update the model weights
Browse files- README.md +12 -6
- configs/evaluate.json +2 -4
- configs/inference.json +3 -2
- configs/metadata.json +3 -2
- configs/train.json +16 -5
- docs/README.md +12 -6
- models/model.pt +2 -2
- models/model.ts +2 -2
README.md
CHANGED
|
@@ -6,19 +6,25 @@ library_name: monai
|
|
| 6 |
license: apache-2.0
|
| 7 |
---
|
| 8 |
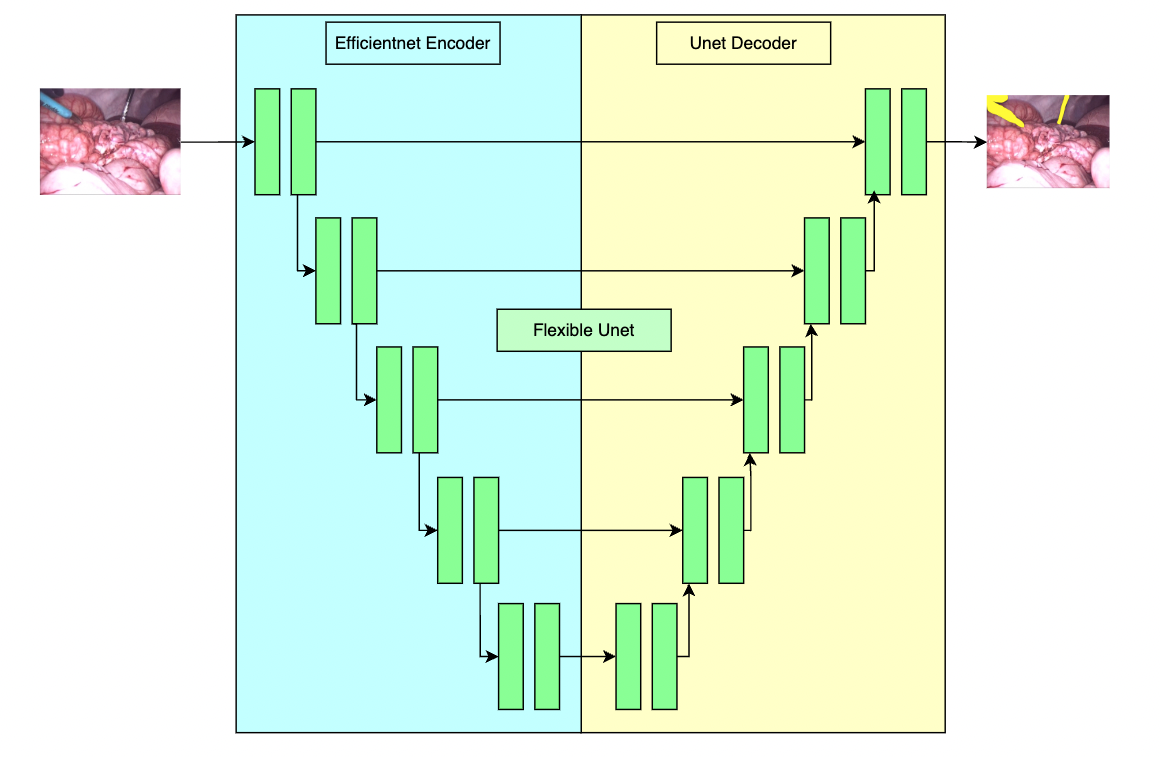
# Model Overview
|
| 9 |
-
A pre-trained model for the endoscopic tool segmentation task
|
| 10 |
|
| 11 |
-
The [PyTorch model](https://drive.google.com/file/d/
|
| 12 |
|
| 13 |
|
| 14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 15 |
## Data
|
| 16 |
Datasets used in this work were provided by [Activ Surgical](https://www.activsurgical.com/).
|
| 17 |
|
| 18 |
Since datasets are private, existing public datasets like [EndoVis 2017](https://endovissub2017-roboticinstrumentsegmentation.grand-challenge.org/Data/) can be used to train a similar model.
|
| 19 |
|
| 20 |
### Preprocessing
|
| 21 |
-
When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in
|
| 22 |
|
| 23 |
Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem.
