Spaces:
Sleeping
Sleeping
Upload 6 files
Browse files- app.py +105 -0
- confussion_matrix.png +0 -0
- final_model.h5 +3 -0
- model.h5 +3 -0
- plant disease classification.ipynb +0 -0
- requirements.txt +8 -0
app.py
ADDED
|
@@ -0,0 +1,105 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
from PIL import Image
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import numpy as np
|
| 5 |
+
from keras.models import load_model
|
| 6 |
+
model = load_model('./model.h5')
|
| 7 |
+
|
| 8 |
+
# Classification report data
|
| 9 |
+
data = {
|
| 10 |
+
"classes": ['Pepper__bell___Bacterial_spot',
|
| 11 |
+
'Pepper__bell___healthy',
|
| 12 |
+
'Potato___Early_blight',
|
| 13 |
+
'Potato___Late_blight',
|
| 14 |
+
'Potato___healthy',
|
| 15 |
+
'Tomato_Bacterial_spot',
|
| 16 |
+
'Tomato_Early_blight',
|
| 17 |
+
'Tomato_Late_blight',
|
| 18 |
+
'Tomato_Leaf_Mold',
|
| 19 |
+
'Tomato_Septoria_leaf_spot',
|
| 20 |
+
'Tomato_Spider_mites_Two_spotted_spider_mite',
|
| 21 |
+
'Tomato__Target_Spot',
|
| 22 |
+
'Tomato__Tomato_YellowLeaf__Curl_Virus',
|
| 23 |
+
'Tomato__Tomato_mosaic_virus',
|
| 24 |
+
'Tomato_healthy'],
|
| 25 |
+
"precision": [0.99, 0.99, 0.98, 0.83, 0.41, 0.99, 0.74, 1.00, 1.00, 0.91, 0.95, 0.60, 1.00, 0.87, 0.91],
|
| 26 |
+
"recall": [0.99, 0.86, 1.00, 0.99, 1.00, 0.74, 0.90, 0.83, 0.71, 0.97, 0.64, 0.99, 0.98, 0.98, 1.00],
|
| 27 |
+
"f1-score": [0.99, 0.92, 0.99, 0.90, 0.58, 0.85, 0.82, 0.90, 0.83, 0.94, 0.77, 0.74, 0.99, 0.92, 0.95],
|
| 28 |
+
"support": [199, 296, 213, 205, 28, 399, 206, 379, 187, 359, 326, 290, 629, 81, 331]
|
| 29 |
+
}
|
| 30 |
+
|
| 31 |
+
# Create a DataFrame
|
| 32 |
+
df = pd.DataFrame(data)
|
| 33 |
+
|
| 34 |
+
def get_predictions(immg):
|
| 35 |
+
# from skimage import io
|
| 36 |
+
# from keras.preprocessing import image
|
| 37 |
+
import keras.utils as image
|
| 38 |
+
#path='imbalanced/Scratch/Scratch_400.jpg'
|
| 39 |
+
# "C:\Users\prati\Desktop\AML 3104 final project\PlantVillage\Pepper__bell___Bacterial_spot\00f2e69a-1e56-412d-8a79-fdce794a17e4___JR_B.Spot 3132.JPG"
|
| 40 |
+
# pth = immg
|
| 41 |
+
# show_img=image.load_img(pth, grayscale=False, target_size=(200, 200))
|
| 42 |
+
disease_class = data["classes"]
|
| 43 |
+
|
| 44 |
+
x = image.img_to_array(immg)
|
| 45 |
+
x = np.expand_dims(x, axis = 0)
|
| 46 |
+
x = np.array(x, 'float32')
|
| 47 |
+
x /= 255
|
| 48 |
+
|
| 49 |
+
custom = model.predict(x)
|
| 50 |
+
print(custom[0])
|
| 51 |
+
|
| 52 |
+
# x = x.reshape([64, 64])
|
| 53 |
+
|
| 54 |
+
#plt.gray()
|
| 55 |
+
# plt.imshow(show_img)
|
| 56 |
+
# plt.show()
|
| 57 |
+
|
| 58 |
+
a=custom[0]
