New model version initial commit
Browse files- README.md +227 -0
- catboost_without_hospital_number_01.pkl +3 -0
- config.json +174 -0
- confusion_matrix.png +0 -0
README.md
ADDED
|
@@ -0,0 +1,227 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
license: mit
|
| 3 |
+
library_name: sklearn
|
| 4 |
+
tags:
|
| 5 |
+
- sklearn
|
| 6 |
+
- skops
|
| 7 |
+
- tabular-classification
|
| 8 |
+
model_format: pickle
|
| 9 |
+
model_file: catboost_without_hospital_number_01.pkl
|
| 10 |
+
widget:
|
| 11 |
+
structuredData:
|
| 12 |
+
abdomen:
|
| 13 |
+
- distend_small
|
| 14 |
+
- distend_small
|
| 15 |
+
- distend_large
|
| 16 |
+
abdominal_distention:
|
| 17 |
+
- none
|
| 18 |
+
- none
|
| 19 |
+
- moderate
|
| 20 |
+
abdomo_appearance:
|
| 21 |
+
- serosanguious
|
| 22 |
+
- cloudy
|
| 23 |
+
- serosanguious
|
| 24 |
+
abdomo_protein:
|
| 25 |
+
- 4.1
|
| 26 |
+
- 4.3
|
| 27 |
+
- 2.0
|
| 28 |
+
age:
|
| 29 |
+
- adult
|
| 30 |
+
- adult
|
| 31 |
+
- adult
|
| 32 |
+
capillary_refill_time:
|
| 33 |
+
- less_3_sec
|
| 34 |
+
- less_3_sec
|
| 35 |
+
- more_3_sec
|
| 36 |
+
cp_data:
|
| 37 |
+
- 'yes'
|
| 38 |
+
- 'yes'
|
| 39 |
+
- 'no'
|
| 40 |
+
lesion_1:
|
| 41 |
+
- 7209
|
| 42 |
+
- 2112
|
| 43 |
+
- 5400
|
| 44 |
+
lesion_2:
|
| 45 |
+
- 0
|
| 46 |
+
- 0
|
| 47 |
+
- 0
|
| 48 |
+
lesion_3:
|
| 49 |
+
- 0
|
| 50 |
+
- 0
|
| 51 |
+
- 0
|
| 52 |
+
mucous_membrane:
|
| 53 |
+
- bright_pink
|
| 54 |
+
- bright_pink
|
| 55 |
+
- dark_cyanotic
|
| 56 |
+
nasogastric_reflux:
|
| 57 |
+
- none
|
| 58 |
+
- none
|
| 59 |
+
- more_1_liter
|
| 60 |
+
nasogastric_reflux_ph:
|
| 61 |
+
- 7.0
|
| 62 |
+
- 3.5
|
| 63 |
+
- 2.0
|
| 64 |
+
nasogastric_tube:
|
| 65 |
+
- slight
|
| 66 |
+
- none
|
| 67 |
+
- significant
|
| 68 |
+
packed_cell_volume:
|
| 69 |
+
- 37.0
|
| 70 |
+
- 44.0
|
| 71 |
+
- 65.0
|
| 72 |
+
pain:
|
| 73 |
+
- depressed
|
| 74 |
+
- mild_pain
|
| 75 |
+
- extreme_pain
|
| 76 |
+
peripheral_pulse:
|
| 77 |
+
- normal
|
| 78 |
+
- normal
|
| 79 |
+
- reduced
|
| 80 |
+
peristalsis:
|
| 81 |
+
- hypermotile
|
| 82 |
+
- hypomotile
|
| 83 |
+
- absent
|
| 84 |
+
pulse:
|
| 85 |
+
- 84.0
|
| 86 |
+
- 66.0
|
| 87 |
+
- 72.0
|
| 88 |
+
rectal_exam_feces:
|
| 89 |
+
- absent
|
| 90 |
+
- decreased
|
| 91 |
+
- absent
|
| 92 |
+
rectal_temp:
|
| 93 |
+
- 39.0
|
| 94 |
+
- 38.5
|
| 95 |
+
- 37.3
|
| 96 |
+
respiratory_rate:
|
| 97 |
+
- 24.0
|
| 98 |
+
- 21.0
|
| 99 |
+
- 30.0
|
| 100 |
+
surgery:
|
| 101 |
+
- 'yes'
|
| 102 |
+
- 'yes'
|
| 103 |
+
- 'yes'
|
| 104 |
+
surgical_lesion:
|
| 105 |
+
- 'yes'
|
| 106 |
+
- 'yes'
|
| 107 |
+
- 'yes'
|
| 108 |
+
temp_of_extremities:
|
| 109 |
+
- cool
|
| 110 |
+
- normal
|
| 111 |
+
- cool
|
| 112 |
+
total_protein:
|
| 113 |
+
- 6.5
|
| 114 |
+
- 7.6
|
| 115 |
+
- 13.0
|
| 116 |
+
---
|
| 117 |
+
|
| 118 |
+
# Model description
|
| 119 |
+
|
| 120 |
+
This is a `Catboost` model trained on horse health outcome data from Kaggle.
|
| 121 |
+
|
| 122 |
+
## Intended uses & limitations
|
| 123 |
+
|
| 124 |
+
This model is not ready to be used in production.
|
| 125 |
+
|
| 126 |
+
## Training Procedure
|
| 127 |
+
|
| 128 |
+
[More Information Needed]
