Spaces:
Runtime error
Runtime error
add new files
Browse files- .gitattributes +1 -0
- app.py +137 -0
- assets/style.css +19 -0
- breast-cancer-wisconsin.ipynb +0 -0
- charts/DecisionTreeClassifier(random_state=42)_confusion_matrix.png +0 -0
- charts/DecisionTreeClassifier(random_state=42)_normalized_confusion_matrix.png +0 -0
- charts/Features_with_Correlation.png +0 -0
- charts/GaussianNB()_confusion_matrix.png +0 -0
- charts/GaussianNB()_normalized_confusion_matrix.png +0 -0
- charts/KNeighborsClassifier(n_neighbors=2)_confusion_matrix.png +0 -0
- charts/KNeighborsClassifier(n_neighbors=2)_normalized_confusion_matrix.png +0 -0
- charts/LogisticRegression()_confusion_matrix.png +0 -0
- charts/LogisticRegression()_normalized_confusion_matrix.png +0 -0
- charts/RandomForestClassifier(random_state=0)_confusion_matrix.png +0 -0
- charts/RandomForestClassifier(random_state=0)_normalized_confusion_matrix.png +0 -0
- charts/clustermap_high_correlation.png +0 -0
- charts/correlation_heatmap.png +0 -0
- charts/model_Accuracy.png +0 -0
- charts/pairplot_high_correlation.png +0 -0
- charts/target_count.png +0 -0
- data/data.csv +0 -0
- full code.py +196 -0
- images/1.jpg +0 -0
- images/2.jpg +0 -0
- images/3.jpg +0 -0
- images/4.gif +3 -0
- model/main.py +57 -0
- model/model.pkl +3 -0
- model/scaler.pkl +3 -0
- pages/📈_Analysis.py +227 -0
- pages/📊_Predict.py +195 -0
- requirements.txt +54 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
images/4.gif filter=lfs diff=lfs merge=lfs -text
|
app.py
ADDED
|
@@ -0,0 +1,137 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
|
| 3 |
+
st.set_page_config(
|
| 4 |
+
page_title="Breast Cancer Introduction",
|
| 5 |
+
page_icon=":female-doctor:",
|
| 6 |
+
layout="wide",
|
| 7 |
+
initial_sidebar_state="expanded"
|
| 8 |
+
)
|
| 9 |
+
|
| 10 |
+
# Title of the page
|
| 11 |
+
st.image('./images/1.jpg')
|
| 12 |
+
st.title("Breast Cancer Overview")
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
# Overview section
|
| 16 |
+
st.header("Overview")
|
| 17 |
+
st.write("""
|
| 18 |
+
Breast cancer occurs when breast cells mutate and become cancerous cells that multiply and form tumors. It typically affects women and people assigned female at birth (AFAB) age 50 and older, but it can also affect men and people assigned male at birth (AMAB), as well as younger women.
|
| 19 |
+
Healthcare providers may treat breast cancer with surgery to remove tumors or treatment to kill cancerous cells.
|
| 20 |
+
""")
|
| 21 |
+
|
| 22 |
+
st.subheader("What is breast cancer?")
|
| 23 |
+
st.image('./images/2.jpg')
|
| 24 |
+
st.write("""
|
| 25 |
+
Breast cancer is one of the most common cancers that affects women and people AFAB. It happens when cancerous cells in the breasts multiply and become tumors. About 80% of breast cancer cases are invasive, meaning a tumor may spread from your breast to other areas of your body.
|
| 26 |
+
Breast cancer typically affects women age 50 and older, but it can also affect women and people AFAB who are younger than 50. Men and people assigned male at birth (AMAB) may also develop breast cancer.
|
| 27 |
+
""")
|
| 28 |
+
|
| 29 |
+
st.subheader("Breast cancer types")
|
| 30 |
+
st.write("""
|
| 31 |
+
Healthcare providers determine cancer types and subtypes so they can tailor treatment to be as effective as possible with the fewest possible side effects. Common types of breast cancer include:
|
| 32 |
+
|
| 33 |
+
- Invasive (infiltrating) ductal carcinoma (IDC): This cancer starts in your milk ducts and spreads to nearby breast tissue. It’s the most common type of breast cancer in the United States.
|
| 34 |
+
- Lobular breast cancer: This cancer starts in the milk-producing glands (lobules) and often spreads to nearby breast tissue. It’s the second most common breast cancer in the United States.
|
| 35 |
+
- Ductal carcinoma in situ (DCIS): Like IDC, this cancer starts in your milk ducts but doesn’t spread beyond the ducts.
|
| 36 |
+
|
| 37 |
+
Other types include triple-negative breast cancer (TNBC), inflammatory breast cancer (IBC), and Paget’s disease of the breast.
|
| 38 |
+
""")
|
| 39 |
+
|
| 40 |
+
st.subheader("Breast cancer subtypes")
|
| 41 |
+
st.write("""
|
| 42 |
+
Breast cancer subtypes are classified by receptor cell status:
|
| 43 |
+
|
| 44 |
+
- ER-positive (ER+): Has estrogen receptors.
|
| 45 |
+
- PR-positive (PR+): Has progesterone receptors.
|
| 46 |
+
- HR-positive (HR+): Has both estrogen and progesterone receptors.
|
| 47 |
+
- HR-negative (HR-): Lacks these hormone receptors.
|
| 48 |
+
- HER2-positive (HER2+): Has higher than normal HER2 protein levels. About 15% to 20% of all breast cancers are HER2-positive.
|
| 49 |
+
""")
|
| 50 |
+
|
| 51 |
+
# Symptoms and Causes section
|
| 52 |
+
st.header("Symptoms and Causes")
|
| 53 |
+
st.image('./images/3.jpg')
|
| 54 |
+
st.subheader("What are breast cancer symptoms?")
|
| 55 |
+
st.write("""
|
| 56 |
+
The condition can affect your breasts in different ways. Symptoms include:
|
| 57 |
+
|
| 58 |
+
- A change in the size, shape, or contour of your breast.
|
| 59 |
+
- A lump or thickening near your breast or in your underarm.
|
| 60 |
+
- Skin changes (dimpling, puckering, or inflamed skin).
|
| 61 |
+
- Discharge from your nipple, which could be clear or blood-stained.
|
| 62 |
+
""")
|
| 63 |
+
|
| 64 |
+
st.subheader("What causes breast cancer?")
|
| 65 |
+
st.write("""
|
| 66 |
+
Breast cancer occurs when breast cells mutate, but the exact cause is unknown. Risk factors include:
|
| 67 |
+
|
| 68 |
+
- Age: Being 55 or older.
|
| 69 |
+
- Sex: Women and people AFAB are at higher risk.
|
| 70 |
+
- Family history: A close family member with breast cancer increases your risk.
|
| 71 |
+
- Genetics: Inherited mutations, especially BRCA1 and BRCA2 genes.
|
| 72 |
+
- Smoking, alcohol use, obesity, and radiation exposure.
|
| 73 |
+
""")
|
| 74 |
+
|
| 75 |
+
# Diagnosis and Tests section
|
| 76 |
+
st.header("Diagnosis and Tests")
|
| 77 |
+
st.image('./images/4.gif')
|
| 78 |
+
st.write("""
|
| 79 |
+
To diagnose breast cancer, healthcare providers may perform:
|
| 80 |
+
|
| 81 |
+
- Breast ultrasound or MRI scans.
|
| 82 |
+
- Breast biopsy.
|
| 83 |
+
- Immunohistochemistry tests to check for hormone receptors.
|
| 84 |
+
- Genetic tests to identify mutations.
|
| 85 |
+
|
| 86 |
+
Breast cancer is classified into different stages:
|
| 87 |
+
|
| 88 |
+
- Stage 0: Noninvasive, meaning it hasn’t spread.
