initialize release of the bundle
Browse files- .gitattributes +2 -0
- README.md +112 -0
- configs/inference.json +130 -0
- configs/logging.conf +21 -0
- configs/metadata.json +74 -0
- configs/multi_gpu_train.json +36 -0
- configs/train.json +377 -0
- docs/README.md +105 -0
- docs/license.txt +128 -0
- models/model.pt +3 -0
- models/model.ts +3 -0
- testing.csv +48 -0
- training.csv +3 -0
- validation.csv +0 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,5 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
models/model.ts filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
training.csv filter=lfs diff=lfs merge=lfs -text
|
README.md
ADDED
|
@@ -0,0 +1,112 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
tags:
|
| 3 |
+
- monai
|
| 4 |
+
- medical
|
| 5 |
+
library_name: monai
|
| 6 |
+
license: unknown
|
| 7 |
+
---
|
| 8 |
+
# Model Overview
|
| 9 |
+
|
| 10 |
+
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 11 |
+
|
| 12 |
+
## Workflow
|
| 13 |
+
|
| 14 |
+
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 15 |
+
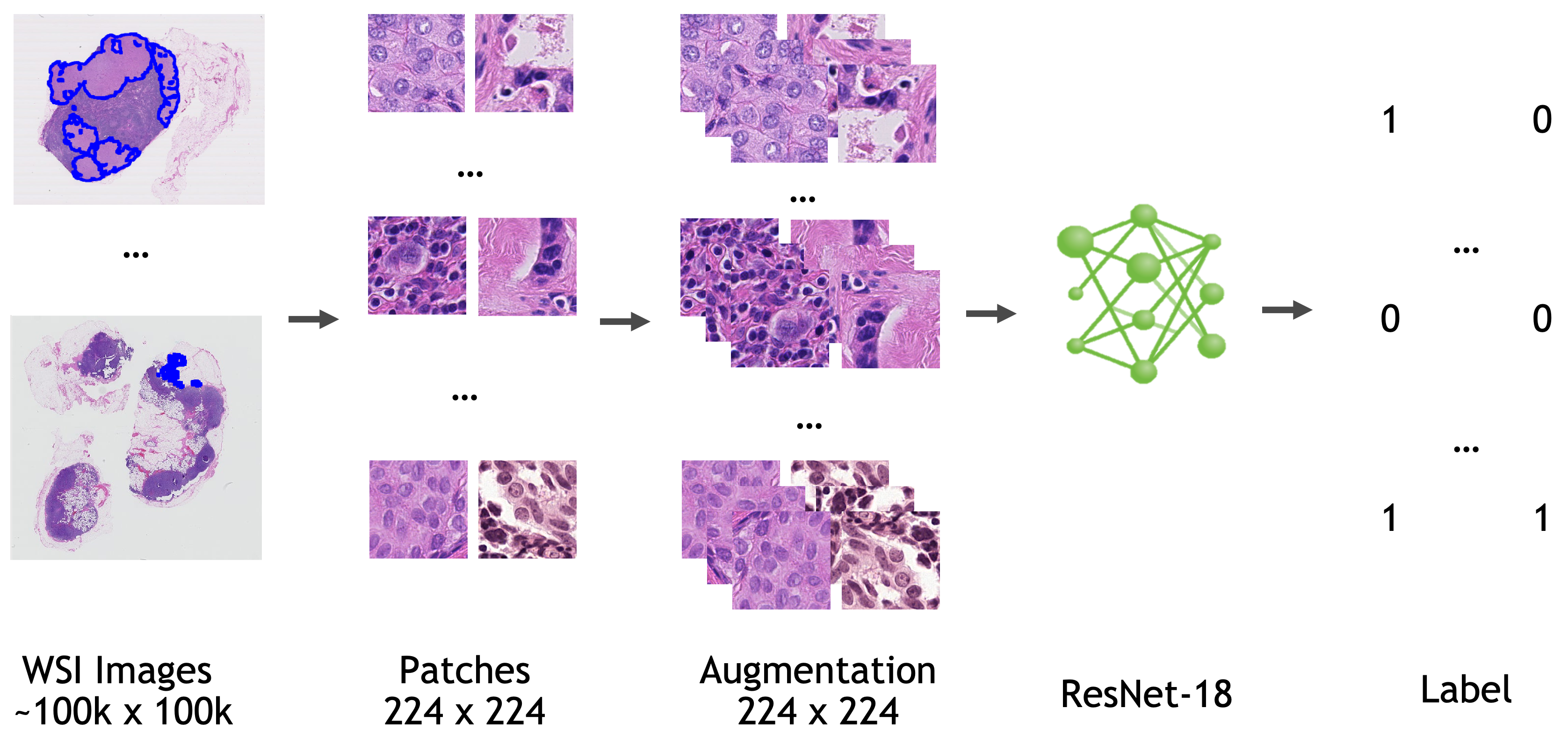
|
| 16 |
+
|
| 17 |
+
## Data
|
| 18 |
+
|
| 19 |
+
All the data used to train, validate, and test this model is from [Camelyon-16 Challenge](https://camelyon16.grand-challenge.org/). You can download all the images for "CAMELYON16" data set from various sources listed [here](https://camelyon17.grand-challenge.org/Data/).
|
| 20 |
+
|
| 21 |
+
Location information for training/validation patches (the location on the whole slide image where patches are extracted) are adopted from [NCRF/coords](https://github.com/baidu-research/NCRF/tree/master/coords).
|
| 22 |
+
|
| 23 |
+
Annotation information are adopted from [NCRF/jsons](https://github.com/baidu-research/NCRF/tree/master/jsons).
|
| 24 |
+
|
| 25 |
+
- Target: Tumor
|
| 26 |
+
- Task: Detection
|
| 27 |
+
- Modality: Histopathology
|
| 28 |
+
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 29 |
+
|
| 30 |
+
### Data Preparation
|
| 31 |
+
|
| 32 |
+
This MMAR expects the training/validation data (whole slide images) reside in `$DATA_ROOT/training/images`. By default `$DATA_ROOT` is pointing to `/workspace/data/medical/pathology/` You can easily modify `$DATA_ROOT` to point to a different directory in `config/environment.json`.
|
| 33 |
+
|
| 34 |
+
To reduce the computation burden during the inference, patches are extracted only where there is tissue and ignoring the background according to a tissue mask. You should run `prepare_inference_data.sh` prior to the inference to generate foreground masks, where the input is the whole slide test images and the output is the foreground masks. Please also create a directory for prediction output, aligning with the one specified with `$MMAR_EVAL_OUTPUT_PATH` in `config/environment.json` (e.g. `/eval`)
|
| 35 |
+
|
| 36 |
+
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 37 |
+
|
| 38 |
+
# Training configuration
|
| 39 |
+
|
| 40 |
+
The training was performed with the following:
|
| 41 |
+
|
| 42 |
+
- Script: train.sh
|
| 43 |
+
- GPU: at least 16 GB of GPU memory.
