complete the model package
Browse files- README.md +86 -0
- configs/evaluate.json +77 -0
- configs/inference.json +141 -0
- configs/logging.conf +21 -0
- configs/metadata.json +91 -0
- configs/multi_gpu_train.json +36 -0
- configs/train.json +324 -0
- docs/README.md +79 -0
- docs/license.txt +6 -0
- docs/val_dice.png +0 -0
- models/model.pt +3 -0
README.md
ADDED
|
@@ -0,0 +1,86 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
tags:
|
| 3 |
+
- monai
|
| 4 |
+
- medical
|
| 5 |
+
library_name: monai
|
| 6 |
+
license: unknown
|
| 7 |
+
---
|
| 8 |
+
# Description
|
| 9 |
+
A pre-trained model for volumetric (3D) multi-organ segmentation from CT image.
|
| 10 |
+
|
| 11 |
+
# Model Overview
|
| 12 |
+
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
| 13 |
+
## Data
|
| 14 |
+
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 15 |
+
The dataset format needs to be redefined using the following commands:
|
| 16 |
+
|
| 17 |
+
```
|
| 18 |
+
unzip RawData.zip
|
| 19 |
+
mv RawData/Training/img/ RawData/imagesTr
|
| 20 |
+
mv RawData/Training/label/ RawData/labelsTr
|
| 21 |
+
mv RawData/Testing/img/ RawData/imagesTs
|
| 22 |
+
```
|
| 23 |
+
|
| 24 |
+
- Target: Multi-organs
|
| 25 |
+
- Task: Segmentation
|
| 26 |
+
- Modality: CT
|
| 27 |
+
- Size: 30 3D volumes (24 Training + 6 Testing)
|
| 28 |
+
|
| 29 |
+
## Training configuration
|
| 30 |
+
The training was performed with at least 32GB-memory GPUs.
|
| 31 |
+
|
| 32 |
+
Actual Model Input: 96 x 96 x 96
|
| 33 |
+
|
| 34 |
+
## Input and output formats
|
| 35 |
+
Input: 1 channel CT image
|
| 36 |
+
|
| 37 |
+
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left Kideny, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland
|
| 38 |
+
|
| 39 |
+
## Performance
|
| 40 |
+
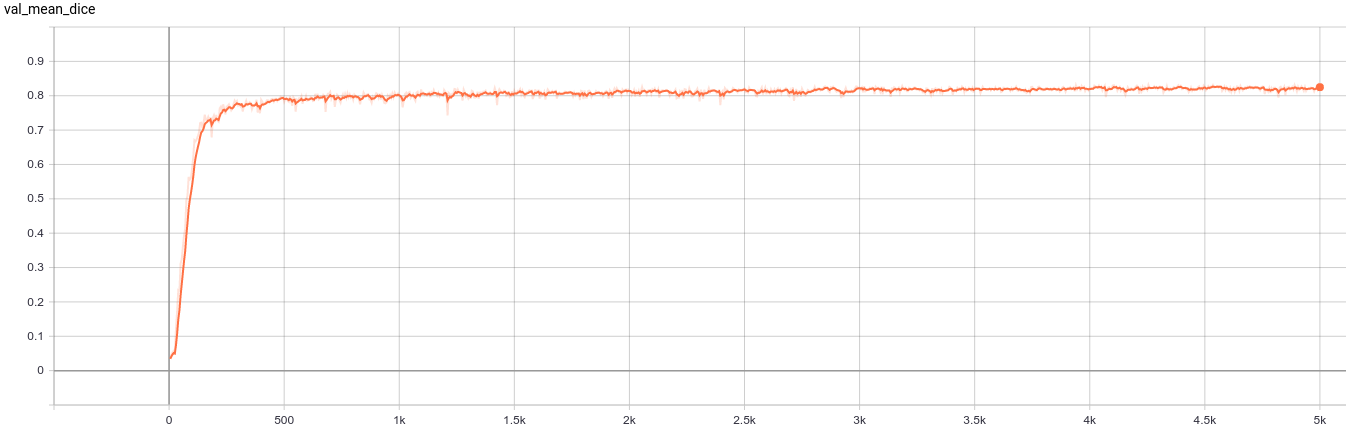
A graph showing the validation mean Dice for 5000 epochs.
|
| 41 |
+
|
| 42 |
+
 <br>
|
| 43 |
+
|
| 44 |
+
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 45 |
+
|
| 46 |
+
Mean Dice = 0.8283
|
| 47 |
+
|
| 48 |
+
Note that mean dice is computed in the original spacing of the input data.
|
| 49 |
+
## commands example
|
| 50 |
+
Execute training:
|
| 51 |
+
|
| 52 |
+
```
|
| 53 |
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 54 |
+
```
|
| 55 |
+
|
| 56 |
+
Override the `train` config to execute multi-GPU training:
|
| 57 |
+
|
| 58 |
+
```
|
| 59 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 60 |
+
```
|
| 61 |
+
|
| 62 |
+
Override the `train` config to execute evaluation with the trained model:
|
| 63 |
+
|
| 64 |
+
```
|
| 65 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 66 |
+
```
|
| 67 |
+
|
| 68 |
+
Execute inference:
|
| 69 |
+
|
| 70 |
+
```
|
| 71 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 72 |
+
```
|
| 73 |
+
|
| 74 |
+
Export checkpoint to TorchScript file:
|
| 75 |
+
|
| 76 |
+
TorchScript conversion is currently not supported.
