Spaces:
Sleeping
Sleeping
Upload 61 files
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitattributes +22 -0
- wetransfer_data-zip_2024-05-17_1431/1_qc_eda-Copy1.ipynb +0 -0
- wetransfer_data-zip_2024-05-17_1431/1_qc_eda.ipynb +0 -0
- wetransfer_data-zip_2024-05-17_1431/2_background_substraction.ipynb +0 -0
- wetransfer_data-zip_2024-05-17_1431/3_z_scores.ipynb +0 -0
- wetransfer_data-zip_2024-05-17_1431/4_markers_tresholds.ipynb +3 -0
- wetransfer_data-zip_2024-05-17_1431/5_cells_quant_class.ipynb +0 -0
- wetransfer_data-zip_2024-05-17_1431/__pycache__/my_modules.cpython-311.pyc +0 -0
- wetransfer_data-zip_2024-05-17_1431/data.zip +3 -0
- wetransfer_data-zip_2024-05-17_1431/my_modules.py +468 -0
- wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S1_bs.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S2_bs.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S3_bs.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_bs/TMA_bs.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_cqc/test_cell_subtypes_number_by_scenes.csv +1 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/Ashlar_Exposure_Time.csv +91 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/DD3S1.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/DD3S2.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/DD3S3.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/TMA.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/new_data.csv +46 -0
- wetransfer_data-zip_2024-05-17_1431/test_data/stored_variables.json +1 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/Exposure_Time.csv +37 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/Set_B_unique_ROIs.csv +468 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/Slide_B_DD1s1.one_1.tif.csv +46 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/Slide_B_DD1s1.one_2.tif.csv +46 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/TMA_Clinical_Data_187-OC.csv +188 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/cellsubtype_color_data.csv +13 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/celltype_color_data.csv +5 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/channel_color_data.csv +5 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/combined_metadata.csv +91 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/full_to_short_column_names.csv +109 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Cellsubtype_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Celltype_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Channel_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Round_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Sample_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/images/immune_checkpoint_legend.png +0 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/immunecheckpoint_color_data.csv +6 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/marker_intensity_metadata.csv +109 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/not_intensities.csv +12 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/round_color_data.csv +10 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/sample_color_data.csv +5 -0
- wetransfer_data-zip_2024-05-17_1431/test_metadata/short_to_full_column_names.csv +109 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S1_mt.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S2_mt.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S3_mt.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/TMA_mt.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/all_Samples_Set_A.csv +3 -0
- wetransfer_data-zip_2024-05-17_1431/test_mt/images/set_a.png +0 -0
.gitattributes
CHANGED
|
@@ -32,3 +32,25 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 32 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 33 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 32 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 33 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 35 |
+
wetransfer_data-zip_2024-05-17_1431/4_markers_tresholds.ipynb filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S1_bs.csv filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S2_bs.csv filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S3_bs.csv filter=lfs diff=lfs merge=lfs -text
|
| 39 |
+
wetransfer_data-zip_2024-05-17_1431/test_bs/TMA_bs.csv filter=lfs diff=lfs merge=lfs -text
|
| 40 |
+
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S1.csv filter=lfs diff=lfs merge=lfs -text
|
| 41 |
+
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S2.csv filter=lfs diff=lfs merge=lfs -text
|
| 42 |
+
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S3.csv filter=lfs diff=lfs merge=lfs -text
|
| 43 |
+
wetransfer_data-zip_2024-05-17_1431/test_data/TMA.csv filter=lfs diff=lfs merge=lfs -text
|
| 44 |
+
wetransfer_data-zip_2024-05-17_1431/test_mt/all_Samples_Set_A.csv filter=lfs diff=lfs merge=lfs -text
|
| 45 |
+
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S1_mt.csv filter=lfs diff=lfs merge=lfs -text
|
| 46 |
+
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S2_mt.csv filter=lfs diff=lfs merge=lfs -text
|
| 47 |
+
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S3_mt.csv filter=lfs diff=lfs merge=lfs -text
|
| 48 |
+
wetransfer_data-zip_2024-05-17_1431/test_mt/TMA_mt.csv filter=lfs diff=lfs merge=lfs -text
|
| 49 |
+
wetransfer_data-zip_2024-05-17_1431/test_qc_eda/DD3S1_qc_eda.csv filter=lfs diff=lfs merge=lfs -text
|
| 50 |
+
wetransfer_data-zip_2024-05-17_1431/test_qc_eda/DD3S2_qc_eda.csv filter=lfs diff=lfs merge=lfs -text
|
| 51 |
+
wetransfer_data-zip_2024-05-17_1431/test_qc_eda/DD3S3_qc_eda.csv filter=lfs diff=lfs merge=lfs -text
|
| 52 |
+
wetransfer_data-zip_2024-05-17_1431/test_qc_eda/TMA_qc_eda.csv filter=lfs diff=lfs merge=lfs -text
|
| 53 |
+
wetransfer_data-zip_2024-05-17_1431/test_zscore/DD3S1_zscore.csv filter=lfs diff=lfs merge=lfs -text
|
| 54 |
+
wetransfer_data-zip_2024-05-17_1431/test_zscore/DD3S2_zscore.csv filter=lfs diff=lfs merge=lfs -text
|
| 55 |
+
wetransfer_data-zip_2024-05-17_1431/test_zscore/DD3S3_zscore.csv filter=lfs diff=lfs merge=lfs -text
|
| 56 |
+
wetransfer_data-zip_2024-05-17_1431/test_zscore/TMA_zscore.csv filter=lfs diff=lfs merge=lfs -text
|
wetransfer_data-zip_2024-05-17_1431/1_qc_eda-Copy1.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
wetransfer_data-zip_2024-05-17_1431/1_qc_eda.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
wetransfer_data-zip_2024-05-17_1431/2_background_substraction.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
wetransfer_data-zip_2024-05-17_1431/3_z_scores.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
wetransfer_data-zip_2024-05-17_1431/4_markers_tresholds.ipynb
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:df84f15a0e72c1a84daae210d6334f49150c5d2aa03aae2688f1f839fdb1aeb1
|
| 3 |
+
size 49760040
|
wetransfer_data-zip_2024-05-17_1431/5_cells_quant_class.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
wetransfer_data-zip_2024-05-17_1431/__pycache__/my_modules.cpython-311.pyc
ADDED
|
Binary file (20.9 kB). View file
|
|
|
wetransfer_data-zip_2024-05-17_1431/data.zip
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:72623169e7fa754d5892a19670be3a1f4cd2bec5fb4736db5e8de103d70c5fe4
|
| 3 |
+
size 326996556
|
wetransfer_data-zip_2024-05-17_1431/my_modules.py
ADDED
|
@@ -0,0 +1,468 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import numpy as np
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import subprocess
|
| 5 |
+
import os
|
| 6 |
+
import random
|
| 7 |
+
import re
|
| 8 |
+
import pandas as pd
|
| 9 |
+
import numpy as np
|
| 10 |
+
import seaborn as sb
|
| 11 |
+
import matplotlib.pyplot as plt
|
| 12 |
+
import matplotlib.colors as mplc
|
| 13 |
+
import subprocess
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
from scipy import signal
|
| 17 |
+
|
| 18 |
+
import plotly.figure_factory as ff
|
| 19 |
+
import plotly
|
| 20 |
+
import plotly.graph_objs as go
|
| 21 |
+
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
# This function takes in a dataframe, changes the names
|
| 25 |
+
# of the column in various ways, and returns the dataframe.
|
| 26 |
+
# For best accuracy and generalizability, the code uses
|
| 27 |
+
# regular expressions (regex) to find strings for replacement.
|
| 28 |
+
def apply_header_changes(df):
|
| 29 |
+
# remove lowercase x at beginning of name
|
| 30 |
+
df.columns = df.columns.str.replace("^x","")
|
| 31 |
+
# remove space at beginning of name
|
| 32 |
+
df.columns = df.columns.str.replace("^ ","")
|
| 33 |
+
# replace space with underscore
|
| 34 |
+
df.columns = df.columns.str.replace(" ","_")
|
| 35 |
+
# fix typos
|
| 36 |
+
df.columns = df.columns.str.replace("AF_AF","AF")
|
| 37 |
+
# change "Cell Id" into "ID"
|
| 38 |
+
df.columns = df.columns.str.replace("Cell Id","ID")
|
| 39 |
+
# if the ID is the index, change "Cell Id" into "ID"
|
| 40 |
+
df.index.name = "ID"
|
| 41 |
+
#
|
| 42 |
+
df.columns = df.columns.str.replace("","")
|
| 43 |
+
return df
|
| 44 |
+
|
| 45 |
+
def apply_df_changes(df):
|
| 46 |
+
# Remove "@1" after the ID in the index
|
| 47 |
+
df.index = df.index.str.replace(r'@1$', '')
|
| 48 |
+
return df
|
| 49 |
+
|
| 50 |
+
def compare_headers(expected, actual, name):
|
| 51 |
+
missing_actual = np.setdiff1d(expected, actual)
|
| 52 |
+
extra_actual = np.setdiff1d(actual, expected)
|
| 53 |
+
if len(missing_actual) > 0:
|
| 54 |
+
#print("WARNING: File '" + name + "' lacks the following expected header(s) after import header reformatting: \n"
|
| 55 |
+
# + str(missing_actual))
|
| 56 |
+
print("WARNING: File '" + name + "' lacks the following expected item(s): \n" + str(missing_actual))
|
| 57 |
+
if len(extra_actual) > 0:
|
| 58 |
+
#print("WARNING: '" + name + "' has the following unexpected header(s) after import header reformatting: \n"
|
| 59 |
+
# + str(extra_actual))
|
| 60 |
+
print("WARNING: '" + name + "' has the following unexpected item(s): \n" + str(extra_actual))
|
| 61 |
+
|
| 62 |
+
return None
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
def add_metadata_location(row):
|
| 66 |
+
fc = row['full_column'].lower()
|
| 67 |
+
if 'cytoplasm' in fc and 'cell' not in fc and 'nucleus' not in fc:
|
| 68 |
+
return 'cytoplasm'
|
| 69 |
+
elif 'cell' in fc and 'cytoplasm' not in fc and 'nucleus' not in fc:
|
| 70 |
+
return 'cell'
|
| 71 |
+
elif 'nucleus' in fc and 'cell' not in fc and 'cytoplasm' not in fc:
|
| 72 |
+
return 'nucleus'
|
| 73 |
+
else:
|
| 74 |
+
return 'unknown'
|
| 75 |
+
|
| 76 |
+
|
| 77 |
+
def get_perc(row, cell_type):
|
| 78 |
+
total = row['stroma'] + row['immune'] + row['cancer']+row['endothelial']
|
| 79 |
+
return round(row[cell_type]/total *100,1)
|
| 80 |
+
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
# Divide each marker (and its localisation) by the right exposure setting for each group of samples
|
| 84 |
+
def divide_exp_time(col, exp_col, metadata):
|
| 85 |
+
exp_time = metadata.loc[metadata['full_column'] == col.name, exp_col].values[0]
|
| 86 |
+
return col/exp_time
|
| 87 |
+
|
| 88 |
+
|
| 89 |
+
def do_background_sub(col, df, metadata):
|
| 90 |
+
#print(col.name)
|
| 91 |
+
location = metadata.loc[metadata['full_column'] == col.name, 'localisation'].values[0]
|
| 92 |
+
#print('location = ' + location)
|
| 93 |
+
channel = metadata.loc[metadata['full_column'] == col.name, 'Channel'].values[0]
|
| 94 |
+
#print('channel = ' + channel)
|
| 95 |
+
af_target = metadata.loc[
|
| 96 |
+
(metadata['Channel']==channel) \
|
| 97 |
+
& (metadata['localisation']==location) \
|
| 98 |
+
& (metadata['target_lower'].str.contains(r'^af\d{3}$')),\
|
| 99 |
+
'full_column'].values[0]
|
| 100 |
+
return col - df.loc[:,af_target]
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
"""
|
| 104 |
+
This function plots distributions. It takes in a string title (title), a list of
|
| 105 |
+
dataframes from which to plot (dfs), a list of dataframe names for the legend
|
| 106 |
+
(names), a list of the desired colors for the plotted samples (colors),
|
| 107 |
+
a string for the x-axis label (x_label), ```a float binwidth for histrogram (bin_size)```,
|
| 108 |
+
a boolean to show the legend or not (legend),
|
| 109 |
+
and the names of the marker(s) to plot (input_labels). If not specified,
|
| 110 |
+
the function will plot all markers in one plot. input_labels can either be a
|
| 111 |
+
single string, e.g., 'my_marker', or a list, e.g., ['my_marker1','my_marker2'].
|
| 112 |
+
|
| 113 |
+
The function will create a distribution plot and save it to png. It requires
|
| 114 |
+
a list of items not to be considered as markers when evaluating column names
|
| 115 |
+
(not_markers) to be in memory. It also requires a desired output location of
|
| 116 |
+
the files (output_dir) to already be in memory.
|
| 117 |
+
"""
|
| 118 |
+
|
| 119 |
+
|
| 120 |
+
|
| 121 |
+
def make_distr_plot_per_sample(title, location, dfs, df_names, colors, x_label, legend, xlims = None, markers = ['all'],not_intensities = None):
|
| 122 |
+
### GET LIST OF MARKERS TO PLOT ###
|
| 123 |
+
# Get list of markers to plot if not specified by user, using columns in first df
|
| 124 |
+
# Writing function(parameter = FILLER) makes that parameter optional when user calls function,
|
| 125 |
+
# since it is given a default value!
|
| 126 |
+
if markers == ["all"]:
|
| 127 |
+
markers = [c for c in dfs[0].columns.values if c not in not_intensities]
|
| 128 |
+
elif not isinstance(markers, list):
|
| 129 |
+
markers = [markers]
|
| 130 |
+
# Make input labels a set to get only unique values, then put back into list
|
| 131 |
+
markers = list(set(markers))
|
| 132 |
+
|
| 133 |
+
### GET XLIMS ###
|
| 134 |
+
if xlims == None:
|
| 135 |
+
mins = [df.loc[:,markers].min().min() for df in dfs]
|
| 136 |
+
maxes = [df.loc[:,markers].max().max() for df in dfs]
|
| 137 |
+
xlims = [min(mins), max(maxes)]
|
| 138 |
+
if not isinstance(xlims, list):
|
| 139 |
+
print("Problem - xlmis not list. Exiting method...")
|
| 140 |
+
return None
|
| 141 |
+
### CHECK DATA CAN BE PLOTTED ###
|
| 142 |
+
# Check for data with only 1 unique value - this will cause error if plotted
|
| 143 |
+
group_labels = []
|
| 144 |
+
hist_data = []
|
| 145 |
+
# Iterate through all dataframes (dfs)
|
| 146 |
+
for i in range(len(dfs)):
|
| 147 |
+
# Iterate through all marker labels
|
| 148 |
+
for f in markers:
|
| 149 |
+
# If there is only one unique value in the marker data for this dataframe,
|
| 150 |
+
# you cannot plot a distribution plot. It gives you a linear algebra
|
| 151 |
+
# singular value matrix error
|
| 152 |
+
if dfs[i][f].nunique() != 1:
|
| 153 |
+
# Add df name and marker name to labels list
|
| 154 |
+
# If we have >1 df, we want to make clear
|
| 155 |
+
# which legend label is associated with which df
|
| 156 |
+
if len(df_names) > 1:
|
| 157 |
+
group_labels.append(df_names[i]+"_"+f)
|
| 158 |
+
else:
|
| 159 |
+
group_labels.append(f)
|
| 160 |
+
# add the data to the data list
|
| 161 |
+
hist_data.append(dfs[i][f])
|
| 162 |
+
# if no data had >1 unique values, there is nothing to plot
|
| 163 |
+
if len(group_labels) < 1:
|
| 164 |
+
print("No markers plotted - all were singular value. Names and markers were " + str(df_names) + ", " + str(markers))
|
| 165 |
+
return None
|
| 166 |
+
|
| 167 |
+
### TRANSFORM COLOR ITEMS TO CORRECT TYPE ###
|
| 168 |
+
if isinstance(colors[0], tuple):
|
| 169 |
+
colors = ['rgb' + str(color) for color in colors]
|
| 170 |
+
|
| 171 |
+
### PLOT DATA ###
|
| 172 |
+
# Create plot
|
| 173 |
+
fig = ff.create_distplot(hist_data, group_labels, bin_size=0.1,
|
| 174 |
+
#colors=colors, bin_size=bin_size, show_rug=False)#show_hist=False,
|
| 175 |
+
colors=colors, show_rug=False)
|
| 176 |
+
# Adjust title, font, background color, legend...
|
| 177 |
+
fig.update_layout(title_text=title, font=dict(size=18),
|
| 178 |
+
plot_bgcolor = 'white', showlegend = legend)#, legend_x = 3)
|
| 179 |
+
# Adjust opacity
|
| 180 |
+
fig.update_traces(opacity=0.6)
|
| 181 |
+
# Adjust x-axis parameters
|
| 182 |
+
fig.update_xaxes(title_text = x_label, showline=True, linewidth=2, linecolor='black',
|
| 183 |
+
tickfont=dict(size=18), range = xlims) # x lims was here
|
| 184 |
+
# Adjust y-axis parameters
|
| 185 |
+
fig.update_yaxes(title_text = "Kernel density estimate",showline=True, linewidth=1, linecolor='black',
|
| 186 |
+
tickfont=dict(size=18))
|
| 187 |
+
|
| 188 |
+
|
| 189 |
+
### SAVE/DISPLAY PLOT ###
|
| 190 |
+
# Save plot to HTML
|
| 191 |
+
# plotly.io.write_html(fig, file = output_dir + "/" + title + ".html")
|
| 192 |
+
# Plot in new tab
|
| 193 |
+
#plot(fig)
|
| 194 |
+
# Save to png
|
| 195 |
+
filename = os.path.join(location, title.replace(" ","_") + ".png")
|
| 196 |
+
fig.write_image(filename)
|
| 197 |
+
return None
|
| 198 |
+
|
| 199 |
+
|
| 200 |
+
|
| 201 |
+
|
| 202 |
+
|
| 203 |
+
# this could be changed to use recursion and make it 'smarter'
|
| 204 |
+
|
| 205 |
+
def shorten_feature_names(long_names):
|
| 206 |
+
name_dict = dict(zip(long_names,[n.split('_')[0] for n in long_names]))
|
| 207 |
+
names_lts, long_names, iteration = shorten_feature_names_helper(name_dict, long_names, 1)
|
| 208 |
+
# names_lts = names long-to-short
|
| 209 |
+
# names_stl = names stl
|
| 210 |
+
names_stl = {}
|
| 211 |
+
for n in names_lts.items():
|
| 212 |
+
names_stl[n[1]] = n[0]
|
| 213 |
+
return names_lts, names_stl
|
| 214 |
+
|
| 215 |
+
|
| 216 |
+
def shorten_feature_names_helper(name_dict, long_names, iteration):
|
| 217 |
+
#print("\nThis is iteration #"+str(iteration))
|
| 218 |
+
#print("name_dict is: " + str(name_dict))
|
| 219 |
+
#print("long_names is: " + str(long_names))
|
| 220 |
+
## If the number of unique nicknames == number of long names
|
| 221 |
+
## then the work here is done
|
| 222 |
+
#print('\nCompare lengths: ' + str(len(set(name_dict.values()))) + ", " + str(len(long_names)))
|
| 223 |
+
#print('set(name_dict.values()): ' + str(set(name_dict.values())))
|
| 224 |
+
#print('long_names: ' + str(long_names))
|
| 225 |
+
if len(set(name_dict.values())) == len(long_names):
|
| 226 |
+
#print('All done!')
