Spaces:
Sleeping
Sleeping
Hamza Salhi
commited on
Commit
•
50bf100
1
Parent(s):
12b6218
Upload 92 files
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +3 -0
- data/Worksheet COGs.csv +33 -0
- data/readme.txt +1 -0
- data/sim_pts.csv +0 -0
- exercises/__pycache__/about_course.cpython-39.pyc +0 -0
- exercises/__pycache__/capping_ex.cpython-39.pyc +0 -0
- exercises/__pycache__/variograms.cpython-39.pyc +0 -0
- exercises/about_course.py +32 -0
- exercises/block_modelling.py +331 -0
- exercises/capping_ex.py +289 -0
- exercises/cut_off.py +142 -0
- exercises/geo_interp.py +81 -0
- exercises/interp.py +167 -0
- exercises/reporting.py +10 -0
- exercises/variograms.py +89 -0
- headshots/Ian_Weir.jpg +3 -0
- headshots/Pierre_Landry.jpg +0 -0
- headshots/Sean_Horan.jpg +3 -0
- headshots/Valerie_Wilson.jpg +3 -0
- headshots/notes.txt +1 -0
- images/Domains123.jpg +0 -0
- images/Domains123_Caps.jpg +0 -0
- images/Domains123_Caps2.jpg +0 -0
- images/Fe_sect.jpg +0 -0
- images/HG1_Decile.jpg +0 -0
- images/HG1_HISTO.jpg +0 -0
- images/HG1_ObliqueCaps.jpg +0 -0
- images/HG1_PP.jpg +0 -0
- images/HG1_PlanCaps.jpg +0 -0
- images/HG_2_7_Decile.jpg +0 -0
- images/HG_2_7_HISTO.jpg +0 -0
- images/HG_2_7_ObliqueCaps.jpg +0 -0
- images/HG_2_7_PP.jpg +0 -0
- images/HG_2_7_PlanCaps.jpg +0 -0
- images/HG_Decile.jpg +0 -0
- images/HG_HISTO.jpg +0 -0
- images/HG_LG_Decile.jpg +0 -0
- images/HG_LG_HISTO.jpg +0 -0
- images/HG_LG_ObliqueCaps.jpg +0 -0
- images/HG_LG_PP.jpg +0 -0
- images/HG_LG_PlanCaps.jpg +0 -0
- images/HG_ObliqueCaps.jpg +0 -0
- images/HG_PP.jpg +0 -0
- images/HG_PlanCaps.jpg +0 -0
- images/LG_Decile.jpg +0 -0
- images/LG_HISTO.jpg +0 -0
- images/LG_ObliqueCaps.jpg +0 -0
- images/LG_PP.jpg +0 -0
- images/LG_PlanCaps.jpg +0 -0
- images/P_sect.jpg +0 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,6 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
headshots/Ian_Weir.jpg filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
headshots/Sean_Horan.jpg filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
headshots/Valerie_Wilson.jpg filter=lfs diff=lfs merge=lfs -text
|
data/Worksheet COGs.csv
ADDED
|
@@ -0,0 +1,33 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
PRODUCTION,Units,Resource COG,Comments
|
| 2 |
+
Production,tonnes,"5,000,000",Resources
|
| 3 |
+
Cu,%,1,
|
| 4 |
+
Au,g/t,0.5,
|
| 5 |
+
Metallurgical Recovery,,,
|
| 6 |
+
Cu,%,87%,Studies
|
| 7 |
+
Au,%,90%,
|
| 8 |
+
Metal Production,,,
|
| 9 |
+
Cu,000 lbs,"95,901",
|
| 10 |
+
Au,oz,"72,340",
|
| 11 |
+
Concentrate,dry tonnes,"174,000",
|
| 12 |
+
Cu,%,25,Studies
|
| 13 |
+
Au,g/t,12.93124236,
|
| 14 |
+
Payable Metal,,,Payable %
|
| 15 |
+
Cu,000 lbs,"86,311",90%
|
| 16 |
+
Au,oz,"71,617",99%
|
| 17 |
+
REVENUE,,,
|
| 18 |
+
Metal Price - Cu,US$/lb,3.25,
|
| 19 |
+
Metal Price - Au,US$/oz,1500,
|
| 20 |
+
Revenue - Cu,US$ '000,"280,510",
|
| 21 |
+
Revenue - Au,US$ '000,"107,425",
|
| 22 |
+
Gross Revenue,US$ '000,"387,936",
|
| 23 |
+
SMELTER CHARGES,,,
|
| 24 |
+
Treatment Charges,US$ '000,"1,566",US$09.00 / dmt conc
|
| 25 |
+
Refining Charges - Cu,US$ '000,"7,768",US$0.09 / lb Cu
|
| 26 |
+
Refining Charges - Au,US$ '000,358.0844292,US$5.00 / oz Au
|
| 27 |
+
Net Revenue,US$ '000,"378,244",
|
| 28 |
+
Revenue by Metal,,,
|
| 29 |
+
Cu,%,74%,
|
| 30 |
+
Au,%,28%,
|
| 31 |
+
Revenue by Metal unit,,,
|
| 32 |
+
Cu,US$ per %,55.78731385,
|
| 33 |
+
Au,US$ per g,42.82689773,
|
data/readme.txt
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
|
data/sim_pts.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
exercises/__pycache__/about_course.cpython-39.pyc
ADDED
|
Binary file (1.2 kB). View file
|
|
|
exercises/__pycache__/capping_ex.cpython-39.pyc
ADDED
|
Binary file (8.7 kB). View file
|
|
|
exercises/__pycache__/variograms.cpython-39.pyc
ADDED
|
Binary file (3.08 kB). View file
|
|
|
exercises/about_course.py
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import itertools
|
| 3 |
+
import os
|
| 4 |
+
import funcs
|
| 5 |
+
|
| 6 |
+
def about_course():
|
| 7 |
+
|
| 8 |
+
st.image("..//pdac2021_res_est_course_link2//images//wireframe_header.jpg", use_column_width=True)
|
| 9 |
+
st.title("Fundamentals of Resource Estimation")
|
| 10 |
+
|
| 11 |
+
# get text from text_blocks:
|
| 12 |
+
text = funcs.get_text_block("about_course.txt")
|
| 13 |
+
st.write(text)
|
| 14 |
+
|
| 15 |
+
st.write("")
|
| 16 |
+
st.image("..//pdac2021_res_est_course_link2//images//wireframe_header.jpg", use_column_width=True)
|
| 17 |
+
st.markdown("# Presenters")
|
| 18 |
+
# presenter = st.selectbox("", options=['Sean Horan', 'Valerie Wilson', 'Pierre Landry', 'Ian Weir'])
|
| 19 |
+
for presenter in ['Sean Horan', 'Valerie Wilson', 'Pierre Landry', 'Ian Weir']:
|
| 20 |
+
split_name = presenter.split(" ")
|
| 21 |
+
lcase_name = (split_name[0] + "_" + split_name[1]).lower()
|
| 22 |
+
ucase_name = (split_name[0] + "_" + split_name[1])
|
| 23 |
+
|
| 24 |
+
col1, col2 = st.beta_columns([1,2.5])
|
| 25 |
+
with col1:
|
| 26 |
+
st.image("..//pdac2021_res_est_course_link2//headshots//" + ucase_name + ".jpg", use_column_width=True)
|
| 27 |
+
text = funcs.get_text_block("title_" + lcase_name + ".txt")
|
| 28 |
+
st.markdown(text)
|
| 29 |
+
with col2:
|
| 30 |
+
text = funcs.get_text_block("resume_" + lcase_name + ".txt")
|
| 31 |
+
st.write(text)
|
| 32 |
+
|
exercises/block_modelling.py
ADDED
|
@@ -0,0 +1,331 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import streamlit as st
|
| 4 |
+
import plotly.express as px
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import scipy.spatial as spatial
|
| 7 |
+
from sklearn.neighbors import KDTree
|
| 8 |
+
import warnings
|
| 9 |
+
from multiprocessing.pool import ThreadPool as Pool
|
| 10 |
+
from matplotlib.patches import Ellipse
|
| 11 |
+
warnings.simplefilter("ignore")
|
| 12 |
+
from scipy.interpolate import Rbf
|
| 13 |
+
import matplotlib as mpl
|
| 14 |
+
import plotly.graph_objects as go
|
| 15 |
+
from plotly.subplots import make_subplots
|
| 16 |
+
|
| 17 |
+
color_seq = np.array(['grey', 'blue', 'green', 'yellow', 'orange', 'red', 'purple', 'purple'])
|
| 18 |
+
cog = np.array([-99, 0., 1.0, 1.5, 2.0, 2.5, 3.0, 3.0001])
|
| 19 |
+
cmap = mpl.colors.ListedColormap(color_seq)
|
| 20 |
+
norm = mpl.colors.BoundaryNorm(cog, cmap.N)
|
| 21 |
+
|
| 22 |
+
#@st.cache
|
| 23 |
+
|
| 24 |
+
def get_text_block(fname):
|
| 25 |
+
# this is how to read a block of text:
|
| 26 |
+
path = ""
|
| 27 |
+
f = open(fname, "r")
|
| 28 |
+
# and then write it to the app
|
| 29 |
+
return f.read();
|
| 30 |
+
|
| 31 |
+
|
| 32 |
+
def pad_matrix(mat, dim=2):
|
| 33 |
+
mat = np.array(mat)
|
| 34 |
+
if dim == 2:
|
| 35 |
+
mat = np.pad(mat, (0, 1), 'constant', constant_values=(1))
|
| 36 |
+
mat[-1, -1] = 0.
|
| 37 |
+
else:
|
| 38 |
+
mat = np.pad(mat, (0, 1), 'constant', constant_values=(1, 1))
|
| 39 |
+
return mat;
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
def variogram(h, var, nugget):
|
| 43 |
+
gamma = nugget
|
| 44 |
+
for i in range(2):
|
| 45 |
+
gam = (var[i, 0]) * ((3 * h) / (2 * var[i, 1]) - (h ** 3) / (2 * var[i, 1] ** 3))
|
| 46 |
+
gam[h > var[i, 1]] = var[i, 0]
|
| 47 |
+
gamma += gam
|
| 48 |
+
gamma[h == 0] = 0.
|
| 49 |
+
return gamma;
|
| 50 |
+
|
| 51 |
+
|
| 52 |
+
def OK(x, y, var, nugget):
|
| 53 |
+
# x is samples
|
| 54 |
+
# y in blocks
|
| 55 |
+
x = np.array(x)
|
| 56 |
+
y = np.array(y)
|
| 57 |
+
xx = spatial.distance_matrix(x, x)
|
| 58 |
+
xx_gamma = variogram(xx, var, nugget)
|
| 59 |
+
xx_gamma = pad_matrix(xx_gamma)
|
| 60 |
+
xy_gamma = variogram(y, var, nugget)
|
| 61 |
+
xy_gamma = pad_matrix(xy_gamma, dim=1)
|
| 62 |
+
xx_inv = np.linalg.inv(xx_gamma)
|
| 63 |
+
return np.dot(xy_gamma, xx_inv)[:-1];
|
| 64 |
+
|
| 65 |
+
|
| 66 |
+
def rotate(pts, rot):
|
| 67 |
+
c = np.cos(np.radians(rot))
|
| 68 |
+
s = np.sin(np.radians(rot))
|
| 69 |
+
rotmat = np.array([[c, -s], [s, c]])
|
| 70 |
+
pts = np.dot(pts, rotmat)
|
| 71 |
+
return pts;
|
| 72 |
+
|
| 73 |
+
def plot_samps(df):
|
| 74 |
+
aniso = (300.) / (750.)
|
| 75 |
+
fig, ax = plt.subplots(figsize=(15, 15 * aniso))
|
| 76 |
+
xx, yy = dgrid(1.)
|
| 77 |
+
rbfi = Rbf(df.YPT, df.ZPT, df.AU_G_T, function='cubic')
|
| 78 |
+
zz = rbfi(xx, yy)
|
| 79 |
+
ax.contour(xx, yy, zz, cog, colors=color_seq, alpha=0.5)
|
| 80 |
+
ax.imshow(zz, origin='lower', extent=(0., 750, 0., 300.), alpha=0.2, cmap=cmap, norm=norm)
|
| 81 |
+
scat = ax.scatter(df.YPT, df.ZPT, c=df.AU_G_T, cmap=cmap, norm=norm, edgecolor="black", s=40)
|
| 82 |
+
cbar = fig.colorbar(scat, ticks=cog)
|
| 83 |
+
cbar.set_label('Au g/t', rotation=0)
|
| 84 |
+
plt.xlim((0,750))
|
| 85 |
+
plt.ylim((0, 300))
|
| 86 |
+
plt.xlabel('X')
|
| 87 |
+
plt.ylabel('Y')
|
| 88 |
+
return fig, ax;
|
| 89 |
+
|
| 90 |
+
def plot_blocks(block_size, grades, df):
|
| 91 |
+
aniso = (300. + block_size) / (750. + block_size)
|
| 92 |
+
fig, ax = plt.subplots(figsize=(15, 15 * aniso))
|
| 93 |
+
xx, yy = dgrid(block_size)
|
| 94 |
+
extents = (0., 750 + block_size, 0., 300. + block_size)
|
| 95 |
+
ax.imshow(np.reshape(grades, xx.shape), origin='lower', extent=extents, alpha=0.8, cmap=cmap, norm=norm)
|
| 96 |
+
scat = ax.scatter(df.YPT, df.ZPT, c=df.AU_G_T, cmap=cmap, norm=norm, edgecolor="black", s=40)
|
| 97 |
+
cbar = fig.colorbar(scat, ticks=cog)
|
| 98 |
+
cbar.set_label('Au g/t', rotation=0)
|
| 99 |
+
plt.xlim((0, 750))
|
| 100 |
+
plt.ylim((0, 300))
|
| 101 |
+
plt.xlabel('X')
|
| 102 |
+
plt.ylabel('Y')
|
| 103 |
+
return fig, ax;
|
| 104 |
+
|
| 105 |
+
|
| 106 |
+
def dgrid(block_size=5.):
|
| 107 |
+
x = np.arange(0., 750. + block_size, block_size)
|
| 108 |
+
y = np.arange(0., 300. + block_size, block_size)
|
| 109 |
+
return np.meshgrid(x, y);
|
| 110 |
+
|
| 111 |
+
def gtcurve(grades, block_size):
|
| 112 |
+
|
| 113 |
+
cogs = [0., 0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 1.75, 2.0]
|
| 114 |
+
grades[grades<0.] = 0.
|
| 115 |
+
grades[np.isnan(grades)]=-99.
|
| 116 |
+
bt = block_size*2.7
|
| 117 |
+
|
| 118 |
+
g = []
|
| 119 |
+
t = []
|
| 120 |
+
c = []
|
| 121 |
+
|
| 122 |
+
for cog in cogs:
|
| 123 |
+
filt = grades>cog
|
| 124 |
+
if np.sum(filt) > 0:
|
| 125 |
+
g.append(np.average(grades[filt]))
|
| 126 |
+
t.append(np.sum(filt)*bt)
|
| 127 |
+
c.append(cog)
|
| 128 |
+
|
| 129 |
+
return pd.DataFrame({'COG':c, 'Tonnes':t, 'Grade':g});
|
| 130 |
+
|
| 131 |
+
def block_modelling():
|
| 132 |
+
|
| 133 |
+
st.title("Block Modelling Exercise")
|
| 134 |
+
st.markdown("The figure below is an orthogonal projection of full width intercepts within a narrow vein.")
