694 -11.565 1.00 78.77 A O
+ATOM 2163 N ARG A 336 13.387 -91.302 -11.645 1.00 63.13 A N
+ATOM 2164 CA ARG A 336 13.853 -89.955 -11.971 1.00 70.26 A C
+ATOM 2165 C ARG A 336 13.029 -89.366 -13.106 1.00 72.94 A C
+ATOM 2166 O ARG A 336 12.734 -90.041 -14.091 1.00 61.11 A O
+ATOM 2167 CB ARG A 336 15.313 -89.955 -12.399 1.00 62.41 A C
+ATOM 2168 CG ARG A 336 16.321 -90.448 -11.434 1.00 63.44 A C
+ATOM 2169 CD ARG A 336 17.664 -90.407 -12.137 1.00 68.16 A C
+ATOM 2170 NE ARG A 336 18.016 -89.051 -12.563 1.00 68.92 A N
+ATOM 2171 CZ ARG A 336 19.092 -88.733 -13.287 1.00 60.10 A C
+ATOM 2172 NH1 ARG A 336 19.941 -89.670 -13.690 1.00 65.52 A N1+
+ATOM 2173 NH2 ARG A 336 19.322 -87.463 -13.598 1.00 67.04 A N
+ATOM 2174 N THR A 337 12.647 -88.105 -12.974 1.00 68.06 A N
+ATOM 2175 CA THR A 337 11.811 -87.473 -13.983 1.00 65.45 A C
+ATOM 2176 C THR A 337 12.560 -86.536 -14.941 1.00 74.66 A C
+ATOM 2177 O THR A 337 11.934 -85.938 -15.826 1.00 66.45 A O
+ATOM 2178 CB THR A 337 10.668 -86.764 -13.282 1.00 66.57 A C
+ATOM 2179 CG2 THR A 337 9.806 -87.812 -12.600 1.00 64.50 A C
+ATOM 2180 OG1 THR A 337 11.204 -85.893 -12.284 1.00 69.07 A O
+ATOM 2181 N ASP A 338 13.880 -86.433 -14.837 1.00 67.29 A N
+ATOM 2182 CA ASP A 338 14.688 -85.519 -15.653 1.00 62.79 A C
+ATOM 2183 C ASP A 338 14.998 -86.067 -17.081 1.00 65.64 A C
+ATOM 2184 O ASP A 338 16.016 -85.699 -17.681 1.00 74.18 A O
+ATOM 2185 CB ASP A 338 15.945 -85.045 -14.881 1.00 64.29 A C
+ATOM 2186 CG ASP A 338 16.578 -86.122 -13.969 1.00 75.12 A C
+ATOM 2187 OD1 ASP A 338 16.150 -87.295 -14.007 1.00 72.94 A O
+ATOM 2188 OD2 ASP A 338 17.529 -85.780 -13.210 1.00 71.80 A O1-
+ATOM 2189 N ARG A 339 14.139 -86.968 -17.580 1.00 66.63 A N
+ATOM 2190 CA ARG A 339 14.235 -87.556 -18.920 1.00 63.23 A C
+ATOM 2191 C ARG A 339 14.526 -86.499 -19.971 1.00 61.44 A C
+ATOM 2192 O ARG A 339 14.092 -85.347 -19.847 1.00 59.55 A O
+ATOM 2193 CB ARG A 339 12.900 -88.213 -19.314 1.00 61.15 A C
+ATOM 2194 CG ARG A 339 12.736 -89.670 -18.988 1.00 74.43 A C
+ATOM 2195 CD ARG A 339 11.260 -90.093 -19.054 1.00 89.28 A C
+ATOM 2196 NE ARG A 339 11.075 -91.459 -18.547 1.00121.85 A N
+ATOM 2197 CZ ARG A 339 9.913 -92.111 -18.504 1.00116.95 A C
+ATOM 2198 NH1 ARG A 339 8.802 -91.534 -18.948 1.00104.58 A N1+
+ATOM 2199 NH2 ARG A 339 9.865 -93.347 -18.014 1.00114.90 A N
+ATOM 2200 N GLY A 340 15.254 -86.913 -21.027 1.00 57.14 A N
+ATOM 2201 CA GLY A 340 15.383 -86.120 -22.242 1.00 56.44 A C
+ATOM 2202 C GLY A 340 16.768 -85.533 -22.462 1.00 64.00 A C
+ATOM 2203 O GLY A 340 17.763 -85.957 -21.867 1.00 69.87 A O
+ATOM 2204 N TRP A 341 16.818 -84.571 -23.393 1.00 62.26 A N
+ATOM 2205 CA TRP A 341 18.067 -84.048 -23.947 1.00 59.86 A C
+ATOM 2206 C TRP A 341 18.640 -82.870 -23.158 1.00 67.97 A C
+ATOM 2207 O TRP A 341 17.902 -82.010 -22.671 1.00 63.25 A O
+ATOM 2208 CB TRP A 341 17.846 -83.613 -25.395 1.00 60.63 A C
+ATOM 2209 CG TRP A 341 17.892 -84.745 -26.360 1.00 58.55 A C
+ATOM 2210 CD1 TRP A 341 16.831 -85.384 -26.921 1.00 53.94 A C
+ATOM 2211 CD2 TRP A 341 19.065 -85.391 -26.866 1.00 55.12 A C
+ATOM 2212 CE2 TRP A 341 18.642 -86.409 -27.730 1.00 60.32 A C
+ATOM 2213 CE3 TRP A 341 20.432 -85.203 -26.670 1.00 66.73 A C
+ATOM 2214 NE1 TRP A 341 17.273 -86.382 -27.757 1.00 65.00 A N
+ATOM 2215 CZ2 TRP A 341 19.535 -87.242 -28.393 1.00 60.31 A C
+ATOM 2216 CZ3 TRP A 341 21.319 -86.032 -27.337 1.00 71.40 A C
+ATOM 2217 CH2 TRP A 341 20.867 -87.038 -28.184 1.00 67.23 A C
+ATOM 2218 N TYR A 342 19.973 -82.824 -23.060 1.00 66.93 A N
+ATOM 2219 CA TYR A 342 20.690 -81.711 -22.443 1.00 55.35 A C
+ATOM 2220 C TYR A 342 21.827 -81.270 -23.354 1.00 71.68 A C
+ATOM 2221 O TYR A 342 22.589 -82.108 -23.850 1.00 65.66 A O
+ATOM 2222 CB TYR A 342 21.254 -82.083 -21.058 1.00 55.37 A C
+ATOM 2223 CG TYR A 342 20.212 -82.450 -20.006 1.00 65.55 A C
+ATOM 2224 CD1 TYR A 342 19.661 -83.722 -19.949 1.00 61.78 A C
+ATOM 2225 CD2 TYR A 342 19.783 -81.523 -19.072 1.00 57.18 A C
+ATOM 2226 CE1 TYR A 342 18.709 -84.052 -19.005 1.00 62.08 A C
+ATOM 2227 CE2 TYR A 342 18.832 -81.845 -18.133 1.00 60.80 A C
+ATOM 2228 CZ TYR A 342 18.304 -83.108 -18.106 1.00 66.76 A C
+ATOM 2229 OH TYR A 342 17.361 -83.429 -17.163 1.00 67.70 A O
+ATOM 2230 N CYS A 343 21.947 -79.953 -23.571 1.00 60.79 A N
+ATOM 2231 CA CYS A 343 23.069 -79.405 -24.326 1.00 57.89 A C
+ATOM 2232 C CYS A 343 23.692 -78.225 -23.602 1.00 63.76 A C
+ATOM 2233 O CYS A 343 22.992 -77.406 -22.993 1.00 73.42 A O
+ATOM 2234 CB CYS A 343 22.666 -78.940 -25.719 1.00 58.63 A C
+ATOM 2235 SG CYS A 343 22.147 -80.248 -26.795 1.00 73.67 A S
+ATOM 2236 N ASP A 344 25.014 -78.129 -23.701 1.00 66.33 A N
+ATOM 2237 CA ASP A 344 25.692 -76.936 -23.212 1.00 67.40 A C
+ATOM 2238 C ASP A 344 25.243 -75.731 -24.044 1.00 73.06 A C
+ATOM 2239 O ASP A 344 25.349 -75.743 -25.276 1.00 69.88 A O
+ATOM 2240 CB ASP A 344 27.223 -77.108 -23.300 1.00 65.32 A C
+ATOM 2241 CG ASP A 344 27.804 -78.116 -22.282 1.00 76.00 A C
+ATOM 2242 OD1 ASP A 344 27.042 -78.820 -21.581 1.00 80.85 A O
+ATOM 2243 OD2 ASP A 344 29.052 -78.189 -22.193 1.00 89.96 A O1-
+ATOM 2244 N ASN A 345 24.716 -74.700 -23.387 1.00 73.34 A N
+ATOM 2245 CA ASN A 345 24.368 -73.456 -24.073 1.00 72.89 A C
+ATOM 2246 C ASN A 345 24.821 -72.293 -23.197 1.00 71.39 A C
+ATOM 2247 O ASN A 345 24.230 -72.054 -22.137 1.00 74.39 A O
+ATOM 2248 CB ASN A 345 22.853 -73.409 -24.329 1.00 63.46 A C
+ATOM 2249 CG ASN A 345 22.420 -72.307 -25.304 1.00 72.17 A C
+ATOM 2250 ND2 ASN A 345 21.417 -72.614 -26.111 1.00 61.89 A N
+ATOM 2251 OD1 ASN A 345 22.934 -71.190 -25.292 1.00 66.49 A O
+ATOM 2252 N ALA A 346 25.832 -71.545 -23.649 1.00 76.67 A N
+ATOM 2253 CA ALA A 346 26.171 -70.228 -23.077 1.00 71.85 A C
+ATOM 2254 C ALA A 346 26.448 -70.277 -21.569 1.00 77.34 A C
+ATOM 2255 O ALA A 346 25.903 -69.488 -20.789 1.00 76.88 A O
+ATOM 2256 CB ALA A 346 25.066 -69.208 -23.379 1.00 73.36 A C
+ATOM 2257 N GLY A 347 27.306 -71.214 -21.154 1.00 77.96 A N
+ATOM 2258 CA GLY A 347 27.613 -71.396 -19.747 1.00 75.51 A C
+ATOM 2259 C GLY A 347 26.526 -72.058 -18.925 1.00 75.00 A C
+ATOM 2260 O GLY A 347 26.795 -72.481 -17.790 1.00 75.74 A O
+ATOM 2261 N SER A 348 25.315 -72.171 -19.449 1.00 70.91 A N
+ATOM 2262 CA SER A 348 24.267 -72.940 -18.810 1.00 70.17 A C
+ATOM 2263 C SER A 348 24.092 -74.260 -19.544 1.00 74.41 A C
+ATOM 2264 O SER A 348 24.785 -74.573 -20.513 1.00 73.01 A O
+ATOM 2265 CB SER A 348 22.959 -72.153 -18.787 1.00 70.18 A C
+ATOM 2266 OG SER A 348 23.028 -71.111 -17.831 1.00 75.82 A O
+ATOM 2267 N VAL A 349 23.155 -75.056 -19.062 1.00 67.57 A N
+ATOM 2268 CA VAL A 349 22.700 -76.240 -19.770 1.00 66.77 A C
+ATOM 2269 C VAL A 349 21.238 -76.016 -20.091 1.00 71.41 A C
+ATOM 2270 O VAL A 349 20.466 -75.620 -19.208 1.00 67.22 A O
+ATOM 2271 CB VAL A 349 22.916 -77.519 -18.943 1.00 70.92 A C
+ATOM 2272 CG1 VAL A 349 21.916 -78.584 -19.307 1.00 69.82 A C
+ATOM 2273 CG2 VAL A 349 24.294 -78.042 -19.219 1.00 65.79 A C
+ATOM 2274 N SER A 350 20.885 -76.249 -21.346 1.00 61.88 A N
+ATOM 2275 CA SER A 350 19.518 -76.116 -21.776 1.00 59.55 A C
+ATOM 2276 C SER A 350 18.941 -77.449 -21.364 1.00 70.07 A C
+ATOM 2277 O SER A 350 19.641 -78.276 -20.779 1.00 67.22 A O
+ATOM 2278 CB SER A 350 19.437 -75.941 -23.290 1.00 62.15 A C
+ATOM 2279 OG SER A 350 20.730 -75.844 -23.862 1.00 67.95 A O
+ATOM 2280 N PHE A 351 17.675 -77.672 -21.662 1.00 61.92 A N
+ATOM 2281 CA PHE A 351 17.053 -78.931 -21.301 1.00 62.12 A C
+ATOM 2282 C PHE A 351 15.804 -79.084 -22.145 1.00 72.03 A C
+ATOM 2283 O PHE A 351 14.905 -78.229 -22.097 1.00 78.00 A O
+ATOM 2284 CB PHE A 351 16.694 -78.934 -19.813 1.00 58.25 A C
+ATOM 2285 CG PHE A 351 15.884 -80.123 -19.365 1.00 66.63 A C
+ATOM 2286 CD1 PHE A 351 15.986 -81.346 -19.994 1.00 67.41 A C
+ATOM 2287 CD2 PHE A 351 14.990 -79.995 -18.321 1.00 66.14 A C
+ATOM 2288 CE1 PHE A 351 15.214 -82.425 -19.575 1.00 72.41 A C
+ATOM 2289 CE2 PHE A 351 14.228 -81.067 -17.901 1.00 69.33 A C
+ATOM 2290 CZ PHE A 351 14.335 -82.286 -18.527 1.00 71.60 A C
+ATOM 2291 N PHE A 352 15.755 -80.175 -22.903 1.00 62.99 A N
+ATOM 2292 CA PHE A 352 14.604 -80.532 -23.715 1.00 61.93 A C
+ATOM 2293 C PHE A 352 13.925 -81.737 -23.090 1.00 68.66 A C
+ATOM 2294 O PHE A 352 14.453 -82.862 -23.146 1.00 70.38 A O
+ATOM 2295 CB PHE A 352 15.065 -80.809 -25.139 1.00 62.07 A C
+ATOM 2296 CG PHE A 352 15.986 -79.757 -25.640 1.00 66.79 A C
+ATOM 2297 CD1 PHE A 352 15.484 -78.551 -26.087 1.00 66.49 A C
+ATOM 2298 CD2 PHE A 352 17.351 -79.937 -25.603 1.00 65.48 A C
+ATOM 2299 CE1 PHE A 352 16.323 -77.558 -26.530 1.00 66.06 A C
+ATOM 2300 CE2 PHE A 352 18.206 -78.944 -26.050 1.00 70.83 A C
+ATOM 2301 CZ PHE A 352 17.691 -77.757 -26.517 1.00 69.43 A C
+ATOM 2302 N PRO A 353 12.797 -81.525 -22.414 1.00 66.34 A N
+ATOM 2303 CA PRO A 353 12.119 -82.618 -21.721 1.00 67.45 A C
+ATOM 2304 C PRO A 353 11.352 -83.542 -22.644 1.00 68.13 A C
+ATOM 2305 O PRO A 353 10.917 -84.614 -22.201 1.00 71.99 A O
+ATOM 2306 CB PRO A 353 11.188 -81.872 -20.759 1.00 60.83 A C
+ATOM 2307 CG PRO A 353 10.823 -80.643 -21.522 1.00 58.61 A C
+ATOM 2308 CD PRO A 353 11.967 -80.311 -22.464 1.00 59.26 A C
+ATOM 2309 N GLN A 354 11.113 -83.132 -23.884 1.00 63.86 A N
+ATOM 2310 CA GLN A 354 10.196 -83.815 -24.788 1.00 69.82 A C
+ATOM 2311 C GLN A 354 10.983 -84.362 -25.972 1.00 74.43 A C
+ATOM 2312 O GLN A 354 11.630 -83.594 -26.690 1.00 74.72 A O
+ATOM 2313 CB GLN A 354 9.111 -82.839 -25.259 1.00 60.01 A C
+ATOM 2314 CG GLN A 354 8.459 -82.031 -24.144 1.00 84.61 A C
+ATOM 2315 CD GLN A 354 6.952 -82.038 -24.226 1.00120.59 A C
+ATOM 2316 NE2 GLN A 354 6.309 -82.868 -23.405 1.00136.37 A N
+ATOM 2317 OE1 GLN A 354 6.369 -81.306 -25.024 1.00125.24 A O
+ATOM 2318 N ALA A 355 10.930 -85.682 -26.188 1.00 89.59 A N
+ATOM 2319 CA ALA A 355 11.707 -86.250 -27.293 1.00 86.31 A C
+ATOM 2320 C ALA A 355 11.236 -85.741 -28.652 1.00 85.54 A C
+ATOM 2321 O ALA A 355 11.994 -85.824 -29.625 1.00 91.19 A O
+ATOM 2322 CB ALA A 355 11.669 -87.787 -27.274 1.00 72.10 A C
+ATOM 2323 N GLU A 356 10.012 -85.210 -28.737 1.00 79.04 A N
+ATOM 2324 CA GLU A 356 9.546 -84.632 -29.993 1.00 83.17 A C
+ATOM 2325 C GLU A 356 10.329 -83.370 -30.364 1.00 78.45 A C
+ATOM 2326 O GLU A 356 10.556 -83.113 -31.555 1.00 92.89 A O
+ATOM 2327 CB GLU A 356 8.045 -84.329 -29.904 1.00 94.89 A C
+ATOM 2328 CG GLU A 356 7.537 -83.382 -30.996 1.00121.49 A C
+ATOM 2329 CD GLU A 356 6.189 -82.759 -30.673 1.00134.61 A C
+ATOM 2330 OE1 GLU A 356 5.520 -83.220 -29.718 1.00134.99 A O
+ATOM 2331 OE2 GLU A 356 5.808 -81.797 -31.377 1.00136.96 A O1-
+ATOM 2332 N THR A 357 10.746 -82.571 -29.371 1.00 74.82 A N
+ATOM 2333 CA THR A 357 11.484 -81.347 -29.676 1.00 76.60 A C
+ATOM 2334 C THR A 357 12.803 -81.652 -30.386 1.00 79.68 A C
+ATOM 2335 O THR A 357 13.258 -80.859 -31.224 1.00 72.32 A O
+ATOM 2336 CB THR A 357 11.731 -80.527 -28.398 1.00 72.07 A C
+ATOM 2337 CG2 THR A 357 12.598 -79.307 -28.689 1.00 73.61 A C
+ATOM 2338 OG1 THR A 357 10.483 -80.093 -27.840 1.00 83.65 A O
+ATOM 2339 N CYS A 358 13.405 -82.805 -30.113 1.00 73.26 A N
+ATOM 2340 CA CYS A 358 14.732 -83.112 -30.616 1.00 66.18 A C
+ATOM 2341 C CYS A 358 14.681 -84.179 -31.692 1.00 71.12 A C
+ATOM 2342 O CYS A 358 13.980 -85.186 -31.556 1.00 74.15 A O
+ATOM 2343 CB CYS A 358 15.631 -83.565 -29.487 1.00 59.81 A C
+ATOM 2344 SG CYS A 358 15.952 -82.237 -28.361 1.00 68.83 A S
+ATOM 2345 N LYS A 359 15.432 -83.949 -32.760 1.00 59.86 A N
+ATOM 2346 CA LYS A 359 15.603 -84.909 -33.837 1.00 58.91 A C
+ATOM 2347 C LYS A 359 17.093 -85.182 -33.972 1.00 63.56 A C
+ATOM 2348 O LYS A 359 17.916 -84.271 -33.833 1.00 61.06 A O
+ATOM 2349 CB LYS A 359 14.998 -84.368 -35.138 1.00 75.26 A C
+ATOM 2350 CG LYS A 359 13.611 -83.785 -34.911 1.00 95.07 A C
+ATOM 2351 CD LYS A 359 13.203 -82.792 -35.973 1.00 97.02 A C
+ATOM 2352 CE LYS A 359 11.979 -82.015 -35.509 1.00 81.18 A C
+ATOM 2353 NZ LYS A 359 12.321 -81.050 -34.400 1.00 84.62 A N1+
+ATOM 2354 N VAL A 360 17.439 -86.437 -34.214 1.00 68.13 A N
+ATOM 2355 CA VAL A 360 18.817 -86.890 -34.108 1.00 59.62 A C
+ATOM 2356 C VAL A 360 19.320 -87.258 -35.497 1.00 64.38 A C
+ATOM 2357 O VAL A 360 18.997 -88.343 -35.995 1.00 64.89 A O
+ATOM 2358 CB VAL A 360 18.908 -88.105 -33.181 1.00 65.07 A C
+ATOM 2359 CG1 VAL A 360 20.309 -88.256 -32.668 1.00 64.15 A C
+ATOM 2360 CG2 VAL A 360 17.876 -88.005 -32.079 1.00 59.72 A C
+ATOM 2361 N GLN A 361 20.179 -86.438 -36.116 1.00 57.94 A N
+ATOM 2362 CA GLN A 361 20.818 -86.913 -37.344 1.00 65.10 A C
+ATOM 2363 C GLN A 361 22.276 -87.230 -37.015 1.00 70.07 A C
+ATOM 2364 O GLN A 361 23.153 -86.361 -36.995 1.00 74.35 A O
+ATOM 2365 CB GLN A 361 20.678 -85.915 -38.489 1.00 59.61 A C
+ATOM 2366 CG GLN A 361 19.235 -85.582 -38.833 1.00 71.58 A C
+ATOM 2367 CD GLN A 361 18.531 -86.610 -39.724 1.00 84.62 A C
+ATOM 2368 NE2 GLN A 361 17.583 -87.358 -39.147 1.00 80.13 A N
+ATOM 2369 OE1 GLN A 361 18.793 -86.687 -40.926 1.00 87.02 A O
+ATOM 2370 N SER A 362 22.487 -88.546 -36.970 1.00 78.63 A N
+ATOM 2371 CA SER A 362 23.776 -89.174 -36.773 1.00 79.81 A C
+ATOM 2372 C SER A 362 24.308 -88.714 -35.464 1.00 71.90 A C
+ATOM 2373 O SER A 362 23.865 -89.164 -34.407 1.00 71.84 A O
+ATOM 2374 CB SER A 362 24.738 -88.789 -37.898 1.00 84.83 A C
+ATOM 2375 OG SER A 362 25.304 -89.940 -38.502 1.00 91.81 A O
+ATOM 2376 N ASN A 363 25.259 -87.801 -35.518 1.00 76.23 A N
+ATOM 2377 CA ASN A 363 25.776 -87.315 -34.238 1.00 77.73 A C
+ATOM 2378 C ASN A 363 25.289 -85.924 -33.844 1.00 72.04 A C
+ATOM 2379 O ASN A 363 25.599 -85.480 -32.732 1.00 62.25 A O
+ATOM 2380 CB ASN A 363 27.320 -87.391 -34.172 1.00 88.22 A C
+ATOM 2381 CG ASN A 363 28.008 -87.123 -35.504 1.00 96.97 A C
+ATOM 2382 ND2 ASN A 363 29.077 -87.877 -35.763 1.00 97.18 A N
+ATOM 2383 OD1 ASN A 363 27.597 -86.262 -36.289 1.00 99.26 A O
+ATOM 2384 N ARG A 364 24.587 -85.206 -34.723 1.00 59.75 A N
+ATOM 2385 CA ARG A 364 24.064 -83.891 -34.388 1.00 56.48 A C
+ATOM 2386 C ARG A 364 22.640 -84.090 -33.860 1.00 53.27 A C
+ATOM 2387 O ARG A 364 21.917 -84.992 -34.297 1.00 65.34 A O
+ATOM 2388 CB ARG A 364 24.122 -82.927 -35.605 1.00 61.76 A C
+ATOM 2389 CG ARG A 364 25.533 -82.235 -35.990 1.00 66.75 A C
+ATOM 2390 CD ARG A 364 25.498 -81.403 -37.345 1.00 75.86 A C
+ATOM 2391 NE ARG A 364 26.635 -81.524 -38.316 1.00 73.83 A N
+ATOM 2392 CZ ARG A 364 26.561 -81.909 -39.613 1.00 68.36 A C
+ATOM 2393 NH1 ARG A 364 25.437 -82.318 -40.151 1.00 66.58 A N1+
+ATOM 2394 NH2 ARG A 364 27.633 -81.933 -40.391 1.00 66.91 A N
+ATOM 2395 N VAL A 365 22.285 -83.308 -32.850 1.00 56.17 A N
+ATOM 2396 CA VAL A 365 20.967 -83.296 -32.231 1.00 53.14 A C
+ATOM 2397 C VAL A 365 20.363 -81.931 -32.500 1.00 60.48 A C
+ATOM 2398 O VAL A 365 20.763 -80.934 -31.886 1.00 60.45 A O
+ATOM 2399 CB VAL A 365 21.055 -83.558 -30.726 1.00 58.03 A C
+ATOM 2400 CG1 VAL A 365 19.692 -83.716 -30.128 1.00 64.18 A C
+ATOM 2401 CG2 VAL A 365 21.864 -84.794 -30.489 1.00 72.48 A C
+ATOM 2402 N PHE A 366 19.395 -81.879 -33.395 1.00 58.52 A N
+ATOM 2403 CA PHE A 366 18.695 -80.645 -33.697 1.00 57.73 A C
+ATOM 2404 C PHE A 366 17.548 -80.494 -32.707 1.00 62.93 A C
+ATOM 2405 O PHE A 366 16.648 -81.343 -32.678 1.00 67.62 A O
+ATOM 2406 CB PHE A 366 18.149 -80.683 -35.121 1.00 51.33 A C
+ATOM 2407 CG PHE A 366 19.194 -80.573 -36.190 1.00 52.72 A C
+ATOM 2408 CD1 PHE A 366 19.941 -81.678 -36.568 1.00 53.22 A C
+ATOM 2409 CD2 PHE A 366 19.396 -79.374 -36.851 1.00 53.72 A C
+ATOM 2410 CE1 PHE A 366 20.886 -81.590 -37.571 1.00 58.79 A C
+ATOM 2411 CE2 PHE A 366 20.353 -79.277 -37.850 1.00 56.89 A C
+ATOM 2412 CZ PHE A 366 21.097 -80.391 -38.211 1.00 58.07 A C
+ATOM 2413 N CYS A 367 17.562 -79.422 -31.906 1.00 65.25 A N
+ATOM 2414 CA CYS A 367 16.473 -79.197 -30.954 1.00 61.61 A C
+ATOM 2415 C CYS A 367 15.944 -77.785 -31.082 1.00 70.28 A C
+ATOM 2416 O CYS A 367 16.560 -76.921 -31.704 1.00 77.54 A O
+ATOM 2417 CB CYS A 367 16.894 -79.456 -29.506 1.00 58.06 A C
+ATOM 2418 SG CYS A 367 17.468 -81.154 -29.249 1.00 68.16 A S
+ATOM 2419 N ASP A 368 14.780 -77.565 -30.492 1.00 66.07 A N
+ATOM 2420 CA ASP A 368 14.118 -76.274 -30.566 1.00 55.30 A C
+ATOM 2421 C ASP A 368 14.109 -75.643 -29.182 1.00 64.09 A C
+ATOM 2422 O ASP A 368 13.437 -76.140 -28.273 1.00 65.16 A O
+ATOM 2423 CB ASP A 368 12.707 -76.416 -31.114 1.00 52.77 A C
+ATOM 2424 CG ASP A 368 12.173 -75.106 -31.661 1.00 53.95 A C
+ATOM 2425 OD1 ASP A 368 12.854 -74.077 -31.448 1.00 66.55 A O
+ATOM 2426 OD2 ASP A 368 11.091 -75.096 -32.300 1.00 70.59 A O1-
+ATOM 2427 N THR A 369 14.838 -74.535 -29.025 1.00 55.10 A N
+ATOM 2428 CA THR A 369 14.933 -73.928 -27.707 1.00 64.77 A C
+ATOM 2429 C THR A 369 13.583 -73.449 -27.216 1.00 58.83 A C
+ATOM 2430 O THR A 369 13.406 -73.312 -25.999 1.00 64.60 A O
+ATOM 2431 CB THR A 369 15.895 -72.746 -27.707 1.00 65.27 A C
+ATOM 2432 CG2 THR A 369 17.235 -73.164 -28.307 1.00 63.76 A C
+ATOM 2433 OG1 THR A 369 15.342 -71.675 -28.480 1.00 68.44 A O
+ATOM 2434 N MET A 370 12.631 -73.191 -28.130 1.00 63.44 A N
+ATOM 2435 CA MET A 370 11.313 -72.696 -27.719 1.00 65.09 A C
+ATOM 2436 C MET A 370 10.661 -73.619 -26.692 1.00 66.96 A C
+ATOM 2437 O MET A 370 10.116 -73.145 -25.686 1.00 70.03 A O
+ATOM 2438 CB MET A 370 10.384 -72.512 -28.915 1.00 63.66 A C
+ATOM 2439 CG MET A 370 9.175 -71.674 -28.538 1.00 82.88 A C
+ATOM 2440 SD MET A 370 7.902 -71.577 -29.810 1.00 93.00 A S
+ATOM 2441 CE MET A 370 8.840 -72.014 -31.276 1.00100.38 A C
+ATOM 2442 N ASN A 371 10.711 -74.943 -26.912 1.00 65.40 A N
+ATOM 2443 CA ASN A 371 10.315 -75.861 -25.846 1.00 65.08 A C
+ATOM 2444 C ASN A 371 11.577 -76.424 -25.197 1.00 68.11 A C
+ATOM 2445 O ASN A 371 12.038 -77.520 -25.521 1.00 67.40 A O
+ATOM 2446 CB ASN A 371 9.429 -76.960 -26.410 1.00 77.00 A C
+ATOM 2447 CG ASN A 371 7.968 -76.553 -26.451 1.00 92.45 A C
+ATOM 2448 ND2 ASN A 371 7.059 -77.541 -26.417 1.00 79.91 A N
+ATOM 2449 OD1 ASN A 371 7.657 -75.359 -26.471 1.00108.09 A O
+ATOM 2450 N SER A 372 12.043 -75.714 -24.172 1.00 65.04 A N
+ATOM 2451 CA SER A 372 13.265 -76.004 -23.436 1.00 61.37 A C
+ATOM 2452 C SER A 372 13.253 -75.163 -22.177 1.00 62.69 A C
+ATOM 2453 O SER A 372 12.595 -74.120 -22.125 1.00 67.83 A O
+ATOM 2454 CB SER A 372 14.511 -75.672 -24.253 1.00 62.98 A C
+ATOM 2455 OG SER A 372 14.805 -74.289 -24.131 1.00 62.96 A O
+ATOM 2456 N LEU A 373 14.050 -75.578 -21.195 1.00 69.45 A N
+ATOM 2457 CA LEU A 373 14.382 -74.753 -20.035 1.00 60.25 A C
+ATOM 2458 C LEU A 373 15.891 -74.564 -19.990 1.00 62.43 A C
+ATOM 2459 O LEU A 373 16.639 -75.428 -20.441 1.00 67.43 A O
+ATOM 2460 CB LEU A 373 13.922 -75.400 -18.728 1.00 60.05 A C
+ATOM 2461 CG LEU A 373 12.456 -75.804 -18.681 1.00 70.84 A C
+ATOM 2462 CD1 LEU A 373 12.137 -76.434 -17.348 1.00 68.20 A C
+ATOM 2463 CD2 LEU A 373 11.544 -74.616 -18.948 1.00 65.04 A C
+ATOM 2464 N THR A 374 16.366 -73.454 -19.446 1.00 57.59 A N
+ATOM 2465 CA THR A 374 17.809 -73.314 -19.263 1.00 56.60 A C
+ATOM 2466 C THR A 374 18.110 -73.266 -17.770 1.00 62.79 A C
+ATOM 2467 O THR A 374 17.798 -72.280 -17.091 1.00 65.68 A O
+ATOM 2468 CB THR A 374 18.343 -72.096 -19.994 1.00 64.05 A C
+ATOM 2469 CG2 THR A 374 18.034 -72.233 -21.452 1.00 71.85 A C
+ATOM 2470 OG1 THR A 374 17.658 -70.945 -19.524 1.00 66.08 A O
+ATOM 2471 N LEU A 375 18.714 -74.365 -17.261 1.00 61.49 A N
+ATOM 2472 CA LEU A 375 19.175 -74.956 -16.014 1.00 65.45 A C
+ATOM 2473 C LEU A 375 20.651 -74.663 -15.776 1.00 64.43 A C
+ATOM 2474 O LEU A 375 21.470 -74.750 -16.705 1.00 64.35 A O
+ATOM 2475 CB LEU A 375 18.966 -76.458 -16.015 1.00 58.50 A C
+ATOM 2476 CG LEU A 375 17.610 -76.924 -16.514 1.00 68.21 A C
+ATOM 2477 CD1 LEU A 375 17.390 -78.366 -16.093 1.00 57.18 A C
+ATOM 2478 CD2 LEU A 375 16.499 -76.037 -16.004 1.00 61.97 A C
+ATOM 2479 N PRO A 376 20.975 -74.318 -14.534 1.00 76.23 A N
+ATOM 2480 CA PRO A 376 22.381 -74.141 -14.163 1.00 70.49 A C
+ATOM 2481 C PRO A 376 23.134 -75.459 -14.251 1.00 74.94 A C
+ATOM 2482 O PRO A 376 22.598 -76.529 -13.943 1.00 75.23 A O
+ATOM 2483 CB PRO A 376 22.302 -73.628 -12.721 1.00 71.44 A C
+ATOM 2484 CG PRO A 376 20.842 -73.121 -12.557 1.00 85.28 A C
+ATOM 2485 CD PRO A 376 20.058 -74.057 -13.406 1.00 76.77 A C
+ATOM 2486 N SER A 377 24.411 -75.355 -14.644 1.00 70.61 A N
+ATOM 2487 CA SER A 377 25.198 -76.519 -15.056 1.00 68.53 A C
+ATOM 2488 C SER A 377 25.149 -77.668 -14.052 1.00 67.60 A C
+ATOM 2489 O SER A 377 25.252 -78.840 -14.445 1.00 72.12 A O
+ATOM 2490 CB SER A 377 26.650 -76.094 -15.289 1.00 68.56 A C
+ATOM 2491 OG SER A 377 27.456 -77.215 -15.611 1.00 84.66 A O
+ATOM 2492 N GLU A 378 24.948 -77.356 -12.763 1.00 70.43 A N
+ATOM 2493 CA GLU A 378 25.018 -78.340 -11.683 1.00 74.35 A C
+ATOM 2494 C GLU A 378 23.984 -79.451 -11.822 1.00 77.12 A C
+ATOM 2495 O GLU A 378 24.027 -80.425 -11.060 1.00 85.42 A O
+ATOM 2496 CB GLU A 378 24.848 -77.659 -10.318 1.00 72.15 A C
+ATOM 2497 CG GLU A 378 26.068 -76.879 -9.833 1.00 81.54 A C
+ATOM 2498 CD GLU A 378 26.329 -75.633 -10.654 1.00 90.49 A C
+ATOM 2499 OE1 GLU A 378 25.345 -75.031 -11.131 1.00 89.02 A O
+ATOM 2500 OE2 GLU A 378 27.508 -75.264 -10.842 1.00 96.87 A O1-
+ATOM 2501 N VAL A 379 23.045 -79.327 -12.756 1.00 75.84 A N
+ATOM 2502 CA VAL A 379 22.144 -80.440 -13.009 1.00 74.33 A C
+ATOM 2503 C VAL A 379 22.906 -81.675 -13.478 1.00 76.35 A C
+ATOM 2504 O VAL A 379 22.460 -82.807 -13.243 1.00 85.41 A O
+ATOM 2505 CB VAL A 379 21.064 -80.002 -14.004 1.00 77.36 A C
+ATOM 2506 CG1 VAL A 379 21.714 -79.405 -15.215 1.00 64.19 A C
+ATOM 2507 CG2 VAL A 379 20.198 -81.186 -14.388 1.00 73.25 A C
+ATOM 2508 N ASN A 380 24.094 -81.501 -14.078 1.00 74.44 A N
+ATOM 2509 CA ASN A 380 24.853 -82.695 -14.450 1.00 77.46 A C
+ATOM 2510 C ASN A 380 25.262 -83.495 -13.228 1.00 75.75 A C
+ATOM 2511 O ASN A 380 25.825 -84.586 -13.382 1.00 76.88 A O
+ATOM 2512 CB ASN A 380 26.101 -82.370 -15.297 1.00 78.37 A C
+ATOM 2513 CG ASN A 380 26.628 -83.608 -16.104 1.00 89.96 A C
+ATOM 2514 ND2 ASN A 380 26.513 -84.797 -15.523 1.00 93.91 A N
+ATOM 2515 OD1 ASN A 380 27.129 -83.474 -17.228 1.00 91.63 A O
+ATOM 2516 N LEU A 381 24.995 -82.988 -12.022 1.00 81.16 A N
+ATOM 2517 CA LEU A 381 25.340 -83.744 -10.827 1.00 76.16 A C
+ATOM 2518 C LEU A 381 24.568 -85.061 -10.749 1.00 78.46 A C
+ATOM 2519 O LEU A 381 25.161 -86.119 -10.512 1.00 82.39 A O
+ATOM 2520 CB LEU A 381 25.126 -82.888 -9.579 1.00 73.12 A C
+ATOM 2521 CG LEU A 381 26.136 -81.739 -9.450 1.00 84.24 A C
+ATOM 2522 CD1 LEU A 381 26.186 -81.275 -8.028 1.00 76.64 A C
+ATOM 2523 CD2 LEU A 381 27.539 -82.076 -9.959 1.00 75.44 A C
+ATOM 2524 N CYS A 382 23.262 -85.034 -11.005 1.00 73.23 A N
+ATOM 2525 CA CYS A 382 22.442 -86.186 -10.632 1.00 81.63 A C
+ATOM 2526 C CYS A 382 22.746 -87.453 -11.389 1.00 82.45 A C
+ATOM 2527 O CYS A 382 22.193 -88.512 -11.062 1.00 79.78 A O
+ATOM 2528 CB CYS A 382 21.002 -85.823 -10.785 1.00 75.03 A C
+ATOM 2529 SG CYS A 382 20.843 -84.578 -9.588 1.00 81.78 A S
+ATOM 2530 N ASN A 383 23.590 -87.374 -12.387 1.00 73.44 A N
+ATOM 2531 CA ASN A 383 24.120 -88.592 -12.946 1.00 79.10 A C
+ATOM 2532 C ASN A 383 25.240 -89.172 -12.076 1.00 78.74 A C
+ATOM 2533 O ASN A 383 25.513 -90.375 -12.177 1.00 95.67 A O
+ATOM 2534 CB ASN A 383 24.507 -88.272 -14.389 1.00 73.60 A C
+ATOM 2535 CG ASN A 383 23.370 -87.524 -15.128 1.00 73.82 A C
+ATOM 2536 ND2 ASN A 383 23.710 -86.739 -16.151 1.00 71.45 A N
+ATOM 2537 OD1 ASN A 383 22.205 -87.638 -14.740 1.00 77.12 A O
+ATOM 2538 N VAL A 384 25.821 -88.366 -11.160 1.00 70.56 A N
+ATOM 2539 CA VAL A 384 26.795 -88.829 -10.160 1.00 84.52 A C
+ATOM 2540 C VAL A 384 26.230 -88.883 -8.732 1.00 85.95 A C
+ATOM 2541 O VAL A 384 25.827 -89.955 -8.258 1.00 87.44 A O
+ATOM 2542 CB VAL A 384 28.039 -87.931 -10.194 1.00100.53 A C
+ATOM 2543 CG1 VAL A 384 29.191 -88.614 -9.476 1.00107.46 A C
+ATOM 2544 CG2 VAL A 384 28.381 -87.574 -11.632 1.00102.53 A C
+ATOM 2545 N ASP A 385 26.149 -87.734 -8.039 1.00101.92 A N
+ATOM 2546 CA ASP A 385 25.519 -87.685 -6.710 1.00 94.28 A C
+ATOM 2547 C ASP A 385 24.118 -87.150 -6.889 1.00 86.66 A C
+ATOM 2548 O ASP A 385 23.912 -85.932 -6.946 1.00 90.59 A O
+ATOM 2549 CB ASP A 385 26.237 -86.780 -5.709 1.00 98.21 A C
+ATOM 2550 CG ASP A 385 27.702 -87.088 -5.566 1.00111.22 A C
+ATOM 2551 OD1 ASP A 385 28.149 -88.140 -6.080 1.00113.40 A O
+ATOM 2552 OD2 ASP A 385 28.402 -86.274 -4.913 1.00128.49 A O1-
+ATOM 2553 N ILE A 386 23.145 -88.060 -6.847 1.00 88.93 A N
+ATOM 2554 CA ILE A 386 21.757 -87.645 -6.974 1.00 81.01 A C
+ATOM 2555 C ILE A 386 21.303 -86.995 -5.679 1.00 98.06 A C
+ATOM 2556 O ILE A 386 20.319 -86.232 -5.657 1.00 89.74 A O
+ATOM 2557 CB ILE A 386 20.904 -88.858 -7.389 1.00 82.28 A C
+ATOM 2558 CG1 ILE A 386 19.425 -88.493 -7.472 1.00 85.52 A C
+ATOM 2559 CG2 ILE A 386 21.135 -90.009 -6.446 1.00 83.72 A C
+ATOM 2560 CD1 ILE A 386 18.620 -89.511 -8.232 1.00 83.31 A C
+ATOM 2561 N PHE A 387 22.055 -87.241 -4.610 1.00 77.36 A N
+ATOM 2562 CA PHE A 387 21.797 -86.694 -3.295 1.00 76.30 A C
+ATOM 2563 C PHE A 387 22.571 -85.411 -3.035 1.00 83.70 A C
+ATOM 2564 O PHE A 387 22.457 -84.846 -1.943 1.00 90.15 A O
+ATOM 2565 CB PHE A 387 22.134 -87.752 -2.255 1.00 75.39 A C
+ATOM 2566 CG PHE A 387 21.130 -88.848 -2.190 1.00 80.16 A C
+ATOM 2567 CD1 PHE A 387 19.786 -88.557 -2.051 1.00 81.56 A C
+ATOM 2568 CD2 PHE A 387 21.514 -90.168 -2.311 1.00 85.36 A C
+ATOM 2569 CE1 PHE A 387 18.832 -89.577 -1.997 1.00 82.37 A C
+ATOM 2570 CE2 PHE A 387 20.566 -91.194 -2.261 1.00 82.24 A C
+ATOM 2571 CZ PHE A 387 19.222 -90.895 -2.105 1.00 82.90 A C
+ATOM 2572 N ASN A 388 23.332 -84.928 -4.012 1.00 92.68 A N
+ATOM 2573 CA ASN A 388 24.123 -83.726 -3.799 1.00 99.33 A C
+ATOM 2574 C ASN A 388 23.237 -82.501 -3.598 1.00 97.39 A C
+ATOM 2575 O ASN A 388 22.391 -82.208 -4.447 1.00 97.01 A O
+ATOM 2576 CB ASN A 388 25.038 -83.469 -4.964 1.00103.89 A C
+ATOM 2577 CG ASN A 388 26.306 -82.836 -4.520 1.00110.07 A C
+ATOM 2578 ND2 ASN A 388 27.248 -83.666 -4.079 1.00102.30 A N
+ATOM 2579 OD1 ASN A 388 26.457 -81.609 -4.571 1.00108.86 A O
+ATOM 2580 N PRO A 389 23.430 -81.738 -2.522 1.00 99.64 A N
+ATOM 2581 CA PRO A 389 22.496 -80.642 -2.216 1.00 97.28 A C
+ATOM 2582 C PRO A 389 22.632 -79.426 -3.123 1.00 94.14 A C
+ATOM 2583 O PRO A 389 21.712 -78.597 -3.139 1.00111.27 A O
+ATOM 2584 CB PRO A 389 22.832 -80.295 -0.760 1.00 94.36 A C
+ATOM 2585 CG PRO A 389 24.310 -80.587 -0.662 1.00 94.50 A C
+ATOM 2586 CD PRO A 389 24.633 -81.694 -1.673 1.00 95.26 A C
+ATOM 2587 N LYS A 390 23.748 -79.271 -3.850 1.00 90.27 A N
+ATOM 2588 CA LYS A 390 23.922 -78.076 -4.679 1.00 93.76 A C
+ATOM 2589 C LYS A 390 22.883 -77.994 -5.789 1.00100.43 A C
+ATOM 2590 O LYS A 390 22.523 -76.890 -6.224 1.00 95.24 A O
+ATOM 2591 CB LYS A 390 25.332 -78.023 -5.277 1.00 89.84 A C
+ATOM 2592 CG LYS A 390 26.384 -77.512 -4.304 1.00 95.10 A C
+ATOM 2593 CD LYS A 390 27.557 -76.847 -5.009 1.00108.32 A C
+ATOM 2594 CE LYS A 390 28.394 -77.849 -5.760 1.00121.02 A C
+ATOM 2595 NZ LYS A 390 29.551 -77.166 -6.383 1.00140.55 A N1+
+ATOM 2596 N TYR A 391 22.403 -79.140 -6.267 1.00 96.27 A N
+ATOM 2597 CA TYR A 391 21.299 -79.201 -7.215 1.00 88.66 A C
+ATOM 2598 C TYR A 391 20.318 -80.261 -6.756 1.00 88.24 A C
+ATOM 2599 O TYR A 391 20.697 -81.424 -6.589 1.00 94.83 A O
+ATOM 2600 CB TYR A 391 21.787 -79.518 -8.625 1.00 83.71 A C
+ATOM 2601 CG TYR A 391 20.685 -79.400 -9.649 1.00 89.91 A C
+ATOM 2602 CD1 TYR A 391 20.278 -78.152 -10.109 1.00 86.71 A C
+ATOM 2603 CD2 TYR A 391 20.055 -80.531 -10.163 1.00 87.92 A C
+ATOM 2604 CE1 TYR A 391 19.266 -78.026 -11.044 1.00 89.10 A C
+ATOM 2605 CE2 TYR A 391 19.046 -80.418 -11.103 1.00 86.51 A C
+ATOM 2606 CZ TYR A 391 18.656 -79.159 -11.542 1.00 91.53 A C
+ATOM 2607 OH TYR A 391 17.652 -79.014 -12.478 1.00 94.45 A O
+ATOM 2608 N ASP A 392 19.060 -79.873 -6.587 1.00 80.46 A N
+ATOM 2609 CA ASP A 392 18.050 -80.760 -6.017 1.00 82.24 A C
+ATOM 2610 C ASP A 392 17.329 -81.481 -7.159 1.00 85.32 A C
+ATOM 2611 O ASP A 392 16.521 -80.873 -7.866 1.00 83.59 A O
+ATOM 2612 CB ASP A 392 17.092 -79.937 -5.150 1.00 87.92 A C
+ATOM 2613 CG ASP A 392 15.806 -80.677 -4.782 1.00 91.75 A C
+ATOM 2614 OD1 ASP A 392 14.753 -80.013 -4.813 1.00 93.11 A O
+ATOM 2615 OD2 ASP A 392 15.831 -81.885 -4.447 1.00 94.00 A O1-
+ATOM 2616 N CYS A 393 17.578 -82.782 -7.326 1.00 80.71 A N
+ATOM 2617 CA CYS A 393 17.017 -83.473 -8.478 1.00 77.49 A C
+ATOM 2618 C CYS A 393 15.600 -83.951 -8.329 1.00 88.37 A C
+ATOM 2619 O CYS A 393 15.173 -84.410 -7.271 1.00 83.49 A O
+ATOM 2620 CB CYS A 393 17.894 -84.622 -8.933 1.00 78.32 A C
+ATOM 2621 SG CYS A 393 19.046 -83.829 -10.033 1.00 89.64 A S
+ATOM 2622 N LYS A 394 14.912 -83.908 -9.454 1.00 69.44 A N
+ATOM 2623 CA LYS A 394 13.497 -84.142 -9.530 1.00 71.71 A C
+ATOM 2624 C LYS A 394 13.243 -85.615 -9.780 1.00 74.36 A C
+ATOM 2625 O LYS A 394 13.931 -86.246 -10.589 1.00 68.37 A O
+ATOM 2626 CB LYS A 394 12.908 -83.273 -10.633 1.00 66.33 A C
+ATOM 2627 CG LYS A 394 13.204 -81.800 -10.416 1.00 68.64 A C
+ATOM 2628 CD LYS A 394 12.074 -80.966 -10.932 1.00 90.23 A C
+ATOM 2629 CE LYS A 394 10.749 -81.497 -10.426 1.00 82.78 A C
+ATOM 2630 NZ LYS A 394 10.511 -81.190 -9.005 1.00 85.83 A N1+
+ATOM 2631 N ILE A 395 12.264 -86.155 -9.047 1.00 77.76 A N
+ATOM 2632 CA ILE A 395 11.899 -87.569 -9.070 1.00 75.92 A C
+ATOM 2633 C ILE A 395 10.417 -87.698 -8.746 1.00 75.47 A C
+ATOM 2634 O ILE A 395 9.834 -86.865 -8.042 1.00 75.79 A O
+ATOM 2635 CB ILE A 395 12.717 -88.401 -8.057 1.00 73.33 A C
+ATOM 2636 CG1 ILE A 395 12.687 -87.703 -6.696 1.00 76.95 A C
+ATOM 2637 CG2 ILE A 395 14.127 -88.645 -8.555 1.00 78.31 A C
+ATOM 2638 CD1 ILE A 395 13.198 -88.541 -5.586 1.00 77.25 A C
+ATOM 2639 N MET A 396 9.816 -88.774 -9.239 1.00 77.69 A N
+ATOM 2640 CA MET A 396 8.471 -89.155 -8.852 1.00 73.87 A C
+ATOM 2641 C MET A 396 8.585 -90.397 -7.982 1.00 71.77 A C
+ATOM 2642 O MET A 396 9.553 -91.165 -8.083 1.00 73.81 A O
+ATOM 2643 CB MET A 396 7.584 -89.469 -10.057 1.00 70.15 A C
+ATOM 2644 CG MET A 396 7.444 -90.957 -10.347 1.00 86.86 A C
+ATOM 2645 SD MET A 396 6.310 -91.196 -11.695 1.00109.63 A S
+ATOM 2646 CE MET A 396 5.015 -90.094 -11.164 1.00 82.59 A C
+ATOM 2647 N THR A 397 7.605 -90.574 -7.108 1.00 75.02 A N
+ATOM 2648 CA THR A 397 7.546 -91.716 -6.216 1.00 79.84 A C
+ATOM 2649 C THR A 397 6.170 -92.350 -6.306 1.00 79.89 A C
+ATOM 2650 O THR A 397 5.168 -91.654 -6.491 1.00 90.08 A O
+ATOM 2651 CB THR A 397 7.824 -91.279 -4.796 1.00 82.77 A C
+ATOM 2652 CG2 THR A 397 9.199 -90.647 -4.697 1.00 83.38 A C
+ATOM 2653 OG1 THR A 397 6.815 -90.336 -4.396 1.00 84.90 A O
+ATOM 2654 N SER A 398 6.120 -93.667 -6.177 1.00 80.66 A N
+ATOM 2655 CA SER A 398 4.828 -94.336 -6.128 1.00 78.44 A C
+ATOM 2656 C SER A 398 5.022 -95.769 -5.663 1.00 78.53 A C
+ATOM 2657 O SER A 398 6.144 -96.264 -5.524 1.00 75.55 A O
+ATOM 2658 CB SER A 398 4.121 -94.304 -7.483 1.00 74.74 A C
+ATOM 2659 OG SER A 398 2.917 -95.053 -7.443 1.00 83.70 A O
+ATOM 2660 N LYS A 399 3.892 -96.441 -5.490 1.00 83.69 A N
+ATOM 2661 CA LYS A 399 3.772 -97.773 -4.920 1.00 79.39 A C
+ATOM 2662 C LYS A 399 3.722 -98.869 -5.974 1.00 87.12 A C
+ATOM 2663 O LYS A 399 3.697-100.052 -5.616 1.00 95.66 A O
+ATOM 2664 CB LYS A 399 2.493 -97.830 -4.063 1.00 88.77 A C
+ATOM 2665 CG LYS A 399 2.316 -99.029 -3.142 1.00 99.22 A C
+ATOM 2666 CD LYS A 399 0.966 -98.938 -2.440 1.00111.54 A C
+ATOM 2667 CE LYS A 399 0.884 -99.865 -1.245 1.00116.69 A C
+ATOM 2668 NZ LYS A 399 -0.326 -99.626 -0.414 1.00107.13 A N1+
+ATOM 2669 N THR A 400 3.706 -98.514 -7.259 1.00 78.79 A N
+ATOM 2670 CA THR A 400 3.276 -99.443 -8.304 1.00 85.84 A C
+ATOM 2671 C THR A 400 4.517-100.005 -8.989 1.00 78.88 A C
+ATOM 2672 O THR A 400 5.133 -99.345 -9.827 1.00 77.36 A O
+ATOM 2673 CB THR A 400 2.362 -98.747 -9.312 1.00 98.44 A C
+ATOM 2674 CG2 THR A 400 1.516 -99.776 -10.021 1.00 97.63 A C
+ATOM 2675 OG1 THR A 400 1.476 -97.851 -8.632 1.00106.26 A O
+ATOM 2676 N ASP A 401 4.846-101.253 -8.684 1.00 83.28 A N
+ATOM 2677 CA ASP A 401 5.999-101.907 -9.285 1.00 80.06 A C
+ATOM 2678 C ASP A 401 5.488-102.663 -10.501 1.00 86.85 A C
+ATOM 2679 O ASP A 401 4.784-103.673 -10.363 1.00 90.00 A O
+ATOM 2680 CB ASP A 401 6.687-102.834 -8.280 1.00 85.74 A C
+ATOM 2681 CG ASP A 401 5.705-103.685 -7.470 1.00 90.66 A C
+ATOM 2682 OD1 ASP A 401 4.930-103.128 -6.665 1.00 83.73 A O
+ATOM 2683 OD2 ASP A 401 5.750-104.926 -7.585 1.00 91.67 A O1-
+ATOM 2684 N VAL A 402 5.823-102.167 -11.695 1.00 76.80 A N
+ATOM 2685 CA VAL A 402 5.358-102.746 -12.953 1.00 67.23 A C
+ATOM 2686 C VAL A 402 6.550-102.895 -13.879 1.00 64.23 A C
+ATOM 2687 O VAL A 402 7.376-101.980 -13.994 1.00 72.47 A O
+ATOM 2688 CB VAL A 402 4.276-101.885 -13.626 1.00 78.16 A C
+ATOM 2689 CG1 VAL A 402 3.525-102.710 -14.627 1.00 92.97 A C
+ATOM 2690 CG2 VAL A 402 3.352-101.291 -12.597 1.00 79.09 A C
+ATOM 2691 N SER A 403 6.630-104.032 -14.561 1.00 71.61 A N
+ATOM 2692 CA SER A 403 7.791-104.291 -15.401 1.00 68.58 A C
+ATOM 2693 C SER A 403 7.809-103.340 -16.596 1.00 69.88 A C
+ATOM 2694 O SER A 403 6.869-103.280 -17.394 1.00 73.54 A O
+ATOM 2695 CB SER A 403 7.804-105.744 -15.853 1.00 66.41 A C
+ATOM 2696 OG SER A 403 7.718-106.595 -14.729 1.00 68.69 A O
+ATOM 2697 N SER A 404 8.891-102.600 -16.708 1.00 69.86 A N
+ATOM 2698 CA SER A 404 9.068-101.615 -17.746 1.00 73.43 A C
+ATOM 2699 C SER A 404 10.417-101.883 -18.395 1.00 74.92 A C
+ATOM 2700 O SER A 404 11.232-102.657 -17.892 1.00 73.63 A O
+ATOM 2701 CB SER A 404 8.976-100.195 -17.161 1.00 69.53 A C
+ATOM 2702 OG SER A 404 9.210 -99.195 -18.144 1.00 75.34 A O
+ATOM 2703 N SER A 405 10.647-101.272 -19.542 1.00 77.53 A N
+ATOM 2704 CA SER A 405 11.977-101.313 -20.111 1.00 67.97 A C
+ATOM 2705 C SER A 405 12.154-100.081 -20.963 1.00 73.00 A C
+ATOM 2706 O SER A 405 11.180 -99.528 -21.469 1.00 68.54 A O
+ATOM 2707 CB SER A 405 12.210-102.560 -20.952 1.00 66.13 A C
+ATOM 2708 OG SER A 405 13.472-102.456 -21.585 1.00 70.62 A O
+ATOM 2709 N VAL A 406 13.395 -99.643 -21.118 1.00 64.24 A N
+ATOM 2710 CA VAL A 406 13.652 -98.447 -21.904 1.00 60.80 A C
+ATOM 2711 C VAL A 406 14.859 -98.700 -22.789 1.00 68.47 A C
+ATOM 2712 O VAL A 406 15.960 -98.941 -22.277 1.00 71.86 A O
+ATOM 2713 CB VAL A 406 13.873 -97.216 -21.014 1.00 66.58 A C
+ATOM 2714 CG1 VAL A 406 14.623 -96.150 -21.754 1.00 57.18 A C
+ATOM 2715 CG2 VAL A 406 12.538 -96.670 -20.578 1.00 56.51 A C
+ATOM 2716 N ILE A 407 14.662 -98.637 -24.111 1.00 61.64 A N
+ATOM 2717 CA ILE A 407 15.766 -98.818 -25.047 1.00 58.31 A C
+ATOM 2718 C ILE A 407 16.503 -97.496 -25.136 1.00 61.30 A C
+ATOM 2719 O ILE A 407 15.914 -96.476 -25.510 1.00 67.14 A O
+ATOM 2720 CB ILE A 407 15.287 -99.251 -26.435 1.00 64.53 A C
+ATOM 2721 CG1 ILE A 407 14.208-100.320 -26.350 1.00 58.99 A C
+ATOM 2722 CG2 ILE A 407 16.471 -99.838 -27.178 1.00 57.13 A C
+ATOM 2723 CD1 ILE A 407 14.657-101.555 -25.675 1.00 56.87 A C
+ATOM 2724 N THR A 408 17.782 -97.510 -24.790 1.00 64.86 A N
+ATOM 2725 CA THR A 408 18.607 -96.321 -24.826 1.00 60.14 A C
+ATOM 2726 C THR A 408 19.395 -96.311 -26.125 1.00 64.92 A C
+ATOM 2727 O THR A 408 19.227 -97.190 -26.976 1.00 64.60 A O
+ATOM 2728 CB THR A 408 19.537 -96.271 -23.619 1.00 60.68 A C
+ATOM 2729 CG2 THR A 408 18.771 -96.615 -22.358 1.00 59.62 A C
+ATOM 2730 OG1 THR A 408 20.612 -97.204 -23.814 1.00 67.55 A O
+ATOM 2731 N SER A 409 20.281 -95.317 -26.265 1.00 62.57 A N
+ATOM 2732 CA SER A 409 21.088 -95.175 -27.477 1.00 66.13 A C
+ATOM 2733 C SER A 409 21.991 -96.385 -27.684 1.00 68.31 A C
+ATOM 2734 O SER A 409 21.942 -97.045 -28.730 1.00 67.21 A O
+ATOM 2735 CB SER A 409 21.909 -93.887 -27.409 1.00 60.20 A C
+ATOM 2736 OG SER A 409 21.052 -92.778 -27.236 1.00 70.65 A O
+ATOM 2737 N LEU A 410 22.830 -96.687 -26.697 1.00 62.39 A N
+ATOM 2738 CA LEU A 410 23.757 -97.802 -26.789 1.00 65.02 A C
+ATOM 2739 C LEU A 410 23.283 -99.069 -26.087 1.00 73.87 A C
+ATOM 2740 O LEU A 410 24.040-100.042 -26.034 1.00 68.95 A O
+ATOM 2741 CB LEU A 410 25.121 -97.383 -26.250 1.00 65.18 A C
+ATOM 2742 CG LEU A 410 26.147 -97.073 -27.332 1.00 68.41 A C
+ATOM 2743 CD1 LEU A 410 25.558 -96.165 -28.362 1.00 69.39 A C
+ATOM 2744 CD2 LEU A 410 27.336 -96.397 -26.697 1.00 71.93 A C
+ATOM 2745 N GLY A 411 22.078 -99.098 -25.539 1.00 64.35 A N
+ATOM 2746 CA GLY A 411 21.658-100.292 -24.838 1.00 58.08 A C
+ATOM 2747 C GLY A 411 20.240-100.197 -24.338 1.00 64.67 A C
+ATOM 2748 O GLY A 411 19.432 -99.419 -24.848 1.00 66.98 A O
+ATOM 2749 N ALA A 412 19.943-101.029 -23.337 1.00 66.89 A N
+ATOM 2750 CA ALA A 412 18.597-101.140 -22.788 1.00 63.72 A C
+ATOM 2751 C ALA A 412 18.632-101.248 -21.269 1.00 61.83 A C
+ATOM 2752 O ALA A 412 19.388-102.047 -20.707 1.00 65.65 A O
+ATOM 2753 CB ALA A 412 17.855-102.348 -23.371 1.00 60.07 A C
+ATOM 2754 N ILE A 413 17.823-100.427 -20.624 1.00 59.69 A N
+ATOM 2755 CA ILE A 413 17.506-100.539 -19.213 1.00 59.06 A C
+ATOM 2756 C ILE A 413 16.298-101.445 -19.079 1.00 64.62 A C
+ATOM 2757 O ILE A 413 15.389-101.406 -19.916 1.00 60.51 A O
+ATOM 2758 CB ILE A 413 17.213 -99.156 -18.621 1.00 59.92 A C
+ATOM 2759 CG1 ILE A 413 18.379 -98.203 -18.859 1.00 59.66 A C
+ATOM 2760 CG2 ILE A 413 16.894 -99.290 -17.177 1.00 51.09 A C
+ATOM 2761 CD1 ILE A 413 18.147 -96.818 -18.302 1.00 55.32 A C
+ATOM 2762 N VAL A 414 16.267-102.259 -18.032 1.00 60.01 A N
+ATOM 2763 CA VAL A 414 15.136-103.148 -17.796 1.00 59.48 A C
+ATOM 2764 C VAL A 414 14.748-103.099 -16.319 1.00 61.90 A C
+ATOM 2765 O VAL A 414 15.597-103.242 -15.435 1.00 64.68 A O
+ATOM 2766 CB VAL A 414 15.459-104.584 -18.245 1.00 67.62 A C
+ATOM 2767 CG1 VAL A 414 14.437-105.552 -17.687 1.00 64.90 A C
+ATOM 2768 CG2 VAL A 414 15.483-104.641 -19.746 1.00 63.07 A C
+ATOM 2769 N SER A 415 13.469-102.888 -16.051 1.00 64.28 A N
+ATOM 2770 CA SER A 415 12.942-102.926 -14.696 1.00 68.71 A C
+ATOM 2771 C SER A 415 12.008-104.124 -14.593 1.00 77.19 A C
+ATOM 2772 O SER A 415 10.862-104.065 -15.059 1.00 72.68 A O
+ATOM 2773 CB SER A 415 12.214-101.625 -14.370 1.00 62.45 A C
+ATOM 2774 OG SER A 415 13.075-100.525 -14.571 1.00 74.16 A O
+ATOM 2775 N CYS A 416 12.471-105.181 -13.917 1.00 80.02 A N
+ATOM 2776 CA CYS A 416 11.753-106.450 -13.837 1.00 80.17 A C
+ATOM 2777 C CYS A 416 11.239-106.655 -12.419 1.00 82.72 A C
+ATOM 2778 O CYS A 416 12.016-106.970 -11.512 1.00 84.32 A O
+ATOM 2779 CB CYS A 416 12.665-107.601 -14.238 1.00 77.48 A C
+ATOM 2780 SG CYS A 416 11.749-108.975 -14.921 1.00 91.59 A S
+ATOM 2781 N TYR A 417 9.928-106.517 -12.241 1.00 72.19 A N
+ATOM 2782 CA TYR A 417 9.289-106.605 -10.935 1.00 77.24 A C
+ATOM 2783 C TYR A 417 8.126-107.586 -11.006 1.00 79.83 A C
+ATOM 2784 O TYR A 417 7.397-107.631 -12.004 1.00 74.85 A O
+ATOM 2785 CB TYR A 417 8.793-105.239 -10.487 1.00 77.72 A C
+ATOM 2786 CG TYR A 417 9.873-104.192 -10.514 1.00 84.18 A C
+ATOM 2787 CD1 TYR A 417 11.183-104.510 -10.170 1.00 79.46 A C
+ATOM 2788 CD2 TYR A 417 9.587-102.879 -10.869 1.00 79.48 A C
+ATOM 2789 CE1 TYR A 417 12.187-103.564 -10.202 1.00 82.90 A C
+ATOM 2790 CE2 TYR A 417 10.586-101.913 -10.905 1.00 78.21 A C
+ATOM 2791 CZ TYR A 417 11.888-102.263 -10.564 1.00 82.46 A C
+ATOM 2792 OH TYR A 417 12.890-101.313 -10.585 1.00 82.25 A O
+ATOM 2793 N GLY A 418 7.947-108.372 -9.947 1.00 75.57 A N
+ATOM 2794 CA GLY A 418 6.903-109.381 -9.966 1.00 71.99 A C
+ATOM 2795 C GLY A 418 7.342-110.634 -10.705 1.00 82.51 A C
+ATOM 2796 O GLY A 418 8.529-110.874 -10.962 1.00 79.60 A O
+ATOM 2797 N LYS A 419 6.359-111.434 -11.081 1.00 81.78 A N
+ATOM 2798 CA LYS A 419 6.701-112.679 -11.745 1.00 81.69 A C
+ATOM 2799 C LYS A 419 6.993-112.486 -13.226 1.00 79.13 A C
+ATOM 2800 O LYS A 419 7.106-113.482 -13.952 1.00 77.07 A O
+ATOM 2801 CB LYS A 419 5.610-113.742 -11.551 1.00 77.35 A C
+ATOM 2802 CG LYS A 419 5.818-114.601 -10.282 1.00 78.17 A C
+ATOM 2803 CD LYS A 419 5.282-116.038 -10.429 1.00 87.47 A C
+ATOM 2804 CE LYS A 419 3.848-116.188 -9.949 1.00 84.00 A C
+ATOM 2805 NZ LYS A 419 3.718-115.794 -8.514 1.00 84.60 A N1+
+ATOM 2806 N THR A 420 7.062-111.236 -13.691 1.00 76.14 A N
+ATOM 2807 CA THR A 420 7.272-110.957 -15.107 1.00 81.85 A C
+ATOM 2808 C THR A 420 8.609-111.497 -15.588 1.00 76.62 A C
+ATOM 2809 O THR A 420 9.623-111.383 -14.894 1.00 74.80 A O
+ATOM 2810 CB THR A 420 7.232-109.460 -15.363 1.00 79.71 A C
+ATOM 2811 CG2 THR A 420 7.149-109.202 -16.841 1.00 72.61 A C
+ATOM 2812 OG1 THR A 420 6.109-108.871 -14.693 1.00 73.57 A O
+ATOM 2813 N LYS A 421 8.612-112.064 -16.799 1.00 75.45 A N
+ATOM 2814 CA LYS A 421 9.821-112.651 -17.371 1.00 80.75 A C
+ATOM 2815 C LYS A 421 10.544-111.610 -18.224 1.00 80.40 A C
+ATOM 2816 O LYS A 421 9.963-111.068 -19.174 1.00 76.79 A O
+ATOM 2817 CB LYS A 421 9.501-113.900 -18.201 1.00 73.94 A C
+ATOM 2818 CG LYS A 421 10.715-114.848 -18.345 1.00100.87 A C
+ATOM 2819 CD LYS A 421 11.137-115.417 -16.966 1.00111.93 A C
+ATOM 2820 CE LYS A 421 12.572-115.964 -16.934 1.00120.42 A C
+ATOM 2821 NZ LYS A 421 12.754-117.328 -17.489 1.00115.98 A N1+
+ATOM 2822 N CYS A 422 11.807-111.336 -17.888 1.00 73.03 A N
+ATOM 2823 CA CYS A 422 12.624-110.363 -18.600 1.00 70.60 A C
+ATOM 2824 C CYS A 422 13.901-111.032 -19.069 1.00 80.73 A C
+ATOM 2825 O CYS A 422 14.610-111.653 -18.276 1.00 73.53 A O
+ATOM 2826 CB CYS A 422 12.961-109.159 -17.722 1.00 69.70 A C
+ATOM 2827 SG CYS A 422 11.528-108.450 -16.889 1.00 76.17 A S
+ATOM 2828 N THR A 423 14.187-110.907 -20.358 1.00 61.89 A N
+ATOM 2829 CA THR A 423 15.309-111.603 -20.960 1.00 63.03 A C
+ATOM 2830 C THR A 423 15.889-110.744 -22.070 1.00 71.76 A C
+ATOM 2831 O THR A 423 15.222-109.866 -22.619 1.00 69.63 A O
+ATOM 2832 CB THR A 423 14.897-112.968 -21.537 1.00 64.32 A C
+ATOM 2833 CG2 THR A 423 14.289-113.877 -20.470 1.00 64.69 A C
+ATOM 2834 OG1 THR A 423 13.955-112.784 -22.604 1.00 71.50 A O
+ATOM 2835 N ALA A 424 17.143-111.015 -22.399 1.00 61.15 A N
+ATOM 2836 CA ALA A 424 17.794-110.431 -23.556 1.00 61.44 A C
+ATOM 2837 C ALA A 424 18.336-111.568 -24.398 1.00 68.43 A C
+ATOM 2838 O ALA A 424 18.922-112.518 -23.861 1.00 66.55 A O
+ATOM 2839 CB ALA A 424 18.920-109.471 -23.162 1.00 58.29 A C
+ATOM 2840 N SER A 425 18.131-111.465 -25.714 1.00 60.10 A N
+ATOM 2841 CA SER A 425 18.546-112.482 -26.672 1.00 65.90 A C
+ATOM 2842 C SER A 425 19.284-111.879 -27.872 1.00 70.84 A C
+ATOM 2843 O SER A 425 19.180-110.685 -28.189 1.00 71.68 A O
+ATOM 2844 CB SER A 425 17.340-113.329 -27.137 1.00 67.14 A C
+ATOM 2845 OG SER A 425 16.148-112.570 -27.190 1.00 84.86 A O
+ATOM 2846 N ASN A 426 20.052-112.757 -28.517 1.00 66.30 A N
+ATOM 2847 CA ASN A 426 20.841-112.519 -29.718 1.00 69.09 A C
+ATOM 2848 C ASN A 426 20.197-113.252 -30.887 1.00 85.38 A C
+ATOM 2849 O ASN A 426 19.555-114.290 -30.708 1.00 93.65 A O
+ATOM 2850 CB ASN A 426 22.277-113.025 -29.531 1.00 74.55 A C
+ATOM 2851 CG ASN A 426 23.206-112.610 -30.662 1.00 92.90 A C
+ATOM 2852 ND2 ASN A 426 24.069-111.637 -30.384 1.00 95.59 A N
+ATOM 2853 OD1 ASN A 426 23.174-113.179 -31.762 1.00103.76 A O
+ATOM 2854 N LYS A 427 20.374-112.713 -32.092 1.00 79.42 A N
+ATOM 2855 CA LYS A 427 19.685-113.299 -33.234 1.00 78.66 A C
+ATOM 2856 C LYS A 427 20.134-114.729 -33.516 1.00 68.00 A C
+ATOM 2857 O LYS A 427 19.394-115.471 -34.178 1.00 70.27 A O
+ATOM 2858 CB LYS A 427 19.865-112.423 -34.485 1.00 81.32 A C
+ATOM 2859 CG LYS A 427 21.191-112.565 -35.206 1.00 95.98 A C
+ATOM 2860 CD LYS A 427 21.286-111.625 -36.398 1.00 97.49 A C
+ATOM 2861 CE LYS A 427 21.304-110.178 -35.950 1.00 96.05 A C
+ATOM 2862 NZ LYS A 427 21.662-109.268 -37.056 1.00 97.37 A N1+
+ATOM 2863 N ASN A 428 21.315-115.135 -33.015 1.00 74.69 A N
+ATOM 2864 CA ASN A 428 21.885-116.468 -33.234 1.00 80.66 A C
+ATOM 2865 C ASN A 428 21.778-117.343 -31.992 1.00 90.93 A C
+ATOM 2866 O ASN A 428 21.115-118.383 -32.005 1.00103.56 A O
+ATOM 2867 CB ASN A 428 23.363-116.375 -33.627 1.00 84.68 A C
+ATOM 2868 CG ASN A 428 23.585-115.531 -34.839 1.00 94.59 A C
+ATOM 2869 ND2 ASN A 428 23.215-116.052 -36.011 1.00 97.48 A N
+ATOM 2870 OD1 ASN A 428 24.073-114.400 -34.726 1.00 95.79 A O
+ATOM 2871 N ARG A 429 22.440-116.935 -30.913 1.00 91.11 A N
+ATOM 2872 CA ARG A 429 22.593-117.741 -29.716 1.00 95.45 A C
+ATOM 2873 C ARG A 429 21.285-117.902 -28.926 1.00 82.40 A C
+ATOM 2874 O ARG A 429 21.289-118.629 -27.924 1.00 84.68 A O
+ATOM 2875 CB ARG A 429 23.710-117.141 -28.833 1.00 99.34 A C
+ATOM 2876 CG ARG A 429 25.167-117.412 -29.317 1.00105.62 A C
+ATOM 2877 CD ARG A 429 26.201-116.368 -28.811 1.00115.76 A C
+ATOM 2878 NE ARG A 429 26.203-115.101 -29.565 1.00114.69 A N
+ATOM 2879 CZ ARG A 429 27.068-114.761 -30.532 1.00112.04 A C
+ATOM 2880 NH1 ARG A 429 28.049-115.583 -30.892 1.00 96.93 A N1+
+ATOM 2881 NH2 ARG A 429 26.956-113.583 -31.143 1.00118.07 A N
+ATOM 2882 N GLY A 430 20.175-117.267 -29.329 1.00 93.68 A N
+ATOM 2883 CA GLY A 430 18.955-117.324 -28.524 1.00 72.48 A C
+ATOM 2884 C GLY A 430 19.035-116.391 -27.318 1.00 81.65 A C
+ATOM 2885 O GLY A 430 19.808-115.420 -27.307 1.00 79.39 A O
+ATOM 2886 N ILE A 431 18.237-116.683 -26.283 1.00 69.20 A N
+ATOM 2887 CA ILE A 431 18.350-115.887 -25.063 1.00 72.67 A C
+ATOM 2888 C ILE A 431 19.759-116.028 -24.515 1.00 65.34 A C
+ATOM 2889 O ILE A 431 20.223-117.145 -24.267 1.00 70.01 A O
+ATOM 2890 CB ILE A 431 17.341-116.337 -24.015 1.00 63.89 A C
+ATOM 2891 CG1 ILE A 431 15.915-116.032 -24.449 1.00 71.11 A C
+ATOM 2892 CG2 ILE A 431 17.664-115.672 -22.686 1.00 62.63 A C
+ATOM 2893 CD1 ILE A 431 14.878-116.269 -23.334 1.00 64.70 A C
+ATOM 2894 N ILE A 432 20.470-114.911 -24.362 1.00 59.14 A N
+ATOM 2895 CA ILE A 432 21.775-114.950 -23.710 1.00 69.51 A C
+ATOM 2896 C ILE A 432 21.799-114.366 -22.303 1.00 68.97 A C
+ATOM 2897 O ILE A 432 22.850-114.444 -21.649 1.00 79.37 A O
+ATOM 2898 CB ILE A 432 22.862-114.291 -24.577 1.00 75.48 A C
+ATOM 2899 CG1 ILE A 432 22.477-112.865 -24.935 1.00 66.90 A C
+ATOM 2900 CG2 ILE A 432 23.056-115.108 -25.841 1.00 79.72 A C
+ATOM 2901 CD1 ILE A 432 23.477-112.198 -25.879 1.00 72.61 A C
+ATOM 2902 N LYS A 433 20.717-113.770 -21.812 1.00 69.95 A N
+ATOM 2903 CA LYS A 433 20.704-113.415 -20.395 1.00 69.82 A C
+ATOM 2904 C LYS A 433 19.272-113.387 -19.886 1.00 69.36 A C
+ATOM 2905 O LYS A 433 18.372-112.921 -20.593 1.00 71.24 A O
+ATOM 2906 CB LYS A 433 21.380-112.060 -20.125 1.00 68.44 A C
+ATOM 2907 CG LYS A 433 21.261-111.597 -18.670 1.00 67.11 A C
+ATOM 2908 CD LYS A 433 22.172-110.409 -18.330 1.00 81.17 A C
+ATOM 2909 CE LYS A 433 22.253-110.191 -16.797 1.00 85.57 A C
+ATOM 2910 NZ LYS A 433 23.276-109.188 -16.320 1.00 84.85 A N1+
+ATOM 2911 N THR A 434 19.066-113.863 -18.656 1.00 67.96 A N
+ATOM 2912 CA THR A 434 17.787-113.735 -17.963 1.00 66.15 A C
+ATOM 2913 C THR A 434 17.964-112.806 -16.774 1.00 69.56 A C
+ATOM 2914 O THR A 434 18.920-112.949 -16.006 1.00 74.70 A O
+ATOM 2915 CB THR A 434 17.244-115.087 -17.499 1.00 67.11 A C
+ATOM 2916 CG2 THR A 434 15.917-114.909 -16.783 1.00 66.18 A C
+ATOM 2917 OG1 THR A 434 17.059-115.945 -18.630 1.00 68.05 A O
+ATOM 2918 N PHE A 435 17.051-111.854 -16.640 1.00 69.20 A N
+ATOM 2919 CA PHE A 435 17.146-110.822 -15.623 1.00 73.90 A C
+ATOM 2920 C PHE A 435 16.410-111.246 -14.362 1.00 79.34 A C
+ATOM 2921 O PHE A 435 15.316-111.823 -14.428 1.00 82.34 A O
+ATOM 2922 CB PHE A 435 16.550-109.506 -16.118 1.00 69.65 A C
+ATOM 2923 CG PHE A 435 17.236-108.918 -17.319 1.00 78.10 A C
+ATOM 2924 CD1 PHE A 435 18.484-108.325 -17.200 1.00 77.88 A C
+ATOM 2925 CD2 PHE A 435 16.598-108.886 -18.547 1.00 77.52 A C
+ATOM 2926 CE1 PHE A 435 19.103-107.752 -18.300 1.00 80.25 A C
+ATOM 2927 CE2 PHE A 435 17.208-108.316 -19.645 1.00 69.70 A C
+ATOM 2928 CZ PHE A 435 18.460-107.746 -19.525 1.00 77.67 A C
+ATOM 2929 N SER A 436 17.012-110.934 -13.216 1.00 79.87 A N
+ATOM 2930 CA SER A 436 16.375-111.137 -11.921 1.00 87.84 A C
+ATOM 2931 C SER A 436 15.459-109.947 -11.630 1.00 85.25 A C
+ATOM 2932 O SER A 436 15.221-109.089 -12.484 1.00 84.19 A O
+ATOM 2933 CB SER A 436 17.428-111.342 -10.832 1.00 77.18 A C
+ATOM 2934 OG SER A 436 18.185-112.516 -11.070 1.00124.51 A O
+ATOM 2935 N ASN A 437 14.920-109.886 -10.417 1.00 75.25 A N
+ATOM 2936 CA ASN A 437 14.049-108.781 -10.051 1.00 76.16 A C
+ATOM 2937 C ASN A 437 14.897-107.576 -9.660 1.00 73.76 A C
+ATOM 2938 O ASN A 437 15.829-107.690 -8.859 1.00 79.84 A O
+ATOM 2939 CB ASN A 437 13.100-109.195 -8.924 1.00 73.56 A C
+ATOM 2940 CG ASN A 437 11.922-110.043 -9.427 1.00 89.27 A C
+ATOM 2941 ND2 ASN A 437 12.149-111.347 -9.596 1.00118.62 A N
+ATOM 2942 OD1 ASN A 437 10.822-109.528 -9.647 1.00 85.51 A O
+ATOM 2943 N GLY A 438 14.611-106.439 -10.272 1.00 74.49 A N
+ATOM 2944 CA GLY A 438 15.370-105.232 -10.027 1.00 73.48 A C
+ATOM 2945 C GLY A 438 15.457-104.410 -11.296 1.00 77.75 A C
+ATOM 2946 O GLY A 438 14.794-104.692 -12.287 1.00 68.46 A O
+ATOM 2947 N CYS A 439 16.302-103.387 -11.244 1.00 64.49 A N
+ATOM 2948 CA CYS A 439 16.609-102.555 -12.401 1.00 65.51 A C
+ATOM 2949 C CYS A 439 18.021-102.881 -12.888 1.00 68.04 A C
+ATOM 2950 O CYS A 439 18.999-102.651 -12.167 1.00 70.32 A O
+ATOM 2951 CB CYS A 439 16.473-101.073 -12.048 1.00 57.45 A C
+ATOM 2952 SG CYS A 439 17.184 -99.968 -13.267 1.00147.39 A S
+ATOM 2953 N ASP A 440 18.127-103.421 -14.104 1.00 65.94 A N
+ATOM 2954 CA ASP A 440 19.402-103.804 -14.699 1.00 66.78 A C
+ATOM 2955 C ASP A 440 19.565-103.109 -16.054 1.00 73.16 A C
+ATOM 2956 O ASP A 440 18.715-102.331 -16.497 1.00 67.24 A O
+ATOM 2957 CB ASP A 440 19.504-105.328 -14.878 1.00 70.41 A C
+ATOM 2958 CG ASP A 440 20.942-105.840 -14.785 1.00 73.02 A C
+ATOM 2959 OD1 ASP A 440 21.863-105.004 -14.673 1.00 78.88 A O
+ATOM 2960 OD2 ASP A 440 21.157-107.071 -14.875 1.00 86.92 A O1-
+ATOM 2961 N TYR A 441 20.668-103.422 -16.730 1.00 67.79 A N
+ATOM 2962 CA TYR A 441 21.001-102.837 -18.020 1.00 67.13 A C
+ATOM 2963 C TYR A 441 21.788-103.857 -18.836 1.00 65.92 A C
+ATOM 2964 O TYR A 441 22.421-104.767 -18.290 1.00 68.03 A O
+ATOM 2965 CB TYR A 441 21.798-101.544 -17.834 1.00 66.95 A C
+ATOM 2966 CG TYR A 441 22.409-100.975 -19.093 1.00 69.82 A C
+ATOM 2967 CD1 TYR A 441 21.807 -99.928 -19.762 1.00 72.57 A C
+ATOM 2968 CD2 TYR A 441 23.607-101.470 -19.602 1.00 71.52 A C
+ATOM 2969 CE1 TYR A 441 22.369 -99.395 -20.907 1.00 62.47 A C
+ATOM 2970 CE2 TYR A 441 24.165-100.955 -20.750 1.00 67.28 A C
+ATOM 2971 CZ TYR A 441 23.544 -99.910 -21.390 1.00 69.14 A C
+ATOM 2972 OH TYR A 441 24.098 -99.376 -22.524 1.00 72.64 A O
+ATOM 2973 N VAL A 442 21.731-103.699 -20.158 1.00 59.95 A N
+ATOM 2974 CA VAL A 442 22.526-104.513 -21.070 1.00 61.34 A C
+ATOM 2975 C VAL A 442 22.931-103.660 -22.269 1.00 68.59 A C
+ATOM 2976 O VAL A 442 22.144-102.855 -22.776 1.00 62.64 A O
+ATOM 2977 CB VAL A 442 21.770-105.799 -21.479 1.00 68.62 A C
+ATOM 2978 CG1 VAL A 442 20.365-105.503 -21.962 1.00 60.86 A C
+ATOM 2979 CG2 VAL A 442 22.529-106.501 -22.547 1.00 69.29 A C
+ATOM 2980 N SER A 443 24.179-103.821 -22.702 1.00 67.36 A N
+ATOM 2981 CA SER A 443 24.693-103.073 -23.837 1.00 79.20 A C
+ATOM 2982 C SER A 443 24.091-103.588 -25.137 1.00 73.91 A C
+ATOM 2983 O SER A 443 23.584-104.712 -25.210 1.00 75.19 A O
+ATOM 2984 CB SER A 443 26.209-103.189 -23.924 1.00 69.77 A C
+ATOM 2985 OG SER A 443 26.570-104.503 -24.322 1.00 81.54 A O
+ATOM 2986 N ASN A 444 24.168-102.747 -26.180 1.00 73.11 A N
+ATOM 2987 CA ASN A 444 23.744-103.148 -27.523 1.00 71.64 A C
+ATOM 2988 C ASN A 444 24.743-104.074 -28.202 1.00 70.88 A C
+ATOM 2989 O ASN A 444 24.388-104.709 -29.203 1.00 85.04 A O
+ATOM 2990 CB ASN A 444 23.512-101.921 -28.406 1.00 66.49 A C
+ATOM 2991 CG ASN A 444 24.808-101.301 -28.900 1.00 68.43 A C
+ATOM 2992 ND2 ASN A 444 25.551-100.676 -27.986 1.00 74.40 A N
+ATOM 2993 OD1 ASN A 444 25.137-101.373 -30.091 1.00 88.58 A O
+ATOM 2994 N LYS A 445 25.982-104.156 -27.698 1.00 65.85 A N
+ATOM 2995 CA LYS A 445 26.960-105.083 -28.263 1.00 77.34 A C
+ATOM 2996 C LYS A 445 26.516-106.511 -27.980 1.00 79.77 A C
+ATOM 2997 O LYS A 445 26.213-106.863 -26.836 1.00 81.22 A O
+ATOM 2998 CB LYS A 445 28.363-104.848 -27.687 1.00 74.63 A C
+ATOM 2999 CG LYS A 445 28.801-103.375 -27.521 1.00 90.64 A C
+ATOM 3000 CD LYS A 445 30.175-103.286 -26.805 1.00110.45 A C
+ATOM 3001 CE LYS A 445 30.622-101.851 -26.482 1.00125.19 A C
+ATOM 3002 NZ LYS A 445 29.738-101.179 -25.491 1.00130.94 A N1+
+ATOM 3003 N GLY A 446 26.446-107.325 -29.031 1.00 82.56 A N
+ATOM 3004 CA GLY A 446 26.082-108.721 -28.890 1.00 79.74 A C
+ATOM 3005 C GLY A 446 24.630-108.990 -28.563 1.00 84.67 A C
+ATOM 3006 O GLY A 446 24.234-110.163 -28.480 1.00 90.41 A O
+ATOM 3007 N VAL A 447 23.821-107.954 -28.372 1.00 80.82 A N
+ATOM 3008 CA VAL A 447 22.398-108.114 -28.116 1.00 81.00 A C
+ATOM 3009 C VAL A 447 21.632-107.530 -29.285 1.00 83.88 A C
+ATOM 3010 O VAL A 447 22.029-106.514 -29.872 1.00 87.21 A O
+ATOM 3011 CB VAL A 447 21.959-107.439 -26.806 1.00 77.64 A C
+ATOM 3012 CG1 VAL A 447 20.572-107.936 -26.390 1.00 76.04 A C
+ATOM 3013 CG2 VAL A 447 22.991-107.692 -25.751 1.00 72.58 A C
+ATOM 3014 N ASP A 448 20.534-108.187 -29.624 1.00 71.38 A N
+ATOM 3015 CA ASP A 448 19.579-107.674 -30.586 1.00 72.88 A C
+ATOM 3016 C ASP A 448 18.255-107.361 -29.897 1.00 70.25 A C
+ATOM 3017 O ASP A 448 17.793-106.213 -29.881 1.00 70.86 A O
+ATOM 3018 CB ASP A 448 19.439-108.687 -31.729 1.00 78.81 A C
+ATOM 3019 CG ASP A 448 20.751-108.877 -32.493 1.00 89.17 A C
+ATOM 3020 OD1 ASP A 448 21.460-107.878 -32.731 1.00 92.04 A O
+ATOM 3021 OD2 ASP A 448 21.090-110.027 -32.833 1.00 97.51 A O1-
+ATOM 3022 N THR A 449 17.639-108.351 -29.278 1.00 71.64 A N
+ATOM 3023 CA THR A 449 16.318-108.157 -28.721 1.00 68.52 A C
+ATOM 3024 C THR A 449 16.322-108.145 -27.201 1.00 73.88 A C
+ATOM 3025 O THR A 449 17.098-108.850 -26.554 1.00 70.23 A O
+ATOM 3026 CB THR A 449 15.418-109.254 -29.220 1.00 74.12 A C
+ATOM 3027 CG2 THR A 449 14.016-108.869 -29.012 1.00 73.18 A C
+ATOM 3028 OG1 THR A 449 15.699-109.464 -30.612 1.00 78.05 A O
+ATOM 3029 N VAL A 450 15.430-107.346 -26.634 1.00 67.35 A N
+ATOM 3030 CA VAL A 450 15.175-107.338 -25.202 1.00 62.73 A C
+ATOM 3031 C VAL A 450 13.680-107.546 -25.009 1.00 56.73 A C
+ATOM 3032 O VAL A 450 12.873-106.682 -25.374 1.00 63.82 A O
+ATOM 3033 CB VAL A 450 15.646-106.042 -24.544 1.00 67.22 A C
+ATOM 3034 CG1 VAL A 450 15.012-105.886 -23.195 1.00 75.50 A C
+ATOM 3035 CG2 VAL A 450 17.142-106.082 -24.414 1.00 70.08 A C
+ATOM 3036 N SER A 451 13.307-108.687 -24.441 1.00 62.57 A N
+ATOM 3037 CA SER A 451 11.913-109.075 -24.290 1.00 64.64 A C
+ATOM 3038 C SER A 451 11.529-108.944 -22.822 1.00 67.52 A C
+ATOM 3039 O SER A 451 12.250-109.423 -21.941 1.00 67.49 A O
+ATOM 3040 CB SER A 451 11.689-110.501 -24.813 1.00 64.74 A C
+ATOM 3041 OG SER A 451 12.491-110.763 -25.972 1.00 83.91 A O
+ATOM 3042 N VAL A 452 10.425-108.248 -22.561 1.00 62.63 A N
+ATOM 3043 CA VAL A 452 9.960-107.972 -21.202 1.00 58.83 A C
+ATOM 3044 C VAL A 452 8.458-108.219 -21.151 1.00 59.59 A C
+ATOM 3045 O VAL A 452 7.686-107.560 -21.864 1.00 74.13 A O
+ATOM 3046 CB VAL A 452 10.267-106.535 -20.751 1.00 71.20 A C
+ATOM 3047 CG1 VAL A 452 9.846-106.355 -19.329 1.00 65.54 A C
+ATOM 3048 CG2 VAL A 452 11.735-106.228 -20.903 1.00 74.49 A C
+ATOM 3049 N GLY A 453 8.038-109.146 -20.309 1.00 59.02 A N
+ATOM 3050 CA GLY A 453 6.639-109.521 -20.357 1.00 62.75 A C
+ATOM 3051 C GLY A 453 6.303-110.039 -21.735 1.00 71.86 A C
+ATOM 3052 O GLY A 453 7.004-110.896 -22.278 1.00 72.47 A O
+ATOM 3053 N ASN A 454 5.232-109.520 -22.325 1.00 68.36 A N
+ATOM 3054 CA ASN A 454 4.874-109.920 -23.676 1.00 64.65 A C
+ATOM 3055 C ASN A 454 5.355-108.953 -24.753 1.00 64.15 A C
+ATOM 3056 O ASN A 454 5.081-109.188 -25.936 1.00 70.84 A O
+ATOM 3057 CB ASN A 454 3.360-110.109 -23.800 1.00 64.54 A C
+ATOM 3058 CG ASN A 454 2.838-111.212 -22.906 1.00 76.11 A C
+ATOM 3059 ND2 ASN A 454 1.645-111.007 -22.365 1.00 71.80 A N
+ATOM 3060 OD1 ASN A 454 3.501-112.235 -22.693 1.00 77.06 A O
+ATOM 3061 N THR A 455 6.057-107.881 -24.397 1.00 52.75 A N
+ATOM 3062 CA THR A 455 6.491-106.923 -25.402 1.00 60.11 A C
+ATOM 3063 C THR A 455 7.937-107.233 -25.778 1.00 66.25 A C
+ATOM 3064 O THR A 455 8.786-107.431 -24.900 1.00 67.38 A O
+ATOM 3065 CB THR A 455 6.351-105.493 -24.885 1.00 63.33 A C
+ATOM 3066 CG2 THR A 455 5.197-105.395 -23.887 1.00 61.51 A C
+ATOM 3067 OG1 THR A 455 7.547-105.127 -24.207 1.00 63.72 A O
+ATOM 3068 N LEU A 456 8.209-107.308 -27.081 1.00 66.20 A N
+ATOM 3069 CA LEU A 456 9.545-107.598 -27.605 1.00 59.11 A C
+ATOM 3070 C LEU A 456 10.174-106.349 -28.209 1.00 63.81 A C
+ATOM 3071 O LEU A 456 9.758-105.905 -29.283 1.00 66.08 A O
+ATOM 3072 CB LEU A 456 9.454-108.716 -28.632 1.00 68.50 A C
+ATOM 3073 CG LEU A 456 10.791-109.115 -29.219 1.00 64.01 A C
+ATOM 3074 CD1 LEU A 456 10.914-110.617 -29.329 1.00 67.54 A C
+ATOM 3075 CD2 LEU A 456 10.883-108.523 -30.601 1.00 81.85 A C
+ATOM 3076 N TYR A 457 11.194-105.802 -27.542 1.00 60.67 A N
+ATOM 3077 CA TYR A 457 11.855-104.574 -27.993 1.00 59.19 A C
+ATOM 3078 C TYR A 457 13.097-104.912 -28.825 1.00 60.19 A C
+ATOM 3079 O TYR A 457 13.845-105.838 -28.491 1.00 63.36 A O
+ATOM 3080 CB TYR A 457 12.264-103.689 -26.800 1.00 54.51 A C
+ATOM 3081 CG TYR A 457 11.134-103.128 -25.939 1.00 68.18 A C
+ATOM 3082 CD1 TYR A 457 10.478-101.957 -26.290 1.00 75.41 A C
+ATOM 3083 CD2 TYR A 457 10.758-103.744 -24.759 1.00 78.10 A C
+ATOM 3084 CE1 TYR A 457 9.447-101.440 -25.517 1.00 88.58 A C
+ATOM 3085 CE2 TYR A 457 9.742-103.222 -23.970 1.00 89.16 A C
+ATOM 3086 CZ TYR A 457 9.085-102.074 -24.354 1.00 97.30 A C
+ATOM 3087 OH TYR A 457 8.065-101.562 -23.570 1.00101.83 A O
+ATOM 3088 N TYR A 458 13.318-104.179 -29.915 1.00 64.10 A N
+ATOM 3089 CA TYR A 458 14.610-104.266 -30.590 1.00 66.28 A C
+ATOM 3090 C TYR A 458 15.459-103.102 -30.102 1.00 61.30 A C
+ATOM 3091 O TYR A 458 14.931-102.024 -29.810 1.00 59.93 A O
+ATOM 3092 CB TYR A 458 14.487-104.198 -32.117 1.00 64.78 A C
+ATOM 3093 CG TYR A 458 13.992-105.445 -32.838 1.00 67.84 A C
+ATOM 3094 CD1 TYR A 458 12.671-105.842 -32.735 1.00 57.58 A C
+ATOM 3095 CD2 TYR A 458 14.809-106.156 -33.709 1.00 61.41 A C
+ATOM 3096 CE1 TYR A 458 12.193-106.961 -33.418 1.00 60.43 A C
+ATOM 3097 CE2 TYR A 458 14.326-107.282 -34.399 1.00 63.09 A C
+ATOM 3098 CZ TYR A 458 13.012-107.670 -34.242 1.00 67.86 A C
+ATOM 3099 OH TYR A 458 12.474-108.755 -34.895 1.00 70.83 A O
+ATOM 3100 N VAL A 459 16.780-103.332 -29.981 1.00 56.83 A N
+ATOM 3101 CA VAL A 459 17.711-102.295 -29.529 1.00 64.15 A C
+ATOM 3102 C VAL A 459 18.373-101.667 -30.743 1.00 66.90 A C
+ATOM 3103 O VAL A 459 18.601-102.338 -31.754 1.00 66.61 A O
+ATOM 3104 CB VAL A 459 18.779-102.833 -28.555 1.00 69.93 A C
+ATOM 3105 CG1 VAL A 459 18.129-103.406 -27.345 1.00 64.61 A C
+ATOM 3106 CG2 VAL A 459 19.650-103.872 -29.236 1.00 70.02 A C
+ATOM 3107 N ASN A 460 18.661-100.363 -30.658 1.00 63.16 A N
+ATOM 3108 CA ASN A 460 19.425 -99.706 -31.714 1.00 56.42 A C
+ATOM 3109 C ASN A 460 20.795-100.355 -31.822 1.00 64.49 A C
+ATOM 3110 O ASN A 460 21.405-100.713 -30.813 1.00 66.83 A O
+ATOM 3111 CB ASN A 460 19.582 -98.214 -31.435 1.00 70.48 A C
+ATOM 3112 CG ASN A 460 18.388 -97.643 -30.765 1.00 70.61 A C
+ATOM 3113 ND2 ASN A 460 17.475 -97.079 -31.524 1.00 63.26 A N
+ATOM 3114 OD1 ASN A 460 18.270 -97.746 -29.564 1.00 69.31 A O
+ATOM 3115 N LYS A 461 21.262-100.555 -33.049 1.00 61.51 A N
+ATOM 3116 CA LYS A 461 22.553-101.188 -33.239 1.00 69.64 A C
+ATOM 3117 C LYS A 461 23.687-100.195 -33.491 1.00 73.86 A C
+ATOM 3118 O LYS A 461 24.797-100.633 -33.795 1.00 91.95 A O
+ATOM 3119 CB LYS A 461 22.469-102.251 -34.337 1.00 68.86 A C
+ATOM 3120 CG LYS A 461 21.683-103.513 -33.882 1.00 69.19 A C
+ATOM 3121 CD LYS A 461 22.322-104.261 -32.669 1.00 82.40 A C
+ATOM 3122 CE LYS A 461 23.338-105.364 -33.093 1.00 81.52 A C
+ATOM 3123 NZ LYS A 461 24.136-106.101 -32.020 1.00 68.05 A N1+
+ATOM 3124 N GLN A 462 23.455 -98.880 -33.377 1.00 88.07 A N
+ATOM 3125 CA GLN A 462 24.549 -97.934 -33.609 1.00 87.64 A C
+ATOM 3126 C GLN A 462 25.646 -98.080 -32.555 1.00 81.42 A C
+ATOM 3127 O GLN A 462 25.390 -98.381 -31.388 1.00 85.53 A O
+ATOM 3128 CB GLN A 462 24.062 -96.478 -33.653 1.00 94.29 A C
+ATOM 3129 CG GLN A 462 25.156 -95.454 -34.188 1.00 98.48 A C
+ATOM 3130 CD GLN A 462 25.568 -95.603 -35.707 1.00113.79 A C
+ATOM 3131 NE2 GLN A 462 25.261 -94.574 -36.510 1.00116.60 A N
+ATOM 3132 OE1 GLN A 462 26.186 -96.601 -36.119 1.00125.60 A O
+ATOM 3133 N GLU A 463 26.885 -97.860 -32.999 1.00 85.35 A N
+ATOM 3134 CA GLU A 463 28.096 -98.264 -32.302 1.00 95.23 A C
+ATOM 3135 C GLU A 463 28.473 -97.287 -31.193 1.00 91.56 A C
+ATOM 3136 O GLU A 463 28.095 -96.112 -31.203 1.00 88.41 A O
+ATOM 3137 CB GLU A 463 29.259 -98.396 -33.287 1.00 95.60 A C
+ATOM 3138 CG GLU A 463 29.063 -99.443 -34.400 1.00116.35 A C
+ATOM 3139 CD GLU A 463 29.002-100.900 -33.896 1.00128.88 A C
+ATOM 3140 OE1 GLU A 463 29.342-101.160 -32.719 1.00138.11 A O
+ATOM 3141 OE2 GLU A 463 28.623-101.791 -34.695 1.00132.91 A O1-
+ATOM 3142 N GLY A 464 29.230 -97.801 -30.230 1.00 93.41 A N
+ATOM 3143 CA GLY A 464 29.799 -97.003 -29.159 1.00 83.31 A C
+ATOM 3144 C GLY A 464 30.130 -97.861 -27.958 1.00 87.26 A C
+ATOM 3145 O GLY A 464 29.743 -99.033 -27.865 1.00 87.85 A O
+ATOM 3146 N LYS A 465 30.845 -97.251 -27.008 1.00 83.34 A N
+ATOM 3147 CA LYS A 465 31.296 -97.976 -25.826 1.00 79.32 A C
+ATOM 3148 C LYS A 465 30.374 -97.697 -24.646 1.00 83.58 A C
+ATOM 3149 O LYS A 465 29.963 -96.552 -24.399 1.00 83.52 A O
+ATOM 3150 CB LYS A 465 32.749 -97.649 -25.453 1.00 83.05 A C
+ATOM 3151 CG LYS A 465 33.397 -98.741 -24.567 1.00 97.14 A C
+ATOM 3152 CD LYS A 465 34.884 -98.496 -24.300 1.00118.38 A C
+ATOM 3153 CE LYS A 465 35.450 -99.523 -23.326 1.00118.61 A C
+ATOM 3154 NZ LYS A 465 35.206-100.917 -23.794 1.00126.44 A N1+
+ATOM 3155 N SER A 466 30.063 -98.766 -23.917 1.00 81.39 A N
+ATOM 3156 CA SER A 466 29.034 -98.763 -22.892 1.00 79.95 A C
+ATOM 3157 C SER A 466 29.649 -99.210 -21.572 1.00 78.51 A C
+ATOM 3158 O SER A 466 30.272-100.276 -21.505 1.00 88.71 A O
+ATOM 3159 CB SER A 466 27.882 -99.683 -23.309 1.00 74.29 A C
+ATOM 3160 OG SER A 466 26.669 -99.297 -22.697 1.00 98.19 A O
+ATOM 3161 N LEU A 467 29.471 -98.400 -20.528 1.00 77.90 A N
+ATOM 3162 CA LEU A 467 30.048 -98.648 -19.213 1.00 74.17 A C
+ATOM 3163 C LEU A 467 28.943 -98.783 -18.171 1.00 79.08 A C
+ATOM 3164 O LEU A 467 28.133 -97.859 -17.993 1.00 71.79 A O
+ATOM 3165 CB LEU A 467 31.001 -97.522 -18.814 1.00 78.16 A C
+ATOM 3166 CG LEU A 467 31.686 -97.734 -17.457 1.00 90.99 A C
+ATOM 3167 CD1 LEU A 467 32.817 -98.754 -17.554 1.00 83.80 A C
+ATOM 3168 CD2 LEU A 467 32.174 -96.419 -16.862 1.00 78.67 A C
+ATOM 3169 N TYR A 468 28.931 -99.927 -17.476 1.00 86.53 A N
+ATOM 3170 CA TYR A 468 28.017-100.199 -16.365 1.00 79.19 A C
+ATOM 3171 C TYR A 468 28.625 -99.687 -15.066 1.00 76.53 A C
+ATOM 3172 O TYR A 468 29.743-100.074 -14.716 1.00 83.19 A O
+ATOM 3173 CB TYR A 468 27.770-101.700 -16.259 1.00 75.62 A C
+ATOM 3174 CG TYR A 468 26.557-102.129 -15.464 1.00 73.81 A C
+ATOM 3175 CD1 TYR A 468 25.575-102.926 -16.052 1.00 76.06 A C
+ATOM 3176 CD2 TYR A 468 26.411-101.784 -14.123 1.00 66.81 A C
+ATOM 3177 CE1 TYR A 468 24.474-103.358 -15.342 1.00 68.11 A C
+ATOM 3178 CE2 TYR A 468 25.306-102.211 -13.394 1.00 72.59 A C
+ATOM 3179 CZ TYR A 468 24.333-103.001 -14.014 1.00 75.00 A C
+ATOM 3180 OH TYR A 468 23.214-103.432 -13.314 1.00 77.50 A O
+ATOM 3181 N VAL A 469 27.898 -98.839 -14.342 1.00 72.66 A N
+ATOM 3182 CA VAL A 469 28.403 -98.236 -13.110 1.00 79.42 A C
+ATOM 3183 C VAL A 469 27.552 -98.724 -11.940 1.00 74.95 A C
+ATOM 3184 O VAL A 469 26.433 -98.231 -11.720 1.00 73.60 A O
+ATOM 3185 CB VAL A 469 28.421 -96.706 -13.197 1.00 81.21 A C
+ATOM 3186 CG1 VAL A 469 28.764 -96.111 -11.871 1.00 68.36 A C
+ATOM 3187 CG2 VAL A 469 29.449 -96.285 -14.202 1.00 74.21 A C
+ATOM 3188 N LYS A 470 28.121 -99.627 -11.138 1.00 73.05 A N
+ATOM 3189 CA LYS A 470 27.386-100.244 -10.044 1.00 78.53 A C
+ATOM 3190 C LYS A 470 27.030 -99.197 -8.989 1.00 74.93 A C
+ATOM 3191 O LYS A 470 27.549 -98.079 -8.973 1.00 77.36 A O
+ATOM 3192 CB LYS A 470 28.195-101.389 -9.432 1.00 77.83 A C
+ATOM 3193 CG LYS A 470 28.671-102.400 -10.463 1.00 79.45 A C
+ATOM 3194 CD LYS A 470 29.627-103.467 -9.896 1.00 96.69 A C
+ATOM 3195 CE LYS A 470 28.885-104.635 -9.218 1.00105.54 A C
+ATOM 3196 NZ LYS A 470 29.786-105.773 -8.830 1.00108.31 A N1+
+ATOM 3197 N GLY A 471 26.073 -99.544 -8.148 1.00 80.95 A N
+ATOM 3198 CA GLY A 471 25.658 -98.640 -7.096 1.00 73.33 A C
+ATOM 3199 C GLY A 471 24.641 -99.304 -6.197 1.00 77.25 A C
+ATOM 3200 O GLY A 471 24.006-100.304 -6.567 1.00 86.28 A O
+ATOM 3201 N GLU A 472 24.503 -98.717 -4.938 1.00 83.41 A N
+ATOM 3202 CA GLU A 472 23.379 -99.454 -4.389 1.00 77.61 A C
+ATOM 3203 C GLU A 472 22.082 -98.747 -4.770 1.00 74.57 A C
+ATOM 3204 O GLU A 472 22.027 -97.507 -4.763 1.00 72.95 A O
+ATOM 3205 CB GLU A 472 23.483 -99.568 -2.874 1.00 75.98 A C
+ATOM 3206 CG GLU A 472 22.627-100.687 -2.303 1.00 90.66 A C
+ATOM 3207 CD GLU A 472 22.338-100.510 -0.831 1.00104.29 A C
+ATOM 3208 OE1 GLU A 472 23.074 -99.745 -0.168 1.00107.68 A O
+ATOM 3209 OE2 GLU A 472 21.378-101.138 -0.335 1.00112.09 A O1-
+ATOM 3210 N PRO A 473 21.052 -99.511 -5.138 1.00 78.23 A N
+ATOM 3211 CA PRO A 473 19.807 -98.880 -5.591 1.00 75.68 A C
+ATOM 3212 C PRO A 473 19.252 -97.979 -4.504 1.00 76.77 A C
+ATOM 3213 O PRO A 473 18.863 -98.437 -3.432 1.00 74.95 A O
+ATOM 3214 CB PRO A 473 18.888-100.075 -5.856 1.00 73.75 A C
+ATOM 3215 CG PRO A 473 19.820-101.203 -6.166 1.00 73.68 A C
+ATOM 3216 CD PRO A 473 21.043-100.973 -5.334 1.00 76.47 A C
+ATOM 3217 N ILE A 474 19.121 -96.697 -4.833 1.00 72.91 A N
+ATOM 3218 CA ILE A 474 18.826 -95.706 -3.820 1.00 80.68 A C
+ATOM 3219 C ILE A 474 17.462 -95.927 -3.213 1.00 77.82 A C
+ATOM 3220 O ILE A 474 17.162 -95.360 -2.164 1.00 88.19 A O
+ATOM 3221 CB ILE A 474 18.930 -94.285 -4.409 1.00 78.52 A C
+ATOM 3222 CG1 ILE A 474 17.832 -94.063 -5.446 1.00 74.34 A C
+ATOM 3223 CG2 ILE A 474 20.274 -94.097 -5.072 1.00 73.93 A C
+ATOM 3224 CD1 ILE A 474 17.863 -92.702 -6.064 1.00 79.87 A C
+ATOM 3225 N ILE A 475 16.621 -96.746 -3.846 1.00 86.44 A N
+ATOM 3226 CA ILE A 475 15.371 -97.116 -3.205 1.00 82.71 A C
+ATOM 3227 C ILE A 475 15.625 -97.814 -1.883 1.00 82.41 A C
+ATOM 3228 O ILE A 475 14.739 -97.846 -1.021 1.00 87.04 A O
+ATOM 3229 CB ILE A 475 14.510 -98.011 -4.122 1.00 82.53 A C
+ATOM 3230 CG1 ILE A 475 13.048 -97.997 -3.658 1.00 80.78 A C
+ATOM 3231 CG2 ILE A 475 15.048 -99.435 -4.150 1.00 84.22 A C
+ATOM 3232 CD1 ILE A 475 12.495 -96.598 -3.499 1.00 74.11 A C
+ATOM 3233 N ASN A 476 16.823 -98.375 -1.691 1.00 77.48 A N
+ATOM 3234 CA ASN A 476 17.097 -99.123 -0.471 1.00 77.64 A C
+ATOM 3235 C ASN A 476 17.334 -98.222 0.740 1.00 83.03 A C
+ATOM 3236 O ASN A 476 17.077 -98.659 1.868 1.00 86.37 A O
+ATOM 3237 CB ASN A 476 18.280-100.066 -0.705 1.00 82.21 A C
+ATOM 3238 CG ASN A 476 17.911-101.246 -1.591 1.00 93.07 A C
+ATOM 3239 ND2 ASN A 476 18.870-102.116 -1.850 1.00 95.81 A N
+ATOM 3240 OD1 ASN A 476 16.779-101.359 -2.049 1.00 89.57 A O
+ATOM 3241 N PHE A 477 17.729 -96.961 0.535 1.00 89.53 A N
+ATOM 3242 CA PHE A 477 18.058 -96.057 1.637 1.00 83.21 A C
+ATOM 3243 C PHE A 477 16.824 -95.457 2.290 1.00 83.86 A C
+ATOM 3244 O PHE A 477 16.956 -94.594 3.159 1.00 91.18 A O
+ATOM 3245 CB PHE A 477 18.951 -94.893 1.181 1.00 84.30 A C
+ATOM 3246 CG PHE A 477 20.216 -95.300 0.460 1.00 82.45 A C
+ATOM 3247 CD1 PHE A 477 21.059 -94.317 -0.048 1.00 81.34 A C
+ATOM 3248 CD2 PHE A 477 20.575 -96.628 0.287 1.00 78.83 A C
+ATOM 3249 CE1 PHE A 477 22.231 -94.644 -0.723 1.00 88.06 A C
+ATOM 3250 CE2 PHE A 477 21.746 -96.963 -0.387 1.00 78.05 A C
+ATOM 3251 CZ PHE A 477 22.576 -95.967 -0.895 1.00 76.63 A C
+ATOM 3252 N TYR A 478 15.638 -95.839 1.858 1.00 90.36 A N
+ATOM 3253 CA TYR A 478 14.408 -95.233 2.331 1.00 83.53 A C
+ATOM 3254 C TYR A 478 13.692 -96.194 3.272 1.00 86.44 A C
+ATOM 3255 O TYR A 478 13.660 -97.407 3.034 1.00 82.42 A O
+ATOM 3256 CB TYR A 478 13.517 -94.844 1.130 1.00 73.56 A C
+ATOM 3257 CG TYR A 478 13.952 -93.554 0.439 1.00 75.81 A C
+ATOM 3258 CD1 TYR A 478 13.429 -92.336 0.844 1.00 74.36 A C
+ATOM 3259 CD2 TYR A 478 14.913 -93.552 -0.576 1.00 73.88 A C
+ATOM 3260 CE1 TYR A 478 13.826 -91.152 0.258 1.00 76.73 A C
+ATOM 3261 CE2 TYR A 478 15.327 -92.362 -1.167 1.00 71.50 A C
+ATOM 3262 CZ TYR A 478 14.773 -91.159 -0.739 1.00 76.29 A C
+ATOM 3263 OH TYR A 478 15.138 -89.939 -1.284 1.00 82.33 A O
+ATOM 3264 N ASP A 479 13.140 -95.656 4.352 1.00 80.73 A N
+ATOM 3265 CA ASP A 479 12.340 -96.471 5.260 1.00 82.71 A C
+ATOM 3266 C ASP A 479 10.922 -96.581 4.722 1.00 91.45 A C
+ATOM 3267 O ASP A 479 10.225 -95.562 4.644 1.00 93.94 A O
+ATOM 3268 CB ASP A 479 12.323 -95.866 6.668 1.00 84.13 A C
+ATOM 3269 CG ASP A 479 12.065 -96.917 7.785 1.00 98.86 A C
+ATOM 3270 OD1 ASP A 479 11.260 -97.862 7.578 1.00 99.53 A O
+ATOM 3271 OD2 ASP A 479 12.680 -96.794 8.877 1.00105.26 A O1-
+ATOM 3272 N PRO A 480 10.451 -97.784 4.366 1.00 82.33 A N
+ATOM 3273 CA PRO A 480 9.097 -97.922 3.808 1.00 82.49 A C
+ATOM 3274 C PRO A 480 8.040 -97.186 4.588 1.00 83.36 A C
+ATOM 3275 O PRO A 480 7.057 -96.720 3.998 1.00 88.45 A O
+ATOM 3276 CB PRO A 480 8.851 -99.438 3.850 1.00 78.53 A C
+ATOM 3277 CG PRO A 480 10.176-100.017 3.743 1.00 80.25 A C
+ATOM 3278 CD PRO A 480 11.147 -99.074 4.426 1.00 78.35 A C
+ATOM 3279 N LEU A 481 8.214 -97.042 5.898 1.00 82.76 A N
+ATOM 3280 CA LEU A 481 7.113 -96.560 6.721 1.00 87.87 A C
+ATOM 3281 C LEU A 481 6.937 -95.053 6.592 1.00 90.34 A C
+ATOM 3282 O LEU A 481 5.807 -94.566 6.505 1.00 93.37 A O
+ATOM 3283 CB LEU A 481 7.326 -96.995 8.177 1.00 95.15 A C
+ATOM 3284 CG LEU A 481 7.160 -98.511 8.457 1.00111.94 A C
+ATOM 3285 CD1 LEU A 481 8.408 -99.363 8.135 1.00107.82 A C
+ATOM 3286 CD2 LEU A 481 6.709 -98.764 9.885 1.00120.66 A C
+ATOM 3287 N VAL A 482 8.038 -94.303 6.539 1.00 99.57 A N
+ATOM 3288 CA VAL A 482 7.970 -92.849 6.408 1.00100.32 A C
+ATOM 3289 C VAL A 482 8.108 -92.376 4.966 1.00 99.57 A C
+ATOM 3290 O VAL A 482 8.140 -91.160 4.720 1.00106.15 A O
+ATOM 3291 CB VAL A 482 9.044 -92.159 7.270 1.00101.95 A C
+ATOM 3292 CG1 VAL A 482 8.604 -92.111 8.734 1.00100.96 A C
+ATOM 3293 CG2 VAL A 482 10.406 -92.849 7.073 1.00 96.43 A C
+ATOM 3294 N PHE A 483 8.201 -93.297 4.009 1.00 96.53 A N
+ATOM 3295 CA PHE A 483 8.496 -92.951 2.625 1.00100.03 A C
+ATOM 3296 C PHE A 483 7.196 -92.665 1.877 1.00105.17 A C
+ATOM 3297 O PHE A 483 6.377 -93.584 1.703 1.00106.91 A O
+ATOM 3298 CB PHE A 483 9.260 -94.096 1.973 1.00 91.20 A C
+ATOM 3299 CG PHE A 483 9.312 -94.028 0.491 1.00 97.52 A C
+ATOM 3300 CD1 PHE A 483 10.321 -93.324 -0.138 1.00 94.79 A C
+ATOM 3301 CD2 PHE A 483 8.357 -94.678 -0.278 1.00 86.83 A C
+ATOM 3302 CE1 PHE A 483 10.374 -93.265 -1.506 1.00 94.47 A C
+ATOM 3303 CE2 PHE A 483 8.396 -94.615 -1.640 1.00 93.51 A C
+ATOM 3304 CZ PHE A 483 9.409 -93.913 -2.261 1.00 97.98 A C
+ATOM 3305 N PRO A 484 6.937 -91.416 1.457 1.00 96.90 A N
+ATOM 3306 CA PRO A 484 5.686 -91.116 0.737 1.00 92.62 A C
+ATOM 3307 C PRO A 484 5.719 -91.626 -0.699 1.00 96.43 A C
+ATOM 3308 O PRO A 484 6.688 -91.415 -1.428 1.00 98.25 A O
+ATOM 3309 CB PRO A 484 5.603 -89.587 0.790 1.00 89.61 A C
+ATOM 3310 CG PRO A 484 7.035 -89.155 0.844 1.00 96.99 A C
+ATOM 3311 CD PRO A 484 7.835 -90.250 1.514 1.00 95.91 A C
+ATOM 3312 N SER A 485 4.654 -92.311 -1.094 1.00 88.12 A N
+ATOM 3313 CA SER A 485 4.403 -92.683 -2.477 1.00 95.86 A C
+ATOM 3314 C SER A 485 3.382 -91.779 -3.158 1.00 99.16 A C
+ATOM 3315 O SER A 485 3.038 -92.016 -4.318 1.00100.46 A O
+ATOM 3316 CB SER A 485 3.955 -94.145 -2.542 1.00 92.93 A C
+ATOM 3317 OG SER A 485 2.818 -94.366 -1.718 1.00110.52 A O
+ATOM 3318 N ASP A 486 2.875 -90.766 -2.464 1.00101.59 A N
+ATOM 3319 CA ASP A 486 1.861 -89.885 -3.023 1.00 97.65 A C
+ATOM 3320 C ASP A 486 2.467 -88.727 -3.799 1.00103.90 A C
+ATOM 3321 O ASP A 486 1.729 -87.830 -4.231 1.00113.10 A O
+ATOM 3322 CB ASP A 486 0.923 -89.344 -1.924 1.00 97.24 A C
+ATOM 3323 CG ASP A 486 1.659 -88.569 -0.813 1.00106.10 A C
+ATOM 3324 OD1 ASP A 486 2.905 -88.403 -0.892 1.00 97.68 A O
+ATOM 3325 OD2 ASP A 486 0.966 -88.087 0.123 1.00100.08 A O1-
+ATOM 3326 N GLN A 487 3.788 -88.720 -3.984 1.00 90.44 A N
+ATOM 3327 CA GLN A 487 4.458 -87.618 -4.662 1.00 92.67 A C
+ATOM 3328 C GLN A 487 4.543 -87.920 -6.156 1.00 92.72 A C
+ATOM 3329 O GLN A 487 5.326 -88.777 -6.585 1.00 96.61 A O
+ATOM 3330 CB GLN A 487 5.835 -87.384 -4.048 1.00 89.19 A C
+ATOM 3331 CG GLN A 487 5.776 -87.127 -2.555 1.00 89.19 A C
+ATOM 3332 CD GLN A 487 4.981 -85.859 -2.206 1.00 94.87 A C
+ATOM 3333 NE2 GLN A 487 4.022 -85.983 -1.283 1.00 92.43 A N
+ATOM 3334 OE1 GLN A 487 5.231 -84.784 -2.766 1.00100.86 A O
+ATOM 3335 N PHE A 488 3.755 -87.183 -6.949 1.00 83.71 A N
+ATOM 3336 CA PHE A 488 3.739 -87.357 -8.402 1.00 90.40 A C
+ATOM 3337 C PHE A 488 4.871 -86.549 -9.030 1.00 81.72 A C
+ATOM 3338 O PHE A 488 5.833 -87.113 -9.560 1.00 82.18 A O
+ATOM 3339 CB PHE A 488 2.367 -86.945 -8.949 1.00 88.42 A C
+ATOM 3340 CG PHE A 488 2.088 -87.424 -10.348 1.00 90.23 A C
+ATOM 3341 CD1 PHE A 488 1.983 -88.772 -10.625 1.00 86.03 A C
+ATOM 3342 CD2 PHE A 488 1.870 -86.518 -11.375 1.00 97.38 A C
+ATOM 3343 CE1 PHE A 488 1.713 -89.215 -11.911 1.00 94.78 A C
+ATOM 3344 CE2 PHE A 488 1.592 -86.952 -12.667 1.00 90.03 A C
+ATOM 3345 CZ PHE A 488 1.515 -88.304 -12.933 1.00 84.69 A C
+ATOM 3346 N ASP A 489 4.774 -85.218 -8.952 1.00 82.03 A N
+ATOM 3347 CA ASP A 489 5.874 -84.307 -9.267 1.00 84.27 A C
+ATOM 3348 C ASP A 489 6.584 -83.926 -7.973 1.00 84.32 A C
+ATOM 3349 O ASP A 489 5.978 -83.301 -7.089 1.00 94.67 A O
+ATOM 3350 CB ASP A 489 5.378 -83.041 -9.962 1.00 94.20 A C
+ATOM 3351 CG ASP A 489 6.379 -81.910 -9.870 1.00100.20 A C
+ATOM 3352 OD1 ASP A 489 7.552 -82.127 -10.239 1.00 94.00 A O
+ATOM 3353 OD2 ASP A 489 6.007 -80.824 -9.384 1.00112.31 A O1-
+ATOM 3354 N ALA A 490 7.867 -84.271 -7.876 1.00 83.95 A N
+ATOM 3355 CA ALA A 490 8.586 -84.141 -6.616 1.00 85.30 A C
+ATOM 3356 C ALA A 490 10.078 -84.015 -6.889 1.00 85.53 A C
+ATOM 3357 O ALA A 490 10.545 -84.220 -8.010 1.00 76.06 A O
+ATOM 3358 CB ALA A 490 8.287 -85.324 -5.695 1.00 81.71 A C
+ATOM 3359 N SER A 491 10.813 -83.629 -5.851 1.00 78.80 A N
+ATOM 3360 CA SER A 491 12.273 -83.581 -5.845 1.00 80.39 A C
+ATOM 3361 C SER A 491 12.827 -84.588 -4.837 1.00 82.80 A C
+ATOM 3362 O SER A 491 12.083 -85.321 -4.184 1.00 85.14 A O
+ATOM 3363 CB SER A 491 12.766 -82.175 -5.513 1.00 83.18 A C
+ATOM 3364 OG SER A 491 12.003 -81.617 -4.461 1.00 85.74 A O
+ATOM 3365 N ILE A 492 14.158 -84.629 -4.722 1.00 78.59 A N
+ATOM 3366 CA ILE A 492 14.777 -85.442 -3.678 1.00 75.88 A C
+ATOM 3367 C ILE A 492 14.502 -84.831 -2.315 1.00 87.26 A C
+ATOM 3368 O ILE A 492 14.086 -85.520 -1.369 1.00 84.99 A O
+ATOM 3369 CB ILE A 492 16.289 -85.592 -3.922 1.00 76.26 A C
+ATOM 3370 CG1 ILE A 492 16.564 -86.628 -5.000 1.00 82.25 A C
+ATOM 3371 CG2 ILE A 492 16.986 -86.006 -2.658 1.00 72.65 A C
+ATOM 3372 CD1 ILE A 492 16.525 -88.035 -4.492 1.00 81.82 A C
+ATOM 3373 N SER A 493 14.704 -83.517 -2.211 1.00 76.46 A N
+ATOM 3374 CA SER A 493 14.561 -82.835 -0.934 1.00 94.87 A C
+ATOM 3375 C SER A 493 13.098 -82.700 -0.506 1.00 89.58 A C
+ATOM 3376 O SER A 493 12.819 -82.635 0.694 1.00 92.61 A O
+ATOM 3377 CB SER A 493 15.240 -81.468 -1.013 1.00 77.86 A C
+ATOM 3378 OG SER A 493 14.516 -80.582 -1.853 1.00107.73 A O
+ATOM 3379 N GLN A 494 12.156 -82.639 -1.455 1.00 79.64 A N
+ATOM 3380 CA GLN A 494 10.741 -82.609 -1.084 1.00 83.34 A C
+ATOM 3381 C GLN A 494 10.305 -83.941 -0.483 1.00 83.12 A C
+ATOM 3382 O GLN A 494 9.475 -83.969 0.437 1.00 81.93 A O
+ATOM 3383 CB GLN A 494 9.879 -82.256 -2.304 1.00 85.97 A C
+ATOM 3384 CG GLN A 494 8.417 -81.873 -2.017 1.00106.44 A C
+ATOM 3385 CD GLN A 494 7.663 -81.439 -3.278 1.00128.44 A C
+ATOM 3386 NE2 GLN A 494 6.388 -81.825 -3.376 1.00140.60 A N
+ATOM 3387 OE1 GLN A 494 8.224 -80.765 -4.150 1.00134.93 A O
+ATOM 3388 N VAL A 495 10.856 -85.049 -0.986 1.00 83.18 A N
+ATOM 3389 CA VAL A 495 10.634 -86.347 -0.360 1.00 84.75 A C
+ATOM 3390 C VAL A 495 11.279 -86.384 1.018 1.00 94.32 A C
+ATOM 3391 O VAL A 495 10.659 -86.819 2.001 1.00 97.39 A O
+ATOM 3392 CB VAL A 495 11.162 -87.471 -1.261 1.00 87.30 A C
+ATOM 3393 CG1 VAL A 495 11.554 -88.660 -0.431 1.00 80.74 A C
+ATOM 3394 CG2 VAL A 495 10.090 -87.865 -2.233 1.00 78.39 A C
+ATOM 3395 N ASN A 496 12.534 -85.914 1.113 1.00 87.03 A N
+ATOM 3396 CA ASN A 496 13.166 -85.762 2.422 1.00101.65 A C
+ATOM 3397 C ASN A 496 12.302 -84.948 3.372 1.00101.37 A C
+ATOM 3398 O ASN A 496 12.321 -85.191 4.579 1.00 98.60 A O
+ATOM 3399 CB ASN A 496 14.529 -85.087 2.292 1.00 89.80 A C
+ATOM 3400 CG ASN A 496 15.621 -86.060 1.934 1.00100.02 A C
+ATOM 3401 ND2 ASN A 496 16.860 -85.555 1.821 1.00 91.41 A N
+ATOM 3402 OD1 ASN A 496 15.364 -87.265 1.773 1.00101.56 A O
+ATOM 3403 N GLU A 497 11.542 -83.985 2.841 1.00 81.00 A N
+ATOM 3404 CA GLU A 497 10.744 -83.064 3.641 1.00 83.56 A C
+ATOM 3405 C GLU A 497 9.458 -83.721 4.135 1.00 87.49 A C
+ATOM 3406 O GLU A 497 9.111 -83.606 5.318 1.00 89.74 A O
+ATOM 3407 CB GLU A 497 10.438 -81.812 2.813 1.00 85.32 A C
+ATOM 3408 CG GLU A 497 9.921 -80.610 3.594 1.00118.00 A C
+ATOM 3409 CD GLU A 497 9.762 -79.370 2.719 1.00144.91 A C
+ATOM 3410 OE1 GLU A 497 9.770 -79.504 1.474 1.00157.59 A O
+ATOM 3411 OE2 GLU A 497 9.642 -78.259 3.279 1.00146.63 A O1-
+ATOM 3412 N LYS A 498 8.731 -84.408 3.244 1.00 85.99 A N
+ATOM 3413 CA LYS A 498 7.555 -85.151 3.694 1.00 88.45 A C
+ATOM 3414 C LYS A 498 7.968 -86.256 4.673 1.00 91.19 A C
+ATOM 3415 O LYS A 498 7.217 -86.594 5.610 1.00 92.08 A O
+ATOM 3416 CB LYS A 498 6.776 -85.685 2.470 1.00 86.50 A C
+ATOM 3417 CG LYS A 498 5.324 -86.188 2.726 1.00 94.22 A C
+ATOM 3418 CD LYS A 498 4.195 -85.130 2.587 1.00 94.59 A C
+ATOM 3419 CE LYS A 498 2.812 -85.781 2.911 1.00104.46 A C
+ATOM 3420 NZ LYS A 498 1.591 -84.902 2.988 1.00105.82 A N1+
+ATOM 3421 N ILE A 499 9.193 -86.771 4.522 1.00 83.81 A N
