Upload 7 files
Browse files- Model_arch.png +0 -0
- SkimLit.ipynb +0 -0
- Untitled.ipynb +123 -0
- app.py +132 -0
- model.png +0 -0
- skim.png +0 -0
- skimlit_example_abstracts.json +1 -0
Model_arch.png
ADDED
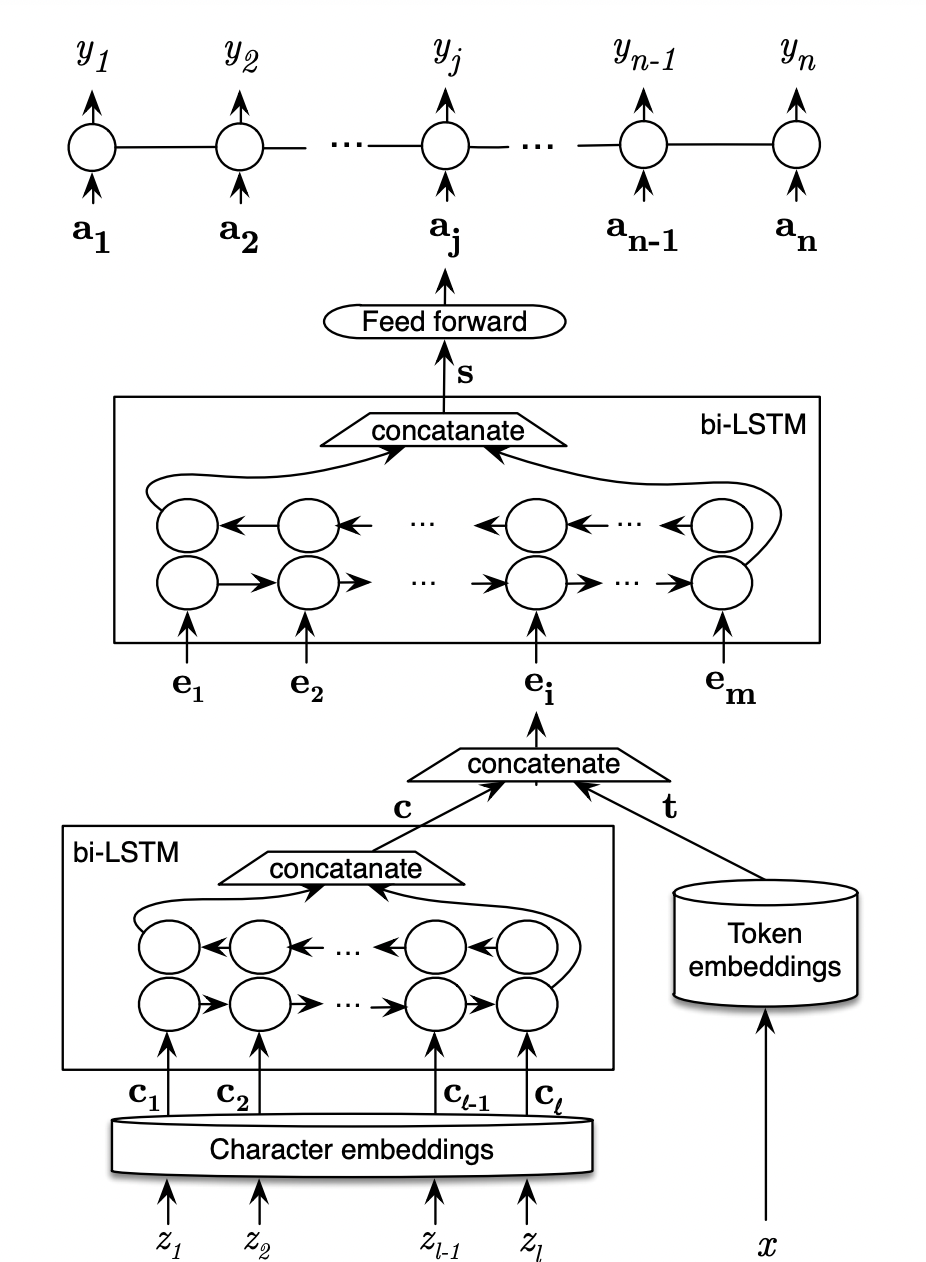
|
SkimLit.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
Untitled.ipynb
ADDED
|
@@ -0,0 +1,123 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 4,
|
| 6 |
+
"id": "3d72f70e",
|
| 7 |
+
"metadata": {},
|
| 8 |
+
"outputs": [],
|
| 9 |
+
"source": [
|
| 10 |
+
"import cv2\n",
|
| 11 |
+
"import os\n",
|
| 12 |
+
"import numpy as np\n",
|
| 13 |
+
"import pickle\n",
|
| 14 |
+
"import tensorflow as tf\n",
|
| 15 |
+
"from tensorflow.keras import layers\n",
|
| 16 |
+
"\n",
|
| 17 |
+
"from tensorflow.keras import models,utils\n",
|
| 18 |
+
"\n",
|
| 19 |
+
"import pandas as pd\n",
|
| 20 |
+
"\n",
|
| 21 |
+
"from tensorflow.keras.models import load_model\n",
|
| 22 |
+
"\n",
|
| 23 |
+
"from tensorflow.keras.preprocessing.image import load_img,img_to_array\n",
|
| 24 |
+
"\n",
|
| 25 |
+
"from tensorflow.python.keras import utils\n",
|
| 26 |
+
"current_path = os.getcwd()\n"
|
| 27 |
+
]
|
| 28 |
+
},
|
| 29 |
+
{
|
| 30 |
+
"cell_type": "code",
|
| 31 |
+
"execution_count": 3,
|
| 32 |
+
"id": "d0655482",
|
| 33 |
+
"metadata": {},
|
| 34 |
+
"outputs": [
|
| 35 |
+
{
|
| 36 |
+
"name": "stdout",
|
| 37 |
+
"output_type": "stream",
|
| 38 |
+
"text": [
|
| 39 |
+
"Collecting opencv-python\n",
|
| 40 |
+
" Downloading opencv_python-4.7.0.72-cp37-abi3-macosx_11_0_arm64.whl (32.6 MB)\n",
|
| 41 |
+
"\u001b[2K \u001b[90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\u001b[0m \u001b[32m32.6/32.6 MB\u001b[0m \u001b[31m1.4 MB/s\u001b[0m eta \u001b[36m0:00:00\u001b[0m00:01\u001b[0m00:01\u001b[0m\n",
|
| 42 |
+
"\u001b[?25hRequirement already satisfied: numpy>=1.21.2 in /Users/ujjwalbansal/anaconda3/lib/python3.10/site-packages (from opencv-python) (1.24.3)\n",
|
| 43 |
+
"Installing collected packages: opencv-python\n",
|
| 44 |
+
"Successfully installed opencv-python-4.7.0.72\n",
|
| 45 |
+
"Note: you may need to restart the kernel to use updated packages.\n"
|
| 46 |
+
]
|
| 47 |
+
}
|
| 48 |
+
],
|
| 49 |
+
"source": [
|
| 50 |
+
"pip install opencv-python"
|
| 51 |
+
]
|
| 52 |
+
},
|
| 53 |
+
{
|
| 54 |
+
"cell_type": "code",
|
| 55 |
+
"execution_count": 10,