|
| 89 |
+
- Stage I-IV: Based on tumor size, spread to lymph nodes, and metastasis.
|
| 90 |
+
""")
|
| 91 |
+
|
| 92 |
+
# Management and Treatment section
|
| 93 |
+
st.header("Management and Treatment")
|
| 94 |
+
st.write("""
|
| 95 |
+
Surgery is often the primary treatment, such as a mastectomy or lumpectomy. Other treatments include:
|
| 96 |
+
|
| 97 |
+
- Chemotherapy
|
| 98 |
+
- Radiation therapy
|
| 99 |
+
- Immunotherapy
|
| 100 |
+
- Hormone therapy (e.g., selective estrogen receptor modulators)
|
| 101 |
+
- Targeted therapy
|
| 102 |
+
|
| 103 |
+
Side effects of treatment vary but may include nausea, fatigue, gastrointestinal issues, and infection risk from surgery.
|
| 104 |
+
""")
|
| 105 |
+
|
| 106 |
+
# Prevention section
|
| 107 |
+
st.header("Prevention")
|
| 108 |
+
st.write("""
|
| 109 |
+
While breast cancer can’t always be prevented, these steps can reduce risk:
|
| 110 |
+
|
| 111 |
+
- Maintain a healthy weight.
|
| 112 |
+
- Eat a balanced diet rich in fruits and vegetables.
|
| 113 |
+
- Stay physically active.
|
| 114 |
+
- Limit alcohol consumption.
|
| 115 |
+
- Regular screenings, including mammograms and self-exams.
|
| 116 |
+
""")
|
| 117 |
+
|
| 118 |
+
# Outlook and Prognosis section
|
| 119 |
+
st.header("Outlook / Prognosis")
|
| 120 |
+
st.write("""
|
| 121 |
+
Survival rates depend on the cancer’s stage and type. Generally, the five-year survival rate for localized cancer is 99%. Early detection and treatment improve outcomes significantly. However, metastatic breast cancer, which spreads to other organs, has a five-year survival rate of about 30%.
|
| 122 |
+
""")
|
| 123 |
+
|
| 124 |
+
# Living with Breast Cancer
|
| 125 |
+
st.header("Living With")
|
| 126 |
+
st.write("""
|
| 127 |
+
Managing breast cancer can be challenging. It’s important to get enough rest, eat a balanced diet, manage stress, and seek support through survivorship programs.
|
| 128 |
+
|
| 129 |
+
Regular check-ins with healthcare providers can help monitor for any new symptoms or complications.
|
| 130 |
+
""")
|
| 131 |
+
|
| 132 |
+
# Additional questions section
|
| 133 |
+
st.header("Additional Common Questions")
|
| 134 |
+
st.write("""
|
| 135 |
+
- How long can you have breast cancer without knowing? You can have breast cancer for years before noticing symptoms.
|
| 136 |
+
- Can men get breast cancer? Yes, although it's rare, men and people AMAB can develop breast cancer.
|
| 137 |
+
""")
|
assets/style.css
ADDED
|
@@ -0,0 +1,19 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
.css-j5r0tf {
|
| 2 |
+
padding: 1rem;
|
| 3 |
+
border-radius: 0.5rem;
|
| 4 |
+
background-color: #7E99AB;
|
| 5 |
+
}
|
| 6 |
+
|
| 7 |
+
.diagnosis {
|
| 8 |
+
color: #fff;
|
| 9 |
+
padding: 0.2em 0.5em;
|
| 10 |
+
border-radius: 0.5em;
|
| 11 |
+
}
|
| 12 |
+
|
| 13 |
+
.diagnosis.benign {
|
| 14 |
+
background-color: #01DB4B
|
| 15 |
+
}
|
| 16 |
+
|
| 17 |
+
.diagnosis.malicious {
|
| 18 |
+
background-color: #ff4b4b
|
| 19 |
+
}
|
breast-cancer-wisconsin.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
charts/DecisionTreeClassifier(random_state=42)_confusion_matrix.png
ADDED
_confusion_matrix.png)
|
charts/DecisionTreeClassifier(random_state=42)_normalized_confusion_matrix.png
ADDED
_normalized_confusion_matrix.png)
|
charts/Features_with_Correlation.png
ADDED
|
charts/GaussianNB()_confusion_matrix.png
ADDED
_confusion_matrix.png)
|
charts/GaussianNB()_normalized_confusion_matrix.png
ADDED
_normalized_confusion_matrix.png)
|
charts/KNeighborsClassifier(n_neighbors=2)_confusion_matrix.png
ADDED
_confusion_matrix.png)
|
charts/KNeighborsClassifier(n_neighbors=2)_normalized_confusion_matrix.png
ADDED
_normalized_confusion_matrix.png)
|
charts/LogisticRegression()_confusion_matrix.png
ADDED
_confusion_matrix.png)
|
charts/LogisticRegression()_normalized_confusion_matrix.png
ADDED
_normalized_confusion_matrix.png)
|
charts/RandomForestClassifier(random_state=0)_confusion_matrix.png
ADDED
_confusion_matrix.png)
|
charts/RandomForestClassifier(random_state=0)_normalized_confusion_matrix.png
ADDED
_normalized_confusion_matrix.png)
|
charts/clustermap_high_correlation.png
ADDED

|
charts/correlation_heatmap.png
ADDED
|
charts/model_Accuracy.png
ADDED
|
charts/pairplot_high_correlation.png
ADDED

|
charts/target_count.png
ADDED

|
data/data.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
full code.py
ADDED
|
@@ -0,0 +1,196 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Data preparation and import libraries
|
| 2 |
+
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import numpy as np
|
| 5 |
+
import datetime as dt
|
| 6 |
+
# import matplotlib.pyplot as plt
|
| 7 |
+
# import seaborn as sns
|
| 8 |
+
import plotly.express as px
|
| 9 |
+
|
| 10 |
+
# %matplotlib inline
|
| 11 |
+
|
| 12 |
+
import warnings
|
| 13 |
+
warnings.filterwarnings('ignore')
|
| 14 |
+
|
| 15 |
+
from sklearn.model_selection import train_test_split
|
| 16 |
+
from sklearn.preprocessing import StandardScaler
|
| 17 |
+
from sklearn.linear_model import LogisticRegression
|
| 18 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 19 |
+
from sklearn.ensemble import RandomForestClassifier
|
| 20 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 21 |
+
from sklearn.naive_bayes import GaussianNB
|
| 22 |
+
|
| 23 |
+
from sklearn.metrics import classification_report, accuracy_score
|
| 24 |
+
from sklearn.metrics import confusion_matrix
|
| 25 |
+
|
| 26 |
+
data = pd.read_csv('/kaggle/input/breast-cancer-wisconsin-data/data.csv')
|
| 27 |
+
|
| 28 |
+
# Data Exploration and Data Cleaning
|
| 29 |
+
|
| 30 |
+
data.head()
|
| 31 |
+
|
| 32 |
+
data.shape
|
| 33 |
+
|
| 34 |
+
data.info()
|
| 35 |
+
|
| 36 |
+
# Check Duplication
|
| 37 |
+
data.duplicated().sum()
|
| 38 |
+
|
| 39 |
+
# There aren't duplicate values
|
| 40 |
+
|
| 41 |
+
# check Missing value
|
| 42 |
+
data.isna().sum()
|
| 43 |
+
|
| 44 |
+
#- No Missing Value is avalible
|
| 45 |
+
|
| 46 |
+
# Check the number of unique values of each column
|
| 47 |
+
data.nunique()
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
#- Dropping the id and Unnamed: 32 columns which will not provide any information for our model
|
| 51 |
+
data = data.drop(['id','Unnamed: 32'], axis= 1)
|
| 52 |
+
|
| 53 |
+
#- Changed the name of the diagnostic column to target!
