Spaces:
Running
on
T4
Running
on
T4
Simon Duerr
commited on
Commit
•
e1a6cd9
1
Parent(s):
4aee85f
add server
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- LICENSE +21 -0
- ProteinMPNN/.gitignore +129 -0
- ProteinMPNN/LICENSE +21 -0
- ProteinMPNN/README.md +151 -0
- ProteinMPNN/colab_notebooks/README.md +1 -0
- ProteinMPNN/colab_notebooks/quickdemo.ipynb +322 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/assigned_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/fixed_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/seqs/4YOW.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_3_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/assigned_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/fixed_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/seqs/4YOW.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/assigned_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/fixed_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/seqs/4YOW.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/tied_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/bias_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/seqs/4YOW.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/pdbs/3HTN.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/pdbs/4YOW.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/seqs/4GYT.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/seqs/6EHB.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/tied_pdbs.jsonl +1 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/pdbs/4GYT.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/pdbs/6EHB.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/parsed_pdbs.jsonl +2 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/seqs/5L33.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/seqs/6MRR.fa +6 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/pdbs/5L33.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/pdbs/6MRR.pdb +0 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_1.sh +27 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_2.sh +32 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_3.sh +26 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_4.sh +39 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_5.sh +43 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_6.sh +33 -0
- ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_7.sh +40 -0
- ProteinMPNN/vanilla_proteinmpnn/helper_scripts/assign_fixed_chains.py +39 -0
- ProteinMPNN/vanilla_proteinmpnn/helper_scripts/make_bias_AA.py +27 -0
- ProteinMPNN/vanilla_proteinmpnn/helper_scripts/make_bias_per_res_dict.py +53 -0
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2022 Justas Dauparas, Simon Duerr
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
ProteinMPNN/.gitignore
ADDED
|
@@ -0,0 +1,129 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Byte-compiled / optimized / DLL files
|
| 2 |
+
__pycache__/
|
| 3 |
+
*.py[cod]
|
| 4 |
+
*$py.class
|
| 5 |
+
|
| 6 |
+
# C extensions
|
| 7 |
+
*.so
|
| 8 |
+
|
| 9 |
+
# Distribution / packaging
|
| 10 |
+
.Python
|
| 11 |
+
build/
|
| 12 |
+
develop-eggs/
|
| 13 |
+
dist/
|
| 14 |
+
downloads/
|
| 15 |
+
eggs/
|
| 16 |
+
.eggs/
|
| 17 |
+
lib/
|
| 18 |
+
lib64/
|
| 19 |
+
parts/
|
| 20 |
+
sdist/
|
| 21 |
+
var/
|
| 22 |
+
wheels/
|
| 23 |
+
pip-wheel-metadata/
|
| 24 |
+
share/python-wheels/
|
| 25 |
+
*.egg-info/
|
| 26 |
+
.installed.cfg
|
| 27 |
+
*.egg
|
| 28 |
+
MANIFEST
|
| 29 |
+
|
| 30 |
+
# PyInstaller
|
| 31 |
+
# Usually these files are written by a python script from a template
|
| 32 |
+
# before PyInstaller builds the exe, so as to inject date/other infos into it.
|
| 33 |
+
*.manifest
|
| 34 |
+
*.spec
|
| 35 |
+
|
| 36 |
+
# Installer logs
|
| 37 |
+
pip-log.txt
|
| 38 |
+
pip-delete-this-directory.txt
|
| 39 |
+
|
| 40 |
+
# Unit test / coverage reports
|
| 41 |
+
htmlcov/
|
| 42 |
+
.tox/
|
| 43 |
+
.nox/
|
| 44 |
+
.coverage
|
| 45 |
+
.coverage.*
|
| 46 |
+
.cache
|
| 47 |
+
nosetests.xml
|
| 48 |
+
coverage.xml
|
| 49 |
+
*.cover
|
| 50 |
+
*.py,cover
|
| 51 |
+
.hypothesis/
|
| 52 |
+
.pytest_cache/
|
| 53 |
+
|
| 54 |
+
# Translations
|
| 55 |
+
*.mo
|
| 56 |
+
*.pot
|
| 57 |
+
|
| 58 |
+
# Django stuff:
|
| 59 |
+
*.log
|
| 60 |
+
local_settings.py
|
| 61 |
+
db.sqlite3
|
| 62 |
+
db.sqlite3-journal
|
| 63 |
+
|
| 64 |
+
# Flask stuff:
|
| 65 |
+
instance/
|
| 66 |
+
.webassets-cache
|
| 67 |
+
|
| 68 |
+
# Scrapy stuff:
|
| 69 |
+
.scrapy
|
| 70 |
+
|
| 71 |
+
# Sphinx documentation
|
| 72 |
+
docs/_build/
|
| 73 |
+
|
| 74 |
+
# PyBuilder
|
| 75 |
+
target/
|
| 76 |
+
|
| 77 |
+
# Jupyter Notebook
|
| 78 |
+
.ipynb_checkpoints
|
| 79 |
+
|
| 80 |
+
# IPython
|
| 81 |
+
profile_default/
|
| 82 |
+
ipython_config.py
|
| 83 |
+
|
| 84 |
+
# pyenv
|
| 85 |
+
.python-version
|
| 86 |
+
|
| 87 |
+
# pipenv
|
| 88 |
+
# According to pypa/pipenv#598, it is recommended to include Pipfile.lock in version control.
|
| 89 |
+
# However, in case of collaboration, if having platform-specific dependencies or dependencies
|
| 90 |
+
# having no cross-platform support, pipenv may install dependencies that don't work, or not
|
| 91 |
+
# install all needed dependencies.
|
| 92 |
+
#Pipfile.lock
|
| 93 |
+
|
| 94 |
+
# PEP 582; used by e.g. github.com/David-OConnor/pyflow
|
| 95 |
+
__pypackages__/
|
| 96 |
+
|
| 97 |
+
# Celery stuff
|
| 98 |
+
celerybeat-schedule
|
| 99 |
+
celerybeat.pid
|
| 100 |
+
|
| 101 |
+
# SageMath parsed files
|
| 102 |
+
*.sage.py
|
| 103 |
+
|
| 104 |
+
# Environments
|
| 105 |
+
.env
|
| 106 |
+
.venv
|
| 107 |
+
env/
|
| 108 |
+
venv/
|
| 109 |
+
ENV/
|
| 110 |
+
env.bak/
|
| 111 |
+
venv.bak/
|
| 112 |
+
|
| 113 |
+
# Spyder project settings
|
| 114 |
+
.spyderproject
|
| 115 |
+
.spyproject
|
| 116 |
+
|
| 117 |
+
# Rope project settings
|
| 118 |
+
.ropeproject
|
| 119 |
+
|
| 120 |
+
# mkdocs documentation
|
| 121 |
+
/site
|
| 122 |
+
|
| 123 |
+
# mypy
|
| 124 |
+
.mypy_cache/
|
| 125 |
+
.dmypy.json
|
| 126 |
+
dmypy.json
|
| 127 |
+
|
| 128 |
+
# Pyre type checker
|
| 129 |
+
.pyre/
|
ProteinMPNN/LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2022 Justas Dauparas
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
ProteinMPNN/README.md
ADDED
|
@@ -0,0 +1,151 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# ProteinMPNN
|
| 2 |
+
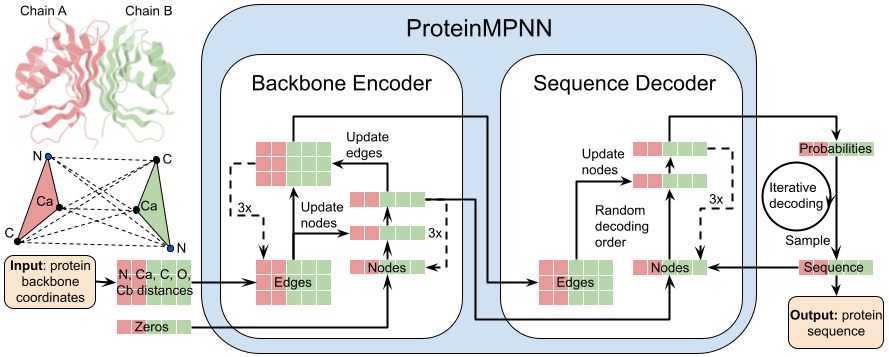
|
| 3 |
+
Read [ProteinMPNN paper](https://www.biorxiv.org/content/10.1101/2022.06.03.494563v1).
|
| 4 |
+
|
| 5 |
+
To run ProteinMPNN clone this github repo and install Python>=3.0, PyTorch, Numpy.
|
| 6 |
+
|
| 7 |
+
Full protein backbone models: `vanilla_proteinmpnn`.
|
| 8 |
+
|
| 9 |
+
Code organization:
|
| 10 |
+
* `vanilla_proteinmpnn/protein_mpnn_run.py` - the main script to initialialize and run the model.
|
| 11 |
+
* `vanilla_proteinmpnn/protein_mpnn_utils.py` - utility functions for the main script.
|
| 12 |
+
* `vanilla_proteinmpnn/helper_scripts/` - helper functions to parse PDBs, assign which chains to design, which residues to fix, adding AA bias, tying residues etc.
|
| 13 |
+
* `vanilla_proteinmpnn/examples/` - simple code examples.
|
| 14 |
+
-----------------------------------------------------------------------------------------------------
|
| 15 |
+
Input flags:
|
| 16 |
+
```
|
| 17 |
+
argparser.add_argument("--path_to_model_weights", type=str, default="", help="Path to model weights folder;")
|
| 18 |
+
argparser.add_argument("--model_name", type=str, default="v_48_020", help="ProteinMPNN model name: v_48_002, v_48_010, v_48_020, v_48_030; v_48_010=version with 48 edges 0.10A noise")
|
| 19 |
+
|
| 20 |
+
argparser.add_argument("--save_score", type=int, default=0, help="0 for False, 1 for True; save score=-mean[log_probs] to npy files")
|
| 21 |
+
argparser.add_argument("--save_probs", type=int, default=0, help="0 for False, 1 for True; save MPNN predicted probabilites per position")
|
| 22 |
+
argparser.add_argument("--score_only", type=int, default=0, help="0 for False, 1 for True; score input backbone-sequence pairs")
|
| 23 |
+
argparser.add_argument("--conditional_probs_only", type=int, default=0, help="0 for False, 1 for True; output conditional probabilities p(s_i given the rest of the sequence and backbone)")
|
| 24 |
+
argparser.add_argument("--conditional_probs_only_backbone", type=int, default=0, help="0 for False, 1 for True; if true output conditional probabilities p(s_i given backbone)")
|
| 25 |
+
|
| 26 |
+
argparser.add_argument("--backbone_noise", type=float, default=0.00, help="Standard deviation of Gaussian noise to add to backbone atoms during the inference.")
|
| 27 |
+
argparser.add_argument("--num_seq_per_target", type=int, default=1, help="Number of sequences to generate per target.")
|
| 28 |
+
argparser.add_argument("--batch_size", type=int, default=1, help="Batch size when using GPUs.")
|
| 29 |
+
argparser.add_argument("--max_length", type=int, default=20000, help="Maximum sequence length.")
|
| 30 |
+
argparser.add_argument("--sampling_temp", type=str, default="0.1", help="A string of temperatures, 0.1 0.3 0.5. Sampling temperature for amino acids, T=0.0 means taking argmax, T>>1.0 means sampling randomly.")
|
| 31 |
+
|
| 32 |
+
argparser.add_argument("--out_folder", type=str, help="Path to a folder to output sequences, e.g. /home/out/")
|
| 33 |
+
argparser.add_argument("--pdb_path", type=str, default='', help="Path to a single PDB to be designed.")
|
| 34 |
+
argparser.add_argument("--pdb_path_chains", type=str, default='', help="Define which chains need to be designed for a single PDB.")
|
| 35 |
+
argparser.add_argument("--jsonl_path", type=str, help="Path to a folder with parsed PDBs into jsonl.")
|
| 36 |
+
argparser.add_argument("--chain_id_jsonl",type=str, default='', help="Path to a dictionary specifying which chains need to be designed and which ones are fixed, if not specied all chains will be designed.")
|
| 37 |
+
argparser.add_argument("--fixed_positions_jsonl", type=str, default='', help="Path to a dictionary with fixed positions.")
|
| 38 |
+
argparser.add_argument("--omit_AAs", type=list, default='X', help="Specify which amino acids should be omitted in the generated sequence, e.g. 'AC' would omit alanine and cystine.")
|
| 39 |
+
argparser.add_argument("--bias_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies AA composion bias, e.g. {A: -1.1, F: 0.7} would make A less likely and F more likely.")
|
| 40 |
+
argparser.add_argument("--bias_by_res_jsonl", default='', help="Path to dictionary with per position bias.")
|
| 41 |
+
argparser.add_argument("--omit_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies which amino acids need to be omited from design at specific chain indices.")
|
| 42 |
+
argparser.add_argument("--pssm_jsonl", type=str, default='', help="Path to a dictionary with pssm.")
|
| 43 |
+
argparser.add_argument("--pssm_multi", type=float, default=0.0, help="A value between [0.0, 1.0], 0.0 means do not use pssm, 1.0 ignore MPNN predictions.")
|
| 44 |
+
argparser.add_argument("--pssm_threshold", type=float, default=0.0, help="A value between -inf + inf to restric per position AAs.")
|
| 45 |
+
argparser.add_argument("--pssm_log_odds_flag", type=int, default=0, help="0 for False, 1 for True.")
|
| 46 |
+
argparser.add_argument("--pssm_bias_flag", type=int, default=0, help="0 for False, 1 for True.")
|
| 47 |
+
argparser.add_argument("--tied_positions_jsonl", type=str, default='', help="Path to a dictionary with tied positions for symmetric design.")
|
| 48 |
+
```
|
| 49 |
+
-----------------------------------------------------------------------------------------------------
|
| 50 |
+
Example from `vanilla_proteinmpnn/examples/` to design a single PDB file:
|
| 51 |
+
```
|
| 52 |
+
path_to_PDB="../PDB_complexes/pdbs/3HTN.pdb"
|
| 53 |
+
|
| 54 |
+
output_dir="../PDB_complexes/example_3_outputs"
|
| 55 |
+
if [ ! -d $output_dir ]
|
| 56 |
+
then
|
| 57 |
+
mkdir -p $output_dir
|
| 58 |
+
fi
|
| 59 |
+
|
| 60 |
+
chains_to_design="A B" #design only chains A and B while using the context of other chains
|
| 61 |
+
|
| 62 |
+
python ../protein_mpnn_run.py \
|
| 63 |
+
--pdb_path $path_to_PDB \
|
| 64 |
+
--pdb_path_chains "$chains_to_design" \
|
| 65 |
+
--out_folder $output_dir \
|
| 66 |
+
--num_seq_per_target 2 \
|
| 67 |
+
--sampling_temp "0.1" \
|
| 68 |
+
--batch_size 1
|
| 69 |
+
```
|
| 70 |
+
-----------------------------------------------------------------------------------------------------
|
| 71 |
+
Example from `vanilla_proteinmpnn/examples/` to design some monomers:
|
| 72 |
+
```
|
| 73 |
+
folder_with_pdbs="../PDB_monomers/pdbs/"
|
| 74 |
+
|
| 75 |
+
output_dir="../PDB_monomers/example_1_outputs"
|
| 76 |
+
if [ ! -d $output_dir ]
|
| 77 |
+
then
|
| 78 |
+
mkdir -p $output_dir
|
| 79 |
+
fi
|
| 80 |
+
|
| 81 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 82 |
+
|
| 83 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 84 |
+
|
| 85 |
+
python ../protein_mpnn_run.py \
|
| 86 |
+
--jsonl_path $path_for_parsed_chains \
|
| 87 |
+
--out_folder $output_dir \
|
| 88 |
+
--num_seq_per_target 2 \
|
| 89 |
+
--sampling_temp "0.1" \
|
| 90 |
+
--batch_size 1
|
| 91 |
+
```
|
| 92 |
+
-----------------------------------------------------------------------------------------------------
|
| 93 |
+
Example from `vanilla_proteinmpnn/examples/` to design some homomers:
|
| 94 |
+
```
|
| 95 |
+
folder_with_pdbs="../PDB_homooligomers/pdbs/"
|
| 96 |
+
|
| 97 |
+
output_dir="../PDB_homooligomers/example_6_outputs"
|
| 98 |
+
if [ ! -d $output_dir ]
|
| 99 |
+
then
|
| 100 |
+
mkdir -p $output_dir
|
| 101 |
+
fi
|
| 102 |
+
|
| 103 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 104 |
+
path_for_tied_positions=$output_dir"/tied_pdbs.jsonl"
|
| 105 |
+
|
| 106 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 107 |
+
|
| 108 |
+
python ../helper_scripts/make_tied_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_tied_positions --homooligomer 1
|
| 109 |
+
|
| 110 |
+
python ../protein_mpnn_run.py \
|
| 111 |
+
--jsonl_path $path_for_parsed_chains \
|
| 112 |
+
--tied_positions_jsonl $path_for_tied_positions \
|
| 113 |
+
--out_folder $output_dir \
|
| 114 |
+
--num_seq_per_target 2 \
|
| 115 |
+
--sampling_temp "0.2" \
|
| 116 |
+
--batch_size 1
|
| 117 |
+
```
|
| 118 |
+
-----------------------------------------------------------------------------------------------------
|
| 119 |
+
Example from `vanilla_proteinmpnn/examples/` to design some complexes:
|
| 120 |
+
```
|
| 121 |
+
folder_with_pdbs="../PDB_complexes/pdbs/"
|
| 122 |
+
|
| 123 |
+
output_dir="../PDB_complexes/example_4_outputs"
|
| 124 |
+
if [ ! -d $output_dir ]
|
| 125 |
+
then
|
| 126 |
+
mkdir -p $output_dir
|
| 127 |
+
fi
|
| 128 |
+
|
| 129 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 130 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 131 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 132 |
+
chains_to_design="A C"
|
| 133 |
+
#The first amino acid in the chain corresponds to 1 and not PDB residues index for now.