|
| 135 |
+
st.markdown("## **Visual Trend Analysis**")
|
| 136 |
+
df = pd.read_csv("..//pdac2021_res_est_course//data//sim_pts.csv")
|
| 137 |
+
df = df[df.use==1].copy().reset_index(drop=True)
|
| 138 |
+
fig, ax = plot_samps(df)
|
| 139 |
+
st.pyplot(fig)
|
| 140 |
+
xx = spatial.distance_matrix(df[['YPT', 'ZPT']], df[['YPT', 'ZPT']])
|
| 141 |
+
xx = np.array(xx)
|
| 142 |
+
|
| 143 |
+
#-----------------------------------------------------------------------------------------------------------------#
|
| 144 |
+
# Variogram
|
| 145 |
+
# ----------------------------------------------------------------------------------------------------------------#
|
| 146 |
+
|
| 147 |
+
st.markdown("## **Variogram**")
|
| 148 |
+
st.markdown("The omni-directional variogram is given in the chart that follows." +
|
| 149 |
+
" Keep in mind that no direction has been chosen and that the range shown will be shorter than" +
|
| 150 |
+
" the longest direction and longer than the shortest direction. Your job is to estimate the range" +
|
| 151 |
+
" in the longest direction given your observations from the plot above.")
|
| 152 |
+
|
| 153 |
+
g1, g2 = np.meshgrid(df.AU_G_T, df.AU_G_T)
|
| 154 |
+
col1, col2, col3 = st.beta_columns((1,1,1))
|
| 155 |
+
|
| 156 |
+
with col1:
|
| 157 |
+
st.markdown('#### Experimental Variogram')
|
| 158 |
+
lag_dist = st.slider('Lag Distance', min_value=5., max_value=50., value=10., step=5.,key='var_lag')
|
| 159 |
+
vartype = st.selectbox('Select Experimental Variogram Type',
|
| 160 |
+
options=['Traditional Variogram', 'Correlogram'],
|
| 161 |
+
index=0)
|
| 162 |
+
with col2:
|
| 163 |
+
st.markdown('#### Variogram Model (Variances)')
|
| 164 |
+
nugget = st.slider('Nugget Effect',min_value=0.0, max_value=1.0, value=0.1, step=0.05)
|
| 165 |
+
c1 = st.slider('C1', min_value=0.0, max_value=1.0-nugget, value=0.0, step=0.05)
|
| 166 |
+
c2 = 1.0 - (c1 + nugget)
|
| 167 |
+
with col3:
|
| 168 |
+
st.markdown('#### Variogram Model (Ranges)')
|
| 169 |
+
r1 = st.slider('Range s1', min_value=0.0, max_value=200.0, value=10., step=5., key='k1')
|
| 170 |
+
r2 = st.slider('Range s2', min_value=0.0, max_value=200.0, value=10., step=5., key='k2')
|
| 171 |
+
|
| 172 |
+
var = np.array([[c1, r1], [c2, r2]])
|
| 173 |
+
h = np.arange(0.,200., 1.)
|
| 174 |
+
vmod = variogram(h, var, nugget)
|
| 175 |
+
lags = np.arange(lag_dist, 200., lag_dist)
|
| 176 |
+
gammas = np.zeros(len(lags))
|
| 177 |
+
numpairs = np.zeros(len(lags))
|
| 178 |
+
fig, ax = plt.subplots()
|
| 179 |
+
|
| 180 |
+
for i, lag in enumerate(lags):
|
| 181 |
+
filt = (xx>=lag-lag_dist*0.75)&(xx<lag+lag_dist*0.75)
|
| 182 |
+
sq_dif = np.sum((g1[filt]-g2[filt])**2)
|
| 183 |
+
m = np.average(g1[filt])
|
| 184 |
+
s = np.std(g1[filt])
|
| 185 |
+
ns = np.sum(filt)
|
| 186 |
+
numpairs[i] = ns
|
| 187 |
+
|
| 188 |
+
if vartype == 'Traditional Variogram':
|
| 189 |
+
gammas[i] = sq_dif/(2*float(np.sum(filt))) / np.var(df.AU_G_T)
|
| 190 |
+
else:
|
| 191 |
+
gammas[i] = (np.sum(g1[filt]*g2[filt]) - ns*m**2)/(ns*s**2)
|
| 192 |
+
gammas[i] = 1.0 - gammas[i]
|
| 193 |
+
ax.annotate(str(ns), (lag, gammas[i]+0.05), size=5)
|
| 194 |
+
|
| 195 |
+
ax.bar(lags, numpairs/np.max(numpairs), width=lag_dist/2)
|
| 196 |
+
ax.plot(lags, gammas, '-or', markeredgecolor='k', markersize=4, markeredgewidth=0.5)
|
| 197 |
+
ax.plot(h, vmod, '-g', markeredgecolor='k', markersize=4, markeredgewidth=0.5)
|
| 198 |
+
|
| 199 |
+
plt.xlim((0, 200))
|
| 200 |
+
plt.ylim((0, 1.5))
|
| 201 |
+
ax.plot([0., 200], [1.0, 1.0], '--k')
|
| 202 |
+
plt.xlabel("Separation Distance/Lag Distance")
|
| 203 |
+
plt.ylabel("Gamma")
|
| 204 |
+
st.pyplot(fig)
|
| 205 |
+
|
| 206 |
+
#-----------------------------------------------------------------------------------------------------------------#
|
| 207 |
+
# Set up search ellipse
|
| 208 |
+
# ----------------------------------------------------------------------------------------------------------------#
|
| 209 |
+
st.markdown("## **Search Ellipse**")
|
| 210 |
+
|
| 211 |
+
scol1, scol2 = st.beta_columns((1, 1))
|
| 212 |
+
with scol1:
|
| 213 |
+
st.markdown('#### Ellipse Shape')
|
| 214 |
+
rot = st.number_input('Pick a Rotation (-360 to 360)', min_value=-360., max_value=360., value=0., step=5.)
|
| 215 |
+
rot = (360. - rot)
|
| 216 |
+
srange_major = st.number_input('Major Axis Range', min_value=10., max_value=500., value=100., step=5.)
|
| 217 |
+
srange_minor = st.number_input('Minor Axis Range', min_value=10., max_value=500., value=100., step=5.)
|
| 218 |
+
with scol2:
|
| 219 |
+
st.markdown('#### Sample Selection')
|
| 220 |
+
min_samps = st.number_input("Minimum Samples", min_value=1, max_value=40, value=2, step=1)
|
| 221 |
+
max_samps = st.number_input("Maximum Samples", min_value=1, max_value=40, value=10, step=1)
|
| 222 |
+
fig, ax = plot_samps(df)
|
| 223 |
+
e = Ellipse(xy=[350, 150], width=srange_minor * 2, height=srange_major * 2, angle=rot, linewidth=2)
|
| 224 |
+
ax.add_artist(e)
|
| 225 |
+
e.set_facecolor('None')
|
| 226 |
+
e.set_edgecolor('black')
|
| 227 |
+
st.pyplot(fig)
|
| 228 |
+
|
| 229 |
+
#-----------------------------------------------------------------------------------------------------------------#
|
| 230 |
+
# Block modelling parameters
|
| 231 |
+
# ----------------------------------------------------------------------------------------------------------------#
|
| 232 |
+
|
| 233 |
+
st.markdown("## **Additional Parameters**")
|
| 234 |
+
|
| 235 |
+
bcol1, bcol2 = st.beta_columns((1, 1))
|
| 236 |
+
with bcol1:
|
| 237 |
+
block_size = st.number_input('Block Size', min_value=5., max_value=100., value=10., step=5.)
|
| 238 |
+
with bcol2:
|
| 239 |
+
id_exponent = st.number_input('ID Exponent', min_value=0.0, max_value=10.0, value=2., step=1.)
|
| 240 |
+
|
| 241 |
+
if st.button("Run Interpolation"):
|
| 242 |
+
|
| 243 |
+
xx, yy = dgrid(block_size)
|
| 244 |
+
grid = np.array([xx.flatten(), yy.flatten()]).transpose()
|
| 245 |
+
points = np.array(df.loc[:, ['YPT', 'ZPT']])
|
| 246 |
+
|
| 247 |
+
AUID = np.zeros(len(grid))
|
| 248 |
+
AUNN = np.zeros(len(grid))
|
| 249 |
+
AUOK = np.zeros(len(grid))
|
| 250 |
+
NDIST = np.zeros(len(grid))
|
| 251 |
+
|
| 252 |
+
AUID[:] = np.nan
|
| 253 |
+
AUID[:] = np.nan
|
| 254 |
+
AUNN[:] = np.nan
|
| 255 |
+
AUOK[:] = np.nan
|
| 256 |
+
NDIST[:] = np.nan
|
| 257 |
+
|
| 258 |
+
# rotate
|
| 259 |
+
# grid2 = grid.copy()
|
| 260 |
+
grid = rotate(grid, rot)
|
| 261 |
+
points = rotate(points, rot)
|
| 262 |
+
# apply anisotropy
|
| 263 |
+
grid[:, 0] *= srange_major / srange_minor
|
| 264 |
+
points[:, 0] *= srange_major / srange_minor
|
| 265 |
+
|
| 266 |
+
point_tree = KDTree(points)
|
| 267 |
+
idx, dist = point_tree.query_radius(grid, r=srange_major, return_distance=True, sort_results=True)
|
| 268 |
+
|
| 269 |
+
for i, ix in enumerate(idx):
|
| 270 |
+
mx = max_samps
|
| 271 |
+
if len(ix) <= mx:
|
| 272 |
+
mx = len(ix)
|
| 273 |
+
dists = dist[i][:mx]
|
| 274 |
+
grades = np.array(df.loc[ix[:mx], 'AU_G_T'])
|
| 275 |
+
if len(ix) >= min_samps:
|
| 276 |
+
AUID[i] = (np.average(grades, weights=1.0 / dists ** id_exponent))
|
| 277 |
+
AUNN[i] = (grades[0])
|
| 278 |
+
NDIST[i] = (dists[0])
|
| 279 |
+
OK_weights = OK(x=points[ix[:mx]], y=dists, var=var, nugget=nugget)
|
| 280 |
+
AUOK[i] = (np.sum(OK_weights * grades))
|
| 281 |
+
|
| 282 |
+
|
| 283 |
+
fig, ax = plot_blocks(block_size, AUNN, df)
|
| 284 |
+
plt.title("Nearest Neighbour Interpolation")
|
| 285 |
+
st.pyplot(fig)
|
| 286 |
+
|
| 287 |
+
fig, ax = plot_blocks(block_size, AUID, df)
|
| 288 |
+
plt.title("Inverse Distance Interpolation")
|
| 289 |
+
st.pyplot(fig)
|
| 290 |
+
|
| 291 |
+
fig, ax = plot_blocks(block_size, AUOK, df)
|
| 292 |
+
plt.title("Ordinary Kriging Interpolation")
|
| 293 |
+
st.pyplot(fig)
|
| 294 |
+
|
| 295 |
+
st.write("Grade Tonnage Curve:")
|
| 296 |
+
fig = make_subplots(specs=[[{"secondary_y": True}]])
|
| 297 |
+
curve = gtcurve(AUNN, block_size)
|
| 298 |
+
fig.add_trace(
|
| 299 |
+
go.Scatter(x=curve.COG, y=curve.Tonnes, name="NN Tonnes"),
|
| 300 |
+
secondary_y=False,
|
| 301 |
+
)
|
| 302 |
+
fig.add_trace(
|
| 303 |
+
go.Scatter(x=curve.COG, y=curve.Grade, name="NN Grade"),
|
| 304 |
+
secondary_y=True,
|
| 305 |
+
)
|
| 306 |
+
|
| 307 |
+
curve = gtcurve(AUID, block_size)
|
| 308 |
+
fig.add_trace(
|
| 309 |
+
go.Scatter(x=curve.COG, y=curve.Tonnes, name="ID Tonnes"),
|
| 310 |
+
secondary_y=False,
|
| 311 |
+
)
|
| 312 |
+
fig.add_trace(
|
| 313 |
+
go.Scatter(x=curve.COG, y=curve.Grade, name="ID Grade"),
|
| 314 |
+
secondary_y=True,
|
| 315 |
+
)
|
| 316 |
+
|
| 317 |
+
curve = gtcurve(AUOK, block_size)
|
| 318 |
+
fig.add_trace(
|
| 319 |
+
go.Scatter(x=curve.COG, y=curve.Tonnes, name="OK Tonnes"),
|
| 320 |
+
secondary_y=False,
|
| 321 |
+
)
|
| 322 |
+
fig.add_trace(
|
| 323 |
+
go.Scatter(x=curve.COG, y=curve.Grade, name="OK Grade"),
|
| 324 |
+
secondary_y=True,
|
| 325 |
+
)
|
| 326 |
+
|
| 327 |
+
fig.update_xaxes(title_text="Cut-off (Au g/t)")
|
| 328 |
+
fig.update_yaxes(title_text="Tonnes", secondary_y=False)
|
| 329 |
+
fig.update_yaxes(title_text="Grade (Au g/t)", secondary_y=True)
|
| 330 |
+
|
| 331 |
+
st.plotly_chart(fig)
|
exercises/capping_ex.py
ADDED
|
@@ -0,0 +1,289 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
|
| 3 |
+
def capping_ex():
|
| 4 |
+
|
| 5 |
+
st.title("Capping Exercise")
|
| 6 |
+
|
| 7 |
+
count_correct = 0
|
| 8 |
+
|
| 9 |
+
st.write("The grade restriction module introduced some of the tools that can help you identify whether high grade restraining is required. This exercise uses real data and shows how the high grade restraining workflow is iterative, and that your decision to domain, cap, or otherwise restrict outlier samples should be informed by a group of tools. Lastly, it is important to note that you may elect to revisit your high grade restraining workflow after validating your estimate visually and statistically since the impact of high grade samples on the final resource estimate can be significant.")