|
| 227 |
+
return name_dict, long_names, iteration
|
| 228 |
+
|
| 229 |
+
## otherwise, if the number of unique nicknames is not
|
| 230 |
+
## equal to the number of long names (must be shorter than),
|
| 231 |
+
## then we need to find more unique names
|
| 232 |
+
iteration += 1
|
| 233 |
+
nicknames_set = set()
|
| 234 |
+
non_unique_nicknames = set()
|
| 235 |
+
# construct set of current nicknames
|
| 236 |
+
for long_name in long_names:
|
| 237 |
+
#print('long_name is ' + long_name + ' and non_unique_nicknames set is ' + str(non_unique_nicknames))
|
| 238 |
+
short_name = name_dict[long_name]
|
| 239 |
+
if short_name in nicknames_set:
|
| 240 |
+
non_unique_nicknames.add(short_name)
|
| 241 |
+
else:
|
| 242 |
+
nicknames_set.add(short_name)
|
| 243 |
+
#print('non_unique_nicknames are: ' + str(non_unique_nicknames))
|
| 244 |
+
|
| 245 |
+
# figure out all long names associated
|
| 246 |
+
# with the non-unique short names
|
| 247 |
+
trouble_long_names = set()
|
| 248 |
+
for long_name in long_names:
|
| 249 |
+
short_name = name_dict[long_name]
|
| 250 |
+
if short_name in non_unique_nicknames:
|
| 251 |
+
trouble_long_names.add(long_name)
|
| 252 |
+
|
| 253 |
+
#print('troublesome long names are: ' + str(trouble_long_names))
|
| 254 |
+
#print('name_dict: ' + str(name_dict))
|
| 255 |
+
# operate on all names that are associated with
|
| 256 |
+
# the non-unique short nicknames
|
| 257 |
+
for long_name in trouble_long_names:
|
| 258 |
+
#print('trouble long name is: ' + long_name)
|
| 259 |
+
#print('old nickname is: ' + name_dict[long_name])
|
| 260 |
+
name_dict[long_name] = '_'.join(long_name.split('_')[0:iteration])
|
| 261 |
+
#print('new nickname is: ' + name_dict[long_name])
|
| 262 |
+
shorten_feature_names_helper(name_dict, long_names, iteration)
|
| 263 |
+
return name_dict, long_names, iteration
|
| 264 |
+
|
| 265 |
+
|
| 266 |
+
def heatmap_function2(title,
|
| 267 |
+
data,
|
| 268 |
+
method, metric, cmap,
|
| 269 |
+
cbar_kws, xticklabels, save_loc,
|
| 270 |
+
row_cluster, col_cluster,
|
| 271 |
+
annotations = {'rows':[],'cols':[]}):
|
| 272 |
+
|
| 273 |
+
sb.set(font_scale= 6.0)
|
| 274 |
+
|
| 275 |
+
# Extract row and column mappings
|
| 276 |
+
row_mappings = []
|
| 277 |
+
col_mappings = []
|
| 278 |
+
for ann in annotations['rows']:
|
| 279 |
+
row_mappings.append(ann['mapping'])
|
| 280 |
+
for ann in annotations['cols']:
|
| 281 |
+
col_mappings.append(ann['mapping'])
|
| 282 |
+
# If empty lists, convert to None so seaborn accepts
|
| 283 |
+
# as the row_colors or col_colors objects
|
| 284 |
+
if len(row_mappings) == 0:
|
| 285 |
+
row_mappings = None
|
| 286 |
+
if len(col_mappings) == 0:
|
| 287 |
+
col_mappings = None
|
| 288 |
+
|
| 289 |
+
def heatmap_function(title,
|
| 290 |
+
data,
|
| 291 |
+
method, metric, cmap,
|
| 292 |
+
cbar_kws, xticklabels, save_loc,
|
| 293 |
+
row_cluster, col_cluster,
|
| 294 |
+
annotations = {'rows':[],'cols':[]}):
|
| 295 |
+
|
| 296 |
+
sb.set(font_scale= 2.0)
|
| 297 |
+
|
| 298 |
+
# Extract row and column mappings
|
| 299 |
+
row_mappings = []
|
| 300 |
+
col_mappings = []
|
| 301 |
+
for ann in annotations['rows']:
|
| 302 |
+
row_mappings.append(ann['mapping'])
|
| 303 |
+
for ann in annotations['cols']:
|
| 304 |
+
col_mappings.append(ann['mapping'])
|
| 305 |
+
# If empty lists, convert to None so seaborn accepts
|
| 306 |
+
# as the row_colors or col_colors objects
|
| 307 |
+
if len(row_mappings) == 0:
|
| 308 |
+
row_mappings = None
|
| 309 |
+
if len(col_mappings) == 0:
|
| 310 |
+
col_mappings = None
|
| 311 |
+
|
| 312 |
+
# Create clustermap
|
| 313 |
+
g = sb.clustermap(data = data,
|
| 314 |
+
robust = True,
|
| 315 |
+
method = method, metric = metric,
|
| 316 |
+
cmap = cmap,
|
| 317 |
+
row_cluster = row_cluster, col_cluster = col_cluster,
|
| 318 |
+
figsize = (40,30),
|
| 319 |
+
row_colors=row_mappings, col_colors=col_mappings,
|
| 320 |
+
yticklabels = False,
|
| 321 |
+
cbar_kws = cbar_kws,
|
| 322 |
+
xticklabels = xticklabels)
|
| 323 |
+
|
| 324 |
+
# To rotate slightly the x labels
|
| 325 |
+
plt.setp(g.ax_heatmap.xaxis.get_majorticklabels(), rotation=45)
|
| 326 |
+
|
| 327 |
+
# Add title
|
| 328 |
+
g.fig.suptitle(title, fontsize = 60.0)
|
| 329 |
+
|
| 330 |
+
#And now for the legends:
|
| 331 |
+
# iterate through 'rows', 'cols'
|
| 332 |
+
for ann_type in annotations.keys():
|
| 333 |
+
# iterate through each individual annotation feature
|
| 334 |
+
for ann in annotations[ann_type]:
|
| 335 |
+
color_dict = ann['dict']
|
| 336 |
+
handles = []
|
| 337 |
+
for item in color_dict.keys():
|
| 338 |
+
h = g.ax_col_dendrogram.bar(0,0, color = color_dict[item], label = item,
|
| 339 |
+
linewidth = 0)
|
| 340 |
+
handles.append(h)
|
| 341 |
+
legend = plt.legend(handles = handles, loc = ann['location'], title = ann['label'],
|
| 342 |
+
bbox_to_anchor=ann['bbox_to_anchor'],
|
| 343 |
+
bbox_transform=plt.gcf().transFigure)
|
| 344 |
+
ax = plt.gca().add_artist(legend)
|
| 345 |
+
|
| 346 |
+
# Save image
|
| 347 |
+
filename = os.path.join(save_loc, title.lower().replace(" ","_") + ".png")
|
| 348 |
+
g.savefig(filename)
|
| 349 |
+
|
| 350 |
+
return None
|
| 351 |
+
|
| 352 |
+
|
| 353 |
+
|
| 354 |
+
# sources -
|
| 355 |
+
#https://stackoverflow.com/questions/27988846/how-to-express-classes-on-the-axis-of-a-heatmap-in-seaborn
|
| 356 |
+
# https://matplotlib.org/3.1.1/tutorials/intermediate/legend_guide.html
|
| 357 |
+
|
| 358 |
+
|
| 359 |
+
def verify_line_no(filename, lines_read):
|
| 360 |
+
# Use Linux "wc -l" command to get the number of lines in the unopened file
|
| 361 |
+
wc = subprocess.check_output(['wc', '-l', filename]).decode("utf-8")
|
| 362 |
+
# Take that string, turn it into a list, extract the first item,
|
| 363 |
+
# and make that an int - this is the number of lines in the file
|
| 364 |
+
wc = int(wc.split()[0])
|
| 365 |
+
if lines_read != wc:
|
| 366 |
+
print("WARNING: '" + filename + "' has " + str(wc) +
|
| 367 |
+
" lines, but imported dataframe has "
|
| 368 |
+
+ str(lines_read) + " (including header).")
|
| 369 |
+
return None
|
| 370 |
+
|
| 371 |
+
|
| 372 |
+
def rgb_tuple_from_str(rgb_str):
|
| 373 |
+
rgb_str = rgb_str.replace("(","").replace(")","").replace(" ","")
|
| 374 |
+
rgb = list(map(float,rgb_str.split(",")))
|
| 375 |
+
return tuple(rgb)
|
| 376 |
+
|
| 377 |
+
def color_dict_to_df(cd, column_name):
|
| 378 |
+
df = pd.DataFrame.from_dict(cd, orient = 'index')
|
| 379 |
+
df['rgb'] = df.apply(lambda row: (np.float64(row[0]), np.float64(row[1]), np.float64(row[2])), axis = 1)
|
| 380 |
+
df = df.drop(columns = [0,1,2])
|
| 381 |
+
df['hex'] = df.apply(lambda row: mplc.to_hex(row['rgb']), axis = 1)
|
| 382 |
+
df[column_name] = df.index
|
| 383 |
+
return df
|
| 384 |
+
|
| 385 |
+
|
| 386 |
+
# p-values that are less than or equal to 0.05
|
| 387 |
+
def p_add_star(row):
|
| 388 |
+
m = [str('{:0.3e}'.format(m)) + "*"
|
| 389 |
+
if m <= 0.05 \
|
| 390 |
+
else str('{:0.3e}'.format(m))
|
| 391 |
+
for m in row ]
|
| 392 |
+
return pd.Series(m)
|
| 393 |
+
|
| 394 |
+
# assigns a specific number of asterisks based on the thresholds
|
| 395 |
+
def p_to_star(row):
|
| 396 |
+
output = []
|
| 397 |
+
for item in row:
|
| 398 |
+
if item <= 0.001:
|
| 399 |
+
stars = 3
|
| 400 |
+
elif item <= 0.01:
|
| 401 |
+
stars = 2
|
| 402 |
+
elif item <= 0.05:
|
| 403 |
+
stars = 1
|
| 404 |
+
else:
|
| 405 |
+
stars = 0
|
| 406 |
+
value = ''
|
| 407 |
+
for i in range(stars):
|
| 408 |
+
value += '*'
|
| 409 |
+
output.append(value)
|
| 410 |
+
return pd.Series(output)
|
| 411 |
+
|
| 412 |
+
|
| 413 |
+
|
| 414 |
+
def plot_gaussian_distributions(df):
|
| 415 |
+
# Initialize thresholds list to store all calculated thresholds
|
| 416 |
+
all_thresholds = []
|
| 417 |
+
|
| 418 |
+
# Iterate over all columns except the first one (assuming the first one is non-numeric or an index)
|
| 419 |
+
for column in df.columns:
|
| 420 |
+
# Extract the marker data
|
| 421 |
+
marker_data = df[column]
|
| 422 |
+
|
| 423 |
+
# Calculating mean and standard deviation for each marker
|
| 424 |
+
m_mean, m_std = np.mean(marker_data), np.std(marker_data)
|
| 425 |
+
|
| 426 |
+
# Generating x values for the Gaussian curve
|
| 427 |
+
x_vals = np.linspace(marker_data.min(), marker_data.max(), 100)
|
| 428 |
+
|
| 429 |
+
# Calculating Gaussian distribution curve
|
| 430 |
+
gaussian_curve = (1 / (m_std * np.sqrt(2 * np.pi))) * np.exp(-(x_vals - m_mean) ** 2 / (2 * m_std ** 2))
|
| 431 |
+
|
| 432 |
+
# Creating figure for Gaussian distribution for each marker
|
| 433 |
+
fig = go.Figure()
|
| 434 |
+
fig.add_trace(go.Scatter(x=x_vals, y=gaussian_curve, mode='lines', name=f'{column} Gaussian Distribution'))
|
| 435 |
+
fig.update_layout(title=f'Gaussian Distribution for {column} Marker')
|
| 436 |
+
|
| 437 |
+
# Calculating thresholds based on each marker's distribution
|
| 438 |
+
seuil_1sigma = m_mean + m_std
|
| 439 |
+
seuil_2sigma = m_mean + 2 * m_std
|
| 440 |
+
seuil_3sigma = m_mean + 3 * m_std
|
| 441 |
+
|
| 442 |
+
# Display the figures with thresholds
|
| 443 |
+
fig.add_shape(type='line', x0=seuil_1sigma, y0=0, x1=seuil_1sigma, y1=np.max(gaussian_curve),
|
| 444 |
+
line=dict(color='red', dash='dash'), name=f'Seuil 1σ: {seuil_1sigma:.2f}')
|
| 445 |
+
fig.add_shape(type='line', x0=seuil_2sigma, y0=0, x1=seuil_2sigma, y1=np.max(gaussian_curve),
|
| 446 |
+
line=dict(color='green', dash='dash'), name=f'Seuil 2σ: {seuil_2sigma:.2f}')
|
| 447 |
+
fig.add_shape(type='line', x0=seuil_3sigma, y0=0, x1=seuil_3sigma, y1=np.max(gaussian_curve),
|
| 448 |
+
line=dict(color='blue', dash='dash'), name=f'Seuil 3σ: {seuil_3sigma:.2f}')
|
| 449 |
+
|
| 450 |
+
# Add markers and values to the plot
|
| 451 |
+
fig.add_trace(go.Scatter(x=[seuil_1sigma, seuil_2sigma, seuil_3sigma],
|
| 452 |
+
y=[0, 0, 0],
|
| 453 |
+
mode='markers+text',
|
| 454 |
+
text=[f'{seuil_1sigma:.2f}', f'{seuil_2sigma:.2f}', f'{seuil_3sigma:.2f}'],
|
| 455 |
+
textposition="top center",
|
| 456 |
+
marker=dict(size=10, color=['red', 'green', 'blue']),
|
| 457 |
+
name='Threshold Values'))
|
| 458 |
+
|
| 459 |
+
fig.show()
|
| 460 |
+
|
| 461 |
+
# Append thresholds for each marker to the list
|
| 462 |
+
all_thresholds.append((column, seuil_1sigma, seuil_2sigma, seuil_3sigma)) # Include the column name
|
| 463 |
+
|
| 464 |
+
# Return thresholds for all markers
|
| 465 |
+
return all_thresholds
|
| 466 |
+
|
| 467 |
+
|
| 468 |
+
|
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S1_bs.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e554b52d689c4163abcc6256d41c996caff2eb545a6c6dceef7a4ab66f9541db
|
| 3 |
+
size 133327656
|
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S2_bs.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:8e7b08b8eda9e9c78adec651f40aa373646543a1425dcbff5394f5e83459460c
|
| 3 |
+
size 138615691
|
wetransfer_data-zip_2024-05-17_1431/test_bs/DD3S3_bs.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:92a152e8e267d5d835d84edb1e93ae8b940b655bcde7a1ffda639ce21d19fcce
|
| 3 |
+
size 225018985
|
wetransfer_data-zip_2024-05-17_1431/test_bs/TMA_bs.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e19792f19207ff5f7c039c79527c5fc9a6ad3590f56caebe5a52bd30b585bf78
|
| 3 |
+
size 181932794
|
wetransfer_data-zip_2024-05-17_1431/test_cqc/test_cell_subtypes_number_by_scenes.csv
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
dc,b,tcd4,tcd8,m1,m2,treg,immune_other,cancer,αsma_mycaf,stroma_other,endothelial,total_cells
|
wetransfer_data-zip_2024-05-17_1431/test_data/Ashlar_Exposure_Time.csv
ADDED
|
@@ -0,0 +1,91 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Target,Round,Channel,ExposureTime
|
| 2 |
+
DAPI0,R0,c0,30.0
|
| 3 |
+
AF488,R0,c1,300.0
|
| 4 |
+
AF555,R0,c2,1500.0
|
| 5 |
+
AF647,R0,c3,1500.0
|
| 6 |
+
AF750,R0,c4,1500.0
|
| 7 |
+
DAPI1,R1,c0,20.0
|
| 8 |
+
ColVI,R1,c1,300.0
|
| 9 |
+
CD31,R1,c2,1200.0
|
| 10 |
+
CD4,R1,c3,1500.0
|
| 11 |
+
Ecad,R1,c4,800.0
|
| 12 |
+
DAPI2,R2,c0,15.0
|
| 13 |
+
Desmin,R2,c1,300.0
|
| 14 |
+
B7H4,R2,c2,1500.0
|
| 15 |
+
CD8,R2,c3,1500.0
|
| 16 |
+
CD20,R2,c4,1500.0
|
| 17 |
+
DAPI3,R3,c0,20.0
|
| 18 |
+
aSMA,R3,c1,50.0
|
| 19 |
+
CD68,R3,c2,1500.0
|
| 20 |
+
PD1,R3,c3,1500.0
|
| 21 |
+
CD45,R3,c4,1500.0
|
| 22 |
+
DAPI4,R4,c0,10.0
|
| 23 |
+
Vimentin,R4,c1,150.0
|
| 24 |
+
AXL,R4,c2,1500.0
|
| 25 |
+
PDL1,R4,c3,1500.0
|
| 26 |
+
FOXP3,R4,c4,1500.0
|
| 27 |
+
DAPI5,R5,c0,10.0
|
| 28 |
+
r5c2,R5,c1,20.0
|
| 29 |
+
CA9,R5,c2,1500.0
|
| 30 |
+
CD163,R5,c3,1500.0
|
| 31 |
+
Ki67,R5,c4,1000.0
|
| 32 |
+
DAPI6,R6,c0,10.0
|
| 33 |
+
CKs,R6,c1,200.0
|
| 34 |
+
Fibronectin,R6,c2,1500.0
|
| 35 |
+
CD44,R6,c3,1200.0
|
| 36 |
+
HLA,R6,c4,500.0
|
| 37 |
+
DAPI7,R7,c0,10.0
|
| 38 |
+
r7c2,R7,c1,20.0
|
| 39 |
+
PDGFR,R7,c2,1500.0
|
| 40 |
+
MMP9,R7,c3,1500.0
|
| 41 |
+
GATA3,R7,c4,1500.0
|
| 42 |
+
DAPI8,R8,c0,10.0
|
| 43 |
