Spaces:
Sleeping
Sleeping
Jeysshon
commited on
Commit
•
3cc72c9
1
Parent(s):
489b816
Upload 19 files
Browse files- README.md +6 -5
- ada_f1_skin.png +0 -0
- ada_f1_skin_be.png +0 -0
- ada_learn_malben.pkl +3 -0
- ada_learn_skin_norm2000.pkl +3 -0
- akiec1.jpg +0 -0
- app.py +173 -0
- bcc1.jpg +0 -0
- benign1.jpg +0 -0
- benign3.jpg +0 -0
- bkl1.jpg +0 -0
- df1.jpg +0 -0
- gitattributes.txt +29 -0
- mel1.jpg +0 -0
- nevi1.jpg +0 -0
- requirements.txt +2 -0
- scc1.jpg +0 -0
- unk1.jpg +0 -0
- vl1.jpg +0 -0
README.md
CHANGED
|
@@ -1,12 +1,13 @@
|
|
| 1 |
---
|
| 2 |
-
title:
|
| 3 |
-
emoji:
|
| 4 |
-
colorFrom:
|
| 5 |
-
colorTo:
|
| 6 |
sdk: gradio
|
| 7 |
-
sdk_version: 3.
|
| 8 |
app_file: app.py
|
| 9 |
pinned: false
|
|
|
|
| 10 |
---
|
| 11 |
|
| 12 |
Check out the configuration reference at https://huggingface.co/docs/hub/spaces-config-reference
|
|
|
|
| 1 |
---
|
| 2 |
+
title: Skin Cancer Diagnose
|
| 3 |
+
emoji: 🐠
|
| 4 |
+
colorFrom: yellow
|
| 5 |
+
colorTo: indigo
|
| 6 |
sdk: gradio
|
| 7 |
+
sdk_version: 3.0.22
|
| 8 |
app_file: app.py
|
| 9 |
pinned: false
|
| 10 |
+
license: gpl-3.0
|
| 11 |
---
|
| 12 |
|
| 13 |
Check out the configuration reference at https://huggingface.co/docs/hub/spaces-config-reference
|
ada_f1_skin.png
ADDED
|
ada_f1_skin_be.png
ADDED
|
ada_learn_malben.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:441760abd9a3a6b143917c68cf1709a1d96323fa334b4952009f50a34ddb57c2
|
| 3 |
+
size 87548073
|
ada_learn_skin_norm2000.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:820252747a54e3e7494fdc86c998d1d67996cb31bde244ddd4260307e80dd819
|
| 3 |
+
size 87739881
|
akiec1.jpg
ADDED
|
app.py
ADDED
|
@@ -0,0 +1,173 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
|
| 2 |
+
import fastai
|
| 3 |
+
import fastai.vision
|
| 4 |
+
import PIL
|
| 5 |
+
import gradio
|
| 6 |
+
import matplotlib
|
| 7 |
+
import numpy
|
| 8 |
+
import pandas
|
| 9 |
+
from fastai.vision.all import *
|
| 10 |
+
#
|
| 11 |
+
# create class
|
| 12 |
+
class ADA_SKIN(object):
|
| 13 |
+
#
|
| 14 |
+
# initialize the object
|
| 15 |
+
def __init__(self, name="Wallaby",verbose=True,*args, **kwargs):
|
| 16 |
+
super(ADA_SKIN, self).__init__(*args, **kwargs)
|
| 17 |
+
self.author = "Duc Haba"
|
| 18 |
+
self.name = name
|
| 19 |
+
if (verbose):
|
| 20 |
+
self._ph()
|
| 21 |
+
self._pp("Hello from class", str(self.__class__) + " Class: " + str(self.__class__.__name__))
|
| 22 |
+
self._pp("Code name", self.name)
|
| 23 |
+
self._pp("Author is", self.author)
|
| 24 |
+
self._ph()
|
| 25 |
+
#
|
| 26 |
+
self.article = '<div><h3>Warning:</h3>Do NOT use this for any medical diagnosis.<br>'
|
| 27 |
+
self.article += 'I am not a dermatologist, and NO dermatologist has endorsed it. '
|
| 28 |
+
self.article += 'This DL model is for my independent research. <br>Please refer to the GPL 3.0 for usage and license.'