|
| 54 |
+
data = data.rename(columns={'diagnosis' : 'target'})
|
| 55 |
+
|
| 56 |
+
#- changed taget data in the dataset. I changed malignant to 1 and benign to 0.
|
| 57 |
+
data.target.replace({'M' : '1','B': '0'},inplace=True)
|
| 58 |
+
|
| 59 |
+
# Converting target type to int64
|
| 60 |
+
data.target = data.target.astype('float64')
|
| 61 |
+
|
| 62 |
+
### Data processing result
|
| 63 |
+
data.head()
|
| 64 |
+
|
| 65 |
+
data.tail()
|
| 66 |
+
|
| 67 |
+
data.info()
|
| 68 |
+
|
| 69 |
+
# Check statistic of dataset
|
| 70 |
+
data.describe().T
|
| 71 |
+
|
| 72 |
+
|
| 73 |
+
# Analysis📝 & EDA📊
|
| 74 |
+
|
| 75 |
+
# I looked at how many benign and malignant yields there are.
|
| 76 |
+
data.target.value_counts()
|
| 77 |
+
|
| 78 |
+
# visualized target data in the dataset.
|
| 79 |
+
data['target'].value_counts().plot(kind='bar',edgecolor='black',color=['lightsteelblue','navajowhite'])
|
| 80 |
+
plt.title(" Target",fontsize=20)
|
| 81 |
+
plt.show()
|
| 82 |
+
|
| 83 |
+
#- 1-->Malignant
|
| 84 |
+
#- 0-->Benign
|
| 85 |
+
|
| 86 |
+
|
| 87 |
+
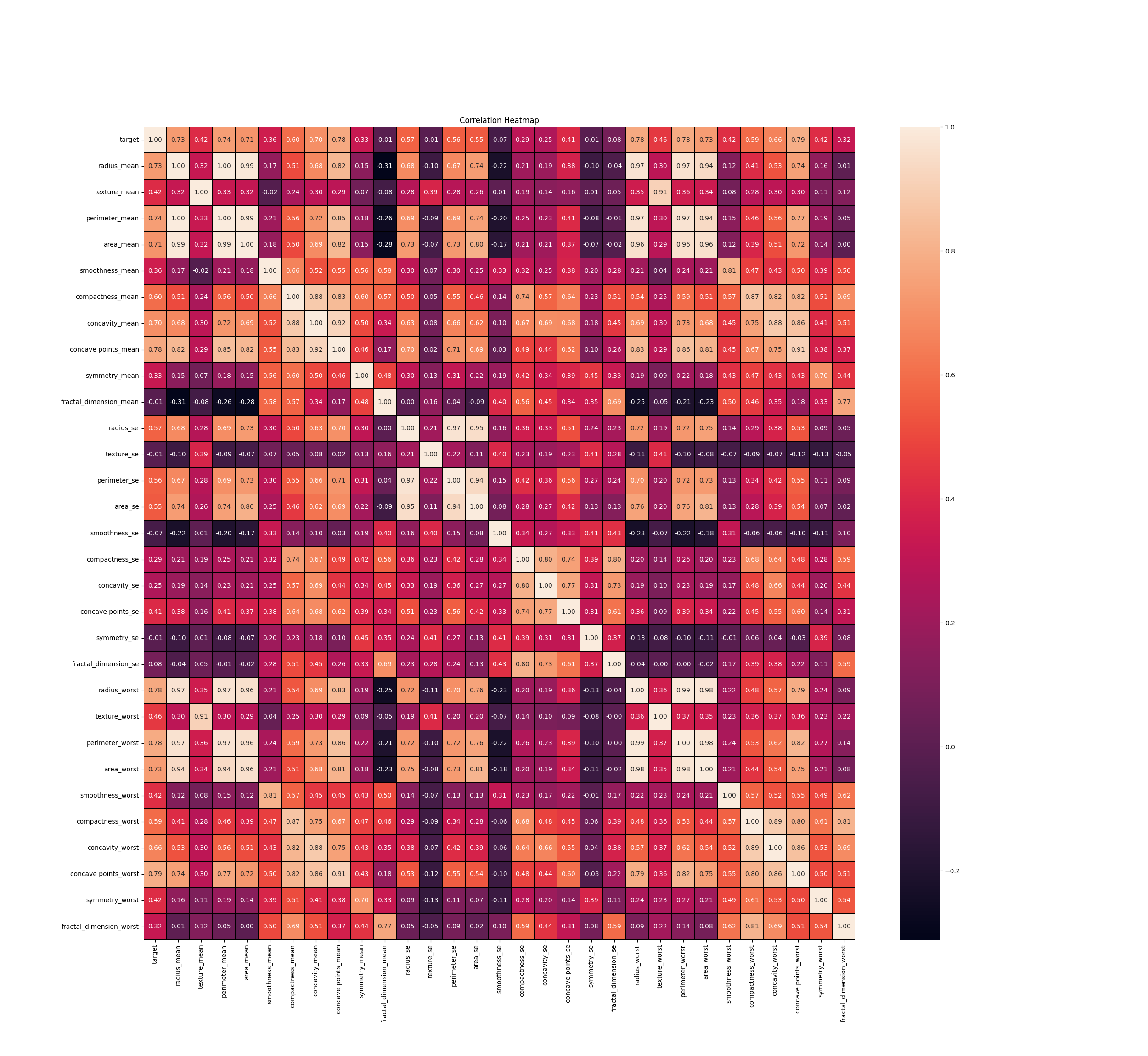
# Correlation Analysis
|
| 88 |
+
cor = data.corr()
|
| 89 |
+
cor
|
| 90 |
+
|
| 91 |
+
plt.figure(figsize=(25,23))
|
| 92 |
+
sns.heatmap(cor, annot= True, linewidths= 0.3 ,linecolor = "black", fmt = ".2f")
|
| 93 |
+
plt.title('Correlation Heatmap')
|
| 94 |
+
plt.show()
|
| 95 |
+
|
| 96 |
+
#- Data with a correlation greater than 0.75.