+
r8c2,R8,c1,25.0
|
| 44 |
+
CD11c,R8,c2,1500.0
|
| 45 |
+
Sting,R8,c3,1000.0
|
| 46 |
+
CD11b,R8,c4,1500.0
|
| 47 |
+
DAPI0,R0,c0,30.0
|
| 48 |
+
AF488,R0,c1,300.0
|
| 49 |
+
AF555,R0,c2,1500.0
|
| 50 |
+
AF647,R0,c3,1500.0
|
| 51 |
+
AF750,R0,c4,1500.0
|
| 52 |
+
DAPI1,R1,c0,20.0
|
| 53 |
+
ColVI,R1,c1,300.0
|
| 54 |
+
CD31,R1,c2,1200.0
|
| 55 |
+
CD4,R1,c3,1500.0
|
| 56 |
+
Ecad,R1,c4,800.0
|
| 57 |
+
DAPI2,R2,c0,15.0
|
| 58 |
+
Desmin,R2,c1,300.0
|
| 59 |
+
B7H4,R2,c2,1500.0
|
| 60 |
+
CD8,R2,c3,1500.0
|
| 61 |
+
CD20,R2,c4,1500.0
|
| 62 |
+
DAPI3,R3,c0,20.0
|
| 63 |
+
aSMA,R3,c1,50.0
|
| 64 |
+
CD68,R3,c2,1500.0
|
| 65 |
+
PD1,R3,c3,1500.0
|
| 66 |
+
CD45,R3,c4,1500.0
|
| 67 |
+
DAPI4,R4,c0,10.0
|
| 68 |
+
Vimentin,R4,c1,150.0
|
| 69 |
+
AXL,R4,c2,1500.0
|
| 70 |
+
PDL1,R4,c3,1500.0
|
| 71 |
+
FOXP3,R4,c4,1500.0
|
| 72 |
+
DAPI5,R5,c0,10.0
|
| 73 |
+
r5c2,R5,c1,20.0
|
| 74 |
+
CA9,R5,c2,1500.0
|
| 75 |
+
CD163,R5,c3,1500.0
|
| 76 |
+
Ki67,R5,c4,1000.0
|
| 77 |
+
DAPI6,R6,c0,10.0
|
| 78 |
+
CKs,R6,c1,200.0
|
| 79 |
+
Fibronectin,R6,c2,1500.0
|
| 80 |
+
CD44,R6,c3,1200.0
|
| 81 |
+
HLA,R6,c4,500.0
|
| 82 |
+
DAPI7,R7,c0,10.0
|
| 83 |
+
r7c2,R7,c1,20.0
|
| 84 |
+
PDGFR,R7,c2,1500.0
|
| 85 |
+
MMP9,R7,c3,1500.0
|
| 86 |
+
GATA3,R7,c4,1500.0
|
| 87 |
+
DAPI8,R8,c0,10.0
|
| 88 |
+
r8c2,R8,c1,25.0
|
| 89 |
+
CD11c,R8,c2,1500.0
|
| 90 |
+
Sting,R8,c3,1000.0
|
| 91 |
+
CD11b,R8,c4,1500.0
|
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S1.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c26fb09be4db360c1ca0490bd0624b0b14123537e66aa05ae7a4eb95e0ca512f
|
| 3 |
+
size 208337030
|
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S2.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e2ed3706f14d9284987b77e3826a0aa0e97f4f15a96863eaea1ecda7ae6aea6a
|
| 3 |
+
size 216509787
|
wetransfer_data-zip_2024-05-17_1431/test_data/DD3S3.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:b29edf8f829be7c8d8450e94492705e04febd85839c6c57a9f9a3db21bd32b94
|
| 3 |
+
size 329608939
|
wetransfer_data-zip_2024-05-17_1431/test_data/TMA.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cee32a38d4883b8980cd07a16b8e209a3a0b9ebe92948c9d7fd33562f97ff627
|
| 3 |
+
size 275328609
|
wetransfer_data-zip_2024-05-17_1431/test_data/new_data.csv
ADDED
|
@@ -0,0 +1,46 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Target,Round,Channel,ExposureTime
|
| 2 |
+
DAPI0,R0,c0,30.0
|
| 3 |
+
AF488,R0,c1,300.0
|
| 4 |
+
AF555,R0,c2,1500.0
|
| 5 |
+
AF647,R0,c3,1500.0
|
| 6 |
+
AF750,R0,c4,1500.0
|
| 7 |
+
DAPI1,R1,c0,20.0
|
| 8 |
+
ColVI,R1,c1,300.0
|
| 9 |
+
CD31,R1,c2,1200.0
|
| 10 |
+
CD4,R1,c3,1500.0
|
| 11 |
+
Ecad,R1,c4,800.0
|
| 12 |
+
DAPI2,R2,c0,15.0
|
| 13 |
+
Desmin,R2,c1,300.0
|
| 14 |
+
B7H4,R2,c2,1500.0
|
| 15 |
+
CD8,R2,c3,1500.0
|
| 16 |
+
CD20,R2,c4,1500.0
|
| 17 |
+
DAPI3,R3,c0,20.0
|
| 18 |
+
aSMA,R3,c1,50.0
|
| 19 |
+
CD68,R3,c2,1500.0
|
| 20 |
+
PD1,R3,c3,1500.0
|
| 21 |
+
CD45,R3,c4,1500.0
|
| 22 |
+
DAPI4,R4,c0,10.0
|
| 23 |
+
Vimentin,R4,c1,150.0
|
| 24 |
+
AXL,R4,c2,1500.0
|
| 25 |
+
PDL1,R4,c3,1500.0
|
| 26 |
+
FOXP3,R4,c4,1500.0
|
| 27 |
+
DAPI5,R5,c0,10.0
|
| 28 |
+
r5c2,R5,c1,20.0
|
| 29 |
+
CA9,R5,c2,1500.0
|
| 30 |
+
CD163,R5,c3,1500.0
|
| 31 |
+
Ki67,R5,c4,1000.0
|
| 32 |
+
DAPI6,R6,c0,10.0
|
| 33 |
+
CKs,R6,c1,200.0
|
| 34 |
+
Fibronectin,R6,c2,1500.0
|
| 35 |
+
CD44,R6,c3,1200.0
|
| 36 |
+
HLA,R6,c4,500.0
|
| 37 |
+
DAPI7,R7,c0,10.0
|
| 38 |
+
r7c2,R7,c1,20.0
|
| 39 |
+
PDGFR,R7,c2,1500.0
|
| 40 |
+
MMP9,R7,c3,1500.0
|
| 41 |
+
GATA3,R7,c4,1500.0
|
| 42 |
+
DAPI8,R8,c0,10.0
|
| 43 |
+
r8c2,R8,c1,25.0
|
| 44 |
+
CD11c,R8,c2,1500.0
|
| 45 |
+
Sting,R8,c3,1000.0
|
| 46 |
+
CD11b,R8,c4,1500.0
|
wetransfer_data-zip_2024-05-17_1431/test_data/stored_variables.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"selected_metadata_files": ["Slide_B_DD1s1.one_1.tif.csv", "Slide_B_DD1s1.one_2.tif.csv"]}
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/Exposure_Time.csv
ADDED
|
@@ -0,0 +1,37 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Round,Target,Exp,Channel
|
| 2 |
+
R0,AF488,300,c2
|
| 3 |
+
R0,AF555,1500,c3
|
| 4 |
+
R0,AF647,1500,c4
|
| 5 |
+
R0,AF750,1500,c5
|
| 6 |
+
R1,ColVI,300,c2
|
| 7 |
+
R1,CD31,1200,c3
|
| 8 |
+
R1,CD4,1500,c4
|
| 9 |
+
R1,Ecad,800,c5
|
| 10 |
+
R2,Desmin,300,c2
|
| 11 |
+
R2,B7H4,1500,c3
|
| 12 |
+
R2,CD8,1500,c4
|
| 13 |
+
R2,CD20,1500,c5
|
| 14 |
+
R3,aSMA,50,c2
|
| 15 |
+
R3,CD68,1500,c3
|
| 16 |
+
R3,PD1,1500,c4
|
| 17 |
+
R3,CD45,1500,c5
|
| 18 |
+
R4,Vimentin,150,c2
|
| 19 |
+
R4,AXL,1500,c3
|
| 20 |
+
R4,PDL1,1500,c4
|
| 21 |
+
R4,FOXP3,1500,c5
|
| 22 |
+
R5,r5c2,20,c2
|
| 23 |
+
R5,CA9,1500,c3
|
| 24 |
+
R5,CD163,1500,c4
|
| 25 |
+
R5,Ki67,1000,c5
|
| 26 |
+
R6,CKs,200,c2
|
| 27 |
+
R6,Fibronectin,1500,c3
|
| 28 |
+
R6,CD44,1200,c4
|
| 29 |
+
R6,HLA,500,c5
|
| 30 |
+
R7,r7c2,20,c2
|
| 31 |
+
R7,PDGFR,1500,c3
|
| 32 |
+
R7,MMP9,1500,c4
|
| 33 |
+
R7,GATA3,1500,c5
|
| 34 |
+
R8,r8c2,25,c2
|
| 35 |
+
R8,CD11c,1500,c3
|
| 36 |
+
R8,Sting,1000,c4
|
| 37 |
+
R8,CD11b,1500,c5
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/Set_B_unique_ROIs.csv
ADDED
|
@@ -0,0 +1,468 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Sample_ID;ROI_index;Patient;Unique_ROI_index
|
| 2 |
+
DD3S1.csv;0;61;61a
|
| 3 |
+
DD3S1.csv;1;62;62a
|
| 4 |
+
DD3S1.csv;2;63;63a
|
| 5 |
+
DD3S1.csv;3;59;59a
|
| 6 |
+
DD3S1.csv;4;60;60a
|
| 7 |
+
DD3S1.csv;5;33;33a
|
| 8 |
+
DD3S1.csv;6;35;35a
|
| 9 |
+
DD3S1.csv;7;36;36a
|
| 10 |
+
DD3S1.csv;8;37;37a
|
| 11 |
+
DD3S1.csv;9;38;38a
|
| 12 |
+
DD3S1.csv;10;30;30a
|
| 13 |
+
DD3S1.csv;11;32;32a
|
| 14 |
+
DD3S1.csv;12;25;25a
|
| 15 |
+
DD3S1.csv;13;26;26a
|
| 16 |
+
DD3S1.csv;14;27;27a
|
| 17 |
+
DD3S1.csv;15;29;29a
|
| 18 |
+
DD3S1.csv;16;20;20a
|
| 19 |
+
DD3S1.csv;17;21;21a
|
| 20 |
+
DD3S1.csv;18;22;22a
|
| 21 |
+
DD3S1.csv;19;23;23a
|
| 22 |
+
DD3S1.csv;20;24;24a
|
| 23 |
+
DD3S1.csv;21;14;14a
|
| 24 |
+
DD3S1.csv;22;15;15a
|
| 25 |
+
DD3S1.csv;23;16;16a
|
| 26 |
+
DD3S1.csv;24;17;17a
|
| 27 |
+
DD3S1.csv;25;18;18a
|
| 28 |
+
DD3S1.csv;26;19;19a
|
| 29 |
+
DD3S1.csv;27;11;11a
|
| 30 |
+
DD3S1.csv;28;12;12a
|
| 31 |
+
DD3S1.csv;29;5;5a
|
| 32 |
+
DD3S1.csv;30;8;8a
|
| 33 |
+
DD3S1.csv;31;9;9a
|
| 34 |
+
DD3S1.csv;32;10;10a
|
| 35 |
+
DD3S1.csv;33;4;4a
|
| 36 |
+
DD3S1.csv;34;59;59b
|
| 37 |
+
DD3S1.csv;35;60;60b
|
| 38 |
+
DD3S1.csv;36;61;61b
|
| 39 |
+
DD3S1.csv;37;62;62b
|
| 40 |
+
DD3S1.csv;38;63;63b
|
| 41 |
+
DD3S1.csv;39;59;59c
|
| 42 |
+
DD3S2.csv;0;52;52a
|
| 43 |
+
DD3S2.csv;1;53;53a
|
| 44 |
+
DD3S2.csv;2;54;54a
|
| 45 |
+
DD3S2.csv;3;55;55a
|
| 46 |
+
DD3S2.csv;4;56;56a
|
| 47 |
+
DD3S2.csv;5;57;57a
|
| 48 |
+
DD3S2.csv;6;50;50a
|
| 49 |
+
DD3S2.csv;7;51;51a
|
| 50 |
+
DD3S2.csv;8;42;42a
|
| 51 |
+
DD3S2.csv;9;43;43a
|
| 52 |
+
DD3S2.csv;10;44;44a
|
| 53 |
+
DD3S2.csv;11;45;45a
|
| 54 |
+
DD3S2.csv;12;47;47a
|
| 55 |
+
DD3S2.csv;13;39;39a
|
| 56 |
+
DD3S2.csv;14;40;40a
|
| 57 |
+
DD3S2.csv;15;41;41a
|
| 58 |
+
DD3S2.csv;16;54;54b
|
| 59 |
+
DD3S2.csv;17;55;55b
|
| 60 |
+
DD3S2.csv;18;57;57b
|
| 61 |
+
DD3S2.csv;19;49;49a
|
| 62 |
+
DD3S2.csv;20;50;50b
|
| 63 |
+
DD3S2.csv;21;51;51b
|
| 64 |
+
DD3S2.csv;22;52;52b
|
| 65 |
+
DD3S2.csv;23;53;53b
|
| 66 |
+
DD3S2.csv;24;45;45b
|
| 67 |
+
DD3S2.csv;25;40;40b
|
| 68 |
+
DD3S2.csv;26;41;41b
|
| 69 |
+
DD3S2.csv;27;42;42b
|
| 70 |
+
DD3S2.csv;28;43;43b
|
| 71 |
+
DD3S2.csv;29;57;57c
|
| 72 |
+
DD3S2.csv;30;50;50c
|
| 73 |
+
DD3S2.csv;31;51;51c
|
| 74 |
+
DD3S2.csv;32;39;39b
|
| 75 |
+
DD3S2.csv;33;52;52c
|
| 76 |
+
DD3S2.csv;34;53;53c
|
| 77 |
+
DD3S2.csv;35;54;54c
|
| 78 |
+
DD3S2.csv;36;55;55c
|
| 79 |
+
DD3S2.csv;37;56;56b
|
| 80 |
+
DD3S2.csv;38;45;45c
|
| 81 |
+
DD3S2.csv;39;46;46a
|
| 82 |
+
DD3S2.csv;40;47;47b
|
| 83 |
+
DD3S2.csv;41;48;48a
|
| 84 |
+
DD3S2.csv;42;40;40c
|
| 85 |
+
DD3S2.csv;43;41;41c
|
| 86 |
+
DD3S2.csv;44;42;42c
|
| 87 |
+
DD3S2.csv;45;44;44b
|
| 88 |
+
DD3S3.csv;0;38;38b
|
| 89 |
+
DD3S3.csv;1;58;58a
|
| 90 |
+
DD3S3.csv;2;30;30b
|
| 91 |
+
DD3S3.csv;3;32;32b
|
| 92 |
+
DD3S3.csv;4;33;33a
|
| 93 |
+
DD3S3.csv;5;36;36a
|
| 94 |
+
DD3S3.csv;6;37;37b
|
| 95 |
+
DD3S3.csv;7;28;28a
|
| 96 |
+
DD3S3.csv;8;29;29b
|
| 97 |
+
DD3S3.csv;9;20;20b
|
| 98 |
+
DD3S3.csv;10;21;21b
|
| 99 |
+
DD3S3.csv;11;22;22b
|
| 100 |
+
DD3S3.csv;12;23;23b
|
| 101 |
+
DD3S3.csv;13;25;25b
|
| 102 |
+
DD3S3.csv;14;26;26b
|
| 103 |
+
DD3S3.csv;15;27;27b
|
| 104 |
+
DD3S3.csv;16;17;17b
|
| 105 |
+
DD3S3.csv;17;18;18b
|
| 106 |
+
DD3S3.csv;18;19;19b
|
| 107 |
+
DD3S3.csv;19;11;11b
|
| 108 |
+
DD3S3.csv;20;12;12b
|
| 109 |
+
DD3S3.csv;21;13;13a
|
| 110 |
+
DD3S3.csv;22;14;14b
|
| 111 |
+
DD3S3.csv;23;15;15b
|
| 112 |
+
DD3S3.csv;24;16;16b
|
| 113 |
+
DD3S3.csv;25;8;8b
|
| 114 |
+
DD3S3.csv;26;9;9b
|
| 115 |
+
DD3S3.csv;27;10;10b
|
| 116 |
+
DD3S3.csv;28;4;4b
|
| 117 |
+
DD3S3.csv;29;5;5b
|
| 118 |
+
DD3S3.csv;30;6;6a
|
| 119 |
+
DD3S3.csv;31;30;30c
|
| 120 |
+
DD3S3.csv;32;32;32c
|
| 121 |
+
DD3S3.csv;33;33;33b
|
| 122 |
+
DD3S3.csv;34;34;34a
|
| 123 |
+
DD3S3.csv;35;36;36b
|
| 124 |
+
DD3S3.csv;36;37;37c
|
| 125 |
+
DD3S3.csv;37;38;38c
|
| 126 |
+
DD3S3.csv;38;20;20c
|
| 127 |
+
DD3S3.csv;39;21;21c
|
| 128 |
+
DD3S3.csv;40;22;22c
|
| 129 |
+
DD3S3.csv;41;23;23c
|
| 130 |
+
DD3S3.csv;42;24;24b
|
| 131 |
+
DD3S3.csv;43;25;25c
|
| 132 |
+
DD3S3.csv;44;26;26c
|
| 133 |
+
DD3S3.csv;45;27;27c
|
| 134 |
+
DD3S3.csv;46;28;28a
|
| 135 |
+
DD3S3.csv;47;29;29c
|
| 136 |
+
DD3S3.csv;48;11;11c
|
| 137 |
+
DD3S3.csv;49;12;12c
|
| 138 |
+
DD3S3.csv;50;13;13b
|
| 139 |
+
DD3S3.csv;51;14;14c
|
| 140 |
+
DD3S3.csv;52;15;15c
|
| 141 |
+
DD3S3.csv;53;16;16c
|
| 142 |
+
DD3S3.csv;54;17;17c
|
| 143 |
+
DD3S3.csv;55;18;18c
|
| 144 |
+
DD3S3.csv;56;19;19c
|
| 145 |
+
DD3S3.csv;57;3;3a
|
| 146 |
+
DD3S3.csv;58;5;5c
|
| 147 |
+
DD3S3.csv;59;6;6b
|
| 148 |
+
DD3S3.csv;60;7;7a
|
| 149 |
+
DD3S3.csv;61;8;8c
|
| 150 |
+
DD3S3.csv;62;9;9c
|
| 151 |
+
DD3S3.csv;63;10;10c
|
| 152 |
+
DD4S1.csv;0;95;95a
|
| 153 |
+
DD4S1.csv;1;122;122a
|
| 154 |
+
DD4S1.csv;2;121;121a
|
| 155 |
+
DD4S1.csv;3;125;125a
|
| 156 |
+
DD4S1.csv;4;124;124a
|
| 157 |
+
DD4S1.csv;5;94;94a
|
| 158 |
+
DD4S1.csv;6;101;101a
|
| 159 |
+
DD4S1.csv;7;86;86a
|
| 160 |
+
DD4S1.csv;8;85;85a
|
| 161 |
+
DD4S1.csv;9;84;84a
|
| 162 |
+
DD4S1.csv;10;83;83a
|
| 163 |
+
DD4S1.csv;11;91;91a
|
| 164 |
+
DD4S1.csv;12;88;88a
|
| 165 |
+
DD4S1.csv;13;87;87a
|
| 166 |
+
DD4S1.csv;14;75;75a
|
| 167 |
+
DD4S1.csv;15;74;74a
|
| 168 |
+
DD4S1.csv;16;82;82a
|
| 169 |
+
DD4S1.csv;17;81;81a
|
| 170 |
+
DD4S1.csv;18;80;80a
|
| 171 |
+
DD4S1.csv;19;79;79a
|
| 172 |
+
DD4S1.csv;20;67;67a
|
| 173 |
+
DD4S1.csv;21;66;66a
|
| 174 |
+
DD4S1.csv;22;65;65a
|
| 175 |
+
DD4S1.csv;23;64;64a
|
| 176 |
+
DD4S1.csv;24;73;73a
|
| 177 |
+
DD4S1.csv;25;71;71a
|
| 178 |
+
DD4S1.csv;26;69;69a
|
| 179 |
+
DD4S1.csv;27;68;68a
|
| 180 |
+
DD4S1.csv;28;125;125b
|
| 181 |
+
DD4S1.csv;29;124;124b
|
| 182 |
+
DD4S1.csv;30;123;123a
|
| 183 |
+
DD4S1.csv;31;122;122b
|
| 184 |
+
DD4S1.csv;32;121;121b
|
| 185 |
+
DD4S1.csv;33;125;125c
|
| 186 |
+
DD4S1.csv;34;124;124c
|
| 187 |
+
DD4S1.csv;35;123;123b
|
| 188 |
+
DD4S1.csv;36;121;121c
|
| 189 |
+
DD4S1.csv;37;115;115a
|
| 190 |
+
DD4S1.csv;38;114;114a
|
| 191 |
+
DD4S1.csv;39;113;113a
|
| 192 |
+
DD4S1.csv;40;111;111a
|
| 193 |
+
DD4S1.csv;41;120;120a
|
| 194 |
+
DD4S1.csv;42;117;117a
|
| 195 |
+
DD4S1.csv;43;116;116a
|
| 196 |
+
DD4S1.csv;44;98;98a
|
| 197 |
+
DD4S1.csv;45;126;126a
|
| 198 |
+
DD4S2.csv;0;105;105a
|
| 199 |
+
DD4S2.csv;1;103;103a
|
| 200 |
+
DD4S2.csv;2;102;102a
|
| 201 |
+
DD4S2.csv;3;110;110a
|
| 202 |
+
DD4S2.csv;4;109;109a
|
| 203 |
+
DD4S2.csv;5;106;106a
|
| 204 |
+
DD4S2.csv;6;117;117b
|
| 205 |
+
DD4S2.csv;7;166;116b
|
| 206 |
+
DD4S2.csv;8;114;114b
|
| 207 |
+
DD4S2.csv;9;113;113b
|
| 208 |
+
DD4S2.csv;10;112;112a
|
| 209 |
+
DD4S2.csv;11;119;119a
|
| 210 |
+
DD4S2.csv;12;106;106b
|
| 211 |
+
DD4S2.csv;13;105;105b
|
| 212 |
+
DD4S2.csv;14;104;104a
|
| 213 |
+
DD4S2.csv;15;110;110b
|
| 214 |
+
DD4S2.csv;16;109;109b
|
| 215 |
+
DD4S2.csv;17;119;119b
|
| 216 |
+
DD4S2.csv;18;115;115b
|
| 217 |
+
DD4S2.csv;19;114;114c
|
| 218 |
+
DD4S2.csv;20;113;113c
|
| 219 |
+
DD4S2.csv;21;112;112b
|
| 220 |
+
DD4S2.csv;22;111;111b
|
| 221 |
+
DD4S2.csv;23;110;110c
|
| 222 |
+
DD4S2.csv;24;108;108a
|
| 223 |
+
DD4S2.csv;25;107;107a
|
| 224 |
+
DD4S2.csv;26;106;106c
|
| 225 |
+
DD4S2.csv;27;105;105c
|
| 226 |
+
DD4S2.csv;28;103;103b
|
| 227 |
+
DD4S2.csv;29;102;102b
|
| 228 |
+
DD4S2.csv;30;98;98b
|
| 229 |
+
DD4S2.csv;31;97;97a
|
| 230 |
+
DD4S2.csv;32;96;96a
|
| 231 |
+
DD4S2.csv;33;95;95b
|
| 232 |
+
DD4S2.csv;34;126;126b
|
| 233 |
+
DD4S2.csv;35;100;100a
|
| 234 |
+
DD4S3.csv;0;89;89a
|
| 235 |
+
DD4S3.csv;1;88;88b
|
| 236 |
+
DD4S3.csv;2;87;87b
|
| 237 |
+
DD4S3.csv;3;85;85b
|
| 238 |
+
DD4S3.csv;4;84;84b
|
| 239 |
+
DD4S3.csv;5;83;83b
|
| 240 |
+
DD4S3.csv;6;91;91b
|
| 241 |
+
DD4S3.csv;7;78;78a
|
| 242 |
+
DD4S3.csv;8;77;77a
|
| 243 |
+