|
| 77 |
+
|
| 78 |
+
# Disclaimer
|
| 79 |
+
This is an example, not to be used for diagnostic purposes.
|
| 80 |
+
|
| 81 |
+
# References
|
| 82 |
+
[1] Hatamizadeh, Ali, et al. "Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images." arXiv preprint arXiv:2201.01266 (2022). https://arxiv.org/abs/2201.01266.
|
| 83 |
+
|
| 84 |
+
[2] Tang, Yucheng, et al. "Self-supervised pre-training of swin transformers for 3d medical image analysis." arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791.
|
| 85 |
+
|
| 86 |
+
[3] Landman B, et al. "MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge." In Proc. of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge 2015 Oct (Vol. 5, p. 12).
|
configs/evaluate.json
ADDED
|
@@ -0,0 +1,77 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"validate#postprocessing": {
|
| 3 |
+
"_target_": "Compose",
|
| 4 |
+
"transforms": [
|
| 5 |
+
{
|
| 6 |
+
"_target_": "Activationsd",
|
| 7 |
+
"keys": "pred",
|
| 8 |
+
"softmax": true
|
| 9 |
+
},
|
| 10 |
+
{
|
| 11 |
+
"_target_": "Invertd",
|
| 12 |
+
"keys": [
|
| 13 |
+
"pred",
|
| 14 |
+
"label"
|
| 15 |
+
],
|
| 16 |
+
"transform": "@validate#preprocessing",
|
| 17 |
+
"orig_keys": "image",
|
| 18 |
+
"meta_key_postfix": "meta_dict",
|
| 19 |
+
"nearest_interp": [
|
| 20 |
+
false,
|
| 21 |
+
true
|
| 22 |
+
],
|
| 23 |
+
"to_tensor": true
|
| 24 |
+
},
|
| 25 |
+
{
|
| 26 |
+
"_target_": "AsDiscreted",
|
| 27 |
+
"keys": [
|
| 28 |
+
"pred",
|
| 29 |
+
"label"
|
| 30 |
+
],
|
| 31 |
+
"argmax": [
|
| 32 |
+
true,
|
| 33 |
+
false
|
| 34 |
+
],
|
| 35 |
+
"to_onehot": 14
|
| 36 |
+
},
|
| 37 |
+
{
|
| 38 |
+
"_target_": "SaveImaged",
|
| 39 |
+
"keys": "pred",
|
| 40 |
+
"meta_keys": "pred_meta_dict",
|
| 41 |
+
"output_dir": "@output_dir",
|
| 42 |
+
"resample": false,
|
| 43 |
+
"squeeze_end_dims": true
|
| 44 |
+
}
|
| 45 |
+
]
|
| 46 |
+
},
|
| 47 |
+
"validate#handlers": [
|
| 48 |
+
{
|
| 49 |
+
"_target_": "CheckpointLoader",
|
| 50 |
+
"load_path": "$@ckpt_dir + '/model.pt'",
|
| 51 |
+
"load_dict": {
|
| 52 |
+
"model": "@network"
|
| 53 |
+
}
|
| 54 |
+
},
|
| 55 |
+
{
|
| 56 |
+
"_target_": "StatsHandler",
|
| 57 |
+
"iteration_log": false
|
| 58 |
+
},
|
| 59 |
+
{
|
| 60 |
+
"_target_": "MetricsSaver",
|
| 61 |
+
"save_dir": "@output_dir",
|
| 62 |
+
"metrics": [
|
| 63 |
+
"val_mean_dice",
|
| 64 |
+
"val_acc"
|
| 65 |
+
],
|
| 66 |
+
"metric_details": [
|
| 67 |
+
"val_mean_dice"
|
| 68 |
+
],
|
| 69 |
+
"batch_transform": "$monai.handlers.from_engine(['image_meta_dict'])",
|
| 70 |
+
"summary_ops": "*"
|
| 71 |
+
}
|
| 72 |
+
],
|
| 73 |
+
"evaluating": [
|
| 74 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 75 |
+
"$@validate#evaluator.run()"
|
| 76 |
+
]
|
| 77 |
+
}
|
configs/inference.json
ADDED
|
@@ -0,0 +1,141 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import os"
|
| 5 |
+
],
|
| 6 |
+
"bundle_root": "/workspace/MONAI_Bundle/swin_unetr_btcv_segmentation/",
|
| 7 |
+
"output_dir": "$@bundle_root + '/eval'",
|
| 8 |
+
"dataset_dir": "/dataset/dataset0",
|
| 9 |
+
"datalist": "$list(sorted(glob.glob(@dataset_dir + '/imagesTs/*.nii.gz')))",
|
| 10 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 11 |
+
"network_def": {
|
| 12 |
+
"_target_": "SwinUNETR",
|
| 13 |
+
"spatial_dims": 3,
|
| 14 |
+
"img_size": 96,
|
| 15 |
+
"in_channels": 1,
|
| 16 |
+
"out_channels": 14,
|
| 17 |
+
"feature_size": 48,
|
| 18 |
+
"use_checkpoint": true
|
| 19 |
+
},
|
| 20 |
+
"network": "$@network_def.to(@device)",
|
| 21 |
+
"preprocessing": {
|
| 22 |
+
"_target_": "Compose",
|
| 23 |
+