|
| 97 |
+
|
| 98 |
+
threshold = 0.75
|
| 99 |
+
filtre = np.abs(cor["target"] > threshold)
|
| 100 |
+
corr_features = cor.columns[filtre].tolist()
|
| 101 |
+
plt.figure(figsize=(10,8))
|
| 102 |
+
sns.clustermap(data[corr_features].corr(), annot = True, fmt = ".2f")
|
| 103 |
+
plt.title("\n Correlation Between Features with Cor Thresgold [0.75]\n",fontsize=20)
|
| 104 |
+
plt.show()
|
| 105 |
+
|
| 106 |
+
### visualized the data with a correlation greater than 0.75
|
| 107 |
+
|
| 108 |
+
sns.pairplot(data[corr_features], diag_kind = "kde" , markers = "*", hue="target")
|
| 109 |
+
plt.show()
|
| 110 |
+
|
| 111 |
+
# Machine Learning Model Evaluation
|
| 112 |
+
|
| 113 |
+
# Splitting data
|
| 114 |
+
x= data.drop('target',axis=1)
|
| 115 |
+
y= data['target']
|
| 116 |
+
|
| 117 |
+
|
| 118 |
+
x_train, x_test, y_train, y_test = train_test_split(x,y,test_size=0.30,random_state=101)
|
| 119 |
+
|
| 120 |
+
s= StandardScaler()
|
| 121 |
+
x_train = s.fit_transform(x_train)
|
| 122 |
+
x_test = s.fit_transform(x_test)
|
| 123 |
+
|
| 124 |
+
algorithm = ['KNeighborsClassifier','RandomForestClassifier','DecisionTreeClassifier','GaussianNB','LogisticRegression']
|
| 125 |
+
Accuracy=[]
|
| 126 |
+
|
| 127 |
+
def all(model):
|
| 128 |
+
model.fit(x_train,y_train)
|
| 129 |
+
pred = model.predict(x_test)
|
| 130 |
+
|
| 131 |
+
acc=accuracy_score(y_test,pred)
|
| 132 |
+
Accuracy.append(acc)
|
| 133 |
+
|
| 134 |
+
# confusion matrix without Normalization
|
| 135 |
+
print('confusion matrix')
|
| 136 |
+
# Calculate confusion matrix
|
| 137 |
+
cm = confusion_matrix(y_test,pred)
|
| 138 |
+
# Plot the confusion matrix
|
| 139 |
+
sns.heatmap(cm, annot=True, fmt='d',cmap=['lightsteelblue','navajowhite'])
|
| 140 |
+
plt.title('Confusion matrix')
|
| 141 |
+
plt.xlabel('Predcted lablel')
|
| 142 |
+
plt.ylabel('True lable')
|
| 143 |
+
plt.show()
|
| 144 |
+
|
| 145 |
+
# confusion matrix without Normalization
|
| 146 |
+
print('Normalized confusion matrix')
|
| 147 |
+
# Calculate confusion matrix
|
| 148 |
+
cm1 = confusion_matrix(y_test,pred, normalize='true')
|
| 149 |
+
# Plot the confusion matrix
|
| 150 |
+
sns.heatmap(cm1, annot=True,cmap=['lightsteelblue','navajowhite'])
|
| 151 |
+
plt.title('Normalized Confusion matrix')
|
| 152 |
+
plt.xlabel('Predcted lablel')
|
| 153 |
+
plt.ylabel('True lable')
|
| 154 |
+
plt.show()
|
| 155 |
+
|
| 156 |
+
# print Confusion matrix, Classification report and accuracy report
|
| 157 |
+
print(cm)
|
| 158 |
+
print(classification_report(y_test,pred))
|
| 159 |
+
print('accuracy_score : ' , acc)
|
| 160 |
+
|
| 161 |
+
|
| 162 |
+
### KNN machine learning model Evaluation for breast cancer
|
| 163 |
+
|
| 164 |
+
model_1 =KNeighborsClassifier(n_neighbors=2)
|
| 165 |
+
all(model_1)
|
| 166 |
+
|
| 167 |
+
|
| 168 |
+
### RandomForest
|
| 169 |
+
model_2= RandomForestClassifier(n_estimators=100,random_state=0)
|
| 170 |
+
all(model_2)
|
| 171 |
+
|
| 172 |
+
### DecisionTree
|
| 173 |
+
|
| 174 |
+
model_3 = DecisionTreeClassifier(random_state=42)
|
| 175 |
+
all(model_3)
|
| 176 |
+
|
| 177 |
+
|
| 178 |
+
### Naive_bayes
|
| 179 |
+
model_4 = GaussianNB()
|
| 180 |
+
all(model_4)
|
| 181 |
+
|
| 182 |
+
### Logistic Regression
|
| 183 |
+
|
| 184 |
+
model_5 = LogisticRegression()
|
| 185 |
+
all(model_5)
|
| 186 |
+
|
| 187 |
+
df = pd.DataFrame({'Algorithm':algorithm,'Accuracy':Accuracy })
|
| 188 |
+
df
|
| 189 |
+
|
| 190 |
+
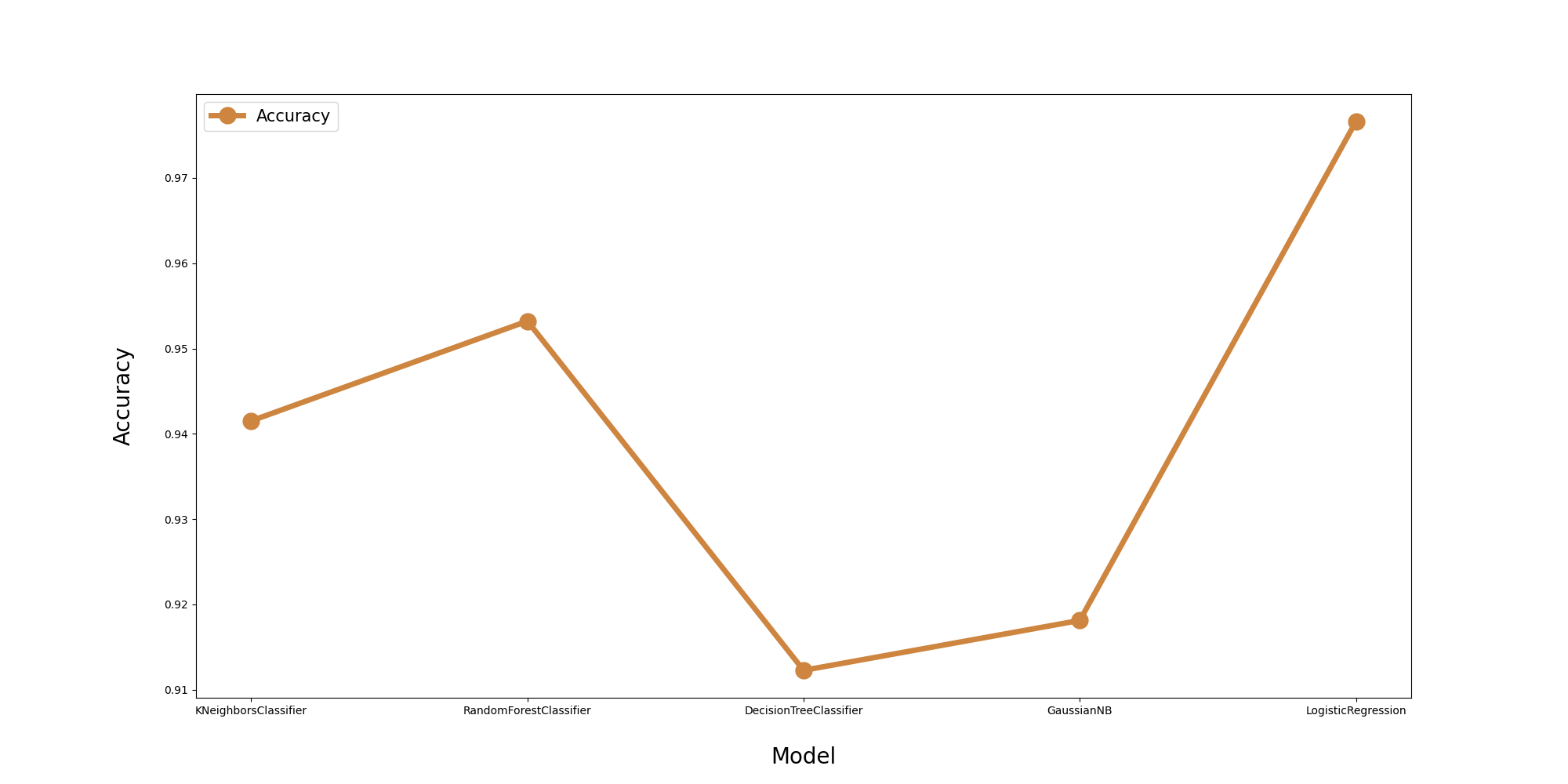
fig = plt.figure(figsize=(20,10))
|
| 191 |
+
plt.plot(df.Algorithm,df.Accuracy,label='Accuracy',lw=5,color='peru',marker='o',markersize = 15)
|
| 192 |
+
plt.legend(fontsize=15)
|
| 193 |
+
plt.xlabel('\nModel',fontsize= 20)
|
| 194 |
+
plt.ylabel('Accuracy\n',fontsize= 20)
|
| 195 |
+
plt.show()
|
| 196 |
+
|
images/1.jpg
ADDED

|
images/2.jpg
ADDED

|
images/3.jpg
ADDED
|
images/4.gif
ADDED

|
Git LFS Details
|
model/main.py
ADDED
|
@@ -0,0 +1,57 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
from sklearn.preprocessing import StandardScaler
|
| 3 |
+
from sklearn.model_selection import train_test_split
|
| 4 |
+
from sklearn.linear_model import LogisticRegression
|
| 5 |
+
from sklearn.metrics import accuracy_score, classification_report
|
| 6 |
+
import pickle5 as pickle
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
def create_model(data):
|
| 10 |
+
X = data.drop(['diagnosis'], axis=1)
|
| 11 |
+
y = data['diagnosis']
|
| 12 |
+
|
| 13 |
+
# scale the data
|
| 14 |
+
scaler = StandardScaler()
|
| 15 |
+
X = scaler.fit_transform(X)
|
| 16 |
+
|
| 17 |
+
# split the data
|
| 18 |
+
X_train, X_test, y_train, y_test = train_test_split(
|
| 19 |
+
X, y, test_size=0.2, random_state=42
|
| 20 |
+
)
|
| 21 |
+
|
| 22 |
+
