Upload 17 files
Browse files- .gitattributes +1 -0
- bin/foldseek +3 -0
- data/demo_input.csv +0 -0
- environment.yml +191 -0
- model-image.png +0 -0
- scripts/apply_foldseek_to_pdb.py +61 -0
- scripts/augmentation.py +61 -0
- scripts/calculate_shap.py +160 -0
- scripts/convert_to_PreTrainedModel.py +61 -0
- scripts/datasets.py +127 -0
- scripts/foldseek_util.py +106 -0
- scripts/get_aa_from_uniprot_accession.py +93 -0
- scripts/models.py +120 -0
- scripts/predict.py +168 -0
- scripts/predict_with_PreTrainedModel.py +149 -0
- scripts/train.py +493 -0
- scripts/use_foldseek_for_uniprot.py +106 -0
- utils.py +88 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
bin/foldseek filter=lfs diff=lfs merge=lfs -text
|
bin/foldseek
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0f903f1e906d2a7b38335caf2cb65323d40c0740825e2b2ab122cc7787d7e22b
|
| 3 |
+
size 100165624
|
data/demo_input.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
environment.yml
ADDED
|
@@ -0,0 +1,191 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
name: pltnum
|
| 2 |
+
channels:
|
| 3 |
+
- anaconda
|
| 4 |
+
- pytorch
|
| 5 |
+
- nvidia
|
| 6 |
+
- conda-forge
|
| 7 |
+
- defaults
|
| 8 |
+
dependencies:
|
| 9 |
+
- _libgcc_mutex=0.1=conda_forge
|
| 10 |
+
- _openmp_mutex=4.5=2_kmp_llvm
|
| 11 |
+
- abseil-cpp=20211102.0=hd4dd3e8_0
|
| 12 |
+
- aiohttp=3.9.5=py311h5eee18b_0
|
| 13 |
+
- aiosignal=1.2.0=pyhd3eb1b0_0
|
| 14 |
+
- arrow-cpp=14.0.2=h374c478_1
|
| 15 |
+
- attrs=23.1.0=py311h06a4308_0
|
| 16 |
+
- aws-c-auth=0.6.19=h5eee18b_0
|
| 17 |
+
- aws-c-cal=0.5.20=hdbd6064_0
|
| 18 |
+
- aws-c-common=0.8.5=h5eee18b_0
|
| 19 |
+
- aws-c-compression=0.2.16=h5eee18b_0
|
| 20 |
+
- aws-c-event-stream=0.2.15=h6a678d5_0
|
| 21 |
+
- aws-c-http=0.6.25=h5eee18b_0
|
| 22 |
+
- aws-c-io=0.13.10=h5eee18b_0
|
| 23 |
+
- aws-c-mqtt=0.7.13=h5eee18b_0
|
| 24 |
+
- aws-c-s3=0.1.51=hdbd6064_0
|
| 25 |
+
- aws-c-sdkutils=0.1.6=h5eee18b_0
|
| 26 |
+
- aws-checksums=0.1.13=h5eee18b_0
|
| 27 |
+
- aws-crt-cpp=0.18.16=h6a678d5_0
|
| 28 |
+
- aws-sdk-cpp=1.10.55=h721c034_0
|
| 29 |
+
- biopython=1.84=py311h331c9d8_0
|
| 30 |
+
- blas=1.0=mkl
|
| 31 |
+
- boost-cpp=1.82.0=hdb19cb5_2
|
| 32 |
+
- bottleneck=1.3.7=py311hf4808d0_0
|
| 33 |
+
- brotli-python=1.0.9=py311h6a678d5_8
|
| 34 |
+
- bzip2=1.0.8=h5eee18b_6
|
| 35 |
+
- c-ares=1.19.1=h5eee18b_0
|
| 36 |
+
- ca-certificates=2024.7.4=hbcca054_0
|
| 37 |
+
- certifi=2024.7.4=pyhd8ed1ab_0
|
| 38 |
+
- cffi=1.16.0=py311h5eee18b_1
|
| 39 |
+
- charset-normalizer=2.0.4=pyhd3eb1b0_0
|
| 40 |
+
- cloudpickle=3.0.0=pyhd8ed1ab_0
|
| 41 |
+
- cryptography=42.0.5=py311hdda0065_1
|
| 42 |
+
- cuda-cudart=12.1.105=0
|
| 43 |
+
- cuda-cupti=12.1.105=0
|
| 44 |
+
- cuda-libraries=12.1.0=0
|
| 45 |
+
- cuda-nvrtc=12.1.105=0
|
| 46 |
+
- cuda-nvtx=12.1.105=0
|
| 47 |
+
- cuda-opencl=12.5.39=0
|
| 48 |
+
- cuda-runtime=12.1.0=0
|
| 49 |
+
- cuda-version=12.5=3
|
| 50 |
+
- datasets=2.19.1=py311h06a4308_0
|
| 51 |
+
- dill=0.3.8=py311h06a4308_0
|
| 52 |
+
- ffmpeg=4.3=hf484d3e_0
|
| 53 |
+
- filelock=3.13.1=py311h06a4308_0
|
| 54 |
+
- freetype=2.12.1=h4a9f257_0
|
| 55 |
+
- frozenlist=1.4.0=py311h5eee18b_0
|
| 56 |
+
- fsspec=2024.3.1=py311h06a4308_0
|
| 57 |
+
- gflags=2.2.2=h6a678d5_1
|
| 58 |
+
- glog=0.5.0=h6a678d5_1
|
| 59 |
+
- gmp=6.2.1=h295c915_3
|
| 60 |
+
- gmpy2=2.1.2=py311hc9b5ff0_0
|
| 61 |
+
- gnutls=3.6.15=he1e5248_0
|
| 62 |
+
- grpc-cpp=1.48.2=he1ff14a_1
|
| 63 |
+
- huggingface_hub=0.23.1=py311h06a4308_0
|
| 64 |
+
- icu=73.1=h6a678d5_0
|
| 65 |
+
- idna=3.7=py311h06a4308_0
|
| 66 |
+
- intel-openmp=2023.1.0=hdb19cb5_46306
|
| 67 |
+
- jinja2=3.1.4=py311h06a4308_0
|
| 68 |
+
- joblib=1.4.2=py311h06a4308_0
|
| 69 |
+
- jpeg=9e=h5eee18b_1
|
| 70 |
+
- krb5=1.20.1=h143b758_1
|
| 71 |
+
- lame=3.100=h7b6447c_0
|
| 72 |
+
- lcms2=2.12=h3be6417_0
|
| 73 |
+
- ld_impl_linux-64=2.38=h1181459_1
|
| 74 |
+
- lerc=3.0=h295c915_0
|
| 75 |
+
- libboost=1.82.0=h109eef0_2
|
| 76 |
+
- libbrotlicommon=1.0.9=h5eee18b_8
|
| 77 |
+
- libbrotlidec=1.0.9=h5eee18b_8
|
| 78 |
+
- libbrotlienc=1.0.9=h5eee18b_8
|
| 79 |
+
- libcublas=12.1.0.26=0
|
| 80 |
+
- libcufft=11.0.2.4=0
|
| 81 |
+
- libcufile=1.10.1.7=0
|
| 82 |
+
- libcurand=10.3.6.82=0
|
| 83 |
+
- libcurl=8.7.1=h251f7ec_0
|
| 84 |
+
- libcusolver=11.4.4.55=0
|
| 85 |
+
- libcusparse=12.0.2.55=0
|
| 86 |
+
- libdeflate=1.17=h5eee18b_1
|
| 87 |
+
- libedit=3.1.20230828=h5eee18b_0
|
| 88 |
+
- libev=4.33=h7f8727e_1
|
| 89 |
+
- libevent=2.1.12=hdbd6064_1
|
| 90 |
+
- libffi=3.4.4=h6a678d5_1
|
| 91 |
+
- libgcc-ng=14.1.0=h77fa898_0
|
| 92 |
+
- libgfortran-ng=11.2.0=h00389a5_1
|
| 93 |
+
- libgfortran5=11.2.0=h1234567_1
|
| 94 |
+
- libiconv=1.16=h5eee18b_3
|
| 95 |
+
- libidn2=2.3.4=h5eee18b_0
|
| 96 |
+
- libjpeg-turbo=2.0.0=h9bf148f_0
|
| 97 |
+
- libllvm14=14.0.6=hdb19cb5_3
|
| 98 |
+
- libnghttp2=1.57.0=h2d74bed_0
|
| 99 |
+
- libnpp=12.0.2.50=0
|
| 100 |
+
- libnvjitlink=12.1.105=0
|
| 101 |
+
- libnvjpeg=12.1.1.14=0
|
| 102 |
+
- libpng=1.6.39=h5eee18b_0
|
| 103 |
+
- libprotobuf=3.20.3=he621ea3_0
|
| 104 |
+
- libssh2=1.11.0=h251f7ec_0
|
| 105 |
+
- libstdcxx-ng=14.1.0=hc0a3c3a_0
|
| 106 |
+
- libtasn1=4.19.0=h5eee18b_0
|
| 107 |
+
- libthrift=0.15.0=h1795dd8_2
|
| 108 |
+
- libtiff=4.5.1=h6a678d5_0
|
| 109 |
+
- libunistring=0.9.10=h27cfd23_0
|
| 110 |
+
- libuuid=1.41.5=h5eee18b_0
|
| 111 |
+
- libwebp-base=1.3.2=h5eee18b_0
|
| 112 |
+
- llvm-openmp=14.0.6=h9e868ea_0
|
| 113 |
+
- llvmlite=0.43.0=py311h6a678d5_0
|
| 114 |
+
- lz4-c=1.9.4=h6a678d5_1
|
| 115 |
+
- markupsafe=2.1.3=py311h5eee18b_0
|
| 116 |
+
- mkl=2023.1.0=h213fc3f_46344
|
| 117 |
+
- mkl-service=2.4.0=py311h5eee18b_1
|
| 118 |
+
- mkl_fft=1.3.8=py311h5eee18b_0
|
| 119 |
+
- mkl_random=1.2.4=py311hdb19cb5_0
|
| 120 |
+
- mpc=1.1.0=h10f8cd9_1
|
| 121 |
+
- mpfr=4.0.2=hb69a4c5_1
|
| 122 |
+
- mpmath=1.3.0=py311h06a4308_0
|
| 123 |
+
- multidict=6.0.4=py311h5eee18b_0
|
| 124 |
+
- multiprocess=0.70.15=py311h06a4308_0
|
| 125 |
+
- ncurses=6.4=h6a678d5_0
|
| 126 |
+
- nettle=3.7.3=hbbd107a_1
|
| 127 |
+
- networkx=3.3=py311h06a4308_0
|
| 128 |
+
- numba=0.60.0=py311h4bc866e_0
|
| 129 |
+
- numexpr=2.8.7=py311h65dcdc2_0
|
| 130 |
+
- numpy=1.26.0=py311h08b1b3b_0
|
| 131 |
+
- numpy-base=1.26.0=py311hf175353_0
|
| 132 |
+
- openh264=2.1.1=h4ff587b_0
|
| 133 |
+
- openjpeg=2.4.0=h9ca470c_1
|
| 134 |
+
- openssl=3.3.1=h4bc722e_2
|
| 135 |
+
- orc=1.7.4=hb3bc3d3_1
|
| 136 |
+
- packaging=24.1=py311h06a4308_0
|
| 137 |
+
- pandas=2.1.1=py311ha02d727_0
|
| 138 |
+
- pillow=10.3.0=py311h5eee18b_0
|
| 139 |
+
- pip=24.0=py311h06a4308_0
|
| 140 |
+
- pyarrow=14.0.2=py311hb6e97c4_0
|
| 141 |
+
- pybind11-abi=4=hd3eb1b0_1
|
| 142 |
+
- pycparser=2.21=pyhd3eb1b0_0
|
| 143 |
+
- pyopenssl=24.0.0=py311h06a4308_0
|
| 144 |
+
- pysocks=1.7.1=py311h06a4308_0
|
| 145 |
+
- python=3.11.8=h955ad1f_0
|
| 146 |
+
- python-dateutil=2.9.0post0=py311h06a4308_2
|
| 147 |
+
- python-tzdata=2023.3=pyhd3eb1b0_0
|
| 148 |
+
- python-xxhash=2.0.2=py311h5eee18b_1
|
| 149 |
+
- python_abi=3.11=2_cp311
|
| 150 |
+
- pytorch=2.3.1=py3.11_cuda12.1_cudnn8.9.2_0
|
| 151 |
+
- pytorch-cuda=12.1=ha16c6d3_5
|
| 152 |
+
- pytorch-mutex=1.0=cuda
|
| 153 |
+
- pytz=2024.1=py311h06a4308_0
|
| 154 |
+
- pyyaml=6.0.1=py311h5eee18b_0
|
| 155 |
+
- re2=2022.04.01=h295c915_0
|
| 156 |
+
- readline=8.2=h5eee18b_0
|
| 157 |
+
- regex=2023.10.3=py311h5eee18b_0
|
| 158 |
+
- requests=2.31.0=py311h06a4308_0
|
| 159 |
+
- s2n=1.3.27=hdbd6064_0
|
| 160 |
+
- safetensors=0.4.2=py311h24d97f6_1
|
| 161 |
+
- scikit-learn=1.2.2=py311h6a678d5_1
|
| 162 |
+
- scipy=1.13.1=py311h08b1b3b_0
|
| 163 |
+