|
| 56 |
+
"id": "867fc892",
|
| 57 |
+
"metadata": {},
|
| 58 |
+
"outputs": [],
|
| 59 |
+
"source": [
|
| 60 |
+
"import tensorflow_hub as hub"
|
| 61 |
+
]
|
| 62 |
+
},
|
| 63 |
+
{
|
| 64 |
+
"cell_type": "code",
|
| 65 |
+
"execution_count": 13,
|
| 66 |
+
"id": "0fb1cb14",
|
| 67 |
+
"metadata": {},
|
| 68 |
+
"outputs": [],
|
| 69 |
+
"source": [
|
| 70 |
+
"model_path=\"skimlit_final_model\""
|
| 71 |
+
]
|
| 72 |
+
},
|
| 73 |
+
{
|
| 74 |
+
"cell_type": "code",
|
| 75 |
+
"execution_count": 14,
|
| 76 |
+
"id": "20ad6fa1",
|
| 77 |
+
"metadata": {},
|
| 78 |
+
"outputs": [
|
| 79 |
+
{
|
| 80 |
+
"name": "stderr",
|
| 81 |
+
"output_type": "stream",
|
| 82 |
+
"text": [
|
| 83 |
+
"2023-06-14 12:49:03.226973: W tensorflow/tsl/platform/profile_utils/cpu_utils.cc:128] Failed to get CPU frequency: 0 Hz\n"
|
| 84 |
+
]
|
| 85 |
+
}
|
| 86 |
+
],
|
| 87 |
+
"source": [
|
| 88 |
+
"my_reloaded_model = tf.keras.models.load_model(\n",
|
| 89 |
+
" (model_path),\n",
|
| 90 |
+
" custom_objects={'KerasLayer':hub.KerasLayer})"
|
| 91 |
+
]
|
| 92 |
+
},
|
| 93 |
+
{
|
| 94 |
+
"cell_type": "code",
|
| 95 |
+
"execution_count": null,
|
| 96 |
+
"id": "ece84ef9",
|
| 97 |
+
"metadata": {},
|
| 98 |
+
"outputs": [],
|
| 99 |
+
"source": []
|
| 100 |
+
}
|
| 101 |
+
],
|
| 102 |
+
"metadata": {
|
| 103 |
+
"kernelspec": {
|
| 104 |
+
"display_name": "Python 3.10 (tensorflow)",
|
| 105 |
+
"language": "python",
|
| 106 |
+
"name": "tensorflow"
|
| 107 |
+
},
|
| 108 |
+
"language_info": {
|
| 109 |
+
"codemirror_mode": {
|
| 110 |
+
"name": "ipython",
|
| 111 |
+
"version": 3
|
| 112 |
+
},
|
| 113 |
+
"file_extension": ".py",
|
| 114 |
+
"mimetype": "text/x-python",
|
| 115 |
+
"name": "python",
|
| 116 |
+
"nbconvert_exporter": "python",
|
| 117 |
+
"pygments_lexer": "ipython3",
|
| 118 |
+
"version": "3.10.9"
|
| 119 |
+
}
|
| 120 |
+
},
|
| 121 |
+
"nbformat": 4,
|
| 122 |
+
"nbformat_minor": 5
|
| 123 |
+
}
|
app.py
ADDED
|
@@ -0,0 +1,132 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import tensorflow as tf
|
| 3 |
+
from spacy.lang.en import English
|
| 4 |
+
from PIL import Image
|
| 5 |
+
|
| 6 |
+
def load_image(file):
|
| 7 |
+
img = Image.open(file)
|
| 8 |
+
return img
|
| 9 |
+
|
| 10 |
+
def preprocess_text(text):
|
| 11 |
+
nlp = English()
|
| 12 |
+
nlp.add_pipe('sentencizer')
|
