language:
- en
license: llama3
tags:
- llama-3
- llama
- Mixtral
- instruct
- finetune
- chatml
- DPO
- RLHF
- gpt4
- distillation
- GGUF
base_model: meta-llama/Meta-Llama-3-70B-Instruct
widget:
- example_title: OpenBioLLM-70B
messages:
- role: system
content: >-
You are an expert and experienced from the healthcare and biomedical
domain with extensive medical knowledge and practical experience.
- role: user
content: How long does it take for newborn jaundice to go away?
output:
text: >-
Newborn jaundice, also known as neonatal jaundice, is a common condition
in newborns where the yellowing of the skin and eyes occurs due to an
elevated level of bilirubin in the blood. Bilirubin is a yellow pigment
that forms when red blood cells break down. In most cases, newborn
jaundice resolves on its own without any specific treatment.
The duration of newborn jaundice can vary depending on several factors
such as the underlying cause, gestational age at birth, and individual
variations in bilirubin metabolism. Here are some general guidelines:
1. Physiological jaundice: This is the most common type of newborn
jaundice and usually appears within 24-72 hours after birth. It tends to
peak between the second and fifth day of life and gradually improves
over the next week or two. By the time the baby is one week old, the
jaundice should have mostly resolved. 2. Breast milk jaundice: This type
of jaundice occurs in breastfed babies and may appear later than
physiological jaundice, typically between the fifth and fourteenth day
of life. It tends to persist for a longer duration but usually resolves
within six weeks after birth. 3. Pathological jaundice: This type of
jaundice is less common and occurs due to an underlying medical
condition that affects bilirubin metabolism or liver function. The
duration of pathological jaundice depends on the specific cause and may
require treatment.
It's important for parents to monitor their newborn's jaundice closely
and seek medical advice if the jaundice progresses rapidly, becomes
severe, or is accompanied by other symptoms such as poor feeding,
lethargy, or excessive sleepiness. In these cases, further evaluation
and management may be necessary. Remember that each baby is unique, and
the timing of jaundice resolution can vary. If you have concerns about
your newborn's jaundice, it's always best to consult with a healthcare
professional for personalized advice and guidance.
model-index:
- name: OpenBioLLM-70B
results: []
quantized_by: andrijdavid
Llama3-OpenBioLLM-70B-GGUF
- Original model: Llama3-OpenBioLLM-70B
Description
This repo contains GGUF format model files for Llama3-OpenBioLLM-70B.
About GGUF
GGUF is a new format introduced by the llama.cpp team on August 21st 2023. It is a replacement for GGML, which is no longer supported by llama.cpp. Here is an incomplete list of clients and libraries that are known to support GGUF:
- llama.cpp. This is the source project for GGUF, providing both a Command Line Interface (CLI) and a server option.
- text-generation-webui, Known as the most widely used web UI, this project boasts numerous features and powerful extensions, and supports GPU acceleration.
- Ollama Ollama is a lightweight and extensible framework designed for building and running language models locally. It features a simple API for creating, managing, and executing models, along with a library of pre-built models for use in various applications
- KoboldCpp, A comprehensive web UI offering GPU acceleration across all platforms and architectures, particularly renowned for storytelling.
- GPT4All, This is a free and open source GUI that runs locally, supporting Windows, Linux, and macOS with full GPU acceleration.
- LM Studio An intuitive and powerful local GUI for Windows and macOS (Silicon), featuring GPU acceleration.
- LoLLMS Web UI. A notable web UI with a variety of unique features, including a comprehensive model library for easy model selection.
- Faraday.dev, An attractive, user-friendly character-based chat GUI for Windows and macOS (both Silicon and Intel), also offering GPU acceleration.
- llama-cpp-python, A Python library equipped with GPU acceleration, LangChain support, and an OpenAI-compatible API server.
- candle, A Rust-based ML framework focusing on performance, including GPU support, and designed for ease of use.
