Spaces:
Running
Running
cleaning up beginning of documentation
Browse files- documentation.html +95 -84
- img/install_orthanc.png +0 -0
documentation.html
CHANGED
|
@@ -32,102 +32,51 @@
|
|
| 32 |
<div class="col-lg-11">
|
| 33 |
<h1 class="mb-4 mt-4">Documentation</h1>
|
| 34 |
|
| 35 |
-
<section id="
|
| 36 |
-
<h2>
|
| 37 |
-
<p>Instructions for installing PARROT on your system... Warning: The Orthanc server will run on port 8042 of your local computer and the nginx server on port 2000.</p>
|
| 38 |
-
<p>The installation package includes all servers. However, the Orthanc server is a manual installation. The installation instructions can be found below.</p>
|
| 39 |
-
</section>
|
| 40 |
-
|
| 41 |
-
<section id="PARROT-local-application">
|
| 42 |
-
<h2>PARROT Local Application</h2>
|
| 43 |
<p>
|
| 44 |
-
|
| 45 |
-
The application provides different modules that allow you to
|
| 46 |
</p>
|
| 47 |
<ul>
|
| 48 |
<li>
|
| 49 |
-
<p>
|
| 50 |
</li>
|
| 51 |
|
| 52 |
<li>
|
| 53 |
-
<p>
|
| 54 |
</li>
|
| 55 |
|
| 56 |
<li>
|
| 57 |
-
<p>
|
| 58 |
</li>
|
| 59 |
|
| 60 |
<li>
|
| 61 |
-
<p>
|
| 62 |
</li>
|
| 63 |
</ul>
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 64 |
<p>
|
| 65 |
-
|
| 66 |
-
prediction of the integrated trained AI model, and an Orthanc server for the storage of the study data. The installation package includes all servers. However, the Orthanc server is a
|
| 67 |
-
manual installation.
|
| 68 |
</p>
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 69 |
<p class="warning">
|
| 70 |
-
⚠ The Orthanc server will run on <a target="_blank" href="http://localhost:8042/">port 8042</a>
|
| 71 |
-
the nginx server on <a target="_blank" href="http://localhost:2000/">port 2000</a>.
|
| 72 |
</p>
|
| 73 |
</section>
|
| 74 |
|
| 75 |
-
|
| 76 |
-
<h2>Stopping the application</h2>
|
| 77 |
-
<ul>
|
| 78 |
-
<li>
|
| 79 |
-
<p>
|
| 80 |
-
The nginx server will continue running, even when you close the web browser window. To completely stop the server, double click the <strong>'Stop_PARROT.bat'</strong> file in the <strong>'parrot'</strong> folder (or stop the nginx processes in the Task Manager).
|
| 81 |
-
</p>
|
| 82 |
-
</li>
|
| 83 |
-
<li>
|
| 84 |
-
<p>
|
| 85 |
-
The Orthanc server runs as a Windows service and has to be stopped in the <strong>'Task Manager'</strong> (in the tab <strong>'Services'</strong>). Don't forget to start the service again when you want to use the <strong>'PARROT application'</strong>.