|
| 134 |
+
fixed_positions="1 2 3 4 5 6 7 8 23 25, 10 11 12 13 14 15 16 17 18 19 20 40" #fixing/not designing residues 1 2 3...25 in chain A and residues 10 11 12...40 in chain C
|
| 135 |
+
|
| 136 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 137 |
+
|
| 138 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 139 |
+
|
| 140 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$fixed_positions"
|
| 141 |
+
|
| 142 |
+
python ../protein_mpnn_run.py \
|
| 143 |
+
--jsonl_path $path_for_parsed_chains \
|
| 144 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 145 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 146 |
+
--out_folder $output_dir \
|
| 147 |
+
--num_seq_per_target 2 \
|
| 148 |
+
--sampling_temp "0.1" \
|
| 149 |
+
--batch_size 1
|
| 150 |
+
```
|
| 151 |
+
|
ProteinMPNN/colab_notebooks/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
<a href="https://colab.research.google.com/github/dauparas/ProteinMPNN/blob/main/colab_notebooks/quickdemo.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
|
ProteinMPNN/colab_notebooks/quickdemo.ipynb
ADDED
|
@@ -0,0 +1,322 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"nbformat": 4,
|
| 3 |
+
"nbformat_minor": 0,
|
| 4 |
+
"metadata": {
|
| 5 |
+
"colab": {
|
| 6 |
+
"name": "quickdemo.ipynb",
|
| 7 |
+
"provenance": [],
|
| 8 |
+
"include_colab_link": true
|
| 9 |
+
},
|
| 10 |
+
"kernelspec": {
|
| 11 |
+
"name": "python3",
|
| 12 |
+
"display_name": "Python 3"
|
| 13 |
+
},
|
| 14 |
+
"language_info": {
|
| 15 |
+
"name": "python"
|
| 16 |
+
}
|
| 17 |
+
},
|
| 18 |
+
"cells": [
|
| 19 |
+
{
|
| 20 |
+
"cell_type": "markdown",
|
| 21 |
+
"metadata": {
|
| 22 |
+
"id": "view-in-github",
|
| 23 |
+
"colab_type": "text"
|
| 24 |
+
},
|
| 25 |
+
"source": [
|
| 26 |
+
"<a href=\"https://colab.research.google.com/github/dauparas/ProteinMPNN/blob/main/colab_notebooks/quickdemo.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>"
|
| 27 |
+
]
|
| 28 |
+
},
|
| 29 |
+
{
|
| 30 |
+
"cell_type": "markdown",
|
| 31 |
+
"source": [
|
| 32 |
+
"#ProteinMPNN\n",
|
| 33 |
+
"This notebook is intended as a quick demo, more features to come!"
|
| 34 |
+
],
|
| 35 |
+
"metadata": {
|
| 36 |
+
"id": "AYZebfKn8gef"
|
| 37 |
+
}
|
| 38 |
+
},
|
| 39 |
+
{
|
| 40 |
+
"cell_type": "code",
|
| 41 |
+
"source": [
|
| 42 |
+
"#@title Setup Model\n",
|
| 43 |
+
"import json, time, os, sys, glob\n",
|
| 44 |
+
"\n",
|
| 45 |
+
"if not os.path.isdir(\"ProteinMPNN\"):\n",
|
| 46 |
+
" os.system(\"git clone -q https://github.com/dauparas/ProteinMPNN.git\")\n",
|
| 47 |
+
"sys.path.append('/content/ProteinMPNN/vanilla_proteinmpnn')\n",
|
| 48 |
+
"\n",
|
| 49 |
+
"import matplotlib.pyplot as plt\n",
|
| 50 |
+
"import shutil\n",
|
| 51 |
+
"import warnings\n",
|
| 52 |
+
"import numpy as np\n",
|
| 53 |
+
"import torch\n",
|
| 54 |
+
"from torch import optim\n",
|
| 55 |
+
"from torch.utils.data import DataLoader\n",
|
| 56 |
+
"from torch.utils.data.dataset import random_split, Subset\n",
|
| 57 |
+
"import copy\n",
|
| 58 |
+
"import torch.nn as nn\n",
|
| 59 |
+
"import torch.nn.functional as F\n",
|
| 60 |
+
"import random\n",
|
| 61 |
+
"import os.path\n",
|
| 62 |
+
"from protein_mpnn_utils import loss_nll, loss_smoothed, gather_edges, gather_nodes, gather_nodes_t, cat_neighbors_nodes, _scores, _S_to_seq, tied_featurize, parse_PDB\n",
|
| 63 |
+
"from protein_mpnn_utils import StructureDataset, StructureDatasetPDB, ProteinMPNN\n",
|
| 64 |
+
"\n",
|
| 65 |
+
"device = torch.device(\"cuda:0\" if (torch.cuda.is_available()) else \"cpu\")\n",
|
| 66 |
+
"model_name=\"v_48_020\" # ProteinMPNN model name: v_48_002, v_48_010, v_48_020, v_48_030, v_32_002, v_32_010; v_32_020, v_32_030; v_48_010=version with 48 edges 0.10A noise\n",
|
| 67 |
+
"backbone_noise=0.00 # Standard deviation of Gaussian noise to add to backbone atoms\n",
|
| 68 |
+
"\n",
|
| 69 |
+
"path_to_model_weights='/content/ProteinMPNN/vanilla_proteinmpnn/vanilla_model_weights' \n",
|
| 70 |
+
"hidden_dim = 128\n",
|
| 71 |
+
"num_layers = 3 \n",
|
| 72 |
+
"model_folder_path = path_to_model_weights\n",
|
| 73 |
+
"if model_folder_path[-1] != '/':\n",
|
| 74 |
+
" model_folder_path = model_folder_path + '/'\n",
|
| 75 |
+
"checkpoint_path = model_folder_path + f'{model_name}.pt'\n",
|
| 76 |
+
"\n",
|
| 77 |
+
"checkpoint = torch.load(checkpoint_path, map_location=device) \n",
|
| 78 |
+
"print('Number of edges:', checkpoint['num_edges'])\n",
|
| 79 |
+
"noise_level_print = checkpoint['noise_level']\n",
|
| 80 |
+
"print(f'Training noise level: {noise_level_print}A')\n",
|
| 81 |
+
"model = ProteinMPNN(num_letters=21, node_features=hidden_dim, edge_features=hidden_dim, hidden_dim=hidden_dim, num_encoder_layers=num_layers, num_decoder_layers=num_layers, augment_eps=backbone_noise, k_neighbors=checkpoint['num_edges'])\n",
|
| 82 |
+
"model.to(device)\n",
|
| 83 |
+
"model.load_state_dict(checkpoint['model_state_dict'])\n",
|
| 84 |
+
"model.eval()\n",
|
| 85 |
+
"print(\"Model loaded\")"
|
| 86 |
+
],
|
| 87 |
+
"metadata": {
|
| 88 |
+
"id": "iYDU3ftml2k5",
|
| 89 |
+
"cellView": "form"
|
| 90 |
+
},
|
| 91 |
+
"execution_count": null,
|
| 92 |
+
"outputs": []
|
| 93 |
+
},
|
| 94 |
+
{
|
| 95 |
+
"cell_type": "code",
|
| 96 |
+
"source": [
|
| 97 |
+
"import re\n",
|
| 98 |
+
"from google.colab import files\n",
|
| 99 |
+
"import numpy as np\n",
|
| 100 |
+
"\n",
|
| 101 |
+
"#########################\n",
|
| 102 |
+
"def get_pdb(pdb_code=\"\"):\n",
|
| 103 |
+
" if pdb_code is None or pdb_code == \"\":\n",
|
| 104 |
+
" upload_dict = files.upload()\n",
|
| 105 |
+
" pdb_string = upload_dict[list(upload_dict.keys())[0]]\n",
|
| 106 |
+
" with open(\"tmp.pdb\",\"wb\") as out: out.write(pdb_string)\n",
|
| 107 |
+
" return \"tmp.pdb\"\n",
|
| 108 |
+
" else:\n",
|
| 109 |
+
" os.system(f\"wget -qnc https://files.rcsb.org/view/{pdb_code}.pdb\")\n",
|
| 110 |
+
" return f\"{pdb_code}.pdb\"\n",
|
| 111 |
+
"\n",
|
| 112 |
+
"#@markdown ### Input Options\n",
|
| 113 |
+
"pdb='6MRR' #@param {type:\"string\"}\n",
|
| 114 |
+
"pdb_path = get_pdb(pdb)\n",
|
| 115 |
+
"#@markdown - pdb code (leave blank to get an upload prompt)\n",
|
| 116 |
+
"\n",
|
| 117 |
+
"designed_chain = \"A\" #@param {type:\"string\"}\n",
|
| 118 |
+
"fixed_chain = \"\" #@param {type:\"string\"}\n",
|
| 119 |
+
"\n",
|
| 120 |
+
"if designed_chain == \"\":\n",
|
| 121 |
+
" designed_chain_list = []\n",
|
| 122 |
+
"else:\n",
|
| 123 |
+
" designed_chain_list = re.sub(\"[^A-Za-z]+\",\",\", designed_chain).split(\",\")\n",
|
| 124 |
+
"\n",
|
| 125 |
+
"if fixed_chain == \"\":\n",
|
| 126 |
+
" fixed_chain_list = []\n",
|
| 127 |
+
"else:\n",
|
| 128 |
+
" fixed_chain_list = re.sub(\"[^A-Za-z]+\",\",\", fixed_chain).split(\",\")\n",
|
| 129 |
+
"\n",
|
| 130 |
+
"chain_list = list(set(designed_chain_list + fixed_chain_list))\n",
|
| 131 |
+
"\n",
|
| 132 |
+
"#@markdown - specified which chain(s) to design and which chain(s) to keep fixed. \n",
|
| 133 |
+
"#@markdown Use comma:`A,B` to specifiy more than one chain\n",
|
| 134 |
+
"\n",
|
| 135 |
+
"#chain = \"A\" #@param {type:\"string\"}\n",
|
| 136 |
+
"#pdb_path_chains = chain\n",
|
| 137 |
+
"##@markdown - Define which chain to redesign\n",
|
| 138 |
+
"\n",
|
| 139 |
+
"#@markdown ### Design Options\n",
|
| 140 |
+
"num_seqs = 1 #@param [\"1\", \"2\", \"4\", \"8\", \"16\", \"32\", \"64\"] {type:\"raw\"}\n",
|
| 141 |
+
"num_seq_per_target = num_seqs\n",
|
| 142 |
+
"sampling_temp = \"0.1\" #@param [\"0.1\", \"0.15\", \"0.2\", \"0.25\", \"0.3\"]\n",
|
| 143 |
+
"#@markdown - Sampling temperature for amino acids, T=0.0 means taking \n",
|
| 144 |
+
"#@markdown argmax, T>>1.0 means sample randomly. Suggested values \n",
|
| 145 |
+
"#@markdown 0.1, 0.15, 0.2, 0.25, 0.3. Higher values will lead to more diversity.\n",
|
| 146 |
+
"\n",
|
| 147 |
+
"\n",
|
| 148 |
+
"save_score=0 # 0 for False, 1 for True; save score=-log_prob to npy files\n",
|
| 149 |
+
"save_probs=0 # 0 for False, 1 for True; save MPNN predicted probabilites per position\n",
|
| 150 |
+
"score_only=0 # 0 for False, 1 for True; score input backbone-sequence pairs\n",
|
| 151 |
+
"conditional_probs_only=0 # 0 for False, 1 for True; output conditional probabilities p(s_i given the rest of the sequence and backbone)\n",
|
| 152 |
+
"conditional_probs_only_backbone=0 # 0 for False, 1 for True; if true output conditional probabilities p(s_i given backbone)\n",
|
| 153 |
+
" \n",
|
| 154 |
+
"batch_size=1 # Batch size; can set higher for titan, quadro GPUs, reduce this if running out of GPU memory\n",
|
| 155 |
+
"max_length=20000 # Max sequence length\n",
|
| 156 |
+
" \n",
|
| 157 |
+
"out_folder='.' # Path to a folder to output sequences, e.g. /home/out/\n",
|
| 158 |
+
"jsonl_path='' # Path to a folder with parsed pdb into jsonl\n",
|
| 159 |
+
"omit_AAs='X' # Specify which amino acids should be omitted in the generated sequence, e.g. 'AC' would omit alanine and cystine.\n",
|
| 160 |
+
" \n",
|
| 161 |
+
"pssm_multi=0.0 # A value between [0.0, 1.0], 0.0 means do not use pssm, 1.0 ignore MPNN predictions\n",
|
| 162 |
+
"pssm_threshold=0.0 # A value between -inf + inf to restric per position AAs\n",
|
| 163 |
+
"pssm_log_odds_flag=0 # 0 for False, 1 for True\n",
|
| 164 |
+
"pssm_bias_flag=0 # 0 for False, 1 for True\n",
|
| 165 |
+
"\n",
|
| 166 |
+
"\n",
|
| 167 |
+
"##############################################################\n",
|
| 168 |
+
"\n",
|
| 169 |
+
"folder_for_outputs = out_folder\n",
|
| 170 |
+
"\n",
|
| 171 |
+
"NUM_BATCHES = num_seq_per_target//batch_size\n",
|
| 172 |
+
"BATCH_COPIES = batch_size\n",
|
| 173 |
+
"temperatures = [float(item) for item in sampling_temp.split()]\n",
|
| 174 |
+
"omit_AAs_list = omit_AAs\n",
|
| 175 |
+
"alphabet = 'ACDEFGHIKLMNPQRSTVWYX'\n",
|
| 176 |
+
"\n",
|
| 177 |
+
"omit_AAs_np = np.array([AA in omit_AAs_list for AA in alphabet]).astype(np.float32)\n",
|
| 178 |
+
"\n",
|
| 179 |
+
"chain_id_dict = None\n",
|
| 180 |
+
"fixed_positions_dict = None\n",
|
| 181 |
+
"pssm_dict = None\n",
|
| 182 |
+
"omit_AA_dict = None\n",
|
| 183 |
+
"bias_AA_dict = None\n",
|
| 184 |
+
"tied_positions_dict = None\n",
|
| 185 |
+
"bias_by_res_dict = None\n",
|
| 186 |
+
"bias_AAs_np = np.zeros(len(alphabet))\n",
|
| 187 |
+
"\n",
|
| 188 |
+
"\n",
|
| 189 |
+
"###############################################################\n",
|
| 190 |
+
"pdb_dict_list = parse_PDB(pdb_path, input_chain_list=chain_list)\n",
|
| 191 |
+
"dataset_valid = StructureDatasetPDB(pdb_dict_list, truncate=None, max_length=max_length)\n",
|
| 192 |
+
"\n",
|
| 193 |
+
"chain_id_dict = {}\n",
|
| 194 |
+
"chain_id_dict[pdb_dict_list[0]['name']]= (designed_chain_list, fixed_chain_list)\n",
|
| 195 |
+
"\n",
|
| 196 |
+
"print(chain_id_dict)"
|
| 197 |
+
],
|
| 198 |
+
"metadata": {
|
| 199 |
+
"cellView": "form",
|
| 200 |
+
"id": "k4o6w2Y23wxO"
|
| 201 |
+
},
|
| 202 |
+
"execution_count": null,
|
| 203 |
+
"outputs": []
|
| 204 |
+
},
|
| 205 |
+
{
|
| 206 |
+
"cell_type": "code",
|
| 207 |
+
"source": [
|
| 208 |
+
"#@title RUN\n",
|
| 209 |
+
"with torch.no_grad():\n",
|
| 210 |
+
" print('Generating sequences...')\n",
|
| 211 |
+
" for ix, protein in enumerate(dataset_valid):\n",
|
| 212 |
+
" score_list = []\n",
|
| 213 |
+
" all_probs_list = []\n",
|
| 214 |
+
" all_log_probs_list = []\n",
|
| 215 |
+
" S_sample_list = []\n",
|
| 216 |
+
" batch_clones = [copy.deepcopy(protein) for i in range(BATCH_COPIES)]\n",
|
| 217 |
+
" X, S, mask, lengths, chain_M, chain_encoding_all, chain_list_list, visible_list_list, masked_list_list, masked_chain_length_list_list, chain_M_pos, omit_AA_mask, residue_idx, dihedral_mask, tied_pos_list_of_lists_list, pssm_coef, pssm_bias, pssm_log_odds_all, bias_by_res_all, tied_beta = tied_featurize(batch_clones, device, chain_id_dict, fixed_positions_dict, omit_AA_dict, tied_positions_dict, pssm_dict, bias_by_res_dict)\n",