|
| 24 |
|
|
@@ -38,13 +44,13 @@ Two channels:
|
|
| 38 |
- Label 0: everything else
|
| 39 |
|
| 40 |
## Performance
|
| 41 |
-
IoU was used for evaluating the performance of the model. This model achieves a mean IoU score of 0.
|
| 42 |
|
| 43 |
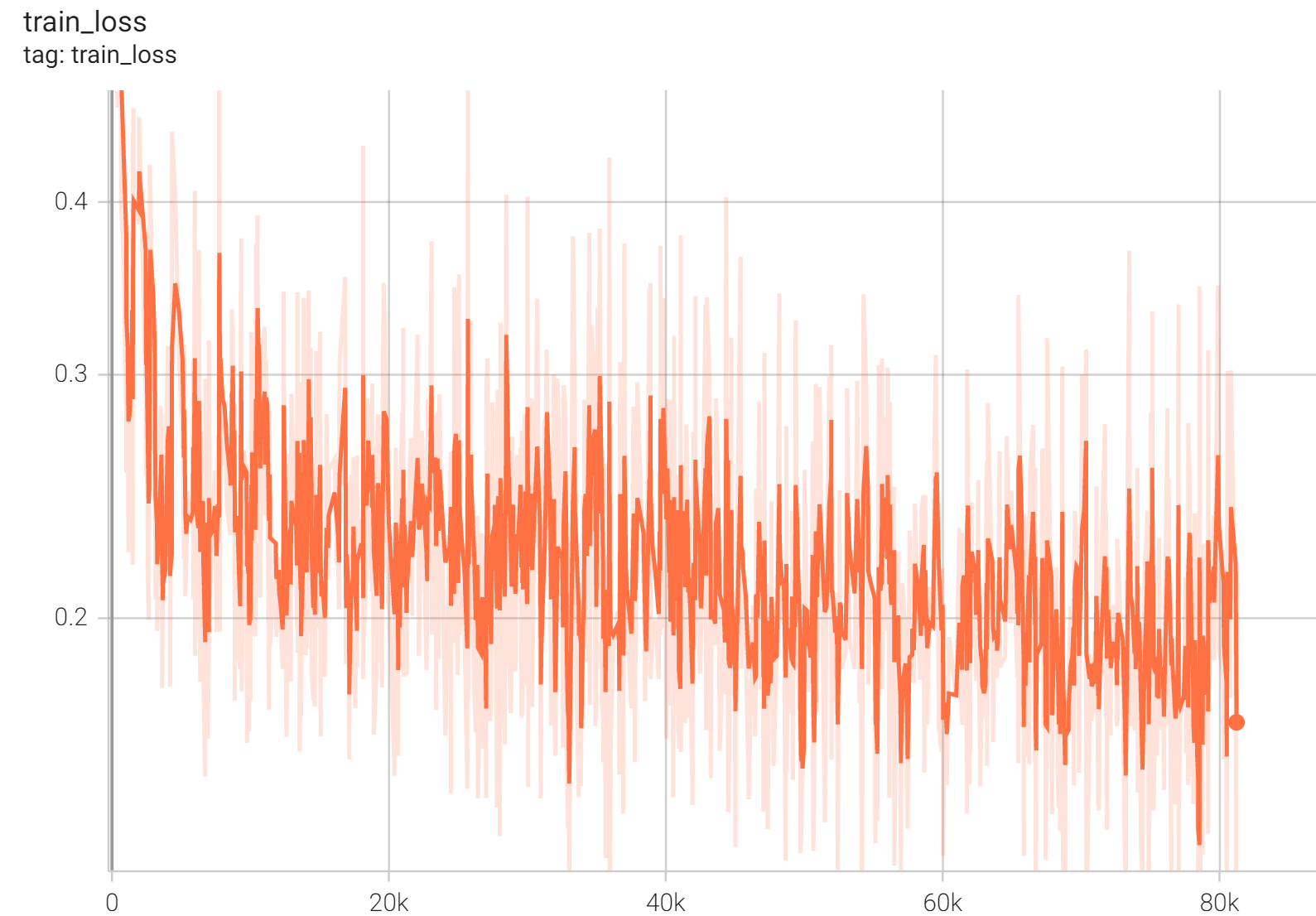
#### Training Loss
|
| 44 |
-
.
