code
stringlengths 235
11.6M
| repo_path
stringlengths 3
263
|
|---|---|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="Sz3j5dlzYWEM" colab_type="text"
# # Clique Cover
#
# In graph theory, a clique cover or partition into cliques of a given undirected graph is a partition of the vertices of the graph into cliques, subsets of vertices within which every two vertices are adjacent. A minimum clique cover is a clique cover that uses as few cliques as possible. The minimum k for which a clique cover exists is called the clique cover number of the given graph.
#
# https://en.wikipedia.org/wiki/Clique_cover
# + [markdown] id="FPWEQpF3YWEN" colab_type="text"
# # Cost Function
# When we color vertex $v$ with color $i$ we show using qubit as $x_{v,i}$.
# Now we have the cost function
#
# $ \displaystyle H = A \sum_v \left( 1 - \sum_{i = 1}^n x_{v,i} \right)^2 + B \sum_{i=1}^n \left[ \frac {1}{2} \left( -1 + \sum_v x_{v,i} \right) \sum_v x_{v,i} - \sum_{(uv) \in E} x_{u,i}x_{v.i} \right]$
#
# Expand it and we have,
#
# $ \displaystyle H = A \sum_v \left\{ -2 \sum_{i=1}^n x_{v,i} + \left(\sum_{i=1}^n x_{v,i}\right)^2 \right\} + B \sum_{i=1}^n \left\{ -\frac{1}{2} \sum_v x_{v,i} + \frac{1}{2}\left( \sum_v x_{v,i}\right)^2 - \sum_{(u,v) \in E} x_{u,i}x_{v,i}\right\}+ Const. $
# $ \displaystyle = A \sum_v \left( -2 \sum_{i=1}^n x_{v,i} + \sum_{i=1}^n x_{v,i}^2 + 2\mathop{ \sum \sum }_{i \neq j }^{n} x_{v,i}x_{v,j} \right) + B \sum_{i=1}^n \left\{ \frac{1}{2} \left(-\sum_v x_{v,i} + \sum_v x_{v,i}^2 + \mathop{\sum \sum}_{u \neq v}^{n} x_{u,i}x_{v,i} \right) - \sum_{(u,v) \in E} x_{u,i}x_{v,i}\right\}+ Const. $
# $ \displaystyle = A \sum_v \left( - \sum_{i=1}^n x_{v,i}^2 + 2\mathop { \sum \sum }_{i \neq j }^{n} x_{v,i}x_{v,j} \right) + B \sum_{i=1}^n \left( \frac{1}{2} \mathop{\sum \sum}_{u \neq v}^{n}x_{u,i}x_{v,i} - \sum_{(u,v) \in E} x_{u,i}x_{v,i}\right)+ Const. $
# + [markdown] id="-P_h2fICYWEO" colab_type="text"
# # Solving QUBO
# + id="E9k5tuN7Zd0C" colab_type="code" colab={}
# !pip install blueqat
# + id="biPn2HdLYWEP" colab_type="code" colab={}
import blueqat.opt as wq
import numpy as np
def get_qubo(adjacency_matrix, n_color, A, B):
graph_size = len(adjacency_matrix)
qubo_size = graph_size * n_color
qubo = np.zeros((qubo_size, qubo_size))
indices = [(u,v,i,j) for u in range(graph_size) for v in range(graph_size) for i in range(n_color) for j in range(n_color)]
for u,v,i,j in indices:
ui = u * n_color + i
vj = v * n_color + j
if ui > vj:
continue
if ui == vj:
qubo[ui][vj] -= A
if u == v and i != j:
qubo[ui][vj] += A * 2
if u != v and i == j:
qubo[ui][vj] += B * 0.5
if adjacency_matrix[u][v] > 0:
qubo[ui][vj] -= B
return qubo
# + id="bXHjnVQMYWES" colab_type="code" colab={}
def show_answer(q, graph_size, n_color):
print(q)
for v in range(graph_size):
color = []
for i in range(n_color):
index = v * n_color + i
if q[index] > 0:
color.append(i)
print(f"vertex{v}'s color is {color}")
# + id="2k9isCHQYWEV" colab_type="code" colab={}
def calculate_H(q, adjacency_matrix, n_color, A, B):
graph_size = len(adjacency_matrix)
h_a = calculate_H_A(q, graph_size, n_color, A)
h_b = calculate_H_B(q, adjacency_matrix, n_color, B)
print(f"H = {h_a + h_b}")
return h_a + h_b
def calculate_H_A(q, graph_size, n_color, A):
hamiltonian = 0
for v in range(graph_size):
sum_x = 0
for i in range(n_color):
index = v * n_color + i
sum_x += q[index]
hamiltonian += (1 - sum_x) ** 2
hamiltonian *= A
print(f"H_A = {hamiltonian}")
return hamiltonian
def calculate_H_B(q, adjacency_matrix, n_color, B):
graph_size = len(adjacency_matrix)
hamiltonian = 0
for i in range(n_color):
sum_x = 0
for v in range(graph_size):
vi = v * n_color + i
sum_x += q[vi]
for u in range(graph_size):
if u >= v:
continue
ui = u * n_color + i
hamiltonian -= adjacency_matrix[u][v] * q[ui] * q[vi]
hamiltonian += 0.5 * (-1 + sum_x) * sum_x
hamiltonian *= B
print(f"H_B = {hamiltonian}")
return hamiltonian
# + [markdown] id="jfxYM3GMYWEa" colab_type="text"
# This time we have an example like below,
#
# 
# + id="pTcc263zYWEc" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 1717} outputId="e2650336-9354-43e5-fcb7-d324f785eeaa"
adjacency_matrix = \
[ \
[0,1,1,0,0], \
[1,0,1,1,1], \
[1,1,0,1,0], \
[0,1,1,0,1], \
[0,1,0,1,0], \
]
n_color = 2
A = 0.1
B = 0.1
annealer = wq.opt()
annealer.qubo = get_qubo(adjacency_matrix, n_color, A, B)
for _ in range(10):
q = annealer.sa()
calculate_H(q, adjacency_matrix, n_color, A, B)
show_answer(q, len(adjacency_matrix), n_color)
print()
# + [markdown] id="2L8nX_R9YWEi" colab_type="text"
# When we have $H = 0$ these as answers.
| tutorial/305_cliquecover_en.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Environment (conda_datatest)
# language: python
# name: conda_datatest
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# # Data Checks
# + [markdown] slideshow={"slide_type": "fragment"}
# - Schema checks:
# - Making sure that only the columns that are expected are provided.
# - Making sure the data types are correct: `str`/`object`, `int`, `float32`, `float64`, `datetime`.
# + [markdown] slideshow={"slide_type": "subslide"}
# - Datum checks:
# - Looking for missing values
# - Ensuring that expected value ranges are correct
# + [markdown] slideshow={"slide_type": "fragment"}
# - Statistical checks:
# - Visual check of data distributions.
# - Correlations between columns.
# - Statistical distribution checks.
# + [markdown] slideshow={"slide_type": "slide"}
# # Schema Checks
#
# Schema checks are all about making sure that the data columns that you want to have are all present, and that they have the expecte data types.
#
# We're going to use a few datasets from Boston's open data repository. Let's first take a look at Boston's annual budget data.
# + slideshow={"slide_type": "subslide"}
# %load_ext autoreload
# %autoreload 2
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
# + [markdown] slideshow={"slide_type": "subslide"}
# ## A bit of basic `pandas`
# + slideshow={"slide_type": "fragment"}
import pandas as pd
df = pd.read_csv('data/boston_budget.csv')
df.head()
# + [markdown] slideshow={"slide_type": "subslide"}
# To get the columns of a DataFrame object `df`, call `df.columns`. This is a list-like object that can be iterated over.
# + slideshow={"slide_type": "fragment"}
df.columns
# + [markdown] slideshow={"slide_type": "subslide"}
# ## YAML Files
# + [markdown] slideshow={"slide_type": "fragment"}
# Describe data in a human-friendly & computer-readable format.
# + [markdown] slideshow={"slide_type": "fragment"}
# Structure:
#
# ```yaml
# key1: value
# key2:
# - value1
# - value2
# - subkey1:
# - value3
# ```
# + [markdown] slideshow={"slide_type": "subslide"}
# Example YAML-formatted schema:
#
# ```yaml
# filename: boston_budget.csv
# columns:
# - "Fiscal Year"
# - "Service (cabinet)"
# - "Department"
# - "Program #"
# ...
# - "Fund"
# - "Amount"
# #```
# + [markdown] slideshow={"slide_type": "subslide"}
# YAML-formatted text can be read as dictionaries.
# + slideshow={"slide_type": "-"}
spec = """
filename: boston_budget.csv
columns:
- "Fiscal Year"
- "Service (Cabinet)"
- "Department"
- "Program #"
- "Program"
- "Expense Type"
- "ACCT #"
- "Expense Category (Account)"
- "Fund"
- "Amount"
"""
# + slideshow={"slide_type": "subslide"}
import yaml
metadata = yaml.load(spec)
metadata
# + [markdown] slideshow={"slide_type": "subslide"}
# By having things YAML formatted, you preserve human-readability and computer-readability simultaneously.
# + [markdown] slideshow={"slide_type": "subslide"}
# Let's now switch roles, and pretend that we're on side of the "analyst" and are no longer the "data provider".
#
# How would you check that the columns match the spec? Basically, check that every element in `df.columns` is present inside the `metadata['columns']` list.
# + slideshow={"slide_type": "subslide"}
for col in df.columns:
# print(col)
try:
assert col in metadata['columns']
except AssertionError:
print(f'"{col}" not in metadata columns')
# + [markdown] slideshow={"slide_type": "subslide"}
# If there is even a slight mis-spelling, this kind of check will help you pinpoint where that is. Note how the "Amount" column is spelled with an extra space. Where would be the most human-oriented place to correct this? At the data provider stage.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise
#
# Encode the aforementioned test into a test function named `test_data_columns`. It should only be concerned with the Boston Budget dataset, and should only test whether the columns match the YAML spec.
# + slideshow={"slide_type": "subslide"}
# Copy to test_datafuncs.py
import yaml
import pandas as pd
def read_metadata(handle):
with open(handle, 'r+') as f:
metadata_str = ''.join(l for l in f.readlines())
return yaml.load(metadata_str)
def test_data_columns():
metadata = read_metadata('data/metadata_budget.yml')
df = pd.read_csv('data/boston_budget.csv')
for col in df.columns:
assert col in metadata['columns'], f'"{col}" not on metadata spec.'
# + [markdown] slideshow={"slide_type": "subslide"}
# It is a logical practice to keep one schema spec file per table provided to you. However, it is also possible to take advantage of YAML "documents" to keep multiple schema specs inside a single YAML file.
#
# The choice is yours - in cases where there are a lot of data files, it may make sense (for the sake of file-system sanity) to keep all of the specs in multiple files that represent logical groupings of data.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise: Write `YAML` metadata spec.
#
# Put yourself in the shoes of a data provider. Take any file in the `data/` directory, and make a schema spec file for that file.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise: Write test for metadata spec.
#
# Next, put yourself in the shoes of a data analyst. Take the schema spec file and write a test for it.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise: Write meta-test.
#
# Now, let's go "meta". Write a "meta-test" that ensures that every CSV file in the `data/` directory has a schema file associated with it. (The function need not check each schema.)
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Notes
#
# - Point: have trusted copy of schema apart from data file. YAML not necessarily only way.
# - If no schema provided, manually create one; this is exploratory data analysis anyways - no effort wasted!
# + [markdown] slideshow={"slide_type": "slide"}
# # Datum Checks
#
# Now that we're done with the schema checks, let's do some sanity checks on the data as well. This is my personal favourite too, as some of the activities here overlap with the early stages of exploratory data analysis.
#
# We're going to switch datasets here, and move to a 'corrupted' version of the Boston Economic Indicators dataset. Its file path is: `./data/boston_ei-corrupt.csv`.
# + slideshow={"slide_type": "subslide"}
import pandas as pd
import seaborn as sns
sns.set_style('white')
# %matplotlib inline
df = pd.read_csv('data/boston_ei-corrupt.csv')
df.head()
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Demo: Visual Diagnostics
#
# We can use a package called `missingno`, which gives us a quick visual view of the completeness of the data. This is a good starting point for deciding whether you need to manually comb through the data or not.
# + slideshow={"slide_type": "subslide"}
# First, we check for missing data.
import missingno as msno
msno.matrix(df)
# + [markdown] slideshow={"slide_type": "subslide"}
# Immediately it's clear that there's a number of rows with empty values! Nothing beats a quick visual check like this one.
# + [markdown] slideshow={"slide_type": "fragment"}
# We can get a table version of this using another package called `pandas_summary`.
# + slideshow={"slide_type": "subslide"}
# We can do the same using pandas-summary.
from pandas_summary import DataFrameSummary
dfs = DataFrameSummary(df)
dfs.summary()
# + [markdown] slideshow={"slide_type": "subslide"}
# `dfs.summary()` returns a Pandas DataFrame; can write tests for data completeness.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise: Test for data completeness.
#
# Write a test that confirms that there's no missing data.
# + slideshow={"slide_type": "fragment"}
# Add this to test_datafuncs.py
from pandas_summary import DataFrameSummary
def test_data_completeness(df):
df_summary = DataFrameSummary(df).summary()
for col in df_summary.columns:
assert df_summary.loc['missing', col] == 0, f'{col} has missing values'
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Exercise: Test for value correctness.
#
# Next, we can sanity-check our data values.
#
# Basic checks:
# - `val >= 0` (positive real numbers)
# - `0 <= val <= 1` (fraction/rates/percentages)
#
# Let's write one test that encompasses the [0, 1] scenario.
# + slideshow={"slide_type": "subslide"}
def test_data_range(df, col):
if col == 'labor_force_part_rate': # hard code one condition per column
assert df[col].min() >= 0, "minimum value less than zero"
assert df[col].max() <= 1, "maximum value greater than zero"
test_data_range(df, 'labor_force_part_rate')
# + [markdown] slideshow={"slide_type": "subslide"}
# We can take the EDA portion further, by doing an empirical cumulative distribution plot for each data column.
# + slideshow={"slide_type": "skip"}
import numpy as np
def compute_dimensions(length):
"""
Given an integer, compute the "square-est" pair of dimensions for plotting.
Examples:
- length: 17 => rows: 4, cols: 5
- length: 14 => rows: 4, cols: 4
"""
sqrt = np.sqrt(length)
floor = int(np.floor(sqrt))
ceil = int(np.ceil(sqrt))
if floor ** 2 >= length:
return (floor, floor)
elif floor * ceil >= length:
return (floor, ceil)
else:
return (ceil, ceil)
compute_dimensions(length=17)
assert compute_dimensions(17) == (4, 5)
assert compute_dimensions(16) == (4, 4)
assert compute_dimensions(15) == (4, 4)
assert compute_dimensions(11) == (3, 4)
# + slideshow={"slide_type": "subslide"}
# Next, let's visualize the empirical CDF for each column of data.
import matplotlib.pyplot as plt
def empirical_cumdist(data, ax, title=None):
"""
Plots the empirical cumulative distribution of values.
"""
x, y = np.sort(data), np.arange(1, len(data)+1) / len(data)
ax.scatter(x, y)
ax.set_title(title)
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
empirical_cumdist(df[col], ax, title=col)
plt.tight_layout()
plt.show()
# + [markdown] slideshow={"slide_type": "slide"}
# # Statistical Checks
#
# - Report on deviations from normality.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## Normality?!
#
# - The Gaussian (Normal) distribution is commonly assumed in downstream statistical procedures, e.g. outlier detection.
# - We can test for normality by using a K-S test.
# + [markdown] slideshow={"slide_type": "subslide"}
# ## K-S test
#
# From Wikipedia:
#
# > In statistics, the Kolmogorov–Smirnov test (K–S test or KS test) is a nonparametric test of the equality of continuous, one-dimensional probability distributions that can be used to compare a sample with a reference probability distribution (one-sample K–S test), or to compare two samples (two-sample K–S test). It is named after <NAME> and <NAME>.
# + [markdown] slideshow={"slide_type": "subslide"}
# 
# + slideshow={"slide_type": "subslide"}
from scipy.stats import ks_2samp
import numpy.random as npr
# Simulate a normal distribution with 10000 draws.
normal_rvs = npr.normal(size=10000)
result = ks_2samp(normal_rvs, df['labor_force_part_rate'].dropna())
result.pvalue < 0.05
# + slideshow={"slide_type": "subslide"}
fig = plt.figure()
ax = fig.add_subplot(111)
ecdf_scatter(normal_rvs, ax=ax)
ecdf_scatter(df['hotel_occup_rate'], ax=ax)
# -
| 4-data-checks.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (TensorFlow 2.1 Python 3.6 CPU Optimized)
# language: python
# name: python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:us-east-1:081325390199:image/tensorflow-2.1-cpu-py36
# ---
# # Lab: Bring your own custom container with Amazon SageMaker
# ## Overview
# Here, we’ll show how to package a simple Python example which showcases the decision tree algorithm from the widely used scikit-learn machine learning package. The example is purposefully fairly trivial since the point is to show the surrounding structure that you’ll want to add to your own code so you can train and host it in Amazon SageMaker.
#
# The ideas shown here will work in any language or environment. You’ll need to choose the right tools for your environment to serve HTTP requests for inference, but good HTTP environments are available in every language these days.
#
# In this example, we use a single image to support training and hosting. This is easy because it means that we only need to manage one image and we can set it up to do everything. Sometimes you’ll want separate images for training and hosting because they have different requirements. Just separate the parts discussed below into separate Dockerfiles and build two images. Choosing whether to have a single image or two images is really a matter of which is more convenient for you to develop and manage.
#
# If you’re only using Amazon SageMaker for training or hosting, but not both, there is no need to build the unused functionality into your container.
#
#
# ## Building the container
# Docker provides a simple way to package arbitrary code into an image that is totally self-contained完全独立的. Once you have an image, you can use Docker to run a container based on that image. Running a container is just like running a program on the machine except that the container creates a fully self-contained environment for the program to run. Containers are isolated from each other and from the host environment, so the way you set up your program is the way it runs, no matter where you run it.
#
# Amazon SageMaker uses Docker to allow users to train and deploy arbitrary algorithms.
# ### Parts of the container
# In the container directory are all the components you need to package the sample algorithm for Amazon SageMager:
# ```
# .
# |-- Dockerfile
# |-- build_and_push.sh
# `-- decision_trees
# |-- nginx.conf
# |-- predictor.py
# |-- serve
# |-- train
# `-- wsgi.py
# ```
# Let’s discuss each of these in turn:
#
# - Dockerfile describes how to build your Docker container image. More details below:
# - build_and_push.sh is a script that uses the Dockerfile to build your container images and then pushes it to ECR. We’ll invoke the commands directly later in this notebook, but you can just copy and run the script for your own algorithms.
# - decision_trees is the directory which contains the files that will be installed in the container.
# - local_test is a directory that shows how to test your new container on any computer that can run Docker, including an Amazon SageMaker notebook instance. Using this method, you can quickly iterate using small datasets to eliminate any structural bugs before you use the container with Amazon SageMaker. We’ll walk through local testing later in this notebook.
#
# In this simple application, we only install five files in the container.
#
# The files that we’ll put in the container are:
#
# - nginx.conf is the configuration file for the nginx front-end. Generally, you should be able to take this file as-is保持原样.
# - predictor.py is the program that actually implements the Flask web server and the decision tree predictions for this app. You’ll want to customize the actual prediction parts to your application. Since this algorithm is simple, we do all the processing here in this file, but you may choose to have separate files for implementing your custom logic.
# - serve is the program started when the container is started for hosting. It simply launches the gunicorn server(Gunicorn“ Green Unicorn”是Python Web服务器网关接口HTTP服务器。它是从Ruby的Unicorn项目移植而来的pre-fork worker模型。Gunicorn服务器与许多Web框架广泛兼容,这些Web框架实现简单,占用服务器资源少且速度相当快。) which runs multiple instances of the Flask app defined in predictor.py. You should be able to take this file as-is.
# - train is the program that is invoked when the container is run for training. You will modify this program to implement your training algorithm.
# - wsgi.py is a small wrapper used to invoke the Flask app. You should be able to take this file as-is. Web服务器网关接口 (Web Server Gateway Interface)是为Python语言定义的Web服务器和Web应用程序或框架之间的一种简单而通用的接口。
#
# In summary, the two files you will probably want to change for your application are train and predictor.py
# ### The Dockerfile
# The Dockerfile describes the image that we want to build. You can think of it as describing the complete operating system installation of the system that you want to run. A Docker container running is quite a bit lighter than a full operating system, because it takes advantage of Linux on the host machine for the basic operations.
#
# For the Python science stack, we will start from a standard Ubuntu installation and run the normal tools to install the things needed by scikit-learn. Finally, we add the code that implements our specific algorithm to the container and set up the right environment to run under.
# cell 00
# Async client for amazon services using botocore and aiohttp/asyncio.
# https://github.com/aio-libs/aiobotocore
# !pip install --upgrade aiobotocore
# +
import zipfile
file_dir = 'scikit_bring_your_own.zip'
# 解压文件
zip_File = zipfile.ZipFile(file_dir,'r')
# 获取压缩文件中的内容
zip_list = zip_File.namelist()
#打印详细信息
# zzz = zip_File.printdir()
#打印列表信息
# print(zip_list)
#解压到当前文件夹中
zip_File.extractall(r'.')
# +
# cell 01
# # !unzip scikit_bring_your_own.zip
# 移动以下两个文件 【scikit_bring_your_own/data/】 【scikit_bring_your_own/container/】 到指定的位置
# !mv scikit_bring_your_own/data/ ./lab03_data/
# !mv scikit_bring_your_own/container/ ./lab03_container/
# 删除 scikit_bring_your_own
# !rm -rf scikit_bring_your_own
# 查看 Dockerfile 文件内容
# !cat lab03_container/Dockerfile
# -
# ## Building and registering the container
# cell 02
# !pip install sagemaker-studio-image-build
# > *In the next cell, if you run into IAM permission issue related to CodeBuild, make sure that you follow the steps outlined in the instructions*
# !pwd
# +
# # %%sh
# cell 03
# !cd lab03_container
# 为以下两个文件【train】 【serve】 加入可执行权限
# !chmod +x lab03_container/decision_trees/train
# !chmod +x lab03_container/decision_trees/serve
# 构建一个docker镜像
# !sm-docker build ./lab03_container --repository sagemaker-decision-trees:latest
# -
# ## Using the container
# Here we specify a bucket to use and the role that will be used for working with SageMaker.
# +
# cell 04
# S3 prefix
prefix = 'DEMO-scikit-byo-iris'
# Define IAM role
import boto3
import re
import os
import numpy as np
import pandas as pd
from sagemaker import get_execution_role
role = get_execution_role()
role
# -
# The session remembers our connection parameters to SageMaker. We’ll use it to perform all of our SageMaker operations.
# +
# cell 05
import sagemaker as sage
from time import gmtime, strftime
# 创建一个Session, 管理与 Amazon SageMaker API 和任何其他所需 AWS 服务的交互。
sess = sage.Session()
# -
# When training large models with huge amounts of data, you’ll typically use big data tools, like Amazon Athena, AWS Glue, or Amazon EMR, to create your data in S3. For the purposes of this example, we’re using some the classic Iris dataset, which we have included.
#
# We can use use the tools provided by the SageMaker Python SDK to upload the data to a default bucket.
# +
# cell 06
WORK_DIRECTORY = 'lab03_data'
# 上传数据到指定的目录
data_location = sess.upload_data(WORK_DIRECTORY, key_prefix=prefix)
data_location
# -
# In order to use SageMaker to fit our algorithm, we’ll create an Estimator that defines how to use the container to train. This includes the configuration we need to invoke SageMaker training:
#
# - The container name. This is constructed as in the shell commands above.
# - The role. As defined above.
# - The instance count which is the number of machines to use for training.
# - The instance type which is the type of machine to use for training.
# - The output path determines where the model artifact will be written.
# - The session is the SageMaker session object that we defined above.
#
# Then we use `fit()` on the estimator to train against the data that we uploaded above.
# +
# cell 07
account = sess.boto_session.client('sts').get_caller_identity()['Account']
region = sess.boto_session.region_name
image = '{}.dkr.ecr.{}.amazonaws.com/sagemaker-decision-trees:latest'.format(account, region)
tree = sage.estimator.Estimator(image,
role, instance_count=1, instance_type='ml.c4.2xlarge',
output_path="s3://{}/output".format(sess.default_bucket()),
sagemaker_session=sess)
file_location = data_location + '/iris.csv'
tree.fit(file_location)
# -
# ## Hosting your model
# You can use a trained model to get real time predictions using HTTP endpoint. Follow these steps to walk you through the process.
#
# Deploying the model to SageMaker hosting just requires a deploy call on the fitted model. This call takes an instance count, instance type, and optionally serializer and deserializer functions. These are used when the resulting predictor is created on the endpoint.
# cell 08
from sagemaker.serializers import CSVSerializer
predictor = tree.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge', serializer=CSVSerializer())
# ## Preparing test data to run inferences
# In order to do some predictions, we’ll extract some of the data we used for training and do predictions against it. This is, of course, bad statistical practice, but a good way to see how the mechanism works.
# +
# cell 09
# 利用pandas读取数据csv数据, 不带header
shape=pd.read_csv("lab03_data/iris.csv", header=None)
# 随机显示3条样本数据
shape.sample(3)
# +
# cell 10
# drop the label column in the training set
# 除去数据中的第0列代表的标签数据。inplace=True表示在原有数据上直接进行删除操作。
shape.drop(shape.columns[[0]],axis=1,inplace=True)
shape.sample(3)
# +
# cell 11
import itertools
# 利用列表生成器a,b两组数据
a = [50*i for i in range(3)]
b = [40+i for i in range(10)]
# 利用双层循环生成 40~49, 90~99, 140~149范围数据
indices = [i+j for i,j in itertools.product(a,b)]
# iloc基于行索引进行索引数据, 作为测试数据
test_data=shape.iloc[indices[:-1]]
test_data
# -
# ## Run predictions
#
# Prediction is as easy as calling predict with the predictor we got back from deploy and the data we want to do predictions with. The serializers take care of doing the data conversions for us.
# +
# cell 12
# 执行预测,并打印结果。
print(predictor.predict(test_data.values).decode('utf-8'))
# -
# ## Cleanup
# After completing the lab, use these steps to [delete the endpoint](https://docs.aws.amazon.com/sagemaker/latest/dg/ex1-cleanup.html) or run the following code
#
# cell 13
# 删除指定的endpoint
sess.delete_endpoint(predictor.endpoint_name)
# +
# cell 14
# 删除【lab03_container】 【lab03_data】目录
# # !rm -rf lab03_container lab03_data
| lab3/bring-custom-container.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="cedf868076a2"
# ##### Copyright 2020 The Cirq Developers
# + cellView="form" id="906e07f6e562"
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] id="166af89a7bc3"
# # Neutral Atom Device Class
# + [markdown] id="416c50754585"
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.example.org/cirq/tutorials/educators/neutral_atom"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on QuantumLib</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/quantumlib/Cirq/blob/master/docs/tutorials/educators/neutral_atom.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/quantumlib/Cirq/blob/master/docs/tutorials/educators/neutral_atom.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/Cirq/docs/tutorials/educators/neutral_atom.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# </table>
# + [markdown] id="bNlUyYQcgMF-"
# This notebook provides an introductiong to making circuits that are compatable with neutral atom devices in Cirq. The NeutralAtomDevice class is available starting with release 0.5.0.
# + id="GTjMbjyAfJCK"
# install release containing NeutralAtomDevice and IonDevice classes
# !pip install cirq~=0.5.0 --quiet
# + [markdown] id="Z0X2AWrhrcHR"
# Let's get some imports out of the way and construct an instance of the NeutralAtomDevice class. We won't worry about the details of the constructor arguments just yet, we just want an instance of the class for demonstration purposes.
# + id="On6Wrh3XhSPO"
import cirq
import numpy as np
from matplotlib import pyplot as plt
ms = cirq.Duration(nanos=10**6)
us = cirq.Duration(nanos=10**3)
neutral_device = cirq.NeutralAtomDevice(measurement_duration = 5*ms,
gate_duration = 100*us,
control_radius = 2,
max_parallel_z = 3,
max_parallel_xy = 3,
max_parallel_c = 3,
qubits = [cirq.GridQubit(row, col)
for col in range(3)
for row in range(3)])
# + [markdown] id="v3YMaPw1hfV_"
# ## NeutralAtomDevice
# Disclaimer: As with any proposed architecture for quantum computing, several research groups around the world are working towards a device based on neutral atom qubits. Each research group has a different approach, such as using different atomic species or working with a different number of dimensions of atomic qubit arrays. As such, the NeutralAtomDevice class will not accurately reflect all such devices. The class is based on the two dimensional cesium array at the University of Wisconsin-Madison in the research group of <NAME>. Development of this device is being pursued as part of a strategic partnershp between the University of Wisconsin-Madison and ColdQuanta.
#
#
# ---
#
# ### Native Gate Set
# The gates supported by the NeutralAtomDevice class can be placed in three categories:
#
#
# 1. Single qubit rotations about the Z axis
# 2. Single qubit rotations about an arbitrary axis in the X-Y plane (I refer to these as XY gates in this tutorial)
# 3. Controlled gates: CZ, CNOT, CCZ, CCNOT (TOFFOLI)
#
#
# For the single qubit rotations, any rotation angle is fine. However, for the controlled gates, the rotation must be a multiple of $\pi$ due to the physical implementation of the gates. Cirq supports raising gates to arbitrary exponents. In order to satisfy the restriction of the controlled gate rotation, the exponent of the controlled gate must be an integer.
#
#
# You are allowed to specify the connectivity of the controlled gates via the control_radius argument of the NeutralAtomDevice constructor. This argument specifies the maximum distance between pairs of atoms acted on by a controlled gate.
#
#
# The neutral atom scheme for controlled gates also allows for multiple controls so long as every pair of atoms acted on by the controlled gate are close enough to eachother. The NeutralAtomDevice class does not currently support gates with greater than two controls but we hope to implement support for them in the near term.
#
#
# ---
#
#
# Some examples of gates in Cirq that the device supports are given below.
# + id="Hotk4cHCpXCV"
# Single Qubit Z rotation by Pi/5 radians
neutral_device.validate_gate(cirq.Rz(np.pi/5))
# Single Qubit rotation about axis in X-Y plane Pi/3 radians from X axis by angle of Pi/7
neutral_device.validate_gate(cirq.PhasedXPowGate(phase_exponent=np.pi/3,exponent=np.pi/7))
# Controlled gate with integer exponent
neutral_device.validate_gate(cirq.CNOT)
# Controlled Not gate with two controls
neutral_device.validate_gate(cirq.TOFFOLI)
# + [markdown] id="nc4zLydMsrkV"
# Some examples of gates in Cirq the device does not support are given below. These code blocks will yield errors describing since the gates are not valid for NeutralAtomDevice.
# + id="ChT4QK7TsabR"
#Controlled gate with non-integer exponent (rotation angle must be a multiple of pi)
# neutral_device.validate_gate(cirq.CZ**1.5)
# + id="UPKPh0XMs7zh"
# Hadamard gates rotate about the X-Z axis, which isn't compatable with our single qubit rotations
# neutral_device.validate_gate(cirq.H)
# + [markdown] id="7QvsNStFtXll"
# ### Constructor
#
# Let's take a look at the constructor for the NeutralAtomDevice class. There are a number of arguments needed to fully parametarize a device. The specific values for these numbers are expected improve over time for any physical device. Instead of constraining ourselves to how the device(s) look at a specific moment in time, the user is allowed to free or constrain themselves as they see fit.
#
# ---
#
# Gate duration variables: Depending on the specific implementation of the quantum gate, there may be a different amount of time needed to perform various actions. These arguments accept Duration and timedelta objects.
#
# * measurement_duration: How long it takes for the device to measure a qubit
#
# * gate_duration: The maximum amount of time it takes to execute a quantum gate
#
# ---
#
# Parallelism limitations: Neutral atom devices implement quantum gates in one of two ways. One method is by hitting the entire qubit array with microwaves to simultaneously act on every qubit. This method implements global XY gates which take up to 100 microseconds to perform. Alternatively, we can shine laser light on some fraction of the array. Gates of this type typically take around 1 microsecond to perform. This method can act on one or more qubits at a time up to some limit dictated by the available laser power and the beam steering system used to address the qubits. Each category in the native gate set has its own limit.
#
#
# * max_parallel_z: The maximum number of single qubit Z-axis rotations that can be applied in parallel
#
# * max_parallel_xy: The maximum number of single qubit XY rotations that can be applied in parallel
#
# * max_parallel_c: The maximum number of atoms that can be affected by controlled gates simulataneously
#
# Z-axis rotations and XY rotations use different light, so they have independent constraints. However, controlled gates make use of the light used to perform single qubit rotations in addition to extra laser frequencies. As such, the max_parallel_c argument is bounded above by the lesser of max_parallel_z and max_parallel_xy.
#
# ---
#
# Other variables:
#
# * qubits: A list of the qubits on the device. The only supported qubit type is GridQubit.
#
# * control_radius: Every pair of atoms acted on by a controlled gate must be within this maximum distance of eachother for the gate to succeed. The distance is between qubits is measured by using their row and column values as coordinates.
#
#
# ---
#
# The example device below has the following properties:
#
# * The device is a 3x3 grid of qubits
# * Measurements take 5ms
# * Gates may take as long as 100us if we utilize global microwave gates. Otherwise, a more reasonable bound would be 1us.
# * Controlled gates have next-nearest neighbor connectivity (control_radius of 2)
# * A maximum of 3 qubits may be simultaneously acted on by any gate category
#
# + id="PPYYsBZr2UTD"
ms = cirq.Duration(nanos=10**6)
us = cirq.Duration(nanos=10**3)
neutral_device = cirq.NeutralAtomDevice(measurement_duration = 5*ms,
gate_duration = 100*us,
control_radius = 2,
max_parallel_z = 3,
max_parallel_xy = 3,
max_parallel_c = 3,
qubits = [cirq.GridQubit(row, col)
for col in range(3)
for row in range(3)])
# + [markdown] id="Ug-2oBPPtZzU"
# ### Moment/Circuit Rules
#
# Now that we know how to parametarize a NeutralAtomDevice, we can discuss some examples of valid and invalid moments and circuits. Each operation in a moment is treated as if they are performed simultaneously. This leaves us with a few constraints we need to abide by.
#
#
# 1. We need to respect the maximum number of parallel gates for any gate type as discussed above
# 2. All instances of gates in the same category in the same moment must be identical
# 3. Since controlled gates make use of all types of light used to make gates, controlled gates cannot be applied in parallel with other gate types
# 4. Qubits acted on by different controlled gates in parallel must be farther apart than control_radius so that the entanglement mechanism doesn't cause the gates to interfere with one another
# 5. Measurements must be terminal
#
#
#
# ---
#
# Some examples of valid moments are given below. We know the moments we constructed were valid because the circuit allowed us to add them knowing that they needed to pass the neutral_device validation methods.
# + id="Nr-rfUgOtDxE"
# Moment/Circuit Examples
moment_circ = cirq.Circuit(device=neutral_device)
qubits = [cirq.GridQubit(row, col) for col in range(3) for row in range(3)]
# Three qubits affected by a Z gate in parallel with Three qubits affected
# by an XY gate
operation_list_one = cirq.Z.on_each(*qubits[:3])+cirq.X.on_each(*qubits[3:6])
valid_moment_one = cirq.Moment(operation_list_one)
moment_circ.append(valid_moment_one)
# A TOFFOLI gate on three qubits that are close enough to eachother
operation_list_two = [cirq.TOFFOLI.on(*qubits[:3])]
valid_moment_two = cirq.Moment(operation_list_two)
moment_circ.append(valid_moment_two)
print(moment_circ)
# + [markdown] id="l6jE7NnKtjQU"
# Frequently, you aren't explicitly constructing moments and adding them into circuits. You may more frequently find yourself directly appending a list of operations into the circuit. Let's look at the result if we attempt this method with a global operation.
# + id="u8oA9R5aCbEP"
global_circuit = cirq.Circuit(device=neutral_device)
global_list_of_operations = cirq.X.on_each(*qubits)
global_circuit.append(global_list_of_operations)
print(global_circuit)
# + [markdown] id="ZPd3emooFqa5"
# That doesn't look right! It looks like the insertion method tried to place the gates in one at a time and decided it needed three moments to satisfy the request. There are two ways of getting around this in Cirq. One is to manually construct the moment as above.
# + id="FxmKkfCEF744"
global_moment_circuit = cirq.Circuit(device=neutral_device)
global_moment = cirq.Moment(cirq.X.on_each(*qubits))
global_moment_circuit.append(global_moment)
print(global_moment_circuit)
# + [markdown] id="3E-tqsmlGKjD"
# Another way to achieve this task is the ParallelGateOperation class. Most Operation objects correspond to a single gate acting on some qubits. The ParallelGateOperation class corresponds to multiple copies of a single gate acting on some qubits in parallel. Since it is a single operation, the default insertion methods won't break it up into multiple moments when adding it into the circuit!
# + id="OIZux5rFGrLs"
parallel_gate_op_circuit = cirq.Circuit(device=neutral_device)
parallel_gate_op = cirq.ParallelGateOperation(cirq.X,qubits)
parallel_gate_op_circuit.append(parallel_gate_op)
print(parallel_gate_op_circuit)
# + [markdown] id="1byvHcWvHx-D"
# ###Grover's Algorithm
# Now that we have the details out of the way, we can take a look at what implementing a real quantum algorithm might look like on a NeutralAtomDevice. Since the current limit on having multiple controls on a gate is two, lets look at the three qubit Grover search. For this problem, there is some special target state out of the $N = 2^n$ possible basis states that we'd like to identify. We have access to a quantum oracle that applies a phase to this state relative to every other basis state. The algorithm succeeds if we measure the desired state when we make a measurement on all the qubits after the circuit is run.
#
#
# ---
#
# Classically, this corresponds to an unordered database search with a database of size $N=2^n$. On average, it will take $N/2$ applications of a classical oracle to find the desired state in the database. With Grover's algorithm, it is possible to succeed with high probability in $O(\sqrt N)$ applications of the quantum oracle.
#
#
# ---
#
# Grover's algorithm works by initially preparing the quantum state
# $$|s\rangle = \frac{1}{\sqrt{N}}\sum_{x=0}^{N-1}{|x\rangle}_.$$
#
# This is an even superposition of each state in the computational basis and can be obtained by applying a Hadamard gate or a $Y^{1/2}$ gate on a register with each qubit initialized to $|0\rangle$. After this initial state is prepared, Grover's algorithm consists of applying the quantum oracle followed by a diffusion operator.
#
# The quantum oracle places a relative phase on the target state we want to measure and the diffusion operator utilizes that phase difference to amplify the probability of measuring the target state relative to other states in the computational basis.
# The diffusion operator is given by $U_s = 2 |s\rangle \langle s| - I.$
#
#
# ---
#
#
# We can calculate the probability of success as a function of the number of repititions R that we apply the oracle and diffusion operator. The result is $$P_{success} = \sin^2 ((2R+1)\arcsin(\frac{1}{\sqrt{N}})_,$$
# with the maximum probability of success will occur when $R \approx \pi \sqrt{N}/4.$
#
#
# ---
#
# We know what these operators look like in Dirac notation, but now we have to compile them into the language of quantum gates. We'll start with the oracle. We can construct the oracle out of controlled-Z gates and NOT (X) gates. A Controlled-Z gate applies a $\pi$ phase shift to the state where each qubit is in the $|1 \rangle $ state. We can select a different state to recieve the phase shift by sandwiching the controlled-Z gate with X gates on any of the qubits whose desired measurement outcome is $|0 \rangle$. We can implement a generator for the oracle operator with the below code in Cirq.
# + id="uQ50bqZKZXVf"
def oracle(qubits, key_bits):
yield (cirq.X(q) for (q, bit) in zip(qubits, key_bits) if not bit)
yield cirq.CCZ(*qubits)
yield (cirq.X(q) for (q, bit) in zip(qubits, key_bits) if not bit)
# Try changing the key to see the relationship between
# the placement of the X gates and the key
key = (1, 0, 1)
qubits = [cirq.GridQubit(0,col) for col in range(3)]
oracle_example_circuit = cirq.Circuit().from_ops(oracle(qubits,key))
print(oracle_example_circuit)
# + [markdown] id="hOCGBzJ9dviO"
# A quick Google search of how to implement the diffusion operator will provide us with the below implementation.
# + id="63Y-GsLiH8Nm"
def diffusion_operator(qubits):
yield cirq.H.on_each(*qubits)
yield cirq.X.on_each(*qubits)
yield cirq.CCZ(*qubits)
yield cirq.X.on_each(*qubits)
yield cirq.H.on_each(*qubits)
qubits = [cirq.GridQubit(0,col) for col in range(3)]
diffusion_circuit = cirq.Circuit().from_ops(diffusion_operator(qubits))
print(diffusion_circuit)
# + [markdown] id="yo4E2JSoe00x"
# If we put it all together and note that for the three qubit case, the optimal number of applications of the oracle and diffusion operator is two, we get the below circuit.
# + id="TSfFAmk2e_Q8"
def initial_hadamards(qubits):
yield cirq.H.on_each(*qubits)
uncompiled_circuit = cirq.Circuit()
key = (1,0,1)
qubits = [cirq.GridQubit(0,0),cirq.GridQubit(0,1),cirq.GridQubit(0,2)]
uncompiled_circuit.append(initial_hadamards(qubits))
uncompiled_circuit.append(oracle(qubits,key))
uncompiled_circuit.append(diffusion_operator(qubits))
uncompiled_circuit.append(oracle(qubits,key))
uncompiled_circuit.append(diffusion_operator(qubits))
print(uncompiled_circuit)
# + [markdown] id="iTmPFGsahCh-"
# This circuit does implement Grover's algorithm, but it doesn't work for our NeutralAtomDevice class because of the Hadamard gates. The Hadamard gates show up in the Diffusion Operator steps and the initial "Hadamard Everything" step. We can simplify the Diffusion Operator definition and the initial Hadamard step by decomposing the Hadamard gate into products of XY gates.
#
#
# * $H=X Y^{1/2}$
# * $H=Y^{-1/2} X$
# * $H=Y^{1/2}Z$
#
# Using these two rules, we can instead make a neutral atom comptable version of the circuit given below.
# + id="HsjMeVNkie2b"
def neutral_atom_initial_step(qubits):
yield cirq.ParallelGateOperation(cirq.Y**(1/2), qubits)
def neutral_atom_diffusion_operator(qubits):
yield cirq.ParallelGateOperation(cirq.Y**(1/2), qubits)
yield cirq.CCZ(*qubits)
yield cirq.ParallelGateOperation(cirq.Y**(-1/2), qubits)
ms = cirq.Duration(nanos=10**6)
us = cirq.Duration(nanos=10**3)
qubits = [cirq.GridQubit(row, col) for col in range(3) for row in range(1)]
three_qubit_device = cirq.NeutralAtomDevice(measurement_duration = 5*ms,
gate_duration = us,
control_radius = 2,
max_parallel_z = 3,
max_parallel_xy = 3,
max_parallel_c = 3,
qubits=qubits)
key = (0,1,0)
compiled_grover_circuit = cirq.Circuit(device=three_qubit_device)
compiled_grover_circuit.append(neutral_atom_initial_step(qubits))
compiled_grover_circuit.append(oracle(qubits,key))
compiled_grover_circuit.append(neutral_atom_diffusion_operator(qubits))
compiled_grover_circuit.append(oracle(qubits,key))
compiled_grover_circuit.append(neutral_atom_diffusion_operator(qubits))
print(compiled_grover_circuit)
# + [markdown] id="zBfHYu7lfwbY"
# Let's quickly verify that this implementation of the algorithm on our device actually works by altering the number of times we apply the oracle and diffusion operators. We would expect to reproduce the analytical result given above.
# + id="MUC33tMYgJQV"
def grover_circuit_with_n_repetitions(n, key):
ms = cirq.Duration(nanos=10**6)
us = cirq.Duration(nanos=10**3)
qubits = [cirq.GridQubit(row, col) for col in range(3) for row in range(1)]
three_qubit_device = cirq.NeutralAtomDevice(measurement_duration = 5*ms,
gate_duration = us,
control_radius = 2,
max_parallel_z = 3,
max_parallel_xy = 3,
max_parallel_c = 3,
qubits=qubits)
grover_circuit = cirq.Circuit(device=three_qubit_device)
grover_circuit.append(neutral_atom_initial_step(qubits))
for repetition in range(n):
grover_circuit.append(oracle(qubits,key))
grover_circuit.append(neutral_atom_diffusion_operator(qubits))
return grover_circuit
success_probabilities = []
key = (0,1,1)
N = 2**3
#Convert key from binary to a base 10 number
diag = sum(2**(2-count) for (count, val) in enumerate(key) if val)
num_points = 10
for repetitions in range(num_points):
test_circuit = grover_circuit_with_n_repetitions(repetitions, key)
sim = cirq.Simulator()
result = sim.simulate(test_circuit)
rho = result.density_matrix_of(qubits)
success_probabilities.append(np.real(rho[diag][diag]))
plt.scatter(range(num_points), success_probabilities, label="Simulation")
x = np.linspace(0, num_points, 1000)
y = np.sin((2*x+1)*np.arcsin(1/np.sqrt(N)))**2
plt.plot(x, y, label="Theoretical Curve")
plt.title("Probability of Success Vs. Number of Oracle-Diffusion Operators")
plt.ylabel("Probability of Success")
plt.xlabel("Number of Times Oracle and Diffusion Operators are Applied")
plt.legend(loc='upper right')
plt.show()
# + [markdown] id="oSPDCZPCmugO"
# The results match! And we actually see that if we can afford to increase the number of times we apply the oracle and diffusion operators to six, we can get an improved probability of success.
| docs/tutorials/educators/neutral_atom.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Exploring the voicing of two repeated roman numeral annotations
# *If you are asked to harmonize a progression like*
#
# ```
# I IV | V V | I ||
# ```
#
# *Should you change the voicing of the two* `V` *chords, or can you just repeat the same one*?
import music21
from urllib.request import urlopen
import re
# # Getting the data
# The analyzed Bach chorales are located in the [KernScores](http://kern.ccarh.org/) website.
#
# They can be visualized interactively in the [Verovio Humdrum Viewer](https://verovio.humdrum.org/).
#
# They can also be scraped with a single query to the Kern Scores server: https://kern.humdrum.org/data?l=BachChoralesAnalyzed
#
# I used the last approach to collect the data for this experiment.
bachChoralesURL = 'https://kern.humdrum.org/data?l=BachChoralesAnalyzed'
choralesData = urlopen(bachChoralesURL).read().decode('iso-8859-1')
# This is a hack to force music21 into reading the **harm spines
choralesData2 = re.sub(r'(\*MM[0-9]+\n)', r"\1*staff1/2/3/4\t*\t*staff1\t*staff2\t*staff3\t*staff4\n", choralesData)
# Semicolons confuse the **harm parser
choralesData2 = choralesData2.replace(';', '')
chorales = re.split(r'!!!!SEGMENT: (chor[0-9]+)\.krn\n', choralesData2)
# The first string is empty
chorales = chorales[1:]
# +
out = music21.stream.Stream()
sameVoicing = []
for name, chorale in zip(chorales[0::2], chorales[1::2]):
print(name)
s = music21.converter.parseData(chorale, format='humdrum')
rna = {rn.offset: rn for rn in s.flat.getElementsByClass("RomanNumeral")}
offsets = list(rna.keys())
pairs = zip(list(rna.values())[:-1], list(rna.values())[1:])
filename = name
for idx, pair in enumerate(pairs):
rn1, rn2 = pair
offs1 = offsets[idx]
offs2 = offsets[idx + 1]
if rn1.key == rn2.key and rn1.figure == rn2.figure:
notes1 = list(
s.flat.getElementsByOffset(
offs1, mustBeginInSpan=False, classList="Note"
)
)
notes1 = [n.pitch.nameWithOctave for n in notes1]
notes2 = list(
s.flat.getElementsByOffset(
offs2, mustBeginInSpan=False, classList="Note"
)
)
notes2 = [n.pitch.nameWithOctave for n in notes2]
isSameVoicing = notes1 == notes2
sameVoicing.append(isSameVoicing)
print(
"\t{}\t{}\t{}\t{}\t{}".format(
offsets[idx],
notes1,
offsets[idx + 1],
notes2,
isSameVoicing,
)
)
chord1 = music21.chord.Chord(notes1, quarterLength=2)
chord1.style.color = "green" if isSameVoicing else "red"
chord1.addLyric(f"{offs1}")
chord1.addLyric(filename)
chord2 = music21.chord.Chord(notes2, quarterLength=2)
chord2.addLyric(f"{offs2}")
chord2.style.color = "green" if isSameVoicing else "red"
out.append(chord1)
out.append(chord2)
filename = ""
same = sum(sameVoicing) / len(sameVoicing)
print(
"""
Number of consecutive identical roman numerals: {}
Same voicing: {:.2f}%
Different voicing: {:.2f}%
""".format(
len(sameVoicing), same * 100.0, (1 - same) * 100.0
)
)
out.insert(0, music21.clef.Treble8vbClef())
# -
# In the table above, offsets are given in quarter notes from the beginning of the score (starting from 0.0).
#
# In all instances, `chord1` and `chord2` are contiguous, express the same chord (i.e., same roman numeral in the same key context), and have been annotated by a human expert.
# Show some of the scores
out.show()
# In the figure above, red-colored pairs represent a change in the voicing between the two contiguous annotations of the same roman numeral.
#
# Green-colored pairs show examples in which the voicing was identical in the two contiguous annotations.
# Generally, when a voice-leading algorithm finds two contiguous instances of the same roman numeral, it should be safe to assume that the voicing should change. For example, with a change of position in the three upper voices and/or changing the octave of the bass.
| content/post/repeatedharmonies/.ipynb_checkpoints/index-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Random Forest
# A single decision tree - tasked to learn a dataset - might not be able to perform well due to the outliers and the breadth and depth complexity of the data.
# So instead of relying on a single tree, random forests rely on a multitude of cleverly grown decision trees.
# Each tree within the forest is allowed to become highly specialised in a specific area but still retains some general knowledge about most areas. When a random forest classifies, it is actualy each tree in the forest working together to cast votes on what label they think a specific sample should be assigned.
#
# Instead of sharing the entire dataset with each decision tree, the forest performs an operation which is essential a train / test split of the training data. Each decision tree in the forest randomly samples from the overall training data set. Through doing so, each tree exist in an independent subspace and the variation between trees is controlled. This technique is known as **tree bagging, or bootstrap aggregating**.
#
# In addition to the tree bagging of training samples at the forest level, each individual decision tree further 'feature bags' at each node-branch split. This is helpful because some datasets contain a feature that is very correlated to the target (the 'y'-label). By selecting a random sampling of features every split - if such a feature were to exist - it wouldn't show up on as many branches of the tree and there would be more diversity of the features examined.
#
# Check [my post](https://mashimo.wordpress.com/2020/04/26/random-forest/) to see more details about Random Forests!
# # Human activity prediction
# As an example, we will predict human activity by looking at data from wearables.
# For this , we train a random forest against a public domain Human Activity Dataset titled *Wearable Computing: Accelerometers' Data Classification of Body Postures and Movements*, containing 165633 data points.
#
# Within the dataset, there are five target activities:
# - Sitting
# - Sitting Down
# - Standing
# - Standing Up
# - Walking
#
# These activities were captured from 30 people wearing accelerometers mounted on their waist, left thigh, right arm, and right ankle.
# ## Read the data
# The original dataset can be found on the [UCI MachineLearning Repository](https://archive.ics.uci.edu/ml/datasets/human+activity+recognition+using+smartphones)
#
# A copy can be found also here on GitHub (URL is below) and on [Kaggle](https://www.kaggle.com/uciml/human-activity-recognition-with-smartphones)
import pandas as pd
import time
# +
# Grab the DLA HAR dataset from the links above
# we assume that is stored in a dataset folder
#
# Load up the dataset into dataframe 'X'
#
X = pd.read_csv("../datasets/dataset-har-PUC-rio-ugulino.csv", sep=';', low_memory=False)
# -
X.head(2)
X.describe()
# ## Pre-processing the data
#
# What we want to do is to predict the activity class based on the accelerometer's data from the wearables.
#
# An easy way to show which rows have NaNs in them:
print (X[pd.isnull(X).any(axis=1)])
# Great, no NaNs here. Let's go on.
# +
#
# Encode the gender column: 0 as male, 1 as female
#
X.gender = X.gender.map({'Woman':1, 'Man':0})
#
# Clean up any column with commas in it
# so that they're properly represented as decimals instead
#
X.how_tall_in_meters = X.how_tall_in_meters.str.replace(',','.').astype(float)
X.body_mass_index = X.body_mass_index.str.replace(',','.').astype(float)
#
# Check data types
print (X.dtypes)
# +
# column z4 is type "object". Something is wrong with the dataset.
#
# Convert that column into numeric
# Use errors='raise'. This will alert you if something ends up being
# problematic
#
#
# INFO: There is an error raised ... you will find it if you try the method
#
# print (X[pd.isnull(X).any(axis=1)])
# 122076 --> z4 = -14420-11-2011 04:50:23.713
#
# !! The data point #122076 is a wrong coded record,
# change it or drop it before calling the to_numeric methods:
#
#X.at[122076, 'z4'] = -144 // change to correct value
# I keep this value for later and drop it from the dataset
wrongRow = X.loc[122076]
X.drop(X.index[[122076]], inplace=True)
X.z4 = pd.to_numeric(X.z4, errors='raise')
print (X.dtypes)
# everything ok now
# -
# ## Extract the target values
# +
# Activity to predict is in "class" column
# Encode 'y' value as a dummies version of dataset's "class" column
#
y = pd.get_dummies(X['class'].copy())
# this produces a 5 column wide dummies dataframe as the y value
#
# Get rid of the user and class columns in X
#
X.drop(['class','user'], axis=1, inplace=True)
print (X.head(2))
# -
print (y.head())
# ## Split the dataset into training and test
# +
#
# Split data into test / train sets
#
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3,
random_state=7)
# -
# ## Train the Random Forest model
# +
#
# Create an RForest classifier 'model'
#
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=30, max_depth= 20, random_state=0)
# -
# You can check the [SKlearn documentation](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html) to see all possible parameters.
#
# The ones used here:
#
# - n_estimators: integer, optional (default=100)
# The number of trees in the forest. Note that this number changed from 10 to 100 (following the progress in computing performance and memory)
# - max_depth: integer or None, optional (default=None)
# The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples.
# Setting a limit helps with the computing time and memory needed.Not setting a max depth will lead to have unpruned and fully grown trees which - depending on the dataset - will require large memory footprint.
# - oob_score: bool (default=False)
# Whether to use out-of-bag samples to estimate the generalization accuracy.
# - random_state: int, RandomState instance or None, optional (default=None)
# Controls both the randomness of the bootstrapping of the samples used when building trees (if bootstrap=True) and the sampling of the features to consider
#
# And other useful / important:
#
# - criterion: string, optional (default=”gini”)
# The function to measure the quality of a split. Supported criteria are “gini” for the Gini impurity and “entropy” for the information gain.
# Same as for the Trees.
# - bootstrap: boolean, optional (default=True)
# Whether bootstrap samples are used when building trees. If False, the whole datset is used to build each tree.
# +
print ("Fitting...")
s = time.time()
model.fit(X_train, y_train)
print("completed in: ", time.time() - s, "seconds")
# -
# Note that it takes a much longer time to train a forest than a single decision tree.
#
# This is the score based on the test dataset that we split earlier. Note how good it is.
# +
print ("Scoring...")
s = time.time()
score = model.score(X_test, y_test)
print ("Score: ", round(score*100, 3))
print ("Scoring completed in: ", time.time() - s)
# -
# These are the top 5 features used in the classification.
# They are all related to the movements, no gender or age.
# +
# Extract feature importances
fi = pd.DataFrame({'feature': list(X_train.columns),
'importance': model.feature_importances_}).\
sort_values('importance', ascending = False)
# Display
fi.head()
# -
# ## Example prediction
#
# Let's use the wrong row - that we extracted earlier from the dataset - as a prediction example.
# but first we need to correct it:
# +
outputClassPredictionExample = wrongRow['class']
forPredictionExample = wrongRow.drop(labels=['class','user']) # remove class and user
forPredictionExample.z4 = -144 # correct the value
print("We use this example for prediction later:")
print(forPredictionExample)
print("The class shall be: ", outputClassPredictionExample)
# -
model.predict(forPredictionExample.values.reshape(1, -1))
# Remember that these were the categories for the classes:
y_test.iloc[0]
# The fourth one is "standing up". Seems that the model predicted correctly.
# ## OutOfBag error instead of splitting into train and test
# Since each tree within the forest is only trained using a subset of the overall training set, the forest ensemble has the ability to error test itself.
# It does this by scoring each tree's predictions against that tree's out-of-bag samples. A tree's out of bag samples are those forest training samples that were withheld from a specific tree during training.
#
# One of the advantages of using the out of bag (OOB) error is that eliminates the need to split your data into a training / testing before feeding it into the forest model, since that's part of the forest algorithm. However using the OOB error metric often underestimates the actual performance improvement and the optimal number of training iterations.
modelOOB = RandomForestClassifier(n_estimators=30, max_depth= 20, random_state=0,
oob_score=True)
# +
print ("Fitting...")
s = time.time()
modelOOB.fit(X, y)
print("completed in: ", time.time() - s, "seconds")
# -
# Time needed is similar.
# Let's check the score:
# Display the OOB Score of data
scoreOOB = modelOOB.oob_score_
print ("OOB Score: ", round(scoreOOB*100, 3))
# The out-of-bag estimation is not far away from the more precise score estimated from the test dataset.
#
# And now we predict the same user's movement. Class output shall be "standing up", the fourth one
modelOOB.predict(forPredictionExample.values.reshape(1, -1))
# Yup!
| 02-Classification/forest.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# # Parameter selection, Validation, and Testing
# Most models have parameters that influence how complex a model they can learn. Remember using `KNeighborsRegressor`.
# If we change the number of neighbors we consider, we get a smoother and smoother prediction:
# <img src="figures/plot_kneigbors_regularization.png" width="100%">
# In the above figure, we see fits for three different values of ``n_neighbors``.
# For ``n_neighbors=2``, the data is overfit, the model is too flexible and can adjust too much to the noise in the training data. For ``n_neighbors=20``, the model is not flexible enough, and can not model the variation in the data appropriately.
#
# In the middle, for ``n_neighbors = 5``, we have found a good mid-point. It fits
# the data fairly well, and does not suffer from the overfit or underfit
# problems seen in the figures on either side. What we would like is a
# way to quantitatively identify overfit and underfit, and optimize the
# hyperparameters (in this case, the polynomial degree d) in order to
# determine the best algorithm.
#
# We trade off remembering too much about the particularities and noise of the training data vs. not modeling enough of the variability. This is a trade-off that needs to be made in basically every machine learning application and is a central concept, called bias-variance-tradeoff or "overfitting vs underfitting".
# <img src="figures/overfitting_underfitting_cartoon.svg" width="100%">
#
# ## Hyperparameters, Over-fitting, and Under-fitting
#
# Unfortunately, there is no general rule how to find the sweet spot, and so machine learning practitioners have to find the best trade-off of model-complexity and generalization by trying several hyperparameter settings. Hyperparameters are the internal knobs or tuning parameters of a machine learning algorithm (in contrast to model parameters that the algorithm learns from the training data -- for example, the weight coefficients of a linear regression model); the number of *k* in K-nearest neighbors is such a hyperparameter.
#
# Most commonly this "hyperparameter tuning" is done using a brute force search, for example over multiple values of ``n_neighbors``:
#
# +
from sklearn.model_selection import cross_val_score, KFold
from sklearn.neighbors import KNeighborsRegressor
# generate toy dataset:
x = np.linspace(-3, 3, 100)
rng = np.random.RandomState(42)
y = np.sin(4 * x) + x + rng.normal(size=len(x))
X = x[:, np.newaxis]
cv = KFold(shuffle=True)
# for each parameter setting do cross-validation:
for n_neighbors in [1, 3, 5, 10, 20]:
scores = cross_val_score(KNeighborsRegressor(n_neighbors=n_neighbors), X, y, cv=cv)
print("n_neighbors: %d, average score: %f" % (n_neighbors, np.mean(scores)))
# +
rng = np.random.RandomState(seed=1234)
x = np.linspace(-3,3,100)
y = np.sin(4*x) + x + rng.normal(size=len(x))
cv = KFold(shuffle=True)
for n in [1,3,5,10]:
clf = KNeighborsRegressor(n)
score = cross_val_score(clf,X,y,cv=cv)
print("for neigh: %d score %f"%(n,np.mean(score)))
# -
# There is a function in scikit-learn, called ``validation_plot`` to reproduce the cartoon figure above. It plots one parameter, such as the number of neighbors, against training and validation error (using cross-validation):
from sklearn.model_selection import validation_curve
n_neighbors = [1, 3, 5, 10, 20, 50]
train_scores, test_scores = validation_curve(KNeighborsRegressor(), X, y, param_name="n_neighbors",
param_range=n_neighbors, cv=cv)
plt.plot(n_neighbors, train_scores.mean(axis=1), 'b', label="train accuracy")
plt.plot(n_neighbors, test_scores.mean(axis=1), 'g', label="test accuracy")
plt.ylabel('Accuracy')
plt.xlabel('Number of neighbors')
plt.xlim([50, 0])
plt.legend(loc="best");
# <div class="alert alert-warning">
# Note that many neighbors mean a "smooth" or "simple" model, so the plot uses a reverted x axis.
# </div>
# If multiple parameters are important, like the parameters ``C`` and ``gamma`` in an ``SVM`` (more about that later), all possible combinations are tried:
# +
from sklearn.model_selection import cross_val_score, KFold
from sklearn.svm import SVR
# each parameter setting do cross-validation:
for C in [0.001, 0.01, 0.1, 1, 10]:
for gamma in [0.001, 0.01, 0.1, 1]:
scores = cross_val_score(SVR(C=C, gamma=gamma), X, y, cv=cv)
print("C: %f, gamma: %f, average score: %f" % (C, gamma, np.mean(scores)))
# -
# As this is such a very common pattern, there is a built-in class for this in scikit-learn, ``GridSearchCV``. ``GridSearchCV`` takes a dictionary that describes the parameters that should be tried and a model to train.
#
# The grid of parameters is defined as a dictionary, where the keys are the parameters and the values are the settings to be tested.
# +
from sklearn.model_selection import GridSearchCV
param_grid = {'C': [0.001, 0.01, 0.1, 1, 10], 'gamma': [0.001, 0.01, 0.1, 1]}
grid = GridSearchCV(SVR(), param_grid=param_grid, cv=cv,verbose=3)
# -
# One of the great things about GridSearchCV is that it is a *meta-estimator*. It takes an estimator like SVR above, and creates a new estimator, that behaves exactly the same - in this case, like a regressor.
# So we can call ``fit`` on it, to train it:
grid.fit(X, y)
# What ``fit`` does is a bit more involved then what we did above. First, it runs the same loop with cross-validation, to find the best parameter combination.
# Once it has the best combination, it runs fit again on all data passed to fit (without cross-validation), to built a single new model using the best parameter setting.
# Then, as with all models, we can use ``predict`` or ``score``:
#
grid.predict(X)
# You can inspect the best parameters found by ``GridSearchCV`` in the ``best_params_`` attribute, and the best score in the ``best_score_`` attribute:
print(grid.best_score_)
print(grid.best_params_)
# But you can investigate the performance and much more for each set of parameter values by accessing the `cv_results_` attributes. The `cv_results_` attribute is a dictionary where each key is a string and each value is array. It can therefore be used to make a pandas DataFrame.
type(grid.cv_results_)
print(grid.cv_results_.keys())
# +
import pandas as pd
cv_results = pd.DataFrame(grid.cv_results_)
cv_results.head()
# -
cv_results_tiny = cv_results[['param_C', 'param_gamma', 'mean_test_score']]
cv_results_tiny.sort_values(by='mean_test_score', ascending=False).head()
# There is a problem with using this score for evaluation, however. You might be making what is called a multiple hypothesis testing error. If you try very many parameter settings, some of them will work better just by chance, and the score that you obtained might not reflect how your model would perform on new unseen data.
# Therefore, it is good to split off a separate test-set before performing grid-search. This pattern can be seen as a training-validation-test split, and is common in machine learning:
# <img src="figures/grid_search_cross_validation.svg" width="100%">
# We can do this very easily by splitting of some test data using ``train_test_split``, training ``GridSearchCV`` on the training set, and applying the ``score`` method to the test set:
# +
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
param_grid = {'C': [0.001, 0.01, 0.1, 1, 10], 'gamma': [0.001, 0.01, 0.1, 1]}
cv = KFold(n_splits=10, shuffle=True)
grid = GridSearchCV(SVR(), param_grid=param_grid, cv=cv)
grid.fit(X_train, y_train)
grid.score(X_test, y_test)
# -
# We can also look at the parameters that were selected:
grid.best_params_
# Some practitioners go for an easier scheme, splitting the data simply into three parts, training, validation and testing. This is a possible alternative if your training set is very large, or it is infeasible to train many models using cross-validation because training a model takes very long.
# You can do this with scikit-learn for example by splitting of a test-set and then applying GridSearchCV with ShuffleSplit cross-validation with a single iteration:
#
# <img src="figures/train_validation_test2.svg" width="100%">
# +
from sklearn.model_selection import train_test_split, ShuffleSplit
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
param_grid = {'C': [0.001, 0.01, 0.1, 1, 10], 'gamma': [0.001, 0.01, 0.1, 1]}
single_split_cv = ShuffleSplit(n_splits=1)
grid = GridSearchCV(SVR(), param_grid=param_grid, cv=single_split_cv, verbose=3)
grid.fit(X_train, y_train)
grid.score(X_test, y_test)
# -
# This is much faster, but might result in worse hyperparameters and therefore worse results.
clf = GridSearchCV(SVR(), param_grid=param_grid)
clf.fit(X_train, y_train)
clf.score(X_test, y_test)
# <div class="alert alert-success">
# <b>EXERCISE</b>:
# <ul>
# <li>
# Apply grid-search to find the best setting for the number of neighbors in ``KNeighborsClassifier``, and apply it to the digits dataset.
# </li>
# </ul>
# </div>
# +
from sklearn.neighbors import KNeighborsClassifier
from sklearn.grid_search import GridSearchCV
from sklearn.datasets import load_digits
digits = load_digits()
X,y = digits.data, digits.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1234)
# -
params = {'n_neighbors':[1,2,5,8,12,20]}
clf = GridSearchCV(KNeighborsClassifier(),params,cv=5,verbose=3)
clf.fit(X_train,y_train)
print(clf.best_params_, clf.best_score_)
# +
# # %load solutions/14_grid_search.py
from sklearn.datasets import load_digits
from sklearn.neighbors import KNeighborsClassifier
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target, random_state=0)
param_grid = {'n_neighbors': [1, 3, 5, 10, 50]}
gs = GridSearchCV(KNeighborsClassifier(), param_grid=param_grid, cv=5, verbose=3)
gs.fit(X_train, y_train)
print("Score on test set: %f" % gs.score(X_test, y_test))
print("Best parameters: %s" % gs.best_params_)
# -
print(gs.best_params_, gs.best_score_)
| notebooks/14.Model_Complexity_and_GridSearchCV.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="yitlha1UimxW"
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
# + [markdown] id="dDjOKyerisZe"
# ### Building a dataset
# + colab={"base_uri": "https://localhost:8080/", "height": 296} id="3FKdszaVirdC" outputId="eaa3073f-b85a-4186-f755-0a1888bc2090"
Sx = np.array([0, 1, 2.5, 3, 4, 5])
Sy = np.array([0.6, 0, 2, 2.2, 4.7, 5])
# Plotting in graph
plt.scatter(Sx, Sy)
# Graph axis names and grids
plt.grid(True)
plt.xlabel('Sx')
plt.ylabel('Sy')
# + [markdown] id="HyQCP2XokTqq"
# Lets assume a line
#
# $$y = mx + c$$
#
# Where $m$ and $c$ are unknown, which we are trying to find.
#
# We assume a random value for $m$ and $c$ ($m = 2$ and $c = 2$)
# + id="TXXK2p4Gi9-0"
m = tf.Variable(2, dtype=tf.float32)
c = tf.Variable(2, dtype=tf.float32)
def line_fn(x):
return m*x + c
# + colab={"base_uri": "https://localhost:8080/", "height": 296} id="fOmpTrMpoTPV" outputId="650ae5a6-123c-4a17-ca47-0747d46f6c50"
p = np.arange(0, 5, 0.1)
plt.plot(p, line_fn(p).numpy())
# Plotting in graph
plt.scatter(Sx, Sy)
# Graph axis names and grids
plt.grid(True)
plt.xlabel('Sx')
plt.ylabel('Sy')
# + [markdown] id="uIYUfoD88l3F"
# ## Gradient descending algorithm:
# $$m_{t} = m_{t-1} - lr \; \frac{\partial \;\; loss(l(x), y)}{\partial m} $$
#
# $$loss(l(x), y) = (l(x) - y)^2$$
#
# #### Here,
#
# * $t$ = Time step
# * $x$ = Input
# * $y$ = Output
# * $m$ = Updatable variable
# * $loss(\cdot, \cdot)$ = Loss function
# * $lr$ = Learning rate
# * $l(\cdot)$ = Line function
# + [markdown] id="c5xNkNpTAVm0"
# #### Partial derivatives:
#
# $\frac{\partial \;\; loss(l(x), y)}{\partial m} = (l(x) - y)^2$
# $ = (mx+c-y)^2$
# $ = 2(mx+c-y)x$
#
# $\frac{\partial \;\; loss(l(x), y)}{\partial c} = (l(x) - y)^2$
# $ = (mx+c-y)^2$
# $ = 2(mx+c-y)$
# + colab={"base_uri": "https://localhost:8080/"} id="n3HPcI68kPXS" outputId="86c6a274-fc0b-4aeb-cd04-a92c75b2ff6a"
# learning rate
lr = 0.01
total_steps = 100
for step in range(total_steps):
print(f"Step {step+1:2}:")
print("-"*30)
with tf.GradientTape() as tape:
# Printing value of the variables
print(f"M: {m.numpy():.4f}, C: {c.numpy():.4f}")
# Stating what variables need to be partially differentiated and calibrated
tape.watch([m, c])
# Passing the points to the line function
pred_y = line_fn(Sx)
# Calculating the difference/loss of the output (pred_y) of the function
# w.r.t. the known output (Sy)
loss = (pred_y - Sy) * (pred_y - Sy)
# Calculating the gradients w.r.t. the partially diff. parameters
# and the generated output loss
grads = tape.gradient(loss, [m, c])
# Showing the output just for educational purposs
print(f"M_grad:, {grads[0].numpy():.2f}, C_grad: {grads[1].numpy():.2f}")
# Updating the gradients
m = m - lr * grads[0]
c = c - lr * grads[1]
print()
# + [markdown] id="TxHoUb4VBARF"
# ## Lets check the final result
# + colab={"base_uri": "https://localhost:8080/", "height": 296} id="rh5HNbytmd6W" outputId="df383f61-eba2-4a76-ab0e-4e029036e00b"
p = np.arange(0, 5, 0.1)
plt.plot(p, line_fn(p).numpy())
# Plotting in graph
plt.scatter(Sx, Sy)
# Graph axis names and grids
plt.grid(True)
plt.xlabel('Sx')
plt.ylabel('Sy')
# + id="HDsw4mslo2CC"
| Day 1/LineAssumption.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python
# language: python
# name: conda-env-python-py
# ---
# <a href="https://www.bigdatauniversity.com"><img src="https://ibm.box.com/shared/static/cw2c7r3o20w9zn8gkecaeyjhgw3xdgbj.png" width="400" align="center"></a>
#
# <h1 align=center><font size="5"> SVM (Support Vector Machines)</font></h1>
# In this notebook, you will use SVM (Support Vector Machines) to build and train a model using human cell records, and classify cells to whether the samples are benign or malignant.
#
# SVM works by mapping data to a high-dimensional feature space so that data points can be categorized, even when the data are not otherwise linearly separable. A separator between the categories is found, then the data is transformed in such a way that the separator could be drawn as a hyperplane. Following this, characteristics of new data can be used to predict the group to which a new record should belong.
# <h1>Table of contents</h1>
#
# <div class="alert alert-block alert-info" style="margin-top: 20px">
# <ol>
# <li><a href="#load_dataset">Load the Cancer data</a></li>
# <li><a href="#modeling">Modeling</a></li>
# <li><a href="#evaluation">Evaluation</a></li>
# <li><a href="#practice">Practice</a></li>
# </ol>
# </div>
# <br>
# <hr>
import pandas as pd
import pylab as pl
import numpy as np
import scipy.optimize as opt
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
# %matplotlib inline
import matplotlib.pyplot as plt
# + [markdown] button=false new_sheet=false run_control={"read_only": false}
# <h2 id="load_dataset">Load the Cancer data</h2>
# The example is based on a dataset that is publicly available from the UCI Machine Learning Repository (Asuncion and Newman, 2007)[http://mlearn.ics.uci.edu/MLRepository.html]. The dataset consists of several hundred human cell sample records, each of which contains the values of a set of cell characteristics. The fields in each record are:
#
# |Field name|Description|
# |--- |--- |
# |ID|Clump thickness|
# |Clump|Clump thickness|
# |UnifSize|Uniformity of cell size|
# |UnifShape|Uniformity of cell shape|
# |MargAdh|Marginal adhesion|
# |SingEpiSize|Single epithelial cell size|
# |BareNuc|Bare nuclei|
# |BlandChrom|Bland chromatin|
# |NormNucl|Normal nucleoli|
# |Mit|Mitoses|
# |Class|Benign or malignant|
#
# <br>
# <br>
#
# For the purposes of this example, we're using a dataset that has a relatively small number of predictors in each record. To download the data, we will use `!wget` to download it from IBM Object Storage.
# __Did you know?__ When it comes to Machine Learning, you will likely be working with large datasets. As a business, where can you host your data? IBM is offering a unique opportunity for businesses, with 10 Tb of IBM Cloud Object Storage: [Sign up now for free](http://cocl.us/ML0101EN-IBM-Offer-CC)
# + button=false new_sheet=false run_control={"read_only": false}
#Click here and press Shift+Enter
# !wget -O cell_samples.csv https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/ML0101ENv3/labs/cell_samples.csv
# + [markdown] button=false new_sheet=false run_control={"read_only": false}
# ### Load Data From CSV File
# + button=false new_sheet=false run_control={"read_only": false}
cell_df = pd.read_csv("cell_samples.csv")
cell_df.head()
# -
# The ID field contains the patient identifiers. The characteristics of the cell samples from each patient are contained in fields Clump to Mit. The values are graded from 1 to 10, with 1 being the closest to benign.
#
# The Class field contains the diagnosis, as confirmed by separate medical procedures, as to whether the samples are benign (value = 2) or malignant (value = 4).
#
# Lets look at the distribution of the classes based on Clump thickness and Uniformity of cell size:
ax = cell_df[cell_df['Class'] == 4][0:50].plot(kind='scatter', x='Clump', y='UnifSize', color='DarkBlue', label='malignant');
cell_df[cell_df['Class'] == 2][0:50].plot(kind='scatter', x='Clump', y='UnifSize', color='Yellow', label='benign', ax=ax);
plt.show()
# ## Data pre-processing and selection
# Lets first look at columns data types:
cell_df.dtypes
# It looks like the __BareNuc__ column includes some values that are not numerical. We can drop those rows:
cell_df = cell_df[pd.to_numeric(cell_df['BareNuc'], errors='coerce').notnull()]
cell_df['BareNuc'] = cell_df['BareNuc'].astype('int')
cell_df.dtypes
feature_df = cell_df[['Clump', 'UnifSize', 'UnifShape', 'MargAdh', 'SingEpiSize', 'BareNuc', 'BlandChrom', 'NormNucl', 'Mit']]
X = np.asarray(feature_df)
X[0:5]
# We want the model to predict the value of Class (that is, benign (=2) or malignant (=4)). As this field can have one of only two possible values, we need to change its measurement level to reflect this.
cell_df['Class'] = cell_df['Class'].astype('int')
y = np.asarray(cell_df['Class'])
y [0:5]
# ## Train/Test dataset
# Okay, we split our dataset into train and test set:
X_train, X_test, y_train, y_test = train_test_split( X, y, test_size=0.2, random_state=4)
print ('Train set:', X_train.shape, y_train.shape)
print ('Test set:', X_test.shape, y_test.shape)
# <h2 id="modeling">Modeling (SVM with Scikit-learn)</h2>
# The SVM algorithm offers a choice of kernel functions for performing its processing. Basically, mapping data into a higher dimensional space is called kernelling. The mathematical function used for the transformation is known as the kernel function, and can be of different types, such as:
#
# 1.Linear
# 2.Polynomial
# 3.Radial basis function (RBF)
# 4.Sigmoid
# Each of these functions has its characteristics, its pros and cons, and its equation, but as there's no easy way of knowing which function performs best with any given dataset, we usually choose different functions in turn and compare the results. Let's just use the default, RBF (Radial Basis Function) for this lab.
from sklearn import svm
clf = svm.SVC(kernel='rbf')
clf.fit(X_train, y_train)
# After being fitted, the model can then be used to predict new values:
yhat = clf.predict(X_test)
yhat [0:5]
# <h2 id="evaluation">Evaluation</h2>
from sklearn.metrics import classification_report, confusion_matrix
import itertools
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print('Confusion matrix, without normalization')
print(cm)
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
fmt = '.2f' if normalize else 'd'
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, format(cm[i, j], fmt),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
# +
# Compute confusion matrix
cnf_matrix = confusion_matrix(y_test, yhat, labels=[2,4])
np.set_printoptions(precision=2)
print (classification_report(y_test, yhat))
# Plot non-normalized confusion matrix
plt.figure()
plot_confusion_matrix(cnf_matrix, classes=['Benign(2)','Malignant(4)'],normalize= False, title='Confusion matrix')
# -
# You can also easily use the __f1_score__ from sklearn library:
from sklearn.metrics import f1_score
f1_score(y_test, yhat, average='weighted')
# Lets try jaccard index for accuracy:
from sklearn.metrics import jaccard_similarity_score
jaccard_similarity_score(y_test, yhat)
# <h2 id="practice">Practice</h2>
# Can you rebuild the model, but this time with a __linear__ kernel? You can use __kernel='linear'__ option, when you define the svm. How the accuracy changes with the new kernel function?
# write your code here
# Double-click __here__ for the solution.
#
# <!-- Your answer is below:
#
# clf2 = svm.SVC(kernel='linear')
# clf2.fit(X_train, y_train)
# yhat2 = clf2.predict(X_test)
# print("Avg F1-score: %.4f" % f1_score(y_test, yhat2, average='weighted'))
# print("Jaccard score: %.4f" % jaccard_similarity_score(y_test, yhat2))
#
# -->
# + [markdown] button=false new_sheet=false run_control={"read_only": false}
# <h2>Want to learn more?</h2>
#
# IBM SPSS Modeler is a comprehensive analytics platform that has many machine learning algorithms. It has been designed to bring predictive intelligence to decisions made by individuals, by groups, by systems – by your enterprise as a whole. A free trial is available through this course, available here: <a href="http://cocl.us/ML0101EN-SPSSModeler">SPSS Modeler</a>
#
# Also, you can use Watson Studio to run these notebooks faster with bigger datasets. Watson Studio is IBM's leading cloud solution for data scientists, built by data scientists. With Jupyter notebooks, RStudio, Apache Spark and popular libraries pre-packaged in the cloud, Watson Studio enables data scientists to collaborate on their projects without having to install anything. Join the fast-growing community of Watson Studio users today with a free account at <a href="https://cocl.us/ML0101EN_DSX">Watson Studio</a>
#
# <h3>Thanks for completing this lesson!</h3>
#
# <h4>Author: <a href="https://ca.linkedin.com/in/saeedaghabozorgi"><NAME></a></h4>
# <p><a href="https://ca.linkedin.com/in/saeedaghabozorgi"><NAME></a>, PhD is a Data Scientist in IBM with a track record of developing enterprise level applications that substantially increases clients’ ability to turn data into actionable knowledge. He is a researcher in data mining field and expert in developing advanced analytic methods like machine learning and statistical modelling on large datasets.</p>
#
# <hr>
#
# <p>Copyright © 2018 <a href="https://cocl.us/DX0108EN_CC">Cognitive Class</a>. This notebook and its source code are released under the terms of the <a href="https://bigdatauniversity.com/mit-license/">MIT License</a>.</p>
| SVM-cancer.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Introduction
# Loading files into FSLeyes and adjusting settings manually is cumbersom and error prone. Luckily, which FSLeyes it is possible to script the display of files, which saves time and makes the result figures more reproducible.
#
# There are different ways how we can interact with FSLeyes programatically:
# * We can open a Python shell in the FSLeyes GUI from `View` > `Python shell`
# * We can open FSLeyes from the command line and run a script at the same time: `fsleyes -r myscript.py`
# * for an interactive mode, one can also use a Jupyter Notebook `fsleyes --notebookFile my_notebook.ipynb`
#
# Before we continue, here two links with the relevant User Guides:
# * [FSLeyes documentation](https://users.fmrib.ox.ac.uk/~paulmc/fsleyes/userdoc/latest/index.html)
# * [FSLeyes Python API documentation](https://users.fmrib.ox.ac.uk/~paulmc/fsleyes/apidoc/latest/)
#
# Please note that I put together this tutorial based on my own understanding of the software and this might or not be the best way to use it. If in doubt, please refer to these User Guides or contact the FSLeyes developers to get advice.
# # Installation
# If you installed FSLeyes as part of FSL (prior to version 6), you have a standalone version. If you want to use it via the interactive mode, however, it is recommended to use it as Python package. From FSL version 6 onwards, this is automatically included (I think...).
#
# Here are the steps to install FSLeyes for this tutorial:
# * Download and install miniconda (https://docs.conda.io/en/latest/miniconda.html)
#
# * In your terminal create a new environment:
#
# `$ conda create -n fsleyes_tutorial python=3.7`
#
# * When the environment is installed, activate it:
#
# `$ conda activate fsleyes_tutorial`
#
#
# * Install the FSLeyes Python package to the new environment:
#
# `$ conda install -c conda-forge fsleyes`
#
#
# * Check that the path to FSLeyes is inside your environment
#
# `$ which fsleyes`
#
#
# # Start tutorial
#
# If you haven't done so already, load the fsleyes_tutorial conda environment (see above)
# Then navigate to the retreat folder:
#
# `cd ~/myPath/tutorial`
#
# Launch the tutorial notebook together with FSLeyes:
#
# `fsleyes --notebookFile scripts/fsleyes_tutorial.ipynb`
# # Basic example
# As a first example to see how the interactive mode is working, we will load the human standard MNI brain using the command below. You might notice that you don't need to import the 'load' function, because the some useful packages are already impored when FSLeyes is launched. You might also notice that that the variable `FSLDIR` is acessible within FSLeyes, but only if it was defined within the terminal session, where we launched FSLeyes.
import os
load(os.path.expandvars('$FSLDIR/data/standard/MNI152_T1_2mm'))
# All files that we load are stored automatically in a list called `overlayList`, which holds all the Image objects. We can access the first element, which is the MNI brain, using regular indexing:
# first element of list:
overlayList[0]
# As a simple manipulation we can change the colour map from greyscale to `Render3`. Note that the default colour maps are directly accessible in this way, but we will other custom colour maps can be included (see more below).
displayCtx.getOpts(overlayList[0]).cmap = 'Render3'
# We can remove this file again, because we don't need it for now:
overlayList.remove(overlayList[0])
# # Goal for this session
# The goal of this session is to have a script that automatically creates the display of a structural brain scan together with tractography results from two tracts. The cursor will be centered on the voxel of maximal probability of one tract. The script will contain one variable to define, which subject group we want to display (for example controls, patient group 1, patient group 2, etc. Changing this variable change all the settings adaptively so that a comparable display is created.
# # Input files and settings
# We need to provide the filenames of interest in an organized way, where Pandas data frames can be handy.
# In the tutorial example the filenames have very convenient names, and are organized in a neat way, which will most likely not be the case in a real-life example
# +
import pandas as pd
# folder where data is stored
mydir = os.path.join('mypath', 'tutorial', 'data')
# filenames
df = pd.DataFrame(columns=['subject_group', 'structural', 'CST', 'MDLF'])
df.loc[len(df)] = ['control', 'structural', 'cst', 'mdlf']
df.loc[len(df)] = ['patient-group1', 'structural', 'cst', 'mdlf']
df.loc[len(df)] = ['patient-group2', 'structural', 'cst', 'mdlf']
# -
# Here we define the variable for 'subject-group':
subject_group = 'control'
# In the following lines we define two different colours for the two tracts that we will display. We will later access the correct colour based on the index of the tract. In a similar way, we could define settings that differ for the three subject groups.
# +
# colour for the two tracts
my_colours = np.array([[0. , 0.6 , 1. ], # blue
[1. , 0.33, 0.68]]) # pink
# -
# The tractography results that we load have been normalized with intensities franging from 0 to 1. Therefore, we can apply a comparable threshold of 0.2 to both tracts. In a different situation we might want to have this setting variable depending on tract type or subject group.
# display range for thresholding the tracts
display_range = (0.2, 1)
# # Generate Display
# +
# import package for colour map (should be at top of script)
from matplotlib.colors import LinearSegmentedColormap
# make sure all previous overlays are removed
overlayList.clear()
# load structural
structural_fname = f'{df[df.subject_group == subject_group].structural.values[0]}.nii.gz'
load(os.path.join(os.sep, mydir, structural_fname))
# load tractograms
# loop over hemispheres
for hemi in ['l', 'r']:
for i_t, tract in enumerate(['CST', 'MDLF']):
tract_fname = os.path.join(os.sep, mydir, f'{df[df.subject_group == subject_group][tract].values[0]}_{hemi}.nii.gz')
load(os.path.join(os.sep, mydir, tract_fname))
# set display range and clipping range
displayCtx.getOpts(overlayList[-1]).clippingRange = display_range
displayCtx.getOpts(overlayList[-1]).displayRange = display_range
# set colour map specific for tract type
# use a colour map where luminance linearly increases from black to white
displayCtx.getOpts(overlayList[-1]).cmap = LinearSegmentedColormap.from_list('mycmap', ['black', my_colours[i_t], 'white'])
# determine max voxel for MDLF tractogram in left hemisphere
if (hemi == 'l') & (tract == 'MDLF'):
max_voxel = np.unravel_index(np.argmax(overlayList[-1].data, axis=None), overlayList[-1].data.shape)
# place cross hair on maximal voxel for MDLF_L
displayCtx = frame.viewPanels[0].displayCtx
displayCtx.location = displayCtx.getOpts(overlayList[-1]).transformCoords(max_voxel, 'voxel', 'display')
# -
# # Importing Atlases
# It is possible to include custom atlases to FSLeyes and they will be included in the Atlas panel in the GUI.
# Note that any custom atlas files must be described by an XML specification file as outlined [here](https://users.fmrib.ox.ac.uk/~paulmc/fsleyes/userdoc/latest/customising.html#atlases).
#
# It's just a single line of code:
# + pycharm={"name": "#%%\n"}
import fsl
fsl.data.atlases.addAtlas('/myPath/tutorial/myatlas.xml')
| data_visualization/fsleyes_tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Kernel Shape Example
# Spring 2019 AME-70790 Final Project
#
# <NAME> (<EMAIL>)
#
# Reference: <NAME>., & <NAME>. (1994). Kernel smoothing. Chapman and Hall/CRC.
# ___
# In the previous example we looked at how the bandwidth of a kernel in significantly influence the kernel smoother prediction.
# Now let us look at the impact of the shape of the kernel function $K$.
# Again consider this arbitrary density:
# $$f_{1}(x)=\frac{3}{4}\phi\left(x | 0,1\right) + \frac{1}{4}\phi\left(x | 3/2, 1/3\right),$$
# where $\phi(x)$ is the normal PDF making this a mixture of two Gaussians. We will use 1000 training data points to approximate this density function with the kernel density estimator.
import numpy as np
from scipy.stats import norm
import matplotlib.pyplot as plt
# +
plt.close("all")
np.random.seed(123)
ntrain = 1000
h = 0.25 # band width
# Target data
x_test = np.linspace(-3,3,200)
y_test = 0.75*norm.pdf(x_test, loc=0, scale=1) + 0.75*norm.pdf(x_test, loc=3/2, scale=1/3)
# Training data
c1 = np.sum(np.random.rand(ntrain) < 0.75)
c2 = ntrain - c1
x_train0 = np.concatenate([np.random.randn(c1), (1/3.)*np.random.randn(c1)+1.5], axis=0)
# -
# Here we will consider three different kernels.
# For kernels to be comparable, three constraints are imposed:
# $$\int K(x)dx = 1, \quad \int xK(x)dx=0, \quad \int x^{2}K(x)dx=a^{2}<\infty,$$
# which impose normalization, symmetry and the variance respectively.
# Although kernel symmetry is not required, symmetric kernels are commonly used since they are easier to interpret.
# Its important to note that the variance is **not** always the bandwidth.
# To illustrate this point and the impact of kernel shape on the KDE prediction, we will compare kernels using the same *bandwidth* but different standard deviation versus kernels using the same *standard deviation* (a=h).
#
# The first will be the standard Gaussian kernel we used in the previous example:
# $$f(x,h)=(nh)^{-1}\sum_{i=1}^{n}K\left(\frac{x-x_{i}}{h}\right), \quad K(x)=\frac{1}{\sqrt{2\pi} a}\exp\left\{\frac{-x^{2}}{2 a^{2}}\right\},$$
# where $a$ is the standard deviation.
# The normal kernel is unique in the sense that the bandwidth and standard deviation are the interchangeable.
# +
# Set-up prediction points
x_pred0 = np.linspace(-3, 3, 500)
# Expand array dims and repeat
x_pred = np.expand_dims(x_pred0, axis=1).repeat(x_train0.shape[0], axis=1)
x_train = np.expand_dims(x_train0, axis=0).repeat(x_pred0.shape[0], axis=0)
x0 = x_pred-x_train
normal_pred = []
normal_mass = []
# Compute normal kernel using set bandwidth
x0_scaled = x0/h
y_pred0 = (1/(ntrain*h))*np.sum(norm.pdf(x0_scaled), axis=1)
normal_pred.append(y_pred0)
# Compute normal kernel using set variance
y_pred0 = (1/ntrain)*np.sum(norm.pdf(x0, scale=h), axis=1)
normal_pred.append(y_pred0)
normal_mass.append(norm.pdf(x_pred0/h))
normal_mass.append(norm.pdf(x_pred0, scale=h))
# -
# For the second kernel we will use the *Epanechnikov kernel*, which is given by:
# $$K(x)=\frac{3}{4}\left[\frac{1-x^{2}/(5a^{2})}{a\sqrt{5}}\right]\mathbf{1}_{|x|<a\sqrt{5}},$$
# where $\mathbf{1}_{A}$ is the indicator function and $a$ is its scale parameter.
# The most commonly used standard deviation is $a^2=1/5$ as this clearly simplifies the kernel.
# This kernel is particularly unique in the sense that there are theoretical arguments that the Epanechnikov kernel is the optimal kernel based on data efficiency, although the Gaussian is more frequently used.
# Additionally, we note that both the Gaussian kernel and Epanechnikov kernel are derived from the same family:
# $$K(x,p)=\left[2^{2p+1}B(p+1,p+1)\right]^{-1}\left(1-x^{2}\right)^{p}\mathbf{1}_{|x|<1},$$
# where $B(a,b)$ is the beta function.
# The Gaussian kernel and Epanechnikov kernel can be recovered when $p\rightarrow \infty$ and $p=1$ respectively.
# Additional kernels in this family include the Bi-weight and Tri-weight which are when $p=2$ and $p=3$ respectively.
# +
epan_pred = []
epan_mass = []
# Compute Epanechnikov kernel using set bandwidth (a^2 = 1/5)
x0_scaled = x0/h
y_pred0 = (1/(ntrain*h))*np.sum(0.75*(1-x0_scaled**2)*(np.abs(x0_scaled) < 1), axis=1)
epan_pred.append(y_pred0)
# Compute Epanechnikov kernel using set variance
y_pred0 = (1/ntrain)*np.sum(0.75*((1-x0**2/(5*h**2))/(h*np.sqrt(5)))*(np.abs(x0) < h*np.sqrt(5)), axis=1)
epan_pred.append(y_pred0)
epan_mass.append(0.75*(1-(x_pred0/h)**2)*(np.abs(x_pred0/h) < 1))
epan_mass.append(0.75*((1-x_pred0**2/(5*h**2))/(h*np.sqrt(5)))*(np.abs(x_pred0) < h*np.sqrt(5)))
# -
# The third kernel we will consider is the *triangular kernel* which is defined as:
# $$K(x)=\frac{1}{a\sqrt{6}}\left(1-\frac{|x|}{a\sqrt{6}}\right)\mathbf{1}_{|x|<a\sqrt{6}}.$$
# Similar to the Epanechnikov kernel the most commonly used standard deviation is $a^2 = 1/6$ due to the obvious simplifications.
# These and other kernels can be reference on [Wikipedia](https://en.wikipedia.org/wiki/Kernel_%28statistics%29).
# +
tri_pred = []
tri_mass = []
# Compute Triangular kernel using set bandwidth (a^2 = 1/6)
x0_scaled = x0/h
y_pred0 = (1/(ntrain*h))*np.sum((1-np.abs(x0_scaled)) * (np.abs(x0_scaled) < 1), axis=1)
tri_pred.append(y_pred0)
# Compute Triangular kernel using set variance
h0 = h*np.sqrt(6)
y_pred0 = (1/ntrain)*np.sum((1-np.abs(x0)/h0)/h0 * (np.abs(x0) < h0), axis=1)
tri_pred.append(y_pred0)
tri_mass.append((1-np.abs(x_pred0/h)) * (np.abs(x_pred0/h) < 1))
tri_mass.append((1-np.abs(x_pred0)/h0)/h0 * (np.abs(x_pred0) < h0))
# +
fig = plt.figure(figsize=(15,10))
ax = []
ax.append(plt.subplot2grid((2, 2), (0, 0)))
ax.append(plt.subplot2grid((2, 2), (0, 1)))
ax.append(plt.subplot2grid((2, 2), (1, 0)))
ax.append(plt.subplot2grid((2, 2), (1, 1)))
for i in range(2):
# Normal prediction
ax[i].plot(x_pred0, normal_pred[i], '--', c='r', label='Normal Kernel')
# Epanechnikov prediction
ax[i].plot(x_pred0, epan_pred[i], '--', c='b', label='Epanechnikov Kernel')
# Triangular prediction
ax[i].plot(x_pred0, tri_pred[i], '--', c='g', label='Triangular Kernel')
# Target density
ax[i].plot(x_test, y_test, c='k', label='Target')
ax[i].set_xlabel('x')
ax[i].set_ylabel('Density')
# Kernel Mass
ax[i + 2].plot(x_pred0, normal_mass[i], c='r')
ax[i + 2].plot(x_pred0, epan_mass[i], c='b')
ax[i + 2].plot(x_pred0, tri_mass[i], c='g')
ax[i + 2].set_xlim([-1,1])
ax[i + 2].set_title('Kernel Mass')
ax[i + 2].set_xlabel('x')
ax[i + 2].set_ylabel('Density')
ax[0].set_title('KDE same bandwidth')
ax[1].set_title('KDE same variance')
ax[0].legend(loc=2)
# Save and show figure
plt.savefig('figs/02_kernel_shape.pdf')
plt.savefig('figs/02_kernel_shape.png')
plt.show()
# -
# (Left to right) The KDE using kernels with the same bandwidth but different variance and the KDE using kernels with the same variance. (Top to bottom) The KDE and the kernel mass. We can see that when the kernels have roughly the shape (or standard deviation) the KDE is approximately the same. Thus bandwidth **and** kernel shape have strong influence over the density estimates.
| 02_kernel_shape.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Sensitivity: Algebraic explanation
# ## Changes in the coefficients of the objective function
# Let us start with the production Mix example from class:
# The final Tableau is:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|---------|---------|---------|---------|---------|------|-------|
# | - | 1 | 0 | 0 | 0 | 250/3 | 650/3 | 6350 | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 | 1 | 12 | - |
#
# Let us assume we want to make a change in coefficient $c_{1}$ of $\Delta c_{1}$. The objective function becomes:
#
# $max Z = (300 +\Delta c_{1})·x_{1} + 250x_{2}$
#
# It can be proved that the final Tableau now becomes:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|-----------------|---------|---------|---------|---------|------|-------|
# | - | 1 | $-\Delta c_{1}$ | 0 | 0 | 250/3 | 650/3 | 6350 | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 | 1 | 12 | - |
#
# The Simplex method requires that the Tableau is in canonical form. Thus we need to add the last row times $\Delta c_{1}$ to the first row to fulfill this:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|---------|---------|---------|---------|------------------------|------|-------|
# | - | 1 | 0 | 0 | 0 | 250/3 | 650/3 + $\Delta c_{1}$ | 6350 + 12·$\Delta c_{1}$ | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 | 1 | 12 | - |
# And taking into account that the coefficient of $s_{3}$ needs to be positive for $x_{1}$ to remain in the basis, $\Delta c_{1}$ cannot be lower than -650/3 and can increase up to infinity.
# Let us now assume we want to make a change in coefficient $c_{2}$ of $\Delta c_{2}$. The objective function becomes:
#
# $max Z = 300·x_{1} + (250 +\Delta c_{2})·x_{2}$
#
# The final Tableau now becomes:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|-----------------|---------|---------|---------|---------|------|-------|
# | - | 1 | 0 | $-\Delta c_{2}$ | 0 | 250/3 | 650/3 | 6350 | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 | 1 | 12 | - |
#
# Again, the Simplex method requires that the Tableau is in canonical form. Thus we need to add the second last row times $\Delta c_{2}$ to the first row to fulfill this:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|---------|---------|---------|---------|------------------------|------|-------|
# | - | 1 | 0 | 0 | 0 | 250/3 + $\Delta c_{2}/3$ | 650/3 - $\Delta c_{2}/3$ | 6350 + 11·$\Delta c_{2}$ | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 | 1 | 12 | - |
# And taking into account that the coefficients of $s_{2}$ and $s_{3}$ need to be positive for $x_{2}$ to remain in the basis, $\Delta c_{2}$ cannot be lower than -250 nor higher than 650.
# ## Changes in the constraint independent terms
# Now let us see what is the effect of changing b, for instance decreasing $b_{1}$ an amount equal to $\Delta b_{1}$. In the original problem formulation, this means:
#
# $2x_{1} + x_{2} + s_{1} = 40 - \Delta b_{1} $
# $x_{1} + 3x_{2} + s_{2} = 45$
# $x_{1} + s_{3} = 12$
# Changing the Right Hand Side by $\Delta b_{1}$ is equivalent to changing $s_{1}$ an amount of $\Delta b_{1}$. Note that $s_{1}$ is in the basis, thus for it to remain in the basis, it must satisfy:
#
# $s_{1}=5 + \Delta b_{1} \geq 0$
#
# $\Delta b_{1} \geq -5$
#
# Now, take the second constraint and let us apply the same change:
#
# $2x_{1} + x_{2} + s_{1} = 40 $
# $x_{1} + 3x_{2} + s_{2} = 45 - \Delta b_{2} $
# $x_{1} + s_{3} = 12$
#
# Again, this is equivalent to:
#
# $2x_{1} + x_{2} + s_{1} = 40 $
# $x_{1} + 3x_{2} + s_{2} + \Delta b_{2} = 45 $
# $x_{1} + s_{3} = 12$
#
# Which can be regarded as a change of $\Delta b_{2}$ in $s_{2}$. Now, if we change $s_{2}$ from $s_{2}=0$ to $s_{2} =\Delta b_{2}$ in the final Tableau:
#
# | Basic | z | $x_{1}$ | $x_{2}$ | $s_{1}$ | $s_{2}$ | $s_{3}$ | RHS | Ratio |
# |---------|---|---------|---------|---------|------------------------|---------|------|-------|
# | - | 1 | 0 | 0 | 0 | 250/3·$\Delta b_{2}$ | 650/3 | 6350 | - |
# | $s_{1}$ | 0 | 0 | 0 | 1 | -1/3 ·$\Delta b_{2}$ | -5/3 | 5 | - |
# | $x_{2}$ | 0 | 0 | 1 | 0 | 1/3 ·$\Delta b_{2}$ | -1/3 | 11 | - |
# | $x_{1}$ | 0 | 1 | 0 | 0 | 0 ·$\Delta b_{2}$ | 1 | 12 | - |
#
# The basis remains unchanged as long as all the constraints are still met:
#
# $s_{1} - 1/3 · \Delta b_{2} = 5 $
# $x_{2} + 1/3· \Delta b_{2} = 11 $
# $x_{1} + 0 ·\Delta b_{2} = 12$
#
# $s_{1} = 5 + 1/3 · \Delta b_{2} \geq 0 (\Delta b_{2} \geq -15)$
# $x_{2} = 11 - 1/3· \Delta b_{2} \geq 0 (\Delta b_{2} \leq 33)$
# $x_{1} = 12 - 0·\Delta b_{2} \geq 0 (\Delta b_{2} \leq Inf)$
| docs/source/CLP/tutorials/Sensitivity - Algebraic explanation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # GCP Dataflow Component Sample
# A Kubeflow Pipeline component that prepares data by submitting an Apache Beam job (authored in Python) to Cloud Dataflow for execution. The Python Beam code is run with Cloud Dataflow Runner.
#
# ## Intended use
#
# Use this component to run a Python Beam code to submit a Cloud Dataflow job as a step of a Kubeflow pipeline.
#
# ## Runtime arguments
# Name | Description | Optional | Data type| Accepted values | Default |
# :--- | :----------| :----------| :----------| :----------| :---------- |
# python_file_path | The path to the Cloud Storage bucket or local directory containing the Python file to be run. | | GCSPath | | |
# project_id | The ID of the Google Cloud Platform (GCP) project containing the Cloud Dataflow job.| | String | | |
# region | The Google Cloud Platform (GCP) region to run the Cloud Dataflow job.| | String | | |
# staging_dir | The path to the Cloud Storage directory where the staging files are stored. A random subdirectory will be created under the staging directory to keep the job information.This is done so that you can resume the job in case of failure. `staging_dir` is passed as the command line arguments (`staging_location` and `temp_location`) of the Beam code. | Yes | GCSPath | | None |
# requirements_file_path | The path to the Cloud Storage bucket or local directory containing the pip requirements file. | Yes | GCSPath | | None |
# args | The list of arguments to pass to the Python file. | No | List | A list of string arguments | None |
# wait_interval | The number of seconds to wait between calls to get the status of the job. | Yes | Integer | | 30 |
#
# ## Input data schema
#
# Before you use the component, the following files must be ready in a Cloud Storage bucket:
# - A Beam Python code file.
# - A `requirements.txt` file which includes a list of dependent packages.
#
# The Beam Python code should follow the [Beam programming guide](https://beam.apache.org/documentation/programming-guide/) as well as the following additional requirements to be compatible with this component:
# - It accepts the command line arguments `--project`, `--region`, `--temp_location`, `--staging_location`, which are [standard Dataflow Runner options](https://cloud.google.com/dataflow/docs/guides/specifying-exec-params#setting-other-cloud-pipeline-options).
# - It enables `info logging` before the start of a Cloud Dataflow job in the Python code. This is important to allow the component to track the status and ID of the job that is created. For example, calling `logging.getLogger().setLevel(logging.INFO)` before any other code.
#
#
# ## Output
# Name | Description
# :--- | :----------
# job_id | The id of the Cloud Dataflow job that is created.
#
# ## Cautions & requirements
# To use the components, the following requirements must be met:
# - Cloud Dataflow API is enabled.
# - The component is running under a secret Kubeflow user service account in a Kubeflow Pipeline cluster. For example:
# ```
# component_op(...)
# ```
# The Kubeflow user service account is a member of:
# - `roles/dataflow.developer` role of the project.
# - `roles/storage.objectViewer` role of the Cloud Storage Objects `python_file_path` and `requirements_file_path`.
# - `roles/storage.objectCreator` role of the Cloud Storage Object `staging_dir`.
#
# ## Detailed description
# The component does several things during the execution:
# - Downloads `python_file_path` and `requirements_file_path` to local files.
# - Starts a subprocess to launch the Python program.
# - Monitors the logs produced from the subprocess to extract the Cloud Dataflow job information.
# - Stores the Cloud Dataflow job information in `staging_dir` so the job can be resumed in case of failure.
# - Waits for the job to finish.
#
# # Setup
# + pycharm={"name": "#%%\n"} tags=["parameters"]
project = 'Input your PROJECT ID'
region = 'Input GCP region' # For example, 'us-central1'
output = 'Input your GCS bucket name' # No ending slash
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Install Pipeline SDK
# -
# !python3 -m pip install 'kfp>=0.1.31' --quiet
# + [markdown] pycharm={"name": "#%%\n"}
#
# ## Load the component using KFP SDK
#
# + pycharm={"name": "#%%\n"}
import kfp.components as comp
dataflow_python_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/1.4.0/components/gcp/dataflow/launch_python/component.yaml')
help(dataflow_python_op)
# -
# ## Use the wordcount python sample
# In this sample, we run a wordcount sample code in a Kubeflow Pipeline. The output will be stored in a Cloud Storage bucket. Here is the sample code:
# !gsutil cat gs://ml-pipeline-playground/samples/dataflow/wc/wc.py
# ## Example pipeline that uses the component
import kfp
import kfp.dsl as dsl
import json
output_file = '{}/wc/wordcount.out'.format(output)
@dsl.pipeline(
name='Dataflow launch python pipeline',
description='Dataflow launch python pipeline'
)
def pipeline(
python_file_path = 'gs://ml-pipeline/sample-pipeline/word-count/wc.py',
project_id = project,
region = region,
staging_dir = output,
requirements_file_path = 'gs://ml-pipeline/sample-pipeline/word-count/requirements.txt',
args = json.dumps([
'--output', output_file
]),
wait_interval = 30
):
dataflow_python_op(
python_file_path = python_file_path,
project_id = project_id,
region = region,
staging_dir = staging_dir,
requirements_file_path = requirements_file_path,
args = args,
wait_interval = wait_interval)
# ## Submit the pipeline for execution
kfp.Client().create_run_from_pipeline_func(pipeline, arguments={})
# #### Inspect the output
# !gsutil cat $output_file
# ## References
# * [Component python code](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/component_sdk/python/kfp_component/google/dataflow/_launch_python.py)
# * [Component docker file](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/Dockerfile)
# * [Sample notebook](https://github.com/kubeflow/pipelines/blob/master/components/gcp/dataflow/launch_python/sample.ipynb)
# * [Dataflow Python Quickstart](https://cloud.google.com/dataflow/docs/quickstarts/quickstart-python)
| courses/machine_learning/deepdive2/production_ml/labs/samples/core/dataflow/dataflow.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# This example shows how to:
# 1. Load a counts matrix (10X Chromium data from human peripheral blood cells)
# 2. Run the default Scrublet pipeline
# 3. Check that doublet predictions make sense
# %matplotlib inline
import scrublet as scr
import scipy.io
import matplotlib.pyplot as plt
import numpy as np
import os
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.sans-serif'] = 'Arial'
plt.rc('font', size=14)
plt.rcParams['pdf.fonttype'] = 42
# #### Download 8k PBMC data set from 10X Genomics
# Download raw data from this link:
# http://cf.10xgenomics.com/samples/cell-exp/2.1.0/pbmc8k/pbmc8k_filtered_gene_bc_matrices.tar.gz
#
#
# Or use wget:
# !wget http://cf.10xgenomics.com/samples/cell-exp/2.1.0/pbmc8k/pbmc8k_filtered_gene_bc_matrices.tar.gz
# Uncompress:
# !tar xfz pbmc8k_filtered_gene_bc_matrices.tar.gz
# #### Load counts matrix and gene list
# Load the raw counts matrix as a scipy sparse matrix with cells as rows and genes as columns.
# +
input_dir = 'filtered_gene_bc_matrices/GRCh38/'
counts_matrix = scipy.io.mmread(input_dir + '/matrix.mtx').T.tocsc()
genes = np.array(scr.load_genes(input_dir + 'genes.tsv', delimiter='\t', column=1))
print('Counts matrix shape: {} rows, {} columns'.format(counts_matrix.shape[0], counts_matrix.shape[1]))
print('Number of genes in gene list: {}'.format(len(genes)))
# -
# #### Initialize Scrublet object
# The relevant parameters are:
# - *expected_doublet_rate*: the expected fraction of transcriptomes that are doublets, typically 0.05-0.1. Results are not particularly sensitive to this parameter. For this example, the expected doublet rate comes from the Chromium User Guide: https://support.10xgenomics.com/permalink/3vzDu3zQjY0o2AqkkkI4CC
# - *sim_doublet_ratio*: the number of doublets to simulate, relative to the number of observed transcriptomes. This should be high enough that all doublet states are well-represented by simulated doublets. Setting it too high is computationally expensive. The default value is 2, though values as low as 0.5 give very similar results for the datasets that have been tested.
# - *n_neighbors*: Number of neighbors used to construct the KNN classifier of observed transcriptomes and simulated doublets. The default value of `round(0.5*sqrt(n_cells))` generally works well.
#
scrub = scr.Scrublet(counts_matrix, expected_doublet_rate=0.06)
# #### Run the default pipeline, which includes:
# 1. Doublet simulation
# 2. Normalization, gene filtering, rescaling, PCA
# 3. Doublet score calculation
# 4. Doublet score threshold detection and doublet calling
#
doublet_scores, predicted_doublets = scrub.scrub_doublets(min_counts=2,
min_cells=3,
min_gene_variability_pctl=85,
n_prin_comps=30)
# #### Plot doublet score histograms for observed transcriptomes and simulated doublets
# The simulated doublet histogram is typically bimodal. The left mode corresponds to "embedded" doublets generated by two cells with similar gene expression. The right mode corresponds to "neotypic" doublets, which are generated by cells with distinct gene expression (e.g., different cell types) and are expected to introduce more artifacts in downstream analyses. Scrublet can only detect neotypic doublets.
#
# To call doublets vs. singlets, we must set a threshold doublet score, ideally at the minimum between the two modes of the simulated doublet histogram. `scrub_doublets()` attempts to identify this point automatically and has done a good job in this example. However, if automatic threshold detection doesn't work well, you can adjust the threshold with the `call_doublets()` function. For example:
# ```python
# scrub.call_doublets(threshold=0.25)
# ```
scrub.plot_histogram();
# #### Get 2-D embedding to visualize the results
# +
print('Running UMAP...')
scrub.set_embedding('UMAP', scr.get_umap(scrub.manifold_obs_, 10, min_dist=0.3))
# # Uncomment to run tSNE - slow
# print('Running tSNE...')
# scrub.set_embedding('tSNE', scr.get_tsne(scrub.manifold_obs_, angle=0.9))
# # Uncomment to run force layout - slow
# print('Running ForceAtlas2...')
# scrub.set_embedding('FA', scr.get_force_layout(scrub.manifold_obs_, n_neighbors=5. n_iter=1000))
print('Done.')
# -
# #### Plot doublet predictions on 2-D embedding
# Predicted doublets should co-localize in distinct states.
# +
scrub.plot_embedding('UMAP', order_points=True);
# scrub.plot_embedding('tSNE', order_points=True);
# scrub.plot_embedding('FA', order_points=True);
| examples/scrublet_basics.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.5
# language: python
# name: py35
# ---
# # Welcome to Algosoc, the soceity for algorithmic trading and quantitative finance at Imperial
# ## Who are We?
#
# We are founded in March 2018 with the aim to provide education and networking opportunities for students interested in algorithmic trading and quantitative finance. In 2018 and 2019 we organise the Algothon with Blackrock which is the flagship event of our soceity. Algothon 2020 was cancelled this year due to covid-19. This year we will be running courses online.
#
# http://www.algosoc.com/
#
# Please join the society by subscribing on the union website. Membership is free. Becoming a member of the soceity allows you to join our exclusive networking events and receive up-to-date news through our mailing list.
#
# https://www.imperialcollegeunion.org/activities/a-to-z/algorithmic-trading
#
#
# ## Algosoc Portfolio
#
# We will be running trading strategies on different platforms, such as Quantopian. We welcome students to contribute by developing trading strategies and signals. We will guide you through the steps to write your first algo. Details in how to contribute will be annouced through our mailing list.
#
# Our asset universe includes US Equities and Futures. Most stratgies running are at daily resolution
#
#
# ## Algosoc Toolbox
#
# We are building a collection of tools useful for algorithmic trading which are avaliable on Github. If you would like to contribute, please email us.
#
# https://github.com/algotradingsoc/pedlar_2020
#
#
#
# Plan for today
#
# We will go through the really basics of (quantitative) finance for the rest of the session followed by an Q&A. Experienced members are wellcomed to check out the tools we are building on Github.
#
#
# The notebook uses data from Quantopian which could only be run on their platform due to data licensing. Please create an account at Quantopian https://www.quantopian.com/ as most teaching is done on Quantopian
# Lecture 1: Understanding financial data
# ## Price data
#
# The most basic data to be dealt with in quantitative finance is price data, which represent how much a financial asset is worth at a given time. Traditionally (90s), time series models such as ARIMA model are used widely in trading to capture trends in the market. Recently, the focus has moved to using deep learning models such as LSTM and CNN to learn more complicated behaviour.
#
# https://reference.wolfram.com/language/ref/ARIMAProcess.html
# https://www.tensorflow.org/api_docs/python/tf/keras/layers/LSTM
#
# Price data is determined by two charactereistics, frequency of data and types of data. Frequency of data can ranged from microseconds data (tick level) to end-of-data data (daily resolution). We will focus on data that have a minute resoluion or above. Major types of data include bar data, trade data and quote data.
#
# In an exchange, quote data (Orderbook) is displayed for a stock in real time which has the bid and ask price (with size) submitted by different market participants. A trade is made if someone is willing the buy the security at the ask price (or sell the security at the bid price). Bar data is then aggregrated using the trade price and volume over an interval, which usually is a minute, an hour and a day.
#
# https://iextrading.com/apps/tops/
#
#
#
#
#
#
# ### Bar data on Quantopian
#
# Bar data for stocks are provided in the following format (OHLCV), which summarise transactions conducted within the time period. 4 representative prices are provided, namely Open, High, Low, Close. Volume is the sum of the number of shares traded over the time period.
#
# https://images.app.goo.gl/7QKVKYWw9jpC4jVx5
#
#
# Example of daily bar data of SPY, SPDR S&P 500, the most popular ETF to track the Standard & Poor's 500 Index.
from quantopian.research.experimental import history
from quantopian.research import symbols
SPY = history('SPY', fields=['open_price','high','low','close_price','volume'],frequency='daily',start='2020-01-01',end='2020-06-30')
SPY.tail(10)
SPY['close_price'].plot()
# ## Pipeline on Quantopian
#
# Data on quantopian are organised into different datasets which can be aggregated using Pipelines in research notebook and trading algos. Pipelines can be regarded as pandas dataframes with index by time and assets.
#
# Data Reference: https://www.quantopian.com/docs/data-reference/overview
# Pipeline: https://www.quantopian.com/tutorials/pipeline
#
#
# +
import datetime
from quantopian.pipeline import CustomFactor, Pipeline
from quantopian.pipeline.data.morningstar import Fundamentals
from quantopian.pipeline.domain import US_EQUITIES, GB_EQUITIES, HK_EQUITIES, DE_EQUITIES
from quantopian.research import run_pipeline, symbols
from quantopian.pipeline.data import EquityPricing
from quantopian.pipeline.factors import Returns
from quantopian.pipeline.filters import StaticAssets, QTradableStocksUS
# Get the latest daily close price for all equities.
yesterday_close = EquityPricing.close.latest
# Get the latest daily trading volume for all equities.
yesterday_volume = EquityPricing.volume.latest
Top_350 = Fundamentals.market_cap.latest.rank(ascending=False) <= 350
Stocks = ['HSY','MSFT','GLW','AXP','FB','NVR','RL','WPM','KKR','FCX']
# Add the factor to the pipeline.
pipe = Pipeline({
'close': yesterday_close,
'volume': yesterday_volume,
'return': Returns([EquityPricing.close],window_length=252),
'cap': Fundamentals.market_cap.latest,
'eps': Fundamentals.normalized_diluted_eps_earnings_reports.latest
},
domain = US_EQUITIES,
# screen = Top_350 & (EquityPricing.volume.latest>1000),
screen = StaticAssets(symbols(Stocks)),
)
refday = datetime.datetime.now() + datetime.timedelta(days=-1)
today = refday.replace(year=refday.year-1).date()
yesterday = today.replace(year=today.year-2)
print('Start date {} End date {}'.format(yesterday,today))
df = run_pipeline(pipe, yesterday, refday)
# Run the pipeline over a year and print the result.
df.tail(20)
# -
df.tail(20)
# Demo: Sample algo provided by Quantopian
#
# We will run a sample algorithm provided by Quantopian to show how to use the backtest environment and how do we evaluate the perfomance of a trading algorithm
# ### Future plans
#
# Lecture 2: How to write a simple algo on Quantopian
# Lecture 3: Non-stationaity of time-series data
# Lecture 4: Using decision trees on fundamentals data
| Notebook/2020_21/Lecture 1 Understanding financial data.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# **[HWR-01]** 必要なモジュールをインポートします。
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
# **[HWR-02]** 1段目の畳み込みフィルターとプーリング層を定義します。
# +
num_filters1 = 32
x = tf.placeholder(tf.float32, [None, 784])
x_image = tf.reshape(x, [-1,28,28,1])
W_conv1 = tf.Variable(tf.truncated_normal([5,5,1,num_filters1],
stddev=0.1))
h_conv1 = tf.nn.conv2d(x_image, W_conv1,
strides=[1,1,1,1], padding='SAME')
b_conv1 = tf.Variable(tf.constant(0.1, shape=[num_filters1]))
h_conv1_cutoff = tf.nn.relu(h_conv1 + b_conv1)
h_pool1 =tf.nn.max_pool(h_conv1_cutoff, ksize=[1,2,2,1],
strides=[1,2,2,1], padding='SAME')
# -
# **[HWR-03]** 2段目の畳み込みフィルターとプーリング層を定義します。
# +
num_filters2 = 64
W_conv2 = tf.Variable(
tf.truncated_normal([5,5,num_filters1,num_filters2],
stddev=0.1))
h_conv2 = tf.nn.conv2d(h_pool1, W_conv2,
strides=[1,1,1,1], padding='SAME')
b_conv2 = tf.Variable(tf.constant(0.1, shape=[num_filters2]))
h_conv2_cutoff = tf.nn.relu(h_conv2 + b_conv2)
h_pool2 =tf.nn.max_pool(h_conv2_cutoff, ksize=[1,2,2,1],
strides=[1,2,2,1], padding='SAME')
# -
# **[HWR-04]** 全結合層、ドロップアウト層、ソフトマックス関数を定義します。
# +
h_pool2_flat = tf.reshape(h_pool2, [-1, 7*7*num_filters2])
num_units1 = 7*7*num_filters2
num_units2 = 1024
w2 = tf.Variable(tf.truncated_normal([num_units1, num_units2]))
b2 = tf.Variable(tf.constant(0.1, shape=[num_units2]))
hidden2 = tf.nn.relu(tf.matmul(h_pool2_flat, w2) + b2)
keep_prob = tf.placeholder(tf.float32)
hidden2_drop = tf.nn.dropout(hidden2, keep_prob)
w0 = tf.Variable(tf.zeros([num_units2, 10]))
b0 = tf.Variable(tf.zeros([10]))
p = tf.nn.softmax(tf.matmul(hidden2_drop, w0) + b0)
# -
# **[HWR-05]** セッションを用意して Variable を初期化した後、最適化処理を実施済みのセッションを復元します。
sess = tf.Session()
sess.run(tf.initialize_all_variables())
saver = tf.train.Saver()
saver.restore(sess, 'cnn_session-20000')
# **[HWR-06]** 手書き文字を入力するためのJavaScriptのコードを用意します。
# +
input_form = """
<table>
<td style="border-style: none;">
<div style="border: solid 2px #666; width: 143px; height: 144px;">
<canvas width="140" height="140"></canvas>
</div></td>
<td style="border-style: none;">
<button onclick="clear_value()">Clear</button>
</td>
</table>
"""
javascript = """
<script type="text/Javascript">
var pixels = [];
for (var i = 0; i < 28*28; i++) pixels[i] = 0
var click = 0;
var canvas = document.querySelector("canvas");
canvas.addEventListener("mousemove", function(e){
if (e.buttons == 1) {
click = 1;
canvas.getContext("2d").fillStyle = "rgb(0,0,0)";
canvas.getContext("2d").fillRect(e.offsetX, e.offsetY, 8, 8);
x = Math.floor(e.offsetY * 0.2)
y = Math.floor(e.offsetX * 0.2) + 1
for (var dy = 0; dy < 2; dy++){
for (var dx = 0; dx < 2; dx++){
if ((x + dx < 28) && (y + dy < 28)){
pixels[(y+dy)+(x+dx)*28] = 1
}
}
}
} else {
if (click == 1) set_value()
click = 0;
}
});
function set_value(){
var result = ""
for (var i = 0; i < 28*28; i++) result += pixels[i] + ","
var kernel = IPython.notebook.kernel;
kernel.execute("image = [" + result + "]");
}
function clear_value(){
canvas.getContext("2d").fillStyle = "rgb(255,255,255)";
canvas.getContext("2d").fillRect(0, 0, 140, 140);
for (var i = 0; i < 28*28; i++) pixels[i] = 0
}
</script>
"""
# -
# **[HWR-07]** JavaScriptを実行して、手書き文字を入力します。入力結果は変数 image に格納されます。
from IPython.display import HTML
HTML(input_form + javascript)
# **[HWR-08]** 入力した文字に対して、CNNで確率を計算して表示します。
# +
p_val = sess.run(p, feed_dict={x:[image], keep_prob:1.0})
fig = plt.figure(figsize=(4,2))
pred = p_val[0]
subplot = fig.add_subplot(1,1,1)
subplot.set_xticks(range(10))
subplot.set_xlim(-0.5,9.5)
subplot.set_ylim(0,1)
subplot.bar(range(10), pred, align='center')
# -
# **[HWR-09]** 1段目のフィルターを適用した画像を表示します。
#
# ここでは、小さなピクセル値をカットする前と後のそれぞれの画像を表示します。
# +
conv1_vals, cutoff1_vals = sess.run(
[h_conv1, h_conv1_cutoff], feed_dict={x:[image], keep_prob:1.0})
fig = plt.figure(figsize=(16,4))
for f in range(num_filters1):
subplot = fig.add_subplot(4, 16, f+1)
subplot.set_xticks([])
subplot.set_yticks([])
subplot.imshow(conv1_vals[0,:,:,f],
cmap=plt.cm.gray_r, interpolation='nearest')
for f in range(num_filters1):
subplot = fig.add_subplot(4, 16, num_filters1+f+1)
subplot.set_xticks([])
subplot.set_yticks([])
subplot.imshow(cutoff1_vals[0,:,:,f],
cmap=plt.cm.gray_r, interpolation='nearest')
# -
# **[HWR-10]** 2段目のフィルターを適用した画像を表示します。
#
# ここでは、小さなピクセル値をカットする前と後のそれぞれの画像を表示します。
# +
conv2_vals, cutoff2_vals = sess.run(
[h_conv2, h_conv2_cutoff], feed_dict={x:[image], keep_prob:1.0})
fig = plt.figure(figsize=(16,8))
for f in range(num_filters2):
subplot = fig.add_subplot(8, 16, f+1)
subplot.set_xticks([])
subplot.set_yticks([])
subplot.imshow(conv2_vals[0,:,:,f],
cmap=plt.cm.gray_r, interpolation='nearest')
for f in range(num_filters2):
subplot = fig.add_subplot(8, 16, num_filters2+f+1)
subplot.set_xticks([])
subplot.set_yticks([])
subplot.imshow(cutoff2_vals[0,:,:,f],
cmap=plt.cm.gray_r, interpolation='nearest')
| Chapter05/Handwriting recognizer.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Homework 7
# In this homework, we will implement a simplified version of object detection process. Note that the tests on the notebook are not comprehensive, autograder will contain more tests.
# +
from __future__ import print_function
import random
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from skimage import io
from skimage.feature import hog
from skimage import data, color, exposure
from skimage.transform import rescale, resize, downscale_local_mean
import glob, os
import fnmatch
import time
import warnings
warnings.filterwarnings('ignore')
from detection import *
from visualization import *
from utils import *
# This code is to make matplotlib figures appear inline in the
# notebook rather than in a new window.
# %matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
# %load_ext autoreload
# %autoreload 2
# %reload_ext autoreload
# -
# # Part 1: Hog Representation (5 points)
#
# In this section, we will compute the average hog representation of human faces.<br>
# There are 31 aligned face images provided in the `\face` folder. They are all aligned and have the same size. We will get an average face from these images and compute a hog feature representation for the averaged face. <br>
# Use the hog function provided by skimage library, and implement a hog representation of objects.
# Implement **`hog_feature`** function in `detection.py`
# +
image_paths = fnmatch.filter(os.listdir('./face'), '*.jpg')
list.sort(image_paths)
n = len(image_paths)
face_shape, avg_face = load_faces(image_paths, n)
(face_feature, hog_image) = hog_feature(avg_face)
print("Sum of face feature = ", np.sum(face_feature))
assert np.abs(np.sum(face_feature) - 499.970465079) < 1e-2
plot_part1(avg_face, hog_image)
# -
# # Part 2: Sliding Window (20 points)
# Implement **`sliding_window`** function to have windows slide across an image with a specific window size. The window slides through the image and checks if an object is detected with a high similarity score with the template at every location. We compute these scores as the dot product of the HoG features of the template and the HoG features of each window as the windows slide through the image. These scores will generate a response map and you will be able to find the location of the window with the highest hog score.
#
# +
image_path = 'image_0001.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 0.8)
(winH, winW) = face_shape
(score, r, c, response_map_resized, response_map) = \
sliding_window(image, face_feature, step_size=30, window_size=face_shape, return_unresized_response=True)
print("Maximum HOG face feature score over sliding window = ", score)
print("Maximum score location = row {}, col {}".format(r, c))
crop = image[r:r+winH, c:c+winW]
plot_part2(image, r, c, response_map_resized, response_map, winW, winH)
# -
# Sliding window successfully found the human face in the above example. However, in the cell below, we are only changing the scale of the image, and you can see that sliding window does not work once the scale of the image is changed.
# +
image_path = 'image_0001.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 1.2)
(winH, winW) = face_shape
(score, r, c, response_map_resized, response_map) = \
sliding_window(image, face_feature, step_size=30, window_size=face_shape, return_unresized_response=True)
print("Maximum HoG face feature score over sliding window = ", score)
print("Maximum score location = row {}, col {}".format(r, c))
crop = image[r:r+winH, c:c+winW]
plot_part2(image, r, c, response_map_resized, response_map, winW, winH)
# -
# # Part 3: Image Pyramids (20 points)
# In order to make sliding window work for different scales of images, you need to implement image pyramids where you resize the image to different scales and run the sliding window method on each resized image. This way you scale the objects and can detect both small and large objects.
#
# ### 3.1 Image Pyramid (5 points)
#
# Implement **`pyramid`** function in `detection.py`, this will create pyramid of images at different scales. Run the following code, and you will see the shape of the original image gets smaller until it reaches a minimum size.
#
# +
image_path = 'image_0001.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 1.2)
images = pyramid(image, scale = 0.9)
plot_part3_1(images)
# -
# ### 3.2 Pyramid Score (15 points)
#
# After getting the image pyramid, we will run sliding window on all the images to find a place that gets the highest score. Implement **`pyramid_score`** function in `detection.py`. It will return the highest score and its related information in the image pyramids.
# +
image_path = 'image_0001.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 1.2)
(winH, winW) = face_shape
max_score, maxr, maxc, max_scale, max_response_map = pyramid_score \
(image, face_feature, face_shape, step_size = 30, scale=0.8)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(maxr, maxc))
plot_part3_2(image, max_scale, winW, winH, maxc, maxr, max_response_map)
# -
# From the above example, we can see that image pyramid has fixed the problem of scaling. Then in the example below, we will try another image and implement a deformable parts model.
# +
image_path = 'image_0338.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 1.0)
(winH, winW) = face_shape
max_score, maxr, maxc, max_scale, max_response_map = pyramid_score \
(image, face_feature, face_shape, step_size = 30, scale=0.8)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(maxr, maxc))
plot_part3_2(image, max_scale, winW, winH, maxc, maxr, max_response_map)
# -
# # Part 4: Deformable Parts Detection (15 Points)
# In order to solve the problem above, you will implement deformable parts model in this section, and apply it on human faces. <br>
# The first step is to get a detector for each part of the face, including left eye, right eye, nose and mouth. <br>
# For example for the left eye, we have provided the groundtruth location of left eyes for each image in the `\face` directory. This is stored in the `lefteyes` array with shape `(n,2)`, each row is the `(r,c)` location of the center of left eye. You will then find the average hog representation of the left eyes in the images.
# Run through the following code to get a detector for left eyes.
# +
image_paths = fnmatch.filter(os.listdir('./face'), '*.jpg')
parts = read_facial_labels(image_paths)
lefteyes, righteyes, noses, mouths = parts
# Typical shape for left eye
lefteye_h = 10
lefteye_w = 20
lefteye_shape = (lefteye_h, lefteye_w)
avg_lefteye = get_detector(lefteye_h, lefteye_w, lefteyes, image_paths)
(lefteye_feature, lefteye_hog) = hog_feature(avg_lefteye, pixel_per_cell=2)
plot_part4(avg_lefteye, lefteye_hog, 'left eye')
# -
# Run through the following code to get a detector for right eye.
# +
righteye_h = 10
righteye_w = 20
righteye_shape = (righteye_h, righteye_w)
avg_righteye = get_detector(righteye_h, righteye_w, righteyes, image_paths)
(righteye_feature, righteye_hog) = hog_feature(avg_righteye, pixel_per_cell=2)
plot_part4(avg_righteye, righteye_hog, 'right eye')
# -
# Run through the following code to get a detector for nose.
# +
nose_h = 30
nose_w = 26
nose_shape = (nose_h, nose_w)
avg_nose = get_detector(nose_h, nose_w, noses, image_paths)
(nose_feature, nose_hog) = hog_feature(avg_nose, pixel_per_cell=2)
plot_part4(avg_nose, nose_hog, 'nose')
# -
# Run through the following code to get a detector for mouth
# +
mouth_h = 20
mouth_w = 36
mouth_shape = (mouth_h, mouth_w)
avg_mouth = get_detector(mouth_h, mouth_w, mouths, image_paths)
(mouth_feature, mouth_hog) = hog_feature(avg_mouth, pixel_per_cell=2)
detectors_list = [lefteye_feature, righteye_feature, nose_feature, mouth_feature]
plot_part4(avg_mouth, mouth_hog, 'mouth')
# -
# ### 4.1 Compute displacement (10 points)
#
# Implement **`compute_displacement`** to get an average shift vector mu and standard deviation sigma for each part of the face. The vector mu is the distance from the main center, i.e the center of the face, to the center of the part. Note that you can and should leave mu as a decimal instead of rounding to integers, because our next step of applying the shift in **`shift_heatmap`** will interpolate the shift, which is valid for decimal shifts.
#
# test for compute_displacement
test_array = np.array([[0,1],[1,2],[2,3],[3,4]])
test_shape = (6,6)
mu, std = compute_displacement(test_array, test_shape)
assert(np.all(mu == [1.5,0.5]))
assert(np.sum(std-[ 1.11803399, 1.11803399])<1e-5)
print("Your implementation is correct!")
# +
lefteye_mu, lefteye_std = compute_displacement(lefteyes, face_shape)
righteye_mu, righteye_std = compute_displacement(righteyes, face_shape)
nose_mu, nose_std = compute_displacement(noses, face_shape)
mouth_mu, mouth_std = compute_displacement(mouths, face_shape)
print("Left eye shift = ", lefteye_mu)
print("Right eye shift = ", righteye_mu)
print("Nose shift = ", nose_mu)
print("Mouth shift = ", mouth_mu)
print("\nLeft eye std = ", lefteye_std)
print("Right eye std = ", righteye_std)
print("Nose std = ", nose_std)
print("Mouth std = ", mouth_std)
# -
# After getting the shift vectors, we can run our detector on a test image. We will first run the following code to detect each part of left eye, right eye, nose and mouth in the image. You will see a response map for each of them.
# +
image_path = 'image_0338.jpg'
image = io.imread(image_path, as_gray=True)
image = rescale(image, 1.0)
(face_H, face_W) = face_shape
max_score, face_r, face_c, face_scale, face_response_map = pyramid_score\
(image, face_feature, face_shape,step_size = 30, scale=0.8)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(face_r, face_c))
plot_part5_1(face_response_map)
# +
max_score, lefteye_r, lefteye_c, lefteye_scale, lefteye_response_map = \
pyramid_score(image, lefteye_feature,lefteye_shape, step_size = 20,scale=0.9, pixel_per_cell = 2)
lefteye_response_map = resize(lefteye_response_map, face_response_map.shape)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(lefteye_r, lefteye_c))
plot_part5_1(lefteye_response_map)
# +
max_score, righteye_r, righteye_c, righteye_scale, righteye_response_map = \
pyramid_score (image, righteye_feature, righteye_shape, step_size = 20,scale=0.9, pixel_per_cell=2)
righteye_response_map = resize(righteye_response_map, face_response_map.shape)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(righteye_r, righteye_c))
plot_part5_1(righteye_response_map)
# +
max_score, nose_r, nose_c, nose_scale, nose_response_map = \
pyramid_score (image, nose_feature, nose_shape, step_size = 20,scale=0.9, pixel_per_cell = 2)
nose_response_map = resize(nose_response_map, face_response_map.shape)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(nose_r, nose_c))
plot_part5_1(nose_response_map)
# +
max_score, mouth_r, mouth_c, mouth_scale, mouth_response_map =\
pyramid_score (image, mouth_feature, mouth_shape, step_size = 20,scale=0.9, pixel_per_cell = 2)
mouth_response_map = resize(mouth_response_map, face_response_map.shape)
print("Maximum HoG face feature score over pyramid and sliding window = ", max_score)
print("Maximum score location = row {}, col {}".format(mouth_r, mouth_c))
plot_part5_1(mouth_response_map)
# -
# ### 4.2 Shift heatmap (5 points)
#
# After getting the response maps for each part of the face, we will shift these maps so that they all have the same center as the face. We have calculated the shift vector mu in `compute_displacement`, so we are shifting based on vector mu. Implement `shift_heatmap` function in `detection.py`.
face_heatmap_shifted = shift_heatmap(face_response_map, [0,0])
print("Heatmap face max and min = ", face_heatmap_shifted.max(), face_heatmap_shifted.min())
print("Heatmap face max location = ", np.unravel_index(face_heatmap_shifted.argmax(), face_heatmap_shifted.shape))
plot_part5_2_face(face_heatmap_shifted)
# +
lefteye_heatmap_shifted = shift_heatmap(lefteye_response_map, lefteye_mu)
righteye_heatmap_shifted = shift_heatmap(righteye_response_map, righteye_mu)
nose_heatmap_shifted = shift_heatmap(nose_response_map, nose_mu)
mouth_heatmap_shifted = shift_heatmap(mouth_response_map, mouth_mu)
print("Heatmap left eye max and min = ",
lefteye_heatmap_shifted.max(), lefteye_heatmap_shifted.min())
print("Heatmap left eye max location = ",
np.unravel_index(lefteye_heatmap_shifted.argmax(), lefteye_heatmap_shifted.shape))
print("Heatmap right eye max and min = ",
righteye_heatmap_shifted.max(), righteye_heatmap_shifted.min())
print("Heatmap right eye max location = ",
np.unravel_index(righteye_heatmap_shifted.argmax(), righteye_heatmap_shifted.shape))
print("Heatmap nose max and min = ",
nose_heatmap_shifted.max(), nose_heatmap_shifted.min())
print("Heatmap nose max location = ",
np.unravel_index(nose_heatmap_shifted.argmax(), nose_heatmap_shifted.shape))
print("Heatmap mouth max and min = ",
mouth_heatmap_shifted.max(), mouth_heatmap_shifted.min())
print("Heatmap mouth max location = ",
np.unravel_index(mouth_heatmap_shifted.argmax(), mouth_heatmap_shifted.shape))
plot_part5_2_parts(lefteye_heatmap_shifted, righteye_heatmap_shifted,
nose_heatmap_shifted, mouth_heatmap_shifted)
# -
# # Part 5: Gaussian Filter (15 points)
#
# ## Part 5.1 Gaussian Filter (10 points)
# In this part, apply gaussian filter convolution to each heatmap. Blur by kernel of standard deviation sigma, and then add the heatmaps of the parts with the heatmap of the face. On the combined heatmap, find the maximum value and its location. You can use function provided by skimage to implement **`gaussian_heatmap`**.
#
# +
heatmap_face = face_heatmap_shifted
heatmaps = [lefteye_heatmap_shifted,
righteye_heatmap_shifted,
nose_heatmap_shifted,
mouth_heatmap_shifted]
sigmas = [lefteye_std, righteye_std, nose_std, mouth_std]
heatmap, i , j = gaussian_heatmap(heatmap_face, heatmaps, sigmas)
print("Heatmap shape = ", heatmap.shape)
print("Image shape = ", image.shape)
print("Gaussian heatmap max and min = ", heatmap.max(), heatmap.min())
print("Gaussian heatmap max location = ", np.unravel_index(heatmap.argmax(), heatmap.shape))
print("Resizing heatmap to image shape ...")
plot_part6_1(winH, winW, heatmap, image, i, j)
# -
# ## 5.2 Result Analysis (5 points)
#
# Does your DPM work on detecting human faces? Can you think of a case where DPM may work better than the detector we had in part 3 (sliding window + image pyramid)? You can also have examples that are not faces.
# **Your Answer:** Write your answer in this markdown cell.
# ## Extra Credit
# You have tried detecting one face from the image, and the next step is to extend it to detecting multiple occurences of the object. For example in the following image, how do you detect more than one face from your response map? Implement the function **`detect_multiple`**, and write code to visualize your detected faces in the cell below.
image_path = 'image_0002.jpg'
image = io.imread(image_path, as_gray=True)
plt.imshow(image)
plt.show()
# +
image_path = 'image_0002.jpg'
image = io.imread(image_path, as_gray=True)
heatmap = get_heatmap(image, face_feature, face_shape, detectors_list, parts)
plt.imshow(heatmap, cmap='viridis', interpolation='nearest')
plt.show()
# +
detected_faces = detect_multiple(image, heatmap)
# Visualize your detected faces
### YOUR CODE HERE
pass
### END YOUR CODE
# -
# ---
# # Part 6: K-Nearest Neighbors Classification (25 points)
#
# ## Face Dataset
#
# We will use a dataset of faces of celebrities. Download the dataset using the following command:
#
# sh get_dataset.sh
#
# The face dataset for CS131 assignment.
# The directory containing the dataset has the following structure:
#
# faces/
# train/
# angelina jolie/
# anne hathaway/
# ...
# test/
# angelina jolie/
# anne hathaway/
# ...
#
# Each class has 50 training images and 10 testing images.
# +
from utils_knn import load_dataset
X_train, y_train, classes_train = load_dataset('faces', train=True, as_gray=True)
X_test, y_test, classes_test = load_dataset('faces', train=False, as_gray=True)
assert classes_train == classes_test
classes = classes_train
print('Class names:', classes)
print('Training data shape:', X_train.shape)
print('Training labels shape: ', y_train.shape)
print('Test data shape:', X_test.shape)
print('Test labels shape: ', y_test.shape)
# -
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
num_classes = len(classes)
samples_per_class = 10
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y)
idxs = np.random.choice(idxs, samples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt_idx = i * num_classes + y + 1
plt.subplot(samples_per_class, num_classes, plt_idx)
plt.imshow(X_train[idx])
plt.axis('off')
if i == 0:
plt.title(y)
plt.show()
# Flatten the image data into rows
# we now have one 4096 dimensional featue vector for each example
X_train_flat = np.reshape(X_train, (X_train.shape[0], -1))
X_test_flat = np.reshape(X_test, (X_test.shape[0], -1))
print("New training data shape:", X_train_flat.shape)
print("New test data shape:", X_test_flat.shape)
# ## Part 6.1: Cross Validation on Raw Pixel Features (15 Points)
#
# We're now going to try to classify the test images using the k-nearest neighbors algorithm on the **raw features of the images** (i.e. the pixel values themselves). We will see later how we can use kNN on better features.
#
# The gist of the k-nearest neighbors algorithm is to predict a test image's class based on which classes the k nearest train images belong to. For example, using k = 3, if we found that for test image X, the three nearest train images were 2 pictures of <NAME>, and one picture of <NAME>, we would predict that the test image X is a picture of <NAME>.
