code
stringlengths 235
11.6M
| repo_path
stringlengths 3
263
|
|---|---|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Example 5: Laplace equation
#
# In this tutorial we will look constructing the steady-state heat example using the Laplace equation. In contrast to the previous tutorials this example is entirely driven by the prescribed Dirichlet and Neumann boundary conditions, instead of an initial condition. We will also demonstrate how to use Devito to solve a steady-state problem without time derivatives and how to switch buffers explicitly without having to re-compile the kernel.
#
# First, we again define our governing equation:
# $$\frac{\partial ^2 p}{\partial x^2} + \frac{\partial ^2 p}{\partial y^2} = 0$$
#
# We are again discretizing second-order derivatives using a central difference scheme to construct a diffusion problem (see tutorial 3). This time we have no time-dependent term in our equation though, since there is no term $p_{i,j}^{n+1}$. This means that we are simply updating our field variable $p$ over and over again, until we have reached an equilibrium state. In a discretised form, after rearranging to update the central point $p_{i,j}^n$ we have
# $$p_{i,j}^n = \frac{\Delta y^2(p_{i+1,j}^n+p_{i-1,j}^n)+\Delta x^2(p_{i,j+1}^n + p_{i,j-1}^n)}{2(\Delta x^2 + \Delta y^2)}$$
#
# And, as always, we first re-create the original implementation to see what we are aiming for. Here we initialise the field $p$ to $0$ and apply the following bounday conditions:
#
# $p=0$ at $x=0$
#
# $p=y$ at $x=2$
#
# $\frac{\partial p}{\partial y}=0$ at $y=0, \ 1$
#
# **Developer note:**
# The original tutorial stores the field data in the layout `(ny, nx)`. Until now we have used `(x, y)` notation for creating our Devito examples, but for this one we will adopt the `(y, x)` layout for compatibility reasons.
# +
from examples.cfd import plot_field
import numpy as np
# %matplotlib inline
# Some variable declarations
nx = 31
ny = 31
c = 1
dx = 2. / (nx - 1)
dy = 1. / (ny - 1)
# -
def laplace2d(p, bc_y, dx, dy, l1norm_target):
l1norm = 1
pn = np.empty_like(p)
while l1norm > l1norm_target:
pn = p.copy()
p[1:-1, 1:-1] = ((dy**2 * (pn[1:-1, 2:] + pn[1:-1, 0:-2]) +
dx**2 * (pn[2:, 1:-1] + pn[0:-2, 1:-1])) /
(2 * (dx**2 + dy**2)))
p[:, 0] = 0 # p = 0 @ x = 0
p[:, -1] = bc_right # p = y @ x = 2
p[0, :] = p[1, :] # dp/dy = 0 @ y = 0
p[-1, :] = p[-2, :] # dp/dy = 0 @ y = 1
l1norm = (np.sum(np.abs(p[:]) - np.abs(pn[:])) /
np.sum(np.abs(pn[:])))
return p
# +
#NBVAL_IGNORE_OUTPUT
# Out initial condition is 0 everywhere,except at the boundary
p = np.zeros((ny, nx))
# Boundary conditions
bc_right = np.linspace(0, 1, ny)
p[:, 0] = 0 # p = 0 @ x = 0
p[:, -1] = bc_right # p = y @ x = 2
p[0, :] = p[1, :] # dp/dy = 0 @ y = 0
p[-1, :] = p[-2, :] # dp/dy = 0 @ y = 1
plot_field(p, ymax=1.0, view=(30, 225))
# +
#NBVAL_IGNORE_OUTPUT
p = laplace2d(p, bc_right, dx, dy, 1e-4)
plot_field(p, ymax=1.0, view=(30, 225))
# -
# Ok, nice. Now, to re-create this example in Devito we need to look a little bit further under the hood. There are two things that make this different to the examples we covered so far:
# * We have no time dependence in the `p` field, but we still need to advance the state of p in between buffers. So, instead of using `TimeFunction` objects that provide multiple data buffers for timestepping schemes, we will use `Function` objects that have no time dimension and only allocate a single buffer according to the space dimensions. However, since we are still implementing a pseudo-timestepping loop, we will need two objects, say `p` and `pn`, to act as alternating buffers.
# * If we're using two different symbols to denote our buffers, any operator we create will only perform a single timestep. This is desired though, since we need to check a convergence criteria outside of the main stencil update to determine when we stop iterating. As a result we will need to call the operator repeatedly after instantiating it outside the convergence loop.
#
# So, how do we make sure our operator doesn't accidentally overwrite values in the same buffer? Well, we can again let SymPy reorganise our Laplace equation based on `pn` to generate the stencil, but when we create the update expression, we set the LHS to our second buffer variable `p`.
# +
from devito import Grid, Function, Eq, INTERIOR, solve
# Create two explicit buffers for pseudo-timestepping
grid = Grid(shape=(nx, ny), extent=(1., 2.))
p = Function(name='p', grid=grid, space_order=2)
pn = Function(name='pn', grid=grid, space_order=2)
# Create Laplace equation base on `pn`
eqn = Eq(pn.laplace, region=INTERIOR)
# Let SymPy solve for the central stencil point
stencil = solve(eqn, pn)
# Now we let our stencil populate our second buffer `p`
eq_stencil = Eq(p, stencil)
# In the resulting stencil `pn` is exclusively used on the RHS
# and `p` on the LHS is the grid the kernel will update
print("Update stencil:\n%s\n" % eq_stencil)
# -
# Now we can add our boundary conditions. We have already seen how to prescribe constant Dirichlet BCs by simply setting values using the low-level notation. This time we will go a little further by setting a prescribed profile, which we create first as a custom 1D symbol and supply with the BC values. For this we need to create a `Function` object that has a different shape than our general `grid`, so instead of the grid we provide an explicit pair of dimension symbols and the according shape for the data.
x, y = grid.dimensions
bc_right = Function(name='bc_right', shape=(nx, ), dimensions=(x, ))
bc_right.data[:] = np.linspace(0, 1, nx)
# Now we can create a set of expressions for the BCs again, where we wet prescribed values on the right and left of our grid. For the Neuman BCs along the top and bottom boundaries we simply copy the second rwo from the outside into the outermost row, just as the original tutorial did. Using these expressions and our stencil update we can now create an operator.
# +
#NBVAL_IGNORE_OUTPUT
from devito import Operator
# Create boundary condition expressions
bc = [Eq(p[x, 0], 0.)] # p = 0 @ x = 0
bc += [Eq(p[x, ny-1], bc_right[x])] # p = y @ x = 2
bc += [Eq(p[0, y], p[1, y])] # dp/dy = 0 @ y = 0
bc += [Eq(p[nx-1, y], p[nx-2, y])] # dp/dy = 0 @ y = 1
# Now we can build the operator that we need
op = Operator(expressions=[eq_stencil] + bc)
# -
# We can now use this single-step operator repeatedly in a Python loop, where we can arbitrarily execute other code in between invocations. This allows us to update our L1 norm and check for convergence. Using our pre0compiled operator now comes down to a single function call that supplies the relevant data symbols. One thing to note is that we now do exactly the same thing as the original NumPy loop, in that we deep-copy the data between each iteration of the loop, which we will look at after this.
# +
#NBVAL_IGNORE_OUTPUT
# Silence the runtime performance logging
from devito import configuration
configuration['log_level'] = 'ERROR'
# Initialise the two buffer fields
p.data[:] = 0.
p.data[:, -1] = np.linspace(0, 1, ny)
pn.data[:] = 0.
pn.data[:, -1] = np.linspace(0, 1, ny)
# Visualize the initial condition
plot_field(p.data, ymax=1.0, view=(30, 225))
# Run the convergence loop with deep data copies
l1norm_target = 1.e-4
l1norm = 1
while l1norm > l1norm_target:
# This call implies a deep data copy
pn.data[:] = p.data[:]
op(p=p, pn=pn)
l1norm = (np.sum(np.abs(p.data[:]) - np.abs(pn.data[:])) /
np.sum(np.abs(pn.data[:])))
# Visualize the converged steady-state
plot_field(p.data, ymax=1.0, view=(30, 225))
# -
# One crucial detail about the code above is that the deep data copy between iterations will really hurt performance if we were to run this on a large grid. However, we have already seen how we can match data symbols to symbolic names when calling the pre-compiled operator, which we can now use to actually switch the roles of `pn` and `p` between iterations, eg. `op(p=pn, pn=p)`. Thus, we can implement a simple buffer-switching scheme by simply testing for odd and even time-steps, without ever having to shuffle data around.
# +
#NBVAL_IGNORE_OUTPUT
# Initialise the two buffer fields
p.data[:] = 0.
p.data[:, -1] = np.linspace(0, 1, ny)
pn.data[:] = 0.
pn.data[:, -1] = np.linspace(0, 1, ny)
# Visualize the initial condition
plot_field(p.data, ymax=1.0, view=(30, 225))
# Run the convergence loop by explicitly flipping buffers
l1norm_target = 1.e-4
l1norm = 1
counter = 0
while l1norm > l1norm_target:
# Determine buffer order
if counter % 2 == 0:
_p = p
_pn = pn
else:
_p = pn
_pn = p
# Apply operator
op(p=_p, pn=_pn)
# Compute L1 norm
l1norm = (np.sum(np.abs(_p.data[:]) - np.abs(_pn.data[:])) /
np.sum(np.abs(_pn.data[:])))
counter += 1
plot_field(p.data, ymax=1.0, view=(30, 225))
| examples/cfd/05_laplace.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# **[Machine Learning Course Home Page](https://www.kaggle.com/learn/machine-learning)**
#
# ---
#
# This exercise will test your ability to read a data file and understand statistics about the data.
#
# In later exercises, you will apply techniques to filter the data, build a machine learning model, and iteratively improve your model.
#
# The course examples use data from Melbourne. To ensure you can apply these techniques on your own, you will have to apply them to a new dataset (with house prices from Iowa).
#
# The exercises use a "notebook" coding environment. In case you are unfamiliar with notebooks, we have a [90-second intro video](https://www.youtube.com/watch?v=4C2qMnaIKL4).
#
# # Exercises
#
# Run the following cell to set up code-checking, which will verify your work as you go.
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.machine_learning.ex2 import *
print("Setup Complete")
# ## Step 1: Loading Data
# Read the Iowa data file into a Pandas DataFrame called `home_data`.
# +
import pandas as pd
# Path of the file to read
iowa_file_path = '../input/home-data-for-ml-course/train.csv'
# Fill in the line below to read the file into a variable home_data
home_data = pd.read_csv(iowa_file_path)
# Call line below with no argument to check that you've loaded the data correctly
step_1.check()
# +
# Lines below will give you a hint or solution code
#step_1.hint()
#step_1.solution()
# -
# ## Step 2: Review The Data
# Use the command you learned to view summary statistics of the data. Then fill in variables to answer the following questions
# Print summary statistics in next line
home_data.describe()
# +
# What is the average lot size (rounded to nearest integer)?
avg_lot_size = 10517
# As of today, how old is the newest home (current year - the date in which it was built)
newest_home_age = 2021-2010
# Checks your answers
step_2.check()
# -
step_2.hint()
step_2.solution()
# ## Think About Your Data
#
# The newest house in your data isn't that new. A few potential explanations for this:
# 1. They haven't built new houses where this data was collected.
# 1. The data was collected a long time ago. Houses built after the data publication wouldn't show up.
#
# If the reason is explanation #1 above, does that affect your trust in the model you build with this data? What about if it is reason #2?
#
# How could you dig into the data to see which explanation is more plausible?
#
# Check out this **[discussion thread](https://www.kaggle.com/learn-forum/60581)** to see what others think or to add your ideas.
#
# # Keep Going
#
# You are ready for **[Your First Machine Learning Model](https://www.kaggle.com/dansbecker/your-first-machine-learning-model).**
#
# ---
# **[Machine Learning Course Home Page](https://www.kaggle.com/learn/machine-learning)**
#
#
| Intro to Machine Learning/exercise-2-explore-your-data-preet-mehta.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.6.2
# language: julia
# name: julia-1.6
# ---
# # Exercises: Control flow
# ### if...
#
# Write a conditional statement that prints the number itself if it is smaller than zero, and the string "positive" if the number is larger than or equal to zero.
#
# ### for-loops
#
# Loop over integers between 1 and 100 and print their squares.
# ### while
#
# Do the same with a `while` statement
# ### arrays
#
# Use an array comprehension to create an an array that stores the squares for all integers between 1 and 100.
# ## Epidemic simulation
#
# Fill in the missing pieces to the second for loop below so that the infection spreads horizontally as well.
# The following two lines load the epidemic functions from a file
include("../epidemic_simple.jl")
cells = make_cells()
"Update the simulation one time step"
function update!(cells)
# Create a copy to remember the old state
old_cells = deepcopy(cells)
# Loop over pairs of cells in the same row. There are size(cells)[1] columns, and size(cells)[1]-1 pairs.
for i in 1:size(cells)[1]-1
# loop over all columns
for j in 1:size(cells)[2]
# So the cells are (i+1,j) and (i,j). Each will interact with the other.
cells[i,j] = interact(cells[i,j], old_cells[i+1,j])
cells[i+1,j] = interact(cells[i+1,j], old_cells[i,j])
end
end
# Loop over pairs of cells in the same row. There are size(cells)[1] columns, and size(cells)[1]-1 pairs.
for i in 1:size(cells)[1]
# loop over all columns
for j in BLANK
# The cells are (i+1,j) and (i,j). Each will interact with the other.
BLANK
BLANK
end
end
end
update!(cells)
cells
# ### Advanced: FizzBuzz
#
# Implement the (infamous) FizzBuzz test using Julia:
#
# Loop over numbers between 1 and 100. For every element:
# - given a number, N, print "Fizz" if N is divisible by 3,
# - "Buzz" if N is divisible by 5,
# - and "FizzBuzz" if N is divisible by 3 and 5.
# - Otherwise just print the number itself
#
# You can check the remainder of division using the `%` symbol, i.e., `3 % 2 = 1`
| exercises/04_Exercises-control-flow.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Making multipanel plots with matplot lib
# first we import numpy and matplot lib as usual
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# then we define an array of angles an their sins and coss using numpy. this time we will use linspace
# +
x= np.linspace(0,2*np.pi,100)
print(x[-1],2*np.pi)
y=np.sin(x)
z=np.cos(x)
w=np.sin(4*x)
v=np.cos(4*x)
# -
# Now lets make a 2 panel plot side-by-side
# +
#call subplots to ger=nerate a multipanel figure. This means 1 row 2 columns of figures
f, axarr = plt.subplots(1 , 2)
#treat axarr as an array, from left to right
#first panel
axarr[0].plot(x ,y)
axarr[0].set_xlabel('x')
axarr[0].set_ylabel('y')
axarr[0].set_title(r'$\cos(x)$')
#second panel
axarr[1].plot(x ,z)
axarr[1].set_xlabel('x')
axarr[1].set_ylabel('cos(x)')
axarr[1].set_title(r'$\cos(x)$')
#add more space between figures
f.subplots_adjust(wspace=0.4)
#fis the axis ratio
#here are 2 options
axarr[0].set_aspect('equal') #make the ratio of the thick units equal, a bit counter intuitive
axarr[1].set_aspect(np.pi) #make a sqare by setting aspect to be the ratio of the tick unit change
# +
#adjust size of figure
fig=plt.figure(figsize=(6,6))
plt.plot(x,y, label=r'$y=\sin(x)$')
plt.plot(x,z, label=r'$y=\cos(x)$')
plt.plot(x,w, label=r'$y=\sin(4x)$')
plt.plot(x,v, label=r'$y=\cos(4x)$')
plt.xlabel(r'$x$')
plt.ylabel(r'$y(x)$')
plt.xlim([0,2*np.pi])
plt.ylim([-1.2,1.2])
plt.legend(loc=1,framealpha=0.95)
plt.gca().set_aspect(np.pi/1.2)
# -
| Multipanel Figures.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Using a CNN model
import torch
from torch import nn
from torch.utils.data import DataLoader
from torchvision import datasets
from torchvision.transforms import ToTensor, Lambda
import torch.nn.functional as F
import matplotlib.pyplot as plt
device = 'cuda' if torch.cuda.is_available() else 'cpu'
print(f'Using {device} device')
# +
# Following the same code as in training_a_model notebook, but using a CNN model
training_data = datasets.FashionMNIST(
root="../data",
train=True,
download=True,
transform=ToTensor()
)
test_data = datasets.FashionMNIST(
root="../data",
train=False,
download=True,
transform=ToTensor()
)
labels_map = {
0: "T-Shirt",
1: "Trouser",
2: "Pullover",
3: "Dress",
4: "Coat",
5: "Sandal",
6: "Shirt",
7: "Sneaker",
8: "Bag",
9: "Ankle Boot",
}
# +
# Look at a sample of the data
x, y = next(iter(training_data))
plt.figure()
plt.imshow(x.squeeze(), cmap="gray")
plt.title(labels_map[y])
plt.show()
# +
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# 1 input image channel, 6 output channels, 3x3 square convolution kernel
self.relu = nn.ReLU()