|
| 86 |
-
</p>
|
| 87 |
-
</li>
|
| 88 |
-
</ul>
|
| 89 |
-
</section>
|
| 90 |
-
|
| 91 |
-
<section id="folder-structure">
|
| 92 |
-
<h2>Folder Structure</h2>
|
| 93 |
-
<p>The <strong>'parrot'</strong> folder contains the following files:</p>
|
| 94 |
-
<ul>
|
| 95 |
-
<li>
|
| 96 |
-
<p><strong>'aiIntegration'</strong> contains the integrated trained AI models, the Python server environment and the API server directories.</p>
|
| 97 |
-
</li>
|
| 98 |
-
<li>
|
| 99 |
-
<p><strong>'dist'</strong> contains the PARROT web application.</p>
|
| 100 |
-
</li>
|
| 101 |
-
<li>
|
| 102 |
-
<p><strong>'logs'</strong> contains the log files of the nginx server.</p>
|
| 103 |
-
</li>
|
| 104 |
-
<li>
|
| 105 |
-
<p><strong>'temp'</strong> contains the temporary files of nginx.</p>
|
| 106 |
-
</li>
|
| 107 |
-
|
| 108 |
-
<li>
|
| 109 |
-
<p><strong>'tempai'</strong> contains the results of running the Python code. The user can configure the path for AI integration.</p>
|
| 110 |
-
</li>
|
| 111 |
-
<li>
|
| 112 |
-
<p><strong>'License'</strong> contains the License of the application.</p>
|
| 113 |
-
</li>
|
| 114 |
-
<li>
|
| 115 |
-
<p><strong>'nginx.exe'</strong> starts the nginx server (see the instructions above).</p>
|
| 116 |
-
</li>
|
| 117 |
-
<li>
|
| 118 |
-
<p><strong>'Start_PARROT.bat'</strong> start the nginx server (see the instructions above).</p>
|
| 119 |
-
</li>
|
| 120 |
-
<li>
|
| 121 |
-
<p><strong>'Stop_PARROT.bat'</strong> stop the ngnix server (see the instructions above).</p>
|
| 122 |
-
</li>
|
| 123 |
-
<li>
|
| 124 |
-
<p><strong>'unins000.exe'</strong> uninstalls the application from the computer.</p>
|
| 125 |
-
</li>
|
| 126 |
-
</ul>
|
| 127 |
-
</section>
|
| 128 |
-
|
| 129 |
<section id="patient-management">
|
| 130 |
-
<
|
| 131 |
<p>Users can import patient data (files or zip) from this screen to a local Orthanc daemon—a lightweight, standalone DICOM server, which ensures the confidentiality of patient files. A list of patient data present in the Orthanc server is displayed with summarized information. Users can load the patient into the app by clicking on the Load button. Users can delete a patient from the server with the red arrow on the right.</p>
|
| 132 |
<figure>
|
| 133 |
<img src="img/screen_patient_management.PNG" alt="Patient Management Image" class="img-fluid mb-3">
|
|
@@ -136,7 +85,7 @@
|
|
| 136 |
</section>
|
| 137 |
|
| 138 |
<section id="study-management">
|
| 139 |
-
<
|
| 140 |
<p>A list of studies of the loaded patient present in the Orthanc server is displayed with summarized information. Users can load the data into the app by clicking on the Load button. The left icon displays more complete information from DICOM tags and users can delete a patient study from the server with the red arrow on the right.</p>
|
| 141 |
<figure>
|
| 142 |
<img src="img/screen_study_management.png" alt="Study Management Image" class="img-fluid mb-3">
|
|
@@ -145,7 +94,7 @@
|
|
| 145 |
</section>
|
| 146 |
|
| 147 |
<section id="ai-models-management">
|
| 148 |
-
<
|
| 149 |
<figure>
|
| 150 |
<img src="img/screen_ai_models_management.png" alt="AI Models Management Image" class="img-fluid mb-3">
|
| 151 |
<figcaption>Figure ?: AI Models Management screen.</figcaption>
|
|
@@ -156,7 +105,7 @@
|
|
| 156 |
</section>
|
| 157 |
|
| 158 |
<section id="patient-modeling">
|
| 159 |
-
<
|
| 160 |
<figure>
|
| 161 |
<img src="img/screen_patient_modeling.png" alt="Patient Modeling Image" class="img-fluid mb-3">
|
| 162 |
<figcaption>Figure ?: Patient Modeling screen.</figcaption>
|
|
@@ -170,7 +119,7 @@
|
|
| 170 |
</section>
|
| 171 |
|
| 172 |
<section id="plan-evaluation">
|
| 173 |
-
<
|
| 174 |
<figure>
|
| 175 |
<img src="img/screen_plan_evaluation.png" alt="Plan Evaluation Image" class="img-fluid mb-3">
|
| 176 |
<figcaption>Figure ?: Plan Evaluation screen.</figcaption>
|
|
@@ -178,14 +127,76 @@
|
|
| 178 |
<p>A comprehensive display of two dose distributions comparison is presented on the last screen. The two dose distributions can originate from the importation step or result from the inference of ai models. We provide a set of tools for comparison, such as DVH curves, dose statistics, and the fulfillment of clinical goals. Clinical goals can be uploaded following an Excel template so that it fits any clinical requirements of the users institutions. The platform incorporates treatment indications based on NTCP protocols. It has been shown in multiple studies that predicted dose distributions yield meaningful output when used with NTCP models for treatment selection decision support [5] [6]. This is a useful decision support tool for the clinical community.
