Spaces:
Runtime error
Runtime error
anonymous
commited on
Commit
•
a2dba58
1
Parent(s):
9ac2c3a
first commit without models
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- README.md +65 -5
- app.py +192 -0
- auxiliary/notebooks_and_reporting/generate_figures.py +175 -0
- auxiliary/notebooks_and_reporting/print_table_results.py +0 -0
- auxiliary/notebooks_and_reporting/print_tests_shared_weights.py +222 -0
- auxiliary/notebooks_and_reporting/results_per_timestep.pdf +0 -0
- auxiliary/notebooks_and_reporting/results_per_timestep_dice.pdf +0 -0
- auxiliary/notebooks_and_reporting/results_per_timestep_prec_recall.pdf +0 -0
- auxiliary/notebooks_and_reporting/results_shared_weights.pdf +0 -0
- auxiliary/notebooks_and_reporting/visualisations.pdf +0 -0
- auxiliary/notebooks_and_reporting/visualisations.py +162 -0
- auxiliary/notebooks_and_reporting/visualisations2.pdf +0 -0
- auxiliary/postprocessing/run_tests.py +162 -0
- auxiliary/postprocessing/testing_shared_weights.py +145 -0
- auxiliary/preprocessing/CXR14_preprocessing_separate_data.py +31 -0
- auxiliary/preprocessing/JSRT_preprocessing_separate_data.py +26 -0
- config.py +84 -0
- data/JSRT_test_split.csv +26 -0
- data/JSRT_train_split.csv +198 -0
- data/JSRT_val_split.csv +26 -0
- data/correspondence_with_chestXray8.csv +101 -0
- data/test_split.csv +0 -0
- data/train_split.csv +0 -0
- data/val_split.csv +0 -0
- dataloaders/CXR14.py +74 -0
- dataloaders/JSRT.py +94 -0
- dataloaders/Montgomery.py +61 -0
- dataloaders/NIH.py +50 -0
- img_examples/00015548_000.png +0 -0
- img_examples/00016568_041.png +0 -0
- img_examples/NIH_0006.png +0 -0
- img_examples/NIH_0012.png +0 -0
- img_examples/NIH_0014.png +0 -0
- img_examples/NIH_0019.png +0 -0
- img_examples/NIH_0024.png +0 -0
- img_examples/NIH_0035.png +0 -0
- img_examples/NIH_0051.png +0 -0
- img_examples/NIH_0055.png +0 -0
- img_examples/NIH_0076.png +0 -0
- img_examples/NIH_0094.png +0 -0
- img_examples/TEDM-model-visualisation.png +0 -0
- models/datasetDM_model.py +88 -0
- models/diffusion_model.py +301 -0
- models/global_local_cl.py +111 -0
- models/unet_model.py +375 -0
- requirements.txt +16 -0
- train.py +56 -0
- trainers/datasetDM_per_step.py +115 -0
- trainers/finetune_glob_cl.py +172 -0
- trainers/finetune_glob_loc_cl.py +172 -0
README.md
CHANGED
|
@@ -1,8 +1,8 @@
|
|
| 1 |
---
|
| 2 |
-
title: TEDM
|
| 3 |
-
emoji:
|
| 4 |
-
colorFrom:
|
| 5 |
-
colorTo:
|
| 6 |
sdk: gradio
|
| 7 |
sdk_version: 3.35.2
|
| 8 |
app_file: app.py
|
|
@@ -10,4 +10,64 @@ pinned: false
|
|
| 10 |
license: mit
|
| 11 |
---
|
| 12 |
|
| 13 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
---
|
| 2 |
+
title: TEDM
|
| 3 |
+
emoji: 🐨
|
| 4 |
+
colorFrom: purple
|
| 5 |
+
colorTo: yellow
|
| 6 |
sdk: gradio
|
| 7 |
sdk_version: 3.35.2
|
| 8 |
app_file: app.py
|
|
|
|
| 10 |
license: mit
|
| 11 |
---
|
| 12 |
|
| 13 |
+
# Timestep ensembling diffusion models for semi-supervised image segmentation
|
| 14 |
+
|
| 15 |
+
Results
|
| 16 |
+
|
| 17 |
+
| Training data size | 1 (1\%) | 3 (2\%) | 6 (3\%) | 12 (96\%) | 197 (100\%) |
|
| 18 |
+
|:------------------|:----------------------------------------------:|:-----------------------:|:-----------------------:|:-----------------------:|:-----------------------:|
|
| 19 |
+
| |JSRT (labelled in-domain) |
|
| 20 |
+
| Baseline | 84.4 $\pm$ 5.4 | 91.7 $\pm$ 3.7 | 93.3 $\pm$ 2.9 | 95.3 $\pm$ 2.3 | 97.3 $\pm$ 1.2 |
|
| 21 |
+
| LEDM | 90.8 $\pm$ 3.5 | 94.1 $\pm$ 1.6 | 95.5 $\pm$ 1.4 | 96.4 $\pm$ 1.4 | 97.0 $\pm$ 1.3 |
|
| 22 |
+
| LEDMe | **93.7 $\pm$ 2.6** | **95.5 $\pm$ 1.5** | **96.7 $\pm$ 1.5** | **97.0 $\pm$ 1.1** | **97.6 $\pm$ 1.2** |
|
| 23 |
+
| Ours | **93.1 $\pm$ 3.4** | 94.8 $\pm$ 1.4 | 95.8 $\pm$ 1.2 | 96.6 $\pm$ 1.1 | 97.3 $\pm$ 1.2 |
|
| 24 |
+
| |NIH (unlabelled in-domain) |
|
| 25 |
+
| Baseline | 68.5 $\pm$ 12.8 | 71.2 $\pm$ 15.1 | 71.4 $\pm$ 15.9 | 77.8 $\pm$ 14.0 | 81.5 $\pm$ 12.7 |
|
| 26 |
+
| LEDM | 63.3 $\pm$ 12.2 | 78.0 $\pm$ 10.1 | 81.2 $\pm$ 9.3 | 85.9 $\pm$ 7.4 | 88.9 $\pm$ 5.9 |
|
| 27 |
+
| LEDMe | 70.3 $\pm$ 11.4 | 78.3 $\pm$ 9.8 | 83.0 $\pm$ 8.6 | 84.4 $\pm$ 8.1 | 90.1 $\pm$ 5.3 |
|
| 28 |
+
| Ours | **80.3 $\pm$ 9.0** | **86.4 $\pm$ 6.2** | **89.2 $\pm$ 5.5** | **91.3 $\pm$ 4.1** | **92.9 $\pm$ 3.2** |
|
| 29 |
+
| | Montgomery (out-of-domain) |
|
| 30 |
+
| Baseline | 77.1 $\pm$ 12.0 | 83.0 $\pm$ 12.2 | 80.9 $\pm$ 14.7 | 83.8 $\pm$ 14.9 | 94.1 $\pm$ 6.6 |
|
| 31 |
+
| LEDM | 79.3 $\pm$ 8.1 | 85.9 $\pm$ 7.4 | 89.4 $\pm$ 6.7 | 92.3 $\pm$ 7.2 | 94.4 $\pm$ 7.2 |
|
| 32 |
+
| LEDMe | 80.7 $\pm$ 6.6 | 86.3 $\pm$ 6.5 | 89.5 $\pm$ 5.9 | 91.2 $\pm$ 5.6 | **95.3 $\pm$ 4.0** |
|
| 33 |
+
| Ours | **90.5 $\pm$ 5.3** | **91.4 $\pm$ 6.1** | **93.3 $\pm$ 6.0** | **94.6 $\pm$ 6.0** | 95.1 $\pm$ 6.9 |
|
| 34 |
+
|
| 35 |
+
## Training
|
| 36 |
+
|
| 37 |
+
- training the backbone
|
| 38 |
+
|
| 39 |
+
```python train.py --dataset CXR14 --data_dir <PATH TO CXR14 DATASET>```
|
| 40 |
+
|
| 41 |
+
- our method
|
| 42 |
+
|
| 43 |
+
```python train.py --experiment TEDM --data_dir <PATH TO JSRT DATASET> --n_labelled_images <TRAINING SET SIZE>```
|
| 44 |
+
|
| 45 |
+
- LEDM method
|
| 46 |
+
|
| 47 |
+
```python train.py --experiment LEDM --data_dir <PATH TO JSRT DATASET> --n_labelled_images <TRAINING SET SIZE>```
|
| 48 |
+
|
| 49 |
+
- LEDMe method
|
| 50 |
+
|
| 51 |
+
```python train.py --experiment LEDMe --data_dir <PATH TO JSRT DATASET> --n_labelled_images <TRAINING SET SIZE>```
|
| 52 |
+
|
| 53 |
+
- baseline method
|
| 54 |
+
|
| 55 |
+
```python train.py --experiment JSRT_baseline --data_dir <PATH TO JSRT DATASET> --n_labelled_images <TRAINING SET SIZE>```
|
| 56 |
+
|
| 57 |
+
## Testing
|
| 58 |
+
|
| 59 |
+
- update
|
| 60 |
+
- `DATADIR` in paths `dataloaders/JSRT.py`, `dataloaders/NIH.py` and `dataloaders/Montgomery.py`
|
| 61 |
+
- `NIHPATH`, `NIHFILE`, `MONPATH` and `MONFILE` in paths `auxiliary/postprocessing/run_tests.py` and `auxiliary/postprocessing/testing_shared_weights.py`
|
| 62 |
+
|
| 63 |
+
- for baseline and LEDM methods, run
|
| 64 |
+
|
| 65 |
+
```python auxiliary/postprocessing/run_tests.py --experiment <PATH TO LOG FOLDER>```
|
| 66 |
+
|
| 67 |
+
- for our method, run
|
| 68 |
+
|
| 69 |
+
```python auxiliary/postprocessing/testing_shared_weights.py --experiment <PATH TO LOG FOLDER>```
|
| 70 |
+
|
| 71 |
+
## Figures and reporting
|
| 72 |
+
|
| 73 |
+
VS Code notebooks can be found in `auxiliary/notebooks_and_reporting`.
|
app.py
ADDED
|
@@ -0,0 +1,192 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import gradio as gr
|
| 3 |
+
from PIL import Image
|
| 4 |
+
import torch
|
| 5 |
+
from torch import nn
|
| 6 |
+
from einops.layers.torch import Rearrange
|
| 7 |
+
from torchvision import transforms
|
| 8 |
+
from models.unet_model import Unet
|
| 9 |
+
from models.datasetDM_model import DatasetDM
|
| 10 |
+
from skimage import measure, segmentation
|
| 11 |
+
import cv2
|
| 12 |
+
from tqdm import tqdm
|
| 13 |
+
from einops import repeat
|
| 14 |
+
|
| 15 |
+
img_size = 128
|
| 16 |
+
font = cv2.FONT_HERSHEY_SIMPLEX
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
## %%
|
| 20 |
+
def load_img(img_file):
|
| 21 |
+
# assert type of input
|
| 22 |
+
if isinstance(img_file, np.ndarray):
|
| 23 |
+
img = torch.Tensor(img_file).float()
|
| 24 |
+
# make sure img is between 0 and 1
|
| 25 |
+
if img.max() > 1:
|
| 26 |
+
img /= 255
|
| 27 |
+
# resize
|
| 28 |
+
img = transforms.Resize(img_size)(img)
|
| 29 |
+
elif isinstance(img_file, str):
|
| 30 |
+
img = Image.open(img_file).convert('L').resize((img_size, img_size))
|
| 31 |
+
img = transforms.ToTensor()(img).float()
|
| 32 |
+
elif isinstance(img_file, Image.Image):
|
| 33 |
+
img = img_file.convert('L').resize((img_size, img_size))
|
| 34 |
+
img = transforms.ToTensor()(img).float()
|
| 35 |
+
else:
|
| 36 |
+
raise TypeError("Input must be a numpy array, PIL image, or filepath")
|
| 37 |
+
if len(img.shape) == 2:
|
| 38 |
+
img = img[None, None]
|
| 39 |
+
elif len(img.shape) == 3:
|
| 40 |
+
img = img[None]
|
| 41 |
+
else:
|
| 42 |
+
raise ValueError("Input must be a 2D or 3D array")
|
| 43 |
+
return img
|
| 44 |
+
|
| 45 |
+
def predict_baseline(img, checkpoint_path):
|
| 46 |
+
checkpoint = torch.load(checkpoint_path, map_location=torch.device("cpu"))
|
| 47 |
+
config = checkpoint["config"]
|
| 48 |
+
baseline = Unet(**vars(config))
|
| 49 |
+
baseline.load_state_dict(checkpoint["model_state_dict"])
|
| 50 |
+
baseline.eval()
|
| 51 |
+
return (torch.sigmoid(baseline(img)) > .5).float().squeeze().numpy()
|
| 52 |
+
|
| 53 |
+
def predict_LEDM(img, checkpoint_path):
|
| 54 |
+
checkpoint = torch.load(checkpoint_path, map_location=torch.device("cpu"))
|
| 55 |
+
config = checkpoint["config"]
|
| 56 |
+
config.verbose = False
|
| 57 |
+
LEDM = DatasetDM(config)
|
| 58 |
+
LEDM.load_state_dict(checkpoint["model_state_dict"])
|
| 59 |
+
LEDM.eval()
|
| 60 |
+
return (torch.sigmoid(LEDM(img)) > .5).float().squeeze().numpy()
|
| 61 |
+
|
| 62 |
+
def predict_TEDM(img, checkpoint_path):
|
| 63 |
+
checkpoint = torch.load(checkpoint_path, map_location=torch.device("cpu"))
|
| 64 |
+
config = checkpoint["config"]
|
| 65 |
+
config.verbose = False
|
| 66 |
+
TEDM = DatasetDM(config)
|
| 67 |
+
TEDM.classifier = nn.Sequential(
|
| 68 |
+
Rearrange('b (step act) h w -> (b step) act h w', step=len(TEDM.steps)),
|
| 69 |
+
nn.Conv2d(960, 128, 1),
|
| 70 |
+
nn.ReLU(),
|
| 71 |
+
nn.BatchNorm2d(128),
|
| 72 |
+
nn.Conv2d(128, 32, 1),
|
| 73 |
+
nn.ReLU(),
|
| 74 |
+
nn.BatchNorm2d(32),
|
| 75 |
+
nn.Conv2d(32, 1, config.out_channels)
|
| 76 |
+
)
|
| 77 |
+
TEDM.load_state_dict(checkpoint["model_state_dict"])
|
| 78 |
+
TEDM.eval()
|
| 79 |
+
return (torch.sigmoid(TEDM(img)).mean(0) > .5).float().squeeze().numpy()
|
| 80 |
+
|
| 81 |
+
predictors = {'Baseline': predict_baseline,
|
| 82 |
+
'Global CL': predict_baseline,
|
| 83 |
+
'Global & Local CL': predict_baseline,
|
| 84 |
+
'LEDM': predict_LEDM,
|
| 85 |
+
'LEDMe': predict_LEDM,
|
| 86 |
+
'TEDM': predict_TEDM}
|
| 87 |
+
model_folders = {
|
| 88 |
+
'Baseline': 'baseline',
|
| 89 |
+
'Global CL': 'global_finetune',
|
| 90 |
+
'Global & Local CL': 'glob_loc_finetune',
|
| 91 |
+
'LEDM': 'LEDM',
|
| 92 |
+
'LEDMe': 'LEDMe',
|
| 93 |
+
'TEDM': 'TEDM'
|
| 94 |
+
}
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
def postprocess(pred, img):
|
| 98 |
+
all_labels = measure.label(pred, background=0)
|
| 99 |
+
_, cn = np.unique(all_labels, return_counts=True)
|
| 100 |
+
# find the two largest connected components that are not the background
|
| 101 |
+
if len(cn) >= 3:
|
| 102 |
+
lungs = np.argsort(cn[1:])[-2:] + 1
|
| 103 |
+
all_labels[(all_labels!=lungs[0]) & (all_labels!=lungs[1])] = 0
|
| 104 |
+
all_labels[(all_labels==lungs[0]) | (all_labels==lungs[1])] = 1
|
| 105 |
+
# put all_labels into a cv2 object
|
| 106 |
+
if len(cn) > 1:
|
| 107 |
+
img = segmentation.mark_boundaries(img, all_labels, color=(1,0,0), mode='outer', background_label=0)
|
| 108 |
+
else:
|
| 109 |
+
img = repeat(img, 'h w -> h w c', c=3)
|
| 110 |
+
return img
|
| 111 |
+
|
| 112 |
+
|
| 113 |
+
|
| 114 |
+
def predict(img_file, models:list, training_sizes:list, seg_img=False, progress=gr.Progress()):
|
| 115 |
+
max_progress = len(models) * len(training_sizes)
|
| 116 |
+
n_progress = 0
|
| 117 |
+
progress((n_progress, max_progress), desc="Starting")
|
| 118 |
+
img = load_img(img_file)
|
| 119 |
+
print(img.shape)
|
| 120 |
+
preds = []
|
| 121 |
+
# sorting models so that they show as baseline - LEDM - LEDMe - TEDM
|
| 122 |
+
models = sorted(models, key=lambda x: 0 if x == 'Baseline' else 1 if x == 'Global CL' else 2 if x == 'Global & Local CL' else 3 if x == 'LEDM' else 4 if x == 'LEDMe' else 5)
|
| 123 |
+
|
| 124 |
+
for model in models:
|
| 125 |
+
print(model)
|
| 126 |
+
model_preds = []
|
| 127 |
+
for training_size in sorted(training_sizes):
|
| 128 |
+
#if n_progress < max_progress:
|
| 129 |
+
progress((n_progress, max_progress) , desc=f"Predicting {model} {training_size}")
|
| 130 |
+
n_progress += 1
|
| 131 |
+
print(training_size)
|
| 132 |
+
out = predictors[model](img, f"logs/{model_folders[model]}/{training_size}/best_model.pt")
|
| 133 |
+
writing_colour = (.5,.5,.5)
|
| 134 |
+
if seg_img:
|
| 135 |
+
out = postprocess(out, img.squeeze().numpy())
|
| 136 |
+
writing_colour = (1,1,1)
|
| 137 |
+
out = cv2.putText(np.array(out),f"{model} {training_size}",(5,125), font, .5, writing_colour,1, cv2.LINE_AA)
|
| 138 |
+
#ImageDraw.Draw(out).text((0,128), f"{model} {training_size}", fill=(255,0,0))
|
| 139 |
+
model_preds.append(np.asarray(out))
|
| 140 |
+
preds.append(np.concatenate(model_preds, axis=1))
|
| 141 |
+
prediction = np.concatenate(preds, axis=0)
|
| 142 |
+
if (prediction.shape[1] <=128*2):
|
| 143 |
+
pad = (330 - prediction.shape[1])//2
|
| 144 |
+
if len(prediction.shape) == 2:
|
| 145 |
+
prediction = np.pad(prediction, ((0,0), (pad, pad)), 'constant', constant_values=1)
|
| 146 |
+
else:
|
| 147 |
+
prediction = np.pad(prediction, ((0,0), (pad, pad), (0,0)), 'constant', constant_values=1)
|
| 148 |
+
return prediction
|
| 149 |
+
|
| 150 |
+
|
| 151 |
+
## %%
|
| 152 |
+
input = gr.Image( label="Chest X-ray", shape=(img_size, img_size), type="pil")
|
| 153 |
+
output = gr.Image(label="Segmentation", shape=(img_size, img_size))
|
| 154 |
+
## %%
|
| 155 |
+
demo = gr.Interface(
|
| 156 |
+
fn=predict,
|
| 157 |
+
inputs=[input,
|
| 158 |
+
gr.CheckboxGroup(["Baseline", "Global CL", "Global & Local CL", "LEDM", "LEDMe", "TEDM"], label="Model", value=["Baseline", "LEDM", "LEDMe", "TEDM"]),
|
| 159 |
+
gr.CheckboxGroup([1,3,6,12,197], label="Training size", value=[1,3,6,12,197]),
|
| 160 |
+
gr.Checkbox(label="Show masked image (otherwise show binary segmentation)", value=True),],
|
| 161 |
+
|
| 162 |
+
outputs=output,
|
| 163 |
+
examples = [
|
| 164 |
+
['img_examples/NIH_0006.png'],
|
| 165 |
+
['img_examples/NIH_0076.png'],
|
| 166 |
+
["img_examples/00016568_041.png"],
|
| 167 |
+
['img_examples/NIH_0024.png'],
|
| 168 |
+
['img_examples/00015548_000.png'],
|
| 169 |
+
['img_examples/NIH_0019.png'],
|
| 170 |
+
['img_examples/NIH_0094.png'],
|
| 171 |
+
['img_examples/NIH_0051.png'],
|
| 172 |
+
['img_examples/NIH_0012.png'],
|
| 173 |
+
['img_examples/NIH_0014.png'],
|
| 174 |
+
['img_examples/NIH_0055.png'],
|
| 175 |
+
['img_examples/NIH_0035.png'],
|
| 176 |
+
],
|
| 177 |
+
title="Chest X-ray Segmentation with TEDM.",
|
| 178 |
+
description="""<img src="file/img_examples/TEDM-model-visualisation.png"
|
| 179 |
+
alt="Markdown Monster icon"
|
| 180 |
+
style="margin-right: 10px;" />"""+
|
| 181 |
+
"\nMedical image segmentation is a challenging task, made more difficult by many datasets' limited size and annotations. Denoising diffusion probabilistic models (DDPM) have recently shown promise in modelling " +
|
| 182 |
+
"the distribution of natural images and were successfully applied to various medical imaging tasks. This work focuses on semi-supervised image segmentation using diffusion models, particularly addressing domain " +
|
| 183 |
+
"generalisation. Firstly, we demonstrate that smaller diffusion steps generate latent representations that are more robust for downstream tasks than larger steps. Secondly, we use this insight to propose an improved " +
|
| 184 |
+
"esembling scheme that leverages information-dense small steps and the regularising effect of larger steps to generate predictions. Our model shows significantly better performance in domain-shifted settings while " +
|
| 185 |
+
"retaining competitive performance in-domain. Overall, this work highlights the potential of DDPMs for semi-supervised medical image segmentation and provides insights into optimising their performance under domain shift."+
|
| 186 |
+
"\n\n\n When choosing 'Show masked image', we post-process the segmentation by choosing up to two largest connected components and drawing their outline. "+
|
| 187 |
+
"\nNote that each model takes 10-35 seconds to run on CPU. Choosing all models and all training sizes will take some time. "+
|
| 188 |
+
"We noticed that gradio sometimes fails on the first try. If it doesn't work, try again.",
|
| 189 |
+
cache_examples=False,
|
| 190 |
+
)
|
| 191 |
+
demo.queue().launch(debug=True)
|
| 192 |
+
#demo.queue().launch(share=True)
|
auxiliary/notebooks_and_reporting/generate_figures.py
ADDED
|
@@ -0,0 +1,175 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# %%
|
| 2 |
+
import numpy as np
|
| 3 |
+
import torch
|
| 4 |
+
from pathlib import Path
|
| 5 |
+
import os
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import seaborn as sns
|
| 8 |
+
import matplotlib.pyplot as plt
|
| 9 |
+
HEAD = Path(os.getcwd()).parent.parent
|
| 10 |
+
|
| 11 |
+
if __name__=="__main__":
|
| 12 |
+
# load baseline and LEDM data
|
| 13 |
+
metrics = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[]}
|
| 14 |
+
files_needed = ["JSRT_val_predictions.pt", "JSRT_test_predictions.pt", "NIH_predictions.pt", "Montgomery_predictions.pt",]
|
| 15 |
+
head = HEAD / 'logs'
|
| 16 |
+
for exp in ['baseline', 'LEDM']:
|
| 17 |
+
for datasize in [1, 3, 6, 12, 24, 49, 98, 197]:
|
| 18 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize)))) == 0:
|
| 19 |
+
print(f"Experiment {exp} {datasize}")
|
| 20 |
+
output = torch.load(head / exp / str(datasize) / "JSRT_val_predictions.pt")
|
| 21 |
+
print(f"{output['dice'].mean()}\t{output['dice'].std()}")
|
| 22 |
+
for file in files_needed[1:]:
|
| 23 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 24 |
+
metrics_datasize = 197 if datasize == "None" else int(datasize)
|
| 25 |
+
metrics["dice"].append(output["dice"].numpy())
|
| 26 |
+
metrics["precision"].append(output["precision"].numpy())
|
| 27 |
+
metrics["recall"].append(output["recall"].numpy())
|
| 28 |
+
metrics["exp"].append(np.array([exp] * len(output["dice"])))
|
| 29 |
+
metrics["datasize"].append(np.array([int(datasize)] * len(output["dice"])))
|
| 30 |
+
metrics["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 31 |
+
else:
|
| 32 |
+
print(f"Experiment {exp} is missing files")
|
| 33 |
+
|
| 34 |
+
for key in metrics:
|
| 35 |
+
metrics[key] = np.concatenate([el.squeeze() for el in metrics[key]])
|
| 36 |
+
df = pd.DataFrame(metrics)
|
| 37 |
+
df.head()
|
| 38 |
+
|
| 39 |
+
|
| 40 |
+
# %% Load step data
|
| 41 |
+
metrics2 = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[], 'timestep':[]}
|
| 42 |
+
for timestep in [1, 10, 25, 50, 500, 950]:
|
| 43 |
+
exp = f"Step_{timestep}"
|
| 44 |
+
for datasize in [197, 98, 49, 24, 12, 6, 3, 1]:
|
| 45 |
+
if os.path.isdir(head / exp / str(datasize)):
|
| 46 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize)))) == 0:
|
| 47 |
+
print(f"Experiment {datasize} {timestep}")
|
| 48 |
+
output = torch.load(head / exp / str(datasize)/ "JSRT_val_predictions.pt")
|
| 49 |
+
print(f"{output['dice'].mean()}\t{output['dice'].std()}")
|
| 50 |
+
for file in files_needed[1:]:
|
| 51 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 52 |
+
metrics_datasize = datasize if datasize is not None else 197
|
| 53 |
+
metrics2["dice"].append(output["dice"].numpy())
|
| 54 |
+
metrics2["precision"].append(output["precision"].numpy())
|
| 55 |
+
metrics2["recall"].append(output["recall"].numpy())
|
| 56 |
+
metrics2["exp"].append(np.array([exp] * len(output["dice"])))
|
| 57 |
+
metrics2["datasize"].append(np.array([metrics_datasize] * len(output["dice"])))
|
| 58 |
+
metrics2["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 59 |
+
metrics2["timestep"].append(np.array([timestep] * len(output["dice"])))
|
| 60 |
+
else:
|
| 61 |
+
print(f"Experiment {datasize} is missing files")
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
for key in metrics2:
|
| 65 |
+
metrics2[key] = np.concatenate(metrics2[key]).squeeze()
|
| 66 |
+
print(key, metrics2[key].shape)
|
| 67 |
+
df2 = pd.DataFrame(metrics2)
|
| 68 |
+
|
| 69 |
+
# %% figure with line for baseline and datasetDM and boxplots for the rest
|
| 70 |
+
# separating dice from precision and recall
|
| 71 |
+
font = 16
|
| 72 |
+
x = [1, 1, 3, 3, 6, 6, 12, 12, 24, 24, 49, 49, 197, 197]
|
| 73 |
+
plot_x = np.concatenate([np.array([-.4, .4]) + i for i in range(len(x)//2)]).flatten()
|
| 74 |
+
fig, axs = plt.subplots(3, 1, figsize=[12, 10])
|
| 75 |
+
sns.set_style("whitegrid")
|
| 76 |
+
m = 'dice'
|
| 77 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 78 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'baseline') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 79 |
+
ys_std = np.quantile(ys, (.25, .75), axis=1, )
|
| 80 |
+
axs[i ].fill_between(plot_x, ys_std[0], ys_std[1], alpha=.2, zorder=0, color='C6')
|
| 81 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'LEDM') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 82 |
+
ys_std = np.quantile(ys, (.25, .75), axis=1, )
|
| 83 |
+
axs[i ].fill_between(plot_x, ys_std[0], ys_std[1], alpha=.2, zorder=0, color='C8')
|
| 84 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'baseline') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 85 |
+
ys_mean = np.quantile(ys, .5, axis=1)
|
| 86 |
+
axs[i ].plot(plot_x, ys_mean, label="baseline", c='C6', zorder=0)
|
| 87 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'LEDM') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 88 |
+
ys_mean = np.quantile(ys, .5, axis=1)
|
| 89 |
+
axs[i ].plot(plot_x, ys_mean, label="LEDM" , c='C7', zorder=0)
|
| 90 |
+
|
| 91 |
+
|
| 92 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 93 |
+
temp_df = df2[(df2.dataset == dataset) & (df2.datasize != 98)]
|
| 94 |
+
out = sns.boxplot(data=temp_df, x="datasize", y=m, hue="timestep", ax=axs[i ], showfliers=False, saturation=1,)
|
| 95 |
+
axs[i ].set_title(f"{dataset}", fontsize=font)
|
| 96 |
+
axs[i ].set_xlabel("" )
|
| 97 |
+
y_min, _ = axs[i ].get_ylim()
|
| 98 |
+
axs[i ].set_ylim(y_min, 1)
|
| 99 |
+
h, l = axs[i].get_legend_handles_labels()
|
| 100 |
+
axs[i].get_legend().remove()
|
| 101 |
+
axs[i].set_ylabel("Dice", fontsize=font)
|
| 102 |
+
sns.despine(ax=axs[0 ], offset=10, trim=True, bottom=True)
|
| 103 |
+
sns.despine(ax=axs[1 ], offset=10, trim=True, bottom=True)
|
| 104 |
+
sns.despine(ax=axs[2 ], offset=10, trim=True)
|
| 105 |
+
axs[0].set_xticks([])
|
| 106 |
+
axs[1].set_xticks([])
|
| 107 |
+
axs[-1 ].set_xlabel("Training dataset size", fontsize=font)
|
| 108 |
+
# Shrink current axis by 20%
|
| 109 |
+
for i, ax in enumerate(axs):
|
| 110 |
+
box = ax.get_position()
|
| 111 |
+
ax.tick_params(axis='both', labelsize=font)
|
| 112 |
+
ax.set_position([box.x0, box.y0, box.width , box.height])
|
| 113 |
+
|
| 114 |
+
# Put a legend to the right of the current axis
|
| 115 |
+
|
| 116 |
+
fig.legend(h, ['baseline', 'LEDM'] + ['step ' + _l for _l in l[2:]], title="", ncol=4,
|
| 117 |
+
loc='center left', bbox_to_anchor=(0.2, -0.03), fontsize=font)
|
| 118 |
+
plt.tight_layout()
|
| 119 |
+