- setuptools=69.5.1=py311h06a4308_0
|
| 164 |
+
- shap=0.45.1=cpu_py311h9c1f9ec_0
|
| 165 |
+
- six=1.16.0=pyhd3eb1b0_1
|
| 166 |
+
- slicer=0.0.8=pyhd8ed1ab_0
|
| 167 |
+
- snappy=1.1.10=h6a678d5_1
|
| 168 |
+
- sqlite=3.45.3=h5eee18b_0
|
| 169 |
+
- sympy=1.12=py311h06a4308_0
|
| 170 |
+
- tbb=2021.8.0=hdb19cb5_0
|
| 171 |
+
- threadpoolctl=3.5.0=py311h92b7b1e_0
|
| 172 |
+
- tk=8.6.14=h39e8969_0
|
| 173 |
+
- tokenizers=0.15.1=py311h22610ee_0
|
| 174 |
+
- torchaudio=2.3.1=py311_cu121
|
| 175 |
+
- torchtriton=2.3.1=py311
|
| 176 |
+
- torchvision=0.18.1=py311_cu121
|
| 177 |
+
- tqdm=4.66.4=py311h92b7b1e_0
|
| 178 |
+
- transformers=4.38.2=pyhd8ed1ab_0
|
| 179 |
+
- typing-extensions=4.11.0=py311h06a4308_0
|
| 180 |
+
- typing_extensions=4.11.0=py311h06a4308_0
|
| 181 |
+
- tzdata=2024a=h04d1e81_0
|
| 182 |
+
- urllib3=1.26.19=py311h06a4308_0
|
| 183 |
+
- utf8proc=2.6.1=h5eee18b_1
|
| 184 |
+
- wheel=0.43.0=py311h06a4308_0
|
| 185 |
+
- xlrd=2.0.1=pyhd3eb1b0_1
|
| 186 |
+
- xxhash=0.8.0=h7f8727e_3
|
| 187 |
+
- xz=5.4.6=h5eee18b_1
|
| 188 |
+
- yaml=0.2.5=h7b6447c_0
|
| 189 |
+
- yarl=1.9.3=py311h5eee18b_0
|
| 190 |
+
- zlib=1.2.13=h5eee18b_1
|
| 191 |
+
- zstd=1.5.5=hc292b87_2
|
model-image.png
ADDED
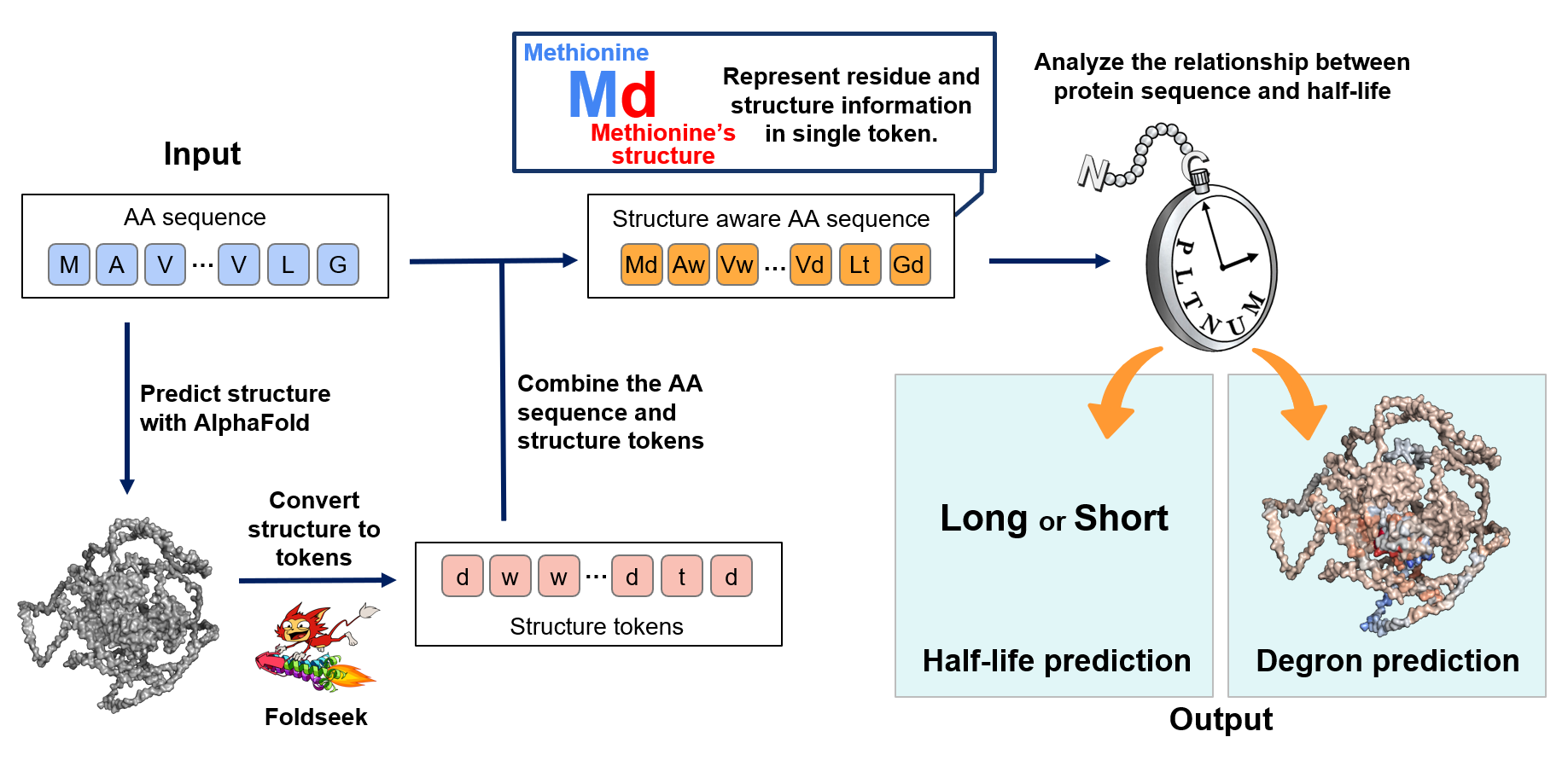
|
scripts/apply_foldseek_to_pdb.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import random
|
| 3 |
+
import argparse
|
| 4 |
+
import glob
|
| 5 |
+
import pandas as pd
|
| 6 |
+
import multiprocessing as mp
|
| 7 |
+
from foldseek_util import get_struc_seq
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
def parse_args():
|
| 11 |
+
parser = argparse.ArgumentParser()
|
| 12 |
+
parser.add_argument(
|
| 13 |
+
"--pdb_dir",
|
| 14 |
+
type=str,
|
| 15 |
+
default="./pdb_files",
|
| 16 |
+
help="Directory containing PDB files.",
|
| 17 |
+
)
|
| 18 |
+
parser.add_argument(
|
| 19 |
+
"--num_processes",
|
| 20 |
+
type=int,
|
| 21 |
+
default=2,
|
| 22 |
+
help="Number of processes to use for multiprocessing. Default is 2.",
|
| 23 |
+
)
|
| 24 |
+
parser.add_argument(
|
| 25 |
+
"--output_dir",
|
| 26 |
+
type=str,
|
| 27 |
+
default="./data",
|
| 28 |
+
help="Output directory.",
|
| 29 |
+
)
|
| 30 |
+
return parser.parse_args()
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
def get_foldseek_seq(pdb_path):
|
| 34 |
+
parsed_seqs = get_struc_seq(
|
| 35 |
+
"bin/foldseek",
|
| 36 |
+
pdb_path,
|
| 37 |
+
["A"],
|
| 38 |
+
process_id=random.randint(0, 10000000),
|
| 39 |
+
)["A"]
|
| 40 |
+
return parsed_seqs
|
| 41 |
+
|
| 42 |
+
|
| 43 |
+
if __name__ == "__main__":
|
| 44 |
+
config = parse_args()
|
| 45 |
+
|
| 46 |
+
pdb_files = glob.glob(os.path.join(config.pdb_dir, "*.pdb"))
|
| 47 |
+
|
| 48 |
+
with mp.Pool(config.num_processes) as pool:
|
| 49 |
+
output = pool.map(get_foldseek_seq, pdb_files)
|
| 50 |
+
|
| 51 |
+
aa, foldseek, aa_foldseek = zip(*output)
|
| 52 |
+
|
| 53 |
+
result = {}
|
| 54 |
+
result["file"] = pdb_files
|
| 55 |
+
result["aa"] = aa
|
| 56 |
+
result["foldseek"] = foldseek
|
| 57 |
+
result["aa_foldseek"] = aa_foldseek
|
| 58 |
+
|
| 59 |
+
df = pd.DataFrame(result)
|
| 60 |
+
|
| 61 |
+
df.to_csv(os.path.join(config.output_dir, "foldseek_result.csv"), index=False)
|
scripts/augmentation.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import random
|
| 2 |
+
|
| 3 |
+
def random_change_augmentation(aas, cfg):
|
| 4 |
+
residue_tokens = ("A", "C", "D", "E", "F", "G", "H", "I", "K", "L", "M", "N", "P", "Q", "R", "S", "T", "V", "W", "Y")
|
| 5 |
+
stracture_aware_tokens = ("a", "c", "d", "e", "f", "g", "h", "i", "k", "l", "m", "n", "p", "q", "r", "s", "t", "v", "w", "y")
|
| 6 |
+
length = len(aas)
|
| 7 |
+
swap_indices = random.sample(
|
| 8 |
+
range(length), int(length * cfg.random_change_ratio)
|
| 9 |
+
)
|
| 10 |
+
new_aas = ""
|
| 11 |
+
for i, aa in enumerate(aas):
|
| 12 |
+
if i in swap_indices:
|
| 13 |
+
if aas[i] in residue_tokens:
|
| 14 |
+
new_aas += random.choice(residue_tokens)
|
| 15 |
+
elif aas[i] in stracture_aware_tokens:
|
| 16 |
+
new_aas += random.choice(stracture_aware_tokens)
|
| 17 |
+
else:
|
| 18 |
+
new_aas += aa
|
| 19 |
+
return new_aas
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
def mask_augmentation(aas, cfg):
|
| 23 |
+
length = len(aas)
|
| 24 |
+
swap_indices = random.sample(
|
| 25 |
+
range(0, length // cfg.token_length),
|
| 26 |
+
int(length // cfg.token_length * cfg.mask_ratio),
|
| 27 |
+
)
|
| 28 |
+
for ith in swap_indices:
|
| 29 |
+
aas = (
|
| 30 |
+
aas[: ith * cfg.token_length]
|
| 31 |
+
+ "@" * cfg.token_length
|
| 32 |
+
+ aas[(ith + 1) * cfg.token_length :]
|
| 33 |
+
)
|
| 34 |
+
aas = aas.replace("@@", "<mask>").replace("@", "<mask>")
|
| 35 |
+
return aas
|
| 36 |
+
|
| 37 |
+
|
| 38 |
+
def random_delete_augmentation(aas, cfg):
|
| 39 |
+
length = len(aas)
|
| 40 |
+
swap_indices = random.sample(
|
| 41 |
+
range(0, length // cfg.token_length),
|
| 42 |
+
int(length // cfg.token_length * cfg.random_delete_ratio),
|
| 43 |
+
)
|
| 44 |
+
for ith in swap_indices:
|
| 45 |
+
aas = (
|
| 46 |
+
aas[: ith * cfg.token_length]
|
| 47 |
+
+ "@" * cfg.token_length
|
| 48 |
+
+ aas[(ith + 1) * cfg.token_length :]
|
| 49 |
+
)
|
| 50 |
+
aas = aas.replace("@@", "").replace("@", "")
|
| 51 |
+
return aas
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
def truncate_augmentation(aas, cfg):
|
| 55 |
+
length = len(aas)
|
| 56 |
+
if length > cfg.max_length:
|
| 57 |
+
diff = length - cfg.max_length
|
| 58 |
+
start = random.randint(0, diff)
|
| 59 |
+
return aas[start : start + cfg.max_length]
|
| 60 |
+
else:
|
| 61 |
+
return aas
|
scripts/calculate_shap.py
ADDED
|
@@ -0,0 +1,160 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import glob
|
| 3 |
+
import sys
|
| 4 |
+
import argparse
|
| 5 |
+
import pandas as pd
|
| 6 |
+
import torch
|
| 7 |
+
from transformers import AutoTokenizer
|
| 8 |
+
import shap
|
| 9 |
+
|
| 10 |
+
sys.path.append(".")
|
| 11 |
+
from utils import seed_everything, save_pickle
|
| 12 |
+
from models import PLTNUM, PLTNUM_PreTrainedModel
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
def parse_args():
|
| 16 |
+
parser = argparse.ArgumentParser(
|
| 17 |
+
description="Calculate SHAP values with a pretrained protein half-life prediction model."
|
| 18 |
+
)
|
| 19 |
+
parser.add_argument(
|
| 20 |
+
"--data_path",
|
| 21 |
+
type=str,
|
| 22 |
+
required=True,
|
| 23 |
+
help="Path to the input data.",
|
| 24 |
+
)
|
| 25 |
+
parser.add_argument(
|
| 26 |
+
"--model",
|
| 27 |
+
type=str,
|
| 28 |
+
default="westlake-repl/SaProt_650M_AF2",
|
| 29 |
+
help="Pretrained model name or path.",
|
| 30 |
+
)
|
| 31 |
+
parser.add_argument(
|
| 32 |
+
"--architecture",
|
| 33 |
+
type=str,
|
| 34 |
+
default="SaProt",
|
| 35 |
+
help="Model architecture: 'ESM2', 'SaProt', or 'LSTM'.",
|
| 36 |
+
)
|
| 37 |
+
parser.add_argument(
|
| 38 |
+
"--folds",
|
| 39 |
+
type=int,
|
| 40 |
+
default=10,
|
| 41 |
+
help="The number of folds for prediction.",
|
| 42 |
+
)
|
| 43 |
+
parser.add_argument(
|
| 44 |
+
"--do_cross_validation",
|
| 45 |
+
action="store_true",
|
| 46 |
+
default=False,
|
| 47 |
+
help="Use cross validation for prediction. If True, you have to specify the 'data_path' that contanins fold information, 'folds' for the number of folds, and 'model_path' for the directory of the model weights.",
|
| 48 |
+
)
|
| 49 |
+
parser.add_argument(
|
| 50 |
+
"--model_path",
|
| 51 |
+
type=str,
|
| 52 |
+
required=False,
|
| 53 |
+
help="Path to the model weight(s).",
|
| 54 |
+
)
|
| 55 |
+
parser.add_argument("--batch_size", type=int, default=4, help="Batch size.")