+ATOM 3422 CA ILE A 499 9.714 -87.729 5.489 1.00 88.55 A C
+ATOM 3423 C ILE A 499 10.015 -87.049 6.816 1.00 94.00 A C
+ATOM 3424 O ILE A 499 9.629 -87.542 7.875 1.00101.03 A O
+ATOM 3425 CB ILE A 499 10.952 -88.454 4.939 1.00 82.42 A C
+ATOM 3426 CG1 ILE A 499 10.545 -89.421 3.825 1.00 74.87 A C
+ATOM 3427 CG2 ILE A 499 11.663 -89.150 6.061 1.00 78.83 A C
+ATOM 3428 CD1 ILE A 499 11.530 -90.543 3.553 1.00 78.25 A C
+ATOM 3429 N ASN A 500 10.712 -85.914 6.782 1.00 83.29 A N
+ATOM 3430 CA ASN A 500 11.054 -85.218 8.022 1.00 87.07 A C
+ATOM 3431 C ASN A 500 9.803 -84.802 8.796 1.00 90.24 A C
+ATOM 3432 O ASN A 500 9.827 -84.794 10.030 1.00 96.78 A O
+ATOM 3433 CB ASN A 500 11.945 -83.997 7.732 1.00 94.17 A C
+ATOM 3434 CG ASN A 500 13.446 -84.295 7.889 1.00107.46 A C
+ATOM 3435 ND2 ASN A 500 14.232 -83.262 8.194 1.00119.87 A N
+ATOM 3436 OD1 ASN A 500 13.887 -85.434 7.720 1.00109.37 A O
+ATOM 3437 N GLN A 501 8.698 -84.476 8.104 1.00 86.90 A N
+ATOM 3438 CA GLN A 501 7.444 -84.196 8.817 1.00 92.35 A C
+ATOM 3439 C GLN A 501 6.838 -85.477 9.415 1.00 98.27 A C
+ATOM 3440 O GLN A 501 6.393 -85.482 10.581 1.00103.05 A O
+ATOM 3441 CB GLN A 501 6.430 -83.475 7.893 1.00 94.84 A C
+ATOM 3442 CG GLN A 501 5.263 -84.356 7.306 1.00106.95 A C
+ATOM 3443 CD GLN A 501 4.225 -83.590 6.461 1.00128.48 A C
+ATOM 3444 NE2 GLN A 501 3.271 -84.326 5.886 1.00136.92 A N
+ATOM 3445 OE1 GLN A 501 4.277 -82.365 6.338 1.00132.66 A O
+ATOM 3446 N SER A 502 6.828 -86.584 8.651 1.00 91.83 A N
+ATOM 3447 CA SER A 502 6.194 -87.788 9.180 1.00 91.85 A C
+ATOM 3448 C SER A 502 7.044 -88.488 10.245 1.00 97.09 A C
+ATOM 3449 O SER A 502 6.517 -89.328 10.987 1.00104.65 A O
+ATOM 3450 CB SER A 502 5.857 -88.751 8.041 1.00 93.50 A C
+ATOM 3451 OG SER A 502 4.924 -89.725 8.477 1.00 92.32 A O
+ATOM 3452 N LEU A 503 8.346 -88.181 10.319 1.00 90.03 A N
+ATOM 3453 CA LEU A 503 9.189 -88.576 11.448 1.00 94.62 A C
+ATOM 3454 C LEU A 503 9.025 -87.610 12.611 1.00106.09 A C
+ATOM 3455 O LEU A 503 9.054 -88.022 13.780 1.00106.30 A O
+ATOM 3456 CB LEU A 503 10.664 -88.631 11.034 1.00 82.19 A C
+ATOM 3457 CG LEU A 503 11.196 -89.852 10.285 1.00 94.86 A C
+ATOM 3458 CD1 LEU A 503 12.631 -89.613 9.904 1.00 95.86 A C
+ATOM 3459 CD2 LEU A 503 11.078 -91.107 11.115 1.00 81.33 A C
+ATOM 3460 N ALA A 504 8.877 -86.319 12.298 1.00 99.08 A N
+ATOM 3461 CA ALA A 504 8.663 -85.317 13.332 1.00100.32 A C
+ATOM 3462 C ALA A 504 7.476 -85.686 14.205 1.00106.78 A C
+ATOM 3463 O ALA A 504 7.610 -85.784 15.430 1.00 98.93 A O
+ATOM 3464 CB ALA A 504 8.463 -83.939 12.703 1.00100.52 A C
+ATOM 3465 N PHE A 505 6.304 -85.931 13.598 1.00101.97 A N
+ATOM 3466 CA PHE A 505 5.148 -86.193 14.458 1.00106.89 A C
+ATOM 3467 C PHE A 505 5.309 -87.479 15.271 1.00109.70 A C
+ATOM 3468 O PHE A 505 4.676 -87.616 16.329 1.00118.00 A O
+ATOM 3469 CB PHE A 505 3.839 -86.169 13.649 1.00110.85 A C
+ATOM 3470 CG PHE A 505 3.154 -84.792 13.637 1.00116.60 A C
+ATOM 3471 CD1 PHE A 505 3.255 -83.932 14.750 1.00122.94 A C
+ATOM 3472 CD2 PHE A 505 2.394 -84.362 12.547 1.00115.52 A C
+ATOM 3473 CE1 PHE A 505 2.636 -82.665 14.769 1.00124.94 A C
+ATOM 3474 CE2 PHE A 505 1.765 -83.089 12.568 1.00111.73 A C
+ATOM 3475 CZ PHE A 505 1.896 -82.246 13.683 1.00117.10 A C
+ATOM 3476 N ILE A 506 6.206 -88.379 14.856 1.00103.40 A N
+ATOM 3477 CA ILE A 506 6.601 -89.493 15.718 1.00102.28 A C
+ATOM 3478 C ILE A 506 7.436 -88.994 16.890 1.00101.76 A C
+ATOM 3479 O ILE A 506 7.240 -89.418 18.036 1.00116.17 A O
+ATOM 3480 CB ILE A 506 7.371 -90.554 14.914 1.00 95.41 A C
+ATOM 3481 CG1 ILE A 506 6.537 -91.008 13.718 1.00 99.11 A C
+ATOM 3482 CG2 ILE A 506 7.739 -91.725 15.816 1.00 86.15 A C
+ATOM 3483 CD1 ILE A 506 7.242 -91.970 12.823 1.00101.16 A C
+ATOM 3484 N ARG A 507 8.396 -88.106 16.620 1.00101.60 A N
+ATOM 3485 CA ARG A 507 9.229 -87.579 17.696 1.00110.06 A C
+ATOM 3486 C ARG A 507 8.367 -86.876 18.744 1.00127.25 A C
+ATOM 3487 O ARG A 507 8.447 -87.191 19.938 1.00119.03 A O
+ATOM 3488 CB ARG A 507 10.297 -86.636 17.122 1.00 99.96 A C
+ATOM 3489 CG ARG A 507 11.385 -86.202 18.110 1.00102.16 A C
+ATOM 3490 CD ARG A 507 12.214 -85.028 17.578 1.00112.67 A C
+ATOM 3491 NE ARG A 507 11.355 -83.959 17.053 1.00113.23 A N
+ATOM 3492 CZ ARG A 507 10.902 -82.919 17.761 1.00124.85 A C
+ATOM 3493 NH1 ARG A 507 11.226 -82.790 19.044 1.00134.76 A N1+
+ATOM 3494 NH2 ARG A 507 10.120 -82.002 17.187 1.00124.95 A N
+ATOM 3495 N LYS A 508 7.497 -85.954 18.307 1.00109.65 A N
+ATOM 3496 CA LYS A 508 6.631 -85.250 19.250 1.00113.83 A C
+ATOM 3497 C LYS A 508 5.720 -86.208 20.004 1.00113.00 A C
+ATOM 3498 O LYS A 508 5.590 -86.105 21.227 1.00116.37 A O
+ATOM 3499 CB LYS A 508 5.794 -84.177 18.551 1.00115.16 A C
+ATOM 3500 CG LYS A 508 6.538 -82.885 18.273 1.00117.99 A C
+ATOM 3501 CD LYS A 508 5.648 -81.860 17.552 1.00119.78 A C
+ATOM 3502 CE LYS A 508 6.479 -80.781 16.839 1.00116.25 A C
+ATOM 3503 NZ LYS A 508 5.649 -79.920 15.945 1.00111.60 A N1+
+ATOM 3504 N SER A 509 5.068 -87.144 19.307 1.00115.23 A N
+ATOM 3505 CA SER A 509 4.130 -87.997 20.037 1.00109.68 A C
+ATOM 3506 C SER A 509 4.848 -88.908 21.031 1.00113.97 A C
+ATOM 3507 O SER A 509 4.331 -89.158 22.127 1.00122.37 A O
+ATOM 3508 CB SER A 509 3.268 -88.807 19.070 1.00 95.58 A C
+ATOM 3509 OG SER A 509 4.052 -89.558 18.164 1.00 92.75 A O
+ATOM 3510 N ASP A 510 6.048 -89.388 20.689 1.00111.10 A N
+ATOM 3511 CA ASP A 510 6.792 -90.236 21.622 1.00111.10 A C
+ATOM 3512 C ASP A 510 7.334 -89.445 22.804 1.00111.10 A C
+ATOM 3513 O ASP A 510 7.368 -89.952 23.935 1.00111.10 A O
+ATOM 3514 CB ASP A 510 7.937 -90.948 20.912 1.00111.10 A C
+ATOM 3515 CG ASP A 510 7.481 -92.186 20.194 1.00111.10 A C
+ATOM 3516 OD1 ASP A 510 6.351 -92.635 20.487 1.00111.10 A O
+ATOM 3517 OD2 ASP A 510 8.250 -92.713 19.355 1.00111.10 A O1-
+ATOM 3518 N GLU A 511 7.776 -88.210 22.571 1.00111.31 A N
+ATOM 3519 CA GLU A 511 8.256 -87.409 23.687 1.00111.31 A C
+ATOM 3520 C GLU A 511 7.119 -86.936 24.591 1.00111.31 A C
+ATOM 3521 O GLU A 511 7.319 -86.822 25.804 1.00111.31 A O
+ATOM 3522 CB GLU A 511 9.075 -86.240 23.165 1.00111.31 A C
+ATOM 3523 CG GLU A 511 10.362 -86.691 22.499 1.00111.31 A C
+ATOM 3524 CD GLU A 511 11.141 -85.533 21.896 1.00111.31 A C
+ATOM 3525 OE1 GLU A 511 10.557 -84.424 21.782 1.00111.31 A O
+ATOM 3526 OE2 GLU A 511 12.337 -85.727 21.553 1.00111.31 A O1-
+ATOM 3527 N LEU A 512 5.926 -86.663 24.044 1.00 98.81 A N
+ATOM 3528 CA LEU A 512 4.781 -86.372 24.907 1.00 98.81 A C
+ATOM 3529 C LEU A 512 4.388 -87.595 25.718 1.00 98.81 A C
+ATOM 3530 O LEU A 512 4.138 -87.496 26.923 1.00 98.81 A O
+ATOM 3531 CB LEU A 512 3.575 -85.888 24.105 1.00 98.81 A C
+ATOM 3532 CG LEU A 512 3.513 -84.440 23.617 1.00 98.81 A C
+ATOM 3533 CD1 LEU A 512 2.056 -84.052 23.354 1.00 98.81 A C
+ATOM 3534 CD2 LEU A 512 4.171 -83.467 24.572 1.00 98.81 A C
+ATOM 3535 N LEU A 513 4.318 -88.759 25.075 1.00101.12 A N
+ATOM 3536 CA LEU A 513 3.996 -89.963 25.830 1.00101.12 A C
+ATOM 3537 C LEU A 513 5.024 -90.247 26.922 1.00101.12 A C
+ATOM 3538 O LEU A 513 4.659 -90.741 27.993 1.00101.12 A O
+ATOM 3539 CB LEU A 513 3.869 -91.160 24.892 1.00101.12 A C
+ATOM 3540 CG LEU A 513 3.334 -92.460 25.501 1.00101.12 A C
+ATOM 3541 CD1 LEU A 513 2.275 -93.113 24.608 1.00101.12 A C
+ATOM 3542 CD2 LEU A 513 4.493 -93.423 25.767 1.00101.12 A C
+ATOM 3543 N SER A 514 6.307 -89.952 26.688 1.00105.32 A N
+ATOM 3544 CA SER A 514 7.273 -90.159 27.764 1.00105.32 A C
+ATOM 3545 C SER A 514 7.267 -89.021 28.781 1.00105.32 A C
+ATOM 3546 O SER A 514 7.740 -89.218 29.909 1.00105.32 A O
+ATOM 3547 CB SER A 514 8.692 -90.347 27.209 1.00105.32 A C
+ATOM 3548 OG SER A 514 9.419 -89.126 27.197 1.00105.32 A O
+ATOM 3549 N ALA A 515 6.731 -87.846 28.419 1.00102.87 A N
+ATOM 3550 CA ALA A 515 6.725 -86.704 29.332 1.00102.87 A C
+ATOM 3551 C ALA A 515 5.656 -86.834 30.408 1.00102.87 A C
+ATOM 3552 O ALA A 515 5.835 -86.304 31.512 1.00102.87 A O
+ATOM 3553 CB ALA A 515 6.529 -85.391 28.568 1.00102.87 A C
+ATOM 3554 N ILE A 516 4.544 -87.525 30.117 1.00 99.24 A N
+ATOM 3555 CA ILE A 516 3.556 -87.815 31.159 1.00 99.24 A C
+ATOM 3556 C ILE A 516 3.976 -88.979 32.045 1.00 99.24 A C
+ATOM 3557 O ILE A 516 3.285 -89.277 33.029 1.00 99.24 A O
+ATOM 3558 CB ILE A 516 2.155 -88.079 30.568 1.00 99.24 A C
+ATOM 3559 CG1 ILE A 516 2.175 -89.269 29.600 1.00 99.24 A C
+ATOM 3560 CG2 ILE A 516 1.614 -86.821 29.905 1.00 99.24 A C
+ATOM 3561 CD1 ILE A 516 0.819 -89.960 29.438 1.00 99.24 A C
+ATOM 3562 N GLY A 517 5.084 -89.654 31.725 1.00 97.92 A N
+ATOM 3563 CA GLY A 517 5.647 -90.602 32.670 1.00 97.92 A C
+ATOM 3564 C GLY A 517 5.950 -89.946 34.002 1.00 97.92 A C
+ATOM 3565 O GLY A 517 5.539 -90.433 35.060 1.00 97.92 A O
+ATOM 3566 N GLY A 518 6.609 -88.785 33.960 1.00 89.76 A N
+ATOM 3567 CA GLY A 518 7.027 -88.063 35.142 1.00 89.76 A C
+ATOM 3568 C GLY A 518 5.925 -87.392 35.933 1.00 89.76 A C
+ATOM 3569 O GLY A 518 6.213 -86.584 36.820 1.00 89.76 A O
+ATOM 3570 N TYR A 519 4.666 -87.683 35.638 1.00 88.53 A N
+ATOM 3571 CA TYR A 519 3.540 -87.107 36.353 1.00 88.53 A C
+ATOM 3572 C TYR A 519 2.852 -88.211 37.143 1.00 88.53 A C
+ATOM 3573 O TYR A 519 2.939 -89.392 36.787 1.00 88.53 A O
+ATOM 3574 CB TYR A 519 2.593 -86.408 35.369 1.00 88.53 A C
+ATOM 3575 CG TYR A 519 3.225 -85.144 34.831 1.00 88.53 A C
+ATOM 3576 CD1 TYR A 519 4.330 -85.199 33.982 1.00 88.53 A C
+ATOM 3577 CD2 TYR A 519 2.741 -83.895 35.186 1.00 88.53 A C
+ATOM 3578 CE1 TYR A 519 4.941 -84.039 33.505 1.00 88.53 A C
+ATOM 3579 CE2 TYR A 519 3.338 -82.726 34.710 1.00 88.53 A C
+ATOM 3580 CZ TYR A 519 4.435 -82.803 33.870 1.00 88.53 A C
+ATOM 3581 OH TYR A 519 5.015 -81.641 33.404 1.00 88.53 A O
+ATOM 3582 N ILE A 520 2.235 -87.839 38.264 1.00 75.89 A N
+ATOM 3583 CA ILE A 520 1.763 -88.870 39.190 1.00 75.89 A C
+ATOM 3584 C ILE A 520 0.296 -89.234 38.971 1.00 75.89 A C
+ATOM 3585 O ILE A 520 -0.546 -88.346 38.733 1.00 75.89 A O
+ATOM 3586 CB ILE A 520 2.018 -88.474 40.655 1.00 75.89 A C
+ATOM 3587 CG1 ILE A 520 1.333 -87.170 41.016 1.00 75.89 A C
+ATOM 3588 CG2 ILE A 520 3.496 -88.342 40.947 1.00 75.89 A C
+ATOM 3589 CD1 ILE A 520 1.557 -86.857 42.468 1.00 75.89 A C
+ATOM 3590 N PRO A 521 -0.047 -90.546 39.074 1.00 71.26 A N
+ATOM 3591 CA PRO A 521 -1.436 -90.995 38.904 1.00 71.26 A C
+ATOM 3592 C PRO A 521 -2.347 -90.546 40.023 1.00 71.26 A C
+ATOM 3593 O PRO A 521 -1.934 -89.814 40.928 1.00 71.26 A O
+ATOM 3594 CB PRO A 521 -1.312 -92.520 38.935 1.00 71.26 A C
+ATOM 3595 CG PRO A 521 -0.204 -92.731 39.886 1.00 71.26 A C
+ATOM 3596 CD PRO A 521 0.809 -91.654 39.517 1.00 71.26 A C
+ATOM 3597 N GLU A 522 -3.589 -91.011 39.986 1.00 72.58 A N
+ATOM 3598 CA GLU A 522 -4.490 -90.795 41.105 1.00 72.58 A C
+ATOM 3599 C GLU A 522 -4.199 -91.833 42.178 1.00 72.58 A C
+ATOM 3600 O GLU A 522 -3.930 -93.006 41.887 1.00 72.58 A O
+ATOM 3601 CB GLU A 522 -5.954 -90.870 40.657 1.00 72.58 A C
+ATOM 3602 CG GLU A 522 -6.974 -90.769 41.776 1.00 72.58 A C
+ATOM 3603 CD GLU A 522 -6.896 -89.441 42.503 1.00 72.58 A C
+ATOM 3604 OE1 GLU A 522 -6.668 -88.426 41.809 1.00 72.58 A O
+ATOM 3605 OE2 GLU A 522 -7.047 -89.418 43.761 1.00 72.58 A O1-
+ATOM 3606 N ALA A 523 -4.203 -91.377 43.419 1.00 66.35 A N
+ATOM 3607 CA ALA A 523 -4.130 -92.276 44.541 1.00 66.35 A C
+ATOM 3608 C ALA A 523 -5.423 -93.081 44.652 1.00 66.35 A C
+ATOM 3609 O ALA A 523 -6.449 -92.709 44.079 1.00 66.35 A O
+ATOM 3610 CB ALA A 523 -3.879 -91.474 45.812 1.00 66.35 A C
+ATOM 3611 N PRO A 524 -5.405 -94.168 45.415 1.00 61.84 A N
+ATOM 3612 CA PRO A 524 -6.623 -94.960 45.613 1.00 61.84 A C
+ATOM 3613 C PRO A 524 -7.831 -94.113 46.004 1.00 61.84 A C
+ATOM 3614 O PRO A 524 -7.706 -92.990 46.498 1.00 61.84 A O
+ATOM 3615 CB PRO A 524 -6.229 -95.916 46.743 1.00 61.84 A C
+ATOM 3616 CG PRO A 524 -4.769 -96.093 46.576 1.00 61.84 A C
+ATOM 3617 CD PRO A 524 -4.208 -94.902 45.842 1.00 61.84 A C
+ATOM 3618 N ARG A 525 -9.016 -94.636 45.693 1.00 76.81 A N
+ATOM 3619 CA ARG A 525 -10.277 -93.966 45.977 1.00 76.81 A C
+ATOM 3620 C ARG A 525 -10.927 -94.442 47.271 1.00 76.81 A C
+ATOM 3621 O ARG A 525 -12.118 -94.189 47.492 1.00 76.81 A O
+ATOM 3622 CB ARG A 525 -11.221 -94.087 44.790 1.00 76.81 A C
+ATOM 3623 CG ARG A 525 -10.794 -93.135 43.693 1.00 76.81 A C
+ATOM 3624 CD ARG A 525 -11.777 -93.049 42.542 1.00 76.81 A C
+ATOM 3625 NE ARG A 525 -11.179 -92.341 41.409 1.00 76.81 A N
+ATOM 3626 CZ ARG A 525 -11.397 -91.054 41.142 1.00 76.81 A C
+ATOM 3627 NH1 ARG A 525 -12.211 -90.343 41.926 1.00 76.81 A N1+
+ATOM 3628 NH2 ARG A 525 -10.808 -90.478 40.094 1.00 76.81 A N
+ATOM 3629 N ASP A 526 -10.180 -95.134 48.118 1.00 69.18 A N
+ATOM 3630 CA ASP A 526 -10.723 -95.991 49.161 1.00 69.18 A C
+ATOM 3631 C ASP A 526 -11.189 -95.250 50.401 1.00 69.18 A C
+ATOM 3632 O ASP A 526 -11.396 -95.900 51.431 1.00 69.18 A O
+ATOM 3633 CB ASP A 526 -9.668 -96.980 49.599 1.00 69.18 A C
+ATOM 3634 CG ASP A 526 -8.447 -96.287 50.069 1.00 69.18 A C
+ATOM 3635 OD1 ASP A 526 -8.451 -95.030 50.073 1.00 69.18 A O
+ATOM 3636 OD2 ASP A 526 -7.490 -96.987 50.435 1.00 69.18 A O1-
+ATOM 3637 N GLY A 527 -11.230 -93.926 50.399 1.00 68.06 A N
+ATOM 3638 CA GLY A 527 -11.642 -93.257 51.620 1.00 68.06 A C
+ATOM 3639 C GLY A 527 -10.543 -93.095 52.644 1.00 68.06 A C
+ATOM 3640 O GLY A 527 -10.831 -92.818 53.821 1.00 68.06 A O
+ATOM 3641 N GLN A 528 -9.297 -93.274 52.228 1.00 61.82 A N
+ATOM 3642 CA GLN A 528 -8.136 -92.943 53.026 1.00 61.82 A C
+ATOM 3643 C GLN A 528 -7.364 -91.814 52.359 1.00 61.82 A C
+ATOM 3644 O GLN A 528 -7.479 -91.584 51.151 1.00 61.82 A O
+ATOM 3645 CB GLN A 528 -7.245 -94.169 53.225 1.00 61.82 A C
+ATOM 3646 CG GLN A 528 -7.941 -95.253 54.032 1.00 61.82 A C
+ATOM 3647 CD GLN A 528 -7.148 -96.541 54.114 1.00 61.82 A C
+ATOM 3648 NE2 GLN A 528 -7.246 -97.351 53.072 1.00 61.82 A N
+ATOM 3649 OE1 GLN A 528 -6.474 -96.820 55.112 1.00 61.82 A O
+ATOM 3650 N ALA A 529 -6.615 -91.082 53.174 1.00 54.32 A N
+ATOM 3651 CA ALA A 529 -5.753 -90.019 52.689 1.00 54.32 A C
+ATOM 3652 C ALA A 529 -4.381 -90.590 52.389 1.00 54.32 A C
+ATOM 3653 O ALA A 529 -3.857 -91.414 53.147 1.00 54.32 A O
+ATOM 3654 CB ALA A 529 -5.639 -88.896 53.723 1.00 54.32 A C
+ATOM 3655 N TYR A 530 -3.793 -90.137 51.295 1.00 52.96 A N
+ATOM 3656 CA TYR A 530 -2.524 -90.673 50.838 1.00 52.96 A C
+ATOM 3657 C TYR A 530 -1.485 -89.576 50.668 1.00 52.96 A C
+ATOM 3658 O TYR A 530 -1.791 -88.474 50.222 1.00 52.96 A O
+ATOM 3659 CB TYR A 530 -2.715 -91.408 49.528 1.00 52.96 A C
+ATOM 3660 CG TYR A 530 -3.306 -92.768 49.708 1.00 52.96 A C
+ATOM 3661 CD1 TYR A 530 -2.502 -93.838 50.041 1.00 52.96 A C
+ATOM 3662 CD2 TYR A 530 -4.678 -92.983 49.581 1.00 52.96 A C
+ATOM 3663 CE1 TYR A 530 -3.039 -95.099 50.221 1.00 52.96 A C
+ATOM 3664 CE2 TYR A 530 -5.224 -94.244 49.755 1.00 52.96 A C
+ATOM 3665 CZ TYR A 530 -4.394 -95.300 50.077 1.00 52.96 A C
+ATOM 3666 OH TYR A 530 -4.885 -96.568 50.262 1.00 52.96 A O
+ATOM 3667 N VAL A 531 -0.248 -89.904 50.989 1.00 49.90 A N
+ATOM 3668 CA VAL A 531 0.872 -88.998 50.862 1.00 49.90 A C
+ATOM 3669 C VAL A 531 1.851 -89.628 49.871 1.00 49.90 A C
+ATOM 3670 O VAL A 531 1.837 -90.843 49.653 1.00 49.90 A O
+ATOM 3671 CB VAL A 531 1.501 -88.802 52.237 1.00 49.90 A C
+ATOM 3672 CG1 VAL A 531 2.396 -87.621 52.265 1.00 49.90 A C
+ATOM 3673 CG2 VAL A 531 0.422 -88.690 53.240 1.00 49.90 A C
+ATOM 3674 N ARG A 532 2.712 -88.806 49.262 1.00 57.79 A N
+ATOM 3675 CA ARG A 532 3.686 -89.298 48.290 1.00 57.79 A C
+ATOM 3676 C ARG A 532 5.005 -89.607 48.990 1.00 57.79 A C
+ATOM 3677 O ARG A 532 5.550 -88.734 49.676 1.00 57.79 A O
+ATOM 3678 CB ARG A 532 3.899 -88.261 47.197 1.00 57.79 A C
+ATOM 3679 CG ARG A 532 4.450 -88.836 45.922 1.00 57.79 A C
+ATOM 3680 CD ARG A 532 3.454 -89.841 45.360 1.00 57.79 A C
+ATOM 3681 NE ARG A 532 3.836 -90.343 44.045 1.00 57.79 A N
+ATOM 3682 CZ ARG A 532 3.227 -91.351 43.433 1.00 57.79 A C
+ATOM 3683 NH1 ARG A 532 2.218 -91.979 44.020 1.00 57.79 A N1+
+ATOM 3684 NH2 ARG A 532 3.636 -91.742 42.238 1.00 57.79 A N
+ATOM 3685 N LYS A 533 5.501 -90.848 48.848 1.00 56.42 A N
+ATOM 3686 CA LYS A 533 6.831 -91.241 49.345 1.00 56.42 A C
+ATOM 3687 C LYS A 533 7.515 -92.154 48.337 1.00 56.42 A C
+ATOM 3688 O LYS A 533 6.944 -93.192 47.970 1.00 56.42 A O
+ATOM 3689 CB LYS A 533 6.758 -91.964 50.687 1.00 56.42 A C
+ATOM 3690 CG LYS A 533 8.106 -92.349 51.267 1.00 56.42 A C
+ATOM 3691 CD LYS A 533 7.890 -93.204 52.498 1.00 56.42 A C
+ATOM 3692 CE LYS A 533 9.153 -93.422 53.298 1.00 56.42 A C
+ATOM 3693 NZ LYS A 533 8.834 -94.083 54.608 1.00 56.42 A N1+
+ATOM 3694 N ASP A 534 8.714 -91.749 47.872 1.00 66.00 A N
+ATOM 3695 CA ASP A 534 9.548 -92.439 46.867 1.00 66.00 A C
+ATOM 3696 C ASP A 534 8.715 -93.230 45.847 1.00 66.00 A C
+ATOM 3697 O ASP A 534 8.670 -94.471 45.868 1.00 66.00 A O
+ATOM 3698 CB ASP A 534 10.702 -93.318 47.438 1.00 66.00 A C
+ATOM 3699 CG ASP A 534 10.380 -94.064 48.714 1.00 66.00 A C
+ATOM 3700 OD1 ASP A 534 9.246 -94.553 48.842 1.00 66.00 A O
+ATOM 3701 OD2 ASP A 534 11.308 -94.227 49.558 1.00 66.00 A O1-
+ATOM 3702 N GLY A 535 8.011 -92.500 44.981 1.00 68.89 A N
+ATOM 3703 CA GLY A 535 7.323 -93.088 43.861 1.00 68.89 A C
+ATOM 3704 C GLY A 535 5.925 -93.588 44.134 1.00 68.89 A C
+ATOM 3705 O GLY A 535 5.183 -93.838 43.176 1.00 68.89 A O
+ATOM 3706 N GLU A 536 5.521 -93.746 45.391 1.00 72.34 A N
+ATOM 3707 CA GLU A 536 4.215 -94.337 45.625 1.00 72.34 A C
+ATOM 3708 C GLU A 536 3.343 -93.476 46.524 1.00 72.34 A C
+ATOM 3709 O GLU A 536 3.771 -92.458 47.083 1.00 72.34 A O
+ATOM 3710 CB GLU A 536 4.365 -95.722 46.229 1.00 72.34 A C
+ATOM 3711 CG GLU A 536 4.703 -96.731 45.180 1.00 72.34 A C
+ATOM 3712 CD GLU A 536 4.519 -98.147 45.684 1.00 72.34 A C
+ATOM 3713 OE1 GLU A 536 4.457 -98.331 46.944 1.00 72.34 A O
+ATOM 3714 OE2 GLU A 536 4.426 -99.061 44.812 1.00 72.34 A O1-
+ATOM 3715 N TRP A 537 2.102 -93.922 46.666 1.00 57.12 A N
+ATOM 3716 CA TRP A 537 1.234 -93.409 47.705 1.00 57.12 A C
+ATOM 3717 C TRP A 537 1.329 -94.297 48.935 1.00 57.12 A C
+ATOM 3718 O TRP A 537 1.378 -95.525 48.826 1.00 57.12 A O
+ATOM 3719 CB TRP A 537 -0.209 -93.331 47.233 1.00 57.12 A C
+ATOM 3720 CG TRP A 537 -0.422 -92.412 46.098 1.00 57.12 A C
+ATOM 3721 CD1 TRP A 537 -0.637 -92.753 44.804 1.00 57.12 A C
+ATOM 3722 CD2 TRP A 537 -0.462 -90.978 46.146 1.00 57.12 A C
+ATOM 3723 CE2 TRP A 537 -0.712 -90.521 44.840 1.00 57.12 A C
+ATOM 3724 CE3 TRP A 537 -0.314 -90.038 47.169 1.00 57.12 A C
+ATOM 3725 NE1 TRP A 537 -0.817 -91.621 44.033 1.00 57.12 A N
+ATOM 3726 CZ2 TRP A 537 -0.811 -89.174 44.533 1.00 57.12 A C
+ATOM 3727 CZ3 TRP A 537 -0.416 -88.706 46.860 1.00 57.12 A C
+ATOM 3728 CH2 TRP A 537 -0.664 -88.283 45.558 1.00 57.12 A C
+ATOM 3729 N VAL A 538 1.391 -93.666 50.099 1.00 50.16 A N
+ATOM 3730 CA VAL A 538 1.376 -94.336 51.389 1.00 50.16 A C
+ATOM 3731 C VAL A 538 0.203 -93.757 52.158 1.00 50.16 A C
+ATOM 3732 O VAL A 538 -0.250 -92.640 51.886 1.00 50.16 A O
+ATOM 3733 CB VAL A 538 2.672 -94.100 52.158 1.00 50.16 A C
+ATOM 3734 CG1 VAL A 538 3.824 -94.508 51.316 1.00 50.16 A C
+ATOM 3735 CG2 VAL A 538 2.781 -92.644 52.503 1.00 50.16 A C
+ATOM 3736 N LEU A 539 -0.301 -94.499 53.131 1.00 47.24 A N
+ATOM 3737 CA LEU A 539 -1.433 -93.947 53.859 1.00 47.24 A C
+ATOM 3738 C LEU A 539 -0.919 -92.920 54.857 1.00 47.24 A C
+ATOM 3739 O LEU A 539 0.158 -93.085 55.440 1.00 47.24 A O
+ATOM 3740 CB LEU A 539 -2.243 -95.043 54.546 1.00 47.24 A C
+ATOM 3741 CG LEU A 539 -2.432 -96.182 53.544 1.00 47.24 A C
+ATOM 3742 CD1 LEU A 539 -1.463 -97.366 53.843 1.00 47.24 A C
+ATOM 3743 CD2 LEU A 539 -3.878 -96.610 53.358 1.00 47.24 A C
+ATOM 3744 N LEU A 540 -1.652 -91.817 55.000 1.00 41.44 A N
+ATOM 3745 CA LEU A 540 -1.275 -90.875 56.041 1.00 41.44 A C
+ATOM 3746 C LEU A 540 -1.255 -91.590 57.383 1.00 41.44 A C
+ATOM 3747 O LEU A 540 -0.368 -91.364 58.223 1.00 41.44 A O
+ATOM 3748 CB LEU A 540 -2.249 -89.707 56.060 1.00 41.44 A C
+ATOM 3749 CG LEU A 540 -1.904 -88.563 56.996 1.00 41.44 A C
+ATOM 3750 CD1 LEU A 540 -0.476 -88.147 56.807 1.00 41.44 A C
+ATOM 3751 CD2 LEU A 540 -2.854 -87.441 56.717 1.00 41.44 A C
+ATOM 3752 N SER A 541 -2.200 -92.517 57.557 1.00 47.65 A N
+ATOM 3753 CA SER A 541 -2.338 -93.289 58.784 1.00 47.65 A C
+ATOM 3754 C SER A 541 -1.011 -93.860 59.222 1.00 47.65 A C
+ATOM 3755 O SER A 541 -0.714 -93.902 60.415 1.00 47.65 A O
+ATOM 3756 CB SER A 541 -3.341 -94.404 58.531 1.00 47.65 A C
+ATOM 3757 OG SER A 541 -4.339 -93.909 57.637 1.00 47.65 A O
+ATOM 3758 N THR A 542 -0.186 -94.264 58.260 1.00 48.64 A N
+ATOM 3759 CA THR A 542 1.133 -94.799 58.558 1.00 48.64 A C
+ATOM 3760 C THR A 542 1.879 -93.931 59.542 1.00 48.64 A C
+ATOM 3761 O THR A 542 2.591 -94.439 60.422 1.00 48.64 A O
+ATOM 3762 CB THR A 542 1.965 -94.874 57.293 1.00 48.64 A C
+ATOM 3763 CG2 THR A 542 3.164 -95.798 57.519 1.00 48.64 A C
+ATOM 3764 OG1 THR A 542 1.149 -95.355 56.222 1.00 48.64 A O
+ATOM 3765 N PHE A 543 1.761 -92.612 59.384 1.00 48.10 A N
+ATOM 3766 CA PHE A 543 2.598 -91.679 60.116 1.00 48.10 A C
+ATOM 3767 C PHE A 543 1.938 -91.120 61.363 1.00 48.10 A C
+ATOM 3768 O PHE A 543 2.613 -90.429 62.130 1.00 48.10 A O
+ATOM 3769 CB PHE A 543 3.043 -90.550 59.191 1.00 48.10 A C
+ATOM 3770 CG PHE A 543 3.724 -91.042 57.952 1.00 48.10 A C
+ATOM 3771 CD1 PHE A 543 2.994 -91.361 56.825 1.00 48.10 A C
+ATOM 3772 CD2 PHE A 543 5.088 -91.232 57.931 1.00 48.10 A C
+ATOM 3773 CE1 PHE A 543 3.616 -91.824 55.697 1.00 48.10 A C
+ATOM 3774 CE2 PHE A 543 5.721 -91.698 56.796 1.00 48.10 A C
+ATOM 3775 CZ PHE A 543 4.984 -91.995 55.679 1.00 48.10 A C
+ATOM 3776 N LEU A 544 0.670 -91.442 61.625 1.00 38.38 A N
+ATOM 3777 CA LEU A 544 0.005 -90.911 62.822 1.00 38.38 A C
+ATOM 3778 C LEU A 544 -0.484 -92.018 63.785 1.00 38.38 A C
+ATOM 3779 O LEU A 544 0.151 -92.327 64.815 1.00 38.38 A O
+ATOM 3780 CB LEU A 544 -1.171 -90.025 62.404 1.00 38.38 A C
+ATOM 3781 CG LEU A 544 -1.032 -89.143 61.163 1.00 38.38 A C
+ATOM 3782 CD1 LEU A 544 -2.280 -88.341 60.997 1.00 38.38 A C
+ATOM 3783 CD2 LEU A 544 0.130 -88.239 61.294 1.00 38.38 A C
+TER
+ATOM 3784 N VAL H 2 24.712 -62.728 -82.954 1.00 62.22 B N
+ATOM 3785 CA VAL H 2 24.045 -61.988 -81.866 1.00 58.35 B C
+ATOM 3786 C VAL H 2 24.512 -60.545 -81.702 1.00 59.47 B C
+ATOM 3787 O VAL H 2 25.583 -60.299 -81.141 1.00 60.10 B O
+ATOM 3788 CB VAL H 2 24.236 -62.666 -80.519 1.00 61.67 B C
+ATOM 3789 CG1 VAL H 2 23.507 -61.859 -79.436 1.00 61.67 B C
+ATOM 3790 CG2 VAL H 2 23.769 -64.098 -80.588 1.00 61.67 B C
+ATOM 3791 N GLN H 3 23.700 -59.578 -82.121 1.00 67.34 B N
+ATOM 3792 CA GLN H 3 24.203 -58.223 -82.247 1.00 69.24 B C
+ATOM 3793 C GLN H 3 23.197 -57.207 -81.729 1.00 65.06 B C
+ATOM 3794 O GLN H 3 21.980 -57.397 -81.812 1.00 64.92 B O
+ATOM 3795 CB GLN H 3 24.544 -57.927 -83.699 1.00 73.55 B C
+ATOM 3796 CG GLN H 3 25.765 -58.696 -84.144 1.00 73.55 B C
+ATOM 3797 CD GLN H 3 26.842 -57.817 -84.761 1.00 73.55 B C
+ATOM 3798 NE2 GLN H 3 28.122 -58.200 -84.573 1.00 73.55 B N
+ATOM 3799 OE1 GLN H 3 26.532 -56.807 -85.407 1.00 73.55 B O
+ATOM 3800 N LEU H 4 23.744 -56.123 -81.185 1.00 53.68 B N
+ATOM 3801 CA LEU H 4 23.022 -54.910 -80.822 1.00 55.13 B C
+ATOM 3802 C LEU H 4 23.747 -53.744 -81.481 1.00 56.69 B C
+ATOM 3803 O LEU H 4 24.961 -53.574 -81.302 1.00 57.19 B O
+ATOM 3804 CB LEU H 4 22.990 -54.725 -79.309 1.00 63.45 B C
+ATOM 3805 CG LEU H 4 21.674 -55.001 -78.614 1.00 63.45 B C
+ATOM 3806 CD1 LEU H 4 21.213 -56.345 -79.051 1.00 63.45 B C
+ATOM 3807 CD2 LEU H 4 21.887 -54.992 -77.126 1.00 63.45 B C
+ATOM 3808 N VAL H 5 23.030 -52.961 -82.271 1.00 57.51 B N
+ATOM 3809 CA VAL H 5 23.685 -51.927 -83.053 1.00 57.42 B C
+ATOM 3810 C VAL H 5 23.027 -50.599 -82.738 1.00 59.81 B C
+ATOM 3811 O VAL H 5 21.853 -50.378 -83.068 1.00 61.62 B O
+ATOM 3812 CB VAL H 5 23.653 -52.227 -84.552 1.00 67.33 B C
+ATOM 3813 CG1 VAL H 5 24.566 -53.394 -84.824 1.00 67.33 B C
+ATOM 3814 CG2 VAL H 5 22.259 -52.560 -85.017 1.00 67.33 B C
+ATOM 3815 N GLU H 6 23.779 -49.723 -82.084 1.00 57.30 B N
+ATOM 3816 CA GLU H 6 23.312 -48.384 -81.788 1.00 60.63 B C
+ATOM 3817 C GLU H 6 23.420 -47.509 -83.024 1.00 60.38 B C
+ATOM 3818 O GLU H 6 24.299 -47.698 -83.868 1.00 56.38 B O
+ATOM 3819 CB GLU H 6 24.120 -47.783 -80.647 1.00 66.68 B C
+ATOM 3820 CG GLU H 6 24.026 -48.580 -79.375 1.00 66.68 B C
+ATOM 3821 CD GLU H 6 25.124 -49.593 -79.260 1.00 66.68 B C
+ATOM 3822 OE1 GLU H 6 25.723 -49.919 -80.312 1.00 66.68 B O
+ATOM 3823 OE2 GLU H 6 25.422 -50.007 -78.112 1.00 66.68 B O1-
+ATOM 3824 N SER H 7 22.515 -46.545 -83.120 1.00 62.25 B N
+ATOM 3825 CA SER H 7 22.409 -45.671 -84.273 1.00 58.61 B C
+ATOM 3826 C SER H 7 21.862 -44.327 -83.815 1.00 58.51 B C
+ATOM 3827 O SER H 7 21.043 -44.268 -82.893 1.00 57.45 B O
+ATOM 3828 CB SER H 7 21.496 -46.295 -85.326 1.00 67.75 B C
+ATOM 3829 OG SER H 7 21.106 -45.315 -86.259 1.00 67.75 B O
+ATOM 3830 N GLY H 8 22.309 -43.246 -84.447 1.00 63.57 B N
+ATOM 3831 CA GLY H 8 21.642 -41.968 -84.259 1.00 62.26 B C
+ATOM 3832 C GLY H 8 22.295 -40.981 -83.312 1.00 67.14 B C
+ATOM 3833 O GLY H 8 21.774 -39.868 -83.140 1.00 71.88 B O
+ATOM 3834 N GLY H 9 23.397 -41.351 -82.679 1.00 69.25 B N
+ATOM 3835 CA GLY H 9 24.230 -40.353 -82.044 1.00 69.79 B C
+ATOM 3836 C GLY H 9 24.788 -39.369 -83.053 1.00 78.64 B C
+ATOM 3837 O GLY H 9 24.822 -39.607 -84.264 1.00 86.22 B O
+ATOM 3838 N GLY H 11 25.246 -38.244 -82.533 1.00 68.38 B N
+ATOM 3839 CA GLY H 11 25.710 -37.189 -83.397 1.00 64.09 B C
+ATOM 3840 C GLY H 11 26.208 -36.030 -82.573 1.00 64.71 B C
+ATOM 3841 O GLY H 11 26.596 -36.193 -81.415 1.00 65.19 B O
+ATOM 3842 N LEU H 12 26.210 -34.860 -83.205 1.00 53.71 B N
+ATOM 3843 CA LEU H 12 26.625 -33.599 -82.606 1.00 52.35 B C
+ATOM 3844 C LEU H 12 25.398 -32.712 -82.457 1.00 53.60 B C
+ATOM 3845 O LEU H 12 24.776 -32.315 -83.451 1.00 57.55 B O
+ATOM 3846 CB LEU H 12 27.677 -32.931 -83.475 1.00 62.49 B C
+ATOM 3847 CG LEU H 12 28.716 -32.124 -82.759 1.00 62.49 B C
+ATOM 3848 CD1 LEU H 12 29.595 -31.585 -83.837 1.00 62.49 B C
+ATOM 3849 CD2 LEU H 12 28.049 -31.019 -81.996 1.00 62.49 B C
+ATOM 3850 N VAL H 13 25.053 -32.403 -81.227 1.00 55.40 B N
+ATOM 3851 CA VAL H 13 23.791 -31.753 -80.929 1.00 57.90 B C
+ATOM 3852 C VAL H 13 24.072 -30.566 -80.030 1.00 62.89 B C
+ATOM 3853 O VAL H 13 24.869 -30.661 -79.089 1.00 67.15 B O
+ATOM 3854 CB VAL H 13 22.794 -32.729 -80.268 1.00 64.70 B C
+ATOM 3855 CG1 VAL H 13 21.407 -32.099 -80.172 1.00 64.70 B C
+ATOM 3856 CG2 VAL H 13 22.730 -34.031 -81.054 1.00 64.70 B C
+ATOM 3857 N GLN H 14 23.421 -29.445 -80.325 1.00 71.77 B N
+ATOM 3858 CA GLN H 14 23.582 -28.263 -79.499 1.00 72.52 B C
+ATOM 3859 C GLN H 14 22.869 -28.437 -78.159 1.00 71.06 B C
+ATOM 3860 O GLN H 14 21.866 -29.152 -78.064 1.00 68.94 B O
+ATOM 3861 CB GLN H 14 23.042 -27.029 -80.214 1.00 74.20 B C
+ATOM 3862 CG GLN H 14 23.899 -26.573 -81.388 1.00 74.20 B C
+ATOM 3863 CD GLN H 14 24.629 -25.260 -81.119 1.00 74.20 B C
+ATOM 3864 NE2 GLN H 14 25.692 -25.006 -81.890 1.00 74.20 B N
+ATOM 3865 OE1 GLN H 14 24.237 -24.479 -80.235 1.00 74.20 B O
+ATOM 3866 N PRO H 15 23.339 -27.754 -77.130 1.00 67.74 B N
+ATOM 3867 CA PRO H 15 22.711 -27.851 -75.803 1.00 66.40 B C
+ATOM 3868 C PRO H 15 21.220 -27.556 -75.816 1.00 62.57 B C
+ATOM 3869 O PRO H 15 20.706 -27.028 -76.799 1.00 63.65 B O
+ATOM 3870 CB PRO H 15 23.465 -26.795 -74.989 1.00 68.28 B C
+ATOM 3871 CG PRO H 15 24.805 -26.721 -75.649 1.00 68.28 B C
+ATOM 3872 CD PRO H 15 24.628 -27.047 -77.099 1.00 68.28 B C
+ATOM 3873 N ARG H 16 20.494 -27.977 -74.785 1.00 86.46 B N
+ATOM 3874 CA ARG H 16 19.096 -27.620 -74.570 1.00 86.52 B C
+ATOM 3875 C ARG H 16 18.160 -28.188 -75.656 1.00 89.49 B C
+ATOM 3876 O ARG H 16 16.945 -28.036 -75.525 1.00 93.16 B O
+ATOM 3877 CB ARG H 16 18.934 -26.093 -74.441 1.00 92.39 B C
+ATOM 3878 CG ARG H 16 17.743 -25.578 -73.623 1.00 92.39 B C
+ATOM 3879 CD ARG H 16 17.515 -24.030 -73.843 1.00 92.39 B C
+ATOM 3880 NE ARG H 16 17.547 -23.625 -75.272 1.00 92.39 B N
+ATOM 3881 CZ ARG H 16 16.944 -22.552 -75.806 1.00 92.39 B C
+ATOM 3882 NH1 ARG H 16 16.205 -21.744 -75.050 1.00 92.39 B N1+
+ATOM 3883 NH2 ARG H 16 17.062 -22.305 -77.113 1.00 92.39 B N
+ATOM 3884 N ARG H 17 18.676 -28.875 -76.698 1.00 87.08 B N
+ATOM 3885 CA ARG H 17 17.880 -29.413 -77.804 1.00 86.03 B C
+ATOM 3886 C ARG H 17 17.823 -30.953 -77.759 1.00 85.49 B C
+ATOM 3887 O ARG H 17 18.249 -31.581 -76.790 1.00 81.59 B O
+ATOM 3888 CB ARG H 17 18.408 -28.891 -79.147 1.00 88.79 B C
+ATOM 3889 CG ARG H 17 17.313 -28.825 -80.239 1.00 88.79 B C
+ATOM 3890 CD ARG H 17 17.926 -28.674 -81.622 1.00 88.79 B C
+ATOM 3891 NE ARG H 17 17.293 -29.514 -82.641 1.00 88.79 B N
+ATOM 3892 CZ ARG H 17 17.785 -29.687 -83.868 1.00 88.79 B C
+ATOM 3893 NH1 ARG H 17 18.891 -29.058 -84.235 1.00 88.79 B N1+
+ATOM 3894 NH2 ARG H 17 17.166 -30.476 -84.732 1.00 88.79 B N
+ATOM 3895 N SER H 18 17.237 -31.567 -78.792 1.00 78.28 B N
+ATOM 3896 CA SER H 18 16.760 -32.944 -78.724 1.00 78.79 B C
+ATOM 3897 C SER H 18 17.432 -33.860 -79.741 1.00 77.87 B C
+ATOM 3898 O SER H 18 17.766 -33.437 -80.852 1.00 78.69 B O
+ATOM 3899 CB SER H 18 15.253 -32.988 -78.961 1.00 84.10 B C
+ATOM 3900 OG SER H 18 14.805 -34.321 -79.159 1.00 84.10 B O
+ATOM 3901 N LEU H 19 17.558 -35.141 -79.368 1.00 64.34 B N
+ATOM 3902 CA LEU H 19 18.151 -36.184 -80.211 1.00 68.35 B C
+ATOM 3903 C LEU H 19 17.522 -37.548 -79.904 1.00 68.76 B C
+ATOM 3904 O LEU H 19 17.132 -37.812 -78.764 1.00 76.13 B O
+ATOM 3905 CB LEU H 19 19.671 -36.244 -80.006 1.00 74.62 B C
+ATOM 3906 CG LEU H 19 20.359 -37.291 -80.861 1.00 74.62 B C
+ATOM 3907 CD1 LEU H 19 20.082 -36.987 -82.307 1.00 74.62 B C
+ATOM 3908 CD2 LEU H 19 21.833 -37.299 -80.580 1.00 74.62 B C
+ATOM 3909 N ARG H 20 17.425 -38.425 -80.917 1.00 69.11 B N
+ATOM 3910 CA ARG H 20 16.907 -39.785 -80.730 1.00 70.46 B C
+ATOM 3911 C ARG H 20 17.957 -40.821 -81.093 1.00 68.38 B C
+ATOM 3912 O ARG H 20 18.570 -40.740 -82.166 1.00 68.64 B O
+ATOM 3913 CB ARG H 20 15.673 -40.083 -81.576 1.00 76.94 B C
+ATOM 3914 CG ARG H 20 15.669 -41.541 -82.074 1.00 76.94 B C
+ATOM 3915 CD ARG H 20 14.405 -41.960 -82.789 1.00 76.94 B C
+ATOM 3916 NE ARG H 20 13.324 -42.203 -81.843 1.00 76.94 B N
+ATOM 3917 CZ ARG H 20 12.148 -42.735 -82.169 1.00 76.94 B C
+ATOM 3918 NH1 ARG H 20 11.905 -43.110 -83.437 1.00 76.94 B N1+
+ATOM 3919 NH2 ARG H 20 11.226 -42.914 -81.214 1.00 76.94 B N
+ATOM 3920 N LEU H 21 18.121 -41.810 -80.209 1.00 60.07 B N
+ATOM 3921 CA LEU H 21 19.002 -42.957 -80.390 1.00 59.75 B C
+ATOM 3922 C LEU H 21 18.158 -44.202 -80.598 1.00 64.51 B C
+ATOM 3923 O LEU H 21 17.150 -44.397 -79.911 1.00 63.61 B O
+ATOM 3924 CB LEU H 21 19.900 -43.157 -79.164 1.00 69.03 B C
+ATOM 3925 CG LEU H 21 21.166 -42.339 -78.897 1.00 69.03 B C
+ATOM 3926 CD1 LEU H 21 20.973 -40.860 -78.983 1.00 69.03 B C
+ATOM 3927 CD2 LEU H 21 21.549 -42.647 -77.484 1.00 69.03 B C
+ATOM 3928 N SER H 22 18.576 -45.054 -81.529 1.00 62.43 B N
+ATOM 3929 CA SER H 22 17.894 -46.315 -81.777 1.00 58.77 B C
+ATOM 3930 C SER H 22 18.885 -47.477 -81.698 1.00 60.00 B C
+ATOM 3931 O SER H 22 20.092 -47.316 -81.891 1.00 57.65 B O
+ATOM 3932 CB SER H 22 17.197 -46.296 -83.135 1.00 67.92 B C
+ATOM 3933 OG SER H 22 18.165 -46.323 -84.166 1.00 67.92 B O
+ATOM 3934 N CYS H 23 18.359 -48.664 -81.402 1.00 66.01 B N
+ATOM 3935 CA CYS H 23 19.154 -49.879 -81.294 1.00 66.64 B C
+ATOM 3936 C CYS H 23 18.460 -50.977 -82.078 1.00 65.91 B C
+ATOM 3937 O CYS H 23 17.303 -51.288 -81.807 1.00 62.18 B O
+ATOM 3938 CB CYS H 23 19.317 -50.286 -79.832 1.00 73.36 B C
+ATOM 3939 SG CYS H 23 20.273 -51.816 -79.543 1.00 73.36 B S
+ATOM 3940 N ALA H 24 19.142 -51.550 -83.053 1.00 63.59 B N
+ATOM 3941 CA ALA H 24 18.568 -52.660 -83.795 1.00 65.16 B C
+ATOM 3942 C ALA H 24 19.175 -53.949 -83.286 1.00 63.56 B C
+ATOM 3943 O ALA H 24 20.377 -54.007 -83.003 1.00 65.11 B O
+ATOM 3944 CB ALA H 24 18.813 -52.546 -85.296 1.00 63.07 B C
+ATOM 3945 N ALA H 25 18.338 -54.983 -83.182 1.00 65.59 B N
+ATOM 3946 CA ALA H 25 18.696 -56.245 -82.554 1.00 64.49 B C
+ATOM 3947 C ALA H 25 18.815 -57.341 -83.594 1.00 67.14 B C
+ATOM 3948 O ALA H 25 18.097 -57.354 -84.596 1.00 69.41 B O
+ATOM 3949 CB ALA H 25 17.659 -56.650 -81.518 1.00 64.17 B C
+ATOM 3950 N SER H 26 19.702 -58.279 -83.330 1.00 60.73 B N
+ATOM 3951 CA SER H 26 20.034 -59.274 -84.325 1.00 61.26 B C
+ATOM 3952 C SER H 26 20.400 -60.580 -83.650 1.00 60.93 B C
+ATOM 3953 O SER H 26 21.006 -60.569 -82.577 1.00 58.54 B O
+ATOM 3954 CB SER H 26 21.198 -58.783 -85.170 1.00 65.71 B C
+ATOM 3955 OG SER H 26 21.913 -59.889 -85.673 1.00 65.71 B O
+ATOM 3956 N GLY H 27 20.026 -61.699 -84.273 1.00 69.02 B N
+ATOM 3957 CA GLY H 27 20.596 -62.999 -83.926 1.00 64.25 B C
+ATOM 3958 C GLY H 27 20.195 -63.609 -82.593 1.00 66.78 B C
+ATOM 3959 O GLY H 27 20.830 -64.579 -82.149 1.00 70.51 B O
+ATOM 3960 N PHE H 28 19.186 -63.065 -81.923 1.00 63.94 B N
+ATOM 3961 CA PHE H 28 18.634 -63.679 -80.729 1.00 63.94 B C
+ATOM 3962 C PHE H 28 17.188 -63.221 -80.653 1.00 63.94 B C
+ATOM 3963 O PHE H 28 16.845 -62.152 -81.166 1.00 63.94 B O
+ATOM 3964 CB PHE H 28 19.401 -63.285 -79.457 1.00 63.94 B C
+ATOM 3965 CG PHE H 28 19.179 -61.856 -79.046 1.00 63.94 B C
+ATOM 3966 CD1 PHE H 28 18.108 -61.510 -78.223 1.00 63.94 B C
+ATOM 3967 CD2 PHE H 28 20.013 -60.860 -79.493 1.00 63.94 B C
+ATOM 3968 CE1 PHE H 28 17.875 -60.202 -77.850 1.00 63.94 B C
+ATOM 3969 CE2 PHE H 28 19.779 -59.547 -79.121 1.00 63.94 B C
+ATOM 3970 CZ PHE H 28 18.704 -59.221 -78.293 1.00 63.94 B C
+ATOM 3971 N THR H 29 16.340 -64.006 -79.991 1.00 71.07 B N
+ATOM 3972 CA THR H 29 14.916 -63.693 -80.051 1.00 71.07 B C
+ATOM 3973 C THR H 29 14.680 -62.502 -79.129 1.00 71.07 B C
+ATOM 3974 O THR H 29 14.806 -62.605 -77.904 1.00 71.07 B O
+ATOM 3975 CB THR H 29 14.062 -64.914 -79.691 1.00 71.07 B C
+ATOM 3976 CG2 THR H 29 14.325 -66.072 -80.688 1.00 71.07 B C
+ATOM 3977 OG1 THR H 29 14.359 -65.361 -78.363 1.00 71.07 B O
+ATOM 3978 N PHE H 30 14.295 -61.376 -79.728 1.00 69.79 B N
+ATOM 3979 CA PHE H 30 14.451 -60.097 -79.053 1.00 69.79 B C
+ATOM 3980 C PHE H 30 13.492 -59.960 -77.883 1.00 69.79 B C
+ATOM 3981 O PHE H 30 13.894 -59.504 -76.810 1.00 69.79 B O
+ATOM 3982 CB PHE H 30 14.230 -58.961 -80.039 1.00 69.79 B C
+ATOM 3983 CG PHE H 30 14.374 -57.589 -79.441 1.00 69.79 B C
+ATOM 3984 CD1 PHE H 30 15.607 -57.117 -79.055 1.00 69.79 B C
+ATOM 3985 CD2 PHE H 30 13.276 -56.767 -79.283 1.00 69.79 B C
+ATOM 3986 CE1 PHE H 30 15.742 -55.844 -78.542 1.00 69.79 B C
+ATOM 3987 CE2 PHE H 30 13.404 -55.499 -78.766 1.00 69.79 B C
+ATOM 3988 CZ PHE H 30 14.630 -55.037 -78.395 1.00 69.79 B C
+ATOM 3989 N ASP H 35 12.226 -60.355 -78.070 1.00 78.95 B N
+ATOM 3990 CA ASP H 35 11.199 -60.065 -77.072 1.00 78.95 B C
+ATOM 3991 C ASP H 35 11.585 -60.609 -75.710 1.00 78.95 B C
+ATOM 3992 O ASP H 35 11.384 -59.944 -74.688 1.00 78.95 B O
+ATOM 3993 CB ASP H 35 9.854 -60.648 -77.493 1.00 78.95 B C
+ATOM 3994 CG ASP H 35 9.498 -60.317 -78.928 1.00 78.95 B C
+ATOM 3995 OD1 ASP H 35 10.201 -59.478 -79.543 1.00 78.95 B O
+ATOM 3996 OD2 ASP H 35 8.502 -60.886 -79.441 1.00 78.95 B O1-
+ATOM 3997 N ASP H 36 12.197 -61.786 -75.677 1.00 75.30 B N
+ATOM 3998 CA ASP H 36 12.418 -62.463 -74.410 1.00 75.30 B C
+ATOM 3999 C ASP H 36 13.306 -61.696 -73.430 1.00 75.30 B C
+ATOM 4000 O ASP H 36 13.212 -61.940 -72.220 1.00 75.30 B O
+ATOM 4001 CB ASP H 36 12.976 -63.842 -74.706 1.00 75.30 B C
+ATOM 4002 CG ASP H 36 11.977 -64.689 -75.437 1.00 75.30 B C
+ATOM 4003 OD1 ASP H 36 10.779 -64.296 -75.400 1.00 75.30 B O
+ATOM 4004 OD2 ASP H 36 12.374 -65.718 -76.037 1.00 75.30 B O1-
+ATOM 4005 N TYR H 37 14.140 -60.766 -73.891 1.00 67.24 B N
+ATOM 4006 CA TYR H 37 15.091 -60.104 -73.006 1.00 66.21 B C
+ATOM 4007 C TYR H 37 14.668 -58.686 -72.690 1.00 66.21 B C
+ATOM 4008 O TYR H 37 14.009 -58.010 -73.484 1.00 66.65 B O
+ATOM 4009 CB TYR H 37 16.498 -60.079 -73.596 1.00 57.33 B C
+ATOM 4010 CG TYR H 37 16.979 -61.455 -73.806 1.00 57.33 B C
+ATOM 4011 CD1 TYR H 37 17.513 -62.179 -72.767 1.00 57.33 B C
+ATOM 4012 CD2 TYR H 37 16.787 -62.077 -75.006 1.00 57.33 B C
+ATOM 4013 CE1 TYR H 37 17.917 -63.485 -72.949 1.00 57.33 B C
+ATOM 4014 CE2 TYR H 37 17.170 -63.378 -75.197 1.00 57.33 B C
+ATOM 4015 CZ TYR H 37 17.739 -64.080 -74.174 1.00 57.33 B C
+ATOM 4016 OH TYR H 37 18.123 -65.380 -74.400 1.00 57.33 B O
+ATOM 4017 N THR H 38 15.060 -58.252 -71.511 1.00 59.89 B N
+ATOM 4018 CA THR H 38 14.884 -56.859 -71.169 1.00 59.11 B C
+ATOM 4019 C THR H 38 16.177 -56.119 -71.503 1.00 59.11 B C
+ATOM 4020 O THR H 38 17.272 -56.691 -71.456 1.00 59.11 B O
+ATOM 4021 CB THR H 38 14.487 -56.729 -69.697 1.00 54.91 B C
+ATOM 4022 CG2 THR H 38 13.687 -57.995 -69.303 1.00 54.91 B C
+ATOM 4023 OG1 THR H 38 15.653 -56.623 -68.871 1.00 54.91 B O
+ATOM 4024 N ILE H 39 16.041 -54.866 -71.933 1.00 48.59 B N
+ATOM 4025 CA ILE H 39 17.170 -54.147 -72.511 1.00 48.59 B C
+ATOM 4026 C ILE H 39 17.375 -52.828 -71.789 1.00 48.59 B C
+ATOM 4027 O ILE H 39 16.404 -52.167 -71.396 1.00 48.59 B O
+ATOM 4028 CB ILE H 39 16.983 -53.894 -74.011 1.00 51.77 B C
+ATOM 4029 CG1 ILE H 39 16.385 -55.123 -74.689 1.00 51.77 B C
+ATOM 4030 CG2 ILE H 39 18.300 -53.565 -74.609 1.00 51.77 B C
+ATOM 4031 CD1 ILE H 39 17.352 -56.250 -74.893 1.00 51.77 B C
+ATOM 4032 N HIS H 40 18.650 -52.446 -71.624 1.00 47.82 B N
+ATOM 4033 CA HIS H 40 19.064 -51.229 -70.938 1.00 47.82 B C
+ATOM 4034 C HIS H 40 19.874 -50.344 -71.872 1.00 47.82 B C
+ATOM 4035 O HIS H 40 20.586 -50.833 -72.755 1.00 47.82 B O
+ATOM 4036 CB HIS H 40 19.934 -51.501 -69.693 1.00 54.40 B C
+ATOM 4037 CG HIS H 40 19.433 -52.598 -68.806 1.00 54.40 B C
+ATOM 4038 CD2 HIS H 40 20.043 -53.715 -68.340 1.00 54.40 B C
+ATOM 4039 ND1 HIS H 40 18.168 -52.603 -68.265 1.00 54.40 B N
+ATOM 4040 CE1 HIS H 40 18.012 -53.687 -67.523 1.00 54.40 B C
+ATOM 4041 NE2 HIS H 40 19.135 -54.379 -67.552 1.00 54.40 B N
+ATOM 4042 N TRP H 41 19.749 -49.036 -71.668 1.00 54.06 B N
+ATOM 4043 CA TRP H 41 20.686 -48.057 -72.186 1.00 48.76 B C
+ATOM 4044 C TRP H 41 21.646 -47.720 -71.075 1.00 48.76 B C
+ATOM 4045 O TRP H 41 21.207 -47.310 -69.980 1.00 48.76 B O
+ATOM 4046 CB TRP H 41 19.991 -46.788 -72.641 1.00 57.76 B C
+ATOM 4047 CG TRP H 41 19.383 -46.943 -73.937 1.00 57.76 B C
+ATOM 4048 CD1 TRP H 41 18.076 -47.204 -74.203 1.00 57.76 B C
+ATOM 4049 CD2 TRP H 41 20.047 -46.869 -75.188 1.00 57.76 B C
+ATOM 4050 CE2 TRP H 41 19.082 -47.086 -76.185 1.00 57.76 B C
+ATOM 4051 CE3 TRP H 41 21.363 -46.621 -75.568 1.00 57.76 B C
+ATOM 4052 NE1 TRP H 41 17.882 -47.290 -75.556 1.00 57.76 B N
+ATOM 4053 CZ2 TRP H 41 19.393 -47.072 -77.548 1.00 57.76 B C
+ATOM 4054 CZ3 TRP H 41 21.672 -46.608 -76.931 1.00 57.76 B C
+ATOM 4055 CH2 TRP H 41 20.689 -46.837 -77.900 1.00 57.76 B C
+ATOM 4056 N VAL H 42 22.941 -47.905 -71.355 1.00 43.76 B N
+ATOM 4057 CA VAL H 42 24.021 -47.578 -70.427 1.00 43.76 B C
+ATOM 4058 C VAL H 42 25.001 -46.658 -71.137 1.00 46.28 B C
+ATOM 4059 O VAL H 42 25.464 -46.965 -72.240 1.00 45.10 B O
+ATOM 4060 CB VAL H 42 24.749 -48.829 -69.908 1.00 54.19 B C
+ATOM 4061 CG1 VAL H 42 25.972 -48.422 -69.157 1.00 54.19 B C
+ATOM 4062 CG2 VAL H 42 23.870 -49.570 -68.983 1.00 54.19 B C
+ATOM 4063 N ARG H 43 25.335 -45.543 -70.501 1.00 47.65 B N
+ATOM 4064 CA ARG H 43 26.223 -44.574 -71.114 1.00 47.65 B C
+ATOM 4065 C ARG H 43 27.542 -44.556 -70.373 1.00 47.65 B C
+ATOM 4066 O ARG H 43 27.634 -44.969 -69.211 1.00 47.65 B O
+ATOM 4067 CB ARG H 43 25.618 -43.168 -71.129 1.00 58.22 B C
+ATOM 4068 CG ARG H 43 25.498 -42.521 -69.773 1.00 58.22 B C
+ATOM 4069 CD ARG H 43 25.830 -41.062 -69.909 1.00 58.22 B C
+ATOM 4070 NE ARG H 43 25.347 -40.231 -68.816 1.00 58.22 B N
+ATOM 4071 CZ ARG H 43 24.141 -39.676 -68.813 1.00 58.22 B C
+ATOM 4072 NH1 ARG H 43 23.350 -39.866 -69.859 1.00 58.22 B N1+
+ATOM 4073 NH2 ARG H 43 23.736 -38.919 -67.794 1.00 58.22 B N
+ATOM 4074 N GLN H 44 28.575 -44.130 -71.095 1.00 50.16 B N
+ATOM 4075 CA GLN H 44 29.919 -43.975 -70.539 1.00 50.16 B C
+ATOM 4076 C GLN H 44 30.472 -42.643 -71.011 1.00 50.16 B C
+ATOM 4077 O GLN H 44 30.731 -42.483 -72.210 1.00 51.20 B O
+ATOM 4078 CB GLN H 44 30.829 -45.117 -70.990 1.00 60.51 B C
+ATOM 4079 CG GLN H 44 32.216 -45.134 -70.396 1.00 60.51 B C
+ATOM 4080 CD GLN H 44 32.875 -46.491 -70.576 1.00 60.51 B C
+ATOM 4081 NE2 GLN H 44 33.592 -46.930 -69.550 1.00 60.51 B N
+ATOM 4082 OE1 GLN H 44 32.736 -47.144 -71.627 1.00 60.51 B O
+ATOM 4083 N ALA H 45 30.687 -41.704 -70.080 1.00 56.14 B N
+ATOM 4084 CA ALA H 45 31.370 -40.470 -70.431 1.00 61.17 B C
+ATOM 4085 C ALA H 45 32.786 -40.807 -70.909 1.00 63.10 B C
+ATOM 4086 O ALA H 45 33.278 -41.918 -70.668 1.00 66.58 B O
+ATOM 4087 CB ALA H 45 31.399 -39.523 -69.235 1.00 58.01 B C
+ATOM 4088 N PRO H 46 33.445 -39.898 -71.643 1.00 59.76 B N
+ATOM 4089 CA PRO H 46 34.795 -40.211 -72.136 1.00 60.94 B C
+ATOM 4090 C PRO H 46 35.807 -40.208 -70.991 1.00 59.31 B C
+ATOM 4091 O PRO H 46 35.782 -39.334 -70.114 1.00 62.54 B O
+ATOM 4092 CB PRO H 46 35.063 -39.098 -73.146 1.00 69.43 B C
+ATOM 4093 CG PRO H 46 34.301 -37.922 -72.571 1.00 69.43 B C
+ATOM 4094 CD PRO H 46 33.055 -38.509 -71.968 1.00 69.43 B C
+ATOM 4095 N GLY H 47 36.673 -41.228 -70.989 1.00 64.00 B N
+ATOM 4096 CA GLY H 47 37.606 -41.457 -69.901 1.00 62.74 B C
+ATOM 4097 C GLY H 47 36.970 -41.385 -68.522 1.00 65.98 B C
+ATOM 4098 O GLY H 47 37.484 -40.699 -67.630 1.00 66.57 B O
+ATOM 4099 N LYS H 48 35.846 -42.078 -68.343 1.00 55.15 B N
+ATOM 4100 CA LYS H 48 35.039 -42.038 -67.119 1.00 58.10 B C
+ATOM 4101 C LYS H 48 34.378 -43.407 -66.963 1.00 57.43 B C
+ATOM 4102 O LYS H 48 34.730 -44.364 -67.660 1.00 56.07 B O
+ATOM 4103 CB LYS H 48 34.019 -40.888 -67.176 1.00 66.13 B C
+ATOM 4104 CG LYS H 48 34.553 -39.479 -66.905 1.00 66.13 B C
+ATOM 4105 CD LYS H 48 34.540 -39.178 -65.411 1.00 66.13 B C
+ATOM 4106 CE LYS H 48 35.140 -37.811 -65.078 1.00 66.13 B C
+ATOM 4107 NZ LYS H 48 35.192 -37.555 -63.594 1.00 66.13 B N1+
+ATOM 4108 N GLY H 49 33.405 -43.502 -66.055 1.00 57.89 B N
+ATOM 4109 CA GLY H 49 32.783 -44.769 -65.719 1.00 56.80 B C
+ATOM 4110 C GLY H 49 31.520 -45.090 -66.511 1.00 58.45 B C
+ATOM 4111 O GLY H 49 31.200 -44.473 -67.531 1.00 57.95 B O
+ATOM 4112 N LEU H 50 30.789 -46.087 -66.010 1.00 43.33 B N
+ATOM 4113 CA LEU H 50 29.563 -46.559 -66.636 1.00 43.33 B C
+ATOM 4114 C LEU H 50 28.371 -46.050 -65.843 1.00 43.33 B C
+ATOM 4115 O LEU H 50 28.312 -46.220 -64.622 1.00 43.33 B O
+ATOM 4116 CB LEU H 50 29.536 -48.084 -66.680 1.00 54.06 B C
+ATOM 4117 CG LEU H 50 30.600 -48.796 -67.505 1.00 54.06 B C
+ATOM 4118 CD1 LEU H 50 30.850 -50.175 -66.902 1.00 54.06 B C
+ATOM 4119 CD2 LEU H 50 30.172 -48.901 -68.946 1.00 54.06 B C
+ATOM 4120 N GLU H 51 27.423 -45.439 -66.529 1.00 45.00 B N
+ATOM 4121 CA GLU H 51 26.207 -44.965 -65.889 1.00 45.90 B C
+ATOM 4122 C GLU H 51 25.013 -45.634 -66.547 1.00 45.00 B C
+ATOM 4123 O GLU H 51 24.862 -45.589 -67.781 1.00 45.00 B O
+ATOM 4124 CB GLU H 51 26.079 -43.451 -65.986 1.00 55.61 B C
+ATOM 4125 CG GLU H 51 24.843 -42.896 -65.324 1.00 55.61 B C
+ATOM 4126 CD GLU H 51 24.853 -41.382 -65.298 1.00 55.61 B C
+ATOM 4127 OE1 GLU H 51 25.387 -40.784 -66.256 1.00 55.61 B O
+ATOM 4128 OE2 GLU H 51 24.359 -40.790 -64.313 1.00 55.61 B O1-
+ATOM 4129 N TRP H 52 24.184 -46.276 -65.728 1.00 47.28 B N
+ATOM 4130 CA TRP H 52 22.947 -46.847 -66.235 1.00 47.28 B C
+ATOM 4131 C TRP H 52 21.987 -45.729 -66.577 1.00 47.66 B C
+ATOM 4132 O TRP H 52 21.699 -44.867 -65.740 1.00 48.28 B O
+ATOM 4133 CB TRP H 52 22.336 -47.766 -65.196 1.00 57.57 B C
+ATOM 4134 CG TRP H 52 21.065 -48.438 -65.601 1.00 57.57 B C
+ATOM 4135 CD1 TRP H 52 20.922 -49.505 -66.434 1.00 57.57 B C
+ATOM 4136 CD2 TRP H 52 19.752 -48.109 -65.141 1.00 57.57 B C
+ATOM 4137 CE2 TRP H 52 18.860 -49.011 -65.748 1.00 57.57 B C
+ATOM 4138 CE3 TRP H 52 19.245 -47.133 -64.279 1.00 57.57 B C
+ATOM 4139 NE1 TRP H 52 19.600 -49.858 -66.532 1.00 57.57 B N
+ATOM 4140 CZ2 TRP H 52 17.487 -48.962 -65.521 1.00 57.57 B C
+ATOM 4141 CZ3 TRP H 52 17.896 -47.090 -64.049 1.00 57.57 B C
+ATOM 4142 CH2 TRP H 52 17.028 -47.996 -64.667 1.00 57.57 B C
+ATOM 4143 N VAL H 53 21.507 -45.726 -67.810 1.00 43.55 B N
+ATOM 4144 CA VAL H 53 20.660 -44.625 -68.234 1.00 43.69 B C
+ATOM 4145 C VAL H 53 19.215 -44.982 -67.985 1.00 49.67 B C
+ATOM 4146 O VAL H 53 18.511 -44.258 -67.277 1.00 52.72 B O
+ATOM 4147 CB VAL H 53 20.869 -44.249 -69.696 1.00 53.17 B C
+ATOM 4148 CG1 VAL H 53 20.006 -43.079 -69.983 1.00 53.17 B C
+ATOM 4149 CG2 VAL H 53 22.315 -43.904 -69.918 1.00 53.17 B C
+ATOM 4150 N SER H 54 18.749 -46.060 -68.604 1.00 56.97 B N
+ATOM 4151 CA SER H 54 17.390 -46.531 -68.318 1.00 53.84 B C
+ATOM 4152 C SER H 54 17.234 -47.950 -68.853 1.00 54.60 B C
+ATOM 4153 O SER H 54 18.193 -48.555 -69.347 1.00 57.62 B O
+ATOM 4154 CB SER H 54 16.353 -45.610 -68.923 1.00 58.75 B C
+ATOM 4155 OG SER H 54 16.860 -45.128 -70.124 1.00 58.75 B O
+ATOM 4156 N GLY H 55 16.016 -48.488 -68.735 1.00 56.69 B N
+ATOM 4157 CA GLY H 55 15.758 -49.807 -69.286 1.00 56.26 B C
+ATOM 4158 C GLY H 55 14.300 -50.207 -69.307 1.00 56.99 B C
+ATOM 4159 O GLY H 55 13.520 -49.746 -68.461 1.00 58.69 B O
+ATOM 4160 N ILE H 56 13.932 -51.096 -70.237 1.00 66.00 B N
+ATOM 4161 CA ILE H 56 12.568 -51.596 -70.379 1.00 65.76 B C
+ATOM 4162 C ILE H 56 12.599 -53.116 -70.330 1.00 67.25 B C
+ATOM 4163 O ILE H 56 13.651 -53.744 -70.491 1.00 69.80 B O
+ATOM 4164 CB ILE H 56 11.903 -51.134 -71.690 1.00 57.59 B C
+ATOM 4165 CG1 ILE H 56 12.912 -51.207 -72.809 1.00 57.59 B C
+ATOM 4166 CG2 ILE H 56 11.489 -49.710 -71.665 1.00 57.59 B C
+ATOM 4167 CD1 ILE H 56 13.055 -52.576 -73.380 1.00 57.59 B C
+ATOM 4168 N THR H 57 11.454 -53.760 -70.107 1.00 74.89 B N
+ATOM 4169 CA THR H 57 11.533 -55.219 -69.992 1.00 73.95 B C
+ATOM 4170 C THR H 57 10.576 -56.259 -70.612 1.00 73.95 B C
+ATOM 4171 O THR H 57 9.780 -56.872 -69.899 1.00 75.03 B O
+ATOM 4172 CB THR H 57 11.723 -55.613 -68.506 1.00 55.92 B C
+ATOM 4173 CG2 THR H 57 10.494 -55.238 -67.693 1.00 55.92 B C
+ATOM 4174 OG1 THR H 57 11.944 -57.025 -68.408 1.00 55.92 B O
+ATOM 4175 N TRP H 58 10.683 -56.495 -71.918 1.00 71.27 B N
+ATOM 4176 CA TRP H 58 9.931 -57.590 -72.545 1.00 71.27 B C
+ATOM 4177 C TRP H 58 8.533 -57.210 -73.048 1.00 71.27 B C
+ATOM 4178 O TRP H 58 8.035 -57.794 -74.006 1.00 71.27 B O
+ATOM 4179 CB TRP H 58 9.807 -58.766 -71.562 1.00 71.27 B C
+ATOM 4180 CG TRP H 58 8.518 -59.481 -71.736 1.00 71.27 B C
+ATOM 4181 CD1 TRP H 58 7.444 -59.397 -70.931 1.00 71.27 B C
+ATOM 4182 CD2 TRP H 58 8.165 -60.390 -72.781 1.00 71.27 B C
+ATOM 4183 CE2 TRP H 58 6.836 -60.786 -72.555 1.00 71.27 B C
+ATOM 4184 CE3 TRP H 58 8.842 -60.905 -73.899 1.00 71.27 B C
+ATOM 4185 NE1 TRP H 58 6.419 -60.165 -71.414 1.00 71.27 B N
+ATOM 4186 CZ2 TRP H 58 6.168 -61.685 -73.390 1.00 71.27 B C
+ATOM 4187 CZ3 TRP H 58 8.180 -61.802 -74.738 1.00 71.27 B C
+ATOM 4188 CH2 TRP H 58 6.854 -62.178 -74.476 1.00 71.27 B C
+ATOM 4189 N ASN H 59 7.971 -56.180 -72.446 1.00 70.45 B N
+ATOM 4190 CA ASN H 59 6.698 -55.679 -72.871 1.00 70.45 B C
+ATOM 4191 C ASN H 59 6.572 -54.252 -72.368 1.00 70.45 B C
+ATOM 4192 O ASN H 59 5.517 -53.870 -71.861 1.00 70.45 B O
+ATOM 4193 CB ASN H 59 5.544 -56.513 -72.310 1.00 70.45 B C
+ATOM 4194 CG ASN H 59 5.470 -56.462 -70.797 1.00 70.45 B C
+ATOM 4195 ND2 ASN H 59 4.357 -56.929 -70.243 1.00 70.45 B N
+ATOM 4196 OD1 ASN H 59 6.402 -56.008 -70.133 1.00 70.45 B O
+ATOM 4197 N SER H 62 7.627 -53.438 -72.477 1.00 69.75 B N
+ATOM 4198 CA SER H 62 7.378 -52.119 -71.942 1.00 69.75 B C
+ATOM 4199 C SER H 62 6.407 -52.246 -70.776 1.00 69.75 B C
+ATOM 4200 O SER H 62 5.547 -51.405 -70.572 1.00 69.75 B O
+ATOM 4201 CB SER H 62 6.846 -51.166 -73.016 1.00 69.75 B C
+ATOM 4202 OG SER H 62 7.907 -50.544 -73.753 1.00 69.75 B O
+ATOM 4203 N GLY H 63 6.565 -53.305 -69.994 1.00 70.01 B N
+ATOM 4204 CA GLY H 63 5.781 -53.477 -68.791 1.00 70.01 B C
+ATOM 4205 C GLY H 63 6.273 -52.641 -67.627 1.00 70.01 B C
+ATOM 4206 O GLY H 63 5.503 -51.941 -66.960 1.00 70.01 B O
+ATOM 4207 N TYR H 64 7.566 -52.709 -67.367 1.00 75.94 B N
+ATOM 4208 CA TYR H 64 8.195 -51.868 -66.372 1.00 75.94 B C
+ATOM 4209 C TYR H 64 9.327 -51.148 -67.082 1.00 75.94 B C
+ATOM 4210 O TYR H 64 10.082 -51.776 -67.825 1.00 77.82 B O
+ATOM 4211 CB TYR H 64 8.702 -52.709 -65.205 1.00 59.63 B C
+ATOM 4212 CG TYR H 64 9.190 -51.902 -64.020 1.00 59.63 B C
+ATOM 4213 CD1 TYR H 64 8.526 -51.941 -62.783 1.00 59.63 B C
+ATOM 4214 CD2 TYR H 64 10.325 -51.078 -64.130 1.00 59.63 B C
+ATOM 4215 CE1 TYR H 64 8.997 -51.167 -61.671 1.00 59.63 B C
+ATOM 4216 CE2 TYR H 64 10.800 -50.300 -63.038 1.00 59.63 B C
+ATOM 4217 CZ TYR H 64 10.139 -50.346 -61.815 1.00 59.63 B C
+ATOM 4218 OH TYR H 64 10.639 -49.576 -60.772 1.00 59.63 B O
+ATOM 4219 N ILE H 65 9.429 -49.834 -66.910 1.00 63.81 B N
+ATOM 4220 CA ILE H 65 10.583 -49.133 -67.452 1.00 65.70 B C
+ATOM 4221 C ILE H 65 11.073 -48.224 -66.346 1.00 64.48 B C
+ATOM 4222 O ILE H 65 10.289 -47.765 -65.512 1.00 64.71 B O
+ATOM 4223 CB ILE H 65 10.289 -48.285 -68.705 1.00 58.43 B C
+ATOM 4224 CG1 ILE H 65 9.654 -46.956 -68.333 1.00 58.43 B C
+ATOM 4225 CG2 ILE H 65 9.203 -48.956 -69.552 1.00 58.43 B C
+ATOM 4226 CD1 ILE H 65 9.907 -45.870 -69.385 1.00 58.43 B C
+ATOM 4227 N GLY H 66 12.359 -47.930 -66.357 1.00 61.69 B N
+ATOM 4228 CA GLY H 66 12.931 -47.135 -65.285 1.00 61.69 B C
+ATOM 4229 C GLY H 66 14.125 -46.338 -65.746 1.00 61.69 B C
+ATOM 4230 O GLY H 66 14.815 -46.705 -66.701 1.00 61.69 B O
+ATOM 4231 N TYR H 67 14.384 -45.250 -65.040 1.00 71.67 B N
+ATOM 4232 CA TYR H 67 15.326 -44.241 -65.475 1.00 70.38 B C
+ATOM 4233 C TYR H 67 16.328 -43.985 -64.364 1.00 71.35 B C
+ATOM 4234 O TYR H 67 16.089 -44.318 -63.199 1.00 72.26 B O
+ATOM 4235 CB TYR H 67 14.599 -42.944 -65.819 1.00 62.53 B C
+ATOM 4236 CG TYR H 67 13.712 -43.031 -67.036 1.00 62.53 B C
+ATOM 4237 CD1 TYR H 67 12.822 -42.013 -67.358 1.00 62.53 B C
+ATOM 4238 CD2 TYR H 67 13.783 -44.110 -67.879 1.00 62.53 B C
+ATOM 4239 CE1 TYR H 67 12.016 -42.094 -68.489 1.00 62.53 B C
+ATOM 4240 CE2 TYR H 67 12.998 -44.203 -69.006 1.00 62.53 B C
+ATOM 4241 CZ TYR H 67 12.115 -43.197 -69.310 1.00 62.53 B C
+ATOM 4242 OH TYR H 67 11.339 -43.329 -70.437 1.00 62.53 B O
+ATOM 4243 N ALA H 68 17.456 -43.389 -64.728 1.00 67.00 B N
+ATOM 4244 CA ALA H 68 18.338 -42.836 -63.717 1.00 66.55 B C
+ATOM 4245 C ALA H 68 17.763 -41.519 -63.222 1.00 69.80 B C
+ATOM 4246 O ALA H 68 16.988 -40.855 -63.914 1.00 72.28 B O
+ATOM 4247 CB ALA H 68 19.751 -42.633 -64.265 1.00 66.55 B C
+ATOM 4248 N ASP H 69 18.118 -41.155 -61.994 1.00 90.44 B N
+ATOM 4249 CA ASP H 69 17.687 -39.862 -61.482 1.00 97.36 B C
+ATOM 4250 C ASP H 69 18.196 -38.735 -62.365 1.00 96.24 B C
+ATOM 4251 O ASP H 69 17.520 -37.716 -62.520 1.00 92.96 B O
+ATOM 4252 CB ASP H 69 18.167 -39.693 -60.039 1.00 72.01 B C
+ATOM 4253 CG ASP H 69 17.470 -40.670 -59.068 1.00 72.01 B C
+ATOM 4254 OD1 ASP H 69 16.261 -40.953 -59.306 1.00 72.01 B O
+ATOM 4255 OD2 ASP H 69 18.120 -41.158 -58.088 1.00 72.01 B O1-
+ATOM 4256 N SER H 70 19.358 -38.935 -62.989 1.00 81.83 B N
+ATOM 4257 CA SER H 70 19.988 -37.914 -63.819 1.00 82.18 B C
+ATOM 4258 C SER H 70 19.099 -37.502 -64.973 1.00 84.14 B C
+ATOM 4259 O SER H 70 18.877 -36.313 -65.207 1.00 84.02 B O
+ATOM 4260 CB SER H 70 21.301 -38.448 -64.368 1.00 64.37 B C
+ATOM 4261 OG SER H 70 21.926 -39.280 -63.408 1.00 64.37 B O
+ATOM 4262 N VAL H 71 18.631 -38.464 -65.743 1.00 87.73 B N
+ATOM 4263 CA VAL H 71 17.744 -38.195 -66.863 1.00 86.23 B C
+ATOM 4264 C VAL H 71 16.353 -38.617 -66.430 1.00 91.44 B C
+ATOM 4265 O VAL H 71 16.060 -39.811 -66.341 1.00 95.78 B O
+ATOM 4266 CB VAL H 71 18.204 -38.928 -68.139 1.00 65.73 B C
+ATOM 4267 CG1 VAL H 71 19.547 -38.389 -68.575 1.00 65.73 B C
+ATOM 4268 CG2 VAL H 71 18.379 -40.428 -67.928 1.00 65.73 B C
+ATOM 4269 N LYS H 72 15.474 -37.679 -66.129 1.00 98.13 B N
+ATOM 4270 CA LYS H 72 14.139 -38.107 -65.741 1.00106.19 B C
+ATOM 4271 C LYS H 72 13.167 -37.137 -66.371 1.00110.40 B C
+ATOM 4272 O LYS H 72 13.324 -35.923 -66.226 1.00115.16 B O
+ATOM 4273 CB LYS H 72 13.985 -38.185 -64.219 1.00 72.38 B C
+ATOM 4274 CG LYS H 72 12.581 -38.559 -63.752 1.00 72.38 B C
+ATOM 4275 CD LYS H 72 12.507 -39.968 -63.125 1.00 72.38 B C
+ATOM 4276 CE LYS H 72 11.138 -40.201 -62.392 1.00 72.38 B C
+ATOM 4277 NZ LYS H 72 10.932 -41.540 -61.690 1.00 72.38 B N1+
+ATOM 4278 N GLY H 74 12.242 -37.663 -67.165 1.00 99.57 B N
+ATOM 4279 CA GLY H 74 11.403 -36.812 -67.974 1.00 95.47 B C
+ATOM 4280 C GLY H 74 12.152 -36.077 -69.062 1.00 98.76 B C
+ATOM 4281 O GLY H 74 11.521 -35.471 -69.935 1.00100.38 B O
+ATOM 4282 N ARG H 75 13.479 -36.099 -69.030 1.00106.92 B N
+ATOM 4283 CA ARG H 75 14.273 -35.662 -70.161 1.00100.32 B C
+ATOM 4284 C ARG H 75 14.374 -36.761 -71.203 1.00 94.99 B C
+ATOM 4285 O ARG H 75 14.326 -36.491 -72.408 1.00 99.23 B O
+ATOM 4286 CB ARG H 75 15.656 -35.277 -69.676 1.00 68.51 B C
+ATOM 4287 CG ARG H 75 15.795 -33.877 -69.245 1.00 68.51 B C
+ATOM 4288 CD ARG H 75 17.232 -33.488 -69.426 1.00 68.51 B C
+ATOM 4289 NE ARG H 75 18.068 -34.115 -68.412 1.00 68.51 B N
+ATOM 4290 CZ ARG H 75 19.371 -34.322 -68.556 1.00 68.51 B C
+ATOM 4291 NH1 ARG H 75 19.964 -33.972 -69.689 1.00 68.51 B N1+
+ATOM 4292 NH2 ARG H 75 20.073 -34.905 -67.588 1.00 68.51 B N
+ATOM 4293 N PHE H 76 14.512 -37.999 -70.744 1.00 86.73 B N
+ATOM 4294 CA PHE H 76 14.737 -39.166 -71.577 1.00 88.91 B C
+ATOM 4295 C PHE H 76 13.495 -40.043 -71.591 1.00 91.84 B C
+ATOM 4296 O PHE H 76 12.743 -40.089 -70.614 1.00 95.35 B O
+ATOM 4297 CB PHE H 76 15.923 -39.996 -71.058 1.00 62.97 B C
+ATOM 4298 CG PHE H 76 17.293 -39.456 -71.431 1.00 62.97 B C
+ATOM 4299 CD1 PHE H 76 17.463 -38.165 -71.890 1.00 62.97 B C
+ATOM 4300 CD2 PHE H 76 18.404 -40.266 -71.333 1.00 62.97 B C
+ATOM 4301 CE1 PHE H 76 18.720 -37.698 -72.227 1.00 62.97 B C
+ATOM 4302 CE2 PHE H 76 19.642 -39.799 -71.663 1.00 62.97 B C
+ATOM 4303 CZ PHE H 76 19.805 -38.510 -72.099 1.00 62.97 B C
+ATOM 4304 N THR H 77 13.276 -40.730 -72.709 1.00 84.50 B N
+ATOM 4305 CA THR H 77 12.208 -41.720 -72.784 1.00 80.33 B C
+ATOM 4306 C THR H 77 12.556 -43.021 -73.504 1.00 81.75 B C
+ATOM 4307 O THR H 77 12.915 -43.007 -74.686 1.00 78.67 B O
+ATOM 4308 CB THR H 77 10.928 -41.077 -73.326 1.00 57.29 B C
+ATOM 4309 CG2 THR H 77 10.087 -40.442 -72.222 1.00 57.29 B C
+ATOM 4310 OG1 THR H 77 11.279 -40.104 -74.315 1.00 57.29 B O
+ATOM 4311 N ILE H 78 12.430 -44.171 -72.857 1.00 74.60 B N
+ATOM 4312 CA ILE H 78 12.715 -45.406 -73.601 1.00 77.51 B C
+ATOM 4313 C ILE H 78 11.693 -46.465 -74.040 1.00 80.73 B C
+ATOM 4314 O ILE H 78 11.170 -47.210 -73.211 1.00 78.96 B O
+ATOM 4315 CB ILE H 78 13.736 -46.203 -72.763 1.00 56.16 B C
+ATOM 4316 CG1 ILE H 78 14.257 -47.402 -73.557 1.00 56.16 B C
+ATOM 4317 CG2 ILE H 78 13.111 -46.656 -71.452 1.00 56.16 B C
+ATOM 4318 CD1 ILE H 78 15.477 -48.056 -72.946 1.00 56.16 B C
+ATOM 4319 N SER H 79 11.375 -46.505 -75.338 1.00 82.66 B N
+ATOM 4320 CA SER H 79 10.400 -47.484 -75.805 1.00 80.23 B C
+ATOM 4321 C SER H 79 10.932 -48.440 -76.837 1.00 83.60 B C
+ATOM 4322 O SER H 79 11.939 -48.194 -77.493 1.00 82.52 B O
+ATOM 4323 CB SER H 79 9.079 -46.921 -76.304 1.00 59.76 B C
+ATOM 4324 OG SER H 79 8.221 -46.705 -75.188 1.00 59.76 B O
+ATOM 4325 N ARG H 80 10.225 -49.545 -76.967 1.00 81.05 B N
+ATOM 4326 CA ARG H 80 10.618 -50.606 -77.864 1.00 81.38 B C
+ATOM 4327 C ARG H 80 9.481 -50.914 -78.819 1.00 84.96 B C
+ATOM 4328 O ARG H 80 8.325 -50.571 -78.573 1.00 87.16 B O
+ATOM 4329 CB ARG H 80 10.990 -51.848 -77.081 1.00 60.67 B C
+ATOM 4330 CG ARG H 80 9.900 -52.199 -76.099 1.00 60.67 B C
+ATOM 4331 CD ARG H 80 10.281 -53.363 -75.227 1.00 60.67 B C
+ATOM 4332 NE ARG H 80 11.320 -54.171 -75.849 1.00 60.67 B N
+ATOM 4333 CZ ARG H 80 12.000 -55.106 -75.203 1.00 60.67 B C
+ATOM 4334 NH1 ARG H 80 11.749 -55.334 -73.930 1.00 60.67 B N1+
+ATOM 4335 NH2 ARG H 80 12.930 -55.806 -75.825 1.00 60.67 B N
+ATOM 4336 N ASP H 81 9.844 -51.564 -79.920 1.00102.17 B N
+ATOM 4337 CA ASP H 81 8.903 -52.030 -80.926 1.00102.34 B C
+ATOM 4338 C ASP H 81 8.611 -53.513 -80.742 1.00101.33 B C
+ATOM 4339 O ASP H 81 7.447 -53.919 -80.658 1.00101.11 B O
+ATOM 4340 CB ASP H 81 9.443 -51.746 -82.334 1.00 72.20 B C
+ATOM 4341 CG ASP H 81 8.321 -51.528 -83.374 1.00 72.20 B C
+ATOM 4342 OD1 ASP H 81 7.134 -51.394 -82.967 1.00 72.20 B O
+ATOM 4343 OD2 ASP H 81 8.626 -51.456 -84.598 1.00 72.20 B O1-
+ATOM 4344 N ASN H 82 9.661 -54.334 -80.775 1.00 97.20 B N
+ATOM 4345 CA ASN H 82 9.617 -55.792 -80.721 1.00 96.28 B C
+ATOM 4346 C ASN H 82 9.166 -56.282 -82.071 1.00 97.45 B C
+ATOM 4347 O ASN H 82 9.371 -57.438 -82.438 1.00 98.87 B O
+ATOM 4348 CB ASN H 82 8.677 -56.337 -79.645 1.00 69.39 B C
+ATOM 4349 CG ASN H 82 9.214 -56.156 -78.249 1.00 69.39 B C
+ATOM 4350 ND2 ASN H 82 8.310 -56.033 -77.266 1.00 69.39 B N
+ATOM 4351 OD1 ASN H 82 10.429 -56.151 -78.042 1.00 69.39 B O
+ATOM 4352 N ALA H 83 8.567 -55.387 -82.826 1.00111.84 B N
+ATOM 4353 CA ALA H 83 8.275 -55.696 -84.203 1.00113.30 B C
+ATOM 4354 C ALA H 83 9.510 -55.434 -85.041 1.00115.20 B C
+ATOM 4355 O ALA H 83 10.128 -56.361 -85.571 1.00115.16 B O
+ATOM 4356 CB ALA H 83 7.086 -54.860 -84.657 1.00 33.06 B C
+ATOM 4357 N ARG H 84 9.925 -54.173 -85.098 1.00118.66 B N
+ATOM 4358 CA ARG H 84 11.135 -53.778 -85.801 1.00118.44 B C
+ATOM 4359 C ARG H 84 12.349 -54.407 -85.112 1.00116.03 B C
+ATOM 4360 O ARG H 84 13.481 -54.270 -85.592 1.00111.79 B O
+ATOM 4361 CB ARG H 84 11.250 -52.236 -85.875 1.00 80.38 B C
+ATOM 4362 CG ARG H 84 10.524 -51.523 -87.086 1.00 80.38 B C
+ATOM 4363 CD ARG H 84 10.838 -49.966 -87.238 1.00 80.38 B C
+ATOM 4364 NE ARG H 84 12.279 -49.651 -87.417 1.00 80.38 B N
+ATOM 4365 CZ ARG H 84 12.846 -48.434 -87.349 1.00 80.38 B C
+ATOM 4366 NH1 ARG H 84 12.111 -47.343 -87.125 1.00 80.38 B N1+
+ATOM 4367 NH2 ARG H 84 14.168 -48.310 -87.506 1.00 80.38 B N
+ATOM 4368 N ASN H 85 12.130 -55.095 -83.989 1.00107.39 B N
+ATOM 4369 CA ASN H 85 13.218 -55.598 -83.149 1.00100.85 B C
+ATOM 4370 C ASN H 85 14.161 -54.455 -82.809 1.00101.08 B C
+ATOM 4371 O ASN H 85 15.353 -54.473 -83.113 1.00101.32 B O
+ATOM 4372 CB ASN H 85 13.978 -56.755 -83.808 1.00 69.23 B C
+ATOM 4373 CG ASN H 85 13.444 -58.132 -83.396 1.00 69.23 B C
+ATOM 4374 ND2 ASN H 85 14.205 -59.185 -83.713 1.00 69.23 B N
+ATOM 4375 OD1 ASN H 85 12.367 -58.246 -82.797 1.00 69.23 B O
+ATOM 4376 N SER H 86 13.575 -53.423 -82.221 1.00 95.76 B N
+ATOM 4377 CA SER H 86 14.256 -52.155 -82.086 1.00 91.67 B C
+ATOM 4378 C SER H 86 13.993 -51.558 -80.717 1.00 94.66 B C
+ATOM 4379 O SER H 86 13.005 -51.860 -80.048 1.00 97.56 B O
+ATOM 4380 CB SER H 86 13.825 -51.176 -83.179 1.00 64.08 B C
+ATOM 4381 OG SER H 86 14.609 -51.343 -84.358 1.00 64.08 B O
+ATOM 4382 N LEU H 87 14.907 -50.697 -80.323 1.00 88.64 B N
+ATOM 4383 CA LEU H 87 14.871 -50.001 -79.061 1.00 88.64 B C
+ATOM 4384 C LEU H 87 15.117 -48.536 -79.347 1.00 88.64 B C
+ATOM 4385 O LEU H 87 15.839 -48.197 -80.279 1.00 89.43 B O
+ATOM 4386 CB LEU H 87 15.926 -50.556 -78.121 1.00 49.73 B C
+ATOM 4387 CG LEU H 87 15.868 -49.962 -76.717 1.00 49.73 B C
+ATOM 4388 CD1 LEU H 87 14.427 -49.890 -76.256 1.00 49.73 B C
+ATOM 4389 CD2 LEU H 87 16.683 -50.783 -75.755 1.00 49.73 B C
+ATOM 4390 N TYR H 88 14.504 -47.667 -78.560 1.00 88.92 B N
+ATOM 4391 CA TYR H 88 14.565 -46.236 -78.812 1.00 89.47 B C
+ATOM 4392 C TYR H 88 14.734 -45.510 -77.489 1.00 89.00 B C
+ATOM 4393 O TYR H 88 14.025 -45.804 -76.522 1.00 92.98 B O
+ATOM 4394 CB TYR H 88 13.296 -45.752 -79.543 1.00 67.14 B C
+ATOM 4395 CG TYR H 88 12.972 -46.514 -80.827 1.00 67.14 B C
+ATOM 4396 CD1 TYR H 88 13.353 -46.028 -82.078 1.00 67.14 B C
+ATOM 4397 CD2 TYR H 88 12.321 -47.744 -80.786 1.00 67.14 B C
+ATOM 4398 CE1 TYR H 88 13.065 -46.746 -83.255 1.00 67.14 B C
+ATOM 4399 CE2 TYR H 88 12.034 -48.467 -81.948 1.00 67.14 B C
+ATOM 4400 CZ TYR H 88 12.404 -47.969 -83.175 1.00 67.14 B C
+ATOM 4401 OH TYR H 88 12.109 -48.704 -84.308 1.00 67.14 B O
+ATOM 4402 N LEU H 89 15.677 -44.574 -77.447 1.00 79.27 B N
+ATOM 4403 CA LEU H 89 15.835 -43.675 -76.311 1.00 78.57 B C
+ATOM 4404 C LEU H 89 15.818 -42.244 -76.825 1.00 80.71 B C
+ATOM 4405 O LEU H 89 16.531 -41.914 -77.777 1.00 80.73 B O
+ATOM 4406 CB LEU H 89 17.136 -43.951 -75.546 1.00 57.10 B C
+ATOM 4407 CG LEU H 89 17.512 -42.922 -74.474 1.00 57.10 B C
+ATOM 4408 CD1 LEU H 89 16.644 -43.105 -73.288 1.00 57.10 B C
+ATOM 4409 CD2 LEU H 89 18.950 -43.018 -74.066 1.00 57.10 B C
+ATOM 4410 N GLN H 90 15.013 -41.397 -76.198 1.00 94.70 B N
+ATOM 4411 CA GLN H 90 14.836 -40.025 -76.642 1.00 91.07 B C
+ATOM 4412 C GLN H 90 15.365 -39.029 -75.625 1.00 95.01 B C
+ATOM 4413 O GLN H 90 15.105 -39.151 -74.424 1.00 99.63 B O
+ATOM 4414 CB GLN H 90 13.366 -39.735 -76.924 1.00 65.19 B C
+ATOM 4415 CG GLN H 90 13.125 -38.300 -77.334 1.00 65.19 B C
+ATOM 4416 CD GLN H 90 13.027 -38.110 -78.837 1.00 65.19 B C
+ATOM 4417 NE2 GLN H 90 13.358 -36.889 -79.276 1.00 65.19 B N
+ATOM 4418 OE1 GLN H 90 12.646 -39.032 -79.604 1.00 65.19 B O
+ATOM 4419 N MET H 91 16.042 -38.009 -76.140 1.00 97.01 B N
+ATOM 4420 CA MET H 91 16.898 -37.106 -75.382 1.00 98.12 B C
+ATOM 4421 C MET H 91 16.368 -35.690 -75.553 1.00 99.96 B C
+ATOM 4422 O MET H 91 16.477 -35.119 -76.646 1.00100.69 B O
+ATOM 4423 CB MET H 91 18.315 -37.151 -75.937 1.00 67.01 B C
+ATOM 4424 CG MET H 91 19.051 -38.481 -75.878 1.00 67.01 B C
+ATOM 4425 SD MET H 91 20.064 -38.758 -74.437 1.00 67.01 B S
+ATOM 4426 CE MET H 91 21.585 -39.240 -75.251 1.00 67.01 B C
+ATOM 4427 N ASN H 92 15.860 -35.094 -74.479 1.00 98.65 B N
+ATOM 4428 CA ASN H 92 15.269 -33.767 -74.560 1.00101.51 B C
+ATOM 4429 C ASN H 92 15.822 -32.898 -73.447 1.00104.58 B C
+ATOM 4430 O ASN H 92 16.306 -33.402 -72.432 1.00105.96 B O
+ATOM 4431 CB ASN H 92 13.748 -33.837 -74.466 1.00 70.39 B C
+ATOM 4432 CG ASN H 92 13.140 -34.626 -75.612 1.00 70.39 B C
+ATOM 4433 ND2 ASN H 92 13.084 -34.034 -76.805 1.00 70.39 B N
+ATOM 4434 OD1 ASN H 92 12.746 -35.775 -75.425 1.00 70.39 B O
+ATOM 4435 N SER H 93 15.740 -31.580 -73.646 1.00106.04 B N
+ATOM 4436 CA SER H 93 16.436 -30.631 -72.778 1.00107.13 B C
+ATOM 4437 C SER H 93 17.850 -31.126 -72.546 1.00102.60 B C
+ATOM 4438 O SER H 93 18.391 -31.050 -71.446 1.00100.37 B O
+ATOM 4439 CB SER H 93 15.700 -30.415 -71.444 1.00 64.54 B C
+ATOM 4440 OG SER H 93 16.310 -29.408 -70.608 1.00 64.54 B O
+ATOM 4441 N LEU H 94 18.430 -31.680 -73.591 1.00 94.69 B N
+ATOM 4442 CA LEU H 94 19.668 -32.408 -73.441 1.00 94.97 B C
+ATOM 4443 C LEU H 94 20.702 -31.503 -72.809 1.00100.87 B C
+ATOM 4444 O LEU H 94 21.054 -30.470 -73.373 1.00102.56 B O
+ATOM 4445 CB LEU H 94 20.101 -32.865 -74.823 1.00 57.97 B C
+ATOM 4446 CG LEU H 94 21.220 -33.835 -75.157 1.00 57.97 B C
+ATOM 4447 CD1 LEU H 94 21.135 -35.082 -74.298 1.00 57.97 B C
+ATOM 4448 CD2 LEU H 94 21.160 -34.198 -76.648 1.00 57.97 B C
+ATOM 4449 N ARG H 95 21.200 -31.881 -71.643 1.00119.44 B N
+ATOM 4450 CA ARG H 95 22.159 -30.999 -71.011 1.00118.59 B C
+ATOM 4451 C ARG H 95 23.533 -31.373 -71.553 1.00111.02 B C
+ATOM 4452 O ARG H 95 23.683 -32.298 -72.354 1.00108.46 B O
+ATOM 4453 CB ARG H 95 22.054 -31.043 -69.474 1.00 76.27 B C
+ATOM 4454 CG ARG H 95 20.796 -30.290 -68.866 1.00 76.27 B C
+ATOM 4455 CD ARG H 95 20.608 -28.845 -69.510 1.00 76.27 B C
+ATOM 4456 NE ARG H 95 19.265 -28.212 -69.390 1.00 76.27 B N
+ATOM 4457 CZ ARG H 95 19.018 -26.960 -68.953 1.00 76.27 B C
+ATOM 4458 NH1 ARG H 95 20.014 -26.140 -68.592 1.00 76.27 B N1+
+ATOM 4459 NH2 ARG H 95 17.757 -26.512 -68.896 1.00 76.27 B N
+ATOM 4460 N ALA H 96 24.546 -30.632 -71.131 1.00 77.72 B N
+ATOM 4461 CA ALA H 96 25.858 -30.723 -71.753 1.00 81.14 B C
+ATOM 4462 C ALA H 96 26.719 -31.838 -71.185 1.00 80.37 B C
+ATOM 4463 O ALA H 96 27.764 -32.150 -71.759 1.00 83.73 B O
+ATOM 4464 CB ALA H 96 26.575 -29.385 -71.593 1.00 78.76 B C
+ATOM 4465 N GLU H 97 26.329 -32.413 -70.061 1.00 79.55 B N
+ATOM 4466 CA GLU H 97 27.055 -33.515 -69.460 1.00 83.82 B C
+ATOM 4467 C GLU H 97 26.505 -34.854 -69.890 1.00 84.47 B C
+ATOM 4468 O GLU H 97 27.023 -35.887 -69.462 1.00 88.20 B O
+ATOM 4469 CB GLU H 97 27.017 -33.413 -67.937 1.00 89.53 B C
+ATOM 4470 CG GLU H 97 27.604 -32.113 -67.391 1.00 89.53 B C
+ATOM 4471 CD GLU H 97 26.626 -30.943 -67.426 1.00 89.53 B C
+ATOM 4472 OE1 GLU H 97 25.411 -31.173 -67.641 1.00 89.53 B O
+ATOM 4473 OE2 GLU H 97 27.073 -29.790 -67.237 1.00 89.53 B O1-
+ATOM 4474 N ASP H 98 25.459 -34.861 -70.711 1.00 75.56 B N
+ATOM 4475 CA ASP H 98 24.964 -36.122 -71.235 1.00 72.34 B C
+ATOM 4476 C ASP H 98 25.880 -36.695 -72.291 1.00 71.16 B C
+ATOM 4477 O ASP H 98 25.669 -37.838 -72.704 1.00 69.73 B O
+ATOM 4478 CB ASP H 98 23.562 -35.963 -71.825 1.00 79.58 B C
+ATOM 4479 CG ASP H 98 22.481 -35.886 -70.760 1.00 79.58 B C
+ATOM 4480 OD1 ASP H 98 22.694 -36.393 -69.625 1.00 79.58 B O
+ATOM 4481 OD2 ASP H 98 21.407 -35.322 -71.063 1.00 79.58 B O1-
+ATOM 4482 N THR H 99 26.872 -35.934 -72.741 1.00 64.99 B N
+ATOM 4483 CA THR H 99 27.751 -36.429 -73.785 1.00 63.29 B C
+ATOM 4484 C THR H 99 28.507 -37.644 -73.276 1.00 62.88 B C
+ATOM 4485 O THR H 99 29.008 -37.654 -72.148 1.00 62.63 B O
+ATOM 4486 CB THR H 99 28.734 -35.357 -74.198 1.00 71.04 B C
+ATOM 4487 CG2 THR H 99 28.140 -34.037 -73.937 1.00 71.04 B C
+ATOM 4488 OG1 THR H 99 29.876 -35.456 -73.360 1.00 71.04 B O
+ATOM 4489 N ALA H 100 28.576 -38.674 -74.111 1.00 62.73 B N
+ATOM 4490 CA ALA H 100 29.062 -39.983 -73.695 1.00 61.26 B C
+ATOM 4491 C ALA H 100 28.912 -40.915 -74.883 1.00 57.14 B C
+ATOM 4492 O ALA H 100 28.229 -40.603 -75.867 1.00 59.38 B O
+ATOM 4493 CB ALA H 100 28.293 -40.558 -72.506 1.00 57.14 B C
+ATOM 4494 N LEU H 101 29.541 -42.075 -74.773 1.00 47.91 B N
+ATOM 4495 CA LEU H 101 29.265 -43.144 -75.703 1.00 47.79 B C
+ATOM 4496 C LEU H 101 28.080 -43.898 -75.132 1.00 47.79 B C
+ATOM 4497 O LEU H 101 28.089 -44.250 -73.946 1.00 50.20 B O
+ATOM 4498 CB LEU H 101 30.476 -44.041 -75.866 1.00 58.00 B C
+ATOM 4499 CG LEU H 101 30.294 -44.754 -77.181 1.00 58.00 B C
+ATOM 4500 CD1 LEU H 101 31.614 -44.749 -77.896 1.00 58.00 B C
+ATOM 4501 CD2 LEU H 101 29.850 -46.128 -76.913 1.00 58.00 B C
+ATOM 4502 N TYR H 102 27.034 -44.075 -75.944 1.00 46.71 B N
+ATOM 4503 CA TYR H 102 25.773 -44.671 -75.495 1.00 46.71 B C
+ATOM 4504 C TYR H 102 25.671 -46.108 -75.992 1.00 46.71 B C
+ATOM 4505 O TYR H 102 25.774 -46.372 -77.206 1.00 46.71 B O
+ATOM 4506 CB TYR H 102 24.566 -43.860 -75.963 1.00 57.38 B C
+ATOM 4507 CG TYR H 102 24.270 -42.691 -75.066 1.00 57.38 B C
+ATOM 4508 CD1 TYR H 102 23.290 -42.761 -74.102 1.00 57.38 B C
+ATOM 4509 CD2 TYR H 102 25.014 -41.528 -75.162 1.00 57.38 B C
+ATOM 4510 CE1 TYR H 102 23.041 -41.701 -73.283 1.00 57.38 B C
+ATOM 4511 CE2 TYR H 102 24.773 -40.469 -74.354 1.00 57.38 B C
+ATOM 4512 CZ TYR H 102 23.786 -40.559 -73.413 1.00 57.38 B C
+ATOM 4513 OH TYR H 102 23.549 -39.482 -72.598 1.00 57.38 B O
+ATOM 4514 N TYR H 103 25.450 -47.014 -75.036 1.00 48.49 B N
+ATOM 4515 CA TYR H 103 25.559 -48.461 -75.172 1.00 48.49 B C
+ATOM 4516 C TYR H 103 24.183 -49.094 -75.077 1.00 48.49 B C
+ATOM 4517 O TYR H 103 23.433 -48.833 -74.119 1.00 51.52 B O
+ATOM 4518 CB TYR H 103 26.416 -49.048 -74.051 1.00 58.68 B C
+ATOM 4519 CG TYR H 103 27.894 -48.986 -74.238 1.00 58.68 B C
+ATOM 4520 CD1 TYR H 103 28.473 -49.410 -75.422 1.00 58.68 B C
+ATOM 4521 CD2 TYR H 103 28.716 -48.509 -73.224 1.00 58.68 B C
+ATOM 4522 CE1 TYR H 103 29.838 -49.365 -75.598 1.00 58.68 B C
+ATOM 4523 CE2 TYR H 103 30.085 -48.449 -73.385 1.00 58.68 B C
+ATOM 4524 CZ TYR H 103 30.643 -48.876 -74.580 1.00 58.68 B C
+ATOM 4525 OH TYR H 103 32.010 -48.828 -74.771 1.00 58.68 B O
+ATOM 4526 N CYS H 104 23.873 -49.940 -76.047 1.00 57.06 B N
+ATOM 4527 CA CYS H 104 22.735 -50.842 -75.977 1.00 56.66 B C
+ATOM 4528 C CYS H 104 23.176 -52.125 -75.270 1.00 58.26 B C
+ATOM 4529 O CYS H 104 24.161 -52.751 -75.688 1.00 59.14 B O
+ATOM 4530 CB CYS H 104 22.256 -51.124 -77.392 1.00 67.66 B C
+ATOM 4531 SG CYS H 104 20.607 -51.721 -77.518 1.00 67.66 B S
+ATOM 4532 N VAL H 105 22.485 -52.506 -74.187 1.00 49.90 B N
+ATOM 4533 CA VAL H 105 22.970 -53.574 -73.307 1.00 49.90 B C
+ATOM 4534 C VAL H 105 21.842 -54.509 -72.926 1.00 51.74 B C
+ATOM 4535 O VAL H 105 20.905 -54.088 -72.240 1.00 50.00 B O
+ATOM 4536 CB VAL H 105 23.583 -53.039 -72.014 1.00 58.06 B C
+ATOM 4537 CG1 VAL H 105 24.272 -54.170 -71.334 1.00 58.06 B C
+ATOM 4538 CG2 VAL H 105 24.540 -51.909 -72.303 1.00 58.06 B C
+ATOM 4539 N ARG H 106 21.987 -55.794 -73.257 1.00 54.17 B N
+ATOM 4540 CA ARG H 106 20.950 -56.785 -72.992 1.00 54.92 B C
+ATOM 4541 C ARG H 106 21.116 -57.412 -71.614 1.00 51.72 B C
+ATOM 4542 O ARG H 106 22.223 -57.765 -71.202 1.00 51.82 B O
+ATOM 4543 CB ARG H 106 20.983 -57.868 -74.056 1.00 55.79 B C
+ATOM 4544 CG ARG H 106 20.251 -59.125 -73.704 1.00 55.79 B C
+ATOM 4545 CD ARG H 106 21.072 -60.234 -74.234 1.00 55.79 B C
+ATOM 4546 NE ARG H 106 20.316 -61.345 -74.785 1.00 55.79 B N
+ATOM 4547 CZ ARG H 106 20.915 -62.455 -75.196 1.00 55.79 B C
+ATOM 4548 NH1 ARG H 106 22.229 -62.531 -75.102 1.00 55.79 B N1+
+ATOM 4549 NH2 ARG H 106 20.242 -63.474 -75.716 1.00 55.79 B N
+ATOM 4550 N ASP H 107 20.014 -57.545 -70.898 1.00 59.29 B N
+ATOM 4551 CA ASP H 107 20.059 -58.096 -69.554 1.00 59.29 B C
+ATOM 4552 C ASP H 107 19.818 -59.603 -69.620 1.00 59.29 B C
+ATOM 4553 O ASP H 107 18.724 -60.046 -69.993 1.00 59.29 B O
+ATOM 4554 CB ASP H 107 19.031 -57.385 -68.679 1.00 59.29 B C
+ATOM 4555 CG ASP H 107 19.243 -57.644 -67.198 1.00 59.29 B C
+ATOM 4556 OD1 ASP H 107 20.262 -58.282 -66.836 1.00 59.29 B O
+ATOM 4557 OD2 ASP H 107 18.405 -57.192 -66.378 1.00 59.29 B O1-
+ATOM 4558 N ALA H 108 20.836 -60.388 -69.243 1.00 54.40 B N
+ATOM 4559 CA ALA H 108 20.834 -61.842 -69.413 1.00 54.40 B C
+ATOM 4560 C ALA H 108 19.820 -62.569 -68.554 1.00 54.40 B C
+ATOM 4561 O ALA H 108 19.671 -63.780 -68.723 1.00 54.40 B O
+ATOM 4562 CB ALA H 108 22.201 -62.443 -69.095 1.00 54.40 B C
+ATOM 4563 N TYR H 109 19.137 -61.894 -67.639 1.00 64.04 B N
+ATOM 4564 CA TYR H 109 18.170 -62.562 -66.775 1.00 64.04 B C
+ATOM 4565 C TYR H 109 16.814 -62.561 -67.456 1.00 64.04 B C
+ATOM 4566 O TYR H 109 16.161 -61.513 -67.561 1.00 64.04 B O
+ATOM 4567 CB TYR H 109 18.086 -61.884 -65.415 1.00 64.04 B C
+ATOM 4568 CG TYR H 109 16.958 -62.438 -64.597 1.00 64.04 B C
+ATOM 4569 CD1 TYR H 109 17.061 -63.664 -63.976 1.00 64.04 B C
+ATOM 4570 CD2 TYR H 109 15.770 -61.745 -64.470 1.00 64.04 B C
+ATOM 4571 CE1 TYR H 109 16.012 -64.190 -63.214 1.00 64.04 B C
+ATOM 4572 CE2 TYR H 109 14.713 -62.258 -63.712 1.00 64.04 B C
+ATOM 4573 CZ TYR H 109 14.840 -63.487 -63.079 1.00 64.04 B C
+ATOM 4574 OH TYR H 109 13.792 -63.996 -62.322 1.00 64.04 B O
+ATOM 4575 N VAL H 110 16.381 -63.744 -67.894 1.00 68.66 B N
+ATOM 4576 CA VAL H 110 15.201 -63.842 -68.743 1.00 68.66 B C
+ATOM 4577 C VAL H 110 13.891 -63.907 -67.960 1.00 68.66 B C
+ATOM 4578 O VAL H 110 12.837 -63.554 -68.509 1.00 68.66 B O
+ATOM 4579 CB VAL H 110 15.311 -65.078 -69.641 1.00 68.66 B C
+ATOM 4580 CG1 VAL H 110 15.086 -66.334 -68.785 1.00 68.66 B C
+ATOM 4581 CG2 VAL H 110 14.362 -64.964 -70.820 1.00 68.66 B C
+ATOM 4582 N SER H 111 13.929 -64.326 -66.694 1.00 69.05 B N
+ATOM 4583 CA SER H 111 12.722 -64.531 -65.895 1.00 69.05 B C
+ATOM 4584 C SER H 111 11.750 -65.529 -66.493 1.00 69.05 B C
+ATOM 4585 O SER H 111 10.825 -65.161 -67.219 1.00 69.05 B O
+ATOM 4586 CB SER H 111 11.954 -63.249 -65.612 1.00 69.05 B C
+ATOM 4587 OG SER H 111 11.027 -63.536 -64.561 1.00 69.05 B O
+ATOM 4588 N GLY H 111A 11.974 -66.796 -66.227 1.00 67.69 B N
+ATOM 4589 CA GLY H 111A 10.990 -67.775 -66.573 1.00 67.69 B C
+ATOM 4590 C GLY H 111A 11.429 -68.737 -67.614 1.00 67.69 B C
+ATOM 4591 O GLY H 111A 10.670 -69.661 -67.906 1.00 67.69 B O
+ATOM 4592 N SER H 111B 12.606 -68.563 -68.206 1.00 78.31 B N
+ATOM 4593 CA SER H 111B 13.186 -69.729 -68.863 1.00 78.31 B C
+ATOM 4594 C SER H 111B 13.778 -70.673 -67.814 1.00 78.31 B C
+ATOM 4595 O SER H 111B 13.954 -70.318 -66.646 1.00 78.31 B O
+ATOM 4596 CB SER H 111B 14.224 -69.344 -69.929 1.00 78.31 B C
+ATOM 4597 OG SER H 111B 15.553 -69.374 -69.431 1.00 78.31 B O
+ATOM 4598 N ASP H 112C 14.033 -71.910 -68.242 1.00 82.12 B N
+ATOM 4599 CA ASP H 112C 14.343 -72.967 -67.288 1.00 82.12 B C
+ATOM 4600 C ASP H 112C 15.725 -72.794 -66.671 1.00 82.12 B C
+ATOM 4601 O ASP H 112C 15.921 -73.115 -65.493 1.00 82.12 B O
+ATOM 4602 CB ASP H 112C 14.225 -74.329 -67.959 1.00 82.12 B C
+ATOM 4603 CG ASP H 112C 12.790 -74.684 -68.296 1.00 82.12 B C
+ATOM 4604 OD1 ASP H 112C 11.888 -73.883 -67.951 1.00 82.12 B O
+ATOM 4605 OD2 ASP H 112C 12.562 -75.761 -68.897 1.00 82.12 B O1-
+ATOM 4606 N TYR H 112B 16.699 -72.317 -67.438 1.00 85.22 B N
+ATOM 4607 CA TYR H 112B 18.031 -72.021 -66.917 1.00 85.22 B C
+ATOM 4608 C TYR H 112B 18.256 -70.513 -66.965 1.00 85.22 B C
+ATOM 4609 O TYR H 112B 18.200 -69.906 -68.043 1.00 85.22 B O
+ATOM 4610 CB TYR H 112B 19.104 -72.774 -67.705 1.00 85.22 B C
+ATOM 4611 CG TYR H 112B 20.485 -72.928 -67.034 1.00 85.22 B C
+ATOM 4612 CD1 TYR H 112B 21.293 -74.047 -67.304 1.00 85.22 B C
+ATOM 4613 CD2 TYR H 112B 21.021 -71.932 -66.196 1.00 85.22 B C
+ATOM 4614 CE1 TYR H 112B 22.576 -74.191 -66.722 1.00 85.22 B C
+ATOM 4615 CE2 TYR H 112B 22.307 -72.068 -65.611 1.00 85.22 B C
+ATOM 4616 CZ TYR H 112B 23.074 -73.200 -65.882 1.00 85.22 B C
+ATOM 4617 OH TYR H 112B 24.331 -73.349 -65.321 1.00 85.22 B O
+ATOM 4618 N TYR H 112A 18.519 -69.921 -65.791 1.00 61.55 B N
+ATOM 4619 CA TYR H 112A 18.706 -68.483 -65.621 1.00 61.55 B C
+ATOM 4620 C TYR H 112A 20.150 -68.062 -65.881 1.00 61.55 B C
+ATOM 4621 O TYR H 112A 21.095 -68.762 -65.515 1.00 61.55 B O
+ATOM 4622 CB TYR H 112A 18.306 -68.069 -64.205 1.00 61.55 B C
+ATOM 4623 CG TYR H 112A 16.823 -68.080 -63.945 1.00 61.55 B C
+ATOM 4624 CD1 TYR H 112A 15.979 -67.207 -64.598 1.00 61.55 B C
+ATOM 4625 CD2 TYR H 112A 16.269 -68.958 -63.038 1.00 61.55 B C
+ATOM 4626 CE1 TYR H 112A 14.623 -67.203 -64.358 1.00 61.55 B C
+ATOM 4627 CE2 TYR H 112A 14.904 -68.976 -62.789 1.00 61.55 B C
+ATOM 4628 CZ TYR H 112A 14.083 -68.092 -63.454 1.00 61.55 B C
+ATOM 4629 OH TYR H 112A 12.721 -68.094 -63.211 1.00 61.55 B O
+ATOM 4630 N TYR H 112 20.317 -66.916 -66.518 1.00 62.31 B N
+ATOM 4631 CA TYR H 112 21.613 -66.266 -66.589 1.00 62.31 B C
+ATOM 4632 C TYR H 112 21.442 -64.857 -66.049 1.00 62.31 B C
+ATOM 4633 O TYR H 112 20.326 -64.345 -65.990 1.00 62.31 B O
+ATOM 4634 CB TYR H 112 22.171 -66.241 -68.024 1.00 62.31 B C
+ATOM 4635 CG TYR H 112 22.365 -67.627 -68.589 1.00 62.31 B C
+ATOM 4636 CD1 TYR H 112 23.540 -68.331 -68.380 1.00 62.31 B C
+ATOM 4637 CD2 TYR H 112 21.371 -68.234 -69.335 1.00 62.31 B C
+ATOM 4638 CE1 TYR H 112 23.709 -69.619 -68.886 1.00 62.31 B C
+ATOM 4639 CE2 TYR H 112 21.534 -69.518 -69.851 1.00 62.31 B C
+ATOM 4640 CZ TYR H 112 22.702 -70.207 -69.623 1.00 62.31 B C
+ATOM 4641 OH TYR H 112 22.848 -71.482 -70.137 1.00 62.31 B O
+ATOM 4642 N TYR H 113 22.544 -64.227 -65.637 1.00 52.74 B N
+ATOM 4643 CA TYR H 113 22.481 -62.894 -65.036 1.00 52.74 B C
+ATOM 4644 C TYR H 113 23.574 -62.000 -65.618 1.00 52.74 B C
+ATOM 4645 O TYR H 113 24.385 -62.424 -66.447 1.00 52.74 B O
+ATOM 4646 CB TYR H 113 22.604 -62.969 -63.511 1.00 52.74 B C
+ATOM 4647 CG TYR H 113 21.710 -63.995 -62.887 1.00 52.74 B C
+ATOM 4648 CD1 TYR H 113 20.491 -63.650 -62.371 1.00 52.74 B C
+ATOM 4649 CD2 TYR H 113 22.086 -65.316 -62.818 1.00 52.74 B C
+ATOM 4650 CE1 TYR H 113 19.666 -64.608 -61.784 1.00 52.74 B C
+ATOM 4651 CE2 TYR H 113 21.266 -66.278 -62.248 1.00 52.74 B C
+ATOM 4652 CZ TYR H 113 20.057 -65.927 -61.728 1.00 52.74 B C
+ATOM 4653 OH TYR H 113 19.240 -66.890 -61.155 1.00 52.74 B O
+ATOM 4654 N GLY H 114 23.598 -60.747 -65.166 1.00 54.79 B N
+ATOM 4655 CA GLY H 114 24.577 -59.797 -65.648 1.00 54.79 B C
+ATOM 4656 C GLY H 114 24.123 -59.172 -66.951 1.00 54.79 B C
+ATOM 4657 O GLY H 114 22.996 -59.384 -67.416 1.00 54.79 B O
+ATOM 4658 N LEU H 115 25.025 -58.393 -67.558 1.00 46.31 B N
+ATOM 4659 CA LEU H 115 24.774 -57.769 -68.856 1.00 47.65 B C
+ATOM 4660 C LEU H 115 25.683 -58.456 -69.867 1.00 52.78 B C
+ATOM 4661 O LEU H 115 26.890 -58.205 -69.899 1.00 54.63 B O
+ATOM 4662 CB LEU H 115 25.052 -56.279 -68.759 1.00 44.41 B C
+ATOM 4663 CG LEU H 115 24.267 -55.691 -67.600 1.00 44.41 B C
+ATOM 4664 CD1 LEU H 115 24.627 -54.264 -67.395 1.00 44.41 B C
+ATOM 4665 CD2 LEU H 115 22.803 -55.834 -67.881 1.00 44.41 B C
+ATOM 4666 N ASP H 116 25.111 -59.344 -70.672 1.00 54.96 B N
+ATOM 4667 CA ASP H 116 25.940 -60.176 -71.531 1.00 54.96 B C
+ATOM 4668 C ASP H 116 26.238 -59.565 -72.889 1.00 54.96 B C
+ATOM 4669 O ASP H 116 27.245 -59.933 -73.497 1.00 54.96 B O
+ATOM 4670 CB ASP H 116 25.309 -61.549 -71.713 1.00 59.63 B C
+ATOM 4671 CG ASP H 116 23.885 -61.451 -72.097 1.00 59.63 B C
+ATOM 4672 OD1 ASP H 116 23.149 -60.766 -71.364 1.00 59.63 B O
+ATOM 4673 OD2 ASP H 116 23.484 -62.043 -73.115 1.00 59.63 B O1-
+ATOM 4674 N VAL H 117 25.395 -58.682 -73.424 1.00 55.04 B N
+ATOM 4675 CA VAL H 117 25.556 -58.237 -74.813 1.00 55.04 B C
+ATOM 4676 C VAL H 117 25.550 -56.710 -74.910 1.00 55.04 B C
+ATOM 4677 O VAL H 117 24.579 -56.061 -74.488 1.00 55.04 B O
+ATOM 4678 CB VAL H 117 24.475 -58.833 -75.710 1.00 62.31 B C
+ATOM 4679 CG1 VAL H 117 24.483 -58.118 -77.057 1.00 62.31 B C
+ATOM 4680 CG2 VAL H 117 24.727 -60.323 -75.847 1.00 62.31 B C
+ATOM 4681 N TRP H 118 26.621 -56.144 -75.492 1.00 45.98 B N
+ATOM 4682 CA TRP H 118 26.830 -54.701 -75.586 1.00 41.15 B C
+ATOM 4683 C TRP H 118 27.150 -54.318 -77.017 1.00 42.63 B C
+ATOM 4684 O TRP H 118 28.039 -54.913 -77.633 1.00 48.64 B O
+ATOM 4685 CB TRP H 118 28.001 -54.234 -74.717 1.00 51.78 B C
+ATOM 4686 CG TRP H 118 27.863 -54.503 -73.277 1.00 51.78 B C
+ATOM 4687 CD1 TRP H 118 27.728 -55.711 -72.695 1.00 51.78 B C
+ATOM 4688 CD2 TRP H 118 27.877 -53.538 -72.212 1.00 51.78 B C
+ATOM 4689 CE2 TRP H 118 27.724 -54.236 -71.019 1.00 51.78 B C
+ATOM 4690 CE3 TRP H 118 27.999 -52.157 -72.163 1.00 51.78 B C
+ATOM 4691 NE1 TRP H 118 27.627 -55.563 -71.336 1.00 51.78 B N