#plt.savefig("results_per_timestep.png")
|
| 120 |
+
plt.savefig("results_per_timestep_dice.pdf", bbox_inches='tight')
|
| 121 |
+
plt.show()
|
| 122 |
+
# %%
|
| 123 |
+
x = [1, 1, 3, 3, 6, 6, 12, 12, 24, 24, 49, 49, 197, 197]
|
| 124 |
+
plot_x = np.concatenate([np.array([-.4, .4]) + i for i in range(len(x)//2)]).flatten()
|
| 125 |
+
fig, axs = plt.subplots(3, 2, figsize=[15, 15])
|
| 126 |
+
sns.set_style("whitegrid")
|
| 127 |
+
for j, m in enumerate(["precision", "recall"]):
|
| 128 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 129 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'baseline') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 130 |
+
ys_std = np.quantile(ys, (.25, .75), axis=1, )
|
| 131 |
+
axs[i, j].fill_between(plot_x, ys_std[0], ys_std[1], alpha=.2, zorder=0, color='C6')
|
| 132 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'LEDM') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 133 |
+
ys_std = np.quantile(ys, (.25, .75), axis=1, )
|
| 134 |
+
axs[i, j].fill_between(plot_x, ys_std[0], ys_std[1], alpha=.2, zorder=0, color='C8')
|
| 135 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'baseline') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 136 |
+
ys_mean = np.quantile(ys, .5, axis=1)
|
| 137 |
+
axs[i, j].plot(plot_x, ys_mean, label="baseline", c='C6', zorder=0)
|
| 138 |
+
ys = np.stack([df.loc[(df.dataset == dataset)& (df.exp == 'LEDM') & (df.datasize == _x), m].to_numpy() for _x in x])
|
| 139 |
+
ys_mean = np.quantile(ys, .5, axis=1)
|
| 140 |
+
axs[i, j].plot(plot_x, ys_mean, label="LEDM" , c='C7', zorder=0)
|
| 141 |
+
|
| 142 |
+
|
| 143 |
+
##
|
| 144 |
+
temp_df = df2[(df2.dataset == dataset) & (df2.datasize != 98)]
|
| 145 |
+
out = sns.boxplot(data=temp_df, x="datasize", y=m, hue="timestep", ax=axs[i,j], showfliers=False, saturation=1)
|
| 146 |
+
axs[i,j].set_title(f"{dataset}", fontsize=font)
|
| 147 |
+
y_min, _ = axs[i,j].get_ylim()
|
| 148 |
+
axs[i,j].set_ylim(y_min, 1)
|
| 149 |
+
sns.despine(ax=axs[i,j], offset=10, trim=True)
|
| 150 |
+
h, l = axs[i,j].get_legend_handles_labels()
|
| 151 |
+
axs[i,j].get_legend().remove()
|
| 152 |
+
axs[i, 0].set_ylabel("Precison", fontsize=font)
|
| 153 |
+
axs[i, 1].set_ylabel("Recall", fontsize=font)
|
| 154 |
+
axs[i,j].set_xlabel("")
|
| 155 |
+
|
| 156 |
+
for ax in axs.flatten():
|
| 157 |
+
ax.tick_params(axis='both', labelsize=font)
|
| 158 |
+
for ax in [axs[:, 0], axs[:, 1]]:
|
| 159 |
+
sns.despine(ax=ax[0 ], offset=10, trim=True, bottom=True)
|
| 160 |
+
sns.despine(ax=ax[1 ], offset=10, trim=True, bottom=True)
|
| 161 |
+
sns.despine(ax=ax[2 ], offset=10, trim=True)
|
| 162 |
+
ax[0].set_xticks([])
|
| 163 |
+
ax[1].set_xticks([])
|
| 164 |
+
ax[-1 ].set_xlabel("Training dataset size", fontsize=font)
|
| 165 |
+
# Put a legend to the right of the current axis
|
| 166 |
+
|
| 167 |
+
|
| 168 |
+
fig.legend(h, ['baseline', 'LEDM'] + ['step ' + _l for _l in l[2:]], title="", ncol=4,
|
| 169 |
+
loc='center left', bbox_to_anchor=(0.25, -0.03), fontsize=font)
|
| 170 |
+
plt.tight_layout()
|
| 171 |
+
#plt.savefig("results_per_timestep.png")
|
| 172 |
+
plt.savefig("results_per_timestep_prec_recall.pdf", bbox_inches='tight')
|
| 173 |
+
plt.show()
|
| 174 |
+
|
| 175 |
+
# %%
|
auxiliary/notebooks_and_reporting/print_table_results.py
ADDED
|
File without changes
|
auxiliary/notebooks_and_reporting/print_tests_shared_weights.py
ADDED
|
@@ -0,0 +1,222 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# %%
|
| 2 |
+
import numpy as np
|
| 3 |
+
import torch
|
| 4 |
+
from pathlib import Path
|
| 5 |
+
import os
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import seaborn as sns
|
| 8 |
+
import matplotlib.pyplot as plt
|
| 9 |
+
HEAD = Path(os.getcwd()).parent.parent
|
| 10 |
+
|
| 11 |
+
if __name__=="__main__":
|
| 12 |
+
# load baseline and LEDM data
|
| 13 |
+
metrics = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[]}
|
| 14 |
+
files_needed = ["JSRT_val_predictions.pt", "JSRT_test_predictions.pt", "NIH_predictions.pt", "Montgomery_predictions.pt",]
|
| 15 |
+
head = HEAD / 'logs'
|
| 16 |
+
for exp in ['baseline', 'LEDM']:
|
| 17 |
+
for datasize in [1, 3, 6, 12, 24, 49, 98, 197]:
|
| 18 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize)))) == 0:
|
| 19 |
+
print(f"Experiment {exp} {datasize}")
|
| 20 |
+
output = torch.load(head / exp / str(datasize) / "JSRT_val_predictions.pt")
|
| 21 |
+
print(f"{output['dice'].mean()}\t{output['dice'].std()}")
|
| 22 |
+
for file in files_needed[1:]:
|
| 23 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 24 |
+
metrics_datasize = 197 if datasize == "None" else int(datasize)
|
| 25 |
+
metrics["dice"].append(output["dice"].numpy())
|
| 26 |
+
metrics["precision"].append(output["precision"].numpy())
|
| 27 |
+
metrics["recall"].append(output["recall"].numpy())
|
| 28 |
+
metrics["exp"].append(np.array([exp] * len(output["dice"])))
|
| 29 |
+
metrics["datasize"].append(np.array([int(datasize)] * len(output["dice"])))
|
| 30 |
+
metrics["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 31 |
+
else:
|
| 32 |
+
print(f"Experiment {exp} is missing files")
|
| 33 |
+
|
| 34 |
+
for key in metrics:
|
| 35 |
+
metrics[key] = np.concatenate([el.squeeze() for el in metrics[key]])
|
| 36 |
+
df = pd.DataFrame(metrics)
|
| 37 |
+
df.head()
|
| 38 |
+
|
| 39 |
+
# %% load TEDM data
|
| 40 |
+
metrics3 = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[], }
|
| 41 |
+
exp = "TEDM"
|
| 42 |
+
for datasize in [1, 3, 6, 12, 24, 49, 98, 197]:
|
| 43 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize) ))) == 0:
|
| 44 |
+
print(f"Experiment {datasize}")
|
| 45 |
+
output = torch.load(head / exp / str(datasize)/ "JSRT_val_predictions.pt")
|
| 46 |
+
print(f"{output['dice'].mean()}\t{output['dice'].std()}")
|
| 47 |
+
for file in files_needed[1:]:
|
| 48 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 49 |
+
|
| 50 |
+
metrics_datasize = datasize if datasize is not None else 197
|
| 51 |
+
metrics3["dice"].append(output["dice"].numpy())
|
| 52 |
+
metrics3["precision"].append(output["precision"].numpy())
|
| 53 |
+
metrics3["recall"].append(output["recall"].numpy())
|
| 54 |
+
metrics3["exp"].append(np.array(['TEDM'] * len(output["dice"])))
|
| 55 |
+
metrics3["datasize"].append(np.array([metrics_datasize] * len(output["dice"])))
|
| 56 |
+
metrics3["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 57 |
+
|
| 58 |
+
else:
|
| 59 |
+
print(f"Experiment {datasize} is missing files")
|
| 60 |
+
|
| 61 |
+
for key in metrics3:
|
| 62 |
+
metrics3[key] = np.concatenate(metrics3[key]).squeeze()
|
| 63 |
+
print(key, metrics3[key].shape)
|
| 64 |
+
df3 = pd.DataFrame(metrics3)
|
| 65 |
+
# %% Boxplot of TEDM vs LEDM and baseline
|
| 66 |
+
df4 = pd.concat([df, df3])
|
| 67 |
+
df4.datasize = df4.datasize.astype(int)
|
| 68 |
+
m='dice'
|
| 69 |
+
dataset="JSRT"
|
| 70 |
+
fig, axs = plt.subplots(3, 3, figsize=(20, 20))
|
| 71 |
+
for j, m in enumerate(["dice", "precision", "recall"]):
|
| 72 |
+
#axs[0,j].set_ylim(0.8, 1)
|
| 73 |
+
#axs[0,j].set_ylim(0.6, 1)
|
| 74 |
+
#axs[0,j].set_ylim(0.7, 1)
|
| 75 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 76 |
+
temp_df = df4[(df4.dataset == dataset)]
|
| 77 |
+
#sns.lineplot(data=df[df.dataset == dataset], x="datasize", y=m, hue="exp", ax=axs[i,j])
|
| 78 |
+
sns.boxplot(data=temp_df, x="datasize", y=m, ax=axs[i,j], hue="exp", showfliers=False, saturation=1,
|
| 79 |
+
hue_order=['baseline', 'LEDM', 'TEDM'])
|
| 80 |
+
axs[i,j].set_title(f"{dataset} {m}")
|
| 81 |
+
axs[i,j].set_xlabel("Training dataset size")
|
| 82 |
+
h, l = axs[i,j].get_legend_handles_labels()
|
| 83 |
+
axs[i,j].legend(h, ['Baseline', 'LEDM', 'TEDM (ours)'], title="", loc='lower right')
|
| 84 |
+
plt.tight_layout()
|
| 85 |
+
plt.savefig("results_shared_weights.pdf")
|
| 86 |
+
plt.show()
|
| 87 |
+
# %% Load LEDMe and Step 1
|
| 88 |
+
metrics2 = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[], }
|
| 89 |
+
for exp in ["LEDMe", 'Step_1']:
|
| 90 |
+
for datasize in [1, 3, 6, 12, 24, 49, 98, 197]:
|
| 91 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize) ))) == 0:
|
| 92 |
+
print(f"Experiment {exp} {datasize}")
|
| 93 |
+
output = torch.load(head / exp / str(datasize)/ "JSRT_val_predictions.pt")
|
| 94 |
+
print(f"{output['dice'].mean()}\t{output['dice'].std()}")
|
| 95 |
+
for file in files_needed[1:]:
|
| 96 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 97 |
+
#print(f"{output['dice'].mean()*100:.3}\t{output['dice'].std()*100:.3}\t{output['precision'].mean()*100:.3}\t{output['precision'].std()*100:.3}\t{output['recall'].mean()*100:.3}\t{output['recall'].std()*100:.3}",
|
| 98 |
+
# end="\n\n\n\n")
|
| 99 |
+
metrics_datasize = 197 if datasize == "None" else datasize
|
| 100 |
+
metrics2["dice"].append(output["dice"].numpy())
|
| 101 |
+
metrics2["precision"].append(output["precision"].numpy())
|
| 102 |
+
metrics2["recall"].append(output["recall"].numpy())
|
| 103 |
+
metrics2["exp"].append(np.array([exp] * len(output["dice"])))
|
| 104 |
+
metrics2["datasize"].append(np.array([int(metrics_datasize)] * len(output["dice"])))
|
| 105 |
+
metrics2["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 106 |
+
else:
|
| 107 |
+
print(f"Experiment {exp} is missing files")
|
| 108 |
+
|
| 109 |
+
for key in metrics2:
|
| 110 |
+
metrics2[key] = np.concatenate(metrics2[key]).squeeze()
|
| 111 |
+
print(key, metrics2[key].shape)
|
| 112 |
+
df2 = pd.DataFrame(metrics2)
|
| 113 |
+
# %% Boxplot of TEDM vs LEDM and baseline, Step 1 and LEDMe
|
| 114 |
+
df4 = pd.concat([df, df3, df2])
|
| 115 |
+
df4.datasize = df4.datasize.astype(int)
|
| 116 |
+
|
| 117 |
+
|
| 118 |
+
m='dice'
|
| 119 |
+
dataset="JSRT"
|
| 120 |
+
fig, axs = plt.subplots(3, 3, figsize=(20, 20))
|
| 121 |
+
for j, m in enumerate(["dice", "precision", "recall"]):
|
| 122 |
+
|
| 123 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 124 |
+
temp_df = df4[(df4.dataset == dataset)]
|
| 125 |
+
#sns.lineplot(data=df[df.dataset == dataset], x="datasize", y=m, hue="exp", ax=axs[i,j])
|
| 126 |
+
sns.boxplot(data=temp_df, x="datasize", y=m, ax=axs[i,j], hue="exp", showfliers=False, saturation=1,
|
| 127 |
+
hue_order=['baseline', 'LEDM', 'Step_1', 'LEDMe', 'TEDM', ])
|
| 128 |
+
axs[i,j].set_title(f"{dataset} {m}")
|
| 129 |
+
axs[i,j].set_xlabel("Training dataset size")
|
| 130 |
+
h, l = axs[i,j].get_legend_handles_labels()
|
| 131 |
+
axs[i,j].legend(h, ['Baseline', 'LEDM', 'Step 1', 'LEDMe', 'TEDM'], title="", loc='lower right')
|
| 132 |
+
plt.tight_layout()
|
| 133 |
+
plt.savefig("results_shared_weights.pdf")
|
| 134 |
+
plt.show()
|
| 135 |
+
# %% Load TEDM ablation studies
|
| 136 |
+
metrics4 = {"dice": [], "precision": [], "recall": [], "exp": [], "datasize": [], "dataset":[], }
|
| 137 |
+
exp = "TEDM"
|
| 138 |
+
for datasize in [1, 3, 6, 12, 24, 49, 98, 197]:
|
| 139 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize)))) == 0:
|
| 140 |
+
print(f"Experiment {datasize} ")
|
| 141 |
+
for step in [1,10,25]:
|
| 142 |
+
for file in files_needed[1:]:
|
| 143 |
+
output = torch.load(head / exp / str(datasize) / file.replace("predictions", f"timestep{step}_predictions"))
|
| 144 |
+
#print(f"{output['dice'].mean()*100:.3}\t{output['dice'].std()*100:.3}\t{output['precision'].mean()*100:.3}\t{output['precision'].std()*100:.3}\t{output['recall'].mean()*100:.3}\t{output['recall'].std()*100:.3}",
|
| 145 |
+
# end="\n\n\n\n")
|
| 146 |
+
metrics_datasize = datasize if datasize is not None else 197
|
| 147 |
+
metrics4["dice"].append(output["dice"].numpy())
|
| 148 |
+
metrics4["precision"].append(output["precision"].numpy())
|
| 149 |
+
metrics4["recall"].append(output["recall"].numpy())
|
| 150 |
+
metrics4["exp"].append(np.array([f'Step {step} (MLP)'] * len(output["dice"])))
|
| 151 |
+
metrics4["datasize"].append(np.array([metrics_datasize] * len(output["dice"])))
|
| 152 |
+
metrics4["dataset"].append(np.array([file.split("_")[0]]*len(output["dice"])))
|
| 153 |
+
#metrics3["timestep"].append(np.array(timestep * len(output["dice"])))
|
| 154 |
+
else:
|
| 155 |
+
print(f"Experiment {datasize} is missing files")
|
| 156 |
+
|
| 157 |
+
for key in metrics3:
|
| 158 |
+
metrics4[key] = np.concatenate(metrics4[key]).squeeze()
|
| 159 |
+
print(key, metrics4[key].shape)
|
| 160 |
+
df4 = pd.DataFrame(metrics4)
|
| 161 |
+
# %% Print inputs to paper table
|
| 162 |
+
df_all = pd.concat([df, df3, df2, df4])
|
| 163 |
+
df_all.datasize = df_all.datasize.astype(int)
|
| 164 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 165 |
+
temp_df = df_all.loc[(df_all.dataset == dataset) & (df_all.datasize.isin([1, 3, 6, 12, 197])), ["exp", "datasize", "dice"]]
|
| 166 |
+
print(dataset)
|
| 167 |
+
mean = temp_df.groupby(["exp", "datasize"]).mean().unstack() * 100
|
| 168 |
+
std = temp_df.groupby(["exp", "datasize"]).std().unstack() * 100
|
| 169 |
+
for exp, exp_name in zip(['baseline', 'LEDM','Step_1', 'Step 1 (MLP)',
|
| 170 |
+
'Step 10 (MLP)','Step 25 (MLP)', 'LEDMe', 'TEDM'],
|
| 171 |
+
['Baseline', 'DatasetDDPM', 'Step 1 (linear)','Step 1 (MLP)', 'Step 10 (MLP)','Step 25 (MLP)','DatasetDDPMe', 'Ours', ]):
|
| 172 |
+
|
| 173 |
+
print(exp_name, end='&\t')
|
| 174 |
+
print(f"{round(mean.loc[exp, ('dice', 1)],2):.3} $\pm$ {round(std.loc[exp, ('dice', 1)],1)}", end='&\t')
|
| 175 |
+
print(f"{round(mean.loc[exp, ('dice', 3)], 2):.3} $\pm$ {round(std.loc[exp, ('dice', 3)],1)}", end='&\t')
|
| 176 |
+
print(f"{round(mean.loc[exp, ('dice', 6)], 2):.3} $\pm$ {round(std.loc[exp, ('dice', 6)],1)}", end='&\t')
|
| 177 |
+
print(f"{round(mean.loc[exp, ('dice', 12)], 2):.3} $\pm$ {round(std.loc[exp, ('dice', 12)],1)}", end='&\t')
|
| 178 |
+
print(f"{round(mean.loc[exp, ('dice', 197)], 2):.3} $\pm$ {round(std.loc[exp, ('dice', 197)],1)}", end="""\\\\""")
|
| 179 |
+
|
| 180 |
+
print()
|
| 181 |
+
|
| 182 |
+
# %% Print inputs to paper appendix table
|
| 183 |
+
for i, dataset in enumerate(["JSRT", "NIH", "Montgomery"]):
|
| 184 |
+
print("\n" + dataset)
|
| 185 |
+
for m in ["precision", "recall"]:
|
| 186 |
+
temp_df = df_all.loc[(df_all.dataset == dataset) & (df_all.datasize.isin([1, 3, 6, 12, 24, 49, 98, 197])), ["exp", "datasize", m]]
|
| 187 |
+
print("\n"+m)
|
| 188 |
+
mean = temp_df.groupby(["exp", "datasize"]).mean().unstack() * 100
|
| 189 |
+
std = temp_df.groupby(["exp", "datasize"]).std().unstack() * 100
|
| 190 |
+
for exp, exp_name in zip(['baseline', 'LEDM','Step_1', 'LEDMe', 'TEDM'],
|
| 191 |
+
['Baseline', 'LEDM', 'Step 1 (linear)','LEDMe', 'TEDM (ours)',]):
|
| 192 |
+
|
| 193 |
+
print(exp_name, end='&\t')
|
| 194 |
+
print(f"{round(mean.loc[exp, (m, 1)],2):.3} $\pm$ {round(std.loc[exp, (m, 1)],1)}", end='&\t')
|
| 195 |
+
print(f"{round(mean.loc[exp, (m, 3)],2):.3} $\pm$ {round(std.loc[exp, (m, 3)],1)}", end='&\t')
|
| 196 |
+
print(f"{round(mean.loc[exp, (m, 6)],2):.3} $\pm$ {round(std.loc[exp, (m, 6)],1)}", end='&\t')
|
| 197 |
+
print(f"{round(mean.loc[exp, (m, 12)],2):.3} $\pm$ {round(std.loc[exp, (m, 12)],1)}", end='&\t')
|
| 198 |
+
print(f"{round(mean.loc[exp, (m, 197)],2):.3} $\pm$ {round(std.loc[exp, (m, 197)],1)}", end='\\\\')
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
print()
|
| 202 |
+
|
| 203 |
+
# %% Wilcoxon tests - to use interactively
|
| 204 |
+
from scipy.stats import wilcoxon
|
| 205 |
+
m ="precision"
|
| 206 |
+
m='recall'
|
| 207 |
+
dataset ="Montgomery"
|
| 208 |
+
dssize =12
|
| 209 |
+
|
| 210 |
+
exp = "baseline"
|
| 211 |
+
exp = 'Step_1'
|
| 212 |
+
exp = "LEDM"
|
| 213 |
+
exp="TEDM"
|
| 214 |
+
exp_2= 'LEDMe'
|
| 215 |
+
|
| 216 |
+
x = df_all.loc[(df_all.dataset == dataset) & (df_all.exp == exp_2) & (df_all.datasize == dssize), m].to_numpy()
|
| 217 |
+
y = df_all.loc[(df_all.dataset == dataset) & (df_all.exp == exp)& (df_all.datasize == dssize), m].to_numpy()
|
| 218 |
+
print(f"{m} - {dataset} - {dssize} - {exp_2}: {x.mean():.4}+/-{x.std():.3} ")
|
| 219 |
+
print(f"{m} - {dataset} - {dssize} - {exp}: {y.mean():.4}+/-{y.std():.3} ")
|
| 220 |
+
print(f"{m} - {dataset} - {dssize}: {wilcoxon(x, y=y, zero_method='wilcox', correction=False, alternative='two-sided',).pvalue:.3} obs given equal ")
|
| 221 |
+
print(f"{m} - {dataset} - {dssize}: {wilcoxon(x, y=y, zero_method='wilcox', correction=False, alternative='greater',).pvalue:.3} obs given {exp_2} < {exp} ")
|
| 222 |
+
print(f"{m} - {dataset} - {dssize}: {wilcoxon(x, y=y, zero_method='wilcox', correction=False, alternative='less',).pvalue:.3} obs given {exp_2} > {exp} ")
|
auxiliary/notebooks_and_reporting/results_per_timestep.pdf
ADDED
|
Binary file (79.6 kB). View file
|
|
|
auxiliary/notebooks_and_reporting/results_per_timestep_dice.pdf
ADDED
|
Binary file (177 kB). View file
|
|
|
auxiliary/notebooks_and_reporting/results_per_timestep_prec_recall.pdf
ADDED
|
Binary file (197 kB). View file
|
|
|
auxiliary/notebooks_and_reporting/results_shared_weights.pdf
ADDED
|
Binary file (66.2 kB). View file
|
|
|
auxiliary/notebooks_and_reporting/visualisations.pdf
ADDED
|
Binary file (296 kB). View file
|
|
|
auxiliary/notebooks_and_reporting/visualisations.py
ADDED
|
@@ -0,0 +1,162 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# %%
|
| 2 |
+
import numpy as np
|
| 3 |
+
import torch
|
| 4 |
+
from pathlib import Path
|
| 5 |
+
import os, sys
|
| 6 |
+
import pandas as pd
|
| 7 |
+
import seaborn as sns
|
| 8 |
+
import matplotlib.pyplot as plt
|
| 9 |
+
HEAD = Path(os.getcwd()).parent.parent
|
| 10 |
+
head = HEAD / 'logs'
|
| 11 |
+
sys.path.append(HEAD)
|
| 12 |
+
from dataloaders.JSRT import JSRTDataset
|
| 13 |
+
from dataloaders.NIH import NIHDataset
|
| 14 |
+
from dataloaders.Montgomery import MonDataset
|
| 15 |
+
NIHPATH = "<PATH_TO_DATA>/NIH/"
|
| 16 |
+
NIHFILE = "correspondence_with_chestXray8.csv"
|
| 17 |
+
MONPATH = "<PATH_TO_DATA>/MontgomerySet/"
|
| 18 |
+
MONFILE = "patient_data.csv"
|
| 19 |
+
JSRTPATH = "<PATH_TO_DATA>/JSRT"
|
| 20 |
+
|
| 21 |
+
if __name__=="__main__":
|
| 22 |
+
predictions = {'baseline':{'JSRT':{}, 'NIH':{}, 'Montgomery':{}},
|
| 23 |
+
'LEDM':{'JSRT':{}, 'NIH':{}, 'Montgomery':{}},
|
| 24 |
+
'TEDM':{'JSRT':{}, 'NIH':{}, 'Montgomery':{}},}
|
| 25 |
+
files_needed = ["JSRT_val_predictions.pt", "JSRT_test_predictions.pt", "NIH_predictions.pt", "Montgomery_predictions.pt",]
|
| 26 |
+
for exp in ['baseline', 'LEDM', "TEDM"]:
|
| 27 |
+
for datasize in [1,3,6,12,24,49,98,197]:
|
| 28 |
+
if len(set(files_needed) - set(os.listdir(head / exp / str(datasize) ))) == 0:
|
| 29 |
+
for file in files_needed[1:]:
|
| 30 |
+
output = torch.load(head / exp / str(datasize) / file)
|
| 31 |
+
metrics_datasize = 197 if datasize == "None" else int(datasize)
|
| 32 |
+
predictions[exp][file.rsplit("_")[0]][metrics_datasize]= output['y_hat']
|
| 33 |
+
else:
|
| 34 |
+
print(f"Experiment {exp} is missing files")
|
| 35 |
+
# %%
|
| 36 |
+
|
| 37 |
+
img_size = 128
|
| 38 |
+
NIH_dataset = NIHDataset(NIHPATH, NIHPATH, NIHFILE, img_size)
|
| 39 |
+
JSRT_dataset = JSRTDataset(JSRTPATH, HEAD/ "data/", "JSRT_test_split.csv", img_size)
|
| 40 |
+
MON_dataset = MonDataset(MONPATH, MONPATH, MONFILE, img_size)
|
| 41 |
+
|
| 42 |
+
# %%
|
| 43 |
+
loaders = {'JSRT': JSRT_dataset, 'NIH': NIH_dataset, 'Montgomery': MON_dataset}
|
| 44 |
+
m ="dice"
|
| 45 |
+
sz=4
|
| 46 |
+
ftsize= 40
|
| 47 |
+
fig, all_axs = plt.subplots(6, 21, figsize=(21*sz, 6*sz))
|
| 48 |
+
all_patients = [17, 13, 0, 1, 72, 78]
|
| 49 |
+
|
| 50 |
+
# JSRT
|
| 51 |
+
dataset ="JSRT"
|
| 52 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 53 |
+
patient = all_patients[0]
|
| 54 |
+
print("JSRT1 - ", patient)
|
| 55 |
+
out = loaders[dataset][patient]
|
| 56 |
+
axs = all_axs[:3, :7]
|
| 57 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 58 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 59 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 60 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 61 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 62 |
+
axs[0, 0].set_title("JSRT - Image", fontsize=ftsize)
|
| 63 |
+
axs[0, 1].set_title("JSRT - GT", fontsize=ftsize)
|
| 64 |
+
axs[0, 2].set_title("1 (1%)" , fontsize=ftsize)
|
| 65 |
+
axs[0, 3].set_title("3 (2%)", fontsize=ftsize)
|
| 66 |
+
axs[0, 4].set_title("6 (3%)", fontsize=ftsize)
|
| 67 |
+
axs[0, 5].set_title("12 (6%)", fontsize=ftsize)
|
| 68 |
+
axs[0, 6].set_title("197 (100%)", fontsize=ftsize)
|
| 69 |
+
axs[0,0].set_ylabel("Baseline", fontsize=ftsize)
|
| 70 |
+
axs[1,0].set_ylabel("LEDM", fontsize=ftsize)
|
| 71 |
+
axs[2,0].set_ylabel("TEDM", fontsize=ftsize)
|
| 72 |
+
#
|
| 73 |
+
axs = all_axs[3:, :7]
|
| 74 |
+
dataset ="JSRT"
|
| 75 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 76 |
+
patient = all_patients[1]
|
| 77 |
+
print("JSRT2 - ", patient)
|
| 78 |
+
out = loaders[dataset][patient]
|
| 79 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 80 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 81 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 82 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 83 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 84 |
+
axs[0,0].set_ylabel("Baseline", fontsize=ftsize)
|
| 85 |
+
axs[1,0].set_ylabel("LEDM", fontsize=ftsize)
|
| 86 |
+
axs[2,0].set_ylabel("TEDM", fontsize=ftsize)
|
| 87 |
+
#
|
| 88 |
+
axs = all_axs[:3, 7:14]
|
| 89 |
+
dataset ="NIH"
|
| 90 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 91 |
+
patient = all_patients[2]
|
| 92 |
+
print("NIH1 - ", patient)
|
| 93 |
+
out = loaders[dataset][patient]
|
| 94 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 95 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 96 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 97 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 98 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 99 |
+
axs[0, 0].set_title("NIH - Image", fontsize=ftsize)
|
| 100 |
+
axs[0, 1].set_title("NIH - GT", fontsize=ftsize)
|
| 101 |
+
axs[0, 2].set_title("1 (1%)" , fontsize=ftsize)
|
| 102 |
+
axs[0, 3].set_title("3 (2%)", fontsize=ftsize)
|
| 103 |
+
axs[0, 4].set_title("6 (3%)", fontsize=ftsize)
|
| 104 |
+
axs[0, 5].set_title("12 (6%)", fontsize=ftsize)
|
| 105 |
+
axs[0, 6].set_title("197 (100%)", fontsize=ftsize)
|
| 106 |
+
#
|
| 107 |
+
#
|
| 108 |
+
axs = all_axs[3:, 7:14]
|
| 109 |
+
dataset ="NIH"
|
| 110 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 111 |
+
patient = all_patients[3]
|
| 112 |
+
print("NIH2 - ", patient)
|
| 113 |
+
out = loaders[dataset][patient]
|
| 114 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 115 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 116 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 117 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 118 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 119 |
+
#
|
| 120 |
+
#
|
| 121 |
+
axs = all_axs[:3, 14:]
|
| 122 |
+
dataset ="Montgomery"
|
| 123 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 124 |