|
| 129 |
+
|
| 130 |
+
### Hyperparameters
|
| 131 |
+
|
| 132 |
+
<details>
|
| 133 |
+
<summary> Click to expand </summary>
|
| 134 |
+
|
| 135 |
+
| Hyperparameter | Value |
|
| 136 |
+
|--------------------------------------------------|-----------------------------------------------------------------------------------------------------------------------------|
|
| 137 |
+
| memory | |
|
| 138 |
+
| steps | [('preprocessor', ColumnTransformer(remainder='passthrough',<br /> transformers=[('num',<br /> Pipeline(steps=[('imputer',<br /> SimpleImputer(strategy='median')),<br /> ('scaler', StandardScaler())]),<br /> ['rectal_temp', 'pulse', 'respiratory_rate',<br /> 'nasogastric_reflux_ph', 'packed_cell_volume',<br /> 'total_protein', 'abdomo_protein', 'lesion_1',<br /> 'lesion_2', 'lesion_3']),<br /> ('cat',<br /> Pipeline(steps=[('imputer',<br /> SimpleI...='missing',<br /> strategy='constant')),<br /> ('onehot',<br /> OneHotEncoder(handle_unknown='ignore'))]),<br /> ['surgery', 'age', 'temp_of_extremities',<br /> 'peripheral_pulse', 'mucous_membrane',<br /> 'capillary_refill_time', 'pain',<br /> 'peristalsis', 'abdominal_distention',<br /> 'nasogastric_tube', 'nasogastric_reflux',<br /> 'rectal_exam_feces', 'abdomen',<br /> 'abdomo_appearance', 'surgical_lesion',<br /> 'cp_data'])])), ('classifier', <catboost.core.CatBoostClassifier object at 0x000001C4D1219250>)] |
|
| 139 |
+
| verbose | False |
|
| 140 |
+
| preprocessor | ColumnTransformer(remainder='passthrough',<br /> transformers=[('num',<br /> Pipeline(steps=[('imputer',<br /> SimpleImputer(strategy='median')),<br /> ('scaler', StandardScaler())]),<br /> ['rectal_temp', 'pulse', 'respiratory_rate',<br /> 'nasogastric_reflux_ph', 'packed_cell_volume',<br /> 'total_protein', 'abdomo_protein', 'lesion_1',<br /> 'lesion_2', 'lesion_3']),<br /> ('cat',<br /> Pipeline(steps=[('imputer',<br /> SimpleI...='missing',<br /> strategy='constant')),<br /> ('onehot',<br /> OneHotEncoder(handle_unknown='ignore'))]),<br /> ['surgery', 'age', 'temp_of_extremities',<br /> 'peripheral_pulse', 'mucous_membrane',<br /> 'capillary_refill_time', 'pain',<br /> 'peristalsis', 'abdominal_distention',<br /> 'nasogastric_tube', 'nasogastric_reflux',<br /> 'rectal_exam_feces', 'abdomen',<br /> 'abdomo_appearance', 'surgical_lesion',<br /> 'cp_data'])]) |
|
| 141 |
+
| classifier | <catboost.core.CatBoostClassifier object at 0x000001C4D1219250> |
|
| 142 |
+
| preprocessor__n_jobs | |
|
| 143 |
+
| preprocessor__remainder | passthrough |
|
| 144 |
+
| preprocessor__sparse_threshold | 0.3 |
|
| 145 |
+
| preprocessor__transformer_weights | |
|
| 146 |
+
| preprocessor__transformers | [('num', Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),<br /> ('scaler', StandardScaler())]), ['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']), ('cat', Pipeline(steps=[('imputer',<br /> SimpleImputer(fill_value='missing', strategy='constant')),<br /> ('onehot', OneHotEncoder(handle_unknown='ignore'))]), ['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data'])] |
|
| 147 |
+
| preprocessor__verbose | False |
|
| 148 |
+