# The output of a convo layer will be (W - K + 2P) / S + 1, where W is the
# image size, K is the kernel size, P is the padding size, and S is the stride.
# My input images are 1 x 28 x 28 and will be output as 6 x 30 x 30.
self.conv1 = nn.Conv2d(1, 6, kernel_size=3, stride=1, padding=2)
# The output of a pooling layer will be (W - K) / S + 1, where K is the pooling kernel
# and S is the stride. So input 6 x 30 x 30 images will be output as 6 x 14 x 14.
self.pool1 = nn.MaxPool2d(kernel_size=2, stride=2)
# Input: 6 x 14 x 14, Output: 16 x 12 x 12
self.conv2 = nn.Conv2d(6, 16, kernel_size=3, stride=1)
# Input: 16 x 12 x 12, Output: 16 x 6 x 6
self.pool2 = nn.MaxPool2d(kernel_size=2, stride=2)
# Fully connected layers. All 16*6*6 inputs connect to 120 outputs, connecting the
# convolutional layers to the FC layers.
self.fc1 = nn.Linear(16 * 6 * 6, 120)
# 120 input nodes to 84 output nodes
self.fc2 = nn.Linear(120, 84)
# 84 input nodes to 10 output nodes: the number of labels
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
# Max pooling over a (2, 2) window
x = self.relu(self.conv1(x))
x = self.pool1(x)
# If the size is a square you can only specify a single number
x = self.relu(self.conv2(x))
x = self.pool2(x)
x = x.view(-1, self.num_flat_features(x))
x = self.relu(self.fc1(x))
x = self.relu(self.fc2(x))
x = self.fc3(x)
return x
def num_flat_features(self, x):
size = x.size()[1:] # all dimensions except the batch dimension
num_features = 1
for s in size:
num_features *= s
return num_features
net = Net()
print(net)
# -
epochs = 10
batch_size = 64
learning_rate = 1e-3
momentum = 0.9
weight_decay = 0.0005
# +
# Initialize the dataloaders and model
train_dataloader = DataLoader(training_data, batch_size=batch_size)
test_dataloader = DataLoader(test_data, batch_size=batch_size)
model = Net()
model.to(device)
# -
loss_fn = nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate, momentum=momentum, weight_decay=weight_decay)
# +
def train_loop(dataloader, model, loss_fn, optimizer):
size = len(dataloader.dataset)
# Get the input data X and label y from the dataloader
for batch, (X, y) in enumerate(dataloader):
# Compute the model prediction given current model parameters.
pred = model(X.to(device))
# Compute the loss from the prediction and the label
loss = loss_fn(pred, y.to(device))
# Optimization: zero gradients, backpropogation, adjust parameters.
optimizer.zero_grad()
loss.backward()
optimizer.step()
def test_loop(dataloader, model, loss_fn):
size = len(dataloader.dataset)
test_loss, correct = 0, 0
# Turn off grad computation to reduce overhead of forward pass for testing.
with torch.no_grad():
for X, y in dataloader:
pred = model(X.to(device))
# Accumulate the total loss on the test data.
test_loss += loss_fn(pred, y.to(device)).item()
# Count the number of correct answers to calculate the accuracy.
correct += (pred.argmax(1) == y.to(device)).type(torch.float).sum().item()
# Compute average loss and the overall accuracy of the model.
test_loss /= size
correct /= size
print(f"Test Error: \n Accuracy: {(100*correct):>0.1f}%, Avg loss: {test_loss:>8f} \n")
# -
for t in range(epochs):
print(f"Epoch {t+1}\n-------------------------------")
train_loop(train_dataloader, model, loss_fn, optimizer)
test_loop(test_dataloader, model, loss_fn)
print("Done!")
| notebooks/cnn_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="m_3F2cWjR5oY" colab_type="text"
# # MNIST DCGAN Example
# + [markdown] id="dA2m6DVoIMPP" colab_type="text"
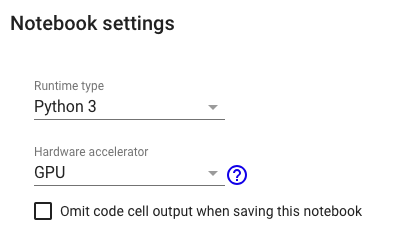
# Note: This notebook is desinged to run with Python3 and GPU runtime.
#
# 
# + [markdown] id="53sGGhqt_00C" colab_type="text"
# This notebook uses TensorFlow 2.x.
# + id="u0PiOopl7MH0" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="a5eec980-4b59-414a-c142-6115766a4646"
# %tensorflow_version 2.x
# + [markdown] id="HqKgjio7IQCa" colab_type="text"
# ####[MDE-01]
# Import modules and set a random seed.
# + id="8uoZRr9eOmwG" colab_type="code" colab={}
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import layers, models, initializers
from tensorflow.keras.datasets import mnist
np.random.seed(20191019)
tf.random.set_seed(20191019)
# + [markdown] id="8NxTNnogIUeV" colab_type="text"
# ####[MDE-02]
# Download the MNIST dataset and store into NumPy arrays.
# + id="0ByKCdhESGpP" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 51} outputId="e4859864-783a-4965-dc99-96ceac51ba5c"
(train_images, train_labels), (test_images, test_labels) = mnist.load_data()
train_images = train_images.reshape(
(len(train_images), 784)).astype('float32') / 255
test_images = test_images.reshape(
(len(test_images), 784)).astype('float32') / 255
train_labels = tf.keras.utils.to_categorical(train_labels, 10)
test_labels = tf.keras.utils.to_categorical(test_labels, 10)
# + [markdown] id="LyJSyjr9JA_G" colab_type="text"
# ####[MDE-03]
# Defina a generator model.
# + id="qKvSYzl9SSkf" colab_type="code" outputId="f525df80-a126-4255-e9d0-d2a2d78d8b42" colab={"base_uri": "https://localhost:8080/", "height": 391}
latent_dim = 64
generator = models.Sequential()
generator.add(
layers.Dense(7*7*128, kernel_initializer=initializers.TruncatedNormal(),
input_shape=(latent_dim,), name='expand'))
generator.add(layers.LeakyReLU(name='leaky_relu1'))
generator.add(layers.Reshape((7, 7, 128), name='reshape'))
generator.add(
layers.Conv2DTranspose(64, 5, strides=2, padding='same',
kernel_initializer=initializers.TruncatedNormal(),
name='deconv1'))
generator.add(layers.LeakyReLU(name='leaky_relu2'))
generator.add(
layers.Conv2DTranspose(1, 5, strides=2, padding='same',
kernel_initializer=initializers.TruncatedNormal(),
activation='sigmoid', name='deconv2'))
generator.add(layers.Flatten(name='flatten'))
generator.summary()
# + [markdown] id="-3DbHA-0Jb5z" colab_type="text"
# ####[MDE-04]
# Defina a discriminator model.
# + id="iyOLjN7kX1Px" colab_type="code" outputId="cd8a3ab3-268e-4f1f-fdbe-2f7ee1506813" colab={"base_uri": "https://localhost:8080/", "height": 425}
discriminator = models.Sequential()
discriminator.add(layers.Reshape((28, 28, 1), input_shape=((28*28,)),
name='reshape'))
discriminator.add(
layers.Conv2D(64, (5, 5), strides=2, padding='same',
kernel_initializer=initializers.TruncatedNormal(),
name='conv1'))
discriminator.add(layers.LeakyReLU(name='leaky_relu1'))
discriminator.add(
layers.Conv2D(128, (5, 5), strides=2, padding='same',
kernel_initializer=initializers.TruncatedNormal(),
name='conv2'))
discriminator.add(layers.LeakyReLU(name='leaky_relu2'))
discriminator.add(layers.Flatten(name='flatten'))
discriminator.add(layers.Dropout(rate=0.4, name='dropout'))
discriminator.add(layers.Dense(1, activation='sigmoid', name='sigmoid'))
discriminator.summary()
# + [markdown] id="9zzzeCD2J-9q" colab_type="text"
# ####[MDE-05]
# Compile the discriminator using the Adam optimizer, and Cross entroy as a loss function.
# + id="XEwdyyX5SeHg" colab_type="code" colab={}
discriminator.compile(optimizer='adam', loss='binary_crossentropy')
# + [markdown] id="is6hVDllKRAi" colab_type="text"
# ####[MDE-06]
# Define an end-to-end GAN model to train the generator.
# + id="VqLQnc3Gd_rR" colab_type="code" outputId="283ee2f0-e6e1-40ca-9dd1-8e0b1fbf7f44" colab={"base_uri": "https://localhost:8080/", "height": 255}
discriminator.trainable = False
gan_input = tf.keras.Input(shape=(latent_dim,))
gan_output = discriminator(generator(gan_input))
gan_model = models.Model(gan_input, gan_output)
gan_model.summary()
# + [markdown] id="PVMUs1WtKrQu" colab_type="text"
# ####[MDE-07]
# Compile the GAN model using the Adam optimizer, and Cross entroy as a loss function.
# + id="2A4dwLwTeywN" colab_type="code" colab={}
gan_model.compile(optimizer='adam', loss='binary_crossentropy')
# + [markdown] id="5FocFzOJK8ac" colab_type="text"
# ####[MDE-08]
# Define some working variables to trace the training process.
# + id="bK7cwT-umD1r" colab_type="code" colab={}
batch_size = 32
image_num = 0
step = 0
examples = []
sample_inputs = np.random.rand(8, latent_dim) * 2.0 - 1.0
examples.append(generator.predict(sample_inputs))
# + [markdown] id="zCGbB86tLNfK" colab_type="text"
# ####[MDE-09]
# Train the model for 40,000 batches.
# + id="33eI8EHqTHgo" colab_type="code" outputId="f13b168b-b917-400d-9a33-614e5a93ef11" colab={"base_uri": "https://localhost:8080/", "height": 187}
for _ in range(40000):
random_inputs = np.random.rand(batch_size, latent_dim) * 2.0 - 1.0
generated_images = generator.predict(random_inputs)
real_images = train_images[image_num : image_num+batch_size]
all_images = np.concatenate([generated_images, real_images])
labels = np.concatenate([np.zeros((batch_size, 1)),
np.ones((batch_size, 1))])
labels += 0.05 * np.random.random(labels.shape)
d_loss = discriminator.train_on_batch(all_images, labels)
random_inputs = np.random.rand(batch_size, latent_dim) * 2.0 - 1.0
fake_labels = np.ones((batch_size, 1))
g_loss = gan_model.train_on_batch(random_inputs, fake_labels)
image_num += batch_size
if image_num + batch_size > len(train_images):
image_num = 0
step += 1
if step % 4000 == 0:
print('step: {}, loss(discriminator, generator): {:6.4f}, {:6.4f}'.format(
step, d_loss, g_loss))
examples.append(generator.predict(sample_inputs))
# + [markdown] id="kIwtyrxVLrWZ" colab_type="text"
# ####[MDE-10]
# Show the progress of sample images.
# + id="yR5SvOxtSu1v" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 755} outputId="7bf14fd8-db1c-4d4d-da26-9353d56fded6"
def show_images(examples):
fig = plt.figure(figsize=(10, 1.2*len(examples)))
c = 1
for images in examples:
for image in images:
subplot = fig.add_subplot(len(examples), 8, c)
subplot.set_xticks([])
subplot.set_yticks([])
subplot.imshow(image.reshape((28, 28)),
vmin=0, vmax=1, cmap=plt.cm.gray_r)
c += 1
show_images(examples)
# + [markdown] id="TBfRGalOL-kq" colab_type="text"
# ####[MDE-10]
# Mount your Google Drive on `/content/gdrive`.
# + id="qwe3Gb8xH-wW" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c461aba8-6059-4996-cf52-c7135b2cf77a"
from google.colab import drive
drive.mount('/content/gdrive')
# + [markdown] id="l1HdULSEMDS9" colab_type="text"
# ####[MDE-11]
# Export the trained model as a file `gan_generator.hd5` on your Google Drive.
# + id="ShhsTXJtH3rg" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="699cbeca-a29e-4bea-d732-cd5118ec575b"
generator.save('/content/gdrive/My Drive/gan_generator.hd5', save_format='h5')
# !ls -lh '/content/gdrive/My Drive/gan_generator.hd5'
| Chapter05/7. MNIST DCGAN example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="view-in-github"
# <a href="https://colab.research.google.com/github/emanbuc/ultrasonic-vision/blob/main/data_visualizzation.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="Ev6kEE8HoC-J"
# # Univariate Plots
# Techniques that you can use to understand each attribute independently.
# + [markdown] id="QcXQHKTfn2zP"
# ## Histograms
# A fast way to get an idea of the distribution of each attribute is to look at histograms.
#
# Histograms group data into bins and provide you a count of the number of observations in each bin. From the shape of the bins you can quickly get a feeling for whether an attribute is Gaussian’, skewed or even has an exponential distribution. It can also help you see possible outliers
# + colab={"base_uri": "https://localhost:8080/", "height": 519} id="IuTL72XokA4J" outputId="72a3f672-d20c-4734-d166-614d24e5627d"
# Univariate Histograms
import matplotlib.pyplot as plt
import pandas
url = "https://raw.githubusercontent.com/emanbuc/ultrasonic-vision/main/sample_acquisitions/7sensors/20210102/20210102_alldata.csv"
names = ['HCSR04_001', 'HCSR04_002', 'HCSR04_003', 'HCSR04_004', 'HCSR04_005', 'HCSR04_006', 'HCSR04_007']
data = pandas.read_csv(url, usecols=names)
print(data)
data.hist()
plt.show()
# + [markdown] id="fXG87Jhxnt0Q"
# ### Density Plots
# Density plots are another way of getting a quick idea of the distribution of each attribute. The plots look like an abstracted histogram with a smooth curve drawn through the top of each bin, much like your eye tried to do with the histograms.
# + colab={"base_uri": "https://localhost:8080/", "height": 266} id="_2EDOu-FnqHb" outputId="2bf92f88-3045-4dc5-a155-4c4874c05e8b"
data.plot(kind='density', subplots=True, layout=(3,3), sharex=False)
plt.show()
# + [markdown] id="4wcPoOzoDQdN"
# La distanza stimata da HCSR04_006, HCSR04_007 è errata: la distanza reale era compresa tra i 20 -70 cm
# + [markdown] id="atbfneCqpAhP"
# ### Box and Whisker Plots
# Another useful way to review the distribution of each attribute is to use Box and Whisker Plots or boxplots for short.
#
# Boxplots summarize the distribution of each attribute, drawing a line for the median (middle value) and a box around the 25th and 75th percentiles (the middle 50% of the data). The whiskers give an idea of the spread of the data and dots outside of the whiskers show candidate outlier values (values that are 1.5 times greater than the size of spread of the middle 50% of the data).
# + colab={"base_uri": "https://localhost:8080/", "height": 266} id="jNOhbNOlo5Jn" outputId="dadcf7c8-2a7f-46c5-9698-206cdb586143"
data.plot(kind='box', subplots=True, layout=(2,4), sharex=False, sharey=False)
plt.show()
# + [markdown] id="qcKDS370pz7m"
# # Multivariate Plots
# Examples of plots with interactions between multiple variables.
# + [markdown] id="ddITQAWEp5q1"
# ## Correlation Matrix Plot
# Correlation gives an indication of how related the changes are between two variables. If two variables change in the same direction they are positively correlated. If the change in opposite directions together (one goes up, one goes down), then they are negatively correlated.
#
# You can calculate the correlation between each pair of attributes. This is called a correlation matrix. You can then plot the correlation matrix and get an idea of which variables have a high correlation with each other.