DD4S3.csv;9;75;75b
|
| 244 |
+
DD4S3.csv;10;81;81b
|
| 245 |
+
DD4S3.csv;11;80;80b
|
| 246 |
+
DD4S3.csv;12;70;70a
|
| 247 |
+
DD4S3.csv;13;69;69b
|
| 248 |
+
DD4S3.csv;14;67;67b
|
| 249 |
+
DD4S3.csv;15;65;65b
|
| 250 |
+
DD4S3.csv;16;64;64b
|
| 251 |
+
DD4S3.csv;17;73;73b
|
| 252 |
+
DD4S3.csv;18;72;72a
|
| 253 |
+
DD4S3.csv;19;71;71b
|
| 254 |
+
DD4S3.csv;20;100;100b
|
| 255 |
+
DD4S3.csv;21;99;99a
|
| 256 |
+
DD4S3.csv;22;95;95c
|
| 257 |
+
DD4S3.csv;23;92;92a
|
| 258 |
+
DD4S3.csv;24;91;91c
|
| 259 |
+
DD4S3.csv;25;90;90b
|
| 260 |
+
DD4S3.csv;26;89;89b
|
| 261 |
+
DD4S3.csv;27;88;88c
|
| 262 |
+
DD4S3.csv;28;87;87c
|
| 263 |
+
DD4S3.csv;29;85;85c
|
| 264 |
+
DD4S3.csv;30;83;83c
|
| 265 |
+
DD4S3.csv;31;81;81c
|
| 266 |
+
DD4S3.csv;32;80;80c
|
| 267 |
+
DD4S3.csv;33;78;78b
|
| 268 |
+
DD4S3.csv;34;75;75c
|
| 269 |
+
DD4S3.csv;35;74;74b
|
| 270 |
+
DD4S3.csv;36;73;73c
|
| 271 |
+
DD4S3.csv;37;71;71c
|
| 272 |
+
DD4S3.csv;38;70;70b
|
| 273 |
+
DD4S3.csv;39;69;69c
|
| 274 |
+
DD4S3.csv;40;68;68b
|
| 275 |
+
DD4S3.csv;41;67;67c
|
| 276 |
+
DD4S3.csv;42;66;66b
|
| 277 |
+
DD4S3.csv;43;65;65c
|
| 278 |
+
DD4S3.csv;44;90;90a
|
| 279 |
+
DD4S3.csv;45;79;79b
|
| 280 |
+
DD5S1.csv;0;136;136a
|
| 281 |
+
DD5S1.csv;1;135;135a
|
| 282 |
+
DD5S1.csv;2;134;134a
|
| 283 |
+
DD5S1.csv;3;133;133a
|
| 284 |
+
DD5S1.csv;4;132;132a
|
| 285 |
+
DD5S1.csv;5;131;131a
|
| 286 |
+
DD5S1.csv;6;130;130a
|
| 287 |
+
DD5S1.csv;7;129;129a
|
| 288 |
+
DD5S1.csv;8;144;144a
|
| 289 |
+
DD5S1.csv;9;143;143a
|
| 290 |
+
DD5S1.csv;10;140;140a
|
| 291 |
+
DD5S1.csv;11;137;137a
|
| 292 |
+
DD5S1.csv;12;155;155a
|
| 293 |
+
DD5S1.csv;13;154;154a
|
| 294 |
+
DD5S1.csv;14;153;153a
|
| 295 |
+
DD5S1.csv;15;150;150a
|
| 296 |
+
DD5S1.csv;16;149;149a
|
| 297 |
+
DD5S1.csv;17;148;148a
|
| 298 |
+
DD5S1.csv;18;147;147a
|
| 299 |
+
DD5S1.csv;19;146;146a
|
| 300 |
+
DD5S1.csv;20;164;164a
|
| 301 |
+
DD5S1.csv;21;162;162a
|
| 302 |
+
DD5S1.csv;22;161;161a
|
| 303 |
+
DD5S1.csv;23;160;160a
|
| 304 |
+
DD5S1.csv;24;159;159a
|
| 305 |
+
DD5S1.csv;25;157;157a
|
| 306 |
+
DD5S1.csv;26;133;133b
|
| 307 |
+
DD5S1.csv;27;132;132b
|
| 308 |
+
DD5S1.csv;28;131;131b
|
| 309 |
+
DD5S1.csv;29;130;130b
|
| 310 |
+
DD5S1.csv;30;129;129b
|
| 311 |
+
DD5S1.csv;31;128;128a
|
| 312 |
+
DD5S1.csv;32;127;127a
|
| 313 |
+
DD5S1.csv;33;136;136b
|
| 314 |
+
DD5S1.csv;34;135;135b
|
| 315 |
+
DD5S1.csv;35;134;134a
|
| 316 |
+
DD5S1.csv;36;142;142a
|
| 317 |
+
DD5S1.csv;37;139;139a
|
| 318 |
+
DD5S1.csv;38;137;137b
|
| 319 |
+
DD5S1.csv;39;144;144b
|
| 320 |
+
DD5S1.csv;40;143;143b
|
| 321 |
+
DD5S1.csv;41;152;152a
|
| 322 |
+
DD5S1.csv;42;150;150b
|
| 323 |
+
DD5S1.csv;43;149;149b
|
| 324 |
+
DD5S1.csv;44;148;148b
|
| 325 |
+
DD5S1.csv;45;147;147b
|
| 326 |
+
DD5S1.csv;46;146;146b
|
| 327 |
+
DD5S1.csv;47;155;155b
|
| 328 |
+
DD5S1.csv;48;154;154b
|
| 329 |
+
DD5S1.csv;49;153;153b
|
| 330 |
+
DD5S2.csv;0;162;162b
|
| 331 |
+
DD5S2.csv;1;161;161b
|
| 332 |
+
DD5S2.csv;2;159;159b
|
| 333 |
+
DD5S2.csv;3;158;158a
|
| 334 |
+
DD5S2.csv;4;157;157b
|
| 335 |
+
DD5S2.csv;5;156;156a
|
| 336 |
+
DD5S2.csv;6;164;164b
|
| 337 |
+
DD5S2.csv;7;172;172a
|
| 338 |
+
DD5S2.csv;8;171;171a
|
| 339 |
+
DD5S2.csv;9;170;170a
|
| 340 |
+
DD5S2.csv;10;169;169a
|
| 341 |
+
DD5S2.csv;11;168;168a
|
| 342 |
+
DD5S2.csv;12;167;167a
|
| 343 |
+
DD5S2.csv;13;165;165a
|
| 344 |
+
DD5S2.csv;14;183;183a
|
| 345 |
+
DD5S2.csv;15;182;182a
|
| 346 |
+
DD5S2.csv;16;181;181a
|
| 347 |
+
DD5S2.csv;17;180;180a
|
| 348 |
+
DD5S2.csv;18;179;179a
|
| 349 |
+
DD5S2.csv;19;177;177a
|
| 350 |
+
DD5S2.csv;20;176;176a
|
| 351 |
+
DD5S2.csv;21;171;171b
|
| 352 |
+
DD5S2.csv;22;170;170b
|
| 353 |
+
DD5S2.csv;23;168;168b
|
| 354 |
+
DD5S2.csv;24;167;167b
|
| 355 |
+
DD5S2.csv;25;166;166a
|
| 356 |
+
DD5S2.csv;26;165;165b
|
| 357 |
+
DD5S2.csv;27;174;174a
|
| 358 |
+
DD5S2.csv;28;173;173a
|
| 359 |
+
DD5S2.csv;29;172;172b
|
| 360 |
+
DD5S2.csv;30;180;180b
|
| 361 |
+
DD5S2.csv;31;179;179b
|
| 362 |
+
DD5S2.csv;32;178;178a
|
| 363 |
+
DD5S2.csv;33;176;176b
|
| 364 |
+
DD5S2.csv;34;175;175a
|
| 365 |
+
DD5S2.csv;35;183;183b
|
| 366 |
+
DD5S2.csv;36;181;181b
|
| 367 |
+
DD5S2.csv;37;168;168c
|
| 368 |
+
DD5S2.csv;38;166;166b
|
| 369 |
+
DD5S2.csv;39;174;174b
|
| 370 |
+
DD5S2.csv;40;171;171c
|
| 371 |
+
DD5S2.csv;41;170;170c
|
| 372 |
+
DD5S2.csv;42;169;169b
|
| 373 |
+
DD5S3.csv;0;178;178b
|
| 374 |
+
DD5S3.csv;1;177;177b
|
| 375 |
+
DD5S3.csv;2;176;176c
|
| 376 |
+
DD5S3.csv;3;175;175b
|
| 377 |
+
DD5S3.csv;4;183;183c
|
| 378 |
+
DD5S3.csv;5;182;182b
|
| 379 |
+
DD5S3.csv;6;180;180c
|
| 380 |
+
DD5S3.csv;7;187;187a
|
| 381 |
+
DD5S3.csv;8;185;185a
|
| 382 |
+
DD5S3.csv;9;184;184a
|
| 383 |
+
DD5S3.csv;10;187;187b
|
| 384 |
+
DD5S3.csv;11;185;185b
|
| 385 |
+
DD5S3.csv;12;130;130c
|
| 386 |
+
DD5S3.csv;13;129;129c
|
| 387 |
+
DD5S3.csv;14;136;136c
|
| 388 |
+
DD5S3.csv;15;134;134b
|
| 389 |
+
DD5S3.csv;16;133;133c
|
| 390 |
+
DD5S3.csv;17;132;132c
|
| 391 |
+
DD5S3.csv;18;144;114c
|
| 392 |
+
DD5S3.csv;19;141;141a
|
| 393 |
+
DD5S3.csv;20;140;140b
|
| 394 |
+
DD5S3.csv;21;149;149c
|
| 395 |
+
DD5S3.csv;22;148;148c
|
| 396 |
+
DD5S3.csv;23;154;154c
|
| 397 |
+
DD5S3.csv;24;153;153c
|
| 398 |
+
DD5S3.csv;25;152;152a
|
| 399 |
+
DD5S3.csv;26;157;157c
|
| 400 |
+
DD5S3.csv;27;155;155c
|
| 401 |
+
DD5S3.csv;28;164;164c
|
| 402 |
+
DD5S3.csv;29;162;162c
|
| 403 |
+
DD5S3.csv;30;161;161c
|
| 404 |
+
DD5S3.csv;31;159;159c
|
| 405 |
+
DD5S3.csv;32;185;185c
|
| 406 |
+
DD5S3.csv;33;184;184b
|
| 407 |
+
DD5S3.csv;34;187;187c
|
| 408 |
+
DD5S3.csv;35;186;186a
|
| 409 |
+
TMA.csv;0;c0;c0a
|
| 410 |
+
TMA.csv;1;c1;c1a
|
| 411 |
+
TMA.csv;2;c2;c2a
|
| 412 |
+
TMA.csv;3;c3;c3a
|
| 413 |
+
TMA.csv;4;c4;c4a
|
| 414 |
+
TMA.csv;5;c5;c5a
|
| 415 |
+
TMA.csv;6;c6;c6a
|
| 416 |
+
TMA.csv;7;c7;c7a
|
| 417 |
+
TMA.csv;8;c8;c8a
|
| 418 |
+
TMA.csv;9;c9;c9a
|
| 419 |
+
TMA.csv;10;c10;c10a
|
| 420 |
+
TMA.csv;11;c11;c11a
|
| 421 |
+
TMA.csv;12;c12;c12a
|
| 422 |
+
TMA.csv;13;c13;c13a
|
| 423 |
+
TMA.csv;14;c14;c14a
|
| 424 |
+
TMA.csv;15;c15;c15a
|
| 425 |
+
TMA.csv;16;c16;c16a
|
| 426 |
+
TMA.csv;17;c17;c17a
|
| 427 |
+
TMA.csv;18;c18;c18a
|
| 428 |
+
TMA.csv;19;c19;c19a
|
| 429 |
+
TMA.csv;20;c20;c20a
|
| 430 |
+
TMA.csv;21;c21;c21a
|
| 431 |
+
TMA.csv;22;c22;c22a
|
| 432 |
+
TMA.csv;23;c23;c23a
|
| 433 |
+
TMA.csv;24;c24;c24a
|
| 434 |
+
TMA.csv;25;c25;c25a
|
| 435 |
+
TMA.csv;26;c26;c26a
|
| 436 |
+
TMA.csv;27;c27;c27a
|
| 437 |
+
TMA.csv;28;c28;c28a
|
| 438 |
+
TMA.csv;29;c29;c29a
|
| 439 |
+
TMA.csv;30;c30;c30a
|
| 440 |
+
TMA.csv;31;c31;c31a
|
| 441 |
+
TMA.csv;32;c32;c32a
|
| 442 |
+
TMA.csv;33;c33;c33a
|
| 443 |
+
TMA.csv;34;c34;c34a
|
| 444 |
+
TMA.csv;35;c35;c35a
|
| 445 |
+
TMA.csv;36;c36;c36a
|
| 446 |
+
TMA.csv;37;c37;c37a
|
| 447 |
+
TMA.csv;38;c38;c38a
|
| 448 |
+
TMA.csv;39;c39;c39a
|
| 449 |
+
TMA.csv;40;c40;c40a
|
| 450 |
+
TMA.csv;41;c41;c41a
|
| 451 |
+
TMA.csv;42;c42;c42a
|
| 452 |
+
TMA.csv;43;c43;c43a
|
| 453 |
+
TMA.csv;44;c44;c44a
|
| 454 |
+
TMA.csv;45;c45;c45a
|
| 455 |
+
TMA.csv;46;c46;c46a
|
| 456 |
+
TMA.csv;47;c47;c47a
|
| 457 |
+
TMA.csv;48;c48;c48a
|
| 458 |
+
TMA.csv;49;c49;c49a
|
| 459 |
+
TMA.csv;50;c50;c50a
|
| 460 |
+
TMA.csv;51;c51;c51a
|
| 461 |
+
TMA.csv;52;c52;c52a
|
| 462 |
+
TMA.csv;53;c53;c53a
|
| 463 |
+
TMA.csv;54;c54;c54a
|
| 464 |
+
TMA.csv;55;c55;c55a
|
| 465 |
+
TMA.csv;56;c56;c56a
|
| 466 |
+
TMA.csv;57;c57;c57a
|
| 467 |
+
TMA.csv;58;c58;c58a
|
| 468 |
+
TMA.csv;59;c59;c59a
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/Slide_B_DD1s1.one_1.tif.csv
ADDED
|
@@ -0,0 +1,46 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Channel,Name,Cycle,ChannelIndex,ExposureTime,ExposureTimeUnit,Fluor,AcquisitionMode,IlluminationType,ContrastMethod,ExcitationWavelength,ExcitationWavelengthUnit,EmissionWavelength,EmissionWavelengthUnit,Color
|
| 2 |
+
0,DAPI0,0,0,30.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 3 |
+
1,AF488,0,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 4 |
+
2,AF555,0,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 5 |
+
3,AF647,0,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 6 |
+
4,AF750,0,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 7 |
+
5,DAPI1,1,0,20.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 8 |
+
6,ColVI,1,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 9 |
+
7,CD31,1,2,1200.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 10 |
+
8,CD4,1,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 11 |
+
9,Ecad,1,4,800.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 12 |
+
10,DAPI2,2,0,15.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 13 |
+
11,Desmin,2,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 14 |
+
12,B7H4,2,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 15 |
+
13,CD8,2,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 16 |
+
14,CD20,2,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 17 |
+
15,DAPI3,3,0,20.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 18 |
+
16,aSMA,3,1,50.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 19 |
+
17,CD68,3,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 20 |
+
18,PD1,3,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 21 |
+
19,CD45,3,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 22 |
+
20,DAPI4,4,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 23 |
+
21,Vimentin,4,1,150.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 24 |
+
22,AXL,4,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 25 |
+
23,PDL1,4,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 26 |
+
24,FOXP3,4,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 27 |
+
25,DAPI5,5,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 28 |
+
26,r5c2,5,1,20.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 29 |
+
27,CA9,5,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 30 |
+
28,CD163,5,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 31 |
+
29,Ki67,5,4,1000.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 32 |
+
30,DAPI6,6,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 33 |
+
31,CKs,6,1,200.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 34 |
+
32,Fibronectin,6,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 35 |
+
33,CD44,6,3,1200.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 36 |
+
34,HLA,6,4,500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 37 |
+
35,DAPI7,7,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 38 |
+
36,r7c2,7,1,20.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 39 |
+
37,PDGFR,7,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 40 |
+
38,MMP9,7,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 41 |
+
39,GATA3,7,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 42 |
+
40,DAPI8,8,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 43 |
+
41,r8c2,8,1,25.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 44 |
+
42,CD11c,8,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 45 |
+
43,Sting,8,3,1000.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 46 |
+
44,CD11b,8,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/Slide_B_DD1s1.one_2.tif.csv
ADDED
|
@@ -0,0 +1,46 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Channel,Name,Cycle,ChannelIndex,ExposureTime,ExposureTimeUnit,Fluor,AcquisitionMode,IlluminationType,ContrastMethod,ExcitationWavelength,ExcitationWavelengthUnit,EmissionWavelength,EmissionWavelengthUnit,Color
|
| 2 |
+
0,DAPI0,0,0,30.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 3 |
+
1,AF488,0,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 4 |
+
2,AF555,0,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 5 |
+
3,AF647,0,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 6 |
+
4,AF750,0,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 7 |
+
5,DAPI1,1,0,20.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 8 |
+
6,ColVI,1,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 9 |
+
7,CD31,1,2,1200.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 10 |
+
8,CD4,1,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 11 |
+
9,Ecad,1,4,800.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 12 |
+
10,DAPI2,2,0,15.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 13 |
+
11,Desmin,2,1,300.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 14 |
+
12,B7H4,2,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 15 |
+
13,CD8,2,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 16 |
+
14,CD20,2,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 17 |
+
15,DAPI3,3,0,20.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 18 |
+
16,aSMA,3,1,50.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 19 |
+
17,CD68,3,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 20 |
+
18,PD1,3,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 21 |
+
19,CD45,3,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 22 |