| 13 |
+
doc = nlp(text)
|
| 14 |
+
sentences = [str(sent) for sent in list(doc.sents)]
|
| 15 |
+
return data_vis(sentences)
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
def data_vis(sentences):
|
| 20 |
+
total_lines_in_sample = len(sentences)
|
| 21 |
+
sample_lines = []
|
| 22 |
+
for i, line in enumerate(sentences):
|
| 23 |
+
sample_dict = {}
|
| 24 |
+
sample_dict["text"] = str(line)
|
| 25 |
+
sample_dict["line_number"] = i
|
| 26 |
+
sample_dict["total_lines"] = total_lines_in_sample - 1
|
| 27 |
+
sample_lines.append(sample_dict)
|
| 28 |
+
return sample_lines
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
def one_hot_encoding_line_numbers(sample_lines):
|
| 32 |
+
test_abstract_line_numbers = [line["line_number"] for line in sample_lines]
|
| 33 |
+
test_abstract_line_numbers_one_hot = tf.one_hot(test_abstract_line_numbers, depth=15)
|
| 34 |
+
return test_abstract_line_numbers_one_hot
|
| 35 |
+
|
| 36 |
+
def one_hot_encoding_total_lines(sample_lines):
|
| 37 |
+
test_abstract_total_lines = [line["total_lines"] for line in sample_lines]
|
| 38 |
+
test_abstract_total_lines_one_hot = tf.one_hot(test_abstract_total_lines, depth=20)
|
| 39 |
+
return test_abstract_total_lines_one_hot
|
| 40 |
+
|
| 41 |
+
def spacing_char(text):
|
| 42 |
+
nlp = English()
|
| 43 |
+
nlp.add_pipe('sentencizer')
|
| 44 |
+
doc=nlp(text)
|
| 45 |
+
sentences = [sent.text for sent in doc.sents]
|
| 46 |
+
abstract_chars = [split_to_char(sentence) for sentence in sentences]
|
| 47 |
+
return sentences,abstract_chars
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
classes=['BACKGROUND', 'CONCLUSIONS', 'METHODS', 'OBJECTIVE', 'RESULTS']
|
| 51 |
+
|
| 52 |
+
loaded_model=tf.keras.models.load_model("skimlit_final_model")
|
| 53 |
+
|
| 54 |
+
|
| 55 |
+
def split_to_char(text):
|
| 56 |
+
return " " .join(list(text))
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
def main():
|
| 60 |
+
st.title('SkimLit 📕')
|
| 61 |
+
st.caption('### An NLP model to classify abstract sentences into the role they play (e.g. objective, methods, results, etc..) to enable researchers to skim through the literature and dive deeper when necessary.')
|
| 62 |
+
st.image(load_image('skim.png'))
|
| 63 |
+
|
| 64 |
+
raw_text=st.text_area('### Enter the text you want to analyse. Please do not leave the space empty')
|