+
patient = all_patients[4]
|
| 125 |
+
print("MON1 - ",patient)
|
| 126 |
+
out = loaders[dataset][patient]
|
| 127 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 128 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 129 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 130 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 131 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 132 |
+
axs[0, 0].set_title("Mont. - Image", fontsize=ftsize)
|
| 133 |
+
axs[0, 1].set_title("Mont. - GT", fontsize=ftsize)
|
| 134 |
+
axs[0, 2].set_title("1 (1%)", fontsize=ftsize)
|
| 135 |
+
axs[0, 3].set_title("3 (2%)", fontsize=ftsize)
|
| 136 |
+
axs[0, 4].set_title("6 (3%)", fontsize=ftsize)
|
| 137 |
+
axs[0, 5].set_title("12 (6%)", fontsize=ftsize)
|
| 138 |
+
axs[0, 6].set_title("197 (100%)", fontsize=ftsize)
|
| 139 |
+
#
|
| 140 |
+
axs = all_axs[3:, 14:]
|
| 141 |
+
dataset ="Montgomery"
|
| 142 |
+
patient = np.random.randint(0, len(loaders[dataset]))
|
| 143 |
+
patient = all_patients[5]
|
| 144 |
+
print("MON2 - ",patient)
|
| 145 |
+
out = loaders[dataset][patient]
|
| 146 |
+
for rowax, exp in zip(axs, ['baseline', 'LEDM', 'TEDM']):
|
| 147 |
+
rowax[0].imshow(out[0][0].numpy(), cmap='gray')
|
| 148 |
+
rowax[1].imshow(out[1][0].numpy(), interpolation='none', cmap='gray')
|
| 149 |
+
for ax, dssize in zip(rowax[2:], [1, 3, 6, 12, 197]):
|
| 150 |
+
ax.imshow(predictions[exp][dataset][dssize][patient].numpy()[0]>.5, interpolation='none')
|
| 151 |
+
|
| 152 |
+
|
| 153 |
+
# remove ticks
|
| 154 |
+
for ax in all_axs.flatten():
|
| 155 |
+
ax.set_xticks([])
|
| 156 |
+
ax.set_yticks([])
|
| 157 |
+
sns.despine(ax=ax, left=True, bottom=True)
|
| 158 |
+
plt.subplots_adjust(wspace=0.00,
|
| 159 |
+
hspace=0.00)
|
| 160 |
+
plt.tight_layout()
|
| 161 |
+
plt.savefig("visualisations2.pdf", bbox_inches='tight')
|
| 162 |
+
plt.show()
|
auxiliary/notebooks_and_reporting/visualisations2.pdf
ADDED
|
Binary file (852 kB). View file
|
|
|
auxiliary/postprocessing/run_tests.py
ADDED
|
@@ -0,0 +1,162 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
import os
|
| 4 |
+
import torch
|
| 5 |
+
from tqdm.auto import tqdm
|
| 6 |
+
from torch import autocast
|
| 7 |
+
from torch.utils.data import DataLoader
|
| 8 |
+
import sys
|
| 9 |
+
HEAD = Path(os.getcwd()).parent.parent
|
| 10 |
+
sys.path.append("/vol/biomedic3/mmr12/projects/TEDM/")
|
| 11 |
+
from models.diffusion_model import DiffusionModel
|
| 12 |
+
from models.unet_model import Unet
|
| 13 |
+
from models.datasetDM_model import DatasetDM
|
| 14 |
+
from trainers.datasetDM_per_step import ModDatasetDM
|
| 15 |
+
from trainers.train_baseline import dice, precision, recall
|
| 16 |
+
from dataloaders.JSRT import build_dataloaders
|
| 17 |
+
from dataloaders.NIH import NIHDataset
|
| 18 |
+
from dataloaders.Montgomery import MonDataset
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
NIHPATH = "/vol/biodata/data/chest_xray/NIH/"
|
| 22 |
+
NIHFILE = "correspondence_with_chestXray8.csv"
|
| 23 |
+
MONPATH = "/vol/biodata/data/chest_xray/NLM/MontgomerySet/"
|
| 24 |
+
MONFILE = "patient_data.csv"
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
if __name__ == "__main__":
|
| 28 |
+
# load config file and parse arguments
|
| 29 |
+
parser = argparse.ArgumentParser()
|
| 30 |
+
parser.add_argument('--experiment', "-e", type=str, help='Experiment path', default="logs/JSRT_conditional/20230213_171633")
|
| 31 |
+
parser.add_argument('--rerun', "-r", help='Run the test again', default=False, action="store_true")
|
| 32 |
+
args = parser.parse_args()
|
| 33 |
+
|
| 34 |
+
if os.path.isdir(args.experiment):
|
| 35 |
+
print("Experiment path identified as a directory")
|
| 36 |
+
else:
|
| 37 |
+
raise ValueError("Experiment path is not a directory")
|
| 38 |
+
files = os.listdir(args.experiment)
|
| 39 |
+
torch_file = None
|
| 40 |
+
if {'JSRT_val_predictions.pt', 'JSRT_test_predictions.pt', 'NIH_predictions.pt', 'Montgomery_predictions.pt'} <= set(files) and not args.rerun:
|
| 41 |
+
print("Experiment already tested")
|
| 42 |
+
for file in ['JSRT_val_predictions.pt', 'JSRT_test_predictions.pt', 'NIH_predictions.pt', 'Montgomery_predictions.pt']:
|
| 43 |
+
output = torch.load(Path(args.experiment) / file)
|
| 44 |
+
dataset_key = file.split("_")[0]
|
| 45 |
+
print(f"{dataset_key} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 46 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 47 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 48 |
+
#torch.save(output, Path(args.experiment) / f'{dataset_key}_predictions.pt')
|
| 49 |
+
exit(0)
|
| 50 |
+
|
| 51 |
+
for f in files:
|
| 52 |
+
if "model" in f:
|
| 53 |
+
torch_file = f
|
| 54 |
+
break
|
| 55 |
+
if torch_file is None:
|
| 56 |
+
raise ValueError("No checkpoint file found in experiment directory")
|
| 57 |
+
|
| 58 |
+
print(f"Loading experiment from {torch_file}")
|
| 59 |
+
data = torch.load(Path(args.experiment) / torch_file)
|
| 60 |
+
config = data["config"]
|
| 61 |
+
|
| 62 |
+
# pick model
|
| 63 |
+
if config.experiment in ["baseline", "global_finetune", "glob_loc_finetune"]:
|
| 64 |
+
model = Unet(**vars(config))
|
| 65 |
+
elif config.experiment == "datasetDM":
|
| 66 |
+
model = DatasetDM(config)
|
| 67 |
+
elif config.experiment == "simple_datasetDM":
|
| 68 |
+
model = ModDatasetDM(config)
|
| 69 |
+
else:
|
| 70 |
+
raise ValueError(f"Experiment {config.experiment} not recognized")
|
| 71 |
+
model.load_state_dict(data['model_state_dict'])
|
| 72 |
+
|
| 73 |
+
# Gather model output
|
| 74 |
+
model.eval().to(config.device)
|
| 75 |
+
|
| 76 |
+
# Load data
|
| 77 |
+
dataloaders = build_dataloaders(
|
| 78 |
+
config.data_dir,
|
| 79 |
+
config.img_size,
|
| 80 |
+
config.batch_size,
|
| 81 |
+
config.num_workers,
|
| 82 |
+
)
|
| 83 |
+
datasets_to_test = {
|
| 84 |
+
"JSRT_val": dataloaders["val"],
|
| 85 |
+
"JSRT_test": dataloaders["test"],
|
| 86 |
+
"NIH": DataLoader(NIHDataset(NIHPATH, NIHPATH, NIHFILE, config.img_size),
|
| 87 |
+
config.batch_size, num_workers=config.num_workers),
|
| 88 |
+
"Montgomery": DataLoader(MonDataset(MONPATH, MONPATH, MONFILE, config.img_size),
|
| 89 |
+
config.batch_size, num_workers=config.num_workers)
|
| 90 |
+
|
| 91 |
+
}
|
| 92 |
+
if config.experiment == "simple_datasetDM":
|
| 93 |
+
# re-calculate mean and var as they were not saved in the model dict
|
| 94 |
+
train_dl = dataloaders["train"]
|
| 95 |
+
for x, _ in tqdm(train_dl, desc="Calculating mean and variance"):
|
| 96 |
+
x = x.to(config.device)
|
| 97 |
+
features = model.extract_features(x)
|
| 98 |
+
model.mean += features.sum(dim=0)
|
| 99 |
+
model.mean_squared += (features ** 2).sum(dim=0)
|
| 100 |
+
model.mean = model.mean / len(train_dl.dataset)
|
| 101 |
+
model.std = (model.mean_squared / len(train_dl.dataset) - model.mean ** 2).sqrt() + 1e-6
|
| 102 |
+
|
| 103 |
+
model.mean = model.mean.to(config.device)
|
| 104 |
+
model.std = model.std.to(config.device)
|
| 105 |
+
|
| 106 |
+
for dataset_key in datasets_to_test:
|
| 107 |
+
if f"{dataset_key}_predictions.pt" in files and not args.rerun:
|
| 108 |
+
print(f"{dataset_key} already tested")
|
| 109 |
+
output = torch.load(Path(args.experiment) / f'{dataset_key}_predictions.pt')
|
| 110 |
+
print(f"{dataset_key} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 111 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 112 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 113 |
+
continue
|
| 114 |
+
|
| 115 |
+
print(f"Testing {dataset_key} set")
|
| 116 |
+
y_hat = []
|
| 117 |
+
y_star = []
|
| 118 |
+
for i, (x, y) in tqdm(enumerate(datasets_to_test[dataset_key]), desc='Validating'):
|
| 119 |
+
x = x.to(config.device)
|
| 120 |
+
|
| 121 |
+
if config.experiment == "conditional":
|
| 122 |
+
# sample n = 5 different segmetations
|
| 123 |
+
y_hats = []
|
| 124 |
+
for _ in range(5):
|
| 125 |
+
img = torch.randn(x.shape, device=config.device)
|
| 126 |
+
for t in tqdm(range(0, config.timesteps)[::-1]):
|
| 127 |
+
# sample next timestep image (x_{t-1})
|
| 128 |
+
with autocast(device_type=config.device, enabled=config.mixed_precision):
|
| 129 |
+
with torch.no_grad():
|
| 130 |
+
img = model.sample_timestep(img, t=t, cond=x)
|
| 131 |
+
y_hats.append(img.detach().cpu() / 2 + .5)
|
| 132 |
+
# take the average over the 5 samples
|
| 133 |
+
y_hats = torch.stack(y_hats, -1).mean(-1)
|
| 134 |
+
|
| 135 |
+
# record
|
| 136 |
+
y_hat.append(y_hats)
|
| 137 |
+
y_star.append(y)
|
| 138 |
+
|
| 139 |
+
elif config.experiment in ["baseline", "datasetDM", "simple_datasetDM", "global_finetune", "glob_loc_finetune"] :
|
| 140 |
+
with autocast(device_type=config.device, enabled=config.mixed_precision):
|
| 141 |
+
with torch.no_grad():
|
| 142 |
+
pred = torch.sigmoid(model(x))
|
| 143 |
+
y_hat.append(pred.detach().cpu())
|
| 144 |
+
y_star.append(y)
|
| 145 |
+
|
| 146 |
+
else:
|
| 147 |
+
raise ValueError(f"Experiment {config.experiment} not recognized")
|
| 148 |
+
|
| 149 |
+
# save predictions
|
| 150 |
+
y_hat = torch.cat(y_hat, 0)
|
| 151 |
+
y_star = torch.cat(y_star, 0)
|
| 152 |
+
output = {
|
| 153 |
+
'y_hat': y_hat,
|
| 154 |
+
'y_star': y_star,
|
| 155 |
+
'dice':dice(y_hat>.5, y_star),
|
| 156 |
+
'precision':precision(y_hat>.5, y_star),
|
| 157 |
+
'recall':recall(y_hat>.5, y_star),}
|
| 158 |
+
|
| 159 |
+
print(f"{dataset_key} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 160 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 161 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 162 |
+
torch.save(output, Path(args.experiment) / f'{dataset_key}_predictions.pt')
|
auxiliary/postprocessing/testing_shared_weights.py
ADDED
|
@@ -0,0 +1,145 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
import os
|
| 4 |
+
import numpy as np
|
| 5 |
+
import pandas as pd
|
| 6 |
+
import torch
|
| 7 |
+
import seaborn as sns
|
| 8 |
+
import matplotlib.pyplot as plt
|
| 9 |
+
from torch import nn
|
| 10 |
+
from tqdm.auto import tqdm
|
| 11 |
+
from torch import autocast
|
| 12 |
+
from torch.utils.data import DataLoader
|
| 13 |
+
from einops.layers.torch import Rearrange
|
| 14 |
+
from einops import rearrange
|
| 15 |
+
import sys
|
| 16 |
+
HEAD = Path(os.getcwd()).parent.parent
|
| 17 |
+
sys.path.append(HEAD)
|
| 18 |
+
from models.datasetDM_model import DatasetDM
|
| 19 |
+
from trainers.train_baseline import dice, precision, recall
|
| 20 |
+
from dataloaders.JSRT import build_dataloaders
|
| 21 |
+
from dataloaders.NIH import NIHDataset
|
| 22 |
+
from dataloaders.Montgomery import MonDataset
|
| 23 |
+
|
| 24 |
+
NIHPATH = "<PATH_TO_DATA>/NIH/"
|
| 25 |
+
NIHFILE = "correspondence_with_chestXray8.csv" # saved in data
|
| 26 |
+
MONPATH = "<PATH_TO_DATA>/NLM/MontgomerySet/"
|
| 27 |
+
MONFILE = "patient_data.csv"
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
if __name__ == "__main__":
|
| 31 |
+
# load config file and parse arguments
|
| 32 |
+
parser = argparse.ArgumentParser()
|
| 33 |
+
parser.add_argument('--experiment', "-e", type=str, help='Experiment path', default="logs/JSRT_conditional/20230213_171633")
|
| 34 |
+
parser.add_argument('--rerun', "-r", help='Run the test again', default=False, action="store_true")
|
| 35 |
+
args = parser.parse_args()
|
| 36 |
+
|
| 37 |
+
if os.path.isdir(args.experiment):
|
| 38 |
+
print("Experiment path identified as a directory")
|
| 39 |
+
else:
|
| 40 |
+
raise ValueError("Experiment path is not a directory")
|
| 41 |
+
files = os.listdir(args.experiment)
|
| 42 |
+
torch_file = None
|
| 43 |
+
if {'JSRT_val_predictions.pt', 'JSRT_test_predictions.pt', 'NIH_predictions.pt', 'Montgomery_predictions.pt'} <= set(files) and not args.rerun:
|
| 44 |
+
print("Experiment already tested")
|
| 45 |
+
sys.exit(0)
|
| 46 |
+
|
| 47 |
+
for f in files:
|
| 48 |
+
if "model" in f:
|
| 49 |
+
torch_file = f
|
| 50 |
+
break
|
| 51 |
+
if torch_file is None:
|
| 52 |
+
raise ValueError("No checkpoint file found in experiment directory")
|
| 53 |
+
|
| 54 |
+
print(f"Loading experiment from {torch_file}")
|
| 55 |
+
data = torch.load(Path(args.experiment) / torch_file)
|
| 56 |
+
config = data["config"]
|
| 57 |
+
|
| 58 |
+
# pick model
|
| 59 |
+
if config.experiment == "datasetDM":
|
| 60 |
+
model = DatasetDM(config)
|
| 61 |
+
model.classifier = nn.Sequential(
|
| 62 |
+
Rearrange('b (step act) h w -> (b step) act h w', step=len(model.steps)),
|
| 63 |
+
nn.Conv2d(960, 128, 1),
|
| 64 |
+
nn.ReLU(),
|
| 65 |
+
nn.BatchNorm2d(128),
|
| 66 |
+
nn.Conv2d(128, 32, 1),
|
| 67 |
+
nn.ReLU(),
|
| 68 |
+
nn.BatchNorm2d(32),
|
| 69 |
+
nn.Conv2d(32, 1, config.out_channels)
|
| 70 |
+
)
|
| 71 |
+
else:
|
| 72 |
+
raise ValueError(f"Experiment {config.experiment} not recognized")
|
| 73 |
+
model.load_state_dict(data['model_state_dict'])
|
| 74 |
+
|
| 75 |
+
# Gather model output
|
| 76 |
+
model.eval().to(config.device)
|
| 77 |
+
|
| 78 |
+
# Load data
|
| 79 |
+
dataloaders = build_dataloaders(
|
| 80 |
+
config.data_dir,
|
| 81 |
+
config.img_size,
|
| 82 |
+
config.batch_size,
|
| 83 |
+
config.num_workers,
|
| 84 |
+
)
|
| 85 |
+
datasets_to_test = {
|
| 86 |
+
"JSRT_val": dataloaders["val"],
|
| 87 |
+
"JSRT_test": dataloaders["test"],
|
| 88 |
+
"NIH": DataLoader(NIHDataset(NIHPATH, NIHPATH, NIHFILE, config.img_size),
|
| 89 |
+
config.batch_size, num_workers=config.num_workers),
|
| 90 |
+
"Montgomery": DataLoader(MonDataset(MONPATH, MONPATH, MONFILE, config.img_size),
|
| 91 |
+
config.batch_size, num_workers=config.num_workers)
|
| 92 |
+
|
| 93 |
+
}
|
| 94 |
+
|
| 95 |
+
for dataset_key in datasets_to_test:
|
| 96 |
+
if f"{dataset_key}_predictions.pt" in files and not args.rerun:
|
| 97 |
+
print(f"{dataset_key} already tested")
|
| 98 |
+
output = torch.load(Path(args.experiment) / f'{dataset_key}_predictions.pt')
|
| 99 |
+
print(f"{dataset_key} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 100 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 101 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 102 |
+
continue
|
| 103 |
+
|
| 104 |
+
print(f"Testing {dataset_key} set")
|
| 105 |
+
y_hats = []
|
| 106 |
+
y_star = []
|
| 107 |
+
for i, (x, y) in tqdm(enumerate(datasets_to_test[dataset_key]), desc='Validating'):
|
| 108 |
+
x = x.to(config.device)
|
| 109 |
+
|
| 110 |
+
with autocast(device_type=config.device, enabled=config.mixed_precision):
|
| 111 |
+
with torch.no_grad():
|
| 112 |
+
# all depths
|
| 113 |
+
pred = torch.sigmoid(model(x))
|
| 114 |
+
y_hats.append(pred.detach().cpu())
|
| 115 |
+
y_star.append(y)
|
| 116 |
+
|
| 117 |
+
# save predictions
|
| 118 |
+
y_star = torch.cat(y_star, 0)
|
| 119 |
+
y_hats = torch.cat(y_hats, 0)
|
| 120 |
+
y_hats = rearrange(y_hats, '(b step) 1 h w -> step b 1 h w', step=len(model.steps))
|
| 121 |
+
for i, y_hat in enumerate(y_hats):
|
| 122 |
+
output = {
|
| 123 |
+
'y_hat': y_hat,
|
| 124 |
+
'y_star': y_star,
|
| 125 |
+
'dice':dice(y_hat>.5, y_star),
|
| 126 |
+
'precision':precision(y_hat>.5, y_star),
|
| 127 |
+
'recall':recall(y_hat>.5, y_star),}
|
| 128 |
+
|
| 129 |
+
print(f"{dataset_key} {model.steps[i]} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 130 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 131 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 132 |
+
torch.save(output, Path(args.experiment) / f'{dataset_key}_timestep{model.steps[i]}_predictions.pt')
|
| 133 |
+
y_hat = y_hats.mean(0)
|
| 134 |
+
output = {
|
| 135 |
+
'y_hat': y_hat,
|
| 136 |
+
'y_star': y_star,
|
| 137 |
+
'dice':dice(y_hat>.5, y_star),
|
| 138 |
+
'precision':precision(y_hat>.5, y_star),
|
| 139 |
+
'recall':recall(y_hat>.5, y_star),}
|
| 140 |
+
|
| 141 |
+
print(f"{dataset_key} metrics: \n\tdice: {output['dice'].mean():.3}+/-{output['dice'].std():.3}")
|
| 142 |
+
print(f"\tprecision: {output['precision'].mean():.3}+/-{output['precision'].std():.3}")
|
| 143 |
+
print(f"\trecall: {output['recall'].mean():.3}+/-{output['recall'].std():.3}")
|
| 144 |
+
torch.save(output, Path(args.experiment) / f'{dataset_key}_predictions.pt')
|
| 145 |
+
|
auxiliary/preprocessing/CXR14_preprocessing_separate_data.py
ADDED
|
@@ -0,0 +1,31 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# %%
|
| 2 |
+
import pandas as pd
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
import numpy as np
|
| 5 |
+
import os
|
| 6 |
+
|
| 7 |
+
CWDIR = Path(os.getcwd()).parent.parent
|
| 8 |
+
DATADIR = Path("<PATH_TO_DATA>/ChestXray-NIHCC")
|
| 9 |
+
if not os.path.isdir(DATADIR):
|
| 10 |
+
print(f"Data directory {DATADIR} not found")
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
df = pd.concat([pd.read_csv(DATADIR / "train_val_list.csv"),pd.read_csv(DATADIR / "test_list.csv")])
|
| 14 |
+
df.reset_index(inplace=True)
|
| 15 |
+
# %%
|
| 16 |
+
from tqdm import tqdm
|
| 17 |
+
items = []
|
| 18 |
+
for el in tqdm(df["Image Index"]):
|
| 19 |
+
items.append(os.path.isfile(DATADIR / "images"/ el))
|
| 20 |
+
# %% Shuffle and remove 20% for test and val
|
| 21 |
+
idx = np.arange(len(df))
|
| 22 |
+
np.random.shuffle(idx)
|
| 23 |
+
n1 = int(len(df)*.8)
|
| 24 |
+
n2 = int(len(df)*.9)
|
| 25 |
+
idxs = [idx[:n1], idx[n1:n2], idx[n2:]]
|
| 26 |
+
for i in range(3):
|
| 27 |
+
print(len(df.loc[idxs[i]]))
|
| 28 |
+
# %%
|
| 29 |
+
df.loc[idxs[0]].to_csv(CWDIR / 'data' / 'train_split.csv', index=False)
|
| 30 |
+
df.loc[idxs[1]].to_csv(CWDIR / 'data' / 'val_split.csv', index=False)
|
| 31 |
+
df.loc[idxs[2]].to_csv(CWDIR / 'data' / 'test_split.csv', index=False)
|
auxiliary/preprocessing/JSRT_preprocessing_separate_data.py
ADDED
|
@@ -0,0 +1,26 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# %%
|
| 2 |
+
import pandas as pd
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
import numpy as np
|
| 5 |
+
import os
|
| 6 |
+
|
| 7 |
+
CWDIR = Path(os.getcwd()).parent.parent
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
head = Path("<PATH_TO_DATA>/JSRT")
|
| 11 |
+
|
| 12 |
+
df = pd.read_csv(head / "jsrt_metadata_with_masks.csv")
|
| 13 |
+
df.reset_index(inplace=True)
|
| 14 |
+
|
| 15 |
+
# %% Shuffle and remove 20% for test and val
|
| 16 |
+
idx = np.arange(len(df))
|
| 17 |
+
np.random.shuffle(idx)
|
| 18 |
+
n1 = int(len(df)*.8)
|
| 19 |
+
n2 = int(len(df)*.9)
|
| 20 |
+
idxs = [idx[:n1], idx[n1:n2], idx[n2:]]
|
| 21 |
+
for i in range(3):
|
| 22 |
+
print(len(df.loc[idxs[i]]))
|
| 23 |
+
# %%
|
| 24 |
+
df.loc[idxs[0]].to_csv(CWDIR / 'data' / 'JSRT_train_split.csv', index=False)
|
| 25 |
+
df.loc[idxs[1]].to_csv(CWDIR / 'data' / 'JSRT_val_split.csv', index=False)
|
| 26 |
+
df.loc[idxs[2]].to_csv(CWDIR / 'data' / 'JSRT_test_split.csv', index=False)
|
config.py
ADDED
|
@@ -0,0 +1,84 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
import os
|
| 3 |
+
|
| 4 |
+
from datetime import datetime
|
| 5 |
+
from pathlib import Path
|
| 6 |
+
import torch
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
this_dir = os.path.dirname(os.path.realpath(__file__))
|
| 10 |
+
default_logdir = os.path.join(this_dir, 'logs', datetime.now().strftime('%Y%m%d_%H%M%S'))
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
parser = argparse.ArgumentParser()
|
| 14 |
+
parser.add_argument('--debug', action='store_true')
|
| 15 |
+
parser.add_argument('--mixed_precision', type=bool, default=False, help='Use mixed precision')
|
| 16 |
+
parser.add_argument('--resume_path', type=str, default=None, help='Path to checkpoint to resume from')
|
| 17 |
+
|
| 18 |
+
# Experiment parameters
|
| 19 |
+
parser.add_argument('--experiment', type=str, default="img_only",choices=[
|
| 20 |
+
"PDDM",
|
| 21 |
+
"baseline",
|
| 22 |
+
"LEDM",
|
| 23 |
+
"LEDMe",
|
| 24 |
+
"TEDM",
|
| 25 |
+
"global_cl",
|
| 26 |
+
"local_cl",
|
| 27 |
+
"global_finetune",
|
| 28 |
+
"glob_loc_finetune"
|
| 29 |
+
], help='Whether to generate only images or images and segmentations')
|
| 30 |
+
parser.add_argument('--dataset', type=str, default="JSRT",choices=["JSRT", "CXR14"], help='Dataset to use')
|
| 31 |
+
|
| 32 |
+
# Data parameters
|
| 33 |
+
parser.add_argument('--img_size', type=int, default=128, help='Height / width of the input image to the network')
|
| 34 |
+
parser.add_argument('--data_dir', type=str, help='Path to the dataset')
|
| 35 |
+
parser.add_argument('--num_workers', type=int, default=4, help='Number of subprocesses to use for data loading')
|
| 36 |
+
|
| 37 |
+
# Model parameters
|
| 38 |
+
parser.add_argument('--dim', type=int, default=64, help='Width of the U-Net')
|
| 39 |
+
parser.add_argument('--dim_mults', nargs='+', type=int, default=(1, 2, 4, 8), help='Dimension multipliers for U-Net levels')
|
| 40 |
+
# SegDiff model parameters
|
| 41 |
+
parser.add_argument('--seg_out_dim', type=int, default=1, help='Dimension of segmentation embedding')
|
| 42 |
+
parser.add_argument('--img_out_dim', type=int, default=4, help='Dimension of image embedding')
|
| 43 |
+
parser.add_argument('--img_inter_dim', type=int, default=32, help='Width of image embedding')
|
| 44 |
+
|
| 45 |
+
# Diffusion parameters
|
| 46 |
+
parser.add_argument('--timesteps', type=int, default=1000, help='Number of diffusion timesteps')
|
| 47 |
+
parser.add_argument('--beta_schedule', type=str, default='cosine', choices=['linear', 'cosine'])
|
| 48 |
+
parser.add_argument('--objective', type=str, default='pred_noise', help='Model output', choices=['pred_noise', 'pred_x_0'])
|
| 49 |
+
|
| 50 |
+
# CL parameters
|
| 51 |
+
parser.add_argument('--tau', type=float, default=0.1, help='Temperature parameter for contrastive loss')
|
| 52 |
+
parser.add_argument('--global_model_path', type=str, default=None, help='Path to global model checkpoint')
|
| 53 |
+
parser.add_argument('--glob_loc_model_path', type=str, default=None, help='Path to global & local CL model checkpoint')
|
| 54 |
+
parser.add_argument('--unfreeze_weights_at_step', type=int, default=0, help='Step at which to unfreeze pretrained weights. If 0, weights are not frozen')
|
| 55 |
+
parser.add_argument('--augment_at_finetuning', default=False, action='store_true', help='Whether to augment images during finetuning')
|
| 56 |
+
|
| 57 |
+
# Training parameters
|
| 58 |
+
parser.add_argument('--batch_size', type=int, default=16, help='Input batch size')
|
| 59 |
+
parser.add_argument('--lr', type=float, default=1e-4, help='Learning rate')
|
| 60 |
+
parser.add_argument('--weight_decay', type=float, default=0, help='Weight decay')
|
| 61 |
+
# parser.add_argument('--adam_betas', nargs=2, type=float, default=(0.9, 0.99), help='Betas for the Adam optimizer')
|
| 62 |
+
parser.add_argument('--max_steps', type=int, default=500000, help='Number of training steps to perform')
|
| 63 |
+
parser.add_argument('--p2_loss_weight_gamma', type=float, default=0., help='p2 loss weight, from https://arxiv.org/abs/2204.00227 - 0 is equivalent to weight of 1 across time - 1. is recommended')
|
| 64 |
+
parser.add_argument('--p2_loss_weight_k', type=float, default=1.)
