|
--- |
|
license: apache-2.0 |
|
metrics: |
|
- accuracy |
|
pipeline_tag: feature-extraction |
|
tags: |
|
- chemistry |
|
- foundation models |
|
- AI4Science |
|
- materials |
|
- molecules |
|
- safetensors |
|
- pytorch |
|
- transformer |
|
- diffusers |
|
library_name: transformers |
|
--- |
|
|
|
# SMILES-based State-Space Encoder-Decoder (SMI-SSED) - MoLMamba |
|
|
|
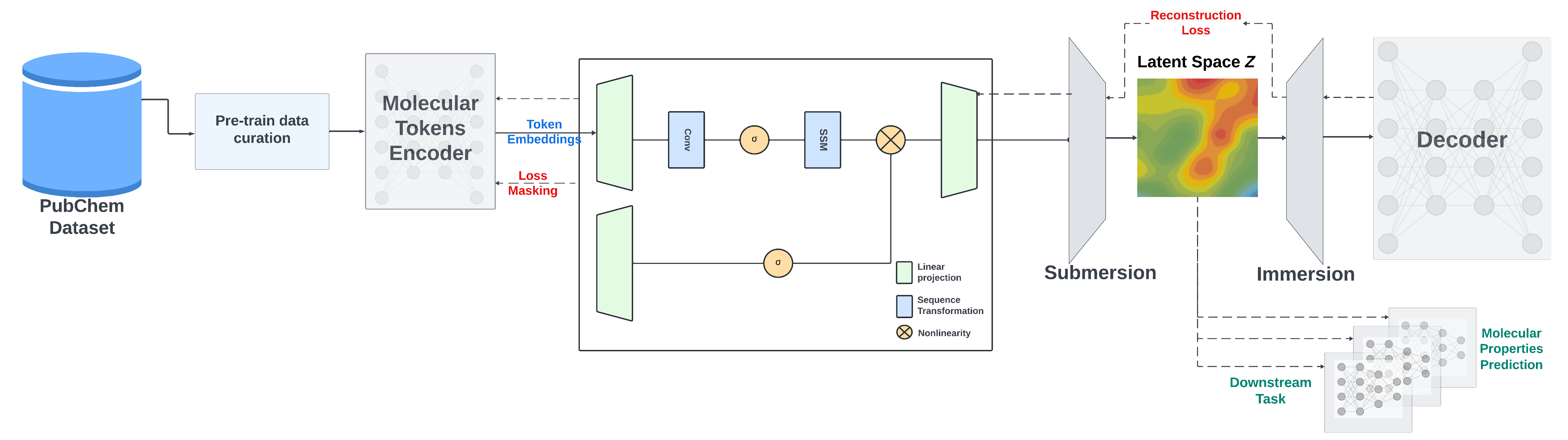
This repository provides PyTorch source code associated with our publication, "A Mamba-Based Foundation Model for Chemistry". |
|
|
|
**Paper NeurIPS AI4Mat 2024:** [Arxiv Link](https://openreview.net/pdf?id=HTgCs0KSTl) |
|
|
|
For more information contact: eduardo.soares@ibm.com or evital@br.ibm.com. |
|
|
|
 |
|
|
|
## Introduction |
|
|
|
We present a Mamba-based encoder-decoder chemical foundation model, SMILES-based State-Space Encoder-Decoder (SMI-SSED), pre-trained on a curated dataset of 91 million SMILES samples sourced from PubChem, equivalent to 4 billion molecular tokens. SMI-SSED supports various complex tasks, including quantum property prediction, with two main variants ($336$ and $8 \times 336M$). Our experiments across multiple benchmark datasets demonstrate state-of-the-art performance for various tasks. |
|
|
|
We provide the model weights in two formats: |
|
|
|
- PyTorch (`.pt`): [smi_ssed_130.pt](smi_ssed_130.pt) |
|
- safetensors (`.bin`): [smi_ssed_130.bin](smi_ssed_130.bin) |
|
|
|
For more information contact: eduardo.soares@ibm.com or evital@br.ibm.com. |
|
|
|
## Table of Contents |
|
|
|
1. [Getting Started](#getting-started) |
|
1. [Pretrained Models and Training Logs](#pretrained-models-and-training-logs) |
|
2. [Replicating Conda Environment](#replicating-conda-environment) |
|
2. [Pretraining](#pretraining) |
|
3. [Finetuning](#finetuning) |
|
4. [Feature Extraction](#feature-extraction) |
|
|
|
## Getting Started |
|
|
|
**This code and environment have been tested on Nvidia V100s and Nvidia A100s** |
|
|
|
### Pretrained Models and Training Logs |
|
|
|
We provide checkpoints of the SMI-SSED model pre-trained on a dataset of ~91M molecules curated from PubChem. The pre-trained model shows competitive performance on classification and regression benchmarks from MoleculeNet. |
|
|
|
Add the SMI-SSED `pre-trained weights.pt` to the `inference/` or `finetune/` directory according to your needs. The directory structure should look like the following: |
|
|
|
``` |
|
inference/ |
|
├── smi_ssed |
|
│ ├── smi_ssed.pt |
|
│ ├── bert_vocab_curated.txt |
|
│ └── load.py |
|
``` |
|
and/or: |
|
|
|
``` |
|
finetune/ |
|
├── smi_ssed |
|
│ ├── smi_ssed.pt |
|
│ ├── bert_vocab_curated.txt |
|
│ └── load.py |
|
``` |
|
|
|
### Replicating Conda Environment |
|
|
|
