enhance readme with details of model training
Browse files- README.md +31 -2
- configs/metadata.json +2 -1
- docs/README.md +31 -2
README.md
CHANGED
|
@@ -11,13 +11,31 @@ A pre-trained model for volumetric (3D) segmentation of the spleen from CT image
|
|
| 11 |
# Model Overview
|
| 12 |
This model is trained using the runner-up [1] awarded pipeline of the "Medical Segmentation Decathlon Challenge 2018" using the UNet architecture [2] with 32 training images and 9 validation images.
|
| 13 |
|
|
|
|
|
|
|
| 14 |
## Data
|
| 15 |
The training dataset is Task09_Spleen.tar from http://medicaldecathlon.com/.
|
| 16 |
|
| 17 |
## Training configuration
|
| 18 |
-
The
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 19 |
|
| 20 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 21 |
|
| 22 |
## Input and output formats
|
| 23 |
Input: 1 channel CT image
|
|
@@ -29,6 +47,17 @@ This model achieves the following Dice score on the validation data (our own spl
|
|
| 29 |
|
| 30 |
Mean Dice = 0.96
|
| 31 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 32 |
## commands example
|
| 33 |
Execute training:
|
| 34 |
|
|
|
|
| 11 |
# Model Overview
|
| 12 |
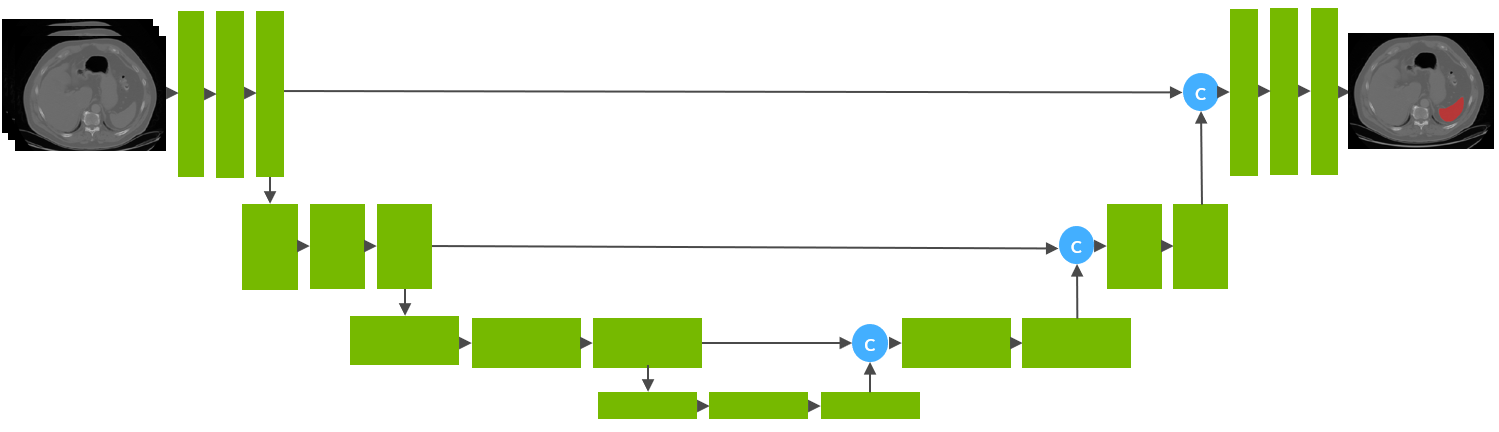
This model is trained using the runner-up [1] awarded pipeline of the "Medical Segmentation Decathlon Challenge 2018" using the UNet architecture [2] with 32 training images and 9 validation images.
|
| 13 |
|
| 14 |
+
|
| 15 |
+
|
| 16 |
## Data
|
| 17 |
The training dataset is Task09_Spleen.tar from http://medicaldecathlon.com/.
|
| 18 |
|
| 19 |
## Training configuration
|
| 20 |
+
The segmentation of spleen region is formulated as the voxel-wise binary classification. Each voxel is predicted as either foreground (spleen) or background. And the model is optimized with gradient descent method minimizing Dice + cross entropy loss between the predicted mask and ground truth segmentation.