|
| 65 |
+
parser.add_argument('--device', type=str, default='cuda' if torch.cuda.is_available() else 'cpu', help='Device to use')
|
| 66 |
+
parser.add_argument('--seed', type=int, default=0, help='Random seed')
|
| 67 |
+
|
| 68 |
+
# Logging parameters
|
| 69 |
+
parser.add_argument('--log_freq', type=int, default=100, help='Frequency of logging')
|
| 70 |
+
parser.add_argument('--val_freq', type=int, default=100, help='Frequency of validation')
|
| 71 |
+
parser.add_argument('--val_steps', type=int, default=250, help='Number of timestep to use for validation')
|
| 72 |
+
parser.add_argument('--log_dir', type=str, default=default_logdir, help='Logging directory')
|
| 73 |
+
parser.add_argument('--n_sampled_imgs', type=int, default=8, help='Number of images to sample during logging')
|
| 74 |
+
parser.add_argument('--max_val_steps', type=int, default=-1, help='Number of validation steps to perform')
|
| 75 |
+
|
| 76 |
+
# datasetGAN like segmentation model parameters
|
| 77 |
+
parser.add_argument("--saved_diffusion_model", type=str, help='Path to checkpoint of trained diffusion model', default="logs/20230127_164150/best_model.pt")
|
| 78 |
+
parser.add_argument("--t_steps_to_save", type=int, nargs='*', choices=range(1000), help='Diffusion steps to be used as features', default=[50, 200, 400, 600, 800])
|
| 79 |
+
parser.add_argument("--n_labelled_images", type=int, help='Number of labelled images to use for semi-supervised training', default=None,
|
| 80 |
+
choices=[197, 98, 49, 24, 12, 6, 3, 1])
|
| 81 |
+
|
| 82 |
+
# other experiments I played with
|
| 83 |
+
parser.add_argument("--shared_weights_over_timesteps", help='In datasetDM, only use last timestep to predict, and intermediate timesteps to train', default=False, action='store_true')
|
| 84 |
+
parser.add_argument("--early_stop", help='In baseline, if validation loss increases by more than 50%, stop', default=False, action='store_true')
|
data/JSRT_test_split.csv
ADDED
|
@@ -0,0 +1,26 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
id,path
|
| 2 |
+
JPCNN017,JSRT/PNG_data/JPCNN017.png
|
| 3 |
+
JPCLN151,JSRT/PNG_data/JPCLN151.png
|
| 4 |
+
JPCNN007,JSRT/PNG_data/JPCNN007.png
|
| 5 |
+
JPCNN089,JSRT/PNG_data/JPCNN089.png
|
| 6 |
+
JPCLN153,JSRT/PNG_data/JPCLN153.png
|
| 7 |
+
JPCNN020,JSRT/PNG_data/JPCNN020.png
|
| 8 |
+
JPCNN093,JSRT/PNG_data/JPCNN093.png
|
| 9 |
+
JPCLN118,JSRT/PNG_data/JPCLN118.png
|
| 10 |
+
JPCLN143,JSRT/PNG_data/JPCLN143.png
|
| 11 |
+
JPCLN073,JSRT/PNG_data/JPCLN073.png
|
| 12 |
+
JPCLN018,JSRT/PNG_data/JPCLN018.png
|
| 13 |
+
JPCLN109,JSRT/PNG_data/JPCLN109.png
|
| 14 |
+
JPCLN095,JSRT/PNG_data/JPCLN095.png
|
| 15 |
+
JPCNN055,JSRT/PNG_data/JPCNN055.png
|
| 16 |
+
JPCLN131,JSRT/PNG_data/JPCLN131.png
|
| 17 |
+
JPCLN130,JSRT/PNG_data/JPCLN130.png
|
| 18 |
+
JPCLN053,JSRT/PNG_data/JPCLN053.png
|
| 19 |
+
JPCLN107,JSRT/PNG_data/JPCLN107.png
|
| 20 |
+
JPCNN081,JSRT/PNG_data/JPCNN081.png
|
| 21 |
+
JPCLN146,JSRT/PNG_data/JPCLN146.png
|
| 22 |
+
JPCLN058,JSRT/PNG_data/JPCLN058.png
|
| 23 |
+
JPCLN010,JSRT/PNG_data/JPCLN010.png
|
| 24 |
+
JPCLN137,JSRT/PNG_data/JPCLN137.png
|
| 25 |
+
JPCLN086,JSRT/PNG_data/JPCLN086.png
|
| 26 |
+
JPCLN114,JSRT/PNG_data/JPCLN114.png
|
data/JSRT_train_split.csv
ADDED
|
@@ -0,0 +1,198 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
id,path
|
| 2 |
+
JPCLN001,JSRT/PNG_data/JPCLN001.png
|
| 3 |
+
JPCLN002,JSRT/PNG_data/JPCLN002.png
|
| 4 |
+
JPCLN003,JSRT/PNG_data/JPCLN003.png
|
| 5 |
+
JPCLN004,JSRT/PNG_data/JPCLN004.png
|
| 6 |
+
JPCLN005,JSRT/PNG_data/JPCLN005.png
|
| 7 |
+
JPCLN006,JSRT/PNG_data/JPCLN006.png
|
| 8 |
+
JPCLN007,JSRT/PNG_data/JPCLN007.png
|
| 9 |
+
JPCLN008,JSRT/PNG_data/JPCLN008.png
|
| 10 |
+
JPCLN009,JSRT/PNG_data/JPCLN009.png
|
| 11 |
+
JPCLN011,JSRT/PNG_data/JPCLN011.png
|
| 12 |
+
JPCLN012,JSRT/PNG_data/JPCLN012.png
|
| 13 |
+
JPCLN013,JSRT/PNG_data/JPCLN013.png
|
| 14 |
+
JPCLN014,JSRT/PNG_data/JPCLN014.png
|
| 15 |
+
JPCLN015,JSRT/PNG_data/JPCLN015.png
|
| 16 |
+
JPCLN016,JSRT/PNG_data/JPCLN016.png
|
| 17 |
+
JPCLN017,JSRT/PNG_data/JPCLN017.png
|
| 18 |
+
JPCLN019,JSRT/PNG_data/JPCLN019.png
|
| 19 |
+
JPCLN020,JSRT/PNG_data/JPCLN020.png
|
| 20 |
+
JPCLN021,JSRT/PNG_data/JPCLN021.png
|
| 21 |
+
JPCLN022,JSRT/PNG_data/JPCLN022.png
|
| 22 |
+
JPCLN023,JSRT/PNG_data/JPCLN023.png
|
| 23 |
+
JPCLN024,JSRT/PNG_data/JPCLN024.png
|
| 24 |
+
JPCLN025,JSRT/PNG_data/JPCLN025.png
|
| 25 |
+
JPCLN026,JSRT/PNG_data/JPCLN026.png
|
| 26 |
+
JPCLN027,JSRT/PNG_data/JPCLN027.png
|
| 27 |
+
JPCLN029,JSRT/PNG_data/JPCLN029.png
|
| 28 |
+
JPCLN031,JSRT/PNG_data/JPCLN031.png
|
| 29 |
+
JPCLN032,JSRT/PNG_data/JPCLN032.png
|
| 30 |
+
JPCLN033,JSRT/PNG_data/JPCLN033.png
|
| 31 |
+
JPCLN034,JSRT/PNG_data/JPCLN034.png
|
| 32 |
+
JPCLN035,JSRT/PNG_data/JPCLN035.png
|
| 33 |
+
JPCLN036,JSRT/PNG_data/JPCLN036.png
|
| 34 |
+
JPCLN038,JSRT/PNG_data/JPCLN038.png
|
| 35 |
+
JPCLN039,JSRT/PNG_data/JPCLN039.png
|
| 36 |
+
JPCLN040,JSRT/PNG_data/JPCLN040.png
|
| 37 |
+
JPCLN041,JSRT/PNG_data/JPCLN041.png
|
| 38 |
+
JPCLN042,JSRT/PNG_data/JPCLN042.png
|
| 39 |
+
JPCLN043,JSRT/PNG_data/JPCLN043.png
|
| 40 |
+
JPCLN044,JSRT/PNG_data/JPCLN044.png
|
| 41 |
+
JPCLN046,JSRT/PNG_data/JPCLN046.png
|
| 42 |
+
JPCLN047,JSRT/PNG_data/JPCLN047.png
|
| 43 |
+
JPCLN048,JSRT/PNG_data/JPCLN048.png
|
| 44 |
+
JPCLN049,JSRT/PNG_data/JPCLN049.png
|
| 45 |
+
JPCLN050,JSRT/PNG_data/JPCLN050.png
|
| 46 |
+
JPCLN051,JSRT/PNG_data/JPCLN051.png
|
| 47 |
+
JPCLN052,JSRT/PNG_data/JPCLN052.png
|
| 48 |
+
JPCLN054,JSRT/PNG_data/JPCLN054.png
|
| 49 |
+
JPCLN056,JSRT/PNG_data/JPCLN056.png
|
| 50 |
+
JPCLN057,JSRT/PNG_data/JPCLN057.png
|
| 51 |
+
JPCLN059,JSRT/PNG_data/JPCLN059.png
|
| 52 |
+
JPCLN060,JSRT/PNG_data/JPCLN060.png
|
| 53 |
+
JPCLN061,JSRT/PNG_data/JPCLN061.png
|
| 54 |
+
JPCLN062,JSRT/PNG_data/JPCLN062.png
|
| 55 |
+
JPCLN063,JSRT/PNG_data/JPCLN063.png
|
| 56 |
+
JPCLN065,JSRT/PNG_data/JPCLN065.png
|
| 57 |
+
JPCLN066,JSRT/PNG_data/JPCLN066.png
|
| 58 |
+
JPCLN067,JSRT/PNG_data/JPCLN067.png
|
| 59 |
+
JPCLN068,JSRT/PNG_data/JPCLN068.png
|
| 60 |
+
JPCLN069,JSRT/PNG_data/JPCLN069.png
|
| 61 |
+
JPCLN070,JSRT/PNG_data/JPCLN070.png
|
| 62 |
+
JPCLN072,JSRT/PNG_data/JPCLN072.png
|
| 63 |
+
JPCLN074,JSRT/PNG_data/JPCLN074.png
|
| 64 |
+
JPCLN075,JSRT/PNG_data/JPCLN075.png
|
| 65 |
+
JPCLN076,JSRT/PNG_data/JPCLN076.png
|
| 66 |
+
JPCLN077,JSRT/PNG_data/JPCLN077.png
|
| 67 |
+
JPCLN078,JSRT/PNG_data/JPCLN078.png
|
| 68 |
+
JPCLN079,JSRT/PNG_data/JPCLN079.png
|
| 69 |
+
JPCLN080,JSRT/PNG_data/JPCLN080.png
|
| 70 |
+
JPCLN081,JSRT/PNG_data/JPCLN081.png
|
| 71 |
+
JPCLN082,JSRT/PNG_data/JPCLN082.png
|
| 72 |
+
JPCLN083,JSRT/PNG_data/JPCLN083.png
|
| 73 |
+
JPCLN084,JSRT/PNG_data/JPCLN084.png
|
| 74 |
+
JPCLN085,JSRT/PNG_data/JPCLN085.png
|
| 75 |
+
JPCLN088,JSRT/PNG_data/JPCLN088.png
|
| 76 |
+
JPCLN089,JSRT/PNG_data/JPCLN089.png
|
| 77 |
+
JPCLN090,JSRT/PNG_data/JPCLN090.png
|
| 78 |
+
JPCLN091,JSRT/PNG_data/JPCLN091.png
|
| 79 |
+
JPCLN092,JSRT/PNG_data/JPCLN092.png
|
| 80 |
+
JPCLN093,JSRT/PNG_data/JPCLN093.png
|
| 81 |
+
JPCLN097,JSRT/PNG_data/JPCLN097.png
|
| 82 |
+
JPCLN098,JSRT/PNG_data/JPCLN098.png
|
| 83 |
+
JPCLN100,JSRT/PNG_data/JPCLN100.png
|
| 84 |
+
JPCLN101,JSRT/PNG_data/JPCLN101.png
|
| 85 |
+
JPCLN102,JSRT/PNG_data/JPCLN102.png
|
| 86 |
+
JPCLN103,JSRT/PNG_data/JPCLN103.png
|
| 87 |
+
JPCLN104,JSRT/PNG_data/JPCLN104.png
|
| 88 |
+
JPCLN105,JSRT/PNG_data/JPCLN105.png
|
| 89 |
+
JPCLN108,JSRT/PNG_data/JPCLN108.png
|
| 90 |
+
JPCLN110,JSRT/PNG_data/JPCLN110.png
|
| 91 |
+
JPCLN111,JSRT/PNG_data/JPCLN111.png
|
| 92 |
+
JPCLN112,JSRT/PNG_data/JPCLN112.png
|
| 93 |
+
JPCLN113,JSRT/PNG_data/JPCLN113.png
|
| 94 |
+
JPCLN115,JSRT/PNG_data/JPCLN115.png
|
| 95 |
+
JPCLN116,JSRT/PNG_data/JPCLN116.png
|
| 96 |
+
JPCLN117,JSRT/PNG_data/JPCLN117.png
|
| 97 |
+
JPCLN120,JSRT/PNG_data/JPCLN120.png
|
| 98 |
+
JPCLN121,JSRT/PNG_data/JPCLN121.png
|
| 99 |
+
JPCLN122,JSRT/PNG_data/JPCLN122.png
|
| 100 |
+
JPCLN123,JSRT/PNG_data/JPCLN123.png
|
| 101 |
+
JPCLN124,JSRT/PNG_data/JPCLN124.png
|
| 102 |
+
JPCLN125,JSRT/PNG_data/JPCLN125.png
|
| 103 |
+
JPCLN126,JSRT/PNG_data/JPCLN126.png
|
| 104 |
+
JPCLN127,JSRT/PNG_data/JPCLN127.png
|
| 105 |
+
JPCLN128,JSRT/PNG_data/JPCLN128.png
|
| 106 |
+
JPCLN129,JSRT/PNG_data/JPCLN129.png
|
| 107 |
+
JPCLN132,JSRT/PNG_data/JPCLN132.png
|
| 108 |
+
JPCLN133,JSRT/PNG_data/JPCLN133.png
|
| 109 |
+
JPCLN134,JSRT/PNG_data/JPCLN134.png
|
| 110 |
+
JPCLN135,JSRT/PNG_data/JPCLN135.png
|
| 111 |
+
JPCLN136,JSRT/PNG_data/JPCLN136.png
|
| 112 |
+
JPCLN138,JSRT/PNG_data/JPCLN138.png
|
| 113 |
+
JPCLN139,JSRT/PNG_data/JPCLN139.png
|
| 114 |
+
JPCLN140,JSRT/PNG_data/JPCLN140.png
|
| 115 |
+
JPCLN141,JSRT/PNG_data/JPCLN141.png
|
| 116 |
+
JPCLN142,JSRT/PNG_data/JPCLN142.png
|
| 117 |
+
JPCLN144,JSRT/PNG_data/JPCLN144.png
|
| 118 |
+
JPCLN145,JSRT/PNG_data/JPCLN145.png
|
| 119 |
+
JPCLN147,JSRT/PNG_data/JPCLN147.png
|
| 120 |
+
JPCLN148,JSRT/PNG_data/JPCLN148.png
|
| 121 |
+
JPCLN149,JSRT/PNG_data/JPCLN149.png
|
| 122 |
+
JPCLN150,JSRT/PNG_data/JPCLN150.png
|
| 123 |
+
JPCLN152,JSRT/PNG_data/JPCLN152.png
|
| 124 |
+
JPCLN154,JSRT/PNG_data/JPCLN154.png
|
| 125 |
+
JPCNN001,JSRT/PNG_data/JPCNN001.png
|
| 126 |
+
JPCNN002,JSRT/PNG_data/JPCNN002.png
|
| 127 |
+
JPCNN004,JSRT/PNG_data/JPCNN004.png
|
| 128 |
+
JPCNN006,JSRT/PNG_data/JPCNN006.png
|
| 129 |
+
JPCNN008,JSRT/PNG_data/JPCNN008.png
|
| 130 |
+
JPCNN009,JSRT/PNG_data/JPCNN009.png
|
| 131 |
+
JPCNN010,JSRT/PNG_data/JPCNN010.png
|
| 132 |
+
JPCNN011,JSRT/PNG_data/JPCNN011.png
|
| 133 |
+
JPCNN014,JSRT/PNG_data/JPCNN014.png
|
| 134 |
+
JPCNN015,JSRT/PNG_data/JPCNN015.png
|
| 135 |
+
JPCNN016,JSRT/PNG_data/JPCNN016.png
|
| 136 |
+
JPCNN018,JSRT/PNG_data/JPCNN018.png
|
| 137 |
+
JPCNN019,JSRT/PNG_data/JPCNN019.png
|
| 138 |
+
JPCNN021,JSRT/PNG_data/JPCNN021.png
|
| 139 |
+
JPCNN022,JSRT/PNG_data/JPCNN022.png
|
| 140 |
+
JPCNN023,JSRT/PNG_data/JPCNN023.png
|
| 141 |
+
JPCNN024,JSRT/PNG_data/JPCNN024.png
|
| 142 |
+
JPCNN025,JSRT/PNG_data/JPCNN025.png
|
| 143 |
+
JPCNN026,JSRT/PNG_data/JPCNN026.png
|
| 144 |
+
JPCNN028,JSRT/PNG_data/JPCNN028.png
|
| 145 |
+
JPCNN029,JSRT/PNG_data/JPCNN029.png
|
| 146 |
+
JPCNN030,JSRT/PNG_data/JPCNN030.png
|
| 147 |
+
JPCNN031,JSRT/PNG_data/JPCNN031.png
|
| 148 |
+
JPCNN032,JSRT/PNG_data/JPCNN032.png
|
| 149 |
+
JPCNN033,JSRT/PNG_data/JPCNN033.png
|
| 150 |
+
JPCNN034,JSRT/PNG_data/JPCNN034.png
|
| 151 |
+
JPCNN035,JSRT/PNG_data/JPCNN035.png
|
| 152 |
+
JPCNN037,JSRT/PNG_data/JPCNN037.png
|
| 153 |
+
JPCNN038,JSRT/PNG_data/JPCNN038.png
|
| 154 |
+
JPCNN039,JSRT/PNG_data/JPCNN039.png
|
| 155 |
+
JPCNN040,JSRT/PNG_data/JPCNN040.png
|
| 156 |
+
JPCNN041,JSRT/PNG_data/JPCNN041.png
|
| 157 |
+
JPCNN042,JSRT/PNG_data/JPCNN042.png
|
| 158 |
+
JPCNN043,JSRT/PNG_data/JPCNN043.png
|
| 159 |
+
JPCNN044,JSRT/PNG_data/JPCNN044.png
|
| 160 |
+
JPCNN045,JSRT/PNG_data/JPCNN045.png
|
| 161 |
+
JPCNN046,JSRT/PNG_data/JPCNN046.png
|
| 162 |
+
JPCNN047,JSRT/PNG_data/JPCNN047.png
|
| 163 |
+
JPCNN048,JSRT/PNG_data/JPCNN048.png
|
| 164 |
+
JPCNN049,JSRT/PNG_data/JPCNN049.png
|
| 165 |
+
JPCNN050,JSRT/PNG_data/JPCNN050.png
|
| 166 |
+
JPCNN051,JSRT/PNG_data/JPCNN051.png
|
| 167 |
+
JPCNN052,JSRT/PNG_data/JPCNN052.png
|
| 168 |
+
JPCNN053,JSRT/PNG_data/JPCNN053.png
|
| 169 |
+
JPCNN056,JSRT/PNG_data/JPCNN056.png
|
| 170 |
+
JPCNN059,JSRT/PNG_data/JPCNN059.png
|
| 171 |
+
JPCNN060,JSRT/PNG_data/JPCNN060.png
|
| 172 |
+
JPCNN062,JSRT/PNG_data/JPCNN062.png
|
| 173 |
+
JPCNN063,JSRT/PNG_data/JPCNN063.png
|
| 174 |
+
JPCNN065,JSRT/PNG_data/JPCNN065.png
|
| 175 |
+
JPCNN067,JSRT/PNG_data/JPCNN067.png
|
| 176 |
+
JPCNN068,JSRT/PNG_data/JPCNN068.png
|
| 177 |
+
JPCNN069,JSRT/PNG_data/JPCNN069.png
|
| 178 |
+
JPCNN070,JSRT/PNG_data/JPCNN070.png
|
| 179 |
+
JPCNN071,JSRT/PNG_data/JPCNN071.png
|
| 180 |
+
JPCNN072,JSRT/PNG_data/JPCNN072.png
|
| 181 |
+
JPCNN073,JSRT/PNG_data/JPCNN073.png
|
| 182 |
+
JPCNN074,JSRT/PNG_data/JPCNN074.png
|
| 183 |
+
JPCNN075,JSRT/PNG_data/JPCNN075.png
|
| 184 |
+
JPCNN076,JSRT/PNG_data/JPCNN076.png
|
| 185 |
+
JPCNN077,JSRT/PNG_data/JPCNN077.png
|
| 186 |
+
JPCNN078,JSRT/PNG_data/JPCNN078.png
|
| 187 |
+
JPCNN079,JSRT/PNG_data/JPCNN079.png
|
| 188 |
+
JPCNN080,JSRT/PNG_data/JPCNN080.png
|
| 189 |
+
JPCNN082,JSRT/PNG_data/JPCNN082.png
|
| 190 |
+
JPCNN083,JSRT/PNG_data/JPCNN083.png
|
| 191 |
+
JPCNN084,JSRT/PNG_data/JPCNN084.png
|
| 192 |
+
JPCNN085,JSRT/PNG_data/JPCNN085.png
|
| 193 |
+
JPCNN086,JSRT/PNG_data/JPCNN086.png
|
| 194 |
+
JPCNN087,JSRT/PNG_data/JPCNN087.png
|
| 195 |
+
JPCNN088,JSRT/PNG_data/JPCNN088.png
|
| 196 |
+
JPCNN090,JSRT/PNG_data/JPCNN090.png
|
| 197 |
+
JPCNN091,JSRT/PNG_data/JPCNN091.png
|
| 198 |
+
JPCNN092,JSRT/PNG_data/JPCNN092.png
|
data/JSRT_val_split.csv
ADDED
|
@@ -0,0 +1,26 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
id,path
|
| 2 |
+
JPCLN028,JSRT/PNG_data/JPCLN028.png
|
| 3 |
+
JPCNN005,JSRT/PNG_data/JPCNN005.png
|
| 4 |
+
JPCNN013,JSRT/PNG_data/JPCNN013.png
|
| 5 |
+
JPCLN064,JSRT/PNG_data/JPCLN064.png
|
| 6 |
+
JPCLN055,JSRT/PNG_data/JPCLN055.png
|
| 7 |
+
JPCNN054,JSRT/PNG_data/JPCNN054.png
|
| 8 |
+
JPCLN096,JSRT/PNG_data/JPCLN096.png
|
| 9 |
+
JPCLN099,JSRT/PNG_data/JPCLN099.png
|
| 10 |
+
JPCNN064,JSRT/PNG_data/JPCNN064.png
|
| 11 |
+
JPCLN030,JSRT/PNG_data/JPCLN030.png
|
| 12 |
+
JPCNN057,JSRT/PNG_data/JPCNN057.png
|
| 13 |
+
JPCLN094,JSRT/PNG_data/JPCLN094.png
|
| 14 |
+
JPCLN087,JSRT/PNG_data/JPCLN087.png
|
| 15 |
+
JPCNN012,JSRT/PNG_data/JPCNN012.png
|
| 16 |
+
JPCNN061,JSRT/PNG_data/JPCNN061.png
|
| 17 |
+
JPCLN071,JSRT/PNG_data/JPCLN071.png
|
| 18 |
+
JPCLN119,JSRT/PNG_data/JPCLN119.png
|
| 19 |
+
JPCNN027,JSRT/PNG_data/JPCNN027.png
|
| 20 |
+
JPCLN037,JSRT/PNG_data/JPCLN037.png
|
| 21 |
+
JPCLN045,JSRT/PNG_data/JPCLN045.png
|
| 22 |
+
JPCNN066,JSRT/PNG_data/JPCNN066.png
|
| 23 |
+
JPCNN003,JSRT/PNG_data/JPCNN003.png
|
| 24 |
+
JPCNN058,JSRT/PNG_data/JPCNN058.png
|
| 25 |
+
JPCNN036,JSRT/PNG_data/JPCNN036.png
|
| 26 |
+
JPCLN106,JSRT/PNG_data/JPCLN106.png
|
data/correspondence_with_chestXray8.csv
ADDED
|
@@ -0,0 +1,101 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
NIH,ChestX-ray14,scan,mask
|
| 2 |
+
NIH_0010,00009863_008.png,images/NIH_0010.png,masks/NIH_0010_mask.png
|
| 3 |
+
NIH_0017,00003028_078.png,images/NIH_0017.png,masks/NIH_0017_mask.png
|
| 4 |
+
NIH_0065,00012045_005.png,images/NIH_0065.png,masks/NIH_0065_mask.png
|
| 5 |
+
NIH_0019,00029481_008.png,images/NIH_0019.png,masks/NIH_0019_mask.png
|
| 6 |
+
NIH_0062,00010805_040.png,images/NIH_0062.png,masks/NIH_0062_mask.png
|
| 7 |
+
NIH_0057,00011950_019.png,images/NIH_0057.png,masks/NIH_0057_mask.png
|
| 8 |
+
NIH_0086,00004648_001.png,images/NIH_0086.png,masks/NIH_0086_mask.png
|
| 9 |
+
NIH_0050,00019499_006.png,images/NIH_0050.png,masks/NIH_0050_mask.png
|
| 10 |
+
NIH_0081,00022572_048.png,images/NIH_0081.png,masks/NIH_0081_mask.png
|
| 11 |
+
NIH_0022,00014398_023.png,images/NIH_0022.png,masks/NIH_0022_mask.png
|
| 12 |
+
NIH_0025,00017753_003.png,images/NIH_0025.png,masks/NIH_0025_mask.png
|
| 13 |
+
NIH_0059,00020289_004.png,images/NIH_0059.png,masks/NIH_0059_mask.png
|
| 14 |
+
NIH_0088,00004832_025.png,images/NIH_0088.png,masks/NIH_0088_mask.png
|
| 15 |
+
NIH_0034,00009863_038.png,images/NIH_0034.png,masks/NIH_0034_mask.png
|
| 16 |
+
NIH_0099,00018840_034.png,images/NIH_0099.png,masks/NIH_0099_mask.png
|
| 17 |
+
NIH_0048,00018984_001.png,images/NIH_0048.png,masks/NIH_0048_mask.png
|
| 18 |
+
NIH_0033,00009863_020.png,images/NIH_0033.png,masks/NIH_0033_mask.png
|
| 19 |
+
NIH_0090,00009218_026.png,images/NIH_0090.png,masks/NIH_0090_mask.png
|
| 20 |
+