#
# Here are the steps that we will follow:
#
# 1. We compute the L2 distances between every element of X_test and every element of X_train in `compute_distances`.
# 2. We split the dataset into 5 folds for cross-validation in `split_folds`.
# 3. For each fold, and for different values of `k`, we predict the labels and measure accuracy.
# 4. Using the best `k` found through cross-validation, we measure accuracy on the test set.
#
# Resources for understanding cross-validation:
# https://towardsdatascience.com/why-and-how-to-cross-validate-a-model-d6424b45261f
# +
from k_nearest_neighbor import compute_distances
# Step 1: compute the distances between all features from X_train and from X_test
dists = compute_distances(X_test_flat, X_train_flat)
assert dists.shape == (160, 800)
print("dists shape:", dists.shape)
# +
from k_nearest_neighbor import predict_labels
# We use k = 1 (which corresponds to only taking the nearest neighbor to decide)
y_test_pred = predict_labels(dists, y_train, k=1)
# Compute and print the fraction of correctly predicted examples
num_test = y_test.shape[0]
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))
# -
# ### Cross-Validation
#
# We don't know the best value for our parameter `k`.
# There is no theory on how to choose an optimal `k`, and the way to choose it is through cross-validation.
#
# We **cannot** compute any metric on the test set to choose the best `k`, because we want our final test accuracy to reflect a real use case. This real use case would be a setting where we have new examples come and we classify them on the go. There is no way to check the accuracy beforehand on that set of test examples to determine `k`.
#
# Cross-validation will make use split the data into different fold (5 here).
# For each fold, if we have a total of 5 folds we will have:
# - 80% of the data as training data
# - 20% of the data as validation data
#
# We will compute the accuracy on the validation accuracy for each fold, and use the mean of these 5 accuracies to determine the best parameter `k`.
# +
from k_nearest_neighbor import split_folds
# Step 2: split the data into 5 folds to perform cross-validation.
num_folds = 5
X_trains, y_trains, X_vals, y_vals = split_folds(X_train_flat, y_train, num_folds)
assert X_trains.shape == (5, 640, 4096)
assert y_trains.shape == (5, 640)
assert X_vals.shape == (5, 160, 4096)
assert y_vals.shape == (5, 160)
# +
# Step 3: Measure the mean accuracy for each value of `k`
# List of k to choose from
k_choices = list(range(5, 101, 5))
# Dictionnary mapping k values to accuracies
# For each k value, we will have `num_folds` accuracies to compute
# k_to_accuracies[1] will be for instance [0.22, 0.23, 0.19, 0.25, 0.20] for 5 folds
k_to_accuracies = {}
for k in k_choices:
print("Running for k=%d" % k)
accuracies = []
for i in range(num_folds):
# Make predictions
fold_dists = compute_distances(X_vals[i], X_trains[i])
y_pred = predict_labels(fold_dists, y_trains[i], k)
# Compute and print the fraction of correctly predicted examples
num_correct = np.sum(y_pred == y_vals[i])
accuracy = float(num_correct) / len(y_vals[i])
accuracies.append(accuracy)
k_to_accuracies[k] = accuracies
# +
# plot the raw observations
plt.figure(figsize=(12,8))
for k in k_choices:
accuracies = k_to_accuracies[k]
plt.scatter([k] * len(accuracies), accuracies)
# plot the trend line with error bars that correspond to standard deviation
accuracies_mean = np.array([np.mean(v) for k,v in sorted(k_to_accuracies.items())])
accuracies_std = np.array([np.std(v) for k,v in sorted(k_to_accuracies.items())])
plt.errorbar(k_choices, accuracies_mean, yerr=accuracies_std)
plt.title('Cross-validation on k')
plt.xlabel('k')
plt.ylabel('Cross-validation accuracy')
plt.show()
# +
# Based on the cross-validation results above, choose the best value for k,
# retrain the classifier using all the training data, and test it on the test
# data. You should be able to get above 26% accuracy on the test data.
best_k = None
### YOUR CODE HERE
# Choose the best k based on the cross validation above
pass
### END YOUR CODE
y_test_pred = predict_labels(dists, y_train, k=best_k)
# Compute and display the accuracy
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('For k = %d, got %d / %d correct => accuracy: %f' % (best_k, num_correct, num_test, accuracy))
# -
# ## Part 6.2: Cross Validation on HOG Features (10 Points)
#
# We're now going to try to classify the test images using the k-nearest neighbors algorithm on HOG features!
# +
# Create HOG datasets
X_train_hog = [hog_feature(x)[0] for x in X_train]
X_test_hog = [hog_feature(x)[0] for x in X_test]
print("Loaded {} HoG features.".format(len(X_train_hog)))
print("Loaded {} HoG features.".format(len(X_test_hog)))
X_train_hog = np.stack(X_train_hog)
X_test_hog = np.stack(X_test_hog)
print("HOG Training data shape:", X_train_hog.shape)
print("HOG Test data shape:", X_test_hog.shape)
# +
# Create Cross Validation datasets
num_folds = 5
X_hog_trains, y_trains, X_hog_vals, y_vals = split_folds(X_train_hog, y_train, num_folds)
# List of k to choose from
k_choices = list(range(5, 101, 5))
k_to_accuracies = {}
for k in k_choices:
print("Running for k=%d" % k)
accuracies = []
for i in range(num_folds):
# Make predictions
fold_dists = compute_distances(X_hog_vals[i], X_hog_trains[i])
y_pred = predict_labels(fold_dists, y_trains[i], k)
# Compute and print the fraction of correctly predicted examples
num_correct = np.sum(y_pred == y_vals[i])
accuracy = float(num_correct) / len(y_vals[i])
accuracies.append(accuracy)
k_to_accuracies[k] = accuracies
# plot the raw observations
plt.figure(figsize=(12,8))
for k in k_choices:
accuracies = k_to_accuracies[k]
plt.scatter([k] * len(accuracies), accuracies)
# plot the trend line with error bars that correspond to standard deviation
accuracies_mean = np.array([np.mean(v) for k,v in sorted(k_to_accuracies.items())])
accuracies_std = np.array([np.std(v) for k,v in sorted(k_to_accuracies.items())])
plt.errorbar(k_choices, accuracies_mean, yerr=accuracies_std)
plt.title('Cross-validation on k')
plt.xlabel('k')
plt.ylabel('Cross-validation accuracy')
plt.show()
# +
# Based on the cross-validation results above, choose the best value for k,
# retrain the classifier using all the training data, and test it on the test
# data. You should be able to get above 50% accuracy on the test data.
best_k = None
### YOUR CODE HERE
# Choose the best k based on the cross validation above
pass
### END YOUR CODE
dists = compute_distances(X_test_hog, X_train_hog)
y_test_pred = predict_labels(dists, y_train, k=best_k)
# Compute and display the accuracy
num_test = X_test_hog.shape[0]
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print('For k = %d, got %d / %d correct => accuracy: %f' % (best_k, num_correct, num_test, accuracy))
# -
# ### Written Questions
# **Guidance on expectations and grading:**
# These are fairly open-ended questions that don't have a black and white, right or wrong answer. Instead, there are many ways of reasoning about these questions, and we're looking for engagement with and understanding of the purpose and mechanics of HOG, K-Nearest Neighbors, Cross Validation, and splitting data into training, validation, and testing sets. As long as you meaningfully engage with these concepts, as they're relevant to each question, and show understanding of them, you'll earn full credit!
#
# **Here's some clarification on Question 3:**
# We mention a variety of performance metrics for each value of k on the cross validation results: mean, standard deviation, maximum, and minimum. Mean is represented by the middle of each error bar that has the horizontal blue line connecting all the means for each value of k, while standard deviation is the size of the error bar. Mean is the average accuracy of that value of k across all of the cross validation splits, and standard deviation is also measured across the cross validation splits. You only need to consider one of these factors to inform your choice of the 'best' k, but you're free to consider multiple or all of them in your reasoning about choosing the 'best' k. You can get full credit for accurately defending the use of any combination of these metrics.
#
# If you're stuck on Question 3, think about these questions to get you started:
# *Hint 1:* for a given value of k, what does the mean of validation set accuracy tell you about expected test set accuracy with that value of k? What does the standard deviation of validation set accuracy tell you about the uncertainty of test set accuracy with that value of k?
# *Hint 2:* you can also similarly think about minimum and maximum accuracy across the splits for a given value of k, which relates to standard deviation.
#
# **Question 1**: Why did HOG features do so much better than raw pixels? You'll notice that even the luckiest high outlier of cross validation on raw pixels is outperformed by the unluckiest low outlier in HOG. Remember that the goal of this classification task is to learn to classify the identity of a profile picture using the selected feature type. How do you think we could improve to do even better?
#
# **Your Answer:** Write your answer in this markdown cell.
# Either of both of these answers is correct, or something new as long as it demonstrates some understanding of HoG features and KNN
#
#
# **Question 2**: Why did we tell you to choose the best k from cross validation, and then evaluate accuracy for that k on the test set, instead of directly evaluating a range of k values on the test set and picking the one with the best accuracy?
#
# **Your Answer:** Write your answer in this markdown cell.
#
#
# **Question 3**: How did you decide which value of k was 'best'? In a real-world scenario, if you were deploying this K-Nearest Neighbors HOG feature classifier, how would you consider the roles of the mean, standard deviation, maximum, and/or minimum of each value of k that you observed in cross validation when choosing the 'best' k?
#
# **Your Answer:** Write your answer in this markdown cell.
#
#
| fall_2021/hw7_release/hw7.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# In this tutorial we will go through some of the most common ways of using pytac. The aim is to give you an understanding of the interface and how to find out what is available.
#
# # Loading the lattice
#
# The central object in pytac is the `lattice`. It holds the information about all of the elements in the accelerator.
#
# All the data about the lattice and its elements is stored in CSV files inside the pytac repository. We use the `load_csv` module to load the data and initialise a `lattice` object; this is the normal starting point for using pytac.
#
# The "ring mode" describes one configuration of the elements in the lattice. There is one set of CSV files for each ring mode. So when we load the lattice, we specify the ring mode we want to load.
#
# At the time of writing the normal ring mode in use at Diamond is "DIAD", so let's load that.
#
# First some required imports.
import sys, os
# Make the pytac package available from this subdirectory
sys.path.append(os.path.join(os.getcwd(), '..'))
import pytac
# Initialize the DIAD mode. The import of the Cothread channel access library will allow us to get some live values from the Diamond accelerators.
import cothread
lattice = pytac.load_csv.load('DIAD')
# The lattice object itself has some fields with its own properties:
lattice.get_fields()
# The name "live" is referring to the data source - Pytac can also be set up with additional data sources for simulation, but that isn't described here.
#
# We can ask for the values of these fields. These commands will try to get the real values from the live machine (so won't work if you're not on a suitable Diamond network).
lattice.get_value("energy")
lattice.get_value("beam_current")
# ## Families, elements and fields
#
# The elements in the lattice are grouped by families, and this is the most common way to choose some to access. We can list the available families:
lattice.get_all_families()
# Let's get all the beam position monitors (BPMs). We do this by using get_elements which takes an argument for family name - in this case we use the family name "BPM".
bpms = lattice.get_elements('BPM')
print("Got {} BPMs".format(len(bpms)))
# Let's look at what we can find out about a single BPM.
#
# Each one has some fields:
one_bpm = bpms[0]
one_bpm.get_fields()
# The fields represent a property of the BPM that can change. For example, x and y are the measured positions.
one_bpm.get_value("x")
#
# ## Devices
#
# Each field has a `device` object associated with it, which knows how to set and get the value.
one_bpm.get_device("x")
# The `device` object knows the PV names for reading and writing the value of the field. Each field might have a "setpoint" or "readback" handle, which could be associated with different PV names.
#
# You can use either strings or pytac constants to specify which handle to use.
readback_pv = one_bpm.get_pv_name("x_sofb_disabled", "readback")
same_readback_pv = one_bpm.get_pv_name("x_sofb_disabled", pytac.RB)
print(readback_pv, same_readback_pv)
# Some fields are read-only, in which case there is no setpoint PV to get.
try:
one_bpm.get_pv_name("x_sofb_disabled", pytac.SP)
except Exception as e:
print(e)
# It's not normally necessary to interact with the `device` directly; you can do most things through methods of the `element` or `lattice`. E.g. element.get_value() above and `lattice.get_element_pv_names`:
lattice.get_element_pv_names('BPM', 'y', 'readback')[:10]
# ## Unit conversions
#
# Many fields can be represented in either engineering units or physics units. For example, for a magnet field, the physics unit would be the field strength and the engineering unit would be the current applied by the magnet power supply controller.
# Get a corrector magnet
corrector = lattice.get_elements("HSTR")[5]
# Request
corrector.get_value("x_kick", units=pytac.ENG)
# In order to get the unit itslef, we have to ask for the `unitconv` object associated with the field.
corrector.get_unitconv("x_kick").eng_units
# ## Magnet fields
#
# This seems like a good time to talk about the names for the magnetic fields of magnets.
#
# In accelerator physics we refer to the different components of magnetic fields as $a_n$ for vertical fields and $b_n$ for horizontal fields, where n is:
#
# | n | Field |
# |-----|------------|
# | 0 | Dipole |
# | 1 | Quadrupole |
# | 2 | Sextupole |
# | ... | ... |
#
# These names are used for the `field`s associated with magnet `element`s in pytac.
#
# For corrector magnets, although the corrector field acts like a dipole, it is given the name `x_kick` or `y_kick` so that it can be easily distinguished. An example of this is when several magnets are combined into the same `element`. The following example shows an element which combines a corrector, a skew quadrupole and a sextupole.
an_element = lattice.get_elements("HSTR")[12]
print("Fields:", an_element.get_fields())
print("Families:", an_element.families)
# ## Other methods of the `lattice`
#
# To finish off for now, let's look at some more of the methods of the `lattice`
#
# `lattice.get_element_values` lets you get all the live values for a field from a while family of elements. E.g. the currents for the horizontal corrector magnets. There is also an analogous command `lattice.set_element_values()`.
lattice.get_element_values("HSTR", "x_kick", "readback")
# `s` position is the position of an element in metres around the ring.
#
# There is a method to get the `s` positions of all elements in a family:
lattice.get_family_s("BPM")[:10]
| jupyter/tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import torch
import torch.nn as nn
from torch.autograd import Variable
import matplotlib.pyplot as plt
import numpy as np
from torch.utils.data import TensorDataset, DataLoader
# ## Convolutional neural networks
import torch
import torchvision
import torchvision.transforms as transforms
# MNIST dataset
transform = transforms.Compose(
[transforms.ToTensor()])
train_dataset = torchvision.datasets.MNIST(root='~', train=True, transform=transform, download=True)
test_dataset = torchvision.datasets.MNIST(root='~', train=False, transform=transform)
# create training and testing data
trainloader = torch.utils.data.DataLoader(train_dataset, batch_size=4,
shuffle=True, num_workers=0)
testloader = torch.utils.data.DataLoader(test_dataset, batch_size=4,
shuffle=False, num_workers=0)
# Flattens the dimensions of a convolutional network. This module does not exist in pytorch so we have
# to create it
class Flatten(nn.Module):
def __init__(self):
super(Flatten, self).__init__()
def forward(self, x):
shape = torch.prod(torch.tensor(x.shape[1:])).item()
return x.view(-1, shape)
model = nn.Sequential(
nn.Conv2d(1, 3, kernel_size=5, stride=1, padding=2),
nn.ReLU(),
nn.MaxPool2d(2, 2),
nn.Conv2d(3, 2, kernel_size=3, stride=1, padding=0),
nn.ReLU(),
Flatten(),
nn.Linear(288, 20),
nn.ReLU(),
nn.Linear(20, 10),
)
# Loss and optimizer
learning_rate = 0.01
num_epochs = 10
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
# Train the model
total_step = len(trainloader)
loss_list = []
acc_list = []
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(trainloader):
# Run the forward pass
outputs = model(images)
loss = criterion(outputs, labels)
loss_list.append(loss.item())
# Backprop and perform Adam optimisation
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Track the accuracy
total = labels.size(0)
_, predicted = torch.max(outputs.data, 1)
correct = (predicted == labels).sum().item()
acc_list.append(correct / total)
if (i + 1) % 1000 == 0:
print('Epoch [{}/{}], Step [{}/{}], Loss: {:.4f}, Accuracy: {:.2f}%'
.format(epoch + 1, num_epochs, i + 1, total_step, loss.item(),
(correct / total) * 100))
# # Recurrent neural networks
# Lets generate the movement of a spring
# +
import numpy as np
import torch
np.random.seed(2)
T = 20
L = 1000
N = 100
x = np.empty((N, L), 'int64')
x[:] = np.array(range(L)) + np.random.randint(-4 * T, 4 * T, N).reshape(N, 1)
data = np.sin(x / 1.0 / T).astype('float32')
# -
data.shape
X0 = data[:, :-10]
Y0 = data[:, 10:]
# randomize the data
dataloader = DataLoader(TensorDataset(
torch.from_numpy(X0).reshape(X0.shape[0], X0.shape[1], -1),
torch.from_numpy(Y0).reshape(Y0.shape[0], Y0.shape[1], -1)), batch_size=10,
shuffle=True)
# Flattens the dimensions of a convolutional network
class CustomLSTM(nn.Module):
def __init__(self, input_size, hidden_size, num_layers=1):
super(CustomLSTM, self).__init__()
self.lstm = nn.LSTM(input_size, hidden_size, num_layers)
def forward(self, x):
output, _ = self.lstm(x)
return output
model = nn.Sequential(CustomLSTM(1, 10), nn.Linear(10, 1))
num_epochs = 10
learning_rate = 0.01
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
# Train the model
total_step = len(dataloader)
loss_list = []
acc_list = []
for epoch in range(num_epochs):
for i, (inputs, labels) in enumerate(dataloader):
outputs = model(inputs)
loss = criterion(outputs, labels)
loss_list.append(loss.item())
# Backprop and perform Adam optimisation
optimizer.zero_grad()
loss.backward()
optimizer.step()
if (i + 1) % 10 == 0:
print('Epoch [{}/{}], Step [{}/{}], Loss: {:.4f}'
.format(epoch + 1, num_epochs, i + 1, total_step, loss.item())
)
plt.plot(model(inputs)[0, :, 0].detach().numpy(), 'r--')
plt.plot(labels[0, :, 0].detach().numpy(), 'b');
plt.figure()
| notebooks/pytorch_cnn_rnn.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:dlnd]
# language: python
# name: conda-env-dlnd-py
# ---
# # Your first neural network
#
# In this project, you'll build your first neural network and use it to predict daily bike rental ridership. We've provided some of the code, but left the implementation of the neural network up to you (for the most part). After you've submitted this project, feel free to explore the data and the model more.
#
#
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# -
# ## Load and prepare the data
#
# A critical step in working with neural networks is preparing the data correctly. Variables on different scales make it difficult for the network to efficiently learn the correct weights. Below, we've written the code to load and prepare the data. You'll learn more about this soon!
# +
data_path = 'Bike-Sharing-Dataset/hour.csv'
rides = pd.read_csv(data_path)
# -
rides.head()
# ## Checking out the data
#
# This dataset has the number of riders for each hour of each day from January 1 2011 to December 31 2012. The number of riders is split between casual and registered, summed up in the `cnt` column. You can see the first few rows of the data above.
#
# Below is a plot showing the number of bike riders over the first 10 days in the data set. You can see the hourly rentals here. This data is pretty complicated! The weekends have lower over all ridership and there are spikes when people are biking to and from work during the week. Looking at the data above, we also have information about temperature, humidity, and windspeed, all of these likely affecting the number of riders. You'll be trying to capture all this with your model.
rides[:24*10].plot(x='dteday', y='cnt')
# ### Dummy variables
# Here we have some categorical variables like season, weather, month. To include these in our model, we'll need to make binary dummy variables. This is simple to do with Pandas thanks to `get_dummies()`.
# +
dummy_fields = ['season', 'weathersit', 'mnth', 'hr', 'weekday']
for each in dummy_fields:
dummies = pd.get_dummies(rides[each], prefix=each, drop_first=False)
rides = pd.concat([rides, dummies], axis=1)
fields_to_drop = ['instant', 'dteday', 'season', 'weathersit',
'weekday', 'atemp', 'mnth', 'workingday', 'hr']
data = rides.drop(fields_to_drop, axis=1)
data.head()
# -
# ### Scaling target variables
# To make training the network easier, we'll standardize each of the continuous variables. That is, we'll shift and scale the variables such that they have zero mean and a standard deviation of 1.
#
# The scaling factors are saved so we can go backwards when we use the network for predictions.
quant_features = ['casual', 'registered', 'cnt', 'temp', 'hum', 'windspeed']
# Store scalings in a dictionary so we can convert back later
scaled_features = {}
for each in quant_features:
mean, std = data[each].mean(), data[each].std()
scaled_features[each] = [mean, std]
data.loc[:, each] = (data[each] - mean)/std
# ### Splitting the data into training, testing, and validation sets
#
# We'll save the last 21 days of the data to use as a test set after we've trained the network. We'll use this set to make predictions and compare them with the actual number of riders.
# +
# Save the last 21 days
test_data = data[-21*24:]
data = data[:-21*24]
# Separate the data into features and targets
target_fields = ['cnt', 'casual', 'registered']
features, targets = data.drop(target_fields, axis=1), data[target_fields]
test_features, test_targets = test_data.drop(target_fields, axis=1), test_data[target_fields]
# -
# We'll split the data into two sets, one for training and one for validating as the network is being trained. Since this is time series data, we'll train on historical data, then try to predict on future data (the validation set).
# Hold out the last 60 days of the remaining data as a validation set
train_features, train_targets = features[:-60*24], targets[:-60*24]
val_features, val_targets = features[-60*24:], targets[-60*24:]
# ## Time to build the network
#
# Below you'll build your network. We've built out the structure and the backwards pass. You'll implement the forward pass through the network. You'll also set the hyperparameters: the learning rate, the number of hidden units, and the number of training passes.
#
# The network has two layers, a hidden layer and an output layer. The hidden layer will use the sigmoid function for activations. The output layer has only one node and is used for the regression, the output of the node is the same as the input of the node. That is, the activation function is $f(x)=x$. A function that takes the input signal and generates an output signal, but takes into account the threshold, is called an activation function. We work through each layer of our network calculating the outputs for each neuron. All of the outputs from one layer become inputs to the neurons on the next layer. This process is called *forward propagation*.
#
# We use the weights to propagate signals forward from the input to the output layers in a neural network. We use the weights to also propagate error backwards from the output back into the network to update our weights. This is called *backpropagation*.
#
# > **Hint:** You'll need the derivative of the output activation function ($f(x) = x$) for the backpropagation implementation. If you aren't familiar with calculus, this function is equivalent to the equation $y = x$. What is the slope of that equation? That is the derivative of $f(x)$.
#
# Below, you have these tasks:
# 1. Implement the sigmoid function to use as the activation function. Set `self.activation_function` in `__init__` to your sigmoid function.
# 2. Implement the forward pass in the `train` method.
# 3. Implement the backpropagation algorithm in the `train` method, including calculating the output error.
# 4. Implement the forward pass in the `run` method.
#
class NeuralNetwork(object):
def __init__(self, input_nodes, hidden_nodes, output_nodes, learning_rate):
# Set number of nodes in input, hidden and output layers.
self.input_nodes = input_nodes
self.hidden_nodes = hidden_nodes
self.output_nodes = output_nodes
# Initialize weights
self.weights_input_to_hidden = np.random.normal(0.0, self.hidden_nodes**-0.5,
(self.hidden_nodes, self.input_nodes))
self.weights_hidden_to_output = np.random.normal(0.0, self.output_nodes**-0.5,
(self.output_nodes, self.hidden_nodes))
self.lr = learning_rate
#### Set this to your implemented sigmoid function ####
# Activation function is the sigmoid function
self.activation_function = self.sigmoid
def sigmoid(self, x):
return 1.0/(1 + np.exp(-x))
def sig_derivative(self, output):
return output * (1-output)
def train(self, inputs_list, targets_list):
# Convert inputs list to 2d array
inputs = np.array(inputs_list, ndmin=2).T
targets = np.array(targets_list, ndmin=2).T
n = float(len(inputs)*2)
#print (n)
#### Implement the forward pass here ####
### Forward pass ###
# TODO: Hidden layer
hidden_inputs = np.dot(self.weights_input_to_hidden, inputs)# signals into hidden layer
hidden_outputs = self.activation_function(hidden_inputs)# signals from hidden layer
# TODO: Output layer
final_inputs = np.dot(self.weights_hidden_to_output, hidden_outputs) # signals into final output layer
final_outputs = final_inputs # signals from final output layer
#### Implement the backward pass here ####
### Backward pass ###
# TODO: Output error
output_errors = targets - final_outputs# Output layer error is the difference between desired target and actual output.
output_gradient = output_errors
hidden_errors = np.dot(self.weights_hidden_to_output.T, output_errors)
# TODO: Backpropagated error
hidden_grad = self.sig_derivative(hidden_outputs) # hidden layer gradients
hidden_gradient_product = hidden_errors * hidden_grad
# TODO: Update the weights
self.weights_hidden_to_output += (self.lr * np.dot(output_gradient,hidden_outputs.T)) # update hidden-to-output weights with gradient descent step
self.weights_input_to_hidden += (self.lr * np.dot(hidden_gradient_product ,inputs.T)) # update input-to-hidden weights with gradient descent step
def run(self, inputs_list):
# Run a forward pass through the network
inputs = np.array(inputs_list, ndmin=2).T
#### Implement the forward pass here ####
# TODO: Hidden layer
hidden_inputs = np.dot(self.weights_input_to_hidden, inputs)# signals into hidden layer
hidden_outputs = self.activation_function(hidden_inputs) # signals from hidden layer
# TODO: Output layer
final_inputs = np.dot(self.weights_hidden_to_output ,hidden_outputs)# signals into final output layer
final_outputs = final_inputs # signals from final output layer
return final_outputs
def MSE(y, Y):
return np.mean((y-Y)**2)
# ## Training the network
#
# Here you'll set the hyperparameters for the network. The strategy here is to find hyperparameters such that the error on the training set is low, but you're not overfitting to the data. If you train the network too long or have too many hidden nodes, it can become overly specific to the training set and will fail to generalize to the validation set. That is, the loss on the validation set will start increasing as the training set loss drops.
#
# You'll also be using a method know as Stochastic Gradient Descent (SGD) to train the network. The idea is that for each training pass, you grab a random sample of the data instead of using the whole data set. You use many more training passes than with normal gradient descent, but each pass is much faster. This ends up training the network more efficiently. You'll learn more about SGD later.
#
# ### Choose the number of epochs
# This is the number of times the dataset will pass through the network, each time updating the weights. As the number of epochs increases, the network becomes better and better at predicting the targets in the training set. You'll need to choose enough epochs to train the network well but not too many or you'll be overfitting.
#
# ### Choose the learning rate
# This scales the size of weight updates. If this is too big, the weights tend to explode and the network fails to fit the data. A good choice to start at is 0.1. If the network has problems fitting the data, try reducing the learning rate. Note that the lower the learning rate, the smaller the steps are in the weight updates and the longer it takes for the neural network to converge.
#
# ### Choose the number of hidden nodes
# The more hidden nodes you have, the more accurate predictions the model will make. Try a few different numbers and see how it affects the performance. You can look at the losses dictionary for a metric of the network performance. If the number of hidden units is too low, then the model won't have enough space to learn and if it is too high there are too many options for the direction that the learning can take. The trick here is to find the right balance in number of hidden units you choose.
# +
import sys
### Set the hyperparameters here ###
epochs = 1200
learning_rate = 0.02 #0.095 0.095 0.05 0.11
hidden_nodes = 20 # 130 5-0.565 10-0.560 30-0.554
output_nodes = 1
N_i = train_features.shape[1]
network = NeuralNetwork(N_i, hidden_nodes, output_nodes, learning_rate)
losses = {'train':[], 'validation':[]}
for e in range(epochs):
# Go through a random batch of 128 records from the training data set
batch = np.random.choice(train_features.index, size=128)
for record, target in zip(train_features.ix[batch].values,
train_targets.ix[batch]['cnt']):
network.train(record, target)
# Printing out the training progress
train_loss = MSE(network.run(train_features), train_targets['cnt'].values)
val_loss = MSE(network.run(val_features), val_targets['cnt'].values)
sys.stdout.write("\rProgress: " + str(100 * e/float(epochs))[:4] \
+ "% ... Training loss: " + str(train_loss)[:5] \
+ " ... Validation loss: " + str(val_loss)[:5])
losses['train'].append(train_loss)
losses['validation'].append(val_loss)
# -
plt.plot(losses['train'], label='Training loss')
plt.plot(losses['validation'], label='Validation loss')
plt.legend()
plt.ylim(ymax=0.5)
# ## Check out your predictions
#
# Here, use the test data to view how well your network is modeling the data. If something is completely wrong here, make sure each step in your network is implemented correctly.
# +
fig, ax = plt.subplots(figsize=(8,4))
mean, std = scaled_features['cnt']
predictions = network.run(test_features)*std + mean
ax.plot(predictions[0], label='Prediction')
ax.plot((test_targets['cnt']*std + mean).values, label='Data')
ax.set_xlim(right=len(predictions))
ax.legend()
dates = pd.to_datetime(rides.ix[test_data.index]['dteday'])
dates = dates.apply(lambda d: d.strftime('%b %d'))
ax.set_xticks(np.arange(len(dates))[12::24])
_ = ax.set_xticklabels(dates[12::24], rotation=45)
# -
# ## Thinking about your results
#
# Answer these questions about your results. How well does the model predict the data? Where does it fail? Why does it fail where it does?
#
# > **Note:** You can edit the text in this cell by double clicking on it. When you want to render the text, press control + enter
#
# #### Your answer below
#
# The model predicts the data very well from December 11 to December 21st. It fails betweeen
# the dates December 22 and December 31, especially around 25th December and 31st December.
#
# The reason for this may be that our training model does not account for holidays where most people will not be commuting to work and spending time at home/recreation. Our model tries to predict the target on the holidays like it will for any other day and hence we observe the failures around those dates.
#
# We can possibly add a binary(0,1) record field to our data, which records whether a day was holiday or not. Then we may be able to fit the data better.
# ## Unit tests
#
# Run these unit tests to check the correctness of your network implementation. These tests must all be successful to pass the project.
# +
import unittest
inputs = [0.5, -0.2, 0.1]
targets = [0.4]
test_w_i_h = np.array([[0.1, 0.4, -0.3],
[-0.2, 0.5, 0.2]])
test_w_h_o = np.array([[0.3, -0.1]])
class TestMethods(unittest.TestCase):
##########
# Unit tests for data loading
##########
def test_data_path(self):
# Test that file path to dataset has been unaltered
self.assertTrue(data_path.lower() == 'bike-sharing-dataset/hour.csv')
def test_data_loaded(self):
# Test that data frame loaded
self.assertTrue(isinstance(rides, pd.DataFrame))
##########
# Unit tests for network functionality
##########
def test_activation(self):
network = NeuralNetwork(3, 2, 1, 0.5)
# Test that the activation function is a sigmoid
self.assertTrue(np.all(network.activation_function(0.5) == 1/(1+np.exp(-0.5))))
def test_train(self):
# Test that weights are updated correctly on training
network = NeuralNetwork(3, 2, 1, 0.5)
network.weights_input_to_hidden = test_w_i_h.copy()
network.weights_hidden_to_output = test_w_h_o.copy()
network.train(inputs, targets)
self.assertTrue(np.allclose(network.weights_hidden_to_output,
np.array([[ 0.37275328, -0.03172939]])))
self.assertTrue(np.allclose(network.weights_input_to_hidden,
np.array([[ 0.10562014, 0.39775194, -0.29887597],
[-0.20185996, 0.50074398, 0.19962801]])))
def test_run(self):
# Test correctness of run method
network = NeuralNetwork(3, 2, 1, 0.5)
network.weights_input_to_hidden = test_w_i_h.copy()
network.weights_hidden_to_output = test_w_h_o.copy()
self.assertTrue(np.allclose(network.run(inputs), 0.09998924))
suite = unittest.TestLoader().loadTestsFromModule(TestMethods())
unittest.TextTestRunner().run(suite)
# -
| dlnd_neural_network_submission.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Deep Neural Network for Image Classification: Application
#
# When you finish this, you will have finished the last programming assignment of Week 4, and also the last programming assignment of this course!
#
# You will use use the functions you'd implemented in the previous assignment to build a deep network, and apply it to cat vs non-cat classification. Hopefully, you will see an improvement in accuracy relative to your previous logistic regression implementation.
#
# **After this assignment you will be able to:**
# - Build and apply a deep neural network to supervised learning.
#
# Let's get started!
# ## 1 - Packages
# Let's first import all the packages that you will need during this assignment.
# - [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.
# - [matplotlib](http://matplotlib.org) is a library to plot graphs in Python.
# - [h5py](http://www.h5py.org) is a common package to interact with a dataset that is stored on an H5 file.
# - [PIL](http://www.pythonware.com/products/pil/) and [scipy](https://www.scipy.org/) are used here to test your model with your own picture at the end.
# - dnn_app_utils provides the functions implemented in the "Building your Deep Neural Network: Step by Step" assignment to this notebook.
# - np.random.seed(1) is used to keep all the random function calls consistent. It will help us grade your work.
# +
import time
import numpy as np
import h5py
import matplotlib.pyplot as plt
import scipy
from PIL import Image
from scipy import ndimage
from dnn_app_utils_v2 import *
# %matplotlib inline
plt.rcParams['figure.figsize'] = (5.0, 4.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# %load_ext autoreload
# %autoreload 2
np.random.seed(1)
# -
# ## 2 - Dataset
#
# You will use the same "Cat vs non-Cat" dataset as in "Logistic Regression as a Neural Network" (Assignment 2). The model you had built had 70% test accuracy on classifying cats vs non-cats images. Hopefully, your new model will perform a better!
#
# **Problem Statement**: You are given a dataset ("data.h5") containing:
# - a training set of m_train images labelled as cat (1) or non-cat (0)
# - a test set of m_test images labelled as cat and non-cat
# - each image is of shape (num_px, num_px, 3) where 3 is for the 3 channels (RGB).
#
# Let's get more familiar with the dataset. Load the data by running the cell below.
train_x_orig, train_y, test_x_orig, test_y, classes = load_data()
# The following code will show you an image in the dataset. Feel free to change the index and re-run the cell multiple times to see other images.
# Example of a picture
index = 7
plt.imshow(train_x_orig[index])
print ("y = " + str(train_y[0,index]) + ". It's a " + classes[train_y[0,index]].decode("utf-8") + " picture.")
# +
# Explore your dataset
m_train = train_x_orig.shape[0]
num_px = train_x_orig.shape[1]
m_test = test_x_orig.shape[0]
print ("Number of training examples: " + str(m_train))
print ("Number of testing examples: " + str(m_test))
print ("Each image is of size: (" + str(num_px) + ", " + str(num_px) + ", 3)")
print ("train_x_orig shape: " + str(train_x_orig.shape))
print ("train_y shape: " + str(train_y.shape))
print ("test_x_orig shape: " + str(test_x_orig.shape))
print ("test_y shape: " + str(test_y.shape))
# -
# As usual, you reshape and standardize the images before feeding them to the network. The code is given in the cell below.
#
# <img src="images/imvectorkiank.png" style="width:450px;height:300px;">
#
# <caption><center> <u>Figure 1</u>: Image to vector conversion. <br> </center></caption>
# +
# Reshape the training and test examples
train_x_flatten = train_x_orig.reshape(train_x_orig.shape[0], -1).T # The "-1" makes reshape flatten the remaining dimensions
test_x_flatten = test_x_orig.reshape(test_x_orig.shape[0], -1).T
# Standardize data to have feature values between 0 and 1.
train_x = train_x_flatten/255.
test_x = test_x_flatten/255.
print ("train_x's shape: " + str(train_x.shape))
print ("test_x's shape: " + str(test_x.shape))
# -
# $12,288$ equals $64 \times 64 \times 3$ which is the size of one reshaped image vector.
# ## 3 - Architecture of your model
# Now that you are familiar with the dataset, it is time to build a deep neural network to distinguish cat images from non-cat images.
#
# You will build two different models:
# - A 2-layer neural network
# - An L-layer deep neural network
#
# You will then compare the performance of these models, and also try out different values for $L$.
#
# Let's look at the two architectures.
#
# ### 3.1 - 2-layer neural network
#
# <img src="images/2layerNN_kiank.png" style="width:650px;height:400px;">
# <caption><center> <u>Figure 2</u>: 2-layer neural network. <br> The model can be summarized as: ***INPUT -> LINEAR -> RELU -> LINEAR -> SIGMOID -> OUTPUT***. </center></caption>
#
# <u>Detailed Architecture of figure 2</u>:
# - The input is a (64,64,3) image which is flattened to a vector of size $(12288,1)$.
# - The corresponding vector: $[x_0,x_1,...,x_{12287}]^T$ is then multiplied by the weight matrix $W^{[1]}$ of size $(n^{[1]}, 12288)$.
# - You then add a bias term and take its relu to get the following vector: $[a_0^{[1]}, a_1^{[1]},..., a_{n^{[1]}-1}^{[1]}]^T$.
# - You then repeat the same process.
# - You multiply the resulting vector by $W^{[2]}$ and add your intercept (bias).
# - Finally, you take the sigmoid of the result. If it is greater than 0.5, you classify it to be a cat.
#
# ### 3.2 - L-layer deep neural network
#
# It is hard to represent an L-layer deep neural network with the above representation. However, here is a simplified network representation:
#
# <img src="images/LlayerNN_kiank.png" style="width:650px;height:400px;">
# <caption><center> <u>Figure 3</u>: L-layer neural network. <br> The model can be summarized as: ***[LINEAR -> RELU] $\times$ (L-1) -> LINEAR -> SIGMOID***</center></caption>
#
# <u>Detailed Architecture of figure 3</u>:
# - The input is a (64,64,3) image which is flattened to a vector of size (12288,1).
# - The corresponding vector: $[x_0,x_1,...,x_{12287}]^T$ is then multiplied by the weight matrix $W^{[1]}$ and then you add the intercept $b^{[1]}$. The result is called the linear unit.
# - Next, you take the relu of the linear unit. This process could be repeated several times for each $(W^{[l]}, b^{[l]})$ depending on the model architecture.
# - Finally, you take the sigmoid of the final linear unit. If it is greater than 0.5, you classify it to be a cat.
#
# ### 3.3 - General methodology
#
# As usual you will follow the Deep Learning methodology to build the model:
# 1. Initialize parameters / Define hyperparameters
# 2. Loop for num_iterations:
# a. Forward propagation
# b. Compute cost function
# c. Backward propagation
# d. Update parameters (using parameters, and grads from backprop)
# 4. Use trained parameters to predict labels
#
# Let's now implement those two models!
# ## 4 - Two-layer neural network
#
# **Question**: Use the helper functions you have implemented in the previous assignment to build a 2-layer neural network with the following structure: *LINEAR -> RELU -> LINEAR -> SIGMOID*. The functions you may need and their inputs are:
# ```python
# def initialize_parameters(n_x, n_h, n_y):
# ...
# return parameters
# def linear_activation_forward(A_prev, W, b, activation):
# ...
# return A, cache
# def compute_cost(AL, Y):
# ...
# return cost
# def linear_activation_backward(dA, cache, activation):
# ...
# return dA_prev, dW, db
# def update_parameters(parameters, grads, learning_rate):
# ...
# return parameters
# ```
### CONSTANTS DEFINING THE MODEL ####
n_x = 12288 # num_px * num_px * 3
n_h = 7
n_y = 1
layers_dims = (n_x, n_h, n_y)
# +
# GRADED FUNCTION: two_layer_model
def two_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 3000, print_cost=False):
"""
Implements a two-layer neural network: LINEAR->RELU->LINEAR->SIGMOID.
Arguments:
X -- input data, of shape (n_x, number of examples)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)
layers_dims -- dimensions of the layers (n_x, n_h, n_y)
num_iterations -- number of iterations of the optimization loop
learning_rate -- learning rate of the gradient descent update rule
print_cost -- If set to True, this will print the cost every 100 iterations
Returns:
parameters -- a dictionary containing W1, W2, b1, and b2
"""
np.random.seed(1)
grads = {}
costs = [] # to keep track of the cost
m = X.shape[1] # number of examples
(n_x, n_h, n_y) = layers_dims
# Initialize parameters dictionary, by calling one of the functions you'd previously implemented
### START CODE HERE ### (≈ 1 line of code)
parameters = initialize_parameters(n_x, n_h, n_y)
### END CODE HERE ###
# Get W1, b1, W2 and b2 from the dictionary parameters.
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: LINEAR -> RELU -> LINEAR -> SIGMOID. Inputs: "X, W1, b1". Output: "A1, cache1, A2, cache2".
### START CODE HERE ### (≈ 2 lines of code)
A1, cache1 = linear_activation_forward(X, W1, b1, 'relu')
A2, cache2 = linear_activation_forward(A1, W2, b2, 'sigmoid')
### END CODE HERE ###
# Compute cost
### START CODE HERE ### (≈ 1 line of code)
cost = compute_cost(A2, Y)
### END CODE HERE ###
# Initializing backward propagation
dA2 = - (np.divide(Y, A2) - np.divide(1 - Y, 1 - A2))
# Backward propagation. Inputs: "dA2, cache2, cache1". Outputs: "dA1, dW2, db2; also dA0 (not used), dW1, db1".
### START CODE HERE ### (≈ 2 lines of code)
dA1, dW2, db2 = linear_activation_backward(dA2, cache2, 'sigmoid')
dA0, dW1, db1 = linear_activation_backward(dA1, cache1, 'relu')
### END CODE HERE ###
# Set grads['dWl'] to dW1, grads['db1'] to db1, grads['dW2'] to dW2, grads['db2'] to db2
grads['dW1'] = dW1
grads['db1'] = db1
grads['dW2'] = dW2
grads['db2'] = db2
# Update parameters.
### START CODE HERE ### (approx. 1 line of code)
parameters = update_parameters(parameters, grads, learning_rate)
### END CODE HERE ###
# Retrieve W1, b1, W2, b2 from parameters
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
# Print the cost every 100 training example
if print_cost and i % 100 == 0:
print("Cost after iteration {}: {}".format(i, np.squeeze(cost)))
if print_cost and i % 100 == 0:
costs.append(cost)
# plot the cost
plt.plot(np.squeeze(costs))
plt.ylabel('cost')
plt.xlabel('iterations (per tens)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
# -
# Run the cell below to train your parameters. See if your model runs. The cost should be decreasing. It may take up to 5 minutes to run 2500 iterations. Check if the "Cost after iteration 0" matches the expected output below, if not click on the square (⬛) on the upper bar of the notebook to stop the cell and try to find your error.
parameters = two_layer_model(train_x, train_y, layers_dims = (n_x, n_h, n_y), num_iterations = 2500, print_cost=True)
# **Expected Output**:
# <table>
# <tr>
# <td> **Cost after iteration 0**</td>
# <td> 0.6930497356599888 </td>
# </tr>
# <tr>
# <td> **Cost after iteration 100**</td>
# <td> 0.6464320953428849 </td>
# </tr>
# <tr>
# <td> **...**</td>
# <td> ... </td>
# </tr>
# <tr>
# <td> **Cost after iteration 2400**</td>
# <td> 0.048554785628770206 </td>
# </tr>
# </table>
# Good thing you built a vectorized implementation! Otherwise it might have taken 10 times longer to train this.
#
# Now, you can use the trained parameters to classify images from the dataset. To see your predictions on the training and test sets, run the cell below.
predictions_train = predict(train_x, train_y, parameters)
# **Expected Output**:
# <table>
# <tr>
# <td> **Accuracy**</td>
# <td> 1.0 </td>
# </tr>
# </table>
predictions_test = predict(test_x, test_y, parameters)
# **Expected Output**:
#
# <table>
# <tr>
# <td> **Accuracy**</td>
# <td> 0.72 </td>
# </tr>
# </table>
# **Note**: You may notice that running the model on fewer iterations (say 1500) gives better accuracy on the test set. This is called "early stopping" and we will talk about it in the next course. Early stopping is a way to prevent overfitting.
#
# Congratulations! It seems that your 2-layer neural network has better performance (72%) than the logistic regression implementation (70%, assignment week 2). Let's see if you can do even better with an $L$-layer model.
# ## 5 - L-layer Neural Network
#
# **Question**: Use the helper functions you have implemented previously to build an $L$-layer neural network with the following structure: *[LINEAR -> RELU]$\times$(L-1) -> LINEAR -> SIGMOID*. The functions you may need and their inputs are:
# ```python
# def initialize_parameters_deep(layer_dims):
# ...
# return parameters
# def L_model_forward(X, parameters):
# ...
# return AL, caches
# def compute_cost(AL, Y):
# ...
# return cost
# def L_model_backward(AL, Y, caches):
# ...
# return grads
# def update_parameters(parameters, grads, learning_rate):
# ...
# return parameters
# ```
### CONSTANTS ###
layers_dims = [12288, 20, 7, 5, 1] # 5-layer model
# +
# GRADED FUNCTION: L_layer_model
def L_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 3000, print_cost=False):#lr was 0.009
"""
Implements a L-layer neural network: [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID.
Arguments:
X -- data, numpy array of shape (number of examples, num_px * num_px * 3)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)
layers_dims -- list containing the input size and each layer size, of length (number of layers + 1).
learning_rate -- learning rate of the gradient descent update rule
num_iterations -- number of iterations of the optimization loop
print_cost -- if True, it prints the cost every 100 steps
Returns:
parameters -- parameters learnt by the model. They can then be used to predict.
"""
np.random.seed(1)
costs = [] # keep track of cost
# Parameters initialization.
### START CODE HERE ###
parameters = initialize_parameters_deep(layers_dims)
### END CODE HERE ###
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: [LINEAR -> RELU]*(L-1) -> LINEAR -> SIGMOID.
### START CODE HERE ### (≈ 1 line of code)
AL, caches = L_model_forward(X, parameters)
### END CODE HERE ###
# Compute cost.
### START CODE HERE ### (≈ 1 line of code)
cost = compute_cost(AL, Y)
### END CODE HERE ###
# Backward propagation.
### START CODE HERE ### (≈ 1 line of code)
grads = L_model_backward(AL, Y, caches)
### END CODE HERE ###
# Update parameters.
### START CODE HERE ### (≈ 1 line of code)
parameters = update_parameters(parameters, grads, learning_rate)
### END CODE HERE ###
# Print the cost every 100 training example
if print_cost and i % 100 == 0:
print ("Cost after iteration %i: %f" %(i, cost))
if print_cost and i % 100 == 0:
costs.append(cost)
# plot the cost
plt.plot(np.squeeze(costs))
plt.ylabel('cost')
plt.xlabel('iterations (per tens)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
# -
# You will now train the model as a 5-layer neural network.
#
# Run the cell below to train your model. The cost should decrease on every iteration. It may take up to 5 minutes to run 2500 iterations. Check if the "Cost after iteration 0" matches the expected output below, if not click on the square (⬛) on the upper bar of the notebook to stop the cell and try to find your error.
parameters = L_layer_model(train_x, train_y, layers_dims, num_iterations = 2500, print_cost = True)
# **Expected Output**:
# <table>
# <tr>
# <td> **Cost after iteration 0**</td>
# <td> 0.771749 </td>
# </tr>
# <tr>
# <td> **Cost after iteration 100**</td>
# <td> 0.672053 </td>
# </tr>
# <tr>
# <td> **...**</td>
# <td> ... </td>
# </tr>
# <tr>
# <td> **Cost after iteration 2400**</td>
# <td> 0.092878 </td>
# </tr>
# </table>
pred_train = predict(train_x, train_y, parameters)
# <table>
# <tr>
# <td>
# **Train Accuracy**
# </td>
# <td>
# 0.985645933014
# </td>
# </tr>
# </table>
pred_test = predict(test_x, test_y, parameters)
# **Expected Output**:
#
# <table>
# <tr>
# <td> **Test Accuracy**</td>
# <td> 0.8 </td>
# </tr>
# </table>
# Congrats! It seems that your 5-layer neural network has better performance (80%) than your 2-layer neural network (72%) on the same test set.
#
# This is good performance for this task. Nice job!
#
# Though in the next course on "Improving deep neural networks" you will learn how to obtain even higher accuracy by systematically searching for better hyperparameters (learning_rate, layers_dims, num_iterations, and others you'll also learn in the next course).
# ## 6) Results Analysis
#
# First, let's take a look at some images the L-layer model labeled incorrectly. This will show a few mislabeled images.
print_mislabeled_images(classes, test_x, test_y, pred_test)
# **A few type of images the model tends to do poorly on include:**
# - Cat body in an unusual position
# - Cat appears against a background of a similar color
# - Unusual cat color and species
# - Camera Angle
# - Brightness of the picture
# - Scale variation (cat is very large or small in image)
# ## 7) Test with your own image (optional/ungraded exercise) ##
#
# Congratulations on finishing this assignment. You can use your own image and see the output of your model. To do that:
# 1. Click on "File" in the upper bar of this notebook, then click "Open" to go on your Coursera Hub.
# 2. Add your image to this Jupyter Notebook's directory, in the "images" folder
# 3. Change your image's name in the following code
# 4. Run the code and check if the algorithm is right (1 = cat, 0 = non-cat)!
# +
## START CODE HERE ##
my_image = "my_image.jpg" # change this to the name of your image file
my_label_y = [1] # the true class of your image (1 -> cat, 0 -> non-cat)
## END CODE HERE ##
fname = "images/" + my_image
image = np.array(ndimage.imread(fname, flatten=False))
my_image = scipy.misc.imresize(image, size=(num_px,num_px)).reshape((num_px*num_px*3,1))
my_predicted_image = predict(my_image, my_label_y, parameters)
plt.imshow(image)
print ("y = " + str(np.squeeze(my_predicted_image)) + ", your L-layer model predicts a \"" + classes[int(np.squeeze(my_predicted_image)),].decode("utf-8") + "\" picture.")
# -
# **References**:
#
# - for auto-reloading external module: http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
| 01-Neural Networks and Deep Learning/week4/Deep Neural Network for Image Classification-Application.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Image Classification Model - Serving Function
#
# This notebook demonstrates how to deploy a Tensorflow model using MLRun & Nuclio.
#
# **In this notebook you will:**
# * Write a Tensorflow-Model class to load and predict on the incoming data
# * Deploy the model as a serverless function
# * Invoke the serving endpoint with data as:
# * URLs to images hosted on S3
# * Direct image send
#
# **Steps:**
# * [Define Nuclio function](#Define-Nuclio-function)
# * [Install dependencies and set config](#Install-dependencies-and-set-config)
# * [Model serving class](#Model-Serving-Class)
# * [Deploy the serving function to the cluster](#Deploy-the-serving-function-to-the-cluster)
# * [Define test parameters](#Define-test-parameters)
# * [Test the deployed function on the cluster](#Test-the-deployed-function-on-the-cluster)
# ## Define Nuclio Function
# To use the magic commands for deploying this jupyter notebook as a nuclio function we must first import nuclio
# Since we do not want to import nuclio in the actual function, the comment annotation `nuclio: ignore` is used. This marks the cell for nuclio, telling it to ignore the cell's values when building the function.
# nuclio: ignore
import nuclio
# ### Install dependencies and set config
# > Note: Since tensorflow is being pulled from the baseimage it is not directly installed as a build command.
# If it is not installed on your system please uninstall and install using the line: `pip install tensorflow`
# +
# %nuclio config kind="nuclio:serving"
# %nuclio env MODEL_CLASS=TF2Model
# tensorflow 2 use the default serving image (or the mlrun/ml-models for a faster build)
# %nuclio config spec.build.baseImage = "mlrun/mlrun"
# -
# Since we are using packages which are not surely installed on our baseimage, or want to verify that a specific version of the package will be installed we use the `%nuclio cmd` annotation.
# >`%nuclio cmd` works both locally and during deployment by default, but can be set with `-c` flag to only run the commands while deploying or `-l` to set the variable for the local environment only.
# %%nuclio cmd -c
pip install tensorflow>=2.1
pip install requests pillow
# ## Function Code
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import json
import numpy as np
import requests
from tensorflow import keras
from tensorflow.keras.models import load_model
from tensorflow.keras.preprocessing import image
from tensorflow.keras.preprocessing.image import load_img
from os import environ, path
from PIL import Image
from io import BytesIO
from urllib.request import urlopen
import mlrun
# ### Model Serving Class
# We define the `TFModel` class which we will use to define data handling and prediction of our model.
#
# The class should consist of:
# * `__init__(name, model_dir)` - Setup the internal parameters
# * `load(self)` - How to load the model and broadcast it's ready for prediction
# * `preprocess(self, body)` - How to handle the incoming event, forming the request to an `{'instances': [<samples>]}` dictionary as requested by the protocol
# * `predict(self, data)` - Receives and `{'instances': [<samples>]}` and returns the model's prediction as a list
# * `postprocess(self, data)` - Does any additional processing needed on the predictions.
class TFModel(mlrun.runtimes.MLModelServer):
def __init__(self, name: str, model_dir: str):
super().__init__(name, model_dir)
self.IMAGE_WIDTH = int(environ.get('IMAGE_WIDTH', '128'))
self.IMAGE_HEIGHT = int(environ.get('IMAGE_HEIGHT', '128'))
try:
with open(environ['classes_map'], 'r') as f:
self.classes = json.load(f)
except:
self.classes = None
def load(self):
model_file, extra_data = self.get_model('.h5')
self.model = load_model(model_file)
def preprocess(self, body):
try:
output = {'instances': []}
instances = body.get('instances', [])
for byte_image in instances:
img = Image.open(byte_image)
img = img.resize((self.IMAGE_WIDTH, self.IMAGE_HEIGHT))
# Load image
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
output['instances'].append(x)
# Format instances list
output['instances'] = [np.vstack(output['instances'])]
return output
except:
raise Exception(f'received: {body}')
def predict(self, data):
images = data.get('instances', [])
# Predict
predicted_probability = self.model.predict(images)
# return prediction
return predicted_probability
def postprocess(self, predicted_probability):
if self.classes:
predicted_classes = np.around(predicted_probability, 1).tolist()[0]
predicted_probabilities = predicted_probability.tolist()[0]
return {
'prediction': [self.classes[str(int(cls))] for cls in predicted_classes],
f'{self.classes["1"]}-probability': predicted_probabilities
}
else:
return predicted_probability.tolist()[0]
# To let our nuclio builder know that our function code ends at this point we will use the comment annotation `nuclio: end-code`.
#
# Any new cell from now on will be treated as if a `nuclio: ignore` comment was set, and will not be added to the funcion.
# +
# nuclio: end-code
# -
# ## Test the function locally
# Make sure your local TF / Keras version is the same as pulled in the nuclio image for accurate testing
#
# Set the served models and their file paths using: `SERVING_MODEL_<name> = <model file path>`
#
# > Note: this notebook assumes the model and categories are under <b>/User/mlrun/examples/</b>
from PIL import Image
from io import BytesIO
import matplotlib.pyplot as plt
import os
# ### Define test parameters
# +
# Testing event
cat_image_url = 'https://s3.amazonaws.com/iguazio-sample-data/images/catanddog/cat.102.jpg'
response = requests.get(cat_image_url)
cat_image = response.content
img = Image.open(BytesIO(cat_image))
print('Test image:')
plt.imshow(img)
# -
# ### Define Function specifications
# +
import os
from mlrun import mlconf
# Model Server variables
model_class = 'TFModel'
model_name = 'cat_vs_dog_tfv2' # Define for later use in tests
models = {model_name: os.path.join(mlconf.artifact_path, 'tf2/cats_n_dogs.h5')}
# Specific model variables
function_envs = {
'IMAGE_HEIGHT': 128,
'IMAGE_WIDTH': 128,
'classes_map': '/User/artifacts/categories_map.json',
}
# -
# ## Deploy the serving function to the cluster
from mlrun import new_model_server, mount_v3io
# +
# Setup the model server function
fn = new_model_server('tf2-serving',
model_class=model_class,
models=models)
fn.set_envs(function_envs)
fn.spec.description = "tf2 image classification server"
fn.metadata.categories = ['serving', 'dl']
fn.metadata.labels = {'author': 'yaronh'}
fn.export("function.yaml")
# -
if "V3IO_HOME" in list(os.environ):
from mlrun import mount_v3io
fn.apply(mount_v3io())
else:
# is you set up mlrun using the instructions at
# https://github.com/mlrun/mlrun/blob/master/hack/local/README.md
from mlrun.platforms import mount_pvc
fn.apply(mount_pvc('nfsvol', 'nfsvol', '/home/joyan/data'))
# Deploy the model server
addr = fn.deploy(project='cat-and-dog-servers')
# ## Test the deployed function on the cluster
# ### Test the deployed function (with URL)
# +
# URL event
event_body = json.dumps({"data_url": cat_image_url})
print(f'Sending event: {event_body}')
headers = {'Content-type': 'application/json'}
response = requests.post(url=addr + f'/{model_name}/predict', data=event_body, headers=headers)
response.content
# -
# ### Test the deployed function (with Jpeg Image)
# +
# URL event
event_body = cat_image
print(f'Sending image from {cat_image_url}')
plt.imshow(img)
headers = {'Content-type': 'image/jpeg'}
response = requests.post(url=addr + f'/{model_name}/predict/', data=event_body, headers=headers)
response.content
| tf2_serving/tf2_serving.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import pandas as pd
from scipy.optimize import curve_fit
import mdtraj as md
def calc_NHVecs(traj_file, top_file, start_snap=0, end_snap=-1):
"""
Uses mdtraj to load the trajectory and get the atomic indices and coordinates to calculate the correlation functions.
For each, trajectory load the trajectory using mdtraj, get the atomic index for the the N-H atoms and calculate the vector between the two.
Append the vector to the NHVecs list for all the trajectories.
NHVecs should return a list of shape: (# Trajectories, # Snapshots, # Residues w/N-H Vectors, 3)
"""
traj = md.load(traj_file, top=top_file)
top = traj.topology
##AtomSelection Indices
Nit = top.select('name N and not resname PRO') ## PRO residue do not have N-H vectors
Hyd = top.select('name H and not resname PRO')
NH_Pair = [[i,j] for i,j in zip(Nit,Hyd)]
NH_Pair_Name = [[top.atom(i),top.atom(j)] for i,j in NH_Pair]
NH_Res = ["{}-{}{}".format(str(i).split('-')[0],str(i).split('-')[1], str(j).split('-')[1]) for i,j in NH_Pair_Name]
##Generate the N-H vectors in Laboratory Frame
NHVecs_tmp = np.take(traj.xyz, Hyd, axis=1) - np.take(traj.xyz, Nit, axis=1)
sh = list(NHVecs_tmp.shape)
sh[2] = 1
NHVecs_tmp = NHVecs_tmp / np.linalg.norm(NHVecs_tmp, axis=2).reshape(sh)
return NHVecs_tmp[start_snap:end_snap]
def split_NHVecs(nhvecs, dt, tau):
"""
This function will split the trajectory in chunks defined by tau.
nhvecs = array of N-H bond vectors,
dt = timestep of the simulation
tau = length of chunks
"""
nFiles = len(nhvecs) ## number of trajectories
nFramesPerChunk = int(tau/dt) ###tau/timestep
used_frames = np.zeros(nFiles,dtype=int)
remainingFrames = np.zeros(nFiles,dtype=int)
for i in range(nFiles):
nFrames = nhvecs[i].shape[0]
used_frames[i] = int(nFrames/nFramesPerChunk)*nFramesPerChunk
remainingFrames[i] = nFrames % nFramesPerChunk
nFramesTot=int(used_frames.sum())
out = np.zeros((nFramesTot,NHVecs[0].shape[1],NHVecs[0].shape[2]), dtype=NHVecs[0].dtype)
start = 0
for i in range(nFiles):
end = int(start+used_frames[i])
endv = int(used_frames[i])
out[start:end,...] = nhvecs[i][0:endv,...]
start = end
sh = out.shape
vecs = out.reshape((int(nFramesTot/nFramesPerChunk), nFramesPerChunk, sh[-2], sh[-1]))
return vecs
def calc_Ct(nhvecs):
"""
Calculates the correlation function of the N-H bond vectors found in nhvecs.
Direct space calculation. This could be changed to Fourier space calculation for increased speed.
LICENSE INFO:
MIT License
Copyright (c) 2017 <NAME>
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
"""
sh = nhvecs.shape
nReplicates=sh[0] ; nDeltas=int(sh[1]/2) ; nResidues=sh[2]
Ct = np.zeros( (nDeltas, nResidues), dtype=nhvecs.dtype )
dCt = np.zeros( (nDeltas, nResidues), dtype=nhvecs.dtype )
for delta in range(1,1+nDeltas):
nVals=sh[1]-delta
# = = Create < vi.v'i > with dimensions (nRep, nFr, nRes, 3) -> (nRep, nFr, nRes) -> ( nRep, nRes ), then average across replicates with SEM.
tmp = -0.5 + 1.5 * np.square( np.einsum( 'ijkl,ijkl->ijk', nhvecs[:,:-delta,...] , nhvecs[:,delta:,...] ) )
tmp = np.einsum( 'ijk->ik', tmp ) / nVals
Ct[delta-1] = np.mean( tmp, axis=0 )
dCt[delta-1] = np.std( tmp, axis=0 ) / ( np.sqrt(nReplicates) - 1.0 )
return Ct, dCt
def _bound_check(func, params):
"""
Checks if the fit returns a sum of the amplitudes greater than 1.
MIT License
Copyright (c) 2017 <NAME>
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
"""
if len(params) == 1:
return False
elif len(params) %2 == 0 :
s = sum(params[0::2])
return (s>1)
else:
s = params[0]+sum(params[1::2])
return (s>1)
def calc_chi(y1, y2, dy=[]):
"""
Calculates the chi^2 difference between the predicted model and the actual data.
LICENSE INFO:
MIT License
Copyright (c) 2017 <NAME>
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
"""
if dy != []:
return np.sum( (y1-y2)**2.0/dy )/len(y1)
else:
return np.sum( (y1-y2)**2.0 )/len(y1)
# +
## Functions 1,3,5,7,9 are the functions that the sum of coefficients are equal to 1. They have one less parameter.
## Functions 2,4,6,8,10 are the functions where the sum of coefficients are not restricted.
def func_exp_decay1(t, tau_a):
return np.exp(-t/tau_a)
def func_exp_decay2(t, A, tau_a):
return A*np.exp(-t/tau_a)
def func_exp_decay3(t, A, tau_a, tau_b):
return A*np.exp(-t/tau_a) + (1-A)*np.exp(-t/tau_b)
def func_exp_decay4(t, A, tau_a, B, tau_b ):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b)
def func_exp_decay5(t, A, tau_a, B, tau_b, tau_g ):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + (1-A-B)*np.exp(-t/tau_g)
def func_exp_decay6(t, A, tau_a, B, tau_b, G, tau_g ):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + G*np.exp(-t/tau_g)
def func_exp_decay7(t, A, tau_a, B, tau_b, G, tau_g, tau_d):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + G*np.exp(-t/tau_g) + (1-A-B-G)*np.exp(-t/tau_d)
def func_exp_decay8(t, A, tau_a, B, tau_b, G, tau_g, D, tau_d):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + G*np.exp(-t/tau_g) + D*np.exp(-t/tau_d)
def func_exp_decay9(t, A, tau_a, B, tau_b, G, tau_g, D, tau_d, tau_e):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + G*np.exp(-t/tau_g) + D*np.exp(-t/tau_d) + (1-A-B-G-D)*np.exp(-t/tau_e)
def func_exp_decay10(t, A, tau_a, B, tau_b, G, tau_g, D, tau_d, E, tau_e):
return A*np.exp(-t/tau_a) + B*np.exp(-t/tau_b) + G*np.exp(-t/tau_g) + D*np.exp(-t/tau_d) + E*np.exp(-t/tau_e)
# -
def _return_parameter_names(num_pars):
"""
Function that returns the names of the parameters for writing to the dataframe after the fit.
num_pars is the number of parameters in the fit. 1,3,5,7,9 are the num_params that constrain the fit.
while the even numbers are the parameters for the functions that don't constrain the fits.
LICENSE INFO:
MIT License
Copyright (c) 2017 <NAME>
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
"""
if num_pars==1:
return ['C_a', 'tau_a']
elif num_pars==2:
return ['C_a', 'tau_a']
elif num_pars==3:
return ['C_a', 'tau_a', 'tau_b']
elif num_pars==4:
return ['C_a', 'tau_a', 'C_b', 'tau_b']
elif num_pars==5:
return ['C_a', 'tau_a', 'C_b', 'tau_b', 'tau_g']
elif num_pars==6:
return ['C_a', 'tau_a', 'C_b', 'tau_b', 'C_g', 'tau_g']
elif num_pars==7:
return ['C_a', 'tau_a', 'C_b', 'tau_b', 'C_g', 'tau_g', 'tau_d']
elif num_pars==8:
return ['C_a', 'tau_a', 'C_b', 'tau_b', 'C_g', 'tau_g', 'C_d', 'tau_d']
elif num_pars==9:
return ['C_a', 'tau_a', 'C_b', 'tau_b', 'C_g', 'tau_g', 'C_d', 'tau_d', 'tau_e']
elif num_pars==10:
return [ 'C_a', 'tau_a', 'C_b', 'tau_b', 'C_g', 'tau_g', 'C_d', 'tau_d', 'C_e', 'tau_e']
return []
def do_Expstyle_fit2(num_pars, x, y, dy=np.empty([]), tau_mem=50.):
"""
Performs the exponential fit on the function defined by num_pars using scipy optimize curve fit.
Provides initial guesses for the amplitudes and the correlation times.
Takes the number of parameters, x values, y values, error in the y (dy), and tau_mem.
Tau_mem to help scale the initial guesses
Can also be set to np.inf if you want no bounds.
Returns, the Chi-squared value of the fit to the model along with the parameter values (popt),
the parameter error (popv) and the model itself.
"""
b1_guess = y[0]/num_pars/2
t1_guess = [tau_mem/1280.0, tau_mem/640.0, tau_mem/64.0, tau_mem/8.0]
if num_pars==1:
func=func_exp_decay1
guess=(t1_guess[2])
bound=(0.,np.inf)
elif num_pars==2:
func=func_exp_decay2
guess=(b1_guess, t1_guess[2])
bound=([0.0, x[0]],[1., np.inf])
elif num_pars==3:
func=func_exp_decay3
guess=(b1_guess, t1_guess[3], t1_guess[2])
bound=([0.0,x[0],x[0]],[1., np.inf, np.inf])
elif num_pars==4:
func=func_exp_decay4
guess=(b1_guess, t1_guess[3], b1_guess, t1_guess[2])
bound=([0.0, x[0], 0.0, x[0]],[1., np.inf, 1., np.inf])
elif num_pars==5:
func=func_exp_decay5
guess=(b1_guess, t1_guess[3], b1_guess, t1_guess[2], t1_guess[1])
bound=([0.0, x[0], 0.0, x[0],x[0]],[1., np.inf, 1., np.inf, np.inf])
elif num_pars==6:
func=func_exp_decay6
guess=(b1_guess, t1_guess[3], b1_guess, t1_guess[2], b1_guess, t1_guess[1])
bound=([0.0, x[0], 0.0, x[0], 0.0, x[0]],[1., np.inf, 1., np.inf, 1., np.inf])
elif num_pars==7:
func=func_exp_decay7
guess=(b1_guess, t1_guess[2], b1_guess, t1_guess[1], b1_guess, t1_guess[0],
t1_guess[3])
bound=([0.0, x[0], 0.0, x[0], 0.0, x[0], x[0]],[1., np.inf, 1., np.inf, 1., np.inf, np.inf])
elif num_pars==8:
func=func_exp_decay8
guess=(b1_guess, t1_guess[3], b1_guess, t1_guess[2], b1_guess, t1_guess[1],
b1_guess, t1_guess[0])
bound=([0.0, x[0], 0.0, x[0], 0.0, x[0], 0.0, x[0]],[1., np.inf, 1., np.inf, 1., np.inf, 1., np.inf])
if dy != []:
popt, popv = curve_fit(func, x, y, p0=guess, sigma=dy, bounds=bound, method='trf', loss='soft_l1')
else:
popt, popv = curve_fit(func, x, y, p0=guess, bounds=bound, loss='soft_l1')
ymodel=[ func(x[i], *popt) for i in range(len(x)) ]
#print ymodel
bExceed=_bound_check(func, popt)
if bExceed:
print >> sys.stderr, "= = = WARNING, curve fitting in do_LSstyle_fit returns a sum>1.//"
return 9999.99, popt, np.sqrt(np.diag(popv)), ymodel
else:
return calc_chi(y, ymodel, dy), popt, popv, ymodel
def findbest_Expstyle_fits2(x, y, taum=150.0, dy=[], bPrint=True, par_list=[2,3,5,7], threshold=1.0):
"""
Function tries to find the best set of parameters to describe the correlation fucntion for each residues
Takes the x,y values for the fit and the errors, dy. par_list is the number of parameters to check,
threshold is the cutoff for the chi2. This is the old way of checking, but can be re-implemented.
Runs the fit for a given parameter by calling do_Expstyle_fit3. The initial fit is chosen, but
subsequent fits are chosen with a strict criteria based on the ratio of the number of parameters from
the current best fit and the latest fit.
Returns the chi^2, names of the parameters, parameters, errors, model, and covariance matrix of the best fit.
"""
chi_min=np.inf
# Search forwards
print('Starting New Fit')
for npars in par_list:
print(npars)
names = _return_parameter_names(npars)
try:
chi, params, covarMat, ymodel = do_Expstyle_fit2(npars, x, y, dy, taum)
except:
print(" ...fit returns an error! Continuing.")
break
bBadFit=False
errors = np.sqrt(np.diag(covarMat))
step_check = 0
while step_check < npars:
## Check the error to make sure there is no overfitting
chkerr = errors[step_check]/params[step_check]
if (chkerr>0.10):
print( " --- fit shows overfitting with %d parameters." % npars)
print( " --- Occurred with parameter %s: %g +- %g " % (names[step_check], params[step_check],
errors[step_check]))
bBadFit=True
break
step_check += 1
## Chi^2 model fitting check.
## SclChk can be increased to make it easier to fit higher order models, or lower for a stronger criteria
## First model check is always set to 1.0 so its accepted
SclChk = 0.5
chi_check = chi/chi_min
if npars == par_list[0]:
threshold = 1.0
else:
threshold = (1-npar_min/npars)*SclChk
print("--- The chi_check for {} parameters is {}".format(npars, chi_check))
print("--- The threshold for this check is {}".format(threshold))
if (not bBadFit) and (chi/chi_min < threshold):
chi_min=chi ; par_min=params ; err_min=errors ; npar_min=npars ; ymod_min=ymodel; covar_min = covarMat;
else:
break;
tau_min = par_min[1::2]
sort_tau = np.argsort(tau_min)[::-1]
nsort_params = np.array([[2*tau_ind, 2*tau_ind+1] for tau_ind in sort_tau]).flatten()
err_min = err_min[nsort_params]
par_min = par_min[nsort_params]
sort_covarMat = covar_min[:,nsort_params][nsort_params]
names = _return_parameter_names(npar_min)
if bPrint:
print( "= = Found %d parameters to be the minimum necessary to describe curve: chi(%d) = %g vs. chi(%d) = %g)" % (npar_min, npar_min, chi_min, npars, chi))
print( "Parameter %d %s: %g +- %g " % (npar_min, len(names), len(par_min), len(err_min)))
for i in range(npar_min):
print( "Parameter %d %s: %g +- %g " % (i, names[i], par_min[i], err_min[i]))
print('\n')
return chi_min, names, par_min, err_min, ymod_min, sort_covarMat
def fitstoDF(resnames, chi_list, pars_list, errs_list, names_list):
## Set Up columns indices and names for the data frame
"""
Function that takes the residue names, chi^2, parameters, errors and names of the fits and returns a data frame
of the parameters.
"""
mparnames = _return_parameter_names(8) ## Always return the longest possible number of
mtau_names = np.array(mparnames)[1::2]
mc_names = np.array(mparnames)[::2]
colnames = np.array(['Resname','NumExp'])
tau_errnames = np.array([[c,"{}_err".format(c)] for c in mtau_names]).flatten()
mc_errnames = np.array([[c, "{}_err".format(c)] for c in mc_names]).flatten()
colnames = np.hstack([colnames,mc_errnames])
colnames = np.hstack([colnames,tau_errnames])
colnames = np.hstack([colnames,np.array(['Chi_Fit'])])
FitDF = pd.DataFrame(index=np.arange(len(pars_list)), columns=colnames).fillna(0.0)
FitDF['Resname'] = resnames
FitDF['Chi_Fit'] = chi_list
for i in range(len(pars_list)):
npar = len(pars_list[i])
if (npar%2)==1:
ccut = npar-2
tau_f, terr = pars_list[i][1:ccut+1:2], errs_list[i][1:ccut+1:2]
tau_f = np.hstack([tau_f, pars_list[i][-1]])
terr = np.hstack([terr, errs_list[i][-1]])
sort_tau = np.argsort(tau_f)
coeff, cerr= pars_list[i][0:ccut:2], errs_list[i][0:ccut:2]
Clast = 1; Clasterr = 0.0;
for n,m in zip(coeff, cerr):
Clast -= n
Clasterr += m
coeff = np.hstack([coeff, np.array(Clast)])
cerr = np.hstack([cerr, np.array(Clasterr)])
tne = np.array([[c,"{}_err".format(c)] for c in mparnames[1:npar+1:2]]).flatten()
cne = np.array([[c, "{}_err".format(c)] for c in mparnames[0:npar:2]]).flatten()
else:
tau_f, terr = pars_list[i][1::2], errs_list[i][1::2]
coeff, cerr= pars_list[i][0::2], errs_list[i][0::2]
sort_tau = np.argsort(tau_f)[::-1]
tne = np.array([[c,"{}_err".format(c)] for c in names_list[i][1::2]]).flatten()
cne = np.array([[c, "{}_err".format(c)] for c in names_list[i][0::2]]).flatten()
NumExp=np.array(len(tau_f))
tau_err = np.array([[t,e] for t,e in zip(tau_f[sort_tau],terr[sort_tau])]).flatten()
c_err = np.array([[c,e] for c,e in zip(coeff[sort_tau], cerr[sort_tau])]).flatten()
namesarr = np.hstack([np.array('NumExp'),cne,tne])
valarr = np.hstack([NumExp,c_err,tau_err])
FitDF.loc[i,namesarr] = valarr
FitDF['AUC_a'] = FitDF.C_a*FitDF.tau_a; FitDF['AUC_b'] = FitDF.C_b*FitDF.tau_b;
FitDF['AUC_g'] = FitDF.C_g*FitDF.tau_g; FitDF['AUC_d'] = FitDF.C_d*FitDF.tau_d;
FitDF['AUC_Total'] = FitDF[['AUC_a','AUC_b','AUC_g','AUC_d']].sum(axis=1)
return FitDF
def fitCorrF(CorrDF, dCorrDF, tau_mem, pars_l, fixfit=False, threshold=1.0):
"""
Input Variables:
CorrDF: Dataframe containing the correlation functions. Columns are the NH-bond vectors, rows are timesteps.
dCorrDF: Error in the correlation function at time t
tau_mem: Cut-Off time to remove noise at the tail of the correlation function
pars_l : parameters list.
fixfit : Boolean to decide if you want to use a specific exponential function
Main function to fit the correlation function.
Loops over all residues with N-H vectors and calculates the fit, appends the best fit from findbest_Expstyle_fits2.
Passes the set of lists to fitstoDF to return a data frame of the best fits for each residue.
Takes the correlation function CorrDF and errors in the correlation function, maximum tau mem to cut correlation
function off from, the list of parameters you want to fit too. If you don't want to test the fit and use
a fixed parameter set, set fixfit to True and pass a list of length 1 into pars_l.
"""
NH_Res = CorrDF.columns
chi_list=[] ; names_list=[] ; pars_list=[] ; errs_list=[] ; ymodel_list=[]; covarMat_list = [];
for i in CorrDF.columns:
tstop = np.where(CorrDF.index.values==tau_mem)[0][0]
x = CorrDF.index.values[:tstop]
y = CorrDF[i].values[:tstop]
dy = dCorrDF[i].values[:tstop]
## If there is no error provided i.e. no std. dev. over correlation functions is provided then set dy to empty set
if np.all(np.isnan(dy)):
dy = []
## if not fixfit then find find the best expstyle fit. Otherwise force the fit to nparams
if (not fixfit)&(len(pars_l)>1):
print("Finding the best fit for residue {}".format(i))
chi, names, pars, errs, ymodel, covarMat = findbest_Expstyle_fits2(x, y, tau_mem, dy,
par_list=pars_l, threshold=threshold)
elif (fixfit)&(len(pars_l)==1):
print("Performing a fixed fit for {} exponentials".format(int(pars_l[0]/2)))
chi, pars, covarMat, ymodel = do_Expstyle_fit2(pars_l[0], x, y, dy, tau_mem)
names = _return_parameter_names(len(pars))
errs = np.sqrt(np.diag(covarMat))
else:
print("The list of parameters is empty. Breaking out.")
break;
chi_list.append(chi)
names_list.append(names)
pars_list.append(pars)
errs_list.append(errs)
ymodel_list.append(ymodel)
FitDF = fitstoDF(NH_Res, chi_list, pars_list, errs_list, names_list)
return FitDF
def J_direct_transform(om, consts, taus):
"""
Calculation of the spectral density from the parameters of the fit by direct fourier transform
"""
## Calculation for the direct spectral density
ndecay=len(consts) ; noms=1;###lnden(om)
Jmat = np.zeros( (ndecay, noms ) )
for i in range(ndecay):
Jmat[i] = consts[i]*(taus[i]*1e-9)/(
1 + np.power((taus[i]*1e-9)*(om),2.))
return Jmat.sum(axis=0)
def calc_NMR_Relax(J, fdd, fcsa, gammaH, gammaN):
"""
Function to calculate the R1, R2 and NOE from the spectral densities and the physical parameters for the
dipole-dipole and csa contributions, fdd and fcsa.
"""
R1 = fdd * (J['Diff'] + 3*J['15N'] + 6*J['Sum']) + fcsa * J['15N']
R2 = (0.5 * fdd * (4*J['0'] + J['Diff'] + 3*J['15N'] + 6*J['1H'] + 6*J['Sum'])
+ (1./6.) * fcsa*(4*J['0'] + 3*J['15N']) )
NOE = 1 + ((fdd*gammaH)/(gammaN*R1))*(6*J['Sum'] - J['Diff'])
return R1, R2, NOE
# # Begin Implementation of Code:
# ## Definition of global file locations
# 1. Notebook can be run in the local directory, in which case, skip over the first cell
# 2. File locations of trajectories to be loaded using mdtraj for calculation of N-H bond vectors. These should be changed by the user.
#
# +
## Global Variables for the calculation of the NH Vecs and the correlation functions
FileLoc = "" ## Main Directory Location
RUN = ["Run{}".format(i) for i in range(1,5)]
JOBS = ['PROD1','PROD2','PROD3']
## For use if replicate trajectories are stored as follows
TRAJLIST_LOC = ["{}/Analysis/{}".format(J,R) for J in JOBS for R in RUN]
FTOPN = "Q15.gro" ## Name of topology for the trajectory
FMDN = "Q15.noH20.xtc" ## Name of the trajectory, should be centered and stripped of solute
# -
# ## Definition of physical constants and parameters
# 1. Several parameters should be changed if necessary
# a. B0 --> Set to experimental magnetic field you want to compare against
# b. dSigmaN --> -170e-6 is a well-established value, but can be changed
# 2. Units are in s in the parameters, but the timesteps should be in ns. Converted in J_direct_transform.
# +
## Parameters and Physical Constants for calculation of Relaxation Rates
H_gyro = 2*np.pi*42.57748*1e6 ## Gyromagnetic Ratio: Hydrogen ([rad]/[s][T])
N_gyro = -2*np.pi*4.317267*1e6 ## Gyromagnetic Ratio: Nitrogen ([rad]/[s][T])
B0 = 18.8 ## Field Strength = 18.8 [Teslas]
## Need 5 Frequencies: ## J[0], J[wH], J[wN], J[wH-wN], J[wH+wN]
Larmor1H = H_gyro*B0 ## Larmor Frequency: Hydrogen ([rad]/[s])
Larmor15N = N_gyro*B0 ## Larmor Frequency: Hydrogen ([rad]/[s])
omDiff = Larmor1H - Larmor15N ## Diff in Larmor Frequencies of Spin IS
omSum = Larmor1H + Larmor15N ## Sum of Larmor Frequencies of Spin IS
mu_0 = 4*np.pi*1e-7 ; ## Permeability of Free Space: ([H]/[m])
hbar = 1.0545718e-34 ; ## Reduced Plank's constant: [J] * [s] = [kg] * [m^2] * [s^-1]
R_NH = 1.02e-10 ## distance between N-H atoms in Angstroms
dSigmaN = -170e-6 ## CSA of the S-spin atom
FDD = (1./10.)*np.power((mu_0*hbar*H_gyro*N_gyro)/(4*np.pi*np.power(R_NH,3)),2)
#FCSA = 498637299.69233465
FCSA = (2.0/15.0)*(Larmor15N**2)*(dSigmaN**2) ## CSA factor
# -
# ## Load trajectories and calculate the NH-Vecs in the laboratory frame
# ### Skip to calculation of correlation functions if already performed
## Change directory to examples to test code
# %cd EXAMPLES
## Calculate the NHVecs; Can be adapted to loop over multiple trajectories using TRAJLIST_LOC
NHVecs = []
start=0; end=-1; ##
NHV = calc_NHVecs(FMDN, FTOPN, start, end)
NHVecs.append(NHV)
# +
dt = 10 ## timestep of simulations: (ps)
tau_split = np.array(NHVecs).shape[1]*dt ## Number of snapshots to calculate the correlation function over.
## Split the vecs based off the tau_split you want and the time step.
vecs_split = split_NHVecs(NHVecs, dt, tau_split)
# -
## Calculate the correlation functions and the standard deviation in the correlation function.
## Save the correlation functions in a dataframe and then to a csv file for later use.
Ct, dCt = calc_Ct(vecs_split)
## Convert to dataframe with index set as timesteps in ns
CtOutFname = 'NH_Ct.csv'
dCtOutFname = 'NH_dCt.csv'
CtDF = pd.DataFrame(Ct, index = np.arange(1, Ct.shape[0]+1)*dt/1000)
dCtDF = pd.DataFrame(dCt, index = np.arange(1, dCt.shape[0]+1)*dt/1000)
CtDF.to_csv(CtOutFname)
dCtDF.to_csv(dCtOutFname)
# ## Begin fitting of the correlation functions
# 1. Load the correlation functions from before
# 2. Calculate the correlation functions
# a. For a single exponential model, fixfit=True
# b. Find the best exponential model, fixfit=False (default)
# 3. Pass the fitted parameters for each residue to calculate the spectral density
# 4. Calculate the NMR relaxation parameters.
## Load the correlation functions from the saved csv files
CtInName = 'NH_Ct.csv'
dCtInName = 'NH_dCt.csv'
CtDF = pd.read_csv(CtInName, index_col=0)
dCtDF = pd.read_csv(dCtInName, index_col=0)
tau_mem=2.5 ## Cut off to remove noise from the tail of the correlation function in the fit (ns)
fixfit = True ## find the best model
parameters_list = [4] ## for fixfit = False
thresh=1.0 ##
FitDF = fitCorrF(CtDF, dCtDF, tau_mem, parameters_list, fixfit, thresh)
# +
## Calculate spectral density from the FitDF by calling the J_direct_transform function for each of the 5 frequencies.
## Loop over the rows of the FitDF dataframe from fitCorrF function and calcuate the spectral densities.
## Save the spectral densities to a dictionary and append to a list.
Jarr = []
for i,fit in FitDF.iterrows():
c = fit[['C_a','C_b','C_g','C_d']].values
t = fit[['tau_a','tau_b','tau_g','tau_d']].values
Jdict = {'0':0, '1H':0,'15N':0,'Sum':0,'Diff':0}
J0 = J_direct_transform(0, c, t)
JH = J_direct_transform(Larmor1H, c, t)
JN = J_direct_transform(Larmor15N, c, t)
JSum = J_direct_transform(omSum, c, t)
JDiff = J_direct_transform(omDiff, c, t)
Jdict['1H'] = JH ; Jdict['15N'] = JN; Jdict['0'] = J0;
Jdict['Sum'] = JSum; Jdict['Diff'] = JDiff;
Jarr.append(Jdict)
# +
## Calculate NMR relaxation parameters for each residue by calling calc_NMR_relax
## Save the T1, T2 and NOE parameters to a dataframe
NMRRelaxDF = pd.DataFrame(np.zeros((len(Jarr),3)),index=range(1,len(Jarr)+1), columns=['T1','T2','NOE'])
for index in range(1,len(Jarr)+1):
r1, r2, noe = calc_NMR_Relax(Jarr[index-1], FDD, FCSA, H_gyro, N_gyro)
NMRRelaxDF.loc[index,'T1'] = 1/r1;
NMRRelaxDF.loc[index,'T2'] = 1/r2;
NMRRelaxDF.loc[index,'NOE'] = noe;
NMRRelaxDF['Resname'] = FitDF['Resname'].values
NMRRelaxDF['RESNUM'] = NMRRelaxDF['Resname'].str.extract('([0-9]+)',expand=False).astype('int')+1
# -
## Merge the NMR relaxation dataframes with the FitDF dataframe
FitRelaxDF = FitDF.merge(NMRRelaxDF, how='left', left_on='Resname',right_on='Resname').set_index(NMRRelaxDF.index)
## Save FitRelaxDF to a csv file
FitRelaxDF.to_csv('NMRRelaxtionDF.csv')
| CorrFunction_NMRRelaxation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Bode Plots
import control
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (5, 5)
s = control.tf([1, 0], [0, 1])
# ## Derivative Terms
#
# $ G(s) = s$
G_D = s
control.bode(G_D, dB=True);
G_D
# ## Integral Terms
#
# $ G(s) = \dfrac{1}{s}$
G_I = 1/s
control.bode(G_I, dB=True);
G_I
# ## First Order Zeros
#
# $ G(s) = \dfrac{s + \omega}{\omega}$
G_FZ = (s+1)/1
control.bode(G_FZ, dB=True);
G_FZ
# ## First Order Poles
#
# $ G(s) = \dfrac{\omega}{s + \omega}$
G_FP = 1/(s+1)
control.bode(G_FP, dB=True);
G_FP
# ## Second Order Poles: Case 1, Real Poles
#
# $G(s) = \dfrac{\omega}{s^2 + 2 \zeta \omega s + \omega_n^2}$
#
# $\zeta >= 1$
#
# Factor into two first order poles.
# +
zeta = 10
wn = 1
G_SR = 1/(s**2 + 2*zeta*wn*s + wn**2)
control.bode(G_SR, dB=True);
print('roots', np.roots([1, 2*zeta*wn, wn**2]))
G_SR
# -
# ## Second Order Poles: Case 2, Imaginary Poles
#
# $G(s) = \dfrac{\omega_n}{s^2 + 2 \zeta \omega_n s + \omega_n^2}$
#
# $\zeta < 1$
# +
zeta = 0.1
wn = 1
G_SR = 1/(s**2 + 2*zeta*wn*s + wn**2)
G_damp = [1/(s**2 + 2*zeta*wn*s + wn**2) for zeta in np.arange(0.01, 0.9, 0.2)]
control.bode(G_damp, dB=True, omega=np.logspace(-2, 2, 1000));
G_SR
# -
# ## Unstable Poles and Zeros
#
# * Same magnitude
# * Oppositive slope
#
# $G(s) = \dfrac{1}{s + 1}$
#
# vs.
#
# $G(s) = \dfrac{1}{s - 1}$
control.bode([1/(s+1), 1/(s-1)], dB=True);
ax = plt.gca()
# $G(s) = s + 1$
#
# vs.
#
# $G(s) = s - 1$
control.bode([(s+1), (s-1)], dB=True);
# ## Leading Negative
#
# * Add 180 deg phase.
#
# $G(s) = -\dfrac{1}{s + 1}$
control.bode([1/(s+1), -1/(s+1)], dB=True);
# ## **Bode Plot Rules Summary**
#
# 1. Find $|G(0)|$, where your magnitude plot starts and $|G(\infty)|$, where it ends.
# 1. Find $\angle G(0)$, where your phase plot starts and $\angle G(\infty)$, where it ends.
# 1. Make table of slope and phase contribution for each factor. If you have a second order pole or zero if it has real roots, treat as two first order sytems. If it has complex roots, find the natural frequency and the damping ratio. The damping ratio determines the size of the resonant peak, and the natural frequency is the corner frequency.
# 1. Draw asymptotes using $|G(0)|$ to start the magnitude plot and $\angle G(0)$ to start the phase plot.
# 1. Interpolate between asymptotes for magnitude and phase.
#
# Zeros:
#
# | factor | corner freq. (rad/s) | phase (deg) | slope (dB/dec) |
# |- -|- -|- -|- -|
# |$s$|0 | +90 | +20|
# |$(s+\omega)$|$\omega$| +90 | +20|
# |$(s-\omega)$|$\omega$| -90 | +20|
# |$(s^2 + 2\zeta \omega_n s + \omega_n^2)$|$\omega_n$| +180 | +40|
#
# Poles:
#
# | factor | corner freq. (rad/s) | phase (deg) | slope (dB/dec) |
# |- -|- -|- -|- -|
# |$1/s$| 0 | -90 |-20|
# |$1/(s+\omega)$|$\omega$| -90 | -20|
# |$1/(s-\omega)$|$\omega$| +90 | -20|
# |$1/(s^2 + 2\zeta \omega_n s + \omega_n^2)$|$\omega_n$| -180 | -40|
#
# ## Combining Factors
#
# $G(s) = \dfrac{-1}{(s-1)(s-2)}$
#
# $|G(0)| = |-1/2| = 1/2 \approx -6 dB$
#
# $\angle G(0) = \angle -1/2 = -180 deg$
#
# | factor | corner freq. (rad/s) | phase (deg) | slope (dB/dec) |
# |- -|- -|- -|- -|
# |$1/(s-1)$|1|+90|-20|
# |$1/(s-2)$|2|+90|-20|
#
# Notice phase contribution of the pole is now positive, since it is an unstable pole.
#
import control
s = control.tf([1, 0], [0, 1])
control.bode(-1/((s-1)*(s-2)), dB=True);
| 09.Bode.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import nltk
paragraph = """I have three visions for India. In 3000 years of our history, people from all over
the world have come and invaded us, captured our lands, conquered our minds.
From Alexander onwards, the Greeks, the Turks, the Moguls, the Portuguese, the British,
the French, the Dutch, all of them came and looted us, took over what was ours.
Yet we have not done this to any other nation. We have not conquered anyone.
We have not grabbed their land, their culture,
their history and tried to enforce our way of life on them.
Why? Because we respect the freedom of others.That is why my
first vision is that of freedom. I believe that India got its first vision of
this in 1857, when we started the War of Independence. It is this freedom that
we must protect and nurture and build on. If we are not free, no one will respect us.
My second vision for India’s development. For fifty years we have been a developing nation.
It is time we see ourselves as a developed nation. We are among the top 5 nations of the world
in terms of GDP. We have a 10 percent growth rate in most areas. Our poverty levels are falling.
Our achievements are being globally recognised today. Yet we lack the self-confidence to
see ourselves as a developed nation, self-reliant and self-assured. Isn’t this incorrect?
I have a third vision. India must stand up to the world. Because I believe that unless India
stands up to the world, no one will respect us. Only strength respects strength. We must be
strong not only as a military power but also as an economic power. Both must go hand-in-hand.
My good fortune was to have worked with three great minds. Dr. <NAME> of the Dept. of
space, Professor <NAME>, who succeeded him and Dr. <NAME>, father of nuclear material.
I was lucky to have worked with all three of them closely and consider this the great opportunity of my life.
I see four milestones in my career"""
# -
# Cleaning the texts
import re
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from nltk.stem import WordNetLemmatizer
ps = PorterStemmer()
wordnet=WordNetLemmatizer()
sentences = nltk.sent_tokenize(paragraph)
corpus = []
for i in range(len(sentences)):
review = re.sub('[^a-zA-Z]', ' ', sentences[i])
review = review.lower()
review = review.split()
review = [wordnet.lemmatize(word) for word in review if not word in set(stopwords.words('english'))]
review = ' '.join(review)
corpus.append(review)
corpus
# Creating the TF-IDF model
from sklearn.feature_extraction.text import TfidfVectorizer
cv = TfidfVectorizer()
X = cv.fit_transform(corpus).toarray()
X.shape
| TF-IDF/Natural Language Processing TF-IDF.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### PyFVCOM plotting tools examples
#
# Here, we demonstrate plotting in three different dimensions: horizontal space, vertical space and time.
#
# First we load some model output into an object which can be passed to a number of plotting objects. These objects have methods for plotting different aspects of the data.
#
# For the horizontal plots, we plot the sea surface elevation across the model domain at a given time.
# For the vertical plots, we take a transect through the water column temperature data and plot it.
# For the time plots, we plot both a simple surface elevation time series and a time-varying water column temperature plot.
#
# %matplotlib inline
# Load an FVCOM model output and plot a surface.
from PyFVCOM.read import FileReader
from PyFVCOM.plot import Plotter, Time, Depth
from PyFVCOM.tide import make_water_column
from cmocean import cm
import matplotlib.pyplot as plt
# Create an object which holds the model outputs. We're only loading
# surface elevation and temperature for the first 200 time steps.
fvcom = FileReader('sample.nc', dims={'time': range(200)}, variables=['zeta', 'temp'])
# Make a plot of the surface elevation.
plot = Plotter(fvcom,
figsize=(20, 20),
res='i',
tick_inc=(4, 2),
cb_label='{} ({})'.format(fvcom.atts.zeta.long_name,
fvcom.atts.zeta.units),
cmap=cm.balance)
plot.plot_field(fvcom.data.zeta[5, :])
plot.axes.set_title(fvcom.time.datetime[5].strftime('%Y-%m-%d %H:%M:%S'))
# Plot a temperature transect between two locations.
positions = np.array(((-5, 50), (-4.5, 49.5)))
indices, distance = fvcom.horizontal_transect_nodes(positions)
plot = Depth(fvcom, figsize=(20, 9),
cb_label='Temperature ({})'.format(fvcom.ds.variables['temp'].units),
cmap=cm.thermal)
# fill_seabed makes the part of the plot below the seabed grey.
plot.plot_slice(distance / 1000, # to kilometres from metres
fvcom.grid.siglay_z[:, indices],
fvcom.data.temp[4, :, indices],
fill_seabed=True)
plot.axes.set_xlim(right=(distance / 1000).max()) # set the x-axis to the data range
plot.axes.set_xlabel('Distance (km)')
plot.axes.set_ylabel('Depth (m)')
# Save the figure.
plot.figure.savefig('temperature_profile.png')
# Do a time series at a specific location.
gauge = (-5, 55) # a sample (lon, lat) position
index = fvcom.closest_node(gauge)
time = Time(fvcom, figsize=(20, 9), title='{} at {}, {}'.format(fvcom.atts.zeta.long_name,
*gauge))
time.plot_line(fvcom.data.zeta[:, index], color='r')
time.axes.set_ylabel('{} ({})'.format(fvcom.atts.zeta.long_name,
fvcom.atts.zeta.units))
# Plot a depth-varying time profile through a water column
fvcom = FileReader('sample.nc', variables=['temp', 'zeta'], dims={'time': range(400), 'node': [5000]})
time = Time(fvcom, figsize=(20, 9), cb_label='{} ({})'.format(fvcom.atts.temp.long_name,
fvcom.atts.temp.units))
z = make_water_column(fvcom.data.zeta, fvcom.grid.h, fvcom.grid.siglay)
# fill_seabed makes the part of the plot below the seabed grey.
# We need to squeeze the data array since we've only extracted a single
# position.
time.plot_surface(z, np.squeeze(fvcom.data.temp), fill_seabed=True)
time.axes.set_ylabel('Depth (m)')
| examples/pyfvcom_plot_example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Quick-Start Guide
# **MLRun** is an end-to-end [open-source](https://github.com/mlrun/mlrun) MLOps solution to manage and automate your
# analytics and machine learning lifecycle, from data ingestion, through model development and full pipeline/model deployment, to model monitoring.
# Its primary goal is to ease the development of machine learning pipeline at scale and help organizations build a
# robust process for moving from the research phase to fully operational production deployments.
#
# MLRun is automating the process of moving code to production by implementing a **serverless** approach, where different tasks or services are executed over elastic **serverless functions** (read more about [MLRun functions](./runtimes/functions.md)), in this quick start guide we will use existing (marketplace) functions, see the [**tutorial**](./tutorial/index.md) with more detailed example of how to create and use functions.
# **Table of Contents**
# * [Working with MLRun](#working-with-mlrun)
# * [Train a Model](#train-a-model)
# * [Test the Model](#test-the-model)
# * [Serve the Model](#serve-the-model)
# ## Working with MLRun
# <a name="working-with-mlrun"></a>
# If you need to install MLRun, refer to the [Installation Guide](install.md).
# >**Note**: If you are using the [Iguazio MLOps Platform](https://www.iguazio.com/), MLRun already comes
# >preinstalled and integrated in your system.
#
# If you are not viewing this quick-start guide from a Jupyter Lab instance, open it on your cluster, create a
# new notebook, and copy the sections below to the notebook to run them.
# ### Set Environment
# Before you begin, initialize MLRun by calling `set_environment` and provide it with the project name. All the work will be saved and tracked under that project.
# +
import mlrun
project = mlrun.new_project('quick-start', user_project=True)
# -
# ## Train a Model
# <a name="train-a-model"></a>
# MLRun introduces the concept of [functions](./runtimes/functions.md). You can run your own code in functions, or use
# functions from the [function marketplace](https://www.mlrun.org/marketplace/). Functions can run locally or over elastic **"serverless"** engines (as containers over [kubernetes](https://kubernetes.io/)).
#
# In the example below, you'll use the [`sklearn_classifier`](https://github.com/mlrun/functions/tree/master/sklearn_classifier)
# from MLRun [function marketplace](https://www.mlrun.org/marketplace/) to train a model and use a sample dataset
# (CSV file) as the input. You can read more on how to [**use data items**](./store/datastore.md) from different data sources
# or from the [**Feature Store**](./feature-store/feature-store.md).
# **Note: When training a model in an air-gapped site** expand the cell below ..
# + [markdown] jupyter={"source_hidden": true} tags=["hide-cell"]
# > If you are working in MLRun:
# > 1. Download your data file and save it locally.
# > 2. Run:</br>
# > `import os`</br>
# > `os.environ["env_name"] = 1`
# > 2. Use the same command for the sample data set path, for example: <br>`source_url = mlrun.get_sample_path("data/iris/iris_dataset.csv")`
# >
# > If your system is integrated with an MLOps Platform:
# > 1. Download your data file and save it locally.
# > 2. In the UI, click the settings icon (<img src="./_static/images/icon-igz-settings.png" alt="Settings"/>) in the top-right of the header in any page to open the **Settings** dialog.
# > 2. Click **Environment variables | Create a new environment variable**, and set an environmental variable: SAMPLE_DATA_SOURCE_URL_PREFIX = the relative path to locally-stored data. For example: <br>`/v3io/bigdata/path/...`
# > 2. Use the same command for the sample data set path, for example: <br>`source_url = mlrun.get_sample_path("data/iris/iris_dataset.csv")`
# +
# import the training function from the marketplace (hub://)
train = mlrun.import_function('hub://sklearn_classifier')
# Get a sample dataset path (points to MLRun data samples repository)
source_url = mlrun.get_sample_path("data/iris/iris_dataset.csv")
# run the function and specify input dataset path and some parameters (algorithm and label column name)
train_run = train.run(name='train',
inputs={'dataset': source_url},
params={'model_pkg_class': 'sklearn.linear_model.LogisticRegression',
'label_column': 'label'})
# -
# The run output above contains a link to the MLRun UI. Click it to inspect the various aspects of the jobs you run:
#
# <img src="./_static/images/mlrun-quick-start/train-info.png" alt="ui-info" width="800"/>
# As well as their artifacts:
#
# <img src="./_static/images/mlrun-quick-start/train-artifacts.png" alt="ui-artifacts" width="800"/>
# When running the function in a Jupyter notebook, the output cell for your function execution contains a table with
# run information — including the state of the execution, all inputs and parameters, and the execution results and artifacts.
#
# 
# ## Test the Model
# <a name="test-the-model"></a>
# Now that you have a trained model, you can test it: run a task that uses the [test_classifier](https://github.com/mlrun/functions/tree/master/test_classifier)
# function from the function marketplace to run the selected trained model against the test dataset. The test dataset
# was returned from the training task (`train_run`) in the previous step.
test = mlrun.import_function('hub://test_classifier')
# You can then run the function as part of your project, just as any other function that you have written yourself.
# To view the function documentation, call the `doc` method:
test.doc()
# Configure parameters for the test function (`params`), and provide the selected trained model from the train task as an input artifact (`inputs`)
test_run = test.run(name="test",
params={"label_column": "label"},
inputs={"models_path": train_run.outputs['model'],
"test_set": train_run.outputs['test_set']})
# ## Serve the Model
# <a name="serve-the-model"></a>
# MLRun serving can take MLRun models or standard model files and produce managed, real-time, serverless functions using
# the [Nuclio real-time serverless framework](https://www.iguazio.com/open-source/nuclio/).
# Nuclio is built around data, I/O, and compute-intensive workloads, and is focused on performance and flexibility.
# Nuclio is also deeply integrated into the MLRun framework.
# See [MLRun Serving documentation](./serving/serving-graph.md) to learn more about the rich serving capabilities
# MLRun has to offer.
#
#
# To deploy your model using the [v2_model_server function](https://github.com/mlrun/functions/tree/master/v2_model_server),
# run the following code:
serve = mlrun.import_function('hub://v2_model_server')
model_name='iris'
serve.add_model(model_name, model_path=train_run.outputs['model'])
addr = serve.deploy()
# The `invoke` method enables to programmatically test the function.
# +
import json
inputs = [[5.1, 3.5, 1.4, 0.2],
[7.7, 3.8, 6.7, 2.2]]
my_data = json.dumps({'inputs': inputs})
serve.invoke(f'v2/models/{model_name}/infer', my_data)
# + [markdown] pycharm={"name": "#%% md\n"}
# Open the Nuclio UI to view the function and test it.
# -
# 
# <br>
#
# For a more detailed walk-through, refer to the [**getting-started tutorial**](tutorial/index.md).
| docs/quick-start.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src="https://raw.githubusercontent.com/melipass/umayor-ui-proyectos/main/logo-escuela.png" width="400" align="left" style="margin-bottom:20px;margin-right:20px;margin-top:25px">
#
# # Inteligencia Artificial
# ## Experiencia de Laboratorio 6: Perceptrón simple
#
# ## Objetivo del laboratorio
# Comprobar el funcionamiento de los modelos de redes neuronales correspondientes al perceptrón simple.
# ## Conceptos
# ### Perceptrón
# El perceptrón es la red neuronal más básica, de una sola capa y con una función de activación de escalón. El perceptrón se utiliza para la clasificación binaria de datos.
# ## Desarrollo
# ### 1. Implementar el perceptrón
#
# >Implementar un perceptrón simple y genérico en Python con una función de activación. Ajustar los pesos en cada iteración según corresponda, y entregar una salida acorde a la entrada recibida.
#
# Como primer paso, inicializamos el notebook con las librerías requeridas para trabajar. De ser necesario, hay que instalarlas.
# +
# #!pip install seaborn
# #!pip install numpy
# #!pip install matplotlib
# #!pip install pandas
# #!pip install graphviz
import seaborn as sn
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
plt.rcParams['figure.figsize'] = [6, 4]
plt.rcParams['figure.dpi'] = 100
import pandas as pd
import itertools
from graphviz import Digraph
from IPython.core.display import HTML
HTML("""
<style>
.output_png {
display: table-cell;
text-align: center;
vertical-align: middle;
}
</style>
""")
# -
# Según lo visto en clases, la función de activación del perceptrón es la de escalón, y será la utilizada en este notebook. Esta está definida de la siguiente manera:
#
# $$ \theta(z)= \left\{
# \begin{array}{ll}
# 1 & z \geq 0 \\
# 0 & z < 0 \\
# \end{array}
# \right. $$
#
# En donde el valor de $z$ corresponde a el valor ingresado en la función de activación $\theta$, y la salida de la función solo tiene dos posibles valores: $0$ y $1$.
# Función de activación del perceptrón
def theta(z):
return 1.0 if (z > 0) else 0.0
# Gráfico que representa la función de activación anterior
x = [-10,0,1,10]
y = [0,0,1,1]
plt.step(x, y)
plt.show()
# Usando esta función de activación, definiremos nuestro perceptrón como:
#
# $$
# y(\boldsymbol{x},\boldsymbol{w},b) = \theta(\boldsymbol{w}\cdot\boldsymbol{x}+b) = \theta(w_1x_1+w_2x_2+\dots+w_nx_n+b)
# $$
#
#
# Donde el vector $\boldsymbol{x}$ corresponde a las entradas del perceptrón y el vector $\boldsymbol{w}$ corresponde al peso asignado a cada uno. Esto se suma para después aplicar la función de activación, y queda nuestro perceptrón de la siguiente manera:
def perceptron(x,w,b):
entrada_theta = np.dot(w,x)+b
return theta(entrada_theta)
# El código de arriba contiene una función de producto punto que suma la multiplicación de cada elemento de los vectores, agregando el valor $b$ al final y almacenando el resultado en la variable ```entrada_theta```:
#
# $$\text{entrada_theta} = \left(\Sigma w_ix_i\right)+b$$
#
# Ese resultado se ingresa en la función de activación que definimos como $\theta$, la que luego nos devolverá el resultado del perceptrón. Visualmente, esto fluye así:
# +
nodos = ['x_1','x_2','⋮','x_n','w_1','w_2','⋮ ', 'w_n','Σ','θ','y']
vertices = {('x_1','w_1'),
('x_2','w_2'),
('⋮','⋮ '),
('x_n','w_n'),
('w_1','Σ'),
('w_2','Σ'),
('⋮ ','Σ'),
('w_n','Σ'),
('Σ','θ'),
('θ','y')}
dot = Digraph(node_attr={'shape':'circle','fixedsize':'true'},
graph_attr={'rankdir':'LR','label':'Perceptrón genérico','labelloc':'t','fontsize':'20'})
for nodo in nodos:
dot.node(nodo)
for vertice in vertices:
dot.edge(vertice[0],vertice[1])
dot
# -
# ### 2. Probar el perceptrón en AND, OR y XOR
# >Probar el perceptrón implementado para una compuerta lógica $\text{AND}$, $\text{OR}$ y $\text{XOR}$ con una entrada de tamaño 4.
#
# Para implementar perceptrones que simulen las compuertas lógicas $\text{AND}$, $\text{OR}$ y $\text{XOR}$ y que tengan cuatro entradas $x_1$, $x_2$, $x_3$ y $x_4$, debemos tomar el perceptrón y ajustarlo para que quede de la siguiente manera:
# $$
# y(\boldsymbol{x},\boldsymbol{w},b)=\theta(w_1x_1+w_2x_2+w_3x_3+w_4x_4+b)
# $$
#
# También, sabemos que existe un total de 16 posibles combinaciones de entradas para las compuertras con cuatro entradas, así que las dejaremos codificadas en un arreglo para probar en la parte 4:
l = [0,1]
x = [list(i) for i in itertools.product(l, repeat=4)]
print(x)
# #### 2.1. Compuerta AND
#
# La compuerta lógica $\text{AND}$ entrega el valor $1$ solo cuando todas sus entradas son $1$. Para que el perceptrón nos entregue el valor $1$ solo cuando todos los valores son $1$, debemos asignar valores a cada peso $w$ y al valor $b$, quedando así:
#
# $$
# \text{AND}(\text{x})=\theta(x_1+x_2+x_3+x_4-3.9)
# $$
#
# Donde decidimos dejar cada uno de los pesos $w_i = 1$ y el valor $b=-3.9$ para que la salida del perceptrón sea mayor a $0$ solo cuando todas las entradas valen $1$, teniendo que $1\cdot4-3.9=0.1$, y que $0.1>0 \Rightarrow y=1$.
def compuerta_AND(x):
w = [1,1,1,1]
b = -3.9
return perceptron(x,w,b)
# #### 2.2. Compuerta OR
# La compuerta lógica $\text{OR}$ también obtiene cuatro valores binarios y devuelve $1$ cuando existe al menos una entrada con valor $1$. El perceptrón queda así:
#
# $$
# \text{OR}(\text{x})=\theta(x_1+x_2+x_3+x_4-0.9)
# $$
#
# Donde decidimos dejar cada uno de los pesos $w_i = 1$ y el valor $b=-0.9$. Así, el perceptrón devolverá $1$ cuando exista al menos una entrada con valor $1$, ya que $1-0.9=0.1$ y sabemos que $0.1>0\Rightarrow y=1$.
def compuerta_OR(x):
w = [1,1,1,1]
b = -0.9
return perceptron(x,w,b)
# #### 2.3. Compuerta XOR
# En el caso de la compuerta $\text{XOR}$, sabemos que es necesario que nuestra compuerta devuelva el valor $0$ cuando todas las entradas son $0$ o $1$. Para ello, tuvimos que codificar la siguiente situación:
#
# - Encontrar la función que nos de $0$ solo cuando $\text{AND}$ nos devuelve $1$ (en otras palabras, hacer la compuerta $\text{NAND}$).
# - Poder combinar el resultado de la compuerta $\text{NAND}$ con el de la compuerta $\text{OR}$, es decir, crear un $\text{AND}$ con dos entradas.
#
# Una vez que identificamos esos dos requisitos creamos una función para cada uno, y así pudimos crear la compuerta $\text{XOR}$ que funciona satisfactoriamente. De esta forma, nos quedó la función $\text{XOR}$ como:
#
# $$
# \text{XOR}(\text{x})=\text{AND}(\text{NAND}(\text{x}),\text{OR}(\text{x})) = \text{AND}(\text{NAND}(x_1,x_2,x_3,x_4),\text{OR}(x_1,x_2,x_3,x_4))
# $$
# +
def compuerta_AND_2_entradas(x):
w = [1,1]
b = -1.9
return perceptron(x,w,b)
def compuerta_NAND(x):
w = [-1,-1,-1,-1]
b = 3.9
return perceptron(x,w,b)
def compuerta_XOR(x):
compuerta_1 = compuerta_NAND(x)
compuerta_2 = compuerta_OR(x)
return compuerta_AND_2_entradas([compuerta_1,compuerta_2])
# -
# Visualmente, las entradas pasan por las compuertas de la siguiente manera:
# +
nodos = ['x_1','x_2','Perceptrón NAND','Perceptrón OR','Perceptrón AND','y']
vertices = {('x_1','Perceptrón NAND'),
('x_2','Perceptrón NAND'),
('x_1','Perceptrón OR'),
('x_2','Perceptrón OR'),
('Perceptrón NAND','Perceptrón AND'),
('Perceptrón OR','Perceptrón AND'),
('Perceptrón AND','y')}
dot = Digraph(node_attr={'fixedsize':'true','width':'1.6'},
graph_attr={'rankdir':'LR','label':'Perceptrón XOR','labelloc':'t','fontsize':'20'})
for nodo in nodos:
dot.node(nodo)
for vertice in vertices:
dot.edge(vertice[0],vertice[1])
dot
# -
# ### 3. Probar el perceptrón con un dataset
# >Probar el perceptrón implementado para clasificar las entradas del dataset "letters.csv" determinando a qué letra corresponde cada entrada según las características recibidas.
#
# Primero que nada, cargamos los datos para ver con qué estamos trabajando.
df = pd.read_csv('letters.csv', header=None)
df
df[2].unique()
plt.scatter(df[0], df[1])#, color="orchid", marker="x", s=50)
plt.title("Dataset 'letters.csv'")
plt.xlabel('Valores $x_1$')
plt.ylabel('Valores $x_2$')
plt.xlim([-0.2, 1.2])
plt.ylim([-0.2, 1.2])
plt.show()
# Todos los valores numéricos de la tabla son positivos y no tenemos información de qué significa cada uno de ellos. Asumiremos que las dos primeras columnas de cada fila $i$ corresponderán a valores de entrada $x_1$ y $x_2$ y la tercera columna será la letra a clasificar, con valor ``o`` o ``l``.
# A continuación pasamos los datos del dataframe a un array:
datos = np.copy(df)
datos = np.array(datos)
# Creamos una función que nos devuelva un dataframe con la letra y la salida del perceptrón. Al perceptrón le daremos las dos columnas numéricas como entradas, un peso $\text{w}$ y un valor $b$ como variables para optimizar la clasificación en la sección 4.
def perceptron_datos(x,w,b):
df_datos = []
for valor in x:
df_datos.append([valor[2],perceptron([valor[0],valor[1]],w,b),valor[0],valor[1]])
return df_datos
# ### 4. Cálculo de error
# >Calcular el error obtenido en los pasos 2 y 3 durante el entrenamiento del perceptrón y expresarlo a través de un gráfico Iteración vs. Error.
# #### 4.1. Perceptrones de compuertas lógicas
# Para poder confirmar visualmente la existencia de un error dentro de nuestros perceptrones que implementan compuertas lógicas, decidimos hacer una función genérica para crear gráficos estilo 'serie de tiempo', donde podemos ver cómo va variando entre $0$ y $1$ el resultado de nuestros perceptrones a medida que vamos ingresando una lista de entradas. De esta manera, tendremos una guía visual de cuándo un perceptrón devuelve $0$ y cuándo devuelve $1$, y le entregaremos una función que marca el momento en el que se detecta un error para cada uno de los perceptrones.
def graficar_compuerta(x,compuerta,detector_error,titulo):
h = [0]
v = [0]
i = 1
error_en_titulo = ""
for posible_valor in x:
h.append(i)
i += 1
v.append(compuerta(posible_valor))
plt.plot(h, v)
plt.scatter(h, v, color="orchid", marker="x", s=50)
plt.xticks(h)
plt.yticks(v)
if detector_error(x,compuerta) != 1:
x_error = int(detector_error(x,compuerta))
plt.axvline(x = x_error+1, color = 'red')
error_en_titulo = " (Error)"
plt.title(titulo+error_en_titulo)
plt.show()
df = pd.DataFrame({'Entrada': x, 'Salida': v[1:]})
print(df)
# ##### 4.1.1. Prueba AND
#
# Sabemos que para un conjunto de entradas $x_n$, donde ninguna combinación de entradas se repite, la suma de todas las posibles salidas de la compuerta $\text{AND}$ jamás superará el valor $1$, cualquiera sea su dimensión. Teniendo esto presente, creamos la siguiente función:
# +
def error_compuerta_AND(x,compuerta):
i = 0
for posible_valor in x:
i += compuerta(posible_valor)
if i > 1:
print('Error: Existen entradas incorrectas o la lógica de la compuerta AND no es la correcta.')
return i
print("No se detectaron errores.")
return 1
error_compuerta_AND(x,compuerta_AND)
# -
# Tomando los valores que asignamos en la sección 2, obtenemos el siguiente gráfico y la siguiente tabla, donde en el eje x tendremos el índice de la combinación de entradas y en el eje y si esta corresponde a 0 o a 1, lo que nos permitirá confirmar si hay o no hay errores.
graficar_compuerta(x,compuerta_AND,error_compuerta_AND,'Salida del perceptrón para la compuerta AND')
# Para poner a prueba nuestro perceptrón que implementa una compuerta, usaremos dos conjuntos: uno con valores que no deberían ser aceptados, y otro con valores repetidos. La línea roja representará la iteración desde la cual comienza a estar erróneo nuestro perceptrón bajo las reglas establecidas de no repetición y admisión exclusiva de entradas $0$ y $1$, teniendo que los valores a la derecha de la línea ya no son correctos pues harán que la suma de todas las salidas sea mayor a $1$.
x_2_3 = [[2,3,3,2],[2,2,2,2],[3,3,3,3]]
x_repetido = [[1,1,1,1],[0,1,1,1],[1,1,1,1],[1,1,0,1],[1,1,1,1],[1,1,1,1],[1,1,1,1]]
error_compuerta_AND(x_2_3,compuerta_AND)
graficar_compuerta(x_2_3,compuerta_AND,error_compuerta_AND,'Salida del perceptrón para la compuerta AND')
error_compuerta_AND(x_repetido,compuerta_AND)
graficar_compuerta(x_repetido,compuerta_AND,error_compuerta_AND,'Salida del perceptrón para la compuerta AND')
# ##### 4.1.2. Prueba OR
# Para la prueba en el caso de la compuerta OR, sabemos que si existen $x^2$ posibles combinaciones de entradas (donde $x$ es la cantidad de entradas), siempre existirá una salida que jamás tendrá el valor $1$. Por lo tanto, haremos un chequeo para detectar un error cuando la suma de los resultados de $\text{OR}(x)$ sea igual o mayor que $x^2$, teniendo la misma regla del caso anterior en la que cada combinación solo puede existir una vez en nuestro arreglo.
#
# En el caso específico del perceptrón donde implementamos la compuerta $\text{OR}$, sabemos que hay cuatro entradas, por lo tanto la cantidad de salidas será $4^2=4\times4=16$.
def error_compuerta_OR(x,compuerta):
i = 0
for posible_valor in x:
i += compuerta(posible_valor)
if i >= (len(x[0])*len(x[0])):
print('Error: Existen entradas incorrectas o la lógica de la compuerta OR no es la correcta.')
return i
print("No se detectaron errores.")
return 1
graficar_compuerta(x,compuerta_OR,error_compuerta_OR,'Salida del perceptrón para la compuerta OR')
# En el ejemplo de arriba, la suma total da 15. Sin embargo, en el ejemplo de abajo, la suma total supera el valor de 15 y nuestro código insertará una línea roja desde el punto en el que el error aparece, considerando incorrecto el gráfico desde esa línea vertical hacia la derecha.
x_error_or = np.copy(x)
x_error_or = x_error_or.tolist()
x_error_or.append([1,2,1,1])
x_error_or.append([1,2,5,1])
graficar_compuerta(x_error_or,compuerta_OR,error_compuerta_OR,'Salida del perceptrón para la compuerta OR')
# ##### 4.1.3. Prueba XOR
# Bajo la misma lógica que la prueba anterior, sabemos que la suma de todas las salidas de $\text{XOR}$ siempre será $x^2-2$, donde $x$ corresponde a la cantidad de entradas. Teniendo esto presente, tomaremos el código para validar $\text{OR}$ y lo ajustaremos a $\text{XOR}$, mostrándonos un error cuando la suma de las salidas supera $x^2-2$.
def error_compuerta_XOR(x,compuerta):
i = 0
for posible_valor in x:
i += compuerta(posible_valor)
if i >= (len(x[0])*len(x[0])-1):
print('Error: Existen entradas incorrectas o la lógica de la compuerta XOR no es la correcta.')
return i
print("No se detectaron errores.")
return 1
graficar_compuerta(x,compuerta_XOR,error_compuerta_XOR,'Salida del perceptrón para la compuerta XOR')
# Si repetimos algunas combinaciones, nos encontraremos con que la suma de todas las salidas superará $x^2-2$ y nos dará error:
x_error_xor = np.copy(x)
x_error_xor = x_error_xor.tolist()
x_error_xor.remove([1,1,1,1])
x_error_xor.append([1,0,1,1])
x_error_xor.append([1,0,0,1])
x_error_xor.append([1,0,1,0])
x_error_xor.append([1,1,1,1])
graficar_compuerta(x_error_xor,compuerta_XOR,error_compuerta_XOR,'Salida del perceptrón para la compuerta XOR')
# #### 4.2. Perceptrón con el dataset 'letters.csv'
# Para trabajar con el perceptrón, usaremos la función definida en la sección 3 para poder armar un dataframe que contenga dos columnas: la letra clasificada, y la salida del perceptrón.
#
# En esa función debemos ingresar el conjunto de datos a usar, los pesos $\text{w}$ y también un valor $b$. Para sistematizar la búsqueda de valores óptimos para los pesos y para el valor $b$, sabemos que idealmente tendremos una clasificación 100% correcta para la letra ``o`` y la letra ``l``, lo que llevado a números equivale a $1.0$ para cada una, por lo tanto, si sumamos esto, sabemos que el valor ideal de la suma de los valores óptimos de ``o`` y ``l`` será $2.0$, e iremos probando con números cercanos a esa cifra:
def evaluacion_perceptron_datos(suma_optima_a_buscar):
for i in range(1,11):
for j in range(1,11):
for k in range(1,11):
for l in [-1,1]:
df_salida = pd.DataFrame(perceptron_datos(datos,[1/i,1/j],l*1/k), columns=['Letra','Salida','x_1','x_2'])
o_en_cero = 0
o_en_uno = 0
l_en_cero = 0
l_en_uno = 0
for valor in df_salida.values:
if (valor[0] == 'o'):
if valor[1] == 0.0:
o_en_cero += 1
else:
o_en_uno += 1
else:
if valor[1] == 0.0:
l_en_cero += 1
else:
l_en_uno += 1
if o_en_cero/(o_en_cero+o_en_uno) + l_en_uno/(l_en_cero+l_en_uno) > suma_optima_a_buscar:
print("¡Valores encontrados!")
print("Peso w_1: " + str(1/i))
print("Peso w_2: " + str(1/j))
print("Valor b: " + str(l*1/k))
# Buscamos primero si existen valores que nos entreguen la clasificación perfecta, es decir, sin errores:
evaluacion_perceptron_datos(2.0)
# Y al no recibir valores de vuelta, bajamos la cifra a $1.8$:
evaluacion_perceptron_datos(1.8)
# Sigue muy alto, así que probaremos con $1.7$:
evaluacion_perceptron_datos(1.7)
# Y en esta ocasión sí encontramos valores que nos darán muy poco error. Revisamos si podemos discriminar entre cuál de las dos combinaciones de valores del perceptrón será mejor y subimos un poco hasta $1.73$:
evaluacion_perceptron_datos(1.73)
# Y ahora que solo tenemos una única combinación de valores, los ingresaremos en nuestro perceptrón para obtener los mejores resultados posibles según los datos y el trabajo previo:
# +
peso_w_1 = 0.3333333333333333
peso_w_2 = 0.1
valor_b = -0.25
df_salida = perceptron_datos(datos,[peso_w_1,peso_w_2],valor_b)
# -
# Creamos la siguiente función para encontrar todos los valores importantes en la evaluación de nuestro modelo:
def checkearErrores(df_salida):
o_en_cero = 0
o_en_uno = 0
l_en_cero = 0
l_en_uno = 0
for valor in df_salida:
if (valor[0] == 'o'):
if valor[1] == 0.0:
o_en_cero += 1
else:
o_en_uno += 1
else:
if valor[1] == 0.0:
l_en_cero += 1
else:
l_en_uno += 1
print('La cantidad de letras "o" clasificadas como 0 es: ' + str(o_en_cero))
print('La cantidad de letras "o" clasificadas como 1 es: ' + str(o_en_uno))
print('La cantidad de letras "l" clasificadas como 0 es: ' + str(l_en_cero))
print('La cantidad de letras "l" clasificadas como 1 es: ' + str(l_en_uno))
precision_o = o_en_cero/(o_en_cero+o_en_uno)
print('La precisión para clasificar "o" como 0 es: ' + str(precision_o))
precision_l = l_en_uno/(l_en_cero+l_en_uno)
print('La precisión para clasificar "l" como 1 es: ' + str(precision_l))
error_o = 1-o_en_cero/(o_en_cero+o_en_uno)
print('El error para clasificar "o" como 0 es: ' + str(error_o))
error_l = 1-l_en_uno/(l_en_cero+l_en_uno)
print('El error para clasificar "l" como 1 es: ' + str(error_l))
return precision_o, precision_l, error_o, error_l
precision_o, precision_l, error_o, error_l = checkearErrores(df_salida)
# Y así obtuvimos que el error para clasificar la letra ``o`` como $0$ es $0.26\dots$, mientras que clasificar ``l`` como $1$ no entrega ningún error, por lo tanto es una clasificación perfecta en base a los datos usados. Gráficamente, podemos ver esto en una matriz de confusión:
array = [[precision_o,error_o],[error_l,precision_l]]
df_cm = pd.DataFrame(array, index = [i for i in "ol"],
columns = [i for i in "01"])
sn.heatmap(df_cm, annot=True, cmap="OrRd")
# Finalmente, graficándo esto en un plano, tenemos:
# +
df_salida = pd.DataFrame(df_salida)
y = df_salida[1]
x1 = df_salida[2]
x2 = df_salida[3]
color = ['r' if value == 1 else 'b' for value in y]
label = [['l'],['o']]
plt.scatter(x1, x2, marker='o', color=color)
plt.xlabel('Valores $x_1$')
plt.ylabel('Valores $x_2$')
plt.title('Clasificación dada por el perceptrón')
a = -peso_w_1/peso_w_2
xx = np.linspace(-5, 5)
yy = a * xx - valor_b/peso_w_2
plt.xlim([-0.2, 1.2])
plt.ylim([-0.2, 1.2])
plt.plot(xx,yy)
plt.show()
# -
# Donde para generar la línea divisora entre los dos tipos de valores clasificados, hubo que hacer el siguiente cálculo en base a los pesos $\text{w}$ y el valor $b$:
#
# $$
# y(x) = \frac{w_1 \cdot b \cdot x}{w_2^2} = \frac{-0.\bar{3}\cdot0.25\cdot x}{0.01}
# $$
#
#
# En base a estos resultados, queremos tener un perceptrón que se auto-corrija iterativamente. El objetivo es tener un algoritmo que encuentre los mismos pesos óptimos que identificamos en los pasos anteriores. Para ello, definiremos una nueva función con *hiperparámetros*. Un hiperparámetro es un parámetro usado para controlar el perceptrón, y que no ingresa en él. En este caso, entregaremos la razón de aprendizaje y la cantidad de iteraciones en las que el perceptrón se irá ajustando:
# +
datos = np.copy(df)
datos = np.array(datos)
def perceptron_iterativo_datos(datos,aprendizaje,iteraciones):
valores = datos[:,:-1]
letras = datos[:,-1]
m, n = valores.shape
w = np.zeros(shape=(n+1,1),dtype=float)
lista_errores = []
for iteracion in range(iteraciones):
numero_errores = 0
for indice, x_i in enumerate(valores):
#insertando 1 para hacer x_0 = 1 como en varios modelos
x_i = np.insert(x_i,0,1).reshape(-1,1)
# llamamos la función de inicialización del principio del cuaderno
y = theta(np.dot(x_i.T, w))
# transformamos las letras a valores numéricos para comparar
letras[indice] = 1.0 if (letras[indice] == 'l') else 0.0
# en caso de haber diferencia entre el valor entregado por la función
# de inicialización y el valor esperado, registramos el error
if(np.squeeze(y) - letras[indice]) != 0:
w += (aprendizaje*(letras[indice] - y*x_i)).astype(np.float64)
numero_errores += 1
lista_errores.append(numero_errores)
return w, lista_errores
# -
w, lista_errores = perceptron_iterativo_datos(datos,0.5,10)
print(w)
print(lista_errores)
# Arriba podemos ver los resultados entregados por el código, que nos entrega los pesos correspondientes a cada entrada (en este caso, también agregamos una columna $x_0 = 1$ y así pudimos encontrar un nuevo $b$). No son los mismos valores que en el método que usamos anteriormente, sin embargo, podemos ver a continuación cómo este nuevo método iterativo comienza en un valor por defecto que da bastantes errores, y con el paso de las iteraciones va acercándose más a cero errores en la clasificación de las letras ```l``` y ```o```, llegando a cero en la tercera iteración.
lista_iteraciones = np.arange(1, 11)
plt.plot(lista_iteraciones, lista_errores)
plt.xlabel('Iteraciones')
plt.ylabel('Errores')
plt.show()
# +
valores = datos[:,:-1]
x1 = np.linspace(-5, 5)
m = -w[1]*3.33333/(w[2])
c = -w[0]/(w[2])
x2 = m*x1 + c*0.906
plt.scatter(valores[:, 0][y==0], valores[:, 1][y==0])
plt.scatter(valores[:, 0][y==1], valores[:, 1][y==1])
plt.xlabel("Valores $x_1$")
plt.ylabel("Valores $x_2$")
plt.title('Clasificación dada por el perceptrón entrenado')
plt.xlim([-0.2, 1.2])
plt.ylim([-0.2, 1.2])
plt.plot(x1, x2, 'y-')
plt.show()
# -
# ### 5. Análisis
# >Analizar y concluír sobre los resultados obtenidos en los pasos 2, 3 y 4.
# <!-- Respecto a la compuerta AND y OR, sabemos que el perceptrón devolverá $1$ para cualquier valor entrante mayor o igual que $0$, así que decidimos asignarle el valor $-1.9$ a $b$ para que el resultado de la suma $x_1w_1+x_2w_2+b$ sea $0.1$ solo cuando se cumpla simultáneamente que $x_1=1$ y $x_2=1$. Al analizarlo con el código dentro del $\text{for}$ en la misma celda donde definimos la función, pudimos comprobar que la compuerta funciona correctamente. -->
# Al realizar cada uno de los pasos anteriores fuimos aprendiendo diferentes características de los perceptrones, y consideramos que tras realizar este laboratorio pudimos entender correctamente su funcionamiento y el porqué es importante que auto-ajuste sus hiperparámetros.
#
# Esta importancia la demostramos al intentar encontrar los pesos $w$ utilizando iteraciones ```for``` en Python para cada uno de los posibles valores que varían dentro de la función de nuestro perceptrón, y luego irnos por el camino correcto de generar un perceptrón que vaya ajustando por sí mismo estos valores en la función ```perceptron_iterativo_datos```, basándonos en información del sitio web *Towards Data Science*. Esta última función tiene muchísimas menos iteraciones para encontrar valores óptimos para una correcta clasificación en comparación con la cantidad de iteraciones que debimos ejecutar en un principio, siendo computacionalmente muy beneficiosa a pesar de llegar al mismo resultado. Eso sí, la función auto-ajustada nos generó una recta que no se condice adecuadamente con lo que realmente debería mostrar, ya que a pesar de obtener una clasificación adecuada tuvimos que realizar un pequeño ajuste para mostrar esa recta en el gráfico, multiplicando por 3.333 el $w_1$. No logramos identificar exactamente por qué ocurre esto, pero al menos identificamos qué pasaba y lo corregimos con el fin de una buena visualización de los resultados.
#
# Un desafío que tuvimos fue interpretar correctamente el conjunto de datos ```letters.csv```, y tras explorar un poco los datos identificamos a qué correspondía cada columna. Esto lo pudimos manejar correctamente en el perceptrón y los resultados van acorde a esos datos.
#
# Por otro lado, para las compuertas lógicas razonamos tal como si hubieramos realizado un circuito en la vida real, donde las entradas y las salidas son binarias y ninguna 'cuenta' más que otra, así que desde un principio tomamos la decisión de dar peso $1$ y un límite específico que debiera ser superado para devolver un $1$. Esto, basándonos en nuestro conocimiento previo adquirido en la asignatura *Arquitectura de Computadoras* de nuestra escuela. Decidimos identificar errores como una serie de entradas de posibles combinaciones porque se nos volvió más práctico de representar visualmente, y estamos muy satisfechos con las gráficas generadas pues conseguimos codificar funciones genéricas para encontrar perturbaciones en nuestro perceptrón que no deberían existir bajo las reglas que definimos.
#
# Para finalizar, nos gustaría mucho implementar varias mejoras y lograr optimizar el código para el siguiente laboratorio en base a lo aprendido en esta experiencia, que sin duda fue muy positiva para complementar con la práctica lo visto en clases y en donde logramos probar nuestras propias teorías e investigar lo que es aceptado por la comunidad y la academia.
| lab-6-perceptron.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"} tags=[] toc-hr-collapsed=false
# # Fuzzing with Grammars
#
# In the chapter on ["Mutation-Based Fuzzing"](MutationFuzzer.ipynb), we have seen how to use extra hints – such as sample input files – to speed up test generation. In this chapter, we take this idea one step further, by providing a _specification_ of the legal inputs to a program. Specifying inputs via a _grammar_ allows for very systematic and efficient test generation, in particular for complex input formats. Grammars also serve as the base for configuration fuzzing, API fuzzing, GUI fuzzing, and many more.
# + slideshow={"slide_type": "skip"}
from bookutils import YouTubeVideo
YouTubeVideo('Jc8Whz0W41o')
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"}
# **Prerequisites**
#
# * You should know how basic fuzzing works, e.g. from the [Chapter introducing fuzzing](Fuzzer.ipynb).
# * Knowledge on [mutation-based fuzzing](MutationFuzzer.ipynb) and [coverage](Coverage.ipynb) is _not_ required yet, but still recommended.
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"}
import bookutils
# + slideshow={"slide_type": "skip"}
from typing import List, Dict, Union, Any, Tuple, Optional
# + slideshow={"slide_type": "skip"}
import Fuzzer
# + [markdown] slideshow={"slide_type": "skip"}
# ## Synopsis
# <!-- Automatically generated. Do not edit. -->
#
# To [use the code provided in this chapter](Importing.ipynb), write
#
# ```python
# >>> from fuzzingbook.Grammars import <identifier>
# ```
#
# and then make use of the following features.
#
#
# This chapter introduces _grammars_ as a simple means to specify input languages, and to use them for testing programs with syntactically valid inputs. A grammar is defined as a mapping of nonterminal symbols to lists of alternative expansions, as in the following example:
#
# ```python
# >>> US_PHONE_GRAMMAR: Grammar = {
# >>> "<start>": ["<phone-number>"],
# >>> "<phone-number>": ["(<area>)<exchange>-<line>"],
# >>> "<area>": ["<lead-digit><digit><digit>"],
# >>> "<exchange>": ["<lead-digit><digit><digit>"],
# >>> "<line>": ["<digit><digit><digit><digit>"],
# >>> "<lead-digit>": ["2", "3", "4", "5", "6", "7", "8", "9"],
# >>> "<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
# >>> }
# >>>
# >>> assert is_valid_grammar(US_PHONE_GRAMMAR)
# ```
# Nonterminal symbols are enclosed in angle brackets (say, `<digit>`). To generate an input string from a grammar, a _producer_ starts with the start symbol (`<start>`) and randomly chooses a random expansion for this symbol. It continues the process until all nonterminal symbols are expanded. The function `simple_grammar_fuzzer()` does just that:
#
# ```python
# >>> [simple_grammar_fuzzer(US_PHONE_GRAMMAR) for i in range(5)]
# ['(692)449-5179',
# '(519)230-7422',
# '(613)761-0853',
# '(979)881-3858',
# '(810)914-5475']
# ```
# In practice, though, instead of `simple_grammar_fuzzer()`, you should use [the `GrammarFuzzer` class](GrammarFuzzer.ipynb) or one of its [coverage-based](GrammarCoverageFuzzer.ipynb), [probabilistic-based](ProbabilisticGrammarFuzzer.ipynb), or [generator-based](GeneratorGrammarFuzzer.ipynb) derivatives; these are more efficient, protect against infinite growth, and provide several additional features.
#
# This chapter also introduces a [grammar toolbox](#A-Grammar-Toolbox) with several helper functions that ease the writing of grammars, such as using shortcut notations for character classes and repetitions, or extending grammars
#
#
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Input Languages
#
# All possible behaviors of a program can be triggered by its input. "Input" here can be a wide range of possible sources: We are talking about data that is read from files, from the environment, or over the network, data input by the user, or data acquired from interaction with other resources. The set of all these inputs determines how the program will behave – including its failures. When testing, it is thus very helpful to think about possible input sources, how to get them under control, and _how to systematically test them_.
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# For the sake of simplicity, we will assume for now that the program has only one source of inputs; this is the same assumption we have been using in the previous chapters, too. The set of valid inputs to a program is called a _language_. Languages range from the simple to the complex: the CSV language denotes the set of valid comma-separated inputs, whereas the Python language denotes the set of valid Python programs. We commonly separate data languages and programming languages, although any program can also be treated as input data (say, to a compiler). The [Wikipedia page on file formats](https://en.wikipedia.org/wiki/List_of_file_formats) lists more than 1,000 different file formats, each of which is its own language.
# + [markdown] slideshow={"slide_type": "subslide"}
# To formally describe languages, the field of *formal languages* has devised a number of *language specifications* that describe a language. *Regular expressions* represent the simplest class of these languages to denote sets of strings: The regular expression `[a-z]*`, for instance, denotes a (possibly empty) sequence of lowercase letters. *Automata theory* connects these languages to automata that accept these inputs; *finite state machines*, for instance, can be used to specify the language of regular expressions.
# + [markdown] slideshow={"slide_type": "subslide"}
# Regular expressions are great for not-too-complex input formats, and the associated finite state machines have many properties that make them great for reasoning. To specify more complex inputs, though, they quickly encounter limitations. At the other end of the language spectrum, we have *universal grammars* that denote the language accepted by *Turing machines*. A Turing machine can compute anything that can be computed; and with Python being Turing-complete, this means that we can also use a Python program $p$ to specify or even enumerate legal inputs. But then, computer science theory also tells us that each such testing program has to be written specifically for the program to be tested, which is not the level of automation we want.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"} toc-hr-collapsed=true toc-nb-collapsed=true
# ## Grammars
#
# The middle ground between regular expressions and Turing machines is covered by *grammars*. Grammars are among the most popular (and best understood) formalisms to formally specify input languages. Using a grammar, one can express a wide range of the properties of an input language. Grammars are particularly great for expressing the *syntactical structure* of an input, and are the formalism of choice to express nested or recursive inputs. The grammars we use are so-called *context-free grammars*, one of the easiest and most popular grammar formalisms.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# ### Rules and Expansions
#
# A grammar consists of a *start symbol* and a set of *expansion rules* (or simply *rules*) which indicate how the start symbol (and other symbols) can be expanded. As an example, consider the following grammar, denoting a sequence of two digits:
#
# ```
# <start> ::= <digit><digit>
# <digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
# ```
#
# To read such a grammar, start with the start symbol (`<start>`). An expansion rule `<A> ::= <B>` means that the symbol on the left side (`<A>`) can be replaced by the string on the right side (`<B>`). In the above grammar, `<start>` would be replaced by `<digit><digit>`.
#
# In this string again, `<digit>` would be replaced by the string on the right side of the `<digit>` rule. The special operator `|` denotes *expansion alternatives* (or simply *alternatives*), meaning that any of the digits can be chosen for an expansion. Each `<digit>` thus would be expanded into one of the given digits, eventually yielding a string between `00` and `99`. There are no further expansions for `0` to `9`, so we are all set.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# The interesting thing about grammars is that they can be *recursive*. That is, expansions can make use of symbols expanded earlier – which would then be expanded again. As an example, consider a grammar that describes integers:
#
# ```
# <start> ::= <integer>
# <integer> ::= <digit> | <digit><integer>
# <digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
# ```
#
# Here, a `<integer>` is either a single digit, or a digit followed by another integer. The number `1234` thus would be represented as a single digit `1`, followed by the integer `234`, which in turn is a digit `2`, followed by the integer `34`.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# If we wanted to express that an integer can be preceded by a sign (`+` or `-`), we would write the grammar as
#
# ```
# <start> ::= <number>
# <number> ::= <integer> | +<integer> | -<integer>
# <integer> ::= <digit> | <digit><integer>
# <digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
# ```
#
# These rules formally define the language: Anything that can be derived from the start symbol is part of the language; anything that cannot is not.
# + slideshow={"slide_type": "skip"}
from bookutils import quiz
# + slideshow={"slide_type": "subslide"}
quiz("Which of these strings cannot be produced "
"from the above `<start>` symbol?",
[
"`007`",
"`-42`",
"`++1`",
"`3.14`"
], "[27 ** (1/3), 256 ** (1/4)]")
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# ### Arithmetic Expressions
#
# Let us expand our grammar to cover full *arithmetic expressions* – a poster child example for a grammar. We see that an expression (`<expr>`) is either a sum, or a difference, or a term; a term is either a product or a division, or a factor; and a factor is either a number or a parenthesized expression. Almost all rules can have recursion, and thus allow arbitrary complex expressions such as `(1 + 2) * (3.4 / 5.6 - 789)`.
#
# ```
# <start> ::= <expr>
# <expr> ::= <term> + <expr> | <term> - <expr> | <term>
# <term> ::= <term> * <factor> | <term> / <factor> | <factor>
# <factor> ::= +<factor> | -<factor> | (<expr>) | <integer> | <integer>.<integer>
# <integer> ::= <digit><integer> | <digit>
# <digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
# ```
#
# In such a grammar, if we start with `<start>` and then expand one symbol after another, randomly choosing alternatives, we can quickly produce one valid arithmetic expression after another. Such *grammar fuzzing* is highly effective as it comes to produce complex inputs, and this is what we will implement in this chapter.