#
# This is useful to know, because some machine learning algorithms like linear and logistic regression can have poor performance if there are highly correlated input variables in your data.
# + colab={"base_uri": "https://localhost:8080/", "height": 272} id="HpJaKUP3p4cl" outputId="8cafd58d-38a7-4e79-bbfb-8ed3fad023f1"
# Correction Matrix Plot
import matplotlib.pyplot as plt
import pandas
import numpy
correlations = data.corr()
# plot correlation matrix
fig = plt.figure()
ax = fig.add_subplot(111)
cax = ax.matshow(correlations, vmin=-1, vmax=1)
fig.colorbar(cax)
ticks = numpy.arange(0,9,1)
ax.set_xticks(ticks)
ax.set_yticks(ticks)
ax.set_xticklabels(names)
ax.set_yticklabels(names)
plt.show()
# + [markdown] id="hBRQD2g9qz_c"
# ### Scatterplot Matrix
# A scatterplot shows the relationship between two variables as dots in two dimensions, one axis for each attribute. You can create a scatterplot for each pair of attributes in your data. Drawing all these scatterplots together is called a scatterplot matrix.
#
# Scatter plots are useful for spotting structured relationships between variables, like whether you could summarize the relationship between two variables with a line. Attributes with structured relationships may also be correlated and good candidates for removal from your dataset.
# + colab={"base_uri": "https://localhost:8080/", "height": 307} id="6RBG2yBorBa7" outputId="61397f5d-5037-4558-9498-bf7294f822ac"
# Scatterplot Matrix
import matplotlib.pyplot as plt
import pandas
from pandas.plotting import scatter_matrix
scatter_matrix(data)
plt.show()
# + [markdown] id="DVU4nzjzsjDT"
# # New Features
# + colab={"base_uri": "https://localhost:8080/"} id="NSrG52NOsiwj" outputId="a4a8874c-fb1b-4725-8f96-636948a6fd1e"
url = "https://raw.githubusercontent.com/emanbuc/ultrasonic-vision/main/sample_acquisitions/7sensors/20210102/20210102_alldata.csv"
names = ['HCSR04_001', 'HCSR04_002', 'HCSR04_003', 'HCSR04_004', 'HCSR04_005', 'HCSR04_006', 'HCSR04_007',"ObjectClass"]
data = pandas.read_csv(url, usecols=names)
# somma delle distanze dai sensori bassi montati sui pannelli verticali
data['distanceSumHi'] = data.HCSR04_001 + data.HCSR04_002
# somma delle distanze dai sensori alti montati sui pannelli orizzontali
data['distanceSumLow'] = data.HCSR04_003 + data.HCSR04_004
# differenza distanza da sensori tetto
data['differentialDistanceFromRoof65'] = data.HCSR04_006 - data.HCSR04_005
data['differentialDistanceFromRoof67'] = data.HCSR04_006 - data.HCSR04_007
data['differentialDistanceFromRoof57'] = data.HCSR04_005 - data.HCSR04_007
print(data)
# + [markdown] id="wO_kaOncJ_-V"
# # Analisi dati di training per le varie classi di oggetti
# + colab={"base_uri": "https://localhost:8080/", "height": 472} id="0qW2u98_tx76" outputId="409e7686-7929-499d-cb95-e043d933e534"
groupedByClass = data.groupby(['ObjectClass'])
groupedByClass.first()
# + [markdown] id="zB5rmUT-PHft"
# # Elimino letture anomale
# + [markdown] id="tgZIVZ4rYB3f"
# Le distanze misurate quando non ci sono oggetti sono i valori massimi che mi aspetto. Valori superiori indicano una anomalia nella misurazione (es. multipah, scattering, ...)
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} id="QACZuA8TXs1V" outputId="e6bb0f84-2625-4f7b-ad8e-76e1fb8ba4a3"
df_EMPTY_SEVEN = groupedByClass.get_group('EMPTY_SEVEN')
df_EMPTY_SEVEN.plot(kind='density', subplots=True, layout=(2,6), sharex=False, figsize=(30,15))
plt.show()
df_EMPTY_SEVEN
# + [markdown] id="L64MUTanZ8Hx"
# La misura del sensore 007 presenta un errore sistematico che sposta il picco da 50cm a 100cm circa
#
# La distanza reale è di circa 50cm
# + id="ogs9b9ypPOQe"
cleanedData= data[(data["distanceSumLow"] <= 200) & (data["HCSR04_006"] <= 100) & (data["HCSR04_005"] <= 100)]
# + colab={"base_uri": "https://localhost:8080/", "height": 606} id="6XwCcjBBLmfS" outputId="3458a7cb-6283-47cf-c991-30941f9e0711"
newdf = cleanedData.query('ObjectClass == "SQUARE_MILK_45" | ObjectClass == "SQUARE_MILK_90" | ObjectClass == "SOAP_BOTTLE_FRONT" | ObjectClass == "SOAP_BOTTLE_SIDE" | ObjectClass == "BEAN_CAN" | ObjectClass=="RECTANGULAR_BOX" | ObjectClass=="RECTANGULAR_BOX_SIDE" | ObjectClass=="GLASS" | ObjectClass=="EMPTY_SEVEN"')
fig, ax = plt.subplots()
colors = {'SQUARE_MILK_45':'red', 'SQUARE_MILK_90':'blue', 'SOAP_BOTTLE_SIDE':'green', 'SOAP_BOTTLE_FRONT':'black','BEAN_CAN':'yellow','RECTANGULAR_BOX':'pink', 'RECTANGULAR_BOX_SIDE': 'orange','GLASS':'brown',"EMPTY_SEVEN":'grey'}
groupedByClass = newdf.groupby(['ObjectClass'])
for key, group in groupedByClass:
group.plot(ax=ax, kind='scatter', x='distanceSumLow', y='distanceSumHi', label=key, legend=True, color=colors[key],figsize=(20,10))
plt.show()
# + colab={"base_uri": "https://localhost:8080/", "height": 606} id="WZaqeGeLUl8x" outputId="4dae3bd3-16d9-4ac9-e498-ae514a533470"
fig, ax = plt.subplots()
for key, group in groupedByClass:
group.plot(ax=ax, kind='scatter', x='differentialDistanceFromRoof65', y='differentialDistanceFromRoof67', label=key, legend=True, color=colors[key],figsize=(20,10))
plt.show()
| notebooks/.ipynb_checkpoints/data_visualizzation-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="Svyum7G3gHcK"
# ## Ungraded Lab: Convolutional Autoencoders
#
# In this lab, you will use convolution layers to build your autoencoder. This usually leads to better results than dense networks and you will see it in action with the [Fashion MNIST dataset](https://www.tensorflow.org/datasets/catalog/fashion_mnist).
# + [markdown] id="Jk0Tld-U5XFD"
# ## Imports
# + id="3EXwoz-KHtWO"
try:
# # %tensorflow_version only exists in Colab.
# %tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
import tensorflow_datasets as tfds
import numpy as np
import matplotlib.pyplot as plt
# + [markdown] id="e0WGuXlw5bK-"
# ## Prepare the Dataset
# + [markdown] id="aTySDKEhLNLY"
# As before, you will load the train and test sets from TFDS. Notice that we don't flatten the image this time. That's because we will be using convolutional layers later that can deal with 2D images.
# + id="t9F7YsCNIKSA"
def map_image(image, label):
'''Normalizes the image. Returns image as input and label.'''
image = tf.cast(image, dtype=tf.float32)
image = image / 255.0
return image, image
# + id="9ZsciqJXL368" colab={"base_uri": "https://localhost:8080/", "height": 371, "referenced_widgets": ["f8e320c0a49b4db1b3cdf79058f40ac2", "5ab1e33e948345e8a8a273524b2c9475", "fbab6969771c4d7fb02ea48bd7fff528", "<KEY>", "d6920ea10b804e8dbb11bb3ec103efa0", "8585bf370a9d4d82afe572197740738b", "518ca65da37545e98355bd61503ba509", "<KEY>", "32f35279c0e34a04a7fc534d6c0e9904", "<KEY>", "eec84037edd7463fb38c35402a148fc8", "1677ef9392f145a29e55611f66c455ae", "0721c372a32f46f1b4b817dea4b8b1aa", "d315f1a68ab540268ee8b4a6b55b933c", "<KEY>", "508d2d59d9ac44f1812e5d6cd745b486", "0db1d963c96549dbb9e26d4b37a8da6d", "<KEY>", "42e10f3503f64cbe8175cc37540e66bf", "b1f376ba8ed748b880e0c4901f3c51ee", "8bd5e7ca69494a6b8a1e964325416d90", "2e2d18b7ed7848489f44bf5b8edb12b5", "0dce67928dea4a6d887c0516ef1cd0d2", "<KEY>", "e9db3ec39871438dac920e55e50c3f99", "2cea6c3762a842e4aa92fa950664e893", "<KEY>", "4dd4abad7c1e4aa4bc501de2596393c4", "<KEY>", "2072d7180d6c4f1db0e7ecea47f3f9fe", "cfee4743a7ee4ffba4924b4e5afea53e", "<KEY>", "<KEY>", "26f452ab1e4f4150b6ef4e408f2739bd", "<KEY>", "dd463e610ec64fa7b1b53c6e49cc21e5", "b66d0d7551b646af8d5cc42aafe1cc00", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "<KEY>", "fbe74dd1a5234135bb9a0bed5502b003", "<KEY>", "35ac7e69a51644b7be94364352e2d65b", "<KEY>", "<KEY>", "<KEY>", "f061b729e27640c09e9469df315a423b", "<KEY>", "84f706d6faaa4ba5af49e0d7b430224a", "<KEY>", "85cbe3fe969b462aa5cfc3e75c4de37d", "<KEY>", "e05190427d2048578b9f30c4a0cc4934", "83be79673ad14998b3f222a7978697ed"]} outputId="41c9bd2d-f6fe-44be-8c54-9957ec5ee999"
BATCH_SIZE = 128
SHUFFLE_BUFFER_SIZE = 1024
train_dataset = tfds.load('fashion_mnist', as_supervised=True, split="train")
train_dataset = train_dataset.map(map_image)
train_dataset = train_dataset.shuffle(SHUFFLE_BUFFER_SIZE).batch(BATCH_SIZE).repeat()
test_dataset = tfds.load('fashion_mnist', as_supervised=True, split="test")
test_dataset = test_dataset.map(map_image)
test_dataset = test_dataset.batch(BATCH_SIZE).repeat()
# + [markdown] id="uoyz09uKMDn5"
# ## Define the Model
# + [markdown] id="V1-Fw_qnZPV7"
# As mentioned, you will use convolutional layers to build the model. This is composed of three main parts: encoder, bottleneck, and decoder. You will follow the configuration shown in the image below.
# + [markdown] id="568W0TYyY9nl"
# <img src="https://drive.google.com/uc?export=view&id=15zh7bst9KKvciRdCvMAH7kXt3nNkABzO" width="75%" height="75%"/>
# + [markdown] id="O2IvtyIoZnb4"
# The encoder, just like in previous labs, will contract with each additional layer. The features are generated with the Conv2D layers while the max pooling layers reduce the dimensionality.
# + id="wxh8h-UMk2iL"
def encoder(inputs):
'''Defines the encoder with two Conv2D and max pooling layers.'''
conv_1 = tf.keras.layers.Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same')(inputs)
max_pool_1 = tf.keras.layers.MaxPooling2D(pool_size=(2,2))(conv_1)
conv_2 = tf.keras.layers.Conv2D(filters=128, kernel_size=(3,3), activation='relu', padding='same')(max_pool_1)
max_pool_2 = tf.keras.layers.MaxPooling2D(pool_size=(2,2))(conv_2)
return max_pool_2
# + [markdown] id="g9KQYnabazLl"
# A bottleneck layer is used to get more features but without further reducing the dimension afterwards. Another layer is inserted here for visualizing the encoder output.
# + id="wRWmLA3VliDr"
def bottle_neck(inputs):
'''Defines the bottleneck.'''
bottle_neck = tf.keras.layers.Conv2D(filters=256, kernel_size=(3,3), activation='relu', padding='same')(inputs)
encoder_visualization = tf.keras.layers.Conv2D(filters=1, kernel_size=(3,3), activation='sigmoid', padding='same')(bottle_neck)
return bottle_neck, encoder_visualization
# + [markdown] id="FayvcE3ebZxk"
# The decoder will upsample the bottleneck output back to the original image size.
# + id="XZgLt5uAmArk"
def decoder(inputs):
'''Defines the decoder path to upsample back to the original image size.'''
conv_1 = tf.keras.layers.Conv2D(filters=128, kernel_size=(3,3), activation='relu', padding='same')(inputs)
up_sample_1 = tf.keras.layers.UpSampling2D(size=(2,2))(conv_1)
conv_2 = tf.keras.layers.Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same')(up_sample_1)
up_sample_2 = tf.keras.layers.UpSampling2D(size=(2,2))(conv_2)
conv_3 = tf.keras.layers.Conv2D(filters=1, kernel_size=(3,3), activation='sigmoid', padding='same')(up_sample_2)
return conv_3
# + [markdown] id="Dvfhvk9qbvCp"
# You can now build the full autoencoder using the functions above.
# + id="fQKwO64iiOYl"
def convolutional_auto_encoder():
'''Builds the entire autoencoder model.'''
inputs = tf.keras.layers.Input(shape=(28, 28, 1,))
encoder_output = encoder(inputs)
bottleneck_output, encoder_visualization = bottle_neck(encoder_output)
decoder_output = decoder(bottleneck_output)
model = tf.keras.Model(inputs =inputs, outputs=decoder_output)
encoder_model = tf.keras.Model(inputs=inputs, outputs=encoder_visualization)
return model, encoder_model
# + id="1MmS7r0tkuIf" colab={"base_uri": "https://localhost:8080/"} outputId="8fd022d8-6030-4805-b2ac-95171d5d0814"
convolutional_model, convolutional_encoder_model = convolutional_auto_encoder()
convolutional_model.summary()
# + [markdown] id="5FRxRr0LMLCs"
# ## Compile and Train the model
# + id="J0Umj_xaiHL_" colab={"base_uri": "https://localhost:8080/"} outputId="dc4a8c0d-8d48-4c91-c243-9b46f8e2ce16"
train_steps = 60000 // BATCH_SIZE
valid_steps = 60000 // BATCH_SIZE
convolutional_model.compile(optimizer=tf.keras.optimizers.Adam(), loss='binary_crossentropy')
conv_model_history = convolutional_model.fit(train_dataset, steps_per_epoch=train_steps, validation_data=test_dataset, validation_steps=valid_steps, epochs=40)
# + [markdown] id="-8zE9OiAMUd7"
# ## Display sample results
# + [markdown] id="DCUOM7F_cf26"
# As usual, let's see some sample results from the trained model.
# + id="A35RlIqKIsQv"
def display_one_row(disp_images, offset, shape=(28, 28)):
'''Display sample outputs in one row.'''
for idx, test_image in enumerate(disp_images):
plt.subplot(3, 10, offset + idx + 1)
plt.xticks([])
plt.yticks([])
test_image = np.reshape(test_image, shape)
plt.imshow(test_image, cmap='gray')
def display_results(disp_input_images, disp_encoded, disp_predicted, enc_shape=(8,4)):
'''Displays the input, encoded, and decoded output values.'''
plt.figure(figsize=(15, 5))
display_one_row(disp_input_images, 0, shape=(28,28,))
display_one_row(disp_encoded, 10, shape=enc_shape)
display_one_row(disp_predicted, 20, shape=(28,28,))
# + id="qtQyQRxRN_hH" colab={"base_uri": "https://localhost:8080/", "height": 297} outputId="46bd3c11-e102-4d9b-8d11-6bea7c7624de"
# take 1 batch of the dataset
test_dataset = test_dataset.take(1)
# take the input images and put them in a list
output_samples = []
for input_image, image in tfds.as_numpy(test_dataset):
output_samples = input_image
# pick 10 indices
idxs = np.array([1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
# prepare test samples as a batch of 10 images
conv_output_samples = np.array(output_samples[idxs])
conv_output_samples = np.reshape(conv_output_samples, (10, 28, 28, 1))
# get the encoder ouput
encoded = convolutional_encoder_model.predict(conv_output_samples)
# get a prediction for some values in the dataset
predicted = convolutional_model.predict(conv_output_samples)
# display the samples, encodings and decoded values!
display_results(conv_output_samples, encoded, predicted, enc_shape=(7,7))
| Generative Deep Learning with TensorFlow/Week 2 AutoEncoders/Lab_4_FashionMNIST_CNNAutoEncoder.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Neural Machine Translation
#
# Welcome to your first programming assignment for this week!
#
# * You will build a Neural Machine Translation (NMT) model to translate human-readable dates ("25th of June, 2009") into machine-readable dates ("2009-06-25").