|
| 10 |
|
| 11 |
+
The [PyTorch model](https://drive.google.com/file/d/1I7UtWDKDEcezMqYiA-i_hsRTCrvWwJ61/view?usp=sharing) and [torchscript model](https://drive.google.com/file/d/1e_wYd1HjJQ0dz_HKdbthRcMOyUL02aLG/view?usp=sharing) are shared in google drive. Details can be found in `large_files.yml` file. Modify the "bundle_root" parameter specified in configs/train.json and configs/inference.json to reflect where models are downloaded. Expected directory path to place downloaded models is "models/" under "bundle_root".
|
| 12 |
|
| 13 |

|
| 14 |
|
| 15 |
+
## Pre-trained weights
|
| 16 |
+
A pre-trained encoder weights would benefit the model training. In this bundle, the encoder is trained with pre-trained weights from some internal data. For convenience, we provide two options in `configs/train.json` to enable users to load pre-trained weights:
|
| 17 |
+
|
| 18 |
+
1. Via setting the `use_imagenet_pretrain` parameter in the config file to `True`, [ImageNet](https://ieeexplore.ieee.org/document/5206848) pre-trained weights from the [EfficientNet-PyTorch repo](https://github.com/lukemelas/EfficientNet-PyTorch) can be loaded. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use.
|
| 19 |
+
2. Via updating the `load_path` parameter of the `CheckpointLoader` in the config file, weights from a local path can be loaded.
|
| 20 |
+
|
| 21 |
## Data
|
| 22 |
Datasets used in this work were provided by [Activ Surgical](https://www.activsurgical.com/).
|
| 23 |
|
| 24 |
Since datasets are private, existing public datasets like [EndoVis 2017](https://endovissub2017-roboticinstrumentsegmentation.grand-challenge.org/Data/) can be used to train a similar model.
|
| 25 |
|
| 26 |
### Preprocessing
|
| 27 |
+
When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in `configs/train.json` and "datalist" in `configs/inference.json` should be modified to fit given dataset. After that, "dataset_dir" parameter in `configs/train.json` and `configs/inference.json` should be changed to root folder which contains previous "train", "valid" and "test" folders.
|
| 28 |
|
| 29 |
Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem.
|
| 30 |
|
|
|
|
| 44 |
- Label 0: everything else
|
| 45 |
|
| 46 |
## Performance
|
| 47 |
+