|
| 44 |
+
- Actual Model Input: 224 x 224 x 3
|
| 45 |
+
- AMP: True
|
| 46 |
+
- Optimizer: Novograd
|
| 47 |
+
- Learning Rate: 1e-3
|
| 48 |
+
- Loss: BCEWithLogitsLoss
|
| 49 |
+
|
| 50 |
+
## Input
|
| 51 |
+
|
| 52 |
+
Input: Input for the training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
| 53 |
+
|
| 54 |
+
1. Extract 224 x 224 x 3 patch from WSI according to the location information from json
|
| 55 |
+
2. Randomly applying color jittering
|
| 56 |
+
3. Randomly applying spatial flipping
|
| 57 |
+
4. Randomly applying spatial rotation
|
| 58 |
+
5. Randomly applying spatial zooming
|
| 59 |
+
6. Randomly applying intensity scaling
|
| 60 |
+
|
| 61 |
+
## Output
|
| 62 |
+
|
| 63 |
+
Output of the network is a probability number of the input patch being tumor or normal.
|
| 64 |
+
|
| 65 |
+
## Inference on a WSI
|
| 66 |
+
|
| 67 |
+
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 68 |
+
|
| 69 |
+
# Model Performance
|
| 70 |
+
|
| 71 |
+
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 72 |
+
This model achieve the ~0.92 accuracy on validation patches, and FROC of ~0.72 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 73 |
+
|
| 74 |
+
# Commands example
|
| 75 |
+
|
| 76 |
+
Execute training:
|
| 77 |
+
|
| 78 |
+
```
|
| 79 |
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 80 |
+
```
|
| 81 |
+
|
| 82 |
+
Override the `train` config to execute multi-GPU training:
|
| 83 |
+
|
| 84 |
+
```
|
| 85 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 86 |
+
```
|
| 87 |
+
|
| 88 |
+
Override the `train` config to execute evaluation with the trained model:
|
| 89 |
+
|
| 90 |
+
```
|
| 91 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 92 |
+
```
|
| 93 |
+
|
| 94 |
+
Execute inference:
|
| 95 |
+
|
| 96 |
+
```
|
| 97 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 98 |
+
```
|
| 99 |
+
|
| 100 |
+
# Intended Use
|
| 101 |
+
|
| 102 |
+
The model needs to be used with NVIDIA hardware and software. For hardware, the model can run on any NVIDIA GPU with memory greater than 16 GB. For software, this model is usable only as part of Transfer Learning & Annotation Tools in Clara Train SDK container. Find out more about Clara Train at the [Clara Train Collections on NGC](https://ngc.nvidia.com/catalog/collections/nvidia:claratrainframework).
|
| 103 |
+
|
| 104 |
+
**The pre-trained models are for developmental purposes only and cannot be used directly for clinical procedures.**
|
| 105 |
+
|
| 106 |
+
# License
|
| 107 |
+
|
| 108 |
+
[End User License Agreement](https://developer.nvidia.com/clara-train-eula) is included with the product. Licenses are also available along with the model application zip file. By pulling and using the Clara Train SDK container and downloading models, you accept the terms and conditions of these licenses.
|
| 109 |
+
|
| 110 |
+
# References
|
| 111 |
+
|
| 112 |
+
[1] He, Kaiming, et al, "Deep Residual Learning for Image Recognition." In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770-778. 2016. <https://arxiv.org/pdf/1512.03385.pdf>
|
configs/inference.json
ADDED
|
@@ -0,0 +1,130 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import os"
|
| 5 |
+
],
|
| 6 |
+
"bundle_root": ".",
|
| 7 |
+
"output_dir": "$os.path.join(@bundle_root, 'eval')",
|
| 8 |
+
"dataset_dir": "/workspace/data/medical/pathology",
|
| 9 |
+
"testing_file": "$os.path.join(@bundle_root, 'testing.csv')",
|
| 10 |
+
"patch_size": [
|
| 11 |
+
224,
|
| 12 |
+
224
|
| 13 |
+
],
|
| 14 |
+
"number_intensity_ch": 3,
|
| 15 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 16 |
+
"network_def": {
|
| 17 |
+
"_target_": "TorchVisionFCModel",
|
| 18 |
+
"model_name": "resnet18",
|
| 19 |
+
"num_classes": 1,
|
| 20 |
+
"use_conv": true,
|
| 21 |
+
"pretrained": true
|
| 22 |
+
},
|
| 23 |
+
"network": "$@network_def.to(@device)",
|
| 24 |
+
"preprocessing": {
|
| 25 |
+
"_target_": "Compose",
|
| 26 |
+
"transforms": [
|
| 27 |
+
{
|
| 28 |
+
"_target_": "CastToTyped",
|
| 29 |
+
"keys": "image",
|
| 30 |
+
"dtype": "float32"
|
| 31 |
+
},
|
| 32 |
+
{
|
| 33 |
+
"_target_": "ScaleIntensityRanged",
|
| 34 |
+
"keys": "image",
|
| 35 |
+
"a_min": 0.0,
|
| 36 |
+
"a_max": 255.0,
|
| 37 |
+
"b_min": -1.0,
|
| 38 |
+
"b_max": 1.0
|
| 39 |
+
},
|
| 40 |
+
{
|
| 41 |
+
"_target_": "ToTensord",
|
| 42 |
+
"keys": "image"
|
| 43 |
+
}
|
| 44 |
+
]
|
| 45 |
+
},
|
| 46 |
+
"datalist": {
|
| 47 |
+
"_target_": "CSVDataset",
|
| 48 |
+
"src": "@testing_file",
|
| 49 |
+
"kwargs_read_csv": {
|
| 50 |
+
"names": [
|
| 51 |
+
"image"
|
| 52 |
+
],
|
| 53 |
+
"header": null
|
| 54 |
+
},
|
| 55 |
+
"transform": {
|
| 56 |
+
"_target_": "Lambdad",
|
| 57 |
+
"keys": "image",
|
| 58 |
+
"func": "$lambda x: os.path.join(@dataset_dir, 'testing/images', x + '.tif')"
|
| 59 |
+
}
|
| 60 |
+
},
|
| 61 |
+
"dataset": {
|
| 62 |
+
"_target_": "MaskedPatchWSIDataset",
|
| 63 |
+
"data": "@datalist",
|
| 64 |
+
"mask_level": 6,
|
| 65 |
+
"patch_size": "@patch_size",
|
| 66 |