|
| 29 |
+
self.article += '<h3>Citation:</h3><ul><li>'
|
| 30 |
+
self.article += 'Author/Dev: Duc Haba, 2022.</li>'
|
| 31 |
+
self.article += '<li><a target="_blank" href="https://linkedin.com/in/duchaba">https://linkedin.com/in/duchaba</a></li>'
|
| 32 |
+
self.article += '<li>The training dataset are from the International Skin Imaging Collaboration (ISIC)</li>'
|
| 33 |
+
self.article += '<li>The Skin Cancer Identification are from 3 seperate datasets.</li>'
|
| 34 |
+
self.article += '<ol>'
|
| 35 |
+
self.article += '<li>https://www.kaggle.com/datasets/surajghuwalewala/ham1000-segmentation-and-classification</li>'
|
| 36 |
+
self.article += '<li>https://www.kaggle.com/datasets/andrewmvd/isic-2019</li>'
|
| 37 |
+
self.article += '<li>https://www.kaggle.com/datasets/jnegrini/skin-lesions-act-keratosis-and-melanoma</li>'
|
| 38 |
+
self.article += '<ul><li>'
|
| 39 |
+
self.article += 'The Malignant versus Benign dataset</li>'
|
| 40 |
+
self.article += '<ol><li>https://www.kaggle.com/datasets/fanconic/skin-cancer-malignant-vs-benign</li>'
|
| 41 |
+
self.article += '</ol></ul>'
|
| 42 |
+
self.article += '<h3>Articles:</h3><ul>'
|
| 43 |
+
self.article += '<li><a target="_blank" href="https://www.linkedin.com/pulse/skin-cancer-diagnose-using-deep-learning-duc-haba/">'
|
| 44 |
+
self.article += '"Skin Cancer Diagnose"</a> on LinkedIn, on <a target="_blank" href='
|
| 45 |
+
self.article += '"https://www.linkedin.com/pulse/skin-cancer-diagnose-using-deep-learning-duc-haba/">Medium.</a></li>'
|
| 46 |
+
self.article += '</ul>'
|
| 47 |
+
self.article += '<h3>Example Images: (left to right)</h3><ol>'
|
| 48 |
+
self.article += '<li>Bowen Disease (AKIEC)</li>'
|
| 49 |
+
self.article += '<li>Basal Cell Carcinoma</li>'
|
| 50 |
+
self.article += '<li>Benign Keratosis-like Lesions</li>'
|
| 51 |
+
self.article += '<li>Dermatofibroma</li>'
|
| 52 |
+
self.article += '<li>Melanoma</li>'
|
| 53 |
+
self.article += '<li>Melanocytic Nevi</li>'