|
| 10 |
+
|
| 11 |
+
# st.image("..//pdac2021_res_est_course_link2//images//wireframe_header.jpg", use_column_width=True)
|
| 12 |
+
|
| 13 |
+
st.header("Exercise 1 - Gold Deposit - Question 1")
|
| 14 |
+
|
| 15 |
+
q1_options = ['Please Select an Answer',
|
| 16 |
+
'10 g/t Au',
|
| 17 |
+
'20 g/t Au',
|
| 18 |
+
'25 g/t Au',
|
| 19 |
+
'Other',
|
| 20 |
+
'Something is wrong']
|
| 21 |
+
|
| 22 |
+
|
| 23 |
+
q1_answer=st.radio('Review the following histogram, probability plot and decile analysis and determine which capping level is most appropriate. Careful review of the Plan and Oblique views should help provide a better sense of the spatial distribution of the pre-selected decile thresholds.', options=q1_options, index=0, key='quest1')
|
| 24 |
+
col1, col2, col3 = st.beta_columns((1,1.5,1))
|
| 25 |
+
with col1:
|
| 26 |
+
st.subheader("Decile Analysis")
|
| 27 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_LG_Decile.jpg", use_column_width=True)
|
| 28 |
+
with col2:
|
| 29 |
+
st.subheader("Histogram")
|
| 30 |
+
st.write("")
|
| 31 |
+
st.write("")
|
| 32 |
+
st.write("")
|
| 33 |
+
st.write("")
|
| 34 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_LG_HISTO.jpg", use_column_width=True)
|
| 35 |
+
with col3:
|
| 36 |
+
st.subheader("Probability Plot")
|
| 37 |
+
st.write("")
|
| 38 |
+
st.write("")
|
| 39 |
+
st.write("")
|
| 40 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_LG_PP.jpg", use_column_width=True)
|
| 41 |
+
|
| 42 |
+
cola1, cola2 = st.beta_columns((1,1.6))
|
| 43 |
+
with cola1:
|
| 44 |
+
st.subheader("Plan View - Looking Down")
|
| 45 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_LG_PlanCaps.jpg", use_column_width=True)
|
| 46 |
+
with cola2:
|
| 47 |
+
st.subheader("Oblique View - Looking Along Strike")
|
| 48 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_LG_ObliqueCaps.jpg", use_column_width=True)
|
| 49 |
+
|
| 50 |
+
|
| 51 |
+
st.header("Exercise 1 - Gold Deposit - Question 2")
|
| 52 |
+
|
| 53 |
+
q2_options = ['Please Select an Answer',
|
| 54 |
+
'5 g/t Au',
|
| 55 |
+
'10 g/t Au',
|
| 56 |
+
'15 g/t Au',
|
| 57 |
+
'Other',
|
| 58 |
+
'Something is wrong']
|
| 59 |
+
|
| 60 |
+
q2_answer=st.radio('Review the following histogram, probability plot and decile analysis and determine which capping level is most appropriate. Careful review of the Plan and Oblique views should help provide a better sense of the spatial distribution of the pre-selected decile thresholds.', options=q2_options, index=0, key='quest2')
|
| 61 |
+
colb1, colb2, colb3 = st.beta_columns((1,1.5,1))
|
| 62 |
+
with colb1:
|
| 63 |
+
st.subheader("Decile Analysis")
|
| 64 |
+
st.image("..//pdac2021_res_est_course_link2//images//LG_Decile.jpg", use_column_width=True)
|
| 65 |
+
with colb2:
|
| 66 |
+
st.subheader("Histogram")
|
| 67 |
+
st.write("")
|
| 68 |
+
st.write("")
|
| 69 |
+
st.write("")
|
| 70 |
+
st.write("")
|
| 71 |
+
st.image("..//pdac2021_res_est_course_link2//images//LG_HISTO.jpg", use_column_width=True)
|
| 72 |
+
with colb3:
|
| 73 |
+
st.subheader("Probability Plot")
|
| 74 |
+
st.write("")
|
| 75 |
+
st.write("")
|
| 76 |
+
st.write("")
|
| 77 |
+
st.image("..//pdac2021_res_est_course_link2//images//LG_PP.jpg", use_column_width=True)
|
| 78 |
+
|
| 79 |
+
colc1, colc2 = st.beta_columns((1,1.6))
|
| 80 |
+
with colc1:
|
| 81 |
+
st.subheader("Plan View - Looking Down")
|
| 82 |
+
st.image("..//pdac2021_res_est_course_link2//images//LG_PlanCaps.jpg", use_column_width=True)
|
| 83 |
+
with colc2:
|
| 84 |
+
st.subheader("Oblique View - Looking Along Strike")
|
| 85 |
+
st.image("..//pdac2021_res_est_course_link2//images//LG_ObliqueCaps.jpg", use_column_width=True)
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
|
| 89 |
+
|
| 90 |
+
st.header("Exercise 1 - Gold Deposit - Question 3")
|
| 91 |
+
|
| 92 |
+
q3_options = ['Please Select an Answer',
|
| 93 |
+
'10 g/t Au',
|
| 94 |
+
'20 g/t Au',
|
| 95 |
+
'25 g/t Au',
|
| 96 |
+
'Other',
|
| 97 |
+
'Something is wrong']
|
| 98 |
+
|
| 99 |
+
q3_answer=st.radio('Review the following histogram, probability plot and decile analysis and determine which capping level is most appropriate. Careful review of the Plan and Oblique views should help provide a better sense of the spatial distribution of the pre-selected decile thresholds.', options=q3_options, index=0, key='quest3')
|
| 100 |
+
cold1, cold2, cold3 = st.beta_columns((1,1.5,1))
|
| 101 |
+
with cold1:
|
| 102 |
+
st.subheader("Decile Analysis")
|
| 103 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_Decile.jpg", use_column_width=True)
|
| 104 |
+
with cold2:
|
| 105 |
+
st.subheader("Histogram")
|
| 106 |
+
st.write("")
|
| 107 |
+
st.write("")
|
| 108 |
+
st.write("")
|
| 109 |
+
st.write("")
|
| 110 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_HISTO.jpg", use_column_width=True)
|
| 111 |
+
with cold3:
|
| 112 |
+
st.subheader("Probability Plot")
|
| 113 |
+
st.write("")
|
| 114 |
+
st.write("")
|
| 115 |
+
st.write("")
|
| 116 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_PP.jpg", use_column_width=True)
|
| 117 |
+
|
| 118 |
+
cole1, cole2 = st.beta_columns((1,1.6))
|
| 119 |
+
with cole1:
|
| 120 |
+
st.subheader("Plan View - Looking Down")
|
| 121 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_PlanCaps.jpg", use_column_width=True)
|
| 122 |
+
with cole2:
|
| 123 |
+
st.subheader("Oblique View - Looking Along Strike")
|
| 124 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_ObliqueCaps.jpg", use_column_width=True)
|
| 125 |
+
|
| 126 |
+
|
| 127 |
+
st.header("Exercise 1 - Gold Deposit - Question 4")
|
| 128 |
+
|
| 129 |
+
q4_options = ['Please Select an Answer',
|
| 130 |
+
'5 g/t Au',
|
| 131 |
+
'10 g/t Au',
|
| 132 |
+
'15 g/t Au',
|
| 133 |
+
'Other',
|
| 134 |
+
'Something is wrong']
|
| 135 |
+
|
| 136 |
+
q4_answer=st.radio('Review the following histogram, probability plot and decile analysis and determine which capping level is most appropriate. Careful review of the Plan and Oblique views should help provide a better sense of the spatial distribution of the pre-selected decile thresholds.', options=q4_options, index=0, key='quest4')
|
| 137 |
+
colf1, colf2, colf3 = st.beta_columns((1,1.5,1))
|
| 138 |
+
with colf1:
|
| 139 |
+
st.subheader("Decile Analysis")
|
| 140 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_2_7_Decile.jpg", use_column_width=True)
|
| 141 |
+
with colf2:
|
| 142 |
+
st.subheader("Histogram")
|
| 143 |
+
st.write("")
|
| 144 |
+
st.write("")
|
| 145 |
+
st.write("")
|
| 146 |
+
st.write("")
|
| 147 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_2_7_HISTO.jpg", use_column_width=True)
|
| 148 |
+
with colf3:
|
| 149 |
+
st.subheader("Probability Plot")
|
| 150 |
+
st.write("")
|
| 151 |
+
st.write("")
|
| 152 |
+
st.write("")
|
| 153 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_2_7_PP.jpg", use_column_width=True)
|
| 154 |
+
|
| 155 |
+
colg1, colg2 = st.beta_columns((1,1.6))
|
| 156 |
+
with colg1:
|
| 157 |
+
st.subheader("Plan View - Looking Down")
|
| 158 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_2_7_PlanCaps.jpg", use_column_width=True)
|
| 159 |
+
with colg2:
|
| 160 |
+
st.subheader("Oblique View - Looking Along Strike")
|
| 161 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG_2_7_ObliqueCaps.jpg", use_column_width=True)
|
| 162 |
+
|
| 163 |
+
|
| 164 |
+
|
| 165 |
+
st.header("Exercise 1 - Gold Deposit - Question 5")
|
| 166 |
+
|
| 167 |
+
q5_options = ['Please Select an Answer',
|
| 168 |
+
'10 g/t Au',
|
| 169 |
+
'20 g/t Au',
|
| 170 |
+
'25 g/t Au',
|
| 171 |
+
'Other',
|
| 172 |
+
'Something is wrong']
|
| 173 |
+
|
| 174 |
+
q5_answer=st.radio('Review the following histogram, probability plot and decile analysis and determine which capping level is most appropriate. Careful review of the Plan and Oblique views should help provide a better sense of the spatial distribution of the pre-selected decile thresholds.', options=q5_options, index=0, key='quest5')
|
| 175 |
+
colh1, colh2, colh3 = st.beta_columns((1,1.5,1))
|
| 176 |
+
with colh1:
|
| 177 |
+
st.subheader("Decile Analysis")
|
| 178 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG1_Decile.jpg", use_column_width=True)
|
| 179 |
+
with colh2:
|
| 180 |
+
st.subheader("Histogram")
|
| 181 |
+
st.write("")
|
| 182 |
+
st.write("")
|
| 183 |
+
st.write("")
|
| 184 |
+
st.write("")
|
| 185 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG1_HISTO.jpg", use_column_width=True)
|
| 186 |
+
with colh3:
|
| 187 |
+
st.subheader("Probability Plot")
|
| 188 |
+
st.write("")
|
| 189 |
+
st.write("")
|
| 190 |
+
st.write("")
|
| 191 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG1_PP.jpg", use_column_width=True)
|
| 192 |
+
|
| 193 |
+
coli1, coli2 = st.beta_columns((1,1.6))
|
| 194 |
+
with coli1:
|
| 195 |
+
st.subheader("Plan View - Looking Down")
|
| 196 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG1_PlanCaps.jpg", use_column_width=True)
|
| 197 |
+
with coli2:
|
| 198 |
+
st.subheader("Oblique View - Looking Along Strike")
|
| 199 |
+
st.image("..//pdac2021_res_est_course_link2//images//HG1_ObliqueCaps.jpg", use_column_width=True)
|
| 200 |
+
|
| 201 |
+
|
| 202 |
+
|
| 203 |
+
st.image("..//pdac2021_res_est_course_link2//images//wireframe_header.jpg", use_column_width=True)
|
| 204 |
+
|
| 205 |
+
st.write("As you might have gathered from the plan and oblique views, the gold deposit dataset from Question 1 contains a high grade and low grade population, where the high grade veins are contained within a lower grade alteration halo. The second exercise is a continuation from the first and requires you to match the domains shown in the images below with the statistics presented in each of the questions from the first exercise. If you had some incorrect responses in the first exercise, consult the information and images below prior to beginning Exercise 2.")
|
| 206 |
+
|
| 207 |
+
st.write("Note: The images below are inclined views and are looking down over the along strike oblique views presented in Exercise 1. Wireframes were constructed for Domains 1 and 2 at a nominal cut-off grade of 1 g/t Au while the Domain 3 wireframes were constructed at a nominal 0.20 g/t Au cut-off grade.")
|
| 208 |
+
|
| 209 |
+
st.subheader("Domains 1, 2 and 3")
|
| 210 |
+
st.image("..//pdac2021_res_est_course_link2//images//Domains123.jpg", use_column_width=True)
|
| 211 |
+
st.subheader("Domains 1, 2 and 3 with Capped Assays")
|
| 212 |
+
st.image("..//pdac2021_res_est_course_link2//images//Domains123_Caps2.jpg", use_column_width=True)
|
| 213 |
+
st.subheader("Capped Assays")
|
| 214 |
+
st.image("..//pdac2021_res_est_course_link2//images//Domains123_Caps.jpg", use_column_width=True)
|
| 215 |
+
|
| 216 |
+
st.header("Exercise 2 - Gold Deposit - Question 1")
|
| 217 |
+
|
| 218 |
+
q6_options = ['Please Select an Answer',
|
| 219 |
+
'Domain 1',
|
| 220 |
+
'Domain 2',
|
| 221 |
+
'Domain 3',
|
| 222 |
+
'Domains 1 and 2',
|
| 223 |
+
'Domains 1 and 3',
|
| 224 |
+
'Domains 1, 2, and 3']
|
| 225 |
+
|
| 226 |
+
q6_answer=st.radio('Which domain(s) best reflect the histogram, probability plot and decile analysis presented in Question 1 from Exercise 1?', options=q6_options, index=0, key='quest6')
|
| 227 |
+
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
|
| 231 |
+
st.header("Exercise 2 - Gold Deposit - Question 2")
|
| 232 |
+
|
| 233 |
+
q7_options = ['Please Select an Answer',
|
| 234 |
+
'Domain 1',
|
| 235 |
+
'Domain 2',
|
| 236 |
+
'Domain 3',
|
| 237 |
+
'Domains 1 and 2',
|
| 238 |
+
'Domains 1 and 3',
|
| 239 |
+
'Domains 1, 2, and 3']
|
| 240 |
+
|
| 241 |
+
q7_answer=st.radio('Which domain(s) best reflect the histogram, probability plot and decile analysis presented in Question 2 from Exercise 1?', options=q7_options, index=0, key='quest7')
|
| 242 |
+
|
| 243 |
+
|
| 244 |
+
|
| 245 |
+
|
| 246 |
+
st.header("Exercise 2 - Gold Deposit - Question 4")
|
| 247 |
+
|
| 248 |
+
q9_options = ['Please Select an Answer',
|
| 249 |
+
'Domain 1',
|
| 250 |
+
'Domain 2',
|
| 251 |
+
'Domain 3',
|
| 252 |
+
'Domains 1 and 2',
|
| 253 |
+
'Domains 1 and 3',
|
| 254 |
+
'Domains 1, 2, and 3']
|
| 255 |
+
|
| 256 |
+
q9_answer=st.radio('Which domain(s) best reflect the histogram, probability plot and decile analysis presented in Question 3 from Exercise 1?', options=q9_options, index=0, key='quest8')
|
| 257 |
+
|
| 258 |
+
|
| 259 |
+
|
| 260 |
+
|
| 261 |
+
st.header("Exercise 2 - Gold Deposit - Question 4")
|
| 262 |
+
|
| 263 |
+
q9_options = ['Please Select an Answer',
|
| 264 |
+
'Domain 1',
|
| 265 |
+
'Domain 2',
|
| 266 |
+
'Domain 3',
|
| 267 |
+
'Domains 1 and 2',
|
| 268 |
+
'Domains 1 and 3',
|
| 269 |
+
'Domains 1, 2, and 3']
|
| 270 |
+
|
| 271 |
+
q9_answer=st.radio('Which domain(s) best reflect the histogram, probability plot and decile analysis presented in Question 4 from Exercise 1?', options=q9_options, index=0, key='quest9')
|
| 272 |
+
|
| 273 |
+
|
| 274 |
+
|
| 275 |
+
|
| 276 |
+
st.header("Exercise 2 - Gold Deposit - Question 5")
|
| 277 |
+
|
| 278 |
+
q10_options = ['Please Select an Answer',
|
| 279 |
+
'Domain 1',
|
| 280 |
+
'Domain 2',
|
| 281 |
+
'Domain 3',
|
| 282 |
+
'Domains 1 and 2',
|
| 283 |
+
'Domains 1 and 3',
|
| 284 |
+
'Domains 1, 2, and 3']
|
| 285 |
+
|
| 286 |
+