+
"transform": "@preprocessing"
|
| 67 |
+
},
|
| 68 |
+
"dataloader": {
|
| 69 |
+
"_target_": "DataLoader",
|
| 70 |
+
"dataset": "@dataset",
|
| 71 |
+
"batch_size": 400,
|
| 72 |
+
"shuffle": false,
|
| 73 |
+
"num_workers": 8
|
| 74 |
+
},
|
| 75 |
+
"inferer": {
|
| 76 |
+
"_target_": "SimpleInferer"
|
| 77 |
+
},
|
| 78 |
+
"postprocessing": {
|
| 79 |
+
"_target_": "Compose",
|
| 80 |
+
"transforms": [
|
| 81 |
+
{
|
| 82 |
+
"_target_": "EnsureTyped",
|
| 83 |
+
"keys": "pred"
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"_target_": "Activationsd",
|
| 87 |
+
"keys": "pred",
|
| 88 |
+
"sigmoid": true
|
| 89 |
+
},
|
| 90 |
+
{
|
| 91 |
+
"_target_": "ToNumpyd",
|
| 92 |
+
"keys": "pred"
|
| 93 |
+
}
|
| 94 |
+
]
|
| 95 |
+
},
|
| 96 |
+
"handlers": [
|
| 97 |
+
{
|
| 98 |
+
"_target_": "CheckpointLoader",
|
| 99 |
+
"load_path": "$@bundle_root + '/models/model.pt'",
|
| 100 |
+
"load_dict": {
|
| 101 |
+
"model": "@network"
|
| 102 |
+
}
|
| 103 |
+
},
|
| 104 |
+
{
|
| 105 |
+
"_target_": "StatsHandler",
|
| 106 |
+
"tag_name": "progress",
|
| 107 |
+
"iteration_print_logger": "$lambda engine: print(f'image: \"{engine.state.batch[\"metadata\"][\"name\"][0]}\", iter: {engine.state.iteration}/{engine.state.epoch_length}') if engine.state.iteration % 100 == 0 else None",
|
| 108 |
+
"output_transform": "$lambda x: None"
|
| 109 |
+
},
|
| 110 |
+
{
|
| 111 |
+
"_target_": "monai.handlers.ProbMapProducer",
|
| 112 |
+
"output_dir": "@output_dir"
|
| 113 |
+
}
|
| 114 |
+
],
|
| 115 |
+
"evaluator": {
|
| 116 |
+
"_target_": "SupervisedEvaluator",
|
| 117 |
+
"device": "@device",
|
| 118 |
+
"val_data_loader": "@dataloader",
|
| 119 |
+
"network": "@network",
|
| 120 |
+
"inferer": "@inferer",
|
| 121 |
+
"postprocessing": "@postprocessing",
|
| 122 |
+
"val_handlers": "@handlers",
|
| 123 |
+
"amp": true,
|
| 124 |
+
"decollate": false
|
| 125 |
+
},
|
| 126 |
+
"evaluating": [
|
| 127 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 128 |
+
"$@evaluator.run()"
|
| 129 |
+
]
|
| 130 |
+
}
|
configs/logging.conf
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[loggers]
|
| 2 |
+
keys=root
|
| 3 |
+
|
| 4 |
+
[handlers]
|
| 5 |
+
keys=consoleHandler
|
| 6 |
+
|
| 7 |
+
[formatters]
|
| 8 |
+
keys=fullFormatter
|
| 9 |
+
|
| 10 |
+
[logger_root]
|
| 11 |
+
level=INFO
|
| 12 |
+
handlers=consoleHandler
|
| 13 |
+
|
| 14 |
+
[handler_consoleHandler]
|
| 15 |
+
class=StreamHandler
|
| 16 |
+
level=INFO
|
| 17 |
+
formatter=fullFormatter
|
| 18 |
+
args=(sys.stdout,)
|
| 19 |
+
|
| 20 |
+
[formatter_fullFormatter]
|
| 21 |
+
format=%(asctime)s - %(name)s - %(levelname)s - %(message)s
|
configs/metadata.json
ADDED
|
@@ -0,0 +1,74 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.1.0",
|
| 4 |
+
"changelog": {
|
| 5 |
+
"0.1.0": "initialize release of the bundle"
|
| 6 |
+
},
|
| 7 |
+
"monai_version": "0.9.1",
|
| 8 |
+
"pytorch_version": "1.12.0",
|
| 9 |
+
"numpy_version": "1.21.2",
|
| 10 |
+
"optional_packages_version": {
|
| 11 |
+
"cucim": "22.04",
|
| 12 |
+
"pandas": "1.3.5",
|
| 13 |
+
"torchvision": "0.13.0"
|
| 14 |
+
},
|
| 15 |
+
"task": "Pathology metastasis detection",
|
| 16 |
+
"description": "A pre-trained model for metastasis detection on Camelyon 16 dataset.",
|
| 17 |
+
"authors": "MONAI team",
|
| 18 |
+
"copyright": "Copyright (c) MONAI Consortium",
|
| 19 |
+
"data_source": "Camelyon dataset",
|
| 20 |
+
"data_type": "tiff",
|
| 21 |
+
"image_classes": "RGB image with intensity between 0 and 255",
|
| 22 |
+
"label_classes": "binary labels for each patch",
|
| 23 |
+
"pred_classes": "scalar probability",
|
| 24 |
+
"eval_metrics": {
|
| 25 |
+
"accuracy": 0,
|
| 26 |
+
"froc": 0
|
| 27 |
+
},
|
| 28 |
+
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 29 |
+
"references": [
|
| 30 |
+
""
|
| 31 |
+
],
|
| 32 |
+
"network_data_format": {
|
| 33 |
+
"inputs": {
|
| 34 |
+
"image": {
|
| 35 |
+
"type": "image",
|
| 36 |
+
"format": "magnitude",
|
| 37 |
+
"num_channels": 3,
|
| 38 |
+
"spatial_shape": [
|
| 39 |
+
224,
|
| 40 |
+
224
|
| 41 |
+
],
|
| 42 |
+
"dtype": "float32",
|
| 43 |
+
"value_range": [
|
| 44 |
+
0,
|
| 45 |
+
255
|
| 46 |
+
],
|
| 47 |
+
"is_patch_data": true,
|
| 48 |
+
"channel_def": {
|
| 49 |
+
"0": "image"
|
| 50 |
+
}
|
| 51 |
+
}
|
| 52 |
+
},
|
| 53 |
+
"outputs": {
|
| 54 |
+
"pred": {
|
| 55 |
+
"type": "probability",
|
| 56 |
+
"format": "classification",
|
| 57 |
+
"num_channels": 1,
|
| 58 |
+
"spatial_shape": [
|
| 59 |
+
1,
|
| 60 |
+
1
|
| 61 |
+
],
|
| 62 |
+
"dtype": "float32",
|
| 63 |
+
"is_patch_data": true,
|
| 64 |
+
"value_range": [
|
| 65 |
+
0,
|
| 66 |
+
1
|
| 67 |
+
],
|
| 68 |
+
"channel_def": {
|
| 69 |
+
"0": "metastasis"
|
| 70 |
+
}
|
| 71 |
+
}
|
| 72 |
+
}
|
| 73 |
+
}
|
| 74 |
+
}
|
configs/multi_gpu_train.json
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"device": "$torch.device(f'cuda:{dist.get_rank()}')",
|
| 3 |
+
"network": {
|
| 4 |
+
"_target_": "torch.nn.parallel.DistributedDataParallel",
