Update with figure links
Browse files- README.md +7 -7
- configs/metadata.json +2 -1
- docs/README.md +7 -7
README.md
CHANGED
|
@@ -21,7 +21,7 @@ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovern
|
|
| 21 |
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
|
| 22 |
unzip -q consep_dataset.zip
|
| 23 |
```
|
| 24 |
-
 based on the centroid given in the dataset.
|
| 60 |
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
|
| 61 |
-
- Update the label index for the target
|
| 62 |
-
- Other cells which are part of the patch are modified to have label
|
| 63 |
|
| 64 |
Example dataset.json
|
| 65 |
```json
|
|
@@ -102,7 +102,7 @@ Example dataset.json
|
|
| 102 |
- 0 = Background
|
| 103 |
- 1 = Nuclei
|
| 104 |
|
| 105 |
-
<br/>
|
| 25 |
|
| 26 |
## Training configuration
|
| 27 |
The training was performed with the following:
|
|
|
|
| 58 |
|
| 59 |
- Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset.
|
| 60 |
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
|
| 61 |
+
- Update the label index for the target nuclei based on the class value
|
| 62 |
+
- Other cells which are part of the patch are modified to have label idx = 255
|
| 63 |
|
| 64 |
Example dataset.json
|
| 65 |
```json
|
|
|
|
| 102 |
- 0 = Background
|
| 103 |
- 1 = Nuclei
|
| 104 |
|
| 105 |
+
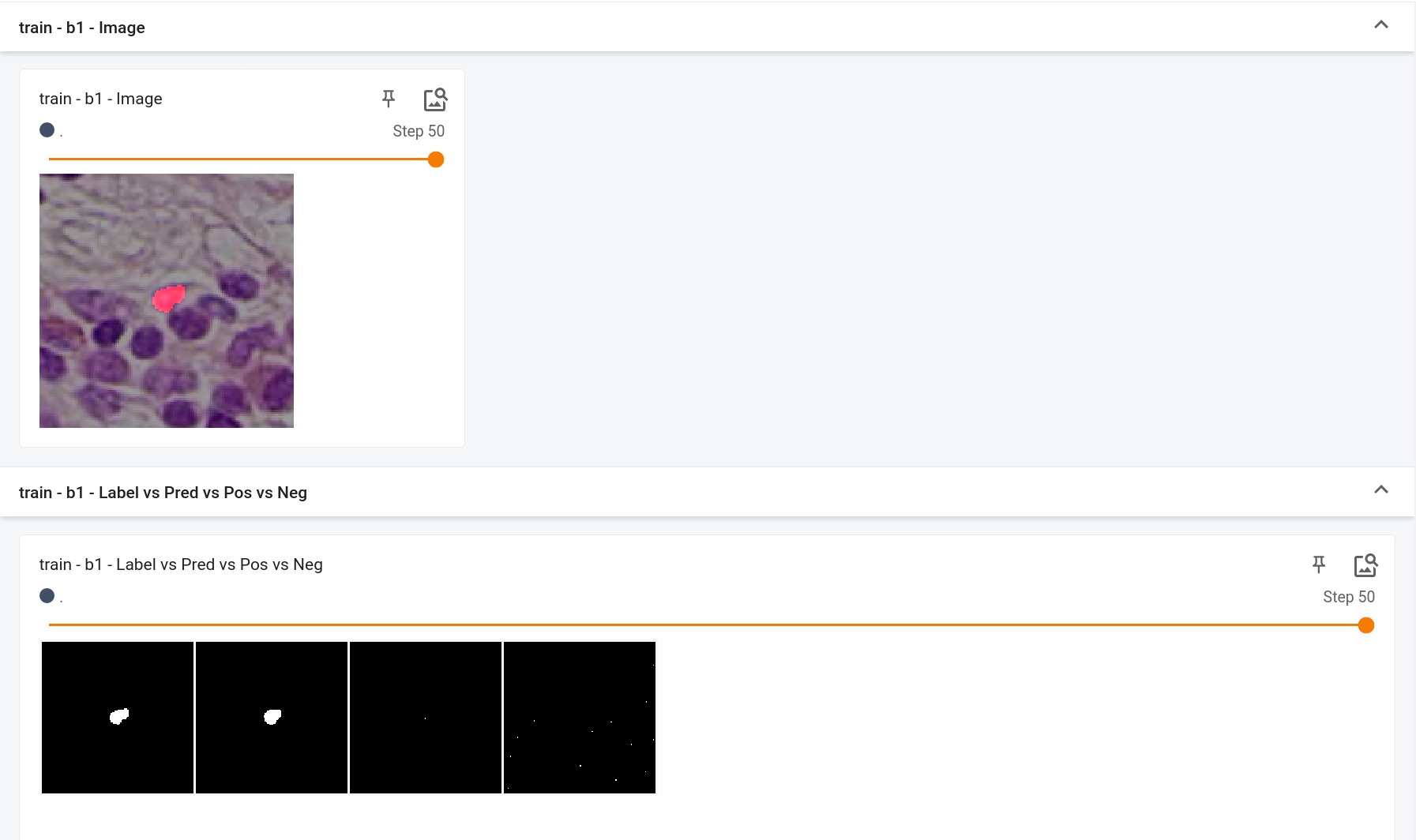
|
| 106 |
|
| 107 |
## Scores
|
| 108 |
This model achieves the following Dice score on the validation data provided as part of the dataset:
|
|
|
|
| 114 |
## Training Performance
|
| 115 |
A graph showing the training Loss and Dice over 50 epochs.