# * You will do this using an attention model, one of the most sophisticated sequence-to-sequence models.
#
# This notebook was produced together with NVIDIA's Deep Learning Institute.
# ## <font color='darkblue'>Updates</font>
#
# #### If you were working on the notebook before this update...
# * The current notebook is version "4a".
# * You can find your original work saved in the notebook with the previous version name ("v4")
# * To view the file directory, go to the menu "File->Open", and this will open a new tab that shows the file directory.
#
# #### List of updates
# * Clarified names of variables to be consistent with the lectures and consistent within the assignment
# - pre-attention bi-directional LSTM: the first LSTM that processes the input data.
# - 'a': the hidden state of the pre-attention LSTM.
# - post-attention LSTM: the LSTM that outputs the translation.
# - 's': the hidden state of the post-attention LSTM.
# - energies "e". The output of the dense function that takes "a" and "s" as inputs.
# - All references to "output activation" are updated to "hidden state".
# - "post-activation" sequence model is updated to "post-attention sequence model".
# - 3.1: "Getting the activations from the Network" renamed to "Getting the attention weights from the network."
# - Appropriate mentions of "activation" replaced "attention weights."
# - Sequence of alphas corrected to be a sequence of "a" hidden states.
# * one_step_attention:
# - Provides sample code for each Keras layer, to show how to call the functions.
# - Reminds students to provide the list of hidden states in a specific order, in order to pause the autograder.
# * model
# - Provides sample code for each Keras layer, to show how to call the functions.
# - Added a troubleshooting note about handling errors.
# - Fixed typo: outputs should be of length 10 and not 11.
# * define optimizer and compile model
# - Provides sample code for each Keras layer, to show how to call the functions.
#
# * Spelling, grammar and wording corrections.
# Let's load all the packages you will need for this assignment.
# +
from keras.layers import Bidirectional, Concatenate, Permute, Dot, Input, LSTM, Multiply
from keras.layers import RepeatVector, Dense, Activation, Lambda
from keras.optimizers import Adam
from keras.utils import to_categorical
from keras.models import load_model, Model
import keras.backend as K
import numpy as np
from faker import Faker
import random
from tqdm import tqdm
from babel.dates import format_date
from nmt_utils import *
import matplotlib.pyplot as plt
# %matplotlib inline
# -
# ## 1 - Translating human readable dates into machine readable dates
#
# * The model you will build here could be used to translate from one language to another, such as translating from English to Hindi.
# * However, language translation requires massive datasets and usually takes days of training on GPUs.
# * To give you a place to experiment with these models without using massive datasets, we will perform a simpler "date translation" task.
# * The network will input a date written in a variety of possible formats (*e.g. "the 29th of August 1958", "03/30/1968", "24 JUNE 1987"*)
# * The network will translate them into standardized, machine readable dates (*e.g. "1958-08-29", "1968-03-30", "1987-06-24"*).
# * We will have the network learn to output dates in the common machine-readable format YYYY-MM-DD.
#
# <!--
# Take a look at [nmt_utils.py](./nmt_utils.py) to see all the formatting. Count and figure out how the formats work, you will need this knowledge later. !-->
# ### 1.1 - Dataset
#
# We will train the model on a dataset of 10,000 human readable dates and their equivalent, standardized, machine readable dates. Let's run the following cells to load the dataset and print some examples.
m = 10000
dataset, human_vocab, machine_vocab, inv_machine_vocab = load_dataset(m)
dataset[:10]
# You've loaded:
# - `dataset`: a list of tuples of (human readable date, machine readable date).
# - `human_vocab`: a python dictionary mapping all characters used in the human readable dates to an integer-valued index.
# - `machine_vocab`: a python dictionary mapping all characters used in machine readable dates to an integer-valued index.
# - **Note**: These indices are not necessarily consistent with `human_vocab`.
# - `inv_machine_vocab`: the inverse dictionary of `machine_vocab`, mapping from indices back to characters.
#
# Let's preprocess the data and map the raw text data into the index values.
# - We will set Tx=30
# - We assume Tx is the maximum length of the human readable date.
# - If we get a longer input, we would have to truncate it.
# - We will set Ty=10
# - "YYYY-MM-DD" is 10 characters long.
# +
Tx = 30
Ty = 10
X, Y, Xoh, Yoh = preprocess_data(dataset, human_vocab, machine_vocab, Tx, Ty)
print("X.shape:", X.shape)
print("Y.shape:", Y.shape)
print("Xoh.shape:", Xoh.shape)
print("Yoh.shape:", Yoh.shape)
# -
# You now have:
# - `X`: a processed version of the human readable dates in the training set.
# - Each character in X is replaced by an index (integer) mapped to the character using `human_vocab`.
# - Each date is padded to ensure a length of $T_x$ using a special character (< pad >).
# - `X.shape = (m, Tx)` where m is the number of training examples in a batch.
# - `Y`: a processed version of the machine readable dates in the training set.
# - Each character is replaced by the index (integer) it is mapped to in `machine_vocab`.
# - `Y.shape = (m, Ty)`.
# - `Xoh`: one-hot version of `X`
# - Each index in `X` is converted to the one-hot representation (if the index is 2, the one-hot version has the index position 2 set to 1, and the remaining positions are 0.
# - `Xoh.shape = (m, Tx, len(human_vocab))`
# - `Yoh`: one-hot version of `Y`
# - Each index in `Y` is converted to the one-hot representation.
# - `Yoh.shape = (m, Tx, len(machine_vocab))`.
# - `len(machine_vocab) = 11` since there are 10 numeric digits (0 to 9) and the `-` symbol.
# * Let's also look at some examples of preprocessed training examples.
# * Feel free to play with `index` in the cell below to navigate the dataset and see how source/target dates are preprocessed.
index = 0
print("Source date:", dataset[index][0])
print("Target date:", dataset[index][1])
print()
print("Source after preprocessing (indices):", X[index])
print("Target after preprocessing (indices):", Y[index])
print()
print("Source after preprocessing (one-hot):", Xoh[index])
print("Target after preprocessing (one-hot):", Yoh[index])
# ## 2 - Neural machine translation with attention
#
# * If you had to translate a book's paragraph from French to English, you would not read the whole paragraph, then close the book and translate.
# * Even during the translation process, you would read/re-read and focus on the parts of the French paragraph corresponding to the parts of the English you are writing down.
# * The attention mechanism tells a Neural Machine Translation model where it should pay attention to at any step.
#
#
# ### 2.1 - Attention mechanism
#
# In this part, you will implement the attention mechanism presented in the lecture videos.
# * Here is a figure to remind you how the model works.
# * The diagram on the left shows the attention model.
# * The diagram on the right shows what one "attention" step does to calculate the attention variables $\alpha^{\langle t, t' \rangle}$.
# * The attention variables $\alpha^{\langle t, t' \rangle}$ are used to compute the context variable $context^{\langle t \rangle}$ for each timestep in the output ($t=1, \ldots, T_y$).
#
# <table>
# <td>
# <img src="images/attn_model.png" style="width:500;height:500px;"> <br>
# </td>
# <td>
# <img src="images/attn_mechanism.png" style="width:500;height:500px;"> <br>
# </td>
# </table>
# <caption><center> **Figure 1**: Neural machine translation with attention</center></caption>
#
# Here are some properties of the model that you may notice:
#
# #### Pre-attention and Post-attention LSTMs on both sides of the attention mechanism
# - There are two separate LSTMs in this model (see diagram on the left): pre-attention and post-attention LSTMs.
# - *Pre-attention* Bi-LSTM is the one at the bottom of the picture is a Bi-directional LSTM and comes *before* the attention mechanism.
# - The attention mechanism is shown in the middle of the left-hand diagram.
# - The pre-attention Bi-LSTM goes through $T_x$ time steps
# - *Post-attention* LSTM: at the top of the diagram comes *after* the attention mechanism.
# - The post-attention LSTM goes through $T_y$ time steps.
#
# - The post-attention LSTM passes the hidden state $s^{\langle t \rangle}$ and cell state $c^{\langle t \rangle}$ from one time step to the next.
# #### An LSTM has both a hidden state and cell state
# * In the lecture videos, we were using only a basic RNN for the post-attention sequence model
# * This means that the state captured by the RNN was outputting only the hidden state $s^{\langle t\rangle}$.
# * In this assignment, we are using an LSTM instead of a basic RNN.
# * So the LSTM has both the hidden state $s^{\langle t\rangle}$ and the cell state $c^{\langle t\rangle}$.
# #### Each time step does not use predictions from the previous time step
# * Unlike previous text generation examples earlier in the course, in this model, the post-attention LSTM at time $t$ does not take the previous time step's prediction $y^{\langle t-1 \rangle}$ as input.
# * The post-attention LSTM at time 't' only takes the hidden state $s^{\langle t\rangle}$ and cell state $c^{\langle t\rangle}$ as input.
# * We have designed the model this way because unlike language generation (where adjacent characters are highly correlated) there isn't as strong a dependency between the previous character and the next character in a YYYY-MM-DD date.
# #### Concatenation of hidden states from the forward and backward pre-attention LSTMs
# - $\overrightarrow{a}^{\langle t \rangle}$: hidden state of the forward-direction, pre-attention LSTM.
# - $\overleftarrow{a}^{\langle t \rangle}$: hidden state of the backward-direction, pre-attention LSTM.
# - $a^{\langle t \rangle} = [\overrightarrow{a}^{\langle t \rangle}, \overleftarrow{a}^{\langle t \rangle}]$: the concatenation of the activations of both the forward-direction $\overrightarrow{a}^{\langle t \rangle}$ and backward-directions $\overleftarrow{a}^{\langle t \rangle}$ of the pre-attention Bi-LSTM.
# #### Computing "energies" $e^{\langle t, t' \rangle}$ as a function of $s^{\langle t-1 \rangle}$ and $a^{\langle t' \rangle}$
# - Recall in the lesson videos "Attention Model", at time 6:45 to 8:16, the definition of "e" as a function of $s^{\langle t-1 \rangle}$ and $a^{\langle t \rangle}$.
# - "e" is called the "energies" variable.
# - $s^{\langle t-1 \rangle}$ is the hidden state of the post-attention LSTM
# - $a^{\langle t' \rangle}$ is the hidden state of the pre-attention LSTM.
# - $s^{\langle t-1 \rangle}$ and $a^{\langle t \rangle}$ are fed into a simple neural network, which learns the function to output $e^{\langle t, t' \rangle}$.
# - $e^{\langle t, t' \rangle}$ is then used when computing the attention $a^{\langle t, t' \rangle}$ that $y^{\langle t \rangle}$ should pay to $a^{\langle t' \rangle}$.
# - The diagram on the right of figure 1 uses a `RepeatVector` node to copy $s^{\langle t-1 \rangle}$'s value $T_x$ times.
# - Then it uses `Concatenation` to concatenate $s^{\langle t-1 \rangle}$ and $a^{\langle t \rangle}$.
# - The concatenation of $s^{\langle t-1 \rangle}$ and $a^{\langle t \rangle}$ is fed into a "Dense" layer, which computes $e^{\langle t, t' \rangle}$.
# - $e^{\langle t, t' \rangle}$ is then passed through a softmax to compute $\alpha^{\langle t, t' \rangle}$.
# - Note that the diagram doesn't explicitly show variable $e^{\langle t, t' \rangle}$, but $e^{\langle t, t' \rangle}$ is above the Dense layer and below the Softmax layer in the diagram in the right half of figure 1.
# - We'll explain how to use `RepeatVector` and `Concatenation` in Keras below.
# ### Implementation Details
#
# Let's implement this neural translator. You will start by implementing two functions: `one_step_attention()` and `model()`.
#
# #### one_step_attention
# * The inputs to the one_step_attention at time step $t$ are:
# - $[a^{<1>},a^{<2>}, ..., a^{<T_x>}]$: all hidden states of the pre-attention Bi-LSTM.
# - $s^{<t-1>}$: the previous hidden state of the post-attention LSTM
# * one_step_attention computes:
# - $[\alpha^{<t,1>},\alpha^{<t,2>}, ..., \alpha^{<t,T_x>}]$: the attention weights
# - $context^{ \langle t \rangle }$: the context vector:
#
# $$context^{<t>} = \sum_{t' = 1}^{T_x} \alpha^{<t,t'>}a^{<t'>}\tag{1}$$
#
# ##### Clarifying 'context' and 'c'
# - In the lecture videos, the context was denoted $c^{\langle t \rangle}$
# - In the assignment, we are calling the context $context^{\langle t \rangle}$.
# - This is to avoid confusion with the post-attention LSTM's internal memory cell variable, which is also denoted $c^{\langle t \rangle}$.
# #### Implement `one_step_attention`
#
# **Exercise**: Implement `one_step_attention()`.
#
# * The function `model()` will call the layers in `one_step_attention()` $T_y$ using a for-loop.
# * It is important that all $T_y$ copies have the same weights.
# * It should not reinitialize the weights every time.
# * In other words, all $T_y$ steps should have shared weights.
# * Here's how you can implement layers with shareable weights in Keras:
# 1. Define the layer objects in a variable scope that is outside of the `one_step_attention` function. For example, defining the objects as global variables would work.
# - Note that defining these variables inside the scope of the function `model` would technically work, since `model` will then call the `one_step_attention` function. For the purposes of making grading and troubleshooting easier, we are defining these as global variables. Note that the automatic grader will expect these to be global variables as well.
# 2. Call these objects when propagating the input.
# * We have defined the layers you need as global variables.
# * Please run the following cells to create them.
# * Please note that the automatic grader expects these global variables with the given variable names. For grading purposes, please do not rename the global variables.
# * Please check the Keras documentation to learn more about these layers. The layers are functions. Below are examples of how to call these functions.
# * [RepeatVector()](https://keras.io/layers/core/#repeatvector)
# ```Python
# var_repeated = repeat_layer(var1)
# ```
# * [Concatenate()](https://keras.io/layers/merge/#concatenate)
# ```Python
# concatenated_vars = concatenate_layer([var1,var2,var3])
# ```
# * [Dense()](https://keras.io/layers/core/#dense)
# ```Python
# var_out = dense_layer(var_in)
# ```
# * [Activation()](https://keras.io/layers/core/#activation)
# ```Python
# activation = activation_layer(var_in)
# ```
# * [Dot()](https://keras.io/layers/merge/#dot)
# ```Python
# dot_product = dot_layer([var1,var2])
# ```
# Defined shared layers as global variables
repeator = RepeatVector(Tx)
concatenator = Concatenate(axis=-1)
densor1 = Dense(10, activation = "tanh")
densor2 = Dense(1, activation = "relu")
activator = Activation(softmax, name='attention_weights') # We are using a custom softmax(axis = 1) loaded in this notebook
dotor = Dot(axes = 1)
# +
# GRADED FUNCTION: one_step_attention
def one_step_attention(a, s_prev):
"""
Performs one step of attention: Outputs a context vector computed as a dot product of the attention weights
"alphas" and the hidden states "a" of the Bi-LSTM.