- ctransformers, A Python library featuring GPU acceleration, LangChain support, and an OpenAI-compatible AI server.
- localGPT An open-source initiative enabling private conversations with documents.
Explanation of quantisation methods
Click to see details
The new methods available are:- GGML_TYPE_Q2_K - "type-1" 2-bit quantization in super-blocks containing 16 blocks, each block having 16 weight. Block scales and mins are quantized with 4 bits. This ends up effectively using 2.5625 bits per weight (bpw)
- GGML_TYPE_Q3_K - "type-0" 3-bit quantization in super-blocks containing 16 blocks, each block having 16 weights. Scales are quantized with 6 bits. This end up using 3.4375 bpw.
- GGML_TYPE_Q4_K - "type-1" 4-bit quantization in super-blocks containing 8 blocks, each block having 32 weights. Scales and mins are quantized with 6 bits. This ends up using 4.5 bpw.
- GGML_TYPE_Q5_K - "type-1" 5-bit quantization. Same super-block structure as GGML_TYPE_Q4_K resulting in 5.5 bpw
- GGML_TYPE_Q6_K - "type-0" 6-bit quantization. Super-blocks with 16 blocks, each block having 16 weights. Scales are quantized with 8 bits. This ends up using 6.5625 bpw.
How to download GGUF files
Note for manual downloaders: You almost never want to clone the entire repo! Multiple different quantisation formats are provided, and most users only want to pick and download a single folder.
The following clients/libraries will automatically download models for you, providing a list of available models to choose from:
- LM Studio
- LoLLMS Web UI
- Faraday.dev
In text-generation-webui
Under Download Model, you can enter the model repo: LiteLLMs/Llama3-OpenBioLLM-70B-GGUF and below it, a specific filename to download, such as: Q4_0/Q4_0-00001-of-00009.gguf.
Then click Download.
On the command line, including multiple files at once
I recommend using the huggingface-hub Python library:
pip3 install huggingface-hub
Then you can download any individual model file to the current directory, at high speed, with a command like this:
huggingface-cli download LiteLLMs/Llama3-OpenBioLLM-70B-GGUF Q4_0/Q4_0-00001-of-00009.gguf --local-dir . --local-dir-use-symlinks False
More advanced huggingface-cli download usage (click to read)
You can also download multiple files at once with a pattern:
huggingface-cli download LiteLLMs/Llama3-OpenBioLLM-70B-GGUF --local-dir . --local-dir-use-symlinks False --include='*Q4_K*gguf'
For more documentation on downloading with huggingface-cli, please see: HF -> Hub Python Library -> Download files -> Download from the CLI.
To accelerate downloads on fast connections (1Gbit/s or higher), install hf_transfer:
pip3 install huggingface_hub[hf_transfer]
And set environment variable HF_HUB_ENABLE_HF_TRANSFER to 1:
HF_HUB_ENABLE_HF_TRANSFER=1 huggingface-cli download LiteLLMs/Llama3-OpenBioLLM-70B-GGUF Q4_0/Q4_0-00001-of-00009.gguf --local-dir . --local-dir-use-symlinks False
Windows Command Line users: You can set the environment variable by running set HF_HUB_ENABLE_HF_TRANSFER=1 before the download command.
Make sure you are using llama.cpp from commit d0cee0d or later.
./main -ngl 35 -m Q4_0/Q4_0-00001-of-00009.gguf --color -c 8192 --temp 0.7 --repeat_penalty 1.1 -n -1 -p "<PROMPT>"
Change -ngl 32 to the number of layers to offload to GPU. Remove it if you don't have GPU acceleration.
Change -c 8192 to the desired sequence length. For extended sequence models - eg 8K, 16K, 32K - the necessary RoPE scaling parameters are read from the GGUF file and set by llama.cpp automatically. Note that longer sequence lengths require much more resources, so you may need to reduce this value.