|
| 56 |
+
parser.add_argument(
|
| 57 |
+
"--seed",
|
| 58 |
+
type=int,
|
| 59 |
+
default=42,
|
| 60 |
+
help="Seed for reproducibility.",
|
| 61 |
+
)
|
| 62 |
+
parser.add_argument(
|
| 63 |
+
"--max_length",
|
| 64 |
+
type=int,
|
| 65 |
+
default=512,
|
| 66 |
+
help="Maximum input sequence length. Two tokens are used fo <cls> and <eos> tokens. So the actual length of input sequence is max_length - 2. Padding or truncation is applied to make the length of input sequence equal to max_length.",
|
| 67 |
+
)
|
| 68 |
+
parser.add_argument(
|
| 69 |
+
"--output_dir",
|
| 70 |
+
type=str,
|
| 71 |
+
default="./output",
|
| 72 |
+
help="Output directory.",
|
| 73 |
+
)
|
| 74 |
+
parser.add_argument(
|
| 75 |
+
"--task",
|
| 76 |
+
type=str,
|
| 77 |
+
default="classification",
|
| 78 |
+
help="Task type: 'classification' or 'regression'.",
|
| 79 |
+
)
|
| 80 |
+
parser.add_argument(
|
| 81 |
+
"--sequence_col",
|
| 82 |
+
type=str,
|
| 83 |
+
default="aa_foldseek",
|
| 84 |
+
help="Column name fot the input sequence.",
|
| 85 |
+
)
|
| 86 |
+
parser.add_argument(
|
| 87 |
+
"--max_evals",
|
| 88 |
+
type=int,
|
| 89 |
+
default=5000,
|
| 90 |
+
help="Number of evaluations for SHAP values calculation.",
|
| 91 |
+
)
|
| 92 |
+
|
| 93 |
+
|
| 94 |
+
return parser.parse_args()
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
def calculate_shap_fn(texts, model, cfg):
|
| 98 |
+
if len(texts) == 1:
|
| 99 |
+
texts = texts[0]
|
| 100 |
+
else:
|
| 101 |
+
texts = texts.tolist()
|
| 102 |
+
|
| 103 |
+
inputs = cfg.tokenizer(
|
| 104 |
+
texts,
|
| 105 |
+
return_tensors="pt",
|
| 106 |
+
padding=True,
|
| 107 |
+
truncation=True,
|
| 108 |
+
max_length=cfg.max_length,
|
| 109 |
+
)
|
| 110 |
+
inputs = {k: v.to(cfg.device) for k, v in inputs.items()}
|
| 111 |
+
with torch.no_grad():
|
| 112 |
+
outputs = model(inputs)
|
| 113 |
+
outputs = torch.sigmoid(outputs).detach().cpu().numpy()
|
| 114 |
+
return outputs
|
| 115 |
+
|
| 116 |
+
|
| 117 |
+
if __name__ == "__main__":
|
| 118 |
+
config = parse_args()
|
| 119 |
+
config.device = "cuda" if torch.cuda.is_available() else "cpu"
|
| 120 |
+
|
| 121 |
+
if not os.path.exists(config.output_dir):
|
| 122 |
+
os.makedirs(config.output_dir)
|
| 123 |
+
seed_everything(config.seed)
|
| 124 |
+
|
| 125 |
+
df = pd.read_csv(config.data_path)
|
| 126 |
+
config.tokenizer = AutoTokenizer.from_pretrained(config.model)
|
| 127 |
+
|
| 128 |
+
if config.do_cross_validation:
|
| 129 |
+
model_weights = glob.glob(os.path.join(config.model_path, "*.pth"))
|
| 130 |
+
for fold in range(config.folds):
|
| 131 |
+
model = PLTNUM(config).to(config.device)
|
| 132 |
+
model_weight = [w for w in model_weights if f"fold{fold}.pth" in w][0]
|
| 133 |
+
model.load_state_dict(torch.load(model_weight, map_location="cpu"))
|
| 134 |
+
model.eval()
|
| 135 |
+
|
| 136 |
+
df_fold = df[df["fold"] == fold].reset_index(drop=True)
|
| 137 |
+
explainer = shap.Explainer(lambda x: calculate_shap_fn(x, model, config), config.tokenizer)
|
| 138 |
+
shap_values = explainer(
|
| 139 |
+
df_fold[config.sequence_col].values.tolist(),
|
| 140 |
+
batch_size=config.batch_size,
|
| 141 |
+
max_evals=config.max_evals,
|
| 142 |
+
)
|
| 143 |
+
|
| 144 |
+
save_pickle(os.path.join(config.output_dir, f"shap_values_fold{fold}.pickle"), shap_values)
|
| 145 |
+
else:
|
| 146 |
+
model = PLTNUM_PreTrainedModel.from_pretrained(config.model_path, cfg=config).to(config.device)
|
| 147 |
+
model.eval()
|
| 148 |
+
|
| 149 |
+
# build an explainer using a token masker
|
| 150 |
+
explainer = shap.Explainer(lambda x: calculate_shap_fn(x, model, config), config.tokenizer)
|
| 151 |
+
|
| 152 |
+
shap_values = explainer(
|
| 153 |
+
df[config.sequence_col].values.tolist(),
|
| 154 |
+
batch_size=config.batch_size,
|
| 155 |
+
max_evals=config.max_evals,
|
| 156 |
+
)
|
| 157 |
+
|
| 158 |
+
save_pickle(
|
| 159 |
+
os.path.join(config.output_dir, "shap_values.pickle"), shap_values
|
| 160 |
+
)
|
scripts/convert_to_PreTrainedModel.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import sys
|
| 3 |
+
import argparse
|
| 4 |
+
|
| 5 |
+
import torch
|
| 6 |
+
import shutil
|
| 7 |
+
|
| 8 |
+
# Append the utils module path
|
| 9 |
+
sys.path.append("../")
|
| 10 |
+
from models import PLTNUM
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
def parse_args():
|
| 14 |
+
"""
|
| 15 |
+
Parse command line arguments.
|
| 16 |
+
"""
|
| 17 |
+
parser = argparse.ArgumentParser(
|
| 18 |
+
description="Convert the model implemented with nn.Module to a model implemented with transformers' PreTrainedModel."
|
| 19 |
+
)
|
| 20 |
+
parser.add_argument(
|
| 21 |
+
"--model_path",
|
| 22 |
+
type=str,
|
| 23 |
+
help="The path to a model weight which you want to convert.",
|
| 24 |
+
)
|
| 25 |
+
parser.add_argument(
|
| 26 |
+
"--config_and_tokenizer_path",
|
| 27 |
+
type=str,
|
| 28 |
+
help="The path to a config and tokenizer of the model which you want to convert.",
|
| 29 |
+
)
|
| 30 |
+
parser.add_argument(
|
| 31 |
+
"--model",
|
| 32 |
+
type=str,
|
| 33 |
+
help="The name of the base model of the finetuned model",
|
| 34 |
+
)
|
| 35 |
+
parser.add_argument(
|
| 36 |
+
"--output_dir",
|
| 37 |
+
type=str,
|
| 38 |
+
default="./",
|
| 39 |
+
help="Directory to save the prediction.",
|
| 40 |
+
)
|
| 41 |
+
parser.add_argument(
|
| 42 |
+
"--task",
|
| 43 |
+
type=str,
|
| 44 |
+
default="classification",
|
| 45 |
+
)
|
| 46 |
+
|
| 47 |
+
return parser.parse_args()
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
if __name__ == "__main__":
|
| 51 |
+
config = parse_args()
|
| 52 |
+
|
| 53 |
+
if not os.path.exists(config.output_dir):
|
| 54 |
+
os.makedirs(config.output_dir)
|
| 55 |
+
|
| 56 |
+
model = PLTNUM(config)
|
| 57 |
+
model.load_state_dict(torch.load(config.model_path, map_location="cpu"))
|
| 58 |
+
|
| 59 |
+
torch.save(model.state_dict(), os.path.join(config.output_dir, "pytorch_model.bin"))
|
| 60 |
+
for filename in ["config.json", "special_tokens_map.json", "tokenizer_config.json", "vocab.txt"]:
|
| 61 |
+
shutil.copy(os.path.join(config.config_and_tokenizer_path, filename), os.path.join(config.output_dir, filename))
|
scripts/datasets.py
ADDED
|
@@ -0,0 +1,127 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import random
|
| 2 |
+
import numpy as np
|
| 3 |
+
import torch
|
| 4 |
+
from torch.utils.data import Dataset
|
| 5 |
+
from augmentation import (
|
| 6 |
+
mask_augmentation,
|
| 7 |
+
random_change_augmentation,
|
| 8 |
+
random_delete_augmentation,
|
| 9 |
+
truncate_augmentation,
|
| 10 |
+
)
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
def tokenize_input(cfg, text):
|
| 14 |
+
inputs = cfg.tokenizer(
|
| 15 |
+
text,
|
| 16 |
+
add_special_tokens=True,
|
| 17 |
+
max_length=cfg.max_length,
|
| 18 |
+
padding="max_length",
|
| 19 |
+
truncation=True,
|
| 20 |
+
return_offsets_mapping=False,
|
| 21 |
+
return_attention_mask=True,
|
| 22 |
+
)
|
| 23 |
+
for k, v in inputs.items():
|
| 24 |
+
inputs[k] = torch.tensor(v, dtype=torch.long)
|
| 25 |
+
return inputs
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
def one_hot_encoding(aa, amino_acids, cfg):
|
| 29 |
+
aa = aa[: cfg.max_length].ljust(cfg.max_length, " ")
|
| 30 |
+
one_hot = np.zeros((len(aa), len(amino_acids)))
|
| 31 |
+
for i, a in enumerate(aa):
|
| 32 |
+
if a in amino_acids:
|
| 33 |
+
one_hot[i, amino_acids.index(a)] = 1
|
| 34 |
+
return one_hot
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
def one_hot_encode_input(text, cfg):
|
| 38 |
+
inputs = one_hot_encoding(text, ("A","C","D","E","F","G","H","I","K","L","M","N","P","Q","R","S","T","V","W","Y"," "), cfg)
|
| 39 |
+
return torch.tensor(inputs, dtype=torch.float)
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
class PLTNUMDataset(Dataset):
|
| 43 |
+
def __init__(self, cfg, df, train=True):
|
| 44 |
+
self.df = df
|
| 45 |
+
self.cfg = cfg
|
| 46 |
+
self.train = train
|
| 47 |
+
|
| 48 |
+
def __len__(self):
|
| 49 |
+
return len(self.df)
|
| 50 |
+
|
| 51 |
+
def __getitem__(self, idx):
|
| 52 |
+
data = self.df.iloc[idx]
|
| 53 |
+
aas = self._adjust_sequence_length(data[self.cfg.sequence_col])
|
| 54 |
+
|
| 55 |
+
if self.train:
|
| 56 |
+
aas = self._apply_augmentation(aas)
|
| 57 |
+
|
| 58 |
+
aas = aas.replace("__", "<pad>")
|
| 59 |
+
|
| 60 |
+
inputs = tokenize_input(self.cfg, aas)
|
| 61 |
+
|
| 62 |
+
if "target" in data:
|
| 63 |
+
return inputs, torch.tensor(data["target"], dtype=torch.float32)
|
| 64 |
+
return inputs, np.nan
|
| 65 |
+
|
| 66 |
+
def _adjust_sequence_length(self, aas):
|
| 67 |
+
max_length = (self.cfg.max_length - 2) * self.cfg.token_length
|
| 68 |
+
if len(aas) > max_length:
|
| 69 |
+
if self.cfg.used_sequence == "left":
|
| 70 |
+
return aas[: max_length]
|
| 71 |
+
elif self.cfg.used_sequence == "right":
|
| 72 |
+
return aas[-max_length:]
|
| 73 |
+
elif self.cfg.used_sequence == "both":
|
| 74 |
+
half_max_len = max_length // 2
|
| 75 |
+
return aas[:half_max_len] + "__" + aas[-half_max_len:]
|
| 76 |
+
elif self.cfg.used_sequence == "internal":
|
| 77 |
+
offset = (len(aas) - max_length) // 2
|
| 78 |
+
return aas[offset:offset + max_length]
|
| 79 |
+
return aas
|
| 80 |
+
|
| 81 |
+
def _apply_augmentation(self, aas):
|
| 82 |
+
if self.cfg.random_change_ratio > 0:
|
| 83 |
+
aas = random_change_augmentation(aas, self.cfg)
|
| 84 |
+
if (
|
| 85 |
+
random.random() <= self.cfg.random_delete_prob
|
| 86 |
+
) and self.cfg.random_delete_ratio > 0:
|
| 87 |
+
aas = random_delete_augmentation(aas, self.cfg)
|
| 88 |
+
if (random.random() <= self.cfg.mask_prob) and self.cfg.mask_ratio > 0:
|
| 89 |
+
aas = mask_augmentation(aas, self.cfg)
|
| 90 |
+
if random.random() <= self.cfg.truncate_augmentation_prob:
|
| 91 |
+
aas = truncate_augmentation(aas, self.cfg)
|
| 92 |
+
return aas
|
| 93 |
+
|
| 94 |
+
|
| 95 |
+
class LSTMDataset(Dataset):
|
| 96 |
+
def __init__(self, cfg, df, train=True):
|
| 97 |
+
self.df = df
|
| 98 |
+
self.cfg = cfg
|
| 99 |
+
self.train = train
|
| 100 |
+
|
| 101 |
+
def __len__(self):
|
| 102 |
+
return len(self.df)
|
| 103 |
+
|
| 104 |
+
def __getitem__(self, idx):
|
| 105 |
+
data = self.df.iloc[idx]
|
| 106 |
+
aas = data[self.cfg.sequence_col]
|
| 107 |
+
aas = self._adjust_sequence_length(aas)
|
| 108 |
+
aas = aas.replace("__", "<pad>")
|
| 109 |
+
|
| 110 |
+
inputs = one_hot_encode_input(aas, self.cfg)
|
| 111 |
+
|
| 112 |
+
return inputs, torch.tensor(data["target"], dtype=torch.float32)
|
| 113 |
+
|
| 114 |
+
def _adjust_sequence_length(self, aas):
|
| 115 |
+
max_length = (self.cfg.max_length - 2) * self.cfg.token_length
|
| 116 |
+
if len(aas) > max_length:
|
| 117 |
+
if self.cfg.used_sequence == "left":
|
| 118 |
+
return aas[:max_length]
|
| 119 |
+
elif self.cfg.used_sequence == "right":
|
| 120 |
+
return aas[-max_length:]
|
| 121 |
+
elif self.cfg.used_sequence == "both":
|
| 122 |
+
half_max_len = max_length // 2
|
| 123 |
+
return aas[:half_max_len] + "__" + aas[-half_max_len:]
|
| 124 |
+
elif self.cfg.used_sequence == "internal":
|
| 125 |
+
offset = (len(aas) - max_length) // 2
|
| 126 |
+
return aas[offset:offset + max_length]
|
| 127 |
+
return aas
|
scripts/foldseek_util.py
ADDED
|
@@ -0,0 +1,106 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# https://github.com/westlake-repl/SaProt/blob/main/utils/foldseek_util.py
