Initial commit
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +3 -0
- .gitignore +4 -0
- Makefile +10 -0
- README.md +323 -3
- assets/__init__.py +0 -0
- assets/body_lvm_med.jpg +0 -0
- assets/lvm_med_compare.jpg +0 -0
- assets/lvm_med_teaser.gif +3 -0
- classification_R50/__init__.py +0 -0
- classification_R50/train_R50_classification.py +349 -0
- dataloader/__init__.py +3 -0
- dataloader/dataloader.py +55 -0
- dataloader/dataset.py +434 -0
- dataloader/dataset_ete.py +267 -0
- dataloader/sam_transforms.py +102 -0
- dataloader/yaml_data/brain_endtoend_R50_frozen_1_fcn.yml +24 -0
- dataloader/yaml_data/brain_endtoend_R50_frozen_fcns.yml +24 -0
- dataloader/yaml_data/brain_endtoend_R50_non_frozen_1_fcn.yml +24 -0
- dataloader/yaml_data/brain_endtoend_R50_non_frozen_fcns.yml +24 -0
- dataloader/yaml_data/buid_endtoend_R50.yml +32 -0
- dataloader/yaml_data/buid_lvm_med_sam.yml +28 -0
- dataloader/yaml_data/buid_sam.yml +28 -0
- dataloader/yaml_data/fgadr_endtoend_R50_frozen_1_fcn.yml +24 -0
- dataloader/yaml_data/fgadr_endtoend_R50_frozen_fcns.yml +24 -0
- dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_1_fcn.yml +24 -0
- dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_fcns.yml +24 -0
- dataloader/yaml_data/kvasir_endtoend_R50.yml +32 -0
- dataloader/yaml_data/kvasir_sam.yml +28 -0
- dataloader/yaml_data/mmwhs_ct_endtoend_R50.yml +32 -0
- dataloader/yaml_data/mmwhs_ct_sam.yml +28 -0
- dataloader/yaml_data/mmwhs_mr_endtoend_R50.yml +32 -0
- dataloader/yaml_data/mmwhs_mri_sam.yml +28 -0
- dataset_demo/__init__.py +1 -0
- datasets_split/BUID_split.py +73 -0
- datasets_split/FGADR_split.py +38 -0
- datasets_split/Kvasir_split.py +19 -0
- datasets_split/MMWHS_CT_Heart_split.py +127 -0
- datasets_split/MMWHS_MR_Heart_split.py +128 -0
- datasets_split/__init__.py +0 -0
- evaluate.py +136 -0
- files_split/BUID.json +1 -0
- files_split/Kvasir.json +1 -0
- files_split/MMWHS_CT_Heart.json +0 -0
- files_split/MMWHS_MR_Heart.json +0 -0
- files_split/__init__.py +0 -0
- files_split/fgadr_pkl_file.pkl +3 -0
- further_training_lvm_med/README.md +20 -0
- further_training_lvm_med/__init__.py +0 -0
- lvm-med-training-data/0_SSL_BraTS2018_Brain/file_list.txt +3 -0
- lvm-med-training-data/0_SSL_HNSCC_Mixed_HeadNeck/file_list.txt +3 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,6 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
*.gif filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
*.txt filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
*.png filter=lfs diff=lfs merge=lfs -text
|
.gitignore
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
__pycache__
|
| 2 |
+
*.ipynb_checkpoints
|
| 3 |
+
.DS_Store
|
| 4 |
+
work_dir
|
Makefile
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
default_target: main
|
| 2 |
+
.PHONY : default_target
|
| 3 |
+
|
| 4 |
+
$(VERBOSE).SILENT:
|
| 5 |
+
|
| 6 |
+
SHELL = /bin/sh
|
| 7 |
+
|
| 8 |
+
sam_2D_example:
|
| 9 |
+
python3 medsam.py -c dataloader/yaml_data/buid_sam.yml -2D
|
| 10 |
+
.PHONY: sam_2D_example
|
README.md
CHANGED
|
@@ -1,3 +1,323 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
## LVM-Med: Learning Large-Scale Self-Supervised Vision Models for Medical Imaging via Second-order Graph Matching (Neurips 2023).
|
| 2 |
+
We release [LVM-Med](https://arxiv.org/abs/2306.11925)'s pre-trained models and demonstrate downstream tasks on 2D-3D segmentations, linear/fully finetuning image classification, and object detection.
|
| 3 |
+
|
| 4 |
+
LVM-Med was trained with ~ 1.3 million medical images collected from 55 datasets using a second-order graph matching formulation unifying
|
| 5 |
+
current contrastive and instance-based SSL.
|
| 6 |
+
|
| 7 |
+
<p align="center">
|
| 8 |
+
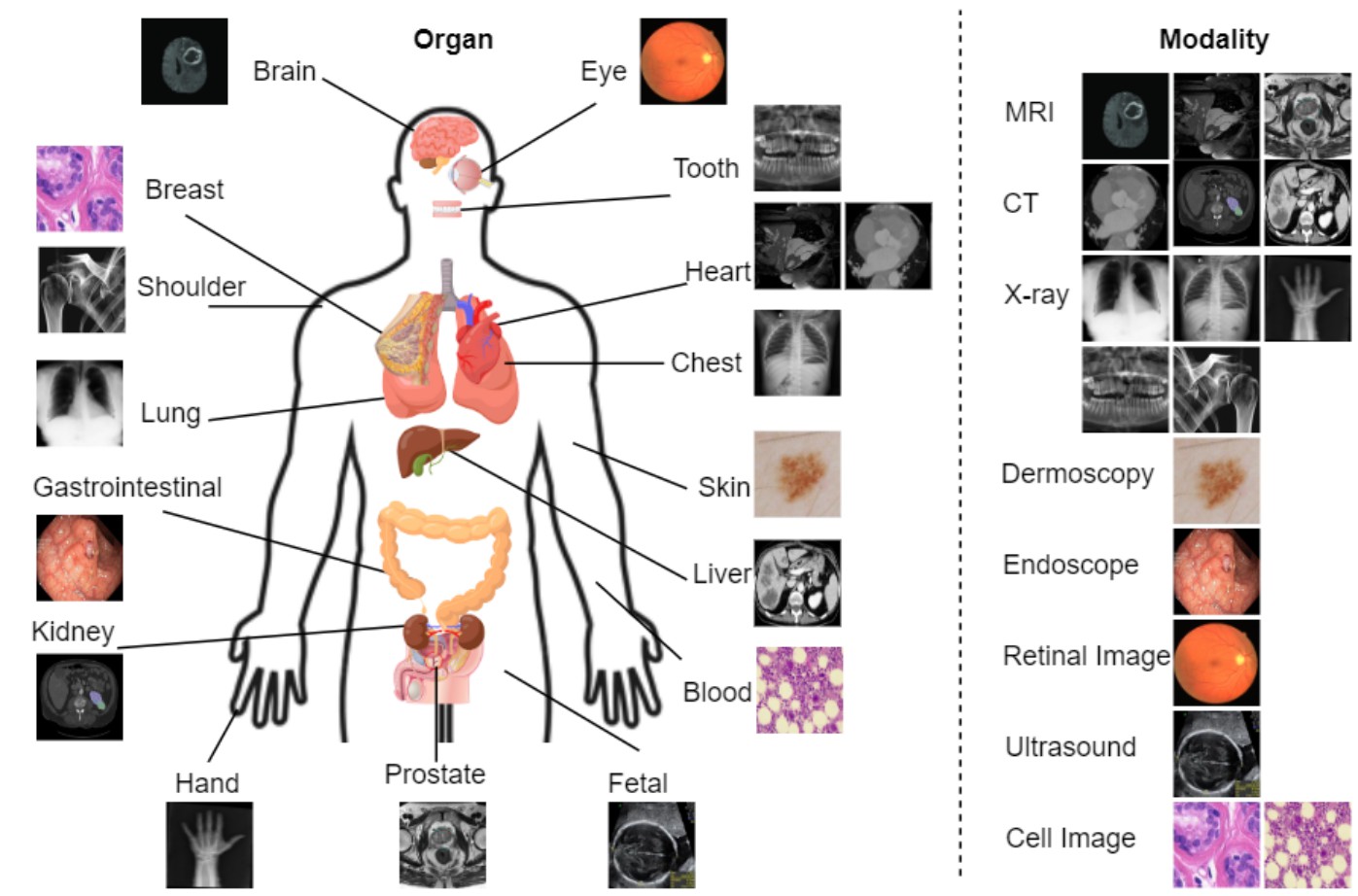
<img src="assets/body_lvm_med.jpg" alt="drawing" width="698.5" heigh="461"/>
|
| 9 |
+
</p>
|
| 10 |
+
|
| 11 |
+
<p align="center">
|
| 12 |
+
<img src="assets/lvm_med_teaser.gif" alt="drawing" width="471.6" heigh="520"/>
|
| 13 |
+
</p>
|
| 14 |
+
|
| 15 |
+
## Table of contents
|
| 16 |
+
* [News](#news)
|
| 17 |
+
* [LVM-Med Pretrained Models](#lvm-med-pretrained-models)
|
| 18 |
+
* [Further Training LVM-Med on Large Dataset](#further-training-lvm-med-on-large-dataset)
|
| 19 |
+
* [Prerequisites](#prerequisites)
|
| 20 |
+
* [Preparing Dataset](#preparing-datasets)
|
| 21 |
+
* [Downstream Tasks](#downstream-tasks)
|
| 22 |
+
* [Segmentation](#segmentation)
|
| 23 |
+
* [Image Classification](#image-classification)
|
| 24 |
+
* [Object Detection](#object-detection)
|
| 25 |
+
* [Citation](#citation)
|
| 26 |
+
* [Related Work](#related-work)
|
| 27 |
+
* [License](#license)
|
| 28 |
+
|
| 29 |
+
## News
|
| 30 |
+
- :mega: **14/12/2023**: The LVM-Med training algorithm is ready to be released! Please send us an email to request!
|
| 31 |
+
- :pencil2: If you want to have other architecture, send us a request by email or create an Issue. If the requests are enough, we will train them.
|
| 32 |
+
- :construction: Coming soon: [ConvNext](https://arxiv.org/abs/2201.03545) architecture trained by LVM-Med.
|
| 33 |
+
- :construction: Coming soon: ViT architectures for end-to-end segmentation with better performance reported in the paper.
|
| 34 |
+
- **31/07/2023**: Release ONNX support for LVM-Med ResNet50 and LVM-Med ViT as backbones in [`onnx_model`](/onnx_model/) folder.
|
| 35 |
+
- **26/07/2023**: We release ViT architectures (**ViT-B** and **ViT-H**) initialized from LVM-Med and further training on the LIVECell dataset with 1.6 million high-quality cells. See at this [table](#further-training-lvm-med-on-large-dataset).
|
| 36 |
+
- **25/06/2023**: We release two pre-trained models of LVM-Med: ResNet-50 and ViT-B. Providing scripts for downstream tasks.
|
| 37 |
+
|
| 38 |
+
## LVM-Med Pretrained Models
|
| 39 |
+
<table>
|
| 40 |
+
<tr>
|
| 41 |
+
<th>Arch</th>
|
| 42 |
+
<th>Params (M)</th>
|
| 43 |
+
<th> 2D Segmentation (Dice) </th>
|
| 44 |
+
<th> 3D Segmentation (3D IoU) </th>
|
| 45 |
+
<th>Weights</th>
|
| 46 |
+
</tr>
|
| 47 |
+
<tr>
|
| 48 |
+
<td>ResNet-50</td>
|
| 49 |
+
<td>25.5M</td>
|
| 50 |
+
<td>83.05</td>
|
| 51 |
+
<td>79.02</td>
|
| 52 |
+
<td> <a href="https://drive.google.com/file/d/11Uamq4bT_AbTf8sigIctIAnQJN4EethW/view?usp=sharing">backbone</a> </td>
|
| 53 |
+
</tr>
|
| 54 |
+
<tr>
|
| 55 |
+
<td>ViT-B</td>
|
| 56 |
+
<td>86.0M</td>
|
| 57 |
+
<td>85.80</td>
|
| 58 |
+
<td>80.90</td>
|
| 59 |
+
<td> <a href="https://drive.google.com/file/d/17WnE34S0ylYiA3tMXobH8uUrK_mCVPT4/view?usp=sharing">backbone</a> </td>
|
| 60 |
+
</tr>
|
| 61 |
+
</table>
|
| 62 |
+
|
| 63 |
+
After downloading the pre-trained models, please place them in [`lvm_med_weights`](/lvm_med_weights/) folder to use.
|
| 64 |
+
|
| 65 |
+
- For **Resnet-50**, we demo **end-to-end** segmentation/classification/object detection.
|
| 66 |
+
- For **ViT-B**, we demo **prompt-based** segmentation using bounding-boxes.
|
| 67 |
+
|
| 68 |
+
**Important Note:** please check[```dataset.md```](https://github.com/duyhominhnguyen/LVM-Med/blob/main/lvm-med-training-data/README.md) to avoid potential leaking testing data when using our model.
|
| 69 |
+
|
| 70 |
+
**Segment Anything Model-related Experiments**
|
| 71 |
+
- For all experiments using [SAM](https://github.com/facebookresearch/segment-anything) model, we use the base architecture of SAM which is `sam_vit_b`. You could browse the [`original repo`](https://github.com/facebookresearch/segment-anything) for this pre-trained weight and put it in [`./working_dir/sam_vit_b_01ec64.pth`](./working_dir/) folder to use yaml properly.
|
| 72 |
+
|
| 73 |
+
## Further Training LVM-Med on Large Dataset
|
| 74 |
+
We release some further pre-trained weight on other large datasets as mentioned in the Table below.
|
| 75 |
+
<table>
|
| 76 |
+
<tr>
|
| 77 |
+
<th>Arch</th>
|
| 78 |
+
<th>Params (M)</th>
|
| 79 |
+
<th>Dataset Name </th>
|
| 80 |
+
<th>Weights</th>
|
| 81 |
+
<th>Descriptions</th>
|
| 82 |
+
</tr>
|
| 83 |
+
<tr>
|
| 84 |
+
<td>ViT-B</td>
|
| 85 |
+
<td>86.0M</td>
|
| 86 |
+
<td> <a href="https://www.nature.com/articles/s41592-021-01249-6">LIVECell</a> </td>
|
| 87 |
+
<td> <a href="https://drive.google.com/file/d/1SxaGXQ4FMbG8pS2zzwTIXXgxF4GdwyEU/view?usp=sharing">backbone</a> </td>
|
| 88 |
+
<td> <a href="https://github.com/duyhominhnguyen/LVM-Med/blob/main/further_training_lvm_med/README.md">Link</a></td>
|
| 89 |
+
</tr>
|
| 90 |
+
<tr>
|
| 91 |
+
<td>ViT-H</td>
|
| 92 |
+
<td>632M</td>
|
| 93 |
+
<td> <a href="https://www.nature.com/articles/s41592-021-01249-6">LIVECell</a> </td>
|
| 94 |
+
<td> <a href="https://drive.google.com/file/d/14IhoyBXI9eP9V2xeOV2-6LlNICKjzBaJ/view?usp=sharing">backbone</a> </td>
|
| 95 |
+
<td> <a href="https://github.com/duyhominhnguyen/LVM-Med/blob/main/further_training_lvm_med/README.md">Link</a></td>
|
| 96 |
+
</tr>
|
| 97 |
+
</table>
|
| 98 |
+
|
| 99 |
+
|
| 100 |
+
|
| 101 |
+
## Prerequisites
|
| 102 |
+
|
| 103 |
+
The code requires `python>=3.8`, as well as `pytorch>=1.7` and `torchvision>=0.8`. Please follow the instructions [here](https://pytorch.org/get-started/locally/) to install both PyTorch and TorchVision dependencies. Installing both PyTorch and TorchVision with CUDA support is strongly recommended.
|
| 104 |
+
|
| 105 |
+
To set up our project, run the following command:
|
| 106 |
+
|
| 107 |
+
```bash
|
| 108 |
+
git clone https://github.com/duyhominhnguyen/LVM-Med
|
| 109 |
+
cd LVM-Med
|
| 110 |
+
conda env create -f lvm_med.yml
|
| 111 |
+
conda activate lvm_med
|
| 112 |
+
```
|
| 113 |
+
|
| 114 |
+
To **fine-tune for [Segmentation](#segmentation) using ResNet-50**, we utilize U-Net from `segmentation-models-pytorch` package. To install this library, you can do the following ones:
|
| 115 |
+
|
| 116 |
+
```bash
|
| 117 |
+
git clone https://github.com/qubvel/segmentation_models.pytorch.git
|
| 118 |
+
cd segmentation_models.pytorch
|
| 119 |
+
pip install -e
|
| 120 |
+
cd ..
|
| 121 |
+
mv segmentation_models_pytorch_example/encoders/__init__.py segmentation_models.pytorch/segmentation_models_pytorch/__init__.py
|
| 122 |
+
mv segmentation_models_pytorch_example/encoders/resnet.py segmentation_models.pytorch/segmentation_models_pytorch/resnet.py
|
| 123 |
+
```
|
| 124 |
+
|
| 125 |
+
<!--
|
| 126 |
+
1. `git clone https://github.com/qubvel/segmentation_models.pytorch.git`
|
| 127 |
+
2. `cd segmentation_models.pytorch; pip install -e .`
|
| 128 |
+
4. Copy file [`__init__.py`](segmentation_models_pytorch_example/encoders/__init__.py) and [`resnet.py`](segmentation_models_pytorch_example/encoders/resnet.py) in [`segmentation_models_pytorch_example`](segmentation_models_pytorch_example) folder
|
| 129 |
+
5. Paste [`__init__.py`](segmentation_models_pytorch_example/encoders/__init__.py) and [`resnet.py`](segmentation_models_pytorch_example/encoders/resnet.py) in the folder `encoders` of clone `segmentation_models.pytorch/segmentation_models_pytorch/` package to configure new pre-trained models
|
| 130 |
+
-->
|
| 131 |
+
|
| 132 |
+
## Preparing datasets
|
| 133 |
+
### For the Brain Tumor Dataset
|
| 134 |
+
You could download the `Brain` dataset via Kaggle's [`Brain Tumor Classification (MRI)`](https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri) and change the name into ```BRAIN```.
|
| 135 |
+
|
| 136 |
+
### For VinDr
|
| 137 |
+
You can download the dataset from this link [`VinDr`](https://www.kaggle.com/datasets/awsaf49/vinbigdata-512-image-dataset) and put the folder ```vinbigdata``` into the folder ```object_detection```. To build the dataset, after downloading the dataset, you can run script ```convert_to_coco.py``` inside the folder object_detection.
|
| 138 |
+
```bash
|
| 139 |
+
python convert_to_coco.py # Note, please check links inside the code in lines 146 and 158 to build dataset correctly
|
| 140 |
+
```
|
| 141 |
+
More information can be found in [```object_detection```](./object_detection).
|
| 142 |
+
|
| 143 |
+
### Others
|
| 144 |
+
First you should download the respective dataset that you need to run to the [`dataset_demo`](/dataset_demo/) folder. To get as close results as your work as possible, you could prepare some of our specific dataset (which are not pre-distributed) the same way as we do:
|
| 145 |
+
```bash
|
| 146 |
+
python prepare_dataset.py -ds [dataset_name]
|
| 147 |
+
```
|
| 148 |
+
such that: `dataset_name` is the name of dataset that you would like to prepare. After that, you should change paths to your loaded dataset on our pre-defined yaml file in [`dataloader/yaml_data`](/dataloader/yaml_data/).
|
| 149 |
+
|
| 150 |
+
Currently support for `Kvasir`, `BUID`, `FGADR`, `MMWHS_MR_Heart` and `MMWHS_CT_Heart`.
|
| 151 |
+
|
| 152 |
+
**Note:** You should change your dataset name into the correct format (i.e., Kvasir, BUID) as our current support dataset name. Or else it won't work as expected.
|
| 153 |
+
|
| 154 |
+
## Downstream Tasks
|
| 155 |
+
### Segmentation
|
| 156 |
+
### 1. End-to-End Segmentation
|
| 157 |
+
**a) Training Phase:**
|
| 158 |
+
|
| 159 |
+
**Fine-tune for downstream tasks using ResNet-50**
|
| 160 |
+
|
| 161 |
+
```bash
|
| 162 |
+
python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_R50.yml
|
| 163 |
+
```
|
| 164 |
+
Changing name of dataset in ``.yml`` configs in [```./dataloader/yaml_data/```](./dataloader/yaml_data/) for other experiments.
|
| 165 |
+
|
| 166 |
+
**Note**: to apply segmentation models (2D or 3D) using ResNet-50, we suggest normalizing gradient for stable training phases by set:
|
| 167 |
+
|
| 168 |
+
```bash
|
| 169 |
+
clip_value = 1
|
| 170 |
+
torch.nn.utils.clip_grad_norm_(net.parameters(), clip_value)
|
| 171 |
+
```
|
| 172 |
+
See examples in file [```/segmentation_2d/train_R50_seg_adam_optimizer_2d.py```](./segmentation_2d/train_R50_seg_adam_optimizer_2d.py) lines 129-130.
|
| 173 |
+
|
| 174 |
+
[//]: # (#### Fine-tune for downstream tasks using SAM's VIT)
|
| 175 |
+
|
| 176 |
+
[//]: # (```bash)
|
| 177 |
+
|
| 178 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_SAM_VIT.yml)
|
| 179 |
+
|
| 180 |
+
[//]: # (```)
|
| 181 |
+
**b) Inference:**
|
| 182 |
+
#### ResNet-50 version
|
| 183 |
+
|
| 184 |
+
```bash
|
| 185 |
+
python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_R50.yml -test
|
| 186 |
+
```
|
| 187 |
+
For the end-to-end version using SAM's ViT, we will soon release a better version than the reported results in the paper.
|
| 188 |
+
|
| 189 |
+
[//]: # (#### SAM's ViT version)
|
| 190 |
+
|
| 191 |
+
[//]: # (```bash)
|
| 192 |
+
|
| 193 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_SAM_VIT.yml -test)
|
| 194 |
+
|
| 195 |
+
[//]: # (```)
|
| 196 |
+
|
| 197 |
+
### 2. Prompt-based Segmentation with ViT-B
|
| 198 |
+
**a. Prompt-based segmentation with fine-tuned decoder of SAM ([MedSAM](https://github.com/bowang-lab/MedSAM)).**
|
| 199 |
+
|
| 200 |
+
We run the MedSAM baseline to compare performance by:
|
| 201 |
+
#### Train
|
| 202 |
+
```bash
|
| 203 |
+
python3 medsam.py -c dataloader/yaml_data/buid_sam.yml
|
| 204 |
+
```
|
| 205 |
+
#### Inference
|
| 206 |
+
```bash
|
| 207 |
+
python3 medsam.py -c dataloader/yaml_data/buid_sam.yml -test
|
| 208 |
+
```
|
| 209 |
+
|
| 210 |
+
**b. Prompt-based segmentation as [MedSAM](https://github.com/bowang-lab/MedSAM) but using LVM-Med's Encoder.**
|
| 211 |
+
|
| 212 |
+
The training script is similar as MedSAM case but specify the weight model by ```-lvm_encoder```.
|
| 213 |
+
#### Train
|
| 214 |
+
```bash
|
| 215 |
+
python3 medsam.py -c dataloader/yaml_data/buid_lvm_med_sam.yml -lvm_encoder ./lvm_med_weights/lvmmed_vit.pth
|
| 216 |
+
```
|
| 217 |
+
|
| 218 |
+
#### Test
|
| 219 |
+
```bash
|
| 220 |
+
python3 medsam.py -c dataloader/yaml_data/buid_lvm_med_sam.yml -lvm_encoder ./lvm_med_weights/lvmmed_vit.pth -test
|
| 221 |
+
```
|
| 222 |
+
|
| 223 |
+
You could also check our example notebook [`Prompt_Demo.ipynb`](/notebook/Prompt_Demo.ipynb) for results visualization using prompt-based MedSAM and prompt-based SAM with LVM-Med's encoder. The pre-trained weights for each SAM decoder model in the demo are [here](https://drive.google.com/drive/u/0/folders/1tjrkyEozE-98HAGEtyHboCT2YHBSW15U). Please download trained models of LVM-Med and MedSAM and put them into [`working_dir/checkpoints`](./working_dir/checkpoints/) folder for running the aforementioned notebook file.
|
| 224 |
+
|
| 225 |
+
**c. Zero-shot prompt-based segmentation with Segment Anything Model (SAM) for downstream tasks**
|
| 226 |
+
|
| 227 |
+
The SAM model without any finetuning using bounding box-based prompts can be done by:
|
| 228 |
+
```bash
|
| 229 |
+
python3 zero_shot_segmentation.py -c dataloader/yaml_data/buid_sam.yml
|
| 230 |
+
```
|
| 231 |
+
### Image Classification
|
| 232 |
+
We provide training and testing scripts using LVM-Med's ResNet-50 models for Brain Tumor Classification and Diabetic Retinopathy Grading in FGADR dataset (Table 5 in main paper and Table 12 in Appendix). The version with ViT models will be updated soon.
|
| 233 |
+
|
| 234 |
+
**a. Training with FGADR**
|
| 235 |
+
```bash
|
| 236 |
+
# Fully fine-tuned with 1 FCN
|
| 237 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_1_fcn.yml
|
| 238 |
+
|
| 239 |
+
# Fully fine-tuned with multiple FCNs
|
| 240 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_fcns.yml
|
| 241 |
+
|
| 242 |
+
# Freeze all and fine-tune 1-layer FCN only
|
| 243 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_frozen_1_fcn.yml
|
| 244 |
+
|
| 245 |
+
# Freeze all and fine-tune multi-layer FCN only
|
| 246 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_frozen_fcns.yml
|
| 247 |
+
```
|
| 248 |
+
To run for ```Brain dataset```, choose other config files ```brain_xyz.yml```in folder [`./dataloader/yaml_data/`](/dataloader/yaml_data).
|
| 249 |
+
|
| 250 |
+
**b. Inference with FGADR**
|
| 251 |
+
```bash
|
| 252 |
+
# Fully fine-tuned with 1 FCN
|
| 253 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_1_fcn.yml -test
|
| 254 |
+
|
| 255 |
+
# Fully fine-tuned with multiple FCNs
|
| 256 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_fcns.yml -test
|
| 257 |
+
|
| 258 |
+
# Freeze all and fine-tune 1-layer FCN only
|
| 259 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_frozen_1_fcn.yml -test
|
| 260 |
+
|
| 261 |
+
# Freeze all and fine-tune multi-layer FCN only
|
| 262 |
+
python train_classification.py -c ./dataloader/yaml_data/fgadr_endtoend_R50_frozen_fcns.yml -test
|
| 263 |
+
```
|
| 264 |
+
### Object Detection
|
| 265 |
+
We demonstrate using LVM-Med ResNet-50 for object detection with Vin-Dr dataset. We use Faster-RCNN for the network backbone.
|
| 266 |
+
You can access [`object_detection`](./object_detection) folder for more details.
|
| 267 |
+
|
| 268 |
+
## Citation
|
| 269 |
+
Please cite this paper if it helps your research:
|
| 270 |
+
```bibtex
|
| 271 |
+
@article{nguyen2023lvm,
|
| 272 |
+
title={LVM-Med: Learning Large-Scale Self-Supervised Vision Models for Medical Imaging via Second-order Graph Matching},
|
| 273 |
+
author={Nguyen, Duy MH and Nguyen, Hoang and Diep, Nghiem T and Pham, Tan N and Cao, Tri and Nguyen, Binh T and Swoboda, Paul and Ho, Nhat and Albarqouni, Shadi and Xie, Pengtao and others},
|
| 274 |
+
journal={arXiv preprint arXiv:2306.11925},
|
| 275 |
+
year={2023}
|
| 276 |
+
}
|
| 277 |
+
```
|
| 278 |
+
|
| 279 |
+
## Related Work
|
| 280 |
+
We use and modify codes from [SAM](https://github.com/facebookresearch/segment-anything) and [MedSAM](https://github.com/bowang-lab/MedSAM) for prompt-based segmentation settings. A part of LVM-Med algorithm adopt data transformations from [Vicregl](https://github.com/facebookresearch/VICRegL), [Deepcluster-v2](https://github.com/facebookresearch/swav?utm_source=catalyzex.com). We also utilize [vissl](https://github.com/facebookresearch/vissl) framework to train 2D self-supervised methods in our collected data. Thank the authors for their great work!
|
| 281 |
+
|
| 282 |
+
## License
|
| 283 |
+
Licensed under the [CC BY-NC-ND 2.0](https://creativecommons.org/licenses/by-nc-nd/2.0/) (**Attribution-NonCommercial-NoDerivs 2.0 Generic**). The code is released for academic research use only. For commercial use, please contact [Ho_Minh_Duy.Nguyen@dfki.de](Ho_Minh_Duy.Nguyen@dfki.de)
|
| 284 |
+
|
| 285 |
+
[//]: # (### f. LVM-Med )
|
| 286 |
+
|
| 287 |
+
[//]: # (#### Training Phase)
|
| 288 |
+
|
| 289 |
+
[//]: # (#### Fine-tune for downstream tasks using ResNet-50)
|
| 290 |
+
|
| 291 |
+
[//]: # ()
|
| 292 |
+
[//]: # (```bash)
|
| 293 |
+
|
| 294 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_R50.yml)
|
| 295 |
+
|
| 296 |
+
[//]: # (```)
|
| 297 |
+
|
| 298 |
+
[//]: # (#### Fine-tune for downstream tasks using SAM's VIT)
|
| 299 |
+
|
| 300 |
+
[//]: # (```bash)
|
| 301 |
+
|
| 302 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_SAM_VIT.yml)
|
| 303 |
+
|
| 304 |
+
[//]: # (```)
|
| 305 |
+
|
| 306 |
+
[//]: # (#### Inference)
|
| 307 |
+
|
| 308 |
+
[//]: # (#### Downstream tasks using ResNet-50)
|
| 309 |
+
|
| 310 |
+
[//]: # ()
|
| 311 |
+
[//]: # (```bash)
|
| 312 |
+
|
| 313 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_R50.yml -test)
|
| 314 |
+
|
| 315 |
+
[//]: # (```)
|
| 316 |
+
|
| 317 |
+
[//]: # (#### Downstream tasks using SAM's VIT)
|
| 318 |
+
|
| 319 |
+
[//]: # (```bash)
|
| 320 |
+
|
| 321 |
+
[//]: # (python train_segmentation.py -c ./dataloader/yaml_data/buid_endtoend_SAM_VIT.yml -test)
|
| 322 |
+
|
| 323 |
+
[//]: # (```)
|
assets/__init__.py
ADDED
|
File without changes
|
assets/body_lvm_med.jpg
ADDED
|
assets/lvm_med_compare.jpg
ADDED

|
assets/lvm_med_teaser.gif
ADDED

|
Git LFS Details
|
classification_R50/__init__.py
ADDED
|
File without changes
|
classification_R50/train_R50_classification.py
ADDED
|
@@ -0,0 +1,349 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""
|
| 2 |
+
Linear and non-linear image classification tasks with and w/o frozen image encoders
|
| 3 |
+
"""
|
| 4 |
+
import torch
|
| 5 |
+
import torch.nn as nn
|
| 6 |
+
import torchvision
|
| 7 |
+
from torchvision import transforms
|
| 8 |
+
from torch.utils.data import DataLoader, Dataset
|
| 9 |
+
from PIL import Image
|
| 10 |
+
import numpy as np
|
| 11 |
+
from sklearn.model_selection import train_test_split
|
| 12 |
+
import torchvision.datasets as datasets
|
| 13 |
+
import argparse
|
| 14 |
+
from tqdm import tqdm
|
| 15 |
+
import os
|
| 16 |
+
import random
|
| 17 |
+
import torch.nn.functional as F
|
| 18 |
+
seed = 42
|
| 19 |
+
torch.manual_seed(seed)
|
| 20 |
+
np.random.seed(seed)
|
| 21 |
+
random.seed(seed)
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
weight_collections = {
|
| 26 |
+
"resnet50": {
|
| 27 |
+
"lvm-med-resnet": "./lvm_med_weights/lvmmed_resnet.torch",
|
| 28 |
+
}
|
| 29 |
+
}
|
| 30 |
+
|
| 31 |
+
def eval(net, dataloader, device, criterion, num_samples):
|
| 32 |
+
# Evaluate the model on the validation set
|
| 33 |
+
val_loss = 0.0
|
| 34 |
+
val_acc = 0.0
|
| 35 |
+
|
| 36 |
+
net.eval()
|
| 37 |
+
with tqdm(total=len(dataloader), desc='Validation round', unit=' img') as pbar:
|
| 38 |
+
for inputs, labels in dataloader:
|
| 39 |
+
inputs = inputs.to(device)
|
| 40 |
+
labels = labels.to(device)
|
| 41 |
+
with torch.no_grad():
|
| 42 |
+
outputs = net(inputs)
|
| 43 |
+
loss = criterion(outputs, labels)
|
| 44 |
+
val_loss += loss.item() * inputs.size(0)
|
| 45 |
+
_, preds = torch.max(outputs, 1)
|
| 46 |
+
val_acc += torch.sum(preds == labels.data)
|
| 47 |
+
pbar.update(inputs.shape[0])
|
| 48 |
+
val_loss /= num_samples
|
| 49 |
+
val_acc /= num_samples
|
| 50 |
+
net.train()
|
| 51 |
+
return val_loss, val_acc
|
| 52 |
+
|
| 53 |
+
def TrainingTesting(cfg, numtry, pretrained_weight_name, data_path, num_classes, data_tranform, device, solver, name_weights,
|
| 54 |
+
frozen_encoder, architecture_type, number_epoch=50, learning_rate=0.001, batch_size=32, test_mode='best_valid',
|
| 55 |
+
valid_rate=0.2):
|
| 56 |
+
# Load the datasets
|
| 57 |
+
train_dir = data_path + "/Training"
|
| 58 |
+
test_dir = data_path + "/Testing"
|
| 59 |
+
if frozen_encoder:
|
| 60 |
+
checkpoint_dir = cfg.base.best_valid_model_checkpoint + cfg.base.dataset_name + "_" + architecture_type + "_" + name_weights + "_frozen/"
|
| 61 |
+
else:
|
| 62 |
+
checkpoint_dir = cfg.base.best_valid_model_checkpoint + cfg.base.dataset_name + "_" + architecture_type + "_" + name_weights + "_non_frozen/"
|
| 63 |
+
|
| 64 |
+
CHECK_FOLDER = os.path.isdir(checkpoint_dir)
|
| 65 |
+
|
| 66 |
+
if not CHECK_FOLDER:
|
| 67 |
+
os.makedirs(checkpoint_dir)
|
| 68 |
+
print("created folder: ", checkpoint_dir)
|
| 69 |
+
|
| 70 |
+
train_dataset = datasets.ImageFolder(root=train_dir, transform=data_tranform)
|
| 71 |
+
test_dataset = datasets.ImageFolder(root=test_dir, transform=data_tranform)
|
| 72 |
+
|
| 73 |
+
print ("valid size is {}".format(valid_rate))
|
| 74 |
+
# Split the training dataset into training and validation sets
|
| 75 |
+
train_indices, val_indices = train_test_split(list(range(len(train_dataset))), test_size=valid_rate, random_state=42)
|
| 76 |
+
train_sampler = torch.utils.data.sampler.SubsetRandomSampler(train_indices)
|
| 77 |
+
val_sampler = torch.utils.data.sampler.SubsetRandomSampler(val_indices)
|
| 78 |
+
|
| 79 |
+
loader_args = dict(num_workers=10, pin_memory=True)
|
| 80 |
+
train_loader = DataLoader(train_dataset, batch_size=batch_size, sampler=train_sampler, **loader_args)
|
| 81 |
+
val_loader = DataLoader(train_dataset, batch_size=batch_size, sampler=val_sampler, **loader_args)
|
| 82 |
+
test_loader = DataLoader(test_dataset, batch_size=batch_size, shuffle=True, **loader_args)
|
| 83 |
+
|
| 84 |
+
n_train = len(train_indices)
|
| 85 |
+
|
| 86 |
+
# Define the ResNet50 model
|
| 87 |
+
model = torchvision.models.resnet50(pretrained=True)
|
| 88 |
+
|
| 89 |
+
# Freeze the layers of the ResNet50 model
|
| 90 |
+
if frozen_encoder:
|
| 91 |
+
print ("Frozen encoder")
|
| 92 |
+
for param in model.parameters():
|
| 93 |
+
param.requires_grad = False
|
| 94 |
+
|
| 95 |
+
num_ftrs = model.fc.in_features
|
| 96 |
+
|
| 97 |
+
# define linear or non-linear architecture
|
| 98 |
+
if architecture_type == '1-fcn':
|
| 99 |
+
print ("Using single fully-connected layer")
|
| 100 |
+
model.fc = nn.Linear(num_ftrs, num_classes)
|
| 101 |
+
elif architecture_type == "fcns":
|
| 102 |
+
print("Using several fully-connected layers")
|
| 103 |
+
if cfg.base.dataset_name == 'brain':
|
| 104 |
+
model.fc = nn.Sequential(
|
| 105 |
+
nn.Linear(num_ftrs, 512),
|
| 106 |
+
nn.ReLU(),
|
| 107 |
+
nn.Linear(512, 256),
|
| 108 |
+
nn.ReLU(),
|
| 109 |
+
nn.Linear(256, num_classes))
|
| 110 |
+
elif cfg.base.dataset_name == 'fgadr':
|
| 111 |
+
model.fc = nn.Sequential(
|
| 112 |
+
nn.Linear(num_ftrs, 512),
|
| 113 |
+
nn.ReLU(),
|
| 114 |
+
nn.Linear(512, 128),
|
| 115 |
+
nn.ReLU(),
|
| 116 |
+
nn.Linear(128, num_classes))
|
| 117 |
+
else:
|
| 118 |
+
print(">>> Not implemented for selected datasets")
|
| 119 |
+
exit()
|
| 120 |
+
else:
|
| 121 |
+
print (">>> No available option for achitecture. Please check 'help' with --linear option")
|
| 122 |
+
exit()
|
| 123 |
+
|
| 124 |
+
pretrained_weight = torch.load(weight_collections["resnet50"][pretrained_weight_name], map_location=device)
|
| 125 |
+
model.load_state_dict(pretrained_weight, strict=False)
|
| 126 |
+
print("Loaded pretrained-weight of ", pretrained_weight_name)
|
| 127 |
+
|
| 128 |
+
model = model.to(device)
|
| 129 |
+
|
| 130 |
+
criterion = nn.CrossEntropyLoss()
|
| 131 |
+
|
| 132 |
+
if solver == "sgd":
|
| 133 |
+
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate, momentum=0.9)
|
| 134 |
+
elif solver == "adam":
|
| 135 |
+
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate, betas=(0.9, 0.999), eps=1e-08,
|
| 136 |
+
weight_decay=0.)