Arguments:
a -- hidden state output of the Bi-LSTM, numpy-array of shape (m, Tx, 2*n_a)
s_prev -- previous hidden state of the (post-attention) LSTM, numpy-array of shape (m, n_s)
Returns:
context -- context vector, input of the next (post-attention) LSTM cell
"""
### START CODE HERE ###
# Use repeator to repeat s_prev to be of shape (m, Tx, n_s) so that you can concatenate it with all hidden states "a" (≈ 1 line)
s_prev = repeator(s_prev)
# Use concatenator to concatenate a and s_prev on the last axis (≈ 1 line)
# For grading purposes, please list 'a' first and 's_prev' second, in this order.
concat = concatenator([a, s_prev])
# Use densor1 to propagate concat through a small fully-connected neural network to compute the "intermediate energies" variable e. (≈1 lines)
e = densor1(concat)
# Use densor2 to propagate e through a small fully-connected neural network to compute the "energies" variable energies. (≈1 lines)
energies = densor2(e)
# Use "activator" on "energies" to compute the attention weights "alphas" (≈ 1 line)
alphas = activator(energies)
# Use dotor together with "alphas" and "a" to compute the context vector to be given to the next (post-attention) LSTM-cell (≈ 1 line)
context = dotor([alphas, a])
### END CODE HERE ###
return context
# -
# You will be able to check the expected output of `one_step_attention()` after you've coded the `model()` function.
# #### model
# * `model` first runs the input through a Bi-LSTM to get $[a^{<1>},a^{<2>}, ..., a^{<T_x>}]$.
# * Then, `model` calls `one_step_attention()` $T_y$ times using a `for` loop. At each iteration of this loop:
# - It gives the computed context vector $context^{<t>}$ to the post-attention LSTM.
# - It runs the output of the post-attention LSTM through a dense layer with softmax activation.
# - The softmax generates a prediction $\hat{y}^{<t>}$.
# **Exercise**: Implement `model()` as explained in figure 1 and the text above. Again, we have defined global layers that will share weights to be used in `model()`.
# +
n_a = 32 # number of units for the pre-attention, bi-directional LSTM's hidden state 'a'
n_s = 64 # number of units for the post-attention LSTM's hidden state "s"
# Please note, this is the post attention LSTM cell.
# For the purposes of passing the automatic grader
# please do not modify this global variable. This will be corrected once the automatic grader is also updated.
post_activation_LSTM_cell = LSTM(n_s, return_state = True) # post-attention LSTM
output_layer = Dense(len(machine_vocab), activation=softmax)
# -
# Now you can use these layers $T_y$ times in a `for` loop to generate the outputs, and their parameters will not be reinitialized. You will have to carry out the following steps:
#
# 1. Propagate the input `X` into a bi-directional LSTM.
# * [Bidirectional](https://keras.io/layers/wrappers/#bidirectional)
# * [LSTM](https://keras.io/layers/recurrent/#lstm)
# * Remember that we want the LSTM to return a full sequence instead of just the last hidden state.
#
# Sample code:
#
# ```Python
# sequence_of_hidden_states = Bidirectional(LSTM(units=..., return_sequences=...))(the_input_X)
# ```
#
# 2. Iterate for $t = 0, \cdots, T_y-1$:
# 1. Call `one_step_attention()`, passing in the sequence of hidden states $[a^{\langle 1 \rangle},a^{\langle 2 \rangle}, ..., a^{ \langle T_x \rangle}]$ from the pre-attention bi-directional LSTM, and the previous hidden state $s^{<t-1>}$ from the post-attention LSTM to calculate the context vector $context^{<t>}$.
# 2. Give $context^{<t>}$ to the post-attention LSTM cell.
# - Remember to pass in the previous hidden-state $s^{\langle t-1\rangle}$ and cell-states $c^{\langle t-1\rangle}$ of this LSTM
# * This outputs the new hidden state $s^{<t>}$ and the new cell state $c^{<t>}$.
#
# Sample code:
# ```Python
# next_hidden_state, _ , next_cell_state =
# post_activation_LSTM_cell(inputs=..., initial_state=[prev_hidden_state, prev_cell_state])
# ```
# Please note that the layer is actually the "post attention LSTM cell". For the purposes of passing the automatic grader, please do not modify the naming of this global variable. This will be fixed when we deploy updates to the automatic grader.
# 3. Apply a dense, softmax layer to $s^{<t>}$, get the output.
# Sample code:
# ```Python
# output = output_layer(inputs=...)
# ```
# 4. Save the output by adding it to the list of outputs.
#
# 3. Create your Keras model instance.
# * It should have three inputs:
# * `X`, the one-hot encoded inputs to the model, of shape ($T_{x}, humanVocabSize)$
# * $s^{\langle 0 \rangle}$, the initial hidden state of the post-attention LSTM
# * $c^{\langle 0 \rangle}$), the initial cell state of the post-attention LSTM
# * The output is the list of outputs.
# Sample code
# ```Python
# model = Model(inputs=[...,...,...], outputs=...)
# ```
# +
# GRADED FUNCTION: model
def model(Tx, Ty, n_a, n_s, human_vocab_size, machine_vocab_size):
"""
Arguments:
Tx -- length of the input sequence
Ty -- length of the output sequence
n_a -- hidden state size of the Bi-LSTM
n_s -- hidden state size of the post-attention LSTM
human_vocab_size -- size of the python dictionary "human_vocab"
machine_vocab_size -- size of the python dictionary "machine_vocab"
Returns:
model -- Keras model instance
"""
# Define the inputs of your model with a shape (Tx,)
# Define s0 (initial hidden state) and c0 (initial cell state)
# for the decoder LSTM with shape (n_s,)
X = Input(shape=(Tx, human_vocab_size))
s0 = Input(shape=(n_s,), name='s0')
c0 = Input(shape=(n_s,), name='c0')
s = s0
c = c0
# Initialize empty list of outputs
outputs = []
### START CODE HERE ###
# Step 1: Define your pre-attention Bi-LSTM. (≈ 1 line)
a = Bidirectional(LSTM(units= n_a, return_sequences= True), input_shape = (m, Tx, 2*n_a))(X)
# Step 2: Iterate for Ty steps
for t in range(Ty):
# Step 2.A: Perform one step of the attention mechanism to get back the context vector at step t (≈ 1 line)
context = one_step_attention(a, s)
# Step 2.B: Apply the post-attention LSTM cell to the "context" vector.
# Don't forget to pass: initial_state = [hidden state, cell state] (≈ 1 line)
s, _, c = post_activation_LSTM_cell(context, initial_state = [s, c])
# Step 2.C: Apply Dense layer to the hidden state output of the post-attention LSTM (≈ 1 line)
out = output_layer(s)
# Step 2.D: Append "out" to the "outputs" list (≈ 1 line)
outputs.append(out)
# Step 3: Create model instance taking three inputs and returning the list of outputs. (≈ 1 line)
model = Model(inputs = [X, s0, c0], output = outputs)
### END CODE HERE ###
return model
# -
# Run the following cell to create your model.
model = model(Tx, Ty, n_a, n_s, len(human_vocab), len(machine_vocab))
# #### Troubleshooting Note
# * If you are getting repeated errors after an initially incorrect implementation of "model", but believe that you have corrected the error, you may still see error messages when building your model.
# * A solution is to save and restart your kernel (or shutdown then restart your notebook), and re-run the cells.
# Let's get a summary of the model to check if it matches the expected output.
model.summary()
# **Expected Output**:
#
# Here is the summary you should see
# <table>
# <tr>
# <td>
# **Total params:**
# </td>
# <td>
# 52,960
# </td>
# </tr>
# <tr>
# <td>
# **Trainable params:**
# </td>
# <td>
# 52,960
# </td>
# </tr>
# <tr>
# <td>
# **Non-trainable params:**
# </td>
# <td>
# 0
# </td>
# </tr>
# <tr>
# <td>
# **bidirectional_1's output shape **
# </td>
# <td>
# (None, 30, 64)
# </td>
# </tr>
# <tr>
# <td>
# **repeat_vector_1's output shape **
# </td>
# <td>
# (None, 30, 64)
# </td>
# </tr>
# <tr>
# <td>
# **concatenate_1's output shape **
# </td>
# <td>
# (None, 30, 128)
# </td>
# </tr>
# <tr>
# <td>
# **attention_weights's output shape **
# </td>
# <td>
# (None, 30, 1)
# </td>
# </tr>
# <tr>
# <td>
# **dot_1's output shape **
# </td>
# <td>
# (None, 1, 64)
# </td>
# </tr>
# <tr>
# <td>
# **dense_3's output shape **
# </td>
# <td>
# (None, 11)
# </td>
# </tr>
# </table>
#
# #### Compile the model
# * After creating your model in Keras, you need to compile it and define the loss function, optimizer and metrics you want to use.
# * Loss function: 'categorical_crossentropy'.
# * Optimizer: [Adam](https://keras.io/optimizers/#adam) [optimizer](https://keras.io/optimizers/#usage-of-optimizers)
# - learning rate = 0.005
# - $\beta_1 = 0.9$
# - $\beta_2 = 0.999$
# - decay = 0.01
# * metric: 'accuracy'
#
# Sample code
# ```Python
# optimizer = Adam(lr=..., beta_1=..., beta_2=..., decay=...)
# model.compile(optimizer=..., loss=..., metrics=[...])
# ```
### START CODE HERE ### (≈2 lines)
opt = Adam(lr=0.005 , beta_1=0.9, beta_2=0.999, decay=0.01)
model.compile(optimizer= opt, loss='categorical_crossentropy', metrics=['accuracy'])
### END CODE HERE ###
# #### Define inputs and outputs, and fit the model
# The last step is to define all your inputs and outputs to fit the model:
# - You have input X of shape $(m = 10000, T_x = 30)$ containing the training examples.
# - You need to create `s0` and `c0` to initialize your `post_attention_LSTM_cell` with zeros.
# - Given the `model()` you coded, you need the "outputs" to be a list of 10 elements of shape (m, T_y).
# - The list `outputs[i][0], ..., outputs[i][Ty]` represents the true labels (characters) corresponding to the $i^{th}$ training example (`X[i]`).
# - `outputs[i][j]` is the true label of the $j^{th}$ character in the $i^{th}$ training example.
s0 = np.zeros((m, n_s))
c0 = np.zeros((m, n_s))
outputs = list(Yoh.swapaxes(0,1))
# Let's now fit the model and run it for one epoch.
model.fit([Xoh, s0, c0], outputs, epochs=1, batch_size=100)
# While training you can see the loss as well as the accuracy on each of the 10 positions of the output. The table below gives you an example of what the accuracies could be if the batch had 2 examples:
#
# <img src="images/table.png" style="width:700;height:200px;"> <br>
# <caption><center>Thus, `dense_2_acc_8: 0.89` means that you are predicting the 7th character of the output correctly 89% of the time in the current batch of data. </center></caption>
#
#
# We have run this model for longer, and saved the weights. Run the next cell to load our weights. (By training a model for several minutes, you should be able to obtain a model of similar accuracy, but loading our model will save you time.)
model.load_weights('models/model.h5')
# You can now see the results on new examples.
EXAMPLES = ['3 May 1979', '5 April 09', '21th of August 2016', 'Tue 10 Jul 2007', 'Saturday May 9 2018', 'March 3 2001', 'March 3rd 2001', '1 March 2001']
for example in EXAMPLES:
source = string_to_int(example, Tx, human_vocab)
source = np.array(list(map(lambda x: to_categorical(x, num_classes=len(human_vocab)), source))).swapaxes(0,1)
prediction = model.predict([source, s0, c0])
prediction = np.argmax(prediction, axis = -1)
output = [inv_machine_vocab[int(i)] for i in prediction]
print("source:", example)
print("output:", ''.join(output),"\n")
# You can also change these examples to test with your own examples. The next part will give you a better sense of what the attention mechanism is doing--i.e., what part of the input the network is paying attention to when generating a particular output character.
# ## 3 - Visualizing Attention (Optional / Ungraded)
#
# Since the problem has a fixed output length of 10, it is also possible to carry out this task using 10 different softmax units to generate the 10 characters of the output. But one advantage of the attention model is that each part of the output (such as the month) knows it needs to depend only on a small part of the input (the characters in the input giving the month). We can visualize what each part of the output is looking at which part of the input.
#
# Consider the task of translating "Saturday 9 May 2018" to "2018-05-09". If we visualize the computed $\alpha^{\langle t, t' \rangle}$ we get this:
#
# <img src="images/date_attention.png" style="width:600;height:300px;"> <br>
# <caption><center> **Figure 8**: Full Attention Map</center></caption>
#
# Notice how the output ignores the "Saturday" portion of the input. None of the output timesteps are paying much attention to that portion of the input. We also see that 9 has been translated as 09 and May has been correctly translated into 05, with the output paying attention to the parts of the input it needs to to make the translation. The year mostly requires it to pay attention to the input's "18" in order to generate "2018."
# ### 3.1 - Getting the attention weights from the network
#
# Lets now visualize the attention values in your network. We'll propagate an example through the network, then visualize the values of $\alpha^{\langle t, t' \rangle}$.
#
# To figure out where the attention values are located, let's start by printing a summary of the model .
model.summary()
# Navigate through the output of `model.summary()` above. You can see that the layer named `attention_weights` outputs the `alphas` of shape (m, 30, 1) before `dot_2` computes the context vector for every time step $t = 0, \ldots, T_y-1$. Let's get the attention weights from this layer.
#
# The function `attention_map()` pulls out the attention values from your model and plots them.
attention_map = plot_attention_map(model, human_vocab, inv_machine_vocab, "Tuesday 09 Oct 1993", num = 7, n_s = 64);
# On the generated plot you can observe the values of the attention weights for each character of the predicted output. Examine this plot and check that the places where the network is paying attention makes sense to you.
#
# In the date translation application, you will observe that most of the time attention helps predict the year, and doesn't have much impact on predicting the day or month.
# ### Congratulations!
#
#
# You have come to the end of this assignment
#
# ## Here's what you should remember
#
# - Machine translation models can be used to map from one sequence to another. They are useful not just for translating human languages (like French->English) but also for tasks like date format translation.
# - An attention mechanism allows a network to focus on the most relevant parts of the input when producing a specific part of the output.
# - A network using an attention mechanism can translate from inputs of length $T_x$ to outputs of length $T_y$, where $T_x$ and $T_y$ can be different.
# - You can visualize attention weights $\alpha^{\langle t,t' \rangle}$ to see what the network is paying attention to while generating each output.
# Congratulations on finishing this assignment! You are now able to implement an attention model and use it to learn complex mappings from one sequence to another.
| Neural_machine_translation_with_attention_v4a.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# +
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams.update({
"text.usetex": True,
"font.family": "sans-serif",
"font.sans-serif": ["Helvetica"]})
# for Palatino and other serif fonts use:
plt.rcParams.update({
"text.usetex": True,
"font.family": "serif",
"font.serif": ["Palatino"],
})
# %matplotlib inline
import pymesh
#https://pymesh.readthedocs.io/en/latest/basic.html
import time
import multiprocessing
import meshplot
import itertools
from multiprocessing import Pool # Process pool
from multiprocessing import sharedctypes
plt.rcParams['xtick.labelsize'] = 14
plt.rcParams['ytick.labelsize'] = 14
from mshmthds import *
from BYORP_YORP import *
import sys
# -
#
# The surface thermal inertia is neglected, so that thermal radiation is re-emitted with no time lag, and the reflected and thermally radiated components are assumed Lambertian (isotropic) and so emitted with flux
# parallel to the local surface normal. We ignore heat conduction. The surface is described with a closed
# triangular mesh.
#
#
# The radiation force from the $i$-th facet is
# $$ {\bf F}_i = - \frac{F_\odot}{c} {S_i} (\hat {\bf n}_i \cdot \hat {\bf s}_\odot) \hat {\bf n}_i $$
# where $S_i$ is the area of the $i$-th facet and $\hat {\bf n}_i$ is its surface normal.
# Here $F_\odot$ is the solar radiation flux and $c$ is the speed of light.
# The direction of the Sun is $\hat {\bf s}_\odot$.
#
# The total Yarkovsky force is a sum over all the facets
# $${\bf F}_Y = \sum_{i: \hat {\bf n}_i \cdot \hat {\bf s}_\odot >0} {\bf F}_i $$
# Only facets on the day side or with $\hat {\bf n}_i \cdot \hat {\bf s}_\odot >0$
# are included in the sum.
#
# The torque affecting the binary orbit from a single facet is
# $$ {\boldsymbol \tau}_{i,B} =
# \begin{cases}
# - \frac{F_\odot}{c} {S_i} (\hat {\bf n}_i \cdot \hat {\bf s}_\odot) ( {\bf a}_B \times \hat {\bf n}_i)
# & \mbox{if } \hat {\bf n}_i \cdot \hat {\bf s}_\odot >0 \\
# 0 & \mbox{otherwise}
# \end{cases}
# $$
# where ${\bf a}_B$ is the secondary's radial vector from the binary center of mass.
#
#
# The torque affecting the binary orbit is the sum of the torques from each facet and should be an average
# over the orbit around the Sun and
# over the binary orbit and spin of the secondary.
# $$ {\boldsymbol \tau}_{BY} = \frac{1}{T} \int_0^T dt\ \sum_{i: \hat {\bf n}_i \cdot \hat {\bf s}_\odot >0}
# {\boldsymbol \tau}_{i,B} $$
#
#
# If $\hat {\bf l}$ is the binary orbit normal then
# $$ {\boldsymbol \tau}_{BY} \cdot \hat {\bf l} $$
# changes the binary's orbital angular momentum and causes binary orbit migration.
#
#
# The torque affecting the spin (also known as YORP) instantaneously depends on
# the radii of each facit ${\bf r}_i$ from the asteroid center of mass
# $$ {\boldsymbol \tau}_{i,s} = \begin{cases}
# - \frac{F_\odot}{c} {S_i} (\hat {\bf n}_i \cdot \hat {\bf s}_\odot) ({\bf r}_i \times \hat{\bf n}_i)
# & \mbox{if } \hat {\bf n}_i \cdot \hat {\bf s}_\odot >0 \\
# 0 & \mbox{otherwise}
# \end{cases}$$
#
#
# $$ {\boldsymbol \tau}_Y = \frac{1}{T} \int_0^T dt \ \sum_{i: \hat {\bf n}_i \cdot \hat {\bf s}_\odot >0} {\boldsymbol \tau}_{i,s} $$
# where the average is done over the orbit about the Sun and the spin of the asteroid.
# If the spin axis is $\hat {\boldsymbol \omega}$ then
# $$ {\boldsymbol \tau}_Y \cdot \hat {\boldsymbol \omega} $$ gives the body spin up or spin down rate.
#
#
# In practice we average over the Sun's directions first and then average over spin (for YORP) or and spin and binary orbit direction (for BYORP) afterward.