|
| 21 |
+
|
| 22 |
+
The training was performed with the following:
|
| 23 |
+
|
| 24 |
+
- GPU: at least 12GB of GPU memory
|
| 25 |
+
- Actual Model Input: 96 x 96 x 96
|
| 26 |
+
- AMP: True
|
| 27 |
+
- Optimizer: Adam
|
| 28 |
+
- Learning Rate: 1e-4
|
| 29 |
+
- Loss: DiceCELoss
|
| 30 |
|
| 31 |
+
Pre-processing transforms:
|
| 32 |
+
|
| 33 |
+
1. Convert data to channel-first
|
| 34 |
+
2. Resample to resolution 1.5 x 1.5 x 2 mm
|
| 35 |
+
3. Scale intensity
|
| 36 |
+
4. Cropping foreground surrounding regions
|
| 37 |
+
5. Cropping random fixed sized regions of size [96,96,96] with the center being a foreground or background voxel at ratio 1 : 1
|
| 38 |
+
6. Randomly shifting intensity of the volume
|
| 39 |
|
| 40 |
## Input and output formats
|
| 41 |
Input: 1 channel CT image
|
|
|
|
| 47 |
|
| 48 |
Mean Dice = 0.96
|
| 49 |
|
| 50 |
+
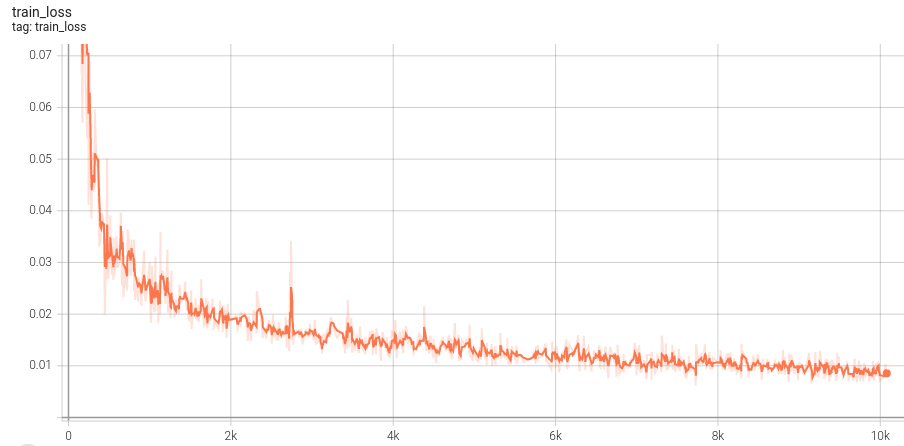
## Training Performance
|
| 51 |
+
A graph showing the training loss over 1260 epochs (10080 iterations).
|
| 52 |
+
|
| 53 |
+
 <br>
|
| 54 |
+
|
| 55 |
+
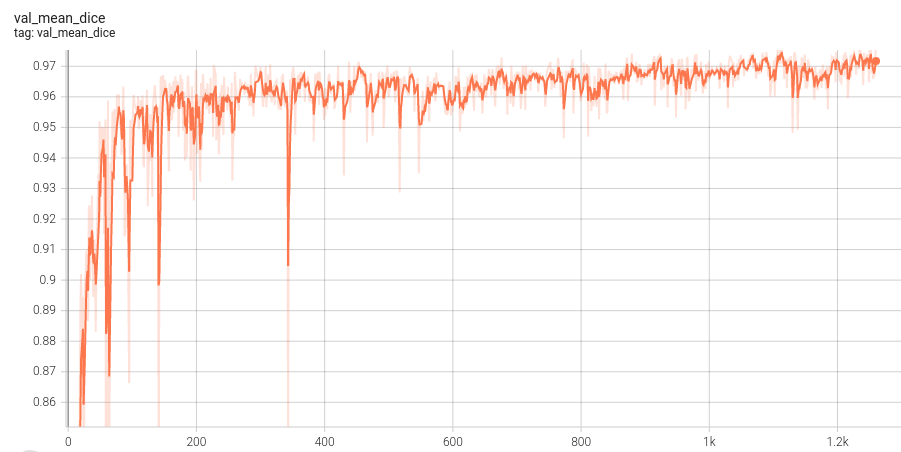
## Validation Performance
|
| 56 |
+
A graph showing the validation mean Dice over 1260 epochs.