"transforms": [
|
| 24 |
+
{
|
| 25 |
+
"_target_": "LoadImaged",
|
| 26 |
+
"keys": "image"
|
| 27 |
+
},
|
| 28 |
+
{
|
| 29 |
+
"_target_": "EnsureChannelFirstd",
|
| 30 |
+
"keys": "image"
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"_target_": "Orientationd",
|
| 34 |
+
"keys": "image",
|
| 35 |
+
"axcodes": "RAS"
|
| 36 |
+
},
|
| 37 |
+
{
|
| 38 |
+
"_target_": "Spacingd",
|
| 39 |
+
"keys": "image",
|
| 40 |
+
"pixdim": [
|
| 41 |
+
1.5,
|
| 42 |
+
1.5,
|
| 43 |
+
2.0
|
| 44 |
+
],
|
| 45 |
+
"mode": "bilinear"
|
| 46 |
+
},
|
| 47 |
+
{
|
| 48 |
+
"_target_": "ScaleIntensityRanged",
|
| 49 |
+
"keys": "image",
|
| 50 |
+
"a_min": -175,
|
| 51 |
+
"a_max": 250,
|
| 52 |
+
"b_min": 0.0,
|
| 53 |
+
"b_max": 1.0,
|
| 54 |
+
"clip": true
|
| 55 |
+
},
|
| 56 |
+
{
|
| 57 |
+
"_target_": "EnsureTyped",
|
| 58 |
+
"keys": "image"
|
| 59 |
+
}
|
| 60 |
+
]
|
| 61 |
+
},
|
| 62 |
+
"dataset": {
|
| 63 |
+
"_target_": "Dataset",
|
| 64 |
+
"data": "$[{'image': i} for i in @datalist]",
|
| 65 |
+
"transform": "@preprocessing"
|
| 66 |
+
},
|
| 67 |
+
"dataloader": {
|
| 68 |
+
"_target_": "DataLoader",
|
| 69 |
+
"dataset": "@dataset",
|
| 70 |
+
"batch_size": 1,
|
| 71 |
+
"shuffle": false,
|
| 72 |
+
"num_workers": 4
|
| 73 |
+
},
|
| 74 |
+
"inferer": {
|
| 75 |
+
"_target_": "SlidingWindowInferer",
|
| 76 |
+
"roi_size": [
|
| 77 |
+
96,
|
| 78 |
+
96,
|
| 79 |
+
96
|
| 80 |
+
],
|
| 81 |
+
"sw_batch_size": 4,
|
| 82 |
+
"overlap": 0.5
|
| 83 |
+
},
|
| 84 |
+
"postprocessing": {
|
| 85 |
+
"_target_": "Compose",
|
| 86 |
+
"transforms": [
|
| 87 |
+
{
|
| 88 |
+
"_target_": "Activationsd",
|
| 89 |
+
"keys": "pred",
|
| 90 |
+
"softmax": true
|
| 91 |
+
},
|
| 92 |
+
{
|
| 93 |
+
"_target_": "Invertd",
|
| 94 |
+
"keys": "pred",
|
| 95 |
+
"transform": "@preprocessing",
|
| 96 |
+
"orig_keys": "image",
|
| 97 |
+
"meta_key_postfix": "meta_dict",
|
| 98 |
+
"nearest_interp": false,
|
| 99 |
+
"to_tensor": true
|
| 100 |
+
},
|
| 101 |
+
{
|
| 102 |
+
"_target_": "AsDiscreted",
|
| 103 |
+
"keys": "pred",
|
| 104 |
+
"argmax": true
|
| 105 |
+
},
|
| 106 |
+
{
|
| 107 |
+
"_target_": "SaveImaged",
|
| 108 |
+
"keys": "pred",
|
| 109 |
+
"meta_keys": "pred_meta_dict",
|
| 110 |
+
"output_dir": "@output_dir"
|
| 111 |
+
}
|
| 112 |
+
]
|
| 113 |
+
},
|
| 114 |
+
"handlers": [
|
| 115 |
+
{
|
| 116 |
+
"_target_": "CheckpointLoader",
|
| 117 |
+
"load_path": "$@bundle_root + '/models/model.pt'",
|
| 118 |
+
"load_dict": {
|
| 119 |
+
"model": "@network"
|
| 120 |
+
}
|
| 121 |
+
},
|
| 122 |
+
{
|
| 123 |
+
"_target_": "StatsHandler",
|
| 124 |
+
"iteration_log": false
|
| 125 |
+
}
|
| 126 |
+
],
|
| 127 |
+
"evaluator": {
|
| 128 |
+
"_target_": "SupervisedEvaluator",
|
| 129 |
+
"device": "@device",
|
| 130 |
+
"val_data_loader": "@dataloader",
|
| 131 |
+
"network": "@network",
|
| 132 |
+
"inferer": "@inferer",
|
| 133 |
+
"postprocessing": "@postprocessing",
|
| 134 |
+
"val_handlers": "@handlers",
|
| 135 |
+
"amp": true
|
| 136 |
+
},
|
| 137 |
+
"evaluating": [
|
| 138 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 139 |
+
"$@evaluator.run()"
|
| 140 |
+
]
|
| 141 |
+
}
|
configs/logging.conf
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[loggers]
|
| 2 |
+
keys=root
|
| 3 |
+
|
| 4 |
+
[handlers]
|
| 5 |
+
keys=consoleHandler
|
| 6 |
+
|
| 7 |
+
[formatters]
|
| 8 |
+
keys=fullFormatter
|
| 9 |
+
|
| 10 |
+
[logger_root]
|
| 11 |
+
level=INFO
|
| 12 |
+
handlers=consoleHandler
|
| 13 |
+
|
| 14 |
+
[handler_consoleHandler]
|
| 15 |
+
class=StreamHandler
|
| 16 |
+
level=INFO
|
