Spaces:
Runtime error
Runtime error
Commit
·
37dec6e
1
Parent(s):
d6e77d3
initial commit
Browse files- .gitignore +1 -0
- README.md +8 -5
- app.py +91 -0
- img1.jpg +0 -0
- img2.jpg +0 -0
- requirements.txt +6 -0
- thumbnail.png +0 -0
.gitignore
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
weights/*
|
README.md
CHANGED
|
@@ -1,11 +1,14 @@
|
|
| 1 |
---
|
| 2 |
title: Echocardiogram Segmentation
|
| 3 |
-
emoji:
|
| 4 |
-
colorFrom:
|
| 5 |
colorTo: red
|
| 6 |
-
sdk:
|
|
|
|
|
|
|
| 7 |
pinned: false
|
| 8 |
-
license: unknown
|
| 9 |
---
|
| 10 |
|
| 11 |
-
|
|
|
|
|
|
|
|
|
| 1 |
---
|
| 2 |
title: Echocardiogram Segmentation
|
| 3 |
+
emoji: 📊
|
| 4 |
+
colorFrom: red
|
| 5 |
colorTo: red
|
| 6 |
+
sdk: gradio
|
| 7 |
+
sdk_version: 3.34.0
|
| 8 |
+
app_file: app.py
|
| 9 |
pinned: false
|
|
|
|
| 10 |
---
|
| 11 |
|
| 12 |
+
Cloned from: https://huggingface.co/spaces/abidlabs/Echocardiogram-Segmentation
|
| 13 |
+
|
| 14 |
+
This is a demo based on a very simplified approach described in the paper, ["High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy with Cardiovascular Deep Learning"](https://arxiv.org/abs/2306.07954)
|
app.py
ADDED
|
@@ -0,0 +1,91 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os, os.path
|
| 2 |
+
from os.path import splitext
|
| 3 |
+
import numpy as np
|
| 4 |
+
import sys
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import torch
|
| 7 |
+
import torchvision
|
| 8 |
+
import wget
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
destination_folder = "output"
|
| 12 |
+
destination_for_weights = "weights"
|
| 13 |
+
|
| 14 |
+
if os.path.exists(destination_for_weights):
|
| 15 |
+
print("The weights are at", destination_for_weights)
|
| 16 |
+
else:
|
| 17 |
+
print("Creating folder at ", destination_for_weights, " to store weights")
|
| 18 |
+
os.mkdir(destination_for_weights)
|
| 19 |
+
|
| 20 |
+
segmentationWeightsURL = 'https://github.com/douyang/EchoNetDynamic/releases/download/v1.0.0/deeplabv3_resnet50_random.pt'
|
| 21 |
+
|
| 22 |
+
if not os.path.exists(os.path.join(destination_for_weights, os.path.basename(segmentationWeightsURL))):
|
| 23 |
+
print("Downloading Segmentation Weights, ", segmentationWeightsURL," to ",os.path.join(destination_for_weights, os.path.basename(segmentationWeightsURL)))
|
| 24 |
+
filename = wget.download(segmentationWeightsURL, out = destination_for_weights)
|
| 25 |
+
else:
|
| 26 |
+
print("Segmentation Weights already present")
|
| 27 |
+
|
| 28 |
+
