metadata
language:
- en
tags:
- summarization
datasets:
- scientific_papers
metrics:
- rouge
model-index:
- name: ccdv/lsg-bart-base-4096-pubmed
results: []
This model relies on a custom modeling file, you need to add trust_remote_code=True
See #13467
from transformers import AutoTokenizer, AutoModelForSeq2SeqLM, pipeline
tokenizer = AutoTokenizer.from_pretrained("ccdv/lsg-bart-base-4096-pubmed", trust_remote_code=True)
model = AutoModelForSeq2SeqLM.from_pretrained("ccdv/lsg-bart-base-4096-pubmed", trust_remote_code=True)
text = "Replace by what you want."
pipe = pipeline("text2text-generation", model=model, tokenizer=tokenizer, device=0)
generated_text = pipe(
text,
truncation=True,
max_length=64,
no_repeat_ngram_size=7,
num_beams=2,
early_stopping=True
)
ccdv/lsg-bart-base-4096-pubmed
This model is a fine-tuned version of ccdv/lsg-bart-base-4096 on the scientific_papers pubmed dataset.
It achieves the following results on the test set:
| Length | Sparse Type | Block Size | Sparsity | Connexions | R1 | R2 | RL | RLsum |
|---|---|---|---|---|---|---|---|---|
| 4096 | Local | 256 | 0 | 768 | 47.37 | 21.74 | 28.59 | 43.67 |
| 4096 | Local | 128 | 0 | 384 | 47.02 | 21.33 | 28.34 | 43.31 |
| 4096 | Pooling | 128 | 4 | 644 | 47.11 | 21.42 | 28.43 | 43.40 |
| 4096 | Stride | 128 | 4 | 644 | 47.16 | 21.49 | 28.38 | 43.44 |
| 4096 | Norm | 128 | 4 | 644 | 47.09 | 21.44 | 28.40 | 43.36 |
| 4096 | LSH | 128 | 4 | 644 | 47.11 | 21.41 | 28.41 | 43.42 |
With blocks of size 32 (lower ressources):
| Length | Sparse Type | Block Size | Sparsity | Connexions | R1 | R2 | RL | RLsum |
|---|---|---|---|---|---|---|---|---|
| 4096 | Pooling | 32 | 4 | 160 | 44.60 | 19.35 | 26.83 | 40.85 |
| 4096 | Stride | 32 | 4 | 160 | 45.52 | 20.07 | 27.39 | 41.75 |
| 4096 | Block Stride | 32 | 4 | 160 | 45.30 | 19.89 | 27.22 | 41.54 |
| 4096 | Norm | 32 | 4 | 160 | 44.30 | 19.05 | 26.57 | 40.47 |
| 4096 | LSH | 32 | 4 | 160 | 44.53 | 19.27 | 26.84 | 40.74 |
Model description
The model relies on Local-Sparse-Global attention to handle long sequences:
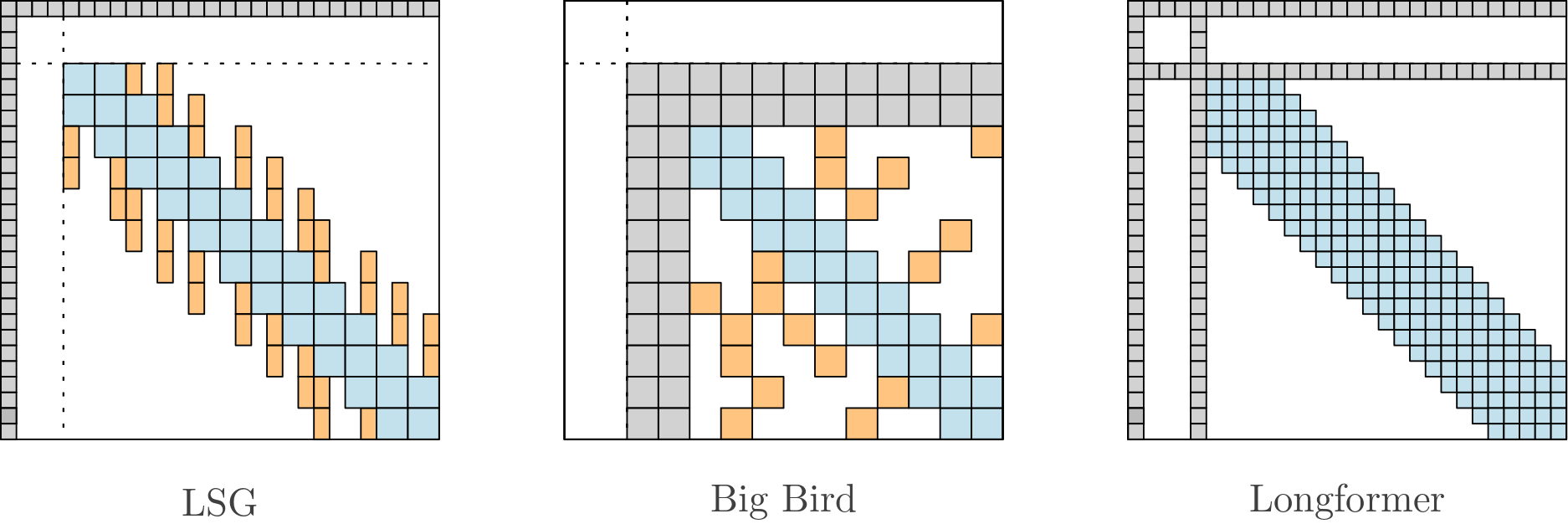
The model has about ~145 millions parameters (6 encoder layers - 6 decoder layers).
The model is warm started from BART-base, converted to handle long sequences (encoder only) and fine tuned. \
Intended uses & limitations
More information needed
Training and evaluation data
More information needed
Training procedure
Training hyperparameters
The following hyperparameters were used during training:
- learning_rate: 8e-05
- train_batch_size: 8
- seed: 42
- gradient_accumulation_steps: 4
- total_train_batch_size: 32
- optimizer: Adam with betas=(0.9,0.999) and epsilon=1e-08
- lr_scheduler_type: linear
- lr_scheduler_warmup_ratio: 0.1
- num_epochs: 8.0
Generate hyperparameters
The following hyperparameters were used during generation:
- dataset_name: scientific_papers
- dataset_config_name: pubmed
- eval_batch_size: 8
- eval_samples: 6658
- early_stopping: True
- ignore_pad_token_for_loss: True
- length_penalty: 2.0
- max_length: 512
- min_length: 128
- num_beams: 5
- no_repeat_ngram_size: None
- seed: 123
Framework versions
- Transformers 4.18.0
- Pytorch 1.10.1+cu102
- Datasets 2.1.0
- Tokenizers 0.11.6