q10_answer=st.radio('Which domain(s) best reflect the histogram, probability plot and decile analysis presented in Question 5 from Exercise 1?', options=q10_options, index=0, key='quest10')
|
| 287 |
+
|
| 288 |
+
|
| 289 |
+
|
exercises/cut_off.py
ADDED
|
@@ -0,0 +1,142 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import funcs
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import numpy as np
|
| 5 |
+
import plotly.express as px
|
| 6 |
+
|
| 7 |
+
def adjust_calculation(df2):
|
| 8 |
+
|
| 9 |
+
colx1, colx2, colx3, colx4 = st.beta_columns((1,1,1,1))
|
| 10 |
+
|
| 11 |
+
with colx1:
|
| 12 |
+
st.markdown("Block Grade")
|
| 13 |
+
df2.loc[0, 'Resource COG'] = 1000000
|
| 14 |
+
df2.loc[1, 'Resource COG'] = st.slider("Cu %", 0., 2.0, 0.9, 0.1, key="cgsl2")
|
| 15 |
+
df2.loc[2, 'Resource COG'] = st.slider("Au g/t", 0., 2.0, 0.7, 0.1, key="cgsl3")
|
| 16 |
+
cug = df2.loc[1, 'Resource COG']
|
| 17 |
+
aug = df2.loc[2, 'Resource COG']
|
| 18 |
+
with colx2:
|
| 19 |
+
st.markdown("Recoveries")
|
| 20 |
+
df2.loc[4, 'Resource COG'] = st.slider("Cu Recovery", 60., 95., 87., 1.0, key="cgsl4")
|
| 21 |
+
df2.loc[5, 'Resource COG'] = st.slider("Au Recover", 60., 95., 90., 1.0, key="cgsl5")
|
| 22 |
+
with colx3:
|
| 23 |
+
st.markdown("Metal Prices")
|
| 24 |
+
df2.loc[16, 'Resource COG'] = st.slider("Cu Price ($/lbs)", 2.5, 10.0, 3.25, 0.1, key="cgsl6")
|
| 25 |
+
df2.loc[17, 'Resource COG'] = float(st.slider("Au Price ($/oz)", 1000.0, 2000.0, 1500.0, 100.0, key="cgsl7"))
|
| 26 |
+
with colx4:
|
| 27 |
+
st.markdown("Other")
|
| 28 |
+
df2.loc[10, 'Resource COG'] = st.slider("Cu Con Grade", 20.0, 35.0, 25.0, 1.0, key="cgsl8")
|
| 29 |
+
df2.loc[13, 'Comments'] = st.slider("Payabe Cu", 85., 100., 90., 1.0, key="cgsl9")
|
| 30 |
+
df2.loc[14, 'Comments'] = st.slider("Payable Au", 85., 100., 99., 1.0, key="cgsl10")
|
| 31 |
+
|
| 32 |
+
df2.loc[7, 'Resource COG'] = np.round(df2.loc[0, 'Resource COG']*(df2.loc[1, 'Resource COG']/100.)*(df2.loc[4, 'Resource COG']/100.)*2.20462,0)
|
| 33 |
+
df2.loc[8, 'Resource COG'] = np.round(df2.loc[0, 'Resource COG']*(df2.loc[2, 'Resource COG'])/31.1035*(df2.loc[5, 'Resource COG']/100.), 0)
|
| 34 |
+
df2.loc[9, 'Resource COG'] = np.round(df2.loc[7, 'Resource COG']*1000./2204.62/(df2.loc[10, 'Resource COG']/100.), 0)
|
| 35 |
+
df2.loc[11, 'Resource COG'] = np.round(df2.loc[8, 'Resource COG']*31.1035/df2.loc[9, 'Resource COG'],2)
|
| 36 |
+
|
| 37 |
+
df2.loc[13, 'Resource COG'] = np.round(df2.loc[7, 'Resource COG']*(df2.loc[13, 'Comments'])/100.,0)
|
| 38 |
+
df2.loc[14, 'Resource COG'] = np.round(df2.loc[8, 'Resource COG']*(df2.loc[14, 'Comments'])/100,0)
|
| 39 |
+
|
| 40 |
+
df2.loc[18, 'Resource COG'] = np.round(df2.loc[13, 'Resource COG']*df2.loc[16, 'Resource COG'], 0)
|
| 41 |
+
df2.loc[19, 'Resource COG'] = np.round(df2.loc[14, 'Resource COG']*float(df2.loc[17, 'Resource COG']/1000.), 0)
|
| 42 |
+
|
| 43 |
+
df2.loc[20, 'Resource COG'] = df2.loc[18, 'Resource COG']+df2.loc[19, 'Resource COG']
|
| 44 |
+
|
| 45 |
+
df2.loc[22, 'Resource COG'] = np.round(df2.loc[9, 'Resource COG']*9.0/1000, 0)
|
| 46 |
+
df2.loc[23, 'Resource COG'] = np.round(df2.loc[13, 'Resource COG']*0.09,0)
|
| 47 |
+
df2.loc[24, 'Resource COG'] = np.round(df2.loc[14, 'Resource COG']*5.0/1000, 0)
|
| 48 |
+
|
| 49 |
+
df2.loc[25, 'Resource COG'] = np.round(df2.loc[20, 'Resource COG']-df2.loc[22, 'Resource COG']-df2.loc[23, 'Resource COG']-df2.loc[24, 'Resource COG'], 0)
|
| 50 |
+
|
| 51 |
+
df2.loc[27, 'Resource COG'] = np.round((df2.loc[18, 'Resource COG']-df2.loc[22, 'Resource COG']-0.09*df2.loc[13, 'Resource COG']/1000.)/df2.loc[25, 'Resource COG'], 2)
|
| 52 |
+
df2.loc[28, 'Resource COG'] = np.round((df2.loc[19, 'Resource COG']-df2.loc[23, 'Resource COG']-5.0*df2.loc[14, 'Resource COG']/1000.)/df2.loc[25, 'Resource COG'], 2)
|
| 53 |
+
|
| 54 |
+
df2.loc[30, 'Resource COG'] = np.round((df2.loc[27, 'Resource COG']*df2.loc[25, 'Resource COG']*1000./(df2.loc[0, 'Resource COG']*df2.loc[1, 'Resource COG'])),0)
|
| 55 |
+
df2.loc[31, 'Resource COG'] = np.round((df2.loc[28, 'Resource COG']*df2.loc[25, 'Resource COG']*1000./(df2.loc[0, 'Resource COG']*df2.loc[2, 'Resource COG'])),0)
|
| 56 |
+
|
| 57 |
+
colz1, colz2, colz3 = st.beta_columns((1,1,1))
|
| 58 |
+
|
| 59 |
+
metal = ['Cu', 'Au']
|
| 60 |
+
grades = np.array([cug, aug])
|
| 61 |
+
nsr_fact = np.array([df2.loc[30, 'Resource COG'], df2.loc[31, 'Resource COG']])
|
| 62 |
+
blk_rev = grades*nsr_fact
|
| 63 |
+
ddf = pd.DataFrame({'Metal':metal, 'Grade': grades, 'Revenue by Metal Unit': nsr_fact, 'Block Revenue per tonne':blk_rev})
|
| 64 |
+
|
| 65 |
+
with colz1:
|
| 66 |
+
st.write("")
|
| 67 |
+
st.write("")
|
| 68 |
+
st.write("")
|
| 69 |
+
st.write("")
|
| 70 |
+
st.write("")
|
| 71 |
+
st.write("")
|
| 72 |
+
st.write("")
|
| 73 |
+
st.write("")
|
| 74 |
+
st.table(ddf)
|
| 75 |
+
with colz2:
|
| 76 |
+
fig = px.bar(ddf, x='Metal', y='Revenue by Metal Unit', color='Metal')
|
| 77 |
+
fig.update_yaxes(range=(0,200))
|
| 78 |
+
st.plotly_chart(fig, use_container_width=True)
|
| 79 |
+
|
| 80 |
+
with colz3:
|
| 81 |
+
fig = px.bar(ddf, x='Metal', y='Block Revenue per tonne', color='Metal')
|
| 82 |
+
fig.update_yaxes(range=(0,200))
|
| 83 |
+
st.plotly_chart(fig, use_container_width=True)
|
| 84 |
+
|
| 85 |
+
# st.table(df2)
|
| 86 |
+
|
| 87 |
+
def cut_off():
|
| 88 |
+
|
| 89 |
+
df = pd.read_csv("..//pdac2021_res_est_course_link2//data//Worksheet COGs.csv")
|
| 90 |
+
df = df.fillna("")
|
| 91 |
+
df = df[:32].copy()
|
| 92 |
+
|
| 93 |
+
st.title("Cut-off Grade Exercise")
|
| 94 |
+
st.write("")
|
| 95 |
+
st.markdown("## Question 1: Do we send this block to the processing plant?")
|
| 96 |
+
st.write("")
|
| 97 |
+
|
| 98 |
+
col1, col2 = st.beta_columns([1, 2])
|
| 99 |
+
|
| 100 |
+
with col1:
|
| 101 |
+
st.image("..//pdac2021_res_est_course_link2//images//cog1_block.jpg", width=300)
|
| 102 |
+
with col2:
|
| 103 |
+
st.write("")
|
| 104 |
+
st.write("")
|
| 105 |
+
st.write("")
|
| 106 |
+
st.write("")
|
| 107 |
+
st.write("")
|
| 108 |
+
st.write("")
|
| 109 |
+
st.markdown('### Operating costs:')
|
| 110 |
+
st.markdown('* Mining: $50/t')
|
| 111 |
+
st.markdown('* Process: $20/t')
|
| 112 |
+
st.markdown('* G&A: $15/t')
|
| 113 |
+
|
| 114 |
+
q1_options = ['yes', 'no']
|
| 115 |
+
cog_q1_answer = st.radio("Do we send this block to the processing plant?", options=q1_options, key='cog_q1')
|
| 116 |
+
|
| 117 |
+
st.write("")
|
| 118 |
+
st.markdown("## Question 2: Complex Cut-off Calculation")
|
| 119 |
+
st.write("")
|
| 120 |
+
st.markdown("### Considering the block grades from Question 1, analyze the NSR calculation and answer the question below.")
|
| 121 |
+
st.write("")
|
| 122 |
+
col3, col4 = st.beta_columns([1, 2])
|
| 123 |
+
with col3:
|
| 124 |
+
text = funcs.get_text_block("cog_q2_intro.txt")
|
| 125 |
+
st.markdown(text)
|
| 126 |
+
with col4:
|
| 127 |
+
st.image("..//pdac2021_res_est_course_link2//images//nsr_table.png", width=500)
|
| 128 |
+
st.write("*Note that the average grade of the deposit is not known until the cut-off is known. This is normally an early approximation and an iterative process. While it does not impact the calculations, once the average grade is known, a simple cash flow analysis can be performed.")
|
| 129 |
+
q2_options = ['A. I have no idea',
|
| 130 |
+
'B. The block value is below all cut-off grades',
|
| 131 |
+
'C. The block value is greater than the marginal but less than the break-even cut-off',
|
| 132 |
+
'D. The block value exceeds all cut-off grades']
|
| 133 |
+
cog_q2_answer = st.radio("Select the appropriate statement:", options=q2_options, key='cog_q2')
|
| 134 |
+
st.write("")
|
| 135 |
+
st.markdown("## Question 3: Sensitivities")
|
| 136 |
+
st.write("")
|
| 137 |
+
st.markdown("### By Adjusting the various input parameters given below, comment on the following:")
|
| 138 |
+
st.text_area("1. What copper price results in a revenue which is double the break-even cut-off grade", height=5, key='tt1')
|
| 139 |
+
st.text_area("2. What copper grade results in a revenue which is double the break-even cut-off grade", height=5, key='tt2')
|
| 140 |
+
st.write("")
|
| 141 |
+
adjust_calculation(df.copy())
|
| 142 |
+
|
exercises/geo_interp.py
ADDED
|
@@ -0,0 +1,81 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import funcs
|
| 3 |
+
|
| 4 |
+
def geo_interp():
|
| 5 |
+
|
| 6 |
+
st.title("Geological Interpretation")
|
| 7 |
+
|
| 8 |
+
st.markdown("## Question 1: Pick a modelling cut-off")
|
| 9 |
+
st.markdown("")
|
| 10 |
+
st.markdown("You are tasked with building a Mineral Resource domain for a typical orogenic gold deposit of Ontario.")
|
| 11 |
+
st.markdown("The mineralization is erratic and, for the most part, is hosted within a fractured basalt.")
|
| 12 |
+
st.markdown("Review the following cross sections showing mineralization at a variety of cut-off grades, as well as lithology and answer the following questions:")
|
| 13 |
+
|
| 14 |
+
q1_options = ["Select an Answer",
|
| 15 |
+
"25% of the economic cut-off",
|
| 16 |
+
"50% of the economic cut-off",
|
| 17 |
+
"75% of the economic cut-off",
|
| 18 |
+
"100% of the economic cut-off",
|
| 19 |
+
"Limit to host lithology irrespective of grade"]
|
| 20 |
+
|
| 21 |
+
st.radio("What cut-off grade will you use to guide your Mineralization Domain?", options=q1_options, key="q1")
|
| 22 |
+
|
| 23 |
+
q2_options = ["Select an Answer",
|
| 24 |
+
"I will need to manage high grades.",
|
| 25 |
+
"I will need to carefully design the interpolation to not over-smooth grades.",
|
| 26 |
+
"I will need to be cautious about how I connect mineralization in less tightly drilled areas.",
|
| 27 |
+
"I will need to confirm this choice using exploratory data analysis (EDA)."]
|
| 28 |
+
|
| 29 |
+
st.radio("How will this choice impact how I design the downstream workflow for Mineral Resource estimation?", options=q2_options, key="q2")
|
| 30 |
+
|
| 31 |
+
col1, col2, col3 = st.beta_columns((1,1,1))
|
| 32 |
+
|
| 33 |
+
with col1:
|
| 34 |
+
st.write("")
|
| 35 |
+
st.write("Litho")
|
| 36 |
+
st.write("")
|
| 37 |
+
st.write("")
|
| 38 |
+
st.image("..//pdac2021_res_est_course_link2//images//litho.jpg", width=400)
|
| 39 |
+
|
| 40 |
+
with col2:
|
| 41 |
+
|
| 42 |
+
cog = st.slider("Select a cut-off (% of economic cut-off)", min_value=0,value=0, step=1, max_value=3, key="sl1")
|
| 43 |
+
st.image("..//pdac2021_res_est_course_link2//images//gt" + str(int(cog)) + ".jpg", width=400)
|
| 44 |
+
|
| 45 |
+
with col3:
|
| 46 |
+
|
| 47 |
+
cog2 = st.slider("Select a cut-off (% of economic cut-off)", min_value=0, value=0, step=1, max_value=4, key="sl2")
|
| 48 |
+
st.image("..//pdac2021_res_est_course_link2//images//gt2" + str(int(cog2))+ ".jpg", width=400)
|
| 49 |
+
|
| 50 |
+
st.markdown("## Question 2: Choose a domaining strategy")
|
| 51 |
+
st.markdown("")
|
| 52 |
+
text = funcs.get_text_block("geo_interp_q2.txt")
|
| 53 |
+
st.markdown(text)
|
| 54 |
+
q3_options = ["Select an Answer",
|
| 55 |
+
"All together.",
|
| 56 |
+
"All separate.",
|
| 57 |
+
"There is no need to model phosphorus.",
|
| 58 |
+
"Silica and iron together, phosphorus separate."]
|
| 59 |
+
|
| 60 |
+
st.radio("How will I domain and estimate these elements?", options=q3_options, key="q3")
|
| 61 |
+
colz1, colz2, colz3 = st.beta_columns((1,1,1))
|
| 62 |
+
|
| 63 |
+
with colz1:
|
| 64 |
+
st.write("")
|
| 65 |
+
st.write("")
|
| 66 |
+
st.write("")
|
| 67 |
+
st.write("")
|
| 68 |
+
st.image("..//pdac2021_res_est_course_link2//images//flow_chart.jpg", width=350)
|
| 69 |
+
with colz2:
|
| 70 |
+
sel_graph = st.selectbox("Select Graph", options=['Fe vs Si', 'P vs Si'], index=0, key='g1')
|
| 71 |
+
if sel_graph == 'Fe vs Si':
|
| 72 |
+
inp = 'fesi.jpg'
|
| 73 |
+
else:
|
| 74 |
+
inp = 'psi.jpg'
|
| 75 |
+
st.image("..//pdac2021_res_est_course_link2//images//" + inp, width=300)
|
| 76 |
+
|
| 77 |
+
with colz3:
|
| 78 |
+
sel_sect = st.selectbox("Display Section", options=['Si', 'Fe', 'P'], index=0, key='g1')
|
| 79 |
+
st.image("..//pdac2021_res_est_course_link2//images//" + sel_sect + "_sect.jpg", width=500)
|
| 80 |
+
|
| 81 |
+
|
exercises/interp.py
ADDED
|
@@ -0,0 +1,167 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import streamlit as st
|
| 4 |
+
import matplotlib.pyplot as plt
|
| 5 |
+
import warnings
|
| 6 |
+
from matplotlib.patches import Ellipse
|
| 7 |
+
warnings.simplefilter("ignore")
|
| 8 |
+
import matplotlib as mpl
|
| 9 |
+
|
| 10 |
+
color_seq = np.array(['grey', 'blue', 'green', 'yellow', 'orange', 'red', 'purple', 'purple'])
|
| 11 |
+
cog = np.array([-99, 0., 1.0, 1.5, 2.0, 2.5, 3.0, 3.0001])
|
| 12 |
+
cmap = mpl.colors.ListedColormap(color_seq)
|
| 13 |
+
norm = mpl.colors.BoundaryNorm(cog, cmap.N)
|
| 14 |
+
|
| 15 |
+
#@st.cache
|
| 16 |
+
|
| 17 |
+
def get_text_block(fname):
|
| 18 |
+
# this is how to read a block of text:
|
| 19 |
+
path = ""
|
| 20 |
+
f = open(fname, "r")
|
| 21 |
+
# and then write it to the app
|
| 22 |
+
return f.read();
|
| 23 |
+
|
| 24 |
+
|
| 25 |
+
def rotate(pts, rot):
|
| 26 |
+
c = np.cos(np.radians(rot))
|
| 27 |
+
s = np.sin(np.radians(rot))
|
| 28 |
+
rotmat = np.array([[c, -s], [s, c]])
|
| 29 |
+
pts = np.dot(pts, rotmat)
|
| 30 |
+
return pts;
|
| 31 |
+
|
| 32 |
+
def plot_samps(df, plot_contour=True):
|
| 33 |
+
aniso = (300.) / (750.)