# + slideshow={"slide_type": "subslide"}
quiz("Which of these strings cannot be produced "
"from the above `<start>` symbol?",
[
"`1 + 1`",
"`1+1`",
"`+1`",
"`+(1)`",
], "4 ** 0.5")
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Representing Grammars in Python
#
# Our first step in building a grammar fuzzer is to find an appropriate format for grammars. To make the writing of grammars as simple as possible, we use a format that is based on strings and lists. Our grammars in Python take the format of a _mapping_ between symbol names and expansions, where expansions are _lists_ of alternatives. A one-rule grammar for digits thus takes the form
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
DIGIT_GRAMMAR = {
"<start>":
["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Excursion: A `Grammar` Type
# + [markdown] slideshow={"slide_type": "fragment"}
# Let us define a type for grammars, such that we can check grammar types statically.
# + [markdown] slideshow={"slide_type": "fragment"}
# A first attempt at a grammar type would be that each symbol (a string) is mapped to a list of expansions (strings):
# + slideshow={"slide_type": "fragment"}
SimpleGrammar = Dict[str, List[str]]
# + [markdown] slideshow={"slide_type": "fragment"}
# However, our `opts()` feature for adding optional attributes, which we will introduce later in this chapter, also allows expansions to be _pairs_ that consist of strings and options, where options are mappings of strings to values:
# + slideshow={"slide_type": "fragment"}
Option = Dict[str, Any]
# + [markdown] slideshow={"slide_type": "fragment"}
# Hence, an expansion is either a string – or a pair of a string and an option.
# + slideshow={"slide_type": "fragment"}
Expansion = Union[str, Tuple[str, Option]]
# + [markdown] slideshow={"slide_type": "subslide"}
# With this, we can now define a `Grammar` as a mapping of strings to `Expansion` lists.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### End of Excursion
# + [markdown] slideshow={"slide_type": "fragment"}
# We can capture the grammar structure in a _`Grammar`_ type, in which each symbol (a string) is mapped to a list of expansions (strings):
# + slideshow={"slide_type": "fragment"}
Grammar = Dict[str, List[Expansion]]
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# With this `Grammar` type, the full grammar for arithmetic expressions looks like this:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
EXPR_GRAMMAR: Grammar = {
"<start>":
["<expr>"],
"<expr>":
["<term> + <expr>", "<term> - <expr>", "<term>"],
"<term>":
["<factor> * <term>", "<factor> / <term>", "<factor>"],
"<factor>":
["+<factor>",
"-<factor>",
"(<expr>)",
"<integer>.<integer>",
"<integer>"],
"<integer>":
["<digit><integer>", "<digit>"],
"<digit>":
["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# In the grammar, every symbol can be defined exactly once. We can access any rule by its symbol...
# + button=false code_folding=[] new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
EXPR_GRAMMAR["<digit>"]
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# ....and we can check whether a symbol is in the grammar:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
"<identifier>" in EXPR_GRAMMAR
# + [markdown] slideshow={"slide_type": "fragment"}
# Note that we assume that on the left hand side of a rule (i.e., the key in the mapping) is always a single symbol. This is the property that gives our grammars the characterization of _context-free_.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Some Definitions
# + [markdown] slideshow={"slide_type": "fragment"}
# We assume that the canonical start symbol is `<start>`:
# + slideshow={"slide_type": "fragment"}
START_SYMBOL = "<start>"
# + [markdown] slideshow={"slide_type": "subslide"}
# The handy `nonterminals()` function extracts the list of nonterminal symbols (i.e., anything between `<` and `>`, except spaces) from an expansion.
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"}
import re
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
RE_NONTERMINAL = re.compile(r'(<[^<> ]*>)')
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
def nonterminals(expansion):
# In later chapters, we allow expansions to be tuples,
# with the expansion being the first element
if isinstance(expansion, tuple):
expansion = expansion[0]
return RE_NONTERMINAL.findall(expansion)
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert nonterminals("<term> * <factor>") == ["<term>", "<factor>"]
assert nonterminals("<digit><integer>") == ["<digit>", "<integer>"]
assert nonterminals("1 < 3 > 2") == []
assert nonterminals("1 <3> 2") == ["<3>"]
assert nonterminals("1 + 2") == []
assert nonterminals(("<1>", {'option': 'value'})) == ["<1>"]
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# Likewise, `is_nonterminal()` checks whether some symbol is a nonterminal:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
def is_nonterminal(s):
return RE_NONTERMINAL.match(s)
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert is_nonterminal("<abc>")
assert is_nonterminal("<symbol-1>")
assert not is_nonterminal("+")
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## A Simple Grammar Fuzzer
#
# Let us now put the above grammars to use. We will build a very simple grammar fuzzer that starts with a start symbol (`<start>`) and then keeps on expanding it. To avoid expansion to infinite inputs, we place a limit (`max_nonterminals`) on the number of nonterminals. Furthermore, to avoid being stuck in a situation where we cannot reduce the number of symbols any further, we also limit the total number of expansion steps.
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"}
import random
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
class ExpansionError(Exception):
pass
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
def simple_grammar_fuzzer(grammar: Grammar,
start_symbol: str = START_SYMBOL,
max_nonterminals: int = 10,
max_expansion_trials: int = 100,
log: bool = False) -> str:
"""Produce a string from `grammar`.
`start_symbol`: use a start symbol other than `<start>` (default).
`max_nonterminals`: the maximum number of nonterminals
still left for expansion
`max_expansion_trials`: maximum # of attempts to produce a string
`log`: print expansion progress if True"""
term = start_symbol
expansion_trials = 0
while len(nonterminals(term)) > 0:
symbol_to_expand = random.choice(nonterminals(term))
expansions = grammar[symbol_to_expand]
expansion = random.choice(expansions)
# In later chapters, we allow expansions to be tuples,
# with the expansion being the first element
if isinstance(expansion, tuple):
expansion = expansion[0]
new_term = term.replace(symbol_to_expand, expansion, 1)
if len(nonterminals(new_term)) < max_nonterminals:
term = new_term
if log:
print("%-40s" % (symbol_to_expand + " -> " + expansion), term)
expansion_trials = 0
else:
expansion_trials += 1
if expansion_trials >= max_expansion_trials:
raise ExpansionError("Cannot expand " + repr(term))
return term
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# Let us see how this simple grammar fuzzer obtains an arithmetic expression from the start symbol:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
simple_grammar_fuzzer(grammar=EXPR_GRAMMAR, max_nonterminals=3, log=True)
# + [markdown] slideshow={"slide_type": "subslide"}
# By increasing the limit of nonterminals, we can quickly get much longer productions:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
for i in range(10):
print(simple_grammar_fuzzer(grammar=EXPR_GRAMMAR, max_nonterminals=5))
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# Note that while fuzzer does the job in most cases, it has a number of drawbacks.
# + slideshow={"slide_type": "fragment"}
quiz("What drawbacks does `simple_grammar_fuzzer()` have?",
[
"It has a large number of string search and replace operations",
"It may fail to produce a string (`ExpansionError`)",
"It often picks some symbol to expand "
"that does not even occur in the string",
"All of the above"
], "1 << 2")
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# Indeed, `simple_grammar_fuzzer()` is rather inefficient due to the large number of search and replace operations, and it may even fail to produce a string. On the other hand, the implementation is straightforward and does the job in most cases. For this chapter, we'll stick to it; in the [next chapter](GrammarFuzzer.ipynb), we'll show how to build a more efficient one.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Visualizing Grammars as Railroad Diagrams
# + [markdown] slideshow={"slide_type": "fragment"}
# With grammars, we can easily specify the format for several of the examples we discussed earlier. The above arithmetic expressions, for instance, can be directly sent into `bc` (or any other program that takes arithmetic expressions). Before we introduce a few additional grammars, let us give a means to _visualize_ them, giving an alternate view to aid their understanding.
# + [markdown] slideshow={"slide_type": "fragment"}
# _Railroad diagrams_, also called _syntax diagrams_, are a graphical representation of context-free grammars. They are read left to right, following possible "rail" tracks; the sequence of symbols encountered on the track defines the language. To produce railroad diagrams, we implement a function `syntax_diagram()`.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Excursion: Implementing `syntax_diagram()`
# + [markdown] slideshow={"slide_type": "fragment"}
# We use [RailroadDiagrams](RailroadDiagrams.ipynb), an external library for visualization.
# + slideshow={"slide_type": "skip"}
from RailroadDiagrams import NonTerminal, Terminal, Choice, HorizontalChoice, Sequence
from RailroadDiagrams import show_diagram
# + slideshow={"slide_type": "skip"}
from IPython.display import SVG
# + [markdown] slideshow={"slide_type": "fragment"}
# We first define the method `syntax_diagram_symbol()` to visualize a given symbol. Terminal symbols are denoted as ovals, whereas nonterminal symbols (such as `<term>`) are denoted as rectangles.
# + slideshow={"slide_type": "fragment"}
def syntax_diagram_symbol(symbol: str) -> Any:
if is_nonterminal(symbol):
return NonTerminal(symbol[1:-1])
else:
return Terminal(symbol)
# + slideshow={"slide_type": "fragment"}
SVG(show_diagram(syntax_diagram_symbol('<term>')))
# + [markdown] slideshow={"slide_type": "fragment"}
# We define `syntax_diagram_expr()` to visualize expansion alternatives.
# + slideshow={"slide_type": "subslide"}
def syntax_diagram_expr(expansion: Expansion) -> Any:
# In later chapters, we allow expansions to be tuples,
# with the expansion being the first element
if isinstance(expansion, tuple):
expansion = expansion[0]
symbols = [sym for sym in re.split(RE_NONTERMINAL, expansion) if sym != ""]
if len(symbols) == 0:
symbols = [""] # special case: empty expansion
return Sequence(*[syntax_diagram_symbol(sym) for sym in symbols])
# + slideshow={"slide_type": "fragment"}
SVG(show_diagram(syntax_diagram_expr(EXPR_GRAMMAR['<term>'][0])))
# + [markdown] slideshow={"slide_type": "fragment"}
# This is the first alternative of `<term>` – a `<factor>` followed by `*` and a `<term>`.
# + [markdown] slideshow={"slide_type": "subslide"}
# Next, we define `syntax_diagram_alt()` for displaying alternate expressions.
# + slideshow={"slide_type": "skip"}
from itertools import zip_longest
# + slideshow={"slide_type": "fragment"}
def syntax_diagram_alt(alt: List[Expansion]) -> Any:
max_len = 5
alt_len = len(alt)
if alt_len > max_len:
iter_len = alt_len // max_len
alts = list(zip_longest(*[alt[i::iter_len] for i in range(iter_len)]))
exprs = [[syntax_diagram_expr(expr) for expr in alt
if expr is not None] for alt in alts]
choices = [Choice(len(expr) // 2, *expr) for expr in exprs]
return HorizontalChoice(*choices)
else:
return Choice(alt_len // 2, *[syntax_diagram_expr(expr) for expr in alt])
# + slideshow={"slide_type": "subslide"}
SVG(show_diagram(syntax_diagram_alt(EXPR_GRAMMAR['<digit>'])))
# + [markdown] slideshow={"slide_type": "fragment"}
# We see that a `<digit>` can be any single digit from `0` to `9`.
# + [markdown] slideshow={"slide_type": "fragment"}
# Finally, we define `syntax_diagram()` which given a grammar, displays the syntax diagram of its rules.
# + slideshow={"slide_type": "fragment"}
def syntax_diagram(grammar: Grammar) -> None:
from IPython.display import SVG, display
for key in grammar:
print("%s" % key[1:-1])
display(SVG(show_diagram(syntax_diagram_alt(grammar[key]))))
# + [markdown] slideshow={"slide_type": "subslide"}
# ### End of Excursion
# + [markdown] slideshow={"slide_type": "fragment"}
# Let us use `syntax_diagram()` to produce a railroad diagram of our expression grammar:
# + slideshow={"slide_type": "subslide"}
syntax_diagram(EXPR_GRAMMAR)
# + [markdown] slideshow={"slide_type": "subslide"}
# This railroad representation will come in handy as it comes to visualizing the structure of grammars – especially for more complex grammars.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"} toc-hr-collapsed=false
# ## Some Grammars
#
# Let us create (and visualize) some more grammars and use them for fuzzing.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# ### A CGI Grammar
#
# Here's a grammar for `cgi_decode()` introduced in the [chapter on coverage](Coverage.ipynb).
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
CGI_GRAMMAR: Grammar = {
"<start>":
["<string>"],
"<string>":
["<letter>", "<letter><string>"],
"<letter>":
["<plus>", "<percent>", "<other>"],
"<plus>":
["+"],
"<percent>":
["%<hexdigit><hexdigit>"],
"<hexdigit>":
["0", "1", "2", "3", "4", "5", "6", "7",
"8", "9", "a", "b", "c", "d", "e", "f"],
"<other>": # Actually, could be _all_ letters
["0", "1", "2", "3", "4", "5", "a", "b", "c", "d", "e", "-", "_"],
}
# + slideshow={"slide_type": "subslide"}
syntax_diagram(CGI_GRAMMAR)
# + [markdown] slideshow={"slide_type": "subslide"}
# In contrast to [basic fuzzing](Fuzzer.ipynb) or [mutation-based fuzzing](MutationFuzzer.ipynb), the grammar quickly produces all sorts of combinations:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
for i in range(10):
print(simple_grammar_fuzzer(grammar=CGI_GRAMMAR, max_nonterminals=10))
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# ### A URL Grammar
#
# The same properties we have seen for CGI input also hold for more complex inputs. Let us use a grammar to produce a large number of valid URLs:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
URL_GRAMMAR: Grammar = {
"<start>":
["<url>"],
"<url>":
["<scheme>://<authority><path><query>"],
"<scheme>":
["http", "https", "ftp", "ftps"],
"<authority>":
["<host>", "<host>:<port>", "<userinfo>@<host>", "<userinfo>@<host>:<port>"],
"<host>": # Just a few
["cispa.saarland", "www.google.com", "fuzzingbook.com"],
"<port>":
["80", "8080", "<nat>"],
"<nat>":
["<digit>", "<digit><digit>"],
"<digit>":
["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
"<userinfo>": # Just one
["user:password"],
"<path>": # Just a few
["", "/", "/<id>"],
"<id>": # Just a few
["abc", "def", "x<digit><digit>"],
"<query>":
["", "?<params>"],
"<params>":
["<param>", "<param>&<params>"],
"<param>": # Just a few
["<id>=<id>", "<id>=<nat>"],
}
# + slideshow={"slide_type": "subslide"}
syntax_diagram(URL_GRAMMAR)
# + [markdown] slideshow={"slide_type": "subslide"}
# Again, within milliseconds, we can produce plenty of valid inputs.
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
for i in range(10):
print(simple_grammar_fuzzer(grammar=URL_GRAMMAR, max_nonterminals=10))
# + [markdown] slideshow={"slide_type": "subslide"}
# ### A Natural Language Grammar
#
# Finally, grammars are not limited to *formal languages* such as computer inputs, but can also be used to produce *natural language*. This is the grammar we used to pick a title for this book:
# + slideshow={"slide_type": "subslide"}
TITLE_GRAMMAR: Grammar = {
"<start>": ["<title>"],
"<title>": ["<topic>: <subtopic>"],
"<topic>": ["Generating Software Tests", "<fuzzing-prefix>Fuzzing", "The Fuzzing Book"],
"<fuzzing-prefix>": ["", "The Art of ", "The Joy of "],
"<subtopic>": ["<subtopic-main>",
"<subtopic-prefix><subtopic-main>",
"<subtopic-main><subtopic-suffix>"],
"<subtopic-main>": ["Breaking Software",
"Generating Software Tests",
"Principles, Techniques and Tools"],
"<subtopic-prefix>": ["", "Tools and Techniques for "],
"<subtopic-suffix>": [" for <reader-property> and <reader-property>",
" for <software-property> and <software-property>"],
"<reader-property>": ["Fun", "Profit"],
"<software-property>": ["Robustness", "Reliability", "Security"],
}
# + slideshow={"slide_type": "subslide"}
syntax_diagram(TITLE_GRAMMAR)
# + slideshow={"slide_type": "skip"}
from typing import Set
# + slideshow={"slide_type": "subslide"}
titles: Set[str] = set()
while len(titles) < 10:
titles.add(simple_grammar_fuzzer(
grammar=TITLE_GRAMMAR, max_nonterminals=10))
titles
# + [markdown] slideshow={"slide_type": "subslide"}
# (If you find that there is redundancy ("Robustness and Robustness") in here: In [our chapter on coverage-based fuzzing](GrammarCoverageFuzzer.ipynb), we will show how to cover each expansion only once. And if you like some alternatives more than others, [probabilistic grammar fuzzing](ProbabilisticGrammarFuzzer.ipynb) will be there for you.)
# + [markdown] slideshow={"slide_type": "slide"}
# ## Grammars as Mutation Seeds
# + [markdown] slideshow={"slide_type": "slide"}
# One very useful property of grammars is that they produce mostly valid inputs. From a syntactical standpoint, the inputs are actually _always_ valid, as they satisfy the constraints of the given grammar. (Of course, one needs a valid grammar in the first place.) However, there are also _semantical_ properties that cannot be easily expressed in a grammar. If, say, for a URL, the port range is supposed to be between 1024 and 2048, this is hard to write in a grammar. If one has to satisfy more complex constraints, one quickly reaches the limits of what a grammar can express.
# + [markdown] slideshow={"slide_type": "fragment"}
# One way around this is to attach constraints to grammars, as we will discuss [later in this book](ConstraintFuzzer.ipynb). Another possibility is to put together the strengths of grammar-based fuzzing and [mutation-based fuzzing](MutationFuzzer.ipynb). The idea is to use the grammar-generated inputs as *seeds* for further mutation-based fuzzing. This way, we can explore not only _valid_ inputs, but also check out the _boundaries_ between valid and invalid inputs. This is particularly interesting as slightly invalid inputs allow to find parser errors (which are often abundant). As with fuzzing in general, it is the unexpected which reveals errors in programs.
# + [markdown] slideshow={"slide_type": "subslide"}
# To use our generated inputs as seeds, we can feed them directly into the mutation fuzzers introduced earlier:
# + slideshow={"slide_type": "skip"}
from MutationFuzzer import MutationFuzzer # minor dependency
# + slideshow={"slide_type": "fragment"}
number_of_seeds = 10
seeds = [
simple_grammar_fuzzer(
grammar=URL_GRAMMAR,
max_nonterminals=10) for i in range(number_of_seeds)]
seeds
# + slideshow={"slide_type": "subslide"}
m = MutationFuzzer(seeds)
# + slideshow={"slide_type": "fragment"}
[m.fuzz() for i in range(20)]
# + [markdown] slideshow={"slide_type": "subslide"}
# While the first 10 `fuzz()` calls return the seeded inputs (as designed), the later ones again create arbitrary mutations. Using `MutationCoverageFuzzer` instead of `MutationFuzzer`, we could again have our search guided by coverage – and thus bring together the best of multiple worlds.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"} toc-hr-collapsed=false
# ## A Grammar Toolbox
#
# Let us now introduce a few techniques that help us writing grammars.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# ### Escapes
#
# With `<` and `>` delimiting nonterminals in our grammars, how can we actually express that some input should contain `<` and `>`? The answer is simple: Just introduce a symbol for them.
# + slideshow={"slide_type": "fragment"}
simple_nonterminal_grammar: Grammar = {
"<start>": ["<nonterminal>"],
"<nonterminal>": ["<left-angle><identifier><right-angle>"],
"<left-angle>": ["<"],
"<right-angle>": [">"],
"<identifier>": ["id"] # for now
}
# + [markdown] slideshow={"slide_type": "fragment"}
# In `simple_nonterminal_grammar`, neither the expansion for `<left-angle>` nor the expansion for `<right-angle>` can be mistaken as a nonterminal. Hence, we can produce as many as we want.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Extending Grammars
#
# In the course of this book, we frequently run into the issue of creating a grammar by _extending_ an existing grammar with new features. Such an extension is very much like subclassing in object-oriented programming.
# + [markdown] slideshow={"slide_type": "fragment"}
# To create a new grammar $g'$ from an existing grammar $g$, we first copy $g$ into $g'$, and then go and extend existing rules with new alternatives and/or add new symbols. Here's an example, extending the above `nonterminal` grammar with a better rule for identifiers:
# + slideshow={"slide_type": "skip"}
import copy
# + slideshow={"slide_type": "fragment"}
nonterminal_grammar = copy.deepcopy(simple_nonterminal_grammar)
nonterminal_grammar["<identifier>"] = ["<idchar>", "<identifier><idchar>"]
nonterminal_grammar["<idchar>"] = ['a', 'b', 'c', 'd'] # for now
# + slideshow={"slide_type": "subslide"}
nonterminal_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# Since such an extension of grammars is a common operation, we introduce a custom function `extend_grammar()` which first copies the given grammar and then updates it from a dictionary, using the Python dictionary `update()` method:
# + slideshow={"slide_type": "fragment"}
def extend_grammar(grammar: Grammar, extension: Grammar = {}) -> Grammar:
new_grammar = copy.deepcopy(grammar)
new_grammar.update(extension)
return new_grammar
# + [markdown] slideshow={"slide_type": "subslide"}
# This call to `extend_grammar()` extends `simple_nonterminal_grammar` to `nonterminal_grammar` just like the "manual" example above:
# + slideshow={"slide_type": "fragment"}
nonterminal_grammar = extend_grammar(simple_nonterminal_grammar,
{
"<identifier>": ["<idchar>", "<identifier><idchar>"],
# for now
"<idchar>": ['a', 'b', 'c', 'd']
}
)
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Character Classes
# + [markdown] slideshow={"slide_type": "fragment"}
# In the above `nonterminal_grammar`, we have enumerated only the first few letters; indeed, enumerating all letters or digits in a grammar manually, as in `<idchar> ::= 'a' | 'b' | 'c' ...` is a bit painful.
# + [markdown] slideshow={"slide_type": "fragment"}
# However, remember that grammars are part of a program, and can thus also be constructed programmatically. We introduce a function `srange()` which constructs a list of characters in a string:
# + slideshow={"slide_type": "fragment"}
import string
# + slideshow={"slide_type": "fragment"}
def srange(characters: str) -> List[Expansion]:
"""Construct a list with all characters in the string"""
return [c for c in characters]
# + [markdown] slideshow={"slide_type": "fragment"}
# If we pass it the constant `string.ascii_letters`, which holds all ASCII letters, `srange()` returns a list of all ASCII letters:
# + slideshow={"slide_type": "fragment"}
string.ascii_letters
# + slideshow={"slide_type": "fragment"}
srange(string.ascii_letters)[:10]
# + [markdown] slideshow={"slide_type": "subslide"}
# We can use such constants in our grammar to quickly define identifiers:
# + slideshow={"slide_type": "fragment"}
nonterminal_grammar = extend_grammar(nonterminal_grammar,
{
"<idchar>": (srange(string.ascii_letters) +
srange(string.digits) +
srange("-_"))
}
)
# + slideshow={"slide_type": "fragment"}
[simple_grammar_fuzzer(nonterminal_grammar, "<identifier>") for i in range(10)]
# + [markdown] slideshow={"slide_type": "subslide"}
# The shortcut `crange(start, end)` returns a list of all characters in the ASCII range of `start` to (including) `end`:
# + slideshow={"slide_type": "fragment"}
def crange(character_start: str, character_end: str) -> List[Expansion]:
return [chr(i)
for i in range(ord(character_start), ord(character_end) + 1)]
# + [markdown] slideshow={"slide_type": "fragment"}
# We can use this to express ranges of characters:
# + slideshow={"slide_type": "fragment"}
crange('0', '9')
# + slideshow={"slide_type": "fragment"}
assert crange('a', 'z') == srange(string.ascii_lowercase)
# + [markdown] slideshow={"slide_type": "subslide"} toc-hr-collapsed=false
# ### Grammar Shortcuts
# + [markdown] slideshow={"slide_type": "fragment"}
# In the above `nonterminal_grammar`, as in other grammars, we have to express repetitions of characters using _recursion_, that is, by referring to the original definition:
# + slideshow={"slide_type": "fragment"}
nonterminal_grammar["<identifier>"]
# + [markdown] slideshow={"slide_type": "fragment"}
# It could be a bit easier if we simply could state that a nonterminal should be a non-empty sequence of letters – for instance, as in
#
# ```
# <identifier> = <idchar>+
# ```
#
# where `+` denotes a non-empty repetition of the symbol it follows.
# + [markdown] slideshow={"slide_type": "subslide"}
# Operators such as `+` are frequently introduced as handy _shortcuts_ in grammars. Formally, our grammars come in the so-called [Backus-Naur form](https://en.wikipedia.org/wiki/Backus-Naur_form), or *BNF* for short. Operators _extend_ BNF to so-called _extended BNF*, or *EBNF* for short:
#
# * The form `<symbol>?` indicates that `<symbol>` is optional – that is, it can occur 0 or 1 times.
# * The form `<symbol>+` indicates that `<symbol>` can occur 1 or more times repeatedly.
# * The form `<symbol>*` indicates that `<symbol>` can occur 0 or more times. (In other words, it is an optional repetition.)
#
# To make matters even more interesting, we would like to use _parentheses_ with the above shortcuts. Thus, `(<foo><bar>)?` indicates that the sequence of `<foo>` and `<bar>` is optional.
# + [markdown] slideshow={"slide_type": "subslide"}
# Using such operators, we can define the identifier rule in a simpler way. To this end, let us create a copy of the original grammar and modify the `<identifier>` rule:
# + slideshow={"slide_type": "fragment"}
nonterminal_ebnf_grammar = extend_grammar(nonterminal_grammar,
{
"<identifier>": ["<idchar>+"]
}
)
# + [markdown] slideshow={"slide_type": "subslide"}
# Likewise, we can simplify the expression grammar. Consider how signs are optional, and how integers can be expressed as sequences of digits.
# + slideshow={"slide_type": "fragment"}
EXPR_EBNF_GRAMMAR: Grammar = {
"<start>":
["<expr>"],
"<expr>":
["<term> + <expr>", "<term> - <expr>", "<term>"],
"<term>":
["<factor> * <term>", "<factor> / <term>", "<factor>"],
"<factor>":
["<sign>?<factor>", "(<expr>)", "<integer>(.<integer>)?"],
"<sign>":
["+", "-"],
"<integer>":
["<digit>+"],
"<digit>":
srange(string.digits)
}
# + [markdown] slideshow={"slide_type": "subslide"}
# Let us implement a function `convert_ebnf_grammar()` that takes such an EBNF grammar and automatically translates it into a BNF grammar.
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Excursion: Implementing `convert_ebnf_grammar()`
# + [markdown] slideshow={"slide_type": "subslide"}
# Our aim is to convert EBNF grammars such as the ones above into a regular BNF grammar. This is done by four rules:
#
# 1. An expression `(content)op`, where `op` is one of `?`, `+`, `*`, becomes `<new-symbol>op`, with a new rule `<new-symbol> ::= content`.
# 2. An expression `<symbol>?` becomes `<new-symbol>`, where `<new-symbol> ::= <empty> | <symbol>`.
# 3. An expression `<symbol>+` becomes `<new-symbol>`, where `<new-symbol> ::= <symbol> | <symbol><new-symbol>`.
# 4. An expression `<symbol>*` becomes `<new-symbol>`, where `<new-symbol> ::= <empty> | <symbol><new-symbol>`.
#
# Here, `<empty>` expands to the empty string, as in `<empty> ::= `. (This is also called an *epsilon expansion*.)
# + [markdown] slideshow={"slide_type": "fragment"}
# If these operators remind you of _regular expressions_, this is not by accident: Actually, any basic regular expression can be converted into a grammar using the above rules (and character classes with `crange()`, as defined above).
# + [markdown] slideshow={"slide_type": "subslide"}
# Applying these rules on the examples above yields the following results:
#
# * `<idchar>+` becomes `<idchar><new-symbol>` with `<new-symbol> ::= <idchar> | <idchar><new-symbol>`.
# * `<integer>(.<integer>)?` becomes `<integer><new-symbol>` with `<new-symbol> ::= <empty> | .<integer>`.
# + [markdown] slideshow={"slide_type": "skip"}
# Let us implement these rules in three steps.
# + [markdown] slideshow={"slide_type": "subslide"}
# ##### Creating New Symbols
#
# First, we need a mechanism to create new symbols. This is fairly straightforward.
# + slideshow={"slide_type": "fragment"}
def new_symbol(grammar: Grammar, symbol_name: str = "<symbol>") -> str:
"""Return a new symbol for `grammar` based on `symbol_name`"""
if symbol_name not in grammar:
return symbol_name
count = 1
while True:
tentative_symbol_name = symbol_name[:-1] + "-" + repr(count) + ">"
if tentative_symbol_name not in grammar:
return tentative_symbol_name
count += 1
# + slideshow={"slide_type": "fragment"}
assert new_symbol(EXPR_EBNF_GRAMMAR, '<expr>') == '<expr-1>'
# + [markdown] slideshow={"slide_type": "subslide"}
# ##### Expanding Parenthesized Expressions
# + [markdown] slideshow={"slide_type": "fragment"}
# Next, we need a means to extract parenthesized expressions from our expansions and expand them according to the rules above. Let's start with extracting expressions:
# + slideshow={"slide_type": "fragment"}
RE_PARENTHESIZED_EXPR = re.compile(r'\([^()]*\)[?+*]')
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
def parenthesized_expressions(expansion: Expansion) -> List[str]:
# In later chapters, we allow expansions to be tuples,
# with the expansion being the first element
if isinstance(expansion, tuple):
expansion = expansion[0]
return re.findall(RE_PARENTHESIZED_EXPR, expansion)
# + slideshow={"slide_type": "fragment"}
assert parenthesized_expressions("(<foo>)* (<foo><bar>)+ (+<foo>)? <integer>(.<integer>)?") == [
'(<foo>)*', '(<foo><bar>)+', '(+<foo>)?', '(.<integer>)?']
# + [markdown] slideshow={"slide_type": "subslide"}
# We can now use these to apply rule number 1, above, introducing new symbols for expressions in parentheses.
# + slideshow={"slide_type": "fragment"}
def convert_ebnf_parentheses(ebnf_grammar: Grammar) -> Grammar:
"""Convert a grammar in extended BNF to BNF"""
grammar = extend_grammar(ebnf_grammar)
for nonterminal in ebnf_grammar:
expansions = ebnf_grammar[nonterminal]
for i in range(len(expansions)):
expansion = expansions[i]
if not isinstance(expansion, str):
expansion = expansion[0]
while True:
parenthesized_exprs = parenthesized_expressions(expansion)
if len(parenthesized_exprs) == 0:
break
for expr in parenthesized_exprs:
operator = expr[-1:]
contents = expr[1:-2]
new_sym = new_symbol(grammar)
exp = grammar[nonterminal][i]
opts = None
if isinstance(exp, tuple):
(exp, opts) = exp
assert isinstance(exp, str)
expansion = exp.replace(expr, new_sym + operator, 1)
if opts:
grammar[nonterminal][i] = (expansion, opts)
else:
grammar[nonterminal][i] = expansion
grammar[new_sym] = [contents]
return grammar
# + [markdown] slideshow={"slide_type": "subslide"}
# This does the conversion as sketched above:
# + slideshow={"slide_type": "fragment"}
convert_ebnf_parentheses({"<number>": ["<integer>(.<integer>)?"]})
# + [markdown] slideshow={"slide_type": "fragment"}
# It even works for nested parenthesized expressions:
# + slideshow={"slide_type": "fragment"}
convert_ebnf_parentheses({"<foo>": ["((<foo>)?)+"]})
# + [markdown] slideshow={"slide_type": "subslide"}
# ##### Expanding Operators
#
# After expanding parenthesized expressions, we now need to take care of symbols followed by operators (`?`, `*`, `+`). As with `convert_ebnf_parentheses()`, above, we first extract all symbols followed by an operator.
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
RE_EXTENDED_NONTERMINAL = re.compile(r'(<[^<> ]*>[?+*])')
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
def extended_nonterminals(expansion: Expansion) -> List[str]:
# In later chapters, we allow expansions to be tuples,
# with the expansion being the first element
if isinstance(expansion, tuple):
expansion = expansion[0]
return re.findall(RE_EXTENDED_NONTERMINAL, expansion)
# + slideshow={"slide_type": "fragment"}
assert extended_nonterminals(
"<foo>* <bar>+ <elem>? <none>") == ['<foo>*', '<bar>+', '<elem>?']
# + [markdown] slideshow={"slide_type": "subslide"}
# Our converter extracts the symbol and the operator, and adds new symbols according to the rules laid out above.
# + slideshow={"slide_type": "fragment"}
def convert_ebnf_operators(ebnf_grammar: Grammar) -> Grammar:
"""Convert a grammar in extended BNF to BNF"""
grammar = extend_grammar(ebnf_grammar)
for nonterminal in ebnf_grammar:
expansions = ebnf_grammar[nonterminal]
for i in range(len(expansions)):
expansion = expansions[i]
extended_symbols = extended_nonterminals(expansion)
for extended_symbol in extended_symbols:
operator = extended_symbol[-1:]
original_symbol = extended_symbol[:-1]
assert original_symbol in ebnf_grammar, \
f"{original_symbol} is not defined in grammar"
new_sym = new_symbol(grammar, original_symbol)
exp = grammar[nonterminal][i]
opts = None
if isinstance(exp, tuple):
(exp, opts) = exp
assert isinstance(exp, str)
new_exp = exp.replace(extended_symbol, new_sym, 1)
if opts:
grammar[nonterminal][i] = (new_exp, opts)
else:
grammar[nonterminal][i] = new_exp
if operator == '?':
grammar[new_sym] = ["", original_symbol]
elif operator == '*':
grammar[new_sym] = ["", original_symbol + new_sym]
elif operator == '+':
grammar[new_sym] = [
original_symbol, original_symbol + new_sym]
return grammar
# + slideshow={"slide_type": "subslide"}
convert_ebnf_operators({"<integer>": ["<digit>+"], "<digit>": ["0"]})
# + [markdown] slideshow={"slide_type": "subslide"} tags=[]
# ##### All Together
#
# We can combine the two, first extending parentheses and then operators:
# + slideshow={"slide_type": "fragment"}
def convert_ebnf_grammar(ebnf_grammar: Grammar) -> Grammar:
return convert_ebnf_operators(convert_ebnf_parentheses(ebnf_grammar))
# + [markdown] slideshow={"slide_type": "subslide"}
# #### End of Excursion
# + [markdown] slideshow={"slide_type": "fragment"}
# Here's an example of using `convert_ebnf_grammar()`:
# + slideshow={"slide_type": "fragment"}
convert_ebnf_grammar({"<authority>": ["(<userinfo>@)?<host>(:<port>)?"]})
# + slideshow={"slide_type": "subslide"}
expr_grammar = convert_ebnf_grammar(EXPR_EBNF_GRAMMAR)
expr_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# Success! We have nicely converted the EBNF grammar into BNF.
# + [markdown] slideshow={"slide_type": "fragment"}
# With character classes and EBNF grammar conversion, we have two powerful tools that make the writing of grammars easier. We will use these again and again as it comes to working with grammars.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Grammar Extensions
# + [markdown] slideshow={"slide_type": "fragment"}
# During the course of this book, we frequently want to specify _additional information_ for grammars, such as [_probabilities_](ProbabilisticGrammarFuzzer.ipynb) or [_constraints_](GeneratorGrammarFuzzer.ipynb). To support these extensions, as well as possibly others, we define an _annotation_ mechanism.
# + [markdown] slideshow={"slide_type": "subslide"}
# Our concept for annotating grammars is to add _annotations_ to individual expansions. To this end, we allow that an expansion cannot only be a string, but also a _pair_ of a string and a set of attributes, as in
#
# ```python
# "<expr>":
# [("<term> + <expr>", opts(min_depth=10)),
# ("<term> - <expr>", opts(max_depth=2)),
# "<term>"]
# ```
#
# Here, the `opts()` function would allow us to express annotations that apply to the individual expansions; in this case, the addition would be annotated with a `min_depth` value of 10, and the subtraction with a `max_depth` value of 2. The meaning of these annotations is left to the individual algorithms dealing with the grammars; the general idea, though, is that they can be ignored.
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Excursion: Implementing `opts()`
# + [markdown] slideshow={"slide_type": "fragment"}
# Our `opts()` helper function returns a mapping of its arguments to values:
# + slideshow={"slide_type": "fragment"}
def opts(**kwargs: Any) -> Dict[str, Any]:
return kwargs
# + slideshow={"slide_type": "fragment"}
opts(min_depth=10)
# + [markdown] slideshow={"slide_type": "fragment"}
# To deal with both expansion strings and pairs of expansions and annotations, we access the expansion string and the associated annotations via designated helper functions, `exp_string()` and `exp_opts()`:
# + slideshow={"slide_type": "fragment"}
def exp_string(expansion: Expansion) -> str:
"""Return the string to be expanded"""
if isinstance(expansion, str):
return expansion
return expansion[0]
# + slideshow={"slide_type": "subslide"}
exp_string(("<term> + <expr>", opts(min_depth=10)))
# + slideshow={"slide_type": "fragment"}
def exp_opts(expansion: Expansion) -> Dict[str, Any]:
"""Return the options of an expansion. If options are not defined, return {}"""
if isinstance(expansion, str):
return {}
return expansion[1]
# + slideshow={"slide_type": "fragment"}
def exp_opt(expansion: Expansion, attribute: str) -> Any:
"""Return the given attribution of an expansion.
If attribute is not defined, return None"""
return exp_opts(expansion).get(attribute, None)
# + slideshow={"slide_type": "fragment"}
exp_opts(("<term> + <expr>", opts(min_depth=10)))
# + slideshow={"slide_type": "fragment"}
exp_opt(("<term> - <expr>", opts(max_depth=2)), 'max_depth')
# + [markdown] slideshow={"slide_type": "subslide"}
# Finally, we define a helper function that sets a particular option:
# + slideshow={"slide_type": "subslide"}
def set_opts(grammar: Grammar, symbol: str, expansion: Expansion,
opts: Option = {}) -> None:
"""Set the options of the given expansion of grammar[symbol] to opts"""
expansions = grammar[symbol]
for i, exp in enumerate(expansions):
if exp_string(exp) != exp_string(expansion):
continue
new_opts = exp_opts(exp)
if opts == {} or new_opts == {}:
new_opts = opts
else:
for key in opts:
new_opts[key] = opts[key]
if new_opts == {}:
grammar[symbol][i] = exp_string(exp)
else:
grammar[symbol][i] = (exp_string(exp), new_opts)
return
raise KeyError(
"no expansion " +
repr(symbol) +
" -> " +
repr(
exp_string(expansion)))
# + [markdown] slideshow={"slide_type": "subslide"}
# #### End of Excursion
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Checking Grammars
#
# Since grammars are represented as strings, it is fairly easy to introduce errors. So let us introduce a helper function that checks a grammar for consistency.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# The helper function `is_valid_grammar()` iterates over a grammar to check whether all used symbols are defined, and vice versa, which is very useful for debugging; it also checks whether all symbols are reachable from the start symbol. You don't have to delve into details here, but as always, it is important to get the input data straight before we make use of it.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Excursion: Implementing `is_valid_grammar()`
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"}
import sys
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
def def_used_nonterminals(grammar: Grammar, start_symbol:
str = START_SYMBOL) -> Tuple[Optional[Set[str]],
Optional[Set[str]]]:
"""Return a pair (`defined_nonterminals`, `used_nonterminals`) in `grammar`.
In case of error, return (`None`, `None`)."""
defined_nonterminals = set()
used_nonterminals = {start_symbol}
for defined_nonterminal in grammar:
defined_nonterminals.add(defined_nonterminal)
expansions = grammar[defined_nonterminal]
if not isinstance(expansions, list):
print(repr(defined_nonterminal) + ": expansion is not a list",
file=sys.stderr)
return None, None
if len(expansions) == 0:
print(repr(defined_nonterminal) + ": expansion list empty",
file=sys.stderr)
return None, None
for expansion in expansions:
if isinstance(expansion, tuple):
expansion = expansion[0]
if not isinstance(expansion, str):
print(repr(defined_nonterminal) + ": "
+ repr(expansion) + ": not a string",
file=sys.stderr)
return None, None
for used_nonterminal in nonterminals(expansion):
used_nonterminals.add(used_nonterminal)
return defined_nonterminals, used_nonterminals
# + slideshow={"slide_type": "fragment"}
def reachable_nonterminals(grammar: Grammar,
start_symbol: str = START_SYMBOL) -> Set[str]:
reachable = set()
def _find_reachable_nonterminals(grammar, symbol):
nonlocal reachable
reachable.add(symbol)
for expansion in grammar.get(symbol, []):
for nonterminal in nonterminals(expansion):
if nonterminal not in reachable:
_find_reachable_nonterminals(grammar, nonterminal)
_find_reachable_nonterminals(grammar, start_symbol)
return reachable
# + slideshow={"slide_type": "fragment"}
def unreachable_nonterminals(grammar: Grammar,
start_symbol=START_SYMBOL) -> Set[str]:
return grammar.keys() - reachable_nonterminals(grammar, start_symbol)
# + slideshow={"slide_type": "fragment"}
def opts_used(grammar: Grammar) -> Set[str]:
used_opts = set()
for symbol in grammar:
for expansion in grammar[symbol]:
used_opts |= set(exp_opts(expansion).keys())
return used_opts
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
def is_valid_grammar(grammar: Grammar,
start_symbol: str = START_SYMBOL,
supported_opts: Set[str] = set()) -> bool:
"""Check if the given `grammar` is valid.
`start_symbol`: optional start symbol (default: `<start>`)
`supported_opts`: options supported (default: none)"""
defined_nonterminals, used_nonterminals = \
def_used_nonterminals(grammar, start_symbol)
if defined_nonterminals is None or used_nonterminals is None:
return False
# Do not complain about '<start>' being not used,
# even if start_symbol is different
if START_SYMBOL in grammar:
used_nonterminals.add(START_SYMBOL)
for unused_nonterminal in defined_nonterminals - used_nonterminals:
print(repr(unused_nonterminal) + ": defined, but not used",
file=sys.stderr)
for undefined_nonterminal in used_nonterminals - defined_nonterminals:
print(repr(undefined_nonterminal) + ": used, but not defined",
file=sys.stderr)
# Symbols must be reachable either from <start> or given start symbol
unreachable = unreachable_nonterminals(grammar, start_symbol)
msg_start_symbol = start_symbol
if START_SYMBOL in grammar:
unreachable = unreachable - \
reachable_nonterminals(grammar, START_SYMBOL)
if start_symbol != START_SYMBOL:
msg_start_symbol += " or " + START_SYMBOL
for unreachable_nonterminal in unreachable:
print(repr(unreachable_nonterminal) + ": unreachable from " + msg_start_symbol,
file=sys.stderr)
used_but_not_supported_opts = set()
if len(supported_opts) > 0:
used_but_not_supported_opts = opts_used(
grammar).difference(supported_opts)
for opt in used_but_not_supported_opts:
print(
"warning: option " +
repr(opt) +
" is not supported",
file=sys.stderr)
return used_nonterminals == defined_nonterminals and len(unreachable) == 0
# + [markdown] slideshow={"slide_type": "subslide"}
# ### End of Excursion
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# Let us make use of `is_valid_grammar()`. Our grammars defined above pass the test:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert is_valid_grammar(EXPR_GRAMMAR)
assert is_valid_grammar(CGI_GRAMMAR)
assert is_valid_grammar(URL_GRAMMAR)
# + [markdown] slideshow={"slide_type": "fragment"}
# The check can also be applied to EBNF grammars:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert is_valid_grammar(EXPR_EBNF_GRAMMAR)
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# These ones do not pass the test, though:
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert not is_valid_grammar({"<start>": ["<x>"], "<y>": ["1"]}) # type: ignore
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert not is_valid_grammar({"<start>": "123"}) # type: ignore
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert not is_valid_grammar({"<start>": []}) # type: ignore
# + button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
assert not is_valid_grammar({"<start>": [1, 2, 3]}) # type: ignore
# + [markdown] slideshow={"slide_type": "fragment"}
# (The `#type: ignore` annotations avoid static checkers flagging the above as errors).
# + [markdown] slideshow={"slide_type": "fragment"}
# From here on, we will always use `is_valid_grammar()` when defining a grammar.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Synopsis
#
# This chapter introduces _grammars_ as a simple means to specify input languages, and to use them for testing programs with syntactically valid inputs. A grammar is defined as a mapping of nonterminal symbols to lists of alternative expansions, as in the following example:
# + slideshow={"slide_type": "subslide"}
US_PHONE_GRAMMAR: Grammar = {
"<start>": ["<phone-number>"],
"<phone-number>": ["(<area>)<exchange>-<line>"],
"<area>": ["<lead-digit><digit><digit>"],
"<exchange>": ["<lead-digit><digit><digit>"],
"<line>": ["<digit><digit><digit><digit>"],
"<lead-digit>": ["2", "3", "4", "5", "6", "7", "8", "9"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
assert is_valid_grammar(US_PHONE_GRAMMAR)
# + [markdown] slideshow={"slide_type": "subslide"}
# Nonterminal symbols are enclosed in angle brackets (say, `<digit>`). To generate an input string from a grammar, a _producer_ starts with the start symbol (`<start>`) and randomly chooses a random expansion for this symbol. It continues the process until all nonterminal symbols are expanded. The function `simple_grammar_fuzzer()` does just that:
# + slideshow={"slide_type": "fragment"}
[simple_grammar_fuzzer(US_PHONE_GRAMMAR) for i in range(5)]
# + [markdown] slideshow={"slide_type": "subslide"}
# In practice, though, instead of `simple_grammar_fuzzer()`, you should use [the `GrammarFuzzer` class](GrammarFuzzer.ipynb) or one of its [coverage-based](GrammarCoverageFuzzer.ipynb), [probabilistic-based](ProbabilisticGrammarFuzzer.ipynb), or [generator-based](GeneratorGrammarFuzzer.ipynb) derivatives; these are more efficient, protect against infinite growth, and provide several additional features.
# + [markdown] slideshow={"slide_type": "fragment"}
# This chapter also introduces a [grammar toolbox](#A-Grammar-Toolbox) with several helper functions that ease the writing of grammars, such as using shortcut notations for character classes and repetitions, or extending grammars
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Lessons Learned
#
# * Grammars are powerful tools to express and produce syntactically valid inputs.
# * Inputs produced from grammars can be used as is, or used as seeds for mutation-based fuzzing.
# * Grammars can be extended with character classes and operators to make writing easier.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Next Steps
#
# As they make a great foundation for generating software tests, we use grammars again and again in this book. As a sneak preview, we can use grammars to [fuzz configurations](ConfigurationFuzzer.ipynb):
#
# ```
# <options> ::= <option>*
# <option> ::= -h | --version | -v | -d | -i | --global-config <filename>
# ```
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# We can use grammars for [fuzzing functions and APIs](APIFuzzer.ipynb) and [fuzzing graphical user interfaces](WebFuzzer.ipynb):
#
# ```
# <call-sequence> ::= <call>*
# <call> ::= urlparse(<url>) | urlsplit(<url>)
# ```
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# We can assign [probabilities](ProbabilisticGrammarFuzzer.ipynb) and [constraints](GeneratorGrammarFuzzer.ipynb) to individual expansions:
#
# ```
# <term>: 50% <factor> * <term> | 30% <factor> / <term> | 20% <factor>
# <integer>: <digit>+ { <integer> >= 100 }
# ```
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# All these extras become especially valuable as we can
#
# 1. _infer grammars automatically_, dropping the need to specify them manually, and
# 2. _guide them towards specific goals_ such as coverage or critical functions;
#
# which we also discuss for all techniques in this book.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# To get there, however, we still have bit of homework to do. In particular, we first have to learn how to
#
# * [create an efficient grammar fuzzer](GrammarFuzzer.ipynb)
# + [markdown] slideshow={"slide_type": "slide"}
# ## Background
#
# As one of the foundations of human language, grammars have been around as long as human language existed. The first _formalization_ of generative grammars was by <NAME> in 350 BC \cite{Panini350bce}. As a general means to express formal languages for both data and programs, their role in computer science cannot be overstated. The seminal work by Chomsky \cite{Chomsky1956} introduced the central models of regular languages, context-free grammars, context-sensitive grammars, and universal grammars as they are used (and taught) in computer science as a means to specify input and programming languages ever since.
# + [markdown] slideshow={"slide_type": "subslide"}
# The use of grammars for _producing_ test inputs goes back to Burkhardt \cite{Burkhardt1967}, to be later rediscovered and applied by Hanford \cite{Hanford1970} and Purdom \cite{Purdom1972}. The most important use of grammar testing since then has been *compiler testing*. Actually, grammar-based testing is one important reason why compilers and Web browsers work as they should:
#
# * The [CSmith](https://embed.cs.utah.edu/csmith/) tool \cite{Yang2011} specifically targets C programs, starting with a C grammar and then applying additional steps, such as referring to variables and functions defined earlier or ensuring integer and type safety. Their authors have used it "to find and report more than 400 previously unknown compiler bugs."
#
# * The [LangFuzz](http://issta2016.cispa.saarland/interview-with-christian-holler/) work \cite{Holler2012}, which shares two authors with this book, uses a generic grammar to produce outputs, and is used day and night to generate JavaScript programs and test their interpreters; as of today, it has found more than 2,600 bugs in browsers such as Mozilla Firefox, Google Chrome, and Microsoft Edge.
#
# * The [EMI Project](http://web.cs.ucdavis.edu/~su/emi-project/) \cite{Le2014} uses grammars to stress-test C compilers, transforming known tests into alternative programs that should be semantically equivalent over all inputs. Again, this has led to more than 100 bugs in C compilers being fixed.
#
# * [Grammarinator](https://github.com/renatahodovan/grammarinator) \cite{Hodovan2018} is an open-source grammar fuzzer (written in Python!), using the popular ANTLR format as grammar specification. Like LangFuzz, it uses the grammar for both parsing and producing, and has found more than 100 issues in the *JerryScript* lightweight JavaScript engine and an associated platform.
#
# * [Domato](https://github.com/googleprojectzero/domato) is a generic grammar generation engine that is specifically used for fuzzing DOM input. It has revealed a number of security issues in popular Web browsers.
# + [markdown] slideshow={"slide_type": "subslide"}
# Compilers and Web browsers, of course, are not only domains where grammars are needed for testing, but also domains where grammars are well-known. Our claim in this book is that grammars can be used to generate almost _any_ input, and our aim is to empower you to do precisely that.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"} toc-hr-collapsed=true toc-nb-collapsed=true
# ## Exercises
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# ### Exercise 1: A JSON Grammar
#
# Take a look at the [JSON specification](http://www.json.org) and derive a grammar from it:
#
# * Use _character classes_ to express valid characters
# * Use EBNF to express repetitions and optional parts
# * Assume that
# - a string is a sequence of digits, ASCII letters, punctuation and space characters without quotes or escapes
# - whitespace is just a single space.
# * Use `is_valid_grammar()` to ensure the grammar is valid.
#
# Feed the grammar into `simple_grammar_fuzzer()`. Do you encounter any errors, and why?
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** This is a fairly straightforward translation:
# + slideshow={"slide_type": "skip"} solution2="hidden"
CHARACTERS_WITHOUT_QUOTE = (string.digits
+ string.ascii_letters
+ string.punctuation.replace('"', '').replace('\\', '')
+ ' ')
# + slideshow={"slide_type": "skip"} solution2="hidden"
JSON_EBNF_GRAMMAR: Grammar = {
"<start>": ["<json>"],
"<json>": ["<element>"],
"<element>": ["<ws><value><ws>"],
"<value>": ["<object>", "<array>", "<string>", "<number>",
"true", "false", "null", "'; DROP TABLE STUDENTS"],
"<object>": ["{<ws>}", "{<members>}"],
"<members>": ["<member>(,<members>)*"],
"<member>": ["<ws><string><ws>:<element>"],
"<array>": ["[<ws>]", "[<elements>]"],
"<elements>": ["<element>(,<elements>)*"],
"<element>": ["<ws><value><ws>"],
"<string>": ['"' + "<characters>" + '"'],
"<characters>": ["<character>*"],
"<character>": srange(CHARACTERS_WITHOUT_QUOTE),
"<number>": ["<int><frac><exp>"],
"<int>": ["<digit>", "<onenine><digits>", "-<digit>", "-<onenine><digits>"],
"<digits>": ["<digit>+"],
"<digit>": ['0', "<onenine>"],
"<onenine>": crange('1', '9'),
"<frac>": ["", ".<digits>"],
"<exp>": ["", "E<sign><digits>", "e<sign><digits>"],
"<sign>": ["", '+', '-'],
# "<ws>": srange(string.whitespace)
"<ws>": [" "]
}
assert is_valid_grammar(JSON_EBNF_GRAMMAR)
# + slideshow={"slide_type": "skip"} solution2="hidden"
JSON_GRAMMAR = convert_ebnf_grammar(JSON_EBNF_GRAMMAR)
# + slideshow={"slide_type": "skip"} solution2="hidden"
from ExpectError import ExpectError
# + slideshow={"slide_type": "skip"} solution2="hidden"
for i in range(50):
with ExpectError():
print(simple_grammar_fuzzer(JSON_GRAMMAR, '<object>'))
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# We get these errors because `simple_grammar_fuzzer()` first expands to a maximum number of elements, and then is limited because every further expansion would _increase_ the number of nonterminals, even though these may eventually reduce the string length. This issue is addressed in the [next chapter](GrammarFuzzer.ipynb), introducing a more solid algorithm for producing strings from grammars.
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# ### Exercise 2: Finding Bugs
#
# The name `simple_grammar_fuzzer()` does not come by accident: The way it expands grammars is limited in several ways. What happens if you apply `simple_grammar_fuzzer()` on `nonterminal_grammar` and `expr_grammar`, as defined above, and why?
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution**. `nonterminal_grammar` does not work because `simple_grammar_fuzzer()` eventually tries to expand the just generated nonterminal:
# + slideshow={"slide_type": "skip"} solution2="hidden"
from ExpectError import ExpectError, ExpectTimeout
# + slideshow={"slide_type": "skip"} solution2="hidden"
with ExpectError():
simple_grammar_fuzzer(nonterminal_grammar, log=True)
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# For `expr_grammar`, things are even worse, as `simple_grammar_fuzzer()` can start a series of infinite expansions:
# + slideshow={"slide_type": "skip"} solution2="hidden"
with ExpectTimeout(1):
for i in range(10):
print(simple_grammar_fuzzer(expr_grammar))
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# Both issues are addressed and discussed in the [next chapter](GrammarFuzzer.ipynb), introducing a more solid algorithm for producing strings from grammars.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Exercise 3: Grammars with Regular Expressions
#
# In a _grammar extended with regular expressions_, we can use the special form
# ```
# /regex/
# ```
# to include regular expressions in expansions. For instance, we can have a rule
# ```
# <integer> ::= /[+-]?[0-9]+/
# ```
# to quickly express that an integer is an optional sign, followed by a sequence of digits.
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# #### Part 1: Convert regular expressions
#
# Write a converter `convert_regex(r)` that takes a regular expression `r` and creates an equivalent grammar. Support the following regular expression constructs:
#
# * `*`, `+`, `?`, `()` should work just in EBNFs, above.
# * `a|b` should translate into a list of alternatives `[a, b]`.
# * `.` should match any character except newline.
# * `[abc]` should translate into `srange("abc")`
# * `[^abc]` should translate into the set of ASCII characters _except_ `srange("abc")`.
# * `[a-b]` should translate into `crange(a, b)`
# * `[^a-b]` should translate into the set of ASCII characters _except_ `crange(a, b)`.
#
# Example: `convert_regex(r"[0-9]+")` should yield a grammar such as
# ```python
# {
# "<start>": ["<s1>"],
# "<s1>": [ "<s2>", "<s1><s2>" ],
# "<s2>": crange('0', '9')
# }
# ```
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** Left as exercise to the reader.
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Part 2: Identify and expand regular expressions
#
# Write a converter `convert_regex_grammar(g)` that takes a EBNF grammar `g` containing regular expressions in the form `/.../` and creates an equivalent BNF grammar. Support the regular expression constructs as above.
#
# Example: `convert_regex_grammar({ "<integer>" : "/[+-]?[0-9]+/" })` should yield a grammar such as
# ```python
# {
# "<integer>": ["<s1><s3>"],
# "<s1>": [ "", "<s2>" ],
# "<s2>": srange("+-"),
# "<s3>": [ "<s4>", "<s4><s3>" ],
# "<s4>": crange('0', '9')
# }
# ```
# + [markdown] slideshow={"slide_type": "fragment"} solution2="hidden" solution2_first=true
# Optional: Support _escapes_ in regular expressions: `\c` translates to the literal character `c`; `\/` translates to `/` (and thus does not end the regular expression); `\\` translates to `\`.
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** Left as exercise to the reader.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"} solution="hidden" solution2="hidden" solution2_first=true solution_first=true
# ### Exercise 4: Defining Grammars as Functions (Advanced)
#
# To obtain a nicer syntax for specifying grammars, one can make use of Python constructs which then will be _parsed_ by an additional function. For instance, we can imagine a grammar definition which uses `|` as a means to separate alternatives:
# + slideshow={"slide_type": "fragment"}
def expression_grammar_fn():
start = "<expr>"
expr = "<term> + <expr>" | "<term> - <expr>"
term = "<factor> * <term>" | "<factor> / <term>" | "<factor>"
factor = "+<factor>" | "-<factor>" | "(<expr>)" | "<integer>.<integer>" | "<integer>"
integer = "<digit><integer>" | "<digit>"
digit = '0' | '1' | '2' | '3' | '4' | '5' | '6' | '7' | '8' | '9'
# + [markdown] slideshow={"slide_type": "subslide"}
# If we execute `expression_grammar_fn()`, this will yield an error. Yet, the purpose of `expression_grammar_fn()` is not to be executed, but to be used as _data_ from which the grammar will be constructed.
# + slideshow={"slide_type": "fragment"}
with ExpectError():
expression_grammar_fn()
# + [markdown] slideshow={"slide_type": "fragment"}
# To this end, we make use of the `ast` (abstract syntax tree) and `inspect` (code inspection) modules.
# + slideshow={"slide_type": "skip"}
import ast
import inspect
# + [markdown] slideshow={"slide_type": "fragment"}
# First, we obtain the source code of `expression_grammar_fn()`...
# + slideshow={"slide_type": "subslide"}
source = inspect.getsource(expression_grammar_fn)
source
# + [markdown] slideshow={"slide_type": "fragment"}
# ... which we then parse into an abstract syntax tree:
# + slideshow={"slide_type": "fragment"}
tree = ast.parse(source)
# + [markdown] slideshow={"slide_type": "fragment"}
# We can now parse the tree to find operators and alternatives. `get_alternatives()` iterates over all nodes `op` of the tree; If the node looks like a binary _or_ (`|` ) operation, we drill deeper and recurse. If not, we have reached a single production, and we try to get the expression from the production. We define the `to_expr` parameter depending on how we want to represent the production. In this case, we represent a single production by a single string.
# + slideshow={"slide_type": "subslide"}
def get_alternatives(op, to_expr=lambda o: o.s):
if isinstance(op, ast.BinOp) and isinstance(op.op, ast.BitOr):
return get_alternatives(op.left, to_expr) + [to_expr(op.right)]
return [to_expr(op)]
# + [markdown] slideshow={"slide_type": "fragment"}
# `funct_parser()` takes the abstract syntax tree of a function (say, `expression_grammar_fn()`) and iterates over all assignments:
# + slideshow={"slide_type": "fragment"}
def funct_parser(tree, to_expr=lambda o: o.s):
return {assign.targets[0].id: get_alternatives(assign.value, to_expr)
for assign in tree.body[0].body}
# + [markdown] slideshow={"slide_type": "fragment"}
# The result is a grammar in our regular format:
# + slideshow={"slide_type": "subslide"}
grammar = funct_parser(tree)
for symbol in grammar:
print(symbol, "::=", grammar[symbol])
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# #### Part 1 (a): One Single Function
#
# Write a single function `define_grammar(fn)` that takes a grammar defined as function (such as `expression_grammar_fn()`) and returns a regular grammar.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"} solution="hidden" solution2="hidden"
# **Solution**. This is straightforward:
# + slideshow={"slide_type": "skip"} solution2="hidden"
def define_grammar(fn, to_expr=lambda o: o.s):
source = inspect.getsource(fn)
tree = ast.parse(source)
grammar = funct_parser(tree, to_expr)
return grammar
# + slideshow={"slide_type": "skip"} solution2="hidden"
define_grammar(expression_grammar_fn)
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Note.** Python allows us to directly bind the generated grammar to the name `expression_grammar_fn` using function decorators. This can be used to ensure that we do not have a faulty function lying around:
#
# ```python
# @define_grammar
# def expression_grammar():
# start = "<expr>"
# expr = "<term> + <expr>" | "<term> - <expr>"
# #...
# ```
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Part 1 (b): Alternative representations
# + [markdown] slideshow={"slide_type": "fragment"}
# We note that the grammar representation we designed previously does not allow simple generation of alternatives such as `srange()` and `crange()`. Further, one may find the string representation of expressions limiting. It turns out that it is simple to extend our grammar definition to support grammars such as below:
# + slideshow={"slide_type": "subslide"}
def define_name(o):
return o.id if isinstance(o, ast.Name) else o.s
# + slideshow={"slide_type": "subslide"}
def define_expr(op):
if isinstance(op, ast.BinOp) and isinstance(op.op, ast.Add):
return (*define_expr(op.left), define_name(op.right))
return (define_name(op),)
# + slideshow={"slide_type": "subslide"}
def define_ex_grammar(fn):
return define_grammar(fn, define_expr)
# + [markdown] slideshow={"slide_type": "subslide"}
# The grammar:
#
# ```python
# @define_ex_grammar
# def expression_grammar():
# start = expr
# expr = (term + '+' + expr
# | term + '-' + expr)
# term = (factor + '*' + term
# | factor + '/' + term
# | factor)
# factor = ('+' + factor
# | '-' + factor
# | '(' + expr + ')'
# | integer + '.' + integer
# | integer)
# integer = (digit + integer
# | digit)
# digit = '0' | '1' | '2' | '3' | '4' | '5' | '6' | '7' | '8' | '9'
#
# for symbol in expression_grammar:
# print(symbol, "::=", expression_grammar[symbol])
# ```
# + [markdown] slideshow={"slide_type": "subslide"}
# **Note.** The grammar data structure thus obtained is a little more detailed than the standard data structure. It represents each production as a tuple.
# + [markdown] slideshow={"slide_type": "fragment"}
# We note that we have not enabled `srange()` or `crange()` in the above grammar. How would you go about adding these? (*Hint:* wrap `define_expr()` to look for `ast.Call`)
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Part 2: Extended Grammars
#
# Introduce an operator `*` that takes a pair `(min, max)` where `min` and `max` are the minimum and maximum number of repetitions, respectively. A missing value `min` stands for zero; a missing value `max` for infinity.
# + slideshow={"slide_type": "fragment"}
def identifier_grammar_fn():
identifier = idchar * (1,)
# + [markdown] slideshow={"slide_type": "fragment"} solution2="hidden" solution2_first=true
# With the `*` operator, we can generalize the EBNF operators – `?` becomes (0,1), `*` becomes (0,), and `+` becomes (1,). Write a converter that takes an extended grammar defined using `*`, parse it, and convert it into BNF.
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** No solution yet :-)
| docs/notebooks/Grammars.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# In this last part of the dissertation, we will price an Interest rate swap (IRS), a Credit default swap (CDS), CVA and Credit Insurance in Python.
import numpy as np
import pandas as pd
from scipy.interpolate import interp1d
import matplotlib.pyplot as plt
import math
import unittest
# First, we start by writing a code to find the present value of an Interest rate swap.
#
# By defintion, an IRS is an agreement between two parties to exchange future interest rate payments of a set period of time. In this case we will consider Vanilla IRS which involves exchange of a fixed rate for a floating rate or viceversa.
# In order to find the present value of an IRS, we start with a set of data $years1$ which is a set of matuirties and $zero$_$rates1$ which are the zero rates at each maturity date. Then we use this data set to construct a zero yield curve ($zero$ _$yield$ _$curve$) using linear interpolation.
years1 = np.array([0, 1, 2, 4, 5, 10, 20])
zero_rates1 = np.array([0.01, 0.01, 0.011, 0.012, 0.012, 0.015, 0.015])
zero_yield_curve = interp1d(years1, zero_rates1)
xnew = np.linspace(0, 20, num=21, endpoint=True)
plt.title('Zero yield curve through linear interpolation.')
plt.xlabel('maturities')
plt.ylabel('zero rates')
plt.plot(years1, zero_rates1, 'o', xnew, zero_yield_curve(xnew))
plt.legend(['data', 'linear'], loc='best')
plt.show()
# A vanilla IRS is made of a fixed and a floating leg.
#
# The present value of a fixed rate leg is given by:
# $$PV_{fixed}(t)=RN \sum^{n}_{i=1} \tau_i D_i$$
# and the present value of a floating leg is given by:
# $$PV_{float}(t)=N \sum^{n}_{i=1} (F_i +s) \tau_i D_i$$
# where:
#
# $D_i=D(t,T_i)$ is the discount factor,
#
# $s$ is the floating spread,
#
# $N$ is the notional,
#
# $\tau_i$ is $(T_i-T_{i-1})$,
#
# $R$ is the fixed rate,and
#
# $F_i$ is the forward rate.
# In order the evaluate the present value of the fixed leg, we start by computing the discount factor $D_i=D(t,T_i)= e^{-(T_i-t)*zero \_ yield \_ curve(i)}$. We then use some known results and the unittest to check the code.
# +
def discount_factor1(t, Ti, zero_yield_curve):
return np.exp(-(Ti - t) * zero_yield_curve(Ti))
class TestDiscountFactor(unittest.TestCase):
def test_discountfactor(self):
self.assertAlmostEqual(discount_factor1(0, years1[1], zero_yield_curve), np.exp(-0.01))
self.assertAlmostEqual(discount_factor1(0, years1[2], zero_yield_curve), np.exp(-0.022))
unittest.main(argv=[''], verbosity=2, exit=False)
# -
# Next we compute the forward rate $F_i=\frac{(\frac{D_{i-1}}{D_i}-1)}{\tau_i}$.
# +
def forward_rates(
t, time1, time2, zero_yield_curve):
#time1 is the time used to evalaute D_{i-1} whilst time2 is used to evaluate D_i or viceversa
if time1 == time2:
tau = 0
elif time1 > time2:
tau = (time1 - time2)
else:
tau = (time2 - time1)
y1 = discount_factor1(t, time1, zero_yield_curve)
y2 = discount_factor1(t, time2, zero_yield_curve)
if time1 == time2:
forward_rate = zero_yield_curve(time1)
elif time1 > time2:
forward_rate = ((y2 / y1) - 1) / tau
else:
forward_rate = ((y1 / y2) - 1) / tau
return forward_rate
class TestForwardRates(unittest.TestCase):
def test_forwardrates(self):
self.assertAlmostEqual(forward_rates(0, years1[2], years1[1], zero_yield_curve), np.exp(-0.01) / np.exp(-0.022) - 1)
unittest.main(argv=[''], verbosity=2, exit=False)
# -
# We evaluate the present value of the fixed leg by using the formula we stated above. The variables in the formula below are:
#
# $t:$ time at which the fixed leg is evaluated,
#
# $coupon:$ coupon rate, frequency of payments in a year,
#
# $end\_ date:$ maturity of the swap,
#
# $k:$ fixed rate,
#
# $n:$ notional.
#
#
#
#
def fixed_leg(t, coupon, end_date, k, n, zero_yield_curve):
q = end_date * coupon #number of payments until end_date of the swap
s = 0
times = [0] * (q + 1)
tau = 1 / coupon
df = [0] * (q + 1)
for i in range(1, q + 1):
times[i] += (t + (1 / coupon) * i)
df[i] += discount_factor1(t, times[i], zero_yield_curve)
s += tau * df[i]
return s * n * k
fixed_leg(0, 2, 5, 0.05, 100, zero_yield_curve)
# We now evaluate the floating leg using the formula stated above. The variables used are the same as for the fixed leg.
def floating_leg(t, n, coupon, end_date, zero_yield_curve, forward_rates, spread):
s1 = 0
q = end_date *coupon
times = [0] * (q + 1)
tau = 1 / coupon
y = [0] * (q + 1)
z = [0] * (q + 1)
for k in range(1, q + 1):
times[k] += (t + (1 / coupon) * k)
y[k] += discount_factor1(t, times[k], zero_yield_curve)
z[k] += forward_rates(t, times[k - 1], times[k], zero_yield_curve)
s1 += (z[k]+ spread) * tau * y[k]
return n * s1
floating_leg(0, 100, 2, 5, zero_yield_curve, forward_rates,0.01)
# The present value of IRS from the fixed rate receiver perspective = Present value of the fixed leg - Present value of the floating leg.
def IRS(coupon, end_date, n, t, zero_yield_curve, k,spread):
return fixed_leg(t, coupon, end_date, k, n, zero_yield_curve) - floating_leg(t, n, coupon, end_date, zero_yield_curve, forward_rates,spread)
IRS(2, 5, 100, 0, zero_yield_curve, 0.05, 0.01)
# The present value of an interest rate swap from the fixed rate payer perspective is equal to the present value of the floating leg - present value of the fixed leg.
def IRS1(coupon, end_date, n, t, zero_yield_curve, k,spread):
return floating_leg(t, n, coupon, end_date, zero_yield_curve, forward_rates,spread)-fixed_leg(t, coupon, end_date, k, n, zero_yield_curve)
IRS1(2, 5, 100, 0, zero_yield_curve, 0.05, 0.01)
# We can now compute the Par swap rate, which is the value of the fixed rate that, at time $t$, makes the present value of the interest rate swap equal to $0$.
# $$par\_ rate(t)=\frac{\sum^{n}_{i=1}(F_i +s)D_i \tau_i}{\sum^{n}_{i=1}D_i \tau_i}$$
def par_rate(coupon,end_date, t,zero_yield_curve, spread):
q = coupon * end_date
df = [0] * (q + 1)
fr = [0] * (q + 1)
times = [0] * (q + 1)
tau = 1 / coupon
s = 0
k = 0
for i in range(1, q + 1):
times[i] = (t + (1 / coupon) * i)
df[i] += discount_factor1(t, times[i], zero_yield_curve)
fr[i] += forward_rates(t, times[i], times[i - 1], zero_yield_curve)
s += (fr[i]+spread) * df[i] * tau
k += df[i] * tau
return s / k
par_rate(2, 5, 0, zero_yield_curve, 0.01)
# We can also compute the annuity $$A(t)=\sum^{n}_{i=1}D_i \tau_i$$
def annuity(coupon, end_date, t, zero_yield_curve):
q = coupon * end_date
df = [0] * (q + 1)
times = [0] * (q + 1)
tau = 1 / coupon
s1 = 0
for i in range(1, q + 1):
times[i] = (t + (1 / coupon) * i)
df[i] += discount_factor1(t, times[i], zero_yield_curve)
s1 += df[i] * tau
return s1
annuity(2, 5, 0, zero_yield_curve)
# Then, the present value of the interest rate swap in terms of the annuity is given by:
# $$IRS(t)=notional*(fixed\_ rate - par\; swap\; rate(t))* A(t)$$
# $$=n*(k - par\; swap\; rate(t))*A(t)$$
def pv_swap(n, coupon, end_date, t, zero_yield_curve, k, par_rate,annuity,spread):
a = par_rate(coupon, end_date, t, zero_yield_curve, spread)
b = annuity(coupon, end_date, t, zero_yield_curve)
return n * (k - a) * b
# +
class TestIRS(unittest.TestCase):
def test_IRS(self):
self.assertAlmostEqual(pv_swap(100, 1, 5, 0, zero_yield_curve, 0.05, par_rate,annuity,0.01), IRS(1, 5, 100, 0, zero_yield_curve, 0.05, 0.01))
unittest.main(argv=[''], verbosity=2, exit=False)
# -
# The present value of the interest rate calculated using the annuity agrees with the present value of the interest rate swap calculated using the fixed and the floating leg.
# We want to price a CDS which is also made of two legs:
#
# The premium leg which can be calculated as follows:
# $$R \sum^{b}_{i=a+1} P(0,T_i) \alpha_i Q(\tau \geq T_i),$$
# and the floating leg which can be calculated as:
# $$LGD \int^{T_b}_{T_a} P(0,t) d_t Q(\tau \geq t)$$
# where:
#
# $\alpha_i= T_{i}-T_{i-1}$,
#
# $R$ is the fixed rate,and
#
# $LGD=(1-Recovery\; rate)$ loss-given-default.
#
# The premium leg does include another term, called the accrual term (see Equation 90 in the dissertation), however for simplicity, we are going to ignore it.
#
#
# Our set of data in this case consists of: maturities $y$, hazard rates $hazardrates$, and zero rates $zerorates$ at each maturity date.
# +
y = [0, 1, 3, 5, 7, 10]
hazardrates = [0.03199, 0.03199, 0.03780, 0.04033, 0.04458, 0.03891]
zerorates = [0.01, 0.014, 0.011, 0.01, 0.001, 0.012]
# -
# We use the $hazardrates$ and $y$ to write a function for the hazard rate using constant interpolation.
#
# +
def hazard_curve(x, years, hazard_rates):
hz_rate = 0
if years[0] <= x < years[1]:
hz_rate += hazard_rates[1]
elif years[1] <= x < years[2]:
hz_rate += hazard_rates[2]
elif years[2] <= x < years[3]:
hz_rate += hazard_rates[3]
elif years[3] <= x < years[4]:
hz_rate += hazard_rates[4]
else:
hz_rate += hazard_rates[5]
return hz_rate
class Testhazardratecurve(unittest.TestCase):
def test_hzrates(self):
self.assertAlmostEqual(hazard_curve(1.5, y, hazardrates), 0.0378)
unittest.main(argv=[''], verbosity=2, exit=False)
# -
plt.title('Constant interpolation')
plt.xlabel('years')
plt.ylabel('hazard rates')
plt.step(y, hazardrates)
plt.show()
print(y)
print(hazardrates)
# Next, we write a function to evaluate the survival probability $Q(\tau \geq t)$ and we check the results by using a unittest and the data in table 22.1 and 22.3 in Brigo Mercurio's book.
# +
def survival_probability(t, years, hazard_rates, hazard_curve):
y = np.linspace(0, t, 1000)
d = 0
for j in range(0, len(y)):
if j == 0:
d += 0
else:
d += (y[j] - y[j - 1]) * hazard_curve(y[j], years, hazard_rates)
return np.exp(-d)
class Testsurvivalprob(unittest.TestCase):
def test_survivalprob(self):
self.assertAlmostEqual(survival_probability(y[1],y, hazardrates, hazard_curve), 0.968, places=2)
unittest.main(argv=[''], verbosity=2, exit=False)
# -
# We use linear interpolation on $y$ and $zerorates$ to find the zero yield curve on this set of data.
zero_rate_curve = interp1d(y, zerorates)
# We now write a function to compute $P(0,t).$
def curve(t, years, zero_rates, zero_rate_curve):
z = np.linspace(0, t, 100)
d1 = 0
for j in range(0, len(z)):
if j == 0:
d1 += 0
else:
d1 += (z[j] - z[j - 1]) * zero_rate_curve(z[j])
return np.exp(-d1)
# Now, we write a function to compute the premium leg using the formula:
# $$Premium\_ Leg(t)=\sum^{b}_{i=a+1}(R*Q(t\geq T_i)*P(0,T_i)*(T_i -T_{i-1}))$$
def prem_leg(t, end_date,coupon, k, zero_rates, hazard_rates, years):
#end_date=maturity of the CDS
s2 = 0
q = (end_date-t) * coupon
times = [0] * (q + 1)
for i in range(1, q + 1):
times[i] += (t + (1 / coupon) * i)
s2 += (survival_probability(times[i], years, hazard_rates, hazard_curve)) * curve(times[i], years, zero_rates, zero_rate_curve) * (
times[i] - times[i - 1])
return k * s2
prem_leg(0, 3, 2, 0.05, zerorates, hazardrates, y)
# The formula to compute the protection leg is:
# $$LGD\sum^{n}_{i=a+1}\frac{1}{2} (Q(t \geq T_{i-1})-Q(\tau \geq T_i))*(P(0,T_{i-1})+ P(0,T_i))$$
def protect_leg(t, end_date,coupon, zero_rates, hazard_rates, years, LGD):
q = (end_date-t) * coupon
times = [0] * (q + 1)
s4 = 0
for i in range(1, (q + 1)):
times[i] += (t + (1 / coupon) * i)
s4 += (survival_probability(times[i - 1], years, hazard_rates, hazard_curve) - survival_probability(times[i], years, hazard_rates,
hazard_curve)) * (curve(times[i - 1], years, zero_rates, zero_rate_curve) + curve(times[i], years, zero_rates, zero_rate_curve)) / 2
return (LGD * s4)
protect_leg(0, 3, 2, zerorates, hazardrates, y, 0.6)
# Then the value of the CDS from the protection seller point of view= Premium leg - Protection leg.
def credit_default_swap(t, end_date, coupon, k, zero_rates, hazard_rates, years, LGD):
return prem_leg(t, end_date, coupon, k, zero_rates, hazard_rates, years) - protect_leg(t, end_date, coupon, zero_rates, hazard_rates, years, LGD)
credit_default_swap(0, 3, 2, 0.05, zerorates, hazardrates, y, 0.6)
# Premium leg(t)= $R*Risky\_ Annuity(t)$ where the $$Risky\_ Annuity(t)=\frac{1}{2}(T_i -T_{1-i})*(P(0,T_i))*(Q(\tau \geq T_{i-1})+Q(\tau \geq T_i)$$
def risky_annuity(t,end_date,coupon,years,hazard_rates,zero_rates):
q=(end_date-t)*coupon
z=[0]*(q+1)
summ=0
for i in range(1,(q+1)):
z[i]+=(t+(1/coupon)*i)
summ+=(z[i]-z[i-1])*curve(z[i],years,zero_rates,zero_rate_curve)*(survival_probability(z[i-1],years,hazard_rates,hazard_curve)+survival_probability(z[i],years,hazard_rates,hazard_curve))
return summ/2
risky_annuity(0, 3, 2, y, hazardrates, zerorates)
risky_annuity(0, 3, 2, y, hazardrates, zerorates) * 0.05
# $R*Risky\_ Annuity(t)= Premium\; Leg (t)$ is satisfied.
# The par credit swap for a CDS is defined as $\frac{Protect\; leg(t)}{Annuity(t)}$. We can compute it and then check the result using the test data in Brigo-Mercurio's book (Interest Rate Models Theory and Practice (2001, Springer)) and unittest. Here we did not use bootstrapping instead we are using the hazard rates in the book to show that we get the same CDS spreads.
def par_credit_swap(t, end_date, coupon, zero_rates, hazard_rates, years, LGD):
return protect_leg(t, end_date, coupon, zero_rates, hazard_rates, years, LGD) / risky_annuity(t, end_date, coupon, years, hazard_rates, zero_rates)
class Testparcreditswap(unittest.TestCase):
def test_parcreditswap(self):
self.assertAlmostEqual(par_credit_swap(0, 1, 2, zerorates, hazardrates, y, 0.6)
, 0.01925,places=3)
self.assertAlmostEqual(par_credit_swap(0, 3, 2, zerorates, hazardrates, y, 0.6)
, 0.0215,places=3)
self.assertAlmostEqual(par_credit_swap(0, 5, 2, zerorates, hazardrates, y, 0.6)
, 0.0225,places=3)
self.assertAlmostEqual(par_credit_swap(0, 10, 2, zerorates, hazardrates, y, 0.6)
, 0.0235,places=3)
unittest.main(argv=[''], verbosity=2, exit=False)
# Next, we want to price a CVA. From here on, no test data were available.
#
# In the dissertation, we have mentioned that CVA can be expressed as sum of swaptions. Here, we are going to use this fact, by evaluating first the price of the nromal swaption both from the payer and receiver perspectives.
# Then we use those to price the CVA.
from scipy.stats import norm
# $\text{Price of normal swaption payer at time 0} = \text{notional}* A(0) *\sigma *(T_0)^{1/2}(d_1*\Phi (d_1)+\phi(d_1))$
# $A(0)=\sum^{b}_{i=a}\tau_i P(0,T_i): annuity\; at\; time\; 0$,
#
# $\sigma:$ implied volatility,
# $d_1=\frac{s(0)-K}{\sigma * (T_0)^{1/2}}$,
# $s(0)=\frac{P(0,T_a)-P(0,T_b)}{A(0)}$,
#
# $K: fixed\; rate,$
#
# $T_0=T_a:$ maturity of the swaption,
#
# $T_b:$ maturity of the swap,
#
# $\Phi:$ cdf of standard normal distribution,and
#
# $\phi:$ pdf of standard normal distribution.
def annuity_0(end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
s5 = 0
q = (end_date-initial_date) * coupon
times = [0] * (q + 1)
for k in range(1, (q + 1)):
times[k] += initial_date + (1 / coupon) * k
s5 += (times[k] - times[k - 1]) * curve(times[k], years, zero_rates, zero_rate_curve)
return s5
annuity_0(8,1,zerorates,zero_rate_curve,5,y)
#s(0)
def rate(end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
return (curve(initial_date, years, zero_rates, zero_rate_curve) - curve(end_date, years, zero_rates,
zero_rate_curve)) / annuity_0(end_date,
coupon,
zero_rates,
zero_rate_curve,
initial_date,
years)
rate(8, 1, zerorates, zero_rate_curve, 5, y)
def d1(strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
return (rate(end_date, coupon, zero_rates, zero_rate_curve, initial_date, years) - strike) / (sigma * np.sqrt(
initial_date))
d1(0.01, 0.2, 8, 1, zerorates, zero_rate_curve, 5, y)
def d2(strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
return - d1(strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years)
d2(0.01, 0.2, 8, 1, zerorates, zero_rate_curve, 5, y)
# Now, using the above functions we can evaluate the price of a normal swaption from the payer perspective:
#the notional is taken to be 1
def norm_swaption_payer(notional, strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
d1_new = d1(strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years)
return notional * annuity_0(end_date, coupon, zero_rates, zero_rate_curve, initial_date, years)*sigma * np.sqrt(
initial_date) * (d1_new * norm.cdf(d1_new) + norm.pdf(d1_new))
norm_swaption_payer(1, 0.002, 0.02, 8, 1, zerorates, zero_rate_curve, 5, y)
# and also the price of the normal swaption from the receiver perspective:
def norm_swaption_receiver(notional, strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years):
d2_new = d2(strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years)
return notional * sigma *annuity_0(end_date, coupon, zero_rates, zero_rate_curve, initial_date, years)* np.sqrt(
initial_date) * (d2_new * norm.cdf(d2_new) + norm.pdf(d2_new))
norm_swaption_receiver(1, 0.002, 0.02, 8, 2, zerorates, zero_rate_curve,5, y)
# Then the CVA can be evaluated using the following formula:
#
# $CVA=LGD*\sum^{b}_{i=a+1}(Q(t_{i-1})-Q(t_i))*Swaption\; Payer_t$,
#
# where the $swaption\; payer_t$ is the price of a normal swaption with expiry $t$. In the case of CVA the strike of the swaption is taken to be $0$.
def cva(LGD, notional, strike, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years, hazard_rates, hazard_curve):
h = 0
q = (end_date-initial_date) * coupon
s=[0]*(q+1)
m = [0] * (q + 1)
for k in range(1, q + 1):
m[k] += initial_date +int(1/coupon)* k
s[k]+=norm_swaption_payer(notional, strike, sigma,m[k], coupon, zero_rates, zero_rate_curve,initial_date, years)
h += (survival_probability(m[k-1], years, hazard_rates, hazard_curve)-survival_probability(m[k], years, hazard_rates, hazard_curve)) * s[k]
return LGD * h
cva(0.6,1,0,0.02,8,1,zerorates,zero_rate_curve,5,y,hazardrates, hazard_curve)
# Lastly, we are going to price Credit Insurance.
#
# In Chapter 7, we found that the fair value of the price of Credit Insurance can be calculated as:
# $$CI= Premium\;Leg - Protection\;Leg$$
# $$=\sum^{b}_{i=a+1} \mathbb{E}[D(0, T_i) \cdot \alpha_i \cdot R ]-LGD \int_{t}^{T}\lambda_C(s) \cdot e^{-\int_{t}^{s}r_F(u)+\lambda_C(u) du}\mathbb{E}[min((1-p)V^+,K)]ds $$
#
# where:
#
# $(1-p)$ is the participation percentage,
#
# $\alpha_i=(T_i-T_{i-1})$, and the rest of the variables have already been defined above.
#
# $CI= \text{premium leg} - \left(CVA(\text{with strike 0})-CVA \left(\text{with strike} \frac{K}{1-p}\right)\right)$
def prem_leg_CI(end_date, coupon, initial_date, k, zero_rates, years):
s6 = 0
q = (end_date-initial_date) * coupon
times = [0] * (q + 1)
for i in range(1, q + 1):
times[i] += (initial_date + (1 / coupon) * i)
s6 += (times[i] - times[i - 1]) * curve(times[i], years, zero_rates, zero_rate_curve)
return k * s6
prem_leg_CI(8, 1, 5, 0.1, zerorates, y)
def ci_swaption(LGD, notional, strike, p, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years,hazard_rates,hazard_curve):
c = strike / (1 - p)
return cva(LGD, notional, 0, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years,hazard_rates, hazard_curve) - cva(LGD,notional,c,sigma,end_date,coupon, zero_rates,zero_rate_curve,initial_date,years,hazard_rates,hazard_curve)
ci_swaption(0.6,1,0.1,0.3,0.02,8,1,zerorates,zero_rate_curve,5,y,hazardrates, hazard_curve)
def ci(LGD, notional, strike, p, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years, k,hazard_rates,hazard_curve):
return prem_leg_CI(end_date, coupon, initial_date, k, zero_rates, years) - ci_swaption(LGD, notional, strike, p, sigma, end_date, coupon, zero_rates, zero_rate_curve, initial_date, years,hazard_rates,hazard_curve)
ci(0.6,1,0.1,0.4,0.02,8,1,zerorates,zero_rate_curve,1,y,0.1,hazardrates,hazard_curve)
| MSc Dissertation-UCL.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # [Assignment #2: NPFL067 Statistical NLP II](http://ufal.mff.cuni.cz/~hajic/courses/npfl067/assign2.html)
#
# ## Words and The Company They Keep
#
# ### Author: <NAME>
#
# ### March 28, 2018
#
# ---
# This Python notebook examines the role of mutual information in natural language processing.
#
# Code and explanation of results is fully viewable within this webpage.
#
# ## Files
#
# - [index.html](./index.html) - Contains all veiwable code and a summary of results
# - [README.md](./README.md) - Instructions on how to run the code with Python
# - [nlp-assignment-2.ipynb](./nlp-assignment-2.ipynb) - Jupyter notebook where code can be run
# - [brown_cluster.py](./brown_cluster.py) - Code defining the Brown clustering algorithm
# - [requirements.txt](./requirements.txt) - Required python packages for running
#
# - *.csv - CSV output of results
# ## 1. Best Friends
#
# #### Problem Statement
# > In this task you will do a simple exercise to find out the best word association pairs using the pointwise mutual information method.
#
# > First, you will have to prepare the data: take the same texts as in the previous assignment, i.e.
#
# > `TEXTEN1.txt` and `TEXTCZ1.txt`
#
# > (For this part of Assignment 2, there is no need to split the data in any way.)
#
# > Compute the pointwise mutual information for all the possible word pairs appearing consecutively in the data, **disregarding pairs in which one or both words appear less than 10 times in the corpus**, and sort the results from the best to the worst (did you get any negative values? Why?) Tabulate the results, and show the best 20 pairs for both data sets.
#
# > Do the same now but for distant words, i.e. words which are at least 1 word apart, but not farther than 50 words (both directions). Again, tabulate the results, and show the best 20 pairs for both data sets.
# ### Process Text
# The first step is to process the frequency distribution of the unigrams and bigrams and define a function to calculate the pointwise mutual information between two words. The class `LanguageModel` will handle this.
# +
# Import Python packages
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
# # %load_ext autoreload
# # %autoreload 2
from collections import defaultdict, Counter, Iterable
import itertools
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from tqdm import tqdm_notebook as tqdm, tnrange as trange
from scipy.special import comb
# Configure Plots
plt.rcParams['lines.linewidth'] = 4
pd.set_option('max_colwidth', 150)
np.random.seed(200) # Set a seed so that this notebook has the same output each time
# -
def open_text(filename):
"""Reads a text line by line, applies light preprocessing, and returns an array of words"""
with open(filename, encoding='iso-8859-2') as f:
content = f.readlines()
preprocess = lambda word: word.strip()
return np.array([preprocess(word) for word in content])
class LanguageModel:
"""Counts words and calculates the probabilities of a language model"""
def __init__(self, words, min_words=10):
self.min_words = min_words
# Unigrams
self.unigrams = words
self.unigram_set = list(set(self.unigrams))
self.total_unigram_count = len(self.unigrams)
self.unigram_dist = Counter(self.unigrams)
self.unigram_pdist = defaultdict(float)
for w in self.unigram_dist:
self.unigram_pdist[w] = self.unigram_dist[w] / self.total_unigram_count
# Bigrams
self.bigrams = list(zip(words, words[1:]))
self.bigram_set = list(set(self.bigrams))
self.total_bigram_count = len(self.bigrams)
self.bigram_dist = Counter(self.bigrams)
self.bigram_pdist = defaultdict(float)
for w in self.bigram_dist:
self.bigram_pdist[w] = self.bigram_dist[w] / self.total_bigram_count
def p_unigram(self, w):
"""Calculates the probability a unigram appears in the distribution"""
return self.unigram_pdist[w]
def p_bigram(self, wprev, w):
"""Calculates the probability a bigram appears in the distribution"""
return self.bigram_pdist[(wprev, w)]
def pointwise_mi(self, wprev, w, p_bigram_func=None):
"""Calculates the pointwise mutual information in a word pair"""
p_bigram_func = self.p_bigram if p_bigram_func is None else p_bigram_func
joint = p_bigram_func(wprev, w)
independent = self.p_unigram(wprev) * self.p_unigram(w)
return np.log2(joint / independent) if independent != 0 else 0
# +
# Read the texts into memory
english = './TEXTEN1.txt'
czech = './TEXTCZ1.txt'
words_en = open_text(english)
words_cz = open_text(czech)
# -
lm_en = LanguageModel(words_en)
lm_cz = LanguageModel(words_cz)
# Loop over all pairs of bigrams and calculate their pointwise mutual information, collecting them into a table.
def mutual_information(lm):
# Obtain all word pairs in the word list, disregarding pairs in which one or both words appear less than 10 times in the corpus
pairs = [pair for pair in lm.bigram_set
if lm.unigram_dist[pair[0]] >= lm.min_words
and lm.unigram_dist[pair[1]] >= lm.min_words]
mi = [(' '.join(pair), lm.pointwise_mi(*pair)) for pair in pairs]
return pd.DataFrame(mi, columns=['pair', 'mutual_information'])
mi_en = mutual_information(lm_en).sort_values(by='mutual_information', ascending=False)
mi_cz = mutual_information(lm_cz).sort_values(by='mutual_information', ascending=False)
# ### Results - Consecutive Pairs
# The two tables below show the pointwise mutual information (sorted descending) between pairs of words appearing consecutively in the English and Czech texts respectively.
#
# We see that proper names like Great Britain and Tomáš Ježek provide a lot of mutual information, as those words are frequently seen together and rarely seen apart from each other. However, some of these values are negative (see below).
mi_en[:20] # English
mi_cz[:20] # Czech
# Sorting in ascending order, there are pairs of words that provide negative mutual information. This can be explained by the definition of pointwise mutual information (PMI):
#
# $$PMI(w_t,w_{t+1}) = \log \frac{p(w_t,w_{t+1})}{p(w_t)p(w_{t+1})}$$
#
# where $w_t,w_{t+1}$ are consecutive words (in this instance). The `log` is negative when its input is less than 1, which is to say that
#
# $$p(w_t,w_{t+1}) < p(w_t)p(w_{t+1})$$
#
# i.e., the probability of the pair appearing consecutively in the text is less than the probability of them appearing independently from each other.
#
# This can be verified by the data below. For instance, '_the_' and '_,_' both appear very frequently in the text. However, they are unlikely to be seen consecutively, since 'the ,' is ungrammatical. Therefore, their pointwise mutual information must be negative.
mi_en[:-5:-1]
# Now define a function to calculate pointwise mutual information on all pairs of words a constant distance apart (up to 50) and store the results in a table.
def mutual_information_dist(lm):
def mi_step(distance):
# Get all pairs in the word list a certain distance apart
pair_list = list(zip(lm.unigrams, lm.unigrams[distance+1:]))
dist = Counter(pair_list)
# Obtain all word pairs in the word list, disregarding pairs in which one or both words appear less than 10 times in the corpus
pairs = [pair for pair in list(set(pair_list))
if lm.unigram_dist[pair[0]] >= lm.min_words
and lm.unigram_dist[pair[1]] >= lm.min_words]
p_bigram = lambda wprev, w: dist[(wprev, w)] / lm.total_bigram_count
yield ((distance, wprev, w, lm.pointwise_mi(wprev, w, p_bigram)) for wprev,w in pairs)
max_distance = 50
results = [m for distance in tqdm(range(1, max_distance+1)) for mi in mi_step(distance) for m in mi]
return pd.DataFrame(results, columns=['distance', 'word_1', 'word_2', 'mutual_information'])
mi_dist_en = mutual_information_dist(lm_en).sort_values(by='mutual_information', ascending=False)
mi_dist_cz = mutual_information_dist(lm_cz).sort_values(by='mutual_information', ascending=False)
# ### Results - Distant Pairs
# As before, the two tables below show the pointwise mutual information (sorted descending) between pairs of words appearing in the English and Czech texts. There is an added column called `distance` which indicates the number of words between the two words of interest.
#
# Expectedly, pairs of words with high pointwise mutual information appear close together. For example 'survival \_ \_ fittest' can be filled in as 'survival _of the_ fittest', which is a common phrase in the text. More surprisingly, some words appearing far apart from each other provide a lot of mutual information. It is likely pairs like 'Nastaseho \_ [x25] Newcomba' is a part of multiple quotations in the text such that the word pair appears infrequently outside of them.
mi_dist_en[:20] # English
mi_dist_cz[:20] # Czech
# ## 2. Best Friends
#
# #### Word Classes
#
# > **The Data**
#
# > Get `TEXTEN1.ptg`, `TEXTCZ1.ptg`. These are your data. They are almost the same as the .txt data you have used so far, except they now contain the part of speech tags in the following form:
#
# > `rady/NNFS2-----A----`
# `,/Z:-------------`
#
# > where the tag is separated from the word by a slash ('/'). Be careful: the tags might contain everything (including slashes, dollar signs and other weird characters). It is guaranteed however that there is no slash-word.
#
# > Similarly for the English texts (except the tags are shorter of course).
#
# > **The Task**
#
# > Compute a full class hierarchy of **words** using the first 8,000 words of those data, and only for words occurring 10 times or more (use the same setting for both languages). Ignore the other words for building the classes, but keep them in the data for the bigram counts. For details on the algorithm, use the Brown et al. paper distributed in the class; some formulas are wrong, however, so please see the corrections on the web (Class 12, formulas for Trick \#4). Note the history of the merges, and attach it to your homework. Now run the same algorithm again, but stop when reaching 15 classes. Print out all the members of your 15 classes and attach them too.
#
# > **Hints:**
#
# > The initial mutual information is (English, words, limit 8000):
#
# > `4.99726326162518` (if you add one extra word at the beginning of the data)
# > `4.99633675507535` (if you use the data as they are and are carefull at the beginning and end).
#
# > NB: the above numbers are finally confirmed from an independent source :-).
#
# > The first 5 merges you get on the English data should be:
#
# > `case subject`
# > `cannot may`
# > `individuals structure`
# > `It there`
# > `even less`
#
# > The loss of Mutual Information when merging the words "case" and "subject":
#
# > Minimal loss: `0.00219656653357569` for `case+subject`
# ### Process Text
# Process the text using the `LmCluster` class defined in `brown_cluster.py`. The code will perform the Brown clustering algorithm on the given texts.
from brown_cluster import LmCluster
def open_text(filename):
"""Reads a text line by line, applies light preprocessing, and returns an array of words and tags"""
with open(filename, encoding='iso-8859-2') as f:
content = f.readlines()
preprocess = lambda word: word.strip().rsplit('/', 1)
return [preprocess(word) for word in content]
# +
# Read the texts into memory
english = './TEXTEN1.ptg'
czech = './TEXTCZ1.ptg'
words_en, tags_en = zip(*open_text(english))
words_cz, tags_cz = zip(*open_text(czech))
# -
# ### Cluster the word classes
text_size = 8000
lm_en = LmCluster(words_en[:text_size])
lm_cz = LmCluster(words_cz[:text_size])
lm_en.cluster()
lm_cz.cluster()
def history(cluster):
return pd.DataFrame(cluster.merge_history, columns=['class 1', 'class 2', 'cluster id', 'mutual_information_loss'])
# ### History of Merges
# The tables below show the history of merges in the English and Czech texts respectively. The class (cluster) id is displayed by its corresponding word (if the class contains just one word).
#
# According to the Brown clustering algorithm, words appearing in the most similar contexts (and hence reducing the text's total mutual information the least) get clustered first. For instance, helper verbs 'may' and 'cannot' can be interchanged in the text without reducing the text's mutual information much.
history(lm_en) # English
history(lm_cz) # Czech
# As before, do the clustering, this time stopping at 15 clusters.
clusters = 15
lm_en_15 = LmCluster(words_en[:text_size])
lm_cz_15 = LmCluster(words_cz[:text_size])
lm_en_15.cluster(clusters)
lm_cz_15.cluster(clusters)
def class_cluster(lm):
classes = lm.get_classes()
return pd.DataFrame([(x, [lm.class_name(c) for c in classes[x] if c < len(lm.int2word)]) for x in classes], columns=['class', 'words'])
# ### Cluster Distribution with 15 Classes
# The tables below display the contents of each of the 15 classes merged with the clustering algorithm.
#
# Words that appear very frequently with other words like 'the' and 'of' will reduce the mutual information a lot if clustered with any other class, and so are left over. Class 1721 shows quantifiers like 'several' and 'one' are in similar contexts and hence in their own cluster. This is similar for articles in class 1758.
class_cluster(lm_en_15) # English
class_cluster(lm_cz_15) # Czech
# ## 3. Tag Classes
#
# > Use the same original data as above, but this time, you will compute the classes for tags (the strings after slashes). Compute tag classes for all tags appearing 5 times or more in the data. Use as much data as time allows. You will be graded relative to the other student's results. Again, note the full history of merges, and attach it to your homework. Pick three interesting classes as the algorithm goes (English data only; Czech optional), and comment on them (why you think you see those tags there together (or not), etc.).
cluster_en_tag = LmCluster(tags_en, word_cutoff=5)
cluster_en_tag.cluster()
# The tables below display the history of merges with regards to part-of-speech tags in the texts.
#
# Some interesting classes include:
#
# - 'JJ' (adjective) and 'JJR' (comparative adjective). These tags are both denote slightly different types of adjectives, so it makes sense that they would get merged into their own cluster.
# - 'TO' (to) and 'RBS' (superlative adverb). Likewise, the infinitive 'to' and adverbs like 'best' most frequently appear before a verb, and so get merged due to the similar context.
# - 'IN' (preposition), 'WP$' (posessive wh-pronoun), '(', and '"' all appear in a single class, likely due to the fact that all of these tags appear frequently at the beginning of a clause and break up sentences into phrases. For instance, 'the chair _which_ is ...' or 'the chair _in_ the ...'.
history(cluster_en_tag) # English
cluster_cz_tag = LmCluster(tags_cz, word_cutoff=5)
cluster_cz_tag.cluster()
history(cluster_cz_tag) # Czech
# #### Save all results to text files
history(lm_en).to_csv('merge_english.csv', index=False)
history(lm_cz).to_csv('merge_czech.csv', index=False)
class_cluster(lm_en_15).to_csv('classes_english_15.csv', index=False)
class_cluster(lm_cz_15).to_csv('classes_czech_15.csv', index=False)
history(cluster_en_tag).to_csv('cluster_english_tag.csv', index=False)
history(cluster_cz_tag).to_csv('cluster_czech_tag.csv', index=False)
| charles-university/statistical-nlp/assignment-2/kondrad.assign2/nlp-assignment-2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Tensor images
# This notebook gives an overview of the concept of tensor images, and demonstrates how to use this feature.
import diplib as dip
# After reading the "*PyDIP* basics" notebook, you should be familiar with the concepts of scalar images and color images. We remind the reader that an image can have any number of values associated to each pixel. An image with a single value per pixel is a scalar image. Multiple values can be arranged in one or two dimensions, as a vector image or a matrix image. A color image is an example of a vector image, for example in the RGB color space the vector for each pixel has 3 values, it is a 3D vector.
#
# The generalization of vectors and matrices is a tensor. A rank 0 tensor is a scalar, a rank 1 tensor is a vector, and a rank 2 tensor is a matrix.
#
# This is a scalar image:
img = dip.ImageRead('../trui.ics')
img.Show()
# We can compute its gradient, which is a vector image:
g = dip.Gradient(img)
g.Show()
# The vector image is displayed by showing the first vector component in the red channel, and the second one in the green channel. `g` has two components:
print(g.TensorElements())
print(g.TensorShape())
# Multiplying a vector with its transposed leads to a symmetric matrix:
S = g * dip.Transpose(g)
print("Tensor size:", S.TensorSizes())
print("Tensor shape:", S.TensorShape())
print("Tensor elements:", S.TensorElements())
# Note how the 2x2 symmetric matrix stores only 3 elements per pixel. Because of the symmetry, the `[0,1]` and the `[1,0]` elements are identical, and need not be both stored. See [the documentation](https://diplib.org/diplib-docs/classdip_1_1Tensor.html#aa803a3cb47468de269ee5467f60af457) for details on how the individual elements are stored.
#
# Local averaging of this matrix image (i.e. applying a low-pass filter) leads to the structure tensor:
S = dip.Gauss(S, [5])
S.Show()
# We can still display this tensor image, because it has only 3 tensor elements, which can be mapped to the three RGB channels of the display.
#
# The structure tensor is one of the more important applications for the concept of the tensor image. In [this documentation page](https://diplib.org/diplib-docs/why_tensors.html) there are some example applications of the structure tensor. Here we show how to get the local orientation from it using the eigenvalue decomposition.
eigenvalues, eigenvectors = dip.EigenDecomposition(S)
print(eigenvalues.TensorShape())
print(eigenvectors.TensorShape())
# The eigendecomposition is such that `S * eigenvectors == eigenvectors * eigenvalues`. `eigenvectors` is a full 2x2 matrix, and hence has 4 tensor elements. These are stored in column-major order. The first column is the eigenvector that corresponds to the first eigenvalue. Eigenvalues are sorted in descending order, and hence the first eigenvector is perpendicular to the edges in the image.
v1 = eigenvectors.TensorColumn(0)
angle = dip.Angle(v1)
angle.Show('orientation')
# Note that extracting a column from the tensor yields a vector image, and that this vector image shares data with the column-major matrix image. Transposing a matrix is a cheap operation that just changes the storage order of the matrix, without a need to copy or reorder the data:
tmp = dip.Transpose(eigenvectors)
print(tmp.TensorShape())
print(tmp.SharesData(eigenvectors))
# A second important matrix image is the Hessian matrix, which contains all second order derivatives. Just like the strucutre tensor, it is a symmetric 2x2 matrix:
H = dip.Hessian(img)
print("Tensor size:", S.TensorSizes())
print("Tensor shape:", S.TensorShape())
print("Tensor elements:", S.TensorElements())
H.Show()
| examples/python/tensor_images.ipynb |
# ### Creating multi-panel plots using `facets`.
#
# #### Problem
#
# You want to see more aspects of your data and it's not practcal to use the regular `aesthetics` approach for that.
#
# #### Solution - `facets`
#
# You can add one or more new dimentions to your plot using `faceting`.
#
# This approach allows you to split up your data by one or more variables and plot the subsets of data together.
#
#
# In this demo we will explore how various faceting functions work, as well as the built-in `sorting` and `formatting` options.
#
# To learn more about formatting templates see: [Formatting](https://github.com/JetBrains/lets-plot-kotlin/blob/master/docs/formats.md).
%useLatestDescriptors
%use lets-plot
%use krangl
var data = DataFrame.readCSV("https://raw.githubusercontent.com/JetBrains/lets-plot-kotlin/master/docs/examples/data/mpg2.csv")
data.head(3)
# ### One plot
#
# Create a scatter plot to show how `mpg` is related to a car's `engine horsepower`.
#
# Also use the `color` aesthetic to vizualise the region where a car was designed.
val p = (letsPlot(data.toMap()) {x="engine horsepower"; y="miles per gallon"} +
geomPoint {color="origin of car"})
p + ggsize(800, 350)
# ### More dimentions
#
# There are two functions for faceting:
#
# - facetGrid()
# - facetWrap()
#
# The former creates 2-D matrix of plot panels and latter creates 1-D strip of plot panels.
#
# We'll be using the `number of cylinders` variable as 1st fatceting variable, and sometimes the `origin of car` as a 2nd fatceting variable.
# ### facetGrid()
#
# The data can be split up by one or two variables that vary on the X and/or Y direction.
# #### One facet
#
# Let's split up the data by `number of cylinders`.
p + facetGrid(x="number of cylinders")
# #### Two facets
#
# Split up the data by two faceting variables: `number of cylinders` and `origin of car`.
p + facetGrid(x="number of cylinders", y="origin of car")
# #### Formatting and sorting.
#
# Apply a formatting template to the `number of cylinders` and
# sort the `origin of car` values in discending order.
#
# To learn more about formatting templates see: [Formatting](https://github.com/JetBrains/lets-plot-kotlin/blob/master/docs/formats.md).
p + facetGrid(x="number of cylinders", y="origin of car", xFormat="{d} cyl", yOrder=-1)
# ### facetWrap()
#
# The data can be split up by one or more variables.
# The panels layout is flexible and controlled by `ncol`, `nrow` and `dir` options.
# #### One facet
#
# Split data by the `number of cylinders` variable and arrange tiles in two rows.
p + facetWrap(facets="number of cylinders", nrow=2)
# #### Two facets
#
# Split data by `origin of car` and `number of cylinders` and arrange tiles in 5 columns.
p + facetWrap(facets=listOf("origin of car", "number of cylinders"), ncol=5)
# #### Arrange panels vertically.
#
# Use the `dir` parameter to arrange tiles by columns, in 3 columns (the default tile arrangment is "by row").
#
# Also, format `number of cylinders` labels and reverse the sorting direction for this facetting variable.
p + facetWrap(facets=listOf("origin of car", "number of cylinders"),
ncol=3,
format=listOf(null, "{} cyl"),
order=listOf(1, -1),
dir="v")
| docs/examples/jupyter-notebooks/facets.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Binary classification
# -----------------------
#
# This example shows how to use ATOM to solve a binary classification problem. Additonnaly, we'll perform a variety of data cleaning steps to prepare the data for modelling.
#
# The data used is a variation on the [Australian weather dataset](https://www.kaggle.com/jsphyg/weather-dataset-rattle-package) from Kaggle. You can download it from [here](https://github.com/tvdboom/ATOM/blob/master/examples/datasets/weatherAUS.csv). The goal of this dataset is to predict whether or not it will rain tomorrow training a binary classifier on target `RainTomorrow`.
# ## Load the data
# Import packages
import pandas as pd
from atom import ATOMClassifier
# +
# Load data
X = pd.read_csv("./datasets/weatherAUS.csv")
# Let's have a look
X.head()
# -
# ## Run the pipeline
# Call atom using only 5% of the complete dataset (for explanatory purposes)
atom = ATOMClassifier(X, "RainTomorrow", n_rows=0.05, n_jobs=8, warnings=False, verbose=2)
# Impute missing values
atom.impute(strat_num="median", strat_cat="drop", max_nan_rows=0.8)
# Encode the categorical features
atom.encode(strategy="Target", max_onehot=10, frac_to_other=0.04)
# Train an Extra-Trees and a Random Forest model
atom.run(models=["ET", "RF"], metric="f1", n_bootstrap=5)
# ## Analyze the results
# Let's have a look at the final results
atom.results
# Visualize the bootstrap results
atom.plot_results(title="RF vs ET performance")
# Print the results of some common metrics
atom.evaluate()
# The winner attribute calls the best model (atom.winner == atom.rf)
print(f"The winner is the {atom.winner.fullname} model!!")
# Visualize the distribution of predicted probabilities
atom.winner.plot_probabilities()
# + pycharm={"name": "#%%\n"}
# Compare how different metrics perform for different thresholds
atom.winner.plot_threshold(metric=["f1", "accuracy", "average_precision"], steps=50)
| docs_sources/examples/binary_classification.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Relationships in Data pt.1
# ## Variance
# Measures how far a set of numbers is spread out from their average.
# ### Compute the variance of an array of numbers
# +
import numpy as np
data = np.array([1, 3, 5, 2, 3, 7, 8, 4, 10, 0, 6, 7, 3, 0, 3, 0, 5, 7, 10, 1, 4, 9, 3])
# first we have a function to calculate the mean
def mean(data):
return sum(data) / len(data)
def variance(data):
m = mean(data)
S = 0
for xi in data:
S += xi
return S / float(len(data) - 1)
print(variance(data))
print(mean(data))
#To check your work you can use the built in numpy variance method (np.var())
print(np.var(data, ddof=1))
# -
# ### Application of variance
# +
import numpy as np
import matplotlib.pyplot as plt
# Running Distance in Mile
X = np.array([3.3,4.4,5.5,6.71,6.93,4.168,9.779,6.182,7.59,2.167,
7.042,10.791,5.313,7.997,5.654,9.27,3.1])
# Water Drinks in Litre
Y = np.array([1.7,2.76,2.09,3.19,1.694,1.573,3.366,2.596,2.53,1.221,
2.827,3.465,1.65,2.904,2.42,2.94,1.3])
plt.scatter(X, Y)
plt.xlabel('Running Distance (Mile)')
plt.ylabel('Water Drinks (Litre)')
# -
predicted_y_values = list(map(lambda x: 0.7*x + 0.3, X))
plt.scatter(X, Y)
plt.plot(X, predicted_y_values, 'ro-')
# ## Percentile
# Percentile is defined as the value below which a percentage of the data falls. Percentiles can help us interpret the standing of a particular value within a data set. Given a dataset we can calculate the nth percentile using the steps below:
#
# * Arrange the data in ascending order
# * Find the index of the (ordinal rank) of the percentile value by calculating index = ceiling((percent/100) * len(data))
# * Find the value that is located at the index
#
# +
data = np.array([1, 3, 5, 2, 3, 7, 8, 4, 10, 0, 6, 7, 3, 0, 3, 0, 5, 7, 10, 1, 4, 9, 3])
def percentile(data, percent):
#first we want to sort the data in ascending order
data = np.sort(data)
#then we will get the index
index = (percent/100)*len(data) #TODO: finish this
#we will have to round up to the nearest whole number using the ceiling method and covert to an int
index = int(np.ceil(index))
return data[index-1] #adjust by -1 since indices start with 0
print(percentile(data, 44))
#check your work by comparing to numpy.percentile()
print(int(np.percentile(data, 44)))
# -
# ## Covariance and Correlation
# #### Obtain the correlation between two columns in Titanic, Fare and Siblings/Spouses Aboard
# * We want to know if we have large famility size then can we conclude we paid more
# +
import pandas as pd
import scipy.stats
df = pd.read_csv('titanic.csv')
#here is a function to calculate pearson's correlation coefficient
def pearson_corr(x, y):
x_mean = np.mean(x)
y_mean = np.mean(y)
num = [(i - x_mean)*(j - y_mean) for i,j in zip(x,y)]
den_1 = [(i - x_mean)**2 for i in x]
den_2 = [(j - y_mean)**2 for j in y]
correlation_x_y = np.sum(num)/np.sqrt(np.sum(den_1))/np.sqrt(np.sum(den_2))
return correlation_x_y
print(pearson_corr(df['Fare'])# , df['Siblings/Spouses Aboard']))
print(scipy.stats.pearsonr(df['Fare'])#, df['Siblings/Spouses Aboard']))
| Class 2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .r
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: R
# language: R
# name: ir
# ---
# # Part 3 Twitter Data Analysis
# ## Installing and importing R packages
# Inorder to help with the efficient running of our R scripts, it is essential to install the necessary packages to help support the corresponding functions that help with data manipulation. Here, the packages installed are:
# 1. rlang - Supports the basic R functionalities
# 2. usmap - Allows plotting the map of USA
# 3. dplyr - Helps with manipulation and working with dataframes i.e filter(), select()
# 4. tidyverse - Helps with the easy installation and loading of other 'tidyverse' packages
# 5. gridExtra - Aids to work with grid-based plots and drawing tables
# +
.libPaths()
#install.packages("rlang",repos='http://cran.us.r-project.org')
#install.packages('usmap',repos='http://cran.us.r-project.org')
#install.packages("dplyr",repos='http://cran.us.r-project.org',versions="0.3.1")
#install.packages("tidyverse",repos='http://cran.us.r-project.org',versions="0.3.1")
#install.packages("gridExtra",repos='http://cran.us.r-project.org')
library(gridExtra)
# -
library(usmap)
library(dplyr)
library(tidyverse)
library(rtweet)
# ## Tweet Collection
# Here, we have used the rtweet package to help with Twitter data collection and processing. It allows the use of functions which after the authentication of your twitter key credentials, helps collect streaming tweets using the Twitter Search API. Here, the process followed is,
# 1. Used 'search_tweets()' function is collect recent tweets. This is done using multiple keywords related to flu. One of the parameters we use while searching is lookup_coords()' which makes use of the Google API key; it basically helps getting the latitude/longitude coordinate information for the specified location.
# 2. Everytime, the tweets are collected and stored in dataframes. Now, all the dataframes are combined into one to hold all the data together. For this, we used the 'bind_rows()' function.
# 3. From the collected data, we select only the necessary fields using the 'select()' function.
# 4. From this dataframe, we retrieve the location of the tweets.
# 5. Now, we write the collected data into a CSV file for storage.
token <-create_token(
app = "InforRetrieve",
consumer_key = "-",
consumer_secret = "-",
access_token = "-",
access_secret = "-")
flu <- search_tweets("influenza", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
flu1 <- search_tweets("flu", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
dim(flu1)
dim(twt_df1)
flu2 <- search_tweets("flu shot", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
dim(twt_df2)
flu3 <- search_tweets("flu virus", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
flu4 <- search_tweets("flu virus", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
flu5 <- search_tweets("H1N1", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
flu6 <- search_tweets("#H1N1", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
flu7 <- search_tweets("flu awareness", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
combo = bind_rows(flu,flu1)
dim(combo)
combo = bind_rows(combo,flu2)
combo = bind_rows(combo,flu3)
combo = bind_rows(combo,flu4)
combo = bind_rows(combo,flu5)
combo = bind_rows(combo,flu6)
combo = bind_rows(combo,flu7)
combo = bind_rows(combo,flu8)
combo = bind_rows(combo,flu9)
head(combo)
colnames(combo)
tweets <-select(combo, user_id, status_id, created_at, screen_name, text, location ,source,retweet_count,lang, verified,country,country_code,url )
write.csv(tweets, file="dic_tweets_initial.csv")
locat <-select(combo, screen_name, text, location)
write.csv(locat, file="dic_tweets.csv")
# ## Tweet data Manipulation
# Now, we continute the procedure from above. The data stored in the CSV is read using the 'read.csv()'. Before moving ahead with the map creation, it is essential to clean the data. We first strip the data of retweets which is done by setting the 'include_rts' parameter to false. Then we check for duplicates. If duplicate tweets exist, we need to remove those data rows. Once data has been filtered and cleaned, we need to retrieve the states assigned to each tweet and the count from each state.
locat1 <- read.csv("tweets_with_location.csv", header=T)
head(locat1)
duplicated(locat1)
head(locat1[duplicated(locat1),])
dim(locat1)
# +
draw_map <- function(locat){
states <- c()
names(locat)
lst <- as.vector(locat$location)
for(i in lst){
x <- strsplit(i, "," )[[1]][2] #%>%
#sapply(tail, 1 )
states <- c(states,x)
}
rs <- as.data.frame(table(states))
class(rs)
names(rs)
#rs
result <- merge(x = statepop, y = rs, x.by = "abbr", y.by = "states", all.x = T)
result <- result[trimws(result$states) == trimws(result$abbr),]
#write.csv(result, file="flu_processed.csv")
#sum(result$Freq)
#print(result)
plt <- plot_usmap(data = result, values = "Freq", lines = "black") +
scale_fill_continuous(name = "Freq", label = scales::comma) +
theme(legend.position = "right")
return(plt)
}
# -
# ## Map Creation
# We now plot the graphs for the diffrent datasets. For plotting purposes , we use 'plot_usmap()'. We have three maps generated below:
# 1. Heatmap for the total number of tweets collected
# 2. Heatmap for the tweets collected corresponding to two specific keywords
total_plt <- draw_map(locat1)
total_plt + ggtitle("Twitter Data HeatChart")
search_flu <- locat1[grepl("flu",locat1$text),]
#write.csv(search_flu, file="flu_tweets.csv")
flu_plt <- draw_map(search_flu)
flu_plt + ggtitle("HeatMap for Keyword FLU")
search_hn <- locat1[grepl("H1N1",locat1$text),]
#write.csv(search_hn, file="hn_tweets.csv")
hn_plt <- draw_map(search_hn)
hn_plt + ggtitle("HeatMap for Keyword H1N1")
flu8 <- search_tweets("fight flu", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
dim(flu8)
flu9 <- search_tweets("flu 2019", geocode = lookup_coords("usa"), n = 1000, include_rts = FALSE)
dim(flu9)
# ## Compare Maps
# The next task is to compare the heatmap obtained in part 2 i.e the map generated from the CDC data and the heatmap generated in part 3 from the collected tweet data. To help with this, we use the 'grid.arrange()' function.
# +
library(ggplot2)
library(usmap)
data3 <- read.csv(file='StateDatabyWeekforMap_2018-19week40-8.csv', header=T)
colfunc <- colorRampPalette(c("red", "yellow", "green"))
usmapdata <- merge(x=data3, y=statepop, x.by=STATENAME, y.by=full, x.all= TRUE)
usmapdata <- usmapdata[ usmapdata$STATENAME == usmapdata$full & usmapdata$WEEK == 8,]
unique(usmapdata$ACTIVITY.LEVEL)
usmapdata$ACTIVITY.LEVEL <- factor(usmapdata$ACTIVITY.LEVEL, levels = c("Level 10", "Level 9", "Level 8", "Level 7", "Level 6", "Level 5", "Level 4", "Level 1"))
#write.csv(usmapdata,file="cdcshiny.csv")
part2_plt <- plot_usmap(data = usmapdata, values = "ACTIVITY.LEVEL", lines = "black") +
scale_fill_manual(values = c("#FF0000", "#FF3800" ,"#FF7100" ,"#FFAA00" ,"#FFE200" ,"#E2FF00" ,"#AAFF00", "#71FF00", "#38FF00" ,"#00FF00")) +
theme(legend.position = "right",
legend.title = element_text("ILI Activity Level", face = "bold"),
plot.title = element_text(hjust = 0.5, face="bold", size=6.5)) +
ggtitle("2018-19 Influenza Season Week 8 ending Feb 23, 2019")
# -
# ## Twitter Data VS CDC HeatMap
# We display the heatmap generated for the CDC data and the total collected tweets against each other. The graphs illustrate the intensity of the tweet count from each of the corresponding states. With respect to the Twitter Data HeatChart, the intensity goes lighter with increase in count whereas for the CDC chart, darker the color greater the count. Twitter Chart shows light blue for the state of California depicting that it contains the largest number of tweets i.e about 215; a slightly darker shade of blue for New York with about 150 tweets. With respect to the CDC graph, the red shades potray the greater count.
grid.arrange(total_plt + ggtitle("Twitter Data HeatChart"),part2_plt,nrow=1)
# ## CDC HeatMap VS Flu Data
# We display the heatmap generated for the CDC data and the tweets collected for the keyword 'flu' against each other. Similar to the previous depiction, for Flu keyword DataChart, the intensity goes lighter with increase in count whereas for the CDC chart, darker the color greater the count. Flu Chart shows light blue for the state of California depicting that it contains the largest number of tweets; a slightly darker shade of blue for the state of New York, Texas. With respect to the CDC graph, the red shades potray the greater count.
grid.arrange(flu_plt + ggtitle("HeatMap for Keyword FLU"),part2_plt,nrow=1)
# ## CDC HeatMap VS H1N1 Data
# We display the heatmap generated for the CDC data and the tweets collected for the keyword 'H1N1' against each other. H1N1 Chart shows light blue for the state of Kansas depicting that it contains the largest number of tweets; a slightly darker shade of blue for the state of Florida, California. With respect to the CDC graph, the red shades potray the greater count.
grid.arrange(hn_plt + ggtitle("HeatMap for Keyword H1N1"),part2_plt,nrow=1)
| part3 - Flu twitter data exploration/Part3.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <h3>Simulación matemática 2018 </h3>
# <div style="background-color:#0099cc;">
# <font color = white>
# <ul>
# <li><NAME> </li>
# <li>Email: `<EMAIL>, <EMAIL>`</li>
# </ul>
# </font>
# </div>
# <!--NAVIGATION-->
# < [Programación Lineal](Clase5_ProgramacionLineal.ipynb) | [Guía](Clase0_GuiaSimulacionM.ipynb) | [Clasificación Binaria](Clase7_ClasificacionBinaria.ipynb) >
# ___
# # Ajuste de curvas
#
# <img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/a/a8/Regression_pic_assymetrique.gif" width="400px" height="125px" />
#
# > El **ajuste de curvas** es el proceso de construir una curva (función), que sea el mejor ajuste a una serie de puntos. Las curvas ajustadas pueden ser usadas como asistencia en la visualización de datos, para inferir valores de una función donde no hay datos disponibles, y para resumir la relación entre variables.
#
# **Referencia**:
# - https://en.wikipedia.org/wiki/Curve_fitting
# ___
# ## Introducción
#
# Consideremos un polinomio de grado uno:
#
# $$y = \beta_1 x + \beta_0.$$
#
# Esta es una **línea recta** que tiene pendiente $\beta_1$. Sabemos que habrá una línea conectando dos puntos cualesquiera. Por tanto, *una ecuación polinómica de primer grado es un ajuste perfecto entre dos puntos*.
#
# Si consideramos ahora un polinomio de segundo grado,
#
# $$y = \beta_2 x^2 + \beta_1 x + \beta_0,$$
#
# este se ajustará exactamente a tres puntos. Si aumentamos el grado de la función a la de un polinomio de tercer grado, obtenemos:
#
# $$y = \beta_3 x^3 + \beta_2 x^2 + \beta_1 x + \beta_0,$$
#
# que se ajustará a cuatro puntos.
#
# **Ejemplos**
# 1. Encontrar la línea recta que pasa exactamente por los puntos $(0,1)$ y $(1,0)$.
# 2. Encontrar la parábola que pasa exactamente por los puntos $(-1,1)$, $(0,0)$ y $(1,1)$.
#
# **Solución**
# 1. Consideramos $y=\beta_1 x + \beta_0$. Evaluando en el punto $(0,1)$, obtenemos $\beta_1(0) + \beta_0 = 1$. Ahora, evaluando en el punto $(1,0)$, obtenemos $\beta_1(1) + \beta_0 = 0$. De esta manera,
# $$\left[\begin{array}{cc} 1 & 0 \\ 1 & 1\end{array}\right]\left[\begin{array}{c} \beta_0 \\ \beta_1\end{array}\right]=\left[\begin{array}{c} 1 \\ 0\end{array}\right].$$
# Resolviendo, $\beta_0=-\beta_1=1$.
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
# %matplotlib inline
# +
P1 = [0, 1]
P2 = [1, 0]
X = np.array([[1, 0], [1, 1]])
y = np.array([1, 0])
b0, b1 = np.linalg.inv(X).dot(y)
b0, b1
# +
x = np.linspace(-0.2, 1.2, 100)
y = b1*x + b0
plt.figure(figsize=(6,6))
plt.scatter([0, 1], [1, 0], c = "r", s = 50);
plt.plot(x, y, 'b', label = 'recta ajustada')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.legend(loc = 'best')
plt.grid(True)
plt.show()
# -
# 2. Consideramos $y=\beta_2 x^2 + \beta_1 x + \beta_0$. Evaluando en el punto $(-1,1)$, obtenemos $\beta_2(-1)^2 + \beta_1(-1) + \beta_0 = 1$. Ahora, evaluando en el punto $(0,0)$, obtenemos $\beta_2(0)^2 + \beta_1(0) + \beta_0 = 0$. Finalmente, evaluando en el punto $(1,1)$, obtenemos $\beta_2(1)^2 + \beta_1(1) + \beta_0 = 1$. De esta manera,
# $$\left[\begin{array}{ccc} 1 & -1 & 1 \\ 1 & 0 & 0 \\ 1 & 1 & 1 \end{array}\right]\left[\begin{array}{c} \beta_0 \\ \beta_1 \\ \beta_2 \end{array}\right]=\left[\begin{array}{c} 1 \\ 0 \\ 1 \end{array}\right].$$
# Resolviendo, $\beta_0=\beta_1=0$ y $\beta_2=1$.
# +
P1 = [-1, 1]
P2 = [0, 0]
P3 = [1, 1]
X = np.array([[1, -1, 1], [1, 0, 0], [1, 1, 1]])
y = np.array([1, 0, 1])
b0, b1, b2 = np.linalg.inv(X).dot(y)
b0, b1, b2
# +
x = np.linspace(-1.2, 1.2, 100)
y = b2*x**2+b1*x+b0
plt.figure(figsize=(6,6))
plt.scatter([-1,0,1],[1,0,1], s = 100, label = 'puntos')
plt.plot(x, y, 'b', label = 'parábola ajustada')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.legend(loc = 'best')
plt.grid(True)
plt.show()
# -
# ### ¿Qué tienen en común los anteriores problemas?
# Las curvas están completamente determinadas por los puntos (datos limpios, suficientes y necesarios).
#
# Esto se traduce en que, al llevar el problema a un sistema de ecuaciones lineales, existe una única solución: **no hay necesidad, ni se puede optimizar nada**.
#
# ¿Tendremos datos así de '*bonitos*' en la vida real?
#
# La realidad es que los datos que encontraremos en nuestra vida profesional se parecen más a esto...
# +
x = np.linspace(0, 1, 30)
y = 10*x + 2 + np.random.randn(30)
plt.figure(figsize=(6,6))
plt.scatter(x, y)
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# ### ¿Cómo ajustamos una curva a esto?
# ## Problema básico
#
# <img style="float: right; margin: 0px 0px 15px 15px;" src="https://upload.wikimedia.org/wikipedia/commons/3/3a/Linear_regression.svg" width="400px" height="125px" />
#
# Consideramos que tenemos un conjunto de n pares ordenados de datos $(x_i,y_i)$, para $i=1,2,3,\dots,n$.
#
# ### ¿Cuál es la recta que mejor se ajusta a estos datos?
# Consideramos entonces ajustes de la forma $\hat{f}(x) = \beta_0+\beta_1 x = \left[1 \quad x\right]\left[\begin{array}{c} \beta_0 \\ \beta_1 \end{array}\right]=\left[1 \quad x\right]\boldsymbol{\beta}$ (lineas rectas).
#
# Para decir '*mejor*', tenemos que definir algún sentido en que una recta se ajuste *mejor* que otra.
#
# **Mínimos cuadrados**: el objetivo es seleccionar los coeficientes $\boldsymbol{\beta}=\left[\beta_0 \quad \beta_1 \right]^T$, de forma que la función evaluada en los puntos $x_i$ ($\hat{f}(x_i)$) aproxime los valores correspondientes $y_i$.
#
# La formulación por mínimos cuadrados, encuentra los $\boldsymbol{\beta}=\left[\beta_0 \quad \beta_1 \right]^T$ que minimiza
# $$\sum_{i=1}^{n}(y_i-\hat{f}(x_i))^2=\sum_{i=1}^{n}(y_i-\left[1 \quad x_i\right]\boldsymbol{\beta})^2=\left|\left|\boldsymbol{y}-\boldsymbol{X}\boldsymbol{\beta}\right|\right|^2,$$
#
# donde $\boldsymbol{y}=\left[y_1\quad\dots\quad y_n\right]^T$, y $\boldsymbol{X}=\left[\begin{array}{ccc}1 & x_1\\ \vdots & \vdots \\ 1 & x_n\end{array}\right].$ Esto es,
#
# $$\boldsymbol{\beta}^{ls} = \arg \min_{\boldsymbol{\beta}} \left|\left|\boldsymbol{y}-\boldsymbol{X}\boldsymbol{\beta}\right|\right|^2$$
# Para llevar a cabo la anterior minimización, la librería `SciPy` en su módulo `optimize` contiene la función `minimize`.
import scipy.optimize as opt
def fun_obj1(b, x, y):
return np.sum((y-b[0]-b[1]*x)**2)
b0 = np.array([1, 5])
res = opt.minimize(fun_obj1, b0, args = (x, y))
res
# +
yhat = res.x[0]+res.x[1]*x
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'Datos')
plt.plot(x, yhat, '-r', label = 'Ajuste')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# ### Ajuste polinomial
#
# Ahora, considere el siguiente conjunto de datos...
# +
n = 100
x = np.linspace(np.pi/6, 5*np.pi/3, n)
y = 4*np.sin(x) + 0.5*np.random.randn(n)
plt.figure(figsize=(6,6))
plt.scatter(x, y)
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# #### Ajustando una línea recta ?
# +
def obj1(b, x, y):
return np.sum((y-b[0]-b[1]*x)**2)
b0 = np.random.random((2,))
# -
res = opt.minimize(obj1, b0, args=(x,y))
res
# +
yhat1 = res.x[0]+res.x[1]*x
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'datos')
plt.plot(x, yhat1, '-r', label = 'ajuste 1')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# #### Ajustando una parábola?
# +
def obj2(b, x, y):
return np.sum((y-b[0]-b[1]*x-b[2]*x**2)**2)
b0 = np.random.random((3,))
# +
res = opt.minimize(obj2, b0, args=(x,y))
yhat2 = res.x[0]+res.x[1]*x+res.x[2]*x**2
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'datos')
plt.plot(x, yhat1, '-r', label = 'ajuste 1')
plt.plot(x, yhat2, '-g', label = 'ajuste 2')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# #### Quizá un polinomio cúbico...
# +
def obj3(b, x, y):
return np.sum((y-b[0]-b[1]*x-b[2]*x**2-b[3]*x**3)**2)
b0 = np.random.random((4,))
# +
res = opt.minimize(obj3, b0, args=(x,y))
yhat3 = res.x[0]+res.x[1]*x+res.x[2]*x**2+res.x[3]*x**3
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'datos')
plt.plot(x, yhat1, '-r', label = 'ajuste 1')
plt.plot(x, yhat2, '-g', label = 'ajuste 2')
plt.plot(x, yhat3, '-k', label = 'ajuste 3')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# #### Entonces, ¿mientras más se suba el orden mejor la aproximación?
#
# ## <font color = red > ¡Cuidado! OVERFITTING... </font>
def obj7(b, x, y):
return np.sum((y-np.array([x**i for i in range(8)]).T.dot(b))**2)
b0 = np.random.random((8,))
res = opt.minimize(obj7, b0, args=(x,y))
yhat7 = np.array([x**i for i in range(8)]).T.dot(res.x)
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'datos')
plt.plot(x, yhat1, '-r', label = 'ajuste 1')
plt.plot(x, yhat2, '-g', label = 'ajuste 2')
plt.plot(x, yhat3, '-k', label = 'ajuste 3')
plt.plot(x, yhat7, '-c', label = 'ajuste 7')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# #### Es conveniente ver el error como función del orden del polinomio... <font color = red> selección de modelos </font>
# +
e_ms = []
def obj(b, x, y, n):
return np.sum((y - np.array([x**i for i in range(n + 1)]).T.dot(b))**2)
for i in range(7):
b0 = np.random.random((i + 2,))
res = opt.minimize(obj, b0, args=(x,y,i + 1))
yhat = np.array([x**j for j in range(i + 2)]).T.dot(res.x)
e_ms.append(sum((y - yhat)**2))
plt.figure(figsize=(6,6))
plt.plot(np.arange(7) + 1, e_ms, 'o')
plt.xlabel('orden', fontsize = 18)
plt.ylabel('error', fontsize = 18)
plt.show()
# -
# ### ¿Cómo prevenir el <font color = red > *overfitting* </font> sin importar el orden del modelo?
# ## Regularización
#
# Vimos que la solución de mínimos cuadrados es:
# $$\boldsymbol{\beta}^{ls} = \arg \min_{\boldsymbol{\beta}} \left|\left|\boldsymbol{y}-\boldsymbol{X}\boldsymbol{\beta}\right|\right|^2.$$
#
# Sin embargo, si crecemos el orden del modelo hay overfitting y algunos coeficientes óptimos $\boldsymbol{\beta}$ crecen muchísimo. Que un coeficiente sea muy grande, significa que se le da mucha importancia a alguna característica (que quizá sea ruido... no sirve para predecir).
#
# La regularización consiste en penalizar la magnitud de los coeficientes $\boldsymbol{\beta}$ en el problema de optimización, para que no crezcan tanto.
# - [Ridge](Ridge.ipynb)
# - [Lasso](Lasso.ipynb)
# - [Ajuste robusto](Ajuste_robusto.ipynb)
#
# ___
# ### Actividad
#
# 1. Ajustar polinomios de grado 1 hasta grado 7 a los siguientes datos.
# 2. Graficar el error cuadrático acumulado contra el número de términos, y elegir un polinomio que ajuste bien y su grado no sea muy alto.