|
| 218 |
+
" pssm_log_odds_mask = (pssm_log_odds_all > pssm_threshold).float() #1.0 for true, 0.0 for false\n",
|
| 219 |
+
" name_ = batch_clones[0]['name']\n",
|
| 220 |
+
"\n",
|
| 221 |
+
" randn_1 = torch.randn(chain_M.shape, device=X.device)\n",
|
| 222 |
+
" log_probs = model(X, S, mask, chain_M*chain_M_pos, residue_idx, chain_encoding_all, randn_1)\n",
|
| 223 |
+
" mask_for_loss = mask*chain_M*chain_M_pos\n",
|
| 224 |
+
" scores = _scores(S, log_probs, mask_for_loss)\n",
|
| 225 |
+
" native_score = scores.cpu().data.numpy()\n",
|
| 226 |
+
"\n",
|
| 227 |
+
" for temp in temperatures:\n",
|
| 228 |
+
" for j in range(NUM_BATCHES):\n",
|
| 229 |
+
" randn_2 = torch.randn(chain_M.shape, device=X.device)\n",
|
| 230 |
+
" if tied_positions_dict == None:\n",
|
| 231 |
+
" sample_dict = model.sample(X, randn_2, S, chain_M, chain_encoding_all, residue_idx, mask=mask, temperature=temp, omit_AAs_np=omit_AAs_np, bias_AAs_np=bias_AAs_np, chain_M_pos=chain_M_pos, omit_AA_mask=omit_AA_mask, pssm_coef=pssm_coef, pssm_bias=pssm_bias, pssm_multi=pssm_multi, pssm_log_odds_flag=bool(pssm_log_odds_flag), pssm_log_odds_mask=pssm_log_odds_mask, pssm_bias_flag=bool(pssm_bias_flag), bias_by_res=bias_by_res_all)\n",
|
| 232 |
+
" S_sample = sample_dict[\"S\"] \n",
|
| 233 |
+
" else:\n",
|
| 234 |
+
" sample_dict = model.tied_sample(X, randn_2, S, chain_M, chain_encoding_all, residue_idx, mask=mask, temperature=temp, omit_AAs_np=omit_AAs_np, bias_AAs_np=bias_AAs_np, chain_M_pos=chain_M_pos, omit_AA_mask=omit_AA_mask, pssm_coef=pssm_coef, pssm_bias=pssm_bias, pssm_multi=pssm_multi, pssm_log_odds_flag=bool(pssm_log_odds_flag), pssm_log_odds_mask=pssm_log_odds_mask, pssm_bias_flag=bool(pssm_bias_flag), tied_pos=tied_pos_list_of_lists_list[0], tied_beta=tied_beta, bias_by_res=bias_by_res_all)\n",
|
| 235 |
+
" # Compute scores\n",
|
| 236 |
+
" S_sample = sample_dict[\"S\"]\n",
|
| 237 |
+
" log_probs = model(X, S_sample, mask, chain_M*chain_M_pos, residue_idx, chain_encoding_all, randn_2, use_input_decoding_order=True, decoding_order=sample_dict[\"decoding_order\"])\n",
|
| 238 |
+
" mask_for_loss = mask*chain_M*chain_M_pos\n",
|
| 239 |
+
" scores = _scores(S_sample, log_probs, mask_for_loss)\n",
|
| 240 |
+
" scores = scores.cpu().data.numpy()\n",
|
| 241 |
+
" all_probs_list.append(sample_dict[\"probs\"].cpu().data.numpy())\n",
|
| 242 |
+
" all_log_probs_list.append(log_probs.cpu().data.numpy())\n",
|
| 243 |
+
" S_sample_list.append(S_sample.cpu().data.numpy())\n",
|
| 244 |
+
" for b_ix in range(BATCH_COPIES):\n",
|
| 245 |
+
" masked_chain_length_list = masked_chain_length_list_list[b_ix]\n",
|
| 246 |
+
" masked_list = masked_list_list[b_ix]\n",
|
| 247 |
+
" seq_recovery_rate = torch.sum(torch.sum(torch.nn.functional.one_hot(S[b_ix], 21)*torch.nn.functional.one_hot(S_sample[b_ix], 21),axis=-1)*mask_for_loss[b_ix])/torch.sum(mask_for_loss[b_ix])\n",
|
| 248 |
+
" seq = _S_to_seq(S_sample[b_ix], chain_M[b_ix])\n",
|
| 249 |
+
" score = scores[b_ix]\n",
|
| 250 |
+
" score_list.append(score)\n",
|
| 251 |
+
" native_seq = _S_to_seq(S[b_ix], chain_M[b_ix])\n",
|
| 252 |
+
" if b_ix == 0 and j==0 and temp==temperatures[0]:\n",
|
| 253 |
+
" start = 0\n",
|
| 254 |
+
" end = 0\n",
|
| 255 |
+
" list_of_AAs = []\n",
|
| 256 |
+
" for mask_l in masked_chain_length_list:\n",
|
| 257 |
+
" end += mask_l\n",
|
| 258 |
+
" list_of_AAs.append(native_seq[start:end])\n",
|
| 259 |
+
" start = end\n",
|
| 260 |
+
" native_seq = \"\".join(list(np.array(list_of_AAs)[np.argsort(masked_list)]))\n",
|
| 261 |
+
" l0 = 0\n",
|
| 262 |
+
" for mc_length in list(np.array(masked_chain_length_list)[np.argsort(masked_list)])[:-1]:\n",
|
| 263 |
+
" l0 += mc_length\n",
|
| 264 |
+
" native_seq = native_seq[:l0] + '/' + native_seq[l0:]\n",
|
| 265 |
+
" l0 += 1\n",
|
| 266 |
+
" sorted_masked_chain_letters = np.argsort(masked_list_list[0])\n",
|
| 267 |
+
" print_masked_chains = [masked_list_list[0][i] for i in sorted_masked_chain_letters]\n",
|
| 268 |
+
" sorted_visible_chain_letters = np.argsort(visible_list_list[0])\n",
|
| 269 |
+
" print_visible_chains = [visible_list_list[0][i] for i in sorted_visible_chain_letters]\n",
|
| 270 |
+
" native_score_print = np.format_float_positional(np.float32(native_score.mean()), unique=False, precision=4)\n",
|
| 271 |
+
" line = '>{}, score={}, fixed_chains={}, designed_chains={}, model_name={}\\n{}\\n'.format(name_, native_score_print, print_visible_chains, print_masked_chains, model_name, native_seq)\n",
|
| 272 |
+
" print(line.rstrip())\n",
|
| 273 |
+
" start = 0\n",
|
| 274 |
+
" end = 0\n",
|
| 275 |
+
" list_of_AAs = []\n",
|
| 276 |
+
" for mask_l in masked_chain_length_list:\n",
|
| 277 |
+
" end += mask_l\n",
|
| 278 |
+
" list_of_AAs.append(seq[start:end])\n",
|
| 279 |
+
" start = end\n",
|
| 280 |
+
"\n",
|
| 281 |
+
" seq = \"\".join(list(np.array(list_of_AAs)[np.argsort(masked_list)]))\n",
|
| 282 |
+
" l0 = 0\n",
|
| 283 |
+
" for mc_length in list(np.array(masked_chain_length_list)[np.argsort(masked_list)])[:-1]:\n",
|
| 284 |
+
" l0 += mc_length\n",
|
| 285 |
+
" seq = seq[:l0] + '/' + seq[l0:]\n",
|
| 286 |
+
" l0 += 1\n",
|
| 287 |
+
" score_print = np.format_float_positional(np.float32(score), unique=False, precision=4)\n",
|
| 288 |
+
" seq_rec_print = np.format_float_positional(np.float32(seq_recovery_rate.detach().cpu().numpy()), unique=False, precision=4)\n",
|
| 289 |
+
" line = '>T={}, sample={}, score={}, seq_recovery={}\\n{}\\n'.format(temp,b_ix,score_print,seq_rec_print,seq)\n",
|
| 290 |
+
" print(line.rstrip())\n",
|
| 291 |
+
"\n",
|
| 292 |
+
"\n",
|
| 293 |
+
"all_probs_concat = np.concatenate(all_probs_list)\n",
|
| 294 |
+
"all_log_probs_concat = np.concatenate(all_log_probs_list)\n",
|
| 295 |
+
"S_sample_concat = np.concatenate(S_sample_list)"
|
| 296 |
+
],
|
| 297 |
+
"metadata": {
|
| 298 |
+
"id": "xMVlYh8Fv2of",
|
| 299 |
+
"cellView": "form"
|
| 300 |
+
},
|
| 301 |
+
"execution_count": null,
|
| 302 |
+
"outputs": []
|
| 303 |
+
},
|
| 304 |
+
{
|
| 305 |
+
"cell_type": "code",
|
| 306 |
+
"source": [
|
| 307 |
+
"# experimental output\n",
|
| 308 |
+
"plt.figure(figsize=(20,5), dpi=100)\n",
|
| 309 |
+
"plt.imshow(all_probs_concat.mean(0).T,vmin=0,vmax=1)\n",
|
| 310 |
+
"plt.xlabel(\"positions\")\n",
|
| 311 |
+
"plt.ylabel(\"amino acids\")\n",
|
| 312 |
+
"plt.yticks(range(21),list(alphabet))\n",
|
| 313 |
+
"plt.show()"
|
| 314 |
+
],
|
| 315 |
+
"metadata": {
|
| 316 |
+
"id": "4jSKLU3L17Sf"
|
| 317 |
+
},
|
| 318 |
+
"execution_count": null,
|
| 319 |
+
"outputs": []
|
| 320 |
+
}
|
| 321 |
+
]
|
| 322 |
+
}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/assigned_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": [["A", "B"], ["C", "D", "E", "F"]], "3HTN": [["A", "B"], ["C"]]}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/fixed_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": {"A": [1, 2, 3, 4, 5, 6, 7, 8, 23, 25], "C": [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 40], "B": [], "D": [], "E": [], "F": []}, "3HTN": {"A": [1, 2, 3, 4, 5, 6, 7, 8, 23, 25], "C": [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 40], "B": []}}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.1538, fixed_chains=['C'], designed_chains=['A', 'B'], model_name=v_48_020
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKXXXXDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=0, score=0.7465, seq_recovery=0.5426
|
| 4 |
+
KMYEYKKIGNDYIVSIKNNTDLVTAIKEFCKEKKIKSGTINGIGQVKQVTLRFYNFETKEYEEKTFNENLDISNLTGIISTHNNEIFLHLHGTFGKENFSALAGHLISAIVNGKGILKVEDFKEEISTKYDEKLGLYLLDFNK/SMYKYKKIGNDYIVKINNGKNLVESLLEFVKDKNIKSGTINGTGSVSKVTLEFFDPEXXXXKTKTFNDNFDISNLTGFISTKDGKPLIDLHITIGDSDFSALAGHLIDAIVNGEANIKVEDYNVEINVRYDEELGLWLLDFNL
|
| 5 |
+
>T=0.1, sample=0, score=0.7500, seq_recovery=0.5851
|
| 6 |
+
NMYTYEKIGNKYIVSINNNTELITAIKNFCKEKKIKSGTINGIGQVSSVTLRFYNYETKTYENKTFNAQFTISNLTGIISTYNNEIFLHLHITIGDSNFSALAGHLLSAVVNGTCILIVEDFKEKISRKYNEELGLYLLDFNK/NMYKYKKIGNKYIVSINNGKELYEALLDFVKDKNIKSGSVNGTGMISKLTLSFFDPNXXXXTTKTFNMNMDISNLTGFISTKNGEPLLDLHVTVGDSDFSALAGHLVSAVVNGEADVIIENFNKEINVKYNEELGLWLLDFNL
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_2_outputs/seqs/4YOW.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4YOW, score=1.1024, fixed_chains=['C', 'D', 'E', 'F'], designed_chains=['A', 'B'], model_name=v_48_020
|
| 2 |
+
MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/XXXX
|
| 3 |
+
>T=0.1, sample=0, score=0.7444, seq_recovery=0.5903
|
| 4 |
+
MKIVAADTGGALADENYNPIGKIATVAVLVTKPYRTSDTFLVEYLDPTKYDLSSHEGIKRELELAIELAEQVKPDLIHLDINLGGVPVAELNPEVIDKLQISEETKKILKELAKTLTPLAQAYLAKTGIPILATGDDSVPVHIAHIYASGAAVKWALENVKELKGLRVLLEEATSVEIKEDSIVVRSLDPRDGGLYGEIKTEIPEGITTELYPNPLRSNHMIFEVKT/XXXX
|
| 5 |
+
>T=0.1, sample=0, score=0.7336, seq_recovery=0.5551
|
| 6 |
+
MKIVAADTGGYLLDENYRPIGRIATVAVLVEKPYRTSDVFLVEYLDPTNYDLSSHEGILREFRLAVELAERVKPDLIHLDIDLGGVPVAELTPEVIEALQISEETKATLKELAKTLTPAAQAFLARTGIPVLAMGSRSVPVRIADIAASVAAVKYALENVKKRKGLRVGLEEAVSVEIEEKSIVGRSLDPRDGGLYFRIETEIPEGVTYELYPNPLRTNHLVFEVKV/XXXX
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_3_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.1501, fixed_chains=['C'], designed_chains=['A', 'B'], model_name=v_48_020
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKXXXXDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=0, score=0.7254, seq_recovery=0.5355
|
| 4 |
+
HMYEYKEIGNKYIVSINNNTDIVEAIKKFCEEKNIKSGTINGIGQVKSVTLRFYNFETKESKEVTINDNLTISNLTGIISTYNNEIFLDLHITIGDSNFSALAGHLLSAIVNGDCILIIEDYKENISKKYDKELGLWLLDFNK/KMYSYKKIGNKYIVNINNGKDLVTSILKFCEDKKIKSGTINGTGMISKLTLEFFDPEXXXXTTKTFNDILDISNLTGFISTKDGKVFVKLYITAGKKDFSALAGKLVSAIVNGEMNLTIEDFNVEINVEYNEELGLYLLNFNK
|
| 5 |
+
>T=0.1, sample=0, score=0.7494, seq_recovery=0.5745
|
| 6 |
+
SMYEYKKIGNSYIVSVNNNTELVEALTAFCTEKGIKSGTVNGIGQVKSVTLRFYNFETKEYKEKTFEENLEISNLTGIISTYNNKVFLDIHGTFGKSDFSALAGHLVSAIVNGKAILKVEDYKEEISRTYNEETGLWLLDFNK/KMYKYKKIGNDYIVSIKNGKNLVEAIKKFCEDKNIKSGSVNGTGQISKVTLRFFDPEXXXXTTKTFNENMDISNLTGFISTKNGEVLVDLHITVGKSNFSALAGHLVDAIVNGEADLKIEDYNVEINVEYDEKTGLWLLNFNK
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/assigned_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": [["A", "C"], ["B", "D", "E", "F"]], "3HTN": [["A", "C"], ["B"]]}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/fixed_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": {"A": [1, 2, 3, 4, 5, 6, 7, 8, 23, 25], "C": [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 40], "B": [], "D": [], "E": [], "F": []}, "3HTN": {"A": [1, 2, 3, 4, 5, 6, 7, 8, 23, 25], "C": [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 40], "B": []}}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.1682, fixed_chains=['B'], designed_chains=['A', 'C'], model_name=v_48_020
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=0, score=0.7425, seq_recovery=0.5606
|
| 4 |
+
NMYSYKKIGNKYIVNINNNTELVEAIKKFCKDENIKSGSINGTGQVSKVTLRFYNPETKEYKETTFNDNFDISNLTGFISTYNNEVFLDLHITIGKSNFSALAGHLLSAVVNGEMTLVVEDYNELLSMKYNEELGLYLLDFNK/NLYSYKKIGNKYIVSINNHTDIVTALKTFCEDKNIKSGTINGIGQVSSVTLRFFNIETKEVKEVTFNENLEISNLTGIISEKDGKVFLHLHGTFGKSDFSALAGHLLSAVVNGKALFEIEDFKEKVNVEYDEELGLWLLNFNK
|
| 5 |
+
>T=0.1, sample=0, score=0.7474, seq_recovery=0.5644
|
| 6 |
+
NMYSYKKIGNKYIVSINNNTNLVTAIKKFCEDKNIKSGTINGTGQVSKVTLRFYNPETKTYTDKTFNDNFDISNLTGFISTYNGKIFLHLHITIGDSNFSALAGHLIDAIVNGTADLVIEDYNENISMKYDEELGLYLLDFNK/NLYSYKKIGNKYIVSINNHTDIVEALKTFCEEKNIKSGTINGIGTVSSLTLAFRNIETGEVDRKTFNQNLEISNLTGIISTKNGKVFLDLHVTFGDSNFSALAGHLESAIVNGTALVVVEDYNEEVNVEYDEKLGLNLLNFNK