|
| 5 |
+
"module": "$@network_def.to(@device)",
|
| 6 |
+
"device_ids": [
|
| 7 |
+
"@device"
|
| 8 |
+
]
|
| 9 |
+
},
|
| 10 |
+
"train#sampler": {
|
| 11 |
+
"_target_": "DistributedSampler",
|
| 12 |
+
"dataset": "@train#dataset",
|
| 13 |
+
"even_divisible": true,
|
| 14 |
+
"shuffle": true
|
| 15 |
+
},
|
| 16 |
+
"train#dataloader#sampler": "@train#sampler",
|
| 17 |
+
"train#dataloader#shuffle": false,
|
| 18 |
+
"train#trainer#train_handlers": "$@train#handlers[: -2 if dist.get_rank() > 0 else None]",
|
| 19 |
+
"validate#sampler": {
|
| 20 |
+
"_target_": "DistributedSampler",
|
| 21 |
+
"dataset": "@validate#dataset",
|
| 22 |
+
"even_divisible": false,
|
| 23 |
+
"shuffle": false
|
| 24 |
+
},
|
| 25 |
+
"validate#dataloader#sampler": "@validate#sampler",
|
| 26 |
+
"validate#evaluator#val_handlers": "$None if dist.get_rank() > 0 else @validate#handlers",
|
| 27 |
+
"training": [
|
| 28 |
+
"$import torch.distributed as dist",
|
| 29 |
+
"$dist.init_process_group(backend='nccl')",
|
| 30 |
+
"$torch.cuda.set_device(@device)",
|
| 31 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 32 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 33 |
+
"$@train#trainer.run()",
|
| 34 |
+
"$dist.destroy_process_group()"
|
| 35 |
+
]
|
| 36 |
+
}
|
configs/train.json
ADDED
|
@@ -0,0 +1,377 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import os",
|
| 4 |
+
"$import ignite"
|
| 5 |
+
],
|
| 6 |
+
"lr": 0.001,
|
| 7 |
+
"num_epochs": 4,
|
| 8 |
+
"bundle_root": ".",
|
| 9 |
+
"ckpt_dir": "$os.path.join(@bundle_root, 'models')",
|
| 10 |
+
"output_dir": "$os.path.join(@bundle_root, 'log')",
|
| 11 |
+
"training_file": "$os.path.join(@bundle_root, 'training.csv')",
|
| 12 |
+
"validation_file": "$os.path.join(@bundle_root, 'validation.csv')",
|
| 13 |
+
"data_root": "/workspace/data/medical/pathology",
|
| 14 |
+
"region_size": [
|
| 15 |
+
768,
|
| 16 |
+
768
|
| 17 |
+
],
|
| 18 |
+
"patch_size": [
|
| 19 |
+
224,
|
| 20 |
+
224
|
| 21 |
+
],
|
| 22 |
+
"grid_shape": [
|
| 23 |
+
3,
|
| 24 |
+
3
|
| 25 |
+
],
|
| 26 |
+
"number_intensity_ch": 3,
|
| 27 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 28 |
+
"network_def": {
|
| 29 |
+
"_target_": "TorchVisionFCModel",
|
| 30 |
+
"model_name": "resnet18",
|
| 31 |
+
"num_classes": 1,
|
| 32 |
+
"use_conv": true,
|
| 33 |
+
"pretrained": true
|
| 34 |
+
},
|
| 35 |
+
"network": "$@network_def.to(@device)",
|
| 36 |
+
"loss": {
|
| 37 |
+
"_target_": "torch.nn.BCEWithLogitsLoss"
|
| 38 |
+
},
|
| 39 |
+
"optimizer": {
|
| 40 |
+
"_target_": "Novograd",
|
| 41 |
+
"params": "$@network.parameters()",
|
| 42 |
+
"lr": "@lr"
|
| 43 |
+
},
|
| 44 |
+
"lr_scheduler": {
|
| 45 |
+
"_target_": "torch.optim.lr_scheduler.CosineAnnealingLR",
|
| 46 |
+
"optimizer": "@optimizer",
|
| 47 |
+
"T_max": "@num_epochs"
|
| 48 |
+
},
|
| 49 |
+
"train": {
|
| 50 |
+
"preprocessing": {
|
| 51 |
+
"_target_": "Compose",
|
| 52 |
+
"transforms": [
|
| 53 |
+
{
|
| 54 |
+
"_target_": "Lambdad",
|
| 55 |
+
"keys": [
|
| 56 |
+
"label"
|
| 57 |
+
],
|
| 58 |
+
"func": "$lambda x: x.reshape((1, *@grid_shape))"
|
| 59 |
+
},
|
| 60 |
+
{
|
| 61 |
+
"_target_": "GridSplitd",
|
| 62 |
+
"keys": [
|
| 63 |
+
"image",
|
| 64 |
+
"label"
|
| 65 |
+
],
|
| 66 |
+
"grid": "@grid_shape",
|
| 67 |
+
"size": {
|
| 68 |
+
"image": "@patch_size",
|
| 69 |
+
"label": 1
|
| 70 |
+
}
|
| 71 |
+
},
|
| 72 |
+
{
|
| 73 |
+
"_target_": "ToTensord",
|
| 74 |
+
"keys": "image"
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"_target_": "TorchVisiond",
|
| 78 |
+
"keys": "image",
|
| 79 |
+
"name": "ColorJitter",
|
| 80 |
+
"brightness": 0.25,
|
| 81 |
+
"contrast": 0.75,
|
| 82 |
+
"saturation": 0.25,
|
| 83 |
+
"hue": 0.04
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"_target_": "ToNumpyd",
|
| 87 |
+
"keys": "image"
|
| 88 |
+
},
|
| 89 |
+
{
|
| 90 |
+
"_target_": "RandFlipd",
|
| 91 |
+
"keys": "image",
|
| 92 |
+
"prob": 0.5
|
| 93 |
+
},
|
| 94 |
+
{
|
| 95 |
+
"_target_": "RandRotate90d",
|
| 96 |
+
"keys": "image",
|
| 97 |
+
"prob": 0.5,
|
| 98 |
+
"max_k": 3,
|
| 99 |
+
"spatial_axes": [
|
| 100 |
+
-2,
|
| 101 |
+
-1
|
| 102 |
+
]
|
| 103 |
+
},
|
| 104 |
+
{
|
| 105 |
+
"_target_": "CastToTyped",
|
| 106 |
+
"keys": "image",
|
| 107 |
+
"dtype": "float32"
|
| 108 |
+
},
|
| 109 |
+
{
|
| 110 |
+
"_target_": "RandZoomd",
|
| 111 |
+
"keys": "image",
|
| 112 |
+
"prob": 0.5,
|
| 113 |
+
"min_zoom": 0.9,
|
| 114 |
+
"max_zoom": 1.1
|
| 115 |
+
},
|
| 116 |
+
{
|
| 117 |
+
"_target_": "ScaleIntensityRanged",
|
| 118 |
+
"keys": "image",
|
| 119 |
+
"a_min": 0.0,
|
| 120 |
+
"a_max": 255.0,
|
| 121 |
+
"b_min": -1.0,
|
| 122 |
+
"b_max": 1.0
|
| 123 |
+
},
|
| 124 |
+
{
|
| 125 |
+
"_target_": "ToTensord",
|
| 126 |
+
"keys": [
|
| 127 |
+
"image",
|
| 128 |
+
"label"
|
| 129 |
+
]
|
| 130 |
+
}
|
| 131 |
+
]
|
| 132 |
+
},
|
| 133 |
+
"datalist": {
|
| 134 |
+
"_target_": "CSVDataset",
|
| 135 |
+
"src": "@training_file",
|
| 136 |
+
"col_groups": {
|
| 137 |
+
"image": 0,