+ATOM 4692 CZ2 TRP H 118 27.680 -53.609 -69.797 1.00 51.78 B C
+ATOM 4693 CZ3 TRP H 118 27.958 -51.539 -70.951 1.00 51.78 B C
+ATOM 4694 CH2 TRP H 118 27.803 -52.260 -69.786 1.00 51.78 B C
+ATOM 4695 N GLY H 119 26.486 -53.300 -77.530 1.00 57.81 B N
+ATOM 4696 CA GLY H 119 26.886 -52.780 -78.820 1.00 57.81 B C
+ATOM 4697 C GLY H 119 28.246 -52.096 -78.762 1.00 57.81 B C
+ATOM 4698 O GLY H 119 28.824 -51.864 -77.694 1.00 57.81 B O
+ATOM 4699 N ARG H 120 28.780 -51.794 -79.957 1.00 66.11 B N
+ATOM 4700 CA ARG H 120 29.911 -50.875 -80.048 1.00 70.13 B C
+ATOM 4701 C ARG H 120 29.572 -49.534 -79.424 1.00 68.50 B C
+ATOM 4702 O ARG H 120 30.450 -48.852 -78.890 1.00 71.08 B O
+ATOM 4703 CB ARG H 120 30.323 -50.660 -81.505 1.00 77.75 B C
+ATOM 4704 CG ARG H 120 30.706 -51.889 -82.306 1.00 77.75 B C
+ATOM 4705 CD ARG H 120 30.776 -51.550 -83.828 1.00 77.75 B C
+ATOM 4706 NE ARG H 120 32.095 -51.086 -84.318 1.00 77.75 B N
+ATOM 4707 CZ ARG H 120 32.373 -49.860 -84.802 1.00 77.75 B C
+ATOM 4708 NH1 ARG H 120 31.419 -48.925 -84.917 1.00 77.75 B N1+
+ATOM 4709 NH2 ARG H 120 33.617 -49.578 -85.209 1.00 77.75 B N
+ATOM 4710 N GLY H 121 28.320 -49.132 -79.521 1.00 61.49 B N
+ATOM 4711 CA GLY H 121 27.770 -47.914 -78.956 1.00 63.37 B C
+ATOM 4712 C GLY H 121 27.973 -46.728 -79.867 1.00 64.20 B C
+ATOM 4713 O GLY H 121 28.975 -46.615 -80.568 1.00 67.54 B O
+ATOM 4714 N THR H 122 27.007 -45.814 -79.851 1.00 62.63 B N
+ATOM 4715 CA THR H 122 27.089 -44.604 -80.659 1.00 62.63 B C
+ATOM 4716 C THR H 122 27.941 -43.600 -79.894 1.00 63.25 B C
+ATOM 4717 O THR H 122 28.634 -43.976 -78.948 1.00 67.22 B O
+ATOM 4718 CB THR H 122 25.698 -44.000 -80.923 1.00 73.01 B C
+ATOM 4719 CG2 THR H 122 25.099 -43.460 -79.633 1.00 73.01 B C
+ATOM 4720 OG1 THR H 122 25.810 -42.936 -81.875 1.00 73.01 B O
+ATOM 4721 N LEU H 123 27.901 -42.330 -80.284 1.00 60.34 B N
+ATOM 4722 CA LEU H 123 28.696 -41.335 -79.572 1.00 60.34 B C
+ATOM 4723 C LEU H 123 28.029 -39.958 -79.640 1.00 61.59 B C
+ATOM 4724 O LEU H 123 28.052 -39.291 -80.677 1.00 65.66 B O
+ATOM 4725 CB LEU H 123 30.103 -41.382 -80.171 1.00 70.34 B C
+ATOM 4726 CG LEU H 123 30.156 -41.251 -81.720 1.00 70.34 B C
+ATOM 4727 CD1 LEU H 123 31.420 -40.586 -82.194 1.00 70.34 B C
+ATOM 4728 CD2 LEU H 123 29.888 -42.530 -82.582 1.00 70.34 B C
+ATOM 4729 N VAL H 124 27.459 -39.524 -78.516 1.00 58.80 B N
+ATOM 4730 CA VAL H 124 26.602 -38.340 -78.434 1.00 55.96 B C
+ATOM 4731 C VAL H 124 27.359 -37.219 -77.746 1.00 62.40 B C
+ATOM 4732 O VAL H 124 27.766 -37.374 -76.587 1.00 63.92 B O
+ATOM 4733 CB VAL H 124 25.328 -38.636 -77.643 1.00 66.55 B C
+ATOM 4734 CG1 VAL H 124 24.480 -37.392 -77.580 1.00 66.55 B C
+ATOM 4735 CG2 VAL H 124 24.595 -39.780 -78.270 1.00 66.55 B C
+ATOM 4736 N THR H 125 27.516 -36.072 -78.412 1.00 57.12 B N
+ATOM 4737 CA THR H 125 28.274 -34.976 -77.812 1.00 53.18 B C
+ATOM 4738 C THR H 125 27.442 -33.702 -77.829 1.00 54.85 B C
+ATOM 4739 O THR H 125 27.104 -33.189 -78.895 1.00 56.08 B O
+ATOM 4740 CB THR H 125 29.607 -34.738 -78.526 1.00 62.04 B C
+ATOM 4741 CG2 THR H 125 30.197 -36.040 -79.027 1.00 62.04 B C
+ATOM 4742 OG1 THR H 125 29.376 -33.919 -79.662 1.00 62.04 B O
+ATOM 4743 N VAL H 126 27.157 -33.174 -76.651 1.00 60.27 B N
+ATOM 4744 CA VAL H 126 26.325 -31.994 -76.479 1.00 57.07 B C
+ATOM 4745 C VAL H 126 27.246 -30.817 -76.217 1.00 61.62 B C
+ATOM 4746 O VAL H 126 27.703 -30.615 -75.092 1.00 62.07 B O
+ATOM 4747 CB VAL H 126 25.336 -32.194 -75.337 1.00 61.95 B C
+ATOM 4748 CG1 VAL H 126 24.451 -31.003 -75.177 1.00 61.95 B C
+ATOM 4749 CG2 VAL H 126 24.533 -33.424 -75.651 1.00 61.95 B C
+ATOM 4750 N SER H 127 27.425 -29.990 -77.240 1.00 65.99 B N
+ATOM 4751 CA SER H 127 28.364 -28.880 -77.218 1.00 64.73 B C
+ATOM 4752 C SER H 127 27.852 -27.802 -78.160 1.00 69.92 B C
+ATOM 4753 O SER H 127 27.191 -28.090 -79.162 1.00 73.21 B O
+ATOM 4754 CB SER H 127 29.780 -29.330 -77.629 1.00 60.38 B C
+ATOM 4755 OG SER H 127 30.732 -28.268 -77.607 1.00 60.38 B O
+ATOM 4756 N SER H 128 28.152 -26.559 -77.824 1.00 78.66 B N
+ATOM 4757 CA SER H 128 27.788 -25.443 -78.681 1.00 80.49 B C
+ATOM 4758 C SER H 128 28.900 -25.030 -79.639 1.00 79.87 B C
+ATOM 4759 O SER H 128 28.665 -24.215 -80.536 1.00 83.23 B O
+ATOM 4760 CB SER H 128 27.351 -24.264 -77.806 1.00 53.40 B C
+ATOM 4761 OG SER H 128 28.305 -24.031 -76.775 1.00 53.40 B O
+ATOM 4762 N ALA H 129 30.091 -25.579 -79.493 1.00 63.84 B N
+ATOM 4763 CA ALA H 129 31.135 -25.257 -80.443 1.00 65.44 B C
+ATOM 4764 C ALA H 129 30.770 -25.776 -81.827 1.00 69.35 B C
+ATOM 4765 O ALA H 129 30.000 -26.726 -81.980 1.00 71.52 B O
+ATOM 4766 CB ALA H 129 32.464 -25.863 -80.002 1.00 62.31 B C
+ATOM 4767 N SER H 130 31.351 -25.153 -82.842 1.00 56.39 B N
+ATOM 4768 CA SER H 130 31.091 -25.544 -84.218 1.00 62.50 B C
+ATOM 4769 C SER H 130 32.235 -26.388 -84.752 1.00 63.20 B C
+ATOM 4770 O SER H 130 33.391 -26.177 -84.395 1.00 66.74 B O
+ATOM 4771 CB SER H 130 30.889 -24.317 -85.108 1.00 65.60 B C
+ATOM 4772 OG SER H 130 29.853 -23.485 -84.602 1.00 70.32 B O
+ATOM 4773 N THR H 131 31.902 -27.340 -85.619 1.00 65.02 B N
+ATOM 4774 CA THR H 131 32.921 -28.179 -86.242 1.00 60.59 B C
+ATOM 4775 C THR H 131 34.025 -27.304 -86.808 1.00 60.41 B C
+ATOM 4776 O THR H 131 33.748 -26.293 -87.449 1.00 63.37 B O
+ATOM 4777 CB THR H 131 32.302 -29.024 -87.365 1.00 58.81 B C
+ATOM 4778 CG2 THR H 131 33.375 -29.828 -88.133 1.00 54.78 B C
+ATOM 4779 OG1 THR H 131 31.308 -29.909 -86.821 1.00 59.71 B O
+ATOM 4780 N LYS H 132 35.276 -27.660 -86.532 1.00 64.47 B N
+ATOM 4781 CA LYS H 132 36.396 -26.853 -86.996 1.00 62.71 B C
+ATOM 4782 C LYS H 132 37.571 -27.732 -87.384 1.00 58.67 B C
+ATOM 4783 O LYS H 132 37.960 -28.623 -86.625 1.00 57.26 B O
+ATOM 4784 CB LYS H 132 36.848 -25.860 -85.924 1.00 64.12 B C
+ATOM 4785 CG LYS H 132 37.876 -24.886 -86.444 1.00 66.54 B C
+ATOM 4786 CD LYS H 132 38.216 -23.820 -85.427 1.00 70.23 B C
+ATOM 4787 CE LYS H 132 39.350 -22.922 -85.934 1.00 71.47 B C
+ATOM 4788 NZ LYS H 132 40.608 -23.666 -86.270 1.00 73.18 B N1+
+ATOM 4789 N GLY H 133 38.166 -27.457 -88.538 1.00 65.97 B N
+ATOM 4790 CA GLY H 133 39.433 -28.063 -88.864 1.00 64.64 B C
+ATOM 4791 C GLY H 133 40.493 -27.691 -87.840 1.00 65.66 B C
+ATOM 4792 O GLY H 133 40.424 -26.640 -87.194 1.00 66.29 B O
+ATOM 4793 N PRO H 134 41.479 -28.565 -87.650 1.00 59.03 B N
+ATOM 4794 CA PRO H 134 42.598 -28.244 -86.757 1.00 57.88 B C
+ATOM 4795 C PRO H 134 43.594 -27.291 -87.391 1.00 62.36 B C
+ATOM 4796 O PRO H 134 43.909 -27.401 -88.577 1.00 65.21 B O
+ATOM 4797 CB PRO H 134 43.259 -29.604 -86.536 1.00 56.67 B C
+ATOM 4798 CG PRO H 134 42.971 -30.345 -87.767 1.00 59.97 B C
+ATOM 4799 CD PRO H 134 41.619 -29.897 -88.250 1.00 61.39 B C
+ATOM 4800 N SER H 135 44.123 -26.376 -86.580 1.00 61.83 B N
+ATOM 4801 CA SER H 135 45.355 -25.669 -86.921 1.00 62.14 B C
+ATOM 4802 C SER H 135 46.522 -26.512 -86.421 1.00 64.53 B C
+ATOM 4803 O SER H 135 46.516 -26.944 -85.271 1.00 69.21 B O
+ATOM 4804 CB SER H 135 45.398 -24.270 -86.292 1.00 60.14 B C
+ATOM 4805 OG SER H 135 44.348 -23.429 -86.750 1.00 62.09 B O
+ATOM 4806 N VAL H 136 47.512 -26.755 -87.278 1.00 65.66 B N
+ATOM 4807 CA VAL H 136 48.575 -27.719 -86.990 1.00 64.24 B C
+ATOM 4808 C VAL H 136 49.919 -27.007 -87.027 1.00 70.64 B C
+ATOM 4809 O VAL H 136 50.385 -26.606 -88.101 1.00 77.68 B O
+ATOM 4810 CB VAL H 136 48.559 -28.888 -87.978 1.00 60.36 B C
+ATOM 4811 CG1 VAL H 136 49.853 -29.680 -87.883 1.00 58.24 B C
+ATOM 4812 CG2 VAL H 136 47.333 -29.753 -87.739 1.00 58.66 B C
+ATOM 4813 N PHE H 137 50.575 -26.905 -85.874 1.00 80.01 B N
+ATOM 4814 CA PHE H 137 51.816 -26.158 -85.791 1.00 75.10 B C
+ATOM 4815 C PHE H 137 52.960 -27.092 -85.450 1.00 76.23 B C
+ATOM 4816 O PHE H 137 52.760 -28.050 -84.705 1.00 76.45 B O
+ATOM 4817 CB PHE H 137 51.718 -25.096 -84.709 1.00 70.79 B C
+ATOM 4818 CG PHE H 137 50.477 -24.330 -84.776 1.00 70.97 B C
+ATOM 4819 CD1 PHE H 137 50.272 -23.426 -85.790 1.00 71.56 B C
+ATOM 4820 CD2 PHE H 137 49.496 -24.512 -83.829 1.00 71.78 B C
+ATOM 4821 CE1 PHE H 137 49.097 -22.708 -85.868 1.00 71.82 B C
+ATOM 4822 CE2 PHE H 137 48.313 -23.801 -83.890 1.00 72.15 B C
+ATOM 4823 CZ PHE H 137 48.113 -22.891 -84.909 1.00 71.90 B C
+ATOM 4824 N PRO H 138 54.166 -26.847 -85.965 1.00 81.08 B N
+ATOM 4825 CA PRO H 138 55.266 -27.803 -85.757 1.00 81.77 B C
+ATOM 4826 C PRO H 138 55.916 -27.685 -84.380 1.00 79.81 B C
+ATOM 4827 O PRO H 138 56.091 -26.589 -83.840 1.00 80.25 B O
+ATOM 4828 CB PRO H 138 56.259 -27.438 -86.865 1.00 82.19 B C
+ATOM 4829 CG PRO H 138 56.034 -25.981 -87.066 1.00 82.69 B C
+ATOM 4830 CD PRO H 138 54.574 -25.703 -86.792 1.00 81.71 B C
+ATOM 4831 N LEU H 139 56.306 -28.845 -83.827 1.00 78.61 B N
+ATOM 4832 CA LEU H 139 57.067 -28.947 -82.581 1.00 75.70 B C
+ATOM 4833 C LEU H 139 58.511 -29.310 -82.934 1.00 81.07 B C
+ATOM 4834 O LEU H 139 58.801 -30.448 -83.332 1.00 85.99 B O
+ATOM 4835 CB LEU H 139 56.435 -29.970 -81.646 1.00 76.00 B C
+ATOM 4836 CG LEU H 139 55.067 -29.491 -81.175 1.00 75.16 B C
+ATOM 4837 CD1 LEU H 139 54.447 -30.518 -80.266 1.00 67.93 B C
+ATOM 4838 CD2 LEU H 139 55.164 -28.135 -80.488 1.00 71.46 B C
+ATOM 4839 N ALA H 140 59.423 -28.329 -82.756 1.00 81.43 B N
+ATOM 4840 CA ALA H 140 60.730 -28.221 -83.392 1.00 81.43 B C
+ATOM 4841 C ALA H 140 61.851 -28.710 -82.487 1.00 81.43 B C
+ATOM 4842 O ALA H 140 61.898 -28.320 -81.306 1.00 81.43 B O
+ATOM 4843 CB ALA H 140 60.977 -26.770 -83.782 1.00 81.43 B C
+ATOM 4844 N PRO H 141 62.772 -29.555 -83.006 1.00 72.68 B N
+ATOM 4845 CA PRO H 141 64.018 -29.990 -82.331 1.00 76.72 B C
+ATOM 4846 C PRO H 141 64.984 -28.846 -81.865 1.00 82.91 B C
+ATOM 4847 O PRO H 141 65.647 -28.860 -80.795 1.00 82.58 B O
+ATOM 4848 CB PRO H 141 64.696 -30.888 -83.397 1.00 75.85 B C
+ATOM 4849 CG PRO H 141 63.872 -30.775 -84.650 1.00 74.63 B C
+ATOM 4850 CD PRO H 141 62.518 -30.310 -84.248 1.00 71.60 B C
+ATOM 4851 N GLY H 149 70.926 -37.392 -76.822 1.00109.78 B N
+ATOM 4852 CA GLY H 149 71.668 -38.016 -77.907 1.00103.08 B C
+ATOM 4853 C GLY H 149 70.772 -38.397 -79.069 1.00 92.70 B C
+ATOM 4854 O GLY H 149 71.222 -38.597 -80.202 1.00 92.48 B O
+ATOM 4855 N THR H 150 69.483 -38.513 -78.767 1.00119.37 B N
+ATOM 4856 CA THR H 150 68.435 -38.645 -79.766 1.00106.19 B C
+ATOM 4857 C THR H 150 67.416 -37.527 -79.561 1.00 92.35 B C
+ATOM 4858 O THR H 150 67.213 -37.046 -78.436 1.00 91.24 B O
+ATOM 4859 CB THR H 150 67.764 -40.009 -79.682 1.00105.69 B C
+ATOM 4860 CG2 THR H 150 68.741 -41.103 -80.125 1.00106.85 B C
+ATOM 4861 OG1 THR H 150 67.343 -40.242 -78.332 1.00101.91 B O
+ATOM 4862 N ALA H 151 66.785 -37.101 -80.659 1.00102.35 B N
+ATOM 4863 CA ALA H 151 66.041 -35.847 -80.692 1.00 92.41 B C
+ATOM 4864 C ALA H 151 64.541 -36.084 -80.819 1.00 90.57 B C
+ATOM 4865 O ALA H 151 64.091 -36.946 -81.587 1.00 90.88 B O
+ATOM 4866 CB ALA H 151 66.529 -34.959 -81.840 1.00 82.20 B C
+ATOM 4867 N ALA H 152 63.773 -35.297 -80.066 1.00 83.29 B N
+ATOM 4868 CA ALA H 152 62.325 -35.415 -80.024 1.00 78.15 B C
+ATOM 4869 C ALA H 152 61.691 -34.224 -80.725 1.00 78.87 B C
+ATOM 4870 O ALA H 152 62.039 -33.065 -80.444 1.00 78.14 B O
+ATOM 4871 CB ALA H 152 61.820 -35.510 -78.586 1.00 79.12 B C
+ATOM 4872 N LEU H 153 60.766 -34.528 -81.641 1.00 75.92 B N
+ATOM 4873 CA LEU H 153 60.034 -33.541 -82.426 1.00 76.60 B C
+ATOM 4874 C LEU H 153 58.618 -34.050 -82.626 1.00 78.77 B C
+ATOM 4875 O LEU H 153 58.318 -35.215 -82.379 1.00 79.77 B O
+ATOM 4876 CB LEU H 153 60.703 -33.266 -83.779 1.00 77.15 B C
+ATOM 4877 CG LEU H 153 60.910 -34.451 -84.730 1.00 81.93 B C
+ATOM 4878 CD1 LEU H 153 59.654 -34.863 -85.504 1.00 83.31 B C
+ATOM 4879 CD2 LEU H 153 62.080 -34.148 -85.670 1.00 82.68 B C
+ATOM 4880 N GLY H 154 57.749 -33.172 -83.092 1.00 78.87 B N
+ATOM 4881 CA GLY H 154 56.355 -33.531 -83.198 1.00 78.35 B C
+ATOM 4882 C GLY H 154 55.582 -32.438 -83.880 1.00 80.24 B C
+ATOM 4883 O GLY H 154 56.151 -31.615 -84.595 1.00 82.86 B O
+ATOM 4884 N CYS H 155 54.269 -32.466 -83.687 1.00 79.99 B N
+ATOM 4885 CA CYS H 155 53.404 -31.403 -84.168 1.00 80.51 B C
+ATOM 4886 C CYS H 155 52.177 -31.355 -83.276 1.00 79.30 B C
+ATOM 4887 O CYS H 155 51.779 -32.354 -82.670 1.00 79.36 B O
+ATOM 4888 CB CYS H 155 53.026 -31.556 -85.661 1.00 70.11 B C
+ATOM 4889 SG CYS H 155 52.387 -33.195 -86.136 1.00 77.29 B S
+ATOM 4890 N LEU H 156 51.616 -30.160 -83.186 1.00 68.08 B N
+ATOM 4891 CA LEU H 156 50.587 -29.784 -82.232 1.00 65.67 B C
+ATOM 4892 C LEU H 156 49.330 -29.573 -83.041 1.00 67.35 B C
+ATOM 4893 O LEU H 156 49.283 -28.666 -83.873 1.00 68.98 B O
+ATOM 4894 CB LEU H 156 50.976 -28.494 -81.515 1.00 63.54 B C
+ATOM 4895 CG LEU H 156 50.266 -27.992 -80.278 1.00 61.13 B C
+ATOM 4896 CD1 LEU H 156 50.914 -26.685 -79.879 1.00 61.81 B C
+ATOM 4897 CD2 LEU H 156 48.827 -27.771 -80.535 1.00 62.89 B C
+ATOM 4898 N VAL H 157 48.309 -30.356 -82.769 1.00 58.58 B N
+ATOM 4899 CA VAL H 157 47.038 -30.255 -83.467 1.00 57.54 B C
+ATOM 4900 C VAL H 157 46.092 -29.477 -82.558 1.00 58.89 B C
+ATOM 4901 O VAL H 157 45.584 -30.015 -81.566 1.00 58.60 B O
+ATOM 4902 CB VAL H 157 46.498 -31.646 -83.799 1.00 59.57 B C
+ATOM 4903 CG1 VAL H 157 45.224 -31.552 -84.585 1.00 61.64 B C
+ATOM 4904 CG2 VAL H 157 47.561 -32.435 -84.553 1.00 57.02 B C
+ATOM 4905 N LYS H 158 45.823 -28.211 -82.890 1.00 59.02 B N
+ATOM 4906 CA LYS H 158 45.200 -27.283 -81.956 1.00 58.21 B C
+ATOM 4907 C LYS H 158 43.885 -26.767 -82.502 1.00 58.35 B C
+ATOM 4908 O LYS H 158 43.721 -26.601 -83.718 1.00 58.93 B O
+ATOM 4909 CB LYS H 158 46.099 -26.093 -81.657 1.00 63.15 B C
+ATOM 4910 CG LYS H 158 45.687 -25.335 -80.425 1.00 67.45 B C
+ATOM 4911 CD LYS H 158 46.612 -24.165 -80.226 1.00 67.02 B C
+ATOM 4912 CE LYS H 158 46.368 -23.461 -78.895 1.00 68.15 B C
+ATOM 4913 NZ LYS H 158 44.943 -23.149 -78.610 1.00 69.26 B N1+
+ATOM 4914 N ASP H 159 42.956 -26.514 -81.579 1.00 65.08 B N
+ATOM 4915 CA ASP H 159 41.676 -25.881 -81.893 1.00 64.52 B C
+ATOM 4916 C ASP H 159 40.993 -26.589 -83.065 1.00 68.57 B C
+ATOM 4917 O ASP H 159 40.992 -26.126 -84.209 1.00 70.44 B O
+ATOM 4918 CB ASP H 159 41.866 -24.386 -82.162 1.00 63.95 B C
+ATOM 4919 CG ASP H 159 42.289 -23.629 -80.918 1.00 69.25 B C
+ATOM 4920 OD1 ASP H 159 41.792 -23.959 -79.809 1.00 66.50 B O
+ATOM 4921 OD2 ASP H 159 43.127 -22.713 -81.054 1.00 69.97 B O1-
+ATOM 4922 N TYR H 160 40.482 -27.779 -82.742 1.00 65.85 B N
+ATOM 4923 CA TYR H 160 39.686 -28.586 -83.653 1.00 66.07 B C
+ATOM 4924 C TYR H 160 38.513 -29.160 -82.870 1.00 64.57 B C
+ATOM 4925 O TYR H 160 38.482 -29.140 -81.633 1.00 63.17 B O
+ATOM 4926 CB TYR H 160 40.515 -29.708 -84.295 1.00 69.59 B C
+ATOM 4927 CG TYR H 160 40.766 -30.860 -83.355 1.00 68.33 B C
+ATOM 4928 CD1 TYR H 160 39.959 -31.988 -83.392 1.00 67.67 B C
+ATOM 4929 CD2 TYR H 160 41.769 -30.796 -82.389 1.00 64.37 B C
+ATOM 4930 CE1 TYR H 160 40.160 -33.032 -82.522 1.00 64.79 B C
+ATOM 4931 CE2 TYR H 160 41.974 -31.838 -81.510 1.00 66.70 B C
+ATOM 4932 CZ TYR H 160 41.159 -32.953 -81.580 1.00 66.86 B C
+ATOM 4933 OH TYR H 160 41.335 -34.012 -80.722 1.00 69.17 B O
+ATOM 4934 N PHE H 161 37.557 -29.701 -83.609 1.00 59.92 B N
+ATOM 4935 CA PHE H 161 36.274 -30.094 -83.050 1.00 60.09 B C
+ATOM 4936 C PHE H 161 35.426 -30.732 -84.143 1.00 62.29 B C
+ATOM 4937 O PHE H 161 35.524 -30.352 -85.296 1.00 65.88 B O
+ATOM 4938 CB PHE H 161 35.556 -28.881 -82.469 1.00 61.46 B C
+ATOM 4939 CG PHE H 161 34.299 -29.217 -81.759 1.00 64.04 B C
+ATOM 4940 CD1 PHE H 161 34.296 -29.396 -80.391 1.00 61.37 B C
+ATOM 4941 CD2 PHE H 161 33.118 -29.368 -82.453 1.00 65.56 B C
+ATOM 4942 CE1 PHE H 161 33.122 -29.706 -79.725 1.00 61.02 B C
+ATOM 4943 CE2 PHE H 161 31.962 -29.685 -81.802 1.00 65.29 B C
+ATOM 4944 CZ PHE H 161 31.959 -29.854 -80.440 1.00 63.11 B C
+ATOM 4945 N PRO H 162 34.626 -31.739 -83.796 1.00 63.84 B N
+ATOM 4946 CA PRO H 162 34.733 -32.431 -82.520 1.00 62.16 B C
+ATOM 4947 C PRO H 162 35.706 -33.594 -82.718 1.00 62.88 B C
+ATOM 4948 O PRO H 162 36.155 -33.797 -83.869 1.00 64.91 B O
+ATOM 4949 CB PRO H 162 33.307 -32.889 -82.268 1.00 64.42 B C
+ATOM 4950 CG PRO H 162 32.807 -33.195 -83.667 1.00 59.66 B C
+ATOM 4951 CD PRO H 162 33.532 -32.281 -84.618 1.00 64.20 B C
+ATOM 4952 N GLU H 163 36.028 -34.339 -81.660 1.00 60.86 B N
+ATOM 4953 CA GLU H 163 36.986 -35.419 -81.820 1.00 61.42 B C
+ATOM 4954 C GLU H 163 36.313 -36.444 -82.682 1.00 64.14 B C
+ATOM 4955 O GLU H 163 35.105 -36.458 -82.754 1.00 65.11 B O
+ATOM 4956 CB GLU H 163 37.416 -35.978 -80.474 1.00 58.64 B C
+ATOM 4957 CG GLU H 163 38.915 -35.861 -80.283 1.00 62.88 B C
+ATOM 4958 CD GLU H 163 39.422 -36.562 -79.045 1.00 64.52 B C
+ATOM 4959 OE1 GLU H 163 38.686 -36.545 -78.021 1.00 64.85 B O
+ATOM 4960 OE2 GLU H 163 40.557 -37.119 -79.100 1.00 66.16 B O1-
+ATOM 4961 N PRO H 164 37.064 -37.337 -83.319 1.00 64.90 B N
+ATOM 4962 CA PRO H 164 38.476 -37.706 -83.323 1.00 63.13 B C
+ATOM 4963 C PRO H 164 39.354 -37.069 -84.367 1.00 65.37 B C
+ATOM 4964 O PRO H 164 38.890 -36.523 -85.359 1.00 64.88 B O
+ATOM 4965 CB PRO H 164 38.413 -39.175 -83.667 1.00 57.21 B C
+ATOM 4966 CG PRO H 164 37.330 -39.185 -84.710 1.00 58.63 B C
+ATOM 4967 CD PRO H 164 36.305 -38.181 -84.248 1.00 60.52 B C
+ATOM 4968 N VAL H 165 40.644 -37.343 -84.222 1.00 62.38 B N
+ATOM 4969 CA VAL H 165 41.653 -36.936 -85.184 1.00 62.54 B C
+ATOM 4970 C VAL H 165 42.726 -38.007 -85.164 1.00 63.05 B C
+ATOM 4971 O VAL H 165 42.961 -38.647 -84.138 1.00 59.05 B O
+ATOM 4972 CB VAL H 165 42.196 -35.524 -84.848 1.00 62.36 B C
+ATOM 4973 CG1 VAL H 165 42.506 -35.429 -83.386 1.00 62.69 B C
+ATOM 4974 CG2 VAL H 165 43.457 -35.228 -85.643 1.00 64.27 B C
+ATOM 4975 N THR H 166 43.334 -38.252 -86.318 1.00 62.46 B N
+ATOM 4976 CA THR H 166 44.372 -39.269 -86.423 1.00 61.34 B C
+ATOM 4977 C THR H 166 45.628 -38.623 -86.983 1.00 61.54 B C
+ATOM 4978 O THR H 166 45.566 -37.933 -88.004 1.00 65.42 B O
+ATOM 4979 CB THR H 166 43.932 -40.449 -87.314 1.00 62.62 B C
+ATOM 4980 CG2 THR H 166 42.629 -41.055 -86.803 1.00 60.91 B C
+ATOM 4981 OG1 THR H 166 43.748 -40.013 -88.671 1.00 67.16 B O
+ATOM 4982 N VAL H 167 46.759 -38.824 -86.311 1.00 59.69 B N
+ATOM 4983 CA VAL H 167 48.059 -38.417 -86.827 1.00 61.82 B C
+ATOM 4984 C VAL H 167 48.822 -39.680 -87.171 1.00 66.72 B C
+ATOM 4985 O VAL H 167 48.865 -40.621 -86.365 1.00 70.63 B O
+ATOM 4986 CB VAL H 167 48.853 -37.561 -85.826 1.00 61.32 B C
+ATOM 4987 CG1 VAL H 167 50.207 -37.247 -86.393 1.00 59.80 B C
+ATOM 4988 CG2 VAL H 167 48.122 -36.269 -85.513 1.00 57.19 B C
+ATOM 4989 N SER H 168 49.370 -39.721 -88.385 1.00 74.26 B N
+ATOM 4990 CA SER H 168 50.345 -40.718 -88.800 1.00 74.98 B C
+ATOM 4991 C SER H 168 51.484 -39.978 -89.467 1.00 76.59 B C
+ATOM 4992 O SER H 168 51.261 -38.958 -90.130 1.00 75.21 B O
+ATOM 4993 CB SER H 168 49.754 -41.760 -89.752 1.00 76.10 B C
+ATOM 4994 OG SER H 168 49.006 -42.736 -89.033 1.00 79.37 B O
+ATOM 4995 N TRP H 169 52.701 -40.476 -89.270 1.00 78.76 B N
+ATOM 4996 CA TRP H 169 53.906 -39.725 -89.599 1.00 78.81 B C
+ATOM 4997 C TRP H 169 54.538 -40.216 -90.894 1.00 84.92 B C
+ATOM 4998 O TRP H 169 54.526 -41.416 -91.189 1.00 88.37 B O
+ATOM 4999 CB TRP H 169 54.920 -39.811 -88.462 1.00 73.06 B C
+ATOM 5000 CG TRP H 169 54.620 -38.853 -87.387 1.00 73.73 B C
+ATOM 5001 CD1 TRP H 169 53.817 -39.061 -86.315 1.00 73.84 B C
+ATOM 5002 CD2 TRP H 169 55.105 -37.518 -87.274 1.00 74.31 B C
+ATOM 5003 CE2 TRP H 169 54.555 -36.973 -86.101 1.00 73.21 B C
+ATOM 5004 CE3 TRP H 169 55.955 -36.732 -88.045 1.00 73.33 B C
+ATOM 5005 NE1 TRP H 169 53.768 -37.938 -85.534 1.00 72.65 B N
+ATOM 5006 CZ2 TRP H 169 54.832 -35.677 -85.675 1.00 71.32 B C
+ATOM 5007 CZ3 TRP H 169 56.231 -35.441 -87.618 1.00 73.20 B C
+ATOM 5008 CH2 TRP H 169 55.670 -34.928 -86.443 1.00 72.93 B C
+ATOM 5009 N ASN H 170 55.109 -39.273 -91.653 1.00 85.74 B N
+ATOM 5010 CA ASN H 170 55.732 -39.573 -92.937 1.00 88.17 B C
+ATOM 5011 C ASN H 170 54.812 -40.459 -93.777 1.00 88.64 B C
+ATOM 5012 O ASN H 170 55.218 -41.483 -94.331 1.00 92.14 B O
+ATOM 5013 CB ASN H 170 57.110 -40.212 -92.742 1.00 87.29 B C
+ATOM 5014 CG ASN H 170 58.204 -39.191 -92.449 1.00 86.17 B C
+ATOM 5015 ND2 ASN H 170 59.460 -39.596 -92.654 1.00 81.81 B N
+ATOM 5016 OD1 ASN H 170 57.929 -38.064 -92.025 1.00 89.68 B O
+ATOM 5017 N SER H 171 53.534 -40.062 -93.833 1.00 95.11 B N
+ATOM 5018 CA SER H 171 52.515 -40.743 -94.642 1.00 94.67 B C
+ATOM 5019 C SER H 171 52.434 -42.224 -94.293 1.00 95.22 B C
+ATOM 5020 O SER H 171 52.147 -43.064 -95.148 1.00 99.89 B O
+ATOM 5021 CB SER H 171 52.772 -40.547 -96.143 1.00 95.01 B C
+ATOM 5022 OG SER H 171 52.884 -39.166 -96.487 1.00 97.47 B O
+ATOM 5023 N GLY H 172 52.693 -42.545 -93.030 1.00 86.75 B N
+ATOM 5024 CA GLY H 172 52.718 -43.913 -92.566 1.00 86.87 B C
+ATOM 5025 C GLY H 172 54.087 -44.553 -92.479 1.00 91.01 B C
+ATOM 5026 O GLY H 172 54.178 -45.721 -92.069 1.00 95.01 B O
+ATOM 5027 N ALA H 173 55.158 -43.826 -92.816 1.00 91.27 B N
+ATOM 5028 CA ALA H 173 56.492 -44.427 -92.807 1.00 93.52 B C
+ATOM 5029 C ALA H 173 56.957 -44.676 -91.382 1.00 95.32 B C
+ATOM 5030 O ALA H 173 57.136 -45.829 -90.965 1.00 98.48 B O
+ATOM 5031 CB ALA H 173 57.502 -43.539 -93.542 1.00 95.26 B C
+ATOM 5032 N LEU H 174 57.086 -43.602 -90.601 1.00 86.83 B N
+ATOM 5033 CA LEU H 174 57.727 -43.638 -89.290 1.00 84.86 B C
+ATOM 5034 C LEU H 174 56.663 -44.064 -88.292 1.00 80.68 B C
+ATOM 5035 O LEU H 174 55.790 -43.278 -87.921 1.00 81.02 B O
+ATOM 5036 CB LEU H 174 58.293 -42.254 -88.988 1.00 85.62 B C
+ATOM 5037 CG LEU H 174 58.601 -41.717 -87.600 1.00 85.33 B C
+ATOM 5038 CD1 LEU H 174 59.643 -42.546 -86.866 1.00 86.68 B C
+ATOM 5039 CD2 LEU H 174 59.070 -40.294 -87.769 1.00 81.02 B C
+ATOM 5040 N THR H 175 56.721 -45.331 -87.879 1.00 82.11 B N
+ATOM 5041 CA THR H 175 55.716 -45.900 -86.994 1.00 82.56 B C
+ATOM 5042 C THR H 175 56.179 -46.061 -85.550 1.00 85.35 B C
+ATOM 5043 O THR H 175 55.353 -46.375 -84.685 1.00 89.01 B O
+ATOM 5044 CB THR H 175 55.261 -47.256 -87.542 1.00 84.23 B C
+ATOM 5045 CG2 THR H 175 54.658 -47.069 -88.919 1.00 87.11 B C
+ATOM 5046 OG1 THR H 175 56.388 -48.131 -87.652 1.00 84.69 B O
+ATOM 5047 N SER H 176 57.451 -45.812 -85.258 1.00 89.05 B N
+ATOM 5048 CA SER H 176 58.060 -46.227 -84.003 1.00 91.47 B C
+ATOM 5049 C SER H 176 58.356 -45.033 -83.102 1.00 91.48 B C
+ATOM 5050 O SER H 176 58.766 -43.961 -83.568 1.00 92.84 B O
+ATOM 5051 CB SER H 176 59.359 -46.979 -84.261 1.00 94.99 B C
+ATOM 5052 OG SER H 176 60.411 -46.035 -84.393 1.00 99.63 B O
+ATOM 5053 N GLY H 177 58.196 -45.252 -81.800 1.00 88.93 B N
+ATOM 5054 CA GLY H 177 58.414 -44.179 -80.846 1.00 84.09 B C
+ATOM 5055 C GLY H 177 57.556 -42.982 -81.166 1.00 83.02 B C
+ATOM 5056 O GLY H 177 58.038 -41.845 -81.137 1.00 83.11 B O
+ATOM 5057 N VAL H 178 56.299 -43.224 -81.516 1.00 76.19 B N
+ATOM 5058 CA VAL H 178 55.354 -42.183 -81.877 1.00 73.31 B C
+ATOM 5059 C VAL H 178 54.277 -42.168 -80.805 1.00 74.36 B C
+ATOM 5060 O VAL H 178 53.435 -43.070 -80.741 1.00 78.67 B O
+ATOM 5061 CB VAL H 178 54.746 -42.438 -83.258 1.00 71.87 B C
+ATOM 5062 CG1 VAL H 178 53.970 -41.224 -83.719 1.00 70.10 B C
+ATOM 5063 CG2 VAL H 178 55.818 -42.833 -84.247 1.00 72.22 B C
+ATOM 5064 N HIS H 179 54.279 -41.150 -79.971 1.00 66.99 B N
+ATOM 5065 CA HIS H 179 53.277 -41.004 -78.926 1.00 66.66 B C
+ATOM 5066 C HIS H 179 52.365 -39.850 -79.323 1.00 67.84 B C
+ATOM 5067 O HIS H 179 52.811 -38.698 -79.362 1.00 68.98 B O
+ATOM 5068 CB HIS H 179 53.917 -40.705 -77.571 1.00 70.79 B C
+ATOM 5069 CG HIS H 179 54.938 -41.703 -77.116 1.00 74.55 B C
+ATOM 5070 CD2 HIS H 179 56.271 -41.775 -77.341 1.00 75.97 B C
+ATOM 5071 ND1 HIS H 179 54.636 -42.737 -76.255 1.00 77.18 B N
+ATOM 5072 CE1 HIS H 179 55.735 -43.420 -75.994 1.00 75.46 B C
+ATOM 5073 NE2 HIS H 179 56.741 -42.856 -76.638 1.00 73.22 B N
+ATOM 5074 N THR H 180 51.092 -40.136 -79.583 1.00 64.14 B N
+ATOM 5075 CA THR H 180 50.123 -39.088 -79.882 1.00 62.06 B C
+ATOM 5076 C THR H 180 49.089 -39.094 -78.770 1.00 58.18 B C
+ATOM 5077 O THR H 180 48.446 -40.112 -78.514 1.00 60.44 B O
+ATOM 5078 CB THR H 180 49.458 -39.320 -81.229 1.00 61.12 B C
+ATOM 5079 CG2 THR H 180 50.497 -39.722 -82.275 1.00 60.12 B C
+ATOM 5080 OG1 THR H 180 48.579 -40.433 -81.092 1.00 62.34 B O
+ATOM 5081 N PHE H 181 48.928 -37.978 -78.131 1.00 59.10 B N
+ATOM 5082 CA PHE H 181 48.331 -37.943 -76.811 1.00 58.77 B C
+ATOM 5083 C PHE H 181 46.814 -37.927 -76.859 1.00 57.50 B C
+ATOM 5084 O PHE H 181 46.217 -37.732 -77.916 1.00 59.96 B O
+ATOM 5085 CB PHE H 181 48.900 -36.743 -76.074 1.00 53.42 B C
+ATOM 5086 CG PHE H 181 50.313 -36.976 -75.661 1.00 56.14 B C
+ATOM 5087 CD1 PHE H 181 51.331 -36.926 -76.594 1.00 56.99 B C
+ATOM 5088 CD2 PHE H 181 50.635 -37.216 -74.348 1.00 55.63 B C
+ATOM 5089 CE1 PHE H 181 52.628 -37.177 -76.240 1.00 57.36 B C
+ATOM 5090 CE2 PHE H 181 51.948 -37.437 -73.990 1.00 57.37 B C
+ATOM 5091 CZ PHE H 181 52.941 -37.416 -74.945 1.00 57.84 B C
+ATOM 5092 N PRO H 182 46.166 -38.231 -75.740 1.00 51.82 B N
+ATOM 5093 CA PRO H 182 44.748 -37.916 -75.616 1.00 52.31 B C
+ATOM 5094 C PRO H 182 44.529 -36.451 -75.892 1.00 56.63 B C
+ATOM 5095 O PRO H 182 45.297 -35.594 -75.442 1.00 63.29 B O
+ATOM 5096 CB PRO H 182 44.434 -38.259 -74.163 1.00 53.63 B C
+ATOM 5097 CG PRO H 182 45.312 -39.388 -73.896 1.00 56.47 B C
+ATOM 5098 CD PRO H 182 46.593 -39.164 -74.693 1.00 54.79 B C
+ATOM 5099 N ALA H 183 43.494 -36.172 -76.673 1.00 57.26 B N
+ATOM 5100 CA ALA H 183 43.046 -34.799 -76.776 1.00 53.28 B C
+ATOM 5101 C ALA H 183 42.704 -34.286 -75.391 1.00 57.82 B C
+ATOM 5102 O ALA H 183 42.301 -35.035 -74.503 1.00 61.59 B O
+ATOM 5103 CB ALA H 183 41.833 -34.687 -77.691 1.00 56.89 B C
+ATOM 5104 N VAL H 184 42.912 -33.017 -75.195 1.00 59.11 B N
+ATOM 5105 CA VAL H 184 42.407 -32.373 -74.006 1.00 57.60 B C
+ATOM 5106 C VAL H 184 41.394 -31.341 -74.468 1.00 54.06 B C
+ATOM 5107 O VAL H 184 41.431 -30.899 -75.619 1.00 54.82 B O
+ATOM 5108 CB VAL H 184 43.554 -31.761 -73.189 1.00 59.76 B C
+ATOM 5109 CG1 VAL H 184 44.240 -30.669 -73.966 1.00 65.00 B C
+ATOM 5110 CG2 VAL H 184 43.016 -31.231 -71.910 1.00 53.00 B C
+ATOM 5111 N LEU H 185 40.447 -31.001 -73.606 1.00 58.17 B N
+ATOM 5112 CA LEU H 185 39.352 -30.119 -73.994 1.00 60.36 B C
+ATOM 5113 C LEU H 185 39.638 -28.730 -73.459 1.00 64.65 B C
+ATOM 5114 O LEU H 185 39.588 -28.488 -72.248 1.00 65.63 B O
+ATOM 5115 CB LEU H 185 38.004 -30.635 -73.499 1.00 59.83 B C
+ATOM 5116 CG LEU H 185 36.776 -29.730 -73.632 1.00 61.51 B C
+ATOM 5117 CD1 LEU H 185 36.769 -28.937 -74.906 1.00 60.88 B C
+ATOM 5118 CD2 LEU H 185 35.572 -30.606 -73.612 1.00 65.58 B C
+ATOM 5119 N GLN H 186 39.897 -27.818 -74.373 1.00 72.48 B N
+ATOM 5120 CA GLN H 186 40.328 -26.503 -73.978 1.00 73.64 B C
+ATOM 5121 C GLN H 186 39.133 -25.756 -73.396 1.00 78.38 B C
+ATOM 5122 O GLN H 186 37.982 -26.167 -73.567 1.00 81.19 B O
+ATOM 5123 CB GLN H 186 40.949 -25.847 -75.198 1.00 70.25 B C
+ATOM 5124 CG GLN H 186 42.083 -24.929 -74.951 1.00 70.81 B C
+ATOM 5125 CD GLN H 186 42.977 -24.922 -76.190 1.00 72.18 B C
+ATOM 5126 NE2 GLN H 186 42.596 -25.740 -77.198 1.00 68.43 B N
+ATOM 5127 OE1 GLN H 186 44.033 -24.260 -76.221 1.00 73.89 B O
+ATOM 5128 N SER H 187 39.410 -24.694 -72.632 1.00 77.52 B N
+ATOM 5129 CA SER H 187 38.333 -23.904 -72.025 1.00 73.65 B C
+ATOM 5130 C SER H 187 37.403 -23.288 -73.065 1.00 72.96 B C
+ATOM 5131 O SER H 187 36.301 -22.860 -72.721 1.00 76.07 B O
+ATOM 5132 CB SER H 187 38.941 -22.820 -71.138 1.00 74.17 B C
+ATOM 5133 OG SER H 187 40.158 -22.348 -71.701 1.00 75.29 B O
+ATOM 5134 N SER H 188 37.810 -23.305 -74.328 1.00 71.55 B N
+ATOM 5135 CA SER H 188 37.140 -22.728 -75.480 1.00 69.94 B C
+ATOM 5136 C SER H 188 36.132 -23.667 -76.122 1.00 74.49 B C
+ATOM 5137 O SER H 188 35.718 -23.416 -77.256 1.00 76.87 B O
+ATOM 5138 CB SER H 188 38.183 -22.354 -76.532 1.00 65.98 B C
+ATOM 5139 OG SER H 188 38.777 -23.528 -77.097 1.00 68.37 B O
+ATOM 5140 N GLY H 189 35.824 -24.797 -75.494 1.00 68.36 B N
+ATOM 5141 CA GLY H 189 35.017 -25.803 -76.159 1.00 66.75 B C
+ATOM 5142 C GLY H 189 35.712 -26.518 -77.290 1.00 68.49 B C
+ATOM 5143 O GLY H 189 35.069 -27.305 -77.989 1.00 66.50 B O
+ATOM 5144 N LEU H 190 37.012 -26.286 -77.472 1.00 76.30 B N
+ATOM 5145 CA LEU H 190 37.785 -26.806 -78.595 1.00 71.34 B C
+ATOM 5146 C LEU H 190 38.869 -27.772 -78.123 1.00 69.37 B C
+ATOM 5147 O LEU H 190 39.551 -27.524 -77.119 1.00 71.08 B O
+ATOM 5148 CB LEU H 190 38.434 -25.659 -79.366 1.00 69.72 B C
+ATOM 5149 CG LEU H 190 37.643 -25.203 -80.579 1.00 70.43 B C
+ATOM 5150 CD1 LEU H 190 37.856 -26.174 -81.698 1.00 65.65 B C
+ATOM 5151 CD2 LEU H 190 36.150 -25.091 -80.255 1.00 66.83 B C
+ATOM 5152 N TYR H 191 39.050 -28.863 -78.859 1.00 61.72 B N
+ATOM 5153 CA TYR H 191 40.106 -29.782 -78.475 1.00 64.58 B C
+ATOM 5154 C TYR H 191 41.474 -29.296 -78.936 1.00 65.30 B C
+ATOM 5155 O TYR H 191 41.613 -28.501 -79.866 1.00 65.49 B O
+ATOM 5156 CB TYR H 191 39.852 -31.175 -79.031 1.00 62.69 B C
+ATOM 5157 CG TYR H 191 38.647 -31.807 -78.408 1.00 65.84 B C
+ATOM 5158 CD1 TYR H 191 38.216 -31.399 -77.154 1.00 63.53 B C
+ATOM 5159 CD2 TYR H 191 37.946 -32.821 -79.052 1.00 66.17 B C
+ATOM 5160 CE1 TYR H 191 37.115 -31.957 -76.572 1.00 60.69 B C
+ATOM 5161 CE2 TYR H 191 36.837 -33.392 -78.471 1.00 64.45 B C
+ATOM 5162 CZ TYR H 191 36.431 -32.950 -77.230 1.00 60.48 B C
+ATOM 5163 OH TYR H 191 35.335 -33.500 -76.621 1.00 62.60 B O
+ATOM 5164 N SER H 192 42.499 -29.786 -78.257 1.00 56.62 B N
+ATOM 5165 CA SER H 192 43.853 -29.665 -78.759 1.00 55.53 B C
+ATOM 5166 C SER H 192 44.664 -30.849 -78.250 1.00 55.25 B C
+ATOM 5167 O SER H 192 44.440 -31.333 -77.138 1.00 60.27 B O
+ATOM 5168 CB SER H 192 44.471 -28.348 -78.318 1.00 62.29 B C
+ATOM 5169 OG SER H 192 45.868 -28.505 -78.269 1.00 68.00 B O
+ATOM 5170 N LEU H 193 45.587 -31.340 -79.072 1.00 53.08 B N
+ATOM 5171 CA LEU H 193 46.439 -32.444 -78.642 1.00 52.00 B C
+ATOM 5172 C LEU H 193 47.749 -32.400 -79.406 1.00 52.98 B C
+ATOM 5173 O LEU H 193 47.805 -31.939 -80.545 1.00 55.52 B O
+ATOM 5174 CB LEU H 193 45.803 -33.816 -78.871 1.00 56.12 B C
+ATOM 5175 CG LEU H 193 45.582 -34.258 -80.319 1.00 57.55 B C
+ATOM 5176 CD1 LEU H 193 46.790 -34.985 -80.915 1.00 53.83 B C
+ATOM 5177 CD2 LEU H 193 44.375 -35.141 -80.358 1.00 57.32 B C
+ATOM 5178 N SER H 194 48.786 -32.969 -78.802 1.00 55.97 B N
+ATOM 5179 CA SER H 194 50.110 -33.014 -79.396 1.00 55.45 B C
+ATOM 5180 C SER H 194 50.428 -34.438 -79.818 1.00 54.04 B C
+ATOM 5181 O SER H 194 49.892 -35.399 -79.261 1.00 56.48 B O
+ATOM 5182 CB SER H 194 51.140 -32.509 -78.404 1.00 62.07 B C
+ATOM 5183 OG SER H 194 50.727 -32.897 -77.107 1.00 76.59 B O
+ATOM 5184 N SER H 195 51.255 -34.569 -80.850 1.00 60.69 B N
+ATOM 5185 CA SER H 195 51.711 -35.877 -81.320 1.00 61.38 B C
+ATOM 5186 C SER H 195 53.198 -35.766 -81.584 1.00 63.91 B C
+ATOM 5187 O SER H 195 53.615 -34.934 -82.397 1.00 64.66 B O
+ATOM 5188 CB SER H 195 50.985 -36.329 -82.586 1.00 62.29 B C
+ATOM 5189 OG SER H 195 51.777 -37.270 -83.278 1.00 68.66 B O
+ATOM 5190 N VAL H 196 53.992 -36.590 -80.899 1.00 62.49 B N
+ATOM 5191 CA VAL H 196 55.441 -36.469 -80.928 1.00 64.11 B C
+ATOM 5192 C VAL H 196 56.067 -37.787 -81.342 1.00 69.44 B C
+ATOM 5193 O VAL H 196 55.464 -38.858 -81.262 1.00 73.82 B O
+ATOM 5194 CB VAL H 196 55.996 -36.038 -79.573 1.00 60.12 B C
+ATOM 5195 CG1 VAL H 196 55.390 -34.720 -79.188 1.00 55.88 B C
+ATOM 5196 CG2 VAL H 196 55.670 -37.085 -78.555 1.00 58.26 B C
+ATOM 5197 N VAL H 197 57.300 -37.684 -81.791 1.00 74.25 B N
+ATOM 5198 CA VAL H 197 58.103 -38.831 -82.158 1.00 73.63 B C
+ATOM 5199 C VAL H 197 59.537 -38.554 -81.727 1.00 77.75 B C
+ATOM 5200 O VAL H 197 60.036 -37.422 -81.818 1.00 80.53 B O
+ATOM 5201 CB VAL H 197 58.010 -39.150 -83.664 1.00 74.30 B C
+ATOM 5202 CG1 VAL H 197 58.760 -38.110 -84.483 1.00 74.86 B C
+ATOM 5203 CG2 VAL H 197 58.546 -40.543 -83.947 1.00 73.57 B C
+ATOM 5204 N THR H 198 60.160 -39.590 -81.187 1.00 84.18 B N
+ATOM 5205 CA THR H 198 61.575 -39.610 -80.876 1.00 86.54 B C
+ATOM 5206 C THR H 198 62.304 -40.204 -82.068 1.00 92.12 B C
+ATOM 5207 O THR H 198 61.880 -41.230 -82.610 1.00 92.77 B O
+ATOM 5208 CB THR H 198 61.809 -40.444 -79.617 1.00 84.58 B C
+ATOM 5209 CG2 THR H 198 61.287 -39.722 -78.403 1.00 81.62 B C
+ATOM 5210 OG1 THR H 198 61.133 -41.707 -79.749 1.00 88.54 B O
+ATOM 5211 N VAL H 199 63.375 -39.550 -82.498 1.00101.70 B N
+ATOM 5212 CA VAL H 199 64.054 -40.006 -83.705 1.00102.48 B C
+ATOM 5213 C VAL H 199 65.556 -39.795 -83.546 1.00103.83 B C
+ATOM 5214 O VAL H 199 65.968 -38.858 -82.838 1.00107.37 B O
+ATOM 5215 CB VAL H 199 63.487 -39.298 -84.947 1.00102.50 B C
+ATOM 5216 CG1 VAL H 199 64.166 -37.940 -85.200 1.00101.38 B C
+ATOM 5217 CG2 VAL H 199 63.554 -40.217 -86.154 1.00 99.30 B C
+ATOM 5218 N PRO H 200 66.400 -40.656 -84.134 1.00103.56 B N
+ATOM 5219 CA PRO H 200 67.857 -40.476 -84.000 1.00106.66 B C
+ATOM 5220 C PRO H 200 68.335 -39.139 -84.562 1.00107.40 B C
+ATOM 5221 O PRO H 200 68.008 -38.764 -85.695 1.00109.66 B O
+ATOM 5222 CB PRO H 200 68.424 -41.664 -84.784 1.00109.22 B C
+ATOM 5223 CG PRO H 200 67.403 -42.733 -84.567 1.00106.95 B C
+ATOM 5224 CD PRO H 200 66.070 -42.017 -84.606 1.00101.81 B C
+ATOM 5225 N SER H 201 69.130 -38.424 -83.749 1.00 96.53 B N
+ATOM 5226 CA SER H 201 69.491 -37.042 -84.045 1.00101.23 B C
+ATOM 5227 C SER H 201 70.499 -36.915 -85.179 1.00101.06 B C
+ATOM 5228 O SER H 201 70.690 -35.804 -85.692 1.00104.47 B O
+ATOM 5229 CB SER H 201 70.036 -36.351 -82.794 1.00107.54 B C
+ATOM 5230 OG SER H 201 70.330 -34.983 -83.057 1.00113.70 B O
+ATOM 5231 N SER H 202 71.151 -38.010 -85.577 1.00111.85 B N
+ATOM 5232 CA SER H 202 72.003 -37.964 -86.760 1.00115.33 B C
+ATOM 5233 C SER H 202 71.185 -37.930 -88.052 1.00117.18 B C
+ATOM 5234 O SER H 202 71.732 -37.609 -89.113 1.00120.51 B O
+ATOM 5235 CB SER H 202 72.985 -39.150 -86.757 1.00117.50 B C
+ATOM 5236 OG SER H 202 72.347 -40.392 -86.497 1.00118.52 B O
+ATOM 5237 N SER H 203 69.893 -38.246 -87.993 1.00112.78 B N
+ATOM 5238 CA SER H 203 69.028 -38.166 -89.158 1.00115.57 B C
+ATOM 5239 C SER H 203 68.232 -36.869 -89.228 1.00112.54 B C
+ATOM 5240 O SER H 203 67.377 -36.733 -90.108 1.00111.51 B O
+ATOM 5241 CB SER H 203 68.094 -39.369 -89.205 1.00120.80 B C
+ATOM 5242 OG SER H 203 68.736 -40.431 -89.885 1.00122.25 B O
+ATOM 5243 N LEU H 204 68.478 -35.921 -88.321 1.00109.00 B N
+ATOM 5244 CA LEU H 204 67.765 -34.645 -88.355 1.00109.73 B C
+ATOM 5245 C LEU H 204 67.897 -33.957 -89.712 1.00115.46 B C
+ATOM 5246 O LEU H 204 66.893 -33.656 -90.367 1.00117.90 B O
+ATOM 5247 CB LEU H 204 68.262 -33.747 -87.221 1.00105.63 B C
+ATOM 5248 CG LEU H 204 67.316 -33.670 -86.011 1.00101.62 B C
+ATOM 5249 CD1 LEU H 204 66.963 -35.066 -85.507 1.00 98.61 B C
+ATOM 5250 CD2 LEU H 204 67.932 -32.820 -84.875 1.00100.32 B C
+ATOM 5251 N GLY H 205 69.130 -33.716 -90.159 1.00 96.74 B N
+ATOM 5252 CA GLY H 205 69.350 -33.141 -91.476 1.00102.97 B C
+ATOM 5253 C GLY H 205 69.336 -34.131 -92.625 1.00107.20 B C
+ATOM 5254 O GLY H 205 69.346 -33.712 -93.785 1.00108.83 B O
+ATOM 5255 N THR H 206 69.314 -35.435 -92.329 1.00112.64 B N
+ATOM 5256 CA THR H 206 69.235 -36.461 -93.373 1.00115.03 B C
+ATOM 5257 C THR H 206 67.790 -36.639 -93.829 1.00112.25 B C
+ATOM 5258 O THR H 206 67.410 -36.202 -94.922 1.00113.55 B O
+ATOM 5259 CB THR H 206 69.797 -37.799 -92.860 1.00119.28 B C
+ATOM 5260 CG2 THR H 206 69.478 -38.948 -93.836 1.00123.48 B C
+ATOM 5261 OG1 THR H 206 71.218 -37.710 -92.703 1.00122.11 B O
+ATOM 5262 N GLN H 207 66.968 -37.267 -92.986 1.00114.92 B N
+ATOM 5263 CA GLN H 207 65.579 -37.504 -93.338 1.00107.19 B C
+ATOM 5264 C GLN H 207 64.718 -36.280 -93.015 1.00101.71 B C
+ATOM 5265 O GLN H 207 65.007 -35.500 -92.095 1.00 99.19 B O
+ATOM 5266 CB GLN H 207 65.040 -38.764 -92.649 1.00106.87 B C
+ATOM 5267 CG GLN H 207 63.607 -39.045 -93.058 1.00110.32 B C
+ATOM 5268 CD GLN H 207 63.330 -40.497 -93.270 1.00114.75 B C
+ATOM 5269 NE2 GLN H 207 62.336 -40.792 -94.107 1.00116.98 B N
+ATOM 5270 OE1 GLN H 207 64.019 -41.356 -92.724 1.00117.29 B O
+ATOM 5271 N THR H 208 63.682 -36.094 -93.836 1.00102.83 B N
+ATOM 5272 CA THR H 208 62.698 -35.030 -93.694 1.00103.62 B C
+ATOM 5273 C THR H 208 61.430 -35.602 -93.066 1.00100.32 B C
+ATOM 5274 O THR H 208 60.872 -36.582 -93.575 1.00 99.12 B O
+ATOM 5275 CB THR H 208 62.396 -34.432 -95.062 1.00107.86 B C
+ATOM 5276 CG2 THR H 208 63.634 -33.711 -95.595 1.00110.07 B C
+ATOM 5277 OG1 THR H 208 62.065 -35.500 -95.956 1.00108.00 B O
+ATOM 5278 N TYR H 209 60.982 -34.993 -91.956 1.00100.46 B N
+ATOM 5279 CA TYR H 209 59.895 -35.519 -91.128 1.00 93.84 B C
+ATOM 5280 C TYR H 209 58.620 -34.703 -91.343 1.00 90.95 B C
+ATOM 5281 O TYR H 209 58.574 -33.509 -91.009 1.00 90.63 B O
+ATOM 5282 CB TYR H 209 60.307 -35.528 -89.655 1.00 91.64 B C
+ATOM 5283 CG TYR H 209 61.430 -36.499 -89.391 1.00 92.11 B C
+ATOM 5284 CD1 TYR H 209 61.235 -37.866 -89.562 1.00 91.31 B C
+ATOM 5285 CD2 TYR H 209 62.684 -36.063 -88.986 1.00 92.77 B C
+ATOM 5286 CE1 TYR H 209 62.256 -38.780 -89.347 1.00 93.58 B C
+ATOM 5287 CE2 TYR H 209 63.717 -36.974 -88.763 1.00 93.43 B C
+ATOM 5288 CZ TYR H 209 63.495 -38.335 -88.948 1.00 95.05 B C
+ATOM 5289 OH TYR H 209 64.508 -39.254 -88.740 1.00 95.87 B O
+ATOM 5290 N ILE H 210 57.581 -35.360 -91.876 1.00 87.54 B N
+ATOM 5291 CA ILE H 210 56.293 -34.729 -92.177 1.00 87.28 B C
+ATOM 5292 C ILE H 210 55.189 -35.364 -91.328 1.00 85.61 B C
+ATOM 5293 O ILE H 210 54.911 -36.573 -91.408 1.00 87.70 B O
+ATOM 5294 CB ILE H 210 55.966 -34.750 -93.685 1.00 88.54 B C
+ATOM 5295 CG1 ILE H 210 55.884 -36.189 -94.243 1.00 92.11 B C
+ATOM 5296 CG2 ILE H 210 56.949 -33.840 -94.462 1.00 85.42 B C
+ATOM 5297 CD1 ILE H 210 55.257 -36.336 -95.656 1.00 94.37 B C
+ATOM 5298 N CYS H 211 54.636 -34.528 -90.461 1.00 83.39 B N
+ATOM 5299 CA CYS H 211 53.411 -34.729 -89.709 1.00 81.99 B C
+ATOM 5300 C CYS H 211 52.217 -34.944 -90.633 1.00 81.29 B C
+ATOM 5301 O CYS H 211 52.026 -34.181 -91.587 1.00 81.11 B O
+ATOM 5302 CB CYS H 211 53.256 -33.470 -88.856 1.00 76.90 B C
+ATOM 5303 SG CYS H 211 51.766 -33.173 -88.053 1.00 83.11 B S
+ATOM 5304 N ASN H 212 51.385 -35.942 -90.339 1.00 76.50 B N
+ATOM 5305 CA ASN H 212 50.274 -36.240 -91.233 1.00 80.33 B C
+ATOM 5306 C ASN H 212 49.002 -36.253 -90.399 1.00 81.29 B C
+ATOM 5307 O ASN H 212 48.764 -37.216 -89.661 1.00 84.61 B O
+ATOM 5308 CB ASN H 212 50.508 -37.596 -91.891 1.00 81.53 B C
+ATOM 5309 CG ASN H 212 50.251 -37.583 -93.352 1.00 82.03 B C
+ATOM 5310 ND2 ASN H 212 51.326 -37.687 -94.138 1.00 81.35 B N
+ATOM 5311 OD1 ASN H 212 49.108 -37.479 -93.793 1.00 81.58 B O
+ATOM 5312 N VAL H 213 48.146 -35.234 -90.546 1.00 69.94 B N
+ATOM 5313 CA VAL H 213 46.934 -35.149 -89.722 1.00 68.91 B C
+ATOM 5314 C VAL H 213 45.746 -35.647 -90.534 1.00 71.22 B C
+ATOM 5315 O VAL H 213 45.788 -35.735 -91.764 1.00 75.21 B O
+ATOM 5316 CB VAL H 213 46.679 -33.702 -89.217 1.00 64.42 B C
+ATOM 5317 CG1 VAL H 213 45.628 -33.660 -88.101 1.00 66.61 B C
+ATOM 5318 CG2 VAL H 213 47.955 -33.082 -88.747 1.00 58.44 B C
+ATOM 5319 N ASN H 214 44.646 -35.937 -89.842 1.00 65.95 B N
+ATOM 5320 CA ASN H 214 43.377 -36.143 -90.513 1.00 68.48 B C
+ATOM 5321 C ASN H 214 42.254 -35.916 -89.525 1.00 69.43 B C
+ATOM 5322 O ASN H 214 42.299 -36.427 -88.405 1.00 70.52 B O
+ATOM 5323 CB ASN H 214 43.287 -37.542 -91.109 1.00 71.01 B C
+ATOM 5324 CG ASN H 214 42.434 -37.568 -92.339 1.00 76.90 B C
+ATOM 5325 ND2 ASN H 214 42.942 -38.189 -93.406 1.00 82.35 B N
+ATOM 5326 OD1 ASN H 214 41.339 -36.991 -92.354 1.00 77.99 B O
+ATOM 5327 N HIS H 215 41.222 -35.212 -89.968 1.00 72.38 B N
+ATOM 5328 CA HIS H 215 40.032 -34.958 -89.161 1.00 68.48 B C
+ATOM 5329 C HIS H 215 38.860 -35.334 -90.050 1.00 70.78 B C
+ATOM 5330 O HIS H 215 38.552 -34.608 -91.000 1.00 71.11 B O
+ATOM 5331 CB HIS H 215 39.962 -33.479 -88.752 1.00 59.05 B C
+ATOM 5332 CG HIS H 215 38.942 -33.152 -87.700 1.00 63.87 B C
+ATOM 5333 CD2 HIS H 215 38.409 -33.890 -86.699 1.00 63.13 B C
+ATOM 5334 ND1 HIS H 215 38.426 -31.882 -87.547 1.00 64.99 B N
+ATOM 5335 CE1 HIS H 215 37.587 -31.864 -86.526 1.00 65.35 B C
+ATOM 5336 NE2 HIS H 215 37.559 -33.068 -85.990 1.00 64.70 B N
+ATOM 5337 N LYS H 216 38.184 -36.440 -89.755 1.00 60.18 B N
+ATOM 5338 CA LYS H 216 37.046 -36.772 -90.604 1.00 65.81 B C
+ATOM 5339 C LYS H 216 35.927 -35.758 -90.425 1.00 66.32 B C
+ATOM 5340 O LYS H 216 35.431 -35.240 -91.441 1.00 70.73 B O
+ATOM 5341 CB LYS H 216 36.597 -38.210 -90.357 1.00 67.62 B C
+ATOM 5342 CG LYS H 216 36.912 -39.152 -91.521 1.00 72.03 B C
+ATOM 5343 CD LYS H 216 38.284 -39.834 -91.377 1.00 77.11 B C
+ATOM 5344 CE LYS H 216 38.427 -41.084 -92.263 1.00 80.72 B C
+ATOM 5345 NZ LYS H 216 39.794 -41.677 -92.158 1.00 82.88 B N1+
+ATOM 5346 N PRO H 217 35.523 -35.378 -89.205 1.00 68.49 B N
+ATOM 5347 CA PRO H 217 34.373 -34.458 -89.069 1.00 64.67 B C
+ATOM 5348 C PRO H 217 34.486 -33.150 -89.832 1.00 68.98 B C
+ATOM 5349 O PRO H 217 33.448 -32.532 -90.085 1.00 71.64 B O
+ATOM 5350 CB PRO H 217 34.297 -34.212 -87.558 1.00 63.41 B C
+ATOM 5351 CG PRO H 217 34.770 -35.480 -86.976 1.00 63.24 B C
+ATOM 5352 CD PRO H 217 35.869 -35.986 -87.908 1.00 64.81 B C
+ATOM 5353 N SER H 218 35.682 -32.661 -90.147 1.00 65.04 B N
+ATOM 5354 CA SER H 218 35.789 -31.480 -90.998 1.00 66.81 B C
+ATOM 5355 C SER H 218 36.171 -31.794 -92.440 1.00 71.57 B C
+ATOM 5356 O SER H 218 36.259 -30.868 -93.248 1.00 71.01 B O
+ATOM 5357 CB SER H 218 36.777 -30.461 -90.398 1.00 65.01 B C
+ATOM 5358 OG SER H 218 38.074 -31.004 -90.218 1.00 66.13 B O
+ATOM 5359 N ASN H 219 36.390 -33.064 -92.780 1.00 58.50 B N
+ATOM 5360 CA ASN H 219 37.005 -33.464 -94.055 1.00 62.40 B C
+ATOM 5361 C ASN H 219 38.324 -32.744 -94.310 1.00 61.31 B C
+ATOM 5362 O ASN H 219 38.674 -32.420 -95.447 1.00 65.79 B O
+ATOM 5363 CB ASN H 219 36.068 -33.266 -95.241 1.00 67.20 B C
+ATOM 5364 CG ASN H 219 36.432 -34.166 -96.407 1.00 71.76 B C
+ATOM 5365 ND2 ASN H 219 36.451 -33.598 -97.611 1.00 73.58 B N
+ATOM 5366 OD1 ASN H 219 36.725 -35.356 -96.224 1.00 74.85 B O
+ATOM 5367 N THR H 220 39.082 -32.519 -93.250 1.00 68.52 B N
+ATOM 5368 CA THR H 220 40.388 -31.899 -93.363 1.00 65.23 B C
+ATOM 5369 C THR H 220 41.486 -32.947 -93.368 1.00 62.54 B C
+ATOM 5370 O THR H 220 41.450 -33.904 -92.593 1.00 62.78 B O
+ATOM 5371 CB THR H 220 40.625 -30.937 -92.211 1.00 64.97 B C
+ATOM 5372 CG2 THR H 220 41.760 -29.974 -92.563 1.00 58.15 B C
+ATOM 5373 OG1 THR H 220 39.416 -30.213 -91.958 1.00 67.91 B O
+ATOM 5374 N LYS H 221 42.460 -32.762 -94.250 1.00 63.35 B N
+ATOM 5375 CA LYS H 221 43.732 -33.467 -94.172 1.00 66.50 B C
+ATOM 5376 C LYS H 221 44.829 -32.419 -94.275 1.00 69.30 B C
+ATOM 5377 O LYS H 221 44.953 -31.750 -95.304 1.00 72.29 B O
+ATOM 5378 CB LYS H 221 43.864 -34.529 -95.263 1.00 70.35 B C
+ATOM 5379 CG LYS H 221 45.141 -35.359 -95.156 1.00 74.80 B C
+ATOM 5380 CD LYS H 221 44.970 -36.780 -95.737 1.00 79.54 B C
+ATOM 5381 CE LYS H 221 46.250 -37.633 -95.540 1.00 82.48 B C
+ATOM 5382 NZ LYS H 221 46.102 -39.079 -95.927 1.00 84.24 B N1+
+ATOM 5383 N VAL H 222 45.600 -32.266 -93.199 1.00 66.71 B N
+ATOM 5384 CA VAL H 222 46.728 -31.344 -93.121 1.00 66.07 B C
+ATOM 5385 C VAL H 222 48.009 -32.164 -93.071 1.00 66.28 B C
+ATOM 5386 O VAL H 222 48.098 -33.135 -92.308 1.00 67.38 B O
+ATOM 5387 CB VAL H 222 46.629 -30.440 -91.884 1.00 65.77 B C
+ATOM 5388 CG1 VAL H 222 47.845 -29.563 -91.782 1.00 65.98 B C
+ATOM 5389 CG2 VAL H 222 45.383 -29.610 -91.952 1.00 67.87 B C
+ATOM 5390 N ASP H 223 48.990 -31.795 -93.893 1.00 69.93 B N
+ATOM 5391 CA ASP H 223 50.350 -32.305 -93.781 1.00 72.29 B C
+ATOM 5392 C ASP H 223 51.235 -31.145 -93.365 1.00 76.99 B C
+ATOM 5393 O ASP H 223 51.216 -30.092 -94.011 1.00 81.24 B O
+ATOM 5394 CB ASP H 223 50.851 -32.897 -95.099 1.00 74.34 B C
+ATOM 5395 CG ASP H 223 50.208 -34.231 -95.427 1.00 77.66 B C
+ATOM 5396 OD1 ASP H 223 49.084 -34.490 -94.938 1.00 77.77 B O
+ATOM 5397 OD2 ASP H 223 50.819 -35.016 -96.197 1.00 80.69 B O1-
+ATOM 5398 N LYS H 224 51.984 -31.323 -92.279 1.00 85.12 B N
+ATOM 5399 CA LYS H 224 52.875 -30.282 -91.788 1.00 81.58 B C
+ATOM 5400 C LYS H 224 54.320 -30.762 -91.886 1.00 75.42 B C
+ATOM 5401 O LYS H 224 54.624 -31.931 -91.618 1.00 74.24 B O
+ATOM 5402 CB LYS H 224 52.531 -29.893 -90.356 1.00 84.88 B C
+ATOM 5403 CG LYS H 224 53.141 -28.584 -89.912 1.00 89.53 B C
+ATOM 5404 CD LYS H 224 52.737 -27.451 -90.833 1.00 95.72 B C
+ATOM 5405 CE LYS H 224 53.511 -26.171 -90.519 1.00100.38 B C
+ATOM 5406 NZ LYS H 224 53.131 -25.041 -91.422 1.00103.64 B N1+
+ATOM 5407 N LYS H 225 55.205 -29.871 -92.317 1.00 83.71 B N
+ATOM 5408 CA LYS H 225 56.619 -30.188 -92.390 1.00 84.81 B C
+ATOM 5409 C LYS H 225 57.267 -29.783 -91.075 1.00 82.44 B C
+ATOM 5410 O LYS H 225 56.939 -28.733 -90.508 1.00 85.19 B O
+ATOM 5411 CB LYS H 225 57.269 -29.471 -93.573 1.00 90.18 B C
+ATOM 5412 CG LYS H 225 58.275 -30.331 -94.345 1.00 96.96 B C
+ATOM 5413 CD LYS H 225 58.799 -29.641 -95.619 1.00103.49 B C
+ATOM 5414 CE LYS H 225 60.239 -30.044 -95.902 1.00109.87 B C
+ATOM 5415 NZ LYS H 225 60.401 -31.520 -95.984 1.00113.63 B N1+
+ATOM 5416 N VAL H 226 58.158 -30.640 -90.568 1.00 86.22 B N
+ATOM 5417 CA VAL H 226 58.788 -30.428 -89.265 1.00 86.59 B C
+ATOM 5418 C VAL H 226 60.306 -30.432 -89.430 1.00 90.11 B C
+ATOM 5419 O VAL H 226 60.899 -31.460 -89.792 1.00 90.77 B O
+ATOM 5420 CB VAL H 226 58.341 -31.473 -88.233 1.00 84.08 B C
+ATOM 5421 CG1 VAL H 226 58.831 -31.071 -86.839 1.00 81.80 B C
+ATOM 5422 CG2 VAL H 226 56.810 -31.634 -88.262 1.00 82.68 B C
+ATOM 5423 N GLU H 227 60.918 -29.284 -89.147 1.00 78.62 B N
+ATOM 5424 CA GLU H 227 62.331 -28.976 -89.305 1.00 85.24 B C
+ATOM 5425 C GLU H 227 62.786 -28.164 -88.101 1.00 81.46 B C
+ATOM 5426 O GLU H 227 61.968 -27.507 -87.445 1.00 82.90 B O
+ATOM 5427 CB GLU H 227 62.600 -28.162 -90.581 1.00 94.96 B C
+ATOM 5428 CG GLU H 227 61.806 -28.588 -91.805 1.00104.99 B C
+ATOM 5429 CD GLU H 227 61.648 -27.465 -92.822 1.00112.53 B C
+ATOM 5430 OE1 GLU H 227 61.925 -26.294 -92.467 1.00113.70 B O
+ATOM 5431 OE2 GLU H 227 61.253 -27.758 -93.974 1.00116.02 B O1-
+ATOM 5432 N PRO H 228 64.097 -28.175 -87.792 1.00 78.37 B N
+ATOM 5433 CA PRO H 228 64.690 -27.390 -86.684 1.00 84.15 B C
+ATOM 5434 C PRO H 228 64.552 -25.861 -86.793 1.00 92.08 B C
+ATOM 5435 O PRO H 228 64.914 -25.156 -85.843 1.00 93.07 B O
+ATOM 5436 CB PRO H 228 66.176 -27.784 -86.730 1.00 81.53 B C
+ATOM 5437 CG PRO H 228 66.363 -28.513 -88.060 1.00 80.67 B C
+ATOM 5438 CD PRO H 228 65.053 -29.146 -88.355 1.00 78.16 B C
+TER
+ATOM 5439 N GLU L 1 19.563 -45.257 -54.751 1.00122.44 C N
+ATOM 5440 CA GLU L 1 20.329 -46.343 -55.361 1.00110.26 C C
+ATOM 5441 C GLU L 1 21.521 -46.709 -54.507 1.00 98.29 C C
+ATOM 5442 O GLU L 1 22.020 -45.880 -53.746 1.00 98.59 C O
+ATOM 5443 CB GLU L 1 20.810 -45.960 -56.767 1.00 90.62 C C
+ATOM 5444 CG GLU L 1 22.245 -45.424 -56.897 1.00 90.62 C C
+ATOM 5445 CD GLU L 1 22.393 -44.034 -56.326 1.00 90.62 C C
+ATOM 5446 OE1 GLU L 1 21.344 -43.415 -56.004 1.00 90.62 C O
+ATOM 5447 OE2 GLU L 1 23.549 -43.553 -56.234 1.00 90.62 C O1-
+ATOM 5448 N ILE L 2 21.994 -47.942 -54.633 1.00 80.45 C N
+ATOM 5449 CA ILE L 2 23.112 -48.387 -53.815 1.00 72.28 C C
+ATOM 5450 C ILE L 2 24.372 -48.150 -54.607 1.00 72.52 C C
+ATOM 5451 O ILE L 2 24.387 -48.321 -55.826 1.00 74.39 C O
+ATOM 5452 CB ILE L 2 22.976 -49.864 -53.415 1.00 66.75 C C
+ATOM 5453 CG1 ILE L 2 22.773 -50.723 -54.629 1.00 66.75 C C
+ATOM 5454 CG2 ILE L 2 21.746 -50.068 -52.531 1.00 66.75 C C
+ATOM 5455 CD1 ILE L 2 22.403 -52.090 -54.251 1.00 66.75 C C
+ATOM 5456 N VAL L 3 25.428 -47.754 -53.918 1.00 63.33 C N
+ATOM 5457 CA VAL L 3 26.638 -47.250 -54.548 1.00 64.77 C C
+ATOM 5458 C VAL L 3 27.697 -48.330 -54.527 1.00 63.76 C C
+ATOM 5459 O VAL L 3 28.182 -48.698 -53.454 1.00 65.06 C O
+ATOM 5460 CB VAL L 3 27.153 -46.001 -53.826 1.00 50.28 C C
+ATOM 5461 CG1 VAL L 3 28.485 -45.591 -54.402 1.00 50.28 C C
+ATOM 5462 CG2 VAL L 3 26.135 -44.879 -53.920 1.00 50.28 C C
+ATOM 5463 N MET L 4 28.098 -48.815 -55.689 1.00 59.21 C N
+ATOM 5464 CA MET L 4 29.291 -49.637 -55.705 1.00 59.21 C C
+ATOM 5465 C MET L 4 30.511 -48.767 -55.513 1.00 63.13 C C
+ATOM 5466 O MET L 4 30.631 -47.712 -56.132 1.00 66.98 C O
+ATOM 5467 CB MET L 4 29.412 -50.405 -56.998 1.00 50.04 C C
+ATOM 5468 CG MET L 4 28.241 -51.292 -57.259 1.00 50.04 C C
+ATOM 5469 SD MET L 4 27.877 -52.313 -55.845 1.00 50.04 C S
+ATOM 5470 CE MET L 4 26.430 -51.454 -55.213 1.00 50.04 C C
+ATOM 5471 N THR L 5 31.407 -49.203 -54.646 1.00 57.01 C N
+ATOM 5472 CA THR L 5 32.651 -48.503 -54.380 1.00 53.42 C C
+ATOM 5473 C THR L 5 33.791 -49.398 -54.842 1.00 53.68 C C
+ATOM 5474 O THR L 5 34.162 -50.365 -54.162 1.00 53.36 C O
+ATOM 5475 CB THR L 5 32.757 -48.193 -52.911 1.00 48.57 C C
+ATOM 5476 CG2 THR L 5 33.761 -47.104 -52.715 1.00 48.57 C C
+ATOM 5477 OG1 THR L 5 31.478 -47.753 -52.438 1.00 48.57 C O
+ATOM 5478 N SER L 6 34.365 -49.045 -55.981 1.00 53.67 C N
+ATOM 5479 CA SER L 6 35.446 -49.825 -56.540 1.00 53.67 C C
+ATOM 5480 C SER L 6 36.692 -49.563 -55.705 1.00 55.07 C C
+ATOM 5481 O SER L 6 36.787 -48.537 -55.037 1.00 63.61 C O
+ATOM 5482 CB SER L 6 35.628 -49.441 -58.014 1.00 51.39 C C
+ATOM 5483 OG SER L 6 36.191 -50.494 -58.780 1.00 51.39 C O
+ATOM 5484 N SER L 7 37.638 -50.501 -55.712 1.00 61.71 C N
+ATOM 5485 CA SER L 7 38.904 -50.278 -54.978 1.00 62.11 C C
+ATOM 5486 C SER L 7 39.907 -51.404 -55.110 1.00 61.27 C C
+ATOM 5487 O SER L 7 39.545 -52.563 -55.079 1.00 61.11 C O
+ATOM 5488 CB SER L 7 38.626 -50.050 -53.490 1.00 52.39 C C
+ATOM 5489 OG SER L 7 38.198 -51.255 -52.865 1.00 52.39 C O
+ATOM 5490 N PRO L 8 41.186 -51.082 -55.259 1.00 58.15 C N
+ATOM 5491 CA PRO L 8 41.908 -49.821 -55.334 1.00 57.03 C C
+ATOM 5492 C PRO L 8 41.604 -48.981 -56.576 1.00 57.15 C C
+ATOM 5493 O PRO L 8 41.081 -49.481 -57.569 1.00 56.72 C O
+ATOM 5494 CB PRO L 8 43.351 -50.285 -55.406 1.00 50.72 C C
+ATOM 5495 CG PRO L 8 43.260 -51.527 -56.228 1.00 50.72 C C
+ATOM 5496 CD PRO L 8 42.054 -52.204 -55.652 1.00 50.72 C C
+ATOM 5497 N ALA L 9 41.971 -47.705 -56.518 1.00 63.77 C N
+ATOM 5498 CA ALA L 9 41.970 -46.848 -57.696 1.00 65.47 C C
+ATOM 5499 C ALA L 9 42.644 -47.551 -58.868 1.00 62.93 C C
+ATOM 5500 O ALA L 9 41.996 -47.929 -59.851 1.00 67.49 C O
+ATOM 5501 CB ALA L 9 42.701 -45.537 -57.375 1.00 59.88 C C
+ATOM 5502 N THR L 10 43.958 -47.721 -58.753 1.00 62.28 C N
+ATOM 5503 CA THR L 10 44.787 -48.467 -59.682 1.00 65.20 C C
+ATOM 5504 C THR L 10 45.408 -49.614 -58.920 1.00 63.97 C C
+ATOM 5505 O THR L 10 45.488 -49.591 -57.691 1.00 66.57 C O
+ATOM 5506 CB THR L 10 45.901 -47.606 -60.274 1.00 63.44 C C
+ATOM 5507 CG2 THR L 10 46.420 -48.204 -61.560 1.00 63.44 C C
+ATOM 5508 OG1 THR L 10 45.395 -46.293 -60.534 1.00 63.44 C O
+ATOM 5509 N LEU L 11 45.830 -50.627 -59.655 1.00 61.57 C N
+ATOM 5510 CA LEU L 11 46.546 -51.738 -59.058 1.00 60.71 C C
+ATOM 5511 C LEU L 11 47.626 -52.165 -60.029 1.00 60.71 C C
+ATOM 5512 O LEU L 11 47.357 -52.368 -61.216 1.00 65.86 C O
+ATOM 5513 CB LEU L 11 45.605 -52.898 -58.740 1.00 55.74 C C
+ATOM 5514 CG LEU L 11 46.271 -54.105 -58.112 1.00 55.74 C C
+ATOM 5515 CD1 LEU L 11 45.447 -54.603 -56.938 1.00 55.74 C C
+ATOM 5516 CD2 LEU L 11 46.388 -55.166 -59.176 1.00 55.74 C C
+ATOM 5517 N SER L 12 48.845 -52.295 -59.535 1.00 64.93 C N
+ATOM 5518 CA SER L 12 49.979 -52.623 -60.384 1.00 64.93 C C
+ATOM 5519 C SER L 12 50.526 -53.962 -59.928 1.00 69.01 C C
+ATOM 5520 O SER L 12 50.906 -54.117 -58.764 1.00 73.52 C O
+ATOM 5521 CB SER L 12 51.027 -51.493 -60.355 1.00 57.27 C C
+ATOM 5522 OG SER L 12 51.094 -50.842 -59.087 1.00 57.27 C O
+ATOM 5523 N VAL L 13 50.506 -54.944 -60.818 1.00 66.97 C N
+ATOM 5524 CA VAL L 13 50.995 -56.273 -60.484 1.00 70.16 C C
+ATOM 5525 C VAL L 13 51.740 -56.828 -61.677 1.00 66.40 C C
+ATOM 5526 O VAL L 13 51.301 -56.674 -62.819 1.00 65.70 C O
+ATOM 5527 CB VAL L 13 49.863 -57.232 -60.063 1.00 55.85 C C
+ATOM 5528 CG1 VAL L 13 49.421 -56.962 -58.596 1.00 55.85 C C
+ATOM 5529 CG2 VAL L 13 48.722 -57.114 -60.974 1.00 55.85 C C
+ATOM 5530 N SER L 14 52.857 -57.491 -61.402 1.00 72.38 C N
+ATOM 5531 CA SER L 14 53.761 -57.917 -62.454 1.00 71.84 C C
+ATOM 5532 C SER L 14 53.188 -59.117 -63.196 1.00 74.14 C C
+ATOM 5533 O SER L 14 52.470 -59.924 -62.608 1.00 72.08 C O
+ATOM 5534 CB SER L 14 55.108 -58.283 -61.856 1.00 57.00 C C
+ATOM 5535 OG SER L 14 55.679 -57.145 -61.247 1.00 57.00 C O
+ATOM 5536 N PRO L 15 53.505 -59.270 -64.480 1.00 73.36 C N
+ATOM 5537 CA PRO L 15 52.907 -60.364 -65.255 1.00 73.42 C C
+ATOM 5538 C PRO L 15 53.091 -61.705 -64.556 1.00 75.20 C C
+ATOM 5539 O PRO L 15 54.111 -61.964 -63.915 1.00 75.84 C O
+ATOM 5540 CB PRO L 15 53.645 -60.301 -66.598 1.00 55.96 C C
+ATOM 5541 CG PRO L 15 54.913 -59.573 -66.291 1.00 55.96 C C
+ATOM 5542 CD PRO L 15 54.545 -58.566 -65.238 1.00 55.96 C C
+ATOM 5543 N GLY L 16 52.055 -62.531 -64.622 1.00 77.05 C N
+ATOM 5544 CA GLY L 16 52.031 -63.766 -63.871 1.00 77.05 C C
+ATOM 5545 C GLY L 16 52.004 -63.581 -62.369 1.00 77.05 C C
+ATOM 5546 O GLY L 16 52.926 -64.029 -61.692 1.00 77.24 C O
+ATOM 5547 N GLU L 17 51.007 -62.896 -61.825 1.00 91.57 C N
+ATOM 5548 CA GLU L 17 50.899 -62.783 -60.379 1.00 91.57 C C
+ATOM 5549 C GLU L 17 49.436 -62.629 -60.027 1.00 91.57 C C
+ATOM 5550 O GLU L 17 48.664 -62.058 -60.800 1.00 91.57 C O
+ATOM 5551 CB GLU L 17 51.660 -61.584 -59.823 1.00 73.14 C C
+ATOM 5552 CG GLU L 17 53.139 -61.576 -60.130 1.00 73.14 C C
+ATOM 5553 CD GLU L 17 53.975 -62.368 -59.140 1.00 73.14 C C
+ATOM 5554 OE1 GLU L 17 53.532 -62.539 -57.966 1.00 73.14 C O
+ATOM 5555 OE2 GLU L 17 55.075 -62.829 -59.562 1.00 73.14 C O1-
+ATOM 5556 N ARG L 18 49.056 -63.126 -58.854 1.00 97.70 C N
+ATOM 5557 CA ARG L 18 47.674 -62.958 -58.437 1.00 96.86 C C
+ATOM 5558 C ARG L 18 47.399 -61.481 -58.206 1.00101.41 C C
+ATOM 5559 O ARG L 18 48.255 -60.729 -57.729 1.00103.63 C O
+ATOM 5560 CB ARG L 18 47.336 -63.791 -57.183 1.00 74.93 C C
+ATOM 5561 CG ARG L 18 46.544 -65.108 -57.471 1.00 74.93 C C
+ATOM 5562 CD ARG L 18 46.067 -65.859 -56.221 1.00 74.93 C C
+ATOM 5563 NE ARG L 18 47.043 -65.784 -55.133 1.00 74.93 C N
+ATOM 5564 CZ ARG L 18 48.200 -66.473 -55.085 1.00 74.93 C C
+ATOM 5565 NH1 ARG L 18 48.548 -67.301 -56.089 1.00 74.93 C N1+
+ATOM 5566 NH2 ARG L 18 49.036 -66.321 -54.039 1.00 74.93 C N
+ATOM 5567 N VAL L 19 46.214 -61.063 -58.617 1.00 90.22 C N
+ATOM 5568 CA VAL L 19 45.743 -59.713 -58.409 1.00 90.06 C C
+ATOM 5569 C VAL L 19 44.352 -59.800 -57.830 1.00 87.46 C C
+ATOM 5570 O VAL L 19 43.565 -60.673 -58.209 1.00 87.12 C O
+ATOM 5571 CB VAL L 19 45.727 -58.932 -59.705 1.00 32.96 C C
+ATOM 5572 N THR L 20 44.060 -58.910 -56.905 1.00 70.03 C N
+ATOM 5573 CA THR L 20 42.768 -58.932 -56.248 1.00 71.42 C C
+ATOM 5574 C THR L 20 42.181 -57.536 -56.232 1.00 74.19 C C
+ATOM 5575 O THR L 20 42.858 -56.565 -55.880 1.00 76.66 C O
+ATOM 5576 CB THR L 20 42.847 -59.446 -54.813 1.00 58.82 C C
+ATOM 5577 CG2 THR L 20 43.088 -60.955 -54.798 1.00 58.82 C C
+ATOM 5578 OG1 THR L 20 43.896 -58.757 -54.122 1.00 58.82 C O
+ATOM 5579 N LEU L 21 40.908 -57.464 -56.595 1.00 58.22 C N
+ATOM 5580 CA LEU L 21 40.161 -56.237 -56.752 1.00 57.32 C C
+ATOM 5581 C LEU L 21 38.977 -56.326 -55.814 1.00 58.21 C C
+ATOM 5582 O LEU L 21 38.480 -57.418 -55.542 1.00 58.38 C O
+ATOM 5583 CB LEU L 21 39.694 -56.080 -58.206 1.00 49.87 C C
+ATOM 5584 CG LEU L 21 40.729 -55.611 -59.249 1.00 49.87 C C
+ATOM 5585 CD1 LEU L 21 42.049 -56.397 -59.257 1.00 49.87 C C
+ATOM 5586 CD2 LEU L 21 40.132 -55.646 -60.612 1.00 49.87 C C
+ATOM 5587 N PHE L 22 38.525 -55.193 -55.322 1.00 66.94 C N
+ATOM 5588 CA PHE L 22 37.528 -55.170 -54.263 1.00 63.76 C C
+ATOM 5589 C PHE L 22 36.334 -54.287 -54.624 1.00 65.33 C C
+ATOM 5590 O PHE L 22 36.495 -53.226 -55.242 1.00 68.86 C O
+ATOM 5591 CB PHE L 22 38.159 -54.672 -52.956 1.00 55.08 C C
+ATOM 5592 CG PHE L 22 39.418 -55.434 -52.519 1.00 55.08 C C
+ATOM 5593 CD1 PHE L 22 39.585 -56.787 -52.796 1.00 55.08 C C
+ATOM 5594 CD2 PHE L 22 40.422 -54.780 -51.797 1.00 55.08 C C
+ATOM 5595 CE1 PHE L 22 40.723 -57.463 -52.385 1.00 55.08 C C
+ATOM 5596 CE2 PHE L 22 41.555 -55.462 -51.379 1.00 55.08 C C
+ATOM 5597 CZ PHE L 22 41.705 -56.804 -51.679 1.00 55.08 C C
+ATOM 5598 N CYS L 23 35.135 -54.714 -54.213 1.00 61.19 C N
+ATOM 5599 CA CYS L 23 33.904 -53.956 -54.455 1.00 65.85 C C
+ATOM 5600 C CYS L 23 33.089 -53.885 -53.162 1.00 63.24 C C
+ATOM 5601 O CYS L 23 32.600 -54.907 -52.684 1.00 62.02 C O
+ATOM 5602 CB CYS L 23 33.105 -54.616 -55.585 1.00 53.02 C C
+ATOM 5603 SG CYS L 23 31.573 -53.796 -56.217 1.00 53.02 C S
+ATOM 5604 N ARG L 24 32.934 -52.691 -52.589 1.00 63.26 C N
+ATOM 5605 CA ARG L 24 32.074 -52.488 -51.423 1.00 63.26 C C
+ATOM 5606 C ARG L 24 30.729 -52.007 -51.946 1.00 63.26 C C
+ATOM 5607 O ARG L 24 30.661 -51.366 -52.988 1.00 63.26 C O
+ATOM 5608 CB ARG L 24 32.684 -51.467 -50.436 1.00 56.88 C C
+ATOM 5609 CG ARG L 24 31.766 -50.870 -49.320 1.00 56.88 C C
+ATOM 5610 CD ARG L 24 32.367 -49.593 -48.549 1.00 56.88 C C
+ATOM 5611 NE ARG L 24 31.873 -48.258 -49.019 1.00 56.88 C N
+ATOM 5612 CZ ARG L 24 30.889 -47.494 -48.467 1.00 56.88 C C
+ATOM 5613 NH1 ARG L 24 30.220 -47.868 -47.363 1.00 56.88 C N1+
+ATOM 5614 NH2 ARG L 24 30.554 -46.323 -49.036 1.00 56.88 C N
+ATOM 5615 N ALA L 25 29.648 -52.371 -51.279 1.00 62.91 C N
+ATOM 5616 CA ALA L 25 28.327 -51.943 -51.723 1.00 62.91 C C
+ATOM 5617 C ALA L 25 27.618 -51.155 -50.624 1.00 63.97 C C
+ATOM 5618 O ALA L 25 27.748 -51.459 -49.438 1.00 65.10 C O
+ATOM 5619 CB ALA L 25 27.486 -53.139 -52.166 1.00 62.91 C C
+ATOM 5620 N SER L 26 26.881 -50.128 -51.023 1.00 70.46 C N
+ATOM 5621 CA SER L 26 26.226 -49.249 -50.067 1.00 70.46 C C
+ATOM 5622 C SER L 26 25.362 -50.048 -49.098 1.00 72.18 C C
+ATOM 5623 O SER L 26 25.721 -50.193 -47.931 1.00 75.25 C O
+ATOM 5624 CB SER L 26 25.411 -48.198 -50.834 1.00 69.00 C C
+ATOM 5625 OG SER L 26 24.335 -47.635 -50.080 1.00 69.00 C O
+ATOM 5626 N GLN L 27 24.240 -50.584 -49.559 1.00 80.90 C N
+ATOM 5627 CA GLN L 27 23.516 -51.572 -48.783 1.00 81.29 C C
+ATOM 5628 C GLN L 27 23.795 -52.953 -49.371 1.00 84.03 C C
+ATOM 5629 O GLN L 27 24.443 -53.090 -50.409 1.00 82.95 C O
+ATOM 5630 CB GLN L 27 22.008 -51.243 -48.734 1.00 77.85 C C
+ATOM 5631 CG GLN L 27 21.135 -52.178 -47.769 1.00 77.85 C C
+ATOM 5632 CD GLN L 27 21.605 -52.277 -46.240 1.00 77.85 C C
+ATOM 5633 NE2 GLN L 27 21.222 -51.267 -45.432 1.00 77.85 C N
+ATOM 5634 OE1 GLN L 27 22.271 -53.267 -45.816 1.00 77.85 C O
+ATOM 5635 N SER L 28 23.298 -53.982 -48.685 1.00 67.73 C N
+ATOM 5636 CA SER L 28 23.635 -55.351 -49.028 1.00 67.73 C C
+ATOM 5637 C SER L 28 23.107 -55.714 -50.385 1.00 67.73 C C
+ATOM 5638 O SER L 28 21.928 -55.533 -50.679 1.00 67.73 C O
+ATOM 5639 CB SER L 28 23.042 -56.336 -48.042 1.00 67.73 C C
+ATOM 5640 OG SER L 28 22.885 -57.573 -48.730 1.00 67.73 C O
+ATOM 5641 N VAL L 29 23.989 -56.247 -51.224 1.00 61.03 C N
+ATOM 5642 CA VAL L 29 23.617 -56.662 -52.570 1.00 62.01 C C
+ATOM 5643 C VAL L 29 23.483 -58.180 -52.626 1.00 61.40 C C
+ATOM 5644 O VAL L 29 23.149 -58.748 -53.666 1.00 63.45 C O
+ATOM 5645 CB VAL L 29 24.655 -56.206 -53.611 1.00 62.12 C C
+ATOM 5646 CG1 VAL L 29 24.033 -56.165 -54.998 1.00 62.12 C C
+ATOM 5647 CG2 VAL L 29 25.222 -54.846 -53.233 1.00 62.12 C C
+ATOM 5648 N ILE L 36 23.747 -58.829 -51.497 1.00 61.88 C N
+ATOM 5649 CA ILE L 36 23.660 -60.283 -51.399 1.00 60.82 C C
+ATOM 5650 C ILE L 36 24.726 -60.965 -52.257 1.00 59.08 C C
+ATOM 5651 O ILE L 36 25.886 -61.060 -51.857 1.00 67.25 C O
+ATOM 5652 CB ILE L 36 22.257 -60.794 -51.785 1.00 67.10 C C
+ATOM 5653 CG1 ILE L 36 21.188 -60.123 -50.921 1.00 67.10 C C
+ATOM 5654 CG2 ILE L 36 22.186 -62.308 -51.651 1.00 67.10 C C
+ATOM 5655 CD1 ILE L 36 21.030 -60.747 -49.552 1.00 67.10 C C
+ATOM 5656 N SER L 37 24.330 -61.435 -53.436 1.00 58.75 C N
+ATOM 5657 CA SER L 37 25.250 -62.103 -54.347 1.00 59.92 C C
+ATOM 5658 C SER L 37 25.087 -61.548 -55.756 1.00 59.07 C C
+ATOM 5659 O SER L 37 26.037 -61.513 -56.538 1.00 58.75 C O
+ATOM 5660 CB SER L 37 25.010 -63.613 -54.344 1.00 57.65 C C
+ATOM 5661 OG SER L 37 23.832 -63.944 -55.059 1.00 57.65 C O
+ATOM 5662 N ASN L 38 23.871 -61.113 -56.070 1.00 58.34 C N
+ATOM 5663 CA ASN L 38 23.565 -60.549 -57.383 1.00 58.34 C C
+ATOM 5664 C ASN L 38 24.545 -59.419 -57.670 1.00 58.34 C C
+ATOM 5665 O ASN L 38 24.242 -58.246 -57.502 1.00 58.93 C O
+ATOM 5666 CB ASN L 38 22.152 -59.973 -57.384 1.00 54.26 C C
+ATOM 5667 CG ASN L 38 21.097 -61.019 -57.306 1.00 54.26 C C
+ATOM 5668 ND2 ASN L 38 20.021 -60.727 -56.564 1.00 54.26 C N
+ATOM 5669 OD1 ASN L 38 21.236 -62.097 -57.910 1.00 54.26 C O
+ATOM 5670 N LEU L 39 25.645 -59.768 -58.322 1.00 53.74 C N
+ATOM 5671 CA LEU L 39 26.718 -58.834 -58.607 1.00 53.74 C C
+ATOM 5672 C LEU L 39 27.513 -59.452 -59.731 1.00 53.74 C C
+ATOM 5673 O LEU L 39 27.512 -60.669 -59.895 1.00 53.74 C O
+ATOM 5674 CB LEU L 39 27.590 -58.600 -57.383 1.00 47.75 C C
+ATOM 5675 CG LEU L 39 28.798 -57.691 -57.568 1.00 47.75 C C
+ATOM 5676 CD1 LEU L 39 28.906 -56.657 -56.427 1.00 47.75 C C
+ATOM 5677 CD2 LEU L 39 30.077 -58.526 -57.731 1.00 47.75 C C
+ATOM 5678 N ALA L 40 28.212 -58.618 -60.487 1.00 45.49 C N
+ATOM 5679 CA ALA L 40 28.880 -59.081 -61.687 1.00 45.49 C C
+ATOM 5680 C ALA L 40 30.164 -58.303 -61.816 1.00 45.49 C C
+ATOM 5681 O ALA L 40 30.299 -57.214 -61.257 1.00 45.49 C O
+ATOM 5682 CB ALA L 40 28.015 -58.899 -62.933 1.00 45.49 C C
+ATOM 5683 N TRP L 41 31.110 -58.882 -62.541 1.00 47.38 C N
+ATOM 5684 CA TRP L 41 32.396 -58.264 -62.803 1.00 47.38 C C
+ATOM 5685 C TRP L 41 32.582 -58.192 -64.300 1.00 47.38 C C
+ATOM 5686 O TRP L 41 32.364 -59.197 -64.988 1.00 50.08 C O
+ATOM 5687 CB TRP L 41 33.550 -59.065 -62.197 1.00 45.54 C C
+ATOM 5688 CG TRP L 41 33.604 -58.987 -60.751 1.00 45.54 C C
+ATOM 5689 CD1 TRP L 41 33.027 -59.839 -59.857 1.00 45.54 C C
+ATOM 5690 CD2 TRP L 41 34.227 -57.968 -59.995 1.00 45.54 C C
+ATOM 5691 CE2 TRP L 41 34.006 -58.255 -58.641 1.00 45.54 C C
+ATOM 5692 CE3 TRP L 41 34.944 -56.832 -60.328 1.00 45.54 C C
+ATOM 5693 NE1 TRP L 41 33.275 -59.413 -58.576 1.00 45.54 C N
+ATOM 5694 CZ2 TRP L 41 34.480 -57.439 -57.624 1.00 45.54 C C
+ATOM 5695 CZ3 TRP L 41 35.420 -56.025 -59.315 1.00 45.54 C C
+ATOM 5696 CH2 TRP L 41 35.187 -56.328 -57.982 1.00 45.54 C C
+ATOM 5697 N TYR L 42 33.028 -57.032 -64.792 1.00 44.77 C N
+ATOM 5698 CA TYR L 42 33.263 -56.770 -66.206 1.00 44.77 C C
+ATOM 5699 C TYR L 42 34.713 -56.393 -66.445 1.00 44.77 C C
+ATOM 5700 O TYR L 42 35.336 -55.723 -65.618 1.00 44.77 C O
+ATOM 5701 CB TYR L 42 32.374 -55.652 -66.721 1.00 45.15 C C
+ATOM 5702 CG TYR L 42 30.935 -55.965 -66.549 1.00 45.15 C C
+ATOM 5703 CD1 TYR L 42 30.163 -56.313 -67.632 1.00 45.15 C C
+ATOM 5704 CD2 TYR L 42 30.345 -55.954 -65.300 1.00 45.15 C C
+ATOM 5705 CE1 TYR L 42 28.814 -56.624 -67.482 1.00 45.15 C C
+ATOM 5706 CE2 TYR L 42 29.008 -56.261 -65.133 1.00 45.15 C C
+ATOM 5707 CZ TYR L 42 28.241 -56.592 -66.227 1.00 45.15 C C
+ATOM 5708 OH TYR L 42 26.904 -56.902 -66.088 1.00 45.15 C O
+ATOM 5709 N GLN L 43 35.241 -56.826 -67.579 1.00 53.14 C N
+ATOM 5710 CA GLN L 43 36.555 -56.420 -68.050 1.00 53.14 C C
+ATOM 5711 C GLN L 43 36.334 -55.499 -69.242 1.00 53.98 C C
+ATOM 5712 O GLN L 43 35.461 -55.755 -70.078 1.00 58.30 C O
+ATOM 5713 CB GLN L 43 37.402 -57.650 -68.434 1.00 51.34 C C
+ATOM 5714 CG GLN L 43 38.800 -57.412 -69.003 1.00 51.34 C C
+ATOM 5715 CD GLN L 43 39.152 -58.408 -70.112 1.00 51.34 C C
+ATOM 5716 NE2 GLN L 43 40.005 -59.392 -69.799 1.00 51.34 C N
+ATOM 5717 OE1 GLN L 43 38.658 -58.290 -71.240 1.00 51.34 C O
+ATOM 5718 N GLN L 44 37.061 -54.388 -69.278 1.00 58.73 C N
+ATOM 5719 CA GLN L 44 36.992 -53.453 -70.393 1.00 58.73 C C
+ATOM 5720 C GLN L 44 38.409 -53.142 -70.833 1.00 58.89 C C
+ATOM 5721 O GLN L 44 39.139 -52.442 -70.119 1.00 60.66 C O
+ATOM 5722 CB GLN L 44 36.259 -52.176 -69.996 1.00 52.75 C C
+ATOM 5723 CG GLN L 44 35.796 -51.337 -71.166 1.00 52.75 C C
+ATOM 5724 CD GLN L 44 35.118 -50.039 -70.732 1.00 52.75 C C
+ATOM 5725 NE2 GLN L 44 34.035 -49.669 -71.436 1.00 52.75 C N
+ATOM 5726 OE1 GLN L 44 35.575 -49.362 -69.795 1.00 52.75 C O
+ATOM 5727 N LYS L 45 38.781 -53.647 -72.012 1.00 73.48 C N
+ATOM 5728 CA LYS L 45 40.059 -53.315 -72.622 1.00 73.48 C C
+ATOM 5729 C LYS L 45 39.878 -51.984 -73.332 1.00 74.86 C C
+ATOM 5730 O LYS L 45 38.785 -51.672 -73.813 1.00 77.04 C O
+ATOM 5731 CB LYS L 45 40.511 -54.397 -73.617 1.00 66.99 C C
+ATOM 5732 CG LYS L 45 40.161 -55.856 -73.234 1.00 66.99 C C
+ATOM 5733 CD LYS L 45 41.037 -56.917 -73.959 1.00 66.99 C C
+ATOM 5734 CE LYS L 45 40.404 -58.354 -73.940 1.00 66.99 C C
+ATOM 5735 NZ LYS L 45 39.197 -58.560 -74.871 1.00 66.99 C N1+
+ATOM 5736 N SER L 46 40.941 -51.181 -73.367 1.00 73.55 C N
+ATOM 5737 CA SER L 46 40.787 -49.799 -73.813 1.00 75.05 C C
+ATOM 5738 C SER L 46 40.198 -49.735 -75.218 1.00 81.10 C C
+ATOM 5739 O SER L 46 40.624 -50.451 -76.130 1.00 81.93 C O
+ATOM 5740 CB SER L 46 42.123 -49.056 -73.748 1.00 67.40 C C
+ATOM 5741 OG SER L 46 43.078 -49.622 -74.627 1.00 67.40 C O
+ATOM 5742 N GLY L 47 39.179 -48.901 -75.365 1.00 70.02 C N
+ATOM 5743 CA GLY L 47 38.567 -48.696 -76.652 1.00 68.29 C C
+ATOM 5744 C GLY L 47 37.695 -49.832 -77.120 1.00 69.65 C C
+ATOM 5745 O GLY L 47 37.701 -50.153 -78.313 1.00 70.72 C O
+ATOM 5746 N GLN L 48 36.945 -50.460 -76.222 1.00 73.92 C N
+ATOM 5747 CA GLN L 48 35.901 -51.389 -76.644 1.00 75.92 C C
+ATOM 5748 C GLN L 48 34.901 -51.520 -75.508 1.00 75.33 C C
+ATOM 5749 O GLN L 48 35.040 -50.892 -74.456 1.00 75.82 C O
+ATOM 5750 CB GLN L 48 36.486 -52.736 -77.087 1.00 66.17 C C
+ATOM 5751 CG GLN L 48 37.532 -53.331 -76.163 1.00 66.17 C C
+ATOM 5752 CD GLN L 48 38.342 -54.440 -76.846 1.00 66.17 C C
+ATOM 5753 NE2 GLN L 48 39.049 -54.081 -77.920 1.00 66.17 C N
+ATOM 5754 OE1 GLN L 48 38.363 -55.592 -76.392 1.00 66.17 C O
+ATOM 5755 N ALA L 49 33.868 -52.309 -75.741 1.00 63.85 C N
+ATOM 5756 CA ALA L 49 32.789 -52.428 -74.780 1.00 61.21 C C
+ATOM 5757 C ALA L 49 33.230 -53.292 -73.608 1.00 59.70 C C
+ATOM 5758 O ALA L 49 34.321 -53.868 -73.606 1.00 59.70 C O
+ATOM 5759 CB ALA L 49 31.548 -52.998 -75.460 1.00 59.70 C C
+ATOM 5760 N PRO L 50 32.428 -53.350 -72.555 1.00 53.48 C N
+ATOM 5761 CA PRO L 50 32.659 -54.363 -71.519 1.00 53.48 C C
+ATOM 5762 C PRO L 50 32.514 -55.790 -72.045 1.00 53.48 C C
+ATOM 5763 O PRO L 50 31.663 -56.085 -72.884 1.00 57.15 C O
+ATOM 5764 CB PRO L 50 31.584 -54.038 -70.479 1.00 47.84 C C
+ATOM 5765 CG PRO L 50 31.337 -52.555 -70.662 1.00 47.84 C C
+ATOM 5766 CD PRO L 50 31.510 -52.285 -72.118 1.00 47.84 C C
+ATOM 5767 N ARG L 51 33.387 -56.669 -71.568 1.00 67.46 C N
+ATOM 5768 CA ARG L 51 33.163 -58.110 -71.578 1.00 67.22 C C
+ATOM 5769 C ARG L 51 32.639 -58.458 -70.194 1.00 66.83 C C
+ATOM 5770 O ARG L 51 33.083 -57.873 -69.199 1.00 67.22 C O
+ATOM 5771 CB ARG L 51 34.461 -58.884 -71.882 1.00 61.05 C C
+ATOM 5772 CG ARG L 51 34.359 -60.336 -72.452 1.00 61.05 C C
+ATOM 5773 CD ARG L 51 35.709 -60.835 -73.146 1.00 61.05 C C
+ATOM 5774 NE ARG L 51 36.722 -61.398 -72.198 1.00 61.05 C N
+ATOM 5775 CZ ARG L 51 38.008 -61.716 -72.497 1.00 61.05 C C
+ATOM 5776 NH1 ARG L 51 38.457 -61.534 -73.750 1.00 61.05 C N1+
+ATOM 5777 NH2 ARG L 51 38.860 -62.212 -71.558 1.00 61.05 C N
+ATOM 5778 N LEU L 52 31.655 -59.358 -70.127 1.00 53.55 C N
+ATOM 5779 CA LEU L 52 31.109 -59.781 -68.838 1.00 53.55 C C
+ATOM 5780 C LEU L 52 31.943 -60.941 -68.316 1.00 53.55 C C
+ATOM 5781 O LEU L 52 32.045 -61.981 -68.972 1.00 53.55 C O
+ATOM 5782 CB LEU L 52 29.638 -60.184 -68.933 1.00 47.72 C C
+ATOM 5783 CG LEU L 52 29.187 -61.005 -67.715 1.00 47.72 C C
+ATOM 5784 CD1 LEU L 52 29.103 -60.159 -66.483 1.00 47.72 C C
+ATOM 5785 CD2 LEU L 52 27.860 -61.668 -67.944 1.00 47.72 C C
+ATOM 5786 N LEU L 53 32.538 -60.760 -67.142 1.00 52.17 C N
+ATOM 5787 CA LEU L 53 33.435 -61.757 -66.568 1.00 52.17 C C
+ATOM 5788 C LEU L 53 32.715 -62.717 -65.630 1.00 52.58 C C
+ATOM 5789 O LEU L 53 32.687 -63.928 -65.869 1.00 52.17 C O
+ATOM 5790 CB LEU L 53 34.570 -61.066 -65.821 1.00 45.82 C C
+ATOM 5791 CG LEU L 53 35.852 -61.060 -66.608 1.00 45.82 C C
+ATOM 5792 CD1 LEU L 53 35.568 -60.593 -68.018 1.00 45.82 C C
+ATOM 5793 CD2 LEU L 53 36.755 -60.119 -65.881 1.00 45.82 C C
+ATOM 5794 N ILE L 54 32.176 -62.203 -64.533 1.00 51.09 C N
+ATOM 5795 CA ILE L 54 31.509 -63.056 -63.569 1.00 51.09 C C
+ATOM 5796 C ILE L 54 30.113 -62.511 -63.314 1.00 51.09 C C
+ATOM 5797 O ILE L 54 29.838 -61.326 -63.508 1.00 51.09 C O
+ATOM 5798 CB ILE L 54 32.286 -63.117 -62.260 1.00 43.43 C C
+ATOM 5799 CG1 ILE L 54 33.778 -63.148 -62.543 1.00 43.43 C C
+ATOM 5800 CG2 ILE L 54 31.846 -64.306 -61.438 1.00 43.43 C C
+ATOM 5801 CD1 ILE L 54 34.197 -64.393 -63.155 1.00 43.43 C C
+ATOM 5802 N TYR L 55 29.231 -63.386 -62.843 1.00 52.24 C N
+ATOM 5803 CA TYR L 55 27.926 -62.950 -62.378 1.00 52.24 C C
+ATOM 5804 C TYR L 55 27.437 -63.896 -61.294 1.00 52.24 C C
+ATOM 5805 O TYR L 55 27.923 -65.019 -61.165 1.00 53.95 C O
+ATOM 5806 CB TYR L 55 26.942 -62.860 -63.529 1.00 50.27 C C
+ATOM 5807 CG TYR L 55 26.589 -64.191 -64.114 1.00 50.27 C C
+ATOM 5808 CD1 TYR L 55 25.528 -64.934 -63.625 1.00 50.27 C C
+ATOM 5809 CD2 TYR L 55 27.303 -64.695 -65.190 1.00 50.27 C C
+ATOM 5810 CE1 TYR L 55 25.199 -66.154 -64.185 1.00 50.27 C C
+ATOM 5811 CE2 TYR L 55 26.981 -65.913 -65.766 1.00 50.27 C C
+ATOM 5812 CZ TYR L 55 25.929 -66.639 -65.261 1.00 50.27 C C
+ATOM 5813 OH TYR L 55 25.610 -67.856 -65.822 1.00 50.27 C O
+ATOM 5814 N GLY L 56 26.472 -63.427 -60.502 1.00 51.20 C N
+ATOM 5815 CA GLY L 56 26.164 -64.111 -59.260 1.00 51.20 C C
+ATOM 5816 C GLY L 56 27.320 -64.087 -58.298 1.00 51.20 C C
+ATOM 5817 O GLY L 56 27.375 -64.892 -57.364 1.00 51.20 C O
+ATOM 5818 N ALA L 57 28.290 -63.223 -58.573 1.00 57.17 C N
+ATOM 5819 CA ALA L 57 29.514 -62.961 -57.829 1.00 57.17 C C
+ATOM 5820 C ALA L 57 30.498 -64.114 -57.948 1.00 57.35 C C
+ATOM 5821 O ALA L 57 31.716 -63.922 -57.875 1.00 57.20 C O
+ATOM 5822 CB ALA L 57 29.200 -62.702 -56.355 1.00 57.17 C C
+ATOM 5823 N SER L 65 29.975 -65.309 -58.150 1.00 61.10 C N
+ATOM 5824 CA SER L 65 30.787 -66.499 -58.067 1.00 60.00 C C
+ATOM 5825 C SER L 65 30.965 -67.281 -59.361 1.00 59.91 C C
+ATOM 5826 O SER L 65 31.771 -68.210 -59.380 1.00 62.40 C O
+ATOM 5827 CB SER L 65 30.179 -67.407 -56.998 1.00 57.75 C C
+ATOM 5828 OG SER L 65 28.759 -67.254 -57.028 1.00 57.75 C O
+ATOM 5829 N THR L 66 30.242 -66.985 -60.428 1.00 56.21 C N
+ATOM 5830 CA THR L 66 30.110 -67.966 -61.499 1.00 56.21 C C
+ATOM 5831 C THR L 66 30.587 -67.329 -62.790 1.00 57.54 C C
+ATOM 5832 O THR L 66 29.998 -66.347 -63.248 1.00 56.32 C O
+ATOM 5833 CB THR L 66 28.658 -68.432 -61.647 1.00 50.35 C C
+ATOM 5834 CG2 THR L 66 27.891 -68.338 -60.312 1.00 50.35 C C
+ATOM 5835 OG1 THR L 66 27.989 -67.566 -62.559 1.00 50.35 C O
+ATOM 5836 N ARG L 67 31.645 -67.856 -63.381 1.00 68.08 C N
+ATOM 5837 CA ARG L 67 32.187 -67.162 -64.538 1.00 66.16 C C
+ATOM 5838 C ARG L 67 31.343 -67.438 -65.768 1.00 67.24 C C
+ATOM 5839 O ARG L 67 30.789 -68.523 -65.926 1.00 71.18 C O
+ATOM 5840 CB ARG L 67 33.654 -67.517 -64.778 1.00 58.71 C C
+ATOM 5841 CG ARG L 67 34.058 -68.910 -64.454 1.00 58.71 C C
+ATOM 5842 CD ARG L 67 35.570 -69.070 -64.510 1.00 58.71 C C
+ATOM 5843 NE ARG L 67 35.820 -70.452 -64.889 1.00 58.71 C N
+ATOM 5844 CZ ARG L 67 35.762 -71.482 -64.040 1.00 58.71 C C
+ATOM 5845 NH1 ARG L 67 35.521 -71.252 -62.729 1.00 58.71 C N1+
+ATOM 5846 NH2 ARG L 67 35.968 -72.738 -64.501 1.00 58.71 C N
+ATOM 5847 N ALA L 68 31.194 -66.412 -66.598 1.00 67.82 C N
+ATOM 5848 CA ALA L 68 30.370 -66.508 -67.790 1.00 68.33 C C
+ATOM 5849 C ALA L 68 31.085 -67.349 -68.825 1.00 69.01 C C
+ATOM 5850 O ALA L 68 32.302 -67.538 -68.765 1.00 68.23 C O
+ATOM 5851 CB ALA L 68 30.065 -65.125 -68.371 1.00 46.80 C C
+ATOM 5852 N THR L 69 30.320 -67.865 -69.781 1.00 76.89 C N
+ATOM 5853 CA THR L 69 30.920 -68.772 -70.747 1.00 80.55 C C
+ATOM 5854 C THR L 69 31.946 -68.018 -71.584 1.00 79.65 C C
+ATOM 5855 O THR L 69 31.698 -66.909 -72.066 1.00 76.54 C O
+ATOM 5856 CB THR L 69 29.860 -69.441 -71.631 1.00 56.08 C C
+ATOM 5857 CG2 THR L 69 29.005 -70.467 -70.815 1.00 56.08 C C
+ATOM 5858 OG1 THR L 69 29.011 -68.447 -72.218 1.00 56.08 C O
+ATOM 5859 N GLY L 70 33.115 -68.628 -71.717 1.00 65.00 C N
+ATOM 5860 CA GLY L 70 34.280 -68.054 -72.318 1.00 63.75 C C
+ATOM 5861 C GLY L 70 35.329 -67.592 -71.325 1.00 67.32 C C
+ATOM 5862 O GLY L 70 36.522 -67.637 -71.637 1.00 69.93 C O
+ATOM 5863 N ILE L 71 34.935 -67.238 -70.118 1.00 62.89 C N
+ATOM 5864 CA ILE L 71 35.930 -66.656 -69.220 1.00 63.18 C C
+ATOM 5865 C ILE L 71 36.852 -67.760 -68.726 1.00 64.86 C C
+ATOM 5866 O ILE L 71 36.363 -68.808 -68.279 1.00 69.24 C O
+ATOM 5867 CB ILE L 71 35.247 -65.952 -68.055 1.00 51.06 C C
+ATOM 5868 CG1 ILE L 71 34.801 -64.540 -68.444 1.00 51.06 C C
+ATOM 5869 CG2 ILE L 71 36.189 -65.880 -66.870 1.00 51.06 C C
+ATOM 5870 CD1 ILE L 71 33.658 -64.507 -69.380 1.00 51.06 C C
+ATOM 5871 N PRO L 72 38.169 -67.590 -68.791 1.00 62.53 C N
+ATOM 5872 CA PRO L 72 39.083 -68.630 -68.294 1.00 62.53 C C
+ATOM 5873 C PRO L 72 39.043 -68.732 -66.775 1.00 62.53 C C
+ATOM 5874 O PRO L 72 38.443 -67.916 -66.074 1.00 62.88 C O
+ATOM 5875 CB PRO L 72 40.459 -68.181 -68.798 1.00 58.36 C C
+ATOM 5876 CG PRO L 72 40.157 -67.202 -69.912 1.00 58.36 C C
+ATOM 5877 CD PRO L 72 38.895 -66.511 -69.468 1.00 58.36 C C
+ATOM 5878 N SER L 74 39.662 -69.787 -66.260 1.00 69.94 C N
+ATOM 5879 CA SER L 74 39.529 -70.031 -64.831 1.00 69.94 C C
+ATOM 5880 C SER L 74 40.496 -69.209 -63.987 1.00 69.94 C C
+ATOM 5881 O SER L 74 40.537 -69.382 -62.761 1.00 73.70 C O
+ATOM 5882 CB SER L 74 39.697 -71.521 -64.530 1.00 27.51 C C
+ATOM 5883 N ARG L 75 41.289 -68.332 -64.595 1.00 69.94 C N
+ATOM 5884 CA ARG L 75 41.983 -67.353 -63.773 1.00 69.94 C C
+ATOM 5885 C ARG L 75 40.986 -66.476 -63.028 1.00 71.35 C C
+ATOM 5886 O ARG L 75 41.155 -66.201 -61.837 1.00 73.40 C O
+ATOM 5887 CB ARG L 75 42.947 -66.501 -64.610 1.00 59.35 C C
+ATOM 5888 CG ARG L 75 43.622 -67.195 -65.783 1.00 59.35 C C
+ATOM 5889 CD ARG L 75 44.402 -66.190 -66.613 1.00 59.35 C C
+ATOM 5890 NE ARG L 75 44.340 -66.394 -68.054 1.00 59.35 C N
+ATOM 5891 CZ ARG L 75 43.604 -65.672 -68.882 1.00 59.35 C C
+ATOM 5892 NH1 ARG L 75 42.898 -64.679 -68.392 1.00 59.35 C N1+
+ATOM 5893 NH2 ARG L 75 43.602 -65.921 -70.194 1.00 59.35 C N
+ATOM 5894 N PHE L 76 39.915 -66.072 -63.697 1.00 67.88 C N
+ATOM 5895 CA PHE L 76 38.990 -65.106 -63.125 1.00 67.05 C C
+ATOM 5896 C PHE L 76 38.002 -65.775 -62.174 1.00 68.58 C C
+ATOM 5897 O PHE L 76 37.332 -66.755 -62.529 1.00 70.54 C O
+ATOM 5898 CB PHE L 76 38.251 -64.366 -64.235 1.00 55.56 C C
+ATOM 5899 CG PHE L 76 39.159 -63.701 -65.208 1.00 55.56 C C
+ATOM 5900 CD1 PHE L 76 39.636 -62.426 -64.962 1.00 55.56 C C
+ATOM 5901 CD2 PHE L 76 39.528 -64.341 -66.370 1.00 55.56 C C
+ATOM 5902 CE1 PHE L 76 40.482 -61.811 -65.847 1.00 55.56 C C
+ATOM 5903 CE2 PHE L 76 40.361 -63.730 -67.260 1.00 55.56 C C
+ATOM 5904 CZ PHE L 76 40.846 -62.462 -67.000 1.00 55.56 C C
+ATOM 5905 N SER L 77 37.916 -65.236 -60.963 1.00 64.58 C N
+ATOM 5906 CA SER L 77 37.131 -65.870 -59.921 1.00 64.46 C C
+ATOM 5907 C SER L 77 36.681 -64.819 -58.922 1.00 64.46 C C
+ATOM 5908 O SER L 77 37.426 -63.888 -58.613 1.00 64.46 C O
+ATOM 5909 CB SER L 77 37.972 -66.932 -59.233 1.00 59.92 C C
+ATOM 5910 OG SER L 77 39.314 -66.465 -59.193 1.00 59.92 C O
+ATOM 5911 N GLY L 78 35.502 -65.023 -58.338 1.00 65.01 C N
+ATOM 5912 CA GLY L 78 34.909 -63.995 -57.511 1.00 65.01 C C
+ATOM 5913 C GLY L 78 34.185 -64.572 -56.313 1.00 65.01 C C
+ATOM 5914 O GLY L 78 33.865 -65.766 -56.253 1.00 68.74 C O
+ATOM 5915 N SER L 79 34.003 -63.749 -55.287 1.00 69.63 C N
+ATOM 5916 CA SER L 79 33.350 -64.189 -54.056 1.00 69.63 C C
+ATOM 5917 C SER L 79 32.188 -63.252 -53.862 1.00 69.83 C C
+ATOM 5918 O SER L 79 31.815 -62.561 -54.810 1.00 69.63 C O
+ATOM 5919 CB SER L 79 34.311 -64.101 -52.871 1.00 60.57 C C
+ATOM 5920 OG SER L 79 35.626 -63.796 -53.302 1.00 60.57 C O
+ATOM 5921 N GLY L 80 31.589 -63.181 -52.676 1.00 59.39 C N
+ATOM 5922 CA GLY L 80 30.496 -62.228 -52.640 1.00 59.39 C C
+ATOM 5923 C GLY L 80 29.341 -62.450 -51.688 1.00 59.39 C C
+ATOM 5924 O GLY L 80 29.514 -63.020 -50.609 1.00 59.39 C O
+ATOM 5925 N SER L 83 28.162 -61.927 -52.055 1.00 70.19 C N
+ATOM 5926 CA SER L 83 26.931 -62.023 -51.255 1.00 70.19 C C
+ATOM 5927 C SER L 83 27.093 -61.294 -49.919 1.00 70.19 C C
+ATOM 5928 O SER L 83 27.302 -61.880 -48.863 1.00 70.19 C O
+ATOM 5929 CB SER L 83 26.506 -63.488 -51.040 1.00 70.19 C C
+ATOM 5930 OG SER L 83 25.478 -63.611 -50.060 1.00 70.19 C O
+ATOM 5931 N GLY L 84 27.017 -59.976 -50.015 1.00 65.17 C N
+ATOM 5932 CA GLY L 84 27.199 -59.150 -48.837 1.00 65.17 C C
+ATOM 5933 C GLY L 84 27.265 -57.683 -49.202 1.00 65.17 C C
+ATOM 5934 O GLY L 84 26.554 -57.213 -50.093 1.00 65.17 C O
+ATOM 5935 N THR L 85 27.991 -56.947 -48.386 1.00 67.74 C N
+ATOM 5936 CA THR L 85 28.504 -55.659 -48.798 1.00 67.74 C C
+ATOM 5937 C THR L 85 29.850 -55.740 -49.509 1.00 67.74 C C
+ATOM 5938 O THR L 85 30.081 -54.995 -50.458 1.00 67.74 C O
+ATOM 5939 CB THR L 85 28.549 -54.763 -47.576 1.00 67.74 C C
+ATOM 5940 CG2 THR L 85 29.391 -53.504 -47.801 1.00 67.74 C C
+ATOM 5941 OG1 THR L 85 27.198 -54.425 -47.241 1.00 67.74 C O
+ATOM 5942 N GLU L 86 30.765 -56.601 -49.084 1.00 75.19 C N
+ATOM 5943 CA GLU L 86 32.101 -56.595 -49.659 1.00 75.19 C C
+ATOM 5944 C GLU L 86 32.296 -57.793 -50.583 1.00 75.19 C C
+ATOM 5945 O GLU L 86 31.734 -58.868 -50.360 1.00 78.73 C O
+ATOM 5946 CB GLU L 86 33.172 -56.579 -48.570 1.00 61.05 C C
+ATOM 5947 CG GLU L 86 34.406 -55.737 -48.940 1.00 61.05 C C
+ATOM 5948 CD GLU L 86 34.278 -54.222 -48.575 1.00 61.05 C C
+ATOM 5949 OE1 GLU L 86 33.270 -53.848 -47.894 1.00 61.05 C O
+ATOM 5950 OE2 GLU L 86 35.198 -53.419 -48.963 1.00 61.05 C O1-