| preprocessor__verbose_feature_names_out | True |
|
| 149 |
+
| preprocessor__num | Pipeline(steps=[('imputer', SimpleImputer(strategy='median')),<br /> ('scaler', StandardScaler())]) |
|
| 150 |
+
| preprocessor__cat | Pipeline(steps=[('imputer',<br /> SimpleImputer(fill_value='missing', strategy='constant')),<br /> ('onehot', OneHotEncoder(handle_unknown='ignore'))]) |
|
| 151 |
+
| preprocessor__num__memory | |
|
| 152 |
+
| preprocessor__num__steps | [('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())] |
|
| 153 |
+
| preprocessor__num__verbose | False |
|
| 154 |
+
| preprocessor__num__imputer | SimpleImputer(strategy='median') |
|
| 155 |
+
| preprocessor__num__scaler | StandardScaler() |
|
| 156 |
+
| preprocessor__num__imputer__add_indicator | False |
|
| 157 |
+
| preprocessor__num__imputer__copy | True |
|
| 158 |
+
| preprocessor__num__imputer__fill_value | |
|
| 159 |
+
| preprocessor__num__imputer__keep_empty_features | False |
|
| 160 |
+
| preprocessor__num__imputer__missing_values | nan |
|
| 161 |
+
| preprocessor__num__imputer__strategy | median |
|
| 162 |
+
| preprocessor__num__scaler__copy | True |
|
| 163 |
+
| preprocessor__num__scaler__with_mean | True |
|
| 164 |
+
| preprocessor__num__scaler__with_std | True |
|
| 165 |
+
| preprocessor__cat__memory | |
|
| 166 |
+
| preprocessor__cat__steps | [('imputer', SimpleImputer(fill_value='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))] |
|
| 167 |
+
| preprocessor__cat__verbose | False |
|
| 168 |
+
| preprocessor__cat__imputer | SimpleImputer(fill_value='missing', strategy='constant') |
|
| 169 |
+
| preprocessor__cat__onehot | OneHotEncoder(handle_unknown='ignore') |
|
| 170 |
+
| preprocessor__cat__imputer__add_indicator | False |
|
| 171 |
+
| preprocessor__cat__imputer__copy | True |
|
| 172 |
+
| preprocessor__cat__imputer__fill_value | missing |
|
| 173 |
+
| preprocessor__cat__imputer__keep_empty_features | False |
|
| 174 |
+
| preprocessor__cat__imputer__missing_values | nan |
|
| 175 |
+
| preprocessor__cat__imputer__strategy | constant |
|
| 176 |
+
| preprocessor__cat__onehot__categories | auto |
|
| 177 |
+
| preprocessor__cat__onehot__drop | |
|
| 178 |
+
| preprocessor__cat__onehot__dtype | <class 'numpy.float64'> |
|
| 179 |
+
| preprocessor__cat__onehot__feature_name_combiner | concat |
|
| 180 |
+
| preprocessor__cat__onehot__handle_unknown | ignore |
|
| 181 |
+
| preprocessor__cat__onehot__max_categories | |
|
| 182 |
+
| preprocessor__cat__onehot__min_frequency | |
|
| 183 |
+
| preprocessor__cat__onehot__sparse | deprecated |
|
| 184 |
+
| preprocessor__cat__onehot__sparse_output | True |
|
| 185 |
+
| classifier__learning_rate | 0.2 |
|
| 186 |
+
| classifier__silent | True |
|
| 187 |
+
| classifier__max_depth | 4 |
|
| 188 |
+
| classifier__n_estimators | 300 |
|
| 189 |
+
|
| 190 |
+
</details>
|
| 191 |
+
|
| 192 |
+
### Model Plot
|
| 193 |
+
|
| 194 |
+