If you want to have a chat-style conversation, replace the -p <PROMPT> argument with -i -ins
For other parameters and how to use them, please refer to the llama.cpp documentation
How to run in text-generation-webui
Further instructions can be found in the text-generation-webui documentation, here: text-generation-webui/docs/04 ‐ Model Tab.md.
How to run from Python code
You can use GGUF models from Python using the llama-cpp-python or ctransformers libraries. Note that at the time of writing (Nov 27th 2023), ctransformers has not been updated for some time and is not compatible with some recent models. Therefore I recommend you use llama-cpp-python.
How to load this model in Python code, using llama-cpp-python
For full documentation, please see: llama-cpp-python docs.
First install the package
Run one of the following commands, according to your system:
# Base ctransformers with no GPU acceleration
pip install llama-cpp-python
# With NVidia CUDA acceleration
CMAKE_ARGS="-DLLAMA_CUBLAS=on" pip install llama-cpp-python
# Or with OpenBLAS acceleration
CMAKE_ARGS="-DLLAMA_BLAS=ON -DLLAMA_BLAS_VENDOR=OpenBLAS" pip install llama-cpp-python
# Or with CLBLast acceleration
CMAKE_ARGS="-DLLAMA_CLBLAST=on" pip install llama-cpp-python
# Or with AMD ROCm GPU acceleration (Linux only)
CMAKE_ARGS="-DLLAMA_HIPBLAS=on" pip install llama-cpp-python
# Or with Metal GPU acceleration for macOS systems only
CMAKE_ARGS="-DLLAMA_METAL=on" pip install llama-cpp-python
# In windows, to set the variables CMAKE_ARGS in PowerShell, follow this format; eg for NVidia CUDA:
$env:CMAKE_ARGS = "-DLLAMA_OPENBLAS=on"
pip install llama-cpp-python
Simple llama-cpp-python example code
from llama_cpp import Llama
# Set gpu_layers to the number of layers to offload to GPU. Set to 0 if no GPU acceleration is available on your system.
llm = Llama(
model_path="./Q4_0/Q4_0-00001-of-00009.gguf", # Download the model file first
n_ctx=32768, # The max sequence length to use - note that longer sequence lengths require much more resources
n_threads=8, # The number of CPU threads to use, tailor to your system and the resulting performance
n_gpu_layers=35 # The number of layers to offload to GPU, if you have GPU acceleration available
)
# Simple inference example
output = llm(
"<PROMPT>", # Prompt
max_tokens=512, # Generate up to 512 tokens
stop=["</s>"], # Example stop token - not necessarily correct for this specific model! Please check before using.
echo=True # Whether to echo the prompt
)
# Chat Completion API
llm = Llama(model_path="./Q4_0/Q4_0-00001-of-00009.gguf", chat_format="llama-2") # Set chat_format according to the model you are using
llm.create_chat_completion(
messages = [
{"role": "system", "content": "You are a story writing assistant."},
{
"role": "user",
"content": "Write a story about llamas."
}
]
)
How to use with LangChain
Here are guides on using llama-cpp-python and ctransformers with LangChain:
Original model card: Llama3-OpenBioLLM-70B

Advancing Open-source Large Language Models in Medical Domain
Online Demo
|
 GitHub
|
GitHub
|
 Paper
|
Paper
|
 Discord
Discord
Introducing OpenBioLLM-70B: A State-of-the-Art Open Source Biomedical Large Language Model
OpenBioLLM-70B is an advanced open source language model designed specifically for the biomedical domain. Developed by Saama AI Labs, this model leverages cutting-edge techniques to achieve state-of-the-art performance on a wide range of biomedical tasks.
🏥 Biomedical Specialization: OpenBioLLM-70B is tailored for the unique language and knowledge requirements of the medical and life sciences fields. It was fine-tuned on a vast corpus of high-quality biomedical data, enabling it to understand and generate text with domain-specific accuracy and fluency.