|
| 54 |
+
self.article += '<li>Squamous Cell Carcinoma</li>'
|
| 55 |
+
self.article += '<li>Vascular Lesions</li>'
|
| 56 |
+
self.article += '<li>Benign</li>'
|
| 57 |
+
self.article += '<li>Benign 2</li></ol>'
|
| 58 |
+
self.article += '<h3>Train Result:</h3><ul>'
|
| 59 |
+
self.article += '<li>Skin Cancer Classificaiton: F1-Score, Precision, and Recall Graph</li>'
|
| 60 |
+
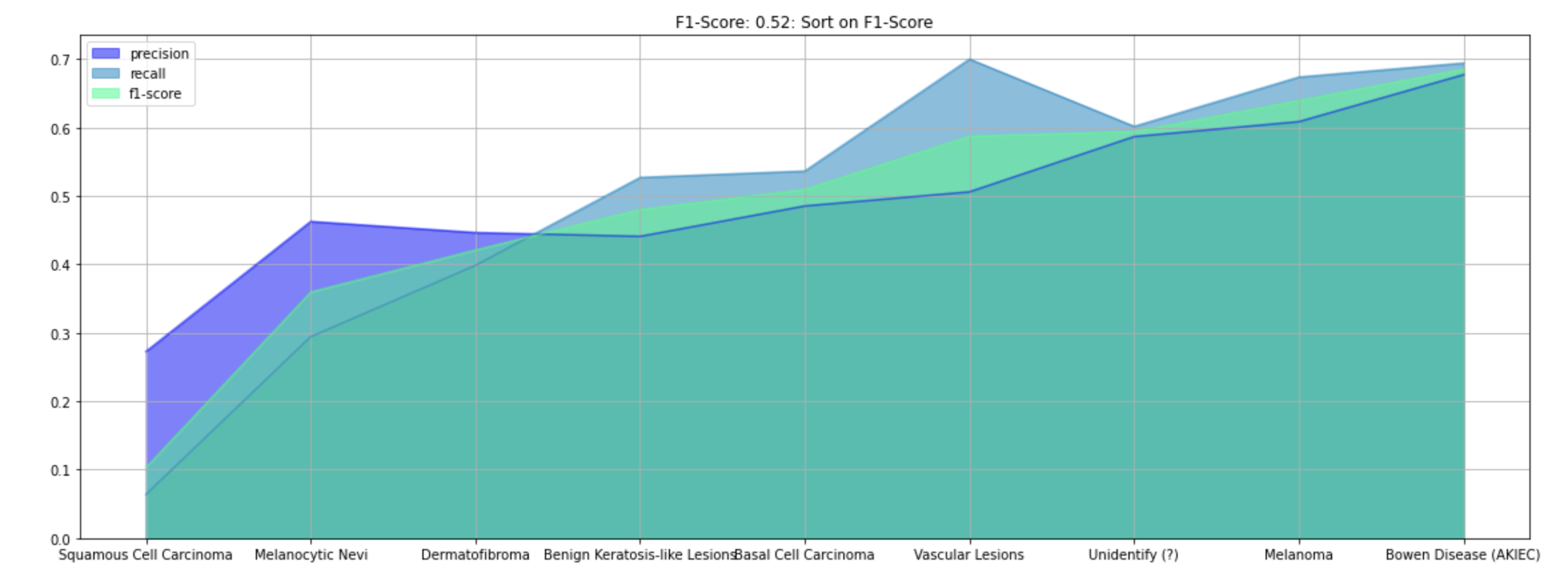
self.article += '<li><img src="file/ada_f1_skin.png" alt="F1-Score, Precision, and Recall Graph" width="640"</li>'
|
| 61 |
+
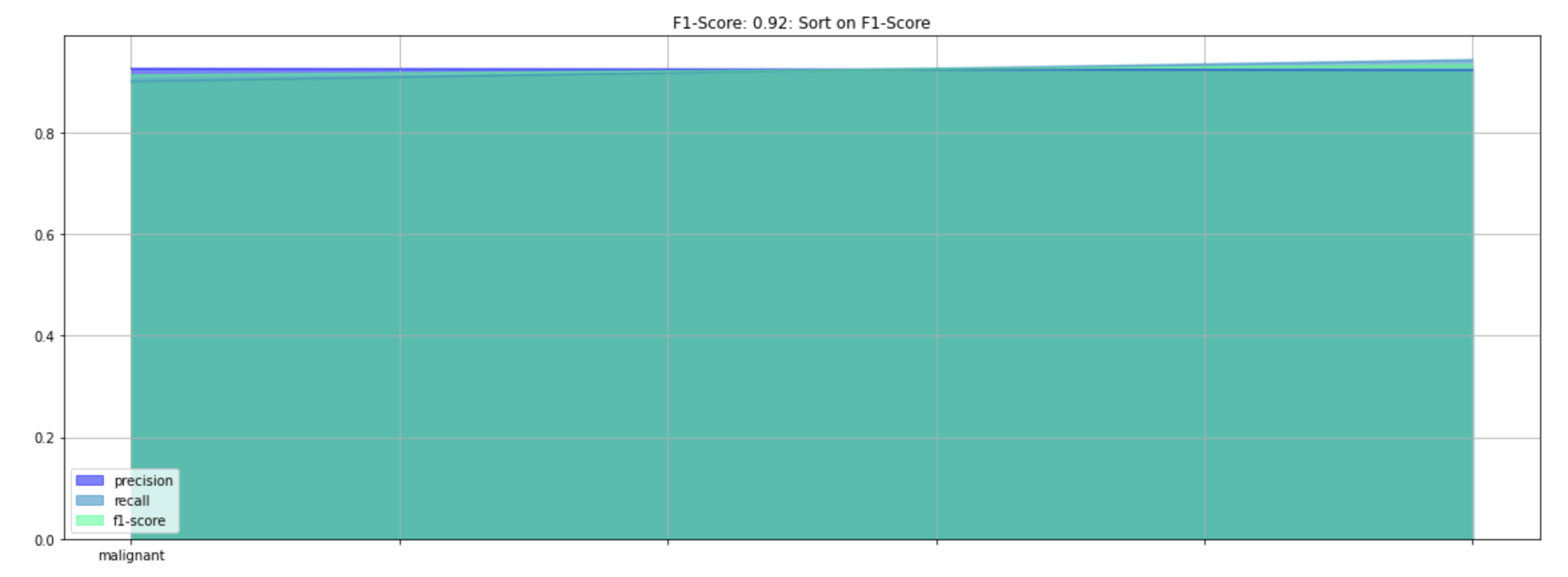
self.article += '<li>Skin Cancer Malignant or Benign: F1-Score, Precision, and Recall Graph</li>'
|
| 62 |
+
self.article += '<li><img src="file/ada_f1_skin_be.png" alt="F1-Score, Precision, and Recall Graph" width="640"</li>'
|
| 63 |
+
self.article += '</ul>'
|
| 64 |
+
self.article += '<h3>Dev Stack:</h3><ul>'
|
| 65 |
+
self.article += '<li>Jupyter Notebook, Python, Pandas, Matplotlib, Sklearn</li>'
|
| 66 |
+
self.article += '<li>Fast.ai, PyTorch</li>'
|
| 67 |
+
self.article += '</ul>'
|
| 68 |
+
self.article += '<h3>Licenses:</h3><ul>'
|
| 69 |
+
self.article += '<li><a target="_blank" href="https://www.gnu.org/licenses/gpl-3.0.txt">GNU GPL 3.0</a></li>'
|
| 70 |
+
self.article += '</ul></div>'
|
| 71 |
+
self.examples = ['akiec1.jpg','bcc1.jpg','bkl1.jpg','df1.jpg','mel1.jpg',
|
| 72 |
+
'nevi1.jpg','scc1.jpg','vl1.jpg','benign1.jpg','benign3.jpg']
|
| 73 |
+
self.title = "Skin Cancer Diagnose"
|
| 74 |
+
return
|
| 75 |
+
#
|
| 76 |
+
# pretty print output name-value line
|
| 77 |
+
def _pp(self, a, b):
|
| 78 |
+
print("%34s : %s" % (str(a), str(b)))
|
| 79 |
+
return
|
| 80 |
+
#
|
| 81 |
+
# pretty print the header or footer lines
|
| 82 |
+
def _ph(self):
|
| 83 |
+
print("-" * 34, ":", "-" * 34)
|
| 84 |
+
return
|
| 85 |
+
#
|
| 86 |
+
def _predict_image(self,img,cat):
|
| 87 |
+
pred,idx,probs = learn.predict(img)
|
| 88 |
+