IoU was used for evaluating the performance of the model. This model achieves a mean IoU score of 0.86.
|
| 48 |
|
| 49 |
#### Training Loss
|
| 50 |
+
|
| 51 |
|
| 52 |
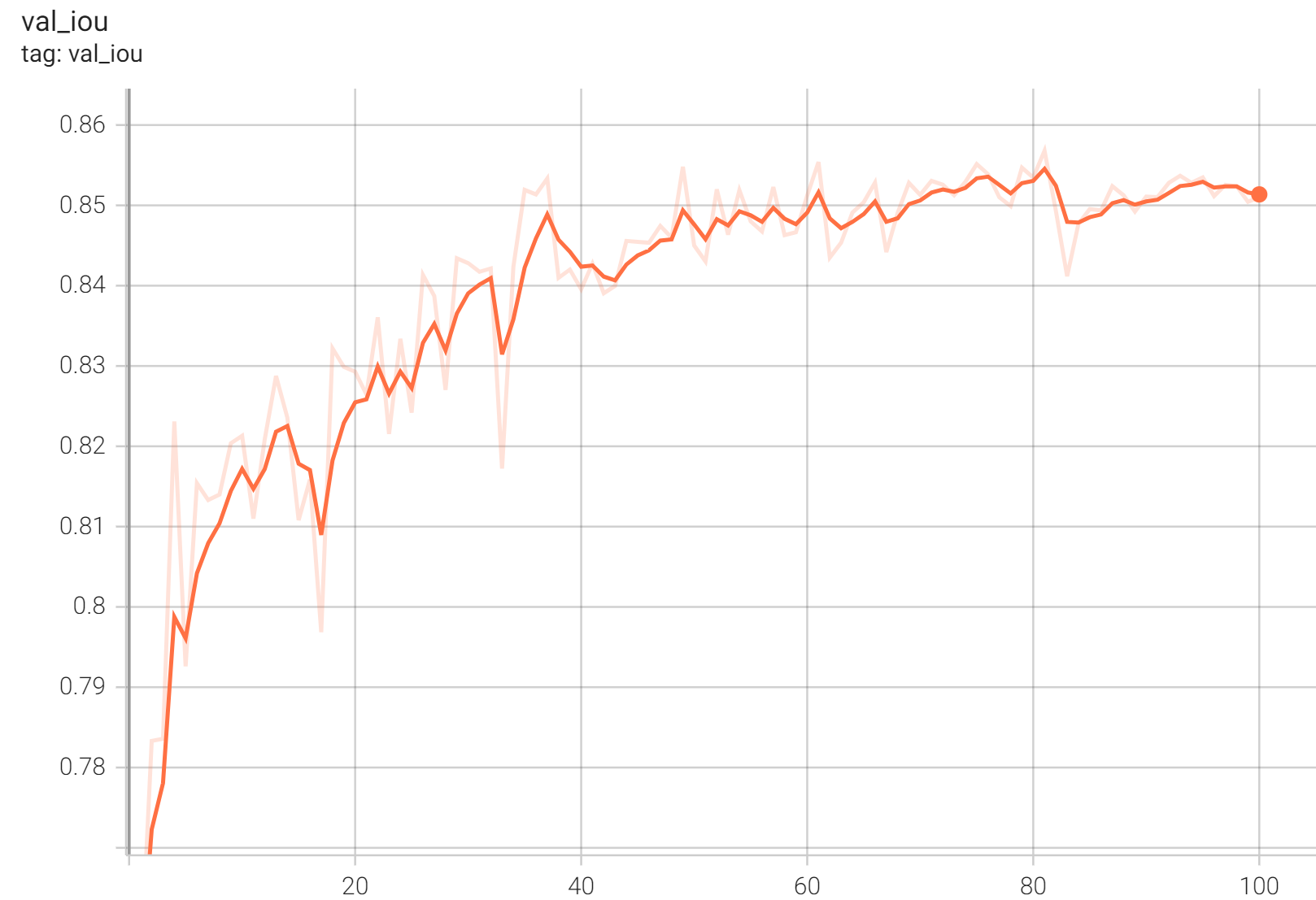
#### Validation IoU
|
| 53 |
+
|
| 54 |
|
| 55 |
#### TensorRT speedup
|
| 56 |
The `endoscopic_tool_segmentation` bundle supports the TensorRT acceleration. The table below shows the speedup ratios benchmarked on an A100 80G GPU.
|
configs/evaluate.json
CHANGED
|
@@ -47,7 +47,8 @@
|
|
| 47 |
"load_path": "$@ckpt_dir + '/model.pt'",
|
| 48 |
"load_dict": {
|
| 49 |
"model": "@network"
|
| 50 |
-
}
|
|
|
|
| 51 |
},
|
| 52 |
{
|
| 53 |
"_target_": "StatsHandler",
|
|
@@ -68,9 +69,6 @@
|
|
| 68 |
"summary_ops": "*"
|
| 69 |
}
|
| 70 |
],
|
| 71 |
-
"initialize": [
|
| 72 |
-
"$setattr(torch.backends.cudnn, 'benchmark', True)"
|
| 73 |
-
],
|
| 74 |
"run": [
|
| 75 |
"$@validate#evaluator.run()"
|
| 76 |
]
|
|
|
|
| 47 |
"load_path": "$@ckpt_dir + '/model.pt'",
|
| 48 |
"load_dict": {
|
| 49 |
"model": "@network"
|
| 50 |
+
},
|
| 51 |
+
"map_location": "@device"
|
| 52 |
},
|
| 53 |
{
|
| 54 |
"_target_": "StatsHandler",
|
|
|
|
| 69 |
"summary_ops": "*"
|
| 70 |
}
|
| 71 |
],
|
|
|
|
|
|
|
|
|
|
| 72 |
"run": [
|
| 73 |
"$@validate#evaluator.run()"
|
| 74 |
]
|
configs/inference.json
CHANGED
|
@@ -114,7 +114,8 @@
|
|
| 114 |
"load_path": "$@bundle_root + '/models/model.pt'",
|
| 115 |
"load_dict": {
|
| 116 |
"model": "@network"
|
| 117 |
-
}
|
|
|
|
| 118 |
},
|
| 119 |
{
|
| 120 |
"_target_": "StatsHandler",
|
|
@@ -131,7 +132,7 @@
|
|
| 131 |
"val_handlers": "@handlers"
|
| 132 |
},
|
| 133 |
"initialize": [
|
| 134 |
-
"$
|
| 135 |
],
|
| 136 |
"run": [
|
| 137 |
"$@evaluator.run()"
|
|
|
|
| 114 |
"load_path": "$@bundle_root + '/models/model.pt'",
|
| 115 |
"load_dict": {
|
| 116 |
"model": "@network"
|
| 117 |
+
},
|
| 118 |
+
"map_location": "@device"
|
| 119 |
},
|
| 120 |
{
|
| 121 |
"_target_": "StatsHandler",
|
|
|
|
| 132 |
"val_handlers": "@handlers"
|
| 133 |
},
|
| 134 |
"initialize": [
|
| 135 |
+
"$monai.utils.set_determinism(seed=123)"
|
| 136 |
],
|
| 137 |
"run": [
|
| 138 |
"$@evaluator.run()"
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.4.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.4.8": "update the TensorRT part in the README file",