| 65 |
+
button= st.button('Extract')
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
if button:
|
| 69 |
+
if raw_text is None:
|
| 70 |
+
st.write("Error occurred during preprocessing. Please check your input.")
|
| 71 |
+
return
|
| 72 |
+
else:
|
| 73 |
+
lines=preprocess_text(raw_text)
|
| 74 |
+
test_abstract_line_numbers_one_hot=one_hot_encoding_line_numbers(lines)
|
| 75 |
+
test_abstract_total_lines_one_hot=one_hot_encoding_total_lines(lines)
|
| 76 |
+
abstract_lines,abstract_chars=spacing_char(raw_text)
|
| 77 |
+
|
| 78 |
+
tf.config.run_functions_eagerly(True)
|
| 79 |
+
|
| 80 |
+
test_abstract_pred_probs=loaded_model.predict(x=(test_abstract_line_numbers_one_hot,
|
| 81 |
+
test_abstract_total_lines_one_hot,
|
| 82 |
+
tf.constant(abstract_lines),
|
| 83 |
+
tf.constant(abstract_chars)))
|
| 84 |
+
test_abstract_preds = tf.argmax(test_abstract_pred_probs, axis=1)
|
| 85 |
+
with st.expander('Original Text'):
|
| 86 |
+
st.write(raw_text)
|
| 87 |
+
text_abstract_pred_classes = [classes[i] for i in test_abstract_preds]
|
| 88 |
+
|
| 89 |
+
objective = ''
|
| 90 |
+
background = ''
|
| 91 |
+
method = ''
|
| 92 |
+
conclusion = ''
|
| 93 |
+
result = ''
|
| 94 |
+
for i, line in enumerate(abstract_lines):
|
| 95 |
+
if text_abstract_pred_classes[i]=='OBJECTIVE':
|
| 96 |
+
objective=objective+line
|
| 97 |
+
if text_abstract_pred_classes[i]=='BACKGROUND':
|
| 98 |
+
background=background+line
|
| 99 |
+
if text_abstract_pred_classes[i]=='METHODS':
|
| 100 |
+
method=method+line
|
| 101 |
+
if text_abstract_pred_classes[i]=='RESULTS':
|
| 102 |
+
result =result+line
|
| 103 |
+
if text_abstract_pred_classes[i]=='CONCLUSIONS':
|
| 104 |
+
conclusion =conclusion+line
|
| 105 |
+
|
| 106 |
+
with st.spinner('Wait for prediction....'):
|
| 107 |
+
st.markdown(f'### Objective : ')
|
| 108 |
+
st.write(f'{objective}')
|
| 109 |
+
st.markdown(f'### Background : ')
|
| 110 |
+
st.write(f'{background}')
|
| 111 |
+
st.markdown(f'### Methods : ')
|
| 112 |
+
st.write(f'{method}')
|
| 113 |
+
st.markdown(f'### Result : ')
|
| 114 |
+
st.write(f'{result}')
|
| 115 |
+
st.markdown(f'### Conclusion : ')
|
| 116 |
+
st.write(f'{conclusion}')
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
|
| 120 |
+
|
| 121 |
+
|
| 122 |
+
|
| 123 |
+
# st.write(text_abstract_pred_classes[i])
|
| 124 |
+
# st.write(line)
|