+ATOM 5951 N PHE L 87 33.089 -57.586 -51.636 1.00 61.05 C N
+ATOM 5952 CA PHE L 87 33.194 -58.520 -52.752 1.00 61.05 C C
+ATOM 5953 C PHE L 87 34.607 -58.505 -53.301 1.00 63.26 C C
+ATOM 5954 O PHE L 87 35.340 -57.523 -53.155 1.00 67.91 C O
+ATOM 5955 CB PHE L 87 32.267 -58.164 -53.916 1.00 49.75 C C
+ATOM 5956 CG PHE L 87 30.809 -58.250 -53.609 1.00 49.75 C C
+ATOM 5957 CD1 PHE L 87 30.059 -59.292 -54.085 1.00 49.75 C C
+ATOM 5958 CD2 PHE L 87 30.171 -57.260 -52.897 1.00 49.75 C C
+ATOM 5959 CE1 PHE L 87 28.701 -59.369 -53.823 1.00 49.75 C C
+ATOM 5960 CE2 PHE L 87 28.817 -57.327 -52.634 1.00 49.75 C C
+ATOM 5961 CZ PHE L 87 28.083 -58.380 -53.101 1.00 49.75 C C
+ATOM 5962 N THR L 88 34.945 -59.571 -54.022 1.00 57.55 C N
+ATOM 5963 CA THR L 88 36.302 -59.771 -54.512 1.00 57.55 C C
+ATOM 5964 C THR L 88 36.347 -60.356 -55.906 1.00 60.92 C C
+ATOM 5965 O THR L 88 35.702 -61.380 -56.167 1.00 65.60 C O
+ATOM 5966 CB THR L 88 37.067 -60.715 -53.612 1.00 49.83 C C
+ATOM 5967 CG2 THR L 88 38.343 -61.151 -54.291 1.00 49.83 C C
+ATOM 5968 OG1 THR L 88 37.358 -60.047 -52.388 1.00 49.83 C O
+ATOM 5969 N LEU L 89 37.157 -59.741 -56.769 1.00 57.05 C N
+ATOM 5970 CA LEU L 89 37.557 -60.326 -58.037 1.00 54.34 C C
+ATOM 5971 C LEU L 89 39.022 -60.697 -57.949 1.00 54.51 C C
+ATOM 5972 O LEU L 89 39.824 -59.932 -57.419 1.00 56.50 C O
+ATOM 5973 CB LEU L 89 37.330 -59.371 -59.215 1.00 51.13 C C
+ATOM 5974 CG LEU L 89 37.894 -59.856 -60.565 1.00 51.13 C C
+ATOM 5975 CD1 LEU L 89 37.365 -61.215 -60.989 1.00 51.13 C C
+ATOM 5976 CD2 LEU L 89 37.576 -58.869 -61.651 1.00 51.13 C C
+ATOM 5977 N THR L 90 39.362 -61.873 -58.462 1.00 62.73 C N
+ATOM 5978 CA THR L 90 40.701 -62.438 -58.355 1.00 62.73 C C
+ATOM 5979 C THR L 90 41.139 -62.964 -59.699 1.00 65.41 C C
+ATOM 5980 O THR L 90 40.475 -63.840 -60.270 1.00 70.60 C O
+ATOM 5981 CB THR L 90 40.769 -63.580 -57.351 1.00 59.58 C C
+ATOM 5982 CG2 THR L 90 42.108 -64.251 -57.428 1.00 59.58 C C
+ATOM 5983 OG1 THR L 90 40.526 -63.088 -56.033 1.00 59.58 C O
+ATOM 5984 N ILE L 91 42.274 -62.460 -60.173 1.00 75.82 C N
+ATOM 5985 CA ILE L 91 42.901 -62.940 -61.392 1.00 74.55 C C
+ATOM 5986 C ILE L 91 44.146 -63.701 -60.984 1.00 75.41 C C
+ATOM 5987 O ILE L 91 45.021 -63.138 -60.316 1.00 80.16 C O
+ATOM 5988 CB ILE L 91 43.260 -61.795 -62.344 1.00 62.15 C C
+ATOM 5989 CG1 ILE L 91 42.069 -60.874 -62.539 1.00 62.15 C C
+ATOM 5990 CG2 ILE L 91 43.551 -62.371 -63.682 1.00 62.15 C C
+ATOM 5991 CD1 ILE L 91 42.301 -59.774 -63.565 1.00 62.15 C C
+ATOM 5992 N SER L 92 44.249 -64.952 -61.443 1.00 94.39 C N
+ATOM 5993 CA SER L 92 45.185 -65.919 -60.872 1.00 91.50 C C
+ATOM 5994 C SER L 92 46.619 -65.609 -61.255 1.00 91.50 C C
+ATOM 5995 O SER L 92 47.479 -65.426 -60.392 1.00 94.47 C O
+ATOM 5996 CB SER L 92 44.828 -67.324 -61.334 1.00 67.47 C C
+ATOM 5997 OG SER L 92 43.474 -67.600 -61.030 1.00 67.47 C O
+ATOM 5998 N SER L 93 46.921 -65.637 -62.540 1.00 83.18 C N
+ATOM 5999 CA SER L 93 48.213 -65.171 -63.013 1.00 83.18 C C
+ATOM 6000 C SER L 93 47.925 -64.163 -64.106 1.00 83.18 C C
+ATOM 6001 O SER L 93 47.414 -64.524 -65.172 1.00 83.18 C O
+ATOM 6002 CB SER L 93 49.067 -66.323 -63.527 1.00 60.32 C C
+ATOM 6003 OG SER L 93 48.727 -66.604 -64.878 1.00 60.32 C O
+ATOM 6004 N LEU L 94 48.246 -62.906 -63.843 1.00 84.53 C N
+ATOM 6005 CA LEU L 94 47.906 -61.869 -64.797 1.00 84.53 C C
+ATOM 6006 C LEU L 94 48.563 -62.156 -66.131 1.00 84.53 C C
+ATOM 6007 O LEU L 94 49.755 -62.440 -66.203 1.00 86.87 C O
+ATOM 6008 CB LEU L 94 48.340 -60.502 -64.284 1.00 62.54 C C
+ATOM 6009 CG LEU L 94 47.182 -59.758 -63.628 1.00 62.54 C C
+ATOM 6010 CD1 LEU L 94 47.406 -58.272 -63.613 1.00 62.54 C C
+ATOM 6011 CD2 LEU L 94 45.929 -60.080 -64.387 1.00 62.54 C C
+ATOM 6012 N GLN L 95 47.792 -62.088 -67.190 1.00100.29 C N
+ATOM 6013 CA GLN L 95 48.355 -62.353 -68.492 1.00100.29 C C
+ATOM 6014 C GLN L 95 48.121 -61.136 -69.383 1.00101.20 C C
+ATOM 6015 O GLN L 95 47.313 -60.261 -69.069 1.00100.29 C O
+ATOM 6016 CB GLN L 95 47.747 -63.615 -69.091 1.00 69.87 C C
+ATOM 6017 CG GLN L 95 48.560 -64.154 -70.246 1.00 69.87 C C
+ATOM 6018 CD GLN L 95 47.675 -64.760 -71.330 1.00 69.87 C C
+ATOM 6019 NE2 GLN L 95 48.082 -64.597 -72.613 1.00 69.87 C N
+ATOM 6020 OE1 GLN L 95 46.629 -65.365 -71.022 1.00 69.87 C O
+ATOM 6021 N SER L 96 48.829 -61.103 -70.516 1.00 96.11 C N
+ATOM 6022 CA SER L 96 48.963 -59.893 -71.326 1.00 96.11 C C
+ATOM 6023 C SER L 96 47.639 -59.180 -71.520 1.00101.08 C C
+ATOM 6024 O SER L 96 47.531 -57.968 -71.324 1.00107.79 C O
+ATOM 6025 CB SER L 96 49.531 -60.259 -72.693 1.00 67.77 C C
+ATOM 6026 OG SER L 96 50.483 -61.314 -72.611 1.00 67.77 C O
+ATOM 6027 N GLU L 97 46.617 -59.922 -71.885 1.00103.20 C N
+ATOM 6028 CA GLU L 97 45.334 -59.347 -72.230 1.00103.19 C C
+ATOM 6029 C GLU L 97 44.420 -59.187 -71.027 1.00100.63 C C
+ATOM 6030 O GLU L 97 43.227 -58.918 -71.201 1.00102.77 C O
+ATOM 6031 CB GLU L 97 44.675 -60.202 -73.307 1.00 74.91 C C
+ATOM 6032 CG GLU L 97 44.235 -61.591 -72.819 1.00 74.91 C C
+ATOM 6033 CD GLU L 97 45.298 -62.693 -73.034 1.00 74.91 C C
+ATOM 6034 OE1 GLU L 97 46.483 -62.356 -73.320 1.00 74.91 C O
+ATOM 6035 OE2 GLU L 97 44.922 -63.895 -72.943 1.00 74.91 C O1-
+ATOM 6036 N ASP L 98 44.928 -59.419 -69.822 1.00 83.07 C N
+ATOM 6037 CA ASP L 98 44.156 -59.157 -68.619 1.00 83.06 C C
+ATOM 6038 C ASP L 98 44.409 -57.783 -68.038 1.00 83.93 C C
+ATOM 6039 O ASP L 98 43.700 -57.383 -67.115 1.00 83.81 C O
+ATOM 6040 CB ASP L 98 44.452 -60.193 -67.543 1.00 65.01 C C
+ATOM 6041 CG ASP L 98 44.331 -61.594 -68.061 1.00 65.01 C C
+ATOM 6042 OD1 ASP L 98 43.707 -61.752 -69.141 1.00 65.01 C O
+ATOM 6043 OD2 ASP L 98 44.835 -62.530 -67.394 1.00 65.01 C O1-
+ATOM 6044 N PHE L 99 45.381 -57.045 -68.551 1.00 80.82 C N
+ATOM 6045 CA PHE L 99 45.604 -55.703 -68.040 1.00 80.35 C C
+ATOM 6046 C PHE L 99 44.609 -54.801 -68.740 1.00 80.35 C C
+ATOM 6047 O PHE L 99 44.728 -54.543 -69.939 1.00 80.35 C O
+ATOM 6048 CB PHE L 99 47.037 -55.248 -68.304 1.00 60.07 C C
+ATOM 6049 CG PHE L 99 48.087 -56.065 -67.589 1.00 60.07 C C
+ATOM 6050 CD1 PHE L 99 48.493 -55.731 -66.314 1.00 60.07 C C
+ATOM 6051 CD2 PHE L 99 48.669 -57.153 -68.196 1.00 60.07 C C
+ATOM 6052 CE1 PHE L 99 49.445 -56.479 -65.664 1.00 60.07 C C
+ATOM 6053 CE2 PHE L 99 49.625 -57.894 -67.542 1.00 60.07 C C
+ATOM 6054 CZ PHE L 99 50.011 -57.556 -66.278 1.00 60.07 C C
+ATOM 6055 N ALA L 100 43.655 -54.297 -67.988 1.00 61.28 C N
+ATOM 6056 CA ALA L 100 42.508 -53.574 -68.525 1.00 61.28 C C
+ATOM 6057 C ALA L 100 41.777 -52.981 -67.333 1.00 61.28 C C
+ATOM 6058 O ALA L 100 42.256 -53.076 -66.199 1.00 61.28 C O
+ATOM 6059 CB ALA L 100 41.621 -54.491 -69.357 1.00 62.23 C C
+ATOM 6060 N VAL L 101 40.637 -52.368 -67.545 1.00 50.32 C N
+ATOM 6061 CA VAL L 101 39.910 -51.846 -66.396 1.00 50.32 C C
+ATOM 6062 C VAL L 101 38.798 -52.802 -66.006 1.00 51.87 C C
+ATOM 6063 O VAL L 101 38.150 -53.398 -66.869 1.00 52.47 C O
+ATOM 6064 CB VAL L 101 39.369 -50.447 -66.679 1.00 51.95 C C
+ATOM 6065 CG1 VAL L 101 38.487 -49.989 -65.513 1.00 51.95 C C
+ATOM 6066 CG2 VAL L 101 40.554 -49.535 -66.876 1.00 51.95 C C
+ATOM 6067 N TYR L 102 38.578 -52.966 -64.707 1.00 49.04 C N
+ATOM 6068 CA TYR L 102 37.560 -53.884 -64.225 1.00 49.04 C C
+ATOM 6069 C TYR L 102 36.482 -53.103 -63.502 1.00 49.04 C C
+ATOM 6070 O TYR L 102 36.779 -52.128 -62.813 1.00 49.15 C O
+ATOM 6071 CB TYR L 102 38.169 -54.953 -63.325 1.00 50.47 C C
+ATOM 6072 CG TYR L 102 39.026 -55.899 -64.125 1.00 50.47 C C
+ATOM 6073 CD1 TYR L 102 38.522 -57.101 -64.606 1.00 50.47 C C
+ATOM 6074 CD2 TYR L 102 40.319 -55.561 -64.474 1.00 50.47 C C
+ATOM 6075 CE1 TYR L 102 39.310 -57.960 -65.374 1.00 50.47 C C
+ATOM 6076 CE2 TYR L 102 41.100 -56.411 -65.246 1.00 50.47 C C
+ATOM 6077 CZ TYR L 102 40.592 -57.603 -65.688 1.00 50.47 C C
+ATOM 6078 OH TYR L 102 41.376 -58.433 -66.443 1.00 50.47 C O
+ATOM 6079 N PHE L 103 35.230 -53.511 -63.706 1.00 45.86 C N
+ATOM 6080 CA PHE L 103 34.069 -52.839 -63.148 1.00 45.86 C C
+ATOM 6081 C PHE L 103 33.208 -53.867 -62.454 1.00 46.09 C C
+ATOM 6082 O PHE L 103 33.048 -54.972 -62.964 1.00 47.99 C O
+ATOM 6083 CB PHE L 103 33.214 -52.170 -64.211 1.00 49.42 C C
+ATOM 6084 CG PHE L 103 33.850 -50.985 -64.831 1.00 49.42 C C
+ATOM 6085 CD1 PHE L 103 33.901 -49.771 -64.146 1.00 49.42 C C
+ATOM 6086 CD2 PHE L 103 34.364 -51.062 -66.118 1.00 49.42 C C
+ATOM 6087 CE1 PHE L 103 34.493 -48.659 -64.725 1.00 49.42 C C
+ATOM 6088 CE2 PHE L 103 34.951 -49.969 -66.716 1.00 49.42 C C
+ATOM 6089 CZ PHE L 103 35.021 -48.761 -66.022 1.00 49.42 C C
+ATOM 6090 N CYS L 104 32.635 -53.504 -61.310 1.00 49.22 C N
+ATOM 6091 CA CYS L 104 31.645 -54.340 -60.654 1.00 49.22 C C
+ATOM 6092 C CYS L 104 30.279 -53.691 -60.751 1.00 49.22 C C
+ATOM 6093 O CYS L 104 30.161 -52.470 -60.818 1.00 49.22 C O
+ATOM 6094 CB CYS L 104 31.999 -54.586 -59.199 1.00 49.62 C C
+ATOM 6095 SG CYS L 104 31.989 -53.115 -58.170 1.00 49.62 C S
+ATOM 6096 N GLN L 105 29.251 -54.522 -60.789 1.00 53.52 C N
+ATOM 6097 CA GLN L 105 27.882 -54.038 -60.888 1.00 53.52 C C
+ATOM 6098 C GLN L 105 27.002 -54.868 -59.982 1.00 53.52 C C
+ATOM 6099 O GLN L 105 27.227 -56.070 -59.874 1.00 53.93 C O
+ATOM 6100 CB GLN L 105 27.363 -54.150 -62.315 1.00 51.60 C C
+ATOM 6101 CG GLN L 105 25.878 -53.987 -62.405 1.00 51.60 C C
+ATOM 6102 CD GLN L 105 25.271 -54.928 -63.395 1.00 51.60 C C
+ATOM 6103 NE2 GLN L 105 23.983 -55.249 -63.221 1.00 51.60 C N
+ATOM 6104 OE1 GLN L 105 25.952 -55.360 -64.324 1.00 51.60 C O
+ATOM 6105 N GLN L 106 26.006 -54.276 -59.331 1.00 60.80 C N
+ATOM 6106 CA GLN L 106 25.018 -55.170 -58.754 1.00 60.80 C C
+ATOM 6107 C GLN L 106 23.741 -55.066 -59.576 1.00 60.80 C C
+ATOM 6108 O GLN L 106 23.480 -54.050 -60.213 1.00 60.80 C O
+ATOM 6109 CB GLN L 106 24.680 -54.904 -57.279 1.00 60.80 C C
+ATOM 6110 CG GLN L 106 23.456 -54.023 -57.022 1.00 60.80 C C
+ATOM 6111 CD GLN L 106 23.460 -52.716 -57.696 1.00 60.80 C C
+ATOM 6112 NE2 GLN L 106 22.333 -52.025 -57.581 1.00 60.80 C N
+ATOM 6113 OE1 GLN L 106 24.371 -52.389 -58.466 1.00 60.80 C O
+ATOM 6114 N TYR L 107 23.011 -56.164 -59.669 1.00 59.53 C N
+ATOM 6115 CA TYR L 107 21.665 -56.148 -60.225 1.00 59.53 C C
+ATOM 6116 C TYR L 107 20.564 -56.311 -59.177 1.00 59.53 C C
+ATOM 6117 O TYR L 107 19.422 -56.618 -59.543 1.00 59.53 C O
+ATOM 6118 CB TYR L 107 21.535 -57.158 -61.374 1.00 53.20 C C
+ATOM 6119 CG TYR L 107 22.271 -58.416 -61.156 1.00 53.20 C C
+ATOM 6120 CD1 TYR L 107 21.601 -59.549 -60.803 1.00 53.20 C C
+ATOM 6121 CD2 TYR L 107 23.633 -58.476 -61.321 1.00 53.20 C C
+ATOM 6122 CE1 TYR L 107 22.258 -60.711 -60.591 1.00 53.20 C C
+ATOM 6123 CE2 TYR L 107 24.308 -59.625 -61.109 1.00 53.20 C C
+ATOM 6124 CZ TYR L 107 23.615 -60.752 -60.741 1.00 53.20 C C
+ATOM 6125 OH TYR L 107 24.260 -61.950 -60.517 1.00 53.20 C O
+ATOM 6126 N ASN L 108 20.882 -56.214 -57.888 1.00 68.38 C N
+ATOM 6127 CA ASN L 108 19.869 -56.507 -56.876 1.00 67.95 C C
+ATOM 6128 C ASN L 108 18.667 -55.590 -56.992 1.00 68.84 C C
+ATOM 6129 O ASN L 108 17.522 -56.038 -56.863 1.00 77.93 C O
+ATOM 6130 CB ASN L 108 20.454 -56.385 -55.480 1.00 61.24 C C
+ATOM 6131 CG ASN L 108 19.688 -57.193 -54.460 1.00 61.24 C C
+ATOM 6132 ND2 ASN L 108 18.697 -56.553 -53.805 1.00 61.24 C N
+ATOM 6133 OD1 ASN L 108 20.000 -58.381 -54.236 1.00 61.24 C O
+ATOM 6134 N ASN L 109 18.906 -54.298 -57.188 1.00 75.43 C N
+ATOM 6135 CA ASN L 109 17.836 -53.350 -57.461 1.00 78.14 C C
+ATOM 6136 C ASN L 109 18.117 -52.579 -58.731 1.00 79.52 C C
+ATOM 6137 O ASN L 109 19.262 -52.459 -59.180 1.00 78.63 C O
+ATOM 6138 CB ASN L 109 17.639 -52.343 -56.341 1.00 69.45 C C
+ATOM 6139 CG ASN L 109 16.725 -52.853 -55.285 1.00 69.45 C C
+ATOM 6140 ND2 ASN L 109 15.426 -52.550 -55.434 1.00 69.45 C N
+ATOM 6141 OD1 ASN L 109 17.157 -53.553 -54.352 1.00 69.45 C O
+ATOM 6142 N TRP L 110 17.051 -52.055 -59.306 1.00 63.52 C N
+ATOM 6143 CA TRP L 110 17.238 -50.991 -60.251 1.00 64.14 C C
+ATOM 6144 C TRP L 110 17.339 -49.745 -59.378 1.00 62.01 C C
+ATOM 6145 O TRP L 110 16.697 -49.708 -58.333 1.00 65.77 C O
+ATOM 6146 CB TRP L 110 16.085 -50.924 -61.239 1.00 54.69 C C
+ATOM 6147 CG TRP L 110 16.140 -51.984 -62.308 1.00 54.69 C C
+ATOM 6148 CD1 TRP L 110 16.896 -53.127 -62.310 1.00 54.69 C C
+ATOM 6149 CD2 TRP L 110 15.424 -51.979 -63.545 1.00 54.69 C C
+ATOM 6150 CE2 TRP L 110 15.775 -53.149 -64.240 1.00 54.69 C C
+ATOM 6151 CE3 TRP L 110 14.513 -51.096 -64.130 1.00 54.69 C C
+ATOM 6152 NE1 TRP L 110 16.677 -53.837 -63.469 1.00 54.69 C N
+ATOM 6153 CZ2 TRP L 110 15.251 -53.451 -65.488 1.00 54.69 C C
+ATOM 6154 CZ3 TRP L 110 13.998 -51.400 -65.362 1.00 54.69 C C
+ATOM 6155 CH2 TRP L 110 14.364 -52.566 -66.029 1.00 54.69 C C
+ATOM 6156 N PRO L 111 18.147 -48.735 -59.770 1.00 58.63 C N
+ATOM 6157 CA PRO L 111 18.969 -48.643 -60.963 1.00 58.31 C C
+ATOM 6158 C PRO L 111 20.257 -49.448 -60.826 1.00 61.47 C C
+ATOM 6159 O PRO L 111 20.856 -49.514 -59.750 1.00 62.71 C O
+ATOM 6160 CB PRO L 111 19.254 -47.153 -61.046 1.00 49.03 C C
+ATOM 6161 CG PRO L 111 19.364 -46.754 -59.689 1.00 49.03 C C
+ATOM 6162 CD PRO L 111 18.424 -47.598 -58.884 1.00 49.03 C C
+ATOM 6163 N LEU L 112 20.667 -50.056 -61.933 1.00 29.48 C N
+ATOM 6164 CA LEU L 112 21.923 -50.761 -61.975 1.00169.14 C C
+ATOM 6165 C LEU L 112 23.025 -49.753 -61.834 1.00169.72 C C
+ATOM 6166 O LEU L 112 23.045 -48.720 -62.501 1.00168.67 C O
+ATOM 6167 CB LEU L 112 22.077 -51.515 -63.276 1.00 46.33 C C
+ATOM 6168 CG LEU L 112 20.972 -52.521 -63.509 1.00 46.33 C C
+ATOM 6169 CD1 LEU L 112 21.168 -53.181 -64.847 1.00 46.33 C C
+ATOM 6170 CD2 LEU L 112 20.934 -53.515 -62.399 1.00 46.33 C C
+ATOM 6171 N THR L 113 23.911 -50.031 -60.936 1.00 74.08 C N
+ATOM 6172 CA THR L 113 25.004 -49.147 -60.704 1.00 75.36 C C
+ATOM 6173 C THR L 113 26.298 -49.885 -60.973 1.00 74.90 C C
+ATOM 6174 O THR L 113 26.322 -51.091 -61.214 1.00 76.29 C O
+ATOM 6175 CB THR L 113 24.967 -48.667 -59.273 1.00 50.85 C C
+ATOM 6176 CG2 THR L 113 23.554 -48.406 -58.885 1.00 50.85 C C
+ATOM 6177 OG1 THR L 113 25.454 -49.725 -58.448 1.00 50.85 C O
+ATOM 6178 N PHE L 114 27.389 -49.154 -60.871 1.00 56.89 C N
+ATOM 6179 CA PHE L 114 28.696 -49.713 -61.095 1.00 56.89 C C
+ATOM 6180 C PHE L 114 29.651 -49.055 -60.133 1.00 59.09 C C
+ATOM 6181 O PHE L 114 29.340 -48.049 -59.501 1.00 56.89 C O
+ATOM 6182 CB PHE L 114 29.178 -49.469 -62.515 1.00 49.74 C C
+ATOM 6183 CG PHE L 114 28.299 -50.060 -63.598 1.00 49.74 C C
+ATOM 6184 CD1 PHE L 114 28.459 -51.372 -64.006 1.00 49.74 C C
+ATOM 6185 CD2 PHE L 114 27.367 -49.277 -64.258 1.00 49.74 C C
+ATOM 6186 CE1 PHE L 114 27.688 -51.894 -65.022 1.00 49.74 C C
+ATOM 6187 CE2 PHE L 114 26.594 -49.803 -65.286 1.00 49.74 C C
+ATOM 6188 CZ PHE L 114 26.755 -51.105 -65.661 1.00 49.74 C C
+ATOM 6189 N GLY L 115 30.817 -49.643 -60.018 1.00 49.54 C N
+ATOM 6190 CA GLY L 115 31.921 -48.970 -59.389 1.00 48.13 C C
+ATOM 6191 C GLY L 115 32.587 -48.014 -60.357 1.00 48.63 C C
+ATOM 6192 O GLY L 115 32.203 -47.866 -61.523 1.00 50.71 C O
+ATOM 6193 N GLY L 116 33.615 -47.341 -59.836 1.00 52.58 C N
+ATOM 6194 CA GLY L 116 34.415 -46.477 -60.678 1.00 56.71 C C
+ATOM 6195 C GLY L 116 35.379 -47.263 -61.533 1.00 56.69 C C
+ATOM 6196 O GLY L 116 35.738 -46.839 -62.640 1.00 61.94 C O
+ATOM 6197 N GLY L 117 35.784 -48.421 -61.057 1.00 54.22 C N
+ATOM 6198 CA GLY L 117 36.707 -49.229 -61.816 1.00 52.34 C C
+ATOM 6199 C GLY L 117 38.077 -49.262 -61.175 1.00 54.54 C C
+ATOM 6200 O GLY L 117 38.454 -48.394 -60.388 1.00 60.61 C O
+ATOM 6201 N THR L 118 38.820 -50.301 -61.522 1.00 56.81 C N
+ATOM 6202 CA THR L 118 40.172 -50.511 -61.036 1.00 53.10 C C
+ATOM 6203 C THR L 118 41.043 -50.719 -62.266 1.00 54.65 C C
+ATOM 6204 O THR L 118 40.827 -51.674 -63.015 1.00 54.05 C O
+ATOM 6205 CB THR L 118 40.218 -51.729 -60.095 1.00 49.89 C C
+ATOM 6206 CG2 THR L 118 41.593 -51.925 -59.492 1.00 49.89 C C
+ATOM 6207 OG1 THR L 118 39.250 -51.581 -59.044 1.00 49.89 C O
+ATOM 6208 N GLN L 119 41.981 -49.813 -62.520 1.00 67.42 C N
+ATOM 6209 CA GLN L 119 42.937 -50.069 -63.588 1.00 68.58 C C
+ATOM 6210 C GLN L 119 43.964 -51.046 -63.040 1.00 67.82 C C
+ATOM 6211 O GLN L 119 44.469 -50.841 -61.934 1.00 73.88 C O
+ATOM 6212 CB GLN L 119 43.627 -48.779 -64.043 1.00 62.55 C C
+ATOM 6213 CG GLN L 119 44.549 -48.919 -65.298 1.00 62.55 C C
+ATOM 6214 CD GLN L 119 45.369 -47.626 -65.658 1.00 62.55 C C
+ATOM 6215 NE2 GLN L 119 45.807 -47.520 -66.929 1.00 62.55 C N
+ATOM 6216 OE1 GLN L 119 45.583 -46.742 -64.808 1.00 62.55 C O
+ATOM 6217 N VAL L 120 44.261 -52.116 -63.773 1.00 60.98 C N
+ATOM 6218 CA VAL L 120 45.419 -52.947 -63.447 1.00 60.98 C C
+ATOM 6219 C VAL L 120 46.519 -52.608 -64.439 1.00 60.98 C C
+ATOM 6220 O VAL L 120 46.412 -52.915 -65.631 1.00 64.63 C O
+ATOM 6221 CB VAL L 120 45.107 -54.449 -63.460 1.00 54.33 C C
+ATOM 6222 CG1 VAL L 120 44.382 -54.845 -62.211 1.00 54.33 C C
+ATOM 6223 CG2 VAL L 120 44.321 -54.816 -64.654 1.00 54.33 C C
+ATOM 6224 N ASN L 121 47.581 -51.964 -63.978 1.00 73.12 C N
+ATOM 6225 CA ASN L 121 48.631 -51.661 -64.928 1.00 73.12 C C
+ATOM 6226 C ASN L 121 49.766 -52.627 -64.668 1.00 73.41 C C
+ATOM 6227 O ASN L 121 49.943 -53.124 -63.552 1.00 73.12 C O
+ATOM 6228 CB ASN L 121 49.111 -50.185 -64.900 1.00 59.98 C C
+ATOM 6229 CG ASN L 121 50.015 -49.795 -63.688 1.00 59.98 C C
+ATOM 6230 ND2 ASN L 121 51.235 -49.312 -63.997 1.00 59.98 C N
+ATOM 6231 OD1 ASN L 121 49.596 -49.828 -62.525 1.00 59.98 C O
+ATOM 6232 N VAL L 122 50.472 -52.957 -65.745 1.00 65.88 C N
+ATOM 6233 CA VAL L 122 51.603 -53.865 -65.659 1.00 68.94 C C
+ATOM 6234 C VAL L 122 52.752 -53.159 -64.937 1.00 73.04 C C
+ATOM 6235 O VAL L 122 53.070 -52.001 -65.236 1.00 76.16 C O
+ATOM 6236 CB VAL L 122 51.954 -54.384 -67.070 1.00 49.22 C C
+ATOM 6237 CG1 VAL L 122 52.113 -53.256 -68.069 1.00 49.22 C C
+ATOM 6238 CG2 VAL L 122 53.196 -55.262 -67.041 1.00 49.22 C C
+ATOM 6239 N GLN L 123 53.326 -53.829 -63.921 1.00 65.32 C N
+ATOM 6240 CA GLN L 123 54.294 -53.205 -63.017 1.00 65.32 C C
+ATOM 6241 C GLN L 123 55.723 -53.434 -63.495 1.00 65.32 C C
+ATOM 6242 O GLN L 123 56.099 -54.542 -63.883 1.00 65.32 C O
+ATOM 6243 CB GLN L 123 54.154 -53.709 -61.578 1.00 65.32 C C
+ATOM 6244 CG GLN L 123 54.931 -52.847 -60.575 1.00 65.32 C C
+ATOM 6245 CD GLN L 123 54.864 -53.334 -59.103 1.00 65.32 C C
+ATOM 6246 NE2 GLN L 123 55.033 -52.402 -58.165 1.00 65.32 C N
+ATOM 6247 OE1 GLN L 123 54.670 -54.521 -58.819 1.00 65.32 C O
+ATOM 6248 N ARG L 124 56.516 -52.364 -63.426 1.00 63.16 C N
+ATOM 6249 CA ARG L 124 57.797 -52.186 -64.093 1.00 60.56 C C
+ATOM 6250 C ARG L 124 58.800 -51.687 -63.066 1.00 59.12 C C
+ATOM 6251 O ARG L 124 58.425 -51.246 -61.978 1.00 58.05 C O
+ATOM 6252 CB ARG L 124 57.678 -51.170 -65.240 1.00 60.53 C C
+ATOM 6253 CG ARG L 124 58.861 -51.136 -66.145 1.00 61.35 C C
+ATOM 6254 CD ARG L 124 59.039 -49.788 -66.755 1.00 62.56 C C
+ATOM 6255 NE ARG L 124 59.976 -49.857 -67.873 1.00 66.82 C N
+ATOM 6256 CZ ARG L 124 61.306 -49.778 -67.751 1.00 68.50 C C
+ATOM 6257 NH1 ARG L 124 61.850 -49.620 -66.541 1.00 67.78 C N1+
+ATOM 6258 NH2 ARG L 124 62.101 -49.857 -68.833 1.00 71.80 C N
+ATOM 6259 N THR L 125 60.085 -51.753 -63.415 1.00 57.81 C N
+ATOM 6260 CA THR L 125 61.113 -51.078 -62.628 1.00 59.77 C C
+ATOM 6261 C THR L 125 61.004 -49.572 -62.848 1.00 65.47 C C
+ATOM 6262 O THR L 125 60.730 -49.123 -63.964 1.00 67.25 C O
+ATOM 6263 CB THR L 125 62.494 -51.566 -63.046 1.00 62.62 C C
+ATOM 6264 CG2 THR L 125 62.392 -53.007 -63.555 1.00 62.40 C C
+ATOM 6265 OG1 THR L 125 62.976 -50.761 -64.126 1.00 64.91 C O
+ATOM 6266 N VAL L 126 61.208 -48.785 -61.788 1.00 56.60 C N
+ATOM 6267 CA VAL L 126 61.042 -47.341 -61.921 1.00 50.72 C C
+ATOM 6268 C VAL L 126 61.899 -46.834 -63.066 1.00 52.02 C C
+ATOM 6269 O VAL L 126 63.056 -47.235 -63.220 1.00 58.98 C O
+ATOM 6270 CB VAL L 126 61.416 -46.625 -60.614 1.00 50.52 C C
+ATOM 6271 CG1 VAL L 126 61.091 -45.156 -60.717 1.00 45.87 C C
+ATOM 6272 CG2 VAL L 126 60.696 -47.235 -59.462 1.00 48.98 C C
+ATOM 6273 N ALA L 127 61.332 -45.964 -63.895 1.00 49.16 C N
+ATOM 6274 CA ALA L 127 62.046 -45.432 -65.051 1.00 49.70 C C
+ATOM 6275 C ALA L 127 61.845 -43.928 -65.117 1.00 54.24 C C
+ATOM 6276 O ALA L 127 60.707 -43.459 -65.212 1.00 54.36 C O
+ATOM 6277 CB ALA L 127 61.563 -46.089 -66.336 1.00 47.87 C C
+ATOM 6278 N ALA L 128 62.941 -43.173 -65.073 1.00 49.94 C N
+ATOM 6279 CA ALA L 128 62.814 -41.726 -64.941 1.00 47.10 C C
+ATOM 6280 C ALA L 128 62.477 -41.113 -66.294 1.00 44.86 C C
+ATOM 6281 O ALA L 128 62.926 -41.617 -67.317 1.00 45.25 C O
+ATOM 6282 CB ALA L 128 64.101 -41.126 -64.397 1.00 46.93 C C
+ATOM 6283 N PRO L 129 61.676 -40.054 -66.338 1.00 45.99 C N
+ATOM 6284 CA PRO L 129 61.202 -39.532 -67.623 1.00 46.47 C C
+ATOM 6285 C PRO L 129 62.240 -38.663 -68.309 1.00 47.05 C C
+ATOM 6286 O PRO L 129 63.047 -38.001 -67.656 1.00 46.76 C O
+ATOM 6287 CB PRO L 129 59.991 -38.692 -67.211 1.00 45.22 C C
+ATOM 6288 CG PRO L 129 60.419 -38.119 -65.887 1.00 44.96 C C
+ATOM 6289 CD PRO L 129 61.306 -39.163 -65.228 1.00 46.98 C C
+ATOM 6290 N SER L 130 62.196 -38.628 -69.642 1.00 52.98 C N
+ATOM 6291 CA SER L 130 63.075 -37.720 -70.389 1.00 49.80 C C
+ATOM 6292 C SER L 130 62.328 -36.446 -70.761 1.00 53.40 C C
+ATOM 6293 O SER L 130 61.275 -36.506 -71.398 1.00 54.87 C O
+ATOM 6294 CB SER L 130 63.643 -38.376 -71.651 1.00 52.12 C C
+ATOM 6295 OG SER L 130 64.286 -39.606 -71.353 1.00 63.19 C O
+ATOM 6296 N VAL L 131 62.881 -35.298 -70.393 1.00 57.41 C N
+ATOM 6297 CA VAL L 131 62.173 -34.030 -70.510 1.00 53.50 C C
+ATOM 6298 C VAL L 131 62.611 -33.280 -71.760 1.00 58.37 C C
+ATOM 6299 O VAL L 131 63.805 -33.104 -72.015 1.00 60.24 C O
+ATOM 6300 CB VAL L 131 62.383 -33.170 -69.263 1.00 52.09 C C
+ATOM 6301 CG1 VAL L 131 61.690 -31.836 -69.438 1.00 49.82 C C
+ATOM 6302 CG2 VAL L 131 61.857 -33.909 -68.059 1.00 53.16 C C
+ATOM 6303 N PHE L 132 61.646 -32.824 -72.532 1.00 69.23 C N
+ATOM 6304 CA PHE L 132 61.909 -31.961 -73.664 1.00 65.58 C C
+ATOM 6305 C PHE L 132 60.945 -30.808 -73.607 1.00 66.20 C C
+ATOM 6306 O PHE L 132 59.760 -31.006 -73.344 1.00 65.48 C O
+ATOM 6307 CB PHE L 132 61.688 -32.639 -74.961 1.00 64.75 C C
+ATOM 6308 CG PHE L 132 62.506 -33.825 -75.151 1.00 64.68 C C
+ATOM 6309 CD1 PHE L 132 62.232 -34.984 -74.454 1.00 62.17 C C
+ATOM 6310 CD2 PHE L 132 63.533 -33.803 -76.077 1.00 63.99 C C
+ATOM 6311 CE1 PHE L 132 62.988 -36.109 -74.640 1.00 61.44 C C
+ATOM 6312 CE2 PHE L 132 64.300 -34.922 -76.292 1.00 67.40 C C
+ATOM 6313 CZ PHE L 132 64.027 -36.091 -75.560 1.00 65.54 C C
+ATOM 6314 N ILE L 133 61.428 -29.616 -73.887 1.00 67.39 C N
+ATOM 6315 CA ILE L 133 60.553 -28.465 -73.982 1.00 61.57 C C
+ATOM 6316 C ILE L 133 60.505 -28.037 -75.437 1.00 61.76 C C
+ATOM 6317 O ILE L 133 61.488 -28.174 -76.176 1.00 61.02 C O
+ATOM 6318 CB ILE L 133 61.000 -27.329 -73.048 1.00 60.98 C C
+ATOM 6319 CG1 ILE L 133 59.894 -26.305 -72.934 1.00 60.25 C C
+ATOM 6320 CG2 ILE L 133 62.265 -26.714 -73.522 1.00 58.39 C C
+ATOM 6321 CD1 ILE L 133 59.725 -25.797 -71.541 1.00 60.05 C C
+ATOM 6322 N PHE L 134 59.336 -27.589 -75.864 1.00 72.22 C N
+ATOM 6323 CA PHE L 134 59.164 -26.993 -77.164 1.00 73.11 C C
+ATOM 6324 C PHE L 134 58.576 -25.600 -76.983 1.00 75.13 C C
+ATOM 6325 O PHE L 134 57.665 -25.409 -76.153 1.00 76.78 C O
+ATOM 6326 CB PHE L 134 58.229 -27.836 -78.033 1.00 68.51 C C
+ATOM 6327 CG PHE L 134 58.685 -29.256 -78.214 1.00 70.82 C C
+ATOM 6328 CD1 PHE L 134 59.646 -29.579 -79.163 1.00 73.43 C C
+ATOM 6329 CD2 PHE L 134 58.145 -30.271 -77.449 1.00 72.32 C C
+ATOM 6330 CE1 PHE L 134 60.057 -30.895 -79.344 1.00 76.10 C C
+ATOM 6331 CE2 PHE L 134 58.555 -31.582 -77.621 1.00 75.69 C C
+ATOM 6332 CZ PHE L 134 59.509 -31.894 -78.567 1.00 75.54 C C
+ATOM 6333 N PRO L 135 59.077 -24.612 -77.723 1.00 73.15 C N
+ATOM 6334 CA PRO L 135 58.525 -23.267 -77.629 1.00 77.08 C C
+ATOM 6335 C PRO L 135 57.470 -23.080 -78.701 1.00 78.43 C C
+ATOM 6336 O PRO L 135 57.395 -23.893 -79.634 1.00 78.87 C O
+ATOM 6337 CB PRO L 135 59.752 -22.380 -77.883 1.00 78.92 C C
+ATOM 6338 CG PRO L 135 60.856 -23.357 -78.430 1.00 78.18 C C
+ATOM 6339 CD PRO L 135 60.185 -24.671 -78.688 1.00 74.57 C C
+ATOM 6340 N PRO L 136 56.701 -21.995 -78.669 1.00 79.85 C N
+ATOM 6341 CA PRO L 136 55.579 -21.881 -79.609 1.00 78.75 C C
+ATOM 6342 C PRO L 136 56.070 -21.828 -81.050 1.00 81.62 C C
+ATOM 6343 O PRO L 136 57.261 -21.672 -81.326 1.00 83.39 C O
+ATOM 6344 CB PRO L 136 54.863 -20.589 -79.183 1.00 76.77 C C
+ATOM 6345 CG PRO L 136 55.788 -19.907 -78.253 1.00 78.94 C C
+ATOM 6346 CD PRO L 136 56.622 -20.977 -77.612 1.00 78.98 C C
+ATOM 6347 N SER L 137 55.136 -22.035 -81.980 1.00 88.79 C N
+ATOM 6348 CA SER L 137 55.481 -22.097 -83.393 1.00 88.46 C C
+ATOM 6349 C SER L 137 55.623 -20.706 -83.979 1.00 89.15 C C
+ATOM 6350 O SER L 137 54.859 -19.792 -83.643 1.00 87.98 C O
+ATOM 6351 CB SER L 137 54.429 -22.867 -84.182 1.00 89.05 C C
+ATOM 6352 OG SER L 137 54.461 -24.235 -83.837 1.00 89.34 C O
+ATOM 6353 N ASP L 138 56.617 -20.548 -84.859 1.00 87.77 C N
+ATOM 6354 CA ASP L 138 56.780 -19.274 -85.543 1.00 93.67 C C
+ATOM 6355 C ASP L 138 55.497 -18.908 -86.265 1.00 93.69 C C
+ATOM 6356 O ASP L 138 54.988 -17.789 -86.120 1.00 91.43 C O
+ATOM 6357 CB ASP L 138 57.966 -19.338 -86.499 1.00 98.34 C C
+ATOM 6358 CG ASP L 138 59.265 -18.997 -85.809 1.00102.52 C C
+ATOM 6359 OD1 ASP L 138 59.254 -18.121 -84.909 1.00101.74 C O
+ATOM 6360 OD2 ASP L 138 60.292 -19.612 -86.156 1.00104.94 C O1-
+ATOM 6361 N GLU L 139 54.918 -19.874 -86.984 1.00 88.00 C N
+ATOM 6362 CA GLU L 139 53.601 -19.676 -87.584 1.00 87.61 C C
+ATOM 6363 C GLU L 139 52.556 -19.337 -86.525 1.00 82.77 C C
+ATOM 6364 O GLU L 139 51.937 -18.267 -86.564 1.00 83.77 C O
+ATOM 6365 CB GLU L 139 53.189 -20.927 -88.359 1.00 95.07 C C
+ATOM 6366 CG GLU L 139 51.714 -20.927 -88.734 1.00102.94 C C
+ATOM 6367 CD GLU L 139 51.303 -22.134 -89.556 1.00110.49 C C
+ATOM 6368 OE1 GLU L 139 52.186 -22.957 -89.907 1.00113.87 C O
+ATOM 6369 OE2 GLU L 139 50.088 -22.253 -89.842 1.00113.32 C O1-
+ATOM 6370 N GLN L 140 52.371 -20.232 -85.548 1.00 88.65 C N
+ATOM 6371 CA GLN L 140 51.340 -20.040 -84.531 1.00 87.44 C C
+ATOM 6372 C GLN L 140 51.443 -18.677 -83.869 1.00 87.59 C C
+ATOM 6373 O GLN L 140 50.442 -18.133 -83.380 1.00 88.77 C O
+ATOM 6374 CB GLN L 140 51.442 -21.125 -83.452 1.00 83.26 C C
+ATOM 6375 CG GLN L 140 50.464 -20.924 -82.294 1.00 80.48 C C
+ATOM 6376 CD GLN L 140 50.840 -21.704 -81.062 1.00 80.66 C C
+ATOM 6377 NE2 GLN L 140 51.644 -22.752 -81.243 1.00 78.45 C N
+ATOM 6378 OE1 GLN L 140 50.421 -21.366 -79.953 1.00 80.90 C O
+ATOM 6379 N LEU L 141 52.644 -18.113 -83.830 1.00 81.84 C N
+ATOM 6380 CA LEU L 141 52.798 -16.850 -83.139 1.00 85.35 C C
+ATOM 6381 C LEU L 141 52.036 -15.744 -83.848 1.00 91.16 C C
+ATOM 6382 O LEU L 141 51.581 -14.792 -83.199 1.00 94.38 C O
+ATOM 6383 CB LEU L 141 54.277 -16.533 -82.994 1.00 82.67 C C
+ATOM 6384 CG LEU L 141 54.717 -17.241 -81.716 1.00 82.93 C C
+ATOM 6385 CD1 LEU L 141 56.202 -17.133 -81.545 1.00 83.32 C C
+ATOM 6386 CD2 LEU L 141 53.981 -16.682 -80.500 1.00 83.99 C C
+ATOM 6387 N LYS L 142 51.837 -15.878 -85.164 1.00 80.28 C N
+ATOM 6388 CA LYS L 142 50.997 -14.916 -85.873 1.00 88.59 C C
+ATOM 6389 C LYS L 142 49.574 -14.915 -85.315 1.00 91.80 C C
+ATOM 6390 O LYS L 142 48.947 -13.855 -85.197 1.00 96.76 C O
+ATOM 6391 CB LYS L 142 51.008 -15.210 -87.380 1.00 95.37 C C
+ATOM 6392 CG LYS L 142 52.404 -15.161 -88.005 1.00102.36 C C
+ATOM 6393 CD LYS L 142 52.366 -15.231 -89.523 1.00106.93 C C
+ATOM 6394 CE LYS L 142 52.733 -13.885 -90.143 1.00107.84 C C
+ATOM 6395 NZ LYS L 142 52.893 -13.957 -91.621 1.00109.88 C N1+
+ATOM 6396 N SER L 143 49.079 -16.085 -84.898 1.00 89.68 C N
+ATOM 6397 CA SER L 143 47.685 -16.266 -84.503 1.00 89.28 C C
+ATOM 6398 C SER L 143 47.307 -15.549 -83.207 1.00 83.83 C C
+ATOM 6399 O SER L 143 46.114 -15.491 -82.876 1.00 87.87 C O
+ATOM 6400 CB SER L 143 47.389 -17.759 -84.363 1.00 94.84 C C
+ATOM 6401 OG SER L 143 47.809 -18.457 -85.517 1.00100.25 C O
+ATOM 6402 N GLY L 144 48.267 -15.014 -82.462 1.00 91.93 C N
+ATOM 6403 CA GLY L 144 47.928 -14.349 -81.225 1.00 89.80 C C
+ATOM 6404 C GLY L 144 47.829 -15.246 -80.010 1.00 87.82 C C
+ATOM 6405 O GLY L 144 47.457 -14.757 -78.932 1.00 88.28 C O
+ATOM 6406 N THR L 145 48.137 -16.545 -80.147 1.00 91.12 C N
+ATOM 6407 CA THR L 145 48.304 -17.450 -79.011 1.00 89.06 C C
+ATOM 6408 C THR L 145 49.634 -18.180 -79.134 1.00 86.43 C C
+ATOM 6409 O THR L 145 50.047 -18.559 -80.240 1.00 85.39 C O
+ATOM 6410 CB THR L 145 47.184 -18.512 -78.890 1.00 89.73 C C
+ATOM 6411 CG2 THR L 145 45.876 -17.891 -78.389 1.00 86.80 C C
+ATOM 6412 OG1 THR L 145 46.998 -19.189 -80.145 1.00 92.25 C O
+ATOM 6413 N ALA L 146 50.284 -18.378 -77.983 1.00 84.67 C N
+ATOM 6414 CA ALA L 146 51.526 -19.136 -77.864 1.00 80.83 C C
+ATOM 6415 C ALA L 146 51.272 -20.383 -77.024 1.00 79.09 C C
+ATOM 6416 O ALA L 146 50.799 -20.289 -75.881 1.00 80.78 C O
+ATOM 6417 CB ALA L 146 52.636 -18.283 -77.245 1.00 80.34 C C
+ATOM 6418 N SER L 147 51.574 -21.546 -77.596 1.00 75.37 C N
+ATOM 6419 CA SER L 147 51.498 -22.820 -76.895 1.00 70.83 C C
+ATOM 6420 C SER L 147 52.910 -23.307 -76.638 1.00 70.35 C C
+ATOM 6421 O SER L 147 53.655 -23.573 -77.586 1.00 74.97 C O
+ATOM 6422 CB SER L 147 50.724 -23.864 -77.702 1.00 74.20 C C
+ATOM 6423 OG SER L 147 49.326 -23.635 -77.666 1.00 75.21 C O
+ATOM 6424 N VAL L 148 53.275 -23.422 -75.369 1.00 69.31 C N
+ATOM 6425 CA VAL L 148 54.533 -24.039 -74.973 1.00 67.27 C C
+ATOM 6426 C VAL L 148 54.245 -25.467 -74.555 1.00 69.08 C C
+ATOM 6427 O VAL L 148 53.286 -25.722 -73.812 1.00 69.50 C O
+ATOM 6428 CB VAL L 148 55.190 -23.275 -73.824 1.00 67.19 C C
+ATOM 6429 CG1 VAL L 148 56.614 -23.756 -73.641 1.00 64.45 C C
+ATOM 6430 CG2 VAL L 148 55.121 -21.822 -74.117 1.00 65.75 C C
+ATOM 6431 N VAL L 149 55.072 -26.405 -75.002 1.00 64.98 C N
+ATOM 6432 CA VAL L 149 54.762 -27.811 -74.785 1.00 63.76 C C
+ATOM 6433 C VAL L 149 55.935 -28.496 -74.100 1.00 66.57 C C
+ATOM 6434 O VAL L 149 57.012 -28.629 -74.686 1.00 66.50 C O
+ATOM 6435 CB VAL L 149 54.383 -28.519 -76.090 1.00 65.78 C C
+ATOM 6436 CG1 VAL L 149 54.540 -30.021 -75.946 1.00 62.38 C C
+ATOM 6437 CG2 VAL L 149 52.929 -28.188 -76.433 1.00 61.66 C C
+ATOM 6438 N CYS L 150 55.719 -28.946 -72.868 1.00 54.24 C N
+ATOM 6439 CA CYS L 150 56.676 -29.779 -72.157 1.00 56.83 C C
+ATOM 6440 C CYS L 150 56.287 -31.250 -72.273 1.00 60.70 C C
+ATOM 6441 O CYS L 150 55.128 -31.616 -72.043 1.00 60.50 C O
+ATOM 6442 CB CYS L 150 56.761 -29.375 -70.691 1.00 55.53 C C
+ATOM 6443 SG CYS L 150 58.188 -30.087 -69.890 1.00 67.63 C S
+ATOM 6444 N LEU L 151 57.269 -32.081 -72.608 1.00 56.81 C N
+ATOM 6445 CA LEU L 151 57.107 -33.506 -72.860 1.00 53.94 C C
+ATOM 6446 C LEU L 151 57.935 -34.307 -71.867 1.00 53.38 C C
+ATOM 6447 O LEU L 151 59.100 -33.982 -71.624 1.00 57.50 C O
+ATOM 6448 CB LEU L 151 57.536 -33.846 -74.281 1.00 56.15 C C
+ATOM 6449 CG LEU L 151 57.966 -35.282 -74.552 1.00 54.85 C C
+ATOM 6450 CD1 LEU L 151 56.813 -36.207 -74.334 1.00 53.32 C C
+ATOM 6451 CD2 LEU L 151 58.441 -35.418 -75.992 1.00 53.45 C C
+ATOM 6452 N LEU L 152 57.328 -35.344 -71.292 1.00 40.29 C N
+ATOM 6453 CA LEU L 152 57.994 -36.294 -70.413 1.00 44.04 C C
+ATOM 6454 C LEU L 152 57.858 -37.654 -71.068 1.00 49.15 C C
+ATOM 6455 O LEU L 152 56.740 -38.144 -71.259 1.00 49.05 C O
+ATOM 6456 CB LEU L 152 57.379 -36.306 -69.015 1.00 43.88 C C
+ATOM 6457 CG LEU L 152 57.364 -35.028 -68.165 1.00 49.60 C C
+ATOM 6458 CD1 LEU L 152 56.577 -33.888 -68.776 1.00 51.51 C C
+ATOM 6459 CD2 LEU L 152 56.861 -35.326 -66.770 1.00 48.38 C C
+ATOM 6460 N ASN L 153 58.979 -38.257 -71.438 1.00 54.58 C N
+ATOM 6461 CA ASN L 153 58.939 -39.435 -72.289 1.00 52.84 C C
+ATOM 6462 C ASN L 153 59.379 -40.697 -71.562 1.00 56.44 C C
+ATOM 6463 O ASN L 153 60.464 -40.741 -70.950 1.00 58.11 C O
+ATOM 6464 CB ASN L 153 59.774 -39.247 -73.544 1.00 54.47 C C
+ATOM 6465 CG ASN L 153 59.082 -39.806 -74.756 1.00 57.51 C C
+ATOM 6466 ND2 ASN L 153 59.433 -39.295 -75.926 1.00 56.95 C N
+ATOM 6467 OD1 ASN L 153 58.200 -40.665 -74.639 1.00 60.68 C O
+ATOM 6468 N ASN L 154 58.486 -41.695 -71.622 1.00 62.60 C N
+ATOM 6469 CA ASN L 154 58.740 -43.104 -71.336 1.00 61.26 C C
+ATOM 6470 C ASN L 154 59.209 -43.301 -69.893 1.00 61.31 C C
+ATOM 6471 O ASN L 154 60.355 -43.634 -69.611 1.00 65.47 C O
+ATOM 6472 CB ASN L 154 59.728 -43.656 -72.373 1.00 60.48 C C
+ATOM 6473 CG ASN L 154 59.068 -43.913 -73.729 1.00 64.43 C C
+ATOM 6474 ND2 ASN L 154 59.876 -44.244 -74.728 1.00 72.38 C N
+ATOM 6475 OD1 ASN L 154 57.854 -43.778 -73.878 1.00 62.49 C O
+ATOM 6476 N PHE L 155 58.280 -43.078 -68.976 1.00 59.63 C N
+ATOM 6477 CA PHE L 155 58.606 -43.156 -67.565 1.00 55.45 C C
+ATOM 6478 C PHE L 155 57.670 -44.145 -66.894 1.00 56.91 C C
+ATOM 6479 O PHE L 155 56.778 -44.709 -67.534 1.00 56.56 C O
+ATOM 6480 CB PHE L 155 58.520 -41.782 -66.901 1.00 53.65 C C
+ATOM 6481 CG PHE L 155 57.171 -41.139 -67.005 1.00 53.58 C C
+ATOM 6482 CD1 PHE L 155 56.891 -40.245 -68.025 1.00 52.65 C C
+ATOM 6483 CD2 PHE L 155 56.189 -41.394 -66.054 1.00 51.06 C C
+ATOM 6484 CE1 PHE L 155 55.649 -39.637 -68.108 1.00 49.77 C C
+ATOM 6485 CE2 PHE L 155 54.945 -40.791 -66.134 1.00 51.29 C C
+ATOM 6486 CZ PHE L 155 54.675 -39.911 -67.163 1.00 50.47 C C
+ATOM 6487 N TYR L 156 57.850 -44.302 -65.580 1.00 55.88 C N
+ATOM 6488 CA TYR L 156 57.087 -45.221 -64.744 1.00 51.50 C C
+ATOM 6489 C TYR L 156 57.524 -44.947 -63.332 1.00 55.18 C C
+ATOM 6490 O TYR L 156 58.712 -44.818 -63.077 1.00 58.87 C O
+ATOM 6491 CB TYR L 156 57.352 -46.698 -65.084 1.00 53.64 C C
+ATOM 6492 CG TYR L 156 56.417 -47.608 -64.350 1.00 51.07 C C
+ATOM 6493 CD1 TYR L 156 56.587 -47.846 -63.010 1.00 51.51 C C
+ATOM 6494 CD2 TYR L 156 55.356 -48.210 -64.987 1.00 47.95 C C
+ATOM 6495 CE1 TYR L 156 55.721 -48.618 -62.289 1.00 55.22 C C
+ATOM 6496 CE2 TYR L 156 54.469 -49.017 -64.280 1.00 47.04 C C
+ATOM 6497 CZ TYR L 156 54.660 -49.221 -62.906 1.00 53.11 C C
+ATOM 6498 OH TYR L 156 53.811 -50.017 -62.119 1.00 59.24 C O
+ATOM 6499 N PRO L 157 56.573 -44.886 -62.394 1.00 57.81 C N
+ATOM 6500 CA PRO L 157 55.114 -44.965 -62.579 1.00 56.71 C C
+ATOM 6501 C PRO L 157 54.452 -43.663 -63.090 1.00 55.01 C C
+ATOM 6502 O PRO L 157 55.156 -42.660 -63.256 1.00 54.11 C O
+ATOM 6503 CB PRO L 157 54.612 -45.314 -61.179 1.00 58.08 C C
+ATOM 6504 CG PRO L 157 55.670 -44.806 -60.261 1.00 58.63 C C
+ATOM 6505 CD PRO L 157 56.965 -44.937 -60.976 1.00 56.96 C C
+ATOM 6506 N ARG L 158 53.127 -43.688 -63.323 1.00 55.40 C N
+ATOM 6507 CA ARG L 158 52.440 -42.572 -63.983 1.00 59.54 C C
+ATOM 6508 C ARG L 158 52.581 -41.263 -63.194 1.00 64.01 C C
+ATOM 6509 O ARG L 158 52.741 -40.189 -63.789 1.00 65.29 C O
+ATOM 6510 CB ARG L 158 50.965 -42.945 -64.212 1.00 61.82 C C
+ATOM 6511 CG ARG L 158 50.007 -41.799 -64.589 1.00 69.97 C C
+ATOM 6512 CD ARG L 158 48.575 -42.303 -64.923 1.00 79.15 C C
+ATOM 6513 NE ARG L 158 47.614 -41.215 -65.184 1.00 85.09 C N
+ATOM 6514 CZ ARG L 158 46.332 -41.385 -65.532 1.00 89.37 C C
+ATOM 6515 NH1 ARG L 158 45.841 -42.614 -65.682 1.00 91.79 C N1+
+ATOM 6516 NH2 ARG L 158 45.537 -40.327 -65.733 1.00 91.49 C N
+ATOM 6517 N GLU L 159 52.574 -41.331 -61.861 1.00 60.26 C N
+ATOM 6518 CA GLU L 159 52.538 -40.121 -61.044 1.00 61.22 C C
+ATOM 6519 C GLU L 159 53.781 -39.279 -61.282 1.00 62.42 C C
+ATOM 6520 O GLU L 159 54.902 -39.754 -61.093 1.00 61.31 C O
+ATOM 6521 CB GLU L 159 52.429 -40.501 -59.569 1.00 67.91 C C
+ATOM 6522 CG GLU L 159 51.181 -41.305 -59.246 1.00 76.15 C C
+ATOM 6523 CD GLU L 159 51.244 -42.774 -59.695 1.00 77.54 C C
+ATOM 6524 OE1 GLU L 159 51.895 -43.072 -60.727 1.00 78.79 C O
+ATOM 6525 OE2 GLU L 159 50.593 -43.631 -59.044 1.00 76.49 C O1-
+ATOM 6526 N ALA L 160 53.586 -38.030 -61.705 1.00 58.15 C N
+ATOM 6527 CA ALA L 160 54.685 -37.084 -61.861 1.00 54.78 C C
+ATOM 6528 C ALA L 160 54.135 -35.675 -61.736 1.00 56.70 C C
+ATOM 6529 O ALA L 160 52.999 -35.412 -62.132 1.00 57.72 C O
+ATOM 6530 CB ALA L 160 55.391 -37.257 -63.203 1.00 51.38 C C
+ATOM 6531 N LYS L 161 54.936 -34.762 -61.192 1.00 58.93 C N
+ATOM 6532 CA LYS L 161 54.513 -33.369 -61.074 1.00 62.59 C C
+ATOM 6533 C LYS L 161 55.238 -32.548 -62.131 1.00 57.75 C C
+ATOM 6534 O LYS L 161 56.472 -32.434 -62.098 1.00 53.73 C O
+ATOM 6535 CB LYS L 161 54.755 -32.801 -59.669 1.00 71.17 C C
+ATOM 6536 CG LYS L 161 53.439 -32.646 -58.862 1.00 79.91 C C
+ATOM 6537 CD LYS L 161 53.560 -32.053 -57.430 1.00 88.90 C C
+ATOM 6538 CE LYS L 161 52.209 -32.220 -56.679 1.00 95.67 C C
+ATOM 6539 NZ LYS L 161 52.206 -31.795 -55.247 1.00100.21 C N1+
+ATOM 6540 N VAL L 162 54.475 -32.005 -63.083 1.00 55.15 C N
+ATOM 6541 CA VAL L 162 54.986 -31.019 -64.028 1.00 57.76 C C
+ATOM 6542 C VAL L 162 54.622 -29.639 -63.511 1.00 56.99 C C
+ATOM 6543 O VAL L 162 53.471 -29.395 -63.127 1.00 64.47 C O
+ATOM 6544 CB VAL L 162 54.431 -31.239 -65.440 1.00 55.82 C C
+ATOM 6545 CG1 VAL L 162 54.788 -30.071 -66.294 1.00 55.38 C C
+ATOM 6546 CG2 VAL L 162 55.039 -32.474 -66.039 1.00 57.51 C C
+ATOM 6547 N GLN L 163 55.599 -28.743 -63.497 1.00 65.52 C N
+ATOM 6548 CA GLN L 163 55.456 -27.402 -62.961 1.00 65.82 C C
+ATOM 6549 C GLN L 163 55.869 -26.397 -64.030 1.00 61.33 C C
+ATOM 6550 O GLN L 163 56.763 -26.676 -64.838 1.00 57.85 C O
+ATOM 6551 CB GLN L 163 56.321 -27.250 -61.734 1.00 61.48 C C
+ATOM 6552 CG GLN L 163 55.892 -26.171 -60.822 1.00 58.82 C C
+ATOM 6553 CD GLN L 163 55.129 -26.688 -59.618 1.00 68.00 C C
+ATOM 6554 NE2 GLN L 163 53.791 -26.655 -59.688 1.00 70.59 C N
+ATOM 6555 OE1 GLN L 163 55.740 -27.069 -58.610 1.00 74.20 C O
+ATOM 6556 N TRP L 164 55.206 -25.233 -64.064 1.00 63.57 C N
+ATOM 6557 CA TRP L 164 55.552 -24.191 -65.027 1.00 62.00 C C
+ATOM 6558 C TRP L 164 55.978 -22.936 -64.292 1.00 65.09 C C
+ATOM 6559 O TRP L 164 55.327 -22.520 -63.327 1.00 68.66 C O
+ATOM 6560 CB TRP L 164 54.396 -23.839 -65.948 1.00 57.49 C C
+ATOM 6561 CG TRP L 164 54.143 -24.814 -67.010 1.00 57.19 C C
+ATOM 6562 CD1 TRP L 164 53.206 -25.791 -67.000 1.00 57.38 C C
+ATOM 6563 CD2 TRP L 164 54.812 -24.908 -68.263 1.00 56.45 C C
+ATOM 6564 CE2 TRP L 164 54.229 -25.979 -68.967 1.00 60.17 C C
+ATOM 6565 CE3 TRP L 164 55.844 -24.195 -68.861 1.00 56.80 C C
+ATOM 6566 NE1 TRP L 164 53.249 -26.501 -68.170 1.00 59.87 C N
+ATOM 6567 CZ2 TRP L 164 54.650 -26.363 -70.245 1.00 62.64 C C
+ATOM 6568 CZ3 TRP L 164 56.264 -24.573 -70.143 1.00 58.70 C C
+ATOM 6569 CH2 TRP L 164 55.666 -25.649 -70.816 1.00 60.49 C C
+ATOM 6570 N LYS L 165 57.072 -22.333 -64.752 1.00 72.93 C N
+ATOM 6571 CA LYS L 165 57.493 -21.027 -64.256 1.00 71.37 C C
+ATOM 6572 C LYS L 165 57.913 -20.158 -65.431 1.00 73.57 C C
+ATOM 6573 O LYS L 165 58.833 -20.512 -66.181 1.00 72.15 C O
+ATOM 6574 CB LYS L 165 58.627 -21.156 -63.235 1.00 70.05 C C
+ATOM 6575 CG LYS L 165 58.225 -21.915 -61.970 1.00 71.20 C C
+ATOM 6576 CD LYS L 165 59.355 -21.965 -60.957 1.00 77.05 C C
+ATOM 6577 CE LYS L 165 59.804 -23.384 -60.721 1.00 88.31 C C
+ATOM 6578 NZ LYS L 165 60.492 -23.500 -59.420 1.00 92.93 C N1+
+ATOM 6579 N VAL L 166 57.218 -19.039 -65.610 1.00 84.56 C N
+ATOM 6580 CA VAL L 166 57.632 -18.028 -66.574 1.00 81.90 C C
+ATOM 6581 C VAL L 166 58.434 -16.974 -65.819 1.00 84.43 C C
+ATOM 6582 O VAL L 166 58.011 -16.502 -64.748 1.00 91.03 C O
+ATOM 6583 CB VAL L 166 56.425 -17.417 -67.324 1.00 81.87 C C
+ATOM 6584 CG1 VAL L 166 55.536 -18.499 -67.902 1.00 83.98 C C
+ATOM 6585 CG2 VAL L 166 55.612 -16.476 -66.448 1.00 75.78 C C
+ATOM 6586 N ASP L 167 59.625 -16.648 -66.347 1.00 84.82 C N
+ATOM 6587 CA ASP L 167 60.571 -15.765 -65.663 1.00 85.31 C C
+ATOM 6588 C ASP L 167 60.661 -16.166 -64.204 1.00 84.99 C C
+ATOM 6589 O ASP L 167 60.681 -15.331 -63.297 1.00 88.22 C O
+ATOM 6590 CB ASP L 167 60.197 -14.290 -65.815 1.00 85.01 C C
+ATOM 6591 CG ASP L 167 60.560 -13.739 -67.183 1.00 88.74 C C
+ATOM 6592 OD1 ASP L 167 61.425 -14.341 -67.868 1.00 87.84 C O
+ATOM 6593 OD2 ASP L 167 59.966 -12.709 -67.577 1.00 90.62 C O1-
+ATOM 6594 N ASN L 168 60.628 -17.478 -63.996 1.00 83.48 C N
+ATOM 6595 CA ASN L 168 60.882 -18.121 -62.725 1.00 87.61 C C
+ATOM 6596 C ASN L 168 59.819 -17.793 -61.670 1.00 90.86 C C
+ATOM 6597 O ASN L 168 60.075 -17.937 -60.471 1.00 91.57 C O
+ATOM 6598 CB ASN L 168 62.291 -17.730 -62.257 1.00 95.78 C C
+ATOM 6599 CG ASN L 168 63.120 -18.915 -61.837 1.00101.13 C C
+ATOM 6600 ND2 ASN L 168 64.406 -18.897 -62.212 1.00100.16 C N
+ATOM 6601 OD1 ASN L 168 62.613 -19.856 -61.211 1.00105.13 C O
+ATOM 6602 N ALA L 169 58.615 -17.383 -62.089 1.00 80.25 C N
+ATOM 6603 CA ALA L 169 57.456 -17.248 -61.206 1.00 89.58 C C
+ATOM 6604 C ALA L 169 56.416 -18.309 -61.563 1.00 86.20 C C
+ATOM 6605 O ALA L 169 56.210 -18.622 -62.744 1.00 85.08 C O
+ATOM 6606 CB ALA L 169 56.846 -15.849 -61.298 1.00 90.42 C C
+ATOM 6607 N LEU L 170 55.757 -18.858 -60.541 1.00 78.33 C N
+ATOM 6608 CA LEU L 170 55.029 -20.117 -60.689 1.00 75.96 C C
+ATOM 6609 C LEU L 170 53.658 -19.907 -61.336 1.00 79.19 C C
+ATOM 6610 O LEU L 170 52.789 -19.235 -60.767 1.00 80.68 C O
+ATOM 6611 CB LEU L 170 54.865 -20.783 -59.327 1.00 77.16 C C
+ATOM 6612 CG LEU L 170 54.175 -22.127 -59.486 1.00 75.23 C C
+ATOM 6613 CD1 LEU L 170 55.075 -22.971 -60.315 1.00 74.75 C C
+ATOM 6614 CD2 LEU L 170 53.877 -22.775 -58.154 1.00 73.52 C C
+ATOM 6615 N GLN L 171 53.439 -20.533 -62.492 1.00 81.00 C N
+ATOM 6616 CA GLN L 171 52.193 -20.372 -63.230 1.00 80.76 C C
+ATOM 6617 C GLN L 171 51.147 -21.343 -62.713 1.00 81.75 C C
+ATOM 6618 O GLN L 171 51.459 -22.495 -62.398 1.00 81.42 C O
+ATOM 6619 CB GLN L 171 52.418 -20.621 -64.716 1.00 77.44 C C
+ATOM 6620 CG GLN L 171 53.477 -19.744 -65.292 1.00 81.09 C C
+ATOM 6621 CD GLN L 171 53.030 -18.316 -65.365 1.00 83.95 C C
+ATOM 6622 NE2 GLN L 171 52.174 -18.014 -66.336 1.00 79.29 C N
+ATOM 6623 OE1 GLN L 171 53.447 -17.481 -64.561 1.00 89.03 C O
+ATOM 6624 N SER L 172 49.898 -20.882 -62.644 1.00 83.45 C N
+ATOM 6625 CA SER L 172 48.804 -21.705 -62.133 1.00 85.46 C C
+ATOM 6626 C SER L 172 47.555 -21.517 -62.989 1.00 85.69 C C
+ATOM 6627 O SER L 172 47.046 -20.400 -63.119 1.00 82.50 C O
+ATOM 6628 CB SER L 172 48.509 -21.366 -60.664 1.00 83.63 C C
+ATOM 6629 OG SER L 172 47.715 -22.365 -60.049 1.00 83.79 C O
+ATOM 6630 N GLY L 173 47.050 -22.609 -63.554 1.00 80.76 C N
+ATOM 6631 CA GLY L 173 45.739 -22.625 -64.161 1.00 69.41 C C
+ATOM 6632 C GLY L 173 45.719 -22.504 -65.664 1.00 73.80 C C
+ATOM 6633 O GLY L 173 44.655 -22.689 -66.262 1.00 78.98 C O
+ATOM 6634 N ASN L 174 46.840 -22.165 -66.285 1.00 73.00 C N
+ATOM 6635 CA ASN L 174 46.935 -22.007 -67.731 1.00 73.41 C C
+ATOM 6636 C ASN L 174 47.511 -23.226 -68.465 1.00 71.25 C C
+ATOM 6637 O ASN L 174 47.785 -23.145 -69.672 1.00 78.44 C O
+ATOM 6638 CB ASN L 174 47.732 -20.744 -68.045 1.00 65.27 C C
+ATOM 6639 CG ASN L 174 48.893 -20.556 -67.099 1.00 73.90 C C
+ATOM 6640 ND2 ASN L 174 49.840 -19.684 -67.475 1.00 70.92 C N
+ATOM 6641 OD1 ASN L 174 48.966 -21.231 -66.057 1.00 76.90 C O
+ATOM 6642 N SER L 175 47.721 -24.338 -67.767 1.00 65.87 C N
+ATOM 6643 CA SER L 175 48.299 -25.542 -68.346 1.00 66.75 C C
+ATOM 6644 C SER L 175 47.301 -26.696 -68.361 1.00 67.96 C C
+ATOM 6645 O SER L 175 46.433 -26.801 -67.486 1.00 68.44 C O
+ATOM 6646 CB SER L 175 49.523 -25.973 -67.545 1.00 65.79 C C
+ATOM 6647 OG SER L 175 49.093 -26.664 -66.380 1.00 68.62 C O
+ATOM 6648 N GLN L 176 47.459 -27.591 -69.344 1.00 57.40 C N
+ATOM 6649 CA GLN L 176 46.699 -28.840 -69.403 1.00 59.71 C C
+ATOM 6650 C GLN L 176 47.620 -30.032 -69.635 1.00 58.95 C C
+ATOM 6651 O GLN L 176 48.604 -29.939 -70.378 1.00 59.30 C O
+ATOM 6652 CB GLN L 176 45.651 -28.811 -70.502 1.00 62.72 C C
+ATOM 6653 CG GLN L 176 44.266 -28.468 -70.024 1.00 62.19 C C
+ATOM 6654 CD GLN L 176 43.424 -27.921 -71.153 1.00 66.07 C C
+ATOM 6655 NE2 GLN L 176 42.199 -27.502 -70.840 1.00 67.37 C N
+ATOM 6656 OE1 GLN L 176 43.872 -27.880 -72.302 1.00 71.31 C O
+ATOM 6657 N GLU L 177 47.279 -31.158 -69.007 1.00 57.54 C N
+ATOM 6658 CA GLU L 177 48.076 -32.379 -69.070 1.00 55.42 C C
+ATOM 6659 C GLU L 177 47.263 -33.489 -69.718 1.00 56.57 C C
+ATOM 6660 O GLU L 177 46.116 -33.735 -69.329 1.00 57.04 C O
+ATOM 6661 CB GLU L 177 48.534 -32.822 -67.666 1.00 56.24 C C
+ATOM 6662 CG GLU L 177 49.709 -32.033 -67.074 1.00 66.12 C C
+ATOM 6663 CD GLU L 177 50.239 -32.634 -65.765 1.00 70.38 C C
+ATOM 6664 OE1 GLU L 177 49.999 -33.842 -65.512 1.00 73.83 C O
+ATOM 6665 OE2 GLU L 177 50.899 -31.891 -64.990 1.00 69.86 C O1-
+ATOM 6666 N SER L 178 47.861 -34.160 -70.687 1.00 57.18 C N
+ATOM 6667 CA SER L 178 47.337 -35.406 -71.220 1.00 53.57 C C
+ATOM 6668 C SER L 178 48.407 -36.478 -71.048 1.00 55.52 C C
+ATOM 6669 O SER L 178 49.594 -36.156 -70.986 1.00 57.67 C O
+ATOM 6670 CB SER L 178 46.954 -35.233 -72.691 1.00 54.89 C C
+ATOM 6671 OG SER L 178 46.741 -36.468 -73.354 1.00 60.85 C O
+ATOM 6672 N VAL L 179 48.015 -37.754 -70.948 1.00 50.81 C N
+ATOM 6673 CA VAL L 179 48.983 -38.799 -70.616 1.00 47.83 C C
+ATOM 6674 C VAL L 179 48.703 -40.063 -71.428 1.00 52.96 C C
+ATOM 6675 O VAL L 179 47.548 -40.463 -71.607 1.00 55.58 C O
+ATOM 6676 CB VAL L 179 48.979 -39.084 -69.097 1.00 52.82 C C
+ATOM 6677 CG1 VAL L 179 47.767 -39.916 -68.676 1.00 59.38 C C
+ATOM 6678 CG2 VAL L 179 50.261 -39.723 -68.681 1.00 53.60 C C
+ATOM 6679 N THR L 180 49.775 -40.712 -71.888 1.00 59.86 C N
+ATOM 6680 CA THR L 180 49.681 -41.941 -72.671 1.00 60.14 C C
+ATOM 6681 C THR L 180 48.992 -43.027 -71.871 1.00 58.81 C C
+ATOM 6682 O THR L 180 48.967 -43.009 -70.640 1.00 58.53 C O
+ATOM 6683 CB THR L 180 51.070 -42.485 -73.031 1.00 59.73 C C
+ATOM 6684 CG2 THR L 180 51.022 -43.399 -74.245 1.00 58.56 C C
+ATOM 6685 OG1 THR L 180 51.962 -41.408 -73.305 1.00 61.76 C O
+ATOM 6686 N GLU L 181 48.444 -43.994 -72.577 1.00 72.48 C N
+ATOM 6687 CA GLU L 181 48.127 -45.242 -71.915 1.00 74.40 C C
+ATOM 6688 C GLU L 181 49.369 -46.120 -71.941 1.00 73.16 C C
+ATOM 6689 O GLU L 181 50.162 -46.066 -72.886 1.00 72.90 C O
+ATOM 6690 CB GLU L 181 46.932 -45.926 -72.579 1.00 73.33 C C
+ATOM 6691 CG GLU L 181 45.646 -45.073 -72.563 1.00 76.81 C C
+ATOM 6692 CD GLU L 181 45.251 -44.523 -71.143 1.00 83.32 C C
+ATOM 6693 OE1 GLU L 181 45.618 -45.151 -70.091 1.00 86.28 C O
+ATOM 6694 OE2 GLU L 181 44.567 -43.447 -71.099 1.00 87.66 C O1-
+ATOM 6695 N GLN L 182 49.557 -46.900 -70.876 1.00 63.48 C N
+ATOM 6696 CA GLN L 182 50.826 -47.593 -70.660 1.00 65.25 C C
+ATOM 6697 C GLN L 182 51.217 -48.428 -71.865 1.00 69.06 C C
+ATOM 6698 O GLN L 182 50.447 -49.268 -72.326 1.00 72.85 C O
+ATOM 6699 CB GLN L 182 50.716 -48.471 -69.418 1.00 64.08 C C
+ATOM 6700 CG GLN L 182 52.019 -49.127 -69.004 1.00 69.45 C C
+ATOM 6701 CD GLN L 182 51.861 -49.986 -67.753 1.00 68.38 C C
+ATOM 6702 NE2 GLN L 182 52.964 -50.215 -67.043 1.00 70.49 C N
+ATOM 6703 OE1 GLN L 182 50.759 -50.421 -67.422 1.00 67.83 C O
+ATOM 6704 N ASP L 183 52.414 -48.196 -72.381 1.00 67.64 C N
+ATOM 6705 CA ASP L 183 52.788 -48.811 -73.647 1.00 73.20 C C
+ATOM 6706 C ASP L 183 52.917 -50.325 -73.523 1.00 81.44 C C
+ATOM 6707 O ASP L 183 53.289 -50.859 -72.478 1.00 82.56 C O
+ATOM 6708 CB ASP L 183 54.098 -48.224 -74.165 1.00 72.53 C C
+ATOM 6709 CG ASP L 183 54.544 -48.865 -75.467 1.00 78.54 C C
+ATOM 6710 OD1 ASP L 183 53.698 -49.504 -76.137 1.00 78.98 C O
+ATOM 6711 OD2 ASP L 183 55.727 -48.724 -75.840 1.00 83.34 C O1-
+ATOM 6712 N SER L 184 52.618 -51.015 -74.626 1.00 78.05 C N
+ATOM 6713 CA SER L 184 52.684 -52.476 -74.651 1.00 81.42 C C
+ATOM 6714 C SER L 184 54.114 -52.989 -74.457 1.00 82.87 C C
+ATOM 6715 O SER L 184 54.361 -53.849 -73.600 1.00 88.74 C O
+ATOM 6716 CB SER L 184 52.114 -52.987 -75.969 1.00 81.52 C C
+ATOM 6717 OG SER L 184 52.891 -52.500 -77.050 1.00 83.64 C O
+ATOM 6718 N LYS L 185 55.065 -52.488 -75.258 1.00 82.92 C N
+ATOM 6719 CA LYS L 185 56.428 -53.019 -75.236 1.00 85.93 C C
+ATOM 6720 C LYS L 185 57.134 -52.741 -73.911 1.00 79.41 C C
+ATOM 6721 O LYS L 185 57.562 -53.674 -73.223 1.00 82.24 C O
+ATOM 6722 CB LYS L 185 57.243 -52.444 -76.404 1.00 94.41 C C
+ATOM 6723 CG LYS L 185 57.151 -53.185 -77.763 1.00104.03 C C
+ATOM 6724 CD LYS L 185 57.929 -54.521 -77.700 1.00111.84 C C
+ATOM 6725 CE LYS L 185 57.918 -55.309 -79.015 1.00116.41 C C
+ATOM 6726 NZ LYS L 185 57.976 -56.799 -78.807 1.00120.21 C N1+
+ATOM 6727 N ASP L 186 57.290 -51.463 -73.545 1.00 74.79 C N
+ATOM 6728 CA ASP L 186 58.119 -51.075 -72.397 1.00 72.91 C C
+ATOM 6729 C ASP L 186 57.343 -50.731 -71.132 1.00 73.07 C C
+ATOM 6730 O ASP L 186 57.961 -50.366 -70.135 1.00 75.39 C O
+ATOM 6731 CB ASP L 186 59.015 -49.890 -72.763 1.00 73.30 C C
+ATOM 6732 CG ASP L 186 58.232 -48.715 -73.277 1.00 76.16 C C
+ATOM 6733 OD1 ASP L 186 57.028 -48.902 -73.549 1.00 72.17 C O
+ATOM 6734 OD2 ASP L 186 58.826 -47.623 -73.442 1.00 82.31 C O1-
+ATOM 6735 N SER L 187 56.019 -50.790 -71.160 1.00 68.17 C N
+ATOM 6736 CA SER L 187 55.184 -50.492 -69.997 1.00 66.19 C C
+ATOM 6737 C SER L 187 55.515 -49.131 -69.393 1.00 68.82 C C
+ATOM 6738 O SER L 187 55.657 -48.983 -68.175 1.00 71.69 C O
+ATOM 6739 CB SER L 187 55.300 -51.592 -68.947 1.00 67.09 C C
+ATOM 6740 OG SER L 187 54.913 -52.833 -69.495 1.00 75.41 C O
+ATOM 6741 N THR L 188 55.620 -48.117 -70.254 1.00 66.91 C N
+ATOM 6742 CA THR L 188 55.916 -46.759 -69.815 1.00 65.15 C C
+ATOM 6743 C THR L 188 54.830 -45.780 -70.234 1.00 67.36 C C
+ATOM 6744 O THR L 188 54.465 -45.694 -71.417 1.00 69.35 C O
+ATOM 6745 CB THR L 188 57.244 -46.262 -70.369 1.00 67.46 C C
+ATOM 6746 CG2 THR L 188 58.315 -47.324 -70.197 1.00 72.49 C C
+ATOM 6747 OG1 THR L 188 57.077 -45.937 -71.759 1.00 63.36 C O
+ATOM 6748 N TYR L 189 54.362 -45.011 -69.264 1.00 61.68 C N
+ATOM 6749 CA TYR L 189 53.599 -43.812 -69.551 1.00 58.86 C C
+ATOM 6750 C TYR L 189 54.502 -42.732 -70.163 1.00 58.34 C C
+ATOM 6751 O TYR L 189 55.722 -42.720 -69.953 1.00 64.10 C O
+ATOM 6752 CB TYR L 189 52.955 -43.315 -68.260 1.00 55.85 C C
+ATOM 6753 CG TYR L 189 52.132 -44.380 -67.593 1.00 55.81 C C
+ATOM 6754 CD1 TYR L 189 50.780 -44.493 -67.846 1.00 58.57 C C
+ATOM 6755 CD2 TYR L 189 52.716 -45.301 -66.748 1.00 57.76 C C
+ATOM 6756 CE1 TYR L 189 50.021 -45.484 -67.249 1.00 59.40 C C
+ATOM 6757 CE2 TYR L 189 51.977 -46.295 -66.149 1.00 57.51 C C
+ATOM 6758 CZ TYR L 189 50.629 -46.384 -66.399 1.00 64.66 C C
+ATOM 6759 OH TYR L 189 49.881 -47.373 -65.804 1.00 74.23 C O
+ATOM 6760 N SER L 190 53.892 -41.840 -70.964 1.00 55.97 C N
+ATOM 6761 CA SER L 190 54.480 -40.554 -71.347 1.00 57.42 C C
+ATOM 6762 C SER L 190 53.413 -39.468 -71.214 1.00 54.95 C C
+ATOM 6763 O SER L 190 52.211 -39.742 -71.307 1.00 58.50 C O
+ATOM 6764 CB SER L 190 55.039 -40.570 -72.767 1.00 56.85 C C
+ATOM 6765 OG SER L 190 55.585 -41.842 -73.057 1.00 57.96 C O
+ATOM 6766 N LEU L 191 53.848 -38.231 -70.978 1.00 51.38 C N
+ATOM 6767 CA LEU L 191 52.933 -37.173 -70.560 1.00 51.76 C C
+ATOM 6768 C LEU L 191 53.238 -35.894 -71.321 1.00 52.11 C C
+ATOM 6769 O LEU L 191 54.402 -35.547 -71.525 1.00 52.75 C O
+ATOM 6770 CB LEU L 191 53.028 -36.912 -69.047 1.00 51.94 C C
+ATOM 6771 CG LEU L 191 52.212 -35.798 -68.393 1.00 50.40 C C
+ATOM 6772 CD1 LEU L 191 52.132 -36.044 -66.900 1.00 42.45 C C
+ATOM 6773 CD2 LEU L 191 52.802 -34.440 -68.617 1.00 50.23 C C
+ATOM 6774 N SER L 192 52.181 -35.185 -71.706 1.00 52.96 C N
+ATOM 6775 CA SER L 192 52.263 -33.877 -72.338 1.00 56.20 C C
+ATOM 6776 C SER L 192 51.635 -32.824 -71.433 1.00 60.09 C C
+ATOM 6777 O SER L 192 50.549 -33.036 -70.871 1.00 60.82 C O
+ATOM 6778 CB SER L 192 51.558 -33.877 -73.691 1.00 55.50 C C
+ATOM 6779 OG SER L 192 51.107 -32.564 -73.993 1.00 64.21 C O
+ATOM 6780 N SER L 193 52.322 -31.697 -71.289 1.00 65.75 C N
+ATOM 6781 CA SER L 193 51.760 -30.540 -70.608 1.00 62.75 C C
+ATOM 6782 C SER L 193 51.886 -29.344 -71.532 1.00 62.41 C C
+ATOM 6783 O SER L 193 52.940 -29.129 -72.143 1.00 64.01 C O
+ATOM 6784 CB SER L 193 52.453 -30.246 -69.279 1.00 62.87 C C
+ATOM 6785 OG SER L 193 51.727 -29.266 -68.558 1.00 65.19 C O
+ATOM 6786 N THR L 194 50.815 -28.569 -71.634 1.00 62.14 C N
+ATOM 6787 CA THR L 194 50.768 -27.439 -72.543 1.00 59.68 C C
+ATOM 6788 C THR L 194 50.419 -26.192 -71.739 1.00 62.33 C C
+ATOM 6789 O THR L 194 49.412 -26.165 -71.013 1.00 65.86 C O
+ATOM 6790 CB THR L 194 49.766 -27.706 -73.662 1.00 65.03 C C
+ATOM 6791 CG2 THR L 194 49.678 -26.523 -74.541 1.00 62.60 C C
+ATOM 6792 OG1 THR L 194 50.207 -28.835 -74.432 1.00 68.56 C O
+ATOM 6793 N LEU L 195 51.289 -25.188 -71.825 1.00 67.38 C N
+ATOM 6794 CA LEU L 195 51.044 -23.872 -71.257 1.00 65.13 C C
+ATOM 6795 C LEU L 195 50.601 -22.975 -72.397 1.00 69.28 C C
+ATOM 6796 O LEU L 195 51.403 -22.628 -73.274 1.00 72.22 C O
+ATOM 6797 CB LEU L 195 52.291 -23.309 -70.584 1.00 67.86 C C
+ATOM 6798 CG LEU L 195 51.954 -22.049 -69.788 1.00 70.27 C C
+ATOM 6799 CD1 LEU L 195 50.820 -22.397 -68.867 1.00 68.07 C C
+ATOM 6800 CD2 LEU L 195 53.145 -21.524 -68.992 1.00 73.32 C C
+ATOM 6801 N THR L 196 49.322 -22.616 -72.389 1.00 70.74 C N
+ATOM 6802 CA THR L 196 48.715 -21.814 -73.445 1.00 76.23 C C
+ATOM 6803 C THR L 196 48.583 -20.394 -72.926 1.00 78.99 C C
+ATOM 6804 O THR L 196 47.817 -20.148 -71.988 1.00 82.49 C O
+ATOM 6805 CB THR L 196 47.353 -22.380 -73.846 1.00 80.39 C C
+ATOM 6806 CG2 THR L 196 46.548 -21.363 -74.618 1.00 78.72 C C
+ATOM 6807 OG1 THR L 196 47.518 -23.563 -74.645 1.00 84.97 C O
+ATOM 6808 N LEU L 197 49.337 -19.462 -73.517 1.00 80.42 C N
+ATOM 6809 CA LEU L 197 49.277 -18.072 -73.081 1.00 80.47 C C
+ATOM 6810 C LEU L 197 49.194 -17.106 -74.255 1.00 78.74 C C
+ATOM 6811 O LEU L 197 49.715 -17.358 -75.348 1.00 81.58 C O
+ATOM 6812 CB LEU L 197 50.458 -17.687 -72.176 1.00 83.22 C C
+ATOM 6813 CG LEU L 197 49.992 -17.602 -70.711 1.00 86.73 C C
+ATOM 6814 CD1 LEU L 197 51.014 -16.898 -69.831 1.00 85.59 C C
+ATOM 6815 CD2 LEU L 197 48.620 -16.915 -70.587 1.00 91.40 C C
+ATOM 6816 N SER L 198 48.501 -15.998 -73.991 1.00 84.31 C N
+ATOM 6817 CA SER L 198 48.355 -14.904 -74.936 1.00 87.88 C C
+ATOM 6818 C SER L 198 49.712 -14.461 -75.461 1.00 87.91 C C
+ATOM 6819 O SER L 198 50.656 -14.281 -74.685 1.00 86.23 C O
+ATOM 6820 CB SER L 198 47.655 -13.745 -74.224 1.00 94.62 C C
+ATOM 6821 OG SER L 198 48.061 -12.490 -74.734 1.00 99.09 C O
+ATOM 6822 N LYS L 199 49.808 -14.291 -76.785 1.00 77.12 C N
+ATOM 6823 CA LYS L 199 51.042 -13.783 -77.389 1.00 84.05 C C
+ATOM 6824 C LYS L 199 51.521 -12.511 -76.685 1.00 84.29 C C
+ATOM 6825 O LYS L 199 52.728 -12.337 -76.435 1.00 84.01 C O
+ATOM 6826 CB LYS L 199 50.812 -13.557 -78.882 1.00 88.38 C C
+ATOM 6827 CG LYS L 199 51.959 -12.980 -79.682 1.00 90.00 C C
+ATOM 6828 CD LYS L 199 51.455 -12.674 -81.096 1.00 98.26 C C
+ATOM 6829 CE LYS L 199 52.489 -12.017 -81.995 1.00107.82 C C
+ATOM 6830 NZ LYS L 199 52.657 -10.570 -81.707 1.00112.96 C N1+
+ATOM 6831 N ALA L 200 50.574 -11.643 -76.302 1.00 90.60 C N
+ATOM 6832 CA ALA L 200 50.875 -10.470 -75.482 1.00 88.79 C C
+ATOM 6833 C ALA L 200 51.672 -10.849 -74.237 1.00 91.37 C C
+ATOM 6834 O ALA L 200 52.783 -10.348 -74.014 1.00 91.96 C O
+ATOM 6835 CB ALA L 200 49.574 -9.765 -75.084 1.00 84.64 C C
+ATOM 6836 N ASP L 201 51.121 -11.755 -73.420 1.00 83.64 C N
+ATOM 6837 CA ASP L 201 51.831 -12.192 -72.223 1.00 85.37 C C
+ATOM 6838 C ASP L 201 53.139 -12.878 -72.578 1.00 83.68 C C
+ATOM 6839 O ASP L 201 54.102 -12.801 -71.809 1.00 84.67 C O
+ATOM 6840 CB ASP L 201 50.956 -13.140 -71.409 1.00 86.79 C C
+ATOM 6841 CG ASP L 201 49.742 -12.455 -70.829 1.00 89.71 C C
+ATOM 6842 OD1 ASP L 201 49.907 -11.470 -70.069 1.00 94.19 C O
+ATOM 6843 OD2 ASP L 201 48.619 -12.901 -71.159 1.00 89.12 C O1-
+ATOM 6844 N TYR L 202 53.195 -13.545 -73.736 1.00 80.51 C N
+ATOM 6845 CA TYR L 202 54.370 -14.342 -74.067 1.00 80.81 C C
+ATOM 6846 C TYR L 202 55.585 -13.462 -74.325 1.00 83.36 C C
+ATOM 6847 O TYR L 202 56.712 -13.833 -73.971 1.00 85.51 C O
+ATOM 6848 CB TYR L 202 54.096 -15.253 -75.268 1.00 81.10 C C
+ATOM 6849 CG TYR L 202 55.325 -16.020 -75.760 1.00 79.31 C C
+ATOM 6850 CD1 TYR L 202 55.618 -17.292 -75.297 1.00 76.92 C C
+ATOM 6851 CD2 TYR L 202 56.179 -15.471 -76.712 1.00 81.58 C C
+ATOM 6852 CE1 TYR L 202 56.734 -17.982 -75.756 1.00 77.06 C C
+ATOM 6853 CE2 TYR L 202 57.293 -16.156 -77.166 1.00 79.71 C C
+ATOM 6854 CZ TYR L 202 57.566 -17.406 -76.685 1.00 77.96 C C
+ATOM 6855 OH TYR L 202 58.673 -18.072 -77.150 1.00 77.26 C O
+ATOM 6856 N GLU L 203 55.395 -12.302 -74.947 1.00 85.03 C N
+ATOM 6857 CA GLU L 203 56.553 -11.425 -75.097 1.00 89.15 C C
+ATOM 6858 C GLU L 203 56.908 -10.696 -73.798 1.00 90.80 C C
+ATOM 6859 O GLU L 203 58.080 -10.342 -73.598 1.00 90.06 C O
+ATOM 6860 CB GLU L 203 56.297 -10.447 -76.239 1.00 92.54 C C
+ATOM 6861 CG GLU L 203 56.034 -11.143 -77.565 1.00 99.37 C C
+ATOM 6862 CD GLU L 203 54.845 -10.539 -78.309 1.00106.89 C C
+ATOM 6863 OE1 GLU L 203 54.175 -9.648 -77.736 1.00108.77 C O
+ATOM 6864 OE2 GLU L 203 54.573 -10.952 -79.459 1.00109.81 C O1-
+ATOM 6865 N LYS L 204 55.931 -10.507 -72.897 1.00 90.88 C N
+ATOM 6866 CA LYS L 204 56.183 -9.910 -71.584 1.00 90.53 C C
+ATOM 6867 C LYS L 204 57.259 -10.649 -70.792 1.00 88.94 C C
+ATOM 6868 O LYS L 204 57.788 -10.095 -69.819 1.00 94.83 C O
+ATOM 6869 CB LYS L 204 54.887 -9.876 -70.760 1.00 96.00 C C
+ATOM 6870 CG LYS L 204 53.922 -8.754 -71.096 1.00102.59 C C
+ATOM 6871 CD LYS L 204 52.666 -8.827 -70.217 1.00108.90 C C
+ATOM 6872 CE LYS L 204 52.925 -8.547 -68.739 1.00116.09 C C
+ATOM 6873 NZ LYS L 204 51.663 -8.706 -67.937 1.00119.52 C N1+
+ATOM 6874 N HIS L 205 57.582 -11.888 -71.166 1.00 89.25 C N
+ATOM 6875 CA HIS L 205 58.463 -12.736 -70.385 1.00 85.84 C C
+ATOM 6876 C HIS L 205 59.541 -13.324 -71.282 1.00 83.49 C C
+ATOM 6877 O HIS L 205 59.426 -13.331 -72.514 1.00 85.66 C O
+ATOM 6878 CB HIS L 205 57.654 -13.825 -69.682 1.00 85.88 C C
+ATOM 6879 CG HIS L 205 56.543 -13.282 -68.835 1.00 91.76 C C
+ATOM 6880 CD2 HIS L 205 55.254 -13.674 -68.690 1.00 94.05 C C
+ATOM 6881 ND1 HIS L 205 56.692 -12.160 -68.046 1.00 93.74 C N
+ATOM 6882 CE1 HIS L 205 55.550 -11.900 -67.433 1.00 94.66 C C
+ATOM 6883 NE2 HIS L 205 54.661 -12.804 -67.806 1.00 97.45 C N
+ATOM 6884 N LYS L 206 60.608 -13.805 -70.640 1.00 85.17 C N
+ATOM 6885 CA LYS L 206 61.829 -14.213 -71.334 1.00 86.96 C C
+ATOM 6886 C LYS L 206 62.175 -15.676 -71.079 1.00 85.75 C C
+ATOM 6887 O LYS L 206 62.330 -16.435 -72.043 1.00 89.56 C O
+ATOM 6888 CB LYS L 206 62.992 -13.287 -70.939 1.00 85.38 C C
+ATOM 6889 CG LYS L 206 64.154 -13.210 -71.961 1.00 92.04 C C
+ATOM 6890 CD LYS L 206 65.263 -12.245 -71.469 1.00103.57 C C
+ATOM 6891 CE LYS L 206 66.623 -12.470 -72.145 1.00109.92 C C
+ATOM 6892 NZ LYS L 206 66.645 -12.116 -73.594 1.00113.31 C N1+
+ATOM 6893 N VAL L 207 62.363 -16.082 -69.818 1.00 83.78 C N
+ATOM 6894 CA VAL L 207 62.753 -17.451 -69.471 1.00 84.69 C C
+ATOM 6895 C VAL L 207 61.512 -18.289 -69.183 1.00 89.50 C C
+ATOM 6896 O VAL L 207 60.764 -18.011 -68.235 1.00 95.12 C O
+ATOM 6897 CB VAL L 207 63.704 -17.478 -68.265 1.00 84.02 C C
+ATOM 6898 CG1 VAL L 207 63.836 -18.906 -67.727 1.00 83.00 C C
+ATOM 6899 CG2 VAL L 207 65.071 -16.918 -68.635 1.00 81.72 C C
+ATOM 6900 N TYR L 208 61.326 -19.345 -69.976 1.00 82.70 C N
+ATOM 6901 CA TYR L 208 60.217 -20.282 -69.841 1.00 79.34 C C
+ATOM 6902 C TYR L 208 60.744 -21.624 -69.359 1.00 80.74 C C
+ATOM 6903 O TYR L 208 61.714 -22.155 -69.918 1.00 80.46 C O
+ATOM 6904 CB TYR L 208 59.474 -20.437 -71.171 1.00 78.70 C C
+ATOM 6905 CG TYR L 208 58.654 -19.218 -71.488 1.00 80.65 C C
+ATOM 6906 CD1 TYR L 208 58.692 -18.638 -72.742 1.00 77.60 C C
+ATOM 6907 CD2 TYR L 208 57.867 -18.617 -70.502 1.00 79.49 C C
+ATOM 6908 CE1 TYR L 208 57.969 -17.509 -73.007 1.00 79.00 C C
+ATOM 6909 CE2 TYR L 208 57.134 -17.489 -70.765 1.00 81.88 C C
+ATOM 6910 CZ TYR L 208 57.186 -16.946 -72.024 1.00 84.04 C C
+ATOM 6911 OH TYR L 208 56.469 -15.818 -72.306 1.00 88.50 C O
+ATOM 6912 N ALA L 209 60.111 -22.166 -68.319 1.00 70.28 C N
+ATOM 6913 CA ALA L 209 60.673 -23.337 -67.664 1.00 71.32 C C
+ATOM 6914 C ALA L 209 59.602 -24.331 -67.236 1.00 71.12 C C
+ATOM 6915 O ALA L 209 58.609 -23.973 -66.585 1.00 70.12 C O
+ATOM 6916 CB ALA L 209 61.515 -22.924 -66.454 1.00 69.57 C C
+ATOM 6917 N CYS L 210 59.853 -25.586 -67.591 1.00 57.61 C N
+ATOM 6918 CA CYS L 210 59.052 -26.740 -67.218 1.00 61.04 C C
+ATOM 6919 C CYS L 210 59.864 -27.575 -66.232 1.00 58.67 C C
+ATOM 6920 O CYS L 210 60.951 -28.055 -66.580 1.00 55.81 C O
+ATOM 6921 CB CYS L 210 58.702 -27.547 -68.465 1.00 69.29 C C
+ATOM 6922 SG CYS L 210 58.164 -29.164 -68.103 1.00 84.38 C S
+ATOM 6923 N GLU L 211 59.352 -27.732 -65.006 1.00 56.22 C N
+ATOM 6924 CA GLU L 211 60.070 -28.381 -63.914 1.00 55.41 C C
+ATOM 6925 C GLU L 211 59.346 -29.664 -63.524 1.00 52.92 C C
+ATOM 6926 O GLU L 211 58.253 -29.618 -62.955 1.00 51.95 C O
+ATOM 6927 CB GLU L 211 60.185 -27.435 -62.727 1.00 61.07 C C
+ATOM 6928 CG GLU L 211 60.971 -27.977 -61.553 1.00 73.83 C C
+ATOM 6929 CD GLU L 211 60.909 -27.045 -60.353 1.00 77.54 C C
+ATOM 6930 OE1 GLU L 211 59.780 -26.624 -60.010 1.00 72.34 C O
+ATOM 6931 OE2 GLU L 211 61.978 -26.714 -59.777 1.00 87.61 C O1-
+ATOM 6932 N VAL L 212 59.970 -30.802 -63.814 1.00 51.24 C N
+ATOM 6933 CA VAL L 212 59.419 -32.131 -63.583 1.00 52.00 C C
+ATOM 6934 C VAL L 212 59.990 -32.692 -62.297 1.00 55.09 C C
+ATOM 6935 O VAL L 212 61.199 -32.594 -62.053 1.00 52.97 C O
+ATOM 6936 CB VAL L 212 59.770 -33.075 -64.741 1.00 50.38 C C
+ATOM 6937 CG1 VAL L 212 59.355 -34.479 -64.391 1.00 50.16 C C
+ATOM 6938 CG2 VAL L 212 59.134 -32.621 -66.019 1.00 51.15 C C
+ATOM 6939 N THR L 213 59.147 -33.317 -61.488 1.00 55.05 C N
+ATOM 6940 CA THR L 213 59.634 -34.119 -60.375 1.00 58.21 C C
+ATOM 6941 C THR L 213 58.971 -35.494 -60.449 1.00 54.84 C C
+ATOM 6942 O THR L 213 57.761 -35.598 -60.687 1.00 55.45 C O
+ATOM 6943 CB THR L 213 59.337 -33.459 -59.034 1.00 61.05 C C
+ATOM 6944 CG2 THR L 213 59.276 -31.944 -59.187 1.00 59.10 C C
+ATOM 6945 OG1 THR L 213 58.056 -33.890 -58.592 1.00 67.78 C O
+ATOM 6946 N HIS L 214 59.762 -36.548 -60.256 1.00 48.82 C N
+ATOM 6947 CA HIS L 214 59.274 -37.906 -60.451 1.00 51.30 C C
+ATOM 6948 C HIS L 214 60.114 -38.850 -59.613 1.00 56.22 C C
+ATOM 6949 O HIS L 214 61.231 -38.517 -59.202 1.00 55.73 C O
+ATOM 6950 CB HIS L 214 59.323 -38.313 -61.919 1.00 47.34 C C
+ATOM 6951 CG HIS L 214 58.852 -39.710 -62.172 1.00 48.37 C C
+ATOM 6952 CD2 HIS L 214 57.845 -40.427 -61.621 1.00 51.07 C C
+ATOM 6953 ND1 HIS L 214 59.442 -40.539 -63.105 1.00 46.46 C N
+ATOM 6954 CE1 HIS L 214 58.822 -41.704 -63.118 1.00 51.42 C C
+ATOM 6955 NE2 HIS L 214 57.851 -41.664 -62.224 1.00 50.12 C N
+ATOM 6956 N GLN L 215 59.557 -40.047 -59.380 1.00 50.83 C N
+ATOM 6957 CA GLN L 215 60.151 -40.988 -58.434 1.00 49.42 C C
+ATOM 6958 C GLN L 215 61.538 -41.445 -58.879 1.00 47.37 C C
+ATOM 6959 O GLN L 215 62.383 -41.776 -58.027 1.00 53.14 C O
+ATOM 6960 CB GLN L 215 59.218 -42.187 -58.229 1.00 57.07 C C
+ATOM 6961 CG GLN L 215 59.763 -43.196 -57.225 1.00 67.22 C C
+ATOM 6962 CD GLN L 215 58.832 -44.368 -56.961 1.00 75.19 C C
+ATOM 6963 NE2 GLN L 215 59.343 -45.387 -56.272 1.00 79.18 C N
+ATOM 6964 OE1 GLN L 215 57.668 -44.353 -57.353 1.00 76.59 C O
+ATOM 6965 N GLY L 216 61.801 -41.441 -60.198 1.00 40.93 C N
+ATOM 6966 CA GLY L 216 63.093 -41.798 -60.753 1.00 45.85 C C
+ATOM 6967 C GLY L 216 64.119 -40.681 -60.869 1.00 52.80 C C
+ATOM 6968 O GLY L 216 65.268 -40.938 -61.260 1.00 51.90 C O
+ATOM 6969 N LEU L 217 63.743 -39.444 -60.551 1.00 48.23 C N
+ATOM 6970 CA LEU L 217 64.665 -38.320 -60.582 1.00 48.64 C C
+ATOM 6971 C LEU L 217 65.167 -38.033 -59.168 1.00 53.33 C C
+ATOM 6972 O LEU L 217 64.370 -37.863 -58.233 1.00 46.74 C O
+ATOM 6973 CB LEU L 217 63.998 -37.088 -61.189 1.00 44.55 C C
+ATOM 6974 CG LEU L 217 63.224 -37.311 -62.494 1.00 43.21 C C
+ATOM 6975 CD1 LEU L 217 62.492 -36.061 -62.962 1.00 39.90 C C
+ATOM 6976 CD2 LEU L 217 64.118 -37.781 -63.584 1.00 40.40 C C
+ATOM 6977 N SER L 218 66.499 -37.988 -59.019 1.00 49.97 C N
+ATOM 6978 CA SER L 218 67.153 -37.784 -57.730 1.00 59.61 C C
+ATOM 6979 C SER L 218 66.964 -36.379 -57.228 1.00 52.70 C C
+ATOM 6980 O SER L 218 67.385 -36.076 -56.102 1.00 53.74 C O
+ATOM 6981 CB SER L 218 68.645 -38.050 -57.842 1.00 70.07 C C
+ATOM 6982 OG SER L 218 69.091 -37.562 -59.090 1.00 76.07 C O
+ATOM 6983 N SER L 219 66.405 -35.525 -58.075 1.00 65.47 C N
+ATOM 6984 CA SER L 219 65.916 -34.195 -57.769 1.00 60.20 C C
+ATOM 6985 C SER L 219 65.319 -33.600 -59.043 1.00 51.75 C C
+ATOM 6986 O SER L 219 65.707 -33.981 -60.155 1.00 43.43 C O
+ATOM 6987 CB SER L 219 67.037 -33.340 -57.185 1.00 64.91 C C
+ATOM 6988 OG SER L 219 67.640 -32.484 -58.116 1.00 69.18 C O
+ATOM 6989 N PRO L 220 64.365 -32.688 -58.920 1.00 54.84 C N
+ATOM 6990 CA PRO L 220 63.557 -32.294 -60.077 1.00 50.91 C C
+ATOM 6991 C PRO L 220 64.386 -31.831 -61.263 1.00 49.01 C C
+ATOM 6992 O PRO L 220 65.216 -30.931 -61.152 1.00 50.40 C O
+ATOM 6993 CB PRO L 220 62.708 -31.150 -59.525 1.00 53.34 C C
+ATOM 6994 CG PRO L 220 62.678 -31.370 -58.059 1.00 56.90 C C
+ATOM 6995 CD PRO L 220 63.959 -32.000 -57.688 1.00 59.03 C C
+ATOM 6996 N VAL L 221 64.115 -32.423 -62.416 1.00 53.88 C N
+ATOM 6997 CA VAL L 221 64.729 -31.995 -63.667 1.00 50.49 C C
+ATOM 6998 C VAL L 221 63.979 -30.793 -64.205 1.00 50.92 C C
+ATOM 6999 O VAL L 221 62.747 -30.812 -64.291 1.00 48.14 C O
+ATOM 7000 CB VAL L 221 64.697 -33.127 -64.691 1.00 48.86 C C
+ATOM 7001 CG1 VAL L 221 65.312 -32.663 -65.970 1.00 48.54 C C
+ATOM 7002 CG2 VAL L 221 65.396 -34.324 -64.133 1.00 53.74 C C
+ATOM 7003 N THR L 222 64.708 -29.763 -64.621 1.00 61.50 C N
+ATOM 7004 CA THR L 222 64.082 -28.566 -65.171 1.00 56.84 C C
+ATOM 7005 C THR L 222 64.622 -28.272 -66.564 1.00 57.48 C C
+ATOM 7006 O THR L 222 65.839 -28.280 -66.787 1.00 61.73 C O
+ATOM 7007 CB THR L 222 64.293 -27.365 -64.257 1.00 53.02 C C
+ATOM 7008 CG2 THR L 222 63.723 -26.125 -64.914 1.00 53.87 C C
+ATOM 7009 OG1 THR L 222 63.643 -27.603 -62.997 1.00 58.05 C O
+ATOM 7010 N LYS L 223 63.712 -28.027 -67.495 1.00 63.76 C N
+ATOM 7011 CA LYS L 223 64.045 -27.705 -68.870 1.00 60.37 C C
+ATOM 7012 C LYS L 223 63.541 -26.300 -69.150 1.00 60.24 C C
+ATOM 7013 O LYS L 223 62.506 -25.901 -68.608 1.00 60.82 C O
+ATOM 7014 CB LYS L 223 63.419 -28.705 -69.810 1.00 64.98 C C
+ATOM 7015 CG LYS L 223 64.413 -29.352 -70.726 1.00 69.95 C C
+ATOM 7016 CD LYS L 223 65.260 -30.428 -70.048 1.00 72.65 C C
+ATOM 7017 CE LYS L 223 66.213 -31.062 -71.097 1.00 76.45 C C
+ATOM 7018 NZ LYS L 223 66.829 -32.383 -70.709 1.00 76.60 C N1+
+ATOM 7019 N SER L 224 64.281 -25.530 -69.956 1.00 71.97 C N
+ATOM 7020 CA SER L 224 63.955 -24.112 -70.114 1.00 69.10 C C
+ATOM 7021 C SER L 224 64.493 -23.569 -71.434 1.00 71.83 C C
+ATOM 7022 O SER L 224 65.321 -24.191 -72.108 1.00 74.75 C O
+ATOM 7023 CB SER L 224 64.490 -23.273 -68.946 1.00 74.95 C C
+ATOM 7024 OG SER L 224 65.838 -23.598 -68.667 1.00 80.77 C O
+ATOM 7025 N PHE L 225 63.983 -22.389 -71.796 1.00 82.30 C N
+ATOM 7026 CA PHE L 225 64.436 -21.654 -72.972 1.00 84.83 C C
+ATOM 7027 C PHE L 225 64.195 -20.162 -72.753 1.00 88.78 C C
+ATOM 7028 O PHE L 225 63.402 -19.765 -71.890 1.00 87.65 C O
+ATOM 7029 CB PHE L 225 63.736 -22.145 -74.254 1.00 82.25 C C
+ATOM 7030 CG PHE L 225 62.260 -21.785 -74.351 1.00 83.51 C C
+ATOM 7031 CD1 PHE L 225 61.845 -20.649 -75.018 1.00 82.48 C C
+ATOM 7032 CD2 PHE L 225 61.290 -22.609 -73.819 1.00 84.84 C C
+ATOM 7033 CE1 PHE L 225 60.500 -20.335 -75.117 1.00 86.99 C C
+ATOM 7034 CE2 PHE L 225 59.945 -22.297 -73.933 1.00 87.54 C C
+ATOM 7035 CZ PHE L 225 59.557 -21.166 -74.577 1.00 88.46 C C
+ATOM 7036 N ASN L 226 64.886 -19.329 -73.541 1.00 93.89 C N
+ATOM 7037 CA ASN L 226 64.684 -17.884 -73.492 1.00 93.65 C C
+ATOM 7038 C ASN L 226 63.970 -17.382 -74.742 1.00 92.39 C C
+ATOM 7039 O ASN L 226 64.301 -17.794 -75.862 1.00 94.64 C O
+ATOM 7040 CB ASN L 226 66.005 -17.139 -73.297 1.00 95.67 C C
+ATOM 7041 CG ASN L 226 66.395 -17.037 -71.830 1.00102.99 C C
+ATOM 7042 ND2 ASN L 226 66.773 -15.836 -71.391 1.00107.52 C N
+ATOM 7043 OD1 ASN L 226 66.315 -18.021 -71.091 1.00105.09 C O
+ATOM 7044 N ARG L 227 63.032 -16.440 -74.530 1.00 88.77 C N
+ATOM 7045 CA ARG L 227 62.014 -16.104 -75.529 1.00 97.54 C C
+ATOM 7046 C ARG L 227 62.617 -15.911 -76.911 1.00111.28 C C
+ATOM 7047 O ARG L 227 62.193 -16.549 -77.881 1.00120.87 C O
+ATOM 7048 CB ARG L 227 61.258 -14.833 -75.107 1.00 91.09 C C
+ATOM 7049 CG ARG L 227 60.443 -14.181 -76.230 1.00 89.35 C C
+ATOM 7050 CD ARG L 227 59.767 -12.877 -75.790 1.00 91.59 C C
+ATOM 7051 NE ARG L 227 60.727 -11.814 -75.411 1.00 93.21 C N
+ATOM 7052 CZ ARG L 227 61.174 -10.821 -76.212 1.00 97.39 C C
+ATOM 7053 NH1 ARG L 227 60.777 -10.755 -77.483 1.00 99.05 C N1+
+ATOM 7054 NH2 ARG L 227 62.046 -9.896 -75.760 1.00103.01 C N
+ATOM 7055 N GLY L 228 63.613 -15.037 -77.014 1.00 98.05 C N
+ATOM 7056 CA GLY L 228 64.191 -14.702 -78.298 1.00109.87 C C
+ATOM 7057 C GLY L 228 65.318 -15.598 -78.764 1.00117.73 C C
+ATOM 7058 O GLY L 228 65.477 -15.820 -79.970 1.00121.38 C O
+ATOM 7059 N GLU L 229 66.092 -16.142 -77.818 1.00107.10 C N
+ATOM 7060 CA GLU L 229 67.339 -16.842 -78.131 1.00109.83 C C
+ATOM 7061 C GLU L 229 67.124 -18.230 -78.736 1.00118.76 C C
+ATOM 7062 O GLU L 229 68.119 -18.870 -79.126 1.00119.81 C O
+ATOM 7063 CB GLU L 229 68.204 -16.907 -76.872 1.00105.02 C C
+ATOM 7064 CG GLU L 229 68.631 -15.512 -76.418 1.00103.20 C C
+ATOM 7065 CD GLU L 229 68.935 -15.415 -74.933 1.00102.82 C C
+ATOM 7066 OE1 GLU L 229 69.771 -16.199 -74.433 1.00105.21 C O
+ATOM 7067 OE2 GLU L 229 68.341 -14.535 -74.265 1.00100.61 C O1-
+ATOM 7068 N CYS L 230 65.866 -18.689 -78.818 1.00109.79 C N
+ATOM 7069 CA CYS L 230 65.472 -19.909 -79.544 1.00120.44 C C
+ATOM 7070 C CYS L 230 63.992 -19.849 -79.942 1.00119.61 C C
+ATOM 7071 O CYS L 230 63.288 -18.889 -79.622 1.00121.00 C O
+ATOM 7072 CB CYS L 230 65.735 -21.168 -78.702 1.00124.82 C C
+ATOM 7073 SG CYS L 230 65.858 -22.707 -79.653 1.00205.08 C S
+TER
+END
diff --git a/utils/examples/secondary_structure.sh b/utils/examples/secondary_structure.sh
new file mode 100644
index 0000000000000000000000000000000000000000..bab74c9f80d34c962d7379c431f2ae63750c9559
--- /dev/null
+++ b/utils/examples/secondary_structure.sh
@@ -0,0 +1,21 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --secondary_structure XXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXXXXXXXHHHHXXXLLLXXXXX
+
+# FOR SECONDARY STRUCTURE:
+# X - mask
+# H - helix
+# E - strand
+# L - loop
diff --git a/utils/examples/secondary_structure_bias.sh b/utils/examples/secondary_structure_bias.sh
new file mode 100644
index 0000000000000000000000000000000000000000..c76d6747e1508af285efc18331e266e456d842b6
--- /dev/null
+++ b/utils/examples/secondary_structure_bias.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --helix_bias 0.01 --strand_bias 0.01 --loop_bias 0.0
diff --git a/utils/examples/secondary_structure_from_pdb.sh b/utils/examples/secondary_structure_from_pdb.sh
new file mode 100644
index 0000000000000000000000000000000000000000..b548870e8eade138a4c59ebada89fb4ab2a9ec68
--- /dev/null
+++ b/utils/examples/secondary_structure_from_pdb.sh
@@ -0,0 +1,21 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 110 \
+ --T 25 --save_best_plddt \
+ --dssp_pdb ./pdbs/cd86.pdb
+
+# FOR SECONDARY STRUCTURE:
+# X - mask
+# H - helix
+# E - strand
+# L - loop
diff --git a/utils/examples/symmetric_design.sh b/utils/examples/symmetric_design.sh
new file mode 100644
index 0000000000000000000000000000000000000000..cca1065f1aa383c16f7f5338de0a6a44c6109479
--- /dev/null
+++ b/utils/examples/symmetric_design.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/symmetric_design \
+ --contigs 25,0 25,0 25,0 \
+ --T 50 \
+ --save_best_plddt \
+ --symmetry 3
diff --git a/utils/examples/weighted_sequence.sh b/utils/examples/weighted_sequence.sh
new file mode 100644
index 0000000000000000000000000000000000000000..779abda1d21255ed8d68198f72ac55729839be26
--- /dev/null
+++ b/utils/examples/weighted_sequence.sh
@@ -0,0 +1,15 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 100 \
+ --T 25 --save_best_plddt \
+ --aa_composition W0.2 --potential_scale 1.75
diff --git a/utils/examples/weighted_sequence_json.sh b/utils/examples/weighted_sequence_json.sh
new file mode 100644
index 0000000000000000000000000000000000000000..701d7d8d8c2352e2cc95255732d7de341979083a
--- /dev/null
+++ b/utils/examples/weighted_sequence_json.sh
@@ -0,0 +1,16 @@
+#!/bin/bash
+#SBATCH -J seq_diff
+#SBATCH -p gpu
+#SBATCH --mem=8g
+#SBATCH --gres=gpu:a6000:1
+#SBATCH -o ./out/slurm/slurm_%j.out
+
+source activate /software/conda/envs/SE3nv
+
+srun python ../inference.py \
+ --num_designs 10 \
+ --out out/design \
+ --contigs 75 \
+ --aa_weights_json aa_weights.json \
+ --add_weight_every_n 5 --add_weight_every_n \
+ --T 25 --save_best_plddt
diff --git a/utils/figs/.ipynb_checkpoints/fig3-checkpoint.jpg b/utils/figs/.ipynb_checkpoints/fig3-checkpoint.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..d4bce6dfae5115b075ef199b592d3b32e4e3ca4a
Binary files /dev/null and b/utils/figs/.ipynb_checkpoints/fig3-checkpoint.jpg differ
diff --git a/utils/figs/fig1.jpg b/utils/figs/fig1.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..a514787d2c75eeede8bfab24f78440746debd28e
Binary files /dev/null and b/utils/figs/fig1.jpg differ
diff --git a/utils/figs/fig1.png b/utils/figs/fig1.png
new file mode 100644
index 0000000000000000000000000000000000000000..10498fc9eb1c295e95e146db0e76bb1e0d0dd599
Binary files /dev/null and b/utils/figs/fig1.png differ
diff --git a/utils/figs/fig100.jpg b/utils/figs/fig100.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..087283e846bb5fec84353b626b42f98c7dda295c
Binary files /dev/null and b/utils/figs/fig100.jpg differ
diff --git a/utils/figs/fig100.png b/utils/figs/fig100.png
new file mode 100644
index 0000000000000000000000000000000000000000..2064be727a0c2c0cfe1c1ac05ca664d7c55e894a
Binary files /dev/null and b/utils/figs/fig100.png differ
diff --git a/utils/figs/fig2.jpg b/utils/figs/fig2.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..053bafd7c530d636e94fb6e3c58ee0c1231b45c9
Binary files /dev/null and b/utils/figs/fig2.jpg differ
diff --git a/utils/figs/fig3.jpg b/utils/figs/fig3.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..d4bce6dfae5115b075ef199b592d3b32e4e3ca4a
Binary files /dev/null and b/utils/figs/fig3.jpg differ
diff --git a/utils/figs/thumbnail_github.png b/utils/figs/thumbnail_github.png
new file mode 100644
index 0000000000000000000000000000000000000000..4d0a57f9aa3790200492d3bda12147ab3cbc1abf
Binary files /dev/null and b/utils/figs/thumbnail_github.png differ
diff --git a/utils/geometry.py b/utils/geometry.py
new file mode 100644
index 0000000000000000000000000000000000000000..58edab102102bf5650d11c72a7d5a76bb1abfb33
--- /dev/null
+++ b/utils/geometry.py
@@ -0,0 +1,200 @@
+import numpy as np
+import torch
+
+# ============================================================
+def get_pair_dist(a, b):
+ """calculate pair distances between two sets of points
+
+ Parameters
+ ----------
+ a,b : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of two sets of atoms
+ Returns
+ -------
+ dist : pytorch tensor of shape [batch,nres,nres]
+ stores paitwise distances between atoms in a and b
+ """
+
+ dist = torch.cdist(a, b, p=2)
+ return dist
+
+# ============================================================
+def get_ang(a, b, c):
+ """calculate planar angles for all consecutive triples (a[i],b[i],c[i])
+ from Cartesian coordinates of three sets of atoms a,b,c
+
+ Parameters
+ ----------
+ a,b,c : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of three sets of atoms
+ Returns
+ -------
+ ang : pytorch tensor of shape [batch,nres]
+ stores resulting planar angles
+ """
+ v = a - b
+ w = c - b
+ v = v / torch.norm(v, dim=-1, keepdim=True)
+ w = w / torch.norm(w, dim=-1, keepdim=True)
+
+ # this is not stable at the poles
+ #vw = torch.sum(v*w, dim=-1)
+ #ang = torch.acos(vw)
+
+ # this is better
+ # https://math.stackexchange.com/questions/1143354/numerically-stable-method-for-angle-between-3d-vectors/1782769
+ y = torch.norm(v-w,dim=-1)
+ x = torch.norm(v+w,dim=-1)
+ ang = 2*torch.atan2(y, x)
+
+ return ang
+
+# ============================================================
+def get_dih(a, b, c, d):
+ """calculate dihedral angles for all consecutive quadruples (a[i],b[i],c[i],d[i])
+ given Cartesian coordinates of four sets of atoms a,b,c,d
+
+ Parameters
+ ----------
+ a,b,c,d : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of four sets of atoms
+ Returns
+ -------
+ dih : pytorch tensor of shape [batch,nres]
+ stores resulting dihedrals
+ """
+ b0 = a - b
+ b1r = c - b
+ b2 = d - c
+
+ b1 = b1r/torch.norm(b1r, dim=-1, keepdim=True)
+
+ v = b0 - torch.sum(b0*b1, dim=-1, keepdim=True)*b1
+ w = b2 - torch.sum(b2*b1, dim=-1, keepdim=True)*b1
+
+ x = torch.sum(v*w, dim=-1)
+ y = torch.sum(torch.cross(b1,v,dim=-1)*w, dim=-1)
+ ang = torch.atan2(y, x)
+
+ return ang
+
+
+# ============================================================
+def xyz_to_c6d(xyz, params):
+ """convert cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz : pytorch tensor of shape [batch,3,nres,3]
+ stores Cartesian coordinates of backbone N,Ca,C atoms
+ Returns
+ -------
+ c6d : pytorch tensor of shape [batch,nres,nres,4]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+
+ batch = xyz.shape[0]
+ nres = xyz.shape[2]
+
+ # three anchor atoms
+ N = xyz[:,0]
+ Ca = xyz[:,1]
+ C = xyz[:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # 6d coordinates order: (dist,omega,theta,phi)
+ c6d = torch.zeros([batch,nres,nres,4],dtype=xyz.dtype,device=xyz.device)
+
+ dist = get_pair_dist(Cb,Cb)
+ dist[torch.isnan(dist)] = 999.9
+ c6d[...,0] = dist + 999.9*torch.eye(nres,device=xyz.device)[None,...]