# train the model
|
| 23 |
+
model = LogisticRegression()
|
| 24 |
+
model.fit(X_train, y_train)
|
| 25 |
+
|
| 26 |
+
# test model
|
| 27 |
+
y_pred = model.predict(X_test)
|
| 28 |
+
print('Accuracy of our model: ', accuracy_score(y_test, y_pred))
|
| 29 |
+
print("Classification report: \n", classification_report(y_test, y_pred))
|
| 30 |
+
|
| 31 |
+
return model, scaler
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
def get_clean_data():
|
| 35 |
+
data = pd.read_csv("data/data.csv")
|
| 36 |
+
|
| 37 |
+
data = data.drop(['Unnamed: 32', 'id'], axis=1)
|
| 38 |
+
|
| 39 |
+
data['diagnosis'] = data['diagnosis'].map({ 'M': 1, 'B': 0 })
|
| 40 |
+
|
| 41 |
+
return data
|
| 42 |
+
|
| 43 |
+
|
| 44 |
+
def main():
|
| 45 |
+
data = get_clean_data()
|
| 46 |
+
|
| 47 |
+
model, scaler = create_model(data)
|
| 48 |
+
|
| 49 |
+
with open('model/model.pkl', 'wb') as f:
|
| 50 |
+
pickle.dump(model, f)
|
| 51 |
+
|
| 52 |
+
with open('model/scaler.pkl', 'wb') as f:
|
| 53 |
+
pickle.dump(scaler, f)
|
| 54 |
+
|
| 55 |
+
|
| 56 |
+
if __name__ == '__main__':
|
| 57 |
+
main()
|
model/model.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a2bc2faeb5cc85f2c6b5bd1a8acb5c21c6eaa363617416f370aa874a2dda4205
|
| 3 |
+
size 948
|
model/scaler.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:18c463cb78613d8d7721b49e42264018613fda151a19d8ceb160645c2e2c0e0b
|
| 3 |
+
size 1771
|
pages/📈_Analysis.py
ADDED
|
@@ -0,0 +1,227 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import numpy as np
|
| 4 |
+
import seaborn as sns
|
| 5 |
+
import matplotlib.pyplot as plt
|
| 6 |
+
import plotly.express as px
|
| 7 |
+
from sklearn.model_selection import train_test_split
|
| 8 |
+
from sklearn.preprocessing import StandardScaler
|
| 9 |
+
from sklearn.linear_model import LogisticRegression
|
| 10 |
+
from sklearn.neighbors import KNeighborsClassifier
|
| 11 |
+
from sklearn.ensemble import RandomForestClassifier
|
| 12 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 13 |
+
from sklearn.naive_bayes import GaussianNB
|
| 14 |
+
from sklearn.metrics import classification_report, accuracy_score, confusion_matrix
|
| 15 |
+
import os
|
| 16 |
+
|
| 17 |
+
# Create 'charts' folder if it doesn't exist
|
| 18 |
+
if not os.path.exists('charts'):
|
| 19 |
+
os.makedirs('charts')
|
| 20 |
+
|
| 21 |
+
# Function to save and show charts
|
| 22 |
+
def save_and_show_chart(fig, filename):
|
| 23 |
+
filepath = os.path.join('charts', filename)
|
| 24 |
+
fig.savefig(filepath)
|
| 25 |
+
st.image(filepath)
|
| 26 |
+
|
| 27 |
+
st.set_page_config(
|
| 28 |
+
page_title="Breast Cancer Analysis",
|
| 29 |
+
page_icon=":female-doctor:",
|
| 30 |
+
layout="wide",
|
| 31 |
+
initial_sidebar_state="expanded"
|
| 32 |
+
)
|
| 33 |
+
|
| 34 |
+
# Set page title
|
| 35 |
+
st.title('Breast Cancer Diagnosis - Machine Learning Model Evaluation')
|
| 36 |
+
st.divider()
|
| 37 |
+
|
| 38 |
+
# Breast Cancer Wisconsin Dataset Information
|
| 39 |
+
st.header("Breast Cancer Wisconsin (Diagnostic) Data Set")
|
| 40 |
+
st.write("""
|
| 41 |
+
The **Breast Cancer Wisconsin (Diagnostic) Data Set** is a collection of clinical breast cancer diagnostic data. The data includes features that describe characteristics of cell nuclei from breast cancer biopsies, which are used to predict whether a tumor is benign or malignant.
|
| 42 |
+
Key features in the dataset include:
|
| 43 |
+
- **Radius:** The average distance from the center to points on the perimeter
|
| 44 |
+
- **Texture:** The standard deviation of grayscale values
|
| 45 |
+
- **Perimeter, Area, Smoothness:** Other morphological features describing cell shapes
|
| 46 |
+
The dataset is commonly used for classification tasks in machine learning to predict the likelihood of breast cancer malignancy.
|
| 47 |
+
""")
|
| 48 |
+
|
| 49 |
+
# Footer or additional information
|
| 50 |
+
st.write("This application aims to provide insights into breast cancer through data analysis and prediction based on the Breast Cancer Wisconsin dataset.")
|
| 51 |
+
st.divider()
|
| 52 |
+
|
| 53 |
+
# Data Preparation and Import Libraries
|
| 54 |
+
st.header('Data Preparation')
|
| 55 |
+
data = pd.read_csv('./data/data.csv')
|
| 56 |
+
|
| 57 |
+
st.subheader('Data Preview (show only first 5 rows)')
|
| 58 |
+
st.write(data.head())
|
| 59 |
+
|
| 60 |
+
st.subheader('Data Shape')
|
| 61 |
+
st.success(f"There are {data.shape[0]} rows and {data.shape[1]} columns in this dataset.")
|
| 62 |
+
|
| 63 |
+
# st.subheader('Data Info')
|
| 64 |
+
# st.table(data.info())
|
| 65 |
+
st.divider()
|
| 66 |
+
st.subheader('Checking Duplicates and Missing Values')
|
| 67 |
+
st.success(f"There are {data.duplicated().sum()} duplicate rows in this dataset.")