| 17 |
+
formatter=fullFormatter
|
| 18 |
+
args=(sys.stdout,)
|
| 19 |
+
|
| 20 |
+
[formatter_fullFormatter]
|
| 21 |
+
format=%(asctime)s - %(name)s - %(levelname)s - %(message)s
|
configs/metadata.json
ADDED
|
@@ -0,0 +1,91 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.1.0",
|
| 4 |
+
"changelog": {
|
| 5 |
+
"0.1.0": "complete the model package",
|
| 6 |
+
"0.0.1": "initialize the model package structure"
|
| 7 |
+
},
|
| 8 |
+
"monai_version": "0.9.0",
|
| 9 |
+
"pytorch_version": "1.10.0",
|
| 10 |
+
"numpy_version": "1.21.2",
|
| 11 |
+
"optional_packages_version": {
|
| 12 |
+
"nibabel": "3.2.1",
|
| 13 |
+
"pytorch-ignite": "0.4.8",
|
| 14 |
+
"einops": "0.4.1"
|
| 15 |
+
},
|
| 16 |
+
"task": "BTCV multi-organ segmentation",
|
| 17 |
+
"description": "A pre-trained model for volumetric (3D) multi-organ segmentation from CT image",
|
| 18 |
+
"authors": "MONAI team",
|
| 19 |
+
"copyright": "Copyright (c) MONAI Consortium",
|
| 20 |
+
"data_source": "RawData.zip from https://www.synapse.org/#!Synapse:syn3193805/wiki/217752/",
|
| 21 |
+
"data_type": "nibabel",
|
| 22 |
+
"image_classes": "single channel data, intensity scaled to [0, 1]",
|
| 23 |
+
"label_classes": "multi-channel data,0:background,1:spleen, 2:Right Kidney, 3:Left Kideny, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland",
|
| 24 |
+
"pred_classes": "14 channels OneHot data, 0:background,1:spleen, 2:Right Kidney, 3:Left Kideny, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland",
|
| 25 |
+
"eval_metrics": {
|
| 26 |
+
"mean_dice": 0.8283
|
| 27 |
+
},
|
| 28 |
+
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 29 |
+
"references": [
|
| 30 |
+
"Hatamizadeh, Ali, et al. 'Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv preprint arXiv:2201.01266 (2022). https://arxiv.org/abs/2201.01266.",
|
| 31 |
+
"Tang, Yucheng, et al. 'Self-supervised pre-training of swin transformers for 3d medical image analysis. arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791."
|
| 32 |
+
],
|
| 33 |
+
"network_data_format": {
|
| 34 |
+
"inputs": {
|
| 35 |
+
"image": {
|
| 36 |
+
"type": "image",
|
| 37 |
+
"format": "hounsfield",
|
| 38 |
+
"modality": "CT",
|
| 39 |
+
"num_channels": 1,
|
| 40 |
+
"spatial_shape": [
|
| 41 |
+
96,
|
| 42 |
+
96,
|
| 43 |
+
96
|
| 44 |
+
],
|
| 45 |
+
"dtype": "float32",
|
| 46 |
+
"value_range": [
|
| 47 |
+
0,
|
| 48 |
+
1
|
| 49 |
+
],
|
| 50 |
+
"is_patch_data": true,
|
| 51 |
+
"channel_def": {
|
| 52 |
+
"0": "image"
|
| 53 |
+
}
|
| 54 |
+
}
|
| 55 |
+
},
|
| 56 |
+
"outputs": {
|
| 57 |
+
"pred": {
|
| 58 |
+
"type": "image",
|
| 59 |
+
"format": "segmentation",
|
| 60 |
+
"num_channels": 14,
|
| 61 |
+
"spatial_shape": [
|
| 62 |
+
96,
|
| 63 |
+
96,
|
| 64 |
+
96
|
| 65 |
+
],
|
| 66 |
+
"dtype": "float32",
|
| 67 |
+
"value_range": [
|
| 68 |
+
0,
|
| 69 |
+
1
|
| 70 |
+
],
|
| 71 |
+
"is_patch_data": true,
|
| 72 |
+
"channel_def": {
|
| 73 |
+
"0": "background",
|
| 74 |
+
"1": "spleen",
|
| 75 |
+
"2": "Right Kidney",
|
| 76 |
+
"3": "Left Kideny",
|
| 77 |
+
"4": "Gallbladder",
|
| 78 |
+
"5": "Esophagus",
|
| 79 |
+
"6": "Liver",
|
| 80 |
+
"7": "Stomach",
|
| 81 |
+
"8": "Aorta",
|
| 82 |
+
"9": "IVC",
|
| 83 |
+
"10": "Portal and Splenic Veins",
|
| 84 |
+
"11": "Pancreas",
|
| 85 |
+
"12": "Right adrenal gland",
|
| 86 |
+
"13": "Left adrenal gland"
|
| 87 |
+
}
|
| 88 |
+
}
|
| 89 |
+
}
|
| 90 |
+
}
|
| 91 |
+
}
|
configs/multi_gpu_train.json
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"device": "$torch.device(f'cuda:{dist.get_rank()}')",
|
| 3 |
+
"network": {