|
| 116 |
|
| 117 |
+
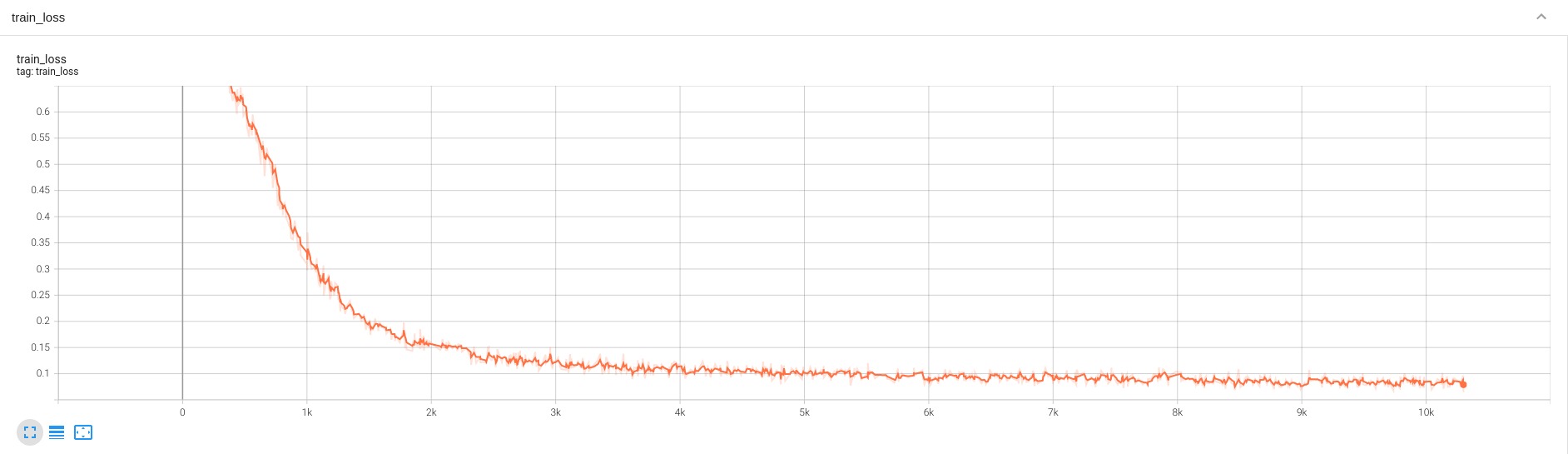 <br>
|
| 118 |
+
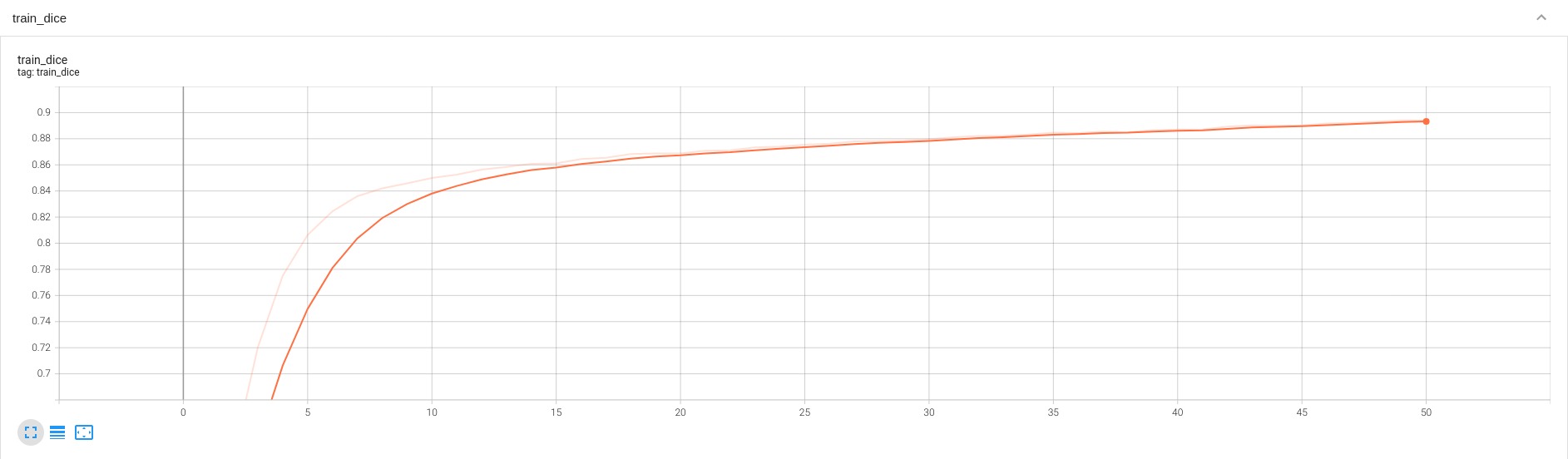 <br>
|
| 119 |
|
| 120 |
## Validation Performance
|
| 121 |
A graph showing the validation mean Dice over 50 epochs.