# 4. Comparar los beta.
#
# Abrir un nuevo notebook, llamado `ActividadClase6_nombreApellido`.
def f(x):
return np.exp(-x**2/2)/np.sqrt(2*np.pi)
# +
x = np.linspace(-3, 3)
y = f(x) + 0.04*np.random.randn(50)
plt.figure(figsize=(6,6))
plt.scatter(x, y, label = 'datos')
plt.legend(loc = 'best')
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.grid(True)
plt.show()
# -
# ___
# <!--NAVIGATION-->
# < [Programación Lineal](Clase5_ProgramacionLineal.ipynb) | [Guía](Clase0_GuiaSimulacionM.ipynb) | [Clasificación Binaria](Clase7_ClasificacionBinaria.ipynb) >
# <script>
# $(document).ready(function(){
# $('div.prompt').hide();
# $('div.back-to-top').hide();
# $('nav#menubar').hide();
# $('.breadcrumb').hide();
# $('.hidden-print').hide();
# });
# </script>
#
# <footer id="attribution" style="float:right; color:#808080; background:#fff;">
# Created with Jupyter by <NAME>.
# <Strong> Copyright: </Strong> Public Domain como en [CC](https://creativecommons.org/licenses/by/2.0/) (Exepto donde se indique lo contrario)
#
#
# </footer>
| Modulo1/Clase6_AjusteCurvas.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python
# language: python3
# name: python3
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# # Foundations of Computational Economics #8
#
# by <NAME>, ANU
#
# <img src="_static/img/dag3logo.png" style="width:256px;">
# + [markdown] slideshow={"slide_type": "fragment"}
# ## Bundle goods market
#
# <img src="_static/img/lab.png" style="width:64px;">
# + [markdown] slideshow={"slide_type": "subslide"}
# <img src="_static/img/youtube.png" style="width:65px;">
#
# [https://youtu.be/Y6CtsI8X914](https://youtu.be/Y6CtsI8X914)
#
# Description: Object oriented programming in modeling consumer choice model.
# + [markdown] slideshow={"slide_type": "slide"}
# Consider the following model of a bundle goods market. A
# bundle of goods is a collection of particular items offered at a
# specified price. For example, Happy Meals at McDonalds is a set
# of items in a meal sold for a particular price.
#
# One other example of bundled goods - subscription packages in theaters,
# for example [La Scala in
# Milan](http://www.teatroallascala.org/en/box-office/subscriptions/types/subscription-types-2018-2019.html)
# or [Mariinsky in
# St.Petersburg](https://www.mariinsky.ru/playbill/subscriptions/2018_2019).
#
# In this task you will write code to implement and operationalize this
# setup.
# + [markdown] slideshow={"slide_type": "slide"}
# ### Bundle_good class
#
# Develop the Python class to represent a bundle good with the following specifications:
#
# - The class attribute (common to all objects of this class) is a
# list of goods
# - The public property is a vector of integers defining how many of each goods are in the bundle
# - The other property is the price for that bundle
#
#
# The following arithmetic operations are defined for the bungles:
#
# - addition:
# 1. sum of two bundles is a bundle with added up items and prices
# 1. sum of a bundle and a number (float or int) increases the price
# - subtraction:
# 1. difference between two bundles should produce a bundle with
# difference in items and difference in prices
# 1. subtracting a number (float or int) from a bundle should only
# decrease its price
# - multiplication is only defined for bundle and an integers, and results in the bundle with all items multiplied by this number, and price increased by the same number
# - devision is only defined for integers, and only such that the all quantities are divisible by this integer, the resulting bundle is a fraction of the original, with the price also decreased by the same number
#
#
# Complete the class definition code, and run the tests in the next cell.
# + hide-output=false slideshow={"slide_type": "slide"}
class bundle_good():
'''Class of bundled goods with well defined arithmetics'''
items = ('Opera A', 'Opera B', \
'Ballet A', 'Ballet B', \
'Symphonic orchestra concert', \
'Rock opera', \
'Operetta') # 7 different goods
def __init__(self,quantities=[0,],price=0.0):
'''Creates the bundle good object, empty by default'''
pass
# ignore extra quantities if passed
# add zeros for the unspecified items
# ensure all quantities are integers
def __repr__(@@@):
'''String representation of the object'''
pass
def __add__(self,other):
'''Addition for bundle goods'''
pass
# if wrong type pass, raise the TypeError
# raise TypeError('Can only add bundle to bundle, or number to bundle price')
def __sub__(self,other):
'''Subtraction for bundles: subtract items and prices, or decrease price'''
pass
def __mul__(self,num):
'''Multiplication for bundles: proportional increase in nomenclature and price'''
pass
def __truediv__(self,num):
'''Division for bundles: fraction of the original bundle, only if quantities are divisable'''
pass
# + [markdown] slideshow={"slide_type": "slide"}
# ### Tests
#
# To make sure the class is running as it is supposed to, run all the
# tests below and confirm that the output is as expected.
# + hide-output=false slideshow={"slide_type": "slide"}
# Tests
x=bundle_good([1,2,3,4,5,6,7],11.43)
print(x) #should print "Bundle object [1, 2, 3, 4, 5, 6, 7] with price 11.43"
# + hide-output=false slideshow={"slide_type": "slide"}
x=bundle_good([1,2])
print(x) #should print "Bundle object [1, 2, 0, 0, 0, 0, 0] with price 0.00"
# + hide-output=false slideshow={"slide_type": "slide"}
x=bundle_good(range(25),100.2)
print(x) #should print "Bundle object [0, 1, 2, 3, 4, 5, 6] with price 100.20"
# + hide-output=false slideshow={"slide_type": "slide"}
x=bundle_good([1.5,2.3,3.2,4.1,5.75,6.86,7.97],1.43)
print(x) #should print "Bundle object [1, 2, 3, 4, 5, 6, 7] with price 1.43"
# + hide-output=false slideshow={"slide_type": "slide"}
x=bundle_good([1,2,3,4,5,6,7],11.43)
y=bundle_good([7,6,5,4,3,2,1],77.45)
z=x+y
print(z) #should print "Bundle object [8, 8, 8, 8, 8, 8, 8] with price 88.88"
# + hide-output=false slideshow={"slide_type": "slide"}
z=y-x
print(z) #should print "Bundle object [6, 4, 2, 0, -2, -4, -6] with price 66.02"
# + hide-output=false slideshow={"slide_type": "slide"}
z=x+4.531
print(z) #should print "Bundle object [1, 2, 3, 4, 5, 6, 7] with price 15.96"
# + hide-output=false slideshow={"slide_type": "slide"}
z=y-77
print(z) #should print "Bundle object [7, 6, 5, 4, 3, 2, 1] with price 0.45"
# + hide-output=false slideshow={"slide_type": "slide"}
z=x*11
print(z) #should print "Bundle object [11, 22, 33, 44, 55, 66, 77] with price 125.73"
# + hide-output=false slideshow={"slide_type": "slide"}
try:
z=x*11.5 #should raise a TypeError
except TypeError:
print("Ok 1") #should print "Ok 1"
# + hide-output=false slideshow={"slide_type": "slide"}
try:
z=x*y #should raise a TypeError
except TypeError:
print("Ok 2") #should print "Ok 2"
# + hide-output=false slideshow={"slide_type": "slide"}
try:
z=x/y #should raise a TypeError
except TypeError:
print("Ok 3") #should print "Ok 3"
# + hide-output=false slideshow={"slide_type": "slide"}
z=(x+y)/8
print(z) #should print "Bundle object [1, 1, 1, 1, 1, 1, 1] with price 11.11"
# + hide-output=false slideshow={"slide_type": "slide"}
try:
(x+y)/7 #should raise a ValueError
except ValueError:
print("Ok 4") #should print "Ok 4"
# + hide-output=false slideshow={"slide_type": "slide"}
z=x*15-y*2
print(z) #should print "Bundle object [1, 18, 35, 52, 69, 86, 103] with price 16.55"
# + [markdown] slideshow={"slide_type": "slide"}
# ### Solution
# + hide-output=false slideshow={"slide_type": "slide"}
class bundle_good():
'''Class of bundled goods with well defined arithmetics'''
items = ('Opera A', 'Opera B', \
'Ballet A', 'Ballet B', \
'Symphonic orchestra concert', \
'Rock opera', \
'Operetta') # 7 different goods
def __init__(self,quantities=[0,],price=0.0):
'''Creates the bundle good object
'''
n = len(bundle_good.items) # number of available items
if len(quantities)<n:
# add zeros for the unspecified items
quantities += [0,]*(n-len(quantities))
elif len(quantities)>n:
# ignore extra numbers
quantities = quantities[0:n]
# create public attributes
# ensure the quantities in the object are integer
self.quantities=[int(x) for x in quantities]
self.price=price
def __repr__(self):
'''String representation of the object
'''
return 'Bundle object %r with price %1.2f' % (self.quantities,self.price)
def __add__(self,other):
'''Addition for bundles: add items and sum prices, or increase price
'''
if type(other) is bundle_good:
# add the quantities using list comprehension with one-to-one matching (zip)
q1 = [x+y for x,y in zip(self.quantities, other.quantities)]
# sum of the prices
p1 = self.price + other.price
# return new bundle
return bundle_good(quantities=q1,price=p1)
elif type(other) in (float,int):
# increase the price
p1 = self.price + other
# return new bundle
return bundle_good(quantities=self.quantities,price=p1)
else:
raise TypeError('Can only add bundle to bundle, or number to bundle price')
def __sub__(self,other):
'''Subtraction for bundles: subtract items and prices, or decrease price
'''
if type(other) is bundle_good:
# subtract the quantities using list comprehension with one-to-one matching (zip)
q1 = [x-y for x,y in zip(self.quantities, other.quantities)]
# sum of the prices
p1 = self.price - other.price
# return new bundle
return bundle_good(quantities=q1,price=p1)
elif type(other) in (float,int):
# decrease the price
p1 = self.price - other
# return new bundle
return bundle_good(quantities=self.quantities,price=p1)
else:
raise TypeError('Can only subtract bundle from bundle, or number from bundle price')
def __mul__(self,num):
'''Multiplication for bundles: repetition of the original bundle
'''
if type(num) is int:
# multiply quantities using list comprehension
q1 = [x * num for x in self.quantities]
# multiply the price
p1 = self.price * num
# return new bundle
return bundle_good(price=p1,quantities=q1)
else:
raise TypeError('Can only multiply bundle by an integer')
def __truediv__(self,num):
'''Division for bundles: fraction of the original bundle, only if quantities are devisable
'''
if type(num) is int:
# divide quantities and check for divisibility
q1 = [q//num for q in self.quantities]
if not all(q%num==0 for q in self.quantities):
# if can not be devided without a remainder, raise ValueError
raise ValueError('Can not divide bundle into fractional parts')
# divide the price
p1=self.price / num
# return new bundle
return bundle_good(price=p1,quantities=q1)
else:
raise TypeError('Can only divide bundle by an integer')
| 08_bundles_ex2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
# %matplotlib inline
from tqdm import tqdm_notebook
import concurrent.futures
from multiprocessing import Pool
def mean_list(inp):
l = len(inp)
return sum(inp)/l
# # 梯度下降
# ## 各个损失函数的偏导
# ### **MSE (回归)**
# MSE计算公式:
# - $ MSE = (y-y_{true})^2 $
# - <font color=gray>[batch 累加形式] $ MSE = \frac{1}{m}\sum_{i=1}^{m}(y-y_i)^2$ </font>
#
# MSE偏导数:$\frac{\partial MSE}{\partial x} = 2(y-y_{true})\frac{\partial y}{\partial x}$
#
# 例如 $y=ax$ 带入到MSE计算**(a的)**偏导为 $\frac{\partial MSE}{\partial a} = 2(y-y_{true})\frac{\partial y}{\partial a} = 2(y-y_{true})x$
# ### Logloss (分类)
# - y、y_true 都是0、1类别(二分类)
# - 底数是e
#
# Logloss计算公式:
# - $ logloss = \frac{y_{true}}{n}log(y)+\frac{1-y_{true}}{n}log(1-y)$
# - <font color=gray>[batch 累加形式] $ logloss = -\sum_{i=1}^{n}(\frac{y_i}{n}log(p_i)+\frac{(1-y_i)}{n}log(1-p_i)) $ </font>
#
# Logloss偏导数:$\frac{\partial logloss}{\partial x} = \frac{y_{true}}{n}\frac{1}{y}\frac{\partial y}{\partial x}+\frac{1-y_{true}}{n}\frac{1}{1-y}(-\frac{\partial y}{\partial x})$
#
# 例如 $y=ax$ 带入到Logloss计算**(a的)**偏导为 $\frac{\partial MSE}{\partial a} = 2(y-y_{true})\frac{\partial y}{\partial a} = 2(y-y_{true})x$
# ### 直接误差
# 直接误差计算公式:$ Direct = y-y_{true} $
#
# 直接误差偏导数:$\frac{\partial Direct}{\partial x} = \frac{\partial y}{\partial x}$
#
# 例如 $y=ax$ 带入到直接误差计算**(a的)**偏导为 $\frac{\partial Direct}{\partial a} = \frac{\partial y}{\partial a} = x$
#
# ## 解一元一次方程(无常数项)
# ### SGD
# +
# 目标函数 y=ax,构造一批样本
import random
a_true=9
def y_true(x):
return a_true*x
allSamples = [[i,y_true(i)] for i in range(150)]
samples = allSamples[:100]
verify_samples = allSamples[100:]
a=0.05 # 初始化a
n = 0.001 # 定义学习率为0.01
# 损失函数设计为均方误差[mse = (y-y_true)^2]
# 参数更新方式为 param_new = param - 学习率*损失函数对param的(在x处的)偏导数
print(f"[true]: y={a_true}x")
print(f"[initial]: y={a}x")
it = 0
while it <= 2:
print(f"\n\n[第 {it} 次迭代]")
cnt = 0
for (x,y_true) in samples:
y = a*x
grad_a = (y-y_true)*x
a = a - n*grad_a #
verify_list = [pow((a*x-y_true),2) for (x,y_true) in verify_samples]
verify_mse = sum(verify_list)/len(verify_list)
if cnt%5==0:
print(f" x:{x}, y={a:.4f}x, verify_mse:{verify_mse:.4f}, grad_a:{grad_a:.4f}")
cnt += 1
if verify_mse<=0.001:
print(" [已完成]:")
print(f" x:{x}, y={a:.4f}x, verify_mse:{verify_mse:.4f}, grad_a:{grad_a:.4f}")
it = 2
break
it += 1
# assert False
# mse_list = [pow((a*x+b-y_true),2) for (x,y_true) in samples]
# new_mse = sum(mse_list)/len(mse_list)
# print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse}")
# -
# ### SGD (mini-batch)
# +
import itertools
# 目标函数 y=ax,构造一批样本
import random
a_true=9
def y_true(x):
return a_true*x
allSamples = [[i,y_true(i)] for i in range(150)]
samples = allSamples[:100]
verify_samples = allSamples[100:]
a=0.05 # 初始化a
n = 0.001 # 定义学习率为0.01
# 损失函数设计为均方误差[mse = (y-y_true)^2]
# 参数更新方式为 param_new = param - 学习率*损失函数对param的(在x处的)偏导数
print(f"[true]: y={a_true}x")
print(f"[initial]: y={a}x")
it = 0
while it <= 2:
print(f"\n\n[第 {it} 次迭代]")
cnt = 0
for (x,y_true) in samples:
y = a*x
grad_a = (y-y_true)*x
a = a - n*grad_a #
verify_list = [pow((a*x-y_true),2) for (x,y_true) in verify_samples]
verify_mse = sum(verify_list)/len(verify_list)
if cnt%5==0:
print(f" x:{x}, y={a:.4f}x, verify_mse:{verify_mse:.4f}, grad_a:{grad_a:.4f}")
cnt += 1
if verify_mse<=0.001:
print(" [已完成]:")
print(f" x:{x}, y={a:.4f}x, verify_mse:{verify_mse:.4f}, grad_a:{grad_a:.4f}")
it = 2
break
it += 1
# assert False
# mse_list = [pow((a*x+b-y_true),2) for (x,y_true) in samples]
# new_mse = sum(mse_list)/len(mse_list)
# print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse}")
# -
# ### GD
# # 二元一次
#
# +
# 目标函数 2x1+3x2+5,构造一批样本
import random
(a_true,b_true,c_true) = (2,3,0)
def y_true(x1,x2):
return a_true*x1+b_true*x2+c_true
samples = [[i,i+1,y_true(i,i+1)] for i in range(100)]
samples[:2]
(a,b,c) = (0.5,0.5,0) # 0.5初始化,或随机初始化a,b,c
n = 0.001 # 定义学习率为0.1
# 损失函数设计为均方误差
# 参数更新方式为 param_new = param - 学习率*损失函数对param的(在x处的)偏导数
print(f"[true]: y={a_true}x1+{b_true}x2+{c_true}")
print(f"[initial]: y={a}x1+{b}x2+{c}")
for _ in range(2):
print(f"第 {_} 次迭代")
for (x1,x2,y_true) in tqdm_notebook(samples):
y = a*x1+b*x2+c
grad_a = (y-y_true)*x1
grad_b = (y-y_true)*x2
grad_c = (y-y_true)*1
a = a - n*grad_a #
b = b - n*grad_b #
# c = c - n*grad_c
mse_list = [pow((a*x1+b*x2+c-y_true),2) for (x1,x2,y_true) in samples]
new_mse = sum(mse_list)/len(mse_list)
print(f"y={a:.4f}x1+{b:.4f}x2+{c:.4f}, new_mse:{new_mse:.2f}, x1:{x1},x2:{x2},grad_a:{grad_a:.4f},grad_b:{grad_b:.4f}")
if new_mse<=0.01:
break
print(f"y={a:.4f}x1+{b:.4f}x2+{c:.4f}, new_mse:{new_mse:.2f}")
# assert False
# mse_list = [pow((a*x+b-y_true),2) for (x,y_true) in samples]
# new_mse = sum(mse_list)/len(mse_list)
# print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse}")
# -
# # 一元二次方程
# +
# 目标函数 2x+5,构造一批样本
import random
(a_true,b_true) = (9,2)
def y_true(x):
return pow(a_true*x,2)+b_true
samples = [[i,y_true(i)] for i in range(100)]
samples[:2]
(a,b) = (0.5,0.5) # 0.5初始化,或随机初始化a,b
n = 0.001 # 定义学习率为0.1
# 损失函数设计为均方误差
# 参数更新方式为 param_new = param - 学习率*损失函数对param的(在x处的)偏导数
print(f"[true]: y={a_true}x+{b_true}")
print(f"[initial]: y={a}x+{b}")
for _ in range(10):
print(f"第 {_} 次迭代")
for (x,y_true) in tqdm_notebook(samples):
y = a*x+b
grad_a = (y-y_true)*x
grad_b = (y-y_true)*1
a = a - n*grad_a #
b = b - n*grad_b #
mse_list = [pow((a*x+b-y_true),2) for (x,y_true) in samples]
new_mse = sum(mse_list)/len(mse_list)
print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse:.2f}, grad_a:{grad_a:.4f},grad_b:{grad_b:.4f}")
if new_mse<=0.01:
break
print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse:.2f}")
# assert False
# mse_list = [pow((a*x+b-y_true),2) for (x,y_true) in samples]
# new_mse = sum(mse_list)/len(mse_list)
# print(f"y={a:.4f}x+{b:.4f}, new_mse:{new_mse}")
# -
| note_books/Basic.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Binary classification single feature
#
# Classification using "raw" python or libraries (SciKit Learn, Tensorflow).
#
# The classification is first on a single boundary defined by a continuous univariate function and added white noise
# +
import math
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as pltcolors
from sklearn import metrics as metrics
from sklearn.linear_model import LogisticRegression as SkLinReg
import scipy as sy
import seaborn as sns
import pandas
import tensorflow as tf
# -
# ## Model
#
# We want to measure or predict a value y to be above a threshold. E.g.: y is a temperature.
#
# We know a feature x, y is related to x through a quadratic function we do not a priori know and some unknown
#
# This unknown is modeled by a Gaussian noise
# Single feature, Gaussian noise
nFeatures = 1
def generateBatch(N):
#
xMin = 0
xMax = 1
b = 0.2
std = 0.2
# Threshold from 0 to 1
threshold = 1
#
x = np.random.uniform(xMin, xMax, N)
# 4th degree relation between y and x
yClean = 2*(x**4 + (x-0.3)**3 + b)
labels = yClean + np.random.normal(0, std, N) > threshold
return (x, yClean, labels)
# The values of X are uniformly distributed and independent
# +
N = 2000
# x and y have 1 dim in R, label has 1 dim in B
xTrain, yCleanTrain, labelTrain = generateBatch(N)
colors = ['blue','red']
fig = plt.figure(figsize=(8,4))
plt.subplot(1,2,1)
plt.scatter(xTrain, yCleanTrain, c=labelTrain, cmap=pltcolors.ListedColormap(colors), marker=',', alpha=0.01)
plt.xlabel('x')
plt.ylabel('y')
plt.grid()
plt.subplot(1,2,2)
plt.scatter(xTrain, labelTrain, marker=',', alpha=0.01)
plt.xlabel('x')
plt.ylabel('label')
plt.grid()
# -
count, bins, ignored = plt.hist(labelTrain*1.0, 10, density=True, alpha=0.5)
p = np.mean(labelTrain)
print('Bernouilli parameter of the distribution:', p)
# Note: The two values are not a priori equi probable. In theory, ressampling of the training values would be required to balance the a priori distribution.
xTest, yTest, labelTest = generateBatch(N)
# ## Helpers
# +
def plotHeatMap(X, classes, title=None, fmt='.2g', ax=None, xlabel=None, ylabel=None):
""" Fix heatmap plot from Seaborn with pyplot 3.1.0, 3.1.1
https://stackoverflow.com/questions/56942670/matplotlib-seaborn-first-and-last-row-cut-in-half-of-heatmap-plot
"""
ax = sns.heatmap(X, xticklabels=classes, yticklabels=classes, annot=True, fmt=fmt, cmap=plt.cm.Blues, ax=ax) #notation: "annot" not "annote"
bottom, top = ax.get_ylim()
ax.set_ylim(bottom + 0.5, top - 0.5)
if title:
ax.set_title(title)
if xlabel:
ax.set_xlabel(xlabel)
if ylabel:
ax.set_ylabel(ylabel)
def plotConfusionMatrix(yTrue, yEst, classes, title=None, fmt='.2g', ax=None):
plotHeatMap(metrics.confusion_matrix(yTrue, yEst), classes, title, fmt, ax, xlabel='Estimations', \
ylabel='True values');
# -
# ### Logistic and log of Logistic functions
def logistic(X):
return (1+(np.exp(-(X))))**-1
xx = np.linspace(-10, 10)
xlogistic = logistic(xx)
plt.figure(figsize=(10,5))
plt.subplot(1, 2, 1)
plt.plot(xx, xlogistic)
plt.grid()
plt.subplot(1, 2, 2)
plt.plot(xx, np.log(xlogistic))
plt.grid()
# # Logistic regression
#
# \begin{align}
# y \in \left\{ 0, 1 \right\}
# \end{align}
#
# \begin{align}
# p(Y=1 \mid x) & = \frac{1}{1+e^{-f_\theta(x)}} \\
# f_\theta(x) & = b + w x \\
# \theta &= \{b, w\}
# \end{align}
#
# We are looking for the value of w that maximize the likelyhood:
# \begin{align}
# \hat{\theta} & = \max_{\theta}{\prod_{i=0}^N{p(y_i \mid x_i, w)}} \\
# & = \max_{\theta}{\sum_{i=0}^N{log \left(p(y_i \mid x_i, w)\right)} } \\
# & = \max_{\theta}{\sum_{i=0}^N{log \left(\left(\frac{1}{1+e^{-f_\theta(x_i)}}\right)^{y_i}\left(1-\frac{1}{1+e^{-f_\theta(x_i)}}\right)^{1-y_i}\right)} } \\
# & = \max_{\theta}{\sum_{i=0}^N{log \left(y_i * \left(\frac{1}{1+e^{-f_\theta(x_i)}}\right) + \left(1-y_i\right) * \left(1-\frac{1}{1+e^{-f_\theta(x_I)}}\right) \right)} } \\
# \end{align}
#
# Using the fact that $y_i$ is either 0 or 1. The last formulation is avoiding logarithm of zero as one of the two terms within the sum is null.
#
# Since the number of classes is 2, the maximum log likelyhood is also called binary cross entropy.
#
# Reference:
# - https://en.wikipedia.org/wiki/Logistic_regression
#
# ## Fitting of $b$ and then $w$
#
#
# Suboptimal fitting:
# - Taking some assumption on $w$ to fit $b$ as $\hat{b}$
# - and then fitting $w$ with the $\hat{b}$ estimate
b = np.linspace(-5, 5)
w = 1
px = np.zeros(len(b))
for i in range(len(b)):
fx = logistic(b[i] + w*xTrain)
px[i] = 1/N * np.sum(np.log(labelTrain*fx + (1-labelTrain)*(1-fx)))
plt.plot(b, px);
plt.xlabel('$b$')
plt.ylabel('l(b, X)')
plt.grid()
bHat = b[np.argmax(px)]
print('Estimate b =', bHat)
w = np.linspace(-20, 20)
px = np.zeros(len(w))
for i in range(len(w)):
fx = logistic(bHat + w[i]*xTrain)
px[i] = 1/N * np.sum(np.log(labelTrain*fx + (1-labelTrain)*(1-fx)))
plt.plot(w, px);
plt.xlabel('w')
plt.ylabel('l(w, X)')
plt.grid()
wHat = w[np.argmax(px)]
print('Estimate w =', wHat)
pXTest0 = logistic(bHat + wHat * xTest)
labelEst0 = pXTest0 > 0.5
plt.scatter(xTest, pXTest0, c=labelEst0, cmap=pltcolors.ListedColormap(colors), marker=',', alpha=0.01);
plt.scatter(xTest, yTest/np.max(yTest), c = labelTest, cmap=pltcolors.ListedColormap(colors), marker='x', alpha=0.01);
plt.xlabel('x')
plt.legend(('Estimated probability', 'Normalized model'));
plt.hist(labelEst0*1.0, 10, density=True)
print('Bernouilli parameter =', np.mean(labelEst0))
accuracy0 = np.sum(labelTest == labelEst0)/N
print('Accuracy =', accuracy0)
# ### Precision
# $p(y = 1 \mid \hat{y} = 1)$
print('Precision =', np.sum(labelTest[labelEst0 == 1])/np.sum(labelEst0))
# ### Recall
# $p(\hat{y} = 1 \mid y = 1)$
print('Recall =', np.sum(labelTest[labelEst0 == 1])/np.sum(labelTest))
# ### Confusion matrix
plotConfusionMatrix(labelTest, labelEst0, np.array(['Blue', 'Red']));
print(metrics.classification_report(labelTest, labelEst0))
# # SciKit Learn
#
# References:
# - SciKit documentation
# - https://www.geeksforgeeks.org/ml-logistic-regression-using-python/
model1 = SkLinReg(solver='lbfgs')
model1.fit(xTrain.reshape(-1,1), labelTrain)
model1.coef_
labelEst1 = model1.predict(xTest.reshape(-1,1))
print('Accuracy =',model1.score(xTest.reshape(-1,1), labelTest))
plt.hist(labelEst1*1.0, 10, density=True)
print('Bernouilli parameter =', np.mean(labelEst1))
# ### Confusion matrix (plot)
plotConfusionMatrix(labelTest, labelEst1, np.array(['Blue', 'Red']))
# ### Classification report
print(metrics.classification_report(labelTest, labelEst1))
# References :
# - https://towardsdatascience.com/building-a-logistic-regression-in-python-step-by-step-becd4d56c9c8
# - https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html#sklearn.linear_model.LogisticRegression.get_params
# ### ROC curve
logit_roc_auc = metrics.roc_auc_score(labelTest, labelEst1)
fpr, tpr, thresholds = metrics.roc_curve(labelTest, model1.predict_proba(xTest.reshape(-1,1))[:,1])
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right");
# # Using TensorFlow 2.0
#
# In TensorFlow 2.0 many possibilities are available to design a sequential layer. It could be based on high level API using Keras, down to function code close to the syntax of Tensorflow 1.0.
#
# Following design is showing how to implement a custom layer within a Sequential pipeline of Keras, and how to implement a custom metric. This is the favoured method to implement custom code in TensorFlow 2.0.
# Labels as floats {0., 1.}
labelTrainF = np.multiply(labelTrain, 1.0)
labelTrainF.dtype, labelTrainF.shape
# (Mini) Batch size
nBatch = 100
# Number of batches per Epoch
nBatchPerEpoch =20
# Number of epochs
nEpochMax = 1000
# Simple custom layer exposing the linear regression model
class MyLogisticRegressionLayer(tf.keras.layers.Layer):
def __init__(self, *args, **kwargs):
super(MyLogisticRegressionLayer, self).__init__(*args, **kwargs)
def build(self, input_shape):
self.w = self.add_weight(
shape=input_shape[0],
dtype=self.dtype,
initializer=tf.keras.initializers.ones(),
#regularizer=tf.keras.regularizers.l2(0.02),
trainable=True)
self.b = self.add_weight(
shape=1,
dtype=self.dtype,
initializer=tf.keras.initializers.ones(),
#regularizer=tf.keras.regularizers.l2(0.02),
trainable=True)
@tf.function
def call(self, x, training=None):
return tf.math.sigmoid(tf.math.add(tf.math.multiply(x, self.w), self.b))
# Using TensorFlow 2.0 style of metrics to implement accuracy
class MyBinaryAccuracy(tf.keras.metrics.Metric):
def __init__(self, name='my_accuracy', **kwargs):
super(MyBinaryAccuracy, self).__init__(name=name, **kwargs)
self.accuracySum = self.add_weight(name='accuracySum',
initializer='zeros')
self.accuracyCount = self.add_weight(name='accuracyCount',
initializer='zeros')
def update_state(self, labels, yEst):
labels = tf.cast(labels, tf.bool)
labelEst = tf.greater(yEst, 0.5)
values = tf.cast(tf.equal(labels, labelEst), self.dtype)
self.accuracySum.assign_add(tf.reduce_sum(values))
self.accuracyCount.assign_add(values.get_shape()[0])
def result(self):
return self.accuracySum / self.accuracyCount
# +
# Model 1, instantiate the custom layer
model1 = tf.keras.Sequential([MyLogisticRegressionLayer(input_shape=[nFeatures], dtype="float64")])
# Stochastic Gradient Descent Optimizer
optim1 = tf.keras.optimizers.SGD(0.01)
# Perform a train step on a mini-batch
# This function's code is rewritten by TensorFlow 2.0 and shall be compiled at every execution of the optimizer
@tf.function
def trainStep1(x, labels):
with tf.GradientTape() as tape:
predictions = model1(x, training=True)
loss = -tf.reduce_sum(tf.math.log((labels * predictions) + ((1 - labels) * (1 - predictions))))
#loss = tf.keras.losses.categorical_crossentropy(labels, predictions)
gradients = tape.gradient(loss, model1.trainable_variables)
optim1.apply_gradients(zip(gradients, model1.trainable_variables))
return loss, predictions
# Initialize values and loop on epochs and mini batch
epoch = 0
cost_epoch = 1
histo = []
accuracy = MyBinaryAccuracy()
for epoch in range(nEpochMax):
cost_cumul = 0
accuracy.reset_states()
for b in range(0, nBatchPerEpoch*nBatch, nBatch):
cost, predictions = trainStep1(xTrain[b : b + nBatch], labelTrainF[b : b + nBatch])
cost_cumul += cost
accuracy.update_state(labelTrainF[b : b + nBatch], predictions)
cost_epoch = cost_cumul / nBatchPerEpoch
W = model1.get_weights()
histo.append((cost_epoch.numpy(), accuracy.result().numpy(), W[1][0], W[0]))
print("Predicted model: {b:.3f} + {w:.3f} x, num epochs={c}".format(w=W[0], b=W[1][0], c=len(histo)))
# Save history as a Panda Data Frame
df = pandas.DataFrame(histo, columns = ('cost', 'accuracy', 'b', 'w0'))
# -
# SGD shows that there is not a single optimal value for b+w (intercept + slope) but a straight line as shown on the graph below.
# This is explained by the single feature: the decision boundary does not need to be a straight line, a single intercept point would be enough.
plt.scatter(df['b'], df['w0'], marker='.', alpha=0.2);
plt.xlabel('intercept')
plt.ylabel('weight');
fig, ax = plt.subplots(1,2, figsize=(16, 4))
ax[0].plot(df['cost'])
ax[0].grid()
ax[1].plot(df['accuracy'])
ax[1].grid()
# # Where to go from here ?
#
# __More complex models__ with the 2 feature [binary classification](ClassificationContinuous2Features.html) ([Notebook](ClassificationContinuous2Features.ipynb)) or the [K Nearest Neighbors classifier](ClassificationContinuous2Features-KNN.html) ([Notebook](ClassificationContinuous2Features-KNN.ipynb))
#
# __Compare with the single feature linear regression__ [using simple algorithms](../linear/LinearRegressionUnivariate.html) ([Notebook](LinearRegressionUnivariate.ipynb])), [or using Tensorflow](LinearRegressionUnivariate-TensorFlow.html) ([Notebook](LinearRegressionUnivariate-TensorFlow.ipynb))
| classification/ClassificationContinuousSingleFeature.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] tags=["pdf-title"]
# # Multiclass Support Vector Machine exercise
#
# *Complete and hand in this completed worksheet (including its outputs and any supporting code outside of the worksheet) with your assignment submission. For more details see the [assignments page](http://vision.stanford.edu/teaching/cs231n/assignments.html) on the course website.*
#
# In this exercise you will:
#
# - implement a fully-vectorized **loss function** for the SVM
# - implement the fully-vectorized expression for its **analytic gradient**
# - **check your implementation** using numerical gradient
# - use a validation set to **tune the learning rate and regularization** strength
# - **optimize** the loss function with **SGD**
# - **visualize** the final learned weights
#
# + tags=["pdf-ignore"]
# Run some setup code for this notebook.
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
# This is a bit of magic to make matplotlib figures appear inline in the
# notebook rather than in a new window.
# %matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
# %load_ext autoreload
# %autoreload 2
# + [markdown] tags=["pdf-ignore"]
# ## CIFAR-10 Data Loading and Preprocessing
# + tags=["pdf-ignore"]
# Load the raw CIFAR-10 data.
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
# Cleaning up variables to prevent loading data multiple times (which may cause memory issue)
try:
del X_train, y_train
del X_test, y_test
print('Clear previously loaded data.')
except:
pass
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# As a sanity check, we print out the size of the training and test data.
print('Training data shape: ', X_train.shape)
print('Training labels shape: ', y_train.shape)
print('Test data shape: ', X_test.shape)
print('Test labels shape: ', y_test.shape)
# + tags=["pdf-ignore"]
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
num_classes = len(classes)
samples_per_class = 7
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y)
idxs = np.random.choice(idxs, samples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt_idx = i * num_classes + y + 1
plt.subplot(samples_per_class, num_classes, plt_idx)
plt.imshow(X_train[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls)
plt.show()
# + tags=["pdf-ignore"]
# Split the data into train, val, and test sets. In addition we will
# create a small development set as a subset of the training data;
# we can use this for development so our code runs faster.
num_training = 49000
num_validation = 1000
num_test = 1000
num_dev = 500
# Our validation set will be num_validation points from the original
# training set.
mask = range(num_training, num_training + num_validation)
X_val = X_train[mask]
y_val = y_train[mask]
# Our training set will be the first num_train points from the original
# training set.
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask]
# We will also make a development set, which is a small subset of
# the training set.
mask = np.random.choice(num_training, num_dev, replace=False)
X_dev = X_train[mask]
y_dev = y_train[mask]
# We use the first num_test points of the original test set as our
# test set.
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask]
print('Train data shape: ', X_train.shape)
print('Train labels shape: ', y_train.shape)
print('Validation data shape: ', X_val.shape)
print('Validation labels shape: ', y_val.shape)
print('Test data shape: ', X_test.shape)
print('Test labels shape: ', y_test.shape)
# + tags=["pdf-ignore"]
# Preprocessing: reshape the image data into rows
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_val = np.reshape(X_val, (X_val.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
X_dev = np.reshape(X_dev, (X_dev.shape[0], -1))
# As a sanity check, print out the shapes of the data
print('Training data shape: ', X_train.shape)
print('Validation data shape: ', X_val.shape)
print('Test data shape: ', X_test.shape)
print('dev data shape: ', X_dev.shape)
# + tags=["pdf-ignore-input"]
# Preprocessing: subtract the mean image
# first: compute the image mean based on the training data
mean_image = np.mean(X_train, axis=0)
print(mean_image[:10]) # print a few of the elements
plt.figure(figsize=(4,4))
plt.imshow(mean_image.reshape((32,32,3)).astype('uint8')) # visualize the mean image
plt.show()
# second: subtract the mean image from train and test data
X_train -= mean_image
X_val -= mean_image
X_test -= mean_image
X_dev -= mean_image
# third: append the bias dimension of ones (i.e. bias trick) so that our SVM
# only has to worry about optimizing a single weight matrix W.
X_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])
X_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])
X_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])
X_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))])
print(X_train.shape, X_val.shape, X_test.shape, X_dev.shape)
# -
# ## SVM Classifier
#
# Your code for this section will all be written inside `cs231n/classifiers/linear_svm.py`.
#
# As you can see, we have prefilled the function `svm_loss_naive` which uses for loops to evaluate the multiclass SVM loss function.
# +
# Evaluate the naive implementation of the loss we provided for you:
from cs231n.classifiers.linear_svm import svm_loss_naive
import time
# generate a random SVM weight matrix of small numbers
W = np.random.randn(3073, 10) * 0.0001
loss, grad = svm_loss_naive(W, X_dev, y_dev, 0.000005)
print('loss: %f' % (loss, ))
# -
# The `grad` returned from the function above is right now all zero. Derive and implement the gradient for the SVM cost function and implement it inline inside the function `svm_loss_naive`. You will find it helpful to interleave your new code inside the existing function.
#
# To check that you have correctly implemented the gradient correctly, you can numerically estimate the gradient of the loss function and compare the numeric estimate to the gradient that you computed. We have provided code that does this for you:
# +
# Once you've implemented the gradient, recompute it with the code below
# and gradient check it with the function we provided for you
# Compute the loss and its gradient at W.
loss, grad = svm_loss_naive(W, X_dev, y_dev, 0.0)
# Numerically compute the gradient along several randomly chosen dimensions, and
# compare them with your analytically computed gradient. The numbers should match
# almost exactly along all dimensions.
from cs231n.gradient_check import grad_check_sparse
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 0.0)[0]
grad_numerical = grad_check_sparse(f, W, grad)
# do the gradient check once again with regularization turned on
# you didn't forget the regularization gra|ient did you?
loss, grad = svm_loss_naive(W, X_dev, y_dev, 5e1)
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 5e1)[0]
grad_numerical = grad_check_sparse(f, W, grad)
# + [markdown] tags=["pdf-inline"]
# **Inline Question 1**
#
# It is possible that once in a while a dimension in the gradcheck will not match exactly. What could such a discrepancy be caused by? Is it a reason for concern? What is a simple example in one dimension where a gradient check could fail? How would change the margin affect of the frequency of this happening? *Hint: the SVM loss function is not strictly speaking differentiable*
#
# $\color{blue}{\textit Your Answer:}$ Non-linear function, e.g. max() has some kink points. No. y=max(0, x) when x = -1e-10, and step = 1e-4. Use only a few datapoints.
#
# + id="vectorized_time_1"
# Next implement the function svm_loss_vectorized; for now only compute the loss;
# we will implement the gradient in a moment.
tic = time.time()
loss_naive, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('Naive loss: %e computed in %fs' % (loss_naive, toc - tic))
from cs231n.classifiers.linear_svm import svm_loss_vectorized
tic = time.time()
loss_vectorized, _ = svm_loss_vectorized(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('Vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic))
# The losses should match but your vectorized implementation should be much faster.
print('difference: %f' % (loss_naive - loss_vectorized))
# + id="vectorized_time_2"
# Complete the implementation of svm_loss_vectorized, and compute the gradient
# of the loss function in a vectorized way.
# The naive implementation and the vectorized implementation should match, but
# the vectorized version should still be much faster.
tic = time.time()
_, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('Naive loss and gradient: computed in %fs' % (toc - tic))
tic = time.time()
_, grad_vectorized = svm_loss_vectorized(W, X_dev, y_dev, 0.000005)
toc = time.time()
print('Vectorized loss and gradient: computed in %fs' % (toc - tic))
# The loss is a single number, so it is easy to compare the values computed
# by the two implementations. The gradient on the other hand is a matrix, so
# we use the Frobenius norm to compare them.
difference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')
print('difference: %f' % difference)
# -
# ### Stochastic Gradient Descent
#
# We now have vectorized and efficient expressions for the loss, the gradient and our gradient matches the numerical gradient. We are therefore ready to do SGD to minimize the loss. Your code for this part will be written inside `cs231n/classifiers/linear_classifier.py`.
# + id="sgd"
# In the file linear_classifier.py, implement SGD in the function
# LinearClassifier.train() and then run it with the code below.
from cs231n.classifiers import LinearSVM
svm = LinearSVM()
tic = time.time()
loss_hist = svm.train(X_train, y_train, learning_rate=1e-7, reg=2.5e4,
num_iters=1500, verbose=True)
toc = time.time()
print('That took %fs' % (toc - tic))
# -
# A useful debugging strategy is to plot the loss as a function of
# iteration number:
plt.plot(loss_hist)
plt.xlabel('Iteration number')
plt.ylabel('Loss value')
plt.show()
# + id="validate"
# Write the LinearSVM.predict function and evaluate the performance on both the
# training and validation set
y_train_pred = svm.predict(X_train)
print('training accuracy: %f' % (np.mean(y_train == y_train_pred), ))
y_val_pred = svm.predict(X_val)
print('validation accuracy: %f' % (np.mean(y_val == y_val_pred), ))
# + id="tuning" tags=["code"]
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of about 0.39 on the validation set.
# Note: you may see runtime/overflow warnings during hyper-parameter search.
# This may be caused by extreme values, and is not a bug.
# results is dictionary mapping tuples of the form
# (learning_rate, regularization_strength) to tuples of the form
# (training_accuracy, validation_accuracy). The accuracy is simply the fraction
# of data points that are correctly classified.
results = {}
best_val = -1 # The highest validation accuracy that we have seen so far.
best_svm = None # The LinearSVM object that achieved the highest validation rate.
################################################################################
# TODO: #
# Write code that chooses the best hyperparameters by tuning on the validation #
# set. For each combination of hyperparameters, train a linear SVM on the #
# training set, compute its accuracy on the training and validation sets, and #
# store these numbers in the results dictionary. In addition, store the best #
# validation accuracy in best_val and the LinearSVM object that achieves this #
# accuracy in best_svm. #
# #
# Hint: You should use a small value for num_iters as you develop your #
# validation code so that the SVMs don't take much time to train; once you are #
# confident that your validation code works, you should rerun the validation #
# code with a larger value for num_iters. #
################################################################################
# Provided as a reference. You may or may not want to change these hyperparameters
# learning_rates = [1e-7, 5e-5]
# regularization_strengths = [2.5e4, 5e4]
# These settings comes from https://github.com/lightaime/cs231n/blob/master/assignment1/svm.ipynb.
learning_rates = [1.4e-7, 1.5e-7, 1.6e-7]
regularization_strengths = [(1+i*0.1)*1e4 for i in range(-3,3)] + [(2+0.1*i)*1e4 for i in range(-3,3)]
# *****START OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
for lr in learning_rates:
for reg in regularization_strengths:
svm = LinearSVM()
svm.train(X_train, y_train, learning_rate = lr, reg = reg, num_iters = 1500)
tr_acc, val_acc = np.mean(svm.predict(X_train)==y_train), np.mean(svm.predict(X_val)==y_val)
if(val_acc > best_val):
best_svm = svm
best_val = val_acc
results[(lr, reg)] = (tr_acc, val_acc)
# *****END OF YOUR CODE (DO NOT DELETE/MODIFY THIS LINE)*****
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)
# + tags=["pdf-ignore-input"]
# Visualize the cross-validation results
import math
import pdb
# pdb.set_trace()
x_scatter = [math.log10(x[0]) for x in results]
y_scatter = [math.log10(x[1]) for x in results]
# plot training accuracy
marker_size = 100
colors = [results[x][0] for x in results]
plt.subplot(2, 1, 1)
plt.tight_layout(pad=3)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors, cmap=plt.cm.coolwarm)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 training accuracy')
# plot validation accuracy
colors = [results[x][1] for x in results] # default size of markers is 20
plt.subplot(2, 1, 2)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors, cmap=plt.cm.coolwarm)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 validation accuracy')
plt.show()
# + id="test"
# Evaluate the best svm on test set
y_test_pred = best_svm.predict(X_test)
test_accuracy = np.mean(y_test == y_test_pred)
print('linear SVM on raw pixels final test set accuracy: %f' % test_accuracy)
# + tags=["pdf-ignore-input"]
# Visualize the learned weights for each class.
# Depending on your choice of learning rate and regularization strength, these may
# or may not be nice to look at.
w = best_svm.W[:-1,:] # strip out the bias
w = w.reshape(32, 32, 3, 10)
w_min, w_max = np.min(w), np.max(w)
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
for i in range(10):
plt.subplot(2, 5, i + 1)
# Rescale the weights to be between 0 and 255
wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)
plt.imshow(wimg.astype('uint8'))
plt.axis('off')
plt.title(classes[i])
# + [markdown] tags=["pdf-inline"]
# **Inline question 2**
#
# Describe what your visualized SVM weights look like, and offer a brief explanation for why they look they way that they do.
#
# $\color{blue}{\textit Your Answer:}$ *The SVM weights look like random noise. However, with careful observation, we can know the weights at a certain point is relative to the average of the pixel value of the point. Since the goal of SVM is to minimize the loss, that is, maximize the score of the correct class, the picture of the weights must be as similar to all the pictures in that corresponing catagory as possible.*
#
| assignment1/svm.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [anaconda3]
# language: python
# name: Python [anaconda3]
# ---
# # RHT example workflow
# ### by <NAME>
#
# Imports. Note we are importing `rht` and `RHT_tools` from this repo.
from astropy.io import fits
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import rht, RHT_tools
# %matplotlib inline
# Load some test data. Let's use a fits version of a tesla coil image from <a href="https://commons.wikimedia.org/wiki/File:225W_Zeus_Tesla_coil_-_arcs2_(cropped).jpg">Wikimedia commons</a>.
data_fn = "testim_tesla_small"
tesla_data = fits.getdata(data_fn+".fits")
# Let's take a look at the original image.
fig = plt.figure(figsize=(6,6))
plt.imshow(tesla_data, cmap="Greys")
# Run the RHT! It's as simple as this. Note that depending on your setup, this may run quite slowly in a Jupyter notebook. The following should only take a few seconds from the command line. From the command line, simply do
#
# ~~~
# python rht.py data_fn --wlen=21 --smr=2
# ~~~
#
# Where wlen is the window length and smr is the unsharp mask smoothing radius. For details please refer to <a href="http://adsabs.harvard.edu/abs/2014ApJ...789...82C">the RHT paper</a>.
rht.main(data_fn, smr=2, wlen=21)
# By default, the data are saved as a fits file of the same name, with "_xytNN" appended, where NN is the RHT run number.
rht_data_fn = data_fn+"_xyt01.fits"
rht_tesla = fits.getdata(rht_data_fn)
# The backprojection is stored as the first hdu. This is total RHT linear intensity integrated over orientation. More prominent features in the backprojection indicate regions with greater total linear power.
fig = plt.figure(figsize=(6,6))
plt.imshow(rht_tesla, cmap="Greys")
# Some helper functions are provided in `RHT_tools.py`. Let's use them to grab the total RHT output (pixel indices and R(x, y, theta)) from the second header object.
ipoints, jpoints, hthets, naxis1, naxis2, wlen, smr, thresh = RHT_tools.get_RHT_data(rht_data_fn)
# Just to demonstrate, let's grab a random point. We'll also get the array of theta bins using `RHT_tools`.
indx = 20000
ipoint_example = ipoints[indx]
jpoint_example = jpoints[indx]
hthets_example = hthets[indx]
thets_arr = RHT_tools.get_thets(wlen, save=False)
# Plot the RHT spectrum at this random point.
# +
fig=plt.figure(figsize=(12,6))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122)
ax1.plot(np.degrees(thets_arr), hthets_example)
ax1.set_xlabel("theta [degrees]")
ax1.set_ylabel("RHT intensity")
ax1.set_title("RHT spectrum at point ({}, {})".format(ipoint_example, jpoint_example))
ax2.imshow(rht_tesla, cmap="Greys")
ax2.plot(ipoint_example, jpoint_example, '+', color="pink", ms=15, mew=3)
# -
# Let's now plot all of the RHT spectra that lie in a given row in our image.
# +
row_js = jpoints[np.where(jpoints == 250)]
row_is = ipoints[np.where(jpoints == 250)]
row_hthets = hthets[np.where(jpoints == 250)]
cmap = matplotlib.cm.get_cmap('Reds_r')
fig=plt.figure(figsize=(12,6))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122)
for _i in range(len(row_js)):
ax1.plot(np.degrees(thets_arr), row_hthets[_i, :,], color=cmap(_i*1./len(row_js)))
ax1.set_xlabel("theta [degrees]")
ax1.set_ylabel("RHT intensity")
ax1.set_title("RHT spectra where jpoint = {}".format(250))
ax2.imshow(rht_tesla, cmap="Greys")
plt.scatter(row_is, row_js, color=cmap(np.arange(len(row_js)*1.)/len(row_js)))
# -
| .ipynb_checkpoints/RHT_example_workflow-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # 02-1데이터 집합 불러오기
# ## 데이터 분석의 시작은 데이터 불러오기부터
# 데이터 분석을 위해 가장 먼저 해야 할 일은 무엇일까요? 바로 데이터를 불러오는 것입니다. 이때 불러오는 데이터를 '데이터 집합'이라고 합니다. 그러면 데이터 집합을 불러오는 방법과 데이터를 간단히 살펴보는 방법에 대해 알아보겠습니다. 우리가 처음 불러올 데이터 집합은 갭마인더입니다. '02_practice'를 주피터 노트북으로 열어 실습을 시작해 볼까요?
# ## 갭마인더 데이터 집합 불러오기
#
# 1. 판다스의 여러 기능을 사용하려면 판다스 라이브러리를 불러와야 합니다. 다음과 같이 입력하여 판다스 라이브러리를 불러오세요.
import pandas
# 2. 갭마인더 데이터 집합을 불러오려면 read_csv메서드를 사용해야 합니다. read_csv메서드는 기본적으로 쉽표(,)로 열어 구분되어 있는 데이터를 불러옵니다. 하지만 갭마인더는 열이 탭으로 구분되어 있기 때문에 read_csv 메서드를 호 출할 때 열이 탭으로 구분되어 있따고 미리 알려주어야 합니다. sep 속성값으로 \t를 지정하세요
df = pandas.read_csv('data/gapminder.tsv',sep='\t')
# 3. 판다스에 있는 메서드를 호출하려면 pandas와 점(.) 연산자를 사용해야 합니다. 그런데 매번 pandas라고 입력하려면 번거롭겠죠. 그래서 이를 해결하기 위해 관습적으로 pandas를 pd로 줄여 사용합니다. 다음과 같이 입력하면 pandas를 pd로 줄여 사용할 수 있습니다. 앞으로는 이 방법을 사용하겠습니다.
import pandas as pd
df = pd.read_csv('data/gapminder.tsv',sep='\t')
# ## 시리즈와 데이터프레임
# 갭마인더 데이터 집합을 잘 불러왔나요? 이번에는 판다스에서 사용되는 자료형을 알아볼 차례입니다. 판다스는 데이터를 효율적으로 다루기 위해 시리즈와 데이터프레임이라는 자료형을 사용합니다. 데이터프레임은 엑셀에서 볼 수 있는 시트와 동일한 개념이며 시리즈는 시트의 열 1개를 의미합니다. 파이썬으로 비유하여 설명하면 데이터프레임은 시리즈들이 각 요소가 되는 딕셔너리라고 생각하면 됩니다.
# ### 불러온 데이터 집합 살펴보기.
# 1. rdad_csv 메서드는 데이터 집합을 읽어 들여와 데이터프레임이라는 자료형으로 반환합니다. 데이터프레임에는 데이터 분석에 유용한 여러 메서드가 미리 정의되어 있습니다. 데이터 프레임의 데이터를 확인하는 용도로 자주 사용하는 head 메서드에 대해 먼저 알아보겠습니다. head 메서드는 데이터프레임에서 가장 앞에 있는 5개의 행을 출력하므로 내가 불러온 데이터가 어떤 값을 가지고 있는지 살펴보기에 안성맞춤이죠.
print(df.head())
# 2. 이번에는 df에 저장된 값이 정말 데이터프레임이라는 자료형인지 확인해 보겠습니다. 실행 결과를 보면 판다스의 데이터프레임이라는 것을 알 수 있습니다. type 메서드는 자료형을 출력해 줍니다. 앞으로 자주 사용할 메서드이므로 꼭 기억해 두기 바랍니다.
print(type(df))
# 3. 데이터프레임은 자신이 가지고 있는 데이터의 행과 열의 크기에 대한 정보를 shape라는 속성에 저장하고 있습니다. 다음을 입력하여 실행하면 갭마인더의 행과 열의 크기를 확인할 수 있습니다. 1번째 값은 행의 크기이고 2번째 값은 열의 크기 입니다.
print(df.shape)
# 4.이번에는 갭마인더에 어떤 정보가 들어 있는지 알아보겠습니다. 먼저 열을 살펴보겠습니다. 과정 3에서 shape 속성을 사용했던 것처럼 columns속성을 사용하면 데이터 프레임의 열 이름을 확인할 수 있습니다. 갭마인더를 구성하는 열 이름은 각각 country,continent,year,lifeExp,pop, gdpPercap 입니다.
print(df.columns)
# 5. 데이터프레임을 구성하는 값의 자료형은 데이터프레임의 dtypes 속성이나 info 메서드로 쉽게 확인할 수 있습니다.
print(df.dtypes)
print(df.info())
# ## 판다스와 파이썬 자료형 비교
# 다음 표에 앞으로 판다스를 공부하며 자주 다루게 될 자료형을 정리했습니다. 그런데 판다스와 파이썬은 같은 자료형도 다르게 인식합니다. 예를 들어 판다스는 문자열 자료형을 dbect라는 이름으로 인식하고 파이썬은 string이라는 이름으로 인식합니다. 같은 자료형이라도 판다스, 파이썬이 서로 다른 이름으로 인식한다는 점을 주의 깊게 살펴보고 다음으로 넘어가세요.
# 판다스 자료형$\qquad$파이썬 자료형$\qquad$$\qquad$설명<br>
# object$\qquad$$\qquad$$\quad$string$\qquad$$\qquad$$\qquad$문자열<br>
# int64$\qquad$$\qquad$$\quad$$\;$$\;$$\;$int$\qquad$$\qquad$$\qquad$$\;$$\;$정수<br>
# float64$\qquad$$\qquad$$\quad$float$\qquad$$\qquad$$\qquad$수소점을 가진숫자<br>
# datetime64$\qquad$$\;$$\;$$\;$$\;$datetime$\qquad$$\qquad$$\;$$\;$$\;$$\;$$\;$파이썬 표준 라이브러리인 datetime이 반환하는 자료형
# # 02-2 데이터 추출하기
# 지금까지 데이터프레임의 크기와 자료형을 살펴보는 방법에 대해 알아보았습니다. 앞에서 haed 메서드를 이용해 데이터프레임에서 가장 앞에 있는 5개의 데이터를 추출하여 출력했던 것을 기억하나요? 이번에는 데이터프레임에서 데이터를 열 단위로 추출하는 방법과 행 단위로 추출하는 방법을 알아보겠습니다. 먼저 열 단위로 데이터를 추출하는 방법을 알아보겠습니다.
# ### 열 단위 데이터 추출하기
# 데이터프레임에서 데이터를 열 단위로 추출하려면 대괄호와 열 이름을 사용해야 합니다. 이때 열 이름은 꼭 작은따옴표를 사용해서 지정해야 하고 추출한 열은 변수에 저장해서 사용할 수도 있습니다. 이때 1개의 열만 추출하면 시리즈를 얻을 수 있고 2개 이상의 열을 추출하면 데이터프레임을 얻을 수 있습니다.
# #### 열 단위로 데이터 추출하기
# 1. 다음은 데이터프레임에서 열 이름이 country인 열을 추출하여 country_df에 저장한 것입니다. type 메서드를 사용하면 country_df에 저장된 데이터의 자료형이 시리즈라는 것을 확인할 수 있습니다. 시리즈도 head,tail 메서드를 가지고 있기 때문에 gead,tail메서드로 가장 앞이나 뒤에 있는 5개의 데이터를 출력할 수 있습니다.
country_df=df['country']
print(type(country_df))
print(country_df.head())
print(country_df.tail())
# 2.리스트에 열 이름을 전달하면 여러 개의 열을 한 번에 추출할 수 있습니다. 다음은 열 이름이 country,continent,year인 열을 추출하여 변수 subset에 저장한 것입니다. 이때 1개의 열이 아니라 2개 이상의 열을 추출했기 때문에 시리즈가 아니라 데이터프레임을 얻을 수 있습니다.
subset=df[['country','continent','year']]
print(type(subset))
print(subset.head())
print(subset.tail)
# ## 행단위 데이터 추출하기
# 이번에는 데이터를 행 당위로 추출하는 방법에 대해 알아보겠습니다. 데이터를 행 단위로 추출하려면 loc,iloc 속성을 사용해야 합니다. 밑에 두 속성을 간단하게 정리한 표입니다.<br>
# 속성$\quad$$\quad$$\quad$설명<br>
# loc$\quad$$\quad$$\quad$인덱스를 기준으로 행 데이터 추출<br>
# iloc$\quad$$\quad$$\quad$행 번호를 기준으로 행 데이터 추출
# 표의 설명을 보면 인덱스와 행 번호라는 것이 있습니다. 파이썬을 공부한 독자라면 리스트같은 자료형에 저장된 데이터의 순서를 인덱스라고 알고 있을 것입니다. 하지만 판다스에서는 이런 개념을 행 번호라고 부릅니다. 다음예제를 실습하면서 판다스에서 말하는 인덱스와 행 번호가 무엇인지 알아보겠습니다.
# ## 인덱스와 행 번호 개념 알아보기
# 다음은 갭마인더 데이터 집합을 불러온 다음 head메서드를 실행한 결과입니다.
print(df.head())
# 왼쪽에 번호가 보이나요? 바로 이것이 인덱스입니다. 인덱스는 보통 0부터 시작하지만 행 데이터가 추가, 삭제되면 언제든지 변할 수 있으며 숫자가 아니라 문자열을 사용할 수도 있습니다. 즉, 인덱스는 first, second,third와 같은 문자열로 지정할 수도 있습니다. 반면에 행 번호는 데이터의 순서를 따라가기 때문에 정수만으로 데이터를 조회하거나 추출할 수 있으며 실제 데이터프레임에서는 확인할 수 없는 값입니다.
print(df.loc[0])
print(df.loc[99])
# 2. 만약 데이터프레임의 마지막 행 데이터를 추출하려면 어떻게 해야 할까요? 마지막 행데이터의 인덱스를 알아내야 합니다. shape[0]에 행 크기(1704)가 저장되어 있다는 점을 이용하여 마지막 행의 인덱스를 구하면 됩니다. 다음은 shape[0]에서 1을 뺀 값으로(1704-1=1703)마지막 행 데이터를 추출한 것입니다.
number_of_rows=df.shape[0]
last_row_index=number_of_rows -1
print(df.loc[last_row_index])
# 3. 데이터프레임의 마지막 행 데이터를 추출하는 또 다른 방법으로는 tail메서드를 사용하는 방법이 있습니다. 다음과 같이 tail 메서드의 인자 n에 1을 전달하면 마지막 행의 데이터를 출출할 수 있습니다. 이방법이 조금 더 유용하겠죠?
print(df.tail(n=1))
# 4. 만약 인덱스가 0,99,999인 데이터를 한 번에 추출하려면 리스트에 원하는 인덱스를 담아 loc 속성에 전달하면 됩니다.
print(df.loc[[0,99,999]])
# ### tail메서드와 loc 속성이 반환하는 자료형은 서로 달라요!
# tail 메서드와 loc 속성이 반환하는 데이터의 자료형은 다릅니다. 다음은 tail 메서드와 lic속성으로 추출한 데이터의 자료형을 type메서드로 확인한 것입니다. loc속성이 반환한 데이터 자료형은 시리즈이고 tail 메서드가 반환한 데이터 자료형은 데이터프레임입니다.
# +
subset_loc=df.loc[0]
subset_tail=df.tail(n=1)
print(type(subset_loc))
print(type(subset_tail))
# -
# ### iloc 속성으로 행 데이터 추출하기
# 1. 이번에는 iloc속성으로 행 데이터를 추출하는 방법에 대해 알아보겠습니다. loc속성은 데이터프레임의 인덱스를 사용하여 데이터를 추출했지만 iloc 속성은 데이터 순서를 의미하는 행 번호를 사용하여 데이터를 추출합니다.지금은 인덱스와 행 번호가 동일하여 동일한 결괏값이 출력됩니다. 다음은 iloc속성에 1을 전달하여 데이터를 추출한 것입니다.
print(df.iloc[1])
print(df.iloc[99])
# 2. iloc 속성은 음수를 사용해도 데이터를 추출할 수 있습니다. 다음은 -1을 전달하여 마지막 행 데이터를 추출한 것입니다. 하지만 데이터프레임에 아예 존재하지 않는 행 번호를 전달하면 오류가 발생합니다.
print(df.iloc[-1])
# 3. iloc 속성도 여러 데이터를 한 번에 추출할 수 있습니다. loc 속성을 사용했던 것처럼 원하는 데이터의 행 번호를 리스트에 담아 전달하면 됩니다.
print(df.iloc[[0,99,999]])
# ## loc, iloc 속성 자유자재로 사용하기
# loc, iloc속성을 좀더 자유자재로 사용하려면 추출할 데이터의 행과 열을 지정하는 방법을 알아야 합니다. 두속성 모두 추출할 데이터의 행을 먼저 지정하고 그런 다음 열을 지정하는 방법으로 데이터를 추출합니다. 즉 df.loc[[행],[열]]이나 df.iloc[[행],[열]]과 같은 방법으로 코드를 작성하면 됩니다. <br> 이때 행과 열을 지정하는 방법은 슬라이싱 구문을 사용하는 방법과 range 메서드를 사용하는 방법이 있습니다. 먼저 슬라이싱 구문으로 원하는 데이터를 추출하는 방법을 알아보겠습니다.
# ### 데이터 추출하기--슬라이싱 구문, range메서드
# #### 1.슬라이싱 구문으로 데이터 추출하기
# 다음은 모든 행(:)의 데이터에 대해 tear,pop열을 추출하는 방법입니다. 이때 loc와 iloc속성에 전달하는 열 지정값은 반드시 형식에 맞게 전달해야 합니다. 예를 들어 loc 속성의 열 지정값에 정수 리스트를 전달하면 오류가 발생합니다.
subset=df.loc[:,['year','pop']]
print(subset.head())
subset=df.iloc[:,[2,4,-1]]
print(subset.head())
# #### 2. range메서드로 데이터 추출하기
# 이번에는 iloc 속성과 파이썬 내장 메서드인 range를 응용하는 방법을 알아보겠습니다. range 메서드는 지정한 구간의 정수 리스트를 반환해 줍니다. iloc속성의 열 지정값에는 정수 리스트를 전달해야 한다는 점과 range메서드의 반환값이 정수 리스트인 점을 이용하여 원하는 데이터를 추출하는 것이죠<br> 그런데 range 메서드는 조금 더 정확하게 말하면 지정한 범위의 정수 리스트를 반환하는 것이 아니라 제네레이터를 반환합니다. iloc속성은 제네레이터로 데이터 추출을 할 수 없죠. 다행이 제네레이터는 간단하게 리스트로 변환할 수 있습니다. 다음은 range(5)가 반환한 제네레이터를 정숫값을 가진 리스트 [0,1,2,3,4]로 변환하여 iloc의 열 지정값에 전달한 것입니다. 자주 사용하는 방법은 아니지만 알아두면 유용할 것입니다.|
small_range=list(range(5))
print(small_range)
print(type(small_range))
subset=df.iloc[:,small_range]
print(subset.head())
small_range=list(range(3,6))
print(small_range)
subset=df.iloc[:,small_range]
print(subset.head())
# #### 3. range 메서드에 대해 조금 더 알아볼까요? range 메서드에 range(0,6,2)와 같은 방법으로 3개의 인자를 전달하면 어떻게 될까요? 0부터 5까지 2만큼 건너뛰는 제네레이터를 생성합니다. 이 네네레이터를 리스트로 변환하면 번위는 0~5이고 짝수로 된 정수 리스트를 얻을 수 있죠.
small_range=list(range(0,6,2))
subset=df.iloc[:,small_range]
print(subset.head())
# #### 4.슬라이싱 구문과 range 메서드 비교하기
# 그런데 실무에서는 range 메서드보다는 간편하게 사용할 수 있는 파이썬 슬라이싱 구문을 더 선호합니다. range메서드가 반환한 제네레이터를 리스트로 변환하는 등의 과정을 거치지 않아도 되기 때문이죠. 예를 들어 list(range(3))과 [:3]의 결괏값은 동일합니다.
subset=df.iloc[:,:3]
print(subset.head())
# #### 5. 0:6:2를 열징정값에 전달하면 과정 3에서 얻은 결괏값과 동일한 결괏값을 얻을수 있습니다. range메서드와 슬라이싱 구문을 비교해 보세요.
subset=df.iloc[:,0:6:2]
print(subset.head())
# #### 6. loc,iloc 속성 자유자재로 사용하기
# 만약 iloc 속성으로 0,99,999번째 행의 0,3,5번째 열 데이터를 추출하려면 다음과 같이 코드를 작성하면 됩니다.
print(df.iloc[[0,99,999],[0,3,5]])
# #### 7. iloc 속성의 열 지정값으로 정수 리스트를 전달하는 것이 간편해 보일 수 있지만 이렇게 작성한 코드는 나중에 어떤 데이터를 추출하기 위한 코드인지 파악하지 못 할 수도 있습니다. 그래서 보통은 다음과 같은 방법으로 loc 속성을 이용하여 열 지정값으로 열 이름을 전달합니다.
print(df.loc[[0,99,999],['country','lifeExp','gdpPercap']])
# #### 8. 앞에서 배운 내용을 모두 응용하여 데이터를 추출해 볼까요? 다음은 인덱스가 10인 행부터 13인 행의 country,lifeExp,gdpPercap열 데이터를 추출하는 코드입니다.
print(df.loc[10:13,['country','lifeExp','gdpPercap']])
# # 02-3 기초적인 통계 계산하기
# 지금까지는 데이터를 추출하는 방법에 대해 알아보았습니다. 이번에는 추출한 데이터를 가지고 몇 가지 기초적인 통계 계산을 해보겠습니다. 다음은 갭마인더 데이터 집합에서 0~9번째 데이터를 추출하여 출력한 것입니다.
print(df.head(n=10))
# ### 그룹화한 데이터의 평균 구하기
# #### 1. lifeExp열을 연도별로 그룹화하여 평균 계산하기
# 예를 들어 연도별 lifeExp 열의 평균을 계산하려면 어떻게 해야 할까요? 데이터를 year열로 그룹화하고 lifeExp 열의 평균을 구하면 됩니다. 다음은 데이터프레임의 groupby 메서드에 year 열을 전달하여 연도별로 그룹화한 다음 lifeExp 열을 지정하여 mean 메서드로 평균을 구한 것입니다.
print(df.groupby('year')['lifeExp'].mean())
# #### 2. 과정 1에서 작성한 코드가 조금 복잡해서 어리둥절할 수도 있을 것입니다. 어떤 일이 벌어진 것일까요? 과정 1에서 작성한 코드를 작은 단위로 나누어 살펴보겠습니다. 먼저 데이터프레임을 연도별로 그룹화한 결과를 살펴보겠습니다. groupby 메서드에 year열 이름을 전달하면 연도별로 그룹화한 country, continent,.....gdpPercap 열을 모은 데이터프레임을 얻을 수 있습니다.
grouped_year_df=df.groupby('year')
print(type(grouped_year_df))
# #### 3. groupde_year_df를 출력하면 과정 2에서 얻은 데이터프레임이 저장된 메모리의 위치를 알수 있습니다. 이결과를 통해 연도별로 그룹화한 데이터는 데이터프레임 형태로 현재 메모리의 0x7fa9f012e700이라는 위치에 저장되어 있음을 알 수 있습니다.
print(grouped_year_df)
# #### 4. 이어서 lifeExp 열을 추출한 결과를 살펴보겠습니다. 그룹화한 데이터프레임에서 lifeExp 열을 추출하면 그룹화한 시리즈를 얻을 수 있습니다. 즉, 연도별로 그룹화한 lifeExp 열을 얻을 수 있습니다.
grouped_year_df_lifeExp=grouped_year_df['lifeExp']
print(type(grouped_year_df_lifeExp))
# #### 5. 마지막으로 평군을 구하는 mean 메서드를 사용한 결과를 살펴보겠습니다. 과정 4에서 연도별로 그룹화한 lifeExp에 mean 메서드를 사용했기 때문에 각 연도별 lifeExp 열의 평균값을 얻을 수 있습니다.
mean_lifeExp_by_year=grouped_year_df_lifeExp.mean()
print(mean_lifeExp_by_year)
# #### 6. lifeExpm gdpPercap 열의 평균값을 연도, 지역별로 그룹화하여 한 번에 계산하기.
# 다음은 과정 1~4를 응용한 코드입니다. year, continent 열로 그룹화한 그룹 데이터프레임에서 lifeExp, gdpPercap 열만 추출하여 평균값을 구한 것입니다.
multi_group_var=df.groupby(['year','continent'])[['lifeExp','gdpPercap']].mean()
print(multi_group_var)
print(type(multi_group_var))
# #### 7. 그룹화한 데이터 개수 세기
# 이번에는 그룹화한 데이터의 개수가 몇 개인지 알아보겠습니다. 이를 통계에서는 '빈도수'라고 부릅니다. 데이터의 빈도수는 nunique 메서드를 사용하면 쉽게 구할 수 있습니다. 다음은 continent를 기준으로 데이터프레임을 만들고 country 열만 추출하여 데이터의 빈도수를 계산할 것입니다.
print(df.groupby('continent')['country'].nunique())
# # 02-4 그래프 그리기
# 그래프와 같은 데이터의 시각화는 데이터 분석 과정에서 가장 중요한 요소입니다. 데이터를 시각화하면 데이터를 이해하거나 추이를 파악하는 등의 작업을 할 때 많은 도움이 됩니다. 여기에서는 간단한 그래프를 그려보고 데이터 시각화가 무엇인지 알아보겠습니다. 자세한 내용은 04장에서 더 자세히 설명하겠습니다.
# ### 그래프 그리기
# #### 1.먼저 그래프와 연관된 라이브러리를 불러옵니다.
# %matplotlib inline
import matplotlib.pyplot as plt
# #### 2. 그런 다음 year 열을 기준으로 그릅화한 데이터프레임에서 lifeExp 열만 추출하여 평균 값을 구합니다.
global_yearly_life_expectancy=df.groupby('year')['lifeExp'].mean()
print(global_yearly_life_expectancy)
# #### 3. 과정 2에서 구한 값에 plot메서드를 사용하면 다음과 같은 그래프가 그려집니다.
global_yearly_life_expectancy.plot()
# ### 마무리하며
# 이 장에서는 데이터 집합을 불러오는 방법과 데이터를 추출하는 방법 등을 알아보았습니다. 판다스가 무엇인지 감이 좀 잡혔나요? 다음 장에서는 판다스의 기본 자료형인 데이터프레임과 시리즈를 좀 더 자세히 알아보겠습니다.
# 출처 : "판다스"
| chapter_02-1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Как писать быстрый код на Python
# ## <NAME>
# Язык Python обладает многими необходимыми для вычислений функциями.
# Целые числа хранятся со знаком и имеют произвольную длину
n = 1 # Целое число
for _ in range(500):
n *= 10
print(n)
print(type(n))
# Арифметика на целых определенна обычным образом.
print(f"1+2={1+2}")
print(f"1-2={1-2}")
print(f"1*2={1*2}")
# Обратите однако внимание, что целочисленное деление обозначается //
print(f"1/2={1/2}")
print(f"1//2={1//2}")
# Часто бывает полезен остаток от деления.
print(f"4%3={4%3}")
# Обратете внимание, что остаток от отрицательного числа положителен.
# Остаток определен таким образом, чтобы согласовываться с арифметикой по данному модулю.
print(f"(-1)%3={(-1)%3}")
assert ((-1)%3 + 1%3)%3 == (1-1)%3
# Вещественные числа имеют в своей записи точку или экспоненту
print(f"type(1)={type(1)}")
print(f"type(1.0)={type(1.0)}")
print(f"type(1e1)={type(1e1)}")
# Научная форма записи чисел указывает показатель после символа `e`:
# $$\textrm{314e-2}=e14\cdot 10^{-2}=3.14.$$
# Вещественные числа хранятся в виде чисел с плавающей запятой двойной точности.
print(f"1.0 + 1e-15 = {1.0 + 1e-15}")
print(f"1.0 + 1e-16 = {1.0 + 1e-16}")
print(f"1e307 * 10 = {1e307 * 10}")
print(f"1e308 * 10 = {1e308 * 10}")
print(f"1e309 = {1e309}")
print(f"1e-323 = {1e-323}")
print(f"1e-324 = {1e-324}")
# Также питон ествественно поддерживает комплексные числа.
# Чисто мнимое число получается добавлением символа j после вещественного числа.
print(f"1+2i = {1+2j}")
print(f"i*(1+2i) = {1j*(1+2j)}")
# Не во всех языках общего назначения в стандартной библиотеке есть рациональные числа, но в питоне они есть.
from fractions import Fraction
pi = Fraction(355, 113)
print(f"pi ~ {pi} ~ {float(pi)}")
print(f"pi*2/5 = {pi*2/5}")
# Обратите внимание, что типы конвертируются между собой вызовом конструктора.
print(f"int(3.14) = {int(3.14)}")
print(f"float('3.14') = {float('3.14')}")
# +
# Для хранения векторов на питоне есть две возможности: списки и кортежи.
a = [1,2,3] # Список
print(f"a = {a}")
a[1] = 5 # Списки можно изменять.
print(f"a[1]=5; a = {a}")
a.insert(1, 6) # Можно даже менять длину списка.
print(f"a.insert(1, 5); a = {a}")
b = (1,2,3) # Кортеж
print(f"b = {b}")
# b[1] = 5 # Кортежи нельзя изменять.
# И списки, и кортежи могут содержать любые объекты.
a1 = [1, 1.0, 'a']
b1 = (1, 1.0, 'a')
# -
# Универсальность списков и кортежей не позволяет хранить в них вектора чисел максимально плотно,
# и работать с ними максимально быстро.
# Магия IPython/Jupyter позволяет нам измерить время выполнения команды.
# В данном случае мы создаем список чисел до 1 000 000
# %timeit a = list(x for x in range(1000000))
# %%timeit
# Аналогично можно было создать список в цикле
a = []
for x in range(1000000):
a.append(x)
# В этом варианте несколько большие затраты на интерпретацию.
# Однако оба этих варианта работают слишком медленно.
# Кортежи дают аналогичный результат.
# %timeit a = tuple(x for x in range(1000000))
# В таком духе можно делать операции над векторами, но это медленно.
# Например, сложим два вектора.
# %time a = list(x for x in range(1000000))
# %time b = list(x*x for x in range(1000000))
# Интересно, что хотя во втором случае мы возвели числа в квадрат, на скорость вычислений это не повлияло.
# В данном случае основные расходы на интерпретацию, а остальное на выделение памяти, сами вычисления на этом фоне теряются.
# Правда можно сделать еще хуже, если добавить вызов функции.
# %time b = list(x**2 for x in range(1000000))
# Складываем вектора, используя list comprehension.
# %time c = list(x+y for x,y in zip(a,b))
# %%time
# А теперь сложим вектора без выделения новой памяти, сохраняя результат в существующий вектор.
for n in range(len(a)):
c[n] = a[n] + b[n]
# ## NumPy
# Как мы видим, на питоне можно считать, но он плохо подходит для численного моделирования, так как
# 1. Мало типов данных, невозможно контролировать точность, нет поддержки массивов, матриц и т.п.
# 2. Слишком малая скорость вычислений из-за интерпретируемости языка.
#
# Проблемы с хранением могуть быть решены создания специального типа, в котором хранятся числа только одного типа,
# тогда их можно хранить подряд друг за другом, что уменьшает требуемый обьем памяти.
# Такой класс определен в пакете NumPy.
import numpy as np # Далее пакет NumPy доступен по сокращению np.
# Снова создадим вектор из 1 000 000 первых целых чисел, но теперь в типе numpy.NDArray
# %time a = np.arange(1000000)
print(f"type a = {type(a)}")
# Время выполнения на порядок сохранилось, для больших массивов разница будет еще больше.
# Также тип NDArray удобен для хранения многомерных массивов.
m = np.array([[1,2,3],[4,5,6]])
# Здесь мы преобразовали матрицу в виде списка списков в NDArray
print(f"m = {m}")
# Теперь матрицу можно транспонировать
print(f"m.T = {m.T}")
# В виде списков это было бы сделать гораздо сложнее.
# +
# Над массивами естественным образом определены арифметические операции
# %time b = a**2
# %time b = a*a
# %time b = a**2
# Теперь время работы гораздо более разумное, так как арифметика над массивами написана
# на низкоуровневых языках и использует векторные команды процессора.
# Иногда инструкции NumPy работают быстрее наивного кода на C.
# %time c=a+b
# %time c+=a
# %time c=a+b
# Обратите внимание, что вторая команда работает чуть быстрее первой,
# так как в ней не выделяется память.
# Интересная особенность Jupyter, что третья команда выполняется на порядок быстрее первой,
# хотя команды буквально совпадают.
# Видимо, если переменная уже существовала, она переиспользуется.
# -
# %%time
# Вычисления в цикле работают значительно медленнее.
for n in range(len(a)):
c[n] += a[n]
# +
# Главный вывод: если вы делает операции над многими элементами, то пусть цикл будет внутри функции numpy,
# а не в коде на python.
# -
# ## Numba
#
# Если вам привычнее думать в терминах циклов, то вам может помочь Numba.
# С помощью этой библиотеке функция на python компилируется во время выполнения в весьма эффективный код.
# +
import numba as nb # Теперь Numba доступна под именем nb
# Для примера создадим функцию, которая складывает вектора.
@nb.njit(nb.int64[:](nb.int64[:],nb.int64[:]))
def add(a, b):
c = np.empty_like(a)
for n in range(a.shape[0]):
c[n] = a[n] + b[n]
return c
# Декоратор @nb.njit говорит, что следующая функция должна быть откомпилирована.
# Здесь нам пришлось задать типы входных и выходных значений, чтобы компилятор мог заменить сложение
# на машинную инструкцию.
# %time c=add(a,b)
# Производительность почти как у функции из NumPy.
# Не все функции можно использовать из Numba, см. поддерживаемые команды в документации.
# +
# Кроме эффективного преобразования циклов, Numba может быть полезно, если над одним элементом
# массива производится много операций.
# Так как в наше время основные затраты при вычислениях приходятся на доступ к памяти,
# то выполняя больше операций над одним элементом сразу, мы значительно ускоряем работу программы.
# Создадим массив чисел с плавающей запятой двойной точности
a=np.arange(10000000,dtype=np.float64)
# # %timeit c=np.sin(a)
# # %timeit c=np.sin(np.sin(a))
# %timeit c=a*a
# %timeit c=(a+3.14)*a
# Две операции занимают в два раза больше времени, что кажется логичным.
# +
@nb.njit(nb.float64[:](nb.float64[:]))
def f1(x):
y = np.empty_like(x)
for n in nb.prange(x.shape[0]):
y[n] = x[n]*x[n]
return y
@nb.njit(nb.float64[:](nb.float64[:]))
def f2(x):
y = np.empty_like(x)
for n in range(x.shape[0]):
y[n] = (x[n]+3.14)*x[n]
return y
# %timeit c=f1(a)
# %timeit c=f2(a)
# Магическим образом получили время работы f2 почти идентичное f1, хотя операций делалось две, вместо одной.
# Видим, что основное время работы занимал доступ к памяти, а не арифметика.
# Для дорогих операций, вроде np.sin, такой разницы во времени не будет.
# +
# Функция f1 выше работала медленнее, чем умножение в Numpy, но мы можем ускорить функцию, использую несколько потоков.
# Обратите внимание на использование numba.prange вместо range.
@nb.njit(nb.float64[:](nb.float64[:]), parallel=True)
def f1(x):
y = np.empty_like(x)
for n in nb.prange(x.shape[0]):
y[n] = x[n]*x[n]
return y
@nb.njit(nb.float64[:](nb.float64[:]), parallel=True)
def f2(x):
y = np.empty_like(x)
for n in nb.prange(x.shape[0]):
y[n] = (x[n]+3.14)*x[n]
return y
# %timeit c=f1(a)
# %timeit c=f2(a)
# +
# Если массивы заведомо непрерывные (т.е. не результат индексации),
# то можно это явно указать, включив дополнительные оптимизации.
# @nb.njit(nb.float64[::1](nb.float64[::1]), parallel=True)
# +
# Еще сильнее можно ускорить вычисления, исключив проверки чисел с плавающей запятой на нечисловые значения,
# и разрешив оптимизации, которые могут незначительно повлиять на ответ.
# В большинстве случаев безопасно использовать
# @nb.njit(..., parallel=True, nogil=True, fastmath=True)
# +
# Для получения оптимальной производительности нужно всегда учитывать работу кеша.
# Сравним два варианта сложения матриц, отличающихся порядком суммирования элементов.
a = np.arange(9000000, dtype=np.float64).reshape((3000,3000))
b = a.copy() # Чтобы создать копию массива, мало сделать присваивание, нужно вызвать copy.
@nb.njit(nb.float64[:,:](nb.float64[:,:],nb.float64[:,:]))
def sum1(a,b):
c = np.empty_like(a)
for n in range(a.shape[0]):
for m in range(a.shape[1]):
c[n,m] = a[n,m]+b[n,m]
return c
@nb.njit(nb.float64[:,:](nb.float64[:,:],nb.float64[:,:]))
def sum2(a,b):
c = np.empty_like(a)
for m in range(a.shape[1]):
for n in range(a.shape[0]):
c[n,m] = a[n,m]+b[n,m]
return c
# %timeit c = sum1(a,b)
# %timeit c = sum2(a,b)
# Вариант с внутренним циклом по столбцам на порядок быстрее.
# Это объясняется тем, что при чтении одного значения из памяти сразу целый набор последовательных
# значений загружаются в кеш, из которого чтение затем идем на порядок быстрее.
# Для максимальной производительности нужно максимально использовать записанные в кеш значения.
# +
# Чтобы получить максимальную производительность, нужно четко представлять,
# во что преобразуется ваш код, что часто не очевидно.
# Например, сравним следующие коды, вычисляющие конечную разность.
@nb.njit(nb.float64[::1](nb.float64[::1]))
def f0(a):
c = np.empty_like(a)
for n in range(1,a.shape[0]):
c[n] = a[n]-a[n-1]
c[0] = a[0] - a[-1]
return c
@nb.njit(nb.void(nb.float64[::1], nb.float64[::1]))
def f1(a, c):
for n in range(1, a.shape[0]):
c[n] = a[n] - a[n-1]
c[0] = a[0] - a[-1]
@nb.njit(nb.void(nb.float64[::1], nb.float64[::1]))
def f2(a, c):
sx, = a.shape
for n in range(sx):
c[n] = a[n]-a[(n-1)%sx]
a = np.arange(10000000,dtype=np.float64)
c = np.empty_like(a)
# %timeit c=f0(a)
# %timeit f1(a, c)
# %timeit f2(a, c)
# Вариант f0 отличается от f1 только выделением памяти в f0, что делает этот вариант самым медленным.
# Варианты f1 и f2 не выделяют памяти, но время их выполнения отличается в разы.
# В варианте f2 вычисляется остаток от деления %, который компилятор не может эффективно векторизовать.
# -
| practice/FastPython.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python3.6
# language: python
# name: python3
# ---
# # Online Trading Customer Attrition Risk Prediction using SparkML
#
# There are many users of online trading platforms and these companies would like to run analytics on and predict churn based on user activity on the platform. Since competition is rife, keeping customers happy so they do not move their investments elsewhere is key to maintaining profitability.
#
# In this notebook, we will leverage IBM Cloud Private for Data to do the following:
#
# 1. Ingest merged customer demographics and trading activity data
# 2. Visualize the merged dataset to get a better understanding of the data and build hypotheses for prediction
# 3. Leverage the SparkML library to build a classification model that predicts whether a customer has a propensity to churn
# 4. Expose the SparkML classification model as a RESTful API endpoint for the end-to-end customer churn risk prediction and risk remediation application
#
# <a id="top"></a>
# ## Table of Contents
#
# 1. [Load the customer demographics and trading activity data](#load_data)
# 2. [Load libraries](#load_libraries)
# 3. [Visualize the customer demographics and trading activity data](#visualize)
# 4. [Prepare data for building SparkML classification model](#prepare_data)
# 5. [Train classification model and test model performance](#build_model)
# 6. [Save model to ML repository and expose it as REST API endpoint](#save_model)
# 7. [Summary](#summary)
# ### Quick set of instructions to work through the notebook
#
# If you are new to Notebooks, here's a quick overview of how to work in this environment.
#
# 1. The notebook has 2 types of cells - markdown (text) such as this and code such as the one below.
# 2. Each cell with code can be executed independently or together (see options under the Cell menu). When working in this notebook, we will be running one cell at a time because we need to make code changes to some of the cells.
# 3. To run the cell, position cursor in the code cell and click the Run (arrow) icon. The cell is running when you see the * next to it. Some cells have printable output.
# 4. Work through this notebook by reading the instructions and executing code cell by cell. Some cells will require modifications before you run them.
# <a id="load_data"></a>
# ## 1. Load the customer and trading activity data
# [Top](#top)
#
# Data can be easily loaded within IBM Cloud Private for Data using point-and-click functionality. The following image illustrates how to load the data from a database. The data set can be located by its name and inserted into the notebook as a Spark DataFrame as shown below.
#
# 
#
# The generated code comes up with a generic name and it is good practice to rename the dataframe to match the use case context.
# +
# Use the find data 10/01 icon and under your remote data set
# use "Insert to code" and "Insert Spark DataFrame in Python"
# here.
import dsx_core_utils, requests, os, io
from pyspark.sql import SparkSession
# Add asset from remote connection
df4 = None
dataSet = dsx_core_utils.get_remote_data_set_info('merge2')
dataSource = dsx_core_utils.get_data_source_info(dataSet['datasource'])
sparkSession = SparkSession(sc).builder.getOrCreate()
# Load JDBC data to Spark dataframe
dbTableOrQuery = ('"' + dataSet['schema'] + '"."' if(len(dataSet['schema'].strip()) != 0) else '') + dataSet['table'] + '"'
if (dataSet['query']):
dbTableOrQuery = "(" + dataSet['query'] + ") TBL"
df4 = sparkSession.read.format("jdbc").option("url", dataSource['URL']).option("dbtable", dbTableOrQuery).option("user",dataSource['user']).option("password",dataSource['password']).load()
df4.show(5)
# -
# After inserting the Spark DataFrame code above, change the following
# df# to match the variable used in the above code. df_churn is used
# later in the notebook.
#df_churn = df#
df_churn = df4
# <a id="load_libraries"></a>
# ## 2. Load libraries
# [Top](#top)
#
# Running the following cell will load all libraries needed to load, visualize, prepare the data and build ML models for our use case
import os
from pyspark.sql import SQLContext
from pyspark.sql.types import DoubleType
from pyspark.ml.feature import OneHotEncoder, StringIndexer, VectorIndexer, IndexToString
from pyspark.sql.types import IntegerType
from pyspark.ml import Pipeline
from pyspark.ml.feature import VectorAssembler
from pyspark.ml.classification import RandomForestClassifier, NaiveBayes
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.mllib.evaluation import MulticlassMetrics
import brunel
from dsx_ml.ml import save
import pandas as pd, numpy as np
import matplotlib.pyplot as plt
import dsx_core_utils, requests, os, io
from pyspark.sql import SparkSession
% matplotlib inline
# <a id="visualize"></a>
# ## 2. Visualize the customer demographics and trading activity data
# [Top](#top)
#
# Data visualization is a key step in the data mining process that helps to better understand the data before it can be prepared for building ML models.
#
# We will use the Brunel visualization which comes preloaded in IBM Cloud Private for Data analytics projects.
#
# The Brunel Visualization Language is a highly succinct and novel language that defines interactive data visualizations based on tabular data. The language is well suited for both data scientists and business users. More information about Brunel Visualization: https://github.com/Brunel-Visualization/Brunel/wiki
#
# Load the Spark DataFrame in to a pandas DataFrame
df_churn = df_churn.filter("ChurnRisk!='ChurnR'") # Filter out CSV header.
df_churn_pd = df_churn.toPandas()
df_churn_pd.head(5)
# %brunel data('df_churn_pd') stack polar bar x(CHURNRISK) y(#count) color(CHURNRISK) bar tooltip(#all)
# %brunel data('df_churn_pd') bar x(STATUS) y(#count) color(STATUS) tooltip(#all) | stack bar x(STATUS) y(#count) color(CHURNRISK: pink-orange-yellow) bin(STATUS) sort(STATUS) percent(#count) label(#count) tooltip(#all) :: width=1200, height=350
# %brunel data('df_churn_pd') bar x(TOTALUNITSTRADED) y(#count) color(CHURNRISK: pink-gray-orange) sort(STATUS) percent(#count) label(#count) tooltip(#all) :: width=1200, height=350
# %brunel data('df_churn_pd') bar x(DAYSSINCELASTTRADE) y(#count) color(CHURNRISK: pink-gray-orange) sort(STATUS) percent(#count) label(#count) tooltip(#all) :: width=1200, height=350
# <a id="prepare_data"></a>
# ## 3. Data preparation
# [Top](#top)
#
# Data preparation is a very important step in machine learning model building. This is because the model can perform well only when the data it is trained on is good and well prepared. Hence, this step consumes bulk of data scientist's time spent building models.
#
# During this process, we identify categorical columns in the dataset. Categories needed to be indexed, which means the string labels are converted to label indices. These label indices and encoded using One-hot encoding to a binary vector with at most a single one-value indicating the presence of a specific feature value from among the set of all feature values. This encoding allows algorithms which expect continuous features to use categorical features.
#
# Final step in the data preparation process is to assemble all the categorical and non-categorical columns into a feature vector. We use VectorAssembler for this. VectorAssembler is a transformer that combines a given list of columns into a single vector column. It is useful for combining raw features and features generated by different feature transformers into a single feature vector, in order to train ML models.
# Defining the categorical columns
categoricalColumns = ['Gender', 'Status', 'HomeOwner']
non_categoricalColumns = df_churn.select([c for c in df_churn.columns if c not in categoricalColumns]).columns
print(non_categoricalColumns)
non_categoricalColumns.remove('ChurnRisk')
stages = []
for categoricalCol in categoricalColumns:
# Category Indexing with StringIndexer
stringIndexer = StringIndexer(inputCol=categoricalCol, outputCol=categoricalCol + "Index")
#Use OneHotEncoder to convert categorical variables into binary SparseVectors
encoder = OneHotEncoder(inputCol=categoricalCol + "Index", outputCol=categoricalCol + "classVec")
stages += [stringIndexer, encoder]
labelIndexer = StringIndexer(inputCol='ChurnRisk', outputCol='label').fit(df_churn)
for colnum in non_categoricalColumns:
df_churn = df_churn.withColumn(colnum, df_churn[colnum].cast(IntegerType()))
# Transform all features into a vector using VectorAssembler
assemblerInputs = [c + "classVec" for c in categoricalColumns] + non_categoricalColumns
assembler = VectorAssembler(inputCols=assemblerInputs, outputCol="features")
# <a id="build_model"></a>
# ## 4. Build SparkML Random Forest classification model
# [Top](#top)
# We instantiate a decision-tree based classification algorithm, namely, RandomForestClassifier. Next we define a pipeline to chain together the various transformers and estimaters defined during the data preparation step before. MLlib standardizes APIs for machine learning algorithms to make it easier to combine multiple algorithms into a single pipeline, or workflow.
#
# We split original dataset into train and test datasets. We fit the pipeline to training data and apply the trained model to transform test data and generate churn risk class prediction
# +
# instantiate a random forest classifier, take the default settings
rf=RandomForestClassifier(labelCol="label", featuresCol="features")
# Convert indexed labels back to original labels.
labelConverter = IndexToString(inputCol="prediction", outputCol="predictedLabel", labels=labelIndexer.labels)
stages += [labelIndexer, assembler, rf, labelConverter]
pipeline = Pipeline(stages=stages)
# -
# Split data into train and test datasets
train, test = df_churn.randomSplit([0.7,0.3], seed=100)
train.cache()
test.cache()
print(train)
# Build models
model = pipeline.fit(train)
model.transform(test)
results = model.transform(test)
results=results.select(results["ID"],results["ChurnRisk"],results["label"],results["predictedLabel"],results["prediction"],results["probability"])
results.toPandas().head(6)
# ### Model results
#
# In a supervised classification problem such as churn risk classification, we have a true output and a model-generated predicted output for each data point. For this reason, the results for each data point can be assigned to one of four categories:
#
# 1. True Positive (TP) - label is positive and prediction is also positive
# 2. True Negative (TN) - label is negative and prediction is also negative
# 3. False Positive (FP) - label is negative but prediction is positive
# 4. False Negative (FN) - label is positive but prediction is negative
#
# These four numbers are the building blocks for most classifier evaluation metrics. A fundamental point when considering classifier evaluation is that pure accuracy (i.e. was the prediction correct or incorrect) is not generally a good metric. The reason for this is because a dataset may be highly unbalanced. For example, if a model is designed to predict fraud from a dataset where 95% of the data points are not fraud and 5% of the data points are fraud, then a naive classifier that predicts not fraud, regardless of input, will be 95% accurate. For this reason, metrics like precision and recall are typically used because they take into account the type of error. In most applications there is some desired balance between precision and recall, which can be captured by combining the two into a single metric, called the F-measure.
#
#
print('Model Precision = {:.2f}.'.format(results.filter(results.label == results.prediction).count() / float(results.count())))
# An added advantage of such tree-based classifiers is we can study feature importances and learn further about relative importances of features in the classification decision.
# +
# Evaluate model
# Compute raw scores on the test set
#predictionAndLabels = results.rdd.map(lambda lp: (results.prediction, results.label))
res = model.transform(test)
predictions = res.rdd.map(lambda pr: pr.prediction)
labels = res.rdd.map(lambda pr: pr.label)
predictionAndLabels = sc.parallelize(zip(predictions.collect(), labels.collect()))
# Instantiate metrics object
metrics = MulticlassMetrics(predictionAndLabels)
# Overall statistics
print("Overall Statistics")
f_measure = metrics.accuracy
print("Model F-measure = %s\n" % f_measure)
# statistics by class
print("Statistics by Class")
labels_itr = labels.distinct().collect()
for label in sorted(labels_itr):
print("Class %s F-Measure = %s" % (label, metrics.fMeasure(label)))
# +
# Feature importance
rfModel = model.stages[-2]
features = df_churn.columns
importances = rfModel.featureImportances.values
indices = np.argsort(importances)
# -
plt.figure(1)
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b',align='center')
plt.yticks(range(len(indices)), (np.array(features))[indices])
plt.xlabel('Relative Importance')
# Before we save the random forest classifier to repository, let us first evaluate the performance of a simple Naive Bayes classifier trained on the training dataset.
# +
nb = NaiveBayes(labelCol="label", featuresCol="features")
stages_nb = stages
stages_nb[-2] = nb
pipeline_nb = Pipeline(stages = stages_nb)
# Build models
model_nb = pipeline_nb.fit(train)
results_nb = model_nb.transform(test)
print('Naive Bayes Model Precision = {:.2f}.'.format(results_nb.filter(results_nb.label == results_nb.prediction).count() / float(results_nb.count())))
# -
# As you can see from the results above, Naive Bayes classifier does not perform well. Random forest classifier shows high F-measure upon evaluation and shows strong performance. Hence, we will save this model to the repository.
# <a id="save_model"></a>
# ## 5. Save the model into ML repository
# [Top](#top)
# +
# save(name='TradingChurnRiskClassificationSparkML',
# model=model,
# test_data = test,
# algorithm_type='Classification',
# description='This is a SparkML Model to Classify Trading Customer Churn Risk')
# -
# Write the test data without label to a .csv so that we can later use it for batch scoring
write_score_CSV=test.toPandas().drop(['ChurnRisk'], axis=1)
write_score_CSV.to_csv('../datasets/TradingCustomerSparkMLBatchScore.csv', sep=',', index=False)
# Write the test data to a .csv so that we can later use it for Evaluation
write_eval_CSV=test.toPandas()
write_eval_CSV.to_csv('../datasets/TradingCustomerSparkMLEval.csv', sep=',', index=False)
# <a id="summary"></a>
# ## 6. Summary
# [Top](#top)
# You have finished working on this hands-on lab. In this notebook you created a model using SparkML API, deployed it in Machine Learning service for online (real time) scoring and tested it using a test client.
# Created by **<NAME>** and **<NAME>**
#
# <EMAIL><br/>
# <EMAIL><br/>
#
# August 2018
| examples/TradingCustomerChurnClassifierSparkML.jupyter-py36.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Ungraded lab 1: Linear algebra in Python with numpy
#
# *Copyrighted material*
#
# **Objectives:** Use numpy function to apply the most common linear algebra in Python
#
# **Steps:**
# * Create numpy arrays from lists
# * Create numpy matrix
# * Element wise multiplication
# * Transpose
# * The norm of a vector
# * All the dot products flavors
# * Sum by rows and sum by columns with numpy
# * Normalize
#
# In this ungraded lab, you will have the oportunity to remember somer basic concept abouts linear algebra and how use them in Python.
#
# Numpy is one of the most used libraries in Python for arrays manipulation. It adds to Python a set of functions that allows to operate on large multidimensional arrays with few lines. So forget about writing nested loops for adding matrices!. In numpy this is as simple as adding numbers.
#
# Let's start importing the numpy library and creating the alias np for it. You will see this line almost in every python code from here in advance.
import numpy as np # The swiss knife of the data scientist.
# ## Defining lists and numpy arrays
alist = [1, 2, 3, 4, 5] # Define a python list. It looks like an np array
narray = np.array([1, 2, 3, 4]) # Define a numpy array
# Note the difference between a python list and a numpy array
# +
print(alist)
print(narray)
print(type(alist))
print(type(narray))
# -
# ## Algebraic opertators on numpy arrays vs python lists
#
# One of the most common beginers mistakes in Python is mixing up the concepts of numpy array and python arrays. Just observe the next example, where you try to "add" two objects of different types. Note that the '+' operator on numpy arrays perform a sum element wise, while the same operator is used to apply a list concatenation. Be carefull while coding. Knowing this can save you a lot of headeachs.
print(narray + narray)
print(alist + alist)
# And the same with the product operator. In the first case you scale the vector and in the second case you concatenate the same list 3 times.
print(narray * 3)
print(alist * 3)
# Be aware of the difference, because within a function you can have both types.
# Nparrays are designed for numerical and matrix operations, while lists are for more general purposes.
# ## Matrix or Array of Arrays
#
# In linear algebra, a matrix is structure composed of n rows by m columns. That means each row, must have exactly the same number of columns. With Numpy, we have 2 ways to create a matrix:
# * Creating an array of arrays: This is the recomended way, although you cannot ensure that all the row has the same amount of columns
# * Creating a matrix using np.matrix. However this is not recomended since this class will desapear from numpy in the near future.
#
# You can use to initialize a matrix nparrays or lists, and the resulting matrix will contain only nparrays inside.
npmatrix1 = np.array([narray, narray, narray])
npmatrix2 = np.array([alist, alist, alist])
npmatrix3 = np.array([narray, [1, 1, 1, 1], narray])
print(npmatrix1)
print(npmatrix2)
print(npmatrix3)
# However, if you want to define a matrix be sure that all the rows contains the same number of elements. Otherwise you will end up with something that cannot be operated using the linear algebra operators. Analize the following 2 examples:
# +
# Example 1:
okmatrix = np.array([[1, 2], [3, 4]]) # Define a 2x2 matrix
print(okmatrix) # Print okmatrix
print(okmatrix * 2) # Print a scaled version of okmatrix
# +
# Example 2:
badmatrix = np.array([[1, 2], [3, 4], [5, 6, 7]]) # Define a matrix. Note the third row contains 3 elements
print(badmatrix) # Print the weird matrix
print(badmatrix * 2) # It is supposed to scale the whole matrix
# -
# ## Scaling and translating matrices
#
# So, now that you know how to build correct nparrays and matrices, let's see how easy is to operate with them in Python.
# Nparrays can be operated using the normal algebraic operator like '+-'. You can operate between nparrays and nparrays or between nparrays and scalars.
# Scale by 2 and translate 1 unit the matrix
result = okmatrix * 2 + 1 # For each element in the matrix, multiply by 2 and add 1
print(result)
# +
# Add two sum compatible matrices
result1 = okmatrix + okmatrix
print(result1)
# Substract two sum compatible matrices. This is called the difference vector
result2 = okmatrix - okmatrix
print(result2)
# -
# The product operator '*' when used on nparrays or matrices indicates element wise multiplications.
# Don't miss it with the dot product.
result = okmatrix * okmatrix # Multiply each element by itself
print(result)
# ## Transpose a matrix
#
# In linear algebra, the transpose of a matrix is an operator which flips a matrix over its diagonal, that is it switches the row and column indices of the matrix by producing another matrix. It original matrix dimension was n by m, the resulting transposed matrix will be m by n.
# With numpy matrices, the trasnpose operations is denoted by .T
matrix3x2 = np.array([[1, 2], [3, 4], [5, 6]]) # Define a 3x2 matrix
print('Original matrix 3 x 2')
print(matrix3x2)
print('Transposed matrix 2 x 3')
print(matrix3x2.T)
# However notice that the transpose operation does not have effect on 1D nparrays
nparray = np.array([1, 2, 3, 4]) # Define an array
print('Original array')
print(nparray)
print('Transposed array')
print(nparray.T)
# perhaps in this case you wanted to do:
nparray = np.array([[1, 2, 3, 4]]) # Define a 1 x 4 matrix. Note the 2 level of square brackets
print('Original array')
print(nparray)
print('Transposed array')
print(nparray.T)
# ## Get the norm of a nparray or matrix
#
# In linear algebra, the norm of a nD vector $\vec a$ is defined as:
#
# $$ norm(\vec a) = ||\vec a|| = \sqrt {\sum_{i=1}^{n} a_i ^ 2}$$
# Calculating the norm of vector or even of a matrix is a very common operation when dealing with data. Numpy has a set of functions for linear algebra in the subpackage linalg, including the norm function. Let's see how to get the norm a given nparray or matrix:
# +
nparray1 = np.array([1, 2, 3, 4]) # Define an array
norm1 = np.linalg.norm(nparray1)
nparray2 = np.array([[1, 2], [3, 4]]) # Define a 2 x 2 matrix. Note the 2 level of square brackets
norm2 = np.linalg.norm(nparray2)
print(norm1)
print(norm2)
# -
# Note that without any other parameter, the norm function assume you want treat your matrix as being just an array of numbers.
#
# But you can get the norm by rows or by columns. You control the dimension of the operation with the __axis__ parammeter. axis=0 means get the norm of each column. axis=1 means get the norm of each row. Let's see how
# +
nparray2 = np.array([[1, 1], [2, 2], [3, 3]]) # Define a 3 x 2 matrix.
normByCols = np.linalg.norm(nparray2, axis=0) # Get the norm for each column. Returns 2 elements
normByRows = np.linalg.norm(nparray2, axis=1) # get the norm for each row. Returns 3 elements
print(normByCols)
print(normByRows)
# -
# However, there is more ways to get the norm of matrix in Python.
# For that let's see all different ways as you can get the dot product between 2 nparrays
# ## Dot product between nparrays: All the flavors
#
# The dot product or scalar product or inner product between two vectors $\vec a$ and $\vec a$ of the same size is defined as:
# $$\vec a \cdot \vec b = \sum_{i=1}^{n} a_i b_i$$
#
# The dot product takes 2 vectors and returns a single number
# +
nparray1 = np.array([0, 1, 2, 3]) # Define an array
nparray2 = np.array([4, 5, 6, 7]) # Define an array
flavor1 = np.dot(nparray1, nparray2) # Recommended way
print(flavor1)
flavor2 = np.sum(nparray1 * nparray2) # Ok way
print(flavor2)
flavor3 = nparray1 @ nparray2 # Geeks way
print(flavor3)
# As you never should do: # Noobs way
flavor4 = 0
for a, b in zip(nparray1, nparray2):
flavor4 += a * b
print(flavor4)
# -
# **We strongly recommend you to use np.dot, since it is the only method that accepts nparrays and lists without problems**
# +
norm1 = np.dot(np.array([1, 2]), np.array([3, 4])) # Dot product on nparrays
norm2 = np.dot([1, 2], [3, 4]) # Dot product on python lists
print(norm1, '=', norm2 )
# -
# And finally note that the norm of a vector is defined as the dot product of the vector with it self. Thus, you can write the norm of a vector using any of the flavors of the dot product:
# $$ norm(\vec a) = ||\vec a|| = \sqrt {\sum_{i=1}^{n} a_i ^ 2} = \sqrt {a \cdot a}$$
#
# ## Sums by rows or columns
#
# Another very common operation that you must perform on data is to get the sum of the elements of the matrix by rows or columns.
#
# This is very similar as you already did for the norm function. You control the dimension of the operation with the axis parammeter. axis=0 means sum the elements of each column together. axis=1 means sum the elements of each row together.
# +
nparray2 = np.array([[1, -1], [2, -2], [3, -3]]) # Define a 3 x 2 matrix.
sumByCols = np.sum(nparray2, axis=0) # Get the sum for each column. Returns 2 elements
sumByRows = np.sum(nparray2, axis=1) # get the sum for each row. Returns 3 elements
print('Sum by columns: ')
print(sumByCols)
print('Sum by rows:')
print(sumByRows)
# -
# ## Get the mean by rows or columns
#
# As with the sums, you can use Numpy functions to get the mean of a vector or matrix. You can specify the axis of the operation as well. Just remember that the mean of a vector is the sum of its elements divided by the length of the vector
# $$ mean(\vec a) = \frac {\sqrt {\sum_{i=1}^{n} a_i }}{n}$$
# +
nparray2 = np.array([[1, -1], [2, -2], [3, -3]]) # Define a 3 x 2 matrix. Chosen to be a matrix with 0 mean
mean = np.mean(nparray2) # Get the mean for the whole matrix
meanByCols = np.mean(nparray2, axis=0) # Get the mean for each column. Returns 2 elements
meanByRows = np.mean(nparray2, axis=1) # get the mean for each row. Returns 3 elements
print('Matrix mean: ')
print(mean)
print('Mean by columns: ')
print(meanByCols)
print('Mean by rows:')
print(meanByRows)
# -
# ## Center the columns of a matrix
#
# Centering the attributes of a data matrix is a very important preprocessing step. Centering means removing the mean
# of each column of the matrix, such that the mean by columns of the resulting matrix is always 0.
#
# With numpy this process is as simple as this:
# +
nparray2 = np.array([[1, 1], [2, 2], [3, 3]]) # Define a 3 x 2 matrix.
nparrayCentered = nparray2 - np.mean(nparray2, axis=0) # Remove the mean for each column
print('Original matrix')
print(nparray2)
print('Centered by columns matrix')
print(nparrayCentered)
print('New mean by column')
print(nparrayCentered.mean(axis=0))
# -
# Warning!!. This process does not apply for centering rows. If you want to do so, consider trasposing the matrix, centering by columns and the transpose back the result.
# +
nparray2 = np.array([[1, 3], [2, 4], [3, 5]]) # Define a 3 x 2 matrix.
nparrayCentered = nparray2.T - np.mean(nparray2, axis=1) # Remove the mean for each row
nparrayCentered = nparrayCentered.T # Transpose back the result
print('Original matrix')
print(nparray2)
print('Centered by columns matrix')
print(nparrayCentered)
# -
# Note that some operations can be performed by the static function **np.sum** or **np.mean**, or rather by the inner function of the array
# +
nparray2 = np.array([[1, 3], [2, 4], [3, 5]]) # Define a 3 x 2 matrix.
mean1 = np.mean(nparray2) # Static way
mean2 = nparray2.mean() # Dinamic way
print(mean1, ' == ', mean2)
# -
# Even if they are equivalent, we encourage you to use the static way always.
#
#
# **Congratulations!!** you have now the tools to operate vector and matrices in Python with Numpy.
| Natural Language Processing with Classification and Vector Spaces/Week 3/NLP_C1_W3_lecture_nb_01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Our First CNN in Keras
# ### Creating a model based on the MNIST Dataset of Handwrittent Digits
# ### Step 1: Lets load our dataset
# +
from keras.datasets import mnist
# loads the MNIST dataset
(x_train, y_train), (x_test, y_test) = mnist.load_data()
print (x_train.shape)
# -
# ### Step 2A: Examine the size and image dimenions (not required but good practice)
# - Check the number of samples, dimenions and whether images are color or grayscale
# - We see that our training data consist of **60,000** samples of training data, **10,000** samples of test data
# - Our labels are appropriately sized as well
# - Our Image dimenions are **28 x 28**, with **no color channels** (i.e. they are grayscale, so no BGR channels)
# +
# printing the number of samples in x_train, x_test, y_train, y_test
print("Initial shape or dimensions of x_train", str(x_train.shape))
print ("Number of samples in our training data: " + str(len(x_train)))
print ("Number of labels in our training data: " + str(len(y_train)))
print ("Number of samples in our test data: " + str(len(x_test)))
print ("Number of labels in our test data: " + str(len(y_test)))
print()
print ("Dimensions of x_train:" + str(x_train[0].shape))
print ("Labels in x_train:" + str(y_train.shape))
print()
print ("Dimensions of x_test:" + str(x_test[0].shape))
print ("Labels in y_test:" + str(y_test.shape))
# -
# ### Step 2B - Let's take a look at some of images in this dataset
# - Using OpenCV
# - Using Matplotlib
# +
# Using OpenCV
# import opencv and numpy
import cv2
import numpy as np
# Use OpenCV to display 6 random images from our dataset
for i in range(0,6):
random_num = np.random.randint(0, len(x_train))
img = x_train[random_num]
window_name = 'Random Sample #' + str(i)
cv2.imshow(window_name, img)
cv2.waitKey(0)
cv2.destroyAllWindows()
# -
# ### Let's do the same thing but using matplotlib to plot 6 images
# +
# importing matplot lib
import matplotlib.pyplot as plt
# Plots 6 images, note subplot's arugments are nrows,ncols,index
# we set the color map to grey since our image dataset is grayscale
plt.subplot(331)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
plt.subplot(332)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
plt.subplot(333)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
plt.subplot(334)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
plt.subplot(335)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
plt.subplot(336)
random_num = np.random.randint(0,len(x_train))
plt.imshow(x_train[random_num], cmap=plt.get_cmap('gray'))
# Display out plots
plt.show()
# -
# ### Step 3A - Prepare our dataset for training
# +
# Lets store the number of rows and columns
img_rows = x_train[0].shape[0]
img_cols = x_train[0].shape[1]
# Getting our date in the right 'shape' needed for Keras
# We need to add a 4th dimenion to our date thereby changing our
# Our original image shape of (60000,28,28) to (60000,28,28,1)
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
# store the shape of a single image
input_shape = (img_rows, img_cols, 1)
# change our image type to float32 data type
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# Normalize our data by changing the range from (0 to 255) to (0 to 1)
x_train /= 255
x_test /= 255
print('x_train shape:', x_train.shape)
print(x_train.shape[0], 'train samples')
print(x_test.shape[0], 'test samples')
# -
# ### Step 3B - One Hot Encode Our Labels (Y)
# +
from keras.utils import np_utils
# Now we one hot encode outputs
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
# Let's count the number columns in our hot encoded matrix
print ("Number of Classes: " + str(y_test.shape[1]))
num_classes = y_test.shape[1]
num_pixels = x_train.shape[1] * x_train.shape[2]
# -
y_train[0]
# ### Step 4 - Create Our Model
# - We're constructing a simple but effective CNN that uses 32 filters of size 3x3
# - We've added a 2nd CONV layer of 64 filters of the same size 3x2
# - We then downsample our data to 2x2, here he apply a dropout where p is set to 0.25
# - We then flatten our Max Pool output that is connected to a Dense/FC layer that has an output size of 128
# - How we apply a dropout where P is set to 0.5
# - Thus 128 output is connected to another FC/Dense layer that outputs to the 10 categorical units
# +
import keras
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras import backend as K
from keras.optimizers import SGD
# create model
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3),
activation='relu',
input_shape=input_shape))
model.add(Conv2D(64, (3, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(num_classes, activation='softmax'))
model.compile(loss = 'categorical_crossentropy',
optimizer = SGD(0.01),
metrics = ['accuracy'])
print(model.summary())
# -
# ### Step 5 - Train our Model
# - We place our formatted data as the inputs and set the batch size, number of epochs
# - We store our model's training results for plotting in future
# - We then use Kera's molel.evaluate function to output the model's fina performance. Here we are examing Test Loss and Test Accuracy
# +
batch_size = 32
epochs = 10
history = model.fit(x_train,
y_train,
batch_size = batch_size,
epochs = epochs,
verbose = 1,
validation_data = (x_test, y_test))
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
# -
# ### Step 6 - Ploting our Loss and Accuracy Charts
# +
# Plotting our loss charts
import matplotlib.pyplot as plt
history_dict = history.history
loss_values = history_dict['loss']
val_loss_values = history_dict['val_loss']
epochs = range(1, len(loss_values) + 1)
line1 = plt.plot(epochs, val_loss_values, label='Validation/Test Loss')
line2 = plt.plot(epochs, loss_values, label='Training Loss')
plt.setp(line1, linewidth=2.0, marker = '+', markersize=10.0)
plt.setp(line2, linewidth=2.0, marker = '4', markersize=10.0)
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.grid(True)
plt.legend()
plt.show()
# +
# Plotting our accuracy charts
import matplotlib.pyplot as plt
history_dict = history.history
acc_values = history_dict['acc']
val_acc_values = history_dict['val_acc']
epochs = range(1, len(loss_values) + 1)
line1 = plt.plot(epochs, val_acc_values, label='Validation/Test Accuracy')
line2 = plt.plot(epochs, acc_values, label='Training Accuracy')
plt.setp(line1, linewidth=2.0, marker = '+', markersize=10.0)
plt.setp(line2, linewidth=2.0, marker = '4', markersize=10.0)
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.grid(True)
plt.legend()
plt.show()
# -
# ### Step 7A - Saving our Model
model.save("/home/deeplearningcv/DeepLearningCV/Trained Models/8_mnist_simple_cnn_10_Epochs.h5")
print("Model Saved")
# ### Step 7B - Loading our Model
# +
from keras.models import load_model
classifier = load_model('/home/deeplearningcv/DeepLearningCV/Trained Models/8_mnist_simple_cnn_10_Epochs.h5')
# -
# ### Step 8 - Lets input some of our test data into our classifer
# +
import cv2
import numpy as np
def draw_test(name, pred, input_im):
BLACK = [0,0,0]
expanded_image = cv2.copyMakeBorder(input_im, 0, 0, 0, imageL.shape[0] ,cv2.BORDER_CONSTANT,value=BLACK)
expanded_image = cv2.cvtColor(expanded_image, cv2.COLOR_GRAY2BGR)
cv2.putText(expanded_image, str(pred), (152, 70) , cv2.FONT_HERSHEY_COMPLEX_SMALL,4, (0,255,0), 2)
cv2.imshow(name, expanded_image)
for i in range(0,10):
rand = np.random.randint(0,len(x_test))
input_im = x_test[rand]
imageL = cv2.resize(input_im, None, fx=4, fy=4, interpolation = cv2.INTER_CUBIC)
input_im = input_im.reshape(1,28,28,1)
## Get Prediction
res = str(classifier.predict_classes(input_im, 1, verbose = 0)[0])
draw_test("Prediction", res, imageL)
cv2.waitKey(0)
cv2.destroyAllWindows()
# -
# ### Putting All Together!
# We don't need to run each section of code separately. Once we know it all works as it's supposed to, we can put all te pieces together and start training our model
# +
from keras.datasets import mnist
from keras.utils import np_utils
import keras
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras import backend as K
from keras.optimizers import SGD
# Training Parameters
batch_size = 128
epochs = 10
# loads the MNIST dataset
(x_train, y_train), (x_test, y_test) = mnist.load_data()
# Lets store the number of rows and columns
img_rows = x_train[0].shape[0]
img_cols = x_train[1].shape[0]
# Getting our date in the right 'shape' needed for Keras
# We need to add a 4th dimenion to our date thereby changing our
# Our original image shape of (60000,28,28) to (60000,28,28,1)
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
# store the shape of a single image
input_shape = (img_rows, img_cols, 1)
# change our image type to float32 data type
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# Normalize our data by changing the range from (0 to 255) to (0 to 1)
x_train /= 255
x_test /= 255
print('x_train shape:', x_train.shape)
print(x_train.shape[0], 'train samples')
print(x_test.shape[0], 'test samples')
# Now we one hot encode outputs
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
# Let's count the number columns in our hot encoded matrix
print ("Number of Classes: " + str(y_test.shape[1]))
num_classes = y_test.shape[1]
num_pixels = x_train.shape[1] * x_train.shape[2]
# create model
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3),
activation='relu',
input_shape=input_shape))
model.add(Conv2D(64, (3, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(num_classes, activation='softmax'))
model.compile(loss = 'categorical_crossentropy',
optimizer = SGD(0.01),
metrics = ['accuracy'])
print(model.summary())
history = model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(x_test, y_test))
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
# -
# ### Visualizing Our Model
# - First let's re-create our model
# +
# %matplotlib inline
import keras
from keras.models import Sequential
from keras.utils.vis_utils import plot_model
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import numpy as np
from keras.utils import np_utils
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras import backend as K
input_shape = (28,28,1)
num_classes = 10
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3),
activation='relu',
input_shape=input_shape))
model.add(Conv2D(64, (3, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(num_classes, activation='softmax'))
model.compile(loss=keras.losses.categorical_crossentropy,
optimizer=keras.optimizers.Adadelta(),
metrics=['accuracy'])
print(model.summary())
# -
# ### Generating the diagram of the model architecture
# +
# Save our model diagrams to this path
model_diagrams_path = '/home/deeplearningcv/DeeplearningCV/Trained Models/'
# Generate the plot
plot_model(model, to_file = model_diagrams_path + 'model_plot.png',
show_shapes = True,
show_layer_names = True)
# Show the plot here
img = mpimg.imread(model_diagrams_path + 'model_plot.png')
plt.figure(figsize=(30,15))
imgplot = plt.imshow(img)
| 8. Making a CNN in Keras/8.3 to 8.10 - Building a CNN for handwritten digits - MNIST.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.8.5 64-bit (''base'': conda)'
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/JonasRigo/AD-Numerical-Renormalization-Group/blob/main/dNRG.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="x_2eVyMfIjgS"
# # Differentiable Numerical Renormalization Group for a single Anderson impurity model
# by <NAME> and <NAME>
#
# In this notebook we follow the implementation of NRG for a single Anderson Impurity Model (siAM) laid out by *<NAME>, <NAME> and <NAME>* in *PRB 21, 3 1980*.
#
# ## Libraries
#
# We use the `jax` library for efficient GPU backed implementation and easy automatic differentiation.
# + id="bar_ONbWIjgW"
from jax.config import config # enable for 64 precision
config.update("jax_enable_x64",True)
import jax
import jax.numpy as jp
from jax import ops as jops
import jax.scipy.linalg as la
from jax import jacrev, grad, jacfwd
from jax import make_jaxpr
from jax import custom_vjp
from jax import custom_jvp
from functools import partial
# + [markdown] id="Nfjqr_9IIujW"
# # Global variables
#
# The NRG solver is based on iteratively expanding the Hilbert space of the Hamiltonian and then diagonalizing the grown Hamiltonian. The Hilbert space of the Hamiltonain at a step $N$ is always multiplied by the same $4$ dimensional Hilbert space of an $\uparrow$ and a $\downarrow$ flavoured fermions. The matrix elements of the grown Hamailtonian can be obtained as follows:
#
# $\langle i',l',N,\vert H_{N+1}\vert i,l,N\rangle = \Lambda^{1/2} E(l,N)\delta_{i i'}\delta_{l l'}+ t_N(\langle l',N\vert f^\dagger_{N\sigma}\vert l,N\rangle \langle i' \vert f_{N+1\sigma}\vert i\rangle + \rm h.c. )$
#
# where $l$ enumerates the eigenbasis of $H_N$ at iteration $N$ and $i$ enumerates the basis of a single Wilson chain site in the following way
#
# $\vert i = 0 \rangle = \vert vac \rangle \\ \vert i = 1 \rangle = f^{\dagger}_{\uparrow}\vert vac\rangle \\
# \vert i = 2 \rangle = f^{\dagger}_{\uparrow}f^{\dagger}_{\downarrow}\vert vac\rangle \\
# \vert i = 3 \rangle = f^{\dagger}_{\downarrow}\vert vac\rangle $
#
# Since the basis of a single Wilson chain site never changes, we can introduce
#
# $\eta_{\sigma i i’} = \langle i' \vert f_{N+1\sigma}\vert i\rangle$,
#
# which is saved as a global tensor called `elemaddedsite`. In the following we refer to `elemaddedsite` as “transfer tensor”.
#
# + colab={"base_uri": "https://localhost:8080/"} id="KlZkBkT8T0fq" outputId="602d51ce-6161-4d30-869d-ada0bf398956"
elemaddedsite = jp.zeros((2,4,4))
elemaddedsite = jops.index_update(elemaddedsite,jops.index[0,0,3],1.)
elemaddedsite = jops.index_update(elemaddedsite,jops.index[0,1,2],-1.)
elemaddedsite = jops.index_update(elemaddedsite,jops.index[1,0,1],1.)
elemaddedsite = jops.index_update(elemaddedsite,jops.index[1,3,2],1.)
elemaddedsite_index = [[1,3,2],[1,0,1],[0,1,2],[0,0,3]]
print("elemaddedsite: \n", elemaddedsite)
# + [markdown] id="6WeWzBsWp1pd"
# # Hamiltonian class
#
# The Hamiltonain has to undergo two important steps:
#
# * Initialize: set up $H_{-1} = H_{imp}$
# * Grow: $H_N \rightarrow H_{N+1}$
#
# For the *initialization* we need to define the impurity Hamiltonian (includes no bath or hybridization) and bring it in diagonal form. For the siAM the occupation basis is already an eigenbasis.
#
# To *grow* the Hamiltonian means to add a Wilson chain site to the Hamiltonian
# $\langle i',l',N,\vert H_{N+1}\vert i,l,N\rangle = \Lambda^{1/2} E(l,N)\delta_{i i'}\delta_{l l'}+ t_N(\langle l',N\vert f^\dagger_{N\sigma}\vert l,N\rangle \langle i' \vert f_{N+1\sigma}\vert i\rangle + \rm h.c. )$,
#
# where $t_N$ is the Wislon chain hopping and $\vert i,l,N\rangle$ an eigenvector of $H_N$. This equation can be rewritten as
# $\langle i',l',N,\vert H_{N+1}\vert i,l,N\rangle = \Lambda^{1/2} E(l,N)\delta_{i i'}\delta_{l l'}+ t_N((-1)^{l'}\eta^{N+1}_{\sigma l l’} \eta_{\sigma i’ i} + \rm h.c. )$,
#
# where $\eta^{N+1}_{\sigma l l’}$ is the complex transposed transfer tensor in the eigenbasis of $H_N$. How to obtain the transfer tensor in the basis of $H_N$ will be explained in detail later. The last crucial step performed in `grow` is to truncate the eigenbasis of $H_N$. No more than `rlim` states are kept.
# + id="fdWlYwgcgJt0"
class Hamiltonian(object):
def __init__(self, eps, U, V,lmax=40,rlim=200,Lam=3.,Himp_dim=4):
"""
Initialize the Hamiltonian class with all impurity couplings and the
parameters required by the NRG.
===========================================================
input:
eps: on-site energy
U: on-site interaction
V: hybridization strength
lmax: chain length
rlim: maximum number of kept states
Lam: discretization parameter
Himp_dim: dimension of a Wilson chain site
"""
print("Anderson impurity model")
print("Lambda = ",Lam)
print("max number of states kept at each iteration = ",rlim)
print("\n")
self.Lam = Lam
self.rlim = rlim
self.dim = Himp_dim
self.aux_dim = 4
self.eps = eps
self.U = U
self.V = V
def initialize(self):
"""
Create the `-1` Hamiltonian, it contains only the impurity without hybridization.
"""
print("U/D = ", self.U)
print("epsilon/D = ",self.eps)
print("V/D = ",self.V)
print("\n")
"""Renormalize the impurity parameters"""
alambda = (self.Lam + 1.)/(self.Lam - 1.)*jp.log(self.Lam)/2. # this factor accounts for the discretation
self.eps /= self.Lam
self.U /= self.Lam
"""
The impurity Hamiltonian is diagonal in the occupation basis.
We exploit that to write it in terms of its eigenvalues and use the occupation
basis as eigenbasis.
"""
energies = jp.zeros(self.dim)
energies = jops.index_update(energies,jp.array([0,1,2,3]),[0.,self.eps,(2.*self.eps + self.U),self.eps])
"""
The first hopping term is the hybridization between impurity and bath.
"""
self.wilson_t = jp.sqrt(alambda/self.Lam) * self.V # * we account for the descrete bath
"""
The transfer tensor in the basis of H_N becomes `elemlastsite`.
"""
elemlastsite = jp.zeros((2,self.aux_dim,self.aux_dim))
elemlastsite = jops.index_update(elemlastsite,0,jp.transpose(elemaddedsite[0]))
elemlastsite = jops.index_update(elemlastsite,1,jp.transpose(elemaddedsite[1]))
return energies, elemlastsite
def grow(self,wilson_site,energies,elemlastsite):
"""
The Hilbert space grows by an up and a down spin. We keep the
old dimension locally in `dim` and update the dimension in `self` by
multiplying it with the dimension of the added Hilbert space
===========================================================
input:
wilson_site: integer number defining the number of sites attached to the impurity
energies: array containing eigenvalues of the Hamiltonian
"""
dim = self.dim
"""
This step is crucial: we limit the maximum amount of states by truncating the
dimension of the Hilbert space.
"""
self.dim = self.dim*self.aux_dim
H = jp.zeros((self.dim,self.dim))
"""
The diagonal consists only of the beforehand calculated energies. Mind the
tesnor product! The factor `0.5` accounts for a factor `2` that the diagonal acquires
after summing up the trasnpose of `H`
"""
id = lambda x: dim*x
for i in jp.arange(4):
H = jops.index_update(H,jops.index[id(i):id(i+1),id(i):id(i+1)], 0.5 * jp.diag(jp.sqrt(self.Lam) * energies[0:dim]))
"""
We exploit the tensor nature of of `elementlastside` and `elementaddedstite` and
express the sum over the flavour (in this case spin) as a scalar product. Note that
this corresponds to taking a kronecker tensorproduct of the type: |N,l> x |i>.
"""
for idx in elemaddedsite_index:
sign, k, kp = idx
H = jops.index_update(H,jops.index[id(k):id(k+1),id(kp):id(kp+1)],self.wilson_t * elemlastsite[sign]*elemaddedsite[sign,k,kp] * (-1.)**(k))
"""
Add the hermitian conjugate that was neglected earlier
"""
H += H.T
"""
The wilson chain has now grown, so we calculate the next hoping paramter that links the
new Hamiltonian to next Hilbert space in the following iteration.
"""
self.wilson_t = .5*(1.+self.Lam**(-1.))*(1.-self.Lam**(-wilson_site-1.))*((1.-self.Lam**(-2.*wilson_site-1.))*(1.-self.Lam**(-2.*wilson_site-3.)))**(-.5)
return H
# + [markdown] id="FBuipGFokGuq"
# ## Differentiable Hamiltonian class
#
# Also the differentiable Hamitlonian has two functions
#
# * Initialize
# * Grow
#
# For the initialization we need to define the derivative of the impurity Hamiltonian with respect to a coupling parameter.
#
# To grow the Hamiltonian means in this case to propagate it forward. Where in the `Hamiltonian` routine we add sites to Hamiltonian, here we simply expand the Hilbert space and transform the differentiated Hamiltonian in the eigenbasis of the Hamiltonian obtained in `Hamiltonian`.
# + id="bGO2BVxCtSNX"
class dHamiltonian(object):
def __init__(self, eps, U, V,lmax=40,rlim=200,Lam=3.,Himp_dim=4):
"""
Initialize the Hamiltonian class with all impurity couplings and the
parameters required by the NRG.
===========================================================
input:
eps: on-site energy
U: on-site interaction
V: hybridization strength
lmax: chain length
rlim: maximum number of kept states
Lam: discretization parameter
Himp_dim: dimension of a Wilson chain site
"""
print("Anderson impurity model")
print("Lambda = ",Lam)
print("max number of states kept at each iteration = ",rlim)
print("\n")
self.Lam = Lam
self.rlim = rlim
self.dim = Himp_dim
self.aux_dim = 4
self.eps = eps
self.U = U
self.V = V
self.lmax = lmax
def initialize(self):
"""
Here we create the derivitative of the `-1` Hamiltonian.
"""
print("U/D = ", self.U)
print("epsilon/D = ",self.eps)
print("V/D = ",self.V)
print("\n")
"""Renormalize the impurity parameters"""
alambda = (self.Lam + 1.)/(self.Lam - 1.)*jp.log(self.Lam)/2. # this factor accounts for the discretation error
self.eps /= self.Lam
self.U /= self.Lam
"""
The impurity Hamiltonian is diagonal in the occupation basis.
We exploit that to write it in terms of its eigenvalues and use the occupation
basis as eigenbasis.
"""
energies = jp.zeros(self.dim)
energies = jops.index_update(energies,jp.array([0,1,2,3]),[0.,self.eps,(2.*self.eps + self.U),self.eps])
self.ham = jp.diag(energies)
"""
The first hopping term is the hybridization between impurity and bath:
"""
self.wilson_t = jp.sqrt(alambda/self.Lam) * self.V
def grow(self,eigsrset,rkept,trafoset):
"""
The Hilbert space grows now for an up and a down spin. We keep the
old dimension locally in `dim` and update the dimension in `self` by
multiplying it with the dimension of the added Hilbert space
===========================================================
input:
eigsrset: list of oll eigenstates of all iterative diagonalization of the
Hamiltonian.
rkept: number of kept states in every iteration
trafoset: list containing the `elemaddedsite` tensor in the eigenbasis of
of all iterations of the Hamiltonian.
"""
for i in range(self.lmax):
print("i: ", i)
eigs = eigsrset[i]
dim = rkept[i]
elemlastsite = trafoset[i]
print("dim: ",dim)
self.dim = rkept[i]*self.aux_dim
H = jp.zeros((self.dim,self.dim))
id = lambda x: dim*x
self.ham = jp.dot(eigs.T,jp.dot(self.ham,eigs))
for i in jp.arange(4):
H = jops.index_update(H,jops.index[id(i):id(i+1),id(i):id(i+1)], 0.5 * jp.sqrt(self.Lam) * self.ham[0:dim,0:dim])
for idx in elemaddedsite_index:
sign, k, kp = idx
H = jops.index_update(H,jops.index[id(k):id(k+1),id(kp):id(kp+1)],self.wilson_t * elemlastsite[sign]*elemaddedsite[sign,k,kp] * (-1.)**(k))
H += H.T
self.ham = H
"""
The wilson chain paramter is set to 0 becuase it vanishes through the derivative.
"""
self.wilson_t = 0.
return H
# + [markdown] id="x5hB6MOgsyCQ"
# ## Solver
#
# The solver handles two main routines
#
#
# * Basis change of the transfer tensor
# * Diagonalization of the Hamiltonian
# * Computing the thermodynamics
#
# ### Transfer tensor
#
# The basis change of the transfer tensor is a clever way to avoid high computational cost. We wrote the transfer tensor as follows
# $ \eta_{\sigma i j} = \langle j \vert f_\sigma\vert i\rangle $, now we express it in the basis of $H_N$. Let $\vert i \rangle \otimes \vert l, N \rangle$ be the basis of $H_N$ and $U$ the unitary transformation that diagonalizes $H_{N+1}$. Then
#
# $ \eta^{N+1}_{\sigma i j} = \sum_{l,l'}\sum_{k,k'}[U(i;lk)]^\dagger U(j;l'k')\eta_{\sigma k k'}$
# + id="PpqKQ6PF1Tfb"
def transfertensor(eigen_system,dim,trunc_dim):
"""
We use the knowledge about the transfer tensor `elemaddedsite` and calculate it
in the eigenbasis of the Hamitlonian. Thus, conceptually we transform
`elemaddesite` into `elemlastsite`.
"""
id = lambda x: dim*x
elemlastsite = jp.zeros((2,trunc_dim,trunc_dim))
for idx in elemaddedsite_index:
sigma, k, kp = idx
elemlastsite = jops.index_add(elemlastsite,jops.index[sigma],elemaddedsite[sigma,k,kp]*jp.matmul(jp.transpose(eigen_system[id(k):id(k)+dim,0:trunc_dim]),eigen_system[id(kp):id(kp)+dim,0:trunc_dim]))
return elemlastsite
# + [markdown] id="r-HtJ0FONROU"
# ### Thermodynamics
#
# To extract some physical insight from the simulation we compute thermodynamic quantities. Since we have the diagonalized Hamiltonian from NRG it is straightforward to calculate the full density matrix
#
# $\rho = \frac{1}{Z}e^{-\beta H_N},~Z=\sum_i e^{-\beta \lambda_i}$,
#
# where $\lambda_i$ are the eigenvalues of $H_N$. The temperature in this definition comes from the Hamiltonian itself. In the NRG scheme one can relate a certain chain length $N$ to a temperature $T$. The inverse temperature is given defined as
# $ \beta_N = 1/T_N$
#
# $\beta_N \Lambda^{-(N-1)/2} = \bar{\beta},~\bar{\beta} \propto \mathcal{O}(1)$
#
# For $\bar{\beta}$ we choose $\bar{\beta} = 0.9$. With this we know at which temperature the density matrix is calculated. Now we can compute is the system entropy
#
# $F = -T \ln(Z) \\
# S = - \frac{\partial F}{\partial T} \\
# \Rightarrow S = \bar{\beta}\langle H_N \rangle + \ln(Z),~\rm{with}~\langle H_N \rangle = \rm{tr}[\rho H ]$
# + id="G1wueUMT1b0t"
def thermo(i,energies,Lam):
"""
We calculate the density matrix for the momentary temperature
=====================================================
input:
i: wilson chain length
energies: eigen energies of the Hamiltonian
Lam: discretization parameter
"""
beta_bar = 0.9
"""the temperature is realted to the lenght of the wilson chain"""
temperature = np.power(Lam,-0.5*(i - 1.))/beta_bar
print("Temperature = ", temperature)
"""
from the diagonal hamiltonian we can directly calcualte the
partition funciton, desnity matrix and entropy
"""
rho = np.exp(-beta_bar*energies) # desnity vector -> diagonal only
exp_H = beta_bar*np.dot(energies,rho) # expectation value of the Hamiltonian
Z = np.sum(rho) # partition function
entropy = exp_H/Z + np.log(Z) # entropy
return temperature, entropy
# + [markdown] id="LVxKTAsT1czU"
# ### Solver class
#
# The iterative diagonalization routine.
# + id="JJj78PwPd3dK"
class Solver(object):
def __init__(self,ham,lmax):
"""
input:
ham: initialized hamiltonian of class type `Hamiltonian`
lmax: maximum chain length
"""
self.ham = ham
self.lmax = lmax
self.eigsrset = []
self.rkept = []
self.trafoset = []
self.rseed = rseed
def solve(self):
energies, elemlastsite = self.ham.initialize()
eigs = jp.eye(4,dtype=float)
entropy = []
for i in jp.arange(self.lmax):
"""
We save the dimension before growing, then pass it to the
trans fertensor and truncating:
"""
dim = self.ham.dim
self.eigsrset += [eigs]
self.trafoset += [elemlastsite]
self.rkept += [self.ham.dim]
""" Update for user: """
print("Interation: ", i)
print("States kept at this iteration = ", dim)
""" iterate the wilson chain and diagonalize the Hamiltonian"""
ham = self.ham.grow(i,energies,elemlastsite)
energies, eigs = la.eigh(ham,turbo=True)
""" set the groundstate to zero energy with a variable shift """
#energies -= energies[0]
""" update dimension and truncate """
self.ham.dim = jp.minimum(self.ham.dim, self.ham.rlim)
elemlastsite = transfertensor(eigs,dim,self.ham.dim)
print("\n")
self.eigsrset += [eigs]
self.trafoset += [elemlastsite]
self.rkept += [self.ham.dim]
print("Calculation complete.")
return energies, eigs
# + [markdown] id="daTybADwkT-3"
# ## Differentiable Solver Routine
#
# Other than the non-differentiable solver routine `Solver` the differentiable solver routine does not return the eigenstates and eigenenergies. The `dSolver` routine returns the final Hamiltonian matrix $H_N$.
# + id="3SfVggJ1kTFb"
class dSolver(object):
def __init__(self,ham,lmax,rseed):
"""
input:
ham: initialized hamiltonian of class type Hamiltonian
lmax: maximum chain length
rseed: numpy array containing ranodm values
"""
self.ham = ham
self.lmax = lmax
self.eigsrset = []
self.rkept = []
self.trafoset = []
self.rseed = rseed
def solve(self):
energies, elemlastsite = self.ham.initialize()
eigs = jp.eye(4,dtype=float)
entropy = []
for i in jp.arange(self.lmax):
dim = self.ham.dim
"""
Saving the eigensystem, the `elemaddedsite` tensor in the Hamiltonian
eigenbasis and the number of kept states.
"""
self.eigsrset += [eigs]
self.trafoset += [elemlastsite]
self.rkept += [self.ham.dim]
""" Update for user:"""
print("Interation: ", i)
print("States kept at this iteration = ", dim)
""" iterate the wilson chain and diagonalize the Hamiltonian"""
ham = self.ham.grow(i,energies,elemlastsite)
""" we add random noise to the diagonal to lift the degeneracy of eigenvalues """
energies, eigs = la.eigh(ham+jp.diag(rseed[:ham.shape[0]]),turbo=True)
""" set the groundstate to zero energy with a variable shift """
#energies -= energies[0]
""" updated dimension and truncate """
self.ham.dim = jp.minimum(self.ham.dim, self.ham.rlim)
elemlastsite = transfertensor(eigs,dim,self.ham.dim)
print("\n")
self.eigsrset += [eigs]
self.trafoset += [elemlastsite]
self.rkept += [self.ham.dim]
print("Calculation complete.")
return ham
# + [markdown] id="jLSV4SKHxzwp"
# ## Automatic Differentiation
#
# ### Differentiable `jax` NRG primitive
#
# The derivative of the whole NRG code with respect to impurity coupling constants, is given through the derivative of the Hamiltonian $H_N$. The the derivative of $H_N$ with respect to impurity coupling constants can be obtained through propagating forward $\text{d} H_{\rm imp}$, such that it can act on the $N^{\rm th}$ Hilbert space associated to $H_N$.
# + id="fz6EMfcrps8l"
@custom_jvp
def dH(eps,U,V,l,rlim,Lam):
"""
Initialize the Hamiltonian and obtain the Hamiltonian matrix
of maximal length.
Note: Derivatives can only be obtained with resepect to
eps, U or V.
===========================================================
input:
eps: on-site energy
U: on-site interaction
V: hybridization strength
l: chain length
rlim: maximum number of kept states
Lam: discretization parameter
"""
H1 = Hamiltonian(eps,U,V,l,rlim,Lam)
S = dSolver(H1,l,rseed)
return S.solve()
@dH.defjvp
def dH_jvp(primals, tangents):
"""
Returns the derivative of the Hamiltonian with respect to the
input parameters `eps`, `U` or `V`.
"""
eps_dot, U_dot, V_dot,adot,bdot,cdot = tangents
eps,U,V,l,rlim,Lam = primals
H1 = Hamiltonian(eps,U,V,l,rlim,Lam)
S = dSolver(H1,l,rseed)
ham = S.solve()
"""derivative wrt eps """
H = dHamiltonian(1.,0.,0.,l,rlim,Lam)
H.initialize()
dH1 = H.grow(S.eigsrset,S.rkept,S.trafoset)
"""derivative wrt U """
H = dHamiltonian(0.,1.,0.,l,rlim,Lam)
H.initialize()
dH2 = H.grow(S.eigsrset,S.rkept,S.trafoset)
"""derivative wrt V """
H = dHamiltonian(0.,0.,1.,l,rlim,Lam)
H.initialize()
dH3 = H.grow(S.eigsrset,S.rkept,S.trafoset)
""" accumulating derivative """
primal_out = ham
tangent_out = dH1*eps_dot + U_dot*dH2 + V_dot*dH3 + 0.*adot+0.*bdot+0.*cdot
return primal_out, tangent_out
# + [markdown] id="vLa2bgQzisqM"
# ### Examples of possible derivatives
# + id="mouTE36ySXTl"
import numpy as np
"""
The random values generated here are used the lift the
degeneracies of the Hamiltonian. This allows the computation of the
derivatives of the eigenvalues and eigenvectors.
"""
rseed = np.random.randn(4000)*1e-12
# + id="OvIABiaouxuD"
def free_energy(eps,U,V,l,rlim,Lam):
"""
Calculate the free energy of an Anderson impurity model
===========================================================
input:
eps: on-site energy
U: on-site interaction
V: hybridization strength
l: chain length
rlim: maximum number of kept states
Lam: discretization parameter
"""
ham = dH(eps,U,V,l,rlim,Lam)
energies, eivecs = la.eigh(ham + jp.diag(rseed[:ham.shape[0]]),turbo=True)
gs = energies[0]
energies -= energies[0]
beta_bar = 0.9
rho = jp.exp(-energies*beta_bar)
F = -(jp.log(jp.sum(rho))-beta_bar*gs)/(beta_bar*Lam**((l-2.)/2.))
return F
def nexp(eps,U,V,l,rlim,Lam):
"""
Calculate the occupation of an Anderson impurity model.
===========================================================
input:
eps: on-site energy
U: on-site interaction
V: hybridization strength
l: chain length
rlim: maximum number of kept states
Lam: discretization parameter
"""
H1 = Hamiltonian(eps,U,V,l,rlim,Lam)
S = Solver(H1,l)
eivals, eivecs = S.solve()
eivals -= eivals[0]
""" obtaining the occupation operator """
H = dHamiltonian(1.,0.,0.,l,rlim,Lam)
H.initialize()
dH1 = H.grow(S.eigsrset,S.rkept,S.trafoset)
beta_bar = 0.9
""" constructing the density matrix """
rho = np.dot(eivecs,np.dot(np.diag(np.exp(-eivals*beta_bar)),eivecs.conj().T))
""" calculating the thermodynamic expectation value """
nrho = np.trace(np.dot(rho,dH1))/jp.trace(rho)
return Lam**(-(l-2.)/2.)*nrho
""" backwards derivative of the free energy with respect to eps and U """
dFde = jacrev(free_energy,[0,1])
# + [markdown] id="JQZEeA16ZYbR"
# Here we calculate $\partial_\epsilon F$, where $F$ is the free energy and $\epsilon$ the on-site energy.
# + colab={"base_uri": "https://localhost:8080/"} id="jSMSgcubuxuE" outputId="fd3df802-9cb8-481d-891c-b958f63c7827"
eps,U,V,l,rlim,Lam = -0.15,0.3,0.1,2,200,3.
""" finite difference value to approximate the derivative """
dfs = 1e-6
""" finite difference derivative """
a = (free_energy(eps+dfs,U,V,l,rlim,Lam)/dfs-free_energy(eps,U,V,l,rlim,Lam)/dfs)
""" forward pass of the free energy """
b = free_energy(eps,U,V,l,rlim,Lam)
""" backward pass of the free energy """
c = dFde(eps,U,V,l,rlim,Lam)[0]
""" thermodynamic expectation value of the occupation """
d = nexp(eps,U,V,l,rlim,Lam)
print("\n")
print("free energy: ",b)
print("\n")
print("automatic derivative: ",c)
print("\n")
print("finite difference derivative: ",a)
print("\n")
print("expectation value: ",d)
# + [markdown] id="WpW1lAiaZtif"
# Here we calculate $\frac{\partial^2}{\partial\epsilon \partial U} F$, where $F$ is the free energy and $\epsilon$ the on-site energy and $U$ the on-site interaction.
# + id="6_N1WgtZuxuE"
""" calculating the second order derivative of the free energy"""
dFdedU = jacrev(jacfwd(free_energy,[0]),[1])
# + colab={"base_uri": "https://localhost:8080/"} id="UJpaJlPEuxuF" outputId="62eb6108-4798-4c23-ab19-f084ee10849a"
""" finite difference value to approximate derivative """
dfs = 0.00390625
print("AD")
a = dFdedU(eps,U,V,l,rlim,Lam)
print("\n")
print("FD")
""" finite difference derivative """
b = nexp(eps,U+dfs,V,l,rlim,Lam)/dfs-nexp(eps,U,V,l,rlim,Lam)/dfs
print("\n")
print("automatic derivative: ",a[0][0])
print("finite difference derivative: ",b)
# + id="PGC5RmbIfWN1"
| dNRG.ipynb |
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Q#
# language: qsharp
# name: iqsharp
# ---
# # Superdense Coding Kata
#
# **Superdense Coding** quantum kata is a series of exercises designed to get you familiar with programming in Q#.
#
# It covers the superdense coding protocol which allows us to transmit two bits of classical information by sending just one qubit using previously shared quantum entanglement.
#
# - A good description can be found in [the Wikipedia article](https://en.wikipedia.org/wiki/Superdense_coding).
# - A great interactive demonstration can be found [on the Wolfram Demonstrations Project](http://demonstrations.wolfram.com/SuperdenseCoding/).
# - Superdense coding protocol is described in Nielsen & Chuang, section 2.3 (pp. 97-98).
#
# Each task is wrapped in one operation preceded by the description of the task. Your goal is to fill in the blank (marked with `// ...` comment) with some Q# code that solves the task. To verify your answer, run the cell using Ctrl/⌘+Enter.
#
# Each task defines an operation that can be used in subsequent tasks to simplify implementations and build on existing code. We split the superdense coding protocol into several steps, following the description in the [Wikipedia article](https://en.wikipedia.org/wiki/Superdense_coding):
#
# * Preparation (creating the entangled pair of qubits that are sent to Alice and Bob).
# * Encoding the message (Alice's task): Encoding the classical bits of the message into the state of Alice's qubit which then is sent to Bob.
# * Decoding the message (Bob's task): Using Bob's original qubit and the qubit he received from Alice to decode the classical message sent.
# * Finally, we compose those steps into the complete superdense coding protocol.
# To begin, first prepare this notebook for execution (if you skip this step, you'll get "Syntax does not match any known patterns" error when you try to execute Q# code in the next cells):
%package Microsoft.Quantum.Katas::0.8.1907.1701
# > The package versions in the output of the cell above should always match. If you are running the Notebooks locally and the versions do not match, please install the IQ# version that matches the version of the `Microsoft.Quantum.Katas` package.