|
| 57 |
+
|
| 58 |
+
 <br>
|
| 59 |
+
|
| 60 |
+
|
| 61 |
## commands example
|
| 62 |
Execute training:
|
| 63 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.3.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.3.5": "update to use monai 1.0.1",
|
| 6 |
"0.3.4": "enhance readme on commands example",
|
| 7 |
"0.3.3": "fix license Copyright error",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.3.6",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.3.6": "enhance readme with details of model training",
|
| 6 |
"0.3.5": "update to use monai 1.0.1",
|
| 7 |
"0.3.4": "enhance readme on commands example",
|
| 8 |
"0.3.3": "fix license Copyright error",
|
docs/README.md
CHANGED
|
@@ -4,13 +4,31 @@ A pre-trained model for volumetric (3D) segmentation of the spleen from CT image
|
|
| 4 |
# Model Overview
|
| 5 |
This model is trained using the runner-up [1] awarded pipeline of the "Medical Segmentation Decathlon Challenge 2018" using the UNet architecture [2] with 32 training images and 9 validation images.
|
| 6 |
|
|
|
|
|
|
|
| 7 |
## Data
|
| 8 |
The training dataset is Task09_Spleen.tar from http://medicaldecathlon.com/.
|
| 9 |
|
| 10 |
## Training configuration
|
| 11 |
-
The
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 12 |
|
| 13 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 14 |
|
| 15 |
## Input and output formats
|
| 16 |
Input: 1 channel CT image
|
|
@@ -22,6 +40,17 @@ This model achieves the following Dice score on the validation data (our own spl
|
|
| 22 |
|
| 23 |
Mean Dice = 0.96
|
| 24 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 25 |
## commands example
|
| 26 |
Execute training:
|
| 27 |
|
|
|
|
| 4 |
# Model Overview
|
| 5 |
This model is trained using the runner-up [1] awarded pipeline of the "Medical Segmentation Decathlon Challenge 2018" using the UNet architecture [2] with 32 training images and 9 validation images.
|
| 6 |
|
| 7 |
+

|
| 8 |
+
|
| 9 |
## Data
|
| 10 |
The training dataset is Task09_Spleen.tar from http://medicaldecathlon.com/.
|
| 11 |
|
| 12 |
## Training configuration
|
| 13 |
+
The segmentation of spleen region is formulated as the voxel-wise binary classification. Each voxel is predicted as either foreground (spleen) or background. And the model is optimized with gradient descent method minimizing Dice + cross entropy loss between the predicted mask and ground truth segmentation.
|
| 14 |
+
|
| 15 |
+
The training was performed with the following:
|
| 16 |
+
|
| 17 |
+
- GPU: at least 12GB of GPU memory
|
| 18 |
+
- Actual Model Input: 96 x 96 x 96
|
| 19 |
+
- AMP: True
|
| 20 |
+
- Optimizer: Adam
|
| 21 |
+
- Learning Rate: 1e-4
|
| 22 |
+
- Loss: DiceCELoss
|
| 23 |
|
| 24 |
+
Pre-processing transforms:
|
| 25 |
+
|
| 26 |
+
1. Convert data to channel-first
|
| 27 |
+
2. Resample to resolution 1.5 x 1.5 x 2 mm
|
| 28 |
+
3. Scale intensity
|
| 29 |
+
4. Cropping foreground surrounding regions
|
| 30 |
+
5. Cropping random fixed sized regions of size [96,96,96] with the center being a foreground or background voxel at ratio 1 : 1
|
| 31 |
+
6. Randomly shifting intensity of the volume
|
| 32 |
|
| 33 |
## Input and output formats
|
| 34 |
Input: 1 channel CT image
|
|
|
|
| 40 |
|
| 41 |
Mean Dice = 0.96
|
| 42 |
|
| 43 |
+
## Training Performance
|
| 44 |
+
A graph showing the training loss over 1260 epochs (10080 iterations).
|
| 45 |
+
|
| 46 |
+
 <br>
|
| 47 |
+
|
| 48 |
+
## Validation Performance
|
| 49 |
+
A graph showing the validation mean Dice over 1260 epochs.
|
| 50 |
+
|
| 51 |
+
 <br>
|
| 52 |
+
|
| 53 |
+
|
| 54 |
## commands example
|
| 55 |
Execute training:
|
| 56 |
|