|
| 6 |
"0.4.7": "fix mgpu finalize issue",
|
| 7 |
"0.4.6": "enable deterministic training",
|
|
@@ -37,7 +38,7 @@
|
|
| 37 |
"label_classes": "single channel data, 1/255 is tool, 0 is background",
|
| 38 |
"pred_classes": "2 channels OneHot data, channel 1 is tool, channel 0 is background",
|
| 39 |
"eval_metrics": {
|
| 40 |
-
"mean_iou": 0.
|
| 41 |
},
|
| 42 |
"references": [
|
| 43 |
"Tan, M. and Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. ICML, 2019a. https://arxiv.org/pdf/1905.11946.pdf",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.4.9",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.4.9": "update the model weights",
|
| 6 |
"0.4.8": "update the TensorRT part in the README file",
|
| 7 |
"0.4.7": "fix mgpu finalize issue",
|
| 8 |
"0.4.6": "enable deterministic training",
|
|
|
|
| 38 |
"label_classes": "single channel data, 1/255 is tool, 0 is background",
|
| 39 |
"pred_classes": "2 channels OneHot data, channel 1 is tool, channel 0 is background",
|
| 40 |
"eval_metrics": {
|
| 41 |
+
"mean_iou": 0.86
|
| 42 |
},
|
| 43 |
"references": [
|
| 44 |
"Tan, M. and Le, Q. V. Efficientnet: Rethinking model scaling for convolutional neural networks. ICML, 2019a. https://arxiv.org/pdf/1905.11946.pdf",
|
configs/train.json
CHANGED
|
@@ -6,6 +6,7 @@
|
|
| 6 |
"$import numpy as np"
|
| 7 |
],
|
| 8 |
"bundle_root": ".",
|
|
|
|
| 9 |
"ckpt_dir": "$@bundle_root + '/models'",
|
| 10 |
"output_dir": "$@bundle_root + '/eval'",
|
| 11 |
"dataset_dir": "/workspace/data/endoscopic_tool_dataset",
|
|
@@ -21,14 +22,14 @@
|
|
| 21 |
"out_channels": 2,
|
| 22 |
"backbone": "efficientnet-b2",
|
| 23 |
"spatial_dims": 2,
|
| 24 |
-
"dropout": 0.
|
| 25 |
-
"pretrained":
|
| 26 |
"is_pad": false,
|
| 27 |
"pre_conv": null
|
| 28 |
},
|
| 29 |
"network": "$@network_def.to(@device)",
|
| 30 |
"loss": {
|
| 31 |
-
"_target_": "
|
| 32 |
"include_background": false,
|
| 33 |
"to_onehot_y": true,
|
| 34 |
"softmax": true,
|
|
@@ -122,7 +123,7 @@
|
|
| 122 |
"_target_": "CacheDataset",
|
| 123 |
"data": "$[{'image': i, 'label': l} for i, l in zip(@images, @labels)]",
|
| 124 |
"transform": "@train#preprocessing",
|
| 125 |
-
"cache_rate": 0.