|
| 34 |
+
fig, ax = plt.subplots(figsize=(15, 15 * aniso * 0.8))
|
| 35 |
+
if plot_contour:
|
| 36 |
+
xx, yy = dgrid(1.)
|
| 37 |
+
rbfi = Rbf(df.YPT, df.ZPT, df.AU_G_T, function='cubic')
|
| 38 |
+
zz = rbfi(xx, yy)
|
| 39 |
+
ax.contour(xx, yy, zz, cog, colors=color_seq, alpha=0.5)
|
| 40 |
+
ax.imshow(zz, origin='lower', extent=(0., 750, 0., 300.), alpha=0.2, cmap=cmap, norm=norm)
|
| 41 |
+
scat = ax.scatter(df.YPT, df.ZPT, c=df.AU_G_T, cmap=cmap, norm=norm, edgecolor="black", s=40)
|
| 42 |
+
cbar = fig.colorbar(scat, ticks=cog)
|
| 43 |
+
cbar.set_label('Au g/t', rotation=0)
|
| 44 |
+
plt.xlim((0,750))
|
| 45 |
+
plt.ylim((0, 300))
|
| 46 |
+
plt.xlabel('X')
|
| 47 |
+
plt.ylabel('Y')
|
| 48 |
+
return fig, ax;
|
| 49 |
+
|
| 50 |
+
def block_modelling():
|
| 51 |
+
|
| 52 |
+
st.title("Block Modelling Exercise")
|
| 53 |
+
|
| 54 |
+
st.markdown("## **Visual Trend Analysis**")
|
| 55 |
+
st.markdown("The figure below is an orthogonal projection of full width intercepts within a narrow vein.")
|
| 56 |
+
st.markdown("Prior to any statistical analysis, it is useful just to look at your data." +
|
| 57 |
+
" Take it for a spin, look to see where high grades are located and what trends you can observe." +
|
| 58 |
+
" When selecting a colour profile, try use a scheme that highlights trends but considers some economic" +
|
| 59 |
+
" criteria too. It is always useful to have one or two colour bins below your cut-off grade." +
|
| 60 |
+
" The last tip is, be consistent with your legend, make sure the samples and contours/blocks use the" +
|
| 61 |
+
" same colour scheme.")
|
| 62 |
+
|
| 63 |
+
df = pd.read_csv(".//data//sim_pts.csv")
|
| 64 |
+
df = df[df.use==1].copy().reset_index(drop=True)
|
| 65 |
+
# fig, ax = plot_samps(df)
|
| 66 |
+
# st.pyplot(fig)
|
| 67 |
+
st.image("..//pdac2021_res_est_course_link2//images//contour.jpg", use_column_width=True)
|
| 68 |
+
|
| 69 |
+
#-----------------------------------------------------------------------------------------------------------------#
|
| 70 |
+
# Variogram
|
| 71 |
+
# ----------------------------------------------------------------------------------------------------------------#
|
| 72 |
+
|
| 73 |
+
st.markdown("## **Variogram Activity**")
|
| 74 |
+
st.markdown("The omni-directional variogram is given in the chart that follows." +
|
| 75 |
+
" Keep in mind that no direction has been chosen and that the range shown will be shorter than" +
|
| 76 |
+
" the longest direction and longer than the shortest direction. Your job is to estimate the range" +
|
| 77 |
+
" in the longest direction given your observations from the plot above.")
|
| 78 |
+
|
| 79 |
+
st.image("..//pdac2021_res_est_course_link2//images//interp_var.jpg")
|
| 80 |
+
|
| 81 |
+
var_options = ["Select and Answer",
|
| 82 |
+
"Major = 75m, Semi-Major = 75m",
|
| 83 |
+
"Major = 125m, Semi-Major = 60m",
|
| 84 |
+
"Major = 75m, Semi-Major = 100m",
|
| 85 |
+
"Major = 150m, Semi-Major = 75m"]
|
| 86 |
+
|
| 87 |
+
st.radio("What is you estimate of the major and semi major direction ranges?", options=var_options, key='vv1')
|
| 88 |
+
|
| 89 |
+
#-----------------------------------------------------------------------------------------------------------------#
|
| 90 |
+
# Set up search ellipse
|
| 91 |
+
# ----------------------------------------------------------------------------------------------------------------#
|
| 92 |
+
st.markdown("## **Search Ellipse Activity**")
|
| 93 |
+
|
| 94 |
+
# scol1, scol2 = st.beta_columns((1, 1))
|
| 95 |
+
# with scol1:
|
| 96 |
+
st.markdown('#### Ellipse Shape')
|
| 97 |
+
rot = st.number_input('Pick a Rotation (-360 to 360)', min_value=-360., max_value=360., value=0., step=5.)
|
| 98 |
+
rot = (360. - rot)
|
| 99 |
+
srange_major = st.number_input('Major Axis Range', min_value=10., max_value=500., value=100., step=5.)
|
| 100 |
+
srange_minor = st.number_input('Semi-Major Axis Range', min_value=10., max_value=500., value=100., step=5.)
|
| 101 |
+
# with scol2:
|
| 102 |
+
# st.markdown('#### Sample Selection')
|
| 103 |
+
# min_samps = st.number_input("Minimum Samples", min_value=1, max_value=40, value=2, step=1)
|
| 104 |
+
# max_samps = st.number_input("Maximum Samples", min_value=1, max_value=40, value=10, step=1)
|
| 105 |
+
fig, ax = plot_samps(df, plot_contour=False)
|
| 106 |
+
e = Ellipse(xy=[350, 150], width=srange_minor * 2, height=srange_major * 2, angle=rot, linewidth=2)
|
| 107 |
+
ax.add_artist(e)
|
| 108 |
+
e.set_facecolor('None')
|
| 109 |
+
e.set_edgecolor('black')
|
| 110 |
+
st.pyplot(fig)
|
| 111 |
+
src_options = ["Select an Answer",
|
| 112 |
+
"Major = 75m, Semi-Major = 75m, Rotated by 90, Min 2 Max 40",
|
| 113 |
+
"Major = 125m, Semi-Major = 60m, Rotated -60 towards South East, Min 3 Max 12",
|
| 114 |
+
"Major = 125m, Semi-Major = 60m, Rotated 20 towards South West, Min 2 Max 12",
|
| 115 |
+
"Major = 150m, Semi-Major = 75m, Min 2 Max 10, No Rotation"]
|
| 116 |
+
|
| 117 |
+
st.radio("Select the appropriate search parameters?", options=src_options, key='ss1')
|
| 118 |
+
|
| 119 |
+
st.markdown("## **Guess the Model: Question 1**")
|
| 120 |
+
st.markdown("The interpolations given below are Nearest Neighbour (NN), Inverse Distance (ID) and Ordinary Kriging (OK) respectively")
|
| 121 |
+
st.markdown("The curves increasing to the right on the grade tonnage curves are the grades foe each technique at different cut-offs.")
|
| 122 |
+
scol1, scol2 = st.beta_columns((1, 1))
|
| 123 |
+
with scol1:
|
| 124 |
+
st.image("..//pdac2021_res_est_course_link2//images//interp_ests_q1.jpg", use_column_width=True)
|
| 125 |
+
with scol2:
|
| 126 |
+
st.write("")
|
| 127 |
+
st.write("")
|
| 128 |
+
st.write("")
|
| 129 |
+
st.write("")
|
| 130 |
+
st.write("")
|
| 131 |
+
st.write("")
|
| 132 |
+
st.write("")
|
| 133 |
+
st.write("")
|
| 134 |
+
st.write("")
|
| 135 |
+
st.write("")
|
| 136 |
+
st.image("..//pdac2021_res_est_course_link2//images//interp_gt_q1.jpg", use_column_width=True)
|
| 137 |
+
|
| 138 |
+
gt_options = ["Select an Answer",
|
| 139 |
+
"Light blue is NN, purple is OK and red is ID",
|
| 140 |
+
"Light blue is NN, purple is ID and red is OK",
|
| 141 |
+
"Light blue is OK, purple is NN and red is ID",
|
| 142 |
+
"Light blue is OK, purple is ID and red is NN"]
|
| 143 |
+
|
| 144 |
+
st.radio("Which lines on the gt_curve belong to which estimate?", options=gt_options, key='gg1')
|
| 145 |
+
st.text("Any additional comments?")
|
| 146 |
+
|
| 147 |
+
st.markdown("## **Guess the Model: Question 2**")
|
| 148 |
+
st.markdown("Use your knowledge gained to decipher the mystery of the images below.")
|
| 149 |
+
st.markdown("What we know:")
|
| 150 |
+
st.markdown("* ID squared was used for one estimate and OK for another")
|
| 151 |
+
st.markdown("* Identical search parameters were used")
|
| 152 |
+
st.markdown("* The variogram has nugget of around 0.3 and most of the variability is accounted for within about 20-30m.")
|
| 153 |
+
|
| 154 |
+
st.image("..//pdac2021_res_est_course_link2//images//interp_OK_q2.jpg")
|
| 155 |
+
st.image("..//pdac2021_res_est_course_link2//images//interp_ID_q2.jpg")
|
| 156 |
+
|
| 157 |
+
est_options = ["Select an Answer",
|
| 158 |
+
"Estimate A is more variable than estimate B. Estimate A is OK and Estimate B is ID",
|
| 159 |
+
"Estimate B is more variable than estimate B. Estimate A is OK and Estimate B is ID",
|
| 160 |
+
"Both estimates have equal variances as they use the same sample data. Estimate A is OK and Estimate B is ID",
|
| 161 |
+
"Estimate B is more variable than estimate A. Estimate A is ID and Estimate B is OK",]
|
| 162 |
+
|
| 163 |
+
st.radio("Select the correct statement", options=est_options, key='est1')
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
|
exercises/reporting.py
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import funcs
|
| 3 |
+
|
| 4 |
+
def reporting():
|
| 5 |
+
st.title("Reporting/Classification Exercise")
|
| 6 |
+
text = funcs.get_text_block("reporting.txt")
|
| 7 |
+
st.markdown(text)
|
| 8 |
+
st.image("..//pdac2021_res_est_course_link2//images//res_table.jpg", use_column_width=True)
|
| 9 |
+
|
| 10 |
+
|
exercises/variograms.py
ADDED
|
@@ -0,0 +1,89 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
import streamlit as st
|
| 3 |
+
import funcs
|
| 4 |
+
import pandas as pd
|
| 5 |
+
import getpass
|
| 6 |
+
# import pymysql
|
| 7 |
+
import matplotlib.pyplot as plt
|
| 8 |
+
from sqlalchemy import create_engine
|
| 9 |
+
|
| 10 |
+
def variogram(nugget=0.0, srange=100., struct_type='Spherical'):
|
| 11 |
+
var = 1.-nugget
|
| 12 |
+
h = np.arange(0,150.,5.)
|
| 13 |
+
gamma = h
|
| 14 |
+
if struct_type == 'Spherical':
|
| 15 |
+
gamma = nugget + var*((3*h)/(2*srange)-(h**3)/(2*srange**3))
|
| 16 |
+
gamma[h>srange]=1.0
|
| 17 |
+
gamma[0] = nugget
|
| 18 |
+
elif struct_type == 'Power':
|
| 19 |
+
gamma = nugget + var*(h/srange)**1.5
|
| 20 |
+
else:
|
| 21 |
+
gamma = var*(1.0-np.cos((h**2/srange**2)*np.pi)*np.exp((-3*h)/(srange*1.2))) + nugget
|
| 22 |
+
return gamma, h;
|
| 23 |
+
|
| 24 |
+
def ex_var(example=1,
|
| 25 |
+
hint='Hint: Contrary to popular flat earther beliefs.',
|
| 26 |
+
key_nugget='nug1', key_range='range1', key_stype='stype1',
|
| 27 |
+
im='sim1.jpg'):
|
| 28 |
+
st.markdown("### Example " + str(example) + ":")
|
| 29 |
+
st.markdown(hint)
|
| 30 |
+
col1, col2 = st.beta_columns([1, 2])
|
| 31 |
+
with col1:
|
| 32 |
+
struct_type = st.selectbox("Structure Type", options=["Spherical", "Power", "Hole Effect"], index=0, key=key_stype)
|
| 33 |
+
nugget = st.slider('Nugget', 0.0, 1.0, 0.1, 0.05, key=key_nugget)
|
| 34 |
+
srange = st.slider('Range', 0.0, 120., 100., 10., key=key_range)
|
| 35 |
+
gamma, h = variogram(nugget, srange, struct_type)
|
| 36 |
+
fig, ax = plt.subplots()
|
| 37 |
+
plt.plot([0, 120.], [1.0, 1.0], '--k')
|
| 38 |
+
plt.plot(h, gamma, '-r')
|
| 39 |
+
plt.xlabel('Range (m)')
|
| 40 |
+
plt.ylabel('Gamma')
|
| 41 |
+
plt.xlim((0.,120.))
|
| 42 |
+
plt.ylim((0.,1.5))
|
| 43 |
+
st.pyplot(fig)
|
| 44 |
+
with col2:
|
| 45 |
+
st.write("")
|
| 46 |
+
st.write("")
|
| 47 |
+
st.write("")
|
| 48 |
+
st.image("..//pdac2021_res_est_course_link2//images//" + im, width=500)
|
| 49 |
+
|
| 50 |
+
return struct_type, nugget, srange
|
| 51 |
+
|
| 52 |
+
def variograms():
|
| 53 |
+
st.title("Variogram Exercise")
|
| 54 |
+
st.write("")
|
| 55 |
+
st.markdown("## Model the appropriate variogram for each grade pattern observed")
|
| 56 |
+
text = funcs.get_text_block("variography_intro.txt")
|
| 57 |
+
st.markdown(text)
|
| 58 |
+
|
| 59 |
+
results = []
|
| 60 |
+
|
| 61 |
+
s,n,r = ex_var(example=1,
|
| 62 |
+
hint='Hint: Contrary to popular flat earther beliefs.',
|
| 63 |
+
key_nugget='nug1', key_range='range1', key_stype='stype1',
|
| 64 |
+
im='sim1.jpg')
|
| 65 |
+
results.append([1, s,n,r])
|
| 66 |
+
s,n,r = ex_var(example=2,
|
| 67 |
+
hint='Hint: Think about the seasons.',
|
| 68 |
+
key_nugget='nug2', key_range='range2', key_stype='stype2',
|
| 69 |
+
im='sim2.jpg')
|
| 70 |
+
results.append([2, s,n,r])
|
| 71 |
+
s,n,r = ex_var(example=3,
|
| 72 |
+
hint='Hint: Think about sand...',
|
| 73 |
+
key_nugget='nug3', key_range='range3', key_stype='stype3',
|
| 74 |
+
im='sim3.jpg')
|
| 75 |
+
results.append([3, s,n,r])
|
| 76 |
+
s,n,r = ex_var(example=4,
|
| 77 |
+
hint='Hint: This is when you wonder why you bother to get out of bed in the morning.',
|
| 78 |
+
key_nugget='nug4', key_range='range4', key_stype='stype4',
|
| 79 |
+
im='sim4.jpg')
|
| 80 |
+
results.append([4, s,n,r])
|
| 81 |
+
df = pd.DataFrame(data=results,
|
| 82 |
+
columns=['Example', 'Structure Type', 'Nugget', 'Range'])
|
| 83 |
+
uname = getpass.getuser()
|
| 84 |
+
st.table(df)
|
| 85 |
+
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
|
| 89 |
+
|
headshots/Ian_Weir.jpg
ADDED

|
Git LFS Details
|
headshots/Pierre_Landry.jpg
ADDED

|
headshots/Sean_Horan.jpg
ADDED

|
Git LFS Details
|
headshots/Valerie_Wilson.jpg
ADDED

|
Git LFS Details
|
headshots/notes.txt
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
Need to agree on consistent headshots for the app.