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_4_outputs/seqs/4YOW.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4YOW, score=1.1328, fixed_chains=['B', 'D', 'E', 'F'], designed_chains=['A', 'C'], model_name=v_48_020
|
| 2 |
+
MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS
|
| 3 |
+
>T=0.1, sample=0, score=0.7592, seq_recovery=0.6088
|
| 4 |
+
MRIVAADTGGALLNENYEPIGKIATVAVLVEKPYRTSKEFLVKYHDPLNYDLSSNQGIRDEVLLAIELARKVRPDMIHLDIDLGGVPLAELTPEVIEALQISEETKATLKELAKELTPLAQAFLAETGIPILCIGSRSVPVHIADIYASAEAVRWALENVKKLKGLLVGLEYATRVEIGKDSIKATSLDPRDGGLYAEVKTKIPEGITYELYPDPLRTGHMVFKITT/MKIVAADTGGAVLDESFQPVGRIATVAVVVEEPYRTSKEFLVKYLDPFKYDLSSHEGILEELELAIELAEKVKPDLIHLDLDLGGVELGELDPEVIDALQISPETRATLKELAKTLAPKARAFKEKTGIPILLTGEASVPVRIAEIYASIASVAWALEHVKELKGLRVLLEEAVSVEIEADKIVGRSLDPRDGGLYQELPTAVPEGITWELFPNPLRANHLVFEVTV
|
| 5 |
+
>T=0.1, sample=0, score=0.7365, seq_recovery=0.5972
|
| 6 |
+
MRIVAADTGGYLLDENYRPIGPIATVAVLVEKPYRTSKEFLVRYHDPENYDLTGNQGLYDEFELAIELAEKVKPDLIHLDIDLGGVPVAELTPEVINKLPISEETKKTLIELSKTLTPKAQAFYKKTGIPILAIGDRSVPVKIADIYASIAAVKWALENVKERKGLRVLLEEGVRVEIKENSIVGTSLDPRDGGLYGEIETEVPEGVTYKLYPNPLRLGHLVFEIST/MKIVAADTGGAVLDESFQPVGRIATVAVVVEEPYTTSKEFLVRYLDPFAYDLSSHEGLREEVELAIELARKVKPDLIHLDIDLGGVDVADLDEAVIDALQISPETKAVLKELAKELAPLAKAFKAETGIPILATGHRSVPVHIAHIAASGAAVKWALEHVKELKGLRVLLEEATAVEIKENKIVVTSLDPRHGGLHFEIETEVPEGIEYELFPNPLDAGHMVFEVTV
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/assigned_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": [["A", "C"], ["B", "D", "E", "F"]], "3HTN": [["A", "C"], ["B"]]}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/fixed_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": {"A": [9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23], "C": [10, 11, 18, 19, 20, 22], "B": [], "D": [], "E": [], "F": []}, "3HTN": {"A": [9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23], "C": [10, 11, 18, 19, 20, 22], "B": []}}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.1439, fixed_chains=['B'], designed_chains=['A', 'C'], model_name=v_48_020
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=0, score=0.7451, seq_recovery=0.5849
|
| 4 |
+
NMYKYKKIGNKYIVSINNHTEIVKAIKEFCKEKNIKSGTVNGTGQISKLTLRFYNMETKTSTDTTFNQNLDISNLTGFISEHENEVFLDLHITAGDSNFSALAGHLISAISNGTCELVVEDFKEKLSTKYDEETGLYLLDFEK/NMYKYKKIGNKYIVSINNHTEIVKAIKKFCEDKNIKSGTINGIGTISSLTLEFYDIKTKKKKLKTFNAQLEISNLTGIISTKNGEVFLDLHVTVGDSNFSALAGHLVSAVVNGTAKLVVEDYKEEVNVKYDEELGLYLLDFNL
|
| 5 |
+
>T=0.1, sample=0, score=0.7454, seq_recovery=0.5887
|
| 6 |
+
NMYKYKKIGNKYIVSINNHTEIVKAIKEFCKEKNIKSGTVNGIGQVSSVTLKFYNPETKESTLKTFNKLLDISNLTGFISTYNNEVFLDLHITFGDSDFSALAGHLVSAIVDGYAELIVEDYNENISMKYDEELGLWKLDFEK/NMYKYKKIGNKYIVSINNHTNIVKAIKKFCEDKKIKSGTINGIGQISKLTLAFRNIETGEVDLKTFNDNYTISNLTGFISTINGKVFLDLHITVGNSNFSALAGHLIDAISNGTVNLVIEDYKEEINKKYNEELGLWLLDFNL
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/seqs/4YOW.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4YOW, score=1.1085, fixed_chains=['B', 'D', 'E', 'F'], designed_chains=['A', 'C'], model_name=v_48_020
|
| 2 |
+
MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS
|
| 3 |
+
>T=0.1, sample=0, score=0.7336, seq_recovery=0.6074
|
| 4 |
+
MKIVAADTGGAVLDESFQPVGLIATVAVVVEKPYRTSERYKVEYLDPFNYDLTGHEGIYREIRLAIELAREVKPDLIHLDIDLGGVNVAELTPEVIDALQISAETKEVLKELAKELTPLAQEFLAETGIPILAIGDRSVPVHIADIAASGAAVKWALEHVKERKGLRVGLVYATEVEIKEDKIIVRSLDPRDGGLYFEIETEIPEGITWELYPNPLELNHMVFEVTV/MKIVAADTGGALLDENYQPVGLIATVAVVVTYPYRTSDVFKVRYLDPLAYDLASDEHLRLELELAIELAKEVKPDEIHLDLDLGGVDVASLTPEVINALQISPETKARLLELAKELAPLAAAFRKETGIPIKAVGERSVAVRIAEIYASAEAVRWALEHVKERGGLRVLLEEAVSVEIGEDSITARSLDPRHGGLYQEVPVEVPEGVTWELYPNPLRANHMIFEVTV
|
| 5 |
+
>T=0.1, sample=0, score=0.7137, seq_recovery=0.6143
|
| 6 |
+
MKIVAADTGGAVLDESFQPVGLIATVAVLVEKPYRTSDEYLVRYHDPYKYDLTGHQDLRDEVELAIELAEKVKPDLIHLDVDLGGVELATLTPEVIDALPISAETKATLKELAKTLTPLAQKFLAKTGIPIRLIGDRSVPVHIADIAASVYAVKWALENVKKHKGLRVRLVEATEVEIGENEIIGRSLDPRDGGLYFRVETKIPEGIEYKLYPDPLRRHHMVFEVTV/MKIVAADTGGALLDENYQPVGLIATVAVVVTYPYTTSDVFKVRYLDPTAYDLSSDEHLRHEVELAIELAKEVNPDEIHLDLDLGGVDVADLTPEVIDALQISPETRARLKELAKELAPLAAAFKAETGIPIKAIGERSVAVHIAEIYASIYSVKWALEHVKERKGLRVGLEEAVSVEIKEDRIIGRSLDPRDGGLYGEVEVEVPEGIEWELYPNPLRSGHMVFEVTV
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_5_outputs/tied_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"4YOW": [{"A": [1], "C": [1]}, {"A": [2], "C": [2]}, {"A": [3], "C": [3]}, {"A": [4], "C": [4]}, {"A": [5], "C": [5]}, {"A": [6], "C": [6]}, {"A": [7], "C": [7]}, {"A": [8], "C": [8]}], "3HTN": [{"A": [1], "C": [1]}, {"A": [2], "C": [2]}, {"A": [3], "C": [3]}, {"A": [4], "C": [4]}, {"A": [5], "C": [5]}, {"A": [6], "C": [6]}, {"A": [7], "C": [7]}, {"A": [8], "C": [8]}]}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/bias_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"G": 40.1, "P": 0.3, "A": -0.05}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.2048, fixed_chains=[], designed_chains=['A', 'B', 'C'], model_name=v_48_020
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKXXXXDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=0, score=2.4061, seq_recovery=0.0847
|
| 4 |
+
GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGXXXXGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG
|
| 5 |
+
>T=0.1, sample=0, score=2.4041, seq_recovery=0.0847
|
| 6 |
+
GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGXXXXGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/example_7_outputs/seqs/4YOW.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4YOW, score=1.1322, fixed_chains=[], designed_chains=['A', 'B', 'C', 'D', 'E', 'F'], model_name=v_48_020
|
| 2 |
+
MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/XXXX/MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/XXXX/MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/XXXX
|
| 3 |
+
>T=0.1, sample=0, score=2.7441, seq_recovery=0.0793
|
| 4 |
+
GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX
|
| 5 |
+
>T=0.1, sample=0, score=2.6962, seq_recovery=0.0793
|
| 6 |
+
GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX/GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG/XXXX
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/pdbs/3HTN.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_complexes/pdbs/4YOW.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/seqs/4GYT.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4GYT, score=1.6782, fixed_chains=[], designed_chains=['A', 'B'], model_name=v_48_020
|
| 2 |
+
SLHLPKYDDFVQSISVLALTMSGSELHGIMCGYLCAGADSQGEAYIRALLNNKKDEQSRNALLSMFSVFSISQQQMNNFDFEFEMLLPDDDESLVTRAQAFSEWCEGFTQGLTIAGVGMEQFYEEESQDALQHLMEFAELDCESLEVGEEDERALMEVSEYTRMAVLRLHSDLVLHE/SLHLPKYDDFVQSISVLALTMSGSELHGIMCGYLCAGADSQGEAYIRALLNNKKDEQSRNALLSMFSVFSISQQQMNNFDFEFEMLLPDDDESLVTRAQAFSEWCEGFTQGLTIAGVGMEQFYEEESQDALQHLMEFAELDCESLEVGEEDERALMEVSEYTRMAVLRLHSDLVLHE
|
| 3 |
+
>T=0.2, sample=0, score=0.8880, seq_recovery=0.4463
|
| 4 |
+
ELRLPPYEEFAAAIAVLQLPVSPSELAGLILGYLAAGKIDLGRAWIRSLLGGRTDAASQAALAALLEVFDILEEQLNNPELELELLLPPPDASLRERARALSEFARGFALGLELAGVDKESFKTEESKEAYERILELARLDASALREGPADRARLAELEEWLREAILQIHDDLVNHG/ELRLPPYEEFAAAIAVLQLPVSPSELAGLILGYLAAGKIDLGRAWIRSLLGGRTDAASQAALAALLEVFDILEEQLNNPELELELLLPPPDASLRERARALSEFARGFALGLELAGVDKESFKTEESKEAYERILELARLDASALREGPADRARLAELEEWLREAILQIHDDLVNHG
|
| 5 |
+
>T=0.2, sample=0, score=0.8673, seq_recovery=0.3898
|
| 6 |
+
SLALPPYDEFAAAVAPLKLPFSASYLAGLILGFIVAGKLELGRAWIKSLAKGKTDAATQAAVAALLDVFEILTRQLNDSSLELELLLPPKDASLKERAKALSEFAKGFVEGLELAGVTEESFSKESSKKAYKEIKELAKMDVSKLKEGEEDEKELEEKKEWLKNSILEIHKDLKENK/SLALPPYDEFAAAVAPLKLPFSASYLAGLILGFIVAGKLELGRAWIKSLAKGKTDAATQAAVAALLDVFEILTRQLNDSSLELELLLPPKDASLKERAKALSEFAKGFVEGLELAGVTEESFSKESSKKAYKEIKELAKMDVSKLKEGEEDEKELEEKKEWLKNSILEIHKDLKENK
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/seqs/6EHB.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>6EHB, score=1.3507, fixed_chains=[], designed_chains=['A', 'B', 'C'], model_name=v_48_020
|
| 2 |
+
DGINQSGDKAGSTVYSAKGTSLEVGGRAEARLSLKDGKAQDNSRVRLNFLGKAEINDSLYGVGFYEGEFTTNDQGKNASNNSLDNRYTYAGIGGTYGEVTYGKNDGALGVITDFTDIMSYHGNTAAEKIAVADRVDNMLAYKGQFGDLGVKASYRFADRNAVDAMGNVVTETNAAKYSDNGEDGYSLSAIYTFGDTGFNVGAGYADQDDQNEYMLAASYRMENLYFAGLFTDGELAKDVDYTGYELAAGYKLGQAAFTATYNNAETAKKTSADNFAIDATYYFKPNFRSYISYQFNLLDSXXASKVASEDELAIGLRYDF/DGINQSGDKAGSTVYSAKGTSLEVGGRAEARLSLKDGKAQDNSRVRLNFLGKAEINDSLYGVGFYEGEFTTNDQGKNASNNSLDNRYTYAGIGGTYGEVTYGKNDGALGVITDFTDIMSYHGNTAAEKIAVADRVDNMLAYKGQFGDLGVKASYRFADRNAVDAMGNVVTETNAAKYSDNGEDGYSLSAIYTFGDTGFNVGAGYADQDDQNEYMLAASYRMENLYFAGLFTDGELAKDVDYTGYELAAGYKLGQAAFTATYNNAETAKKTSADNFAIDATYYFKPNFRSYISYQFNLLDXXXASKVASEDELAIGLRYDF/DGINQSGDKAGSTVYSAKGTSLEVGGRAEARLSLKDGKAQDNSRVRLNFLGKAEINDSLYGVGFYEGEFTTNDQGKNASNNSLDNRYTYAGIGGTYGEVTYGKNDGALGVITDFTDIMSYHGNTAAEKIAVADRVDNMLAYKGQFGDLGVKASYRFADRNAVDAMGNVVTETNAAKYSDNGEDGYSLSAIYTFGDTGFNVGAGYADQDDQNEYMLAASYRMENLYFAGLFTDGELAKDVDYTGYELAAGYKLGQAAFTATYNNAETAKKTSADNFAIDATYYFKPNFRSYISYQFNLLDSDKASKVASEDELAIGLRYDF
|
| 3 |
+
>T=0.2, sample=0, score=0.8934, seq_recovery=0.5435
|
| 4 |
+
GGRNLSGPKPGQTVYSSNGSTLKIGGFADANLDIVDGKAKDNSKGRVSLLRTDKINDDLYGVGYIEVELTTNDNGTNAINNNLNVKKLYAGIGGKWGTVTYGKNDGALQIIADFTDIMPYGGNKAAPLIPVADNVDNTLSYSATYGDLTVRAAYQFATQVAVDSNGNVVDEENAAKYSDNGKDGWSASAVYDFGDSGWSVGAGAAKQGDQWAVAVAASYEKDNFYVAALYVAGDLAEGVPYAGLSLAASYKVGNTTYTVNYDVAFVDGKVSEDVLSYGVTYKFTDRFSTYVEYEDNLLDXXXASYVDSADVLRVGIRTDF/GGRNLSGPKPGQTVYSSNGSTLKIGGFADANLDIVDGKAKDNSKGRVSLLRTDKINDDLYGVGYIEVELTTNDNGTNAINNNLNVKKLYAGIGGKWGTVTYGKNDGALQIIADFTDIMPYGGNKAAPLIPVADNVDNTLSYSATYGDLTVRAAYQFATQVAVDSNGNVVDEENAAKYSDNGKDGWSASAVYDFGDSGWSVGAGAAKQGDQWAVAVAASYEKDNFYVAALYVAGDLAEGVPYAGLSLAASYKVGNTTYTVNYDVAFVDGKVSEDVLSYGVTYKFTDRFSTYVEYEDNLLDXXXASYVDSADVLRVGIRTDF/GGRNLSGPKPGQTVYSSNGSTLKIGGFADANLDIVDGKAKDNSKGRVSLLRTDKINDDLYGVGYIEVELTTNDNGTNAINNNLNVKKLYAGIGGKWGTVTYGKNDGALQIIADFTDIMPYGGNKAAPLIPVADNVDNTLSYSATYGDLTVRAAYQFATQVAVDSNGNVVDEENAAKYSDNGKDGWSASAVYDFGDSGWSVGAGAAKQGDQWAVAVAASYEKDNFYVAALYVAGDLAEGVPYAGLSLAASYKVGNTTYTVNYDVAFVDGKVSEDVLSYGVTYKFTDRFSTYVEYEDNLLDXXXASYVDSADVLRVGIRTDF
|
| 5 |
+
>T=0.2, sample=0, score=0.8911, seq_recovery=0.5497
|
| 6 |
+
GGLNLSSPKAGKTVYESGGSTLELGGRADAILKVVDGKFEDKSYGTVSLLKTDQINDDLYGTGYVELEFTVNDNGTNAVNNNLNNVKLYAGIGGKWGTVTYGKNDGALKPIRDFTDIMPYGGNRAAPLIPVADNIDNTLSYSATYGNLSVRASYRFANRIYVDENGNVVAKEEAARVSDNGNDGWSASAIYDFGDTGISVGAGAAHQGDQWQVALAASYKKDNFYVAALLTAGQLAKDVPYLGLSLAASYDFGNWRFTASYDLALVDGKVSEDRLTYGVTYDFTPNFSVSVEYTDNLLDXXXSSYVDSLDELVLGVRTDF/GGLNLSSPKAGKTVYESGGSTLELGGRADAILKVVDGKFEDKSYGTVSLLKTDQINDDLYGTGYVELEFTVNDNGTNAVNNNLNNVKLYAGIGGKWGTVTYGKNDGALKPIRDFTDIMPYGGNRAAPLIPVADNIDNTLSYSATYGNLSVRASYRFANRIYVDENGNVVAKEEAARVSDNGNDGWSASAIYDFGDTGISVGAGAAHQGDQWQVALAASYKKDNFYVAALLTAGQLAKDVPYLGLSLAASYDFGNWRFTASYDLALVDGKVSEDRLTYGVTYDFTPNFSVSVEYTDNLLDXXXSSYVDSLDELVLGVRTDF/GGLNLSSPKAGKTVYESGGSTLELGGRADAILKVVDGKFEDKSYGTVSLLKTDQINDDLYGTGYVELEFTVNDNGTNAVNNNLNNVKLYAGIGGKWGTVTYGKNDGALKPIRDFTDIMPYGGNRAAPLIPVADNIDNTLSYSATYGNLSVRASYRFANRIYVDENGNVVAKEEAARVSDNGNDGWSASAIYDFGDTGISVGAGAAHQGDQWQVALAASYKKDNFYVAALLTAGQLAKDVPYLGLSLAASYDFGNWRFTASYDLALVDGKVSEDRLTYGVTYDFTPNFSVSVEYTDNLLDXXXSSYVDSLDELVLGVRTDF