|
| 138 |
+
"patch_location": [
|
| 139 |
+
2,
|
| 140 |
+
1
|
| 141 |
+
],
|
| 142 |
+
"label": [
|
| 143 |
+
3,
|
| 144 |
+
6,
|
| 145 |
+
9,
|
| 146 |
+
4,
|
| 147 |
+
7,
|
| 148 |
+
10,
|
| 149 |
+
5,
|
| 150 |
+
8,
|
| 151 |
+
11
|
| 152 |
+
]
|
| 153 |
+
},
|
| 154 |
+
"kwargs_read_csv": {
|
| 155 |
+
"header": null
|
| 156 |
+
},
|
| 157 |
+
"transform": {
|
| 158 |
+
"_target_": "Lambdad",
|
| 159 |
+
"keys": "image",
|
| 160 |
+
"func": "$lambda x: os.path.join(@data_root, 'training/images', x + '.tif')"
|
| 161 |
+
}
|
| 162 |
+
},
|
| 163 |
+
"dataset": {
|
| 164 |
+
"_target_": "monai.data.wsi_datasets.PatchWSIDataset",
|
| 165 |
+
"data": "@train#datalist",
|
| 166 |
+
"patch_level": 0,
|
| 167 |
+
"patch_size": "@region_size",
|
| 168 |
+
"reader": "cucim",
|
| 169 |
+
"transform": "@train#preprocessing"
|
| 170 |
+
},
|
| 171 |
+
"dataloader": {
|
| 172 |
+
"_target_": "DataLoader",
|
| 173 |
+
"dataset": "@train#dataset",
|
| 174 |
+
"batch_size": 128,
|
| 175 |
+
"pin_memory": true,
|
| 176 |
+
"num_workers": 8
|
| 177 |
+
},
|
| 178 |
+
"inferer": {
|
| 179 |
+
"_target_": "SimpleInferer"
|
| 180 |
+
},
|
| 181 |
+
"postprocessing": {
|
| 182 |
+
"_target_": "Compose",
|
| 183 |
+
"transforms": [
|
| 184 |
+
{
|
| 185 |
+
"_target_": "Activationsd",
|
| 186 |
+
"keys": "pred",
|
| 187 |
+
"sigmoid": true
|
| 188 |
+
},
|
| 189 |
+
{
|
| 190 |
+
"_target_": "AsDiscreted",
|
| 191 |
+
"keys": "pred",
|
| 192 |
+
"threshold": 0.5
|
| 193 |
+
}
|
| 194 |
+
]
|
| 195 |
+
},
|
| 196 |
+
"handlers": [
|
| 197 |
+
{
|
| 198 |
+
"_target_": "ValidationHandler",
|
| 199 |
+
"validator": "@validate#evaluator",
|
| 200 |
+
"epoch_level": true,
|
| 201 |
+
"interval": 1
|
| 202 |
+
},
|
| 203 |
+
{
|
| 204 |
+
"_target_": "StatsHandler",
|
| 205 |
+
"tag_name": "train_loss",
|
| 206 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 207 |
+
},
|
| 208 |
+
{
|
| 209 |
+
"_target_": "LrScheduleHandler",
|
| 210 |
+
"lr_scheduler": "@lr_scheduler",
|
| 211 |
+
"print_lr": true
|
| 212 |
+
},
|
| 213 |
+
{
|
| 214 |
+
"_target_": "TensorBoardStatsHandler",
|
| 215 |
+
"log_dir": "@output_dir",
|
| 216 |
+
"tag_name": "train_loss",
|
| 217 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 218 |
+
}
|
| 219 |
+
],
|
| 220 |
+
"key_metric": {
|
| 221 |
+
"train_acc": {
|
| 222 |
+
"_target_": "ignite.metrics.Accuracy",
|
| 223 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 224 |
+
}
|
| 225 |
+
},
|
| 226 |
+
"trainer": {
|
| 227 |
+
"_target_": "SupervisedTrainer",
|
| 228 |
+
"device": "@device",
|
| 229 |
+
"max_epochs": "@num_epochs",
|
| 230 |
+
"train_data_loader": "@train#dataloader",
|
| 231 |
+
"network": "@network",
|
| 232 |
+
"optimizer": "@optimizer",
|
| 233 |
+
"loss_function": "@loss",
|
| 234 |
+
"inferer": "@train#inferer",
|
| 235 |
+
"amp": true,
|
| 236 |
+
"postprocessing": "@train#postprocessing",
|
| 237 |
+
"key_train_metric": "@train#key_metric",
|
| 238 |
+
"train_handlers": "@train#handlers"
|
| 239 |
+
}
|
| 240 |
+
},
|
| 241 |
+
"validate": {
|
| 242 |
+
"preprocessing": {
|
| 243 |
+
"_target_": "Compose",
|
| 244 |
+
"transforms": [
|
| 245 |
+
{
|
| 246 |
+
"_target_": "Lambdad",
|
| 247 |
+
"keys": "label",
|
| 248 |
+
"func": "$lambda x: x.reshape((1, *@grid_shape))"
|
| 249 |
+
},
|
| 250 |
+
{
|
| 251 |
+
"_target_": "GridSplitd",
|
| 252 |
+
"keys": [
|
| 253 |
+
"image",
|
| 254 |
+
"label"
|
| 255 |
+
],
|
| 256 |
+
"grid": "@grid_shape",
|
| 257 |
+
"size": {
|
| 258 |
+
"image": "@patch_size",
|
| 259 |
+
"label": 1
|
| 260 |
+
}
|
| 261 |
+
},
|
| 262 |
+
{
|
| 263 |
+
"_target_": "CastToTyped",
|
| 264 |
+
"keys": "image",
|
| 265 |
+
"dtype": "float32"
|
| 266 |
+
},
|
| 267 |
+
{
|
| 268 |
+
"_target_": "ScaleIntensityRanged",
|
| 269 |
+
"keys": "image",
|
| 270 |
+
"a_min": 0.0,
|
| 271 |
+
"a_max": 255.0,
|
| 272 |
+
"b_min": -1.0,
|
| 273 |
+
"b_max": 1.0
|
| 274 |
+
},
|
| 275 |
+
{
|
| 276 |
+
"_target_": "ToTensord",
|
| 277 |
+
"keys": [
|
| 278 |
+
"image",
|
| 279 |
+
"label"
|
| 280 |
+
]
|
| 281 |
+
}
|
| 282 |
+
]
|
| 283 |
+
},
|
| 284 |
+
"datalist": {
|
| 285 |
+
"_target_": "CSVDataset",
|
| 286 |
+
"src": "@validation_file",
|
| 287 |
+
"col_groups": {
|
| 288 |
+
"image": 0,
|
| 289 |
+
"patch_location": [
|
| 290 |
+
2,
|
| 291 |
+
1
|
| 292 |
+
],
|
| 293 |
+
"label": [
|
| 294 |
+
3,
|
| 295 |
+
6,
|
| 296 |
+
9,
|
| 297 |
+
4,
|
| 298 |
+
7,
|
| 299 |
+
10,
|
| 300 |
+
5,
|
| 301 |
+
8,
|
| 302 |
+
11
|
| 303 |
+
]
|
| 304 |
+
},
|
| 305 |
+
"kwargs_read_csv": {
|
| 306 |
+
"header": null
|
| 307 |
+
},
|
| 308 |
+
"transform": {
|
| 309 |
+
"_target_": "Lambdad",
|
| 310 |
+
"keys": "image",
|
| 311 |
+
"func": "$lambda x: os.path.join(@data_root, 'training/images', x + '.tif')"
|
| 312 |
+
}
|
| 313 |
+
},
|
| 314 |
+
"dataset": {
|
| 315 |
+
"_target_": "monai.data.wsi_datasets.PatchWSIDataset",
|
| 316 |
+
"data": "@validate#datalist",
|
| 317 |
+
"patch_level": 0,
|
| 318 |
+
"patch_size": "@region_size",
|
| 319 |
+