|
| 68 |
+
st.write(data.isna().sum())
|
| 69 |
+
|
| 70 |
+
st.divider()
|
| 71 |
+
st.subheader('Dropping Irrelevant Columns')
|
| 72 |
+
data = data.drop(['id', 'Unnamed: 32'], axis=1)
|
| 73 |
+
st.success("**id** and **Unnamed** columned were deleted because these variables can not be used for classification.")
|
| 74 |
+
|
| 75 |
+
st.divider()
|
| 76 |
+
st.subheader('Renaming Columns')
|
| 77 |
+
st.write("In the dataset, the 'diagnosis' variable was renamed as 'target,' where the value 'M' (Malignant) was renamed to 1 and 'B' (Benign) was renamed to 0 for easier modeling.")
|
| 78 |
+
|
| 79 |
+
data = data.rename(columns={'diagnosis': 'target'})
|
| 80 |
+
df = data.copy()
|
| 81 |
+
data.target.replace({'M': '1', 'B': '0'}, inplace=True)
|
| 82 |
+
data.target = data.target.astype('float64')
|
| 83 |
+
|
| 84 |
+
st.write(data.head())
|
| 85 |
+
st.divider()
|
| 86 |
+
|
| 87 |
+
# Analysis & EDA
|
| 88 |
+
st.header('Analysis & EDA')
|
| 89 |
+
st.subheader('Target Value Counts')
|
| 90 |
+
|
| 91 |
+
y = df.target
|
| 92 |
+
ax = sns.countplot(y,label="Count") # M = 212, B = 357
|
| 93 |
+
B, M = y.value_counts()
|
| 94 |
+
st.write('Number of Benign (0): ',B)
|
| 95 |
+
st.write('Number of Malignant (1): ',M)
|
| 96 |
+
|
| 97 |
+
# Visualizing target data
|
| 98 |
+
st.subheader('Bar Plot of Target Values')
|
| 99 |
+
fig, ax = plt.subplots(figsize=(8, 6))
|
| 100 |
+
# data['target'].value_counts().plot(kind='bar', edgecolor='black', color=['lightsteelblue', 'navajowhite'], ax=ax)
|
| 101 |
+
sns.countplot(y, label="Count", ax=ax)
|
| 102 |
+
ax.set_title("Target Distribution")
|
| 103 |
+
# st.pyplot(fig)
|
| 104 |
+
save_and_show_chart(fig, 'target_count.png')
|
| 105 |
+
st.divider()
|
| 106 |
+
|
| 107 |
+
# Correlation Analysis
|
| 108 |
+
st.subheader('Correlation Analysis')
|
| 109 |
+
cor = data.corr()
|
| 110 |
+
st.write(cor)
|
| 111 |
+
st.divider()
|
| 112 |
+
|
| 113 |
+
st.subheader('Correlation Heatmap')
|
| 114 |
+
fig, ax = plt.subplots(figsize=(25, 23))
|
| 115 |
+
sns.heatmap(cor, annot=True, linewidths=0.3, linecolor="black", fmt=".2f", ax=ax)
|
| 116 |
+
ax.set_title('Correlation Heatmap')
|
| 117 |
+
# st.pyplot(fig)
|
| 118 |
+
save_and_show_chart(fig, 'correlation_heatmap.png')
|
| 119 |
+
st.divider()
|
| 120 |
+
|
| 121 |
+
st.subheader('Features with Correlation > 0.75')
|
| 122 |
+
threshold = 0.75
|
| 123 |
+
filtre = np.abs(cor["target"]) > threshold
|
| 124 |
+
corr_features = cor.columns[filtre].tolist()
|
| 125 |
+
|
| 126 |
+
cluster_map = sns.clustermap(
|
| 127 |
+
data[corr_features].corr(),
|
| 128 |
+
annot=True,
|
| 129 |
+
fmt=".2f",
|
| 130 |
+
figsize=(10, 8) # Adjust the figsize here
|
| 131 |
+
)
|
| 132 |
+
plt.savefig('charts/clustermap_high_correlation.png')
|
| 133 |
+
st.image('charts/clustermap_high_correlation.png')
|
| 134 |
+
# st.pyplot(cluster_map.fig)
|
| 135 |
+
st.divider()
|
| 136 |
+
|
| 137 |
+
# Pairplot for high-correlation features
|
| 138 |
+
st.subheader('Pairplot for Features with High Correlation')
|
| 139 |
+
fig, ax = plt.subplots(figsize=(8, 6))
|
| 140 |
+
pairplot = sns.pairplot(data[corr_features], diag_kind="kde", markers="*", hue="target")
|
| 141 |
+
plt.savefig('charts/pairplot_high_correlation.png')
|
| 142 |
+
st.image('charts/pairplot_high_correlation.png')
|
| 143 |
+
# st.pyplot(pairplot)
|
| 144 |
+
|
| 145 |
+
st.divider()
|
| 146 |
+
# Machine Learning Model Evaluation
|
| 147 |
+
st.header('Machine Learning Model Evaluation')
|
| 148 |
+
|
| 149 |
+
# Splitting the data
|
| 150 |
+
x = data.drop('target', axis=1)
|
| 151 |
+
y = data['target']
|
| 152 |
+
|
| 153 |
+
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.30, random_state=101)
|
| 154 |
+
|
| 155 |
+
scaler = StandardScaler()
|
| 156 |
+
x_train = scaler.fit_transform(x_train)
|
| 157 |
+
x_test = scaler.transform(x_test)
|
| 158 |
+
|
| 159 |
+
algorithm = ['KNeighborsClassifier', 'RandomForestClassifier', 'DecisionTreeClassifier', 'GaussianNB', 'LogisticRegression']
|
| 160 |
+
Accuracy = []
|
| 161 |
+
|
| 162 |
+
def evaluate_model(model):
|
| 163 |
+
model.fit(x_train, y_train)
|
| 164 |
+
pred = model.predict(x_test)
|
| 165 |
+
acc = accuracy_score(y_test, pred)
|
| 166 |
+
Accuracy.append(acc)
|
| 167 |
+
|
| 168 |
+
# Confusion Matrix
|
| 169 |
+
cm = confusion_matrix(y_test, pred)
|
| 170 |
+
st.subheader(f'Confusion Matrix for {model.__class__.__name__}')
|
| 171 |
+
fig, ax = plt.subplots()
|
| 172 |
+
sns.heatmap(cm, annot=True, fmt='d', cmap='Blues', ax=ax)
|
| 173 |
+
save_and_show_chart(fig, f'{model}_confusion_matrix.png')
|
| 174 |
+
# st.pyplot(fig)
|
| 175 |
+
|
| 176 |
+
# Normalized Confusion Matrix
|
| 177 |
+
cm_norm = confusion_matrix(y_test, pred, normalize='true')
|
| 178 |
+
st.subheader(f'Normalized Confusion Matrix for {model.__class__.__name__}')
|
| 179 |
+
fig, ax = plt.subplots()
|
| 180 |
+
sns.heatmap(cm_norm, annot=True, cmap='Blues', ax=ax)
|
| 181 |
+
save_and_show_chart(fig, f'{model}_normalized_confusion_matrix.png')
|
| 182 |
+
# st.pyplot(fig)
|
| 183 |
+
|
| 184 |
+
# Classification Report
|
| 185 |
+
st.subheader(f'Classification Report for {model.__class__.__name__}')
|
| 186 |
+
st.text(classification_report(y_test, pred))
|
| 187 |
+
st.write(f"Accuracy: {acc}")
|
| 188 |
+
|
| 189 |
+
# Evaluating different models
|
| 190 |
+
st.subheader('01. KNeighborsClassifier Evaluation')
|
| 191 |
+
model_knn = KNeighborsClassifier(n_neighbors=2)