|
| 2 |
+
|
| 3 |
+
# MIT License
|
| 4 |
+
|
| 5 |
+
# Copyright (c) 2023 westlake-repl
|
| 6 |
+
|
| 7 |
+
# Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 8 |
+
# of this software and associated documentation files (the "Software"), to deal
|
| 9 |
+
# in the Software without restriction, including without limitation the rights
|
| 10 |
+
# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 11 |
+
# copies of the Software, and to permit persons to whom the Software is
|
| 12 |
+
# furnished to do so, subject to the following conditions:
|
| 13 |
+
|
| 14 |
+
# The above copyright notice and this permission notice shall be included in all
|
| 15 |
+
# copies or substantial portions of the Software.
|
| 16 |
+
|
| 17 |
+
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 18 |
+
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 19 |
+
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 20 |
+
# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 21 |
+
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 22 |
+
# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 23 |
+
# SOFTWARE.
|
| 24 |
+
|
| 25 |
+
import os
|
| 26 |
+
import json
|
| 27 |
+
import numpy as np
|
| 28 |
+
import sys
|
| 29 |
+
|
| 30 |
+
sys.path.append(".")
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
# Get structural seqs from pdb file
|
| 34 |
+
def get_struc_seq(
|
| 35 |
+
foldseek,
|
| 36 |
+
path,
|
| 37 |
+
chains: list = None,
|
| 38 |
+
process_id: int = 0,
|
| 39 |
+
plddt_path: str = None,
|
| 40 |
+
plddt_threshold: float = 70.0,
|
| 41 |
+
) -> dict:
|
| 42 |
+
"""
|
| 43 |
+
|
| 44 |
+
Args:
|
| 45 |
+
foldseek: Binary executable file of foldseek
|
| 46 |
+
path: Path to pdb file
|
| 47 |
+
chains: Chains to be extracted from pdb file. If None, all chains will be extracted.
|
| 48 |
+
process_id: Process ID for temporary files. This is used for parallel processing.
|
| 49 |
+
plddt_path: Path to plddt file. If None, plddt will not be used.
|
| 50 |
+
plddt_threshold: Threshold for plddt. If plddt is lower than this value, the structure will be masked.
|
| 51 |
+
|
| 52 |
+
Returns:
|
| 53 |
+
seq_dict: A dict of structural seqs. The keys are chain IDs. The values are tuples of
|
| 54 |
+
(seq, struc_seq, combined_seq).
|
| 55 |
+
"""
|
| 56 |
+
assert os.path.exists(foldseek), f"Foldseek not found: {foldseek}"
|
| 57 |
+
assert os.path.exists(path), f"Pdb file not found: {path}"
|
| 58 |
+
assert plddt_path is None or os.path.exists(
|
| 59 |
+
plddt_path
|
| 60 |
+
), f"Plddt file not found: {plddt_path}"
|
| 61 |
+
|
| 62 |
+
tmp_save_path = f"get_struc_seq_{process_id}.tsv"
|
| 63 |
+
cmd = f"{foldseek} structureto3didescriptor -v 0 --threads 1 --chain-name-mode 1 {path} {tmp_save_path}"
|
| 64 |
+
os.system(cmd)
|
| 65 |
+
|
| 66 |
+
seq_dict = {}
|
| 67 |
+
name = os.path.basename(path)
|
| 68 |
+
with open(tmp_save_path, "r") as r:
|
| 69 |
+
for i, line in enumerate(r):
|
| 70 |
+
desc, seq, struc_seq = line.split("\t")[:3]
|
| 71 |
+
|
| 72 |
+
# Mask low plddt
|
| 73 |
+
if plddt_path is not None:
|
| 74 |
+
with open(plddt_path, "r") as r:
|
| 75 |
+
plddts = np.array(json.load(r)["confidenceScore"])
|
| 76 |
+
|
| 77 |
+
# Mask regions with plddt < threshold
|
| 78 |
+
indices = np.where(plddts < plddt_threshold)[0]
|
| 79 |
+
np_seq = np.array(list(struc_seq))
|
| 80 |
+
np_seq[indices] = "#"
|
| 81 |
+
struc_seq = "".join(np_seq)
|
| 82 |
+
|
| 83 |
+
name_chain = desc.split(" ")[0]
|
| 84 |
+
chain = name_chain.replace(name, "").split("_")[-1]
|
| 85 |
+
|
| 86 |
+
if chains is None or chain in chains:
|
| 87 |
+
if chain not in seq_dict:
|
| 88 |
+
combined_seq = "".join(
|
| 89 |
+
[a + b.lower() for a, b in zip(seq, struc_seq)]
|
| 90 |
+
)
|
| 91 |
+
seq_dict[chain] = (seq, struc_seq, combined_seq)
|
| 92 |
+
|
| 93 |
+
os.remove(tmp_save_path)
|
| 94 |
+
os.remove(tmp_save_path + ".dbtype")
|
| 95 |
+
return seq_dict
|
| 96 |
+
|
| 97 |
+
|
| 98 |
+
if __name__ == "__main__":
|
| 99 |
+
foldseek = "/sujin/bin/foldseek"
|
| 100 |
+
# test_path = "/sujin/Datasets/PDB/all/6xtd.cif"
|
| 101 |
+
test_path = "/sujin/Datasets/FLIP/meltome/af2_structures/A0A061ACX4.pdb"
|
| 102 |
+
plddt_path = "/sujin/Datasets/FLIP/meltome/af2_plddts/A0A061ACX4.json"
|
| 103 |
+
res = get_struc_seq(
|
| 104 |
+
foldseek, test_path, plddt_path=plddt_path, plddt_threshold=70.0
|
| 105 |
+
)
|
| 106 |
+
print(res["A"][1].lower())
|
scripts/get_aa_from_uniprot_accession.py
ADDED
|
@@ -0,0 +1,93 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import multiprocessing as mp
|
| 2 |
+
import requests as r
|
| 3 |
+
import argparse
|
| 4 |
+
from Bio import SeqIO
|
| 5 |
+
from io import StringIO
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import numpy as np
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
def parse_args():
|
| 11 |
+
parser = argparse.ArgumentParser()
|
| 12 |
+
parser.add_argument(
|
| 13 |
+
"--file_path",
|
| 14 |
+
type=str,
|
| 15 |
+
required=True,
|
| 16 |
+
help="Path to the file that have a column cotaining uniprotid information.",
|
| 17 |
+
)
|
| 18 |
+
parser.add_argument(
|
| 19 |
+
"--sheet_name",
|
| 20 |
+
type=str,
|
| 21 |
+
default="Sheet1",
|
| 22 |
+
help="Name of the sheet to read. Default is Sheet1.",
|
| 23 |
+
)
|
| 24 |
+
parser.add_argument(
|
| 25 |
+
"--uniprotid_column",
|
| 26 |
+
type=str,
|
| 27 |
+
help="Name of the column that have uniprotid information. Default is None.",
|
| 28 |
+
)
|
| 29 |
+
parser.add_argument(
|
| 30 |
+
"--uniprotids_column",
|
| 31 |
+
type=str,
|
| 32 |
+
help="Name of the column that have uniprotids information. Default is None. The ids are expected to be separated by semi-colon, and the first id is used.",
|
| 33 |
+
)
|
| 34 |
+
parser.add_argument(
|
| 35 |
+
"--num_processes",
|
| 36 |
+
type=int,
|
| 37 |
+
default=2,
|
| 38 |
+
help="Number of processes to use.",
|
| 39 |
+
)
|
| 40 |
+
return parser.parse_args()
|
| 41 |
+
|
| 42 |
+
|
| 43 |
+
def fetch_sequence(row, cfg):
|
| 44 |
+
try:
|
| 45 |
+
baseURL = "http://www.uniprot.org/uniprot/"
|
| 46 |
+
uniprot_id = row[cfg.uniprotid_column]
|
| 47 |
+
URL = baseURL + uniprot_id + ".fasta"
|
| 48 |
+
response = r.post(URL)
|
| 49 |
+
Data = "".join(response.text)
|
| 50 |
+
Seq = StringIO(Data)
|
| 51 |
+
pSeq = list(SeqIO.parse(Seq, "fasta"))
|
| 52 |
+
return str(pSeq[0].seq)
|
| 53 |
+
except:
|
| 54 |
+
return None
|
| 55 |
+
|
| 56 |
+
|
| 57 |
+
def process_rows(df_chunk, cfg):
|
| 58 |
+
return [fetch_sequence(row, cfg) for idx, row in df_chunk.iterrows()]
|
| 59 |
+
|
| 60 |
+
|
| 61 |
+
if __name__ == "__main__":
|
| 62 |
+
config = parse_args()
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
if config.file_path.endswith(".xls"):
|
| 66 |
+
df = pd.read_excel(
|
| 67 |
+
config.file_path,
|
| 68 |
+
sheet_name=config.sheet_name,
|
| 69 |
+
)
|
| 70 |
+
else:
|
| 71 |
+
df = pd.read_csv(config.file_path)
|
| 72 |
+
|
| 73 |
+
if config.uniprotid_column is None and config.uniprotids_column is None:
|
| 74 |
+
raise ValueError(
|
| 75 |
+
"Either uniprotid_column or uniprotids_column should be provided."