Follow these steps to replicate our Conda environment and install the necessary libraries: |
|
|
|
#### Create and Activate Conda Environment |
|
|
|
``` |
|
conda create --name smi-ssed-env python=3.10 |
|
conda activate smi-ssed-env |
|
``` |
|
|
|
#### Install Packages with Conda |
|
|
|
``` |
|
conda install pytorch=2.1.0 pytorch-cuda=11.8 -c pytorch -c nvidia |
|
``` |
|
|
|
#### Install Packages with Pip |
|
|
|
``` |
|
pip install -r requirements.txt |
|
``` |
|
|
|
## Pretraining |
|
|
|
For pretraining, we use two strategies: the masked language model method to train the encoder part and an encoder-decoder strategy to refine SMILES reconstruction and improve the generated latent space. |
|
|
|
SMI-SSED is pre-trained on canonicalized and curated 91M SMILES from PubChem with the following constraints: |
|
|
|
- Compounds are filtered to a maximum length of 202 tokens during preprocessing. |
|
- A 95/5/0 split is used for encoder training, with 5% of the data for decoder pretraining. |
|
- A 100/0/0 split is also used to train the encoder and decoder directly, enhancing model performance. |
|
|
|
The pretraining code provides examples of data processing and model training on a smaller dataset, requiring 8 A100 GPUs. |
|
|
|
To pre-train the SMI-SSED model, run: |
|
|
|
``` |
|
bash training/run_model_training.sh |
|
``` |
|
|
|
Use `train_model_D.py` to train only the decoder or `train_model_ED.py` to train both the encoder and decoder. |
|
|
|
## Finetuning |
|
|
|
The finetuning datasets and environment can be found in the [finetune](finetune/) directory. After setting up the environment, you can run a finetuning task with: |
|
|
|
``` |
|
bash finetune/smi_ssed/esol/run_finetune_esol.sh |
|
``` |
|
|
|
Finetuning training/checkpointing resources will be available in directories named `checkpoint_<measure_name>`. |
|
|
|
## Feature Extraction |
|
|
|
The example notebook [smi_ssed_encoder_decoder_example.ipynb](notebooks/smi_ssed_encoder_decoder_example.ipynb) contains code to load checkpoint files and use the pre-trained model for encoder and decoder tasks. It also includes examples of classification and regression tasks. For model weights: [HuggingFace Link](https://huggingface.co/ibm/materials.smi-ted) |
|
|
|
To load smi-ssed, you can simply use: |
|
|
|
```python |
|
model = load_smi_ssed( |
|
folder='../inference/smi_ssed', |
|
ckpt_filename='smi_ssed.pt' |
|
) |
|
``` |
|
|
|
To encode SMILES into embeddings, you can use: |
|
|
|
```python |
|
with torch.no_grad(): |
|
encoded_embeddings = model.encode(df['SMILES'], return_torch=True) |
|
``` |
|
For decoder, you can use the function, so you can return from embeddings to SMILES strings: |
|
|
|
```python |
|
with torch.no_grad(): |
|
decoded_smiles = model.decode(encoded_embeddings) |
|
``` |
|
|
|
|