+ b,i,j = torch.where(c6d[...,0]=params['DMAX']] = 999.9
+
+ return c6d
+
+
+# ============================================================
+def c6d_to_bins(c6d,params):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=c6d.dtype,device=c6d.device)
+ ab360 = torch.linspace(-np.pi+astep, np.pi, params['ABINS'],dtype=c6d.dtype,device=c6d.device)
+ ab180 = torch.linspace(astep, np.pi, params['ABINS']//2,dtype=c6d.dtype,device=c6d.device)
+
+ db = torch.bucketize(c6d[...,0].contiguous(),dbins)
+ ob = torch.bucketize(c6d[...,1].contiguous(),ab360)
+ tb = torch.bucketize(c6d[...,2].contiguous(),ab360)
+ pb = torch.bucketize(c6d[...,3].contiguous(),ab180)
+
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).to(torch.uint8)
+
+
+# ============================================================
+def dist_to_bins(dist,params):
+ """bin 2d distance maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ db = torch.round((dist-params['DMIN']-dstep/2)/dstep)
+
+ db[db<0] = 0
+ db[db>params['DBINS']] = params['DBINS']
+
+ return db.long()
+
+
+# ============================================================
+def c6d_to_bins2(c6d,params):
+ """bin 2d distance and orientation maps
+ (alternative slightly simpler version)
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ db = torch.round((c6d[...,0]-params['DMIN']-dstep/2)/dstep)
+ ob = torch.round((c6d[...,1]+np.pi-astep/2)/astep)
+ tb = torch.round((c6d[...,2]+np.pi-astep/2)/astep)
+ pb = torch.round((c6d[...,3]-astep/2)/astep)
+
+ # put all dparams['DBINS']] = params['DBINS']
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).long()
+
+
+# ============================================================
+def get_cb(N,Ca,C):
+ """recreate Cb given N,Ca,C"""
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+ return Cb
diff --git a/utils/inpainting_util.py b/utils/inpainting_util.py
new file mode 100644
index 0000000000000000000000000000000000000000..9acb5356ac24fba71f65eb09bb777c62ccb97a45
--- /dev/null
+++ b/utils/inpainting_util.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'w+') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/utils/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py b/utils/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..ec42a3cd8a5a5bd8f59cff12963908760eff59b7
--- /dev/null
+++ b/utils/model/.ipynb_checkpoints/RoseTTAFoldModel-checkpoint.py
@@ -0,0 +1,140 @@
+import torch
+import torch.nn as nn
+from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
+from Track_module import IterativeSimulator
+from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
+from util import INIT_CRDS
+from opt_einsum import contract as einsum
+from icecream import ic
+
+class RoseTTAFoldModule(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
+ d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
+ n_head_msa=8, n_head_pair=4, n_head_templ=4,
+ d_hidden=32, d_hidden_templ=64,
+ p_drop=0.15, d_t1d=24, d_t2d=44,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ ):
+ super(RoseTTAFoldModule, self).__init__()
+ #
+ # Input Embeddings
+ d_state = SE3_param_topk['l0_out_features']
+ self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
+ self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
+ self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
+ n_head=n_head_templ,
+ d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
+ # Update inputs with outputs from previous round
+ self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
+ #
+ self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
+ n_main_block=n_main_block,
+ n_ref_block=n_ref_block,
+ d_msa=d_msa, d_msa_full=d_msa_full,
+ d_pair=d_pair, d_hidden=d_hidden,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ SE3_param_full=SE3_param_full,
+ SE3_param_topk=SE3_param_topk,
+ p_drop=p_drop)
+ ##
+ self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
+ self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
+ self.lddt_pred = LDDTNetwork(d_state)
+
+ self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
+
+ def forward(self, msa_latent, msa_full, seq, xyz, idx,
+ seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
+ msa_prev=None, pair_prev=None, state_prev=None,
+ return_raw=False, return_full=False,
+ use_checkpoint=False, return_infer=False):
+ B, N, L = msa_latent.shape[:3]
+ # Get embeddings
+ #ic(seq.shape)
+ #ic(msa_latent.shape)
+ #ic(seq1hot.shape)
+ #ic(idx.shape)
+ #ic(xyz.shape)
+ #ic(seq1hot.shape)
+ #ic(t1d.shape)
+ #ic(t2d.shape)
+
+ idx = idx.long()
+ msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
+
+ msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
+ #
+ # Do recycling
+ if msa_prev == None:
+ msa_prev = torch.zeros_like(msa_latent[:,0])
+ if pair_prev == None:
+ pair_prev = torch.zeros_like(pair)
+ if state_prev == None:
+ state_prev = torch.zeros_like(state)
+
+ #ic(seq.shape)
+ #ic(msa_prev.shape)
+ #ic(pair_prev.shape)
+ #ic(xyz.shape)
+ #ic(state_prev.shape)
+
+
+ msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
+ msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
+ pair = pair + pair_recycle
+ state = state + state_recycle
+ #
+ #ic(t1d.dtype)
+ #ic(t2d.dtype)
+ #ic(alpha_t.dtype)
+ #ic(xyz_t.dtype)
+ #ic(pair.dtype)
+ #ic(state.dtype)
+
+
+ #import pdb; pdb.set_trace()
+
+ # add template embedding
+ pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
+
+ #ic(seq.dtype)
+ #ic(msa_latent.dtype)
+ #ic(msa_full.dtype)
+ #ic(pair.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+ # Predict coordinates from given inputs
+ msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
+ state, idx, use_checkpoint=use_checkpoint)
+
+ if return_raw:
+ # get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return msa[:,0], pair, xyz, state, alpha_s[-1]
+
+ # predict masked amino acids
+ logits_aa = self.aa_pred(msa)
+ #
+ # predict distogram & orientograms
+ logits = self.c6d_pred(pair)
+
+ # Predict LDDT
+ lddt = self.lddt_pred(state)
+
+ # predict experimentally resolved or not
+ logits_exp = self.exp_pred(msa[:,0], state)
+
+ if return_infer:
+ #get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
+
+
+ # get all intermediate bb structures
+ xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
+
+ return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
diff --git a/utils/model/Attention_module.py b/utils/model/Attention_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..44310ebc94cc40f7cca9eda31463e172188a0084
--- /dev/null
+++ b/utils/model/Attention_module.py
@@ -0,0 +1,411 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+import math
+from opt_einsum import contract as einsum
+from util_module import init_lecun_normal
+from icecream import ic
+
+class FeedForwardLayer(nn.Module):
+ def __init__(self, d_model, r_ff, p_drop=0.1):
+ super(FeedForwardLayer, self).__init__()
+ self.norm = nn.LayerNorm(d_model)
+ self.linear1 = nn.Linear(d_model, d_model*r_ff)
+ self.dropout = nn.Dropout(p_drop)
+ self.linear2 = nn.Linear(d_model*r_ff, d_model)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize linear layer right before ReLu: He initializer (kaiming normal)
+ nn.init.kaiming_normal_(self.linear1.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear1.bias)
+
+ # initialize linear layer right before residual connection: zero initialize
+ nn.init.zeros_(self.linear2.weight)
+ nn.init.zeros_(self.linear2.bias)
+
+ def forward(self, src):
+ src = self.norm(src)
+ src = self.linear2(self.dropout(F.relu_(self.linear1(src))))
+ return src
+
+class Attention(nn.Module):
+ # calculate multi-head attention
+ def __init__(self, d_query, d_key, n_head, d_hidden, d_out, p_drop=0.1):
+ super(Attention, self).__init__()
+ self.h = n_head
+ self.dim = d_hidden
+ #
+ self.to_q = nn.Linear(d_query, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_key, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_key, n_head*d_hidden, bias=False)
+ #
+ self.to_out = nn.Linear(n_head*d_hidden, d_out)
+ self.scaling = 1/math.sqrt(d_hidden)
+ #
+ # initialize all parameters properly
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, query, key, value):
+ B, Q = query.shape[:2]
+ B, K = key.shape[:2]
+ #
+ query = self.to_q(query).reshape(B, Q, self.h, self.dim)
+ key = self.to_k(key).reshape(B, K, self.h, self.dim)
+ value = self.to_v(value).reshape(B, K, self.h, self.dim)
+ #
+ query = query * self.scaling
+ attn = einsum('bqhd,bkhd->bhqk', query, key)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bhqk,bkhd->bqhd', attn, value)
+ out = out.reshape(B, Q, self.h*self.dim)
+ #
+ out = self.to_out(out)
+
+ return out
+
+class AttentionWithBias(nn.Module):
+ def __init__(self, d_in=256, d_bias=128, n_head=8, d_hidden=32):
+ super(AttentionWithBias, self).__init__()
+ self.norm_in = nn.LayerNorm(d_in)
+ self.norm_bias = nn.LayerNorm(d_bias)
+ #
+ self.to_q = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_in, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_bias, n_head, bias=False)
+ self.to_g = nn.Linear(d_in, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_in)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, x, bias):
+ B, L = x.shape[:2]
+ #
+ x = self.norm_in(x)
+ bias = self.norm_bias(bias)
+ #
+ query = self.to_q(x).reshape(B, L, self.h, self.dim)
+ key = self.to_k(x).reshape(B, L, self.h, self.dim)
+ value = self.to_v(x).reshape(B, L, self.h, self.dim)
+ bias = self.to_b(bias) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(x))
+ #
+ key = key * self.scaling
+ attn = einsum('bqhd,bkhd->bqkh', query, key)
+ attn = attn + bias
+ attn = F.softmax(attn, dim=-2)
+ #
+ out = einsum('bqkh,bkhd->bqhd', attn, value).reshape(B, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+# MSA Attention (row/column) from AlphaFold architecture
+class SequenceWeight(nn.Module):
+ def __init__(self, d_msa, n_head, d_hidden, p_drop=0.1):
+ super(SequenceWeight, self).__init__()
+ self.h = n_head
+ self.dim = d_hidden
+ self.scale = 1.0 / math.sqrt(self.dim)
+
+ self.to_query = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_key = nn.Linear(d_msa, n_head*d_hidden)
+ self.dropout = nn.Dropout(p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_query.weight)
+ nn.init.xavier_uniform_(self.to_key.weight)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+
+ tar_seq = msa[:,0]
+
+ q = self.to_query(tar_seq).view(B, 1, L, self.h, self.dim)
+ k = self.to_key(msa).view(B, N, L, self.h, self.dim)
+
+ q = q * self.scale
+ attn = einsum('bqihd,bkihd->bkihq', q, k)
+ attn = F.softmax(attn, dim=1)
+ return self.dropout(attn)
+
+class MSARowAttentionWithBias(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, n_head=8, d_hidden=32):
+ super(MSARowAttentionWithBias, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ #
+ self.seq_weight = SequenceWeight(d_msa, n_head, d_hidden, p_drop=0.1)
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_pair, n_head, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa, pair): # TODO: make this as tied-attention
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ pair = self.norm_pair(pair)
+ #
+ seq_weight = self.seq_weight(msa) # (B, N, L, h, 1)
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
+ value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
+ bias = self.to_b(pair) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(msa))
+ #
+ query = query * seq_weight.expand(-1, -1, -1, -1, self.dim)
+ key = key * self.scaling
+ attn = einsum('bsqhd,bskhd->bqkh', query, key)
+ attn = attn + bias
+ attn = F.softmax(attn, dim=-2)
+ #
+ out = einsum('bqkh,bskhd->bsqhd', attn, value).reshape(B, N, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+class MSAColAttention(nn.Module):
+ def __init__(self, d_msa=256, n_head=8, d_hidden=32):
+ super(MSAColAttention, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ #
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ #
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ key = self.to_k(msa).reshape(B, N, L, self.h, self.dim)
+ value = self.to_v(msa).reshape(B, N, L, self.h, self.dim)
+ gate = torch.sigmoid(self.to_g(msa))
+ #
+ query = query * self.scaling
+ attn = einsum('bqihd,bkihd->bihqk', query, key)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bihqk,bkihd->bqihd', attn, value).reshape(B, N, L, -1)
+ out = gate * out
+ #
+ out = self.to_out(out)
+ return out
+
+class MSAColGlobalAttention(nn.Module):
+ def __init__(self, d_msa=64, n_head=8, d_hidden=8):
+ super(MSAColGlobalAttention, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ #
+ self.to_q = nn.Linear(d_msa, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_msa, d_hidden, bias=False)
+ self.to_v = nn.Linear(d_msa, d_hidden, bias=False)
+ self.to_g = nn.Linear(d_msa, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_msa)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, msa):
+ B, N, L = msa.shape[:3]
+ #
+ msa = self.norm_msa(msa)
+ #
+ query = self.to_q(msa).reshape(B, N, L, self.h, self.dim)
+ query = query.mean(dim=1) # (B, L, h, dim)
+ key = self.to_k(msa) # (B, N, L, dim)
+ value = self.to_v(msa) # (B, N, L, dim)
+ gate = torch.sigmoid(self.to_g(msa)) # (B, N, L, h*dim)
+ #
+ query = query * self.scaling
+ attn = einsum('bihd,bkid->bihk', query, key) # (B, L, h, N)
+ attn = F.softmax(attn, dim=-1)
+ #
+ out = einsum('bihk,bkid->bihd', attn, value).reshape(B, 1, L, -1) # (B, 1, L, h*dim)
+ out = gate * out # (B, N, L, h*dim)
+ #
+ out = self.to_out(out)
+ return out
+
+# Instead of triangle attention, use Tied axail attention with bias from coordinates..?
+class BiasedAxialAttention(nn.Module):
+ def __init__(self, d_pair, d_bias, n_head, d_hidden, p_drop=0.1, is_row=True):
+ super(BiasedAxialAttention, self).__init__()
+ #
+ self.is_row = is_row
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_bias = nn.LayerNorm(d_bias)
+
+ self.to_q = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_k = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_v = nn.Linear(d_pair, n_head*d_hidden, bias=False)
+ self.to_b = nn.Linear(d_bias, n_head, bias=False)
+ self.to_g = nn.Linear(d_pair, n_head*d_hidden)
+ self.to_out = nn.Linear(n_head*d_hidden, d_pair)
+
+ self.scaling = 1/math.sqrt(d_hidden)
+ self.h = n_head
+ self.dim = d_hidden
+
+ # initialize all parameters properly
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # query/key/value projection: Glorot uniform / Xavier uniform
+ nn.init.xavier_uniform_(self.to_q.weight)
+ nn.init.xavier_uniform_(self.to_k.weight)
+ nn.init.xavier_uniform_(self.to_v.weight)
+
+ # bias: normal distribution
+ self.to_b = init_lecun_normal(self.to_b)
+
+ # gating: zero weights, one biases (mostly open gate at the begining)
+ nn.init.zeros_(self.to_g.weight)
+ nn.init.ones_(self.to_g.bias)
+
+ # to_out: right before residual connection: zero initialize -- to make it sure residual operation is same to the Identity at the begining
+ nn.init.zeros_(self.to_out.weight)
+ nn.init.zeros_(self.to_out.bias)
+
+ def forward(self, pair, bias, same_chain = None):
+ # pair: (B, L, L, d_pair)
+ B, L = pair.shape[:2]
+
+ if self.is_row:
+ pair = pair.permute(0,2,1,3)
+ bias = bias.permute(0,2,1,3)
+
+ pair = self.norm_pair(pair)
+ bias = self.norm_bias(bias)
+
+ query = self.to_q(pair).reshape(B, L, L, self.h, self.dim)
+ key = self.to_k(pair).reshape(B, L, L, self.h, self.dim)
+ value = self.to_v(pair).reshape(B, L, L, self.h, self.dim)
+ bias = self.to_b(bias) # (B, L, L, h)
+ gate = torch.sigmoid(self.to_g(pair)) # (B, L, L, h*dim)
+
+ query = query * self.scaling
+ key = key / math.sqrt(L) # normalize for tied attention
+ attn = einsum('bnihk,bnjhk->bijh', query, key) # tied attention
+ attn = attn + bias # apply bias
+ attn = F.softmax(attn, dim=-2) # (B, L, L, h)
+
+ if same_chain is not None:
+ ic(same_chain)
+ ic(attn)
+ ic(attn[~same_chain])
+ attn[~same_chain] *= 1.1
+
+ out = einsum('bijh,bkjhd->bikhd', attn, value).reshape(B, L, L, -1)
+ out = gate * out
+
+ out = self.to_out(out)
+ if self.is_row:
+ out = out.permute(0,2,1,3)
+ return out
+
diff --git a/utils/model/AuxiliaryPredictor.py b/utils/model/AuxiliaryPredictor.py
new file mode 100644
index 0000000000000000000000000000000000000000..af392f9a9e8a00775c690fdaec2c3db6b39afc11
--- /dev/null
+++ b/utils/model/AuxiliaryPredictor.py
@@ -0,0 +1,92 @@
+import torch
+import torch.nn as nn
+
+class DistanceNetwork(nn.Module):
+ def __init__(self, n_feat, p_drop=0.1):
+ super(DistanceNetwork, self).__init__()
+ #
+ self.proj_symm = nn.Linear(n_feat, 37*2)
+ self.proj_asymm = nn.Linear(n_feat, 37+19)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize linear layer for final logit prediction
+ nn.init.zeros_(self.proj_symm.weight)
+ nn.init.zeros_(self.proj_asymm.weight)
+ nn.init.zeros_(self.proj_symm.bias)
+ nn.init.zeros_(self.proj_asymm.bias)
+
+ def forward(self, x):
+ # input: pair info (B, L, L, C)
+
+ # predict theta, phi (non-symmetric)
+ logits_asymm = self.proj_asymm(x)
+ logits_theta = logits_asymm[:,:,:,:37].permute(0,3,1,2)
+ logits_phi = logits_asymm[:,:,:,37:].permute(0,3,1,2)
+
+ # predict dist, omega
+ logits_symm = self.proj_symm(x)
+ logits_symm = logits_symm + logits_symm.permute(0,2,1,3)
+ logits_dist = logits_symm[:,:,:,:37].permute(0,3,1,2)
+ logits_omega = logits_symm[:,:,:,37:].permute(0,3,1,2)
+
+ return logits_dist, logits_omega, logits_theta, logits_phi
+
+class MaskedTokenNetwork(nn.Module):
+ def __init__(self, n_feat, p_drop=0.1):
+ super(MaskedTokenNetwork, self).__init__()
+ self.proj = nn.Linear(n_feat, 21)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, x):
+ B, N, L = x.shape[:3]
+ logits = self.proj(x).permute(0,3,1,2).reshape(B, -1, N*L)
+
+ return logits
+
+class LDDTNetwork(nn.Module):
+ def __init__(self, n_feat, n_bin_lddt=50):
+ super(LDDTNetwork, self).__init__()
+ self.proj = nn.Linear(n_feat, n_bin_lddt)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, x):
+ logits = self.proj(x) # (B, L, 50)
+
+ return logits.permute(0,2,1)
+
+class ExpResolvedNetwork(nn.Module):
+ def __init__(self, d_msa, d_state, p_drop=0.1):
+ super(ExpResolvedNetwork, self).__init__()
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.proj = nn.Linear(d_msa+d_state, 1)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.zeros_(self.proj.weight)
+ nn.init.zeros_(self.proj.bias)
+
+ def forward(self, seq, state):
+ B, L = seq.shape[:2]
+
+ seq = self.norm_msa(seq)
+ state = self.norm_state(state)
+ feat = torch.cat((seq, state), dim=-1)
+ logits = self.proj(feat)
+ return logits.reshape(B, L)
+
+
+
diff --git a/utils/model/Embeddings.py b/utils/model/Embeddings.py
new file mode 100644
index 0000000000000000000000000000000000000000..d4558dd4eb9646070d333011d0d8783249c85264
--- /dev/null
+++ b/utils/model/Embeddings.py
@@ -0,0 +1,307 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import torch.utils.checkpoint as checkpoint
+from util import get_tips
+from util_module import Dropout, create_custom_forward, rbf, init_lecun_normal
+from Attention_module import Attention, FeedForwardLayer, AttentionWithBias
+from Track_module import PairStr2Pair
+from icecream import ic
+
+# Module contains classes and functions to generate initial embeddings
+
+class PositionalEncoding2D(nn.Module):
+ # Add relative positional encoding to pair features
+ def __init__(self, d_model, minpos=-32, maxpos=32, p_drop=0.1):
+ super(PositionalEncoding2D, self).__init__()
+ self.minpos = minpos
+ self.maxpos = maxpos
+ self.nbin = abs(minpos)+maxpos+1
+ self.emb = nn.Embedding(self.nbin, d_model)
+ self.drop = nn.Dropout(p_drop)
+
+ def forward(self, x, idx):
+ bins = torch.arange(self.minpos, self.maxpos, device=x.device)
+ seqsep = idx[:,None,:] - idx[:,:,None] # (B, L, L)
+ #
+ ib = torch.bucketize(seqsep, bins).long() # (B, L, L)
+ emb = self.emb(ib) #(B, L, L, d_model)
+ x = x + emb # add relative positional encoding
+ return self.drop(x)
+
+class MSA_emb(nn.Module):
+ # Get initial seed MSA embedding
+ def __init__(self, d_msa=256, d_pair=128, d_state=32, d_init=22+22+2+2,
+ minpos=-32, maxpos=32, p_drop=0.1):
+ super(MSA_emb, self).__init__()
+ self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
+ self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence -- used for MSA embedding
+ self.emb_left = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
+ self.emb_right = nn.Embedding(22, d_pair) # embedding for query sequence -- used for pair embedding
+ self.emb_state = nn.Embedding(22, d_state)
+ self.drop = nn.Dropout(p_drop)
+ self.pos = PositionalEncoding2D(d_pair, minpos=minpos, maxpos=maxpos, p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ self.emb_q = init_lecun_normal(self.emb_q)
+ self.emb_left = init_lecun_normal(self.emb_left)
+ self.emb_right = init_lecun_normal(self.emb_right)
+ self.emb_state = init_lecun_normal(self.emb_state)
+
+ nn.init.zeros_(self.emb.bias)
+
+ def forward(self, msa, seq, idx, seq1hot=None):
+ # Inputs:
+ # - msa: Input MSA (B, N, L, d_init)
+ # - seq: Input Sequence (B, L)
+ # - idx: Residue index
+ # Outputs:
+ # - msa: Initial MSA embedding (B, N, L, d_msa)
+ # - pair: Initial Pair embedding (B, L, L, d_pair)
+
+ N = msa.shape[1] # number of sequenes in MSA
+
+ # msa embedding
+ msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
+ seq = seq.long()
+ tmp = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ msa = msa + tmp.expand(-1, N, -1, -1) # adding query embedding to MSA
+ msa = self.drop(msa)
+
+ # pair embedding
+ if seq1hot is not None:
+ left = (seq1hot @ self.emb_left.weight)[:,None] # (B, 1, L, d_pair)
+ right = (seq1hot @ self.emb_right.weight)[:,:,None] # (B, L, 1, d_pair)
+ else:
+ left = self.emb_left(seq)[:,None] # (B, 1, L, d_pair)
+ right = self.emb_right(seq)[:,:,None] # (B, L, 1, d_pair)
+ #ic(torch.norm(self.emb_left.weight, dim=1))
+ #ic(torch.norm(self.emb_right.weight, dim=1))
+ pair = left + right # (B, L, L, d_pair)
+ pair = self.pos(pair, idx) # add relative position
+
+ # state embedding
+ state = self.drop(self.emb_state(seq))
+
+ return msa, pair, state
+
+class Extra_emb(nn.Module):
+ # Get initial seed MSA embedding
+ def __init__(self, d_msa=256, d_init=22+1+2, p_drop=0.1):
+ super(Extra_emb, self).__init__()
+ self.emb = nn.Linear(d_init, d_msa) # embedding for general MSA
+ self.emb_q = nn.Embedding(22, d_msa) # embedding for query sequence
+ self.drop = nn.Dropout(p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ nn.init.zeros_(self.emb.bias)
+
+ def forward(self, msa, seq, idx, seq1hot=None):
+ # Inputs:
+ # - msa: Input MSA (B, N, L, d_init)
+ # - seq: Input Sequence (B, L)
+ # - idx: Residue index
+ # Outputs:
+ # - msa: Initial MSA embedding (B, N, L, d_msa)
+ N = msa.shape[1] # number of sequenes in MSA
+ msa = self.emb(msa) # (B, N, L, d_model) # MSA embedding
+ if seq1hot is not None:
+ seq = (seq1hot @ self.emb_q.weight).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ else:
+ seq = self.emb_q(seq).unsqueeze(1) # (B, 1, L, d_model) -- query embedding
+ #ic(torch.norm(self.emb_q.weight, dim=1))
+ msa = msa + seq.expand(-1, N, -1, -1) # adding query embedding to MSA
+ return self.drop(msa)
+
+class TemplatePairStack(nn.Module):
+ # process template pairwise features
+ # use structure-biased attention
+ def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.25):
+ super(TemplatePairStack, self).__init__()
+ self.n_block = n_block
+ proc_s = [PairStr2Pair(d_pair=d_templ, n_head=n_head, d_hidden=d_hidden, p_drop=p_drop) for i in range(n_block)]
+ self.block = nn.ModuleList(proc_s)
+ self.norm = nn.LayerNorm(d_templ)
+ def forward(self, templ, rbf_feat, use_checkpoint=False):
+ B, T, L = templ.shape[:3]
+ templ = templ.reshape(B*T, L, L, -1)
+
+ for i_block in range(self.n_block):
+ if use_checkpoint:
+ templ = checkpoint.checkpoint(create_custom_forward(self.block[i_block]), templ, rbf_feat)
+ else:
+ templ = self.block[i_block](templ, rbf_feat)
+ return self.norm(templ).reshape(B, T, L, L, -1)
+
+class TemplateTorsionStack(nn.Module):
+ def __init__(self, n_block=2, d_templ=64, n_head=4, d_hidden=16, p_drop=0.15):
+ super(TemplateTorsionStack, self).__init__()
+ self.n_block=n_block
+ self.proj_pair = nn.Linear(d_templ+36, d_templ)
+ proc_s = [AttentionWithBias(d_in=d_templ, d_bias=d_templ,
+ n_head=n_head, d_hidden=d_hidden) for i in range(n_block)]
+ self.row_attn = nn.ModuleList(proc_s)
+ proc_s = [FeedForwardLayer(d_templ, 4, p_drop=p_drop) for i in range(n_block)]
+ self.ff = nn.ModuleList(proc_s)
+ self.norm = nn.LayerNorm(d_templ)
+
+ def reset_parameter(self):
+ self.proj_pair = init_lecun_normal(self.proj_pair)
+ nn.init.zeros_(self.proj_pair.bias)
+
+ def forward(self, tors, pair, rbf_feat, use_checkpoint=False):
+ B, T, L = tors.shape[:3]
+ tors = tors.reshape(B*T, L, -1)
+ pair = pair.reshape(B*T, L, L, -1)
+ pair = torch.cat((pair, rbf_feat), dim=-1)
+ pair = self.proj_pair(pair)
+
+ for i_block in range(self.n_block):
+ if use_checkpoint:
+ tors = tors + checkpoint.checkpoint(create_custom_forward(self.row_attn[i_block]), tors, pair)
+ else:
+ tors = tors + self.row_attn[i_block](tors, pair)
+ tors = tors + self.ff[i_block](tors)
+ return self.norm(tors).reshape(B, T, L, -1)
+
+class Templ_emb(nn.Module):
+ # Get template embedding
+ # Features are
+ # t2d:
+ # - 37 distogram bins + 6 orientations (43)
+ # - Mask (missing/unaligned) (1)
+ # t1d:
+ # - tiled AA sequence (20 standard aa + gap)
+ # - seq confidence (1)
+ # - global time step (1)
+ # - struc confidence (1)
+ #
+ def __init__(self, d_t1d=21+1+1+1, d_t2d=43+1, d_tor=30, d_pair=128, d_state=32,
+ n_block=2, d_templ=64,
+ n_head=4, d_hidden=16, p_drop=0.25):
+ super(Templ_emb, self).__init__()
+ # process 2D features
+ self.emb = nn.Linear(d_t1d*2+d_t2d, d_templ)
+ self.templ_stack = TemplatePairStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
+ d_hidden=d_hidden, p_drop=p_drop)
+
+ self.attn = Attention(d_pair, d_templ, n_head, d_hidden, d_pair, p_drop=p_drop)
+
+ # process torsion angles
+ self.emb_t1d = nn.Linear(d_t1d+d_tor, d_templ)
+ self.proj_t1d = nn.Linear(d_templ, d_templ)
+ #self.tor_stack = TemplateTorsionStack(n_block=n_block, d_templ=d_templ, n_head=n_head,
+ # d_hidden=d_hidden, p_drop=p_drop)
+ self.attn_tor = Attention(d_state, d_templ, n_head, d_hidden, d_state, p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.emb = init_lecun_normal(self.emb)
+ #nn.init.zeros_(self.emb.weight) #init weights to zero
+ nn.init.zeros_(self.emb.bias)
+
+ nn.init.kaiming_normal_(self.emb_t1d.weight, nonlinearity='relu')
+ #nn.init.zeros_(self.emb_t1d.weight)
+ nn.init.zeros_(self.emb_t1d.bias)
+
+ self.proj_t1d = init_lecun_normal(self.proj_t1d)
+ nn.init.zeros_(self.proj_t1d.bias)
+
+ def forward(self, t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=False):
+ # Input
+ # - t1d: 1D template info (B, T, L, 23) 24 SL
+ # - t2d: 2D template info (B, T, L, L, 44)
+ B, T, L, _ = t1d.shape
+
+ # Prepare 2D template features
+ left = t1d.unsqueeze(3).expand(-1,-1,-1,L,-1)
+ right = t1d.unsqueeze(2).expand(-1,-1,L,-1,-1)
+ #
+ templ = torch.cat((t2d, left, right), -1) # (B, T, L, L, 88)
+
+ #ic(templ.shape)
+ #ic(templ.dtype)
+ #ic(self.emb.weight.dtype)
+ templ = self.emb(templ) # Template templures (B, T, L, L, d_templ)
+ # process each template features
+ xyz_t = xyz_t.reshape(B*T, L, -1, 3)
+ rbf_feat = rbf(torch.cdist(xyz_t[:,:,1], xyz_t[:,:,1]))
+ templ = self.templ_stack(templ, rbf_feat, use_checkpoint=use_checkpoint) # (B, T, L,L, d_templ)
+
+ # Prepare 1D template torsion angle features
+ t1d = torch.cat((t1d, alpha_t), dim=-1) # (B, T, L, 22+30)
+ # process each template features
+ t1d = self.proj_t1d(F.relu_(self.emb_t1d(t1d)))
+
+ # mixing query state features to template state features
+ state = state.reshape(B*L, 1, -1)
+ t1d = t1d.permute(0,2,1,3).reshape(B*L, T, -1)
+ if use_checkpoint:
+ out = checkpoint.checkpoint(create_custom_forward(self.attn_tor), state, t1d, t1d)
+ out = out.reshape(B, L, -1)
+ else:
+ out = self.attn_tor(state, t1d, t1d).reshape(B, L, -1)
+ state = state.reshape(B, L, -1)
+ state = state + out
+
+ # mixing query pair features to template information (Template pointwise attention)
+ pair = pair.reshape(B*L*L, 1, -1)
+ templ = templ.permute(0, 2, 3, 1, 4).reshape(B*L*L, T, -1)
+ if use_checkpoint:
+ out = checkpoint.checkpoint(create_custom_forward(self.attn), pair, templ, templ)
+ out = out.reshape(B, L, L, -1)
+ else:
+ out = self.attn(pair, templ, templ).reshape(B, L, L, -1)
+ #
+ pair = pair.reshape(B, L, L, -1)
+ pair = pair + out
+
+ return pair, state
+
+class Recycling(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_state=32):
+ super(Recycling, self).__init__()
+ self.proj_dist = nn.Linear(36+d_state*2, d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_msa = nn.LayerNorm(d_msa)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ self.proj_dist = init_lecun_normal(self.proj_dist)
+ nn.init.zeros_(self.proj_dist.bias)
+
+ def forward(self, seq, msa, pair, xyz, state):
+ B, L = pair.shape[:2]
+ state = self.norm_state(state)
+ #
+ left = state.unsqueeze(2).expand(-1,-1,L,-1)
+ right = state.unsqueeze(1).expand(-1,L,-1,-1)
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ dist = rbf(torch.cdist(Cb, Cb))
+ dist = torch.cat((dist, left, right), dim=-1)
+ dist = self.proj_dist(dist)
+ pair = dist + self.norm_pair(pair)
+ msa = self.norm_msa(msa)
+ return msa, pair, state
+
diff --git a/utils/model/RoseTTAFoldModel.py b/utils/model/RoseTTAFoldModel.py
new file mode 100644
index 0000000000000000000000000000000000000000..ec42a3cd8a5a5bd8f59cff12963908760eff59b7
--- /dev/null
+++ b/utils/model/RoseTTAFoldModel.py
@@ -0,0 +1,140 @@
+import torch
+import torch.nn as nn
+from Embeddings import MSA_emb, Extra_emb, Templ_emb, Recycling
+from Track_module import IterativeSimulator
+from AuxiliaryPredictor import DistanceNetwork, MaskedTokenNetwork, ExpResolvedNetwork, LDDTNetwork
+from util import INIT_CRDS
+from opt_einsum import contract as einsum
+from icecream import ic
+
+class RoseTTAFoldModule(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=8, n_ref_block=4,\
+ d_msa=256, d_msa_full=64, d_pair=128, d_templ=64,
+ n_head_msa=8, n_head_pair=4, n_head_templ=4,
+ d_hidden=32, d_hidden_templ=64,
+ p_drop=0.15, d_t1d=24, d_t2d=44,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ ):
+ super(RoseTTAFoldModule, self).__init__()
+ #
+ # Input Embeddings
+ d_state = SE3_param_topk['l0_out_features']
+ self.latent_emb = MSA_emb(d_msa=d_msa, d_pair=d_pair, d_state=d_state, p_drop=p_drop)
+ self.full_emb = Extra_emb(d_msa=d_msa_full, d_init=25, p_drop=p_drop)
+ self.templ_emb = Templ_emb(d_pair=d_pair, d_templ=d_templ, d_state=d_state,
+ n_head=n_head_templ,
+ d_hidden=d_hidden_templ, p_drop=0.25, d_t1d=d_t1d, d_t2d=d_t2d)
+ # Update inputs with outputs from previous round
+ self.recycle = Recycling(d_msa=d_msa, d_pair=d_pair, d_state=d_state)
+ #
+ self.simulator = IterativeSimulator(n_extra_block=n_extra_block,
+ n_main_block=n_main_block,
+ n_ref_block=n_ref_block,
+ d_msa=d_msa, d_msa_full=d_msa_full,
+ d_pair=d_pair, d_hidden=d_hidden,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ SE3_param_full=SE3_param_full,
+ SE3_param_topk=SE3_param_topk,
+ p_drop=p_drop)
+ ##
+ self.c6d_pred = DistanceNetwork(d_pair, p_drop=p_drop)
+ self.aa_pred = MaskedTokenNetwork(d_msa, p_drop=p_drop)
+ self.lddt_pred = LDDTNetwork(d_state)
+
+ self.exp_pred = ExpResolvedNetwork(d_msa, d_state)
+
+ def forward(self, msa_latent, msa_full, seq, xyz, idx,
+ seq1hot=None, t1d=None, t2d=None, xyz_t=None, alpha_t=None,
+ msa_prev=None, pair_prev=None, state_prev=None,
+ return_raw=False, return_full=False,
+ use_checkpoint=False, return_infer=False):
+ B, N, L = msa_latent.shape[:3]
+ # Get embeddings
+ #ic(seq.shape)
+ #ic(msa_latent.shape)
+ #ic(seq1hot.shape)
+ #ic(idx.shape)
+ #ic(xyz.shape)
+ #ic(seq1hot.shape)
+ #ic(t1d.shape)
+ #ic(t2d.shape)
+
+ idx = idx.long()
+ msa_latent, pair, state = self.latent_emb(msa_latent, seq, idx, seq1hot=seq1hot)
+
+ msa_full = self.full_emb(msa_full, seq, idx, seq1hot=seq1hot)
+ #
+ # Do recycling
+ if msa_prev == None:
+ msa_prev = torch.zeros_like(msa_latent[:,0])
+ if pair_prev == None:
+ pair_prev = torch.zeros_like(pair)
+ if state_prev == None:
+ state_prev = torch.zeros_like(state)
+
+ #ic(seq.shape)
+ #ic(msa_prev.shape)
+ #ic(pair_prev.shape)
+ #ic(xyz.shape)
+ #ic(state_prev.shape)
+
+
+ msa_recycle, pair_recycle, state_recycle = self.recycle(seq, msa_prev, pair_prev, xyz, state_prev)
+ msa_latent[:,0] = msa_latent[:,0] + msa_recycle.reshape(B,L,-1)
+ pair = pair + pair_recycle
+ state = state + state_recycle
+ #
+ #ic(t1d.dtype)
+ #ic(t2d.dtype)
+ #ic(alpha_t.dtype)
+ #ic(xyz_t.dtype)
+ #ic(pair.dtype)
+ #ic(state.dtype)
+
+
+ #import pdb; pdb.set_trace()
+
+ # add template embedding
+ pair, state = self.templ_emb(t1d, t2d, alpha_t, xyz_t, pair, state, use_checkpoint=use_checkpoint)
+
+ #ic(seq.dtype)
+ #ic(msa_latent.dtype)
+ #ic(msa_full.dtype)
+ #ic(pair.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+ # Predict coordinates from given inputs
+ msa, pair, R, T, alpha_s, state = self.simulator(seq, msa_latent, msa_full.type(torch.float32), pair, xyz[:,:,:3],
+ state, idx, use_checkpoint=use_checkpoint)
+
+ if return_raw:
+ # get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return msa[:,0], pair, xyz, state, alpha_s[-1]
+
+ # predict masked amino acids
+ logits_aa = self.aa_pred(msa)
+ #
+ # predict distogram & orientograms
+ logits = self.c6d_pred(pair)
+
+ # Predict LDDT
+ lddt = self.lddt_pred(state)
+
+ # predict experimentally resolved or not
+ logits_exp = self.exp_pred(msa[:,0], state)
+
+ if return_infer:
+ #get last structure
+ xyz = einsum('bnij,bnaj->bnai', R[-1], xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T[-1].unsqueeze(-2)
+ return logits, logits_aa, logits_exp, xyz, lddt, msa[:,0], pair, state, alpha_s[-1]
+
+
+ # get all intermediate bb structures
+ xyz = einsum('rbnij,bnaj->rbnai', R, xyz[:,:,:3]-xyz[:,:,1].unsqueeze(-2)) + T.unsqueeze(-2)
+
+ return logits, logits_aa, logits_exp, xyz, alpha_s, lddt
diff --git a/utils/model/SE3_network.py b/utils/model/SE3_network.py
new file mode 100644
index 0000000000000000000000000000000000000000..a1f01d116af573f19d84a91432498a68d5b47950
--- /dev/null
+++ b/utils/model/SE3_network.py
@@ -0,0 +1,83 @@
+import torch
+import torch.nn as nn
+
+#from equivariant_attention.modules import get_basis_and_r, GSE3Res, GNormBias
+#from equivariant_attention.modules import GConvSE3, GNormSE3
+#from equivariant_attention.fibers import Fiber
+
+from util_module import init_lecun_normal_param
+from se3_transformer.model import SE3Transformer
+from se3_transformer.model.fiber import Fiber
+
+class SE3TransformerWrapper(nn.Module):
+ """SE(3) equivariant GCN with attention"""
+ def __init__(self, num_layers=2, num_channels=32, num_degrees=3, n_heads=4, div=4,
+ l0_in_features=32, l0_out_features=32,
+ l1_in_features=3, l1_out_features=2,
+ num_edge_features=32):
+ super().__init__()
+ # Build the network
+ self.l1_in = l1_in_features
+ #
+ fiber_edge = Fiber({0: num_edge_features})
+ if l1_out_features > 0:
+ if l1_in_features > 0:
+ fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
+ else:
+ fiber_in = Fiber({0: l0_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features, 1: l1_out_features})
+ else:
+ if l1_in_features > 0:
+ fiber_in = Fiber({0: l0_in_features, 1: l1_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features})
+ else:
+ fiber_in = Fiber({0: l0_in_features})
+ fiber_hidden = Fiber.create(num_degrees, num_channels)
+ fiber_out = Fiber({0: l0_out_features})
+
+ self.se3 = SE3Transformer(num_layers=num_layers,
+ fiber_in=fiber_in,
+ fiber_hidden=fiber_hidden,
+ fiber_out = fiber_out,
+ num_heads=n_heads,
+ channels_div=div,
+ fiber_edge=fiber_edge,
+ use_layer_norm=True)
+ #use_layer_norm=False)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+
+ # make sure linear layer before ReLu are initialized with kaiming_normal_
+ for n, p in self.se3.named_parameters():
+ if "bias" in n:
+ nn.init.zeros_(p)
+ elif len(p.shape) == 1:
+ continue
+ else:
+ if "radial_func" not in n:
+ p = init_lecun_normal_param(p)
+ else:
+ if "net.6" in n:
+ nn.init.zeros_(p)
+ else:
+ nn.init.kaiming_normal_(p, nonlinearity='relu')
+
+ # make last layers to be zero-initialized
+ #self.se3.graph_modules[-1].to_kernel_self['0'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['0'])
+ #self.se3.graph_modules[-1].to_kernel_self['1'] = init_lecun_normal_param(self.se3.graph_modules[-1].to_kernel_self['1'])
+ nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['0'])
+ nn.init.zeros_(self.se3.graph_modules[-1].to_kernel_self['1'])
+
+ def forward(self, G, type_0_features, type_1_features=None, edge_features=None):
+ if self.l1_in > 0:
+ node_features = {'0': type_0_features, '1': type_1_features}
+ else:
+ node_features = {'0': type_0_features}
+ edge_features = {'0': edge_features}
+ return self.se3(G, node_features, edge_features)
diff --git a/utils/model/Track_module.py b/utils/model/Track_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..e710dd3763b9eb6475fc609a1d26ee19063bc13a
--- /dev/null
+++ b/utils/model/Track_module.py
@@ -0,0 +1,476 @@
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import torch.utils.checkpoint as checkpoint
+from util import cross_product_matrix
+from util_module import *
+from Attention_module import *
+from SE3_network import SE3TransformerWrapper
+from icecream import ic
+
+# Components for three-track blocks
+# 1. MSA -> MSA update (biased attention. bias from pair & structure)
+# 2. Pair -> Pair update (biased attention. bias from structure)
+# 3. MSA -> Pair update (extract coevolution signal)
+# 4. Str -> Str update (node from MSA, edge from Pair)
+
+# Update MSA with biased self-attention. bias from Pair & Str
+class MSAPairStr2MSA(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, n_head=8, d_state=16,
+ d_hidden=32, p_drop=0.15, use_global_attn=False):
+ super(MSAPairStr2MSA, self).__init__()
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.proj_pair = nn.Linear(d_pair+36, d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+ self.proj_state = nn.Linear(d_state, d_msa)
+ self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
+ self.row_attn = MSARowAttentionWithBias(d_msa=d_msa, d_pair=d_pair,
+ n_head=n_head, d_hidden=d_hidden)
+ if use_global_attn:
+ self.col_attn = MSAColGlobalAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
+ else:
+ self.col_attn = MSAColAttention(d_msa=d_msa, n_head=n_head, d_hidden=d_hidden)
+ self.ff = FeedForwardLayer(d_msa, 4, p_drop=p_drop)
+
+ # Do proper initialization
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize weights to normal distrib
+ self.proj_pair = init_lecun_normal(self.proj_pair)
+ self.proj_state = init_lecun_normal(self.proj_state)
+
+ # initialize bias to zeros
+ nn.init.zeros_(self.proj_pair.bias)
+ nn.init.zeros_(self.proj_state.bias)
+
+ def forward(self, msa, pair, rbf_feat, state):
+ '''
+ Inputs:
+ - msa: MSA feature (B, N, L, d_msa)
+ - pair: Pair feature (B, L, L, d_pair)
+ - rbf_feat: Ca-Ca distance feature calculated from xyz coordinates (B, L, L, 36)
+ - xyz: xyz coordinates (B, L, n_atom, 3)
+ - state: updated node features after SE(3)-Transformer layer (B, L, d_state)
+ Output:
+ - msa: Updated MSA feature (B, N, L, d_msa)
+ '''
+ B, N, L = msa.shape[:3]
+
+ # prepare input bias feature by combining pair & coordinate info
+ pair = self.norm_pair(pair)
+ pair = torch.cat((pair, rbf_feat), dim=-1)
+ pair = self.proj_pair(pair) # (B, L, L, d_pair)
+ #
+ # update query sequence feature (first sequence in the MSA) with feedbacks (state) from SE3
+ state = self.norm_state(state)
+ state = self.proj_state(state).reshape(B, 1, L, -1)
+
+ msa = msa.index_add(1, torch.tensor([0,], device=state.device), state.type(torch.float32))
+ #
+ # Apply row/column attention to msa & transform
+ msa = msa + self.drop_row(self.row_attn(msa, pair))
+ msa = msa + self.col_attn(msa)
+ msa = msa + self.ff(msa)
+
+ return msa
+
+class PairStr2Pair(nn.Module):
+ def __init__(self, d_pair=128, n_head=4, d_hidden=32, d_rbf=36, p_drop=0.15):
+ super(PairStr2Pair, self).__init__()
+
+ self.emb_rbf = nn.Linear(d_rbf, d_hidden)
+ self.proj_rbf = nn.Linear(d_hidden, d_pair)
+
+ self.drop_row = Dropout(broadcast_dim=1, p_drop=p_drop)
+ self.drop_col = Dropout(broadcast_dim=2, p_drop=p_drop)
+
+ self.row_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=True)
+ self.col_attn = BiasedAxialAttention(d_pair, d_pair, n_head, d_hidden, p_drop=p_drop, is_row=False)
+
+ self.ff = FeedForwardLayer(d_pair, 2)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ nn.init.kaiming_normal_(self.emb_rbf.weight, nonlinearity='relu')
+ nn.init.zeros_(self.emb_rbf.bias)
+
+ self.proj_rbf = init_lecun_normal(self.proj_rbf)
+ nn.init.zeros_(self.proj_rbf.bias)
+
+ def forward(self, pair, rbf_feat):
+ B, L = pair.shape[:2]
+
+ rbf_feat = self.proj_rbf(F.relu_(self.emb_rbf(rbf_feat)))
+
+ pair = pair + self.drop_row(self.row_attn(pair, rbf_feat))
+ pair = pair + self.drop_col(self.col_attn(pair, rbf_feat))
+ pair = pair + self.ff(pair)
+ return pair
+
+class MSA2Pair(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_hidden=32, p_drop=0.15):
+ super(MSA2Pair, self).__init__()
+ self.norm = nn.LayerNorm(d_msa)
+ self.proj_left = nn.Linear(d_msa, d_hidden)
+ self.proj_right = nn.Linear(d_msa, d_hidden)
+ self.proj_out = nn.Linear(d_hidden*d_hidden, d_pair)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # normal initialization
+ self.proj_left = init_lecun_normal(self.proj_left)
+ self.proj_right = init_lecun_normal(self.proj_right)
+ nn.init.zeros_(self.proj_left.bias)
+ nn.init.zeros_(self.proj_right.bias)
+
+ # zero initialize output
+ nn.init.zeros_(self.proj_out.weight)
+ nn.init.zeros_(self.proj_out.bias)
+
+ def forward(self, msa, pair):
+ B, N, L = msa.shape[:3]
+ msa = self.norm(msa)
+ left = self.proj_left(msa)
+ right = self.proj_right(msa)
+ right = right / float(N)
+ out = einsum('bsli,bsmj->blmij', left, right).reshape(B, L, L, -1)
+ out = self.proj_out(out)
+
+ pair = pair + out
+
+ return pair
+
+class SCPred(nn.Module):
+ def __init__(self, d_msa=256, d_state=32, d_hidden=128, p_drop=0.15):
+ super(SCPred, self).__init__()
+ self.norm_s0 = nn.LayerNorm(d_msa)
+ self.norm_si = nn.LayerNorm(d_state)
+ self.linear_s0 = nn.Linear(d_msa, d_hidden)
+ self.linear_si = nn.Linear(d_state, d_hidden)
+
+ # ResNet layers
+ self.linear_1 = nn.Linear(d_hidden, d_hidden)
+ self.linear_2 = nn.Linear(d_hidden, d_hidden)
+ self.linear_3 = nn.Linear(d_hidden, d_hidden)
+ self.linear_4 = nn.Linear(d_hidden, d_hidden)
+
+ # Final outputs
+ self.linear_out = nn.Linear(d_hidden, 20)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # normal initialization
+ self.linear_s0 = init_lecun_normal(self.linear_s0)
+ self.linear_si = init_lecun_normal(self.linear_si)
+ self.linear_out = init_lecun_normal(self.linear_out)
+ nn.init.zeros_(self.linear_s0.bias)
+ nn.init.zeros_(self.linear_si.bias)
+ nn.init.zeros_(self.linear_out.bias)
+
+ # right before relu activation: He initializer (kaiming normal)
+ nn.init.kaiming_normal_(self.linear_1.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear_1.bias)
+ nn.init.kaiming_normal_(self.linear_3.weight, nonlinearity='relu')
+ nn.init.zeros_(self.linear_3.bias)
+
+ # right before residual connection: zero initialize
+ nn.init.zeros_(self.linear_2.weight)
+ nn.init.zeros_(self.linear_2.bias)
+ nn.init.zeros_(self.linear_4.weight)
+ nn.init.zeros_(self.linear_4.bias)
+
+ def forward(self, seq, state):
+ '''
+ Predict side-chain torsion angles along with backbone torsions
+ Inputs:
+ - seq: hidden embeddings corresponding to query sequence (B, L, d_msa)
+ - state: state feature (output l0 feature) from previous SE3 layer (B, L, d_state)
+ Outputs:
+ - si: predicted torsion angles (phi, psi, omega, chi1~4 with cos/sin, Cb bend, Cb twist, CG) (B, L, 10, 2)
+ '''
+ B, L = seq.shape[:2]
+ seq = self.norm_s0(seq)
+ state = self.norm_si(state)
+ si = self.linear_s0(seq) + self.linear_si(state)
+
+ si = si + self.linear_2(F.relu_(self.linear_1(F.relu_(si))))
+ si = si + self.linear_4(F.relu_(self.linear_3(F.relu_(si))))
+
+ si = self.linear_out(F.relu_(si))
+ return si.view(B, L, 10, 2)
+
+
+class Str2Str(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128, d_state=16,
+ SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}, p_drop=0.1):
+ super(Str2Str, self).__init__()
+
+ # initial node & pair feature process
+ self.norm_msa = nn.LayerNorm(d_msa)
+ self.norm_pair = nn.LayerNorm(d_pair)
+ self.norm_state = nn.LayerNorm(d_state)
+
+ self.embed_x = nn.Linear(d_msa+d_state, SE3_param['l0_in_features'])
+ self.embed_e1 = nn.Linear(d_pair, SE3_param['num_edge_features'])
+ self.embed_e2 = nn.Linear(SE3_param['num_edge_features']+36+1, SE3_param['num_edge_features'])
+
+ self.norm_node = nn.LayerNorm(SE3_param['l0_in_features'])
+ self.norm_edge1 = nn.LayerNorm(SE3_param['num_edge_features'])
+ self.norm_edge2 = nn.LayerNorm(SE3_param['num_edge_features'])
+
+ self.se3 = SE3TransformerWrapper(**SE3_param)
+ self.sc_predictor = SCPred(d_msa=d_msa, d_state=SE3_param['l0_out_features'],
+ p_drop=p_drop)
+
+ self.reset_parameter()
+
+ def reset_parameter(self):
+ # initialize weights to normal distribution
+ self.embed_x = init_lecun_normal(self.embed_x)
+ self.embed_e1 = init_lecun_normal(self.embed_e1)
+ self.embed_e2 = init_lecun_normal(self.embed_e2)
+
+ # initialize bias to zeros
+ nn.init.zeros_(self.embed_x.bias)
+ nn.init.zeros_(self.embed_e1.bias)
+ nn.init.zeros_(self.embed_e2.bias)
+
+ @torch.cuda.amp.autocast(enabled=False)
+ def forward(self, msa, pair, R_in, T_in, xyz, state, idx, top_k=64, eps=1e-5):
+ B, N, L = msa.shape[:3]
+
+ state = state.type(torch.float32)
+ mas = msa.type(torch.float32)
+ pair = pair.type(torch.float32)
+ R_in = R_in.type(torch.float32)
+ T_in = T_in.type(torch.float32)
+ xyz = xyz.type(torch.float32)
+
+ #ic(msa.dtype)
+ #ic(pair.dtype)
+ #ic(R_in.dtype)
+ #ic(T_in.dtype)
+ #ic(xyz.dtype)
+ #ic(state.dtype)
+ #ic(idx.dtype)
+
+
+ # process msa & pair features
+ node = self.norm_msa(msa[:,0])
+ pair = self.norm_pair(pair)
+ state = self.norm_state(state)
+
+ node = torch.cat((node, state), dim=-1)
+ node = self.norm_node(self.embed_x(node))
+ pair = self.norm_edge1(self.embed_e1(pair))
+
+ neighbor = get_seqsep(idx)
+ rbf_feat = rbf(torch.cdist(xyz[:,:,1], xyz[:,:,1]))
+ pair = torch.cat((pair, rbf_feat, neighbor), dim=-1)
+ pair = self.norm_edge2(self.embed_e2(pair))
+
+ # define graph
+ if top_k != 0:
+ G, edge_feats = make_topk_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
+ else:
+ G, edge_feats = make_full_graph(xyz[:,:,1,:], pair, idx, top_k=top_k)
+ l1_feats = xyz - xyz[:,:,1,:].unsqueeze(2)
+ l1_feats = l1_feats.reshape(B*L, -1, 3)
+
+ # apply SE(3) Transformer & update coordinates
+ shift = self.se3(G, node.reshape(B*L, -1, 1), l1_feats, edge_feats)
+
+ state = shift['0'].reshape(B, L, -1) # (B, L, C)
+
+ offset = shift['1'].reshape(B, L, 2, 3)
+ delTi = offset[:,:,0,:] / 10.0 # translation
+ R = offset[:,:,1,:] / 100.0 # rotation
+
+ Qnorm = torch.sqrt( 1 + torch.sum(R*R, dim=-1) )
+ qA, qB, qC, qD = 1/Qnorm, R[:,:,0]/Qnorm, R[:,:,1]/Qnorm, R[:,:,2]/Qnorm
+
+ delRi = torch.zeros((B,L,3,3), device=xyz.device)
+ delRi[:,:,0,0] = qA*qA+qB*qB-qC*qC-qD*qD
+ delRi[:,:,0,1] = 2*qB*qC - 2*qA*qD
+ delRi[:,:,0,2] = 2*qB*qD + 2*qA*qC
+ delRi[:,:,1,0] = 2*qB*qC + 2*qA*qD
+ delRi[:,:,1,1] = qA*qA-qB*qB+qC*qC-qD*qD
+ delRi[:,:,1,2] = 2*qC*qD - 2*qA*qB
+ delRi[:,:,2,0] = 2*qB*qD - 2*qA*qC
+ delRi[:,:,2,1] = 2*qC*qD + 2*qA*qB
+ delRi[:,:,2,2] = qA*qA-qB*qB-qC*qC+qD*qD
+ #
+ ## convert vector to rotation matrix
+ #R_angle = torch.norm(R, dim=-1, keepdim=True) # (B, L, 1)
+ #cos_angle = torch.cos(R_angle).unsqueeze(2) # (B, L, 1, 1)
+ #sin_angle = torch.sin(R_angle).unsqueeze(2) # (B, L, 1, 1)
+ #R_vector = R / (R_angle+eps) # (B, L, 3)
+
+ #delRi = cos_angle*torch.eye(3, device=R.device).reshape(1,1,3,3) \
+ # + sin_angle*cross_product_matrix(R_vector) \
+ # + (1.0-cos_angle)*einsum('bni,bnj->bnij', R_vector, R_vector)
+
+ Ri = einsum('bnij,bnjk->bnik', delRi, R_in)
+ Ti = delTi + T_in #einsum('bnij,bnj->bni', delRi, T_in) + delTi
+
+ alpha = self.sc_predictor(msa[:,0], state)
+ return Ri, Ti, state, alpha
+
+class IterBlock(nn.Module):
+ def __init__(self, d_msa=256, d_pair=128,
+ n_head_msa=8, n_head_pair=4,
+ use_global_attn=False,
+ d_hidden=32, d_hidden_msa=None, p_drop=0.15,
+ SE3_param={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32}):
+ super(IterBlock, self).__init__()
+ if d_hidden_msa == None:
+ d_hidden_msa = d_hidden
+
+ self.msa2msa = MSAPairStr2MSA(d_msa=d_msa, d_pair=d_pair,
+ n_head=n_head_msa,
+ d_state=SE3_param['l0_out_features'],
+ use_global_attn=use_global_attn,
+ d_hidden=d_hidden_msa, p_drop=p_drop)
+ self.msa2pair = MSA2Pair(d_msa=d_msa, d_pair=d_pair,
+ d_hidden=d_hidden//2, p_drop=p_drop)
+ #d_hidden=d_hidden, p_drop=p_drop)
+ self.pair2pair = PairStr2Pair(d_pair=d_pair, n_head=n_head_pair,
+ d_hidden=d_hidden, p_drop=p_drop)
+ self.str2str = Str2Str(d_msa=d_msa, d_pair=d_pair,
+ d_state=SE3_param['l0_out_features'],
+ SE3_param=SE3_param,
+ p_drop=p_drop)
+
+ def forward(self, msa, pair, R_in, T_in, xyz, state, idx, use_checkpoint=False):
+ rbf_feat = rbf(torch.cdist(xyz[:,:,1,:], xyz[:,:,1,:]))
+ if use_checkpoint:
+ msa = checkpoint.checkpoint(create_custom_forward(self.msa2msa), msa, pair, rbf_feat, state)
+ pair = checkpoint.checkpoint(create_custom_forward(self.msa2pair), msa, pair)
+ pair = checkpoint.checkpoint(create_custom_forward(self.pair2pair), pair, rbf_feat)
+ R, T, state, alpha = checkpoint.checkpoint(create_custom_forward(self.str2str, top_k=0), msa, pair, R_in, T_in, xyz, state, idx)
+ else:
+ msa = self.msa2msa(msa, pair, rbf_feat, state)
+ pair = self.msa2pair(msa, pair)
+ pair = self.pair2pair(pair, rbf_feat)
+ R, T, state, alpha = self.str2str(msa, pair, R_in, T_in, xyz, state, idx, top_k=0)
+
+ return msa, pair, R, T, state, alpha
+
+class IterativeSimulator(nn.Module):
+ def __init__(self, n_extra_block=4, n_main_block=12, n_ref_block=4,
+ d_msa=256, d_msa_full=64, d_pair=128, d_hidden=32,
+ n_head_msa=8, n_head_pair=4,
+ SE3_param_full={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ SE3_param_topk={'l0_in_features':32, 'l0_out_features':16, 'num_edge_features':32},
+ p_drop=0.15):
+ super(IterativeSimulator, self).__init__()
+ self.n_extra_block = n_extra_block
+ self.n_main_block = n_main_block
+ self.n_ref_block = n_ref_block
+
+ self.proj_state = nn.Linear(SE3_param_topk['l0_out_features'], SE3_param_full['l0_out_features'])
+ # Update with extra sequences
+ if n_extra_block > 0:
+ self.extra_block = nn.ModuleList([IterBlock(d_msa=d_msa_full, d_pair=d_pair,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ d_hidden_msa=8,
+ d_hidden=d_hidden,
+ p_drop=p_drop,
+ use_global_attn=True,
+ SE3_param=SE3_param_full)
+ for i in range(n_extra_block)])
+
+ # Update with seed sequences
+ if n_main_block > 0:
+ self.main_block = nn.ModuleList([IterBlock(d_msa=d_msa, d_pair=d_pair,
+ n_head_msa=n_head_msa,
+ n_head_pair=n_head_pair,
+ d_hidden=d_hidden,
+ p_drop=p_drop,
+ use_global_attn=False,
+ SE3_param=SE3_param_full)
+ for i in range(n_main_block)])
+
+ self.proj_state2 = nn.Linear(SE3_param_full['l0_out_features'], SE3_param_topk['l0_out_features'])
+ # Final SE(3) refinement
+ if n_ref_block > 0:
+ self.str_refiner = Str2Str(d_msa=d_msa, d_pair=d_pair,
+ d_state=SE3_param_topk['l0_out_features'],
+ SE3_param=SE3_param_topk,
+ p_drop=p_drop)
+
+ self.reset_parameter()
+ def reset_parameter(self):
+ self.proj_state = init_lecun_normal(self.proj_state)
+ nn.init.zeros_(self.proj_state.bias)
+ self.proj_state2 = init_lecun_normal(self.proj_state2)
+ nn.init.zeros_(self.proj_state2.bias)
+
+ def forward(self, seq, msa, msa_full, pair, xyz_in, state, idx, use_checkpoint=False):
+ # input:
+ # seq: query sequence (B, L)
+ # msa: seed MSA embeddings (B, N, L, d_msa)
+ # msa_full: extra MSA embeddings (B, N, L, d_msa_full)
+ # pair: initial residue pair embeddings (B, L, L, d_pair)
+ # xyz_in: initial BB coordinates (B, L, n_atom, 3)
+ # state: initial state features containing mixture of query seq, sidechain, accuracy info (B, L, d_state)
+ # idx: residue index
+
+ B, L = pair.shape[:2]
+
+ R_in = torch.eye(3, device=xyz_in.device).reshape(1,1,3,3).expand(B, L, -1, -1)
+ T_in = xyz_in[:,:,1].clone()
+ xyz_in = xyz_in - T_in.unsqueeze(-2)
+
+ state = self.proj_state(state)
+
+ R_s = list()
+ T_s = list()
+ alpha_s = list()
+ for i_m in range(self.n_extra_block):
+ R_in = R_in.detach() # detach rotation (for stability)
+ T_in = T_in.detach()
+ # Get current BB structure
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+
+ msa_full, pair, R_in, T_in, state, alpha = self.extra_block[i_m](msa_full, pair,
+ R_in, T_in, xyz, state, idx,
+ use_checkpoint=use_checkpoint)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ for i_m in range(self.n_main_block):
+ R_in = R_in.detach()
+ T_in = T_in.detach()
+ # Get current BB structure
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+
+ msa, pair, R_in, T_in, state, alpha = self.main_block[i_m](msa, pair,
+ R_in, T_in, xyz, state, idx,
+ use_checkpoint=use_checkpoint)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ state = self.proj_state2(state)
+ for i_m in range(self.n_ref_block):
+ R_in = R_in.detach()
+ T_in = T_in.detach()
+ xyz = einsum('bnij,bnaj->bnai', R_in, xyz_in) + T_in.unsqueeze(-2)
+ R_in, T_in, state, alpha = self.str_refiner(msa, pair, R_in, T_in, xyz, state, idx, top_k=64)
+ R_s.append(R_in)
+ T_s.append(T_in)
+ alpha_s.append(alpha)
+
+ R_s = torch.stack(R_s, dim=0)
+ T_s = torch.stack(T_s, dim=0)
+ alpha_s = torch.stack(alpha_s, dim=0)
+
+ return msa, pair, R_s, T_s, alpha_s, state
diff --git a/utils/model/__pycache__/Attention_module.cpython-310.pyc b/utils/model/__pycache__/Attention_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b3322289b0411833d51fa3ec56561e167490a97c
Binary files /dev/null and b/utils/model/__pycache__/Attention_module.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc b/utils/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0640806f046ee39a07f43474a92e7863eac08249
Binary files /dev/null and b/utils/model/__pycache__/AuxiliaryPredictor.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/Embeddings.cpython-310.pyc b/utils/model/__pycache__/Embeddings.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0e1de9e9bc00e4678d998dca23d1c4937120641a
Binary files /dev/null and b/utils/model/__pycache__/Embeddings.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc b/utils/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5d1316d2c7827ae620c416d0c6dce0ad4384589b
Binary files /dev/null and b/utils/model/__pycache__/RoseTTAFoldModel.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/SE3_network.cpython-310.pyc b/utils/model/__pycache__/SE3_network.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9bd3c3173207da5f95e9fca5350bfefd7b00d763
Binary files /dev/null and b/utils/model/__pycache__/SE3_network.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/Track_module.cpython-310.pyc b/utils/model/__pycache__/Track_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b8bc6b2787f3fbde9891f4aa45c3e78949b10b4b
Binary files /dev/null and b/utils/model/__pycache__/Track_module.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/ab_tools.cpython-310.pyc b/utils/model/__pycache__/ab_tools.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9865c1ff58c72107bf69cfcf86336b8880d0b70e
Binary files /dev/null and b/utils/model/__pycache__/ab_tools.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/apply_masks.cpython-310.pyc b/utils/model/__pycache__/apply_masks.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..fe3081116d10b9f1d83620ff7bc99f231ed5632a
Binary files /dev/null and b/utils/model/__pycache__/apply_masks.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/arguments.cpython-310.pyc b/utils/model/__pycache__/arguments.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..fb972ac10b616e96ee62c25f471b52cfee4fe07a
Binary files /dev/null and b/utils/model/__pycache__/arguments.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/chemical.cpython-310.pyc b/utils/model/__pycache__/chemical.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5150406fe03402a121d1ba5c5057166aca9942dd
Binary files /dev/null and b/utils/model/__pycache__/chemical.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/data_loader.cpython-310.pyc b/utils/model/__pycache__/data_loader.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..8c14615387c282c92028e4ccb224c5e25f3a1141
Binary files /dev/null and b/utils/model/__pycache__/data_loader.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/diffusion.cpython-310.pyc b/utils/model/__pycache__/diffusion.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..184f706857ce375fce3d08ea762e3265d18231d0
Binary files /dev/null and b/utils/model/__pycache__/diffusion.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/kinematics.cpython-310.pyc b/utils/model/__pycache__/kinematics.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..75973ac48c41c032972159fba097a8f07e3e886d
Binary files /dev/null and b/utils/model/__pycache__/kinematics.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/loss.cpython-310.pyc b/utils/model/__pycache__/loss.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..63a1aee0f9ff190da7197bd870322242fb4b0716
Binary files /dev/null and b/utils/model/__pycache__/loss.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/mask_generator.cpython-310.pyc b/utils/model/__pycache__/mask_generator.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..0d07980de4880838df2a2a548d2145b800a09b09
Binary files /dev/null and b/utils/model/__pycache__/mask_generator.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/parsers.cpython-310.pyc b/utils/model/__pycache__/parsers.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..3188471a2c59eb6a67aa63b841e9aef1548a1e4c
Binary files /dev/null and b/utils/model/__pycache__/parsers.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/scheduler.cpython-310.pyc b/utils/model/__pycache__/scheduler.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..5a8f61d5257f9f26cb71049fdc8c68d9352d9aa3
Binary files /dev/null and b/utils/model/__pycache__/scheduler.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/scoring.cpython-310.pyc b/utils/model/__pycache__/scoring.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..dfd273eea8119c39592a933e7dd72b3f642a1371
Binary files /dev/null and b/utils/model/__pycache__/scoring.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/train_multi_deep.cpython-310.pyc b/utils/model/__pycache__/train_multi_deep.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..302fe224c624840c1200a68e22db9990b8e070f9
Binary files /dev/null and b/utils/model/__pycache__/train_multi_deep.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc b/utils/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..d3e9bafc9b0645924d26dfdc4a2dafa9e774705c
Binary files /dev/null and b/utils/model/__pycache__/train_multi_deep_selfcond_nostruc.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/util.cpython-310.pyc b/utils/model/__pycache__/util.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b3d1ff3bb6e5fb456825033bb2e9c9be41a3a6ce
Binary files /dev/null and b/utils/model/__pycache__/util.cpython-310.pyc differ
diff --git a/utils/model/__pycache__/util_module.cpython-310.pyc b/utils/model/__pycache__/util_module.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..bc0740e035a77ef694f6060b13d4683f99e5b4f6
Binary files /dev/null and b/utils/model/__pycache__/util_module.cpython-310.pyc differ
diff --git a/utils/model/add_weights_dimension.py b/utils/model/add_weights_dimension.py
new file mode 100644
index 0000000000000000000000000000000000000000..6f85d75f28a6d46c2c8dfb3dbd1fc340924801e1
--- /dev/null
+++ b/utils/model/add_weights_dimension.py
@@ -0,0 +1,73 @@
+import torch
+
+orig_t1d = 22
+#orig_t1d = 23
+orig_t2d = 44
+orig_dtor = 30
+
+new_t1d = orig_t1d + 2 + 4 + 1 #(+2 time step and seq confidence) + 4 (dssp) + 1 (hot spot)
+new_t2d = orig_t2d + 0
+
+#ckpt = torch.load('/net/scratch/jgershon/models/autofold4_seq2str_base.pt', map_location=torch.device('cpu'))
+#ckpt = torch.load('/home/jgershon/projects/c6d_diff/BFF/autofold4/experiments/2209235seqdiffV4_accum4_str5_aa5_continued/models/BFF_4.pt', map_location=torch.device('cpu'))
+ckpt = torch.load('/net/scratch/lisanza/diffuse_3track_fullcon/models/BFF_last.pt', map_location=torch.device('cpu'))
+
+weights = ckpt['model_state_dict']
+
+print("original weights")
+print('templ_emb.emb.weight', weights['templ_emb.emb.weight'].shape)
+print('templ_emb.emb_t1d.weight', weights['templ_emb.emb_t1d.weight'].shape)
+
+# weights['templ_emb.emb.weight'] # Original shape: (64, 88)
+# weights['templ_emb.emb_t1d.weight'] # Original shape: (64, 52)
+
+# Adding 2D embedding features
+# d_t1d*2+d_t2d
+if True:
+ #pt1_add_dim = new_t2d - orig_t2d
+ pt2_add_dim = new_t1d - orig_t1d
+ pt3_add_dim = new_t1d - orig_t1d
+
+ #pt1_emb_zeros = torch.zeros(64, pt1_add_dim)
+ pt2_emb_zeros = torch.zeros(64, pt2_add_dim)
+ pt3_emb_zeros = torch.zeros(64, pt3_add_dim)
+
+ '''
+ The way that the t2d input to embedding is created is not straightforward
+ It looks like this:
+
+ # Prepare 2D template features
+ left = t1d.unsqueeze(3).expand(-1,-1,-1,L,-1)
+ right = t1d.unsqueeze(2).expand(-1,-1,L,-1,-1)
+
+ templ = torch.cat((t2d, left, right), -1) # (B, T, L, L, 109)
+ templ = self.emb(templ) # Template templures (B, T, L, L, d_templ)
+ '''
+
+ #new_emb_weights = torch.cat( (weights['templ_emb.emb.weight'][:,:orig_t2d], pt1_emb_zeros), dim=-1 )
+ #new_emb_weights = torch.cat( (new_emb_weights, weights['templ_emb.emb.weight'][:,orig_t2d:orig_t2d+orig_t1d], pt2_emb_zeros), dim=-1 )
+ new_emb_weights = torch.cat( (weights['templ_emb.emb.weight'][:,:orig_t2d+orig_t1d], pt2_emb_zeros), dim=-1 )
+ new_emb_weights = torch.cat( (new_emb_weights, weights['templ_emb.emb.weight'][:,orig_t2d+orig_t1d:], pt3_emb_zeros), dim=-1 )
+
+ #new_emb_weights = torch.cat( (pt1_emb_weights, pt2_emb_weights, pt3_emb_weights), dim=-1 )
+
+# Adding 1D embedding features
+# d_t1d+d_tor
+if True:
+ t1d_weights_dim = new_t1d + orig_dtor
+ t1d_add_dim = t1d_weights_dim - weights['templ_emb.emb_t1d.weight'].shape[1] #52
+
+ t1d_zeros = torch.zeros(64, t1d_add_dim)
+ new_t1d_weights = torch.cat( (weights['templ_emb.emb_t1d.weight'][:,:orig_t1d], t1d_zeros), dim=-1 )
+ new_t1d_weights = torch.cat( (new_t1d_weights, weights['templ_emb.emb_t1d.weight'][:,orig_t1d:]), dim=-1 )
+
+weights['templ_emb.emb.weight'] = new_emb_weights
+weights['templ_emb.emb_t1d.weight'] = new_t1d_weights
+print("new t1d weights dim")
+print(new_t1d_weights.shape)
+
+ckpt['model_state_dict'] = weights
+#torch.save(ckpt, './models/t1d_23_t2d_44_BFF_last.pt')
+#torch.save(ckpt, './models/t1d_24_t2d_44_BFF_last.pt')
+torch.save(ckpt, '/net/scratch/lisanza/projects/diffusion/models/t1d_29_t2d_44_BFF_SE3big_2.pt')
+#torch.save(ckpt, './models/t1d_24_t2d_44_BFF_diffV4_accum4_str5_aa5_continued.pt')
diff --git a/utils/model/apply_masks.py b/utils/model/apply_masks.py
new file mode 100755
index 0000000000000000000000000000000000000000..ecb0fd70f8fd973aa4abb992188cd2335db0cc57
--- /dev/null
+++ b/utils/model/apply_masks.py
@@ -0,0 +1,196 @@
+import sys, os
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+sys.path.append('../')
+from utils.calc_dssp import annotate_sse
+
+ic.configureOutput(includeContext=True)
+
+def mask_inputs(seq,
+ msa_masked,
+ msa_full,
+ xyz_t,
+ t1d,
+ mask_msa,
+ input_seq_mask=None,
+ input_str_mask=None,
+ input_floating_mask=None,
+ input_t1dconf_mask=None,
+ loss_seq_mask=None,
+ loss_str_mask=None,
+ loss_str_mask_2d=None,
+ dssp=False,
+ hotspots=False,
+ diffuser=None,
+ t=None,
+ freeze_seq_emb=False,
+ mutate_seq=False,
+ no_clamp_seq=False,
+ norm_input=False,
+ contacts=None,
+ frac_provide_dssp=0.5,
+ dssp_mask_percentage=[0,100],
+ frac_provide_contacts=0.5,
+ struc_cond=False):
+ """
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+
+
+
+ #ic(input_seq_mask.shape)
+ #ic(seq.shape)
+ #ic(msa_masked.shape)
+ #ic(msa_full.shape)
+ #ic(t1d.shape)
+ #ic(xyz_t.shape)
+ #ic(input_str_mask.shape)
+ #ic(mask_msa.shape)
+
+ ###########
+ seq_mask = input_seq_mask
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+
+ str_mask = input_str_mask
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+ seq_diffused = diffuser.q_sample(x_0,t,mask=seq_mask)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t]),mask=msa_seq_mask)
+
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #make msa_full same size as msa_masked
+ #ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ #to do add structure confidence metric; need to expand dimensions of chkpt b4
+ #if t1d_24: JG - changed to be default
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ if dssp:
+ print(f'adding dssp {frac_provide_dssp} of time')
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],4)).float()), -1).to(seq.device)
+ #dssp info
+ #mask some percentage of dssp info in range dssp_mask_percentage[0],dssp_mask_percentage[1]
+ percentage_mask=random.randint(dssp_mask_percentage[0], dssp_mask_percentage[1])
+ dssp=annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=percentage_mask)
+ #dssp_unmasked = annotate_sse(np.array(xyz_t[0,:,1,:].squeeze()), percentage_mask=0)
+ if np.random.rand()>frac_provide_dssp:
+ print('masking dssp')
+ dssp[...]=0 #replace with mask token
+ dssp[:,-1]=1
+ t1d[...,24:]=dssp
+
+ if hotspots:
+ print(f"adding hotspots {frac_provide_contacts} of time")
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ #mask all contacts some fraction of the time
+ if np.random.rand()>frac_provide_contacts:
+ print('masking contacts')
+ contacts = torch.zeros(L)
+ t1d[...,-1] = contacts
+
+ ### xyz_t ###
+ #############
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+ #Sequence masking
+ xyz_t[:,:,3:,:] = float('nan')
+ # Structure masking
+ if struc_cond:
+ print("non-autoregressive structure conditioning")
+ r = diffuser.alphas_cumprod[t]
+ xyz_mask = (torch.rand(xyz_t.shape[1]) > r).to(torch.bool).to(seq.device)
+ xyz_mask = torch.logical_and(xyz_mask,~str_mask)
+ xyz_t[:,xyz_mask,:,:] = float('nan')
+ else:
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ ### mask_msa ###
+ ################
+ # NOTE: this is for loss scoring
+ mask_msa[:,:,~loss_seq_mask] = False
+
+ out=dict(
+ seq= seq,
+ msa_masked= msa_masked,
+ msa_full= msa_full,
+ xyz_t= xyz_t,
+ t1d= t1d,
+ mask_msa= mask_msa,
+ seq_diffused= seq_diffused
+ )
+
+ return out
diff --git a/utils/model/chemical.py b/utils/model/chemical.py
new file mode 100644
index 0000000000000000000000000000000000000000..206adc88ee09010b80e265c2d16b2c6fd447fa43
--- /dev/null
+++ b/utils/model/chemical.py
@@ -0,0 +1,570 @@
+import torch
+import numpy as np
+
+num2aa=[
+ 'ALA','ARG','ASN','ASP','CYS',
+ 'GLN','GLU','GLY','HIS','ILE',
+ 'LEU','LYS','MET','PHE','PRO',
+ 'SER','THR','TRP','TYR','VAL',
+ 'UNK','MAS',
+ ]
+
+aa2num= {x:i for i,x in enumerate(num2aa)}
+
+# full sc atom representation (Nx14)
+aa2long=[
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # ala
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," NE "," CZ "," NH1"," NH2", None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD "," HE ","1HH1","2HH1","1HH2","2HH2"), # arg
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," ND2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HD2","2HD2", None, None, None, None, None, None, None), # asn
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," OD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None, None), # asp
+ (" N "," CA "," C "," O "," CB "," SG ", None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ", None, None, None, None, None, None, None, None), # cys
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," NE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE2","2HE2", None, None, None, None, None), # gln
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," OE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ", None, None, None, None, None, None, None), # glu
+ (" N "," CA "," C "," O ", None, None, None, None, None, None, None, None, None, None," H ","1HA ","2HA ", None, None, None, None, None, None, None, None, None, None), # gly
+ (" N "," CA "," C "," O "," CB "," CG "," ND1"," CD2"," CE1"," NE2", None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1"," HE2", None, None, None, None, None, None), # his
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2"," CD1", None, None, None, None, None, None," H "," HA "," HB ","1HG2","2HG2","3HG2","1HG1","2HG1","1HD1","2HD1","3HD1", None, None), # ile
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ","1HD1","2HD1","3HD1","1HD2","2HD2","3HD2", None, None), # leu
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," CE "," NZ ", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ","1HE ","2HE ","1HZ ","2HZ ","3HZ "), # lys
+ (" N "," CA "," C "," O "," CB "," CG "," SD "," CE ", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE ","2HE ","3HE ", None, None, None, None), # met
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," CE1"," CE2"," CZ ", None, None, None," H "," HA ","1HB ","2HB "," HD1"," HD2"," HE1"," HE2"," HZ ", None, None, None, None), # phe
+ (" N "," CA "," C "," O "," CB "," CG "," CD ", None, None, None, None, None, None, None," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ", None, None, None, None, None, None), # pro
+ (" N "," CA "," C "," O "," CB "," OG ", None, None, None, None, None, None, None, None," H "," HG "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None), # ser
+ (" N "," CA "," C "," O "," CB "," OG1"," CG2", None, None, None, None, None, None, None," H "," HG1"," HA "," HB ","1HG2","2HG2","3HG2", None, None, None, None, None, None), # thr
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," NE1"," CE2"," CE3"," CZ2"," CZ3"," CH2"," H "," HA ","1HB ","2HB "," HD1"," HE1"," HZ2"," HH2"," HZ3"," HE3", None, None, None), # trp
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," CE1"," CE2"," CZ "," OH ", None, None," H "," HA ","1HB ","2HB "," HD1"," HE1"," HE2"," HD2"," HH ", None, None, None, None), # tyr
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2", None, None, None, None, None, None, None," H "," HA "," HB ","1HG1","2HG1","3HG1","1HG2","2HG2","3HG2", None, None, None, None), # val
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # unk
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # mask
+]
+
+# build the "alternate" sc mapping
+aa2longalt=[
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # ala
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," NE "," CZ "," NH1"," NH2", None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD "," HE ","1HH1","2HH1","1HH2","2HH2"), # arg
+ (" N "," CA "," C "," O "," CB "," CG "," OD1"," ND2", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HD2","2HD2", None, None, None, None, None, None, None), # asn
+ (" N "," CA "," C "," O "," CB "," CG "," OD2"," OD1", None, None, None, None, None, None," H "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None, None), # asp
+ (" N "," CA "," C "," O "," CB "," SG ", None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ", None, None, None, None, None, None, None, None), # cys
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE1"," NE2", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE2","2HE2", None, None, None, None, None), # gln
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," OE2"," OE1", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ", None, None, None, None, None, None, None), # glu
+ (" N "," CA "," C "," O ", None, None, None, None, None, None, None, None, None, None," H ","1HA ","2HA ", None, None, None, None, None, None, None, None, None, None), # gly
+ (" N "," CA "," C "," O "," CB "," CG "," NE2"," CD2"," CE1"," ND1", None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1"," HE2", None, None, None, None, None, None), # his
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2"," CD1", None, None, None, None, None, None," H "," HA "," HB ","1HG2","2HG2","3HG2","1HG1","2HG1","1HD1","2HD1","3HD1", None, None), # ile
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2", None, None, None, None, None, None," H "," HA ","1HB ","2HB "," HG ","1HD1","2HD1","3HD1","1HD2","2HD2","3HD2", None, None), # leu
+ (" N "," CA "," C "," O "," CB "," CG "," CD "," CE "," NZ ", None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ","1HE ","2HE ","1HZ ","2HZ ","3HZ "), # lys
+ (" N "," CA "," C "," O "," CB "," CG "," SD "," CE ", None, None, None, None, None, None," H "," HA ","1HB ","2HB ","1HG ","2HG ","1HE ","2HE ","3HE ", None, None, None, None), # met
+ (" N "," CA "," C "," O "," CB "," CG "," CD2"," CD1"," CE2"," CE1"," CZ ", None, None, None," H "," HD2"," HE2"," HZ "," HE1"," HD1"," HA ","1HB ","2HB ", None, None, None, None), # phe
+ (" N "," CA "," C "," O "," CB "," CG "," CD ", None, None, None, None, None, None, None," HA ","1HB ","2HB ","1HG ","2HG ","1HD ","2HD ", None, None, None, None, None, None), # pro
+ (" N "," CA "," C "," O "," CB "," OG ", None, None, None, None, None, None, None, None," H "," HG "," HA ","1HB ","2HB ", None, None, None, None, None, None, None, None), # ser
+ (" N "," CA "," C "," O "," CB "," OG1"," CG2", None, None, None, None, None, None, None," H "," HG1"," HA "," HB ","1HG2","2HG2","3HG2", None, None, None, None, None, None), # thr
+ (" N "," CA "," C "," O "," CB "," CG "," CD1"," CD2"," NE1"," CE2"," CE3"," CZ2"," CZ3"," CH2"," H "," HA ","1HB ","2HB "," HD1"," HE1"," HZ2"," HH2"," HZ3"," HE3", None, None, None), # trp
+ (" N "," CA "," C "," O "," CB "," CG "," CD2"," CD1"," CE2"," CE1"," CZ "," OH ", None, None," H "," HA ","1HB ","2HB "," HD2"," HE2"," HE1"," HD1"," HH ", None, None, None, None), # tyr
+ (" N "," CA "," C "," O "," CB "," CG1"," CG2", None, None, None, None, None, None, None," H "," HA "," HB ","1HG1","2HG1","3HG1","1HG2","2HG2","3HG2", None, None, None, None), # val
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # unk
+ (" N "," CA "," C "," O "," CB ", None, None, None, None, None, None, None, None, None," H "," HA ","1HB ","2HB ","3HB ", None, None, None, None, None, None, None, None), # mask
+]
+
+aabonds=[
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # ala
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," NE "),(" CD ","1HD "),(" CD ","2HD "),(" NE "," CZ "),(" NE "," HE "),(" CZ "," NH1"),(" CZ "," NH2"),(" NH1","1HH1"),(" NH1","2HH1"),(" NH2","1HH2"),(" NH2","2HH2")) , # arg
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," OD1"),(" CG "," ND2"),(" ND2","1HD2"),(" ND2","2HD2")) , # asn
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," OD1"),(" CG "," OD2")) , # asp
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," SG "),(" CB ","1HB "),(" CB ","2HB "),(" SG "," HG ")) , # cys
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," OE1"),(" CD "," NE2"),(" NE2","1HE2"),(" NE2","2HE2")) , # gln
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," OE1"),(" CD "," OE2")) , # glu
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA ","1HA "),(" CA ","2HA "),(" C "," O ")) , # gly
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," ND1"),(" CG "," CD2"),(" ND1"," CE1"),(" CD2"," NE2"),(" CD2"," HD2"),(" CE1"," NE2"),(" CE1"," HE1"),(" NE2"," HE2")) , # his
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG1"),(" CB "," CG2"),(" CB "," HB "),(" CG1"," CD1"),(" CG1","1HG1"),(" CG1","2HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2"),(" CD1","1HD1"),(" CD1","2HD1"),(" CD1","3HD1")) , # ile
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CG "," HG "),(" CD1","1HD1"),(" CD1","2HD1"),(" CD1","3HD1"),(" CD2","1HD2"),(" CD2","2HD2"),(" CD2","3HD2")) , # leu
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD "," CE "),(" CD ","1HD "),(" CD ","2HD "),(" CE "," NZ "),(" CE ","1HE "),(" CE ","2HE "),(" NZ ","1HZ "),(" NZ ","2HZ "),(" NZ ","3HZ ")) , # lys
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," SD "),(" CG ","1HG "),(" CG ","2HG "),(" SD "," CE "),(" CE ","1HE "),(" CE ","2HE "),(" CE ","3HE ")) , # met
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," CE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," HD2"),(" CE1"," CZ "),(" CE1"," HE1"),(" CE2"," CZ "),(" CE2"," HE2"),(" CZ "," HZ ")) , # phe
+ ((" N "," CA "),(" N "," CD "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD "),(" CG ","1HG "),(" CG ","2HG "),(" CD ","1HD "),(" CD ","2HD ")) , # pro
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," OG "),(" CB ","1HB "),(" CB ","2HB "),(" OG "," HG ")) , # ser
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," OG1"),(" CB "," CG2"),(" CB "," HB "),(" OG1"," HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2")) , # thr
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," NE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," CE3"),(" NE1"," CE2"),(" NE1"," HE1"),(" CE2"," CZ2"),(" CE3"," CZ3"),(" CE3"," HE3"),(" CZ2"," CH2"),(" CZ2"," HZ2"),(" CZ3"," CH2"),(" CZ3"," HZ3"),(" CH2"," HH2")) , # trp
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG "),(" CB ","1HB "),(" CB ","2HB "),(" CG "," CD1"),(" CG "," CD2"),(" CD1"," CE1"),(" CD1"," HD1"),(" CD2"," CE2"),(" CD2"," HD2"),(" CE1"," CZ "),(" CE1"," HE1"),(" CE2"," CZ "),(" CE2"," HE2"),(" CZ "," OH "),(" OH "," HH ")) , # tyr
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB "," CG1"),(" CB "," CG2"),(" CB "," HB "),(" CG1","1HG1"),(" CG1","2HG1"),(" CG1","3HG1"),(" CG2","1HG2"),(" CG2","2HG2"),(" CG2","3HG2")), # val
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # unk
+ ((" N "," CA "),(" N "," H "),(" CA "," C "),(" CA "," CB "),(" CA "," HA "),(" C "," O "),(" CB ","1HB "),(" CB ","2HB "),(" CB ","3HB ")) , # mask
+]
+
+aa2type = [
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # ala
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CH2", "NtrR","aroC","Narg","Narg", None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol","Hpol","Hpol","Hpol"), # arg
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CNH2","ONH2","NH2O", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hpol","Hpol", None, None, None, None, None, None, None), # asn
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "COO", "OOC", "OOC", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None, None), # asp
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "SH1", None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","HS", None, None, None, None, None, None, None, None), # cys
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CNH2","ONH2","NH2O", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol", None, None, None, None, None), # gln
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "COO", "OOC", "OOC", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None), # glu
+ ("Nbb", "CAbb","CObb","OCbb", None, None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo", None, None, None, None, None, None, None, None, None, None), # gly
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "Nhis","aroC","aroC","Ntrp", None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hpol","Hapo","Hapo", None, None, None, None, None, None), # his
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "CH2", "CH3", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None), # ile
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH1", "CH3", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None), # leu
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "CH2", "CH2", "Nlys", None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hpol","Hpol","Hpol"), # lys
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH2", "S", "CH3", None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None), # met
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","aroC","aroC","aroC","aroC", None, None, None,"HNbb","Hapo","Hapo","Hapo","Haro","Haro","Haro","Haro","Haro", None, None, None, None), # phe
+ ("Npro","CAbb","CObb","OCbb","CH2", "CH2", "CH2", None, None, None, None, None, None, None,"Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None), # pro
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "OH", None, None, None, None, None, None, None, None,"HNbb","Hpol","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # ser
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "OH", "CH3", None, None, None, None, None, None, None,"HNbb","Hpol","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None), # thr
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","CH0", "Ntrp","CH0", "aroC","aroC","aroC","aroC","HNbb","Haro","Hapo","Hapo","Hapo","Hpol","Haro","Haro","Haro","Haro", None, None, None), # trp
+ ("Nbb", "CAbb","CObb","OCbb","CH2", "CH0", "aroC","aroC","aroC","aroC","CH0", "OHY", None, None,"HNbb","Haro","Haro","Haro","Haro","Hapo","Hapo","Hapo","Hpol", None, None, None, None), # tyr
+ ("Nbb", "CAbb","CObb","OCbb","CH1", "CH3", "CH3", None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo","Hapo", None, None, None, None), # val
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # unk
+ ("Nbb", "CAbb","CObb","OCbb","CH3", None, None, None, None, None, None, None, None, None,"HNbb","Hapo","Hapo","Hapo","Hapo", None, None, None, None, None, None, None, None), # mask
+]
+
+# tip atom
+aa2tip = [
+ " CB ", # ala
+ " CZ ", # arg
+ " ND2", # asn
+ " CG ", # asp
+ " SG ", # cys
+ " NE2", # gln
+ " CD ", # glu
+ " CA ", # gly
+ " NE2", # his
+ " CD1", # ile
+ " CG ", # leu
+ " NZ ", # lys
+ " SD ", # met
+ " CZ ", # phe
+ " CG ", # pro
+ " OG ", # ser
+ " OG1", # thr
+ " CH2", # trp
+ " OH ", # tyr
+ " CB ", # val
+ " CB ", # unknown (gap etc)
+ " CB " # masked
+ ]
+
+
+torsions=[
+ [ None, None, None, None ], # ala
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," NE "], [" CG "," CD "," NE "," CZ "] ], # arg
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," OD1"], None, None ], # asn
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," OD1"], None, None ], # asp
+ [ [" N "," CA "," CB "," SG "], [" CA "," CB "," SG "," HG "], None, None ], # cys
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," OE1"], None ], # gln
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," OE1"], None ], # glu
+ [ None, None, None, None ], # gly
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," ND1"], [" CD2"," CE1"," HE1"," NE2"], None ], # his (protonation handled as a pseudo-torsion)
+ [ [" N "," CA "," CB "," CG1"], [" CA "," CB "," CG1"," CD1"], None, None ], # ile
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # leu
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD "," CE "], [" CG "," CD "," CE "," NZ "] ], # lys
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," SD "], [" CB "," CG "," SD "," CE "], None ], # met
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # phe
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD "], [" CB "," CG "," CD ","1HD "], None ], # pro
+ [ [" N "," CA "," CB "," OG "], [" CA "," CB "," OG "," HG "], None, None ], # ser
+ [ [" N "," CA "," CB "," OG1"], [" CA "," CB "," OG1"," HG1"], None, None ], # thr
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], None, None ], # trp
+ [ [" N "," CA "," CB "," CG "], [" CA "," CB "," CG "," CD1"], [" CE1"," CZ "," OH "," HH "], None ], # tyr
+ [ [" N "," CA "," CB "," CG1"], None, None, None ], # val
+ [ None, None, None, None ], # unk
+ [ None, None, None, None ], # mask
+]
+
+# ideal N, CA, C initial coordinates
+init_N = torch.tensor([-0.5272, 1.3593, 0.000]).float()
+init_CA = torch.zeros_like(init_N)
+init_C = torch.tensor([1.5233, 0.000, 0.000]).float()
+INIT_CRDS = torch.full((27, 3), np.nan)
+INIT_CRDS[:3] = torch.stack((init_N, init_CA, init_C), dim=0) # (3,3)
+
+norm_N = init_N / (torch.norm(init_N, dim=-1, keepdim=True) + 1e-5)
+norm_C = init_C / (torch.norm(init_C, dim=-1, keepdim=True) + 1e-5)
+cos_ideal_NCAC = torch.sum(norm_N*norm_C, dim=-1) # cosine of ideal N-CA-C bond angle
+
+#fd Rosetta ideal coords
+#fd - uses same "frame-building" as AF2
+ideal_coords = [
+ [ # 0 ala
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+ [ # 1 arg
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3467, -0.5055, 0.9018)],
+ [' CB ', 8, (-0.5042,-0.7698,-1.2118)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3639, -0.5323, -0.8789)],
+ [' CG ', 4, (0.6396,1.3794, 0.000)],
+ ['1HG ', 5, (0.3639, -0.5139, 0.8900)],
+ ['2HG ', 5, (0.3641, -0.5140, -0.8903)],
+ [' CD ', 5, (0.5492,1.3801, 0.000)],
+ ['1HD ', 6, (0.3637, -0.5135, 0.8895)],
+ ['2HD ', 6, (0.3636, -0.5134, -0.8893)],
+ [' NE ', 6, (0.5423,1.3491, 0.000)],
+ [' NH1', 7, (0.2012,2.2965, 0.000)],
+ [' NH2', 7, (2.0824,1.0030, 0.000)],
+ [' CZ ', 7, (0.7650,1.1090, 0.000)],
+ [' HE ', 7, (0.4701,-0.8955, 0.000)],
+ ['1HH1', 7, (-0.8059,2.3776, 0.000)],
+ ['1HH2', 7, (2.5160,0.0898, 0.000)],
+ ['2HH1', 7, (0.7745,3.1277, 0.000)],
+ ['2HH2', 7, (2.6554,1.8336, 0.000)],
+ ],
+ [ # 2 asn
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3233, -0.4967, 0.9162)],
+ [' CB ', 8, (-0.5341,-0.7799,-1.1874)],
+ ['1HB ', 4, ( 0.3641, -0.5327, 0.8795)],
+ ['2HB ', 4, ( 0.3639, -0.5323, -0.8789)],
+ [' CG ', 4, (0.5778,1.3881, 0.000)],
+ [' ND2', 5, (0.5839,-1.1711, 0.000)],
+ [' OD1', 5, (0.6331,1.0620, 0.000)],
+ ['1HD2', 5, (1.5825, -1.2322, 0.000)],
+ ['2HD2', 5, (0.0323, -2.0046, 0.000)],
+ ],
+ [ # 3 asp
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3233, -0.4967, 0.9162)],
+ [' CB ', 8, (-0.5162,-0.7757,-1.2144)],
+ ['1HB ', 4, ( 0.3639, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3640, -0.5325, -0.8792)],
+ [' CG ', 4, (0.5926,1.4028, 0.000)],
+ [' OD1', 5, (0.5746,1.0629, 0.000)],
+ [' OD2', 5, (0.5738,-1.0627, 0.000)],
+ ],
+ [ # 4 cys
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3481, -0.5059, 0.9006)],
+ [' CB ', 8, (-0.5046,-0.7727,-1.2189)],
+ ['1HB ', 4, ( 0.3639, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8787)],
+ [' SG ', 4, (0.7386,1.6511, 0.000)],
+ [' HG ', 5, (0.1387,1.3221, 0.000)],
+ ],
+ [ # 5 gln
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3363, -0.5013, 0.9074)],
+ [' CB ', 8, (-0.5226,-0.7776,-1.2109)],
+ ['1HB ', 4, ( 0.3638, -0.5323, 0.8789)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8788)],
+ [' CG ', 4, (0.6225,1.3857, 0.000)],
+ ['1HG ', 5, ( 0.3531, -0.5156, 0.8931)],
+ ['2HG ', 5, ( 0.3531, -0.5156, -0.8931)],
+ [' CD ', 5, (0.5788,1.4021, 0.000)],
+ [' NE2', 6, (0.5908,-1.1895, 0.000)],
+ [' OE1', 6, (0.6347,1.0584, 0.000)],
+ ['1HE2', 6, (1.5825, -1.2525, 0.000)],
+ ['2HE2', 6, (0.0380, -2.0229, 0.000)],
+ ],
+ [ # 6 glu
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3363, -0.5013, 0.9074)],
+ [' CB ', 8, (-0.5197,-0.7737,-1.2137)],
+ ['1HB ', 4, ( 0.3638, -0.5323, 0.8789)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8788)],
+ [' CG ', 4, (0.6287,1.3862, 0.000)],
+ ['1HG ', 5, ( 0.3531, -0.5156, 0.8931)],
+ ['2HG ', 5, ( 0.3531, -0.5156, -0.8931)],
+ [' CD ', 5, (0.5850,1.3849, 0.000)],
+ [' OE1', 6, (0.5752,1.0618, 0.000)],
+ [' OE2', 6, (0.5741,-1.0635, 0.000)],
+ ],
+ [ # 7 gly
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ ['1HA ', 0, ( -0.3676, -0.5329, 0.8771)],
+ ['2HA ', 0, ( -0.3674, -0.5325, -0.8765)],
+ ],
+ [ # 8 his
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3299, -0.5180, 0.9001)],
+ [' CB ', 8, (-0.5163,-0.7809,-1.2129)],
+ ['1HB ', 4, ( 0.3640, -0.5325, 0.8793)],
+ ['2HB ', 4, ( 0.3637, -0.5321, -0.8786)],
+ [' CG ', 4, (0.6016,1.3710, 0.000)],
+ [' CD2', 5, (0.8918,-1.0184, 0.000)],
+ [' CE1', 5, (2.0299,0.8564, 0.000)],
+ [' HE1', 5, (2.8542, 1.5693, 0.000)],
+ [' HD2', 5, ( 0.6584, -2.0835, 0.000) ],
+ [' ND1', 6, (-1.8631, -1.0722, 0.000)],
+ [' NE2', 6, (-1.8625, 1.0707, 0.000)],
+ [' HE2', 6, (-1.5439, 2.0292, 0.000)],
+ ],
+ [ # 9 ile
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3405, -0.5028, 0.9044)],
+ [' CB ', 8, (-0.5140,-0.7885,-1.2184)],
+ [' HB ', 4, (0.3637, -0.4714, 0.9125)],
+ [' CG1', 4, (0.5339,1.4348,0.000)],
+ [' CG2', 4, (0.5319,-0.7693,-1.1994)],
+ ['1HG2', 4, (1.6215, -0.7588, -1.1842)],
+ ['2HG2', 4, (0.1785, -1.7986, -1.1569)],
+ ['3HG2', 4, (0.1773, -0.3016, -2.1180)],
+ [' CD1', 5, (0.6106,1.3829, 0.000)],
+ ['1HG1', 5, (0.3637, -0.5338, 0.8774)],
+ ['2HG1', 5, (0.3640, -0.5322, -0.8793)],
+ ['1HD1', 5, (1.6978, 1.3006, 0.000)],
+ ['2HD1', 5, (0.2873, 1.9236, -0.8902)],
+ ['3HD1', 5, (0.2888, 1.9224, 0.8896)],
+ ],
+ [ # 10 leu
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.525, -0.000, -0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3435, -0.5040, 0.9027)],
+ [' CB ', 8, (-0.5175,-0.7692,-1.2220)],
+ ['1HB ', 4, ( 0.3473, -0.5346, 0.8827)],
+ ['2HB ', 4, ( 0.3476, -0.5351, -0.8836)],
+ [' CG ', 4, (0.6652,1.3823, 0.000)],
+ [' CD1', 5, (0.5083,1.4353, 0.000)],
+ [' CD2', 5, (0.5079,-0.7600,1.2163)],
+ [' HG ', 5, (0.3640, -0.4825, -0.9075)],
+ ['1HD1', 5, (1.5984, 1.4353, 0.000)],
+ ['2HD1', 5, (0.1462, 1.9496, -0.8903)],
+ ['3HD1', 5, (0.1459, 1.9494, 0.8895)],
+ ['1HD2', 5, (1.5983, -0.7606, 1.2158)],
+ ['2HD2', 5, (0.1456, -0.2774, 2.1243)],
+ ['3HD2', 5, (0.1444, -1.7871, 1.1815)],
+ ],
+ [ # 11 lys
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3335, -0.5005, 0.9097)],
+ ['1HB ', 4, ( 0.3640, -0.5324, 0.8791)],
+ ['2HB ', 4, ( 0.3639, -0.5324, -0.8790)],
+ [' CB ', 8, (-0.5259,-0.7785,-1.2069)],
+ ['1HG ', 5, (0.3641, -0.5229, 0.8852)],
+ ['2HG ', 5, (0.3637, -0.5227, -0.8841)],
+ [' CG ', 4, (0.6291,1.3869, 0.000)],
+ [' CD ', 5, (0.5526,1.4174, 0.000)],
+ ['1HD ', 6, (0.3641, -0.5239, 0.8848)],
+ ['2HD ', 6, (0.3638, -0.5219, -0.8850)],
+ [' CE ', 6, (0.5544,1.4170, 0.000)],
+ [' NZ ', 7, (0.5566,1.3801, 0.000)],
+ ['1HE ', 7, (0.4199, -0.4638, 0.9482)],
+ ['2HE ', 7, (0.4202, -0.4631, -0.8172)],
+ ['1HZ ', 7, (1.6223, 1.3980, 0.0658)],
+ ['2HZ ', 7, (0.2970, 1.9326, -0.7584)],
+ ['3HZ ', 7, (0.2981, 1.9319, 0.8909)],
+ ],
+ [ # 12 met
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3303, -0.4990, 0.9108)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3641, -0.5326, -0.8795)],
+ [' CB ', 8, (-0.5331,-0.7727,-1.2048)],
+ ['1HG ', 5, (0.3637, -0.5256, 0.8823)],
+ ['2HG ', 5, (0.3638, -0.5249, -0.8831)],
+ [' CG ', 4, (0.6298,1.3858,0.000)],
+ [' SD ', 5, (0.6953,1.6645,0.000)],
+ [' CE ', 6, (0.3383,1.7581,0.000)],
+ ['1HE ', 6, (1.7054, 2.0532, -0.0063)],
+ ['2HE ', 6, (0.1906, 2.3099, -0.9072)],
+ ['3HE ', 6, (0.1917, 2.3792, 0.8720)],
+ ],
+ [ # 13 phe
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3303, -0.4990, 0.9108)],
+ ['1HB ', 4, ( 0.3635, -0.5318, 0.8781)],
+ ['2HB ', 4, ( 0.3641, -0.5326, -0.8795)],
+ [' CB ', 8, (-0.5150,-0.7729,-1.2156)],
+ [' CG ', 4, (0.6060,1.3746, 0.000)],
+ [' CD1', 5, (0.7078,1.1928, 0.000)],
+ [' CD2', 5, (0.7084,-1.1920, 0.000)],
+ [' CE1', 5, (2.0900,1.1940, 0.000)],
+ [' CE2', 5, (2.0897,-1.1939, 0.000)],
+ [' CZ ', 5, (2.7809, 0.000, 0.000)],
+ [' HD1', 5, (0.1613, 2.1362, 0.000)],
+ [' HD2', 5, (0.1621, -2.1360, 0.000)],
+ [' HE1', 5, (2.6335, 2.1384, 0.000)],
+ [' HE2', 5, (2.6344, -2.1378, 0.000)],
+ [' HZ ', 5, (3.8700, 0.000, 0.000)],
+ ],
+ [ # 14 pro
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' HA ', 0, (-0.3868, -0.5380, 0.8781)],
+ ['1HB ', 4, ( 0.3762, -0.5355, 0.8842)],
+ ['2HB ', 4, ( 0.3762, -0.5355, -0.8842)],
+ [' CB ', 8, (-0.5649,-0.5888,-1.2966)],
+ [' CG ', 4, (0.3657,1.4451,0.0000)],
+ [' CD ', 5, (0.3744,1.4582, 0.0)],
+ ['1HG ', 5, (0.3798, -0.5348, 0.8830)],
+ ['2HG ', 5, (0.3798, -0.5348, -0.8830)],
+ ['1HD ', 6, (0.3798, -0.5348, 0.8830)],
+ ['2HD ', 6, (0.3798, -0.5348, -0.8830)],
+ ],
+ [ # 15 ser
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3425, -0.5041, 0.9048)],
+ ['1HB ', 4, ( 0.3637, -0.5321, 0.8786)],
+ ['2HB ', 4, ( 0.3636, -0.5319, -0.8782)],
+ [' CB ', 8, (-0.5146,-0.7595,-1.2073)],
+ [' OG ', 4, (0.5021,1.3081, 0.000)],
+ [' HG ', 5, (0.2647, 0.9230, 0.000)],
+ ],
+ [ # 16 thr
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3364, -0.5015, 0.9078)],
+ [' HB ', 4, ( 0.3638, -0.5006, 0.8971)],
+ ['1HG2', 4, ( 1.6231, -0.7142, -1.2097)],
+ ['2HG2', 4, ( 0.1792, -1.7546, -1.2237)],
+ ['3HG2', 4, ( 0.1808, -0.2222, -2.1269)],
+ [' CB ', 8, (-0.5172,-0.7952,-1.2130)],
+ [' CG2', 4, (0.5334,-0.7239,-1.2267)],
+ [' OG1', 4, (0.4804,1.3506,0.000)],
+ [' HG1', 5, (0.3194, 0.9056, 0.000)],
+ ],
+ [ # 17 trp
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3436, -0.5042, 0.9031)],
+ ['1HB ', 4, ( 0.3639, -0.5323, 0.8790)],
+ ['2HB ', 4, ( 0.3638, -0.5322, -0.8787)],
+ [' CB ', 8, (-0.5136,-0.7712,-1.2173)],
+ [' CG ', 4, (0.5984,1.3741, 0.000)],
+ [' CD1', 5, (0.8151,1.0921, 0.000)],
+ [' CD2', 5, (0.8753,-1.1538, 0.000)],
+ [' CE2', 5, (2.1865,-0.6707, 0.000)],
+ [' CE3', 5, (0.6541,-2.5366, 0.000)],
+ [' NE1', 5, (2.1309,0.7003, 0.000)],
+ [' CH2', 5, (3.0315,-2.8930, 0.000)],
+ [' CZ2', 5, (3.2813,-1.5205, 0.000)],
+ [' CZ3', 5, (1.7521,-3.3888, 0.000)],
+ [' HD1', 5, (0.4722, 2.1252, 0.000)],
+ [' HE1', 5, ( 2.9291, 1.3191, 0.000)],
+ [' HE3', 5, (-0.3597, -2.9356, 0.000)],
+ [' HZ2', 5, (4.3053, -1.1462, 0.000)],
+ [' HZ3', 5, ( 1.5712, -4.4640, 0.000)],
+ [' HH2', 5, ( 3.8700, -3.5898, 0.000)],
+ ],
+ [ # 18 tyr
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3305, -0.4992, 0.9112)],
+ ['1HB ', 4, ( 0.3642, -0.5327, 0.8797)],
+ ['2HB ', 4, ( 0.3637, -0.5321, -0.8785)],
+ [' CB ', 8, (-0.5305,-0.7799,-1.2051)],
+ [' CG ', 4, (0.6104,1.3840, 0.000)],
+ [' CD1', 5, (0.6936,1.2013, 0.000)],
+ [' CD2', 5, (0.6934,-1.2011, 0.000)],
+ [' CE1', 5, (2.0751,1.2013, 0.000)],
+ [' CE2', 5, (2.0748,-1.2011, 0.000)],
+ [' OH ', 5, (4.1408, 0.000, 0.000)],
+ [' CZ ', 5, (2.7648, 0.000, 0.000)],
+ [' HD1', 5, (0.1485, 2.1455, 0.000)],
+ [' HD2', 5, (0.1484, -2.1451, 0.000)],
+ [' HE1', 5, (2.6200, 2.1450, 0.000)],
+ [' HE2', 5, (2.6199, -2.1453, 0.000)],
+ [' HH ', 6, (0.3190, 0.9057, 0.000)],
+ ],
+ [ # 19 val
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3497, -0.5068, 0.9002)],
+ [' CB ', 8, (-0.5105,-0.7712,-1.2317)],
+ [' CG1', 4, (0.5326,1.4252, 0.000)],
+ [' CG2', 4, (0.5177,-0.7693,1.2057)],
+ [' HB ', 4, (0.3541, -0.4754, -0.9148)],
+ ['1HG1', 4, (1.6228, 1.4063, 0.000)],
+ ['2HG1', 4, (0.1790, 1.9457, -0.8898)],
+ ['3HG1', 4, (0.1798, 1.9453, 0.8903)],
+ ['1HG2', 4, (1.6073, -0.7659, 1.1989)],
+ ['2HG2', 4, (0.1586, -0.2971, 2.1203)],
+ ['3HG2', 4, (0.1582, -1.7976, 1.1631)],
+ ],
+ [ # 20 unk
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+ [ # 21 mask
+ [' N ', 0, (-0.5272, 1.3593, 0.000)],
+ [' CA ', 0, (0.000, 0.000, 0.000)],
+ [' C ', 0, (1.5233, 0.000, 0.000)],
+ [' O ', 3, (0.6303, 1.0574, 0.000)],
+ [' H ', 2, (0.4920,-0.8821, 0.0000)],
+ [' HA ', 0, (-0.3341, -0.4928, 0.9132)],
+ [' CB ', 8, (-0.5289,-0.7734,-1.1991)],
+ ['1HB ', 8, (-0.1265, -1.7863, -1.1851)],
+ ['2HB ', 8, (-1.6173, -0.8147, -1.1541)],
+ ['3HB ', 8, (-0.2229, -0.2744, -2.1172)],
+ ],
+]
diff --git a/utils/model/coords6d.py b/utils/model/coords6d.py
new file mode 100644
index 0000000000000000000000000000000000000000..9f10d593851674d083b9a906c84ee2762839f1d2
--- /dev/null
+++ b/utils/model/coords6d.py
@@ -0,0 +1,80 @@
+import numpy as np
+import scipy
+import scipy.spatial
+
+# calculate dihedral angles defined by 4 sets of points
+def get_dihedrals(a, b, c, d):
+
+ b0 = -1.0*(b - a)
+ b1 = c - b
+ b2 = d - c
+
+ b1 /= np.linalg.norm(b1, axis=-1)[:,None]
+
+ v = b0 - np.sum(b0*b1, axis=-1)[:,None]*b1
+ w = b2 - np.sum(b2*b1, axis=-1)[:,None]*b1
+
+ x = np.sum(v*w, axis=-1)
+ y = np.sum(np.cross(b1, v)*w, axis=-1)
+
+ return np.arctan2(y, x)
+
+# calculate planar angles defined by 3 sets of points
+def get_angles(a, b, c):
+
+ v = a - b
+ v /= np.linalg.norm(v, axis=-1)[:,None]
+
+ w = c - b
+ w /= np.linalg.norm(w, axis=-1)[:,None]
+
+ x = np.sum(v*w, axis=1)
+
+ #return np.arccos(x)
+ return np.arccos(np.clip(x, -1.0, 1.0))
+
+# get 6d coordinates from x,y,z coords of N,Ca,C atoms
+def get_coords6d(xyz, dmax):
+
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[0]
+ Ca = xyz[1]
+ C = xyz[2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = np.cross(b, c)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # fast neighbors search to collect all
+ # Cb-Cb pairs within dmax
+ kdCb = scipy.spatial.cKDTree(Cb)
+ indices = kdCb.query_ball_tree(kdCb, dmax)
+
+ # indices of contacting residues
+ idx = np.array([[i,j] for i in range(len(indices)) for j in indices[i] if i != j]).T
+ idx0 = idx[0]
+ idx1 = idx[1]
+
+ # Cb-Cb distance matrix
+ dist6d = np.full((nres, nres),999.9, dtype=np.float32)
+ dist6d[idx0,idx1] = np.linalg.norm(Cb[idx1]-Cb[idx0], axis=-1)
+
+ # matrix of Ca-Cb-Cb-Ca dihedrals
+ omega6d = np.zeros((nres, nres), dtype=np.float32)
+ omega6d[idx0,idx1] = get_dihedrals(Ca[idx0], Cb[idx0], Cb[idx1], Ca[idx1])
+
+ # matrix of polar coord theta
+ theta6d = np.zeros((nres, nres), dtype=np.float32)
+ theta6d[idx0,idx1] = get_dihedrals(N[idx0], Ca[idx0], Cb[idx0], Cb[idx1])
+
+ # matrix of polar coord phi
+ phi6d = np.zeros((nres, nres), dtype=np.float32)
+ phi6d[idx0,idx1] = get_angles(Ca[idx0], Cb[idx0], Cb[idx1])
+
+ mask = np.zeros((nres, nres), dtype=np.float32)
+ mask[idx0, idx1] = 1.0
+ return dist6d, omega6d, theta6d, phi6d, mask
diff --git a/utils/model/diffusion.py b/utils/model/diffusion.py
new file mode 100644
index 0000000000000000000000000000000000000000..8699f7d8acb3c30d0fed7991f29ab0f27b370861
--- /dev/null
+++ b/utils/model/diffusion.py
@@ -0,0 +1,217 @@
+import enum
+import math
+
+import numpy as np
+import torch as th
+
+
+##########################################################################################
+
+# DIFFUSION CODE BASE FOR PROTEIN SEQUENCE DIFFUSION WAS ADAPTED FROM LM-DIFFUSION #
+
+ # (https://github.com/XiangLi1999/Diffusion-LM) #
+
+##########################################################################################
+
+class GaussianDiffusion_SEQDIFF:
+ """
+ T = number of timesteps to set up diffuser with
+
+ schedule = type of noise schedule to use linear, cosine, gaussian
+
+ noise = type of ditribution to sample from; DEFAULT - normal_gaussian
+
+ """
+
+ def __init__(self,
+ T=1000,
+ schedule='sqrt',
+ sample_distribution='normal',
+ sample_distribution_gmm_means=[-1.0, 1.0],
+ sample_distribution_gmm_variances=[1.0, 1.0],
+ F=1,
+ ):
+
+ # Use float64 for accuracy.
+ betas = np.array(get_named_beta_schedule(schedule, T), dtype=np.float64)
+ self.betas = betas
+ assert len(betas.shape) == 1, "betas must be 1-D"
+ assert (betas > 0).all() and (betas <= 1).all()
+
+ self.num_timesteps = int(betas.shape[0])
+ self.F = F
+
+ alphas = 1.0 - betas
+ self.alphas_cumprod = np.cumprod(alphas, axis=0)
+ self.alphas_cumprod_prev = np.append(1.0, self.alphas_cumprod[:-1])
+ self.alphas_cumprod_next = np.append(self.alphas_cumprod[1:], 0.0)
+ assert self.alphas_cumprod_prev.shape == (self.num_timesteps,)
+
+ # calculations for posterior q(x_{t-1} | x_t, x_0)
+ self.posterior_variance = (betas * (1.0 - self.alphas_cumprod_prev) / (1.0 - self.alphas_cumprod))
+ # log calculation clipped because the posterior variance is 0 at the
+ # beginning of the diffusion chain.
+ self.posterior_log_variance_clipped = np.log(np.append(self.posterior_variance[1], self.posterior_variance[1:]))
+ self.posterior_mean_coef1 = (betas * np.sqrt(self.alphas_cumprod_prev) / (1.0 - self.alphas_cumprod))
+ self.posterior_mean_coef2 = ((1.0 - self.alphas_cumprod_prev) * np.sqrt(alphas) / (1.0 - self.alphas_cumprod))
+
+ # calculations for diffusion q(x_t | x_{t-1}) and others
+ self.sqrt_alphas_cumprod = np.sqrt(self.alphas_cumprod)
+ self.sqrt_one_minus_alphas_cumprod = np.sqrt(1.0 - self.alphas_cumprod)
+ self.log_one_minus_alphas_cumprod = np.log(1.0 - self.alphas_cumprod)
+ self.sqrt_recip_alphas_cumprod = np.sqrt(1.0 / self.alphas_cumprod)
+
+ # sample_distribution_params
+ self.sample_distribution = sample_distribution
+ self.sample_distribution_gmm_means = [float(mean) for mean in sample_distribution_gmm_means]
+ self.sample_distribution_gmm_variances = [float(variance) for variance in sample_distribution_gmm_variances]
+
+ if self.sample_distribution == 'normal':
+ self.noise_function = th.randn_like
+ else:
+ self.noise_function = self.randnmixture_like
+
+
+ def q_mean_variance(self, x_start, t):
+ """
+ Get the distribution q(x_t | x_0).
+ :param x_start: the [N x C x ...] tensor of noiseless inputs.
+ :param t: the number of diffusion steps (minus 1). Here, 0 means one step.
+ :return: A tuple (mean, variance, log_variance), all of x_start's shape.
+ """
+ mean = (
+ _extract(self.sqrt_alphas_cumprod, t, x_start.shape) * x_start
+ )
+ variance = _extract(1.0 - self.alphas_cumprod, t, x_start.shape)
+ log_variance = _extract(
+ self.log_one_minus_alphas_cumprod, t, x_start.shape
+ )
+ return mean, variance, log_variance
+
+ def q_sample(self, x_start, t, mask=None, DEVICE=None):
+ """
+ Diffuse the data for a given number of diffusion steps.
+ In other words, sample from q(x_t | x_0).
+ :param x_start: the initial data batch.
+ :param t: the number of diffusion steps (minus 1). Here, 0 means one step.
+ :param noise: if specified, the split-out normal noise.
+ :return: A noisy version of x_start.
+ """
+
+ # noise_function is determined in init depending on type of noise specified
+ noise = self.noise_function(x_start)*(self.F**2)
+ if DEVICE != None:
+ noise = noise.to(DEVICE)
+
+ assert noise.shape == x_start.shape
+ x_sample = (
+ _extract(self.sqrt_alphas_cumprod, t, x_start.shape) * x_start
+ + _extract(self.sqrt_one_minus_alphas_cumprod, t, x_start.shape)
+ * noise)
+
+ if mask is not None:
+ x_sample[mask]=x_start[mask]
+
+ return x_sample
+
+
+ def q_posterior_mean_variance(self, x_start, x_t, t):
+ """
+ Compute the mean and variance of the diffusion posterior:
+ q(x_{t-1} | x_t, x_0)
+ """
+ assert x_start.shape == x_t.shape
+
+ posterior_mean = (_extract(self.posterior_mean_coef1, t, x_t.shape) * x_start
+ + _extract(self.posterior_mean_coef2, t, x_t.shape) * x_t)
+
+ posterior_variance = _extract(self.posterior_variance, t, x_t.shape)
+
+ posterior_log_variance_clipped = _extract(self.posterior_log_variance_clipped, t, x_t.shape)
+
+ assert (
+ posterior_mean.shape[0]
+ == posterior_variance.shape[0]
+ == posterior_log_variance_clipped.shape[0]
+ == x_start.shape[0]
+ )
+ return posterior_mean, posterior_variance, posterior_log_variance_clipped
+
+
+ def randnmixture_like(self, tensor_like, number_normal=3, weights_normal=None):
+
+ if self.sample_distribution_gmm_means and self.sample_distribution_gmm_variances:
+ assert len(self.sample_distribution_gmm_means) == len(self.sample_distribution_gmm_variances)
+
+ if not weights_normal:
+ mix = th.distributions.Categorical(th.ones(len(self.sample_distribution_gmm_means))) #number_normal
+ else:
+ assert len(weights_normal) == number_normal
+ mix = th.distributions.Categorical(weights_normal)
+ #comp = torch.distributions.Normal(torch.randn(number_normal), torch.rand(number_normal))
+ comp = th.distributions.Normal(th.tensor(self.sample_distribution_gmm_means), th.tensor(self.sample_distribution_gmm_variances))
+ #comp = torch.distributions.Normal([-3, 3], [1, 1])
+ #comp = torch.distributions.Normal([-3, 0, 3], [1, 1, 1])
+ #comp = torch.distributions.Normal([-3, 0, 3], [1, 1, 1])
+ gmm = th.distributions.mixture_same_family.MixtureSameFamily(mix, comp)
+ return th.tensor([gmm.sample() for _ in range(np.prod(tensor_like.shape))]).reshape(tensor_like.shape)
+
+
+
+def get_named_beta_schedule(schedule_name, num_diffusion_timesteps):
+ """
+ Get a pre-defined beta schedule for the given name.
+ The beta schedule library consists of beta schedules which remain similar
+ in the limit of num_diffusion_timesteps.
+ Beta schedules may be added, but should not be removed or changed once
+ they are committed to maintain backwards compatibility.
+ """
+ if schedule_name == "linear":
+ # Linear schedule from Ho et al, extended to work for any number of
+ # diffusion steps.
+ scale = 1000 / num_diffusion_timesteps
+ beta_start = scale * 0.0001
+ beta_end = scale * 0.02
+ return np.linspace(beta_start, beta_end, num_diffusion_timesteps, dtype=np.float64)
+
+ elif schedule_name == "cosine":
+ return betas_for_alpha_bar(num_diffusion_timesteps, lambda t: math.cos((t + 0.008) / 1.008 * math.pi / 2) ** 2,)
+
+ elif schedule_name == 'sqrt':
+ return betas_for_alpha_bar(num_diffusion_timesteps, lambda t: 1-np.sqrt(t + 0.0001),)
+
+ else:
+ raise NotImplementedError(f"unknown beta schedule: {schedule_name}")
+
+def betas_for_alpha_bar(num_diffusion_timesteps, alpha_bar, max_beta=0.999):
+ """
+ Create a beta schedule that discretizes the given alpha_t_bar function,
+ which defines the cumulative product of (1-beta) over time from t = [0,1].
+ :param num_diffusion_timesteps: the number of betas to produce.
+ :param alpha_bar: a lambda that takes an argument t from 0 to 1 and
+ produces the cumulative product of (1-beta) up to that
+ part of the diffusion process.
+ :param max_beta: the maximum beta to use; use values lower than 1 to
+ prevent singularities.
+ """
+ betas = []
+ for i in range(num_diffusion_timesteps):
+ t1 = i / num_diffusion_timesteps
+ t2 = (i + 1) / num_diffusion_timesteps
+ betas.append(min(1 - alpha_bar(t2) / alpha_bar(t1), max_beta))
+ return np.array(betas)
+
+
+def _extract(arr, timesteps, broadcast_shape):
+ """
+ Extract values from a 1-D numpy array for a batch of indices.
+ :param arr: the 1-D numpy array.
+ :param timesteps: a tensor of indices into the array to extract.
+ :param broadcast_shape: a larger shape of K dimensions with the batch
+ dimension equal to the length of timesteps.
+ :return: a tensor of shape [batch_size, 1, ...] where the shape has K dims.
+ """
+ res = th.from_numpy(arr).to(device=timesteps.device)[timesteps].float()
+ while len(res.shape) < len(broadcast_shape):
+ res = res[..., None]
+ return res.expand(broadcast_shape)
diff --git a/utils/model/kinematics.py b/utils/model/kinematics.py
new file mode 100644
index 0000000000000000000000000000000000000000..392dd8ccc124a6c20ed2b4a3d654db331a920d88
--- /dev/null
+++ b/utils/model/kinematics.py
@@ -0,0 +1,307 @@
+import numpy as np
+import torch
+from chemical import INIT_CRDS
+
+PARAMS = {
+ "DMIN" : 2.0,
+ "DMAX" : 20.0,
+ "DBINS" : 36,
+ "ABINS" : 36,
+}
+
+# ============================================================
+def get_pair_dist(a, b):
+ """calculate pair distances between two sets of points
+
+ Parameters
+ ----------
+ a,b : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of two sets of atoms
+ Returns
+ -------
+ dist : pytorch tensor of shape [batch,nres,nres]
+ stores paitwise distances between atoms in a and b
+ """
+
+ dist = torch.cdist(a, b, p=2)
+ return dist
+
+# ============================================================
+def get_ang(a, b, c):
+ """calculate planar angles for all consecutive triples (a[i],b[i],c[i])
+ from Cartesian coordinates of three sets of atoms a,b,c
+
+ Parameters
+ ----------
+ a,b,c : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of three sets of atoms
+ Returns
+ -------
+ ang : pytorch tensor of shape [batch,nres]
+ stores resulting planar angles
+ """
+ v = a - b
+ w = c - b
+ v /= torch.norm(v, dim=-1, keepdim=True)
+ w /= torch.norm(w, dim=-1, keepdim=True)
+ vw = torch.sum(v*w, dim=-1)
+
+ return torch.acos(vw)
+
+# ============================================================
+def get_dih(a, b, c, d):
+ """calculate dihedral angles for all consecutive quadruples (a[i],b[i],c[i],d[i])
+ given Cartesian coordinates of four sets of atoms a,b,c,d
+
+ Parameters
+ ----------
+ a,b,c,d : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of four sets of atoms
+ Returns
+ -------
+ dih : pytorch tensor of shape [batch,nres]
+ stores resulting dihedrals
+ """
+ b0 = a - b
+ b1 = c - b
+ b2 = d - c
+
+ b1 /= torch.norm(b1, dim=-1, keepdim=True)
+
+ v = b0 - torch.sum(b0*b1, dim=-1, keepdim=True)*b1
+ w = b2 - torch.sum(b2*b1, dim=-1, keepdim=True)*b1
+
+ x = torch.sum(v*w, dim=-1)
+ y = torch.sum(torch.cross(b1,v,dim=-1)*w, dim=-1)
+
+ return torch.atan2(y, x)
+
+
+# ============================================================
+def xyz_to_c6d(xyz, params=PARAMS):
+ """convert cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz : pytorch tensor of shape [batch,nres,3,3]
+ stores Cartesian coordinates of backbone N,Ca,C atoms
+ Returns
+ -------
+ c6d : pytorch tensor of shape [batch,nres,nres,4]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+
+ batch = xyz.shape[0]
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # 6d coordinates order: (dist,omega,theta,phi)
+ c6d = torch.zeros([batch,nres,nres,4],dtype=xyz.dtype,device=xyz.device)
+
+ dist = get_pair_dist(Cb,Cb)
+ dist[torch.isnan(dist)] = 999.9
+ c6d[...,0] = dist + 999.9*torch.eye(nres,device=xyz.device)[None,...]