"reader": "cucim",
|
| 320 |
+
"transform": "@validate#preprocessing"
|
| 321 |
+
},
|
| 322 |
+
"dataloader": {
|
| 323 |
+
"_target_": "DataLoader",
|
| 324 |
+
"dataset": "@validate#dataset",
|
| 325 |
+
"batch_size": 128,
|
| 326 |
+
"pin_memory": true,
|
| 327 |
+
"shuffle": false,
|
| 328 |
+
"num_workers": 8
|
| 329 |
+
},
|
| 330 |
+
"inferer": {
|
| 331 |
+
"_target_": "SimpleInferer"
|
| 332 |
+
},
|
| 333 |
+
"postprocessing": "%train#postprocessing",
|
| 334 |
+
"handlers": [
|
| 335 |
+
{
|
| 336 |
+
"_target_": "StatsHandler",
|
| 337 |
+
"iteration_log": false
|
| 338 |
+
},
|
| 339 |
+
{
|
| 340 |
+
"_target_": "TensorBoardStatsHandler",
|
| 341 |
+
"log_dir": "@output_dir",
|
| 342 |
+
"iteration_log": false
|
| 343 |
+
},
|
| 344 |
+
{
|
| 345 |
+
"_target_": "CheckpointSaver",
|
| 346 |
+
"save_dir": "@ckpt_dir",
|
| 347 |
+
"save_dict": {
|
| 348 |
+
"model": "@network"
|
| 349 |
+
},
|
| 350 |
+
"save_key_metric": true,
|
| 351 |
+
"key_metric_filename": "model.pt"
|
| 352 |
+
}
|
| 353 |
+
],
|
| 354 |
+
"key_metric": {
|
| 355 |
+
"valid_acc": {
|
| 356 |
+
"_target_": "ignite.metrics.Accuracy",
|
| 357 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 358 |
+
}
|
| 359 |
+
},
|
| 360 |
+
"evaluator": {
|
| 361 |
+
"_target_": "SupervisedEvaluator",
|
| 362 |
+
"device": "@device",
|
| 363 |
+
"val_data_loader": "@validate#dataloader",
|
| 364 |
+
"network": "@network",
|
| 365 |
+
"inferer": "@validate#inferer",
|
| 366 |
+
"postprocessing": "@validate#postprocessing",
|
| 367 |
+
"key_val_metric": "@validate#key_metric",
|
| 368 |
+
"val_handlers": "@validate#handlers",
|
| 369 |
+
"amp": true
|
| 370 |
+
}
|
| 371 |
+
},
|
| 372 |
+
"training": [
|
| 373 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 374 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 375 |
+
"$@train#trainer.run()"
|
| 376 |
+
]
|
| 377 |
+
}
|
docs/README.md
ADDED
|
@@ -0,0 +1,105 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Model Overview
|
| 2 |
+
|
| 3 |
+
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
|
| 4 |
+
|
| 5 |
+
## Workflow
|
| 6 |
+
|
| 7 |
+
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
|
| 8 |
+

|
| 9 |
+
|
| 10 |
+
## Data
|
| 11 |
+
|
| 12 |
+
All the data used to train, validate, and test this model is from [Camelyon-16 Challenge](https://camelyon16.grand-challenge.org/). You can download all the images for "CAMELYON16" data set from various sources listed [here](https://camelyon17.grand-challenge.org/Data/).
|
| 13 |
+
|
| 14 |
+
Location information for training/validation patches (the location on the whole slide image where patches are extracted) are adopted from [NCRF/coords](https://github.com/baidu-research/NCRF/tree/master/coords).
|
| 15 |
+
|
| 16 |
+
Annotation information are adopted from [NCRF/jsons](https://github.com/baidu-research/NCRF/tree/master/jsons).
|
| 17 |
+
|
| 18 |
+
- Target: Tumor
|
| 19 |
+
- Task: Detection
|
| 20 |
+
- Modality: Histopathology
|
| 21 |
+
- Size: 270 WSIs for training/validation, 48 WSIs for testing
|
| 22 |
+
|
| 23 |
+
### Data Preparation
|
| 24 |
+
|
| 25 |
+
This MMAR expects the training/validation data (whole slide images) reside in `$DATA_ROOT/training/images`. By default `$DATA_ROOT` is pointing to `/workspace/data/medical/pathology/` You can easily modify `$DATA_ROOT` to point to a different directory in `config/environment.json`.
|
| 26 |
+
|
| 27 |
+
To reduce the computation burden during the inference, patches are extracted only where there is tissue and ignoring the background according to a tissue mask. You should run `prepare_inference_data.sh` prior to the inference to generate foreground masks, where the input is the whole slide test images and the output is the foreground masks. Please also create a directory for prediction output, aligning with the one specified with `$MMAR_EVAL_OUTPUT_PATH` in `config/environment.json` (e.g. `/eval`)
|
| 28 |
+
|
| 29 |
+
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
|
| 30 |
+
|
| 31 |
+
# Training configuration
|
| 32 |
+
|
| 33 |
+
The training was performed with the following:
|
| 34 |
+
|
| 35 |
+
- Script: train.sh
|
| 36 |
+
- GPU: at least 16 GB of GPU memory.
|
| 37 |
+
- Actual Model Input: 224 x 224 x 3
|
| 38 |
+
- AMP: True
|
| 39 |
+
- Optimizer: Novograd
|
| 40 |
+
- Learning Rate: 1e-3
|
| 41 |
+
- Loss: BCEWithLogitsLoss
|
| 42 |
+
|
| 43 |
+
## Input
|
| 44 |
+
|
| 45 |
+
Input: Input for the training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
| 46 |
+
|
| 47 |
+
1. Extract 224 x 224 x 3 patch from WSI according to the location information from json
|
| 48 |
+
2. Randomly applying color jittering
|
| 49 |
+
3. Randomly applying spatial flipping
|
| 50 |
+
4. Randomly applying spatial rotation
|
| 51 |
+
5. Randomly applying spatial zooming
|
| 52 |
+
6. Randomly applying intensity scaling
|
| 53 |
+
|
| 54 |
+
## Output
|
| 55 |
+
|
| 56 |
+
Output of the network is a probability number of the input patch being tumor or normal.