|
| 4 |
+
"_target_": "torch.nn.parallel.DistributedDataParallel",
|
| 5 |
+
"module": "$@network_def.to(@device)",
|
| 6 |
+
"device_ids": [
|
| 7 |
+
"@device"
|
| 8 |
+
]
|
| 9 |
+
},
|
| 10 |
+
"train#sampler": {
|
| 11 |
+
"_target_": "DistributedSampler",
|
| 12 |
+
"dataset": "@train#dataset",
|
| 13 |
+
"even_divisible": true,
|
| 14 |
+
"shuffle": true
|
| 15 |
+
},
|
| 16 |
+
"train#dataloader#sampler": "@train#sampler",
|
| 17 |
+
"train#dataloader#shuffle": false,
|
| 18 |
+
"train#trainer#train_handlers": "$@train#handlers[: -2 if dist.get_rank() > 0 else None]",
|
| 19 |
+
"validate#sampler": {
|
| 20 |
+
"_target_": "DistributedSampler",
|
| 21 |
+
"dataset": "@validate#dataset",
|
| 22 |
+
"even_divisible": false,
|
| 23 |
+
"shuffle": false
|
| 24 |
+
},
|
| 25 |
+
"validate#dataloader#sampler": "@validate#sampler",
|
| 26 |
+
"validate#evaluator#val_handlers": "$None if dist.get_rank() > 0 else @validate#handlers",
|
| 27 |
+
"training": [
|
| 28 |
+
"$import torch.distributed as dist",
|
| 29 |
+
"$dist.init_process_group(backend='nccl')",
|
| 30 |
+
"$torch.cuda.set_device(@device)",
|
| 31 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 32 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 33 |
+
"$@train#trainer.run()",
|
| 34 |
+
"$dist.destroy_process_group()"
|
| 35 |
+
]
|
| 36 |
+
}
|
configs/train.json
ADDED
|
@@ -0,0 +1,324 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import os",
|
| 5 |
+
"$import ignite"
|
| 6 |
+
],
|
| 7 |
+
"bundle_root": "/workspace/MONAI_Bundle/swin_unetr_btcv_segmentation/",
|
| 8 |
+
"ckpt_dir": "$@bundle_root + '/models'",
|
| 9 |
+
"output_dir": "$@bundle_root + '/eval'",
|
| 10 |
+
"dataset_dir": "/dataset/dataset0",
|
| 11 |
+
"images": "$list(sorted(glob.glob(@dataset_dir + '/imagesTr/*.nii.gz')))",
|
| 12 |
+
"labels": "$list(sorted(glob.glob(@dataset_dir + '/labelsTr/*.nii.gz')))",
|
| 13 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 14 |
+
"network_def": {
|
| 15 |
+
"_target_": "SwinUNETR",
|
| 16 |
+
"spatial_dims": 3,
|
| 17 |
+
"img_size": 96,
|
| 18 |
+
"in_channels": 1,
|
| 19 |
+
"out_channels": 14,
|
| 20 |
+
"feature_size": 48,
|
| 21 |
+
"use_checkpoint": true
|
| 22 |
+
},
|
| 23 |
+
"network": "$@network_def.to(@device)",
|
| 24 |
+
"loss": {
|
| 25 |
+
"_target_": "DiceCELoss",
|
| 26 |
+
"to_onehot_y": true,
|
| 27 |
+
"softmax": true,
|
| 28 |
+
"squared_pred": true,
|
| 29 |
+
"batch": true
|
| 30 |
+
},
|
| 31 |
+
"optimizer": {
|
| 32 |
+
"_target_": "torch.optim.Adam",
|
| 33 |
+
"params": "$@network.parameters()",
|
| 34 |
+
"lr": 0.0002
|
| 35 |
+
},
|
| 36 |
+
"train": {
|
| 37 |
+
"deterministic_transforms": [
|
| 38 |
+
{
|
| 39 |
+
"_target_": "LoadImaged",
|
| 40 |
+
"keys": [
|
| 41 |
+
"image",
|
| 42 |
+
"label"
|
| 43 |
+
]
|
| 44 |
+
},
|
| 45 |
+
{
|
| 46 |
+
"_target_": "EnsureChannelFirstd",
|
| 47 |
+
"keys": [
|
| 48 |
+
"image",
|
| 49 |
+
"label"
|
| 50 |
+
]
|
| 51 |
+
},
|
| 52 |
+
{
|
| 53 |
+
"_target_": "Orientationd",
|
| 54 |
+
"keys": [
|
| 55 |
+
"image",
|
| 56 |
+
"label"
|
| 57 |
+
],
|
| 58 |
+
"axcodes": "RAS"
|
| 59 |
+
},
|
| 60 |
+
{
|
| 61 |
+
"_target_": "Spacingd",
|
| 62 |
+
"keys": [
|
| 63 |
+
"image",
|
| 64 |
+
"label"
|
| 65 |
+
],
|
| 66 |
+
"pixdim": [
|
| 67 |
+
1.5,
|
| 68 |
+
1.5,
|
| 69 |
+
2.0
|
| 70 |
+
],
|
| 71 |
+
"mode": [
|
| 72 |
+
"bilinear",
|
| 73 |
+
"nearest"
|
| 74 |
+
]
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"_target_": "ScaleIntensityRanged",
|
| 78 |
+
"keys": "image",
|
| 79 |
+
"a_min": -175,
|
| 80 |
+
"a_max": 250,
|
