Update README.md
Browse files
README.md
CHANGED
|
@@ -180,7 +180,6 @@ inference: false
|
|
| 180 |
datasets:
|
| 181 |
- owkin/camelyon16-features
|
| 182 |
- owkin/nct-crc-he
|
| 183 |
-
- 1aurent/NCT-CRC-HE
|
| 184 |
metrics:
|
| 185 |
- roc_auc
|
| 186 |
---
|
|
@@ -188,21 +187,26 @@ metrics:
|
|
| 188 |
# Model card for vit_base_patch16_224.owkin_pancancer
|
| 189 |
|
| 190 |
A Vision Transformer (ViT) image classification model. \
|
| 191 |
-
Trained by Owkin on
|
|
|
|
|
|
|
| 192 |
|
| 193 |
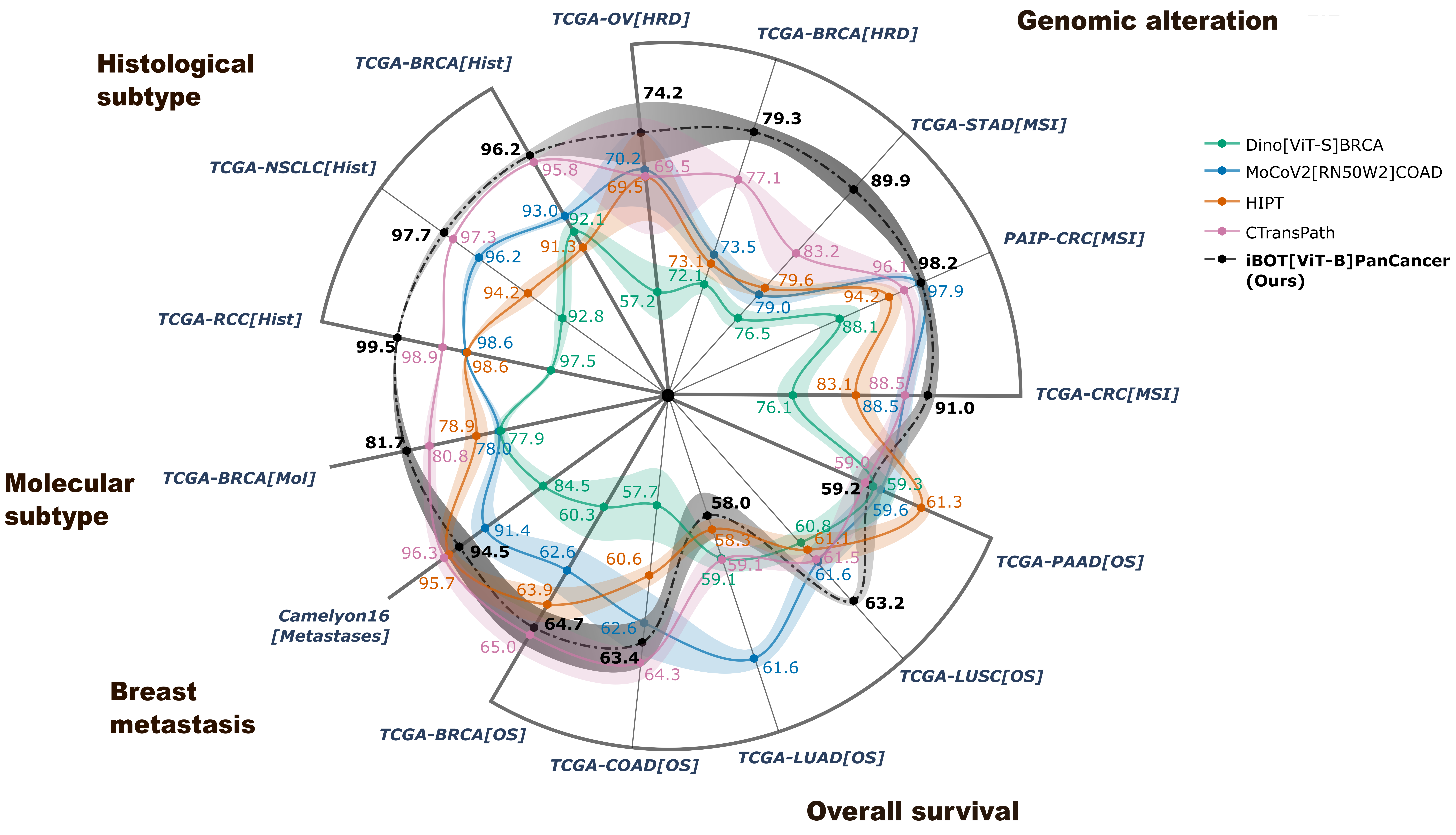
|
| 194 |
|
| 195 |
## Model Details
|
| 196 |
|
| 197 |
- **Model Type:** Feature backbone
|
|
|
|
|
|
|
| 198 |
- **Model Stats:**
|
| 199 |
-
- Params
|
| 200 |
- Image size: 224 x 224 x 3
|
|
|
|
| 201 |
- **Papers:**
|
| 202 |
-
- Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling
|
| 203 |
-
- **Dataset:**
|
| 204 |
-
- **Original:** https://github.com/owkin/HistoSSLscaling
|
| 205 |
-
- **License:** https://github.com/owkin/HistoSSLscaling/blob/main/LICENSE.txt
|
| 206 |
|
| 207 |
## Model Usage
|
| 208 |
|
|
@@ -229,7 +233,8 @@ model = timm.create_model(
|
|
| 229 |
data_config = timm.data.resolve_model_data_config(model)
|
| 230 |
transforms = timm.data.create_transform(**data_config, is_training=False)
|
| 231 |
|
| 232 |
-
|
|
|
|
| 233 |
```
|
| 234 |
|
| 235 |
## Citation
|
|
|
|
| 180 |
datasets:
|
| 181 |
- owkin/camelyon16-features
|
| 182 |
- owkin/nct-crc-he
|
|
|
|
| 183 |
metrics:
|
| 184 |
- roc_auc
|
| 185 |
---
|
|
|
|
| 187 |
# Model card for vit_base_patch16_224.owkin_pancancer
|
| 188 |
|
| 189 |
A Vision Transformer (ViT) image classification model. \
|
| 190 |
+
Trained by Owkin on 40 million pan-cancer histology tiles from TCGA-COAD.
|