# > <details>
# > <summary><u>How to install the right IQ# version</u></summary>
# > For example, if the version of `Microsoft.Quantum.Katas` package above is 0.1.2.3, the installation steps are as follows:
# >
# > 1. Stop the kernel.
# > 2. Uninstall the existing version of IQ#:
# > dotnet tool uninstall microsoft.quantum.iqsharp -g
# > 3. Install the matching version:
# > dotnet tool install microsoft.quantum.iqsharp -g --version 0.1.2.3
# > 4. Reinstall the kernel:
# > dotnet iqsharp install
# > 5. Restart the Notebook.
# > </details>
#
# ### Task 1. Entangled pair
#
# **Input:** Two qubits, each in the $|0\rangle$ state.
#
# **Goal:** Prepare a Bell state $|\Phi^{+}\rangle = \frac{1}{\sqrt{2}} (|00\rangle + |11\rangle)$ on these qubits.
# +
%kata T1_CreateEntangledPair_Test
operation CreateEntangledPair (q1 : Qubit, q2 : Qubit) : Unit is Adj {
// ...
}
# -
# ### Task 2. Send the message (Alice's task)
#
# Encode the message (two classical bits) in the state of Alice's qubit.
#
# **Inputs**:
# 1. Alice's part of the entangled pair of qubits qAlice.
# 2. Two classical bits, stored as ProtocolMessage.
#
# **Goal**: Transform the input qubit to encode the two classical bits.
#
# >`ProtocolMessage` is a custom type that represents the message to be transmitted. It includes two items of type `Bool` called `Bit1` and `Bit2`.
#
# <br/>
# <details>
# <summary>Need a hint? Click here</summary>
# Manipulate Alice's half of the entangled pair to change the joint state of the two qubits to one of the following four states based on the value of message:
#
# * [0; 0]: $|\Phi^{+}\rangle = \frac{1}{\sqrt{2}} (|00\rangle + |11\rangle)$
# * [0; 1]: $|\Psi^{+}\rangle = \frac{1}{\sqrt{2}} (|01\rangle + |10\rangle)$
# * [1; 0]: $|\Phi^{-}\rangle = \frac{1}{\sqrt{2}} (|00\rangle + |11\rangle)$
# * [1; 1]: $|\Psi^{-}\rangle = \frac{1}{\sqrt{2}} (|01\rangle + |10\rangle)$
#
# </details>
# +
%kata T2_EncodeMessageInQubit_Test
open Quantum.Kata.SuperdenseCoding;
operation EncodeMessageInQubit (qAlice : Qubit, message : ProtocolMessage) : Unit {
if (message::Bit1) { // accesses the item 'Bit1' of 'message'
// ...
}
// ...
}
# -
# ### Task 3. Decode the message and reset the qubits (Bob's task)
#
# Decode the message using the qubit received from Alice and reset both qubits to a $|00\rangle$ state.
#
# **Inputs:**
#
# 1. Qubit received from Alice qAlice.
# 2. Bob's part of the entangled pair qBob.
#
# **Goal** : Retrieve two bits of classic data from the qubits and return them as `ProtocolMessage`. The state of the qubits in the end of the operation should be $|00\rangle$.
#
# > You can create an instance of `ProtocolMessage` as `ProtocolMessage(bit1value, bit2value)`.
# +
%kata T3_DecodeMessageFromQubits_Test
open Quantum.Kata.SuperdenseCoding;
operation DecodeMessageFromQubits (qAlice : Qubit, qBob : Qubit) : ProtocolMessage {
// ...
}
# -
# ### Task 4. Superdense coding protocol end-to-end:
#
# Put together the steps performed in tasks 1-3 to implement the full superdense coding protocol.
#
# **Input:** Two classical bits to be transmitted.
#
# **Goal:** Prepare an EPR Pair, encode the two classical bits in the state of the pair by applying quantum gates to one member of the pair, and decode the two classical bits from the state of the pair. Return the result of decoding.
# +
%kata T4_SuperdenseCodingProtocol_Test
open Quantum.Kata.SuperdenseCoding;
operation SuperdenseCodingProtocol (message : ProtocolMessage) : ProtocolMessage {
// ...
}
| SuperdenseCoding/SuperdenseCoding.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Задание 1.2 - Линейный классификатор (Linear classifier)
#
# В этом задании мы реализуем другую модель машинного обучения - линейный классификатор. Линейный классификатор подбирает для каждого класса веса, на которые нужно умножить значение каждого признака и потом сложить вместе.
# Тот класс, у которого эта сумма больше, и является предсказанием модели.
#
# В этом задании вы:
# - потренируетесь считать градиенты различных многомерных функций
# - реализуете подсчет градиентов через линейную модель и функцию потерь softmax
# - реализуете процесс тренировки линейного классификатора
# - подберете параметры тренировки на практике
#
# На всякий случай, еще раз ссылка на туториал по numpy:
# http://cs231n.github.io/python-numpy-tutorial/
# +
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# %load_ext autoreload
# %autoreload 2
# -
from dataset import load_svhn, random_split_train_val
from gradient_check import check_gradient
from metrics import multiclass_accuracy
import linear_classifer
# # Как всегда, первым делом загружаем данные
#
# Мы будем использовать все тот же SVHN.
# +
def prepare_for_linear_classifier(train_X, test_X):
train_flat = train_X.reshape(train_X.shape[0], -1).astype(np.float) / 255.0
test_flat = test_X.reshape(test_X.shape[0], -1).astype(np.float) / 255.0
# Subtract mean
mean_image = np.mean(train_flat, axis = 0)
train_flat -= mean_image
test_flat -= mean_image
# Add another channel with ones as a bias term
train_flat_with_ones = np.hstack([train_flat, np.ones((train_X.shape[0], 1))])
test_flat_with_ones = np.hstack([test_flat, np.ones((test_X.shape[0], 1))])
return train_flat_with_ones, test_flat_with_ones
train_X, train_y, test_X, test_y = load_svhn("data", max_train=10000, max_test=1000)
train_X, test_X = prepare_for_linear_classifier(train_X, test_X)
# Split train into train and val
train_X, train_y, val_X, val_y = random_split_train_val(train_X, train_y, num_val = 1000)
# -
# # Играемся с градиентами!
#
# В этом курсе мы будем писать много функций, которые вычисляют градиенты аналитическим методом.
#
# Все функции, в которых мы будем вычислять градиенты, будут написаны по одной и той же схеме.
# Они будут получать на вход точку, где нужно вычислить значение и градиент функции, а на выходе будут выдавать кортеж (tuple) из двух значений - собственно значения функции в этой точке (всегда одно число) и аналитического значения градиента в той же точке (той же размерности, что и вход).
# ```
# def f(x):
# """
# Computes function and analytic gradient at x
#
# x: np array of float, input to the function
#
# Returns:
# value: float, value of the function
# grad: np array of float, same shape as x
# """
# ...
#
# return value, grad
# ```
#
# Необходимым инструментом во время реализации кода, вычисляющего градиенты, является функция его проверки. Эта функция вычисляет градиент численным методом и сверяет результат с градиентом, вычисленным аналитическим методом.
#
# Мы начнем с того, чтобы реализовать вычисление численного градиента (numeric gradient) в функции `check_gradient` в `gradient_check.py`. Эта функция будет принимать на вход функции формата, заданного выше, использовать значение `value` для вычисления численного градиента и сравнит его с аналитическим - они должны сходиться.
#
# Напишите часть функции, которая вычисляет градиент с помощью численной производной для каждой координаты. Для вычисления производной используйте так называемую two-point formula (https://en.wikipedia.org/wiki/Numerical_differentiation):
#
# 
#
# Все функции приведенные в следующей клетке должны проходить gradient check.
# +
# TODO: Implement check_gradient function in gradient_check.py
# All the functions below should pass the gradient check
def square(x):
return float(x*x), 2*x
check_gradient(square, np.array([3.0]))
def array_sum(x):
assert x.shape == (2,), x.shape
return np.sum(x), np.ones_like(x)
check_gradient(array_sum, np.array([3.0, 2.0]))
def array_2d_sum(x):
assert x.shape == (2,2)
return np.sum(x), np.ones_like(x)
check_gradient(array_2d_sum, np.array([[3.0, 2.0], [1.0, 0.0]]))
# -
# ## Начинаем писать свои функции, считающие аналитический градиент
#
# Теперь реализуем функцию softmax, которая получает на вход оценки для каждого класса и преобразует их в вероятности от 0 до 1:
# 
#
# **Важно:** Практический аспект вычисления этой функции заключается в том, что в ней учавствует вычисление экспоненты от потенциально очень больших чисел - это может привести к очень большим значениям в числителе и знаменателе за пределами диапазона float.
#
# К счастью, у этой проблемы есть простое решение -- перед вычислением softmax вычесть из всех оценок максимальное значение среди всех оценок:
# ```
# predictions -= np.max(predictions)
# ```
# (подробнее здесь - http://cs231n.github.io/linear-classify/#softmax, секция `Practical issues: Numeric stability`)
# +
# TODO Implement softmax and cross-entropy for single sample
probs = linear_classifer.softmax(np.array([-10, 0, 10]))
# Make sure it works for big numbers too!
probs = linear_classifer.softmax(np.array([1000, 0, 0]))
assert np.isclose(probs[0], 1.0)
# -
# Кроме этого, мы реализуем cross-entropy loss, которую мы будем использовать как функцию ошибки (error function).
# В общем виде cross-entropy определена следующим образом:
# 
#
# где x - все классы, p(x) - истинная вероятность принадлежности сэмпла классу x, а q(x) - вероятность принадлежности классу x, предсказанная моделью.
# В нашем случае сэмпл принадлежит только одному классу, индекс которого передается функции. Для него p(x) равна 1, а для остальных классов - 0.
#
# Это позволяет реализовать функцию проще!
probs = linear_classifer.softmax(np.array([-5, 0, 5]))
linear_classifer.cross_entropy_loss(probs, 1)
# После того как мы реализовали сами функции, мы можем реализовать градиент.
#
# Оказывается, что вычисление градиента становится гораздо проще, если объединить эти функции в одну, которая сначала вычисляет вероятности через softmax, а потом использует их для вычисления функции ошибки через cross-entropy loss.
#
# Эта функция `softmax_with_cross_entropy` будет возвращает и значение ошибки, и градиент по входным параметрам. Мы проверим корректность реализации с помощью `check_gradient`.
# TODO Implement combined function or softmax and cross entropy and produces gradient
loss, grad = linear_classifer.softmax_with_cross_entropy(np.array([1, 0, 0]), 1)
check_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, 1), np.array([1, 0, 0], np.float))
# В качестве метода тренировки мы будем использовать стохастический градиентный спуск (stochastic gradient descent или SGD), который работает с батчами сэмплов.
#
# Поэтому все наши фукнции будут получать не один пример, а батч, то есть входом будет не вектор из `num_classes` оценок, а матрица размерности `batch_size, num_classes`. Индекс примера в батче всегда будет первым измерением.
#
# Следующий шаг - переписать наши функции так, чтобы они поддерживали батчи.
#
# Финальное значение функции ошибки должно остаться числом, и оно равно среднему значению ошибки среди всех примеров в батче.
# TODO Extend combined function so it can receive a 2d array with batch of samples
np.random.seed(42)
# Test batch_size = 1
num_classes = 4
batch_size = 1
predictions = np.random.randint(-1, 3, size=(batch_size, num_classes)).astype(np.float)
target_index = np.random.randint(0, num_classes, size=(batch_size, 1)).astype(np.int)
check_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, target_index), predictions)
target_index
print(predictions)
linear_classifer.cross_entropy_loss(predictions, target_index)
predictions.shape[0]
# +
# Test batch_size = 3
num_classes = 4
batch_size = 3
predictions = np.random.randint(-1, 3, size=(batch_size, num_classes)).astype(np.float)
target_index = np.random.randint(0, num_classes, size=(batch_size, 1)).astype(np.int)
check_gradient(lambda x: linear_classifer.softmax_with_cross_entropy(x, target_index), predictions)
# Make sure maximum subtraction for numberic stability is done separately for every sample in the batch
probs = linear_classifer.softmax(np.array([[20,0,0], [1000, 0, 0]]))
assert np.all(np.isclose(probs[:, 0], 1.0))
# -
# ### Наконец, реализуем сам линейный классификатор!
#
# softmax и cross-entropy получают на вход оценки, которые выдает линейный классификатор.
#
# Он делает это очень просто: для каждого класса есть набор весов, на которые надо умножить пиксели картинки и сложить. Получившееся число и является оценкой класса, идущей на вход softmax.
#
# Таким образом, линейный классификатор можно представить как умножение вектора с пикселями на матрицу W размера `num_features, num_classes`. Такой подход легко расширяется на случай батча векторов с пикселями X размера `batch_size, num_features`:
#
# `predictions = X * W`, где `*` - матричное умножение.
#
# Реализуйте функцию подсчета линейного классификатора и градиентов по весам `linear_softmax` в файле `linear_classifer.py`
# +
# TODO Implement linear_softmax function that uses softmax with cross-entropy for linear classifier
batch_size = 2
num_classes = 2
num_features = 3
np.random.seed(42)
W = np.random.randint(-1, 3, size=(num_features, num_classes)).astype(np.float)
X = np.random.randint(-1, 3, size=(batch_size, num_features)).astype(np.float)
target_index = np.ones(batch_size, dtype=np.int)
loss, dW = linear_classifer.linear_softmax(X, W, target_index)
check_gradient(lambda w: linear_classifer.linear_softmax(X, w, target_index), W)
# -
# ### И теперь регуляризация
#
# Мы будем использовать L2 regularization для весов как часть общей функции ошибки.
#
# Напомним, L2 regularization определяется как
#
# l2_reg_loss = regularization_strength * sum<sub>ij</sub> W[i, j]<sup>2</sup>
#
# Реализуйте функцию для его вычисления и вычисления соотвествующих градиентов.
# TODO Implement l2_regularization function that implements loss for L2 regularization
linear_classifer.l2_regularization(W, 0.01)
check_gradient(lambda w: linear_classifer.l2_regularization(w, 0.01), W)
# # Тренировка!
# Градиенты в порядке, реализуем процесс тренировки!
# TODO: Implement LinearSoftmaxClassifier.fit function
classifier = linear_classifer.LinearSoftmaxClassifier()
loss_history = classifier.fit(train_X, train_y, epochs=10, learning_rate=1e-3, batch_size=300, reg=1e1)
# let's look at the loss history!
plt.plot(loss_history)
# +
# Let's check how it performs on validation set
pred = classifier.predict(val_X)
accuracy = multiclass_accuracy(pred, val_y)
print("Accuracy: ", accuracy)
# Now, let's train more and see if it performs better
classifier.fit(train_X, train_y, epochs=100, learning_rate=1e-3, batch_size=300, reg=1e1)
pred = classifier.predict(val_X)
accuracy = multiclass_accuracy(pred, val_y)
print("Accuracy after training for 100 epochs: ", accuracy)
# -
# ### Как и раньше, используем кросс-валидацию для подбора гиперпараметтов.
#
# В этот раз, чтобы тренировка занимала разумное время, мы будем использовать только одно разделение на тренировочные (training) и проверочные (validation) данные.
#
# Теперь нам нужно подобрать не один, а два гиперпараметра! Не ограничивайте себя изначальными значениями в коде.
# Добейтесь точности более чем **20%** на проверочных данных (validation data).
# +
num_epochs = 200
batch_size = 300
learning_rates = [1e-3, 1e-4, 1e-5]
reg_strengths = [1e-4, 1e-5, 1e-6]
best_classifier = None
best_val_accuracy = None
# TODO use validation set to find the best hyperparameters
# hint: for best results, you might need to try more values for learning rate and regularization strength
# than provided initially
print('best validation accuracy achieved: %f' % best_val_accuracy)
# -
# # Какой же точности мы добились на тестовых данных?
test_pred = best_classifier.predict(test_X)
test_accuracy = multiclass_accuracy(test_pred, test_y)
print('Linear softmax classifier test set accuracy: %f' % (test_accuracy, ))
| assignments/assignment1/Linear classifier.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # DL Deep Dive - Training
# In this section, we will be using Keras (https://keras.io/) to build a TensorFlow-based model for recognizing handwritten digits. This walkthough builds the most basic type of deep nerual network, a mulit-layer perceptron (MLP), consisting of:
# * 1 input layer of 784 neurons
# * 2 hidden layers of 512 neurons each, with randomized dropout
# * 1 output layer of 10 neurons
#
# ## Preamble
# The first step will be to load all of the appropriate Python libraries. Keras provides classes and helper functions for building layered neural networks and loading standard datasets, such as the MNIST dataset of handwritten digits. For this exercise, we will be importing keras, the mnist dataset, and the Dense and Dropout layers, as well as the RMSProp optimizer. The optimizer is the function used to update the gradients during backpropagation (when training actually occurs).
# +
from __future__ import print_function
import keras
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout
from keras.optimizers import RMSprop
# -
# ## Hyperparameter selection
# Next, we will select hyperparameter values, such as the number of output classes (in our case 10, 1 for each digit), the batch size, and the number of epochs.
#
# The **batch size** is the number of images to pass through the network simultaneously. This adds an extra dimension to the matrix of stored activations for each layer, which has dimensions corresponding to the width of the prior dimension, the width of the current dimension, and the batch size.
#
# The **epochs** is the number of times we will repeat the entire training dataset. For neural networks, seeing an image once is not enough. Fine tuning of the weights and bias values is slow, and seeing the same images many times can help to fine-tune the model.
num_classes = 10
batch_size = 128
epochs = 20
# ## Collecting the Training and Testing data
# Next, we will use Keras built-in dataset helper functions to download the training and testing datasets. The MNIST dataset consists of 70,000 images, split into 60,000 training images and 10,000 testing images. We will use the testing images to evaluate the performance of the model after each epoch, but we will **not** allow the network to learn from the testing images.
# +
# the data, split between train and test sets
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train = x_train.reshape(60000, 784)
x_test = x_test.reshape(10000, 784)
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
x_train /= 255
x_test /= 255
print(x_train.shape[0], 'train samples')
print(x_test.shape[0], 'test samples')
# -
# ## Set the labels to binary classification
# Since each category is a yes or no (a digit is not part 3, part 6, for example) we use the to_categorical utility function to specify that each category (num_classes) is a binary classification.
y_train = keras.utils.to_categorical(y_train, num_classes)
y_test = keras.utils.to_categorical(y_test, num_classes)
# ## Build the Neural Network
# Now we will actually build the neural network. As we mentioned above, the architecure we are using is a multi-layer perceptron (MLP). Keras calls perceptron (or fully-connected) layers Dense layers.
#
# Each neruon in the Dense layers will be activated by the Rectified Linear Unit (ReLU) function. The ReLU function is a more computationally efficient variant of other traditional non-linear activation functions, such as hyperbolic tangent (tanh) and sigmoid:
#
# 
#
# We will also use a virtual layer after each Dense layer called Dropout. The Dropout layer will periodically zero out the output of a neuron in the preceeding Dense layer. This adds non-linearity to the network, and allows it to explore the function space for more optimal solutions.
#
# The network layout is:
# * *784 input neruons*, corresponding to an image which is 28 x 28 x 1. MNIST images are grayscale, so they contain only 1 color channel. A color (RGB) image would contain 3 channels.
# * 2 *512 neuron* hidden layers, each followed by a virtual layer of *512 Dropout* neurons
# * *10 output neurons*, corresponding to the 10 different digit classifications (0..10). This layer will be activated by the softmax function in order to provide percentage likelihood that an image falls into a particular category.
# +
model = Sequential()
model.add(Dense(512, activation='relu', input_shape=(784,)))
model.add(Dropout(0.2))
model.add(Dense(512, activation='relu'))
model.add(Dropout(0.2))
model.add(Dense(num_classes, activation='softmax'))
model.summary()
# -
# As we can see from the summary above, the network has 669,706 total parameters, all of which are trainable. The number of parameters for the first dense layer is larger because the number of inputs is larger (784 vs 512).
#
# The number of parameters in a layer is the number of neurons in the previous layer, times the number of neurons in the current layer (the connectivity matrix from the previous layer to the current layer), plus the number of neurons in the current layer (the bias values of the current layer).
#
# So for our example:
# * The number of parameters in hidden layer 1 is $(784 \times 512) + 512 = 401,920$.
# * The number of parameters in hidden layer 2 is $(512 \times 512) + 512 = 262,656$.
# * The number of parameters in the output layer is $(512 \times 10) + 10 = 5,130$.
#
# ## Compiling the Model
# The next step is to compile the model. Compilation requires setting:
# * The loss function to optimize
# * The optimization function to be used (RMSProp in this case)
# * The metrics to report (accuracy in this case)
model.compile(loss='categorical_crossentropy',
optimizer=RMSprop(),
metrics=['accuracy'])
# ## Training the Model (finally!)
# Finally, we can fit the model to the training data. the fit() function will allow us to pass the training data to the model, in batches of size batch_size, for a certain number of epochs. We will also provide the validation data (test data), which is the data for which accuracy will be reported.
#
# As the model trains, it will show the progress of each epoch (because verbose is set to 1). It will report the time required per epoch, the time per step (a step is a single batch of images), the loss and accuracy of the last batch, as well as the validation loss and accuracy for that epoch (since we will run the validation after each epoch).
history = model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(x_test, y_test))
# ## Scoring the final model
# Once the model is trained, we can evaluate its performance by using the evaluate function. The evaluate function takes in the evaluation dataset and provides the accuracy and loss for inference on that dataset. In our case, the evaluation set will be the same as a test set, but in a production environment you would likely want a completely separate dataset for final evaluation vs validation.
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
# ## Let's Predict Some Digits!
# Let's play around with different images in the test dataset and see just how accurate our neural network is. In order to look at one of the images in the dataset, let's first pick an image from the dataset. Just grab any range of length 1 from the x_test set ([0:1] ... [9999:10000]).
image = x_test[400:401]
# ## Display the Image of the Handwritten Digit
# Now that we have set the image we want to predict, let's take a look at the actual image. We will use the image object and reshape it as a 28 x 28 grayscale image. MatPlotLib will allow us to do this with the pyplot class.
import matplotlib.pyplot as plt
pixels = image.reshape((28, 28))
plt.imshow(pixels, cmap='gray')
plt.show()
# ## Make the Prediction
# Now we will see if our neural network accurately predicts this digit. First, we get the prediction using model.predict, then we will display the prediction using a bar chart.
# +
prediction = model.predict(image)
print(prediction[0])
plt.bar(range(10),prediction[0])
plt.show()
# -
# ## Try it Out!
# Feel free to change the number of neurons, the dropout rate, the number of epochs, and other parameters and see if you can build a better digit predictor!
| DL_Deep_Dive-Training-keras_mnist_MLP.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Logistic Regression with a Neural Network mindset
#
# Welcome to your first (required) programming assignment! You will build a logistic regression classifier to recognize cats. This assignment will step you through how to do this with a Neural Network mindset, and so will also hone your intuitions about deep learning.
#
# **Instructions:**
# - Do not use loops (for/while) in your code, unless the instructions explicitly ask you to do so.
#
# **You will learn to:**
# - Build the general architecture of a learning algorithm, including:
# - Initializing parameters
# - Calculating the cost function and its gradient
# - Using an optimization algorithm (gradient descent)
# - Gather all three functions above into a main model function, in the right order.
# ## 1 - Packages ##
#
# First, let's run the cell below to import all the packages that you will need during this assignment.
# - [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.
# - [h5py](http://www.h5py.org) is a common package to interact with a dataset that is stored on an H5 file.
# - [matplotlib](http://matplotlib.org) is a famous library to plot graphs in Python.
# - [PIL](http://www.pythonware.com/products/pil/) and [scipy](https://www.scipy.org/) are used here to test your model with your own picture at the end.
# +
import numpy as np
import matplotlib.pyplot as plt
import h5py
import scipy
from PIL import Image
from skimage.transform import resize
from lr_utils import load_dataset
# %matplotlib inline
# -
# ## 2 - Overview of the Problem set ##
#
# **Problem Statement**: You are given a dataset ("data.h5") containing:
# - a training set of m_train images labeled as cat (y=1) or non-cat (y=0)
# - a test set of m_test images labeled as cat or non-cat
# - each image is of shape (num_px, num_px, 3) where 3 is for the 3 channels (RGB). Thus, each image is square (height = num_px) and (width = num_px).
#
# You will build a simple image-recognition algorithm that can correctly classify pictures as cat or non-cat.
#
# Let's get more familiar with the dataset. Load the data by running the following code.
# Loading the data (cat/non-cat)
train_set_x_orig, train_set_y, test_set_x_orig, test_set_y, classes = load_dataset()
# We added "_orig" at the end of image datasets (train and test) because we are going to preprocess them. After preprocessing, we will end up with train_set_x and test_set_x (the labels train_set_y and test_set_y don't need any preprocessing).
#
# Each line of your train_set_x_orig and test_set_x_orig is an array representing an image. You can visualize an example by running the following code. Feel free also to change the `index` value and re-run to see other images.
# Example of a picture
index = 25
plt.imshow(train_set_x_orig[index])
print ("y = " + str(train_set_y[:, index]) + ", it's a '" + classes[np.squeeze(train_set_y[:, index])].decode("utf-8") + "' picture.")
# Many software bugs in deep learning come from having matrix/vector dimensions that don't fit. If you can keep your matrix/vector dimensions straight you will go a long way toward eliminating many bugs.
#
# **Exercise:** Find the values for:
# - m_train (number of training examples)
# - m_test (number of test examples)
# - num_px (= height = width of a training image)
# Remember that `train_set_x_orig` is a numpy-array of shape (m_train, num_px, num_px, 3). For instance, you can access `m_train` by writing `train_set_x_orig.shape[0]`.
# +
### START CODE HERE ### (≈ 3 lines of code)
m_train = len(train_set_x_orig)
m_test = len(test_set_x_orig)
num_px = train_set_x_orig[0].shape[0]
### END CODE HERE ###
print ("Number of training examples: m_train = " + str(m_train))
print ("Number of testing examples: m_test = " + str(m_test))
print ("Height/Width of each image: num_px = " + str(num_px))
print ("Each image is of size: (" + str(num_px) + ", " + str(num_px) + ", 3)")
print ("train_set_x shape: " + str(train_set_x_orig.shape))
print ("train_set_y shape: " + str(train_set_y.shape))
print ("test_set_x shape: " + str(test_set_x_orig.shape))
print ("test_set_y shape: " + str(test_set_y.shape))
# -
# **Expected Output for m_train, m_test and num_px**:
# <table style="width:15%">
# <tr>
# <td>**m_train**</td>
# <td> 209 </td>
# </tr>
#
# <tr>
# <td>**m_test**</td>
# <td> 50 </td>
# </tr>
#
# <tr>
# <td>**num_px**</td>
# <td> 64 </td>
# </tr>
#
# </table>
#
# For convenience, you should now reshape images of shape (num_px, num_px, 3) in a numpy-array of shape (num_px $*$ num_px $*$ 3, 1). After this, our training (and test) dataset is a numpy-array where each column represents a flattened image. There should be m_train (respectively m_test) columns.
#
# **Exercise:** Reshape the training and test data sets so that images of size (num_px, num_px, 3) are flattened into single vectors of shape (num\_px $*$ num\_px $*$ 3, 1).
#
# A trick when you want to flatten a matrix X of shape (a,b,c,d) to a matrix X_flatten of shape (b$*$c$*$d, a) is to use:
# ```python
# X_flatten = X.reshape(X.shape[0], -1).T # X.T is the transpose of X
# ```
# +
# Reshape the training and test examples
### START CODE HERE ### (≈ 2 lines of code)
image_shape = num_px * num_px * 3
train_set_x_flatten = train_set_x_orig.reshape(train_set_x_orig.shape[0], -1).T
test_set_x_flatten = test_set_x_orig.reshape(test_set_x_orig.shape[0], -1).T
### END CODE HERE ###
print ("train_set_x_flatten shape: " + str(train_set_x_flatten.shape))
print ("train_set_y shape: " + str(train_set_y.shape))
print ("test_set_x_flatten shape: " + str(test_set_x_flatten.shape))
print ("test_set_y shape: " + str(test_set_y.shape))
print ("sanity check after reshaping: " + str(train_set_x_flatten[0:5,0]))
# -
# **Expected Output**:
#
# <table style="width:35%">
# <tr>
# <td>**train_set_x_flatten shape**</td>
# <td> (12288, 209)</td>
# </tr>
# <tr>
# <td>**train_set_y shape**</td>
# <td>(1, 209)</td>
# </tr>
# <tr>
# <td>**test_set_x_flatten shape**</td>
# <td>(12288, 50)</td>
# </tr>
# <tr>
# <td>**test_set_y shape**</td>
# <td>(1, 50)</td>
# </tr>
# <tr>
# <td>**sanity check after reshaping**</td>
# <td>[17 31 56 22 33]</td>
# </tr>
# </table>
# To represent color images, the red, green and blue channels (RGB) must be specified for each pixel, and so the pixel value is actually a vector of three numbers ranging from 0 to 255.
#
# One common preprocessing step in machine learning is to center and standardize your dataset, meaning that you substract the mean of the whole numpy array from each example, and then divide each example by the standard deviation of the whole numpy array. But for picture datasets, it is simpler and more convenient and works almost as well to just divide every row of the dataset by 255 (the maximum value of a pixel channel).
#
# <!-- During the training of your model, you're going to multiply weights and add biases to some initial inputs in order to observe neuron activations. Then you backpropogate with the gradients to train the model. But, it is extremely important for each feature to have a similar range such that our gradients don't explode. You will see that more in detail later in the lectures. !-->
#
# Let's standardize our dataset.
train_set_x = train_set_x_flatten/255.
test_set_x = test_set_x_flatten/255.
# <font color='blue'>
# **What you need to remember:**
#
# Common steps for pre-processing a new dataset are:
# - Figure out the dimensions and shapes of the problem (m_train, m_test, num_px, ...)
# - Reshape the datasets such that each example is now a vector of size (num_px \* num_px \* 3, 1)
# - "Standardize" the data
# ## 3 - General Architecture of the learning algorithm ##
#
# It's time to design a simple algorithm to distinguish cat images from non-cat images.
#
# You will build a Logistic Regression, using a Neural Network mindset. The following Figure explains why **Logistic Regression is actually a very simple Neural Network!**
#
# <img src="images/LogReg_kiank.png" style="width:650px;height:400px;">
#
# **Mathematical expression of the algorithm**:
#
# For one example $x^{(i)}$:
# $$z^{(i)} = w^T x^{(i)} + b \tag{1}$$
# $$\hat{y}^{(i)} = a^{(i)} = sigmoid(z^{(i)})\tag{2}$$
# $$ \mathcal{L}(a^{(i)}, y^{(i)}) = - y^{(i)} \log(a^{(i)}) - (1-y^{(i)} ) \log(1-a^{(i)})\tag{3}$$
#
# The cost is then computed by summing over all training examples:
# $$ J = \frac{1}{m} \sum_{i=1}^m \mathcal{L}(a^{(i)}, y^{(i)})\tag{6}$$
#
# **Key steps**:
# In this exercise, you will carry out the following steps:
# - Initialize the parameters of the model
# - Learn the parameters for the model by minimizing the cost
# - Use the learned parameters to make predictions (on the test set)
# - Analyse the results and conclude
# ## 4 - Building the parts of our algorithm ##
#
# The main steps for building a Neural Network are:
# 1. Define the model structure (such as number of input features)
# 2. Initialize the model's parameters
# 3. Loop:
# - Calculate current loss (forward propagation)
# - Calculate current gradient (backward propagation)
# - Update parameters (gradient descent)
#
# You often build 1-3 separately and integrate them into one function we call `model()`.
#
# ### 4.1 - Helper functions
#
# **Exercise**: Using your code from "Python Basics", implement `sigmoid()`. As you've seen in the figure above, you need to compute $sigmoid( w^T x + b) = \frac{1}{1 + e^{-(w^T x + b)}}$ to make predictions. Use np.exp().
# +
# GRADED FUNCTION: sigmoid
def sigmoid(z):
"""
Compute the sigmoid of z
Arguments:
z -- A scalar or numpy array of any size.
Return:
s -- sigmoid(z)
"""
### START CODE HERE ### (≈ 1 line of code)
s = 1 / (1 + np.exp(-z))
### END CODE HERE ###
return s
# -
print ("sigmoid([0, 2]) = " + str(sigmoid(np.array([0,2]))))
# **Expected Output**:
#
# <table>
# <tr>
# <td>**sigmoid([0, 2])**</td>
# <td> [ 0.5 0.88079708]</td>
# </tr>
# </table>
# ### 4.2 - Initializing parameters
#
# **Exercise:** Implement parameter initialization in the cell below. You have to initialize w as a vector of zeros. If you don't know what numpy function to use, look up np.zeros() in the Numpy library's documentation.
# +
# GRADED FUNCTION: initialize_with_zeros
def initialize_with_zeros(dim):
"""
This function creates a vector of zeros of shape (dim, 1) for w and initializes b to 0.
Argument:
dim -- size of the w vector we want (or number of parameters in this case)
Returns:
w -- initialized vector of shape (dim, 1)
b -- initialized scalar (corresponds to the bias)
"""
### START CODE HERE ### (≈ 1 line of code)
w = np.zeros((dim, 1))
b = 0
### END CODE HERE ###
assert(w.shape == (dim, 1))
assert(isinstance(b, float) or isinstance(b, int))
return w, b
# -
dim = 2
w, b = initialize_with_zeros(dim)
print ("w = " + str(w))
print ("b = " + str(b))
# **Expected Output**:
#
#
# <table style="width:15%">
# <tr>
# <td> ** w ** </td>
# <td> [[ 0.]
# [ 0.]] </td>
# </tr>
# <tr>
# <td> ** b ** </td>
# <td> 0 </td>
# </tr>
# </table>
#
# For image inputs, w will be of shape (num_px $\times$ num_px $\times$ 3, 1).
# ### 4.3 - Forward and Backward propagation
#
# Now that your parameters are initialized, you can do the "forward" and "backward" propagation steps for learning the parameters.
#
# **Exercise:** Implement a function `propagate()` that computes the cost function and its gradient.
#
# **Hints**:
#
# Forward Propagation:
# - You get X
# - You compute $A = \sigma(w^T X + b) = (a^{(1)}, a^{(2)}, ..., a^{(m-1)}, a^{(m)})$
# - You calculate the cost function: $J = -\frac{1}{m}\sum_{i=1}^{m}y^{(i)}\log(a^{(i)})+(1-y^{(i)})\log(1-a^{(i)})$
#
# Here are the two formulas you will be using:
#
# $$ \frac{\partial J}{\partial w} = \frac{1}{m}X(A-Y)^T\tag{7}$$
# $$ \frac{\partial J}{\partial b} = \frac{1}{m} \sum_{i=1}^m (a^{(i)}-y^{(i)})\tag{8}$$
# +
# GRADED FUNCTION: propagate
def propagate(w, b, X, Y):
"""
Implement the cost function and its gradient for the propagation explained above
Arguments:
w -- weights, a numpy array of size (num_px * num_px * 3, 1)
b -- bias, a scalar
X -- data of size (num_px * num_px * 3, number of examples)
Y -- true "label" vector (containing 0 if non-cat, 1 if cat) of size (1, number of examples)
Return:
cost -- negative log-likelihood cost for logistic regression
dw -- gradient of the loss with respect to w, thus same shape as w
db -- gradient of the loss with respect to b, thus same shape as b
Tips:
- Write your code step by step for the propagation. np.log(), np.dot()
"""
m = X.shape[1]
# FORWARD PROPAGATION (FROM X TO COST)
### START CODE HERE ### (≈ 2 lines of code)
# compute activation
A = sigmoid(np.dot(np.transpose(w), X) + b)
# compute cost
cost = -1/m * np.sum(Y * np.log(A) + (1 - Y) * np.log(1 - A))
### END CODE HERE ###
# BACKWARD PROPAGATION (TO FIND GRAD)
### START CODE HERE ### (≈ 2 lines of code)
dw = 1/m * np.dot(X, np.transpose(A - Y))
db = 1/m * np.sum(A - Y)
### END CODE HERE ###
assert(dw.shape == w.shape)
assert(db.dtype == float)
cost = np.squeeze(cost)
assert(cost.shape == ())
grads = {"dw": dw,
"db": db}
return grads, cost
# -
w, b, X, Y = np.array([[1.],[2.]]), 2., np.array([[1.,2.,-1.],[3.,4.,-3.2]]), np.array([[1,0,1]])
grads, cost = propagate(w, b, X, Y)
print ("dw = " + str(grads["dw"]))
print ("db = " + str(grads["db"]))
print ("cost = " + str(cost))
# **Expected Output**:
#
# <table style="width:50%">
# <tr>
# <td> ** dw ** </td>
# <td> [[ 0.99845601]
# [ 2.39507239]]</td>
# </tr>
# <tr>
# <td> ** db ** </td>
# <td> 0.00145557813678 </td>
# </tr>
# <tr>
# <td> ** cost ** </td>
# <td> 5.801545319394553 </td>
# </tr>
#
# </table>
# ### 4.4 - Optimization
# - You have initialized your parameters.
# - You are also able to compute a cost function and its gradient.
# - Now, you want to update the parameters using gradient descent.
#
# **Exercise:** Write down the optimization function. The goal is to learn $w$ and $b$ by minimizing the cost function $J$. For a parameter $\theta$, the update rule is $ \theta = \theta - \alpha \text{ } d\theta$, where $\alpha$ is the learning rate.
# +
# GRADED FUNCTION: optimize
def optimize(w, b, X, Y, num_iterations, learning_rate, print_cost = False):
"""
This function optimizes w and b by running a gradient descent algorithm
Arguments:
w -- weights, a numpy array of size (num_px * num_px * 3, 1)
b -- bias, a scalar
X -- data of shape (num_px * num_px * 3, number of examples)
Y -- true "label" vector (containing 0 if non-cat, 1 if cat), of shape (1, number of examples)
num_iterations -- number of iterations of the optimization loop
learning_rate -- learning rate of the gradient descent update rule
print_cost -- True to print the loss every 100 steps
Returns:
params -- dictionary containing the weights w and bias b
grads -- dictionary containing the gradients of the weights and bias with respect to the cost function
costs -- list of all the costs computed during the optimization, this will be used to plot the learning curve.
Tips:
You basically need to write down two steps and iterate through them:
1) Calculate the cost and the gradient for the current parameters. Use propagate().
2) Update the parameters using gradient descent rule for w and b.
"""
costs = []
for i in range(num_iterations):
# Cost and gradient calculation (≈ 1-4 lines of code)
### START CODE HERE ###
grads, cost = propagate(w, b, X, Y)
### END CODE HERE ###
# Retrieve derivatives from grads
dw = grads["dw"]
db = grads["db"]
# update rule (≈ 2 lines of code)
### START CODE HERE ###
w = w - learning_rate * dw
b = b - learning_rate * db
### END CODE HERE ###
# Record the costs
if i % 100 == 0:
costs.append(cost)
# Print the cost every 100 training iterations
if print_cost and i % 100 == 0:
print ("Cost after iteration %i: %f" %(i, cost))
params = {"w": w,
"b": b}
grads = {"dw": dw,
"db": db}
return params, grads, costs
# +
params, grads, costs = optimize(w, b, X, Y, num_iterations= 100, learning_rate = 0.009, print_cost = False)
print ("w = " + str(params["w"]))
print ("b = " + str(params["b"]))
print ("dw = " + str(grads["dw"]))
print ("db = " + str(grads["db"]))
# -
# **Expected Output**:
#
# <table style="width:40%">
# <tr>
# <td> **w** </td>
# <td>[[ 0.19033591]
# [ 0.12259159]] </td>
# </tr>
#
# <tr>
# <td> **b** </td>
# <td> 1.92535983008 </td>
# </tr>
# <tr>
# <td> **dw** </td>
# <td> [[ 0.67752042]
# [ 1.41625495]] </td>
# </tr>
# <tr>
# <td> **db** </td>
# <td> 0.219194504541 </td>
# </tr>
#
# </table>
# **Exercise:** The previous function will output the learned w and b. We are able to use w and b to predict the labels for a dataset X. Implement the `predict()` function. There are two steps to computing predictions:
#
# 1. Calculate $\hat{Y} = A = \sigma(w^T X + b)$
#
# 2. Convert the entries of a into 0 (if activation <= 0.5) or 1 (if activation > 0.5), stores the predictions in a vector `Y_prediction`. If you wish, you can use an `if`/`else` statement in a `for` loop (though there is also a way to vectorize this).
# +
# GRADED FUNCTION: predict
def predict(w, b, X):
'''
Predict whether the label is 0 or 1 using learned logistic regression parameters (w, b)
Arguments:
w -- weights, a numpy array of size (num_px * num_px * 3, 1)
b -- bias, a scalar
X -- data of size (num_px * num_px * 3, number of examples)
Returns:
Y_prediction -- a numpy array (vector) containing all predictions (0/1) for the examples in X
'''
m = X.shape[1]
Y_prediction = np.zeros((1,m))
w = w.reshape(X.shape[0], 1)
# Compute vector "A" predicting the probabilities of a cat being present in the picture
### START CODE HERE ### (≈ 1 line of code)
A = sigmoid(np.dot(np.transpose(w), X) + b)
### END CODE HERE ###
Y_prediction = np.vectorize(lambda x: 1 if x > 0.5 else 0)(A)
return Y_prediction
# -
w = np.array([[0.1124579],[0.23106775]])
b = -0.3
X = np.array([[1.,-1.1,-3.2],[1.2,2.,0.1]])
print ("predictions = " + str(predict(w, b, X)))
# **Expected Output**:
#
# <table style="width:30%">
# <tr>
# <td>
# **predictions**
# </td>
# <td>
# [[ 1. 1. 0.]]
# </td>
# </tr>
#
# </table>
#
# <font color='blue'>
# **What to remember:**
# You've implemented several functions that:
# - Initialize (w,b)
# - Optimize the loss iteratively to learn parameters (w,b):
# - computing the cost and its gradient
# - updating the parameters using gradient descent
# - Use the learned (w,b) to predict the labels for a given set of examples
# ## 5 - Merge all functions into a model ##
#
# You will now see how the overall model is structured by putting together all the building blocks (functions implemented in the previous parts) together, in the right order.
#
# **Exercise:** Implement the model function. Use the following notation:
# - Y_prediction_test for your predictions on the test set
# - Y_prediction_train for your predictions on the train set
# - w, costs, grads for the outputs of optimize()
# +
# GRADED FUNCTION: model
def model(X_train, Y_train, X_test, Y_test, num_iterations = 2000, learning_rate = 0.5, print_cost = False):
"""
Builds the logistic regression model by calling the function you've implemented previously
Arguments:
X_train -- training set represented by a numpy array of shape (num_px * num_px * 3, m_train)
Y_train -- training labels represented by a numpy array (vector) of shape (1, m_train)
X_test -- test set represented by a numpy array of shape (num_px * num_px * 3, m_test)
Y_test -- test labels represented by a numpy array (vector) of shape (1, m_test)
num_iterations -- hyperparameter representing the number of iterations to optimize the parameters
learning_rate -- hyperparameter representing the learning rate used in the update rule of optimize()
print_cost -- Set to true to print the cost every 100 iterations
Returns:
d -- dictionary containing information about the model.
"""
### START CODE HERE ###
dim = X_train.shape[0]
# initialize parameters with zeros (≈ 1 line of code)
w, b = initialize_with_zeros(dim)
# Gradient descent (≈ 1 line of code)
parameters, grads, costs = optimize(w, b, X_train, Y_train, num_iterations, learning_rate, print_cost)
# Retrieve parameters w and b from dictionary "parameters"
w = parameters["w"]
b = parameters["b"]
# Predict test/train set examples (≈ 2 lines of code)
Y_prediction_test = predict(w, b, X_test)
Y_prediction_train = predict(w, b, X_train)
### END CODE HERE ###
# Print train/test Errors
print("train accuracy: {} %".format(100 - np.mean(np.abs(Y_prediction_train - Y_train)) * 100))
print("test accuracy: {} %".format(100 - np.mean(np.abs(Y_prediction_test - Y_test)) * 100))
d = {"costs": costs,
"Y_prediction_test": Y_prediction_test,
"Y_prediction_train" : Y_prediction_train,
"w" : w,
"b" : b,
"learning_rate" : learning_rate,
"num_iterations": num_iterations}
return d
# -
# Run the following cell to train your model.
d = model(train_set_x, train_set_y, test_set_x, test_set_y, num_iterations = 2000, learning_rate = 0.005, print_cost = True)
# **Expected Output**:
#
# <table style="width:40%">
#
# <tr>
# <td> **Cost after iteration 0 ** </td>
# <td> 0.693147 </td>
# </tr>
# <tr>
# <td> <center> $\vdots$ </center> </td>
# <td> <center> $\vdots$ </center> </td>
# </tr>
# <tr>
# <td> **Train Accuracy** </td>
# <td> 99.04306220095694 % </td>
# </tr>
#
# <tr>
# <td>**Test Accuracy** </td>
# <td> 70.0 % </td>
# </tr>
# </table>
#
#
#
# **Comment**: Training accuracy is close to 100%. This is a good sanity check: your model is working and has high enough capacity to fit the training data. Test error is 68%. It is actually not bad for this simple model, given the small dataset we used and that logistic regression is a linear classifier. But no worries, you'll build an even better classifier next week!
#
# Also, you see that the model is clearly overfitting the training data. Later in this specialization you will learn how to reduce overfitting, for example by using regularization. Using the code below (and changing the `index` variable) you can look at predictions on pictures of the test set.
# Example of a picture that was wrongly classified.
index = 1
plt.imshow(test_set_x[:,index].reshape((num_px, num_px, 3)))
print ("y = " + str(test_set_y[0,index]) + ", you predicted that it is a \"" + classes[d["Y_prediction_test"][0,index]].decode("utf-8") + "\" picture.")
# Let's also plot the cost function and the gradients.
# Plot learning curve (with costs)
costs = np.squeeze(d['costs'])
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (per hundreds)')
plt.title("Learning rate =" + str(d["learning_rate"]))
plt.show()
# **Interpretation**:
# You can see the cost decreasing. It shows that the parameters are being learned. However, you see that you could train the model even more on the training set. Try to increase the number of iterations in the cell above and rerun the cells. You might see that the training set accuracy goes up, but the test set accuracy goes down. This is called overfitting.
# ## 6 - Further analysis (optional/ungraded exercise) ##
#
# Congratulations on building your first image classification model. Let's analyze it further, and examine possible choices for the learning rate $\alpha$.
# #### Choice of learning rate ####
#
# **Reminder**:
# In order for Gradient Descent to work you must choose the learning rate wisely. The learning rate $\alpha$ determines how rapidly we update the parameters. If the learning rate is too large we may "overshoot" the optimal value. Similarly, if it is too small we will need too many iterations to converge to the best values. That's why it is crucial to use a well-tuned learning rate.
#
# Let's compare the learning curve of our model with several choices of learning rates. Run the cell below. This should take about 1 minute. Feel free also to try different values than the three we have initialized the `learning_rates` variable to contain, and see what happens.
# +
learning_rates = [0.01, 0.001, 0.0001]
models = {}
for i in learning_rates:
print ("learning rate is: " + str(i))
models[str(i)] = model(train_set_x, train_set_y, test_set_x, test_set_y, num_iterations = 1500, learning_rate = i, print_cost = False)
print ('\n' + "-------------------------------------------------------" + '\n')
for i in learning_rates:
plt.plot(np.squeeze(models[str(i)]["costs"]), label= str(models[str(i)]["learning_rate"]))
plt.ylabel('cost')
plt.xlabel('iterations (hundreds)')
legend = plt.legend(loc='upper center', shadow=True)
frame = legend.get_frame()
frame.set_facecolor('0.90')
plt.show()
# -
# **Interpretation**:
# - Different learning rates give different costs and thus different predictions results.
# - If the learning rate is too large (0.01), the cost may oscillate up and down. It may even diverge (though in this example, using 0.01 still eventually ends up at a good value for the cost).
# - A lower cost doesn't mean a better model. You have to check if there is possibly overfitting. It happens when the training accuracy is a lot higher than the test accuracy.
# - In deep learning, we usually recommend that you:
# - Choose the learning rate that better minimizes the cost function.
# - If your model overfits, use other techniques to reduce overfitting. (We'll talk about this in later videos.)
#
# ## 7 - Test with your own image (optional/ungraded exercise) ##
#
# Congratulations on finishing this assignment. You can use your own image and see the output of your model. To do that:
# 1. Click on "File" in the upper bar of this notebook, then click "Open" to go on your Coursera Hub.
# 2. Add your image to this Jupyter Notebook's directory, in the "images" folder
# 3. Change your image's name in the following code
# 4. Run the code and check if the algorithm is right (1 = cat, 0 = non-cat)!
# +
## START CODE HERE ## (PUT YOUR IMAGE NAME)
my_image = "la_defense.jpg" # change this to the name of your image file
## END CODE HERE ##
# We preprocess the image to fit your algorithm.
fname = "images/" + my_image
image = np.array(plt.imread(fname))
my_image = resize(image, output_shape=(num_px,num_px), mode='constant').reshape((1, num_px*num_px*3)).T
my_predicted_image = predict(d["w"], d["b"], my_image)
plt.imshow(image)
print("y = " + str(np.squeeze(my_predicted_image)) + ", your algorithm predicts a \"" + classes[int(np.squeeze(my_predicted_image)),].decode("utf-8") + "\" picture.")
# -
# <font color='blue'>
# **What to remember from this assignment:**
# 1. Preprocessing the dataset is important.
# 2. You implemented each function separately: initialize(), propagate(), optimize(). Then you built a model().
# 3. Tuning the learning rate (which is an example of a "hyperparameter") can make a big difference to the algorithm. You will see more examples of this later in this course!
# Finally, if you'd like, we invite you to try different things on this Notebook. Make sure you submit before trying anything. Once you submit, things you can play with include:
# - Play with the learning rate and the number of iterations
# - Try different initialization methods and compare the results
# - Test other preprocessings (center the data, or divide each row by its standard deviation)
# Bibliography:
# - http://www.wildml.com/2015/09/implementing-a-neural-network-from-scratch/
# - https://stats.stackexchange.com/questions/211436/why-do-we-normalize-images-by-subtracting-the-datasets-image-mean-and-not-the-c
| 2. Coursera - Neural Networks and Deep Learning/Week 2/Logistic Regression as a Neural Network/2. Logistic+Regression+with+a+Neural+Network+mindset+v5.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <!-- dom:TITLE: Day 3: Homework 2 -->
# # Day 3: Homework 2
# <!-- dom:AUTHOR: Data Analysis and Machine Learning -->
# <!-- Author: -->
# **Data Analysis and Machine Learning**
#
# Date: **May 22, 2020**
#
# ## Day three exercises
#
#
# ### Exercise 1
#
# This exercise is a continuation of exercise 3 from homework 1. We will
# use the same function to generate our data set, still staying with a
# simple function $y(x)$ which we want to fit using linear regression,
# but now extending the analysis to include the Ridge and the Lasso
# regression methods. You can use the code under the Regression as an example on how to use the Ridge and the Lasso methods, see the [regression slides](https://compphysics.github.io/MachineLearning/doc/pub/Regression/html/Regression-bs.html)).
#
# We will thus again generate our own dataset for a function $y(x)$ where
# $x \in [0,1]$ and defined by random numbers computed with the uniform
# distribution. The function $y$ is a quadratic polynomial in $x$ with
# added stochastic noise according to the normal distribution $\cal{N}(0,1)$.
#
# The following simple Python instructions define our $x$ and $y$ values (with 100 data points).
x = np.random.rand(100,1)
y = 5*x*x+0.1*np.random.randn(100,1)
# 1. (1a) Write your own code for the Ridge method (see chapter 3.4 of Hastie *et al.*, equations (3.43) and (3.44)) and compute the parametrization for different values of $\lambda$. Compare and analyze your results with those from exercise 2. Study the dependence on $\lambda$ while also varying the strength of the noise in your expression for $y(x)$.
#
# 2. (1b) Repeat the above but using the functionality of **Scikit-Learn**. Compare your code with the results from **Scikit-Learn**. Remember to run with the same random numbers for generating $x$ and $y$.
#
# 3. (1c) Our next step is to study the variance of the parameters $\beta_1$ and $\beta_2$ (assuming that we are parameterizing our function with a second-order polynomial). We will use standard linear regression and the Ridge regression. You can now opt for either writing your own function or using **Scikit-Learn** to find the parameters $\beta$. From your results calculate the variance of these paramaters (recall that this is equal to the diagonal elements of the matrix $(\hat{X}^T\hat{X})+\lambda\hat{I})^{-1}$). Discuss the results of these variances as functions of $\lambda$. In particular, try to link your discussion with the discussion in Hastie *et al.* and their figure 3.11.
#
# 4. (1d) Repeat the previous step but add now the Lasso method, see equation (3.53) of Hastie *et al.*. Discuss your results and compare with standard regression and the Ridge regression results. You can write your own code or use the functionality of **scikit-learn**. We recommend the latter since we have not yet discussed how to solve the Lasso equations numerically.
#
# 5. (1e) Finally, using **Scikit-Learn** or your own code, compute also the mean square error, a risk metric corresponding to the expected value of the squared (quadratic) error defined as
# $$
# MSE(\hat{y},\hat{\tilde{y}}) = \frac{1}{n}
# \sum_{i=0}^{n-1}(y_i-\tilde{y}_i)^2,
# $$
# and the $R^2$ score function.
# If $\tilde{\hat{y}}_i$ is the predicted value of the $i-th$ sample and $y_i$ is the corresponding true value, then the score $R^2$ is defined as
# $$
# R^2(\hat{y}, \tilde{\hat{y}}) = 1 - \frac{\sum_{i=0}^{n - 1} (y_i - \tilde{y}_i)^2}{\sum_{i=0}^{n - 1} (y_i - \bar{y})^2},
# $$
# where we have defined the mean value of $\hat{y}$ as
# $$
# \bar{y} = \frac{1}{n} \sum_{i=0}^{n - 1} y_i.
# $$
# Discuss these quantities as functions of the variable $\lambda$ in the Ridge and Lasso regression methods.
#
# ## Exercise 2
#
#
# A much used approach before starting to train the data is to preprocess our
# data. Normally the data may need a rescaling and/or may be sensitive
# to extreme values. Scaling the data renders our inputs much more
# suitable for the algorithms we want to employ.
#
# **Scikit-Learn** has several functions which allow us to rescale the
# data, normally resulting in much better results in terms of various
# accuracy scores. The **StandardScaler** function in **Scikit-Learn**
# ensures that for each feature/predictor we study the mean value is
# zero and the variance is one (every column in the design/feature
# matrix). This scaling has the drawback that it does not ensure that
# we have a particular maximum or minimum in our data set. Another
# function included in **Scikit-Learn** is the **MinMaxScaler** which
# ensures that all features are exactly between $0$ and $1$. The
#
#
# The **Normalizer** scales each data
# point such that the feature vector has a euclidean length of one. In other words, it
# projects a data point on the circle (or sphere in the case of higher dimensions) with a
# radius of 1. This means every data point is scaled by a different number (by the
# inverse of it’s length).
# This normalization is often used when only the direction (or angle) of the data matters,
# not the length of the feature vector.
#
# The **RobustScaler** works similarly to the StandardScaler in that it
# ensures statistical properties for each feature that guarantee that
# they are on the same scale. However, the RobustScaler uses the median
# and quartiles, instead of mean and variance. This makes the
# RobustScaler ignore data points that are very different from the rest
# (like measurement errors). These odd data points are also called
# outliers, and might often lead to trouble for other scaling
# techniques.
#
#
# It also common to split the data in a **training** set and a **testing** set. A typical split is to use $80\%$ of the data for training and the rest
# for testing. This can be done as follows with our design matrix $\boldsymbol{X}$ and data $\boldsymbol{y}$ (remember to import **scikit-learn**)
# split in training and test data
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2)
# Then we can use the stadndard scale to scale our data as
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
# In this exercise we want you to to compute the MSE for the training
# data and the test data as function of the complexity of a polynomial,
# that is the degree of a given polynomial. We want you also to compute the $R2$ score as function of the complexity of the model for both training data and test data. You should also run the calculation with and without scaling.
#
# One of
# the aims is to reproduce Figure 2.11 of [Hastie et al](https://github.com/CompPhysics/MLErasmus/blob/master/doc/Textbooks/elementsstat.pdf).
# We will also use Ridge and Lasso regression.
#
#
# Our data is defined by $x\in [-3,3]$ with a total of for example $100$ data points.
np.random.seed()
n = 100
maxdegree = 14
# Make data set.
x = np.linspace(-3, 3, n).reshape(-1, 1)
y = np.exp(-x**2) + 1.5 * np.exp(-(x-2)**2)+ np.random.normal(0, 0.1, x.shape)
# where $y$ is the function we want to fit with a given polynomial.
#
# 1. (2a) Write a first code which sets up a design matrix $X$ defined by a fifth-order polynomial. Scale your data and split it in training and test data.
#
# 2. (2b) Perform an ordinary least squares and compute the means squared error and the $R2$ factor for the training data and the test data, with and without scaling.
#
# 3. (2c) Add now a model which allows you to make polynomials up to degree $15$. Perform a standard OLS fitting of the training data and compute the MSE and $R2$ for the training and test data and plot both test and training data MSE and $R2$ as functions of the polynomial degree. Compare what you see with Figure 2.11 of Hastie et al. Comment your results. For which polynomial degree do you find an optimal MSE (smallest value)?
#
# 4. (2d) Repeat part (2c) but now using Ridge regressions with various hyperparameters $\lambda$. Make the same plots for the optimal $\lambda$ value for each polynomial degree. Compare these results with those from the standard OLS approach.
#
# ## Example of how to solve the previous exercise
# +
# %matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import Ridge
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import mean_squared_error, r2_score, mean_absolute_error
n = 100
# Make data set.
x = np.linspace(-3, 3, n).reshape(-1, 1)
y = np.exp(-x**2) + 1.5 * np.exp(-(x-2)**2)+ np.random.normal(0, 0.1, x.shape)
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Decide which values of lambda to use
nlambdas = 500
lambdas = np.logspace(-3, 5, nlambdas)
estimated_mse_sklearn = np.zeros(nlambdas)
i = 0
for lmb in lambdas:
clf_ridge = Ridge(alpha=lmb).fit(X_train_scaled, y_train)
yridge = clf_ridge.predict(X_test_scaled)
estimated_mse_sklearn[i] = mean_squared_error(y_test, yridge)
i += 1
plt.figure()
plt.plot(np.log10(lambdas), estimated_mse_sklearn, label = 'Ridge MSE')
plt.xlabel('log10(lambda)')
plt.ylabel('MSE')
plt.legend()
plt.show()
# -
# ## And now with OLS only and Bootstrap
# +
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.utils import resample
from sklearn.linear_model import LinearRegression, Ridge, Lasso
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import mean_squared_error, r2_score, mean_absolute_error
n = 100
n_boostraps = 100
maxdegree = 14
# Make data set.
x = np.linspace(-3, 3, n).reshape(-1, 1)
y = np.exp(-x**2) + 1.5 * np.exp(-(x-2)**2)+ np.random.normal(0, 0.1, x.shape)
error = np.zeros(maxdegree)
bias = np.zeros(maxdegree)
variance = np.zeros(maxdegree)
polydegree = np.zeros(maxdegree)
# Make data set.
x = np.linspace(-3, 3, n).reshape(-1, 1)
y = np.exp(-x**2) + 1.5 * np.exp(-(x-2)**2)+ np.random.normal(0, 0.1, x.shape)
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
for degree in range(maxdegree):
model = make_pipeline(PolynomialFeatures(degree=degree), LinearRegression(fit_intercept=False))
y_pred = np.empty((y_test.shape[0], n_boostraps))
for i in range(n_boostraps):
x_, y_ = resample(X_train_scaled, y_train)
y_pred[:, i] = model.fit(x_, y_).predict(X_test_scaled).ravel()
polydegree[degree] = degree
error[degree] = np.mean( np.mean((y_test - y_pred)**2, axis=1, keepdims=True) )
bias[degree] = np.mean( (y_test - np.mean(y_pred, axis=1, keepdims=True))**2 )
variance[degree] = np.mean( np.var(y_pred, axis=1, keepdims=True) )
print('Polynomial degree:', degree)
print('Error:', error[degree])
print('Bias^2:', bias[degree])
print('Var:', variance[degree])
print('{} >= {} + {} = {}'.format(error[degree], bias[degree], variance[degree], bias[degree]+variance[degree]))
plt.plot(polydegree, error, label='Error')
plt.plot(polydegree, bias, label='bias')
plt.plot(polydegree, variance, label='Variance')
plt.legend()
plt.show()
| doc/ProjectsExercises/2020/hw2/ipynb/.ipynb_checkpoints/hw2-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Introdução à Ciência de Dados - UFPB
# Professor: <NAME>
#
# ## NumPy
# Implemente a função `distance` abaixo. Ela deve receber dois pontos e retornar a distância euclidiana entre eles. Cada ponto é representado por um array do NumPy, por exemplo, `p1 = np.array([2,4])` representa o ponto com coordenadas x=2 e y=2.
# +
import numpy as np
def distance(p1, p2):
"""
>>> distance(np.array([0,0]), np.array([1,1]))
1.4142135623730951
>>> distance(np.array([1,2]), np.array([3,4]))
2.8284271247461903
>>> distance(np.array([5,2]), np.array([-2,-1]))
7.615773105863909
"""
# ADICIONE O SEU CÓDIGO AQUI
return ((p2[0]-p1[0])**2 + (p2[1]-p1[1])**2) ** (1/2) #Fórmula distância entre 2 pontos.
# -
distance(np.array([1,2]), np.array([3,4]))
# Implemente a função `n_distances` abaixo. Ela recebe um ponto (`p1`), como um array do NumPy, e uma lista de pontos (`points`), uma matriz do NumPy, onde cada linha representa um ponto.
#
# Esta função deve retornar a distância de `p1` para todos os pontos da matriz `points`. O retorno também é uma matriz, onde cada linha tem a distância de `p1` para o ponto daquela linha.
def n_distances(p1, points):
"""
>>> n_distances(np.array([0,0]), np.array([[1,1]]))
array([1.41421356])
>>> n_distances(np.array([0,0]), np.array([[1,1], [2,2]]))
array([1.41421356, 2.82842712])
>>> n_distances(np.array([1,2]), np.array([[3,-1], [2,1], [5, 2], [10, 1], [-2, -5]]))
array([3.60555128, 1.41421356, 4. , 9.05538514, 7.61577311])
"""
# ADICIONE O SEU CÓDIGO AQUI
z = np.zeros((points.shape[0])) #Criei um array de 0 de acordo com a quantidade de pontos da matriz points.
for i in range(len(points)):
z[i] = distance(p1,points[i]) #For para preencher o array com os resultados da chamada da primeira função.
return z
n_distances(np.array([0,0]), np.array([[1,1], [2,2]]))
# ## Teste
# +
import doctest
doctest.testmod(verbose=True)
# -
| 03.Dist_2_pontos_NumPy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: U4S1DS10 (Python 3.7)
# language: python
# name: u4-s1-nlp-ds10
# ---
# Lambda School Data Science
#
# *Unit 4, Sprint 3, Module 2*
#
# ---
# # Convolutional Neural Networks (Prepare)
#
# > Convolutional networks are simply neural networks that use convolution in place of general matrix multiplication in at least one of their layers. *Goodfellow, et al.*
# ## Learning Objectives
# - <a href="#p1">Part 1: </a>Describe convolution and pooling
# - <a href="#p2">Part 2: </a>Apply a convolutional neural network to a classification task
# - <a href="#p3">Part 3: </a>Use a pre-trained convolution neural network for image classification
#
# Modern __computer vision__ approaches rely heavily on convolutions as both a dimensionality reduction and feature extraction method. Before we dive into convolutions, let's talk about some of the common computer vision applications:
# * Classification [(Hot Dog or Not Dog)](https://www.youtube.com/watch?v=ACmydtFDTGs)
# * Object Detection [(YOLO)](https://www.youtube.com/watch?v=MPU2HistivI)
# * Pose Estimation [(PoseNet)](https://ai.googleblog.com/2019/08/on-device-real-time-hand-tracking-with.html)
# * Facial Recognition [Emotion Detection](https://www.cbronline.com/wp-content/uploads/2018/05/Mona-lIsa-test-570x300.jpg)
# * and *countless* more
#
# We are going to focus on classification and pre-trained classification today. What are some of the applications of image classification?
from IPython.display import YouTubeVideo
YouTubeVideo('MPU2HistivI', width=600, height=400)
# + [markdown] toc-hr-collapsed=false
# # Convolution & Pooling (Learn)
# <a id="p1"></a>
# + [markdown] toc-hr-collapsed=true
# ## Overview
#
# Like neural networks themselves, CNNs are inspired by biology - specifically, the receptive fields of the visual cortex.
#
# Put roughly, in a real brain the neurons in the visual cortex *specialize* to be receptive to certain regions, shapes, colors, orientations, and other common visual features. In a sense, the very structure of our cognitive system transforms raw visual input, and sends it to neurons that specialize in handling particular subsets of it.
#
# CNNs imitate this approach by applying a convolution. A convolution is an operation on two functions that produces a third function, showing how one function modifies another. Convolutions have a [variety of nice mathematical properties](https://en.wikipedia.org/wiki/Convolution#Properties) - commutativity, associativity, distributivity, and more. Applying a convolution effectively transforms the "shape" of the input.
#
# One common confusion - the term "convolution" is used to refer to both the process of computing the third (joint) function and the process of applying it. In our context, it's more useful to think of it as an application, again loosely analogous to the mapping from visual field to receptive areas of the cortex in a real animal.
# -
from IPython.display import YouTubeVideo
YouTubeVideo('IOHayh06LJ4', width=600, height=400)
# + [markdown] toc-hr-collapsed=false
# ## Follow Along
#
# Let's try to do some convolutions and pooling
# -
# ### Convolution
#
# Consider blurring an image - assume the image is represented as a matrix of numbers, where each number corresponds to the color value of a pixel.
#
# 
#
# *Image Credits from __Hands on Machine Learning with Sckit-Learn, Keras & TensorFlow__*
#
#
# Helpful Terms:
# - __Filter__: The weights (parameters) we will apply to our input image.
# - __Stride__: How the filter moves across the image
# - __Padding__: Zeros (or other values) around the the input image border (kind of like a frame of zeros).
# + colab={"base_uri": "https://localhost:8080/", "height": 243} colab_type="code" id="OsAcbKvoeaqU" outputId="dbb28705-36c7-4691-f7df-e9f82e3ee91e"
import imageio
import matplotlib.pyplot as plt
from skimage import color, io
from skimage.exposure import rescale_intensity
austen = io.imread('https://dl.airtable.com/S1InFmIhQBypHBL0BICi_austen.jpg')
austen_grayscale = rescale_intensity(color.rgb2gray(austen))
austen_grayscale.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 351} colab_type="code" id="KN-ibr_DhyaV" outputId="241716ac-3415-4cfd-9602-0dd59a80ed47"
plt.imshow(austen_grayscale, cmap="gray");
# -
austen_grayscale.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 243} colab_type="code" id="QopB0uo6lNxq" outputId="2364bf3d-8fb9-487a-d2db-eb794939c77a"
import numpy as np
import scipy.ndimage as nd
horizontal_edge_convolution = np.array([[1,1,1,1,1],
[0,0,0,0,0],
[-1,-1,-1,-1,-1]])
vertical_edge_convolution = np.array([[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1],
[1, 0, 0, 0, -1]])
austen_edges = nd.convolve(austen_grayscale, vertical_edge_convolution)#horizontal_edge_convolution)
austen_edges.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 368} colab_type="code" id="-LwEpFW1l-6b" outputId="51b9bdf4-dab6-406a-f98b-fd0a7b480859"
plt.imshow(austen_edges, cmap="gray");
# -
# ### Pooling Layer
#
# 
#
# *Image Credits from __Hands on Machine Learning with Sckit-Learn, Keras & TensorFlow__*
#
# We use Pooling Layers to reduce the dimensionality of the feature maps. We get smaller and smaller feature set by apply convolutions and then pooling layers.
#
# Let's take a look very simple example using Austen's pic.
# +
from skimage.measure import block_reduce
reduced = block_reduce(austen_edges,(2,2), np.max) #austen_grayscale, (2,2), np.max)
plt.imshow(reduced, cmap="gray");
# -
reduced.shape
# ## Challenge
#
# You will be expected to be able to describe convolution.
# # CNNs for Classification (Learn)
# + [markdown] toc-hr-collapsed=true
# ## Overview
# + [markdown] colab_type="text" id="OOep4ugw8coa"
# ### Typical CNN Architecture
#
# 
#
# The first stage of a CNN is, unsurprisingly, a convolution - specifically, a transformation that maps regions of the input image to neurons responsible for receiving them. The convolutional layer can be visualized as follows:
#
# 
#
# The red represents the original input image, and the blue the neurons that correspond.
#
# As shown in the first image, a CNN can have multiple rounds of convolutions, [downsampling](https://en.wikipedia.org/wiki/Downsampling_(signal_processing)) (a digital signal processing technique that effectively reduces the information by passing through a filter), and then eventually a fully connected neural network and output layer. Typical output layers for a CNN would be oriented towards classification or detection problems - e.g. "does this picture contain a cat, a dog, or some other animal?"
#
#
# #### A Convolution in Action
#
# 
#
#
#
# Why are CNNs so popular?
# 1. Compared to prior image learning techniques, they require relatively little image preprocessing (cropping/centering, normalizing, etc.)
# 2. Relatedly, they are *robust* to all sorts of common problems in images (shifts, lighting, etc.)
#
# Actually training a cutting edge image classification CNN is nontrivial computationally - the good news is, with transfer learning, we can get one "off-the-shelf"!
# -
# ## Follow Along
from tensorflow.keras import datasets
from tensorflow.keras.models import Sequential, Model # <- May Use
from tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten
# +
(train_images, train_labels), (test_images, test_labels) = datasets.cifar10.load_data()
# Normalize pixel values to be between 0 and 1
train_images, test_images = train_images / 255.0, test_images / 255.0
# +
class_names = ['airplane', 'automobile', 'bird', 'cat', 'deer',
'dog', 'frog', 'horse', 'ship', 'truck']
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(train_images[i], cmap=plt.cm.binary)
# The CIFAR labels happen to be arrays,
# which is why you need the extra index
plt.xlabel(class_names[train_labels[i][0]])
plt.show()
# -
train_images[0].shape
train_labels[1]
32*32*3
# +
# Setup Architecture
model = Sequential()
model.add(Conv2D(32, (3,3), activation='relu', input_shape=(32,32,3)))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(MaxPooling2D((2,2)))
model.add(Conv2D(64, (3,3), activation='relu'))
model.add(Flatten())
model.add(Dense(64, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.summary()
# +
# Compile Model
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# -
# Fit Model
model.fit(train_images, train_labels, epochs=10, validation_data=(test_images, test_labels))
# +
# Evaluate Model
test_loss, test_acc = model.evaluate(test_images, test_labels, verbose=2)
# -
# ## Challenge
#
# You will apply CNNs to a classification task in the module project.
# # Transfer Learning for Image Classification (Learn)
# + [markdown] toc-hr-collapsed=true
# ## Overview
# + [markdown] colab_type="text" id="ic_wzFnprwXI"
# ### Transfer Learning Repositories
#
# #### TensorFlow Hub
#
# "A library for reusable machine learning modules"
#
# This lets you quickly take advantage of a model that was trained with thousands of GPU hours. It also enables transfer learning - reusing a part of a trained model (called a module) that includes weights and assets, but also training the overall model some yourself with your own data. The advantages are fairly clear - you can use less training data, have faster training, and have a model that generalizes better.
#
# https://www.tensorflow.org/hub/
#
# TensorFlow Hub is very bleeding edge, and while there's a good amount of documentation out there, it's not always updated or consistent. You'll have to use your problem-solving skills if you want to use it!
#
# #### Keras API - Applications
#
# > Keras Applications are deep learning models that are made available alongside pre-trained weights. These models can be used for prediction, feature extraction, and fine-tuning.
#
# There is a decent selection of important benchmark models. We'll focus on an image classifier: ResNet50.
# -
# ## Follow Along
# + colab={"base_uri": "https://localhost:8080/", "height": 34} colab_type="code" id="FM_ApKbGYM9S" outputId="4bfd7ce4-47e5-4320-d1b8-2b20e9f66416"
import numpy as np
from tensorflow.keras.applications.resnet50 import ResNet50
from tensorflow.keras.preprocessing import image
from tensorflow.keras.applications.resnet50 import preprocess_input, decode_predictions
def process_img_path(img_path):
return image.load_img(img_path, target_size=(224, 224))
def img_contains_banana(img):
x = image.img_to_array(img)
x = np.expand_dims(x, axis=0)
x = preprocess_input(x)
model = ResNet50(weights='imagenet')
features = model.predict(x)
results = decode_predictions(features, top=3)[0]
print(results)
for entry in results:
if entry[1] == 'banana':
return entry[2]
return 0.0
# + colab={"base_uri": "https://localhost:8080/", "height": 593} colab_type="code" id="_cQ8ZsJF_Z3B" outputId="02545656-8773-4bb2-9ff5-36d8c658dc00"
import requests
image_urls = ["https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/negative_examples/example11.jpeg",
"https://github.com/LambdaSchool/ML-YouOnlyLookOnce/raw/master/sample_data/positive_examples/example0.jpeg"]
for _id,img in enumerate(image_urls):
r = requests.get(img)
with open(f'example{_id}.jpg', 'wb') as f:
f.write(r.content)
# + colab={"base_uri": "https://localhost:8080/", "height": 417} colab_type="code" id="Gxzkai0q_d-4" outputId="a6bd9b95-9665-4df0-c74d-3d4e876eaf48"
from IPython.display import Image
Image(filename='./example0.jpg', width=600)
# + colab={"base_uri": "https://localhost:8080/", "height": 141} colab_type="code" id="X8NIlClb_n8s" outputId="7c9b9f98-073e-4ab0-a336-e3fc89fa8439"
img_contains_banana(process_img_path('example0.jpg'))
# + colab={"base_uri": "https://localhost:8080/", "height": 417} colab_type="code" id="YIwtRazQ_tQr" outputId="7be6599b-253d-4600-e1f5-ac0ab0f2dfbc"
Image(filename='example1.jpg', width=600)
# + colab={"base_uri": "https://localhost:8080/", "height": 52} colab_type="code" id="GDXwkPWOAB14" outputId="6493a0cb-b57b-43be-8a4e-ac06e51bdada"
img_contains_banana(process_img_path('example1.jpg'))
# + [markdown] colab_type="text" id="CdF5A88oPYvX"
# Notice that, while it gets it right, the confidence for the banana image is fairly low. That's because so much of the image is "not-banana"! How can this be improved? Bounding boxes to center on items of interest.
# -
# ## Challenge
#
# You will be expected to apply a pretrained model to a classificaiton problem today.
# # Review
#
# - <a href="#p1">Part 1: </a>Describe convolution and pooling
# * A Convolution is a function applied to another function to produce a third function
# * Convolutional Kernels are typically 'learned' during the process of training a Convolution Neural Network
# * Pooling is a dimensionality reduction technique that uses either Max or Average of a feature map region to downsample data
# - <a href="#p2">Part 2: </a>Apply a convolutional neural network to a classification task
# * Keras has layers for convolutions :)
# - <a href="#p3">Part 3: </a>Transfer Learning for Image Classification
# * Check out both pretinaed models available in Keras & TensorFlow Hub
# # Sources
#
# - *_Deep Learning_*. Goodfellow *et al.*
# - *Hands-on Machine Learnign with Scikit-Learn, Keras & Tensorflow*
# - [Keras CNN Tutorial](https://www.tensorflow.org/tutorials/images/cnn)
# - [Tensorflow + Keras](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)
# - [Convolution Wiki](https://en.wikipedia.org/wiki/Convolution)
# - [Keras Conv2D: Working with CNN 2D Convolutions in Keras](https://missinglink.ai/guides/keras/keras-conv2d-working-cnn-2d-convolutions-keras/)
# - [Intuitively Understanding Convolutions for Deep Learning](https://towardsdatascience.com/intuitively-understanding-convolutions-for-deep-learning-1f6f42faee1)
# - [A Beginner's Guide to Understanding Convolutional Neural Networks Part 2](https://adeshpande3.github.io/A-Beginner%27s-Guide-To-Understanding-Convolutional-Neural-Networks-Part-2/)
| module2-convolutional-neural-networks/LS_DS_432_Convolutional_Neural_Networks_Lecture.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
#
# The :term:`Events <events>` and :class:`~mne.Annotations` data structures
# =========================================================================
#
# :term:`Events <events>` and :term:`annotations` are quite similar.
# This tutorial highlights their differences and similarities, and tries to shed
# some light on which one is preferred to use in different situations when using
# MNE.
#
# Both events and :class:`~mne.Annotations` can be seen as triplets
# where the first element answers to **when** something happens and the last
# element refers to **what** it is.
# The main difference is that events represent the onset in samples taking into
# account the first sample value
# (:attr:`raw.first_samp <mne.io.Raw.first_samp>`), and the description is
# an integer value.
# In contrast, :class:`~mne.Annotations` represents the
# ``onset`` in seconds (relative to the reference ``orig_time``),
# and the ``description`` is an arbitrary string.
# There is no correspondence between the second element of events and
# :class:`~mne.Annotations`.
# For events, the second element corresponds to the previous value on the
# stimulus channel from which events are extracted. In practice, the second
# element is therefore in most cases zero.
# The second element of :class:`~mne.Annotations` is a float
# indicating its duration in seconds.
#
# See `ex-read-events`
# for a complete example of how to read, select, and visualize **events**;
# and `tut-artifact-rejection` to
# learn how :class:`~mne.Annotations` are used to mark bad segments
# of data.
#
# An example of events and annotations
# ------------------------------------
#
# The following example shows the recorded events in `sample_audvis_raw.fif` and
# marks bad segments due to eye blinks.
#
# +
import os.path as op
import numpy as np
import mne
# Load the data
data_path = mne.datasets.sample.data_path()
fname = op.join(data_path, 'MEG', 'sample', 'sample_audvis_raw.fif')
raw = mne.io.read_raw_fif(fname)
# -
# First we'll create and plot events associated with the experimental paradigm:
#
#
# +
# extract the events array from the stim channel
events = mne.find_events(raw)
# Specify event_id dictionary based on the meaning of experimental triggers
event_id = {'Auditory/Left': 1, 'Auditory/Right': 2,
'Visual/Left': 3, 'Visual/Right': 4,
'smiley': 5, 'button': 32}
color = {1: 'green', 2: 'yellow', 3: 'red', 4: 'c', 5: 'black', 32: 'blue'}
mne.viz.plot_events(events, raw.info['sfreq'], raw.first_samp, color=color,
event_id=event_id)
# -
# Next, we're going to detect eye blinks and turn them into
# :class:`~mne.Annotations`:
#
#
# +
# find blinks
annotated_blink_raw = raw.copy()
eog_events = mne.preprocessing.find_eog_events(raw)
n_blinks = len(eog_events)
# Turn blink events into Annotations of 0.5 seconds duration,
# each centered on the blink event:
onset = eog_events[:, 0] / raw.info['sfreq'] - 0.25
duration = np.repeat(0.5, n_blinks)
description = ['bad blink'] * n_blinks
annot = mne.Annotations(onset, duration, description,
orig_time=raw.info['meas_date'])
annotated_blink_raw.set_annotations(annot)
# plot the annotated raw
annotated_blink_raw.plot()
# -
# Add :term:`annotations` to :term:`raw` objects
# ----------------------------------------------
#
# An important element of :class:`~mne.Annotations` is
# ``orig_time`` which is the time reference for the ``onset``.
# It is key to understand that when calling
# :func:`raw.set_annotations <mne.io.Raw.set_annotations>`, given
# annotations are copied and transformed so that
# :class:`raw.annotations.orig_time <mne.Annotations>`
# matches the recording time of the raw object.
# Refer to the documentation of :class:`~mne.Annotations` to see
# the expected behavior depending on ``meas_date`` and ``orig_time``.
# Where ``meas_date`` is the recording time stored in
# :class:`Info <mne.Info>`.
# You can find more information about :class:`Info <mne.Info>` in
# `tut-info-class`.
#
# We'll now manipulate some simulated annotations.
# The first annotations has ``orig_time`` set to ``None`` while the
# second is set to a chosen POSIX timestamp for illustration purposes.
# Note that both annotations have different ``onset`` values.
#
#
# +
# Create an annotation object with orig_time undefined (default)
annot_none = mne.Annotations(onset=[0, 2, 9], duration=[0.5, 4, 0],
description=['foo', 'bar', 'foo'],
orig_time=None)
print(annot_none)
# Create an annotation object with orig_time
orig_time = '2002-12-03 19:01:31.676071'
annot_orig = mne.Annotations(onset=[22, 24, 31], duration=[0.5, 4, 0],
description=['foo', 'bar', 'foo'],
orig_time=orig_time)
print(annot_orig)
# -
# Now we create two raw objects and set each with different annotations.
# Then we plot both raw objects to compare the annotations.
#
#
# +
# Create two cropped copies of raw with the two previous annotations
raw_a = raw.copy().crop(tmax=12).set_annotations(annot_none)
raw_b = raw.copy().crop(tmax=12).set_annotations(annot_orig)
# Plot the raw objects
raw_a.plot()
raw_b.plot()
# -
# Note that although the ``onset`` values of both annotations were different,
# due to complementary ``orig_time`` they are now identical. This is because
# the first one (``annot_none``), once set in raw, adopted its ``orig_time``.
# The second one (``annot_orig``) already had an ``orig_time``, so its
# ``orig_time`` was changed to match the onset time of the raw. Changing an
# already defined ``orig_time`` of annotations caused its ``onset`` to be
# recalibrated with respect to the new ``orig_time``. As a result both
# annotations have now identical ``onset`` and identical ``orig_time``:
#
#
# +
# Show the annotations in the raw objects
print(raw_a.annotations)
print(raw_b.annotations)
# Show that the onsets are the same
np.set_printoptions(precision=6)
print(raw_a.annotations.onset)
print(raw_b.annotations.onset)
# -
# Notice again that for the case where ``orig_time`` is ``None``,
# it is assumed that the ``orig_time`` is the time of the first sample of data.
#
#
raw_delta = (1 / raw.info['sfreq'])
print('raw.first_sample is {}'.format(raw.first_samp * raw_delta))
print('annot_none.onset[0] is {}'.format(annot_none.onset[0]))
print('raw_a.annotations.onset[0] is {}'.format(raw_a.annotations.onset[0]))
# Valid operations in :class:`mne.Annotations`
# --------------------------------------------
#
# Concatenate
# ~~~~~~~~~~~
#
# It is possible to concatenate two annotations with the + operator (just like
# lists) if both share the same ``orig_time``
#
#
annot = mne.Annotations(onset=[10], duration=[0.5],
description=['foobar'],
orig_time=orig_time)
annot = annot_orig + annot # concatenation
print(annot)
# Iterating, Indexing and Slicing :class:`mne.Annotations`
# ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
#
# :class:`~mne.Annotations` supports iterating, indexing and slicing.
# Iterating over :class:`~mne.Annotations` and indexing with an integer returns
# a dictionary. While slicing returns a new :class:`~mne.Annotations` instance.
#
# See the following examples and usages:
#
#
# difference between indexing and slicing a single element
print(annot[0]) # indexing
print(annot[:1]) # slicing
# How about iterations?
#
#
for key, val in annot[0].items(): # iterate on one element which is dictionary
print(key, val)
for idx, my_annot in enumerate(annot): # iterate on the Annotations object
print('annot #{0}: onset={1}'.format(idx, my_annot['onset']))
print('annot #{0}: duration={1}'.format(idx, my_annot['duration']))
print('annot #{0}: description={1}'.format(idx, my_annot['description']))
for idx, my_annot in enumerate(annot[:1]):
for key, val in my_annot.items():
print('annot #{0}: {1} = {2}'.format(idx, key, val))
# Iterating, indexing and slicing return a copy. This has implications like the
# fact that changes are not kept.
#
#
# +
# this change is not kept
annot[0]['onset'] = 42
print(annot[0])
# this change is kept
annot.onset[0] = 42
print(annot[0])
# -
# Save
# ~~~~
#
# Note that you can also save annotations to disk in FIF format::
#
# >>> annot.save('my-annot.fif')
#
# Or as CSV with onsets in (absolute) ISO timestamps::
#
# >>> annot.save('my-annot.csv')
#
# Or in plain text with onsets relative to ``orig_time``::
#
# >>> annot.save('my-annot.txt')
#
#
#
| stable/_downloads/a663da19dfef335563cab63585276d60/plot_object_annotations.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Wide and Deep on TensorFlow (notebook style)
# Copyright 2016 Google Inc. All Rights Reserved. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
# # Introduction
#
# This notebook uses the tf.learn API in TensorFlow to answer a yes/no question. This is called a binary classification problem: Given census data about a person such as age, gender, education and occupation (the features), we will try to predict whether or not the person earns more than 50,000 dollars a year (the target label).
#
# Given an individual's information our model will output a number between 0 and 1, which can be interpreted as the model's certainty that the individual has an annual income of over 50,000 dollars, (1=True, 0=False)
#
# # Imports and constants
# First we'll import our libraries and set up some strings for column names. We also print out the version of TensorFlow we are running.
# +
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import time
import tensorflow as tf
tf.logging.set_verbosity(tf.logging.ERROR) # Set to INFO for tracking training, default is WARN
from tensorflow.contrib.learn.python.learn.datasets import base
print("Using TensorFlow version %s" % (tf.__version__))
CATEGORICAL_COLUMNS = ["workclass", "education", "marital_status", "occupation",
"relationship", "race", "gender", "native_country"]
# Columns of the input csv file
COLUMNS = ["age", "workclass", "fnlwgt", "education", "education_num", "marital_status",
"occupation", "relationship", "race", "gender", "capital_gain", "capital_loss",
"hours_per_week", "native_country", "income_bracket"]
# Feature columns for input into the model
FEATURE_COLUMNS = ["age", "workclass", "education", "education_num", "marital_status",
"occupation", "relationship", "race", "gender", "capital_gain", "capital_loss",
"hours_per_week", "native_country"]
# -
# # Input file parsing
#
# This section puts the file into a `Reader` which reads from the file one batch at a time.
#
# We set up the Tensors to be a dictionary of features mapping from their string name to the tensor value.
#
# Note that the `_input_fn()` function is wrapped, enabling it to be used for different files.
#
# NOTE: This reads from the input file directly via TensorFlow, rather than using an intermediate tool such as pandas to load the entire dataset into memory first. This is done to enable the system to scale to large inputs.
# ## More about input functions
#
# The input function is how we will feed the input data into the model during training and evaluation.
# The structure that must be returned is a pair, where the first element is a dict of the column names (features) mapped to a tensor of values, and the 2nd element is a tensor of values representing the answers (labels). Recall that a tensor is just a general term for an n-dimensional array.
#
# This could be represented as: `map(column_name => [Tensor of values]) , [Tensor of labels])`
#
# More concretely, for this particular dataset, something like this:
#
# {
# 'age': [ 39, 50, 38, 53, 28, … ],
# 'marital_status': [ 'Married-civ-spouse', 'Never-married', 'Widowed', 'Widowed' … ],
# ...
# 'gender': ['Male', 'Female', 'Male', 'Male', 'Female',, … ],
# } ,
# [ 1, 1, 0, 1, 1, 1, 0, 0, 0, 0, 1, 1]
#
# Additionally, we define which columns of the input data we will treat as categorical vs continuous, using the global `CATEGORICAL_COLUMNS`.
#
# You can try different values for `BATCH_SIZE` to see how they impact your results
# +
BATCH_SIZE = 40
def generate_input_fn(filename, batch_size=BATCH_SIZE):
def _input_fn():
filename_queue = tf.train.string_input_producer([filename])
reader = tf.TextLineReader()
# Reads out batch_size number of lines
key, value = reader.read_up_to(filename_queue, num_records=batch_size)
# record_defaults should match the datatypes of each respective column.
record_defaults = [[0], [" "], [0], [" "], [0],
[" "], [" "], [" "], [" "], [" "],
[0], [0], [0], [" "], [" "]]
# Decode CSV data that was just read out.
columns = tf.decode_csv(
value, record_defaults=record_defaults)
# features is a dictionary that maps from column names to tensors of the data.
# income_bracket is the last column of the data. Note that this is NOT a dict.
all_columns = dict(zip(COLUMNS, columns))
# Save the income_bracket column as our labels
# dict.pop() returns the popped array of income_bracket values
income_bracket = all_columns.pop('income_bracket')
# remove the fnlwgt key, which is not used
all_columns.pop('fnlwgt', 'fnlwgt key not found')
# the remaining columns are our features
features = all_columns
# Sparse categorical features must be represented with an additional dimension.
# There is no additional work needed for the Continuous columns; they are the unaltered columns.
# See docs for tf.SparseTensor for more info
for feature_name in CATEGORICAL_COLUMNS:
# Requires tensorflow >= 0.12
features[feature_name] = tf.expand_dims(features[feature_name], -1)
# Convert ">50K" to 1, and "<=50K" to 0
labels = tf.to_int32(tf.equal(income_bracket, " >50K"))
return features, labels
return _input_fn
print('input function configured')
# -
# # Create Feature Columns
# This section configures the model with the information about the model. There are many parameters here to experiment with to see how they affect the accuracy.
#
# This is the bulk of the time and energy that is often spent on making a machine learning model work, called *feature selection* or *feature engineering*. We choose the features (columns) we will use for training, and apply any additional transformations to them as needed.
# ### Sparse Columns
# First we build the sparse columns.
#
# Use `sparse_column_with_keys()` for columns that we know all possible values for.
#
# Use `sparse_column_with_hash_bucket()` for columns that we want the the library to automatically map values for us.
# +
# Sparse base columns.
gender = tf.contrib.layers.sparse_column_with_keys(column_name="gender",
keys=["female", "male"])
race = tf.contrib.layers.sparse_column_with_keys(column_name="race",
keys=["Amer-Indian-Eskimo",
"Asian-Pac-Islander",
"Black", "Other",
"White"])
education = tf.contrib.layers.sparse_column_with_hash_bucket(
"education", hash_bucket_size=1000)
marital_status = tf.contrib.layers.sparse_column_with_hash_bucket(
"marital_status", hash_bucket_size=100)
relationship = tf.contrib.layers.sparse_column_with_hash_bucket(
"relationship", hash_bucket_size=100)
workclass = tf.contrib.layers.sparse_column_with_hash_bucket(
"workclass", hash_bucket_size=100)
occupation = tf.contrib.layers.sparse_column_with_hash_bucket(
"occupation", hash_bucket_size=1000)
native_country = tf.contrib.layers.sparse_column_with_hash_bucket(
"native_country", hash_bucket_size=1000)
print('Sparse columns configured')
# -
# ### Continuous columns
# Second, configure the real-valued columns using `real_valued_column()`.
# +
# Continuous base columns.
age = tf.contrib.layers.real_valued_column("age")
education_num = tf.contrib.layers.real_valued_column("education_num")
capital_gain = tf.contrib.layers.real_valued_column("capital_gain")
capital_loss = tf.contrib.layers.real_valued_column("capital_loss")
hours_per_week = tf.contrib.layers.real_valued_column("hours_per_week")
print('continuous columns configured')
# -
# ### Transformations
# Now for the interesting stuff. We will employ a couple of techniques to get even more out of the data.
#
# * **bucketizing** turns what would have otherwise been a continuous feature into a categorical one.
# * **feature crossing** allows us to compute a model weight for specific pairings across columns, rather than learning them as independently. This essentially encodes related columns together, for situations where having 2 (or more) columns being certain values is meaningful.
#
# Only categorical features can be crossed. This is one reason why age has been bucketized.
#
# For example, crossing education and occupation would enable the model to learn about:
#
# education="Bachelors" AND occupation="Exec-managerial"
#
# or perhaps
#
# education="Bachelors" AND occupation="Craft-repair"
#
# We do a few combined features (feature crosses) here.
#
# Add your own, based on your intuitions about the dataset, to try to improve on the model!
# +
# Transformations.
age_buckets = tf.contrib.layers.bucketized_column(age,
boundaries=[ 18, 25, 30, 35, 40, 45, 50, 55, 60, 65 ])
education_occupation = tf.contrib.layers.crossed_column([education, occupation],
hash_bucket_size=int(1e4))
age_race_occupation = tf.contrib.layers.crossed_column([age_buckets, race, occupation],
hash_bucket_size=int(1e6))
country_occupation = tf.contrib.layers.crossed_column([native_country, occupation],
hash_bucket_size=int(1e4))
print('Transformations complete')
# -
# ### Group feature columns into 2 objects
#
# The wide columns are the sparse, categorical columns that we specified, as well as our hashed, bucket, and feature crossed columns.
#
# The deep columns are composed of embedded categorical columns along with the continuous real-valued columns. **Column embeddings** transform a sparse, categorical tensor into a low-dimensional and dense real-valued vector. The embedding values are also trained along with the rest of the model. For more information about embeddings, see the TensorFlow tutorial on [Vector Representations Words](https://www.tensorflow.org/tutorials/word2vec/), or [Word Embedding](https://en.wikipedia.org/wiki/Word_embedding) on Wikipedia.
#
# The higher the dimension of the embedding is, the more degrees of freedom the model will have to learn the representations of the features. We are starting with an 8-dimension embedding for simplicity, but later you can come back and increase the dimensionality if you wish.
#
#
# +
# Wide columns and deep columns.
wide_columns = [gender, race, native_country,
education, occupation, workclass,
marital_status, relationship,
age_buckets, education_occupation,
age_race_occupation, country_occupation]
deep_columns = [
tf.contrib.layers.embedding_column(workclass, dimension=8),
tf.contrib.layers.embedding_column(education, dimension=8),
tf.contrib.layers.embedding_column(marital_status, dimension=8),
tf.contrib.layers.embedding_column(gender, dimension=8),
tf.contrib.layers.embedding_column(relationship, dimension=8),
tf.contrib.layers.embedding_column(race, dimension=8),
tf.contrib.layers.embedding_column(native_country, dimension=8),
tf.contrib.layers.embedding_column(occupation, dimension=8),
age,
education_num,
capital_gain,
capital_loss,
hours_per_week,
]
print('wide and deep columns configured')
# -
# # Create the model
#
# You can train either a "wide" model, a "deep" model, or a "wide and deep" model, using the classifiers below. Try each one and see what kind of results you get.
#
# * **Wide**: Linear Classifier
# * **Deep**: Deep Neural Net Classifier
# * **Wide & Deep**: Combined Linear and Deep Classifier
#
# The `hidden_units` or `dnn_hidden_units` argument is to specify the size of each layer of the deep portion of the network. For example, `[12, 20, 15]` would create a network with the first layer of size 12, the second layer of size 20, and a third layer of size 15.
# +
def create_model_dir(model_type):
return 'models/model_' + model_type + '_' + str(int(time.time()))
# If new_model=False, pass in the desired model_dir
def get_model(model_type, new_model=True, model_dir=None):
if new_model or model_dir is None:
model_dir = create_model_dir(model_type) # Comment out this line to continue training a existing model
print("Model directory = %s" % model_dir)
m = None
# Linear Classifier
if model_type == 'WIDE':
m = tf.contrib.learn.LinearClassifier(
model_dir=model_dir,
feature_columns=wide_columns)
# Deep Neural Net Classifier
if model_type == 'DEEP':
m = tf.contrib.learn.DNNClassifier(
model_dir=model_dir,
feature_columns=deep_columns,
hidden_units=[100, 50])
# Combined Linear and Deep Classifier
if model_type == 'WIDE_AND_DEEP':
m = tf.contrib.learn.DNNLinearCombinedClassifier(
model_dir=model_dir,
linear_feature_columns=wide_columns,
dnn_feature_columns=deep_columns,
dnn_hidden_units=[100, 70, 50, 25])
print('estimator built')
return m, model_dir
m, model_dir = get_model(model_type = 'WIDE_AND_DEEP')
# -
# # Fit the model (train it)
#
# Run `fit()` to train the model. You can experiment with the `train_steps` and `BATCH_SIZE` parameters.
#
# This can take some time, depending on the values chosen for `train_steps` and `BATCH_SIZE`.
#
# Our datafile is hosted on Google Cloud Storage; the reader we created at the beginning knows how to read from it.
#
# If you don't want to download a new copy of the dataset each time your script runs, you can download it locally using
#
# gsutil cp gs://cloudml-public/census/data/adult.data.csv .
# gsutil cp gs://cloudml-public/census/data/adult.test.csv .
# +
train_file = "adult.data.csv" # "gs://cloudml-public/census/data/adult.data.csv"
test_file = "adult.test.csv" # "gs://cloudml-public/census/data/adult.test.csv"
train_steps = 1000
m.fit(input_fn=generate_input_fn(train_file, BATCH_SIZE), steps=train_steps)
print('fit done')
# -
# # Evaluate the accuracy of the model
# Let's see how the model did. We will evaluate all the test data.
# +
results = m.evaluate(input_fn=generate_input_fn(test_file), steps=100)
print('evaluate done')
print('Accuracy: %s' % results['accuracy'])
# -
# # Export model
# We can upload our trained model to the Cloud Machine Learning Engine's Prediction Service, which will take care of serving our model and scaling it. The code below exports our trained model to a `saved_model.pb` file and a `variables` folder where the trained weights are stored.
#
# The `export_savedmodel()` function expects a `serving_input_fn()`, which returns the mapping from the data that the Prediction Service passes in to the data that should be fed into the trained TensorFlow prediction graph.
# +
from tensorflow.contrib.learn.python.learn.utils import input_fn_utils
def column_to_dtype(column):
if column in CATEGORICAL_COLUMNS:
return tf.string
else:
return tf.float32
def serving_input_fn():
feature_placeholders = {
column: tf.placeholder(column_to_dtype(column), [None])
for column in FEATURE_COLUMNS
}
# DNNCombinedLinearClassifier expects rank 2 Tensors, but inputs should be
# rank 1, so that we can provide scalars to the server
features = {
key: tf.expand_dims(tensor, -1)
for key, tensor in feature_placeholders.items()
}
return input_fn_utils.InputFnOps(
features, # input into graph
None,
feature_placeholders # tensor input converted from request
)
export_folder = m.export_savedmodel(
export_dir_base = model_dir + '/exports',
input_fn=serving_input_fn
)
print('model exported successfully to {}'.format(export_folder))
# -
# # Conclusions
#
# In this Juypter notebook, we have configured, created, and evaluated a Wide & Deep machine learning model, that combines the powers of a Linear Classifier with a Deep Neural Network, using TensorFlow's tf.learn module.
#
# Upon completing training, you exported the trained classifier to a format suitable for running predictions.
#
# With this working example in your toolbelt, you are ready to explore the wide (and deep) world of machine learning with TensorFlow! Some ideas to help you get going:
# * Change the features we used today. Which columns do you think are correlated and should be crossed? Which ones do you think are just adding noise and could be removed to clean up the model?
# * Swap in an entirely new dataset! There are many dataset available on the web, or use a dataset you possess! Check out https://archive.ics.uci.edu/ml to find your own dataset.
| workshop_sections/wide_n_deep/wide_n_deep_flow.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Vanilla RNNs, GRUs and the `scan` function
# In this notebook, you will learn how to define the forward method for vanilla RNNs and GRUs. Additionally, you will see how to define and use the function `scan` to compute forward propagation for RNNs.
#
# By completing this notebook, you will:
#
# - Be able to define the forward method for vanilla RNNs and GRUs
# - Be able to define the `scan` function to perform forward propagation for RNNs
# - Understand how forward propagation is implemented for RNNs.
import numpy as np
from numpy import random
from time import perf_counter
# An implementation of the `sigmoid` function is provided below so you can use it in this notebook.
def sigmoid(x): # Sigmoid function
return 1.0 / (1.0 + np.exp(-x))
# # Part 1: Forward method for vanilla RNNs and GRUs
# In this part of the notebook, you'll see the implementation of the forward method for a vanilla RNN and you'll implement that same method for a GRU. For this excersice you'll use a set of random weights and variables with the following dimensions:
#
# - Embedding size (`emb`) : 128
# - Hidden state size (`h_dim`) : (16,1)
#
# The weights `w_` and biases `b_` are initialized with dimensions (`h_dim`, `emb + h_dim`) and (`h_dim`, 1). We expect the hidden state `h_t` to be a column vector with size (`h_dim`,1) and the initial hidden state `h_0` is a vector of zeros.
random.seed(10) # Random seed, so your results match ours
emb = 128 # Embedding size
T = 256 # Number of variables in the sequences
h_dim = 16 # Hidden state dimension
h_0 = np.zeros((h_dim, 1)) # Initial hidden state
# Random initialization of weights and biases
w1 = random.standard_normal((h_dim, emb+h_dim))
w2 = random.standard_normal((h_dim, emb+h_dim))
w3 = random.standard_normal((h_dim, emb+h_dim))
b1 = random.standard_normal((h_dim, 1))
b2 = random.standard_normal((h_dim, 1))
b3 = random.standard_normal((h_dim, 1))
X = random.standard_normal((T, emb, 1))
weights = [w1, w2, w3, b1, b2, b3]
# ## 1.1 Forward method for vanilla RNNs
# The vanilla RNN cell is quite straight forward. Its most general structure is presented in the next figure:
#
# <img src="RNN.PNG" width="400"/>
#
# As you saw in the lecture videos, the computations made in a vanilla RNN cell are equivalent to the following equations:
#
# \begin{equation}
# h^{<t>}=g(W_{h}[h^{<t-1>},x^{<t>}] + b_h)
# \label{eq: htRNN}
# \end{equation}
#
# \begin{equation}
# \hat{y}^{<t>}=g(W_{yh}h^{<t>} + b_y)
# \label{eq: ytRNN}
# \end{equation}
#
# where $[h^{<t-1>},x^{<t>}]$ means that $h^{<t-1>}$ and $x^{<t>}$ are concatenated together. In the next cell we provide the implementation of the forward method for a vanilla RNN.
def forward_V_RNN(inputs, weights): # Forward propagation for a a single vanilla RNN cell
x, h_t = inputs
# weights.
wh, _, _, bh, _, _ = weights
# new hidden state
h_t = np.dot(wh, np.concatenate([h_t, x])) + bh
h_t = sigmoid(h_t)
return h_t, h_t
# As you can see, we omitted the computation of $\hat{y}^{<t>}$. This was done for the sake of simplicity, so you can focus on the way that hidden states are updated here and in the GRU cell.
# ## 1.2 Forward method for GRUs
# A GRU cell have more computations than the ones that vanilla RNNs have. You can see this visually in the following diagram:
#
# <img src="GRU.PNG" width="400"/>
#
# As you saw in the lecture videos, GRUs have relevance $\Gamma_r$ and update $\Gamma_u$ gates that control how the hidden state $h^{<t>}$ is updated on every time step. With these gates, GRUs are capable of keeping relevant information in the hidden state even for long sequences. The equations needed for the forward method in GRUs are provided below:
#
# \begin{equation}
# \Gamma_r=\sigma{(W_r[h^{<t-1>}, x^{<t>}]+b_r)}
# \end{equation}
#
# \begin{equation}
# \Gamma_u=\sigma{(W_u[h^{<t-1>}, x^{<t>}]+b_u)}
# \end{equation}
#
# \begin{equation}
# c^{<t>}=\tanh{(W_h[\Gamma_r*h^{<t-1>},x^{<t>}]+b_h)}
# \end{equation}
#
# \begin{equation}
# h^{<t>}=\Gamma_u*c^{<t>}+(1-\Gamma_u)*h^{<t-1>}
# \end{equation}
#
# In the next cell, please implement the forward method for a GRU cell by computing the update `u` and relevance `r` gates, and the candidate hidden state `c`.
def forward_GRU(inputs, weights): # Forward propagation for a single GRU cell
x, h_t = inputs
# weights.
wu, wr, wc, bu, br, bc = weights
# Update gate
### START CODE HERE (1-2 lINES) ###
u = np.dot(wu, np.concatenate([h_t, x])) + bu
u = sigmoid(u)
### END CODE HERE ###
# Relevance gate
### START CODE HERE (1-2 lINES) ###
r = np.dot(wr, np.concatenate([h_t, x])) + br
r = sigmoid(u)
### END CODE HERE ###
# Candidate hidden state
### START CODE HERE (1-2 lINES) ###
c = np.dot(wc, np.concatenate([r * h_t, x])) + bc
c = np.tanh(c)
### END CODE HERE ###
# New Hidden state h_t
h_t = u* c + (1 - u)* h_t
return h_t, h_t
# Run the following cell to check your implementation.
forward_GRU([X[1],h_0], weights)[0]
# Expected output:
# <pre>
# array([[ 9.77779014e-01],
# [-9.97986240e-01],
# [-5.19958083e-01],
# [-9.99999886e-01],
# [-9.99707004e-01],
# [-3.02197037e-04],
# [-9.58733503e-01],
# [ 2.10804828e-02],
# [ 9.77365398e-05],
# [ 9.99833090e-01],
# [ 1.63200940e-08],
# [ 8.51874303e-01],
# [ 5.21399924e-02],
# [ 2.15495959e-02],
# [ 9.99878828e-01],
# [ 9.77165472e-01]])
# </pre>
# # Part 2: Implementation of the `scan` function
# In the lectures you saw how the `scan` function is used for forward propagation in RNNs. It takes as inputs:
#
# - `fn` : the function to be called recurrently (i.e. `forward_GRU`)
# - `elems` : the list of inputs for each time step (`X`)
# - `weights` : the parameters needed to compute `fn`
# - `h_0` : the initial hidden state
#
# `scan` goes through all the elements `x` in `elems`, calls the function `fn` with arguments ([`x`, `h_t`],`weights`), stores the computed hidden state `h_t` and appends the result to a list `ys`. Complete the following cell by calling `fn` with arguments ([`x`, `h_t`],`weights`).
def scan(fn, elems, weights, h_0=None): # Forward propagation for RNNs
h_t = h_0
ys = []
for x in elems:
### START CODE HERE (1 lINE) ###
y, h_t = fn([x, h_t], weights)
### END CODE HERE ###
ys.append(y)
return ys, h_t
# # Part 3: Comparison between vanilla RNNs and GRUs
# You have already seen how forward propagation is computed for vanilla RNNs and GRUs. As a quick recap, you need to have a forward method for the recurrent cell and a function like `scan` to go through all the elements from a sequence using a forward method. You saw that GRUs performed more computations than vanilla RNNs, and you can check that they have 3 times more parameters. In the next two cells, we compute forward propagation for a sequence with 256 time steps (`T`) for an RNN and a GRU with the same hidden state `h_t` size (`h_dim`=16).
# vanilla RNNs
tic = perf_counter()
ys, h_T = scan(forward_V_RNN, X, weights, h_0)
toc = perf_counter()
RNN_time=(toc-tic)*1000
print (f"It took {RNN_time:.2f}ms to run the forward method for the vanilla RNN.")
# GRUs
tic = perf_counter()
ys, h_T = scan(forward_GRU, X, weights, h_0)
toc = perf_counter()
GRU_time=(toc-tic)*1000
print (f"It took {GRU_time:.2f}ms to run the forward method for the GRU.")
# As you were told in the lectures, GRUs take more time to compute (However, sometimes, although a rare occurrence, Vanilla RNNs take more time. Can you figure out what might cause this ?). This means that training and prediction would take more time for a GRU than for a vanilla RNN. However, GRUs allow you to propagate relevant information even for long sequences, so when selecting an architecture for NLP you should assess the tradeoff between computational time and performance.
# <b>Congratulations!</b> Now you know how the forward method is implemented for vanilla RNNs and GRUs, and you know how the scan function provides an abstraction for forward propagation in RNNs.
| Natural Language Processing/Course 3 - Natural Language Processing with Sequence Models/Labs/Week 2/Vanilla RNNs, GRUs and the scan function.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + active=""
# .. _registrikood_userguide:
#
# Registrikood Strings
# ============
# + active=""
# Introduction
# ------------
#
# The function :func:`clean_ee_registrikood() <dataprep.clean.clean_ee_registrikood.clean_ee_registrikood>` cleans a column containing Estonian organisation registration code (Registrikood) strings, and standardizes them in a given format. The function :func:`validate_ee_registrikood() <dataprep.clean.clean_ee_registrikood.validate_ee_registrikood>` validates either a single Registrikood strings, a column of Registrikood strings or a DataFrame of Registrikood strings, returning `True` if the value is valid, and `False` otherwise.
# -
# Registrikood strings can be converted to the following formats via the `output_format` parameter:
#
# * `compact`: only number strings without any seperators or whitespace, like "12345678"
# * `standard`: Registrikood strings with proper whitespace in the proper places. Note that in the case of Registrikood, the compact format is the same as the standard one.
#
# Invalid parsing is handled with the `errors` parameter:
#
# * `coerce` (default): invalid parsing will be set to NaN
# * `ignore`: invalid parsing will return the input
# * `raise`: invalid parsing will raise an exception
#
# The following sections demonstrate the functionality of `clean_ee_registrikood()` and `validate_ee_registrikood()`.
# ### An example dataset containing Registrikood strings
import pandas as pd
import numpy as np
df = pd.DataFrame(
{
"registrikood": [
'12345678',
'12345679',
'BE 428759497',
'BE431150351',
"002 724 334",
"hello",
np.nan,
"NULL",
],
"address": [
"123 Pine Ave.",
"main st",
"1234 west main heights 57033",
"apt 1 789 s maple rd manhattan",
"robie house, 789 north main street",
"1111 S Figueroa St, Los Angeles, CA 90015",
"(staples center) 1111 S Figueroa St, Los Angeles",
"hello",
]
}
)
df
# ## 1. Default `clean_ee_registrikood`
#
# By default, `clean_ee_registrikood` will clean registrikood strings and output them in the standard format with proper separators.
from dataprep.clean import clean_ee_registrikood
clean_ee_registrikood(df, column = "registrikood")
# ## 2. Output formats
# This section demonstrates the output parameter.
# ### `standard` (default)
clean_ee_registrikood(df, column = "registrikood", output_format="standard")
# ### `compact`
clean_ee_registrikood(df, column = "registrikood", output_format="compact")
# ## 3. `inplace` parameter
#
# This deletes the given column from the returned DataFrame.
# A new column containing cleaned Registrikood strings is added with a title in the format `"{original title}_clean"`.
clean_ee_registrikood(df, column="registrikood", inplace=True)
# ## 4. `errors` parameter
# ### `coerce` (default)
clean_ee_registrikood(df, "registrikood", errors="coerce")
# ### `ignore`
clean_ee_registrikood(df, "registrikood", errors="ignore")
# ## 4. `validate_ee_registrikood()`
# `validate_ee_registrikood()` returns `True` when the input is a valid Registrikood. Otherwise it returns `False`.
#
# The input of `validate_ee_registrikood()` can be a string, a Pandas DataSeries, a Dask DataSeries, a Pandas DataFrame and a dask DataFrame.
#
# When the input is a string, a Pandas DataSeries or a Dask DataSeries, user doesn't need to specify a column name to be validated.
#
# When the input is a Pandas DataFrame or a dask DataFrame, user can both specify or not specify a column name to be validated. If user specify the column name, `validate_ee_registrikood()` only returns the validation result for the specified column. If user doesn't specify the column name, `validate_ee_registrikood()` returns the validation result for the whole DataFrame.
from dataprep.clean import validate_ee_registrikood
print(validate_ee_registrikood("12345678"))
print(validate_ee_registrikood("12345679"))
print(validate_ee_registrikood('BE 428759497'))
print(validate_ee_registrikood('BE431150351'))
print(validate_ee_registrikood("004085616"))
print(validate_ee_registrikood("hello"))
print(validate_ee_registrikood(np.nan))
print(validate_ee_registrikood("NULL"))
# ### Series
validate_ee_registrikood(df["registrikood"])
# ### DataFrame + Specify Column
validate_ee_registrikood(df, column="registrikood")
# ### Only DataFrame
validate_ee_registrikood(df)
| docs/source/user_guide/clean/clean_ee_registrikood.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Project 3 - Single Inheritance - Solution
# You are writing an inventory application for a budding tech guy who has a video channel featuring computer builds.
# Basically they have a pool of inventory, (for example 5 x AMD Ryzen 2-2700 CPUs) that they use for builds. When they take a CPU from the pool, they will indicate this using the object that tracks that sepcific type of CPU. They may also purchase additional CPUs, or retire some (because they overclocked it too much and burnt them out!).
# Technically we would want a database to back all this data, but here we're just going to build the classes we'll use while our program is running and not worry about retrieving or saving the state of the inventory.
# The base class is going to be a general `Resource`. This class should provide functionality common to all the actual resources (CPU, GPU, Memory, HDD, SSD) - for this exercise we're only going to implement CPU, HDD and SSD.
# It should provide this at a minimum:
# - `name` : user-friendly name of resource instance (e.g.` Intel Core i9-9900K`)
# - `manufacturer` - resource instance manufacturer (e.g. `Nvidia`)
# - `total` : inventory total (how many are in the inventory pool)
# - `allocated` : number allocated (how many are already in use)
# - a `__str__` representation that just returns the resource name
# - a mode detailed `__repr__` implementation
# - `claim(n)` : method to take n resources from the pool (as long as inventory is available)
# - `freeup(n)` : method to return n resources to the pool (e.g. disassembled some builds)
# - `died(n)` : method to return and permanently remove inventory from the pool (e.g. they broke something) - as long as total available allows it
# - `purchased(n)` - method to add inventory to the pool (e.g. they purchased a new CPU)
# - `category` - computed property that returns a lower case version of the class name
# Next we are going to define child classes for each of CPU, HDD and SDD.
# For the `CPU` class:
# - `cores` (e.g. `8`)
# - `socket` (e.g. `AM4`)
# - `power_watts` (e.g. `94`)
# For the HDD and SDD classes, we're going to create an intermediate class called `Storage` with these additional properties:
# - `capacity_GB` (e.g. `120`)
# The `HDD` class extends `Storage` and has these additional properties:
# - `size` (e.g. ``2.5"``)
# - `rpm` (e.g. `7000`)
# The `SSD` class extends `Storage` and has these additional properties:
# - `interface` (e.g. `PCIe NVMe 3.0 x4`)
# For all your classes, implement a full constructor that can be used to initialize all the properties, some form of validation on numeric types, as well as customized `__repr__` as you see fit.
# For the `total` and `allocated` values in the `Resource` init, think of the arguments there as the **current** total and allocated counts. Those `total` and `allocated` attributes should be private **read-only** properties, but they are modifiable through the various methods such as `claim`, `return`, `died` and `purchased`. Other attributes like `name`, `manufacturer_name`, etc should be read-only.
| dd_1/Part 4/Section 07 - Project 3/Project 3 - Description.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: ibaio_dev39
# language: python
# name: ibaio_dev39
# ---
# # Section 3: Homework Exercises
#
# This material provides some hands-on experience using the methods learned from the third day's material.
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import pandas as pd
import scipy.stats as st
import pymc3 as pm
import theano.tensor as tt
import arviz as az
# ## Exercise: Effects of coaching on SAT scores
#
# This example was taken from Gelman *et al.* (2013):
#
# > A study was performed for the Educational Testing Service to analyze the effects of special coaching programs on test scores. Separate randomized experiments were performed to estimate the effects of coaching programs for the SAT-V (Scholastic Aptitude Test- Verbal) in each of eight high schools. The outcome variable in each study was the score on a special administration of the SAT-V, a standardized multiple choice test administered by the Educational Testing Service and used to help colleges make admissions decisions; the scores can vary between 200 and 800, with mean about 500 and standard deviation about 100. The SAT examinations are designed to be resistant to short-term efforts directed specifically toward improving performance on the test; instead they are designed to reflect knowledge acquired and abilities developed over many years of education. Nevertheless, each of the eight schools in this study considered its short-term coaching program to be successful at increasing SAT scores. Also, there was no prior reason to believe that any of the eight programs was more effective than any other or that some were more similar in effect to each other than to any other.
#
# You are given the estimated coaching effects (`d`) and their sampling variances (`s`). The estimates were obtained by independent experiments, with relatively large sample sizes (over thirty students in each school), so you can assume that they have approximately normal sampling distributions with known variances variances.
#
# Here are the data:
J = 8
d = np.array([28., 8., -3., 7., -1., 1., 18., 12.])
s = np.array([15., 10., 16., 11., 9., 11., 10., 18.])
# Construct an appropriate model for estimating whether coaching effects are positive, using a **centered parameterization**, and then compare the diagnostics for this model to that from an **uncentered parameterization**.
#
# Finally, perform goodness-of-fit diagnostics on the better model.
with pm.Model() as centered_schools:
mu = pm.Normal('mu', mu=0, sigma=5)
tau = pm.HalfCauchy('tau', beta=5)
theta = pm.Normal('theta', mu=mu, sigma=tau, shape=J)
effects = pm.Normal('effects', mu=theta, sigma=s, observed=d)
with centered_schools:
trace_centered = pm.sample(1000, tune=1000)
az.plot_trace(trace_centered, var_names=['mu', 'tau']);
az.plot_energy(trace_centered);
# +
def pairplot_divergence(trace, ax=None, divergence=True, color='C3', divergence_color='C2'):
theta = trace.get_values(varname='theta', combine=True)[:, 0]
logtau = trace.get_values(varname='tau_log__', combine=True)
if not ax:
_, ax = plt.subplots(1, 1, figsize=(10, 5))
ax.plot(theta, logtau, 'o', color=color, alpha=.5)
if divergence:
divergent = trace['diverging']
ax.plot(theta[divergent], logtau[divergent], 'o', color=divergence_color)
ax.set_xlabel('theta[0]')
ax.set_ylabel('log(tau)')
ax.set_title('scatter plot between log(tau) and theta[0]');
return ax
pairplot_divergence(trace_centered);
# -
az.plot_parallel(trace_centered);
with pm.Model() as noncentered_schools:
mu = pm.Normal('mu', mu=0, sigma=5)
tau = pm.HalfCauchy('tau', beta=5)
theta_tilde = pm.Normal('theta_t', mu=0, sigma=1, shape=J)
theta = pm.Deterministic('theta', mu + tau * theta_tilde)
effects = pm.Normal('effects', mu=theta, sigma=s, observed=d)
with noncentered_schools:
trace_noncentered = pm.sample(1000, tune=1000)
az.plot_trace(trace_noncentered, var_names=['mu', 'tau']);
az.plot_energy(trace_noncentered);
pairplot_divergence(trace_noncentered);
| solutions/Section3-Homework.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:ds]
# language: python
# name: conda-env-ds-py
# ---
# # Visualizing datasets larger than memory using Datashader with Dask
#
# ## Datashading a 2.7-billion-point Open Street Map database
#
# Most [datashader](https://github.com/bokeh/datashader) examples use "medium-sized" datasets, because they need to be small enough to be distributed over the internet without racking up huge bandwidth charges for the project maintainers. Even though these datasets can be relatively large (such as the [1-billion point Open Street Map example](https://anaconda.org/jbednar/osm-1billion)), they still fit into memory on a 16GB laptop.
#
# Because Datashader supports [Dask](http://dask.pydata.org) dataframes, it also works well with truly large datasets, much bigger than will fit in any one machine's physical memory. On a single machine, Dask will automatically and efficiently page in the data as needed, and you can also easily distribute the data and computation across multiple machines. Here we illustrate how to work "out of core" on a single machine using a 22GB OSM dataset containing 2.7 billion points.
#
# The data is taken from Open Street Map's (OSM) [bulk GPS point data](https://blog.openstreetmap.org/2012/04/01/bulk-gps-point-data/), and is unfortunately too large to distribute with Datashader (7GB compressed). The data was collected by OSM contributors' GPS devices, and was provided as a CSV file of `latitude,longitude` coordinates. The data was downloaded from their website, extracted, converted to use positions in Web Mercator format using `datashader.utils.lnglat_to_meters()`, and then stored in a [parquet](https://github.com/dask/fastparquet) file for [faster disk access](https://github.com/bokeh/datashader/issues/129#issuecomment-300515690). To run this notebook, you would need to do the same process yourself to obtain `osm.snappy.parq`. Once you have it, you can follow the steps below to load and plot the data.
import dask.dataframe as dd
import dask.diagnostics as diag
import datashader as ds
import datashader.transfer_functions as tf
df = dd.io.parquet.read_parquet('data/osm.snappy.parq')
df.head()
# ### Aggregation
#
# First, we create a canvas to provide pixel-shaped bins in which points can be aggregated, and then aggregate the data to produce a fixed-size aggregate array. This process may take up to a minute, so we provide a progress bar using dask:
# +
bound = 20026376.39
bounds = dict(x_range = (-bound, bound), y_range = (int(-bound*0.4), int(bound*0.6)))
plot_width = 1000
plot_height = int(plot_width*0.5)
cvs = ds.Canvas(plot_width=plot_width, plot_height=plot_height, **bounds)
with diag.ProgressBar(), diag.Profiler() as prof, diag.ResourceProfiler(0.5) as rprof:
agg = cvs.points(df, 'x', 'y', ds.count())
# -
# We can now visualize this data very quickly, ignoring low-count noise as described in the [1-billion point OSM version](https://anaconda.org/jbednar/osm-1billion):
tf.shade(agg.where(agg > 15), cmap=["lightblue", "darkblue"])
# ### Performance Profile
#
# Dask offers some tools to visualize how memory and processing power are being used during these calculations:
from bokeh.io import output_notebook
from bokeh.resources import CDN
output_notebook(CDN, hide_banner=True)
diag.visualize([prof, rprof])
None
# Performance notes:
# - On a 16GB machine, most of the time is spent reading the data from disk (the purple rectangles)
# - Reading time includes not just disk I/O, but decompressing chunks of data
# - The disk reads don't release the [Global Interpreter Lock](https://wiki.python.org/moin/GlobalInterpreterLock) (GIL), and so CPU usage (see second chart above) drops to only one core during those periods.
# - During the aggregation steps (the green rectangles), CPU usage on this machine with 8 hyperthreaded cores (4 full cores) spikes to nearly 800%, because the aggregation function is implemented in parallel.
# - The data takes up 22 GB uncompressed, but only a peak of around 6 GB of physical memory is ever used because the data is paged in as needed.
| examples/osm.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <div style="width:1000 px">
#
# <div style="float:right; width:98 px; height:98px;">
# <img src="https://raw.githubusercontent.com/Unidata/MetPy/master/metpy/plots/_static/unidata_150x150.png" alt="Unidata Logo" style="height: 98px;">
# </div>
#
# <h1>Introduction to MetPy</h1>
# <h3>Unidata Python Workshop</h3>
#
# <div style="clear:both"></div>
# </div>
#
# <hr style="height:2px;">
#
#
# ## Overview:
#
# * **Teaching:** 15 minutes
# * **Exercises:** 15 minutes
#
# ### Questions
# 1. What is MetPy?
# 1. How is MetPy structured?
# 1. How are units handled in MetPy?
#
# ### Objectives
# 1. <a href="#whatis">What is MetPy?</a>
# 1. <a href="#units">Units and MetPy</a>
# 1. <a href="#constants">MetPy Constants</a>
# 1. <a href="#calculations">MetPy Calculations</a>
# <a name="whatis"></a>
# ## What is MetPy?
# MetPy is a modern meteorological toolkit for Python. It is now a maintained project of [Unidata](http://www.unidata.ucar.edu) to serve the academic meteorological community. MetPy consists of three major areas of functionality:
#
# 
#
# ### Plots
# As meteorologists, we have many field specific plots that we make. Some of these, such as the Skew-T Log-p require non-standard axes and are difficult to plot in most plotting software. In MetPy we've baked in a lot of this specialized functionality to help you get your plots made and get back to doing science. We will go over making different kinds of plots during the workshop.
#
#
# ### Calculations
# Meteorology also has a common set of calculations that everyone ends up programming themselves. This is error-prone and a huge duplication of work! MetPy contains a set of well tested calculations that is continually growing in an effort to be at feature parity with other legacy packages such as GEMPAK.
#
# ### File I/O
# Finally, there are a number of odd file formats in the meteorological community. MetPy has incorporated a set of readers to help you deal with file formats that you may encounter during your research.
# <a name="units"></a>
# ## Units and MetPy
# Early in our scientific careers we all learn about the importance of paying attention to units in our calculations. Unit conversions can still get the best of us and have caused more than one major technical disaster, including the crash and complete loss of the $327 million [Mars Climate Orbiter](https://en.wikipedia.org/wiki/Mars_Climate_Orbiter).
#
# In MetPy, we use the [pint](https://pint.readthedocs.io/en/latest/) library and a custom unit registry to help prevent unit mistakes in calculations. That means that every quantity you pass to MetPy should have units attached, just like if you were doing the calculation on paper! Attaching units is easy:
# Import the MetPy unit registry
from metpy.units import units
length = 10.4 * units.inches
width = 20 * units.meters
print(length, width)
# Don't forget that you can use tab completion to see what units are available! Just about every imaginable quantity is there, but if you find one that isn't, we're happy to talk about adding it.
#
# While it may seem like a lot of trouble, let's compute the area of a rectangle defined by our length and width variables above. Without units attached, you'd need to remember to perform a unit conversion before multiplying or you would end up with an area in inch-meters and likely forget about it. With units attached, the units are tracked for you.
area = length * width
print(area)
# That's great, now we have an area, but it is not in a very useful unit still. Units can be converted using the `.to()` method. While you won't see m$^2$ in the units list, we can parse complex/compound units as strings:
area.to('m^2')
# ### Exercise
# * Create a variable named `speed` with a value of 25 knots.
# * Create a variable named `time` with a value of 1 fortnight.
# * Calculate how many furlongs you would travel in `time` at `speed`.
# Your code goes here
# #### Solution
# # %load solutions/distance.py
# ### Temperature
# Temperature units are actually relatively tricky (more like absolutely tricky as you'll see). Temperature is a non-multiplicative unit - they are in a system with a reference point. That means that not only is there a scaling factor, but also an offset. This makes the math and unit book-keeping a little more complex. Imagine adding 10 degrees Celsius to 100 degrees Celsius. Is the answer 110 degrees Celsius or 383.15 degrees Celsius (283.15 K + 373.15 K)? That's why there are delta degrees units in the unit registry for offset units. For more examples and explanation you can watch [MetPy Monday #13](https://www.youtube.com/watch?v=iveJCqxe3Z4).
#
# Let's take a look at how this works and fails:
# We would expect this to fail because we cannot add two offset units (and it does fail as an "Ambiguous operation with offset unit").
#
# <pre style='color:#000000;background:#ffffff;'><span style='color:#008c00; '>10</span> <span style='color:#44aadd; '>*</span> units<span style='color:#808030; '>.</span>degC <span style='color:#44aadd; '>+</span> <span style='color:#008c00; '>5</span> <span style='color:#44aadd; '>*</span> units<span style='color:#808030; '>.</span>degC
# </pre>
#
# On the other hand, we can subtract two offset quantities and get a delta:
10 * units.degC - 5 * units.degC
# We can add a delta to an offset unit as well:
25 * units.degC + 5 * units.delta_degF
# Absolute temperature scales like Kelvin and Rankine do not have an offset and therefore can be used in addition/subtraction without the need for a delta verion of the unit.
273 * units.kelvin + 10 * units.kelvin
273 * units.kelvin - 10 * units.kelvin
# ### Exercise
# A cold front is moving through, decreasing the ambient temperature of 25 degC at a rate of 2.3 degF every 10 minutes. What is the temperature after 1.5 hours?
# +
# Your code goes here
# -
# #### Solution
# # %load solutions/temperature_change.py
# <a href="#top">Top</a>
# <hr style="height:2px;">
# <a name="constants"></a>
# ## MetPy Constants
# Another common place that problems creep into scientific code is the value of constants. Can you reproduce someone else's computations from their paper? Probably not unless you know the value of all of their constants. Was the radius of the earth 6000 km, 6300km, 6371 km, or was it actually latitude dependent?
#
# MetPy has a set of constants that can be easily accessed and make your calculations reproducible. You can view a [full table](https://unidata.github.io/MetPy/latest/api/generated/metpy.constants.html#module-metpy.constants) in the docs, look at the module docstring with `metpy.constants?` or checkout what's available with tab completion.
import metpy.constants as mpconst
mpconst.earth_avg_radius
mpconst.dry_air_molecular_weight
# You may also notice in the table that most constants have a short name as well that can be used:
mpconst.Re
mpconst.Md
# <a href="#top">Top</a>
# <hr style="height:2px;">
# <a name="calculations"></a>
# ## MetPy Calculations
# MetPy also encompasses a set of calculations that are common in meteorology (with the goal of have all of the functionality of legacy software like GEMPAK and more). The [calculations documentation](https://unidata.github.io/MetPy/latest/api/generated/metpy.calc.html) has a complete list of the calculations in MetPy.
#
# We'll scratch the surface and show off a few simple calculations here, but will be using many during the workshop.
import metpy.calc as mpcalc
import numpy as np
# +
# Make some fake data for us to work with
np.random.seed(19990503) # So we all have the same data
u = np.random.randint(0, 15, 10) * units('m/s')
v = np.random.randint(0, 15, 10) * units('m/s')
print(u)
print(v)
# -
# Let's use the `wind_direction` function from MetPy to calculate wind direction from these values. Remember you can look at the docstring or the website for help.
direction = mpcalc.wind_direction(u, v)
print(direction)
# ### Exercise
# * Calculate the wind speed using the `wind_speed` function.
# * Print the wind speed in m/s and mph.
# Your code goes here
# #### Solution
# # %load solutions/wind_speed.py
# As one final demonstration, we will calculation the dewpoint given the temperature and relative humidity:
mpcalc.dewpoint_rh(25 * units.degC, 75 * units.percent)
# <a href="#top">Top</a>
# <hr style="height:2px;">
| metpy/Introduction to MetPy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Simple linear regression
# ## Import the relevant libraries
# +
# For these lessons we will need NumPy, pandas, matplotlib and seaborn
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
# and of course the actual regression (machine learning) module
from sklearn.linear_model import LinearRegression
# -
# ## Load the data
# +
# We start by loading the data
data = pd.read_csv('1.01. Simple linear regression.csv')
# Let's explore the top 5 rows of the df
data.head()
# -
# ## Create the regression
# ### Declare the dependent and independent variables
# +
# There is a single independent variable: 'SAT' called input or feature
x = data['SAT']
# and a single depended variable: 'GPA' called output or target
y = data['GPA']
# -
# Often it is useful to check the shapes of the features
x.shape
y.shape
# +
# In order to feed x to sklearn, it should be a 2D array (a matrix)
# Therefore, we must reshape it
# Note that this will not be needed when we've got more than 1 feature (as the inputs will be a 2D array by default)
# x_matrix = x.values.reshape(84,1)
x_matrix = x.values.reshape(-1,1)
# Check the shape just in case
x_matrix.shape
# -
# ### Regression itself
# Full documentation: https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html
# We start by creating a linear regression object
reg = LinearRegression()
# The whole learning process boils down to fitting the regression
# Note that the first argument is the independent variable, while the second - the dependent (unlike with StatsModels)
reg.fit(x_matrix,y)
# ### R-Sqaured
reg.score(x_matrix,y)
# ### Coefficients
reg.coef_
# ### Intercepts
reg.intercept_
# ### Making Predictions
reg.predict([[1740]])
new_data = pd.DataFrame(data=[1740,1760], columns=['SAT'])
new_data
reg.predict(new_data)
new_data['Predicted GPA'] = reg.predict(new_data)
new_data
# Create a scatter plot
plt.scatter(x_matrix,y)
# Define the regression equation, so we can plot it later
y_hat = reg.coef_*x_matrix + reg.intercept_ #formula from above
# Plot the regression line against the independent variable (SAT)
fig = plt.plot(x_matrix,y_hat, lw=4, c='red', label ='regression line')
# Label the axes
plt.xlabel('SAT', fontsize = 25)
plt.ylabel('GPA', fontsize = 25)
plt.show()
# #### Information on Feature Scaling: https://en.wikipedia.org/wiki/Feature_scaling
# #### Information on L1 and L2 Norm: http://www.chioka.in/differences-between-the-l1-norm-and-the-l2-norm-least-absolute-deviations-and-least-squares/
| Part_5_Advanced_Statistical_Methods_(Machine_Learning)/Linear Regression/Sklearn/sklearn - Simple Linear Regression - GPA Problem.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:notebook] *
# language: python
# name: conda-env-notebook-py
# ---
# # Landsat-8
#
#
# <div class="alert-info">
#
# ### Overview
#
# * **teaching:** 30 minutes
# * **exercises:** 0
# * **questions:**
# * How can I find, anaylize, and visualize Landsat8 satellite imagery for an area of interest using Python?
#
# </div>
#
#
# This notebook will focus on accessing public datasets on AWS for a target area affected by Cyclone Kenneth (2019-04-25). Read more about this event and its impact at the [Humanitarian Open Street Map website](https://tasks.hotosm.org/project/5977). We will use a bounding box we will work with covers the island of Nagazidja, including the captial [city of Moroni](https://en.wikipedia.org/wiki/Moroni,_Comoros) - Union of the Comoros, a sovereign archipelago nation in the Indian Ocean.
#
# We will examine raster images from the [Landsat-8 instrument](https://www.usgs.gov/land-resources/nli/landsat). The Landsat program is the longest-running civilian satellite imagery program, with the first satellite launched in 1972 by the US Geological Survey. Landsat 8 is the latest satellite in this program, and was launched in 2013. Landsat observations are processed into “scenes”, each of which is approximately 183 km x 170 km, with a spatial resolution of 30 meters and a temporal resolution of 16 days. The duration of the landsat program makes it an attractive source of medium-scale imagery for land surface change analyses.
#
# Additional code examples for Landsat-8 can be found in Geohackweek 2018 content: https://geohackweek.github.io/raster/04-workingwithrasters/
# ## Table of contents
#
# 1. [**Sat-search**](#Sat-search)
# 1. [**Holoviz visualization**](#Holoviz)
# 1. [**Rasterio and xarray**](#Rasterio-and-xarray)
# +
# Import libraries
import geopandas as gpd
import pandas as pd
import satsearch
from satstac import Items
import holoviews as hv
import hvplot.xarray
import hvplot.pandas
import geoviews as gv
import ipywidgets
import datetime
from ipywidgets import interact
from IPython.display import display, Image
import json
from cartopy import crs as ccrs
import rasterio
import rasterio.mask
from rasterio.session import AWSSession
import xarray as xr
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
# %matplotlib inline
# -
# Set up our bounding box
bbox = [43.16, -11.32, 43.54, -11.96]
west, north, east, south = bbox
bbox_ctr = [0.5*(north+south), 0.5*(west+east)]
# ## Sat-search
#
# [Sat-search](https://github.com/sat-utils/sat-search) is open-source software designed to easily discover public imagery on AWS. It depends upon metadata called Spatio-Temporal Asset Catalogs [STAC catalogs](https://stacspec.org/) to filter scenes. We will use it to search for Landsat-8 data covering our area of interest
# +
# bbox as a python list is great for use in python, but we can instead save to a more interoperable format (GeoJSON)
# Here is a great website for creating and visualizing geojson on a map: http://geojson.io
aoi = { "type": "Polygon",
"coordinates": [[[west, south], [west, north], [east, north], [east, south], [west, south]]]
}
# pretty print formatting
print(json.dumps(aoi, sort_keys=False, indent=2))
# save to file for future use
with open('aoi-5977.geojson', 'w') as f:
json.dump(aoi, f)
# -
# Load results to pandas geodataframe
# now other packages such as geojson can read this file
gfa = gpd.read_file('aoi-5977.geojson')
gfa
# Get results for bbox and time range
results = satsearch.Search(bbox=bbox, datetime='2019-02-01/2019-06-01')
print('%s items' % results.found())
items = results.items()
print('%s collections:' % len(items._collections))
print(items._collections)
# +
# If you are unfamiliar with one of these satellites, we can look at stored metadata
col = items._collections[1]
print('Title:', col.title)
print('Collection Version:', col.version)
print('Keywords: ', col.keywords)
print('License:', col.license)
print('Providers:', col.providers)
print('Extent', col.extent)
# -
# We can delve deeper to see what kind of metadata is available at the scene level
for key in col.properties:
if key == 'eo:bands':
[print(band) for band in col[key]]
else:
print('%s: %s' % (key, col[key]))
# +
# Search for just tier1 Landsat8 scenes, all dates
properties = ["landsat:tier=T1"]
bbox = (west, south, east, north) #(min lon, min lat, max lon, max lat)
results = satsearch.Search.search(collection='landsat-8-l1',
bbox=bbox,
sort=['<datetime'], #earliest scene first
property=properties)
print('%s items' % results.found())
# -
# Save search results for later or to share with others
items = results.items()
items.save('items-landsat8.json')
items = Items.load('items-landsat8.json')
# # Assets correspond to actual images related to a STAC metadata item
# Use pandas to better display python dictionaries!
pd.DataFrame(items[0].assets).T.reset_index()
# Read results into a geopandas GeoDataFrame
gfl = gpd.read_file('items-landsat8.json')
gfl = gfl.sort_values('datetime').reset_index(drop=True)
print('records:', len(gfl))
gfl.head()
# Hack for neat display of band information
import ast
band_info = pd.DataFrame(ast.literal_eval(gfl.iloc[0]['eo:bands']))
band_info
# +
# Note the cloud_cover column, we can narrow our search by any of these properties
properties.extend(["eo:cloud_cover<10"])
test = satsearch.Search.search(collection='landsat-8-l1',
bbox=bbox,
sort=['<datetime'], #earliest scene first
property=properties)
print('%s items' % test.found())
# -
# Or since we can just use geopandas to filter results
subset = gfl[gfl['eo:cloud_cover'] < 10]
print('%s items' % len(subset))
# ## Holoviz
#
# [Holoviz](https://holoviz.org/) is a set of Python visualization libraries that simplify interactive visualizations of data in a web-browser. We'll use several of these libraries including hvplot and geoviews to visualize both vector data (such as image footprints) and raster data (actual raster values).
#
# <div class="alert-warning">
#
# #### Note
#
# the toolbars on the right and side of these plots. We are using a library called Bokeh that gives interactive widgets to zoom in and pan around on maps.
# </div>
# +
# Plot search AOI and frames on a map using Holoviz Libraries
cols = gfl.loc[:,('id','geometry')]
footprints = cols.hvplot(geo=True, line_color='k', alpha=0.1, title='Landsat 8 T1')
aoi = gfa.hvplot(geo=True, line_color='b', fill_color=None)
tiles = gv.tile_sources.CartoEco.options(width=700, height=500)
labels = gv.tile_sources.StamenLabels.options(level='annotation')
tiles * footprints * aoi * labels
# -
# ## ipywidgets
#
# [ipywidgets](https://ipywidgets.readthedocs.io/en/latest/) provide another convenient approach to custom visualizations. The function below allows us to browse through all the image thumbnails for a group of images (more specifically a specific Landsat8 path and row).
def browse_images(items):
n = len(items)
def view_image(i=0):
item = items[i]
print(f"id={item.id}\tdate={item.datetime}\tcloud%={item['eo:cloud_cover']}")
display(Image(item.asset('thumbnail')['href']))
interact(view_image, i=(0,n-1))
# Custom syntax (additional fields, query strings instead of query dict)
properties = ["eo:row=068",
"eo:column=162",
"landsat:tier=T1"]
results = satsearch.Search.search(collection='landsat-8-l1',
bbox=bbox,
sort=['<datetime'], #earliest scene first
property=properties)
print('%s items' % results.found())
items = results.items()
# May not work on Chrome currently, does work on Safari
browse_images(items)
# ## Rasterio and xarray
#
# To actually load full resolution data from a particular Landsat-8 band we'll use rasterio and xarray libraries.
# These are environmnent variable settings for efficiently reading data on AWS S3
env = rasterio.Env(GDAL_DISABLE_READDIR_ON_OPEN='EMPTY_DIR',
CPL_VSIL_CURL_ALLOWED_EXTENSIONS='TIF',
)
item = items[0]
band = 'red'
url = item.asset(band)['href']
print(url)
with env:
with rasterio.open(url) as src:
print(src.profile) # image metadata
width = src.width
blockx = src.profile['blockxsize']
blocky = src.profile['blockysize']
xchunk = int(width/blockx)*blockx
ychunk = blocky
da = xr.open_rasterio(src, chunks={'band': 1, 'x': xchunk, 'y': ychunk})
da
# Nice dask array visualization
da.data
# This will pull raster data over network. if operating in the same AWS region, should be very fast!
# NOTE: seems there is a bug currently with 'logz' for a log-scale colorbar
img = da.hvplot.image(rasterize=True, logz=True, width=700, height=500, cmap='reds', title=f'{item.id} ({band})')
img
# ### Visualize with on-the-fly reprojection
# Display image in latitute, longitude coordinates instead of EPSG:32638 (UTM 38N)
crs = ccrs.UTM(zone='38N')
img = da.hvplot.image(crs=crs, rasterize=True, width=700, height=500, cmap='reds', alpha=0.8, title=f'{item.id} ({band})') # , logz=True not working
aoi = gfa.hvplot(geo=True, line_color='b', fill_color=None)
img * aoi
# ### Image subsets and crop by shapefile
#
# Often we are only interested in small regions of full images. One of the killer features of cloud-optimized data formats stored on the cloud is that we can efficiently pull subsets of an image rather than the whole thing. Here we'll pull only the pixels within a vector polygon in our area of interest.