+
20,DAPI4,4,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 23 |
+
21,Vimentin,4,1,150.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 24 |
+
22,AXL,4,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 25 |
+
23,PDL1,4,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 26 |
+
24,FOXP3,4,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 27 |
+
25,DAPI5,5,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 28 |
+
26,r5c2,5,1,20.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 29 |
+
27,CA9,5,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 30 |
+
28,CD163,5,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 31 |
+
29,Ki67,5,4,1000.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 32 |
+
30,DAPI6,6,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 33 |
+
31,CKs,6,1,200.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 34 |
+
32,Fibronectin,6,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 35 |
+
33,CD44,6,3,1200.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 36 |
+
34,HLA,6,4,500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 37 |
+
35,DAPI7,7,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 38 |
+
36,r7c2,7,1,20.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 39 |
+
37,PDGFR,7,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 40 |
+
38,MMP9,7,3,1500.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 41 |
+
39,GATA3,7,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
| 42 |
+
40,DAPI8,8,0,10.000000,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.000000,nm,465.000000,nm,#FF0000FF
|
| 43 |
+
41,r8c2,8,1,25.000000,ms,FITC,WideField,Epifluorescence,Fluorescence,495.000000,nm,519.000000,nm,#FF00FF28
|
| 44 |
+
42,CD11c,8,2,1500.000000,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.000000,nm,568.000000,nm,#FFFFFF00
|
| 45 |
+
43,Sting,8,3,1000.000000,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.000000,nm,668.000000,nm,#FFFF0000
|
| 46 |
+
44,CD11b,8,4,1500.000000,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.000000,nm,779.000000,nm,#FF00FFFF
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/TMA_Clinical_Data_187-OC.csv
ADDED
|
@@ -0,0 +1,188 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
ID;%_tumor;%_necrosis;Age_Diagnosis;BMI;CA125;Race;Other_Cancers;BRCAStatus;NACT_vs_ACT;Diagnosis_to_Start_Chemo;Diagnosis_to_surgery;Stage;Histo_Type;Optimal_Debulking;Residual_Disease;Platinum_sensitive;Days_surgery_to_recurrence;Avastin;Disease_Stat;Days_from_surgery_to_last_contact_or_death;Recurrence;Vital_Status
|
| 2 |
+
1;80;5;72;35;;C;breast;0;ACT;;8;3C;HGSOC;0;1;1;506;1;DOD;1776;Yes;1
|
| 3 |
+
2;50;0;36;24;;C;breast;1;ACT;;0;3C;HGSOC;1;0;1;669;1;DOD;2223;Yes;1
|
| 4 |
+
3;80;5;55;45;944;C;0;;ACT;;0;3C;HGSOC;1;0;1;596;0;DOD;1355;Yes;1
|
| 5 |
+
4;80;5;59;26;50;C;0;0;ACT;;0;3A;HGtubal;1;0;;;;;1783;;
|
| 6 |
+
5;50;0;70;32;1426;C;0;;NACT;19;90;X;CCC;;0;1;377;1;DOD;977;Yes;1
|
| 7 |
+
6;80;5;44;24;788;C;0;;ACT;;0;3C;HGSOC;0;1;;471;0;;1263;Yes;
|
| 8 |
+
7;80;5;63;29;10965;C;0;;NACT;5;69;X;HGSOC;;0;0;516;0;DOD;814;Yes;1
|
| 9 |
+
8;90;5;54;24;2465;C;0;;ACT;;0;3C;HGStubal;0;1;0;144;1;DOD;693;Yes;1
|
| 10 |
+
9;90;5;57;20;3694;C;0;;ACT;;0;3C;HGSOC;0;1;0;428;0;DOD;475;Yes;1
|
| 11 |
+
10;90;5;50;20;316;C;0;1;ACT;;0;3C;serousfallopian;1;0;1;;0;;2162;No;
|
| 12 |
+
11;90;0;58;20;72;C;0;;NACT;35;133;X;Signetringcell;;0;0;203;0;DOD;263;Yes;1
|
| 13 |
+
12;90;0;73;30;1704;C;breast;;ACT;;0;3C;HGSOC;0;1;;;0;DOD;1448;No;1
|
| 14 |
+
13;60;30;62;31;219;C;0;;ACT;;0;3C;HGSOC;1;0;0;162;0;DOD;2286;Yes;1
|
| 15 |
+
14;80;0;52;27;1222;C;breastuterine;;ACT;;0;2B;ENDOOV ;1;0;1;;0;NED;2933;No;0
|
| 16 |
+
15;60;0;77;35;183;C;0;;NACT;10;57;X;HGSOC;1;0;1;460;0;NED;2585;Yes;0
|
| 17 |
+
16;80;0;36;23;333;C;0;0;ACT;;0;1A;ENDOOV ;1;0;;;0;NED;1442;No;0
|
| 18 |
+
17;80;0;66;19;503;C;0;;ACT;;21;3C;HGSOC;1;0;0;;0;DOD;383;;1
|
| 19 |
+
18;90;0;75;25;55;C;0;0;NACT;7;153;X;HGSOC;1;0;1;359;1;DOD;439;Yes;1
|
| 20 |
+
19;80;0;49;35;1100;C;0;;ACT;;0;3C;HGSOC;0;1;1;892;0;AWD;2543;Yes;0
|
| 21 |
+
20;90;0;49;26;25000;C;0;;ACT;;0;1A;CCC;1;0;1;;0;NED;2217;No;0
|
| 22 |
+
21;90;5;55;20;235;C;0;;ACT;;0;1C;ENDOOV ;1;0;1;;0;NED;2087;No;0
|
| 23 |
+
22;80;5;60;19;11;C;squamousskincarcinoma;0;ACT;;0;1A;mucinouscarcinoma;1;0;;;0;NED;1636;No;0
|
| 24 |
+
23;90;5;60;36;38;C;0;;ACT;;0;3B;HGSOC;1;0;1;960;0;DOD;2352;Yes;1
|
| 25 |
+
24;90;5;68;50;28;C;0;;ACT;;0;1C;CCC;1;0;1;;0;NED;1336;;0
|
| 26 |
+
25;80;0;66;25;168;C;0;;ACT;;0;1C;CCC;1;0;1;;0;NED;1092;No;0
|
| 27 |
+
26;80;0;69;29;1907;C;0;0;NACT;9;50;X;HGSOC;1;0;1;;0;DOD;1085;No;1
|
| 28 |
+
27;90;0;51;22;1692;C;0;;ACT;;0;3C;HGSOC;0;1;0;233;1;DOD;465;Yes;1
|
| 29 |
+
28;50;0;70;43;109;C;0;0;ACT;;0;1A;CCC;1;0;;;0;NED;2423;No;0
|
| 30 |
+
29;90;0;76;23;234;C;0;;ACT;;0;3A;CCC;1;0;1;;0;NED;2036;No;0
|
| 31 |
+
30;80;5;46;31;5632;C;0;;ACT;;0;3C;HGSOC;1;0;;789;0;NED;1846;Yes;0
|
| 32 |
+
31;70;0;62;26;11785;C;0;;NACT;11;105;3C;HGSOC;1;0;1;;1;DOD;328;No;1
|
| 33 |
+
32;90;5;75;41;4532;C;0;;NACT;10;80;X;HGSOC;1;0;1;1157;0;AWD;1864;Yes;0
|
| 34 |
+
33;70;0;65;32;871;C;0;;ACT;;0;3C;HGSOC;1;0;1;967;1;DOD;2299;Yes;1
|
| 35 |
+
34;90;0;59;31;458;C;0;;ACT;;0;3C;HGSOC;1;0;1;249;0;DOD;914;Yes;1
|
| 36 |
+
35;90;0;50;;;C;0;;ACT;;0;X;CCC;1;0;0;58;0;DOD;1003;Yes;1
|
| 37 |
+
36;90;0;46;30;1086;C;0;;ACT;;0;4B;HGSOC;0;1;0;230;0;DOD;916;Yes;1
|
| 38 |
+
37;90;0;67;;200;C;;;ACT;;0;1C;ENDOOV ;1;0;1;;0;;3390;No;
|
| 39 |
+
38;90;5;69;28;39;C;0;;ACT;;0;1A;uterineMMT;1;0;;;0;;475;No;
|
| 40 |
+
39;80;5;53;30;;C;0;;ACT;;0;3C;HGSOC;1;0;1;748;1;DOD;1411;Yes;1
|
| 41 |
+
40;70;5;64;;8393;C;;;NACT;31;101;X;HGSOC;1;0;1;;0;;131;No;
|
| 42 |
+
41;90;0;51;20;78;C;breast;1;ACT;;0;2C;HGSOC;1;0;1;716;1;DOD;1570;Yes;1
|
| 43 |
+
42;90;0;63;29;2760;C;0;;ACT;;0;3C;HGSOC;0;1;0;1321;1;DOD;2909;Yes;1
|
| 44 |
+
43;70;5;64;24;118;C;breast;;ACT;;0;3C;HGSOC;0;1;1;600;1;DOD;1719;Yes;1
|
| 45 |
+
44;90;0;71;27;;C;0;;ACT;;0;1C;HGSOC;1;0;;;;NED;4494;No;0
|
| 46 |
+
45;90;5;65;26;8420;latino;0;;ACT;;0;3C;HGSOC;0;1;0;289;1;DOD;548;Yes;1
|
| 47 |
+
46;90;0;43;34;3769;C;0;;ACT;;0;3C;HGSOC;0;1;0;190;1;DOD;1089;Yes;1
|
| 48 |
+
47;80;0;25;55;83;C;0;;ACT;;0;3C;HGSOC;0;1;;;;;25;No;
|
| 49 |
+
48;90;5;84;27;2927;C;0;;ACT;;0;3C;HGSOC;1;0;1;266;0;DOD;403;Yes;1
|
| 50 |
+
49;90;5;62;44;;C;0;;ACT;;0;3C;HGSOC;1;0;1;433;0;;468;Yes;
|
| 51 |
+
50;90;5;59;32;1040;C;0;;ACT;;0;3C;HGSOC;0;1;0;454;0;DOD;1292;Yes;1
|
| 52 |
+
51;90;0;55;33;1000;C;renalcell;;NACT;;90;X;HGSOC;1;0;1;2103;0;;2124;Yes;
|
| 53 |
+
52;90;0;;;;;;;ACT;;0;;mucinousadenocarcinomag2;;;;293;;DOD;487;Yes;1
|
| 54 |
+
53;80;0;76;21;980;C;0;;ACT;;0;3C;HGSOC;1;0;1;529;0;DOD;670;Yes;1
|
| 55 |
+
54;90;5;64;21;250;C;0;;ACT;;0;3C;HGSOC;1;0;1;297;1;DOD;709;Yes;1
|
| 56 |
+
55;90;0;70;21;1665;C;0;;ACT;;0;1C;CCC;1;0;1;;0;NED;5229;No;0
|
| 57 |
+
56;80;5;71;32;1124;C;0;;ACT;;0;3C;HGSOC;0;1;;;;;966;No;
|
| 58 |
+
57;90;5;48;32;3800;C;breast;;ACT;;0;3C;HGSOC;0;1;;;0;;1006;;
|
| 59 |
+
58;90;0;60;29;18;C;breast;0;ACT;;0;1C;CCC;1;0;1;;0;NED;5143;No;0
|
| 60 |
+
59;90;5;65;22;1927;C;0;;ACT;;0;3C;HGSOC;0;1;0;203;0;AWD;386;Yes;0
|
| 61 |
+
60;90;5;63;18;71;C;0;;ACT;;0;1C;mucinousadenocarcinomag2;1;0;1;;0;NED;4798;No;0
|
| 62 |
+
61;60;10;65;23;281;C;0;;NACT;9;9;X;metastaticadenocarcinoma;;;1;;0;;23;No;
|
| 63 |
+
62;90;0;62;24;675;C;0;0;ACT;;0;3C;HGSOC;0;1;1;1055;1;DOD;1842;Yes;1
|
| 64 |
+
63;80;0;49;32;443;C;0;;ACT;;0;3B;HGSOC;1;0;1;609;1;DOD;1899;Yes;1
|
| 65 |
+
64;90;0;50;29;64;C;0;;ACT;;0;2C;HGSOC;1;0;1;;0;NED;4887;No;0
|
| 66 |
+
65;90;0;79;23;136;C;breast;;ACT;;0;3C;HGSOC;0;1;;;;DOD;23;No;1
|
| 67 |
+
66;90;0;75;26;1703;C;0;0;ACT;;0;3C;HGSOC;1;0;0;295;1;AWD;610;Yes;0
|
| 68 |
+
67;90;0;44;24;529;C;breast;1;ACT;;0;2C;HGSOC;1;0;1;892;1;DOD;4347;Yes;1
|
| 69 |
+
68;90;0;80;22;7861;C;melanoma;;ACT;;0;3C;HGSOC;0;1;;;0;DOD;21;No;1
|
| 70 |
+
69;80;0;60;31;78;C;0;0;ACT;;0;3A;HGSOC;1;0;1;;0;NED;4691;No;0
|
| 71 |
+
70;90;0;62;23;139;C;0;;ACT;;0;3C;serousperitonealg1;1;0;0;238;0;DOD;1439;Yes;1
|
| 72 |
+
71;90;5;75;20;1698;C;0;;ACT;;0;3C;HGSOC;0;1;;;0;DOC;4509;No;1
|
| 73 |
+
72;80;5;;;;C;0;;ACT;;0;;;;;;365;1;;38468;Yes;
|
| 74 |
+
73;70;0;75;21;30;C;0;;ACT;;0;3C;HGSOC;0;1;;;0;DOD;37;No;1
|
| 75 |
+
74;90;0;68;23;1360;C;0;;ACT;;0;3C;HGSOC;1;0;1;335;1;DOD;1309;Yes;1
|
| 76 |
+
75;40;0;53;26;587;C;0;;ACT;;0;1A;mucinousadenocarcinomag1;1;0;;;0;NED;4229;No;0
|
| 77 |
+
76;90;5;74;29;4124;C;breaststomach;;ACT;;0;3C;HGSOC;0;1;;;0;DOD;3164;No;1
|
| 78 |
+
77;80;0;54;28;3757;MiddleEastern;0;;ACT;;0;3A;CCC;1;0;1;;0;NED;4557;No;0
|
| 79 |
+
78;80;5;51;30;176;C;0;0;ACT;;0;2B;ENDOOV ;1;0;1;;0;NED;4470;No;0
|
| 80 |
+
79;80;0;57;21;776;Japanese;0;2;ACT;;0;2C;HGSOC;1;0;0;195;1;NED;4521;Yes;0
|
| 81 |
+
80;70;5;69;24;532;C;0;;ACT;;0;3C;HGSOC;1;0;1;760;0;DOD;1111;Yes;1
|
| 82 |
+
81;90;0;64;28;180;C;0;;ACT;;0;3C;HGSOC;1;0;0;535;1;DOD;3945;Yes;1
|
| 83 |
+
82;80;0;64;42;12;C;0;;ACT;;0;3C;HGSOC;1;0;0;147;0;DOD;379;Yes;1
|
| 84 |
+
83;90;0;72;30;959;C;0;;ACT;;0;X;HGSOC;;1;1;468;0;;1064;Yes;
|
| 85 |
+
84;80;10;66;27;;C;braintumorofovarianorigin;;ACT;;0;4B;HGSOC;;;1;393;0;DOD;463;Yes;1
|
| 86 |
+
85;90;0;61;30;116;hispanic;0;;NACT;8;343;X;HGSOC;1;0;1;572;0;DOD;600;Yes;1
|
| 87 |
+
86;80;10;66;29;406;C;0;;ACT;;0;3C;HGSOC;0;1;0;251;1;DOD;540;Yes;1
|
| 88 |
+
87;80;0;86;27;2779;C;0;;ACT;;0;3C;HGSOC;1;0;0;293;0;AWD;826;Yes;0
|
| 89 |
+
88;90;5;49;41;371;C;breast;2;ACT;;0;3C;HGSOC;1;0;0;132;0;NED;4015;Yes;0
|
| 90 |
+
89;90;0;64;28;855;C;0;0;NACT;48;159;X;HGSOC;1;0;1;540;1;DOD;1820;Yes;1
|
| 91 |
+
90;90;5;61;29;547;C;0;;ACT;;0;3C;HGSOC;0;1;;600;0;DOD;1899;Yes;1
|
| 92 |
+
91;70;5;56;40;877;C;0;2;ACT;;0;3C;HGSOC;1;0;1;1140;0;AWD;1358;Yes;0
|
| 93 |
+
92;70;5;75;38;3083;C;0;0;ACT;;0;3C;HGSOC;1;0;1;615;0;DOD;705;Yes;1
|
| 94 |
+
93;90;5;89;21;2895;C;0;;ACT;;0;3B;HGSOC;0;1;;;0;DOD;297;;1
|
| 95 |
+
94;90;5;51;28;5242;C;0;0;ACT;;0;3C;HGSOC;1;0;1;785;1;DOD;1758;Yes;1
|
| 96 |
+
95;80;5;80;24;2799;C;0;;NACT;7;97;X;HGSOC;1;0;;;0;;96;No;
|
| 97 |
+
96;90;5;68;20;2683;C;0;;ACT;;0;2C;HGSOC;1;0;1;;0;NED;4082;No;0
|
| 98 |
+
97;70;5;57;40;19842;C;0;;ACT;;0;2C;mixedepithelialmucosal;1;0;0;174;0;DOD;206;Yes;1
|
| 99 |
+
98;70;0;79;26;2461;C;0;;NACT;12;135;X;HGSOC;1;0;1;343;0;DOD;923;Yes;1
|
| 100 |
+
99;90;0;55;21;1400;C;breast;1;ACT;;0;2C;HGSOC;1;0;;141;0;;336;Yes;
|
| 101 |
+
100;90;0;62;32;2059;C;0;;ACT;;0;1C;HGSOC;1;0;1;;0;NED;2938;No;0
|
| 102 |
+
101;90;5;75;33;242;C;0;;ACT;;0;3C;HGSOC;0;1;1;460;0;DOD;836;Yes;1
|
| 103 |
+
102;90;0;55;25;1395;C;0;;ACT;;0;3C;HGSOC;1;0;1;797;0;DOD;2554;Yes;1
|
| 104 |
+
103;90;10;46;23;116;C;uterine;PMS2;ACT;;0;1C;HGSOC;1;0;1;;0;NED;3578;No;0
|
| 105 |
+
104;90;0;60;29;63;C;0;;NACT;6;103;X;HGSOC;1;0;1;335;0;DOD;351;Yes;1
|
| 106 |
+
105;80;10;75;26;873;C;0;0;ACT;;0;3B;HGSOC;1;0;1;468;0;DOD;622;Yes;1
|
| 107 |
+
106;80;5;69;29;242;C;skintongue;;ACT;;0;3C;HGSOC;1;0;1;648;1;DOD;1320;Yes;1
|
| 108 |
+
107;90;5;63;26;1928;C;0;MSH6;ACT;;0;3C;HGSOC;1;0;1;;0;NED;3725;No;0
|
| 109 |
+
108;80;5;57;40;3687;C;0;;ACT;;0;4;HGSOC;0;1;0;353;1;DOD;791;Yes;1
|
| 110 |
+
109;80;5;75;35;223;C;0;;ACT;;0;1A;HGSOC;0;1;1;;0;NED;3863;No;0
|
| 111 |
+
110;90;0;56;29;5902;C;0;;ACT;;0;3C;HGSOC;0;1;1;438;1;DOD;1527;Yes;1
|
| 112 |
+
111;90;5;50;32;1960;C;0;0;NACT;15;79;X;HGSOC;1;0;1;533;1;DOD;1888;Yes;1
|
| 113 |
+
112;70;5;64;34;301;C;0;0;ACT;;0;1A;HGSOC;1;0;;896;1;AWD;3656;Yes;0
|
| 114 |
+
113;90;0;62;21;115;C;0;;ACT;;0;3C;mucinousadenocarcinomag2;1;0;0;148;;;161;Yes;
|
| 115 |
+
114;90;0;54;24;8571;C;0;;ACT;;0;3C;HGSOC;1;0;0;175;1;DOD;472;Yes;1
|
| 116 |
+
115;80;5;58;22;942;C;;;ACT;;0;4;HGSOC;1;0;1;;0;NED;3219;No;0
|
| 117 |
+
116;80;0;76;34;7453;C;0;;NACT;8;92;X;HGSOC;1;0;1;;0;;478;No;
|
| 118 |
+
117;90;5;54;27;6910;C;0;;ACT;;0;3C;HGSOC;1;0;1;;0;NED;3672;No;0
|
| 119 |
+
118;90;0;64;45;4;C;0;;ACT;;0;1C;HGSOC;1;0;1;609;0;DOD;688;Yes;1
|
| 120 |
+
119;90;0;57;17;16234;C;0;;NACT;12;95;X;HGSOC;0;1;1;655;0;DOD;374;Yes;1
|
| 121 |
+
120;90;0;77;28;18;C;cervical;;ACT;;0;3C;HGSOC;0;1;1;991;1;DOD;1616;Yes;1
|
| 122 |
+
121;90;5;64;38;2207;C;melanoma;;NACT;6;136;X;HGSOC;1;0;;;1;DOD;2445;;1
|
| 123 |
+
122;90;5;81;25;1884;C;0;;ACT;;0;3C;HGSOC;1;0;;517;0;;877;Yes;
|
| 124 |
+
123;70;0;83;26;471;C;breast;;ACT;;0;3C;HGSOC;1;0;1;538;0;DOD;722;Yes;1
|
| 125 |
+
124;90;0;47;22;1993;C;0;2;ACT;;0;4A;HGSOC;0;1;1;570;1;DOD;1042;Yes;1
|
| 126 |
+
125;90;5;70;36;4078;C;breast;0;ACT;;0;3C;HGSOC;1;0;;;;;1077;;
|
| 127 |
+
126;90;0;72;27;2018;C;0;;ACT;;0;3C;HGSOC;1;0;0;341;;DOD;711;Yes;1
|
| 128 |
+
127;70;0;62;22;547;C;melanoma;;ACT;;0;3C;HGSOC;1;0;1;345;0;DOD;485;Yes;1
|
| 129 |
+
128;80;5;58;34;776;C;0;;ACT;;0;3Bor4B;uterinepapillaryserous;1;0;0;282;0;DOD;582;Yes;1
|
| 130 |
+
129;90;0;48;35;944;C;0;1;ACT;;0;4A;HGSOC;1;0;1;543;0;DOD;1763;Yes;1
|
| 131 |
+
130;90;0;76;26;251;C;0;;ACT;;0;2C;ovarianmucinousgrade2intestinal;1;0;1;;0;NED;3292;No;0
|
| 132 |
+
131;70;5;60;18;109;C;0;;ACT;;0;3C;HGSOC;1;0;1;630;0;AWD;1803;Yes;0
|
| 133 |
+