|
| 122 |
|
| 123 |
+
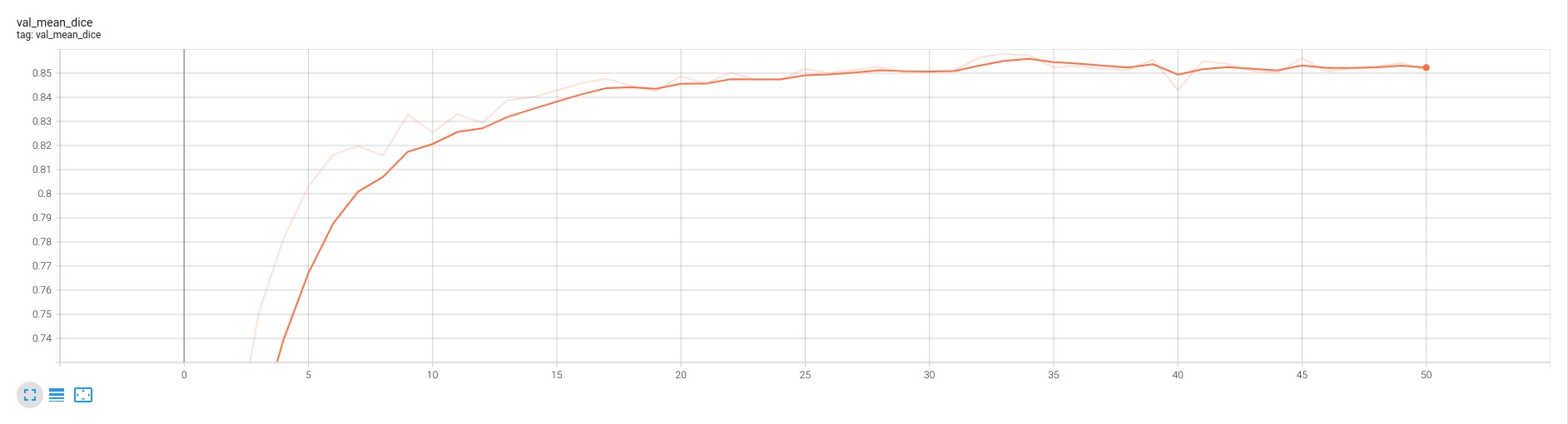 <br>
|
| 124 |
|
| 125 |
|
| 126 |
## commands example
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.0.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.0.6": "adapt to BundleWorkflow interface",
|
| 6 |
"0.0.5": "add name tag",
|
| 7 |
"0.0.4": "Fix evaluation",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.0.7",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.0.7": "Update with figure links",
|
| 6 |
"0.0.6": "adapt to BundleWorkflow interface",
|
| 7 |
"0.0.5": "add name tag",
|
| 8 |
"0.0.4": "Fix evaluation",
|
docs/README.md
CHANGED
|
@@ -14,7 +14,7 @@ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovern
|
|
| 14 |
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
|
| 15 |
unzip -q consep_dataset.zip
|
| 16 |
```
|
| 17 |
-
 based on the centroid given in the dataset.
|
| 53 |
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
|
| 54 |
-
- Update the label index for the target
|
| 55 |
-
- Other cells which are part of the patch are modified to have label
|
| 56 |
|
| 57 |
Example dataset.json
|
| 58 |
```json
|
|
@@ -95,7 +95,7 @@ Example dataset.json
|
|
| 95 |
- 0 = Background
|
| 96 |
- 1 = Nuclei
|
| 97 |
|
| 98 |
-
<br/>
|
| 18 |
|
| 19 |
## Training configuration
|
| 20 |
The training was performed with the following:
|
|
|
|
| 51 |
|
| 52 |
- Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset.
|
| 53 |
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
|
| 54 |
+
- Update the label index for the target nuclei based on the class value
|
| 55 |
+
- Other cells which are part of the patch are modified to have label idx = 255
|
| 56 |
|
| 57 |
Example dataset.json
|
| 58 |
```json
|
|
|
|
| 95 |
- 0 = Background
|
| 96 |
- 1 = Nuclei
|
| 97 |
|
| 98 |
+

|
| 99 |
|
| 100 |
## Scores
|
| 101 |
This model achieves the following Dice score on the validation data provided as part of the dataset:
|
|
|
|
| 107 |
## Training Performance
|
| 108 |
A graph showing the training Loss and Dice over 50 epochs.
|
| 109 |
|
| 110 |
+
 <br>
|
| 111 |
+
 <br>
|
| 112 |
|
| 113 |
## Validation Performance
|
| 114 |
A graph showing the validation mean Dice over 50 epochs.
|
| 115 |
|
| 116 |
+
 <br>
|
| 117 |
|
| 118 |
|
| 119 |
## commands example
|