return dict(zip(cat, map(float,probs)))
|
| 89 |
+
#
|
| 90 |
+
def _predict_image2(self,img,cat):
|
| 91 |
+
pred,idx,probs = learn2.predict(img)
|
| 92 |
+
return dict(zip(cat, map(float,probs)))
|
| 93 |
+
#
|
| 94 |
+
def _draw_pred(self,df_pred, df2):
|
| 95 |
+
canvas, pic = matplotlib.pyplot.subplots(1,2, figsize=(12,6))
|
| 96 |
+
ti = df_pred["vocab"].head(3).values
|
| 97 |
+
ti2 = df2["vocab"].head(2).values
|
| 98 |
+
# special case
|
| 99 |
+
#if (matplotlib.__version__) >= "3.5.2":
|
| 100 |
+
try:
|
| 101 |
+
df_pred["pred"].head(3).plot(ax=pic[0],kind="pie",
|
| 102 |
+
cmap="Set2",labels=ti, explode=(0.02,0,0),
|
| 103 |
+
wedgeprops=dict(width=.4),
|
| 104 |
+
normalize=False)
|
| 105 |
+
df2["pred"].head(2).plot(ax=pic[1],kind="pie",
|
| 106 |
+
colors=["cornflowerblue","darkorange"],labels=ti2, explode=(0.02,0),
|
| 107 |
+
wedgeprops=dict(width=.4),
|
| 108 |
+
normalize=False)
|
| 109 |
+
except:
|
| 110 |
+
df_pred["pred"].head(3).plot(ax=pic[0],kind="pie",
|
| 111 |
+
cmap="Set2",labels=ti, explode=(0.02,0,0),
|
| 112 |
+
wedgeprops=dict(width=.4))
|
| 113 |
+
df2["pred"].head(2).plot(ax=pic[1],kind="pie",
|
| 114 |
+
colors=["cornflowerblue","darkorange"],labels=ti2, explode=(0.02,0),
|
| 115 |
+
wedgeprops=dict(width=.4))
|
| 116 |
+
t = str(ti[0]) + ": " + str(numpy.round(df_pred.head(1).pred.values[0]*100, 2)) + "% Certainty"
|
| 117 |
+
pic[0].set_title(t,fontsize=14.0, fontweight="bold")
|
| 118 |
+
pic[0].axis('off')
|
| 119 |
+
pic[0].legend(ti, loc="lower right",title="Skin Cancers: Top 3")
|
| 120 |
+
#
|
| 121 |
+
k0 = numpy.round(df2.head(1).pred.values[0]*100, 2)
|
| 122 |
+
k1 = numpy.round(df2.tail(1).pred.values[0]*100, 2)
|
| 123 |
+
if (k0 > k1):
|
| 124 |
+
t2 = str(ti2[0]) + ": " + str(k0) + "% Certainty"
|
| 125 |
+
else:
|
| 126 |
+
t2 = str(ti2[1]) + ": " + str(k1) + "% Certainty"
|
| 127 |
+
pic[1].set_title(t2,fontsize=14.0, fontweight="bold")
|
| 128 |
+
pic[1].axis('off')
|
| 129 |
+