| 81 |
+
"b_min": 0.0,
|
| 82 |
+
"b_max": 1.0,
|
| 83 |
+
"clip": true
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"_target_": "EnsureTyped",
|
| 87 |
+
"keys": [
|
| 88 |
+
"image",
|
| 89 |
+
"label"
|
| 90 |
+
]
|
| 91 |
+
}
|
| 92 |
+
],
|
| 93 |
+
"random_transforms": [
|
| 94 |
+
{
|
| 95 |
+
"_target_": "RandCropByPosNegLabeld",
|
| 96 |
+
"keys": [
|
| 97 |
+
"image",
|
| 98 |
+
"label"
|
| 99 |
+
],
|
| 100 |
+
"label_key": "label",
|
| 101 |
+
"spatial_size": [
|
| 102 |
+
96,
|
| 103 |
+
96,
|
| 104 |
+
96
|
| 105 |
+
],
|
| 106 |
+
"pos": 1,
|
| 107 |
+
"neg": 1,
|
| 108 |
+
"num_samples": 4,
|
| 109 |
+
"image_key": "image",
|
| 110 |
+
"image_threshold": 0
|
| 111 |
+
},
|
| 112 |
+
{
|
| 113 |
+
"_target_": "RandFlipd",
|
| 114 |
+
"keys": [
|
| 115 |
+
"image",
|
| 116 |
+
"label"
|
| 117 |
+
],
|
| 118 |
+
"spatial_axis": [
|
| 119 |
+
0
|
| 120 |
+
],
|
| 121 |
+
"prob": 0.1
|
| 122 |
+
},
|
| 123 |
+
{
|
| 124 |
+
"_target_": "RandFlipd",
|
| 125 |
+
"keys": [
|
| 126 |
+
"image",
|
| 127 |
+
"label"
|
| 128 |
+
],
|
| 129 |
+
"spatial_axis": [
|
| 130 |
+
1
|
| 131 |
+
],
|
| 132 |
+
"prob": 0.1
|
| 133 |
+
},
|
| 134 |
+
{
|
| 135 |
+
"_target_": "RandFlipd",
|
| 136 |
+
"keys": [
|
| 137 |
+
"image",
|
| 138 |
+
"label"
|
| 139 |
+
],
|
| 140 |
+
"spatial_axis": [
|
| 141 |
+
2
|
| 142 |
+
],
|
| 143 |
+
"prob": 0.1
|
| 144 |
+
},
|
| 145 |
+
{
|
| 146 |
+
"_target_": "RandRotate90d",
|
| 147 |
+
"keys": [
|
| 148 |
+
"image",
|
| 149 |
+
"label"
|
| 150 |
+
],
|
| 151 |
+
"max_k": 3,
|
| 152 |
+
"prob": 0.1
|
| 153 |
+
},
|
| 154 |
+
{
|
| 155 |
+
"_target_": "RandShiftIntensityd",
|
| 156 |
+
"keys": "image",
|
| 157 |
+
"offsets": 0.1,
|
| 158 |
+
"prob": 0.5
|
| 159 |
+
}
|
| 160 |
+
],
|
| 161 |
+
"preprocessing": {
|
| 162 |
+
"_target_": "Compose",
|
| 163 |
+
"transforms": "$@train#deterministic_transforms + @train#random_transforms"
|
| 164 |
+
},
|
| 165 |
+
"dataset": {
|
| 166 |
+
"_target_": "CacheDataset",
|
| 167 |
+
"data": "$[{'image': i, 'label': l} for i, l in zip(@images[:-9], @labels[:-9])]",
|
| 168 |
+
"transform": "@train#preprocessing",
|
| 169 |
+
"cache_rate": 1.0,
|
| 170 |
+
"num_workers": 4
|
| 171 |
+
},
|
| 172 |
+
"dataloader": {
|
| 173 |
+
"_target_": "DataLoader",
|
| 174 |
+
"dataset": "@train#dataset",
|
| 175 |
+
"batch_size": 2,
|
| 176 |
+
"shuffle": true,
|
| 177 |
+
"num_workers": 4
|
| 178 |
+
},
|
| 179 |
+
"inferer": {
|
| 180 |
+
"_target_": "SimpleInferer"
|
| 181 |
+
},
|
| 182 |
+
"postprocessing": {
|
| 183 |
+
"_target_": "Compose",
|
| 184 |
+
"transforms": [
|
| 185 |
+
{
|
| 186 |
+
"_target_": "Activationsd",
|
| 187 |
+
"keys": "pred",
|
| 188 |
+
"softmax": true
|
| 189 |
+
},
|
| 190 |
+
{
|
| 191 |
+
"_target_": "AsDiscreted",
|
| 192 |
+
"keys": [
|
| 193 |
+
"pred",
|
| 194 |
+
"label"
|
| 195 |
+
],
|
| 196 |
+
"argmax": [
|
| 197 |
+
true,
|
| 198 |
+
false
|
| 199 |
+
],
|
| 200 |
+
"to_onehot": 14
|
| 201 |
+
}
|
| 202 |
+
]
|
| 203 |
+
},
|
| 204 |
+
"handlers": [
|
| 205 |
+
{
|
| 206 |
+
"_target_": "ValidationHandler",
|
| 207 |
+
"validator": "@validate#evaluator",
|
| 208 |
+
"epoch_level": true,
|
| 209 |
+
"interval": 5
|
| 210 |
+
},
|
| 211 |
+
{
|
| 212 |
+
"_target_": "StatsHandler",
|
| 213 |
+
"tag_name": "train_loss",
|
| 214 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 215 |
+
},
|
| 216 |
+
{
|
| 217 |
+
"_target_": "TensorBoardStatsHandler",
|
| 218 |
+
"log_dir": "@output_dir",
|
| 219 |
+
"tag_name": "train_loss",
|
| 220 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 221 |
+
}
|