NIH_0041,00010315_006.png,images/NIH_0041.png,masks/NIH_0041_mask.png
|
| 21 |
+
NIH_0097,00018610_041.png,images/NIH_0097.png,masks/NIH_0097_mask.png
|
| 22 |
+
NIH_0046,00007444_004.png,images/NIH_0046.png,masks/NIH_0046_mask.png
|
| 23 |
+
NIH_0073,00002227_003.png,images/NIH_0073.png,masks/NIH_0073_mask.png
|
| 24 |
+
NIH_0074,00003538_000.png,images/NIH_0074.png,masks/NIH_0074_mask.png
|
| 25 |
+
NIH_0008,00010007_157.png,images/NIH_0008.png,masks/NIH_0008_mask.png
|
| 26 |
+
NIH_0006,00014004_048.png,images/NIH_0006.png,masks/NIH_0006_mask.png
|
| 27 |
+
NIH_0001,00005502_010.png,images/NIH_0001.png,masks/NIH_0001_mask.png
|
| 28 |
+
NIH_0024,00027441_015.png,images/NIH_0024.png,masks/NIH_0024_mask.png
|
| 29 |
+
NIH_0100,00008943_002.png,images/NIH_0100.png,masks/NIH_0100_mask.png
|
| 30 |
+
NIH_0058,00017470_004.png,images/NIH_0058.png,masks/NIH_0058_mask.png
|
| 31 |
+
NIH_0089,00008237_002.png,images/NIH_0089.png,masks/NIH_0089_mask.png
|
| 32 |
+
NIH_0023,00001483_013.png,images/NIH_0023.png,masks/NIH_0023_mask.png
|
| 33 |
+
NIH_0051,00013922_028.png,images/NIH_0051.png,masks/NIH_0051_mask.png
|
| 34 |
+
NIH_0080,00022034_002.png,images/NIH_0080.png,masks/NIH_0080_mask.png
|
| 35 |
+
NIH_0056,00004832_031.png,images/NIH_0056.png,masks/NIH_0056_mask.png
|
| 36 |
+
NIH_0087,00021449_002.png,images/NIH_0087.png,masks/NIH_0087_mask.png
|
| 37 |
+
NIH_0063,00029855_001.png,images/NIH_0063.png,masks/NIH_0063_mask.png
|
| 38 |
+
NIH_0064,00005593_011.png,images/NIH_0064.png,masks/NIH_0064_mask.png
|
| 39 |
+
NIH_0018,00021154_005.png,images/NIH_0018.png,masks/NIH_0018_mask.png
|
| 40 |
+
NIH_0016,00017214_015.png,images/NIH_0016.png,masks/NIH_0016_mask.png
|
| 41 |
+
NIH_0011,00001248_026.png,images/NIH_0011.png,masks/NIH_0011_mask.png
|
| 42 |
+
NIH_0007,00017110_010.png,images/NIH_0007.png,masks/NIH_0007_mask.png
|
| 43 |
+
NIH_0075,00014177_017.png,images/NIH_0075.png,masks/NIH_0075_mask.png
|
| 44 |
+
NIH_0009,00009479_002.png,images/NIH_0009.png,masks/NIH_0009_mask.png
|
| 45 |
+
NIH_0072,00020408_067.png,images/NIH_0072.png,masks/NIH_0072_mask.png
|
| 46 |
+
NIH_0096,00000997_004.png,images/NIH_0096.png,masks/NIH_0096_mask.png
|
| 47 |
+
NIH_0047,00000643_003.png,images/NIH_0047.png,masks/NIH_0047_mask.png
|
| 48 |
+
NIH_0091,00013249_056.png,images/NIH_0091.png,masks/NIH_0091_mask.png
|
| 49 |
+
NIH_0040,00029154_000.png,images/NIH_0040.png,masks/NIH_0040_mask.png
|
| 50 |
+
NIH_0032,00017801_003.png,images/NIH_0032.png,masks/NIH_0032_mask.png
|
| 51 |
+
NIH_0035,00010352_021.png,images/NIH_0035.png,masks/NIH_0035_mask.png
|
| 52 |
+
NIH_0098,00003510_006.png,images/NIH_0098.png,masks/NIH_0098_mask.png
|
| 53 |
+
NIH_0049,00002239_007.png,images/NIH_0049.png,masks/NIH_0049_mask.png
|
| 54 |
+
NIH_0078,00019176_098.png,images/NIH_0078.png,masks/NIH_0078_mask.png
|
| 55 |
+
NIH_0004,00004342_053.png,images/NIH_0004.png,masks/NIH_0004_mask.png
|
| 56 |
+
NIH_0003,00012364_037.png,images/NIH_0003.png,masks/NIH_0003_mask.png
|
| 57 |
+
NIH_0071,00011702_012.png,images/NIH_0071.png,masks/NIH_0071_mask.png
|
| 58 |
+
NIH_0076,00006481_023.png,images/NIH_0076.png,masks/NIH_0076_mask.png
|
| 59 |
+
NIH_0092,00006322_001.png,images/NIH_0092.png,masks/NIH_0092_mask.png
|
| 60 |
+
NIH_0043,00013760_000.png,images/NIH_0043.png,masks/NIH_0043_mask.png
|
| 61 |
+
NIH_0038,00021341_012.png,images/NIH_0038.png,masks/NIH_0038_mask.png
|
| 62 |
+
NIH_0095,00026908_003.png,images/NIH_0095.png,masks/NIH_0095_mask.png
|
| 63 |
+
NIH_0044,00008037_002.png,images/NIH_0044.png,masks/NIH_0044_mask.png
|
| 64 |
+
NIH_0036,00030772_002.png,images/NIH_0036.png,masks/NIH_0036_mask.png
|
| 65 |
+
NIH_0031,00023073_000.png,images/NIH_0031.png,masks/NIH_0031_mask.png
|
| 66 |
+
NIH_0020,00021420_000.png,images/NIH_0020.png,masks/NIH_0020_mask.png
|
| 67 |
+
NIH_0027,00010684_007.png,images/NIH_0027.png,masks/NIH_0027_mask.png
|
| 68 |
+
NIH_0029,00030573_003.png,images/NIH_0029.png,masks/NIH_0029_mask.png
|
| 69 |
+
NIH_0055,00025513_001.png,images/NIH_0055.png,masks/NIH_0055_mask.png
|
| 70 |
+
NIH_0084,00001684_025.png,images/NIH_0084.png,masks/NIH_0084_mask.png
|
| 71 |
+
NIH_0052,00011543_017.png,images/NIH_0052.png,masks/NIH_0052_mask.png
|
| 72 |
+
NIH_0083,00010773_008.png,images/NIH_0083.png,masks/NIH_0083_mask.png
|
| 73 |
+
NIH_0067,00015443_017.png,images/NIH_0067.png,masks/NIH_0067_mask.png
|
| 74 |
+
NIH_0060,00002395_015.png,images/NIH_0060.png,masks/NIH_0060_mask.png
|
| 75 |
+
NIH_0012,00013613_025.png,images/NIH_0012.png,masks/NIH_0012_mask.png
|
| 76 |
+
NIH_0069,00010680_001.png,images/NIH_0069.png,masks/NIH_0069_mask.png
|
| 77 |
+
NIH_0015,00004156_004.png,images/NIH_0015.png,masks/NIH_0015_mask.png
|
| 78 |
+
NIH_0030,00016508_004.png,images/NIH_0030.png,masks/NIH_0030_mask.png
|
| 79 |
+
NIH_0037,00016291_038.png,images/NIH_0037.png,masks/NIH_0037_mask.png
|
| 80 |
+
NIH_0039,00025822_005.png,images/NIH_0039.png,masks/NIH_0039_mask.png
|
| 81 |
+
NIH_0094,00002386_003.png,images/NIH_0094.png,masks/NIH_0094_mask.png
|
| 82 |
+
NIH_0045,00001317_001.png,images/NIH_0045.png,masks/NIH_0045_mask.png
|
| 83 |
+
NIH_0093,00025954_030.png,images/NIH_0093.png,masks/NIH_0093_mask.png
|
| 84 |
+
NIH_0042,00009945_006.png,images/NIH_0042.png,masks/NIH_0042_mask.png
|
| 85 |
+
NIH_0077,00013993_068.png,images/NIH_0077.png,masks/NIH_0077_mask.png
|
| 86 |
+
NIH_0070,00013527_000.png,images/NIH_0070.png,masks/NIH_0070_mask.png
|
| 87 |
+
NIH_0002,00002350_021.png,images/NIH_0002.png,masks/NIH_0002_mask.png
|
| 88 |
+
NIH_0079,00007576_043.png,images/NIH_0079.png,masks/NIH_0079_mask.png
|
| 89 |
+
NIH_0005,00015443_014.png,images/NIH_0005.png,masks/NIH_0005_mask.png
|
| 90 |
+
NIH_0068,00025839_008.png,images/NIH_0068.png,masks/NIH_0068_mask.png
|
| 91 |
+
NIH_0014,00014626_026.png,images/NIH_0014.png,masks/NIH_0014_mask.png
|
| 92 |
+
NIH_0013,00018080_006.png,images/NIH_0013.png,masks/NIH_0013_mask.png
|
| 93 |
+
NIH_0061,00009114_009.png,images/NIH_0061.png,masks/NIH_0061_mask.png
|
| 94 |
+
NIH_0066,00018610_038.png,images/NIH_0066.png,masks/NIH_0066_mask.png
|
| 95 |
+
NIH_0053,00000063_000.png,images/NIH_0053.png,masks/NIH_0053_mask.png
|
| 96 |
+
NIH_0082,00009138_028.png,images/NIH_0082.png,masks/NIH_0082_mask.png
|
| 97 |
+
NIH_0028,00006498_003.png,images/NIH_0028.png,masks/NIH_0028_mask.png
|
| 98 |
+
NIH_0054,00006620_000.png,images/NIH_0054.png,masks/NIH_0054_mask.png
|
| 99 |
+
NIH_0085,00005288_013.png,images/NIH_0085.png,masks/NIH_0085_mask.png
|
| 100 |
+
NIH_0026,00001836_069.png,images/NIH_0026.png,masks/NIH_0026_mask.png
|
| 101 |
+
NIH_0021,00006679_018.png,images/NIH_0021.png,masks/NIH_0021_mask.png
|
data/test_split.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
data/train_split.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
data/val_split.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
dataloaders/CXR14.py
ADDED
|
@@ -0,0 +1,74 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import logging
|
| 2 |
+
import os
|
| 3 |
+
from typing import List, Tuple, TypeVar
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import torch
|
| 6 |
+
from PIL import Image
|
| 7 |
+
from torch.utils.data import Dataset, DataLoader
|
| 8 |
+
from torch import Tensor
|
| 9 |
+
from torchvision import transforms
|
| 10 |
+
from pathlib import Path
|
| 11 |
+
|
| 12 |
+
PathLike = TypeVar("PathLike", str, Path, None)
|
| 13 |
+
log = logging.getLogger(__name__)
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
PROJECT_DIR = Path(os.path.realpath(__file__)).parent.parent
|
| 17 |
+
DATADIR = Path("<PATH_TO_DATA>/ChestXray-NIHCC/images")
|
| 18 |
+
# can be found at https://nihcc.app.box.com/v/ChestXray-NIHCC/folder/36938765345
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
def build_dataloaders(
|
| 22 |
+
data_dir: str=DATADIR,
|
| 23 |
+
img_size: int=128,
|
| 24 |
+
batch_size: int=16,
|
| 25 |
+
num_workers: int=1,
|
| 26 |
+
) -> Tuple[List, List, List]:
|
| 27 |
+
"""
|
| 28 |
+
Build dataloaders for the CXR14 dataset.
|
| 29 |
+
"""
|
| 30 |
+
train_ds = CXR14Dataset(data_dir, PROJECT_DIR / 'data' / 'train_split.csv', img_size)
|
| 31 |
+
val_ds = CXR14Dataset(data_dir, PROJECT_DIR / 'data' / 'train_split.csv', img_size)
|
| 32 |
+
test_ds = CXR14Dataset(data_dir, PROJECT_DIR / 'data' / 'train_split.csv', img_size)
|
| 33 |
+
|
| 34 |
+
dataloaders = {}
|
| 35 |
+
dataloaders['train'] = DataLoader(train_ds, batch_size=batch_size,
|
| 36 |
+
shuffle=True, num_workers=num_workers,
|
| 37 |
+
pin_memory=True)
|
| 38 |
+
dataloaders['val'] = DataLoader(val_ds, batch_size=batch_size,
|
| 39 |
+
shuffle=False, num_workers=num_workers,
|
| 40 |
+
pin_memory=True)
|
| 41 |
+
dataloaders['test'] = DataLoader(test_ds, batch_size=batch_size,
|
| 42 |
+
shuffle=False, num_workers=num_workers,
|
| 43 |
+
pin_memory=True)
|
| 44 |
+
|
| 45 |
+
return dataloaders
|
| 46 |
+
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
class CXR14Dataset(Dataset):
|
| 50 |
+
def __init__(
|
| 51 |
+
self,
|
| 52 |
+
data_path: PathLike,
|
| 53 |
+
csv_path: PathLike,
|
| 54 |
+
img_size: int,
|
| 55 |
+
) -> None:
|
| 56 |
+
super().__init__()
|
| 57 |
+
assert(os.path.isdir(data_path))
|
| 58 |
+
assert(os.path.isfile(csv_path))
|
| 59 |
+
|
| 60 |
+
self.data_path = Path(data_path)
|
| 61 |
+
self.df = pd.read_csv(csv_path)
|
| 62 |
+
self.img_size = img_size
|
| 63 |
+
|
| 64 |
+
def __len__(self) -> int:
|
| 65 |
+
return len(self.df)
|
| 66 |
+
|
| 67 |
+
def load_image(self, fname: str) -> Tensor:
|
| 68 |
+
img = Image.open(self.data_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 69 |
+
img = transforms.ToTensor()(img).float()
|
| 70 |
+
return img
|
| 71 |
+
|
| 72 |
+
def __getitem__(self, index) -> Tuple[Tensor, Tensor]:
|
| 73 |
+
img = self.load_image(self.df.loc[index, "Image Index"])
|
| 74 |
+
return img
|
dataloaders/JSRT.py
ADDED
|
@@ -0,0 +1,94 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from torch.utils.data import Dataset, DataLoader
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from typing import List, Tuple, TypeVar, Optional
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import os
|
| 6 |
+
from PIL import Image
|
| 7 |
+
from torch import Tensor
|
| 8 |
+
from torchvision import transforms
|
| 9 |
+
import torch
|
| 10 |
+
|
| 11 |
+
PathLike = TypeVar("PathLike", str, Path, None)
|
| 12 |
+
|
| 13 |
+
PROJECT_DIR = Path(os.path.realpath(__file__)).parent.parent
|
| 14 |
+
DATADIR = Path("<PATH_TO_DATA>/JSRT")
|
| 15 |
+
# can be found at http://db.jsrt.or.jp/eng.php
|
| 16 |
+
|
| 17 |
+
def build_dataloaders(
|
| 18 |
+
data_dir: str=DATADIR,
|
| 19 |
+
img_size: int=128,
|
| 20 |
+
batch_size: int=16,
|
| 21 |
+
num_workers: int=1,
|
| 22 |
+
n_labelled_images: Optional[int] = None,
|
| 23 |
+
**kwargs
|
| 24 |
+
) -> Tuple[List, List, List]:
|
| 25 |
+
"""
|
| 26 |
+
Build dataloaders for the JSRT dataset.
|
| 27 |
+
"""
|
| 28 |
+
train_ds = JSRTDataset(data_dir, PROJECT_DIR / "data", "JSRT_train_split.csv", img_size)
|
| 29 |
+
if n_labelled_images is not None:
|
| 30 |
+
train_ds = torch.utils.data.Subset(train_ds, range(n_labelled_images))
|
| 31 |
+
print(f"Using {n_labelled_images} labelled images")
|
| 32 |
+
val_ds = JSRTDataset(data_dir, PROJECT_DIR / "data", "JSRT_val_split.csv", img_size)
|
| 33 |
+
test_ds = JSRTDataset(data_dir, PROJECT_DIR / "data", "JSRT_test_split.csv", img_size)
|
| 34 |
+
|
| 35 |
+
dataloaders = {}
|
| 36 |
+
dataloaders['train'] = DataLoader(train_ds, batch_size=batch_size,
|
| 37 |
+
shuffle=True, num_workers=num_workers,
|
| 38 |
+
pin_memory=True)
|
| 39 |
+
dataloaders['val'] = DataLoader(val_ds, batch_size=batch_size,
|
| 40 |
+
shuffle=False, num_workers=num_workers,
|
| 41 |
+
pin_memory=True)
|
| 42 |
+
dataloaders['test'] = DataLoader(test_ds, batch_size=batch_size,
|
| 43 |
+
shuffle=False, num_workers=num_workers,
|
| 44 |
+
pin_memory=True)
|
| 45 |
+
|
| 46 |
+
return dataloaders
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
class JSRTDataset(Dataset):
|
| 50 |
+
def __init__(self, base_path:PathLike,
|
| 51 |
+
csv_path:PathLike,
|
| 52 |
+
csv_name:str,
|
| 53 |
+
img_size:int=128,
|
| 54 |
+
labels:List[str] =('right lung', 'left lung', ),
|
| 55 |
+
**kwargs) -> None:
|
| 56 |
+
self.df = pd.read_csv(os.path.join(csv_path, csv_name))
|
| 57 |
+
self.base_path = Path(base_path)
|
| 58 |
+
self.labels = labels
|
| 59 |
+
self.img_size = img_size
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
def load_image(self, fname: str) -> Tensor:
|
| 63 |
+
img = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 64 |
+
img = transforms.ToTensor()(img).float()
|
| 65 |
+
return img
|
| 66 |
+
|
| 67 |
+
def load_labels(self, fnames: List[str]) -> Tensor:
|
| 68 |
+
labels = []
|
| 69 |
+
for fname in fnames:
|
| 70 |
+
label = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 71 |
+
# convert to tensor
|
| 72 |
+
label = transforms.ToTensor()(label).float()
|
| 73 |
+
# make binary
|
| 74 |
+
label = (label > .5).float()
|
| 75 |
+
labels.append(label)
|
| 76 |
+
# append all labels and merge
|
| 77 |
+
label = torch.stack(labels).sum(0)
|
| 78 |
+
# lungs have no overlap (right?)
|
| 79 |
+
if (label > 1).sum()>0:
|
| 80 |
+
print("overlapping lungs!", fnames)
|
| 81 |
+
label = (label > .5)
|
| 82 |
+
return label
|
| 83 |
+
|
| 84 |
+
def __getitem__(self, index) -> Tuple[Tensor, Tensor]:
|
| 85 |
+
i = self.df.index[index]
|
| 86 |
+
img = self.load_image(self.df.loc[i, "path"])
|
| 87 |
+
|
| 88 |
+
label_paths = ["SCR/masks/" + item + "/" + self.df.loc[i, 'id']+ ".gif" for item in self.labels]
|
| 89 |
+
labels = self.load_labels(label_paths)
|
| 90 |
+
|
| 91 |
+
return img, labels
|
| 92 |
+
|
| 93 |
+
def __len__(self):
|
| 94 |
+
return len(self.df)
|
dataloaders/Montgomery.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from torch.utils.data import Dataset, DataLoader
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from typing import List, Tuple, TypeVar, Optional
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import os
|
| 6 |
+
from PIL import Image
|
| 7 |
+
from torch import Tensor
|
| 8 |
+
from torchvision import transforms
|
| 9 |
+
import torch
|
| 10 |
+
|
| 11 |
+
PathLike = TypeVar("PathLike", str, Path, None)
|
| 12 |
+
# can be found at https://data.lhncbc.nlm.nih.gov/public/Tuberculosis-Chest-X-ray-Datasets/Montgomery-County-CXR-Set/MontgomerySet/index.html
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
class MonDataset(Dataset):
|
| 16 |
+
def __init__(self, base_path:PathLike,
|
| 17 |
+
csv_path:PathLike,
|
| 18 |
+
csv_name:str,
|
| 19 |
+
img_size:int=128,
|
| 20 |
+
labels:List[str] =('right lung', 'left lung', ),
|
| 21 |
+
**kwargs) -> None:
|
| 22 |
+
self.df = pd.read_csv(os.path.join(csv_path, csv_name))
|
| 23 |
+
self.base_path = Path(base_path)
|
| 24 |
+
self.labels = labels
|
| 25 |
+
self.img_size = img_size
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
def load_image(self, fname: str) -> Tensor:
|
| 29 |
+
img = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 30 |
+
img = transforms.ToTensor()(img).float()
|
| 31 |
+
return img
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
def load_labels(self, fnames: List[str]) -> Tensor:
|
| 35 |
+
labels = []
|
| 36 |
+
for fname in fnames:
|
| 37 |
+
label = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 38 |
+
# convert to tensor
|
| 39 |
+
label = transforms.ToTensor()(label).float()
|
| 40 |
+
# make binary
|
| 41 |
+
label = (label > .5).float()
|
| 42 |
+
labels.append(label)
|
| 43 |
+
# append all labels and merge
|
| 44 |
+
label = torch.stack(labels).sum(0)