|
images/Domains123.jpg
ADDED
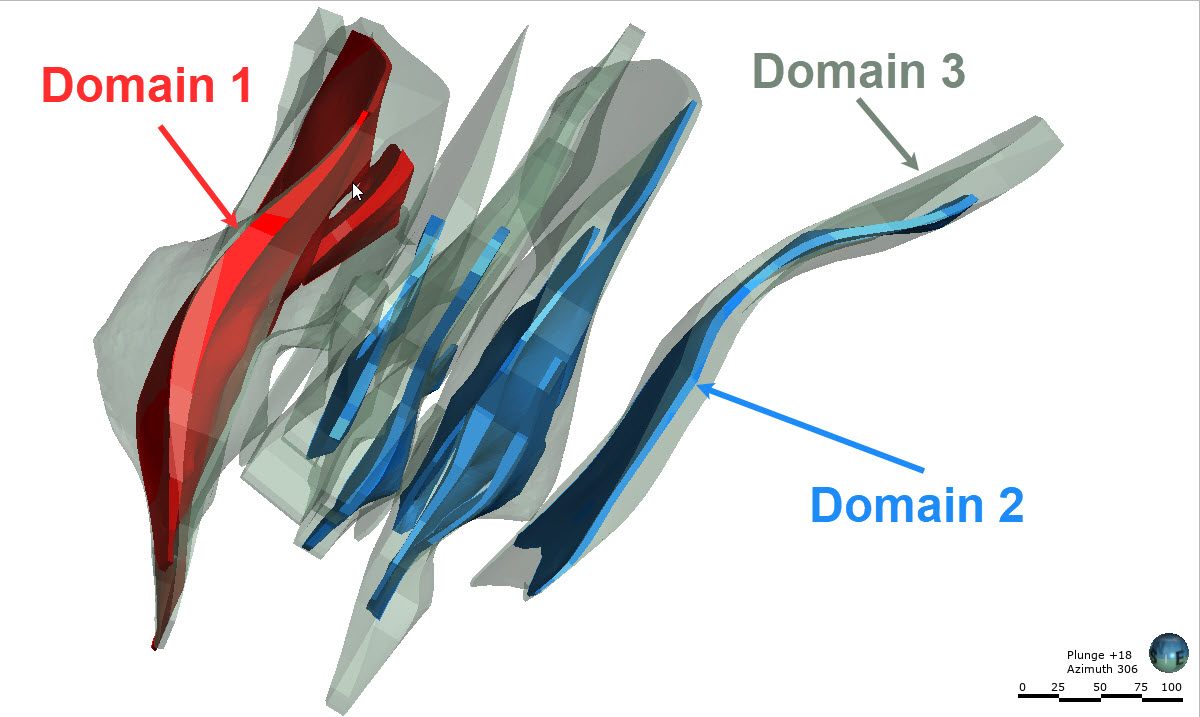
|
images/Domains123_Caps.jpg
ADDED

|
images/Domains123_Caps2.jpg
ADDED
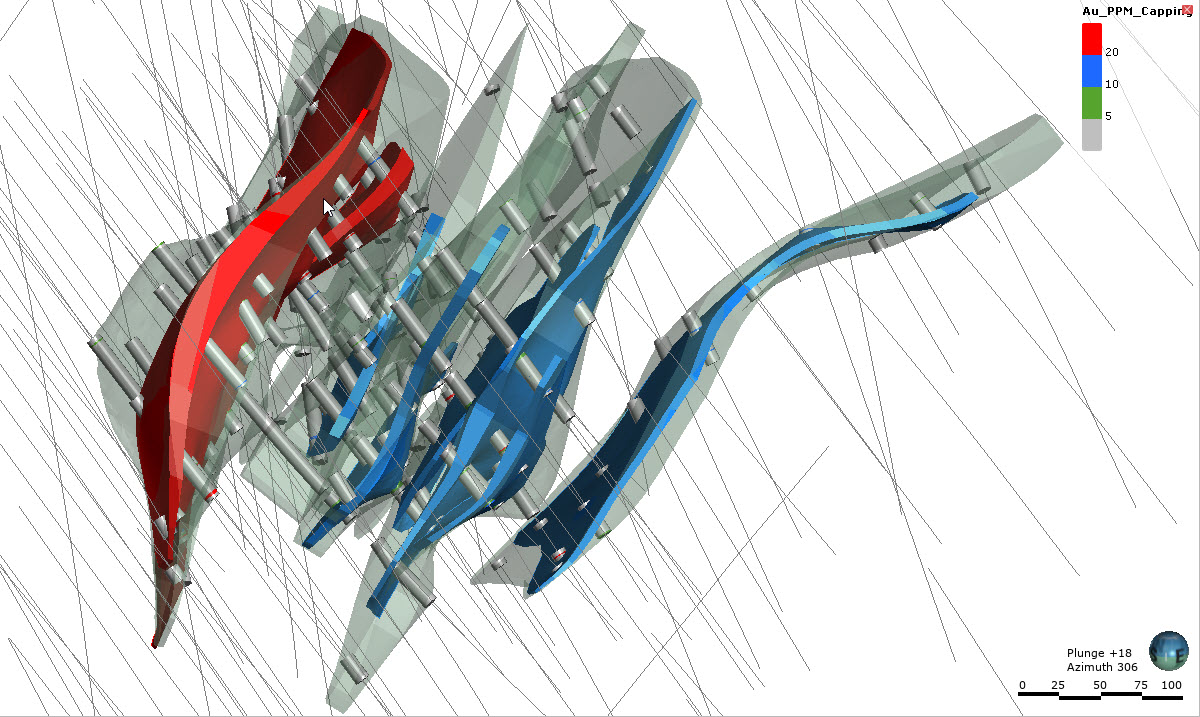
|
images/Fe_sect.jpg
ADDED
|
images/HG1_Decile.jpg
ADDED
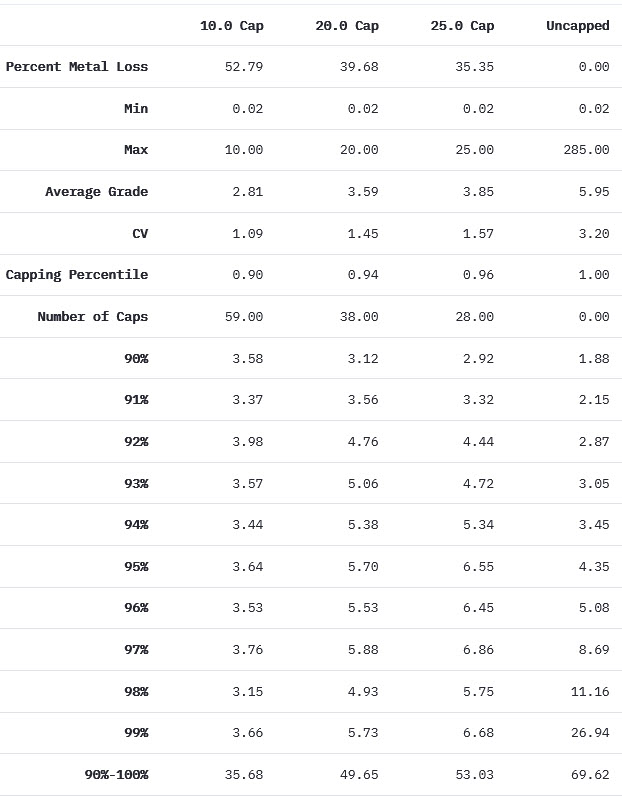
|
images/HG1_HISTO.jpg
ADDED
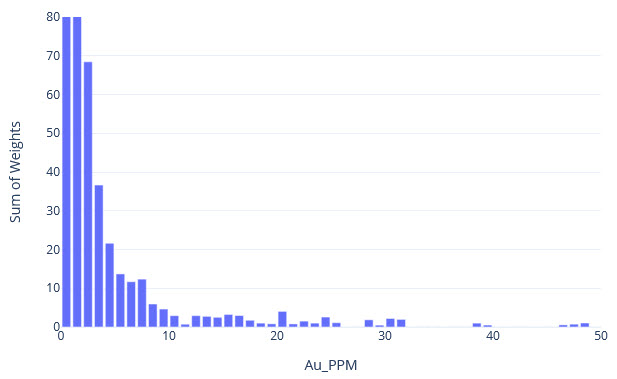
|
images/HG1_ObliqueCaps.jpg
ADDED
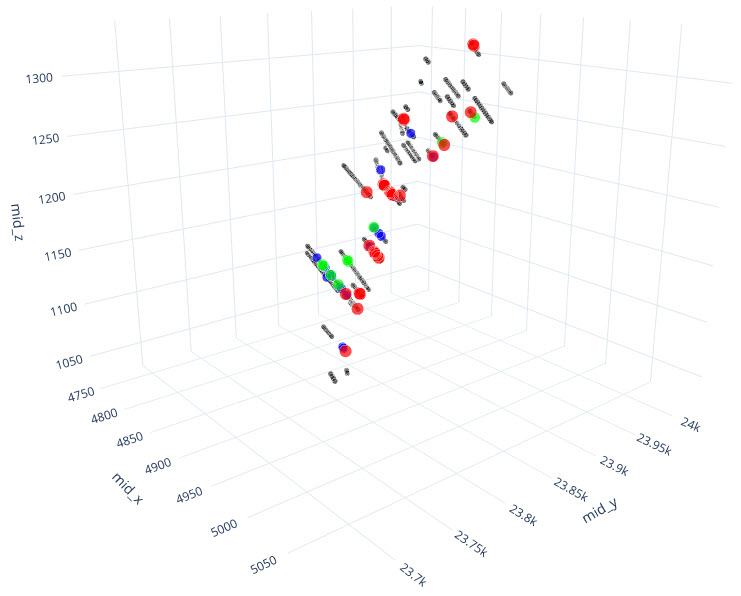
|
images/HG1_PP.jpg
ADDED
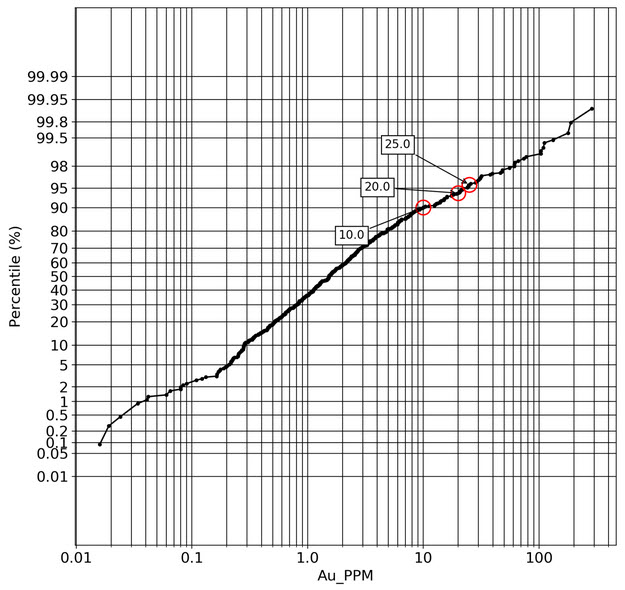
|
images/HG1_PlanCaps.jpg
ADDED
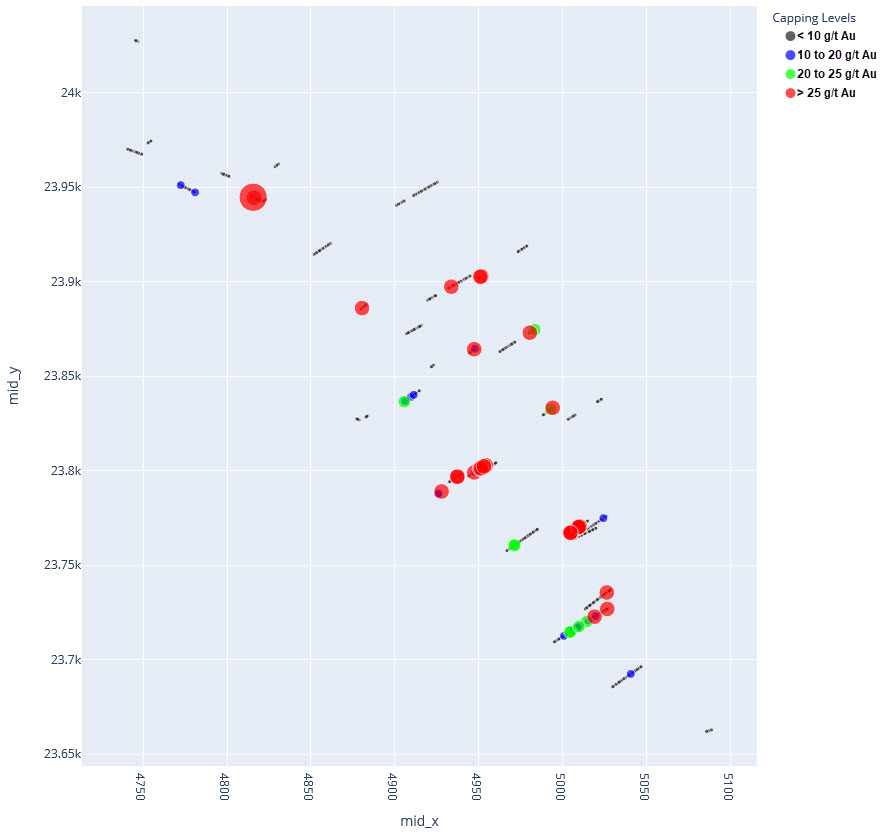
|
images/HG_2_7_Decile.jpg
ADDED

|
images/HG_2_7_HISTO.jpg