<style>#sk-container-id-2 {color: black;}#sk-container-id-2 pre{padding: 0;}#sk-container-id-2 div.sk-toggleable {background-color: white;}#sk-container-id-2 label.sk-toggleable__label {cursor: pointer;display: block;width: 100%;margin-bottom: 0;padding: 0.3em;box-sizing: border-box;text-align: center;}#sk-container-id-2 label.sk-toggleable__label-arrow:before {content: "▸";float: left;margin-right: 0.25em;color: #696969;}#sk-container-id-2 label.sk-toggleable__label-arrow:hover:before {color: black;}#sk-container-id-2 div.sk-estimator:hover label.sk-toggleable__label-arrow:before {color: black;}#sk-container-id-2 div.sk-toggleable__content {max-height: 0;max-width: 0;overflow: hidden;text-align: left;background-color: #f0f8ff;}#sk-container-id-2 div.sk-toggleable__content pre {margin: 0.2em;color: black;border-radius: 0.25em;background-color: #f0f8ff;}#sk-container-id-2 input.sk-toggleable__control:checked~div.sk-toggleable__content {max-height: 200px;max-width: 100%;overflow: auto;}#sk-container-id-2 input.sk-toggleable__control:checked~label.sk-toggleable__label-arrow:before {content: "▾";}#sk-container-id-2 div.sk-estimator input.sk-toggleable__control:checked~label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-2 div.sk-label input.sk-toggleable__control:checked~label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-2 input.sk-hidden--visually {border: 0;clip: rect(1px 1px 1px 1px);clip: rect(1px, 1px, 1px, 1px);height: 1px;margin: -1px;overflow: hidden;padding: 0;position: absolute;width: 1px;}#sk-container-id-2 div.sk-estimator {font-family: monospace;background-color: #f0f8ff;border: 1px dotted black;border-radius: 0.25em;box-sizing: border-box;margin-bottom: 0.5em;}#sk-container-id-2 div.sk-estimator:hover {background-color: #d4ebff;}#sk-container-id-2 div.sk-parallel-item::after {content: "";width: 100%;border-bottom: 1px solid gray;flex-grow: 1;}#sk-container-id-2 div.sk-label:hover label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-2 div.sk-serial::before {content: "";position: absolute;border-left: 1px solid gray;box-sizing: border-box;top: 0;bottom: 0;left: 50%;z-index: 0;}#sk-container-id-2 div.sk-serial {display: flex;flex-direction: column;align-items: center;background-color: white;padding-right: 0.2em;padding-left: 0.2em;position: relative;}#sk-container-id-2 div.sk-item {position: relative;z-index: 1;}#sk-container-id-2 div.sk-parallel {display: flex;align-items: stretch;justify-content: center;background-color: white;position: relative;}#sk-container-id-2 div.sk-item::before, #sk-container-id-2 div.sk-parallel-item::before {content: "";position: absolute;border-left: 1px solid gray;box-sizing: border-box;top: 0;bottom: 0;left: 50%;z-index: -1;}#sk-container-id-2 div.sk-parallel-item {display: flex;flex-direction: column;z-index: 1;position: relative;background-color: white;}#sk-container-id-2 div.sk-parallel-item:first-child::after {align-self: flex-end;width: 50%;}#sk-container-id-2 div.sk-parallel-item:last-child::after {align-self: flex-start;width: 50%;}#sk-container-id-2 div.sk-parallel-item:only-child::after {width: 0;}#sk-container-id-2 div.sk-dashed-wrapped {border: 1px dashed gray;margin: 0 0.4em 0.5em 0.4em;box-sizing: border-box;padding-bottom: 0.4em;background-color: white;}#sk-container-id-2 div.sk-label label {font-family: monospace;font-weight: bold;display: inline-block;line-height: 1.2em;}#sk-container-id-2 div.sk-label-container {text-align: center;}#sk-container-id-2 div.sk-container {/* jupyter's `normalize.less` sets `[hidden] { display: none; }` but bootstrap.min.css set `[hidden] { display: none !important; }` so we also need the `!important` here to be able to override the default hidden behavior on the sphinx rendered scikit-learn.org. See: https://github.com/scikit-learn/scikit-learn/issues/21755 */display: inline-block !important;position: relative;}#sk-container-id-2 div.sk-text-repr-fallback {display: none;}</style><div id="sk-container-id-2" class="sk-top-container" style="overflow: auto;"><div class="sk-text-repr-fallback"><pre>Pipeline(steps=[('preprocessor',ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler',StandardScaler())]),['rectal_temp', 'pulse','respiratory_rate','nasogastric_reflux_ph','packed_cell_volume','total_protein','abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pi...OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age','temp_of_extremities','peripheral_pulse','mucous_membrane','capillary_refill_time','pain', 'peristalsis','abdominal_distention','nasogastric_tube','nasogastric_reflux','rectal_exam_feces','abdomen','abdomo_appearance','surgical_lesion','cp_data'])])),('classifier',<catboost.core.CatBoostClassifier object at 0x000001C4D1219250>)])</pre><b>In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. <br />On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.</b></div><div class="sk-container" hidden><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-12" type="checkbox" ><label for="sk-estimator-id-12" class="sk-toggleable__label sk-toggleable__label-arrow">Pipeline</label><div class="sk-toggleable__content"><pre>Pipeline(steps=[('preprocessor',ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler',StandardScaler())]),['rectal_temp', 'pulse','respiratory_rate','nasogastric_reflux_ph','packed_cell_volume','total_protein','abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pi...OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age','temp_of_extremities','peripheral_pulse','mucous_membrane','capillary_refill_time','pain', 'peristalsis','abdominal_distention','nasogastric_tube','nasogastric_reflux','rectal_exam_feces','abdomen','abdomo_appearance','surgical_lesion','cp_data'])])),('classifier',<catboost.core.CatBoostClassifier object at 0x000001C4D1219250>)])</pre></div></div></div><div class="sk-serial"><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-13" type="checkbox" ><label for="sk-estimator-id-13" class="sk-toggleable__label sk-toggleable__label-arrow">preprocessor: ColumnTransformer</label><div class="sk-toggleable__content"><pre>ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler', StandardScaler())]),['rectal_temp', 'pulse', 'respiratory_rate','nasogastric_reflux_ph', 'packed_cell_volume','total_protein', 'abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pipeline(steps=[('imputer',SimpleI...