|
| 126 |
"num_workers": 4
|
| 127 |
},
|
| 128 |
"dataloader": {
|
|
@@ -136,6 +137,16 @@
|
|
| 136 |
"_target_": "SimpleInferer"
|
| 137 |
},
|
| 138 |
"handlers": [
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 139 |
{
|
| 140 |
"_target_": "ValidationHandler",
|
| 141 |
"validator": "@validate#evaluator",
|
|
@@ -210,7 +221,7 @@
|
|
| 210 |
"_target_": "CacheDataset",
|
| 211 |
"data": "$[{'image': i, 'label': l} for i, l in zip(@val_images, @val_labels)]",
|
| 212 |
"transform": "@validate#preprocessing",
|
| 213 |
-
"cache_rate": 0
|
| 214 |
},
|
| 215 |
"dataloader": {
|
| 216 |
"_target_": "DataLoader",
|
|
|
|
| 6 |
"$import numpy as np"
|
| 7 |
],
|
| 8 |
"bundle_root": ".",
|
| 9 |
+
"use_imagenet_pretrain": false,
|
| 10 |
"ckpt_dir": "$@bundle_root + '/models'",
|
| 11 |
"output_dir": "$@bundle_root + '/eval'",
|
| 12 |
"dataset_dir": "/workspace/data/endoscopic_tool_dataset",
|
|
|
|
| 22 |
"out_channels": 2,
|
| 23 |
"backbone": "efficientnet-b2",
|
| 24 |
"spatial_dims": 2,
|
| 25 |
+
"dropout": 0.6,
|
| 26 |
+
"pretrained": "@use_imagenet_pretrain",
|
| 27 |
"is_pad": false,
|
| 28 |
"pre_conv": null
|
| 29 |
},
|
| 30 |
"network": "$@network_def.to(@device)",
|
| 31 |
"loss": {
|
| 32 |
+
"_target_": "DiceFocalLoss",
|
| 33 |
"include_background": false,
|
| 34 |
"to_onehot_y": true,
|
| 35 |
"softmax": true,
|
|
|
|
| 123 |
"_target_": "CacheDataset",
|
| 124 |
"data": "$[{'image': i, 'label': l} for i, l in zip(@images, @labels)]",
|
| 125 |
"transform": "@train#preprocessing",
|
| 126 |
+
"cache_rate": 0.5,
|
| 127 |
"num_workers": 4
|
| 128 |
},
|
| 129 |
"dataloader": {
|
|
|
|
| 137 |
"_target_": "SimpleInferer"
|
| 138 |
},
|
| 139 |
"handlers": [
|
| 140 |
+
{
|
| 141 |
+
"_target_": "CheckpointLoader",
|
| 142 |
+
"_disabled_": "@use_imagenet_pretrain",
|
| 143 |
+
"load_path": "/path/to/local/weight/model.pt",
|
| 144 |
+
"load_dict": {
|
| 145 |
+
"model": "@network"
|
| 146 |
+
},
|
| 147 |
+
"strict": false,
|
| 148 |
+
"map_location": "@device"
|
| 149 |
+
},
|
| 150 |
{
|
| 151 |
"_target_": "ValidationHandler",
|
| 152 |
"validator": "@validate#evaluator",
|
|
|
|
| 221 |
"_target_": "CacheDataset",
|
| 222 |
"data": "$[{'image': i, 'label': l} for i, l in zip(@val_images, @val_labels)]",
|
| 223 |
"transform": "@validate#preprocessing",
|
| 224 |
+
"cache_rate": 1.0
|
| 225 |
},
|
| 226 |
"dataloader": {
|
| 227 |
"_target_": "DataLoader",
|
docs/README.md
CHANGED
|
@@ -1,17 +1,23 @@
|
|
| 1 |
# Model Overview
|
| 2 |
-
A pre-trained model for the endoscopic tool segmentation task
|
| 3 |
|
| 4 |
-
The [PyTorch model](https://drive.google.com/file/d/
|
| 5 |
|
| 6 |

|
| 7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 8 |
## Data
|
| 9 |
Datasets used in this work were provided by [Activ Surgical](https://www.activsurgical.com/).
|
| 10 |
|
| 11 |
Since datasets are private, existing public datasets like [EndoVis 2017](https://endovissub2017-roboticinstrumentsegmentation.grand-challenge.org/Data/) can be used to train a similar model.