|
| 45 |
+
# lungs have no overlap (right?)
|
| 46 |
+
if (label > 1).sum()>0:
|
| 47 |
+
print("overlapping lungs!", fnames)
|
| 48 |
+
label = (label > .5)
|
| 49 |
+
return label
|
| 50 |
+
|
| 51 |
+
def __getitem__(self, index) -> Tuple[Tensor, Tensor]:
|
| 52 |
+
i = self.df.index[index]
|
| 53 |
+
img = self.load_image(self.df.loc[i, "scan"])
|
| 54 |
+
|
| 55 |
+
fnames = [ self.df.loc[i, l] for l in self.labels]
|
| 56 |
+
labels = self.load_labels(fnames)
|
| 57 |
+
|
| 58 |
+
return img, labels
|
| 59 |
+
|
| 60 |
+
def __len__(self) -> int:
|
| 61 |
+
return len(self.df)
|
dataloaders/NIH.py
ADDED
|
@@ -0,0 +1,50 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from torch.utils.data import Dataset
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from typing import List, Tuple, TypeVar
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import os
|
| 6 |
+
from PIL import Image
|
| 7 |
+
from torch import Tensor
|
| 8 |
+
from torchvision import transforms
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
PathLike = TypeVar("PathLike", str, Path, None)
|
| 12 |
+
# can be found at https://www.kaggle.com/datasets/nih-chest-xrays/data
|
| 13 |
+
|
| 14 |
+
class NIHDataset(Dataset):
|
| 15 |
+
def __init__(self, base_path:PathLike,
|
| 16 |
+
csv_path:PathLike,
|
| 17 |
+
csv_name:str,
|
| 18 |
+
img_size:int=128,
|
| 19 |
+
labels:List[str] =('right lung', 'left lung', ),
|
| 20 |
+
**kwargs) -> None:
|
| 21 |
+
self.df = pd.read_csv(os.path.join(csv_path, csv_name))
|
| 22 |
+
self.base_path = Path(base_path)
|
| 23 |
+
self.labels = labels
|
| 24 |
+
self.img_size = img_size
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
def load_image(self, fname: str) -> Tensor:
|
| 28 |
+
img = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 29 |
+
img = transforms.ToTensor()(img).float()
|
| 30 |
+
return img
|
| 31 |
+
|
| 32 |
+
def load_labels(self, fname: str) -> Tensor:
|
| 33 |
+
|
| 34 |
+
label = Image.open(self.base_path /fname).convert('L').resize((self.img_size, self.img_size))
|
| 35 |
+
# convert to tensor
|
| 36 |
+
label = transforms.ToTensor()(label).float()
|
| 37 |
+
# make binary
|
| 38 |
+
label = (label > .5).float()
|
| 39 |
+
|
| 40 |
+
return label
|
| 41 |
+
|
| 42 |
+
def __getitem__(self, index) -> Tuple[Tensor, Tensor]:
|
| 43 |
+
i = self.df.index[index]
|
| 44 |
+
img = self.load_image(self.df.loc[i, "scan"])
|
| 45 |
+
labels = self.load_labels(self.df.loc[i, "mask"])
|
| 46 |
+
|
| 47 |
+
return img, labels
|
| 48 |
+
|
| 49 |
+
def __len__(self):
|
| 50 |
+
return len(self.df)
|
img_examples/00015548_000.png
ADDED
|
img_examples/00016568_041.png
ADDED
|
img_examples/NIH_0006.png
ADDED
|
img_examples/NIH_0012.png
ADDED
|
img_examples/NIH_0014.png
ADDED
|
img_examples/NIH_0019.png
ADDED
|
img_examples/NIH_0024.png
ADDED
|
img_examples/NIH_0035.png
ADDED
|
img_examples/NIH_0051.png
ADDED
|
img_examples/NIH_0055.png
ADDED
|
img_examples/NIH_0076.png
ADDED
|
img_examples/NIH_0094.png
ADDED
|
img_examples/TEDM-model-visualisation.png
ADDED
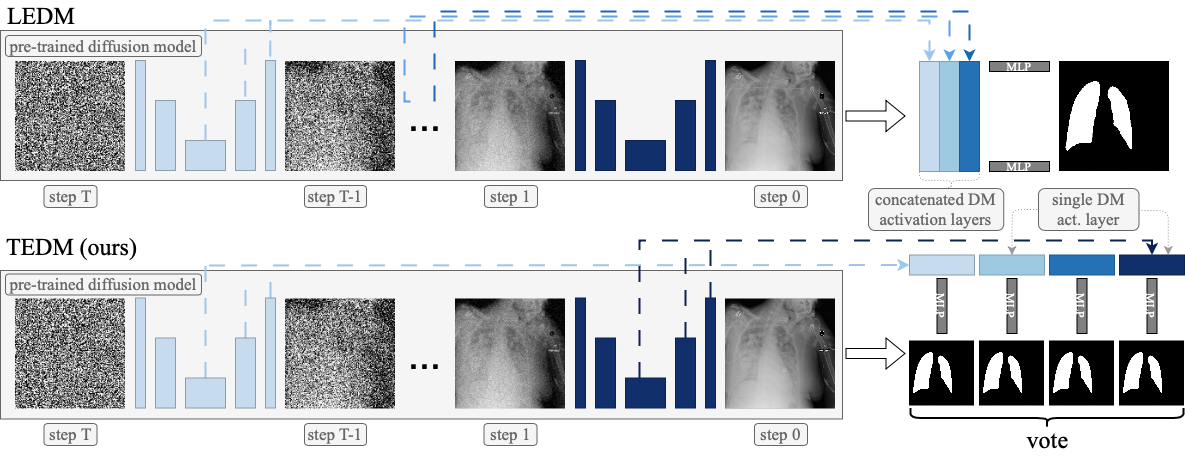
|
models/datasetDM_model.py
ADDED
|
@@ -0,0 +1,88 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import torch
|
| 3 |
+
from torch import nn, Tensor
|
| 4 |
+
from typing import Dict, Tuple, Optional
|
| 5 |
+
from argparse import Namespace
|
| 6 |
+
from einops import repeat
|
| 7 |
+
from einops.layers.torch import Rearrange
|
| 8 |
+
from functools import partial
|
| 9 |
+
from models.diffusion_model import DiffusionModel
|
| 10 |
+
from trainers.utils import compare_configs
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
# Hooks code inspired by https://www.lyndonduong.com/saving-activations/
|
| 15 |
+
# Accessed on 13Feb23
|
| 16 |
+
def save_activations(
|
| 17 |
+
activations: Dict,
|
| 18 |
+
name: str,
|
| 19 |
+
module: nn.Module,
|
| 20 |
+
inp: Tuple,
|
| 21 |
+
out: torch.Tensor
|
| 22 |
+
) -> None:
|
| 23 |
+
"""PyTorch Forward hook to save outputs at each forward
|
| 24 |
+
pass. Mutates specified dict objects with each fwd pass.
|
| 25 |
+
"""
|
| 26 |
+
#activations[name].append(out.detach().cpu())
|
| 27 |
+
activations[name] = out.detach().cpu()
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
class DatasetDM(nn.Module):
|
| 31 |
+
def __init__(self, args: Namespace) -> None:
|
| 32 |
+
super().__init__()
|
| 33 |
+
# Load the model
|
| 34 |
+
if not os.path.isfile(args.saved_diffusion_model):
|
| 35 |
+
self.diffusion_model = DiffusionModel(args)
|
| 36 |
+
if args.verbose:
|
| 37 |
+
print(f'No model found at {args.saved_diffusion_model}. Please load model!')
|
| 38 |
+
else:
|
| 39 |
+
checkpoint = torch.load(args.saved_diffusion_model, map_location=torch.device(args.device))
|
| 40 |
+
old_config = checkpoint['config']
|
| 41 |
+
compare_configs(old_config, args)
|
| 42 |
+
self.diffusion_model = DiffusionModel(old_config)
|
| 43 |
+
self.diffusion_model.load_state_dict(checkpoint['model_state_dict'])
|
| 44 |
+
self.diffusion_model.eval()
|
| 45 |
+
|
| 46 |
+
# storage for saved activations
|
| 47 |
+
self._features = {}
|
| 48 |
+
|
| 49 |
+
# Note that this only works for the model in model.py
|
| 50 |
+
for i, (block1, block2, attn, upsample) in enumerate(self.diffusion_model.model.ups):
|
| 51 |
+
attn.register_forward_hook(
|
| 52 |
+
partial(save_activations, self._features, i)
|
| 53 |
+
)
|
| 54 |
+
|
| 55 |
+
self.steps = args.t_steps_to_save
|
| 56 |
+
|
| 57 |
+
self.classifier = nn.Sequential(
|
| 58 |
+
nn.Conv2d(960 * len(self.steps), 128, 1),
|
| 59 |
+
nn.ReLU(),
|
| 60 |
+
nn.BatchNorm2d(128),
|
| 61 |
+
nn.Conv2d(128, 32, 1),
|
| 62 |
+
nn.ReLU(),
|
| 63 |
+
nn.BatchNorm2d(32),
|
| 64 |
+
nn.Conv2d(32, 1, 1))
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
@torch.no_grad()
|
| 68 |
+
def extract_features(self, x_0: Tensor, noise: Optional[Tensor] = None) -> Dict[int, Tensor]:
|
| 69 |
+
if noise is not None:
|
| 70 |
+
assert(x_0.shape == noise.shape)
|
| 71 |
+
activations=[]
|
| 72 |
+
for t_step in self.steps:
|
| 73 |
+
# Add t_steps of noise to x_0 - forward process
|
| 74 |
+
t_step = torch.Tensor([t_step]).long().to(x_0.device)
|
| 75 |
+
t_step = repeat(t_step, '1 -> b', b=x_0.shape[0])
|
| 76 |
+
x_t, _ = self.diffusion_model.forward_diffusion_model(x_0=x_0, t=t_step, noise=noise)
|
| 77 |
+
# Remove one step of noise from x_t - backward process
|
| 78 |
+
_ = self.diffusion_model.model(x_t, t_step)
|
| 79 |
+
# Resize features so that they all live in the image space
|
| 80 |
+
for idx in self._features:
|
| 81 |
+
activations.append(nn.functional.interpolate(self._features[idx], size=[x_0.shape[-1]] * 2))
|
| 82 |
+
# Return activations
|
| 83 |
+
return torch.cat(activations, dim=1)
|
| 84 |
+
|
| 85 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 86 |
+
features = self.extract_features(x).to(x.device)
|
| 87 |
+
out = self.classifier(features)
|
| 88 |
+
return out
|
models/diffusion_model.py
ADDED
|
@@ -0,0 +1,301 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""Adapted from https://github.com/lucidrains/denoising-diffusion-pytorch"""
|
| 2 |
+
from argparse import Namespace
|
| 3 |
+
import math
|
| 4 |
+
from typing import List, Tuple, Optional
|
| 5 |
+
|
| 6 |
+
import torch
|
| 7 |
+
import torch.nn.functional as F
|
| 8 |
+
|
| 9 |
+
from einops import reduce, rearrange
|
| 10 |
+
from torch import nn, Tensor
|
| 11 |
+
|
| 12 |
+
from models.unet_model import Unet
|
| 13 |
+
from trainers.utils import default, get_index_from_list, normalize_to_neg_one_to_one
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
def linear_beta_schedule(
|
| 17 |
+
timesteps: int,
|
| 18 |
+
start: float = 0.0001,
|
| 19 |
+
end: float = 0.02
|
| 20 |
+
) -> Tensor:
|
| 21 |
+
"""
|
| 22 |
+
:param timesteps: Number of time steps
|
| 23 |
+
|
| 24 |
+
:return schedule: betas at every timestep, (timesteps,)
|
| 25 |
+
"""
|
| 26 |
+
scale = 1000 / timesteps
|
| 27 |
+
beta_start = scale * start
|
| 28 |
+
beta_end = scale * end
|
| 29 |
+
return torch.linspace(beta_start, beta_end, timesteps, dtype=torch.float32)
|
| 30 |
+
|
| 31 |
+
|
| 32 |
+
def cosine_beta_schedule(timesteps: int, s: float = 0.008) -> Tensor:
|
| 33 |
+
"""
|
| 34 |
+
cosine schedule
|
| 35 |
+
as proposed in https://openreview.net/forum?id=-NEXDKk8gZ
|
| 36 |
+
|
| 37 |
+
:param timesteps: Number of time steps
|
| 38 |
+
:param s: scaling factor
|
| 39 |
+
|
| 40 |
+
:return schedule: betas at every timestep, (timesteps,)
|
| 41 |
+
"""
|
| 42 |
+
steps = timesteps + 1
|
| 43 |
+
x = torch.linspace(0, timesteps, steps, dtype=torch.float32)
|
| 44 |
+
alphas_cumprod = torch.cos(((x / timesteps) + s) / (1 + s) * math.pi * 0.5) ** 2
|
| 45 |
+
alphas_cumprod = alphas_cumprod / alphas_cumprod[0]
|
| 46 |
+
betas = 1 - (alphas_cumprod[1:] / alphas_cumprod[:-1])
|
| 47 |
+
return torch.clip(betas, 0, 0.999)
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
class DiffusionModel(nn.Module):
|
| 51 |
+
def __init__(self, config: Namespace):
|
| 52 |
+
super().__init__()
|
| 53 |
+
|
| 54 |
+
# Default parameters
|
| 55 |
+
self.config = config
|
| 56 |
+
dim: int = self.default('dim', 64)
|
| 57 |
+
dim_mults: List[int] = self.default('dim_mults', [1, 2, 4, 8])
|
| 58 |
+
channels: int = self.default('channels', 1)
|
| 59 |
+
timesteps: int = self.default('timesteps', 1000)
|
| 60 |
+
beta_schedule: str = self.default('beta_schedule', 'cosine')
|
| 61 |
+
objective: str = self.default('objective', 'pred_noise') # 'pred_noise' or 'pred_x_0'
|
| 62 |
+
p2_loss_weight_gamma: float = self.default('p2_loss_weight_gamma', 0.) # p2 loss weight, from https://arxiv.org/abs/2204.00227 - 0 is equivalent to weight of 1 across time - 1. is recommended
|
| 63 |
+
p2_loss_weight_k: float = self.default('p2_loss_weight_k', 1.)
|
| 64 |
+
dynamic_threshold_percentile: float = self.default('dynamic_threshold_percentile', 0.995)
|
| 65 |
+
|
| 66 |
+
self.timesteps = timesteps
|
| 67 |
+
self.objective = objective
|
| 68 |
+
self.dynamic_threshold_percentile = dynamic_threshold_percentile
|
| 69 |
+
self.model = Unet(
|
| 70 |
+
dim,
|
| 71 |
+
dim_mults=dim_mults,
|
| 72 |
+
channels=channels
|
| 73 |
+
)
|
| 74 |
+
|
| 75 |
+
if beta_schedule == 'linear':
|
| 76 |
+
betas = linear_beta_schedule(timesteps)
|
| 77 |
+
elif beta_schedule == 'cosine':
|
| 78 |
+
betas = cosine_beta_schedule(timesteps)
|
| 79 |
+
else:
|
| 80 |
+
raise ValueError(f'unknown beta schedule {beta_schedule}')
|
| 81 |
+
|
| 82 |
+
alphas = 1. - betas
|
| 83 |
+
alphas_cumprod = torch.cumprod(alphas, axis=0)
|
| 84 |
+
alphas_cumprod_prev = F.pad(alphas_cumprod[:-1], (1, 0), value=1.)
|
| 85 |
+
|
| 86 |
+
# Calculations for diffusion q(x_t | x_{t-1}) and others
|
| 87 |
+
self.register_buffer('sqrt_alphas_cumprod', torch.sqrt(alphas_cumprod))
|
| 88 |
+
self.register_buffer('sqrt_recip_alphas_cumprod',
|
| 89 |
+
torch.sqrt(1. / alphas_cumprod))
|
| 90 |
+
self.register_buffer('sqrt_recipm1_alphas_cumprod',
|
| 91 |
+
torch.sqrt(1. / alphas_cumprod - 1))
|
| 92 |
+
self.register_buffer('sqrt_one_minus_alphas_cumprod',
|
| 93 |
+
torch.sqrt(1. - alphas_cumprod))
|
| 94 |
+
|
| 95 |
+
# Calculations for posterior q(x_{t-1} | x_t, x_0)
|
| 96 |
+
posterior_variance = betas * (1. - alphas_cumprod_prev) / (1. - alphas_cumprod)
|
| 97 |
+
self.register_buffer('posterior_variance', posterior_variance)
|
| 98 |
+
|
| 99 |
+
self.register_buffer(
|
| 100 |
+
'posterior_log_variance_clipped',
|
| 101 |
+
torch.log(posterior_variance.clamp(min=1e-20))
|
| 102 |
+
)
|
| 103 |
+
self.register_buffer(
|
| 104 |
+
'posterior_mean_coef1',
|
| 105 |
+
betas * torch.sqrt(alphas_cumprod_prev) / (1. - alphas_cumprod)
|
| 106 |
+
)
|
| 107 |
+
self.register_buffer(
|
| 108 |
+
'posterior_mean_coef2',
|
| 109 |
+
(1. - alphas_cumprod_prev) * torch.sqrt(alphas) / (1. - alphas_cumprod)
|
| 110 |
+
)
|
| 111 |
+
|
| 112 |
+
# p2 reweighting
|
| 113 |
+
p2_loss_weight = ((p2_loss_weight_k + alphas_cumprod / (1 - alphas_cumprod))
|
| 114 |
+
** (-p2_loss_weight_gamma))
|
| 115 |
+
self.register_buffer('p2_loss_weight', p2_loss_weight)
|
| 116 |
+
|
| 117 |
+
def default(self, val, d):
|
| 118 |
+
return vars(self.config)[val] if val in self.config else d
|
| 119 |
+
|
| 120 |
+
def train_step(self, x_0: Tensor, cond: Optional[Tensor] = None, t:Optional[Tensor] = None) -> Tensor:
|
| 121 |
+
N, device = x_0.shape[0], x_0.device
|
| 122 |
+
|
| 123 |
+
# If t is not none, use it, otherwise sample from uniform
|
| 124 |
+
if t is not None:
|
| 125 |
+
t = t.long().to(device)
|
| 126 |
+
else:
|
| 127 |
+
t = torch.randint(0, self.timesteps, (N,), device=device).long() # (N)
|
| 128 |
+
|
| 129 |
+
model_out, noise = self(x_0, t, cond=cond)
|
| 130 |
+
|
| 131 |
+
if self.objective == 'pred_noise':
|
| 132 |
+
target = noise # (N, C, H, W)
|
| 133 |
+
elif self.objective == 'pred_x_0':
|
| 134 |
+
target = x_0 # (N, C, H, W)
|
| 135 |
+
else:
|
| 136 |
+
raise ValueError(f'unknown objective {self.objective}')
|
| 137 |
+
|
| 138 |
+
loss = F.l1_loss(model_out, target, reduction='none') # (N, C, H, W)
|
| 139 |
+
loss = reduce(loss, 'b ... -> b (...)', 'mean') # (N, (C x H x W))
|
| 140 |
+
|
| 141 |
+
# p2 reweighting
|
| 142 |
+
loss = loss * get_index_from_list(self.p2_loss_weight, t, loss.shape)
|
| 143 |
+
return loss.mean()
|
| 144 |
+
|
| 145 |
+
def val_step(self, x_0: Tensor, cond: Optional[Tensor] = None, t_steps:Optional[int] = None) -> Tensor:
|
| 146 |
+
if not t_steps:
|
| 147 |
+
t_steps = self.timesteps
|
| 148 |
+
step_size = self.timesteps // t_steps
|
| 149 |
+
N, device = x_0.shape[0], x_0.device
|
| 150 |
+
losses = []
|
| 151 |
+
for t in range(0, self.timesteps, step_size):
|
| 152 |
+
t = torch.ones((N,)) * t
|
| 153 |
+
t = t.long().to(device)
|
| 154 |
+
losses.append(self.train_step(x_0, cond, t))
|
| 155 |
+
|
| 156 |
+
return torch.stack(losses).mean()
|
| 157 |
+
|
| 158 |
+
def forward(self, x_0: Tensor, t: Tensor, cond: Optional[Tensor] = None) -> Tensor:
|
| 159 |
+
"""
|
| 160 |
+
Noise x_0 for t timestep and get the model prediction.
|
| 161 |
+
|
| 162 |
+
:param x_0: Clean image, (N, C, H, W)
|
| 163 |
+
:param t: Timestep, (N,)
|
| 164 |
+
:param cond: element to condition the reconstruction on - eg image when x_0 is a segmentation (N, C', H, W)
|
| 165 |
+
|
| 166 |
+
:return pred: Model output, predicted noise or image, (N, C, H, W)
|
| 167 |
+
:return noise: Added noise, (N, C, H, W)
|
| 168 |
+
"""
|
| 169 |
+
if self.config.normalize:
|
| 170 |
+
x_0 = normalize_to_neg_one_to_one(x_0)
|
| 171 |
+
if cond is not None and self.config.normalize:
|
| 172 |
+
cond = normalize_to_neg_one_to_one(cond)
|
| 173 |
+
x_t, noise = self.forward_diffusion_model(x_0, t)
|
| 174 |
+
return self.model(x_t, t, cond), noise
|
| 175 |
+
|
| 176 |
+
def forward_diffusion_model(
|
| 177 |
+
self,
|
| 178 |
+
x_0: Tensor,
|
| 179 |
+
t: Tensor,
|
| 180 |
+
noise: Optional[Tensor] = None,
|
| 181 |
+
) -> Tuple[Tensor, Tensor]:
|
| 182 |
+
"""
|
| 183 |
+
Takes an image and a timestep as input and returns the noisy version
|
| 184 |
+
of it.
|
| 185 |
+
|
| 186 |
+
:param x_0: Image at timestep 0, (N, C, H, W)
|
| 187 |
+
:param t: Timestep, (N)
|
| 188 |
+
:param cond: element to condition the reconstruction on - eg image when x_0 is a segmentation (N, C', H, W)
|
| 189 |
+
|
| 190 |
+
:return x_t: Noisy image at timestep t, (N, C, H, W)
|
| 191 |
+
:return noise: Noise added to the image, (N, C, H, W)
|
| 192 |
+
"""
|
| 193 |
+
noise = default(noise, lambda: torch.randn_like(x_0))
|
| 194 |
+
|
| 195 |
+
sqrt_alphas_cumprod_t = get_index_from_list(
|
| 196 |
+
self.sqrt_alphas_cumprod, t, x_0.shape)
|
| 197 |
+
sqrt_one_minus_alphas_cumprod_t = get_index_from_list(
|
| 198 |
+
self.sqrt_one_minus_alphas_cumprod, t, x_0.shape)
|
| 199 |
+
|
| 200 |
+
# mean + variance
|
| 201 |
+
x_t = sqrt_alphas_cumprod_t * x_0 + sqrt_one_minus_alphas_cumprod_t * noise
|
| 202 |
+
|
| 203 |
+
return x_t, noise
|
| 204 |
+
|
| 205 |
+
@torch.no_grad()
|
| 206 |
+
def sample_timestep(self, x_t: Tensor, t: int, cond=Optional[Tensor]) -> Tensor:
|
| 207 |
+
"""
|
| 208 |
+
Sample from the model.
|
| 209 |
+
:param x_t: Image noised t times, (N, C, H, W)
|
| 210 |
+
:param t: Timestep
|
| 211 |
+
:return: Sampled image, (N, C, H, W)
|
| 212 |
+
"""
|
| 213 |
+
N = x_t.shape[0]
|
| 214 |
+
device = x_t.device
|
| 215 |
+
batched_t = torch.full((N,), t, device=device, dtype=torch.long) # (N)
|
| 216 |
+
model_mean, model_log_variance, _ = self.p_mean_variance(x_t, batched_t, cond=cond)
|
| 217 |
+
noise = torch.randn_like(x_t) if t > 0 else 0.
|
| 218 |
+
pred_img = model_mean + (0.5 * model_log_variance).exp() * noise
|
| 219 |
+
return pred_img
|
| 220 |
+
|
| 221 |
+
def p_mean_variance(self, x_t: Tensor, t: Tensor, clip_denoised: bool = True, cond:Optional[Tensor] = None) -> Tuple[Tensor, Tensor, Tensor]:
|
| 222 |
+
_, pred_x_0 = self.model_predictions(x_t, t, cond=cond)
|
| 223 |
+
|
| 224 |
+
if clip_denoised:
|
| 225 |
+
# pred_x_0.clamp_(-1., 1.)
|
| 226 |
+
# Dynamic thrsholding
|
| 227 |
+
s = torch.quantile(rearrange(pred_x_0, 'b ... -> b (...)').abs(),
|
| 228 |
+
self.dynamic_threshold_percentile,
|
| 229 |
+
dim=1)
|
| 230 |
+
s = torch.max(s, torch.tensor(1.0))[:, None, None, None]
|
| 231 |
+
pred_x_0 = torch.clip(pred_x_0, -s, s) / s
|
| 232 |
+
|
| 233 |
+
(model_mean,
|
| 234 |
+
posterior_log_variance) = self.q_posterior(pred_x_0, x_t, t)
|
| 235 |
+
return model_mean, posterior_log_variance, pred_x_0
|
| 236 |
+
|
| 237 |
+
def model_predictions(self, x_t: Tensor, t: Tensor, cond:Optional[Tensor] = None) \
|
| 238 |
+
-> Tuple[Tensor, Tensor]:
|
| 239 |
+
"""
|
| 240 |
+
Return the predicted noise and x_0 for a given x_t and t.
|
| 241 |
+
|
| 242 |
+
:param x_t: Noised image at timestep t, (N, C, H, W)
|
| 243 |
+
:param t: Timestep, (N,)
|
| 244 |
+
:return pred_noise: Predicted noise, (N, C, H, W)
|
| 245 |
+
:return pred_x_0: Predicted x_0, (N, C, H, W)
|
| 246 |
+
"""
|
| 247 |
+
model_output = self.model(x_t, t, cond)
|
| 248 |
+
|
| 249 |
+
if self.objective == 'pred_noise':
|
| 250 |
+
pred_noise = model_output
|
| 251 |
+
pred_x_0 = self.predict_x_0_from_noise(x_t, t, model_output)
|
| 252 |
+
|
| 253 |
+
elif self.objective == 'pred_x_start':
|
| 254 |
+
pred_noise = self.predict_noise_from_x_0(x_t, t, model_output)
|
| 255 |
+
pred_x_0 = model_output
|
| 256 |
+
|
| 257 |
+
return pred_noise, pred_x_0
|
| 258 |
+
|
| 259 |
+
def q_posterior(self, x_start: Tensor, x_t: Tensor, t: Tensor) \
|
| 260 |
+
-> Tuple[Tensor, Tensor]:
|
| 261 |
+
posterior_mean = (
|
| 262 |
+
get_index_from_list(self.posterior_mean_coef1, t, x_t.shape) * x_start
|
| 263 |
+
+ get_index_from_list(self.posterior_mean_coef2, t, x_t.shape) * x_t
|
| 264 |
+
)
|
| 265 |
+
posterior_log_variance_clipped = get_index_from_list(
|
| 266 |
+
self.posterior_log_variance_clipped, t, x_t.shape)
|
| 267 |
+
return posterior_mean, posterior_log_variance_clipped
|
| 268 |
+
|
| 269 |
+
def predict_x_0_from_noise(self, x_t: Tensor, t: Tensor, noise: Tensor) \
|
| 270 |
+
-> Tensor:
|
| 271 |
+
"""
|
| 272 |
+
Get x_0 given x_t, t, and the known or predicted noise.