torch.cuda.empty_cache()
|
| 29 |
+
|
| 30 |
+
def collate_fn(x):
|
| 31 |
+
x, f = zip(*x)
|
| 32 |
+
i = list(map(lambda t: t.shape[1], x))
|
| 33 |
+
x = torch.as_tensor(np.swapaxes(np.concatenate(x, 1), 0, 1))
|
| 34 |
+
return x, f, i
|
| 35 |
+
|
| 36 |
+
model = torchvision.models.segmentation.deeplabv3_resnet50(pretrained=False, aux_loss=False)
|
| 37 |
+
model.classifier[-1] = torch.nn.Conv2d(model.classifier[-1].in_channels, 1, kernel_size=model.classifier[-1].kernel_size)
|
| 38 |
+
|
| 39 |
+
print("loading weights from ", os.path.join(destination_for_weights, "deeplabv3_resnet50_random"))
|
| 40 |
+
|
| 41 |
+
if torch.cuda.is_available():
|
| 42 |
+
print("cuda is available, original weights")
|
| 43 |
+
device = torch.device("cuda")
|
| 44 |
+
model = torch.nn.DataParallel(model)
|
| 45 |
+
model.to(device)
|
| 46 |
+
checkpoint = torch.load(os.path.join(destination_for_weights, os.path.basename(segmentationWeightsURL)))
|
| 47 |
+
model.load_state_dict(checkpoint['state_dict'])
|
| 48 |
+
else:
|
| 49 |
+
print("cuda is not available, cpu weights")
|
| 50 |
+
device = torch.device("cpu")
|
| 51 |
+
checkpoint = torch.load(os.path.join(destination_for_weights, os.path.basename(segmentationWeightsURL)), map_location = "cpu")
|
| 52 |
+
state_dict_cpu = {k[7:]: v for (k, v) in checkpoint['state_dict'].items()}
|
| 53 |
+
model.load_state_dict(state_dict_cpu)
|
| 54 |
+
|
| 55 |
+
model.eval()
|
| 56 |
+
|
| 57 |
+
def segment(inp):
|
| 58 |
+
x = inp.transpose([2, 0, 1]) # channels-first
|
| 59 |
+
x = np.expand_dims(x, axis=0) # adding a batch dimension
|
| 60 |
+
|
| 61 |
+
mean = x.mean(axis=(0, 2, 3))
|
| 62 |
+
std = x.std(axis=(0, 2, 3))
|
| 63 |
+
x = x - mean.reshape(1, 3, 1, 1)
|
| 64 |
+
x = x / std.reshape(1, 3, 1, 1)
|
| 65 |
+
|
| 66 |
+
with torch.no_grad():
|
| 67 |
+
x = torch.from_numpy(x).type('torch.FloatTensor').to(device)
|
| 68 |
+
output = model(x)
|
| 69 |
+
|
| 70 |
+
y = output['out'].numpy()
|
| 71 |
+
y = y.squeeze()
|
| 72 |
+
|
| 73 |
+
out = y>0
|
| 74 |
+
|
| 75 |
+
mask = inp.copy()
|
| 76 |
+
mask[out] = np.array([0, 0, 255])
|
| 77 |
+
|
| 78 |
+
return mask
|
| 79 |
+
|
| 80 |
+
import gradio as gr
|
| 81 |
+
|
| 82 |
+
i = gr.Image(shape=(112, 112))
|
| 83 |
+
o = gr.Image()
|
| 84 |
+
|
| 85 |
+
examples = [["img1.jpg"], ["img2.jpg"]]
|
| 86 |
+
title = None #"Left Ventricle Segmentation"
|
| 87 |
+
description = "This semantic segmentation model identifies the left ventricle in echocardiogram images."