|
| 179 |
</p>
|
| 180 |
</section>
|
| 181 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 182 |
|
| 183 |
-
<div class="col-lg-4">
|
| 184 |
-
<nav id="right-menu" class="navbar navbar-light bg-light">
|
| 185 |
-
<nav class="nav flex-column">
|
| 186 |
-
</nav>
|
| 187 |
</nav>
|
| 188 |
-
</
|
| 189 |
</div>
|
| 190 |
</div>
|
| 191 |
|
|
|
|
| 32 |
<div class="col-lg-11">
|
| 33 |
<h1 class="mb-4 mt-4">Documentation</h1>
|
| 34 |
|
| 35 |
+
<section id="PARROT">
|
| 36 |
+
<h2>PARROT: Platform for ARtificial intelligence guided Radiation Oncology Treatment</h2>
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 37 |
<p>
|
| 38 |
+
<strong>PARROT</strong> is a local web application. It provides different modules that allow users to:
|
|
|
|
| 39 |
</p>
|
| 40 |
<ul>
|
| 41 |
<li>
|
| 42 |
+
<p>Run AI models.</p>
|
| 43 |
</li>
|
| 44 |
|
| 45 |
<li>
|
| 46 |
+
<p>Visualize CT and MR images.</p>
|
| 47 |
</li>
|
| 48 |
|
| 49 |
<li>
|
| 50 |
+
<p>Visualize, predict or modify segmentation information.</p>
|
| 51 |
</li>
|
| 52 |
|
| 53 |
<li>
|
| 54 |
+
<p>Visualize, evaluate, and compare dose treatment plans.</p>
|
| 55 |
</li>
|
| 56 |
</ul>
|
| 57 |
+
</section>
|
| 58 |
+
<section>
|
| 59 |
+
<h3>PARROT installation</h3>