|
| 137 |
+
if cfg.base.dataset_name == 'fgadr':
|
| 138 |
+
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate, betas=(0.9, 0.999), eps=1e-08,
|
| 139 |
+
weight_decay=0.005)
|
| 140 |
+
else:
|
| 141 |
+
print ("Non-available solver")
|
| 142 |
+
exit()
|
| 143 |
+
|
| 144 |
+
## ------------ Train the model ------------
|
| 145 |
+
print(" ------------ Training ------------ ")
|
| 146 |
+
num_epochs = number_epoch
|
| 147 |
+
best_acc_val = 0.
|
| 148 |
+
|
| 149 |
+
for epoch in range(num_epochs):
|
| 150 |
+
print(f"Epoch {epoch + 1}:")
|
| 151 |
+
train_loss = 0.0
|
| 152 |
+
train_acc = 0.0
|
| 153 |
+
|
| 154 |
+
# Train the model on the training set
|
| 155 |
+
model.train()
|
| 156 |
+
with tqdm(total=n_train, desc=f'Epoch {epoch + 1}/{num_epochs}', unit='img') as pbar:
|
| 157 |
+
for inputs, labels in train_loader:
|
| 158 |
+
inputs = inputs.to(device)
|
| 159 |
+
labels = labels.to(device)
|
| 160 |
+
optimizer.zero_grad()
|
| 161 |
+
outputs = model(inputs)
|
| 162 |
+
loss = criterion(outputs, labels)
|
| 163 |
+
loss.backward()
|
| 164 |
+
optimizer.step()
|
| 165 |
+
train_loss += loss.item() * inputs.size(0)
|
| 166 |
+
_, preds = torch.max(outputs, 1)
|
| 167 |
+
train_acc += torch.sum(preds == labels.data)
|
| 168 |
+
|
| 169 |
+
# updating progressing bar
|
| 170 |
+
pbar.update(inputs.shape[0])
|
| 171 |
+
pbar.set_postfix(**{'loss (batch)': loss.item()})
|
| 172 |
+
|
| 173 |
+
# Print the results for this epoch
|
| 174 |
+
train_loss /= len(train_indices)
|
| 175 |
+
train_acc /= len(train_indices)
|
| 176 |
+
num_samples = len(val_indices)
|
| 177 |
+
num_samples_test = len(test_dataset)
|
| 178 |
+
|
| 179 |
+
print(" \n >>> Evaluation ")
|
| 180 |
+
val_loss, val_acc = eval(model, val_loader, device, criterion, num_samples)
|
| 181 |
+
|
| 182 |
+
if val_acc >= best_acc_val:
|
| 183 |
+
checkpoint = {
|
| 184 |
+
'epoch': epoch + 1,
|
| 185 |
+
'state_dict': model.state_dict(),
|
| 186 |
+
'optimizer': optimizer.state_dict()
|
| 187 |
+
}
|
| 188 |
+
torch.save(checkpoint, checkpoint_dir
|
| 189 |
+
+ name_weights + "_" + pretrained_weight_name + "_" + str(numtry) + ".pth")
|
| 190 |
+
print("Saved checkpoint at epoch ", epoch + 1)
|
| 191 |
+
best_acc_val = val_acc
|
| 192 |
+
|
| 193 |
+
print(f"Training Loss: {train_loss:.4f}\t Training Accuracy: {train_acc:.4f}")
|
| 194 |
+
print(f"Val Loss: {val_loss:.4f}\tVal Accuracy: {val_acc:.5f}")
|
| 195 |
+
|
| 196 |
+
# print model at last epochs
|
| 197 |
+
checkpoint = {
|
| 198 |
+
'epoch': epoch + 1,
|
| 199 |
+
'state_dict': model.state_dict(),
|
| 200 |
+
'optimizer': optimizer.state_dict()
|
| 201 |
+
}
|
| 202 |
+
torch.save(checkpoint, checkpoint_dir
|
| 203 |
+
+ name_weights + "_" + pretrained_weight_name + "_last_" + str(numtry) + ".pth")
|
| 204 |
+
print("Saved checkpoint at last epoch ", epoch + 1)
|
| 205 |
+
|
| 206 |
+
## ------------ Test the model ------------
|
| 207 |
+
print("------ Testing ------")
|
| 208 |
+
if test_mode == "best_valid":
|
| 209 |
+
print("Loading best models at {}".format(checkpoint_dir))
|
| 210 |
+
ckp = torch.load(checkpoint_dir
|
| 211 |
+
+ name_weights + "_" + pretrained_weight_name + "_" + str(numtry) + ".pth")
|
| 212 |
+
else:
|
| 213 |
+
print("Loading models at last epochs {}".format(checkpoint_dir))
|
| 214 |
+
ckp = torch.load(checkpoint_dir
|
| 215 |
+
+ name_weights + "_" + pretrained_weight_name + "_last_" + str(numtry) + ".pth")
|
| 216 |
+
model.load_state_dict(ckp['state_dict'])
|
| 217 |
+
num_samples_test = len(test_dataset)
|
| 218 |
+
test_loss, test_acc = eval(model, test_loader, device, criterion, num_samples_test)
|
| 219 |
+
print(f"Test Loss: {test_loss:.4f}\tTest Accuracy: {test_acc:.5f}")
|
| 220 |
+
return test_acc
|
| 221 |
+
|
| 222 |
+
def inference(numtry, device, cfg, data_path, data_tranform, name_weights, pretrained_weight_name,
|
| 223 |
+
frozen_encoder, architecture_type, num_classes):
|
| 224 |
+
if frozen_encoder:
|
| 225 |
+
checkpoint_dir = cfg.base.best_valid_model_checkpoint + cfg.base.dataset_name + "_" + architecture_type + "_" + name_weights + "_frozen/"
|
| 226 |
+
else:
|
| 227 |
+
checkpoint_dir = cfg.base.best_valid_model_checkpoint + cfg.base.dataset_name + "_" + architecture_type + "_" + name_weights + "_non_frozen/"
|
| 228 |
+
loader_args = dict(num_workers=10, pin_memory=True)
|
| 229 |
+
test_dir = data_path + "/Testing"
|
| 230 |
+
test_dataset = datasets.ImageFolder(root=test_dir, transform=data_tranform)
|
| 231 |
+
test_loader = DataLoader(test_dataset, batch_size=32, shuffle=False, **loader_args)
|
| 232 |
+
|
| 233 |
+
model = torchvision.models.resnet50(pretrained=True)
|
| 234 |
+
# Freeze the layers of the ResNet50 model
|
| 235 |
+
if frozen_encoder:
|
| 236 |
+
print ("Frozen encoder")
|
| 237 |
+
for param in model.parameters():
|
| 238 |
+
param.requires_grad = False
|
| 239 |
+
|
| 240 |
+
num_ftrs = model.fc.in_features
|
| 241 |
+
if architecture_type == '1-fcn':
|
| 242 |
+
print ("Using single fully-connected layer")
|
| 243 |
+
model.fc = nn.Linear(num_ftrs, num_classes)
|
| 244 |
+
elif architecture_type == "fcns":
|
| 245 |
+
print("Using several fully-connected layers")
|
| 246 |
+
if cfg.base.dataset_name == 'brain':
|
| 247 |
+
model.fc = nn.Sequential(
|
| 248 |
+
nn.Linear(num_ftrs, 512),
|
| 249 |
+
nn.ReLU(),
|
| 250 |
+
nn.Linear(512, 256),
|
| 251 |
+
nn.ReLU(),
|
| 252 |
+
nn.Linear(256, num_classes))
|
| 253 |
+
elif cfg.base.dataset_name == 'fgadr':
|
| 254 |
+
model.fc = nn.Sequential(
|
| 255 |
+
nn.Linear(num_ftrs, 512),
|
| 256 |
+
nn.ReLU(),
|
| 257 |
+
nn.Linear(512, 128),
|
| 258 |
+
nn.ReLU(),
|
| 259 |
+
nn.Linear(128, num_classes))
|
| 260 |
+
else:
|
| 261 |
+
print(">>> Not implemented for selected datasets")
|
| 262 |
+
exit()
|
| 263 |
+
else:
|
| 264 |
+
print (">>> No available option for achitecture. Please check 'help' with --linear option")
|
| 265 |
+
exit()
|
| 266 |
+
|
| 267 |
+
model = model.to(device)
|
| 268 |
+
print("Loading best models at {}".format(checkpoint_dir))
|
| 269 |
+
ckp = torch.load(checkpoint_dir
|
| 270 |
+
+ name_weights + "_" + pretrained_weight_name + "_" + str(numtry) + ".pth")
|
| 271 |
+
model.load_state_dict(ckp['state_dict'])
|
| 272 |
+
num_samples_test = len(test_dataset)
|
| 273 |
+
criterion = nn.CrossEntropyLoss()
|
| 274 |
+
test_loss, test_acc = eval(model, test_loader, device, criterion, num_samples_test)
|
| 275 |
+
print(f"Test Loss: {test_loss:.4f}\tTest Accuracy: {test_acc:.5f}")
|
| 276 |
+
return test_acc
|
| 277 |
+
|
| 278 |
+
def str2bool(v):
|
| 279 |
+
if isinstance(v, bool):
|
| 280 |
+
return v
|
| 281 |
+
if v.lower() in ('yes', 'true', 't', 'y', '1'):
|
| 282 |
+
return True
|
| 283 |
+
elif v.lower() in ('no', 'false', 'f', 'n', '0'):
|
| 284 |
+
return False
|
| 285 |
+
else:
|
| 286 |
+
raise argparse.ArgumentTypeError('Boolean value expected.')
|
| 287 |
+
|
| 288 |
+
|
| 289 |
+
def train_R50(yml_args, cfg):
|
| 290 |
+
|
| 291 |
+
if cfg.base.dataset_name == 'brain':
|
| 292 |
+
data_path = cfg.dataloader.data_path
|
| 293 |
+
num_classes = 4
|
| 294 |
+
data_transforms = transforms.Compose([
|
| 295 |
+
transforms.Resize((224, 224)),
|
| 296 |
+
transforms.ToTensor(),
|
| 297 |
+
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
|
| 298 |
+
])
|
| 299 |
+
elif cfg.base.dataset_name == 'fgadr':
|
| 300 |
+
data_path = cfg.dataloader.data_path
|
| 301 |
+
num_classes = 5
|
| 302 |
+
data_transforms = transforms.Compose([
|
| 303 |
+
transforms.RandomCrop(size=(480, 480)),
|
| 304 |
+
transforms.RandomHorizontalFlip(p=0.5),
|
| 305 |
+
transforms.RandomAutocontrast(p=0.2),
|
| 306 |
+
transforms.ToTensor(),
|
| 307 |
+
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
|
| 308 |
+
])
|
| 309 |
+
else:
|
| 310 |
+
print (">>> No available datasets")
|
| 311 |
+
exit()
|
| 312 |
+
|
| 313 |
+
print ("Using dataset {}".format(cfg.base.dataset_name))
|
| 314 |
+
list_acc = []
|
| 315 |
+
|
| 316 |
+
name_weight = cfg.base.original_checkpoint + "_output"
|
| 317 |
+
cuda_string = 'cuda:' + cfg.base.gpu_id
|
| 318 |
+
devices = torch.device(cuda_string if torch.cuda.is_available() else 'cpu')
|
| 319 |
+
|
| 320 |
+
if not yml_args.use_test_mode:
|
| 321 |
+
# Training model with three trial times
|
| 322 |
+
for numtry in range(3):
|
| 323 |
+
print ("*****"*3 + "\n" + "Trial", numtry)
|
| 324 |
+
test_acc = TrainingTesting(cfg = cfg, numtry=numtry, pretrained_weight_name=cfg.base.original_checkpoint, data_path = data_path,
|
| 325 |
+
num_classes = num_classes,
|
| 326 |
+
data_tranform = data_transforms,
|
| 327 |
+
device=devices,
|
| 328 |
+
solver=cfg.train.solver,
|
| 329 |
+
name_weights=name_weight, frozen_encoder=cfg.base.frozen_eval,
|
| 330 |
+
number_epoch=cfg.train.num_epochs, architecture_type=cfg.base.model,
|
| 331 |
+
learning_rate=cfg.train.learning_rate, batch_size=cfg.train.train_batch_size,
|
| 332 |
+
test_mode=cfg.base.test_mode,
|
| 333 |
+
valid_rate = cfg.base.valid_rate)
|
| 334 |
+
list_acc.append(test_acc.to('cpu'))
|
| 335 |
+
print("==============================================================================")
|
| 336 |
+
print ("*****"*3 + "\n")
|
| 337 |
+
print("Mean Accuracy: ", np.mean(list_acc))
|
| 338 |
+
print("Standard Deviation: ", np.std(list_acc))
|
| 339 |
+
else:
|
| 340 |
+
# Evaluate model with three weights
|
| 341 |
+
for numtry in range(3):
|
| 342 |
+
print ("*****"*3 + "\n" + "weight", numtry+1)
|
| 343 |
+
test_acc = inference(numtry = numtry, device = devices, cfg = cfg, data_path = data_path, data_tranform=data_transforms,
|
| 344 |
+
name_weights=name_weight, pretrained_weight_name=cfg.base.original_checkpoint,
|
| 345 |
+
frozen_encoder=cfg.base.frozen_eval, architecture_type=cfg.base.model, num_classes=num_classes)
|
| 346 |
+
list_acc.append(test_acc.to('cpu'))
|
| 347 |
+
print ("*****"*3 + "\n")
|
| 348 |
+
print("Mean Accuracy: ", np.mean(list_acc))
|
| 349 |
+
print("Standard Deviation: ", np.std(list_acc))
|
dataloader/__init__.py
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from .sam_transforms import (
|
| 2 |
+
ResizeLongestSide
|
| 3 |
+
)
|
dataloader/dataloader.py
ADDED
|
@@ -0,0 +1,55 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from torch.utils.data import (
|
| 2 |
+
DataLoader
|
| 3 |
+
)
|
| 4 |
+
from dataloader.dataset import (
|
| 5 |
+
SegmentationDataset,
|
| 6 |
+
AugmentedSegmentationDataset
|
| 7 |
+
)
|
| 8 |
+
|
| 9 |
+
def sam_dataloader(cfg):
|
| 10 |
+
loader_args = dict(num_workers=cfg.base.num_workers,
|
| 11 |
+
pin_memory=cfg.base.pin_memory)
|
| 12 |
+
"""
|
| 13 |
+
Since the output of SAM's mask decoder is 256 by default (without using a postprocessing step),
|
| 14 |
+
hence, we chose to resize the mask ground truth into 256x256 for a better output (prediction without postprocessing).
|
| 15 |
+
"""
|
| 16 |
+
if cfg.base.dataset_name in ["buidnewprocess", "kvasir", "isiconlytrain", "drive"]:
|
| 17 |
+
train_dataset = SegmentationDataset(cfg.base.dataset_name,
|
| 18 |
+
cfg.dataloader.train_dir_img,
|
| 19 |
+
cfg.dataloader.train_dir_mask,
|
| 20 |
+
scale=(1024, 256))
|
| 21 |
+
elif cfg.base.dataset_name in ["bts", "las_mri", "las_ct"]:
|
| 22 |
+
train_dataset = AugmentedSegmentationDataset(cfg.base.dataset_name,
|
| 23 |
+
cfg.dataloader.train_dir_img,
|
| 24 |
+
cfg.dataloader.train_dir_mask,
|
| 25 |
+
scale=(1024, 256))
|
| 26 |
+
else:
|
| 27 |
+
raise NameError(f"[Error] Dataset {cfg.base.dataset_name} is either in wrong format or not yet implemented!")
|
| 28 |
+
|
| 29 |
+
val_dataset = SegmentationDataset(cfg.base.dataset_name,
|
| 30 |
+
cfg.dataloader.valid_dir_img,
|
| 31 |
+
cfg.dataloader.valid_dir_mask,
|
| 32 |
+
scale=(1024, 256))
|
| 33 |
+
test_dataset = SegmentationDataset(cfg.base.dataset_name,
|
| 34 |
+
cfg.dataloader.test_dir_img,
|
| 35 |
+
cfg.dataloader.test_dir_mask,
|
| 36 |
+
scale=(1024, 256))
|
| 37 |
+
train_loader = DataLoader(train_dataset,
|
| 38 |
+
shuffle=True,
|
| 39 |
+
batch_size=cfg.train.train_batch_size,
|
| 40 |
+
multiprocessing_context="fork",
|
| 41 |
+
**loader_args)
|
| 42 |
+
val_loader = DataLoader(val_dataset,
|
| 43 |
+
shuffle=False,
|
| 44 |
+
drop_last=True,
|
| 45 |
+
batch_size=cfg.train.valid_batch_size,
|
| 46 |
+
multiprocessing_context="fork",
|
| 47 |
+
**loader_args)
|
| 48 |
+
test_loader = DataLoader(test_dataset,
|
| 49 |
+
shuffle=False,
|
| 50 |
+
batch_size=cfg.train.test_batch_size,
|
| 51 |
+
drop_last=True,
|
| 52 |
+
multiprocessing_context="fork",
|
| 53 |
+
**loader_args)
|
| 54 |
+
|
| 55 |
+
return train_loader, val_loader, test_loader, val_dataset, test_dataset
|
dataloader/dataset.py
ADDED
|
@@ -0,0 +1,434 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import logging
|
| 2 |
+
import os
|
| 3 |
+
import numpy as np
|
| 4 |
+
import torch
|
| 5 |
+
import cv2
|
| 6 |
+
from skimage.transform import resize
|
| 7 |
+
from torch.utils.data import Dataset
|
| 8 |
+
|
| 9 |
+
def get_bbox_from_mask(mask):
|
| 10 |
+
'''Returns a bounding box from a mask'''
|
| 11 |
+
bbox = []
|
| 12 |
+
|
| 13 |
+
if len(mask.shape) == 2: #(H, W)
|
| 14 |
+
if np.all(mask == 0):
|
| 15 |
+
y_indices, x_indices = np.random.normal(0, 1024, 2)
|
| 16 |
+
x_min, x_max = np.min(x_indices), np.max(x_indices)
|
| 17 |
+
y_min, y_max = np.min(y_indices), np.max(y_indices)
|
| 18 |
+
x_min = max(0, x_min - np.random.randint(0, 20))
|
| 19 |
+
x_max = min(1024, x_max + np.random.randint(0, 20))
|
| 20 |
+
y_min = max(0, y_min - np.random.randint(0, 20))
|
| 21 |
+
y_max = min(1024, y_max + np.random.randint(0, 20))
|
| 22 |
+
else:
|
| 23 |
+
y_indices, x_indices = np.where(mask > 0)
|
| 24 |
+
x_min, x_max = np.min(x_indices), np.max(x_indices)
|
| 25 |
+
y_min, y_max = np.min(y_indices), np.max(y_indices)
|
| 26 |
+
H, W = mask.shape
|
| 27 |
+
x_min = max(0, x_min - np.random.randint(0, 20))
|
| 28 |
+
x_max = min(W, x_max + np.random.randint(0, 20))
|
| 29 |
+
y_min = max(0, y_min - np.random.randint(0, 20))
|
| 30 |
+
y_max = min(H, y_max + np.random.randint(0, 20))
|
| 31 |
+
return np.array([x_min, y_min, x_max, y_max])
|
| 32 |
+
|
| 33 |
+
for i in range(mask.shape[0]):
|
| 34 |
+
if np.all(mask[i] == 0):
|
| 35 |
+
y_indices, x_indices = np.random.normal(0, 1024, 2)
|
| 36 |
+
x_min, x_max = np.min(x_indices), np.max(x_indices)
|
| 37 |
+
y_min, y_max = np.min(y_indices), np.max(y_indices)
|
| 38 |
+
x_min = max(0, x_min - np.random.randint(0, 20))
|
| 39 |
+
x_max = min(1024, x_max + np.random.randint(0, 20))
|
| 40 |
+
y_min = max(0, y_min - np.random.randint(0, 20))
|
| 41 |
+
y_max = min(1024, y_max + np.random.randint(0, 20))
|
| 42 |
+
else:
|
| 43 |
+
y_indices, x_indices = np.where(mask[i] > 0)
|
| 44 |
+
x_min, x_max = np.min(x_indices), np.max(x_indices)
|
| 45 |
+
y_min, y_max = np.min(y_indices), np.max(y_indices)
|
| 46 |
+
H, W = mask[i].shape
|
| 47 |
+
x_min = max(0, x_min - np.random.randint(0, 20))
|
| 48 |
+
x_max = min(W, x_max + np.random.randint(0, 20))
|
| 49 |
+
y_min = max(0, y_min - np.random.randint(0, 20))
|
| 50 |
+
y_max = min(H, y_max + np.random.randint(0, 20))
|
| 51 |
+
bbox.append([x_min, y_min, x_max, y_max])
|
| 52 |
+
return np.array(bbox)
|
| 53 |
+
|
| 54 |
+
class SegmentationDataset_train(Dataset):
|
| 55 |
+
def __init__(self, nonlabel_path: str, havelabel_path: str, dataset: str, scale = (224, 224)):
|
| 56 |
+
self.nonlabel_path = nonlabel_path
|
| 57 |
+
self.havelabel_path = havelabel_path
|
| 58 |
+
self.name_dataset = dataset
|
| 59 |
+
self.scale = scale
|
| 60 |
+
|
| 61 |
+
with open(self.nonlabel_path, 'r') as nlf:
|
| 62 |
+
lines = nlf.readlines()
|
| 63 |
+
non_label_lines = [line.strip().split(' ')[:2] for line in lines]
|
| 64 |
+
|
| 65 |
+
with open(self.havelabel_path, 'r') as hlf:
|
| 66 |
+
lines = hlf.readlines()
|
| 67 |
+
have_label_lines = [line.strip().split(' ')[:2] for line in lines]
|
| 68 |
+
|
| 69 |
+
if len(non_label_lines) == 0:
|
| 70 |
+
self.ids = np.array(have_label_lines, dtype= object)
|
| 71 |
+
else:
|
| 72 |
+
choose_non_lable_lines = np.random.choice(len(non_label_lines), size = len(have_label_lines))
|
| 73 |
+
non_label_lines = np.array(non_label_lines, dtype= object)
|
| 74 |
+
have_label_lines = np.array(have_label_lines, dtype= object)
|
| 75 |
+
self.ids = np.concatenate([non_label_lines[choose_non_lable_lines], have_label_lines], axis= 0)
|
| 76 |
+
# self.ids = os.listdir(images_dir) #[splitext(file)[0] for file in listdir(images_dir) if not file.startswith('.') and image_type in file]
|
| 77 |
+
# print(len(self.ids))
|
| 78 |
+
# if datasetname == "las_mri":
|
| 79 |
+
# self.ids = [f for f in self.ids if image_type in f]
|
| 80 |
+
if len(self.ids) == 0:
|
| 81 |
+
raise RuntimeError(f'No input file found in {self.images_dir}, make sure you put your images there')
|
| 82 |
+
logging.info(f'Creating dataset with {len(self.ids)} examples')
|
| 83 |
+
self.cache = {}
|
| 84 |
+
|
| 85 |
+
def __len__(self):
|
| 86 |
+
return len(self.ids)
|
| 87 |
+
|
| 88 |
+
@classmethod
|
| 89 |
+
def preprocess(self, img, scale, is_mask):
|
| 90 |
+
img = resize(img,
|
| 91 |
+
(scale[0], scale[0]),
|
| 92 |
+
order=0,
|
| 93 |
+
preserve_range=True,
|
| 94 |
+
anti_aliasing=False).astype('uint8')
|
| 95 |
+
img = np.asarray(img)
|
| 96 |
+
if not is_mask:
|
| 97 |
+
img = ((img - img.min()) * (1/(0.01 + img.max() - img.min()) * 255)).astype('uint8')
|
| 98 |
+
if is_mask:
|
| 99 |
+
img = resize(img,
|
| 100 |
+
(scale[1], scale[1]),
|
| 101 |
+
order=0,
|
| 102 |
+
preserve_range=True,
|
| 103 |
+
anti_aliasing=False).astype('uint8')
|
| 104 |
+
return img
|
| 105 |
+
|
| 106 |
+
@classmethod
|
| 107 |
+
def load(self, filename, is_mask=False):
|
| 108 |
+
if is_mask:
|
| 109 |
+
return cv2.imread(filename, 0)
|
| 110 |
+
else:
|
| 111 |
+
return cv2.imread(filename)
|
| 112 |
+
|
| 113 |
+
def __getitem__(self, idx):
|
| 114 |
+
if idx in self.cache:
|
| 115 |
+
return self.cache[idx]
|
| 116 |
+
|
| 117 |
+
img_file = self.ids[idx][0]
|
| 118 |
+
mask_file = self.ids[idx][1]
|
| 119 |
+
# print(img_file)
|
| 120 |
+
#start = time.time()
|
| 121 |
+
mask = self.load(mask_file, is_mask=True)
|
| 122 |
+
img = self.load(img_file, is_mask=False)
|
| 123 |
+
|
| 124 |
+
assert mask is not None, mask_file
|
| 125 |
+
assert img is not None, img_file
|
| 126 |
+
|
| 127 |
+
img = self.preprocess(img, self.scale, is_mask=False)
|
| 128 |
+
mask = self.preprocess(mask, self.scale, is_mask=True)
|
| 129 |
+
|
| 130 |
+
if self.name_dataset in ["kvasir", "buidnewprocess"]:
|
| 131 |
+
mask[mask < 50] = 0
|
| 132 |
+
mask[mask > 200] = 1
|
| 133 |
+
elif self.name_dataset == "isiconlytrain":
|
| 134 |
+
mask[mask > 1] = 1
|
| 135 |
+
elif self.name_dataset.startswith("las"):
|
| 136 |
+
mask[mask == 30] = 1
|
| 137 |
+
mask[mask == 60] = 2 # main predict
|
| 138 |
+
mask[mask == 90] = 3
|
| 139 |
+
mask[mask == 120] = 4
|
| 140 |
+
mask[mask == 150] = 5
|
| 141 |
+
mask[mask == 180] = 6
|
| 142 |
+
mask[mask == 210] = 7
|
| 143 |
+
mask[mask > 7] = 0
|
| 144 |
+
else:
|
| 145 |
+
mask[mask>0] = 1
|
| 146 |
+
|
| 147 |
+
bboxes = get_bbox_from_mask(mask)
|
| 148 |
+
|
| 149 |
+
data = {
|
| 150 |
+
'image': torch.as_tensor(img.copy()).permute(2, 0, 1).float().contiguous(),
|
| 151 |
+
'mask': torch.tensor(mask[None, :, :]).long(),
|
| 152 |
+
'mask_ete': torch.as_tensor(mask.copy().astype(int)).long().contiguous(),
|
| 153 |
+
'bboxes' : torch.tensor(bboxes).float(),
|
| 154 |
+
'mask_file' : mask_file,
|
| 155 |
+
'img_file' : img_file
|
| 156 |
+
}
|
| 157 |
+
self.cache[idx] = data
|
| 158 |
+
return data
|
| 159 |
+
|
| 160 |
+
def get_3d_iter(self):
|
| 161 |
+
from itertools import groupby
|
| 162 |
+
keyf = lambda idx : self.ids[idx].split("_frame_")[0]
|
| 163 |
+
sorted_ids = sorted(range(len(self.ids)), key=lambda i : self.ids[i])
|
| 164 |
+
for _, items in groupby(sorted_ids, key=keyf):
|
| 165 |
+
images = []
|
| 166 |
+
masks = []
|
| 167 |
+
masks_ete = []
|
| 168 |
+
bboxes = []
|
| 169 |
+
for idx in items:
|
| 170 |
+
d = self.__getitem__(idx)
|
| 171 |
+
images.append(d['image'])
|
| 172 |
+
masks.append(d['mask'])
|
| 173 |
+
masks_ete.append(d['mask_ete'])
|
| 174 |
+
bboxes.append(d['bboxes'])
|
| 175 |
+
# store third dimension in image channels
|
| 176 |
+
images = torch.stack(images, dim=0)
|
| 177 |
+
masks = torch.stack(masks, dim=0)
|
| 178 |
+
masks_ete = torch.stack(masks_ete, dim=0)
|
| 179 |
+
bboxes = torch.stack(bboxes, dim=0)
|
| 180 |
+
_3d_data = {'image': images, 'mask': masks, 'mask_ete': masks_ete, 'bboxes': bboxes}
|
| 181 |
+
yield _3d_data
|
| 182 |
+
|
| 183 |
+
|
| 184 |
+
class SegmentationDataset(Dataset):
|
| 185 |
+
def __init__(self, name_dataset: str, images_dir: str, masks_dir: str, scale = (1024, 256)):
|
| 186 |
+
self.images_dir = images_dir
|
| 187 |
+
self.masks_dir = masks_dir
|
| 188 |
+
self.scale = scale
|
| 189 |
+
self.name_dataset = name_dataset
|
| 190 |
+
self.ids = os.listdir(images_dir)
|
| 191 |
+
if len(self.ids) == 0:
|
| 192 |
+
raise RuntimeError(f'No input file found in {self.images_dir}, make sure you put your images there')
|
| 193 |
+
logging.info(f'Creating dataset with {len(self.ids)} examples')
|
| 194 |
+
self.cache = {}
|
| 195 |
+
|
| 196 |
+
def __len__(self):
|
| 197 |
+
return len(self.ids)
|
| 198 |
+
|
| 199 |
+
@classmethod
|
| 200 |
+
def preprocess(self, img, scale, is_mask):
|
| 201 |
+
img = resize(img,
|
| 202 |
+
(scale[0], scale[0]),
|
| 203 |
+
order=0,
|
| 204 |
+
preserve_range=True,
|
| 205 |
+
anti_aliasing=False).astype('uint8')
|
| 206 |
+
img = np.asarray(img)
|
| 207 |
+
if not is_mask:
|
| 208 |
+
img = ((img - img.min()) * (1/(0.01 + img.max() - img.min()) * 255)).astype('uint8')
|
| 209 |
+
if is_mask:
|
| 210 |
+
img = resize(img,
|
| 211 |
+
(scale[1], scale[1]),
|
| 212 |
+
order=0,
|
| 213 |
+
preserve_range=True,
|
| 214 |
+
anti_aliasing=False).astype('uint8')
|
| 215 |
+
return img
|
| 216 |
+
|
| 217 |
+
@classmethod
|
| 218 |
+
def load(self, filename, is_mask=False):
|
| 219 |
+
if is_mask:
|
| 220 |
+
return cv2.imread(filename, 0)
|
| 221 |
+
else:
|
| 222 |
+
return cv2.imread(filename)
|
| 223 |
+
|
| 224 |
+
def __getitem__(self, idx):
|
| 225 |
+
if idx in self.cache:
|
| 226 |
+
return self.cache[idx]
|
| 227 |
+
|
| 228 |
+
name = self.ids[idx]
|
| 229 |
+
|
| 230 |
+
if self.name_dataset == "isiconlytrain":
|
| 231 |
+
mask_file = os.path.join(self.masks_dir, name).split(".jpg")[0]
|
| 232 |
+
mask_file = mask_file + "_segmentation.png"
|
| 233 |
+
elif self.name_dataset == "buidnewprocess":
|
| 234 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 235 |
+
elif self.name_dataset == "kvasir":
|
| 236 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 237 |
+
elif self.name_dataset == "drive":
|
| 238 |
+
mask_file = os.path.join(self.masks_dir, name).replace("training", "manual1")
|
| 239 |
+
elif self.name_dataset == "bts":
|
| 240 |
+
mask_file = os.path.join(self.masks_dir, name).replace(self.image_type, "_seg_")