|
| 88 |
+
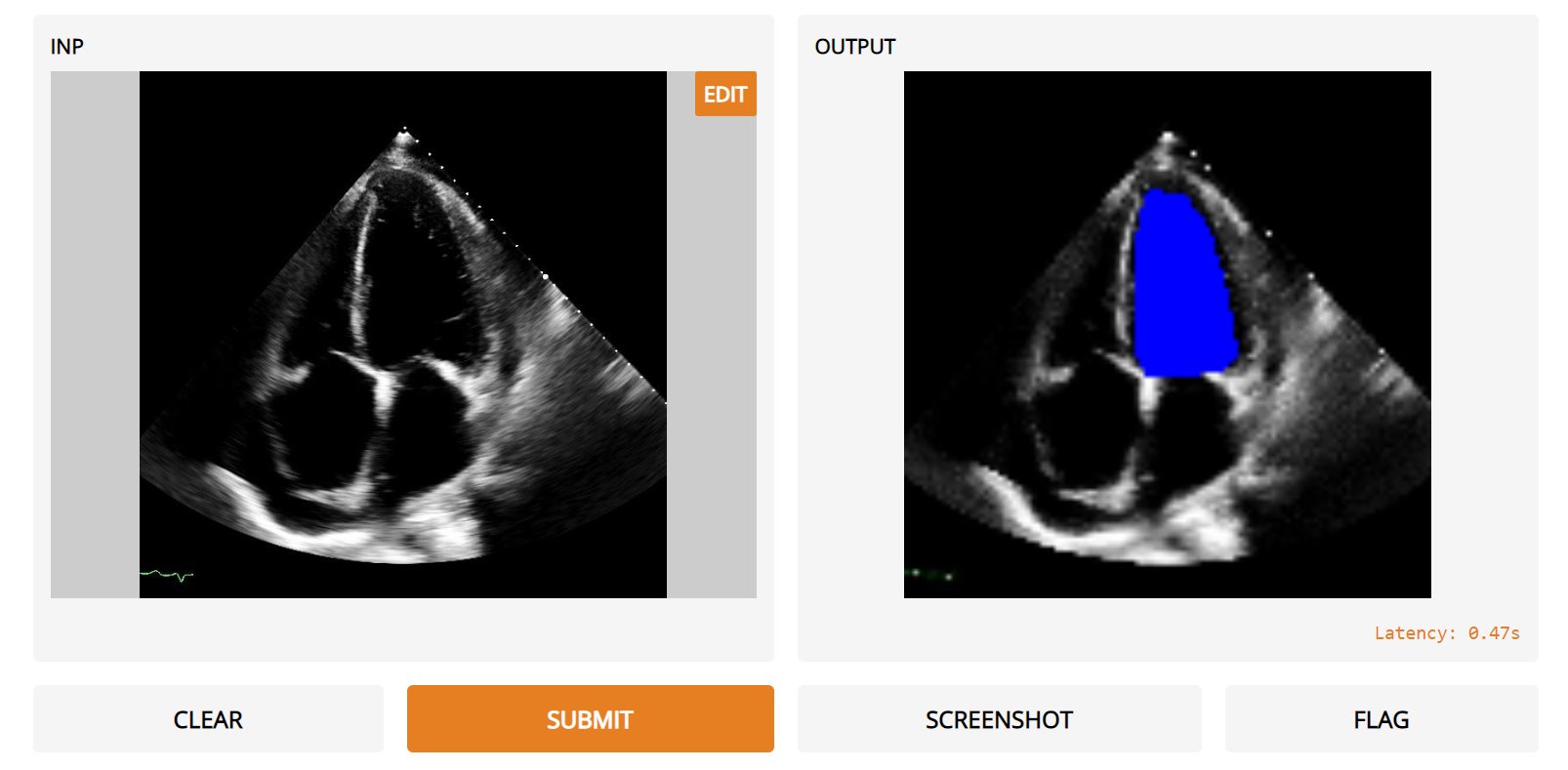
# videos. Accurate evaluation of the motion and size of the left ventricle is crucial for the assessment of cardiac function and ejection fraction. In this interface, the user inputs apical-4-chamber images from echocardiography videos and the model will output a prediction of the localization of the left ventricle in blue. This model was trained on the publicly released EchoNet-Dynamic dataset of 10k echocardiogram videos with 20k expert annotations of the left ventricle and published as part of ‘Video-based AI for beat-to-beat assessment of cardiac function’ by Ouyang et al. in Nature, 2020."
|
| 89 |
+
thumbnail = "https://raw.githubusercontent.com/gradio-app/hub-echonet/master/thumbnail.png"
|
| 90 |
+
gr.Interface(segment, i, o, examples=examples, allow_flagging=False, analytics_enabled=False,
|
| 91 |
+
title=title, description=description, thumbnail=thumbnail).launch()
|
img1.jpg
ADDED
|
img2.jpg
ADDED
|
requirements.txt
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
-f https://download.pytorch.org/whl/torch_stable.html
|
| 2 |
+
numpy
|
| 3 |
+
matplotlib
|
| 4 |
+
wget
|
| 5 |
+
torch
|
| 6 |
+
torchvision
|
thumbnail.png
ADDED
|