|
| 76 |
+
)
|
| 77 |
+
if config.uniprotids_column is not None:
|
| 78 |
+
df = df.dropna(subset=[config.uniprotids_column]).reset_index(drop=True)
|
| 79 |
+
# use the first id and ignore the subunit and domain information
|
| 80 |
+
df["uniprotid"] = df[config.uniprotids_column].apply(
|
| 81 |
+
lambda x: x.split(";")[0].split("-")[0]
|
| 82 |
+
)
|
| 83 |
+
config.uniprotid_column = "uniprotid"
|
| 84 |
+
|
| 85 |
+
df_split = np.array_split(df, config.num_processes)
|
| 86 |
+
|
| 87 |
+
with mp.Pool(processes=config.num_processes) as pool:
|
| 88 |
+
results = pool.map(lambda x: process_rows(x, config), df_split)
|
| 89 |
+
|
| 90 |
+
aas = [seq for result in results for seq in result]
|
| 91 |
+
|
| 92 |
+
df["aa"] = aas
|
| 93 |
+
df.to_csv(f"{config.file_path.split('.')[0]}_with_aa.csv", index=False)
|
scripts/models.py
ADDED
|
@@ -0,0 +1,120 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch.nn as nn
|
| 2 |
+
from transformers import AutoModel, AutoConfig, PreTrainedModel
|
| 3 |
+
|
| 4 |
+
|
| 5 |
+
class PLTNUM(nn.Module):
|
| 6 |
+
def __init__(self, cfg):
|
| 7 |
+
super(PLTNUM, self).__init__()
|
| 8 |
+
self.cfg = cfg
|
| 9 |
+
self.config = AutoConfig.from_pretrained(cfg.model, output_hidden_states=True)
|
| 10 |
+
# self.model = AutoModel.from_pretrained(cfg.model, config=self.config)
|
| 11 |
+
self.model = AutoModel.from_config(config=self.config)
|
| 12 |
+
|
| 13 |
+
self.fc_dropout1 = nn.Dropout(0.8)
|
| 14 |
+
self.fc_dropout2 = nn.Dropout(0.4 if cfg.task == "classification" else 0.8)
|
| 15 |
+
self.fc = nn.Linear(self.config.hidden_size, 1)
|
| 16 |
+
self._init_weights(self.fc)
|
| 17 |
+
|
| 18 |
+
def _init_weights(self, module):
|
| 19 |
+
if isinstance(module, nn.Linear):
|
| 20 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 21 |
+
if module.bias is not None:
|
| 22 |
+
nn.init.constant_(module.bias, 0)
|
| 23 |
+
elif isinstance(module, nn.Embedding):
|
| 24 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 25 |
+
if module.padding_idx is not None:
|
| 26 |
+
nn.init.constant_(module.weight[module.padding_idx], 0.0)
|
| 27 |
+
elif isinstance(module, nn.LayerNorm):
|
| 28 |
+
nn.init.constant_(module.bias, 0)
|
| 29 |
+
nn.init.constant_(module.weight, 1.0)
|
| 30 |
+
|
| 31 |
+
def forward(self, inputs):
|
| 32 |
+
outputs = self.model(**inputs)
|
| 33 |
+
last_hidden_state = outputs.last_hidden_state[:, 0]
|
| 34 |
+
output = (
|
| 35 |
+
self.fc(self.fc_dropout1(last_hidden_state))
|
| 36 |
+
+ self.fc(self.fc_dropout2(last_hidden_state))
|
| 37 |
+
) / 2
|
| 38 |
+
return output
|
| 39 |
+
|
| 40 |
+
def create_embedding(self, inputs):
|
| 41 |
+
outputs = self.model(**inputs)
|
| 42 |
+
last_hidden_state = outputs.last_hidden_state[:, 0]
|
| 43 |
+
return last_hidden_state
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
class PLTNUM_PreTrainedModel(PreTrainedModel):
|
| 47 |
+
config_class = AutoConfig
|
| 48 |
+
|
| 49 |
+
def __init__(self, config, cfg):
|
| 50 |
+
super(PLTNUM_PreTrainedModel, self).__init__(config)
|
| 51 |
+
self.cfg = cfg
|
| 52 |
+
self.model = AutoModel.from_pretrained(self.config._name_or_path)
|
| 53 |
+
|
| 54 |
+
self.fc_dropout1 = nn.Dropout(0.8)
|
| 55 |
+
self.fc_dropout2 = nn.Dropout(0.4 if cfg.task == "classification" else 0.8)
|
| 56 |
+
self.fc = nn.Linear(self.config.hidden_size, 1)
|
| 57 |
+
self._init_weights(self.fc)
|
| 58 |
+
|
| 59 |
+
def _init_weights(self, module):
|
| 60 |
+
if isinstance(module, nn.Linear):
|
| 61 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 62 |
+
if module.bias is not None:
|
| 63 |
+
nn.init.constant_(module.bias, 0)
|
| 64 |
+
elif isinstance(module, nn.Embedding):
|
| 65 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 66 |
+
if module.padding_idx is not None:
|
| 67 |
+
nn.init.constant_(module.weight[module.padding_idx], 0.0)
|
| 68 |
+
elif isinstance(module, nn.LayerNorm):
|
| 69 |
+
nn.init.constant_(module.bias, 0)
|
| 70 |
+
nn.init.constant_(module.weight, 1.0)
|
| 71 |
+
|
| 72 |
+
def forward(self, inputs):
|
| 73 |
+
outputs = self.model(**inputs)
|
| 74 |
+
last_hidden_state = outputs.last_hidden_state[:, 0]
|
| 75 |
+
output = (
|
| 76 |
+
self.fc(self.fc_dropout1(last_hidden_state))
|
| 77 |
+
+ self.fc(self.fc_dropout2(last_hidden_state))
|
| 78 |
+
) / 2
|
| 79 |
+
return output
|
| 80 |
+
|
| 81 |
+
def create_embedding(self, inputs):
|
| 82 |
+
outputs = self.model(**inputs)
|
| 83 |
+
last_hidden_state = outputs.last_hidden_state[:, 0]
|
| 84 |
+
return last_hidden_state
|
| 85 |
+
|
| 86 |
+
|
| 87 |
+
class LSTMModel(nn.Module):
|
| 88 |
+
def __init__(self, cfg):
|
| 89 |
+
super(LSTMModel, self).__init__()
|
| 90 |
+
self.cfg = cfg
|
| 91 |
+
self.lstm = nn.LSTM(
|
| 92 |
+
input_size=21,
|
| 93 |
+
hidden_size=256,
|
| 94 |
+
num_layers=2,
|
| 95 |
+
batch_first=True,
|
| 96 |
+
bidirectional=True,
|
| 97 |
+
dropout=0.2,
|
| 98 |
+
)
|
| 99 |
+
self.fc_dropout = nn.Dropout(0.8)
|
| 100 |
+
self.fc = nn.Linear(256 * 2, 1)
|
| 101 |
+
self._init_weights(self.fc)
|
| 102 |
+
|
| 103 |
+
def _init_weights(self, module):
|
| 104 |
+
if isinstance(module, nn.Linear):
|
| 105 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 106 |
+
if module.bias is not None:
|
| 107 |
+
nn.init.constant_(module.bias, 0)
|
| 108 |
+
elif isinstance(module, nn.Embedding):
|
| 109 |
+
nn.init.normal_(module.weight, mean=0.0, std=self.config.initializer_range)
|
| 110 |
+
if module.padding_idx is not None:
|
| 111 |
+
nn.init.constant_(module.weight[module.padding_idx], 0.0)
|
| 112 |
+
elif isinstance(module, nn.LayerNorm):
|
| 113 |
+
nn.init.constant_(module.bias, 0)
|
| 114 |
+
nn.init.constant_(module.weight, 1.0)
|
| 115 |
+
|
| 116 |
+
def forward(self, inputs):
|
| 117 |
+
outputs, _ = self.lstm(inputs)
|
| 118 |
+
last_hidden_state = outputs[:, -1, :]
|
| 119 |
+
output = self.fc(self.fc_dropout(last_hidden_state))
|
| 120 |
+
return output
|
scripts/predict.py
ADDED
|
@@ -0,0 +1,168 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gc
|
| 2 |
+
import os
|
| 3 |
+
import sys
|
| 4 |
+
import argparse
|
| 5 |
+
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import torch
|
| 8 |
+
from torch.utils.data import DataLoader
|
| 9 |
+
from transformers import AutoTokenizer
|
| 10 |
+
|
| 11 |
+
sys.path.append(".")
|
| 12 |
+
from utils import seed_everything
|
| 13 |
+
from models import PLTNUM
|
| 14 |
+
from datasets import PLTNUMDataset
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
def parse_args():
|
| 18 |
+
parser = argparse.ArgumentParser(
|
| 19 |
+
description="Prediction script for protein sequence classification/regression."
|
| 20 |
+
)
|
| 21 |
+
parser.add_argument(
|
| 22 |
+
"--data_path",
|
| 23 |
+
type=str,
|
| 24 |
+
required=True,
|
| 25 |
+
help="Path to the input data.",
|
| 26 |
+
)
|
| 27 |
+
parser.add_argument(
|
| 28 |
+
"--model",
|
| 29 |
+
type=str,
|
| 30 |
+
default="westlake-repl/SaProt_650M_AF2",
|
| 31 |
+
help="Pretrained model name or path.",
|
| 32 |
+
)
|
| 33 |
+
parser.add_argument(
|
| 34 |
+
"--architecture",
|
| 35 |
+
type=str,
|
| 36 |
+
default="SaProt",
|
| 37 |
+
help="Model architecture: 'ESM2', 'SaProt', or 'LSTM'.",
|
| 38 |
+
)
|
| 39 |
+
parser.add_argument(
|
| 40 |
+
"--model_path",
|
| 41 |
+
type=str,
|
| 42 |
+
required=True,
|
| 43 |
+
help="Path to the model for prediction.",
|
| 44 |
+
)
|
| 45 |
+
parser.add_argument("--batch_size", type=int, default=4, help="Batch size.")
|
| 46 |
+
parser.add_argument(
|
| 47 |
+
"--seed",
|
| 48 |
+
type=int,
|
| 49 |
+
default=42,
|
| 50 |
+
help="Seed for reproducibility.",
|
| 51 |
+
)
|
| 52 |
+
parser.add_argument(
|
| 53 |
+
"--use_amp",
|
| 54 |
+
action="store_true",
|
| 55 |
+
default=False,
|
| 56 |
+
help="Use AMP for mixed precision prediction.",
|
| 57 |
+
)
|
| 58 |
+
parser.add_argument(
|
| 59 |
+
"--num_workers",
|
| 60 |
+
type=int,
|
| 61 |
+
default=4,
|
| 62 |
+
help="Number of workers for data loading.",
|
| 63 |
+
)
|
| 64 |
+
parser.add_argument(
|
| 65 |
+
"--max_length",
|
| 66 |
+
type=int,
|
| 67 |
+
default=512,
|
| 68 |
+
help="Maximum input sequence length. Two tokens are used fo <cls> and <eos> tokens. So the actual length of input sequence is max_length - 2. Padding or truncation is applied to make the length of input sequence equal to max_length.",
|
| 69 |
+
)
|
| 70 |
+
parser.add_argument(
|
| 71 |
+
"--used_sequence",
|
| 72 |
+
type=str,
|
| 73 |
+
default="left",
|
| 74 |
+
help="Which part of the sequence to use: 'left', 'right', 'both', or 'internal'.",
|
| 75 |
+
)
|
| 76 |
+
parser.add_argument(
|
| 77 |
+
"--padding_side",
|
| 78 |
+
type=str,
|
| 79 |
+
default="right",
|
| 80 |
+
help="Padding side: 'right' or 'left'.",
|
| 81 |
+
)
|
| 82 |
+
parser.add_argument(
|
| 83 |
+
"--output_dir",
|
| 84 |
+
type=str,
|
| 85 |
+
default="./output",
|
| 86 |
+
help="Output directory.",
|
| 87 |
+
)
|
| 88 |
+
parser.add_argument(
|
| 89 |
+
"--task",
|
| 90 |
+
type=str,
|
| 91 |
+
default="classification",
|
| 92 |
+
help="Task type: 'classification' or 'regression'.",
|
| 93 |
+
)
|
| 94 |
+
parser.add_argument(
|
| 95 |
+
"--sequence_col",
|
| 96 |
+
type=str,
|
| 97 |
+
default="aa_foldseek",
|
| 98 |
+
help="Column name fot the input sequence.",
|
| 99 |
+
)
|
| 100 |
+
|
| 101 |
+
return parser.parse_args()
|
| 102 |
+
|
| 103 |
+
|
| 104 |
+
def predict_fn(valid_loader, model, cfg):
|
| 105 |
+
model.eval()
|
| 106 |
+
predictions = []
|
| 107 |
+
|
| 108 |
+
for inputs, _ in valid_loader:
|
| 109 |
+
inputs = inputs.to(cfg.device)
|
| 110 |
+
with torch.no_grad():
|
| 111 |
+
with torch.cuda.amp.autocast(enabled=cfg.use_amp):
|
| 112 |
+
preds = (
|
| 113 |
+
torch.sigmoid(model(inputs))
|
| 114 |
+
if cfg.task == "classification"
|
| 115 |
+
else model(inputs)
|
| 116 |
+
)
|
| 117 |
+
predictions += preds.cpu().tolist()
|
| 118 |
+
|
| 119 |
+
return predictions
|
| 120 |
+
|
| 121 |
+
|
| 122 |
+
def predict(folds, model_path, cfg):
|
| 123 |
+
dataset = PLTNUMDataset(cfg, folds, train=False)
|
| 124 |
+
loader = DataLoader(
|
| 125 |
+
dataset,
|
| 126 |
+
batch_size=cfg.batch_size,
|
| 127 |
+
shuffle=False,
|
| 128 |
+
num_workers=cfg.num_workers,
|
| 129 |
+
pin_memory=True,
|
| 130 |
+
drop_last=False,
|
| 131 |
+
)
|
| 132 |
+
|
| 133 |
+
model = PLTNUM(cfg)
|
| 134 |
+
model.load_state_dict(torch.load(model_path, map_location=cfg.device))
|
| 135 |
+
model.to(cfg.device)
|
| 136 |
+
|
| 137 |
+
predictions = predict_fn(loader, model, cfg)
|
| 138 |
+
|
| 139 |
+
folds["raw prediction values"] = predictions
|
| 140 |
+
if cfg.task == "classification":
|
| 141 |
+
folds["binary prediction values"] = [1 if x > 0.5 else 0 for x in predictions]
|
| 142 |
+
torch.cuda.empty_cache()
|
| 143 |
+
gc.collect()
|
| 144 |
+
return folds
|
| 145 |
+
|
| 146 |
+
|
| 147 |
+
if __name__ == "__main__":
|
| 148 |
+
config = parse_args()
|
| 149 |
+
config.token_length = 2 if config.architecture == "SaProt" else 1
|
| 150 |
+
config.device = "cuda" if torch.cuda.is_available() else "cpu"
|
| 151 |
+
|
| 152 |
+
if not os.path.exists(config.output_dir):
|
| 153 |
+
os.makedirs(config.output_dir)
|
| 154 |
+
|
| 155 |
+
if config.used_sequence == "both":
|
| 156 |
+
config.max_length += 1
|
| 157 |
+
|
| 158 |
+
seed_everything(config.seed)
|
| 159 |
+
|
| 160 |
+
df = pd.read_csv(config.data_path)
|
| 161 |
+
|
| 162 |
+
tokenizer = AutoTokenizer.from_pretrained(
|
| 163 |
+
config.model, padding_side=config.padding_side
|
| 164 |
+
)
|
| 165 |
+
config.tokenizer = tokenizer
|
| 166 |
+
|
| 167 |
+
result = predict(df, config.model_path, config)
|
| 168 |
+
result.to_csv(os.path.join(config.output_dir, "result.csv"), index=False)
|
scripts/predict_with_PreTrainedModel.py
ADDED
|
@@ -0,0 +1,149 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gc
|
| 2 |
+
import os
|
| 3 |
+
import sys
|
| 4 |
+
import argparse
|
| 5 |
+
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import torch
|
| 8 |
+
from torch.utils.data import DataLoader
|
| 9 |
+
from transformers import AutoTokenizer
|
| 10 |
+
|
| 11 |
+
sys.path.append(".")
|
| 12 |
+
from utils import seed_everything
|
| 13 |
+
from models import PLTNUM_PreTrainedModel
|
| 14 |
+
from datasets import PLTNUMDataset
|
| 15 |
+
from predict import predict_fn
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
def parse_args():
|
| 19 |
+
parser = argparse.ArgumentParser(
|
| 20 |
+
description="Prediction script for protein sequence classification/regression."
|
| 21 |
+
)
|
| 22 |
+
parser.add_argument(
|
| 23 |
+
"--data_path",
|
| 24 |
+
type=str,
|
| 25 |
+
required=True,
|
| 26 |
+
help="Path to the input data.",
|
| 27 |
+
)
|
| 28 |
+
parser.add_argument(
|
| 29 |
+
"--model",
|
| 30 |
+
type=str,
|
| 31 |
+
default="westlake-repl/SaProt_650M_AF2",
|
| 32 |
+
help="Pretrained model name or path.",
|
| 33 |
+
)
|
| 34 |
+
parser.add_argument(
|
| 35 |
+
"--architecture",
|
| 36 |
+
type=str,
|
| 37 |
+
default="SaProt",
|
| 38 |
+
help="Model architecture: 'ESM2', 'SaProt', or 'LSTM'.",
|
| 39 |
+
)
|
| 40 |
+
parser.add_argument(
|
| 41 |
+
"--model_path",
|
| 42 |
+
type=str,
|
| 43 |
+
required=True,
|
| 44 |
+
help="Path to the model for prediction.",
|
| 45 |
+
)
|
| 46 |
+
parser.add_argument("--batch_size", type=int, default=4, help="Batch size.")