| 125 |
+
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
|
| 129 |
+
|
| 130 |
+
|
| 131 |
+
if __name__ == '__main__':
|
| 132 |
+
main()
|
model.png
ADDED
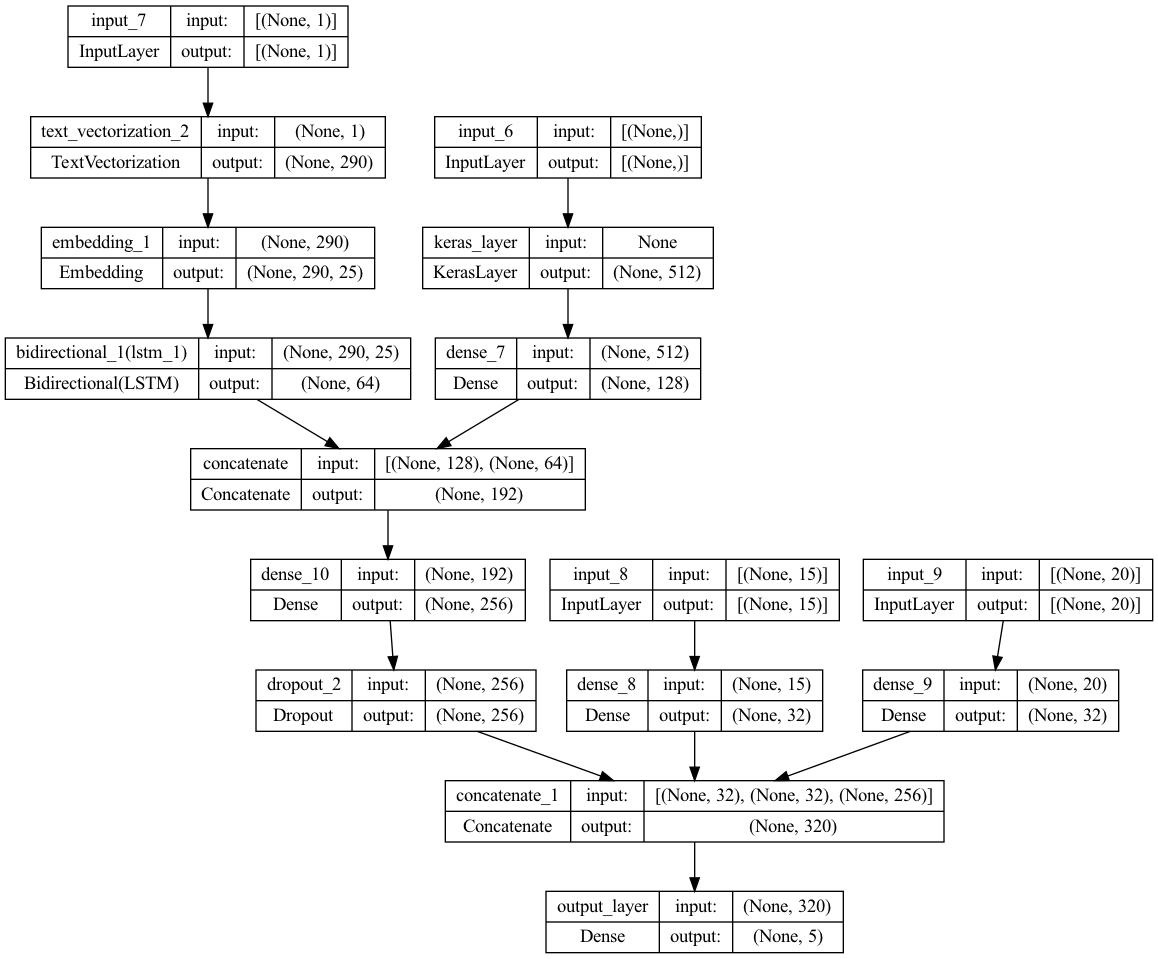
|
skim.png
ADDED

|
skimlit_example_abstracts.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
[{"abstract": "This RCT examined the efficacy of a manualized social intervention for children with HFASDs. Participants were randomly assigned to treatment or wait-list conditions. Treatment included instruction and therapeutic activities targeting social skills, face-emotion recognition, interest expansion, and interpretation of non-literal language. A response-cost program was applied to reduce problem behaviors and foster skills acquisition. Significant treatment effects were found for five of seven primary outcome measures (parent ratings and direct child measures). Secondary measures based on staff ratings (treatment group only) corroborated gains reported by parents. High levels of parent, child and staff satisfaction were reported, along with high levels of treatment fidelity. Standardized effect size estimates were primarily in the medium and large ranges and favored the treatment group.", "source": "https://pubmed.ncbi.nlm.nih.gov/20232240/", "details": "RCT of a manualized social treatment for high-functioning autism spectrum disorders"}, {"abstract": "Postpartum depression (PPD) is the most prevalent mood disorder associated with childbirth. No single cause of PPD has been identified, however the increased risk of nutritional deficiencies incurred through the high nutritional requirements of pregnancy may play a role in the pathology of depressive symptoms. Three nutritional interventions have drawn particular interest as possible non-invasive and cost-effective prevention and/or treatment strategies for PPD; omega-3 (n-3) long chain polyunsaturated fatty acids (LCPUFA), vitamin D and overall diet. We searched for meta-analyses of randomised controlled trials (RCT's) of nutritional interventions during the perinatal period with PPD as an outcome, and checked for any trials published subsequently to the meta-analyses. Fish oil: Eleven RCT's of prenatal fish oil supplementation RCT's show null and positive effects on PPD symptoms. Vitamin D: no relevant RCT's were identified, however seven