#
#
# <b> Units </b>
#
# For our calculation are $F_\odot/c = 1$.
#
# For YORP $R=1$.
# For BYORP $a_B = 1$ and $R=1$ (in the surface area).
#
# Here $R$ is volume equivalent sphere radius.
#
# To put in physical units:
#
# Multiply ${\boldsymbol \tau}_Y$ by $\frac{F_\odot R^3}{c}$.
#
# Multiply ${\boldsymbol \tau}_{BY}$ by $\frac{F_\odot R^2 a_B}{c}$.
#
# Alternatively we are computing:
#
# ${\boldsymbol \tau}_Y \times \frac{c}{F_\odot R^3} $
#
# ${\boldsymbol \tau}_{BY} \times \frac{c}{F_\odot R^2 a_B} $
#
#
# To get the rate the spin changes for YORP
#
# $\dot \omega = \frac{ {\boldsymbol \tau}_Y \cdot \hat {\bf s} }{C} $
#
# where $C$ is the moment of inertia about the spin axis.
#
# To order of magnitude what we are computing can be multiplied by
# $\frac{F_\odot R^3}{c MR^2} $ to estimate $\dot \omega$
# and by $\frac{F_\odot R^3}{c MR^2 \omega} $
# to estimate $\dot \epsilon$.
#
# To get the rate that obliquity changes for YORP
#
# $\dot \epsilon = \frac{ {\boldsymbol \tau}_Y \cdot \hat {\boldsymbol \phi} }{C \omega} $
#
# where unit vector $\hat {\boldsymbol \phi}$ is in the xy plane (ecliptic) and is perpendicular to the spin axis.
#
# To get the semi-major axis drift rate for BYORP
#
# $ \dot a_B = \frac{2 {\boldsymbol \tau}_{BY} \cdot \hat {\bf l}}{M n_Ba_B} $
#
# where $M$ is the secondary mass, $n_B$ and $a_B$ are binary orbit mean motion and semi-major axis.
#
# To order of magnitude to get the drift rate we multiply what we are getting by
# $\frac{F_\odot R^2 a_B}{c} \times \frac{1}{M n_B a_B}$.
#
#
# Dimensionless numbers used by Steiberg+10 (eqns 19,48)
#
# $f_{Y} \equiv \tau_{Y} \frac{3}{2} \frac{c}{\pi R^3 F_\odot}$
#
# $f_{BY} \equiv \tau_{BY} \frac{3}{2} \frac{c}{\pi R^2 a_B F_\odot}$
#
# Our computed values are the same as theirs except for a factor of 3/2
# (but they have a 2/3 in their torque) and a factor of $\pi$.
# We need to divide by $\pi$ to have values consistent with theirs.
#
# <b> Assumptions:</b>
#
# Circular orbit for binary.
#
# Circuilar orbit for binary around Sun.
#
# No shadows.
#
# No conduction. Lambertian isotropic emission. No thermal lag.
#
# We neglect distance of facet centroids from secondary center of mass when computing BYORP.
#
# Coordinate system:
# binary orbit is kept in xy plane
#
# Compare YORP on primary to BYORP on secondary.
#
# $\frac{\tau_{Yp}}{\tau_{BY} }\sim \frac{R_p^2 }{R_s^2 } \frac{R_p }{a_B }\frac{f_Y}{ f_{BY}}$
#
# For Didymos, this is about $8 f_Y/f_{BY}$.
squannit = pymesh.load_mesh("kw4b.obj")
SIZEOFMESH = 0.03
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
folder = 'Squannit'+ str(len(short_squannit.faces))
print(folder)
# +
# compute the BYORP torque on body as a function of obliquity
# for a given inclination and precession angle
# returns obliquity and torque arrays
#Create the mesh
squannit = pymesh.load_mesh("kw4b.obj")
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
vertices = short_squannit.vertices
faces = short_squannit.faces
#Simulation Parameters
size = 20 # Number of Obliquities
block_size = 1 # Obliquities per subprocess
incl = 0; phi_prec=0
tau_s_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_tau_s = sharedctypes.RawArray(tau_s_arr._type_, tau_s_arr)
tau_o_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_tau_o = sharedctypes.RawArray(tau_o_arr._type_, tau_o_arr)
o_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_o = sharedctypes.RawArray(o_arr._type_, o_arr)
# YORP Methods
# compute the YORP torque on body as a function of obliquity
# here obliquity is w.r.t Sun
# returns obliquity and torque arrays
def aj_alt_obliq_Y_fig(nobliq):
body = pymesh.form_mesh(vertices, faces)
body.add_attribute("face_area")
body.add_attribute("face_normal")
nphi_Sun=36 # number of solar positions
nphi = 36 # number of spin positions
# nobliq = 20 # number of obliquities
dobliq = np.pi/20
tau_s_arr = np.ctypeslib.as_array(shared_array_tau_s) # to store torques
tau_o_arr = np.ctypeslib.as_array(shared_array_tau_o) # to store torques
o_arr = np.ctypeslib.as_array(shared_array_o) # to store obliquities in degrees
print(f'Starting {nobliq}')
for i in range(nobliq, nobliq+block_size):
obliquity=i*dobliq
tau_Y_x,tau_Y_y,tau_Y_z,tau_s,tau_o =compute_Y(body,obliquity,nphi,nphi_Sun)
#print(tau_s)
tau_s_arr[i] = tau_s
tau_o_arr[i] = tau_o
o_arr[i] = obliquity*180/np.pi
print(f'Finished {nobliq}')
return o_arr, tau_s_arr, tau_o_arr
start=time.perf_counter()
p=Pool()
# compute YORPs as a function of obliquity (single body, obliquity w.r.t Solar orbit)
print('Starting Analysis')
res = p.map(aj_alt_obliq_Y_fig, range(size))
multi_o_arr = np.ctypeslib.as_array(shared_array_o)
multi_tau_arr_s = np.ctypeslib.as_array(shared_array_tau_s)
multi_tau_arr_o = np.ctypeslib.as_array(shared_array_tau_o)
end = time.perf_counter()
print(f'Time to complete {round(end - start,2)} second(s)')
# o_arr, tau_s_arr, tau_o_arr = obliq_Y_fig(body)
# also check the sphere for YORP
# o_arr2, tau_s_arr2,tau_o_arr2 = obliq_Y_fig(sphere) # note y axis
# compare the two YORPs
fig,ax = plt.subplots(1,1,figsize=(5,4),dpi=150)
# ax.plot(o_arr2,tau_s_arr2,'go-',label='sphere')
#ax.plot(o_arr2,tau_o_arr2,'bo-',label='sphere')
ax.plot(multi_o_arr,multi_tau_arr_s,'rD-',label=r'body, $s$')
ax.plot(multi_o_arr,multi_tau_arr_o,'D:',label='body, $o$', color='orange')
ax.set_xlabel('obliquity (deg)',fontsize=16)
ax.set_ylabel(r'${ \tau}_Y \cdot \hat{ s}, { \tau}_Y \cdot \hat{\phi}$',fontsize=16)
ax.legend()
fig.savefig(folder+'/ObliqY')
# +
# compute the BYORP torque on body as a function of inclination
# for a given obliquity and precession angle
# returns inclination and torque arrays
#Create the mesh
squannit = pymesh.load_mesh("kw4b.obj")
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
vertices = short_squannit.vertices
faces = short_squannit.faces
#Simulation Parameters
size = 20 # Number of Inclinations
block_size = 1 # Obliquities per subprocess
obliquity = 0; phi_prec=0
tau_l_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_tau = sharedctypes.RawArray(tau_l_arr._type_, tau_l_arr)
i_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_i = sharedctypes.RawArray(i_arr._type_, i_arr)
def aj_alt_obliq_BY_fig(nincl):
body = pymesh.form_mesh(vertices, faces)
body.add_attribute("face_area")
body.add_attribute("face_normal")
#mesh.add_attribute("vertex_normal")
body.add_attribute("face_centroid")
f_area = body.get_attribute("face_area")
phi0=0
nphi_Sun=36 # number of solar positions
nphi = 36 # number of spin positions
dincl = np.pi/size
tau_l_arr = np.ctypeslib.as_array(shared_array_tau) # to store torques
i_arr = np.ctypeslib.as_array(shared_array_i)
print(f'Started {nincl}')
for i in range(nincl, nincl+block_size):
incl=i*dincl
tau_BY_x,tau_BY_y,tau_BY_z, tau_l =compute_BY(body,obliquity,nphi,nphi_Sun,incl,phi0,phi_prec)
i_arr[i] = incl*180/np.pi
tau_l_arr[i] = tau_l
print(f'Finished {nincl}')
return i_arr,tau_l_arr
# compute BYORPs as a function of inclination
# i_arr,tau_l_arr = obliq_BY_fig(body,obliquity,phi_prec)
start = time.perf_counter()
p = Pool()
#Complete BYORP analysis
print('Starting analysis')
res = p.map(aj_alt_obliq_BY_fig, range(20))
multi_i_arr = np.ctypeslib.as_array(shared_array_o)
multi_tau_l_arr = np.ctypeslib.as_array(shared_array_tau)
end = time.perf_counter()#Print Time
print(f'D8: time to complete {round(end - start,2)} second(s)')
fig,ax = plt.subplots(1,1,figsize=(5,4),dpi=150)
ax.plot(multi_i_arr,multi_tau_l_arr,'rD-',label='body')
ax.set_xlabel('inclination (deg)',fontsize=16)
ax.set_ylabel(r'${\tau}_{BY} \cdot \hat{l}$',fontsize=16)
ax.legend()
fig.savefig(folder+'/InclBY')
# +
# compute the BYORP torque on body as a function of obliquity
# for a given inclination and precession angle
# returns obliquity and torque arrays
#Create the mesh
squannit = pymesh.load_mesh("kw4b.obj")
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
vertices = short_squannit.vertices
faces = short_squannit.faces
#Simulation Parameters
size = 60 # Number of Obliquities
block_size = 1 # Obliquities per subprocess
incl = 0; phi_prec=0
tau_l_arr = np.ctypeslib.as_ctypes(np.zeros((60)))
shared_array_tau = sharedctypes.RawArray(tau_l_arr._type_, tau_l_arr)
o_arr = np.ctypeslib.as_ctypes(np.zeros((60)))
shared_array_o = sharedctypes.RawArray(o_arr._type_, o_arr)
def aj_alt_obliq_BY_fig2(nobliq):
body = pymesh.form_mesh(vertices, faces)
body.add_attribute("face_area")
body.add_attribute("face_normal")
#mesh.add_attribute("vertex_normal")
body.add_attribute("face_centroid")
f_area = body.get_attribute("face_area")
phi0=0
nphi_Sun=36 # number of solar positions
nphi = 36 # number of spin positions
dobliq = np.pi/60
tau_l_arr = np.ctypeslib.as_array(shared_array_tau) # to store torques
o_arr = np.ctypeslib.as_array(shared_array_o)
for i in range(nobliq, nobliq+block_size):
obliquity=i*dobliq
tau_BY_x,tau_BY_y,tau_BY_z, tau_l = compute_BY(body,obliquity,nphi,nphi_Sun,incl,phi0,phi_prec)
o_arr[i] = obliquity*180/np.pi
tau_l_arr[i] = tau_l
print(f'Finished {nobliq}')
return o_arr,tau_l_arr
start = time.perf_counter()
p = Pool()
#Complete BYORP analysis
print('Starting analysis')
res = p.map(aj_alt_obliq_BY_fig2, range(60))
multi_o_arr = np.ctypeslib.as_array(shared_array_o)
multi_tau_l_arr = np.ctypeslib.as_array(shared_array_tau)
end = time.perf_counter()#Print Time
print(f'D8: time to complete {round(end - start,2)} second(s)')
fig,ax = plt.subplots(1,1,figsize=(5,4),dpi=300)
ax.plot(multi_o_arr,multi_tau_l_arr,'go-',label='sphere')
ax.plot(multi_o_arr,multi_tau_l_arr,'rD-',label='body')
ax.set_xlabel('obliquity (deg)',fontsize=16)
ax.set_ylabel(r'${ \tau}_{BY} \cdot \hat{l}$',fontsize=16)
ax.legend()
fig.savefig(folder+'/ObliqBY')
# +
# compute the BYORP torque on body as a function of precession angle
# for a given obliquity and inclination
# returns precession angle and torque arrays
#Create the mesh
squannit = pymesh.load_mesh("kw4b.obj")
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
vertices = short_squannit.vertices
faces = short_squannit.faces
#Simulation Parameters
size = 30 # Number of Precession Angles
block_size = 1 # Precession < per subprocess
incl = 0; obliquity=np.pi/4
tau_l_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_tau = sharedctypes.RawArray(tau_l_arr._type_, tau_l_arr)
p_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_p = sharedctypes.RawArray(p_arr._type_, p_arr)
def aj_alt_obliq_BY_fig3(nprec):
body = pymesh.form_mesh(vertices, faces)
body.add_attribute("face_area")
body.add_attribute("face_normal")
#mesh.add_attribute("vertex_normal")
body.add_attribute("face_centroid")
f_area = body.get_attribute("face_area")
phi0=0
nphi_Sun=36 # number of solar positions
nphi = 36 # number of spin positions
dprec = np.pi/size # only goes from 0 to pi
tau_l_arr = np.ctypeslib.as_array(shared_array_tau) # to store torques
p_arr = np.ctypeslib.as_array(shared_array_p)
print(f'Starting {nprec}')
for i in range(nprec, nprec+block_size):
phi_prec=i*dprec
tau_BY_x,tau_BY_y,tau_BY_z, tau_l =compute_BY(body,obliquity,nphi,nphi_Sun,incl,phi0,phi_prec)
p_arr[i] = phi_prec*180/np.pi
tau_l_arr[i] = tau_l
print(f'Finished {nprec}')
return p_arr,tau_l_arr
# compute BYORPs as a function of precession angle, seems not sensitive to precession angle
# p_arr,tau_l_arr = obliq_BY_fig3(body,obliquity,incl)
start = time.perf_counter()
p = Pool()
#Complete BYORP analysis
print('Starting analysis')
res = p.map(aj_alt_obliq_BY_fig3, range(size))
multi_p_arr = np.ctypeslib.as_array(shared_array_p)
multi_tau_l_arr = np.ctypeslib.as_array(shared_array_tau)
end = time.perf_counter()#Print Time
print(f'D8: time to complete {round(end - start,2)} second(s)')
fig,ax = plt.subplots(1,1,figsize=(5,4),dpi=150)
ax.plot(multi_p_arr,multi_tau_l_arr,'rD-',label='body')
ax.set_xlabel('precession angle (deg)',fontsize=16)
ax.set_ylabel(r'${ \tau}_{BY} \cdot \hat{l}$',fontsize=16)
ax.legend()
fig.savefig(folder+'/PrecessBY')
# +
# compute the BYORP torque on body as a function of libration angle phi0
# for a given obliquity and inclination and precession angle
# returns libration angle and torque arrays
#Create the mesh
squannit = pymesh.load_mesh("kw4b.obj")
short_squannit, info = pymesh.collapse_short_edges(squannit, SIZEOFMESH)
vertices = short_squannit.vertices
faces = short_squannit.faces
#Simulation Parameters
size = 20 # Number of Libration Angles
block_size = 1 # Precession < per subprocess
incl = 0; phi_prec=0; obliquity = np.pi/4
tau_l_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_tau = sharedctypes.RawArray(tau_l_arr._type_, tau_l_arr)
l_arr = np.ctypeslib.as_ctypes(np.zeros((size)))
shared_array_l = sharedctypes.RawArray(l_arr._type_, l_arr)
def aj_alt_obliq_BY_fig4(nlib):
body = pymesh.form_mesh(vertices, faces)
body.add_attribute("face_area")
body.add_attribute("face_normal")
#mesh.add_attribute("vertex_normal")
body.add_attribute("face_centroid")
f_area = body.get_attribute("face_area")
phi0=0
nphi_Sun=36 # number of solar positions
nphi = 36 # number of spin positions
dlib = 0.5*np.pi/size # going from -pi/4 to pi/4
tau_l_arr = np.ctypeslib.as_array(shared_array_tau) # to store torques
l_arr = np.ctypeslib.as_array(shared_array_l)
print(f'Staring {nlib}')
for i in range(nlib, nlib + block_size):
phi0=i*dlib - np.pi/4
tau_BY_x,tau_BY_y,tau_BY_z, tau_l =compute_BY(body,obliquity,nphi,nphi_Sun,incl,phi0,phi_prec)
l_arr[i] = phi0*180/np.pi
tau_l_arr[i] = tau_l
print(f'Finished {nlib}')
return l_arr,tau_l_arr
# compute BYORPs as a function of libration angle
# l_arr,tau_l_arr=obliq_BY_fig4(body,obliquity,incl,phi_prec)
#plt.savefig('tau_BY_lib.png')
# fairly sensitive to libration angle
start = time.perf_counter()
p = Pool()
#Complete BYORP analysis
print('Starting analysis')
res = p.map(aj_alt_obliq_BY_fig4, range(size))
multi_l_arr = np.ctypeslib.as_array(shared_array_l)
multi_tau_l_arr = np.ctypeslib.as_array(shared_array_tau)
end = time.perf_counter()#Print Time
print(f'D8: time to complete {round(end - start,2)} second(s)')
fig,ax = plt.subplots(1,1,figsize=(5,4),dpi=150)
#ax.plot(o_arr2,tau_l_arr2,'go-',label='sphere')
ax.plot(multi_l_arr,multi_tau_l_arr,'rD-',label='body')
ax.set_xlabel('libration angle (deg)',fontsize=16)
ax.set_ylabel(r'${ \tau}_{BY} \cdot \hat{l}$',fontsize=16)
ax.legend()
fig.savefig(folder+'/LibBY')
# -
| myexamples/pylab/BYORP4.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: cv-homework