|
| 57 |
+
|
| 58 |
+
## Inference on a WSI
|
| 59 |
+
|
| 60 |
+
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 61 |
+
|
| 62 |
+
# Model Performance
|
| 63 |
+
|
| 64 |
+
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 65 |
+
This model achieve the ~0.92 accuracy on validation patches, and FROC of ~0.72 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 66 |
+
|
| 67 |
+
# Commands example
|
| 68 |
+
|
| 69 |
+
Execute training:
|
| 70 |
+
|
| 71 |
+
```
|
| 72 |
+
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
|
| 73 |
+
```
|
| 74 |
+
|
| 75 |
+
Override the `train` config to execute multi-GPU training:
|
| 76 |
+
|
| 77 |
+
```
|
| 78 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
|
| 79 |
+
```
|
| 80 |
+
|
| 81 |
+
Override the `train` config to execute evaluation with the trained model:
|
| 82 |
+
|
| 83 |
+
```
|
| 84 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
|
| 85 |
+
```
|
| 86 |
+
|
| 87 |
+
Execute inference:
|
| 88 |
+
|
| 89 |
+
```
|
| 90 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 91 |
+
```
|
| 92 |
+
|
| 93 |
+
# Intended Use
|
| 94 |
+
|
| 95 |
+
The model needs to be used with NVIDIA hardware and software. For hardware, the model can run on any NVIDIA GPU with memory greater than 16 GB. For software, this model is usable only as part of Transfer Learning & Annotation Tools in Clara Train SDK container. Find out more about Clara Train at the [Clara Train Collections on NGC](https://ngc.nvidia.com/catalog/collections/nvidia:claratrainframework).
|
| 96 |
+
|
| 97 |
+
**The pre-trained models are for developmental purposes only and cannot be used directly for clinical procedures.**
|
| 98 |
+
|
| 99 |
+
# License
|
| 100 |
+
|
| 101 |
+
[End User License Agreement](https://developer.nvidia.com/clara-train-eula) is included with the product. Licenses are also available along with the model application zip file. By pulling and using the Clara Train SDK container and downloading models, you accept the terms and conditions of these licenses.
|
| 102 |
+
|
| 103 |
+
# References
|
| 104 |
+
|
| 105 |
+
[1] He, Kaiming, et al, "Deep Residual Learning for Image Recognition." In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770-778. 2016. <https://arxiv.org/pdf/1512.03385.pdf>
|
docs/license.txt
ADDED
|
@@ -0,0 +1,128 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
CAMELYON16 data set by Computational Pathology Group of Radboud University
|
| 2 |
+
Medical Centre
|
| 3 |
+
|
| 4 |
+
CAMELYON16 data set is available under CC0.
|
| 5 |
+
|
| 6 |
+
==========================================================================
|
| 7 |
+
|
| 8 |
+
Creative Commons Legal Code
|
| 9 |
+
|
| 10 |
+
CC0 1.0 Universal
|
| 11 |
+
|
| 12 |
+
CREATIVE COMMONS CORPORATION IS NOT A LAW FIRM AND DOES NOT PROVIDE
|
| 13 |
+
LEGAL SERVICES. DISTRIBUTION OF THIS DOCUMENT DOES NOT CREATE AN
|
| 14 |
+
ATTORNEY-CLIENT RELATIONSHIP. CREATIVE COMMONS PROVIDES THIS
|
| 15 |
+
INFORMATION ON AN "AS-IS" BASIS. CREATIVE COMMONS MAKES NO WARRANTIES
|
| 16 |
+
REGARDING THE USE OF THIS DOCUMENT OR THE INFORMATION OR WORKS
|
| 17 |
+
PROVIDED HEREUNDER, AND DISCLAIMS LIABILITY FOR DAMAGES RESULTING FROM
|
| 18 |
+
THE USE OF THIS DOCUMENT OR THE INFORMATION OR WORKS PROVIDED
|
| 19 |
+
HEREUNDER.
|
| 20 |
+
|
| 21 |
+
Statement of Purpose
|
| 22 |
+
|
| 23 |
+
The laws of most jurisdictions throughout the world automatically confer
|
| 24 |
+
exclusive Copyright and Related Rights (defined below) upon the creator
|
| 25 |
+
and subsequent owner(s) (each and all, an "owner") of an original work of
|
| 26 |
+
authorship and/or a database (each, a "Work").
|
| 27 |
+
|
| 28 |
+
Certain owners wish to permanently relinquish those rights to a Work for
|
| 29 |
+
the purpose of contributing to a commons of creative, cultural and
|
| 30 |
+
scientific works ("Commons") that the public can reliably and without fear
|
| 31 |
+
of later claims of infringement build upon, modify, incorporate in other
|
| 32 |
+
works, reuse and redistribute as freely as possible in any form whatsoever
|
| 33 |
+
and for any purposes, including without limitation commercial purposes.
|
| 34 |
+
These owners may contribute to the Commons to promote the ideal of a free
|
| 35 |
+
culture and the further production of creative, cultural and scientific
|
| 36 |
+
works, or to gain reputation or greater distribution for their Work in
|
| 37 |
+
part through the use and efforts of others.
|
| 38 |
+
|
| 39 |
+
For these and/or other purposes and motivations, and without any
|
| 40 |
+
expectation of additional consideration or compensation, the person
|
| 41 |
+
associating CC0 with a Work (the "Affirmer"), to the extent that he or she
|
| 42 |
+
is an owner of Copyright and Related Rights in the Work, voluntarily
|
| 43 |
+
elects to apply CC0 to the Work and publicly distribute the Work under its
|
| 44 |
+
terms, with knowledge of his or her Copyright and Related Rights in the
|
| 45 |
+
Work and the meaning and intended legal effect of CC0 on those rights.
|
| 46 |
+
|
| 47 |
+
1. Copyright and Related Rights. A Work made available under CC0 may be
|
| 48 |
+
protected by copyright and related or neighboring rights ("Copyright and
|
| 49 |
+
Related Rights"). Copyright and Related Rights include, but are not
|
| 50 |
+
limited to, the following:
|
| 51 |
+
|
| 52 |
+
i. the right to reproduce, adapt, distribute, perform, display,
|
| 53 |
+
communicate, and translate a Work;
|
| 54 |
+
ii. moral rights retained by the original author(s) and/or performer(s);
|
| 55 |
+
iii. publicity and privacy rights pertaining to a person's image or
|
| 56 |
+
likeness depicted in a Work;
|
| 57 |
+
iv. rights protecting against unfair competition in regards to a Work,
|
| 58 |
+
subject to the limitations in paragraph 4(a), below;
|
| 59 |
+
v. rights protecting the extraction, dissemination, use and reuse of data
|
| 60 |
+
in a Work;
|
| 61 |
+
vi. database rights (such as those arising under Directive 96/9/EC of the
|
| 62 |
+
European Parliament and of the Council of 11 March 1996 on the legal
|
| 63 |
+
protection of databases, and under any national implementation
|
| 64 |
+
thereof, including any amended or successor version of such
|
| 65 |
+
directive); and
|
| 66 |
+
vii. other similar, equivalent or corresponding rights throughout the
|
| 67 |
+
world based on applicable law or treaty, and any national
|
| 68 |
+
implementations thereof.