observational studies of maternal vitamin D levels with PPD outcomes showed inconsistent associations. Diet: Two Australian RCT's with dietary advice interventions in pregnancy had a positive and null result on PPD. With the exception of fish oil, few RCT's with nutritional interventions during pregnancy assess PPD. Further research is needed to determine whether nutritional intervention strategies during pregnancy can protect against symptoms of PPD. Given the prevalence of PPD and ease of administering PPD measures, we recommend future prenatal nutritional RCT's include PPD as an outcome.", "source": "https://pubmed.ncbi.nlm.nih.gov/28012571/", "details": "Formatting removed (can be used to compare model to actual example)"}, {"abstract": "Mental illness, including depression, anxiety and bipolar disorder, accounts for a significant proportion of global disability and poses a substantial social, economic and heath burden. Treatment is presently dominated by pharmacotherapy, such as antidepressants, and psychotherapy, such as cognitive behavioural therapy; however, such treatments avert less than half of the disease burden, suggesting that additional strategies are needed to prevent and treat mental disorders. There are now consistent mechanistic, observational and interventional data to suggest diet quality may be a modifiable risk factor for mental illness. This review provides an overview of the nutritional psychiatry field. It includes a discussion of the neurobiological mechanisms likely modulated by diet, the use of dietary and nutraceutical interventions in mental disorders, and recommendations for further research. Potential biological pathways related to mental disorders include inflammation, oxidative stress, the gut microbiome, epigenetic modifications and neuroplasticity. Consistent epidemiological evidence, particularly for depression, suggests an association between measures of diet quality and mental health, across multiple populations and age groups; these do not appear to be explained by other demographic, lifestyle factors or reverse causality. Our recently published intervention trial provides preliminary clinical evidence that dietary interventions in clinically diagnosed populations are feasible and can provide significant clinical benefit. Furthermore, nutraceuticals including n-3 fatty