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/example_6_outputs/tied_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"6EHB": [{"A": [1], "B": [1], "C": [1]}, {"A": [2], "B": [2], "C": [2]}, {"A": [3], "B": [3], "C": [3]}, {"A": [4], "B": [4], "C": [4]}, {"A": [5], "B": [5], "C": [5]}, {"A": [6], "B": [6], "C": [6]}, {"A": [7], "B": [7], "C": [7]}, {"A": [8], "B": [8], "C": [8]}, {"A": [9], "B": [9], "C": [9]}, {"A": [10], "B": [10], "C": [10]}, {"A": [11], "B": [11], "C": [11]}, {"A": [12], "B": [12], "C": [12]}, {"A": [13], "B": [13], "C": [13]}, {"A": [14], "B": [14], "C": [14]}, {"A": [15], "B": [15], "C": [15]}, {"A": [16], "B": [16], "C": [16]}, {"A": [17], "B": [17], "C": [17]}, {"A": [18], "B": [18], "C": [18]}, {"A": [19], "B": [19], "C": [19]}, {"A": [20], "B": [20], "C": [20]}, {"A": [21], "B": [21], "C": [21]}, {"A": [22], "B": [22], "C": [22]}, {"A": [23], "B": [23], "C": [23]}, {"A": [24], "B": [24], "C": [24]}, {"A": [25], "B": [25], "C": [25]}, {"A": [26], "B": [26], "C": [26]}, {"A": [27], "B": [27], "C": [27]}, {"A": [28], "B": [28], "C": [28]}, {"A": [29], "B": [29], "C": [29]}, {"A": [30], "B": [30], "C": [30]}, {"A": [31], "B": [31], "C": [31]}, {"A": [32], "B": [32], "C": [32]}, {"A": [33], "B": [33], "C": [33]}, {"A": [34], "B": [34], "C": [34]}, {"A": [35], "B": [35], "C": [35]}, {"A": [36], "B": [36], "C": [36]}, {"A": [37], "B": [37], "C": [37]}, {"A": [38], "B": [38], "C": [38]}, {"A": [39], "B": [39], "C": [39]}, {"A": [40], "B": [40], "C": [40]}, {"A": [41], "B": [41], "C": [41]}, {"A": [42], "B": [42], "C": [42]}, {"A": [43], "B": [43], "C": [43]}, {"A": [44], "B": [44], "C": [44]}, {"A": [45], "B": [45], "C": [45]}, {"A": [46], "B": [46], "C": [46]}, {"A": [47], "B": [47], "C": [47]}, {"A": [48], "B": [48], "C": [48]}, {"A": [49], "B": [49], "C": [49]}, {"A": [50], "B": [50], "C": [50]}, {"A": [51], "B": [51], "C": [51]}, {"A": [52], "B": [52], "C": [52]}, {"A": [53], "B": [53], "C": [53]}, {"A": [54], "B": [54], "C": [54]}, {"A": [55], "B": [55], "C": [55]}, {"A": [56], "B": [56], "C": [56]}, {"A": [57], "B": [57], "C": [57]}, {"A": [58], "B": [58], "C": [58]}, {"A": [59], "B": [59], "C": [59]}, {"A": [60], "B": [60], "C": [60]}, {"A": [61], "B": [61], "C": [61]}, {"A": [62], "B": [62], "C": [62]}, {"A": [63], "B": [63], "C": [63]}, {"A": [64], "B": [64], "C": [64]}, {"A": [65], "B": [65], "C": [65]}, {"A": [66], "B": [66], "C": [66]}, {"A": [67], "B": [67], "C": [67]}, {"A": [68], "B": [68], "C": [68]}, {"A": [69], "B": [69], "C": [69]}, {"A": [70], "B": [70], "C": [70]}, {"A": [71], "B": [71], "C": [71]}, {"A": [72], "B": [72], "C": [72]}, {"A": [73], "B": [73], "C": [73]}, {"A": [74], "B": [74], "C": [74]}, {"A": [75], "B": [75], "C": [75]}, {"A": [76], "B": [76], "C": [76]}, {"A": [77], "B": [77], "C": [77]}, {"A": [78], "B": [78], "C": [78]}, {"A": [79], "B": [79], "C": [79]}, {"A": [80], "B": [80], "C": [80]}, {"A": [81], "B": [81], "C": [81]}, {"A": [82], "B": [82], "C": [82]}, {"A": [83], "B": [83], "C": [83]}, {"A": [84], "B": [84], "C": [84]}, {"A": [85], "B": [85], "C": [85]}, {"A": [86], "B": [86], "C": [86]}, {"A": [87], "B": [87], "C": [87]}, {"A": [88], "B": [88], "C": [88]}, {"A": [89], "B": [89], "C": [89]}, {"A": [90], "B": [90], "C": [90]}, {"A": [91], "B": [91], "C": [91]}, {"A": [92], "B": [92], "C": [92]}, {"A": [93], "B": [93], "C": [93]}, {"A": [94], "B": [94], "C": [94]}, {"A": [95], "B": [95], "C": [95]}, {"A": [96], "B": [96], "C": [96]}, {"A": [97], "B": [97], "C": [97]}, {"A": [98], "B": [98], "C": [98]}, {"A": [99], "B": [99], "C": [99]}, {"A": [100], "B": [100], "C": [100]}, {"A": [101], "B": [101], "C": [101]}, {"A": [102], "B": [102], "C": [102]}, {"A": [103], "B": [103], "C": [103]}, {"A": [104], "B": [104], "C": [104]}, {"A": [105], "B": [105], "C": [105]}, {"A": [106], "B": [106], "C": [106]}, {"A": [107], "B": [107], "C": [107]}, {"A": [108], "B": [108], "C": [108]}, {"A": [109], "B": [109], "C": [109]}, {"A": [110], "B": [110], "C": [110]}, {"A": [111], "B": [111], "C": [111]}, {"A": [112], "B": [112], "C": [112]}, {"A": [113], "B": [113], "C": [113]}, {"A": [114], "B": [114], "C": [114]}, {"A": [115], "B": [115], "C": [115]}, {"A": [116], "B": [116], "C": [116]}, {"A": [117], "B": [117], "C": [117]}, {"A": [118], "B": [118], "C": [118]}, {"A": [119], "B": [119], "C": [119]}, {"A": [120], "B": [120], "C": [120]}, {"A": [121], "B": [121], "C": [121]}, {"A": [122], "B": [122], "C": [122]}, {"A": [123], "B": [123], "C": [123]}, {"A": [124], "B": [124], "C": [124]}, {"A": [125], "B": [125], "C": [125]}, {"A": [126], "B": [126], "C": [126]}, {"A": [127], "B": [127], "C": [127]}, {"A": [128], "B": [128], "C": [128]}, {"A": [129], "B": [129], "C": [129]}, {"A": [130], "B": [130], "C": [130]}, {"A": [131], "B": [131], "C": [131]}, {"A": [132], "B": [132], "C": [132]}, {"A": [133], "B": [133], "C": [133]}, {"A": [134], "B": [134], "C": [134]}, {"A": [135], "B": [135], "C": [135]}, {"A": [136], "B": [136], "C": [136]}, {"A": [137], "B": [137], "C": [137]}, {"A": [138], "B": [138], "C": [138]}, {"A": [139], "B": [139], "C": [139]}, {"A": [140], "B": [140], "C": [140]}, {"A": [141], "B": [141], "C": [141]}, {"A": [142], "B": [142], "C": [142]}, {"A": [143], "B": [143], "C": [143]}, {"A": [144], "B": [144], "C": [144]}, {"A": [145], "B": [145], "C": [145]}, {"A": [146], "B": [146], "C": [146]}, {"A": [147], "B": [147], "C": [147]}, {"A": [148], "B": [148], "C": [148]}, {"A": [149], "B": [149], "C": [149]}, {"A": [150], "B": [150], "C": [150]}, {"A": [151], "B": [151], "C": [151]}, {"A": [152], "B": [152], "C": [152]}, {"A": [153], "B": [153], "C": [153]}, {"A": [154], "B": [154], "C": [154]}, {"A": [155], "B": [155], "C": [155]}, {"A": [156], "B": [156], "C": [156]}, {"A": [157], "B": [157], "C": [157]}, {"A": [158], "B": [158], "C": [158]}, {"A": [159], "B": [159], "C": [159]}, {"A": [160], "B": [160], "C": [160]}, {"A": [161], "B": [161], "C": [161]}, {"A": [162], "B": [162], "C": [162]}, {"A": [163], "B": [163], "C": [163]}, {"A": [164], "B": [164], "C": [164]}, {"A": [165], "B": [165], "C": [165]}, {"A": [166], "B": [166], "C": [166]}, {"A": [167], "B": [167], "C": [167]}, {"A": [168], "B": [168], "C": [168]}, {"A": [169], "B": [169], "C": [169]}, {"A": [170], "B": [170], "C": [170]}, {"A": [171], "B": [171], "C": [171]}, {"A": [172], "B": [172], "C": [172]}, {"A": [173], "B": [173], "C": [173]}, {"A": [174], "B": [174], "C": [174]}, {"A": [175], "B": [175], "C": [175]}, {"A": [176], "B": [176], "C": [176]}, {"A": [177], "B": [177], "C": [177]}, {"A": [178], "B": [178], "C": [178]}, {"A": [179], "B": [179], "C": [179]}, {"A": [180], "B": [180], "C": [180]}, {"A": [181], "B": [181], "C": [181]}, {"A": [182], "B": [182], "C": [182]}, {"A": [183], "B": [183], "C": [183]}, {"A": [184], "B": [184], "C": [184]}, {"A": [185], "B": [185], "C": [185]}, {"A": [186], "B": [186], "C": [186]}, {"A": [187], "B": [187], "C": [187]}, {"A": [188], "B": [188], "C": [188]}, {"A": [189], "B": [189], "C": [189]}, {"A": [190], "B": [190], "C": [190]}, {"A": [191], "B": [191], "C": [191]}, {"A": [192], "B": [192], "C": [192]}, {"A": [193], "B": [193], "C": [193]}, {"A": [194], "B": [194], "C": [194]}, {"A": [195], "B": [195], "C": [195]}, {"A": [196], "B": [196], "C": [196]}, {"A": [197], "B": [197], "C": [197]}, {"A": [198], "B": [198], "C": [198]}, {"A": [199], "B": [199], "C": [199]}, {"A": [200], "B": [200], "C": [200]}, {"A": [201], "B": [201], "C": [201]}, {"A": [202], "B": [202], "C": [202]}, {"A": [203], "B": [203], "C": [203]}, {"A": [204], "B": [204], "C": [204]}, {"A": [205], "B": [205], "C": [205]}, {"A": [206], "B": [206], "C": [206]}, {"A": [207], "B": [207], "C": [207]}, {"A": [208], "B": [208], "C": [208]}, {"A": [209], "B": [209], "C": [209]}, {"A": [210], "B": [210], "C": [210]}, {"A": [211], "B": [211], "C": [211]}, {"A": [212], "B": [212], "C": [212]}, {"A": [213], "B": [213], "C": [213]}, {"A": [214], "B": [214], "C": [214]}, {"A": [215], "B": [215], "C": [215]}, {"A": [216], "B": [216], "C": [216]}, {"A": [217], "B": [217], "C": [217]}, {"A": [218], "B": [218], "C": [218]}, {"A": [219], "B": [219], "C": [219]}, {"A": [220], "B": [220], "C": [220]}, {"A": [221], "B": [221], "C": [221]}, {"A": [222], "B": [222], "C": [222]}, {"A": [223], "B": [223], "C": [223]}, {"A": [224], "B": [224], "C": [224]}, {"A": [225], "B": [225], "C": [225]}, {"A": [226], "B": [226], "C": [226]}, {"A": [227], "B": [227], "C": [227]}, {"A": [228], "B": [228], "C": [228]}, {"A": [229], "B": [229], "C": [229]}, {"A": [230], "B": [230], "C": [230]}, {"A": [231], "B": [231], "C": [231]}, {"A": [232], "B": [232], "C": [232]}, {"A": [233], "B": [233], "C": [233]}, {"A": [234], "B": [234], "C": [234]}, {"A": [235], "B": [235], "C": [235]}, {"A": [236], "B": [236], "C": [236]}, {"A": [237], "B": [237], "C": [237]}, {"A": [238], "B": [238], "C": [238]}, {"A": [239], "B": [239], "C": [239]}, {"A": [240], "B": [240], "C": [240]}, {"A": [241], "B": [241], "C": [241]}, {"A": [242], "B": [242], "C": [242]}, {"A": [243], "B": [243], "C": [243]}, {"A": [244], "B": [244], "C": [244]}, {"A": [245], "B": [245], "C": [245]}, {"A": [246], "B": [246], "C": [246]}, {"A": [247], "B": [247], "C": [247]}, {"A": [248], "B": [248], "C": [248]}, {"A": [249], "B": [249], "C": [249]}, {"A": [250], "B": [250], "C": [250]}, {"A": [251], "B": [251], "C": [251]}, {"A": [252], "B": [252], "C": [252]}, {"A": [253], "B": [253], "C": [253]}, {"A": [254], "B": [254], "C": [254]}, {"A": [255], "B": [255], "C": [255]}, {"A": [256], "B": [256], "C": [256]}, {"A": [257], "B": [257], "C": [257]}, {"A": [258], "B": [258], "C": [258]}, {"A": [259], "B": [259], "C": [259]}, {"A": [260], "B": [260], "C": [260]}, {"A": [261], "B": [261], "C": [261]}, {"A": [262], "B": [262], "C": [262]}, {"A": [263], "B": [263], "C": [263]}, {"A": [264], "B": [264], "C": [264]}, {"A": [265], "B": [265], "C": [265]}, {"A": [266], "B": [266], "C": [266]}, {"A": [267], "B": [267], "C": [267]}, {"A": [268], "B": [268], "C": [268]}, {"A": [269], "B": [269], "C": [269]}, {"A": [270], "B": [270], "C": [270]}, {"A": [271], "B": [271], "C": [271]}, {"A": [272], "B": [272], "C": [272]}, {"A": [273], "B": [273], "C": [273]}, {"A": [274], "B": [274], "C": [274]}, {"A": [275], "B": [275], "C": [275]}, {"A": [276], "B": [276], "C": [276]}, {"A": [277], "B": [277], "C": [277]}, {"A": [278], "B": [278], "C": [278]}, {"A": [279], "B": [279], "C": [279]}, {"A": [280], "B": [280], "C": [280]}, {"A": [281], "B": [281], "C": [281]}, {"A": [282], "B": [282], "C": [282]}, {"A": [283], "B": [283], "C": [283]}, {"A": [284], "B": [284], "C": [284]}, {"A": [285], "B": [285], "C": [285]}, {"A": [286], "B": [286], "C": [286]}, {"A": [287], "B": [287], "C": [287]}, {"A": [288], "B": [288], "C": [288]}, {"A": [289], "B": [289], "C": [289]}, {"A": [290], "B": [290], "C": [290]}, {"A": [291], "B": [291], "C": [291]}, {"A": [292], "B": [292], "C": [292]}, {"A": [293], "B": [293], "C": [293]}, {"A": [294], "B": [294], "C": [294]}, {"A": [295], "B": [295], "C": [295]}, {"A": [296], "B": [296], "C": [296]}, {"A": [297], "B": [297], "C": [297]}, {"A": [298], "B": [298], "C": [298]}, {"A": [299], "B": [299], "C": [299]}, {"A": [300], "B": [300], "C": [300]}, {"A": [301], "B": [301], "C": [301]}, {"A": [302], "B": [302], "C": [302]}, {"A": [303], "B": [303], "C": [303]}, {"A": [304], "B": [304], "C": [304]}, {"A": [305], "B": [305], "C": [305]}, {"A": [306], "B": [306], "C": [306]}, {"A": [307], "B": [307], "C": [307]}, {"A": [308], "B": [308], "C": [308]}, {"A": [309], "B": [309], "C": [309]}, {"A": [310], "B": [310], "C": [310]}, {"A": [311], "B": [311], "C": [311]}, {"A": [312], "B": [312], "C": [312]}, {"A": [313], "B": [313], "C": [313]}, {"A": [314], "B": [314], "C": [314]}, {"A": [315], "B": [315], "C": [315]}, {"A": [316], "B": [316], "C": [316]}, {"A": [317], "B": [317], "C": [317]}, {"A": [318], "B": [318], "C": [318]}, {"A": [319], "B": [319], "C": [319]}, {"A": [320], "B": [320], "C": [320]}], "4GYT": [{"A": [1], "B": [1]}, {"A": [2], "B": [2]}, {"A": [3], "B": [3]}, {"A": [4], "B": [4]}, {"A": [5], "B": [5]}, {"A": [6], "B": [6]}, {"A": [7], "B": [7]}, {"A": [8], "B": [8]}, {"A": [9], "B": [9]}, {"A": [10], "B": [10]}, {"A": [11], "B": [11]}, {"A": [12], "B": [12]}, {"A": [13], "B": [13]}, {"A": [14], "B": [14]}, {"A": [15], "B": [15]}, {"A": [16], "B": [16]}, {"A": [17], "B": [17]}, {"A": [18], "B": [18]}, {"A": [19], "B": [19]}, {"A": [20], "B": [20]}, {"A": [21], "B": [21]}, {"A": [22], "B": [22]}, {"A": [23], "B": [23]}, {"A": [24], "B": [24]}, {"A": [25], "B": [25]}, {"