#
# <div class="alert-warning">
#
# #### Note
#
# It's up to you to make sure the vector and raster CRS's match!
# </div>
gfa
with rasterio.open(url) as src:
# re-project vector to match raster CRS
print(src.meta)
shape = gfa.to_crs(epsg=src.crs.to_epsg())
out_image, out_transform = rasterio.mask.mask(src, shape.geometry.values, crop=True)
out_meta = src.meta
out_meta.update({
"height": out_image.shape[1],
"width": out_image.shape[2],
"transform": out_transform})
print(out_meta)
# write small image to local Geotiff file
with rasterio.open('subset.tif', 'w', **out_meta) as dst:
dst.write(out_image)
# Plot just the subset
import rasterio.plot
with rasterio.open('subset.tif') as src:
rasterio.plot.show(src, cmap='Reds')
# +
# Excercise 1) Load and visualize the highest-resolution 15m pancromatic band instead of the red band
# Excercise 2) Calculate a band ratio between any two bands
# -
# # Xarray DataArray
#
# The xarray multidimensional data model works well if you want to perform computations on multiple bands for a single image, and to utilize dask for distributed computations
# Use just 30 meter bands for simplicity
bands = band_info.query('gsd == 30').common_name.to_list()
bands
def load_dataarray(item, bands):
''' Load STAC item into an xarray DataSet '''
data_arrays = []
for band in bands:
url = item.asset(band)['href']
da = xr.open_rasterio(url, chunks={'band': 1, 'x': 1024, 'y': 1024})
data_arrays.append(da.assign_coords(band=[band]))
return xr.concat(data_arrays, dim='band')
da = load_dataarray(item, bands)
da
img = da.hvplot.image(groupby='band', rasterize=True, width=700, height=500, alpha=0.8, title=f'{item.id}') # , logz=True not working
img
# # Xarray DataSets
#
# It is arguable better to think of image bands as observational variables rather than a dimension of the dataset. DataSets are meant for storing multiple variables. This data structure is also useful for timeseries of multiple images.
def load_dataset(item, bands):
''' Load STAC item into an xarray DataSet '''
data_arrays = []
for band in bands:
url = item.asset(band)['href']
da = xr.open_rasterio(url, chunks={'band': 1, 'x': 1024, 'y': 1024})
da = da.expand_dims(time=[pd.to_datetime(item.date)])
ds = da.to_dataset(name=band)
data_arrays.append(ds)
ds = xr.combine_by_coords(data_arrays)
return ds
ds = load_dataset(item, bands)
ds
print(ds)
print('Dataset size (Gb): ', ds.nbytes/1e9)
ds['blue'].hvplot.image(rasterize=True, logz=True, width=700, height=500, cmap='blues', title=f'{item.id} (blue)')
# Lazy computation with dask
NDVI = (ds['nir'] - ds['red']) / (ds['nir'] + ds['red'])
NDVI
# Compute and store in local memory
ndvi = NDVI.compute()
ndvi
# Put together a larger dataset
results = satsearch.Search.search(collection='landsat-8-l1',
bbox=bbox,
datetime='2019-08-15/2019-09-30',
sort=['<datetime']) #earliest scene first
print('%s items' % results.found())
items = results.items()
items.save('set.geojson')
gf = gpd.read_file('set.geojson')
gf
# +
# Plot search AOI and frames on a map using Holoviz Libraries
cols = gf.loc[:,('id','geometry')]
footprints = cols.hvplot(geo=True, line_color='k', alpha=0.1, title='Landsat 8 T1')
tiles = gv.tile_sources.CartoEco.options(width=700, height=500)
labels = gv.tile_sources.StamenLabels.options(level='annotation')
tiles * footprints * labels
# -
# NOTE: this is not a very efficient bit of code, but it works
datasets = []
for item in items:
datasets.append(load_dataset(item, bands))
DS = xr.concat(datasets, dim='band')
print('Dataset size (Gb): ', DS.nbytes/1e9)
DS
# +
from dask.distributed import Client
client = Client("tcp://192.168.14.160:39645")
client
# -
DS = DS.assign_coords(band=range(len(datasets)))
DS
bounds = gfa.to_crs(epsg=32638).bounds #32638 UTM 38N #32738 UTM 38S
bounds
print(bounds.minx[0], bounds.maxx[0], bounds.miny[0], bounds.maxy[0])
DS.sel(x=slice(bounds.minx[0], bounds.maxx[0]), y=slice(bounds.miny[0], bounds.maxy[0]))
mosaic = DS.sel(x=slice(bounds.minx[0], bounds.maxx[0]), y=slice(bounds.miny[0], bounds.maxy[0])).mean(dim='band')
# Can change chunks before computing at dask
mosaic.chunk(chunks=dict(time=3,x=1395,y=2368))
mosaic['nir'].hvplot.image(x='x',y='y',groupby='time', rasterize=True, width=700, height=500)
| notebooks/amazon-web-services/landsat8.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="ADKY4re5Kx-5"
# ##### Copyright 2019 The TensorFlow Probability Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# + cellView="form" id="S2AOrHzjK0_L"
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] id="56dF5DnkKx0a"
# # Approximate inference for STS models with non-Gaussian observations
#
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/probability/examples/STS_approximate_inference_for_models_with_non_Gaussian_observations"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/STS_approximate_inference_for_models_with_non_Gaussian_observations.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/STS_approximate_inference_for_models_with_non_Gaussian_observations.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/probability/tensorflow_probability/examples/jupyter_notebooks/STS_approximate_inference_for_models_with_non_Gaussian_observations.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# </table>
# + [markdown] id="laPe5xoS42ob"
# This notebook demonstrates the use of TFP approximate inference tools to incorporate a (non-Gaussian) observation model when fitting and forecasting with structural time series (STS) models. In this example, we'll use a Poisson observation model to work with discrete count data.
# + id="4YJz-JDu0X9E"
import time
import matplotlib.pyplot as plt
import numpy as np
import tensorflow.compat.v2 as tf
import tensorflow_probability as tfp
from tensorflow_probability import bijectors as tfb
from tensorflow_probability import distributions as tfd
tf.enable_v2_behavior()
# + [markdown] id="YagBskFAO34k"
# ## Synthetic Data
#
# First we'll generate some synthetic count data:
# + id="OKgRbodJ4EuU"
num_timesteps = 30
observed_counts = np.round(3 + np.random.lognormal(np.log(np.linspace(
num_timesteps, 5, num=num_timesteps)), 0.20, size=num_timesteps))
observed_counts = observed_counts.astype(np.float32)
plt.plot(observed_counts)
# + [markdown] id="OH2nvBuOxDrd"
# ## Model
#
# We'll specify a simple model with a randomly walking linear trend:
# + id="hSsekKzIwsg6"
def build_model(approximate_unconstrained_rates):
trend = tfp.sts.LocalLinearTrend(
observed_time_series=approximate_unconstrained_rates)
return tfp.sts.Sum([trend],
observed_time_series=approximate_unconstrained_rates)
# + [markdown] id="iY-pH3hQz0Vp"
# Instead of operating on the observed time series, this model will operate on the series of Poisson rate parameters that govern the observations.
#
# Since Poisson rates must be positive, we'll use a bijector to transform the
# real-valued STS model into a distribution over positive values. The `Softplus`
# transformation $y = \log(1 + \exp(x))$ is a natural choice, since it is nearly linear for positive values, but other choices such as `Exp` (which transforms the normal random walk into a lognormal random walk) are also possible.
# + id="Hg_B4tofzxgc"
positive_bijector = tfb.Softplus() # Or tfb.Exp()
# Approximate the unconstrained Poisson rate just to set heuristic priors.
# We could avoid this by passing explicit priors on all model params.
approximate_unconstrained_rates = positive_bijector.inverse(
tf.convert_to_tensor(observed_counts) + 0.01)
sts_model = build_model(approximate_unconstrained_rates)
# + [markdown] id="Pxua5B2wxIMz"
# To use approximate inference for a non-Gaussian observation model,
# we'll encode the STS model as a TFP JointDistribution. The random variables in this joint distribution are the parameters of the STS model, the time series of latent Poisson rates, and the observed counts.
#
# + id="vquh2LxgBjfy"
Root = tfd.JointDistributionCoroutine.Root
def sts_with_poisson_likelihood_model():
# Encode the parameters of the STS model as random variables.
param_vals = []
for param in sts_model.parameters:
param_val = yield Root(param.prior)
param_vals.append(param_val)
# Use the STS model to encode the log- (or inverse-softplus)
# rate of a Poisson.
unconstrained_rate = yield sts_model.make_state_space_model(
num_timesteps, param_vals)
rate = positive_bijector.forward(unconstrained_rate[..., 0])
observed_counts = yield tfd.Independent(tfd.Poisson(rate),
reinterpreted_batch_ndims=1)
model = tfd.JointDistributionCoroutine(sts_with_poisson_likelihood_model)
# + [markdown] id="R-3amgmKhYn1"
# ### Preparation for inference
#
# We want to infer the unobserved quantities in the model, given the observed counts. First, we condition the joint log density on the observed counts.
# + id="rSj7blvWh1w8"
# Condition a joint log-prob on the observed counts.
target_log_prob_fn = lambda *args: model.log_prob(args + (observed_counts,))
# + [markdown] id="RFeZ7NYt1qnw"
# HMC and VI inference also like to operate over unconstrained real-valued spaces, so we'll construct the list of bijectors that constrains each of the parameters to their respective supports.
# + id="Dyhb06i41qIg"
constraining_bijectors = ([param.bijector for param in sts_model.parameters] +
# `unconstrained_rate` is already unconstrained, but
# we can speed up inference by rescaling it.
[tfb.Scale(positive_bijector.inverse(
np.float32(np.max(observed_counts / 5.))))])
# + [markdown] id="25nJYyx-nW2T"
# ## Inference with HMC
#
# We'll use HMC (specifically, NUTS) to sample from the joint posterior over model parameters and latent rates.
#
# This will be significantly slower than fitting a standard STS model with HMC, since in addition to the model's (relatively small number of) parameters we also have to infer the entire series of Poisson rates. So we'll run for a relatively small number of steps; for applications where inference quality is critical it might make sense to increase these values or to run multiple chains.
# + id="NMPlVBk6PcpT"
#@title Sampler configuration
# Allow external control of sampling to reduce test runtimes.
num_results = 100 # @param { isTemplate: true}
num_results = int(num_results)
num_burnin_steps = 50 # @param { isTemplate: true}
num_burnin_steps = int(num_burnin_steps)
# + [markdown] id="mhSe-GFDPg9o"
# First we specify a sampler, and then use `sample_chain` to run that sampling
# kernel to produce samples.
# + id="15ue-mBGdcmh"
sampler = tfp.mcmc.TransformedTransitionKernel(
tfp.mcmc.NoUTurnSampler(
target_log_prob_fn=target_log_prob_fn,
step_size=0.1),
bijector=constraining_bijectors)
adaptive_sampler = tfp.mcmc.DualAveragingStepSizeAdaptation(
inner_kernel=sampler,
num_adaptation_steps=int(0.8 * num_burnin_steps),
target_accept_prob=0.75,
# NUTS inside of a TTK requires custom getter/setter functions.
step_size_setter_fn=lambda pkr, new_step_size: pkr._replace(
inner_results=pkr.inner_results._replace(step_size=new_step_size)
),
step_size_getter_fn=lambda pkr: pkr.inner_results.step_size,
log_accept_prob_getter_fn=lambda pkr: pkr.inner_results.log_accept_ratio,
)
initial_state = [b.forward(tf.random.normal(part_shape))
for (b, part_shape) in zip(
constraining_bijectors, model.event_shape[:-1])]
# + id="jvriVTPlih3B"
# Speed up sampling by tracing with `tf.function`.
@tf.function(autograph=False, experimental_compile=True)
def do_sampling():
return tfp.mcmc.sample_chain(
kernel=adaptive_sampler,
current_state=initial_state,
num_results=num_results,
num_burnin_steps=num_burnin_steps)
t0 = time.time()
samples, kernel_results = do_sampling()
t1 = time.time()
print("Inference ran in {:.2f}s.".format(t1-t0))
# + [markdown] id="FwE0yWm_2_kE"
# We can sanity-check the inference by examining the parameter traces. In this case they appear to have explored multiple explanations for the data, which is good, although more samples would be helpful to judge how well the chain is mixing.
# + id="LPOVTbboAtGr"
f = plt.figure(figsize=(12, 4))
for i, param in enumerate(sts_model.parameters):
ax = f.add_subplot(1, len(sts_model.parameters), i + 1)
ax.plot(samples[i])
ax.set_title("{} samples".format(param.name))
# + [markdown] id="tZOydxU53oE9"
# Now for the payoff: let's see the posterior over Poisson rates! We'll also plot the 80% predictive interval over observed counts, and can check that this interval appears to contain about 80% of the counts we actually observed.
# + id="56rIH8MCeU9F"
param_samples = samples[:-1]
unconstrained_rate_samples = samples[-1][..., 0]
rate_samples = positive_bijector.forward(unconstrained_rate_samples)
plt.figure(figsize=(10, 4))
mean_lower, mean_upper = np.percentile(rate_samples, [10, 90], axis=0)
pred_lower, pred_upper = np.percentile(np.random.poisson(rate_samples),
[10, 90], axis=0)
_ = plt.plot(observed_counts, color="blue", ls='--', marker='o', label='observed', alpha=0.7)
_ = plt.plot(np.mean(rate_samples, axis=0), label='rate', color="green", ls='dashed', lw=2, alpha=0.7)
_ = plt.fill_between(np.arange(0, 30), mean_lower, mean_upper, color='green', alpha=0.2)
_ = plt.fill_between(np.arange(0, 30), pred_lower, pred_upper, color='grey', label='counts', alpha=0.2)
plt.xlabel("Day")
plt.ylabel("Daily Sample Size")
plt.title("Posterior Mean")
plt.legend()
# + [markdown] id="GuBYar27YZf6"
# ## Forecasting
#
# To forecast the observed counts, we'll use the standard STS tools to build a forecast distribution over the latent rates (in unconstrained space, again since STS is designed to model real-valued data), then pass the sampled forecasts through a Poisson observation model:
# + id="v1HuVuk6Qocm"
def sample_forecasted_counts(sts_model, posterior_latent_rates,
posterior_params, num_steps_forecast,
num_sampled_forecasts):
# Forecast the future latent unconstrained rates, given the inferred latent
# unconstrained rates and parameters.
unconstrained_rates_forecast_dist = tfp.sts.forecast(sts_model,
observed_time_series=unconstrained_rate_samples,
parameter_samples=posterior_params,
num_steps_forecast=num_steps_forecast)
# Transform the forecast to positive-valued Poisson rates.
rates_forecast_dist = tfd.TransformedDistribution(
unconstrained_rates_forecast_dist,
positive_bijector)
# Sample from the forecast model following the chain rule:
# P(counts) = P(counts | latent_rates)P(latent_rates)
sampled_latent_rates = rates_forecast_dist.sample(num_sampled_forecasts)
sampled_forecast_counts = tfd.Poisson(rate=sampled_latent_rates).sample()
return sampled_forecast_counts, sampled_latent_rates
forecast_samples, rate_samples = sample_forecasted_counts(
sts_model,
posterior_latent_rates=unconstrained_rate_samples,
posterior_params=param_samples,
# Days to forecast:
num_steps_forecast=30,
num_sampled_forecasts=100)
# + id="MyPFQzV8SOSs"
forecast_samples = np.squeeze(forecast_samples)
# + id="iD_kLwF1V3m-"
def plot_forecast_helper(data, forecast_samples, CI=90):
"""Plot the observed time series alongside the forecast."""
plt.figure(figsize=(10, 4))
forecast_median = np.median(forecast_samples, axis=0)
num_steps = len(data)
num_steps_forecast = forecast_median.shape[-1]
plt.plot(np.arange(num_steps), data, lw=2, color='blue', linestyle='--', marker='o',
label='Observed Data', alpha=0.7)
forecast_steps = np.arange(num_steps, num_steps+num_steps_forecast)
CI_interval = [(100 - CI)/2, 100 - (100 - CI)/2]
lower, upper = np.percentile(forecast_samples, CI_interval, axis=0)
plt.plot(forecast_steps, forecast_median, lw=2, ls='--', marker='o', color='orange',
label=str(CI) + '% Forecast Interval', alpha=0.7)
plt.fill_between(forecast_steps,
lower,
upper, color='orange', alpha=0.2)
plt.xlim([0, num_steps+num_steps_forecast])
ymin, ymax = min(np.min(forecast_samples), np.min(data)), max(np.max(forecast_samples), np.max(data))
yrange = ymax-ymin
plt.title("{}".format('Observed time series with ' + str(num_steps_forecast) + ' Day Forecast'))
plt.xlabel('Day')
plt.ylabel('Daily Sample Size')
plt.legend()
# + id="IyUp4NnzWOcs"
plot_forecast_helper(observed_counts, forecast_samples, CI=80)
# + [markdown] id="QmS-ybPM903-"
# ## VI inference
#
# Variational inference can be problematic when inferring a full time series, like our approximate counts (as opposed to just
# the *parameters* of a time series, as in standard STS models). The standard assumption that variables have independent posteriors is quite wrong, since each timestep is correlated with its neighbors, which can lead to underestimating uncertainty. For this reason, HMC may be a better choice for approximate inference over full time series. However, VI can be quite a bit faster, and may be useful for model prototyping or in cases where its performance can be empirically shown to be 'good enough'.
#
# To fit our model with VI, we simply build and optimize a surrogate posterior:
# + id="7aZQEnTThgMT"
surrogate_posterior = tfp.experimental.vi.build_factored_surrogate_posterior(
event_shape=model.event_shape[:-1], # Infer everything but the observed counts.
constraining_bijectors=constraining_bijectors)
# + id="65cf0_EiimGq"
# Allow external control of optimization to reduce test runtimes.
num_variational_steps = 200 # @param { isTemplate: true}
num_variational_steps = int(num_variational_steps)
t0 = time.time()
losses = tfp.vi.fit_surrogate_posterior(target_log_prob_fn,
surrogate_posterior,
optimizer=tf.optimizers.Adam(0.1),
num_steps=num_variational_steps)
t1 = time.time()
print("Inference ran in {:.2f}s.".format(t1-t0))
# + id="zX8WtcLmk2mj"
plt.plot(losses)
plt.title("Variational loss")
_ = plt.xlabel("Steps")
# + id="kQoUExeBkpC0"
posterior_samples = surrogate_posterior.sample(50)
param_samples = posterior_samples[:-1]
unconstrained_rate_samples = posterior_samples[-1][..., 0]
rate_samples = positive_bijector.forward(unconstrained_rate_samples)
plt.figure(figsize=(10, 4))
mean_lower, mean_upper = np.percentile(rate_samples, [10, 90], axis=0)
pred_lower, pred_upper = np.percentile(
np.random.poisson(rate_samples), [10, 90], axis=0)
_ = plt.plot(observed_counts, color='blue', ls='--', marker='o',
label='observed', alpha=0.7)
_ = plt.plot(np.mean(rate_samples, axis=0), label='rate', color='green',
ls='dashed', lw=2, alpha=0.7)
_ = plt.fill_between(
np.arange(0, 30), mean_lower, mean_upper, color='green', alpha=0.2)
_ = plt.fill_between(np.arange(0, 30), pred_lower, pred_upper, color='grey',
label='counts', alpha=0.2)
plt.xlabel('Day')
plt.ylabel('Daily Sample Size')
plt.title('Posterior Mean')
plt.legend()
# + id="0aoMoQyf_fWC"
forecast_samples, rate_samples = sample_forecasted_counts(
sts_model,
posterior_latent_rates=unconstrained_rate_samples,
posterior_params=param_samples,
# Days to forecast:
num_steps_forecast=30,
num_sampled_forecasts=100)
# + id="eQ7zJpEr_hHU"
forecast_samples = np.squeeze(forecast_samples)
# + id="lcEpkAEi_jcn"
plot_forecast_helper(observed_counts, forecast_samples, CI=80)
| site/en-snapshot/probability/examples/STS_approximate_inference_for_models_with_non_Gaussian_observations.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Sample CNN for MNIST handwriting recognition
import tensorflow as tf
mnist = tf.keras.datasets.mnist
(train_images, train_labels), (test_images, test_labels) = mnist.load_data()
# Loand and process MNIST data
# reshape and rescale data for the CNN
train_images = train_images.reshape(60000, 28, 28, 1)
test_images = test_images.reshape(10000, 28, 28, 1)
train_images, test_images = train_images/255, test_images/255
# Create LeNet-5 CNN
model = tf.keras.Sequential([
tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(28,28,1)),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Dropout(0.25),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(10, activation='softmax')
])
# Compile
model.compile(optimizer='adam', loss='sparse_categorical_crossentropy', metrics=['accuracy'])
print('Compile complete')
# +
# Set log data to feed to TensorBoard for visual analysis
tensor_board = tf.keras.callbacks.TensorBoard('./logs/sample-LeNet-MNIST-1')
# Train model (with timing)
import time
start_time=time.time()
model.fit(train_images, train_labels, batch_size=128, epochs=15, verbose=1,
validation_data=(test_images, test_labels), callbacks=[tensor_board])
print('Training took {} seconds'.format(time.time()-start_time))
# -
| notebooks/00-sample.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Introduction
#
# #Introduce the concept of sorting and sorting algorithms, discuss the
# relevance of concepts such as complexity (time and space), performance, in-place sorting,
# stable sorting, comparator functions, comparison-based and non-comparison-based sorts,
# etc.
# Sorting algorithms vary greatly in their performance. This benchmarking project is to find out which algorithms will perform the best.
# n
# # Sorting Algorithms
#
# Introduce each of your chosen algorithms in turn,
# discuss their space and time complexity, and explain how each algorithm works using your
# own diagrams and different example input instances.
# ## 1. Bubble Sort (A simple comparison-based sort)
#
# Bubble sort is a simple sorting algorithm. https://en.wikipedia.org/wiki/Bubble_sort
#
# How it works:
# 1. It starts at the beginning of the dataset and compares the first two elements and if the first is greater it will swap them.
# 2. It will continue doing this until no swaps are needed.
#
# #### Performance
# Bubble sort has a worst-case and average complexity of О(n2), where n is the number of items being sorted.
# When the list is already sorted (best-case), the complexity of bubble sort is only O(n).
# In the case of a large dataset, Bubble sort should be avoided. It is not very practical or efficient and rarely used in the real world.
#
# Bubble sort in action https://www.youtube.com/watch?v=lyZQPjUT5B4&feature=youtu.be
#
# #insert a diagram
# +
# code sourced from http://interactivepython.org/runestone/static/pythonds/SortSearch/TheBubbleSort.html
def bubbleSort(alist):
for passnum in range(len(alist)-1,0,-1):
for i in range(passnum):
if alist[i]>alist[i+1]:
temp = alist[i]
alist[i] = alist[i+1]
alist[i+1] = temp
alist = [54,26,93,17,77,31,44,55,20]
bubbleSort(alist)
print(alist)
# +
# import time module
import time
# start timer
start_time = time.time()
# bubble sort function, use the numbers from alist
def bubbleSort(alist):
for passnum in range(len(alist)-1,0,-1):
for i in range(passnum):
if alist[i]>alist[i+1]:
temp = alist[i]
alist[i] = alist[i+1]
alist[i+1] = temp
print(bubbleSort(alist))
#print(bubbleSort(alist1))
# end timer
end_time = time.time()
# calculate time
time_elapsed= end_time - start_time
print(bubbleSort(alist), "alist time: ", time_elapsed)
# -
# ## 2. Merge Sort (An efficient comparison-based sort)
#
# Merge sort is a recursive divide and conquer algorithm that was invented by <NAME> in 1945.(https://en.wikipedia.org/wiki/Merge_sort)
#
# How it works:
# 1. It starts by breaking down the list into sublists until each sublists contains just one element.
# 2. Repeatedly merging the sublists to produce new sorted sublists until there is only one sublist remaining.
#
# #### Performance
# In sorting n objects, merge sort has an average and worst-case performance of O(n log n). It's best, worst and average cases are very similar, making it a good choice for predictable running behaviour. (source from lecture notes)
#
# Merge sort in action:
# https://www.youtube.com/watch?v=XaqR3G_NVoo
#
#
# An efficient sorting algorithm??
#
# ### insert a diagram
# +
# code sourced from http://interactivepython.org/runestone/static/pythonds/SortSearch/TheMergeSort.html
def mergeSort(alist):
#print("Splitting ",alist)
if len(alist)>1:
mid = len(alist)//2
lefthalf = alist[:mid]
righthalf = alist[mid:]
mergeSort(lefthalf)
mergeSort(righthalf)
i=0
j=0
k=0
while i < len(lefthalf) and j < len(righthalf):
if lefthalf[i] < righthalf[j]:
alist[k]=lefthalf[i]
i=i+1
else:
alist[k]=righthalf[j]
j=j+1
k=k+1
while i < len(lefthalf):
alist[k]=lefthalf[i]
i=i+1
k=k+1
while j < len(righthalf):
alist[k]=righthalf[j]
j=j+1
k=k+1
#print("Merging ",alist)
alist = [54,26,93,17,77,31,44,55,20]
mergeSort(alist)
print(alist)
# -
# ## 3. Counting Sort (A non-comparison sort)
# +
# code sourced http://www.learntosolveit.com/python/algorithm_countingsort.html
def counting_sort(array, maxval):
"""in-place counting sort"""
n = len(array)
m = maxval + 1
count = [0] * m # init with zeros
for a in array:
count[a] += 1 # count occurences
i = 0
for a in range(m): # emit
for c in range(count[a]): # - emit 'count[a]' copies of 'a'
array[i] = a
i += 1
return array
print(counting_sort( alist, 93 ))
# -
# ## 4. Quick Sort
#
# Quicksort was developed by British computer scientist <NAME> in 1959. It is a recursive divide and conquer algorithm. Due to it's efficiency, it is still a commonly used algorithm for sorting.(https://en.wikipedia.org/wiki/Quicksort)
#
# How it works (lecture notes referenced):
# 1. Pivot selection: Pick an element, called a “pivot” from the array
# 2. Partioning: reorder the array elements with values < the pivot come beofre it, which all elements the values ≥ than the pivot come after it. After this partioining, the pivot is in its final position.
# 3. Recursion: apply steps 1 and 2 above recursively to each of the two subarrays
#
# #### Performance
# +
# http://interactivepython.org/runestone/static/pythonds/SortSearch/TheQuickSort.html
def quickSort(alist):
quickSortHelper(alist,0,len(alist)-1)
def quickSortHelper(alist,first,last):
if first<last:
splitpoint = partition(alist,first,last)
quickSortHelper(alist,first,splitpoint-1)
quickSortHelper(alist,splitpoint+1,last)
def partition(alist,first,last):
pivotvalue = alist[first]
leftmark = first+1
rightmark = last
done = False
while not done:
while leftmark <= rightmark and alist[leftmark] <= pivotvalue:
leftmark = leftmark + 1
while alist[rightmark] >= pivotvalue and rightmark >= leftmark:
rightmark = rightmark -1
if rightmark < leftmark:
done = True
else:
temp = alist[leftmark]
alist[leftmark] = alist[rightmark]
alist[rightmark] = temp
temp = alist[first]
alist[first] = alist[rightmark]
alist[rightmark] = temp
return rightmark
# alist = [54,26,93,17,77,31,44,55,20]
quickSort(alist)
print(alist)
# -
# ## 5. Insertion Sort
# +
def insertionSort(alist):
for index in range(1,len(alist)):
currentvalue = alist[index]
position = index
while position>0 and alist[position-1]>currentvalue:
alist[position]=alist[position-1]
position = position-1
alist[position]=currentvalue
alist = [54,26,93,17,77,31,44,55,20]
insertionSort(alist)
print(alist)
# -
# # Implementation & Benchmarking
# For this section, a function will be definied to call each sorting function defined above
# 1. Bubble Sort
# 2. Merge Sort
# 3. Counting Sort
# 4. Quick Sort
# 5. Insertion Sort
#
# Firstly, arrays are generated with random numbers using randint from the python's random library (https://docs.python.org/2/library/random.html). These will be used to test the speed of efficiency of the algorithms.
# +
# Creating an array using randint
from random import *
# creating a random array, function takes in n numbers
def random_array(n):
# create an array variable
array = []
# if n = 5, 0,1,2,3,4
for i in range(0,n, 1):
# add to the array random integers between 0 and 100
array.append(randint(0,100))
return array
# assign the random array to alist
alist = random_array(100)
alist1 = random_array(1000)
alist2 = random_array(10000)
# -
# Using the time module (https://docs.python.org/3/library/time.html), a start time and end time for each function will be noted and the elapsed time is what will be noted.
# Above a random arrays were defined. They will be used to test the performance of the
# +
# import time module
import time
# function to call sort functions and time each individually
def callsorts():
# start timer
start_time = time.time()
######## bubblesort
bubbleSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist), "Bubble Sort: ", time_elapsed)
# start timer
start_time = time.time()
bubbleSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist1), "Bubble Sort: ", time_elapsed)
# start timer
start_time = time.time()
bubbleSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist2), "Bubble Sort: ", time_elapsed)
##### Merge Sort
#start timer
start_time = time.time()
mergeSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist), "Merge Sort: ", time_elapsed)
# start timer
start_time = time.time()
mergeSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist1), "Merge Sort: ", time_elapsed)
# start timer
start_time = time.time()
mergeSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist2), "Merge Sort: ", time_elapsed)
##### counting_sort
start_time = time.time()
counting_sort(alist, 100)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
# start timer
start_time = time.time()
counting_sort(alist1, 1000)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
# start timer
start_time = time.time()
counting_sort(alist2, 10000)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
##### quick sort
start_time = time.time()
quickSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
# start timer
start_time = time.time()
quickSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
# start timer
start_time = time.time()
quickSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
##### insertionSort
start_time = time.time()
insertionSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
# start timer
start_time = time.time()
insertionSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
# start timer
start_time = time.time()
insertionSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
callsorts()
# +
# import time module
import time
import pandas as pd
import numpy as np
df = pd.DataFrame(index = ['Bubble Sort', 'Merge Sort', 'Counting sort', 'Quick sort', 'Insertion sort'])
# function to call sort functions and time each individually
def callsorts():
# start timer
start_time = time.time()
######## bubblesort
bubbleSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist), "Bubble Sort: ", time_elapsed)
df.insert(0,'100', time_elapsed)
# start timer
start_time = time.time()
bubbleSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist1), "Bubble Sort: ", time_elapsed)
# start timer
start_time = time.time()
bubbleSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print(bubbleSort(alist2), "Bubble Sort: ", time_elapsed)
##### Merge Sort
#start timer
start_time = time.time()
mergeSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist), "Merge Sort: ", time_elapsed)
df.insert(1,'100', time_elapsed)
# start timer
start_time = time.time()
mergeSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist1), "Merge Sort: ", time_elapsed)
# start timer
start_time = time.time()
mergeSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print(mergeSort(alist2), "Merge Sort: ", time_elapsed)
##### counting_sort
start_time = time.time()
counting_sort(alist, 100)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
# start timer
start_time = time.time()
counting_sort(alist1, 1000)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
# start timer
start_time = time.time()
counting_sort(alist2, 10000)
end_time = time.time()
time_elapsed= end_time - start_time
print("Counting sort: ", time_elapsed)
##### quick sort
start_time = time.time()
quickSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
# start timer
start_time = time.time()
quickSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
# start timer
start_time = time.time()
quickSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print("Quick sort: ", time_elapsed)
##### insertionSort
start_time = time.time()
insertionSort(alist)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
# start timer
start_time = time.time()
insertionSort(alist1)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
# start timer
start_time = time.time()
insertionSort(alist2)
end_time = time.time()
time_elapsed= end_time - start_time
print("Insertion sort: ", time_elapsed)
callsorts()
df
# -
df[2]=2
df
print("Size", '\t', "100") #table column headings
print("---", '\t', "-----")
# generate values for columns
print("Bubble Sort", '\t', bubbleSort(alist2), "BubbleSort: ", time_elapsed)
| Algorithms_problems/Sorting Project/Benchmarking Sorting Algorithms.ipynb |
# ##### Copyright 2020 Google LLC.
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
# # integer_programming_example
# <table align="left">
# <td>
# <a href="https://colab.research.google.com/github/google/or-tools/blob/master/examples/notebook/linear_solver/integer_programming_example.ipynb"><img src="https://raw.githubusercontent.com/google/or-tools/master/tools/colab_32px.png"/>Run in Google Colab</a>
# </td>
# <td>
# <a href="https://github.com/google/or-tools/blob/master/ortools/linear_solver/samples/integer_programming_example.py"><img src="https://raw.githubusercontent.com/google/or-tools/master/tools/github_32px.png"/>View source on GitHub</a>
# </td>
# </table>
# First, you must install [ortools](https://pypi.org/project/ortools/) package in this colab.
# !pip install ortools
# +
# Copyright 2010-2018 Google LLC
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Small example to illustrate solving a MIP problem."""
# [START program]
# [START import]
from ortools.linear_solver import pywraplp
# [END import]
def IntegerProgrammingExample():
"""Integer programming sample."""
# [START solver]
# Create the mip solver with the SCIP backend.
solver = pywraplp.Solver.CreateSolver('SCIP')
# [END solver]
# [START variables]
# x, y, and z are non-negative integer variables.
x = solver.IntVar(0.0, solver.infinity(), 'x')
y = solver.IntVar(0.0, solver.infinity(), 'y')
z = solver.IntVar(0.0, solver.infinity(), 'z')
# [END variables]
# [START constraints]
# 2*x + 7*y + 3*z <= 50
constraint0 = solver.Constraint(-solver.infinity(), 50)
constraint0.SetCoefficient(x, 2)
constraint0.SetCoefficient(y, 7)
constraint0.SetCoefficient(z, 3)
# 3*x - 5*y + 7*z <= 45
constraint1 = solver.Constraint(-solver.infinity(), 45)
constraint1.SetCoefficient(x, 3)
constraint1.SetCoefficient(y, -5)
constraint1.SetCoefficient(z, 7)
# 5*x + 2*y - 6*z <= 37
constraint2 = solver.Constraint(-solver.infinity(), 37)
constraint2.SetCoefficient(x, 5)
constraint2.SetCoefficient(y, 2)
constraint2.SetCoefficient(z, -6)
# [END constraints]
# [START objective]
# Maximize 2*x + 2*y + 3*z
objective = solver.Objective()
objective.SetCoefficient(x, 2)
objective.SetCoefficient(y, 2)
objective.SetCoefficient(z, 3)
objective.SetMaximization()
# [END objective]
# Solve the problem and print the solution.
# [START print_solution]
solver.Solve()
# Print the objective value of the solution.
print('Maximum objective function value = %d' % solver.Objective().Value())
print()
# Print the value of each variable in the solution.
for variable in [x, y, z]:
print('%s = %d' % (variable.name(), variable.solution_value()))
# [END print_solution]
IntegerProgrammingExample()
# [END program]
| examples/notebook/linear_solver/integer_programming_example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Generating Normally Distributed Random Numbers in Power Query
# > Custom function to generate a column with normally distributed random numbers with specified mean and standard deviation
#
# - toc: true
# - badges: true
# - comments: true
# - categories: [Power BI, random number, M, Power Query]
# - hide: false
# ## Power Query Doesn't Have NORMINV()
# In Excel, if you want to generate a column with random numbers that are normally distributed, you can use the `NORMINV()` function like [this](https://support.microsoft.com/en-us/office/norminv-function-87981ab8-2de0-4cb0-b1aa-e21d4cb879b8). You can specify the probability (which is usually a random number drawn from uniform distribution), mean and standard deviation. While DAX has the `NORM.INV()` [function](https://docs.microsoft.com/en-us/dax/norm-inv-dax), M does not. If you create simulations, what-if scenario analyses etc., more than likely you will need to generate a column with random numbers that follow the Gaussian distribution. I have written a blog post on how to generate various distributions using DAX, you can read it [here](https://pawarbi.github.io/blog/power%20bi/statistics/distribution/pert/beta/normal/uniform/lognormal/logistic/weibull/2020/12/24/Statistical-distributions-powerbi.html).
#
# In this blog, I will share a simple formula to generate the normally distributed random numbers using M. It uses the Box-Muller transform to generate the inverse distribution. I won't go into the theory and math, but if you are interested you can read it [here](https://medium.com/mti-technology/how-to-generate-gaussian-samples-3951f2203ab0).
#
# 
# ## Custom Function
# +
#hide-output
// Gaussian Random Number Generator with mean =mean and standard number as sd using Box-Mueller Transform
// Add an index column to the table before invoking this function.
let
gaussianrandom = (mean as number, sd as number) as number=>
(
sd
* (
Number.Sqrt(- 2 * Number.Ln(Number.Random())
)
* Number.Cos( 2.0 * 3.14159265358979323846
* Number.Random()
)
)
+ mean
)
in
gaussianrandom
# -
# ## Steps
#
# - Create a Power Query function using the formula above. In the below example, I named the function `_NormalDist`
#
# 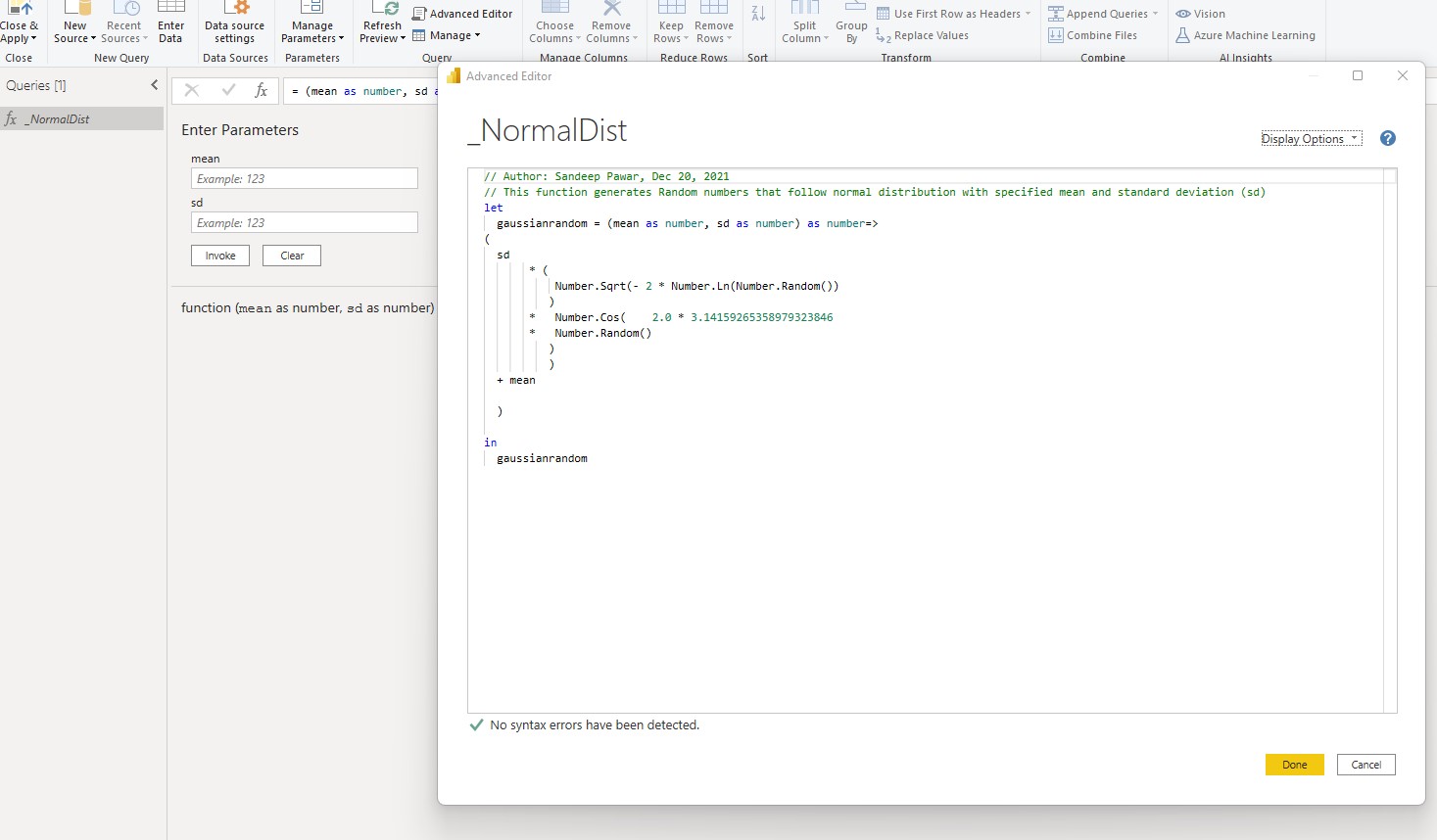
#
# - You will need to have unique rows. If you don't, create an index column (Add Column > Index Column).
# - To create a new column that follows the Gaussian distribution using the above function, go to Add Column and use the above function. In the example below, I created a new column that has mean of 10 and standard deviation of 0.25
#
# 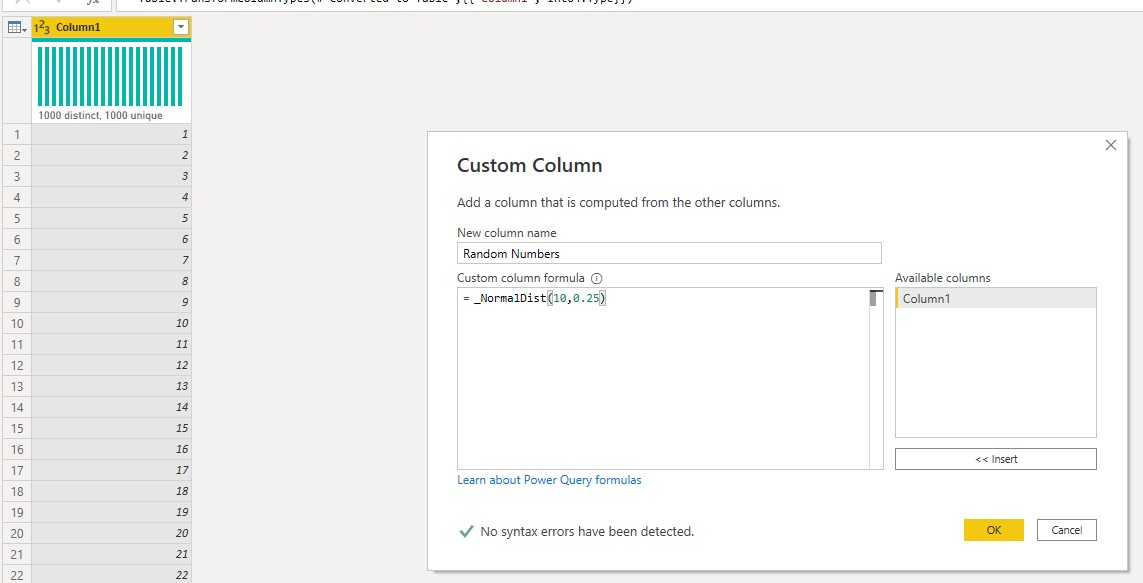
#
#
# - Here is the result:
# 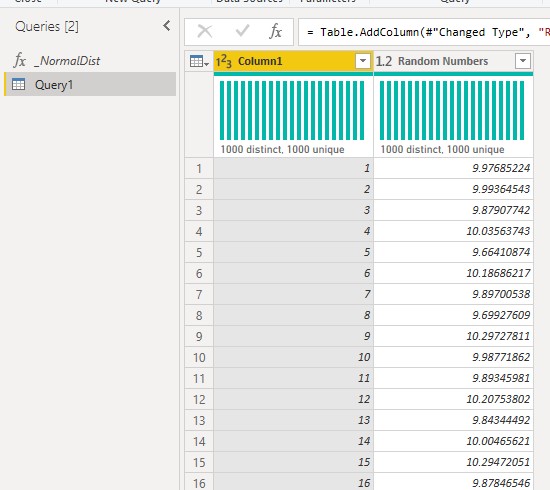
#
#
# Refresh the report and you will see the numbers in your table. If you see same number on all rows, just add another index column and remove it again.
#
# `NORM.INV()` in DAX generates new numbers every time the report is refreshed. In Power Query you can disable the refresh for this table, and hence generated numbers will stay the same even after refreshing the report. If you open the PowerQuery, however, it will generate new numbers. You can use `Table.Buffer()` to freeze it but I haven't had luck with that. If you know how to do it, please let me know.
# Here is the resulting distribution:
#
#hide-input
import pandas as pd
import seaborn as sns
df = pd.read_clipboard().set_index('Column1')
df.head(5)
sns.displot(df['Random Numbers'], rug=True, kde=True);
print("The mean and standard deviation of random numbers : ", round(df['Random Numbers'].mean(),2), round(df['Random Numbers'].std(),2))
# I also wrote another function to generate Traingular Distribution,which is very common to simulate risk profiles. Hope to share that soon.
| _notebooks/2021-12-22-PowerQuery-Normal-distribution.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Preparing the dataset for hippocampus segmentation
#
# In this notebook you will use the skills and methods that we have talked about during our EDA Lesson to prepare the hippocampus dataset using Python. Follow the Notebook, writing snippets of code where directed so using Task comments, similar to the one below, which expects you to put the proper imports in place. Write your code directly in the cell with TASK comment. Feel free to add cells as you see fit, but please make sure that code that performs that tasked activity sits in the same cell as the Task comment.
#
# TASK: Import the following libraries that we will use: nibabel, matplotlib, numpy
import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
import os
import shutil
from glob import glob
import scipy.ndimage as nd
# It will help your understanding of the data a lot if you were able to use a tool that allows you to view NIFTI volumes, like [3D Slicer](https://www.slicer.org/). I will refer to Slicer throughout this Notebook and will be pasting some images showing what your output might look like.
# ## Loading NIFTI images using NiBabel
#
# NiBabel is a python library for working with neuro-imaging formats (including NIFTI) that we have used in some of the exercises throughout the course. Our volumes and labels are in NIFTI format, so we will use nibabel to load and inspect them.
#
# NiBabel documentation could be found here: https://nipy.org/nibabel/
#
# Our dataset sits in two directories - *images* and *labels*. Each image is represented by a single file (we are fortunate to have our data converted to NIFTI) and has a corresponding label file which is named the same as the image file.
#
# Note that our dataset is "dirty". There are a few images and labels that are not quite right. They should be quite obvious to notice, though. The dataset contains an equal amount of "correct" volumes and corresponding labels, and you don't need to alter values of any samples in order to get the clean dataset.
# +
# TASK: Your data sits in directory /data/TrainingSet.
# Load an image and a segmentation mask into variables called image and label
images = glob("/data/TrainingSet/images/*")
labels = glob("/data/TrainingSet/labels/*")
image = nib.load(images[0])
label = nib.load(labels[0])
# +
# Nibabel can present your image data as a Numpy array by calling the method get_fdata()
# The array will contain a multi-dimensional Numpy array with numerical values representing voxel intensities.
# In our case, images and labels are 3-dimensional, so get_fdata will return a 3-dimensional array. You can verify this
# by accessing the .shape attribute. What are the dimensions of the input arrays?
# TASK: using matplotlib, visualize a few slices from the dataset, along with their labels.
# You can adjust plot sizes like so if you find them too small:
# plt.rcParams["figure.figsize"] = (10,10)
image_arr = image.get_fdata()
label_arr = label.get_fdata()
image_arr.shape #3 dimensionsal with dimensions (35, 55, 37)
label_arr.shape #3 dimensionsal with dimensions (35, 55, 37)
#Another volume from the dataset
image2 = nib.load(images[2])
label2 = nib.load(labels[2])
image_arr2 = image2.get_fdata()
label_arr2 = label2.get_fdata()
image_arr2.shape #3 dimensrional with dimensions (37, 45, 46)
label_arr2.shape #3 dimensrional with dimensions (37, 45, 46)
#Another volume from the dataset
image3 = nib.load(images[4])
label3 = nib.load(labels[4])
image_arr3 = image3.get_fdata()
label_arr3 = image3.get_fdata()
image_arr3.shape #3 dimensrional with dimensions (35, 50, 36)
label_arr3.shape #3 dimensrional with dimensions (35, 50, 36)
#Plot of three image slices from image and label loaded
plt.rcParams["figure.figsize"] = (16,16)
plt.subplot(131)
plt.imshow(image_arr[2,:,:] + label_arr[2,:,:], cmap= "gray")
plt.subplot(132)
plt.imshow(image_arr2[4,:,:] + label_arr2[4,:,:], cmap= "gray")
plt.subplot(133)
plt.imshow(image_arr3[6,:,:] + label_arr3[6,:,:], cmap= "gray")
# -
# Load volume into 3D Slicer to validate that your visualization is correct and get a feel for the shape of structures.Try to get a visualization like the one below (hint: while Slicer documentation is not particularly great, there are plenty of YouTube videos available! Just look it up on YouTube if you are not sure how to do something)
#
# 
# Stand out suggestion: use one of the simple Volume Rendering algorithms that we've
# implemented in one of our earlier lessons to visualize some of these volumes
#Plot volumetric rendering using maximum intensity projection for second image
mip = np.zeros((image_arr2.shape[0], image_arr2.shape[2]))
for y in range (image_arr2.shape[1]):
mip = np.maximum(mip,image_arr2[:,y,:])
plt.imshow(nd.rotate(mip, 90), cmap="gray")
# ## Looking at single image data
# In this section we will look closer at the NIFTI representation of our volumes. In order to measure the physical volume of hippocampi, we need to understand the relationship between the sizes of our voxels and the physical world.
# Nibabel supports many imaging formats, NIFTI being just one of them. I told you that our images
# are in NIFTI, but you should confirm if this is indeed the format that we are dealing with
# TASK: using .header_class attribute - what is the format of our images?
img1_type = image.header
img2_type = image2.header
img3_type = image3.header
print("Image type for first image:" + str(img1_type))
print("Image type for second image:" + str(img2_type))
print("Image type for third image:" + str(img3_type))
# Further down we will be inspecting .header attribute that provides access to NIFTI metadata. You can use this resource as a reference for various fields: https://brainder.org/2012/09/23/the-nifti-file-format/
# TASK: How many bits per pixel are used?
'''The first and third images have 32 bits per pixel which can be
inferred from the bitpix field, the seond image has 8 bit per pixel.'''
# +
# TASK: What are the units of measurement?
img1_type.get_xyzt_units() # Units of measurements are millimeters and seconds
img2_type.get_xyzt_units() # Units of measurements are millimeters and seconds
img3_type.get_xyzt_units() # Units of measurements are millimeters and seconds
'''Units of measurement for all three images are millimeters and seconds.'''
# -
# TASK: Do we have a regular grid? What are grid spacings?
'''The pixdim field in the NIFTI header stores an array of spatial and temporal measurements.
The first three bits of the pixdim array indicate spatial measurements or grid spacing in the x, y and z directions,
for all three images the grid spacing is 1, and since it is 1 in the x, y and z directions we have a
regular grid'''
# TASK: What dimensions represent axial, sagittal, and coronal slices? How do you know?
'''The dim field in the NIFTI file header contains an array of dimensions for the given NIFTI volume
,elements 1, 2, 3 in the dim field array contain information on the x, y, z dimensions which would correspond
to the sagittal, coronal, and axial slices in the volume'''
# By now you should have enough information to decide what are dimensions of a single voxel
# TASK: Compute the volume (in mm³) of a hippocampus using one of the labels you've loaded.
# You should get a number between ~2200 and ~4500
# since the grid spacing is 1x1x1 we can simply sum up the number of voxels in the volume
vol = np.sum(label_arr2 > 0)
vol
# ## Plotting some charts
# +
def calc_vol(label_in):
return np.sum(nib.load(label_in).get_fdata() > 0)
volumes = []
def cal_vols(labels):
for label in labels:
vol = calc_vol(label)
volumes.append(vol)
# -
# TASK: Plot a histogram of all volumes that we have in our dataset and see how
# our dataset measures against a slice of a normal population represented by the chart below.
plt.figure(figsize=(16,16))
cal_vols(labels)
plt.hist(volumes)
plt.xlabel('volume mm^3')
plt.ylabel('Number of Images')
#Most hippocampus volumes are between the 2200 mm^3 and 4500 mm^3 range except for the two outliers
# <img src="img/nomogram_fem_right.svg" width=400 align=left>
# Do you see any outliers? Why do you think it's so (might be not immediately obvious, but it's always a good idea to inspect) outliers closer. If you haven't found the images that do not belong, the histogram may help you.
# In the real world we would have precise information about the ages and conditions of our patients, and understanding how our dataset measures against population norm would be the integral part of clinical validation that we talked about in last lesson. Unfortunately, we do not have this information about this dataset, so we can only guess why it measures the way it is. If you would like to explore further, you can use the [calculator from HippoFit project](http://www.smanohar.com/biobank/calculator.html) to see how our dataset compares against different population slices
# Did you notice anything odd about the label files? We hope you did! The mask seems to have two classes, labeled with values `1` and `2` respectively. If you visualized sagittal or axial views, you might have gotten a good guess of what those are. Class 1 is the anterior segment of the hippocampus and class 2 is the posterior one.
#
# For the purpose of volume calculation we do not care about the distinction, however we will still train our network to differentiate between these two classes and the background
def copy_files(labels, images):
for label, image in zip(labels,images):
label_file_name = label[-22:]
image_file_name = label[-22:]
vol = calc_vol(label)
if vol >= 2200 and vol <= 4500:
shutil.copy(label, "/home/workspace/out/labels/"+str(label_file_name))
shutil.copy(label, "/home/workspace/out/images/"+str(image_file_name))
# TASK: Copy the clean dataset to the output folder inside section1/out. You will use it in the next Section
copy_files(labels,images)
# ## Final remarks
#
# Congratulations! You have finished Section 1.
#
# In this section you have inspected a dataset of MRI scans and related segmentations, represented as NIFTI files. We have visualized some slices, and understood the layout of the data. We have inspected file headers to understand what how the image dimensions relate to the physical world and we have understood how to measure our volume. We have then inspected dataset for outliers, and have created a clean set that is ready for consumption by our ML algorithm.
#
# In the next section you will create training and testing pipelines for a UNet-based machine learning model, run and monitor the execution, and will produce test metrics. This will arm you with all you need to use the model in the clinical context and reason about its performance!
| section1/out/Final Project EDA-completed.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import os
import pandas as pd
import numpy as np
from statsmodels.tsa import stattools
# %matplotlib inline
from matplotlib import pyplot as plt
from pandas.plotting import autocorrelation_plot
djia_df = pd.read_excel('datasets/DJIA_Jan2016_Dec2016.xlsx')
djia_df.head(10)
#Let us parse the Date column and use as row index for the DataFrame and drop it as a column
djia_df['Date'] = pd.to_datetime(djia_df['Date'], '%Y-%m-%d')
djia_df.index = djia_df['Date']
djia_df.drop('Date', axis=1, inplace=True)
#Let us see first few rows of the modified DataFrame
djia_df.head(10)
#We would be using the 'Close' values of the DJIA to illustrate Differencing
first_order_diff = djia_df['Close'].diff(1)
#Let us plot the original time series and first-differences
fig, ax = plt.subplots(2, sharex=True)
fig.set_size_inches(5.5, 5.5)
djia_df['Close'].plot(ax=ax[0], color='b')
ax[0].set_title('Close values of DJIA during Jan 2016-Dec 2016')
first_order_diff.plot(ax=ax[1], color='r')
ax[1].set_title('First-order differences of DJIA during Jan 2016-Dec 2016')
#plt.savefig('plots/ch2/B07887_02_06.png', format='png', dpi=300)
#Let us plot the ACFs of original time series and first-differences
fig, ax = plt.subplots(2, sharex=True)
fig.set_size_inches(5.5, 5.5)
autocorrelation_plot(djia_df['Close'], color='b', ax=ax[0])
ax[0].set_title('ACF of DJIA Close values')
autocorrelation_plot(first_order_diff.iloc[1:], color='r', ax=ax[1])
ax[1].set_title('ACF of first differences of DJIA Close values')
plt.tight_layout(pad=0.4, w_pad=0.5, h_pad=2.0)
plt.savefig('plots/ch2/B07887_02_07.png', format='png', dpi=300)
"""
Now we will perform the Ljung-Box test on the ACFs
of the original time series and the first-differences.
For running the test we will limit upto 20 lags
"""
"""
Let us obtain the confidence intervls, Ljung-Box Q-statistics and p-values
for the original DJIA Close values
"""
acf_djia, confint_djia, qstat_djia, pvalues_djia = stattools.acf(djia_df['Close'],
unbiased=True,
nlags=20,
qstat=True,
alpha=0.05)
"""Let us check if at confidence level 95% (alpha=0.05)
if the null hypothesis is rejected at any of the lags
"""
alpha = 0.05
for l, p_val in enumerate(pvalues_djia):
if p_val > alpha:
print('Null hypothesis is accepted at lag = {} for p-val = {}'.format(l, p_val))
else:
print('Null hypothesis is rejected at lag = {} for p-val = {}'.format(l, p_val))
"""
The above results show statistically significant ACF in the original DJIA Close values
"""
"""
Let us obtain the confidence intervls, Ljung-Box Q-statistics and p-values
for the differenced DJIA Close values
"""
acf_first_diff, confint_first_diff,\
qstat_first_diff, pvalues_first_diff = stattools.acf(first_order_diff.iloc[1:],
unbiased=True,
nlags=20,
qstat=True,
alpha=0.05)
"""Let us check if at confidence level of 95% (alpha = 0.05)
if the null hypothesis is rejected at any of the lags
"""
alpha = 0.05
for l, p_val in enumerate(pvalues_first_diff):
if p_val > alpha:
print('Null hypothesis is accepted at lag = {} for p-val = {}'.format(l, p_val))
else:
print('Null hypothesis is rejected at lag = {} for p-val = {}'.format(l, p_val))
"""
The above results show that ACF is essentially random in the differenced DJIA Close values
"""
| time series regression/autocorelation, mov avg etc/First_Order_Differencing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Discretization
#
# ---
#
# In this notebook, you will deal with continuous state and action spaces by discretizing them. This will enable you to apply reinforcement learning algorithms that are only designed to work with discrete spaces.
#
# ### 1. Import the Necessary Packages
# +
import sys
import gym
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# Set plotting options
# %matplotlib inline
plt.style.use('ggplot')
np.set_printoptions(precision=3, linewidth=120)
# -
# ### 2. Specify the Environment, and Explore the State and Action Spaces
#
# We'll use [OpenAI Gym](https://gym.openai.com/) environments to test and develop our algorithms. These simulate a variety of classic as well as contemporary reinforcement learning tasks. Let's use an environment that has a continuous state space, but a discrete action space.
# Create an environment and set random seed
env = gym.make('MountainCar-v0')
env.seed(505);
# Run the next code cell to watch a random agent.
state = env.reset()
score = 0
for t in range(200):
action = env.action_space.sample()
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
# In this notebook, you will train an agent to perform much better! For now, we can explore the state and action spaces, as well as sample them.
# Explore state (observation) space
print("State space:", env.observation_space)
print("- low:", env.observation_space.low)
print("- high:", env.observation_space.high)
# Generate some samples from the state space
print("State space samples:")
print(np.array([env.observation_space.sample() for i in range(10)]))
# +
# Explore the action space
print("Action space:", env.action_space)
# Generate some samples from the action space
print("Action space samples:")
print(np.array([env.action_space.sample() for i in range(10)]))
# -
# ### 3. Discretize the State Space with a Uniform Grid
#
# We will discretize the space using a uniformly-spaced grid. Implement the following function to create such a grid, given the lower bounds (`low`), upper bounds (`high`), and number of desired `bins` along each dimension. It should return the split points for each dimension, which will be 1 less than the number of bins.
#
# For instance, if `low = [-1.0, -5.0]`, `high = [1.0, 5.0]`, and `bins = (10, 10)`, then your function should return the following list of 2 NumPy arrays:
#
# ```
# [array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
# array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
# ```
#
# Note that the ends of `low` and `high` are **not** included in these split points. It is assumed that any value below the lowest split point maps to index `0` and any value above the highest split point maps to index `n-1`, where `n` is the number of bins along that dimension.
# +
def create_uniform_grid(low, high, bins=(10, 10)):
"""Define a uniformly-spaced grid that can be used to discretize a space.
Parameters
----------
low : array_like
Lower bounds for each dimension of the continuous space.
high : array_like
Upper bounds for each dimension of the continuous space.
bins : tuple
Number of bins along each corresponding dimension.
Returns
-------
grid : list of array_like
A list of arrays containing split points for each dimension.
"""
# TODO: Implement this
low_offset = (high[0] - low[0])/bins[0]
high_offset = (high[1] - low[1])/bins[1]
start = low[0] + low_offset
result = list()
result_low = list()
result_high = list()
for i in np.arange(low[0] + low_offset, high[0], low_offset):
result_low.append(round(i, 1))
for i in np.arange(low[1] + high_offset, high[1], high_offset):
result_high.append(round(i, 1))
result.append(result_low)
result.append(result_high)
return result
low = [-1.0, -5.0]
high = [1.0, 5.0]
create_uniform_grid(low, high) # [test]
# -
# Now write a function that can convert samples from a continuous space into its equivalent discretized representation, given a grid like the one you created above. You can use the [`numpy.digitize()`](https://docs.scipy.org/doc/numpy-1.9.3/reference/generated/numpy.digitize.html) function for this purpose.
#
# Assume the grid is a list of NumPy arrays containing the following split points:
# ```
# [array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
# array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
# ```
#
# Here are some potential samples and their corresponding discretized representations:
# ```
# [-1.0 , -5.0] => [0, 0]
# [-0.81, -4.1] => [0, 0]
# [-0.8 , -4.0] => [1, 1]
# [-0.5 , 0.0] => [2, 5]
# [ 0.2 , -1.9] => [6, 3]
# [ 0.8 , 4.0] => [9, 9]
# [ 0.81, 4.1] => [9, 9]
# [ 1.0 , 5.0] => [9, 9]
# ```
#
# **Note**: There may be one-off differences in binning due to floating-point inaccuracies when samples are close to grid boundaries, but that is alright.
# +
def discretize(sample, grid):
"""Discretize a sample as per given grid.
Parameters
----------
sample : array_like
A single sample from the (original) continuous space.
grid : list of array_like
A list of arrays containing split points for each dimension.
Returns
-------
discretized_sample : array_like
A sequence of integers with the same number of dimensions as sample.
"""
# TODO: Implement this
return list(int(np.digitize(s, g)) for s, g in zip(sample, grid))
# Test with a simple grid and some samples
grid = create_uniform_grid([-1.0, -5.0], [1.0, 5.0])
samples = np.array(
[[-1.0 , -5.0],
[-0.81, -4.1],
[-0.8 , -4.0],
[-0.5 , 0.0],
[ 0.2 , -1.9],
[ 0.8 , 4.0],
[ 0.81, 4.1],
[ 1.0 , 5.0]])
discretized_samples = np.array([discretize(sample, grid) for sample in samples])
print("\nSamples:", repr(samples), sep="\n")
print("\nDiscretized samples:", repr(discretized_samples), sep="\n")
# -
# ### 4. Visualization
#
# It might be helpful to visualize the original and discretized samples to get a sense of how much error you are introducing.
# +
import matplotlib.collections as mc
def visualize_samples(samples, discretized_samples, grid, low=None, high=None):
"""Visualize original and discretized samples on a given 2-dimensional grid."""
fig, ax = plt.subplots(figsize=(10, 10))
# Show grid
ax.xaxis.set_major_locator(plt.FixedLocator(grid[0]))
ax.yaxis.set_major_locator(plt.FixedLocator(grid[1]))
ax.grid(True)
# If bounds (low, high) are specified, use them to set axis limits
if low is not None and high is not None:
ax.set_xlim(low[0], high[0])
ax.set_ylim(low[1], high[1])
else:
# Otherwise use first, last grid locations as low, high (for further mapping discretized samples)
low = [splits[0] for splits in grid]
high = [splits[-1] for splits in grid]
# Map each discretized sample (which is really an index) to the center of corresponding grid cell
grid_extended = np.hstack((np.array([low]).T, grid, np.array([high]).T)) # add low and high ends
grid_centers = (grid_extended[:, 1:] + grid_extended[:, :-1]) / 2 # compute center of each grid cell
locs = np.stack(grid_centers[i, discretized_samples[:, i]] for i in range(len(grid))).T # map discretized samples
ax.plot(samples[:, 0], samples[:, 1], 'o') # plot original samples
ax.plot(locs[:, 0], locs[:, 1], 's') # plot discretized samples in mapped locations
ax.add_collection(mc.LineCollection(list(zip(samples, locs)), colors='orange')) # add a line connecting each original-discretized sample
ax.legend(['original', 'discretized'])
visualize_samples(samples, discretized_samples, grid, low, high)
# -
# Now that we have a way to discretize a state space, let's apply it to our reinforcement learning environment.
# Create a grid to discretize the state space
state_grid = create_uniform_grid(env.observation_space.low, env.observation_space.high, bins=(10, 10))
state_grid
# Obtain some samples from the space, discretize them, and then visualize them
state_samples = np.array([env.observation_space.sample() for i in range(10)])
discretized_state_samples = np.array([discretize(sample, state_grid) for sample in state_samples])
visualize_samples(state_samples, discretized_state_samples, state_grid,
env.observation_space.low, env.observation_space.high)
plt.xlabel('position'); plt.ylabel('velocity'); # axis labels for MountainCar-v0 state space
# You might notice that if you have enough bins, the discretization doesn't introduce too much error into your representation. So we may be able to now apply a reinforcement learning algorithm (like Q-Learning) that operates on discrete spaces. Give it a shot to see how well it works!
#
# ### 5. Q-Learning
#
# Provided below is a simple Q-Learning agent. Implement the `preprocess_state()` method to convert each continuous state sample to its corresponding discretized representation.
# +
class QLearningAgent:
"""Q-Learning agent that can act on a continuous state space by discretizing it."""
def __init__(self, env, state_grid, alpha=0.02, gamma=0.99,
epsilon=1.0, epsilon_decay_rate=0.9995, min_epsilon=.01, seed=505):
"""Initialize variables, create grid for discretization."""
# Environment info
self.env = env
self.state_grid = state_grid
self.state_size = tuple(len(splits) + 1 for splits in self.state_grid) # n-dimensional state space
self.action_size = self.env.action_space.n # 1-dimensional discrete action space
self.seed = np.random.seed(seed)
print("Environment:", self.env)
print("State space size:", self.state_size)
print("Action space size:", self.action_size)
# Learning parameters
self.alpha = alpha # learning rate
self.gamma = gamma # discount factor
self.epsilon = self.initial_epsilon = epsilon # initial exploration rate
self.epsilon_decay_rate = epsilon_decay_rate # how quickly should we decrease epsilon
self.min_epsilon = min_epsilon
# Create Q-table
self.q_table = np.zeros(shape=(self.state_size + (self.action_size,)))
print("Q table size:", self.q_table.shape)
def preprocess_state(self, state):
"""Map a continuous state to its discretized representation."""
# TODO: Implement this
pass
def reset_episode(self, state):
"""Reset variables for a new episode."""
# Gradually decrease exploration rate
self.epsilon *= self.epsilon_decay_rate
self.epsilon = max(self.epsilon, self.min_epsilon)
# Decide initial action
self.last_state = self.preprocess_state(state)
self.last_action = np.argmax(self.q_table[self.last_state])
return self.last_action
def reset_exploration(self, epsilon=None):
"""Reset exploration rate used when training."""
self.epsilon = epsilon if epsilon is not None else self.initial_epsilon
def act(self, state, reward=None, done=None, mode='train'):
"""Pick next action and update internal Q table (when mode != 'test')."""
state = self.preprocess_state(state)
if mode == 'test':
# Test mode: Simply produce an action
action = np.argmax(self.q_table[state])
else:
# Train mode (default): Update Q table, pick next action
# Note: We update the Q table entry for the *last* (state, action) pair with current state, reward
self.q_table[self.last_state + (self.last_action,)] += self.alpha * \
(reward + self.gamma * max(self.q_table[state]) - self.q_table[self.last_state + (self.last_action,)])
# Exploration vs. exploitation
do_exploration = np.random.uniform(0, 1) < self.epsilon
if do_exploration:
# Pick a random action
action = np.random.randint(0, self.action_size)
else:
# Pick the best action from Q table
action = np.argmax(self.q_table[state])
# Roll over current state, action for next step
self.last_state = state
self.last_action = action
return action
q_agent = QLearningAgent(env, state_grid)
# -
# Let's also define a convenience function to run an agent on a given environment. When calling this function, you can pass in `mode='test'` to tell the agent not to learn.
# +
def run(agent, env, num_episodes=20000, mode='train'):
"""Run agent in given reinforcement learning environment and return scores."""
scores = []
max_avg_score = -np.inf
for i_episode in range(1, num_episodes+1):
# Initialize episode
state = env.reset()
action = agent.reset_episode(state)
total_reward = 0
done = False
# Roll out steps until done
while not done:
state, reward, done, info = env.step(action)
total_reward += reward
action = agent.act(state, reward, done, mode)
# Save final score
scores.append(total_reward)
# Print episode stats
if mode == 'train':
if len(scores) > 100:
avg_score = np.mean(scores[-100:])
if avg_score > max_avg_score:
max_avg_score = avg_score
if i_episode % 100 == 0:
print("\rEpisode {}/{} | Max Average Score: {}".format(i_episode, num_episodes, max_avg_score), end="")
sys.stdout.flush()
return scores
scores = run(q_agent, env)
# -
# The best way to analyze if your agent was learning the task is to plot the scores. It should generally increase as the agent goes through more episodes.
# Plot scores obtained per episode
plt.plot(scores); plt.title("Scores");
# If the scores are noisy, it might be difficult to tell whether your agent is actually learning. To find the underlying trend, you may want to plot a rolling mean of the scores. Let's write a convenience function to plot both raw scores as well as a rolling mean.
# +
def plot_scores(scores, rolling_window=100):
"""Plot scores and optional rolling mean using specified window."""
plt.plot(scores); plt.title("Scores");
rolling_mean = pd.Series(scores).rolling(rolling_window).mean()
plt.plot(rolling_mean);
return rolling_mean
rolling_mean = plot_scores(scores)
# -
# You should observe the mean episode scores go up over time. Next, you can freeze learning and run the agent in test mode to see how well it performs.
# Run in test mode and analyze scores obtained
test_scores = run(q_agent, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores, rolling_window=10)
# It's also interesting to look at the final Q-table that is learned by the agent. Note that the Q-table is of size MxNxA, where (M, N) is the size of the state space, and A is the size of the action space. We are interested in the maximum Q-value for each state, and the corresponding (best) action associated with that value.
# +
def plot_q_table(q_table):
"""Visualize max Q-value for each state and corresponding action."""
q_image = np.max(q_table, axis=2) # max Q-value for each state
q_actions = np.argmax(q_table, axis=2) # best action for each state
fig, ax = plt.subplots(figsize=(10, 10))
cax = ax.imshow(q_image, cmap='jet');
cbar = fig.colorbar(cax)
for x in range(q_image.shape[0]):
for y in range(q_image.shape[1]):
ax.text(x, y, q_actions[x, y], color='white',
horizontalalignment='center', verticalalignment='center')
ax.grid(False)
ax.set_title("Q-table, size: {}".format(q_table.shape))
ax.set_xlabel('position')
ax.set_ylabel('velocity')
plot_q_table(q_agent.q_table)
# -
# ### 6. Modify the Grid
#
# Now it's your turn to play with the grid definition and see what gives you optimal results. Your agent's final performance is likely to get better if you use a finer grid, with more bins per dimension, at the cost of higher model complexity (more parameters to learn).
# TODO: Create a new agent with a different state space grid
state_grid_new = create_uniform_grid(?, ?, bins=(?, ?))
q_agent_new = QLearningAgent(env, state_grid_new)
q_agent_new.scores = [] # initialize a list to store scores for this agent
# Train it over a desired number of episodes and analyze scores
# Note: This cell can be run multiple times, and scores will get accumulated
q_agent_new.scores += run(q_agent_new, env, num_episodes=50000) # accumulate scores
rolling_mean_new = plot_scores(q_agent_new.scores)
# Run in test mode and analyze scores obtained
test_scores = run(q_agent_new, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores)
# Visualize the learned Q-table
plot_q_table(q_agent_new.q_table)
# ### 7. Watch a Smart Agent
state = env.reset()
score = 0
for t in range(200):
action = q_agent_new.act(state, mode='test')
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
| discretization/Discretization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
# # Gaussian Mixture Model Sine Curve
#
#
# This example demonstrates the behavior of Gaussian mixture models fit on data
# that was not sampled from a mixture of Gaussian random variables. The dataset
# is formed by 100 points loosely spaced following a noisy sine curve. There is
# therefore no ground truth value for the number of Gaussian components.
#
# The first model is a classical Gaussian Mixture Model with 10 components fit
# with the Expectation-Maximization algorithm.
#
# The second model is a Bayesian Gaussian Mixture Model with a Dirichlet process
# prior fit with variational inference. The low value of the concentration prior
# makes the model favor a lower number of active components. This models
# "decides" to focus its modeling power on the big picture of the structure of
# the dataset: groups of points with alternating directions modeled by
# non-diagonal covariance matrices. Those alternating directions roughly capture
# the alternating nature of the original sine signal.
#
# The third model is also a Bayesian Gaussian mixture model with a Dirichlet
# process prior but this time the value of the concentration prior is higher
# giving the model more liberty to model the fine-grained structure of the data.
# The result is a mixture with a larger number of active components that is
# similar to the first model where we arbitrarily decided to fix the number of
# components to 10.
#
# Which model is the best is a matter of subjective judgement: do we want to
# favor models that only capture the big picture to summarize and explain most of
# the structure of the data while ignoring the details or do we prefer models
# that closely follow the high density regions of the signal?
#
# The last two panels show how we can sample from the last two models. The
# resulting samples distributions do not look exactly like the original data
# distribution. The difference primarily stems from the approximation error we
# made by using a model that assumes that the data was generated by a finite
# number of Gaussian components instead of a continuous noisy sine curve.
#
# +
import itertools
import numpy as np
from scipy import linalg
import matplotlib.pyplot as plt
import matplotlib as mpl
from sklearn import mixture
print(__doc__)
color_iter = itertools.cycle(['navy', 'c', 'cornflowerblue', 'gold',
'darkorange'])
def plot_results(X, Y, means, covariances, index, title):
splot = plt.subplot(5, 1, 1 + index)
for i, (mean, covar, color) in enumerate(zip(
means, covariances, color_iter)):
v, w = linalg.eigh(covar)
v = 2. * np.sqrt(2.) * np.sqrt(v)
u = w[0] / linalg.norm(w[0])
# as the DP will not use every component it has access to
# unless it needs it, we shouldn't plot the redundant
# components.
if not np.any(Y == i):
continue
plt.scatter(X[Y == i, 0], X[Y == i, 1], .8, color=color)
# Plot an ellipse to show the Gaussian component
angle = np.arctan(u[1] / u[0])
angle = 180. * angle / np.pi # convert to degrees
ell = mpl.patches.Ellipse(mean, v[0], v[1], 180. + angle, color=color)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.5)
splot.add_artist(ell)
plt.xlim(-6., 4. * np.pi - 6.)
plt.ylim(-5., 5.)
plt.title(title)
plt.xticks(())
plt.yticks(())
def plot_samples(X, Y, n_components, index, title):
plt.subplot(5, 1, 4 + index)
for i, color in zip(range(n_components), color_iter):
# as the DP will not use every component it has access to
# unless it needs it, we shouldn't plot the redundant
# components.
if not np.any(Y == i):
continue
plt.scatter(X[Y == i, 0], X[Y == i, 1], .8, color=color)
plt.xlim(-6., 4. * np.pi - 6.)
plt.ylim(-5., 5.)
plt.title(title)
plt.xticks(())
plt.yticks(())
# Parameters
n_samples = 100
# Generate random sample following a sine curve
np.random.seed(0)
X = np.zeros((n_samples, 2))
step = 4. * np.pi / n_samples
for i in range(X.shape[0]):
x = i * step - 6.
X[i, 0] = x + np.random.normal(0, 0.1)
X[i, 1] = 3. * (np.sin(x) + np.random.normal(0, .2))
plt.figure(figsize=(10, 10))
plt.subplots_adjust(bottom=.04, top=0.95, hspace=.2, wspace=.05,
left=.03, right=.97)
# Fit a Gaussian mixture with EM using ten components
gmm = mixture.GaussianMixture(n_components=10, covariance_type='full',
max_iter=100).fit(X)
plot_results(X, gmm.predict(X), gmm.means_, gmm.covariances_, 0,
'Expectation-maximization')
dpgmm = mixture.BayesianGaussianMixture(
n_components=10, covariance_type='full', weight_concentration_prior=1e-2,
weight_concentration_prior_type='dirichlet_process',
mean_precision_prior=1e-2, covariance_prior=1e0 * np.eye(2),
init_params="random", max_iter=100, random_state=2).fit(X)
plot_results(X, dpgmm.predict(X), dpgmm.means_, dpgmm.covariances_, 1,
"Bayesian Gaussian mixture models with a Dirichlet process prior "
r"for $\gamma_0=0.01$.")
X_s, y_s = dpgmm.sample(n_samples=2000)
plot_samples(X_s, y_s, dpgmm.n_components, 0,
"Gaussian mixture with a Dirichlet process prior "
r"for $\gamma_0=0.01$ sampled with $2000$ samples.")
dpgmm = mixture.BayesianGaussianMixture(
n_components=10, covariance_type='full', weight_concentration_prior=1e+2,
weight_concentration_prior_type='dirichlet_process',
mean_precision_prior=1e-2, covariance_prior=1e0 * np.eye(2),
init_params="kmeans", max_iter=100, random_state=2).fit(X)
plot_results(X, dpgmm.predict(X), dpgmm.means_, dpgmm.covariances_, 2,
"Bayesian Gaussian mixture models with a Dirichlet process prior "
r"for $\gamma_0=100$")
X_s, y_s = dpgmm.sample(n_samples=2000)
plot_samples(X_s, y_s, dpgmm.n_components, 1,
"Gaussian mixture with a Dirichlet process prior "
r"for $\gamma_0=100$ sampled with $2000$ samples.")
plt.show()
| sklearn/sklearn learning/demonstration/auto_examples_jupyter/mixture/plot_gmm_sin.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: TensorFlow 2.4 on Python 3.8 & CUDA 11.1
# language: python
# name: python3
# ---
# **12장 – 텐서플로를 사용한 사용자 정의 모델과 훈련**
# _이 노트북은 12장에 있는 모든 샘플 코드와 연습문제 해답을 가지고 있습니다._
# <table align="left">
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/rickiepark/handson-ml2/blob/master/12_custom_models_and_training_with_tensorflow.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />구글 코랩에서 실행하기</a>
# </td>
# </table>
# # 설정
# 먼저 몇 개의 모듈을 임포트합니다. 맷플롯립 그래프를 인라인으로 출력하도록 만들고 그림을 저장하는 함수를 준비합니다. 또한 파이썬 버전이 3.5 이상인지 확인합니다(파이썬 2.x에서도 동작하지만 곧 지원이 중단되므로 파이썬 3을 사용하는 것이 좋습니다). 사이킷런 버전이 0.20 이상인지와 텐서플로 버전이 2.0 이상인지 확인합니다.
# +
# 파이썬 ≥3.5 필수
import sys
assert sys.version_info >= (3, 5)
# 사이킷런 ≥0.20 필수
import sklearn
assert sklearn.__version__ >= "0.20"
try:
# # %tensorflow_version은 코랩 명령입니다.
# %tensorflow_version 2.x
except Exception:
pass
# 이 노트북은 텐서플로 ≥2.4이 필요합니다
# 2.x 버전은 대부분 동일한 결과를 만들지만 몇 가지 버그가 있습니다.
import tensorflow as tf
from tensorflow import keras
assert tf.__version__ >= "2.4"
# 공통 모듈 임포트
import numpy as np
import os
# 노트북 실행 결과를 동일하게 유지하기 위해
np.random.seed(42)
tf.random.set_seed(42)
# 깔끔한 그래프 출력을 위해
# %matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# 그림을 저장할 위치
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "deep"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("그림 저장:", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# -
# ## 텐서와 연산
# ### 텐서
tf.constant([[1., 2., 3.], [4., 5., 6.]]) # 행렬
tf.constant(42) # 스칼라
t = tf.constant([[1., 2., 3.], [4., 5., 6.]])
t
t.shape
t.dtype
# ### 인덱싱
t[:, 1:]
t[..., 1, tf.newaxis]
# ### 연산
t + 10
tf.square(t)
t @ tf.transpose(t)
# ### `keras.backend` 사용하기
from tensorflow import keras
K = keras.backend
K.square(K.transpose(t)) + 10
# ### 넘파이 변환
a = np.array([2., 4., 5.])
tf.constant(a)
t.numpy()
np.array(t)
tf.square(a)
np.square(t)
# ### 타입 변환
try:
tf.constant(2.0) + tf.constant(40)
except tf.errors.InvalidArgumentError as ex:
print(ex)
try:
tf.constant(2.0) + tf.constant(40., dtype=tf.float64)
except tf.errors.InvalidArgumentError as ex:
print(ex)
t2 = tf.constant(40., dtype=tf.float64)
tf.constant(2.0) + tf.cast(t2, tf.float32)
# ### 문자열
tf.constant(b"hello world")
tf.constant("café")
u = tf.constant([ord(c) for c in "café"])
u
b = tf.strings.unicode_encode(u, "UTF-8")
tf.strings.length(b, unit="UTF8_CHAR")
tf.strings.unicode_decode(b, "UTF-8")
# ### 문자열 배열
p = tf.constant(["Café", "Coffee", "caffè", "咖啡"])
tf.strings.length(p, unit="UTF8_CHAR")
r = tf.strings.unicode_decode(p, "UTF8")
r
print(r)
# ### 래그드 텐서
print(r[1])
print(r[1:3])
r2 = tf.ragged.constant([[65, 66], [], [67]])
print(tf.concat([r, r2], axis=0))
r3 = tf.ragged.constant([[68, 69, 70], [71], [], [72, 73]])
print(tf.concat([r, r3], axis=1))
tf.strings.unicode_encode(r3, "UTF-8")
r.to_tensor()
# ### 희소 텐서
s = tf.SparseTensor(indices=[[0, 1], [1, 0], [2, 3]],
values=[1., 2., 3.],
dense_shape=[3, 4])
print(s)
tf.sparse.to_dense(s)
s2 = s * 2.0
try:
s3 = s + 1.
except TypeError as ex:
print(ex)
s4 = tf.constant([[10., 20.], [30., 40.], [50., 60.], [70., 80.]])
tf.sparse.sparse_dense_matmul(s, s4)
s5 = tf.SparseTensor(indices=[[0, 2], [0, 1]],
values=[1., 2.],
dense_shape=[3, 4])
print(s5)
try:
tf.sparse.to_dense(s5)
except tf.errors.InvalidArgumentError as ex:
print(ex)
s6 = tf.sparse.reorder(s5)
tf.sparse.to_dense(s6)
# ### 집합
set1 = tf.constant([[2, 3, 5, 7], [7, 9, 0, 0]])
set2 = tf.constant([[4, 5, 6], [9, 10, 0]])
tf.sparse.to_dense(tf.sets.union(set1, set2))
tf.sparse.to_dense(tf.sets.difference(set1, set2))
tf.sparse.to_dense(tf.sets.intersection(set1, set2))
# ### 변수
v = tf.Variable([[1., 2., 3.], [4., 5., 6.]])
v.assign(2 * v)
v[0, 1].assign(42)
v[:, 2].assign([0., 1.])
try:
v[1] = [7., 8., 9.]
except TypeError as ex:
print(ex)
v.scatter_nd_update(indices=[[0, 0], [1, 2]],
updates=[100., 200.])
sparse_delta = tf.IndexedSlices(values=[[1., 2., 3.], [4., 5., 6.]],
indices=[1, 0])
v.scatter_update(sparse_delta)
# ### 텐서 배열
array = tf.TensorArray(dtype=tf.float32, size=3)
array = array.write(0, tf.constant([1., 2.]))
array = array.write(1, tf.constant([3., 10.]))
array = array.write(2, tf.constant([5., 7.]))
array.read(1)
array.stack()
mean, variance = tf.nn.moments(array.stack(), axes=0)
mean
variance
# ## 사용자 정의 손실 함수
# 캘리포니아 주택 데이터셋을 로드하여 준비해 보겠습니다. 먼저 이 데이터셋을 로드한 다음 훈련 세트, 검증 세트, 테스트 세트로 나눕니다. 마지막으로 스케일을 변경합니다:
# +
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
housing = fetch_california_housing()
X_train_full, X_test, y_train_full, y_test = train_test_split(
housing.data, housing.target.reshape(-1, 1), random_state=42)
X_train, X_valid, y_train, y_valid = train_test_split(
X_train_full, y_train_full, random_state=42)
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_valid_scaled = scaler.transform(X_valid)
X_test_scaled = scaler.transform(X_test)
# -
def huber_fn(y_true, y_pred):
error = y_true - y_pred
is_small_error = tf.abs(error) < 1
squared_loss = tf.square(error) / 2
linear_loss = tf.abs(error) - 0.5
return tf.where(is_small_error, squared_loss, linear_loss)
plt.figure(figsize=(8, 3.5))
z = np.linspace(-4, 4, 200)
plt.plot(z, huber_fn(0, z), "b-", linewidth=2, label="huber($z$)")
plt.plot(z, z**2 / 2, "b:", linewidth=1, label=r"$\frac{1}{2}z^2$")
plt.plot([-1, -1], [0, huber_fn(0., -1.)], "r--")
plt.plot([1, 1], [0, huber_fn(0., 1.)], "r--")
plt.gca().axhline(y=0, color='k')
plt.gca().axvline(x=0, color='k')
plt.axis([-4, 4, 0, 4])
plt.grid(True)
plt.xlabel("$z$")
plt.legend(fontsize=14)
plt.title("Huber loss", fontsize=14)
plt.show()
# +
input_shape = X_train.shape[1:]
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1),
])
# -
model.compile(loss=huber_fn, optimizer="nadam", metrics=["mae"])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
# ## 사용자 정의 요소를 가진 모델을 저장하고 로드하기
model.save("my_model_with_a_custom_loss.h5")
model = keras.models.load_model("my_model_with_a_custom_loss.h5",
custom_objects={"huber_fn": huber_fn})
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
def create_huber(threshold=1.0):
def huber_fn(y_true, y_pred):
error = y_true - y_pred
is_small_error = tf.abs(error) < threshold
squared_loss = tf.square(error) / 2
linear_loss = threshold * tf.abs(error) - threshold**2 / 2
return tf.where(is_small_error, squared_loss, linear_loss)
return huber_fn
model.compile(loss=create_huber(2.0), optimizer="nadam", metrics=["mae"])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.save("my_model_with_a_custom_loss_threshold_2.h5")
model = keras.models.load_model("my_model_with_a_custom_loss_threshold_2.h5",
custom_objects={"huber_fn": create_huber(2.0)})
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
class HuberLoss(keras.losses.Loss):
def __init__(self, threshold=1.0, **kwargs):
self.threshold = threshold
super().__init__(**kwargs)
def call(self, y_true, y_pred):
error = y_true - y_pred
is_small_error = tf.abs(error) < self.threshold
squared_loss = tf.square(error) / 2
linear_loss = self.threshold * tf.abs(error) - self.threshold**2 / 2
return tf.where(is_small_error, squared_loss, linear_loss)
def get_config(self):
base_config = super().get_config()
return {**base_config, "threshold": self.threshold}
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1),
])
model.compile(loss=HuberLoss(2.), optimizer="nadam", metrics=["mae"])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.save("my_model_with_a_custom_loss_class.h5")
model = keras.models.load_model("my_model_with_a_custom_loss_class.h5",
custom_objects={"HuberLoss": HuberLoss})
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.loss.threshold
# ## 그외 사용자 정의 함수
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
# +
def my_softplus(z): # tf.nn.softplus(z) 값을 반환합니다
return tf.math.log(tf.exp(z) + 1.0)
def my_glorot_initializer(shape, dtype=tf.float32):
stddev = tf.sqrt(2. / (shape[0] + shape[1]))
return tf.random.normal(shape, stddev=stddev, dtype=dtype)
def my_l1_regularizer(weights):
return tf.reduce_sum(tf.abs(0.01 * weights))
def my_positive_weights(weights): # tf.nn.relu(weights) 값을 반환합니다
return tf.where(weights < 0., tf.zeros_like(weights), weights)
# -
layer = keras.layers.Dense(1, activation=my_softplus,
kernel_initializer=my_glorot_initializer,
kernel_regularizer=my_l1_regularizer,
kernel_constraint=my_positive_weights)
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1, activation=my_softplus,
kernel_regularizer=my_l1_regularizer,
kernel_constraint=my_positive_weights,
kernel_initializer=my_glorot_initializer),
])
model.compile(loss="mse", optimizer="nadam", metrics=["mae"])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.save("my_model_with_many_custom_parts.h5")
model = keras.models.load_model(
"my_model_with_many_custom_parts.h5",
custom_objects={
"my_l1_regularizer": my_l1_regularizer,
"my_positive_weights": my_positive_weights,
"my_glorot_initializer": my_glorot_initializer,
"my_softplus": my_softplus,
})
class MyL1Regularizer(keras.regularizers.Regularizer):
def __init__(self, factor):
self.factor = factor
def __call__(self, weights):
return tf.reduce_sum(tf.abs(self.factor * weights))
def get_config(self):
return {"factor": self.factor}
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1, activation=my_softplus,
kernel_regularizer=MyL1Regularizer(0.01),
kernel_constraint=my_positive_weights,
kernel_initializer=my_glorot_initializer),
])
model.compile(loss="mse", optimizer="nadam", metrics=["mae"])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.save("my_model_with_many_custom_parts.h5")
model = keras.models.load_model(
"my_model_with_many_custom_parts.h5",
custom_objects={
"MyL1Regularizer": MyL1Regularizer,
"my_positive_weights": my_positive_weights,
"my_glorot_initializer": my_glorot_initializer,
"my_softplus": my_softplus,
})
# ## 사용자 정의 지표
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1),
])
model.compile(loss="mse", optimizer="nadam", metrics=[create_huber(2.0)])
model.fit(X_train_scaled, y_train, epochs=2)
# **노트**: 손실과 지표에 같은 함수를 사용하면 다른 결과가 나올 수 있습니다. 이는 일반적으로 부동 소수점 정밀도 오차 때문입니다. 수학 식이 동일하더라도 연산은 동일한 순서대로 실행되지 않습니다. 이로 인해 작은 차이가 발생합니다. 또한 샘플 가중치를 사용하면 정밀도보다 더 큰 오차가 생깁니다:
#
# * 에포크에서 손실은 지금까지 본 모든 배치 손실의 평균입니다. 각 배치 손실은 가중치가 적용된 샘플 손실의 합을 _배치 크기_ 로 나눈 것입니다(샘플 가중치의 합으로 나눈 것이 아닙니다. 따라서 배치 손실은 손실의 가중 평균이 아닙니다).
# * 에포크에서 지표는 가중치가 적용된 샘플 손실의 합을 지금까지 본 모든 샘플 가중치의 합으로 나눈 것입니다. 다른 말로하면 모든 샘플 손실의 가중 평균입니다. 따라서 위와 같지 않습니다.
#
# 수학적으로 말하면 손실 = 지표 * 샘플 가중치의 평균(더하기 약간의 부동 소수점 정밀도 오차)입니다.
model.compile(loss=create_huber(2.0), optimizer="nadam", metrics=[create_huber(2.0)])
sample_weight = np.random.rand(len(y_train))
history = model.fit(X_train_scaled, y_train, epochs=2, sample_weight=sample_weight)
history.history["loss"][0], history.history["huber_fn"][0] * sample_weight.mean()
# ### 스트리밍 지표
precision = keras.metrics.Precision()
precision([0, 1, 1, 1, 0, 1, 0, 1], [1, 1, 0, 1, 0, 1, 0, 1])
precision([0, 1, 0, 0, 1, 0, 1, 1], [1, 0, 1, 1, 0, 0, 0, 0])
precision.result()
precision.variables
precision.reset_states()
# 스트리밍 지표 만들기:
class HuberMetric(keras.metrics.Metric):
def __init__(self, threshold=1.0, **kwargs):
super().__init__(**kwargs) # 기본 매개변수 처리 (예를 들면, dtype)
self.threshold = threshold
self.huber_fn = create_huber(threshold)
self.total = self.add_weight("total", initializer="zeros")
self.count = self.add_weight("count", initializer="zeros")
def update_state(self, y_true, y_pred, sample_weight=None):
metric = self.huber_fn(y_true, y_pred)
self.total.assign_add(tf.reduce_sum(metric))
self.count.assign_add(tf.cast(tf.size(y_true), tf.float32))
def result(self):
return self.total / self.count
def get_config(self):
base_config = super().get_config()
return {**base_config, "threshold": self.threshold}
# +
m = HuberMetric(2.)
# total = 2 * |10 - 2| - 2²/2 = 14
# count = 1
# result = 14 / 1 = 14
m(tf.constant([[2.]]), tf.constant([[10.]]))
# +
# total = total + (|1 - 0|² / 2) + (2 * |9.25 - 5| - 2² / 2) = 14 + 7 = 21
# count = count + 2 = 3
# result = total / count = 21 / 3 = 7
m(tf.constant([[0.], [5.]]), tf.constant([[1.], [9.25]]))
m.result()
# -
m.variables
m.reset_states()
m.variables
# `HuberMetric` 클래스가 잘 동작하는지 확인해 보죠:
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1),
])
model.compile(loss=create_huber(2.0), optimizer="nadam", metrics=[HuberMetric(2.0)])
model.fit(X_train_scaled.astype(np.float32), y_train.astype(np.float32), epochs=2)
model.save("my_model_with_a_custom_metric.h5")
model = keras.models.load_model("my_model_with_a_custom_metric.h5",
custom_objects={"huber_fn": create_huber(2.0),
"HuberMetric": HuberMetric})
model.fit(X_train_scaled.astype(np.float32), y_train.astype(np.float32), epochs=2)
# **경고**: 텐서플로 2.2에서 tf.keras가 `model.metrics`의 0번째 위치에 지표를 추가합니다([텐서플로 이슈 #38150](https://github.com/tensorflow/tensorflow/issues/38150) 참조). 따라서 `HuberMetric`에 접근하려면 `model.metrics[0]` 대신 `model.metrics[-1]`를 사용해야 합니다.
model.metrics[-1].threshold
# 잘 동작하는군요! 다음처럼 더 간단하게 클래스를 만들 수 있습니다:
class HuberMetric(keras.metrics.Mean):
def __init__(self, threshold=1.0, name='HuberMetric', dtype=None):
self.threshold = threshold
self.huber_fn = create_huber(threshold)
super().__init__(name=name, dtype=dtype)
def update_state(self, y_true, y_pred, sample_weight=None):
metric = self.huber_fn(y_true, y_pred)
super(HuberMetric, self).update_state(metric, sample_weight)
def get_config(self):
base_config = super().get_config()
return {**base_config, "threshold": self.threshold}
# 이 클래스는 크기를 잘 처리하고 샘플 가중치도 지원합니다.
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="selu", kernel_initializer="lecun_normal",
input_shape=input_shape),
keras.layers.Dense(1),
])
model.compile(loss=keras.losses.Huber(2.0), optimizer="nadam", weighted_metrics=[HuberMetric(2.0)])
sample_weight = np.random.rand(len(y_train))
history = model.fit(X_train_scaled.astype(np.float32), y_train.astype(np.float32),
epochs=2, sample_weight=sample_weight)
history.history["loss"][0], history.history["HuberMetric"][0] * sample_weight.mean()
model.save("my_model_with_a_custom_metric_v2.h5")
model = keras.models.load_model("my_model_with_a_custom_metric_v2.h5",
custom_objects={"HuberMetric": HuberMetric})
model.fit(X_train_scaled.astype(np.float32), y_train.astype(np.float32), epochs=2)
model.metrics[-1].threshold
# ## 사용자 정의 층
exponential_layer = keras.layers.Lambda(lambda x: tf.exp(x))
exponential_layer([-1., 0., 1.])
# 회귀 모델이 예측할 값이 양수이고 스케일이 매우 다른 경우 (예를 들어, 0.001, 10., 10000) 출력층에 지수 함수를 추가하면 유용할 수 있습니다:
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="relu", input_shape=input_shape),
keras.layers.Dense(1),
exponential_layer
])
model.compile(loss="mse", optimizer="sgd")
model.fit(X_train_scaled, y_train, epochs=5,
validation_data=(X_valid_scaled, y_valid))
model.evaluate(X_test_scaled, y_test)
class MyDense(keras.layers.Layer):
def __init__(self, units, activation=None, **kwargs):
super().__init__(**kwargs)
self.units = units
self.activation = keras.activations.get(activation)
def build(self, batch_input_shape):
self.kernel = self.add_weight(
name="kernel", shape=[batch_input_shape[-1], self.units],
initializer="glorot_normal")
self.bias = self.add_weight(
name="bias", shape=[self.units], initializer="zeros")
super().build(batch_input_shape) # must be at the end
def call(self, X):
return self.activation(X @ self.kernel + self.bias)
def compute_output_shape(self, batch_input_shape):
return tf.TensorShape(batch_input_shape.as_list()[:-1] + [self.units])
def get_config(self):
base_config = super().get_config()
return {**base_config, "units": self.units,
"activation": keras.activations.serialize(self.activation)}
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
MyDense(30, activation="relu", input_shape=input_shape),
MyDense(1)
])
model.compile(loss="mse", optimizer="nadam")
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.evaluate(X_test_scaled, y_test)
model.save("my_model_with_a_custom_layer.h5")
model = keras.models.load_model("my_model_with_a_custom_layer.h5",
custom_objects={"MyDense": MyDense})
class MyMultiLayer(keras.layers.Layer):
def call(self, X):
X1, X2 = X
print("X1.shape: ", X1.shape ," X2.shape: ", X2.shape) # 사용자 정의 층 디버깅
return X1 + X2, X1 * X2
def compute_output_shape(self, batch_input_shape):
batch_input_shape1, batch_input_shape2 = batch_input_shape
return [batch_input_shape1, batch_input_shape2]
# 사용자 정의 층은 다음처럼 함수형 API를 사용해 호출할 수 있습니다:
inputs1 = keras.layers.Input(shape=[2])
inputs2 = keras.layers.Input(shape=[2])
outputs1, outputs2 = MyMultiLayer()((inputs1, inputs2))
# `call()` 메서드는 심볼릭 입력을 받습니다. 이 입력의 크기는 부분적으로만 지정되어 있습니다(이 시점에서는 배치 크기를 모릅니다. 그래서 첫 번째 차원이 None입니다):
#
# 사용자 층에 실제 데이터를 전달할 수도 있습니다. 이를 테스트하기 위해 각 데이터셋의 입력을 각각 네 개의 특성을 가진 두 부분으로 나누겠습니다:
# +
def split_data(data):
columns_count = data.shape[-1]
half = columns_count // 2
return data[:, :half], data[:, half:]
X_train_scaled_A, X_train_scaled_B = split_data(X_train_scaled)
X_valid_scaled_A, X_valid_scaled_B = split_data(X_valid_scaled)
X_test_scaled_A, X_test_scaled_B = split_data(X_test_scaled)
# 분할된 데이터 크기 출력
X_train_scaled_A.shape, X_train_scaled_B.shape
# -
# 크기가 완전하게 지정된 것을 볼 수 있습니다:
outputs1, outputs2 = MyMultiLayer()((X_train_scaled_A, X_train_scaled_B))
# 함수형 API를 사용해 완전한 모델을 만들어 보겠습니다(이 모델은 간단한 예제이므로 놀라운 성능을 기대하지 마세요):
# +
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
input_A = keras.layers.Input(shape=X_train_scaled_A.shape[-1])
input_B = keras.layers.Input(shape=X_train_scaled_B.shape[-1])
hidden_A, hidden_B = MyMultiLayer()((input_A, input_B))
hidden_A = keras.layers.Dense(30, activation='selu')(hidden_A)
hidden_B = keras.layers.Dense(30, activation='selu')(hidden_B)
concat = keras.layers.Concatenate()((hidden_A, hidden_B))
output = keras.layers.Dense(1)(concat)
model = keras.models.Model(inputs=[input_A, input_B], outputs=[output])
# -
model.compile(loss='mse', optimizer='nadam')
model.fit((X_train_scaled_A, X_train_scaled_B), y_train, epochs=2,
validation_data=((X_valid_scaled_A, X_valid_scaled_B), y_valid))
# 훈련과 테스트에서 다르게 동작하는 층을 만들어 보죠:
class AddGaussianNoise(keras.layers.Layer):
def __init__(self, stddev, **kwargs):
super().__init__(**kwargs)
self.stddev = stddev
def call(self, X, training=None):
if training:
noise = tf.random.normal(tf.shape(X), stddev=self.stddev)
return X + noise
else:
return X
def compute_output_shape(self, batch_input_shape):
return batch_input_shape
# 다음은 사용자 정의 층을 사용하는 간단한 모델입니다:
# +
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
AddGaussianNoise(stddev=1.0),
keras.layers.Dense(30, activation="selu"),
keras.layers.Dense(1)
])
# -
model.compile(loss="mse", optimizer="nadam")
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.evaluate(X_test_scaled, y_test)
# ## 사용자 정의 모델
X_new_scaled = X_test_scaled
class ResidualBlock(keras.layers.Layer):
def __init__(self, n_layers, n_neurons, **kwargs):
super().__init__(**kwargs)
self.hidden = [keras.layers.Dense(n_neurons, activation="elu",
kernel_initializer="he_normal")
for _ in range(n_layers)]
def call(self, inputs):
Z = inputs
for layer in self.hidden:
Z = layer(Z)
return inputs + Z
class ResidualRegressor(keras.models.Model):
def __init__(self, output_dim, **kwargs):
super().__init__(**kwargs)
self.hidden1 = keras.layers.Dense(30, activation="elu",
kernel_initializer="he_normal")
self.block1 = ResidualBlock(2, 30)
self.block2 = ResidualBlock(2, 30)
self.out = keras.layers.Dense(output_dim)
def call(self, inputs):
Z = self.hidden1(inputs)
for _ in range(1 + 3):
Z = self.block1(Z)
Z = self.block2(Z)
return self.out(Z)
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = ResidualRegressor(1)
model.compile(loss="mse", optimizer="nadam")
history = model.fit(X_train_scaled, y_train, epochs=5)
score = model.evaluate(X_test_scaled, y_test)
y_pred = model.predict(X_new_scaled)
model.save("my_custom_model.ckpt")
model = keras.models.load_model("my_custom_model.ckpt")
history = model.fit(X_train_scaled, y_train, epochs=5)
# 대신 시퀀셜 API를 사용하는 모델을 정의할 수 있습니다:
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
block1 = ResidualBlock(2, 30)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="elu", kernel_initializer="he_normal"),
block1, block1, block1, block1,
ResidualBlock(2, 30),
keras.layers.Dense(1)
])
model.compile(loss="mse", optimizer="nadam")
history = model.fit(X_train_scaled, y_train, epochs=5)
score = model.evaluate(X_test_scaled, y_test)
y_pred = model.predict(X_new_scaled)
# ## 모델 구성 요소에 기반한 손실과 지표
# **노트**: TF 2.2에 있는 이슈([#46858](https://github.com/tensorflow/tensorflow/issues/46858)) 때문에 `build()` 메서드와 함께 `add_loss()`를 사용할 수 없습니다. 따라서 다음 코드는 책과 다릅니다. `build()` 메서드 대신 생성자에 `reconstruct` 층을 만듭니다. 이 때문에 이 층의 유닛 개수를 하드코딩해야 합니다(또는 생성자 매개변수로 전달해야 합니다).
# +
class ReconstructingRegressor(keras.models.Model):
def __init__(self, output_dim, **kwargs):
super().__init__(**kwargs)
self.hidden = [keras.layers.Dense(30, activation="selu",
kernel_initializer="lecun_normal")
for _ in range(5)]
self.out = keras.layers.Dense(output_dim)
self.reconstruct = keras.layers.Dense(8) # TF 이슈 #46858에 대한 대책
self.reconstruction_mean = keras.metrics.Mean(name="reconstruction_error")
# TF 이슈 #46858 때문에 주석 처리
# def build(self, batch_input_shape):
# n_inputs = batch_input_shape[-1]
# self.reconstruct = keras.layers.Dense(n_inputs, name='recon')
# super().build(batch_input_shape)
def call(self, inputs, training=None):
Z = inputs
for layer in self.hidden:
Z = layer(Z)
reconstruction = self.reconstruct(Z)
self.recon_loss = 0.05 * tf.reduce_mean(tf.square(reconstruction - inputs))
if training:
result = self.reconstruction_mean(recon_loss)
self.add_metric(result)
return self.out(Z)
def train_step(self, data):
x, y = data
with tf.GradientTape() as tape:
y_pred = self(x)
loss = self.compiled_loss(y, y_pred, regularization_losses=[self.recon_loss])
gradients = tape.gradient(loss, self.trainable_variables)
self.optimizer.apply_gradients(zip(gradients, self.trainable_variables))
return {m.name: m.result() for m in self.metrics}
# -
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = ReconstructingRegressor(1)
model.compile(loss="mse", optimizer="nadam")
history = model.fit(X_train_scaled, y_train, epochs=2)
y_pred = model.predict(X_test_scaled)
# ## 자동 미분을 사용하여 그레이디언트 계산하기
def f(w1, w2):
return 3 * w1 ** 2 + 2 * w1 * w2
w1, w2 = 5, 3
eps = 1e-6
(f(w1 + eps, w2) - f(w1, w2)) / eps
(f(w1, w2 + eps) - f(w1, w2)) / eps
# +
w1, w2 = tf.Variable(5.), tf.Variable(3.)
with tf.GradientTape() as tape:
z = f(w1, w2)
gradients = tape.gradient(z, [w1, w2])
# -
gradients
# +
with tf.GradientTape() as tape:
z = f(w1, w2)
dz_dw1 = tape.gradient(z, w1)
try:
dz_dw2 = tape.gradient(z, w2)
except RuntimeError as ex:
print(ex)
# +
with tf.GradientTape(persistent=True) as tape:
z = f(w1, w2)
dz_dw1 = tape.gradient(z, w1)
dz_dw2 = tape.gradient(z, w2) # works now!
del tape
# -
dz_dw1, dz_dw2
# +
c1, c2 = tf.constant(5.), tf.constant(3.)
with tf.GradientTape() as tape:
z = f(c1, c2)
gradients = tape.gradient(z, [c1, c2])
# -
gradients
# +
with tf.GradientTape() as tape:
tape.watch(c1)
tape.watch(c2)
z = f(c1, c2)
gradients = tape.gradient(z, [c1, c2])
# -
gradients
# +
with tf.GradientTape() as tape:
z1 = f(w1, w2 + 2.)
z2 = f(w1, w2 + 5.)
z3 = f(w1, w2 + 7.)
tape.gradient([z1, z2, z3], [w1, w2])
# +
with tf.GradientTape(persistent=True) as tape:
z1 = f(w1, w2 + 2.)
z2 = f(w1, w2 + 5.)
z3 = f(w1, w2 + 7.)
tf.reduce_sum(tf.stack([tape.gradient(z, [w1, w2]) for z in (z1, z2, z3)]), axis=0)
del tape
# -
with tf.GradientTape(persistent=True) as hessian_tape:
with tf.GradientTape() as jacobian_tape:
z = f(w1, w2)
jacobians = jacobian_tape.gradient(z, [w1, w2])
hessians = [hessian_tape.gradient(jacobian, [w1, w2])
for jacobian in jacobians]
del hessian_tape
jacobians
hessians
# +
def f(w1, w2):
return 3 * w1 ** 2 + tf.stop_gradient(2 * w1 * w2)
with tf.GradientTape() as tape:
z = f(w1, w2)
tape.gradient(z, [w1, w2])
# +
x = tf.Variable(100.)
with tf.GradientTape() as tape:
z = my_softplus(x)
tape.gradient(z, [x])
# -
tf.math.log(tf.exp(tf.constant(30., dtype=tf.float32)) + 1.)
# +
x = tf.Variable([100.])
with tf.GradientTape() as tape:
z = my_softplus(x)
tape.gradient(z, [x])
# -
@tf.custom_gradient
def my_better_softplus(z):
exp = tf.exp(z)
def my_softplus_gradients(grad):
return grad / (1 + 1 / exp)
return tf.math.log(exp + 1), my_softplus_gradients
def my_better_softplus(z):
return tf.where(z > 30., z, tf.math.log(tf.exp(z) + 1.))
# +
x = tf.Variable([1000.])
with tf.GradientTape() as tape:
z = my_better_softplus(x)
z, tape.gradient(z, [x])
# -
# # 사용자 정의 훈련 반복
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
l2_reg = keras.regularizers.l2(0.05)
model = keras.models.Sequential([
keras.layers.Dense(30, activation="elu", kernel_initializer="he_normal",
kernel_regularizer=l2_reg),
keras.layers.Dense(1, kernel_regularizer=l2_reg)
])
def random_batch(X, y, batch_size=32):
idx = np.random.randint(len(X), size=batch_size)
return X[idx], y[idx]
def print_status_bar(iteration, total, loss, metrics=None):
metrics = " - ".join(["{}: {:.4f}".format(m.name, m.result())
for m in [loss] + (metrics or [])])
end = "" if iteration < total else "\n"
print("\r{}/{} - ".format(iteration, total) + metrics,
end=end)
# +
import time
mean_loss = keras.metrics.Mean(name="loss")
mean_square = keras.metrics.Mean(name="mean_square")
for i in range(1, 50 + 1):
loss = 1 / i
mean_loss(loss)
mean_square(i ** 2)
print_status_bar(i, 50, mean_loss, [mean_square])
time.sleep(0.05)
# -
# A fancier version with a progress bar:
def progress_bar(iteration, total, size=30):
running = iteration < total
c = ">" if running else "="
p = (size - 1) * iteration // total
fmt = "{{:-{}d}}/{{}} [{{}}]".format(len(str(total)))
params = [iteration, total, "=" * p + c + "." * (size - p - 1)]
return fmt.format(*params)
progress_bar(3500, 10000, size=6)
def print_status_bar(iteration, total, loss, metrics=None, size=30):
metrics = " - ".join(["{}: {:.4f}".format(m.name, m.result())
for m in [loss] + (metrics or [])])
end = "" if iteration < total else "\n"
print("\r{} - {}".format(progress_bar(iteration, total), metrics), end=end)
mean_loss = keras.metrics.Mean(name="loss")
mean_square = keras.metrics.Mean(name="mean_square")
for i in range(1, 50 + 1):
loss = 1 / i
mean_loss(loss)
mean_square(i ** 2)
print_status_bar(i, 50, mean_loss, [mean_square])
time.sleep(0.05)
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
n_epochs = 5
batch_size = 32
n_steps = len(X_train) // batch_size
optimizer = keras.optimizers.Nadam(lr=0.01)
loss_fn = keras.losses.mean_squared_error
mean_loss = keras.metrics.Mean()
metrics = [keras.metrics.MeanAbsoluteError()]
for epoch in range(1, n_epochs + 1):
print("Epoch {}/{}".format(epoch, n_epochs))
for step in range(1, n_steps + 1):
X_batch, y_batch = random_batch(X_train_scaled, y_train)
with tf.GradientTape() as tape:
y_pred = model(X_batch)
main_loss = tf.reduce_mean(loss_fn(y_batch, y_pred))
loss = tf.add_n([main_loss] + model.losses)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
for variable in model.variables:
if variable.constraint is not None:
variable.assign(variable.constraint(variable))
mean_loss(loss)
for metric in metrics:
metric(y_batch, y_pred)
print_status_bar(step * batch_size, len(y_train), mean_loss, metrics)
print_status_bar(len(y_train), len(y_train), mean_loss, metrics)
for metric in [mean_loss] + metrics:
metric.reset_states()
try:
from tqdm.notebook import trange
from collections import OrderedDict
with trange(1, n_epochs + 1, desc="All epochs") as epochs:
for epoch in epochs:
with trange(1, n_steps + 1, desc="Epoch {}/{}".format(epoch, n_epochs)) as steps:
for step in steps:
X_batch, y_batch = random_batch(X_train_scaled, y_train)
with tf.GradientTape() as tape:
y_pred = model(X_batch)
main_loss = tf.reduce_mean(loss_fn(y_batch, y_pred))
loss = tf.add_n([main_loss] + model.losses)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
for variable in model.variables:
if variable.constraint is not None:
variable.assign(variable.constraint(variable))
status = OrderedDict()
mean_loss(loss)
status["loss"] = mean_loss.result().numpy()
for metric in metrics:
metric(y_batch, y_pred)
status[metric.name] = metric.result().numpy()
steps.set_postfix(status)
for metric in [mean_loss] + metrics:
metric.reset_states()
except ImportError as ex:
print("To run this cell, please install tqdm, ipywidgets and restart Jupyter")
# ## 텐서플로 함수
def cube(x):
return x ** 3
cube(2)
cube(tf.constant(2.0))
tf_cube = tf.function(cube)
tf_cube
tf_cube(2)
tf_cube(tf.constant(2.0))
# ### TF 함수와 콘크리트 함수
concrete_function = tf_cube.get_concrete_function(tf.constant(2.0))
concrete_function.graph
concrete_function(tf.constant(2.0))
concrete_function is tf_cube.get_concrete_function(tf.constant(2.0))
# ### 함수 정의와 그래프
concrete_function.graph
ops = concrete_function.graph.get_operations()
ops
pow_op = ops[2]
list(pow_op.inputs)
pow_op.outputs
concrete_function.graph.get_operation_by_name('x')
concrete_function.graph.get_tensor_by_name('Identity:0')
concrete_function.function_def.signature
# ### TF 함수가 계산 그래프를 추출하기 위해 파이썬 함수를 트레이싱하는 방법
@tf.function
def tf_cube(x):
print("print:", x)
return x ** 3
result = tf_cube(tf.constant(2.0))
result
result = tf_cube(2)
result = tf_cube(3)
result = tf_cube(tf.constant([[1., 2.]])) # New shape: trace!
result = tf_cube(tf.constant([[3., 4.], [5., 6.]])) # New shape: trace!
result = tf_cube(tf.constant([[7., 8.], [9., 10.], [11., 12.]])) # New shape: trace!
# 특정 입력 시그니처를 지정하는 것도 가능합니다:
@tf.function(input_signature=[tf.TensorSpec([None, 28, 28], tf.float32)])
def shrink(images):
print("트레이싱", images)
return images[:, fd00:c2b6:b24b:be67:2827:688d:e6a1:6a3b, ::2] # 행과 열의 절반을 버립니다
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
img_batch_1 = tf.random.uniform(shape=[100, 28, 28])
img_batch_2 = tf.random.uniform(shape=[50, 28, 28])
preprocessed_images = shrink(img_batch_1) # 함수 트레이싱
preprocessed_images = shrink(img_batch_2) # 동일한 콘크리트 함수 재사용
img_batch_3 = tf.random.uniform(shape=[2, 2, 2])
try:
preprocessed_images = shrink(img_batch_3) # 다른 타입이나 크기 거부
except ValueError as ex:
print(ex)
# ### 오토그래프를 사용해 제어 흐름 나타내기
# `range()`를 사용한 정적인 `for` 반복:
@tf.function
def add_10(x):
for i in range(10):
x += 1
return x
add_10(tf.constant(5))
add_10.get_concrete_function(tf.constant(5)).graph.get_operations()
# `tf.while_loop()`를 사용한 동적인 반복:
@tf.function
def add_10(x):
condition = lambda i, x: tf.less(i, 10)
body = lambda i, x: (tf.add(i, 1), tf.add(x, 1))
final_i, final_x = tf.while_loop(condition, body, [tf.constant(0), x])
return final_x
add_10(tf.constant(5))
add_10.get_concrete_function(tf.constant(5)).graph.get_operations()
# (오토그래프에 의한) `tf.range()`를 사용한 동적인 `for` 반복:
@tf.function
def add_10(x):
for i in tf.range(10):
x = x + 1
return x
add_10.get_concrete_function(tf.constant(0)).graph.get_operations()
# ### TF 함수에서 변수와 다른 자원 다루기
# +
counter = tf.Variable(0)
@tf.function
def increment(counter, c=1):
return counter.assign_add(c)
# -
increment(counter)
increment(counter)
function_def = increment.get_concrete_function(counter).function_def
function_def.signature.input_arg[0]
# +
counter = tf.Variable(0)
@tf.function
def increment(c=1):
return counter.assign_add(c)
# -
increment()
increment()
function_def = increment.get_concrete_function().function_def
function_def.signature.input_arg[0]
class Counter:
def __init__(self):
self.counter = tf.Variable(0)
@tf.function
def increment(self, c=1):
return self.counter.assign_add(c)
c = Counter()
c.increment()
c.increment()
# +
@tf.function
def add_10(x):
for i in tf.range(10):
x += 1
return x
print(tf.autograph.to_code(add_10.python_function))
# -
def display_tf_code(func):
from IPython.display import display, Markdown
if hasattr(func, "python_function"):
func = func.python_function
code = tf.autograph.to_code(func)
display(Markdown('```python\n{}\n```'.format(code)))
display_tf_code(add_10)
# ## tf.keras와 TF 함수를 함께 사용하거나 사용하지 않기
# 기본적으로 tf.keras는 자동으로 사용자 정의 코드를 TF 함수로 변환하기 때문에 `tf.function()`을 사용할 필요가 없습니다:
# 사용자 손실 함수
def my_mse(y_true, y_pred):
print("my_mse() 손실 트레이싱")
return tf.reduce_mean(tf.square(y_pred - y_true))
# 사용자 지표 함수
def my_mae(y_true, y_pred):
print("my_mae() 지표 트레이싱")
return tf.reduce_mean(tf.abs(y_pred - y_true))
# 사용자 정의 층
class MyDense(keras.layers.Layer):
def __init__(self, units, activation=None, **kwargs):
super().__init__(**kwargs)
self.units = units
self.activation = keras.activations.get(activation)
def build(self, input_shape):
self.kernel = self.add_weight(name='kernel',
shape=(input_shape[1], self.units),
initializer='uniform',
trainable=True)
self.biases = self.add_weight(name='bias',
shape=(self.units,),
initializer='zeros',
trainable=True)
super().build(input_shape)
def call(self, X):
print("MyDense.call() 트레이싱")
return self.activation(X @ self.kernel + self.biases)
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
# +
# 사용자 정의 모델
class MyModel(keras.models.Model):
def __init__(self, **kwargs):
super().__init__(**kwargs)
self.hidden1 = MyDense(30, activation="relu")
self.hidden2 = MyDense(30, activation="relu")
self.output_ = MyDense(1)
def call(self, input):
print("MyModel.call() 트레이싱")
hidden1 = self.hidden1(input)
hidden2 = self.hidden2(hidden1)
concat = keras.layers.concatenate([input, hidden2])
output = self.output_(concat)
return output
model = MyModel()
# -
model.compile(loss=my_mse, optimizer="nadam", metrics=[my_mae])
model.fit(X_train_scaled, y_train, epochs=2,
validation_data=(X_valid_scaled, y_valid))
model.evaluate(X_test_scaled, y_test)
# `dynamic=True`로 모델을 만들어 이 기능을 끌 수 있습니다(또는 모델의 생성자에서 `super().__init__(dynamic=True, **kwargs)`를 호출합니다):
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = MyModel(dynamic=True)
model.compile(loss=my_mse, optimizer="nadam", metrics=[my_mae])
# 사용자 정의 코드는 반복마다 호출됩니다. 너무 많이 출력되는 것을 피하기 위해 작은 데이터셋으로 훈련, 검증, 평가해 보겠습니다:
model.fit(X_train_scaled[:64], y_train[:64], epochs=1,
validation_data=(X_valid_scaled[:64], y_valid[:64]), verbose=0)
model.evaluate(X_test_scaled[:64], y_test[:64], verbose=0)
# 또는 모델을 컴파일할 때 `run_eagerly=True`를 지정합니다:
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = MyModel()
model.compile(loss=my_mse, optimizer="nadam", metrics=[my_mae], run_eagerly=True)
model.fit(X_train_scaled[:64], y_train[:64], epochs=1,
validation_data=(X_valid_scaled[:64], y_valid[:64]), verbose=0)
model.evaluate(X_test_scaled[:64], y_test[:64], verbose=0)
# ## 사용자 정의 옵티마이저
# 사용자 정의 옵티마이저를 정의하는 것은 일반적이지 않습니다. 하지만 어쩔 수 없이 만들어야 하는 상황이라면 다음 예를 참고하세요:
class MyMomentumOptimizer(keras.optimizers.Optimizer):
def __init__(self, learning_rate=0.001, momentum=0.9, name="MyMomentumOptimizer", **kwargs):
"""super().__init__()를 호출하고 _set_hyper()를 사용해 하이퍼파라미터를 저장합니다"""
super().__init__(name, **kwargs)
self._set_hyper("learning_rate", kwargs.get("lr", learning_rate)) # lr=learning_rate을 처리
self._set_hyper("decay", self._initial_decay) #
self._set_hyper("momentum", momentum)
def _create_slots(self, var_list):
"""모델 파라미터마다 연관된 옵티마이저 변수를 만듭니다.
텐서플로는 이런 옵티마이저 변수를 '슬롯'이라고 부릅니다.
모멘텀 옵티마이저에서는 모델 파라미터마다 하나의 모멘텀 슬롯이 필요합니다.
"""
for var in var_list:
self.add_slot(var, "momentum")
@tf.function
def _resource_apply_dense(self, grad, var):
"""슬롯을 업데이트하고 모델 파라미터에 대한 옵티마이저 스텝을 수행합니다.
"""
var_dtype = var.dtype.base_dtype
lr_t = self._decayed_lr(var_dtype) # 학습률 감쇠 처리
momentum_var = self.get_slot(var, "momentum")
momentum_hyper = self._get_hyper("momentum", var_dtype)
momentum_var.assign(momentum_var * momentum_hyper - (1. - momentum_hyper)* grad)
var.assign_add(momentum_var * lr_t)
def _resource_apply_sparse(self, grad, var):
raise NotImplementedError
def get_config(self):
base_config = super().get_config()
return {
**base_config,
"learning_rate": self._serialize_hyperparameter("learning_rate"),
"decay": self._serialize_hyperparameter("decay"),
"momentum": self._serialize_hyperparameter("momentum"),
}
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([keras.layers.Dense(1, input_shape=[8])])
model.compile(loss="mse", optimizer=MyMomentumOptimizer())
model.fit(X_train_scaled, y_train, epochs=5)
# # 연습문제
# ## 1. to 11.
# 부록 A 참조.
# # 12. _층 정규화_ 를 수행하는 사용자 정의 층을 구현하세요.
#
# _15장에서 순환 신경망을 사용할 때 이런 종류의 층을 사용합니다._
# ### a.
# _문제: `build()` 메서드에서 두 개의 훈련 가능한 가중치 *α*와 *β*를 정의합니다. 두 가중치 모두 크기가 `input_shape[-1:]`이고 데이터 타입은 `tf.float32`입니다. *α*는 1로 초기화되고 *β*는 0으로 초기화되어야 합니다._
# 솔루션: 아래 참조.
# ### b.
# _문제: `call()` 메서드는 샘플의 특성마다 평균 μ와 표준편차 σ를 계산해야 합니다. 이를 위해 전체 샘플의 평균 μ와 분산 σ<sup>2</sup>을 반환하는 `tf.nn.moments(inputs, axes=-1, keepdims=True)`을 사용할 수 있습니다(분산의 제곱근으로 표준편차를 계산합니다). 그다음 *α*⊗(*X* - μ)/(σ + ε) + *β*를 계산하여 반환합니다. 여기에서 ⊗는 원소별
# 곱셈(`*`)을 나타냅니다. ε은 안전을 위한 항입니다(0으로 나누어지는 것을 막기 위한 작은 상수. 예를 들면 0.001)._
class LayerNormalization(keras.layers.Layer):
def __init__(self, eps=0.001, **kwargs):
super().__init__(**kwargs)
self.eps = eps
def build(self, batch_input_shape):
self.alpha = self.add_weight(
name="alpha", shape=batch_input_shape[-1:],
initializer="ones")
self.beta = self.add_weight(
name="beta", shape=batch_input_shape[-1:],
initializer="zeros")
super().build(batch_input_shape) # 반드시 끝에 와야 합니다
def call(self, X):
mean, variance = tf.nn.moments(X, axes=-1, keepdims=True)
return self.alpha * (X - mean) / (tf.sqrt(variance + self.eps)) + self.beta
def compute_output_shape(self, batch_input_shape):
return batch_input_shape
def get_config(self):
base_config = super().get_config()
return {**base_config, "eps": self.eps}
# _ε_ 하이퍼파라미터(`eps`)는 필수가 아닙니다. 또한 `tf.sqrt(variance) + self.eps` 보다 `tf.sqrt(variance + self.eps)`를 계산하는 것이 좋습니다. sqrt(z)의 도함수는 z=0에서 정의되지 않기 때문에 분산 벡터의 한 원소가 0에 가까우면 훈련이 이리저리 널뜁니다. 제곱근 안에 _ε_를 넣으면 이런 현상을 방지할 수 있습니다.
# ### c.
# _문제: 사용자 정의 층이 `keras.layers.LayerNormalization` 층과 동일한(또는 거의 동일한) 출력을 만드는지 확인하세요._
# 각 클래스의 객체를 만들고 데이터(예를 들면, 훈련 세트)를 적용해 보죠. 차이는 무시할 수 있는 수준입니다.
# +
X = X_train.astype(np.float32)
custom_layer_norm = LayerNormalization()
keras_layer_norm = keras.layers.LayerNormalization()
tf.reduce_mean(keras.losses.mean_absolute_error(
keras_layer_norm(X), custom_layer_norm(X)))
# -
# 네 충분히 가깝네요. 조금 더 확실하게 알파와 베타를 완전히 랜덤하게 지정하고 다시 비교해 보죠:
# +
random_alpha = np.random.rand(X.shape[-1])
random_beta = np.random.rand(X.shape[-1])
custom_layer_norm.set_weights([random_alpha, random_beta])
keras_layer_norm.set_weights([random_alpha, random_beta])
tf.reduce_mean(keras.losses.mean_absolute_error(
keras_layer_norm(X), custom_layer_norm(X)))
# -
# 여전히 무시할 수 있는 수준입니다! 사용자 정의 층이 잘 동작합니다.
# ## 13. 사용자 정의 훈련 반복을 사용해 패션 MNIST 데이터셋으로 모델을 훈련해보세요.
#
# _패션 MNIST 데이터셋은 10장에서 소개했습니다._
# ### a.
# _문제: 에포크, 반복, 평균 훈련 손실, (반복마다 업데이트되는) 에포크의 평균 정확도는 물론 에포크 끝에서 검증 손실과 정확도를 출력하세요._
(X_train_full, y_train_full), (X_test, y_test) = keras.datasets.fashion_mnist.load_data()
X_train_full = X_train_full.astype(np.float32) / 255.
X_valid, X_train = X_train_full[:5000], X_train_full[5000:]
y_valid, y_train = y_train_full[:5000], y_train_full[5000:]
X_test = X_test.astype(np.float32) / 255.
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
model = keras.models.Sequential([
keras.layers.Flatten(input_shape=[28, 28]),
keras.layers.Dense(100, activation="relu"),
keras.layers.Dense(10, activation="softmax"),
])
n_epochs = 5
batch_size = 32
n_steps = len(X_train) // batch_size
optimizer = keras.optimizers.Nadam(lr=0.01)
loss_fn = keras.losses.sparse_categorical_crossentropy
mean_loss = keras.metrics.Mean()
metrics = [keras.metrics.SparseCategoricalAccuracy()]
with trange(1, n_epochs + 1, desc="All epochs") as epochs:
for epoch in epochs:
with trange(1, n_steps + 1, desc="Epoch {}/{}".format(epoch, n_epochs)) as steps:
for step in steps:
X_batch, y_batch = random_batch(X_train, y_train)
with tf.GradientTape() as tape:
y_pred = model(X_batch)
main_loss = tf.reduce_mean(loss_fn(y_batch, y_pred))
loss = tf.add_n([main_loss] + model.losses)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
for variable in model.variables:
if variable.constraint is not None:
variable.assign(variable.constraint(variable))
status = OrderedDict()
mean_loss(loss)
status["loss"] = mean_loss.result().numpy()
for metric in metrics:
metric(y_batch, y_pred)
status[metric.name] = metric.result().numpy()
steps.set_postfix(status)
y_pred = model(X_valid)
status["val_loss"] = np.mean(loss_fn(y_valid, y_pred))
status["val_accuracy"] = np.mean(keras.metrics.sparse_categorical_accuracy(
tf.constant(y_valid, dtype=np.float32), y_pred))
steps.set_postfix(status)
for metric in [mean_loss] + metrics:
metric.reset_states()
# ### b.
# _문제: 상위 층과 하위 층에 학습률이 다른 옵티마이저를 따로 사용해보세요._
keras.backend.clear_session()
np.random.seed(42)
tf.random.set_seed(42)
lower_layers = keras.models.Sequential([
keras.layers.Flatten(input_shape=[28, 28]),
keras.layers.Dense(100, activation="relu"),
])
upper_layers = keras.models.Sequential([
keras.layers.Dense(10, activation="softmax"),
])
model = keras.models.Sequential([
lower_layers, upper_layers
])
lower_optimizer = keras.optimizers.SGD(lr=1e-4)
upper_optimizer = keras.optimizers.Nadam(lr=1e-3)
n_epochs = 5
batch_size = 32
n_steps = len(X_train) // batch_size
loss_fn = keras.losses.sparse_categorical_crossentropy
mean_loss = keras.metrics.Mean()
metrics = [keras.metrics.SparseCategoricalAccuracy()]
with trange(1, n_epochs + 1, desc="All epochs") as epochs:
for epoch in epochs:
with trange(1, n_steps + 1, desc="Epoch {}/{}".format(epoch, n_epochs)) as steps:
for step in steps:
X_batch, y_batch = random_batch(X_train, y_train)
with tf.GradientTape(persistent=True) as tape:
y_pred = model(X_batch)
main_loss = tf.reduce_mean(loss_fn(y_batch, y_pred))
loss = tf.add_n([main_loss] + model.losses)
for layers, optimizer in ((lower_layers, lower_optimizer),
(upper_layers, upper_optimizer)):
gradients = tape.gradient(loss, layers.trainable_variables)
optimizer.apply_gradients(zip(gradients, layers.trainable_variables))
del tape
for variable in model.variables:
if variable.constraint is not None:
variable.assign(variable.constraint(variable))
status = OrderedDict()
mean_loss(loss)
status["loss"] = mean_loss.result().numpy()
for metric in metrics:
metric(y_batch, y_pred)
status[metric.name] = metric.result().numpy()
steps.set_postfix(status)
y_pred = model(X_valid)
status["val_loss"] = np.mean(loss_fn(y_valid, y_pred))
status["val_accuracy"] = np.mean(keras.metrics.sparse_categorical_accuracy(
tf.constant(y_valid, dtype=np.float32), y_pred))
steps.set_postfix(status)
for metric in [mean_loss] + metrics:
metric.reset_states()
| 12_custom_models_and_training_with_tensorflow.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # 머신 러닝 교과서 3판
# # 4장 - 좋은 훈련 데이터셋 만들기 – 데이터 전처리
# **아래 링크를 통해 이 노트북을 주피터 노트북 뷰어(nbviewer.jupyter.org)로 보거나 구글 코랩(colab.research.google.com)에서 실행할 수 있습니다.**
#
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://nbviewer.jupyter.org/github/rickiepark/python-machine-learning-book-3rd-edition/blob/master/ch04/SequentialFeatureSelector.ipynb"><img src="https://jupyter.org/assets/main-logo.svg" width="28" />주피터 노트북 뷰어로 보기</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/rickiepark/python-machine-learning-book-3rd-edition/blob/master/ch04/SequentialFeatureSelector.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />구글 코랩(Colab)에서 실행하기</a>
# </td>
# </table>
# + [markdown] id="8Os-4JDvScgV"
# ## 4.5.4 순차 특성 선택 알고리즘
# + colab={"base_uri": "https://localhost:8080/"} id="1KNgp1IyScgZ" outputId="b9e09add-044a-48e1-c0d7-12040d113b0a"
# 이 노트북은 사이킷런 0.24 이상에서 실행할 수 있습니다.
# 코랩에서 실행할 경우 최신 버전의 사이킷런을 설치하세요.
# !pip install --upgrade scikit-learn
# + [markdown] id="B2ogTR9QScgZ"
# 4.5절에서 사용하는 데이터셋을 로드합니다.
# + id="LKqSVFwhScga"
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# + id="l6Z2jGnUScga"
df_wine = pd.read_csv('https://archive.ics.uci.edu/'
'ml/machine-learning-databases/wine/wine.data',
header=None)
df_wine.columns = ['Class label', 'Alcohol', 'Malic acid', 'Ash',
'Alcalinity of ash', 'Magnesium', 'Total phenols',
'Flavanoids', 'Nonflavanoid phenols', 'Proanthocyanins',
'Color intensity', 'Hue', 'OD280/OD315 of diluted wines',
'Proline']
# + id="-drOG_MMScga"
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
X, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values
X_train, X_test, y_train, y_test =\
train_test_split(X, y,
test_size=0.3,
random_state=0,
stratify=y)
stdsc = StandardScaler()
X_train_std = stdsc.fit_transform(X_train)
X_test_std = stdsc.transform(X_test)
# + [markdown] id="Gj1VsXpgScga"
# `SequentialFeatureSelector`를 임포트하고 최근접 이웃 분류기 객체를 준비합니다.
# + id="vGEcLkkUScgb"
from sklearn.feature_selection import SequentialFeatureSelector
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=5)
# + [markdown] id="0ghi7Y2DScgb"
# 사용할 모델 객체를 첫 번째 매개변수로 전달합니다. 선택할 특성의 개수는 `n_features_to_select`에서 지정합니다. 기본값은 입력 특성의 절반입니다. 0~1 사이 실수를 지정하면 선택할 특성의 비율로 인식합니다.
#
# `direction` 매개변수로 특성 선택 방향(전진 또는 후진)을 선택합니다. 기본값은 전진을 의미하는 `'forward'`이고 후진을 선택하려면 `'backward'`로 지정합니다.
#
# 이 클래스는 특성을 선택하기 위해 교차 검증을 사용합니다. `cv` 매개변수에서 교차 검증 횟수를 지정할 수 있습니다. 기본값은 5입니다. 회귀 모델일 경우 `KFold`, 분류 모델일 경우 `StratifiedKFold`를 사용하여 폴드를 나눕니다.
#
# 이 클래스는 하나의 특성을 선택할 때마다 현재 남은 특성 개수(m)에 대해 교차 검증을 수행하므로 `m * cv`개의 모델을 만듭니다. 이렇게 단계마다 많은 모델을 만들기 때문에 일반적으로 `RFE`나 `SelectFromModel`보다 느립니다. `n_jobs` 매개변수를 1 이상으로 지정하여 여러 코어를 사용하는 것이 좋습니다.
# + id="SPaLDZt6Scgb"
scores = []
for n_features in range(1, 13):
sfs = SequentialFeatureSelector(knn, n_features_to_select=n_features, n_jobs=-1)
sfs.fit(X_train_std, y_train)
f_mask = sfs.support_
knn.fit(X_train_std[:, f_mask], y_train)
scores.append(knn.score(X_train_std[:, f_mask], y_train))
# + colab={"base_uri": "https://localhost:8080/", "height": 297} id="o7y0RoNAScgb" outputId="235b0195-8f90-40cc-a030-474fc22976ba"
plt.plot(range(1, 13), scores, marker='o')
plt.ylim([0.7, 1.02])
plt.ylabel('Accuracy')
plt.xlabel('Number of features')
plt.grid()
plt.tight_layout()
# plt.savefig('images/04_sfs.png', dpi=300)
plt.show()
# + colab={"base_uri": "https://localhost:8080/"} id="0Bt-QBgtScgc" outputId="e523911a-cc8e-4d21-de1b-3479213c75c6"
sfs = SequentialFeatureSelector(knn, n_features_to_select=7, n_jobs=-1)
sfs.fit(X_train_std, y_train)
# + colab={"base_uri": "https://localhost:8080/"} id="xHezF2ZrScgd" outputId="a19cdd1c-64d3-43cc-dca6-e90e2eb393d2"
print(sfs.n_features_to_select_)
f_mask = sfs.support_
df_wine.columns[1:][f_mask]
# + colab={"base_uri": "https://localhost:8080/"} id="76DLeStyScgd" outputId="216e01d3-7ea7-470f-865b-ddfeeeafa1d3"
knn.fit(X_train_std[:, f_mask], y_train)
print('훈련 정확도:', knn.score(X_train_std[:, f_mask], y_train))
print('테스트 정확도:', knn.score(X_test_std[:, f_mask], y_test))
| ch04/SequentialFeatureSelector.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# # Session 5: Generative Networks
# ## Assignment: Generative Adversarial Networks, Variational Autoencoders, and Recurrent Neural Networks
# <p class="lead">
# <a href="https://www.kadenze.com/courses/creative-applications-of-deep-learning-with-tensorflow/info">Creative Applications of Deep Learning with Google's Tensorflow</a><br />
# <a href="http://pkmital.com"><NAME></a><br />
# <a href="https://www.kadenze.com">Kadenze, Inc.</a>
# </p>
#
# Continued from [session-5-part-1.ipynb](session-5-part-1.ipynb)...
#
# # Table of Contents
#
# <!-- MarkdownTOC autolink="true" autoanchor="true" bracket="round" -->
# - [Overview](session-5-part-1.ipynb#overview)
# - [Learning Goals](session-5-part-1.ipynb#learning-goals)
# - [Part 1 - Generative Adversarial Networks \(GAN\) / Deep Convolutional GAN \(DCGAN\)](#part-1---generative-adversarial-networks-gan--deep-convolutional-gan-dcgan)
# - [Introduction](session-5-part-1.ipynb#introduction)
# - [Building the Encoder](session-5-part-1.ipynb#building-the-encoder)
# - [Building the Discriminator for the Training Samples](session-5-part-1.ipynb#building-the-discriminator-for-the-training-samples)
# - [Building the Decoder](session-5-part-1.ipynb#building-the-decoder)
# - [Building the Generator](session-5-part-1.ipynb#building-the-generator)
# - [Building the Discriminator for the Generated Samples](session-5-part-1.ipynb#building-the-discriminator-for-the-generated-samples)
# - [GAN Loss Functions](session-5-part-1.ipynb#gan-loss-functions)
# - [Building the Optimizers w/ Regularization](session-5-part-1.ipynb#building-the-optimizers-w-regularization)
# - [Loading a Dataset](session-5-part-1.ipynb#loading-a-dataset)
# - [Training](session-5-part-1.ipynb#training)
# - [Equilibrium](session-5-part-1.ipynb#equilibrium)
# - [Part 2 - Variational Auto-Encoding Generative Adversarial Network \(VAEGAN\)](#part-2---variational-auto-encoding-generative-adversarial-network-vaegan)
# - [Batch Normalization](session-5-part-1.ipynb#batch-normalization)
# - [Building the Encoder](session-5-part-1.ipynb#building-the-encoder-1)
# - [Building the Variational Layer](session-5-part-1.ipynb#building-the-variational-layer)
# - [Building the Decoder](session-5-part-1.ipynb#building-the-decoder-1)
# - [Building VAE/GAN Loss Functions](session-5-part-1.ipynb#building-vaegan-loss-functions)
# - [Creating the Optimizers](session-5-part-1.ipynb#creating-the-optimizers)
# - [Loading the Dataset](session-5-part-1.ipynb#loading-the-dataset)
# - [Training](session-5-part-1.ipynb#training-1)
# - [Part 3 - Latent-Space Arithmetic](session-5-part-1.ipynb#part-3---latent-space-arithmetic)
# - [Loading the Pre-Trained Model](session-5-part-1.ipynb#loading-the-pre-trained-model)
# - [Exploring the Celeb Net Attributes](session-5-part-1.ipynb#exploring-the-celeb-net-attributes)
# - [Find the Latent Encoding for an Attribute](session-5-part-1.ipynb#find-the-latent-encoding-for-an-attribute)
# - [Latent Feature Arithmetic](session-5-part-1.ipynb#latent-feature-arithmetic)
# - [Extensions](session-5-part-1.ipynb#extensions)
# - [Part 4 - Character-Level Language Model](session-5-part-2.ipynb#part-4---character-level-language-model)
# - [Part 5 - Pretrained Char-RNN of Donald Trump](session-5-part-2.ipynb#part-5---pretrained-char-rnn-of-donald-trump)
# - [Getting the Trump Data](session-5-part-2.ipynb#getting-the-trump-data)
# - [Basic Text Analysis](session-5-part-2.ipynb#basic-text-analysis)
# - [Loading the Pre-trained Trump Model](session-5-part-2.ipynb#loading-the-pre-trained-trump-model)
# - [Inference: Keeping Track of the State](session-5-part-2.ipynb#inference-keeping-track-of-the-state)
# - [Probabilistic Sampling](session-5-part-2.ipynb#probabilistic-sampling)
# - [Inference: Temperature](session-5-part-2.ipynb#inference-temperature)
# - [Inference: Priming](session-5-part-2.ipynb#inference-priming)
# - [Assignment Submission](session-5-part-2.ipynb#assignment-submission)
#
# <!-- /MarkdownTOC -->
#
# +
# First check the Python version
import sys
if sys.version_info < (3,4):
print('You are running an older version of Python!\n\n',
'You should consider updating to Python 3.4.0 or',
'higher as the libraries built for this course',
'have only been tested in Python 3.4 and higher.\n')
print('Try installing the Python 3.5 version of anaconda'
'and then restart `jupyter notebook`:\n',
'https://www.continuum.io/downloads\n\n')
# Now get necessary libraries
try:
import os
import numpy as np
import matplotlib.pyplot as plt
from skimage.transform import resize
from skimage import data
from scipy.misc import imresize
from scipy.ndimage.filters import gaussian_filter
import IPython.display as ipyd
import tensorflow as tf
from libs import utils, gif, datasets, dataset_utils, nb_utils
except ImportError as e:
print("Make sure you have started notebook in the same directory",
"as the provided zip file which includes the 'libs' folder",
"and the file 'utils.py' inside of it. You will NOT be able",
"to complete this assignment unless you restart jupyter",
"notebook inside the directory created by extracting",
"the zip file or cloning the github repo.")
print(e)
# We'll tell matplotlib to inline any drawn figures like so:
# %matplotlib inline
plt.style.use('ggplot')
# -
# Bit of formatting because I don't like the default inline code style:
from IPython.core.display import HTML
HTML("""<style> .rendered_html code {
padding: 2px 4px;
color: #c7254e;
background-color: #f9f2f4;
border-radius: 4px;
} </style>""")
# <style> .rendered_html code {
# padding: 2px 4px;
# color: #c7254e;
# background-color: #f9f2f4;
# border-radius: 4px;
# } </style>
#
#
#
# <a name="part-4---character-level-language-model"></a>
# # Part 4 - Character-Level Language Model
#
# We'll now continue onto the second half of the homework and explore recurrent neural networks. We saw one potential application of a recurrent neural network which learns letter by letter the content of a text file. We were then able to synthesize from the model to produce new phrases. Let's try to build one. Replace the code below with something that loads your own text file or one from the internet. Be creative with this!