='missing',strategy='constant')),('onehot',OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age', 'temp_of_extremities','peripheral_pulse', 'mucous_membrane','capillary_refill_time', 'pain','peristalsis', 'abdominal_distention','nasogastric_tube', 'nasogastric_reflux','rectal_exam_feces', 'abdomen','abdomo_appearance', 'surgical_lesion','cp_data'])])</pre></div></div></div><div class="sk-parallel"><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-14" type="checkbox" ><label for="sk-estimator-id-14" class="sk-toggleable__label sk-toggleable__label-arrow">num</label><div class="sk-toggleable__content"><pre>['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-15" type="checkbox" ><label for="sk-estimator-id-15" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer(strategy='median')</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-16" type="checkbox" ><label for="sk-estimator-id-16" class="sk-toggleable__label sk-toggleable__label-arrow">StandardScaler</label><div class="sk-toggleable__content"><pre>StandardScaler()</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-17" type="checkbox" ><label for="sk-estimator-id-17" class="sk-toggleable__label sk-toggleable__label-arrow">cat</label><div class="sk-toggleable__content"><pre>['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-18" type="checkbox" ><label for="sk-estimator-id-18" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer(fill_value='missing', strategy='constant')</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-19" type="checkbox" ><label for="sk-estimator-id-19" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(handle_unknown='ignore')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-20" type="checkbox" ><label for="sk-estimator-id-20" class="sk-toggleable__label sk-toggleable__label-arrow">remainder</label><div class="sk-toggleable__content"><pre>[]</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-21" type="checkbox" ><label for="sk-estimator-id-21" class="sk-toggleable__label sk-toggleable__label-arrow">passthrough</label><div class="sk-toggleable__content"><pre>passthrough</pre></div></div></div></div></div></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-22" type="checkbox" ><label for="sk-estimator-id-22" class="sk-toggleable__label sk-toggleable__label-arrow">CatBoostClassifier</label><div class="sk-toggleable__content"><pre><catboost.core.CatBoostClassifier object at 0x000001C4D1219250></pre></div></div></div></div></div></div></div>
|
| 195 |
+
|
| 196 |
+
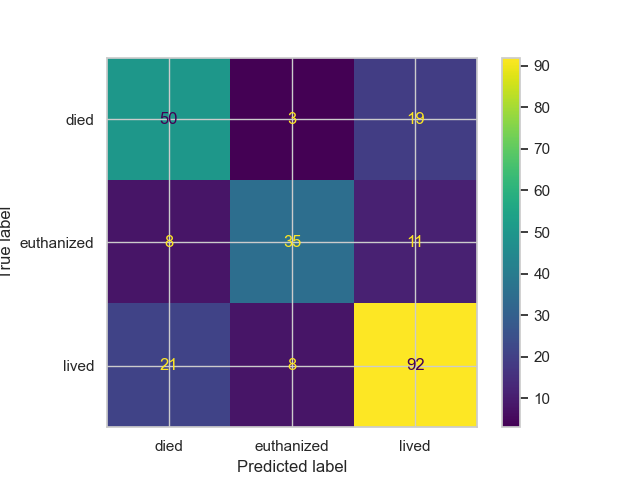
## Evaluation Results
|
| 197 |
+
|
| 198 |
+
| Metric | Value |
|
| 199 |
+
|----------|----------|
|
| 200 |
+
| accuracy | 0.716599 |
|
| 201 |
+
| f1 score | 0.716599 |
|
| 202 |
+
|
| 203 |
+
### Confusion Matrix
|
| 204 |
+
|
| 205 |
+

|
| 206 |
+
|
| 207 |
+
# How to Get Started with the Model
|
| 208 |
+
|
| 209 |
+
[More Information Needed]
|
| 210 |
+
|
| 211 |
+
# Model Card Authors
|
| 212 |
+
|
| 213 |
+
kmposkid
|
| 214 |
+
|
| 215 |
+
# Model Card Contact
|
| 216 |
+
|
| 217 |
+
You can contact the model card authors through following channels:
|
| 218 |
+
[More Information Needed]
|
| 219 |
+
|
| 220 |
+
# Citation
|
| 221 |
+
|
| 222 |
+
Below you can find information related to citation.