A": [26], "B": [26]}, {"A": [27], "B": [27]}, {"A": [28], "B": [28]}, {"A": [29], "B": [29]}, {"A": [30], "B": [30]}, {"A": [31], "B": [31]}, {"A": [32], "B": [32]}, {"A": [33], "B": [33]}, {"A": [34], "B": [34]}, {"A": [35], "B": [35]}, {"A": [36], "B": [36]}, {"A": [37], "B": [37]}, {"A": [38], "B": [38]}, {"A": [39], "B": [39]}, {"A": [40], "B": [40]}, {"A": [41], "B": [41]}, {"A": [42], "B": [42]}, {"A": [43], "B": [43]}, {"A": [44], "B": [44]}, {"A": [45], "B": [45]}, {"A": [46], "B": [46]}, {"A": [47], "B": [47]}, {"A": [48], "B": [48]}, {"A": [49], "B": [49]}, {"A": [50], "B": [50]}, {"A": [51], "B": [51]}, {"A": [52], "B": [52]}, {"A": [53], "B": [53]}, {"A": [54], "B": [54]}, {"A": [55], "B": [55]}, {"A": [56], "B": [56]}, {"A": [57], "B": [57]}, {"A": [58], "B": [58]}, {"A": [59], "B": [59]}, {"A": [60], "B": [60]}, {"A": [61], "B": [61]}, {"A": [62], "B": [62]}, {"A": [63], "B": [63]}, {"A": [64], "B": [64]}, {"A": [65], "B": [65]}, {"A": [66], "B": [66]}, {"A": [67], "B": [67]}, {"A": [68], "B": [68]}, {"A": [69], "B": [69]}, {"A": [70], "B": [70]}, {"A": [71], "B": [71]}, {"A": [72], "B": [72]}, {"A": [73], "B": [73]}, {"A": [74], "B": [74]}, {"A": [75], "B": [75]}, {"A": [76], "B": [76]}, {"A": [77], "B": [77]}, {"A": [78], "B": [78]}, {"A": [79], "B": [79]}, {"A": [80], "B": [80]}, {"A": [81], "B": [81]}, {"A": [82], "B": [82]}, {"A": [83], "B": [83]}, {"A": [84], "B": [84]}, {"A": [85], "B": [85]}, {"A": [86], "B": [86]}, {"A": [87], "B": [87]}, {"A": [88], "B": [88]}, {"A": [89], "B": [89]}, {"A": [90], "B": [90]}, {"A": [91], "B": [91]}, {"A": [92], "B": [92]}, {"A": [93], "B": [93]}, {"A": [94], "B": [94]}, {"A": [95], "B": [95]}, {"A": [96], "B": [96]}, {"A": [97], "B": [97]}, {"A": [98], "B": [98]}, {"A": [99], "B": [99]}, {"A": [100], "B": [100]}, {"A": [101], "B": [101]}, {"A": [102], "B": [102]}, {"A": [103], "B": [103]}, {"A": [104], "B": [104]}, {"A": [105], "B": [105]}, {"A": [106], "B": [106]}, {"A": [107], "B": [107]}, {"A": [108], "B": [108]}, {"A": [109], "B": [109]}, {"A": [110], "B": [110]}, {"A": [111], "B": [111]}, {"A": [112], "B": [112]}, {"A": [113], "B": [113]}, {"A": [114], "B": [114]}, {"A": [115], "B": [115]}, {"A": [116], "B": [116]}, {"A": [117], "B": [117]}, {"A": [118], "B": [118]}, {"A": [119], "B": [119]}, {"A": [120], "B": [120]}, {"A": [121], "B": [121]}, {"A": [122], "B": [122]}, {"A": [123], "B": [123]}, {"A": [124], "B": [124]}, {"A": [125], "B": [125]}, {"A": [126], "B": [126]}, {"A": [127], "B": [127]}, {"A": [128], "B": [128]}, {"A": [129], "B": [129]}, {"A": [130], "B": [130]}, {"A": [131], "B": [131]}, {"A": [132], "B": [132]}, {"A": [133], "B": [133]}, {"A": [134], "B": [134]}, {"A": [135], "B": [135]}, {"A": [136], "B": [136]}, {"A": [137], "B": [137]}, {"A": [138], "B": [138]}, {"A": [139], "B": [139]}, {"A": [140], "B": [140]}, {"A": [141], "B": [141]}, {"A": [142], "B": [142]}, {"A": [143], "B": [143]}, {"A": [144], "B": [144]}, {"A": [145], "B": [145]}, {"A": [146], "B": [146]}, {"A": [147], "B": [147]}, {"A": [148], "B": [148]}, {"A": [149], "B": [149]}, {"A": [150], "B": [150]}, {"A": [151], "B": [151]}, {"A": [152], "B": [152]}, {"A": [153], "B": [153]}, {"A": [154], "B": [154]}, {"A": [155], "B": [155]}, {"A": [156], "B": [156]}, {"A": [157], "B": [157]}, {"A": [158], "B": [158]}, {"A": [159], "B": [159]}, {"A": [160], "B": [160]}, {"A": [161], "B": [161]}, {"A": [162], "B": [162]}, {"A": [163], "B": [163]}, {"A": [164], "B": [164]}, {"A": [165], "B": [165]}, {"A": [166], "B": [166]}, {"A": [167], "B": [167]}, {"A": [168], "B": [168]}, {"A": [169], "B": [169]}, {"A": [170], "B": [170]}, {"A": [171], "B": [171]}, {"A": [172], "B": [172]}, {"A": [173], "B": [173]}, {"A": [174], "B": [174]}, {"A": [175], "B": [175]}, {"A": [176], "B": [176]}, {"A": [177], "B": [177]}]}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/pdbs/4GYT.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_homooligomers/pdbs/6EHB.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/parsed_pdbs.jsonl
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{"seq_chain_A": "GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE", "coords_chain_A": {"N_chain_A": [[-15.113, 4.641, 12.533], [-13.275, 3.42, 10.93], [-10.741, 1.675, 9.445], [-7.432, 1.448, 9.871], [-5.644, -0.548, 8.854], [-7.205, -1.96, 6.899], [-7.793, 0.183, 5.237], [-5.26, 0.685, 4.162], [-4.958, -1.588, 2.516], [-7.177, -1.19, 0.885], [-6.291, 1.085, -0.528], [-4.142, -0.079, -1.945], [-5.518, -1.938, -3.517], [-7.137, -0.17, -5.019], [-5.151, 1.232, -6.404], [-4.087, -0.835, -8.02], [-6.444, -1.411, -9.542], [-6.711, 1.026, -10.874], [-4.295, 1.105, -12.41], [-4.894, -1.039, -14.08], [-7.091, -0.121, -15.482], [-9.668, -0.781, -14.596], [-12.955, -0.348, -13.741], [-15.301, -1.272, -12.622], [-15.546, -2.334, -10.044], [-16.758, -2.673, -6.77], [-16.165, -1.622, -3.469], [-16.378, -2.183, -0.036], [-15.967, -1.433, 3.409], [-16.551, -2.631, 6.58], [-16.843, -2.78, 9.914], [-14.464, -4.361, 10.3], [-14.612, -6.107, 8.198], [-13.921, -6.735, 4.835], [-15.439, -6.161, 1.804], [-15.834, -7.017, -1.536], [-17.223, -6.523, -4.647], [-17.77, -7.513, -7.955], [-18.767, -7.543, -11.345], [-17.695, -9.931, -13.085], [-14.398, -10.398, -14.406], [-10.967, -10.184, -15.446], [-8.395, -12.722, -15.59], [-6.205, -11.462, -14.409], [-7.298, -9.643, -12.523], [-8.704, -11.528, -10.949], [-6.649, -13.146, -9.873], [-5.274, -11.233, -8.368], [-7.233, -10.585, -6.439], [-7.641, -13.078, -5.203], [-5.146, -13.427, -3.951], [-5.083, -11.169, -2.282], [-7.277, -11.766, -0.57], [-6.278, -14.12, 0.645], [-4.045, -13.155, 2.06], [-5.234, -11.366, 3.89], [-7.155, -12.951, 5.25], [-5.415, -14.589, 6.647], [-4.611, -12.942, 8.681], [-6.891, -12.779, 10.155], [-9.004, -11.198, 9.278], [-12.305, -10.775, 7.928], [-13.164, -11.51, 4.65], [-15.046, -10.898, 1.956], [-15.534, -11.745, -1.147], [-17.085, -11.4, -4.207], [-17.565, -12.423, -7.43], [-19.448, -11.812, -10.016]], "CA_chain_A": [[-15.455, 3.353, 11.854], [-12.239, 3.522, 9.924], [-9.735, 0.662, 9.74], [-6.128, 1.8, 9.322], [-5.074, -1.624, 8.054], [-7.991, -2.219, 5.697], [-7.623, 1.317, 4.337], [-4.025, 0.475, 3.411], [-5.233, -2.549, 1.457], [-8.065, -0.527, -0.059], [-5.465, 1.902, -1.408], [-3.396, -0.941, -2.853], [-6.467, -2.459, -4.49], [-7.527, 0.902, -5.927], [-4.022, 1.506, -7.283], [-4.098, -1.901, -9.02], [-7.565, -1.214, -10.455], [-6.381, 2.179, -11.705], [-3.302, 0.671, -13.388], [-5.533, -1.961, -15.007], [-8.251, 0.462, -16.125], [-10.865, -1.492, -14.176], [-13.808, 0.635, -13.093], [-16.571, -1.877, -12.233], [-15.4, -3.01, -8.758], [-17.257, -1.959, -5.603], [-15.423, -1.976, -2.265], [-16.955, -1.707, 1.214], [-15.316, -1.837, 4.653], [-17.596, -2.568, 7.598], [-16.558, -3.484, 11.165], [-13.399, -5.329, 10.144], [-14.87, -6.942, 7.044], [-13.72, -6.182, 3.503], [-16.342, -6.715, 0.811], [-15.531, -6.62, -2.905], [-18.17, -7.079, -5.604], [-17.534, -7.115, -9.33], [-19.369, -8.381, -12.371], [-16.73, -10.481, -13.989], [-13.092, -9.792, -14.467], [-9.913, -10.939, -16.082], [-7.519, -13.504, -14.741], [-5.339, -10.574, -13.637], [-8.066, -9.217, -11.357], [-9.037, -12.676, -10.118], [-5.482, -13.557, -9.098], [-5.007, -10.2, -7.373], [-8.209, -10.695, -5.362], [-7.356, -14.301, -4.461], [-3.977, -13.177, -3.117], [-5.501, -10.237, -1.239], [-8.087, -12.495, 0.396], [-5.447, -15.036, 1.423], [-3.208, -12.371, 2.957], [-6.035, -10.78, 4.959], [-7.726, -14.004, 6.078], [-4.371, -15.076, 7.539], [-4.587, -12.019, 9.803], [-8.15, -12.851, 10.866], [-10.051, -10.372, 8.703], [-13.312, -11.38, 7.069], [-13.029, -10.997, 3.289], [-16.162, -11.398, 1.161], [-15.314, -11.41, -2.547], [-18.077, -11.972, -5.106], [-17.355, -12.096, -8.833], [-20.724, -12.228, -10.578]], "C_chain_A": [[-14.525, 3.068, 10.696], [-11.128, 2.581, 10.337], [-8.423, 1.057, 9.074], [-5.594, 0.705, 8.401], [-5.884, -1.859, 6.782], [-7.943, -1.043, 4.732], [-6.325, 1.21, 3.548], [-4.256, -0.489, 2.257], [-6.223, -2.003, 0.447], [-7.273, 0.337, -1.032], [-4.696, 1.044, -2.401], [-4.311, -1.534, -3.911], [-6.855, -1.387, -5.493], [-6.396, 1.25, -6.885], [-3.907, 0.448, -8.373], [-5.226, -1.69, -10.026], [-7.297, -0.052, -11.4], [-5.369, 1.804, -12.788], [-3.949, -0.183, -14.469], [-6.661, -1.332, -15.813], [-9.553, -0.226, -15.794], [-11.707, -0.553, -13.334], [-15.118, 0.051, -12.584], [-16.509, -2.642, -10.91], [-15.935, -2.111, -7.648], [-16.587, -2.519, -4.359], [-16.155, -1.378, -1.072], [-16.124, -2.248, 2.369], [-16.312, -1.604, 5.773], [-17.173, -3.42, 8.787], [-15.579, -4.644, 10.974], [-13.57, -6.281, 8.988], [-14.622, -6.128, 5.784], [-14.64, -6.92, 2.547], [-15.981, -6.143, -0.547], [-16.543, -7.305, -3.811], [-17.806, -6.597, -6.999], [-18.266, -8.076, -10.237], [-18.258, -8.767, -13.326], [-15.43, -9.727, -13.946], [-12.137, -10.73, -15.157], [-9.201, -11.812, -15.06], [-6.628, -12.616, -13.888], [-6.046, -10.077, -12.38], [-8.321, -10.379, -10.406], [-7.846, -13.1, -9.277], [-5.133, -12.518, -8.043], [-5.947, -10.345, -6.178], [-8.016, -11.971, -4.552], [-6.204, -14.094, -3.491], [-4.309, -12.212, -1.986], [-6.31, -10.947, -0.159], [-7.225, -13.41, 1.257], [-4.622, -14.283, 2.46], [-4.01, -11.836, 4.136], [-6.693, -11.844, 5.834], [-6.69, -14.56, 7.045], [-4.246, -14.212, 8.781], [-5.907, -11.982, 10.559], [-9.234, -11.922, 10.372], [-11.067, -11.248, 7.98], [-13.236, -10.706, 5.708], [-14.076, -11.68, 2.422], [-15.976, -10.869, -0.254], [-16.369, -12.137, -3.367], [-17.771, -11.49, -6.511], [-18.538, -12.681, -9.594], [-20.658, -12.365, -12.09]], "O_chain_A": [[-14.897, 2.519, 9.662], [-10.68, 2.634, 11.485], [-8.304, 0.991, 7.855], [-5.143, 0.977, 7.279], [-5.323, -1.971, 5.685], [-8.0, -1.245, 3.513], [-6.273, 1.603, 2.377], [-3.814, -0.247, 1.129], [-6.118, -2.31, -0.74], [-7.536, 0.331, -2.241], [-4.583, 1.398, -3.577], [-3.94, -1.609, -5.083], [-6.892, -1.646, -6.703], [-6.638, 1.55, -8.059], [-3.651, 0.78, -9.537], [-5.01, -1.776, -11.239], [-7.634, -0.111, -12.591], [-5.549, 2.134, -13.966], [-3.596, -0.085, -15.651], [-7.156, -1.972, -16.745], [-10.461, -0.263, -16.626], [-11.246, -0.047, -12.305], [-15.961, 0.814, -12.103], [-17.363, -3.495, -10.656], [-15.586, -0.928, -7.571], [-16.44, -3.737, -4.225], [-16.472, -0.181, -1.077], [-15.638, -3.382, 2.317], [-16.864, -0.507, 5.887], [-17.148, -4.645, 8.685], [-15.802, -5.764, 11.436], [-12.747, -7.189, 8.815], [-15.045, -4.971, 5.688], [-14.627, -8.155, 2.491], [-15.862, -4.925, -0.694], [-16.72, -8.526, -3.736], [-17.604, -5.4, -7.224], [-18.383, -9.265, -9.953], [-17.93, -8.033, -14.267], [-15.345, -8.577, -13.499], [-12.441, -11.902, -15.409], [-9.352, -11.668, -13.836], [-6.32, -12.97, -12.748], [-5.482, -10.097, -11.272], [-8.202, -10.239, -9.186], [-7.993, -13.366, -8.078], [-4.776, -12.872, -6.917], [-5.52, -10.263, -5.018], [-8.189, -11.956, -3.329], [-6.257, -14.545, -2.341], [-3.883, -12.411, -0.84], [-6.071, -10.762, 1.042], [-7.393, -13.461, 2.478], [-4.512, -14.709, 3.621], [-3.538, -11.859, 5.278], [-6.787, -11.672, 7.055], [-7.035, -14.941, 8.167], [-3.788, -14.689, 9.822], [-6.022, -11.231, 11.533], [-10.311, -11.879, 10.981], [-10.746, -12.329, 7.484], [-13.222, -9.474, 5.623], [-14.005, -12.897, 2.196], [-16.231, -9.692, -0.528], [-16.5, -13.357, -3.266], [-17.731, -10.283, -6.76], [-18.639, -13.904, -9.761], [-21.628, -12.801, -12.712]]}, "name": "6MRR", "num_of_chains": 1, "seq": "GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE"}
|
| 2 |
+