132;90;5;67;37;428;C;0;;ACT;;0;3C;HGSOC;0;1;1;;0;DOC;731;No;1
|
| 134 |
+
133;90;0;75;24;27;C;basalcellcarcinomauterineorcervical;;ACT;;0;3C;HGSOC;1;0;1;2351;0;AWD;2700;Yes;0
|
| 135 |
+
134;80;5;48;17;718;C;0;;ACT;;0;3C;HGSOC;1;0;;;;DOD;977;;1
|
| 136 |
+
135;80;5;44;35;976;C;0;1;ACT;;0;3C;HGSOC;1;0;1;795;0;DOC;2414;Yes;1
|
| 137 |
+
136;90;0;43;38;1000;C;0;;ACT;;0;1C;ENDOOV ;1;0;1;;0;NED;3080;No;0
|
| 138 |
+
137;90;0;54;19;2964;C;neurofibromatosisremotebreast;1;NACT;2;68;X;HGSOC;1;0;1;575;1;DOD;1330;Yes;1
|
| 139 |
+
138;70;5;70;26;1210;Asian;0;;NACT;14;88;X;HGSOC;1;0;0;268;0;DOD;471;Yes;1
|
| 140 |
+
139;80;5;58;33;2162;C;0;;ACT;;0;3C;HGSOC;0;1;0;242;1;DOD;657;Yes;1
|
| 141 |
+
140;70;5;71;21;1046;C;skincancer;;NACT;4;80;X;HGSOC;1;0;;;0;;206;;
|
| 142 |
+
141;70;5;31;31;575;C;0;1;NACT;12;64;X;HGSOC;1;0;1;613;1;DOD;1628;Yes;1
|
| 143 |
+
142;90;0;75;22;95;C;0;0;ACT;;0;1B;HGSOC;1;0;1;1273;0;DOD;1983;Yes;1
|
| 144 |
+
143;80;10;64;28;22;C;0;;ACT;;0;1A;HGSOC;1;0;;;0;NED;3096;No;0
|
| 145 |
+
144;90;0;44;35;377;hispanic;0;1;ACT;;0;4A;HGSOC;1;0;1;455;0;DOD;949;Yes;1
|
| 146 |
+
145;70;0;58;42;222;C;0;0;ACT;;0;3C;highgradetubal;0;1;0;510;1;DOD;849;Yes;1
|
| 147 |
+
146;70;0;71;21;10824;C;0;;NACT;37;136;X;HGSOC;1;0;0;263;0;DOD;352;Yes;1
|
| 148 |
+
147;90;0;51;31;16;C;0;;ACT;;0;2A;HGSOC;1;0;;;;NED;2854;;0
|
| 149 |
+
148;90;5;66;37;1586;C;0;1;ACT;;0;2C;HGSOC;1;0;1;617;0;AWD;1925;Yes;0
|
| 150 |
+
149;70;0;52;26;7363;C;papillarythyroid;;NACT;2;50;X;HGSOC;1;0;1;;0;NED;2706;No;0
|
| 151 |
+
150;90;0;51;26;;C;0;0;ACT;;0;3A;HGSOC;1;0;1;630;0;DOD;2551;Yes;1
|
| 152 |
+
151;80;0;64;;1929;C;0;0;ACT;;0;3C;HGSOC;0;1;1;884;0;DOD;1420;Yes;1
|
| 153 |
+
152;70;5;67;28;15;C;0;0;ACT;;0;2B;ENDOOV ;1;0;1;;0;NED;2867;No;0
|
| 154 |
+
153;80;5;64;23;968;C;othermalignantneoplasmsitenotspecified;2;NACT;19;172;X;HGSOC;1;0;1;361;1;DOD;2336;Yes;1
|
| 155 |
+
154;80;5;54;34;324;C;0;0;ACT;;0;3A;HGSOC;1;0;1;465;1;DOD;2645;Yes;1
|
| 156 |
+
155;90;0;50;32;1218;C;0;;ACT;;0;1C;ENDOOV ;;;;;0;DOC;2743;No;1
|
| 157 |
+
156;90;5;43;38;;C;0;0;ACT;;0;;HGSOC;;;1;6940;0;DOD;9303;Yes;1
|
| 158 |
+
157;80;5;56;36;1971;C;0;2;NACT;8;94;X;ENDOOV ;1;0;1;;0;DOD;1667;No;1
|
| 159 |
+
158;90;5;46;31;853;C;0;0;ACT;;0;3C;HGSOC;1;0;1;1892;0;AWD;2778;Yes;0
|
| 160 |
+
159;90;0;66;23;14365;C;0;;ACT;;0;3C;HGSOC;1;0;1;1209;0;DOD;2073;Yes;1
|
| 161 |
+
160;90;5;59;27;89;C;breast;0;NACT;22;278;X;HGSOC;0;1;1;160;1;DOD;328;Yes;1
|
| 162 |
+
161;90;0;59;28;12;C;0;MSH6;ACT;;0;1C;ENDOOV ;1;0;1;;0;NED;2642;No;0
|
| 163 |
+
162;80;0;75;20;7482;C;0;0;NACT;49;143;X;serousofMullerianorigin;1;0;1;536;0;DOD;2353;Yes;1
|
| 164 |
+
163;80;10;24;21;;hispanic;thymiccarcinoma;MSH2;ACT;;0;1C;mucinouscarcinoma;1;0;;;0;DOD;585;No;1
|
| 165 |
+
164;80;5;68;21;249;C;0;0;ACT;;0;1B;HGSOC;1;0;1;;0;NED;2699;No;0
|
| 166 |
+
165;90;5;51;37;;C;0;;ACT;;0;1A;HGSOC;1;0;1;;0;NED;2674;No;0
|
| 167 |
+
166;90;5;63;23;;C;0;FBXW7;ACT;;0;3C;HGSOC;1;0;0;464;1;DOD;1184;Yes;1
|
| 168 |
+
167;90;5;64;21;624;C;breast;0;ACT;;0;3C;HGSOC;0;1;1;;0;DOC;2593;No;1
|
| 169 |
+
168;90;5;80;30;;C;0;0;ACT;;0;3B;HGSOC;1;0;1;;0;NED;2631;No;0
|
| 170 |
+
169;80;5;75;35;245;C;0;;NACT;10;78;X;HGSOC;1;0;1;1003;0;DOD;1442;Yes;1
|
| 171 |
+
170;70;5;55;21;3071;C;0;;ACT;;0;3C;HGSOC;1;0;1;;0;NED;2345;No;0
|
| 172 |
+
171;80;5;60;34;500;C;0;;NACT;14;145;X;HGSOC;1;0;1;302;0;DOD;456;Yes;1
|
| 173 |
+
172;90;0;56;25;371;C;concomitantmetastaticpancreaticcanceridentifiedatsurgery;;ACT;;0;1A;HGSOC;0;1;;;0;DOD;619;No;1
|
| 174 |
+
173;90;5;72;33;;C;0;;ACT;;0;3C;HGSOC;1;0;1;1038;0;DOD;2404;Yes;1
|
| 175 |
+
174;50;0;54;36;8529;C;0;0;NACT;15;127;X;HGSPC;1;0;1;387;0;DOD;660;Yes;1
|
| 176 |
+
175;80;5;59;44;378;C;0;;ACT;;0;3C;HGSOC;1;0;1;947;0;DOD;1930;Yes;1
|
| 177 |
+
176;70;0;56;29;1812;C;0;0;ACT;;0;3C;HGSOC;1;0;1;2101;0;DOD;2223;Yes;1
|
| 178 |
+
177;90;0;60;32;919;C;colon;0;NACT;10;70;X;HGSOC;1;0;1;450;1;DOD;1743;Yes;1
|
| 179 |
+
178;80;0;67;27;107;C;basalcellcarcinoma;0;NACT;50;603;3C;HGSOC;0;1;1;416;0;DOD;977;Yes;1
|
| 180 |
+
179;80;0;83;25;;C;0;;NACT;15;62;X;HGSOC;1;0;1;;;DOD;1070;;1
|
| 181 |
+
180;70;10;64;25;88;C;0;0;ACT;;0;1A;1Aendometrialadenocarcinomag31Aclearcellofovary;1;0;1;;0;NED;2274;No;0
|
| 182 |
+
181;90;5;20;27;229;C;0;0;ACT;;0;3C;HGSOC;1;0;1;1170;0;AWD;4810;Yes;0
|
| 183 |
+
182;80;0;81;28;990;C;0;;NACT;19;102;X;HGSOC;1;0;0;381;0;DOD;1552;Yes;1
|
| 184 |
+
183;70;5;72;22;597;C;breast;0;NACT;8;88;X;HGSOC;1;0;1;;;DOD;1036;;1
|
| 185 |
+
184;90;0;72;26;3788;C;breast;;NACT;22;91;X;HGSOC;1;0;1;282;0;;468;Yes;
|
| 186 |
+
185;90;0;70;29;1330;C;0;;ACT;;0;3C;HGSOC;1;0;;;;DOD;1020;;1
|
| 187 |
+
186;80;0;84;17;564;C;squamouscellcarcinoma;;ACT;;0;2B;HGSOC;1;0;;;;DOC;1869;No;1
|
| 188 |
+
187;80;0;27;22;922;C;0;0;NACT;11;91;X;serousprimarilylowgradewfociofhighgrade;1;1;1;540;0;DOD;1279;Yes;1
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/cellsubtype_color_data.csv
ADDED
|
@@ -0,0 +1,13 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,cell_subtype
|
| 2 |
+
"(0.6509803921568628, 0.807843137254902, 0.8901960784313725)",#a6cee3,DC
|
| 3 |
+
"(0.12156862745098039, 0.47058823529411764, 0.7058823529411765)",#1f78b4,B
|
| 4 |
+
"(0.6980392156862745, 0.8745098039215686, 0.5411764705882353)",#b2df8a,TCD4
|
| 5 |
+
"(0.2, 0.6274509803921569, 0.17254901960784313)",#33a02c,TCD8
|
| 6 |
+
"(0.984313725490196, 0.6039215686274509, 0.6)",#fb9a99,M1
|
| 7 |
+
"(0.8901960784313725, 0.10196078431372549, 0.10980392156862745)",#e31a1c,M2
|
| 8 |
+
"(0.9921568627450981, 0.7490196078431373, 0.43529411764705883)",#fdbf6f,Treg
|
| 9 |
+
"(1.0, 0.4980392156862745, 0.0)",#ff7f00,IMMUNE_OTHER
|
| 10 |
+
"(0.792156862745098, 0.6980392156862745, 0.8392156862745098)",#cab2d6,CANCER
|
| 11 |
+
"(0.41568627450980394, 0.23921568627450981, 0.6039215686274509)",#6a3d9a,αSMA_myCAF
|
| 12 |
+
"(1.0, 1.0, 0.6)",#ffff99,STROMA_OTHER
|
| 13 |
+
"(0.6941176470588235, 0.34901960784313724, 0.1568627450980392)",#b15928,ENDOTHELIAL
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/celltype_color_data.csv
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,cell_type
|
| 2 |
+
"(0.1333, 0.5451, 0.1333)",#228b22,CANCER
|
| 3 |
+
"(0.4, 0.4, 0.4)",#666666,STROMA
|
| 4 |
+
"(1.0, 1.0, 0.0)",#ffff00,IMMUNE
|
| 5 |
+
"(0.502, 0.0, 0.502)",#800080,ENDOTHELIAL
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/channel_color_data.csv
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,Channel
|
| 2 |
+
"(0.00784313725490196, 0.24313725490196078, 1.0)",#023eff,c2
|
| 3 |
+
"(1.0, 0.48627450980392156, 0.0)",#ff7c00,c3
|
| 4 |
+
"(0.10196078431372549, 0.788235294117647, 0.2196078431372549)",#1ac938,c4
|
| 5 |
+
"(0.9098039215686274, 0.0, 0.043137254901960784)",#e8000b,c5
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/combined_metadata.csv
ADDED
|
@@ -0,0 +1,91 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Channel,Name,Cycle,ChannelIndex,ExposureTime,ExposureTimeUnit,Fluor,AcquisitionMode,IlluminationType,ContrastMethod,ExcitationWavelength,ExcitationWavelengthUnit,EmissionWavelength,EmissionWavelengthUnit,Color
|
| 2 |
+
0,DAPI0,0,0,30.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 3 |
+
1,AF488,0,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 4 |
+
2,AF555,0,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 5 |
+
3,AF647,0,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 6 |
+
4,AF750,0,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 7 |
+
5,DAPI1,1,0,20.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 8 |
+
6,ColVI,1,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 9 |
+
7,CD31,1,2,1200.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 10 |
+
8,CD4,1,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 11 |
+
9,Ecad,1,4,800.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 12 |
+
10,DAPI2,2,0,15.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 13 |
+
11,Desmin,2,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 14 |
+
12,B7H4,2,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 15 |
+
13,CD8,2,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 16 |
+
14,CD20,2,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 17 |
+
15,DAPI3,3,0,20.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 18 |
+
16,aSMA,3,1,50.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 19 |
+
17,CD68,3,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 20 |
+
18,PD1,3,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 21 |
+
19,CD45,3,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 22 |
+
20,DAPI4,4,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 23 |
+
21,Vimentin,4,1,150.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 24 |
+
22,AXL,4,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 25 |
+
23,PDL1,4,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 26 |
+
24,FOXP3,4,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 27 |
+
25,DAPI5,5,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 28 |
+
26,r5c2,5,1,20.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 29 |
+
27,CA9,5,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 30 |
+
28,CD163,5,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 31 |
+
29,Ki67,5,4,1000.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 32 |
+
30,DAPI6,6,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 33 |
+
31,CKs,6,1,200.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 34 |
+
32,Fibronectin,6,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 35 |
+
33,CD44,6,3,1200.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 36 |
+
34,HLA,6,4,500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 37 |
+
35,DAPI7,7,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 38 |
+
36,r7c2,7,1,20.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 39 |
+
37,PDGFR,7,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 40 |
+
38,MMP9,7,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 41 |
+
39,GATA3,7,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 42 |
+
40,DAPI8,8,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 43 |
+
41,r8c2,8,1,25.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 44 |
+
42,CD11c,8,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 45 |
+
43,Sting,8,3,1000.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 46 |
+
44,CD11b,8,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 47 |
+
0,DAPI0,0,0,30.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 48 |
+
1,AF488,0,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 49 |
+
2,AF555,0,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 50 |
+
3,AF647,0,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 51 |
+
4,AF750,0,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 52 |
+
5,DAPI1,1,0,20.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 53 |
+
6,ColVI,1,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 54 |
+
7,CD31,1,2,1200.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 55 |
+
8,CD4,1,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 56 |
+
9,Ecad,1,4,800.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 57 |
+
10,DAPI2,2,0,15.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 58 |
+
11,Desmin,2,1,300.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 59 |
+
12,B7H4,2,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 60 |
+
13,CD8,2,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 61 |
+
14,CD20,2,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 62 |
+
15,DAPI3,3,0,20.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 63 |
+
16,aSMA,3,1,50.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 64 |
+
17,CD68,3,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 65 |
+
18,PD1,3,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 66 |
+
19,CD45,3,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 67 |
+
20,DAPI4,4,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 68 |
+
21,Vimentin,4,1,150.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 69 |
+
22,AXL,4,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 70 |
+
23,PDL1,4,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 71 |
+
24,FOXP3,4,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 72 |
+
25,DAPI5,5,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 73 |
+
26,r5c2,5,1,20.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 74 |
+
27,CA9,5,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 75 |
+
28,CD163,5,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 76 |
+
29,Ki67,5,4,1000.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 77 |
+
30,DAPI6,6,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 78 |
+
31,CKs,6,1,200.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 79 |
+
32,Fibronectin,6,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 80 |
+
33,CD44,6,3,1200.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 81 |
+
34,HLA,6,4,500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 82 |