🎓 Superior Performance: With 70 billion parameters, OpenBioLLM-70B outperforms other open source biomedical language models of similar scale. It has also demonstrated better results compared to larger proprietary & open-source models like GPT-4, Gemini, Meditron-70B, Med-PaLM-1 & Med-PaLM-2 on biomedical benchmarks.
🧠 Advanced Training Techniques: OpenBioLLM-70B builds upon the powerful foundations of the Meta-Llama-3-70B-Instruct and Meta-Llama-3-70B-Instruct models. It incorporates the DPO dataset and fine-tuning recipe along with a custom diverse medical instruction dataset. Key components of the training pipeline include:

- Policy Optimization: Direct Preference Optimization: Your Language Model is Secretly a Reward Model (DPO)
- Fine-tuning dataset: Custom Medical Instruct dataset (We plan to release a sample training dataset in our upcoming paper; please stay updated)
This combination of cutting-edge techniques enables OpenBioLLM-70B to align with key capabilities and preferences for biomedical applications.
⚙️ Release Details:
- Model Size: 70 billion parameters
- Quantization: Optimized quantized versions available Here
- Language(s) (NLP): en
- Developed By: Ankit Pal (Aaditya Ura) from Saama AI Labs
- License: Meta-Llama License
- Fine-tuned from models: Meta-Llama-3-70B-Instruct
- Resources for more information:
- Paper: Coming soon
The model can be fine-tuned for more specialized tasks and datasets as needed.
OpenBioLLM-70B represents an important step forward in democratizing advanced language AI for the biomedical community. By leveraging state-of-the-art architectures and training techniques from leading open source efforts like Llama-3, we have created a powerful tool to accelerate innovation and discovery in healthcare and the life sciences.
We are excited to share OpenBioLLM-70B with researchers and developers around the world.
Use with transformers
Important: Please use the exact chat template provided by Llama-3 instruct version. Otherwise there will be a degradation in the performance. The model output can be verbose in rare cases. Please consider setting temperature = 0 to make this happen less.
See the snippet below for usage with Transformers:
import transformers
import torch
model_id = "aaditya/OpenBioLLM-Llama3-70B"
pipeline = transformers.pipeline(
"text-generation",
model=model_id,