| 222 |
+
],
|
| 223 |
+
"key_metric": {
|
| 224 |
+
"train_accuracy": {
|
| 225 |
+
"_target_": "ignite.metrics.Accuracy",
|
| 226 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 227 |
+
}
|
| 228 |
+
},
|
| 229 |
+
"trainer": {
|
| 230 |
+
"_target_": "SupervisedTrainer",
|
| 231 |
+
"max_epochs": 500,
|
| 232 |
+
"device": "@device",
|
| 233 |
+
"train_data_loader": "@train#dataloader",
|
| 234 |
+
"network": "@network",
|
| 235 |
+
"loss_function": "@loss",
|
| 236 |
+
"optimizer": "@optimizer",
|
| 237 |
+
"inferer": "@train#inferer",
|
| 238 |
+
"postprocessing": "@train#postprocessing",
|
| 239 |
+
"key_train_metric": "@train#key_metric",
|
| 240 |
+
"train_handlers": "@train#handlers",
|
| 241 |
+
"amp": true
|
| 242 |
+
}
|
| 243 |
+
},
|
| 244 |
+
"validate": {
|
| 245 |
+
"preprocessing": {
|
| 246 |
+
"_target_": "Compose",
|
| 247 |
+
"transforms": "%train#deterministic_transforms"
|
| 248 |
+
},
|
| 249 |
+
"dataset": {
|
| 250 |
+
"_target_": "CacheDataset",
|
| 251 |
+
"data": "$[{'image': i, 'label': l} for i, l in zip(@images[-9:], @labels[-9:])]",
|
| 252 |
+
"transform": "@validate#preprocessing",
|
| 253 |
+
"cache_rate": 1.0
|
| 254 |
+
},
|
| 255 |
+
"dataloader": {
|
| 256 |
+
"_target_": "DataLoader",
|
| 257 |
+
"dataset": "@validate#dataset",
|
| 258 |
+
"batch_size": 1,
|
| 259 |
+
"shuffle": false,
|
| 260 |
+
"num_workers": 4
|
| 261 |
+
},
|
| 262 |
+
"inferer": {
|
| 263 |
+
"_target_": "SlidingWindowInferer",
|
| 264 |
+
"roi_size": [
|
| 265 |
+
96,
|
| 266 |
+
96,
|
| 267 |
+
96
|
| 268 |
+
],
|
| 269 |
+
"sw_batch_size": 4,
|
| 270 |
+
"overlap": 0.5
|
| 271 |
+
},
|
| 272 |
+
"postprocessing": "%train#postprocessing",
|
| 273 |
+
"handlers": [
|
| 274 |
+
{
|
| 275 |
+
"_target_": "StatsHandler",
|
| 276 |
+
"iteration_log": false
|
| 277 |
+
},
|
| 278 |
+
{
|
| 279 |
+
"_target_": "TensorBoardStatsHandler",
|
| 280 |
+
"log_dir": "@output_dir",
|
| 281 |
+
"iteration_log": false
|
| 282 |
+
},
|
| 283 |
+
{
|
| 284 |
+
"_target_": "CheckpointSaver",
|
| 285 |
+
"save_dir": "@ckpt_dir",
|
| 286 |
+
"save_dict": {
|
| 287 |
+
"model": "@network"
|
| 288 |
+
},
|
| 289 |
+
"save_key_metric": true,
|
| 290 |
+
"key_metric_filename": "model.pt"
|
| 291 |
+
}
|
| 292 |
+
],
|
| 293 |
+
"key_metric": {
|
| 294 |
+
"val_mean_dice": {
|
| 295 |
+
"_target_": "MeanDice",
|
| 296 |
+
"include_background": false,
|
| 297 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 298 |
+
}
|
| 299 |
+
},
|
| 300 |
+
"additional_metrics": {
|
| 301 |
+
"val_accuracy": {
|
| 302 |
+
"_target_": "ignite.metrics.Accuracy",
|
| 303 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 304 |
+
}
|
| 305 |
+
},
|
| 306 |
+
"evaluator": {
|
| 307 |
+
"_target_": "SupervisedEvaluator",
|
| 308 |
+
"device": "@device",
|
| 309 |
+
"val_data_loader": "@validate#dataloader",
|
| 310 |
+
"network": "@network",
|
| 311 |
+
"inferer": "@validate#inferer",
|
| 312 |
+
"postprocessing": "@validate#postprocessing",
|
| 313 |
+
"key_val_metric": "@validate#key_metric",
|
| 314 |
+
"additional_metrics": "@validate#additional_metrics",
|
| 315 |
+
"val_handlers": "@validate#handlers",
|
| 316 |
+
"amp": true
|
| 317 |
+
}
|
| 318 |
+
},
|
| 319 |
+
"training": [
|
| 320 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 321 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 322 |
+
"$@train#trainer.run()"
|
| 323 |
+
]
|
| 324 |
+
}
|
docs/README.md
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Description
|
| 2 |
+
A pre-trained model for volumetric (3D) multi-organ segmentation from CT image.