+
35,DAPI7,7,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 83 |
+
36,r7c2,7,1,20.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 84 |
+
37,PDGFR,7,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 85 |
+
38,MMP9,7,3,1500.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 86 |
+
39,GATA3,7,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
| 87 |
+
40,DAPI8,8,0,10.0,ms,DAPI,WideField,Epifluorescence,Fluorescence,353.0,nm,465.0,nm,#FF0000FF
|
| 88 |
+
41,r8c2,8,1,25.0,ms,FITC,WideField,Epifluorescence,Fluorescence,495.0,nm,519.0,nm,#FF00FF28
|
| 89 |
+
42,CD11c,8,2,1500.0,ms,Alexa Fluor 555,WideField,Epifluorescence,Fluorescence,553.0,nm,568.0,nm,#FFFFFF00
|
| 90 |
+
43,Sting,8,3,1000.0,ms,Alexa Fluor 647,WideField,Epifluorescence,Fluorescence,653.0,nm,668.0,nm,#FFFF0000
|
| 91 |
+
44,CD11b,8,4,1500.0,ms,Alexa Fluor 750,WideField,Epifluorescence,Fluorescence,752.0,nm,779.0,nm,#FF00FFFF
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/full_to_short_column_names.csv
ADDED
|
@@ -0,0 +1,109 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
full_name,short_name
|
| 2 |
+
AF488_Cell_Intensity_Average,AF488_Cell
|
| 3 |
+
AF488_Cytoplasm_Intensity_Average,AF488_Cytoplasm
|
| 4 |
+
AF488_Nucleus_Intensity_Average,AF488_Nucleus
|
| 5 |
+
AF555_Cell_Intensity_Average,AF555_Cell
|
| 6 |
+
AF555_Cytoplasm_Intensity_Average,AF555_Cytoplasm
|
| 7 |
+
AF555_Nucleus_Intensity_Average,AF555_Nucleus
|
| 8 |
+
AF647_Cell_Intensity_Average,AF647_Cell
|
| 9 |
+
AF647_Cytoplasm_Intensity_Average,AF647_Cytoplasm
|
| 10 |
+
AF647_Nucleus_Intensity_Average,AF647_Nucleus
|
| 11 |
+
AF750_Cell_Intensity_Average,AF750_Cell
|
| 12 |
+
AF750_Cytoplasm_Intensity_Average,AF750_Cytoplasm
|
| 13 |
+
AF750_Nucleus_Intensity_Average,AF750_Nucleus
|
| 14 |
+
aSMA_Cell_Intensity_Average,aSMA_Cell
|
| 15 |
+
aSMA_Cytoplasm_Intensity_Average,aSMA_Cytoplasm
|
| 16 |
+
aSMA_Nucleus_Intensity_Average,aSMA_Nucleus
|
| 17 |
+
AXL_Cell_Intensity_Average,AXL_Cell
|
| 18 |
+
AXL_Cytoplasm_Intensity_Average,AXL_Cytoplasm
|
| 19 |
+
AXL_Nucleus_Intensity_Average,AXL_Nucleus
|
| 20 |
+
B7H4_Cell_Intensity_Average,B7H4_Cell
|
| 21 |
+
B7H4_Cytoplasm_Intensity_Average,B7H4_Cytoplasm
|
| 22 |
+
B7H4_Nucleus_Intensity_Average,B7H4_Nucleus
|
| 23 |
+
CA9_Cell_Intensity_Average,CA9_Cell
|
| 24 |
+
CA9_Cytoplasm_Intensity_Average,CA9_Cytoplasm
|
| 25 |
+
CA9_Nucleus_Intensity_Average,CA9_Nucleus
|
| 26 |
+
CD4_Cell_Intensity_Average,CD4_Cell
|
| 27 |
+
CD4_Cytoplasm_Intensity_Average,CD4_Cytoplasm
|
| 28 |
+
CD4_Nucleus_Intensity_Average,CD4_Nucleus
|
| 29 |
+
CD8_Cell_Intensity_Average,CD8_Cell
|
| 30 |
+
CD8_Cytoplasm_Intensity_Average,CD8_Cytoplasm
|
| 31 |
+
CD8_Nucleus_Intensity_Average,CD8_Nucleus
|
| 32 |
+
CD11b_Cell_Intensity_Average,CD11b_Cell
|
| 33 |
+
CD11b_Cytoplasm_Intensity_Average,CD11b_Cytoplasm
|
| 34 |
+
CD11b_Nucleus_Intensity_Average,CD11b_Nucleus
|
| 35 |
+
CD11c_Cell_Intensity_Average,CD11c_Cell
|
| 36 |
+
CD11c_Cytoplasm_Intensity_Average,CD11c_Cytoplasm
|
| 37 |
+
CD11c_Nucleus_Intensity_Average,CD11c_Nucleus
|
| 38 |
+
CD20_Cell_Intensity_Average,CD20_Cell
|
| 39 |
+
CD20_Cytoplasm_Intensity_Average,CD20_Cytoplasm
|
| 40 |
+
CD20_Nucleus_Intensity_Average,CD20_Nucleus
|
| 41 |
+
CD31_Cell_Intensity_Average,CD31_Cell
|
| 42 |
+
CD31_Cytoplasm_Intensity_Average,CD31_Cytoplasm
|
| 43 |
+
CD31_Nucleus_Intensity_Average,CD31_Nucleus
|
| 44 |
+
CD44_Cell_Intensity_Average,CD44_Cell
|
| 45 |
+
CD44_Cytoplasm_Intensity_Average,CD44_Cytoplasm
|
| 46 |
+
CD44_Nucleus_Intensity_Average,CD44_Nucleus
|
| 47 |
+
CD45_Cell_Intensity_Average,CD45_Cell
|
| 48 |
+
CD45_Cytoplasm_Intensity_Average,CD45_Cytoplasm
|
| 49 |
+
CD45_Nucleus_Intensity_Average,CD45_Nucleus
|
| 50 |
+
CD68_Cell_Intensity_Average,CD68_Cell
|
| 51 |
+
CD68_Cytoplasm_Intensity_Average,CD68_Cytoplasm
|
| 52 |
+
CD68_Nucleus_Intensity_Average,CD68_Nucleus
|
| 53 |
+
CD163_Cell_Intensity_Average,CD163_Cell
|
| 54 |
+
CD163_Cytoplasm_Intensity_Average,CD163_Cytoplasm
|
| 55 |
+
CD163_Nucleus_Intensity_Average,CD163_Nucleus
|
| 56 |
+
CKs_Cell_Intensity_Average,CKs_Cell
|
| 57 |
+
CKs_Cytoplasm_Intensity_Average,CKs_Cytoplasm
|
| 58 |
+
CKs_Nucleus_Intensity_Average,CKs_Nucleus
|
| 59 |
+
ColVI_Cell_Intensity_Average,ColVI_Cell
|
| 60 |
+
ColVI_Cytoplasm_Intensity_Average,ColVI_Cytoplasm
|
| 61 |
+
ColVI_Nucleus_Intensity_Average,ColVI_Nucleus
|
| 62 |
+
Desmin_Cell_Intensity_Average,Desmin_Cell
|
| 63 |
+
Desmin_Cytoplasm_Intensity_Average,Desmin_Cytoplasm
|
| 64 |
+
Desmin_Nucleus_Intensity_Average,Desmin_Nucleus
|
| 65 |
+
Ecad_Cell_Intensity_Average,Ecad_Cell
|
| 66 |
+
Ecad_Cytoplasm_Intensity_Average,Ecad_Cytoplasm
|
| 67 |
+
Ecad_Nucleus_Intensity_Average,Ecad_Nucleus
|
| 68 |
+
Fibronectin_Cell_Intensity_Average,Fibronectin_Cell
|
| 69 |
+
Fibronectin_Cytoplasm_Intensity_Average,Fibronectin_Cytoplasm
|
| 70 |
+
Fibronectin_Nucleus_Intensity_Average,Fibronectin_Nucleus
|
| 71 |
+
FOXP3_Cell_Intensity_Average,FOXP3_Cell
|
| 72 |
+
FOXP3_Cytoplasm_Intensity_Average,FOXP3_Cytoplasm
|
| 73 |
+
FOXP3_Nucleus_Intensity_Average,FOXP3_Nucleus
|
| 74 |
+
GATA3_Cell_Intensity_Average,GATA3_Cell
|
| 75 |
+
GATA3_Cytoplasm_Intensity_Average,GATA3_Cytoplasm
|
| 76 |
+
GATA3_Nucleus_Intensity_Average,GATA3_Nucleus
|
| 77 |
+
HLA_Cell_Intensity_Average,HLA_Cell
|
| 78 |
+
HLA_Cytoplasm_Intensity_Average,HLA_Cytoplasm
|
| 79 |
+
HLA_Nucleus_Intensity_Average,HLA_Nucleus
|
| 80 |
+
Ki67_Cell_Intensity_Average,Ki67_Cell
|
| 81 |
+
Ki67_Cytoplasm_Intensity_Average,Ki67_Cytoplasm
|
| 82 |
+
Ki67_Nucleus_Intensity_Average,Ki67_Nucleus
|
| 83 |
+
MMP9_Cell_Intensity_Average,MMP9_Cell
|
| 84 |
+
MMP9_Cytoplasm_Intensity_Average,MMP9_Cytoplasm
|
| 85 |
+
MMP9_Nucleus_Intensity_Average,MMP9_Nucleus
|
| 86 |
+
PD1_Cell_Intensity_Average,PD1_Cell
|
| 87 |
+
PD1_Cytoplasm_Intensity_Average,PD1_Cytoplasm
|
| 88 |
+
PD1_Nucleus_Intensity_Average,PD1_Nucleus
|
| 89 |
+
PDGFR_Cell_Intensity_Average,PDGFR_Cell
|
| 90 |
+
PDGFR_Cytoplasm_Intensity_Average,PDGFR_Cytoplasm
|
| 91 |
+
PDGFR_Nucleus_Intensity_Average,PDGFR_Nucleus
|
| 92 |
+
PDL1_Cell_Intensity_Average,PDL1_Cell
|
| 93 |
+
PDL1_Cytoplasm_Intensity_Average,PDL1_Cytoplasm
|
| 94 |
+
PDL1_Nucleus_Intensity_Average,PDL1_Nucleus
|
| 95 |
+
r5c2_Cell_Intensity_Average,r5c2_Cell
|
| 96 |
+
r5c2_Cytoplasm_Intensity_Average,r5c2_Cytoplasm
|
| 97 |
+
r5c2_Nucleus_Intensity_Average,r5c2_Nucleus
|
| 98 |
+
r7c2_Cell_Intensity_Average,r7c2_Cell
|
| 99 |
+
r7c2_Cytoplasm_Intensity_Average,r7c2_Cytoplasm
|
| 100 |
+
r7c2_Nucleus_Intensity_Average,r7c2_Nucleus
|
| 101 |
+
r8c2_Cell_Intensity_Average,r8c2_Cell
|
| 102 |
+
r8c2_Cytoplasm_Intensity_Average,r8c2_Cytoplasm
|
| 103 |
+
r8c2_Nucleus_Intensity_Average,r8c2_Nucleus
|
| 104 |
+
Sting_Cell_Intensity_Average,Sting_Cell
|
| 105 |
+
Sting_Cytoplasm_Intensity_Average,Sting_Cytoplasm
|
| 106 |
+
Sting_Nucleus_Intensity_Average,Sting_Nucleus
|
| 107 |
+
Vimentin_Cell_Intensity_Average,Vimentin_Cell
|
| 108 |
+
Vimentin_Cytoplasm_Intensity_Average,Vimentin_Cytoplasm
|
| 109 |
+
Vimentin_Nucleus_Intensity_Average,Vimentin_Nucleus
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Cellsubtype_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Celltype_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Channel_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Round_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/Sample_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/images/immune_checkpoint_legend.png
ADDED

|
wetransfer_data-zip_2024-05-17_1431/test_metadata/immunecheckpoint_color_data.csv
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,immune_checkpoint
|
| 2 |
+
"(0.9677975592919913, 0.44127456009157356, 0.5358103155058701)",#f77189,B7H4
|
| 3 |
+
"(0.3126890019504329, 0.6928754610296064, 0.1923704830330379)",#50b131,PDL1
|
| 4 |
+
"(0.23299120924703914, 0.639586552066035, 0.9260706093977744)",#3ba3ec,PD1
|
| 5 |
+
"(0.6402432806212122, 0.56707501056059, 0.36409039926945397)",#a3915d,B7H4_PDL1
|
| 6 |
+
"(0.5044925901631545, 0.5912455243957383, 0.5514171359788941)",#81978d,None
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/marker_intensity_metadata.csv
ADDED
|
@@ -0,0 +1,109 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Round,Target,Exp,Channel,target_lower,full_column,marker,localisation
|
| 2 |
+
R0,AF488,300,c2,af488,AF488_Cell_Intensity_Average,AF488,cell
|
| 3 |
+
R0,AF488,300,c2,af488,AF488_Cytoplasm_Intensity_Average,AF488,cytoplasm
|
| 4 |
+
R0,AF488,300,c2,af488,AF488_Nucleus_Intensity_Average,AF488,nucleus
|
| 5 |
+
R0,AF555,1500,c3,af555,AF555_Cell_Intensity_Average,AF555,cell
|
| 6 |
+
R0,AF555,1500,c3,af555,AF555_Cytoplasm_Intensity_Average,AF555,cytoplasm
|
| 7 |
+
R0,AF555,1500,c3,af555,AF555_Nucleus_Intensity_Average,AF555,nucleus
|
| 8 |
+
R0,AF647,1500,c4,af647,AF647_Cell_Intensity_Average,AF647,cell
|
| 9 |
+
R0,AF647,1500,c4,af647,AF647_Cytoplasm_Intensity_Average,AF647,cytoplasm
|
| 10 |
+
R0,AF647,1500,c4,af647,AF647_Nucleus_Intensity_Average,AF647,nucleus
|
| 11 |
+
R0,AF750,1500,c5,af750,AF750_Cell_Intensity_Average,AF750,cell
|
| 12 |
+
R0,AF750,1500,c5,af750,AF750_Cytoplasm_Intensity_Average,AF750,cytoplasm
|
| 13 |
+
R0,AF750,1500,c5,af750,AF750_Nucleus_Intensity_Average,AF750,nucleus
|
| 14 |
+
R1,ColVI,300,c2,colvi,ColVI_Cell_Intensity_Average,ColVI,cell
|
| 15 |
+
R1,ColVI,300,c2,colvi,ColVI_Cytoplasm_Intensity_Average,ColVI,cytoplasm
|
| 16 |
+
R1,ColVI,300,c2,colvi,ColVI_Nucleus_Intensity_Average,ColVI,nucleus
|
| 17 |
+
R1,CD31,1200,c3,cd31,CD31_Cell_Intensity_Average,CD31,cell
|
| 18 |
+
R1,CD31,1200,c3,cd31,CD31_Cytoplasm_Intensity_Average,CD31,cytoplasm
|
| 19 |
+
R1,CD31,1200,c3,cd31,CD31_Nucleus_Intensity_Average,CD31,nucleus
|
| 20 |
+
R1,CD4,1500,c4,cd4,CD4_Cell_Intensity_Average,CD4,cell
|
| 21 |
+
R1,CD4,1500,c4,cd4,CD4_Cytoplasm_Intensity_Average,CD4,cytoplasm
|
| 22 |
+
R1,CD4,1500,c4,cd4,CD4_Nucleus_Intensity_Average,CD4,nucleus
|
| 23 |
+
R1,Ecad,800,c5,ecad,Ecad_Cell_Intensity_Average,Ecad,cell
|
| 24 |
+
R1,Ecad,800,c5,ecad,Ecad_Cytoplasm_Intensity_Average,Ecad,cytoplasm
|
| 25 |
+
R1,Ecad,800,c5,ecad,Ecad_Nucleus_Intensity_Average,Ecad,nucleus
|
| 26 |
+
R2,Desmin,300,c2,desmin,Desmin_Cell_Intensity_Average,Desmin,cell
|
| 27 |
+
R2,Desmin,300,c2,desmin,Desmin_Cytoplasm_Intensity_Average,Desmin,cytoplasm
|
| 28 |
+
R2,Desmin,300,c2,desmin,Desmin_Nucleus_Intensity_Average,Desmin,nucleus
|
| 29 |
+
R2,B7H4,1500,c3,b7h4,B7H4_Cell_Intensity_Average,B7H4,cell
|
| 30 |
+
R2,B7H4,1500,c3,b7h4,B7H4_Cytoplasm_Intensity_Average,B7H4,cytoplasm
|
| 31 |
+
R2,B7H4,1500,c3,b7h4,B7H4_Nucleus_Intensity_Average,B7H4,nucleus
|
| 32 |
+
R2,CD8,1500,c4,cd8,CD8_Cell_Intensity_Average,CD8,cell
|
| 33 |
+
R2,CD8,1500,c4,cd8,CD8_Cytoplasm_Intensity_Average,CD8,cytoplasm
|
| 34 |
+
R2,CD8,1500,c4,cd8,CD8_Nucleus_Intensity_Average,CD8,nucleus
|
| 35 |
+
R2,CD20,1500,c5,cd20,CD20_Cell_Intensity_Average,CD20,cell
|
| 36 |
+
R2,CD20,1500,c5,cd20,CD20_Cytoplasm_Intensity_Average,CD20,cytoplasm
|
| 37 |
+
R2,CD20,1500,c5,cd20,CD20_Nucleus_Intensity_Average,CD20,nucleus
|
| 38 |
+
R3,aSMA,50,c2,asma,aSMA_Cell_Intensity_Average,aSMA,cell
|
| 39 |
+
R3,aSMA,50,c2,asma,aSMA_Cytoplasm_Intensity_Average,aSMA,cytoplasm
|
| 40 |
+
R3,aSMA,50,c2,asma,aSMA_Nucleus_Intensity_Average,aSMA,nucleus
|
| 41 |
+
R3,CD68,1500,c3,cd68,CD68_Cell_Intensity_Average,CD68,cell
|
| 42 |
+
R3,CD68,1500,c3,cd68,CD68_Cytoplasm_Intensity_Average,CD68,cytoplasm
|
| 43 |
+
R3,CD68,1500,c3,cd68,CD68_Nucleus_Intensity_Average,CD68,nucleus
|
| 44 |
+
R3,PD1,1500,c4,pd1,PD1_Cell_Intensity_Average,PD1,cell
|
| 45 |
+
R3,PD1,1500,c4,pd1,PD1_Cytoplasm_Intensity_Average,PD1,cytoplasm
|
| 46 |
+
R3,PD1,1500,c4,pd1,PD1_Nucleus_Intensity_Average,PD1,nucleus
|
| 47 |
+
R3,CD45,1500,c5,cd45,CD45_Cell_Intensity_Average,CD45,cell
|
| 48 |
+
R3,CD45,1500,c5,cd45,CD45_Cytoplasm_Intensity_Average,CD45,cytoplasm
|
| 49 |
+
R3,CD45,1500,c5,cd45,CD45_Nucleus_Intensity_Average,CD45,nucleus
|
| 50 |
+
R4,Vimentin,150,c2,vimentin,Vimentin_Cell_Intensity_Average,Vimentin,cell
|
| 51 |
+
R4,Vimentin,150,c2,vimentin,Vimentin_Cytoplasm_Intensity_Average,Vimentin,cytoplasm
|
| 52 |
+
R4,Vimentin,150,c2,vimentin,Vimentin_Nucleus_Intensity_Average,Vimentin,nucleus
|
| 53 |
+
R4,AXL,1500,c3,axl,AXL_Cell_Intensity_Average,AXL,cell
|
| 54 |
+
R4,AXL,1500,c3,axl,AXL_Cytoplasm_Intensity_Average,AXL,cytoplasm
|
| 55 |
+
R4,AXL,1500,c3,axl,AXL_Nucleus_Intensity_Average,AXL,nucleus
|
| 56 |
+
R4,PDL1,1500,c4,pdl1,PDL1_Cell_Intensity_Average,PDL1,cell
|
| 57 |
+
R4,PDL1,1500,c4,pdl1,PDL1_Cytoplasm_Intensity_Average,PDL1,cytoplasm
|
| 58 |
+
R4,PDL1,1500,c4,pdl1,PDL1_Nucleus_Intensity_Average,PDL1,nucleus
|
| 59 |
+
R4,FOXP3,1500,c5,foxp3,FOXP3_Cell_Intensity_Average,FOXP3,cell
|
| 60 |
+
R4,FOXP3,1500,c5,foxp3,FOXP3_Cytoplasm_Intensity_Average,FOXP3,cytoplasm