ADDED
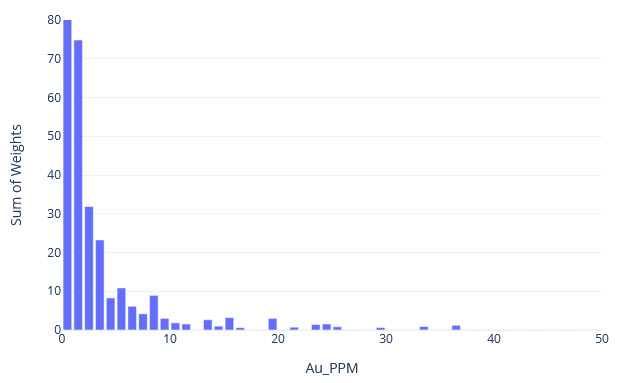
|
images/HG_2_7_ObliqueCaps.jpg
ADDED
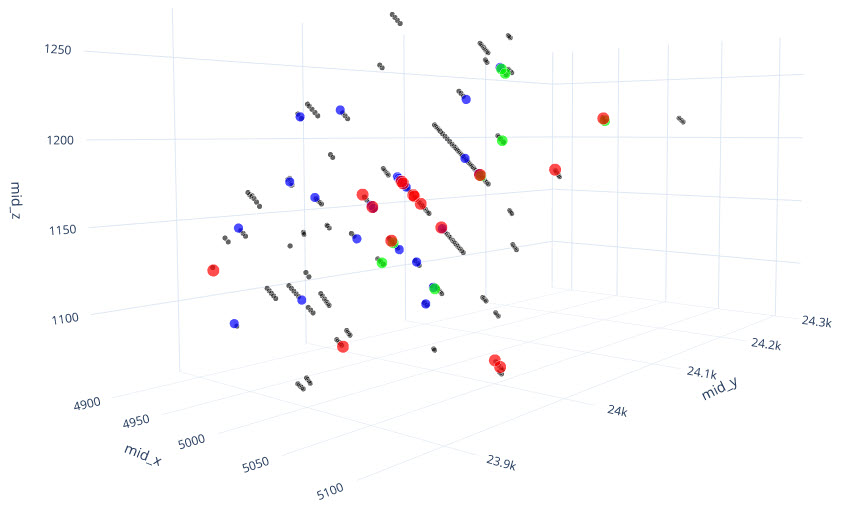
|
images/HG_2_7_PP.jpg
ADDED
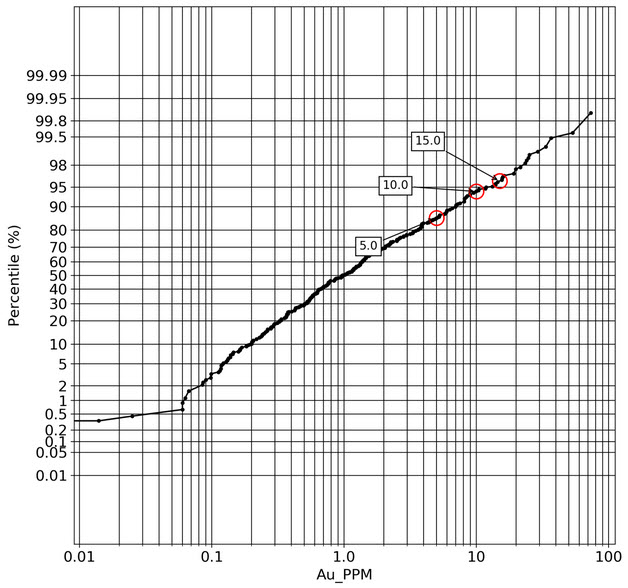
|
images/HG_2_7_PlanCaps.jpg
ADDED
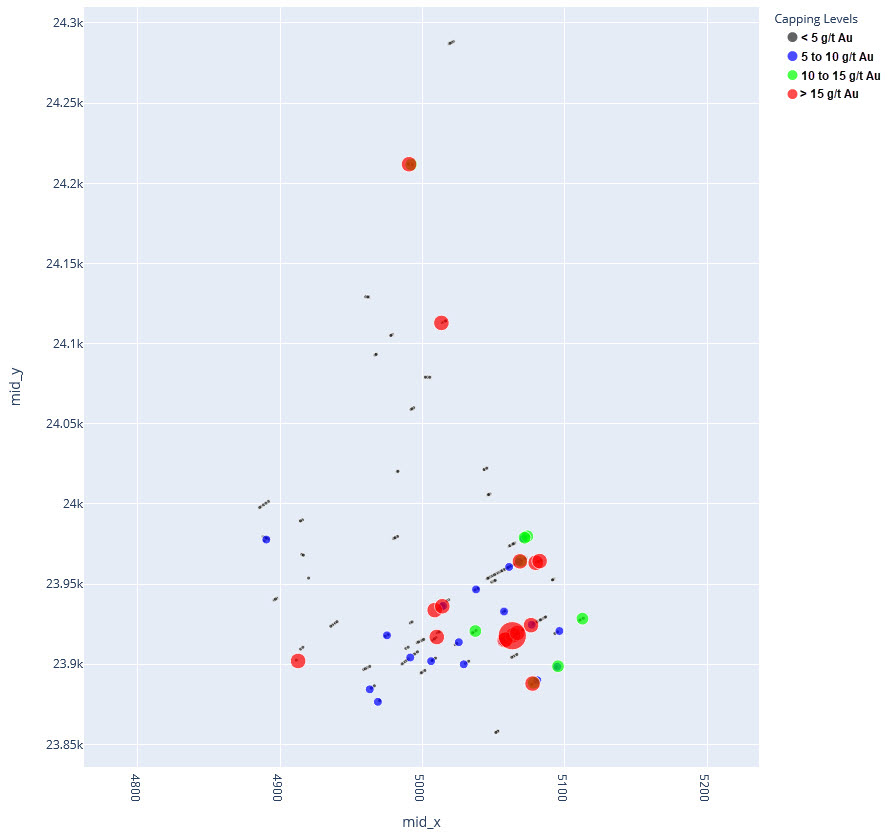
|
images/HG_Decile.jpg
ADDED
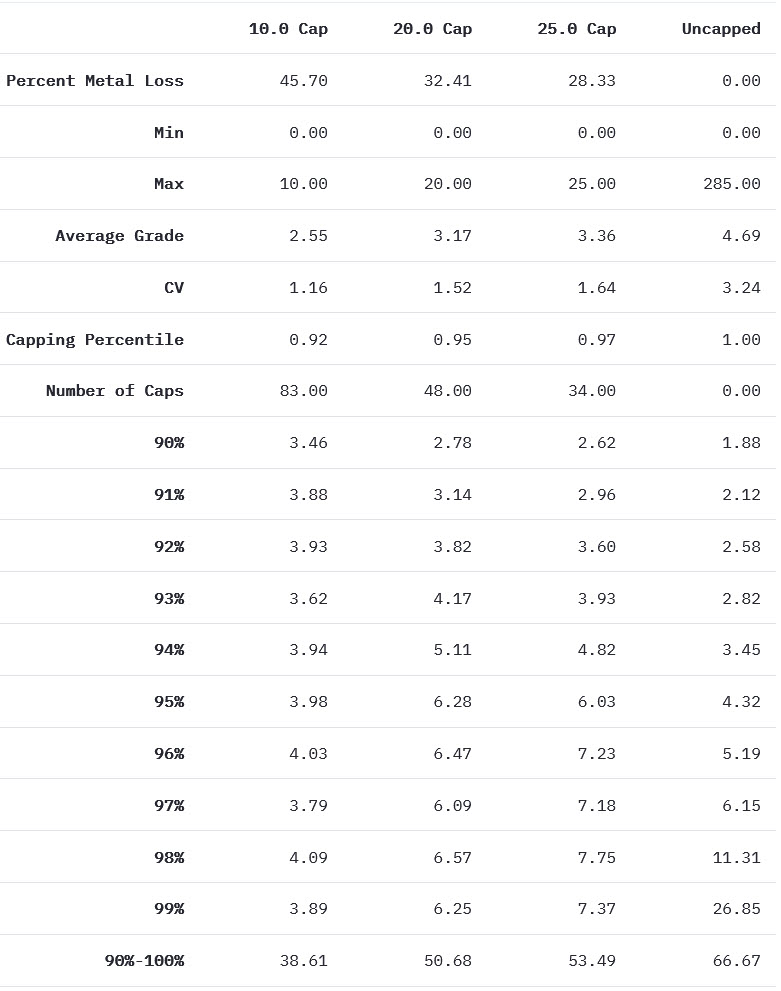
|
images/HG_HISTO.jpg
ADDED

|
images/HG_LG_Decile.jpg
ADDED
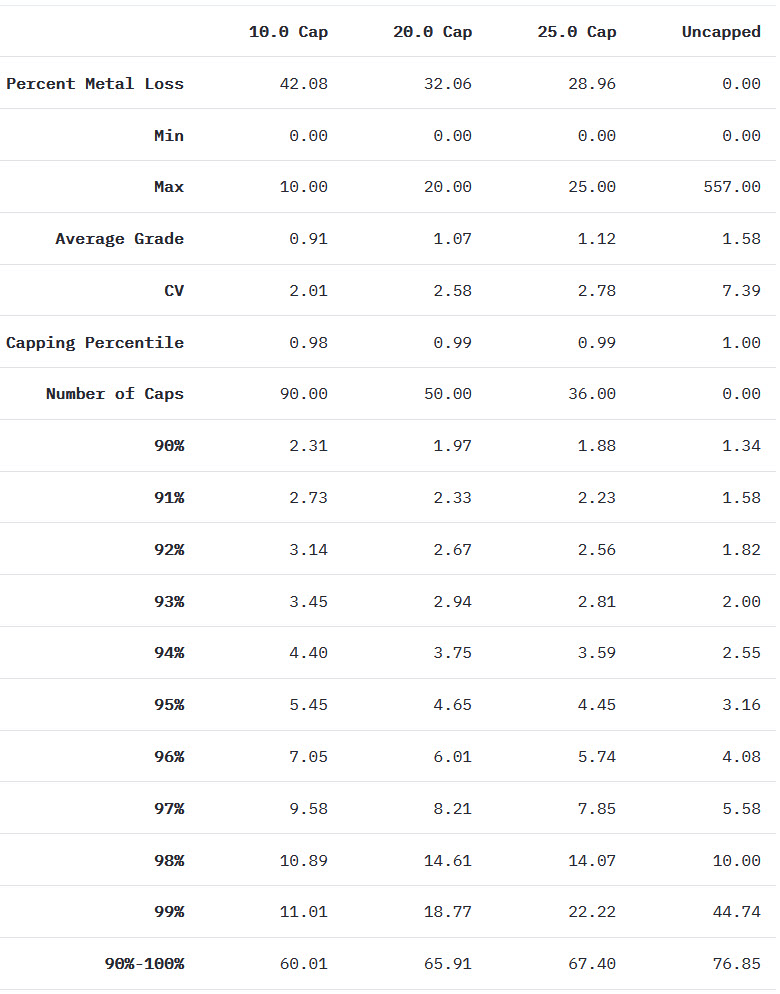
|
images/HG_LG_HISTO.jpg
ADDED
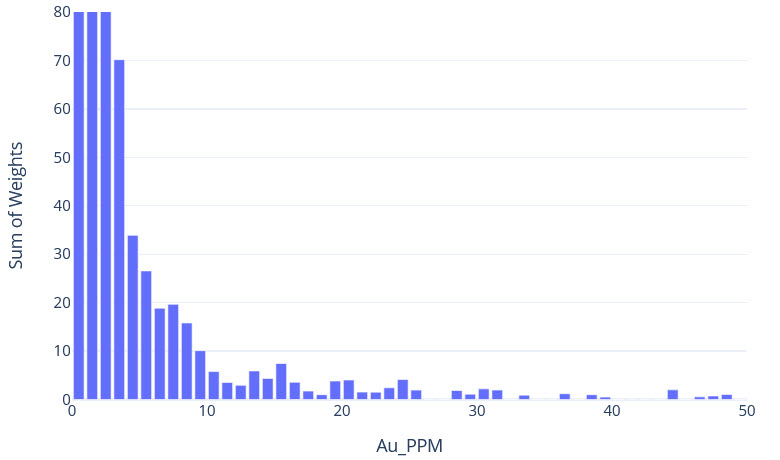
|
images/HG_LG_ObliqueCaps.jpg
ADDED
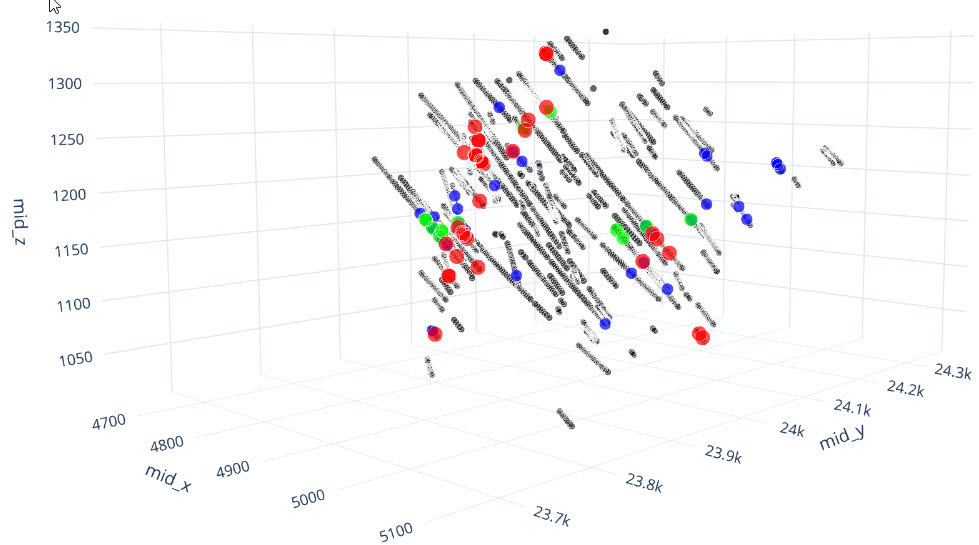
|
images/HG_LG_PP.jpg
ADDED