model_kwargs={"torch_dtype": torch.bfloat16},
device="auto",
)
messages = [
{"role": "system", "content": "You are an expert and experienced from the healthcare and biomedical domain with extensive medical knowledge and practical experience. Your name is OpenBioLLM, and you were developed by Saama AI Labs. who's willing to help answer the user's query with explanation. In your explanation, leverage your deep medical expertise such as relevant anatomical structures, physiological processes, diagnostic criteria, treatment guidelines, or other pertinent medical concepts. Use precise medical terminology while still aiming to make the explanation clear and accessible to a general audience."},
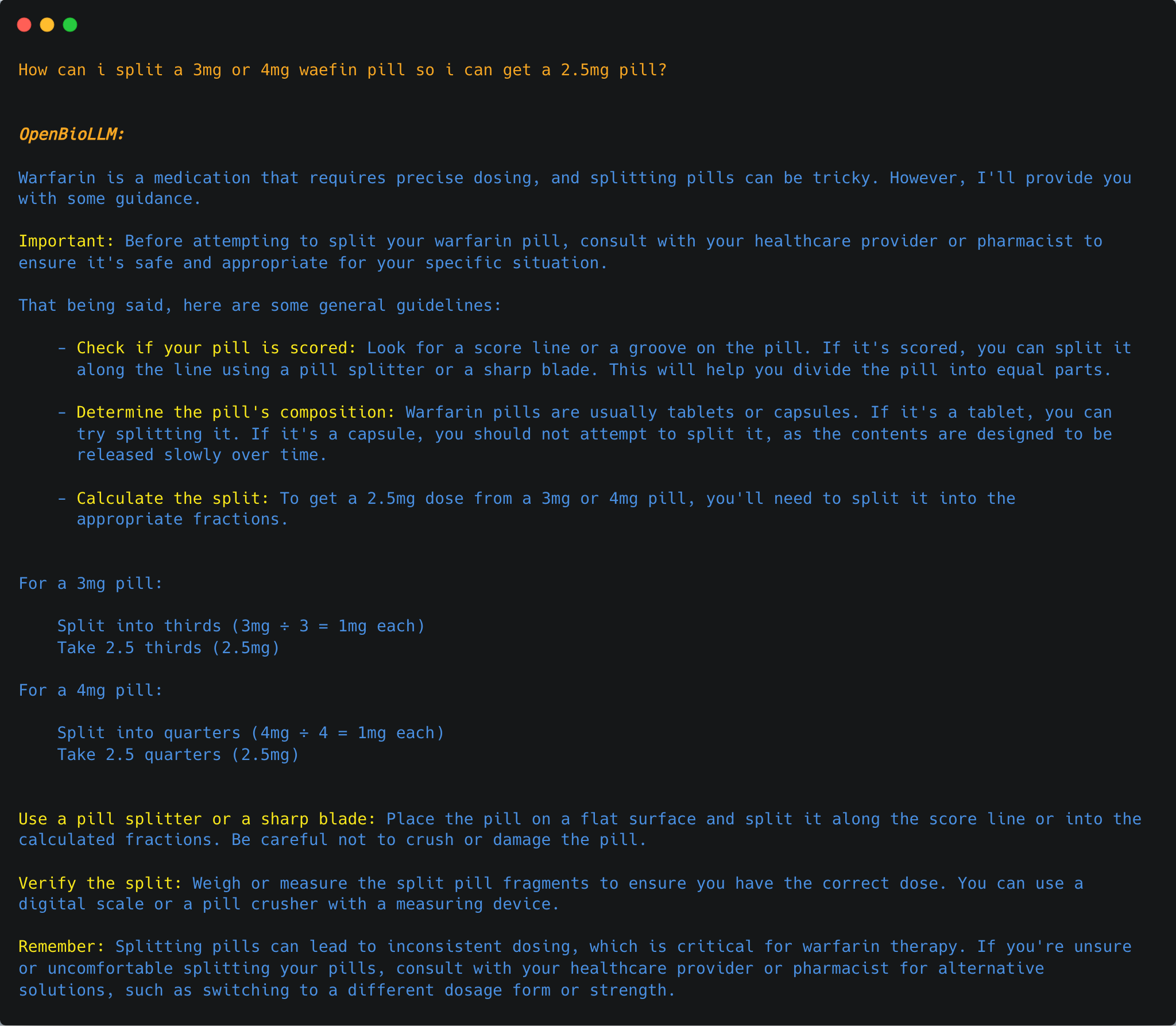
{"role": "user", "content": "How can i split a 3mg or 4mg waefin pill so i can get a 2.5mg pill?"},
]
prompt = pipeline.tokenizer.apply_chat_template(
messages,
tokenize=False,
add_generation_prompt=True
)
terminators = [
pipeline.tokenizer.eos_token_id,
pipeline.tokenizer.convert_tokens_to_ids("<|eot_id|>")
]
outputs = pipeline(
prompt,
max_new_tokens=256,
eos_token_id=terminators,
do_sample=True,
temperature=0.0,
top_p=0.9,
)
print(outputs[0]["generated_text"][len(prompt):])
Training procedure
Training hyperparameters
Click to see details
- learning_rate: 0.0002
- lr_scheduler: cosine
- train_batch_size: 12
- eval_batch_size: 8
- GPU: H100 80GB SXM5
- num_devices: 8
- optimizer: adamw_bnb_8bit
- lr_scheduler_warmup_steps: 100
- num_epochs: 4
Peft hyperparameters
Click to see details
- adapter: qlora
- lora_r: 128
- lora_alpha: 256
- lora_dropout: 0.05
- lora_target_linear: true
-lora_target_modules:
- q_proj
- v_proj
- k_proj
- o_proj
- gate_proj
- down_proj
- up_proj
Training results
Framework versions
- Transformers 4.39.3
- Pytorch 2.1.2+cu121
- Datasets 2.18.0
- Tokenizers 0.15.1
- Axolotl
- Lm harness for evaluation
Benchmark Results
🔥 OpenBioLLM-70B demonstrates superior performance compared to larger models, such as GPT-4, Gemini, Meditron-70B, Med-PaLM-1 & Med-PaLM-2 across 9 diverse biomedical datasets, achieving state-of-the-art results with an average score of 86.06%, despite having a significantly smaller parameter count. The model's strong performance in domain-specific tasks, such as Clinical KG, Medical Genetics, and PubMedQA, highlights its ability to effectively capture and apply biomedical knowledge.