|
| 47 |
+
parser.add_argument(
|
| 48 |
+
"--seed",
|
| 49 |
+
type=int,
|
| 50 |
+
default=42,
|
| 51 |
+
help="Seed for reproducibility.",
|
| 52 |
+
)
|
| 53 |
+
parser.add_argument(
|
| 54 |
+
"--use_amp",
|
| 55 |
+
action="store_true",
|
| 56 |
+
default=False,
|
| 57 |
+
help="Use AMP for mixed precision prediction.",
|
| 58 |
+
)
|
| 59 |
+
parser.add_argument(
|
| 60 |
+
"--num_workers",
|
| 61 |
+
type=int,
|
| 62 |
+
default=4,
|
| 63 |
+
help="Number of workers for data loading.",
|
| 64 |
+
)
|
| 65 |
+
parser.add_argument(
|
| 66 |
+
"--max_length",
|
| 67 |
+
type=int,
|
| 68 |
+
default=512,
|
| 69 |
+
help="Maximum input sequence length. Two tokens are used fo <cls> and <eos> tokens. So the actual length of input sequence is max_length - 2. Padding or truncation is applied to make the length of input sequence equal to max_length.",
|
| 70 |
+
)
|
| 71 |
+
parser.add_argument(
|
| 72 |
+
"--used_sequence",
|
| 73 |
+
type=str,
|
| 74 |
+
default="left",
|
| 75 |
+
help="Which part of the sequence to use: 'left', 'right', 'both', or 'internal'.",
|
| 76 |
+
)
|
| 77 |
+
parser.add_argument(
|
| 78 |
+
"--padding_side",
|
| 79 |
+
type=str,
|
| 80 |
+
default="right",
|
| 81 |
+
help="Padding side: 'right' or 'left'.",
|
| 82 |
+
)
|
| 83 |
+
parser.add_argument(
|
| 84 |
+
"--output_dir",
|
| 85 |
+
type=str,
|
| 86 |
+
default="./output",
|
| 87 |
+
help="Output directory.",
|
| 88 |
+
)
|
| 89 |
+
parser.add_argument(
|
| 90 |
+
"--task",
|
| 91 |
+
type=str,
|
| 92 |
+
default="classification",
|
| 93 |
+
help="Task type: 'classification' or 'regression'.",
|
| 94 |
+
)
|
| 95 |
+
parser.add_argument(
|
| 96 |
+
"--sequence_col",
|
| 97 |
+
type=str,
|
| 98 |
+
default="aa_foldseek",
|
| 99 |
+
help="Column name fot the input sequence.",
|
| 100 |
+
)
|
| 101 |
+
|
| 102 |
+
return parser.parse_args()
|
| 103 |
+
|
| 104 |
+
|
| 105 |
+
def predict(folds, model_path, cfg):
|
| 106 |
+
dataset = PLTNUMDataset(cfg, folds, train=False)
|
| 107 |
+
loader = DataLoader(
|
| 108 |
+
dataset,
|
| 109 |
+
batch_size=cfg.batch_size,
|
| 110 |
+
shuffle=False,
|
| 111 |
+
num_workers=cfg.num_workers,
|
| 112 |
+
pin_memory=True,
|
| 113 |
+
drop_last=False,
|
| 114 |
+
)
|
| 115 |
+
|
| 116 |
+
model = PLTNUM_PreTrainedModel.from_pretrained(model_path, cfg=cfg)
|
| 117 |
+
# model.load_state_dict(torch.load(os.path.join(model_path, "pytorch_model.bin"), map_location=cfg.device))
|
| 118 |
+
model.to(cfg.device)
|
| 119 |
+
|
| 120 |
+
predictions = predict_fn(loader, model, cfg)
|
| 121 |
+
|
| 122 |
+
folds["prediction"] = predictions
|
| 123 |
+
torch.cuda.empty_cache()
|
| 124 |
+
gc.collect()
|
| 125 |
+
return folds
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
if __name__ == "__main__":
|
| 129 |
+
config = parse_args()
|
| 130 |
+
config.token_length = 2 if config.architecture == "SaProt" else 1
|
| 131 |
+
config.device = "cuda" if torch.cuda.is_available() else "cpu"
|
| 132 |
+
|
| 133 |
+
if not os.path.exists(config.output_dir):
|
| 134 |
+
os.makedirs(config.output_dir)
|
| 135 |
+
|
| 136 |
+
if config.used_sequence == "both":
|
| 137 |
+
config.max_length += 1
|
| 138 |
+
|
| 139 |
+
seed_everything(config.seed)
|
| 140 |
+
|
| 141 |
+
df = pd.read_csv(config.data_path)
|
| 142 |
+
|
| 143 |
+
tokenizer = AutoTokenizer.from_pretrained(
|
| 144 |
+
config.model_path, padding_side=config.padding_side
|
| 145 |
+
)
|
| 146 |
+
config.tokenizer = tokenizer
|
| 147 |
+
|
| 148 |
+
result = predict(df, config.model_path, config)
|
| 149 |
+
result.to_csv(os.path.join(config.output_dir, "result.csv"), index=False)
|
scripts/train.py
ADDED
|
@@ -0,0 +1,493 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gc
|
| 2 |
+
import os
|
| 3 |
+
import sys
|
| 4 |
+
import time
|
| 5 |
+
import argparse
|
| 6 |
+
|
| 7 |
+
import numpy as np
|
| 8 |
+
import pandas as pd
|
| 9 |
+
import torch
|
| 10 |
+
import torch.nn as nn
|
| 11 |
+
from sklearn.metrics import accuracy_score, f1_score, r2_score
|
| 12 |
+
from sklearn.model_selection import StratifiedKFold
|
| 13 |
+
from torch.optim import Adam
|
| 14 |
+
from torch.optim.lr_scheduler import CosineAnnealingLR
|
| 15 |
+
from torch.utils.data import DataLoader
|
| 16 |
+
from transformers import AutoTokenizer, get_cosine_schedule_with_warmup
|
| 17 |
+
|
| 18 |
+
sys.path.append(".")
|
| 19 |
+
from utils import AverageMeter, get_logger, seed_everything, timeSince
|
| 20 |
+
from datasets import PLTNUMDataset, LSTMDataset
|
| 21 |
+
from models import PLTNUM, LSTMModel
|
| 22 |
+
|
| 23 |
+
device = "cuda" if torch.cuda.is_available() else "cpu"
|
| 24 |
+
|
| 25 |
+
print("device:", device)
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
def parse_args():
|
| 29 |
+
parser = argparse.ArgumentParser(
|
| 30 |
+
description="Training script for protein half-life prediction."
|
| 31 |
+
)
|
| 32 |
+
parser.add_argument(
|
| 33 |
+
"--data_path",
|
| 34 |
+
type=str,
|
| 35 |
+
required=True,
|
| 36 |
+
help="Path to the training data.",
|
| 37 |
+
)
|
| 38 |
+
parser.add_argument(
|
| 39 |
+
"--model",
|
| 40 |
+
type=str,
|
| 41 |
+
default="westlake-repl/SaProt_650M_AF2",
|
| 42 |
+
help="Pretrained model name or path.",
|
| 43 |
+
)
|
| 44 |
+
parser.add_argument(
|
| 45 |
+
"--architecture",
|
| 46 |
+
type=str,
|
| 47 |
+
default="SaProt",
|
| 48 |
+
help="Model architecture: 'ESM2', 'SaProt', or 'LSTM'.",
|
| 49 |
+
)
|
| 50 |
+
parser.add_argument("--lr", type=float, default=2e-5, help="Learning rate.")
|
| 51 |
+
parser.add_argument(
|
| 52 |
+
"--epochs",
|
| 53 |
+
type=int,
|
| 54 |
+
default=5,
|
| 55 |
+
help="Number of training epochs.",
|
| 56 |
+
)
|
| 57 |
+
parser.add_argument("--batch_size", type=int, default=4, help="Batch size.")
|
| 58 |
+
parser.add_argument(
|
| 59 |
+
"--seed",
|
| 60 |
+
type=int,
|
| 61 |
+
default=42,
|
| 62 |
+
help="Seed for reproducibility.",
|
| 63 |
+
)
|
| 64 |
+
parser.add_argument(
|
| 65 |
+
"--use_amp",
|
| 66 |
+
action="store_true",
|
| 67 |
+
default=False,
|
| 68 |
+
help="Use AMP for mixed precision training.",
|
| 69 |
+
)
|
| 70 |
+
parser.add_argument(
|
| 71 |
+
"--num_workers",
|
| 72 |
+
type=int,
|
| 73 |
+
default=4,
|
| 74 |
+
help="Number of workers for data loading.",
|
| 75 |
+
)
|
| 76 |
+
parser.add_argument(
|
| 77 |
+
"--max_length",
|
| 78 |
+
type=int,
|
| 79 |
+
default=512,
|
| 80 |
+
help="Maximum input sequence length. Two tokens are used fo <cls> and <eos> tokens. So the actual length of input sequence is max_length - 2. Padding or truncation is applied to make the length of input sequence equal to max_length.",
|
| 81 |
+
)
|
| 82 |
+
parser.add_argument(
|
| 83 |
+
"--used_sequence",
|
| 84 |
+
type=str,
|
| 85 |
+
default="left",
|
| 86 |
+
help="Which part of the sequence to use: 'left', 'right', 'both', or 'internal'.",
|
| 87 |
+
)
|
| 88 |
+
parser.add_argument(
|
| 89 |
+
"--padding_side",
|
| 90 |
+
type=str,
|
| 91 |
+
default="right",
|
| 92 |
+
help="Padding side: 'right' or 'left'.",
|
| 93 |
+
)
|
| 94 |
+
parser.add_argument(
|
| 95 |
+
"--mask_ratio",
|
| 96 |
+
type=float,
|
| 97 |
+
default=0.05,
|
| 98 |
+
help="Ratio of mask tokens for augmentation.",
|
| 99 |
+
)
|
| 100 |
+
parser.add_argument(
|
| 101 |
+
"--mask_prob",
|
| 102 |
+
type=float,
|
| 103 |
+
default=0.2,
|
| 104 |
+
help="Probability to apply mask augmentation",
|
| 105 |
+
)
|
| 106 |
+
parser.add_argument(
|
| 107 |
+
"--random_delete_ratio",
|
| 108 |
+
type=float,
|
| 109 |
+
default=0.1,
|
| 110 |
+
help="Ratio of deleting tokens in augmentation.",
|
| 111 |
+
)
|
| 112 |
+
parser.add_argument(
|
| 113 |
+
"--random_delete_prob",
|
| 114 |
+
type=float,
|
| 115 |
+
default=-1,
|
| 116 |
+
help="Probability to apply random delete augmentation.",
|
| 117 |
+
)
|
| 118 |
+
parser.add_argument(
|
| 119 |
+
"--random_change_ratio",
|
| 120 |
+
type=float,
|
| 121 |
+
default=0,
|
| 122 |
+
help="Ratio of changing tokens in augmentation.",
|
| 123 |
+
)
|
| 124 |
+
parser.add_argument(
|
| 125 |
+
"--truncate_augmentation_prob",
|
| 126 |
+
type=float,
|
| 127 |
+
default=-1,
|
| 128 |
+
help="Probability to apply truncate augmentation.",
|
| 129 |
+
)
|
| 130 |
+
parser.add_argument(
|
| 131 |
+
"--n_folds",
|
| 132 |
+
type=int,
|
| 133 |
+
default=10,
|
| 134 |
+
help="Number of folds for cross-validation.",
|
| 135 |
+
)
|
| 136 |
+
parser.add_argument(
|
| 137 |
+
"--print_freq",
|
| 138 |
+
type=int,
|
| 139 |
+
default=300,
|
| 140 |
+
help="Log print frequency.",
|
| 141 |
+
)
|
| 142 |
+
parser.add_argument(
|
| 143 |
+
"--freeze_layer",
|
| 144 |
+
type=int,
|
| 145 |
+
default=-1,
|
| 146 |
+
help="Freeze layers of the model. -1 means no layers are frozen.",
|
| 147 |
+
)
|
| 148 |
+
parser.add_argument(
|
| 149 |
+
"--output_dir",
|
| 150 |
+
type=str,
|
| 151 |
+
default="./output",
|
| 152 |
+
help="Output directory.",
|
| 153 |
+
)
|
| 154 |
+
parser.add_argument(
|
| 155 |
+
"--task",
|
| 156 |
+
type=str,
|
| 157 |
+
default="classification",
|
| 158 |
+
help="Task type: 'classification' or 'regression'.",
|
| 159 |
+
)
|
| 160 |
+
parser.add_argument(
|
| 161 |
+
"--target_col",
|
| 162 |
+
type=str,
|
| 163 |
+
default="Protein half-life average [h]",
|
| 164 |
+
help="Column name of the target.",
|
| 165 |
+
)
|
| 166 |
+
parser.add_argument(
|
| 167 |
+
"--sequence_col",
|
| 168 |
+
type=str,
|
| 169 |
+
default="aa_foldseek",
|
| 170 |
+
help="Column name fot the input sequence.",
|
| 171 |
+
)
|
| 172 |
+
|
| 173 |
+
return parser.parse_args()
|
| 174 |
+
|
| 175 |
+
|
| 176 |
+
def train_fn(train_loader, model, criterion, optimizer, epoch, cfg):
|
| 177 |
+
model.train()
|
| 178 |
+
scaler = torch.cuda.amp.GradScaler(enabled=cfg.use_amp)
|
| 179 |
+
losses = AverageMeter()
|
| 180 |
+
label_list, pred_list = [], []
|
| 181 |
+
start = time.time()
|
| 182 |
+
|
| 183 |
+
for step, (inputs, labels) in enumerate(train_loader):
|
| 184 |
+
inputs, labels = inputs.to(cfg.device), labels.to(cfg.device)
|
| 185 |
+
labels = (
|
| 186 |
+