# language: python
# name: cv-homework
# ---
# # Excercise 2
# ## Import packages
# +
import numpy as np
import scipy.ndimage
import matplotlib.pyplot as plt
import skimage.io
import skimage.color
import skimage.exposure
import time
import math
import random
# -
# ## Task 1
# (2 points)
# 1. Use $f = loss(100)$ which creates a 1D array that mimics a loss curve of some neural network.
# 2. Implement the naive $\mathcal{O}(n w)$ mean filter to smooth the loss.
# 3. Implement the improved $\mathcal{O}(n)$ mean filter to smooth the loss.
# 4. Show all three curves in a single plot with a legend.
# 5. Compare the runtime between the naive and improved implementation.
# +
# 1.
def loss(n):
return 1.0 / (0.01 * (np.arange(0, n) + 1)) + 3.0 * np.random.uniform(-1.00, 1.00, n)
# TODO: generate f with loss helper function
f = loss(100)
# 2.
def naive_running_mean(f, w):
'''
Apply the naive running mean filter and return the smoothed values.
f -- contains the values
w -- window size
'''
g = np.zeros(f.shape)
for x in range(w, len(f) - w):
sum = 0
for xp in range(x-w, x+w+1):
sum += f[xp]
g[x] = sum / (2 * w + 1)
return g
# 3.
def improved_running_mean(f, w):
'''
Apply the improved running mean filter and return the smoothed values.
f -- contains the values
w -- window size
'''
f_summed = np.zeros(f.shape)
f_summed[0] = f[0]
for i in range(1, len(f)):
f_summed[i] = f_summed[i-1] + f[i]
g = np.zeros(f.shape)
g[w] = f_summed[2*w] / (2 * w + 1)
for x in range(w + 1, len(f) - w):
g[x] = (f_summed[x+w] - f_summed[x-w-1]) / (2 * w + 1)
return g
# 4.
ws = 5
# TODO: Plot all three curves in a single plot
g_naive = naive_running_mean(f, ws)
g_improved = improved_running_mean(f, ws)
plt.plot(f, label="original f")
plt.plot(g_naive, label="naive running mean")
plt.plot(g_improved, label = "improved running mean")
plt.legend()
plt.show()
# 5.
n = 1000000
f = loss(n)
# TODO: Measure and print the runtime of both methods in Milliseconds
print("Measuring running time for " + str(n) + " data points")
start = time.time()
naive_running_mean(f, ws)
end = time.time()
print("Running time of naive running mean: " + str(end - start))
start = time.time()
improved_running_mean(f, ws)
end = time.time()
print("Running time of impoved running mean: " + str(end - start))
# -
# ## Task 2
# (4 points)
# 1. Create numpy arrays for the horizontal and vertical Sobel kernel.
# $$
# S_x =\begin{bmatrix}
# -1 & -2 & -1 \\
# 0 & 0 & 0 \\
# 1 & 2 & 1
# \end{bmatrix}
# $$
#
# $$
# S_y =\begin{bmatrix}
# -1 & 0 & 1 \\
# -2 & 0 & 2 \\
# -1 & 0 & 1
# \end{bmatrix}
# $$
#
# 2. Implement the $\textit{convolve}$ function with four nested loops.
# 3. Load the image from the last exercise and detect its edges using the Sobel kernels and the $\textit{convolve}$ function.
# 4. Implement the 1D $\textit{convolve_fast}$ function. Apply the seperated Sobel operators $u$ and $v$. Plot the result together with $\textit{convolve(S_x)}$ and compare the runtime.
# 5. Implement a function that seperates a 2D kernel into 2 1D kernels.
# 6. (Bonus) Use the function for a gaussian blur kernel and apply it on the image.
# +
# 1
# TODO Create S_x and S_y
S_x = np.array([[-1, 0, 1], [-2, 0, 2], [-1, 0, 1]])
S_y = np.array([[-1, -2, -1], [0, 0, 0], [1, 2, 1]])
print('Horizontal Sobel kernel:\n', S_x)
print('Vertical Sobel kernel:\n', S_y)
# -
# 2
def convolve(arr, ker):
'''
Convolve the array using kernel K with four nested for loops.
arr -- 2D array that gets convolved
ker -- kernel
'''
w = math.floor(ker.shape[0] / 2)
arr_conv = np.zeros(arr.shape)
for y in range(w, arr.shape[1] - w):
for x in range(w, arr.shape[0] - w):
conv_sum = 0
for y_ker in range(-w, w + 1):
for x_ker in range(-w, w + 1):
conv_sum += arr[x + x_ker, y + y_ker] * ker[x_ker + w, y_ker + w]
arr_conv[x, y] = conv_sum
return arr_conv
# +
# 3
# TODO: load ./data/pepo.jpg as a grayscale image
image = skimage.io.imread("./data/pepo.jpg")
image = skimage.color.rgb2gray(image)
# TODO: Use the Sobel filter for edge detection:
# Compute the Gradient Magnitude using both Sobel kernels
edges_x = convolve(image, S_x)
edges_y = convolve(image, S_y)
edges = edges_x + edges_y
# Show results
_, axis = plt.subplots(1, 2)
axis[0].imshow(image, cmap='gray')
axis[1].imshow(edges, cmap='gray')
plt.show()
# +
# 4
u = np.array([[1], [2], [1]])
v = np.array([[-1, 0, 1]])
S_x = np.dot(u, v)
print('u =\n', u)
print('v =\n', v)
print('S_x =\n', S_x)
def convolve_fast(arr, K_u, K_v):
'''
Convolve the array using kernel K_u and K_v.
arr -- 2D array that gets convolved
K_u -- kernel u
K_v -- kernel v
'''
w = math.floor(K_u.shape[0] / 2)
arr_conv = np.zeros(arr.shape)
for x in range(w, arr.shape[0] - w):
for y in range(w, arr.shape[1] - w):
conv_sum = 0
for y_ker in range(-w, w + 1):
conv_sum += arr[x, y + y_ker] * K_u[y_ker + w][0]
arr_conv[x, y] = conv_sum
arr = arr_conv
arr_conv = np.zeros(arr.shape)
for y in range(w, arr.shape[1] - w):
for x in range(w, arr.shape[0] - w):
conv_sum = 0
for x_ker in range(-w, w + 1):
conv_sum += arr[x + x_ker, y] * K_v[0][x_ker + w]
arr_conv[x, y] = conv_sum
return arr_conv
# TODO: Run both methods and compare their runtime
start_naive = time.time()
edges_naive = convolve(image, S_y)
end_naive = time.time()
start_improved = time.time()
edges_improved = convolve_fast(image, u, v)
end_improved = time.time()
print("Time naive: " + str(end_naive - start_naive))
print("Time improved: " + str(end_improved - start_improved))
# Show results
f, axis = plt.subplots(1, 2)
f.set_figheight(15)
f.set_figwidth(15)
axis[0].imshow(edges_naive, cmap='gray')
axis[1].imshow(edges_improved, cmap='gray')
plt.show()
# +
# 5
from scipy.linalg import svd
print('rank of s_x:', np.linalg.matrix_rank(S_x))
print('rank of s_y:', np.linalg.matrix_rank(S_y))
def separate(K):
'''
Seperate the 2D kernel into 2 1D kernels.
K -- 2D kernel
'''
U, s, VT = svd(K)
U *= -1
VT *= -1
K_u = np.array([U[:,0] * s[0]]).T
K_v = np.array([VT[0]])
return K_u, K_v
K_u, K_v = separate(S_x)
print('K_u =\n', K_u)
print('K_v =\n', K_v)
print('K =\n', np.dot(K_u, K_v))
# +
# 6 Bonus
from scipy import signal
def gaussian_kernel(kernel_size, sigma):
'''
Return a 2D gaussian kernel.
kernel_size -- size of the kernel
sigma -- sigma of the gaussian blur
'''
kernel_1d = signal.gaussian(kernel_size, std=sigma).reshape(kernel_size, 1)
h = np.outer(kernel_1d, kernel_1d)
return h
K = gaussian_kernel(15, 15)
K_u, K_v = separate(K)
# TODO: Run both methods and compare their runtime
start_naive = time.time()
conv_x_naive = convolve(image, K)
end_naive = time.time()
start_improved = time.time()
conv_x_improved = convolve_fast(image, K_u, K_v)
end_improved = time.time()
print("Time naive: " + str(end_naive - start_naive))
print("Time improved: " + str(end_improved - start_improved))
# Plot the results
_, axis = plt.subplots(1, 2)
axis[0].imshow(conv_x_naive, cmap='gray')
axis[1].imshow(conv_x_improved, cmap='gray')
plt.show()
# -
# ## Task 3
# (4 points)
#
# 1. Implement the naive max function using nested loops.
# 2. Implement the $\mathcal{O}(n log w)$ faster max function using a binary tree.
# +
#1
def naive_max(arr, ws):
'''
Return the maximum-filtered array
arr -- 2D array
ws -- window size
'''
g = np.zeros(len(arr))
for x in range(len(arr)):
max = 0
for xp in range(x-ws, x+ws+1):
if xp < 0 or xp > len(arr) - 1:
continue
if(arr[xp] > max):
max = arr[xp]
g[x] = max
return g
# Generate some random data to filter
f = []
for i in range(25):
f.append(random.randint(0, 100))
print('input = ', f)
print('naive_max =', naive_max(f, 3))
# +
#2
class Node:
def __init__(self):
self.max = -np.inf # value of this node
self.top = None # reference to parent node
self.left = None # left child node
self.right = None # right child node
# HINT: this list will hold a reference to all leaf nodes that
# are children of this node.
# You will need later to add new and replace old values
# in the tree.
self.elems = [] # list of all child leaf nodes
def build_tree(self, depth, top=None):
'''
Build up a tree of certain depth
depth -- tree depth
top -- parent node
'''
self.top = top
if depth > 0:
# TODO: recursively initialize all children
self.left = Node()
self.left.build_tree(depth-1, self)
self.right = Node()
self.right.build_tree(depth-1, self)
# TODO: concatenate elems lists of children
# HINT: nodes that are not leafes should not be added
# to self.elems
self.elems += self.left.elems
self.elems += self.right.elems
else:
# TODO: this is a leaf node
# HINT: this node has to be added to the elems list
self.elems.append(self)
return self.elems
def update(self):
'''
Update the value (self.max) of this node
and its parent nodes recursively
'''
if self.top is None:
return
if (self.top.left.max > self.top.right.max):
self.top.max = self.top.left.max
else:
self.top.max = self.top.right.max
self.top.update()
class MaxTree:
def __init__(self, ws):
'''
ws -- window size
'''
# TODO: compute number of leafes and tree depth (length = 2 * ws + 1)
self.leafes = 2 * ws + 1
self.depth = math.floor(np.log2(2 * self.leafes))
# TODO: initialize root node
self.root = Node()
self.root.build_tree(self.depth)
# init pointer to next element to be replaced
self.ptr = 0
def replace_elem(self, value):
'''
Replace an element (value of leaf node) in the tree
value -- value of new element
'''
# HINT: use self.ptr as a pointer to the next
# element that has to be replaced
leaf_node = self.root.elems[self.ptr % self.leafes]
leaf_node.max = value
leaf_node.update()
self.ptr += 1
pass
def get_max(self):
return self.root.max
def fast_max(arr, ws):
'''
Return the maximum-filtered array
arr -- 2D array
ws -- window size
'''
max_arr = []
# TODO: initialize tree
tree = MaxTree(ws)
# TODO: compute maximum-filtered output array
for x in range(len(arr)):
tree.replace_elem(arr[x])
max_arr.append(tree.get_max())
return max_arr
print('input = ', f)
print('fast_max =', fast_max(f, 3))
# -
# We now use your implementation to filter an image with different window sizes.
# Note, that for small windows, the naive method is indeed faster due to the lower overhead.
# However, as it has a much better time complexity, the tree method is much faster for large windows.
# +
# Nothing to do here ;)
def image_max(arr, ws, method):
'''
Apply a max-filter to an image
arr -- input image
ws -- window size
method -- filter function
'''
out = np.zeros_like(arr)
for y in range(arr.shape[0]):
out[y] = method(arr[y], ws)
for x in range(arr.shape[1]):
out[:, x] = method(out[:, x], ws)
return out
for ws in [2, 4, 8, 16, 32]:
print('\nFilter with window size {}'.format(ws))
start_time = time.time()
max_img_naive = image_max(image, ws, naive_max)
naive_time = time.time() - start_time
print('Naive implementation took {:.3f} ms'.format(1000 * naive_time))
start_time = time.time()
max_img_improved = image_max(image, ws, fast_max)
improved_time = time.time() - start_time
print('Improved implementation took {:.3f} ms'.format(1000 * improved_time))
_, axis = plt.subplots(1, 2)
axis[0].imshow(max_img_naive, cmap='gray')
axis[1].imshow(max_img_improved,cmap='gray')
plt.show()
# -
| 2.0-tl-image-filter.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Convolutional Neural Networks
# ---
# In this notebook, we train a **CNN** to classify images from the CIFAR-10 database.
#
# The images in this database are small color images that fall into one of ten classes; some example images are pictured below.