#
# <h3><font color='red'>TODO! COMPLETE THIS SECTION!</font></h3>
import tensorflow as tf
from six.moves import urllib
script = 'http://www.awesomefilm.com/script/biglebowski.txt'
txts = []
f, _ = urllib.request.urlretrieve(script, script.split('/')[-1])
with open(f, 'r') as fp:
txt = fp.read()
# Let's take a look at the first part of this:
txt[:100]
# We'll just clean up the text a little. This isn't necessary, but can help the training along a little. In the example text I provided, there is a lot of white space (those \t's are tabs). I'll remove them. There are also repetitions of \n, new lines, which are not necessary. The code below will remove the tabs, ending whitespace, and any repeating newlines. Replace this with any preprocessing that makes sense for your dataset. Try to boil it down to just the possible letters for what you want to learn/synthesize while retaining any meaningful patterns:
txt = "\n".join([txt_i.strip()
for txt_i in txt.replace('\t', '').split('\n')
if len(txt_i)])
# Now we can see how much text we have:
len(txt)
# In general, we'll want as much text as possible. But I'm including this just as a minimal example so you can explore your own. Try making a text file and seeing the size of it. You'll want about 1 MB at least.
#
# Let's now take a look at the different characters we have in our file:
vocab = list(set(txt))
vocab.sort()
print(len(vocab))
print(vocab)
# And then create a mapping which can take us from the letter to an integer look up table of that letter (and vice-versa). To do this, we'll use an `OrderedDict` from the `collections` library. In Python 3.6, this is the default behavior of dict, but in earlier versions of Python, we'll need to be explicit by using OrderedDict.
# +
from collections import OrderedDict
encoder = OrderedDict(zip(vocab, range(len(vocab))))
decoder = OrderedDict(zip(range(len(vocab)), vocab))
# -
encoder
# We'll store a few variables that will determine the size of our network. First, `batch_size` determines how many sequences at a time we'll train on. The `seqence_length` parameter defines the maximum length to unroll our recurrent network for. This is effectively the depth of our network during training to help guide gradients along. Within each layer, we'll have `n_cell` LSTM units, and `n_layers` layers worth of LSTM units. Finally, we'll store the total number of possible characters in our data, which will determine the size of our one hot encoding (like we had for MNIST in Session 3).
# +
# Number of sequences in a mini batch
batch_size = 100
# Number of characters in a sequence
sequence_length = 50
# Number of cells in our LSTM layer
n_cells = 128
# Number of LSTM layers
n_layers = 3
# Total number of characters in the one-hot encoding
n_chars = len(vocab)
# -
# Let's now create the input and output to our network. We'll use placeholders and feed these in later. The size of these need to be [`batch_size`, `sequence_length`]. We'll then see how to build the network in between.
#
# <h3><font color='red'>TODO! COMPLETE THIS SECTION!</font></h3>
# +
X = tf.placeholder(tf.int32, shape=..., name='X')
# We'll have a placeholder for our true outputs
Y = tf.placeholder(tf.int32, shape=..., name='Y')
# -
# The first thing we need to do is convert each of our `sequence_length` vectors in our batch to `n_cells` LSTM cells. We use a lookup table to find the value in `X` and use this as the input to `n_cells` LSTM cells. Our lookup table has `n_chars` possible elements and connects each character to `n_cells` cells. We create our lookup table using `tf.get_variable` and then the function `tf.nn.embedding_lookup` to connect our `X` placeholder to `n_cells` number of neurons.
# +
# we first create a variable to take us from our one-hot representation to our LSTM cells
embedding = tf.get_variable("embedding", [n_chars, n_cells])
# And then use tensorflow's embedding lookup to look up the ids in X
Xs = tf.nn.embedding_lookup(embedding, X)
# The resulting lookups are concatenated into a dense tensor
print(Xs.get_shape().as_list())
# -
# Now recall from the lecture that recurrent neural networks share their weights across timesteps. So we don't want to have one large matrix with every timestep, but instead separate them. We'll use `tf.split` to split our `[batch_size, sequence_length, n_cells]` array in `Xs` into a list of `sequence_length` elements each composed of `[batch_size, n_cells]` arrays. This gives us `sequence_length` number of arrays of `[batch_size, 1, n_cells]`. We then use `tf.squeeze` to remove the 1st index corresponding to the singleton `sequence_length` index, resulting in simply `[batch_size, n_cells]`.
with tf.name_scope('reslice'):
Xs = [tf.squeeze(seq, [1])
for seq in tf.split(Xs, sequence_length, 1)]
# With each of our timesteps split up, we can now connect them to a set of LSTM recurrent cells. We tell the `tf.contrib.rnn.BasicLSTMCell` method how many cells we want, i.e. how many neurons there are, and we also specify that our state will be stored as a tuple. This state defines the internal state of the cells as well as the connection from the previous timestep. We can also pass a value for the `forget_bias`. Be sure to experiment with this parameter as it can significantly effect performance (e.g. Gers, <NAME>, Schmidhuber, Jurgen, and Cummins, Fred. Learning to forget: Continual prediction with lstm. Neural computation, 12(10):2451–2471, 2000).
cells = tf.contrib.rnn.BasicLSTMCell(num_units=n_cells, state_is_tuple=True, forget_bias=1.0)
# Let's take a look at the cell's state size:
cells.state_size
# `c` defines the internal memory and `h` the output. We'll have as part of our `cells`, both an `initial_state` and a `final_state`. These will become important during inference and we'll see how these work more then. For now, we'll set the `initial_state` to all zeros using the convenience function provided inside our `cells` object, `zero_state`:
initial_state = cells.zero_state(tf.shape(X)[0], tf.float32)
# Looking at what this does, we can see that it creates a `tf.Tensor` of zeros for our `c` and `h` states for each of our `n_cells` and stores this as a tuple inside the `LSTMStateTuple` object:
initial_state
# So far, we have created a single layer of LSTM cells composed of `n_cells` number of cells. If we want another layer, we can use the `tf.contrib.rnn.MultiRNNCell` method, giving it our current cells, and a bit of pythonery to multiply our cells by the number of layers we want. We'll then update our `initial_state` variable to include the additional cells:
cells = tf.contrib.rnn.MultiRNNCell(
[cells] * n_layers, state_is_tuple=True)
initial_state = cells.zero_state(tf.shape(X)[0], tf.float32)
# Now if we take a look at our `initial_state`, we should see one `LSTMStateTuple` for each of our layers:
initial_state
# So far, we haven't connected our recurrent cells to anything. Let's do this now using the `tf.contrib.rnn.static_rnn` method. We also pass it our `initial_state` variables. It gives us the `outputs` of the rnn, as well as their states after having been computed. Contrast that with the `initial_state`, which set the LSTM cells to zeros. After having computed something, the cells will all have a different value somehow reflecting the temporal dynamics and expectations of the next input. These will be stored in the `state` tensors for each of our LSTM layers inside a `LSTMStateTuple` just like the `initial_state` variable.
# ```python
# help(tf.contrib.rnn.static_rnn)
#
# Help on function static_rnn in module tensorflow.contrib.rnn.python.ops.core_rnn:
#
# static_rnn(cell, inputs, initial_state=None, dtype=None, sequence_length=None, scope=None)
# Creates a recurrent neural network specified by RNNCell `cell`.
#
# The simplest form of RNN network generated is:
#
# state = cell.zero_state(...)
# outputs = []
# for input_ in inputs:
# output, state = cell(input_, state)
# outputs.append(output)
# return (outputs, state)
#
# However, a few other options are available:
#
# An initial state can be provided.
# If the sequence_length vector is provided, dynamic calculation is performed.
# This method of calculation does not compute the RNN steps past the maximum
# sequence length of the minibatch (thus saving computational time),
# and properly propagates the state at an example's sequence length
# to the final state output.
#
# The dynamic calculation performed is, at time t for batch row b,
# (output, state)(b, t) =
# (t >= sequence_length(b))
# ? (zeros(cell.output_size), states(b, sequence_length(b) - 1))
# : cell(input(b, t), state(b, t - 1))
#
# Args:
# cell: An instance of RNNCell.
# inputs: A length T list of inputs, each a `Tensor` of shape
# `[batch_size, input_size]`, or a nested tuple of such elements.
# initial_state: (optional) An initial state for the RNN.
# If `cell.state_size` is an integer, this must be
# a `Tensor` of appropriate type and shape `[batch_size, cell.state_size]`.
# If `cell.state_size` is a tuple, this should be a tuple of
# tensors having shapes `[batch_size, s] for s in cell.state_size`.
# dtype: (optional) The data type for the initial state and expected output.
# Required if initial_state is not provided or RNN state has a heterogeneous
# dtype.
# sequence_length: Specifies the length of each sequence in inputs.
# An int32 or int64 vector (tensor) size `[batch_size]`, values in `[0, T)`.
# scope: VariableScope for the created subgraph; defaults to "RNN".
#
# Returns:
# A pair (outputs, state) where:
# - outputs is a length T list of outputs (one for each input), or a nested
# tuple of such elements.
# - state is the final state
#
# Raises:
# TypeError: If `cell` is not an instance of RNNCell.
# ValueError: If `inputs` is `None` or an empty list, or if the input depth
# (column size) cannot be inferred from inputs via shape inference.
# ```
# Use the help on the function `tf.contrib.rnn.static_rnn` to create the `outputs` and `states` variable as below. We've already created each of the variable you need to use:
#
# <h3><font color='red'>TODO! COMPLETE THIS SECTION!</font></h3>
outputs, state = tf.contrib.rnn.static_rnn(cell=..., inputs=..., initial_state=...)
# Let's take a look at the state now:
state
# Our outputs are returned as a list for each of our timesteps:
outputs
# We'll now stack all our outputs for every timestep. We can treat every observation at each timestep and for each batch using the same weight matrices going forward, since these should all have shared weights. Each timstep for each batch is its own observation. So we'll stack these in a 2d matrix so that we can create our softmax layer:
outputs_flat = tf.reshape(tf.concat(values=outputs, axis=1), [-1, n_cells])
# Our outputs are now concatenated so that we have [`batch_size * timesteps`, `n_cells`]
outputs_flat
# We now create a softmax layer just like we did in Session 3 and in Session 3's homework. We multiply our final LSTM layer's `n_cells` outputs by a weight matrix to give us `n_chars` outputs. We then scale this output using a `tf.nn.softmax` layer so that they become a probability by exponentially scaling its value and dividing by its sum. We store the softmax probabilities in `probs` as well as keep track of the maximum index in `Y_pred`:
with tf.variable_scope('prediction'):
W = tf.get_variable(
"W",
shape=[n_cells, n_chars],
initializer=tf.random_normal_initializer(stddev=0.1))
b = tf.get_variable(
"b",
shape=[n_chars],
initializer=tf.random_normal_initializer(stddev=0.1))
# Find the output prediction of every single character in our minibatch
# we denote the pre-activation prediction, logits.
logits = tf.matmul(outputs_flat, W) + b
# We get the probabilistic version by calculating the softmax of this
probs = tf.nn.softmax(logits)
# And then we can find the index of maximum probability
Y_pred = tf.argmax(probs, 1)
# To train the network, we'll measure the loss between our predicted outputs and true outputs. We could use the `probs` variable, but we can also make use of `tf.nn.softmax_cross_entropy_with_logits` which will compute the softmax for us. We therefore need to pass in the variable just before the softmax layer, denoted as `logits` (unscaled values). This takes our variable `logits`, the unscaled predicted outputs, as well as our true outputs, `Y`. Before we give it `Y`, we'll need to reshape our true outputs in the same way, [`batch_size` x `timesteps`, `n_chars`]. Luckily, tensorflow provides a convenience for doing this, the `tf.nn.sparse_softmax_cross_entropy_with_logits` function:
# ```python
# help(tf.nn.sparse_softmax_cross_entropy_with_logits)
#
# Help on function sparse_softmax_cross_entropy_with_logits in module tensorflow.python.ops.nn_ops:
#
# sparse_softmax_cross_entropy_with_logits(logits, labels, name=None)
# Computes sparse softmax cross entropy between `logits` and `labels`.
#
# Measures the probability error in discrete classification tasks in which the
# classes are mutually exclusive (each entry is in exactly one class). For
# example, each CIFAR-10 image is labeled with one and only one label: an image
# can be a dog or a truck, but not both.
#
# **NOTE:** For this operation, the probability of a given label is considered
# exclusive. That is, soft classes are not allowed, and the `labels` vector
# must provide a single specific index for the true class for each row of
# `logits` (each minibatch entry). For soft softmax classification with
# a probability distribution for each entry, see
# `softmax_cross_entropy_with_logits`.
#
# **WARNING:** This op expects unscaled logits, since it performs a softmax
# on `logits` internally for efficiency. Do not call this op with the
# output of `softmax`, as it will produce incorrect results.
#
# A common use case is to have logits of shape `[batch_size, num_classes]` and
# labels of shape `[batch_size]`. But higher dimensions are supported.
#
# Args:
# logits: Unscaled log probabilities of rank `r` and shape
# `[d_0, d_1, ..., d_{r-2}, num_classes]` and dtype `float32` or `float64`.
# labels: `Tensor` of shape `[d_0, d_1, ..., d_{r-2}]` and dtype `int32` or
# `int64`. Each entry in `labels` must be an index in `[0, num_classes)`.
# Other values will result in a loss of 0, but incorrect gradient
# computations.
# name: A name for the operation (optional).
#
# Returns:
# A `Tensor` of the same shape as `labels` and of the same type as `logits`
# with the softmax cross entropy loss.
#
# Raises:
# ValueError: If logits are scalars (need to have rank >= 1) or if the rank
# of the labels is not equal to the rank of the labels minus one.
# ```
with tf.variable_scope('loss'):
# Compute mean cross entropy loss for each output.
Y_true_flat = tf.reshape(tf.concat(values=Y, axis=1), [-1])
# logits are [batch_size x timesteps, n_chars] and
# Y_true_flat are [batch_size x timesteps]
loss = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=Y_true_flat, logits=logits)
# Compute the mean over our `batch_size` x `timesteps` number of observations
mean_loss = tf.reduce_mean(loss)
# Finally, we can create an optimizer in much the same way as we've done with every other network. Except, we will also "clip" the gradients of every trainable parameter. This is a hacky way to ensure that the gradients do not grow too large (the literature calls this the "exploding gradient problem"). However, note that the LSTM is built to help ensure this does not happen by allowing the gradient to be "gated". To learn more about this, please consider reading the following material:
#
# http://www.felixgers.de/papers/phd.pdf
# https://colah.github.io/posts/2015-08-Understanding-LSTMs/
with tf.name_scope('optimizer'):
optimizer = tf.train.AdamOptimizer(learning_rate=0.001)
gradients = []
clip = tf.constant(5.0, name="clip")
for grad, var in optimizer.compute_gradients(mean_loss):
gradients.append((tf.clip_by_value(grad, -clip, clip), var))
updates = optimizer.apply_gradients(gradients)
# Let's take a look at the graph:
nb_utils.show_graph(tf.get_default_graph().as_graph_def())
# Below is the rest of code we'll need to train the network. I do not recommend running this inside Jupyter Notebook for the entire length of the training because the network can take 1-2 days at least to train, and your browser may very likely complain. Instead, you should write a python script containing the necessary bits of code and run it using the Terminal. We didn't go over how to do this, so I'll leave it for you as an exercise. The next part of this notebook will have you load a pre-trained network.
with tf.Session() as sess:
init = tf.global_variables_initializer()
sess.run(init)
cursor = 0
it_i = 0
while it_i < 500:
Xs, Ys = [], []
for batch_i in range(batch_size):
if (cursor + sequence_length) >= len(txt) - sequence_length - 1:
cursor = 0
Xs.append([encoder[ch]
for ch in txt[cursor:cursor + sequence_length]])
Ys.append([encoder[ch]
for ch in txt[cursor + 1: cursor + sequence_length + 1]])
cursor = (cursor + sequence_length)
Xs = np.array(Xs).astype(np.int32)
Ys = np.array(Ys).astype(np.int32)
loss_val, _ = sess.run([mean_loss, updates],
feed_dict={X: Xs, Y: Ys})
if it_i % 100 == 0:
print(it_i, loss_val)
if it_i % 500 == 0:
p = sess.run(probs, feed_dict={X: np.array(Xs[-1])[np.newaxis]})
ps = [np.random.choice(range(n_chars), p=p_i.ravel())
for p_i in p]
p = [np.argmax(p_i) for p_i in p]
if isinstance(txt[0], str):
print('original:', "".join(
[decoder[ch] for ch in Xs[-1]]))
print('synth(samp):', "".join(
[decoder[ch] for ch in ps]))
print('synth(amax):', "".join(
[decoder[ch] for ch in p]))
else:
print([decoder[ch] for ch in ps])
it_i += 1
# <a name="part-5---pretrained-char-rnn-of-donald-trump"></a>
# # Part 5 - Pretrained Char-RNN of Donald Trump
#
# Rather than stick around to let a model train, let's now explore one I've trained for you Donald Trump. If you've trained your own model on your own text corpus then great! You should be able to use that in place of the one I've provided and still continue with the rest of the notebook.
#
# For the Donald Trump corpus, there are a lot of video transcripts that you can find online. I've searched for a few of these, put them in a giant text file, made everything lowercase, and removed any extraneous letters/symbols to help reduce the vocabulary (not that it's not very large to begin with, ha).
#
# I used the code exactly as above to train on the text I gathered and left it to train for about 2 days. The only modification is that I also used "dropout" which you can see in the libs/charrnn.py file. Let's explore it now and we'll see how we can play with "sampling" the model to generate new phrases, and how to "prime" the model (a psychological term referring to when someone is exposed to something shortly before another event).
#
# First, let's clean up any existing graph:
tf.reset_default_graph()
# <a name="getting-the-trump-data"></a>
# ## Getting the Trump Data
#
# Now let's load the text. This is included in the repo or can be downloaded from:
with open('trump.txt', 'r') as fp:
txt = fp.read()
# Let's take a look at what's going on in here:
txt[:100]
# <a name="basic-text-analysis"></a>
# ## Basic Text Analysis
#
# We can do some basic data analysis to get a sense of what kind of vocabulary we're working with. It's really important to look at your data in as many ways as possible. This helps ensure there isn't anything unexpected going on. Let's find every unique word he uses:
words = set(txt.split(' '))
words
# Now let's count their occurrences:
counts = {word_i: 0 for word_i in words}
for word_i in txt.split(' '):
counts[word_i] += 1
counts
# We can sort this like so:
[(word_i, counts[word_i]) for word_i in sorted(counts, key=counts.get, reverse=True)]
# As we should expect, "the" is the most common word, as it is in the English language: https://en.wikipedia.org/wiki/Most_common_words_in_English
#
# <a name="loading-the-pre-trained-trump-model"></a>
# ## Loading the Pre-trained Trump Model
#
# Let's load the pretrained model. Rather than provide a tfmodel export, I've provided the checkpoint so you can also experiment with training it more if you wish. We'll rebuild the graph using the `charrnn` module in the `libs` directory:
from libs import charrnn
# Let's get the checkpoint and build the model then restore the variables from the checkpoint. The only parameters of consequence are `n_layers` and `n_cells` which define the total size and layout of the model. The rest are flexible. We'll set the `batch_size` and `sequence_length` to 1, meaning we can feed in a single character at a time only, and get back 1 character denoting the very next character's prediction.
ckpt_name = './trump.ckpt'
g = tf.Graph()
n_layers = 3
n_cells = 512
with tf.Session(graph=g) as sess:
model = charrnn.build_model(txt=txt,
batch_size=1,
sequence_length=1,
n_layers=n_layers,
n_cells=n_cells,
gradient_clip=10.0)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
if os.path.exists(ckpt_name):
saver.restore(sess, ckpt_name)
print("Model restored.")
# Let's now take a look at the model:
nb_utils.show_graph(g.as_graph_def())
n_iterations = 100
# <a name="inference-keeping-track-of-the-state"></a>
# ## Inference: Keeping Track of the State
#
# Now recall from Part 4 when we created our LSTM network, we had an `initial_state` variable which would set the LSTM's `c` and `h` state vectors, as well as the final output state which was the output of the `c` and `h` state vectors after having passed through the network. When we input to the network some letter, say 'n', we can set the `initial_state` to zeros, but then after having input the letter `n`, we'll have as output a new state vector for `c` and `h`. On the next letter, we'll then want to set the `initial_state` to this new state, and set the input to the previous letter's output. That is how we ensure the network keeps track of time and knows what has happened in the past, and let it continually generate.
curr_states = None
g = tf.Graph()
with tf.Session(graph=g) as sess:
model = charrnn.build_model(txt=txt,
batch_size=1,
sequence_length=1,
n_layers=n_layers,
n_cells=n_cells,
gradient_clip=10.0)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
if os.path.exists(ckpt_name):
saver.restore(sess, ckpt_name)
print("Model restored.")
# Get every tf.Tensor for the initial state
init_states = []
for s_i in model['initial_state']:
init_states.append(s_i.c)
init_states.append(s_i.h)
# Similarly, for every state after inference
final_states = []
for s_i in model['final_state']:
final_states.append(s_i.c)
final_states.append(s_i.h)
# Let's start with the letter 't' and see what comes out:
synth = [[encoder[' ']]]
for i in range(n_iterations):
# We'll create a feed_dict parameter which includes what to
# input to the network, model['X'], as well as setting
# dropout to 1.0, meaning no dropout.
feed_dict = {model['X']: [synth[-1]],
model['keep_prob']: 1.0}
# Now we'll check if we currently have a state as a result
# of a previous inference, and if so, add to our feed_dict
# parameter the mapping of the init_state to the previous
# output state stored in "curr_states".
if curr_states:
feed_dict.update(
{init_state_i: curr_state_i
for (init_state_i, curr_state_i) in
zip(init_states, curr_states)})
# Now we can infer and see what letter we get
p = sess.run(model['probs'], feed_dict=feed_dict)[0]
# And make sure we also keep track of the new state
curr_states = sess.run(final_states, feed_dict=feed_dict)
# Find the most likely character
p = np.argmax(p)
# Append to string
synth.append([p])
# Print out the decoded letter
print(model['decoder'][p], end='')
sys.stdout.flush()
# <a name="probabilistic-sampling"></a>
# ## Probabilistic Sampling
#
# Run the above cell a couple times. What you should find is that it is deterministic. We always pick *the* most likely character. But we can do something else which will make things less deterministic and a bit more interesting: we can sample from our probabilistic measure from our softmax layer. This means if we have the letter 'a' as 0.4, and the letter 'o' as 0.2, we'll have a 40% chance of picking the letter 'a', and 20% chance of picking the letter 'o', rather than simply always picking the letter 'a' since it is the most probable.
curr_states = None
g = tf.Graph()
with tf.Session(graph=g) as sess:
model = charrnn.build_model(txt=txt,
batch_size=1,
sequence_length=1,
n_layers=n_layers,
n_cells=n_cells,
gradient_clip=10.0)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
if os.path.exists(ckpt_name):
saver.restore(sess, ckpt_name)
print("Model restored.")
# Get every tf.Tensor for the initial state
init_states = []
for s_i in model['initial_state']:
init_states.append(s_i.c)
init_states.append(s_i.h)
# Similarly, for every state after inference
final_states = []
for s_i in model['final_state']:
final_states.append(s_i.c)
final_states.append(s_i.h)
# Let's start with the letter 't' and see what comes out:
synth = [[encoder[' ']]]
for i in range(n_iterations):
# We'll create a feed_dict parameter which includes what to
# input to the network, model['X'], as well as setting
# dropout to 1.0, meaning no dropout.
feed_dict = {model['X']: [synth[-1]],
model['keep_prob']: 1.0}
# Now we'll check if we currently have a state as a result
# of a previous inference, and if so, add to our feed_dict
# parameter the mapping of the init_state to the previous
# output state stored in "curr_states".
if curr_states:
feed_dict.update(
{init_state_i: curr_state_i
for (init_state_i, curr_state_i) in
zip(init_states, curr_states)})
# Now we can infer and see what letter we get
p = sess.run(model['probs'], feed_dict=feed_dict)[0]
# And make sure we also keep track of the new state
curr_states = sess.run(final_states, feed_dict=feed_dict)
# Now instead of finding the most likely character,
# we'll sample with the probabilities of each letter
p = p.astype(np.float64)
p = np.random.multinomial(1, p.ravel() / p.sum())
p = np.argmax(p)
# Append to string
synth.append([p])
# Print out the decoded letter
print(model['decoder'][p], end='')
sys.stdout.flush()
# <a name="inference-temperature"></a>
# ## Inference: Temperature
#
# When performing probabilistic sampling, we can also use a parameter known as temperature which comes from simulated annealing. The basic idea is that as the temperature is high and very hot, we have a lot more free energy to use to jump around more, and as we cool down, we have less energy and then become more deterministic. We can use temperature by scaling our log probabilities like so:
temperature = 0.5
curr_states = None
g = tf.Graph()
with tf.Session(graph=g) as sess:
model = charrnn.build_model(txt=txt,
batch_size=1,
sequence_length=1,
n_layers=n_layers,
n_cells=n_cells,
gradient_clip=10.0)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
if os.path.exists(ckpt_name):
saver.restore(sess, ckpt_name)
print("Model restored.")
# Get every tf.Tensor for the initial state
init_states = []
for s_i in model['initial_state']:
init_states.append(s_i.c)
init_states.append(s_i.h)
# Similarly, for every state after inference
final_states = []
for s_i in model['final_state']:
final_states.append(s_i.c)
final_states.append(s_i.h)
# Let's start with the letter 't' and see what comes out:
synth = [[encoder[' ']]]
for i in range(n_iterations):
# We'll create a feed_dict parameter which includes what to
# input to the network, model['X'], as well as setting
# dropout to 1.0, meaning no dropout.
feed_dict = {model['X']: [synth[-1]],
model['keep_prob']: 1.0}
# Now we'll check if we currently have a state as a result
# of a previous inference, and if so, add to our feed_dict
# parameter the mapping of the init_state to the previous
# output state stored in "curr_states".
if curr_states:
feed_dict.update(
{init_state_i: curr_state_i
for (init_state_i, curr_state_i) in
zip(init_states, curr_states)})
# Now we can infer and see what letter we get
p = sess.run(model['probs'], feed_dict=feed_dict)[0]
# And make sure we also keep track of the new state
curr_states = sess.run(final_states, feed_dict=feed_dict)
# Now instead of finding the most likely character,
# we'll sample with the probabilities of each letter
p = p.astype(np.float64)
p = np.log(p) / temperature
p = np.exp(p) / np.sum(np.exp(p))
p = np.random.multinomial(1, p.ravel() / p.sum())
p = np.argmax(p)
# Append to string
synth.append([p])
# Print out the decoded letter
print(model['decoder'][p], end='')
sys.stdout.flush()
# <a name="inference-priming"></a>
# ## Inference: Priming
#
# Let's now work on "priming" the model with some text, and see what kind of state it is in and leave it to synthesize from there. We'll do more or less what we did before, but feed in our own text instead of the last letter of the synthesis from the model.
prime = "obama"
temperature = 1.0
curr_states = None
n_iterations = 500
g = tf.Graph()
with tf.Session(graph=g) as sess:
model = charrnn.build_model(txt=txt,
batch_size=1,
sequence_length=1,
n_layers=n_layers,
n_cells=n_cells,
gradient_clip=10.0)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver()
if os.path.exists(ckpt_name):
saver.restore(sess, ckpt_name)
print("Model restored.")
# Get every tf.Tensor for the initial state
init_states = []
for s_i in model['initial_state']:
init_states.append(s_i.c)
init_states.append(s_i.h)
# Similarly, for every state after inference
final_states = []
for s_i in model['final_state']:
final_states.append(s_i.c)
final_states.append(s_i.h)
# Now we'll keep track of the state as we feed it one
# letter at a time.
curr_states = None
for ch in prime:
feed_dict = {model['X']: [[model['encoder'][ch]]],
model['keep_prob']: 1.0}
if curr_states:
feed_dict.update(
{init_state_i: curr_state_i
for (init_state_i, curr_state_i) in
zip(init_states, curr_states)})
# Now we can infer and see what letter we get
p = sess.run(model['probs'], feed_dict=feed_dict)[0]
p = p.astype(np.float64)
p = np.log(p) / temperature
p = np.exp(p) / np.sum(np.exp(p))
p = np.random.multinomial(1, p.ravel() / p.sum())
p = np.argmax(p)
# And make sure we also keep track of the new state
curr_states = sess.run(final_states, feed_dict=feed_dict)
# Now we're ready to do what we were doing before but with the
# last predicted output stored in `p`, and the current state of
# the model.
synth = [[p]]
print(prime + model['decoder'][p], end='')
for i in range(n_iterations):
# Input to the network
feed_dict = {model['X']: [synth[-1]],
model['keep_prob']: 1.0}
# Also feed our current state
feed_dict.update(
{init_state_i: curr_state_i
for (init_state_i, curr_state_i) in
zip(init_states, curr_states)})
# Inference
p = sess.run(model['probs'], feed_dict=feed_dict)[0]
# Keep track of the new state
curr_states = sess.run(final_states, feed_dict=feed_dict)
# Sample
p = p.astype(np.float64)
p = np.log(p) / temperature
p = np.exp(p) / np.sum(np.exp(p))
p = np.random.multinomial(1, p.ravel() / p.sum())
p = np.argmax(p)
# Append to string
synth.append([p])
# Print out the decoded letter
print(model['decoder'][p], end='')
sys.stdout.flush()
# <a name="assignment-submission"></a>
# # Assignment Submission
# After you've completed both notebooks, create a zip file of the current directory using the code below. This code will make sure you have included this completed ipython notebook and the following files named exactly as:
#
# session-5/
# session-5-part-1.ipynb
# session-5-part-2.ipynb
# vaegan.gif
# You'll then submit this zip file for your third assignment on Kadenze for "Assignment 5: Generative Adversarial Networks and Recurrent Neural Networks"! If you have any questions, remember to reach out on the forums and connect with your peers or with me.
#
# To get assessed, you'll need to be a premium student! This will allow you to build an online portfolio of all of your work and receive grades. If you aren't already enrolled as a student, register now at http://www.kadenze.com/ and join the #CADL community to see what your peers are doing! https://www.kadenze.com/courses/creative-applications-of-deep-learning-with-tensorflow/info
#
# Also, if you share any of the GIFs on Facebook/Twitter/Instagram/etc..., be sure to use the #CADL hashtag so that other students can find your work!
utils.build_submission('session-5.zip',
('vaegan.gif',
'session-5-part-1.ipynb',
'session-5-part-2.ipynb'))
| session-5/session-5-part-2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Repositorio GCM-TFG
# ## Problemas de optimización en el modelado de materia oscura galáctica
#
# En este repositorio se implementan las funciones necesarias para tratar el problema de modelado de componentes galácticas, en particular el de la componente de materia oscura.
# A continuación se detallan los ficheros y sus respectivas funciones, así como los tipos y estructuras de datos que usan.
# ### `data.py`
# Contiene la lista `galaxlist` de galaxias a explorar. Para cada galaxia se abre y lee su respectivo archivo `.arff`, en el que cada fila corresponde a una partícula que se ha observado que gira en torno a la galaxia. Para cada una de estas partículas se tienen los siguientes datos:
# * Radio de giro en torno a la galaxia
# * Velocidad rotacional de la partícula
# * Errores
# * Velocidad debida a la materia bariónica bariónica
#
# A partir de estos datos construye el diccionario `galaxies` donde se asocia cada galaxia con los datos que se tienen de ella:
# * `R`: Vector de radios de giro de las diferentes partículas observadas
# * `vrot`: Vector de velocidades rotacionales
# * `errs`: Vector de errores
# * `vbary`: Vector de velocidades debidas a la materia bariónica
#
# A continuación se muestra un ejemplo de lectura y recogida de datos de una de las 23 galaxias estudiadas.
# +
from scipy.io import arff
import numpy as np
galaxlist = ["DDO43"]
galaxies = {}
for i in galaxlist:
fp = open("galaxies/"+i+".arff")
dt, metadt = arff.loadarff(fp)
data = []
for d in dt.tolist():
data.append(np.asarray(d))
data = np.asarray(data)
galaxies[i] = {
"R": data[:, 0] * 1000,
"vrot": abs(data[:, 1]),
"errs": data[:, 3],
"vbary": np.sqrt(data[:, 4] ** 2 + data[:, 5] ** 2)
}
fp.close()
# -
# Vector de radios de la galaxia DDO43:
print(galaxies["DDO43"]["R"])
# Vector de velocidades rotacionales de la galaxia DDO43:
print(galaxies["DDO43"]["vrot"])
# Vector de errores de la galaxia DDO43:
print(galaxies["DDO43"]["errs"])
# Vector de velocidades debidas a la materia bariónica de la galaxia DDO43:
print(galaxies["DDO43"]["vbary"])
# En `data.py` también se declaran las constantes $\nu$ (número de parámetros libres) y $CteDim$ (constante de adimensionalización).
# * Puesto que sólo trabajaremos con los perfiles ISO, BUR y NFW, $\nu = 2$.
# * $CteDim = \frac{10000}{4.51697\times3.0856776^ 2}$.
#
# A continuación creamos el diccionario `galaxdata`, donde almacenaremos los datos que más usaremos, en este caso de la galaxia DDO43.
# +
import commonFunctions as cf
import data as dt
galaxdata = {
"radii": np.array([]),
"vrot": np.array([]),
"vbary": np.array([]),
"weights": np.array([]),
"CteDim": dt.CteDim,
"totalnullvbary": False,
"somenullvbary": False,
"vones": np.array([]),
"vv": np.array([]),
"vvbary": np.array([]),
"profile": '',
"graphic": False
}
for i in galaxlist:
radii = galaxies[i]["R"]
galaxdata["radii"] = radii
vrot = galaxies[i]["vrot"]
galaxdata["vrot"] = vrot
vbary = galaxies[i]["vbary"]
galaxdata["vbary"] = vbary
n = len(radii)
vones = np.ones(n)
galaxdata["vones"] = vones
weights = 1 / ((n - dt.nu) * galaxies[i]["errs"] ** 2)
galaxdata["weights"] = weights
totalnullvbary = np.sum(vbary) == 0
galaxdata["totalnullvbary"] = totalnullvbary
somenullvbary = round(np.prod(vbary)) == 0
galaxdata["somenullvbary"] = somenullvbary
vv = cf.vv(galaxdata)
galaxdata["vv"] = vv
vvbary = cf.vvbary(galaxdata)
galaxdata["vvbary"] = vvbary
# galaxdata["graphic"] = True
# -
# ### `commonFunctions.py`
# Aquí se definen algunas funciones comunes a todas las galaxias y para cualquiera de los perfiles ISO, BUR y NFW.
# * `WeighProd(x, y, sigmas)`: Dados los arrays `x` e `y`, y los pesos `sigmas` devuelve el producto escalar pesado definido en (15).
# * `ginf(x, model)`: Dados un array `x` y un perfil de densidad `model`, devuelve el valor de g cuando s tiende a infinito, definida en la Tabla 2.
# * `eqVLimInf(t, ginf, galaxdata)`: Dados el parámetro `t`, el valor de g definida en la Tabla 2 cuando s tiende a infinito y el diccionario `galaxdata` de datos de la galaxia, devuelve la ecuación definida en (33).
# * `g0(x, model)`: Dado un array `x` y un perfil de densidad `model`, devuelve el valor de g cuando s tiende a cero, definida en la Tabla 2.
# * `eqVLim0(t, g0, galaxdata)`: Dados el parámetro `t`, el valor de g definida en la Tabla 2 cuando s tiende a cero y el diccionario `galaxdata` de datos de la galaxia, devuelve la ecuación definida en (35).
# * `v(r, s, model)`: Dado un array de radios `r`, un array de inversos de parámetros de escalas `s` y un perfil de densidad de materia oscura `model`, devuelve el valor de la ecuación definida en (18) para estos parámetros.
# * `chiquad(rho, s, galaxdata)`: Dados un array de parámetro de densidad central `rho`, un array de de inversos de parámetros de escala `s` y un diccionario de datos de una galaxia `galaxdata`, devuelve el valor de la ecuación definida en (16) para estos parámetros.
# * `rho(s, galaxdata)`: Dados un array de inversos de parámetros de escala `s` y un diccionario de datos de una galaxia `galaxdata`, devuelve el valor de rho estudiado en la Proposición 1.
# * `alphaMV(s, galaxdata)`: Dados un array de inversos de parámetros de escala `s` y un diccionario de datos de una galaxia `galaxdata`, devuelve el valor de la ecuación (24) para estos parámetros.
# * `vv(galaxdata)`: Dado un diccionario de datos de una galaxia `galaxdata`, devuelve el producto escalar pesado de la velocidad rotacional.
# * `vvbary(galaxdata)`: Dado un diccionario de datos de una galaxia `galaxdata`, devuelve el el producto escalar pesado de la velocidad debida a la materia bariónica.
# * `phi(s, galaxdata)`: Dados un array de inversos de parámetros de escala `s` y un diccionario de datos de una galaxia `galaxdata`, devuelve el valor de la función varphi y el valor de la función rho para estos parámetros.
#
# ### `calLimits.py`
# Aquí se define la función que calcula los límites de varphi en cero y en infinito, usando el Lema 1.
# * `calLimits(galaxdata)`: Dado un diccionario de datos de una galaxia `galaxdata`, devuelve un array con los valores de los límites de varphi en cero y en infinito.
# Calculamos los límites para la galaxia DDO43 con el perfil ISO.
# +
from calLimits import *
galaxdata["profile"] = "ISO"
varphiLim0, varphiLimInf = calLimits(galaxdata)
# -
# El límite de varphi cuando s tiende a 0 para la galaxia DDO43 con el perfil ISO es
print(varphiLim0)
# El límite de varphi cuando s tiende a infinito para la galaxia DDO43 con el perfil ISO es
print(varphiLimInf)
# Calculamos los límites para la galaxia DDO43 con el perfil BUR.
galaxdata["profile"] = "BUR"
varphiLim0, varphiLimInf = calLimits(galaxdata)
# El límite de varphi cuando s tiende a 0 para la galaxia DDO43 con el perfil BUR es
print(varphiLim0)
# El límite de varphi cuando s tiende a infinito para la galaxia DDO43 con el perfil BUR es
print(varphiLimInf)
# Calculamos los límites para la galaxia DDO43 con el perfil NFW.
galaxdata["profile"] = "NFW"
varphiLim0, varphiLimInf = calLimits(galaxdata)
# El límite de varphi cuando s tiende a 0 para la galaxia DDO43 con el perfil NFW es
print(varphiLim0)
# + active=""
# El límite de varphi cuando s tiende a infinito para la galaxia DDO43 con el perfil NFW es
# -
print(varphiLimInf)
# ### `intervalMinim.py`
# Aquí se definen las distintas funciones que forman el algoritmo de reducción del intervalo de búsqueda. Se especifica una tolerancia `tol`$=10^{-2}$ y se fija la semilla del random a 1.
# * `inftestElementwise(eval)`: Dado un array de puntos vecinos al candidato a extremo inferior del intervalo `eval`, devuelve dos booleanos. El primero indica si los puntos vecinos de la derecha cumplen la ecuación (40) y el segundo indica si la cumplen los vecinos de la izquierda.
# * `suptestElementwise(eval)`: Dado un array de puntos vecinos al candidato a extremo superior del intervalo `eval`, devuelve dos booleanos. El primero indica si los puntos vecinos de la izquierda cumplen la ecuación (39) y el segundo indica si la cumplen los vecinos de la derecha.
# * `inftestElementsum(eval)`: Dado un array de puntos vecinos al candidato a extremo inferior del intervalo `eval`, devuelve dos booleanos. El primero indica si la suma de los puntos vecinos de la derecha cumplen la ecuación (40) y el segundo indica si la cumple la suma de los vecinos de la izquierda.
# * `suptestElementsum(eval)`: Dado un array de puntos vecinos al candidato a extremo superior del intervalo `eval`, devuelve dos booleanos. El primero indica si la suma de los puntos vecinos de la izquierda cumplen la ecuación (39) y el segundo indica si la cumple la suma de los vecinos de la derecha.
# * `infConditions(test1, test2, intervalinf, stop, i)`: Dados un booleano `test1` indicando si los puntos de la derecha (o su suma) cumplen (40), un booleano `test2` indicando si los puntos de la izquierda (o su suma) cumplen (40), un candidato a extremo inferior del intervalo `intervalinf`, un parámetro que controla la condición de parada del algoritmo `stop` y un parámetro `i` que almacena el anterior candidato a extremo inferior en caso de que estemos acercándonos a estar en condición de parada, la función decide si el candidato cumple la condición óptima y en qué dirección moverse.
#
# Supongamos que estamos evaluando la situación de un candidato `intervalinf = 1.5` a extremo inferior. Sus vecinos de la derecha no cumplen (40), y sus vecinos de la izquierda tampoco, es decir, `test1 = False` y `test2 = False`. No estamos en condición de parada, `stop = False`, y el candidato anterior a extremo inferior `i` es cualquiera, supongamos `i=2.0`.
# +
from intervalMinim import *
test1 = False
test2 = False
stop = False
i = 2.0
intervalinf = 1.5
new_intervalinf, direction, stop, i = infConditions(test1, test2, intervalinf, stop, i)
# -
# ¿En qué dirección debemos movernos?
print(direction)
# El nuevo candidato a extremo inferior es
print(new_intervalinf)
# ¿Estamos en condición de parada?
print(stop)
# El candidato anterior no ha cambiado, el valor de i sigue siendo
print(i)
# Supongamos ahora que estamos evaluando la situación de un candidato `intervalinf = 1.2` a extremo inferior que no está en condición de parada, `stop = False`. Sus vecinos de la izquierda cumplen (40), pero sus vecinos de la derecha no, es decir, `test2 = True` y `test1 = False`. El candidato anterior a extremo inferior `i`es cualquiera, supongamos `i=1.25`.
# +
test1 = False
test2 = True
stop = False
i = 1.28
intervalinf = 1.2
new_intervalinf, direction, stop, i = infConditions(test1, test2, intervalinf, stop, i)
# -
# ¿En qué dirección debemos movernos?
print(direction)
# Nos movemos para comprobar en la siguiente iteración que los puntos en esta dirección siguen cumpliendo (40). En la siguiente iteración estudiaremos el punto
print(new_intervalinf)
# ¿Estamos en condición de parada?
print(stop)
# El candidato anterior ha cambiado, ahora el valor de i es
print(i)
# Ahora, para alcanzar por completo la condicón óptima, los valores a la izquierda del candidato `i = 1.2` deberían cumplir (40). Supongamos que sí: `test1 = True` y `test2 = True`.
# +
test1 = True
test2 = True
intervalinf, direction, stop, i = infConditions(test1, test2, new_intervalinf, stop, i)
# -
# Ahora la dirección es
print(direction)
# Esta dirección indica que hemos alcanzaco la condición de parada. Recuperamos de i el que era nuestro candidato. Ahora intervalinf tiene el valor
print(intervalinf)
# * `supConditions(test1, test2, intervalsup, stop, i)`: Dados un booleano `test1` indicando si los puntos de la izquierda (o su suma) cumplen (39), un booleano `test2` indicando si los puntos de la derecha (o su suma) cumplen (39), un candidato a extremo superior del intervalo `intervalsup`, un parámetro que controla la condición de parada del algoritmo `stop` y un parámetro `i` que almacena el anterior candidato a extremo superior, la función decide si el candidato cumple la condición óptima y en qué dirección moverse.
# * `jumpCondition(twoclosevar, varLimdistance, interval, direction, k)`: Dados un booleano `twoclosevar` indicando si los dos últimos candidatos están "cerca", un valor `varLimdistance` indicando a qué distancia está el candidato del valor del límite, un candidato `interval`, una dirección (-1, 0 o 1) `direction` y un contador de la condición de salto `k`, la función devuelve si ha habido salto y, en caso de que sí, cuál es el nuevo candidato.
# * `intervalMin(varphiLim0, varphiLimInf, galaxdata)`: Dados el valor del límite de varphi en cero `varphiLim0`, el valor del límite de varphi en infinito `varphiLimInf` y un diccionario de datos de una galaxia `galaxdata`, la función realiza la reducción del intervalo de búsqueda. Primero busca el extremo inferior que cumple alguna condición satisfactoria y luego el extremo superior, análogamente. Finalmente devuelve los valores propuestos como extremos, el valor mínimo de varphi encontrado en la búsqueda del extremo inferior y el valor mínimo de varphi encontrado en la búsqueda del extremo superior. También puede devolver datos para la elaboración de gráficas.
#
# A continuación hacemos la minimización del intervalo de búsqueda para la galaxia DDO43 con el perfil ISO.
# +
galaxdata["profile"] = "ISO"
varphiLim0, varphiLimInf = calLimits(galaxdata)
interval, intinfmin, intsupmin = intervalMin(varphiLim0, varphiLimInf, galaxdata)
intervalinf = interval[0]
intervalsup = interval[1]
# -
# Así, el extremo inferior del intervalo de búsqueda tras su exploración es
print(intervalinf)
# El extremo superior del intervalo de búsqueda tras su exploración es
print(intervalsup)
# El valor mínimo de varphi encontrado en la exploración del extremo inferior es
print(intinfmin[1])
# para el valor de s
print(intinfmin[0])
# El valor mínimo de varphi encontrado en la exploración del extremo superior es
print(intsupmin[1])
# para el valor de s
print(intsupmin[0])
# A continuación se muestra la exploración del intervalo de búsqueda para su reducción para la galaxia DDO43 con perfil ISO. Los puntos rojos representan los puntos explorados y la línea negra el intervalo deducido en el algoritmo.
# +
import matplotlib.pyplot as plt
# %matplotlib inline
galaxdata["graphic"] = True
res = intervalMin(varphiLim0, varphiLimInf, galaxdata)
intervalinf = res[0][0]
intervalsup = res[0][1]
Xi = res[1]
Yi = res[2]
intinfmin = res[3]
intsupmin = res[4]
plt.semilogx()
plt.title("Galaxia DDO43 con perfil ISO")
plt.xlabel("s (parámetro de escala)")
plt.ylabel(r"$\varphi(s)$")
plt.scatter(intervalinf, 0, c='black', marker=3)
plt.scatter(intervalsup, 0, c='black', marker=3)
plt.hlines(0, intervalinf, intervalsup)
plt.scatter(Xi, Yi, c='r', marker='.')
plt.show()
# -
# ### `varphiMinim.py`
# Aquí se definen las distintas funciones que forman el algoritmo de minimización de la función varphi.
# * `getIMD(intizq, intder, galaxdata)`: Dados el extremo inferior del intervalo `intizq`, el extremo superior del intervalo `intder`y un diccionario de datos de una galaxia `galaxdata`, la función devuelve el valor medio `m`y su evaluación en varphi, así como un punto aleatorio a la derecha y otro a la izquierda, con sus respectivas evaluaciones en varphi.
# * `reductionInterval(varphiLim0, varphiLimInf, intinfmin, intsupmin, intervalinf, intervalsup)`: Realiza la mejora propuesta en la memoria para el algoritmo de minimización de varphi. Dados el límite de varphi en 0 `varphiLim0`, el límite de varphi en infinito `varphiLimInf`, el punto mínimo encontrado en la exploración del intervalo inferior `intinfmin`, el punto mínimo encontrado en la exploración del intervalo superior `intsupmin`, el extremo inferior del intervalo calculado en intervalMinim.py `intervalinf` y el extremo superior del intervalo calculado en intervalMinim.py `intervalsup`, la función devuelve el intervalo de búsqueda nuevamente reducido (en caso de que haya sido posible reducirlo).
# * `varphiMin(varphiLim0, varphiLimInf, intinfmin, intsupmin, intervalinf, intervalsup, galaxdata)`: Dados el límite de varphi cuando s tiende a 0 `varphiLim0`, el límite de varphi cuando s tiende a infinito `varphiLimInf`, el punto mínimo encontrado en la exploración del intervalo inferior `intinfmin`, el punto mínimo encontrado en la exploración del intervalo superior `intsupmin`, el extremo inferior del intervalo calculado en intervalMinim.py `intervalinf`, el extremo superior del intervalo calculado en intervalMinim.py `intervalsup` y el diccionario de datos de una galaxia `galaxdata`, la función realiza la exploración de varphi y devuelve el mínimo valor encontrado.
#
# A continuación realizamos la minimización de varphi para la galaxia DDO43 con el perfil ISO.
# +
from varphiMinim import *
res = varphiMin(varphiLim0, varphiLimInf, intinfmin, intsupmin, intervalinf, intervalsup, galaxdata)
minvarphi = res[0]
minrho = res[1]
minvarphiX = res[2]
Xj = res[3]
Yj = res[4]
forkpoints = res[5]
X = res[6]
intervalinf = res[7]
intervalsup = res[8]
# -
# Así, el intervalo de búsqueda tras aplicar la mejora propuesta en el algoritmo de minimización de varphi es
print("[", intervalinf, ", ", intervalsup, "]")
# El valor mínimo de varphi encontrado es
print(minvarphi)
# para el valor de s
print(minvarphiX)
# Mientras que el valor de la función rho (para este valor de s) definida en la Proposición 1 es
print(minrho)
# A continuación se muestra la exploración del intervalo de búsqueda para la minimización de varphi para la galaxia DDO43 con perfil ISO. Los puntos rojos representan los puntos explorados en el algoritmo de reducción del intervalo, la línea negra el intervalo deducido en el algoritmo y los puntos azules los puntos explorados en la minimización de varphi.
plt.semilogx()
plt.title("Galaxia DDO43 con perfil ISO")
plt.xlabel("s (parámetro de escala)")
plt.ylabel(r"$\varphi(s)$")
plt.scatter(intervalinf, 0, c='black', marker=3)
plt.scatter(intervalsup, 0, c='black', marker=3)
plt.hlines(0, intervalinf, intervalsup)
plt.scatter(Xi, Yi, c='r', marker='.')
plt.scatter(X, np.zeros(len(X)), color='black', marker=3)
plt.scatter(Xj, Yj, c='b', marker='.', linewidths=0.01)
plt.show()
# ### `redMethRotCurveFitting.py`
# Aquí es donde se realiza todo el proceso de ajuste de curvas de rotación, acudiendo a las funciones mencionadas anteriormente. Consta de tres partes: cálculo de límites, reducción del intervalo de búsqueda y minimización de la función varphi. A continuación, incluimos una galaxia más en nuestro conjunto de galaxias y repetimos el proceso desarrollado anteriormente para los perfiles ISO, BUR y NFW, a modo de ejemplo.
# +
import data as dt
galaxlist = ["DDO43", "DDO46"]
fp = open("galaxies/DDO46.arff")
dat, metadt = arff.loadarff(fp)
data = []
for d in dat.tolist():
data.append(np.asarray(d))
data = np.asarray(data)
galaxies["DDO46"] = {
"R": data[:, 0] * 1000,
"vrot": abs(data[:, 1]),
"errs": data[:, 3],
"vbary": np.sqrt(data[:, 4] ** 2 + data[:, 5] ** 2)
}
fp.close()
radii = galaxies["DDO46"]["R"]
galaxdata["radii"] = radii
vrot = galaxies["DDO46"]["vrot"]
galaxdata["vrot"] = vrot
vbary = galaxies["DDO46"]["vbary"]
galaxdata["vbary"] = vbary
n = len(radii)
vones = np.ones(n)
galaxdata["vones"] = vones
weights = 1 / ((n - dt.nu) * galaxies["DDO46"]["errs"] ** 2)
galaxdata["weights"] = weights
totalnullvbary = np.sum(vbary) == 0
galaxdata["totalnullvbary"] = totalnullvbary
somenullvbary = round(np.prod(vbary)) == 0
galaxdata["somenullvbary"] = somenullvbary
vv = cf.vv(galaxdata)
galaxdata["vv"] = vv
vvbary = cf.vvbary(galaxdata)
galaxdata["vvbary"] = vvbary
galaxdata["graphic"] = False
profiles = ["ISO", "BUR", "NFW"]
for g in galaxies:
print("\n")
print("GALAXIA ", g)
for p in profiles:
galaxdata["profile"] = p
print("Para el perfil ", p)
"""
Cálculo de límites
"""
limits = calLimits(galaxdata)
varphiLim0 = limits[0]
varphiLimInf = limits[1]
print("El límite de varphi cuando s tiende a cero es ", varphiLim0)
print("El límite de varphi cuando s tiende a infinito es ", varphiLimInf)
"""
Minimización del intervalo de búsqueda
"""
interval = intervalMin(varphiLim0, varphiLimInf, galaxdata)
intervalinf = interval[0][0]
intervalsup = interval[0][1]
print("El intervalo de búsqueda deducido es [", intervalinf, ", ", intervalsup, "]")
intinfmin = interval[1]
intsupmin = interval[2]
print("Mínimo encontrado en la exploración del intervalo inferior: ", intinfmin)
print("Mínimo encontrado en la exploración del intervalo superior: ", intsupmin)
"""
Minimización de la función varphi
"""
pmin = varphiMin(varphiLim0, varphiLimInf, intinfmin, intsupmin, intervalinf, intervalsup, galaxdata)
minvarphi = pmin[0]
minrho = pmin[1]
minvarphiX = pmin[2]
intervalinf = pmin[5]
intervalsup = pmin[6]
print("Tras la mejora propuesta en. el algoritmo de minimización de varphi, el intervalo de búsqueda es [", intervalinf, ", ", intervalsup, "]")
print("El intervalo de búsqueda tras aplicar la mejora propuesta en el algoritmo de minimización de varphi es [", intervalinf, ", ", intervalsup, "]")
print("El valor mínimo de varphi encontrado es ", minvarphi, ", para s = ", minvarphiX)
print("El valor de rho(", minvarphiX, ") = ", minrho)
# -
| Tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Emojify!
#
# Welcome to the second assignment of Week 2. You are going to use word vector representations to build an Emojifier.
#
# Have you ever wanted to make your text messages more expressive? Your emojifier app will help you do that. So rather than writing "Congratulations on the promotion! Lets get coffee and talk. Love you!" the emojifier can automatically turn this into "Congratulations on the promotion! 👍 Lets get coffee and talk. ☕️ Love you! ❤️"
#
# You will implement a model which inputs a sentence (such as "Let's go see the baseball game tonight!") and finds the most appropriate emoji to be used with this sentence (⚾️). In many emoji interfaces, you need to remember that ❤️ is the "heart" symbol rather than the "love" symbol. But using word vectors, you'll see that even if your training set explicitly relates only a few words to a particular emoji, your algorithm will be able to generalize and associate words in the test set to the same emoji even if those words don't even appear in the training set. This allows you to build an accurate classifier mapping from sentences to emojis, even using a small training set.
#
# In this exercise, you'll start with a baseline model (Emojifier-V1) using word embeddings, then build a more sophisticated model (Emojifier-V2) that further incorporates an LSTM.
#
# Lets get started! Run the following cell to load the package you are going to use.
# +
import numpy as np
from emo_utils import *
import emoji
import matplotlib.pyplot as plt
# %matplotlib inline
# -
# ## 1 - Baseline model: Emojifier-V1
#
# ### 1.1 - Dataset EMOJISET
#
# Let's start by building a simple baseline classifier.
#
# You have a tiny dataset (X, Y) where:
# - X contains 127 sentences (strings)
# - Y contains a integer label between 0 and 4 corresponding to an emoji for each sentence
#
# <img src="images/data_set.png" style="width:700px;height:300px;">
# <caption><center> **Figure 1**: EMOJISET - a classification problem with 5 classes. A few examples of sentences are given here. </center></caption>
#
# Let's load the dataset using the code below. We split the dataset between training (127 examples) and testing (56 examples).
X_train, Y_train = read_csv('data/train_emoji.csv')
X_test, Y_test = read_csv('data/tesss.csv')
maxLen = len(max(X_train, key=len).split())
# Run the following cell to print sentences from X_train and corresponding labels from Y_train. Change `index` to see different examples. Because of the font the iPython notebook uses, the heart emoji may be colored black rather than red.
index = 18
print(X_train[index], label_to_emoji(Y_train[index]))
# ### 1.2 - Overview of the Emojifier-V1
#
# In this part, you are going to implement a baseline model called "Emojifier-v1".
#
# <center>
# <img src="images/image_1.png" style="width:900px;height:300px;">
# <caption><center> **Figure 2**: Baseline model (Emojifier-V1).</center></caption>
# </center>
#
# The input of the model is a string corresponding to a sentence (e.g. "I love you). In the code, the output will be a probability vector of shape (1,5), that you then pass in an argmax layer to extract the index of the most likely emoji output.
# To get our labels into a format suitable for training a softmax classifier, lets convert $Y$ from its current shape current shape $(m, 1)$ into a "one-hot representation" $(m, 5)$, where each row is a one-hot vector giving the label of one example, You can do so using this next code snipper. Here, `Y_oh` stands for "Y-one-hot" in the variable names `Y_oh_train` and `Y_oh_test`:
#
Y_oh_train = convert_to_one_hot(Y_train, C = 5)
Y_oh_test = convert_to_one_hot(Y_test, C = 5)
# Let's see what `convert_to_one_hot()` did. Feel free to change `index` to print out different values.
index = 50
print(Y_train[index], "is converted into one hot", Y_oh_train[index])
# All the data is now ready to be fed into the Emojify-V1 model. Let's implement the model!
# ### 1.3 - Implementing Emojifier-V1
#
# As shown in Figure (2), the first step is to convert an input sentence into the word vector representation, which then get averaged together. Similar to the previous exercise, we will use pretrained 50-dimensional GloVe embeddings. Run the following cell to load the `word_to_vec_map`, which contains all the vector representations.
word_to_index, index_to_word, word_to_vec_map = read_glove_vecs('../../readonly/glove.6B.50d.txt')
# You've loaded:
# - `word_to_index`: dictionary mapping from words to their indices in the vocabulary (400,001 words, with the valid indices ranging from 0 to 400,000)
# - `index_to_word`: dictionary mapping from indices to their corresponding words in the vocabulary
# - `word_to_vec_map`: dictionary mapping words to their GloVe vector representation.
#
# Run the following cell to check if it works.
word = "cucumber"
index = 289846
print("the index of", word, "in the vocabulary is", word_to_index[word])
print("the", str(index) + "th word in the vocabulary is", index_to_word[index])
# **Exercise**: Implement `sentence_to_avg()`. You will need to carry out two steps:
# 1. Convert every sentence to lower-case, then split the sentence into a list of words. `X.lower()` and `X.split()` might be useful.
# 2. For each word in the sentence, access its GloVe representation. Then, average all these values.
# +
# GRADED FUNCTION: sentence_to_avg
def sentence_to_avg(sentence, word_to_vec_map):
"""
Converts a sentence (string) into a list of words (strings). Extracts the GloVe representation of each word
and averages its value into a single vector encoding the meaning of the sentence.
Arguments:
sentence -- string, one training example from X
word_to_vec_map -- dictionary mapping every word in a vocabulary into its 50-dimensional vector representation
Returns:
avg -- average vector encoding information about the sentence, numpy-array of shape (50,)
"""
### START CODE HERE ###
# Step 1: Split sentence into list of lower case words (≈ 1 line)
words = (sentence.lower()).split()
# Initialize the average word vector, should have the same shape as your word vectors.
avg = np.zeros((50, ))
# Step 2: average the word vectors. You can loop over the words in the list "words".
for w in words:
avg += word_to_vec_map[w]
avg = avg / len(words)
### END CODE HERE ###
return avg
# -
avg = sentence_to_avg("Morrocan couscous is my favorite dish", word_to_vec_map)
print("avg = ", avg)
# **Expected Output**:
#
# <table>
# <tr>
# <td>
# **avg= **
# </td>
# <td>
# [-0.008005 0.56370833 -0.50427333 0.258865 0.55131103 0.03104983
# -0.21013718 0.16893933 -0.09590267 0.141784 -0.15708967 0.18525867
# 0.6495785 0.38371117 0.21102167 0.11301667 0.02613967 0.26037767
# 0.05820667 -0.01578167 -0.12078833 -0.02471267 0.4128455 0.5152061
# 0.38756167 -0.898661 -0.535145 0.33501167 0.68806933 -0.2156265
# 1.797155 0.10476933 -0.36775333 0.750785 0.10282583 0.348925
# -0.27262833 0.66768 -0.10706167 -0.283635 0.59580117 0.28747333
# -0.3366635 0.23393817 0.34349183 0.178405 0.1166155 -0.076433
# 0.1445417 0.09808667]
# </td>
# </tr>
# </table>
# #### Model
#
# You now have all the pieces to finish implementing the `model()` function. After using `sentence_to_avg()` you need to pass the average through forward propagation, compute the cost, and then backpropagate to update the softmax's parameters.
#
# **Exercise**: Implement the `model()` function described in Figure (2). Assuming here that $Yoh$ ("Y one hot") is the one-hot encoding of the output labels, the equations you need to implement in the forward pass and to compute the cross-entropy cost are:
# $$ z^{(i)} = W . avg^{(i)} + b$$
# $$ a^{(i)} = softmax(z^{(i)})$$
# $$ \mathcal{L}^{(i)} = - \sum_{k = 0}^{n_y - 1} Yoh^{(i)}_k * log(a^{(i)}_k)$$
#
# It is possible to come up with a more efficient vectorized implementation. But since we are using a for-loop to convert the sentences one at a time into the avg^{(i)} representation anyway, let's not bother this time.
#
# We provided you a function `softmax()`.
# +
# GRADED FUNCTION: model
def model(X, Y, word_to_vec_map, learning_rate = 0.01, num_iterations = 400):
"""
Model to train word vector representations in numpy.
Arguments:
X -- input data, numpy array of sentences as strings, of shape (m, 1)
Y -- labels, numpy array of integers between 0 and 7, numpy-array of shape (m, 1)
word_to_vec_map -- dictionary mapping every word in a vocabulary into its 50-dimensional vector representation
learning_rate -- learning_rate for the stochastic gradient descent algorithm
num_iterations -- number of iterations
Returns:
pred -- vector of predictions, numpy-array of shape (m, 1)
W -- weight matrix of the softmax layer, of shape (n_y, n_h)
b -- bias of the softmax layer, of shape (n_y,)
"""
np.random.seed(1)
# Define number of training examples
m = Y.shape[0] # number of training examples
n_y = 5 # number of classes
n_h = 50 # dimensions of the GloVe vectors
# Initialize parameters using Xavier initialization
W = np.random.randn(n_y, n_h) / np.sqrt(n_h)
b = np.zeros((n_y,))
# Convert Y to Y_onehot with n_y classes
Y_oh = convert_to_one_hot(Y, C = n_y)
# Optimization loop
for t in range(num_iterations): # Loop over the number of iterations
for i in range(m): # Loop over the training examples
### START CODE HERE ### (≈ 4 lines of code)
# Average the word vectors of the words from the i'th training example
avg = sentence_to_avg(X[i], word_to_vec_map)
# Forward propagate the avg through the softmax layer
z = np.dot(W, avg) + b
a = softmax(z)
# Compute cost using the i'th training label's one hot representation and "A" (the output of the softmax)
cost = -np.sum(Y_oh[i] * np.log(a))
### END CODE HERE ###
# Compute gradients
dz = a - Y_oh[i]
dW = np.dot(dz.reshape(n_y,1), avg.reshape(1, n_h))
db = dz
# Update parameters with Stochastic Gradient Descent
W = W - learning_rate * dW
b = b - learning_rate * db
if t % 100 == 0:
print("Epoch: " + str(t) + " --- cost = " + str(cost))
pred = predict(X, Y, W, b, word_to_vec_map)
return pred, W, b
# +
print(X_train.shape)
print(Y_train.shape)
print(np.eye(5)[Y_train.reshape(-1)].shape)
print(X_train[0])
print(type(X_train))
Y = np.asarray([5,0,0,5, 4, 4, 4, 6, 6, 4, 1, 1, 5, 6, 6, 3, 6, 3, 4, 4])
print(Y.shape)
X = np.asarray(['I am going to the bar tonight', 'I love you', 'miss you my dear',
'Lets go party and drinks','Congrats on the new job','Congratulations',
'I am so happy for you', 'Why are you feeling bad', 'What is wrong with you',
'You totally deserve this prize', 'Let us go play football',
'Are you down for football this afternoon', 'Work hard play harder',
'It is suprising how people can be dumb sometimes',
'I am very disappointed','It is the best day in my life',
'I think I will end up alone','My life is so boring','Good job',
'Great so awesome'])
print(X.shape)
print(np.eye(5)[Y_train.reshape(-1)].shape)
print(type(X_train))
# -
# Run the next cell to train your model and learn the softmax parameters (W,b).
pred, W, b = model(X_train, Y_train, word_to_vec_map)
print(pred)
# **Expected Output** (on a subset of iterations):
#
# <table>
# <tr>
# <td>
# **Epoch: 0**
# </td>
# <td>
# cost = 1.95204988128
# </td>
# <td>
# Accuracy: 0.348484848485
# </td>
# </tr>
#
#
# <tr>
# <td>
# **Epoch: 100**
# </td>
# <td>
# cost = 0.0797181872601
# </td>
# <td>
# Accuracy: 0.931818181818
# </td>
# </tr>
#
# <tr>
# <td>
# **Epoch: 200**
# </td>
# <td>
# cost = 0.0445636924368
# </td>
# <td>
# Accuracy: 0.954545454545
# </td>
# </tr>
#
# <tr>
# <td>
# **Epoch: 300**
# </td>
# <td>
# cost = 0.0343226737879
# </td>
# <td>
# Accuracy: 0.969696969697
# </td>
# </tr>
# </table>
# Great! Your model has pretty high accuracy on the training set. Lets now see how it does on the test set.
# ### 1.4 - Examining test set performance
#
print("Training set:")
pred_train = predict(X_train, Y_train, W, b, word_to_vec_map)
print('Test set:')
pred_test = predict(X_test, Y_test, W, b, word_to_vec_map)
# **Expected Output**:
#
# <table>
# <tr>
# <td>
# **Train set accuracy**
# </td>
# <td>
# 97.7
# </td>
# </tr>
# <tr>
# <td>
# **Test set accuracy**
# </td>
# <td>
# 85.7
# </td>
# </tr>
# </table>
# Random guessing would have had 20% accuracy given that there are 5 classes. This is pretty good performance after training on only 127 examples.
#
# In the training set, the algorithm saw the sentence "*I love you*" with the label ❤️. You can check however that the word "adore" does not appear in the training set. Nonetheless, lets see what happens if you write "*I adore you*."
#
#
# +
X_my_sentences = np.array(["i adore you", "i love you", "funny lol", "lets play with a ball", "food is ready", "not feeling happy"])
Y_my_labels = np.array([[0], [0], [2], [1], [4],[3]])
pred = predict(X_my_sentences, Y_my_labels , W, b, word_to_vec_map)
print_predictions(X_my_sentences, pred)
# -
# Amazing! Because *adore* has a similar embedding as *love*, the algorithm has generalized correctly even to a word it has never seen before. Words such as *heart*, *dear*, *beloved* or *adore* have embedding vectors similar to *love*, and so might work too---feel free to modify the inputs above and try out a variety of input sentences. How well does it work?
#
# Note though that it doesn't get "not feeling happy" correct. This algorithm ignores word ordering, so is not good at understanding phrases like "not happy."
#
# Printing the confusion matrix can also help understand which classes are more difficult for your model. A confusion matrix shows how often an example whose label is one class ("actual" class) is mislabeled by the algorithm with a different class ("predicted" class).
#
#
#
print(Y_test.shape)
print(' '+ label_to_emoji(0)+ ' ' + label_to_emoji(1) + ' ' + label_to_emoji(2)+ ' ' + label_to_emoji(3)+' ' + label_to_emoji(4))
print(pd.crosstab(Y_test, pred_test.reshape(56,), rownames=['Actual'], colnames=['Predicted'], margins=True))
plot_confusion_matrix(Y_test, pred_test)
# <font color='blue'>
# **What you should remember from this part**:
# - Even with a 127 training examples, you can get a reasonably good model for Emojifying. This is due to the generalization power word vectors gives you.
# - Emojify-V1 will perform poorly on sentences such as *"This movie is not good and not enjoyable"* because it doesn't understand combinations of words--it just averages all the words' embedding vectors together, without paying attention to the ordering of words. You will build a better algorithm in the next part.
#
# ## 2 - Emojifier-V2: Using LSTMs in Keras:
#
# Let's build an LSTM model that takes as input word sequences. This model will be able to take word ordering into account. Emojifier-V2 will continue to use pre-trained word embeddings to represent words, but will feed them into an LSTM, whose job it is to predict the most appropriate emoji.
#
# Run the following cell to load the Keras packages.
import numpy as np
np.random.seed(0)
from keras.models import Model
from keras.layers import Dense, Input, Dropout, LSTM, Activation
from keras.layers.embeddings import Embedding
from keras.preprocessing import sequence
from keras.initializers import glorot_uniform
np.random.seed(1)
# ### 2.1 - Overview of the model
#
# Here is the Emojifier-v2 you will implement:
#
# <img src="images/emojifier-v2.png" style="width:700px;height:400px;"> <br>
# <caption><center> **Figure 3**: Emojifier-V2. A 2-layer LSTM sequence classifier. </center></caption>
#
#
# ### 2.2 Keras and mini-batching
#
# In this exercise, we want to train Keras using mini-batches. However, most deep learning frameworks require that all sequences in the same mini-batch have the same length. This is what allows vectorization to work: If you had a 3-word sentence and a 4-word sentence, then the computations needed for them are different (one takes 3 steps of an LSTM, one takes 4 steps) so it's just not possible to do them both at the same time.
#
# The common solution to this is to use padding. Specifically, set a maximum sequence length, and pad all sequences to the same length. For example, of the maximum sequence length is 20, we could pad every sentence with "0"s so that each input sentence is of length 20. Thus, a sentence "i love you" would be represented as $(e_{i}, e_{love}, e_{you}, \vec{0}, \vec{0}, \ldots, \vec{0})$. In this example, any sentences longer than 20 words would have to be truncated. One simple way to choose the maximum sequence length is to just pick the length of the longest sentence in the training set.
#
# ### 2.3 - The Embedding layer
#
# In Keras, the embedding matrix is represented as a "layer", and maps positive integers (indices corresponding to words) into dense vectors of fixed size (the embedding vectors). It can be trained or initialized with a pretrained embedding. In this part, you will learn how to create an [Embedding()](https://keras.io/layers/embeddings/) layer in Keras, initialize it with the GloVe 50-dimensional vectors loaded earlier in the notebook. Because our training set is quite small, we will not update the word embeddings but will instead leave their values fixed. But in the code below, we'll show you how Keras allows you to either train or leave fixed this layer.
#
# The `Embedding()` layer takes an integer matrix of size (batch size, max input length) as input. This corresponds to sentences converted into lists of indices (integers), as shown in the figure below.
#
# <img src="images/embedding1.png" style="width:700px;height:250px;">
# <caption><center> **Figure 4**: Embedding layer. This example shows the propagation of two examples through the embedding layer. Both have been zero-padded to a length of `max_len=5`. The final dimension of the representation is `(2,max_len,50)` because the word embeddings we are using are 50 dimensional. </center></caption>
#
# The largest integer (i.e. word index) in the input should be no larger than the vocabulary size. The layer outputs an array of shape (batch size, max input length, dimension of word vectors).
#
# The first step is to convert all your training sentences into lists of indices, and then zero-pad all these lists so that their length is the length of the longest sentence.
#
# **Exercise**: Implement the function below to convert X (array of sentences as strings) into an array of indices corresponding to words in the sentences. The output shape should be such that it can be given to `Embedding()` (described in Figure 4).
# +
# GRADED FUNCTION: sentences_to_indices
def sentences_to_indices(X, word_to_index, max_len):
"""
Converts an array of sentences (strings) into an array of indices corresponding to words in the sentences.
The output shape should be such that it can be given to `Embedding()` (described in Figure 4).
Arguments:
X -- array of sentences (strings), of shape (m, 1)
word_to_index -- a dictionary containing the each word mapped to its index
max_len -- maximum number of words in a sentence. You can assume every sentence in X is no longer than this.
Returns:
X_indices -- array of indices corresponding to words in the sentences from X, of shape (m, max_len)
"""
m = X.shape[0] # number of training examples
### START CODE HERE ###
# Initialize X_indices as a numpy matrix of zeros and the correct shape (≈ 1 line)
X_indices = np.zeros((m, max_len))
for i in range(m): # loop over training examples
# Convert the ith training sentence in lower case and split is into words. You should get a list of words.
sentence_words = (X[i].lower()).split()
# Initialize j to 0
j = 0
# Loop over the words of sentence_words
for w in sentence_words:
# Set the (i,j)th entry of X_indices to the index of the correct word.
X_indices[i, j] = word_to_index[w]
# Increment j to j + 1
j = j + 1
### END CODE HERE ###
return X_indices
# -
# Run the following cell to check what `sentences_to_indices()` does, and check your results.
X1 = np.array(["funny lol", "lets play baseball", "food is ready for you"])
X1_indices = sentences_to_indices(X1,word_to_index, max_len = 5)
print("X1 =", X1)
print("X1_indices =", X1_indices)
# **Expected Output**:
#
# <table>
# <tr>
# <td>
# **X1 =**
# </td>
# <td>
# ['funny lol' 'lets play football' 'food is ready for you']
# </td>
# </tr>
# <tr>
# <td>
# **X1_indices =**
# </td>
# <td>
# [[ 155345. 225122. 0. 0. 0.] <br>
# [ 220930. 286375. 151266. 0. 0.] <br>
# [ 151204. 192973. 302254. 151349. 394475.]]
# </td>
# </tr>
# </table>
# Let's build the `Embedding()` layer in Keras, using pre-trained word vectors. After this layer is built, you will pass the output of `sentences_to_indices()` to it as an input, and the `Embedding()` layer will return the word embeddings for a sentence.
#
# **Exercise**: Implement `pretrained_embedding_layer()`. You will need to carry out the following steps:
# 1. Initialize the embedding matrix as a numpy array of zeroes with the correct shape.
# 2. Fill in the embedding matrix with all the word embeddings extracted from `word_to_vec_map`.
# 3. Define Keras embedding layer. Use [Embedding()](https://keras.io/layers/embeddings/). Be sure to make this layer non-trainable, by setting `trainable = False` when calling `Embedding()`. If you were to set `trainable = True`, then it will allow the optimization algorithm to modify the values of the word embeddings.
# 4. Set the embedding weights to be equal to the embedding matrix
# +
# GRADED FUNCTION: pretrained_embedding_layer
def pretrained_embedding_layer(word_to_vec_map, word_to_index):
"""
Creates a Keras Embedding() layer and loads in pre-trained GloVe 50-dimensional vectors.
Arguments:
word_to_vec_map -- dictionary mapping words to their GloVe vector representation.
word_to_index -- dictionary mapping from words to their indices in the vocabulary (400,001 words)
Returns:
embedding_layer -- pretrained layer Keras instance
"""
vocab_len = len(word_to_index) + 1 # adding 1 to fit Keras embedding (requirement)
emb_dim = word_to_vec_map["cucumber"].shape[0] # define dimensionality of your GloVe word vectors (= 50)
### START CODE HERE ###
# Initialize the embedding matrix as a numpy array of zeros of shape (vocab_len, dimensions of word vectors = emb_dim)
emb_matrix = np.zeros((vocab_len, emb_dim))
# Set each row "index" of the embedding matrix to be the word vector representation of the "index"th word of the vocabulary
for word, index in word_to_index.items():
emb_matrix[index, :] = word_to_vec_map[word]
# Define Keras embedding layer with the correct output/input sizes, make it trainable. Use Embedding(...). Make sure to set trainable=False.
embedding_layer = Embedding(vocab_len, emb_dim, trainable = False)
### END CODE HERE ###
# Build the embedding layer, it is required before setting the weights of the embedding layer. Do not modify the "None".
embedding_layer.build((None,))
# Set the weights of the embedding layer to the embedding matrix. Your layer is now pretrained.
embedding_layer.set_weights([emb_matrix])
return embedding_layer
# -
embedding_layer = pretrained_embedding_layer(word_to_vec_map, word_to_index)
print("weights[0][1][3] =", embedding_layer.get_weights()[0][1][3])
# **Expected Output**:
#
# <table>
# <tr>
# <td>
# **weights[0][1][3] =**
# </td>
# <td>
# -0.3403
# </td>
# </tr>
# </table>
# ## 2.3 Building the Emojifier-V2
#
# Lets now build the Emojifier-V2 model. You will do so using the embedding layer you have built, and feed its output to an LSTM network.
#
# <img src="images/emojifier-v2.png" style="width:700px;height:400px;"> <br>
# <caption><center> **Figure 3**: Emojifier-v2. A 2-layer LSTM sequence classifier. </center></caption>
#
#
# **Exercise:** Implement `Emojify_V2()`, which builds a Keras graph of the architecture shown in Figure 3. The model takes as input an array of sentences of shape (`m`, `max_len`, ) defined by `input_shape`. It should output a softmax probability vector of shape (`m`, `C = 5`). You may need `Input(shape = ..., dtype = '...')`, [LSTM()](https://keras.io/layers/recurrent/#lstm), [Dropout()](https://keras.io/layers/core/#dropout), [Dense()](https://keras.io/layers/core/#dense), and [Activation()](https://keras.io/activations/).
# +
# GRADED FUNCTION: Emojify_V2
def Emojify_V2(input_shape, word_to_vec_map, word_to_index):
"""
Function creating the Emojify-v2 model's graph.
Arguments:
input_shape -- shape of the input, usually (max_len,)
word_to_vec_map -- dictionary mapping every word in a vocabulary into its 50-dimensional vector representation
word_to_index -- dictionary mapping from words to their indices in the vocabulary (400,001 words)
Returns:
model -- a model instance in Keras
"""
### START CODE HERE ###
# Define sentence_indices as the input of the graph, it should be of shape input_shape and dtype 'int32' (as it contains indices).
sentence_indices = Input(shape = input_shape, dtype = 'int32')
# Create the embedding layer pretrained with GloVe Vectors (≈1 line)
embedding_layer = pretrained_embedding_layer(word_to_vec_map, word_to_index)
# Propagate sentence_indices through your embedding layer, you get back the embeddings
embeddings = embedding_layer(sentence_indices)
# Propagate the embeddings through an LSTM layer with 128-dimensional hidden state
# Be careful, the returned output should be a batch of sequences.
X = LSTM(units = 128, return_sequences = True)(embeddings)
# Add dropout with a probability of 0.5
X = Dropout(0.5)(X)
# Propagate X trough another LSTM layer with 128-dimensional hidden state
# Be careful, the returned output should be a single hidden state, not a batch of sequences.
X = LSTM(units = 128)(X)
# Add dropout with a probability of 0.5
X = Dropout(0.5)(X)
# Propagate X through a Dense layer with softmax activation to get back a batch of 5-dimensional vectors.
X = Dense(5)(X)
# Add a softmax activation
X = Activation('softmax')(X)
# Create Model instance which converts sentence_indices into X.
model = Model(inputs = sentence_indices, outputs = X)
### END CODE HERE ###
return model
# -
# Run the following cell to create your model and check its summary. Because all sentences in the dataset are less than 10 words, we chose `max_len = 10`. You should see your architecture, it uses "20,223,927" parameters, of which 20,000,050 (the word embeddings) are non-trainable, and the remaining 223,877 are. Because our vocabulary size has 400,001 words (with valid indices from 0 to 400,000) there are 400,001\*50 = 20,000,050 non-trainable parameters.
model = Emojify_V2((maxLen,), word_to_vec_map, word_to_index)
model.summary()
# As usual, after creating your model in Keras, you need to compile it and define what loss, optimizer and metrics your are want to use. Compile your model using `categorical_crossentropy` loss, `adam` optimizer and `['accuracy']` metrics:
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
# It's time to train your model. Your Emojifier-V2 `model` takes as input an array of shape (`m`, `max_len`) and outputs probability vectors of shape (`m`, `number of classes`). We thus have to convert X_train (array of sentences as strings) to X_train_indices (array of sentences as list of word indices), and Y_train (labels as indices) to Y_train_oh (labels as one-hot vectors).
X_train_indices = sentences_to_indices(X_train, word_to_index, maxLen)
Y_train_oh = convert_to_one_hot(Y_train, C = 5)
# Fit the Keras model on `X_train_indices` and `Y_train_oh`. We will use `epochs = 50` and `batch_size = 32`.
model.fit(X_train_indices, Y_train_oh, epochs = 50, batch_size = 32, shuffle=True)
# Your model should perform close to **100% accuracy** on the training set. The exact accuracy you get may be a little different. Run the following cell to evaluate your model on the test set.
X_test_indices = sentences_to_indices(X_test, word_to_index, max_len = maxLen)
Y_test_oh = convert_to_one_hot(Y_test, C = 5)
loss, acc = model.evaluate(X_test_indices, Y_test_oh)
print()
print("Test accuracy = ", acc)
# You should get a test accuracy between 80% and 95%. Run the cell below to see the mislabelled examples.
# This code allows you to see the mislabelled examples
C = 5
y_test_oh = np.eye(C)[Y_test.reshape(-1)]
X_test_indices = sentences_to_indices(X_test, word_to_index, maxLen)
pred = model.predict(X_test_indices)
for i in range(len(X_test)):
x = X_test_indices
num = np.argmax(pred[i])
if(num != Y_test[i]):
print('Expected emoji:'+ label_to_emoji(Y_test[i]) + ' prediction: '+ X_test[i] + label_to_emoji(num).strip())
# Now you can try it on your own example. Write your own sentence below.
# Change the sentence below to see your prediction. Make sure all the words are in the Glove embeddings.
x_test = np.array(['not feeling happy'])
X_test_indices = sentences_to_indices(x_test, word_to_index, maxLen)
print(x_test[0] +' '+ label_to_emoji(np.argmax(model.predict(X_test_indices))))
# Previously, Emojify-V1 model did not correctly label "not feeling happy," but our implementation of Emojiy-V2 got it right. (Keras' outputs are slightly random each time, so you may not have obtained the same result.) The current model still isn't very robust at understanding negation (like "not happy") because the training set is small and so doesn't have a lot of examples of negation. But if the training set were larger, the LSTM model would be much better than the Emojify-V1 model at understanding such complex sentences.
#
# ### Congratulations!
#
# You have completed this notebook! ❤️❤️❤️
#
# <font color='blue'>
# **What you should remember**:
# - If you have an NLP task where the training set is small, using word embeddings can help your algorithm significantly. Word embeddings allow your model to work on words in the test set that may not even have appeared in your training set.
# - Training sequence models in Keras (and in most other deep learning frameworks) requires a few important details:
# - To use mini-batches, the sequences need to be padded so that all the examples in a mini-batch have the same length.
# - An `Embedding()` layer can be initialized with pretrained values. These values can be either fixed or trained further on your dataset. If however your labeled dataset is small, it's usually not worth trying to train a large pre-trained set of embeddings.
# - `LSTM()` has a flag called `return_sequences` to decide if you would like to return every hidden states or only the last one.
# - You can use `Dropout()` right after `LSTM()` to regularize your network.
#
# Congratulations on finishing this assignment and building an Emojifier. We hope you're happy with what you've accomplished in this notebook!
#
# # 😀😀😀😀😀😀
#
#
#
# ## Acknowledgments
#
# Thanks to <NAME> and the Woebot team for their advice on the creation of this assignment. Woebot is a chatbot friend that is ready to speak with you 24/7. As part of Woebot's technology, it uses word embeddings to understand the emotions of what you say. You can play with it by going to http://woebot.io
#
# <img src="images/woebot.png" style="width:600px;height:300px;">
#
#
#
| Emojify - v2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Exploration of RISE with mnist binary
#
# Function : Exploration of RISE mnist binary
# Author : Team DIANNA
# Contributor :
# First Built : 2021.08.25
# Last Update : 2021.08.25
# Note : We ran the method using the our own trained model on mnist and various instances from mnist dataset. Results look random. There is no sense that we can make of the heatmaps.
import dianna
import onnx
import onnxruntime
import numpy as np
# %matplotlib inline
from matplotlib import pyplot as plt
from scipy.special import softmax
import pandas as pd
from dianna.methods import RISE
from dianna import visualization
data = np.load('./binary-mnist.npz')
X_test = data['X_test'].astype(np.float32).reshape([-1, 28, 28, 1])/255
y_test = data['y_test']
# # Predict classes for test data
# +
def run_model(data):
data = data.reshape([-1, 1, 28, 28]).astype(np.float32)*255
fname = './mnist_model.onnx'
# get ONNX predictions
sess = onnxruntime.InferenceSession(fname)
input_name = sess.get_inputs()[0].name
output_name = sess.get_outputs()[0].name
onnx_input = {input_name: data}
pred_onnx = sess.run([output_name], onnx_input)
return softmax(pred_onnx[0], axis=1)
pred_onnx = run_model(X_test)
# -
# Print class and image of a single instance in the test data
i_instance = 3
print(pred_onnx[i_instance])
plt.imshow(X_test[i_instance][...,0]) # 0 for channel
# +
# heatmaps = dianna.explain(run_model, X_test[[i_instance]], method="RISE", n_masks=2000, feature_res=8, p_keep=0.5)
# +
# investigate which value for p_keep works best by looking at the stddev of the probabilities for the target class,
def print_stats(p_keep):
n_masks = 500
feature_res = 8
explainer = RISE(n_masks=n_masks, feature_res=feature_res, p_keep=p_keep)
explainer(run_model, X_test[[i_instance]])
preds = explainer.predictions[:, y_test[i_instance]]
df = pd.DataFrame(preds)
display(df.describe())
# print_stats(.5) # stddev = .006 -> too low
# print_stats(.3) # .1 -> still a bit low
print_stats(.1) # .26, with minimum probability of .56 and max of 1.0. This may be ok
# -
explainer = RISE(n_masks=5000, feature_res=8, p_keep=.1)
heatmaps = explainer(run_model, X_test[[i_instance]])
visualization.plot_image(heatmaps[0], X_test[i_instance], data_cmap='gray', heatmap_cmap='bwr')
visualization.plot_image(heatmaps[0], heatmap_cmap='gray')
visualization.plot_image(heatmaps[1])
# # Conclusion
# We see that for this zero, the left and right parts of it are most important to determine the class. This makes sense, as a one would not have signal in those regions. For higher values of p_keep, the probability does not change enough for RISE to give sensible results, so this parameter needs to be checked/tuned. With proper values for p_keep, RISE thus seems to work.
# +
def describe(arr):
print('shape:',arr.shape, 'min:',np.min(arr), 'max:',np.max(arr), 'std:',np.std(arr))
describe(heatmaps[0])
describe(heatmaps[1])
# -
for i in range(10):
plt.imshow(explainer.masks[i])
plt.show()
| example_data/xai_method_study/RISE/rise_mnist.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # GDC June 2021 Webinar: GDC Data Submission Overview
#
# ### Monday, June 28, 2021<br>2:00 PM - 3:00 PM (EST)<br><NAME>, Lead for GDC User Services <br>University of Chicago
#
#
# # <a id='overview'>Notebook Overview</a>
#
#
# ### <a id='about_notebook'>About this notebook</a>
#
# - This notebook functions as a step-by-step set of instructions to submit a BAM file to the GDC using Python. Submitters who have a completely empty project or who have just started submitting with python might find this useful.
#
# - Commands and functions in this notebook will rely on the following Python packages:
# - `requests` - if not already installed on your system, can install with command `pip install requests` from command line or using a new code cell in this notebook
# - `json` - part of Python standard library, should already be installed on system
# - To execute code in a code cell, press either 'Cmd + Enter' or 'Control + Enter' depending on operating system and keyboard layout
# - A token file will need to be downloaded from the [GDC Submission Portal](https://docs.gdc.cancer.gov/Data_Submission_Portal/Users_Guide/Data_Submission_Process/#authentication)
# ### Overview
#
# - For projects that have been approved to be included in the GDC, submitters can make use of the `submission` GDC API endpoint to submit node entities to submission projects
# - Submission will require a token downloaded from the [GDC Submission Portal](https://docs.gdc.cancer.gov/Data_Submission_Portal/Users_Guide/Data_Submission_Process/#authentication)
# - Data can be submitted in `JSON` or `TSV` format; depending on the data format, users will need to edit the `"Content-Type"` in the request command (see below)
# - Additionally, `JSON` and `TSV` templates for nodes to be submitted can be downloaded from the GDC Data Dictionary Viewer webpage: https://docs.gdc.cancer.gov/Data_Dictionary/viewer/#?_top=1
# - Submittable files (such as FASTQ or BAM files) should be uploaded with the [GDC Data Transfer Tool](https://gdc.cancer.gov/access-data/gdc-data-transfer-tool)
# - Additional features and more information regarding submission using the GDC API can be found here: https://docs.gdc.cancer.gov/API/Users_Guide/Submission/
# - [Strategies for Submitting in Bulk](https://docs.gdc.cancer.gov/Data_Submission_Portal/Users_Guide/Data_Submission_Walkthrough/#strategies-for-submitting-in-bulk)
#
# ### Endpoint
#
# - The format for using the GDC API Submission endpoint uses the project information, i.e. `https://api.gdc.cancer.gov/submission/<program_name>/<project_code>`
# - For example: https://api.gdc.cancer.gov/submission/TCGA/LUAD or https://api.gdc.cancer.gov/submission/CPTAC/3
#
# ### Steps
#
# 1. Read in token file
# 2. Read in submission file
# 3. Edit endpoint with project ID information and submit data using `POST` (JSON file submission) or `PUT` (TSV file submission) request
#
# ### 1. Submitting a Case (JSON)
# +
#1. Import Python packages and read in token file
import json
import requests
token = open("../gdc-user-token.txt").read().strip()
# +
#2. Read in submission file
case_json = json.load(open("case.json"))
print(json.dumps(case_json, indent=4))
# +
#3. Edit endpoint and submit data using PUT request
ENDPT = "https://api.gdc.cancer.gov/submission/GDC/INTERNAL/_dry_run"
#submission request if data is in JSON format
response = requests.put(url = ENDPT, json = case_json, headers={'X-Auth-Token': token, "Content-Type": "application/json"})
print(json.dumps(json.loads(response.text), indent = 4))
# -
# ### 2: Submitting a Sample
# +
#1. Read in submission file
sample_tsv = open("sample.tsv", "rb")
sample_tsv_display = open("sample.tsv", "r")
for x in sample_tsv_display.readlines():
print(x.strip().split("\t"))
# +
#2. Edit endpoint and submit data using PUT request
ENDPT = "https://api.gdc.cancer.gov/submission/GDC/INTERNAL/"
#submission request if data is in TSV format
response = requests.put(url = ENDPT, data = sample_tsv, headers={'X-Auth-Token': token, "Content-Type": "text/tsv"})
print(json.dumps(json.loads(response.text), indent = 4))
# -
# ### 3: Submitting the Aliquot and Read_Group
# +
#1. Read in submission file
aliquot_rg_json = json.load(open("aliquot_readgroup.json"))
print(json.dumps(aliquot_rg_json, indent=4))
# +
#2. Submit data using PUT request
ENDPT = "https://api.gdc.cancer.gov/submission/GDC/INTERNAL"
#submission request if data is in JSON format
response = requests.put(url = ENDPT, json = aliquot_rg_json, headers={'X-Auth-Token': token, "Content-Type": "application/json"})
print(json.dumps(json.loads(response.text), indent = 4))
# -
# ### 4: Register the Submitted Aligned Reads File
# +
#1. Read in submission file
sar_json = json.load(open("SAR.json"))
print(json.dumps(sar_json, indent=4))
# +
#2. Submit data using PUT request
ENDPT = "https://api.gdc.cancer.gov/submission/GDC/INTERNAL"
#submission request if data is in JSON format
response = requests.put(url = ENDPT, json = sar_json, headers={'X-Auth-Token': token, "Content-Type": "application/json"})
print(json.dumps(json.loads(response.text), indent = 4))
# -
# ### 5: Upload the Submitted Aligned Reads Data File Using Data Transfer Tool
#
# +
## ./gdc-client upload <UUID> -t token_file.txt
| Notebooks/Submission_June_2021/Webinar_June_2021.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import torch
from torch import nn, optim
from torch.utils.data import DataLoader
import torch.nn.functional as F
# for loading MNIST data
from torchvision import transforms, datasets
# -
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm_notebook as tqdm
# %matplotlib inline
# +
# if cuda device is available then run model on gpu
if torch.cuda.is_available():
cuda_flag=True
else:
cuda_flag=False
torch.manual_seed(3120)
# -
# ### Setting up data loader
# +
batch_size=64
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=[0.5], std=[0.5])])
train_loader = torch.utils.data.DataLoader(
datasets.MNIST('./dataset/', train=True, download=True,transform=transforms.ToTensor()),
batch_size=batch_size, shuffle=True)
test_loader = torch.utils.data.DataLoader(
datasets.MNIST('./dataset/', train=False, download=True, transform=transforms.ToTensor()),
batch_size=batch_size)
# -
# ### Class definition for encoder decoder networks
# +
class Encoder(nn.Module):
def __init__(self, input_dim, z_dim):
super(Encoder, self).__init__()
self.conv1 = nn.Conv2d(in_channels=1, out_channels=16, kernel_size=3, padding=1)
self.conv2 = nn.Conv2d(in_channels=16, out_channels=32, kernel_size=3, padding=1)
self.shared_fc1 = nn.Linear(in_features=2*input_dim, out_features=z_dim*8)
self.shared_fc2 = nn.Linear(in_features=z_dim*8, out_features=z_dim*4)
self.pool = nn.MaxPool2d(kernel_size=2)
self.mu = nn.Linear(z_dim*4, z_dim)
self.var = nn.Linear(z_dim*4, z_dim)
def forward(self, x):
x = x.view(len(x),1,28,28)
x = torch.relu(self.conv1(x))
x = self.pool(x)
x = torch.relu(self.conv2(x))
x = self.pool(x)
x = x.view(len(x),-1)
x = F.relu(self.shared_fc1(x))
x = F.relu(self.shared_fc2(x))
z_mu = self.mu(x)
z_var = self.var(x)
return z_mu, z_var
class Decoder(nn.Module):
def __init__(self, output_dim, z_dim):
super(Decoder, self).__init__()
self.fc1 = nn.Linear(in_features=z_dim, out_features=z_dim*4)
self.fc2 = nn.Linear(in_features=z_dim*4, out_features=z_dim*16)
self.fc3 = nn.Linear(in_features=z_dim*16, out_features=output_dim)
def forward(self, x):
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = torch.sigmoid(self.fc3(x))
return x
# -
class VAE(nn.Module):
def __init__(self, enc, dec, z_dim):
super(VAE, self).__init__()
self.enc = enc
self.dec = dec
def forward(self,x):
z_mu,z_logvar = self.enc(x)
# sample z using the mean and variance obtained from encoder
std = torch.exp(0.5*z_logvar)
eps = torch.randn_like(std)
z = z_mu + eps*std
predicted = self.dec(z)
return predicted, z_mu, z_logvar
def crit(y_pred, y, z_mu, z_logvar):
reconstruction_loss = F.binary_cross_entropy(y_pred, y, reduction='sum')
kl_divergence = -0.5 * torch.sum(1 + z_logvar - z_mu.pow(2) - z_logvar.exp())
return reconstruction_loss+kl_divergence
def plot_tensor_img(x):
fig = plt.figure(figsize=(10,1))
generated_images = x.detach().cpu().numpy().reshape(len(x),28,28)
for i in range(10):
plt.subplot(1,10,i+1)
plt.imshow(generated_images[i])
plt.show()
# +
# instantiate the models
enc = Encoder(input_dim=784, z_dim=32)
dec = Decoder(output_dim=784, z_dim=32)
model = VAE(enc, dec, 32)
if cuda_flag:
model = model.cuda()
# -
lr = 1e-3
opt = optim.Adam(model.parameters(), lr=lr)
# +
epochs = 15
loss_history = []
sample_images = next(iter(test_loader))[0]
# +
print("True Images")
plot_tensor_img(sample_images)
for epoch in tqdm(range(epochs)):
train_loss=0
# iterate over dataset
for x,_ in train_loader:
x = x.view(len(x),-1)
# move data to gpu if cuda_flag is set
if cuda_flag:
x = x.cuda()
# zero_grad to ensure no unaccounted calculation creeps in while calculating gradients
opt.zero_grad()
# forward propogation and loss computation
x_gen, z_mu, z_logvar = model(x)
loss = crit(x_gen,x, z_mu, z_logvar)
train_loss+=loss.item()
# backpropogate gradients
loss.backward()
# update weights
opt.step()
train_loss/=len(train_loader)*batch_size
print ("Epoch:{} Train Loss:{:.6}".format(epoch,train_loss))
loss_history.append(train_loss)
if epoch%3==0:
print("Reconstructed Images after Epoch ",epoch)
with torch.no_grad():
x = sample_images.view(len(sample_images),-1)
# move data to gpu if cuda_flag is set
if cuda_flag:
x = x.cuda()
# forward propogation and loss computation
x_gen, _, _ = model(x)
plot_tensor_img(x_gen)
# -
# ### New Image Generation using Decoder
# +
# sample random tensors using standard multivariate normal distribution
z = torch.randn(64,32)
if cuda_flag:
z = z.cuda()
# pass the random tensors through decoder to generate the new images
with torch.no_grad():
generated_images = model.dec(z).cpu().numpy().reshape(len(z),28,28)
fig = plt.figure(figsize=(10,10))
for i in range(64):
plt.subplot(8,8,i+1)
plt.imshow(generated_images[i])
plt.show()
# -
| vision/VAE.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Day and Night Image Classifier
# ---
#
# The day/night image dataset consists of 200 RGB color images in two categories: day and night. There are equal numbers of each example: 100 day images and 100 night images.
#
# We'd like to build a classifier that can accurately label these images as day or night, and that relies on finding distinguishing features between the two types of images!
#
# *Note: All images come from the [AMOS dataset](http://cs.uky.edu/~jacobs/datasets/amos/) (Archive of Many Outdoor Scenes).*
#
# ### Import resources
#
# Before you get started on the project code, import the libraries and resources that you'll need.
# +
import cv2 # computer vision library
import helpers
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
# %matplotlib inline
# -
# ## Training and Testing Data
# The 200 day/night images are separated into training and testing datasets.
#
# * 60% of these images are training images, for you to use as you create a classifier.
# * 40% are test images, which will be used to test the accuracy of your classifier.
#
# First, we set some variables to keep track of some where our images are stored:
#
# image_dir_training: the directory where our training image data is stored
# image_dir_test: the directory where our test image data is stored
# Image data directories
image_dir_training = "day_night_images/training/"
image_dir_test = "day_night_images/test/"
# ## Load the datasets
#
# These first few lines of code will load the training day/night images and store all of them in a variable, `IMAGE_LIST`. This list contains the images and their associated label ("day" or "night").
#
# For example, the first image-label pair in `IMAGE_LIST` can be accessed by index:
# ``` IMAGE_LIST[0][:]```.
#
# Using the load_dataset function in helpers.py
# Load training data
IMAGE_LIST = helpers.load_dataset(image_dir_training)
# ## Construct a `STANDARDIZED_LIST` of input images and output labels.
#
# This function takes in a list of image-label pairs and outputs a **standardized** list of resized images and numerical labels.
# Standardize all training images
STANDARDIZED_LIST = helpers.standardize(IMAGE_LIST)
# ## Visualize the standardized data
#
# Display a standardized image from STANDARDIZED_LIST.
# +
# Display a standardized image and its label
# Select an image by index
image_num = 0
selected_image = STANDARDIZED_LIST[image_num][0]
selected_label = STANDARDIZED_LIST[image_num][1]
# Display image and data about it
plt.imshow(selected_image)
print("Shape: "+str(selected_image.shape))
print("Label [1 = day, 0 = night]: " + str(selected_label))
# -
# # Feature Extraction
#
# Create a feature that represents the brightness in an image. We'll be extracting the **average brightness** using HSV colorspace. Specifically, we'll use the V channel (a measure of brightness), add up the pixel values in the V channel, then divide that sum by the area of the image to get the average Value of the image.
#
# ---
# ### Find the average brightness using the V channel
#
# This function takes in a **standardized** RGB image and returns a feature (a single value) that represent the average level of brightness in the image. We'll use this value to classify the image as day or night.
# Find the average Value or brightness of an image
def avg_brightness(rgb_image):
# Convert image to HSV
hsv = cv2.cvtColor(rgb_image, cv2.COLOR_RGB2HSV)
# Add up all the pixel values in the V channel
sum_brightness = np.sum(hsv[:,:,2])
area = 600*1100.0 # pixels
# find the avg
avg = sum_brightness/area
return avg
# +
# Testing average brightness levels
# Look at a number of different day and night images and think about
# what average brightness value separates the two types of images
# As an example, a "night" image is loaded in and its avg brightness is displayed
image_num = 200
test_im = STANDARDIZED_LIST[image_num][0]
avg = avg_brightness(test_im)
print('Avg brightness: ' + str(avg))
plt.imshow(test_im)
# -
# # Classification and Visualizing Error
#
# In this section, we'll turn our average brightness feature into a classifier that takes in a standardized image and returns a `predicted_label` for that image. This `estimate_label` function should return a value: 0 or 1 (night or day, respectively).
# ---
# ### TODO: Build a complete classifier
#
# Set a threshold that you think will separate the day and night images by average brightness.
# This function should take in RGB image input
def estimate_label(rgb_image):
## TODO: extract average brightness feature from an RGB image
# Use the avg brightness feature to predict a label (0, 1)
avg = avg_brightness(rgb_image)
## TODO: set the value of a threshold that will separate day and night images
threshold = 108.5
## TODO: Return the predicted_label (0 or 1) based on whether the avg is
# above or below the threshold
predicted_label = 0
if(avg > threshold):
predicted_label = 1
return predicted_label
## Test out your code by calling the above function and seeing
# how some of your training data is classified
estimate_label(test_im)
| 1_1_Image_Representation/6_4. Classification.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ###### Текст распространяется на условиях лицензии Creative Commons Attribution license CC-BY 4.0, код — на условиях лицензии MIT license. (c)2015 <NAME>, <NAME>.
# # Упражнение: Вывод панельного метода вихрей-источников
# Потенциал в точке $(x, y)$, создаваемый равномерным потоком, слоем источников и вихревым слоем, может быть записан в виде:
# \begin{equation}
# \begin{split}
# \phi(x, y)
# &= \phi_{uniform\ flow}(x, y) \\
# &+ \phi_{source\ sheet}(x, y) + \phi_{vortex\ sheet}(x, y)
# \end{split}
# \end{equation}
# То есть
# \begin{equation}
# \begin{split}
# \phi(x, y) &= xU_{\infty}\cos(\alpha) + yU_{\infty}\sin(\alpha) \\
# &+
# \frac{1}{2\pi} \int_{sheet} \sigma(s)\ln\left[(x-\xi(s))^2+(y-\eta(s))^2\right]^{\frac{1}{2}}ds \\
# &-
# \frac{1}{2\pi} \int_{sheet} \gamma(s)\tan^{-1} \frac{y-\eta(s)}{x-\xi(s)}ds
# \end{split}
# \end{equation}
# где $s$ — локальная координата слоя, а $\xi(s)$ и $\eta(s)$ — координаты бесконечного ряда источников и вихрей, из которых состоят слои. В записанном выше уравнении мы предполагаем, что слои источников и вихрей пересекаются.
# ------------------------------------------------------
# ### Вопрос 1:
# Пусть слой разбит на $N$ панелей, перепешите уравнение, полученное выше, в дискретном виде. Предположим, что $l_j$ — длина панели $j$. И что
#
# \begin{equation}
# \left\{
# \begin{array}{l}
# \xi_j(s)=x_j-s\sin\beta_j \\
# \eta_j(s)=y_j+s\cos\beta_j
# \end{array}
# ,\ \ \
# 0\le s \le l_j
# \right.
# \end{equation}
#
# На следующей картинке показана панель $j$:
#
# <center> <img src="resources/Lesson11_Exercise_Fig.1.png" width=360> </center>
#
# Подсказка: например, рассмотрим интеграл $\int_0^L f(x) dx$. Если разбить отрезок $0\sim L$ на три панели, его можно записать в виде:
# $$\int_0^L f(x) dx = \int_0^{L/3} f(x)dx+\int_{L/3}^{2L/3} f(x)dx+\int_{2L/3}^{L} f(x)dx \\=
# \sum_{j=1}^3 \int_{l_j}f(x)dx$$
# ----------------------------
# Теперь предположим, что
#
# 1. $\sigma_j(s) = constant = \sigma_j$
# 2. $\gamma_1(s) = \gamma_2(s) = ... = \gamma_N(s) = \gamma$
# ------------------------------------------------
# ### Вопрос 2:
# Примените изложенные выше предположения к уравнению для $\phi(x, y)$, полученному в Вопросе 1.
# ---------------------------
# Нормальную компоненту скорости $U_n$ можно получить, воспользовавшись правилом дифференцирования сложной функции:
# \begin{equation}
# \begin{split}
# U_n &= \frac{\partial \phi}{\partial \vec{n}} \\
# &=
# \frac{\partial \phi}{\partial x}\frac{\partial x}{\partial \vec{n}}
# +
# \frac{\partial \phi}{\partial y}\frac{\partial y}{\partial \vec{n}} \\
# &=
# \frac{\partial \phi}{\partial x}\nabla x\cdot \vec{n}
# +
# \frac{\partial \phi}{\partial y}\nabla y\cdot \vec{n} \\
# &=
# \frac{\partial \phi}{\partial x}n_x
# +
# \frac{\partial \phi}{\partial y}n_y
# \end{split}
# \end{equation}
# Касательную компоненту можно получить, используя тот же подход. Таким образом, в точке $(x, y)$ можно записать выражения для компонент скорости:
# \begin{equation}
# \left\{
# \begin{array}{l}
# U_n(x, y)=\frac{\partial \phi}{\partial x}(x, y) n_x(x, y)+\frac{\partial \phi}{\partial y}(x, y) n_y(x, y) \\
# U_t(x, y)=\frac{\partial \phi}{\partial x}(x, y) t_x(x, y)+\frac{\partial \phi}{\partial y}(x, y) t_y(x, y)
# \end{array}
# \right.
# \end{equation}
# -------------------------------------
# ### Вопрос 3:
# Используя выписанные выше уравнения, выведите соотношения для $U_n(x,y)$ и $U_t(x,y)$ из уравнения, полученного в Вопросе 2.
# -----------------------------------------
# ### Вопрос 4:
# Рассмотрим нормальную компоненту скорости в центре $i$-ой панели, то есть в точке $(x_{c,i}, y_{c,i})$. Подставив $(x_{c,i}, y_{c,i})$ вместо $(x, y)$ в уравнении, выведенном в Вопросе 3, можно переписать его в матричном виде:
# \begin{equation}
# \begin{split}
# U_n(x_{c,i}, y_{c,i}) &= U_{n,i} \\
# &=
# b^n_i
# +
# \left[\begin{matrix} A^n_{i1} && A^n_{i2} && ... && A^n_{iN}\end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \end{matrix}\right]
# +
# \left(\sum_{j=1}^N B^n_{ij}\right)\gamma \\
# &=
# b^n_i
# +
# \left[\begin{matrix} A^n_{i1} && A^n_{i2} && ... && A^n_{iN} && \left(\sum_{j=1}^N B^n_{ij}\right) \end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \\ \gamma \end{matrix}\right]
# \end{split}
# \end{equation}
# \begin{equation}
# \begin{split}
# U_t(x_{c,i}, y_{c,i}) &= U_{t,i} \\
# &=
# b^t_i
# +
# \left[\begin{matrix} A^t_{i1} && A^t_{i2} && ... && A^t_{iN}\end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \end{matrix}\right]
# +
# \left(\sum_{j=1}^N B^t_{ij}\right)\gamma \\
# &=
# b^t_i
# +
# \left[\begin{matrix} A^t_{i1} && A^t_{i2} && ... && A^t_{iN} && \left(\sum_{j=1}^N B^t_{ij}\right) \end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \\ \gamma \end{matrix}\right]
# \end{split}
# \end{equation}
# Чему равны $b^n_i$, $A^n_{ij}$, $B^n_{ij}$, $b^t_i$, $A^t_{ij}$ и $B^t_{ij}$?
# -----------------------
# Учитывая, что (согласно Рис. 1)
#
# \begin{equation}
# \left\{\begin{matrix} \vec{n}_i=n_{x,i}\vec{i}+n_{y,i}\vec{j} = \cos(\beta_i)\vec{i}+\sin(\beta_i)\vec{j} \\ \vec{t}_i=t_{x,i}\vec{i}+t_{y,i}\vec{j} = -\sin(\beta_i)\vec{i}+\cos(\beta_i)\vec{j} \end{matrix}\right.
# \end{equation}
#
# получим
#
# \begin{equation}
# \left\{
# \begin{matrix}
# n_{x,i}=t_{y,i} \\
# n_{y,i}=-t_{x,i}
# \end{matrix}
# \right.
# ,\ or\
# \left\{
# \begin{matrix}
# t_{x,i}=-n_{y,i} \\
# t_{y,i}=n_{x,i}
# \end{matrix}
# \right.
# \end{equation}
# -----------------------
# ### Вопрос 5:
# Применив вышеуказанные соотношения между $\vec{n}_i$ и $\vec{t}_i$ к ответу на Вопрос 4, найдите соотношения между $B^n_{ij}$ и $A^t_{ij}$, а также между $B^t_{ij}$ и $A^n_{ij}$. Наличие таких связей означает, что в коде не нужно вычислять значения $B^n_{ij}$ и $B^t_{ij}$. Какие это соотношения?
# -------------------------
# Теперь, обратите внимание, что при $i=j$, в области интегрирования имеется особенность при вычислении $A^n_{ii}$ и $A^t_{ii}$. Эта особенность возникает при $s=l_i/2$, то есть при $\xi_i(l_i/2)=x_{c,i}$ и $\eta_i(l_i/2)=y_{c,i}$. Это означает, что нужно вывести $A^n_{ii}$ и $A^t_{ii}$ аналитически.
# --------------------------
# ### Вопрос 6:
# Каковы точные значения $A^n_{ii}$ и $A^t_{ii}$?
# ------------------------------
# В нашей задаче есть $N+1$ неизвестных, то есть $\sigma_1, \sigma_2, ..., \sigma_N, \gamma$. Нам понадобится $N+1$ линейных уравнений для определения неизвестных. Первые $N$ уравнений можно получить из условия непротекания на каждой панели. То есть
#
# \begin{equation}
# \begin{split}
# U_{n,i} &= 0 \\
# &=
# b^n_i
# +
# \left[\begin{matrix} A^n_{i1} && A^n_{i2} && ... && A^n_{iN} && \left(\sum_{j=1}^N B^n_{ij}\right) \end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \\ \gamma \end{matrix}\right] \\
# &,\ \ for\
# i=1\sim N
# \end{split}
# \end{equation}
#
# или
#
# \begin{equation}
# \begin{split}
# &\left[\begin{matrix} A^n_{i1} && A^n_{i2} && ... && A^n_{iN} && \left(\sum_{j=1}^N B^n_{ij}\right) \end{matrix}\right]\left[\begin{matrix} \sigma_1 \\ \sigma_2 \\ \vdots \\ \sigma_N \\ \gamma \end{matrix}\right]
# =-b^n_i \\
# &,\ \ for\
# i=1\sim N
# \end{split}
# \end{equation}
# Для последнего уравнения воспользуемся условием Кутты-Жуковского.
#
# \begin{equation}
# U_{t,1} = - U_{t,N}
# \end{equation}
# ----------------------
# ### Вопрос 7:
# Используйте матрицы из соотношений для $U_{t,i}$ и $U_{t,N}$ для записи условия Кутты-Жуковского и получите последнее линейное уравнение. Перегруппируйте уравнения так, чтобы неизвестные оказались в левой части, а известные величины — в правой.
# ---------------------
# ### Вопрос 8:
# Теперь у вас есть $N+1$ линейных уравнений для нахождения $N+1$ неизвестной. Попробуйте скомбинировать первые $N$ линейных уравнений и последнее (то, что получилось при наложении условия Кутты-Жуковского) из ответа на Вопрос 7, чтобы получить полную систему линейных уравнений в матричной форме.
# ----------------------------
# Теперь можно решать уравнения! Это и есть панельный метод вихрей и источников.
# --------------------
# + active=""
# Пожалуйста, не обращайте внимания на ячейку ниже. При её исполнении загружаются стили для отображения блокнота.
# -
from IPython.core.display import HTML
def css_styling():
styles = open('../styles/custom.css', 'r').read()
return HTML(styles)
css_styling()
| lessons/11_Lesson11_Exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Text models, data, and training
# + hide_input=true
from fastai.gen_doc.nbdoc import *
# -
# The [`text`](/text.html#text) module of the fastai library contains all the necessary functions to define a Dataset suitable for the various NLP (Natural Language Processing) tasks and quickly generate models you can use for them. Specifically:
# - [`text.transform`](/text.transform.html#text.transform) contains all the scripts to preprocess your data, from raw text to token ids,
# - [`text.data`](/text.data.html#text.data) contains the definition of [`TextDataset`](/text.data.html#TextDataset), which the main class you'll need in NLP,
# - [`text.learner`](/text.learner.html#text.learner) contains helper functions to quickly create a language model or an RNN classifier.
#
# Have a look at the links above for full details of the API of each module, of read on for a quick overview.
# ## Quick Start: Training an IMDb sentiment model with *ULMFiT*
# Let's start with a quick end-to-end example of training a model. We'll train a sentiment classifier on a sample of the popular IMDb data, showing 4 steps:
#
# 1. Reading and viewing the IMDb data
# 1. Getting your data ready for modeling
# 1. Fine-tuning a language model
# 1. Building a classifier
# ### Reading and viewing the IMDb data
# First let's import everything we need for text.
from fastai.text import *
# Contrary to images in Computer Vision, text can't directly be transformed into numbers to be fed into a model. The first thing we need to do is to preprocess our data so that we change the raw texts to lists of words, or tokens (a step that is called tokenization) then transform these tokens into numbers (a step that is called numericalization). These numbers are then passed to embedding layers that will convert them in arrays of floats before passing them through a model.
#
# You can find on the web plenty of [Word Embeddings](https://en.wikipedia.org/wiki/Word_embedding) to directly convert your tokens into floats. Those word embeddings have generally be trained on a large corpus such as wikipedia. Following the work of [ULMFiT](https://arxiv.org/abs/1801.06146), the fastai library is more focused on using pre-trained Language Models and fine-tuning them. Word embeddings are just vectors of 300 or 400 floats that represent different words, but a pretrained language model not only has those, but has also been trained to get a representation of full sentences and documents.
#
# That's why the library is structured around three steps:
#
# 1. Get your data preprocessed and ready to use in a minimum amount of code,
# 1. Create a language model with pretrained weights that you can fine-tune to your dataset,
# 1. Create other models such as classifiers on top of the encoder of the language model.
#
# To show examples, we have provided a small sample of the [IMDB dataset](https://www.imdb.com/interfaces/) which contains 1,000 reviews of movies with labels (positive or negative).
path = untar_data(URLs.IMDB_SAMPLE)
path
# Creating a dataset from your raw texts is very simple if you have it in one of those ways
# - organized it in folders in an ImageNet style
# - organized in a csv file with labels columns and a text columns
#
# Here, the sample from imdb is in a texts csv files that looks like this:
df = pd.read_csv(path/'texts.csv')
df.head()
# ### Getting your data ready for modeling
# + hide_input=true
for file in ['train_tok.npy', 'valid_tok.npy']:
if os.path.exists(path/'tmp'/file): os.remove(path/'tmp'/file)
# -
# To get a [`DataBunch`](/basic_data.html#DataBunch) quickly, there are also several factory methods depending on how our data is structured. They are all detailed in [`text.data`](/text.data.html#text.data), here we'll use the method <code>from_csv</code> of the [`TextLMDataBunch`](/text.data.html#TextLMDataBunch) (to get the data ready for a language model) and [`TextClasDataBunch`](/text.data.html#TextClasDataBunch) (to get the data ready for a text classifier) classes.
# Language model data
data_lm = TextLMDataBunch.from_csv(path, 'texts.csv')
# Classifier model data
data_clas = TextClasDataBunch.from_csv(path, 'texts.csv', vocab=data_lm.train_ds.vocab, bs=32)
# This does all the necessary preprocessing behind the scene. For the classifier, we also pass the vocabulary (mapping from ids to words) that we want to use: this is to ensure that `data_clas` will use the same dictionary as `data_lm`.
#
# Since this step can be a bit time-consuming, it's best to save the result with:
data_lm.save()
data_clas.save()
# This will create a 'tmp' directory where all the computed stuff will be stored. You can then reload those results with:
data_lm = TextLMDataBunch.load(path)
data_clas = TextClasDataBunch.load(path, bs=32)
# Note that you can load the data with different [`DataBunch`](/basic_data.html#DataBunch) parameters (batch size, `bptt`,...)
# ### Fine-tuning a language model
# We can use the `data_lm` object we created earlier to fine-tune a pretrained language model. [fast.ai](http://www.fast.ai/) has an English model available that we can download. We can create a learner object that will directly create a model, download the pretrained weights and be ready for fine-tuning.
learn = language_model_learner(data_lm, AWD_LSTM, drop_mult=0.5)
learn.fit_one_cycle(1, 1e-2)
# Like a computer vision model, we can then unfreeze the model and fine-tune it.
learn.unfreeze()
learn.fit_one_cycle(1, 1e-3)
# To evaluate your language model, you can run the [`Learner.predict`](/basic_train.html#Learner.predict) method and specify the number of words you want it to guess.
learn.predict("This is a review about", n_words=10)
# It doesn't make much sense (we have a tiny vocabulary here and didn't train much on it) but note that it respects basic grammar (which comes from the pretrained model).
#
# Finally we save the encoder to be able to use it for classification in the next section.
learn.save_encoder('ft_enc')
# ### Building a classifier
# We now use the `data_clas` object we created earlier to build a classifier with our fine-tuned encoder. The learner object can be done in a single line.
learn = text_classifier_learner(data_clas, AWD_LSTM, drop_mult=0.5)
learn.load_encoder('ft_enc')
data_clas.show_batch()
learn.fit_one_cycle(1, 1e-2)
# Again, we can unfreeze the model and fine-tune it.
learn.freeze_to(-2)
learn.fit_one_cycle(1, slice(5e-3/2., 5e-3))
learn.unfreeze()
learn.fit_one_cycle(1, slice(2e-3/100, 2e-3))
# Again, we can predict on a raw text by using the [`Learner.predict`](/basic_train.html#Learner.predict) method.
learn.predict("This was a great movie!")
| docs_src/text.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/Innarticles/Data-Science-Resources/blob/master/Copy_of_S%2BP_Week_2_Exercise_Answer.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + id="D1J15Vh_1Jih" colab_type="code" cellView="both" colab={}
# !pip install tf-nightly-2.0-preview
# + id="BOjujz601HcS" colab_type="code" colab={}
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
# + colab_type="code" id="Zswl7jRtGzkk" colab={}
def plot_series(time, series, format="-", start=0, end=None):
plt.plot(time[start:end], series[start:end], format)
plt.xlabel("Time")
plt.ylabel("Value")
plt.grid(False)
def trend(time, slope=0):
return slope * time
def seasonal_pattern(season_time):
"""Just an arbitrary pattern, you can change it if you wish"""
return np.where(season_time < 0.1,
np.cos(season_time * 6 * np.pi),
2 / np.exp(9 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
"""Repeats the same pattern at each period"""
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
def noise(time, noise_level=1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
time = np.arange(10 * 365 + 1, dtype="float32")
baseline = 10
series = trend(time, 0.1)
baseline = 10
amplitude = 40
slope = 0.005
noise_level = 3
# Create the series
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
# Update with noise
series += noise(time, noise_level, seed=51)
split_time = 3000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
window_size = 20
batch_size = 32
shuffle_buffer_size = 1000
plot_series(time, series)
# + id="4sTTIOCbyShY" colab_type="code" colab={}
def windowed_dataset(series, window_size, batch_size, shuffle_buffer):
dataset = tf.data.Dataset.from_tensor_slices(series)
dataset = dataset.window(window_size + 1, shift=1, drop_remainder=True)
dataset = dataset.flat_map(lambda window: window.batch(window_size + 1))
dataset = dataset.shuffle(shuffle_buffer).map(lambda window: (window[:-1], window[-1]))
dataset = dataset.batch(batch_size).prefetch(1)
return dataset
# + id="TW-vT7eLYAdb" colab_type="code" colab={}
dataset = windowed_dataset(x_train, window_size, batch_size, shuffle_buffer_size)
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(100, input_shape=[window_size], activation="relu"),
tf.keras.layers.Dense(10, activation="relu"),
tf.keras.layers.Dense(1)
])
model.compile(loss="mse", optimizer=tf.keras.optimizers.SGD(lr=1e-6, momentum=0.9))
model.fit(dataset,epochs=100,verbose=0)
# + id="efhco2rYyIFF" colab_type="code" colab={}
forecast = []
for time in range(len(series) - window_size):
forecast.append(model.predict(series[time:time + window_size][np.newaxis]))
forecast = forecast[split_time-window_size:]
results = np.array(forecast)[:, 0, 0]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid)
plot_series(time_valid, results)
# + id="-kT6j186YO6K" colab_type="code" colab={}
tf.keras.metrics.mean_absolute_error(x_valid, results).numpy()
| Copy_of_S+P_Week_2_Exercise_Answer.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Introducción a Jupyter
# ## Expresiones aritmeticas y algebraicas
# Empezaremos esta práctica con algo de conocimientos previos de programación. Se que muchos de ustedes no han tenido la oportunidad de utilizar Python como lenguaje de programación y mucho menos Jupyter como ambiente de desarrollo para computo cientifico, asi que el primer objetivo de esta práctica será acostumbrarnos a la sintaxis del lenguaje y a las funciones que hacen especial a Jupyter.
#
# Primero tratemos de evaluar una expresión aritmetica. Para correr el código en la siguiente celda, tan solo tienes que hacer clic en cualquier punto de ella y presionar las teclas Shift + Return.
2 + 3
2*3
2**3
sin(pi)
# Sin embargo no existen funciones trigonométricas cargadas por default. Para esto tenemos que importarlas de la libreria ```math```:
from math import sin, pi
sin(pi)
# ## Variables
# Las variables pueden ser utilizadas en cualquier momento, sin necesidad de declararlas, tan solo usalas!
a = 10
a
# ### Ejercicio
# Ejecuta el siguiente calculo y guardalo en una variable:
#
# $$
# c = \pi *10^2
# $$
#
# > Nota: Una vez que hayas concluido el calculo y guardado el valor en una variable, puedes desplegar el valor de cualquier variable al ejecutar en una celda el nombre de la variable
c =
# Ejecuta la prueba de abajo para saber si has creado el codigo correcto
from pruebas_1 import prueba_1_1
prueba_1_1(_, c)
# ## Listas
# Las listas son una manera de guardar varios datos en un mismo arreglo. Podemos tener por ejemplo:
A = [2, 4, 8, 10]
A
# Pero si intentamos multiplicar estos datos por un numero, no tendrá el comportamiento esperado.
A*2
# ## Funciones
# Podemos definir funciones propias de la siguiente manera:
f = lambda x: x**2 + 1
# Esta linea de codigo es equivalente a definir una función matemática de la siguiente manera:
#
# $$
# f(x) = x^2 + 1
# $$
#
# Por lo que si la evaluamos con $x = 2$, obviamente obtendremos como resultado $5$.
f(2)
# Esta notación que introducimos es muy util para funciones matemáticas, pero esto nos obliga a pensar en las definiciones de una manera funcional, lo cual no siempre es la solución (sobre todo en un lenguaje con un paradigma de programación orientado a objetos).
#
# Esta función tambien puede ser escrita de la siguiente manera:
def g(x):
y = x**2 + 1
return y
# Con los mismos resultados:
g(2)
# ### Ejercicio
# Define una función que convierta grados Celsius a grados Farenheit, de acuerdo a la siguiente formula:
#
# $$
# F = \frac{9}{5} C + 32
# $$
def cel_a_faren(grados_cel):
grados_faren = # Escribe el codigo para hacer el calculo aqui
return grados_faren
# Y para probar trata de convertir algunos datos:
cel_a_faren(10)
cel_a_faren(50)
# ## Ciclos de control
# Cuando queremos ejecutar código varias veces tenemos varias opciones, vamos a explorar rapidamente el ciclo for.
# ```python
# for paso in pasos:
# ...
# codigo_a_ejecutar(paso)
# ...
# ```
# En este caso el codigo se ejecutará tantas veces sean necesarias para usar todos los elementos que hay en pasos.
#
# Por ejemplo, pordemos ejecutar la multiplicacion por 2 en cada uno de los datos:
for dato in A:
print dato*2
# ó agregarlo en una lista nueva:
# +
B = []
for dato in A:
B.append(dato*2)
B
# -
# y aun muchas cosas mas, pero por ahora es momento de empezar con la práctica.
# ### Ejercicio
# * Crea una lista ```C``` con los enteros positivos de un solo digito, es decir: $\left\{ x \in \mathbb{Z} \mid 0 \leq x < 10\right\}$
# * Crea una segunda lista ```D``` con los cuadrados de cada elemento de ```C```
# +
C = [] # Escribe el codigo para declarar el primer arreglo adentro de los corchetes
C
# +
D = []
# Escribe el codigo de tu ciclo for aqui
D
# -
# Ejecuta las pruebas de abajo
from pruebas_1 import prueba_1_3
prueba_1_3(C, D)
# ## Método de bisección
# Para obtener una raiz real de un polinomio $f(x) = x^3 + 2 x^2 + 10 x - 20$ por el metodo de bisección, tenemos que primero definir dos puntos, uno que evaluado en el polinomio nos de positivo, y otro que nos de negativo. Propondremos $x_1 = 1$ y $x_2 = 2$, y los evaluaremos para asegurarnos de que cumplan lo que acabamos de pedir.
f = lambda x: x**3 + 2*x**2 + 10*x - 20
f(1.0)
f(2.0)
# Una vez que tenemos dos puntos de los que sabemos que definen el intervalo donde se encuetra una raiz, podemos empezar a iterar para descubrir el punto medio.
#
# $$x_M = \frac{x_1 + x_2}{2}$$
#
# Si hacemos esto ingenuamente y lo evaluamos en la función, podremos iterar manualmente:
x_1, x_2 = 1.0, 2.0
xm1 = (x_1 + x_2)/2.0
f(xm1)
# Y de aqui podemos notar que el resultado que nos dio esto es positivo, es decir que la raiz tiene que estar entre $x_1$ y $x_M$. Por lo que para nuestra siguiente iteración usaremos el nuevo intervalo $x_1 = 1$ y $x_2 = 2.875$, es decir que ahora asignaremos el valor de $x_M$ a $x_2$.
x_1, x_2 = x_1, xm1
xm2 = (x_1 + x_2)/2.0
f(xm2)
# Y podriamos seguir haciendo esto hasta que tengamos la exactitud que queremos, pero esa no seria una manera muy inteligente de hacerlo (tenemos una maquina a la que le gusta hacer tareas repetitivas y no la aprovechamos?).
#
# En vez de eso, notemos que la formula no cambia absolutamente en nada, por lo que la podemos hacer una funcion y olvidarnos de ella.
def biseccion(x1, x2):
return (x1 + x2)/2.0
# Si volvemos a ejecutar el codigo que teniamos, sustituyendo esta función, obtendremos exactamente el mismo resultado:
x_1, x_2 = x_1, xm1
xm2 = biseccion(x_1, x_2)
f(xm2)
# Y ahora lo que tenemos que hacer es poner una condicion para que $x_M$ se intercambie con $x_1$ ó $x_2$ dependiendo del signo.
x_1, x_2 = 1.0, 2.0
xm1 = biseccion(x_1, x_2)
f(xm1)
# +
if x_2*xm1 > 0:
x_2 = xm1
else:
x_1 = xm1
xm2 = biseccion(x_1, x_2)
f(xm2)
# +
if x_2*xm2 > 0:
x_2 = xm2
else:
x_1 = xm2
xm3 = biseccion(x_1, x_2)
f(xm3)
# -
# Si, yo se que parece raro, pero si lo revisas con calma te daras cuenta que funciona.
#
# Ya casi llegamos, tan solo tenemos que ir guardando cada una de las aproximaciones en un arreglo, y calcularemos el numero de aproximaciones necesarias para llegar a la precisión requerida. Tomemos en cuenta $\varepsilon = 0.001$. La formula para el numero de aproximaciones necesarias es:
#
# $$n = \frac{\ln{a} - \ln{\varepsilon}}{\ln{2}}$$
#
# donde $a$ es el tamaño del intervalo original.
n = (log(1) - log(0.001))/(log(2))
n
# Es decir, $n = 10$.
def metodo_biseccion(funcion, x1, x2, n):
xs = []
for i in range(n):
xs.append(biseccion(x1, x2))
if funcion(x2)*funcion(xs[-1]) > 0:
x2 = xs[-1]
else:
x1 = xs[-1]
return xs[-1]
metodo_biseccion(f, 1.0, 2.0, 10)
# Y asi obtenemos la aproximación de nuestro ejemplo.
# ## Problemas
# 1. Copia y pega el codigo necesario (no mas, no menos) para calcular una raiz real de un polinomio en el reporte de practica.
# 2. Calcule por el metodo de bisección la raiz real del siguiente polinomio (se espera un error no mayor a $0.001$) $f(x) = x^5 + 4 x^4 + 10 x^3 + x^2 + 20 x - 10$.
# 3. Modifique el codigo para que en lugar de aceptar el numero de iteraciones ($n$), acepte el error maximo ($\varepsilon$).
| Practicas/P1/.ipynb_checkpoints/Practica 1 - Introduccion a Jupyter-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Word2Vec model widgets
#
# This notebook introduces several examples of linking word2vec model in `ml5_ipynb` with jupyter widget `ipywidgets` to produce outputs.
# The model used contains 300-dimension embeddings for 10000 most common English words. There are smaller models in data folder.
#
# This example can refer to an word2vec [example](https://github.com/ml5js/ml5-library/tree/main/examples/p5js/Word2Vec/Word2Vec_Interactive/data) in ml5.js
#
# **Note:** Using words not in the model embeddings will result in errors.
from ml5_ipynb import ml5_text
import ipywidgets as widgets
w2v = ml5_text.word2Vec('data/wordvecs10000.json')
# ## What are the Top 3 nearest words?
#
# The following uses `nearest(word)` function to calculate the cosine distance and output the top 3 words with smallest distance.
nearest = widgets.Textarea(
value='',
placeholder='Type a word',
# description='Please type a word',
disabled=False
)
nearest_output = widgets.HTML(
value="",
# placeholder='Some HTML',
# description='Some HTML',
)
nearest_button = widgets.ToggleButton(
value=False,
description='is nearest to',
disabled=False,
button_style='', # 'success', 'info', 'warning', 'danger' or ''
tooltip='Description',
# icon='check'
)
def get_nearest(val):
if val:
word = nearest.value
if not word:
print('Empty word!')
return
w2v.nearest(word)
nearest_list = w2v.nearest_results[-1]
if not nearest_list:
print('No nearest word!')
return
nearest_words = [i['word'] for i in nearest_list[:3]]
w_str = '<br>'.join(nearest_words)
nearest_output.value = w_str
nearest_button.value = False
out = widgets.interactive_output(get_nearest,{'val':nearest_button})
widgets.VBox([nearest,nearest_button,nearest_output,out])
# ## What's the Top 3 words between two words?
#
# The following uses `average([word1,word2])` function to calculate the average of embedding of two words and output the top 3 words similar to the average embedding.
w1 = widgets.Text(
value='',
placeholder='Type a word',
disabled=False
)
w2 = widgets.Text(
value='',
placeholder='Type a word',
disabled=False
)
btw_output = widgets.HTML(
value="",
)
btw_button = widgets.ToggleButton(
value=False,
description='is',
disabled=False,
button_style='', # 'success', 'info', 'warning', 'danger' or ''
tooltip='Description',
)
def get_btw(val):
if val:
word1 = w1.value
word2 = w2.value
if not word1 or not word2:
print('Please type in both!')
return
w2v.average([word1,word2])
btw_list = w2v.average_results[-1]
if not btw_list:
print('No between word!')
return
btw_words = [i['word'] for i in btw_list[:3]]
w_str = '<br>'.join(btw_words)
btw_output.value = w_str
btw_button.value = False
btw_out = widgets.interactive_output(get_btw,{'val':btw_button})
widgets.VBox([widgets.HBox([widgets.HTML(value="Between "),
w1,
widgets.HTML(value=" and "),
w2,btw_button]),
btw_output,btw_out])
# ## Analogy
#
# Analogy is to show how two things are similar to each other. Analogy of word embedding can refer to element-wise addition and subtraction. It is a "word algebra".