🚨 The GPT-4, Med-PaLM-1, and Med-PaLM-2 results are taken from their official papers. Since Med-PaLM doesn't provide zero-shot accuracy, we are using 5-shot accuracy from their paper for comparison. All results presented are in the zero-shot setting, except for Med-PaLM-2 and Med-PaLM-1, which use 5-shot accuracy.
| | Clinical KG | Medical Genetics | Anatomy | Pro Medicine | College Biology | College Medicine | MedQA 4 opts | PubMedQA | MedMCQA | Avg | | | - | | - | | | OpenBioLLM-70B | 92.93 | 93.197 | 83.904 | 93.75 | 93.827 | 85.749 | 78.162 | 78.97 | 74.014 | 86.05588 | | Med-PaLM-2 (5-shot) | 88.3 | 90 | 77.8 | 95.2 | 94.4 | 80.9 | 79.7 | 79.2 | 71.3 | 84.08 | | GPT-4 | 86.04 | 91 | 80 | 93.01 | 95.14 | 76.88 | 78.87 | 75.2 | 69.52 | 82.85 | | Med-PaLM-1 (Flan-PaLM, 5-shot) | 80.4 | 75 | 63.7 | 83.8 | 88.9 | 76.3 | 67.6 | 79 | 57.6 | 74.7 | | OpenBioLLM-8B | 76.101 | 86.1 | 69.829 | 78.21 | 84.213 | 68.042 | 58.993 | 74.12 | 56.913 | 72.502 | | Gemini-1.0 | 76.7 | 75.8 | 66.7 | 77.7 | 88 | 69.2 | 58 | 70.7 | 54.3 | 70.79 | | GPT-3.5 Turbo 1106 | 74.71 | 74 | 72.79 | 72.79 | 72.91 | 64.73 | 57.71 | 72.66 | 53.79 | 66 | | Meditron-70B | 66.79 | 69 | 53.33 | 71.69 | 76.38 | 63 | 57.1 | 76.6 | 46.85 | 64.52 | | gemma-7b | 69.81 | 70 | 59.26 | 66.18 | 79.86 | 60.12 | 47.21 | 76.2 | 48.96 | 64.18 | | Mistral-7B-v0.1 | 68.68 | 71 | 55.56 | 68.38 | 68.06 | 59.54 | 50.82 | 75.4 | 48.2 | 62.85 | | Apollo-7B | 62.26 | 72 | 61.48 | 69.12 | 70.83 | 55.49 | 55.22 | 39.8 | 53.77 | 60 | | MedAlpaca-7b | 57.36 | 69 | 57.04 | 67.28 | 65.28 | 54.34 | 41.71 | 72.8 | 37.51 | 58.03 | | BioMistral-7B | 59.9 | 64 | 56.5 | 60.4 | 59 | 54.7 | 50.6 | 77.5 | 48.1 | 57.3 | | AlpaCare-llama2-7b | 49.81 | 49 | 45.92 | 33.82 | 50 | 43.35 | 29.77 | 72.2 | 34.42 | 45.36 | | ClinicalGPT | 30.56 | 27 | 30.37 | 19.48 | 25 | 24.27 | 26.08 | 63.8 | 28.18 | 30.52 |
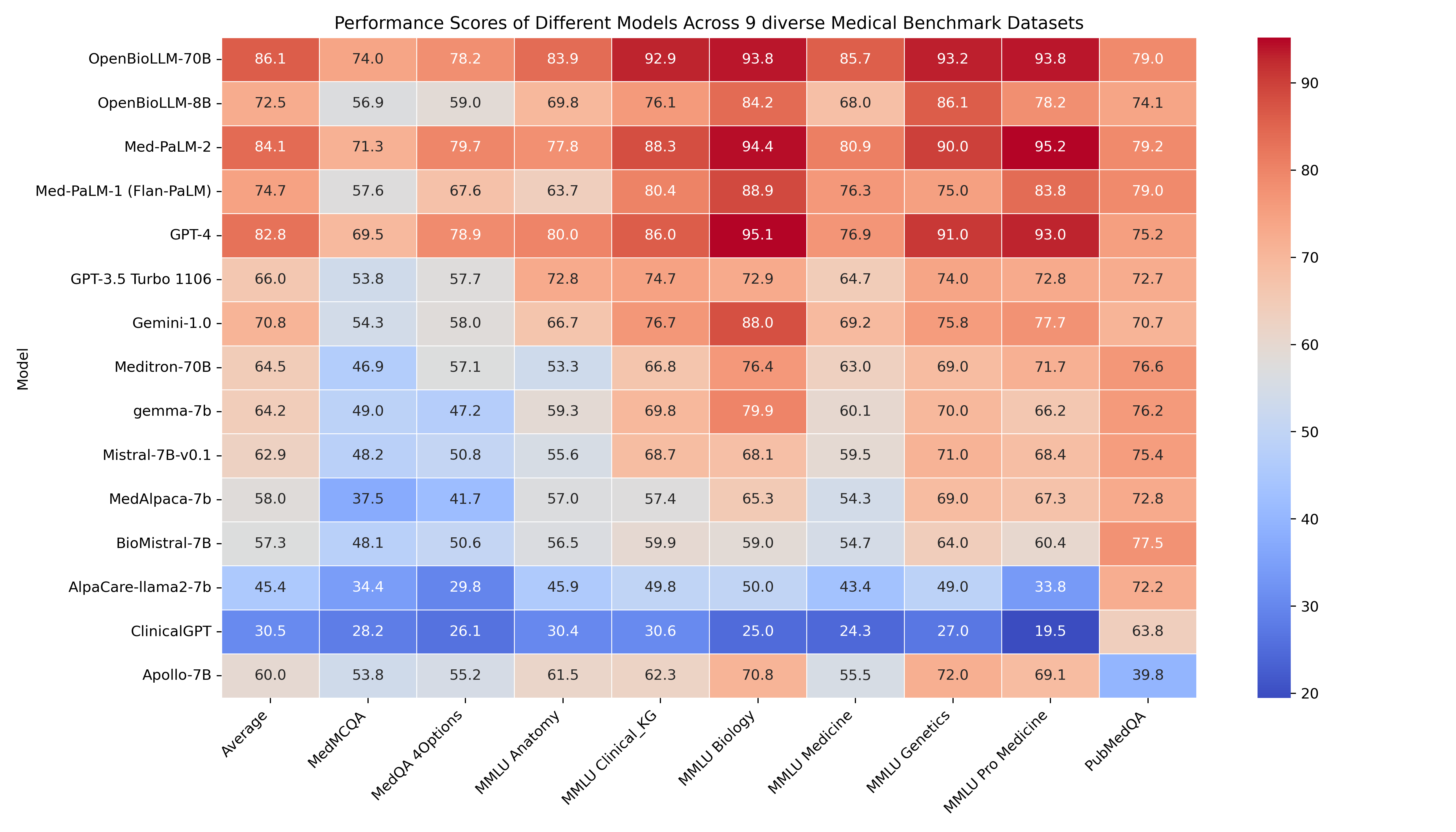
Detailed Medical Subjectwise accuracy
Use Cases & Examples
🚨 **Below results are from the quantized version of OpenBioLLM-70B
Summarize Clinical Notes
OpenBioLLM-70B can efficiently analyze and summarize complex clinical notes, EHR data, and discharge summaries, extracting key information and generating concise, structured summaries
Answer Medical Questions
OpenBioLLM-70B can provide answers to a wide range of medical questions.