|
| 223 |
+
|
| 224 |
+
**BibTeX:**
|
| 225 |
+
```
|
| 226 |
+
[More Information Needed]
|
| 227 |
+
```
|
catboost_without_hospital_number_01.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d43eab8b706b2e4f7cca9f853b30b5f577e413f9d7df14c5c6d0644d40dd63a3
|
| 3 |
+
size 181365
|
config.json
ADDED
|
@@ -0,0 +1,174 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"sklearn": {
|
| 3 |
+
"columns": [
|
| 4 |
+
"surgery",
|
| 5 |
+
"age",
|
| 6 |
+
"rectal_temp",
|
| 7 |
+
"pulse",
|
| 8 |
+
"respiratory_rate",
|
| 9 |
+
"temp_of_extremities",
|
| 10 |
+
"peripheral_pulse",
|
| 11 |
+
"mucous_membrane",
|
| 12 |
+
"capillary_refill_time",
|
| 13 |
+
"pain",
|
| 14 |
+
"peristalsis",
|
| 15 |
+
"abdominal_distention",
|
| 16 |
+
"nasogastric_tube",
|
| 17 |
+
"nasogastric_reflux",
|
| 18 |
+
"nasogastric_reflux_ph",
|
| 19 |
+
"rectal_exam_feces",
|
| 20 |
+
"abdomen",
|
| 21 |
+
"packed_cell_volume",
|
| 22 |
+
"total_protein",
|
| 23 |
+
"abdomo_appearance",
|
| 24 |
+
"abdomo_protein",
|
| 25 |
+
"surgical_lesion",
|
| 26 |
+
"lesion_1",
|
| 27 |
+
"lesion_2",
|
| 28 |
+
"lesion_3",
|
| 29 |
+
"cp_data"
|
| 30 |
+
],
|
| 31 |
+
"environment": [
|
| 32 |
+
"scikit-learn=1.3.0",
|
| 33 |
+
"catboost==1.2.1"
|
| 34 |
+
],
|
| 35 |
+
"example_input": {
|
| 36 |
+
"abdomen": [
|
| 37 |
+
"distend_small",
|
| 38 |
+
"distend_small",
|
| 39 |
+
"distend_large"
|
| 40 |
+
],
|
| 41 |
+
"abdominal_distention": [
|
| 42 |
+
"none",
|
| 43 |
+
"none",
|
| 44 |
+
"moderate"
|
| 45 |
+
],
|
| 46 |
+
"abdomo_appearance": [
|
| 47 |
+
"serosanguious",
|
| 48 |
+
"cloudy",
|
| 49 |
+
"serosanguious"
|
| 50 |
+
],
|
| 51 |
+
"abdomo_protein": [
|
| 52 |
+
4.1,
|
| 53 |
+
4.3,
|
| 54 |
+
2.0
|
| 55 |
+
],
|
| 56 |
+
"age": [
|
| 57 |
+
"adult",
|
| 58 |
+
"adult",
|
| 59 |
+
"adult"
|
| 60 |
+
],
|
| 61 |
+
"capillary_refill_time": [
|
| 62 |
+
"less_3_sec",
|
| 63 |
+
"less_3_sec",
|
| 64 |
+
"more_3_sec"
|
| 65 |
+
],
|
| 66 |
+
"cp_data": [
|
| 67 |
+
"yes",
|
| 68 |
+
"yes",
|
| 69 |
+
"no"
|
| 70 |
+
],
|
| 71 |
+
"lesion_1": [
|
| 72 |
+
7209,
|
| 73 |
+
2112,
|
| 74 |
+
5400
|
| 75 |
+
],
|
| 76 |
+
"lesion_2": [