labels.float()
|
| 187 |
+
if cfg.task == "classification"
|
| 188 |
+
else labels.to(dtype=torch.half)
|
| 189 |
+
)
|
| 190 |
+
batch_size = labels.size(0)
|
| 191 |
+
|
| 192 |
+
with torch.cuda.amp.autocast(enabled=cfg.use_amp):
|
| 193 |
+
y_preds = model(inputs)
|
| 194 |
+
loss = criterion(y_preds, labels.view(-1, 1))
|
| 195 |
+
losses.update(loss.item(), batch_size)
|
| 196 |
+
|
| 197 |
+
scaler.scale(loss).backward()
|
| 198 |
+
scaler.step(optimizer)
|
| 199 |
+
scaler.update()
|
| 200 |
+
optimizer.zero_grad()
|
| 201 |
+
|
| 202 |
+
label_list += labels.tolist()
|
| 203 |
+
pred_list += y_preds.tolist()
|
| 204 |
+
|
| 205 |
+
if step % cfg.print_freq == 0 or step == len(train_loader) - 1:
|
| 206 |
+
if cfg.task == "classification":
|
| 207 |
+
pred_list_new = (torch.Tensor(pred_list) > 0.5).to(dtype=torch.long)
|
| 208 |
+
acc = accuracy_score(label_list, pred_list_new > 0.5)
|
| 209 |
+
cfg.logger.info(
|
| 210 |
+
f"Epoch: [{epoch + 1}][{step}/{len(train_loader)}] "
|
| 211 |
+
f"Elapsed {timeSince(start, float(step + 1) / len(train_loader))} "
|
| 212 |
+
f"Loss: {losses.val:.4f}({losses.avg:.4f}) "
|
| 213 |
+
f"LR: {optimizer.param_groups[0]['lr']:.8f} "
|
| 214 |
+
f"Accuracy: {acc:.4f}"
|
| 215 |
+
)
|
| 216 |
+
elif cfg.task == "regression":
|
| 217 |
+
r2 = r2_score(label_list, pred_list)
|
| 218 |
+
cfg.logger.info(
|
| 219 |
+
f"Epoch: [{epoch + 1}][{step}/{len(train_loader)}] "
|
| 220 |
+
f"Elapsed {timeSince(start, float(step + 1) / len(train_loader))} "
|
| 221 |
+
f"Loss: {losses.val:.4f}({losses.avg:.4f}) "
|
| 222 |
+
f"R2 Score: {r2:.4f} "
|
| 223 |
+
f"LR: {optimizer.param_groups[0]['lr']:.8f}"
|
| 224 |
+
)
|
| 225 |
+
if cfg.task == "classification":
|
| 226 |
+
pred_list_new = (torch.Tensor(pred_list) > 0.5).to(dtype=torch.long)
|
| 227 |
+
acc = accuracy_score(label_list, pred_list_new)
|
| 228 |
+
return losses.avg, acc
|
| 229 |
+
elif cfg.task == "regression":
|
| 230 |
+
return losses.avg, r2_score(label_list, pred_list)
|
| 231 |
+
|
| 232 |
+
|
| 233 |
+
def valid_fn(valid_loader, model, criterion, cfg):
|
| 234 |
+
losses = AverageMeter()
|
| 235 |
+
model.eval()
|
| 236 |
+
label_list, pred_list = [], []
|
| 237 |
+
start = time.time()
|
| 238 |
+
|
| 239 |
+
for step, (inputs, labels) in enumerate(valid_loader):
|
| 240 |
+
inputs, labels = inputs.to(cfg.device), labels.to(cfg.device)
|
| 241 |
+
labels = (
|
| 242 |
+
labels.float()
|
| 243 |
+
if cfg.task == "classification"
|
| 244 |
+
else labels.to(dtype=torch.half)
|
| 245 |
+
)
|
| 246 |
+
|
| 247 |
+
with torch.no_grad():
|
| 248 |
+
with torch.cuda.amp.autocast(enabled=cfg.use_amp):
|
| 249 |
+
y_preds = (
|
| 250 |
+
torch.sigmoid(model(inputs))
|
| 251 |
+
if cfg.task == "classification"
|
| 252 |
+
else model(inputs)
|
| 253 |
+
)
|
| 254 |
+
loss = criterion(y_preds, labels.view(-1, 1))
|
| 255 |
+
losses.update(loss.item(), labels.size(0))
|
| 256 |
+
|
| 257 |
+
label_list += labels.tolist()
|
| 258 |
+
pred_list += y_preds.tolist()
|
| 259 |
+
|
| 260 |
+
if step % cfg.print_freq == 0 or step == len(valid_loader) - 1:
|
| 261 |
+
if cfg.task == "classification":
|
| 262 |
+
pred_list_new = (torch.Tensor(pred_list) > 0.5).to(dtype=torch.long)
|
| 263 |
+
acc = accuracy_score(label_list, pred_list_new > 0.5)
|
| 264 |
+
f1 = f1_score(label_list, pred_list_new, average="macro")
|
| 265 |
+
cfg.logger.info(
|
| 266 |
+
f"EVAL: [{step}/{len(valid_loader)}] "
|
| 267 |
+
f"Elapsed {timeSince(start, float(step + 1) / len(valid_loader))} "
|
| 268 |
+
f"Loss: {losses.val:.4f}({losses.avg:.4f}) "
|
| 269 |
+
f"Accuracy: {acc:.4f} "
|
| 270 |
+
f"F1 Score: {f1:.4f}"
|
| 271 |
+
)
|
| 272 |
+
elif cfg.task == "regression":
|
| 273 |
+
r2 = r2_score(label_list, pred_list)
|
| 274 |
+
cfg.logger.info(
|
| 275 |
+
f"EVAL: [{step}/{len(valid_loader)}] "
|
| 276 |
+
f"Elapsed {timeSince(start, float(step + 1) / len(valid_loader))} "
|
| 277 |
+
f"Loss: {losses.val:.4f}({losses.avg:.4f}) "
|
| 278 |
+
f"R2 Score: {r2:.4f}"
|
| 279 |
+
)
|
| 280 |
+
|
| 281 |
+
if cfg.task == "classification":
|
| 282 |
+
pred_list_new = (torch.Tensor(pred_list) > 0.5).to(dtype=torch.long)
|
| 283 |
+
return (
|
| 284 |
+
f1_score(label_list, pred_list_new, average="macro"),
|
| 285 |
+
accuracy_score(label_list, pred_list_new),
|
| 286 |
+
pred_list,
|
| 287 |
+
)
|
| 288 |
+
elif cfg.task == "regression":
|
| 289 |
+
return losses.avg, r2_score(label_list, pred_list), np.array(pred_list)
|
| 290 |
+
|
| 291 |
+
|
| 292 |
+
def train_loop(folds, fold, cfg):
|
| 293 |
+
cfg.logger.info(f"================== fold: {fold} training ======================")
|
| 294 |
+
train_folds = folds[folds["fold"] != fold].reset_index(drop=True)
|
| 295 |
+
valid_folds = folds[folds["fold"] == fold].reset_index(drop=True)
|
| 296 |
+
|
| 297 |
+
if cfg.architecture in ["ESM2", "SaProt"]:
|
| 298 |
+
train_dataset = PLTNUMDataset(cfg, train_folds, train=True)
|
| 299 |
+
valid_dataset = PLTNUMDataset(cfg, valid_folds, train=False)
|
| 300 |
+
elif cfg.architecture == "LSTM":
|
| 301 |
+
train_dataset = LSTMDataset(cfg, train_folds, train=True)
|
| 302 |
+
valid_dataset = LSTMDataset(cfg, valid_folds, train=False)
|
| 303 |
+
|
| 304 |
+
train_loader = DataLoader(
|
| 305 |
+
train_dataset,
|
| 306 |
+
batch_size=cfg.batch_size,
|
| 307 |
+
shuffle=True,
|
| 308 |
+
num_workers=cfg.num_workers,
|
| 309 |
+
pin_memory=True,
|
| 310 |
+
drop_last=True,
|
| 311 |
+
)
|
| 312 |
+
valid_loader = DataLoader(
|
| 313 |
+
valid_dataset,
|
| 314 |
+
batch_size=cfg.batch_size,
|
| 315 |
+
shuffle=False,
|
| 316 |
+
num_workers=cfg.num_workers,
|
| 317 |
+
pin_memory=True,
|
| 318 |
+
drop_last=False,
|
| 319 |
+
)
|
| 320 |
+
|
| 321 |
+
if cfg.architecture in ["ESM2", "SaProt"]:
|
| 322 |
+
model = PLTNUM(cfg)
|
| 323 |
+
if cfg.freeze_layer >= 0:
|
| 324 |
+
for name, param in model.named_parameters():
|
| 325 |
+
if f"model.encoder.layer.{cfg.freeze_layer}" in name:
|
| 326 |
+
break
|
| 327 |
+
param.requires_grad = False
|
| 328 |
+
model.config.save_pretrained(cfg.output_dir)
|
| 329 |
+
elif cfg.architecture == "LSTM":
|
| 330 |
+
model = LSTMModel(cfg)
|
| 331 |
+
|
| 332 |
+
model.to(cfg.device)
|
| 333 |
+
|
| 334 |
+
optimizer = Adam(model.parameters(), lr=cfg.lr)
|
| 335 |
+
if cfg.architecture in ["ESM2", "SaProt"]:
|
| 336 |
+
scheduler = CosineAnnealingLR(
|
| 337 |
+
optimizer,
|
| 338 |
+
**{"T_max": 2, "eta_min": 1.0e-6, "last_epoch": -1},
|
| 339 |
+
)
|
| 340 |
+
elif cfg.architecture == "LSTM":
|
| 341 |
+
scheduler = get_cosine_schedule_with_warmup(
|
| 342 |
+
optimizer, num_warmup_steps=0, num_training_steps=cfg.epochs, num_cycles=0.5
|
| 343 |
+
)
|
| 344 |
+
|
| 345 |
+
criterion = nn.BCEWithLogitsLoss() if cfg.task == "classification" else nn.MSELoss()
|
| 346 |
+
best_score = 0 if cfg.task == "classification" else float("inf")
|
| 347 |
+
|
| 348 |
+
for epoch in range(cfg.epochs):
|
| 349 |
+
start_time = time.time()
|
| 350 |
+
# train
|
| 351 |
+
avg_loss, train_score = train_fn(
|
| 352 |
+
train_loader, model, criterion, optimizer, epoch, cfg
|
| 353 |
+
)
|
| 354 |
+
scheduler.step()
|
| 355 |
+
|
| 356 |
+
# eval
|
| 357 |
+
val_score, val_score2, predictions = valid_fn(
|
| 358 |
+
valid_loader, model, criterion, cfg
|
| 359 |
+
)
|
| 360 |
+
|
| 361 |
+
elapsed = time.time() - start_time
|
| 362 |
+
|
| 363 |
+
if cfg.task == "classification":
|
| 364 |
+
cfg.logger.info(
|
| 365 |
+
f"Epoch {epoch+1} - avg_train_loss: {avg_loss:.4f} train_acc: {train_score:.4f} valid_acc: {val_score2:.4f} valid_f1: {val_score:.4f} time: {elapsed:.0f}s"
|
| 366 |
+
)
|
| 367 |
+
elif cfg.task == "regression":
|
| 368 |
+
cfg.logger.info(
|
| 369 |
+
f"Epoch {epoch+1} - avg_train_loss: {avg_loss:.4f} train_r2: {train_score:.4f} valid_r2: {val_score2:.4f} valid_loss: {val_score:.4f} time: {elapsed:.0f}s"
|
| 370 |
+
)
|
| 371 |
+
|
| 372 |
+
if (cfg.task == "classification" and best_score < val_score) or (
|
| 373 |
+
cfg.task == "regression" and best_score > val_score
|
| 374 |
+
):
|
| 375 |
+
best_score = val_score
|
| 376 |
+
cfg.logger.info(f"Epoch {epoch+1} - Save Best Score: {val_score:.4f} Model")
|
| 377 |
+
torch.save(
|
| 378 |
+
predictions,
|
| 379 |
+
os.path.join(cfg.output_dir, f"predictions.pth"),
|
| 380 |
+
)
|
| 381 |
+
torch.save(
|
| 382 |
+
model.state_dict(),
|
| 383 |
+
os.path.join(cfg.output_dir, f"model_fold{fold}.pth"),
|
| 384 |
+
)
|
| 385 |
+
|
| 386 |
+
predictions = torch.load(
|
| 387 |
+
os.path.join(cfg.output_dir, f"predictions.pth"), map_location="cpu"
|
| 388 |
+
)
|
| 389 |
+
valid_folds["prediction"] = predictions
|
| 390 |
+
cfg.logger.info(f"[Fold{fold}] Best score: {best_score}")
|
| 391 |
+
torch.cuda.empty_cache()
|
| 392 |
+
gc.collect()
|
| 393 |
+
return valid_folds
|
| 394 |
+
|
| 395 |
+
|
| 396 |
+
def get_embedding(folds, fold, path, cfg):
|
| 397 |
+
valid_folds = folds[folds["fold"] == fold].reset_index(drop=True)
|
| 398 |
+
valid_dataset = PLTNUMDataset(cfg, valid_folds, train=False)
|
| 399 |
+
|
| 400 |
+
valid_loader = DataLoader(
|
| 401 |
+
valid_dataset,
|
| 402 |
+
batch_size=cfg.batch_size,
|
| 403 |
+
shuffle=False,
|
| 404 |
+
num_workers=cfg.num_workers,
|
| 405 |
+
pin_memory=True,
|
| 406 |
+
drop_last=False,
|
| 407 |
+
)
|
| 408 |
+
|
| 409 |
+
model = PLTNUM(cfg)
|
| 410 |
+
model.load_state_dict(torch.load(path, map_location=torch.device("cpu")))
|
| 411 |
+
model.to(device)
|
| 412 |
+
|
| 413 |
+
model.eval()
|
| 414 |
+
embedding_list = []
|
| 415 |
+
for inputs, _ in valid_loader:
|
| 416 |
+
inputs = inputs.to(device)
|
| 417 |
+
with torch.no_grad():
|
| 418 |
+
with torch.cuda.amp.autocast(enabled=cfg.use_amp):
|
| 419 |
+
embedding = model.create_embedding(inputs)
|
| 420 |
+
embedding_list += embedding.tolist()
|
| 421 |
+
|
| 422 |
+
torch.cuda.empty_cache()
|
| 423 |
+
gc.collect()
|
| 424 |
+
return embedding_list
|
| 425 |
+
|
| 426 |
+
|
| 427 |
+
if __name__ == "__main__":
|
| 428 |
+
config = parse_args()
|
| 429 |
+