Clinical Entity Recognition
OpenBioLLM-70B can perform advanced clinical entity recognition by identifying and extracting key medical concepts, such as diseases, symptoms, medications, procedures, and anatomical structures, from unstructured clinical text. By leveraging its deep understanding of medical terminology and context, the model can accurately annotate and categorize clinical entities, enabling more efficient information retrieval, data analysis, and knowledge discovery from electronic health records, research articles, and other biomedical text sources. This capability can support various downstream applications, such as clinical decision support, pharmacovigilance, and medical research.
Biomarkers Extraction
Classification
OpenBioLLM-70B can perform various biomedical classification tasks, such as disease prediction, sentiment analysis, medical document categorization
De-Identification
OpenBioLLM-70B can detect and remove personally identifiable information (PII) from medical records, ensuring patient privacy and compliance with data protection regulations like HIPAA.
Advisory Notice!
While OpenBioLLM-70B leverages high-quality data sources, its outputs may still contain inaccuracies, biases, or misalignments that could pose risks if relied upon for medical decision-making without further testing and refinement. The model's performance has not yet been rigorously evaluated in randomized controlled trials or real-world healthcare environments.
Therefore, we strongly advise against using OpenBioLLM-70B for any direct patient care, clinical decision support, or other professional medical purposes at this time. Its use should be limited to research, development, and exploratory applications by qualified individuals who understand its limitations. OpenBioLLM-70B is intended solely as a research tool to assist healthcare professionals and should never be considered a replacement for the professional judgment and expertise of a qualified medical doctor.
Appropriately adapting and validating OpenBioLLM-70B for specific medical use cases would require significant additional work, potentially including:
- Thorough testing and evaluation in relevant clinical scenarios
- Alignment with evidence-based guidelines and best practices
- Mitigation of potential biases and failure modes
- Integration with human oversight and interpretation
- Compliance with regulatory and ethical standards
Always consult a qualified healthcare provider for personal medical needs.
Citation
If you find OpenBioLLM-70B & 8B useful in your work, please cite the model as follows:
@misc{OpenBioLLMs,
author = {Ankit Pal, Malaikannan Sankarasubbu},
title = {OpenBioLLMs: Advancing Open-Source Large Language Models for Healthcare and Life Sciences},
year = {2024},
publisher = {Hugging Face},
journal = {Hugging Face repository},
howpublished = {\url{https://huggingface.co/aaditya/OpenBioLLM-Llama3-70B}}
}
The accompanying paper is currently in progress and will be released soon.
💌 Contact
We look forward to hearing you and collaborating on this exciting project!
Contributors:
- Ankit Pal (Aaditya Ura) [aadityaura at gmail dot com]
- Saama AI Labs
- Note: I am looking for a funded PhD opportunity, especially if it fits my Responsible Generative AI, Multimodal LLMs, Geometric Deep Learning, and Healthcare AI skillset.
References
We thank the Meta Team for their amazing models!
Result sources
- [1] GPT-4 [Capabilities of GPT-4 on Medical Challenge Problems] (https://arxiv.org/abs/2303.13375)
- [2] Med-PaLM-1 Large Language Models Encode Clinical Knowledge
- [3] Med-PaLM-2 Towards Expert-Level Medical Question Answering with Large Language Models
- [4] Gemini-1.0 Gemini Goes to Med School