|
| 241 |
+
elif self.name_dataset in ["las_mri", "las_ct"]:
|
| 242 |
+
mask_file = os.path.join(self.masks_dir, name).replace("image", "label")
|
| 243 |
+
else:
|
| 244 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 245 |
+
|
| 246 |
+
img_file = os.path.join(self.images_dir, name)
|
| 247 |
+
|
| 248 |
+
mask = self.load(mask_file, is_mask=True)
|
| 249 |
+
img = self.load(img_file, is_mask=False)
|
| 250 |
+
|
| 251 |
+
assert mask is not None, mask_file
|
| 252 |
+
assert img is not None, img_file
|
| 253 |
+
|
| 254 |
+
img = self.preprocess(img, self.scale, is_mask=False)
|
| 255 |
+
mask = self.preprocess(mask, self.scale, is_mask=True)
|
| 256 |
+
|
| 257 |
+
if self.name_dataset in ["kvasir", "buidnewprocess"]:
|
| 258 |
+
mask[mask < 50] = 0
|
| 259 |
+
mask[mask > 200] = 1
|
| 260 |
+
elif self.name_dataset == "isiconlytrain":
|
| 261 |
+
mask[mask > 1] = 1
|
| 262 |
+
elif self.name_dataset.startswith("las"):
|
| 263 |
+
mask[mask == 30] = 1
|
| 264 |
+
mask[mask == 60] = 2 # main predict
|
| 265 |
+
mask[mask == 90] = 3
|
| 266 |
+
mask[mask == 120] = 4
|
| 267 |
+
mask[mask == 150] = 5
|
| 268 |
+
mask[mask == 180] = 6
|
| 269 |
+
mask[mask == 210] = 7
|
| 270 |
+
mask[mask > 7] = 0
|
| 271 |
+
else:
|
| 272 |
+
mask[mask>0] = 1
|
| 273 |
+
|
| 274 |
+
bboxes = get_bbox_from_mask(mask)
|
| 275 |
+
|
| 276 |
+
data = {
|
| 277 |
+
'image': torch.as_tensor(img.copy()).permute(2, 0, 1).float().contiguous(),
|
| 278 |
+
'mask': torch.tensor(mask[None, :, :]).long(),
|
| 279 |
+
'mask_ete': torch.as_tensor(mask.copy().astype(int)).long().contiguous(),
|
| 280 |
+
'bboxes' : torch.tensor(bboxes).float(),
|
| 281 |
+
'mask_file' : mask_file,
|
| 282 |
+
'img_file' : img_file
|
| 283 |
+
}
|
| 284 |
+
self.cache[idx] = data
|
| 285 |
+
return data
|
| 286 |
+
|
| 287 |
+
def get_3d_iter(self):
|
| 288 |
+
from itertools import groupby
|
| 289 |
+
keyf = lambda idx : self.ids[idx].split("_frame_")[0]
|
| 290 |
+
sorted_ids = sorted(range(len(self.ids)), key=lambda i : self.ids[i])
|
| 291 |
+
for _, items in groupby(sorted_ids, key=keyf):
|
| 292 |
+
images = []
|
| 293 |
+
masks = []
|
| 294 |
+
masks_ete = []
|
| 295 |
+
bboxes = []
|
| 296 |
+
for idx in items:
|
| 297 |
+
d = self.__getitem__(idx)
|
| 298 |
+
images.append(d['image'])
|
| 299 |
+
masks.append(d['mask'])
|
| 300 |
+
masks_ete.append(d['mask_ete'])
|
| 301 |
+
bboxes.append(d['bboxes'])
|
| 302 |
+
# store third dimension in image channels
|
| 303 |
+
images = torch.stack(images, dim=0)
|
| 304 |
+
masks = torch.stack(masks, dim=0)
|
| 305 |
+
masks_ete = torch.stack(masks_ete, dim=0)
|
| 306 |
+
bboxes = torch.stack(bboxes, dim=0)
|
| 307 |
+
_3d_data = {'image': images, 'mask': masks, 'mask_ete': masks_ete, 'bboxes': bboxes}
|
| 308 |
+
yield _3d_data
|
| 309 |
+
|
| 310 |
+
class AugmentedSegmentationDataset(Dataset):
|
| 311 |
+
def __init__(self, name_dataset: str, images_dir: str, masks_dir: str, scale = (1024, 256), transform=True):
|
| 312 |
+
|
| 313 |
+
self.images_dir = images_dir
|
| 314 |
+
self.masks_dir = masks_dir
|
| 315 |
+
self.scale = scale
|
| 316 |
+
self.transform = transform
|
| 317 |
+
self.name_dataset = name_dataset
|
| 318 |
+
self.ids = os.listdir(images_dir)
|
| 319 |
+
if len(self.ids) == 0:
|
| 320 |
+
raise RuntimeError(f'No input file found in {self.images_dir}, make sure you put your images there')
|
| 321 |
+
logging.info(f'Creating dataset with {len(self.ids)} examples')
|
| 322 |
+
self.cache = {}
|
| 323 |
+
|
| 324 |
+
def __len__(self):
|
| 325 |
+
return len(self.ids)
|
| 326 |
+
|
| 327 |
+
@classmethod
|
| 328 |
+
def preprocess(self, img, scale, is_mask, transform):
|
| 329 |
+
img = resize(img,
|
| 330 |
+
(scale[0], scale[0]),
|
| 331 |
+
order=0,
|
| 332 |
+
preserve_range=True,
|
| 333 |
+
anti_aliasing=False).astype('uint8')
|
| 334 |
+
img = np.asarray(img)
|
| 335 |
+
if (not is_mask) and transform:
|
| 336 |
+
img = ((img - img.min()) * (1/(0.01 + img.max() - img.min()) * 255)).astype('uint8')
|
| 337 |
+
if is_mask:
|
| 338 |
+
img = resize(img,
|
| 339 |
+
(scale[1], scale[1]),
|
| 340 |
+
order=0,
|
| 341 |
+
preserve_range=True,
|
| 342 |
+
anti_aliasing=False).astype('uint8')
|
| 343 |
+
return img
|
| 344 |
+
|
| 345 |
+
@classmethod
|
| 346 |
+
def preprocess_non_expand(self, img, scale, is_mask, transform):
|
| 347 |
+
img = resize(img,
|
| 348 |
+
(scale[0], scale[0]),
|
| 349 |
+
order=0,
|
| 350 |
+
preserve_range=True,
|
| 351 |
+
anti_aliasing=False).astype('uint8')
|
| 352 |
+
img = np.asarray(img)
|
| 353 |
+
if (not is_mask) and transform:
|
| 354 |
+
img = ((img - img.min()) * (1/(0.01 + img.max() - img.min()) * 255)).astype('uint8')
|
| 355 |
+
if is_mask:
|
| 356 |
+
img = resize(img,
|
| 357 |
+
(scale[1], scale[1]),
|
| 358 |
+
order=0,
|
| 359 |
+
preserve_range=True,
|
| 360 |
+
anti_aliasing=False).astype('uint8')
|
| 361 |
+
return img
|
| 362 |
+
|
| 363 |
+
@classmethod
|
| 364 |
+
def load(self, filename, is_mask=False):
|
| 365 |
+
if is_mask:
|
| 366 |
+
return cv2.imread(filename, 0)
|
| 367 |
+
else:
|
| 368 |
+
return cv2.imread(filename)
|
| 369 |
+
|
| 370 |
+
def __getitem__(self, idx):
|
| 371 |
+
if idx in self.cache:
|
| 372 |
+
return self.cache[idx]
|
| 373 |
+
|
| 374 |
+
name = self.ids[idx]
|
| 375 |
+
|
| 376 |
+
if self.name_dataset == "bts":
|
| 377 |
+
mask_file = os.path.join(self.masks_dir, name).replace(self.image_type, "_seg_")
|
| 378 |
+
elif self.name_dataset in ["las_mri", "las_ct"]:
|
| 379 |
+
mask_file = os.path.join(self.masks_dir, name).replace("image", "label")
|
| 380 |
+
|
| 381 |
+
img_file = os.path.join(self.images_dir, name)
|
| 382 |
+
|
| 383 |
+
mask = self.load(mask_file, is_mask=True)
|
| 384 |
+
img = self.load(img_file, is_mask=False)
|
| 385 |
+
|
| 386 |
+
assert mask is not None, mask_file
|
| 387 |
+
assert img is not None, img_file
|
| 388 |
+
|
| 389 |
+
img = self.preprocess_non_expand(img, self.scale, False, self.transform)
|
| 390 |
+
mask = self.preprocess(mask, self.scale, True, self.transform)
|
| 391 |
+
|
| 392 |
+
if self.name_dataset.startswith("las"):
|
| 393 |
+
mask[mask == 30] = 1
|
| 394 |
+
mask[mask == 60] = 2 # main predict
|
| 395 |
+
mask[mask == 90] = 3
|
| 396 |
+
mask[mask == 120] = 4
|
| 397 |
+
mask[mask == 150] = 5
|
| 398 |
+
mask[mask == 180] = 6
|
| 399 |
+
mask[mask == 210] = 7
|
| 400 |
+
mask[mask > 7] = 0
|
| 401 |
+
else:
|
| 402 |
+
mask[mask>0]=1
|
| 403 |
+
|
| 404 |
+
bboxes = get_bbox_from_mask(mask)
|
| 405 |
+
|
| 406 |
+
data = {
|
| 407 |
+
'image': torch.as_tensor(img.copy()).permute(2, 0, 1).float().contiguous(),
|
| 408 |
+
'mask': torch.tensor(mask[None, :, :]).long(),
|
| 409 |
+
'bboxes' : torch.tensor(bboxes).float(),
|
| 410 |
+
'mask_file' : mask_file,
|
| 411 |
+
'img_file' : img_file
|
| 412 |
+
}
|
| 413 |
+
self.cache[idx] = data
|
| 414 |
+
return data
|
| 415 |
+
|
| 416 |
+
def get_3d_iter(self):
|
| 417 |
+
from itertools import groupby
|
| 418 |
+
keyf = lambda idx : self.ids[idx].split("_frame_")[0]
|
| 419 |
+
sorted_ids = sorted(range(len(self.ids)), key=lambda i : self.ids[i])
|
| 420 |
+
for _, items in groupby(sorted_ids, key=keyf):
|
| 421 |
+
images = []
|
| 422 |
+
masks = []
|
| 423 |
+
bboxes = []
|
| 424 |
+
for idx in items:
|
| 425 |
+
d = self.__getitem__(idx)
|
| 426 |
+
images.append(d['image'])
|
| 427 |
+
masks.append(d['mask'])
|
| 428 |
+
bboxes.append(d['bboxes'])
|
| 429 |
+
# store third dimension in image channels
|
| 430 |
+
images = torch.stack(images, dim=0)
|
| 431 |
+
masks = torch.stack(masks, dim=0)
|
| 432 |
+
bboxes = torch.stack(bboxes, dim=0)
|
| 433 |
+
_3d_data = {'image': images, 'mask': masks, 'bboxes': bboxes}
|
| 434 |
+
yield _3d_data
|
dataloader/dataset_ete.py
ADDED
|
@@ -0,0 +1,267 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import logging
|
| 2 |
+
import os
|
| 3 |
+
import numpy as np
|
| 4 |
+
import torch
|
| 5 |
+
import cv2
|
| 6 |
+
from skimage.transform import resize
|
| 7 |
+
from torch.utils.data import Dataset
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
class SegmentationDataset_train(Dataset):
|
| 12 |
+
def __init__(self, nonlabel_path: str, havelabel_path: str, dataset: str, scale = (224, 224)):
|
| 13 |
+
self.nonlabel_path = nonlabel_path
|
| 14 |
+
self.havelabel_path = havelabel_path
|
| 15 |
+
self.name_dataset = dataset
|
| 16 |
+
self.scale = scale
|
| 17 |
+
|
| 18 |
+
with open(self.nonlabel_path, 'r') as nlf:
|
| 19 |
+
lines = nlf.readlines()
|
| 20 |
+
non_label_lines = [line.strip().split(' ')[:2] for line in lines]
|
| 21 |
+
|
| 22 |
+
with open(self.havelabel_path, 'r') as hlf:
|
| 23 |
+
lines = hlf.readlines()
|
| 24 |
+
have_label_lines = [line.strip().split(' ')[:2] for line in lines]
|
| 25 |
+
|
| 26 |
+
if len(non_label_lines) == 0:
|
| 27 |
+
self.ids = np.array(have_label_lines, dtype= object)
|
| 28 |
+
else:
|
| 29 |
+
choose_non_lable_lines = np.random.choice(len(non_label_lines), size = len(have_label_lines))
|
| 30 |
+
non_label_lines = np.array(non_label_lines, dtype= object)
|
| 31 |
+
have_label_lines = np.array(have_label_lines, dtype= object)
|
| 32 |
+
self.ids = np.concatenate([non_label_lines[choose_non_lable_lines], have_label_lines], axis= 0)
|
| 33 |
+
# self.ids = os.listdir(images_dir) #[splitext(file)[0] for file in listdir(images_dir) if not file.startswith('.') and image_type in file]
|
| 34 |
+
# print(len(self.ids))
|
| 35 |
+
# if datasetname == "las_mri":
|
| 36 |
+
# self.ids = [f for f in self.ids if image_type in f]
|
| 37 |
+
if len(self.ids) == 0:
|
| 38 |
+
raise RuntimeError(f'No input file found in {self.images_dir}, make sure you put your images there')
|
| 39 |
+
logging.info(f'Creating dataset with {len(self.ids)} examples')
|
| 40 |
+
self.cache = {}
|
| 41 |
+
|
| 42 |
+
def __len__(self):
|
| 43 |
+
return len(self.ids)
|
| 44 |
+
|
| 45 |
+
@classmethod
|
| 46 |
+
def preprocess(self, img, scale, is_mask):
|
| 47 |
+
img = resize(img,
|
| 48 |
+
(scale[0], scale[0]),
|
| 49 |
+
order=0,
|
| 50 |
+
preserve_range=True,
|
| 51 |
+
anti_aliasing=False).astype('uint8')
|
| 52 |
+
img = np.asarray(img)
|
| 53 |
+
if not is_mask:
|
| 54 |
+
img = ((img - img.min()) * (1/(0.01 + img.max() - img.min()) * 255)).astype('uint8')
|
| 55 |
+
if len(img.shape) != 3:
|
| 56 |
+
img = np.expand_dims(img, axis=2) #(1, 224, 224)
|
| 57 |
+
if is_mask:
|
| 58 |
+
img = resize(img,
|
| 59 |
+
(scale[1], scale[1]),
|
| 60 |
+
order=0,
|
| 61 |
+
preserve_range=True,
|
| 62 |
+
anti_aliasing=False).astype('uint8')
|
| 63 |
+
return img
|
| 64 |
+
|
| 65 |
+
@classmethod
|
| 66 |
+
def load(self, filename, name_dataset, is_mask=False):
|
| 67 |
+
if name_dataset.startswith("las"):
|
| 68 |
+
if is_mask:
|
| 69 |
+
return cv2.imread(filename, cv2.IMREAD_UNCHANGED)
|
| 70 |
+
else:
|
| 71 |
+
img = cv2.imread(filename, 0)
|
| 72 |
+
return img
|
| 73 |
+
else:
|
| 74 |
+
if is_mask:
|
| 75 |
+
return cv2.imread(filename, 0)
|
| 76 |
+
else:
|
| 77 |
+
return cv2.imread(filename)
|
| 78 |
+
|
| 79 |
+
def __getitem__(self, idx):
|
| 80 |
+
if idx in self.cache:
|
| 81 |
+
return self.cache[idx]
|
| 82 |
+
|
| 83 |
+
img_file = self.ids[idx][0]
|
| 84 |
+
mask_file = self.ids[idx][1]
|
| 85 |
+
# print(img_file)
|
| 86 |
+
#start = time.time()
|
| 87 |
+
mask = self.load(mask_file, self.name_dataset, is_mask=True)
|
| 88 |
+
img = self.load(img_file, self.name_dataset, is_mask=False)
|
| 89 |
+
|
| 90 |
+
assert mask is not None, mask_file
|
| 91 |
+
assert img is not None, img_file
|
| 92 |
+
|
| 93 |
+
|
| 94 |
+
if self.name_dataset in ["kvasir", "buidnewprocess"]:
|
| 95 |
+
mask[mask < 50] = 0
|
| 96 |
+
mask[mask > 200] = 1
|
| 97 |
+
elif self.name_dataset == "isiconlytrain":
|
| 98 |
+
mask[mask > 1] = 1
|
| 99 |
+
elif self.name_dataset.startswith("las"):
|
| 100 |
+
mask[mask == 30] = 1
|
| 101 |
+
mask[mask == 60] = 2 # main predict
|
| 102 |
+
mask[mask == 90] = 3
|
| 103 |
+
mask[mask == 120] = 4
|
| 104 |
+
mask[mask == 150] = 5
|
| 105 |
+
mask[mask == 180] = 6
|
| 106 |
+
mask[mask == 210] = 7
|
| 107 |
+
mask[mask > 7] = 0
|
| 108 |
+
else:
|
| 109 |
+
mask[mask>0] = 1
|
| 110 |
+
|
| 111 |
+
img = self.preprocess(img, self.scale, is_mask=False)
|
| 112 |
+
mask = self.preprocess(mask, self.scale, is_mask=True)
|
| 113 |
+
|
| 114 |
+
data = {
|
| 115 |
+
'image': torch.as_tensor(img.copy()).permute(2, 0, 1).float().contiguous(),
|
| 116 |
+
'mask_ete': torch.as_tensor(mask.copy().astype(int)).long().contiguous(),
|
| 117 |
+
'mask_file' : mask_file,
|
| 118 |
+
'img_file' : img_file
|
| 119 |
+
}
|
| 120 |
+
self.cache[idx] = data
|
| 121 |
+
return data
|
| 122 |
+
|
| 123 |
+
def get_3d_iter(self):
|
| 124 |
+
from itertools import groupby
|
| 125 |
+
keyf = lambda idx : self.ids[idx].split("_frame_")[0]
|
| 126 |
+
sorted_ids = sorted(range(len(self.ids)), key=lambda i : self.ids[i])
|
| 127 |
+
for _, items in groupby(sorted_ids, key=keyf):
|
| 128 |
+
images = []
|
| 129 |
+
masks_ete = []
|
| 130 |
+
for idx in items:
|
| 131 |
+
d = self.__getitem__(idx)
|
| 132 |
+
images.append(d['image'])
|
| 133 |
+
masks_ete.append(d['mask_ete'])
|
| 134 |
+
# store third dimension in image channels
|
| 135 |
+
images = torch.stack(images, dim=0)
|
| 136 |
+
masks_ete = torch.stack(masks_ete, dim=0)
|
| 137 |
+
_3d_data = {'image': images, 'mask_ete': masks_ete}
|
| 138 |
+
yield _3d_data
|
| 139 |
+
|
| 140 |
+
|
| 141 |
+
class SegmentationDataset(Dataset):
|
| 142 |
+
def __init__(self, name_dataset: str, images_dir: str, masks_dir: str, scale = (1024, 256)):
|
| 143 |
+
self.images_dir = images_dir
|
| 144 |
+
self.masks_dir = masks_dir
|
| 145 |
+
self.scale = scale
|
| 146 |
+
self.name_dataset = name_dataset
|
| 147 |
+
self.ids = os.listdir(images_dir)
|
| 148 |
+
if len(self.ids) == 0:
|
| 149 |
+
raise RuntimeError(f'No input file found in {self.images_dir}, make sure you put your images there')
|
| 150 |
+
logging.info(f'Creating dataset with {len(self.ids)} examples')
|
| 151 |
+
self.cache = {}
|
| 152 |
+
|
| 153 |
+
def __len__(self):
|
| 154 |
+
return len(self.ids)
|
| 155 |
+
|
| 156 |
+
@classmethod
|
| 157 |
+
def preprocess(self, img, scale, is_mask):
|
| 158 |
+
img = resize(img,
|
| 159 |
+
(scale[0], scale[0]),
|
| 160 |
+
order=0,
|
| 161 |
+
preserve_range=True,
|
| 162 |
+
anti_aliasing=False).astype('uint8')
|
| 163 |
+
img = np.asarray(img)
|
| 164 |
+
if not is_mask:
|
| 165 |
+
img = ((img - img.min()) * (1/(img.max() - img.min()) * 255)).astype('uint8')
|
| 166 |
+
if len(img.shape) != 3:
|
| 167 |
+
img = np.expand_dims(img, axis=2) #(1, 224, 224)
|
| 168 |
+
if is_mask:
|
| 169 |
+
img = resize(img,
|
| 170 |
+
(scale[1], scale[1]),
|
| 171 |
+
order=0,
|
| 172 |
+
preserve_range=True,
|
| 173 |
+
anti_aliasing=False).astype('uint8')
|
| 174 |
+
return img
|
| 175 |
+
|
| 176 |
+
@classmethod
|
| 177 |
+
def load(self, filename, name_dataset, is_mask=False):
|
| 178 |
+
if name_dataset.startswith("las"):
|
| 179 |
+
if is_mask:
|
| 180 |
+
return cv2.imread(filename, cv2.IMREAD_UNCHANGED)
|
| 181 |
+
else:
|
| 182 |
+
img = cv2.imread(filename, 0)
|
| 183 |
+
return img
|
| 184 |
+
else:
|
| 185 |
+
if is_mask:
|
| 186 |
+
return cv2.imread(filename, 0)
|
| 187 |
+
else:
|
| 188 |
+
return cv2.imread(filename)
|
| 189 |
+
|
| 190 |
+
def __getitem__(self, idx):
|
| 191 |
+
if idx in self.cache:
|
| 192 |
+
return self.cache[idx]
|
| 193 |
+
|
| 194 |
+
name = self.ids[idx]
|
| 195 |
+
|
| 196 |
+
if self.name_dataset == "isiconlytrain":
|
| 197 |
+
mask_file = os.path.join(self.masks_dir, name).split(".jpg")[0]
|
| 198 |
+
mask_file = mask_file + "_segmentation.png"
|
| 199 |
+
elif self.name_dataset == "buidnewprocess":
|
| 200 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 201 |
+
elif self.name_dataset == "kvasir":
|
| 202 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 203 |
+
elif self.name_dataset == "drive":
|
| 204 |
+
mask_file = os.path.join(self.masks_dir, name).replace("training", "manual1")
|
| 205 |
+
elif self.name_dataset == "bts":
|
| 206 |
+
mask_file = os.path.join(self.masks_dir, name).replace(self.image_type, "_seg_")
|
| 207 |
+
elif self.name_dataset in ["las_mri", "las_ct"]:
|
| 208 |
+
mask_file = os.path.join(self.masks_dir, name).replace("image", "label")
|
| 209 |
+
else:
|
| 210 |
+
mask_file = os.path.join(self.masks_dir, name)
|
| 211 |
+
|
| 212 |
+
img_file = os.path.join(self.images_dir, name)
|
| 213 |
+
|
| 214 |
+
mask = self.load(mask_file, self.name_dataset, is_mask=True)
|
| 215 |
+
img = self.load(img_file, self.name_dataset, is_mask=False)
|
| 216 |
+
|
| 217 |
+
assert mask is not None, mask_file
|
| 218 |
+
assert img is not None, img_file
|
| 219 |
+
|
| 220 |
+
|
| 221 |
+
|
| 222 |
+
if self.name_dataset in ["kvasir", "buidnewprocess"]:
|
| 223 |
+
mask[mask < 50] = 0
|
| 224 |
+
mask[mask > 200] = 1
|
| 225 |
+
elif self.name_dataset == "isiconlytrain":
|
| 226 |
+
mask[mask > 1] = 1
|
| 227 |
+
elif self.name_dataset.startswith("las"):
|
| 228 |
+
mask[mask == 30] = 1
|
| 229 |
+
mask[mask == 60] = 2 # main predict
|
| 230 |
+
mask[mask == 90] = 3
|
| 231 |
+
mask[mask == 120] = 4
|
| 232 |
+
mask[mask == 150] = 5
|
| 233 |
+
mask[mask == 180] = 6
|
| 234 |
+
mask[mask == 210] = 7
|
| 235 |
+
mask[mask > 7] = 0
|
| 236 |
+
else:
|
| 237 |
+
mask[mask>0] = 1
|
| 238 |
+
|
| 239 |
+
img = self.preprocess(img, self.scale, is_mask=False)
|
| 240 |
+
mask = self.preprocess(mask, self.scale, is_mask=True)
|
| 241 |
+
|
| 242 |
+
data = {
|
| 243 |
+
'image': torch.as_tensor(img.copy()).permute(2, 0, 1).float().contiguous(),
|
| 244 |
+
'mask_ete': torch.as_tensor(mask.copy().astype(int)).long().contiguous(),
|
| 245 |
+
'mask_file' : mask_file,
|
| 246 |
+
'img_file' : img_file
|
| 247 |
+
}
|
| 248 |
+
self.cache[idx] = data
|
| 249 |
+
return data
|
| 250 |
+
|
| 251 |
+
def get_3d_iter(self):
|
| 252 |
+
from itertools import groupby
|
| 253 |
+
keyf = lambda idx : self.ids[idx].split("_frame_")[0]
|
| 254 |
+
sorted_ids = sorted(range(len(self.ids)), key=lambda i : self.ids[i])
|
| 255 |
+
for _, items in groupby(sorted_ids, key=keyf):
|
| 256 |
+
images = []
|
| 257 |
+
masks_ete = []
|
| 258 |
+
for idx in items:
|
| 259 |
+
d = self.__getitem__(idx)
|
| 260 |
+
images.append(d['image'])
|
| 261 |
+
masks_ete.append(d['mask_ete'])
|
| 262 |
+
# store third dimension in image channels
|
| 263 |
+
images = torch.stack(images, dim=0)
|
| 264 |
+
masks_ete = torch.stack(masks_ete, dim=0)
|
| 265 |
+
_3d_data = {'image': images, 'mask_ete': masks_ete}
|
| 266 |
+
yield _3d_data
|
| 267 |
+
|
dataloader/sam_transforms.py
ADDED
|
@@ -0,0 +1,102 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) Meta Platforms, Inc. and affiliates.
|
| 2 |
+
# All rights reserved.
|
| 3 |
+
|
| 4 |
+
# This source code is licensed under the license found in the
|
| 5 |
+
# LICENSE file in the root directory of this source tree.
|
| 6 |
+
|
| 7 |
+
import numpy as np
|
| 8 |
+
import torch
|
| 9 |
+
from torch.nn import functional as F
|
| 10 |
+
from torchvision.transforms.functional import resize, to_pil_image # type: ignore
|
| 11 |
+
|
| 12 |
+
from copy import deepcopy
|
| 13 |
+
from typing import Tuple
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
class ResizeLongestSide:
|
| 17 |
+
"""
|
| 18 |
+
Resizes images to the longest side 'target_length', as well as provides
|
| 19 |
+
methods for resizing coordinates and boxes. Provides methods for
|
| 20 |
+
transforming both numpy array and batched torch tensors.
|
| 21 |
+
"""
|
| 22 |
+
|
| 23 |
+
def __init__(self, target_length: int) -> None:
|
| 24 |
+
self.target_length = target_length
|
| 25 |
+
|
| 26 |
+
def apply_image(self, image: np.ndarray) -> np.ndarray:
|
| 27 |
+
"""
|
| 28 |
+
Expects a numpy array with shape HxWxC in uint8 format.
|
| 29 |
+
"""
|
| 30 |
+
target_size = self.get_preprocess_shape(image.shape[0], image.shape[1], self.target_length)
|
| 31 |
+
return np.array(resize(to_pil_image(image), target_size))
|
| 32 |
+
|
| 33 |
+
def apply_coords(self, coords: np.ndarray, original_size: Tuple[int, ...]) -> np.ndarray:
|
| 34 |
+
"""
|
| 35 |
+
Expects a numpy array of length 2 in the final dimension. Requires the
|
| 36 |
+
original image size in (H, W) format.
|
| 37 |
+
"""
|
| 38 |
+
old_h, old_w = original_size
|
| 39 |
+
new_h, new_w = self.get_preprocess_shape(
|
| 40 |
+
original_size[0], original_size[1], self.target_length
|
| 41 |
+
)
|
| 42 |
+
coords = deepcopy(coords).astype(float)
|
| 43 |
+
coords[..., 0] = coords[..., 0] * (new_w / old_w)
|
| 44 |
+
coords[..., 1] = coords[..., 1] * (new_h / old_h)
|
| 45 |
+
return coords
|
| 46 |
+
|
| 47 |
+
def apply_boxes(self, boxes: np.ndarray, original_size: Tuple[int, ...]) -> np.ndarray:
|
| 48 |
+
"""
|
| 49 |
+
Expects a numpy array shape Bx4. Requires the original image size
|
| 50 |
+
in (H, W) format.
|
| 51 |
+
"""
|
| 52 |
+
boxes = self.apply_coords(boxes.reshape(-1, 2, 2), original_size)
|
| 53 |
+
return boxes.reshape(-1, 4)
|
| 54 |
+
|
| 55 |
+
def apply_image_torch(self, image: torch.Tensor) -> torch.Tensor:
|
| 56 |
+
"""
|
| 57 |
+
Expects batched images with shape BxCxHxW and float format. This
|
| 58 |
+
transformation may not exactly match apply_image. apply_image is
|
| 59 |
+
the transformation expected by the model.
|
| 60 |
+
"""
|
| 61 |
+
# Expects an image in BCHW format. May not exactly match apply_image.
|
| 62 |
+
target_size = self.get_preprocess_shape(image.shape[2], image.shape[3], self.target_length)
|
| 63 |
+
return F.interpolate(
|
| 64 |
+
image, target_size, mode="bilinear", align_corners=False, antialias=True
|
| 65 |
+
)
|
| 66 |
+
|
| 67 |
+
def apply_coords_torch(
|
| 68 |
+
self, coords: torch.Tensor, original_size: Tuple[int, ...]
|
| 69 |
+
) -> torch.Tensor:
|
| 70 |
+
"""
|
| 71 |
+
Expects a torch tensor with length 2 in the last dimension. Requires the
|
| 72 |
+
original image size in (H, W) format.
|
| 73 |
+
"""
|
| 74 |
+
old_h, old_w = original_size
|
| 75 |
+
new_h, new_w = self.get_preprocess_shape(
|
| 76 |
+
original_size[0], original_size[1], self.target_length
|
| 77 |
+
)
|
| 78 |
+
coords = deepcopy(coords).to(torch.float)
|
| 79 |
+
coords[..., 0] = coords[..., 0] * (new_w / old_w)
|
| 80 |
+
coords[..., 1] = coords[..., 1] * (new_h / old_h)
|
| 81 |
+
return coords
|
| 82 |
+
|
| 83 |
+
def apply_boxes_torch(
|
| 84 |
+
self, boxes: torch.Tensor, original_size: Tuple[int, ...]
|
| 85 |
+
) -> torch.Tensor:
|
| 86 |
+
"""
|
| 87 |
+
Expects a torch tensor with shape Bx4. Requires the original image
|
| 88 |
+
size in (H, W) format.
|
| 89 |
+
"""
|
| 90 |
+
boxes = self.apply_coords_torch(boxes.reshape(-1, 2, 2), original_size)
|
| 91 |
+
return boxes.reshape(-1, 4)
|
| 92 |
+
|
| 93 |
+
@staticmethod
|
| 94 |
+
def get_preprocess_shape(oldh: int, oldw: int, long_side_length: int) -> Tuple[int, int]:
|
| 95 |
+
"""
|
| 96 |
+
Compute the output size given input size and target long side length.