|
| 12 |
|
| 13 |
### Preprocessing
|
| 14 |
-
When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in
|
| 15 |
|
| 16 |
Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem.
|
| 17 |
|
|
@@ -31,13 +37,13 @@ Two channels:
|
|
| 31 |
- Label 0: everything else
|
| 32 |
|
| 33 |
## Performance
|
| 34 |
-
IoU was used for evaluating the performance of the model. This model achieves a mean IoU score of 0.
|
| 35 |
|
| 36 |
#### Training Loss
|
| 37 |
-
.
|
| 3 |
|
| 4 |
+
The [PyTorch model](https://drive.google.com/file/d/1I7UtWDKDEcezMqYiA-i_hsRTCrvWwJ61/view?usp=sharing) and [torchscript model](https://drive.google.com/file/d/1e_wYd1HjJQ0dz_HKdbthRcMOyUL02aLG/view?usp=sharing) are shared in google drive. Details can be found in `large_files.yml` file. Modify the "bundle_root" parameter specified in configs/train.json and configs/inference.json to reflect where models are downloaded. Expected directory path to place downloaded models is "models/" under "bundle_root".
|
| 5 |
|
| 6 |

|
| 7 |
|
| 8 |
+
## Pre-trained weights
|
| 9 |
+
A pre-trained encoder weights would benefit the model training. In this bundle, the encoder is trained with pre-trained weights from some internal data. For convenience, we provide two options in `configs/train.json` to enable users to load pre-trained weights:
|
| 10 |
+
|
| 11 |
+
1. Via setting the `use_imagenet_pretrain` parameter in the config file to `True`, [ImageNet](https://ieeexplore.ieee.org/document/5206848) pre-trained weights from the [EfficientNet-PyTorch repo](https://github.com/lukemelas/EfficientNet-PyTorch) can be loaded. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use.
|
| 12 |
+
2. Via updating the `load_path` parameter of the `CheckpointLoader` in the config file, weights from a local path can be loaded.
|
| 13 |
+
|
| 14 |
## Data
|
| 15 |
Datasets used in this work were provided by [Activ Surgical](https://www.activsurgical.com/).
|
| 16 |
|
| 17 |
Since datasets are private, existing public datasets like [EndoVis 2017](https://endovissub2017-roboticinstrumentsegmentation.grand-challenge.org/Data/) can be used to train a similar model.
|
| 18 |
|
| 19 |
### Preprocessing
|
| 20 |
+
When using EndoVis or any other dataset, it should be divided into "train", "valid" and "test" folders. Samples in each folder would better be images and converted to jpg format. Otherwise, "images", "labels", "val_images" and "val_labels" parameters in `configs/train.json` and "datalist" in `configs/inference.json` should be modified to fit given dataset. After that, "dataset_dir" parameter in `configs/train.json` and `configs/inference.json` should be changed to root folder which contains previous "train", "valid" and "test" folders.
|
| 21 |
|
| 22 |
Please notice that loading data operation in this bundle is adaptive. If images and labels are not in the same format, it may lead to a mismatching problem. For example, if images are in jpg format and labels are in npy format, PIL and Numpy readers will be used separately to load images and labels. Since these two readers have their own way to parse file's shape, loaded labels will be transpose of the correct ones and incur a missmatching problem.
|
| 23 |
|
|
|
|
| 37 |
- Label 0: everything else
|
| 38 |
|
| 39 |
## Performance
|
| 40 |
+
IoU was used for evaluating the performance of the model. This model achieves a mean IoU score of 0.86.
|
| 41 |
|
| 42 |
#### Training Loss
|
| 43 |
+

|
| 44 |
|
| 45 |
#### Validation IoU
|
| 46 |
+

|
| 47 |
|
| 48 |
#### TensorRT speedup
|
| 49 |
The `endoscopic_tool_segmentation` bundle supports the TensorRT acceleration. The table below shows the speedup ratios benchmarked on an A100 80G GPU.
|
models/model.pt
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:1a24d5f1d30a44474e8bb87abb62f289add2c40a291eac67138242679e928729
|
| 3 |
+
size 46290573
|
models/model.ts
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:149ac9473dee60737957505a59a24c6d816092c6aa2ff89e13c8bfc8e9831059
|
| 3 |
+
size 46500237
|