# For example, king is to queen as man is to woman. The resulting word is determined by the following formula.
# ```
# vector('queen') - vector('king') + vector('man')
# ```
is_word = widgets.Text(
value='',
placeholder='Type a word',
disabled=False
)
to_word = widgets.Text(
value='',
placeholder='Type a word',
disabled=False
)
is_word2 = widgets.Text(
value='',
placeholder='Type a word',
disabled=False
)
analogy_output = widgets.HTML(
value="",
)
analogy_button = widgets.ToggleButton(
value=False,
description='is to',
disabled=False,
button_style='', # 'success', 'info', 'warning', 'danger' or ''
tooltip='Description',
)
def get_analogy(val):
if val:
iw = is_word.value
tw = to_word.value
iw2 = is_word2.value
if not iw or not tw or not iw2:
print('Please finish typing!')
return
w2v.subtract([tw,iw])
sub_list = w2v.subtract_results[-1]
if not sub_list:
print('Oops! Please type in other words!')
return
sub_w = sub_list[0]['word']
w2v.add([sub_w,iw2])
add_list = w2v.add_results[-1]
if not add_list:
print('Oops! No analogy for this example!')
add_word = [i['word']+"("+ str(round(i['distance'],2))+")" for i in add_list[:3]]
analogy_output.value = " , ".join(add_word)
analogy_button.value = False
analogy_out = widgets.interactive_output(get_analogy,{'val':analogy_button})
widgets.VBox([widgets.HBox([is_word,
widgets.HTML(value=" is to "),
to_word,
widgets.HTML(value=" as "),
is_word2,analogy_button]),
analogy_output,analogy_out])
| examples/Word2vec widget.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# #### Parallel Cluster Initialization with MPI4py: This could only be run on a HPC cluster
# The is only relevant to running mpi4py in a Jupyter notebook.
import ipyparallel
cluster=ipyparallel.Client(profile='mpi_tutorial')
print("IDs:",cluster.ids)
# %%px
from mpi4py import MPI
# %%px
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
size = comm.Get_size()
print ("I'm rank %d of %d on %s" %(rank,size,MPI.Get_processor_name()))
# #### Packages Import
# %%px
import numpy as np
from numpy import math
from scipy.stats import norm
from scipy import stats
import matplotlib.pyplot as plt
import progressbar
import time
import datetime
# #### Model Specification: OU Process
# 1. $dX_{t} = \theta_{1}(\theta_{2} - X_{t})dt + \sigma dW_{t}$, $Y_{t}|X_{t} \sim \mathcal{N}(X_{t}, \theta_{3}^2)$
# 2. $\mathbb{E}[X_{t}] = x_{0} e^{-\theta_1t} + \theta_{2} (1-e^{-\theta_{1}t})$, $Var[X_{t}] = \frac{\sigma^{2}}{2\theta_{1}}(1-e^{-2t\theta_1})$
# 3. $Y_{1},Y_{2},...$ mutually independent, $Y_{t} \sim_{i.i.d.} \mathcal{N}(\mathbb{E}[X_{t}], \theta_{3}^2 + Var[X_{t}])$, for $t \in \mathbb{N}_{0}$
# + jupyter={"source_hidden": true}
# %%px
initial_val = 1
sigma = 0.5
theta = np.array([1,0,np.sqrt(0.2)])
def diff_coef(x, dt, dw):
return sigma*np.math.sqrt(dt)*dw
def drift_coef(x, dt):
return theta[0]*(theta[1]-x)*dt
# Log-scaled unnormalized likelihood function p(y|x)
def likelihood_logscale(y, x):
d = (y-x)
gn = -1/2*(d**2/(theta[2]**2))
return gn
def likelihood_update(y,un,unormal_weight):
gamma = math.sqrt(0.2)
d = (y-un)
gn1 = -1/2*(d**2/(theta[2]**2)) + unormal_weight
return gn1
def sig_mean(t,theta):
return initial_val*np.exp(-theta[0]*t) + theta[1]*(1-np.exp(-theta[0]*t))
## Used only when theta[0] != 0
def sig_var(t,theta):
return (sigma**2 / (2*theta[0])) * (1-np.exp(-2*theta[0]*t))
def gen_data(T):
Y = np.zeros(T+1)
for t in range(T+1):
std = np.sqrt(sig_var(t,theta) + theta[2]**2)
Y[t] = sig_mean(t,theta) + std * np.random.randn(1)
return Y
def Kalmanfilter(T,Y):
m = np.zeros((T+1))
mhat = np.zeros((T+1))
c = np.zeros((T+1))
a = theta[0]
s = sigma
# observational noise variance is gam^2*I
gam = theta[2]
# dynamics noise variance is sig^2*I
sig = np.sqrt(s**2/2/a*(1-np.exp(-2*a)))
# dynamics determined by A
A = np.exp(-a)
# initial mean&covariance
m[0] = initial_val
c[0] = 0
H = 1
# solution & assimilate!
for t in range(T):
mhat[t] = A*m[t] + theta[1]*(1-A)
chat = A*c[t]*A + sig**2
########################
d = Y[t+1] - H*mhat[t]
# Kalmab Gain
K = (chat*H) / (H*chat*H + gam**2)
# Mean Update
m[t+1] = mhat[t] + K*d
# Covariance update
c[t+1] = (1-K*H)*chat
tv = m[T]
return tv
def Kalmanfilter_path(T,Y):
m = np.zeros((T+1))
mhat = np.zeros((T+1))
c = np.zeros((T+1))
a = theta[0]
s = sigma
# observational noise variance is gam^2*I
gam = theta[2]
# dynamics noise variance is sig^2*I
sig = np.sqrt(s**2/2/a*(1-np.exp(-2*a)))
# dynamics determined by A
A = np.exp(-a)
# initial mean&covariance
m[0] = initial_val
c[0] = 0
H = 1
# solution & assimilate!
for t in range(T):
mhat[t] = A*m[t] + theta[1]*(1-A)
chat = A*c[t]*A + sig**2
########################
d = Y[t+1] - H*mhat[t]
# Kalmab Gain
K = (chat*H) / (H*chat*H + gam**2)
# Mean Update
m[t+1] = mhat[t] + K*d
# Covariance update
c[t+1] = (1-K*H)*chat
return m
# -
# #### Main Function
# + jupyter={"source_hidden": true}
# %%px
# Resampling - input one-dimensional particle x
def resampling(weight, gn, x, N):
ess = 1/((weight**2).sum())
if ess <= (N/2):
## Sample with uniform dice
dice = np.random.random_sample(N)
## np.cumsum obtains CDF out of PMF
bins = np.cumsum(weight)
## np.digitize gets the indice of the bins where the dice belongs to
x_hat = x[np.digitize(dice,bins)]
## after resampling we reset the accumulating weight
gn = np.zeros(N)
if ess > (N/2):
x_hat = x
return x_hat, gn
# Coupled Wasserstein Resampling
def coupled_wasserstein(fine_weight, coarse_weight, gn, gc, fine_par, coarse_par, N):
ess = 1/((fine_weight**2).sum())
fine_hat = fine_par
coarse_hat = coarse_par
if ess <= (N/2):
# Sort in ascending order of particles
ind = np.argsort(fine_par[:])
inc = np.argsort(coarse_par[:])
fine_par = fine_par[ind]
fine_weight = fine_weight[ind]
coarse_par = coarse_par[inc]
coarse_weight = coarse_weight[inc]
# Sample with uniform dice
dice = np.random.random_sample(N)
# CDF
bins = np.cumsum(fine_weight)
bins1 = np.cumsum(coarse_weight)
# get the indices of the bins where the dice belongs to
fine_hat = fine_par[np.digitize(dice, bins)]
coarse_hat = coarse_par[np.digitize(dice, bins1)]
# reset accumulating weight after resampling
gn = np.zeros(N)
gc = np.zeros(N)
if ess > (N/2):
fine_hat = fine_par
coarse_hat = coarse_par
return fine_hat, gn, coarse_hat, gc
# Maixmally Coupled Resampling
def coupled_maximal(fine_weight, coarse_weight, gn, gc, fine_par, coarse_par, N):
ess = 1/((fine_weight**2).sum())
if ess <= (N/2):
# Maximal coupled resampling
fine_hat, coarse_hat = maximal_resample(fine_weight, coarse_weight, fine_par, coarse_par, N)
# reset accumulating weight after resampling
gn = np.zeros(N)
gc = np.zeros(N)
if ess > (N/2):
fine_hat = fine_par
coarse_hat = coarse_par
return fine_hat, gn, coarse_hat, gc
def maximal_resample(weight1,weight2,x1,x2,N):
# Initialize
x1_hat = np.zeros(N)
x2_hat = np.zeros(N)
# Calculating many weights
unormal_min_weight = np.minimum(weight1, weight2)
min_weight_sum = np.sum(unormal_min_weight)
min_weight = unormal_min_weight / min_weight_sum
unormal_reduce_weight1 = weight1 - unormal_min_weight
unormal_reduce_weight2 = weight2 - unormal_min_weight
## Sample with uniform dice
dice = np.random.random_sample(N)
## [0] takes out the numpy array which is suitable afterwards
coupled = np.where(dice <= min_weight_sum)[0]
independ = np.where(dice > min_weight_sum)[0]
ncoupled = np.sum(dice <= min_weight_sum)
nindepend = np.sum(dice > min_weight_sum)
if ncoupled>=0:
dice1 = np.random.random_sample(ncoupled)
bins = np.cumsum(min_weight)
x1_hat[coupled] = x1[np.digitize(dice1,bins)]
x2_hat[coupled] = x2[np.digitize(dice1,bins)]
## nindepend>0 implies min_weight_sum>0 imples np.sum(unormal_reduce_weight*) is positive, thus the division won't report error
if nindepend>0:
reduce_weight1 = unormal_reduce_weight1 / np.sum(unormal_reduce_weight1)
reduce_weight2 = unormal_reduce_weight2 / np.sum(unormal_reduce_weight2)
dice2 = np.random.random_sample(nindepend)
bins1 = np.cumsum(reduce_weight1)
bins2 = np.cumsum(reduce_weight2)
x1_hat[independ] = x1[np.digitize(dice2,bins1)]
x2_hat[independ] = x2[np.digitize(dice2,bins2)]
return x1_hat, x2_hat
def Particle_filter(l,T,N,Y):
hl = 2**(-l)
un = np.zeros(N)+initial_val
un_hat = un
gn = np.zeros(N)
for t in range(T):
un_hat = un
for dt in range(2**l):
dw = np.random.randn(N)
un = un + drift_coef(un, hl) + diff_coef(un, hl, dw)
# Cumulating weight function
gn = likelihood_logscale(Y[t+1], un) + gn
what = np.exp(gn-np.max(gn))
wn = what/np.sum(what)
# Wasserstein resampling
un_hat, gn = resampling(wn, gn, un, N)
return(np.sum(un*wn))
def Coupled_particle_filter_wasserstein(l,T,N,Y):
hl = 2**(-l)
## Initial value
un1 = np.zeros(N) + initial_val
cn1 = np.zeros(N) + initial_val
gn = np.ones(N)
gc = np.ones(N)
for t in range(T):
un = un1
cn = cn1
for dt in range(2**(l-1)):
dw = np.random.randn(2,N)
for s in range(2):
un = un + drift_coef(un, hl) + diff_coef(un, hl, dw[s,:])
cn = cn + drift_coef(cn, hl*2) + diff_coef(cn, hl, (dw[0,:] + dw[1,:]))
## Accumulating Weight Function
gn = likelihood_update(Y[t+1], un, gn)
what = np.exp(gn-np.max(gn))
wn = what/np.sum(what)
gc = likelihood_update(Y[t+1], cn, gc)
wchat = np.exp(gc-np.max(gc))
wc = wchat/np.sum(wchat)
## Wassersteing Resampling
un1, gn, cn1, gc = coupled_wasserstein(wn,wc,gn,gc,un,cn,N)
return(np.sum(un*wn-cn*wc))
def Coupled_particle_filter_maximal(l,T,N,Y):
hl = 2**(-l)
## Initial value
un1 = np.zeros(N) + initial_val
cn1 = np.zeros(N) + initial_val
gn = np.ones(N)
gc = np.ones(N)
for t in range(T):
un = un1
cn = cn1
for dt in range(2**(l-1)):
dw = np.random.randn(2,N)
for s in range(2):
un = un + drift_coef(un, hl) + diff_coef(un, hl, dw[s,:])
cn = cn + drift_coef(cn, hl*2) + diff_coef(cn, hl, (dw[0,:] + dw[1,:]))
## Accumulating Weight Function
gn = likelihood_update(Y[t+1], un, gn)
what = np.exp(gn-np.max(gn))
wn = what/np.sum(what)
gc = likelihood_update(Y[t+1], cn, gc)
wchat = np.exp(gc-np.max(gc))
wc = wchat/np.sum(wchat)
## Wassersteing Resampling
un1, gn, cn1, gc = coupled_maximal(wn,wc,gn,gc,un,cn,N)
return(np.sum(un*wn-cn*wc))
def coef(x, y):
# number of observations/points
n = np.size(x)
# mean of x and y vector
m_x, m_y = np.mean(x), np.mean(y)
# calculating cross-deviation and deviation about x
SS_xy = np.sum(y*x) - n*m_y*m_x
SS_xx = np.sum(x*x) - n*m_x*m_x
# calculating regression coefficients
b_1 = SS_xy / SS_xx
b_0 = m_y - b_1*m_x
return(b_0, b_1)
def num_coupled_par(p, p_max, const):
return int(2**(p+2*p_max) * (p_max**2) * const * c3)
def num_par(p, p_max, const):
return int(2**(p+2*p_max) * (p_max**2) * const * c2)
def prob_l_func(max_val):
prob = np.zeros(max_val)
for l in range(max_val):
prob[l] = 2**(-l*beta)
prob = prob / np.sum(prob)
return prob
def prob_p_func(max_val):
prob = np.zeros(max_val)
for p in range(max_val):
prob[p] = 2**(-p)
prob = prob / np.sum(prob)
return prob
def Xi_zero(T,p_prob,p_max,const,Y):
# sample the variable P
p = int(np.random.choice(p_max, 1, p=p_prob)[0])
#print('p_val is',p)
# construct the estimator
Xi_zero = (Particle_filter(0,T,num_par(p, p_max, const),Y) - Particle_filter(0,T,num_par(p-1, p_max, const),Y)) / p_prob[p]
return Xi_zero
def Xi_nonzero(l,T,p_prob,p_max,const,Y):
# sample the variable P
p = int(np.random.choice(p_max, 1, p=p_prob)[0])
#print('p_val is',p)
# construct the estimator
Xi = (Coupled_particle_filter_maximal(l,T,num_coupled_par(p,p_max,const),Y) - Coupled_particle_filter_maximal(l,T,num_coupled_par(p-1,p_max,const),Y)) / p_prob[p]
return Xi
def Xi(T,l_prob,l_max,p_prob,p_max,const,Y):
l = int(np.random.choice(l_max, 1, p=l_prob)[0])
#print('value of l is',l)
if l==0:
Xi = Xi_zero(T,p_prob,p_max,const,Y)
if l!=0:
Xi = Xi_nonzero(l,T,p_prob,p_max,const,Y)
est = Xi / l_prob[l]
return est
def parallel_particle_filter(M,T,max_val,const,Y):
l_max = max_val
p_max = max_val
l_prob = prob_l_func(l_max)
p_prob = prob_p_func(p_max)
est_summand = np.zeros(M)
for m in range(M):
est_summand[m] = Xi(T,l_prob,l_max,p_prob,p_max,const,Y)
return (np.mean(est_summand))
def parallel_particle_filter_record_progbar(M,T,max_val,const,Y):
l_max = max_val
p_max = max_val
l_prob = prob_l_func(l_max)
p_prob = prob_p_func(p_max)
est_summand = np.zeros(M)
pr = progressbar.ProgressBar(max_value=M).start()
for m in range(M):
est_summand[m] = Xi(T,l_prob,l_max,p_prob,p_max,const,Y)
pr.update(m+1)
pr.finish()
return est_summand
def parallel_particle_filter_record(M,T,max_val,const,Y):
l_max = max_val
p_max = max_val
l_prob = prob_l_func(l_max)
p_prob = prob_p_func(p_max)
est_summand = np.zeros(M)
for m in range(M):
est_summand[m] = Xi(T,l_prob,l_max,p_prob,p_max,const,Y)
return est_summand
# For OU process, beta=2
def num_ml_coupled(l,lmax,const):
return 2**(2*lmax-1.5*l) * const * c3
def num_ml_single(l,lmax,const):
return 2**(2*lmax-1.5*l) * const * c2
def mlpf(T,max_val,const,Y):
L = max_val
level_est = np.zeros(L)
level_est[0] = Particle_filter(0,T,int(num_ml_single(0,L,const)),Y)
for l in range(1,L):
level_est[l] = Coupled_particle_filter_maximal(l,T,int(num_ml_coupled(l,L,const)),Y)
return np.sum(level_est)
# -
# #### Simulation Setup Example
# 1. At discretization level $l=2$, aim at variance level of 10^{-7} for ppf (parallel particle filter), this is so that the variance is banlanced with the square bias, which we have already obtained. This is done by using $C=10^6$ on a single processor, with $M=1$.
#
# 2. Note that the PPF estimator has variance $Var(\sum_{i=1}^{M}\Xi_{i}) = \mathcal{O}(C^{-1}M^{-1})$, this means we can achieve the same variance level by using $C=10^3$ and $M=10^3$. We use $10^3$ parallel cores to obtain $i.i.d.$ realizations of $\Xi$ at the same time, this will give us a giant speed up. The simulation is set out to find how much is the speed up, at the same time ensuring $Var(\sum_{i=1}^{M}\Xi_{i}) \approx Bias(\sum_{i=1}^{M}\Xi_{i}) \approx 10^{-7}$.
# + jupyter={"source_hidden": true}
# %%px
T = 100
data_path = np.load('ou_model_data_path.npy')
c2, c3, beta = np.load('ou_fit_values.npy')
max_val=2
M=1000
const=1000
true_val = Kalmanfilter(T,data_path)
# -
# #### Parallel Implementaion of PPF
# 1. We need to parallel compute the $M$ realizations. We record the time needed for such one parallel realization.
# 2. We check the MSE of such PPF with $M$ values, this can be done in any fashion.
# 3. We can then compare MLPF with PPF 's cost for similar MSE targets.
# + jupyter={"source_hidden": true}
# %%px
# Used to construct a parallel - PPF: evaluate the cost of it
# Use M cores to get M repe of it and record the time
def multi_xi(seed_val):
l_max = max_val
np.random.seed(seed_val)
l = int(np.random.choice(l_max, 1, p=l_prob)[0])
#print('value of l is',l)
if l==0:
Xi = Xi_zero(T,p_prob,p_max,const,Y)
if l!=0:
Xi = Xi_nonzero(l,T,p_prob,p_max,const,Y)
est = Xi / l_prob[l]
return est
# Used to obtain MSE of PPF with M.
# Use Rep_num of cores to get repetition of it and compute the (sample) MSE.
def multi_ppf(seed_val):
np.random.seed(seed_val)
l_max = max_val
p_max = max_val
l_prob = prob_l_func(l_max)
p_prob = prob_p_func(p_max)
est_summand = np.zeros(M)
for m in range(M):
est_summand[m] = Xi(T,l_prob,l_max,p_prob,p_max,const,Y)
return (np.mean(est_summand))
# -
# #### MPI4py HPC Implementation
# + jupyter={"source_hidden": true}
# %%px
iter_num = 0
rank = comm.Get_rank()
size = comm.Get_size()
## Every iteration should have different initial_seed values
initial_seed = iter_num*(size)
seed_val_rankwise = initial_seed + rank
# -
# #### (I) Cost record of M parallel implementations for PPF estimate
# + jupyter={"source_hidden": true}
# %%px
stime = time.time()
xi_reptition = np.zeros(1)
xi_reptition = multi_xi(seed_val_rankwise)
result = np.zeros(size)
comm.Gather(xi_reptition,result,root=0)
if rank == 0 :
x = np.asarray(result)
ppf_estimate = np.mean(x)
print('HPC-PPF outputs:',ppf_estimate)
etime = time.time()
time_len = str(datetime.timedelta(seconds=etime-stime))
print("Time cost for HPC-PPF is:",time_len)
# -
# #### (II) MSE compuation for PPF estimate
# + jupyter={"source_hidden": true}
# %%px
ppf_reptition = np.zeros(1)
ppf_reptition = multi_ppf(seed_val_rankwise)
result = np.zeros(size)
comm.Gather(xi_reptition,result,root=0)
if rank == 0 :
x = np.asarray(result)
mse_ppf = np.mean((x-true_val)**2)
var_ppf = np.var(x)
square_bias_ppf = mse_ppf - var_ppf
print('HPC-PPF has MSE:',mse_ppf, 'Variance:',var_ppf, 'Square Bias:',square_bias_ppf)
| HPC UPF.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Poission GLM (Attempt 1)
#
# Below are the different GLM methods possible. Initially we take Xell's code and clean it a little.
#
# First all package imports have been collated for ease.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import statsmodels.api as sm
import seaborn as sns
from sklearn.linear_model import PoissonRegressor
from sklearn.model_selection import train_test_split
# Next we simply import our data and get it into the format required. We are firslty looking to tackle the 31-day data to start off with.
# +
#Read the data into the file.
fullData = pd.read_csv("31DayDataUpdate.csv")
#Create a dataframe from the relevant columns.
df = fullData[['Quarter', 'HB', 'CancerType','NumberOfEligibleReferrals31DayStandard','NumberOfEligibleReferralsTreatedWithin31Days']]
#Remove the 'all cancer types' rows from the dataframe.
df = df[df['CancerType'] != 'All Cancer Types']
df = df[df['NumberOfEligibleReferrals31DayStandard'].notna()]
# -
# The following may be of use when looking at the correlation between the two sets of numerical values which we have here. We can also identify all the data types within our frame.
# +
#Correlation of the Eligible Referrals with the Eligible Referalls Treated.
rho = df.corr()
#print(rho)
#View of the whole data frame.
types = df.dtypes
#print(types)
# -
# Next we take an overview of the different cancers within the data we have in our set. We firstly achieve four different graphs which are explained below the output, before also looking at the boxplot in the situation.
sns.pairplot(df, hue='CancerType', height=2.5, aspect=1)
sns.boxplot(y='NumberOfEligibleReferrals31DayStandard', x='CancerType',data=df)
# Next we look to form the variables in the correct way. This is the first part of building our GLM.
#
# ## 0. Correct form of data for a GLM
# +
# choose explanatory variables - note we can also include 'Quarter', 'CancerType' and 'Sex' here.
X = df[['HB', 'CancerType']]
# turn our catergories into dummies.
X = pd.get_dummies(data=X, drop_first=True)
#X.head()
# choose which column is the targeted output data.
Y = df['NumberOfEligibleReferrals31DayStandard']
#Y.head()
# -
# ## 1. SKLearn
# Below we create our GLM model using the SKLearn method.
# +
#Build the model using SKLearn.
prSKLearn = PoissonRegressor(alpha=0, fit_intercept=True)
#Fit the model using the build above.
prSKLearn.fit(X, Y)
parameters = prSKLearn.get_params()
score = prSKLearn.score(X, Y)
#Should we want to see the values of each of the intercepts we can do so by uncommenting the below.
#print(prSKLearn.intercept_)
print(score)
#I need to check what the below actually does...
coeff_parameter = pd.DataFrame(prSKLearn.coef_, X.columns,columns=['Coefficient'])
# -
# Next we use instead use our data in a split manner in order to try and see whether the GLM achieves a suitable outcome (from https://medium.com/analytics-vidhya/implementing-linear-regression-using-sklearn-76264a3c073c).
#
# +
#Split our data into test and train.
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.5)
#Use our training data to let the SKLearn model learn
prSKLearn.fit(X_train, y_train)
#Get predictions for the test data using the SKLearn model.
predictions = prSKLearn.predict(X_test)
# -
#Quantify the useful nature of the model using the R2 value.
print(prSKLearn.score(X_train, y_train))
#Create a regressional plot to show
sns.regplot(y_test, predictions)
# ## 2. StatsModels
# Next we use the StatsModels package to create the GLM.
# +
#Add the X to the constants.
XSM = sm.add_constant(X)
#Create the model.
ls=sm.OLS(Y,XSM).fit()
#Print the summary of the model.
print(ls.summary())
# -
# ----------------------------------------------------------------------
#
# # Poisson GLM (Attempt 2)
# Firstly below we read our data in different ways, and furthermore edit the data to a certain extent to make it easier for handling. We also then provide some summary statistics and graphs.
# +
#Import the 31 day standard data.
Data31Read = pd.read_csv("31DayDataUpdate.csv")
Data31 = Data31Read[['Quarter', 'HB', 'CancerType','NumberOfEligibleReferrals31DayStandard','NumberOfEligibleReferralsTreatedWithin31Days']]
#Import the 62 day standard data.
Data62Read = pd.read_csv("62DayDataUpdate.csv")
Data62 = Data62Read[['Quarter', 'HB', 'CancerType','NumberOfEligibleReferrals62DayStandard','NumberOfEligibleReferralsTreatedWithin62Days']]
#Import the weekly cancer data.
WeekData = pd.read_csv("cancerdata.csv")
# +
#Make edits to the data
##31Day Edits start here.
df31 = Data31
#Remove NaNs and 'all cancer types'
df31 = df31[df31['NumberOfEligibleReferrals31DayStandard'].notna()]
df31 = df31[df31['NumberOfEligibleReferralsTreatedWithin31Days'].notna()]
df31 = df31[df31['CancerType'] != 'All Cancer Types']
# add a numerical variable 1-1 to quarters and an index variable before/after pandemic
quarters = df31['Quarter']
date = np.zeros(len(quarters))
quars = df31['Quarter'].unique()
dates = np.arange(len(quars))
pandemic = np.zeros(len(quarters))
x = np.where(quars == '2020Q2') # change to decide in which quarter the pandemic begins
pandemic = np.zeros(len(quarters))
x = np.where(quars == '2020Q2')
j=0
for i in np.arange(len(quarters)):
y = np.where(quars == quarters[i])
date[i] = dates[y]
if dates[y] > x:
pandemic[i] = dates[y]-x
df31['Date'] = date.tolist()
df31['Pandemic'] = pandemic.tolist()
##Do the same with 62 days data
df62 = Data62
#Remove NaNs and 'all cancer types'
df62 = df62[df62['NumberOfEligibleReferrals62DayStandard'].notna()]
df62 = df62[df62['NumberOfEligibleReferralsTreatedWithin62Days'].notna()]
df62 = df62[df62['CancerType'] != 'All Cancer Types']
# add a numerical variable 1-1 to quarters and an index variable before/after pandemic
quarters = df62['Quarter']
date = np.zeros(len(quarters))
quars = df62['Quarter'].unique()
dates = np.arange(len(quars))
pandemic = np.zeros(len(quarters))
x = np.where(quars == '2020Q2') # change to decide in which quarter the pandemic begins
pandemic = np.zeros(len(quarters))
x = np.where(quars == '2020Q2')
j=0
for i in np.arange(len(quarters)):
y = np.where(quars == quarters[i])
date[i] = dates[y]
if dates[y] > x:
pandemic[i] = dates[y]-x
df62['Date'] = date.tolist()
df62['Pandemic'] = pandemic.tolist()
#Edit weekly data
dfW = WeekData
#Remove NaNs and cumulative data.
dfW = dfW[dfW['CancerType'] != 'All Cancers']
dfW = dfW[dfW['Age Group'] != 'All Ages']
dfW = dfW[dfW['Sex'] != 'All']
dfW = dfW[dfW['Count'].notna()]
months = np.asarray(dfW['Month'])
date = np.zeros(len(months))
quars = dfW['Month'].unique()
dates = np.arange(len(quars))
pandemic = np.zeros(len(months))
x = np.where(quars == 202003)
for i in np.arange(len(months)):
y = np.where(quars == months[i])
date[i] = dates[y]
if dates[y] > x:
pandemic[i] = dates[y]-x
dfW['Date'] = date.tolist()
dfW['Pandemic'] = pandemic.tolist()
# +
# take a look at what we have
Cor31 = df31.corr()
Cor62 = df62.corr()
CorW = dfW.corr()
# plot eligibles per cancer type
plt.figure(figsize=(15,8))
ax =sns.boxplot(y='NumberOfEligibleReferrals31DayStandard', x='CancerType', data=df31)
plt.show()
plt.figure(figsize=(15,8))
ax =sns.boxplot(y='NumberOfEligibleReferrals62DayStandard', x='CancerType', data=df62)
plt.show()
plt.figure(figsize=(15,8))
ax =sns.boxplot(y='Count', x='CancerType', data=dfW)
plt.show()
# -
# ## 3. Hybrid Inputs
# Next we take Xell's code which is in more of a functional mode.
#Define a function that fits poisson regression.
def poissonGLM(X, Y, Train):
'''
inputs:
X: explanatory variables (dataframe)
Y: output variable (dataframe)
train: do training or not (boolean)
outputs:
parameters: possion regressor parameters
intercept: value of independetn term (y intercept),
coefficients: coefficients of explanatory vairables (all linera)
y_test: testing data
predictions: predicted data
stats : summary of statistics of the predictive power of the model
'''
#Turn categorical into dummies.
X = pd.get_dummies(data=X, drop_first=True)
#Build model.
prFunction = PoissonRegressor(alpha=0, fit_intercept=True, max_iter = 10000)
#Fit model.
prFunction.fit(X, Y)
#Recover model information.
parameters = prFunction.get_params()
intercept = prFunction.intercept_
coefficients = pd.DataFrame(prFunction.coef_,X.columns, columns=['Coefficient'])
#Initialise training parameters.
y_train = 0
predictions = 0
stats = 0
if Train==1:
#Split data into train and test (test_size specifies %).
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
#Fit training data.
prFunction.fit(X_train, y_train)
#Use model to predict test data outputs.
predictions = prFunction.predict(X_test)
#Also now run the Statsmodels version of prediction.
XSM = sm.add_constant(X)
ls = sm.OLS(Y, X).fit()
stats = ls.summary()
return parameters, intercept, coefficients, y_test, predictions, stats
# +
#Create the options for explanatory and output data.
#FOR 31 DAYS DATA
#Choose explanatory variables
X31 = df31[['Date','CancerType','HB']] #no pandemic
X31_p = df31[['Pandemic','CancerType','HB']] #pandemic, no date
X31_dp = df31[['Date','Pandemic','CancerType','HB']] #both
#Choose output data
Y31 = df31['NumberOfEligibleReferrals31DayStandard']
Y31t = df31['NumberOfEligibleReferralsTreatedWithin31Days']
#FOR 62 DAYS DATA
#Choose explanatory variables
X62 = df62[['Date','HB','CancerType']] #no pandemic
X62_p = df62[['Pandemic','CancerType','HB']] #pandemic, no date
X62_dp = df62[['Date','Pandemic','CancerType','HB']] #both
#Choose output data
Y62 = df62['NumberOfEligibleReferrals62DayStandard']
Y62t = df62['NumberOfEligibleReferralsTreatedWithin62Days']
## FOR WEEKLY DIAGNOSIS DATA
XW = dfW[['Date','HB', 'Sex','Age Group']]
XW_p = dfW[['Pandemic','HB', 'Sex','Age Group']]
XW_dp = dfW[['Pandemic','Date','HB', 'Sex','Age Group']]
#Choose output data
YW = dfW['Count']
# +
#Choose explanatory and output variables from above sets.
X=X31_dp
Y=Y31
#Run Poisson regression for chosen data
parameters, intercept, coefficients, y_test, predictions, stats = poissonGLM(X, Y, 1)
# +
# take a look at results of Poisson Regressor
print(intercept)
print(coefficients[1:2])
# check predictive power
sns.regplot(y_test,predictions)
# take a look at results of Statsmodels
print(stats)
# -
# -----------
# # Analysis of vartiation of $\epsilon$
#
# +
##31Day Edits start here.
df31e = Data31
#Remove NaNs and 'all cancer types' and add a numerical variable 1-1 to quarters
df31e = df31e[df31e['NumberOfEligibleReferrals31DayStandard'].notna()]
df31e = df31e[df31e['NumberOfEligibleReferralsTreatedWithin31Days'].notna()]
df31e = df31e[df31e['CancerType'] != 'All Cancer Types']
quarters = df31e['Quarter']
date = np.zeros(len(quarters))
quars = df31e['Quarter'].unique()
dates = np.arange(len(quars))
CovQuarters = ['2020Q1', '2020Q2', '2020Q3', '2020Q4']
CoefHold = []
for i in CovQuarters:
#index variable before/after pandemic
pandemic = np.zeros(len(quarters))
x = np.where(quars == i) # change to decide in which quarter the pandemic begins
pandemic = np.zeros(len(quarters))
x = np.where(quars == i)
j=0
for j in np.arange(len(quarters)):
y = np.where(quars == quarters[j])
date[j] = dates[y]
if dates[y] > x:
pandemic[j] = dates[y]-x
df31e['Date'] = date.tolist()
df31e['Pandemic'] = pandemic.tolist()
X31_dpe = df31e[['Date','Pandemic','CancerType','HB']]
Y31e = df31e['NumberOfEligibleReferrals31DayStandard']
X=X31_dpe
Y=Y31e
parameters, intercept, coefficients, y_test, predictions, stats = poissonGLM(X, Y, 1)
newCoef = coefficients[1:2].to_numpy()
CoefHold.append(newCoef[0][0])
print(CoefHold)
# +
##31Day Edits start here.
df62e = Data62
#Remove NaNs and 'all cancer types' and add a numerical variable 1-1 to quarters
df62e = df62e[df62e['NumberOfEligibleReferrals62DayStandard'].notna()]
df62e = df62e[df62e['NumberOfEligibleReferralsTreatedWithin62Days'].notna()]
df62e = df62e[df62e['CancerType'] != 'All Cancer Types']
quarters = df62e['Quarter']
date = np.zeros(len(quarters))
quars = df62e['Quarter'].unique()
dates = np.arange(len(quars))
CovQuarters = ['2020Q1', '2020Q2', '2020Q3', '2020Q4']
CoefHold = []
for i in CovQuarters:
#index variable before/after pandemic
pandemic = np.zeros(len(quarters))
x = np.where(quars == i) # change to decide in which quarter the pandemic begins
pandemic = np.zeros(len(quarters))
x = np.where(quars == i)
j=0
for j in np.arange(len(quarters)):
y = np.where(quars == quarters[j])
date[j] = dates[y]
if dates[y] > x:
pandemic[j] = dates[y]-x
df62e['Date'] = date.tolist()
df62e['Pandemic'] = pandemic.tolist()
X62_dpe = df62e[['Date','Pandemic','CancerType','HB']]
Y62e = df62e['NumberOfEligibleReferrals62DayStandard']
X=X62_dpe
Y=Y62e
parameters, intercept, coefficients, y_test, predictions, stats = poissonGLM(X, Y, 1)
newCoef = coefficients[1:2].to_numpy()
CoefHold.append(newCoef[0][0])
print(CoefHold)
| .ipynb_checkpoints/COPoissionGLM-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # _*Pricing Asian Barrier Spreads*_
# ### Introduction
# <br>
# An Asian barrier spread is a combination of 3 different option types, and as such, combines multiple possible features that the Qiskit Finance option pricing framework supports:
#
# - <a href="https://www.investopedia.com/terms/a/asianoption.asp">Asian option</a>: The payoff depends on the average price over the considered time horizon.
# - <a href="https://www.investopedia.com/terms/b/barrieroption.asp">Barrier Option</a>: The payoff is zero if a certain threshold is exceeded at any time within the considered time horizon.
# - <a href="https://www.investopedia.com/terms/b/bullspread.asp">(Bull) Spread</a>: The payoff follows a piecewise linear function (depending on the average price) starting at zero, increasing linear, staying constant.
#
# Suppose strike prices $K_1 < K_2$ and time periods $t=1,2$, with corresponding spot prices $(S_1, S_2)$ following a given multivariate distribution (e.g. generated by some stochastic process), and a barrier threshold $B>0$.
# The corresponding payoff function is defined as
#
#
# $$
# P(S_1, S_2) =
# \begin{cases}
# \min\left\{\max\left\{\frac{1}{2}(S_1 + S_2) - K_1, 0\right\}, K_2 - K_1\right\}, & \text{ if } S_1, S_2 \leq B \\
# 0, & \text{otherwise.}
# \end{cases}
# $$
#
#
# In the following, a quantum algorithm based on amplitude estimation is used to estimate the expected payoff, i.e., the fair price before discounting, for the option
#
#
# $$\mathbb{E}\left[ P(S_1, S_2) \right].$$
#
#
# The approximation of the objective function and a general introduction to option pricing and risk analysis on quantum computers are given in the following papers:
#
# - <a href="https://arxiv.org/abs/1806.06893">Quantum Risk Analysis. Woerner, Egger. 2018.</a>
# - <a href="https://arxiv.org/abs/1905.02666">Option Pricing using Quantum Computers. Stamatopoulos et al. 2019.</a>
# +
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.interpolate import griddata
# %matplotlib inline
import numpy as np
from qiskit import QuantumRegister, QuantumCircuit, Aer, execute
from qiskit.circuit.library import IntegerComparator
from qiskit.aqua.algorithms import IterativeAmplitudeEstimation
from qiskit.aqua.circuits import WeightedSumOperator
from qiskit.aqua.components.uncertainty_problems import UnivariatePiecewiseLinearObjective as PwlObjective
from qiskit.aqua.components.uncertainty_problems import MultivariateProblem
from qiskit.aqua.components.uncertainty_models import MultivariateLogNormalDistribution
# MultivariateProblem internally still needs the methods in the Comparator custom class below.
# The code will be changed so that an IntegerComparator can be passed directly without the need
# of a custom class. Until it happens the custom class is necessary.
class Comparator(IntegerComparator):
def required_ancillas(self):
return self.num_ancilla_qubits
def build(self, qc, q, q_ancillas=None, params=None):
i_state = range(self.num_state_qubits)
i_target = self.num_state_qubits
instr = self.to_instruction()
qr = [q[i] for i in i_state] + [q[i_target]]
if q_ancillas:
qr += [qi for qi in q_ancillas[:self.required_ancillas()]]
qc.append(instr, qr)
def build_inverse(self, qc, q, q_ancillas=None):
qc_ = QuantumCircuit(*qc.qregs)
self.build(qc_, q, q_ancillas)
qc.extend(qc_.inverse())
# -
# ### Uncertainty Model
#
# We construct a circuit factory to load a multivariate log-normal random distribution into a quantum state on $n$ qubits.
# For every dimension $j = 1,\ldots,d$, the distribution is truncated to a given interval $[low_j, high_j]$ and discretized using $2^{n_j}$ grid points, where $n_j$ denotes the number of qubits used to represent dimension $j$, i.e., $n_1+\ldots+n_d = n$.
# The unitary operator corresponding to the circuit factory implements the following:
#
# $$\big|0\rangle_{n} \mapsto \big|\psi\rangle_{n} = \sum_{i_1,\ldots,i_d} \sqrt{p_{i_1\ldots i_d}}\big|i_1\rangle_{n_1}\ldots\big|i_d\rangle_{n_d},$$
#
# where $p_{i_1\ldots i_d}$ denote the probabilities corresponding to the truncated and discretized distribution and where $i_j$ is mapped to the right interval using the affine map:
#
# $$ \{0, \ldots, 2^{n_j}-1\} \ni i_j \mapsto \frac{high_j - low_j}{2^{n_j} - 1} * i_j + low_j \in [low_j, high_j].$$
#
# For simplicity, we assume both stock prices are independent and identically distributed.
# This assumption just simplifies the parametrization below and can be easily relaxed to more complex and also correlated multivariate distributions.
# The only important assumption for the current implementation is that the discretization grid of the different dimensions has the same step size.
# +
# number of qubits per dimension to represent the uncertainty
num_uncertainty_qubits = 2
# parameters for considered random distribution
S = 2.0 # initial spot price
vol = 0.4 # volatility of 40%
r = 0.05 # annual interest rate of 4%
T = 40 / 365 # 40 days to maturity
# resulting parameters for log-normal distribution
mu = ((r - 0.5 * vol**2) * T + np.log(S))
sigma = vol * np.sqrt(T)
mean = np.exp(mu + sigma**2/2)
variance = (np.exp(sigma**2) - 1) * np.exp(2*mu + sigma**2)
stddev = np.sqrt(variance)
# lowest and highest value considered for the spot price; in between, an equidistant discretization is considered.
low = np.maximum(0, mean - 3*stddev)
high = mean + 3*stddev
# map to higher dimensional distribution
# for simplicity assuming dimensions are independent and identically distributed)
dimension = 2
num_qubits=[num_uncertainty_qubits]*dimension
low=low*np.ones(dimension)
high=high*np.ones(dimension)
mu=mu*np.ones(dimension)
cov=sigma**2*np.eye(dimension)
# construct circuit factory
u = MultivariateLogNormalDistribution(num_qubits=num_qubits, low=low, high=high, mu=mu, cov=cov)
# -
# plot PDF of uncertainty model
x = [ v[0] for v in u.values ]
y = [ v[1] for v in u.values ]
z = u.probabilities
#z = map(float, z)
#z = list(map(float, z))
resolution = np.array([2**n for n in num_qubits])*1j
grid_x, grid_y = np.mgrid[min(x):max(x):resolution[0], min(y):max(y):resolution[1]]
grid_z = griddata((x, y), z, (grid_x, grid_y))
fig = plt.figure(figsize=(10, 8))
ax = fig.gca(projection='3d')
ax.plot_surface(grid_x, grid_y, grid_z, cmap=plt.cm.Spectral)
ax.set_xlabel('Spot Price $S_1$ (\$)', size=15)
ax.set_ylabel('Spot Price $S_2$ (\$)', size=15)
ax.set_zlabel('Probability (\%)', size=15)
plt.show()
# ### Payoff Function
#
# For simplicity, we consider the sum of the spot prices instead of their average.
# The result can be transformed to the average by just dividing it by 2.
#
# The payoff function equals zero as long as the sum of the spot prices $(S_1 + S_2)$ is less than the strike price $K_1$ and then increases linearly until the sum of the spot prices reaches $K_2$.
# Then payoff stays constant to $K_2 - K_1$ unless any of the two spot prices exceeds the barrier threshold $B$, then the payoff goes immediately down to zero.
# The implementation first uses a weighted sum operator to compute the sum of the spot prices into an ancilla register, and then uses a comparator, that flips an ancilla qubit from $\big|0\rangle$ to $\big|1\rangle$ if $(S_1 + S_2) \geq K_1$ and another comparator/ancilla to capture the case that $(S_1 + S_2) \geq K_2$.
# These ancillas are used to control the linear part of the payoff function.
#
# In addition, we add another ancilla variable for each time step and use additional comparators to check whether $S_1$, respectively $S_2$, exceed the barrier threshold $B$. The payoff function is only applied if $S_1, S_2 \leq B$.
#
# The linear part itself is approximated as follows.
# We exploit the fact that $\sin^2(y + \pi/4) \approx y + 1/2$ for small $|y|$.
# Thus, for a given approximation scaling factor $c_{approx} \in [0, 1]$ and $x \in [0, 1]$ we consider
#
# $$ \sin^2( \pi/2 * c_{approx} * ( x - 1/2 ) + \pi/4) \approx \pi/2 * c_{approx} * ( x - 1/2 ) + 1/2 $$ for small $c_{approx}$.
#
# We can easily construct an operator that acts as
#
# $$\big|x\rangle \big|0\rangle \mapsto \big|x\rangle \left( \cos(a*x+b) \big|0\rangle + \sin(a*x+b) \big|1\rangle \right),$$
#
# using controlled Y-rotations.
#
# Eventually, we are interested in the probability of measuring $\big|1\rangle$ in the last qubit, which corresponds to
# $\sin^2(a*x+b)$.
# Together with the approximation above, this allows to approximate the values of interest.
# The smaller we choose $c_{approx}$, the better the approximation.
# However, since we are then estimating a property scaled by $c_{approx}$, the number of evaluation qubits $m$ needs to be adjusted accordingly.
#
# For more details on the approximation, we refer to:
# <a href="https://arxiv.org/abs/1806.06893">Quantum Risk Analysis. <NAME>. 2018.</a>
#
# Since the weighted sum operator (in its current implementation) can only sum up integers, we need to map from the original ranges to the representable range to estimate the result, and reverse this mapping before interpreting the result. The mapping essentially corresponds to the affine mapping described in the context of the uncertainty model above.
# +
# determine number of qubits required to represent total loss
weights = []
for n in num_qubits:
for i in range(n):
weights += [2**i]
n_s = WeightedSumOperator.get_required_sum_qubits(weights)
# create circuit factory
agg = WeightedSumOperator(sum(num_qubits), weights)
# +
# set the strike price (should be within the low and the high value of the uncertainty)
strike_price_1 = 3
strike_price_2 = 4
# set the barrier threshold
barrier = 2.5
# map strike prices and barrier threshold from [low, high] to {0, ..., 2^n-1}
max_value = 2**n_s - 1
low_ = low[0]
high_ = high[0]
mapped_strike_price_1 = (strike_price_1 - dimension*low_) / (high_ - low_) * (2**num_uncertainty_qubits - 1)
mapped_strike_price_2 = (strike_price_2 - dimension*low_) / (high_ - low_) * (2**num_uncertainty_qubits - 1)
mapped_barrier = (barrier - low) / (high - low) * (2**num_uncertainty_qubits - 1)
# -
# condition and condition result
conditions = []
barrier_thresholds = [2]*dimension
for i in range(dimension):
# target dimension of random distribution and corresponding condition (which is required to be True)
conditions += [(i, Comparator(num_qubits[i], mapped_barrier[i] + 1, geq=False))]
# +
# set the approximation scaling for the payoff function
c_approx = 0.25
# setup piecewise linear objective fcuntion
breakpoints = [0, mapped_strike_price_1, mapped_strike_price_2]
slopes = [0, 1, 0]
offsets = [0, 0, mapped_strike_price_2 - mapped_strike_price_1]
f_min = 0
f_max = mapped_strike_price_2 - mapped_strike_price_1
bull_spread_objective = PwlObjective(
n_s,
0,
max_value,
breakpoints,
slopes,
offsets,
f_min,
f_max,
c_approx
)
# define overall multivariate problem
asian_barrier_spread = MultivariateProblem(u, agg, bull_spread_objective, conditions=conditions)
# -
# plot exact payoff function
plt.figure(figsize=(7,5))
x = np.linspace(sum(low), sum(high))
y = (x <= 5)*np.minimum(np.maximum(0, x - strike_price_1), strike_price_2 - strike_price_1)
plt.plot(x, y, 'r-')
plt.grid()
plt.title('Payoff Function (for $S_1 = S_2$)', size=15)
plt.xlabel('Sum of Spot Prices ($S_1 + S_2)$', size=15)
plt.ylabel('Payoff', size=15)
plt.xticks(size=15, rotation=90)
plt.yticks(size=15)
plt.show()
# + tags=["nbsphinx-thumbnail"]
# plot contour of payoff function with respect to both time steps, including barrier
plt.figure(figsize=(7,5))
z = np.zeros((17, 17))
x = np.linspace(low[0], high[0], 17)
y = np.linspace(low[1], high[1], 17)
for i, x_ in enumerate(x):
for j, y_ in enumerate(y):
z[i, j] = np.minimum(np.maximum(0, x_ + y_ - strike_price_1), strike_price_2 - strike_price_1)
if x_ > barrier or y_ > barrier:
z[i, j] = 0
plt.title('Payoff Function', size=15)
plt.contourf(x, y, z)
plt.colorbar()
plt.xlabel('Spot Price $S_1$', size=15)
plt.ylabel('Spot Price $S_2$', size=15)
plt.xticks(size=15)
plt.yticks(size=15)
plt.show()
# -
# evaluate exact expected value
sum_values = np.sum(u.values, axis=1)
payoff = np.minimum(np.maximum(sum_values - strike_price_1, 0), strike_price_2 - strike_price_1)
leq_barrier = [ np.max(v) <= barrier for v in u.values ]
exact_value = np.dot(u.probabilities[leq_barrier], payoff[leq_barrier])
print('exact expected value:\t%.4f' % exact_value)
# ### Evaluate Expected Payoff
#
# We first verify the quantum circuit by simulating it and analyzing the resulting probability to measure the $|1\rangle$ state in the objective qubit.
# +
num_req_qubits = asian_barrier_spread.num_target_qubits
num_req_ancillas = asian_barrier_spread.required_ancillas()
q = QuantumRegister(num_req_qubits, name='q')
q_a = QuantumRegister(num_req_ancillas, name='q_a')
qc = QuantumCircuit(q, q_a)
asian_barrier_spread.build(qc, q, q_a)
print('state qubits: ', num_req_qubits)
print('circuit width:', qc.width())
print('circuit depth:', qc.depth())
# -
job = execute(qc, backend=Aer.get_backend('statevector_simulator'))
# +
# evaluate resulting statevector
value = 0
for i, a in enumerate(job.result().get_statevector()):
b = ('{0:0%sb}' % asian_barrier_spread.num_target_qubits).format(i)[-asian_barrier_spread.num_target_qubits:]
prob = np.abs(a)**2
if prob > 1e-4 and b[0] == '1':
value += prob
# all other states should have zero probability due to ancilla qubits
if i > 2**num_req_qubits:
break
# map value to original range
mapped_value = asian_barrier_spread.value_to_estimation(value) / (2**num_uncertainty_qubits - 1) * (high_ - low_)
print('Exact Operator Value: %.4f' % value)
print('Mapped Operator value: %.4f' % mapped_value)
print('Exact Expected Payoff: %.4f' % exact_value)
# -
# Next we use amplitude estimation to estimate the expected payoff.
# Note that this can take a while since we are simulating a large number of qubits. The way we designed the operator (asian_barrier_spread) implies that the number of actual state qubits is significantly smaller, thus, helping to reduce the overall simulation time a bit.
# +
# set target precision and confidence level
epsilon = 0.01
alpha = 0.05
# construct amplitude estimation
ae = IterativeAmplitudeEstimation(epsilon=epsilon, alpha=alpha, a_factory=asian_barrier_spread)
# -
result = ae.run(quantum_instance=Aer.get_backend('qasm_simulator'), shots=100)
conf_int = np.array(result['confidence_interval']) / (2**num_uncertainty_qubits - 1) * (high_ - low_)
print('Exact value: \t%.4f' % exact_value)
print('Estimated value:\t%.4f' % (result['estimation'] / (2**num_uncertainty_qubits - 1) * (high_ - low_)))
print('Confidence interval: \t[%.4f, %.4f]' % tuple(conf_int))
import qiskit.tools.jupyter
# %qiskit_version_table
# %qiskit_copyright
| tutorials/finance/07_asian_barrier_spread_pricing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Mini-Project 2: Source extraction
#
# **Context**
#
# This notebook illustrates the basics of distributing image data, and process them separately. In this notebook, we will load [MegaCam](https://www.cfht.hawaii.edu/Instruments/Imaging/Megacam/) data, and extract source positions in the images.
# + [markdown] slideshow={"slide_type": "slide"}
# **Learning objectives**
#
# After going through this notebook, you should be able to:
#
# - Load and efficiently access astronomical images with Apache Spark
# - Interface and use your favourite image processing package.
#
# For this project, you will use the data at `data/images` (see the `download_data_project.sh` script).
# + slideshow={"slide_type": "slide"}
# We use the custom spark-fits connector
df = spark.read.format("fits").option("hdu", 1).load("../data/images/*.fits")
# + slideshow={"slide_type": "fragment"}
df.show(3)
# + [markdown] slideshow={"slide_type": "subslide"}
# By default, spark-fits will assign one image line per row, without specifying the data provenance. But you can retrieve it easily:
# + slideshow={"slide_type": "fragment"}
from pyspark.sql.functions import input_file_name
df = df.withColumn('ImgIndex', input_file_name())
df.show(3)
# + [markdown] slideshow={"slide_type": "subslide"}
# We have 4 images with 4644 lines each:
# + slideshow={"slide_type": "fragment"}
df.groupBy('ImgIndex').count().show()
# + [markdown] slideshow={"slide_type": "subslide"}
# and for each image, we have 2112 columns:
# + slideshow={"slide_type": "fragment"}
import pandas as pd
from pyspark.sql.functions import pandas_udf
@pandas_udf('int')
def count_col(col: pd.Series) -> pd.Series:
return pd.Series(col.apply(lambda x: len(x)))
df.withColumn('ncolumns', count_col(df['Image'])).select('ncolumns').distinct().show()
# + [markdown] slideshow={"slide_type": "subslide"}
# **Exercise:** Using [photutils](https://photutils.readthedocs.io/en/stable/index.html) (or whatever you prefer), extract the position of sources in the images whose peak value is 50-sigma above the background.
# + slideshow={"slide_type": "subslide"}
from astropy.stats import sigma_clipped_stats
from astropy.visualization import SqrtStretch
from astropy.visualization.mpl_normalize import ImageNormalize
import numpy as np
import pandas as pd
from photutils import DAOStarFinder
from photutils import CircularAperture
from pyspark.sql.types import FloatType, ArrayType
from pyspark.sql.functions import pandas_udf, PandasUDFType
from typing import Iterator, Generator
def get_stat(data, sigma=3.0, iters=3):
""" Estimate the background and background noise using
sigma-clipped statistics.
Parameters
----------
data : 2D array
2d array containing the data.
sigma : float
sigma.
iters : int
Number of iteration to perform to get accurate estimate.
The higher the better, but it will be longer.
"""
mean, median, std = sigma_clipped_stats(data, sigma=sigma, maxiters=iters)
return mean, median, std
def extract_catalog(pdf: pd.DataFrame) -> pd.DataFrame:
""" Use photutils to extract source information
from image (one image per partition).
"""
# Reshape images for photutils
image = np.array(
[np.array(j, dtype=float) for j in pdf['Image'].values],
dtype=float
)
# Get background statistics
mean, median, std = get_stat(image)
# Use star finder
sf = DAOStarFinder(fwhm=10.0, threshold=50.*std)
cat = sf(image - median)
pdf_to_return = cat.to_pandas()
pdf_to_return['ImgIndex'] = pdf['ImgIndex'].values[0]
return pdf_to_return
# + slideshow={"slide_type": "subslide"}
from pyspark.sql.types import StructField, StructType, IntegerType, FloatType, StringType
# Define the output schema (catalog from photutils)
schema = StructType(
[
StructField('id', IntegerType(), True),
StructField('xcentroid', FloatType(), True),
StructField('ycentroid', FloatType(), True),
StructField('sharpness', FloatType(), True),
StructField('roundness1', FloatType(), True),
StructField('roundness2', FloatType(), True),
StructField('npix', IntegerType(), True),
StructField('sky', FloatType(), True),
StructField('peak', FloatType(), True),
StructField('flux', FloatType(), True),
StructField('mag', FloatType(), True),
StructField('ImgIndex', StringType(), True)
]
)
catalog = df.groupBy('ImgIndex').applyInPandas(extract_catalog, schema=schema)
# + slideshow={"slide_type": "subslide"}
catalog.show()
# + [markdown] slideshow={"slide_type": "slide"}
# ## Plotting results
# + slideshow={"slide_type": "subslide"}
# retrieve image indices
indices = df.select('ImgIndex').distinct().collect()[0]
# retrieve the first image to the driver
first_image = df.filter(df['ImgIndex'] == indices[0]).select('Image').collect()
# retrieve the first catalog of sources to the driver
first_catalog = catalog.filter(catalog['ImgIndex'] == indices[0]).toPandas()
# + slideshow={"slide_type": "subslide"}
# Overplot detections on the images
from astropy.visualization import AsinhStretch
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_context('talk')
data = np.transpose(first_image).reshape((2112, 4644))
# only for visualisation purposes!
data = np.log(data)
norm = ImageNormalize(stretch=AsinhStretch())
fig = plt.figure(0, (15, 15))
plt.imshow(data, cmap='binary', origin="lower", norm=norm)
plt.show()
fig = plt.figure(0, (15, 15))
positions = [[y, x] for y, x in zip(first_catalog['ycentroid'].values, first_catalog['xcentroid'].values)]
apertures = CircularAperture(positions, r=30.)
plt.imshow(data, cmap='binary', origin="lower", norm=norm)
apertures.plot(color='blue', lw=1.0, alpha=0.5);
# -
| spark/notebooks/mini-projects-images.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # <img style="float: left; padding-right: 10px; width: 45px" src="https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png"> CS109B Data Science 2: Advanced Topics in Data Science
# ## Lab 2 - Smoothers and Generalized Additive Models - Model Fitting
#
# <div class="discussion"><b>Spring 2020</b></div>
#
# **Harvard University**<br>
# **Spring 2020**<br>
# **Instructors:** <NAME>, <NAME>, and <NAME><br>
# **Lab Instructors:** <NAME> and <NAME><br>
# **Content:** <NAME> and <NAME>
#
# ---
## RUN THIS CELL TO PROPERLY HIGHLIGHT THE EXERCISES
import requests
from IPython.core.display import HTML
styles = requests.get("https://raw.githubusercontent.com/Harvard-IACS/2019-CS109B/master/content/styles/cs109.css").text
HTML(styles)
# +
import numpy as np
from scipy.interpolate import interp1d
import matplotlib.pyplot as plt
import pandas as pd
# %matplotlib inline
# -
# ## Learning Goals
#
# By the end of this lab, you should be able to:
# * Understand how to implement GAMs with the Python package `pyGAM`
# * Learn about the practical aspects of Splines and how to use them.
#
# **This lab corresponds to lectures 1, 2, and 3 and maps to homework 1.**
# ## Table of Contents
#
# * 1 - Overview - A Top View of LMs, GLMs, and GAMs to set the stage
# * 2 - A review of Linear Regression with `statsmodels`. What are those weird formulas?
# * 3 - Splines
# * 4 - Generative Additive Models with pyGAM
# * 5 - Smooting Splines using pyGAM
# ## Overview
#
# Linear Models (LM), Generalized Linear Models (GLMs), Generalized Additive Models (GAMs), Splines, Natural Splines, Smoothing Splines! So many definitions. Let's try and work through an example for each of them so we can better understand them.
#
# 
# *image source: <NAME> (one of the developers of pyGAM)*
# ### A - Linear Models
#
# First we have the **Linear Models** which you know from 109a. These models are linear in the coefficients. Very *interpretable* but suffer from high bias because let's face it, few relationships in life are linear. Simple Linear Regression (defined as a model with one predictor) as well as Multiple Linear Regression (more than one predictors) are examples of LMs. Polynomial Regression extends the linear model by adding terms that are still linear for the coefficients but non-linear when it somes to the predictiors which are now raised in a power or multiplied between them.
#
# 
#
# $$
# \begin{aligned}
# y = \beta{_0} + \beta{_1}{x_1} & \mbox{(simple linear regression)}\\
# y = \beta{_0} + \beta{_1}{x_1} + \beta{_2}{x_2} + \beta{_3}{x_3} & \mbox{(multiple linear regression)}\\
# y = \beta{_0} + \beta{_1}{x_1} + \beta{_2}{x_1^2} + \beta{_3}{x_3^3} & \mbox{(polynomial regression)}\\
# \end{aligned}
# $$
# <div class="discussion"><b>Discussion</b></div>
#
# - What does it mean for a model to be **interpretable**?
# - Are linear regression models interpretable? Are random forests? What about Neural Networks such as FFNs and CNNs?
# - Do we always want interpretability? Describe cases where we do and cases where we do not care.
# - interpretable: easily understand how each predictors affect the response variable
# - linear models are more interpretable than NN
# - It depends on the context. We don't want interpretability when users don't care about how the model works.
# ### B - Generalized Linear Models (GLMs)
#
# 
#
# $$
# \begin{aligned}
# y = \beta{_0} + \beta{_1}{x_1} + \beta{_2}{x_2} + \beta{_3}{x_3}
# \end{aligned}
# $$
#
#
# **Generalized Linear Models** is a term coined in the early 1970s by Nelder and Wedderburn for a class of models that includes both Linear Regression and Logistic Regression. A GLM fits one coefficient per feature (predictor).
# ### C - Generalized Additive Models (GAMs)
#
# Hastie and Tidshirani coined the term **Generalized Additive Models** in 1986 for a class of non-linear extensions to Generalized Linear Models.
#
# 
#
# $$
# \begin{aligned}
# y = \beta{_0} + f_1\left(x_1\right) + f_2\left(x_2\right) + f_3\left(x_3\right) \\
# y = \beta{_0} + f_1\left(x_1\right) + f_2\left(x_2, x_3\right) + f_3\left(x_3\right) & \mbox{(with interaction terms)}
# \end{aligned}
# $$
#
# In practice we add splines and regularization via smoothing penalties to our GLMs. Decision Trees also fit in this category.
#
# *image source: <NAME>*
# ### D - Basis Functions
#
# In our models we can use various types of functions as "basis".
# - Monomials such as $x^2$, $x^4$ (**Polynomial Regression**)
# - Sigmoid functions (neural networks)
# - Fourier functions
# - Wavelets
# - **Regression splines** which we will look at shortly.
# <div class="discussion"><b>Discussion</b></div>
#
# - Where does polynomial regression fit in all this? Linear model: linear with respect to beta coefficients here
# Answer: GLMs include Polynomial Regression so the graphic above should really include curved lines, not just straight...
# ## Implementation
#
# ### 1 - Linear/Polynomial Regression
#
# We will use the `diabetes` dataset.
#
# Variables are:
# - subject: subject ID number
# - age: age diagnosed with diabetes
# - acidity: a measure of acidity called base deficit
# Response:
# - y: natural log of serum C-peptide concentration
#
# *Original source is Sockett et al. (1987) mentioned in Hastie and Tibshirani's book
# "Generalized Additive Models".*
#
#
#
# Reading data and (some) exploring in Pandas:
diab = pd.read_csv("../data/diabetes.csv")
diab.head()
diab.dtypes
diab.describe()
# Plotting with matplotlib:
ax0 = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data") #plotting direclty from pandas!
ax0.set_xlabel("Age at Diagnosis")
ax0.set_ylabel("Log C-Peptide Concentration");
# ### Linear/Polynomial regression with statsmodels.
#
# As you remember from 109a, we have two tools for Linear Regression:
# - `statsmodels` [https://www.statsmodels.org/stable/regression.html](https://www.statsmodels.org/stable/regression.html), and
# - `sklearn`[https://scikit-learn.org/stable/index.html](https://scikit-learn.org/stable/index.html)
#
# Previously, we worked from a vector of target values and a design matrix we built ourself (e.g. using `sklearn`'s PolynomialFeatures). `statsmodels` allows users to fit statistical models using R-style **formulas**. They build the target value and design matrix for you.
#
# ```
# # our target variable is 'Lottery', while 'Region' is a categorical predictor
# df = dta.data[['Lottery', 'Literacy', 'Wealth', 'Region']]
#
# formula='Lottery ~ Literacy + Wealth + C(Region) + Literacy * Wealth'
# ```
#
# For more on these formulas see:
#
# - https://www.statsmodels.org/stable/examples/notebooks/generated/formulas.html
# - https://patsy.readthedocs.io/en/latest/overview.html
# +
import statsmodels.formula.api as sm
model1 = sm.ols('y ~ age',data=diab)
fit1_lm = model1.fit()
# -
# Let's build a dataframe to predict values on (sometimes this is just the test or validation set). Very useful for making pretty plots of the model predictions - predict for TONS of values, not just whatever's in the training set.
# +
x_pred = np.linspace(0,16,100)
predict_df = pd.DataFrame(data={"age":x_pred})
predict_df.head()
# -
# Use `get_prediction(<data>).summary_frame()` to get the model's prediction (and error bars!)
prediction_output = fit1_lm.get_prediction(predict_df).summary_frame()
prediction_output.head()
# Plot the model and error bars
# +
ax1 = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data with least-squares linear fit")
ax1.set_xlabel("Age at Diagnosis")
ax1.set_ylabel("Log C-Peptide Concentration")
ax1.plot(predict_df.age, prediction_output['mean'],color="green")
ax1.plot(predict_df.age, prediction_output['mean_ci_lower'], color="blue",linestyle="dashed")
ax1.plot(predict_df.age, prediction_output['mean_ci_upper'], color="blue",linestyle="dashed");
# -
# <div class="exercise"><b>Exercise 1</b></div>
#
# - Fit a 3rd degree polynomial model and
# - plot the model+error bars.
#
# You can either take
# - **Route1**: Build a design df with a column for each of `age`, `age**2`, `age**3`, or
# - **Route2**: Just edit the formula
# your answer here
poly_model = sm.ols('y ~ age + I(age**2) + I(age**3)',data=diab).fit()
# +
# # %load ../solutions/exercise1-1.py
fit2_lm = sm.ols(formula="y ~ age + np.power(age, 2) + np.power(age, 3)",data=diab).fit()
poly_predictions = fit2_lm.get_prediction(predict_df).summary_frame()
poly_predictions.head()
# +
# # %load ../solutions/exercise1-2.py
ax2 = diab.plot.scatter(x='age',y='y',c='Red',title="Diabetes data with least-squares cubic fit")
ax2.set_xlabel("Age at Diagnosis")
ax2.set_ylabel("Log C-Peptide Concentration")
ax2.plot(predict_df.age, poly_predictions['mean'],color="green")
ax2.plot(predict_df.age, poly_predictions['mean_ci_lower'], color="blue",linestyle="dashed")
ax2.plot(predict_df.age, poly_predictions['mean_ci_upper'], color="blue",linestyle="dashed");
# -
# <div class="discussion"><b>Ed exercise</b></div>
#
# This example was similar with the Ed exercise. [Open it in Ed](https://us.edstem.org/courses/172/lessons/656/slides/2916) and let's go though it.
# ### 2 - Piecewise Polynomials a.k.a. Splines
#
# Splines are a type of piecewise polynomial interpolant. A spline of degree k is a piecewise polynomial that is continuously differentiable k − 1 times.
#
# Splines are the basis of CAD software and vector graphics including a lot of the fonts used in your computer. The name “spline” comes from a tool used by ship designers to draw smooth curves. Here is the letter $epsilon$ written with splines:
#
# 
#
# *font idea inspired by <NAME> (AM205)*
#
# If the degree is 1 then we have a Linear Spline. If it is 3 then we have a Cubic spline. It turns out that cubic splines because they have a continous 2nd derivative at the knots are very smoothly looking to the eye. We do not need higher order than that. The Cubic Splines are usually Natural Cubic Splines which means they have the added constrain of the end points' second derivative = 0.
#
# We will use the CubicSpline and the B-Spline as well as the Linear Spline.
#
# #### scipy.interpolate
#
# See all the different splines that scipy.interpolate has to offer: https://docs.scipy.org/doc/scipy/reference/interpolate.html
#
# Let's use the simplest form which is interpolate on a set of points and then find the points between them.
# +
from scipy.interpolate import splrep, splev
from scipy.interpolate import BSpline, CubicSpline
from scipy.interpolate import interp1d
# define the range of the function
a = -1
b = 1
# define the number of knots
num_knots = 10
x = np.linspace(a,b,num_knots)
# define the function we want to approximate
y = 1/(1+25*(x**2))
# make a linear spline
linspline = interp1d(x, y)
# sample at these points to plot
xx = np.linspace(a,b,1000)
yy = 1/(1+25*(xx**2))
plt.plot(x,y,'*')
plt.plot(xx, yy, label='true function')
plt.plot(xx, linspline(xx), label='linear spline');
plt.legend();
# -
# <div class="exercise"><b>Exercise 2</b></div>
#
# The Linear interpolation does not look very good. Fit a Cubic Spline and plot along the Linear to compare.
# +
# your answer here
cub_spline = CubicSpline(x, y)
plt.plot(x,y,'*')
plt.plot(xx, yy, label='true function')
plt.plot(xx, linspline(xx), label='linear spline');
plt.plot(xx, cub_spline(xx), label='cubic spline');
plt.legend();
# +
# # %load ../solutions/exercise2.py
# define the range of the function
a = -1
b = 1
# define the knots
num_knots = 10
x = np.linspace(a,b,num_knots)
# define the function we want to approximate
y = 1/(1+25*(x**2))
# make the Cubic spline
cubspline = CubicSpline(x, y)
# OR make a linear spline
linspline = interp1d(x, y)
# plot
xx = np.linspace(a,b,1000)
yy = 1/(1+25*(xx**2))
plt.plot(xx, yy, label='true function')
plt.plot(x,y,'*')
plt.plot(xx, linspline(xx), label='linear');
plt.plot(xx, cubspline(xx), label='cubic');
plt.legend();
# -
# <div class="discussion"><b>Discussion</b></div>
#
# - Change the number of knots to 100 and see what happens. What would happen if we run a polynomial model of degree equal to the number of knots (a global one as in polynomial regression, not a spline)?
# - What makes a spline 'Natural'?
# change num_knots to 100 will make the interpolation curve fit much better.
# #### B-Splines
#
# A B-splines (Basis Splines) is defined by a set of **control points** and a set of **basis functions** that intepolate (fit) the function between these points. By choosing to have no smoothing factor we forces the final B-spline to pass though all the points. If, on the other hand, we set a smothing factor, our function is more of an approximation with the control points as "guidance". The latter produced a smoother curve which is prefferable for drawing software. For more on Splines see: https://en.wikipedia.org/wiki/B-spline)
#
# 
#
# We will use [`scipy.splrep`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.interpolate.splrep.html#scipy.interpolate.splrep) to calulate the coefficients for the B-Spline and draw it.
# #### B-Spline with no smooting
# +
from scipy.interpolate import splev, splrep
x = np.linspace(0, 10, 10)
y = np.sin(x)
t,c,k = splrep(x, y) # (tck) is a tuple containing the vector of knots, coefficients, degree of the spline
print(t,c,k)
# define the points to plot on (x2)
x2 = np.linspace(0, 10, 200)
y2 = BSpline(t, c, k)
plt.plot(x, y, 'o', x2, y2(x2))
plt.show()
# -
# #### B-Spline with smooting factor s
# +
from scipy.interpolate import splev, splrep
x = np.linspace(0, 10, 10)
y = np.sin(x)
s = 0.5 # add smooting factor
task = 0 # task needs to be set to 0, which represents:
# we are specifying a smoothing factor and thus only want
# splrep() to find the optimal t and c
t,c,k = splrep(x, y, task=task, s=s)
# define the points to plot on (x2)
x2 = np.linspace(0, 10, 200)
y2 = BSpline(t, c, k)
plt.plot(x, y, 'o', x2, y2(x2))
plt.show()
# -
# #### B-Spline with given knots
x = np.linspace(0, 10, 100)
y = np.sin(x)
knots = np.quantile(x, [0.25, 0.5, 0.75])
print(knots)
# calculate the B-Spline
t,c,k = splrep(x, y, t=knots)
curve = BSpline(t,c,k)
curve
plt.scatter(x=x,y=y,c='grey', alpha=0.4)
yknots = np.sin(knots)
plt.scatter(knots, yknots, c='r')
plt.plot(x,curve(x))
plt.show()
# <div class="discussion"><b>Ed exercise</b></div>
#
# This example was similar with the Ed exercise. [Open it in Ed](https://us.edstem.org/courses/172/lessons/656/slides/2917) and let's go though it.
# ### 3 - GAMs
#
# https://readthedocs.org/projects/pygam/downloads/pdf/latest/
#
# #### A - Classification in `pyGAM`
#
# Let's get our (multivariate!) data, the `kyphosis` dataset, and the `LogisticGAM` model from `pyGAM` to do binary classification.
#
# - kyphosis - wherther a particular deformation was present post-operation
# - age - patient's age in months
# - number - the number of vertebrae involved in the operation
# - start - the number of the topmost vertebrae operated on
# +
kyphosis = pd.read_csv("../data/kyphosis.csv")
display(kyphosis.head())
display(kyphosis.describe(include='all'))
display(kyphosis.dtypes)
# -
# convert the outcome in a binary form, 1 or 0
kyphosis = pd.read_csv("../data/kyphosis.csv")
kyphosis["outcome"] = 1*(kyphosis["Kyphosis"] == "present")
kyphosis.describe()
# +
from pygam import LogisticGAM, s, f, l
X = kyphosis[["Age","Number","Start"]]
y = kyphosis["outcome"]
kyph_gam = LogisticGAM().fit(X,y)
# -
# #### Outcome dependence on features
#
# To help us see how the outcome depends on each feature, `pyGAM` has the `partial_dependence()` function.
# ```
# pdep, confi = kyph_gam.partial_dependence(term=i, X=XX, width=0.95)
# ```
# For more on this see the : https://pygam.readthedocs.io/en/latest/api/logisticgam.html
#
res = kyph_gam.deviance_residuals(X,y)
for i, term in enumerate(kyph_gam.terms):
if term.isintercept:
continue
XX = kyph_gam.generate_X_grid(term=i)
pdep, confi = kyph_gam.partial_dependence(term=i, X=XX, width=0.95)
pdep2, _ = kyph_gam.partial_dependence(term=i, X=X, width=0.95)
plt.figure()
plt.scatter(X.iloc[:,term.feature], pdep2 + res)
plt.plot(XX[:, term.feature], pdep)
plt.plot(XX[:, term.feature], confi, c='r', ls='--')
plt.title(X.columns.values[term.feature])
plt.show()
# Notice that we did not specify the basis functions in the .fit(). Cool. `pyGAM` figures them out for us by using $s()$ (splines) for numerical variables and $f()$ for categorical features. If this is not what we want we can manually specify the basis functions, as follows:
kyph_gam = LogisticGAM(s(0)+s(1)+s(2)).fit(X,y)
res = kyph_gam.deviance_residuals(X,y)
for i, term in enumerate(kyph_gam.terms):
if term.isintercept:
continue
XX = kyph_gam.generate_X_grid(term=i)
pdep, confi = kyph_gam.partial_dependence(term=i, X=XX, width=0.95)
pdep2, _ = kyph_gam.partial_dependence(term=i, X=X, width=0.95)
plt.figure()
plt.scatter(X.iloc[:,term.feature], pdep2 + res)
plt.plot(XX[:, term.feature], pdep)
plt.plot(XX[:, term.feature], confi, c='r', ls='--')
plt.title(X.columns.values[term.feature])
plt.show()
# #### B - Regression in `pyGAM`
#
# For regression problems, we can use a `linearGAM` model. For this part we will use the `wages` dataset.
#
# https://pygam.readthedocs.io/en/latest/api/lineargam.html
# #### The `wages` dataset
#
# Let's inspect another dataset that is included in `pyGAM` that notes the wages of people based on their age, year of employment and education.
# +
# from the pyGAM documentation
from pygam import LinearGAM, s, f
from pygam.datasets import wage
X, y = wage(return_X_y=True)
## model
gam = LinearGAM(s(0) + s(1) + f(2))
gam.gridsearch(X, y)
## plotting
plt.figure();
fig, axs = plt.subplots(1,3);
titles = ['year', 'age', 'education']
for i, ax in enumerate(axs):
XX = gam.generate_X_grid(term=i)
ax.plot(XX[:, i], gam.partial_dependence(term=i, X=XX))
ax.plot(XX[:, i], gam.partial_dependence(term=i, X=XX, width=.95)[1], c='r', ls='--')
if i == 0:
ax.set_ylim(-30,30)
ax.set_title(titles[i]);
# -
# <div class="discussion"><b>Discussion</b></div>
#
# What are your observations from the plots above?
# ### 4 - Smoothing Splines using pyGAM
#
# For clarity: this is the fancy spline model that minimizes $MSE - \lambda\cdot\text{wiggle penalty}$ $=$ $\sum_{i=1}^N \left(y_i - f(x_i)\right)^2 - \lambda \int \left(f''(x)\right)^2$, across all possible functions $f$. The winner will always be a continuous, cubic polynomial with a knot at each data point.
# Let's see how this smoothing works in `pyGAM`. We start by creating some arbitrary data and fitting them with a GAM.
# +
X = np.linspace(0,10,500)
y = np.sin(X*2*np.pi)*X + np.random.randn(len(X))
plt.scatter(X,y);
# -
# let's try a large lambda first and lots of splines
gam = LinearGAM(lam=1e6, n_splines=50). fit(X,y)
XX = gam.generate_X_grid(term=0)
plt.scatter(X,y,alpha=0.3);
plt.plot(XX, gam.predict(XX));
# We see that the large $\lambda$ forces a straight line, no flexibility. Let's see now what happens if we make it smaller.
# let's try a smaller lambda
gam = LinearGAM(lam=1e2, n_splines=50). fit(X,y)
XX = gam.generate_X_grid(term=0)
plt.scatter(X,y,alpha=0.3);
plt.plot(XX, gam.predict(XX));
# There is some curvature there but still not a good fit. Let's try no penalty. That should have the line fit exactly.
# no penalty, let's try a 0 lambda
gam = LinearGAM(lam=0, n_splines=50). fit(X,y)
XX = gam.generate_X_grid(term=0)
plt.scatter(X,y,alpha=0.3)
plt.plot(XX, gam.predict(XX))
# Yes, that is good. Now let's see what happens if we lessen the number of splines. The fit should not be as good.
# no penalty, let's try a 0 lambda
gam = LinearGAM(lam=0, n_splines=10). fit(X,y)
XX = gam.generate_X_grid(term=0)
plt.scatter(X,y,alpha=0.3);
plt.plot(XX, gam.predict(XX));
# Indeed.
| content/labs/lab2 smoothing/cs109b_lab2_smooths_and_GAMs.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] nbgrader={}
# # Interact Exercise 01
# + [markdown] nbgrader={}
# ## Import
# + nbgrader={}
# %matplotlib inline
from matplotlib import pyplot as plt
import numpy as np
# + nbgrader={}
from IPython.html.widgets import interact, interactive, fixed
from IPython.display import display
# + [markdown] nbgrader={}
# ## Interact basics
# + [markdown] nbgrader={}
# Write a `print_sum` function that `prints` the sum of its arguments `a` and `b`.
# + nbgrader={"checksum": "4d7fa34d285413499aa7359dda2a2dcc", "solution": true}
def print_sum(a, b):
return (a+b)
# + [markdown] nbgrader={}
# Use the `interact` function to interact with the `print_sum` function.
#
# * `a` should be a floating point slider over the interval `[-10., 10.]` with step sizes of `0.1`
# * `b` should be an integer slider the interval [-8, 8] with step sizes of `2`.
# + deletable=false nbgrader={"checksum": "6cff4e8e53b15273846c3aecaea84a3d", "solution": true}
w = interactive(print_sum, a = (-10.0,10.0,0.1), b = (-8, 8, 2))
# -
display(w)
w.result
# + deletable=false nbgrader={"checksum": "42c776e2480b70e6a45ee325285f2977", "grade": true, "grade_id": "interactex01a", "points": 5}
assert True # leave this for grading the print_sum exercise
# + [markdown] nbgrader={}
# Write a function named `print_string` that prints a string and additionally prints the length of that string if a boolean parameter is `True`.
# + nbgrader={"checksum": "0a454725f1214af3f65e36c5bc4123e9", "solution": true}
def print_string(s, length=False):
print (s)
if length == True:
print(len(s))
# + [markdown] nbgrader={}
# Use the `interact` function to interact with the `print_string` function.
#
# * `s` should be a textbox with the initial value `"Hello World!"`.
# * `length` should be a checkbox with an initial value of `True`.
# + deletable=false nbgrader={"checksum": "6cff4e8e53b15273846c3aecaea84a3d", "solution": true}
w = interactive(print_string, s = "Hello, World!")
# -
w
# + deletable=false nbgrader={"checksum": "414350009853ea9cb00917ef3bec7b10", "grade": true, "grade_id": "interactex01b", "points": 5}
assert True # leave this for grading the print_string exercise
# -
| assignments/assignment05/InteractEx01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python (pytorch)
# language: python
# name: pytorch
# ---
# + [markdown] code_folding=[1] slideshow={"slide_type": "slide"}
# # Lecture 3: Matvecs and matmuls, memory hierarchy, Strassen algorithm
# + [markdown] slideshow={"slide_type": "slide"}
# ## Recap of the previous lectures
#
# - Floating point arithmetics and related issues
# - Stable algorithms: backward and forward stability
# - Most important matrix norms: spectral and Frobenius
# - Unitary matrices preserve these norms
# - There are two "basic" classes of unitary matrices: Householder and Givens matrices
# + [markdown] slideshow={"slide_type": "slide"}
# ## Examples of peak performance
#
# **Flops** –– floating point operations per second.
#
# Giga = $2^{30} \approx 10^9$,
# Tera = $2^{40} \approx 10^{12}$,
# Peta = $2^{50} \approx 10^{15}$,
# Exa = $2^{60} \approx 10^{18}$
#
# What is the **peak perfomance** of:
#
# 1. Modern CPU
# 2. Modern GPU
# 3. Largest supercomputer of the world?
# + [markdown] slideshow={"slide_type": "slide"}
# ### Clock frequency of CPU vs. performance in flops
#
# FLOPS = sockets * (cores per socket) * (number of clock cycles per second) * (number of floating point operations per cycle).
#
# - Typically sockets = 1
# - Number of cores is typically 2 or 4
# - Number of ticks per second is familiar clock frequency
# - Number of floating point operations per tick depends on the particular CPU
# + [markdown] slideshow={"slide_type": "slide"}
#
# 1. Modern CPU (Intel Core i7) –– 400 Gflops
# 2. Modern GPU (Nvidia Quadro RTX 8000) –– 16.3 Tflops single precision
# 3. [Largest supercomputer in the world](https://www.top500.org/lists/2019/06/) –– 513.85 Pflops –– peak performanse
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# ## Matrix-by-vector multiplication (matvec)
#
# Multiplication of an $n\times n$ matrix $A$ by a vector $x$ of size $n\times 1$ ($y=Ax$):
#
# $$
# y_{i} = \sum_{i=1}^n a_{ij} x_j
# $$
#
# requires $n^2$ mutliplications and $n(n-1)$ additions. Thus, the overall complexity is $2n^2 - n =$ <font color='red'> $\mathcal{O}(n^2)$ </font>
# + [markdown] slideshow={"slide_type": "slide"}
# ## How bad is $\mathcal{O}(n^2)$?
#
# - Let $A$ be the matrix of pairwise gravitational interaction between planets in a galaxy.
#
# - The number of planets in an average galaxy is $10^{11}$, so the size of this matrix is $10^{11} \times 10^{11}$.
#
# - To model evolution in time we have to multiply this matrix by vector at each time step.
#
# - Top supercomputers do around $10^{16}$ floating point operations per second (flops), so the time required to multiply the matrix $A$ by a vector is approximately
#
# \begin{align*}
# \frac{(10^{11})^2 \text{ operations}}{10^{16} \text{ flops}} = 10^6 \text{ sec} \approx 11.5 \text{ days}
# \end{align*}
#
# for one time step. If we could multiply it with $\mathcal{O}(n)$ complexity, we would get
#
# \begin{align*}
# \frac{10^{11} \text{ operations}}{10^{16} \text{ flops}} = 10^{-5} \text{ sec}.
# \end{align*}
#
# Here is the YouTube video that illustrates collision of two galaxisies which was modelled by $\mathcal{O}(n \log n)$ algorithm:
# + slideshow={"slide_type": "slide"}
from IPython.display import YouTubeVideo
YouTubeVideo("7HF5Oy8IMoM")
# + [markdown] slideshow={"slide_type": "slide"}
# ## Can we beat $\mathcal{O}(n^2)$?
#
# - Generally speaking **NO**.
# - The point is that we have $\mathcal{O}(n^2)$ input data, so there is no way to be faster for a general matrix.
# - Fortunately, we can be faster <font color='red'>for certain types of matrices</font>.
# Here are some examples:
#
# * The simplest example may be a matrix of all ones, which can be easily multiplied with only $n-1$ additions. This matrix is of rank one. More generally we can multiply fast by <font color='red'>low-rank </font> matrices (or by matrices that have low-rank blocks)
#
# * <font color='red'>Sparse</font> matrices (contain $\mathcal{O}(n)$ nonzero elements)
#
# * <font color='red'>Structured</font> matrices:
# * Fourier
# * Circulant
# * Toeplitz
# * Hankel
# + [markdown] slideshow={"slide_type": "slide"}
# ## Matrix-by-matrix product
#
# Consider composition of two linear operators:
#
# 1. $y = Bx$
# 2. $z = Ay$
#
# Then, $z = Ay = A B x = C x$, where $C$ is the **matrix-by-matrix product**.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Matrix-by-matrix product (MM): classics
#
# **Definition**. A product of an $n \times k$ matrix $A$ and a $k \times m$ matrix $B$ is a $n \times m$ matrix $C$ with the elements
# $$
# c_{ij} = \sum_{s=1}^k a_{is} b_{sj}, \quad i = 1, \ldots, n, \quad j = 1, \ldots, m
# $$
#
# For $m=k=n$ complexity of a naïve algorithm is $2n^3 - n^2 =$ <font color='red'>$\mathcal{O}(n^3)$</font>.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Discussion of MM
#
# - Matrix-by-matrix product is the **core** for almost all efficient algorithms in numerical linear algebra.
#
# - Basically, all the dense NLA algorithms are reduced to a sequence of matrix-by-matrix products.
#
# - Efficient implementation of MM reduces the complexity of numerical algorithms by the same factor.
#
# - However, implementing MM is not easy at all!
# + [markdown] slideshow={"slide_type": "slide"}
# ## Efficient implementation for MM
# **Q1**: Is it easy to multiply a matrix by a matrix in the most efficient way?
# + [markdown] slideshow={"slide_type": "slide"}
# ## Answer: no, it is not easy
#
# If you want it as fast as possible, using the computers that are at hand.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Demo
# Let us do a short demo and compare a `np.dot()` procedure which in my case uses MKL with a hand-written matrix-by-matrix routine in Python and also its numba version.
# + code_folding=[] slideshow={"slide_type": "slide"}
import numpy as np
def matmul(a, b):
n = a.shape[0]
k = a.shape[1]
m = b.shape[1]
c = np.zeros((n, m))
for i in range(n):
for j in range(m):
for s in range(k):
c[i, j] += a[i, s] * b[s, j]
return c
# + slideshow={"slide_type": "slide"}
import numpy as np
from numba import jit # Just-in-time compiler for Python, see http://numba.pydata.org
@jit(nopython=True)
def numba_matmul(a, b):
n = a.shape[0]
k = a.shape[1]
m = b.shape[1]
c = np.zeros((n, m))
for i in range(n):
for j in range(m):
for s in range(k):
c[i, j] += a[i, s] * b[s, j]
return c
# + [markdown] slideshow={"slide_type": "slide"}
# Then we just compare computational times.
#
# Guess the answer.
# + slideshow={"slide_type": "slide"}
import jax.numpy as jnp
from jax.config import config
config.update("jax_enable_x64", True)
n = 100
a = np.random.randn(n, n)
b = np.random.randn(n, n)
a_jax = jnp.array(a)
b_jax = jnp.array(b)
# %timeit matmul(a, b)
# %timeit numba_matmul(a, b)
# %timeit a @ b
# %timeit (a_jax @ b_jax).block_until_ready()
# + [markdown] slideshow={"slide_type": "slide"}
# Is this answer correct for any dimensions of matrices?
# + slideshow={"slide_type": "slide"}
import matplotlib.pyplot as plt
# %matplotlib inline
dim_range = [10*i for i in range(1, 11)]
time_range_matmul = []
time_range_numba_matmul = []
time_range_np = []
for n in dim_range:
print("Dimension = {}".format(n))
a = np.random.randn(n, n)
b = np.random.randn(n, n)
# t = %timeit -o -q matmul(a, b)
time_range_matmul.append(t.best)
# t = %timeit -o -q numba_matmul(a, b)
time_range_numba_matmul.append(t.best)
# t = %timeit -o -q np.dot(a, b)
time_range_np.append(t.best)
# + slideshow={"slide_type": "slide"}
plt.plot(dim_range, time_range_matmul, label="Matmul")
plt.plot(dim_range, time_range_numba_matmul, label="Matmul Numba")
plt.plot(dim_range, time_range_np, label="Numpy")
plt.legend(fontsize=18)
plt.xlabel("Dimension", fontsize=18)
plt.ylabel("Time", fontsize=18)
plt.yscale("log")
# + [markdown] slideshow={"slide_type": "slide"}
# ## Why is naïve implementation slow?
# It is slow due to two issues:
#
# - It does not use the benefits of fast memory (cache) and in general memory architecture
# - It does not use available parallelization ability (especially important for GPU)
# + [markdown] slideshow={"slide_type": "slide"}
# ## Memory architecture
# <img width=80% src="Memory-Hierarchy.jpg">
#
# - Fast memory is small
# - Bigger memory is slow
# + [markdown] slideshow={"slide_type": "slide"}
# ## Making algorithms more computationally intensive
#
# <font color='red'>**Implementation in NLA**</font>: use block version of algorithms. <br>
#
# This approach is a core of **[BLAS (Basic Linear Algebra Subroutines)](http://www.netlib.org/blas/)**, written in Fortran many years ago, and still rules the computational world.
# + [markdown] slideshow={"slide_type": "slide"}
# Split the matrix into blocks! For illustration consider splitting in $2 \times 2$ block matrix:
#
# $$
# A = \begin{bmatrix}
# A_{11} & A_{12} \\
# A_{21} & A_{22}
# \end{bmatrix}, \quad B = \begin{bmatrix}
# B_{11} & B_{12} \\
# B_{21} & B_{22}
# \end{bmatrix}$$
#
# Then,
#
# $$AB = \begin{bmatrix}A_{11} B_{11} + A_{12} B_{21} & A_{11} B_{12} + A_{12} B_{22} \\
# A_{21} B_{11} + A_{22} B_{21} & A_{21} B_{12} + A_{22} B_{22}\end{bmatrix}.$$
# + [markdown] slideshow={"slide_type": "fragment"}
# If $A_{11}, B_{11}$ and their product fit into the cache memory (which is 12 Mb (L3) for the [recent Intel Chip](https://en.wikipedia.org/wiki/List_of_Intel_microprocessors#Desktop)), then we load them only once into the memory.
# + [markdown] slideshow={"slide_type": "slide"}
# ## BLAS
# BLAS has three levels:
# 1. BLAS-1, operations like $c = a + b$
# 2. BLAS-2, operations like matrix-by-vector product
# 3. BLAS-3, matrix-by-matrix product
#
# What is the principal differences between them?
# + [markdown] slideshow={"slide_type": "slide"}
# The main difference is the number of operations vs. the number of input data!
#
# 1. BLAS-1: $\mathcal{O}(n)$ data, $\mathcal{O}(n)$ operations
# 2. BLAS-2: $\mathcal{O}(n^2)$ data, $\mathcal{O}(n^2)$ operations
# 3. BLAS-3: $\mathcal{O}(n^2)$ data, $\mathcal{O}(n^3)$ operations
# + [markdown] slideshow={"slide_type": "slide"}
# ## Why BLAS is so important and actual?
#
# 1. The state-of-the-art implementation of the basic linear algebra operations
# 2. Provides standard names for operations in any new implementations (e.g. [ATLAS](https://www.netlib.org/atlas/), [OpenBLAS](https://www.openblas.net/), [MKL](https://software.intel.com/en-us/mkl)). You can call matrix-by-matrix multiplication function (GEMM), link your code with any BLAS implementation and it will work correctly
# 3. Formulate new algorithms in terms of BLAS operations
# 4. There are wrappers for the most popular languages
# + [markdown] slideshow={"slide_type": "slide"}
# ## Packages related to BLAS
#
# 1. [ATLAS](http://math-atlas.sourceforge.net) - Automatic Tuned Linear Algebra Software. It automatically adapts to a particular system architechture.
# 2. [LAPACK](http://www.netlib.org/lapack/) - Linear Algebra Package. It provides high-level linear algebra operations (e.g. matrix factorizations), which are based on calls of BLAS subroutines.
# 3. [Intel MKL](https://software.intel.com/en-us/intel-mkl) - Math Kernel Library. It provides re-implementation of BLAS and LAPACK, optimized for Intel processors. Available in Anaconda Python distribution:
# ```
# conda install mkl
# ```
# MATLAB uses Intel MKL by default.
#
# 4. OpenBLAS is an optimized BLAS library based on [GotoBLAS](https://en.wikipedia.org/wiki/GotoBLAS).
#
# 5. PyTorch [supports](https://pytorch.org/docs/stable/torch.html#blas-and-lapack-operations) some calls from BLAS and LAPACK
#
# 6. For GPU it was implemented special [cuBLAS](https://docs.nvidia.com/cuda/cublas/index.html).
#
#
# For comparison of OpenBLAS and Intel MKL, see this [review](https://software.intel.com/en-us/articles/performance-comparison-of-openblas-and-intel-math-kernel-library-in-r)
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# ## Faster algorithms for matrix multiplication
#
# Recall that matrix-matrix multiplication costs $\mathcal{O}(n^3)$ operations.
# However, storage is $\mathcal{O}(n^2)$.
#
# **Question:** is it possible to reduce number operations down to $\mathcal{O}(n^2)$?
# + [markdown] slideshow={"slide_type": "fragment"}
# **Answer**: a quest for $\mathcal{O}(n^2)$ matrix-by-matrix multiplication algorithm is not yet done.
# + [markdown] slideshow={"slide_type": "slide"}
# * Strassen gives $\mathcal{O}(n^{2.807\dots})$ –– sometimes used in practice
#
# * [Current world record](http://arxiv.org/pdf/1401.7714v1.pdf) $\mathcal{O}(n^{2.37\dots})$ –– big constant, not practical, based on [Coppersmith-Winograd_algorithm](https://en.wikipedia.org/wiki/Coppersmith%E2%80%93Winograd_algorithm).
# - It improved the previous record (Williams 2012) by $3\cdot 10^{-7}$
# - The papers still study multiplication of $3 \times 3$ matrices and interpret it from different sides ([Heule, et. al. 2019](https://arxiv.org/pdf/1905.10192.pdf))
#
# Consider Strassen in more details.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Naïve multiplication
#
# Let $A$ and $B$ be two $2\times 2$ matrices. Naïve multiplication $C = AB$
#
# $$
# \begin{bmatrix} c_{11} & c_{12} \\ c_{21} & c_{22} \end{bmatrix} =
# \begin{bmatrix} a_{11} & a_{12} \\ a_{21} & a_{22} \end{bmatrix}
# \begin{bmatrix} b_{11} & b_{12} \\ b_{21} & b_{22} \end{bmatrix} =
# \begin{bmatrix}
# a_{11}b_{11} + a_{12}b_{21} & a_{11}b_{21} + a_{12}b_{22} \\
# a_{21}b_{11} + a_{22}b_{21} & a_{21}b_{21} + a_{22}b_{22}
# \end{bmatrix}
# $$
#
# contains $8$ multiplications and $4$ additions.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Strassen algorithm
#
# In the work [Gaussian elimination is not optimal](http://link.springer.com/article/10.1007%2FBF02165411?LI=true) (1969) Strassen found that one can calculate $C$ using 18 additions and only 7 multiplications:
# $$
# \begin{split}
# c_{11} &= f_1 + f_4 - f_5 + f_7, \\
# c_{12} &= f_3 + f_5, \\
# c_{21} &= f_2 + f_4, \\
# c_{22} &= f_1 - f_2 + f_3 + f_6,
# \end{split}
# $$
# where
# $$
# \begin{split}
# f_1 &= (a_{11} + a_{22}) (b_{11} + b_{22}), \\
# f_2 &= (a_{21} + a_{22}) b_{11}, \\
# f_3 &= a_{11} (b_{12} - b_{22}), \\
# f_4 &= a_{22} (b_{21} - b_{11}), \\
# f_5 &= (a_{11} + a_{12}) b_{22}, \\
# f_6 &= (a_{21} - a_{11}) (b_{11} + b_{12}), \\
# f_7 &= (a_{12} - a_{22}) (b_{21} + b_{22}).
# \end{split}
# $$
#
# Fortunately, these formulas hold even if $a_{ij}$ and $b_{ij}$, $i,j=1,2$ are block matrices.
#
# Thus, Strassen algorithm looks as follows.
# - First of all we <font color='red'>split</font> matrices $A$ and $B$ of sizes $n\times n$, $n=2^d$ <font color='red'> into 4 blocks</font> of size $\frac{n}{2}\times \frac{n}{2}$
# - Then we <font color='red'>calculate multiplications</font> in the described formulas <font color='red'>recursively</font>
#
# This leads us again to the **divide and conquer** idea.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Complexity of the Strassen algorithm
#
# #### Number of multiplications
#
# Calculation of number of multiplications is a trivial task. Let us denote by $M(n)$ number of multiplications used to multiply 2 matrices of sizes $n\times n$ using the divide and conquer concept.
# Then for naïve algorithm we have number of multiplications
#
# $$ M_\text{naive}(n) = 8 M_\text{naive}\left(\frac{n}{2} \right) = 8^2 M_\text{naive}\left(\frac{n}{4} \right)
# = \dots = 8^{d-1} M(1) = 8^{d} = 8^{\log_2 n} = n^{\log_2 8} = n^3 $$
#
# So, even when using divide and coquer idea we can not be better than $n^3$.
#
# Let us calculate number of multiplications for the Strassen algorithm:
#
# $$ M_\text{strassen}(n) = 7 M_\text{strassen}\left(\frac{n}{2} \right) = 7^2 M_\text{strassen}\left(\frac{n}{4} \right)
# = \dots = 7^{d-1} M(1) = 7^{d} = 7^{\log_2 n} = n^{\log_2 7} $$
# + [markdown] slideshow={"slide_type": "slide"}
# #### Number of additions
#
# There is no point to estimate number of addtitions $A(n)$ for naive algorithm, as we already got $n^3$ multiplications.
# For the Strassen algorithm we have:
#
# $$ A_\text{strassen}(n) = 7 A_\text{strassen}\left( \frac{n}{2} \right) + 18 \left( \frac{n}{2} \right)^2 $$
#
# since on the first level we have to add $\frac{n}{2}\times \frac{n}{2}$ matrices 18 times and then go deeper for each of the 7 multiplications. Thus,
#
# <font size=2.0>
#
# $$
# \begin{split}
# A_\text{strassen}(n) =& 7 A_\text{strassen}\left( \frac{n}{2} \right) + 18 \left( \frac{n}{2} \right)^2 = 7 \left(7 A_\text{strassen}\left( \frac{n}{4} \right) + 18 \left( \frac{n}{4} \right)^2 \right) + 18 \left( \frac{n}{2} \right)^2 =
# 7^2 A_\text{strassen}\left( \frac{n}{4} \right) + 7\cdot 18 \left( \frac{n}{4} \right)^2 + 18 \left( \frac{n}{2} \right)^2 = \\
# =& \dots = 18 \sum_{k=1}^d 7^{k-1} \left( \frac{n}{2^k} \right)^2 = \frac{18}{4} n^2 \sum_{k=1}^d \left(\frac{7}{4} \right)^{k-1} = \frac{18}{4} n^2 \frac{\left(\frac{7}{4} \right)^d - 1}{\frac{7}{4} - 1} = 6 n^2 \left( \left(\frac{7}{4} \right)^d - 1\right) \leqslant 6 n^2 \left(\frac{7}{4} \right)^d = 6 n^{\log_2 7}
# \end{split}
# $$
# </font>
#
# (since $4^d = n^2$ and $7^d = n^{\log_2 7}$).
#
#
# Asymptotic behavior of $A(n)$ could be also found from the [master theorem](https://en.wikipedia.org/wiki/Master_theorem).
# + [markdown] slideshow={"slide_type": "slide"}
# #### Total complexity
#
# Total complexity is $M_\text{strassen}(n) + A_\text{strassen}(n)=$ <font color='red'>$7 n^{\log_2 7}$</font>. Strassen algorithm becomes faster
# when
#
# \begin{align*}
# 2n^3 &> 7 n^{\log_2 7}, \\
# n &> 667,
# \end{align*}
#
# so it is not a good idea to get to the bottom level of recursion.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Strassen algorithm reloaded
#
# - Recent paper [Strassen algorithm reloaded](http://jianyuhuang.com/papers/sc16.pdf)
# claim to **break conventional wisdom** that Strassen algorithm is not very practical.
# + [markdown] slideshow={"slide_type": "slide"}
# - Conventional wisdom: it is only
# practical for very large matrices. The proposed implementation is practical
# for small matrices.
# - Conventional wisdom: the matrices being
# multiplied should be relatively square. The proposed implementation is
# practical for rank-$k$ updates, where $k$ is relatively small (a shape
# of importance for libraries like LAPACK).
# - Conventional wisdom:
# it inherently requires substantial workspace. The proposed implementation
# requires no workspace beyond buffers already incorporated
# into conventional high-performance DGEMM implementations.
# - Conventional wisdom: a Strassen DGEMM interface must pass
# in workspace. The proposed implementation requires no such workspace
# and can be plug-compatible with the standard DGEMM interface.
# - Conventional wisdom: it is hard to demonstrate speedup
# on multi-core architectures. The proposed implementation demonstrates
# speedup over conventional DGEMM even on an IntelR Xeon
# PhiTM coprocessor utilizing 240 threads. It is shown how a distributed
# memory matrix-matrix multiplication also benefits from
# these advances.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Strassen algorithm and tensor rank (advanced topic)
#
# - It is not clear how Strassen found these formulas.
# - However, now we can see that they are not artificial.
# - There is a general approach based on the so-called tensor decomposition technique.
# - Here by tensor we imply a multidimensional array - generalization of the matrix concept to many dimensions.
#
# Let us enumerate elements in the $2\times 2$ matrices as follows
#
# $$
# \begin{bmatrix} c_{1} & c_{3} \\ c_{2} & c_{4} \end{bmatrix} =
# \begin{bmatrix} a_{1} & a_{3} \\ a_{2} & a_{4} \end{bmatrix}
# \begin{bmatrix} b_{1} & b_{3} \\ b_{2} & b_{4} \end{bmatrix}=
# \begin{bmatrix}
# a_{1}b_{1} + a_{3}b_{2} & a_{1}b_{3} + a_{3}b_{4} \\
# a_{2}b_{1} + a_{4}b_{2} & a_{2}b_{3} + a_{4}b_{4}
# \end{bmatrix}
# $$
#
# This can be written as
#
# $$ c_k = \sum_{i=1}^4 \sum_{j=1}^4 x_{ijk} a_i b_j, \quad k=1,2,3,4 $$
# + [markdown] slideshow={"slide_type": "slide"}
# $x_{ijk}$ is a 3-dimensional array, that consists of zeros and ones:
#
# $$
# \begin{split}
# x_{\ :,\ :,\ 1} =
# \begin{pmatrix}
# 1 & 0 & 0 & 0 \\
# 0 & 0 & 0 & 0 \\
# 0 & 1 & 0 & 0 \\
# 0 & 0 & 0 & 0 \\
# \end{pmatrix}
# \quad
# x_{\ :,\ :,\ 2} =
# \begin{pmatrix}
# 0 & 0 & 0 & 0 \\
# 1 & 0 & 0 & 0 \\
# 0 & 0 & 0 & 0 \\
# 0 & 1 & 0 & 0 \\
# \end{pmatrix} \\
# x_{\ :,\ :,\ 3} =
# \begin{pmatrix}
# 0 & 0 & 1 & 0 \\
# 0 & 0 & 0 & 0 \\
# 0 & 0 & 0 & 1 \\
# 0 & 0 & 0 & 0 \\
# \end{pmatrix}
# \quad
# x_{\ :,\ :,\ 4} =
# \begin{pmatrix}
# 0 & 0 & 1 & 0 \\
# 0 & 0 & 0 & 0 \\
# 0 & 0 & 0 & 0 \\
# 0 & 0 & 0 & 1 \\
# \end{pmatrix}
# \end{split}
# $$
#
#
# + [markdown] slideshow={"slide_type": "slide"}
# #### Trilinear decomposition
#
# To get Strassen algorithm we should do the following trick –– decompose $x_{ijk}$ in the following way
#
# $$ x_{ijk} = \sum_{\alpha=1}^r u_{i\alpha} v_{j\alpha} w_{k\alpha}. $$
#
# This decomposition is called **trilinear tensor decomposition** and has a meaning of separation of variables: we have a sum of $r$ (called rank) summands with separated $i$, $j$ and $k$.
# + [markdown] slideshow={"slide_type": "slide"}
# #### Strassen via trilinear
#
# Now we have
#
# $$ c_k = \sum_{\alpha=1}^r w_{k\alpha} \left(\sum_{i=1}^4 u_{i\alpha} a_i \right) \left( \sum_{j=1}^4 v_{j\alpha} b_j\right), \quad k=1,2,3,4. $$
#
# Multiplications by $u_{i\alpha}$ or $v_{j\alpha}$ or $w_{k\alpha}$ do not require recursion since $u, v$ and $w$ are known precomputed matrices. Therefore, we have only $r$ multiplications of $\left(\sum_{i=1}^4 u_{i\alpha} a_i \right)$ $\left( \sum_{j=1}^4 v_{j\alpha} b_j\right)$ where both factors depend on the input data.
#
# As you might guess array $x_{ijk}$ has rank $r=7$, which leads us to $7$ multiplications and to the Strassen algorithm!
# + [markdown] slideshow={"slide_type": "slide"}
# ## Summary of MM part
# - MM is the core of NLA. You have to think in block terms, if you want high efficiency
# - This is all about computer memory hierarchy
# - Concept of block algorithms
# - (Advanced topic) Strassen and trilinear form
# + slideshow={"slide_type": "skip"}
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
| lectures/lecture3/lecture-3.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Mini project-------Analysis of student preference
#
# ### Data
#
# I use part of the [student preference](http://archive.ics.uci.edu/ml/datasets/Student+Performance) data from UCI ML repo.
#
# #### Data Set Information:
# This data approach student achievement in secondary education of two Portuguese schools. The data attributes include student grades, demographic, social and school related features) and it was collected by using school reports and questionnaires. Two datasets are provided regarding the performance in two distinct subjects: Mathematics (mat) and Portuguese language (por). In [Cortez and Silva, 2008], the two datasets were modeled under binary/five-level classification and regression tasks. Important note: the target attribute G3 has a strong correlation with attributes G2 and G1. This occurs because G3 is the final year grade (issued at the 3rd period), while G1 and G2 correspond to the 1st and 2nd period grades. It is more difficult to predict G3 without G2 and G1, but such prediction is much more useful (see paper source for more details).
#
#
# #### Attribute Information:
# Attributes for both student-mat.csv (Math course) and student-por.csv (Portuguese language course) datasets:
# 1. school - student's school (binary: 'GP' - Gabriel Pereira or 'MS' - Mousinho da Silveira)
# 2. sex - student's sex (binary: 'F' - female or 'M' - male)
# 3. age - student's age (numeric: from 15 to 22)
# 4. address - student's home address type (binary: 'U' - urban or 'R' - rural)
# 5. famsize - family size (binary: 'LE3' - less or equal to 3 or 'GT3' - greater than 3)
# 6. Pstatus - parent's cohabitation status (binary: 'T' - living together or 'A' - apart)
# 7. Medu - mother's education (numeric: 0 - none, 1 - primary education (4th grade), 2 – 5th to 9th grade, 3 – secondary education or 4 – higher education)
# 8. Fedu - father's education (numeric: 0 - none, 1 - primary education (4th grade), 2 – 5th to 9th grade, 3 – secondary education or 4 – higher education)
# 9. Mjob - mother's job (nominal: 'teacher', 'health' care related, civil 'services' (e.g. administrative or police), 'at_home' or 'other')
# 10. Fjob - father's job (nominal: 'teacher', 'health' care related, civil 'services' (e.g. administrative or police), 'at_home' or 'other')
# 11. reason - reason to choose this school (nominal: close to 'home', school 'reputation', 'course' preference or 'other')
# 12. guardian - student's guardian (nominal: 'mother', 'father' or 'other')
# 13. traveltime - home to school travel time (numeric: 1 - <15 min., 2 - 15 to 30 min., 3 - 30 min. to 1 hour, or 4 - >1 hour)
# 14. studytime - weekly study time (numeric: 1 - <2 hours, 2 - 2 to 5 hours, 3 - 5 to 10 hours, or 4 - >10 hours)
# 15. failures - number of past class failures (numeric: n if 1<=n<3, else 4)
# 16. schoolsup - extra educational support (binary: yes or no)
# 17. famsup - family educational support (binary: yes or no)
# 18. paid - extra paid classes within the course subject (Math or Portuguese) (binary: yes or no)
# 19. activities - extra-curricular activities (binary: yes or no)
# 20. nursery - attended nursery school (binary: yes or no)
# 21. higher - wants to take higher education (binary: yes or no)
# 22. internet - Internet access at home (binary: yes or no)
# 23. romantic - with a romantic relationship (binary: yes or no)
# 24. famrel - quality of family relationships (numeric: from 1 - very bad to 5 - excellent)
# 25. freetime - free time after school (numeric: from 1 - very low to 5 - very high)
# 26. goout - going out with friends (numeric: from 1 - very low to 5 - very high)
# 27. Dalc - workday alcohol consumption (numeric: from 1 - very low to 5 - very high)
# 28. Walc - weekend alcohol consumption (numeric: from 1 - very low to 5 - very high)
# 29. health - current health status (numeric: from 1 - very bad to 5 - very good)
# 30. absences - number of school absences (numeric: from 0 to 93)
#
# these grades are related with the course subject, Math or Portuguese:
# 31. G1 - first period grade (numeric: from 0 to 20)
# 31. G2 - second period grade (numeric: from 0 to 20)
# 32. G3 - final grade (numeric: from 0 to 20, output target)
#
#
# citation: <NAME> and <NAME>. Using Data Mining to Predict Secondary School Student Performance. In A. Brito and <NAME>., Proceedings of 5th FUture BUsiness TEChnology Conference (FUBUTEC 2008) pp. 5-12, Porto, Portugal, April, 2008, EUROSIS, ISBN 978-9077381-39-7.
import os
import pickle
import numpy as np
import pandas as pd
import numpy.linalg as npla
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.metrics import accuracy_score, log_loss
from sklearn.linear_model import LogisticRegression
from sklearn import linear_model
from sklearn.model_selection import train_test_split, GridSearchCV, RandomizedSearchCV, cross_val_score
from sklearn.linear_model import ElasticNet
from sklearn.datasets import make_regression
from sklearn.svm import SVR, LinearSVR
from collections import deque
from sklearn.datasets import load_boston
# %matplotlib inline
# ## Input data
#input raw data
#Due to the whole dataset is a zip file and have several csv file, i can't use URL to download it.
#So i directly download it by hand and put it to my data folder.
student=pd.read_csv('../data/student-mat.csv',sep=";")
student.head()
# Find percentage of missing data in each feature
for i in range(0,33):
if student.iloc[:,i].dtype == "O":
print(student.columns[i])
print(sum(student.iloc[:,i]=="?")/len(student.iloc[:,i]))
#all out put are 0, so there are no missing value
student.shape
# In this case, we want use some features to predict students final grade(G3), so i use G3 as response variable.
y=student["G3"]
X=student.drop(["G3","school"],axis=1)
# ## Data wrangled
# +
# recode all factors as numbers
X["sex"]=X["sex"].map({'F':0,'M':1})
X["address"]=X["address"].map({'U':0,'R':1})
X["famsize"]=X["famsize"].map({'LE3':0,'GT3':1})
X["Pstatus"]=X["Pstatus"].map({'T':0,'A':1})
X["Mjob"]=X["Mjob"].map({'teacher':0,'health':1,'services':2,'at_home':3,'other':4})
X["Fjob"]=X["Fjob"].map({'teacher':0,'health':1,'services':2,'at_home':3,'other':4})
X["reason"]=X["reason"].map({'home':0,'reputation':1,'course':2,'other':3})
X["guardian"]=X["guardian"].map({'mother':0,'father':1,'other':2})
X["schoolsup"]=X["schoolsup"].map({'yes':0,'no':1})
X["famsup"]=X["famsup"].map({'yes':0,'no':1})
X["paid"]=X["paid"].map({'yes':0,'no':1})
X["activities"]=X["activities"].map({'yes':0,'no':1})
X["nursery"]=X["nursery"].map({'yes':0,'no':1})
X["higher"]=X["higher"].map({'yes':0,'no':1})
X["romantic"]=X["romantic"].map({'yes':0,'no':1})
X["internet"]=X["internet"].map({'yes':0,'no':1})
X.head()
# -
#Set training data and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=12345)
X_train.shape
# ## EDA
X.describe()
import seaborn as sns
f,ax = plt.subplots(figsize=(18,18))
sns.heatmap(X.corr(), annot=True, linewidths=.5,ax=ax)
# +
# Plot travel time histogram
plt.hist(X["traveltime"])
plt.title("Frequency of traveltime")
plt.xlabel("traveltime")
plt.ylabel("Frequency")
plt.savefig("../results/traveltime.png")
plt.show()
# +
# Plot study time histogram
plt.hist(X["studytime"])
plt.title("Frequency of study time")
plt.xlabel("study time")
plt.ylabel("Frequency")
plt.savefig("../results/studytime.png")
plt.show()
# +
# Plot failures histogram
plt.hist(X["failures"])
plt.title("Frequency of failures")
plt.xlabel("failures")
plt.ylabel("Frequency")
plt.savefig("../results/failures.png")
plt.show()
# +
# Plot absences histogram
plt.hist(X["absences"])
plt.title("Frequency of absences")
plt.xlabel("absences")
plt.ylabel("Frequency")
plt.savefig("../results/absences.png")
plt.show()
# -
# ## feature selection
# +
#drop features with largr relation ship
#We can see from plot,Dalc(workday alcohol consumption) have large correlation with Walc(weekly alcohol consumption)
#So I delete the Wacl from X.
#G1 and G2 are highly correlated, so i delete G1.
#Mather job, father job, and father education are highly correlated, so I delete Fedu and Mjob.
drop_feature=['Fedu','Mjob','Walc','G1']
X_new=X.drop(drop_feature,axis=1)
X_new.shape
# -
X_new.describe()
#Set training data and test data
X_train, X_test, y_train, y_test = train_test_split(X_new, y, test_size=0.2, random_state=12345)
# ## Model selection
# +
# Model selection
method = {
'linearregression' : LinearRegression(),
'ridge' : Ridge(),
'log regression': LogisticRegression(),
'ElasticNet' : ElasticNet(),
'SVR' : SVR()
}
#MSE in regression
mean_squared_err = lambda y, yhat: np.mean((y-yhat)**2)
for c in method:
clf = method[c]
clf.fit(X_train, y_train)
print(c)
print("Training error: ", mean_squared_err(y_train,clf.predict(X_train)))
print("Validation error: ",mean_squared_err(y_test,clf.predict(X_test)))
print("\n")
# -
# The ridge classifier get small training error and validation error. The ElasticNet classifier can get the lowest validation error but the training error is large. So i choose Ridge method.
# +
#select the parameter
alpha = 10**np.arange(-3,4,0.01)
#L2
train_error=[]
validation_error=[]
for a in alpha:
l2 = Ridge(alpha=a)
l2.fit(X_train,y_train)
train_error.append( mean_squared_err(y_train, l2.predict(X_train)))
validation_error.append(mean_squared_err(y_test, l2.predict(X_test)))
print("Min L2 validation error: %f" % min(validation_error))
print("alpha that Minimun L2 validation error: %f" % alpha[validation_error.index(min(validation_error))])
plt.plot(alpha, validation_error,label='validation error')
plt.plot(alpha, train_error,label='training error')
plt.ylabel("error rate")
plt.xlabel("The value of alpha")
plt.xscale("log")
plt.legend(loc='upper right')
plt.savefig("../results/alpha.png")
plt.show()
# -
# ## Feature selection again (forward selection)
# +
# ForwardSelection
#Reference: lab1 solution in DSCI573
def fit_and_report(model, X, y, Xv, yv):
model.fit(X,y)
mean_squared_err = lambda y, yhat: np.mean((y-yhat)**2)
errors = [mean_squared_err(y, model.predict(X)), mean_squared_err(yv, model.predict(Xv))]
return errors
class ForwardSelection:
def __init__(self, model, min_features=None, max_features=None,
scoring=None, cv=None):
self.max_features = max_features
if min_features is None:
self.min_features = 1
else:
self.min_features = min_features
self.model = model
self.scoring = scoring
self.cv = cv
return
def fit(self, X, y):
if (self.max_features is None) or (self.max_features > X.shape[1]):
self.max_features = X.shape[1]
self.ftr_ = []
idx = np.setdiff1d(range(X.shape[1]), self.ftr_)
best_round_score = deque()
best_round_score.append(np.inf)
X_train, X_val, y_train, y_val = train_test_split(X, y,test_size=0.2,random_state=1245)
for j in range(self.max_features):
round_scores = np.zeros(idx.size)
for i, ii in enumerate(idx):
X_train_s = X_train.iloc[:, self.ftr_ + [ii]]
X_val_s = X_val.iloc[:, self.ftr_ + [ii]]
round_scores[i] = fit_and_report(self.model, X_train_s, y_train,
X_val_s, y_val)[1] # val error only
i_star = np.argmin(round_scores)
ii_star = idx[i_star]
best_round_score.append(round_scores[i_star])
if (len(self.ftr_) > self.min_features) and (best_round_score[-1] >= best_round_score[-2]):
print('found best subset.')
self.best_round_scores_ = np.array(best_round_score)[1:-1]
self.score_ = best_round_score[-2]
return
elif (len(self.ftr_) >= self.max_features):
print('reached max features.')
self.best_round_scores_ = np.array(best_round_score)[1:-1]
self.score_ = best_round_score[-2]
return
else:
self.ftr_ += [ii_star]
idx = np.setdiff1d(range(X.shape[1]), self.ftr_)
return
def transform(self, X, y=None):
return X.iloc[:, self.ftr_]
# +
fs = ForwardSelection(Ridge(alpha=125))
fs.fit(X_train,y_train)
print('features: {}'.format(fs.ftr_))
print('final mean xval error: {}'.format(fs.score_))
# -
#set new training data
X_new = X[X.columns[fs.ftr_]]
X_new
# ## Fit with model and feature i selected
# +
#new train and test set
X_train_new, X_test_new, y_train_new, y_test_new = train_test_split(X, y,test_size=0.2,random_state=1245)
##My best model
model = Ridge(alpha=125)
model.fit(X_train_new,y_train_new)
print("The MSE of train set ", mean_squared_err(y_train_new, model.predict(X_train_new)))
print("The MSE of test set", mean_squared_err(y_test_new, model.predict(X_test_new)))
# -
| src/Student_preference_analysis.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# In order to successfully complete this assignment you must do the required reading, watch the provided videos and complete all instructions. The embedded Google form must be entirely filled out and submitted on or before **11:59pm on Sunday February 16**. Students must come to class the next day prepared to discuss the material covered in this assignment. answer
# # Pre-class assignment: Unit Testing
#
# ### Goals for today's pre-class assignment
#
# </p>
#
#
#
# 2. [2D Array Indexing](#2D_Array_Indexing)
# 1. [Introduction to Unit Testing](#Introduction_to_Unit_Testing)
#
# ---
#
# <a name="2D_Array_Indexing"></a>
#
# # 1. 2D Array Indexing
#
# All memory in a computer is stored linearly with each location in memory given a sequential address. I want you to think for a moment to understand how we take a linear list of numbers and turn it into a 2D array:
#
# IN the following image you can see the relationship between a linear index and a imposed structure that we give it to represent the row and column of the array. We will set up the problem similar to a checker board, such that each grid has a linear label, an index of row, and an index of column.
#
# <img src="https://lh5.googleusercontent.com/tpc1tFPULkPDt1q03E1nPkllHRO60jYyL-rZWmiJLM0AQtVkm1-EF6Pf0yKrm6S5ifVxJ1knHLpVRv7Fsu_pP-rzrauVa2dp2DLrAx2iya9Jbqs2gRWJJ3J-uBqE=w740" alt = "Visual relationship between linear index and a 2D index" width=600 />
#
# First, we will make functions that convert the incremental label of a grid point to it's corresponding row and column index. For example, in the above ($99 \times 49$) grid the 99-th item is at row = 2 and col = 1 .
# ✅ **<font color=red>DO THIS:</font>** Write a function named ```LabelToIndex``` which takes in three arguments (Number of rows, Number of column and the linear index). The function should then return the row and column for that index. The following is a stub function to get you started:
def LabelToIndices(n_row, n_col, lab):
"""function for converting linear label of grid to indices of row and column
Run the function with number of rows, number of columns and the index as input:
>>> LabelToIndices(99, 49, 3675)
(75, 0)
"""
return 2,1
help(LabelToIndices)
# Let's test the ```LabelToIndices``` function for 3675-th grid. Print the index of row and column.
LabelToIndices(99, 49, 3675)
# ✅ **<font color=red>DO THIS:</font>** Using the following stub function as a guild write a function named ```LabelToIndex``` that converts the indices of row and column of a grid to the linear label. For example, the grid at row = 2 and col = 1 is labeled as the 99-th point.
def IndicesToLabel(n_row, n_col, row, col):
"""function for converting row and column indices of a grid to linear label
Usage:
>>> IndicesToLabel(99, 49, 2,1)
99
"""
return 42
IndicesToLabel(99, 49, 2, 1)
# ---
#
# <a name="Introduction_to_Unit_Testing"></a>
#
# # 2. Introduction to Unit Testing
#
# Unit tests are small tests of individual parts of your code. Effective unit testing is absolutely necessary to grow a project past a few developers. Ideally unit tests run after every major/minor change and provide a reality check that nothing is broken. Good unit tests are hard to do and can take practice and time (which is not often where you want to spend your time). That being said, if you know the basic format/syntax of some of the most common testing programs you can format your code to be ready for unit testing.
#
# ## unittest
# Lets start with the most basic unit test program built with python; ```unittest```.
import unittest
help(unittest)
from IPython.display import YouTubeVideo
YouTubeVideo("1Lfv5tUGsn8",width=640,height=360)
# The following are a couple of unit tests for the ```LabelToIndices``` and ```LabelToIndices``` functions. **Note:** It is standard practice to name a unit test ```test_ + <function being tested>```. This naming standard allows for automated test using some libraries.
#
#
# ✅ **<font color=red>DO THIS:</font>** Modify the code to add a few more tests.
#
#
# ✅ **<font color=red>DO THIS:</font>** Temporarily modify the code to make a test fail.
# +
import unittest
# Create a test case
class TestLableToIndeces(unittest.TestCase):
# Create the unit test
def test_LabelToIndices(self):
# Test if 122 equals the output of (6,2)
self.assertEqual((6,2), LabelToIndices(10, 20, 122))
def test_LabelToIndices(self):
# Test if 122 equals the output of (6,2)
self.assertEqual(3110, IndicesToLabel(99, 49, 63, 23))
# -
# We can run all of the unittests in a notebook using the following command:
# +
if __name__ == '__main__':
unittest.main(argv=['first-arg-is-ignored'], exit=False)
# -
# ## doctest
#
# Another type of unit tester is ```doctest```. This is a clever solution that includes the tests inside a function's docstring. See the docstrings for ```LabelToIndices``` and ```LabelToIndices``` and note the "usage" section is one such test.
#
# We can run all of the tests in a jupyter notebook using the following command:
import doctest
doctest.testmod(verbose=True)
# ✅ **<font color=red>DO THIS:</font>** Modify the docstrings for ```makeSchellingGrid``` and ```visualizeGrid``` functions to include some unit testing.
# ## pytest
#
# There are many (Many!) other unit testers out there. Fortunately, most of them work nicely together. One of the best is ```pytest```. Unfortunately, I have not found a clean way to get ```pytest``` to work inside a jupyter notebook. THe options include:
#
# - Export the ipynb as a py file and run pytest on the resulting file.
# - Use one of the many jupyter plug-ins to enable pytest in jupyter (requires an install).
#
# Here is an brief introduction video to ```pytest``` (included with anaconda) to give you a basic idea.
#
from IPython.display import YouTubeVideo
YouTubeVideo("_xoCujgdFgk",width=640,height=360, start=14)
# Although it dosn't work great with Jupyter notebooks PyTest is my prefered unit testing platform and should be used for your class projects.
# ----
# <a name="T5"></a>
# # 5. Assignment wrap-up
#
# Please fill out the form that appears when you run the code below. **You must completely fill this out in order to receive credit for the assignment!**
#
# [Direct Link to Google Form](https://cmse.msu.edu/cmse802-pc-survey)
#
#
# If you have trouble with the embedded form, please make sure you log on with your MSU google account at [googleapps.msu.edu](https://googleapps.msu.edu) and then click on the direct link above.
# ✅ **<font color=red>Assignment-Specific QUESTION:</font>** What are the pros/cons for using unittest, doctest and pytest? Does it make sense to use more than one in your project?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** Summarize what you did in this assignment.
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** What questions do you have, if any, about any of the topics discussed in this assignment after working through the jupyter notebook?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** How well do you feel this assignment helped you to achieve a better understanding of the above mentioned topic(s)?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** What was the **most** challenging part of this assignment for you?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** What was the **least** challenging part of this assignment for you?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** What kind of additional questions or support, if any, do you feel you need to have a better understanding of the content in this assignment?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** Do you have any further questions or comments about this material, or anything else that's going on in class?
# Put your answer to the above question here
# ✅ **<font color=red>QUESTION:</font>** Approximately how long did this pre-class assignment take?
# Put your answer to the above question here
from IPython.display import HTML
HTML(
"""
<iframe
src="https://cmse.msu.edu/cmse802-pc-survey?embedded=true"
width="100%"
height="1200px"
frameborder="0"
marginheight="0"
marginwidth="0">
Loading...
</iframe>
"""
)
# ---------
# ### Congratulations, we're done!
#
# To get credit for this assignment you must fill out and submit the above Google From on or before the assignment due date.
# ### Course Resources:
#
# - [Syllabus](https://docs.google.com/document/d/e/2PACX-1vTW4OzeUNhsuG_zvh06MT4r1tguxLFXGFCiMVN49XJJRYfekb7E6LyfGLP5tyLcHqcUNJjH2Vk-Isd8/pub)
# - [Preliminary Schedule](https://docs.google.com/spreadsheets/d/e/2PACX-1vRsQcyH1nlbSD4x7zvHWAbAcLrGWRo_RqeFyt2loQPgt3MxirrI5ADVFW9IoeLGSBSu_Uo6e8BE4IQc/pubhtml?gid=2142090757&single=true)
# - [Course D2L Page](https://d2l.msu.edu/d2l/home/912152)
# © Copyright 2020, Michigan State University Board of Trustees
| cmse802-s20/0216-Unit_Testing-pre-class-assignment.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text"
# This is a companion notebook for the book [Deep Learning with Python, Second Edition](https://www.manning.com/books/deep-learning-with-python-second-edition?a_aid=keras&a_bid=76564dff). For readability, it only contains runnable code blocks and section titles, and omits everything else in the book: text paragraphs, figures, and pseudocode.
#
# **If you want to be able to follow what's going on, I recommend reading the notebook side by side with your copy of the book.**
#
# This notebook was generated for TensorFlow 2.6.
# + [markdown] colab_type="text"
# ## Interpreting what convnets learn
# + [markdown] colab_type="text"
# ### Visualizing intermediate activations
# + colab_type="code"
# You can use this to load the file "convnet_from_scratch_with_augmentation.keras"
# you obtained in the last chapter.
from google.colab import files
files.upload()
# + colab_type="code"
from tensorflow import keras
model = keras.models.load_model("convnet_from_scratch_with_augmentation.keras")
model.summary()
# + [markdown] colab_type="text"
# **Preprocessing a single image**
# + colab_type="code"
from tensorflow import keras
import numpy as np
img_path = keras.utils.get_file(
fname="cat.jpg",
origin="https://img-datasets.s3.amazonaws.com/cat.jpg")
def get_img_array(img_path, target_size):
img = keras.utils.load_img(
img_path, target_size=target_size)
array = keras.utils.img_to_array(img)
array = np.expand_dims(array, axis=0)
return array
img_tensor = get_img_array(img_path, target_size=(180, 180))
# + [markdown] colab_type="text"
# **Displaying the test picture**
# + colab_type="code"
import matplotlib.pyplot as plt
plt.axis("off")
plt.imshow(img_tensor[0].astype("uint8"))
plt.show()
# + [markdown] colab_type="text"
# **Instantiating a model that returns layer activations**
# + colab_type="code"
from tensorflow.keras import layers
layer_outputs = []
layer_names = []
for layer in model.layers:
if isinstance(layer, (layers.Conv2D, layers.MaxPooling2D)):
layer_outputs.append(layer.output)
layer_names.append(layer.name)
activation_model = keras.Model(inputs=model.input, outputs=layer_outputs)
# + [markdown] colab_type="text"
# **Using the model to compute layer activations**
# + colab_type="code"
activations = activation_model.predict(img_tensor)
# + colab_type="code"
first_layer_activation = activations[0]
print(first_layer_activation.shape)
# + [markdown] colab_type="text"
# **Visualizing the fifth channel**
# + colab_type="code"
import matplotlib.pyplot as plt
plt.matshow(first_layer_activation[0, :, :, 5], cmap="viridis")
# + [markdown] colab_type="text"
# **Visualizing every channel in every intermediate activation**
# + colab_type="code"
images_per_row = 16
for layer_name, layer_activation in zip(layer_names, activations):
n_features = layer_activation.shape[-1]
size = layer_activation.shape[1]
n_cols = n_features // images_per_row
display_grid = np.zeros(((size + 1) * n_cols - 1,
images_per_row * (size + 1) - 1))
for col in range(n_cols):
for row in range(images_per_row):
channel_index = col * images_per_row + row
channel_image = layer_activation[0, :, :, channel_index].copy()
if channel_image.sum() != 0:
channel_image -= channel_image.mean()
channel_image /= channel_image.std()
channel_image *= 64
channel_image += 128
channel_image = np.clip(channel_image, 0, 255).astype("uint8")
display_grid[
col * (size + 1): (col + 1) * size + col,
row * (size + 1) : (row + 1) * size + row] = channel_image
scale = 1. / size
plt.figure(figsize=(scale * display_grid.shape[1],
scale * display_grid.shape[0]))
plt.title(layer_name)
plt.grid(False)
plt.axis("off")
plt.imshow(display_grid, aspect="auto", cmap="viridis")
# + [markdown] colab_type="text"
# ### Visualizing convnet filters
# + [markdown] colab_type="text"
# **Instantiating the Xception convolutional base**
# + colab_type="code"
model = keras.applications.xception.Xception(
weights="imagenet",
include_top=False)
# + [markdown] colab_type="text"
# **Printing the names of all convolutional layers in Xception**
# + colab_type="code"
for layer in model.layers:
if isinstance(layer, (keras.layers.Conv2D, keras.layers.SeparableConv2D)):
print(layer.name)
# + [markdown] colab_type="text"
# **Creating a "feature extractor" model that returns the output of a specific layer**
# + colab_type="code"
layer_name = "block3_sepconv1"
layer = model.get_layer(name=layer_name)
feature_extractor = keras.Model(inputs=model.input, outputs=layer.output)
# + [markdown] colab_type="text"
# **Using the feature extractor**
# + colab_type="code"
activation = feature_extractor(
keras.applications.xception.preprocess_input(img_tensor)
)
# + colab_type="code"
import tensorflow as tf
def compute_loss(image, filter_index):
activation = feature_extractor(image)
filter_activation = activation[:, 2:-2, 2:-2, filter_index]
return tf.reduce_mean(filter_activation)
# + [markdown] colab_type="text"
# **Loss maximization via stochastic gradient ascent**
# + colab_type="code"
@tf.function
def gradient_ascent_step(image, filter_index, learning_rate):
with tf.GradientTape() as tape:
tape.watch(image)
loss = compute_loss(image, filter_index)
grads = tape.gradient(loss, image)
grads = tf.math.l2_normalize(grads)
image += learning_rate * grads
return image
# + [markdown] colab_type="text"
# **Function to generate filter visualizations**
# + colab_type="code"
img_width = 200
img_height = 200
def generate_filter_pattern(filter_index):
iterations = 30
learning_rate = 10.
image = tf.random.uniform(
minval=0.4,
maxval=0.6,
shape=(1, img_width, img_height, 3))
for i in range(iterations):
image = gradient_ascent_step(image, filter_index, learning_rate)
return image[0].numpy()
# + [markdown] colab_type="text"
# **Utility function to convert a tensor into a valid image**
# + colab_type="code"
def deprocess_image(image):
image -= image.mean()
image /= image.std()
image *= 64
image += 128
image = np.clip(image, 0, 255).astype("uint8")
image = image[25:-25, 25:-25, :]
return image
# + colab_type="code"
plt.axis("off")
plt.imshow(deprocess_image(generate_filter_pattern(filter_index=2)))
# + [markdown] colab_type="text"
# **Generating a grid of all filter response patterns in a layer**
# + colab_type="code"
all_images = []
for filter_index in range(64):
print(f"Processing filter {filter_index}")
image = deprocess_image(
generate_filter_pattern(filter_index)
)
all_images.append(image)
margin = 5
n = 8
cropped_width = img_width - 25 * 2
cropped_height = img_height - 25 * 2
width = n * cropped_width + (n - 1) * margin
height = n * cropped_height + (n - 1) * margin
stitched_filters = np.zeros((width, height, 3))
for i in range(n):
for j in range(n):
image = all_images[i * n + j]
stitched_filters[
(cropped_width + margin) * i : (cropped_width + margin) * i + cropped_width,
(cropped_height + margin) * j : (cropped_height + margin) * j
+ cropped_height,
:,
] = image
keras.utils.save_img(
f"filters_for_layer_{layer_name}.png", stitched_filters)
# + [markdown] colab_type="text"
# ### Visualizing heatmaps of class activation
# + [markdown] colab_type="text"
# **Loading the Xception network with pretrained weights**
# + colab_type="code"
model = keras.applications.xception.Xception(weights="imagenet")
# + [markdown] colab_type="text"
# **Preprocessing an input image for Xception**
# + colab_type="code"
img_path = keras.utils.get_file(
fname="elephant.jpg",
origin="https://img-datasets.s3.amazonaws.com/elephant.jpg")
def get_img_array(img_path, target_size):
img = keras.utils.load_img(img_path, target_size=target_size)
array = keras.utils.img_to_array(img)
array = np.expand_dims(array, axis=0)
array = keras.applications.xception.preprocess_input(array)
return array
img_array = get_img_array(img_path, target_size=(299, 299))
# + colab_type="code"
preds = model.predict(img_array)
print(keras.applications.xception.decode_predictions(preds, top=3)[0])
# + colab_type="code"
np.argmax(preds[0])
# + [markdown] colab_type="text"
# **Setting up a model that returns the last convolutional output**
# + colab_type="code"
last_conv_layer_name = "block14_sepconv2_act"
classifier_layer_names = [
"avg_pool",
"predictions",
]
last_conv_layer = model.get_layer(last_conv_layer_name)
last_conv_layer_model = keras.Model(model.inputs, last_conv_layer.output)
# + [markdown] colab_type="text"
# **Setting up a model that goes from the last convolutional output to the final predictions**
# + colab_type="code"
classifier_input = keras.Input(shape=last_conv_layer.output.shape[1:])
x = classifier_input
for layer_name in classifier_layer_names:
x = model.get_layer(layer_name)(x)
classifier_model = keras.Model(classifier_input, x)
# + [markdown] colab_type="text"
# **Retrieving the gradients of the top predicted class with regard to the last convolutional output**
# + colab_type="code"
import tensorflow as tf
with tf.GradientTape() as tape:
last_conv_layer_output = last_conv_layer_model(img_array)
tape.watch(last_conv_layer_output)
preds = classifier_model(last_conv_layer_output)
top_pred_index = tf.argmax(preds[0])
top_class_channel = preds[:, top_pred_index]
grads = tape.gradient(top_class_channel, last_conv_layer_output)
# + [markdown] colab_type="text"
# **Gradient pooling and channel importance weighting**
# + colab_type="code"
pooled_grads = tf.reduce_mean(grads, axis=(0, 1, 2)).numpy()
last_conv_layer_output = last_conv_layer_output.numpy()[0]
for i in range(pooled_grads.shape[-1]):
last_conv_layer_output[:, :, i] *= pooled_grads[i]
heatmap = np.mean(last_conv_layer_output, axis=-1)
# + [markdown] colab_type="text"
# **Heatmap post-processing**
# + colab_type="code"
heatmap = np.maximum(heatmap, 0)
heatmap /= np.max(heatmap)
plt.matshow(heatmap)
# + [markdown] colab_type="text"
# **Superimposing the heatmap with the original picture**
# + colab_type="code"
import matplotlib.cm as cm
img = keras.utils.load_img(img_path)
img = keras.utils.img_to_array(img)
heatmap = np.uint8(255 * heatmap)
jet = cm.get_cmap("jet")
jet_colors = jet(np.arange(256))[:, :3]
jet_heatmap = jet_colors[heatmap]
jet_heatmap = keras.utils.array_to_img(jet_heatmap)
jet_heatmap = jet_heatmap.resize((img.shape[1], img.shape[0]))
jet_heatmap = keras.utils.img_to_array(jet_heatmap)
superimposed_img = jet_heatmap * 0.4 + img
superimposed_img = keras.utils.array_to_img(superimposed_img)
save_path = "elephant_cam.jpg"
superimposed_img.save(save_path)
# + [markdown] colab_type="text"
# ## Chapter summary
| notebooks/dlp09_part03_interpreting_what_convnets_learn.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Training Models - Ch4
# We'll do more than just treat models as black boxes in this chapter.
#
#
# +
#Linear Algebra micro review
import pandas as pd
#Dot Product of vector
vec = pd.DataFrame([1,3,-5])
vec
# -
vec.T
vec.T.dot(pd.DataFrame([4,-2,-1]))
# ## Linear Regression
# Generalized, it is an equation for a line where the "mx" part (of y = mx + b) is a weighted sum of input features.
#
# To train a linear model, you find values of theta that minimize error metric. Most commonly for linear that is RMSE (root mean square error). Or use MSE, since it is easier.
#
# The **Normal Equation** is solved for theta^ directly, and is written in terms of X and y. It is "closed-form".
# We'll generate data to play with this idea.
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
X = 2 * np.random.rand(100,1)
y = 4 + 3 * X + np.random.randn(100,1)
# our random generated linear dataset.
X, y
plt.plot(X, y, "b.")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.axis([0, 2, 0, 15])
plt.show()
X_b = np.c_[np.ones((100,1)), X] #adds x0 = 1 to each instance, for dot product and matrix sizing i presume.
theta_best = np.linalg.inv(X_b.T.dot(X_b)).dot(X_b.T).dot(y)
theta_best
np.linalg.inv(X_b.T.dot(X_b))
X_b.T.dot(X_b)
# Here we used the normal equation to solve for theta. Ideally theta would be 4 and 3, since that is what we used to generate the noise.
#Make predictions against our model and plot.
X_new = np.array([[0],[2]])
X_new_b = np.c_[np.ones((2,1)), X_new]
y_predict = X_new_b.dot(theta_best)
y_predict
plt.plot(X_new, y_predict, "r-")
plt.plot(X,y, "b.")
plt.axis([0,2,0,15])
plt.show()
#Same regression with Sklearn
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(X,y)
lin_reg.intercept_, lin_reg.coef_
lin_reg.predict(X_new)
# ## Gradient Descent
# Tweak parameters iteratively in order to minimize a cost function. (finds local/global mins)
#
# Practically speaking, you can fill theta (your model parameters) with random numbers and start changing them gradually to minimize the MSE. The number of steps you take is the learning rate hyperparam. learning rate too low and it takes too long, too high and you might jump across a valley and miss the minimum.
#
# ### Tip
# For gradient descent, ensure you've used something like `sklearn StandardScaler` to ensure features are similar scale. This helps speed up convergence.
#
# ### Batch Gradient Descent
# We are essentially computing partial derivatives with respect to each model parameter (thetas). AKA how much will the cost function change, if I change this input param just a little bit.
#
# It is called "Batch" because the entire training set is used in the computation for each gradient step. (to compute all the partial derivatives)
#
# The gradients point "up hill" which is why we subtract.
#Example gradient Descent.
eta = 0.1 #learning rate
n_iterations = 1000
m = 100
theta = np.random.randn(2,1) #random initialization.
for iteration in range(n_iterations):
gradients = 2/m * X_b.T.dot(X_b.dot(theta) - y)
theta = theta - eta * gradients
theta # It worked!
# ### Stochastic Gradient Descent
# Much faster than batch since it doesn't use the entire traning set at each step. Once the algorithm stops, the parameter values it found are simply "good", but not guaranteed to be optimal. The randomness also helps it find the global minimum, if the cost function is not convex.
#
# This is also called "simulated annealing".
#
# The "learning schedule" determines what the learning rate will be at each iteration.
# +
n_epochs = 50
t0, t1 = 5, 50 #learning schedule hyperparams.
def learning_schedule(t):
return t0 / (t + t1)
theta = np.random.randn(2,1) #random init
for epoch in range(n_epochs):
for i in range(m):
random_index = np.random.randint(m)
xi = X_b[random_index:random_index+1]
yi = y[random_index:random_index+1]
gradients = 2 * xi.T.dot(xi.dot(theta) - yi)
eta = learning_schedule(epoch * m + i)
theta = theta - eta * gradients
# -
theta
# Similar good results to batch but we only had to iterate 50 times (50 "epochs").
# Stochastic Gradient Descent using sklearn
from sklearn.linear_model import SGDRegressor
sgd_reg = SGDRegressor(n_iter=50, penalty=None, eta0=0.1)
sgd_reg.fit(X,y.ravel())
sgd_reg.intercept_, sgd_reg.coef_
# ### Mini Batch
# sort of a combo of Batch and Stochastic. compute gradient at each step by randomly selecting small subset of instances from training set.
# ## Polynomial Regression
m = 100
X = 6 * np.random.rand(m,1) -3
y = 0.5 * X**2 + X + 2 + np.random.randn(m, 1)
plt.plot(X,y, "b.")
plt.show()
# A simple straight line won't fit this, so lets use PolynomialFeatures
from sklearn.preprocessing import PolynomialFeatures
poly_features = PolynomialFeatures(degree=2, include_bias=False)
X_poly = poly_features.fit_transform(X)
X[0]
X_poly[0] #added the square of X to the dataset.
#Now we can fit a standard linear regression onto this polynomial data.
lin_reg = LinearRegression()
lin_reg.fit(X_poly, y)
lin_reg.intercept_, lin_reg.coef_
# Not too bad, since the original function was `y = 0.5 * X**2 + X + 2 + np.random.randn(m, 1)`
# ### Learning Curves
#
# How do you determine what type of curve to fit your data? (aka which degree polynomial).
#
# Another way to look at model performance is to look at the "learning curves". These are plots of the models performance on training and validation sets, as a fu nction of the training set size (aka training iteration). You train the model several times on different sized subsets of the training set. Aka how does the model respond as it gets more and more data to use?
# +
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
def plot_learning_curves(model, x, y, y_lim=False):
X_train, X_val, y_train, y_val = train_test_split(X,y, test_size=0.2)
train_errors, val_errors = [],[]
for m in range(1, len(X_train)):
model.fit(X_train[:m], y_train[:m])
y_train_predict = model.predict(X_train[:m])
y_val_predict = model.predict(X_val)
train_errors.append(mean_squared_error(y_train_predict, y_train[:m]))
val_errors.append(mean_squared_error(y_val, y_val_predict))
plt.plot(np.sqrt(train_errors), "r-+", linewidth=2, label="train")
plt.plot(np.sqrt(val_errors), "b-", linewidth=3, label="val")
plt.legend(loc="upper right", fontsize=14) # not shown in the book
plt.xlabel("Training set size", fontsize=14) # not shown
plt.ylabel("RMSE", fontsize=14) # not shown
if y_lim:
plt.ylim((0, y_lim))
# -
lin_reg = LinearRegression()
plot_learning_curves(lin_reg, X,y, 3)
# Tip: If your model is underfitting your model (such as this graph suggests) adding more training data will not help, you need a more complex model, or better features in your data.
# +
# Plot learning curves for complex 10th degree polynomial
from sklearn.pipeline import Pipeline
polynomial_regression = Pipeline([
("poly_features", PolynomialFeatures(degree=10, include_bias=False)),
("lin_reg", LinearRegression())
])
plot_learning_curves(polynomial_regression, X,y, 2)
# -
# We notice that the error is lower than with the linear model.
# ## Regularized Linear Models
# To avoid overfitting a model (i.e. constrain it) you can limit the degrees of freedom. A way to do this for polynomial models is to limit the degree.
#
# For linear models, you can constrain the weights of the model. What follors is Ridge Regression, Lasso Regression, and Elastic Net, three weights of constraining linear weights.
# Ridge Regression is a "regularized" version of linear regression. A regularization term is added to the cost function. Hyperparameter alpha (a) controls how much to regularize. alpha of 0 is just plain linear regression.
# ## Logistic Regression
# Uses probablilities to determine if something is class 1 or 0 (aka binary classifier).
from sklearn import datasets
iris = datasets.load_iris() #Famous Iris flower data set.
list(iris.keys())
X = iris["data"][:, 3:] #petal width
y = (iris["target"] == 2).astype(np.int) #1 if Iris-Virginica, else 0
#train logistic regression model.
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression()
log_reg.fit(X,y)
#look at estimated probabilities for flowers with petal widths from 0 to 3cm
X_new = np.linspace(0,3,1000).reshape(-1,1)
y_proba = log_reg.predict_proba(X_new)
plt.plot(X_new, y_proba[:, 1], "g-", label="Iris-Virginica")
plt.plot(X_new, y_proba[:, 0], "b--", label="Not Iris Virginica")
# X axis is petal width in cm.
| handsOn/ch4/TrainingModels.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
N_trials = 100000
x = np.random.uniform(size=N_trials)
y = np.random.uniform(size=N_trials)
z = x+y
_ = plt.hist(z, bins=np.linspace(0,2,100), density=True)
plt.scatter(x,y)
import numpy as np
import matplotlib.pyplot as plt
N_trials = 100000
N_vars = 10
x_array = np.random.uniform(0,1, size=N_trials*N_vars).reshape(N_trials,N_vars)
z = np.sum(x_array,axis=1)
_ = plt.hist(z, bins=np.linspace(0,N_vars,100), density=True)
import numpy as np
import matplotlib.pyplot as plt
N_trials = 100000
def cos_conv(N_vars):
x_array = np.random.uniform(-np.pi,np.pi, size=N_trials*N_vars).reshape(N_trials,N_vars)
cos_array = np.cos(x_array)
z = np.sum(cos_array,axis=1)
return z
bins = np.linspace(-5,5,100)
_ = plt.hist(cos_conv(1), bins=bins, density=True, alpha=.2, label='N=1')
_ = plt.hist(cos_conv(2), bins=bins, density=True, alpha=.2, label='N=2')
_ = plt.hist(cos_conv(5), bins=bins, density=True, alpha=.2, label='N=5')
plt.legend();
| book/error-propagation/convolution-demo.ipynb |