|
| 192 |
+
evaluate_model(model_knn)
|
| 193 |
+
st.divider()
|
| 194 |
+
|
| 195 |
+
st.subheader('02. RandomForestClassifier Evaluation')
|
| 196 |
+
model_rf = RandomForestClassifier(n_estimators=100, random_state=0)
|
| 197 |
+
evaluate_model(model_rf)
|
| 198 |
+
st.divider()
|
| 199 |
+
|
| 200 |
+
st.subheader('03. DecisionTreeClassifier Evaluation')
|
| 201 |
+
model_dt = DecisionTreeClassifier(random_state=42)
|
| 202 |
+
evaluate_model(model_dt)
|
| 203 |
+
st.divider()
|
| 204 |
+
|
| 205 |
+
st.subheader('04. GaussianNB Evaluation')
|
| 206 |
+
model_nb = GaussianNB()
|
| 207 |
+
evaluate_model(model_nb)
|
| 208 |
+
st.divider()
|
| 209 |
+
|
| 210 |
+
st.subheader('05. LogisticRegression Evaluation')
|
| 211 |
+
model_lr = LogisticRegression()
|
| 212 |
+
evaluate_model(model_lr)
|
| 213 |
+
st.divider()
|
| 214 |
+
|
| 215 |
+
# Final Accuracy Plot
|
| 216 |
+
st.header('Model Accuracy Comparison')
|
| 217 |
+
df = pd.DataFrame({'Algorithm': algorithm, 'Accuracy': Accuracy})
|
| 218 |
+
|
| 219 |
+
fig, ax = plt.subplots(figsize=(20, 10))
|
| 220 |
+
ax.plot(df.Algorithm, df.Accuracy, label='Accuracy', lw=5, color='peru', marker='o', markersize=15)
|
| 221 |
+
ax.legend(fontsize=15)
|
| 222 |
+
ax.set_xlabel('\nModel', fontsize=20)
|
| 223 |
+
ax.set_ylabel('Accuracy\n', fontsize=20)
|
| 224 |
+
save_and_show_chart(fig, 'model_Accuracy.png')
|
| 225 |
+
# st.pyplot(fig)
|
| 226 |
+
|
| 227 |
+
# End of Streamlit app
|
pages/📊_Predict.py
ADDED
|
@@ -0,0 +1,195 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import streamlit as st
|
| 2 |
+
import pickle
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import plotly.graph_objects as go
|
| 5 |
+
import numpy as np
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
def get_clean_data():
|
| 9 |
+
data = pd.read_csv("data/data.csv")
|
| 10 |
+
|
| 11 |
+
data = data.drop(['Unnamed: 32', 'id'], axis=1)
|
| 12 |
+
|
| 13 |
+
data['diagnosis'] = data['diagnosis'].map({ 'M': 1, 'B': 0 })
|
| 14 |
+
|
| 15 |
+
return data
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
def add_sidebar():
|
| 19 |
+
st.sidebar.header("Cell Nuclei Measurements")
|
| 20 |
+
|
| 21 |
+
data = get_clean_data()
|
| 22 |
+
|
| 23 |
+
slider_labels = [
|
| 24 |
+
("Radius (mean)", "radius_mean"),
|
| 25 |
+
("Texture (mean)", "texture_mean"),
|
| 26 |
+
("Perimeter (mean)", "perimeter_mean"),
|
| 27 |
+
("Area (mean)", "area_mean"),
|
| 28 |
+
("Smoothness (mean)", "smoothness_mean"),
|
| 29 |
+
("Compactness (mean)", "compactness_mean"),
|
| 30 |
+
("Concavity (mean)", "concavity_mean"),
|
| 31 |
+
("Concave points (mean)", "concave points_mean"),
|
| 32 |
+
("Symmetry (mean)", "symmetry_mean"),
|
| 33 |
+
("Fractal dimension (mean)", "fractal_dimension_mean"),
|
| 34 |
+
("Radius (se)", "radius_se"),
|
| 35 |
+
("Texture (se)", "texture_se"),
|
| 36 |
+
("Perimeter (se)", "perimeter_se"),
|
| 37 |
+
("Area (se)", "area_se"),
|
| 38 |
+
("Smoothness (se)", "smoothness_se"),
|
| 39 |
+
("Compactness (se)", "compactness_se"),
|
| 40 |
+
("Concavity (se)", "concavity_se"),
|
| 41 |
+
("Concave points (se)", "concave points_se"),
|
| 42 |
+
("Symmetry (se)", "symmetry_se"),
|
| 43 |
+
("Fractal dimension (se)", "fractal_dimension_se"),
|
| 44 |
+
("Radius (worst)", "radius_worst"),
|
| 45 |
+
("Texture (worst)", "texture_worst"),
|
| 46 |
+
("Perimeter (worst)", "perimeter_worst"),
|
| 47 |
+
("Area (worst)", "area_worst"),
|
| 48 |
+
("Smoothness (worst)", "smoothness_worst"),
|
| 49 |
+
("Compactness (worst)", "compactness_worst"),
|
| 50 |
+
("Concavity (worst)", "concavity_worst"),
|
| 51 |
+
("Concave points (worst)", "concave points_worst"),
|
| 52 |
+
("Symmetry (worst)", "symmetry_worst"),
|
| 53 |
+
("Fractal dimension (worst)", "fractal_dimension_worst"),
|
| 54 |
+
]
|
| 55 |
+
|
| 56 |
+
input_dict = {}
|
| 57 |
+
|
| 58 |
+
for label, key in slider_labels:
|
| 59 |
+
input_dict[key] = st.sidebar.slider(
|
| 60 |
+
label,
|
| 61 |
+
min_value=float(0),
|
| 62 |
+
max_value=float(data[key].max()),
|
| 63 |
+
value=float(data[key].mean())
|
| 64 |
+
)
|
| 65 |
+
|
| 66 |
+
return input_dict
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
def get_scaled_values(input_dict):
|
| 70 |
+
data = get_clean_data()
|
| 71 |
+
|
| 72 |
+
X = data.drop(['diagnosis'], axis=1)
|
| 73 |
+
|
| 74 |
+
scaled_dict = {}
|
| 75 |
+
|
| 76 |
+
for key, value in input_dict.items():
|
| 77 |
+
max_val = X[key].max()
|
| 78 |
+
min_val = X[key].min()
|
| 79 |
+
scaled_value = (value - min_val) / (max_val - min_val)
|
| 80 |
+
scaled_dict[key] = scaled_value
|
| 81 |
+
|
| 82 |
+
return scaled_dict
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
def get_radar_chart(input_data):
|
| 86 |
+
|
| 87 |
+
input_data = get_scaled_values(input_data)
|
| 88 |
+
|
| 89 |
+
categories = ['Radius', 'Texture', 'Perimeter', 'Area',
|
| 90 |
+
'Smoothness', 'Compactness',
|
| 91 |
+
'Concavity', 'Concave Points',
|
| 92 |
+
'Symmetry', 'Fractal Dimension']
|
| 93 |
+
|
| 94 |
+
fig = go.Figure()
|
| 95 |
+
|
| 96 |
+
fig.add_trace(go.Scatterpolar(
|
| 97 |
+
r=[
|
| 98 |
+
input_data['radius_mean'], input_data['texture_mean'], input_data['perimeter_mean'],
|
| 99 |
+
input_data['area_mean'], input_data['smoothness_mean'], input_data['compactness_mean'],
|
| 100 |
+
input_data['concavity_mean'], input_data['concave points_mean'], input_data['symmetry_mean'],
|
| 101 |
+
input_data['fractal_dimension_mean']
|
| 102 |