pic[1].legend(ti2, loc="lower right",title="Skin Cancers:")
|
| 130 |
+
#
|
| 131 |
+
# # draw circle
|
| 132 |
+
# centre_circle = matplotlib.pyplot.Circle((0, 0), 0.6, fc='white')
|
| 133 |
+
# p = matplotlib.pyplot.gcf()
|
| 134 |
+
# # Adding Circle in Pie chart
|
| 135 |
+
# p.gca().add_artist(centre_circle)
|
| 136 |
+
#
|
| 137 |
+
#p=plt.gcf()
|
| 138 |
+
#p.gca().add_artist(my_circle)
|
| 139 |
+
#
|
| 140 |
+
canvas.tight_layout()
|
| 141 |
+
return canvas
|
| 142 |
+
#
|
| 143 |
+
def predict_donut(self,img):
|
| 144 |
+
d = self._predict_image(img,self.categories)
|
| 145 |
+
df = pandas.DataFrame(d, index=[0])
|
| 146 |
+
df = df.transpose().reset_index()
|
| 147 |
+
df.columns = ["vocab", "pred"]
|
| 148 |
+
df.sort_values("pred", inplace=True,ascending=False, ignore_index=True)
|
| 149 |
+
#
|
| 150 |
+
d2 = self._predict_image2(img,self.categories2)
|
| 151 |
+
df2 = pandas.DataFrame(d2, index=[0])
|
| 152 |
+
df2 = df2.transpose().reset_index()
|
| 153 |
+
df2.columns = ["vocab", "pred"]
|
| 154 |
+
#
|
| 155 |
+
canvas = self._draw_pred(df,df2)
|
| 156 |
+
return canvas
|
| 157 |
+
#
|
| 158 |
+
maxi = ADA_SKIN(verbose=False)
|
| 159 |
+
#
|
| 160 |
+
learn = fastai.learner.load_learner('ada_learn_skin_norm2000.pkl')
|
| 161 |
+
learn2 = fastai.learner.load_learner('ada_learn_malben.pkl')
|
| 162 |
+
maxi.categories = learn.dls.vocab
|
| 163 |
+
maxi.categories2 = learn2.dls.vocab
|
| 164 |
+
hf_image = gradio.inputs.Image(shape=(192, 192))
|
| 165 |
+
hf_label = gradio.outputs.Label()
|
| 166 |
+
intf = gradio.Interface(fn=maxi.predict_donut,
|
| 167 |
+
inputs=hf_image,
|
| 168 |
+
outputs=["plot"],
|
| 169 |
+
examples=maxi.examples,
|
| 170 |
+
title=maxi.title,
|
| 171 |
+
live=True,
|
| 172 |
+
article=maxi.article)
|
| 173 |
+
intf.launch(inline=False,share=True)
|
bcc1.jpg
ADDED
|
benign1.jpg
ADDED
|
benign3.jpg
ADDED
|
bkl1.jpg
ADDED

|
df1.jpg
ADDED
|
gitattributes.txt
ADDED
|