|
| 97 |
+
"""
|
| 98 |
+
scale = long_side_length * 1.0 / max(oldh, oldw)
|
| 99 |
+
newh, neww = oldh * scale, oldw * scale
|
| 100 |
+
neww = int(neww + 0.5)
|
| 101 |
+
newh = int(newh + 0.5)
|
| 102 |
+
return (newh, neww)
|
dataloader/yaml_data/brain_endtoend_R50_frozen_1_fcn.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: brain
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 1
|
| 9 |
+
model: '1-fcn'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.3
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 64
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 20
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/BRAIN'
|
| 24 |
+
|
dataloader/yaml_data/brain_endtoend_R50_frozen_fcns.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: brain
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 1
|
| 9 |
+
model: 'fcns'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.3
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 64
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 20
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/BRAIN'
|
| 24 |
+
|
dataloader/yaml_data/brain_endtoend_R50_non_frozen_1_fcn.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: brain
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 0
|
| 9 |
+
model: '1-fcn'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.3
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 64
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 30
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/BRAIN'
|
| 24 |
+
|
dataloader/yaml_data/brain_endtoend_R50_non_frozen_fcns.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: brain
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 0
|
| 9 |
+
model: 'fcns'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.3
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 64
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 30
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/BRAIN'
|
| 24 |
+
|
dataloader/yaml_data/buid_endtoend_R50.yml
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: 1
|
| 3 |
+
is_3D: 0
|
| 4 |
+
is_R50: 1
|
| 5 |
+
is_SAMVIT: 0
|
| 6 |
+
dataset_name: buidnewprocess
|
| 7 |
+
gpu_id: '5'
|
| 8 |
+
original_checkpoint: 'lvm-med-resnet' # change checkpoints here
|
| 9 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/BUID_R50'
|
| 10 |
+
image_shape: 256
|
| 11 |
+
|
| 12 |
+
train:
|
| 13 |
+
train_batch_size: 8
|
| 14 |
+
valid_batch_size: 1
|
| 15 |
+
test_batch_size: 1
|
| 16 |
+
optimizer: 'adam'
|
| 17 |
+
learning_rate: 0.0001
|
| 18 |
+
num_epochs: 50
|
| 19 |
+
beta1: 0.9
|
| 20 |
+
beta2: 0.999
|
| 21 |
+
weight_decay: 0
|
| 22 |
+
scheduler: 0
|
| 23 |
+
|
| 24 |
+
dataloader:
|
| 25 |
+
train_dir_img: './dataset_demo/BUID/train'
|
| 26 |
+
train_dir_mask: './dataset_demo/BUID/train_labels'
|
| 27 |
+
valid_dir_img: './dataset_demo/BUID/valid'
|
| 28 |
+
valid_dir_mask: './dataset_demo/BUID/valid_labels'
|
| 29 |
+
test_dir_img: './dataset_demo/BUID/test'
|
| 30 |
+
test_dir_mask: './dataset_demo/BUID/test_labels'
|
| 31 |
+
have_label: './dataset_demo/BUID/have_label.txt'
|
| 32 |
+
non_label: './dataset_demo/BUID/non_label.txt'
|
dataloader/yaml_data/buid_lvm_med_sam.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: True
|
| 3 |
+
is_3D: False
|
| 4 |
+
dataset_name: buidnewprocess
|
| 5 |
+
random_seed: 100
|
| 6 |
+
gpu_id: 3
|
| 7 |
+
num_workers: 40
|
| 8 |
+
pin_memory: True
|
| 9 |
+
original_checkpoint: './working_dir/sam_vit_b_01ec64.pth' # Original checkpoint from segment-anything project
|
| 10 |
+
best_valid_model_checkpoint: 'lvm_med_sam_model_best_dice_original_buidnewprocess_seed' # I just change the path to save model here
|
| 11 |
+
image_shape: (3, 1024)
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
task: sam
|
| 15 |
+
train_batch_size: 20
|
| 16 |
+
valid_batch_size: 1
|
| 17 |
+
test_batch_size: 1
|
| 18 |
+
optimizer: 'adam'
|
| 19 |
+
learning_rate: 1e-4
|
| 20 |
+
num_epochs: 20
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
train_dir_img: './dataset_demo/BUID/train'
|
| 24 |
+
train_dir_mask: './dataset_demo/BUID/train_labels'
|
| 25 |
+
valid_dir_img: './dataset_demo/BUID/valid'
|
| 26 |
+
valid_dir_mask: './dataset_demo/BUID/valid_labels'
|
| 27 |
+
test_dir_img: './dataset_demo/BUID/test'
|
| 28 |
+
test_dir_mask: './dataset_demo/BUID/test_labels'
|
dataloader/yaml_data/buid_sam.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: True
|
| 3 |
+
is_3D: False
|
| 4 |
+
dataset_name: buidnewprocess
|
| 5 |
+
random_seed: 100
|
| 6 |
+
gpu_id: 3
|
| 7 |
+
num_workers: 40
|
| 8 |
+
pin_memory: True
|
| 9 |
+
original_checkpoint: './working_dir/sam_vit_b_01ec64.pth' # Original checkpoint from segment-anything project
|
| 10 |
+
best_valid_model_checkpoint: 'sam_model_best_dice_original_buidnewprocess_seed'
|
| 11 |
+
image_shape: (3, 1024)
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
task: sam
|
| 15 |
+
train_batch_size: 20
|
| 16 |
+
valid_batch_size: 1
|
| 17 |
+
test_batch_size: 1
|
| 18 |
+
optimizer: 'adam'
|
| 19 |
+
learning_rate: 1e-4
|
| 20 |
+
num_epochs: 20
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
train_dir_img: './dataset_demo/BUID/train'
|
| 24 |
+
train_dir_mask: './dataset_demo/BUID/train_labels'
|
| 25 |
+
valid_dir_img: './dataset_demo/BUID/valid'
|
| 26 |
+
valid_dir_mask: './dataset_demo/BUID/valid_labels'
|
| 27 |
+
test_dir_img: './dataset_demo/BUID/test'
|
| 28 |
+
test_dir_mask: './dataset_demo/BUID/test_labels'
|
dataloader/yaml_data/fgadr_endtoend_R50_frozen_1_fcn.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: fgadr
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 1
|
| 9 |
+
model: '1-fcn'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.2
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 32
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 20
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/FGADR'
|
| 24 |
+
|
dataloader/yaml_data/fgadr_endtoend_R50_frozen_fcns.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: fgadr
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 1
|
| 9 |
+
model: 'fcns'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.2
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 32
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0005
|
| 18 |
+
num_epochs: 20
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/FGADR'
|
| 24 |
+
|
dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_1_fcn.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: fgadr
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 0
|
| 9 |
+
model: '1-fcn'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.2
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 32
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0001
|
| 18 |
+
num_epochs: 30
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/FGADR'
|
| 24 |
+
|
dataloader/yaml_data/fgadr_endtoend_R50_non_frozen_fcns.yml
ADDED
|
@@ -0,0 +1,24 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_R50: 1
|
| 3 |
+
is_SAMVIT: 0
|
| 4 |
+
dataset_name: fgadr
|
| 5 |
+
gpu_id: '3'
|
| 6 |
+
original_checkpoint: 'lvm-med-resnet'
|
| 7 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/'
|
| 8 |
+
frozen_eval: 0
|
| 9 |
+
model: 'fcns'
|
| 10 |
+
test_mode: 'best_valid'
|
| 11 |
+
valid_rate: 0.2
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
train_batch_size: 32
|
| 15 |
+
valid_batch_size: 1
|
| 16 |
+
test_batch_size: 1
|
| 17 |
+
learning_rate: 0.0001
|
| 18 |
+
num_epochs: 30
|
| 19 |
+
linear_eval: 0
|
| 20 |
+
solver: 'adam'
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
data_path: './dataset_demo/FGADR'
|
| 24 |
+
|
dataloader/yaml_data/kvasir_endtoend_R50.yml
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: 1
|
| 3 |
+
is_3D: 0
|
| 4 |
+
is_R50: 1
|
| 5 |
+
is_SAMVIT: 0
|
| 6 |
+
dataset_name: kvasir
|
| 7 |
+
gpu_id: '1'
|
| 8 |
+
original_checkpoint: 'lvm-med-resnet' # Original checkpoint from segment-anything project
|
| 9 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/Kvasir_R50'
|
| 10 |
+
image_shape: 224
|
| 11 |
+
|
| 12 |
+
train:
|
| 13 |
+
train_batch_size: 64
|
| 14 |
+
valid_batch_size: 1
|
| 15 |
+
test_batch_size: 1
|
| 16 |
+
optimizer: 'adam'
|
| 17 |
+
learning_rate: 0.001
|
| 18 |
+
num_epochs: 35
|
| 19 |
+
beta1: 0.9
|
| 20 |
+
beta2: 0.999
|
| 21 |
+
weight_decay: 0
|
| 22 |
+
scheduler: 0
|
| 23 |
+
|
| 24 |
+
dataloader:
|
| 25 |
+
train_dir_img: './dataset_demo/Kvasir/train'
|
| 26 |
+
train_dir_mask: './dataset_demo/Kvasir/train_labels'
|
| 27 |
+
valid_dir_img: './dataset_demo/Kvasir/valid'
|
| 28 |
+
valid_dir_mask: './dataset_demo/Kvasir/valid_labels'
|
| 29 |
+
test_dir_img: './dataset_demo/Kvasir/test'
|
| 30 |
+
test_dir_mask: './dataset_demo/Kvasir/test_labels'
|
| 31 |
+
have_label: './dataset_demo/Kvasir/have_label.txt'
|
| 32 |
+
non_label: './dataset_demo/Kvasir/non_label.txt'
|
dataloader/yaml_data/kvasir_sam.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: True
|
| 3 |
+
is_3D: False
|
| 4 |
+
dataset_name: kvasir
|
| 5 |
+
random_seed: 100
|
| 6 |
+
gpu_id: 3
|
| 7 |
+
num_workers: 40
|
| 8 |
+
pin_memory: True
|
| 9 |
+
original_checkpoint: ./working_dir/sam_vit_b_01ec64.pth' # Original checkpoint from segment-anything project
|
| 10 |
+
best_valid_model_checkpoint: 'sam_model_best_dice_original_kvasir_seed'
|
| 11 |
+
image_shape: (3, 1024)
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
task: sam
|
| 15 |
+
train_batch_size: 20
|
| 16 |
+
valid_batch_size: 1
|
| 17 |
+
test_batch_size: 1
|
| 18 |
+
optimizer: 'adam'
|
| 19 |
+
learning_rate: 3e-4
|
| 20 |
+
num_epochs: 20
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
train_dir_img: './dataset_demo/Kvasir/train'
|
| 24 |
+
train_dir_mask: './dataset_demo/Kvasir/train_labels'
|
| 25 |
+
valid_dir_img: './dataset_demo/Kvasir/valid'
|
| 26 |
+
valid_dir_mask: './dataset_demo/Kvasir/valid_labels'
|
| 27 |
+
test_dir_img: './dataset_demo/Kvasir/test'
|
| 28 |
+
test_dir_mask: './dataset_demo/Kvasir/test_labels'
|
dataloader/yaml_data/mmwhs_ct_endtoend_R50.yml
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: 0
|
| 3 |
+
is_3D: 1
|
| 4 |
+
is_R50: 1
|
| 5 |
+
is_SAMVIT: 0
|
| 6 |
+
dataset_name: las_ct
|
| 7 |
+
gpu_id: '2'
|
| 8 |
+
original_checkpoint: 'lvm-med-resnet' # Original checkpoint from segment-anything project
|
| 9 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/MMWHS_CT_R50'
|
| 10 |
+
image_shape: 224
|
| 11 |
+
|
| 12 |
+
train:
|
| 13 |
+
train_batch_size: 64
|
| 14 |
+
valid_batch_size: 1
|
| 15 |
+
test_batch_size: 1
|
| 16 |
+
optimizer: 'adam'
|
| 17 |
+
learning_rate: 0.001
|
| 18 |
+
num_epochs: 20
|
| 19 |
+
beta1: 0.9
|
| 20 |
+
beta2: 0.999
|
| 21 |
+
weight_decay: 0
|
| 22 |
+
scheduler: 1
|
| 23 |
+
|
| 24 |
+
dataloader:
|
| 25 |
+
train_dir_img: './dataset_demo/MMWHS_CT_Heart/train'
|
| 26 |
+
train_dir_mask: './dataset_demo/MMWHS_CT_Heart/train_labels'
|
| 27 |
+
valid_dir_img: './dataset_demo/MMWHS_CT_Heart/valid'
|
| 28 |
+
valid_dir_mask: './dataset_demo/MMWHS_CT_Heart/valid_labels'
|
| 29 |
+
test_dir_img: './dataset_demo/MMWHS_CT_Heart/test'
|
| 30 |
+
test_dir_mask: './dataset_demo/MMWHS_CT_Heart/test_labels'
|
| 31 |
+
have_label: './dataset_demo/MMWHS_CT_Heart/have_label.txt'
|
| 32 |
+
non_label: './dataset_demo/MMWHS_CT_Heart/non_label.txt'
|
dataloader/yaml_data/mmwhs_ct_sam.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: False
|
| 3 |
+
is_3D: True
|
| 4 |
+
dataset_name: las_ct
|
| 5 |
+
random_seed: 100
|
| 6 |
+
gpu_id: 3
|
| 7 |
+
num_workers: 40
|
| 8 |
+
pin_memory: True
|
| 9 |
+
original_checkpoint: './working_dir/sam_vit_b_01ec64.pth' # Original checkpoint from segment-anything project
|
| 10 |
+
best_valid_model_checkpoint: 'sam_model_best_original_las_ct_seed'
|
| 11 |
+
image_shape: (3, 1024)
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
task: sam
|
| 15 |
+
train_batch_size: 20
|
| 16 |
+
valid_batch_size: 1
|
| 17 |
+
test_batch_size: 1
|
| 18 |
+
optimizer: 'adam'
|
| 19 |
+
learning_rate: 3e-5
|
| 20 |
+
num_epochs: 30
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
train_dir_img: './dataset_demo/MMWHS_CT_Heart/train'
|
| 24 |
+
train_dir_mask: './dataset_demo/MMWHS_CT_Heart/train_labels'
|
| 25 |
+
valid_dir_img: './dataset_demo/MMWHS_CT_Heart/valid'
|
| 26 |
+
valid_dir_mask: './dataset_demo/MMWHS_CT_Heart/valid_labels'
|
| 27 |
+
test_dir_img: './dataset_demo/MMWHS_CT_Heart/test'
|
| 28 |
+
test_dir_mask: './dataset_demo/MMWHS_CT_Heart/test_labels'
|
dataloader/yaml_data/mmwhs_mr_endtoend_R50.yml
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: 0
|
| 3 |
+
is_3D: 1
|
| 4 |
+
is_R50: 1
|
| 5 |
+
is_SAMVIT: 0
|
| 6 |
+
dataset_name: las_mri
|
| 7 |
+
gpu_id: '4'
|
| 8 |
+
original_checkpoint: 'lvm-med-resnet' # Original checkpoint from segment-anything project
|
| 9 |
+
best_valid_model_checkpoint: './working_dir/checkpoint/MMWHS_MR_R50'
|
| 10 |
+
image_shape: 224
|
| 11 |
+
|
| 12 |
+
train:
|
| 13 |
+
train_batch_size: 64
|
| 14 |
+
valid_batch_size: 32
|
| 15 |
+
test_batch_size: 1
|
| 16 |
+
optimizer: 'adam'
|
| 17 |
+
learning_rate: 0.0015
|
| 18 |
+
num_epochs: 30
|
| 19 |
+
beta1: 0.9
|
| 20 |
+
beta2: 0.999
|
| 21 |
+
weight_decay: 0
|
| 22 |
+
scheduler: 1
|
| 23 |
+
|
| 24 |
+
dataloader:
|
| 25 |
+
train_dir_img: './dataset_demo/MMWHS_MR_Heart/train'
|
| 26 |
+
train_dir_mask: './dataset_demo/MMWHS_MR_Heart/train_labels'
|
| 27 |
+
valid_dir_img: './dataset_demo/MMWHS_MR_Heart/valid'
|
| 28 |
+
valid_dir_mask: './dataset_demo/MMWHS_MR_Heart/valid_labels'
|
| 29 |
+
test_dir_img: './dataset_demo/MMWHS_MR_Heart/test'
|
| 30 |
+
test_dir_mask: './dataset_demo/MMWHS_MR_Heart/test_labels'
|
| 31 |
+
have_label: './dataset_demo/MMWHS_MR_Heart/have_label.txt'
|
| 32 |
+
non_label: './dataset_demo/MMWHS_MR_Heart/non_label.txt'
|
dataloader/yaml_data/mmwhs_mri_sam.yml
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
base:
|
| 2 |
+
is_2D: False
|
| 3 |
+
is_3D: True
|
| 4 |
+
dataset_name: las_mri
|
| 5 |
+
random_seed: 100
|
| 6 |
+
gpu_id: 3
|
| 7 |
+
num_workers: 40
|
| 8 |
+
pin_memory: True
|
| 9 |
+
original_checkpoint: './working_dir/sam_vit_b_01ec64.pth' # Original checkpoint from segment-anything project
|
| 10 |
+
best_valid_model_checkpoint: 'sam_model_best_original_las_mri_seed'
|
| 11 |
+
image_shape: (3, 1024)
|
| 12 |
+
|
| 13 |
+
train:
|
| 14 |
+
task: sam
|
| 15 |
+
train_batch_size: 20
|
| 16 |
+
valid_batch_size: 1
|
| 17 |
+
test_batch_size: 1
|
| 18 |
+
optimizer: 'adam'
|
| 19 |
+
learning_rate: 5e-5
|
| 20 |
+
num_epochs: 30
|
| 21 |
+
|
| 22 |
+
dataloader:
|
| 23 |
+
train_dir_img: './dataset_demo/MMWHS_MR_Heart/train'
|
| 24 |
+
train_dir_mask: './dataset_demo/MMWHS_MR_Heart/train_labels'
|
| 25 |
+
valid_dir_img: './dataset_demo/MMWHS_MR_Heart/valid'
|
| 26 |
+
valid_dir_mask: './dataset_demo/MMWHS_MR_Heart/valid_labels'
|
| 27 |
+
test_dir_img: './dataset_demo/MMWHS_MR_Heart/test'
|
| 28 |
+
test_dir_mask: './dataset_demo/MMWHS_MR_Heart/test_labels'
|
dataset_demo/__init__.py
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
datasets_split/BUID_split.py
ADDED
|
@@ -0,0 +1,73 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import torch
|
| 3 |
+
import shutil
|
| 4 |
+
import os
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import cv2
|
| 7 |
+
import json
|
| 8 |
+
from PIL import Image
|
| 9 |
+
import pickle
|
| 10 |
+
from skimage.transform import resize
|
| 11 |
+
from utils.dataset_prepare import split_data, save_fileLabel
|
| 12 |
+
|
| 13 |
+
def BUID_split():
|
| 14 |
+
dataset_name = "BUID"
|
| 15 |
+
file = "./dataset_demo/BUID"
|
| 16 |
+
if os.path.exists(os.path.join(file, "normal")):
|
| 17 |
+
shutil.rmtree(os.path.join(file, "normal"))
|
| 18 |
+
|
| 19 |
+
dir_data = "./dataset_demo/BUID/images"
|
| 20 |
+
dir_label = "./dataset_demo/BUID/labels"
|
| 21 |
+
|
| 22 |
+
if os.path.exists(dir_data):
|
| 23 |
+
shutil.rmtree(dir_data)
|
| 24 |
+
os.mkdir(dir_data)
|
| 25 |
+
|
| 26 |
+
if os.path.exists(dir_label):
|
| 27 |
+
shutil.rmtree(dir_label)
|
| 28 |
+
os.mkdir(dir_label)
|
| 29 |
+
|
| 30 |
+
for i in os.listdir(file):
|
| 31 |
+
if i == "labels" or i == "images" or "label" in i:
|
| 32 |
+
continue
|
| 33 |
+
file_label = os.path.join(file, i)
|
| 34 |
+
for img in os.listdir(file_label):
|
| 35 |
+
img_file = os.path.join(file_label, img)
|
| 36 |
+
if "mask" in img:
|
| 37 |
+
shutil.copy(img_file, os.path.join(dir_label, img))
|
| 38 |
+
else:
|
| 39 |
+
shutil.copy(img_file, os.path.join(dir_data, img))
|
| 40 |
+
|
| 41 |
+
file = os.listdir(dir_label)
|
| 42 |
+
label_uni = -1
|
| 43 |
+
check = False
|
| 44 |
+
b = 0
|
| 45 |
+
for i in os.listdir(dir_data):
|
| 46 |
+
for k in range(10):
|
| 47 |
+
if k == 0:
|
| 48 |
+
mask = "_mask"
|
| 49 |
+
else:
|
| 50 |
+
mask = "_mask_" + str(k)
|
| 51 |
+
a = i.replace(".png", mask+".png")
|
| 52 |
+
|
| 53 |
+
if a in file:
|
| 54 |
+
b = k
|
| 55 |
+
if not check:
|
| 56 |
+
label_uni = cv2.imread(os.path.join(dir_label, a))
|
| 57 |
+
check = True
|
| 58 |
+
else:
|
| 59 |
+
img = cv2.imread(os.path.join(dir_label, a))
|
| 60 |
+
label_uni = label_uni + img
|
| 61 |
+
os.remove(os.path.join(dir_label, a))
|
| 62 |
+
else:
|
| 63 |
+
check = False
|
| 64 |
+
break
|
| 65 |
+
|
| 66 |
+
#print(i)
|
| 67 |
+
cv2.imwrite(os.path.join(dir_label, i), label_uni)
|
| 68 |
+
label_uni = -1
|
| 69 |
+
check = False
|
| 70 |
+
b = 0
|
| 71 |
+
|
| 72 |
+
split_data(dataset_name)
|
| 73 |
+
save_fileLabel(dataset_name)
|
datasets_split/FGADR_split.py
ADDED
|
@@ -0,0 +1,38 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import torch
|
| 3 |
+
import shutil
|
| 4 |
+
import os
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import cv2
|
| 7 |
+
import json
|
| 8 |
+
from PIL import Image
|
| 9 |
+
import pickle
|
| 10 |
+
from skimage.transform import resize
|
| 11 |
+
from utils.dataset_prepare import split_data, save_fileLabel
|
| 12 |
+
|
| 13 |
+
def FGADR_split():
|
| 14 |
+
pkl_path = './files_split/fgadr_pkl_file.pkl' # change your path here
|
| 15 |
+
path = "./dataset_demo/FGADR"
|
| 16 |
+
f = open(pkl_path, 'rb')
|
| 17 |
+
a = pickle.load(f)
|
| 18 |
+
a_key = a.keys()
|
| 19 |
+
B = ["train", "test"]
|
| 20 |
+
C = ["Training", "Testing"]
|
| 21 |
+
for index, i in enumerate(B):
|
| 22 |
+
print(i)
|
| 23 |
+
print(len(a[i]))
|
| 24 |
+
folder_type = os.path.join(path, i)
|
| 25 |
+
if os.path.exists(folder_type.replace(i, C[index])):
|
| 26 |
+
shutil.rmtree(os.path.join(path, C[index]))
|
| 27 |
+
os.mkdir(os.path.join(path, C[index]))
|
| 28 |
+
for j in a[i]:
|
| 29 |
+
folder_class = os.path.join(folder_type, str(j[1]))
|
| 30 |
+
if not os.path.exists(folder_class.replace(i, C[index])):
|
| 31 |
+
os.mkdir(folder_class.replace(i, C[index]))
|
| 32 |
+
file = j[0].replace("/mnt/sda/haal02-data/FGADR-Seg-Set", "./dataset_demo/FGADR")
|
| 33 |
+
img = cv2.imread(file)
|
| 34 |
+
img = resize(img, (512, 512), order=0, preserve_range=True, anti_aliasing=False).astype('uint8')
|
| 35 |
+
#/home/caduser/Foundmed_Experiment/Classification/FGADR/Seg-set/Original_Images/0001_2.png
|
| 36 |
+
name_img = file.split("/")[-1]
|
| 37 |
+
#print(os.path.join(folder_class.replace(i, C[index])))
|
| 38 |
+
cv2.imwrite(os.path.join(folder_class.replace(i, C[index]), name_img), img)
|
datasets_split/Kvasir_split.py
ADDED
|
@@ -0,0 +1,19 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import torch
|
| 3 |
+
import shutil
|
| 4 |
+
import os
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import cv2
|
| 7 |
+
import json
|
| 8 |
+
from PIL import Image
|
| 9 |
+
import pickle
|
| 10 |
+
from skimage.transform import resize
|
| 11 |
+
from utils.dataset_prepare import split_data, save_fileLabel
|
| 12 |
+
|
| 13 |
+
def Kvasir_split():
|
| 14 |
+
dataset_name = "Kvasir"
|
| 15 |
+
file = "./dataset_demo/Kvasir"
|
| 16 |
+
if os.path.exists(os.path.join(file, "masks")):
|
| 17 |
+
os.rename(os.path.join(file, "masks"), os.path.join(file, "labels"))
|
| 18 |
+
split_data(dataset_name)
|
| 19 |
+
save_fileLabel(dataset_name)
|
datasets_split/MMWHS_CT_Heart_split.py
ADDED
|
@@ -0,0 +1,127 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import math
|
| 2 |
+
import os
|
| 3 |
+
import random
|
| 4 |
+
import numpy as np
|
| 5 |
+
from torch.utils.data import Dataset
|
| 6 |
+
import nibabel
|
| 7 |
+
from scipy import ndimage
|
| 8 |
+
import glob
|
| 9 |
+
from skimage.io import imsave
|
| 10 |
+
from utils.dataset_prepare import split_data, save_fileLabel_3D
|
| 11 |
+
|
| 12 |
+
# input image is the volume
|
| 13 |
+
def __itensity_normalize_one_volume__(image):
|
| 14 |
+
# normalization following Med3D
|
| 15 |
+
top_per = np.percentile(image, 99.5)
|
| 16 |
+
bot_per = np.percentile(image, 0.5)
|
| 17 |
+
image[image > top_per] = top_per
|
| 18 |
+
image[image < bot_per] = bot_per
|
| 19 |
+
image = (image - np.mean(image)) / np.std(image)
|
| 20 |
+
image = image / 10.0
|
| 21 |
+
image[image < 0] = 0.0
|
| 22 |
+
image[image > 1] = 1.0
|
| 23 |
+
return image
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
def __training_data_process__(data, label):
|
| 27 |
+
# crop data according net input size
|
| 28 |
+
data = data.get_fdata()
|
| 29 |
+
label = label.get_fdata()
|
| 30 |
+
|
| 31 |
+
# normalization datas
|
| 32 |
+
data = __itensity_normalize_one_volume__(data)
|
| 33 |
+
|
| 34 |
+
# changing label values
|
| 35 |
+
label[label == 205] = 30
|
| 36 |
+
label[label == 420] = 60
|
| 37 |
+
label[label == 500] = 90
|
| 38 |
+
label[label == 550] = 120
|
| 39 |
+
label[label == 600] = 150
|
| 40 |
+
label[label == 820] = 180
|
| 41 |
+
label[label == 850] = 210
|
| 42 |
+
|
| 43 |
+
return data, label
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
def preprocess_vol(img_name, label_name):
|
| 47 |
+
|
| 48 |
+
assert os.path.isfile(img_name)
|
| 49 |
+
assert os.path.isfile(label_name)
|
| 50 |
+
|
| 51 |
+
img = nibabel.load(img_name) # We have transposed the data from WHD format to DHW
|
| 52 |
+
assert img is not None
|
| 53 |
+
mask = nibabel.load(label_name)
|
| 54 |
+
assert mask is not None
|
| 55 |
+
|
| 56 |
+
img_array, mask_array = __training_data_process__(img, mask)
|
| 57 |
+
assert img_array.shape == mask_array.shape, "img shape:{} is not equal to mask shape:{}".format(img_array.shape, mask_array.shape)
|
| 58 |
+
|
| 59 |
+
return (img_array*255).astype('uint8'), mask_array.astype('uint8')
|
| 60 |
+
|
| 61 |
+
#if __name__ == '__main__':
|
| 62 |
+
def MMWHS_CT_Heart_split():
|
| 63 |
+
dataset_name = "MMWHS_CT_Heart"
|
| 64 |
+
### Training set
|
| 65 |
+
data_dir = './dataset_demo/MMWHS_CT_Heart/Raw/train/'
|
| 66 |
+
img_fold_list = os.listdir(data_dir)
|
| 67 |
+
dest_dir = './dataset_demo/MMWHS_CT_Heart/train/' # dir for saving train images
|
| 68 |
+
dest_dir_label = './dataset_demo/MMWHS_CT_Heart/train_labels/'
|
| 69 |
+
if not os.path.exists(dest_dir):
|
| 70 |
+
os.makedirs(dest_dir)
|
| 71 |
+
if not os.path.exists(dest_dir_label):
|
| 72 |
+
os.makedirs(dest_dir_label)
|
| 73 |
+
|
| 74 |
+
for vol_name in img_fold_list:
|
| 75 |
+
if 'label' in vol_name:
|
| 76 |
+
continue
|
| 77 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 78 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 79 |
+
print(img_flair.shape, mask.shape)
|
| 80 |
+
# img_array.shape[2] is the length of depth dimension
|
| 81 |
+
for depth in range(0, img_flair.shape[2]):
|
| 82 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 83 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 84 |
+
|
| 85 |
+
### Validation set
|
| 86 |
+
data_dir = './dataset_demo/MMWHS_CT_Heart/Raw/valid/'
|
| 87 |
+
img_fold_list = os.listdir(data_dir)
|
| 88 |
+
dest_dir = './dataset_demo/MMWHS_CT_Heart/valid/'
|
| 89 |
+
dest_dir_label = './dataset_demo/MMWHS_CT_Heart/valid_labels/'
|
| 90 |
+
if not os.path.exists(dest_dir):
|
| 91 |
+
os.makedirs(dest_dir)
|
| 92 |
+
if not os.path.exists(dest_dir_label):
|
| 93 |
+
os.makedirs(dest_dir_label)
|
| 94 |
+
|
| 95 |
+
for vol_name in img_fold_list:
|
| 96 |
+
if 'label' in vol_name:
|
| 97 |
+
continue
|
| 98 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 99 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 100 |
+
print(img_flair.shape, mask.shape)
|
| 101 |
+
# img_array.shape[2] is the length of depth dimension
|
| 102 |
+
for depth in range(0, img_flair.shape[2]):
|
| 103 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 104 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 105 |
+
|
| 106 |
+
### Testing set
|
| 107 |
+
data_dir = './dataset_demo/MMWHS_CT_Heart/Raw/test/'
|
| 108 |
+
img_fold_list = os.listdir(data_dir)
|
| 109 |
+
dest_dir = './dataset_demo/MMWHS_CT_Heart/test/'
|
| 110 |
+
dest_dir_label = './dataset_demo/MMWHS_CT_Heart/test_labels/'
|
| 111 |
+
if not os.path.exists(dest_dir):
|
| 112 |
+
os.makedirs(dest_dir)
|
| 113 |
+
if not os.path.exists(dest_dir_label):
|
| 114 |
+
os.makedirs(dest_dir_label)
|
| 115 |
+
|
| 116 |
+
for vol_name in img_fold_list:
|
| 117 |
+
if 'label' in vol_name:
|
| 118 |
+
continue
|
| 119 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 120 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 121 |
+
print(img_flair.shape, mask.shape)
|
| 122 |
+
# img_array.shape[2] is the length of depth dimension
|
| 123 |
+
for depth in range(0, img_flair.shape[2]):
|
| 124 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 125 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 126 |
+
|
| 127 |
+
save_fileLabel_3D(dataset_name)
|
datasets_split/MMWHS_MR_Heart_split.py
ADDED
|
@@ -0,0 +1,128 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import math
|
| 2 |
+
import os
|
| 3 |
+
import random
|
| 4 |
+
import numpy as np
|
| 5 |
+
from torch.utils.data import Dataset
|
| 6 |
+
import nibabel
|
| 7 |
+
from scipy import ndimage
|
| 8 |
+
import glob
|
| 9 |
+
from skimage.io import imsave
|
| 10 |
+
from utils.dataset_prepare import split_data, save_fileLabel_3D
|
| 11 |
+
|
| 12 |
+
# input image is the volume
|
| 13 |
+
def __itensity_normalize_one_volume__(image):
|
| 14 |
+
# normalization following Med3D
|
| 15 |
+
top_per = np.percentile(image, 99.5)
|
| 16 |
+
bot_per = np.percentile(image, 0.5)
|
| 17 |
+
image[image > top_per] = top_per
|
| 18 |
+
image[image < bot_per] = bot_per
|
| 19 |
+
image = (image - np.mean(image)) / np.std(image)
|
| 20 |
+
image = image / 10.0
|
| 21 |
+
image[image < 0] = 0.0
|
| 22 |
+
image[image > 1] = 1.0
|
| 23 |
+
return image
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
def __training_data_process__(data, label):
|
| 27 |
+
# crop data according net input size
|
| 28 |
+
data = data.get_fdata()
|
| 29 |
+
label = label.get_fdata()
|
| 30 |
+
|
| 31 |
+
# normalization datas
|
| 32 |
+
data = __itensity_normalize_one_volume__(data)
|
| 33 |
+
|
| 34 |
+
# changing label values
|
| 35 |
+
label[label == 205] = 30
|
| 36 |
+
label[label == 420] = 60
|
| 37 |
+
label[label == 500] = 90
|
| 38 |
+
label[label == 550] = 120
|
| 39 |
+
label[label == 600] = 150
|
| 40 |
+
label[label == 820] = 180
|
| 41 |
+
label[label == 850] = 210
|
| 42 |
+
|
| 43 |
+
return data, label
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
def preprocess_vol(img_name, label_name):
|
| 47 |
+
|
| 48 |
+
assert os.path.isfile(img_name)
|
| 49 |
+
assert os.path.isfile(label_name)
|
| 50 |
+
|
| 51 |
+
img = nibabel.load(img_name) # We have transposed the data from WHD format to DHW
|
| 52 |
+
assert img is not None
|
| 53 |
+
mask = nibabel.load(label_name)
|
| 54 |
+
assert mask is not None
|
| 55 |
+
|
| 56 |
+
img_array, mask_array = __training_data_process__(img, mask)
|
| 57 |
+
assert img_array.shape == mask_array.shape, "img shape:{} is not equal to mask shape:{}".format(img_array.shape, mask_array.shape)
|
| 58 |
+
|
| 59 |
+
return (img_array*255).astype('uint8'), mask_array.astype('uint8')
|
| 60 |
+
|
| 61 |
+
#if __name__ == '__main__':
|
| 62 |
+
def MMWHS_MR_Heart_split():
|
| 63 |
+
dataset_name = "MMWHS_MR_Heart"
|
| 64 |
+
### Training set
|
| 65 |
+
data_dir = './dataset_demo/MMWHS_MR_Heart/Raw/train/'
|
| 66 |
+
img_fold_list = os.listdir(data_dir)
|
| 67 |
+
dest_dir = './dataset_demo/MMWHS_MR_Heart/train/' # dir for saving train images
|
| 68 |
+
dest_dir_label = './dataset_demo/MMWHS_MR_Heart/train_labels/'
|
| 69 |
+
if not os.path.exists(dest_dir):
|
| 70 |
+
os.makedirs(dest_dir)
|
| 71 |
+
if not os.path.exists(dest_dir_label):
|
| 72 |
+
os.makedirs(dest_dir_label)
|
| 73 |
+
|
| 74 |
+
for vol_name in img_fold_list:
|
| 75 |
+
if 'label' in vol_name:
|
| 76 |
+
continue
|
| 77 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 78 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 79 |
+
print(img_flair.shape, mask.shape)
|
| 80 |
+
# img_array.shape[2] is the length of depth dimension
|
| 81 |
+
for depth in range(0, img_flair.shape[2]):
|
| 82 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 83 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 84 |
+
|
| 85 |
+
### Validation set
|
| 86 |
+
data_dir = './dataset_demo/MMWHS_MR_Heart/Raw/valid/'
|
| 87 |
+
img_fold_list = os.listdir(data_dir)
|
| 88 |
+
dest_dir = './dataset_demo/MMWHS_MR_Heart/valid/'
|
| 89 |
+
dest_dir_label = './dataset_demo/MMWHS_MR_Heart/valid_labels/'
|
| 90 |
+
if not os.path.exists(dest_dir):
|
| 91 |
+
os.makedirs(dest_dir)
|
| 92 |
+
if not os.path.exists(dest_dir_label):
|
| 93 |
+
os.makedirs(dest_dir_label)
|
| 94 |
+
|
| 95 |
+
for vol_name in img_fold_list:
|
| 96 |
+
if 'label' in vol_name:
|
| 97 |
+
continue
|
| 98 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 99 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 100 |
+
print(img_flair.shape, mask.shape)
|
| 101 |
+
# img_array.shape[2] is the length of depth dimension
|
| 102 |
+
for depth in range(0, img_flair.shape[2]):
|
| 103 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 104 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 105 |
+
|
| 106 |
+
### Testing set
|
| 107 |
+
data_dir = './dataset_demo/MMWHS_MR_Heart/Raw/test/'
|
| 108 |
+
img_fold_list = os.listdir(data_dir)
|
| 109 |
+
dest_dir = './dataset_demo/MMWHS_MR_Heart/test/'
|
| 110 |
+
dest_dir_label = './dataset_demo/MMWHS_MR_Heart/test_labels/'
|
| 111 |
+
if not os.path.exists(dest_dir):
|
| 112 |
+
os.makedirs(dest_dir)
|
| 113 |
+
if not os.path.exists(dest_dir_label):
|
| 114 |
+
os.makedirs(dest_dir_label)
|
| 115 |
+
|
| 116 |
+
for vol_name in img_fold_list:
|
| 117 |
+
if 'label' in vol_name:
|
| 118 |
+
continue
|
| 119 |
+
mask_name = os.path.join(data_dir, vol_name).replace('image','label')
|
| 120 |
+
img_flair, mask = preprocess_vol(os.path.join(data_dir, vol_name), mask_name)
|
| 121 |
+
print(img_flair.shape, mask.shape)
|
| 122 |
+
# img_array.shape[2] is the length of depth dimension
|
| 123 |
+
for depth in range(0, img_flair.shape[2]):
|
| 124 |
+
imsave(os.path.join(dest_dir, vol_name.split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), img_flair[:, :, depth], check_contrast=False)
|
| 125 |
+
imsave(os.path.join(dest_dir_label, vol_name.replace('image','label').split('.')[0] + '_frame_' + str(depth).zfill(3) + '.png'), mask[:, :, depth], check_contrast=False)
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
save_fileLabel_3D(dataset_name)
|
datasets_split/__init__.py
ADDED
|
File without changes
|
evaluate.py
ADDED
|
@@ -0,0 +1,136 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn.functional as F
|
| 3 |
+
from tqdm import tqdm
|
| 4 |
+
|
| 5 |
+
from utils.endtoend import multiclass_dice_coeff, multiclass_iou
|
| 6 |
+
num_classes = 8
|
| 7 |
+
|
| 8 |
+
def evaluate(net, dataloader, device, eval_class):
|
| 9 |
+
net.eval()
|
| 10 |
+
num_val_batches = len(dataloader)
|
| 11 |
+
dice_score = 0
|
| 12 |
+
iou_score = 0
|
| 13 |
+
# iterate over the validation set
|
| 14 |
+
for batch in tqdm(dataloader, desc='Validation round', unit='batch', leave=False):
|
| 15 |
+
image, mask_true = batch['image'], batch['mask_ete']
|
| 16 |
+
# move images and labels to correct device and type
|
| 17 |
+
image = image.to(device=device, dtype=torch.float32)
|
| 18 |
+
mask_true = mask_true.to(device=device, dtype=torch.long)
|
| 19 |
+
#mask_true[mask_true == 4] = 3
|
| 20 |
+
#mask_true[mask_true > 4] = 0
|
| 21 |
+
mask_true_vector = F.one_hot(mask_true, num_classes).permute(0, 3 , 1, 2).float()
|
| 22 |
+
with torch.no_grad():
|
| 23 |
+
# predict the mask
|
| 24 |
+
mask_pred = net(image)
|
| 25 |
+
mask_pred = mask_pred.argmax(dim=1)
|
| 26 |
+
mask_pred_vector = F.one_hot(mask_pred, num_classes).permute(0, 3 , 1, 2).float()
|
| 27 |
+
# compute the Dice score, ignoring background
|
| 28 |
+
dice_score += multiclass_dice_coeff(mask_pred_vector[:, eval_class, ...], mask_true_vector[:, eval_class, ...],
|
| 29 |
+
reduce_batch_first=False)
|
| 30 |
+
iou_score += multiclass_iou(mask_pred_vector[:,eval_class, ...], mask_true_vector[:, eval_class, ...])