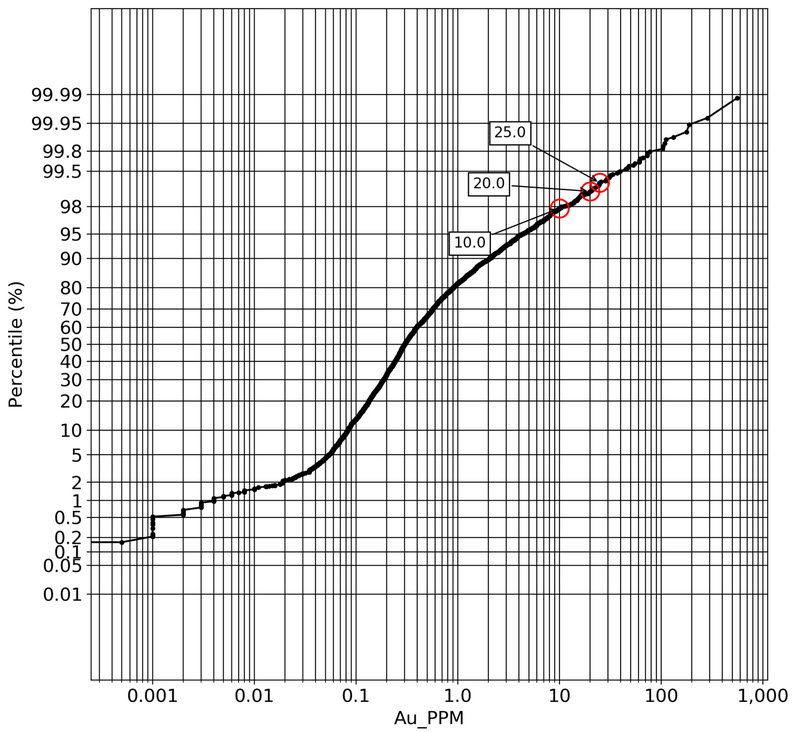
|
images/HG_LG_PlanCaps.jpg
ADDED

|
images/HG_ObliqueCaps.jpg
ADDED
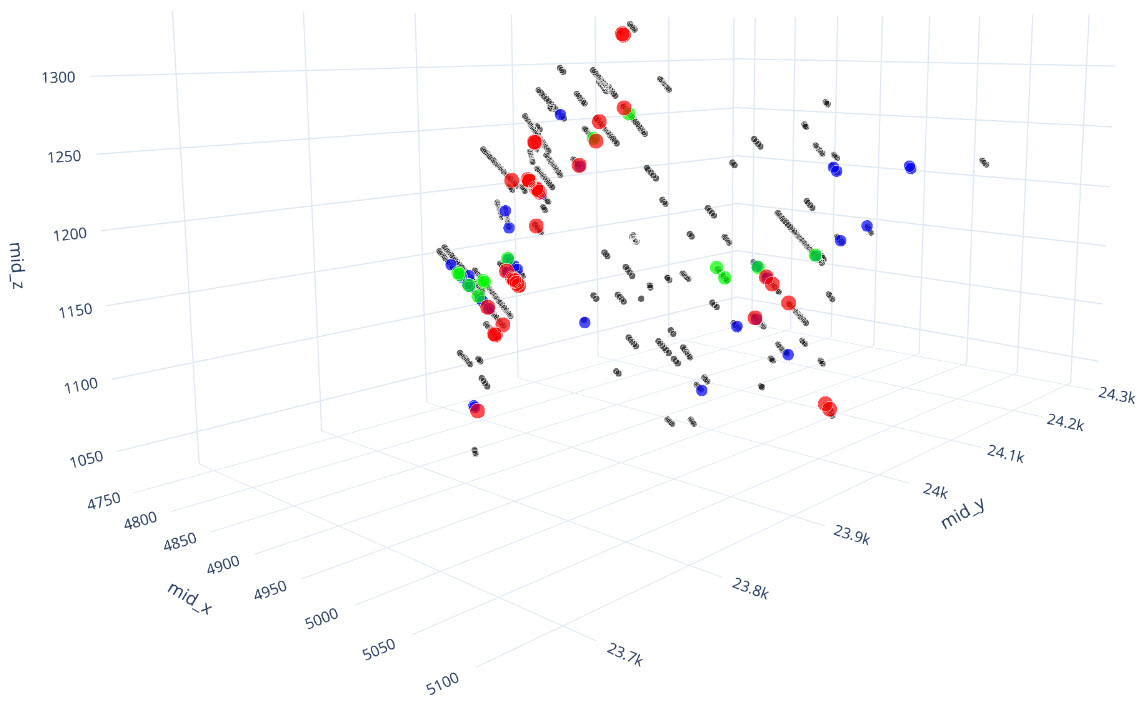
|
images/HG_PP.jpg
ADDED
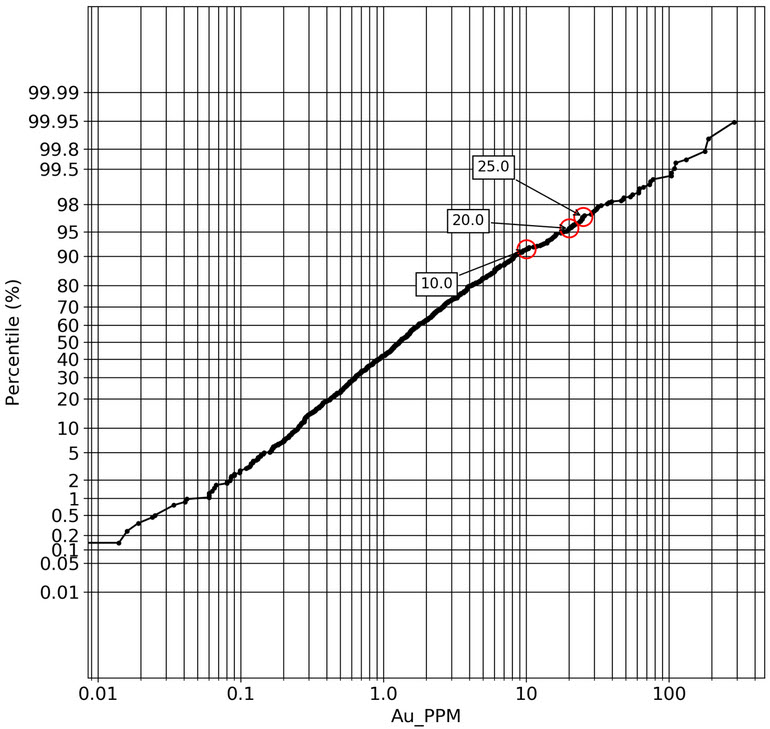
|
images/HG_PlanCaps.jpg
ADDED
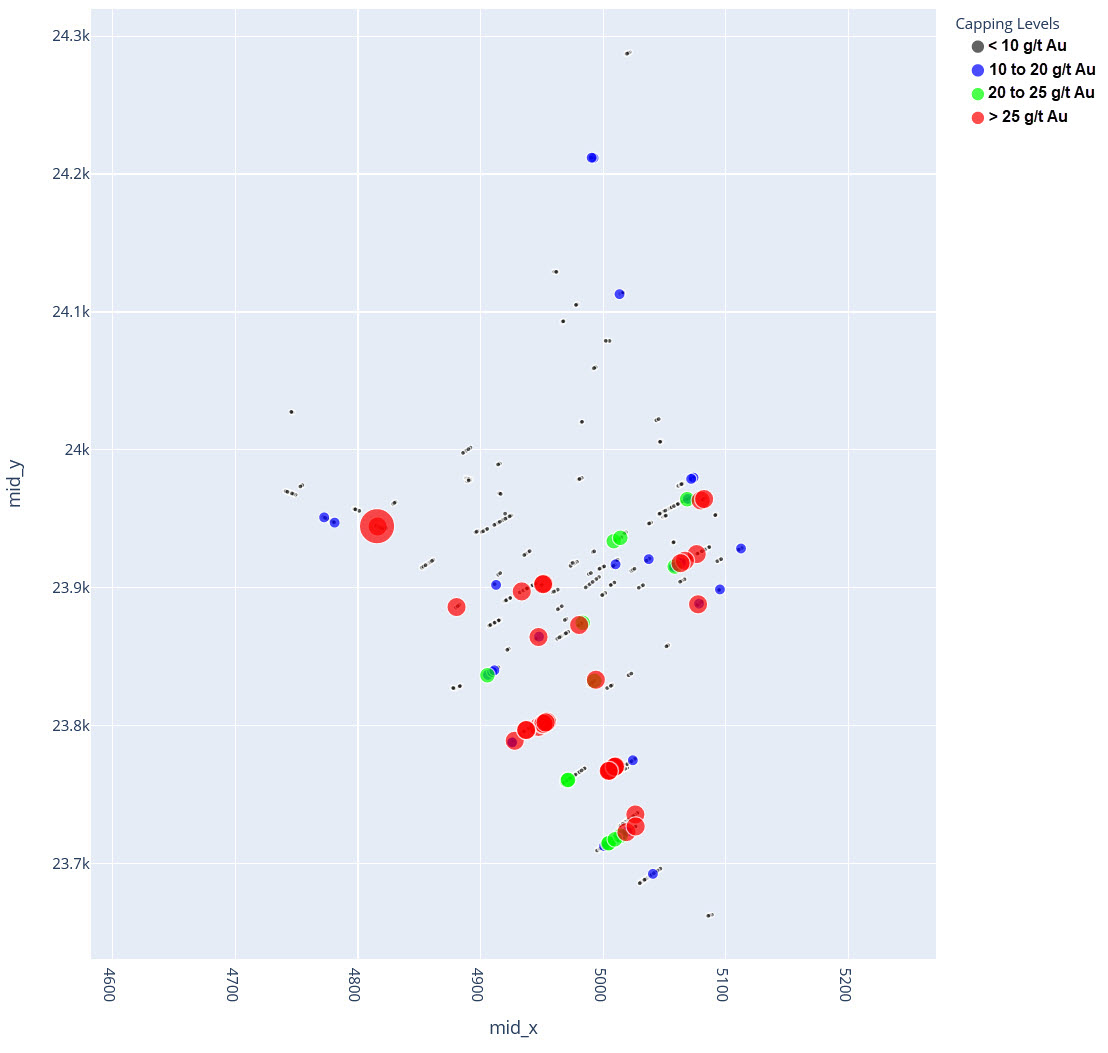
|
images/LG_Decile.jpg
ADDED

|
images/LG_HISTO.jpg
ADDED
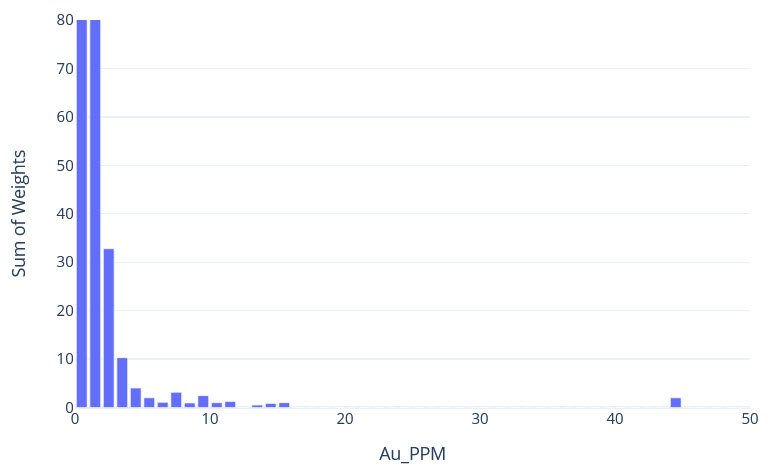
|
images/LG_ObliqueCaps.jpg
ADDED
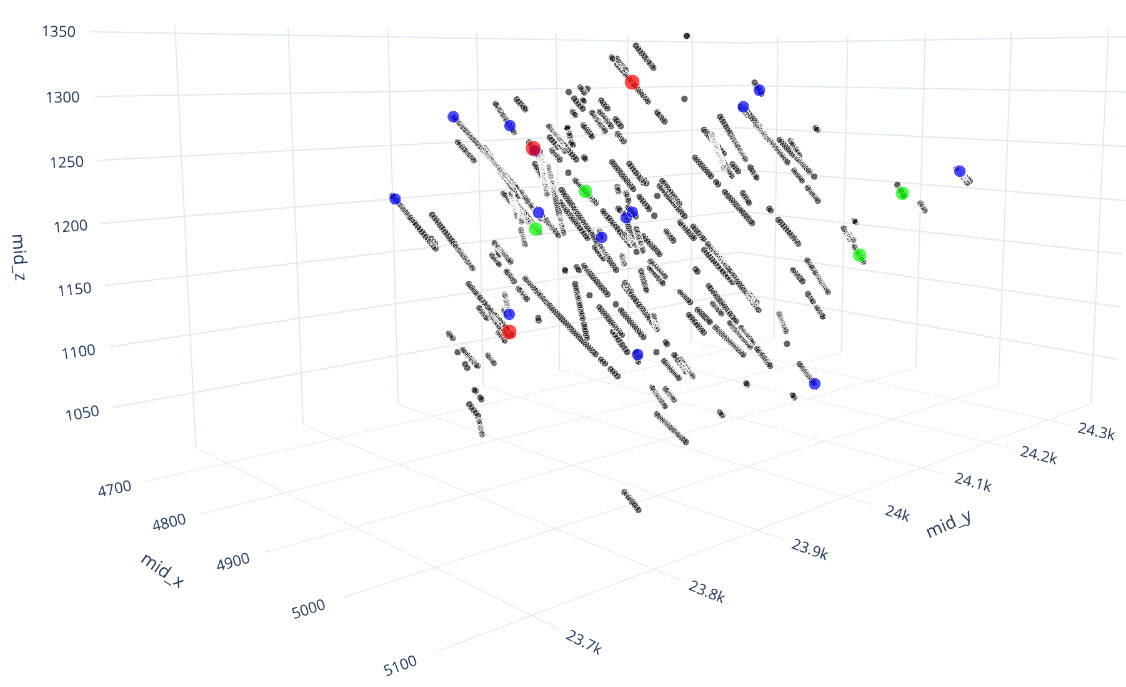
|
images/LG_PP.jpg
ADDED
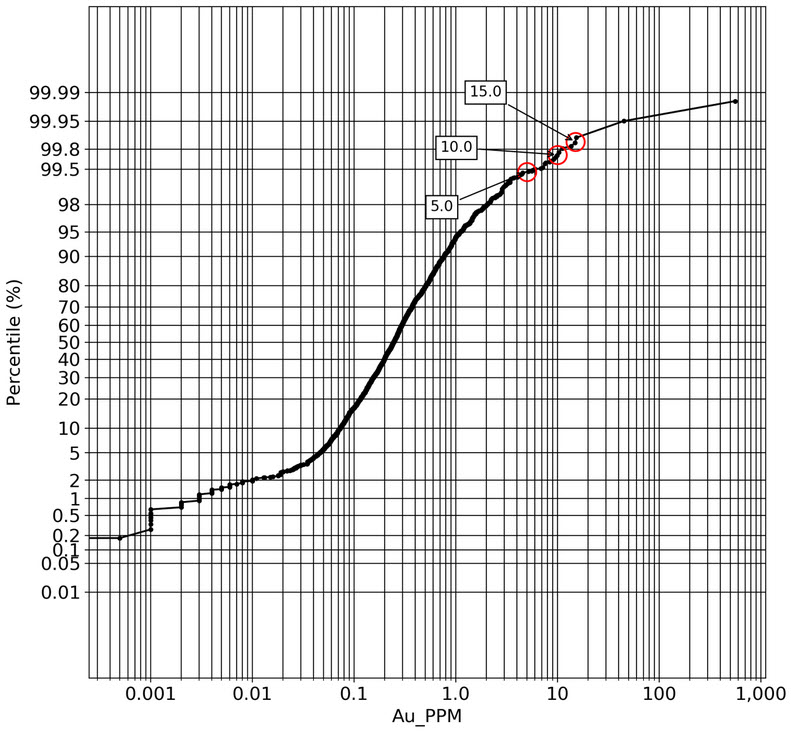
|
images/LG_PlanCaps.jpg
ADDED
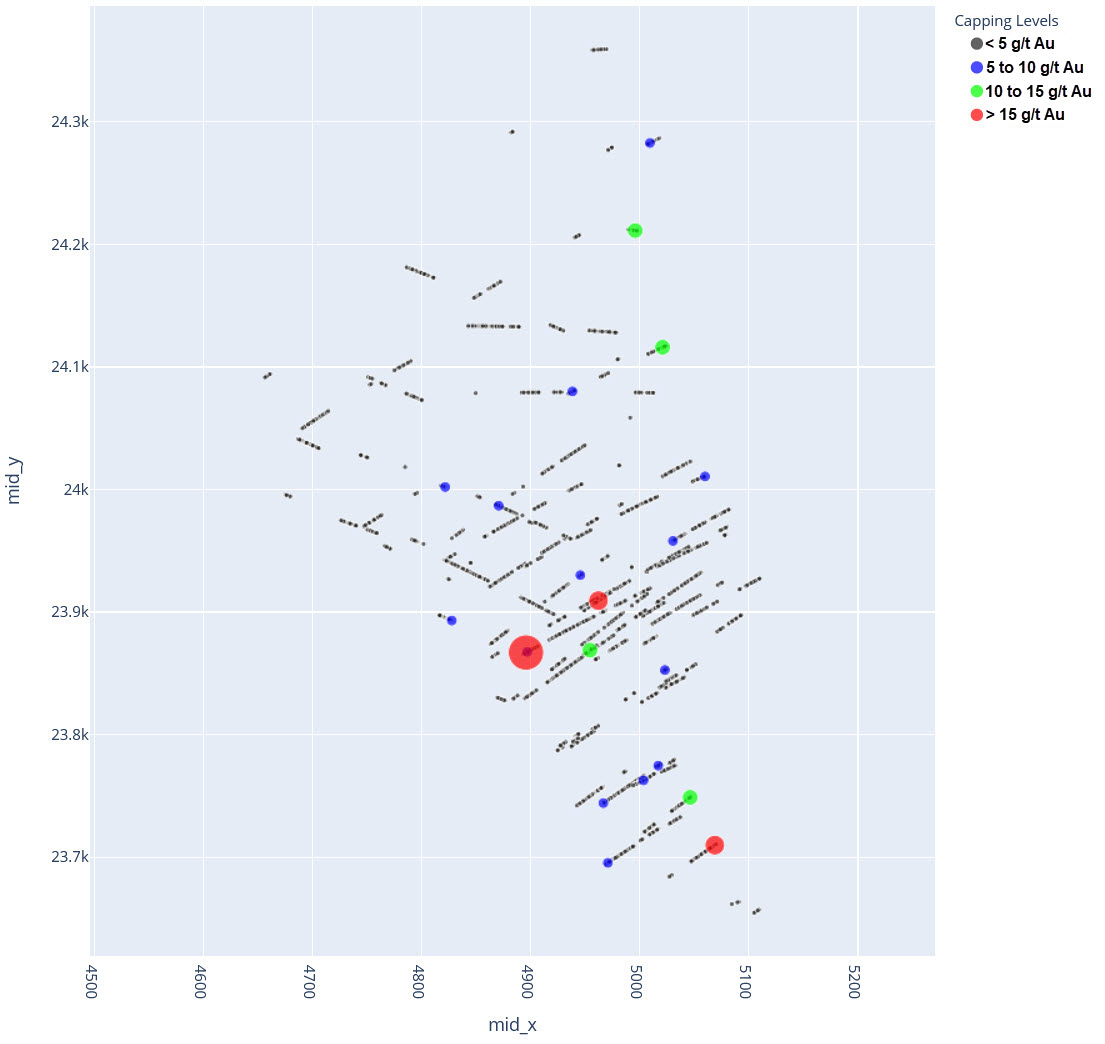
|
images/P_sect.jpg
ADDED
|