|
| 60 |
+
<p>There are two ways to install PARROT, downloading the executable file of the last release from the <a href="index.html">Home</a> page is recommended. Otherwise, the code with building instructions are also available on our <a href="https://gitlab.com/ai4miro/parrot">Gitlab repository</a>.</p>
|
| 61 |
+
</section>
|
| 62 |
+
<section>
|
| 63 |
+
<h3>Orthanc installation</h3>
|
| 64 |
<p>
|
| 65 |
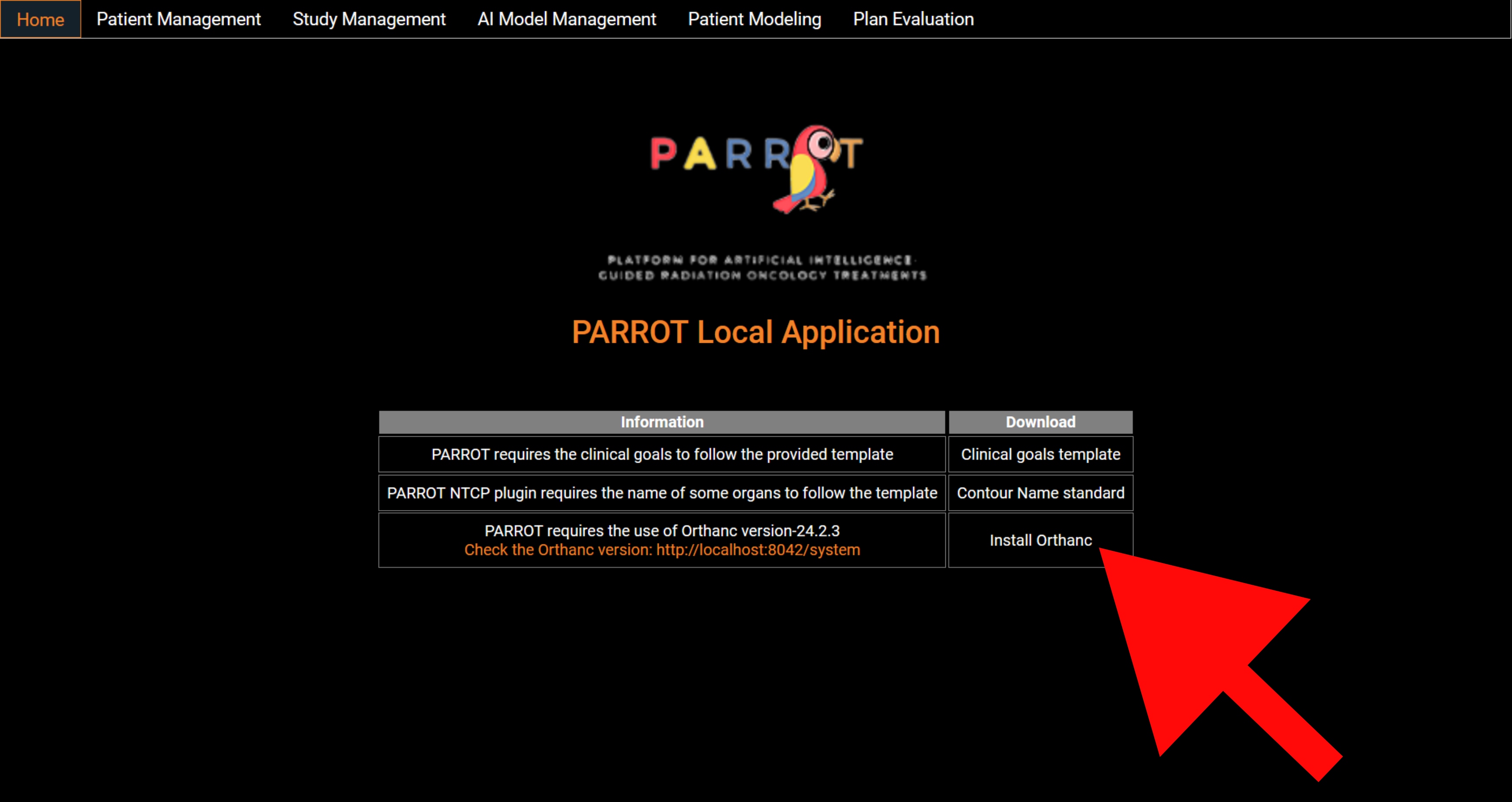
+
Patient data are stored using a Orthanc server. To install Orthanc, either click on the link on PARROT's home page or click on the following link: <a href="https://orthanc.uclouvain.be/downloads/windows-64/installers/OrthancInstaller-Win64-24.2.3.exe">Install Orthanc</a>.
|
|
|
|
|
|
|
| 66 |
</p>
|
| 67 |
+
<figure>
|
| 68 |
+
<img src="img/install_orthanc.png" alt="Install orthanc" class="img-fluid mb-3">
|
| 69 |
+
<figcaption>Figure 1: Home screen, link to install Orthanc.</figcaption>
|
| 70 |
+
</figure>
|
| 71 |
+
|
| 72 |
<p class="warning">
|
| 73 |
+
⚠ The Orthanc server will run on <a target="_blank" href="http://localhost:8042/">port 8042</a> while nginx server for the application will run on <a target="_blank" href="http://localhost:2000/">port 2000</a>.