|
| 59 |
+
ind=np.argmax(a)
|
| 60 |
+
|
| 61 |
+
print('Prediction:',disease_class[ind])
|
| 62 |
+
return(disease_class[ind])
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
#User Interface---------------------------------------------------------
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
pred_flag = False
|
| 69 |
+
def main():
|
| 70 |
+
st.label_visibility='collapse'
|
| 71 |
+
st.title('Plant desise identification')
|
| 72 |
+
st.write('# Detecting plant desise using deep learnings dense net model')
|
| 73 |
+
# Display the DataFrame as a table in Streamlit
|
| 74 |
+
st.header('Evaluation Report:')
|
| 75 |
+
|
| 76 |
+
|
| 77 |
+
# Create two columns
|
| 78 |
+
col1, col2 = st.columns(2)
|
| 79 |
+
|
| 80 |
+
# Add content to the first column
|
| 81 |
+
with col1:
|
| 82 |
+
st.write('Classification Report:')
|
| 83 |
+
st.table(df)
|
| 84 |
+
|
| 85 |
+
# Add content to the second column
|
| 86 |
+
with col2:
|
| 87 |
+
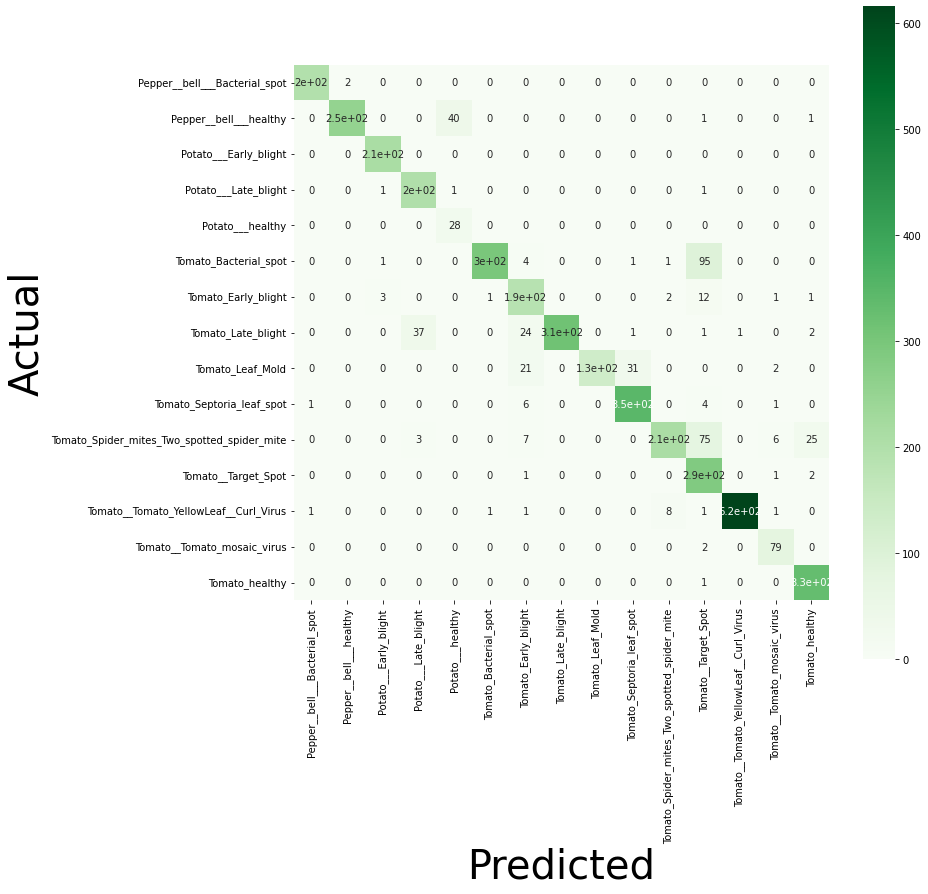
st.write('Confussion matrix')
|
| 88 |
+
st.image("./confussion_matrix.png",width=650)
|
| 89 |
+
|
| 90 |
+
uploaded_file = st.file_uploader("", type=['jpg','png','jpeg'])
|
| 91 |
+
if uploaded_file is not None:
|
| 92 |
+
image = Image.open(uploaded_file)
|
| 93 |
+
st.markdown('<p style="text-align: center;"><label>Image : </label></p>',unsafe_allow_html=True)
|
| 94 |
+
st.image(image,width=500)
|
| 95 |
+
# image = Image.open()
|
| 96 |
+
if st.button("Predict"):
|
| 97 |
+
resized_image = image.resize((64, 64))
|
| 98 |
+
pred_cls = get_predictions(resized_image)
|
| 99 |
+
st.markdown('<p style="text-align: center;"><label>Prediction : </label></p>',unsafe_allow_html=True)
|
| 100 |
+
# st.image(image,width=900)
|
| 101 |
+
st.markdown(pred_cls)
|
| 102 |
+
# result =''
|
| 103 |
+
# st.success('The output is {}'.format(result))
|
| 104 |
+
if __name__ == '__main__': #
|
| 105 |
+
main()
|
confussion_matrix.png
ADDED
|
final_model.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:7a37375a2e143e19e773290e5541d3c6426726674a937fa76697e35a037914fa
|
| 3 |
+
size 88637056
|
model.h5
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:1ad27f74d08d9864cd3574826841909ddc8a4dc902b1e72069f27888b75dfa2a
|
| 3 |
+
size 88637056
|
plant disease classification.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
requirements.txt
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
tensorflow==2.11.0
|
| 2 |
+
keras==2.11.0
|
| 3 |
+
sklearn==0.0
|
| 4 |
+
Pillow==9.2.0
|
| 5 |
+
pandas==1.4.3
|
| 6 |
+
streamlit==1.15.1
|
| 7 |
+
protobuf==3.19.0
|
| 8 |
+
numpy
|