acids, folate, S-adenosylmethionine, N-acetyl cysteine and probiotics, among others, are promising avenues for future research. Continued research is now required to investigate the efficacy of intervention studies in large cohorts and within clinically relevant populations, particularly in patients with schizophrenia, bipolar and anxiety disorders.", "source": "https://pubmed.ncbi.nlm.nih.gov/28942748/", "details": "Effect of nutrition on mental health"}, {"abstract": "Hepatitis C virus (HCV) and alcoholic liver disease (ALD), either alone or in combination, count for more than two thirds of all liver diseases in the Western world. There is no safe level of drinking in HCV-infected patients and the most effective goal for these patients is total abstinence. Baclofen, a GABA(B) receptor agonist, represents a promising pharmacotherapy for alcohol dependence (AD). Previously, we performed a randomized clinical trial (RCT), which demonstrated the safety and efficacy of baclofen in patients affected by AD and cirrhosis. The goal of this post-hoc analysis was to explore baclofen's effect in a subgroup of alcohol-dependent HCV-infected cirrhotic patients. Any patient with HCV infection was selected for this analysis. Among the 84 subjects randomized in the main trial, 24 alcohol-dependent cirrhotic patients had a HCV infection; 12 received baclofen 10mg t.i.d. and 12 received placebo for 12-weeks. With respect to the placebo group (3/12, 25.0%), a significantly higher number of patients who achieved and maintained total alcohol abstinence was found in the baclofen group (10/12, 83.3%; p=0.0123). Furthermore, in the baclofen group, compared to placebo, there was a significantly higher increase in albumin values from baseline (p=0.0132) and a trend toward a significant reduction in INR levels from baseline (p=0.0716). In conclusion, baclofen was safe and significantly more effective than placebo in promoting alcohol abstinence, and improving some Liver Function Tests (LFTs) (i.e. albumin, INR) in alcohol-dependent HCV-infected cirrhotic patients. Baclofen may represent a clinically relevant alcohol pharmacotherapy for these patients.", "source": "https://pubmed.ncbi.nlm.nih.gov/22244707/", "details": "Baclofen promotes alcohol abstinence in alcohol dependent cirrhotic patients with hepatitis C virus (HCV) infection"}]
|