+ b,i,j = torch.where(c6d[...,0]=params['DMAX']] = 999.9
+
+ mask = torch.zeros((batch, nres,nres), dtype=xyz.dtype, device=xyz.device)
+ mask[b,i,j] = 1.0
+ return c6d, mask
+
+def xyz_to_t2d(xyz_t, params=PARAMS):
+ """convert template cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz_t : pytorch tensor of shape [batch,templ,nres,3,3]
+ stores Cartesian coordinates of template backbone N,Ca,C atoms
+
+ Returns
+ -------
+ t2d : pytorch tensor of shape [batch,nres,nres,37+6+3]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+ B, T, L = xyz_t.shape[:3]
+ c6d, mask = xyz_to_c6d(xyz_t[:,:,:,:3].view(B*T,L,3,3), params=params)
+ c6d = c6d.view(B, T, L, L, 4)
+ mask = mask.view(B, T, L, L, 1)
+ #
+ # dist to one-hot encoded
+ dist = dist_to_onehot(c6d[...,0], params)
+ orien = torch.cat((torch.sin(c6d[...,1:]), torch.cos(c6d[...,1:])), dim=-1)*mask # (B, T, L, L, 6)
+ #
+ mask = ~torch.isnan(c6d[:,:,:,:,0]) # (B, T, L, L)
+ t2d = torch.cat((dist, orien, mask.unsqueeze(-1)), dim=-1)
+ t2d[torch.isnan(t2d)] = 0.0
+ return t2d
+
+def xyz_to_chi1(xyz_t):
+ '''convert template cartesian coordinates into chi1 angles
+
+ Parameters
+ ----------
+ xyz_t: pytorch tensor of shape [batch, templ, nres, 14, 3]
+ stores Cartesian coordinates of template atoms. For missing atoms, it should be NaN
+
+ Returns
+ -------
+ chi1 : pytorch tensor of shape [batch, templ, nres, 2]
+ stores cos and sin chi1 angle
+ '''
+ B, T, L = xyz_t.shape[:3]
+ xyz_t = xyz_t.reshape(B*T, L, 14, 3)
+
+ # chi1 angle: N, CA, CB, CG
+ chi1 = get_dih(xyz_t[:,:,0], xyz_t[:,:,1], xyz_t[:,:,4], xyz_t[:,:,5]) # (B*T, L)
+ cos_chi1 = torch.cos(chi1)
+ sin_chi1 = torch.sin(chi1)
+ mask_chi1 = ~torch.isnan(chi1)
+ chi1 = torch.stack((cos_chi1, sin_chi1, mask_chi1), dim=-1) # (B*T, L, 3)
+ chi1[torch.isnan(chi1)] = 0.0
+ chi1 = chi1.reshape(B, T, L, 3)
+ return chi1
+
+def xyz_to_bbtor(xyz, params=PARAMS):
+ batch = xyz.shape[0]
+ nres = xyz.shape[1]
+
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ next_N = torch.roll(N, -1, dims=1)
+ prev_C = torch.roll(C, 1, dims=1)
+ phi = get_dih(prev_C, N, Ca, C)
+ psi = get_dih(N, Ca, C, next_N)
+ #
+ phi[:,0] = 0.0
+ psi[:,-1] = 0.0
+ #
+ astep = 2.0*np.pi / params['ABINS']
+ phi_bin = torch.round((phi+np.pi-astep/2)/astep)
+ psi_bin = torch.round((psi+np.pi-astep/2)/astep)
+ return torch.stack([phi_bin, psi_bin], axis=-1).long()
+
+# ============================================================
+def dist_to_onehot(dist, params=PARAMS):
+ dist[torch.isnan(dist)] = 999.9
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=dist.dtype,device=dist.device)
+ db = torch.bucketize(dist.contiguous(),dbins).long()
+ dist = torch.nn.functional.one_hot(db, num_classes=params['DBINS']+1).float()
+ return dist
+
+def c6d_to_bins(c6d,params=PARAMS):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=c6d.dtype,device=c6d.device)
+ ab360 = torch.linspace(-np.pi+astep, np.pi, params['ABINS'],dtype=c6d.dtype,device=c6d.device)
+ ab180 = torch.linspace(astep, np.pi, params['ABINS']//2,dtype=c6d.dtype,device=c6d.device)
+
+ db = torch.bucketize(c6d[...,0].contiguous(),dbins)
+ ob = torch.bucketize(c6d[...,1].contiguous(),ab360)
+ tb = torch.bucketize(c6d[...,2].contiguous(),ab360)
+ pb = torch.bucketize(c6d[...,3].contiguous(),ab180)
+
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).to(torch.uint8)
+
+
+# ============================================================
+def dist_to_bins(dist,params=PARAMS):
+ """bin 2d distance maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ db = torch.round((dist-params['DMIN']-dstep/2)/dstep)
+
+ db[db<0] = 0
+ db[db>params['DBINS']] = params['DBINS']
+
+ return db.long()
+
+
+# ============================================================
+def c6d_to_bins2(c6d, same_chain, negative=False, params=PARAMS):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ db = torch.round((c6d[...,0]-params['DMIN']-dstep/2)/dstep)
+ ob = torch.round((c6d[...,1]+np.pi-astep/2)/astep)
+ tb = torch.round((c6d[...,2]+np.pi-astep/2)/astep)
+ pb = torch.round((c6d[...,3]-astep/2)/astep)
+
+ # put all dparams['DBINS']] = params['DBINS']
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ if negative:
+ db = torch.where(same_chain.bool(), db.long(), params['DBINS'])
+ ob = torch.where(same_chain.bool(), ob.long(), params['ABINS'])
+ tb = torch.where(same_chain.bool(), tb.long(), params['ABINS'])
+ pb = torch.where(same_chain.bool(), pb.long(), params['ABINS']//2)
+
+ return torch.stack([db,ob,tb,pb],axis=-1).long()
+
+def get_init_xyz(xyz_t):
+ # input: xyz_t (B, T, L, 14, 3)
+ # ouput: xyz (B, T, L, 14, 3)
+ B, T, L = xyz_t.shape[:3]
+ init = INIT_CRDS.to(xyz_t.device).reshape(1,1,1,27,3).repeat(B,T,L,1,1)
+ if torch.isnan(xyz_t).all():
+ return init
+
+ mask = torch.isnan(xyz_t[:,:,:,:3]).any(dim=-1).any(dim=-1) # (B, T, L)
+ #
+ center_CA = ((~mask[:,:,:,None]) * torch.nan_to_num(xyz_t[:,:,:,1,:])).sum(dim=2) / ((~mask[:,:,:,None]).sum(dim=2)+1e-4) # (B, T, 3)
+ xyz_t = xyz_t - center_CA.view(B,T,1,1,3)
+ #
+ idx_s = list()
+ for i_b in range(B):
+ for i_T in range(T):
+ if mask[i_b, i_T].all():
+ continue
+ exist_in_templ = torch.where(~mask[i_b, i_T])[0] # (L_sub)
+ seqmap = (torch.arange(L, device=xyz_t.device)[:,None] - exist_in_templ[None,:]).abs() # (L, L_sub)
+ seqmap = torch.argmin(seqmap, dim=-1) # (L)
+ idx = torch.gather(exist_in_templ, -1, seqmap) # (L)
+ offset_CA = torch.gather(xyz_t[i_b, i_T, :, 1, :], 0, idx.reshape(L,1).expand(-1,3))
+ init[i_b,i_T] += offset_CA.reshape(L,1,3)
+ #
+ xyz = torch.where(mask.view(B, T, L, 1, 1), init, xyz_t)
+ return xyz
diff --git a/utils/model/parsers.py b/utils/model/parsers.py
new file mode 100644
index 0000000000000000000000000000000000000000..001a086b66cb60df21085a0295874ecf3606a1f2
--- /dev/null
+++ b/utils/model/parsers.py
@@ -0,0 +1,219 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+import gzip
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+# read A3M and convert letters into
+# integers in the 0..20 range,
+# also keep track of insertions
+def parse_a3m(filename):
+
+ msa = []
+ ins = []
+
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+
+ #print(filename)
+
+ if filename.split('.')[-1] == 'gz':
+ fp = gzip.open(filename, 'rt')
+ else:
+ fp = open(filename, 'r')
+
+ # read file line by line
+ for line in fp:
+
+ # skip labels
+ if line[0] == '>':
+ continue
+
+ # remove right whitespaces
+ line = line.rstrip()
+
+ if len(line) == 0:
+ continue
+
+ # remove lowercase letters and append to MSA
+ msa.append(line.translate(table))
+
+ # sequence length
+ L = len(msa[-1])
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in line])
+ i = np.zeros((L))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+
+ # append to the matrix of insetions
+ i[pos] = num
+
+ ins.append(i)
+ if len(msa) == 10000:
+ break
+
+ # convert letters into numbers
+ alphabet = np.array(list("ARNDCQEGHILKMFPSTWYV-"), dtype='|S1').view(np.uint8)
+ msa = np.array([list(s) for s in msa], dtype='|S1').view(np.uint8)
+ for i in range(alphabet.shape[0]):
+ msa[msa == alphabet[i]] = i
+
+ # treat all unknown characters as gaps
+ msa[msa > 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return msa,ins
+
+
+# read and extract xyz coords of N,Ca,C atoms
+# from a PDB file
+def parse_pdb(filename):
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines)
+
+#'''
+def parse_pdb_lines(lines):
+
+ # indices of residues observed in the structure
+ idx_s = [int(l[22:26]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(idx_s), 14, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ resNo, atom, aa = int(l[22:26]), l[12:16], l[17:20]
+ idx = idx_s.index(resNo)
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm == atom:
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ return xyz,mask,np.array(idx_s)
+#'''
+
+'''
+def parse_pdb_lines(lines):
+
+ # indices of residues observed in the structure
+ #idx_s = [int(l[22:26]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ idx_s = [int(r[0]) for r in res]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(idx_s), 14, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ resNo, atom, aa = int(l[22:26]), l[12:16], l[17:20]
+ idx = idx_s.index(resNo)
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm == atom:
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ return xyz,mask,np.array(idx_s), np.array(seq)
+'''
+
+
+def parse_templates(item, params):
+
+ # init FFindexDB of templates
+ ### and extract template IDs
+ ### present in the DB
+ ffdb = FFindexDB(read_index(params['FFDB']+'_pdb.ffindex'),
+ read_data(params['FFDB']+'_pdb.ffdata'))
+ #ffids = set([i.name for i in ffdb.index])
+
+ # process tabulated hhsearch output to get
+ # matched positions and positional scores
+ infile = params['DIR']+'/hhr/'+item[-2:]+'/'+item+'.atab'
+ hits = []
+ for l in open(infile, "r").readlines():
+ if l[0]=='>':
+ key = l[1:].split()[0]
+ hits.append([key,[],[]])
+ elif "score" in l or "dssp" in l:
+ continue
+ else:
+ hi = l.split()[:5]+[0.0,0.0,0.0]
+ hits[-1][1].append([int(hi[0]),int(hi[1])])
+ hits[-1][2].append([float(hi[2]),float(hi[3]),float(hi[4])])
+
+ # get per-hit statistics from an .hhr file
+ # (!!! assume that .hhr and .atab have the same hits !!!)
+ # [Probab, E-value, Score, Aligned_cols,
+ # Identities, Similarity, Sum_probs, Template_Neff]
+ lines = open(infile[:-4]+'hhr', "r").readlines()
+ pos = [i+1 for i,l in enumerate(lines) if l[0]=='>']
+ for i,posi in enumerate(pos):
+ hits[i].append([float(s) for s in re.sub('[=%]',' ',lines[posi]).split()[1::2]])
+
+ # parse templates from FFDB
+ for hi in hits:
+ #if hi[0] not in ffids:
+ # continue
+ entry = get_entry_by_name(hi[0], ffdb.index)
+ if entry == None:
+ continue
+ data = read_entry_lines(entry, ffdb.data)
+ hi += list(parse_pdb_lines(data))
+
+ # process hits
+ counter = 0
+ xyz,qmap,mask,f0d,f1d,ids = [],[],[],[],[],[]
+ for data in hits:
+ if len(data)<7:
+ continue
+
+ qi,ti = np.array(data[1]).T
+ _,sel1,sel2 = np.intersect1d(ti, data[6], return_indices=True)
+ ncol = sel1.shape[0]
+ if ncol < 10:
+ continue
+
+ ids.append(data[0])
+ f0d.append(data[3])
+ f1d.append(np.array(data[2])[sel1])
+ xyz.append(data[4][sel2])
+ mask.append(data[5][sel2])
+ qmap.append(np.stack([qi[sel1]-1,[counter]*ncol],axis=-1))
+ counter += 1
+
+ xyz = np.vstack(xyz).astype(np.float32)
+ mask = np.vstack(mask).astype(np.bool)
+ qmap = np.vstack(qmap).astype(np.long)
+ f0d = np.vstack(f0d).astype(np.float32)
+ f1d = np.vstack(f1d).astype(np.float32)
+ ids = ids
+
+ return xyz,mask,qmap,f0d,f1d,ids
diff --git a/utils/model/scoring.py b/utils/model/scoring.py
new file mode 100644
index 0000000000000000000000000000000000000000..21377f66cec15b7b01c23031f9b5b5357cf38e38
--- /dev/null
+++ b/utils/model/scoring.py
@@ -0,0 +1,300 @@
+
+##
+## lk and lk term
+#(LJ_RADIUS LJ_WDEPTH LK_DGFREE LK_LAMBDA LK_VOLUME)
+type2ljlk = {
+ "CNH2":(1.968297,0.094638,3.077030,3.5000,13.500000),
+ "COO":(1.916661,0.141799,-3.332648,3.5000,14.653000),
+ "CH0":(2.011760,0.062642,1.409284,3.5000,8.998000),
+ "CH1":(2.011760,0.062642,-3.538387,3.5000,10.686000),
+ "CH2":(2.011760,0.062642,-1.854658,3.5000,18.331000),
+ "CH3":(2.011760,0.062642,7.292929,3.5000,25.855000),
+ "aroC":(2.016441,0.068775,1.797950,3.5000,16.704000),
+ "Ntrp":(1.802452,0.161725,-8.413116,3.5000,9.522100),
+ "Nhis":(1.802452,0.161725,-9.739606,3.5000,9.317700),
+ "NtrR":(1.802452,0.161725,-5.158080,3.5000,9.779200),
+ "NH2O":(1.802452,0.161725,-8.101638,3.5000,15.689000),
+ "Nlys":(1.802452,0.161725,-20.864641,3.5000,16.514000),
+ "Narg":(1.802452,0.161725,-8.968351,3.5000,15.717000),
+ "Npro":(1.802452,0.161725,-0.984585,3.5000,3.718100),
+ "OH":(1.542743,0.161947,-8.133520,3.5000,10.722000),
+ "OHY":(1.542743,0.161947,-8.133520,3.5000,10.722000),
+ "ONH2":(1.548662,0.182924,-6.591644,3.5000,10.102000),
+ "OOC":(1.492871,0.099873,-9.239832,3.5000,9.995600),
+ "S":(1.975967,0.455970,-1.707229,3.5000,17.640000),
+ "SH1":(1.975967,0.455970,3.291643,3.5000,23.240000),
+ "Nbb":(1.802452,0.161725,-9.969494,3.5000,15.992000),
+ "CAbb":(2.011760,0.062642,2.533791,3.5000,12.137000),
+ "CObb":(1.916661,0.141799,3.104248,3.5000,13.221000),
+ "OCbb":(1.540580,0.142417,-8.006829,3.5000,12.196000),
+ "HNbb":(0.901681,0.005000,0.0000,3.5000,0.0000),
+ "Hapo":(1.421272,0.021808,0.0000,3.5000,0.0000),
+ "Haro":(1.374914,0.015909,0.0000,3.5000,0.0000),
+ "Hpol":(0.901681,0.005000,0.0000,3.5000,0.0000),
+ "HS":(0.363887,0.050836,0.0000,3.5000,0.0000),
+}
+
+# hbond donor/acceptors
+class HbAtom:
+ NO = 0
+ DO = 1 # donor
+ AC = 2 # acceptor
+ DA = 3 # donor & acceptor
+ HP = 4 # polar H
+
+type2hb = {
+ "CNH2":HbAtom.NO, "COO":HbAtom.NO, "CH0":HbAtom.NO, "CH1":HbAtom.NO,
+ "CH2":HbAtom.NO, "CH3":HbAtom.NO, "aroC":HbAtom.NO, "Ntrp":HbAtom.DO,
+ "Nhis":HbAtom.AC, "NtrR":HbAtom.DO, "NH2O":HbAtom.DO, "Nlys":HbAtom.DO,
+ "Narg":HbAtom.DO, "Npro":HbAtom.NO, "OH":HbAtom.DA, "OHY":HbAtom.DA,
+ "ONH2":HbAtom.AC, "OOC":HbAtom.AC, "S":HbAtom.NO, "SH1":HbAtom.NO,
+ "Nbb":HbAtom.DO, "CAbb":HbAtom.NO, "CObb":HbAtom.NO, "OCbb":HbAtom.AC,
+ "HNbb":HbAtom.HP, "Hapo":HbAtom.NO, "Haro":HbAtom.NO, "Hpol":HbAtom.HP,
+ "HS":HbAtom.HP, # HP in rosetta(?)
+}
+
+##
+## hbond term
+class HbDonType:
+ PBA = 0
+ IND = 1
+ IME = 2
+ GDE = 3
+ CXA = 4
+ AMO = 5
+ HXL = 6
+ AHX = 7
+ NTYPES = 8
+
+class HbAccType:
+ PBA = 0
+ CXA = 1
+ CXL = 2
+ HXL = 3
+ AHX = 4
+ IME = 5
+ NTYPES = 6
+
+class HbHybType:
+ SP2 = 0
+ SP3 = 1
+ RING = 2
+ NTYPES = 3
+
+type2dontype = {
+ "Nbb": HbDonType.PBA,
+ "Ntrp": HbDonType.IND,
+ "NtrR": HbDonType.GDE,
+ "Narg": HbDonType.GDE,
+ "NH2O": HbDonType.CXA,
+ "Nlys": HbDonType.AMO,
+ "OH": HbDonType.HXL,
+ "OHY": HbDonType.AHX,
+}
+
+type2acctype = {
+ "OCbb": HbAccType.PBA,
+ "ONH2": HbAccType.CXA,
+ "OOC": HbAccType.CXL,
+ "OH": HbAccType.HXL,
+ "OHY": HbAccType.AHX,
+ "Nhis": HbAccType.IME,
+}
+
+type2hybtype = {
+ "OCbb": HbHybType.SP2,
+ "ONH2": HbHybType.SP2,
+ "OOC": HbHybType.SP2,
+ "OHY": HbHybType.SP3,
+ "OH": HbHybType.SP3,
+ "Nhis": HbHybType.RING,
+}
+
+dontype2wt = {
+ HbDonType.PBA: 1.45,
+ HbDonType.IND: 1.15,
+ HbDonType.IME: 1.42,
+ HbDonType.GDE: 1.11,
+ HbDonType.CXA: 1.29,
+ HbDonType.AMO: 1.17,
+ HbDonType.HXL: 0.99,
+ HbDonType.AHX: 1.00,
+}
+
+acctype2wt = {
+ HbAccType.PBA: 1.19,
+ HbAccType.CXA: 1.21,
+ HbAccType.CXL: 1.10,
+ HbAccType.HXL: 1.15,
+ HbAccType.AHX: 1.15,
+ HbAccType.IME: 1.17,
+}
+
+class HbPolyType:
+ ahdist_aASN_dARG = 0
+ ahdist_aASN_dASN = 1
+ ahdist_aASN_dGLY = 2
+ ahdist_aASN_dHIS = 3
+ ahdist_aASN_dLYS = 4
+ ahdist_aASN_dSER = 5
+ ahdist_aASN_dTRP = 6
+ ahdist_aASN_dTYR = 7
+ ahdist_aASP_dARG = 8
+ ahdist_aASP_dASN = 9
+ ahdist_aASP_dGLY = 10
+ ahdist_aASP_dHIS = 11
+ ahdist_aASP_dLYS = 12
+ ahdist_aASP_dSER = 13
+ ahdist_aASP_dTRP = 14
+ ahdist_aASP_dTYR = 15
+ ahdist_aGLY_dARG = 16
+ ahdist_aGLY_dASN = 17
+ ahdist_aGLY_dGLY = 18
+ ahdist_aGLY_dHIS = 19
+ ahdist_aGLY_dLYS = 20
+ ahdist_aGLY_dSER = 21
+ ahdist_aGLY_dTRP = 22
+ ahdist_aGLY_dTYR = 23
+ ahdist_aHIS_dARG = 24
+ ahdist_aHIS_dASN = 25
+ ahdist_aHIS_dGLY = 26
+ ahdist_aHIS_dHIS = 27
+ ahdist_aHIS_dLYS = 28
+ ahdist_aHIS_dSER = 29
+ ahdist_aHIS_dTRP = 30
+ ahdist_aHIS_dTYR = 31
+ ahdist_aSER_dARG = 32
+ ahdist_aSER_dASN = 33
+ ahdist_aSER_dGLY = 34
+ ahdist_aSER_dHIS = 35
+ ahdist_aSER_dLYS = 36
+ ahdist_aSER_dSER = 37
+ ahdist_aSER_dTRP = 38
+ ahdist_aSER_dTYR = 39
+ ahdist_aTYR_dARG = 40
+ ahdist_aTYR_dASN = 41
+ ahdist_aTYR_dGLY = 42
+ ahdist_aTYR_dHIS = 43
+ ahdist_aTYR_dLYS = 44
+ ahdist_aTYR_dSER = 45
+ ahdist_aTYR_dTRP = 46
+ ahdist_aTYR_dTYR = 47
+ cosBAH_off = 48
+ cosBAH_7 = 49
+ cosBAH_6i = 50
+ AHD_1h = 51
+ AHD_1i = 52
+ AHD_1j = 53
+ AHD_1k = 54
+
+# map donor:acceptor pairs to polynomials
+hbtypepair2poly = {
+ (HbDonType.PBA,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.CXA,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.IME,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.IND,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.AMO,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1j),
+ (HbDonType.AHX,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.PBA): (HbPolyType.ahdist_aGLY_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.CXA): (HbPolyType.ahdist_aASN_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.CXA,HbAccType.CXA): (HbPolyType.ahdist_aASN_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IME,HbAccType.CXA): (HbPolyType.ahdist_aASN_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IND,HbAccType.CXA): (HbPolyType.ahdist_aASN_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AMO,HbAccType.CXA): (HbPolyType.ahdist_aASN_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.CXA): (HbPolyType.ahdist_aASN_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AHX,HbAccType.CXA): (HbPolyType.ahdist_aASN_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.CXA): (HbPolyType.ahdist_aASN_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.CXL): (HbPolyType.ahdist_aASP_dGLY,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.CXA,HbAccType.CXL): (HbPolyType.ahdist_aASP_dASN,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IME,HbAccType.CXL): (HbPolyType.ahdist_aASP_dHIS,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.IND,HbAccType.CXL): (HbPolyType.ahdist_aASP_dTRP,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AMO,HbAccType.CXL): (HbPolyType.ahdist_aASP_dLYS,HbPolyType.cosBAH_off,HbPolyType.AHD_1h),
+ (HbDonType.GDE,HbAccType.CXL): (HbPolyType.ahdist_aASP_dARG,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.AHX,HbAccType.CXL): (HbPolyType.ahdist_aASP_dTYR,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.HXL,HbAccType.CXL): (HbPolyType.ahdist_aASP_dSER,HbPolyType.cosBAH_off,HbPolyType.AHD_1k),
+ (HbDonType.PBA,HbAccType.IME): (HbPolyType.ahdist_aHIS_dGLY,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.IME): (HbPolyType.ahdist_aHIS_dASN,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.IME): (HbPolyType.ahdist_aHIS_dHIS,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.IND,HbAccType.IME): (HbPolyType.ahdist_aHIS_dTRP,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.IME): (HbPolyType.ahdist_aHIS_dLYS,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.IME): (HbPolyType.ahdist_aHIS_dARG,HbPolyType.cosBAH_7,HbPolyType.AHD_1h),
+ (HbDonType.AHX,HbAccType.IME): (HbPolyType.ahdist_aHIS_dTYR,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.IME): (HbPolyType.ahdist_aHIS_dSER,HbPolyType.cosBAH_7,HbPolyType.AHD_1i),
+ (HbDonType.PBA,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dGLY,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dASN,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dHIS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IND,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dTRP,HbPolyType.cosBAH_6i,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dLYS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dARG,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.AHX,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dTYR,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.AHX): (HbPolyType.ahdist_aTYR_dSER,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.PBA,HbAccType.HXL): (HbPolyType.ahdist_aSER_dGLY,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.CXA,HbAccType.HXL): (HbPolyType.ahdist_aSER_dASN,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IME,HbAccType.HXL): (HbPolyType.ahdist_aSER_dHIS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.IND,HbAccType.HXL): (HbPolyType.ahdist_aSER_dTRP,HbPolyType.cosBAH_6i,HbPolyType.AHD_1h),
+ (HbDonType.AMO,HbAccType.HXL): (HbPolyType.ahdist_aSER_dLYS,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.GDE,HbAccType.HXL): (HbPolyType.ahdist_aSER_dARG,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.AHX,HbAccType.HXL): (HbPolyType.ahdist_aSER_dTYR,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+ (HbDonType.HXL,HbAccType.HXL): (HbPolyType.ahdist_aSER_dSER,HbPolyType.cosBAH_6i,HbPolyType.AHD_1i),
+}
+
+
+# polynomials are triplets, (x_min, x_max), (y[xx_max]), (c_9,...,c_0)
+hbpolytype2coeffs = { # Parameters imported from rosetta sp2_elec_params @v2017.48-dev59886
+ HbPolyType.ahdist_aASN_dARG: ((0.7019094761929999, 2.86820307153,),(1.1, 1.1,),( 0.58376113, -9.29345473, 64.86270904, -260.3946711, 661.43138077, -1098.01378958, 1183.58371466, -790.82929582, 291.33125475, -43.01629727,)),
+ HbPolyType.ahdist_aASN_dASN: ((0.625841094801, 2.75107708444,),(1.1, 1.1,),( -1.31243015, 18.6745072, -112.63858313, 373.32878091, -734.99145504, 861.38324861, -556.21026097, 143.5626977, 20.03238394, -11.52167705,)),
+ HbPolyType.ahdist_aASN_dGLY: ((0.7477341047139999, 2.6796350782799996,),(1.1, 1.1,),( -1.61294554, 23.3150793, -144.11313069, 496.13575, -1037.83809166, 1348.76826073, -1065.14368678, 473.89008925, -100.41142701, 7.44453515,)),
+ HbPolyType.ahdist_aASN_dHIS: ((0.344789524346, 2.8303582266000005,),(1.1, 1.1,),( -0.2657122, 4.1073775, -26.9099632, 97.10486507, -209.96002602, 277.33057268, -218.74766996, 97.42852213, -24.07382402, 3.73962807,)),
+ HbPolyType.ahdist_aASN_dLYS: ((0.542905671869, 2.45259389314,),(1.1, 1.1,),( 1.38531754, -18.48733797, 106.14444613, -344.70585054, 698.91577956, -917.0879402, 775.32787908, -403.09588787, 113.65054778, -11.66516403,)),
+ HbPolyType.ahdist_aASN_dSER: ((1.0812774602500002, 2.6832123582599996,),(1.1, 1.1,),( -3.51524353, 47.54032873, -254.40168577, 617.84606386, -255.49935027, -2361.56230539, 6426.85797934, -7760.4403891, 4694.08106855, -1149.83549068,)),
+ HbPolyType.ahdist_aASN_dTRP: ((0.6689984999999999, 3.0704254,),(1.1, 1.1,),( -0.5284840422, 8.3510150838, -56.4100479414, 212.4884326254, -488.3178610608, 703.7762350506, -628.9936994633999, 331.4294356146, -93.265817571, 11.9691623698,)),
+ HbPolyType.ahdist_aASN_dTYR: ((1.08950268805, 2.6887046709400004,),(1.1, 1.1,),( -4.4488705, 63.27696281, -371.44187037, 1121.71921621, -1638.11394306, 142.99988401, 3436.65879147, -5496.07011787, 3709.30505237, -962.79669688,)),
+ HbPolyType.ahdist_aASP_dARG: ((0.8100404642229999, 2.9851230124799994,),(1.1, 1.1,),( -0.66430344, 10.41343145, -70.12656205, 265.12578414, -617.05849171, 911.39378582, -847.25013928, 472.09090981, -141.71513167, 18.57721132,)),
+ HbPolyType.ahdist_aASP_dASN: ((1.05401125073, 3.11129675908,),(1.1, 1.1,),( 0.02090728, -0.24144928, -0.19578075, 16.80904547, -117.70216251, 407.18551288, -809.95195924, 939.83137947, -593.94527692, 159.57610528,)),
+ HbPolyType.ahdist_aASP_dGLY: ((0.886260952629, 2.66843608743,),(1.1, 1.1,),( -7.00699267, 107.33021779, -713.45752385, 2694.43092298, -6353.05100287, 9667.94098394, -9461.9261027, 5721.0086877, -1933.97818198, 279.47763789,)),
+ HbPolyType.ahdist_aASP_dHIS: ((1.03597611139, 2.78208509117,),(1.1, 1.1,),( -1.34823406, 17.08925926, -78.75087193, 106.32795459, 400.18459698, -2041.04320193, 4033.83557387, -4239.60530204, 2324.00877252, -519.38410941,)),
+ HbPolyType.ahdist_aASP_dLYS: ((0.97789485082, 2.50496946108,),(1.1, 1.1,),( -0.41300315, 6.59243438, -44.44525308, 163.11796012, -351.2307798, 443.2463146, -297.84582856, 62.38600547, 33.77496227, -14.11652182,)),
+ HbPolyType.ahdist_aASP_dSER: ((0.542905671869, 2.45259389314,),(1.1, 1.1,),( 1.38531754, -18.48733797, 106.14444613, -344.70585054, 698.91577956, -917.0879402, 775.32787908, -403.09588787, 113.65054778, -11.66516403,)),
+ HbPolyType.ahdist_aASP_dTRP: ((0.419155746414, 3.0486938610500003,),(1.1, 1.1,),( -0.24563471, 3.85598551, -25.75176874, 95.36525025, -214.13175785, 299.76133553, -259.0691378, 132.06975835, -37.15612683, 5.60445773,)),
+ HbPolyType.ahdist_aASP_dTYR: ((1.01057521468, 2.7207545786900003,),(1.1, 1.1,),( -0.15808672, -10.21398871, 178.80080949, -1238.0583801, 4736.25248274, -11071.96777725, 16239.07550047, -14593.21092621, 7335.66765017, -1575.08145078,)),
+ HbPolyType.ahdist_aGLY_dARG: ((0.499016667857, 2.9377031027599996,),(1.1, 1.1,),( -0.15923533, 2.5526639, -17.38788803, 65.71046957, -151.13491186, 218.78048387, -199.15882919, 110.56568974, -35.95143745, 6.47580213,)),
+ HbPolyType.ahdist_aGLY_dASN: ((0.7194388032060001, 2.9303772333599998,),(1.1, 1.1,),( -1.40718342, 23.65929694, -172.97144348, 720.64417348, -1882.85420815, 3194.87197776, -3515.52467458, 2415.75238278, -941.47705161, 159.84784277,)),
+ HbPolyType.ahdist_aGLY_dGLY: ((1.38403812683, 2.9981039433,),(1.1, 1.1,),( -0.5307601, 6.47949946, -22.39522814, -55.14303544, 708.30945242, -2619.49318162, 5227.8805795, -6043.31211632, 3806.04676175, -1007.66024144,)),
+ HbPolyType.ahdist_aGLY_dHIS: ((0.47406840932899996, 2.9234200830400003,),(1.1, 1.1,),( -0.12881679, 1.933838, -12.03134888, 39.92691227, -75.41519959, 78.87968016, -37.82769801, -0.13178679, 4.50193019, 0.45408359,)),
+ HbPolyType.ahdist_aGLY_dLYS: ((0.545347533475, 2.42624380351,),(1.1, 1.1,),( -0.22921901, 2.07015714, -6.2947417, 0.66645697, 45.21805416, -130.26668981, 176.32401031, -126.68226346, 43.96744431, -4.40105281,)),
+ HbPolyType.ahdist_aGLY_dSER: ((1.2803349239700001, 2.2465996077400003,),(1.1, 1.1,),( 6.72508613, -86.98495585, 454.18518444, -1119.89141452, 715.624663, 3172.36852982, -9455.49113097, 11797.38766934, -7363.28302948, 1885.50119665,)),
+ HbPolyType.ahdist_aGLY_dTRP: ((0.686512740494, 3.02901351815,),(1.1, 1.1,),( -0.1051487, 1.41597708, -7.42149173, 17.31830704, -6.98293652, -54.76605063, 130.95272289, -132.77575305, 62.75460448, -9.89110842,)),
+ HbPolyType.ahdist_aGLY_dTYR: ((1.28894687639, 2.26335316892,),(1.1, 1.1,),( 13.84536925, -169.40579865, 893.79467505, -2670.60617561, 5016.46234701, -6293.79378818, 5585.1049063, -3683.50722701, 1709.48661405, -399.5712153,)),
+ HbPolyType.ahdist_aHIS_dARG: ((0.8967400957230001, 2.96809434226,),(1.1, 1.1,),( 0.43460495, -10.52727665, 103.16979807, -551.42887412, 1793.25378923, -3701.08304991, 4861.05155388, -3922.4285529, 1763.82137881, -335.43441944,)),
+ HbPolyType.ahdist_aHIS_dASN: ((0.887120931718, 2.59166903153,),(1.1, 1.1,),( -3.50289894, 54.42813924, -368.14395507, 1418.90186454, -3425.60485859, 5360.92334837, -5428.54462336, 3424.68800187, -1221.49631986, 189.27122436,)),
+ HbPolyType.ahdist_aHIS_dGLY: ((1.01629363411, 2.58523052904,),(1.1, 1.1,),( -1.68095217, 21.31894078, -107.72203494, 251.81021758, -134.07465831, -707.64527046, 1894.6282743, -2156.85951846, 1216.83585872, -275.48078944,)),
+ HbPolyType.ahdist_aHIS_dHIS: ((0.9773010778919999, 2.72533796329,),(1.1, 1.1,),( -2.33350626, 35.66072412, -233.98966111, 859.13714961, -1925.30958567, 2685.35293578, -2257.48067507, 1021.49796136, -169.36082523, -12.1348055,)),
+ HbPolyType.ahdist_aHIS_dLYS: ((0.7080936539849999, 2.47191718632,),(1.1, 1.1,),( -1.88479369, 28.38084382, -185.74039957, 690.81875917, -1605.11404391, 2414.83545623, -2355.9723201, 1442.24496229, -506.45880637, 79.47512505,)),
+ HbPolyType.ahdist_aHIS_dSER: ((0.90846809159, 2.5477956147,),(1.1, 1.1,),( -0.92004641, 15.91841533, -117.83979251, 488.22211296, -1244.13047376, 2017.43704053, -2076.04468019, 1302.42621488, -451.29138643, 67.15812575,)),
+ HbPolyType.ahdist_aHIS_dTRP: ((0.991999676806, 2.81296584506,),(1.1, 1.1,),( -1.29358587, 19.97152857, -131.89796017, 485.29199356, -1084.0466445, 1497.3352889, -1234.58042682, 535.8048197, -75.58951691, -9.91148332,)),
+ HbPolyType.ahdist_aHIS_dTYR: ((0.882661836357, 2.5469016429900004,),(1.1, 1.1,),( -6.94700143, 109.07997256, -747.64035726, 2929.83959536, -7220.15788571, 11583.34170519, -12078.443492, 7881.85479715, -2918.19482068, 468.23988622,)),
+ HbPolyType.ahdist_aSER_dARG: ((1.0204658147399999, 2.8899566041900004,),(1.1, 1.1,),( 0.33887327, -7.54511361, 70.87316645, -371.88263665, 1206.67454443, -2516.82084076, 3379.45432693, -2819.73384601, 1325.33307517, -265.54533008,)),
+ HbPolyType.ahdist_aSER_dASN: ((1.01393052233, 3.0024434159299997,),(1.1, 1.1,),( 0.37012361, -7.46486204, 64.85775924, -318.6047209, 974.66322243, -1924.37334018, 2451.63840629, -1943.1915675, 867.07870559, -163.83771761,)),
+ HbPolyType.ahdist_aSER_dGLY: ((1.3856562156299999, 2.74160605537,),(1.1, 1.1,),( -1.32847415, 22.67528654, -172.53450064, 770.79034865, -2233.48829652, 4354.38807288, -5697.35144236, 4803.38686157, -2361.48028857, 518.28202382,)),
+ HbPolyType.ahdist_aSER_dHIS: ((0.550992321207, 2.68549261999,),(1.1, 1.1,),( -1.98041793, 29.59668639, -190.36751773, 688.43324385, -1534.68894765, 2175.66568976, -1952.07622113, 1066.28943929, -324.23381388, 43.41006168,)),
+ HbPolyType.ahdist_aSER_dLYS: ((0.8603189393170001, 2.77729502744,),(1.1, 1.1,),( 0.90884741, -17.24690746, 141.78469099, -661.85989315, 1929.7674992, -3636.43392779, 4419.00727923, -3332.43482061, 1410.78913266, -253.53829424,)),
+ HbPolyType.ahdist_aSER_dSER: ((1.10866545921, 2.61727781204,),(1.1, 1.1,),( -0.38264308, 4.41779675, -10.7016645, -81.91314845, 668.91174735, -2187.50684758, 3983.56103269, -4213.32320546, 2418.41531442, -580.28918569,)),
+ HbPolyType.ahdist_aSER_dTRP: ((1.4092077245899999, 2.8066121197099996,),(1.1, 1.1,),( 0.73762477, -11.70741276, 73.05154232, -205.00144794, 89.58794368, 1082.94541375, -3343.98293188, 4601.70815729, -3178.53568678, 896.59487831,)),
+ HbPolyType.ahdist_aSER_dTYR: ((1.10773547919, 2.60403567341,),(1.1, 1.1,),( -1.13249925, 14.66643161, -69.01708791, 93.96846742, 380.56063898, -1984.56675689, 4074.08891127, -4492.76927139, 2613.13168054, -627.71933508,)),
+ HbPolyType.ahdist_aTYR_dARG: ((1.05581400627, 2.85499888099,),(1.1, 1.1,),( -0.30396592, 5.30288548, -39.75788579, 167.5416547, -435.15958911, 716.52357586, -735.95195083, 439.76284677, -130.00400085, 13.23827556,)),
+ HbPolyType.ahdist_aTYR_dASN: ((1.0994919065200002, 2.8400869077900004,),(1.1, 1.1,),( 0.33548259, -3.5890451, 8.97769025, 48.1492734, -400.5983616, 1269.89613211, -2238.03101675, 2298.33009115, -1290.42961162, 308.43185147,)),
+ HbPolyType.ahdist_aTYR_dGLY: ((1.36546155066, 2.7303075916400004,),(1.1, 1.1,),( -1.55312915, 18.62092487, -70.91365499, -41.83066505, 1248.88835245, -4719.81948329, 9186.09528168, -10266.11434548, 6266.21959533, -1622.19652457,)),
+ HbPolyType.ahdist_aTYR_dHIS: ((0.5955982461899999, 2.6643551317500003,),(1.1, 1.1,),( -0.47442788, 7.16629863, -46.71287553, 171.46128947, -388.17484011, 558.45202337, -506.35587481, 276.46237273, -83.52554392, 12.05709329,)),
+ HbPolyType.ahdist_aTYR_dLYS: ((0.7978598238760001, 2.7620933782,),(1.1, 1.1,),( -0.20201464, 1.69684984, 0.27677515, -55.05786347, 286.29918332, -725.92372531, 1054.771746, -889.33602341, 401.11342256, -73.02221189,)),
+ HbPolyType.ahdist_aTYR_dSER: ((0.7083554962559999, 2.7032011990599996,),(1.1, 1.1,),( -0.70764192, 11.67978065, -82.80447482, 329.83401367, -810.58976486, 1269.57613941, -1261.04047117, 761.72890446, -254.37526011, 37.24301861,)),
+ HbPolyType.ahdist_aTYR_dTRP: ((1.10934023051, 2.8819112108,),(1.1, 1.1,),( -11.58453967, 204.88308091, -1589.77384548, 7100.84791905, -20113.61354433, 37457.83646055, -45850.02969172, 35559.8805122, -15854.78726237, 3098.04931146,)),
+ HbPolyType.ahdist_aTYR_dTYR: ((1.1105954899400001, 2.60081798685,),(1.1, 1.1,),( -1.63120628, 19.48493187, -81.0332905, 56.80517706, 687.42717782, -2842.77799908, 5385.52231471, -5656.74159307, 3178.83470588, -744.70042777,)),
+ HbPolyType.AHD_1h: ((1.76555274367, 3.1416,),(1.1, 1.1,),( 0.62725838, -9.98558225, 59.39060071, -120.82930213, -333.26536028, 2603.13082592, -6895.51207142, 9651.25238056, -7127.13394872, 2194.77244026,)),
+ HbPolyType.AHD_1i: ((1.59914724347, 3.1416,),(1.1, 1.1,),( -0.18888801, 3.48241679, -25.65508662, 89.57085435, -95.91708218, -367.93452341, 1589.6904702, -2662.3582135, 2184.40194483, -723.28383545,)),
+ HbPolyType.AHD_1j: ((1.1435646388, 3.1416,),(1.1, 1.1,),( 0.47683259, -9.54524724, 83.62557693, -420.55867774, 1337.19354878, -2786.26265686, 3803.178227, -3278.62879901, 1619.04116204, -347.50157909,)),
+ HbPolyType.AHD_1k: ((1.15651981164, 3.1416,),(1.1, 1.1,),( -0.10757999, 2.0276542, -16.51949978, 75.83866839, -214.18025678, 380.55117567, -415.47847283, 255.66998474, -69.94662165, 3.21313428,)),
+ HbPolyType.cosBAH_off: ((-1234.0, 1.1,),(1.1, 1.1,),( 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0,)),
+ HbPolyType.cosBAH_6i: ((-0.23538144897100002, 1.1,),(1.1, 1.1,),( -0.822093, -3.75364636, 46.88852157, -129.5440564, 146.69151428, -67.60598792, 2.91683129, 9.26673173, -3.84488178, 0.05706659,)),
+ HbPolyType.cosBAH_7: ((-0.019373850666900002, 1.1,),(1.1, 1.1,),( 0.0, -27.942923450028, 136.039920253368, -268.06959056747, 275.400462507919, -153.502076215949, 39.741591385461, 0.693861510121, -3.885952320499, 1.024765090788892)),
+}
\ No newline at end of file
diff --git a/utils/model/se3_transformer/__init__.py b/utils/model/se3_transformer/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..e69de29bb2d1d6434b8b29ae775ad8c2e48c5391
diff --git a/utils/model/se3_transformer/__pycache__/__init__.cpython-310.pyc b/utils/model/se3_transformer/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..01f8e2d61996d7fe7617616463cf01fb6b19dee2
Binary files /dev/null and b/utils/model/se3_transformer/__pycache__/__init__.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/data_loading/__init__.py b/utils/model/se3_transformer/data_loading/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..67a9e0ed7b9c787830f1b99cc0132a14bbcaee43
--- /dev/null
+++ b/utils/model/se3_transformer/data_loading/__init__.py
@@ -0,0 +1 @@
+from .qm9 import QM9DataModule
diff --git a/utils/model/se3_transformer/data_loading/data_module.py b/utils/model/se3_transformer/data_loading/data_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..1047d41d16b81c05e2f14130311b42ab5b36eb36
--- /dev/null
+++ b/utils/model/se3_transformer/data_loading/data_module.py
@@ -0,0 +1,63 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import torch.distributed as dist
+from abc import ABC
+from torch.utils.data import DataLoader, DistributedSampler, Dataset
+
+from se3_transformer.runtime.utils import get_local_rank
+
+
+def _get_dataloader(dataset: Dataset, shuffle: bool, **kwargs) -> DataLoader:
+ # Classic or distributed dataloader depending on the context
+ sampler = DistributedSampler(dataset, shuffle=shuffle) if dist.is_initialized() else None
+ return DataLoader(dataset, shuffle=(shuffle and sampler is None), sampler=sampler, **kwargs)
+
+
+class DataModule(ABC):
+ """ Abstract DataModule. Children must define self.ds_{train | val | test}. """
+
+ def __init__(self, **dataloader_kwargs):
+ super().__init__()
+ if get_local_rank() == 0:
+ self.prepare_data()
+
+ # Wait until rank zero has prepared the data (download, preprocessing, ...)
+ if dist.is_initialized():
+ dist.barrier(device_ids=[get_local_rank()])
+
+ self.dataloader_kwargs = {'pin_memory': True, 'persistent_workers': True, **dataloader_kwargs}
+ self.ds_train, self.ds_val, self.ds_test = None, None, None
+
+ def prepare_data(self):
+ """ Method called only once per node. Put here any downloading or preprocessing """
+ pass
+
+ def train_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_train, shuffle=True, **self.dataloader_kwargs)
+
+ def val_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_val, shuffle=False, **self.dataloader_kwargs)
+
+ def test_dataloader(self) -> DataLoader:
+ return _get_dataloader(self.ds_test, shuffle=False, **self.dataloader_kwargs)
diff --git a/utils/model/se3_transformer/data_loading/qm9.py b/utils/model/se3_transformer/data_loading/qm9.py
new file mode 100644
index 0000000000000000000000000000000000000000..b45839868626f56d3a42ce859b2033ce1526373e
--- /dev/null
+++ b/utils/model/se3_transformer/data_loading/qm9.py
@@ -0,0 +1,171 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+from typing import Tuple
+
+import dgl
+import pathlib
+import torch
+from dgl.data import QM9EdgeDataset
+from dgl import DGLGraph
+from torch import Tensor
+from torch.utils.data import random_split, DataLoader, Dataset
+from tqdm import tqdm
+
+from se3_transformer.data_loading.data_module import DataModule
+from se3_transformer.model.basis import get_basis
+from se3_transformer.runtime.utils import get_local_rank, str2bool, using_tensor_cores
+
+
+def _get_relative_pos(qm9_graph: DGLGraph) -> Tensor:
+ x = qm9_graph.ndata['pos']
+ src, dst = qm9_graph.edges()
+ rel_pos = x[dst] - x[src]
+ return rel_pos
+
+
+def _get_split_sizes(full_dataset: Dataset) -> Tuple[int, int, int]:
+ len_full = len(full_dataset)
+ len_train = 100_000
+ len_test = int(0.1 * len_full)
+ len_val = len_full - len_train - len_test
+ return len_train, len_val, len_test
+
+
+class QM9DataModule(DataModule):
+ """
+ Datamodule wrapping https://docs.dgl.ai/en/latest/api/python/dgl.data.html#qm9edge-dataset
+ Training set is 100k molecules. Test set is 10% of the dataset. Validation set is the rest.
+ This includes all the molecules from QM9 except the ones that are uncharacterized.
+ """
+
+ NODE_FEATURE_DIM = 6
+ EDGE_FEATURE_DIM = 4
+
+ def __init__(self,
+ data_dir: pathlib.Path,
+ task: str = 'homo',
+ batch_size: int = 240,
+ num_workers: int = 8,
+ num_degrees: int = 4,
+ amp: bool = False,
+ precompute_bases: bool = False,
+ **kwargs):
+ self.data_dir = data_dir # This needs to be before __init__ so that prepare_data has access to it
+ super().__init__(batch_size=batch_size, num_workers=num_workers, collate_fn=self._collate)
+ self.amp = amp
+ self.task = task
+ self.batch_size = batch_size
+ self.num_degrees = num_degrees
+
+ qm9_kwargs = dict(label_keys=[self.task], verbose=False, raw_dir=str(data_dir))
+ if precompute_bases:
+ bases_kwargs = dict(max_degree=num_degrees - 1, use_pad_trick=using_tensor_cores(amp), amp=amp)
+ full_dataset = CachedBasesQM9EdgeDataset(bases_kwargs=bases_kwargs, batch_size=batch_size, **qm9_kwargs)
+ else:
+ full_dataset = QM9EdgeDataset(**qm9_kwargs)
+
+ self.ds_train, self.ds_val, self.ds_test = random_split(full_dataset, _get_split_sizes(full_dataset),
+ generator=torch.Generator().manual_seed(0))
+
+ train_targets = full_dataset.targets[self.ds_train.indices, full_dataset.label_keys[0]]
+ self.targets_mean = train_targets.mean()
+ self.targets_std = train_targets.std()
+
+ def prepare_data(self):
+ # Download the QM9 preprocessed data
+ QM9EdgeDataset(verbose=True, raw_dir=str(self.data_dir))
+
+ def _collate(self, samples):
+ graphs, y, *bases = map(list, zip(*samples))
+ batched_graph = dgl.batch(graphs)
+ edge_feats = {'0': batched_graph.edata['edge_attr'][..., None]}
+ batched_graph.edata['rel_pos'] = _get_relative_pos(batched_graph)
+ # get node features
+ node_feats = {'0': batched_graph.ndata['attr'][:, :6, None]}
+ targets = (torch.cat(y) - self.targets_mean) / self.targets_std
+
+ if bases:
+ # collate bases
+ all_bases = {
+ key: torch.cat([b[key] for b in bases[0]], dim=0)
+ for key in bases[0][0].keys()
+ }
+
+ return batched_graph, node_feats, edge_feats, all_bases, targets
+ else:
+ return batched_graph, node_feats, edge_feats, targets
+
+ @staticmethod
+ def add_argparse_args(parent_parser):
+ parser = parent_parser.add_argument_group("QM9 dataset")
+ parser.add_argument('--task', type=str, default='homo', const='homo', nargs='?',
+ choices=['mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'U0', 'U', 'H', 'G', 'Cv',
+ 'U0_atom', 'U_atom', 'H_atom', 'G_atom', 'A', 'B', 'C'],
+ help='Regression task to train on')
+ parser.add_argument('--precompute_bases', type=str2bool, nargs='?', const=True, default=False,
+ help='Precompute bases at the beginning of the script during dataset initialization,'
+ ' instead of computing them at the beginning of each forward pass.')
+ return parent_parser
+
+ def __repr__(self):
+ return f'QM9({self.task})'
+
+
+class CachedBasesQM9EdgeDataset(QM9EdgeDataset):
+ """ Dataset extending the QM9 dataset from DGL with precomputed (cached in RAM) pairwise bases """
+
+ def __init__(self, bases_kwargs: dict, batch_size: int, *args, **kwargs):
+ """
+ :param bases_kwargs: Arguments to feed the bases computation function
+ :param batch_size: Batch size to use when iterating over the dataset for computing bases
+ """
+ self.bases_kwargs = bases_kwargs
+ self.batch_size = batch_size
+ self.bases = None
+ super().__init__(*args, **kwargs)
+
+ def load(self):
+ super().load()
+ # Iterate through the dataset and compute bases (pairwise only)
+ # Potential improvement: use multi-GPU and reduction
+ dataloader = DataLoader(self, shuffle=False, batch_size=self.batch_size,
+ collate_fn=lambda samples: dgl.batch([sample[0] for sample in samples]))
+ bases = []
+ for i, graph in tqdm(enumerate(dataloader), total=len(dataloader), desc='Precomputing QM9 bases',
+ disable=get_local_rank() != 0):
+ rel_pos = _get_relative_pos(graph)
+ # Compute the bases with the GPU but convert the result to CPU to store in RAM
+ bases.append({k: v.cpu() for k, v in get_basis(rel_pos.cuda(), **self.bases_kwargs).items()})
+ self.bases = bases # Assign at the end so that __getitem__ isn't confused
+
+ def __getitem__(self, idx: int):
+ graph, label = super().__getitem__(idx)
+
+ if self.bases:
+ bases_idx = idx // self.batch_size
+ bases_cumsum_idx = self.ne_cumsum[idx] - self.ne_cumsum[bases_idx * self.batch_size]
+ bases_cumsum_next_idx = self.ne_cumsum[idx + 1] - self.ne_cumsum[bases_idx * self.batch_size]
+ return graph, label, {key: basis[bases_cumsum_idx:bases_cumsum_next_idx] for key, basis in
+ self.bases[bases_idx].items()}
+ else:
+ return graph, label
diff --git a/utils/model/se3_transformer/model/__init__.py b/utils/model/se3_transformer/model/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..628d01e98d45d360187fb7fcce63f33ebf05d3b7
--- /dev/null
+++ b/utils/model/se3_transformer/model/__init__.py
@@ -0,0 +1,2 @@
+from .transformer import SE3Transformer, SE3TransformerPooled
+from .fiber import Fiber
diff --git a/utils/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc b/utils/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b86f94e8876762ea1a611443581478df39a4dbc9
Binary files /dev/null and b/utils/model/se3_transformer/model/__pycache__/__init__.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc b/utils/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..86e5265ee87961d2d03093c0756a829b408edde9
Binary files /dev/null and b/utils/model/se3_transformer/model/__pycache__/basis.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc b/utils/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..f40874b4b024c1305a9a92ea21435108db7b1271
Binary files /dev/null and b/utils/model/se3_transformer/model/__pycache__/fiber.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc b/utils/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..3472eec408c815c3f5ea1dd15437135eaa4c7f5e
Binary files /dev/null and b/utils/model/se3_transformer/model/__pycache__/transformer.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/basis.py b/utils/model/se3_transformer/model/basis.py
new file mode 100644
index 0000000000000000000000000000000000000000..74f04a0fada04ad63f9d9fc17f4fe7bb038d806b
--- /dev/null
+++ b/utils/model/se3_transformer/model/basis.py
@@ -0,0 +1,178 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from functools import lru_cache
+from typing import Dict, List
+
+import e3nn.o3 as o3
+import torch
+import torch.nn.functional as F
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.runtime.utils import degree_to_dim
+
+
+@lru_cache(maxsize=None)
+def get_clebsch_gordon(J: int, d_in: int, d_out: int, device) -> Tensor:
+ """ Get the (cached) Q^{d_out,d_in}_J matrices from equation (8) """
+ return o3.wigner_3j(J, d_in, d_out, dtype=torch.float64, device=device).permute(2, 1, 0)
+
+
+@lru_cache(maxsize=None)
+def get_all_clebsch_gordon(max_degree: int, device) -> List[List[Tensor]]:
+ all_cb = []
+ for d_in in range(max_degree + 1):
+ for d_out in range(max_degree + 1):
+ K_Js = []
+ for J in range(abs(d_in - d_out), d_in + d_out + 1):
+ K_Js.append(get_clebsch_gordon(J, d_in, d_out, device))
+ all_cb.append(K_Js)
+ return all_cb
+
+
+def get_spherical_harmonics(relative_pos: Tensor, max_degree: int) -> List[Tensor]:
+ all_degrees = list(range(2 * max_degree + 1))
+ with nvtx_range('spherical harmonics'):
+ sh = o3.spherical_harmonics(all_degrees, relative_pos, normalize=True)
+ return torch.split(sh, [degree_to_dim(d) for d in all_degrees], dim=1)
+
+
+@torch.jit.script
+def get_basis_script(max_degree: int,
+ use_pad_trick: bool,
+ spherical_harmonics: List[Tensor],
+ clebsch_gordon: List[List[Tensor]],
+ amp: bool) -> Dict[str, Tensor]:
+ """
+ Compute pairwise bases matrices for degrees up to max_degree
+ :param max_degree: Maximum input or output degree
+ :param use_pad_trick: Pad some of the odd dimensions for a better use of Tensor Cores
+ :param spherical_harmonics: List of computed spherical harmonics
+ :param clebsch_gordon: List of computed CB-coefficients
+ :param amp: When true, return bases in FP16 precision
+ """
+ basis = {}
+ idx = 0
+ # Double for loop instead of product() because of JIT script
+ for d_in in range(max_degree + 1):
+ for d_out in range(max_degree + 1):
+ key = f'{d_in},{d_out}'
+ K_Js = []
+ for freq_idx, J in enumerate(range(abs(d_in - d_out), d_in + d_out + 1)):
+ Q_J = clebsch_gordon[idx][freq_idx]
+ K_Js.append(torch.einsum('n f, k l f -> n l k', spherical_harmonics[J].float(), Q_J.float()))
+
+ basis[key] = torch.stack(K_Js, 2) # Stack on second dim so order is n l f k
+ if amp:
+ basis[key] = basis[key].half()
+ if use_pad_trick:
+ basis[key] = F.pad(basis[key], (0, 1)) # Pad the k dimension, that can be sliced later
+
+ idx += 1
+
+ return basis
+
+
+@torch.jit.script
+def update_basis_with_fused(basis: Dict[str, Tensor],
+ max_degree: int,
+ use_pad_trick: bool,
+ fully_fused: bool) -> Dict[str, Tensor]:
+ """ Update the basis dict with partially and optionally fully fused bases """
+ num_edges = basis['0,0'].shape[0]
+ device = basis['0,0'].device
+ dtype = basis['0,0'].dtype
+ sum_dim = sum([degree_to_dim(d) for d in range(max_degree + 1)])
+
+ # Fused per output degree
+ for d_out in range(max_degree + 1):
+ sum_freq = sum([degree_to_dim(min(d, d_out)) for d in range(max_degree + 1)])
+ basis_fused = torch.zeros(num_edges, sum_dim, sum_freq, degree_to_dim(d_out) + int(use_pad_trick),
+ device=device, dtype=dtype)
+ acc_d, acc_f = 0, 0
+ for d_in in range(max_degree + 1):
+ basis_fused[:, acc_d:acc_d + degree_to_dim(d_in), acc_f:acc_f + degree_to_dim(min(d_out, d_in)),
+ :degree_to_dim(d_out)] = basis[f'{d_in},{d_out}'][:, :, :, :degree_to_dim(d_out)]
+
+ acc_d += degree_to_dim(d_in)
+ acc_f += degree_to_dim(min(d_out, d_in))
+
+ basis[f'out{d_out}_fused'] = basis_fused
+
+ # Fused per input degree
+ for d_in in range(max_degree + 1):
+ sum_freq = sum([degree_to_dim(min(d, d_in)) for d in range(max_degree + 1)])
+ basis_fused = torch.zeros(num_edges, degree_to_dim(d_in), sum_freq, sum_dim,
+ device=device, dtype=dtype)
+ acc_d, acc_f = 0, 0
+ for d_out in range(max_degree + 1):
+ basis_fused[:, :, acc_f:acc_f + degree_to_dim(min(d_out, d_in)), acc_d:acc_d + degree_to_dim(d_out)] \
+ = basis[f'{d_in},{d_out}'][:, :, :, :degree_to_dim(d_out)]
+
+ acc_d += degree_to_dim(d_out)
+ acc_f += degree_to_dim(min(d_out, d_in))
+
+ basis[f'in{d_in}_fused'] = basis_fused
+
+ if fully_fused:
+ # Fully fused
+ # Double sum this way because of JIT script
+ sum_freq = sum([
+ sum([degree_to_dim(min(d_in, d_out)) for d_in in range(max_degree + 1)]) for d_out in range(max_degree + 1)
+ ])
+ basis_fused = torch.zeros(num_edges, sum_dim, sum_freq, sum_dim, device=device, dtype=dtype)
+
+ acc_d, acc_f = 0, 0
+ for d_out in range(max_degree + 1):
+ b = basis[f'out{d_out}_fused']
+ basis_fused[:, :, acc_f:acc_f + b.shape[2], acc_d:acc_d + degree_to_dim(d_out)] = b[:, :, :,
+ :degree_to_dim(d_out)]
+ acc_f += b.shape[2]
+ acc_d += degree_to_dim(d_out)
+
+ basis['fully_fused'] = basis_fused
+
+ del basis['0,0'] # We know that the basis for l = k = 0 is filled with a constant
+ return basis
+
+
+def get_basis(relative_pos: Tensor,
+ max_degree: int = 4,
+ compute_gradients: bool = False,
+ use_pad_trick: bool = False,
+ amp: bool = False) -> Dict[str, Tensor]:
+ with nvtx_range('spherical harmonics'):
+ spherical_harmonics = get_spherical_harmonics(relative_pos, max_degree)
+ with nvtx_range('CB coefficients'):
+ clebsch_gordon = get_all_clebsch_gordon(max_degree, relative_pos.device)
+
+ with torch.autograd.set_grad_enabled(compute_gradients):
+ with nvtx_range('bases'):
+ basis = get_basis_script(max_degree=max_degree,
+ use_pad_trick=use_pad_trick,
+ spherical_harmonics=spherical_harmonics,
+ clebsch_gordon=clebsch_gordon,
+ amp=amp)
+ return basis
diff --git a/utils/model/se3_transformer/model/fiber.py b/utils/model/se3_transformer/model/fiber.py
new file mode 100644
index 0000000000000000000000000000000000000000..38db33b0d27d70116c92650176170e9b3cf9a9c7
--- /dev/null
+++ b/utils/model/se3_transformer/model/fiber.py
@@ -0,0 +1,144 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from collections import namedtuple
+from itertools import product
+from typing import Dict
+
+import torch
+from torch import Tensor
+
+from se3_transformer.runtime.utils import degree_to_dim
+
+FiberEl = namedtuple('FiberEl', ['degree', 'channels'])
+
+
+class Fiber(dict):
+ """
+ Describes the structure of some set of features.
+ Features are split into types (0, 1, 2, 3, ...). A feature of type k has a dimension of 2k+1.
+ Type-0 features: invariant scalars
+ Type-1 features: equivariant 3D vectors
+ Type-2 features: equivariant symmetric traceless matrices
+ ...
+
+ As inputs to a SE3 layer, there can be many features of the same types, and many features of different types.
+ The 'multiplicity' or 'number of channels' is the number of features of a given type.
+ This class puts together all the degrees and their multiplicities in order to describe
+ the inputs, outputs or hidden features of SE3 layers.
+ """
+
+ def __init__(self, structure):
+ if isinstance(structure, dict):
+ structure = [FiberEl(int(d), int(m)) for d, m in sorted(structure.items(), key=lambda x: x[1])]
+ elif not isinstance(structure[0], FiberEl):
+ structure = list(map(lambda t: FiberEl(*t), sorted(structure, key=lambda x: x[1])))
+ self.structure = structure
+ super().__init__({d: m for d, m in self.structure})
+
+ @property
+ def degrees(self):
+ return sorted([t.degree for t in self.structure])
+
+ @property
+ def channels(self):
+ return [self[d] for d in self.degrees]
+
+ @property
+ def num_features(self):
+ """ Size of the resulting tensor if all features were concatenated together """
+ return sum(t.channels * degree_to_dim(t.degree) for t in self.structure)
+
+ @staticmethod
+ def create(num_degrees: int, num_channels: int):
+ """ Create a Fiber with degrees 0..num_degrees-1, all with the same multiplicity """
+ return Fiber([(degree, num_channels) for degree in range(num_degrees)])
+
+ @staticmethod
+ def from_features(feats: Dict[str, Tensor]):
+ """ Infer the Fiber structure from a feature dict """
+ structure = {}
+ for k, v in feats.items():
+ degree = int(k)
+ assert len(v.shape) == 3, 'Feature shape should be (N, C, 2D+1)'
+ assert v.shape[-1] == degree_to_dim(degree)
+ structure[degree] = v.shape[-2]
+ return Fiber(structure)
+
+ def __getitem__(self, degree: int):
+ """ fiber[degree] returns the multiplicity for this degree """
+ return dict(self.structure).get(degree, 0)
+
+ def __iter__(self):
+ """ Iterate over namedtuples (degree, channels) """
+ return iter(self.structure)
+
+ def __mul__(self, other):
+ """
+ If other in an int, multiplies all the multiplicities by other.
+ If other is a fiber, returns the cartesian product.
+ """
+ if isinstance(other, Fiber):
+ return product(self.structure, other.structure)
+ elif isinstance(other, int):
+ return Fiber({t.degree: t.channels * other for t in self.structure})
+
+ def __add__(self, other):
+ """
+ If other in an int, add other to all the multiplicities.
+ If other is a fiber, add the multiplicities of the fibers together.
+ """
+ if isinstance(other, Fiber):
+ return Fiber({t.degree: t.channels + other[t.degree] for t in self.structure})
+ elif isinstance(other, int):
+ return Fiber({t.degree: t.channels + other for t in self.structure})
+
+ def __repr__(self):
+ return str(self.structure)
+
+ @staticmethod
+ def combine_max(f1, f2):
+ """ Combine two fiber by taking the maximum multiplicity for each degree in both fibers """
+ new_dict = dict(f1.structure)
+ for k, m in f2.structure:
+ new_dict[k] = max(new_dict.get(k, 0), m)
+
+ return Fiber(list(new_dict.items()))
+
+ @staticmethod
+ def combine_selectively(f1, f2):
+ """ Combine two fiber by taking the sum of multiplicities for each degree in the first fiber """
+ # only use orders which occur in fiber f1
+ new_dict = dict(f1.structure)
+ for k in f1.degrees:
+ if k in f2.degrees:
+ new_dict[k] += f2[k]
+ return Fiber(list(new_dict.items()))
+
+ def to_attention_heads(self, tensors: Dict[str, Tensor], num_heads: int):
+ # dict(N, num_channels, 2d+1) -> (N, num_heads, -1)
+ fibers = [tensors[str(degree)].reshape(*tensors[str(degree)].shape[:-2], num_heads, -1) for degree in
+ self.degrees]
+ fibers = torch.cat(fibers, -1)
+ return fibers
diff --git a/utils/model/se3_transformer/model/layers/._attention.py b/utils/model/se3_transformer/model/layers/._attention.py
new file mode 100644
index 0000000000000000000000000000000000000000..e104c6317b4c2bd3aed2efb7c5db8556cd8d985e
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/._attention.py differ
diff --git a/utils/model/se3_transformer/model/layers/__init__.py b/utils/model/se3_transformer/model/layers/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..9eb9e3ced5ef94e9a5c5be5883cc14ebaabe31f1
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/__init__.py
@@ -0,0 +1,5 @@
+from .linear import LinearSE3
+from .norm import NormSE3
+from .pooling import GPooling
+from .convolution import ConvSE3
+from .attention import AttentionBlockSE3
\ No newline at end of file
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..b290d94285440c9f97f61ca3c6ea68c7ebcf1644
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/__init__.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..9da62209e5b13745f7e89a936519b085fd9a4e05
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/attention.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..e42942454c78b14df9935261fe61eb959245849d
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/convolution.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..1c09095ac7efe0bda53fef2fd11ccf8475d0e99f
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/linear.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..e4288c55b9b01dc2b17e125898fc76928bc729ae
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/norm.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc b/utils/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..90f9fc414b3be6c849f369d2ea9a0fd4053f201e
Binary files /dev/null and b/utils/model/se3_transformer/model/layers/__pycache__/pooling.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/model/layers/attention.py b/utils/model/se3_transformer/model/layers/attention.py
new file mode 100644
index 0000000000000000000000000000000000000000..1dc68fe95faaee99f362678399fa7e47010e1af1
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/attention.py
@@ -0,0 +1,180 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import dgl
+import numpy as np
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from dgl.ops import edge_softmax
+from torch import Tensor
+from typing import Dict, Optional, Union
+
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.model.layers.convolution import ConvSE3, ConvSE3FuseLevel
+from se3_transformer.model.layers.linear import LinearSE3
+from se3_transformer.runtime.utils import degree_to_dim, aggregate_residual, unfuse_features
+from torch.cuda.nvtx import range as nvtx_range
+
+
+class AttentionSE3(nn.Module):
+ """ Multi-headed sparse graph self-attention (SE(3)-equivariant) """
+
+ def __init__(
+ self,
+ num_heads: int,
+ key_fiber: Fiber,
+ value_fiber: Fiber
+ ):
+ """
+ :param num_heads: Number of attention heads
+ :param key_fiber: Fiber for the keys (and also for the queries)
+ :param value_fiber: Fiber for the values
+ """
+ super().__init__()
+ self.num_heads = num_heads
+ self.key_fiber = key_fiber
+ self.value_fiber = value_fiber
+
+ def forward(
+ self,
+ value: Union[Tensor, Dict[str, Tensor]], # edge features (may be fused)
+ key: Union[Tensor, Dict[str, Tensor]], # edge features (may be fused)
+ query: Dict[str, Tensor], # node features
+ graph: DGLGraph
+ ):
+ with nvtx_range('AttentionSE3'):
+ with nvtx_range('reshape keys and queries'):
+ if isinstance(key, Tensor):
+ # case where features of all types are fused
+ key = key.reshape(key.shape[0], self.num_heads, -1)
+ # need to reshape queries that way to keep the same layout as keys
+ out = torch.cat([query[str(d)] for d in self.key_fiber.degrees], dim=-1)
+ query = out.reshape(list(query.values())[0].shape[0], self.num_heads, -1)
+ else:
+ # features are not fused, need to fuse and reshape them
+ key = self.key_fiber.to_attention_heads(key, self.num_heads)
+ query = self.key_fiber.to_attention_heads(query, self.num_heads)
+
+ with nvtx_range('attention dot product + softmax'):
+ # Compute attention weights (softmax of inner product between key and query)
+ edge_weights = dgl.ops.e_dot_v(graph, key, query).squeeze(-1)
+ edge_weights /= np.sqrt(self.key_fiber.num_features)
+ edge_weights = edge_softmax(graph, edge_weights)
+ edge_weights = edge_weights[..., None, None]
+
+ with nvtx_range('weighted sum'):
+ if isinstance(value, Tensor):
+ # features of all types are fused
+ v = value.view(value.shape[0], self.num_heads, -1, value.shape[-1])
+ weights = edge_weights * v
+ feat_out = dgl.ops.copy_e_sum(graph, weights)
+ feat_out = feat_out.view(feat_out.shape[0], -1, feat_out.shape[-1]) # merge heads
+ out = unfuse_features(feat_out, self.value_fiber.degrees)
+ else:
+ out = {}
+ for degree, channels in self.value_fiber:
+ v = value[str(degree)].view(-1, self.num_heads, channels // self.num_heads,
+ degree_to_dim(degree))
+ weights = edge_weights * v
+ res = dgl.ops.copy_e_sum(graph, weights)
+ out[str(degree)] = res.view(-1, channels, degree_to_dim(degree)) # merge heads
+
+ return out
+
+
+class AttentionBlockSE3(nn.Module):
+ """ Multi-headed sparse graph self-attention block with skip connection, linear projection (SE(3)-equivariant) """
+
+ def __init__(
+ self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Optional[Fiber] = None,
+ num_heads: int = 4,
+ channels_div: int = 2,
+ use_layer_norm: bool = False,
+ max_degree: bool = 4,
+ fuse_level: ConvSE3FuseLevel = ConvSE3FuseLevel.FULL,
+ **kwargs
+ ):
+ """
+ :param fiber_in: Fiber describing the input features
+ :param fiber_out: Fiber describing the output features
+ :param fiber_edge: Fiber describing the edge features (node distances excluded)
+ :param num_heads: Number of attention heads
+ :param channels_div: Divide the channels by this integer for computing values
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param max_degree: Maximum degree used in the bases computation
+ :param fuse_level: Maximum fuse level to use in TFN convolutions
+ """
+ super().__init__()
+ if fiber_edge is None:
+ fiber_edge = Fiber({})
+ self.fiber_in = fiber_in
+ # value_fiber has same structure as fiber_out but #channels divided by 'channels_div'
+ value_fiber = Fiber([(degree, channels // channels_div) for degree, channels in fiber_out])
+ # key_query_fiber has the same structure as fiber_out, but only degrees which are in in_fiber
+ # (queries are merely projected, hence degrees have to match input)
+ key_query_fiber = Fiber([(fe.degree, fe.channels) for fe in value_fiber if fe.degree in fiber_in.degrees])
+
+ self.to_key_value = ConvSE3(fiber_in, value_fiber + key_query_fiber, pool=False, fiber_edge=fiber_edge,
+ use_layer_norm=use_layer_norm, max_degree=max_degree, fuse_level=fuse_level,
+ allow_fused_output=True)
+ self.to_query = LinearSE3(fiber_in, key_query_fiber)
+ self.attention = AttentionSE3(num_heads, key_query_fiber, value_fiber)
+ self.project = LinearSE3(value_fiber + fiber_in, fiber_out)
+
+ def forward(
+ self,
+ node_features: Dict[str, Tensor],
+ edge_features: Dict[str, Tensor],
+ graph: DGLGraph,
+ basis: Dict[str, Tensor]
+ ):
+ with nvtx_range('AttentionBlockSE3'):
+ with nvtx_range('keys / values'):
+ fused_key_value = self.to_key_value(node_features, edge_features, graph, basis)
+ key, value = self._get_key_value_from_fused(fused_key_value)
+
+ with nvtx_range('queries'):
+ query = self.to_query(node_features)
+
+ z = self.attention(value, key, query, graph)
+ z_concat = aggregate_residual(node_features, z, 'cat')
+ return self.project(z_concat)
+
+ def _get_key_value_from_fused(self, fused_key_value):
+ # Extract keys and queries features from fused features
+ if isinstance(fused_key_value, Tensor):
+ # Previous layer was a fully fused convolution
+ value, key = torch.chunk(fused_key_value, chunks=2, dim=-2)
+ else:
+ key, value = {}, {}
+ for degree, feat in fused_key_value.items():
+ if int(degree) in self.fiber_in.degrees:
+ value[degree], key[degree] = torch.chunk(feat, chunks=2, dim=-2)
+ else:
+ value[degree] = feat
+
+ return key, value
diff --git a/utils/model/se3_transformer/model/layers/convolution.py b/utils/model/se3_transformer/model/layers/convolution.py
new file mode 100644
index 0000000000000000000000000000000000000000..fcbb1c22dd8e7b14ab61e717ddbe6e0a19164137
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/convolution.py
@@ -0,0 +1,335 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from enum import Enum
+from itertools import product
+from typing import Dict
+
+import dgl
+import numpy as np
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.runtime.utils import degree_to_dim, unfuse_features
+
+
+class ConvSE3FuseLevel(Enum):
+ """
+ Enum to select a maximum level of fusing optimizations that will be applied when certain conditions are met.
+ If a desired level L is picked and the level L cannot be applied to a level, other fused ops < L are considered.
+ A higher level means faster training, but also more memory usage.
+ If you are tight on memory and want to feed large inputs to the network, choose a low value.
+ If you want to train fast, choose a high value.
+ Recommended value is FULL with AMP.
+
+ Fully fused TFN convolutions requirements:
+ - all input channels are the same
+ - all output channels are the same
+ - input degrees span the range [0, ..., max_degree]
+ - output degrees span the range [0, ..., max_degree]
+
+ Partially fused TFN convolutions requirements:
+ * For fusing by output degree:
+ - all input channels are the same
+ - input degrees span the range [0, ..., max_degree]
+ * For fusing by input degree:
+ - all output channels are the same
+ - output degrees span the range [0, ..., max_degree]
+
+ Original TFN pairwise convolutions: no requirements
+ """
+
+ FULL = 2
+ PARTIAL = 1
+ NONE = 0
+
+
+class RadialProfile(nn.Module):
+ """
+ Radial profile function.
+ Outputs weights used to weigh basis matrices in order to get convolution kernels.
+ In TFN notation: $R^{l,k}$
+ In SE(3)-Transformer notation: $\phi^{l,k}$
+
+ Note:
+ In the original papers, this function only depends on relative node distances ||x||.
+ Here, we allow this function to also take as input additional invariant edge features.
+ This does not break equivariance and adds expressive power to the model.
+
+ Diagram:
+ invariant edge features (node distances included) ───> MLP layer (shared across edges) ───> radial weights
+ """
+
+ def __init__(
+ self,
+ num_freq: int,
+ channels_in: int,
+ channels_out: int,
+ edge_dim: int = 1,
+ mid_dim: int = 32,
+ use_layer_norm: bool = False
+ ):
+ """
+ :param num_freq: Number of frequencies
+ :param channels_in: Number of input channels
+ :param channels_out: Number of output channels
+ :param edge_dim: Number of invariant edge features (input to the radial function)
+ :param mid_dim: Size of the hidden MLP layers
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ """
+ super().__init__()
+ modules = [
+ nn.Linear(edge_dim, mid_dim),
+ nn.LayerNorm(mid_dim) if use_layer_norm else None,
+ nn.ReLU(),
+ nn.Linear(mid_dim, mid_dim),
+ nn.LayerNorm(mid_dim) if use_layer_norm else None,
+ nn.ReLU(),
+ nn.Linear(mid_dim, num_freq * channels_in * channels_out, bias=False)
+ ]
+
+ self.net = nn.Sequential(*[m for m in modules if m is not None])
+
+ def forward(self, features: Tensor) -> Tensor:
+ return self.net(features)
+
+
+class VersatileConvSE3(nn.Module):
+ """
+ Building block for TFN convolutions.
+ This single module can be used for fully fused convolutions, partially fused convolutions, or pairwise convolutions.
+ """
+
+ def __init__(self,
+ freq_sum: int,
+ channels_in: int,
+ channels_out: int,
+ edge_dim: int,
+ use_layer_norm: bool,
+ fuse_level: ConvSE3FuseLevel):
+ super().__init__()
+ self.freq_sum = freq_sum
+ self.channels_out = channels_out
+ self.channels_in = channels_in
+ self.fuse_level = fuse_level
+ self.radial_func = RadialProfile(num_freq=freq_sum,
+ channels_in=channels_in,
+ channels_out=channels_out,
+ edge_dim=edge_dim,
+ use_layer_norm=use_layer_norm)
+
+ def forward(self, features: Tensor, invariant_edge_feats: Tensor, basis: Tensor):
+ with nvtx_range(f'VersatileConvSE3'):
+ num_edges = features.shape[0]
+ in_dim = features.shape[2]
+ with nvtx_range(f'RadialProfile'):
+ radial_weights = self.radial_func(invariant_edge_feats) \
+ .view(-1, self.channels_out, self.channels_in * self.freq_sum)
+
+ if basis is not None:
+ # This block performs the einsum n i l, n o i f, n l f k -> n o k
+ out_dim = basis.shape[-1]
+ if self.fuse_level != ConvSE3FuseLevel.FULL:
+ out_dim += out_dim % 2 - 1 # Account for padded basis
+ basis_view = basis.view(num_edges, in_dim, -1)
+ tmp = (features @ basis_view).view(num_edges, -1, basis.shape[-1])
+ return (radial_weights @ tmp)[:, :, :out_dim]
+ else:
+ # k = l = 0 non-fused case
+ return radial_weights @ features
+
+
+class ConvSE3(nn.Module):
+ """
+ SE(3)-equivariant graph convolution (Tensor Field Network convolution).
+ This convolution can map an arbitrary input Fiber to an arbitrary output Fiber, while preserving equivariance.
+ Features of different degrees interact together to produce output features.
+
+ Note 1:
+ The option is given to not pool the output. This means that the convolution sum over neighbors will not be
+ done, and the returned features will be edge features instead of node features.
+
+ Note 2:
+ Unlike the original paper and implementation, this convolution can handle edge feature of degree greater than 0.
+ Input edge features are concatenated with input source node features before the kernel is applied.
+ """
+
+ def __init__(
+ self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Fiber,
+ pool: bool = True,
+ use_layer_norm: bool = False,
+ self_interaction: bool = False,
+ max_degree: int = 4,
+ fuse_level: ConvSE3FuseLevel = ConvSE3FuseLevel.FULL,
+ allow_fused_output: bool = False
+ ):
+ """
+ :param fiber_in: Fiber describing the input features
+ :param fiber_out: Fiber describing the output features
+ :param fiber_edge: Fiber describing the edge features (node distances excluded)
+ :param pool: If True, compute final node features by averaging incoming edge features
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param self_interaction: Apply self-interaction of nodes
+ :param max_degree: Maximum degree used in the bases computation
+ :param fuse_level: Maximum fuse level to use in TFN convolutions
+ :param allow_fused_output: Allow the module to output a fused representation of features
+ """
+ super().__init__()
+ self.pool = pool
+ self.fiber_in = fiber_in
+ self.fiber_out = fiber_out
+ self.self_interaction = self_interaction
+ self.max_degree = max_degree
+ self.allow_fused_output = allow_fused_output
+
+ # channels_in: account for the concatenation of edge features
+ channels_in_set = set([f.channels + fiber_edge[f.degree] * (f.degree > 0) for f in self.fiber_in])
+ channels_out_set = set([f.channels for f in self.fiber_out])
+ unique_channels_in = (len(channels_in_set) == 1)
+ unique_channels_out = (len(channels_out_set) == 1)
+ degrees_up_to_max = list(range(max_degree + 1))
+ common_args = dict(edge_dim=fiber_edge[0] + 1, use_layer_norm=use_layer_norm)
+
+ if fuse_level.value >= ConvSE3FuseLevel.FULL.value and \
+ unique_channels_in and fiber_in.degrees == degrees_up_to_max and \
+ unique_channels_out and fiber_out.degrees == degrees_up_to_max:
+ # Single fused convolution
+ self.used_fuse_level = ConvSE3FuseLevel.FULL
+
+ sum_freq = sum([
+ degree_to_dim(min(d_in, d_out))
+ for d_in, d_out in product(degrees_up_to_max, degrees_up_to_max)
+ ])
+
+ self.conv = VersatileConvSE3(sum_freq, list(channels_in_set)[0], list(channels_out_set)[0],
+ fuse_level=self.used_fuse_level, **common_args)
+
+ elif fuse_level.value >= ConvSE3FuseLevel.PARTIAL.value and \
+ unique_channels_in and fiber_in.degrees == degrees_up_to_max:
+ # Convolutions fused per output degree
+ self.used_fuse_level = ConvSE3FuseLevel.PARTIAL
+ self.conv_out = nn.ModuleDict()
+ for d_out, c_out in fiber_out:
+ sum_freq = sum([degree_to_dim(min(d_out, d)) for d in fiber_in.degrees])
+ self.conv_out[str(d_out)] = VersatileConvSE3(sum_freq, list(channels_in_set)[0], c_out,
+ fuse_level=self.used_fuse_level, **common_args)
+
+ elif fuse_level.value >= ConvSE3FuseLevel.PARTIAL.value and \
+ unique_channels_out and fiber_out.degrees == degrees_up_to_max:
+ # Convolutions fused per input degree
+ self.used_fuse_level = ConvSE3FuseLevel.PARTIAL
+ self.conv_in = nn.ModuleDict()
+ for d_in, c_in in fiber_in:
+ sum_freq = sum([degree_to_dim(min(d_in, d)) for d in fiber_out.degrees])
+ self.conv_in[str(d_in)] = VersatileConvSE3(sum_freq, c_in, list(channels_out_set)[0],
+ fuse_level=ConvSE3FuseLevel.FULL, **common_args)
+ else:
+ # Use pairwise TFN convolutions
+ self.used_fuse_level = ConvSE3FuseLevel.NONE
+ self.conv = nn.ModuleDict()
+ for (degree_in, channels_in), (degree_out, channels_out) in (self.fiber_in * self.fiber_out):
+ dict_key = f'{degree_in},{degree_out}'
+ channels_in_new = channels_in + fiber_edge[degree_in] * (degree_in > 0)
+ sum_freq = degree_to_dim(min(degree_in, degree_out))
+ self.conv[dict_key] = VersatileConvSE3(sum_freq, channels_in_new, channels_out,
+ fuse_level=self.used_fuse_level, **common_args)
+
+ if self_interaction:
+ self.to_kernel_self = nn.ParameterDict()
+ for degree_out, channels_out in fiber_out:
+ if fiber_in[degree_out]:
+ self.to_kernel_self[str(degree_out)] = nn.Parameter(
+ torch.randn(channels_out, fiber_in[degree_out]) / np.sqrt(fiber_in[degree_out]))
+
+ def forward(
+ self,
+ node_feats: Dict[str, Tensor],
+ edge_feats: Dict[str, Tensor],
+ graph: DGLGraph,
+ basis: Dict[str, Tensor]
+ ):
+ with nvtx_range(f'ConvSE3'):
+ invariant_edge_feats = edge_feats['0'].squeeze(-1)
+ src, dst = graph.edges()
+ out = {}
+ in_features = []
+
+ # Fetch all input features from edge and node features
+ for degree_in in self.fiber_in.degrees:
+ src_node_features = node_feats[str(degree_in)][src]
+ if degree_in > 0 and str(degree_in) in edge_feats:
+ # Handle edge features of any type by concatenating them to node features
+ src_node_features = torch.cat([src_node_features, edge_feats[str(degree_in)]], dim=1)
+ in_features.append(src_node_features)
+
+ if self.used_fuse_level == ConvSE3FuseLevel.FULL:
+ in_features_fused = torch.cat(in_features, dim=-1)
+ out = self.conv(in_features_fused, invariant_edge_feats, basis['fully_fused'])
+
+ if not self.allow_fused_output or self.self_interaction or self.pool:
+ out = unfuse_features(out, self.fiber_out.degrees)
+
+ elif self.used_fuse_level == ConvSE3FuseLevel.PARTIAL and hasattr(self, 'conv_out'):
+ in_features_fused = torch.cat(in_features, dim=-1)
+ for degree_out in self.fiber_out.degrees:
+ out[str(degree_out)] = self.conv_out[str(degree_out)](in_features_fused, invariant_edge_feats,
+ basis[f'out{degree_out}_fused'])
+
+ elif self.used_fuse_level == ConvSE3FuseLevel.PARTIAL and hasattr(self, 'conv_in'):
+ out = 0
+ for degree_in, feature in zip(self.fiber_in.degrees, in_features):
+ out += self.conv_in[str(degree_in)](feature, invariant_edge_feats,
+ basis[f'in{degree_in}_fused'])
+ if not self.allow_fused_output or self.self_interaction or self.pool:
+ out = unfuse_features(out, self.fiber_out.degrees)
+ else:
+ # Fallback to pairwise TFN convolutions
+ for degree_out in self.fiber_out.degrees:
+ out_feature = 0
+ for degree_in, feature in zip(self.fiber_in.degrees, in_features):
+ dict_key = f'{degree_in},{degree_out}'
+ out_feature = out_feature + self.conv[dict_key](feature, invariant_edge_feats,
+ basis.get(dict_key, None))
+ out[str(degree_out)] = out_feature
+
+ for degree_out in self.fiber_out.degrees:
+ if self.self_interaction and str(degree_out) in self.to_kernel_self:
+ with nvtx_range(f'self interaction'):
+ dst_features = node_feats[str(degree_out)][dst]
+ kernel_self = self.to_kernel_self[str(degree_out)]
+ out[str(degree_out)] += kernel_self @ dst_features
+
+ if self.pool:
+ with nvtx_range(f'pooling'):
+ if isinstance(out, dict):
+ out[str(degree_out)] = dgl.ops.copy_e_sum(graph, out[str(degree_out)])
+ else:
+ out = dgl.ops.copy_e_sum(graph, out)
+ return out
diff --git a/utils/model/se3_transformer/model/layers/linear.py b/utils/model/se3_transformer/model/layers/linear.py
new file mode 100644
index 0000000000000000000000000000000000000000..f720d77ecc540423a6a6545f9e50c117ad1c08db
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/linear.py
@@ -0,0 +1,59 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from typing import Dict
+
+import numpy as np
+import torch
+import torch.nn as nn
+from torch import Tensor
+
+from se3_transformer.model.fiber import Fiber
+
+
+class LinearSE3(nn.Module):
+ """
+ Graph Linear SE(3)-equivariant layer, equivalent to a 1x1 convolution.
+ Maps a fiber to a fiber with the same degrees (channels may be different).
+ No interaction between degrees, but interaction between channels.
+
+ type-0 features (C_0 channels) ────> Linear(bias=False) ────> type-0 features (C'_0 channels)
+ type-1 features (C_1 channels) ────> Linear(bias=False) ────> type-1 features (C'_1 channels)
+ :
+ type-k features (C_k channels) ────> Linear(bias=False) ────> type-k features (C'_k channels)
+ """
+
+ def __init__(self, fiber_in: Fiber, fiber_out: Fiber):
+ super().__init__()
+ self.weights = nn.ParameterDict({
+ str(degree_out): nn.Parameter(
+ torch.randn(channels_out, fiber_in[degree_out]) / np.sqrt(fiber_in[degree_out]))
+ for degree_out, channels_out in fiber_out
+ })
+
+ def forward(self, features: Dict[str, Tensor], *args, **kwargs) -> Dict[str, Tensor]:
+ return {
+ degree: self.weights[degree] @ features[degree]
+ for degree, weight in self.weights.items()
+ }
diff --git a/utils/model/se3_transformer/model/layers/norm.py b/utils/model/se3_transformer/model/layers/norm.py
new file mode 100644
index 0000000000000000000000000000000000000000..acbe23d774d50c26e22a60336a3d05b66a164eb3
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/norm.py
@@ -0,0 +1,83 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+
+from typing import Dict
+
+import torch
+import torch.nn as nn
+from torch import Tensor
+from torch.cuda.nvtx import range as nvtx_range
+
+from se3_transformer.model.fiber import Fiber
+
+
+class NormSE3(nn.Module):
+ """
+ Norm-based SE(3)-equivariant nonlinearity.
+
+ ┌──> feature_norm ──> LayerNorm() ──> ReLU() ──┐
+ feature_in ──┤ * ──> feature_out
+ └──> feature_phase ────────────────────────────┘
+ """
+
+ NORM_CLAMP = 2 ** -24 # Minimum positive subnormal for FP16
+
+ def __init__(self, fiber: Fiber, nonlinearity: nn.Module = nn.ReLU()):
+ super().__init__()
+ self.fiber = fiber
+ self.nonlinearity = nonlinearity
+
+ if len(set(fiber.channels)) == 1:
+ # Fuse all the layer normalizations into a group normalization
+ self.group_norm = nn.GroupNorm(num_groups=len(fiber.degrees), num_channels=sum(fiber.channels))
+ else:
+ # Use multiple layer normalizations
+ self.layer_norms = nn.ModuleDict({
+ str(degree): nn.LayerNorm(channels)
+ for degree, channels in fiber
+ })
+
+ def forward(self, features: Dict[str, Tensor], *args, **kwargs) -> Dict[str, Tensor]:
+ with nvtx_range('NormSE3'):
+ output = {}
+ if hasattr(self, 'group_norm'):
+ # Compute per-degree norms of features
+ norms = [features[str(d)].norm(dim=-1, keepdim=True).clamp(min=self.NORM_CLAMP)
+ for d in self.fiber.degrees]
+ fused_norms = torch.cat(norms, dim=-2)
+
+ # Transform the norms only
+ new_norms = self.nonlinearity(self.group_norm(fused_norms.squeeze(-1))).unsqueeze(-1)
+ new_norms = torch.chunk(new_norms, chunks=len(self.fiber.degrees), dim=-2)
+
+ # Scale features to the new norms
+ for norm, new_norm, d in zip(norms, new_norms, self.fiber.degrees):
+ output[str(d)] = features[str(d)] / norm * new_norm
+ else:
+ for degree, feat in features.items():
+ norm = feat.norm(dim=-1, keepdim=True).clamp(min=self.NORM_CLAMP)
+ new_norm = self.nonlinearity(self.layer_norms[degree](norm.squeeze(-1)).unsqueeze(-1))
+ output[degree] = new_norm * feat / norm
+
+ return output
diff --git a/utils/model/se3_transformer/model/layers/pooling.py b/utils/model/se3_transformer/model/layers/pooling.py
new file mode 100644
index 0000000000000000000000000000000000000000..e42c5383ba3239e3d93c928fa83a61a9e19b9437
--- /dev/null
+++ b/utils/model/se3_transformer/model/layers/pooling.py
@@ -0,0 +1,53 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from typing import Dict, Literal
+
+import torch.nn as nn
+from dgl import DGLGraph
+from dgl.nn.pytorch import AvgPooling, MaxPooling
+from torch import Tensor
+
+
+class GPooling(nn.Module):
+ """
+ Graph max/average pooling on a given feature type.
+ The average can be taken for any feature type, and equivariance will be maintained.
+ The maximum can only be taken for invariant features (type 0).
+ If you want max-pooling for type > 0 features, look into Vector Neurons.
+ """
+
+ def __init__(self, feat_type: int = 0, pool: Literal['max', 'avg'] = 'max'):
+ """
+ :param feat_type: Feature type to pool
+ :param pool: Type of pooling: max or avg
+ """
+ super().__init__()
+ assert pool in ['max', 'avg'], f'Unknown pooling: {pool}'
+ assert feat_type == 0 or pool == 'avg', 'Max pooling on type > 0 features will break equivariance'
+ self.feat_type = feat_type
+ self.pool = MaxPooling() if pool == 'max' else AvgPooling()
+
+ def forward(self, features: Dict[str, Tensor], graph: DGLGraph, **kwargs) -> Tensor:
+ pooled = self.pool(graph, features[str(self.feat_type)])
+ return pooled.squeeze(dim=-1)
diff --git a/utils/model/se3_transformer/model/transformer.py b/utils/model/se3_transformer/model/transformer.py
new file mode 100644
index 0000000000000000000000000000000000000000..ab89f77a12b8b40b16c402647059260606c98c60
--- /dev/null
+++ b/utils/model/se3_transformer/model/transformer.py
@@ -0,0 +1,222 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+from typing import Optional, Literal, Dict
+
+import torch
+import torch.nn as nn
+from dgl import DGLGraph
+from torch import Tensor
+
+from se3_transformer.model.basis import get_basis, update_basis_with_fused
+from se3_transformer.model.layers.attention import AttentionBlockSE3
+from se3_transformer.model.layers.convolution import ConvSE3, ConvSE3FuseLevel
+from se3_transformer.model.layers.norm import NormSE3
+from se3_transformer.model.layers.pooling import GPooling
+from se3_transformer.runtime.utils import str2bool
+from se3_transformer.model.fiber import Fiber
+
+
+class Sequential(nn.Sequential):
+ """ Sequential module with arbitrary forward args and kwargs. Used to pass graph, basis and edge features. """
+
+ def forward(self, input, *args, **kwargs):
+ for module in self:
+ input = module(input, *args, **kwargs)
+ return input
+
+
+def get_populated_edge_features(relative_pos: Tensor, edge_features: Optional[Dict[str, Tensor]] = None):
+ """ Add relative positions to existing edge features """
+ edge_features = edge_features.copy() if edge_features else {}
+ r = relative_pos.norm(dim=-1, keepdim=True)
+ if '0' in edge_features:
+ edge_features['0'] = torch.cat([edge_features['0'], r[..., None]], dim=1)
+ else:
+ edge_features['0'] = r[..., None]
+
+ return edge_features
+
+
+class SE3Transformer(nn.Module):
+ def __init__(self,
+ num_layers: int,
+ fiber_in: Fiber,
+ fiber_hidden: Fiber,
+ fiber_out: Fiber,
+ num_heads: int,
+ channels_div: int,
+ fiber_edge: Fiber = Fiber({}),
+ return_type: Optional[int] = None,
+ pooling: Optional[Literal['avg', 'max']] = None,
+ norm: bool = True,
+ use_layer_norm: bool = True,
+ tensor_cores: bool = False,
+ low_memory: bool = False,
+ **kwargs):
+ """
+ :param num_layers: Number of attention layers
+ :param fiber_in: Input fiber description
+ :param fiber_hidden: Hidden fiber description
+ :param fiber_out: Output fiber description
+ :param fiber_edge: Input edge fiber description
+ :param num_heads: Number of attention heads
+ :param channels_div: Channels division before feeding to attention layer
+ :param return_type: Return only features of this type
+ :param pooling: 'avg' or 'max' graph pooling before MLP layers
+ :param norm: Apply a normalization layer after each attention block
+ :param use_layer_norm: Apply layer normalization between MLP layers
+ :param tensor_cores: True if using Tensor Cores (affects the use of fully fused convs, and padded bases)
+ :param low_memory: If True, will use slower ops that use less memory
+ """
+ super().__init__()
+ self.num_layers = num_layers
+ self.fiber_edge = fiber_edge
+ self.num_heads = num_heads
+ self.channels_div = channels_div
+ self.return_type = return_type
+ self.pooling = pooling
+ self.max_degree = max(*fiber_in.degrees, *fiber_hidden.degrees, *fiber_out.degrees)
+ self.tensor_cores = tensor_cores
+ self.low_memory = low_memory
+
+ if low_memory and not tensor_cores:
+ logging.warning('Low memory mode will have no effect with no Tensor Cores')
+
+ # Fully fused convolutions when using Tensor Cores (and not low memory mode)
+ fuse_level = ConvSE3FuseLevel.FULL if tensor_cores and not low_memory else ConvSE3FuseLevel.PARTIAL
+
+ graph_modules = []
+ for i in range(num_layers):
+ graph_modules.append(AttentionBlockSE3(fiber_in=fiber_in,
+ fiber_out=fiber_hidden,
+ fiber_edge=fiber_edge,
+ num_heads=num_heads,
+ channels_div=channels_div,
+ use_layer_norm=use_layer_norm,
+ max_degree=self.max_degree,
+ fuse_level=fuse_level))
+ if norm:
+ graph_modules.append(NormSE3(fiber_hidden))
+ fiber_in = fiber_hidden
+
+ graph_modules.append(ConvSE3(fiber_in=fiber_in,
+ fiber_out=fiber_out,
+ fiber_edge=fiber_edge,
+ self_interaction=True,
+ use_layer_norm=use_layer_norm,
+ max_degree=self.max_degree))
+ self.graph_modules = Sequential(*graph_modules)
+
+ if pooling is not None:
+ assert return_type is not None, 'return_type must be specified when pooling'
+ self.pooling_module = GPooling(pool=pooling, feat_type=return_type)
+
+ def forward(self, graph: DGLGraph, node_feats: Dict[str, Tensor],
+ edge_feats: Optional[Dict[str, Tensor]] = None,
+ basis: Optional[Dict[str, Tensor]] = None):
+ # Compute bases in case they weren't precomputed as part of the data loading
+ basis = basis or get_basis(graph.edata['rel_pos'], max_degree=self.max_degree, compute_gradients=False,
+ use_pad_trick=self.tensor_cores and not self.low_memory,
+ amp=torch.is_autocast_enabled())
+
+ # Add fused bases (per output degree, per input degree, and fully fused) to the dict
+ basis = update_basis_with_fused(basis, self.max_degree, use_pad_trick=self.tensor_cores and not self.low_memory,
+ fully_fused=self.tensor_cores and not self.low_memory)
+
+ edge_feats = get_populated_edge_features(graph.edata['rel_pos'], edge_feats)
+
+ node_feats = self.graph_modules(node_feats, edge_feats, graph=graph, basis=basis)
+
+ if self.pooling is not None:
+ return self.pooling_module(node_feats, graph=graph)
+
+ if self.return_type is not None:
+ return node_feats[str(self.return_type)]
+
+ return node_feats
+
+ @staticmethod
+ def add_argparse_args(parser):
+ parser.add_argument('--num_layers', type=int, default=7,
+ help='Number of stacked Transformer layers')
+ parser.add_argument('--num_heads', type=int, default=8,
+ help='Number of heads in self-attention')
+ parser.add_argument('--channels_div', type=int, default=2,
+ help='Channels division before feeding to attention layer')
+ parser.add_argument('--pooling', type=str, default=None, const=None, nargs='?', choices=['max', 'avg'],
+ help='Type of graph pooling')
+ parser.add_argument('--norm', type=str2bool, nargs='?', const=True, default=False,
+ help='Apply a normalization layer after each attention block')
+ parser.add_argument('--use_layer_norm', type=str2bool, nargs='?', const=True, default=False,
+ help='Apply layer normalization between MLP layers')
+ parser.add_argument('--low_memory', type=str2bool, nargs='?', const=True, default=False,
+ help='If true, will use fused ops that are slower but that use less memory '
+ '(expect 25 percent less memory). '
+ 'Only has an effect if AMP is enabled on Volta GPUs, or if running on Ampere GPUs')
+
+ return parser
+
+
+class SE3TransformerPooled(nn.Module):
+ def __init__(self,
+ fiber_in: Fiber,
+ fiber_out: Fiber,
+ fiber_edge: Fiber,
+ num_degrees: int,
+ num_channels: int,
+ output_dim: int,
+ **kwargs):
+ super().__init__()
+ kwargs['pooling'] = kwargs['pooling'] or 'max'
+ self.transformer = SE3Transformer(
+ fiber_in=fiber_in,
+ fiber_hidden=Fiber.create(num_degrees, num_channels),
+ fiber_out=fiber_out,
+ fiber_edge=fiber_edge,
+ return_type=0,
+ **kwargs
+ )
+
+ n_out_features = fiber_out.num_features
+ self.mlp = nn.Sequential(
+ nn.Linear(n_out_features, n_out_features),
+ nn.ReLU(),
+ nn.Linear(n_out_features, output_dim)
+ )
+
+ def forward(self, graph, node_feats, edge_feats, basis=None):
+ feats = self.transformer(graph, node_feats, edge_feats, basis).squeeze(-1)
+ y = self.mlp(feats).squeeze(-1)
+ return y
+
+ @staticmethod
+ def add_argparse_args(parent_parser):
+ parser = parent_parser.add_argument_group("Model architecture")
+ SE3Transformer.add_argparse_args(parser)
+ parser.add_argument('--num_degrees',
+ help='Number of degrees to use. Hidden features will have types [0, ..., num_degrees - 1]',
+ type=int, default=4)
+ parser.add_argument('--num_channels', help='Number of channels for the hidden features', type=int, default=32)
+ return parent_parser
diff --git a/utils/model/se3_transformer/runtime/__init__.py b/utils/model/se3_transformer/runtime/__init__.py
new file mode 100644
index 0000000000000000000000000000000000000000..e69de29bb2d1d6434b8b29ae775ad8c2e48c5391
diff --git a/utils/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc b/utils/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..2df9df1f20c3e458717d3abe34924e313d57d1b3
Binary files /dev/null and b/utils/model/se3_transformer/runtime/__pycache__/__init__.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc b/utils/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..c77046360b569d12450ee583b5b0338545860ba1
Binary files /dev/null and b/utils/model/se3_transformer/runtime/__pycache__/utils.cpython-310.pyc differ
diff --git a/utils/model/se3_transformer/runtime/arguments.py b/utils/model/se3_transformer/runtime/arguments.py
new file mode 100644
index 0000000000000000000000000000000000000000..d35d5ee1d05a150ea40c411ec47c633213ca9d9f
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/arguments.py
@@ -0,0 +1,70 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import argparse
+import pathlib
+
+from se3_transformer.data_loading import QM9DataModule
+from se3_transformer.model import SE3TransformerPooled
+from se3_transformer.runtime.utils import str2bool
+
+PARSER = argparse.ArgumentParser(description='SE(3)-Transformer')
+
+paths = PARSER.add_argument_group('Paths')
+paths.add_argument('--data_dir', type=pathlib.Path, default=pathlib.Path('./data'),
+ help='Directory where the data is located or should be downloaded')
+paths.add_argument('--log_dir', type=pathlib.Path, default=pathlib.Path('/results'),
+ help='Directory where the results logs should be saved')
+paths.add_argument('--dllogger_name', type=str, default='dllogger_results.json',
+ help='Name for the resulting DLLogger JSON file')
+paths.add_argument('--save_ckpt_path', type=pathlib.Path, default=None,
+ help='File where the checkpoint should be saved')
+paths.add_argument('--load_ckpt_path', type=pathlib.Path, default=None,
+ help='File of the checkpoint to be loaded')
+
+optimizer = PARSER.add_argument_group('Optimizer')
+optimizer.add_argument('--optimizer', choices=['adam', 'sgd', 'lamb'], default='adam')
+optimizer.add_argument('--learning_rate', '--lr', dest='learning_rate', type=float, default=0.002)
+optimizer.add_argument('--min_learning_rate', '--min_lr', dest='min_learning_rate', type=float, default=None)
+optimizer.add_argument('--momentum', type=float, default=0.9)
+optimizer.add_argument('--weight_decay', type=float, default=0.1)
+
+PARSER.add_argument('--epochs', type=int, default=100, help='Number of training epochs')
+PARSER.add_argument('--batch_size', type=int, default=240, help='Batch size')
+PARSER.add_argument('--seed', type=int, default=None, help='Set a seed globally')
+PARSER.add_argument('--num_workers', type=int, default=8, help='Number of dataloading workers')
+
+PARSER.add_argument('--amp', type=str2bool, nargs='?', const=True, default=False, help='Use Automatic Mixed Precision')
+PARSER.add_argument('--gradient_clip', type=float, default=None, help='Clipping of the gradient norms')
+PARSER.add_argument('--accumulate_grad_batches', type=int, default=1, help='Gradient accumulation')
+PARSER.add_argument('--ckpt_interval', type=int, default=-1, help='Save a checkpoint every N epochs')
+PARSER.add_argument('--eval_interval', dest='eval_interval', type=int, default=1,
+ help='Do an evaluation round every N epochs')
+PARSER.add_argument('--silent', type=str2bool, nargs='?', const=True, default=False,
+ help='Minimize stdout output')
+
+PARSER.add_argument('--benchmark', type=str2bool, nargs='?', const=True, default=False,
+ help='Benchmark mode')
+
+QM9DataModule.add_argparse_args(PARSER)
+SE3TransformerPooled.add_argparse_args(PARSER)
diff --git a/utils/model/se3_transformer/runtime/callbacks.py b/utils/model/se3_transformer/runtime/callbacks.py
new file mode 100644
index 0000000000000000000000000000000000000000..b49d670c8305b3c4dea53ebf5c428cac628f2cf7
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/callbacks.py
@@ -0,0 +1,160 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+import time
+from abc import ABC, abstractmethod
+from typing import Optional
+
+import numpy as np
+import torch
+
+from se3_transformer.runtime.loggers import Logger
+from se3_transformer.runtime.metrics import MeanAbsoluteError
+
+
+class BaseCallback(ABC):
+ def on_fit_start(self, optimizer, args):
+ pass
+
+ def on_fit_end(self):
+ pass
+
+ def on_epoch_end(self):
+ pass
+
+ def on_batch_start(self):
+ pass
+
+ def on_validation_step(self, input, target, pred):
+ pass
+
+ def on_validation_end(self, epoch=None):
+ pass
+
+ def on_checkpoint_load(self, checkpoint):
+ pass
+
+ def on_checkpoint_save(self, checkpoint):
+ pass
+
+
+class LRSchedulerCallback(BaseCallback):
+ def __init__(self, logger: Optional[Logger] = None):
+ self.logger = logger
+ self.scheduler = None
+
+ @abstractmethod
+ def get_scheduler(self, optimizer, args):
+ pass
+
+ def on_fit_start(self, optimizer, args):
+ self.scheduler = self.get_scheduler(optimizer, args)
+
+ def on_checkpoint_load(self, checkpoint):
+ self.scheduler.load_state_dict(checkpoint['scheduler_state_dict'])
+
+ def on_checkpoint_save(self, checkpoint):
+ checkpoint['scheduler_state_dict'] = self.scheduler.state_dict()
+
+ def on_epoch_end(self):
+ if self.logger is not None:
+ self.logger.log_metrics({'learning rate': self.scheduler.get_last_lr()[0]}, step=self.scheduler.last_epoch)
+ self.scheduler.step()
+
+
+class QM9MetricCallback(BaseCallback):
+ """ Logs the rescaled mean absolute error for QM9 regression tasks """
+
+ def __init__(self, logger, targets_std, prefix=''):
+ self.mae = MeanAbsoluteError()
+ self.logger = logger
+ self.targets_std = targets_std
+ self.prefix = prefix
+ self.best_mae = float('inf')
+
+ def on_validation_step(self, input, target, pred):
+ self.mae(pred.detach(), target.detach())
+
+ def on_validation_end(self, epoch=None):
+ mae = self.mae.compute() * self.targets_std
+ logging.info(f'{self.prefix} MAE: {mae}')
+ self.logger.log_metrics({f'{self.prefix} MAE': mae}, epoch)
+ self.best_mae = min(self.best_mae, mae)
+
+ def on_fit_end(self):
+ if self.best_mae != float('inf'):
+ self.logger.log_metrics({f'{self.prefix} best MAE': self.best_mae})
+
+
+class QM9LRSchedulerCallback(LRSchedulerCallback):
+ def __init__(self, logger, epochs):
+ super().__init__(logger)
+ self.epochs = epochs
+
+ def get_scheduler(self, optimizer, args):
+ min_lr = args.min_learning_rate if args.min_learning_rate else args.learning_rate / 10.0
+ return torch.optim.lr_scheduler.CosineAnnealingWarmRestarts(optimizer, self.epochs, eta_min=min_lr)
+
+
+class PerformanceCallback(BaseCallback):
+ def __init__(self, logger, batch_size: int, warmup_epochs: int = 1, mode: str = 'train'):
+ self.batch_size = batch_size
+ self.warmup_epochs = warmup_epochs
+ self.epoch = 0
+ self.timestamps = []
+ self.mode = mode
+ self.logger = logger
+
+ def on_batch_start(self):
+ if self.epoch >= self.warmup_epochs:
+ self.timestamps.append(time.time() * 1000.0)
+
+ def _log_perf(self):
+ stats = self.process_performance_stats()
+ for k, v in stats.items():
+ logging.info(f'performance {k}: {v}')
+
+ self.logger.log_metrics(stats)
+
+ def on_epoch_end(self):
+ self.epoch += 1
+
+ def on_fit_end(self):
+ if self.epoch > self.warmup_epochs:
+ self._log_perf()
+ self.timestamps = []
+
+ def process_performance_stats(self):
+ timestamps = np.asarray(self.timestamps)
+ deltas = np.diff(timestamps)
+ throughput = (self.batch_size / deltas).mean()
+ stats = {
+ f"throughput_{self.mode}": throughput,
+ f"latency_{self.mode}_mean": deltas.mean(),
+ f"total_time_{self.mode}": timestamps[-1] - timestamps[0],
+ }
+ for level in [90, 95, 99]:
+ stats.update({f"latency_{self.mode}_{level}": np.percentile(deltas, level)})
+
+ return stats
diff --git a/utils/model/se3_transformer/runtime/gpu_affinity.py b/utils/model/se3_transformer/runtime/gpu_affinity.py
new file mode 100644
index 0000000000000000000000000000000000000000..9d46215bbbcb04f87e4c8250e08b575f8e529693
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/gpu_affinity.py
@@ -0,0 +1,325 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import collections
+import itertools
+import math
+import os
+import pathlib
+import re
+
+import pynvml
+
+
+class Device:
+ # assumes nvml returns list of 64 bit ints
+ _nvml_affinity_elements = math.ceil(os.cpu_count() / 64)
+
+ def __init__(self, device_idx):
+ super().__init__()
+ self.handle = pynvml.nvmlDeviceGetHandleByIndex(device_idx)
+
+ def get_name(self):
+ return pynvml.nvmlDeviceGetName(self.handle)
+
+ def get_uuid(self):
+ return pynvml.nvmlDeviceGetUUID(self.handle)
+
+ def get_cpu_affinity(self):
+ affinity_string = ""
+ for j in pynvml.nvmlDeviceGetCpuAffinity(self.handle, Device._nvml_affinity_elements):
+ # assume nvml returns list of 64 bit ints
+ affinity_string = "{:064b}".format(j) + affinity_string
+
+ affinity_list = [int(x) for x in affinity_string]
+ affinity_list.reverse() # so core 0 is in 0th element of list
+
+ ret = [i for i, e in enumerate(affinity_list) if e != 0]
+ return ret
+
+
+def get_thread_siblings_list():
+ """
+ Returns a list of 2-element integer tuples representing pairs of
+ hyperthreading cores.
+ """
+ path = "/sys/devices/system/cpu/cpu*/topology/thread_siblings_list"
+ thread_siblings_list = []
+ pattern = re.compile(r"(\d+)\D(\d+)")
+ for fname in pathlib.Path(path[0]).glob(path[1:]):
+ with open(fname) as f:
+ content = f.read().strip()
+ res = pattern.findall(content)
+ if res:
+ pair = tuple(map(int, res[0]))
+ thread_siblings_list.append(pair)
+ return thread_siblings_list
+
+
+def check_socket_affinities(socket_affinities):
+ # sets of cores should be either identical or disjoint
+ for i, j in itertools.product(socket_affinities, socket_affinities):
+ if not set(i) == set(j) and not set(i).isdisjoint(set(j)):
+ raise RuntimeError(f"Sets of cores should be either identical or disjoint, " f"but got {i} and {j}.")
+
+
+def get_socket_affinities(nproc_per_node, exclude_unavailable_cores=True):
+ devices = [Device(i) for i in range(nproc_per_node)]
+ socket_affinities = [dev.get_cpu_affinity() for dev in devices]
+
+ if exclude_unavailable_cores:
+ available_cores = os.sched_getaffinity(0)
+ socket_affinities = [list(set(affinity) & available_cores) for affinity in socket_affinities]
+
+ check_socket_affinities(socket_affinities)
+
+ return socket_affinities
+
+
+def set_socket_affinity(gpu_id):
+ """
+ The process is assigned with all available logical CPU cores from the CPU
+ socket connected to the GPU with a given id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ dev = Device(gpu_id)
+ affinity = dev.get_cpu_affinity()
+ os.sched_setaffinity(0, affinity)
+
+
+def set_single_affinity(gpu_id):
+ """
+ The process is assigned with the first available logical CPU core from the
+ list of all CPU cores from the CPU socket connected to the GPU with a given
+ id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ dev = Device(gpu_id)
+ affinity = dev.get_cpu_affinity()
+
+ # exclude unavailable cores
+ available_cores = os.sched_getaffinity(0)
+ affinity = list(set(affinity) & available_cores)
+ os.sched_setaffinity(0, affinity[:1])
+
+
+def set_single_unique_affinity(gpu_id, nproc_per_node):
+ """
+ The process is assigned with a single unique available physical CPU core
+ from the list of all CPU cores from the CPU socket connected to the GPU with
+ a given id.
+
+ Args:
+ gpu_id: index of a GPU
+ """
+ socket_affinities = get_socket_affinities(nproc_per_node)
+
+ siblings_list = get_thread_siblings_list()
+ siblings_dict = dict(siblings_list)
+
+ # remove siblings
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities[idx] = list(set(socket_affinity) - set(siblings_dict.values()))
+
+ affinities = []
+ assigned = []
+
+ for socket_affinity in socket_affinities:
+ for core in socket_affinity:
+ if core not in assigned:
+ affinities.append([core])
+ assigned.append(core)
+ break
+ os.sched_setaffinity(0, affinities[gpu_id])
+
+
+def set_socket_unique_affinity(gpu_id, nproc_per_node, mode, balanced=True):
+ """
+ The process is assigned with an unique subset of available physical CPU
+ cores from the CPU socket connected to a GPU with a given id.
+ Assignment automatically includes hyperthreading siblings (if siblings are
+ available).
+
+ Args:
+ gpu_id: index of a GPU
+ nproc_per_node: total number of processes per node
+ mode: mode
+ balanced: assign an equal number of physical cores to each process
+ """
+ socket_affinities = get_socket_affinities(nproc_per_node)
+
+ siblings_list = get_thread_siblings_list()
+ siblings_dict = dict(siblings_list)
+
+ # remove hyperthreading siblings
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities[idx] = list(set(socket_affinity) - set(siblings_dict.values()))
+
+ socket_affinities_to_device_ids = collections.defaultdict(list)
+
+ for idx, socket_affinity in enumerate(socket_affinities):
+ socket_affinities_to_device_ids[tuple(socket_affinity)].append(idx)
+
+ # compute minimal number of physical cores per GPU across all GPUs and
+ # sockets, code assigns this number of cores per GPU if balanced == True
+ min_physical_cores_per_gpu = min(
+ [len(cores) // len(gpus) for cores, gpus in socket_affinities_to_device_ids.items()]
+ )
+
+ for socket_affinity, device_ids in socket_affinities_to_device_ids.items():
+ devices_per_group = len(device_ids)
+ if balanced:
+ cores_per_device = min_physical_cores_per_gpu
+ socket_affinity = socket_affinity[: devices_per_group * min_physical_cores_per_gpu]
+ else:
+ cores_per_device = len(socket_affinity) // devices_per_group
+
+ for group_id, device_id in enumerate(device_ids):
+ if device_id == gpu_id:
+
+ # In theory there should be no difference in performance between
+ # 'interleaved' and 'continuous' pattern on Intel-based DGX-1,
+ # but 'continuous' should be better for DGX A100 because on AMD
+ # Rome 4 consecutive cores are sharing L3 cache.
+ # TODO: code doesn't attempt to automatically detect layout of
+ # L3 cache, also external environment may already exclude some
+ # cores, this code makes no attempt to detect it and to align
+ # mapping to multiples of 4.
+
+ if mode == "interleaved":
+ affinity = list(socket_affinity[group_id::devices_per_group])
+ elif mode == "continuous":
+ affinity = list(socket_affinity[group_id * cores_per_device: (group_id + 1) * cores_per_device])
+ else:
+ raise RuntimeError("Unknown set_socket_unique_affinity mode")
+
+ # unconditionally reintroduce hyperthreading siblings, this step
+ # may result in a different numbers of logical cores assigned to
+ # each GPU even if balanced == True (if hyperthreading siblings
+ # aren't available for a subset of cores due to some external
+ # constraints, siblings are re-added unconditionally, in the
+ # worst case unavailable logical core will be ignored by
+ # os.sched_setaffinity().
+ affinity += [siblings_dict[aff] for aff in affinity if aff in siblings_dict]
+ os.sched_setaffinity(0, affinity)
+
+
+def set_affinity(gpu_id, nproc_per_node, mode="socket_unique_continuous", balanced=True):
+ """
+ The process is assigned with a proper CPU affinity which matches hardware
+ architecture on a given platform. Usually it improves and stabilizes
+ performance of deep learning training workloads.
+
+ This function assumes that the workload is running in multi-process
+ single-device mode (there are multiple training processes and each process
+ is running on a single GPU), which is typical for multi-GPU training
+ workloads using `torch.nn.parallel.DistributedDataParallel`.
+
+ Available affinity modes:
+ * 'socket' - the process is assigned with all available logical CPU cores
+ from the CPU socket connected to the GPU with a given id.
+ * 'single' - the process is assigned with the first available logical CPU
+ core from the list of all CPU cores from the CPU socket connected to the GPU
+ with a given id (multiple GPUs could be assigned with the same CPU core).
+ * 'single_unique' - the process is assigned with a single unique available
+ physical CPU core from the list of all CPU cores from the CPU socket
+ connected to the GPU with a given id.
+ * 'socket_unique_interleaved' - the process is assigned with an unique
+ subset of available physical CPU cores from the CPU socket connected to a
+ GPU with a given id, hyperthreading siblings are included automatically,
+ cores are assigned with interleaved indexing pattern
+ * 'socket_unique_continuous' - (the default) the process is assigned with an
+ unique subset of available physical CPU cores from the CPU socket connected
+ to a GPU with a given id, hyperthreading siblings are included
+ automatically, cores are assigned with continuous indexing pattern
+
+ 'socket_unique_continuous' is the recommended mode for deep learning
+ training workloads on NVIDIA DGX machines.
+
+ Args:
+ gpu_id: integer index of a GPU
+ nproc_per_node: number of processes per node
+ mode: affinity mode
+ balanced: assign an equal number of physical cores to each process,
+ affects only 'socket_unique_interleaved' and
+ 'socket_unique_continuous' affinity modes
+
+ Returns a set of logical CPU cores on which the process is eligible to run.
+
+ Example:
+
+ import argparse
+ import os
+
+ import gpu_affinity
+ import torch
+
+
+ def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument(
+ '--local_rank',
+ type=int,
+ default=os.getenv('LOCAL_RANK', 0),
+ )
+ args = parser.parse_args()
+
+ nproc_per_node = torch.cuda.device_count()
+
+ affinity = gpu_affinity.set_affinity(args.local_rank, nproc_per_node)
+ print(f'{args.local_rank}: core affinity: {affinity}')
+
+
+ if __name__ == "__main__":
+ main()
+
+ Launch the example with:
+ python -m torch.distributed.launch --nproc_per_node <#GPUs> example.py
+
+
+ WARNING: On DGX A100 only a half of CPU cores have direct access to GPUs.
+ This function restricts execution only to the CPU cores directly connected
+ to GPUs, so on DGX A100 it will limit the code to half of CPU cores and half
+ of CPU memory bandwidth (which may be fine for many DL models).