config.token_length = 2 if config.architecture == "SaProt" else 1
|
| 430 |
+
config.device = device
|
| 431 |
+
|
| 432 |
+
if not os.path.exists(config.output_dir):
|
| 433 |
+
os.makedirs(config.output_dir)
|
| 434 |
+
|
| 435 |
+
if config.used_sequence == "both":
|
| 436 |
+
config.max_length += 1
|
| 437 |
+
|
| 438 |
+
LOGGER = get_logger(os.path.join(config.output_dir, "output"))
|
| 439 |
+
config.logger = LOGGER
|
| 440 |
+
|
| 441 |
+
seed_everything(config.seed)
|
| 442 |
+
|
| 443 |
+
train_df = (
|
| 444 |
+
pd.read_csv(config.data_path)
|
| 445 |
+
.drop_duplicates(subset=[config.sequence_col], keep="first")
|
| 446 |
+
.reset_index(drop=True)
|
| 447 |
+
)
|
| 448 |
+
train_df["T1/2 [h]"] = train_df[config.target_col]
|
| 449 |
+
|
| 450 |
+
if config.task == "classification":
|
| 451 |
+
train_df["target"] = (
|
| 452 |
+
train_df["T1/2 [h]"] > np.median(train_df["T1/2 [h]"])
|
| 453 |
+
).astype(int)
|
| 454 |
+
train_df["class"] = train_df["target"]
|
| 455 |
+
elif config.task == "regression":
|
| 456 |
+
train_df["log1p(T1/2 [h])"] = np.log1p(train_df["T1/2 [h]"])
|
| 457 |
+
train_df["log1p(T1/2 [h])"] = (
|
| 458 |
+
train_df["log1p(T1/2 [h])"] - min(train_df["log1p(T1/2 [h])"])
|
| 459 |
+
) / (max(train_df["log1p(T1/2 [h])"]) - min(train_df["log1p(T1/2 [h])"]))
|
| 460 |
+
train_df["target"] = train_df["log1p(T1/2 [h])"]
|
| 461 |
+
|
| 462 |
+
def get_class(row, class_num=5):
|
| 463 |
+
denom = 1 / class_num
|
| 464 |
+
num = row["log1p(T1/2 [h])"]
|
| 465 |
+
for target in range(class_num):
|
| 466 |
+
if denom * target <= num and num < denom * (target + 1):
|
| 467 |
+
break
|
| 468 |
+
row["class"] = target
|
| 469 |
+
return row
|
| 470 |
+
|
| 471 |
+
train_df = train_df.apply(get_class, axis=1)
|
| 472 |
+
|
| 473 |
+
train_df["fold"] = -1
|
| 474 |
+
kf = StratifiedKFold(
|
| 475 |
+
n_splits=config.n_folds, shuffle=True, random_state=config.seed
|
| 476 |
+
)
|
| 477 |
+
for fold, (trn_ind, val_ind) in enumerate(kf.split(train_df, train_df["class"])):
|
| 478 |
+
train_df.loc[val_ind, "fold"] = int(fold)
|
| 479 |
+
|
| 480 |
+
if config.architecture in ["ESM2", "SaProt"]:
|
| 481 |
+
tokenizer = AutoTokenizer.from_pretrained(
|
| 482 |
+
config.model, padding_side=config.padding_side
|
| 483 |
+
)
|
| 484 |
+
tokenizer.save_pretrained(config.output_dir)
|
| 485 |
+
config.tokenizer = tokenizer
|
| 486 |
+
|
| 487 |
+
oof_df = pd.DataFrame()
|
| 488 |
+
for fold in range(config.n_folds):
|
| 489 |
+
_oof_df = train_loop(train_df, fold, config)
|
| 490 |
+
oof_df = pd.concat([oof_df, _oof_df], axis=0)
|
| 491 |
+
|
| 492 |
+
oof_df = oof_df.reset_index(drop=True)
|
| 493 |
+
oof_df.to_csv(os.path.join(config.output_dir, "oof_df.csv"), index=False)
|
scripts/use_foldseek_for_uniprot.py
ADDED
|
@@ -0,0 +1,106 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import random
|
| 3 |
+
import argparse
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import multiprocessing as mp
|
| 6 |
+
from foldseek_util import get_struc_seq
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
def parse_args():
|
| 10 |
+
parser = argparse.ArgumentParser()
|
| 11 |
+
parser.add_argument(
|
| 12 |
+
"--file_path",
|
| 13 |
+
type=str,
|
| 14 |
+
required=True,
|
| 15 |
+
help="Path to the file containing uniprotid information.",
|
| 16 |
+
)
|
| 17 |
+
parser.add_argument(
|
| 18 |
+
"--sheet_name",
|
| 19 |
+
type=str,
|
| 20 |
+
default="Sheet1",
|
| 21 |
+
help="Name of the sheet to read (for Excel files). Default is 'Sheet1'.",
|
| 22 |
+
)
|
| 23 |
+
parser.add_argument(
|
| 24 |
+
"--pdb_dir",
|
| 25 |
+
type=str,
|
| 26 |
+
default="pdb_files/UP000000589_10090_MOUSE_v4",
|
| 27 |
+
help="Directory containing PDB files.",
|
| 28 |
+
)
|
| 29 |
+
parser.add_argument(
|
| 30 |
+
"--uniprotid_column",
|
| 31 |
+
type=str,
|
| 32 |
+
help="Name of the column containing UniprotID information.",
|
| 33 |
+
)
|
| 34 |
+
parser.add_argument(
|
| 35 |
+
"--uniprotids_column",
|
| 36 |
+
type=str,
|
| 37 |
+
help="Name of the column containing multiple UniprotIDs (separated by semicolons). The first ID will be used.",
|
| 38 |
+
)
|
| 39 |
+
parser.add_argument(
|
| 40 |
+
"--num_processes",
|
| 41 |
+
type=int,
|
| 42 |
+
default=2,
|
| 43 |
+
help="Number of processes to use for multiprocessing. Default is 2.",
|
| 44 |
+
)
|
| 45 |
+
return parser.parse_args()
|
| 46 |
+
|
| 47 |
+
|
| 48 |
+
def validate_columns(cfg, df):
|
| 49 |
+
if cfg.uniprotid_column is None and cfg.uniprotids_column is None:
|
| 50 |
+
raise ValueError("Either --uniprotid_column or --uniprotids_column must be provided.")
|
| 51 |
+
if cfg.uniprotids_column:
|
| 52 |
+
df = df.dropna(subset=[cfg.uniprotids_column]).reset_index(drop=True)
|
| 53 |
+
df["uniprotid"] = df[cfg.uniprotids_column].apply(lambda x: x.split(";")[0].split("-")[0])
|
| 54 |
+
cfg.uniprotid_column = "uniprotid"
|
| 55 |
+
return df.dropna(subset=[cfg.uniprotid_column]).reset_index(drop=True)
|
| 56 |
+
|
| 57 |
+
|
| 58 |
+
def find_pdb_files(pdb_dir, uniprot_ids):
|
| 59 |
+
pdf_files = os.listdir(pdb_dir)
|
| 60 |
+
pdb_paths = []
|
| 61 |
+
for uniprot_id in uniprot_ids:
|
| 62 |
+
matches = [pdf_file for pdf_file in sorted(pdf_files) if uniprot_id in pdf_file]
|
| 63 |
+
pdb_paths.append(matches[0] if matches else None)
|
| 64 |
+
return pdb_paths
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
def get_foldseek_seq(pdb_path, cfg):
|
| 68 |
+
parsed_seqs = get_struc_seq(
|
| 69 |
+
"bin/foldseek",
|
| 70 |
+
os.path.join(cfg.pdb_dir, pdb_path),
|
| 71 |
+
["A"],
|
| 72 |
+
process_id=random.randint(0, 10000000),
|
| 73 |
+
)["A"]
|
| 74 |
+
return parsed_seqs
|
| 75 |
+
|
| 76 |
+
|
| 77 |
+
if __name__ == "__main__":
|
| 78 |
+
|
| 79 |
+
config = parse_args()
|
| 80 |
+
|
| 81 |
+
if config.file_path.endswith(".xls") or config.file_path.endswith(".xlsx"):
|
| 82 |
+
df = pd.read_excel(
|
| 83 |
+
config.file_path,
|
| 84 |
+
sheet_name=config.sheet_name,
|
| 85 |
+
)
|
| 86 |
+
else:
|
| 87 |
+
df = pd.read_csv(config.file_path)
|
| 88 |
+
df = validate_columns(config, df)
|
| 89 |
+
|
| 90 |
+
df = df.dropna(subset=[config.uniprotid_column]).reset_index(drop=True)
|
| 91 |
+
|
| 92 |
+
uniprot_ids = df[config.uniprotid_column].tolist()
|
| 93 |
+
pdb_paths = find_pdb_files(config.pdb_dir, uniprot_ids)
|
| 94 |
+
df["pdb_path"] = pdb_paths
|
| 95 |
+
df = df.dropna(subset=["pdb_path"]).reset_index(drop=True)
|
| 96 |
+
df = df.drop_duplicates(subset=[config.uniprotid_column]).reset_index(drop=True)
|
| 97 |
+
|
| 98 |
+
with mp.Pool(config.num_processes) as pool:
|
| 99 |
+
output = pool.map(lambda x: get_foldseek_seq(x, config), df["pdb_path"].tolist())
|
| 100 |
+
|
| 101 |
+
aa, foldseek, aa_foldseek = zip(*output)
|
| 102 |
+
|
| 103 |
+
df["aa"] = aa
|
| 104 |
+
df["foldseek"] = foldseek
|
| 105 |
+
df["aa_foldseek"] = aa_foldseek
|
| 106 |
+
df.to_csv(f"{config.file_path.split('.')[0]}_foldseek.csv", index=False)
|
utils.py
ADDED
|
@@ -0,0 +1,88 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import random
|
| 2 |
+
import os
|
| 3 |
+
import math
|
| 4 |
+
import time
|
| 5 |
+
import numpy as np
|
| 6 |
+
import pickle
|
| 7 |
+
import torch
|
| 8 |
+
import logging
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
def get_logger(filename: str):
|
| 12 |
+
"""Creates and returns a logger that logs to both the console and a file."""
|
| 13 |
+
logger = logging.getLogger(__name__)
|
| 14 |
+
logger.setLevel(logging.INFO)
|
| 15 |
+
|
| 16 |
+
# Console handler
|
| 17 |
+
stream_handler = logging.StreamHandler()
|
| 18 |
+
stream_handler.setFormatter(logging.Formatter("%(message)s"))
|
| 19 |
+
logger.addHandler(stream_handler)
|
| 20 |
+
|
| 21 |
+
# File handler
|
| 22 |
+
file_handler = logging.FileHandler(f"{filename}.log")
|
| 23 |
+
file_handler.setFormatter(logging.Formatter("%(message)s"))
|
| 24 |
+
logger.addHandler(file_handler)
|
| 25 |
+
|
| 26 |
+
return logger
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
def seed_everything(seed: int):
|
| 30 |
+
"""Sets random seed for reproducibility across various libraries."""
|
| 31 |
+
random.seed(seed)
|
| 32 |
+
os.environ["PYTHONHASHSEED"] = str(seed)
|
| 33 |
+
np.random.seed(seed)
|
| 34 |
+
torch.manual_seed(seed)
|
| 35 |
+
torch.cuda.manual_seed(seed)
|
| 36 |
+
torch.backends.cudnn.deterministic = True
|
| 37 |
+
torch.backends.cudnn.benchmark = False
|
| 38 |
+
|
| 39 |
+
|
| 40 |
+
class AverageMeter:
|
| 41 |
+
"""Tracks and stores the average and current values."""
|
| 42 |
+
|
| 43 |
+
def __init__(self):
|
| 44 |
+
self.reset()
|
| 45 |
+
|
| 46 |
+
def reset(self):
|
| 47 |
+
self.val = 0
|
| 48 |
+
self.avg = 0
|
| 49 |
+
self.sum = 0
|
| 50 |
+
self.count = 0
|
| 51 |
+
|
| 52 |
+
def update(self, val, n=1):
|
| 53 |
+
self.val = val
|
| 54 |
+
self.sum += val * n
|
| 55 |
+
self.count += n
|
| 56 |
+
self.avg = self.sum / self.count
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
def as_minutes(s: int) -> str:
|
| 60 |
+
"""Converts seconds to a string in minutes and seconds."""
|
| 61 |
+
m = math.floor(s / 60)
|
| 62 |
+
s -= m * 60
|
| 63 |
+
return "%dm %ds" % (m, s)
|
| 64 |
+
|
| 65 |
+
|
| 66 |
+
def timeSince(since: float, percent: float) -> str:
|
| 67 |
+
now = time.time()
|
| 68 |
+
s = now - since
|
| 69 |
+
es = s / (percent)
|
| 70 |
+
rs = es - s
|
| 71 |
+
return "%s (remain %s)" % (as_minutes(s), as_minutes(rs))
|
| 72 |
+
|
| 73 |
+
|
| 74 |
+
def convert_all_1d(array: list) -> list:
|
| 75 |
+
"""Converts 0-dimensional arrays in a list to 1-dimensional arrays."""
|
| 76 |
+
return [np.array([item]) if item.ndim == 0 else item for item in array]
|
| 77 |
+
|
| 78 |
+
|
| 79 |
+
def save_pickle(path: str, contents):
|
| 80 |
+
"""Saves contents to a pickle file."""
|
| 81 |
+
with open(path, "wb") as f:
|
| 82 |
+
pickle.dump(contents, f)
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
def load_pickle(path: str):
|
| 86 |
+
"""Loads contents from a pickle file."""
|
| 87 |
+
with open(path, "rb") as f:
|
| 88 |
+
return pickle.load(f)
|