|
| 61 |
+
R4,FOXP3,1500,c5,foxp3,FOXP3_Nucleus_Intensity_Average,FOXP3,nucleus
|
| 62 |
+
R5,r5c2,20,c2,r5c2,r5c2_Cell_Intensity_Average,r5c2,cell
|
| 63 |
+
R5,r5c2,20,c2,r5c2,r5c2_Cytoplasm_Intensity_Average,r5c2,cytoplasm
|
| 64 |
+
R5,r5c2,20,c2,r5c2,r5c2_Nucleus_Intensity_Average,r5c2,nucleus
|
| 65 |
+
R5,CA9,1500,c3,ca9,CA9_Cell_Intensity_Average,CA9,cell
|
| 66 |
+
R5,CA9,1500,c3,ca9,CA9_Cytoplasm_Intensity_Average,CA9,cytoplasm
|
| 67 |
+
R5,CA9,1500,c3,ca9,CA9_Nucleus_Intensity_Average,CA9,nucleus
|
| 68 |
+
R5,CD163,1500,c4,cd163,CD163_Cell_Intensity_Average,CD163,cell
|
| 69 |
+
R5,CD163,1500,c4,cd163,CD163_Cytoplasm_Intensity_Average,CD163,cytoplasm
|
| 70 |
+
R5,CD163,1500,c4,cd163,CD163_Nucleus_Intensity_Average,CD163,nucleus
|
| 71 |
+
R5,Ki67,1000,c5,ki67,Ki67_Cell_Intensity_Average,Ki67,cell
|
| 72 |
+
R5,Ki67,1000,c5,ki67,Ki67_Cytoplasm_Intensity_Average,Ki67,cytoplasm
|
| 73 |
+
R5,Ki67,1000,c5,ki67,Ki67_Nucleus_Intensity_Average,Ki67,nucleus
|
| 74 |
+
R6,CKs,200,c2,cks,CKs_Cell_Intensity_Average,CKs,cell
|
| 75 |
+
R6,CKs,200,c2,cks,CKs_Cytoplasm_Intensity_Average,CKs,cytoplasm
|
| 76 |
+
R6,CKs,200,c2,cks,CKs_Nucleus_Intensity_Average,CKs,nucleus
|
| 77 |
+
R6,Fibronectin,1500,c3,fibronectin,Fibronectin_Cell_Intensity_Average,Fibronectin,cell
|
| 78 |
+
R6,Fibronectin,1500,c3,fibronectin,Fibronectin_Cytoplasm_Intensity_Average,Fibronectin,cytoplasm
|
| 79 |
+
R6,Fibronectin,1500,c3,fibronectin,Fibronectin_Nucleus_Intensity_Average,Fibronectin,nucleus
|
| 80 |
+
R6,CD44,1200,c4,cd44,CD44_Cell_Intensity_Average,CD44,cell
|
| 81 |
+
R6,CD44,1200,c4,cd44,CD44_Cytoplasm_Intensity_Average,CD44,cytoplasm
|
| 82 |
+
R6,CD44,1200,c4,cd44,CD44_Nucleus_Intensity_Average,CD44,nucleus
|
| 83 |
+
R6,HLA,500,c5,hla,HLA_Cell_Intensity_Average,HLA,cell
|
| 84 |
+
R6,HLA,500,c5,hla,HLA_Cytoplasm_Intensity_Average,HLA,cytoplasm
|
| 85 |
+
R6,HLA,500,c5,hla,HLA_Nucleus_Intensity_Average,HLA,nucleus
|
| 86 |
+
R7,r7c2,20,c2,r7c2,r7c2_Cell_Intensity_Average,r7c2,cell
|
| 87 |
+
R7,r7c2,20,c2,r7c2,r7c2_Cytoplasm_Intensity_Average,r7c2,cytoplasm
|
| 88 |
+
R7,r7c2,20,c2,r7c2,r7c2_Nucleus_Intensity_Average,r7c2,nucleus
|
| 89 |
+
R7,PDGFR,1500,c3,pdgfr,PDGFR_Cell_Intensity_Average,PDGFR,cell
|
| 90 |
+
R7,PDGFR,1500,c3,pdgfr,PDGFR_Cytoplasm_Intensity_Average,PDGFR,cytoplasm
|
| 91 |
+
R7,PDGFR,1500,c3,pdgfr,PDGFR_Nucleus_Intensity_Average,PDGFR,nucleus
|
| 92 |
+
R7,MMP9,1500,c4,mmp9,MMP9_Cell_Intensity_Average,MMP9,cell
|
| 93 |
+
R7,MMP9,1500,c4,mmp9,MMP9_Cytoplasm_Intensity_Average,MMP9,cytoplasm
|
| 94 |
+
R7,MMP9,1500,c4,mmp9,MMP9_Nucleus_Intensity_Average,MMP9,nucleus
|
| 95 |
+
R7,GATA3,1500,c5,gata3,GATA3_Cell_Intensity_Average,GATA3,cell
|
| 96 |
+
R7,GATA3,1500,c5,gata3,GATA3_Cytoplasm_Intensity_Average,GATA3,cytoplasm
|
| 97 |
+
R7,GATA3,1500,c5,gata3,GATA3_Nucleus_Intensity_Average,GATA3,nucleus
|
| 98 |
+
R8,r8c2,25,c2,r8c2,r8c2_Cell_Intensity_Average,r8c2,cell
|
| 99 |
+
R8,r8c2,25,c2,r8c2,r8c2_Cytoplasm_Intensity_Average,r8c2,cytoplasm
|
| 100 |
+
R8,r8c2,25,c2,r8c2,r8c2_Nucleus_Intensity_Average,r8c2,nucleus
|
| 101 |
+
R8,CD11c,1500,c3,cd11c,CD11c_Cell_Intensity_Average,CD11c,cell
|
| 102 |
+
R8,CD11c,1500,c3,cd11c,CD11c_Cytoplasm_Intensity_Average,CD11c,cytoplasm
|
| 103 |
+
R8,CD11c,1500,c3,cd11c,CD11c_Nucleus_Intensity_Average,CD11c,nucleus
|
| 104 |
+
R8,Sting,1000,c4,sting,Sting_Cell_Intensity_Average,Sting,cell
|
| 105 |
+
R8,Sting,1000,c4,sting,Sting_Cytoplasm_Intensity_Average,Sting,cytoplasm
|
| 106 |
+
R8,Sting,1000,c4,sting,Sting_Nucleus_Intensity_Average,Sting,nucleus
|
| 107 |
+
R8,CD11b,1500,c5,cd11b,CD11b_Cell_Intensity_Average,CD11b,cell
|
| 108 |
+
R8,CD11b,1500,c5,cd11b,CD11b_Cytoplasm_Intensity_Average,CD11b,cytoplasm
|
| 109 |
+
R8,CD11b,1500,c5,cd11b,CD11b_Nucleus_Intensity_Average,CD11b,nucleus
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/not_intensities.csv
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Marker
|
| 2 |
+
Cell_Size
|
| 3 |
+
Nuc_X
|
| 4 |
+
Nuc_Y_Inv
|
| 5 |
+
ROI_index
|
| 6 |
+
Nucleus_Size
|
| 7 |
+
Nucleus_Roundness
|
| 8 |
+
Sample_ID
|
| 9 |
+
Round
|
| 10 |
+
Exp
|
| 11 |
+
Channel
|
| 12 |
+
Target
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/round_color_data.csv
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,Round
|
| 2 |
+
"(0.28685356234627135, 0.13009829239513535, 0.23110332132624437)",#49213b,R0
|
| 3 |
+
"(0.36541462435986094, 0.2025447048359916, 0.37693310021636883)",#5d3460,R1
|
| 4 |
+
"(0.40867533458903105, 0.2940761173840091, 0.5166711878800253)",#684b84,R2
|
| 5 |
+
"(0.42890613750051265, 0.4082290173220481, 0.6335348887063806)",#6d68a2,R3
|
| 6 |
+
"(0.4444462906865238, 0.5264664993764805, 0.7056321892616532)",#7186b4,R4
|
| 7 |
+
"(0.47707206309601013, 0.6427061780374552, 0.7418477948908153)",#7aa4bd,R5
|
| 8 |
+
"(0.5414454866716836, 0.7466759172596551, 0.7572905778378964)",#8abec1,R6
|
| 9 |
+
"(0.6414710091647722, 0.8321551072276492, 0.7746773027952071)",#a4d4c6,R7
|
| 10 |
+
"(0.7684256891219349, 0.8992667116749021, 0.8171383269422353)",#c4e5d0,R8
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/sample_color_data.csv
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rgb,hex,Sample_ID
|
| 2 |
+
"(0.9677975592919913, 0.44127456009157356, 0.5358103155058701)",#f77189,DD3S1.csv
|
| 3 |
+
"(0.5920891529639701, 0.6418467016378244, 0.1935069134991043)",#97a431,DD3S2.csv
|
| 4 |
+
"(0.21044753832183283, 0.6773105080456748, 0.6433941168468681)",#36ada4,DD3S3.csv
|
| 5 |
+
"(0.5019607843137255, 0.5019607843137255, 0.5019607843137255)",#808080,TMA.csv
|
wetransfer_data-zip_2024-05-17_1431/test_metadata/short_to_full_column_names.csv
ADDED
|
@@ -0,0 +1,109 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
short_name,full_name
|
| 2 |
+
AF488_Cell,AF488_Cell_Intensity_Average
|
| 3 |
+
AF488_Cytoplasm,AF488_Cytoplasm_Intensity_Average
|
| 4 |
+
AF488_Nucleus,AF488_Nucleus_Intensity_Average
|
| 5 |
+
AF555_Cell,AF555_Cell_Intensity_Average
|
| 6 |
+
AF555_Cytoplasm,AF555_Cytoplasm_Intensity_Average
|
| 7 |
+
AF555_Nucleus,AF555_Nucleus_Intensity_Average
|
| 8 |
+
AF647_Cell,AF647_Cell_Intensity_Average
|
| 9 |
+
AF647_Cytoplasm,AF647_Cytoplasm_Intensity_Average
|
| 10 |
+
AF647_Nucleus,AF647_Nucleus_Intensity_Average
|
| 11 |
+
AF750_Cell,AF750_Cell_Intensity_Average
|
| 12 |
+
AF750_Cytoplasm,AF750_Cytoplasm_Intensity_Average
|
| 13 |
+
AF750_Nucleus,AF750_Nucleus_Intensity_Average
|
| 14 |
+
aSMA_Cell,aSMA_Cell_Intensity_Average
|
| 15 |
+
aSMA_Cytoplasm,aSMA_Cytoplasm_Intensity_Average
|
| 16 |
+
aSMA_Nucleus,aSMA_Nucleus_Intensity_Average
|
| 17 |
+
AXL_Cell,AXL_Cell_Intensity_Average
|
| 18 |
+
AXL_Cytoplasm,AXL_Cytoplasm_Intensity_Average
|
| 19 |
+
AXL_Nucleus,AXL_Nucleus_Intensity_Average
|
| 20 |
+
B7H4_Cell,B7H4_Cell_Intensity_Average
|
| 21 |
+
B7H4_Cytoplasm,B7H4_Cytoplasm_Intensity_Average
|
| 22 |
+
B7H4_Nucleus,B7H4_Nucleus_Intensity_Average
|
| 23 |
+
CA9_Cell,CA9_Cell_Intensity_Average
|
| 24 |
+
CA9_Cytoplasm,CA9_Cytoplasm_Intensity_Average
|
| 25 |
+
CA9_Nucleus,CA9_Nucleus_Intensity_Average
|
| 26 |
+
CD4_Cell,CD4_Cell_Intensity_Average
|
| 27 |
+
CD4_Cytoplasm,CD4_Cytoplasm_Intensity_Average
|
| 28 |
+
CD4_Nucleus,CD4_Nucleus_Intensity_Average
|
| 29 |
+
CD8_Cell,CD8_Cell_Intensity_Average
|
| 30 |
+
CD8_Cytoplasm,CD8_Cytoplasm_Intensity_Average
|
| 31 |
+
CD8_Nucleus,CD8_Nucleus_Intensity_Average
|
| 32 |
+
CD11b_Cell,CD11b_Cell_Intensity_Average
|
| 33 |
+
CD11b_Cytoplasm,CD11b_Cytoplasm_Intensity_Average
|
| 34 |
+
CD11b_Nucleus,CD11b_Nucleus_Intensity_Average
|
| 35 |
+
CD11c_Cell,CD11c_Cell_Intensity_Average
|
| 36 |
+
CD11c_Cytoplasm,CD11c_Cytoplasm_Intensity_Average
|
| 37 |
+
CD11c_Nucleus,CD11c_Nucleus_Intensity_Average
|
| 38 |
+
CD20_Cell,CD20_Cell_Intensity_Average
|
| 39 |
+
CD20_Cytoplasm,CD20_Cytoplasm_Intensity_Average
|
| 40 |
+
CD20_Nucleus,CD20_Nucleus_Intensity_Average
|
| 41 |
+
CD31_Cell,CD31_Cell_Intensity_Average
|
| 42 |
+
CD31_Cytoplasm,CD31_Cytoplasm_Intensity_Average
|
| 43 |
+
CD31_Nucleus,CD31_Nucleus_Intensity_Average
|
| 44 |
+
CD44_Cell,CD44_Cell_Intensity_Average
|
| 45 |
+
CD44_Cytoplasm,CD44_Cytoplasm_Intensity_Average
|
| 46 |
+
CD44_Nucleus,CD44_Nucleus_Intensity_Average
|
| 47 |
+
CD45_Cell,CD45_Cell_Intensity_Average
|
| 48 |
+
CD45_Cytoplasm,CD45_Cytoplasm_Intensity_Average
|
| 49 |
+
CD45_Nucleus,CD45_Nucleus_Intensity_Average
|
| 50 |
+
CD68_Cell,CD68_Cell_Intensity_Average
|
| 51 |
+
CD68_Cytoplasm,CD68_Cytoplasm_Intensity_Average
|
| 52 |
+
CD68_Nucleus,CD68_Nucleus_Intensity_Average
|
| 53 |
+
CD163_Cell,CD163_Cell_Intensity_Average
|
| 54 |
+
CD163_Cytoplasm,CD163_Cytoplasm_Intensity_Average
|
| 55 |
+
CD163_Nucleus,CD163_Nucleus_Intensity_Average
|
| 56 |
+
CKs_Cell,CKs_Cell_Intensity_Average
|
| 57 |
+
CKs_Cytoplasm,CKs_Cytoplasm_Intensity_Average
|
| 58 |
+
CKs_Nucleus,CKs_Nucleus_Intensity_Average
|
| 59 |
+
ColVI_Cell,ColVI_Cell_Intensity_Average
|
| 60 |
+
ColVI_Cytoplasm,ColVI_Cytoplasm_Intensity_Average
|
| 61 |
+
ColVI_Nucleus,ColVI_Nucleus_Intensity_Average
|
| 62 |
+
Desmin_Cell,Desmin_Cell_Intensity_Average
|
| 63 |
+
Desmin_Cytoplasm,Desmin_Cytoplasm_Intensity_Average
|
| 64 |
+
Desmin_Nucleus,Desmin_Nucleus_Intensity_Average
|
| 65 |
+
Ecad_Cell,Ecad_Cell_Intensity_Average
|
| 66 |
+
Ecad_Cytoplasm,Ecad_Cytoplasm_Intensity_Average
|
| 67 |
+
Ecad_Nucleus,Ecad_Nucleus_Intensity_Average
|
| 68 |
+
Fibronectin_Cell,Fibronectin_Cell_Intensity_Average
|
| 69 |
+
Fibronectin_Cytoplasm,Fibronectin_Cytoplasm_Intensity_Average
|
| 70 |
+
Fibronectin_Nucleus,Fibronectin_Nucleus_Intensity_Average
|
| 71 |
+
FOXP3_Cell,FOXP3_Cell_Intensity_Average
|
| 72 |
+
FOXP3_Cytoplasm,FOXP3_Cytoplasm_Intensity_Average
|
| 73 |
+
FOXP3_Nucleus,FOXP3_Nucleus_Intensity_Average
|
| 74 |
+
GATA3_Cell,GATA3_Cell_Intensity_Average
|
| 75 |
+
GATA3_Cytoplasm,GATA3_Cytoplasm_Intensity_Average
|
| 76 |
+
GATA3_Nucleus,GATA3_Nucleus_Intensity_Average
|
| 77 |
+
HLA_Cell,HLA_Cell_Intensity_Average
|
| 78 |
+
HLA_Cytoplasm,HLA_Cytoplasm_Intensity_Average
|
| 79 |
+
HLA_Nucleus,HLA_Nucleus_Intensity_Average
|
| 80 |
+
Ki67_Cell,Ki67_Cell_Intensity_Average
|
| 81 |
+
Ki67_Cytoplasm,Ki67_Cytoplasm_Intensity_Average
|
| 82 |
+
Ki67_Nucleus,Ki67_Nucleus_Intensity_Average
|
| 83 |
+
MMP9_Cell,MMP9_Cell_Intensity_Average
|
| 84 |
+
MMP9_Cytoplasm,MMP9_Cytoplasm_Intensity_Average
|
| 85 |
+
MMP9_Nucleus,MMP9_Nucleus_Intensity_Average
|
| 86 |
+
PD1_Cell,PD1_Cell_Intensity_Average
|
| 87 |
+
PD1_Cytoplasm,PD1_Cytoplasm_Intensity_Average
|
| 88 |
+
PD1_Nucleus,PD1_Nucleus_Intensity_Average
|
| 89 |
+
PDGFR_Cell,PDGFR_Cell_Intensity_Average
|
| 90 |
+
PDGFR_Cytoplasm,PDGFR_Cytoplasm_Intensity_Average
|
| 91 |
+
PDGFR_Nucleus,PDGFR_Nucleus_Intensity_Average
|
| 92 |
+
PDL1_Cell,PDL1_Cell_Intensity_Average
|
| 93 |
+
PDL1_Cytoplasm,PDL1_Cytoplasm_Intensity_Average
|
| 94 |
+
PDL1_Nucleus,PDL1_Nucleus_Intensity_Average
|
| 95 |
+
r5c2_Cell,r5c2_Cell_Intensity_Average
|
| 96 |
+
r5c2_Cytoplasm,r5c2_Cytoplasm_Intensity_Average
|
| 97 |
+
r5c2_Nucleus,r5c2_Nucleus_Intensity_Average
|
| 98 |
+
r7c2_Cell,r7c2_Cell_Intensity_Average
|
| 99 |
+
r7c2_Cytoplasm,r7c2_Cytoplasm_Intensity_Average
|
| 100 |
+
r7c2_Nucleus,r7c2_Nucleus_Intensity_Average
|
| 101 |
+
r8c2_Cell,r8c2_Cell_Intensity_Average
|
| 102 |
+
r8c2_Cytoplasm,r8c2_Cytoplasm_Intensity_Average
|
| 103 |
+
r8c2_Nucleus,r8c2_Nucleus_Intensity_Average
|
| 104 |
+
Sting_Cell,Sting_Cell_Intensity_Average
|
| 105 |
+
Sting_Cytoplasm,Sting_Cytoplasm_Intensity_Average
|
| 106 |
+
Sting_Nucleus,Sting_Nucleus_Intensity_Average
|
| 107 |
+
Vimentin_Cell,Vimentin_Cell_Intensity_Average
|
| 108 |
+
Vimentin_Cytoplasm,Vimentin_Cytoplasm_Intensity_Average
|
| 109 |
+
Vimentin_Nucleus,Vimentin_Nucleus_Intensity_Average
|
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S1_mt.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c5e5d712093b48a1b53b42e9b21a7c5b0a98946f70cd4729a54511329af6b8c9
|
| 3 |
+
size 42782100
|
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S2_mt.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:6430f2839ce44437a4016c73fb532e8bc704cf9b92b514c0ec771c8a0fee74cb
|
| 3 |
+
size 38749776
|
wetransfer_data-zip_2024-05-17_1431/test_mt/DD3S3_mt.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:797b5ec539dc0f32407330fd8aedfac86135664ce8387e17ba71b4da8ace1859
|
| 3 |
+
size 72513073
|
wetransfer_data-zip_2024-05-17_1431/test_mt/TMA_mt.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:598bb6d17e4b2a7c1de59b43aa0e289f406ba0d409f90b1dbade6cc46ebbaffe
|
| 3 |
+
size 58806993
|
wetransfer_data-zip_2024-05-17_1431/test_mt/all_Samples_Set_A.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a47760a85a3142b7b31f8fa8dbce6a0b0348b5315361c19254e56ac60a828261
|
| 3 |
+
size 212848897
|
wetransfer_data-zip_2024-05-17_1431/test_mt/images/set_a.png
ADDED
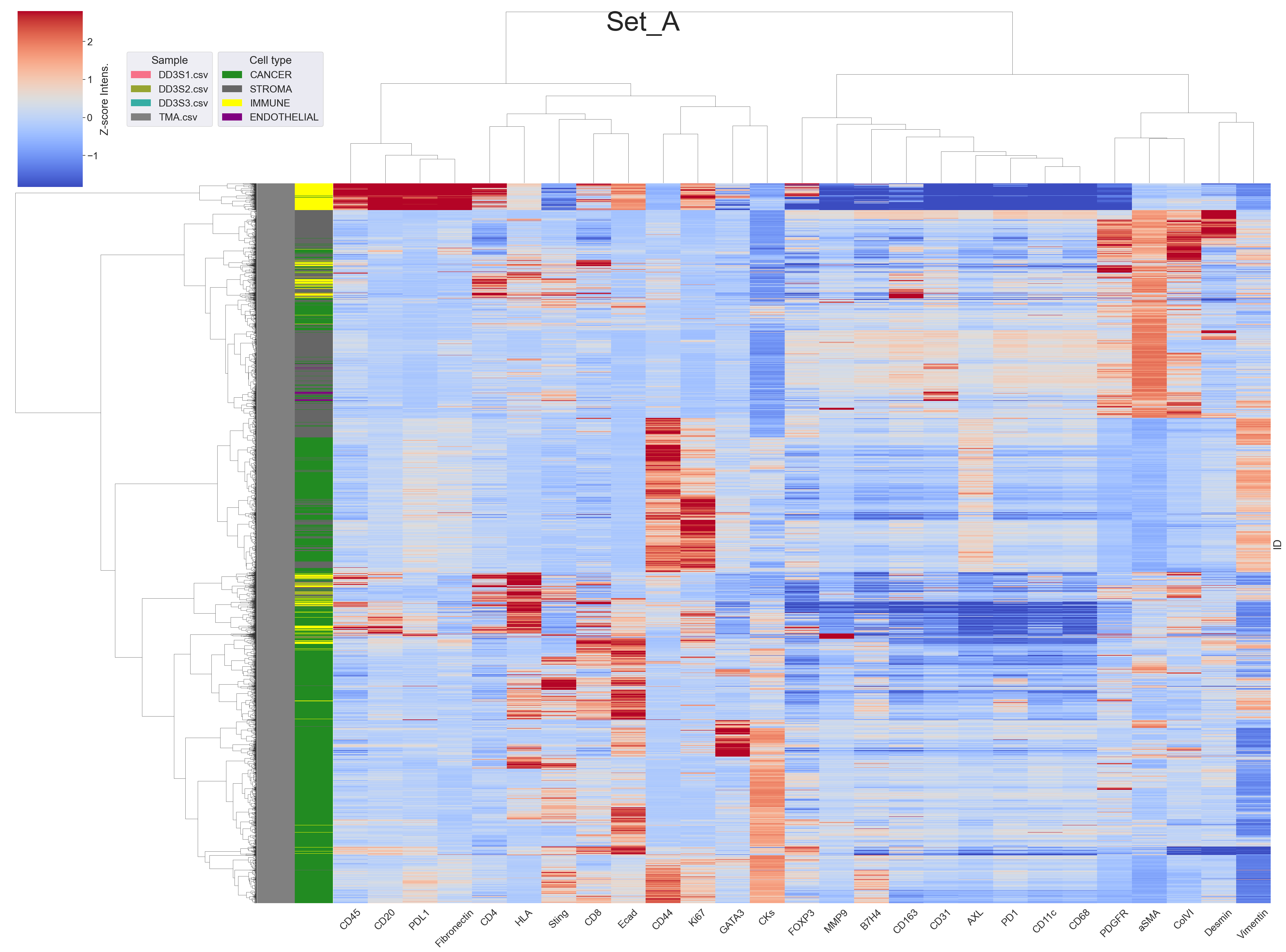
|