+
],
|
| 103 |
+
theta=categories,
|
| 104 |
+
fill='toself',
|
| 105 |
+
name='Mean Value'
|
| 106 |
+
))
|
| 107 |
+
fig.add_trace(go.Scatterpolar(
|
| 108 |
+
r=[
|
| 109 |
+
input_data['radius_se'], input_data['texture_se'], input_data['perimeter_se'], input_data['area_se'],
|
| 110 |
+
input_data['smoothness_se'], input_data['compactness_se'], input_data['concavity_se'],
|
| 111 |
+
input_data['concave points_se'], input_data['symmetry_se'],input_data['fractal_dimension_se']
|
| 112 |
+
],
|
| 113 |
+
theta=categories,
|
| 114 |
+
fill='toself',
|
| 115 |
+
name='Standard Error'
|
| 116 |
+
))
|
| 117 |
+
fig.add_trace(go.Scatterpolar(
|
| 118 |
+
r=[
|
| 119 |
+
input_data['radius_worst'], input_data['texture_worst'], input_data['perimeter_worst'],
|
| 120 |
+
input_data['area_worst'], input_data['smoothness_worst'], input_data['compactness_worst'],
|
| 121 |
+
input_data['concavity_worst'], input_data['concave points_worst'], input_data['symmetry_worst'],
|
| 122 |
+
input_data['fractal_dimension_worst']
|
| 123 |
+
],
|
| 124 |
+
theta=categories,
|
| 125 |
+
fill='toself',
|
| 126 |
+
name='Worst Value'
|
| 127 |
+
))
|
| 128 |
+
|
| 129 |
+
fig.update_layout(
|
| 130 |
+
polar=dict(
|
| 131 |
+
radialaxis=dict(
|
| 132 |
+
visible=True,
|
| 133 |
+
range=[0, 1]
|
| 134 |
+
)),
|
| 135 |
+
showlegend=True
|
| 136 |
+
)
|
| 137 |
+
|
| 138 |
+
return fig
|
| 139 |
+
|
| 140 |
+
|
| 141 |
+
def add_predictions(input_data):
|
| 142 |
+
model = pickle.load(open("model/model.pkl", "rb"))
|
| 143 |
+
scaler = pickle.load(open("model/scaler.pkl", "rb"))
|
| 144 |
+
|
| 145 |
+
input_array = np.array(list(input_data.values())).reshape(1, -1)
|
| 146 |
+
|
| 147 |
+
input_array_scaled = scaler.transform(input_array)
|
| 148 |
+
|
| 149 |
+
prediction = model.predict(input_array_scaled)
|
| 150 |
+
|
| 151 |
+
st.subheader("Cell cluster prediction")
|
| 152 |
+
st.write("The cell cluster is:")
|
| 153 |
+
|
| 154 |
+
if prediction[0] == 0:
|
| 155 |
+
st.write("<span class='diagnosis benign'>Benign</span>", unsafe_allow_html=True)
|
| 156 |
+
else:
|
| 157 |
+
st.write("<span class='diagnosis malicious'>Malicious</span>", unsafe_allow_html=True)
|
| 158 |
+
|
| 159 |
+
|
| 160 |
+
st.write("Probability of being benign: ", model.predict_proba(input_array_scaled)[0][0])
|
| 161 |
+
st.write("Probability of being malicious: ", model.predict_proba(input_array_scaled)[0][1])
|
| 162 |
+
|
| 163 |
+
st.write("This app can assist medical professionals in making a diagnosis, but should not be used as a substitute for a professional diagnosis.")
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
def main():
|
| 168 |
+
st.set_page_config(
|
| 169 |
+
page_title="Breast Cancer Predictor",
|
| 170 |
+
page_icon=":female-doctor:",
|
| 171 |
+
layout="wide",
|
| 172 |
+
initial_sidebar_state="expanded"
|
| 173 |
+
)
|
| 174 |
+
|
| 175 |
+
with open("assets/style.css") as f:
|
| 176 |
+
st.markdown("<style>{}</style>".format(f.read()), unsafe_allow_html=True)
|
| 177 |
+
|
| 178 |
+
input_data = add_sidebar()
|
| 179 |
+
|
| 180 |
+
with st.container():
|
| 181 |
+
st.title("Breast Cancer Predictor")
|
| 182 |
+
st.write("Please connect this app to your cytology lab to help diagnose breast cancer form your tissue sample. This app predicts using a machine learning model whether a breast mass is benign or malignant based on the measurements it receives from your cytosis lab. You can also update the measurements by hand using the sliders in the sidebar. ")
|
| 183 |
+
|
| 184 |
+
col1, col2 = st.columns([4,1])
|
| 185 |
+
|
| 186 |
+
with col1:
|
| 187 |
+
radar_chart = get_radar_chart(input_data)
|
| 188 |
+
st.plotly_chart(radar_chart)
|
| 189 |
+
with col2:
|
| 190 |
+
add_predictions(input_data)
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
|
| 194 |
+
if __name__ == '__main__':
|
| 195 |
+
main()
|
requirements.txt
ADDED
|
@@ -0,0 +1,54 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
altair==5.4.1
|
| 2 |
+
attrs==24.2.0
|
| 3 |
+
blinker==1.8.2
|
| 4 |
+
cachetools==5.5.0
|
| 5 |
+
certifi==2024.8.30
|
| 6 |
+
charset-normalizer==3.3.2
|
| 7 |
+
click==8.1.7
|
| 8 |
+
colorama==0.4.6
|
| 9 |
+
contourpy==1.3.0
|
| 10 |
+
cycler==0.12.1
|
| 11 |
+
fonttools==4.53.1
|
| 12 |
+
gitdb==4.0.11
|
| 13 |
+
GitPython==3.1.43
|
| 14 |
+
idna==3.8
|
| 15 |
+
Jinja2==3.1.4
|
| 16 |
+
joblib==1.4.2
|
| 17 |
+
jsonschema==4.23.0
|
| 18 |
+
jsonschema-specifications==2023.12.1
|
| 19 |
+
kiwisolver==1.4.7
|
| 20 |
+
markdown-it-py==3.0.0
|
| 21 |
+
MarkupSafe==2.1.5
|
| 22 |
+
matplotlib==3.9.2
|
| 23 |
+
mdurl==0.1.2
|
| 24 |
+
narwhals==1.6.4
|
| 25 |
+
numpy==2.1.1
|
| 26 |
+
packaging==24.1
|
| 27 |
+
pandas==2.2.2
|
| 28 |
+
pillow==10.4.0
|
| 29 |
+
plotly==5.24.0
|
| 30 |
+
protobuf==5.28.0
|
| 31 |
+
pyarrow==17.0.0
|
| 32 |
+
pydeck==0.9.1
|
| 33 |
+
Pygments==2.18.0
|
| 34 |
+
pyparsing==3.1.4
|
| 35 |
+
python-dateutil==2.9.0.post0
|
| 36 |
+
pytz==2024.1
|
| 37 |
+
referencing==0.35.1
|
| 38 |
+
requests==2.32.3
|
| 39 |
+
rich==13.8.1
|
| 40 |
+
rpds-py==0.20.0
|
| 41 |
+
scikit-learn==1.5.1
|
| 42 |
+
scipy==1.14.1
|
| 43 |
+
seaborn==0.13.2
|
| 44 |
+
six==1.16.0
|
| 45 |
+
smmap==5.0.1
|
| 46 |
+
streamlit==1.38.0
|
| 47 |
+
tenacity==8.5.0
|
| 48 |
+
threadpoolctl==3.5.0
|
| 49 |
+
toml==0.10.2
|
| 50 |
+
tornado==6.4.1
|
| 51 |
+
typing_extensions==4.12.2
|
| 52 |
+
tzdata==2024.1
|
| 53 |
+
urllib3==2.2.2
|
| 54 |
+
watchdog==4.0.2
|