|
| 273 |
+
|
| 274 |
+
:param x_t: Noised image at timestep t, (N, C, H, W)
|
| 275 |
+
:param t: Timestep, (N,)
|
| 276 |
+
:param noise: Noise, (N, C, H, W)
|
| 277 |
+
:return: Predicted x_0, (N, C, H, W)
|
| 278 |
+
"""
|
| 279 |
+
return (
|
| 280 |
+
get_index_from_list(
|
| 281 |
+
self.sqrt_recip_alphas_cumprod, t, x_t.shape)
|
| 282 |
+
* x_t
|
| 283 |
+
- get_index_from_list(
|
| 284 |
+
self.sqrt_recipm1_alphas_cumprod, t, x_t.shape)
|
| 285 |
+
* noise
|
| 286 |
+
)
|
| 287 |
+
|
| 288 |
+
def predict_noise_from_x_0(self, x_t: Tensor, t: Tensor, x_0: Tensor) \
|
| 289 |
+
-> Tensor:
|
| 290 |
+
"""
|
| 291 |
+
Get noise given the known or predicted x_0, x_t, and t
|
| 292 |
+
|
| 293 |
+
:param x_t: Noised image at timestep t, (N, C, H, W)
|
| 294 |
+
:param t: Timestep, (N,)
|
| 295 |
+
:param noise: Noise, (N, C, H, W)
|
| 296 |
+
:return: Predicted noise, (N, C, H, W)
|
| 297 |
+
"""
|
| 298 |
+
return (
|
| 299 |
+
(get_index_from_list(self.sqrt_recip_alphas_cumprod, t, x_t.shape) * x_t - x_0)
|
| 300 |
+
/ get_index_from_list(self.sqrt_recipm1_alphas_cumprod, t, x_t.shape)
|
| 301 |
+
)
|
models/global_local_cl.py
ADDED
|
@@ -0,0 +1,111 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from models.unet_model import Unet, default
|
| 2 |
+
from torch import Tensor, nn
|
| 3 |
+
import torch
|
| 4 |
+
from typing import Optional, List
|
| 5 |
+
from einops.layers.torch import Rearrange
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
class GlobalCL(Unet):
|
| 9 |
+
def __init__(self,
|
| 10 |
+
img_size,
|
| 11 |
+
dim: int = 64,
|
| 12 |
+
init_dim: Optional[int] = None,
|
| 13 |
+
dim_mults: List[int] = [1, 2, 4, 8],
|
| 14 |
+
**kwargs):
|
| 15 |
+
super().__init__(**kwargs)
|
| 16 |
+
init_dim = default(init_dim, dim)
|
| 17 |
+
# from the paper
|
| 18 |
+
g_emb= 1024
|
| 19 |
+
g_out = 128
|
| 20 |
+
dims = [init_dim, *map(lambda m: dim * m, dim_mults)]
|
| 21 |
+
mid_dim = dims[-1]
|
| 22 |
+
mid_img_size = img_size
|
| 23 |
+
for _ in range(len(dims)-2):
|
| 24 |
+
mid_img_size = int((mid_img_size -1) / 2) + 1
|
| 25 |
+
self.g1 = nn.Sequential(
|
| 26 |
+
Rearrange('b c h w -> b (c h w)'),
|
| 27 |
+
nn.Linear(mid_dim * mid_img_size ** 2, g_emb, bias=False),
|
| 28 |
+
nn.ReLU(),
|
| 29 |
+
nn.Linear(g_emb, g_out, bias=False),
|
| 30 |
+
)
|
| 31 |
+
|
| 32 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 33 |
+
x = self.init_conv(x)
|
| 34 |
+
|
| 35 |
+
t = None
|
| 36 |
+
|
| 37 |
+
for block1, block2, attn, downsample in self.downs:
|
| 38 |
+
x = block1(x, t)
|
| 39 |
+
|
| 40 |
+
x = block2(x, t)
|
| 41 |
+
x = attn(x)
|
| 42 |
+
|
| 43 |
+
x = downsample(x)
|
| 44 |
+
|
| 45 |
+
x = self.mid_block1(x, t)
|
| 46 |
+
x = self.mid_attn(x)
|
| 47 |
+
x = self.mid_block2(x, t)
|
| 48 |
+
|
| 49 |
+
x = self.g1(x)
|
| 50 |
+
return x
|
| 51 |
+
|
| 52 |
+
|
| 53 |
+
class LocalCL(Unet):
|
| 54 |
+
def __init__(self,
|
| 55 |
+
img_size,
|
| 56 |
+
dim: int = 64,
|
| 57 |
+
init_dim: Optional[int] = None,
|
| 58 |
+
dim_mults: List[int] = [1, 2, 4, 8],
|
| 59 |
+
**kwargs):
|
| 60 |
+
super().__init__(**kwargs)
|
| 61 |
+
init_dim = default(init_dim, dim)
|
| 62 |
+
# from the paper
|
| 63 |
+
dims = [init_dim, *map(lambda m: dim * m, dim_mults)]
|
| 64 |
+
#g_2 small network with two 1x1 convolutions
|
| 65 |
+
self.l = 2
|
| 66 |
+
mid_dim = dims[-self.l-1]
|
| 67 |
+
self.g2 = nn.Sequential(
|
| 68 |
+
nn.Conv2d(mid_dim, mid_dim, 1, bias=False),
|
| 69 |
+
nn.ReLU(),
|
| 70 |
+
nn.BatchNorm2d(mid_dim),
|
| 71 |
+
nn.Conv2d(mid_dim, mid_dim, 1, bias=False),
|
| 72 |
+
)
|
| 73 |
+
|
| 74 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 75 |
+
x = self.init_conv(x)
|
| 76 |
+
r = x.clone()
|
| 77 |
+
|
| 78 |
+
t = None
|
| 79 |
+
|
| 80 |
+
h = []
|
| 81 |
+
|
| 82 |
+
for block1, block2, attn, downsample in self.downs:
|
| 83 |
+
x = block1(x, t)
|
| 84 |
+
h.append(x)
|
| 85 |
+
|
| 86 |
+
x = block2(x, t)
|
| 87 |
+
x = attn(x)
|
| 88 |
+
h.append(x)
|
| 89 |
+
|
| 90 |
+
x = downsample(x)
|
| 91 |
+
|
| 92 |
+
x = self.mid_block1(x, t)
|
| 93 |
+
x = self.mid_attn(x)
|
| 94 |
+
x = self.mid_block2(x, t)
|
| 95 |
+
|
| 96 |
+
for block1, block2, attn, upsample in self.ups[:self.l]:
|
| 97 |
+
x = torch.cat((x, h.pop()), dim=1)
|
| 98 |
+
x = block1(x, t)
|
| 99 |
+
|
| 100 |
+
x = torch.cat((x, h.pop()), dim=1)
|
| 101 |
+
x = block2(x, t)
|
| 102 |
+
x = attn(x)
|
| 103 |
+
|
| 104 |
+
x = upsample(x)
|
| 105 |
+
|
| 106 |
+
x = self.g2(x)
|
| 107 |
+
return x
|
| 108 |
+
|
| 109 |
+
|
| 110 |
+
|
| 111 |
+
|
models/unet_model.py
ADDED
|
@@ -0,0 +1,375 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""Adapted from https://github.com/lucidrains/denoising-diffusion-pytorch"""
|
| 2 |
+
import math
|
| 3 |
+
from collections import namedtuple
|
| 4 |
+
from functools import partial
|
| 5 |
+
from typing import List, Optional
|
| 6 |
+
|
| 7 |
+
import torch
|
| 8 |
+
import torch.nn.functional as F
|
| 9 |
+
from einops import rearrange
|
| 10 |
+
from torch import einsum, nn, Tensor
|
| 11 |
+
|
| 12 |
+
from trainers.utils import default, exists
|
| 13 |
+
|
| 14 |
+
# constants
|
| 15 |
+
|
| 16 |
+
ModelPrediction = namedtuple('ModelPrediction', ['pred_noise', 'pred_x_start'])
|
| 17 |
+
|
| 18 |
+
# helpers functions
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
def l2norm(t: Tensor) -> Tensor:
|
| 22 |
+
"""L2 normalize along last dimension"""
|
| 23 |
+
return F.normalize(t, dim=-1)
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
# small helper modules
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
class Residual(nn.Module):
|
| 30 |
+
"""Residual of any Module -> x' = f(x) + x"""
|
| 31 |
+
def __init__(self, fn: nn.Module):
|
| 32 |
+
super().__init__()
|
| 33 |
+
self.fn = fn
|
| 34 |
+
|
| 35 |
+
def forward(self, x, *args, **kwargs):
|
| 36 |
+
return self.fn(x, *args, **kwargs) + x
|
| 37 |
+
|
| 38 |
+
|
| 39 |
+
def Upsample(dim: int, dim_out: Optional[int] = None) -> nn.Sequential:
|
| 40 |
+
"""UpsampleConv with factor 2"""
|
| 41 |
+
return nn.Sequential(
|
| 42 |
+
nn.Upsample(scale_factor=2, mode='nearest'),
|
| 43 |
+
nn.Conv2d(dim, default(dim_out, dim), 3, padding=1)
|
| 44 |
+
)
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
def Downsample(dim: int, dim_out: Optional[int] = None) -> nn.Conv2d:
|
| 48 |
+
"""Strided Conv2d for downsampling"""
|
| 49 |
+
return nn.Conv2d(dim, default(dim_out, dim), 4, 2, 1)
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
class LayerNorm(nn.Module):
|
| 53 |
+
def __init__(self, dim: int):
|
| 54 |
+
super().__init__()
|
| 55 |
+
self.g = nn.Parameter(torch.ones(1, dim, 1, 1))
|
| 56 |
+
|
| 57 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 58 |
+
eps = 1e-5 if x.dtype == torch.float32 else 1e-3
|
| 59 |
+
var = torch.var(x, dim=1, unbiased=False, keepdim=True)
|
| 60 |
+
mean = torch.mean(x, dim=1, keepdim=True)
|
| 61 |
+
return (x - mean) * (var + eps).rsqrt() * self.g
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
class PreNorm(nn.Module):
|
| 65 |
+
"""Apply LayerNorm before any Module"""
|
| 66 |
+
def __init__(self, dim: int, fn: nn.Module):
|
| 67 |
+
super().__init__()
|
| 68 |
+
self.fn = fn
|
| 69 |
+
self.norm = LayerNorm(dim)
|
| 70 |
+
|
| 71 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 72 |
+
x = self.norm(x)
|
| 73 |
+
return self.fn(x)
|
| 74 |
+
|
| 75 |
+
|
| 76 |
+
class SinusoidalPosEmb(nn.Module):
|
| 77 |
+
"""Classical sinosoidal embedding"""
|
| 78 |
+
def __init__(self, dim: int):
|
| 79 |
+
super().__init__()
|
| 80 |
+
self.dim = dim
|
| 81 |
+
|
| 82 |
+
def forward(self, t: Tensor) -> Tensor:
|
| 83 |
+
"""
|
| 84 |
+
:param t: Batch of time steps (b,)
|
| 85 |
+
:return emb: Sinusoidal time embedding (b, dim)
|
| 86 |
+
"""
|
| 87 |
+
device = t.device
|
| 88 |
+
half_dim = self.dim // 2
|
| 89 |
+
emb = math.log(10000) / (half_dim - 1)
|
| 90 |
+
emb = torch.exp(torch.arange(half_dim, device=device) * -emb)
|
| 91 |
+
emb = t[:, None] * emb[None, :]
|
| 92 |
+
emb = torch.cat((emb.sin(), emb.cos()), dim=-1)
|
| 93 |
+
return emb
|
| 94 |
+
|
| 95 |
+
|
| 96 |
+
class LearnedSinusoidalPosEmb(nn.Module):
|
| 97 |
+
""" following @crowsonkb 's lead with learned sinusoidal pos emb """
|
| 98 |
+
""" https://github.com/crowsonkb/v-diffusion-jax/blob/master/diffusion/models/danbooru_128.py#L8 """
|
| 99 |
+
def __init__(self, dim: int):
|
| 100 |
+
super().__init__()
|
| 101 |
+
assert (dim % 2) == 0
|
| 102 |
+
half_dim = dim // 2
|
| 103 |
+
self.weights = nn.Parameter(torch.randn(half_dim))
|
| 104 |
+
|
| 105 |
+
def forward(self, t: Tensor) -> Tensor:
|
| 106 |
+
"""
|
| 107 |
+
:param t: Batch of time steps (b,)
|
| 108 |
+
:return fouriered: Concatenation of t and time embedding (b, dim + 1)
|
| 109 |
+
"""
|
| 110 |
+
t = rearrange(t, 'b -> b 1')
|
| 111 |
+
freqs = t * rearrange(self.weights, 'd -> 1 d') * 2 * math.pi
|
| 112 |
+
fouriered = torch.cat((freqs.sin(), freqs.cos()), dim=-1)
|
| 113 |
+
fouriered = torch.cat((t, fouriered), dim=-1)
|
| 114 |
+
return fouriered
|
| 115 |
+
|
| 116 |
+
# building block modules
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
class Block(nn.Module):
|
| 120 |
+
def __init__(self, dim: int, dim_out: int, groups: int = 8):
|
| 121 |
+
super().__init__()
|
| 122 |
+
self.proj = nn.Conv2d(dim, dim_out, 3, padding=1)
|
| 123 |
+
self.norm = nn.GroupNorm(groups, dim_out)
|
| 124 |
+
self.act = nn.SiLU()
|
| 125 |
+
|
| 126 |
+
def forward(self, x: Tensor, scale_shift: Optional[Tensor] = None) -> Tensor:
|
| 127 |
+
x = self.proj(x)
|
| 128 |
+
x = self.norm(x)
|
| 129 |
+
|
| 130 |
+
if exists(scale_shift):
|
| 131 |
+
scale, shift = scale_shift
|
| 132 |
+
x = x * (scale + 1) + shift
|
| 133 |
+
|
| 134 |
+
x = self.act(x)
|
| 135 |
+
return x
|
| 136 |
+
|
| 137 |
+
|
| 138 |
+
class ResnetBlock(nn.Module):
|
| 139 |
+
def __init__(
|
| 140 |
+
self,
|
| 141 |
+
dim: int,
|
| 142 |
+
dim_out: int,
|
| 143 |
+
*,
|
| 144 |
+
time_emb_dim: Optional[int] = None,
|
| 145 |
+
groups: int = 8
|
| 146 |
+
):
|
| 147 |
+
super().__init__()
|
| 148 |
+
|
| 149 |
+
self.time_mlp = nn.Sequential(
|
| 150 |
+
nn.SiLU(),
|
| 151 |
+
nn.Linear(time_emb_dim, dim_out * 2)
|
| 152 |
+
) if exists(time_emb_dim) else None
|
| 153 |
+
|
| 154 |
+
self.block1 = Block(dim, dim_out, groups=groups)
|
| 155 |
+
self.block2 = Block(dim_out, dim_out, groups=groups)
|
| 156 |
+
if dim != dim_out:
|
| 157 |
+
self.res_conv = nn.Conv2d(dim, dim_out, 1)
|
| 158 |
+
else:
|
| 159 |
+
self.res_conv = nn.Identity()
|
| 160 |
+
|
| 161 |
+
def forward(self, x: Tensor, time_emb: Optional[Tensor] = None) -> Tensor:
|
| 162 |
+
"""
|
| 163 |
+
:param x: Batch of input images (b, c, h, w)
|
| 164 |
+
:param time_emb: Batch of time embeddings (b, c)
|
| 165 |
+
"""
|
| 166 |
+
scale_shift = None
|
| 167 |
+
|
| 168 |
+
if exists(self.time_mlp) and exists(time_emb):
|
| 169 |
+
time_emb = self.time_mlp(time_emb)
|
| 170 |
+
time_emb = rearrange(time_emb, 'b c -> b c 1 1')
|
| 171 |
+
scale_shift = time_emb.chunk(2, dim=1)
|
| 172 |
+
|
| 173 |
+
h = self.block1(x, scale_shift=scale_shift)
|
| 174 |
+
h = self.block2(h)
|
| 175 |
+
return h + self.res_conv(x)
|
| 176 |
+
|
| 177 |
+
|
| 178 |
+
class LinearAttention(nn.Module):
|
| 179 |
+
"""Attention with linear to_qtv"""
|
| 180 |
+
def __init__(self, dim: int, heads: int = 4, dim_head: int = 32):
|
| 181 |
+
super().__init__()
|
| 182 |
+
self.scale = dim_head ** -0.5
|
| 183 |
+
self.heads = heads
|
| 184 |
+
hidden_dim = dim_head * heads
|
| 185 |
+
self.to_qkv = nn.Conv2d(dim, hidden_dim * 3, 1, bias=False)
|
| 186 |
+
|
| 187 |
+
self.to_out = nn.Sequential(
|
| 188 |
+
nn.Conv2d(hidden_dim, dim, 1),
|
| 189 |
+
LayerNorm(dim)
|
| 190 |
+
)
|
| 191 |
+
|
| 192 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 193 |
+
"""
|
| 194 |
+
:param x: Batch of input images (b, c, h, w)
|
| 195 |
+
"""
|
| 196 |
+
b, c, h, w = x.shape
|
| 197 |
+
qkv = self.to_qkv(x).chunk(3, dim=1)
|
| 198 |
+
q, k, v = map(lambda t: rearrange(t, 'b (h c) x y -> b h c (x y)', h=self.heads), qkv)
|
| 199 |
+
|
| 200 |
+
q = q.softmax(dim=-2)
|
| 201 |
+
k = k.softmax(dim=-1)
|
| 202 |
+
|
| 203 |
+
q = q * self.scale
|
| 204 |
+
v = v / (h * w)
|
| 205 |
+
|
| 206 |
+
context = torch.einsum('b h d n, b h e n -> b h d e', k, v)
|
| 207 |
+
|
| 208 |
+
out = torch.einsum('b h d e, b h d n -> b h e n', context, q)
|
| 209 |
+
out = rearrange(out, 'b h c (x y) -> b (h c) x y', h=self.heads, x=h, y=w)
|
| 210 |
+
return self.to_out(out)
|
| 211 |
+
|
| 212 |
+
|
| 213 |
+
class Attention(nn.Module):
|
| 214 |
+
"""Attention with convolutional to_qtv"""
|
| 215 |
+
def __init__(
|
| 216 |
+
self,
|
| 217 |
+
dim: int,
|
| 218 |
+
heads: int = 4,
|
| 219 |
+
dim_head: int = 32,
|
| 220 |
+
scale: int = 16
|
| 221 |
+
):
|
| 222 |
+
super().__init__()
|
| 223 |
+
self.scale = scale
|
| 224 |
+
self.heads = heads
|
| 225 |
+
hidden_dim = dim_head * heads
|
| 226 |
+
self.to_qkv = nn.Conv2d(dim, hidden_dim * 3, 1, bias=False)
|
| 227 |
+
self.to_out = nn.Conv2d(hidden_dim, dim, 1)
|
| 228 |
+
|
| 229 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 230 |
+
b, c, h, w = x.shape
|
| 231 |
+
qkv = self.to_qkv(x).chunk(3, dim=1)
|
| 232 |
+
q, k, v = map(lambda t: rearrange(t, 'b (h c) x y -> b h c (x y)', h=self.heads), qkv)
|
| 233 |
+
|
| 234 |
+
q, k = map(l2norm, (q, k))
|
| 235 |
+
|
| 236 |
+
sim = einsum('b h d i, b h d j -> b h i j', q, k) * self.scale
|
| 237 |
+
attn = sim.softmax(dim=-1)
|
| 238 |
+
|
| 239 |
+
out = einsum('b h i j, b h d j -> b h i d', attn, v)
|
| 240 |
+
out = rearrange(out, 'b h (x y) d -> b (h d) x y', x=h, y=w)
|
| 241 |
+
return self.to_out(out)
|
| 242 |
+
|
| 243 |
+
# model
|
| 244 |
+
|
| 245 |
+
|
| 246 |
+
class Unet(nn.Module):
|
| 247 |
+
def __init__(
|
| 248 |
+
self,
|
| 249 |
+
dim: int = 64,
|
| 250 |
+
init_dim: Optional[int] = None,
|
| 251 |
+
out_dim: Optional[int] = None,
|
| 252 |
+
dim_mults: List[int] = [1, 2, 4, 8],
|
| 253 |
+
channels: int = 1,
|
| 254 |
+
resnet_block_groups: int = 8,
|
| 255 |
+
learned_variance: bool = False,
|
| 256 |
+
learned_sinusoidal_cond: bool = False,
|
| 257 |
+
learned_sinusoidal_dim: int = 16,
|
| 258 |
+
**kwargs
|
| 259 |
+
):
|
| 260 |
+
super().__init__()
|
| 261 |
+
|
| 262 |
+
# determine dimensions
|
| 263 |
+
|
| 264 |
+
self.channels = channels
|
| 265 |
+
|
| 266 |
+
init_dim = default(init_dim, dim)
|
| 267 |
+
self.init_conv = nn.Conv2d(channels, init_dim, 7, padding=3)
|
| 268 |
+
|
| 269 |
+
dims = [init_dim, *map(lambda m: dim * m, dim_mults)]
|
| 270 |
+
in_out = list(zip(dims[:-1], dims[1:]))
|
| 271 |
+
|
| 272 |
+
block_class = partial(ResnetBlock, groups=resnet_block_groups)
|
| 273 |
+
|
| 274 |
+
# time embeddings
|
| 275 |
+
|
| 276 |
+
time_dim = dim * 4
|
| 277 |
+
|
| 278 |
+
self.learned_sinusoidal_cond = learned_sinusoidal_cond
|
| 279 |
+
|
| 280 |
+
if learned_sinusoidal_cond:
|
| 281 |
+
sinu_pos_emb = LearnedSinusoidalPosEmb(learned_sinusoidal_dim)
|
| 282 |
+
fourier_dim = learned_sinusoidal_dim + 1
|
| 283 |
+
else:
|
| 284 |
+
sinu_pos_emb = SinusoidalPosEmb(dim)
|
| 285 |
+
fourier_dim = dim
|
| 286 |
+
|
| 287 |
+
self.time_mlp = nn.Sequential(
|
| 288 |
+
sinu_pos_emb,
|
| 289 |
+
nn.Linear(fourier_dim, time_dim),
|
| 290 |
+
nn.GELU(),
|
| 291 |
+
nn.Linear(time_dim, time_dim)
|
| 292 |
+
)
|
| 293 |
+
|
| 294 |
+
# layers
|
| 295 |
+
|
| 296 |
+
self.downs = nn.ModuleList([])
|
| 297 |
+
self.ups = nn.ModuleList([])
|
| 298 |
+
num_resolutions = len(in_out)
|
| 299 |
+
|
| 300 |
+
for ind, (dim_in, dim_out) in enumerate(in_out):
|
| 301 |
+
is_last = ind >= (num_resolutions - 1)
|
| 302 |
+
|
| 303 |
+
self.downs.append(nn.ModuleList([
|
| 304 |
+
block_class(dim_in, dim_in, time_emb_dim=time_dim),
|
| 305 |
+
block_class(dim_in, dim_in, time_emb_dim=time_dim),
|
| 306 |
+
Residual(PreNorm(dim_in, LinearAttention(dim_in))),
|
| 307 |
+
Downsample(dim_in, dim_out) if not is_last else nn.Conv2d(
|
| 308 |
+
dim_in, dim_out, 3, padding=1)
|
| 309 |
+
]))
|
| 310 |
+
|
| 311 |
+
mid_dim = dims[-1]
|
| 312 |
+
self.mid_block1 = block_class(mid_dim, mid_dim, time_emb_dim=time_dim)
|
| 313 |
+
self.mid_attn = Residual(PreNorm(mid_dim, Attention(mid_dim)))
|
| 314 |
+
self.mid_block2 = block_class(mid_dim, mid_dim, time_emb_dim=time_dim)
|
| 315 |
+
|
| 316 |
+
for ind, (dim_in, dim_out) in enumerate(reversed(in_out)):
|
| 317 |
+
is_last = ind == (len(in_out) - 1)
|
| 318 |
+
|
| 319 |
+
self.ups.append(nn.ModuleList([
|
| 320 |
+
block_class(dim_out + dim_in, dim_out, time_emb_dim=time_dim),
|
| 321 |
+
block_class(dim_out + dim_in, dim_out, time_emb_dim=time_dim),
|
| 322 |
+
Residual(PreNorm(dim_out, LinearAttention(dim_out))),
|
| 323 |
+
Upsample(dim_out, dim_in) if not is_last else nn.Conv2d(
|
| 324 |
+
dim_out, dim_in, 3, padding=1)
|
| 325 |
+
]))
|
| 326 |
+
|
| 327 |
+
default_out_dim = channels * (1 if not learned_variance else 2)
|
| 328 |
+
self.out_dim = default(out_dim, default_out_dim)
|
| 329 |
+
|
| 330 |
+
self.final_res_block = block_class(dim * 2, dim, time_emb_dim=time_dim)
|
| 331 |
+
self.final_conv = nn.Conv2d(dim, self.out_dim, 1)
|
| 332 |
+
|
| 333 |
+
def forward(self, x: Tensor, timestep: Optional[Tensor]=None, cond: Optional[Tensor]=None) -> Tensor:
|
| 334 |
+
x = self.init_conv(x)
|
| 335 |
+
r = x.clone()
|
| 336 |
+
|
| 337 |
+
t = self.time_mlp(timestep) if timestep is not None else None
|
| 338 |
+
|
| 339 |
+
h = []
|
| 340 |
+
|
| 341 |
+
for block1, block2, attn, downsample in self.downs:
|
| 342 |
+
x = block1(x, t)
|
| 343 |
+
h.append(x)
|
| 344 |
+
|
| 345 |
+
x = block2(x, t)
|
| 346 |
+
x = attn(x)
|
| 347 |
+
h.append(x)
|
| 348 |
+
|
| 349 |
+
x = downsample(x)
|
| 350 |
+
|
| 351 |
+
x = self.mid_block1(x, t)
|
| 352 |
+
x = self.mid_attn(x)
|
| 353 |
+
x = self.mid_block2(x, t)
|
| 354 |
+
|
| 355 |
+
for block1, block2, attn, upsample in self.ups:
|
| 356 |
+
x = torch.cat((x, h.pop()), dim=1)
|
| 357 |
+
x = block1(x, t)
|
| 358 |
+
|
| 359 |
+
x = torch.cat((x, h.pop()), dim=1)
|
| 360 |
+
x = block2(x, t)
|
| 361 |
+
x = attn(x)
|
| 362 |
+
|
| 363 |
+
x = upsample(x)
|
| 364 |
+
|
| 365 |
+
x = torch.cat((x, r), dim=1)
|
| 366 |
+
|
| 367 |
+
x = self.final_res_block(x, t)
|
| 368 |
+
return self.final_conv(x)
|
| 369 |
+
|
| 370 |
+
|
| 371 |
+
if __name__ == '__main__':
|
| 372 |
+
model = Unet(channels=1)
|
| 373 |
+
x = torch.randn(1, 1, 128, 128)
|
| 374 |
+
y = model(x, timestep=torch.tensor([100]))
|
| 375 |
+
print(y.shape)
|
requirements.txt
ADDED
|
@@ -0,0 +1,16 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
torch>=1.13.0
|
| 2 |
+
torchvision
|
| 3 |
+
tensorboard==2.13.0
|
| 4 |
+
einops==0.6.1
|
| 5 |
+
gradio==3.35.2
|
| 6 |
+
matplotlib==3.7.1
|
| 7 |
+
numpy==1.24.1
|
| 8 |
+
opencv_python==4.8.0.74
|
| 9 |
+
pandas==2.0.2
|
| 10 |
+
Pillow==9.5.0
|
| 11 |
+
scipy==1.11.1
|
| 12 |
+
seaborn==0.12.2
|
| 13 |
+
tqdm==4.65.0
|
| 14 |
+
scikit_image==0.21.0
|
| 15 |
+
protobuf==3.20
|
| 16 |
+
fastapi==0.99.0
|
train.py
ADDED
|
@@ -0,0 +1,56 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from config import parser
|
| 2 |
+
import argparse
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
from trainers.train_CXR14 import main as train_CXR14
|
| 5 |
+
from trainers.train_baseline import main as train_baseline
|
| 6 |
+
from trainers.train_base_diffusion import main as train_JSRT
|
| 7 |
+
from trainers.train_datasetDM import main as train_datasetDM
|
| 8 |
+
from trainers.datasetDM_per_step import main as train_simple_datasetDM
|
| 9 |
+
from trainers.train_global_cl import main as train_global_cl
|
| 10 |
+
from trainers.train_local_cl import main as train_local_cl
|
| 11 |
+
from trainers.finetune_glob_cl import main as train_global_finetune
|
| 12 |
+
from trainers.finetune_glob_loc_cl import main as train_global_local_finetune
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
if __name__=="__main__":
|
| 16 |
+
parser = argparse.ArgumentParser(parents=[parser], add_help=False)
|
| 17 |
+
config = parser.parse_args()
|
| 18 |
+
|
| 19 |
+
# catch exeptions
|
| 20 |
+
#if len(config.loss_weights) != 4:
|
| 21 |
+
# raise ValueError('loss_weights must be a list of 4 values')
|
| 22 |
+
|
| 23 |
+
config.normalize = True
|
| 24 |
+
config.log_dir = Path(config.log_dir).parent / config.experiment / str(config.n_labelled_images) / Path(config.log_dir).name
|
| 25 |
+
config.channels = 1
|
| 26 |
+
config.out_channels = 1
|
| 27 |
+
if config.dataset == "CXR14":
|
| 28 |
+
config.data_dir = Path("<PATH_TO_DATA>/ChestXray-NIHCC/images")
|
| 29 |
+
elif config.dataset == "JSRT":
|
| 30 |
+
config.data_dir = Path("<PATH_TO_DATA>/JSRT")
|
| 31 |
+
else:
|
| 32 |
+
raise ValueError(f"Unknown dataset: {config.dataset}")
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
if config.experiment == "img_only":
|
| 36 |
+
train_CXR14(config)
|
| 37 |
+
elif config.experiment == "baseline":
|
| 38 |
+
train_baseline(config)
|
| 39 |
+
elif config.experiment == "LEDM":
|
| 40 |
+
config.t_steps_to_save = [50, 150, 250]
|
| 41 |
+
train_datasetDM(config)
|
| 42 |
+
elif config.experiment == "LEDMe":
|
| 43 |
+
config.t_steps_to_save = [1, 10, 25, 50, 200, 400, 600, 800]
|
| 44 |
+
train_datasetDM(config)
|
| 45 |
+
elif config.experiment == "TEDM":
|
| 46 |
+
config.shared_weights_over_timesteps = True
|
| 47 |
+
config.t_steps_to_save = [1, 10, 25, 50, 200, 400, 600, 800]
|
| 48 |
+
train_datasetDM(config)
|
| 49 |
+
elif config.experiment == 'global_cl':
|
| 50 |
+
train_global_cl(config)
|
| 51 |
+
elif config.experiment == 'local_cl':
|
| 52 |
+
train_local_cl(config)
|
| 53 |
+