| 191 |
+
|
| 192 |
+
A version using the transformers library is also available here: https://huggingface.co/owkin/phikon
|
| 193 |
|
| 194 |

|
| 195 |
|
| 196 |
## Model Details
|
| 197 |
|
| 198 |
- **Model Type:** Feature backbone
|
| 199 |
+
- **Developed by**: Owkin
|
| 200 |
+
- **Funded by**: Owkin and IDRIS
|
| 201 |
- **Model Stats:**
|
| 202 |
+
- Params: 85.8M (base)
|
| 203 |
- Image size: 224 x 224 x 3
|
| 204 |
+
- Patch size: 16 x 16 x 3
|
| 205 |
- **Papers:**
|
| 206 |
+
- [Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling](https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v2)
|
| 207 |
+
- **Dataset:** Pancancer40M, created from [TCGA-COAD](https://portal.gdc.cancer.gov/repository?facetTab=cases&filters=%7B%22content%22%3A%5B%7B%22content%22%3A%7B%22field%22%3A%22cases.project.project_id%22%2C%22value%22%3A%5B%22TCGA-COAD%22%5D%7D%2C%22op%22%3A%22in%22%7D%2C%7B%22content%22%3A%7B%22field%22%3A%22files.experimental_strategy%22%2C%22value%22%3A%5B%22Diagnostic%20Slide%22%5D%7D%2C%22op%22%3A%22in%22%7D%5D%2C%22op%22%3A%22and%22%7D&searchTableTab=cases)
|
| 208 |
+
- **Original:** https://github.com/owkin/HistoSSLscaling
|
| 209 |
+
- **License:** [Owkin non-commercial license](https://github.com/owkin/HistoSSLscaling/blob/main/LICENSE.txt)
|
| 210 |
|
| 211 |
## Model Usage
|
| 212 |
|
|
|
|
| 233 |
data_config = timm.data.resolve_model_data_config(model)
|
| 234 |
transforms = timm.data.create_transform(**data_config, is_training=False)
|
| 235 |
|
| 236 |
+
input = transforms(img).unsqueeze(0) # (batch_size, num_channels, img_size, img_size) shaped tensor
|
| 237 |
+
output = model(input) # (batch_size, num_features) shaped tensor
|
| 238 |
```
|
| 239 |
|
| 240 |
## Citation
|