|
| 31 |
+
|
| 32 |
+
net.train()
|
| 33 |
+
return dice_score / num_val_batches, iou_score/ num_val_batches
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
def evaluate_3d_iou(net, dataset, device, eval_class):
|
| 37 |
+
net.eval()
|
| 38 |
+
iou_score = 0
|
| 39 |
+
# iterate over the validation set
|
| 40 |
+
num_items = 0
|
| 41 |
+
for image_3d in tqdm(dataset.get_3d_iter(), desc='3D Evaluation', unit='image(s)', leave=False):
|
| 42 |
+
image, mask_true = image_3d['image'], image_3d['mask_ete']
|
| 43 |
+
num_items += 1
|
| 44 |
+
# move images and labels to correct device and type
|
| 45 |
+
|
| 46 |
+
image = image.to(device=device, dtype=torch.float32)
|
| 47 |
+
mask_true = mask_true.to(device=device, dtype=torch.long)
|
| 48 |
+
mask_true_vector = F.one_hot(mask_true, num_classes).permute(0, 3, 1, 2).float()
|
| 49 |
+
|
| 50 |
+
with torch.no_grad():
|
| 51 |
+
# predict the mask
|
| 52 |
+
mask_pred = net(image)
|
| 53 |
+
mask_pred = mask_pred.argmax(dim=1)
|
| 54 |
+
mask_pred_vector = F.one_hot(mask_pred, num_classes).permute(0, 3, 1, 2).float()
|
| 55 |
+
iou_score += multiclass_iou(mask_pred_vector[:, eval_class, ...], mask_true_vector[:, eval_class, ...], reduce_batch_first=True)
|
| 56 |
+
net.train()
|
| 57 |
+
return iou_score/num_items
|
| 58 |
+
|
| 59 |
+
def evaluate_3d_iou_large(net, dataset, device, eval_class):
|
| 60 |
+
net.eval()
|
| 61 |
+
iou_score = 0
|
| 62 |
+
# iterate over the validation set
|
| 63 |
+
num_items = 0
|
| 64 |
+
for image_3d in tqdm(dataset.get_3d_iter(), desc='3D Evaluation', unit='image(s)', leave=False):
|
| 65 |
+
image, mask_true = image_3d['image'], image_3d['mask']
|
| 66 |
+
num_items += 1
|
| 67 |
+
# move images and labels to correct device and type
|
| 68 |
+
|
| 69 |
+
image = image.to(device=device)
|
| 70 |
+
mask_true = mask_true.to(device=device)
|
| 71 |
+
mask_true_vector = F.one_hot(mask_true, num_classes).permute(0, 3, 1, 2).float()
|
| 72 |
+
|
| 73 |
+
net.to(device=device)
|
| 74 |
+
with torch.no_grad():
|
| 75 |
+
# predict the mask
|
| 76 |
+
mask_pred = net(image)
|
| 77 |
+
mask_pred = mask_pred.argmax(dim=1)
|
| 78 |
+
mask_pred_vector = F.one_hot(mask_pred, num_classes).permute(0, 3, 1, 2).float()
|
| 79 |
+
iou_score += multiclass_iou(mask_pred_vector[:, eval_class, ...], mask_true_vector[:, eval_class, ...], reduce_batch_first=True)
|
| 80 |
+
net.train()
|
| 81 |
+
return iou_score/num_items
|
| 82 |
+
|
| 83 |
+
def evaluate_3d_iou_fast(net, dataset, device, eval_class):
|
| 84 |
+
"""
|
| 85 |
+
This function is similar as evaluate_3d_iou but get a batch size in shape [batch_size, dimension, W, H]
|
| 86 |
+
:param net:
|
| 87 |
+
:param dataset:
|
| 88 |
+
:param device:
|
| 89 |
+
:param eval_class:
|
| 90 |
+
:return:
|
| 91 |
+
"""
|
| 92 |
+
net.eval()
|
| 93 |
+
iou_score = 0
|
| 94 |
+
# iterate over the validation set
|
| 95 |
+
num_items = 0
|
| 96 |
+
for image_3d in tqdm(dataset, desc='3D Evaluation', unit='image(s)', leave=False):
|
| 97 |
+
image, mask_true = image_3d['image'][0], image_3d['mask'][0]
|
| 98 |
+
# print ("Image and mask shapes in 3D evaluation are {}, {}".format(image.shape, mask_true.shape))
|
| 99 |
+
num_items += 1
|
| 100 |
+
# move images and labels to correct device and type
|
| 101 |
+
|
| 102 |
+
image = image.to(device=device, dtype=torch.float32)
|
| 103 |
+
mask_true = mask_true.to(device=device, dtype=torch.long)
|
| 104 |
+
mask_true_vector = F.one_hot(mask_true, num_classes).permute(0, 3, 1, 2).float()
|
| 105 |
+
|
| 106 |
+
with torch.no_grad():
|
| 107 |
+
# predict the mask
|
| 108 |
+
mask_pred = net(image)
|
| 109 |
+
mask_pred = mask_pred.argmax(dim=1)
|
| 110 |
+
mask_pred_vector = F.one_hot(mask_pred, num_classes).permute(0, 3, 1, 2).float()
|
| 111 |
+
iou_score += multiclass_iou(mask_pred_vector[:, eval_class, ...], mask_true_vector[:, eval_class, ...], reduce_batch_first=True)
|
| 112 |
+
net.train()
|
| 113 |
+
return iou_score/num_items
|
| 114 |
+
|
| 115 |
+
def evaluate_3d_dice(net, dataset, device, eval_class):
|
| 116 |
+
net.eval()
|
| 117 |
+
dice_score = 0
|
| 118 |
+
# iterate over the validation set
|
| 119 |
+
num_items = 0
|
| 120 |
+
for image_3d in tqdm(dataset.get_3d_iter(), desc='3D Evaluation', unit='image(s)', leave=False):
|
| 121 |
+
image, mask_true = image_3d['image'], image_3d['mask_ete']
|
| 122 |
+
num_items += 1
|
| 123 |
+
# move images and labels to correct device and type
|
| 124 |
+
|
| 125 |
+
image = image.to(device=device, dtype=torch.float32)
|
| 126 |
+
mask_true = mask_true.to(device=device, dtype=torch.long)
|
| 127 |
+
mask_true_vector = F.one_hot(mask_true, num_classes).permute(0, 3, 1, 2).float()
|
| 128 |
+
|
| 129 |
+
with torch.no_grad():
|
| 130 |
+
# predict the mask
|
| 131 |
+
mask_pred = net(image)
|
| 132 |
+
mask_pred = mask_pred.argmax(dim=1)
|
| 133 |
+
mask_pred_vector = F.one_hot(mask_pred, num_classes).permute(0, 3, 1, 2).float()
|
| 134 |
+
dice_score += multiclass_dice_coeff(mask_pred_vector[:, eval_class, ...], mask_true_vector[:, eval_class, ...], reduce_batch_first=True)
|
| 135 |
+
net.train()
|
| 136 |
+
return dice_score/num_items
|
files_split/BUID.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"train": ["benign (298).png", "benign (420).png", "benign (167).png", "benign (102).png", "benign (256).png", "benign (128).png", "benign (405).png", "benign (273).png", "benign (158).png", "benign (265).png", "malignant (113).png", "benign (41).png", "benign (152).png", "benign (304).png", "benign (151).png", "malignant (191).png", "benign (91).png", "benign (46).png", "benign (105).png", "malignant (170).png", "benign (396).png", "benign (261).png", "benign (412).png", "benign (284).png", "benign (71).png", "malignant (116).png", "benign (222).png", "malignant (3).png", "benign (28).png", "malignant (77).png", "benign (185).png", "benign (1).png", "benign (309).png", "benign (361).png", "benign (154).png", "malignant (86).png", "benign (172).png", "benign (372).png", "benign (225).png", "malignant (42).png", "malignant (201).png", "malignant (27).png", "benign (124).png", "malignant (104).png", "benign (43).png", "benign (98).png", "malignant (19).png", "benign (97).png", "malignant (92).png", "benign (136).png", "malignant (8).png", "malignant (74).png", "malignant (136).png", "benign (419).png", "malignant (97).png", "malignant (68).png", "malignant (51).png", "malignant (203).png", "benign (409).png", "malignant (38).png", "benign (242).png", "benign (354).png", "benign (291).png", "benign (162).png", "benign (392).png", "malignant (141).png", "malignant (40).png", "benign (79).png", "malignant (210).png", "benign (266).png", "benign (64).png", "malignant (165).png", "malignant (21).png", "benign (140).png", "benign (369).png", "benign (88).png", "benign (181).png", "malignant (174).png", "malignant (144).png", "benign (399).png", "malignant (87).png", "benign (13).png", "benign (57).png", "benign (130).png", "benign (293).png", "benign (302).png", "malignant (150).png", "benign (76).png", "malignant (186).png", "malignant (151).png", "benign (259).png", "benign (16).png", "malignant (1).png", "benign (320).png", "malignant (16).png", "malignant (70).png", "benign (104).png", "malignant (71).png", "malignant (13).png", "benign (112).png", "benign (192).png", "benign (183).png", "malignant (47).png", "malignant (125).png", "malignant (34).png", "benign (4).png", "benign (282).png", "malignant (188).png", "benign (50).png", "benign (285).png", "malignant (168).png", "benign (235).png", "benign (286).png", "benign (166).png", "malignant (115).png", "malignant (196).png", "benign (237).png", "benign (281).png", "malignant (156).png", "malignant (57).png", "benign (327).png", "benign (338).png", "benign (290).png", "benign (306).png", "malignant (95).png", "malignant (7).png", "malignant (145).png", "benign (328).png", "malignant (181).png", "benign (375).png", "benign (398).png", "benign (173).png", "benign (278).png", "benign (20).png", "benign (109).png", "malignant (157).png", "benign (288).png", "malignant (62).png", "benign (312).png", "benign (153).png", "benign (171).png", "malignant (140).png", "benign (305).png", "benign (418).png", "benign (268).png", "malignant (162).png", "malignant (41).png", "benign (344).png", "malignant (106).png", "malignant (52).png", "benign (160).png", "benign (395).png", "benign (86).png", "malignant (190).png", "benign (100).png", "benign (114).png", "benign (429).png", "benign (201).png", "malignant (6).png", "benign (23).png", "benign (211).png", "benign (299).png", "malignant (207).png", "benign (143).png", "benign (18).png", "benign (287).png", "malignant (209).png", "malignant (202).png", "benign (127).png", "benign (123).png", "benign (103).png", "malignant (14).png", "benign (422).png", "benign (255).png", "benign (49).png", "malignant (114).png", "benign (332).png", "benign (189).png", "benign (209).png", "malignant (131).png", "benign (414).png", "benign (415).png", "benign (31).png", "benign (246).png", "benign (56).png", "malignant (194).png", "benign (339).png", "benign (121).png", "malignant (46).png", "malignant (130).png", "malignant (81).png", "benign (413).png", "benign (406).png", "benign (208).png", "benign (176).png", "malignant (45).png", "benign (221).png", "benign (303).png", "benign (436).png", "benign (257).png", "benign (250).png", "malignant (103).png", "malignant (142).png", "malignant (88).png", "benign (371).png", "benign (424).png", "benign (417).png", "benign (294).png", "benign (411).png", "benign (384).png", "benign (334).png", "benign (126).png", "malignant (180).png", "malignant (25).png", "benign (81).png", "malignant (169).png", "benign (69).png", "benign (270).png", "malignant (96).png", "malignant (155).png", "benign (315).png", "benign (131).png", "benign (25).png", "benign (199).png", "benign (67).png", "malignant (84).png", "malignant (206).png", "benign (2).png", "benign (39).png", "benign (99).png", "benign (343).png", "benign (358).png", "benign (7).png", "benign (226).png", "benign (212).png", "benign (425).png", "benign (427).png", "benign (279).png", "malignant (161).png", "benign (89).png", "benign (219).png", "malignant (152).png", "benign (317).png", "benign (231).png", "benign (8).png", "malignant (138).png", "benign (17).png", "benign (170).png", "benign (32).png", "benign (297).png", "benign (117).png", "benign (416).png", "malignant (5).png", "benign (397).png", "benign (108).png", "benign (434).png", "benign (355).png", "malignant (184).png", "benign (408).png", "benign (230).png", "benign (323).png", "malignant (65).png", "benign (421).png", "benign (80).png", "benign (83).png", "benign (274).png", "benign (94).png", "benign (179).png", "malignant (54).png", "benign (352).png", "benign (391).png", "benign (65).png", "benign (296).png", "malignant (189).png", "benign (129).png", "benign (85).png", "benign (254).png", "malignant (83).png", "malignant (60).png", "benign (163).png", "malignant (35).png", "benign (161).png", "benign (340).png", "benign (262).png", "benign (356).png", "malignant (39).png", "benign (264).png", "benign (370).png", "benign (349).png", "benign (142).png", "benign (321).png", "benign (435).png", "malignant (199).png", "benign (74).png", "malignant (166).png", "malignant (59).png", "benign (14).png", "benign (203).png", "benign (324).png", "benign (55).png", "benign (186).png", "malignant (171).png", "malignant (99).png", "malignant (111).png", "malignant (107).png", "malignant (153).png", "benign (150).png", "malignant (26).png", "benign (267).png", "malignant (56).png", "malignant (73).png", "malignant (11).png", "malignant (90).png", "benign (47).png", "malignant (37).png", "malignant (101).png", "benign (386).png", "benign (301).png", "benign (336).png", "malignant (124).png", "benign (195).png", "benign (24).png", "malignant (167).png", "malignant (94).png", "malignant (204).png", "benign (59).png", "benign (138).png", "benign (190).png", "benign (144).png", "malignant (160).png", "benign (238).png", "malignant (159).png", "malignant (175).png", "malignant (67).png", "benign (70).png", "malignant (43).png", "benign (44).png", "benign (373).png", "benign (118).png", "benign (248).png", "malignant (102).png", "benign (178).png", "benign (93).png", "benign (45).png", "benign (394).png", "malignant (18).png", "malignant (55).png", "benign (271).png", "benign (229).png", "benign (58).png", "benign (300).png", "benign (423).png", "malignant (28).png", "malignant (183).png", "benign (27).png", "benign (381).png", "benign (132).png", "benign (430).png", "benign (342).png", "malignant (128).png", "malignant (164).png", "malignant (4).png", "benign (310).png", "malignant (64).png", "benign (29).png", "malignant (89).png", "benign (137).png", "benign (390).png", "benign (330).png", "benign (387).png", "malignant (72).png", "malignant (121).png", "benign (364).png", "malignant (20).png", "benign (116).png", "benign (359).png", "malignant (147).png", "benign (251).png", "benign (164).png", "benign (146).png", "benign (35).png", "benign (107).png", "malignant (137).png", "malignant (78).png", "benign (345).png", "benign (62).png", "benign (15).png", "benign (84).png", "benign (252).png", "benign (42).png", "benign (311).png", "benign (34).png", "benign (210).png", "malignant (143).png", "benign (33).png", "benign (68).png", "malignant (200).png", "benign (77).png", "malignant (33).png", "benign (245).png", "benign (96).png", "benign (110).png", "benign (407).png", "benign (234).png", "malignant (100).png", "benign (403).png", "benign (9).png", "malignant (22).png", "benign (223).png", "benign (61).png", "benign (63).png", "benign (155).png", "benign (240).png"], "valid": ["benign (205).png", "benign (115).png", "benign (382).png", "benign (214).png", "malignant (50).png", "malignant (172).png", "benign (433).png", "benign (37).png", "benign (215).png", "malignant (61).png", "benign (182).png", "benign (308).png", "malignant (176).png", "benign (125).png", "malignant (17).png", "benign (159).png", "benign (12).png", "malignant (135).png", "benign (277).png", "malignant (119).png", "benign (187).png", "malignant (192).png", "benign (53).png", "malignant (105).png", "benign (202).png", "benign (147).png", "benign (21).png", "benign (206).png", "benign (329).png", "benign (101).png", "benign (200).png", "benign (404).png", "malignant (126).png", "benign (236).png", "malignant (44).png", "benign (360).png", "benign (135).png", "malignant (98).png", "benign (184).png", "benign (307).png", "benign (60).png", "malignant (146).png", "benign (145).png", "benign (36).png", "benign (243).png", "malignant (80).png", "benign (289).png", "benign (11).png", "malignant (49).png", "benign (95).png", "benign (48).png", "benign (260).png", "malignant (69).png", "malignant (197).png", "malignant (66).png", "malignant (129).png", "benign (52).png", "malignant (29).png", "benign (353).png", "benign (139).png", "benign (314).png", "benign (224).png", "benign (428).png", "benign (175).png", "malignant (10).png", "benign (180).png", "benign (122).png", "benign (220).png", "malignant (117).png", "malignant (53).png", "benign (51).png", "malignant (63).png", "malignant (9).png", "benign (362).png", "benign (87).png", "malignant (108).png", "benign (377).png", "malignant (12).png", "benign (233).png", "malignant (149).png", "benign (383).png", "malignant (177).png", "benign (295).png", "benign (258).png", "malignant (139).png", "malignant (112).png", "benign (432).png", "benign (319).png", "malignant (173).png", "malignant (122).png", "malignant (127).png", "malignant (195).png", "benign (196).png", "benign (111).png", "malignant (123).png", "benign (40).png", "benign (241).png", "benign (325).png", "benign (204).png", "malignant (75).png", "benign (197).png", "benign (272).png", "benign (400).png", "benign (3).png"], "test": ["malignant (109).png", "benign (275).png", "benign (188).png", "benign (90).png", "benign (431).png", "malignant (2).png", "benign (141).png", "benign (263).png", "benign (75).png", "malignant (179).png", "benign (10).png", "malignant (133).png", "benign (244).png", "benign (385).png", "malignant (118).png", "malignant (163).png", "benign (388).png", "benign (376).png", "benign (276).png", "benign (156).png", "benign (26).png", "malignant (198).png", "benign (78).png", "benign (380).png", "benign (326).png", "benign (38).png", "benign (357).png", "benign (227).png", "benign (217).png", "benign (351).png", "malignant (110).png", "benign (393).png", "benign (366).png", "malignant (32).png", "malignant (148).png", "benign (347).png", "benign (368).png", "benign (113).png", "benign (149).png", "benign (216).png", "malignant (79).png", "benign (247).png", "benign (6).png", "malignant (178).png", "benign (280).png", "benign (316).png", "benign (66).png", "benign (378).png", "malignant (15).png", "benign (92).png", "benign (341).png", "benign (249).png", "benign (269).png", "benign (198).png", "malignant (23).png", "benign (333).png", "benign (374).png", "benign (22).png", "benign (318).png", "benign (30).png", "benign (133).png", "benign (213).png", "benign (120).png", "benign (19).png", "benign (218).png", "benign (191).png", "benign (350).png", "malignant (58).png", "benign (207).png", "benign (348).png", "malignant (82).png", "benign (379).png", "malignant (31).png", "benign (157).png", "benign (401).png", "malignant (24).png", "malignant (85).png", "benign (363).png", "malignant (36).png", "benign (193).png", "benign (313).png", "malignant (120).png", "benign (5).png", "benign (335).png", "benign (426).png", "benign (174).png", "benign (410).png", "benign (148).png", "benign (253).png", "benign (346).png", "benign (331).png", "malignant (30).png", "benign (194).png", "benign (134).png", "malignant (91).png", "malignant (187).png", "benign (54).png", "benign (389).png", "benign (402).png", "malignant (205).png", "benign (73).png", "malignant (154).png", "benign (232).png", "benign (169).png", "benign (437).png", "malignant (93).png", "benign (168).png", "benign (365).png", "benign (106).png", "benign (292).png", "malignant (208).png", "benign (367).png", "malignant (48).png", "malignant (158).png", "benign (82).png", "benign (239).png", "malignant (185).png", "benign (119).png", "malignant (182).png", "malignant (132).png", "benign (283).png", "benign (322).png", "malignant (193).png", "malignant (134).png", "benign (177).png", "benign (337).png", "benign (228).png", "benign (165).png", "benign (72).png", "malignant (76).png"], "train_labels": ["benign (298).png", "benign (420).png", "benign (167).png", "benign (102).png", "benign (256).png", "benign (128).png", "benign (405).png", "benign (273).png", "benign (158).png", "benign (265).png", "malignant (113).png", "benign (41).png", "benign (152).png", "benign (304).png", "benign (151).png", "malignant (191).png", "benign (91).png", "benign (46).png", "benign (105).png", "malignant (170).png", "benign (396).png", "benign (261).png", "benign (412).png", "benign (284).png", "benign (71).png", "malignant (116).png", "benign (222).png", "malignant (3).png", "benign (28).png", "malignant (77).png", "benign (185).png", "benign (1).png", "benign (309).png", "benign (361).png", "benign (154).png", "malignant (86).png", "benign (172).png", "benign (372).png", "benign (225).png", "malignant (42).png", "malignant (201).png", "malignant (27).png", "benign (124).png", "malignant (104).png", "benign (43).png", "benign (98).png", "malignant (19).png", "benign (97).png", "malignant (92).png", "benign (136).png", "malignant (8).png", "malignant (74).png", "malignant (136).png", "benign (419).png", "malignant (97).png", "malignant (68).png", "malignant (51).png", "malignant (203).png", "benign (409).png", "malignant (38).png", "benign (242).png", "benign (354).png", "benign (291).png", "benign (162).png", "benign (392).png", "malignant (141).png", "malignant (40).png", "benign (79).png", "malignant (210).png", "benign (266).png", "benign (64).png", "malignant (165).png", "malignant (21).png", "benign (140).png", "benign (369).png", "benign (88).png", "benign (181).png", "malignant (174).png", "malignant (144).png", "benign (399).png", "malignant (87).png", "benign (13).png", "benign (57).png", "benign (130).png", "benign (293).png", "benign (302).png", "malignant (150).png", "benign (76).png", "malignant (186).png", "malignant (151).png", "benign (259).png", "benign (16).png", "malignant (1).png", "benign (320).png", "malignant (16).png", "malignant (70).png", "benign (104).png", "malignant (71).png", "malignant (13).png", "benign (112).png", "benign (192).png", "benign (183).png", "malignant (47).png", "malignant (125).png", "malignant (34).png", "benign (4).png", "benign (282).png", "malignant (188).png", "benign (50).png", "benign (285).png", "malignant (168).png", "benign (235).png", "benign (286).png", "benign (166).png", "malignant (115).png", "malignant (196).png", "benign (237).png", "benign (281).png", "malignant (156).png", "malignant (57).png", "benign (327).png", "benign (338).png", "benign (290).png", "benign (306).png", "malignant (95).png", "malignant (7).png", "malignant (145).png", "benign (328).png", "malignant (181).png", "benign (375).png", "benign (398).png", "benign (173).png", "benign (278).png", "benign (20).png", "benign (109).png", "malignant (157).png", "benign (288).png", "malignant (62).png", "benign (312).png", "benign (153).png", "benign (171).png", "malignant (140).png", "benign (305).png", "benign (418).png", "benign (268).png", "malignant (162).png", "malignant (41).png", "benign (344).png", "malignant (106).png", "malignant (52).png", "benign (160).png", "benign (395).png", "benign (86).png", "malignant (190).png", "benign (100).png", "benign (114).png", "benign (429).png", "benign (201).png", "malignant (6).png", "benign (23).png", "benign (211).png", "benign (299).png", "malignant (207).png", "benign (143).png", "benign (18).png", "benign (287).png", "malignant (209).png", "malignant (202).png", "benign (127).png", "benign (123).png", "benign (103).png", "malignant (14).png", "benign (422).png", "benign (255).png", "benign (49).png", "malignant (114).png", "benign (332).png", "benign (189).png", "benign (209).png", "malignant (131).png", "benign (414).png", "benign (415).png", "benign (31).png", "benign (246).png", "benign (56).png", "malignant (194).png", "benign (339).png", "benign (121).png", "malignant (46).png", "malignant (130).png", "malignant (81).png", "benign (413).png", "benign (406).png", "benign (208).png", "benign (176).png", "malignant (45).png", "benign (221).png", "benign (303).png", "benign (436).png", "benign (257).png", "benign (250).png", "malignant (103).png", "malignant (142).png", "malignant (88).png", "benign (371).png", "benign (424).png", "benign (417).png", "benign (294).png", "benign (411).png", "benign (384).png", "benign (334).png", "benign (126).png", "malignant (180).png", "malignant (25).png", "benign (81).png", "malignant (169).png", "benign (69).png", "benign (270).png", "malignant (96).png", "malignant (155).png", "benign (315).png", "benign (131).png", "benign (25).png", "benign (199).png", "benign (67).png", "malignant (84).png", "malignant (206).png", "benign (2).png", "benign (39).png", "benign (99).png", "benign (343).png", "benign (358).png", "benign (7).png", "benign (226).png", "benign (212).png", "benign (425).png", "benign (427).png", "benign (279).png", "malignant (161).png", "benign (89).png", "benign (219).png", "malignant (152).png", "benign (317).png", "benign (231).png", "benign (8).png", "malignant (138).png", "benign (17).png", "benign (170).png", "benign (32).png", "benign (297).png", "benign (117).png", "benign (416).png", "malignant (5).png", "benign (397).png", "benign (108).png", "benign (434).png", "benign (355).png", "malignant (184).png", "benign (408).png", "benign (230).png", "benign (323).png", "malignant (65).png", "benign (421).png", "benign (80).png", "benign (83).png", "benign (274).png", "benign (94).png", "benign (179).png", "malignant (54).png", "benign (352).png", "benign (391).png", "benign (65).png", "benign (296).png", "malignant (189).png", "benign (129).png", "benign (85).png", "benign (254).png", "malignant (83).png", "malignant (60).png", "benign (163).png", "malignant (35).png", "benign (161).png", "benign (340).png", "benign (262).png", "benign (356).png", "malignant (39).png", "benign (264).png", "benign (370).png", "benign (349).png", "benign (142).png", "benign (321).png", "benign (435).png", "malignant (199).png", "benign (74).png", "malignant (166).png", "malignant (59).png", "benign (14).png", "benign (203).png", "benign (324).png", "benign (55).png", "benign (186).png", "malignant (171).png", "malignant (99).png", "malignant (111).png", "malignant (107).png", "malignant (153).png", "benign (150).png", "malignant (26).png", "benign (267).png", "malignant (56).png", "malignant (73).png", "malignant (11).png", "malignant (90).png", "benign (47).png", "malignant (37).png", "malignant (101).png", "benign (386).png", "benign (301).png", "benign (336).png", "malignant (124).png", "benign (195).png", "benign (24).png", "malignant (167).png", "malignant (94).png", "malignant (204).png", "benign (59).png", "benign (138).png", "benign (190).png", "benign (144).png", "malignant (160).png", "benign (238).png", "malignant (159).png", "malignant (175).png", "malignant (67).png", "benign (70).png", "malignant (43).png", "benign (44).png", "benign (373).png", "benign (118).png", "benign (248).png", "malignant (102).png", "benign (178).png", "benign (93).png", "benign (45).png", "benign (394).png", "malignant (18).png", "malignant (55).png", "benign (271).png", "benign (229).png", "benign (58).png", "benign (300).png", "benign (423).png", "malignant (28).png", "malignant (183).png", "benign (27).png", "benign (381).png", "benign (132).png", "benign (430).png", "benign (342).png", "malignant (128).png", "malignant (164).png", "malignant (4).png", "benign (310).png", "malignant (64).png", "benign (29).png", "malignant (89).png", "benign (137).png", "benign (390).png", "benign (330).png", "benign (387).png", "malignant (72).png", "malignant (121).png", "benign (364).png", "malignant (20).png", "benign (116).png", "benign (359).png", "malignant (147).png", "benign (251).png", "benign (164).png", "benign (146).png", "benign (35).png", "benign (107).png", "malignant (137).png", "malignant (78).png", "benign (345).png", "benign (62).png", "benign (15).png", "benign (84).png", "benign (252).png", "benign (42).png", "benign (311).png", "benign (34).png", "benign (210).png", "malignant (143).png", "benign (33).png", "benign (68).png", "malignant (200).png", "benign (77).png", "malignant (33).png", "benign (245).png", "benign (96).png", "benign (110).png", "benign (407).png", "benign (234).png", "malignant (100).png", "benign (403).png", "benign (9).png", "malignant (22).png", "benign (223).png", "benign (61).png", "benign (63).png", "benign (155).png", "benign (240).png"], "valid_labels": ["benign (205).png", "benign (115).png", "benign (382).png", "benign (214).png", "malignant (50).png", "malignant (172).png", "benign (433).png", "benign (37).png", "benign (215).png", "malignant (61).png", "benign (182).png", "benign (308).png", "malignant (176).png", "benign (125).png", "malignant (17).png", "benign (159).png", "benign (12).png", "malignant (135).png", "benign (277).png", "malignant (119).png", "benign (187).png", "malignant (192).png", "benign (53).png", "malignant (105).png", "benign (202).png", "benign (147).png", "benign (21).png", "benign (206).png", "benign (329).png", "benign (101).png", "benign (200).png", "benign (404).png", "malignant (126).png", "benign (236).png", "malignant (44).png", "benign (360).png", "benign (135).png", "malignant (98).png", "benign (184).png", "benign (307).png", "benign (60).png", "malignant (146).png", "benign (145).png", "benign (36).png", "benign (243).png", "malignant (80).png", "benign (289).png", "benign (11).png", "malignant (49).png", "benign (95).png", "benign (48).png", "benign (260).png", "malignant (69).png", "malignant (197).png", "malignant (66).png", "malignant (129).png", "benign (52).png", "malignant (29).png", "benign (353).png", "benign (139).png", "benign (314).png", "benign (224).png", "benign (428).png", "benign (175).png", "malignant (10).png", "benign (180).png", "benign (122).png", "benign (220).png", "malignant (117).png", "malignant (53).png", "benign (51).png", "malignant (63).png", "malignant (9).png", "benign (362).png", "benign (87).png", "malignant (108).png", "benign (377).png", "malignant (12).png", "benign (233).png", "malignant (149).png", "benign (383).png", "malignant (177).png", "benign (295).png", "benign (258).png", "malignant (139).png", "malignant (112).png", "benign (432).png", "benign (319).png", "malignant (173).png", "malignant (122).png", "malignant (127).png", "malignant (195).png", "benign (196).png", "benign (111).png", "malignant (123).png", "benign (40).png", "benign (241).png", "benign (325).png", "benign (204).png", "malignant (75).png", "benign (197).png", "benign (272).png", "benign (400).png", "benign (3).png"], "test_labels": ["malignant (109).png", "benign (275).png", "benign (188).png", "benign (90).png", "benign (431).png", "malignant (2).png", "benign (141).png", "benign (263).png", "benign (75).png", "malignant (179).png", "benign (10).png", "malignant (133).png", "benign (244).png", "benign (385).png", "malignant (118).png", "malignant (163).png", "benign (388).png", "benign (376).png", "benign (276).png", "benign (156).png", "benign (26).png", "malignant (198).png", "benign (78).png", "benign (380).png", "benign (326).png", "benign (38).png", "benign (357).png", "benign (227).png", "benign (217).png", "benign (351).png", "malignant (110).png", "benign (393).png", "benign (366).png", "malignant (32).png", "malignant (148).png", "benign (347).png", "benign (368).png", "benign (113).png", "benign (149).png", "benign (216).png", "malignant (79).png", "benign (247).png", "benign (6).png", "malignant (178).png", "benign (280).png", "benign (316).png", "benign (66).png", "benign (378).png", "malignant (15).png", "benign (92).png", "benign (341).png", "benign (249).png", "benign (269).png", "benign (198).png", "malignant (23).png", "benign (333).png", "benign (374).png", "benign (22).png", "benign (318).png", "benign (30).png", "benign (133).png", "benign (213).png", "benign (120).png", "benign (19).png", "benign (218).png", "benign (191).png", "benign (350).png", "malignant (58).png", "benign (207).png", "benign (348).png", "malignant (82).png", "benign (379).png", "malignant (31).png", "benign (157).png", "benign (401).png", "malignant (24).png", "malignant (85).png", "benign (363).png", "malignant (36).png", "benign (193).png", "benign (313).png", "malignant (120).png", "benign (5).png", "benign (335).png", "benign (426).png", "benign (174).png", "benign (410).png", "benign (148).png", "benign (253).png", "benign (346).png", "benign (331).png", "malignant (30).png", "benign (194).png", "benign (134).png", "malignant (91).png", "malignant (187).png", "benign (54).png", "benign (389).png", "benign (402).png", "malignant (205).png", "benign (73).png", "malignant (154).png", "benign (232).png", "benign (169).png", "benign (437).png", "malignant (93).png", "benign (168).png", "benign (365).png", "benign (106).png", "benign (292).png", "malignant (208).png", "benign (367).png", "malignant (48).png", "malignant (158).png", "benign (82).png", "benign (239).png", "malignant (185).png", "benign (119).png", "malignant (182).png", "malignant (132).png", "benign (283).png", "benign (322).png", "malignant (193).png", "malignant (134).png", "benign (177).png", "benign (337).png", "benign (228).png", "benign (165).png", "benign (72).png", "malignant (76).png"]}
|
files_split/Kvasir.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"train": ["cju5wphwwlu3m0987hh3ltg88.jpg", "cju8aj01yqeqm0850lhdz3xdw.jpg", "cju2lz8vqktne0993fuym6drw.jpg", "cju2rxm8rpbaf0993o3qr2oph.jpg", "cju3xga12iixg0817dijbvjxw.jpg", "cju5fb86jd1jp0755b1ukbhq5.jpg", "cju2nsmwjlzyl0993jl80chvz.jpg", "cju7fq7mm2pw508176uk5ugtx.jpg", "cju18kevfrojc0835bn90f1in.jpg", "cju2tvrvm53ws0801a0jfjdxg.jpg", "cju2qqn5ys4uo0988ewrt2ip2.jpg", "cju306x7w05nb0835cunv799x.jpg", "cju5c7oijaqmq09878qwgqv8n.jpg", "cju35oyvd3y850988km12hdz1.jpg", "cju2yljr0yzhw0988ecf271ly.jpg", "cju8c9akjsdjj0850s67uzlxq.jpg", "cju5ccpvqash50850kb4bs22k.jpg", "cju7db7lp2f400755tntd1ohf.jpg", "cju6vgdmivcvb08018fra5lnv.jpg", "cju2syxa93yw40799x2iuwabz.jpg", "cju1cvkfwqrec0993wbp1jlzm.jpg", "cju320gyvbch60801v2amdi2g.jpg", "cju7emdni2py40871ivhxjtut.jpg", "cju5f8hxdcxxn08188obby0ea.jpg", "cju2lcyfgkf5809932fn9gucn.jpg", "cju5i5oh2efg60987ez6cpf72.jpg", "cjyzkmjy8evns070165gf9dmq.jpg", "cju31y80qbawn0801twwm2l5s.jpg", "cju5bwhapakm90987c1v4z46a.jpg", "cju1hyolc7aqu0878rrkfn1lr.jpg", "cju0qx73cjw570799j4n5cjze.jpg", "cju7dbppn28nx085097654msi.jpg", "cju2i6acqvo6l0799u20fift8.jpg", "cju2raxlosl630988jdbfy9b0.jpg", "cju30xqmh0ni00835ix3batv1.jpg", "cju1alwgo30z60855fm3y23sm.jpg", "cju7fpfzq2wyf0818xxd1oziv.jpg", "cju8bk8oirjhw0817hgkua2w8.jpg", "cju34i3qvcyog0855qiejxx5w.jpg", "cju2qozsk20cq0855ugrg3cri.jpg", "cju84kplnl1y30755ropua1b0.jpg", "cju40taxlkrho0987smigg0x0.jpg", "cju18849rrsgr0988p90hkygb.jpg", "cju1cyjb5qtie0993njqne9m3.jpg", "cju33o12x2jm50988944mxq0v.jpg", "cju7cq6su27qv075574dir0r3.jpg", "cju0qoxqj9q6s0835b43399p4.jpg", "cju2igw4gvxds0878808qj398.jpg", "cju2zxja9w1eh09933609ho9z.jpg", "cju2iatlki5u309930zmgkv6h.jpg", "cju83rcnzkbsj0755x5anfrcg.jpg", "cju2xs6na81t20878pt6nkfip.jpg", "cju1fmsyf6gxb0801cimx2gle.jpg", "cju2u73dj53oz0878486k8k4b.jpg", "cju7efffp2ivf0817etg3jehl.jpg", "cju8a1jtvpt9m081712iwkca7.jpg", "cju85je7vlht70817c9jcjwi4.jpg", "cju5wcc90lu020850mjrxppv6.jpg", "cju1f320ewfyu0988ndz6blh5.jpg", "cju2zwg05a0oy0801yr73ig7g.jpg", "cju32phw2bv130801yj7bkouq.jpg", "cju32zhbnc1oy0801iyv1ix6p.jpg", "cju887ftknop008177nnjt46y.jpg", "cju32gzs6xo8x0993r8tedbpb.jpg", "cju0rx1idathl0835detmsp84.jpg", "cju2r7h21sj9608354gzks3ae.jpg", "cju5jz5fff8c50871hbe6108f.jpg", "cju35ldepdtlm0801yv79y8vu.jpg", "cju34uhepd3dd0799hs8782ad.jpg", "cju2otvvv0l7z0855x7we8cb0.jpg", "cju7ajnbo1gvm098749rdouk0.jpg", "cju1dnz61vfp40988e78bkjga.jpg", "cju846ec0kj7z08012o10klrb.jpg", "cju5eyfe9cpk90987laa7tsl3.jpg", "cju2zgbj9zmrw0835nnlzxj4c.jpg", "cju7dizi82h2i0755doucgnt3.jpg", "cju7dhpsc2dnn0818025m6857.jpg", "cju1dia8hvc6v098827mgffnm.jpg", "cju6x35ervu2808015c7eoqe4.jpg", "cju2upu4evw7g08358guwozxv.jpg", "cju77196iyshb0850ycbto50a.jpg", "cju87xn2snfmv0987sc3d9xnq.jpg", "cjyzl833ndne80838pzuq6ila.jpg", "cju8bj2ssrmlm0871gc2ug2rs.jpg", "cju7d5m0p23kn09871rk7pu3v.jpg", "cju8432cmkgq90871cxe4iptl.jpg", "cju34xspwzenf0993cyzajv9n.jpg", "cju5eftctcdbj08712gdp989f.jpg", "cju772304yw5t0818vbw8kkjf.jpg", "cju83u9ftk3ni0987qnhlcinv.jpg", "cju2tqfgw4oat0799rn0g5b2z.jpg", "cju2qfie4rvz508357kad9z5o.jpg", "cju2sszfq3uye0878sucelzk2.jpg", "cju1drnhbrb9409935wi7vkhg.jpg", "cju2i03ptvkiu0799xbbd4det.jpg", "cju2oo0wh0bqy0878biujeyhe.jpg", "cju1haab178i70799tk9z8y8x.jpg", "cju5nyu31gv8e0871zpk74a2n.jpg", "cju3v0fl3gwce0755qkjhzmd4.jpg", "cju2r2obh2bjm08553kng0rh7.jpg", "cju2srvy5440s0801y1ba9akr.jpg", "cju8bm24yrrdp081829mbo8ic.jpg", "cju8bh8surexp0987o5pzklk1.jpg", "cju2okvco06xc0799kxe5n1qh.jpg", "cju8ceacrsqkr0755hdz145es.jpg", "ck2bxpfgxu2mk0748gsh7xelu.jpg", "cju8cdeazsm8h0801jxifmzur.jpg", "cju2hw5gjlr5h0988so2qqres.jpg", "cju2suk42469908015ngmq6f2.jpg", "cju2p4ddkmzxj0993p94o62av.jpg", "cju88vx2uoocy075531lc63n3.jpg", "cju77re6fz5bb0817vp9redjg.jpg", "cju5wmvsdlx1j0871npgj8j4b.jpg", "cju787jnjzjuj0871p94nck9g.jpg", "cju5wkonqlrl409877y8zvnub.jpg", "cju1ffnjn6ctm08015perkg37.jpg", "cju2rpa30t07b0835im0erql0.jpg", "cju5ekty5ckzf07550c9u3ckk.jpg", "cju7awzmu1ncs0871hziy65zx.jpg", "cju7el6xv2k520817qxx9wdr5.jpg", "cju1gv7106qd008784gk603mg.jpg", "cju1dfeupuzlw0835gnxip369.jpg", "cju6xmqd9w0250817l5kxfnsk.jpg", "cju42py9mlqyd0818u3d1d7ga.jpg", "cju7eotqi2qea0871y8yc7tqh.jpg", "cju2ricdv2iys0878sv1adh0u.jpg", "cju7f4sc62xqj075597xpmuoy.jpg", "cju2zdhsczmn50988z64qwg2q.jpg", "cju2s9g11pnra0993gn4eh793.jpg", "cju88k75inzyb0850ccv5x3vk.jpg", "cju2saez63gxl08559ucjq3kt.jpg", "cju3yht87j83m08507yk1u1fg.jpg", "cju336l68y7if0993wf092166.jpg", "cjyztzaqtrv430848l8xgcerw.jpg", "cju8b8yair65w09878pyqtr96.jpg", "cju2z6ez69g4u0801qwt088lw.jpg", "cju1c0qb4tzi308355wtsnp0y.jpg", "cju5ht88gedbu0755xrcuddcx.jpg", "cju7d2q1k27nf08715zshsckt.jpg", "cju7epwj82koz098713apjnzo.jpg", "cju2rzpsmtb0f0835jabkbao1.jpg", "cju1d31sp4d4k0878r3fr02ul.jpg", "cju8bop5jrsid08716i24fqda.jpg", "cju2wve9v7esz0878mxsdcy04.jpg", "cju1b0y2e396p08558ois175d.jpg", "cju2top2ruxxy0988p1svx36g.jpg", "cju1erep75us208553i4ofwwe.jpg", "cju6v4szov55u0871qmqz3v8n.jpg", "cju76erapykj30871x5eaxh4q.jpg", "cju7ey10f2rvf0871bwbi9x82.jpg", "cju6v6g6kvdw007552x6mb0po.jpg", "cju85dx63lic408017f0l0400.jpg", "cju2hjrqcvi2j0801bx1i6gxg.jpg", "cju5huurrecm70801y680y13m.jpg", "cju2trbpkv0c00988hxla5dzz.jpg", "cju16ach3m1da0993r1dq3sn2.jpg", "cju6vifjlv55z0987un6y4zdo.jpg", "cju2qs32r1vys07999conmbvx.jpg", "cju32csyfblyh080170aa3x5p.jpg", "cju32fhnhbds40799broyoptc.jpg", "cju8bljw9rqk20801kr54akrl.jpg", "cju7fmvpk2q170987v6i3ola8.jpg", "cju7d6ux323ze0987xos3srkx.jpg", "cju85l4yjlops0801fvmnwptf.jpg", "cju1egh885m1l0855ci1lt37c.jpg", "cju6v5ilsv8hk0850rb5sgh6o.jpg", "cju8bzzy2s66m08016z6mouqt.jpg", "cju1djtprvd7b0988thwwrg09.jpg", "cju7aifyo1p3n07552nxjx51f.jpg", "cju2ulk385h170799rlklxob0.jpg", "cju424hy5lckr085073fva1ok.jpg", "cju5knbbqfipk080128cggukq.jpg", "cju5bf6hxa6m50817rbwettgu.jpg", "cju2tjrog4jy30878pawyazqc.jpg", "cju88rl5eo94l0850kf5wtrm1.jpg", "cju300m3s04fg0988uzupuf7z.jpg", "cju3u1c8tfyqx08503iedc3mx.jpg", "cju43mkj9m8wb0871qiadahub.jpg", "cju18ibp219ub08783i6o98g7.jpg", "cju5fydrud94708507vo6oy21.jpg", "cju8ashhnquqr0801rwduzt7d.jpg", "cju45ty6zn9oz0850qy4qnck1.jpg", "cju2y40d8ulqo0993q0adtgtb.jpg", "cju16whaj0e7n0855q7b6cjkm.jpg", "cju2wxv0hxs2f09884w48v8fi.jpg", "cju1hmff8tkp809931jps6fbr.jpg", "cju857ad0l88m0817qx4cwxnf.jpg", "cju7ekbo32pft0871fv7kzwb9.jpg", "cju8bff9nrfi10850fmfzbf8v.jpg", "cju85plp7lmkw0850rx42jdpf.jpg", "cju1ewnoh5z030855vpex9uzt.jpg", "cju5tenjojp1j0755ms4949h2.jpg", "cju2ntxtdzlvu0799xl3j9pan.jpg", "cju2zm0axztpe0988r8s9twjr.jpg", "cju3ykamdj9u208503pygyuc8.jpg", "cju15czxqp3lv0835jvhgzurz.jpg", "cju88v2f9oi8w0871hx9auh01.jpg", "cju8828oxnool0801qno9luhr.jpg", "cju2sxf3iqbpv09937iksn8ep.jpg", "cju0sr5ghl0nd08789uzf1raf.jpg", "cju43gfosm63n08714rpih8pe.jpg", "cju3ya7goj6at0818v2l5ay7f.jpg", "cju5hjxaae3i40850h5z2laf5.jpg", "cju30u1hbakn808019g15nb8b.jpg", "cju8bbznkrf5g0871jncffynk.jpg", "cju85citjlnfm0755i4rk5tqj.jpg", "cju7dqcwi2dz00850gcmr2ert.jpg", "cju87q6yoneim0871dl4phvkd.jpg", "cju35c4wzdhow0799h6eq4sgs.jpg", "cju7b1ygu1msd0801hywhy0mc.jpg", "cju8c5223s8j80850b4kealt4.jpg", "cju1bhnfitmge0835ynls0l6b.jpg", "cju5x7iskmad90818frchyfwd.jpg", "cju83vvmik9wa08710yeh7cuk.jpg", "cju87nkyrnb970801q84m47yt.jpg", "cju2oi8sq0i2y0801mektzvw8.jpg", "cju40wto5kxwi0755it190f2k.jpg", "cju5jx7jzf7c90871c2i9aiov.jpg", "cju6vucxvvlda0755j7msqnya.jpg", "cju3xtufwiv9c0818djsc4cqd.jpg", "cju84ffdzkrjn08183jh1fxmb.jpg", "cju8auylgqx0z0871u4o4db7o.jpg", "cju2nbdpmlmcj0993s1cht0dz.jpg", "cju6yywx1whbb0871ksgfgf9f.jpg", "cju5ft6mcd5q40987rhjgbrr6.jpg", "cju7dymur2od30755eg8yv2ht.jpg", "cju14g8o4xui30878gkgbrvqj.jpg", "cju83syhdk6gs0801rf1rekdl.jpg", "cju7do8c72dbo0801vxfzxdc4.jpg", "cju7alcgr1lsr0871riqk84z7.jpg", "cju330ofbc2l30801th5g3hw6.jpg", "cju3xzvnzj0hd0755xprz39nj.jpg", "cju2rga4psq9n09881z519xx0.jpg", "cju18gzrq18zw0878wbf4ftw6.jpg", "cju8apjewqrk00801k5d71gky.jpg", "cju43b8daly4408170e5ev06g.jpg", "cju3xiic0ilzp0850lusrb42j.jpg", "cju3v11mrgwwb0755u242ygye.jpg", "cju77b3wyz4160755qis4ljsb.jpg", "cju3uz4o6gr9z0850lhxyxvsj.jpg", "cju5o1vu9gz8a0818eyy92bns.jpg", "cju8ayeq7r1fb0818z1junacy.jpg", "cjyzk8qieoboa0848ogj51wwm.jpg", "cju3128yi0rpu0988o4oo5n8n.jpg", "cju1d50a94qf50855wsowacrc.jpg", "cju6x0yqbvxqt0755dhxislgb.jpg", "cju5buy2bal250818ipl6fqwv.jpg", "cju7ff97z2ow40817u2r83my5.jpg", "cju1g4nsb6ngy0799l4ezm8ab.jpg", "cju34ds2531520988qjpqt6e3.jpg", "cju6wjm81vgsc0987enk9n3pr.jpg", "cju8dic9mtppa0987swn23wbc.jpg", "cju6vta3kvazg0817qbeppjtm.jpg", "cju15wdt3zla10801odjiw7sy.jpg", "cju8aeei7q8k308173n9y4klv.jpg", "cju7aez2x1jtj0871ztezs3oi.jpg", "cju1f5x1164xv08555654c24r.jpg", "cju8chxndsre008015uisl4si.jpg", "cju5wtdu4m0im0871mix0yvc0.jpg", "cju85c2d4ln1b0755zz1z3onx.jpg", "cju7g7ba42z310987bqzbi2bq.jpg", "cju7f5ghb2r5s0801chwkxxh9.jpg", "cju16jgnyzp970878melv7r25.jpg", "cju5v8pgplg6k0755rvi2t63h.jpg", "cju1euuc65wm00799m4sjdnnn.jpg", "cju1871y11d6r0799k6cw4yze.jpg", "cju8bqxxurs6i0850mu7mtef9.jpg", "cju7ecl9i2i060987xawjp4l0.jpg", "cju7d3oc82cho0755dajlwldz.jpg", "cju326h4v1gxw08352px40p7r.jpg", "cju8bafgqrf4x0818twisk3ea.jpg", "cju84aoa3ktwn0755pfl4gfwd.jpg", "cju6xa0qmvzun0818xjukgncj.jpg", "cju85qefyln6v0850szeb9byi.jpg", "cju8c5zcbsdfz0801o5t6jag1.jpg", "cjyzul1qggwwj07216mhiv5sy.jpg", "cju40jl7skiuo0817p0smlgg8.jpg", "cju2hos57llxm08359g92p6jj.jpg", "cju7dubap2g0w0801fgl42mg9.jpg", "cju85nr8elly209872w9n5m0s.jpg", "cju42wamblrqn098798r2yyok.jpg", "cju7f0ec32txj08184asb8w5f.jpg", "cju1cdxvz48hw0801i0fjwcnk.jpg", "cju3xl264ingx0850rcf0rshj.jpg", "cju17bz250pgd0799u1hqkj5u.jpg", "cju783tmkzkqu081803g7q5vk.jpg", "cju41p90plcsx08018cnzpndc.jpg", "cju32jcdabepz0878d0cznmfe.jpg", "cju1gghyjwxt80835vx0wgxw0.jpg", "cju7arvfe1ldu0850erdmphgj.jpg", "cju1hirfi7ekp0855q0vgm9qq.jpg", "cju30525w04r10835ygp257sb.jpg", "cju5vwbr4lhqn0987a1pji0ux.jpg", "cju42qet0lsq90871e50xbnuv.jpg", "cju8aw9n1qyg10801jkjlmors.jpg", "cju7787c5yy3l080159mwqsnj.jpg", "cju15ptjtppz40988odsm9azx.jpg", "cju34ymm8d6700799uop0cw33.jpg", "cju6uy20suzbl0987rzuhz7z9.jpg", "cju310f6val1v0855xo8tc3gu.jpg", "cju7atnm31if40817pqclnjer.jpg", "cju88aq6vo1ij0755c2ey7z7n.jpg", "cju8dn0c3u2v50801k8rvq02f.jpg", "cju8doa16u5gh0818w1ywda3q.jpg", "cju77t0razbvm080106o56289.jpg", "cju84hibuktj80871u519o71q.jpg", "cju7f900s2o0k08175gl1giid.jpg", "cju422cm8lfxn0818ojicxejb.jpg", "cju8567gdlcbq0801dwwyo2jt.jpg", "cju2lyynuymli0855g7fxgbhe.jpg", "cju3x5u2tiihx0818914gzxy1.jpg", "cju5vcmrqla7i0817x4sp4pqw.jpg", "cju0s2a9ekvms080138tjjpxr.jpg", "cju43kj2pm34f0850l28ahpni.jpg", "cju41lojblbs307555jdci937.jpg", "cju88trl3ogi208716qvti51b.jpg", "cju2ro5jqsy680988pi6qsujw.jpg", "cju8c5mxls96t0850wvkvsity.jpg", "cju890guyoiti098753yg6cdu.jpg", "cju5yimthmlv80850zhoc90c2.jpg", "cju1egx9pvz2n0988eoy8jp23.jpg", "cju7d4jk723eu0817bqz2n39m.jpg", "cju2qh5le1ock0878oahaql7d.jpg", "cju8b3ka8r64u0801fh18hk7l.jpg", "cju5yhgznmkzb0801cji2vi8j.jpg", "cju2nqapmzvk20801f9us40dx.jpg", "cju8djdqztu6408506pzhlo18.jpg", "cju418rckl3ur08012psrx1r1.jpg", "cju2hqt33lmra0988fr5ijv8j.jpg", "cju6vvxsev9y30987kespucdg.jpg", "cju84jdl9kv0i0871eog9b3i9.jpg", "cjz14qsk2wci60794un9ozwmw.jpg", "cju7afqon1ip40850ue2308b6.jpg", "cju8a84g0q76m0818hwiggkod.jpg", "cju2rz4k434s70855wwx3ddtx.jpg", "cju1c8ffau5770835g0g343o8.jpg", "cju41s6nbleqy0755e2mslg0b.jpg", "cju31t8xd17bk0835rnb893jk.jpg", "cju8cbsyssiqj0871gr4jedjp.jpg", "cju5klveuff6w0871wbibgh3m.jpg", "cju2sggy13na70855tbeoqgha.jpg", "cju8dm2cau2km0818jsv9eeq2.jpg", "cju7dtb1e2j0t0818deq51ib3.jpg", "cju2xyd9vyi7m098831qcucse.jpg", "cju2r11x7sdgx0988o8ule0wl.jpg", "cju85omszllp30850b6rm9mi3.jpg", "cju2nfnvxzdkd0878399axlco.jpg", "cju5bhv81abur0850ean02atv.jpg", "cju77j66ez52p08019xygi0co.jpg", "cju33yemn2qb20988wfjxximx.jpg", "cju3518w2d838079939fqztbc.jpg", "cju2t62nq45jl0799odpufwx6.jpg", "cju7fen322ou10817ziqkob4k.jpg", "cju5wqonpm0e60801z88ewmy1.jpg", "cju42romflni20817etb9a0fl.jpg", "cju3381d8bz3h07991xtl7ra0.jpg", "cju33qpdvc9g0087825jhf3s9.jpg", "cju8alhigqn2h0801zksudldd.jpg", "cju8418jhkf7d0818ga2v0xq0.jpg", "cju7fcgbe2z3p07550vaflqdb.jpg", "cju428k5fldt108177s6g6f45.jpg", "cju7fnfv02tt90801djnix9m8.jpg", "cju1cj3f0qi5n0993ut8f49rj.jpg", "cju1dq3x1vgx109889c7wyirg.jpg", "cju784jpdzeae0987q5ypq883.jpg", "cju1ats0y372e08011yazcsxm.jpg", "cju7deifq2fzn0755lc8idyh8.jpg", "cju1c6yfz42md08550zgoz3pw.jpg", "cju2zdvjn9h7r08553cp4eed5.jpg", "cju32upim1z7u0988l883nqp6.jpg", "cju0s690hkp960855tjuaqvv0.jpg", "cju7dglf226g50987ohbthl19.jpg", "cju1ejj7dvqfa0835ra184v5m.jpg", "cju83k8fyjsxr0817d6nxs6r4.jpg", "cju35a77vdj4n08556jj2lgmc.jpg", "cju45v0pungu40871acnwtmu5.jpg", "cju1hs0za7jha0855vj0mdrjt.jpg", "cju5woy82m07m08505dmjg7g1.jpg", "cju34repocy5208780gswillm.jpg", "cju5tgbzhjllu08174ca41eus.jpg", "cju5hqz50e7o90850e0prlpa0.jpg", "cju0t4oil7vzk099370nun5h9.jpg", "cju30qbm1ad3x0855znuhpz9u.jpg", "cju1b3zgj3d8e0801kpolea6c.jpg", "cju17r8il13910799dr2wme2e.jpg", "cju2rnkt22xep0801as160g9t.jpg", "cju88gx09o2vk0818610zody3.jpg", "cju0u2g7pmnux0801vkk47ivj.jpg", "cju5nxkujgscq0817l9gss626.jpg", "cju2hfqnmhisa0993gpleeldd.jpg", "cju5uhrdwkmsu0817ervv91l8.jpg", "cju6vrs1ov8cr098788h8gs6j.jpg", "cju353d1eda8c07992afde611.jpg", "cju6w733bveoz0817e600tw72.jpg", "cju2nyc5f02m40801ojqbtiea.jpg", "cju45rj7ln8980850a7821fov.jpg", "cju2xf8e5y2wm08359vcgk09b.jpg", "cju5yeqiwmkgl0801fzv2douc.jpg", "cju35740hzm0g0993zl5ic246.jpg", "cju13fwthn9mq0835gacxgy01.jpg", "cju76l27oyrw907551ri2a7fl.jpg", "cju7cp6dw244p0818gncdol4m.jpg", "cju8c6hnxsdvr0801wn0vrsa6.jpg", "cju43c92lm5cj0755lorsorfg.jpg", "cju45n0oxn5vu08500yfrt9jn.jpg", "cju30ywtc0oar0835bp2en7ec.jpg", "cju5waeduln160817w0agirve.jpg", "cju5clr68b48r0755cmuvponm.jpg", "cju2zpw4q9vzr0801p0lysjdl.jpg", "cju1b75x63ddl0799sdp0i2j3.jpg", "cju87tyddnnad0755bj0wxahe.jpg", "cju5ymyd8mmdc0801ry3by1xr.jpg", "cju8ando2qqdo0818ck7i1be1.jpg", "cju3tsh4lfsok0987w6x3a0v1.jpg", "cju2z2x3nvd3c099350zgty7w.jpg", "cju3uhb79gcgr0871orbrbi3x.jpg", "cju2omjpeqj5a0988pjdlb8l1.jpg", "cju15mhjczc8z0801kit5c6di.jpg", "cju2ycp1u8g2r0799jslnp7cz.jpg", "cju888fr7nveu0818r9uwtiit.jpg", "cju426tomlhll0818fc0i7nvh.jpg", "cju5ukkg6kv7u08011x2b6zl5.jpg", "ck2bxiswtxuw80838qkisqjwz.jpg", "cju5bj926aiec07559rshy4wa.jpg", "cju2zo0fwzv580988qlijd2xa.jpg", "cju1ddr6p4k5z08780uuuzit2.jpg", "cju85a8h8llwm07559wxg4t5w.jpg", "cju2htabevq9108015qjei0x7.jpg", "cju14hjh2ob2o0835ouz3r5aa.jpg", "cju88y1mwoln50871emyfny1g.jpg", "cju3xeexgii1j0817zs68tb4g.jpg", "cju6xlygpw7bs0818n691jsq4.jpg", "cju7aklv31h4309871m29l4e7.jpg", "cju6z2616wqbk07555bvnuyr1.jpg", "cju1c4fcu40hl07992b8gj0c8.jpg", "cju34eqjpcpm508788b3lhp97.jpg", "cju8b5p40r2c60987ofa0mu03.jpg", "cju77q10sz9ug0801449wu1nu.jpg", "cju6uzxk0v83p0801rcwnexdu.jpg", "cju7csvlb22fr0850lvm45n3x.jpg", "cju1cbokpuiw70988j4lq1fpi.jpg", "cju8b2rmgr52s0801p54eyflx.jpg", "cju357rxxdaz30878y2esjpjt.jpg", "cju34m7h536wq0988xz7gx79v.jpg", "cju3yb47cj1xq0817zfotbni4.jpg", "cju2u4pymvc720988wsxrmi84.jpg", "cju41nz76lcxu0755cya2qefx.jpg", "cju3xhpvvimda0987ygrpzni2.jpg", "cju8b4ja9r2s808509d45ma86.jpg", "cju88nroho44508500129f1nh.jpg", "cju1g20bdwq6u0835e16xugcd.jpg", "cju43in5fm22c08175rxziqrk.jpg", "cju8cgi2kspp308011nxdtjp6.jpg", "cju16b6ynq8e40988m8vx0xnj.jpg", "cju2qu37qobl50993aw7ghcfq.jpg", "cju2spdagu1l50835da1f46fr.jpg", "cju3v664kh0px0818y4y7wolf.jpg", "cju30lncba3ny0878jwnous8n.jpg", "cju1amqw6p8pw0993d9gc5crl.jpg", "cju2lejzcy4pc0878c9rlonot.jpg", "cju2qdj95ru8g09886gfi9rsz.jpg", "cju5eq8c8ck690850vix98hv3.jpg", "cju1fyb1d69et0878muzdak9u.jpg", "cju2wtwj87kys0855kx6mddzw.jpg", "cju88itqbny720987hxizbj5y.jpg", "cju3v3ac9gyz30755hfqwyp1i.jpg", "cju5vzjoslpj708186z2fusmz.jpg", "cju2ij9uiic2l09933ljiv6gm.jpg", "cju2uwz9f5yf1085506cfamfx.jpg", "cju2lberzkdzm09938cl40pog.jpg", "cju7cufm7298k0755j09uf3of.jpg", "cju7b9vcs1luz0987ta60j1dy.jpg", "cju40poe4kt9s0755f9cnm3h5.jpg", "cju7ap09p1kz10850ldccjebj.jpg", "cju5ew4h9cqaf0818rrczkmqh.jpg", "cju88fpm4o0tl0871w1i6a4ds.jpg", "cju7bgnvb1sf808717qa799ir.jpg", "cju2m71z2ywwv080131bcrsd3.jpg", "cju41r6v2lcww0871ps8k8pf5.jpg", "cju30ajhw09sx0988qyahx9s8.jpg", "cju87zv8lni0o0850hbbecbq6.jpg", "cju5yjq1pmlgc0801z0t24bly.jpg", "cjyzuio1qgh040763k56deohv.jpg", "cjyzufihqquiw0a46jatrbwln.jpg", "cju2zrojo9kcd0878ld2epejq.jpg", "cju5vxuc5loxw0818u8xgf45p.jpg", "cju2rlqdnoz9k0993cpjae3x0.jpg", "cju8402x1kcy70801t6kz6bdi.jpg", "cju1h89h6xbnx08352k2790o9.jpg", "cju8at3s1qqqx0850hcq8nmnq.jpg", "cju886ryxnsl50801r93jai7q.jpg", "ck2bxqz3evvg20794iiyv5v2m.jpg", "cju35k2fr3vc50988c85qkrwg.jpg", "cju8c1a0ws7o208181c6lbsom.jpg", "cju7dda8w2br20818zhsuz8s7.jpg", "cju1ftaji6isw0855108yqcse.jpg", "cju8c3xs7sauj0801ieyzezr5.jpg", "cju33za6l2qy70988jhrlp2ev.jpg", "cju30ov1oah920801mi8thuyg.jpg", "cju7ez7r22qbc08015xfoz2wb.jpg", "cju5o4pk9h0720755lgp9jq8m.jpg", "cju410dnfl0960755y8lu8d79.jpg", "cju334jzo261t0835yqudnfs1.jpg", "cju2yi9tz8vky0801yqip0xyl.jpg", "cju45qbf3n9sa0987oonbkly9.jpg", "cju6ut4l8va6y0755tyw3vfqq.jpg", "cju6ur9l9v9jq0755paud9uka.jpg", "cju89z6pqpqfx0817mfv8ixjc.jpg", "cju7b10ce1mnm08011c5bwyr4.jpg", "cju175facms5f0993a5tjikvt.jpg", "cju2txjfzv60w098839dcimys.jpg", "cju1csmlc4ht10799b8ymmghg.jpg", "cju3tp94kfstl08181awh6z49.jpg", "cju2p91qir00k08350ddfif0w.jpg", "cju5hyi9yegob0755ho3do8en.jpg", "cju43h43am1dy08176gwfhmnt.jpg", "cju88msmoo3470817m441j4sg.jpg", "cju13hp5rnbjx0835bf0jowgx.jpg", "cju83ipu3jwpx0801z5pvguf8.jpg", "cju33w4sdcivk0855x879zht7.jpg", "cju2tzypl4wss0799ow05oxb9.jpg", "cju6wll7wvo3y08502pagos8m.jpg", "cju7bc95p1mdm0817yqj5jc6j.jpg", "cju30j1rgadut0801vuyrsnt8.jpg", "cju35mdz73x890835eynq1h9v.jpg", "cju43lcnum9y10755bjs7z87f.jpg", "cju7etr3y2p4t0801cdzjj8ab.jpg", "cju774fmayxif0818u2g79usw.jpg", "cju1cu1u2474n0878tt7v4tdr.jpg", "cju323ypb1fbb0988gx5rzudb.jpg", "cju33x0f22peh0988g0ln7w5v.jpg", "cju2y5zas8m7f0801d34g5owq.jpg", "cju5ddda9bkkt0850enzwatb1.jpg", "cju5ufn3skquf0818dhapnhba.jpg", "cju84gpefknwm098714oq8q61.jpg", "cju2trtjf4qjd0878a2zle9v9.jpg", "cju7fazv92ywx0755xov2erga.jpg", "cju2rn0hasxri0835nfy3buay.jpg", "cju3x4blzieu30850x10uuvbm.jpg", "cju3y9difj6th0801kd1rqm3w.jpg", "cju2osuru0ki00855txo0n3uu.jpg", "cjyzlw7f9faqr070129au64sq.jpg", "cju3u39fog1bo0871lxjrabks.jpg", "cju2yo1j1v0qz09934o0e683p.jpg", "cju33231uy4gi0993qc7b1jch.jpg", "cju2xlcqxy9c60988vjacdznb.jpg", "cju77vvcwzcm50850lzoykuva.jpg", "cju8dpa89u6l80818dj6lldh9.jpg", "cju3xwpgviwlx0871rwm15q7v.jpg", "cju1h5w4wxajx0835mc954kxy.jpg", "cju303j5r062k098835zxfds5.jpg", "cju35fxqyzt5p0993vusm54qz.jpg", "cju31w6goazci0799n014ly1q.jpg", "cju3u815rg4ek0850vvhtcvcm.jpg", "cju8bpctzrqkr0850zeldv9kt.jpg", "cju5f26ebcuai0818xlwh6116.jpg", "cju5g163vd6mt0817uccuga6u.jpg", "cju2zjcvj9qma0801dk71hhi0.jpg", "cju2mfjndoz700988b9lc3zeq.jpg", "cju5uxjnol2r509871qv2yeia.jpg", "cju7dwe282dc309876rco45ts.jpg", "cju5wuhm1lwm40987vugqn3vv.jpg", "cju77u1sjz77b0817ft44r3fk.jpg", "cju2yv4imv6cz099314jveiib.jpg", "cju16d65tzw9d0799ouslsw25.jpg", "cju8b6rp0r5st0850184f79xt.jpg", "cju3umoh1geet0817cmpef5am.jpg", "cju5y7buemcw80987p0r30g9f.jpg", "cju32srle1xfq083575i3fl75.jpg", "cju0roawvklrq0799vmjorwfv.jpg", "cju7b5afm1nfw0801xqm8bf8q.jpg", "cju1fb9236a110801yvg0fwju.jpg", "cju2z45kuzf6d0988nz2c819m.jpg", "cju45lbgznahl08180xz1h7u6.jpg", "cju2tpfa5uyx408359datxqqj.jpg", "cju3v72v5h1qz0818fggilwtq.jpg", "cju2yb31a8e8u0878wdashg7o.jpg", "cju6x97w4vwua0850x0997r0a.jpg", "cju42u5bjlvi10801dc13sskp.jpg", "cju77bvg0yv4r0987yh60xmjo.jpg", "cju5wrapcm2290818jsh26ppb.jpg", "cju17v6ih0u7808783zcbg1jy.jpg", "cju8d2q30tfhs0801n7lx77xl.jpg", "cju2nd7l7z98o0799gfjvyfmw.jpg", "cju5uzmaol56l0817flxh4w9p.jpg", "cju17otoe119u0799nqcbl8n1.jpg", "cju2s16zp317h0799gr67jqc2.jpg", "cju0u82z3cuma0835wlxrnrjv.jpg", "cju2zy1e49pqk0878t6ncqn12.jpg", "cju1f15k3w4ct0835cmde6ypo.jpg", "cju8bgdmqrksy0801tozdmraa.jpg", "cju3521y5d5mq0878t3ezsu4p.jpg", "cju40sdwukv3k0755y99ug1k8.jpg", "cju2pjb9v0ywn0878j5g5n69j.jpg", "cju2i3hzclw3o0988rrgh911i.jpg", "cju6yxyt0wh080871sqpepu47.jpg", "cju5x28nzm7t907558ocq4bt7.jpg", "cju1hp9i2xu8e0988u2dazk7m.jpg", "cju160wshltz10993i1gmqxbe.jpg", "cju34aozyyy830993bn16u32n.jpg", "cju7dxffn2eam0817qxosfwch.jpg", "cju8dqkrqu83i0818ev74qpxq.jpg", "cju8bysfgrzkl081786jwac09.jpg", "cju5gucasds9d0801019axylx.jpg", "cju5vi4nxlc530817uoqm2m7a.jpg", "cju5wj0faly5008187n6530af.jpg", "cju30mm25a53s0799qa5wiqe8.jpg", "cju5ca9hcatkc0801jzwe7tfx.jpg", "cju7bd1qu1mx409877xjxibox.jpg", "cju2yg5ht8i4p087800js8hp4.jpg", "cju5y4hgqmk0i08180rjhbwvp.jpg", "cju1euant5l960878iqj5vvto.jpg", "cju1dg44i4z3w0801nyz4p6zf.jpg", "cju2uy8ox62jo0801g88hh42z.jpg", "cju2t16vuucaq0835xcpsivn2.jpg", "cju5u4pywk81x0817vn9pe14z.jpg", "cju8ca4geseia0850i2ru11hw.jpg", "cju83wwn1k55e0850kw6i2d81.jpg", "cju45ofd9ne1j0801ri8dup7t.jpg", "cju2uzabhs6er0993x3aaf87p.jpg", "cju8a56vxpy780850r45yu4wk.jpg", "cju45pm27n80u08174kyow1gj.jpg", "cju3u4lxmg59o0755rz42b9en.jpg", "cju1gkndf6yi10801o1qnje19.jpg", "cju30gxjq0djk0988jytm49rs.jpg", "cju45jpvfn6c809873pv1i34s.jpg", "cju7adqyj1jcx08712r1ro5gx.jpg", "cju7dmlgf2ebw0871ieqas5fh.jpg", "cju8d4jgatgpj0871q2ophhkm.jpg", "cju2wx0gh7fpz0878wwyd9ep8.jpg", "cju2x7vw87mu30878hye2ca0m.jpg", "cju1hhj6mxfp90835n3wofrap.jpg", "cju30ftgja7170855xl9bkdm0.jpg", "cju43eigtm6ev0801mv0m96t1.jpg", "cju1fjsb4sipq09931lvd8e41.jpg", "cju77g99iyxc00817zqi2ppor.jpg", "cju34zivp3fq80988opxbaqyn.jpg", "cju2t9tdwuk700835kv0ljmtl.jpg", "cju5wi6bqlxy90755bu227nvb.jpg", "cju2rmd2rsw9g09888hh1efu0.jpg"], "valid": ["cju5vgawslbe30987ndeepc1b.jpg", "ck2bxlujamu330725szlc2jdu.jpg", "cju2hx006vidl0799igm81vmh.jpg", "cju7aqkue1i2k09879uzcpt8r.jpg", "cju5uget8krjy0818kvywd0zu.jpg", "cju8dk7eztzup08182yxko5zh.jpg", "cju1c3218411b08014g9f6gig.jpg", "cju7f6cqy2ur20818t1saazbm.jpg", "cju6wi3akvn8r0801px8eligc.jpg", "cju5enq1tcn1i0755hnkon787.jpg", "cju2hewssldzx0835ep795xu0.jpg", "ck2bxskgxxzfv08386xkqtqdy.jpg", "cju2np2k9zi3v079992ypxqkn.jpg", "cju5bycdkalkb09875f7bfrvx.jpg", "cju2hlm19vjjf0801o69qnber.jpg", "cju7amjna1ly40871ugiokehb.jpg", "cju34c1xfyz920993itxkkfad.jpg", "cju88oh0po9gq0801nge4tgr1.jpg", "cju5i39mreass0817au8p22zy.jpg", "cju5f0dezct4q08183ydw11dx.jpg", "cju5cky5xb0ay0801oxet697t.jpg", "cju7ehljc2or70871261br8ai.jpg", "cju183od81ff608017ekzif89.jpg", "cju7evxt12m730987rxivne3x.jpg", "cju8cj10qsrau0871o2dr6ai1.jpg", "cju2y26c588bo07993ksd8eoz.jpg", "cju2zkpdl9h7t0799ix60teqg.jpg", "cju85bf1algsq0871y9gtlq97.jpg", "cju40w3hbkwpn08015rbs3wko.jpg", "cju17hw9hr9c5098800fu4u8e.jpg", "cju6wuojavt740818b5qcv3iw.jpg", "cju2qvuj1s9ok0835tp2k4ozh.jpg", "cju33jon3ygbj0993pu22a4k6.jpg", "cju7dsrtb2f8i085064kwugfk.jpg", "cju77k828z46w0871r0avuoo9.jpg", "cju1aqqv02qwz0878a5cyhr67.jpg", "cju412uwlkva50850d1ps1ww7.jpg", "cju2nguelpmlj0835rojdn097.jpg", "cju35eg0tdmjt085525sb4bua.jpg", "cju7dn24o296i09871qfxb8s2.jpg", "cju2ma647l0nj0993ot4deq2q.jpg", "cju1expq45zst0855rjqwwj4m.jpg", "cju88t4fvokxf07558ymyh281.jpg", "cju83nwu1jxte0987h1krpfmv.jpg", "cju1fj6axwfp30835ukhuzhw9.jpg", "cju8b0jr0r2oi0801jiquetd5.jpg", "cju7bmi1v1pnj0987pa52jjok.jpg", "ck2da7fwcjfis07218r1rvm95.jpg", "cju2zblxw9848087853csbrx1.jpg", "cju5udcufki0s09874ll1dbr5.jpg", "cju2rqo702wpx0855fn7d5cxh.jpg", "cju88l66no10s0850rsda7ej1.jpg", "cju83yddek68q0850d2x7zfkm.jpg", "cju6z1bzbwfq50817b2alatvr.jpg", "cju5hi52odyf90817prvcwg45.jpg", "cju324q101fhe08350wae9cif.jpg", "cju5cetivauok0987ok3e5bre.jpg", "cju5u6wf0kh1t0755bg1ssixv.jpg", "cju8clorgsuwn08714toqb7v6.jpg", "cju5b9oyda4yr0850g9viziyv.jpg", "cju2qtee81yd708787bsjr75d.jpg", "cju3tvffffx5f0818t5ov22al.jpg", "cju1d96gsv62d09881b3wecw2.jpg", "cju5yclrymlgj0818k426ud6z.jpg", "cju2i8br1vqtd08784u6vmcjk.jpg", "cju7dvl5m2n4t0755hlnnjjet.jpg", "cju5hl8nee8a40755fm8qjj0o.jpg", "cju7d8m3b2e210755l8fj1yph.jpg", "cju1f79yhsb5w0993txub59ol.jpg", "cju7frtqu2xa20818wq8r9fzf.jpg", "cju35atpxdjot0855q46aqrd0.jpg", "cju3y0pjrj1c30755nxekxccj.jpg", "cju2ti1du4idn0878giuozonw.jpg", "cju2yyhsp933j0855hp32e012.jpg", "cju2r6mt2om21099352pny5gw.jpg", "cju3ttznuftyf09875t11850w.jpg", "cju42xpi8lw4w0871ve317a1p.jpg", "cju5u8gz4kj5b07552e2wpkwp.jpg", "cju7ezs7g2mxm098787atbran.jpg", "cju17g6ykn1cs0993dww6qdi8.jpg", "cju8arof2qpf20850ifr1bnqj.jpg", "cju3xuj20ivgp0818mij8bjrd.jpg", "cju34sh43d8zm08019xbwhc0o.jpg", "cju2zp89k9q1g0855k1x0f1xa.jpg", "cju7agj961l2r0818z29iq8yn.jpg", "cju3ua8u0g9rg0801uayhdxhu.jpg", "cju2hdr06v2bq0799mbm3bks1.jpg", "cju2qz06823a40878ojcz9ccx.jpg", "cju8azmhcr66e0755t61atz72.jpg", "cju87ox0kncom0801b98hqnd2.jpg", "cju1fuoa4wmc50835qfd11sp9.jpg", "cju88cddensj00987788yotmg.jpg", "cju7azuu31mia0801pf9ib5ed.jpg", "cju5k3j3uf6de0817hszzfr7n.jpg", "cju5x00l6m5j608503k78ptee.jpg", "cju430pm2lz0y0755jkhcc3d1.jpg", "cju0vtox5ain6099360pu62rp.jpg", "cju7dp3dw2k4n0755zhe003ad.jpg", "cju885ikhnmkn09878s2lqtuh.jpg", "cju2zr3c3vwb00993jn06bbaz.jpg", "cju8bw697rwg308177tg8huas.jpg", "cju1efbr0rqxz09931z0lf4vf.jpg", "cju7d7aut2a2p0818z4uxc6cd.jpg", "cju2sevf53lkx08558h5bpaig.jpg", "cju15l5ubz9yh0855b3ivdpse.jpg", "cju2nnqrqzp580855z8mhzgd6.jpg", "cju2mh8t6p07008350e01tx2a.jpg", "cju30nyxe0gfb0835p256yoju.jpg", "cju85rkbnlo1c08503uxcpax1.jpg", "cju3280wv1ir009882jze27tc.jpg", "cju7b2l561oas0871decgslaf.jpg", "cju2m56cryvqd0801gtn2yp8t.jpg", "cju5boicjagt20871b1fotkh4.jpg", "ck2bxknhjvs1x0794iogrq49k.jpg", "cju7crgxa28550755wbsgqkel.jpg", "cju1brhsj3rls0855a1vgdlen.jpg", "cju8b542nr81x0871uxnkm9ih.jpg", "cju87li0zn3yb0817kbwgjiz8.jpg", "cju1f8w0t65en0799m9oacq0q.jpg", "cju8a3nhbpwnb0850d37fo2na.jpg", "cju1bm8063nmh07996rsjjemq.jpg", "cju2z1nxlzaj40835wj81s1iy.jpg", "cju83qd0yjyht0817ktkfl268.jpg", "cju414lf2l1lt0801rl3hjllj.jpg", "cju2yw4s7z7p20988lmf2gdgd.jpg", "cju5fs6j6d8350801vglraq4u.jpg", "cju7er4kc2opa0801anuxc0eb.jpg", "cju5bdwa3aatx0818b79i18zf.jpg", "cjyzurzvohqnr0794es1itzek.jpg", "ck2bxw18mmz1k0725litqq2mc.jpg", "cju41z76wlgbz0801qdetlvby.jpg", "cju84ih17kp5l09876bkooocl.jpg", "cju5vbo6jldrt0871jf6f1700.jpg", "cju7es23b2vcp0755gpbm9s7v.jpg", "cju6ywm40wdbo0987pbftsvtg.jpg", "cju32l161bi1v07990vm376in.jpg", "cju5fi0yxd3ei0801v7u0yudn.jpg", "cju884985nlmx0817vzpax3y4.jpg", "cju32qr9tbvsj08013pkpjenq.jpg", "cju7flevb2wii08188otgs9p2.jpg", "cju16fpvhzypl0799p9phnlx6.jpg", "cju1dhfok4mhe0878jlgrag0h.jpg", "cju5xq3tdm9fn0987pbedxdg5.jpg", "cju7bb3ss1uoo0755pmhyco7t.jpg", "cju6us80mv1b50871ebyq2wxa.jpg", "cju2p0eveqtdc0835gpi3p93i.jpg", "cju7apr0c1qqm0755s7msqot4.jpg", "cju34o6dbd2lo0855aqlcy1hs.jpg", "cju2xd75m82720801q4s4ik3n.jpg", "cju7ebe962hr409872ovibahw.jpg", "cju1fr4etsmrr09933u4t4aql.jpg", "cju0ue769mxii08019zqgdbxn.jpg", "cju7cl8zm1xcu0817ado0jpas.jpg", "cju5ktjwofed70817eg58ef7u.jpg", "cju843yjskhq30818qre4rwm2.jpg", "cju8cwy02t9eq08185qn12c02.jpg", "cju892fesoq2g0801n0e0jyia.jpg", "cju3ul8dogf1z09872y2ecowp.jpg", "cju85fc11ljr40818edpb0inh.jpg", "cju8c2rqzs5t80850d0zky5dy.jpg"], "test": ["cju8abobpqbir08189u01huru.jpg", "cju8axq24r4an0755yhv9d4ly.jpg", "cju5cu8qkb84x08186jwo8yin.jpg", "cju30k2z40ds308353kdew70n.jpg", "cju6vqarjv7yo0987q4b1btk1.jpg", "cju1819curo000988pd5xcqme.jpg", "cju7d9seq29zd0871nzl2uu5m.jpg", "cju31ugmfb3dz0855xtqshki6.jpg", "cju858eswlepn0871pzvdrhj1.jpg", "cju5fw37edaae0801vkwvocn7.jpg", "cju83kxitjv340987z09m0ezy.jpg", "cjyzkpsbjdsjq07211dfi4sru.jpg", "cju2z9vlp9j0w0801oag91sy9.jpg", "cju0tl3uz8blh0993wxvn7ly3.jpg", "cju33eqwbcch208012jikwdky.jpg", "cju8bssulrrcy0987h1vq5060.jpg", "cju2qxxko229x08786gvxxhur.jpg", "cju6wn57mvooj0850rp78hhy7.jpg", "cju7ejm2l2ncl0801wq6y84nw.jpg", "cju7b3f5h1sm40755i572jden.jpg", "cju2pmhtr17a00855cvpelzb0.jpg", "cju2u2b9o4zvp08788qb9nqxj.jpg", "cju85ia1slh220987y7c20sm2.jpg", "cju439oazm2fu0871ma0vvrft.jpg", "cju5hwonqedw10801vsd3w6kk.jpg", "cju1gi7jlwyld0835cdf6g6qz.jpg", "cjyzjzssvd8pq0838f4nolj5l.jpg", "cju17x0j4nfc10993y31pvlgs.jpg", "cju1fm3id6gl50801r3fok20c.jpg", "cju30bmab08bi0835mvlr6e0r.jpg", "cju41kd7yl4nm0850gil5qqwh.jpg", "cju2s2527pfyr0993l3h1149a.jpg", "cju2urqpwvxw70835rvndvtsi.jpg", "cju7et17a2vjk0755e743npl1.jpg", "cju7fob3x301u0755x985pmmq.jpg", "cju17z0qongpa0993de4boim4.jpg", "cju2phaksnahz0993yxogjcpv.jpg", "cju8b1v3br45u087189kku66u.jpg", "cju8aqq8uqmoq0987hphto9gg.jpg", "cju33mirdc8mj0799k33wzoes.jpg", "cju2y0z6g87p10878fpk5d3rq.jpg", "cju6vvb8svhed0801jjcquh5e.jpg", "cju2t3ibkuecr0835o7si16zv.jpg", "cju8914beokbf0850isxpocrk.jpg", "cju8chdlqsu620755azjty1tj.jpg", "cju43o6n7m9nk087191ijwqq9.jpg", "cju7bf1lp1shi081835vs84lc.jpg", "cju2ysg748ru80878sp6j0gm0.jpg", "cju88z8bson4h0871nnd7fdxo.jpg", "cju8brv16rx7f0818uf5n89pv.jpg", "cju7da88w2eod0755wejzynvt.jpg", "cju7eueum2oqn0850rodmx8zo.jpg", "cju773hsyyosz0817pk1e7sjq.jpg", "cju358pwtdby20878cg7nm0np.jpg", "cju2oq5570avm079959o20op1.jpg", "cju3y54kwj3nr0801biidlb4e.jpg", "cju5bmhdcafs909878qfzrqzi.jpg", "cju83h9ysjwe808716nt35oah.jpg", "cju5chrxxawka0871qcj171yz.jpg", "cju8czvnztbf40871b4m7t78w.jpg", "cju3xvoo2iqlc0817eku2r3wl.jpg", "cju7f9umg2olj0987fj5y285w.jpg", "cju87vqa0ndwg0850onjdz7ol.jpg", "cju2ouil2mssu0993hvxsed6d.jpg", "cju2zi4l09f5807991s8do2b2.jpg", "cju5k503sfa5f0871lx0rpu5y.jpg", "cju87z6o6nh73085045bzsx6o.jpg", "cju5xopi0md7q0871r1sjc1av.jpg", "cju40r6jrksyk0871wg98zgho.jpg", "cju5xneamme2p0801qdf7fdwv.jpg", "cju3xjqtpikx50817tppy6g84.jpg", "cju8bn7m2rmm70817hgxpb1uq.jpg", "cju42nm68lpyo0818xvvqmupq.jpg", "cju88q6h6obpd0871ckmiabbo.jpg", "cju3wstckialg0871xs0vevsj.jpg", "cju2y8s56ymqr083541ggdsml.jpg", "cju5ygh1zmmdi0755uod5e17i.jpg", "cju7bfx651qr80801cs7epotb.jpg", "cju7d7tly27h408016fyp5nr7.jpg", "cju77idwfz36d0871tzfzz51i.jpg", "cju7ddtz729960801uazp1knc.jpg", "cju30ia8da2bq0799klnehml2.jpg", "cju3y79ofj3va0871uqfb1mzo.jpg", "cju7cue9b232j0801qdzk1ykj.jpg", "cju5vutu7ll8w0871dfp92n9p.jpg", "cju2xa3i4y0160988i679zsqd.jpg", "cju6x4t13vyw60755gtcf9ndu.jpg", "cju7fd6yt2p740987wkr8exo1.jpg", "cju2xbk0080y80801eghyddi2.jpg", "cju32a52lb9rc0799xi40qs00.jpg", "cju42tauqlo5p08171l3cuo4b.jpg", "cju7ae7bq1f820987toc8si1d.jpg", "cju5y84q3mdv50817eyp82xf3.jpg", "cju893jmdompz0817xn3g1w4h.jpg", "cju2uokeg5jm20799xwgsyz89.jpg", "cju42dwedlmk60871jbgu4ehi.jpg", "cju35i2e63uxr0835h7zgkg9k.jpg", "cju6z9a9kwsl007552s49rx6i.jpg", "cju15jr8jz8sb0855ukmkswkz.jpg", "cjyzu9th0qt4r0a46pyl4zik0.jpg", "cju89y9h0puti0818i5yw29e6.jpg", "cju7bduyq1rjf08719giru9ho.jpg", "cju2xwm1s84l50799i60mq0pu.jpg", "cju7dlk532dsh0871zvr6qz0r.jpg", "cju85mpuglq8k0818d2it6hzb.jpg", "cju5xkwzxmf0z0818gk4xabdm.jpg", "cju8cattbsivm0818p446wgel.jpg", "cju2pag1f0s4r0878h52uq83s.jpg", "cju5x15djm7ae0755h8czf6nt.jpg", "cju5c5xc7algd0817pb1ej5yo.jpg", "cju32pzh9bpw10855q4vaxfhe.jpg", "cju6v3bb2v7xo085090blubyw.jpg", "cju83mki1jv5w0817kubxm31r.jpg", "cju5bbtwsa8cl0987wgfsqpao.jpg", "cju14pxbaoksp0835qzorx6g6.jpg", "cju785htizjzo08017tvlhtg4.jpg", "cju42m60jlpcm08186kqppzqv.jpg", "cju847pxykriq0755268ktrk2.jpg", "cju3x9lttikfb0818a0g104zn.jpg", "cju1cnnziug1l0835yh4ropyg.jpg", "cju76lsehyia10987u54vn8rb.jpg", "cju7fbndk2sl608015ravktum.jpg", "cju5cjh3xattc0817j2vbulzi.jpg", "cju3uwz6ogsp10801h2r3bj5l.jpg", "cju34ouumcznz07996gg1xq7v.jpg", "cju3x2s11ibzi0817kk284k0j.jpg", "cju34fojcctcf0799ebolbvkn.jpg", "cju3v56bwgy8v0871w14pz8fx.jpg", "cju87r56lnkyp0755hz30leew.jpg", "cju2r91dg2k090801bh0xzbxk.jpg", "cju7ev2b12owa08500bpfpwyw.jpg", "cju76o55nymqd0871h31sph9w.jpg", "ck2395w2mb4vu07480otsu6tw.jpg", "cju2pkwt3r8b90988v2ywq1px.jpg", "cju33belnbyhm0878yxl42233.jpg", "cju7ahtkb1jr90801jck4kbds.jpg", "cju8adb60qbiu080188mxpf8d.jpg", "cju5wrrs0m2af0818vmnajbtw.jpg", "cju2wzu8wxtgu09880ku9x1pg.jpg", "cju3y21quj0ir0818kgjagr15.jpg", "cju87mrypnb1e0818scv1mxxg.jpg", "cju0sxqiclckk08551ycbwhno.jpg", "cju5h57xedz5h0755mjpc8694.jpg", "cju1cfhyg48bb0799cl5pr2jh.jpg", "cju8c82iosagu0817l74s4m5g.jpg", "cju30df2j09dd08351ayx2t6w.jpg", "cju2rkjfwoxys0993x768l1j2.jpg", "cjyzu3reghjya0794w7pwoi50.jpg", "cju7druhp2gp308715i6km7be.jpg", "cju5w7xn0lrkq0801f9k0htgx.jpg", "cju40u30gkuzc0871rq7t666d.jpg", "cju6v1m1xv07w09870ah3njy1.jpg", "cju849c23kgnk0817cgv2hw1e.jpg", "cju5k7r0yf98c09878csbxb4d.jpg", "cju8a2itsq4dv0755ntlovpxe.jpg", "cju6z7e4bwgdd0987ogkzq9kt.jpg", "cju7avvi51iox0817ym55y6tt.jpg", "cju0qkwl35piu0993l0dewei2.jpg", "cju85hguellg50818kwu3s8d1.jpg", "cju302fqq9spc0878rrygyzzz.jpg", "cju8amfdtqi4x09871tygrgqe.jpg", "cju6z600qwh4z081700qimgl9.jpg", "cju7ea4om2l910801bohqjccy.jpg", "cju45t5ddnbio0987qtqzx762.jpg", "cju84dsvaklpx098750hp83x4.jpg", "cju1cqc7n4gpy0855jt246k68.jpg", "cju5thdbrjp1108715xdfx356.jpg", "cju87kbcen2av0987usezo8kn.jpg", "cju8b7aqtr4a00987coba14b7.jpg", "cju5xjn5mm78b09871spyqhhr.jpg", "cju3x7xsaijq80818f0psavav.jpg", "cju5fu081d8gc0818l3yylujk.jpg", "cju171py4qiha0835u8sl59ds.jpg", "cju2qn2fzs1vy0988l243cvzy.jpg", "cju7d1tvt25bu08019dvw3uff.jpg", "cju2ueb6j5ado0878vf5md13o.jpg", "cju13cgqmnhwn0988yrainhcp.jpg", "cju3tx1qyg0c907552fglumhc.jpg", "cju31rb7vb6110801p9rhacuw.jpg", "cju77afzlz3kp07550x5nafzs.jpg", "cju2hugv9vget0799hhk7ksvg.jpg", "cju88evxanv9r08176zkeovec.jpg", "cju6xifswvwbo0987nibtdr50.jpg", "cju43jcqim2cp08172dvjvyui.jpg", "cju5es375cnzy0801nkq35ffs.jpg", "cju7dff9529h208503w60lbil.jpg", "cju2hgsptlfam0835o3b59h1o.jpg", "cju8buos5rz9b08715lfr0f4f.jpg", "cju7ctvqn25dy08186g442m1r.jpg", "cju7dz5yy2i7z0801ausi7rna.jpg", "cju7b4mtw1n9n080186209f3d.jpg", "cju42g865lorv07552ytz6xxa.jpg", "cju5von04litr08718j8po40a.jpg", "cju7eea9b2m0z0801ynqv1fqu.jpg", "cju8bi8q7rlmn0871abc5ch8k.jpg", "cju5kre09fhka0850h7b1898j.jpg", "cju40m0rjkpw80871z6n6yg1u.jpg", "cju6wt9jvvn500871hjn3t3g0.jpg", "cju2xjz2ju8pe0993ysv9wg17.jpg", "cju2t2ivz43i10878zeg8r1br.jpg"], "train_labels": ["cju5wphwwlu3m0987hh3ltg88.jpg", "cju8aj01yqeqm0850lhdz3xdw.jpg", "cju2lz8vqktne0993fuym6drw.jpg", "cju2rxm8rpbaf0993o3qr2oph.jpg", "cju3xga12iixg0817dijbvjxw.jpg", "cju5fb86jd1jp0755b1ukbhq5.jpg", "cju2nsmwjlzyl0993jl80chvz.jpg", "cju7fq7mm2pw508176uk5ugtx.jpg", "cju18kevfrojc0835bn90f1in.jpg", "cju2tvrvm53ws0801a0jfjdxg.jpg", "cju2qqn5ys4uo0988ewrt2ip2.jpg", "cju306x7w05nb0835cunv799x.jpg", "cju5c7oijaqmq09878qwgqv8n.jpg", "cju35oyvd3y850988km12hdz1.jpg", "cju2yljr0yzhw0988ecf271ly.jpg", "cju8c9akjsdjj0850s67uzlxq.jpg", "cju5ccpvqash50850kb4bs22k.jpg", "cju7db7lp2f400755tntd1ohf.jpg", "cju6vgdmivcvb08018fra5lnv.jpg", "cju2syxa93yw40799x2iuwabz.jpg", "cju1cvkfwqrec0993wbp1jlzm.jpg", "cju320gyvbch60801v2amdi2g.jpg", "cju7emdni2py40871ivhxjtut.jpg", "cju5f8hxdcxxn08188obby0ea.jpg", "cju2lcyfgkf5809932fn9gucn.jpg", "cju5i5oh2efg60987ez6cpf72.jpg", "cjyzkmjy8evns070165gf9dmq.jpg", "cju31y80qbawn0801twwm2l5s.jpg", "cju5bwhapakm90987c1v4z46a.jpg", "cju1hyolc7aqu0878rrkfn1lr.jpg", "cju0qx73cjw570799j4n5cjze.jpg", "cju7dbppn28nx085097654msi.jpg", "cju2i6acqvo6l0799u20fift8.jpg", "cju2raxlosl630988jdbfy9b0.jpg", "cju30xqmh0ni00835ix3batv1.jpg", "cju1alwgo30z60855fm3y23sm.jpg", "cju7fpfzq2wyf0818xxd1oziv.jpg", "cju8bk8oirjhw0817hgkua2w8.jpg", "cju34i3qvcyog0855qiejxx5w.jpg", "cju2qozsk20cq0855ugrg3cri.jpg", "cju84kplnl1y30755ropua1b0.jpg", "cju40taxlkrho0987smigg0x0.jpg", "cju18849rrsgr0988p90hkygb.jpg", "cju1cyjb5qtie0993njqne9m3.jpg", "cju33o12x2jm50988944mxq0v.jpg", "cju7cq6su27qv075574dir0r3.jpg", "cju0qoxqj9q6s0835b43399p4.jpg", "cju2igw4gvxds0878808qj398.jpg", "cju2zxja9w1eh09933609ho9z.jpg", "cju2iatlki5u309930zmgkv6h.jpg", "cju83rcnzkbsj0755x5anfrcg.jpg", "cju2xs6na81t20878pt6nkfip.jpg", "cju1fmsyf6gxb0801cimx2gle.jpg", "cju2u73dj53oz0878486k8k4b.jpg", "cju7efffp2ivf0817etg3jehl.jpg", "cju8a1jtvpt9m081712iwkca7.jpg", "cju85je7vlht70817c9jcjwi4.jpg", "cju5wcc90lu020850mjrxppv6.jpg", "cju1f320ewfyu0988ndz6blh5.jpg", "cju2zwg05a0oy0801yr73ig7g.jpg", "cju32phw2bv130801yj7bkouq.jpg", "cju32zhbnc1oy0801iyv1ix6p.jpg", "cju887ftknop008177nnjt46y.jpg", "cju32gzs6xo8x0993r8tedbpb.jpg", "cju0rx1idathl0835detmsp84.jpg", "cju2r7h21sj9608354gzks3ae.jpg", "cju5jz5fff8c50871hbe6108f.jpg", "cju35ldepdtlm0801yv79y8vu.jpg", "cju34uhepd3dd0799hs8782ad.jpg", "cju2otvvv0l7z0855x7we8cb0.jpg", "cju7ajnbo1gvm098749rdouk0.jpg", "cju1dnz61vfp40988e78bkjga.jpg", "cju846ec0kj7z08012o10klrb.jpg", "cju5eyfe9cpk90987laa7tsl3.jpg", "cju2zgbj9zmrw0835nnlzxj4c.jpg", "cju7dizi82h2i0755doucgnt3.jpg", "cju7dhpsc2dnn0818025m6857.jpg", "cju1dia8hvc6v098827mgffnm.jpg", "cju6x35ervu2808015c7eoqe4.jpg", "cju2upu4evw7g08358guwozxv.jpg", "cju77196iyshb0850ycbto50a.jpg", "cju87xn2snfmv0987sc3d9xnq.jpg", "cjyzl833ndne80838pzuq6ila.jpg", "cju8bj2ssrmlm0871gc2ug2rs.jpg", "cju7d5m0p23kn09871rk7pu3v.jpg", "cju8432cmkgq90871cxe4iptl.jpg", "cju34xspwzenf0993cyzajv9n.jpg", "cju5eftctcdbj08712gdp989f.jpg", "cju772304yw5t0818vbw8kkjf.jpg", "cju83u9ftk3ni0987qnhlcinv.jpg", "cju2tqfgw4oat0799rn0g5b2z.jpg", "cju2qfie4rvz508357kad9z5o.jpg", "cju2sszfq3uye0878sucelzk2.jpg", "cju1drnhbrb9409935wi7vkhg.jpg", "cju2i03ptvkiu0799xbbd4det.jpg", "cju2oo0wh0bqy0878biujeyhe.jpg", "cju1haab178i70799tk9z8y8x.jpg", "cju5nyu31gv8e0871zpk74a2n.jpg", "cju3v0fl3gwce0755qkjhzmd4.jpg", "cju2r2obh2bjm08553kng0rh7.jpg", "cju2srvy5440s0801y1ba9akr.jpg", "cju8bm24yrrdp081829mbo8ic.jpg", "cju8bh8surexp0987o5pzklk1.jpg", "cju2okvco06xc0799kxe5n1qh.jpg", "cju8ceacrsqkr0755hdz145es.jpg", "ck2bxpfgxu2mk0748gsh7xelu.jpg", "cju8cdeazsm8h0801jxifmzur.jpg", "cju2hw5gjlr5h0988so2qqres.jpg", "cju2suk42469908015ngmq6f2.jpg", "cju2p4ddkmzxj0993p94o62av.jpg", "cju88vx2uoocy075531lc63n3.jpg", "cju77re6fz5bb0817vp9redjg.jpg", "cju5wmvsdlx1j0871npgj8j4b.jpg", "cju787jnjzjuj0871p94nck9g.jpg", "cju5wkonqlrl409877y8zvnub.jpg", "cju1ffnjn6ctm08015perkg37.jpg", "cju2rpa30t07b0835im0erql0.jpg", "cju5ekty5ckzf07550c9u3ckk.jpg", "cju7awzmu1ncs0871hziy65zx.jpg", "cju7el6xv2k520817qxx9wdr5.jpg", "cju1gv7106qd008784gk603mg.jpg", "cju1dfeupuzlw0835gnxip369.jpg", "cju6xmqd9w0250817l5kxfnsk.jpg", "cju42py9mlqyd0818u3d1d7ga.jpg", "cju7eotqi2qea0871y8yc7tqh.jpg", "cju2ricdv2iys0878sv1adh0u.jpg", "cju7f4sc62xqj075597xpmuoy.jpg", "cju2zdhsczmn50988z64qwg2q.jpg", "cju2s9g11pnra0993gn4eh793.jpg", "cju88k75inzyb0850ccv5x3vk.jpg", "cju2saez63gxl08559ucjq3kt.jpg", "cju3yht87j83m08507yk1u1fg.jpg", "cju336l68y7if0993wf092166.jpg", "cjyztzaqtrv430848l8xgcerw.jpg", "cju8b8yair65w09878pyqtr96.jpg", "cju2z6ez69g4u0801qwt088lw.jpg", "cju1c0qb4tzi308355wtsnp0y.jpg", "cju5ht88gedbu0755xrcuddcx.jpg", "cju7d2q1k27nf08715zshsckt.jpg", "cju7epwj82koz098713apjnzo.jpg", "cju2rzpsmtb0f0835jabkbao1.jpg", "cju1d31sp4d4k0878r3fr02ul.jpg", "cju8bop5jrsid08716i24fqda.jpg", "cju2wve9v7esz0878mxsdcy04.jpg", "cju1b0y2e396p08558ois175d.jpg", "cju2top2ruxxy0988p1svx36g.jpg", "cju1erep75us208553i4ofwwe.jpg", "cju6v4szov55u0871qmqz3v8n.jpg", "cju76erapykj30871x5eaxh4q.jpg", "cju7ey10f2rvf0871bwbi9x82.jpg", "cju6v6g6kvdw007552x6mb0po.jpg", "cju85dx63lic408017f0l0400.jpg", "cju2hjrqcvi2j0801bx1i6gxg.jpg", "cju5huurrecm70801y680y13m.jpg", "cju2trbpkv0c00988hxla5dzz.jpg", "cju16ach3m1da0993r1dq3sn2.jpg", "cju6vifjlv55z0987un6y4zdo.jpg", "cju2qs32r1vys07999conmbvx.jpg", "cju32csyfblyh080170aa3x5p.jpg", "cju32fhnhbds40799broyoptc.jpg", "cju8bljw9rqk20801kr54akrl.jpg", "cju7fmvpk2q170987v6i3ola8.jpg", "cju7d6ux323ze0987xos3srkx.jpg", "cju85l4yjlops0801fvmnwptf.jpg", "cju1egh885m1l0855ci1lt37c.jpg", "cju6v5ilsv8hk0850rb5sgh6o.jpg", "cju8bzzy2s66m08016z6mouqt.jpg", "cju1djtprvd7b0988thwwrg09.jpg", "cju7aifyo1p3n07552nxjx51f.jpg", "cju2ulk385h170799rlklxob0.jpg", "cju424hy5lckr085073fva1ok.jpg", "cju5knbbqfipk080128cggukq.jpg", "cju5bf6hxa6m50817rbwettgu.jpg", "cju2tjrog4jy30878pawyazqc.jpg", "cju88rl5eo94l0850kf5wtrm1.jpg", "cju300m3s04fg0988uzupuf7z.jpg", "cju3u1c8tfyqx08503iedc3mx.jpg", "cju43mkj9m8wb0871qiadahub.jpg", "cju18ibp219ub08783i6o98g7.jpg", "cju5fydrud94708507vo6oy21.jpg", "cju8ashhnquqr0801rwduzt7d.jpg", "cju45ty6zn9oz0850qy4qnck1.jpg", "cju2y40d8ulqo0993q0adtgtb.jpg", "cju16whaj0e7n0855q7b6cjkm.jpg", "cju2wxv0hxs2f09884w48v8fi.jpg", "cju1hmff8tkp809931jps6fbr.jpg", "cju857ad0l88m0817qx4cwxnf.jpg", "cju7ekbo32pft0871fv7kzwb9.jpg", "cju8bff9nrfi10850fmfzbf8v.jpg", "cju85plp7lmkw0850rx42jdpf.jpg", "cju1ewnoh5z030855vpex9uzt.jpg", "cju5tenjojp1j0755ms4949h2.jpg", "cju2ntxtdzlvu0799xl3j9pan.jpg", "cju2zm0axztpe0988r8s9twjr.jpg", "cju3ykamdj9u208503pygyuc8.jpg", "cju15czxqp3lv0835jvhgzurz.jpg", "cju88v2f9oi8w0871hx9auh01.jpg", "cju8828oxnool0801qno9luhr.jpg", "cju2sxf3iqbpv09937iksn8ep.jpg", "cju0sr5ghl0nd08789uzf1raf.jpg", "cju43gfosm63n08714rpih8pe.jpg", "cju3ya7goj6at0818v2l5ay7f.jpg", "cju5hjxaae3i40850h5z2laf5.jpg", "cju30u1hbakn808019g15nb8b.jpg", "cju8bbznkrf5g0871jncffynk.jpg", "cju85citjlnfm0755i4rk5tqj.jpg", "cju7dqcwi2dz00850gcmr2ert.jpg", "cju87q6yoneim0871dl4phvkd.jpg", "cju35c4wzdhow0799h6eq4sgs.jpg", "cju7b1ygu1msd0801hywhy0mc.jpg", "cju8c5223s8j80850b4kealt4.jpg", "cju1bhnfitmge0835ynls0l6b.jpg", "cju5x7iskmad90818frchyfwd.jpg", "cju83vvmik9wa08710yeh7cuk.jpg", "cju87nkyrnb970801q84m47yt.jpg", "cju2oi8sq0i2y0801mektzvw8.jpg", "cju40wto5kxwi0755it190f2k.jpg", "cju5jx7jzf7c90871c2i9aiov.jpg", "cju6vucxvvlda0755j7msqnya.jpg", "cju3xtufwiv9c0818djsc4cqd.jpg", "cju84ffdzkrjn08183jh1fxmb.jpg", "cju8auylgqx0z0871u4o4db7o.jpg", "cju2nbdpmlmcj0993s1cht0dz.jpg", "cju6yywx1whbb0871ksgfgf9f.jpg", "cju5ft6mcd5q40987rhjgbrr6.jpg", "cju7dymur2od30755eg8yv2ht.jpg", "cju14g8o4xui30878gkgbrvqj.jpg", "cju83syhdk6gs0801rf1rekdl.jpg", "cju7do8c72dbo0801vxfzxdc4.jpg", "cju7alcgr1lsr0871riqk84z7.jpg", "cju330ofbc2l30801th5g3hw6.jpg", "cju3xzvnzj0hd0755xprz39nj.jpg", "cju2rga4psq9n09881z519xx0.jpg", "cju18gzrq18zw0878wbf4ftw6.jpg", "cju8apjewqrk00801k5d71gky.jpg", "cju43b8daly4408170e5ev06g.jpg", "cju3xiic0ilzp0850lusrb42j.jpg", "cju3v11mrgwwb0755u242ygye.jpg", "cju77b3wyz4160755qis4ljsb.jpg", "cju3uz4o6gr9z0850lhxyxvsj.jpg", "cju5o1vu9gz8a0818eyy92bns.jpg", "cju8ayeq7r1fb0818z1junacy.jpg", "cjyzk8qieoboa0848ogj51wwm.jpg", "cju3128yi0rpu0988o4oo5n8n.jpg", "cju1d50a94qf50855wsowacrc.jpg", "cju6x0yqbvxqt0755dhxislgb.jpg", "cju5buy2bal250818ipl6fqwv.jpg", "cju7ff97z2ow40817u2r83my5.jpg", "cju1g4nsb6ngy0799l4ezm8ab.jpg", "cju34ds2531520988qjpqt6e3.jpg", "cju6wjm81vgsc0987enk9n3pr.jpg", "cju8dic9mtppa0987swn23wbc.jpg", "cju6vta3kvazg0817qbeppjtm.jpg", "cju15wdt3zla10801odjiw7sy.jpg", "cju8aeei7q8k308173n9y4klv.jpg", "cju7aez2x1jtj0871ztezs3oi.jpg", "cju1f5x1164xv08555654c24r.jpg", "cju8chxndsre008015uisl4si.jpg", "cju5wtdu4m0im0871mix0yvc0.jpg", "cju85c2d4ln1b0755zz1z3onx.jpg", "cju7g7ba42z310987bqzbi2bq.jpg", "cju7f5ghb2r5s0801chwkxxh9.jpg", "cju16jgnyzp970878melv7r25.jpg", "cju5v8pgplg6k0755rvi2t63h.jpg", "cju1euuc65wm00799m4sjdnnn.jpg", "cju1871y11d6r0799k6cw4yze.jpg", "cju8bqxxurs6i0850mu7mtef9.jpg", "cju7ecl9i2i060987xawjp4l0.jpg", "cju7d3oc82cho0755dajlwldz.jpg", "cju326h4v1gxw08352px40p7r.jpg", "cju8bafgqrf4x0818twisk3ea.jpg", "cju84aoa3ktwn0755pfl4gfwd.jpg", "cju6xa0qmvzun0818xjukgncj.jpg", "cju85qefyln6v0850szeb9byi.jpg", "cju8c5zcbsdfz0801o5t6jag1.jpg", "cjyzul1qggwwj07216mhiv5sy.jpg", "cju40jl7skiuo0817p0smlgg8.jpg", "cju2hos57llxm08359g92p6jj.jpg", "cju7dubap2g0w0801fgl42mg9.jpg", "cju85nr8elly209872w9n5m0s.jpg", "cju42wamblrqn098798r2yyok.jpg", "cju7f0ec32txj08184asb8w5f.jpg", "cju1cdxvz48hw0801i0fjwcnk.jpg", "cju3xl264ingx0850rcf0rshj.jpg", "cju17bz250pgd0799u1hqkj5u.jpg", "cju783tmkzkqu081803g7q5vk.jpg", "cju41p90plcsx08018cnzpndc.jpg", "cju32jcdabepz0878d0cznmfe.jpg", "cju1gghyjwxt80835vx0wgxw0.jpg", "cju7arvfe1ldu0850erdmphgj.jpg", "cju1hirfi7ekp0855q0vgm9qq.jpg", "cju30525w04r10835ygp257sb.jpg", "cju5vwbr4lhqn0987a1pji0ux.jpg", "cju42qet0lsq90871e50xbnuv.jpg", "cju8aw9n1qyg10801jkjlmors.jpg", "cju7787c5yy3l080159mwqsnj.jpg", "cju15ptjtppz40988odsm9azx.jpg", "cju34ymm8d6700799uop0cw33.jpg", "cju6uy20suzbl0987rzuhz7z9.jpg", "cju310f6val1v0855xo8tc3gu.jpg", "cju7atnm31if40817pqclnjer.jpg", "cju88aq6vo1ij0755c2ey7z7n.jpg", "cju8dn0c3u2v50801k8rvq02f.jpg", "cju8doa16u5gh0818w1ywda3q.jpg", "cju77t0razbvm080106o56289.jpg", "cju84hibuktj80871u519o71q.jpg", "cju7f900s2o0k08175gl1giid.jpg", "cju422cm8lfxn0818ojicxejb.jpg", "cju8567gdlcbq0801dwwyo2jt.jpg", "cju2lyynuymli0855g7fxgbhe.jpg", "cju3x5u2tiihx0818914gzxy1.jpg", "cju5vcmrqla7i0817x4sp4pqw.jpg", "cju0s2a9ekvms080138tjjpxr.jpg", "cju43kj2pm34f0850l28ahpni.jpg", "cju41lojblbs307555jdci937.jpg", "cju88trl3ogi208716qvti51b.jpg", "cju2ro5jqsy680988pi6qsujw.jpg", "cju8c5mxls96t0850wvkvsity.jpg", "cju890guyoiti098753yg6cdu.jpg", "cju5yimthmlv80850zhoc90c2.jpg", "cju1egx9pvz2n0988eoy8jp23.jpg", "cju7d4jk723eu0817bqz2n39m.jpg", "cju2qh5le1ock0878oahaql7d.jpg", "cju8b3ka8r64u0801fh18hk7l.jpg", "cju5yhgznmkzb0801cji2vi8j.jpg", "cju2nqapmzvk20801f9us40dx.jpg", "cju8djdqztu6408506pzhlo18.jpg", "cju418rckl3ur08012psrx1r1.jpg", "cju2hqt33lmra0988fr5ijv8j.jpg", "cju6vvxsev9y30987kespucdg.jpg", "cju84jdl9kv0i0871eog9b3i9.jpg", "cjz14qsk2wci60794un9ozwmw.jpg", "cju7afqon1ip40850ue2308b6.jpg", "cju8a84g0q76m0818hwiggkod.jpg", "cju2rz4k434s70855wwx3ddtx.jpg", "cju1c8ffau5770835g0g343o8.jpg", "cju41s6nbleqy0755e2mslg0b.jpg", "cju31t8xd17bk0835rnb893jk.jpg", "cju8cbsyssiqj0871gr4jedjp.jpg", "cju5klveuff6w0871wbibgh3m.jpg", "cju2sggy13na70855tbeoqgha.jpg", "cju8dm2cau2km0818jsv9eeq2.jpg", "cju7dtb1e2j0t0818deq51ib3.jpg", "cju2xyd9vyi7m098831qcucse.jpg", "cju2r11x7sdgx0988o8ule0wl.jpg", "cju85omszllp30850b6rm9mi3.jpg", "cju2nfnvxzdkd0878399axlco.jpg", "cju5bhv81abur0850ean02atv.jpg", "cju77j66ez52p08019xygi0co.jpg", "cju33yemn2qb20988wfjxximx.jpg", "cju3518w2d838079939fqztbc.jpg", "cju2t62nq45jl0799odpufwx6.jpg", "cju7fen322ou10817ziqkob4k.jpg", "cju5wqonpm0e60801z88ewmy1.jpg", "cju42romflni20817etb9a0fl.jpg", "cju3381d8bz3h07991xtl7ra0.jpg", "cju33qpdvc9g0087825jhf3s9.jpg", "cju8alhigqn2h0801zksudldd.jpg", "cju8418jhkf7d0818ga2v0xq0.jpg", "cju7fcgbe2z3p07550vaflqdb.jpg", "cju428k5fldt108177s6g6f45.jpg", "cju7fnfv02tt90801djnix9m8.jpg", "cju1cj3f0qi5n0993ut8f49rj.jpg", "cju1dq3x1vgx109889c7wyirg.jpg", "cju784jpdzeae0987q5ypq883.jpg", "cju1ats0y372e08011yazcsxm.jpg", "cju7deifq2fzn0755lc8idyh8.jpg", "cju1c6yfz42md08550zgoz3pw.jpg", "cju2zdvjn9h7r08553cp4eed5.jpg", "cju32upim1z7u0988l883nqp6.jpg", "cju0s690hkp960855tjuaqvv0.jpg", "cju7dglf226g50987ohbthl19.jpg", "cju1ejj7dvqfa0835ra184v5m.jpg", "cju83k8fyjsxr0817d6nxs6r4.jpg", "cju35a77vdj4n08556jj2lgmc.jpg", "cju45v0pungu40871acnwtmu5.jpg", "cju1hs0za7jha0855vj0mdrjt.jpg", "cju5woy82m07m08505dmjg7g1.jpg", "cju34repocy5208780gswillm.jpg", "cju5tgbzhjllu08174ca41eus.jpg", "cju5hqz50e7o90850e0prlpa0.jpg", "cju0t4oil7vzk099370nun5h9.jpg", "cju30qbm1ad3x0855znuhpz9u.jpg", "cju1b3zgj3d8e0801kpolea6c.jpg", "cju17r8il13910799dr2wme2e.jpg", "cju2rnkt22xep0801as160g9t.jpg", "cju88gx09o2vk0818610zody3.jpg", "cju0u2g7pmnux0801vkk47ivj.jpg", "cju5nxkujgscq0817l9gss626.jpg", "cju2hfqnmhisa0993gpleeldd.jpg", "cju5uhrdwkmsu0817ervv91l8.jpg", "cju6vrs1ov8cr098788h8gs6j.jpg", "cju353d1eda8c07992afde611.jpg", "cju6w733bveoz0817e600tw72.jpg", "cju2nyc5f02m40801ojqbtiea.jpg", "cju45rj7ln8980850a7821fov.jpg", "cju2xf8e5y2wm08359vcgk09b.jpg", "cju5yeqiwmkgl0801fzv2douc.jpg", "cju35740hzm0g0993zl5ic246.jpg", "cju13fwthn9mq0835gacxgy01.jpg", "cju76l27oyrw907551ri2a7fl.jpg", "cju7cp6dw244p0818gncdol4m.jpg", "cju8c6hnxsdvr0801wn0vrsa6.jpg", "cju43c92lm5cj0755lorsorfg.jpg", "cju45n0oxn5vu08500yfrt9jn.jpg", "cju30ywtc0oar0835bp2en7ec.jpg", "cju5waeduln160817w0agirve.jpg", "cju5clr68b48r0755cmuvponm.jpg", "cju2zpw4q9vzr0801p0lysjdl.jpg", "cju1b75x63ddl0799sdp0i2j3.jpg", "cju87tyddnnad0755bj0wxahe.jpg", "cju5ymyd8mmdc0801ry3by1xr.jpg", "cju8ando2qqdo0818ck7i1be1.jpg", "cju3tsh4lfsok0987w6x3a0v1.jpg", "cju2z2x3nvd3c099350zgty7w.jpg", "cju3uhb79gcgr0871orbrbi3x.jpg", "cju2omjpeqj5a0988pjdlb8l1.jpg", "cju15mhjczc8z0801kit5c6di.jpg", "cju2ycp1u8g2r0799jslnp7cz.jpg", "cju888fr7nveu0818r9uwtiit.jpg", "cju426tomlhll0818fc0i7nvh.jpg", "cju5ukkg6kv7u08011x2b6zl5.jpg", "ck2bxiswtxuw80838qkisqjwz.jpg", "cju5bj926aiec07559rshy4wa.jpg", "cju2zo0fwzv580988qlijd2xa.jpg", "cju1ddr6p4k5z08780uuuzit2.jpg", "cju85a8h8llwm07559wxg4t5w.jpg", "cju2htabevq9108015qjei0x7.jpg", "cju14hjh2ob2o0835ouz3r5aa.jpg", "cju88y1mwoln50871emyfny1g.jpg", "cju3xeexgii1j0817zs68tb4g.jpg", "cju6xlygpw7bs0818n691jsq4.jpg", "cju7aklv31h4309871m29l4e7.jpg", "cju6z2616wqbk07555bvnuyr1.jpg", "cju1c4fcu40hl07992b8gj0c8.jpg", "cju34eqjpcpm508788b3lhp97.jpg", "cju8b5p40r2c60987ofa0mu03.jpg", "cju77q10sz9ug0801449wu1nu.jpg", "cju6uzxk0v83p0801rcwnexdu.jpg", "cju7csvlb22fr0850lvm45n3x.jpg", "cju1cbokpuiw70988j4lq1fpi.jpg", "cju8b2rmgr52s0801p54eyflx.jpg", "cju357rxxdaz30878y2esjpjt.jpg", "cju34m7h536wq0988xz7gx79v.jpg", "cju3yb47cj1xq0817zfotbni4.jpg", "cju2u4pymvc720988wsxrmi84.jpg", "cju41nz76lcxu0755cya2qefx.jpg", "cju3xhpvvimda0987ygrpzni2.jpg", "cju8b4ja9r2s808509d45ma86.jpg", "cju88nroho44508500129f1nh.jpg", "cju1g20bdwq6u0835e16xugcd.jpg", "cju43in5fm22c08175rxziqrk.jpg", "cju8cgi2kspp308011nxdtjp6.jpg", "cju16b6ynq8e40988m8vx0xnj.jpg", "cju2qu37qobl50993aw7ghcfq.jpg", "cju2spdagu1l50835da1f46fr.jpg", "cju3v664kh0px0818y4y7wolf.jpg", "cju30lncba3ny0878jwnous8n.jpg", "cju1amqw6p8pw0993d9gc5crl.jpg", "cju2lejzcy4pc0878c9rlonot.jpg", "cju2qdj95ru8g09886gfi9rsz.jpg", "cju5eq8c8ck690850vix98hv3.jpg", "cju1fyb1d69et0878muzdak9u.jpg", "cju2wtwj87kys0855kx6mddzw.jpg", "cju88itqbny720987hxizbj5y.jpg", "cju3v3ac9gyz30755hfqwyp1i.jpg", "cju5vzjoslpj708186z2fusmz.jpg", "cju2ij9uiic2l09933ljiv6gm.jpg", "cju2uwz9f5yf1085506cfamfx.jpg", "cju2lberzkdzm09938cl40pog.jpg", "cju7cufm7298k0755j09uf3of.jpg", "cju7b9vcs1luz0987ta60j1dy.jpg", "cju40poe4kt9s0755f9cnm3h5.jpg", "cju7ap09p1kz10850ldccjebj.jpg", "cju5ew4h9cqaf0818rrczkmqh.jpg", "cju88fpm4o0tl0871w1i6a4ds.jpg", "cju7bgnvb1sf808717qa799ir.jpg", "cju2m71z2ywwv080131bcrsd3.jpg", "cju41r6v2lcww0871ps8k8pf5.jpg", "cju30ajhw09sx0988qyahx9s8.jpg", "cju87zv8lni0o0850hbbecbq6.jpg", "cju5yjq1pmlgc0801z0t24bly.jpg", "cjyzuio1qgh040763k56deohv.jpg", "cjyzufihqquiw0a46jatrbwln.jpg", "cju2zrojo9kcd0878ld2epejq.jpg", "cju5vxuc5loxw0818u8xgf45p.jpg", "cju2rlqdnoz9k0993cpjae3x0.jpg", "cju8402x1kcy70801t6kz6bdi.jpg", "cju1h89h6xbnx08352k2790o9.jpg", "cju8at3s1qqqx0850hcq8nmnq.jpg", "cju886ryxnsl50801r93jai7q.jpg", "ck2bxqz3evvg20794iiyv5v2m.jpg", "cju35k2fr3vc50988c85qkrwg.jpg", "cju8c1a0ws7o208181c6lbsom.jpg", "cju7dda8w2br20818zhsuz8s7.jpg", "cju1ftaji6isw0855108yqcse.jpg", "cju8c3xs7sauj0801ieyzezr5.jpg", "cju33za6l2qy70988jhrlp2ev.jpg", "cju30ov1oah920801mi8thuyg.jpg", "cju7ez7r22qbc08015xfoz2wb.jpg", "cju5o4pk9h0720755lgp9jq8m.jpg", "cju410dnfl0960755y8lu8d79.jpg", "cju334jzo261t0835yqudnfs1.jpg", "cju2yi9tz8vky0801yqip0xyl.jpg", "cju45qbf3n9sa0987oonbkly9.jpg", "cju6ut4l8va6y0755tyw3vfqq.jpg", "cju6ur9l9v9jq0755paud9uka.jpg", "cju89z6pqpqfx0817mfv8ixjc.jpg", "cju7b10ce1mnm08011c5bwyr4.jpg", "cju175facms5f0993a5tjikvt.jpg", "cju2txjfzv60w098839dcimys.jpg", "cju1csmlc4ht10799b8ymmghg.jpg", "cju3tp94kfstl08181awh6z49.jpg", "cju2p91qir00k08350ddfif0w.jpg", "cju5hyi9yegob0755ho3do8en.jpg", "cju43h43am1dy08176gwfhmnt.jpg", "cju88msmoo3470817m441j4sg.jpg", "cju13hp5rnbjx0835bf0jowgx.jpg", "cju83ipu3jwpx0801z5pvguf8.jpg", "cju33w4sdcivk0855x879zht7.jpg", "cju2tzypl4wss0799ow05oxb9.jpg", "cju6wll7wvo3y08502pagos8m.jpg", "cju7bc95p1mdm0817yqj5jc6j.jpg", "cju30j1rgadut0801vuyrsnt8.jpg", "cju35mdz73x890835eynq1h9v.jpg", "cju43lcnum9y10755bjs7z87f.jpg", "cju7etr3y2p4t0801cdzjj8ab.jpg", "cju774fmayxif0818u2g79usw.jpg", "cju1cu1u2474n0878tt7v4tdr.jpg", "cju323ypb1fbb0988gx5rzudb.jpg", "cju33x0f22peh0988g0ln7w5v.jpg", "cju2y5zas8m7f0801d34g5owq.jpg", "cju5ddda9bkkt0850enzwatb1.jpg", "cju5ufn3skquf0818dhapnhba.jpg", "cju84gpefknwm098714oq8q61.jpg", "cju2trtjf4qjd0878a2zle9v9.jpg", "cju7fazv92ywx0755xov2erga.jpg", "cju2rn0hasxri0835nfy3buay.jpg", "cju3x4blzieu30850x10uuvbm.jpg", "cju3y9difj6th0801kd1rqm3w.jpg", "cju2osuru0ki00855txo0n3uu.jpg", "cjyzlw7f9faqr070129au64sq.jpg", "cju3u39fog1bo0871lxjrabks.jpg", "cju2yo1j1v0qz09934o0e683p.jpg", "cju33231uy4gi0993qc7b1jch.jpg", "cju2xlcqxy9c60988vjacdznb.jpg", "cju77vvcwzcm50850lzoykuva.jpg", "cju8dpa89u6l80818dj6lldh9.jpg", "cju3xwpgviwlx0871rwm15q7v.jpg", "cju1h5w4wxajx0835mc954kxy.jpg", "cju303j5r062k098835zxfds5.jpg", "cju35fxqyzt5p0993vusm54qz.jpg", "cju31w6goazci0799n014ly1q.jpg", "cju3u815rg4ek0850vvhtcvcm.jpg", "cju8bpctzrqkr0850zeldv9kt.jpg", "cju5f26ebcuai0818xlwh6116.jpg", "cju5g163vd6mt0817uccuga6u.jpg", "cju2zjcvj9qma0801dk71hhi0.jpg", "cju2mfjndoz700988b9lc3zeq.jpg", "cju5uxjnol2r509871qv2yeia.jpg", "cju7dwe282dc309876rco45ts.jpg", "cju5wuhm1lwm40987vugqn3vv.jpg", "cju77u1sjz77b0817ft44r3fk.jpg", "cju2yv4imv6cz099314jveiib.jpg", "cju16d65tzw9d0799ouslsw25.jpg", "cju8b6rp0r5st0850184f79xt.jpg", "cju3umoh1geet0817cmpef5am.jpg", "cju5y7buemcw80987p0r30g9f.jpg", "cju32srle1xfq083575i3fl75.jpg", "cju0roawvklrq0799vmjorwfv.jpg", "cju7b5afm1nfw0801xqm8bf8q.jpg", "cju1fb9236a110801yvg0fwju.jpg", "cju2z45kuzf6d0988nz2c819m.jpg", "cju45lbgznahl08180xz1h7u6.jpg", "cju2tpfa5uyx408359datxqqj.jpg", "cju3v72v5h1qz0818fggilwtq.jpg", "cju2yb31a8e8u0878wdashg7o.jpg", "cju6x97w4vwua0850x0997r0a.jpg", "cju42u5bjlvi10801dc13sskp.jpg", "cju77bvg0yv4r0987yh60xmjo.jpg", "cju5wrapcm2290818jsh26ppb.jpg", "cju17v6ih0u7808783zcbg1jy.jpg", "cju8d2q30tfhs0801n7lx77xl.jpg", "cju2nd7l7z98o0799gfjvyfmw.jpg", "cju5uzmaol56l0817flxh4w9p.jpg", "cju17otoe119u0799nqcbl8n1.jpg", "cju2s16zp317h0799gr67jqc2.jpg", "cju0u82z3cuma0835wlxrnrjv.jpg", "cju2zy1e49pqk0878t6ncqn12.jpg", "cju1f15k3w4ct0835cmde6ypo.jpg", "cju8bgdmqrksy0801tozdmraa.jpg", "cju3521y5d5mq0878t3ezsu4p.jpg", "cju40sdwukv3k0755y99ug1k8.jpg", "cju2pjb9v0ywn0878j5g5n69j.jpg", "cju2i3hzclw3o0988rrgh911i.jpg", "cju6yxyt0wh080871sqpepu47.jpg", "cju5x28nzm7t907558ocq4bt7.jpg", "cju1hp9i2xu8e0988u2dazk7m.jpg", "cju160wshltz10993i1gmqxbe.jpg", "cju34aozyyy830993bn16u32n.jpg", "cju7dxffn2eam0817qxosfwch.jpg", "cju8dqkrqu83i0818ev74qpxq.jpg", "cju8bysfgrzkl081786jwac09.jpg", "cju5gucasds9d0801019axylx.jpg", "cju5vi4nxlc530817uoqm2m7a.jpg", "cju5wj0faly5008187n6530af.jpg", "cju30mm25a53s0799qa5wiqe8.jpg", "cju5ca9hcatkc0801jzwe7tfx.jpg", "cju7bd1qu1mx409877xjxibox.jpg", "cju2yg5ht8i4p087800js8hp4.jpg", "cju5y4hgqmk0i08180rjhbwvp.jpg", "cju1euant5l960878iqj5vvto.jpg", "cju1dg44i4z3w0801nyz4p6zf.jpg", "cju2uy8ox62jo0801g88hh42z.jpg", "cju2t16vuucaq0835xcpsivn2.jpg", "cju5u4pywk81x0817vn9pe14z.jpg", "cju8ca4geseia0850i2ru11hw.jpg", "cju83wwn1k55e0850kw6i2d81.jpg", "cju45ofd9ne1j0801ri8dup7t.jpg", "cju2uzabhs6er0993x3aaf87p.jpg", "cju8a56vxpy780850r45yu4wk.jpg", "cju45pm27n80u08174kyow1gj.jpg", "cju3u4lxmg59o0755rz42b9en.jpg", "cju1gkndf6yi10801o1qnje19.jpg", "cju30gxjq0djk0988jytm49rs.jpg", "cju45jpvfn6c809873pv1i34s.jpg", "cju7adqyj1jcx08712r1ro5gx.jpg", "cju7dmlgf2ebw0871ieqas5fh.jpg", "cju8d4jgatgpj0871q2ophhkm.jpg", "cju2wx0gh7fpz0878wwyd9ep8.jpg", "cju2x7vw87mu30878hye2ca0m.jpg", "cju1hhj6mxfp90835n3wofrap.jpg", "cju30ftgja7170855xl9bkdm0.jpg", "cju43eigtm6ev0801mv0m96t1.jpg", "cju1fjsb4sipq09931lvd8e41.jpg", "cju77g99iyxc00817zqi2ppor.jpg", "cju34zivp3fq80988opxbaqyn.jpg", "cju2t9tdwuk700835kv0ljmtl.jpg", "cju5wi6bqlxy90755bu227nvb.jpg", "cju2rmd2rsw9g09888hh1efu0.jpg"], "valid_labels": ["cju5vgawslbe30987ndeepc1b.jpg", "ck2bxlujamu330725szlc2jdu.jpg", "cju2hx006vidl0799igm81vmh.jpg", "cju7aqkue1i2k09879uzcpt8r.jpg", "cju5uget8krjy0818kvywd0zu.jpg", "cju8dk7eztzup08182yxko5zh.jpg", "cju1c3218411b08014g9f6gig.jpg", "cju7f6cqy2ur20818t1saazbm.jpg", "cju6wi3akvn8r0801px8eligc.jpg", "cju5enq1tcn1i0755hnkon787.jpg", "cju2hewssldzx0835ep795xu0.jpg", "ck2bxskgxxzfv08386xkqtqdy.jpg", "cju2np2k9zi3v079992ypxqkn.jpg", "cju5bycdkalkb09875f7bfrvx.jpg", "cju2hlm19vjjf0801o69qnber.jpg", "cju7amjna1ly40871ugiokehb.jpg", "cju34c1xfyz920993itxkkfad.jpg", "cju88oh0po9gq0801nge4tgr1.jpg", "cju5i39mreass0817au8p22zy.jpg", "cju5f0dezct4q08183ydw11dx.jpg", "cju5cky5xb0ay0801oxet697t.jpg", "cju7ehljc2or70871261br8ai.jpg", "cju183od81ff608017ekzif89.jpg", "cju7evxt12m730987rxivne3x.jpg", "cju8cj10qsrau0871o2dr6ai1.jpg", "cju2y26c588bo07993ksd8eoz.jpg", "cju2zkpdl9h7t0799ix60teqg.jpg", "cju85bf1algsq0871y9gtlq97.jpg", "cju40w3hbkwpn08015rbs3wko.jpg", "cju17hw9hr9c5098800fu4u8e.jpg", "cju6wuojavt740818b5qcv3iw.jpg", "cju2qvuj1s9ok0835tp2k4ozh.jpg", "cju33jon3ygbj0993pu22a4k6.jpg", "cju7dsrtb2f8i085064kwugfk.jpg", "cju77k828z46w0871r0avuoo9.jpg", "cju1aqqv02qwz0878a5cyhr67.jpg", "cju412uwlkva50850d1ps1ww7.jpg", "cju2nguelpmlj0835rojdn097.jpg", "cju35eg0tdmjt085525sb4bua.jpg", "cju7dn24o296i09871qfxb8s2.jpg", "cju2ma647l0nj0993ot4deq2q.jpg", "cju1expq45zst0855rjqwwj4m.jpg", "cju88t4fvokxf07558ymyh281.jpg", "cju83nwu1jxte0987h1krpfmv.jpg", "cju1fj6axwfp30835ukhuzhw9.jpg", "cju8b0jr0r2oi0801jiquetd5.jpg", "cju7bmi1v1pnj0987pa52jjok.jpg", "ck2da7fwcjfis07218r1rvm95.jpg", "cju2zblxw9848087853csbrx1.jpg", "cju5udcufki0s09874ll1dbr5.jpg", "cju2rqo702wpx0855fn7d5cxh.jpg", "cju88l66no10s0850rsda7ej1.jpg", "cju83yddek68q0850d2x7zfkm.jpg", "cju6z1bzbwfq50817b2alatvr.jpg", "cju5hi52odyf90817prvcwg45.jpg", "cju324q101fhe08350wae9cif.jpg", "cju5cetivauok0987ok3e5bre.jpg", "cju5u6wf0kh1t0755bg1ssixv.jpg", "cju8clorgsuwn08714toqb7v6.jpg", "cju5b9oyda4yr0850g9viziyv.jpg", "cju2qtee81yd708787bsjr75d.jpg", "cju3tvffffx5f0818t5ov22al.jpg", "cju1d96gsv62d09881b3wecw2.jpg", "cju5yclrymlgj0818k426ud6z.jpg", "cju2i8br1vqtd08784u6vmcjk.jpg", "cju7dvl5m2n4t0755hlnnjjet.jpg", "cju5hl8nee8a40755fm8qjj0o.jpg", "cju7d8m3b2e210755l8fj1yph.jpg", "cju1f79yhsb5w0993txub59ol.jpg", "cju7frtqu2xa20818wq8r9fzf.jpg", "cju35atpxdjot0855q46aqrd0.jpg", "cju3y0pjrj1c30755nxekxccj.jpg", "cju2ti1du4idn0878giuozonw.jpg", "cju2yyhsp933j0855hp32e012.jpg", "cju2r6mt2om21099352pny5gw.jpg", "cju3ttznuftyf09875t11850w.jpg", "cju42xpi8lw4w0871ve317a1p.jpg", "cju5u8gz4kj5b07552e2wpkwp.jpg", "cju7ezs7g2mxm098787atbran.jpg", "cju17g6ykn1cs0993dww6qdi8.jpg", "cju8arof2qpf20850ifr1bnqj.jpg", "cju3xuj20ivgp0818mij8bjrd.jpg", "cju34sh43d8zm08019xbwhc0o.jpg", "cju2zp89k9q1g0855k1x0f1xa.jpg", "cju7agj961l2r0818z29iq8yn.jpg", "cju3ua8u0g9rg0801uayhdxhu.jpg", "cju2hdr06v2bq0799mbm3bks1.jpg", "cju2qz06823a40878ojcz9ccx.jpg", "cju8azmhcr66e0755t61atz72.jpg", "cju87ox0kncom0801b98hqnd2.jpg", "cju1fuoa4wmc50835qfd11sp9.jpg", "cju88cddensj00987788yotmg.jpg", "cju7azuu31mia0801pf9ib5ed.jpg", "cju5k3j3uf6de0817hszzfr7n.jpg", "cju5x00l6m5j608503k78ptee.jpg", "cju430pm2lz0y0755jkhcc3d1.jpg", "cju0vtox5ain6099360pu62rp.jpg", "cju7dp3dw2k4n0755zhe003ad.jpg", "cju885ikhnmkn09878s2lqtuh.jpg", "cju2zr3c3vwb00993jn06bbaz.jpg", "cju8bw697rwg308177tg8huas.jpg", "cju1efbr0rqxz09931z0lf4vf.jpg", "cju7d7aut2a2p0818z4uxc6cd.jpg", "cju2sevf53lkx08558h5bpaig.jpg", "cju15l5ubz9yh0855b3ivdpse.jpg", "cju2nnqrqzp580855z8mhzgd6.jpg", "cju2mh8t6p07008350e01tx2a.jpg", "cju30nyxe0gfb0835p256yoju.jpg", "cju85rkbnlo1c08503uxcpax1.jpg", "cju3280wv1ir009882jze27tc.jpg", "cju7b2l561oas0871decgslaf.jpg", "cju2m56cryvqd0801gtn2yp8t.jpg", "cju5boicjagt20871b1fotkh4.jpg", "ck2bxknhjvs1x0794iogrq49k.jpg", "cju7crgxa28550755wbsgqkel.jpg", "cju1brhsj3rls0855a1vgdlen.jpg", "cju8b542nr81x0871uxnkm9ih.jpg", "cju87li0zn3yb0817kbwgjiz8.jpg", "cju1f8w0t65en0799m9oacq0q.jpg", "cju8a3nhbpwnb0850d37fo2na.jpg", "cju1bm8063nmh07996rsjjemq.jpg", "cju2z1nxlzaj40835wj81s1iy.jpg", "cju83qd0yjyht0817ktkfl268.jpg", "cju414lf2l1lt0801rl3hjllj.jpg", "cju2yw4s7z7p20988lmf2gdgd.jpg", "cju5fs6j6d8350801vglraq4u.jpg", "cju7er4kc2opa0801anuxc0eb.jpg", "cju5bdwa3aatx0818b79i18zf.jpg", "cjyzurzvohqnr0794es1itzek.jpg", "ck2bxw18mmz1k0725litqq2mc.jpg", "cju41z76wlgbz0801qdetlvby.jpg", "cju84ih17kp5l09876bkooocl.jpg", "cju5vbo6jldrt0871jf6f1700.jpg", "cju7es23b2vcp0755gpbm9s7v.jpg", "cju6ywm40wdbo0987pbftsvtg.jpg", "cju32l161bi1v07990vm376in.jpg", "cju5fi0yxd3ei0801v7u0yudn.jpg", "cju884985nlmx0817vzpax3y4.jpg", "cju32qr9tbvsj08013pkpjenq.jpg", "cju7flevb2wii08188otgs9p2.jpg", "cju16fpvhzypl0799p9phnlx6.jpg", "cju1dhfok4mhe0878jlgrag0h.jpg", "cju5xq3tdm9fn0987pbedxdg5.jpg", "cju7bb3ss1uoo0755pmhyco7t.jpg", "cju6us80mv1b50871ebyq2wxa.jpg", "cju2p0eveqtdc0835gpi3p93i.jpg", "cju7apr0c1qqm0755s7msqot4.jpg", "cju34o6dbd2lo0855aqlcy1hs.jpg", "cju2xd75m82720801q4s4ik3n.jpg", "cju7ebe962hr409872ovibahw.jpg", "cju1fr4etsmrr09933u4t4aql.jpg", "cju0ue769mxii08019zqgdbxn.jpg", "cju7cl8zm1xcu0817ado0jpas.jpg", "cju5ktjwofed70817eg58ef7u.jpg", "cju843yjskhq30818qre4rwm2.jpg", "cju8cwy02t9eq08185qn12c02.jpg", "cju892fesoq2g0801n0e0jyia.jpg", "cju3ul8dogf1z09872y2ecowp.jpg", "cju85fc11ljr40818edpb0inh.jpg", "cju8c2rqzs5t80850d0zky5dy.jpg"], "test_labels": ["cju8abobpqbir08189u01huru.jpg", "cju8axq24r4an0755yhv9d4ly.jpg", "cju5cu8qkb84x08186jwo8yin.jpg", "cju30k2z40ds308353kdew70n.jpg", "cju6vqarjv7yo0987q4b1btk1.jpg", "cju1819curo000988pd5xcqme.jpg", "cju7d9seq29zd0871nzl2uu5m.jpg", "cju31ugmfb3dz0855xtqshki6.jpg", "cju858eswlepn0871pzvdrhj1.jpg", "cju5fw37edaae0801vkwvocn7.jpg", "cju83kxitjv340987z09m0ezy.jpg", "cjyzkpsbjdsjq07211dfi4sru.jpg", "cju2z9vlp9j0w0801oag91sy9.jpg", "cju0tl3uz8blh0993wxvn7ly3.jpg", "cju33eqwbcch208012jikwdky.jpg", "cju8bssulrrcy0987h1vq5060.jpg", "cju2qxxko229x08786gvxxhur.jpg", "cju6wn57mvooj0850rp78hhy7.jpg", "cju7ejm2l2ncl0801wq6y84nw.jpg", "cju7b3f5h1sm40755i572jden.jpg", "cju2pmhtr17a00855cvpelzb0.jpg", "cju2u2b9o4zvp08788qb9nqxj.jpg", "cju85ia1slh220987y7c20sm2.jpg", "cju439oazm2fu0871ma0vvrft.jpg", "cju5hwonqedw10801vsd3w6kk.jpg", "cju1gi7jlwyld0835cdf6g6qz.jpg", "cjyzjzssvd8pq0838f4nolj5l.jpg", "cju17x0j4nfc10993y31pvlgs.jpg", "cju1fm3id6gl50801r3fok20c.jpg", "cju30bmab08bi0835mvlr6e0r.jpg", "cju41kd7yl4nm0850gil5qqwh.jpg", "cju2s2527pfyr0993l3h1149a.jpg", "cju2urqpwvxw70835rvndvtsi.jpg", "cju7et17a2vjk0755e743npl1.jpg", "cju7fob3x301u0755x985pmmq.jpg", "cju17z0qongpa0993de4boim4.jpg", "cju2phaksnahz0993yxogjcpv.jpg", "cju8b1v3br45u087189kku66u.jpg", "cju8aqq8uqmoq0987hphto9gg.jpg", "cju33mirdc8mj0799k33wzoes.jpg", "cju2y0z6g87p10878fpk5d3rq.jpg", "cju6vvb8svhed0801jjcquh5e.jpg", "cju2t3ibkuecr0835o7si16zv.jpg", "cju8914beokbf0850isxpocrk.jpg", "cju8chdlqsu620755azjty1tj.jpg", "cju43o6n7m9nk087191ijwqq9.jpg", "cju7bf1lp1shi081835vs84lc.jpg", "cju2ysg748ru80878sp6j0gm0.jpg", "cju88z8bson4h0871nnd7fdxo.jpg", "cju8brv16rx7f0818uf5n89pv.jpg", "cju7da88w2eod0755wejzynvt.jpg", "cju7eueum2oqn0850rodmx8zo.jpg", "cju773hsyyosz0817pk1e7sjq.jpg", "cju358pwtdby20878cg7nm0np.jpg", "cju2oq5570avm079959o20op1.jpg", "cju3y54kwj3nr0801biidlb4e.jpg", "cju5bmhdcafs909878qfzrqzi.jpg", "cju83h9ysjwe808716nt35oah.jpg", "cju5chrxxawka0871qcj171yz.jpg", "cju8czvnztbf40871b4m7t78w.jpg", "cju3xvoo2iqlc0817eku2r3wl.jpg", "cju7f9umg2olj0987fj5y285w.jpg", "cju87vqa0ndwg0850onjdz7ol.jpg", "cju2ouil2mssu0993hvxsed6d.jpg", "cju2zi4l09f5807991s8do2b2.jpg", "cju5k503sfa5f0871lx0rpu5y.jpg", "cju87z6o6nh73085045bzsx6o.jpg", "cju5xopi0md7q0871r1sjc1av.jpg", "cju40r6jrksyk0871wg98zgho.jpg", "cju5xneamme2p0801qdf7fdwv.jpg", "cju3xjqtpikx50817tppy6g84.jpg", "cju8bn7m2rmm70817hgxpb1uq.jpg", "cju42nm68lpyo0818xvvqmupq.jpg", "cju88q6h6obpd0871ckmiabbo.jpg", "cju3wstckialg0871xs0vevsj.jpg", "cju2y8s56ymqr083541ggdsml.jpg", "cju5ygh1zmmdi0755uod5e17i.jpg", "cju7bfx651qr80801cs7epotb.jpg", "cju7d7tly27h408016fyp5nr7.jpg", "cju77idwfz36d0871tzfzz51i.jpg", "cju7ddtz729960801uazp1knc.jpg", "cju30ia8da2bq0799klnehml2.jpg", "cju3y79ofj3va0871uqfb1mzo.jpg", "cju7cue9b232j0801qdzk1ykj.jpg", "cju5vutu7ll8w0871dfp92n9p.jpg", "cju2xa3i4y0160988i679zsqd.jpg", "cju6x4t13vyw60755gtcf9ndu.jpg", "cju7fd6yt2p740987wkr8exo1.jpg", "cju2xbk0080y80801eghyddi2.jpg", "cju32a52lb9rc0799xi40qs00.jpg", "cju42tauqlo5p08171l3cuo4b.jpg", "cju7ae7bq1f820987toc8si1d.jpg", "cju5y84q3mdv50817eyp82xf3.jpg", "cju893jmdompz0817xn3g1w4h.jpg", "cju2uokeg5jm20799xwgsyz89.jpg", "cju42dwedlmk60871jbgu4ehi.jpg", "cju35i2e63uxr0835h7zgkg9k.jpg", "cju6z9a9kwsl007552s49rx6i.jpg", "cju15jr8jz8sb0855ukmkswkz.jpg", "cjyzu9th0qt4r0a46pyl4zik0.jpg", "cju89y9h0puti0818i5yw29e6.jpg", "cju7bduyq1rjf08719giru9ho.jpg", "cju2xwm1s84l50799i60mq0pu.jpg", "cju7dlk532dsh0871zvr6qz0r.jpg", "cju85mpuglq8k0818d2it6hzb.jpg", "cju5xkwzxmf0z0818gk4xabdm.jpg", "cju8cattbsivm0818p446wgel.jpg", "cju2pag1f0s4r0878h52uq83s.jpg", "cju5x15djm7ae0755h8czf6nt.jpg", "cju5c5xc7algd0817pb1ej5yo.jpg", "cju32pzh9bpw10855q4vaxfhe.jpg", "cju6v3bb2v7xo085090blubyw.jpg", "cju83mki1jv5w0817kubxm31r.jpg", "cju5bbtwsa8cl0987wgfsqpao.jpg", "cju14pxbaoksp0835qzorx6g6.jpg", "cju785htizjzo08017tvlhtg4.jpg", "cju42m60jlpcm08186kqppzqv.jpg", "cju847pxykriq0755268ktrk2.jpg", "cju3x9lttikfb0818a0g104zn.jpg", "cju1cnnziug1l0835yh4ropyg.jpg", "cju76lsehyia10987u54vn8rb.jpg", "cju7fbndk2sl608015ravktum.jpg", "cju5cjh3xattc0817j2vbulzi.jpg", "cju3uwz6ogsp10801h2r3bj5l.jpg", "cju34ouumcznz07996gg1xq7v.jpg", "cju3x2s11ibzi0817kk284k0j.jpg", "cju34fojcctcf0799ebolbvkn.jpg", "cju3v56bwgy8v0871w14pz8fx.jpg", "cju87r56lnkyp0755hz30leew.jpg", "cju2r91dg2k090801bh0xzbxk.jpg", "cju7ev2b12owa08500bpfpwyw.jpg", "cju76o55nymqd0871h31sph9w.jpg", "ck2395w2mb4vu07480otsu6tw.jpg", "cju2pkwt3r8b90988v2ywq1px.jpg", "cju33belnbyhm0878yxl42233.jpg", "cju7ahtkb1jr90801jck4kbds.jpg", "cju8adb60qbiu080188mxpf8d.jpg", "cju5wrrs0m2af0818vmnajbtw.jpg", "cju2wzu8wxtgu09880ku9x1pg.jpg", "cju3y21quj0ir0818kgjagr15.jpg", "cju87mrypnb1e0818scv1mxxg.jpg", "cju0sxqiclckk08551ycbwhno.jpg", "cju5h57xedz5h0755mjpc8694.jpg", "cju1cfhyg48bb0799cl5pr2jh.jpg", "cju8c82iosagu0817l74s4m5g.jpg", "cju30df2j09dd08351ayx2t6w.jpg", "cju2rkjfwoxys0993x768l1j2.jpg", "cjyzu3reghjya0794w7pwoi50.jpg", "cju7druhp2gp308715i6km7be.jpg", "cju5w7xn0lrkq0801f9k0htgx.jpg", "cju40u30gkuzc0871rq7t666d.jpg", "cju6v1m1xv07w09870ah3njy1.jpg", "cju849c23kgnk0817cgv2hw1e.jpg", "cju5k7r0yf98c09878csbxb4d.jpg", "cju8a2itsq4dv0755ntlovpxe.jpg", "cju6z7e4bwgdd0987ogkzq9kt.jpg", "cju7avvi51iox0817ym55y6tt.jpg", "cju0qkwl35piu0993l0dewei2.jpg", "cju85hguellg50818kwu3s8d1.jpg", "cju302fqq9spc0878rrygyzzz.jpg", "cju8amfdtqi4x09871tygrgqe.jpg", "cju6z600qwh4z081700qimgl9.jpg", "cju7ea4om2l910801bohqjccy.jpg", "cju45t5ddnbio0987qtqzx762.jpg", "cju84dsvaklpx098750hp83x4.jpg", "cju1cqc7n4gpy0855jt246k68.jpg", "cju5thdbrjp1108715xdfx356.jpg", "cju87kbcen2av0987usezo8kn.jpg", "cju8b7aqtr4a00987coba14b7.jpg", "cju5xjn5mm78b09871spyqhhr.jpg", "cju3x7xsaijq80818f0psavav.jpg", "cju5fu081d8gc0818l3yylujk.jpg", "cju171py4qiha0835u8sl59ds.jpg", "cju2qn2fzs1vy0988l243cvzy.jpg", "cju7d1tvt25bu08019dvw3uff.jpg", "cju2ueb6j5ado0878vf5md13o.jpg", "cju13cgqmnhwn0988yrainhcp.jpg", "cju3tx1qyg0c907552fglumhc.jpg", "cju31rb7vb6110801p9rhacuw.jpg", "cju77afzlz3kp07550x5nafzs.jpg", "cju2hugv9vget0799hhk7ksvg.jpg", "cju88evxanv9r08176zkeovec.jpg", "cju6xifswvwbo0987nibtdr50.jpg", "cju43jcqim2cp08172dvjvyui.jpg", "cju5es375cnzy0801nkq35ffs.jpg", "cju7dff9529h208503w60lbil.jpg", "cju2hgsptlfam0835o3b59h1o.jpg", "cju8buos5rz9b08715lfr0f4f.jpg", "cju7ctvqn25dy08186g442m1r.jpg", "cju7dz5yy2i7z0801ausi7rna.jpg", "cju7b4mtw1n9n080186209f3d.jpg", "cju42g865lorv07552ytz6xxa.jpg", "cju5von04litr08718j8po40a.jpg", "cju7eea9b2m0z0801ynqv1fqu.jpg", "cju8bi8q7rlmn0871abc5ch8k.jpg", "cju5kre09fhka0850h7b1898j.jpg", "cju40m0rjkpw80871z6n6yg1u.jpg", "cju6wt9jvvn500871hjn3t3g0.jpg", "cju2xjz2ju8pe0993ysv9wg17.jpg", "cju2t2ivz43i10878zeg8r1br.jpg"]}
|
files_split/MMWHS_CT_Heart.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
files_split/MMWHS_MR_Heart.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
files_split/__init__.py
ADDED
|
File without changes
|
files_split/fgadr_pkl_file.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d44356d5fc5e30bb200ddb789d70730a4158875a61140171232ab44ac3c98400
|
| 3 |
+
size 159475
|
further_training_lvm_med/README.md
ADDED
|
@@ -0,0 +1,20 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
# Description
|
| 3 |
+
|
| 4 |
+
The LIVECell dataset was published by [Edlund et.al.](https://www.nature.com/articles/s41592-021-01249-6) in 2021. This dataset consists of over 1.6 million high-quality cells from various sets of cell morphologies and
|
| 5 |
+
culture densities which are manually annotated carefully by experts.
|
| 6 |
+
There are 8 types of Cells in this dataset. All of them were merged and then continued pre-trained from LVM-MED weights.
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
```bibtex
|
| 10 |
+
@article{edlund2021livecell,
|
| 11 |
+
title={LIVECell—A large-scale dataset for label-free live cell segmentation},
|
| 12 |
+
author={Edlund, Christoffer and Jackson, Timothy R and Khalid, Nabeel and Bevan, Nicola and Dale, Timothy and Dengel, Andreas and Ahmed, Sheraz and Trygg, Johan and Sj{\"o}gren, Rickard},
|
| 13 |
+
journal={Nature methods},
|
| 14 |
+
volume={18},
|
| 15 |
+
number={9},
|
| 16 |
+
pages={1038--1045},
|
| 17 |
+
year={2021},
|
| 18 |
+
publisher={Nature Publishing Group US New York}
|
| 19 |
+
}
|
| 20 |
+
```
|
further_training_lvm_med/__init__.py
ADDED
|
File without changes
|
lvm-med-training-data/0_SSL_BraTS2018_Brain/file_list.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:afc528cf7ea027d47295dfd92d269b7c28f5a798a11fb8b7741c007199088a40
|
| 3 |
+
size 1239
|
lvm-med-training-data/0_SSL_HNSCC_Mixed_HeadNeck/file_list.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cda6b92937fa473d5f34cdfb4a4f5fb7ac0d499f465a89cea70543dd589172ed
|
| 3 |
+
size 12493214
|