elif config.experiment == 'global_finetune':
|
| 54 |
+
train_global_finetune(config)
|
| 55 |
+
elif config.experiment == 'glob_loc_finetune':
|
| 56 |
+
train_global_local_finetune(config)
|
trainers/datasetDM_per_step.py
ADDED
|
@@ -0,0 +1,115 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from argparse import Namespace
|
| 2 |
+
import os
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
from dataloaders.JSRT import build_dataloaders
|
| 5 |
+
import torch
|
| 6 |
+
from tqdm.auto import tqdm
|
| 7 |
+
from trainers.utils import seed_everything, TensorboardLogger
|
| 8 |
+
from torch.cuda.amp import GradScaler
|
| 9 |
+
from torch import Tensor, nn
|
| 10 |
+
from typing import Dict, Optional
|
| 11 |
+
from trainers.train_baseline import train
|
| 12 |
+
from models.datasetDM_model import DatasetDM
|
| 13 |
+
from einops import repeat
|
| 14 |
+
from einops.layers.torch import Rearrange
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
class ModDatasetDM(DatasetDM):
|
| 18 |
+
# the idea here is to pool info per timestep,
|
| 19 |
+
# so that we can then use the aggregate for feature importance
|
| 20 |
+
|
| 21 |
+
def __init__(self, args: Namespace) -> None:
|
| 22 |
+
super().__init__(args)
|
| 23 |
+
self.mean = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 24 |
+
self.mean_squared = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 25 |
+
self.std = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 26 |
+
self.classifier = nn.Conv2d(len(self.steps) * 960, 1, 1)
|
| 27 |
+
|
| 28 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 29 |
+
features = self.extract_features(x).to(x.device)
|
| 30 |
+
out = (features - self.mean ) / self.std
|
| 31 |
+
out = self.classifier(features)
|
| 32 |
+
return out
|
| 33 |
+
|
| 34 |
+
class OneStepPredDatasetDM(DatasetDM):
|
| 35 |
+
# the idea here is to pool info per timestep,
|
| 36 |
+
# so that we can then use the aggregate for feature importance
|
| 37 |
+
|
| 38 |
+
def __init__(self, args: Namespace) -> None:
|
| 39 |
+
super().__init__(args)
|
| 40 |
+
self.mean = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 41 |
+
self.mean_squared = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 42 |
+
self.std = torch.zeros(len(self.steps) * 960, args.img_size, args.img_size, requires_grad=False)
|
| 43 |
+
self.classifier = nn.Sequential(
|
| 44 |
+
Rearrange('b (step act) h w -> (b step) act h w', step=len(self.steps)),
|
| 45 |
+
nn.Conv2d(960, 128, 1),
|
| 46 |
+
nn.ReLU(),
|
| 47 |
+
nn.BatchNorm2d(128),
|
| 48 |
+
nn.Conv2d(128, 32, 1),
|
| 49 |
+
nn.ReLU(),
|
| 50 |
+
nn.BatchNorm2d(32),
|
| 51 |
+
nn.Conv2d(32, 1, args.out_channels)
|
| 52 |
+
)
|
| 53 |
+
|
| 54 |
+
|
| 55 |
+
def forward(self, x: Tensor) -> Tensor:
|
| 56 |
+
features = self.extract_features(x).to(x.device)
|
| 57 |
+
out = (features - self.mean ) / self.std
|
| 58 |
+
out = self.classifier(features)
|
| 59 |
+
return out
|
| 60 |
+
|
| 61 |
+
|
| 62 |
+
def main(config: Namespace) -> None:
|
| 63 |
+
# adjust logdir to include experiment name
|
| 64 |
+
os.makedirs(config.log_dir, exist_ok=True)
|
| 65 |
+
print('Experiment folder: %s' % (config.log_dir))
|
| 66 |
+
|
| 67 |
+
# save config namespace into logdir
|
| 68 |
+
with open(config.log_dir / 'config.txt', 'w') as f:
|
| 69 |
+
for k, v in vars(config).items():
|
| 70 |
+
if type(v) not in [str, int, float, bool]:
|
| 71 |
+
f.write(f'{k}: {str(v)}\n')
|
| 72 |
+
else:
|
| 73 |
+
f.write(f'{k}: {v}\n')
|
| 74 |
+
|
| 75 |
+
# Random seed
|
| 76 |
+
seed_everything(config.seed)
|
| 77 |
+
|
| 78 |
+
model = ModDatasetDM(config)
|
| 79 |
+
model = model.to(config.device)
|
| 80 |
+
model.train()
|
| 81 |
+
|
| 82 |
+
optimizer = torch.optim.Adam(model.classifier.parameters(), lr=config.lr, weight_decay=config.weight_decay) # , betas=config.adam_betas)
|
| 83 |
+
step = 0
|
| 84 |
+
|
| 85 |
+
scaler = GradScaler()
|
| 86 |
+
|
| 87 |
+
dataloaders = build_dataloaders(
|
| 88 |
+
config.data_dir,
|
| 89 |
+
config.img_size,
|
| 90 |
+
config.batch_size,
|
| 91 |
+
config.num_workers,
|
| 92 |
+
config.n_labelled_images
|
| 93 |
+
)
|
| 94 |
+
train_dl = dataloaders['train']
|
| 95 |
+
val_dl = dataloaders['val']
|
| 96 |
+
|
| 97 |
+
# Logger
|
| 98 |
+
logger = TensorboardLogger(config.log_dir, enabled=not config.debug)
|
| 99 |
+
|
| 100 |
+
|
| 101 |
+
# do a loop to calculate mean and variance of the features
|
| 102 |
+
# then use those to normalize the features
|
| 103 |
+
model.to(config.device)
|
| 104 |
+
for x, _ in tqdm(train_dl, desc="Calculating mean and variance"):
|
| 105 |
+
x = x.to(config.device)
|
| 106 |
+
features = model.extract_features(x)
|
| 107 |
+
model.mean += features.sum(dim=0)
|
| 108 |
+
model.mean_squared += (features ** 2).sum(dim=0)
|
| 109 |
+
model.mean = model.mean / len(train_dl.dataset)
|
| 110 |
+
model.std = (model.mean_squared / len(train_dl.dataset) - model.mean ** 2).sqrt() + 1e-6
|
| 111 |
+
|
| 112 |
+
model.mean = model.mean.to(config.device)
|
| 113 |
+
model.std = model.std.to(config.device)
|
| 114 |
+
|
| 115 |
+
train(config, model, optimizer, train_dl, val_dl, logger, scaler, step)
|
trainers/finetune_glob_cl.py
ADDED
|
@@ -0,0 +1,172 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
import os
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
import torch
|
| 5 |
+
from torch import autocast, Tensor
|
| 6 |
+
from torch.nn.functional import binary_cross_entropy_with_logits
|
| 7 |
+
from torch.cuda.amp import GradScaler
|
| 8 |
+
from tqdm import tqdm
|
| 9 |
+
from config import parser
|
| 10 |
+
from einops import rearrange, reduce, repeat
|
| 11 |
+
from dataloaders.JSRT import build_dataloaders
|
| 12 |
+
from models.unet_model import Unet
|
| 13 |
+
from trainers.train_baseline import validate, save
|
| 14 |
+
from trainers.utils import (TensorboardLogger, compare_configs, seed_everything, crop_batch)
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
def train(config, model, optimizer, train_dl, val_dl, logger, scaler, step):
|
| 18 |
+
best_val_loss = float('inf')
|
| 19 |
+
train_losses = []
|
| 20 |
+
if config.dataset == "BRATS2D":
|
| 21 |
+
train_losses_per_class = []
|
| 22 |
+
elif config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 23 |
+
train_losses_per_timestep = []
|
| 24 |
+
|
| 25 |
+
pbar = tqdm(total=config.val_freq, desc='Training')
|
| 26 |
+
while True:
|
| 27 |
+
for x, y in train_dl:
|
| 28 |
+
if config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 29 |
+
y = repeat(y, 'b c h w -> (b step) c h w', step=len(model.steps))
|
| 30 |
+
if config.augment_at_finetuning:
|
| 31 |
+
x, y = crop_batch([x, y], config.img_size, config.batch_size)
|
| 32 |
+
brightness = torch.rand((config.batch_size, 1, 1, 1), device=x.device)*.6 - .3 # random brightness adjustment between [-.3, .3]
|
| 33 |
+
contrast = torch.rand((config.batch_size, 1, 1, 1), device=x.device)*.6 + .7 # random contrast adjustment between [.7, 1.3]
|
| 34 |
+
x = (x + brightness) * contrast # apply brightness and contrast
|
| 35 |
+
|
| 36 |
+
x = x.to(config.device)
|
| 37 |
+
y = y.to(config.device)
|
| 38 |
+
|
| 39 |
+
optimizer.zero_grad()
|
| 40 |
+
with autocast(device_type=config.device, enabled=config.mixed_precision):
|
| 41 |
+
pred = model(x)
|
| 42 |
+
# cross entropy loss
|
| 43 |
+
#loss = - ((y * torch.log(torch.sigmoid(pred)) + (1 - y) * torch.log(1 - torch.sigmoid(pred)))).mean()
|
| 44 |
+
if config.dataset == "BRATS2D":
|
| 45 |
+
weights = repeat(torch.Tensor(config.loss_weights).to(config.device), 'c -> b c h w', b=y.shape[0], h=y.shape[2], w=y.shape[3])
|
| 46 |
+
else:
|
| 47 |
+
weights = None
|
| 48 |
+
expanded_loss = reduce(binary_cross_entropy_with_logits(pred, y, weight=weights, reduction='none'), 'b c h w -> b c', 'mean')
|
| 49 |
+
loss = expanded_loss.mean()
|
| 50 |
+
scaler.scale(loss).backward()
|
| 51 |
+
optimizer.step()
|
| 52 |
+
|
| 53 |
+
train_losses.append(loss.item())
|
| 54 |
+
if config.dataset == "BRATS2D":
|
| 55 |
+
loss_per_class = expanded_loss.mean(0)
|
| 56 |
+
train_losses_per_class.append(loss_per_class.detach().cpu())
|
| 57 |
+
pbar.set_description(f'Training loss: {loss.item():.4f} - {loss_per_class[0].item():.4f} - {loss_per_class[1].item():.4f} - {loss_per_class[2].item():.4f} - {loss_per_class[3].item():.4f}')
|
| 58 |
+
else:
|
| 59 |
+
pbar.set_description(f'Training loss: {loss.item():.4f}')
|
| 60 |
+
|
| 61 |
+
pbar.update(1)
|
| 62 |
+
step += 1
|
| 63 |
+
|
| 64 |
+
if config.unfreeze_weights_at_step == step:
|
| 65 |
+
for name, param in model.named_parameters():
|
| 66 |
+
if name.startswith('downs') or name.startswith('init_conv') or name.startswith('mid_'):
|
| 67 |
+
param.requires_grad = True
|
| 68 |
+
|
| 69 |
+
if step % config.log_freq == 0 or config.debug:
|
| 70 |
+
avg_train_loss = sum(train_losses) / len(train_losses)
|
| 71 |
+
print(f'Step {step} - Train loss: {avg_train_loss:.4f}')
|
| 72 |
+
logger.log({'train/loss': avg_train_loss}, step=step)
|
| 73 |
+
if config.dataset == "BRATS2D":
|
| 74 |
+
avg_train_loss_per_class = torch.stack(train_losses_per_class).mean(0)
|
| 75 |
+
logger.log({'train_loss/0':avg_train_loss_per_class[0].item()}, step=step)
|
| 76 |
+
logger.log({'train_loss/1':avg_train_loss_per_class[1].item()}, step=step)
|
| 77 |
+
logger.log({'train_loss/2':avg_train_loss_per_class[2].item()}, step=step)
|
| 78 |
+
logger.log({'train_loss/3':avg_train_loss_per_class[3].item()}, step=step)
|
| 79 |
+
if config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 80 |
+
avg_train_loss_per_timestep = torch.stack(train_losses_per_timestep).mean(0)
|
| 81 |
+
for i, model_step in enumerate(model.steps):
|
| 82 |
+
logger.log({'train_loss/step_' + str(model_step): avg_train_loss_per_timestep[i].item()}, step=step)
|
| 83 |
+
|
| 84 |
+
if step % config.val_freq == 0 or config.debug:
|
| 85 |
+
val_results = validate(config, model, val_dl)
|
| 86 |
+
logger.log(val_results, step=step)
|
| 87 |
+
|
| 88 |
+
if val_results['val/loss'] < best_val_loss and not config.debug:
|
| 89 |
+
print(f'Step {step} - New best validation loss: '
|
| 90 |
+
f'{val_results["val/loss"]:.4f}, saving model '
|
| 91 |
+
f'in {config.log_dir}')
|
| 92 |
+
best_val_loss = val_results['val/loss']
|
| 93 |
+
save(
|
| 94 |
+
model,
|
| 95 |
+
optimizer,
|
| 96 |
+
config,
|
| 97 |
+
config.log_dir / 'best_model.pt',
|
| 98 |
+
step
|
| 99 |
+
)
|
| 100 |
+
elif val_results['val/loss'] > best_val_loss * 1.5 and config.early_stop:
|
| 101 |
+
print(f'Step {step} - Validation loss increased by more than 50%')
|
| 102 |
+
return model
|
| 103 |
+
|
| 104 |
+
if step >= config.max_steps or config.debug:
|
| 105 |
+
return model
|
| 106 |
+
|
| 107 |
+
|
| 108 |
+
|
| 109 |
+
|
| 110 |
+
def load(config, path):
|
| 111 |
+
raise NotImplementedError
|
| 112 |
+
|
| 113 |
+
def main(config):
|
| 114 |
+
|
| 115 |
+
os.makedirs(config.log_dir, exist_ok=True)
|
| 116 |
+
|
| 117 |
+
# save config namespace into logdir
|
| 118 |
+
with open(config.log_dir / 'config.txt', 'w') as f:
|
| 119 |
+
for k, v in vars(config).items():
|
| 120 |
+
if type(v) not in [str, int, float, bool]:
|
| 121 |
+
f.write(f'{k}: {str(v)}\n')
|
| 122 |
+
else:
|
| 123 |
+
f.write(f'{k}: {v}\n')
|
| 124 |
+
|
| 125 |
+
# Random seed
|
| 126 |
+
seed_everything(config.seed)
|
| 127 |
+
|
| 128 |
+
# Init model and optimizer
|
| 129 |
+
if config.resume_path is not None:
|
| 130 |
+
print('Loading model from', config.resume_path)
|
| 131 |
+
model, optimizer, step = load(config, config.resume_path)
|
| 132 |
+
else:
|
| 133 |
+
model = Unet(
|
| 134 |
+
img_size=config.img_size,
|
| 135 |
+
dim=config.dim,
|
| 136 |
+
dim_mults=config.dim_mults,
|
| 137 |
+
channels=config.channels,
|
| 138 |
+
out_dim=config.out_channels)
|
| 139 |
+
state_dict = torch.load(config.global_model_path, map_location='cpu')['model_state_dict']
|
| 140 |
+
out = model.load_state_dict(state_dict=state_dict, strict=False)
|
| 141 |
+
print("Loaded state dict. \n\tMissing keys: {}\n\tUnexpected keys: {}".format(out.missing_keys, out.unexpected_keys))
|
| 142 |
+
print('Note that although the state dict of the decoder is loaded, its values are random.')
|
| 143 |
+
if config.unfreeze_weights_at_step !=0:
|
| 144 |
+
for name, param in model.named_parameters():
|
| 145 |
+
if name.startswith('downs') or name.startswith('init_conv') or name.startswith('mid_'):
|
| 146 |
+
param.requires_grad = False
|
| 147 |
+
|
| 148 |
+
optimizer = torch.optim.Adam(model.parameters(), lr=config.lr) # , betas=config.adam_betas)
|
| 149 |
+
|
| 150 |
+
step = 0
|
| 151 |
+
model.to(config.device)
|
| 152 |
+
model.train()
|
| 153 |
+
|
| 154 |
+
scaler = GradScaler()
|
| 155 |
+
|
| 156 |
+
# Load data
|
| 157 |
+
dataloaders = build_dataloaders(
|
| 158 |
+
config.data_dir,
|
| 159 |
+
config.img_size,
|
| 160 |
+
config.batch_size,
|
| 161 |
+
config.num_workers,
|
| 162 |
+
n_labelled_images=config.n_labelled_images,
|
| 163 |
+
)
|
| 164 |
+
train_dl = dataloaders['train']
|
| 165 |
+
val_dl = dataloaders['val']
|
| 166 |
+
print('Train dataset size:', len(train_dl.dataset))
|
| 167 |
+
print('Validation dataset size:', len(val_dl.dataset))
|
| 168 |
+
|
| 169 |
+
# Logger
|
| 170 |
+
logger = TensorboardLogger(config.log_dir, enabled=not config.debug)
|
| 171 |
+
|
| 172 |
+
train(config, model, optimizer, train_dl, val_dl, logger, scaler, step)
|
trainers/finetune_glob_loc_cl.py
ADDED
|
@@ -0,0 +1,172 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
import os
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
import torch
|
| 5 |
+
from torch import autocast, Tensor
|
| 6 |
+
from torch.nn.functional import binary_cross_entropy_with_logits
|
| 7 |
+
from torch.cuda.amp import GradScaler
|
| 8 |
+
from tqdm import tqdm
|
| 9 |
+
from config import parser
|
| 10 |
+
from einops import rearrange, reduce, repeat
|
| 11 |
+
from dataloaders.JSRT import build_dataloaders
|
| 12 |
+
from models.unet_model import Unet
|
| 13 |
+
from trainers.train_baseline import validate, save
|
| 14 |
+
from trainers.utils import (TensorboardLogger, compare_configs, seed_everything, crop_batch)
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
def train(config, model, optimizer, train_dl, val_dl, logger, scaler, step):
|
| 18 |
+
best_val_loss = float('inf')
|
| 19 |
+
train_losses = []
|
| 20 |
+
if config.dataset == "BRATS2D":
|
| 21 |
+
train_losses_per_class = []
|
| 22 |
+
elif config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 23 |
+
train_losses_per_timestep = []
|
| 24 |
+
|
| 25 |
+
pbar = tqdm(total=config.val_freq, desc='Training')
|
| 26 |
+
while True:
|
| 27 |
+
for x, y in train_dl:
|
| 28 |
+
if config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 29 |
+
y = repeat(y, 'b c h w -> (b step) c h w', step=len(model.steps))
|
| 30 |
+
if config.augment_at_finetuning:
|
| 31 |
+
x, y = crop_batch([x, y], config.img_size, config.batch_size)
|
| 32 |
+
brightness = torch.rand((config.batch_size, 1, 1, 1), device=x.device)*.6 - .3 # random brightness adjustment between [-.3, .3]
|
| 33 |
+
contrast = torch.rand((config.batch_size, 1, 1, 1), device=x.device)*.6 + .7 # random contrast adjustment between [.7, 1.3]
|
| 34 |
+
x = (x + brightness) * contrast # apply brightness and contrast
|
| 35 |
+
|
| 36 |
+
x = x.to(config.device)
|
| 37 |
+
y = y.to(config.device)
|
| 38 |
+
|
| 39 |
+
optimizer.zero_grad()
|
| 40 |
+
with autocast(device_type=config.device, enabled=config.mixed_precision):
|
| 41 |
+
pred = model(x)
|
| 42 |
+
# cross entropy loss
|
| 43 |
+
#loss = - ((y * torch.log(torch.sigmoid(pred)) + (1 - y) * torch.log(1 - torch.sigmoid(pred)))).mean()
|
| 44 |
+
if config.dataset == "BRATS2D":
|
| 45 |
+
weights = repeat(torch.Tensor(config.loss_weights).to(config.device), 'c -> b c h w', b=y.shape[0], h=y.shape[2], w=y.shape[3])
|
| 46 |
+
else:
|
| 47 |
+
weights = None
|
| 48 |
+
expanded_loss = reduce(binary_cross_entropy_with_logits(pred, y, weight=weights, reduction='none'), 'b c h w -> b c', 'mean')
|
| 49 |
+
loss = expanded_loss.mean()
|
| 50 |
+
scaler.scale(loss).backward()
|
| 51 |
+
optimizer.step()
|
| 52 |
+
|
| 53 |
+
train_losses.append(loss.item())
|
| 54 |
+
if config.dataset == "BRATS2D":
|
| 55 |
+
loss_per_class = expanded_loss.mean(0)
|
| 56 |
+
train_losses_per_class.append(loss_per_class.detach().cpu())
|
| 57 |
+
pbar.set_description(f'Training loss: {loss.item():.4f} - {loss_per_class[0].item():.4f} - {loss_per_class[1].item():.4f} - {loss_per_class[2].item():.4f} - {loss_per_class[3].item():.4f}')
|
| 58 |
+
else:
|
| 59 |
+
pbar.set_description(f'Training loss: {loss.item():.4f}')
|
| 60 |
+
|
| 61 |
+
pbar.update(1)
|
| 62 |
+
step += 1
|
| 63 |
+
|
| 64 |
+
if config.unfreeze_weights_at_step == step:
|
| 65 |
+
for name, param in model.named_parameters():
|
| 66 |
+
if name.startswith('downs') or name.startswith('init_conv') or name.startswith('mid_'):
|
| 67 |
+
param.requires_grad = True
|
| 68 |
+
|
| 69 |
+
if step % config.log_freq == 0 or config.debug:
|
| 70 |
+
avg_train_loss = sum(train_losses) / len(train_losses)
|
| 71 |
+
print(f'Step {step} - Train loss: {avg_train_loss:.4f}')
|
| 72 |
+
logger.log({'train/loss': avg_train_loss}, step=step)
|
| 73 |
+
if config.dataset == "BRATS2D":
|
| 74 |
+
avg_train_loss_per_class = torch.stack(train_losses_per_class).mean(0)
|
| 75 |
+
logger.log({'train_loss/0':avg_train_loss_per_class[0].item()}, step=step)
|
| 76 |
+
logger.log({'train_loss/1':avg_train_loss_per_class[1].item()}, step=step)
|
| 77 |
+
logger.log({'train_loss/2':avg_train_loss_per_class[2].item()}, step=step)
|
| 78 |
+
logger.log({'train_loss/3':avg_train_loss_per_class[3].item()}, step=step)
|
| 79 |
+
if config.shared_weights_over_timesteps and config.experiment == 'datasetDM':
|
| 80 |
+
avg_train_loss_per_timestep = torch.stack(train_losses_per_timestep).mean(0)
|
| 81 |
+
for i, model_step in enumerate(model.steps):
|
| 82 |
+
logger.log({'train_loss/step_' + str(model_step): avg_train_loss_per_timestep[i].item()}, step=step)
|
| 83 |
+
|
| 84 |
+
if step % config.val_freq == 0 or config.debug:
|
| 85 |
+
val_results = validate(config, model, val_dl)
|
| 86 |
+
logger.log(val_results, step=step)
|
| 87 |
+
|
| 88 |
+
if val_results['val/loss'] < best_val_loss and not config.debug:
|
| 89 |
+
print(f'Step {step} - New best validation loss: '
|
| 90 |
+
f'{val_results["val/loss"]:.4f}, saving model '
|
| 91 |
+
f'in {config.log_dir}')
|
| 92 |
+
best_val_loss = val_results['val/loss']
|
| 93 |
+
save(
|
| 94 |
+
model,
|
| 95 |
+
optimizer,
|
| 96 |
+
config,
|
| 97 |
+
config.log_dir / 'best_model.pt',
|
| 98 |
+
step
|
| 99 |
+
)
|
| 100 |
+
elif val_results['val/loss'] > best_val_loss * 1.5 and config.early_stop:
|
| 101 |
+
print(f'Step {step} - Validation loss increased by more than 50%')
|
| 102 |
+
return model
|
| 103 |
+
|
| 104 |
+
if step >= config.max_steps or config.debug:
|
| 105 |
+
return model
|
| 106 |
+
|
| 107 |
+
|
| 108 |
+
|
| 109 |
+
|
| 110 |
+
def load(config, path):
|
| 111 |
+
raise NotImplementedError
|
| 112 |
+
|
| 113 |
+
def main(config):
|
| 114 |
+
|
| 115 |
+
os.makedirs(config.log_dir, exist_ok=True)
|
| 116 |
+
|
| 117 |
+
# save config namespace into logdir
|
| 118 |
+
with open(config.log_dir / 'config.txt', 'w') as f:
|
| 119 |
+
for k, v in vars(config).items():
|
| 120 |
+
if type(v) not in [str, int, float, bool]:
|
| 121 |
+
f.write(f'{k}: {str(v)}\n')
|
| 122 |
+
else:
|
| 123 |
+
f.write(f'{k}: {v}\n')
|
| 124 |
+
|
| 125 |
+
# Random seed
|
| 126 |
+
seed_everything(config.seed)
|
| 127 |
+
|
| 128 |
+
# Init model and optimizer
|
| 129 |
+
if config.resume_path is not None:
|
| 130 |
+
print('Loading model from', config.resume_path)
|
| 131 |
+
model, optimizer, step = load(config, config.resume_path)
|
| 132 |
+
else:
|
| 133 |
+
model = Unet(
|
| 134 |
+
img_size=config.img_size,
|
| 135 |
+
dim=config.dim,
|
| 136 |
+
dim_mults=config.dim_mults,
|
| 137 |
+
channels=config.channels,
|
| 138 |
+
out_dim=config.out_channels)
|
| 139 |
+
state_dict = torch.load(config.glob_loc_model_path, map_location='cpu')['model_state_dict']
|
| 140 |
+
out = model.load_state_dict(state_dict=state_dict, strict=False)
|
| 141 |
+
print("Loaded state dict. \n\tMissing keys: {}\n\tUnexpected keys: {}".format(out.missing_keys, out.unexpected_keys))
|
| 142 |
+
print('Note that although the state dict of the decoder is loaded, its values are random.')
|
| 143 |
+
if config.unfreeze_weights_at_step !=0:
|
| 144 |
+
for name, param in model.named_parameters():
|
| 145 |
+
if name.startswith('downs') or name.startswith('init_conv') or name.startswith('mid_'):
|
| 146 |
+
param.requires_grad = False
|
| 147 |
+
|
| 148 |
+
optimizer = torch.optim.Adam(model.parameters(), lr=config.lr) # , betas=config.adam_betas)
|
| 149 |
+
|
| 150 |
+
step = 0
|
| 151 |
+
model.to(config.device)
|
| 152 |
+
model.train()
|
| 153 |
+
|
| 154 |
+
scaler = GradScaler()
|
| 155 |
+
|
| 156 |
+
# Load data
|
| 157 |
+
dataloaders = build_dataloaders(
|
| 158 |
+
config.data_dir,
|
| 159 |
+
config.img_size,
|
| 160 |
+
config.batch_size,
|
| 161 |
+
config.num_workers,
|
| 162 |
+
n_labelled_images=config.n_labelled_images,
|
| 163 |
+
)
|
| 164 |
+
train_dl = dataloaders['train']
|
| 165 |
+
val_dl = dataloaders['val']
|
| 166 |
+
print('Train dataset size:', len(train_dl.dataset))
|
| 167 |
+
print('Validation dataset size:', len(val_dl.dataset))
|
| 168 |
+
|
| 169 |
+
# Logger
|
| 170 |
+
logger = TensorboardLogger(config.log_dir, enabled=not config.debug)
|
| 171 |
+
|
| 172 |
+
train(config, model, optimizer, train_dl, val_dl, logger, scaler, step)
|