|
| 3 |
+
|
| 4 |
+
# Model Overview
|
| 5 |
+
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
| 6 |
+
## Data
|
| 7 |
+
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 8 |
+
The dataset format needs to be redefined using the following commands:
|
| 9 |
+
|
| 10 |
+
```
|
| 11 |
+
unzip RawData.zip
|
| 12 |
+
mv RawData/Training/img/ RawData/imagesTr
|
| 13 |
+
mv RawData/Training/label/ RawData/labelsTr
|
| 14 |
+
mv RawData/Testing/img/ RawData/imagesTs
|
| 15 |
+
```
|
| 16 |
+
|
| 17 |
+
- Target: Multi-organs
|
| 18 |
+
- Task: Segmentation
|
| 19 |
+
- Modality: CT
|
| 20 |
+
- Size: 30 3D volumes (24 Training + 6 Testing)
|
| 21 |
+
|
| 22 |
+
## Training configuration
|
| 23 |
+
The training was performed with at least 32GB-memory GPUs.
|
| 24 |
+
|
| 25 |
+
Actual Model Input: 96 x 96 x 96
|
| 26 |
+
|
| 27 |
+
## Input and output formats
|
| 28 |
+
Input: 1 channel CT image
|
| 29 |
+
|
| 30 |
+
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left Kideny, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland
|
| 31 |
+
|
| 32 |
+
## Performance
|
| 33 |
+
A graph showing the validation mean Dice for 5000 epochs.
|
| 34 |
+
|
| 35 |
+
 <br>
|
| 36 |
+
|
| 37 |
+
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 38 |
+
|
| 39 |
+
Mean Dice = 0.8283
|
| 40 |
+
|
| 41 |
+
Note that mean dice is computed in the original spacing of the input data.
|
| 42 |
+
## commands example
|
| 43 |
+
Execute training:
|
| 44 |
+
|
| 45 |
+
```
|
| 46 |
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 47 |
+
```
|
| 48 |
+
|
| 49 |
+
Override the `train` config to execute multi-GPU training:
|
| 50 |
+
|
| 51 |
+
```
|
| 52 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 53 |
+
```
|
| 54 |
+
|
| 55 |
+
Override the `train` config to execute evaluation with the trained model:
|
| 56 |
+
|
| 57 |
+
```
|
| 58 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 59 |
+
```
|
| 60 |
+
|
| 61 |
+
Execute inference:
|
| 62 |
+
|
| 63 |
+
```
|
| 64 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 65 |
+
```
|
| 66 |
+
|
| 67 |
+
Export checkpoint to TorchScript file:
|
| 68 |
+
|
| 69 |
+
TorchScript conversion is currently not supported.
|
| 70 |
+
|
| 71 |
+
# Disclaimer
|
| 72 |
+
This is an example, not to be used for diagnostic purposes.
|
| 73 |
+
|
| 74 |
+
# References
|
| 75 |
+
[1] Hatamizadeh, Ali, et al. "Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images." arXiv preprint arXiv:2201.01266 (2022). https://arxiv.org/abs/2201.01266.
|
| 76 |
+
|
| 77 |
+
[2] Tang, Yucheng, et al. "Self-supervised pre-training of swin transformers for 3d medical image analysis." arXiv preprint arXiv:2111.14791 (2021). https://arxiv.org/abs/2111.14791.
|
| 78 |
+
|
| 79 |
+
[3] Landman B, et al. "MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge." In Proc. of the MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge 2015 Oct (Vol. 5, p. 12).
|
docs/license.txt
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Third Party Licenses
|
| 2 |
+
-----------------------------------------------------------------------
|
| 3 |
+
|
| 4 |
+
/*********************************************************************/
|
| 5 |
+
i. Medical Segmentation Decathlon
|
| 6 |
+
http://medicaldecathlon.com/
|
docs/val_dice.png
ADDED
|
models/model.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5486702e73e4ca3eef492e3b53cf91304302805ae49a9f5159038637da818bda
|
| 3 |
+
size 256345027
|