+ """
+ pynvml.nvmlInit()
+
+ if mode == "socket":
+ set_socket_affinity(gpu_id)
+ elif mode == "single":
+ set_single_affinity(gpu_id)
+ elif mode == "single_unique":
+ set_single_unique_affinity(gpu_id, nproc_per_node)
+ elif mode == "socket_unique_interleaved":
+ set_socket_unique_affinity(gpu_id, nproc_per_node, "interleaved", balanced)
+ elif mode == "socket_unique_continuous":
+ set_socket_unique_affinity(gpu_id, nproc_per_node, "continuous", balanced)
+ else:
+ raise RuntimeError("Unknown affinity mode")
+
+ affinity = os.sched_getaffinity(0)
+ return affinity
diff --git a/utils/model/se3_transformer/runtime/inference.py b/utils/model/se3_transformer/runtime/inference.py
new file mode 100644
index 0000000000000000000000000000000000000000..21e9125b24f9865b89cff29063ad997e77297d21
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/inference.py
@@ -0,0 +1,131 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from typing import List
+
+import torch
+import torch.nn as nn
+from torch.nn.parallel import DistributedDataParallel
+from torch.utils.data import DataLoader
+from tqdm import tqdm
+
+from se3_transformer.runtime import gpu_affinity
+from se3_transformer.runtime.arguments import PARSER
+from se3_transformer.runtime.callbacks import BaseCallback
+from se3_transformer.runtime.loggers import DLLogger
+from se3_transformer.runtime.utils import to_cuda, get_local_rank
+
+
+@torch.inference_mode()
+def evaluate(model: nn.Module,
+ dataloader: DataLoader,
+ callbacks: List[BaseCallback],
+ args):
+ model.eval()
+ for i, batch in tqdm(enumerate(dataloader), total=len(dataloader), unit='batch', desc=f'Evaluation',
+ leave=False, disable=(args.silent or get_local_rank() != 0)):
+ *input, target = to_cuda(batch)
+
+ for callback in callbacks:
+ callback.on_batch_start()
+
+ with torch.cuda.amp.autocast(enabled=args.amp):
+ pred = model(*input)
+
+ for callback in callbacks:
+ callback.on_validation_step(input, target, pred)
+
+
+if __name__ == '__main__':
+ from se3_transformer.runtime.callbacks import QM9MetricCallback, PerformanceCallback
+ from se3_transformer.runtime.utils import init_distributed, seed_everything
+ from se3_transformer.model import SE3TransformerPooled, Fiber
+ from se3_transformer.data_loading import QM9DataModule
+ import torch.distributed as dist
+ import logging
+ import sys
+
+ is_distributed = init_distributed()
+ local_rank = get_local_rank()
+ args = PARSER.parse_args()
+
+ logging.getLogger().setLevel(logging.CRITICAL if local_rank != 0 or args.silent else logging.INFO)
+
+ logging.info('====== SE(3)-Transformer ======')
+ logging.info('| Inference on the test set |')
+ logging.info('===============================')
+
+ if not args.benchmark and args.load_ckpt_path is None:
+ logging.error('No load_ckpt_path provided, you need to provide a saved model to evaluate')
+ sys.exit(1)
+
+ if args.benchmark:
+ logging.info('Running benchmark mode with one warmup pass')
+
+ if args.seed is not None:
+ seed_everything(args.seed)
+
+ major_cc, minor_cc = torch.cuda.get_device_capability()
+
+ logger = DLLogger(args.log_dir, filename=args.dllogger_name)
+ datamodule = QM9DataModule(**vars(args))
+ model = SE3TransformerPooled(
+ fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}),
+ fiber_out=Fiber({0: args.num_degrees * args.num_channels}),
+ fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}),
+ output_dim=1,
+ tensor_cores=(args.amp and major_cc >= 7) or major_cc >= 8, # use Tensor Cores more effectively
+ **vars(args)
+ )
+ callbacks = [QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix='test')]
+
+ model.to(device=torch.cuda.current_device())
+ if args.load_ckpt_path is not None:
+ checkpoint = torch.load(str(args.load_ckpt_path), map_location={'cuda:0': f'cuda:{local_rank}'})
+ model.load_state_dict(checkpoint['state_dict'])
+
+ if is_distributed:
+ nproc_per_node = torch.cuda.device_count()
+ affinity = gpu_affinity.set_affinity(local_rank, nproc_per_node)
+ model = DistributedDataParallel(model, device_ids=[local_rank], output_device=local_rank)
+
+ test_dataloader = datamodule.test_dataloader() if not args.benchmark else datamodule.train_dataloader()
+ evaluate(model,
+ test_dataloader,
+ callbacks,
+ args)
+
+ for callback in callbacks:
+ callback.on_validation_end()
+
+ if args.benchmark:
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+ callbacks = [PerformanceCallback(logger, args.batch_size * world_size, warmup_epochs=1, mode='inference')]
+ for _ in range(6):
+ evaluate(model,
+ test_dataloader,
+ callbacks,
+ args)
+ callbacks[0].on_epoch_end()
+
+ callbacks[0].on_fit_end()
diff --git a/utils/model/se3_transformer/runtime/loggers.py b/utils/model/se3_transformer/runtime/loggers.py
new file mode 100644
index 0000000000000000000000000000000000000000..591486d8814c5dff43b8652823900e05add06c83
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/loggers.py
@@ -0,0 +1,134 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import pathlib
+from abc import ABC, abstractmethod
+from enum import Enum
+from typing import Dict, Any, Callable, Optional
+
+import dllogger
+import torch.distributed as dist
+import wandb
+from dllogger import Verbosity
+
+from se3_transformer.runtime.utils import rank_zero_only
+
+
+class Logger(ABC):
+ @rank_zero_only
+ @abstractmethod
+ def log_hyperparams(self, params):
+ pass
+
+ @rank_zero_only
+ @abstractmethod
+ def log_metrics(self, metrics, step=None):
+ pass
+
+ @staticmethod
+ def _sanitize_params(params):
+ def _sanitize(val):
+ if isinstance(val, Callable):
+ try:
+ _val = val()
+ if isinstance(_val, Callable):
+ return val.__name__
+ return _val
+ except Exception:
+ return getattr(val, "__name__", None)
+ elif isinstance(val, pathlib.Path) or isinstance(val, Enum):
+ return str(val)
+ return val
+
+ return {key: _sanitize(val) for key, val in params.items()}
+
+
+class LoggerCollection(Logger):
+ def __init__(self, loggers):
+ super().__init__()
+ self.loggers = loggers
+
+ def __getitem__(self, index):
+ return [logger for logger in self.loggers][index]
+
+ @rank_zero_only
+ def log_metrics(self, metrics, step=None):
+ for logger in self.loggers:
+ logger.log_metrics(metrics, step)
+
+ @rank_zero_only
+ def log_hyperparams(self, params):
+ for logger in self.loggers:
+ logger.log_hyperparams(params)
+
+
+class DLLogger(Logger):
+ def __init__(self, save_dir: pathlib.Path, filename: str):
+ super().__init__()
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ save_dir.mkdir(parents=True, exist_ok=True)
+ dllogger.init(
+ backends=[dllogger.JSONStreamBackend(Verbosity.DEFAULT, str(save_dir / filename))])
+
+ @rank_zero_only
+ def log_hyperparams(self, params):
+ params = self._sanitize_params(params)
+ dllogger.log(step="PARAMETER", data=params)
+
+ @rank_zero_only
+ def log_metrics(self, metrics, step=None):
+ if step is None:
+ step = tuple()
+
+ dllogger.log(step=step, data=metrics)
+
+
+class WandbLogger(Logger):
+ def __init__(
+ self,
+ name: str,
+ save_dir: pathlib.Path,
+ id: Optional[str] = None,
+ project: Optional[str] = None
+ ):
+ super().__init__()
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ save_dir.mkdir(parents=True, exist_ok=True)
+ self.experiment = wandb.init(name=name,
+ project=project,
+ id=id,
+ dir=str(save_dir),
+ resume='allow',
+ anonymous='must')
+
+ @rank_zero_only
+ def log_hyperparams(self, params: Dict[str, Any]) -> None:
+ params = self._sanitize_params(params)
+ self.experiment.config.update(params, allow_val_change=True)
+
+ @rank_zero_only
+ def log_metrics(self, metrics: Dict[str, float], step: Optional[int] = None) -> None:
+ if step is not None:
+ self.experiment.log({**metrics, 'epoch': step})
+ else:
+ self.experiment.log(metrics)
diff --git a/utils/model/se3_transformer/runtime/metrics.py b/utils/model/se3_transformer/runtime/metrics.py
new file mode 100644
index 0000000000000000000000000000000000000000..63026a71989441603df6abd447555524f2fd1e85
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/metrics.py
@@ -0,0 +1,83 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+from abc import ABC, abstractmethod
+
+import torch
+import torch.distributed as dist
+from torch import Tensor
+
+
+class Metric(ABC):
+ """ Metric class with synchronization capabilities similar to TorchMetrics """
+
+ def __init__(self):
+ self.states = {}
+
+ def add_state(self, name: str, default: Tensor):
+ assert name not in self.states
+ self.states[name] = default.clone()
+ setattr(self, name, default)
+
+ def synchronize(self):
+ if dist.is_initialized():
+ for state in self.states:
+ dist.all_reduce(getattr(self, state), op=dist.ReduceOp.SUM, group=dist.group.WORLD)
+
+ def __call__(self, *args, **kwargs):
+ self.update(*args, **kwargs)
+
+ def reset(self):
+ for name, default in self.states.items():
+ setattr(self, name, default.clone())
+
+ def compute(self):
+ self.synchronize()
+ value = self._compute().item()
+ self.reset()
+ return value
+
+ @abstractmethod
+ def _compute(self):
+ pass
+
+ @abstractmethod
+ def update(self, preds: Tensor, targets: Tensor):
+ pass
+
+
+class MeanAbsoluteError(Metric):
+ def __init__(self):
+ super().__init__()
+ self.add_state('error', torch.tensor(0, dtype=torch.float32, device='cuda'))
+ self.add_state('total', torch.tensor(0, dtype=torch.int32, device='cuda'))
+
+ def update(self, preds: Tensor, targets: Tensor):
+ preds = preds.detach()
+ n = preds.shape[0]
+ error = torch.abs(preds.view(n, -1) - targets.view(n, -1)).sum()
+ self.total += n
+ self.error += error
+
+ def _compute(self):
+ return self.error / self.total
diff --git a/utils/model/se3_transformer/runtime/training.py b/utils/model/se3_transformer/runtime/training.py
new file mode 100644
index 0000000000000000000000000000000000000000..2d66f08f45ce7bc753ac463489fc9a8d14a6b8b6
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/training.py
@@ -0,0 +1,238 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import logging
+import pathlib
+from typing import List
+
+import numpy as np
+import torch
+import torch.distributed as dist
+import torch.nn as nn
+from apex.optimizers import FusedAdam, FusedLAMB
+from torch.nn.modules.loss import _Loss
+from torch.nn.parallel import DistributedDataParallel
+from torch.optim import Optimizer
+from torch.utils.data import DataLoader, DistributedSampler
+from tqdm import tqdm
+
+from se3_transformer.data_loading import QM9DataModule
+from se3_transformer.model import SE3TransformerPooled
+from se3_transformer.model.fiber import Fiber
+from se3_transformer.runtime import gpu_affinity
+from se3_transformer.runtime.arguments import PARSER
+from se3_transformer.runtime.callbacks import QM9MetricCallback, QM9LRSchedulerCallback, BaseCallback, \
+ PerformanceCallback
+from se3_transformer.runtime.inference import evaluate
+from se3_transformer.runtime.loggers import LoggerCollection, DLLogger, WandbLogger, Logger
+from se3_transformer.runtime.utils import to_cuda, get_local_rank, init_distributed, seed_everything, \
+ using_tensor_cores, increase_l2_fetch_granularity
+
+
+def save_state(model: nn.Module, optimizer: Optimizer, epoch: int, path: pathlib.Path, callbacks: List[BaseCallback]):
+ """ Saves model, optimizer and epoch states to path (only once per node) """
+ if get_local_rank() == 0:
+ state_dict = model.module.state_dict() if isinstance(model, DistributedDataParallel) else model.state_dict()
+ checkpoint = {
+ 'state_dict': state_dict,
+ 'optimizer_state_dict': optimizer.state_dict(),
+ 'epoch': epoch
+ }
+ for callback in callbacks:
+ callback.on_checkpoint_save(checkpoint)
+
+ torch.save(checkpoint, str(path))
+ logging.info(f'Saved checkpoint to {str(path)}')
+
+
+def load_state(model: nn.Module, optimizer: Optimizer, path: pathlib.Path, callbacks: List[BaseCallback]):
+ """ Loads model, optimizer and epoch states from path """
+ checkpoint = torch.load(str(path), map_location={'cuda:0': f'cuda:{get_local_rank()}'})
+ if isinstance(model, DistributedDataParallel):
+ model.module.load_state_dict(checkpoint['state_dict'])
+ else:
+ model.load_state_dict(checkpoint['state_dict'])
+ optimizer.load_state_dict(checkpoint['optimizer_state_dict'])
+
+ for callback in callbacks:
+ callback.on_checkpoint_load(checkpoint)
+
+ logging.info(f'Loaded checkpoint from {str(path)}')
+ return checkpoint['epoch']
+
+
+def train_epoch(train_dataloader, loss_fn, epoch_idx, grad_scaler, optimizer, local_rank, callbacks, args):
+ losses = []
+ for i, batch in tqdm(enumerate(train_dataloader), total=len(train_dataloader), unit='batch',
+ desc=f'Epoch {epoch_idx}', disable=(args.silent or local_rank != 0)):
+ *inputs, target = to_cuda(batch)
+
+ for callback in callbacks:
+ callback.on_batch_start()
+
+ with torch.cuda.amp.autocast(enabled=args.amp):
+ pred = model(*inputs)
+ loss = loss_fn(pred, target) / args.accumulate_grad_batches
+
+ grad_scaler.scale(loss).backward()
+
+ # gradient accumulation
+ if (i + 1) % args.accumulate_grad_batches == 0 or (i + 1) == len(train_dataloader):
+ if args.gradient_clip:
+ grad_scaler.unscale_(optimizer)
+ torch.nn.utils.clip_grad_norm_(model.parameters(), args.gradient_clip)
+
+ grad_scaler.step(optimizer)
+ grad_scaler.update()
+ optimizer.zero_grad()
+
+ losses.append(loss.item())
+
+ return np.mean(losses)
+
+
+def train(model: nn.Module,
+ loss_fn: _Loss,
+ train_dataloader: DataLoader,
+ val_dataloader: DataLoader,
+ callbacks: List[BaseCallback],
+ logger: Logger,
+ args):
+ device = torch.cuda.current_device()
+ model.to(device=device)
+ local_rank = get_local_rank()
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+
+ if dist.is_initialized():
+ model = DistributedDataParallel(model, device_ids=[local_rank], output_device=local_rank)
+
+ model.train()
+ grad_scaler = torch.cuda.amp.GradScaler(enabled=args.amp)
+ if args.optimizer == 'adam':
+ optimizer = FusedAdam(model.parameters(), lr=args.learning_rate, betas=(args.momentum, 0.999),
+ weight_decay=args.weight_decay)
+ elif args.optimizer == 'lamb':
+ optimizer = FusedLAMB(model.parameters(), lr=args.learning_rate, betas=(args.momentum, 0.999),
+ weight_decay=args.weight_decay)
+ else:
+ optimizer = torch.optim.SGD(model.parameters(), lr=args.learning_rate, momentum=args.momentum,
+ weight_decay=args.weight_decay)
+
+ epoch_start = load_state(model, optimizer, args.load_ckpt_path, callbacks) if args.load_ckpt_path else 0
+
+ for callback in callbacks:
+ callback.on_fit_start(optimizer, args)
+
+ for epoch_idx in range(epoch_start, args.epochs):
+ if isinstance(train_dataloader.sampler, DistributedSampler):
+ train_dataloader.sampler.set_epoch(epoch_idx)
+
+ loss = train_epoch(train_dataloader, loss_fn, epoch_idx, grad_scaler, optimizer, local_rank, callbacks, args)
+ if dist.is_initialized():
+ loss = torch.tensor(loss, dtype=torch.float, device=device)
+ torch.distributed.all_reduce(loss)
+ loss = (loss / world_size).item()
+
+ logging.info(f'Train loss: {loss}')
+ logger.log_metrics({'train loss': loss}, epoch_idx)
+
+ for callback in callbacks:
+ callback.on_epoch_end()
+
+ if not args.benchmark and args.save_ckpt_path is not None and args.ckpt_interval > 0 \
+ and (epoch_idx + 1) % args.ckpt_interval == 0:
+ save_state(model, optimizer, epoch_idx, args.save_ckpt_path, callbacks)
+
+ if not args.benchmark and args.eval_interval > 0 and (epoch_idx + 1) % args.eval_interval == 0:
+ evaluate(model, val_dataloader, callbacks, args)
+ model.train()
+
+ for callback in callbacks:
+ callback.on_validation_end(epoch_idx)
+
+ if args.save_ckpt_path is not None and not args.benchmark:
+ save_state(model, optimizer, args.epochs, args.save_ckpt_path, callbacks)
+
+ for callback in callbacks:
+ callback.on_fit_end()
+
+
+def print_parameters_count(model):
+ num_params_trainable = sum(p.numel() for p in model.parameters() if p.requires_grad)
+ logging.info(f'Number of trainable parameters: {num_params_trainable}')
+
+
+if __name__ == '__main__':
+ is_distributed = init_distributed()
+ local_rank = get_local_rank()
+ args = PARSER.parse_args()
+
+ logging.getLogger().setLevel(logging.CRITICAL if local_rank != 0 or args.silent else logging.INFO)
+
+ logging.info('====== SE(3)-Transformer ======')
+ logging.info('| Training procedure |')
+ logging.info('===============================')
+
+ if args.seed is not None:
+ logging.info(f'Using seed {args.seed}')
+ seed_everything(args.seed)
+
+ logger = LoggerCollection([
+ DLLogger(save_dir=args.log_dir, filename=args.dllogger_name),
+ WandbLogger(name=f'QM9({args.task})', save_dir=args.log_dir, project='se3-transformer')
+ ])
+
+ datamodule = QM9DataModule(**vars(args))
+ model = SE3TransformerPooled(
+ fiber_in=Fiber({0: datamodule.NODE_FEATURE_DIM}),
+ fiber_out=Fiber({0: args.num_degrees * args.num_channels}),
+ fiber_edge=Fiber({0: datamodule.EDGE_FEATURE_DIM}),
+ output_dim=1,
+ tensor_cores=using_tensor_cores(args.amp), # use Tensor Cores more effectively
+ **vars(args)
+ )
+ loss_fn = nn.L1Loss()
+
+ if args.benchmark:
+ logging.info('Running benchmark mode')
+ world_size = dist.get_world_size() if dist.is_initialized() else 1
+ callbacks = [PerformanceCallback(logger, args.batch_size * world_size)]
+ else:
+ callbacks = [QM9MetricCallback(logger, targets_std=datamodule.targets_std, prefix='validation'),
+ QM9LRSchedulerCallback(logger, epochs=args.epochs)]
+
+ if is_distributed:
+ gpu_affinity.set_affinity(gpu_id=get_local_rank(), nproc_per_node=torch.cuda.device_count())
+
+ print_parameters_count(model)
+ logger.log_hyperparams(vars(args))
+ increase_l2_fetch_granularity()
+ train(model,
+ loss_fn,
+ datamodule.train_dataloader(),
+ datamodule.val_dataloader(),
+ callbacks,
+ logger,
+ args)
+
+ logging.info('Training finished successfully')
diff --git a/utils/model/se3_transformer/runtime/utils.py b/utils/model/se3_transformer/runtime/utils.py
new file mode 100644
index 0000000000000000000000000000000000000000..b6248fa6e5d94a14df6726eb73274a4727281a63
--- /dev/null
+++ b/utils/model/se3_transformer/runtime/utils.py
@@ -0,0 +1,130 @@
+# Copyright (c) 2021, NVIDIA CORPORATION & AFFILIATES. All rights reserved.
+#
+# Permission is hereby granted, free of charge, to any person obtaining a
+# copy of this software and associated documentation files (the "Software"),
+# to deal in the Software without restriction, including without limitation
+# the rights to use, copy, modify, merge, publish, distribute, sublicense,
+# and/or sell copies of the Software, and to permit persons to whom the
+# Software is furnished to do so, subject to the following conditions:
+#
+# The above copyright notice and this permission notice shall be included in
+# all copies or substantial portions of the Software.
+#
+# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
+# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
+# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL
+# THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
+# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING
+# FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER
+# DEALINGS IN THE SOFTWARE.
+#
+# SPDX-FileCopyrightText: Copyright (c) 2021 NVIDIA CORPORATION & AFFILIATES
+# SPDX-License-Identifier: MIT
+
+import argparse
+import ctypes
+import logging
+import os
+import random
+from functools import wraps
+from typing import Union, List, Dict
+
+import numpy as np
+import torch
+import torch.distributed as dist
+from torch import Tensor
+
+
+def aggregate_residual(feats1, feats2, method: str):
+ """ Add or concatenate two fiber features together. If degrees don't match, will use the ones of feats2. """
+ if method in ['add', 'sum']:
+ return {k: (v + feats1[k]) if k in feats1 else v for k, v in feats2.items()}
+ elif method in ['cat', 'concat']:
+ return {k: torch.cat([v, feats1[k]], dim=1) if k in feats1 else v for k, v in feats2.items()}
+ else:
+ raise ValueError('Method must be add/sum or cat/concat')
+
+
+def degree_to_dim(degree: int) -> int:
+ return 2 * degree + 1
+
+
+def unfuse_features(features: Tensor, degrees: List[int]) -> Dict[str, Tensor]:
+ return dict(zip(map(str, degrees), features.split([degree_to_dim(deg) for deg in degrees], dim=-1)))
+
+
+def str2bool(v: Union[bool, str]) -> bool:
+ if isinstance(v, bool):
+ return v
+ if v.lower() in ('yes', 'true', 't', 'y', '1'):
+ return True
+ elif v.lower() in ('no', 'false', 'f', 'n', '0'):
+ return False
+ else:
+ raise argparse.ArgumentTypeError('Boolean value expected.')
+
+
+def to_cuda(x):
+ """ Try to convert a Tensor, a collection of Tensors or a DGLGraph to CUDA """
+ if isinstance(x, Tensor):
+ return x.cuda(non_blocking=True)
+ elif isinstance(x, tuple):
+ return (to_cuda(v) for v in x)
+ elif isinstance(x, list):
+ return [to_cuda(v) for v in x]
+ elif isinstance(x, dict):
+ return {k: to_cuda(v) for k, v in x.items()}
+ else:
+ # DGLGraph or other objects
+ return x.to(device=torch.cuda.current_device())
+
+
+def get_local_rank() -> int:
+ return int(os.environ.get('LOCAL_RANK', 0))
+
+
+def init_distributed() -> bool:
+ world_size = int(os.environ.get('WORLD_SIZE', 1))
+ distributed = world_size > 1
+ if distributed:
+ backend = 'nccl' if torch.cuda.is_available() else 'gloo'
+ dist.init_process_group(backend=backend, init_method='env://')
+ if backend == 'nccl':
+ torch.cuda.set_device(get_local_rank())
+ else:
+ logging.warning('Running on CPU only!')
+ assert torch.distributed.is_initialized()
+ return distributed
+
+
+def increase_l2_fetch_granularity():
+ # maximum fetch granularity of L2: 128 bytes
+ _libcudart = ctypes.CDLL('libcudart.so')
+ # set device limit on the current device
+ # cudaLimitMaxL2FetchGranularity = 0x05
+ pValue = ctypes.cast((ctypes.c_int * 1)(), ctypes.POINTER(ctypes.c_int))
+ _libcudart.cudaDeviceSetLimit(ctypes.c_int(0x05), ctypes.c_int(128))
+ _libcudart.cudaDeviceGetLimit(pValue, ctypes.c_int(0x05))
+ assert pValue.contents.value == 128
+
+
+def seed_everything(seed):
+ seed = int(seed)
+ random.seed(seed)
+ np.random.seed(seed)
+ torch.manual_seed(seed)
+ torch.cuda.manual_seed_all(seed)
+
+
+def rank_zero_only(fn):
+ @wraps(fn)
+ def wrapped_fn(*args, **kwargs):
+ if not dist.is_initialized() or dist.get_rank() == 0:
+ return fn(*args, **kwargs)
+
+ return wrapped_fn
+
+
+def using_tensor_cores(amp: bool) -> bool:
+ major_cc, minor_cc = torch.cuda.get_device_capability()
+ return (amp and major_cc >= 7) or major_cc >= 8
diff --git a/utils/model/util.py b/utils/model/util.py
new file mode 100644
index 0000000000000000000000000000000000000000..017f7028636ce8bf5597294913273fc257c2c844
--- /dev/null
+++ b/utils/model/util.py
@@ -0,0 +1,522 @@
+import sys
+
+import numpy as np
+import torch
+
+import scipy.sparse
+
+from chemical import *
+from scoring import *
+
+def th_ang_v(ab,bc,eps:float=1e-8):
+ def th_norm(x,eps:float=1e-8):
+ return x.square().sum(-1,keepdim=True).add(eps).sqrt()
+ def th_N(x,alpha:float=0):
+ return x/th_norm(x).add(alpha)
+ ab, bc = th_N(ab),th_N(bc)
+ cos_angle = torch.clamp( (ab*bc).sum(-1), -1, 1)
+ sin_angle = torch.sqrt(1-cos_angle.square() + eps)
+ dih = torch.stack((cos_angle,sin_angle),-1)
+ return dih
+
+def th_dih_v(ab,bc,cd):
+ def th_cross(a,b):
+ a,b = torch.broadcast_tensors(a,b)
+ return torch.cross(a,b, dim=-1)
+ def th_norm(x,eps:float=1e-8):
+ return x.square().sum(-1,keepdim=True).add(eps).sqrt()
+ def th_N(x,alpha:float=0):
+ return x/th_norm(x).add(alpha)
+
+ ab, bc, cd = th_N(ab),th_N(bc),th_N(cd)
+ n1 = th_N( th_cross(ab,bc) )
+ n2 = th_N( th_cross(bc,cd) )
+ sin_angle = (th_cross(n1,bc)*n2).sum(-1)
+ cos_angle = (n1*n2).sum(-1)
+ dih = torch.stack((cos_angle,sin_angle),-1)
+ return dih
+
+def th_dih(a,b,c,d):
+ return th_dih_v(a-b,b-c,c-d)
+
+# More complicated version splits error in CA-N and CA-C (giving more accurate CB position)
+# It returns the rigid transformation from local frame to global frame
+def rigid_from_3_points(N, Ca, C, non_ideal=False, eps=1e-8):
+ #N, Ca, C - [B,L, 3]
+ #R - [B,L, 3, 3], det(R)=1, inv(R) = R.T, R is a rotation matrix
+ B,L = N.shape[:2]
+
+ v1 = C-Ca
+ v2 = N-Ca
+ e1 = v1/(torch.norm(v1, dim=-1, keepdim=True)+eps)
+ u2 = v2-(torch.einsum('bli, bli -> bl', e1, v2)[...,None]*e1)
+ e2 = u2/(torch.norm(u2, dim=-1, keepdim=True)+eps)
+ e3 = torch.cross(e1, e2, dim=-1)
+ R = torch.cat([e1[...,None], e2[...,None], e3[...,None]], axis=-1) #[B,L,3,3] - rotation matrix
+
+ if non_ideal:
+ v2 = v2/(torch.norm(v2, dim=-1, keepdim=True)+eps)
+ cosref = torch.sum(e1*v2, dim=-1) # cosine of current N-CA-C bond angle
+ costgt = cos_ideal_NCAC.item()
+ cos2del = torch.clamp( cosref*costgt + torch.sqrt((1-cosref*cosref)*(1-costgt*costgt)+eps), min=-1.0, max=1.0 )
+ cosdel = torch.sqrt(0.5*(1+cos2del)+eps)
+ sindel = torch.sign(costgt-cosref) * torch.sqrt(1-0.5*(1+cos2del)+eps)
+ Rp = torch.eye(3, device=N.device).repeat(B,L,1,1)
+ Rp[:,:,0,0] = cosdel
+ Rp[:,:,0,1] = -sindel
+ Rp[:,:,1,0] = sindel
+ Rp[:,:,1,1] = cosdel
+
+ R = torch.einsum('blij,bljk->blik', R,Rp)
+
+ return R, Ca
+
+def get_tor_mask(seq, torsion_indices, mask_in=None):
+ B,L = seq.shape[:2]
+ tors_mask = torch.ones((B,L,10), dtype=torch.bool, device=seq.device)
+ tors_mask[...,3:7] = torsion_indices[seq,:,-1] > 0
+ tors_mask[:,0,1] = False
+ tors_mask[:,-1,0] = False
+
+ # mask for additional angles
+ tors_mask[:,:,7] = seq!=aa2num['GLY']
+ tors_mask[:,:,8] = seq!=aa2num['GLY']
+ tors_mask[:,:,9] = torch.logical_and( seq!=aa2num['GLY'], seq!=aa2num['ALA'] )
+ tors_mask[:,:,9] = torch.logical_and( tors_mask[:,:,9], seq!=aa2num['UNK'] )
+ tors_mask[:,:,9] = torch.logical_and( tors_mask[:,:,9], seq!=aa2num['MAS'] )
+
+ if mask_in != None:
+ # mask for missing atoms
+ # chis
+ ti0 = torch.gather(mask_in,2,torsion_indices[seq,:,0])
+ ti1 = torch.gather(mask_in,2,torsion_indices[seq,:,1])
+ ti2 = torch.gather(mask_in,2,torsion_indices[seq,:,2])
+ ti3 = torch.gather(mask_in,2,torsion_indices[seq,:,3])
+ is_valid = torch.stack((ti0, ti1, ti2, ti3), dim=-2).all(dim=-1)
+ tors_mask[...,3:7] = torch.logical_and(tors_mask[...,3:7], is_valid)
+ tors_mask[:,:,7] = torch.logical_and(tors_mask[:,:,7], mask_in[:,:,4]) # CB exist?
+ tors_mask[:,:,8] = torch.logical_and(tors_mask[:,:,8], mask_in[:,:,4]) # CB exist?
+ tors_mask[:,:,9] = torch.logical_and(tors_mask[:,:,9], mask_in[:,:,5]) # XG exist?
+
+ return tors_mask
+
+def get_torsions(xyz_in, seq, torsion_indices, torsion_can_flip, ref_angles, mask_in=None):
+ B,L = xyz_in.shape[:2]
+
+ tors_mask = get_tor_mask(seq, torsion_indices, mask_in)
+
+ # torsions to restrain to 0 or 180degree
+ tors_planar = torch.zeros((B, L, 10), dtype=torch.bool, device=xyz_in.device)
+ tors_planar[:,:,5] = seq == aa2num['TYR'] # TYR chi 3 should be planar
+
+ # idealize given xyz coordinates before computing torsion angles
+ xyz = xyz_in.clone()
+ Rs, Ts = rigid_from_3_points(xyz[...,0,:],xyz[...,1,:],xyz[...,2,:])
+ Nideal = torch.tensor([-0.5272, 1.3593, 0.000], device=xyz_in.device)
+ Cideal = torch.tensor([1.5233, 0.000, 0.000], device=xyz_in.device)
+ xyz[...,0,:] = torch.einsum('brij,j->bri', Rs, Nideal) + Ts
+ xyz[...,2,:] = torch.einsum('brij,j->bri', Rs, Cideal) + Ts
+
+ torsions = torch.zeros( (B,L,10,2), device=xyz.device )
+ # avoid undefined angles for H generation
+ torsions[:,0,1,0] = 1.0
+ torsions[:,-1,0,0] = 1.0
+
+ # omega
+ torsions[:,:-1,0,:] = th_dih(xyz[:,:-1,1,:],xyz[:,:-1,2,:],xyz[:,1:,0,:],xyz[:,1:,1,:])
+ # phi
+ torsions[:,1:,1,:] = th_dih(xyz[:,:-1,2,:],xyz[:,1:,0,:],xyz[:,1:,1,:],xyz[:,1:,2,:])
+ # psi
+ torsions[:,:,2,:] = -1 * th_dih(xyz[:,:,0,:],xyz[:,:,1,:],xyz[:,:,2,:],xyz[:,:,3,:])
+
+ # chis
+ ti0 = torch.gather(xyz,2,torsion_indices[seq,:,0,None].repeat(1,1,1,3))
+ ti1 = torch.gather(xyz,2,torsion_indices[seq,:,1,None].repeat(1,1,1,3))
+ ti2 = torch.gather(xyz,2,torsion_indices[seq,:,2,None].repeat(1,1,1,3))
+ ti3 = torch.gather(xyz,2,torsion_indices[seq,:,3,None].repeat(1,1,1,3))
+ torsions[:,:,3:7,:] = th_dih(ti0,ti1,ti2,ti3)
+
+ # CB bend
+ NC = 0.5*( xyz[:,:,0,:3] + xyz[:,:,2,:3] )
+ CA = xyz[:,:,1,:3]
+ CB = xyz[:,:,4,:3]
+ t = th_ang_v(CB-CA,NC-CA)
+ t0 = ref_angles[seq][...,0,:]
+ torsions[:,:,7,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ # CB twist
+ NCCA = NC-CA
+ NCp = xyz[:,:,2,:3] - xyz[:,:,0,:3]
+ NCpp = NCp - torch.sum(NCp*NCCA, dim=-1, keepdim=True)/ torch.sum(NCCA*NCCA, dim=-1, keepdim=True) * NCCA
+ t = th_ang_v(CB-CA,NCpp)
+ t0 = ref_angles[seq][...,1,:]
+ torsions[:,:,8,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ # CG bend
+ CG = xyz[:,:,5,:3]
+ t = th_ang_v(CG-CB,CA-CB)
+ t0 = ref_angles[seq][...,2,:]
+ torsions[:,:,9,:] = torch.stack(
+ (torch.sum(t*t0,dim=-1),t[...,0]*t0[...,1]-t[...,1]*t0[...,0]),
+ dim=-1 )
+
+ mask0 = torch.isnan(torsions[...,0]).nonzero()
+ mask1 = torch.isnan(torsions[...,1]).nonzero()
+ torsions[mask0[:,0],mask0[:,1],mask0[:,2],0] = 1.0
+ torsions[mask1[:,0],mask1[:,1],mask1[:,2],1] = 0.0
+
+ # alt chis
+ torsions_alt = torsions.clone()
+ torsions_alt[torsion_can_flip[seq,:]] *= -1
+
+ return torsions, torsions_alt, tors_mask, tors_planar
+
+def get_tips(xyz, seq):
+ B,L = xyz.shape[:2]
+
+ xyz_tips = torch.gather(xyz, 2, tip_indices.to(xyz.device)[seq][:,:,None,None].expand(-1,-1,-1,3)).reshape(B, L, 3)
+ mask = ~(torch.isnan(xyz_tips[:,:,0]))
+ if torch.isnan(xyz_tips).any(): # replace NaN tip atom with virtual Cb atom
+ # three anchor atoms
+ N = xyz[:,:,0]
+ Ca = xyz[:,:,1]
+ C = xyz[:,:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ xyz_tips = torch.where(torch.isnan(xyz_tips), Cb, xyz_tips)
+ return xyz_tips, mask
+
+# process ideal frames
+def make_frame(X, Y):
+ Xn = X / torch.linalg.norm(X)
+ Y = Y - torch.dot(Y, Xn) * Xn
+ Yn = Y / torch.linalg.norm(Y)
+ Z = torch.cross(Xn,Yn)
+ Zn = Z / torch.linalg.norm(Z)
+
+ return torch.stack((Xn,Yn,Zn), dim=-1)
+
+def cross_product_matrix(u):
+ B, L = u.shape[:2]
+ matrix = torch.zeros((B, L, 3, 3), device=u.device)
+ matrix[:,:,0,1] = -u[...,2]
+ matrix[:,:,0,2] = u[...,1]
+ matrix[:,:,1,0] = u[...,2]
+ matrix[:,:,1,2] = -u[...,0]
+ matrix[:,:,2,0] = -u[...,1]
+ matrix[:,:,2,1] = u[...,0]
+ return matrix
+
+# writepdb
+def writepdb(filename, atoms, seq, idx_pdb=None, bfacts=None):
+ f = open(filename,"w")
+ ctr = 1
+ scpu = seq.cpu().squeeze()
+ atomscpu = atoms.cpu().squeeze()
+ if bfacts is None:
+ bfacts = torch.zeros(atomscpu.shape[0])
+ if idx_pdb is None:
+ idx_pdb = 1 + torch.arange(atomscpu.shape[0])
+
+ Bfacts = torch.clamp( bfacts.cpu(), 0, 1)
+ for i,s in enumerate(scpu):
+ if (len(atomscpu.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", num2aa[s],
+ "A", idx_pdb[i], atomscpu[i,0], atomscpu[i,1], atomscpu[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ elif atomscpu.shape[1]==3:
+ for j,atm_j in enumerate([" N "," CA "," C "]):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ "A", idx_pdb[i], atomscpu[i,j,0], atomscpu[i,j,1], atomscpu[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ natoms = atomscpu.shape[1]
+ if (natoms!=14 and natoms!=27):
+ print ('bad size!', atoms.shape)
+ assert(False)
+ atms = aa2long[s]
+ # his prot hack
+ if (s==8 and torch.linalg.norm( atomscpu[i,9,:]-atomscpu[i,5,:] ) < 1.7):
+ atms = (
+ " N "," CA "," C "," O "," CB "," CG "," NE2"," CD2"," CE1"," ND1",
+ None, None, None, None," H "," HA ","1HB ","2HB "," HD2"," HE1",
+ " HD1", None, None, None, None, None, None) # his_d
+
+ for j,atm_j in enumerate(atms):
+ if (j=4] = 4
+ num_bonds[i,...] = torch.tensor(num_bonds_i)
+
+
+# LJ/LK scoring parameters
+ljlk_parameters = torch.zeros((22,27,5), dtype=torch.float)
+lj_correction_parameters = torch.zeros((22,27,4), dtype=bool) # donor/acceptor/hpol/disulf
+for i in range(22):
+ for j,a in enumerate(aa2type[i]):
+ if (a is not None):
+ ljlk_parameters[i,j,:] = torch.tensor( type2ljlk[a] )
+ lj_correction_parameters[i,j,0] = (type2hb[a]==HbAtom.DO)+(type2hb[a]==HbAtom.DA)
+ lj_correction_parameters[i,j,1] = (type2hb[a]==HbAtom.AC)+(type2hb[a]==HbAtom.DA)
+ lj_correction_parameters[i,j,2] = (type2hb[a]==HbAtom.HP)
+ lj_correction_parameters[i,j,3] = (a=="SH1" or a=="HS")
+
+# hbond scoring parameters
+def donorHs(D,bonds,atoms):
+ dHs = []
+ for (i,j) in bonds:
+ if (i==D):
+ idx_j = atoms.index(j)
+ if (idx_j>=14): # if atom j is a hydrogen
+ dHs.append(idx_j)
+ if (j==D):
+ idx_i = atoms.index(i)
+ if (idx_i>=14): # if atom j is a hydrogen
+ dHs.append(idx_i)
+ assert (len(dHs)>0)
+ return dHs
+
+def acceptorBB0(A,hyb,bonds,atoms):
+ if (hyb == HbHybType.SP2):
+ for (i,j) in bonds:
+ if (i==A):
+ B = atoms.index(j)
+ if (B<14):
+ break
+ if (j==A):
+ B = atoms.index(i)
+ if (B<14):
+ break
+ for (i,j) in bonds:
+ if (i==atoms[B]):
+ B0 = atoms.index(j)
+ if (B0<14):
+ break
+ if (j==atoms[B]):
+ B0 = atoms.index(i)
+ if (B0<14):
+ break
+ elif (hyb == HbHybType.SP3 or hyb == HbHybType.RING):
+ for (i,j) in bonds:
+ if (i==A):
+ B = atoms.index(j)
+ if (B<14):
+ break
+ if (j==A):
+ B = atoms.index(i)
+ if (B<14):
+ break
+ for (i,j) in bonds:
+ if (i==A and j!=atoms[B]):
+ B0 = atoms.index(j)
+ break
+ if (j==A and i!=atoms[B]):
+ B0 = atoms.index(i)
+ break
+
+ return B,B0
+
+
+hbtypes = torch.full((22,27,3),-1, dtype=torch.long) # (donortype, acceptortype, acchybtype)
+hbbaseatoms = torch.full((22,27,2),-1, dtype=torch.long) # (B,B0) for acc; (D,-1) for don
+hbpolys = torch.zeros((HbDonType.NTYPES,HbAccType.NTYPES,3,15)) # weight,xmin,xmax,ymin,ymax,c9,...,c0
+
+for i in range(22):
+ for j,a in enumerate(aa2type[i]):
+ if (a in type2dontype):
+ j_hs = donorHs(aa2long[i][j],aabonds[i],aa2long[i])
+ for j_h in j_hs:
+ hbtypes[i,j_h,0] = type2dontype[a]
+ hbbaseatoms[i,j_h,0] = j
+ if (a in type2acctype):
+ j_b, j_b0 = acceptorBB0(aa2long[i][j],type2hybtype[a],aabonds[i],aa2long[i])
+ hbtypes[i,j,1] = type2acctype[a]
+ hbtypes[i,j,2] = type2hybtype[a]
+ hbbaseatoms[i,j,0] = j_b
+ hbbaseatoms[i,j,1] = j_b0
+
+for i in range(HbDonType.NTYPES):
+ for j in range(HbAccType.NTYPES):
+ weight = dontype2wt[i]*acctype2wt[j]
+
+ pdist,pbah,pahd = hbtypepair2poly[(i,j)]
+ xrange,yrange,coeffs = hbpolytype2coeffs[pdist]
+ hbpolys[i,j,0,0] = weight
+ hbpolys[i,j,0,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,0,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,0,5:] = torch.tensor(coeffs)
+ xrange,yrange,coeffs = hbpolytype2coeffs[pahd]
+ hbpolys[i,j,1,0] = weight
+ hbpolys[i,j,1,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,1,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,1,5:] = torch.tensor(coeffs)
+ xrange,yrange,coeffs = hbpolytype2coeffs[pbah]
+ hbpolys[i,j,2,0] = weight
+ hbpolys[i,j,2,1:3] = torch.tensor(xrange)
+ hbpolys[i,j,2,3:5] = torch.tensor(yrange)
+ hbpolys[i,j,2,5:] = torch.tensor(coeffs)
+
+# kinematic parameters
+base_indices = torch.full((22,27),0, dtype=torch.long)
+xyzs_in_base_frame = torch.ones((22,27,4))
+RTs_by_torsion = torch.eye(4).repeat(22,7,1,1)
+reference_angles = torch.ones((22,3,2))
+
+for i in range(22):
+ i_l = aa2long[i]
+ for name, base, coords in ideal_coords[i]:
+ idx = i_l.index(name)
+ base_indices[i,idx] = base
+ xyzs_in_base_frame[i,idx,:3] = torch.tensor(coords)
+
+ # omega frame
+ RTs_by_torsion[i,0,:3,:3] = torch.eye(3)
+ RTs_by_torsion[i,0,:3,3] = torch.zeros(3)
+
+ # phi frame
+ RTs_by_torsion[i,1,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,0,:3] - xyzs_in_base_frame[i,1,:3],
+ torch.tensor([1.,0.,0.])
+ )
+ RTs_by_torsion[i,1,:3,3] = xyzs_in_base_frame[i,0,:3]
+
+ # psi frame
+ RTs_by_torsion[i,2,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,2,:3] - xyzs_in_base_frame[i,1,:3],
+ xyzs_in_base_frame[i,1,:3] - xyzs_in_base_frame[i,0,:3]
+ )
+ RTs_by_torsion[i,2,:3,3] = xyzs_in_base_frame[i,2,:3]
+
+ # chi1 frame
+ if torsions[i][0] is not None:
+ a0,a1,a2 = torsion_indices[i,0,0:3]
+ RTs_by_torsion[i,3,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3]-xyzs_in_base_frame[i,a1,:3],
+ xyzs_in_base_frame[i,a0,:3]-xyzs_in_base_frame[i,a1,:3],
+ )
+ RTs_by_torsion[i,3,:3,3] = xyzs_in_base_frame[i,a2,:3]
+
+ # chi2~4 frame
+ for j in range(1,4):
+ if torsions[i][j] is not None:
+ a2 = torsion_indices[i,j,2]
+ if ((i==18 and j==2) or (i==8 and j==2)): # TYR CZ-OH & HIS CE1-HE1 a special case
+ a0,a1 = torsion_indices[i,j,0:2]
+ RTs_by_torsion[i,3+j,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3]-xyzs_in_base_frame[i,a1,:3],
+ xyzs_in_base_frame[i,a0,:3]-xyzs_in_base_frame[i,a1,:3] )
+ else:
+ RTs_by_torsion[i,3+j,:3,:3] = make_frame(
+ xyzs_in_base_frame[i,a2,:3],
+ torch.tensor([-1.,0.,0.]), )
+ RTs_by_torsion[i,3+j,:3,3] = xyzs_in_base_frame[i,a2,:3]
+
+
+ # CB/CG angles
+ NCr = 0.5*(xyzs_in_base_frame[i,0,:3]+xyzs_in_base_frame[i,2,:3])
+ CAr = xyzs_in_base_frame[i,1,:3]
+ CBr = xyzs_in_base_frame[i,4,:3]
+ CGr = xyzs_in_base_frame[i,5,:3]
+ reference_angles[i,0,:]=th_ang_v(CBr-CAr,NCr-CAr)
+ NCp = xyzs_in_base_frame[i,2,:3]-xyzs_in_base_frame[i,0,:3]
+ NCpp = NCp - torch.dot(NCp,NCr)/ torch.dot(NCr,NCr) * NCr
+ reference_angles[i,1,:]=th_ang_v(CBr-CAr,NCpp)
+ reference_angles[i,2,:]=th_ang_v(CGr,torch.tensor([-1.,0.,0.]))
+
+def get_rmsd(a, b, eps=1e-6):
+ '''
+ align crds b to a : always use all alphas
+ expexted tensor of shape (L,3)
+ jake's torch adapted version
+ '''
+ assert a.shape == b.shape, 'make sure tensors are the same size'
+ L = a.shape[0]
+ assert a.shape == torch.Size([L,3]), 'make sure tensors are in format [L,3]'
+
+ # center to CA centroid
+ a = a - a.mean(dim=0)
+ b = b - b.mean(dim=0)
+
+ # Computation of the covariance matrix
+ C = torch.einsum('kj,ji->ki', torch.transpose(b.type(torch.float32),0,1), a.type(torch.float32))
+
+ # Compute optimal rotation matrix using SVD
+ V, S, W = torch.linalg.svd(C)
+
+ # get sign to ensure right-handedness
+ d = torch.ones([3,3])
+ d[:,-1] = torch.sign(torch.linalg.det(V)*torch.linalg.det(W))
+
+ # Rotation matrix U
+ U = torch.einsum('kj,ji->ki',(d*V),W)
+
+ # Rotate xyz_hal
+ rP = torch.einsum('kj,ji->ki',b.type(torch.float32),U.type(torch.float32))
+
+ L = rP.shape[0]
+ rmsd = torch.sqrt(torch.sum((rP-a)*(rP-a), axis=(0,1)) / L + eps)
+
+ return rmsd, U
+
diff --git a/utils/model/util_module.py b/utils/model/util_module.py
new file mode 100644
index 0000000000000000000000000000000000000000..4ba2b5723434b195468c0d11473a8a33f295f4cf
--- /dev/null
+++ b/utils/model/util_module.py
@@ -0,0 +1,310 @@
+import numpy as np
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+from opt_einsum import contract as einsum
+import copy
+import dgl
+from util import base_indices, RTs_by_torsion, xyzs_in_base_frame, rigid_from_3_points
+
+def init_lecun_normal(module, scale=1.0):
+ def truncated_normal(uniform, mu=0.0, sigma=1.0, a=-2, b=2):
+ normal = torch.distributions.normal.Normal(0, 1)
+
+ alpha = (a - mu) / sigma
+ beta = (b - mu) / sigma
+
+ alpha_normal_cdf = normal.cdf(torch.tensor(alpha))
+ p = alpha_normal_cdf + (normal.cdf(torch.tensor(beta)) - alpha_normal_cdf) * uniform
+
+ v = torch.clamp(2 * p - 1, -1 + 1e-8, 1 - 1e-8)
+ x = mu + sigma * np.sqrt(2) * torch.erfinv(v)
+ x = torch.clamp(x, a, b)
+
+ return x
+
+ def sample_truncated_normal(shape, scale=1.0):
+ stddev = np.sqrt(scale/shape[-1])/.87962566103423978 # shape[-1] = fan_in
+ return stddev * truncated_normal(torch.rand(shape))
+
+ module.weight = torch.nn.Parameter( (sample_truncated_normal(module.weight.shape)) )
+ return module
+
+def init_lecun_normal_param(weight, scale=1.0):
+ def truncated_normal(uniform, mu=0.0, sigma=1.0, a=-2, b=2):
+ normal = torch.distributions.normal.Normal(0, 1)
+
+ alpha = (a - mu) / sigma
+ beta = (b - mu) / sigma
+
+ alpha_normal_cdf = normal.cdf(torch.tensor(alpha))
+ p = alpha_normal_cdf + (normal.cdf(torch.tensor(beta)) - alpha_normal_cdf) * uniform
+
+ v = torch.clamp(2 * p - 1, -1 + 1e-8, 1 - 1e-8)
+ x = mu + sigma * np.sqrt(2) * torch.erfinv(v)
+ x = torch.clamp(x, a, b)
+
+ return x
+
+ def sample_truncated_normal(shape, scale=1.0):
+ stddev = np.sqrt(scale/shape[-1])/.87962566103423978 # shape[-1] = fan_in
+ return stddev * truncated_normal(torch.rand(shape))
+
+ weight = torch.nn.Parameter( (sample_truncated_normal(weight.shape)) )
+ return weight
+
+# for gradient checkpointing
+def create_custom_forward(module, **kwargs):
+ def custom_forward(*inputs):
+ return module(*inputs, **kwargs)
+ return custom_forward
+
+def get_clones(module, N):
+ return nn.ModuleList([copy.deepcopy(module) for i in range(N)])
+
+class Dropout(nn.Module):
+ # Dropout entire row or column
+ def __init__(self, broadcast_dim=None, p_drop=0.15):
+ super(Dropout, self).__init__()
+ # give ones with probability of 1-p_drop / zeros with p_drop
+ self.sampler = torch.distributions.bernoulli.Bernoulli(torch.tensor([1-p_drop]))
+ self.broadcast_dim=broadcast_dim
+ self.p_drop=p_drop
+ def forward(self, x):
+ if not self.training: # no drophead during evaluation mode
+ return x
+ shape = list(x.shape)
+ if not self.broadcast_dim == None:
+ shape[self.broadcast_dim] = 1
+ mask = self.sampler.sample(shape).to(x.device).view(shape)
+
+ x = mask * x / (1.0 - self.p_drop)
+ return x
+
+def rbf(D):
+ # Distance radial basis function
+ D_min, D_max, D_count = 0., 20., 36
+ D_mu = torch.linspace(D_min, D_max, D_count).to(D.device)
+ D_mu = D_mu[None,:]
+ D_sigma = (D_max - D_min) / D_count
+ D_expand = torch.unsqueeze(D, -1)
+ RBF = torch.exp(-((D_expand - D_mu) / D_sigma)**2)
+ return RBF
+
+def get_seqsep(idx):
+ '''
+ Input:
+ - idx: residue indices of given sequence (B,L)
+ Output:
+ - seqsep: sequence separation feature with sign (B, L, L, 1)
+ Sergey found that having sign in seqsep features helps a little
+ '''
+ seqsep = idx[:,None,:] - idx[:,:,None]
+ sign = torch.sign(seqsep)
+ neigh = torch.abs(seqsep)
+ neigh[neigh > 1] = 0.0 # if bonded -- 1.0 / else 0.0
+ neigh = sign * neigh
+ return neigh.unsqueeze(-1)
+
+def make_full_graph(xyz, pair, idx, top_k=64, kmin=9):
+ '''
+ Input:
+ - xyz: current backbone cooordinates (B, L, 3, 3)
+ - pair: pair features from Trunk (B, L, L, E)
+ - idx: residue index from ground truth pdb
+ Output:
+ - G: defined graph
+ '''
+
+ B, L = xyz.shape[:2]
+ device = xyz.device
+
+ # seq sep
+ sep = idx[:,None,:] - idx[:,:,None]
+ b,i,j = torch.where(sep.abs() > 0)
+
+ src = b*L+i
+ tgt = b*L+j
+ G = dgl.graph((src, tgt), num_nodes=B*L).to(device)
+ G.edata['rel_pos'] = (xyz[b,j,:] - xyz[b,i,:]).detach() # no gradient through basis function
+
+ return G, pair[b,i,j][...,None]
+
+def make_topk_graph(xyz, pair, idx, top_k=64, kmin=32, eps=1e-6):
+ '''
+ Input:
+ - xyz: current backbone cooordinates (B, L, 3, 3)
+ - pair: pair features from Trunk (B, L, L, E)
+ - idx: residue index from ground truth pdb
+ Output:
+ - G: defined graph
+ '''
+
+ B, L = xyz.shape[:2]
+ device = xyz.device
+
+ # distance map from current CA coordinates
+ D = torch.cdist(xyz, xyz) + torch.eye(L, device=device).unsqueeze(0)*999.9 # (B, L, L)
+ # seq sep
+ sep = idx[:,None,:] - idx[:,:,None]
+ sep = sep.abs() + torch.eye(L, device=device).unsqueeze(0)*999.9
+ D = D + sep*eps
+
+ # get top_k neighbors
+ D_neigh, E_idx = torch.topk(D, min(top_k, L), largest=False) # shape of E_idx: (B, L, top_k)
+ topk_matrix = torch.zeros((B, L, L), device=device)
+ topk_matrix.scatter_(2, E_idx, 1.0)
+
+ # put an edge if any of the 3 conditions are met:
+ # 1) |i-j| <= kmin (connect sequentially adjacent residues)
+ # 2) top_k neighbors
+ cond = torch.logical_or(topk_matrix > 0.0, sep < kmin)
+ b,i,j = torch.where(cond)
+
+ src = b*L+i
+ tgt = b*L+j
+ G = dgl.graph((src, tgt), num_nodes=B*L).to(device)
+ G.edata['rel_pos'] = (xyz[b,j,:] - xyz[b,i,:]).detach() # no gradient through basis function
+
+ return G, pair[b,i,j][...,None]
+
+def make_rotX(angs, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ RTs[:,:,1,1] = angs[:,:,0]/NORM
+ RTs[:,:,1,2] = -angs[:,:,1]/NORM
+ RTs[:,:,2,1] = angs[:,:,1]/NORM
+ RTs[:,:,2,2] = angs[:,:,0]/NORM
+ return RTs
+
+# rotate about the z axis
+def make_rotZ(angs, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ RTs[:,:,0,0] = angs[:,:,0]/NORM
+ RTs[:,:,0,1] = -angs[:,:,1]/NORM
+ RTs[:,:,1,0] = angs[:,:,1]/NORM
+ RTs[:,:,1,1] = angs[:,:,0]/NORM
+ return RTs
+
+# rotate about an arbitrary axis
+def make_rot_axis(angs, u, eps=1e-6):
+ B,L = angs.shape[:2]
+ NORM = torch.linalg.norm(angs, dim=-1) + eps
+
+ RTs = torch.eye(4, device=angs.device).repeat(B,L,1,1)
+
+ ct = angs[:,:,0]/NORM
+ st = angs[:,:,1]/NORM
+ u0 = u[:,:,0]
+ u1 = u[:,:,1]
+ u2 = u[:,:,2]
+
+ RTs[:,:,0,0] = ct+u0*u0*(1-ct)
+ RTs[:,:,0,1] = u0*u1*(1-ct)-u2*st
+ RTs[:,:,0,2] = u0*u2*(1-ct)+u1*st
+ RTs[:,:,1,0] = u0*u1*(1-ct)+u2*st
+ RTs[:,:,1,1] = ct+u1*u1*(1-ct)
+ RTs[:,:,1,2] = u1*u2*(1-ct)-u0*st
+ RTs[:,:,2,0] = u0*u2*(1-ct)-u1*st
+ RTs[:,:,2,1] = u1*u2*(1-ct)+u0*st
+ RTs[:,:,2,2] = ct+u2*u2*(1-ct)
+ return RTs
+
+class ComputeAllAtomCoords(nn.Module):
+ def __init__(self):
+ super(ComputeAllAtomCoords, self).__init__()
+
+ self.base_indices = nn.Parameter(base_indices, requires_grad=False)
+ self.RTs_in_base_frame = nn.Parameter(RTs_by_torsion, requires_grad=False)
+ self.xyzs_in_base_frame = nn.Parameter(xyzs_in_base_frame, requires_grad=False)
+
+ def forward(self, seq, xyz, alphas, non_ideal=False, use_H=True):
+ B,L = xyz.shape[:2]
+
+ Rs, Ts = rigid_from_3_points(xyz[...,0,:],xyz[...,1,:],xyz[...,2,:], non_ideal=non_ideal)
+
+ RTF0 = torch.eye(4).repeat(B,L,1,1).to(device=Rs.device)
+
+ # bb
+ RTF0[:,:,:3,:3] = Rs
+ RTF0[:,:,:3,3] = Ts
+
+ # omega
+ RTF1 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,0,:], make_rotX(alphas[:,:,0,:]))
+
+ # phi
+ RTF2 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,1,:], make_rotX(alphas[:,:,1,:]))
+
+ # psi
+ RTF3 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, self.RTs_in_base_frame[seq,2,:], make_rotX(alphas[:,:,2,:]))
+
+ # CB bend
+ basexyzs = self.xyzs_in_base_frame[seq]
+ NCr = 0.5*(basexyzs[:,:,2,:3]+basexyzs[:,:,0,:3])
+ CAr = (basexyzs[:,:,1,:3])
+ CBr = (basexyzs[:,:,4,:3])
+ CBrotaxis1 = (CBr-CAr).cross(NCr-CAr)
+ CBrotaxis1 /= torch.linalg.norm(CBrotaxis1, dim=-1, keepdim=True)+1e-8
+
+ # CB twist
+ NCp = basexyzs[:,:,2,:3] - basexyzs[:,:,0,:3]
+ NCpp = NCp - torch.sum(NCp*NCr, dim=-1, keepdim=True)/ torch.sum(NCr*NCr, dim=-1, keepdim=True) * NCr
+ CBrotaxis2 = (CBr-CAr).cross(NCpp)
+ CBrotaxis2 /= torch.linalg.norm(CBrotaxis2, dim=-1, keepdim=True)+1e-8
+
+ CBrot1 = make_rot_axis(alphas[:,:,7,:], CBrotaxis1 )
+ CBrot2 = make_rot_axis(alphas[:,:,8,:], CBrotaxis2 )
+
+ RTF8 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF0, CBrot1,CBrot2)
+
+ # chi1 + CG bend
+ RTF4 = torch.einsum(
+ 'brij,brjk,brkl,brlm->brim',
+ RTF8,
+ self.RTs_in_base_frame[seq,3,:],
+ make_rotX(alphas[:,:,3,:]),
+ make_rotZ(alphas[:,:,9,:]))
+
+ # chi2
+ RTF5 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF4, self.RTs_in_base_frame[seq,4,:],make_rotX(alphas[:,:,4,:]))
+
+ # chi3
+ RTF6 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF5,self.RTs_in_base_frame[seq,5,:],make_rotX(alphas[:,:,5,:]))
+
+ # chi4
+ RTF7 = torch.einsum(
+ 'brij,brjk,brkl->bril',
+ RTF6,self.RTs_in_base_frame[seq,6,:],make_rotX(alphas[:,:,6,:]))
+
+ RTframes = torch.stack((
+ RTF0,RTF1,RTF2,RTF3,RTF4,RTF5,RTF6,RTF7,RTF8
+ ),dim=2)
+
+ xyzs = torch.einsum(
+ 'brtij,brtj->brti',
+ RTframes.gather(2,self.base_indices[seq][...,None,None].repeat(1,1,1,4,4)), basexyzs
+ )
+
+ if use_H:
+ return RTframes, xyzs[...,:3]
+ else:
+ return RTframes, xyzs[...,:14,:3]
diff --git a/utils/model/utils/.inf_methods.py.swp b/utils/model/utils/.inf_methods.py.swp
new file mode 100644
index 0000000000000000000000000000000000000000..1638aa8c4baf5a8ded77e8d71bda84a9f34fce3c
Binary files /dev/null and b/utils/model/utils/.inf_methods.py.swp differ
diff --git a/utils/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py b/utils/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..fe2b975316f2de89d021d4dff442182192d5b7f8
--- /dev/null
+++ b/utils/model/utils/.ipynb_checkpoints/calc_dssp-checkpoint.py
@@ -0,0 +1,234 @@
+#@title get secondary structure (SSE) from given PDB file
+#@markdown So far it seems the best solution is to steal code from biotite
+#@markdown which calculates the SSE of a peptide chain based on the P-SEA algorithm (Labesse 1997)
+# CODE FROM BIOKITE
+# From Krypton
+import numpy as np
+import random
+import torch
+
+def vector_dot(v1,v2):
+ return (v1*v2).sum(axis=-1)
+
+def norm_vector(v):
+ factor = np.linalg.norm(v, axis=-1)
+ if isinstance(factor, np.ndarray):
+ v /= factor[..., np.newaxis]
+ else:
+ v /= factor
+ return v
+
+def coord(x):
+ return np.asarray(x)
+def displacement(atoms1, atoms2):
+ v1 = coord(atoms1)
+ v2 = coord(atoms2)
+ if len(v1.shape) <= len(v2.shape):
+ diff = v2 - v1
+ else:
+ diff = -(v1 - v2)
+ return diff
+def distance(atoms1, atoms2):
+ diff = displacement(atoms1, atoms2)
+ return np.sqrt(vector_dot(diff, diff))
+
+def angle(atoms1, atoms2, atoms3):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms3, atoms2)
+ norm_vector(v1)
+ norm_vector(v2)
+ return np.arccos(vector_dot(v1,v2))
+
+def dihedral(atoms1, atoms2, atoms3, atoms4):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms2, atoms3)
+ v3 = displacement(atoms3, atoms4)
+ norm_vector(v1)
+ norm_vector(v2)
+ norm_vector(v3)
+
+ n1 = np.cross(v1, v2)
+ n2 = np.cross(v2, v3)
+
+ # Calculation using atan2, to ensure the correct sign of the angle
+ x = vector_dot(n1,n2)
+ y = vector_dot(np.cross(n1,n2), v2)
+ return np.arctan2(y,x)
+
+def replace_letters(arr):
+ # Create a dictionary that maps the letters 'a', 'b', and 'c' to the corresponding numbers
+ letter_to_number = {'a': 0, 'b': 1, 'c': 2}
+
+ # Create a new array that will hold the numbers
+ nums = []
+
+ # Loop through the input array and replace the letters with the corresponding numbers
+ for letter in arr:
+ if letter in letter_to_number:
+ nums.append(letter_to_number[letter])
+ else:
+ nums.append(letter)
+
+ return np.array(nums)
+
+def replace_with_mask(arr, percentage, replace_loops=False):
+ # Make sure the percentage is between 0 and 100
+ percentage = min(max(percentage, 0), 100)
+
+ # Calculate the number of values to replace
+ num_to_replace = int(len(arr) * percentage / 100)
+
+ # Choose a random subset of the array to replace
+ replace_indices = random.sample(range(len(arr)), num_to_replace)
+
+ # Replace the values at the chosen indices with the number 3
+ for i in replace_indices:
+ arr[i] = 3
+
+ if replace_loops:
+ for i in arr:
+ if arr[i] == 2:
+ arr[i] = 3
+
+ return arr
+
+def annotate_sse(ca_coord, percentage_mask=0, replace_loops=False):
+ _radians_to_angle = 2*np.pi/360
+
+ _r_helix = ((89-12)*_radians_to_angle, (89+12)*_radians_to_angle)
+ _a_helix = ((50-20)*_radians_to_angle, (50+20)*_radians_to_angle)
+ _d2_helix = ((5.5-0.5), (5.5+0.5))
+ _d3_helix = ((5.3-0.5), (5.3+0.5))
+ _d4_helix = ((6.4-0.6), (6.4+0.6))
+
+ _r_strand = ((124-14)*_radians_to_angle, (124+14)*_radians_to_angle)
+ _a_strand = ((-180)*_radians_to_angle, (-125)*_radians_to_angle,
+ (145)*_radians_to_angle, (180)*_radians_to_angle)
+ _d2_strand = ((6.7-0.6), (6.7+0.6))
+ _d3_strand = ((9.9-0.9), (9.9+0.9))
+ _d4_strand = ((12.4-1.1), (12.4+1.1))
+
+ # Filter all CA atoms in the relevant chain.
+
+ d2i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d3i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d4i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ ri_coord = np.full(( len(ca_coord), 3, 3 ), np.nan)
+ ai_coord = np.full(( len(ca_coord), 4, 3 ), np.nan)
+
+ # The distances and angles are not defined for the entire interval,
+ # therefore the indices do not have the full range
+ # Values that are not defined are NaN
+ for i in range(1, len(ca_coord)-1):
+ d2i_coord[i] = (ca_coord[i-1], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ d3i_coord[i] = (ca_coord[i-1], ca_coord[i+2])
+ for i in range(1, len(ca_coord)-3):
+ d4i_coord[i] = (ca_coord[i-1], ca_coord[i+3])
+ for i in range(1, len(ca_coord)-1):
+ ri_coord[i] = (ca_coord[i-1], ca_coord[i], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ ai_coord[i] = (ca_coord[i-1], ca_coord[i],
+ ca_coord[i+1], ca_coord[i+2])
+
+ d2i = distance(d2i_coord[:,0], d2i_coord[:,1])
+ d3i = distance(d3i_coord[:,0], d3i_coord[:,1])
+ d4i = distance(d4i_coord[:,0], d4i_coord[:,1])
+ ri = angle(ri_coord[:,0], ri_coord[:,1], ri_coord[:,2])
+ ai = dihedral(ai_coord[:,0], ai_coord[:,1],
+ ai_coord[:,2], ai_coord[:,3])
+
+ sse = np.full(len(ca_coord), "c", dtype="U1")
+
+ # Annotate helices
+ # Find CA that meet criteria for potential helices
+ is_pot_helix = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if (
+ d3i[i] >= _d3_helix[0] and d3i[i] <= _d3_helix[1]
+ and d4i[i] >= _d4_helix[0] and d4i[i] <= _d4_helix[1]
+ ) or (
+ ri[i] >= _r_helix[0] and ri[i] <= _r_helix[1]
+ and ai[i] >= _a_helix[0] and ai[i] <= _a_helix[1]
+ ):
+ is_pot_helix[i] = True
+ # Real helices are 5 consecutive helix elements
+ is_helix = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ for i in range(len(sse)):
+ if is_pot_helix[i]:
+ counter += 1
+ else:
+ if counter >= 5:
+ is_helix[i-counter : i] = True
+ counter = 0
+ # Extend the helices by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_helix[i]:
+ sse[i] = "a"
+ if (
+ d3i[i-1] >= _d3_helix[0] and d3i[i-1] <= _d3_helix[1]
+ ) or (
+ ri[i-1] >= _r_helix[0] and ri[i-1] <= _r_helix[1]
+ ):
+ sse[i-1] = "a"
+ sse[i] = "a"
+ if (
+ d3i[i+1] >= _d3_helix[0] and d3i[i+1] <= _d3_helix[1]
+ ) or (
+ ri[i+1] >= _r_helix[0] and ri[i+1] <= _r_helix[1]
+ ):
+ sse[i+1] = "a"
+ i += 1
+
+ # Annotate sheets
+ # Find CA that meet criteria for potential strands
+ is_pot_strand = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if ( d2i[i] >= _d2_strand[0] and d2i[i] <= _d2_strand[1]
+ and d3i[i] >= _d3_strand[0] and d3i[i] <= _d3_strand[1]
+ and d4i[i] >= _d4_strand[0] and d4i[i] <= _d4_strand[1]
+ ) or (
+ ri[i] >= _r_strand[0] and ri[i] <= _r_strand[1]
+ and ( (ai[i] >= _a_strand[0] and ai[i] <= _a_strand[1])
+ or (ai[i] >= _a_strand[2] and ai[i] <= _a_strand[3]))
+ ):
+ is_pot_strand[i] = True
+ # Real strands are 5 consecutive strand elements,
+ # or shorter fragments of at least 3 consecutive strand residues,
+ # if they are in hydrogen bond proximity to 5 other residues
+ pot_strand_coord = ca_coord[is_pot_strand]
+ is_strand = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ contacts = 0
+ for i in range(len(sse)):
+ if is_pot_strand[i]:
+ counter += 1
+ coord = ca_coord[i]
+ for strand_coord in ca_coord:
+ dist = distance(coord, strand_coord)
+ if dist >= 4.2 and dist <= 5.2:
+ contacts += 1
+ else:
+ if counter >= 4:
+ is_strand[i-counter : i] = True
+ elif counter == 3 and contacts >= 5:
+ is_strand[i-counter : i] = True
+ counter = 0
+ contacts = 0
+ # Extend the strands by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_strand[i]:
+ sse[i] = "b"
+ if d3i[i-1] >= _d3_strand[0] and d3i[i-1] <= _d3_strand[1]:
+ sse[i-1] = "b"
+ sse[i] = "b"
+ if d3i[i+1] >= _d3_strand[0] and d3i[i+1] <= _d3_strand[1]:
+ sse[i+1] = "b"
+ i += 1
+ sse=replace_letters(sse)
+ sse=replace_with_mask(sse, percentage_mask, replace_loops=replace_loops)
+ sse=torch.nn.functional.one_hot(torch.tensor(sse), num_classes=4)
+ return sse
diff --git a/utils/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py b/utils/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..2fde867439dc3a604f03a9af67f88992148db71a
--- /dev/null
+++ b/utils/model/utils/.ipynb_checkpoints/diff_utils-checkpoint.py
@@ -0,0 +1,292 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None):
+ """
+ RFnar inference
+ """
+ seq_mask = ~input_seq_mask
+ str_mask = input_str_mask
+ nar = Nonautoregressive()
+ r = t
+ print(f'USING THIS R: {r}')
+
+ #mask sequence
+ if mask_seq_token:
+ print("MASK SEQ TOKEN")
+ seq_corrupt[seq_mask] = self.masked_token
+ elif mask_seq_random:
+ print("MASK SEQ RANDOM")
+
+
+ ic(seq_corrupt)
+
+ seq=seq_corrupt.repeat(seq.shape[0], 1)
+ seq_corrupt_onehot=torch.nn.functional.one_hot(seq_corrupt,num_classes=22).float()
+
+ ### msa_masked ###
+ ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_corrupt_onehot
+
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ #msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_corrupt_onehot
+ #msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ # msa_full #
+ #make msa_full same size as msa_masked
+ ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_corrupt_onehot
+ #msa_full[:,1:,:,:22] = msa_diffused
+
+ ###########
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_corrupt_onehot[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,seq_mask,21] = 0.0
+ t1d[:,~seq_mask,21] = 1.0
+
+ #to do add structure confidence metric; need to expand dimensions of chkpt b4
+ if t1d_24:
+ print("expanding t1d dim to 24")
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],1)).float()), -1).to(seq.device)
+ t1d[:,str_mask,23] = 0.0
+ t1d[:,~str_mask,23] = 1.0
+
+ t1d[:1,:,22] = r
+
+ ################
+ #mask structure#
+ ################
+ if mask_xyz_hole:
+ print("MASK XYZ BLACK HOLE")
+ ic(xyz_t.shape)
+ xyz_corrupt, xyz_mask = nar.xyz_mask_0(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True, cos=True)
+ ic(xyz_corrupt.shape)
+ elif mask_xyz_random:
+ print("MASK XYZ RANDOM")
+ xyz_corrupt, xyz_mask = nar.xyz_mask_random(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True, cos=True)
+ #only corrupt first template
+ xyz_t[0]=xyz_corrupt
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+ mask_msa[:,:,~loss_seq_mask] = False
+
+ seq_diffused = seq_corrupt_onehot
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, RFnar = False):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = None #seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+#def take_step_nar():
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+
diff --git a/utils/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py b/utils/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..0fc7f7ef6e3b7619fd8902acae4e5d8f69ad993b
--- /dev/null
+++ b/utils/model/utils/.ipynb_checkpoints/diff_utils_sid-checkpoint.py
@@ -0,0 +1,376 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+def mask_inputs_RFnar(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, nar= None, t=None, T= None, mask_seq_token=True, mask_seq_random=False, mask_xyz_hole=True, mask_xyz_random=False):
+ """
+ RFnar inference
+ """
+ ic(seq.shape)
+ seq = seq[0,:1]
+ ic(seq.shape)
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = ~input_seq_mask[0]
+ ic(seq_mask.shape)
+
+ str_mask = ~input_str_mask[0]
+ #nar = Nonautoregressive()
+ r = (T-t)/T
+ print(f'USING THIS R: {r}')
+
+ #mask sequence
+ if mask_seq_token:
+ print("MASK SEQ TOKEN")
+ seq_corrupt = seq.clone()
+ seq_corrupt[:,seq_mask] = 21
+ ic(seq)
+ ic(seq_corrupt)
+ elif mask_seq_random:
+ print("MASK SEQ RANDOM")
+
+
+ ic(seq_corrupt)
+
+ seq=seq_corrupt.repeat(seq.shape[0], 1)
+ ic(seq.shape)
+ seq_corrupt_onehot=torch.nn.functional.one_hot(seq_corrupt,num_classes=22).float()[0]
+ ic(seq_corrupt_onehot.shape)
+
+ ### msa_masked ###
+ ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+ msa_masked[:,0,:,:22] = seq_corrupt_onehot
+
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ #msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_corrupt_onehot
+ #msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ # msa_full #
+ #make msa_full same size as msa_masked
+ ic(msa_full.shape)
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_corrupt_onehot
+ #msa_full[:,1:,:,:22] = msa_diffused
+
+ ###########
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_corrupt_onehot[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,seq_mask,21] = 0.0
+ t1d[:,~seq_mask,21] = 1.0
+
+ t1d[:,str_mask,23] = 0.0
+ t1d[:,~str_mask,23] = 1.0
+
+ t1d[:1,:,22] = r
+
+ ################
+ #mask structure#
+ ################
+ if mask_xyz_hole:
+ print("MASK XYZ BLACK HOLE")
+ ic(xyz_t.shape)
+ xyz_corrupt, xyz_mask = nar.xyz_mask_0(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True)
+ ic(xyz_corrupt.shape)
+ elif mask_xyz_random:
+ print("MASK XYZ RANDOM")
+ xyz_corrupt, xyz_mask = nar.xyz_mask_random(xyz_t[0], r, seq_mask = seq_mask, seq_xyz_mask_same = True)
+ #only corrupt first template
+ xyz_t[0]=xyz_corrupt
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+ seq_diffused = seq_corrupt_onehot
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, T=None, RFnar = False):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ if RFnar:
+
+ return mask_inputs_RFnar(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=input_seq_mask,
+ input_str_mask=input_str_mask, input_t1dconf_mask=input_t1dconf_mask, nar=diffuser, t=t, T=T)
+
+
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape )
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(xyz_prev.shape)
+ #xyz_prev = xyz_prev[-1]
+ #ic(xyz_prev.shape)
+
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ #ic(t1d.shape)
+ t1d[:,:,:,:21] = logit_aa_s[0,:21,:].permute(1,0)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
diff --git a/utils/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py b/utils/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..4350df63bd2ad5f5397d0d032c6cf2f200378c99
--- /dev/null
+++ b/utils/model/utils/.ipynb_checkpoints/inpainting_util-checkpoint.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'wt') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/utils/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py b/utils/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
new file mode 100644
index 0000000000000000000000000000000000000000..1a8bc9aca6de872a4e2b4388937c4d85e6b8d6b1
--- /dev/null
+++ b/utils/model/utils/.ipynb_checkpoints/parsers_inference-checkpoint.py
@@ -0,0 +1,151 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ i[pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/utils/model/utils/__pycache__/calc_dssp.cpython-310.pyc b/utils/model/utils/__pycache__/calc_dssp.cpython-310.pyc
new file mode 100644
index 0000000000000000000000000000000000000000..a8099164d1068e06d85c4b12051a8f548eaaddfc
Binary files /dev/null and b/utils/model/utils/__pycache__/calc_dssp.cpython-310.pyc differ
diff --git a/utils/model/utils/calc_dssp.py b/utils/model/utils/calc_dssp.py
new file mode 100644
index 0000000000000000000000000000000000000000..fe2b975316f2de89d021d4dff442182192d5b7f8
--- /dev/null
+++ b/utils/model/utils/calc_dssp.py
@@ -0,0 +1,234 @@
+#@title get secondary structure (SSE) from given PDB file
+#@markdown So far it seems the best solution is to steal code from biotite
+#@markdown which calculates the SSE of a peptide chain based on the P-SEA algorithm (Labesse 1997)
+# CODE FROM BIOKITE
+# From Krypton
+import numpy as np
+import random
+import torch
+
+def vector_dot(v1,v2):
+ return (v1*v2).sum(axis=-1)
+
+def norm_vector(v):
+ factor = np.linalg.norm(v, axis=-1)
+ if isinstance(factor, np.ndarray):
+ v /= factor[..., np.newaxis]
+ else:
+ v /= factor
+ return v
+
+def coord(x):
+ return np.asarray(x)
+def displacement(atoms1, atoms2):
+ v1 = coord(atoms1)
+ v2 = coord(atoms2)
+ if len(v1.shape) <= len(v2.shape):
+ diff = v2 - v1
+ else:
+ diff = -(v1 - v2)
+ return diff
+def distance(atoms1, atoms2):
+ diff = displacement(atoms1, atoms2)
+ return np.sqrt(vector_dot(diff, diff))
+
+def angle(atoms1, atoms2, atoms3):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms3, atoms2)
+ norm_vector(v1)
+ norm_vector(v2)
+ return np.arccos(vector_dot(v1,v2))
+
+def dihedral(atoms1, atoms2, atoms3, atoms4):
+ v1 = displacement(atoms1, atoms2)
+ v2 = displacement(atoms2, atoms3)
+ v3 = displacement(atoms3, atoms4)
+ norm_vector(v1)
+ norm_vector(v2)
+ norm_vector(v3)
+
+ n1 = np.cross(v1, v2)
+ n2 = np.cross(v2, v3)
+
+ # Calculation using atan2, to ensure the correct sign of the angle
+ x = vector_dot(n1,n2)
+ y = vector_dot(np.cross(n1,n2), v2)
+ return np.arctan2(y,x)
+
+def replace_letters(arr):
+ # Create a dictionary that maps the letters 'a', 'b', and 'c' to the corresponding numbers
+ letter_to_number = {'a': 0, 'b': 1, 'c': 2}
+
+ # Create a new array that will hold the numbers
+ nums = []
+
+ # Loop through the input array and replace the letters with the corresponding numbers
+ for letter in arr:
+ if letter in letter_to_number:
+ nums.append(letter_to_number[letter])
+ else:
+ nums.append(letter)
+
+ return np.array(nums)
+
+def replace_with_mask(arr, percentage, replace_loops=False):
+ # Make sure the percentage is between 0 and 100
+ percentage = min(max(percentage, 0), 100)
+
+ # Calculate the number of values to replace
+ num_to_replace = int(len(arr) * percentage / 100)
+
+ # Choose a random subset of the array to replace
+ replace_indices = random.sample(range(len(arr)), num_to_replace)
+
+ # Replace the values at the chosen indices with the number 3
+ for i in replace_indices:
+ arr[i] = 3
+
+ if replace_loops:
+ for i in arr:
+ if arr[i] == 2:
+ arr[i] = 3
+
+ return arr
+
+def annotate_sse(ca_coord, percentage_mask=0, replace_loops=False):
+ _radians_to_angle = 2*np.pi/360
+
+ _r_helix = ((89-12)*_radians_to_angle, (89+12)*_radians_to_angle)
+ _a_helix = ((50-20)*_radians_to_angle, (50+20)*_radians_to_angle)
+ _d2_helix = ((5.5-0.5), (5.5+0.5))
+ _d3_helix = ((5.3-0.5), (5.3+0.5))
+ _d4_helix = ((6.4-0.6), (6.4+0.6))
+
+ _r_strand = ((124-14)*_radians_to_angle, (124+14)*_radians_to_angle)
+ _a_strand = ((-180)*_radians_to_angle, (-125)*_radians_to_angle,
+ (145)*_radians_to_angle, (180)*_radians_to_angle)
+ _d2_strand = ((6.7-0.6), (6.7+0.6))
+ _d3_strand = ((9.9-0.9), (9.9+0.9))
+ _d4_strand = ((12.4-1.1), (12.4+1.1))
+
+ # Filter all CA atoms in the relevant chain.
+
+ d2i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d3i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ d4i_coord = np.full(( len(ca_coord), 2, 3 ), np.nan)
+ ri_coord = np.full(( len(ca_coord), 3, 3 ), np.nan)
+ ai_coord = np.full(( len(ca_coord), 4, 3 ), np.nan)
+
+ # The distances and angles are not defined for the entire interval,
+ # therefore the indices do not have the full range
+ # Values that are not defined are NaN
+ for i in range(1, len(ca_coord)-1):
+ d2i_coord[i] = (ca_coord[i-1], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ d3i_coord[i] = (ca_coord[i-1], ca_coord[i+2])
+ for i in range(1, len(ca_coord)-3):
+ d4i_coord[i] = (ca_coord[i-1], ca_coord[i+3])
+ for i in range(1, len(ca_coord)-1):
+ ri_coord[i] = (ca_coord[i-1], ca_coord[i], ca_coord[i+1])
+ for i in range(1, len(ca_coord)-2):
+ ai_coord[i] = (ca_coord[i-1], ca_coord[i],
+ ca_coord[i+1], ca_coord[i+2])
+
+ d2i = distance(d2i_coord[:,0], d2i_coord[:,1])
+ d3i = distance(d3i_coord[:,0], d3i_coord[:,1])
+ d4i = distance(d4i_coord[:,0], d4i_coord[:,1])
+ ri = angle(ri_coord[:,0], ri_coord[:,1], ri_coord[:,2])
+ ai = dihedral(ai_coord[:,0], ai_coord[:,1],
+ ai_coord[:,2], ai_coord[:,3])
+
+ sse = np.full(len(ca_coord), "c", dtype="U1")
+
+ # Annotate helices
+ # Find CA that meet criteria for potential helices
+ is_pot_helix = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if (
+ d3i[i] >= _d3_helix[0] and d3i[i] <= _d3_helix[1]
+ and d4i[i] >= _d4_helix[0] and d4i[i] <= _d4_helix[1]
+ ) or (
+ ri[i] >= _r_helix[0] and ri[i] <= _r_helix[1]
+ and ai[i] >= _a_helix[0] and ai[i] <= _a_helix[1]
+ ):
+ is_pot_helix[i] = True
+ # Real helices are 5 consecutive helix elements
+ is_helix = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ for i in range(len(sse)):
+ if is_pot_helix[i]:
+ counter += 1
+ else:
+ if counter >= 5:
+ is_helix[i-counter : i] = True
+ counter = 0
+ # Extend the helices by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_helix[i]:
+ sse[i] = "a"
+ if (
+ d3i[i-1] >= _d3_helix[0] and d3i[i-1] <= _d3_helix[1]
+ ) or (
+ ri[i-1] >= _r_helix[0] and ri[i-1] <= _r_helix[1]
+ ):
+ sse[i-1] = "a"
+ sse[i] = "a"
+ if (
+ d3i[i+1] >= _d3_helix[0] and d3i[i+1] <= _d3_helix[1]
+ ) or (
+ ri[i+1] >= _r_helix[0] and ri[i+1] <= _r_helix[1]
+ ):
+ sse[i+1] = "a"
+ i += 1
+
+ # Annotate sheets
+ # Find CA that meet criteria for potential strands
+ is_pot_strand = np.zeros(len(sse), dtype=bool)
+ for i in range(len(sse)):
+ if ( d2i[i] >= _d2_strand[0] and d2i[i] <= _d2_strand[1]
+ and d3i[i] >= _d3_strand[0] and d3i[i] <= _d3_strand[1]
+ and d4i[i] >= _d4_strand[0] and d4i[i] <= _d4_strand[1]
+ ) or (
+ ri[i] >= _r_strand[0] and ri[i] <= _r_strand[1]
+ and ( (ai[i] >= _a_strand[0] and ai[i] <= _a_strand[1])
+ or (ai[i] >= _a_strand[2] and ai[i] <= _a_strand[3]))
+ ):
+ is_pot_strand[i] = True
+ # Real strands are 5 consecutive strand elements,
+ # or shorter fragments of at least 3 consecutive strand residues,
+ # if they are in hydrogen bond proximity to 5 other residues
+ pot_strand_coord = ca_coord[is_pot_strand]
+ is_strand = np.zeros(len(sse), dtype=bool)
+ counter = 0
+ contacts = 0
+ for i in range(len(sse)):
+ if is_pot_strand[i]:
+ counter += 1
+ coord = ca_coord[i]
+ for strand_coord in ca_coord:
+ dist = distance(coord, strand_coord)
+ if dist >= 4.2 and dist <= 5.2:
+ contacts += 1
+ else:
+ if counter >= 4:
+ is_strand[i-counter : i] = True
+ elif counter == 3 and contacts >= 5:
+ is_strand[i-counter : i] = True
+ counter = 0
+ contacts = 0
+ # Extend the strands by one at each end if CA meets extension criteria
+ i = 0
+ while i < len(sse):
+ if is_strand[i]:
+ sse[i] = "b"
+ if d3i[i-1] >= _d3_strand[0] and d3i[i-1] <= _d3_strand[1]:
+ sse[i-1] = "b"
+ sse[i] = "b"
+ if d3i[i+1] >= _d3_strand[0] and d3i[i+1] <= _d3_strand[1]:
+ sse[i+1] = "b"
+ i += 1
+ sse=replace_letters(sse)
+ sse=replace_with_mask(sse, percentage_mask, replace_loops=replace_loops)
+ sse=torch.nn.functional.one_hot(torch.tensor(sse), num_classes=4)
+ return sse
diff --git a/utils/model/utils/contigs.py b/utils/model/utils/contigs.py
new file mode 100644
index 0000000000000000000000000000000000000000..f0057e729528392d3b297d49aec8a7db901b12f2
--- /dev/null
+++ b/utils/model/utils/contigs.py
@@ -0,0 +1,1415 @@
+# utility functions for dealing with contigs during hallucination
+import numpy as np
+import random, copy, torch, geometry, os, sys
+from kinematics import xyz_to_t2d
+
+def parse_range_string(el):
+ ''' Splits string with integer or integer range into start and end ints. '''
+ if '-' in el:
+ s,e = el.split('-')
+ s,e = int(s), int(e)
+ else:
+ s,e = int(el), int(el)
+ return s,e
+
+def ranges_to_indexes(range_string):
+ '''Converts a string containig comma-separated numeric ranges to a list of integers'''
+ idx = []
+ for x in range_string.split(','):
+ start, end = parse_range_string(x)
+ idx.extend(np.arange(start, end+1))
+ return np.array(idx)
+
+def parse_contigs(contig_input, pdb_id):
+ '''
+ Input: contig start/end by pdb chain and residue number as in the pdb file
+ ex - B12-17
+ Output: corresponding start/end indices of the "features" numpy array (idx0)
+ '''
+ contigs = []
+ for con in contig_input.split(','):
+ pdb_ch = con[0]
+ pdb_s, pdb_e = parse_range_string(con[1:])
+
+ np_s = pdb_id.index((pdb_ch, pdb_s))
+ np_e = pdb_id.index((pdb_ch, pdb_e))
+
+ contigs.append([np_s, np_e])
+ return contigs
+
+
+def mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out):
+ #####################################
+ # rearrange ref features according to hal_2_ref_idx0
+ #####################################
+ #1. find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ #2. rearrange the 6D features
+ hal_len = len(hal_2_ref_idx0)
+ if 'feat' in pdb_out:
+ d_feat = pdb_out['feat'].shape[3:]
+
+ feat_hal = np.zeros((1, hal_len, hal_len) + d_feat)
+ feat_ref = pdb_out['feat'] # (B,L,L,...)
+
+ feat_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ else:
+ feat_hal = None
+
+ #3. make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+
+ #####################################
+ # mappings between hal and ref
+ #####################################
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_out['pdb_idx'][i] for i in ref_idx0],
+ 'mask_1d': mask_1d,
+ }
+
+ return feat_hal, mappings
+
+def scatter_feats(template_mask, feat_1d_ref=None, feat_2d_ref=None, pdb_idx=None):
+ '''
+ Scatters 1D and/or 2D reference features according to mappings in hal_2_ref_idx0
+
+ Inputs
+ ----------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ feat_1d_ref: (np.array; (batch, L_ref, ...))
+ 1D refence features to scatter
+ feat_1d_ref: (np.array; (batch, L_ref, L_ref, ...))
+ pdb_idx: (list)
+ List of pdb chain and residue numbers, in the order that pdb features were read/parsed.
+
+ Outputs
+ ----------
+ feat_1d_hal: (np.array, (batch, L_hal, ...))
+ Scattered 1d reference features. "None" mappings are 0.
+ feat_2d_hal: (np.array, (batch, L_hal, L_hal, ...))
+ Scattered 2d reference features. "None" mappings are 0.
+ mappings: (dict)
+ Keeps track of corresponding possitions in ref and hal proteins.
+ '''
+ hal_2_ref_idx0, _ = contigs.sample_mask(template_mask, pdb_idx)
+ out = {}
+
+ # Find corresponding idx0 in hal and ref
+ hal_idx0 = []
+ ref_idx0 = []
+ hal_len = len(hal_2_ref_idx0)
+
+ for hal, ref in enumerate(hal_2_ref_idx0):
+ if ref is not None:
+ hal_idx0.append(hal)
+ ref_idx0.append(ref)
+
+ hal_idx0 = np.array(hal_idx0, dtype=int)
+ ref_idx0 = np.array(ref_idx0, dtype=int)
+
+ # Make the 1d binary mask, for backwards compatibility
+ hal_2_ref_idx0 = np.array(hal_2_ref_idx0, dtype=np.float32) # convert None to NaN
+ mask_1d = (~np.isnan(hal_2_ref_idx0)).astype(float)
+ mask_1d = mask_1d[None]
+
+ # scatter 2D features
+ if feat_2d_ref is not None:
+ B = feat_2d_ref.shape[0]
+ d_feat = feat_2d_ref.shape[3:]
+ feat_2d_hal = np.zeros((B, hal_len, hal_len)+d_feat)
+ feat_2d_hal[:, hal_idx0[:,None], hal_idx0[None,:]] = feat_2d_ref[:, ref_idx0[:,None], ref_idx0[None,:]]
+ out['feat_2d_hal'] = feat_2d_hal
+
+ # scatter 1D features
+ if feat_1d_ref is not None:
+ B = feat_1d_ref.shape[0]
+ d_feat = feat_1d_ref.shape[2:]
+ feat_1d_hal = np.zeros((B, hal_len)+d_feat)
+ feat_1d_hal[:, hal_idx0] = feat_1d_ref[:, ref_idx0]
+ out['feat_1d_hal'] = feat_1d_hal
+
+ # Mappings between hal and ref
+ mappings = {
+ 'con_hal_idx0': hal_idx0.tolist(),
+ 'con_ref_idx0': ref_idx0.tolist(),
+ 'mask_1d': mask_1d,
+ }
+
+ if pdb_idx is not None:
+ mappings.update({
+ 'con_hal_pdb_idx': [('A',i+1) for i in hal_idx0],
+ 'con_ref_pdb_idx': [pdb_idx[i] for i in ref_idx0],
+ })
+
+ out['mappings'] = mappings
+
+ return out
+
+def scatter_contigs(contigs, pdb_out, L_range, keep_order=False, min_gap=0):
+ '''
+ Randomly places contigs in a protein within the length range.
+
+ Inputs
+ Contig: A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number. Comma separated
+ ex: B6-11,A12-19
+ pdb_out: dictionary from the prep_input function
+ L_range: String range of possible lengths.
+ ex: 90-110
+ ex: 70
+ keep_order: keep contigs in the provided order or randomly permute
+ min_gap: minimum number of amino acids separating contigs
+
+ Outputs
+ feat_hal: target pdb features to hallucinate
+ mappings: dictionary of ways to convert from the hallucinated protein
+ to the reference protein
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #####################################
+ # make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ #####################################
+ #1. Permute contig order
+ contigs = contigs.split(',')
+
+ if not keep_order:
+ random.shuffle(contigs)
+
+ #2. convert to ref_idx0
+ contigs_ref_idx0 = []
+ for con in contigs:
+ chain = con[0]
+ s, e = parse_range_string(con[1:])
+ contigs_ref_idx0.append( [ref_pdb_2_idx0[(chain, i)] for i in range(s, e+1)] )
+
+ #3. Add minimum gap size
+ for i in range(len(contigs_ref_idx0) - 1):
+ contigs_ref_idx0[i] += [None] * min_gap
+
+ #4. Sample protein length
+ L_low, L_high = parse_range_string(L_range)
+ L_hal = np.random.randint(L_low, L_high+1)
+
+ L_con = 0
+ for con in contigs_ref_idx0:
+ L_con += len(con)
+
+ L_gaps = L_hal - L_con
+
+ if L_gaps <= 1:
+ print("Error: The protein isn't long enough to incorporate all the contigs."
+ "Consider reduce the min_gap or increasing L_range")
+ return
+
+ #5. Randomly insert contigs into gaps
+ hal_2_ref_idx0 = np.array([None] * L_gaps, dtype=float) # inserting contigs into this
+ n_contigs = len(contigs_ref_idx0)
+ insertion_idxs = np.random.randint(L_gaps + 1, size=n_contigs)
+ insertion_idxs.sort()
+
+ for idx, con in zip(insertion_idxs[::-1], contigs_ref_idx0[::-1]):
+ hal_2_ref_idx0 = np.insert(hal_2_ref_idx0, idx, con)
+
+ #6. Convert mask to feat_hal and mappings
+ hal_2_ref_idx0 = [int(el) if ~np.isnan(el) else None for el in hal_2_ref_idx0] # convert nan to None
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #7. Generate str of the sampled mask
+ contig_positive = np.array(hal_2_ref_idx0) != None
+ boundaries = np.where(np.diff(contig_positive))[0]
+ start_idx0 = np.concatenate([np.array([0]), boundaries+1])
+ end_idx0 = np.concatenate([boundaries, np.array([contig_positive.shape[0]])-1])
+ lengths = end_idx0 - start_idx0 + 1
+ is_contig = contig_positive[start_idx0]
+
+ sampled_mask = []
+ con_counter = 0
+
+ for i, is_con in enumerate(is_contig):
+ if is_con:
+ sampled_mask.append(contigs[con_counter])
+ con_counter += 1
+ else:
+ len_gap = lengths[i]
+ sampled_mask.append(f'{len_gap}-{len_gap}')
+
+ sampled_mask = ','.join(sampled_mask)
+ mappings['sampled_mask'] = sampled_mask
+
+ return feat_hal, mappings
+
+def get_receptor_contig(ref_pdb_idx):
+ rec_pdb_idx = [idx for idx in ref_pdb_idx if idx[0]=='R']
+ return SampledMask.contract(rec_pdb_idx)
+
+def mk_con_to_set(mask, set_id=None, args=None, ref_pdb_idx=None):
+ '''
+ Maps a mask or list of contigs to a set_id. If no set_id is provided, it treats
+ everything as set 0.
+
+ Input
+ -----------
+ mask (str): Mask or list of contigs. Ex: 3,B6-11,12,A12-19,9 or Ex: B6-11,A12-19
+ ref_pdb_idx (List(ch, res)): pdb idxs of the reference pdb. Ex: [(A, 2), (A, 3), ...]
+ args: Arguments object. Must have args.receptor
+ set_id (list): List of integers. Length must match contigs in mask. Ex: [0,1]
+
+ Output
+ -----------
+ con_to_set (dict): Maps str of contig to integer
+ '''
+
+ # Extract contigs
+ cons = [l for l in mask.split(',') if l[0].isalpha()]
+
+ # Assign all contigs to set 0 if set_id is not passed
+ if set_id is None:
+ set_id = [0] * len(cons)
+
+ con_to_set = dict(zip(cons, set_id))
+
+ # Assign receptor to set 0
+ if args.receptor:
+ receptor_contig = get_receptor_contig(ref_pdb_idx)
+ con_to_set.update({receptor_contig: 0})
+
+ return con_to_set
+
+def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = parse_range(contig)
+
+ return ch, s, e
+
+def mask_as_list(sampled_mask):
+ '''
+ Make a length L_hal list, with each position pointing to a ref_pdb_idx (or None)
+ '''
+ mask_list = []
+ for l in sampled_mask.split(','):
+ ch, s, e = parse_contig(l)
+ # contig
+ if ch is not None:
+ mask_list += [(ch, idx) for idx in range(s, e+1)]
+ # gap
+ else:
+ mask_list += [None for _ in range(s, e+1)]
+
+ return mask_list
+
+def mask_subset(sampled_mask, subset):
+ '''
+ Returns a 1D boolean array of where a subset of the contig is in the hallucinated protein
+
+ Input
+ ---------
+ subset (str): Some chain and residue subset of the contigs. Ex: A10-15
+ Can also just pass chain. All contig residues from that chain are selected. Ex: R
+
+ Ouput
+ ---------
+ m_1d (np.array): Boolean array where subset appears in the hallucinated protein
+
+ '''
+ mask_list = mask_as_list(sampled_mask)
+ m_1d = []
+
+ ch_subset, s, e = parse_contig(subset)
+ assert ch_subset.isalpha(), '"Subset" must include a chain reference'
+
+ if (s is None) or (e is None):
+ s = -np.inf
+ e = np.inf
+
+ for l in mask_list:
+ if l is None:
+ continue
+
+ ch, idx = l
+ if (ch == ch_subset) and (idx >= s) and (idx <= e):
+ m_1d.append(True)
+ else:
+ m_1d.append(False)
+
+ return np.array(m_1d)
+
+def mk_cce_and_hal_mask_2d(sampled_mask, con_to_set=None):
+ '''
+ Makes masks for ij pixels where the cce and hallucination loss should be applied.
+
+ Inputs
+ ---------------
+ sampled_mask (str): String of where contigs should be applied. Ex: 3,B6-11,12,A12-19,9
+ cce_cutoff (float): Apply cce loss to cb-cb distances less than this value. Angstroms.
+ con_to_set (dict): Dictionary mapping the string of a contig (ex: 'B6-11') to an integer.
+ L_rec (int): Length of the receptor, if hallucinating in the context of the receptor.
+
+ Outputs
+ ---------------
+ mask_cce (np.array, (L_hal, L_hal)): Boolean array. True where cce loss should be applied.
+ mask_hal (np.array, (L_hal, L_hal)): Boolean array. True where hallucination loss should be applied.
+ '''
+ if con_to_set is None:
+ con_to_set = mk_con_to_set(sampled_mask)
+
+ # Length of hallucinated protein
+ L_hal, L_max = mask_len(sampled_mask)
+ assert L_hal == L_max, 'A sampled mask must have gaps of a single length.'
+
+ # Map each contig to a 1D boolean mask
+ m_con = dict()
+ start_idx = 0
+ for l in sampled_mask.split(','):
+ if l[0].isalpha():
+ s, e = parse_range_string(l[1:])
+ L_con = e - s + 1
+ m = np.zeros(L_hal, dtype=bool)
+ m[start_idx:start_idx+L_con] = True
+
+ m_con[l] = m
+ start_idx += L_con
+ else:
+ L_gap, _ = parse_range_string(l)
+ start_idx += L_gap
+
+ # Combine contigs masks from each set to make 2D mask
+ mask_cce = np.zeros((L_hal, L_hal), dtype=bool)
+ for set_id in set(con_to_set.values()):
+ # gather all masks from contigs in the same set
+ masks = [m_con[k] for k,v in con_to_set.items() if v == set_id]
+ mask_1D = np.any(masks, axis=0)
+ update = mask_1D[:,None] * mask_1D[None,:]
+ mask_cce = np.any([mask_cce, update], axis=0)
+
+ # Make mask_hal
+ mask_hal = ~mask_cce
+
+ # Don't apply ANY losses on diagonal
+ mask_cce[np.arange(L_hal), np.arange(L_hal)] = False
+ mask_hal[np.arange(L_hal), np.arange(L_hal)] = False
+
+ # Don't apply ANY losses to receptor
+ m_1d_rec = mask_subset(sampled_mask, 'R')
+ m_2d_rec = m_1d_rec[:, None] * m_1d_rec[None, :]
+ mask_cce *= ~m_2d_rec
+ mask_hal *= ~m_2d_rec
+
+ return mask_cce, mask_hal
+
+
+def apply_mask(mask, pdb_out):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ pdb_out: dictionary from the prep_input function
+
+
+ Outputs
+ -------------
+ feat_hal: features from pdb_out scattered according to the sampled mask
+ mappings: dict keeping track of corresponding positions in the ref and hal features
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_idx:i for i, pdb_idx in enumerate(pdb_out['pdb_idx'])}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ #2. Convert mask to feat_hal and mappings
+ feat_hal, mappings = mk_feat_hal_and_mappings(hal_2_ref_idx0, pdb_out)
+
+ #3. Record the mask that was sampled
+ mappings['sampled_mask'] = ','.join(sampled_mask)
+
+ return feat_hal, mappings
+
+
+def sample_mask(mask, pdb_idx):
+ '''
+ Uniformly samples gap lengths, then gathers the ref features
+ into the target hal features
+
+ Inputs
+ --------------
+ mask: specify the order and ranges of contigs and gaps
+ Contig - A continuous range of residues from the pdb.
+ Inclusive of the begining and end
+ Must start with the chain number
+ ex: B6-11
+ Gap - a gap length or a range of gaps lengths the
+ model is free to hallucinate
+ Gap ranges are inclusive of the end
+ ex: 9-21
+
+ ex - '3,B6-11,9-21,A36-42,20-30,A12-24,3-6'
+
+ Outputs
+ -------------
+ hal_2_ref_idx0: (list; length=L_hal)
+ List mapping hal_idx0 positions to ref_idx0 positions.
+ "None" used for indices that do not map to ref.
+ ex: [None, None, 3, 4, 5, None, None, None, 34, 35, 36]
+ sampled_mask: (str)
+ string of the sampled mask, so the transformations can be reapplied
+ ex - '3-3,B6-11,9-9,A36-42,20-20,A12-24,5-5'
+
+ '''
+
+ ref_pdb_2_idx0 = {pdb_i:i for i, pdb_i in enumerate(pdb_idx)}
+
+ #1. make a map from hal_idx0 to ref_idx0. Has None for gap regions
+ hal_2_ref_idx0 = []
+ sampled_mask = []
+ for el in mask.split(','):
+
+ if el[0].isalpha(): # el is a contig
+ sampled_mask.append(el)
+ chain = el[0]
+ s,e = parse_range_string(el[1:])
+
+ for i in range(s, e+1):
+ idx0 = ref_pdb_2_idx0[(chain, i)]
+ hal_2_ref_idx0.append(idx0)
+
+ else: # el is a gap
+ # sample gap length
+ s,e = parse_range_string(el)
+ gap_len = np.random.randint(s, e+1)
+ hal_2_ref_idx0 += [None]*gap_len
+ sampled_mask.append(f'{gap_len}-{gap_len}')
+
+ return hal_2_ref_idx0, sampled_mask
+
+
+class GapResampler():
+ def __init__(self, use_bkg=True):
+ '''
+
+ '''
+
+ self.counts_passed = {} # dictionary for tallying counts of gap lengths for designs passing some threshold
+ self.counts_bkg = {}
+ self.use_bkg = use_bkg
+
+
+ def clean_mask(self, mask):
+ '''
+ Makes mask into a cononical form.
+ Ensures masks always alternate gap, contig and that
+ masks begin and end with a gap (even of length 0)
+
+ Input
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+
+ Output
+ -----------
+ A canonicalized mask. Ex: N,9,A12-19,15,B45-52,0,C
+ '''
+ mask = mask.split(',')
+ mask_out = []
+ was_contig = True
+ was_gap = False
+
+ for i, el in enumerate(mask):
+ is_contig = el[0].isalpha()
+ is_gap = not is_contig
+ is_last = i == len(mask) - 1
+
+ # accepting gaps as either x-x or just x
+ if is_gap:
+ if '-' in el:
+ x1, x2 = el.split('-')
+ if x1 != x2:
+ print(f"Error: Gap must not be a range: {mask}")
+ return None
+ gap = x1
+ else:
+ gap = el
+
+ if is_contig:
+ contig = el
+
+ # gap -> contig: just append new contig
+ if (was_gap and is_contig):
+ mask_out.append(contig)
+
+ # contig -> gap: just append gap
+ elif (was_contig and is_gap):
+ mask_out.append(gap)
+
+ # contig -> contig: insert gap of 0, then add contig
+ elif (was_contig and is_contig):
+ mask_out.append('0')
+ mask_out.append(contig)
+
+ # gap -> gap: add them
+ elif (was_gap and is_gap):
+ combined_len = int(mask_out[-1]) + int(gap)
+ mask_out[-1] = str(combined_len)
+
+ # ensure last mask element is a gap
+ if (is_last and is_contig):
+ mask_out.append('0')
+
+ # update what previous element was
+ was_contig = el[0].isalpha()
+ was_gap = ~is_contig
+
+ # add 'N' and 'C' contigs
+ mask_out.insert(0, 'N')
+ mask_out.append('C')
+
+ return ','.join(mask_out)
+
+
+ def add_mask(self, mask, counting_dict):
+ '''
+ Adds counts of gap lengths to counting_dict
+
+ Inputs
+ -----------
+ masks: list of masks (str). Mask format: comma separted list
+ of alternating gap_length (int or int-int), contig.
+ Ex - 9,A12-19,15,B45-52 OR 9-9,A12-19,15-15,B45-52
+ '''
+ mask = self.clean_mask(mask)
+ mask = mask.split(',')
+ n_gaps = len(mask) // 2
+
+ # count occurances of contig,gap,contig triples
+ for i in range(n_gaps):
+ con1, gap, con2 = mask[2*i : 2*i+3]
+
+ # count gap length
+ if con1 in counting_dict:
+ if (gap, con2) in counting_dict[con1]:
+ counting_dict[con1][(gap, con2)] += 1
+ else:
+ counting_dict[con1][(gap, con2)] = 1
+ else:
+ counting_dict[con1] = {(gap, con2): 1}
+
+
+ def add_mask_pass(self, mask):
+ '''
+ Add a mask that passed to self.counts_passed
+ '''
+ self.add_mask(mask, self.counts_passed)
+
+
+ def add_mask_bkg(self, mask):
+ '''
+ Add a mask that passed to self.counts_bkg
+ '''
+ self.add_mask(mask, self.counts_bkg)
+
+
+ def get_enrichment(self):
+ '''
+ Calculate the ratio of counts_passed / count_bkg
+ Also notes all contigs
+ '''
+ if self.use_bkg is False:
+ print('Please pass in background masks and set self.use_bkg=True')
+ return
+
+ self.counts_enrich = copy.copy(self.counts_passed)
+ self.con_all = set()
+
+ for con1 in self.counts_enrich.keys():
+ self.con_all |= set([con1])
+
+ for gap, con2 in self.counts_enrich[con1].keys():
+ self.con_all |= set([con2])
+ bkg = self.counts_bkg[con1][(gap, con2)]
+ cnt = self.counts_passed[con1][(gap, con2)]
+ self.counts_enrich[con1][(gap, con2)] = cnt / bkg
+
+ def sample_mask(self):
+ '''
+ Sample a mask
+ '''
+ searching = True
+ while searching:
+ n_gaps = len(self.con_all) - 1
+ mask = ['N']
+
+ if self.use_bkg:
+ counts = self.counts_enrich
+ else:
+ counts = self.counts_passed
+
+ for i in range(n_gaps):
+ con_last = mask[-1]
+
+ # only allow jump to C as last option
+ if i == n_gaps - 1:
+ con_used = set(mask[::2])
+ else:
+ con_used = set(mask[::2]+['C'])
+
+ con_free = self.con_all - con_used
+
+ # get available "jumps" (con -> gap, con) you can make
+ jumps_all = counts[con_last]
+ jumps_free = {k:v for k,v in jumps_all.items() if k[1] in con_free}
+
+ if len(jumps_free) == 0:
+ print('No available jumps to continue the mask. Sampling again...')
+ else:
+ # normalize counts and sample move
+ mvs, cnt = zip(*jumps_free.items())
+ cnt = np.array(cnt)
+ prob = cnt / cnt.sum()
+ idx = np.random.choice(len(prob), p=prob)
+ mv = mvs[idx]
+
+ # add to the mask
+ mask.append(mv[0])
+ mask.append(mv[1])
+
+ # check that mask has the right number of elements
+ if len(mask) == 2*n_gaps + 1:
+ searching = False
+ else:
+ searching = True
+
+ return ','.join(mask[1:-1])
+
+
+ def gaps_as_ranges(self, mask):
+ '''
+ Convert gaps of a single int to ranges, for
+ backwards compatibility reasons
+ '''
+
+ mask_out = []
+ for el in mask.split(','):
+ if el[0].isalpha():
+ mask_out.append(el)
+ else:
+ mask_out.append(f'{el}-{el}')
+
+ return ','.join(mask_out)
+
+
+def recover_mask(trb):
+ '''
+ Recover the string of the sampled mask given the trb file
+ '''
+
+ L_hal = trb['mask_contig'].shape[0]
+ mask = []
+
+ for idx0 in range(L_hal):
+ # what is the current idx
+ if idx0 in trb['con_hal_idx0']:
+ is_con = True
+ is_gap = False
+ else:
+ is_con = False
+ is_gap = True
+
+ # dealing with the first entry
+ if idx0 == 0:
+ if is_gap:
+ L_gap = 1
+ elif is_con:
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+
+ # take action based on what happend last time
+ else:
+ if (was_gap) and (is_gap):
+ L_gap +=1
+ #elif (was_con) and (is_con):
+ # continue
+ elif (was_gap) and (is_con):
+ # end gap
+ mask.append(f'{L_gap}-{L_gap}')
+ # start con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ con_start = f'{ch}{idx}'
+ elif (was_con) and (is_gap):
+ # end con
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0) ]
+ mask.append(f'{con_start}-{idx}')
+ # start gap
+ L_gap = 1
+
+ # dealing with last entry
+ if idx0 == L_hal-1:
+ if is_gap:
+ mask.append(f'{L_gap}-{L_gap}')
+ elif is_con: # (edge case not handled: con starts and ends on last idx)
+ ch, idx = trb['con_ref_pdb_idx'][ trb['con_hal_idx0'].tolist().index(idx0-1) ]
+ mask.append(f'{con_start}-{idx}')
+
+ # update what last position was
+ was_con = copy.copy(is_con)
+ was_gap = copy.copy(is_gap)
+
+ return ','.join(mask)
+
+
+def mask_len(mask):
+ '''
+ Calculate the min and max possible length that can
+ be sampled given a mask
+ '''
+ L_min = 0
+ L_max = 0
+
+ for el in mask.split(','):
+ if el[0].isalpha(): # is con
+ con_s, con_e = el[1:].split('-')
+ con_s, con_e = int(con_s), int(con_e)
+ L_con = con_e - con_s + 1
+ L_min += L_con
+ L_max += L_con
+
+ else: # is gap
+ if '-' in el:
+ gap_min, gap_max = el.split('-')
+ gap_min, gap_max = int(gap_min), int(gap_max)
+ L_min += gap_min
+ L_max += gap_max
+ else:
+ L_min += int(el)
+ L_max += int(el)
+
+ return L_min, L_max
+
+class SampledMask():
+ def __init__(self, mask_str, ref_pdb_idx, con_to_set=None):
+ self.str = mask_str
+ self.L_hal = len(self)
+ self.L_ref = len(ref_pdb_idx)
+
+ #################
+ # con indices in hal and ref
+ #################
+ self.ref_pdb_idx = ref_pdb_idx
+ self.hal_pdb_idx = [('A', i) for i in range(1, len(self)+1)]
+
+ hal_idx0 = 0
+ con_ref_pdb_idx = []
+ con_hal_pdb_idx = []
+ con_ref_idx0 = []
+ con_hal_idx0 = []
+
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ for res in range(s, e+1):
+ con_ref_pdb_idx.append((ch, res))
+ con_hal_pdb_idx.append(('A', hal_idx0+1))
+ con_ref_idx0.append(self.ref_pdb_idx.index((ch, res)))
+ con_hal_idx0.append(hal_idx0)
+ hal_idx0 += 1
+ # gap
+ else:
+ for _ in range(s):
+ hal_idx0 += 1
+
+ self.con_mappings = {
+ 'ref_pdb_idx': con_ref_pdb_idx,
+ 'hal_pdb_idx': con_hal_pdb_idx,
+ 'ref_idx0': con_ref_idx0,
+ 'hal_idx0': con_hal_idx0,
+ }
+
+ #################
+ # con_to_set mapping
+ #################
+ if con_to_set:
+ self.con_to_set = con_to_set
+ else:
+ contigs = self.get_contigs()
+ self.con_to_set = dict(zip(contigs, len(contigs)*[0]))
+
+ # set_to_con mapping
+ set_to_con = {}
+ for k, v in self.con_to_set.items():
+ set_to_con[v] = set_to_con.get(v, []) + [k] # invert a dictionary with non-unique values
+ self.set_to_con = set_to_con
+
+ def __len__(self,):
+ _, L_max = self.mask_len(self.str)
+ return L_max
+
+ def map(self, sel, src, dst):
+ '''
+ Convert the contig selection in one indexing scheme to another.
+ Will return None if selection is not in a contig.
+
+ Input
+ ----------
+ sel (str): selection of a contig range or idx0 range. Can take multiple comma separated values of same type. Ex: A5-10,B2-8 or 3-8,14-21
+ src (str): <'ref', 'hal'>
+ dst (str): <'ref_pdb_idx', 'hal_pdb_idx', 'ref_idx0', 'hal_idx0>
+ '''
+ out = []
+ for con in sel.split(','):
+
+ ch, s, e = SampledMask.parse_contig(con)
+
+ # selection type is pdb_idx
+ if ch:
+ src_long = f'{src}_pdb_idx'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get((ch, res)) for res in range(s, e+1)]
+
+ # selection type is idx0
+ else:
+ src_long = f'{src}_idx0'
+ mapping = dict(zip(self.con_mappings[src_long], self.con_mappings[dst]))
+ out += [mapping.get(i) for i in range(s, e+1)]
+
+ return out
+
+ @staticmethod
+ def expand(mask_str):
+ '''
+ Ex: '2,A3-5,3' --> [None, None, (A,3), (A,4), (A,5), None, None, None]
+ '''
+ expanded = []
+ for l in mask_str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ expanded += [(ch, res) for res in range(s, e+1)]
+ # gap
+ else:
+ expanded += [None for _ in range(s)]
+
+ return expanded
+
+ @staticmethod
+ def contract(pdb_idx):
+ '''
+ Inverse of expand
+ Ex: [None, None, (A,3), (A,4), (A,5), None, None, None] --> '2,A3-5,3'
+ '''
+
+ contracted = []
+ l_prev = (None, -200)
+ first_el_written = False
+
+ for l_curr in pdb_idx:
+ if l_curr is None:
+ l_curr = (None, -100)
+
+ # extend gap
+ if l_curr == l_prev:
+ L_gap += 1
+
+ # extend con
+ elif l_curr == (l_prev[0], l_prev[1]+1):
+ con_e = l_curr[1]
+
+ # new gap
+ elif (l_curr != l_prev) and (l_curr[0] is None):
+ # write prev con
+ if 'con_ch' in locals():
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+
+ L_gap = 1
+
+ # new con
+ elif (l_curr != l_prev) and isinstance(l_curr[0], str):
+ # write prev con
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ # write prev gap
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ con_ch = l_curr[0]
+ con_s = l_curr[1]
+ con_e = l_curr[1]
+
+ # update l_prev
+ l_prev = l_curr
+
+ # write last element
+ if isinstance(l_prev[0], str) and ('con_ch' in locals()):
+ contracted.append(f'{con_ch}{con_s}-{con_e}')
+ elif 'L_gap' in locals():
+ contracted.append(str(L_gap))
+
+ return ','.join(contracted)
+
+ def subset(self, sub):
+ '''
+ Make a mask_str that is a subset of the original mask_str
+ Ex: self.mask_str = '2,A5-20,4', sub='A5-10' --> '2,A5-10,14'
+ '''
+
+ # map from hal_idx0 to ref_pdb_idx
+ hal_idx0 = self.map(sub, 'ref', 'hal_idx0')
+ ref_pdb_idx = SampledMask.expand(sub)
+ mapping = dict(zip(hal_idx0, ref_pdb_idx))
+
+ expanded = [mapping.get(idx0) for idx0 in range(len(self))]
+
+ return self.contract(expanded)
+
+ def mask_len(self, mask):
+ '''
+ Technically, can take both sampled and unsampled mask
+ '''
+ L_min = 0
+ L_max = 0
+ for l in self.str.split(','):
+ ch, s, e = SampledMask.parse_contig(l)
+
+ # contig
+ if ch:
+ L_min += e - s + 1
+ L_max += e - s + 1
+ # gap
+ else:
+ L_min += s
+ L_max += e
+
+ return L_min, L_max
+
+ def get_contigs(self, include_receptor=True):
+ '''
+ Get a list of all contigs in the mask
+ '''
+ [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0]]
+
+ contigs = []
+ for con in self.str.split(','):
+ ch = SampledMask.parse_contig(con)[0]
+ if ch == 'R' and include_receptor == False:
+ continue
+ if ch:
+ contigs.append(con)
+
+ return contigs
+
+ def get_gaps(self,):
+ '''
+ Get a list of all gaps in the mask
+ '''
+ return [con for con in self.str.split(',') if SampledMask.parse_contig(con)[0] is None]
+
+ @staticmethod
+ def parse_range(_range):
+ if '-' in _range:
+ s, e = _range.split('-')
+ else:
+ s, e = _range, _range
+
+ return int(s), int(e)
+
+ @staticmethod
+ def parse_contig(contig):
+ '''
+ Return the chain, start and end residue in a contig or gap str.
+
+ Ex:
+ 'A4-8' --> 'A', 4, 8
+ 'A5' --> 'A', 5, 5
+ '4-8' --> None, 4, 8
+ 'A' --> 'A', None, None
+ '''
+
+ # is contig
+ if contig[0].isalpha():
+ ch = contig[0]
+ if len(contig) > 1:
+ s, e = SampledMask.parse_range(contig[1:])
+ else:
+ s, e = None, None
+ # is gap
+ else:
+ ch = None
+ s, e = SampledMask.parse_range(contig)
+
+ return ch, s, e
+
+ def remove_diag(self, m_2d):
+ '''
+ Set the diagonal of a 2D boolean array to False
+ '''
+ L = m_2d.shape[0]
+ m_2d[np.arange(L), np.arange(L)] = False
+
+ return m_2d
+
+ def get_receptor_contig(self,):
+ '''
+ Returns None if there is no chain R in the mask_str
+ '''
+ receptor_contig = [l for l in self.get_contigs() if 'R' in l]
+
+ if len(receptor_contig) == 0:
+ receptor_contig = None
+ else:
+ receptor_contig = ','.join(receptor_contig)
+
+ return receptor_contig
+
+ def remove_receptor(self, m_2d):
+ '''
+ Remove intra-receptor contacts (chain R) from a mask
+ '''
+ receptor_contig = self.get_receptor_contig()
+
+ if receptor_contig: # has chain R
+ m_1d = np.zeros(self.L_hal, dtype=bool)
+ idx = np.array(self.map(receptor_contig, 'ref', 'hal_idx0'))
+ m_1d[idx] = True
+ update = m_1d[:, None] * m_1d[None, :]
+ m_2d = m_2d * ~update
+
+ return m_2d
+
+ def get_mask_con(self, include_receptor=False):
+ # Make a 2D boolean mask for each contig set
+ L = self.L_hal
+ mask_con = np.zeros([L, L], dtype=bool)
+
+ for set_id, contigs in self.set_to_con.items():
+ m_1d = np.zeros(L, dtype=bool)
+ for con in contigs:
+ idx = self.map(con, 'ref', 'hal_idx0')
+ idx = [l for l in idx if l != None]
+ idx = np.array(idx, dtype=int)
+ m_1d[idx] = True
+
+ update = m_1d[:, None] * m_1d[None, :]
+ mask_con = np.any([mask_con, update], axis=0)
+
+ # clean up
+ mask_con = self.remove_diag(mask_con)
+
+ if not include_receptor:
+ mask_con = self.remove_receptor(mask_con)
+
+ return mask_con
+
+ def get_mask_hal(self,):
+ mask_hal = ~self.get_mask_con()
+ mask_hal = self.remove_diag(mask_hal)
+ mask_hal = self.remove_receptor(mask_hal)
+
+ return mask_hal
+
+ def get_mask_cce(self, pdb, cce_cutoff=20., include_receptor=False):
+ '''
+ Remove ij pixels where contig distances are greater than cce_cutoff.
+ '''
+ # start with mask_con
+ mask_con = self.get_mask_con(include_receptor=include_receptor)
+
+ # get ref dists
+ xyz_ref = torch.tensor(pdb['xyz'][:,:3,:]).float()
+ c6d_ref = geometry.xyz_to_c6d(xyz_ref[None].permute(0,2,1,3),{'DMAX':20.0}).numpy()
+ dist = c6d_ref[0,:,:,0] # (L_ref, L_ref)
+
+ # scatter
+ dist_scattered = self.scatter_2d(dist)
+
+ # apply cce cuttoff
+ update = dist_scattered < cce_cutoff
+ mask_cce = np.all([mask_con, update], axis=0)
+
+ return mask_cce
+
+ def scatter_2d(self, ref_feat_2d):
+ '''
+ Inputs
+ ---------
+ ref_feat_2d (np.array; (L_ref, L_ref, ...)): Features to be scattered. The first two leading dimensions must be equal to L_ref.
+ '''
+ assert ref_feat_2d.shape[:2] == (self.L_ref, self.L_ref), 'ERROR: feat_2d must have leading dimensions of (L_ref, L_ref)'
+
+ trailing_dims = ref_feat_2d.shape[2:]
+ dtype = ref_feat_2d.dtype
+ hal_feat_2d = np.zeros((self.L_hal, self.L_hal)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_2d[con_hal_idx0[:, None], con_hal_idx0[None, :]] = ref_feat_2d[ref_hal_idx0[:, None], ref_hal_idx0[None, :]]
+
+ return hal_feat_2d
+
+ def scatter_1d(self, ref_feat_1d):
+ '''
+ Inputs
+ ---------
+ ref_feat_1d (np.array; (L_ref, ...)): Features to be scattered. The first leading dimension must be equal to L_ref.
+ '''
+ assert ref_feat_1d.shape[0] == self.L_ref, 'ERROR: feat_1d must have leading dimensions of (L_ref,)'
+
+ trailing_dims = ref_feat_1d.shape[1:]
+ dtype = ref_feat_1d.dtype
+ hal_feat_1d = np.zeros((self.L_hal,)+trailing_dims, dtype=dtype)
+
+ con_hal_idx0 = np.array(self.con_mappings['hal_idx0'])
+ ref_hal_idx0 = np.array(self.con_mappings['ref_idx0'])
+ hal_feat_1d[con_hal_idx0] = ref_feat_1d[ref_hal_idx0]
+
+ return hal_feat_1d
+
+ def idx_for_template(self, gap=200):
+ '''
+ Essentially return hal_idx0, except have a large jump for chain B,
+ to simulate a chain break. If B contains internal jumps in residue
+ numbering, these are preserved.
+ '''
+
+ is_rec = self.m1d_receptor()
+ resi_rec = np.array([idx[1] for idx in SampledMask.expand(self.str)
+ if idx is not None and idx[0]=='R'])
+ L_binder = sum(~is_rec)
+
+
+ if len(resi_rec)>0:
+ if is_rec[0]:
+ # receptor first
+ idx_tmpl = np.arange(resi_rec[-1]+gap+1, resi_rec[-1]+gap+1+L_binder)
+ idx_tmpl = np.concatenate([resi_rec, idx_tmpl])
+ else:
+ # receptor second
+ idx_tmpl = np.arange(L_binder)
+ if resi_rec[0] <= idx_tmpl[-1]+gap:
+ resi_rec += idx_tmpl[-1] - resi_rec[0] + gap + 1
+ idx_tmpl = np.concatenate([idx_tmpl, resi_rec])
+ else:
+ #when no receptor
+ idx_tmpl = np.arange(L_binder)
+ return idx_tmpl
+
+ def m1d_receptor(self,):
+ '''
+ Get a boolean array, True if the position corresponds to the receptor
+ '''
+ m1d = [(l is not None) and (l[0] == 'R') for l in SampledMask.expand(self.str)]
+ return np.array(m1d)
+
+ def erode(self, N_term=True, C_term=True):
+ '''
+ Reduce non-receptor contigs by 1 residue from the N and/or C terminus.
+ '''
+ x = SampledMask.expand(self.str)
+
+ if N_term:
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ if C_term:
+ x = x[::-1]
+
+ for i, l in enumerate(x):
+ if (l is not None) and (l[0] != 'R'):
+ x[i] = None
+ break
+
+ x = x[::-1]
+
+ self.str = self.contract(x)
+
+ return
+
+ def len_contigs(self, include_receptor=False):
+ con_str = ','.join(self.get_contigs(include_receptor))
+ return len(SampledMask.expand(con_str))
+
+
+def make_template_features(pdb, args, device, hal_2_ref_idx0=None, sm_loss=None):
+ '''
+ Inputs
+ ----------
+ sm_loss: Instance of a contig.SampledMask object used for making the loss masks.
+ '''
+ PARAMS = {
+ "DMIN" : 2.0,
+ "DMAX" : 20.0,
+ "DBINS" : 36,
+ "ABINS" : 36,
+ }
+ if args.use_template:
+ B,T = 1,1 # batch, templates
+
+ # spoof reference features
+ xyz_t = torch.tensor(pdb['xyz'][:, :3][None, None]) # (batch,templ,nres,3,3)
+ t0d = torch.ones((1,1,3)) # (batch, templ, 3)
+
+ t2d_ref = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS) # (B,T,L,L,...)
+ L_ref = t2d_ref.shape[2]
+ #t1d_ref = torch.ones(size=(B,T,L_ref,3), dtype=torch.float32, device=device)
+ a = 2 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ b = 0 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+ c = 1 * torch.ones([B,T,L_ref], dtype=torch.float32, device=device)
+
+ t1d_ref = torch.stack([a,b,c], axis=-1)
+
+ # Get the mask_str for scattering template features
+ #1. Template mask = sampled mask
+ if (args.use_template.lower() == 't') or (args.use_template.lower() == 'true'):
+ sm_tmpl = sm_loss
+ #2. Template mask is a subset of the sampled mask
+ else:
+ subset_contigs = args.use_template
+
+ if args.receptor:
+ receptor_contig = sm_loss.get_receptor_contig()
+ subset_contigs = ','.join([subset_contigs, receptor_contig])
+
+ mask_str_tmpl = sm_loss.subset(subset_contigs)
+ sm_tmpl = SampledMask(mask_str=mask_str_tmpl, ref_pdb_idx=pdb['pdb_idx'])
+
+ # scatter template features
+ # make leading dims (L,(L),...)
+ t1d_ref = t1d_ref.permute(2,3,0,1) # (L, ..., B, T)
+ t2d_ref = t2d_ref.permute(2,3,4,0,1) # (L, L, ..., B, T)
+
+ t1d_tmpl = sm_tmpl.scatter_1d(t1d_ref.cpu().numpy())
+ t2d_tmpl = sm_tmpl.scatter_2d(t2d_ref.cpu().numpy())
+
+ # update t2d_tmpl with mask_con (could update with mask_cce instead?)
+ mask_con = sm_tmpl.get_mask_con(include_receptor=True)
+ t2d_tmpl = (t2d_tmpl.T * mask_con.T).T # trick to broadcast arrays if leading dimensions match
+
+ t1d_tmpl = torch.tensor(t1d_tmpl, device=device)
+ t2d_tmpl = torch.tensor(t2d_tmpl, device=device)
+
+ # Permute B and T dims back to front
+ t1d_tmpl = t1d_tmpl.permute(2,3,0,1)
+ t2d_tmpl = t2d_tmpl.permute(3,4,0,1,2)
+
+ # Make last 3 idx of last dim all 1 to mimick Ivan's template feature
+ t2d_tmpl[..., -3:] = 1.
+
+ idx = torch.tensor(sm_tmpl.idx_for_template(gap=200), device=device)[None]
+
+ net_kwargs = {
+ 'idx': idx,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ elif args.template_pdbs is not None:
+ B,T = 1, len(args.template_pdbs) # batch, templates
+
+ # get xyz features of all templates
+ xyz_t = [torch.tensor(parse_pdb(f_pdb)['xyz'][:, :3]) for f_pdb in args.template_pdbs]
+ xyz_t = torch.stack(xyz_t, axis=0)[None] # (batch, template, nres, 3, 3)
+ t0d = torch.ones(B,T,3)
+
+ t2d_tmpl = xyz_to_t2d(xyz_t=xyz_t, t0d=t0d, params=PARAMS).to(device) # (B,T,L,L,...)
+ L_tmpl = t2d_tmpl.shape[2]
+ t1d_tmpl = torch.ones(size=(B,T,L_tmpl,3), dtype=torch.float32, device=device)
+
+ # spoof pdb idx
+ idx_tmpl = torch.range(0, L_tmpl-1, dtype=torch.long, device=device)[None]
+
+ # Net() kwargs
+ net_kwargs = {
+ 'idx': idx_tmpl,
+ 't1d': t1d_tmpl,
+ 't2d': t2d_tmpl
+ }
+
+ else:
+ net_kwargs = {}
+
+ return net_kwargs
diff --git a/utils/model/utils/diff_utils.py b/utils/model/utils/diff_utils.py
new file mode 100644
index 0000000000000000000000000000000000000000..9a6b71a8cc0af83e027e1d8e7b5ad21ab5ccbbe9
--- /dev/null
+++ b/utils/model/utils/diff_utils.py
@@ -0,0 +1,283 @@
+import torch
+from icecream import ic
+import random
+import numpy as np
+from kinematics import get_init_xyz
+import torch.nn as nn
+from util_module import ComputeAllAtomCoords
+from util import *
+from inpainting_util import MSAFeaturize_fixbb, TemplFeaturizeFixbb, lddt_unbin
+from kinematics import xyz_to_t2d
+
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None,
+ input_str_mask=None, input_t1dconf_mask=None, diffuser=None, t=None, MODEL_PARAM=None, hotspots=None, dssp=None):
+
+
+ """
+ JG - adapted slightly for the inference case
+
+ Parameters:
+ seq (torch.tensor, required): (I,L) integer sequence
+
+ msa_masked (torch.tensor, required): (I,N_short,L,48)
+
+ msa_full (torch,.tensor, required): (I,N_long,L,25)
+
+ xyz_t (torch,tensor): (T,L,27,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ t1d_24: is there an extra dimension to input structure confidence?
+
+ diffuser: diffuser class
+
+ t: time step
+
+ NOTE: in the MSA, the order is 20aa, 1x unknown, 1x mask token. We set the masked region to 22 (masked).
+ For the t1d, this has 20aa, 1x unkown, and 1x template conf. Here, we set the masked region to 21 (unknown).
+ This, we think, makes sense, as the template in normal RF training does not perfectly correspond to the MSA.