|
| 77 |
+
0,
|
| 78 |
+
0,
|
| 79 |
+
0
|
| 80 |
+
],
|
| 81 |
+
"lesion_3": [
|
| 82 |
+
0,
|
| 83 |
+
0,
|
| 84 |
+
0
|
| 85 |
+
],
|
| 86 |
+
"mucous_membrane": [
|
| 87 |
+
"bright_pink",
|
| 88 |
+
"bright_pink",
|
| 89 |
+
"dark_cyanotic"
|
| 90 |
+
],
|
| 91 |
+
"nasogastric_reflux": [
|
| 92 |
+
"none",
|
| 93 |
+
"none",
|
| 94 |
+
"more_1_liter"
|
| 95 |
+
],
|
| 96 |
+
"nasogastric_reflux_ph": [
|
| 97 |
+
7.0,
|
| 98 |
+
3.5,
|
| 99 |
+
2.0
|
| 100 |
+
],
|
| 101 |
+
"nasogastric_tube": [
|
| 102 |
+
"slight",
|
| 103 |
+
"none",
|
| 104 |
+
"significant"
|
| 105 |
+
],
|
| 106 |
+
"packed_cell_volume": [
|
| 107 |
+
37.0,
|
| 108 |
+
44.0,
|
| 109 |
+
65.0
|
| 110 |
+
],
|
| 111 |
+
"pain": [
|
| 112 |
+
"depressed",
|
| 113 |
+
"mild_pain",
|
| 114 |
+
"extreme_pain"
|
| 115 |
+
],
|
| 116 |
+
"peripheral_pulse": [
|
| 117 |
+
"normal",
|
| 118 |
+
"normal",
|
| 119 |
+
"reduced"
|
| 120 |
+
],
|
| 121 |
+
"peristalsis": [
|
| 122 |
+
"hypermotile",
|
| 123 |
+
"hypomotile",
|
| 124 |
+
"absent"
|
| 125 |
+
],
|
| 126 |
+
"pulse": [
|
| 127 |
+
84.0,
|
| 128 |
+
66.0,
|
| 129 |
+
72.0
|
| 130 |
+
],
|
| 131 |
+
"rectal_exam_feces": [
|
| 132 |
+
"absent",
|
| 133 |
+
"decreased",
|
| 134 |
+
"absent"
|
| 135 |
+
],
|
| 136 |
+
"rectal_temp": [
|
| 137 |
+
39.0,
|
| 138 |
+
38.5,
|
| 139 |
+
37.3
|
| 140 |
+
],
|
| 141 |
+
"respiratory_rate": [
|
| 142 |
+
24.0,
|
| 143 |
+
21.0,
|
| 144 |
+
30.0
|
| 145 |
+
],
|
| 146 |
+
"surgery": [
|
| 147 |
+
"yes",
|
| 148 |
+
"yes",
|
| 149 |
+
"yes"
|
| 150 |
+
],
|
| 151 |
+
"surgical_lesion": [
|
| 152 |
+
"yes",
|
| 153 |
+
"yes",
|
| 154 |
+
"yes"
|
| 155 |
+
],
|
| 156 |
+
"temp_of_extremities": [
|
| 157 |
+
"cool",
|
| 158 |
+
"normal",
|
| 159 |
+
"cool"
|
| 160 |
+
],
|
| 161 |
+
"total_protein": [
|
| 162 |
+
6.5,
|
| 163 |
+
7.6,
|
| 164 |
+
13.0
|
| 165 |
+
]
|
| 166 |
+
},
|
| 167 |
+
"model": {
|
| 168 |
+
"file": "catboost_without_hospital_number_01.pkl"
|
| 169 |
+
},
|
| 170 |
+
"model_format": "pickle",
|
| 171 |
+
"task": "tabular-classification",
|
| 172 |
+
"use_intelex": false
|
| 173 |
+
}
|
| 174 |
+
}
|
confusion_matrix.png
ADDED
|