|
|
|
|
| 74 |
</p>
|
| 75 |
</section>
|
| 76 |
|
| 77 |
+
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 78 |
<section id="patient-management">
|
| 79 |
+
<h3>Patient Management</h3>
|
| 80 |
<p>Users can import patient data (files or zip) from this screen to a local Orthanc daemon—a lightweight, standalone DICOM server, which ensures the confidentiality of patient files. A list of patient data present in the Orthanc server is displayed with summarized information. Users can load the patient into the app by clicking on the Load button. Users can delete a patient from the server with the red arrow on the right.</p>
|
| 81 |
<figure>
|
| 82 |
<img src="img/screen_patient_management.PNG" alt="Patient Management Image" class="img-fluid mb-3">
|
|
|
|
| 85 |
</section>
|
| 86 |
|
| 87 |
<section id="study-management">
|
| 88 |
+
<h3>Study Management</h3>
|
| 89 |
<p>A list of studies of the loaded patient present in the Orthanc server is displayed with summarized information. Users can load the data into the app by clicking on the Load button. The left icon displays more complete information from DICOM tags and users can delete a patient study from the server with the red arrow on the right.</p>
|
| 90 |
<figure>
|
| 91 |
<img src="img/screen_study_management.png" alt="Study Management Image" class="img-fluid mb-3">
|
|
|
|
| 94 |
</section>
|
| 95 |
|
| 96 |
<section id="ai-models-management">
|
| 97 |
+
<h3>AI Models Management</h3>
|
| 98 |
<figure>
|
| 99 |
<img src="img/screen_ai_models_management.png" alt="AI Models Management Image" class="img-fluid mb-3">
|
| 100 |
<figcaption>Figure ?: AI Models Management screen.</figcaption>
|
|
|
|
| 105 |
</section>
|
| 106 |
|
| 107 |
<section id="patient-modeling">
|
| 108 |
+
<h3>Patient Modeling</h3>
|
| 109 |
<figure>
|
| 110 |
<img src="img/screen_patient_modeling.png" alt="Patient Modeling Image" class="img-fluid mb-3">
|
| 111 |
<figcaption>Figure ?: Patient Modeling screen.</figcaption>
|
|
|
|
| 119 |
</section>
|
| 120 |
|
| 121 |
<section id="plan-evaluation">
|
| 122 |
+
<h3>Plan Evaluation</h3>
|
| 123 |
<figure>
|
| 124 |
<img src="img/screen_plan_evaluation.png" alt="Plan Evaluation Image" class="img-fluid mb-3">
|
| 125 |
<figcaption>Figure ?: Plan Evaluation screen.</figcaption>
|
|
|
|
| 127 |
<p>A comprehensive display of two dose distributions comparison is presented on the last screen. The two dose distributions can originate from the importation step or result from the inference of ai models. We provide a set of tools for comparison, such as DVH curves, dose statistics, and the fulfillment of clinical goals. Clinical goals can be uploaded following an Excel template so that it fits any clinical requirements of the users institutions. The platform incorporates treatment indications based on NTCP protocols. It has been shown in multiple studies that predicted dose distributions yield meaningful output when used with NTCP models for treatment selection decision support [5] [6]. This is a useful decision support tool for the clinical community.
|
| 128 |
</p>
|
| 129 |
</section>
|
| 130 |
+
|
| 131 |
+
|
| 132 |
+
<section id="stopping-the-application">
|
| 133 |
+
<h3>Stopping the application</h3>
|
| 134 |
+
<ul>
|
| 135 |
+
<li>
|
| 136 |
+
<p>
|
| 137 |
+
The nginx server will continue running, even when you close the web browser window. To completely stop the server, double click the <strong>'Stop_PARROT.bat'</strong> file in the <strong>'parrot'</strong> folder (or stop the nginx processes in the Task Manager).