{"seq_chain_A": "HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV", "coords_chain_A": {"N_chain_A": [[37.0, 18.222, 51.819], [35.18, 19.045, 54.805], [33.142, 21.39, 56.357], [32.697, 22.256, 59.882], [30.075, 22.366, 60.868], [28.465, 21.048, 58.967], [29.669, 18.568, 59.079], [29.059, 17.634, 61.702], [26.271, 17.24, 61.58], [26.225, 15.306, 59.622], [27.541, 13.181, 60.918], [25.603, 12.501, 62.842], [23.621, 11.465, 61.194], [25.073, 9.367, 60.115], [25.367, 7.722, 62.376], [22.785, 6.789, 62.655], [22.499, 5.42, 60.214], [24.449, 3.414, 60.569], [23.344, 2.25, 62.7], [24.374, 2.554, 65.225], [24.763, 2.964, 68.494], [26.944, 3.77, 70.035], [28.442, 5.552, 68.362], [26.446, 7.553, 68.106], [26.246, 8.499, 70.748], [28.563, 9.948, 71.018], [28.108, 12.096, 69.352], [25.861, 13.648, 70.164], [24.504, 16.068, 72.578], [23.777, 16.707, 76.082], [21.518, 18.351, 75.963], [20.455, 18.057, 73.397], [17.593, 17.855, 71.366], [15.114, 15.757, 69.978], [12.531, 15.031, 67.686], [10.309, 12.633, 66.571], [7.796, 11.95, 64.523], [5.869, 12.982, 66.497], [6.728, 11.686, 68.914], [8.927, 12.6, 71.49], [12.386, 13.091, 72.157], [15.152, 13.567, 74.34], [18.607, 14.262, 74.992], [20.249, 11.415, 76.286], [18.136, 9.735, 75.635], [18.012, 9.917, 72.876], [20.144, 8.337, 72.204], [19.126, 5.939, 73.133], [17.028, 5.681, 71.32], [18.585, 4.982, 69.05], [19.592, 2.533, 69.856], [17.261, 1.04, 69.728], [16.741, 1.045, 67.012], [18.819, -0.538, 66.081], [18.18, -2.888, 67.459], [15.617, -3.575, 66.242], [16.392, -4.089, 63.726], [16.097, -2.152, 61.8], [16.701, -0.407, 58.831], [18.613, 2.321, 57.804], [19.364, 4.606, 55.165], [21.224, 7.164, 53.799], [20.201, 9.35, 51.263], [20.133, 11.886, 49.855], [20.945, 15.377, 50.442], [20.462, 18.834, 51.494], [21.245, 22.207, 52.499], [18.911, 23.991, 54.2], [17.94, 27.07, 54.88], [18.965, 27.602, 58.12], [19.227, 24.935, 58.758], [19.98, 21.458, 58.206], [18.725, 19.353, 55.737], [17.945, 16.136, 55.375], [17.279, 13.513, 53.17], [16.307, 10.372, 53.735], [16.304, 6.85, 53.277], [14.36, 4.368, 54.732], [14.218, 1.096, 55.78], [12.396, -1.822, 56.732], [11.509, -5.193, 57.372], [10.625, -5.836, 54.804], [9.326, -3.567, 53.956], [9.713, -0.36, 52.528], [10.495, 2.99, 53.244], [10.35, 6.575, 52.736], [11.862, 9.008, 54.514], [12.232, 12.164, 56.203], [14.836, 14.014, 57.563], [15.572, 16.566, 59.658], [18.051, 18.728, 60.656], [19.208, 20.958, 63.088], [22.156, 22.563, 63.663], [23.645, 24.166, 66.175], [26.134, 26.306, 67.255], [28.264, 24.429, 66.79], [27.157, 22.089, 67.871], [24.652, 19.433, 68.054], [21.475, 20.587, 67.962], [18.615, 20.547, 67.532], [16.573, 18.007, 66.237], [14.476, 16.857, 63.707], [12.331, 14.421, 62.706], [10.453, 13.297, 60.044], [7.846, 11.327, 58.803], [5.177, 10.579, 57.057]], "CA_chain_A": [[36.936, 18.773, 53.168], [33.829, 19.307, 55.268], [33.003, 22.335, 57.475], [32.383, 21.616, 61.147], [28.63, 22.278, 61.041], [27.969, 19.998, 58.095], [30.255, 17.336, 59.605], [28.319, 17.193, 62.883], [24.978, 16.74, 61.124], [26.544, 14.088, 58.891], [27.832, 12.133, 61.893], [24.312, 12.112, 63.413], [23.007, 10.631, 60.175], [25.91, 8.164, 60.045], [25.045, 6.895, 63.536], [21.519, 6.158, 62.308], [22.821, 4.501, 59.135], [25.19, 2.299, 61.114], [22.592, 1.729, 63.824], [25.424, 2.426, 66.209], [24.548, 3.825, 69.667], [28.216, 4.325, 70.466], [28.703, 6.763, 67.587], [25.452, 8.572, 68.459], [26.576, 9.042, 72.062], [29.65, 10.906, 70.821], [27.638, 13.226, 68.553], [24.834, 14.469, 70.801], [24.761, 16.77, 73.826], [22.683, 16.47, 77.021], [20.351, 19.16, 75.595], [19.926, 17.722, 72.07], [16.203, 17.421, 71.347], [14.694, 15.122, 68.734], [11.136, 14.692, 67.52], [10.007, 11.689, 65.501], [6.346, 11.777, 64.428], [5.194, 13.191, 67.767], [7.53, 11.223, 70.04], [10.037, 13.505, 71.773], [13.571, 12.523, 72.816], [16.186, 14.512, 74.768], [19.828, 13.667, 75.507], [20.521, 9.981, 76.157], [17.012, 9.105, 74.937], [18.376, 9.832, 71.466], [20.98, 7.138, 72.128], [18.19, 4.839, 73.298], [16.414, 5.594, 69.984], [19.465, 4.246, 68.135], [19.602, 1.154, 70.325], [16.045, 0.407, 69.247], [16.922, 0.85, 65.574], [19.706, -1.684, 66.003], [17.363, -4.009, 67.906], [14.558, -3.966, 65.32], [17.02, -4.306, 62.438], [15.647, -1.091, 60.918], [17.816, 0.015, 57.986], [18.627, 3.687, 57.284], [20.338, 4.921, 54.124], [21.169, 8.544, 53.342], [20.076, 9.473, 49.814], [20.579, 13.205, 49.457], [20.616, 16.466, 51.352], [20.718, 20.236, 51.207], [21.091, 23.102, 53.628], [17.708, 24.807, 54.042], [18.051, 28.065, 55.926], [20.011, 27.214, 59.057], [19.161, 23.525, 59.1], [20.28, 20.496, 57.163], [17.581, 18.51, 55.443], [18.39, 14.859, 54.825], [16.355, 12.518, 52.643], [16.831, 9.069, 54.073], [15.447, 5.708, 53.01], [14.308, 3.533, 55.936], [13.632, -0.203, 55.453], [12.292, -2.933, 57.665], [10.532, -6.258, 57.193], [10.11, -5.81, 53.444], [8.72, -2.286, 53.633], [10.651, 0.732, 52.332], [9.888, 4.302, 53.432], [11.125, 7.801, 52.577], [11.761, 9.852, 55.689], [12.975, 13.407, 56.146], [15.508, 14.281, 58.831], [15.997, 17.954, 59.618], [18.953, 18.92, 61.785], [19.776, 22.273, 63.355], [23.418, 22.556, 64.378], [23.916, 25.488, 66.721], [27.402, 26.364, 67.981], [28.974, 23.169, 66.639], [26.507, 20.955, 68.531], [23.321, 19.08, 67.562], [20.524, 21.296, 68.816], [17.262, 20.046, 67.347], [16.415, 17.077, 65.126], [13.085, 16.599, 63.409], [12.153, 13.313, 61.776], [9.088, 13.272, 59.537], [7.438, 10.301, 57.858], [3.735, 10.392, 57.071]], "C_chain_A": [[35.516, 19.181, 53.529], [33.835, 20.246, 56.472], [32.424, 21.668, 58.723], [30.875, 21.424, 61.352], [28.045, 21.144, 60.221], [28.428, 18.625, 58.602], [29.459, 16.766, 60.777], [26.977, 16.58, 62.491], [25.152, 15.408, 60.402], [26.83, 12.921, 59.83], [26.562, 11.609, 62.585], [23.641, 11.12, 62.474], [23.742, 9.311, 60.031], [25.597, 7.187, 61.182], [23.732, 6.145, 63.321], [21.733, 5.074, 61.25], [23.514, 3.243, 59.644], [24.422, 1.619, 62.24], [23.493, 1.584, 65.033], [25.271, 3.432, 67.346], [25.829, 4.473, 70.157], [28.568, 5.591, 69.685], [27.729, 7.873, 67.973], [25.765, 9.271, 69.778], [27.63, 10.133, 71.946], [29.225, 12.164, 70.07], [26.725, 14.189, 69.313], [25.32, 15.147, 72.075], [23.586, 16.509, 74.765], [21.4, 17.227, 76.67], [19.883, 19.001, 74.14], [18.479, 17.252, 72.148], [15.814, 16.879, 69.984], [13.215, 14.791, 68.796], [11.019, 13.732, 66.357], [8.511, 11.383, 65.495], [5.617, 11.906, 65.766], [5.97, 12.779, 69.006], [8.693, 12.188, 70.248], [11.14, 12.773, 72.507], [14.571, 13.629, 73.143], [17.412, 13.767, 75.307], [19.981, 12.179, 75.234], [19.346, 9.184, 75.593], [17.272, 8.974, 73.437], [19.264, 8.61, 71.249], [20.105, 5.892, 72.234], [17.42, 4.6, 71.996], [17.271, 4.767, 69.027], [19.569, 2.782, 68.557], [18.408, 0.378, 69.783], [16.076, 0.161, 67.749], [17.732, -0.411, 65.332], [18.974, -2.968, 66.397], [16.379, -4.492, 66.836], [15.096, -4.299, 63.928], [16.594, -3.31, 61.369], [16.837, -0.53, 60.144], [17.671, 1.442, 57.459], [19.7, 3.886, 56.222], [20.172, 6.366, 53.668], [21.132, 8.588, 51.81], [20.646, 10.787, 49.314], [20.267, 14.245, 50.522], [21.046, 17.824, 50.859], [20.686, 21.006, 52.51], [19.918, 24.019, 53.329], [17.639, 25.811, 55.176], [19.214, 27.689, 56.819], [19.807, 25.792, 59.584], [19.526, 22.664, 57.913], [19.11, 19.552, 56.986], [18.028, 17.274, 54.701], [17.2, 14.029, 54.397], [16.961, 11.15, 52.89], [15.812, 8.012, 53.681], [15.513, 4.779, 54.22], [13.552, 2.243, 55.625], [13.475, -1.054, 56.689], [11.196, -3.906, 57.263], [9.861, -6.201, 55.824], [9.414, -4.523, 53.038], [9.757, -1.167, 53.588], [9.951, 2.083, 52.441], [10.884, 5.43, 53.152], [10.835, 8.756, 53.723], [12.571, 11.114, 55.473], [13.554, 13.659, 57.513], [16.084, 15.679, 58.809], [16.836, 18.239, 60.853], [19.393, 20.364, 61.913], [21.031, 22.116, 64.2], [23.671, 23.966, 64.867], [25.19, 25.419, 67.548], [28.163, 25.044, 67.963], [28.334, 21.936, 67.267], [25.122, 20.677, 67.946], [22.274, 19.664, 68.485], [19.119, 20.714, 68.743], [17.234, 19.151, 66.116], [14.943, 16.776, 64.944], [12.96, 15.527, 62.351], [10.703, 13.266, 61.349], [8.891, 12.117, 58.572], [5.949, 10.026, 57.982], [3.267, 9.765, 55.77]], "O_chain_A": [[34.75, 19.627, 52.679], [34.466, 19.951, 57.486], [31.745, 20.64, 58.632], [30.444, 20.43, 61.936], [27.223, 20.37, 60.71], [27.666, 17.644, 58.56], [29.228, 15.556, 60.851], [26.587, 15.54, 63.01], [24.344, 14.49, 60.563], [26.43, 11.793, 59.561], [26.466, 10.42, 62.876], [23.133, 10.086, 62.899], [23.11, 8.253, 59.861], [25.565, 5.978, 60.976], [23.587, 5.004, 63.732], [21.208, 3.961, 61.365], [23.214, 2.143, 59.19], [24.797, 0.534, 62.681], [23.414, 0.602, 65.768], [25.628, 4.597, 67.202], [25.794, 5.601, 70.653], [28.946, 6.599, 70.283], [28.147, 9.011, 68.154], [25.567, 10.478, 69.921], [27.585, 11.13, 72.657], [29.904, 13.182, 70.145], [26.795, 15.401, 69.13], [26.4, 14.846, 72.582], [22.509, 16.12, 74.303], [20.32, 16.76, 77.023], [19.016, 19.744, 73.693], [18.171, 16.339, 72.914], [16.109, 17.485, 68.964], [12.702, 14.379, 69.847], [11.564, 13.984, 65.281], [8.025, 10.649, 66.357], [4.861, 11.02, 66.147], [5.876, 13.441, 70.042], [9.374, 12.559, 69.286], [10.86, 11.923, 73.362], [14.807, 14.512, 72.329], [17.271, 12.751, 75.992], [19.846, 11.725, 74.084], [19.535, 8.068, 75.133], [16.802, 8.045, 72.793], [19.141, 7.904, 70.238], [20.301, 4.917, 71.505], [17.186, 3.453, 71.614], [16.744, 3.928, 68.26], [19.568, 1.88, 67.714], [18.527, -0.796, 69.411], [15.514, -0.823, 67.273], [17.375, -1.244, 64.501], [19.117, -3.994, 65.744], [16.312, -5.687, 66.557], [14.345, -4.733, 63.062], [16.717, -3.592, 60.189], [17.874, -0.25, 60.724], [16.729, 1.724, 56.722], [20.833, 3.459, 56.399], [19.101, 6.761, 53.213], [21.922, 7.924, 51.14], [21.54, 10.807, 48.463], [19.441, 14.03, 51.403], [21.88, 17.95, 49.948], [20.154, 20.521, 53.506], [19.91, 24.707, 52.301], [17.308, 25.452, 56.305], [20.323, 27.467, 56.337], [20.162, 25.477, 60.718], [19.425, 23.084, 56.754], [18.544, 19.031, 57.959], [18.427, 17.355, 53.535], [16.231, 13.89, 55.157], [18.022, 10.811, 52.366], [14.608, 8.257, 53.699], [16.601, 4.461, 54.699], [12.391, 2.3, 55.229], [14.338, -1.043, 57.557], [10.101, -3.503, 56.86], [8.668, -6.468, 55.697], [8.959, -4.394, 51.905], [10.569, -1.042, 54.499], [8.934, 2.298, 51.817], [12.103, 5.252, 53.305], [9.722, 9.241, 53.893], [13.499, 11.135, 54.676], [12.845, 13.511, 58.513], [16.983, 15.964, 58.026], [16.401, 17.994, 61.976], [19.882, 20.939, 60.945], [20.986, 21.581, 65.294], [23.86, 24.873, 64.065], [25.32, 24.562, 68.418], [28.641, 24.59, 68.998], [28.902, 20.843, 67.213], [24.492, 21.583, 67.413], [22.176, 19.279, 69.665], [18.494, 20.423, 69.772], [17.819, 19.495, 65.079], [14.236, 16.503, 65.911], [13.432, 15.7, 61.239], [9.814, 13.225, 62.192], [9.69, 11.926, 57.664], [5.503, 9.336, 58.905], [3.96, 9.844, 54.754]]}, "name": "5L33", "num_of_chains": 1, "seq": "HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV"}
|
ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/seqs/5L33.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>5L33, score=1.5848, fixed_chains=[], designed_chains=['A'], model_name=v_48_020
|
| 2 |
+
HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV
|
| 3 |
+
>T=0.1, sample=0, score=0.8419, seq_recovery=0.4245
|
| 4 |
+
SVDPETAKALAFVKALEKADPELMAKVITPDTEMEVNGKKYKGDEIVEYVKKLKEEGIKYKLLSYKKDGDKYVFTMEKSYKGKTYTVTIEIEVKDGKVAKVVITEK
|
| 5 |
+
>T=0.1, sample=0, score=0.8087, seq_recovery=0.4811
|
| 6 |
+
SINEEEKKALDFIEALEKADPELMKKVIEPDTKMEVNGKKYEGEEIVKFVEELKKSGVKYKLKSYKKEGDKYVFTVEKSENGKTYTVTIEVKVENGKVKEVKITEE
|
ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/example_1_outputs/seqs/6MRR.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>6MRR, score=1.4854, fixed_chains=[], designed_chains=['A'], model_name=v_48_020
|
| 2 |
+
GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE
|
| 3 |
+