+ """
+ assert diffuser != None, 'please choose a diffuser'
+
+ ###########
+ seq = seq[0,:1]
+ msa_masked = msa_masked[0,:1]
+ msa_full = msa_full[0,:1]
+ t1d = t1d[0]
+ xyz_t = xyz_t[0]
+
+ seq_mask = input_seq_mask[0]
+
+
+
+ ######################
+ ###sequence diffusion###
+ ######################
+ """
+ #muate some percentage of sequence to have model be able to mutate residues later in denoising trajectory
+ if True:
+ masked_values=input_seq_mask[0].nonzero()[:,0]
+ print(masked_values)
+ mut_p=math.floor(masked_values.shape[0]*.05)
+ print(mut_p)
+ mutate_indices = torch.randperm(len(masked_values))[:mut_p]
+ print(mutate_indices)
+ for i in range(len(mutate_indices)):
+ seq[0,masked_values[mutate_indices[i]]] = torch.randint(0, 21, (1,))
+ """
+ str_mask = input_str_mask[0]
+
+ x_0 = torch.nn.functional.one_hot(seq[0,...],num_classes=22).float()*2-1
+
+ #ic(seq_mask)
+
+ seq_diffused = diffuser.q_sample(x_0,torch.tensor([t-1]),mask=seq_mask)
+ #seq_diffused = torch.clamp(seq_diffused, min=-1, max=1)
+
+ seq_tmp=torch.argmax(seq_diffused,axis=-1).to(device=seq.device)
+ seq=seq_tmp.repeat(seq.shape[0], 1)
+
+ ###################
+ ###msa diffusion###
+ ###################
+
+ ### msa_masked ###
+ #ic(msa_masked.shape)
+ B,N,L,_=msa_masked.shape
+
+ msa_masked[:,0,:,:22] = seq_diffused
+
+ x_0_msa = msa_masked[0,1:,:,:22].float()*2-1
+ msa_seq_mask = seq_mask.unsqueeze(0).repeat(N-1, 1)
+ msa_diffused = diffuser.q_sample(x_0_msa,torch.tensor([t-1]),mask=msa_seq_mask)
+ #msa_diffused = torch.clamp(msa_diffused, min=-1, max=1)
+ msa_masked[:,1:,:,:22] = torch.clone(msa_diffused)
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the masked token NOTE check this
+ # index 42 is the unknown token
+ msa_masked[:,0,:,22:44] = seq_diffused
+ msa_masked[:,1:,:,22:44] = msa_diffused
+
+ # insertion/deletion stuff
+ msa_masked[:,0,~seq_mask,44:46] = 0
+
+ ### msa_full ###
+ ################
+ #msa_full[:,0,:,:22] = seq_diffused
+ #make msa_full same size as msa_masked
+ msa_full = msa_full[:,:msa_masked.shape[1],:,:]
+ msa_full[:,0,:,:22] = seq_diffused
+ msa_full[:,1:,:,:22] = msa_diffused
+
+ ### t1d ###
+ ###########
+ # NOTE: adjusting t1d last dim (confidence) from sequence mask
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],2)).float()), -1).to(seq.device)
+ t1d[:,:,:21] = seq_diffused[...,:21]
+
+ #t1d[:,:,21] *= input_t1dconf_mask
+ #set diffused conf to 0 and everything else to 1
+ t1d[:,~seq_mask,21] = 0.0
+ t1d[:,seq_mask,21] = 1.0
+
+ t1d[:1,:,22] = 1-t/diffuser.num_timesteps
+
+ t1d[:,~str_mask,23] = 0.0
+ t1d[:,str_mask,23] = 1.0
+
+ # EXPAND t1d to match model params
+ if MODEL_PARAM['d_t1d'] == 29:
+ ## added t1d features ##
+ # 24 -- dssp helix
+ # 25 -- dssp sheet
+ # 26 -- dssp loop
+ # 27 -- dssp mask
+ # 28 -- hotspot resi on target
+ t1d = torch.cat((t1d,torch.zeros(t1d.shape[0],t1d.shape[1],5)),dim=-1)
+ t1d[:,:,24:28] = dssp
+ t1d[:,:,28] = hotspots
+ t1d[:,str_mask,24:27] = 0.0
+ t1d[:,str_mask,27] = 1.0
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ xyz_t[:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ xyz_t[:,~str_mask,:,:] = float('nan')
+
+ xyz_t = get_init_xyz(xyz_t[None])
+ xyz_t = xyz_t[0]
+
+ assert torch.sum(torch.isnan(xyz_t[:,:,:3,:]))==0
+
+
+ return seq, msa_masked, msa_full, xyz_t, t1d, seq_diffused
+
+
+
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+
+def take_step(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape)
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
+
+
+def take_step_nostate(model, msa, msa_extra, seq, t1d, t2d, idx_pdb, N_cycle, xyz_prev, alpha, xyz_t,
+ alpha_t, params, T, diffuser, seq_diffused, msa_prev, pair_prev, state_prev):
+ """
+ Single step in the diffusion process, with no conditioning on state
+ """
+ compute_allatom_coords=ComputeAllAtomCoords().to(seq.device)
+ #ic(msa.shape )
+ msa_prev = None
+ pair_prev = None
+ state_prev = None
+
+ B, _, N, L, _ = msa.shape
+ with torch.no_grad():
+ with torch.cuda.amp.autocast(True):
+ for i_cycle in range(N_cycle-1):
+ msa_prev, pair_prev, xyz_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d,
+ xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_raw=True)
+
+
+ logit_s, logit_aa_s, logits_exp, xyz_prev, pred_lddt, msa_prev, pair_prev, state_prev, alpha = model(msa[:,0],
+ msa_extra[:,0],
+ seq[:,0], xyz_prev,
+ idx_pdb,
+ seq1hot=seq_diffused,
+ t1d=t1d, t2d=t2d, xyz_t=xyz_t, alpha_t=alpha_t,
+ msa_prev=msa_prev,
+ pair_prev=pair_prev,
+ state_prev=state_prev,
+ return_infer=True)
+ #ic(xyz_prev.shape)
+ #xyz_prev = xyz_prev[-1]
+ #ic(xyz_prev.shape)
+
+ #ic(logit_aa_s.shape)
+ logit_aa_s_msa = torch.clone(logit_aa_s)
+ logit_aa_s = logit_aa_s.reshape(B,-1,N,L)[:,:,0,:]
+ #ic(logit_aa_s.shape)
+ logit_aa_s = logit_aa_s.reshape(B,-1,L)
+ #ic(logit_aa_s.shape)
+ #ic(t1d.shape)
+ t1d[:,:,:,:21] = logit_aa_s[0,:21,:].permute(1,0)
+ seq_out = torch.argmax(logit_aa_s, dim=-2)
+ #ic(seq_out.shape)
+ #ic(alpha.shape)
+
+ pred_lddt_unbinned = lddt_unbin(pred_lddt)
+ _, xyz_prev = compute_allatom_coords(seq_out, xyz_prev, alpha)
+
+ if N>1:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, logit_aa_s_msa, alpha, msa_prev, pair_prev, state_prev
+ else:
+ return seq_out, xyz_prev, pred_lddt_unbinned, logit_s, logit_aa_s, alpha, msa_prev, pair_prev, state_prev
diff --git a/utils/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg b/utils/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..a514787d2c75eeede8bfab24f78440746debd28e
Binary files /dev/null and b/utils/model/utils/figs/.ipynb_checkpoints/fig1-checkpoint.jpg differ
diff --git a/utils/model/utils/figs/fig1.jpg b/utils/model/utils/figs/fig1.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..a514787d2c75eeede8bfab24f78440746debd28e
Binary files /dev/null and b/utils/model/utils/figs/fig1.jpg differ
diff --git a/utils/model/utils/figs/fig2.jpg b/utils/model/utils/figs/fig2.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..053bafd7c530d636e94fb6e3c58ee0c1231b45c9
Binary files /dev/null and b/utils/model/utils/figs/fig2.jpg differ
diff --git a/utils/model/utils/figs/fig3.jpg b/utils/model/utils/figs/fig3.jpg
new file mode 100644
index 0000000000000000000000000000000000000000..d4bce6dfae5115b075ef199b592d3b32e4e3ca4a
Binary files /dev/null and b/utils/model/utils/figs/fig3.jpg differ
diff --git a/utils/model/utils/geometry.py b/utils/model/utils/geometry.py
new file mode 100644
index 0000000000000000000000000000000000000000..58edab102102bf5650d11c72a7d5a76bb1abfb33
--- /dev/null
+++ b/utils/model/utils/geometry.py
@@ -0,0 +1,200 @@
+import numpy as np
+import torch
+
+# ============================================================
+def get_pair_dist(a, b):
+ """calculate pair distances between two sets of points
+
+ Parameters
+ ----------
+ a,b : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of two sets of atoms
+ Returns
+ -------
+ dist : pytorch tensor of shape [batch,nres,nres]
+ stores paitwise distances between atoms in a and b
+ """
+
+ dist = torch.cdist(a, b, p=2)
+ return dist
+
+# ============================================================
+def get_ang(a, b, c):
+ """calculate planar angles for all consecutive triples (a[i],b[i],c[i])
+ from Cartesian coordinates of three sets of atoms a,b,c
+
+ Parameters
+ ----------
+ a,b,c : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of three sets of atoms
+ Returns
+ -------
+ ang : pytorch tensor of shape [batch,nres]
+ stores resulting planar angles
+ """
+ v = a - b
+ w = c - b
+ v = v / torch.norm(v, dim=-1, keepdim=True)
+ w = w / torch.norm(w, dim=-1, keepdim=True)
+
+ # this is not stable at the poles
+ #vw = torch.sum(v*w, dim=-1)
+ #ang = torch.acos(vw)
+
+ # this is better
+ # https://math.stackexchange.com/questions/1143354/numerically-stable-method-for-angle-between-3d-vectors/1782769
+ y = torch.norm(v-w,dim=-1)
+ x = torch.norm(v+w,dim=-1)
+ ang = 2*torch.atan2(y, x)
+
+ return ang
+
+# ============================================================
+def get_dih(a, b, c, d):
+ """calculate dihedral angles for all consecutive quadruples (a[i],b[i],c[i],d[i])
+ given Cartesian coordinates of four sets of atoms a,b,c,d
+
+ Parameters
+ ----------
+ a,b,c,d : pytorch tensors of shape [batch,nres,3]
+ store Cartesian coordinates of four sets of atoms
+ Returns
+ -------
+ dih : pytorch tensor of shape [batch,nres]
+ stores resulting dihedrals
+ """
+ b0 = a - b
+ b1r = c - b
+ b2 = d - c
+
+ b1 = b1r/torch.norm(b1r, dim=-1, keepdim=True)
+
+ v = b0 - torch.sum(b0*b1, dim=-1, keepdim=True)*b1
+ w = b2 - torch.sum(b2*b1, dim=-1, keepdim=True)*b1
+
+ x = torch.sum(v*w, dim=-1)
+ y = torch.sum(torch.cross(b1,v,dim=-1)*w, dim=-1)
+ ang = torch.atan2(y, x)
+
+ return ang
+
+
+# ============================================================
+def xyz_to_c6d(xyz, params):
+ """convert cartesian coordinates into 2d distance
+ and orientation maps
+
+ Parameters
+ ----------
+ xyz : pytorch tensor of shape [batch,3,nres,3]
+ stores Cartesian coordinates of backbone N,Ca,C atoms
+ Returns
+ -------
+ c6d : pytorch tensor of shape [batch,nres,nres,4]
+ stores stacked dist,omega,theta,phi 2D maps
+ """
+
+ batch = xyz.shape[0]
+ nres = xyz.shape[2]
+
+ # three anchor atoms
+ N = xyz[:,0]
+ Ca = xyz[:,1]
+ C = xyz[:,2]
+
+ # recreate Cb given N,Ca,C
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+
+ # 6d coordinates order: (dist,omega,theta,phi)
+ c6d = torch.zeros([batch,nres,nres,4],dtype=xyz.dtype,device=xyz.device)
+
+ dist = get_pair_dist(Cb,Cb)
+ dist[torch.isnan(dist)] = 999.9
+ c6d[...,0] = dist + 999.9*torch.eye(nres,device=xyz.device)[None,...]
+ b,i,j = torch.where(c6d[...,0]=params['DMAX']] = 999.9
+
+ return c6d
+
+
+# ============================================================
+def c6d_to_bins(c6d,params):
+ """bin 2d distance and orientation maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ dbins = torch.linspace(params['DMIN']+dstep, params['DMAX'], params['DBINS'],dtype=c6d.dtype,device=c6d.device)
+ ab360 = torch.linspace(-np.pi+astep, np.pi, params['ABINS'],dtype=c6d.dtype,device=c6d.device)
+ ab180 = torch.linspace(astep, np.pi, params['ABINS']//2,dtype=c6d.dtype,device=c6d.device)
+
+ db = torch.bucketize(c6d[...,0].contiguous(),dbins)
+ ob = torch.bucketize(c6d[...,1].contiguous(),ab360)
+ tb = torch.bucketize(c6d[...,2].contiguous(),ab360)
+ pb = torch.bucketize(c6d[...,3].contiguous(),ab180)
+
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).to(torch.uint8)
+
+
+# ============================================================
+def dist_to_bins(dist,params):
+ """bin 2d distance maps
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ db = torch.round((dist-params['DMIN']-dstep/2)/dstep)
+
+ db[db<0] = 0
+ db[db>params['DBINS']] = params['DBINS']
+
+ return db.long()
+
+
+# ============================================================
+def c6d_to_bins2(c6d,params):
+ """bin 2d distance and orientation maps
+ (alternative slightly simpler version)
+ """
+
+ dstep = (params['DMAX'] - params['DMIN']) / params['DBINS']
+ astep = 2.0*np.pi / params['ABINS']
+
+ db = torch.round((c6d[...,0]-params['DMIN']-dstep/2)/dstep)
+ ob = torch.round((c6d[...,1]+np.pi-astep/2)/astep)
+ tb = torch.round((c6d[...,2]+np.pi-astep/2)/astep)
+ pb = torch.round((c6d[...,3]-astep/2)/astep)
+
+ # put all dparams['DBINS']] = params['DBINS']
+ ob[db==params['DBINS']] = params['ABINS']
+ tb[db==params['DBINS']] = params['ABINS']
+ pb[db==params['DBINS']] = params['ABINS']//2
+
+ return torch.stack([db,ob,tb,pb],axis=-1).long()
+
+
+# ============================================================
+def get_cb(N,Ca,C):
+ """recreate Cb given N,Ca,C"""
+ b = Ca - N
+ c = C - Ca
+ a = torch.cross(b, c, dim=-1)
+ Cb = -0.58273431*a + 0.56802827*b - 0.54067466*c + Ca
+ return Cb
diff --git a/utils/model/utils/inpainting_util.py b/utils/model/utils/inpainting_util.py
new file mode 100644
index 0000000000000000000000000000000000000000..4350df63bd2ad5f5397d0d032c6cf2f200378c99
--- /dev/null
+++ b/utils/model/utils/inpainting_util.py
@@ -0,0 +1,807 @@
+import math
+import os
+import csv
+import random
+import torch
+from torch.utils import data
+import numpy as np
+from dateutil import parser
+import contigs
+from util import *
+from kinematics import *
+import pandas as pd
+import sys
+import torch.nn as nn
+from icecream import ic
+def write_pdb(filename, seq, atoms, Bfacts=None, prefix=None, chains=None):
+ L = len(seq)
+ ctr = 1
+ seq = seq.long()
+ with open(filename, 'wt') as f:
+ for i,s in enumerate(seq):
+ if chains is None:
+ chain='A'
+ else:
+ chain=chains[i]
+
+ if (len(atoms.shape)==2):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, " CA ", util.num2aa[s],
+ chain, i+1, atoms[i,0], atoms[i,1], atoms[i,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+ elif atoms.shape[1]==3:
+ for j,atm_j in enumerate((" N "," CA "," C ")):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+ else:
+ atms = aa2long[s]
+ for j,atm_j in enumerate(atms):
+ if (atm_j is not None):
+ f.write ("%-6s%5s %4s %3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f\n"%(
+ "ATOM", ctr, atm_j, num2aa[s],
+ chain, i+1, atoms[i,j,0], atoms[i,j,1], atoms[i,j,2],
+ 1.0, Bfacts[i] ) )
+ ctr += 1
+
+def preprocess(xyz_t, t1d, DEVICE, masks_1d, ti_dev=None, ti_flip=None, ang_ref=None):
+
+ B, _, L, _, _ = xyz_t.shape
+
+ seq_tmp = t1d[...,:-1].argmax(dim=-1).reshape(-1,L).to(DEVICE, non_blocking=True)
+ alpha, _, alpha_mask,_ = get_torsions(xyz_t.reshape(-1,L,27,3), seq_tmp, ti_dev, ti_flip, ang_ref)
+ alpha_mask = torch.logical_and(alpha_mask, ~torch.isnan(alpha[...,0]))
+ alpha[torch.isnan(alpha)] = 0.0
+ alpha = alpha.reshape(B,-1,L,10,2)
+ alpha_mask = alpha_mask.reshape(B,-1,L,10,1)
+ alpha_t = torch.cat((alpha, alpha_mask), dim=-1).reshape(B,-1,L,30)
+ #t1d = torch.cat((t1d, chis.reshape(B,-1,L,30)), dim=-1)
+ xyz_t = get_init_xyz(xyz_t)
+ xyz_prev = xyz_t[:,0]
+ state = t1d[:,0]
+ alpha = alpha[:,0]
+ t2d=xyz_to_t2d(xyz_t)
+ return (t2d, alpha, alpha_mask, alpha_t, t1d, xyz_t, xyz_prev, state)
+
+def TemplFeaturizeFixbb(seq, conf_1d=None):
+ """
+ Template 1D featurizer for fixed BB examples :
+ Parameters:
+ seq (torch.tensor, required): Integer sequence
+ conf_1d (torch.tensor, optional): Precalcualted confidence tensor
+ """
+ L = seq.shape[-1]
+ t1d = torch.nn.functional.one_hot(seq, num_classes=21) # one hot sequence
+ if conf_1d is None:
+ conf = torch.ones_like(seq)[...,None]
+ else:
+ conf = conf_1d[:,None]
+ t1d = torch.cat((t1d, conf), dim=-1)
+ return t1d
+
+def MSAFeaturize_fixbb(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa[:1],num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa[:1]
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot[:1].clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust[:1].clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos[:1].clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def MSAFeaturize(msa, params):
+ '''
+ Input: full msa information
+ Output: Single sequence, with some percentage of amino acids mutated (but no resides 'masked')
+
+ This is modified from autofold2, to remove mutations of the single sequence
+ '''
+ N, L = msa.shape
+ # raw MSA profile
+ raw_profile = torch.nn.functional.one_hot(msa, num_classes=22)
+ raw_profile = raw_profile.float().mean(dim=0)
+
+ b_seq = list()
+ b_msa_clust = list()
+ b_msa_seed = list()
+ b_msa_extra = list()
+ b_mask_pos = list()
+ for i_cycle in range(params['MAXCYCLE']):
+ assert torch.max(msa) < 22
+ msa_onehot = torch.nn.functional.one_hot(msa,num_classes=22)
+ msa_fakeprofile_onehot = torch.nn.functional.one_hot(msa,num_classes=26) #add the extra two indel planes, which will be set to zero
+ msa_full_onehot = torch.cat((msa_onehot, msa_fakeprofile_onehot), dim=-1)
+
+ #make fake msa_extra
+ msa_extra_onehot = torch.nn.functional.one_hot(msa,num_classes=25)
+
+ #make fake msa_clust and mask_pos
+ msa_clust = msa
+ mask_pos = torch.full_like(msa_clust, 1).bool()
+ b_seq.append(msa[0].clone())
+ b_msa_seed.append(msa_full_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_extra.append(msa_extra_onehot.clone()) #masked single sequence onehot (nb no mask so just single sequence onehot)
+ b_msa_clust.append(msa_clust.clone()) #unmasked original single sequence
+ b_mask_pos.append(mask_pos.clone()) #mask positions in single sequence (all zeros)
+
+ b_seq = torch.stack(b_seq)
+ b_msa_clust = torch.stack(b_msa_clust)
+ b_msa_seed = torch.stack(b_msa_seed)
+ b_msa_extra = torch.stack(b_msa_extra)
+ b_mask_pos = torch.stack(b_mask_pos)
+
+ return b_seq, b_msa_clust, b_msa_seed, b_msa_extra, b_mask_pos
+
+def mask_inputs(seq, msa_masked, msa_full, xyz_t, t1d, input_seq_mask=None, input_str_mask=None, input_t1dconf_mask=None, loss_seq_mask=None, loss_str_mask=None):
+ """
+ Parameters:
+ seq (torch.tensor, required): (B,I,L) integer sequence
+ msa_masked (torch.tensor, required): (B,I,N_short,L,46)
+ msa_full (torch,.tensor, required): (B,I,N_long,L,23)
+
+ xyz_t (torch,tensor): (B,T,L,14,3) template crds BEFORE they go into get_init_xyz
+
+ t1d (torch.tensor, required): (B,I,L,22) this is the t1d before tacking on the chi angles
+
+ str_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where structure is masked at False positions
+ seq_mask_1D (torch.tensor, required): Shape (L) rank 1 tensor where seq is masked at False positions
+ """
+
+ ###########
+ B,_,_ = seq.shape
+ assert B == 1, 'batch sizes > 1 not supported'
+ seq_mask = input_seq_mask[0]
+ seq[:,:,~seq_mask] = 21 # mask token categorical value
+
+ ### msa_masked ###
+ ##################
+ msa_masked[:,:,:,~seq_mask,:20] = 0
+ msa_masked[:,:,:,~seq_mask,20] = 0
+ msa_masked[:,:,:,~seq_mask,21] = 1 # set to the unkown char
+
+ # index 44/45 is insertion/deletion
+ # index 43 is the unknown token
+ # index 42 is the masked token
+ msa_masked[:,:,:,~seq_mask,22:42] = 0
+ msa_masked[:,:,:,~seq_mask,43] = 1
+ msa_masked[:,:,:,~seq_mask,42] = 0
+
+ # insertion/deletion stuff
+ msa_masked[:,:,:,~seq_mask,44:] = 0
+
+ ### msa_full ###
+ ################
+ msa_full[:,:,:,~seq_mask,:20] = 0
+ msa_full[:,:,:,~seq_mask,21] = 1
+ msa_full[:,:,:,~seq_mask,20] = 0
+ msa_full[:,:,:,~seq_mask,-1] = 0 #NOTE: double check this is insertions/deletions and 0 makes sense
+
+ ### t1d ###
+ ###########
+ # NOTE: Not adjusting t1d last dim (confidence) from sequence mask
+ t1d[:,:,~seq_mask,:20] = 0
+ t1d[:,:,~seq_mask,20] = 1 # unknown
+
+ t1d[:,:,:,21] *= input_t1dconf_mask
+
+ #JG added in here to make sure everything fits
+ print('expanding t1d to 24 dims')
+
+ t1d = torch.cat((t1d, torch.zeros((t1d.shape[0],t1d.shape[1],t1d.shape[2],2)).float()), -1).to(seq.device)
+
+ xyz_t[:,:,~seq_mask,3:,:] = float('nan')
+
+ # Structure masking
+ str_mask = input_str_mask[0]
+ xyz_t[:,:,~str_mask,:,:] = float('nan')
+
+ return seq, msa_masked, msa_full, xyz_t, t1d
+
+
+###########################################################
+#Functions for randomly translating/rotation input residues
+###########################################################
+
+def get_translated_coords(args):
+ '''
+ Parses args.res_translate
+ '''
+ #get positions to translate
+ res_translate = []
+ for res in args.res_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1])))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1])))
+ start = 0
+
+ output = []
+ for i in res_translate:
+ temp = (i[0], i[1], start)
+ output.append(temp)
+ start += 1
+
+ return output
+
+def get_tied_translated_coords(args, untied_translate=None):
+ '''
+ Parses args.tie_translate
+ '''
+ #pdb_idx = list(parsed_pdb['idx'])
+ #xyz = parsed_pdb['xyz']
+ #get positions to translate
+ res_translate = []
+ block = 0
+ for res in args.tie_translate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(2.0) #set default distance
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ res_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ res_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+
+ #sanity check
+ if untied_translate != None:
+ checker = [i[0] for i in res_translate]
+ untied_check = [i[0] for i in untied_translate]
+ for i in checker:
+ if i in untied_check:
+ print(f'WARNING: residue {i} is specified both in --res_translate and --tie_translate. Residue {i} will be ignored in --res_translate, and instead only moved in a tied block (--tie_translate)')
+
+ final_output = res_translate
+ for i in untied_translate:
+ if i[0] not in checker:
+ final_output.append((i[0],i[1],i[2] + block + 1))
+ else:
+ final_output = res_translate
+
+ return final_output
+
+
+
+def translate_coords(parsed_pdb, res_translate):
+ '''
+ Takes parsed list in format [(chain_residue,distance,tieing_block)] and randomly translates residues accordingly.
+ '''
+
+ pdb_idx = parsed_pdb['pdb_idx']
+ xyz = np.copy(parsed_pdb['xyz'])
+ translated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in res_translate]
+ blocks = np.max(temp)
+
+ for block in range(blocks + 1):
+ init_dist = 1.01
+ while init_dist > 1: #gives equal probability to any direction (as keeps going until init_dist is within unit circle)
+ x = random.uniform(-1,1)
+ y = random.uniform(-1,1)
+ z = random.uniform(-1,1)
+ init_dist = np.sqrt(x**2 + y**2 + z**2)
+ x=x/init_dist
+ y=y/init_dist
+ z=z/init_dist
+ translate_dist = random.uniform(0,1) #now choose distance (as proportion of maximum) that coordinates will be translated
+ for res in res_translate:
+ if res[2] == block:
+ res_idx = pdb_idx.index((res[0][0],int(res[0][1:])))
+ original_coords = np.copy(xyz[res_idx,:,:])
+ for i in range(14):
+ if parsed_pdb['mask'][res_idx, i]:
+ xyz[res_idx,i,0] += np.float32(x * translate_dist * float(res[1]))
+ xyz[res_idx,i,1] += np.float32(y * translate_dist * float(res[1]))
+ xyz[res_idx,i,2] += np.float32(z * translate_dist * float(res[1]))
+ translated_coords = xyz[res_idx,:,:]
+ translated_coord_dict[res[0]] = (original_coords.tolist(), translated_coords.tolist())
+
+ return xyz[:,:,:], translated_coord_dict
+
+def parse_block_rotate(args):
+ block_translate = []
+ block = 0
+ for res in args.block_rotate.split(":"):
+ temp_str = []
+ for i in res.split(','):
+ temp_str.append(i)
+ if temp_str[-1][0].isalpha() is True:
+ temp_str.append(10) #set default angle to 10 degrees
+ for i in temp_str[:-1]:
+ if '-' in i:
+ start = int(i.split('-')[0][1:])
+ while start <= int(i.split('-')[1]):
+ block_translate.append((i.split('-')[0][0] + str(start),float(temp_str[-1]), block))
+ start += 1
+ else:
+ block_translate.append((i, float(temp_str[-1]), block))
+ block += 1
+ return block_translate
+
+def rotate_block(xyz, block_rotate,pdb_index):
+ rotated_coord_dict = {}
+ #get number of blocks
+ temp = [int(i[2]) for i in block_rotate]
+ blocks = np.max(temp)
+ for block in range(blocks + 1):
+ idxs = [pdb_index.index((i[0][0],int(i[0][1:]))) for i in block_rotate if i[2] == block]
+ angle = [i[1] for i in block_rotate if i[2] == block][0]
+ block_xyz = xyz[idxs,:,:]
+ com = [float(torch.mean(block_xyz[:,:,i])) for i in range(3)]
+ origin_xyz = np.copy(block_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ origin_xyz[i,j] = origin_xyz[i,j] - com
+ rotated_xyz = rigid_rotate(origin_xyz,angle,angle,angle)
+ recovered_xyz = np.copy(rotated_xyz)
+ for i in range(np.shape(origin_xyz)[0]):
+ for j in range(14):
+ recovered_xyz[i,j] = rotated_xyz[i,j] + com
+ recovered_xyz=torch.tensor(recovered_xyz)
+ rotated_coord_dict[f'rotated_block_{block}_original'] = block_xyz
+ rotated_coord_dict[f'rotated_block_{block}_rotated'] = recovered_xyz
+ xyz_out = torch.clone(xyz)
+ for i in range(len(idxs)):
+ xyz_out[idxs[i]] = recovered_xyz[i]
+ return xyz_out,rotated_coord_dict
+
+def rigid_rotate(xyz,a=180,b=180,c=180):
+ #TODO fix this to make it truly uniform
+ a=(a/180)*math.pi
+ b=(b/180)*math.pi
+ c=(c/180)*math.pi
+ alpha = random.uniform(-a, a)
+ beta = random.uniform(-b, b)
+ gamma = random.uniform(-c, c)
+ rotated = []
+ for i in range(np.shape(xyz)[0]):
+ for j in range(14):
+ try:
+ x = xyz[i,j,0]
+ y = xyz[i,j,1]
+ z = xyz[i,j,2]
+ x2 = x*math.cos(alpha) - y*math.sin(alpha)
+ y2 = x*math.sin(alpha) + y*math.cos(alpha)
+ x3 = x2*math.cos(beta) - z*math.sin(beta)
+ z2 = x2*math.sin(beta) + z*math.cos(beta)
+ y3 = y2*math.cos(gamma) - z2*math.sin(gamma)
+ z3 = y2*math.sin(gamma) + z2*math.cos(gamma)
+ rotated.append([x3,y3,z3])
+ except:
+ rotated.append([float('nan'),float('nan'),float('nan')])
+ rotated=np.array(rotated)
+ rotated=np.reshape(rotated, [np.shape(xyz)[0],14,3])
+
+ return rotated
+
+
+######## from old pred_util.py
+def find_contigs(mask):
+ """
+ Find contiguous regions in a mask that are True with no False in between
+
+ Parameters:
+ mask (torch.tensor or np.array, required): 1D boolean array
+
+ Returns:
+ contigs (list): List of tuples, each tuple containing the beginning and the
+ """
+ assert len(mask.shape) == 1 # 1D tensor of bools
+
+ contigs = []
+ found_contig = False
+ for i,b in enumerate(mask):
+
+
+ if b and not found_contig: # found the beginning of a contig
+ contig = [i]
+ found_contig = True
+
+ elif b and found_contig: # currently have contig, continuing it
+ pass
+
+ elif not b and found_contig: # found the end, record previous index as end, reset indicator
+ contig.append(i)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ else: # currently don't have a contig, and didn't find one
+ pass
+
+
+ # fence post bug - check if the very last entry was True and we didn't get to finish
+ if b:
+ contig.append(i+1)
+ found_contig = False
+ contigs.append(tuple(contig))
+
+ return contigs
+
+
+def reindex_chains(pdb_idx):
+ """
+ Given a list of (chain, index) tuples, and the indices where chains break, create a reordered indexing
+
+ Parameters:
+
+ pdb_idx (list, required): List of tuples (chainID, index)
+
+ breaks (list, required): List of indices where chains begin
+ """
+
+ new_breaks, new_idx = [],[]
+ current_chain = None
+
+ chain_and_idx_to_torch = {}
+
+ for i,T in enumerate(pdb_idx):
+
+ chain, idx = T
+
+ if chain != current_chain:
+ new_breaks.append(i)
+ current_chain = chain
+
+ # create new space for chain id listings
+ chain_and_idx_to_torch[chain] = {}
+
+ # map original pdb (chain, idx) pair to index in tensor
+ chain_and_idx_to_torch[chain][idx] = i
+
+ # append tensor index to list
+ new_idx.append(i)
+
+ new_idx = np.array(new_idx)
+ # now we have ordered list and know where the chainbreaks are in the new order
+ num_additions = 0
+ for i in new_breaks[1:]: # skip the first trivial one
+ new_idx[np.where(new_idx==(i+ num_additions*500))[0][0]:] += 500
+ num_additions += 1
+
+ return new_idx, chain_and_idx_to_torch,new_breaks[1:]
+
+class ObjectView(object):
+ '''
+ Easy wrapper to access dictionary values with "dot" notiation instead
+ '''
+ def __init__(self, d):
+ self.__dict__ = d
+
+def split_templates(xyz_t, t1d, multi_templates,mappings,multi_tmpl_conf=None):
+ templates = multi_templates.split(":")
+ if multi_tmpl_conf is not None:
+ multi_tmpl_conf = [float(i) for i in multi_tmpl_conf.split(",")]
+ assert len(templates) == len(multi_tmpl_conf), "Number of templates must equal number of confidences specified in --multi_tmpl_conf flag"
+ for idx, template in enumerate(templates):
+ parts = template.split(",")
+ template_mask = torch.zeros(xyz_t.shape[2]).bool()
+ for part in parts:
+ start = int(part.split("-")[0][1:])
+ end = int(part.split("-")[1]) + 1
+ chain = part[0]
+ for i in range(start, end):
+ try:
+ ref_pos = mappings['complex_con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['complex_con_hal_idx0'][ref_pos]
+ except:
+ ref_pos = mappings['con_ref_pdb_idx'].index((chain, i))
+ hal_pos_0 = mappings['con_hal_idx0'][ref_pos]
+ template_mask[hal_pos_0] = True
+
+ xyz_t_temp = torch.clone(xyz_t)
+ xyz_t_temp[:,:,~template_mask,:,:] = float('nan')
+ t1d_temp = torch.clone(t1d)
+ t1d_temp[:,:,~template_mask,:20] =0
+ t1d_temp[:,:,~template_mask,20] = 1
+ if multi_tmpl_conf is not None:
+ t1d_temp[:,:,template_mask,21] = multi_tmpl_conf[idx]
+ if idx != 0:
+ xyz_t_out = torch.cat((xyz_t_out, xyz_t_temp),dim=1)
+ t1d_out = torch.cat((t1d_out, t1d_temp),dim=1)
+ else:
+ xyz_t_out = xyz_t_temp
+ t1d_out = t1d_temp
+ return xyz_t_out, t1d_out
+
+
+class ContigMap():
+ '''
+ New class for doing mapping.
+ Supports multichain or multiple crops from a single receptor chain.
+ Also supports indexing jump (+200) or not, based on contig input.
+ Default chain outputs are inpainted chains as A (and B, C etc if multiple chains), and all fragments of receptor chain on the next one (generally B)
+ Output chains can be specified. Sequence must be the same number of elements as in contig string
+ '''
+ def __init__(self, parsed_pdb, contigs=None, inpaint_seq=None, inpaint_str=None, length=None, ref_idx=None, hal_idx=None, idx_rf=None, inpaint_seq_tensor=None, inpaint_str_tensor=None, topo=False):
+ #sanity checks
+ if contigs is None and ref_idx is None:
+ sys.exit("Must either specify a contig string or precise mapping")
+ if idx_rf is not None or hal_idx is not None or ref_idx is not None:
+ if idx_rf is None or hal_idx is None or ref_idx is None:
+ sys.exit("If you're specifying specific contig mappings, the reference and output positions must be specified, AND the indexing for RoseTTAFold (idx_rf)")
+
+ self.chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ if length is not None:
+ if '-' not in length:
+ self.length = [int(length),int(length)+1]
+ else:
+ self.length = [int(length.split("-")[0]),int(length.split("-")[1])+1]
+ else:
+ self.length = None
+ self.ref_idx = ref_idx
+ self.hal_idx=hal_idx
+ self.idx_rf=idx_rf
+ self.inpaint_seq = ','.join(inpaint_seq).split(",") if inpaint_seq is not None else None
+ self.inpaint_str = ','.join(inpaint_str).split(",") if inpaint_str is not None else None
+ self.inpaint_seq_tensor=inpaint_seq_tensor
+ self.inpaint_str_tensor=inpaint_str_tensor
+ self.parsed_pdb = parsed_pdb
+ self.topo=topo
+ if ref_idx is None:
+ #using default contig generation, which outputs in rosetta-like format
+ self.contigs=contigs
+ self.sampled_mask,self.contig_length,self.n_inpaint_chains = self.get_sampled_mask()
+ self.receptor_chain = self.chain_order[self.n_inpaint_chains]
+ self.receptor, self.receptor_hal, self.receptor_rf, self.inpaint, self.inpaint_hal, self.inpaint_rf= self.expand_sampled_mask()
+ self.ref = self.inpaint + self.receptor
+ self.hal = self.inpaint_hal + self.receptor_hal
+ self.rf = self.inpaint_rf + self.receptor_rf
+ else:
+ #specifying precise mappings
+ self.ref=ref_idx
+ self.hal=hal_idx
+ self.rf = rf_idx
+ self.mask_1d = [False if i == ('_','_') else True for i in self.ref]
+
+ #take care of sequence and structure masking
+ if self.inpaint_seq_tensor is None:
+ if self.inpaint_seq is not None:
+ self.inpaint_seq = self.get_inpaint_seq_str(self.inpaint_seq)
+ else:
+ self.inpaint_seq = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_seq = self.inpaint_seq_tensor
+
+ if self.inpaint_str_tensor is None:
+ if self.inpaint_str is not None:
+ self.inpaint_str = self.get_inpaint_seq_str(self.inpaint_str)
+ else:
+ self.inpaint_str = np.array([True if i != ('_','_') else False for i in self.ref])
+ else:
+ self.inpaint_str = self.inpaint_str_tensor
+ #get 0-indexed input/output (for trb file)
+ self.ref_idx0,self.hal_idx0, self.ref_idx0_inpaint, self.hal_idx0_inpaint, self.ref_idx0_receptor, self.hal_idx0_receptor=self.get_idx0()
+
+ def get_sampled_mask(self):
+ '''
+ Function to get a sampled mask from a contig.
+ '''
+ length_compatible=False
+ count = 0
+ while length_compatible is False:
+ inpaint_chains=0
+ contig_list = self.contigs
+ sampled_mask = []
+ sampled_mask_length = 0
+ #allow receptor chain to be last in contig string
+ if all([i[0].isalpha() for i in contig_list[-1].split(",")]):
+ contig_list[-1] = f'{contig_list[-1]},0'
+ for con in contig_list:
+ if ((all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0')) or self.topo is True:
+ #receptor chain
+ sampled_mask.append(con)
+ else:
+ inpaint_chains += 1
+ #chain to be inpainted. These are the only chains that count towards the length of the contig
+ subcons = con.split(",")
+ subcon_out = []
+ for subcon in subcons:
+ if subcon[0].isalpha():
+ subcon_out.append(subcon)
+ if '-' in subcon:
+ sampled_mask_length += (int(subcon.split("-")[1])-int(subcon.split("-")[0][1:])+1)
+ else:
+ sampled_mask_length += 1
+
+ else:
+ if '-' in subcon:
+ length_inpaint=random.randint(int(subcon.split("-")[0]),int(subcon.split("-")[1]))
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += length_inpaint
+ elif subcon == '0':
+ subcon_out.append('0')
+ else:
+ length_inpaint=int(subcon)
+ subcon_out.append(f'{length_inpaint}-{length_inpaint}')
+ sampled_mask_length += int(subcon)
+ sampled_mask.append(','.join(subcon_out))
+ #check length is compatible
+ if self.length is not None:
+ if sampled_mask_length >= self.length[0] and sampled_mask_length < self.length[1]:
+ length_compatible = True
+ else:
+ length_compatible = True
+ count+=1
+ if count == 100000: #contig string incompatible with this length
+ sys.exit("Contig string incompatible with --length range")
+ return sampled_mask, sampled_mask_length, inpaint_chains
+
+ def expand_sampled_mask(self):
+ chain_order='ABCDEFGHIJKLMNOPQRSTUVWXYZ'
+ receptor = []
+ inpaint = []
+ receptor_hal = []
+ inpaint_hal = []
+ receptor_idx = 1
+ inpaint_idx = 1
+ inpaint_chain_idx=-1
+ receptor_chain_break=[]
+ inpaint_chain_break = []
+ for con in self.sampled_mask:
+ if (all([i[0].isalpha() for i in con.split(",")[:-1]]) and con.split(",")[-1] == '0') or self.topo is True:
+ #receptor chain
+ subcons = con.split(",")[:-1]
+ assert all([i[0] == subcons[0][0] for i in subcons]), "If specifying fragmented receptor in a single block of the contig string, they MUST derive from the same chain"
+ assert all(int(subcons[i].split("-")[0][1:]) < int(subcons[i+1].split("-")[0][1:]) for i in range(len(subcons)-1)), "If specifying multiple fragments from the same chain, pdb indices must be in ascending order!"
+ for idx, subcon in enumerate(subcons):
+ ref_to_add = [(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ receptor.extend(ref_to_add)
+ receptor_hal.extend([(self.receptor_chain,i) for i in np.arange(receptor_idx, receptor_idx+len(ref_to_add))])
+ receptor_idx += len(ref_to_add)
+ if idx != len(subcons)-1:
+ idx_jump = int(subcons[idx+1].split("-")[0][1:]) - int(subcon.split("-")[1]) -1
+ receptor_chain_break.append((receptor_idx-1,idx_jump)) #actual chain break in pdb chain
+ else:
+ receptor_chain_break.append((receptor_idx-1,200)) #200 aa chain break
+ else:
+ inpaint_chain_idx += 1
+ for subcon in con.split(","):
+ if subcon[0].isalpha():
+ ref_to_add=[(subcon[0], i) for i in np.arange(int(subcon.split("-")[0][1:]),int(subcon.split("-")[1])+1)]
+ inpaint.extend(ref_to_add)
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+len(ref_to_add))])
+ inpaint_idx += len(ref_to_add)
+
+ else:
+ inpaint.extend([('_','_')] * int(subcon.split("-")[0]))
+ inpaint_hal.extend([(chain_order[inpaint_chain_idx], i) for i in np.arange(inpaint_idx,inpaint_idx+int(subcon.split("-")[0]))])
+ inpaint_idx += int(subcon.split("-")[0])
+ inpaint_chain_break.append((inpaint_idx-1,200))
+
+ if self.topo is True or inpaint_hal == []:
+ receptor_hal = [(i[0], i[1]) for i in receptor_hal]
+ else:
+ receptor_hal = [(i[0], i[1] + inpaint_hal[-1][1]) for i in receptor_hal] #rosetta-like numbering
+ #get rf indexes, with chain breaks
+ inpaint_rf = np.arange(0,len(inpaint))
+ receptor_rf = np.arange(len(inpaint)+200,len(inpaint)+len(receptor)+200)
+ for ch_break in inpaint_chain_break[:-1]:
+ receptor_rf[:] += 200
+ inpaint_rf[ch_break[0]:] += ch_break[1]
+ for ch_break in receptor_chain_break[:-1]:
+ receptor_rf[ch_break[0]:] += ch_break[1]
+
+ return receptor, receptor_hal, receptor_rf.tolist(), inpaint, inpaint_hal, inpaint_rf.tolist()
+
+ def get_inpaint_seq_str(self, inpaint_s):
+ '''
+ function to generate inpaint_str or inpaint_seq masks specific to this contig
+ '''
+ s_mask = np.copy(self.mask_1d)
+ inpaint_s_list = []
+ for i in inpaint_s:
+ if '-' in i:
+ inpaint_s_list.extend([(i[0],p) for p in range(int(i.split("-")[0][1:]), int(i.split("-")[1])+1)])
+ else:
+ inpaint_s_list.append((i[0],int(i[1:])))
+ for res in inpaint_s_list:
+ if res in self.ref:
+ s_mask[self.ref.index(res)] = False #mask this residue
+
+ return np.array(s_mask)
+
+ def get_idx0(self):
+ ref_idx0=[]
+ hal_idx0=[]
+ ref_idx0_inpaint=[]
+ hal_idx0_inpaint=[]
+ ref_idx0_receptor=[]
+ hal_idx0_receptor=[]
+ for idx, val in enumerate(self.ref):
+ if val != ('_','_'):
+ assert val in self.parsed_pdb['pdb_idx'],f"{val} is not in pdb file!"
+ hal_idx0.append(idx)
+ ref_idx0.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.inpaint):
+ if val != ('_','_'):
+ hal_idx0_inpaint.append(idx)
+ ref_idx0_inpaint.append(self.parsed_pdb['pdb_idx'].index(val))
+ for idx, val in enumerate(self.receptor):
+ if val != ('_','_'):
+ hal_idx0_receptor.append(idx)
+ ref_idx0_receptor.append(self.parsed_pdb['pdb_idx'].index(val))
+
+
+ return ref_idx0, hal_idx0, ref_idx0_inpaint, hal_idx0_inpaint, ref_idx0_receptor, hal_idx0_receptor
+
+def get_mappings(rm):
+ mappings = {}
+ mappings['con_ref_pdb_idx'] = [i for i in rm.inpaint if i != ('_','_')]
+ mappings['con_hal_pdb_idx'] = [rm.inpaint_hal[i] for i in range(len(rm.inpaint_hal)) if rm.inpaint[i] != ("_","_")]
+ mappings['con_ref_idx0'] = rm.ref_idx0_inpaint
+ mappings['con_hal_idx0'] = rm.hal_idx0_inpaint
+ if rm.inpaint != rm.ref:
+ mappings['complex_con_ref_pdb_idx'] = [i for i in rm.ref if i != ("_","_")]
+ mappings['complex_con_hal_pdb_idx'] = [rm.hal[i] for i in range(len(rm.hal)) if rm.ref[i] != ("_","_")]
+ mappings['receptor_con_ref_pdb_idx'] = [i for i in rm.receptor if i != ("_","_")]
+ mappings['receptor_con_hal_pdb_idx'] = [rm.receptor_hal[i] for i in range(len(rm.receptor_hal)) if rm.receptor[i] != ("_","_")]
+ mappings['complex_con_ref_idx0'] = rm.ref_idx0
+ mappings['complex_con_hal_idx0'] = rm.hal_idx0
+ mappings['receptor_con_ref_idx0'] = rm.ref_idx0_receptor
+ mappings['receptor_con_hal_idx0'] = rm.hal_idx0_receptor
+ mappings['inpaint_str'] = rm.inpaint_str
+ mappings['inpaint_seq'] = rm.inpaint_seq
+ mappings['sampled_mask'] = rm.sampled_mask
+ mappings['mask_1d'] = rm.mask_1d
+ return mappings
+
+def lddt_unbin(pred_lddt):
+ nbin = pred_lddt.shape[1]
+ bin_step = 1.0 / nbin
+ lddt_bins = torch.linspace(bin_step, 1.0, nbin, dtype=pred_lddt.dtype, device=pred_lddt.device)
+
+ pred_lddt = nn.Softmax(dim=1)(pred_lddt)
+ return torch.sum(lddt_bins[None,:,None]*pred_lddt, dim=1)
+
diff --git a/utils/model/utils/parsers_inference.py b/utils/model/utils/parsers_inference.py
new file mode 100644
index 0000000000000000000000000000000000000000..1a8bc9aca6de872a4e2b4388937c4d85e6b8d6b1
--- /dev/null
+++ b/utils/model/utils/parsers_inference.py
@@ -0,0 +1,151 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ i[pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/utils/parsers_inference.py b/utils/parsers_inference.py
new file mode 100644
index 0000000000000000000000000000000000000000..691317ccc5be8341b3e16740b8e9d10d82236812
--- /dev/null
+++ b/utils/parsers_inference.py
@@ -0,0 +1,152 @@
+import numpy as np
+import scipy
+import scipy.spatial
+import string
+import os,re
+import random
+import util
+
+
+to1letter = {
+ "ALA":'A', "ARG":'R', "ASN":'N', "ASP":'D', "CYS":'C',
+ "GLN":'Q', "GLU":'E', "GLY":'G', "HIS":'H', "ILE":'I',
+ "LEU":'L', "LYS":'K', "MET":'M', "PHE":'F', "PRO":'P',
+ "SER":'S', "THR":'T', "TRP":'W', "TYR":'Y', "VAL":'V' }
+
+
+def parse_a3m(filename):
+ '''read A3M and convert letters into integers in the 0..20 range,
+ also keep track of insertions
+ '''
+
+ # read A3M file line by line
+ lab,seq = [],[] # labels and sequences
+ for line in open(filename, "r"):
+ if line[0] == '>':
+ lab.append(line.split()[0][1:])
+ seq.append("")
+ else:
+ seq[-1] += line.rstrip()
+
+ # parse sequences
+ msa,ins = [],[]
+ table = str.maketrans(dict.fromkeys(string.ascii_lowercase))
+ nrow,ncol = len(seq),len(seq[0])
+
+ for seqi in seq:
+
+ # remove lowercase letters and append to MSA
+ msa.append(seqi.translate(table))
+
+ # 0 - match or gap; 1 - insertion
+ a = np.array([0 if c.isupper() or c=='-' else 1 for c in seqi])
+ i = np.zeros((ncol))
+
+ if np.sum(a) > 0:
+ # positions of insertions
+ pos = np.where(a==1)[0]
+
+ # shift by occurrence
+ a = pos - np.arange(pos.shape[0])
+
+ # position of insertions in the cleaned sequence
+ # and their length
+ pos,num = np.unique(a, return_counts=True)
+ [pos[pos 20] = 20
+
+ ins = np.array(ins, dtype=np.uint8)
+
+ return {"msa":msa, "labels":lab, "insertions":ins}
+
+
+def parse_pdb(filename, **kwargs):
+ '''extract xyz coords for all heavy atoms'''
+ lines = open(filename,'r').readlines()
+ return parse_pdb_lines(lines, **kwargs)
+
+def parse_pdb_lines(lines, parse_hetatom=False, ignore_het_h=True):
+ # indices of residues observed in the structure
+ res = [(l[22:26],l[17:20]) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"]
+ seq = [util.aa2num[r[1]] if r[1] in util.aa2num.keys() else 20 for r in res]
+ pdb_idx = [( l[21:22].strip(), int(l[22:26].strip()) ) for l in lines if l[:4]=="ATOM" and l[12:16].strip()=="CA"] # chain letter, res num
+
+ # 4 BB + up to 10 SC atoms
+ xyz = np.full((len(res), 27, 3), np.nan, dtype=np.float32)
+ for l in lines:
+ if l[:4] != "ATOM":
+ continue
+ chain, resNo, atom, aa = l[21:22], int(l[22:26]), ' '+l[12:16].strip().ljust(3), l[17:20]
+ idx = pdb_idx.index((chain,resNo))
+ for i_atm, tgtatm in enumerate(util.aa2long[util.aa2num[aa]]):
+ if tgtatm is not None and tgtatm.strip() == atom.strip(): # ignore whitespace
+ xyz[idx,i_atm,:] = [float(l[30:38]), float(l[38:46]), float(l[46:54])]
+ break
+
+ # save atom mask
+ mask = np.logical_not(np.isnan(xyz[...,0]))
+ xyz[np.isnan(xyz[...,0])] = 0.0
+
+ # remove duplicated (chain, resi)
+ new_idx = []
+ i_unique = []
+ for i,idx in enumerate(pdb_idx):
+ if idx not in new_idx:
+ new_idx.append(idx)
+ i_unique.append(i)
+
+ pdb_idx = new_idx
+ xyz = xyz[i_unique]
+ mask = mask[i_unique]
+ seq = np.array(seq)[i_unique]
+
+ out = {'xyz':xyz, # cartesian coordinates, [Lx14]
+ 'mask':mask, # mask showing which atoms are present in the PDB file, [Lx14]
+ 'idx':np.array([i[1] for i in pdb_idx]), # residue numbers in the PDB file, [L]
+ 'seq':np.array(seq), # amino acid sequence, [L]
+ 'pdb_idx': pdb_idx, # list of (chain letter, residue number) in the pdb file, [L]
+ }
+
+ # heteroatoms (ligands, etc)
+ if parse_hetatom:
+ xyz_het, info_het = [], []
+ for l in lines:
+ if l[:6]=='HETATM' and not (ignore_het_h and l[77]=='H'):
+ info_het.append(dict(
+ idx=int(l[7:11]),
+ atom_id=l[12:16],
+ atom_type=l[77],
+ name=l[16:20]
+ ))
+ xyz_het.append([float(l[30:38]), float(l[38:46]), float(l[46:54])])
+
+ out['xyz_het'] = np.array(xyz_het)
+ out['info_het'] = info_het
+
+ return out
+
+def parse_fasta(filename):
+ '''
+ Return dict of name: seq
+ '''
+ out = {}
+ with open(filename, 'r') as f_in:
+ while True:
+ name = f_in.readline().strip()[1:]
+ seq = f_in.readline().strip()
+ if not name: break
+
+ out[name] = seq
+
+ return out
diff --git a/utils/potentials.py b/utils/potentials.py
new file mode 100644
index 0000000000000000000000000000000000000000..5bd4d3f412d7181c4d0c64a353a53c2e275a619d
--- /dev/null
+++ b/utils/potentials.py
@@ -0,0 +1,675 @@
+import os, sys
+import shutil
+import glob
+import torch
+import numpy as np
+import copy
+from itertools import groupby
+from operator import itemgetter
+import json
+import re
+import random
+import matplotlib.pyplot as plt
+import pandas as pd
+from tqdm import tqdm
+import random
+import Bio
+from icecream import ic
+DEVICE = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
+
+conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+
+
+### IF ADDING NEW POTENTIAL MAKE SURE TO ADD TO BOTTOM DICTIONARY ###
+
+
+# TEMPLATE CLASS
+class Potential:
+
+ def get_gradients(seq):
+ '''
+ EVERY POTENTIAL CLASS MUST RETURN GRADIENTS
+ '''
+
+ sys.exit('ERROR POTENTIAL HAS NOT BEEN IMPLEMENTED')
+
+
+class AACompositionalBias(Potential):
+ """
+ T = number of timesteps to set up diffuser with
+
+ schedule = type of noise schedule to use linear, cosine, gaussian
+
+ noise = type of ditribution to sample from; DEFAULT - normal_gaussian
+
+ """
+
+ def __init__(self, args, features, potential_scale, DEVICE):
+
+ self.L = features['L']
+ self.DEVICE = DEVICE
+ self.frac_seq_to_weight = args['frac_seq_to_weight']
+ self.add_weight_every_n = args['add_weight_every_n']
+ self.aa_weights_json = args['aa_weights_json']
+ self.one_weight_per_position = args['one_weight_per_position']
+ self.aa_weight = args['aa_weight']
+ self.aa_spec = args['aa_spec']
+ self.aa_composition = args['aa_composition']
+ self.potential_scale = potential_scale
+
+ self.aa_weights_to_add = [0 for l in range(21)]
+ self.aa_max_potential = None
+
+
+ if self.aa_weights_json != None:
+ with open(self.aa_weights_json, 'r') as f:
+ aa_weights = json.load(f)
+ else:
+ aa_weights = {}
+
+ for k,v in aa_weights.items():
+ aa_weights_to_add[conversion.index(k)] = v
+
+ aa_weights_to_add = [0 for l in range(21)]
+
+ self.aa_weights_to_add = torch.tensor(aa_weights_to_add)[None].repeat(self.L,1).to(self.DEVICE, non_blocking=True)
+
+ # BLOCK TO FIND OUT HOW YOU ARE LOOKING TO PROVIDE AA COMPOSITIONAL BIAS
+ if self.add_weight_every_n > 1 or self.frac_seq_to_weight > 0:
+
+ assert (self.add_weight_every_n > 1) ^ (self.frac_seq_to_weight > 0), 'use either --add_weight_every_n or --frac_seq_to_weight but not both'
+ weight_mask = torch.zeros_like(self.aa_weights_to_add)
+ if add_weight_every_n > 1:
+ idxs_to_unmask = torch.arange(0,self.L,self.add_weight_every_n)
+ else:
+ indexs = np.arange(0,self.L).tolist()
+ idxs_to_unmask = random.sample(indexs,int(self.frac_seq_to_weight*self.L))
+ idxs_to_unmask.sort()
+
+ weight_mask[idxs_to_unmask,:] = 1
+ self.aa_weights_to_add *= weight_mask
+
+ if one_weight_per_position:
+ for p in range(self.aa_weights_to_add.shape[0]):
+ where_ones = torch.where(self.aa_weights_to_add[p,:] > 0)[0].tolist()
+ if len(where_ones) > 0:
+ w_sample = random.sample(where_ones,1)[0]
+ self.aa_weights_to_add[p,:w_sample] = 0
+ self.aa_weights_to_add[p,w_sample+1:] = 0
+
+ elif self.aa_spec != None:
+
+ assert self.aa_weight != None, 'please specify --aa_weight'
+ # Use specified repeat structure to bias sequence
+
+ repeat_len = len(self.aa_spec)
+ weight_split = [float(x) for x in self.aa_weight.split(',')]
+
+ aa_idxs = []
+ for k,c in enumerate(self.aa_spec):
+ if c != 'X':
+ assert c in conversion, f'the letter you have chosen is not an amino acid: {c}'
+ aa_idxs.append((k,conversion.index(c)))
+
+ if len(self.aa_weight) > 1:
+ assert len(aa_idxs) == len(weight_split), f'need to give same number of weights as AAs in weight spec'
+
+ self.aa_weights_to_add = torch.zeros(self.L,21)
+
+ for p,w in zip(aa_idxs,weight_split):
+ x,a = p
+ self.aa_weights_to_add[x,a] = w
+
+ self.aa_weights_to_add = self.aa_weights_to_add[:repeat_len,:].repeat(self.L//repeat_len+1,1)[:self.L].to(self.DEVICE, non_blocking=True)
+
+ elif self.aa_composition != None:
+
+ self.aa_comp = [(x[0],float(x[1:])) for x in self.aa_composition.split(',')]
+ self.aa_max_potential = 0 #just a place holder so not None
+ assert sum([f for aa,f in self.aa_comp]) <= 1, f'total sequence fraction specified in aa_composition is > 1'
+
+ else:
+ sys.exit(f'You are missing an argument to use the aa_bias potential')
+
+ def get_gradients(self, seq):
+ '''
+ seq = L,21
+
+ return gradients to update the sequence with for the next pass
+ '''
+
+ if self.aa_max_potential != None:
+ soft_seq = torch.softmax(seq, dim=1)
+ print('ADDING SOFTMAXED SEQUENCE POTENTIAL')
+
+ aa_weights_to_add_list = []
+ for aa,f in self.aa_comp:
+ aa_weights_to_add_copy = self.aa_weights_to_add.clone()
+
+ soft_seq_tmp = soft_seq.clone().detach().requires_grad_(True)
+ aa_idx = conversion.index(aa)
+
+ # get top-k probability of logit to add to
+ where_add = torch.topk(soft_seq_tmp[:,aa_idx], int(f*self.L))[1]
+
+ # set up aa_potenital
+ aa_potential = torch.zeros(21)
+ aa_potential[conversion.index(aa)] = 1.0
+ aa_potential = aa_potential.repeat(self.L,1).to(self.DEVICE, non_blocking=True)
+
+ # apply "loss"
+ aa_comp_loss = torch.sum(torch.sum((aa_potential - soft_seq_tmp)**2, dim=1)**0.5)
+
+ # get gradients
+ aa_comp_loss.backward()
+ update_grads = soft_seq_tmp.grad
+
+ for k in range(self.L):
+ if k in where_add:
+ aa_weights_to_add_copy[k,:] = -update_grads[k,:]*self.potential_scale
+ else:
+ aa_weights_to_add_copy[k,:] = update_grads[k,:]*self.potential_scale
+ aa_weights_to_add_list.append(aa_weights_to_add_copy)
+
+ aa_weights_to_add_array = torch.stack((aa_weights_to_add_list))
+ self.aa_weights_to_add = torch.mean(aa_weights_to_add_array.float(), 0)
+
+
+ return self.aa_weights_to_add
+
+
+class HydrophobicBias(Potential):
+ """
+ Calculate loss with respect to soft_seq of the sequence hydropathy index (Kyte and Doolittle, 1986).
+
+ T = number of timesteps to set up diffuser with
+
+ schedule = type of noise schedule to use linear, cosine, gaussian
+
+ noise = type of ditribution to sample from; DEFAULT - normal_gaussian
+
+ """
+ def __init__(self, args, features, potential_scale, DEVICE):
+
+ self.target_score = args['hydrophobic_score']
+ self.potential_scale = potential_scale
+ self.loss_type = args['hydrophobic_loss_type']
+ print(f'USING {self.loss_type} LOSS TYPE...')
+
+ # -----------------------------------------------------------------------
+ # ---------------------GRAVY index data structures-----------------------
+ # -----------------------------------------------------------------------
+
+ # AA conversion
+ self.alpha_1 = list("ARNDCQEGHILKMFPSTWYVX")
+
+ # Dictionary to convert amino acids to their hyropathy index
+ self.gravy_dict = {'C': 2.5, 'D': -3.5, 'S': -0.8, 'Q': -3.5, 'K': -3.9,
+ 'I': 4.5, 'P': -1.6, 'T': -0.7, 'F': 2.8, 'N': -3.5,
+ 'G': -0.4, 'H': -3.2, 'L': 3.8, 'R': -4.5, 'W': -0.9,
+ 'A': 1.8, 'V':4.2, 'E': -3.5, 'Y': -1.3, 'M': 1.9, 'X': 0, '-': 0}
+
+ self.gravy_list = [self.gravy_dict[a] for a in self.alpha_1]
+
+ # -----------------------------------------------------------------------
+ # -----------------------------------------------------------------------
+
+ print(f'GUIDING SEQUENCES TO HAVE TARGET GRAVY SCORE OF: {self.target_score}')
+ return None
+
+
+ def get_gradients(self, seq):
+ """
+ Calculate gradients with respect to GRAVY index of input seq.
+ Uses a MSE loss.
+
+ Arguments
+ ---------
+ seq : tensor
+ L X 21 logits after saving seq_out from xt
+
+ Returns
+ -------
+ gradients : list of tensors
+ gradients of soft_seq with respect to loss on partial_charge
+ """
+ # Get GRAVY matrix based on length of seq
+ gravy_matrix = torch.tensor(self.gravy_list)[None].repeat(seq.shape[0],1).to(DEVICE)
+
+ # Get softmax of seq
+ soft_seq = torch.softmax(seq,dim=-1).requires_grad_(requires_grad=True).to(DEVICE)
+
+ # Calculate simple MSE loss on gravy_score
+ if self.loss_type == 'simple':
+ gravy_score = torch.mean(torch.sum(soft_seq*gravy_matrix,dim=-1), dim=0)
+ loss = ((gravy_score - self.target_score)**2)**0.5
+ #print(f'LOSS: {loss}')
+ # Take backward step
+ loss.backward()
+
+ # Get gradients from soft_seq
+ self.gradients = soft_seq.grad
+ # plt.imshow(self.gradients.cpu().detach().numpy())
+ # plt.colorbar()
+ # plt.title('gradients')
+
+ # Calculate MSE loss on gravy_score
+ elif self.loss_type == 'complex':
+ loss = torch.mean((torch.sum(soft_seq*gravy_matrix, dim = -1) - self.target_score)**2)
+ #print(f'LOSS: {loss}')
+ # Take backward step
+ loss.backward()
+
+ # Get gradients from soft_seq
+ self.gradients = soft_seq.grad
+ # plt.imshow(self.gradients.cpu().detach().numpy())
+ # plt.colorbar()
+ # plt.title('gradients')
+
+ return -self.gradients*self.potential_scale
+
+
+class ChargeBias(Potential):
+ """
+ Calculate losses and get gradients with respect to soft_seq for the sequence charge at a given pH.
+
+ T = number of timesteps to set up diffuser with
+
+ schedule = type of noise schedule to use linear, cosine, gaussian
+
+ noise = type of ditribution to sample from; DEFAULT - normal_gaussian
+
+ """
+ def __init__(self, args, features, potential_scale, DEVICE):
+
+ self.target_charge = args['target_charge']
+ self.pH = args['target_pH']
+ self.loss_type = args['charge_loss_type']
+ self.potential_scale = potential_scale
+ self.L = features['L']
+ self.DEVICE = DEVICE
+
+ # -----------------------------------------------------------------------
+ # ------------------------pI data structures-----------------------------
+ # -----------------------------------------------------------------------
+
+ # pKa lists to account for every residue.
+ pos_pKs_list = [[0.0, 12.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 5.98, 0.0, 0.0, 10.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]
+ neg_pKs_list = [[0.0, 0.0, 0.0, 4.05, 9.0, 0.0, 4.45, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 10.0, 0.0, 0.0]]
+ cterm_pKs_list = [[0.0, 0.0, 0.0, 4.55, 0.0, 0.0, 4.75, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]
+ nterm_pKs_list = [[7.59, 0.0, 0.0, 0.0, 0.0, 0.0, 7.7, 0.0, 0.0, 0.0, 0.0, 0.0, 7.0, 0.0, 8.36, 6.93, 6.82, 0.0, 0.0, 7.44, 0.0]]
+
+ # Convert pKa lists to tensors
+ self.cterm_pKs = torch.tensor(cterm_pKs_list)
+ self.nterm_pKs = torch.tensor(nterm_pKs_list)
+ self.pos_pKs = torch.tensor(pos_pKs_list)
+ self.neg_pKs = torch.tensor(neg_pKs_list)
+
+ # Repeat charged pKs L - 2 times to populate in all non-terminal residue indices
+ pos_pKs_repeat = self.pos_pKs.repeat(self.L - 2, 1)
+ neg_pKs_repeat = self.neg_pKs.repeat(self.L - 2, 1)
+
+ # Concatenate all pKs tensors with N-term and C-term pKas to get full L X 21 charge matrix
+ self.pos_pKs_matrix = torch.cat((torch.zeros_like(self.nterm_pKs), pos_pKs_repeat, self.nterm_pKs)).to(DEVICE)
+ self.neg_pKs_matrix = torch.cat((self.cterm_pKs, neg_pKs_repeat, torch.zeros_like(self.cterm_pKs))).to(DEVICE)
+
+ # Get indices of positive, neutral, and negative residues
+ self.cterm_charged_idx = torch.nonzero(self.cterm_pKs)
+ self.cterm_neutral_idx = torch.nonzero(self.cterm_pKs == 0)
+ self.nterm_charged_idx = torch.nonzero(self.nterm_pKs)
+ self.nterm_neutral_idx = torch.nonzero(self.nterm_pKs == 0)
+ self.pos_pKs_idx = torch.tensor([[1, 8, 11]])
+ self.neg_pKs_idx = torch.tensor([[3, 4, 6, 18]])
+ self.neutral_pKs_idx = torch.tensor([[0, 2, 5, 7, 9, 10, 12, 13, 14, 15, 16, 17, 19, 20]])
+
+ # -----------------------------------------------------------------------
+ # -----------------------------------------------------------------------
+
+ print(f'OPTIMIZING SEQUENCE TO HAVE CHARGE = {self.target_charge}\nAT pH = {self.pH}' )
+
+ def sum_tensor_indices(self, indices, tensor):
+ total = 0
+ for idx in indices:
+ i, j = idx[0], idx[1]
+ total += tensor[i][j]
+ return total
+
+ def sum_tensor_indices_2(self, indices, tensor):
+ # Create a tensor with the appropriate dimensions
+ j = indices.clone().detach().long().to(self.DEVICE)
+ # Select the values using advanced indexing and sum along dim=-1
+ row_sums = tensor[:, j].sum(dim=-1)
+
+ # Reshape the result to an L x 1 tensor
+ return row_sums.reshape(-1, 1).clone().detach()
+
+
+ def make_table(self, L):
+ """
+ Make table of all (positive, neutral, negative) charges -> (i, j, k)
+ such that:
+ i + j + k = L
+ (1 * i) + (0 * j) + (-1 * k) = target_charge
+
+ Arguments:
+ L: int
+ - length of sequence, defined as seq.shape[0]
+ target_charge : float
+ - Target charge for the sequence to be guided towards
+
+ Returns:
+ table: N x 3 tensor
+ - All combinations of i, j, k such that the above conditions are satisfied
+ """
+
+ table = []
+ for i in range(L):
+ for j in range(L):
+ for k in range(L):
+ # Check that number of residues = L and that sum of charge (i - k) = target_charge
+ # and that there are no 0 entries, as having no pos, no neg, or no neutral is not realistic
+ if i+j+k == L and i-k == self.target_charge and i != 0 and j != 0 and k != 0:
+ table.append([i,j,k])
+ return torch.tensor(np.array(table))
+
+
+ def classify_resis(self, seq):
+ """
+ Classify each position in seq as either positive, neutral, or negative.
+ Classification = max( [sum(positive residue logits), sum(neutral residue logits), sum(negative residue logits)] )
+
+ Arguments:
+ seq: L x 21 tensor
+ - sequence logits from the model
+
+ Returns:
+ charges: tensor
+ - 1 x 3 tensor counting total # of each charge type in the input sequence
+ - charges[0] = # positive residues
+ - charges[1] = # neutral residues
+ - charges[2] = # negative residues
+ charge_classification: tensor
+ - L x 1 tensor of each position's classification. 1 is positive, 0 is neutral, -1 is negative
+ """
+ L = seq.shape[0]
+ # Get softmax of seq
+ soft_seq = torch.softmax(seq.clone(),dim=-1).requires_grad_(requires_grad=True).to(self.DEVICE)
+
+ # Sum the softmax of all the positive and negative charges along dim = -1 (21 amino acids):
+ # Sum across c-term pKs
+ sum_cterm_charged = self.sum_tensor_indices(self.cterm_charged_idx, soft_seq).item()
+ # print(f'SUM OF CTERM CHARGED RESIS: {sum_cterm_charged}')
+ # print(type(sum_cterm_charged.item()))
+ sum_cterm_neutral = self.sum_tensor_indices(self.cterm_neutral_idx, soft_seq).item()
+ # print(f'SUM OF CTERM NEUTRAL RESIS: {sum_cterm_neutral}')
+
+ # Classify c-term as negative or neutral
+ cterm_max = max(sum_cterm_charged, sum_cterm_neutral)
+ # print(f'CTERM MAX: {cterm_max}')
+ if cterm_max == sum_cterm_charged:
+ cterm_class = torch.tensor([[-1]]).to(self.DEVICE)
+ else:
+ cterm_class = torch.tensor([[0]]).to(self.DEVICE)
+ # Prep cterm dataframe
+ cterm_df = torch.tensor([[0, sum_cterm_neutral, sum_cterm_charged, cterm_max, cterm_class]]).to(self.DEVICE)
+
+ # Sum across positive, neutral, and negative pKs
+ sum_pos = self.sum_tensor_indices_2(self.pos_pKs_idx, soft_seq[1:L-1, ...]).to(self.DEVICE)
+ # print(f'SUM POS: {sum_pos}')
+ sum_neg = self.sum_tensor_indices_2(self.neg_pKs_idx, soft_seq[1:L-1, ...]).to(self.DEVICE)
+ # print(f'SUM NEG: {sum_neg}')
+ sum_neutral = self.sum_tensor_indices_2(self.neutral_pKs_idx, soft_seq[1:L-1, ...]).to(self.DEVICE)
+ # print(f'SUM NEUTRAL: {sum_neutral}')
+
+ # Classify non-terminal residues along dim = -1
+ middle_max, _ = torch.max(torch.stack((sum_pos, sum_neg, sum_neutral), dim=-1), dim=-1)
+ middle_max = middle_max.to(self.DEVICE)
+ # create an L x 1 tensor to store the result
+ middle_class = torch.zeros((L - 2, 1), dtype=torch.long).to(self.DEVICE)
+ # set the values of the result tensor based on which tensor had the maximum value
+ middle_class[sum_neg == middle_max] = -1
+ middle_class[sum_neutral == middle_max] = 0
+ middle_class[sum_pos == middle_max] = 1
+
+ # Prepare df of all middle residue classifications and corresponding values
+ middle_df = pd.DataFrame((torch.cat((sum_pos, sum_neutral, sum_neg, middle_max, middle_class), dim=-1)).detach().cpu().numpy())
+ middle_df.rename(columns={0: 'sum_pos',
+ 1: 'sum_neutral', 2: 'sum_neg', 3: 'middle_max', 4: 'middle_classified'},
+ inplace=True, errors='raise')
+
+ # Sum across n-term pKs
+ sum_nterm_charged = self.sum_tensor_indices(self.nterm_charged_idx, soft_seq).to(self.DEVICE)
+ # print(f'SUM OF NTERM CHARGED RESIS: {sum_nterm_charged}')
+ sum_nterm_neutral = self.sum_tensor_indices(self.nterm_neutral_idx, soft_seq).to(self.DEVICE)
+ # print(f'SUM OF NTERM NEUTRAL RESIS: {sum_nterm_neutral}')
+
+ # Classify n-term as negative or neutral
+ nterm_max = max(sum_nterm_charged, sum_nterm_neutral)
+ if nterm_max == sum_nterm_charged:
+ nterm_class = torch.tensor([[-1]]).to(self.DEVICE)
+ else:
+ nterm_class = torch.tensor([[0]]).to(self.DEVICE)
+ nterm_df = torch.tensor([[sum_nterm_charged, sum_nterm_neutral, 0, nterm_max, nterm_class]]).to(self.DEVICE)
+
+ # Prep data to be concatenated into output df
+ middle_df_2 = (torch.cat((sum_pos, sum_neutral, sum_neg, middle_max, middle_class), dim=-1)).to(self.DEVICE)
+ # Concat cterm, middle, and nterm data into one master df with all summed probs, max, and final classification
+ full_tens_np = torch.cat((cterm_df, middle_df_2, nterm_df), dim = 0).detach().cpu().numpy()
+ classification_df = pd.DataFrame(full_tens_np)
+ classification_df.rename(columns={0: 'sum_pos',
+ 1: 'sum_neutral', 2: 'sum_neg', 3: 'max', 4: 'classification'},
+ inplace=True, errors='raise')
+ # Count number of positive, neutral, and negative charges that are stored in charge_classification as 1, 0, -1 respectively
+ charge_classification = torch.cat((cterm_class, middle_class, nterm_class), dim = 0).to(self.DEVICE)
+ charges = [torch.sum(charge_classification == 1).item(), torch.sum(charge_classification == 0).item(), torch.sum(charge_classification == -1).item()]
+ # print('*'*100)
+ # print(classification_df)
+
+ return torch.tensor(charges), classification_df
+
+ def get_target_charge_ratios(self, table, charges):
+ """
+ Find closest distance between x, y, z in table and i, j, k in charges
+
+ Arguments:
+ table: N x 3 tensor of all combinations of positive, neutral, and negative charges that obey the conditions in make_table
+ charges: 1 x 3 tensor
+ - 1 x 3 tensor counting total # of each charge type in the input sequence
+ - charges[0] = # positive residues
+ - charges[1] = # neutral residues
+ - charges[2] = # negative residues
+
+ Returns:
+ target_charge_tensor: tensor
+ - 1 x 3 tensor of closest row in table that matches charges of input sequence
+ """
+ # Compute the difference between the charges and each row of the table
+ diff = table - charges
+
+ # Compute the square of the Euclidean distance between the charges and each row of the table
+ sq_distance = torch.sum(diff ** 2, dim=-1)
+
+ # Find the index of the row with the smallest distance
+ min_idx = torch.argmin(sq_distance)
+
+ # Return the smallest distance and the corresponding row of the table
+ target_charge_tensor = torch.sqrt(sq_distance[min_idx]), table[min_idx]
+ #print(f'CLOSEST COMBINATION OF VALID RESIDUES: {target_charge_tensor[1]}')
+ return target_charge_tensor[1]
+
+ def draft_resis(self, classification_df, target_charge_tensor):
+ """
+ Based on target_charge_tensor, draft the top (i, j, k) positive, neutral, and negative positions from
+ charge_classification and return the idealized guided_charge_classification.
+ guided_charge_classification will determine whether the gradients should be positive or negative
+
+ Draft pick algorithm for determining gradient guided_charge_classification:
+ 1) Define how many positive, negative, and neutral charges are needed
+ 2) Current charge being drafted = sign of target charge, otherwise opposite charge
+ 3) From the classification_df of the currently sampled sequence, choose the position with the highest probability of being current_charge
+ 4) Make that residue +1, 0, or -1 in guided_charge_classification to dictate the sign of gradients
+ 5) Keep drafting that residue charge until it is used up, then move to the next type
+
+ Arguments:
+ classification_df: tensor
+ - L x 1 tensor of each position's classification. 1 is positive, 0 is neutral, -1 is negative
+ target_charge_tensor: tensor
+ - 1 x 3 tensor of closest row in table that matches charges of input sequence
+
+ Returns:
+ guided_charge_classification: L x 1 tensor
+ - L x 1 tensor populated with 1 = positive, 0 = neutral, -1 = negative
+ - in get_gradients, multiply the gradients by guided_charge_classification to determine which direction
+ the gradients should guide toward based on the current sequence distribution and the target charge
+ """
+ charge_dict = {'pos': 0, 'neutral': 0, 'neg': 0}
+ # Define the target number of positive, neutral, and negative charges
+ charge_dict['pos'] = target_charge_tensor[0].detach().clone()
+ charge_dict['neutral'] = target_charge_tensor[1].detach().clone()
+ charge_dict['neg'] = target_charge_tensor[2].detach().clone()
+ # Determine which charge to start drafting
+ if self.target_charge > 0:
+ start_charge = 'pos'
+ elif self.target_charge < 0:
+ start_charge = 'neg'
+ else:
+ start_charge = 'neutral'
+
+ # Initialize guided_charge_classification
+ guided_charge_classification = torch.zeros((classification_df.shape[0], 1))
+
+ # Start drafting
+ draft_charge = start_charge
+ while charge_dict[draft_charge] > 0:
+ # Find the residue with the max probability for the current draft charge
+ max_residue_idx = classification_df.loc[:, ['sum_' + draft_charge]].idxmax()[0]
+ # print(max_residue_idx[0])
+ # print(type(max_residue_idx))
+ #print(f'MAX RESIDUE INDEX for {draft_charge}: {max_residue_idx}')
+ # Populate guided_charge_classification with the appropriate charge
+ if draft_charge == 'pos':
+ guided_charge_classification[max_residue_idx] = 1
+ elif draft_charge == 'neg':
+ guided_charge_classification[max_residue_idx] = -1
+ else:
+ guided_charge_classification[max_residue_idx] = 0
+ # Remove selected row from classification_df
+ classification_df = classification_df.drop(max_residue_idx)
+ # print(classification_df)
+ # Update charges dictionary
+ charge_dict[draft_charge] -= 1
+ #print(f'{charge_dict[draft_charge]} {draft_charge} residues left to draft...')
+ # Switch to the other charged residue if the starting charge has been depleted
+ if charge_dict[draft_charge] == 0:
+ if draft_charge == start_charge:
+ draft_charge = 'neg' if start_charge == 'pos' else 'pos'
+ elif draft_charge == 'neg':
+ draft_charge = 'pos'
+ elif draft_charge == 'pos':
+ draft_charge = 'neg'
+ else:
+ draft_charge = 'neutral'
+
+ return guided_charge_classification.requires_grad_()
+
+ def get_gradients(self, seq):#, guided_charge_classification):
+ """
+ Calculate gradients with respect to SEQUENCE CHARGE at pH.
+ Uses a MSE loss.
+
+ Arguments
+ ---------
+ seq : tensor
+ L X 21 logits after saving seq_out from xt
+
+ Returns
+ -------
+ gradients : list of tensors
+ gradients of soft_seq with respect to loss on partial_charge
+ """
+ # Get softmax of seq
+ # soft_seq = torch.softmax(seq.clone(),dim=-1).requires_grad_(requires_grad=True).to(DEVICE)
+ soft_seq = torch.softmax(seq,dim=-1).requires_grad_(requires_grad=True).to(DEVICE)
+
+ # Get partial positive charges only for titratable residues
+ pos_charge = torch.where(self.pos_pKs_matrix != 0, ((1) / (((10) ** ((self.pH) - self.pos_pKs_matrix)) + (1.0))), (0.0))
+ neg_charge = torch.where(self.neg_pKs_matrix != 0, ((1) / (((10) ** (self.neg_pKs_matrix - (self.pH))) + (1.0))), (0.0))
+ # partial_charge = torch.sum((soft_seq*(pos_charge - neg_charge)).requires_grad_(requires_grad=True))
+
+
+ if self.loss_type == 'simple':
+ # Calculate net partial charge of soft_seq
+ partial_charge = torch.sum((soft_seq*(pos_charge - neg_charge)).requires_grad_(requires_grad=True))
+
+ print(f'CURRENT PARTIAL CHARGE: {partial_charge.item()}')
+ # Calculate MSE loss on partial_charge
+ loss = ((partial_charge - self.target_charge)**2)**0.5
+ #print(f'LOSS: {loss}')
+ # Take backward step
+ loss.backward()
+ # Get gradients from soft_seq
+ self.gradients = soft_seq.grad
+
+ # plt.imshow(self.gradients)
+ # plt.colorbar()
+ # plt.title('gradients')
+
+ elif self.loss_type == 'simple2':
+ # Calculate net partial charge of soft_seq
+ # partial_charge = torch.sum((soft_seq*(pos_charge - neg_charge)).requires_grad_(requires_grad=True))
+
+ print(f'CURRENT PARTIAL CHARGE: {partial_charge.item()}')
+ # Calculate MSE loss on partial_charge
+ loss = (((torch.sum((soft_seq*(pos_charge - neg_charge)).requires_grad_(requires_grad=True)))
+ - self.target_charge)**2)**0.5
+ #print(f'LOSS: {loss}')
+ # Take backward step
+ loss.backward()
+ # Get gradients from soft_seq
+ self.gradients = soft_seq.grad
+
+ # plt.imshow(self.gradients)
+ # plt.colorbar()
+ # plt.title('gradients')
+
+ elif self.loss_type == 'complex':
+ # Preprocessing using method functions
+ table = self.make_table(seq.shape[0])
+ charges, classification_df = self.classify_resis(seq)
+ target_charge_tensor = self.get_target_charge_ratios(table, charges)
+ guided_charge_classification = self.draft_resis(classification_df, target_charge_tensor)
+
+ # Calculate net partial charge of soft_seq
+ soft_partial_charge = (soft_seq*(pos_charge - neg_charge))
+ # print(f'SOFT PARTIAL CHARGE SHAPE: {soft_partial_charge.shape}')
+ # Define partial charge as the sum of softmax * partial charge matrix
+ partial_charge = torch.sum(soft_partial_charge, dim=-1).requires_grad_()
+ #print(partial_charge)
+ # partial_charge = torch.sum((soft_seq*(pos_charge - neg_charge)).requires_grad_(requires_grad=True))
+ print(f'CURRENT PARTIAL CHARGE: {partial_charge.sum().item()}')
+
+ # print(f'DIFFERENCE BETWEEN TARGET CHARGES AND CURRENT CHARGES: {((guided_charge_classification.to(self.DEVICE) - partial_charge.unsqueeze(1).to(self.DEVICE))**2)**0.5}')
+
+ # Calculate loss on partial_charge
+ loss = torch.mean(((guided_charge_classification.to(self.DEVICE) - partial_charge.unsqueeze(1).to(self.DEVICE))**2)**0.5)
+ # loss = torch.mean((guided_charge_classification.to(self.DEVICE) - partial_charge.to(self.DEVICE))**2)
+ #print(f'LOSS: {loss}')
+ # Take backward step
+ loss.backward()
+ # Get gradients from soft_seq
+ self.gradients = soft_seq.grad
+
+ # print(f'GUIDED CHARGE CLASSIFICATION SHAPE: {guided_charge_classification.shape}')
+ # print(f'PARTIAL CHARGE SHAPE: {partial_charge.unsqueeze(1).shape}')
+ # print(partial_charge)
+ # fig, ax = plt.subplots(1,2, dpi=200)
+ # ax[0].imshow((partial_charge.unsqueeze(1)).detach().numpy())
+ # ax[0].set_title('soft_seq partial charge')
+ # ax[1].imshow(self.gradients)#.detach().numpy())
+ # ax[1].set_title('gradients')
+ # print(seq)
+ return -self.gradients*self.potential_scale
+
+
+### ADD NEW POTENTIALS INTO LIST DOWN BELOW ###
+POTENTIALS = {'aa_bias':AACompositionalBias, 'charge':ChargeBias, 'hydrophobic':HydrophobicBias}
diff --git a/utils/requirements.txt b/utils/requirements.txt
new file mode 100644
index 0000000000000000000000000000000000000000..4381612685cf0b7d085d42b23b6afb294096a74a
--- /dev/null
+++ b/utils/requirements.txt
@@ -0,0 +1,7 @@
+torch==1.9
+e3nn==0.3.3
+pynvml==11.0.0
+decorator==5.1.0
+icecream
+biopython
+py3Dmol
diff --git a/utils/sampler.py b/utils/sampler.py
new file mode 100644
index 0000000000000000000000000000000000000000..169680b343e5ba5b1ac0a49b500294c41d628a2b
--- /dev/null
+++ b/utils/sampler.py
@@ -0,0 +1,1063 @@
+#####################################################################
+############# PROTEIN SEQUENCE DIFFUSION SAMPLER ####################
+#####################################################################
+
+import sys, os, subprocess, pickle, time, json
+script_dir = os.path.dirname(os.path.realpath(__file__))
+sys.path = sys.path + [script_dir+'/../model/'] + [script_dir+'/']
+import shutil
+import glob
+import torch
+import numpy as np
+import copy
+import json
+import matplotlib.pyplot as plt
+from torch import nn
+import math
+import re
+import pickle
+import pandas as pd
+import random
+from copy import deepcopy
+import time
+from collections import namedtuple
+import math
+from torch.nn.parallel import DistributedDataParallel as DDP
+from RoseTTAFoldModel import RoseTTAFoldModule
+from util import *
+from inpainting_util import *
+from kinematics import get_init_xyz, xyz_to_t2d
+import parsers_inference as parsers
+import diff_utils
+import pickle
+import pdb
+from utils.calc_dssp import annotate_sse
+from potentials import POTENTIALS
+from diffusion import GaussianDiffusion_SEQDIFF
+
+MODEL_PARAM ={
+ "n_extra_block" : 4,
+ "n_main_block" : 32,
+ "n_ref_block" : 4,
+ "d_msa" : 256,
+ "d_msa_full" : 64,
+ "d_pair" : 128,
+ "d_templ" : 64,
+ "n_head_msa" : 8,
+ "n_head_pair" : 4,
+ "n_head_templ" : 4,
+ "d_hidden" : 32,
+ "d_hidden_templ" : 32,
+ "p_drop" : 0.0
+ }
+
+SE3_PARAMS = {
+ "num_layers_full" : 1,
+ "num_layers_topk" : 1,
+ "num_channels" : 32,
+ "num_degrees" : 2,
+ "l0_in_features_full": 8,
+ "l0_in_features_topk" : 64,
+ "l0_out_features_full": 8,
+ "l0_out_features_topk" : 64,
+ "l1_in_features": 3,
+ "l1_out_features": 2,
+ "num_edge_features_full": 32,
+ "num_edge_features_topk": 64,
+ "div": 4,
+ "n_heads": 4
+ }
+
+SE3_param_full = {}
+SE3_param_topk = {}
+
+for param, value in SE3_PARAMS.items():
+ if "full" in param:
+ SE3_param_full[param[:-5]] = value
+ elif "topk" in param:
+ SE3_param_topk[param[:-5]] = value
+ else: # common arguments
+ SE3_param_full[param] = value
+ SE3_param_topk[param] = value
+
+MODEL_PARAM['SE3_param_full'] = SE3_param_full
+MODEL_PARAM['SE3_param_topk'] = SE3_param_topk
+
+DEFAULT_CKPT = '/home/jgershon/models/SEQDIFF_221219_equalTASKS_nostrSELFCOND_mod30.pt' #this is the one with good sequences
+LOOP_CHECKPOINT = '/home/jgershon/models/SEQDIFF_221202_AB_NOSTATE_fromBASE_mod30.pt'
+t1d_29_CKPT = '/home/jgershon/models/SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt'
+
+class SEQDIFF_sampler:
+
+ '''
+ MODULAR SAMPLER FOR SEQUENCE DIFFUSION
+
+ - the goal for modularizing this code is to make it as
+ easy as possible to edit and mix functions around
+
+ - in the base implementation here this can handle the standard
+ inference mode with default passes through the model, different
+ forms of partial diffusion, and linear symmetry
+
+ '''
+
+ def __init__(self, args=None):
+ '''
+ set args and DEVICE as well as other default params
+ '''
+ self.args = args
+ self.DEVICE = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
+ self.conversion = 'ARNDCQEGHILKMFPSTWYVX-'
+ self.dssp_dict = {'X':3,'H':0,'E':1,'L':2}
+ self.MODEL_PARAM = MODEL_PARAM
+ self.SE3_PARAMS = SE3_PARAMS
+ self.SE3_param_full = SE3_param_full
+ self.SE3_param_topk = SE3_param_topk
+ self.use_potentials = False
+ self.reset_design_num()
+
+ def set_args(self, args):
+ '''
+ set new arguments if iterating through dictionary of multiple arguments
+
+ # NOTE : args pertaining to the model will not be considered as this is
+ used to sample more efficiently without having to reload model for
+ different sets of args
+ '''
+ self.args = args
+ self.diffuser_init()
+ if self.args['potentials'] not in ['', None]:
+ self.potential_init()
+
+ def reset_design_num(self):
+ '''
+ reset design num to 0
+ '''
+ self.design_num = 0
+
+ def diffuser_init(self):
+ '''
+ set up diffuser object of GaussianDiffusion_SEQDIFF
+ '''
+ self.diffuser = GaussianDiffusion_SEQDIFF(T=self.args['T'],
+ schedule=self.args['noise_schedule'],
+ sample_distribution=self.args['sample_distribution'],
+ sample_distribution_gmm_means=self.args['sample_distribution_gmm_means'],
+ sample_distribution_gmm_variances=self.args['sample_distribution_gmm_variances'],
+ )
+ self.betas = self.diffuser.betas
+ self.alphas = 1-self.betas
+ self.alphas_cumprod = np.cumprod(self.alphas, axis=0)
+
+ def make_hotspot_features(self):
+ '''
+ set up hotspot features
+ '''
+ # initialize hotspot features to all 0s
+ self.features['hotspot_feat'] = torch.zeros(self.features['L'])
+
+ # if hotspots exist in args then make hotspot features
+ if self.args['hotspots'] != None:
+ self.features['hotspots'] = [(x[0],int(x[1:])) for x in self.args['hotspots'].split(',')]
+ for n,x in enumerate(self.features['mappings']['complex_con_ref_pdb_idx']):
+ if x in self.features['hotspots']:
+ self.features['hotspot_feat'][self.features['mappings']['complex_con_hal_idx0'][n]] = 1.0
+
+ def make_dssp_features(self):
+ '''
+ set up dssp features
+ '''
+
+ assert not ((self.args['secondary_structure'] != None) and (self.args['dssp_pdb'] != None)), \
+ f'You are attempting to provide both dssp_pdb and/or secondary_secondary structure, please choose one or the other'
+
+ # initialize with all zeros
+ self.features['dssp_feat'] = torch.zeros(self.features['L'],4)
+
+ if self.args['secondary_structure'] != None:
+
+ self.features['secondary_structure'] = [self.dssp_dict[x.upper()] for x in self.args['secondary_structure']]
+
+ assert len(self.features['secondary_structure']*self.features['sym'])+self.features['cap']*2 == self.features['L'], \
+ f'You have specified a secondary structure string that does not match your design length'
+
+ self.features['dssp_feat'] = torch.nn.functional.one_hot(
+ torch.tensor(self.features['cap_dssp']+self.features['secondary_structure']*self.features['sym']+self.features['cap_dssp']),
+ num_classes=4)
+
+ elif self.args['dssp_pdb'] != None:
+ dssp_xyz = torch.from_numpy(parsers.parse_pdb(self.args['dssp_pdb'])['xyz'][:,:,:])
+ dssp_pdb = annotate_sse(np.array(dssp_xyz[:,1,:].squeeze()), percentage_mask=0)
+ #we assume binder is chain A
+ self.features['dssp_feat'][:dssp_pdb.shape[0]] = dssp_pdb
+
+ elif (self.args['helix_bias'] + self.args['strand_bias'] + self.args['loop_bias']) > 0.0:
+
+ tmp_mask = torch.rand(self.features['L']) < self.args['helix_bias']
+ self.features['dssp_feat'][tmp_mask,0] = 1.0
+
+ tmp_mask = torch.rand(self.features['L']) < self.args['strand_bias']
+ self.features['dssp_feat'][tmp_mask,1] = 1.0
+
+ tmp_mask = torch.rand(self.features['L']) < self.args['loop_bias']
+ self.features['dssp_feat'][tmp_mask,2] = 1.0
+
+ #contigs get mask label
+ self.features['dssp_feat'][self.features['mask_str'][0],3] = 1.0
+ #anything not labeled gets mask label
+ mask_index = torch.where(torch.sum(self.features['dssp_feat'], dim=1) == 0)[0]
+ self.features['dssp_feat'][mask_index,3] = 1.0
+
+ def model_init(self):
+ '''
+ get model set up and choose checkpoint
+ '''
+
+ if self.args['checkpoint'] == None:
+ self.args['checkpoint'] = DEFAULT_CKPT
+
+ self.MODEL_PARAM['d_t1d'] = self.args['d_t1d']
+
+ # decide based on input args what checkpoint to load
+ if self.args['hotspots'] != None or self.args['secondary_structure'] != None \
+ or (self.args['helix_bias'] + self.args['strand_bias'] + self.args['loop_bias']) > 0 \
+ or self.args['dssp_pdb'] != None and self.args['checkpoint'] == DEFAULT_CKPT:
+
+ self.MODEL_PARAM['d_t1d'] = 29
+ print('You are using features only compatible with a newer model, switching checkpoint...')
+ self.args['checkpoint'] = t1d_29_CKPT
+
+ elif self.args['loop_design'] and self.args['checkpoint'] == DEFAULT_CKPT:
+ print('Switched to loop design checkpoint')
+ self.args['checkpoint'] = LOOP_CHECKPOINT
+
+ # check to make sure checkpoint chosen exists
+ if not os.path.exists(self.args['checkpoint']):
+ print('WARNING: couldn\'t find checkpoint')
+
+ self.ckpt = torch.load(self.args['checkpoint'], map_location=self.DEVICE)
+
+ # check to see if [loader_param, model_param, loss_param] is in checkpoint
+ # if so then you are using v2 of inference with t2d bug fixed
+ self.v2_mode = False
+ if 'model_param' in self.ckpt.keys():
+ print('You are running a new v2 model switching into v2 inference mode')
+ self.v2_mode = True
+
+ for k in self.MODEL_PARAM.keys():
+ if k in self.ckpt['model_param'].keys():
+ self.MODEL_PARAM[k] = self.ckpt['model_param'][k]
+ else:
+ print(f'no match for {k} in loaded model params')
+
+ # make model and load checkpoint
+ print('Loading model checkpoint...')
+ self.model = RoseTTAFoldModule(**self.MODEL_PARAM).to(self.DEVICE)
+
+ model_state = self.ckpt['model_state_dict']
+ self.model.load_state_dict(model_state, strict=False)
+ self.model.eval()
+ print('Successfully loaded model checkpoint')
+
+ def feature_init(self):
+ '''
+ featurize pdb and contigs and choose type of diffusion
+ '''
+ # initialize features dictionary for all example features
+ self.features = {}
+
+ # set up params
+ self.loader_params = {'MAXCYCLE':self.args['n_cycle'],'TEMPERATURE':self.args['temperature'], 'DISTANCE':self.args['min_decoding_distance']}
+
+ # symmetry
+ self.features['sym'] = self.args['symmetry']
+ self.features['cap'] = self.args['symmetry_cap']
+ self.features['cap_dssp'] = [self.dssp_dict[x.upper()] for x in 'H'*self.features['cap']]
+ if self.features['sym'] > 1:
+ print(f"Input sequence symmetry {self.features['sym']}")
+
+ assert (self.args['contigs'] in [('0'),(0),['0'],[0]] ) ^ (self.args['sequence'] in ['',None]),\
+ f'You are specifying contigs ({self.args["contigs"]}) and sequence ({self.args["sequence"]}) (or neither), please specify one or the other'
+
+ # initialize trb dictionary
+ self.features['trb_d'] = {}
+
+ if self.args['pdb'] == None and self.args['sequence'] not in ['', None]:
+ print('Preparing sequence input')
+
+ allowable_aas = [x for x in self.conversion[:-1]]
+ for x in self.args['sequence']: assert x in allowable_aas, f'Amino Acid {x} is undefinded, please only use standart 20 AAs'
+ self.features['seq'] = torch.tensor([self.conversion.index(x) for x in self.args['sequence']])
+ self.features['xyz_t'] = torch.full((1,1,len(self.args['sequence']),27,3), np.nan)
+
+ self.features['mask_str'] = torch.zeros(len(self.args['sequence'])).long()[None,:].bool()
+ self.features['mask_seq'] = torch.tensor([0 if x == 'X' else 1 for x in self.args['sequence']]).long()[None,:].bool()
+ self.features['blank_mask'] = torch.ones(self.features['mask_str'].size()[-1])[None,:].bool()
+
+ self.features['idx_pdb'] = torch.tensor([i for i in range(len(self.args['sequence']))])[None,:]
+ conf_1d = torch.ones_like(self.features['seq'])
+ conf_1d[~self.features['mask_str'][0]] = 0
+ self.features['seq_hot'], self.features['msa'], \
+ self.features['msa_hot'], self.features['msa_extra_hot'], _ = MSAFeaturize_fixbb(self.features['seq'][None,:],self.loader_params)
+ self.features['t1d'] = TemplFeaturizeFixbb(self.features['seq'], conf_1d=conf_1d)[None,None,:]
+ self.features['seq_hot'] = self.features['seq_hot'].unsqueeze(dim=0)
+ self.features['msa'] = self.features['msa'].unsqueeze(dim=0)
+ self.features['msa_hot'] = self.features['msa_hot'].unsqueeze(dim=0)
+ self.features['msa_extra_hot'] = self.features['msa_extra_hot'].unsqueeze(dim=0)
+
+ self.max_t = int(self.args['T']*self.args['sampling_temp'])
+
+ self.features['pdb_idx'] = [('A',i+1) for i in range(len(self.args['sequence']))]
+ self.features['trb_d']['inpaint_str'] = self.features['mask_str'][0]
+ self.features['trb_d']['inpaint_seq'] = self.features['mask_seq'][0]
+
+ else:
+
+ assert not (self.args['pdb'] == None and self.args['sampling_temp'] != 1.0),\
+ f'You must specify a pdb if attempting to use contigs with partial diffusion, else partially diffuse sequence input'
+
+ if self.args['pdb'] == None:
+ self.features['parsed_pdb'] = {'seq':np.zeros((1),'int64'),
+ 'xyz':np.zeros((1,27,3),'float32'),
+ 'idx':np.zeros((1),'int64'),
+ 'mask':np.zeros((1,27), bool),
+ 'pdb_idx':['A',1]}
+ else:
+ # parse input pdb
+ self.features['parsed_pdb'] = parsers.parse_pdb(self.args['pdb'])
+
+ # generate contig map
+ self.features['rm'] = ContigMap(self.features['parsed_pdb'], self.args['contigs'],
+ self.args['inpaint_seq'], self.args['inpaint_str'],
+ self.args['length'], self.args['ref_idx'],
+ self.args['hal_idx'], self.args['idx_rf'],
+ self.args['inpaint_seq_tensor'], self.args['inpaint_str_tensor'])
+ self.features['mappings'] = get_mappings(self.features['rm'])
+
+ self.features['pdb_idx'] = self.features['rm'].hal
+
+ ### PREPARE FEATURES DEPENDING ON TYPE OF ARGUMENTS SPECIFIED ###
+
+ # FULL DIFFUSION MODE
+ if self.args['trb'] == None and self.args['sampling_temp'] == 1.0:
+ # process contigs and generate masks
+ self.features['mask_str'] = torch.from_numpy(self.features['rm'].inpaint_str)[None,:]
+ self.features['mask_seq'] = torch.from_numpy(self.features['rm'].inpaint_seq)[None,:]
+ self.features['blank_mask'] = torch.ones(self.features['mask_str'].size()[-1])[None,:].bool()
+
+ seq_input = torch.from_numpy(self.features['parsed_pdb']['seq'])
+ xyz_input = torch.from_numpy(self.features['parsed_pdb']['xyz'][:,:,:])
+
+ self.features['xyz_t'] = torch.full((1,1,len(self.features['rm'].ref),27,3), np.nan)
+ self.features['xyz_t'][:,:,self.features['rm'].hal_idx0,:14,:] = xyz_input[self.features['rm'].ref_idx0,:14,:][None, None,...]
+ self.features['seq'] = torch.full((1,len(self.features['rm'].ref)),20).squeeze()
+ self.features['seq'][self.features['rm'].hal_idx0] = seq_input[self.features['rm'].ref_idx0]
+
+ # template confidence
+ conf_1d = torch.ones_like(self.features['seq'])*float(self.args['tmpl_conf'])
+ conf_1d[~self.features['mask_str'][0]] = 0 # zero confidence for places where structure is masked
+ seq_masktok = torch.where(self.features['seq'] == 20, 21, self.features['seq'])
+
+ # Get sequence and MSA input features
+ self.features['seq_hot'], self.features['msa'], \
+ self.features['msa_hot'], self.features['msa_extra_hot'], _ = MSAFeaturize_fixbb(seq_masktok[None,:],self.loader_params)
+ self.features['t1d'] = TemplFeaturizeFixbb(self.features['seq'], conf_1d=conf_1d)[None,None,:]
+ self.features['idx_pdb'] = torch.from_numpy(np.array(self.features['rm'].rf)).int()[None,:]
+ self.features['seq_hot'] = self.features['seq_hot'].unsqueeze(dim=0)
+ self.features['msa'] = self.features['msa'].unsqueeze(dim=0)
+ self.features['msa_hot'] = self.features['msa_hot'].unsqueeze(dim=0)
+ self.features['msa_extra_hot'] = self.features['msa_extra_hot'].unsqueeze(dim=0)
+
+ self.max_t = int(self.args['T']*self.args['sampling_temp'])
+
+ # PARTIAL DIFFUSION MODE, NO INPUT TRB
+ elif self.args['trb'] != None:
+ print('Running in partial diffusion mode . . .')
+ self.features['trb_d'] = np.load(self.args['trb'], allow_pickle=True)
+ self.features['mask_str'] = torch.from_numpy(self.features['trb_d']['inpaint_str'])[None,:]
+ self.features['mask_seq'] = torch.from_numpy(self.features['trb_d']['inpaint_seq'])[None,:]
+ self.features['blank_mask'] = torch.ones(self.features['mask_str'].size()[-1])[None,:].bool()
+
+ self.features['seq'] = torch.from_numpy(self.features['parsed_pdb']['seq'])
+ self.features['xyz_t'] = torch.from_numpy(self.features['parsed_pdb']['xyz'][:,:,:])[None,None,...]
+
+ if self.features['mask_seq'].shape[1] == 0:
+ self.features['mask_seq'] = torch.zeros(self.features['seq'].shape[0])[None].bool()
+ if self.features['mask_str'].shape[1] == 0:
+ self.features['mask_str'] = torch.zeros(self.features['xyz_t'].shape[2])[None].bool()
+
+ idx_pdb = []
+ chains_used = [self.features['parsed_pdb']['pdb_idx'][0][0]]
+ idx_jump = 0
+ for i,x in enumerate(self.features['parsed_pdb']['pdb_idx']):
+ if x[0] not in chains_used:
+ chains_used.append(x[0])
+ idx_jump += 200
+ idx_pdb.append(idx_jump+i)
+
+ self.features['idx_pdb'] = torch.tensor(idx_pdb)[None,:]
+ conf_1d = torch.ones_like(self.features['seq'])
+ conf_1d[~self.features['mask_str'][0]] = 0
+ self.features['seq_hot'], self.features['msa'], \
+ self.features['msa_hot'], self.features['msa_extra_hot'], _ = MSAFeaturize_fixbb(self.features['seq'][None,:],self.loader_params)
+ self.features['t1d'] = TemplFeaturizeFixbb(self.features['seq'], conf_1d=conf_1d)[None,None,:]
+ self.features['seq_hot'] = self.features['seq_hot'].unsqueeze(dim=0)
+ self.features['msa'] = self.features['msa'].unsqueeze(dim=0)
+ self.features['msa_hot'] = self.features['msa_hot'].unsqueeze(dim=0)
+ self.features['msa_extra_hot'] = self.features['msa_extra_hot'].unsqueeze(dim=0)
+
+ self.max_t = int(self.args['T']*self.args['sampling_temp'])
+
+ else:
+ print('running in partial diffusion mode, with no trb input, diffusing whole input')
+ self.features['seq'] = torch.from_numpy(self.features['parsed_pdb']['seq'])
+ self.features['xyz_t'] = torch.from_numpy(self.features['parsed_pdb']['xyz'][:,:,:])[None,None,...]
+
+ if self.args['contigs'] in [('0'),(0),['0'],[0]]:
+ print('no contigs given partially diffusing everything')
+ self.features['mask_str'] = torch.zeros(self.features['xyz_t'].shape[2]).long()[None,:].bool()
+ self.features['mask_seq'] = torch.zeros(self.features['seq'].shape[0]).long()[None,:].bool()
+ self.features['blank_mask'] = torch.ones(self.features['mask_str'].size()[-1])[None,:].bool()
+ else:
+ print('found contigs setting up masking for partial diffusion')
+ self.features['mask_str'] = torch.from_numpy(self.features['rm'].inpaint_str)[None,:]
+ self.features['mask_seq'] = torch.from_numpy(self.features['rm'].inpaint_seq)[None,:]
+ self.features['blank_mask'] = torch.ones(self.features['mask_str'].size()[-1])[None,:].bool()
+
+ idx_pdb = []
+ chains_used = [self.features['parsed_pdb']['pdb_idx'][0][0]]
+ idx_jump = 0
+ for i,x in enumerate(self.features['parsed_pdb']['pdb_idx']):
+ if x[0] not in chains_used:
+ chains_used.append(x[0])
+ idx_jump += 200
+ idx_pdb.append(idx_jump+i)
+
+ self.features['idx_pdb'] = torch.tensor(idx_pdb)[none,:]
+ conf_1d = torch.ones_like(self.features['seq'])
+ conf_1d[~self.features['mask_str'][0]] = 0
+ self.features['seq_hot'], self.features['msa'], \
+ self.features['msa_hot'], self.features['msa_extra_hot'], _ = msafeaturize_fixbb(self.features['seq'][none,:],self.loader_params)
+ self.features['t1d'] = templfeaturizefixbb(self.features['seq'], conf_1d=conf_1d)[none,none,:]
+ self.features['seq_hot'] = self.features['seq_hot'].unsqueeze(dim=0)
+ self.features['msa'] = self.features['msa'].unsqueeze(dim=0)
+ self.features['msa_hot'] = self.features['msa_hot'].unsqueeze(dim=0)
+ self.features['msa_extra_hot'] = self.features['msa_extra_hot'].unsqueeze(dim=0)
+
+ self.max_t = int(self.args['t']*self.args['sampling_temp'])
+
+ # set L
+ self.features['L'] = self.features['seq'].shape[0]
+
+ def potential_init(self):
+ '''
+ initialize potential functions being used and return list of potentails
+ '''
+
+ potentials = self.args['potentials'].split(',')
+ potential_scale = [float(x) for x in self.args['potential_scale'].split(',')]
+ assert len(potentials) == len(potential_scale), \
+ f'Please make sure number of potentials matches potential scales specified'
+
+ self.potential_list = []
+ for p,s in zip(potentials, potential_scale):
+ assert p in POTENTIALS.keys(), \
+ f'The potential specified: {p} , does not match into POTENTIALS dictionary in potentials.py'
+ print(f'Using potential: {p}')
+ self.potential_list.append(POTENTIALS[p](self.args, self.features, s, self.DEVICE))
+
+ self.use_potentials = True
+
+ def setup(self, init_model=True):
+ '''
+ run init model and init features to get everything prepped to go into model
+ '''
+
+ # initialize features
+ self.feature_init()
+
+ # initialize potential
+ if self.args['potentials'] not in ['', None]:
+ self.potential_init()
+
+ # make hostspot features
+ self.make_hotspot_features()
+
+ # make dssp features
+ self.make_dssp_features()
+
+ # diffuse sequence and mask features
+ self.features['seq'], self.features['msa_masked'], \
+ self.features['msa_full'], self.features['xyz_t'], self.features['t1d'], \
+ self.features['seq_diffused'] = diff_utils.mask_inputs(self.features['seq_hot'],
+ self.features['msa_hot'],
+ self.features['msa_extra_hot'],
+ self.features['xyz_t'],
+ self.features['t1d'],
+ input_seq_mask=self.features['mask_seq'],
+ input_str_mask=self.features['mask_str'],
+ input_t1dconf_mask=self.features['blank_mask'],
+ diffuser=self.diffuser,
+ t=self.max_t,
+ MODEL_PARAM=self.MODEL_PARAM,
+ hotspots=self.features['hotspot_feat'],
+ dssp=self.features['dssp_feat'],
+ v2_mode=self.v2_mode)
+
+
+ # move features to device
+ self.features['idx_pdb'] = self.features['idx_pdb'].long().to(self.DEVICE, non_blocking=True) # (B, L)
+ self.features['mask_str'] = self.features['mask_str'][None].to(self.DEVICE, non_blocking=True) # (B, L)
+ self.features['xyz_t'] = self.features['xyz_t'][None].to(self.DEVICE, non_blocking=True)
+ self.features['t1d'] = self.features['t1d'][None].to(self.DEVICE, non_blocking=True)
+ self.features['seq'] = self.features['seq'][None].type(torch.float32).to(self.DEVICE, non_blocking=True)
+ self.features['msa'] = self.features['msa'].type(torch.float32).to(self.DEVICE, non_blocking=True)
+ self.features['msa_masked'] = self.features['msa_masked'][None].type(torch.float32).to(self.DEVICE, non_blocking=True)
+ self.features['msa_full'] = self.features['msa_full'][None].type(torch.float32).to(self.DEVICE, non_blocking=True)
+ self.ti_dev = torsion_indices.to(self.DEVICE, non_blocking=True)
+ self.ti_flip = torsion_can_flip.to(self.DEVICE, non_blocking=True)
+ self.ang_ref = reference_angles.to(self.DEVICE, non_blocking=True)
+ self.features['xyz_prev'] = torch.clone(self.features['xyz_t'][0])
+ self.features['seq_diffused'] = self.features['seq_diffused'][None].to(self.DEVICE, non_blocking=True)
+ self.features['B'], _, self.features['N'], self.features['L'] = self.features['msa'].shape
+ self.features['t2d'] = xyz_to_t2d(self.features['xyz_t'])
+
+ # get alphas
+ self.features['alpha'], self.features['alpha_t'] = diff_utils.get_alphas(self.features['t1d'], self.features['xyz_t'],
+ self.features['B'], self.features['L'],
+ self.ti_dev, self.ti_flip, self.ang_ref)
+
+ # processing template coordinates
+ self.features['xyz_t'] = get_init_xyz(self.features['xyz_t'])
+ self.features['xyz_prev'] = get_init_xyz(self.features['xyz_prev'][:,None]).reshape(self.features['B'], self.features['L'], 27, 3)
+
+ # initialize extra features to none
+ self.features['xyz'] = None
+ self.features['pred_lddt'] = None
+ self.features['logit_s'] = None
+ self.features['logit_aa_s'] = None
+ self.features['best_plddt'] = 0
+ self.features['best_pred_lddt'] = torch.zeros_like(self.features['mask_str'])[0].float()
+ self.features['msa_prev'] = None
+ self.features['pair_prev'] = None
+ self.features['state_prev'] = None
+
+
+ def symmetrize_seq(self, x):
+ '''
+ symmetrize x according sym in features
+ '''
+ assert (self.features['L']-self.features['cap']*2) % self.features['sym'] == 0, f'symmetry does not match for input length'
+ assert x.shape[0] == self.features['L'], f'make sure that dimension 0 of input matches to L'
+
+ n_cap = torch.clone(x[:self.features['cap']])
+ c_cap = torch.clone(x[-self.features['cap']+1:])
+ sym_x = torch.clone(x[self.features['cap']:self.features['sym']]).repeat(self.features['sym'],1)
+
+ return torch.cat([n_cap,sym_x,c_cap], dim=0)
+
+ def predict_x(self):
+ '''
+ take step using X_t-1 features to predict Xo
+ '''
+ self.features['seq'], \
+ self.features['xyz'], \
+ self.features['pred_lddt'], \
+ self.features['logit_s'], \
+ self.features['logit_aa_s'], \
+ self.features['alpha'], \
+ self.features['msa_prev'], \
+ self.features['pair_prev'], \
+ self.features['state_prev'] \
+ = diff_utils.take_step_nostate(self.model,
+ self.features['msa_masked'],
+ self.features['msa_full'],
+ self.features['seq'],
+ self.features['t1d'],
+ self.features['t2d'],
+ self.features['idx_pdb'],
+ self.args['n_cycle'],
+ self.features['xyz_prev'],
+ self.features['alpha'],
+ self.features['xyz_t'],
+ self.features['alpha_t'],
+ self.features['seq_diffused'],
+ self.features['msa_prev'],
+ self.features['pair_prev'],
+ self.features['state_prev'])
+
+ def self_condition_seq(self):
+ '''
+ get previous logits and set at t1d template
+ '''
+ self.features['t1d'][:,:,:,:21] = self.features['logit_aa_s'][0,:21,:].permute(1,0)
+
+ def self_condition_str_scheduled(self):
+ '''
+ unmask random fraction of residues according to timestep
+ '''
+ print('self_conditioning on strcuture')
+ xyz_prev_template = torch.clone(self.features['xyz'])[None]
+ self_conditioning_mask = torch.rand(self.features['L']) < self.diffuser.alphas_cumprod[t]
+ xyz_prev_template[:,:,~self_conditioning_mask] = float('nan')
+ xyz_prev_template[:,:,self.features['mask_str'][0][0]] = float('nan')
+ xyz_prev_template[:,:,:,3:] = float('nan')
+ t2d_sc = xyz_to_t2d(xyz_prev_template)
+
+ xyz_t_sc = torch.zeros_like(self.features['xyz_t'][:,:1])
+ xyz_t_sc[:,:,:,:3] = xyz_prev_template[:,:,:,:3]
+ xyz_t_sc[:,:,:,3:] = float('nan')
+
+ t1d_sc = torch.clone(self.features['t1d'][:,:1])
+ t1d_sc[:,:,~self_conditioning_mask] = 0
+ t1d_sc[:,:,mask_str[0][0]] = 0
+
+ self.features['t1d'] = torch.cat([self.features['t1d'][:,:1],t1d_sc], dim=1)
+ self.features['t2d'] = torch.cat([self.features['t2d'][:,:1],t2d_sc], dim=1)
+ self.features['xyz_t'] = torch.cat([self.features['xyz_t'][:,:1],xyz_t_sc], dim=1)
+
+ self.features['alpha'], self.features['alpha_t'] = diff_utils.get_alphas(self.features['t1d'], self.features['xyz_t'],
+ self.features['B'], self.features['L'],
+ self.ti_dev, self.ti_flip, self.ang_ref)
+ self.features['xyz_t'] = get_init_xyz(self.features['xyz_t'])
+
+
+ def self_condition_str(self):
+ '''
+ conditioining on strucutre in NAR way
+ '''
+ print("conditioning on structure for NAR structure noising")
+ xyz_t_str_sc = torch.zeros_like(self.features['xyz_t'][:,:1])
+ xyz_t_str_sc[:,:,:,:3] = torch.clone(self.features['xyz'])[None]
+ xyz_t_str_sc[:,:,:,3:] = float('nan')
+ t2d_str_sc = xyz_to_t2d(self.features['xyz_t'])
+ t1d_str_sc = torch.clone(self.features['t1d'])
+
+ self.features['xyz_t'] = torch.cat([self.features['xyz_t'],xyz_t_str_sc], dim=1)
+ self.features['t2d'] = torch.cat([self.features['t2d'],t2d_str_sc], dim=1)
+ self.features['t1d'] = torch.cat([self.features['t1d'],t1d_str_sc], dim=1)
+
+ def save_step(self):
+ '''
+ add step to trajectory dictionary
+ '''
+ self.trajectory[f'step{self.t}'] = (self.features['xyz'].squeeze().detach().cpu(),
+ self.features['logit_aa_s'][0,:21,:].permute(1,0).detach().cpu(),
+ self.features['seq_diffused'][0,:,:21].detach().cpu())
+
+ def noise_x(self):
+ '''
+ get X_t-1 from predicted Xo
+ '''
+ # sample x_t-1
+ self.features['post_mean'] = self.diffuser.q_sample(self.features['seq_out'], self.t, DEVICE=self.DEVICE)
+
+ if self.features['sym'] > 1:
+ self.features['post_mean'] = self.symmetrize_seq(self.features['post_mean'])
+
+ # update seq and masks
+ self.features['seq_diffused'][0,~self.features['mask_seq'][0],:21] = self.features['post_mean'][~self.features['mask_seq'][0],...]
+ self.features['seq_diffused'][0,:,21] = 0.0
+
+ # did not know we were clamping seq
+ self.features['seq_diffused'] = torch.clamp(self.features['seq_diffused'], min=-3, max=3)
+
+ # match other features to seq diffused
+ self.features['seq'] = torch.argmax(self.features['seq_diffused'], dim=-1)[None]
+ self.features['msa_masked'][:,:,:,:,:22] = self.features['seq_diffused']
+ self.features['msa_masked'][:,:,:,:,22:44] = self.features['seq_diffused']
+ self.features['msa_full'][:,:,:,:,:22] = self.features['seq_diffused']
+ self.features['t1d'][:1,:,:,22] = 1-int(self.t)/self.args['T']
+
+
+ def apply_potentials(self):
+ '''
+ apply potentials
+ '''
+
+ grads = torch.zeros_like(self.features['seq_out'])
+ for p in self.potential_list:
+ grads += p.get_gradients(self.features['seq_out'])
+
+ self.features['seq_out'] += (grads/len(self.potential_list))
+
+ def generate_sample(self):
+ '''
+ sample from the model
+
+ this function runs the full sampling loop
+ '''
+ # setup example
+ self.setup()
+
+ # start time
+ self.start_time = time.time()
+
+ # set up dictionary to save at each step in trajectory
+ self.trajectory = {}
+
+ # set out prefix
+ self.out_prefix = self.args['out']+f'_{self.design_num:06}'
+ print(f'Generating sample {self.design_num:06} ...')
+
+ # main sampling loop
+ for j in range(self.max_t):
+ self.t = torch.tensor(self.max_t-j-1).to(self.DEVICE)
+
+ # run features through the model to get X_o prediction
+ self.predict_x()
+
+ # save step
+ if self.args['save_all_steps']:
+ self.save_step()
+
+ # get seq out
+ self.features['seq_out'] = torch.permute(self.features['logit_aa_s'][0], (1,0))
+
+ # save best seq
+ if self.features['pred_lddt'][~self.features['mask_seq']].mean().item() > self.features['best_plddt']:
+ self.features['best_seq'] = torch.argmax(torch.clone(self.features['seq_out']), dim=-1)
+ self.features['best_pred_lddt'] = torch.clone(self.features['pred_lddt'])
+ self.features['best_xyz'] = torch.clone(self.features['xyz'])
+ self.features['best_plddt'] = self.features['pred_lddt'][~self.features['mask_seq']].mean().item()
+
+ # self condition on sequence
+ self.self_condition_seq()
+
+ # self condition on structure
+ if self.args['scheduled_str_cond']:
+ self.self_condition_str_scheduled()
+ if self.args['struc_cond_sc']:
+ self.self_condition_str()
+
+ # sequence alterations
+ if self.args['softmax_seqout']:
+ self.features['seq_out'] = torch.softmax(self.features['seq_out'],dim=-1)*2-1
+ if self.args['clamp_seqout']:
+ self.features['seq_out'] = torch.clamp(self.features['seq_out'],
+ min=-((1/self.diffuser.alphas_cumprod[t])*0.25+5),
+ max=((1/self.diffuser.alphas_cumprod[t])*0.25+5))
+
+ # apply potentials
+ if self.use_potentials:
+ self.apply_potentials()
+
+ # noise to X_t-1
+ if self.t != 0:
+ self.noise_x()
+
+ print(''.join([self.conversion[i] for i in torch.argmax(self.features['seq_out'],dim=-1)]))
+ print (" TIMESTEP [%02d/%02d] | current PLDDT: %.4f << >> best PLDDT: %.4f"%(
+ self.t+1, self.args['T'], self.features['pred_lddt'][~self.features['mask_seq']].mean().item(),
+ self.features['best_pred_lddt'][~self.features['mask_seq']].mean().item()))
+
+ # record time
+ self.delta_time = time.time() - self.start_time
+
+ # save outputs
+ self.save_outputs()
+
+ # increment design num
+ self.design_num += 1
+
+ print(f'Finished design {self.out_prefix} in {self.delta_time/60:.2f} minutes.')
+
+ def save_outputs(self):
+ '''
+ save the outputs from the model
+ '''
+ # save trajectory
+ if self.args['save_all_steps']:
+ fname = f'{self.out_prefix}_trajectory.pt'
+ torch.save(self.trajecotry, fname)
+
+ # get items from best plddt step
+ if self.args['save_best_plddt']:
+ self.features['seq'] = torch.clone(self.features['best_seq'])
+ self.features['pred_lddt'] = torch.clone(self.features['best_pred_lddt'])
+ self.features['xyz'] = torch.clone(self.features['best_xyz'])
+
+ # get chain IDs
+ if (self.args['sampling_temp'] == 1.0 and self.args['trb'] == None) or (self.args['sequence'] not in ['',None]):
+ chain_ids = [i[0] for i in self.features['pdb_idx']]
+ elif self.args['dump_pdb']:
+ chain_ids = [i[0] for i in self.features['parsed_pdb']['pdb_idx']]
+
+ # write output pdb
+ fname = self.out_prefix + '.pdb'
+ if len(self.features['seq'].shape) == 2:
+ self.features['seq'] = self.features['seq'].squeeze()
+ write_pdb(fname,
+ self.features['seq'].type(torch.int64),
+ self.features['xyz'].squeeze(),
+ Bfacts=self.features['pred_lddt'].squeeze(),
+ chains=chain_ids)
+
+ if self.args['dump_trb']:
+ self.save_trb()
+
+ if self.args['save_args']:
+ self.save_args()
+
+ def save_trb(self):
+ '''
+ save trb file
+ '''
+
+ lddt = self.features['pred_lddt'].squeeze().cpu().numpy()
+ strmasktemp = self.features['mask_str'].squeeze().cpu().numpy()
+
+ partial_lddt = [lddt[i] for i in range(np.shape(strmasktemp)[0]) if strmasktemp[i] == 0]
+ trb = {}
+ trb['lddt'] = lddt
+ trb['inpaint_lddt'] = partial_lddt
+ trb['contigs'] = self.args['contigs']
+ trb['device'] = self.DEVICE
+ trb['time'] = self.delta_time
+ trb['args'] = self.args
+
+ if self.args['sequence'] != None:
+ for key, value in self.features['trb_d'].items():
+ trb[key] = value
+ else:
+ for key, value in self.features['mappings'].items():
+ if key in self.features['trb_d'].keys():
+ trb[key] = self.features['trb_d'][key]
+ else:
+ if len(value) > 0:
+ if type(value) == list and type(value[0]) != tuple:
+ value=np.array(value)
+ trb[key] = value
+
+ with open(f'{self.out_prefix}.trb','wb') as f_out:
+ pickle.dump(trb, f_out)
+
+ def save_args(self):
+ '''
+ save args
+ '''
+
+ with open(f'{self.out_prefix}_args.json','w') as f_out:
+ json.dump(self.args, f_out)
+
+#####################################################################
+###################### science is cool ##############################
+#####################################################################
+
+
+# making a custom sampler class for HuggingFace app
+
+class HuggingFace_sampler(SEQDIFF_sampler):
+
+ def model_init(self):
+ '''
+ get model set up and choose checkpoint
+ '''
+
+ if self.args['checkpoint'] == None:
+ self.args['checkpoint'] = DEFAULT_CKPT
+
+ self.MODEL_PARAM['d_t1d'] = self.args['d_t1d']
+
+ # check to make sure checkpoint chosen exists
+ if not os.path.exists(self.args['checkpoint']):
+ print('WARNING: couldn\'t find checkpoint')
+
+ self.ckpt = torch.load(self.args['checkpoint'], map_location=self.DEVICE)
+
+ # check to see if [loader_param, model_param, loss_param] is in checkpoint
+ # if so then you are using v2 of inference with t2d bug fixed
+ self.v2_mode = False
+ if 'model_param' in self.ckpt.keys():
+ print('You are running a new v2 model switching into v2 inference mode')
+ self.v2_mode = True
+
+ for k in self.MODEL_PARAM.keys():
+ if k in self.ckpt['model_param'].keys():
+ self.MODEL_PARAM[k] = self.ckpt['model_param'][k]
+ else:
+ print(f'no match for {k} in loaded model params')
+
+ # make model and load checkpoint
+ print('Loading model checkpoint...')
+ self.model = RoseTTAFoldModule(**self.MODEL_PARAM).to(self.DEVICE)
+
+ model_state = self.ckpt['model_state_dict']
+ self.model.load_state_dict(model_state, strict=False)
+ self.model.eval()
+ print('Successfully loaded model checkpoint')
+
+ def generate_sample(self):
+ '''
+ sample from the model
+
+ this function runs the full sampling loop
+ '''
+ # setup example
+ self.setup()
+
+ # start time
+ self.start_time = time.time()
+
+ # set up dictionary to save at each step in trajectory
+ self.trajectory = {}
+
+ # set out prefix
+ print(f'Generating sample {self.out_prefix} ...')
+
+ # main sampling loop
+ for j in range(self.max_t):
+ self.t = torch.tensor(self.max_t-j-1).to(self.DEVICE)
+
+ # run features through the model to get X_o prediction
+ self.predict_x()
+
+ # save step
+ if self.args['save_all_steps']:
+ self.save_step()
+
+ # get seq out
+ self.features['seq_out'] = torch.permute(self.features['logit_aa_s'][0], (1,0))
+
+ # save best seq
+ if self.features['pred_lddt'].mean().item() > self.features['best_plddt']:
+ self.features['best_seq'] = torch.argmax(torch.clone(self.features['seq_out']), dim=-1)
+ self.features['best_pred_lddt'] = torch.clone(self.features['pred_lddt'])
+ self.features['best_xyz'] = torch.clone(self.features['xyz'])
+ self.features['best_plddt'] = self.features['pred_lddt'][~self.features['mask_seq']].mean().item()
+
+ # self condition on sequence
+ self.self_condition_seq()
+
+ # self condition on structure
+ if self.args['scheduled_str_cond']:
+ self.self_condition_str_scheduled()
+ if self.args['struc_cond_sc']:
+ self.self_condition_str()
+
+ # sequence alterations
+ if self.args['softmax_seqout']:
+ self.features['seq_out'] = torch.softmax(self.features['seq_out'],dim=-1)*2-1
+ if self.args['clamp_seqout']:
+ self.features['seq_out'] = torch.clamp(self.features['seq_out'],
+ min=-((1/self.diffuser.alphas_cumprod[t])*0.25+5),
+ max=((1/self.diffuser.alphas_cumprod[t])*0.25+5))
+
+ # apply potentials
+ if self.use_potentials:
+ self.apply_potentials()
+
+ # noise to X_t-1
+ if self.t != 0:
+ self.noise_x()
+
+ print(''.join([self.conversion[i] for i in torch.argmax(self.features['seq_out'],dim=-1)]))
+ print (" TIMESTEP [%02d/%02d] | current PLDDT: %.4f << >> best PLDDT: %.4f"%(
+ self.t+1, self.args['T'], self.features['pred_lddt'][~self.features['mask_seq']].mean().item(),
+ self.features['best_pred_lddt'][~self.features['mask_seq']].mean().item()))
+
+ # record time
+ self.delta_time = time.time() - self.start_time
+
+ # save outputs
+ self.save_outputs()
+
+ # increment design num
+ self.design_num += 1
+
+ print(f'Finished design {self.out_prefix} in {self.delta_time/60:.2f} minutes.')
+
+ def take_step_get_outputs(self, j):
+
+ self.t = torch.tensor(self.max_t-j-1).to(self.DEVICE)
+
+ # run features through the model to get X_o prediction
+ self.predict_x()
+
+ # save step
+ if self.args['save_all_steps']:
+ self.save_step()
+
+ # get seq out
+ self.features['seq_out'] = torch.permute(self.features['logit_aa_s'][0], (1,0))
+
+ # save best seq
+ if self.features['pred_lddt'].mean().item() > self.features['best_plddt']:
+ self.features['best_seq'] = torch.argmax(torch.clone(self.features['seq_out']), dim=-1)
+ self.features['best_pred_lddt'] = torch.clone(self.features['pred_lddt'])
+ self.features['best_xyz'] = torch.clone(self.features['xyz'])
+ self.features['best_plddt'] = self.features['pred_lddt'].mean().item()
+
+
+ # WRITE OUTPUT TO GET TEMPORARY PDB TO DISPLAY
+ if self.t != 0:
+ self.features['seq'] = torch.argmax(torch.clone(self.features['seq_out']), dim=-1)
+ else:
+ # prepare final output
+ if self.args['save_args']:
+ self.save_args()
+
+ # get items from best plddt step
+ if self.args['save_best_plddt']:
+ self.features['seq'] = torch.clone(self.features['best_seq'])
+ self.features['pred_lddt'] = torch.clone(self.features['best_pred_lddt'])
+ self.features['xyz'] = torch.clone(self.features['best_xyz'])
+
+ # get chain IDs
+ if (self.args['sampling_temp'] == 1.0 and self.args['trb'] == None) or (self.args['sequence'] not in ['',None]):
+ chain_ids = [i[0] for i in self.features['pdb_idx']]
+ elif self.args['dump_pdb']:
+ chain_ids = [i[0] for i in self.features['parsed_pdb']['pdb_idx']]
+
+ # write output pdb
+ if len(self.features['seq'].shape) == 2:
+ self.features['seq'] = self.features['seq'].squeeze()
+
+ fname = f'{self.out_prefix}.pdb'
+
+ write_pdb(fname, self.features['seq'].type(torch.int64),
+ self.features['xyz'].squeeze(),
+ Bfacts=self.features['pred_lddt'].squeeze(),
+ chains=chain_ids)
+
+ aa_seq = ''.join([self.conversion[x] for x in self.features['seq'].tolist()])
+
+
+ # self condition on sequence
+ self.self_condition_seq()
+
+ # self condition on structure
+ if self.args['scheduled_str_cond']:
+ self.self_condition_str_scheduled()
+ if self.args['struc_cond_sc']:
+ self.self_condition_str()
+
+ # sequence alterations
+ if self.args['softmax_seqout']:
+ self.features['seq_out'] = torch.softmax(self.features['seq_out'],dim=-1)*2-1
+ if self.args['clamp_seqout']:
+ self.features['seq_out'] = torch.clamp(self.features['seq_out'],
+ min=-((1/self.diffuser.alphas_cumprod[t])*0.25+5),
+ max=((1/self.diffuser.alphas_cumprod[t])*0.25+5))
+
+ # apply potentials
+ if self.use_potentials:
+ self.apply_potentials()
+
+ # noise to X_t-1
+ if self.t != 0:
+ self.noise_x()
+
+ print(''.join([self.conversion[i] for i in torch.argmax(self.features['seq_out'],dim=-1)]))
+ print (" TIMESTEP [%02d/%02d] | current PLDDT: %.4f << >> best PLDDT: %.4f"%(
+ self.t+1, self.args['T'], self.features['pred_lddt'][~self.features['mask_seq']].mean().item(),
+ self.features['best_pred_lddt'][~self.features['mask_seq']].mean().item()))
+
+
+ return aa_seq, fname, self.features['pred_lddt'].mean().item()
+
+ def get_outputs(self):
+
+ aa_seq = ''.join([self.conversion[x] for x in self.features['seq'].tolist()])
+ path_to_pdb = self.out_prefix+'.pdb'
+ return aa_seq, path_to_pdb, self.features['pred_lddt'].mean().item()
diff --git a/utils/tmp/args.json b/utils/tmp/args.json
new file mode 100644
index 0000000000000000000000000000000000000000..fe157435b9a5d55dff83f0afb4fbfe34be96f777
--- /dev/null
+++ b/utils/tmp/args.json
@@ -0,0 +1 @@
+{"F": 1, "T": 25, "aa_composition": "", "aa_spec": null, "aa_weight": null, "aa_weights_json": null, "add_weight_every_n": 1, "argmax_seq": false, "cautious": false, "checkpoint": "./SEQDIFF_230205_dssp_hotspots_25mask_EQtasks_mod30.pt", "clamp_seqout": false, "contigs": ["0"], "d_t1d": 29, "dssp_pdb": null, "dump_all": false, "dump_npz": false, "dump_pdb": true, "dump_trb": true, "frac_seq_to_weight": 0.0, "hal_idx": null, "helix_bias": 0.0, "hotspots": null, "idx_rf": null, "inpaint_seq": null, "inpaint_seq_tensor": null, "inpaint_str": null, "inpaint_str_tensor": null, "input_json": null, "length": null, "loop_bias": 0.0, "loop_design": false, "min_decoding_distance": 15, "multi_templates": null, "multi_tmpl_conf": null, "n_cycle": 4, "noise_schedule": "sqrt", "num_designs": 500, "one_weight_per_position": false, "out": "./tmp/", "pdb": null, "potential_scale": null, "ref_idx": null, "sampling_temp": 1.0, "save_all_steps": false, "save_best_plddt": true, "save_seqs": false, "scheduled_str_cond": false, "secondary_structure": null, "softmax_seqout": false, "start_num": 0, "strand_bias": 0.01, "struc_cond_sc": false, "symmetry": 1, "symmetry_cap": 0, "temperature": 1.0, "tmpl_conf": "1", "trb": null, "sequence": "", "save_args": true, "potentials": null, "target_charge": -10, "charge_loss_type": "complex", "target_pH": 7.4, "hydrophobic_score": -10, "hydrophobic_loss_type": "complex", "sample_distribution": "normal", "sample_distribution_gmm_means": [-1, 1], "sample_distribution_gmm_variances": [1, 1], "gradio": true}
 +
+