|
| 138 |
+
</p>
|
| 139 |
+
</li>
|
| 140 |
+
<li>
|
| 141 |
+
<p>
|
| 142 |
+
The Orthanc server runs as a Windows service and has to be stopped in the <strong>'Task Manager'</strong> (in the tab <strong>'Services'</strong>). Don't forget to start the service again when you want to use the <strong>'PARROT application'</strong>.
|
| 143 |
+
</p>
|
| 144 |
+
</li>
|
| 145 |
+
</ul>
|
| 146 |
+
</section>
|
| 147 |
+
|
| 148 |
+
<section id="folder-structure">
|
| 149 |
+
<h3>Folder Structure</h3>
|
| 150 |
+
<p>The <strong>'parrot'</strong> folder contains the following files:</p>
|
| 151 |
+
<ul>
|
| 152 |
+
<li>
|
| 153 |
+
<p><strong>'aiIntegration'</strong> contains the integrated trained AI models, the Python server environment and the API server directories.</p>
|
| 154 |
+
</li>
|
| 155 |
+
<li>
|
| 156 |
+
<p><strong>'dist'</strong> contains the PARROT web application.</p>
|
| 157 |
+
</li>
|
| 158 |
+
<li>
|
| 159 |
+
<p><strong>'logs'</strong> contains the log files of the nginx server.</p>
|
| 160 |
+
</li>
|
| 161 |
+
<li>
|
| 162 |
+
<p><strong>'temp'</strong> contains the temporary files of nginx.</p>
|
| 163 |
+
</li>
|
| 164 |
+
|
| 165 |
+
<li>
|
| 166 |
+
<p><strong>'tempai'</strong> contains the results of running the Python code. The user can configure the path for AI integration.</p>
|
| 167 |
+
</li>
|
| 168 |
+
<li>
|
| 169 |
+
<p><strong>'License'</strong> contains the License of the application.</p>
|
| 170 |
+
</li>
|
| 171 |
+
<li>
|
| 172 |
+
<p><strong>'nginx.exe'</strong> starts the nginx server (see the instructions above).</p>
|
| 173 |
+
</li>
|
| 174 |
+
<li>
|
| 175 |
+
<p><strong>'Start_PARROT.bat'</strong> start the nginx server (see the instructions above).</p>
|
| 176 |
+
</li>
|
| 177 |
+
<li>
|
| 178 |
+
<p><strong>'Stop_PARROT.bat'</strong> stop the ngnix server (see the instructions above).</p>
|
| 179 |
+
</li>
|
| 180 |
+
<li>
|
| 181 |
+
<p><strong>'unins000.exe'</strong> uninstalls the application from the computer.</p>
|
| 182 |
+
</li>
|
| 183 |
+
</ul>
|
| 184 |
+
</section>
|
| 185 |
+
</div>
|
| 186 |
+
|
| 187 |
+
|
| 188 |
+
</div>
|
| 189 |
+
<div class="col-lg-4">
|
| 190 |
+
<nav id="right-menu" class="navbar navbar-light bg-light">
|
| 191 |
+
<nav class="nav flex-column">
|
| 192 |
+
<a class="nav-link" href="#installation">Installation</a>
|
| 193 |
+
<a class="nav-link" href="#study-management">Study Management</a>
|
| 194 |
+
<a class="nav-link" href="#ai-models-management">AI Models Management</a>
|
| 195 |
+
<a class="nav-link" href="#patient-modeling">Patient Modeling</a>
|
| 196 |
+
<a class="nav-link" href="#plan-evaluation">Plan Evaluation</a>
|
| 197 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 198 |
</nav>
|
| 199 |
+
</nav>
|
| 200 |
</div>
|
| 201 |
</div>
|
| 202 |
|
img/install_orthanc.png
ADDED
|