>T=0.1, sample=0, score=0.9197, seq_recovery=0.5147
|
| 4 |
+
GIDPELEEKVEELKKFLKEKGIDNVEIEVEDGVLKIKVKGASEELKEFLKKLKEELEEKGYEVEVEIE
|
| 5 |
+
>T=0.1, sample=0, score=0.9356, seq_recovery=0.5588
|
| 6 |
+
GKDPELEKYVKELKEFLKKQGITNVKIEVKDGTLTITTKGASEELKKFLEKLKKELEAKGYKVNVKIE
|
ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/pdbs/5L33.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/PDB_monomers/pdbs/6MRR.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_1.sh
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_1.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_monomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_monomers/example_1_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 19 |
+
|
| 20 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 21 |
+
|
| 22 |
+
python ../protein_mpnn_run.py \
|
| 23 |
+
--jsonl_path $path_for_parsed_chains \
|
| 24 |
+
--out_folder $output_dir \
|
| 25 |
+
--num_seq_per_target 2 \
|
| 26 |
+
--sampling_temp "0.1" \
|
| 27 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_2.sh
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_2.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_complexes/example_2_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 19 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 20 |
+
chains_to_design="A B"
|
| 21 |
+
|
| 22 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 23 |
+
|
| 24 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 25 |
+
|
| 26 |
+
python ../protein_mpnn_run.py \
|
| 27 |
+
--jsonl_path $path_for_parsed_chains \
|
| 28 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 29 |
+
--out_folder $output_dir \
|
| 30 |
+
--num_seq_per_target 2 \
|
| 31 |
+
--sampling_temp "0.1" \
|
| 32 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_3.sh
ADDED
|
@@ -0,0 +1,26 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_3.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
path_to_PDB="../PDB_complexes/pdbs/3HTN.pdb"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_complexes/example_3_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
chains_to_design="A B"
|
| 19 |
+
|
| 20 |
+
python ../protein_mpnn_run.py \
|
| 21 |
+
--pdb_path $path_to_PDB \
|
| 22 |
+
--pdb_path_chains "$chains_to_design" \
|
| 23 |
+
--out_folder $output_dir \
|
| 24 |
+
--num_seq_per_target 2 \
|
| 25 |
+
--sampling_temp "0.1" \
|
| 26 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_4.sh
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_4.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_complexes/example_4_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 22 |
+
chains_to_design="A C"
|
| 23 |
+
#The first amino acid in the chain corresponds to 1 and not PDB residues index for now.
|
| 24 |
+
fixed_positions="1 2 3 4 5 6 7 8 23 25, 10 11 12 13 14 15 16 17 18 19 20 40" #fixing/not designing residues 1 2 3...25 in chain A and residues 10 11 12...40 in chain C
|
| 25 |
+
|
| 26 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 27 |
+
|
| 28 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 29 |
+
|
| 30 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$fixed_positions"
|
| 31 |
+
|
| 32 |
+
python ../protein_mpnn_run.py \
|
| 33 |
+
--jsonl_path $path_for_parsed_chains \
|
| 34 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 35 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 36 |
+
--out_folder $output_dir \
|
| 37 |
+
--num_seq_per_target 2 \
|
| 38 |
+
--sampling_temp "0.1" \
|
| 39 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_5.sh
ADDED
|
@@ -0,0 +1,43 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_5.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_complexes/example_5_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 22 |
+
path_for_tied_positions=$output_dir"/tied_pdbs.jsonl"
|
| 23 |
+
chains_to_design="A C"
|
| 24 |
+
fixed_positions="9 10 11 12 13 14 15 16 17 18 19 20 21 22 23, 10 11 18 19 20 22"
|
| 25 |
+
tied_positions="1 2 3 4 5 6 7 8, 1 2 3 4 5 6 7 8" #two list must match in length; residue 1 in chain A and C will be sampled togther;
|
| 26 |
+
|
| 27 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 28 |
+
|
| 29 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 30 |
+
|
| 31 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$fixed_positions"
|
| 32 |
+
|
| 33 |
+
python ../helper_scripts/make_tied_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_tied_positions --chain_list "$chains_to_design" --position_list "$tied_positions"
|
| 34 |
+
|
| 35 |
+
python ../protein_mpnn_run.py \
|
| 36 |
+
--jsonl_path $path_for_parsed_chains \
|
| 37 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 38 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 39 |
+
--tied_positions_jsonl $path_for_tied_positions \
|
| 40 |
+
--out_folder $output_dir \
|
| 41 |
+
--num_seq_per_target 2 \
|
| 42 |
+
--sampling_temp "0.1" \
|
| 43 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_6.sh
ADDED
|
@@ -0,0 +1,33 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_6.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_homooligomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_homooligomers/example_6_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_tied_positions=$output_dir"/tied_pdbs.jsonl"
|
| 21 |
+
path_for_designed_sequences=$output_dir"/temp_0.1"
|
| 22 |
+
|
| 23 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 24 |
+
|
| 25 |
+
python ../helper_scripts/make_tied_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_tied_positions --homooligomer 1
|
| 26 |
+
|
| 27 |
+
python ../protein_mpnn_run.py \
|
| 28 |
+
--jsonl_path $path_for_parsed_chains \
|
| 29 |
+
--tied_positions_jsonl $path_for_tied_positions \
|
| 30 |
+
--out_folder $output_dir \
|
| 31 |
+
--num_seq_per_target 2 \
|
| 32 |
+
--sampling_temp "0.2" \
|
| 33 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/examples/submit_example_7.sh
ADDED
|
@@ -0,0 +1,40 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_7.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../PDB_complexes/example_7_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/PDB_complexes/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_bias=$output_dir"/bias_pdbs.jsonl"
|
| 22 |
+
AA_list="G P A"
|
| 23 |
+
bias_list="40.1 0.3 -0.05" #for G P A respectively; global AA bias in the logit space
|
| 24 |
+
chains_to_design="A B"
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 28 |
+
|
| 29 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 30 |
+
|
| 31 |
+
python ../helper_scripts/make_bias_AA.py --output_path=$path_for_bias --AA_list="$AA_list" --bias_list="$bias_list"
|
| 32 |
+
|
| 33 |
+
python ../protein_mpnn_run.py \
|
| 34 |
+
--jsonl_path $path_for_parsed_chains \
|
| 35 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 36 |
+
--out_folder $output_dir \
|
| 37 |
+
--bias_AA_jsonl $path_for_bias \
|
| 38 |
+
--num_seq_per_target 2 \
|
| 39 |
+
--sampling_temp "0.1" \
|
| 40 |
+
--batch_size 1
|
ProteinMPNN/vanilla_proteinmpnn/helper_scripts/assign_fixed_chains.py
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import json
|
| 5 |
+
|
| 6 |
+
with open(args.input_path, 'r') as json_file:
|
| 7 |
+
json_list = list(json_file)
|
| 8 |
+
|
| 9 |
+
global_designed_chain_list = []
|
| 10 |
+
if args.chain_list != '':
|
| 11 |
+
global_designed_chain_list = [str(item) for item in args.chain_list.split()]
|
| 12 |
+
my_dict = {}
|
| 13 |
+
for json_str in json_list:
|
| 14 |
+
result = json.loads(json_str)
|
| 15 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain'] #['A','B', 'C',...]
|
| 16 |
+
if len(global_designed_chain_list) > 0:
|
| 17 |
+
designed_chain_list = global_designed_chain_list
|
| 18 |
+
else:
|
| 19 |
+
#manually specify, e.g.
|
| 20 |
+
designed_chain_list = ["A"]
|
| 21 |
+
fixed_chain_list = [letter for letter in all_chain_list if letter not in designed_chain_list] #fix/do not redesign these chains
|
| 22 |
+
my_dict[result['name']]= (designed_chain_list, fixed_chain_list)
|
| 23 |
+
|
| 24 |
+
with open(args.output_path, 'w') as f:
|
| 25 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
if __name__ == "__main__":
|
| 29 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 30 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 31 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 32 |
+
argparser.add_argument("--chain_list", type=str, default='', help="List of the chains that need to be designed")
|
| 33 |
+
|
| 34 |
+
args = argparser.parse_args()
|
| 35 |
+
main(args)
|
| 36 |
+
|
| 37 |
+
# Output looks like this:
|
| 38 |
+
# {"5TTA": [["A"], ["B"]], "3LIS": [["A"], ["B"]]}
|
| 39 |
+
|
ProteinMPNN/vanilla_proteinmpnn/helper_scripts/make_bias_AA.py
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
|
| 5 |
+
import numpy as np
|
| 6 |
+
import json
|
| 7 |
+
|
| 8 |
+
bias_list = [float(item) for item in args.bias_list.split()]
|
| 9 |
+
AA_list = [str(item) for item in args.AA_list.split()]
|
| 10 |
+
|
| 11 |
+
my_dict = dict(zip(AA_list, bias_list))
|
| 12 |
+
|
| 13 |
+
with open(args.output_path, 'w') as f:
|
| 14 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
if __name__ == "__main__":
|
| 18 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 19 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 20 |
+
argparser.add_argument("--AA_list", type=str, default='', help="List of AAs to be biased")
|
| 21 |
+
argparser.add_argument("--bias_list", type=str, default='', help="AA bias strengths")
|
| 22 |
+
|
| 23 |
+
args = argparser.parse_args()
|
| 24 |
+
main(args)
|
| 25 |
+
|
| 26 |
+
#e.g. output
|
| 27 |
+
#{"A": -0.01, "G": 0.02}
|
ProteinMPNN/vanilla_proteinmpnn/helper_scripts/make_bias_per_res_dict.py
ADDED
|
@@ -0,0 +1,53 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import glob
|
| 5 |
+
import random
|
| 6 |
+
import numpy as np
|
| 7 |
+
import json
|
| 8 |
+
|
| 9 |
+
mpnn_alphabet = 'ACDEFGHIKLMNPQRSTVWYX'
|
| 10 |
+
|
| 11 |
+
mpnn_alphabet_dict = {'A': 0,'C': 1,'D': 2,'E': 3,'F': 4,'G': 5,'H': 6,'I': 7,'K': 8,'L': 9,'M': 10,'N': 11,'P': 12,'Q': 13,'R': 14,'S': 15,'T': 16,'V': 17,'W': 18,'Y': 19,'X': 20}
|
| 12 |
+
|
| 13 |
+
with open(args.input_path, 'r') as json_file:
|
| 14 |
+
json_list = list(json_file)
|
| 15 |
+
|
| 16 |
+
my_dict = {}
|
| 17 |
+
for json_str in json_list:
|
| 18 |
+
result = json.loads(json_str)
|
| 19 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:10]=='seq_chain_']
|
| 20 |
+
bias_by_res_dict = {}
|
| 21 |
+
for chain in all_chain_list:
|
| 22 |
+
chain_length = len(result[f'seq_chain_{chain}'])
|
| 23 |
+
bias_per_residue = np.zeros([chain_length, 21])
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
if chain == 'A':
|
| 27 |
+
residues = [0, 1, 2, 3, 4, 5, 11, 12, 13, 14, 15]
|
| 28 |
+
amino_acids = [5, 9] #[G, L]
|
| 29 |
+
for res in residues:
|
| 30 |
+
for aa in amino_acids:
|
| 31 |
+
bias_per_residue[res, aa] = 100.5
|
| 32 |
+
|
| 33 |
+
if chain == 'C':
|
| 34 |
+
residues = [0, 1, 2, 3, 4, 5, 11, 12, 13, 14, 15]
|
| 35 |
+
amino_acids = range(21)[1:] #[G, L]
|
| 36 |
+
for res in residues:
|
| 37 |
+
for aa in amino_acids:
|
| 38 |
+
bias_per_residue[res, aa] = -100.5
|
| 39 |
+
|
| 40 |
+
bias_by_res_dict[chain] = bias_per_residue.tolist()
|
| 41 |
+
my_dict[result['name']] = bias_by_res_dict
|
| 42 |
+
|
| 43 |
+
with open(args.output_path, 'w') as f:
|
| 44 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
if __name__ == "__main__":
|
| 48 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 49 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 50 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 51 |
+
|
| 52 |
+
args = argparser.parse_args()
|
| 53 |
+
main(args)
|