@@ -0,0 +1,29 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
*.7z filter=lfs diff=lfs merge=lfs -text
|
| 2 |
+
*.arrow filter=lfs diff=lfs merge=lfs -text
|
| 3 |
+
*.bin filter=lfs diff=lfs merge=lfs -text
|
| 4 |
+
*.bz2 filter=lfs diff=lfs merge=lfs -text
|
| 5 |
+
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 6 |
+
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
+
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 8 |
+
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 9 |
+
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 10 |
+
*.model filter=lfs diff=lfs merge=lfs -text
|
| 11 |
+
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 12 |
+
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 13 |
+
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 14 |
+
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 15 |
+
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 16 |
+
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 17 |
+
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 18 |
+
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 19 |
+
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 20 |
+
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 21 |
+
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 22 |
+
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 23 |
+
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 24 |
+
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 25 |
+
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 26 |
+
*.zstandard filter=lfs diff=lfs merge=lfs -text
|
| 27 |
+
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 28 |
+
ada_learn_malben.pkl filter=lfs diff=lfs merge=lfs -text
|
| 29 |
+
ada_learn_skin_norm2000.pkl filter=lfs diff=lfs merge=lfs -text
|
mel1.jpg
ADDED
|
nevi1.jpg
ADDED
|
requirements.txt
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
fastai>=2.6.3
|
| 2 |
+
pandas>=1.4.1
|
scc1.jpg
ADDED
|
unk1.jpg
ADDED
|
vl1.jpg
ADDED
|