|
| 69 |
+
|
| 70 |
+
2. Waiver. To the greatest extent permitted by, but not in contravention
|
| 71 |
+
of, applicable law, Affirmer hereby overtly, fully, permanently,
|
| 72 |
+
irrevocably and unconditionally waives, abandons, and surrenders all of
|
| 73 |
+
Affirmer's Copyright and Related Rights and associated claims and causes
|
| 74 |
+
of action, whether now known or unknown (including existing as well as
|
| 75 |
+
future claims and causes of action), in the Work (i) in all territories
|
| 76 |
+
worldwide, (ii) for the maximum duration provided by applicable law or
|
| 77 |
+
treaty (including future time extensions), (iii) in any current or future
|
| 78 |
+
medium and for any number of copies, and (iv) for any purpose whatsoever,
|
| 79 |
+
including without limitation commercial, advertising or promotional
|
| 80 |
+
purposes (the "Waiver"). Affirmer makes the Waiver for the benefit of each
|
| 81 |
+
member of the public at large and to the detriment of Affirmer's heirs and
|
| 82 |
+
successors, fully intending that such Waiver shall not be subject to
|
| 83 |
+
revocation, rescission, cancellation, termination, or any other legal or
|
| 84 |
+
equitable action to disrupt the quiet enjoyment of the Work by the public
|
| 85 |
+
as contemplated by Affirmer's express Statement of Purpose.
|
| 86 |
+
|
| 87 |
+
3. Public License Fallback. Should any part of the Waiver for any reason
|
| 88 |
+
be judged legally invalid or ineffective under applicable law, then the
|
| 89 |
+
Waiver shall be preserved to the maximum extent permitted taking into
|
| 90 |
+
account Affirmer's express Statement of Purpose. In addition, to the
|
| 91 |
+
extent the Waiver is so judged Affirmer hereby grants to each affected
|
| 92 |
+
person a royalty-free, non transferable, non sublicensable, non exclusive,
|
| 93 |
+
irrevocable and unconditional license to exercise Affirmer's Copyright and
|
| 94 |
+
Related Rights in the Work (i) in all territories worldwide, (ii) for the
|
| 95 |
+
maximum duration provided by applicable law or treaty (including future
|
| 96 |
+
time extensions), (iii) in any current or future medium and for any number
|
| 97 |
+
of copies, and (iv) for any purpose whatsoever, including without
|
| 98 |
+
limitation commercial, advertising or promotional purposes (the
|
| 99 |
+
"License"). The License shall be deemed effective as of the date CC0 was
|
| 100 |
+
applied by Affirmer to the Work. Should any part of the License for any
|
| 101 |
+
reason be judged legally invalid or ineffective under applicable law, such
|
| 102 |
+
partial invalidity or ineffectiveness shall not invalidate the remainder
|
| 103 |
+
of the License, and in such case Affirmer hereby affirms that he or she
|
| 104 |
+
will not (i) exercise any of his or her remaining Copyright and Related
|
| 105 |
+
Rights in the Work or (ii) assert any associated claims and causes of
|
| 106 |
+
action with respect to the Work, in either case contrary to Affirmer's
|
| 107 |
+
express Statement of Purpose.
|
| 108 |
+
|
| 109 |
+
4. Limitations and Disclaimers.
|
| 110 |
+
|
| 111 |
+
a. No trademark or patent rights held by Affirmer are waived, abandoned,
|
| 112 |
+
surrendered, licensed or otherwise affected by this document.
|
| 113 |
+
b. Affirmer offers the Work as-is and makes no representations or
|
| 114 |
+
warranties of any kind concerning the Work, express, implied,
|
| 115 |
+
statutory or otherwise, including without limitation warranties of
|
| 116 |
+
title, merchantability, fitness for a particular purpose, non
|
| 117 |
+
infringement, or the absence of latent or other defects, accuracy, or
|
| 118 |
+
the present or absence of errors, whether or not discoverable, all to
|
| 119 |
+
the greatest extent permissible under applicable law.
|
| 120 |
+
c. Affirmer disclaims responsibility for clearing rights of other persons
|
| 121 |
+
that may apply to the Work or any use thereof, including without
|
| 122 |
+
limitation any person's Copyright and Related Rights in the Work.
|
| 123 |
+
Further, Affirmer disclaims responsibility for obtaining any necessary
|
| 124 |
+
consents, permissions or other rights required for any use of the
|
| 125 |
+
Work.
|
| 126 |
+
d. Affirmer understands and acknowledges that Creative Commons is not a
|
| 127 |
+
party to this document and has no duty or obligation with respect to
|
| 128 |
+
this CC0 or use of the Work.
|
models/model.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:90c8ae76f342c6fa5d89b9df9f5af014c9fd547067db5bae00a8e9a94e242e7a
|
| 3 |
+
size 44790201
|
models/model.ts
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:13eeb8fc74eab45b2fea84a05de5e4aaed20df466c6293c538fb061a66fa5e1d
|
| 3 |
+
size 44822748
|
testing.csv
ADDED
|
@@ -0,0 +1,48 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
test_001
|
| 2 |
+
test_002
|
| 3 |
+
test_004
|
| 4 |
+
test_008
|
| 5 |
+
test_010
|
| 6 |
+
test_011
|
| 7 |
+
test_013
|
| 8 |
+
test_016
|
| 9 |
+
test_021
|
| 10 |
+
test_026
|
| 11 |
+
test_027
|
| 12 |
+
test_029
|
| 13 |
+
test_030
|
| 14 |
+
test_033
|
| 15 |
+
test_038
|
| 16 |
+
test_040
|
| 17 |
+
test_046
|
| 18 |
+
test_048
|
| 19 |
+
test_051
|
| 20 |
+
test_052
|
| 21 |
+
test_061
|
| 22 |
+
test_064
|
| 23 |
+
test_065
|
| 24 |
+
test_066
|
| 25 |
+
test_068
|
| 26 |
+
test_069
|
| 27 |
+
test_071
|
| 28 |
+
test_073
|
| 29 |
+
test_074
|
| 30 |
+
test_075
|
| 31 |
+
test_079
|
| 32 |
+
test_082
|
| 33 |
+
test_084
|
| 34 |
+
test_090
|
| 35 |
+
test_092
|
| 36 |
+
test_094
|
| 37 |
+
test_097
|
| 38 |
+
test_099
|
| 39 |
+
test_102
|
| 40 |
+
test_104
|
| 41 |
+
test_105
|
| 42 |
+
test_108
|
| 43 |
+
test_110
|
| 44 |
+
test_113
|
| 45 |
+
test_116
|
| 46 |
+
test_117
|
| 47 |
+
test_121
|
| 48 |
+
test_122
|
training.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e3e51c55b11485c51fea7f9dab7811edc120ab473da0f1349c4bf40c9d15d0b4
|
| 3 |
+
size 20690435
|
validation.csv
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|