#
# <img src='notebook_ims/cifar_data.png' width=70% height=70% />
# ### Test for [CUDA](http://pytorch.org/docs/stable/cuda.html)
#
# Since these are larger (32x32x3) images, it may prove useful to speed up your training time by using a GPU. CUDA is a parallel computing platform and CUDA Tensors are the same as typical Tensors, only they utilize GPU's for computation.
# +
import torch
import numpy as np
# check if CUDA is available
train_on_gpu = torch.cuda.is_available()
if not train_on_gpu:
print("CUDA is not available. Training on CPU ...")
else:
print("CUDA is available! Training on GPU ...")
# -
# ---
# ## Load and Augment the [Data](http://pytorch.org/docs/stable/torchvision/datasets.html)
#
# Downloading may take a minute. We load in the training and test data, split the training data into a training and validation set, then create DataLoaders for each of these sets of data.
#
# #### Augmentation
#
# In this cell, we perform some simple [data augmentation](https://medium.com/nanonets/how-to-use-deep-learning-when-you-have-limited-data-part-2-data-augmentation-c26971dc8ced) by randomly flipping and rotating the given image data. We do this by defining a torchvision `transform`, and you can learn about all the transforms that are used to pre-process and augment data, [here](https://pytorch.org/docs/stable/torchvision/transforms.html).
#
# #### TODO: Look at the [transformation documentation](https://pytorch.org/docs/stable/torchvision/transforms.html); add more augmentation transforms, and see how your model performs.
#
# This type of data augmentation should add some positional variety to these images, so that when we train a model on this data, it will be robust in the face of geometric changes (i.e. it will recognize a ship, no matter which direction it is facing). It's recommended that you choose one or two transforms.
# +
from torchvision import datasets
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
# number of subprocesses to use for data loading
num_workers = 0
# how many samples per batch to load
batch_size = 20
# percentage of training set to use as validation
valid_size = 0.2
# convert data to a normalized torch.FloatTensor
transform = transforms.Compose(
[
transforms.RandomHorizontalFlip(), # randomly flip and rotate
transforms.RandomRotation(10),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# choose the training and test datasets
train_data = datasets.CIFAR10("data", train=True, download=True, transform=transform)
test_data = datasets.CIFAR10("data", train=False, download=True, transform=transform)
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# prepare data loaders (combine dataset and sampler)
train_loader = torch.utils.data.DataLoader(
train_data, batch_size=batch_size, sampler=train_sampler, num_workers=num_workers
)
valid_loader = torch.utils.data.DataLoader(
train_data, batch_size=batch_size, sampler=valid_sampler, num_workers=num_workers
)
test_loader = torch.utils.data.DataLoader(
test_data, batch_size=batch_size, num_workers=num_workers
)
# specify the image classes
classes = [
"airplane",
"automobile",
"bird",
"cat",
"deer",
"dog",
"frog",
"horse",
"ship",
"truck",
]
# -
# ### Visualize a Batch of Training Data
# +
import matplotlib.pyplot as plt
# %matplotlib inline
# helper function to un-normalize and display an image
def imshow(img):
img = img / 2 + 0.5 # unnormalize
plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image
# +
# obtain one batch of training images
dataiter = iter(train_loader)
images, labels = dataiter.next()
images = images.numpy() # convert images to numpy for display
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
# display 20 images
for idx in np.arange(20):
ax = fig.add_subplot(2, 20 / 2, idx + 1, xticks=[], yticks=[])
imshow(images[idx])
ax.set_title(classes[labels[idx]])
# -
# ### View an Image in More Detail
#
# Here, we look at the normalized red, green, and blue (RGB) color channels as three separate, grayscale intensity images.
# +
rgb_img = np.squeeze(images[3])
channels = ["red channel", "green channel", "blue channel"]
fig = plt.figure(figsize=(36, 36))
for idx in np.arange(rgb_img.shape[0]):
ax = fig.add_subplot(1, 3, idx + 1)
img = rgb_img[idx]
ax.imshow(img, cmap="gray")
ax.set_title(channels[idx])
width, height = img.shape
thresh = img.max() / 2.5
for x in range(width):
for y in range(height):
val = round(img[x][y], 2) if img[x][y] != 0 else 0
ax.annotate(
str(val),
xy=(y, x),
horizontalalignment="center",
verticalalignment="center",
size=8,
color="white" if img[x][y] < thresh else "black",
)
# -
# ---
# ## Define the Network [Architecture](http://pytorch.org/docs/stable/nn.html)
#
# This time, you'll define a CNN architecture. Instead of an MLP, which used linear, fully-connected layers, you'll use the following:
# * [Convolutional layers](https://pytorch.org/docs/stable/nn.html#conv2d), which can be thought of as stack of filtered images.
# * [Maxpooling layers](https://pytorch.org/docs/stable/nn.html#maxpool2d), which reduce the x-y size of an input, keeping only the most _active_ pixels from the previous layer.
# * The usual Linear + Dropout layers to avoid overfitting and produce a 10-dim output.
#
# A network with 2 convolutional layers is shown in the image below and in the code, and you've been given starter code with one convolutional and one maxpooling layer.
#
# <img src='notebook_ims/2_layer_conv.png' height=50% width=50% />
#
# #### TODO: Define a model with multiple convolutional layers, and define the feedforward metwork behavior.
#
# The more convolutional layers you include, the more complex patterns in color and shape a model can detect. It's suggested that your final model include 2 or 3 convolutional layers as well as linear layers + dropout in between to avoid overfitting.
#
# It's good practice to look at existing research and implementations of related models as a starting point for defining your own models. You may find it useful to look at [this PyTorch classification example](https://github.com/pytorch/tutorials/blob/master/beginner_source/blitz/cifar10_tutorial.py) or [this, more complex Keras example](https://github.com/keras-team/keras/blob/master/examples/cifar10_cnn.py) to help decide on a final structure.
#
# #### Output volume for a convolutional layer
#
# To compute the output size of a given convolutional layer we can perform the following calculation (taken from [Stanford's cs231n course](http://cs231n.github.io/convolutional-networks/#layers)):
# > We can compute the spatial size of the output volume as a function of the input volume size (W), the kernel/filter size (F), the stride with which they are applied (S), and the amount of zero padding used (P) on the border. The correct formula for calculating how many neurons define the output_W is given by `(W−F+2P)/S+1`.
#
# For example for a 7x7 input and a 3x3 filter with stride 1 and pad 0 we would get a 5x5 output. With stride 2 we would get a 3x3 output.
# +
import torch.nn as nn
import torch.nn.functional as F
# define the CNN architecture
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
# convolutional layer (sees 32x32x3 image tensor)
self.conv1 = nn.Conv2d(3, 16, 3, padding=1)
# convolutional layer (sees 16x16x16 tensor)
self.conv2 = nn.Conv2d(16, 32, 3, padding=1)
# convolutional layer (sees 8x8x32 tensor)
self.conv3 = nn.Conv2d(32, 64, 3, padding=1)
# max pooling layer
self.pool = nn.MaxPool2d(2, 2)
# linear layer (64 * 4 * 4 -> 500)
self.fc1 = nn.Linear(64 * 4 * 4, 500)
# linear layer (500 -> 10)
self.fc2 = nn.Linear(500, 10)
# dropout layer (p=0.25)
self.dropout = nn.Dropout(0.25)
def forward(self, x):
# add sequence of convolutional and max pooling layers
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = self.pool(F.relu(self.conv3(x)))
# flatten image input
x = x.view(-1, 64 * 4 * 4)
# add dropout layer
x = self.dropout(x)
# add 1st hidden layer, with relu activation function
x = F.relu(self.fc1(x))
# add dropout layer
x = self.dropout(x)
# add 2nd hidden layer, with relu activation function
x = self.fc2(x)
return x
# create a complete CNN
model = Net()
print(model)
# move tensors to GPU if CUDA is available
if train_on_gpu:
model.cuda()
# -
# ### Specify [Loss Function](http://pytorch.org/docs/stable/nn.html#loss-functions) and [Optimizer](http://pytorch.org/docs/stable/optim.html)
#
# Decide on a loss and optimization function that is best suited for this classification task. The linked code examples from above, may be a good starting point; [this PyTorch classification example](https://github.com/pytorch/tutorials/blob/master/beginner_source/blitz/cifar10_tutorial.py) or [this, more complex Keras example](https://github.com/keras-team/keras/blob/master/examples/cifar10_cnn.py). Pay close attention to the value for **learning rate** as this value determines how your model converges to a small error.
#
# #### TODO: Define the loss and optimizer and see how these choices change the loss over time.
# +
import torch.optim as optim
# specify loss function (categorical cross-entropy)
criterion = nn.CrossEntropyLoss()
# specify optimizer
optimizer = optim.SGD(model.parameters(), lr=0.01)
# -
# ---
# ## Train the Network
#
# Remember to look at how the training and validation loss decreases over time; if the validation loss ever increases it indicates possible overfitting.
# +
# number of epochs to train the model
n_epochs = 30
valid_loss_min = np.Inf # track change in validation loss
for epoch in range(1, n_epochs + 1):
# keep track of training and validation loss
train_loss = 0.0
valid_loss = 0.0
###################
# train the model #
###################
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
# clear the gradients of all optimized variables
optimizer.zero_grad()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the batch loss
loss = criterion(output, target)
# backward pass: compute gradient of the loss with respect to model parameters
loss.backward()
# perform a single optimization step (parameter update)
optimizer.step()
# update training loss
train_loss += loss.item() * data.size(0)
######################
# validate the model #
######################
model.eval()
for batch_idx, (data, target) in enumerate(valid_loader):
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the batch loss
loss = criterion(output, target)
# update average validation loss
valid_loss += loss.item() * data.size(0)
# calculate average losses
train_loss = train_loss / len(train_loader.sampler)
valid_loss = valid_loss / len(valid_loader.sampler)
# print training/validation statistics
print(
"Epoch: {} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f}".format(
epoch, train_loss, valid_loss
)
)
# save model if validation loss has decreased
if valid_loss <= valid_loss_min:
print(
"Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...".format(
valid_loss_min, valid_loss
)
)
torch.save(model.state_dict(), "model_augmented.pt")
valid_loss_min = valid_loss
# -
# ### Load the Model with the Lowest Validation Loss
model.load_state_dict(torch.load("model_augmented.pt"))
# ---
# ## Test the Trained Network
#
# Test your trained model on previously unseen data! A "good" result will be a CNN that gets around 70% (or more, try your best!) accuracy on these test images.
# +
# track test loss
test_loss = 0.0
class_correct = list(0.0 for i in range(10))
class_total = list(0.0 for i in range(10))
model.eval()
# iterate over test data
for batch_idx, (data, target) in enumerate(test_loader):
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the batch loss
loss = criterion(output, target)
# update test loss
test_loss += loss.item() * data.size(0)
# convert output probabilities to predicted class
_, pred = torch.max(output, 1)
# compare predictions to true label
correct_tensor = pred.eq(target.data.view_as(pred))
correct = (
np.squeeze(correct_tensor.numpy())
if not train_on_gpu
else np.squeeze(correct_tensor.cpu().numpy())
)
# calculate test accuracy for each object class
for i in range(batch_size):
label = target.data[i]
class_correct[label] += correct[i].item()
class_total[label] += 1
# average test loss
test_loss = test_loss / len(test_loader.dataset)
print("Test Loss: {:.6f}\n".format(test_loss))
for i in range(10):
if class_total[i] > 0:
print(
"Test Accuracy of %5s: %2d%% (%2d/%2d)"
% (
classes[i],
100 * class_correct[i] / class_total[i],
np.sum(class_correct[i]),
np.sum(class_total[i]),
)
)
else:
print("Test Accuracy of %5s: N/A (no training examples)" % (classes[i]))
print(
"\nTest Accuracy (Overall): %2d%% (%2d/%2d)"
% (
100.0 * np.sum(class_correct) / np.sum(class_total),
np.sum(class_correct),
np.sum(class_total),
)
)
# -
# ### Visualize Sample Test Results
# +
# obtain one batch of test images
dataiter = iter(test_loader)
images, labels = dataiter.next()
images.numpy()
# move model inputs to cuda, if GPU available
if train_on_gpu:
images = images.cuda()
# get sample outputs
output = model(images)
# convert output probabilities to predicted class
_, preds_tensor = torch.max(output, 1)
preds = (
np.squeeze(preds_tensor.numpy())
if not train_on_gpu
else np.squeeze(preds_tensor.cpu().numpy())
)
# plot the images in the batch, along with predicted and true labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(20):
ax = fig.add_subplot(2, 20 / 2, idx + 1, xticks=[], yticks=[])
imshow(images[idx] if not train_on_gpu else images[idx].cpu())
ax.set_title(
"{} ({})".format(classes[preds[idx]], classes[labels[idx]]),
color=("green" if preds[idx] == labels[idx].item() else "red"),
)
# -
| deep_learning_v2_pytorch/convolutional-neural-networks/cifar-cnn/cifar10_cnn_augmentation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # VARMAX models
#
# This is a brief introduction notebook to VARMAX models in statsmodels. The VARMAX model is generically specified as:
# $$
# y_t = \nu + A_1 y_{t-1} + \dots + A_p y_{t-p} + B x_t + \epsilon_t +
# M_1 \epsilon_{t-1} + \dots M_q \epsilon_{t-q}
# $$
#
# where $y_t$ is a $\text{k_endog} \times 1$ vector.
# %matplotlib inline
import numpy as np
import pandas as pd
import statsmodels.api as sm
import matplotlib.pyplot as plt
dta = sm.datasets.webuse('lutkepohl2', 'https://www.stata-press.com/data/r12/')
dta.index = dta.qtr
endog = dta.loc['1960-04-01':'1978-10-01', ['dln_inv', 'dln_inc', 'dln_consump']]
# ## Model specification
#
# The `VARMAX` class in statsmodels allows estimation of VAR, VMA, and VARMA models (through the `order` argument), optionally with a constant term (via the `trend` argument). Exogenous regressors may also be included (as usual in statsmodels, by the `exog` argument), and in this way a time trend may be added. Finally, the class allows measurement error (via the `measurement_error` argument) and allows specifying either a diagonal or unstructured innovation covariance matrix (via the `error_cov_type` argument).
# ## Example 1: VAR
#
# Below is a simple VARX(2) model in two endogenous variables and an exogenous series, but no constant term. Notice that we needed to allow for more iterations than the default (which is `maxiter=50`) in order for the likelihood estimation to converge. This is not unusual in VAR models which have to estimate a large number of parameters, often on a relatively small number of time series: this model, for example, estimates 27 parameters off of 75 observations of 3 variables.
exog = endog['dln_consump']
mod = sm.tsa.VARMAX(endog[['dln_inv', 'dln_inc']], order=(2,0), trend='nc', exog=exog)
res = mod.fit(maxiter=1000, disp=False)
print(res.summary())
# From the estimated VAR model, we can plot the impulse response functions of the endogenous variables.
ax = res.impulse_responses(10, orthogonalized=True).plot(figsize=(13,3))
ax.set(xlabel='t', title='Responses to a shock to `dln_inv`');
# ## Example 2: VMA
#
# A vector moving average model can also be formulated. Below we show a VMA(2) on the same data, but where the innovations to the process are uncorrelated. In this example we leave out the exogenous regressor but now include the constant term.
mod = sm.tsa.VARMAX(endog[['dln_inv', 'dln_inc']], order=(0,2), error_cov_type='diagonal')
res = mod.fit(maxiter=1000, disp=False)
print(res.summary())
# ## Caution: VARMA(p,q) specifications
#
# Although the model allows estimating VARMA(p,q) specifications, these models are not identified without additional restrictions on the representation matrices, which are not built-in. For this reason, it is recommended that the user proceed with error (and indeed a warning is issued when these models are specified). Nonetheless, they may in some circumstances provide useful information.
mod = sm.tsa.VARMAX(endog[['dln_inv', 'dln_inc']], order=(1,1))
res = mod.fit(maxiter=1000, disp=False)
print(res.summary())
| examples/notebooks/statespace_varmax.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # <font color=Teal>ATOMIC and ASTRING FUNCTIONS (Python Code)</font>
# ### By <NAME>, PhD, Dr.Eng., Professor, Honorary Professor
# - https://www.researchgate.net/profile/Sergei_Eremenko
# - https://www.amazon.com/Sergei-Eremenko/e/B082F3MQ4L
# - https://www.linkedin.com/in/sergei-eremenko-3862079
# - https://www.facebook.com/SergeiEremenko.Author
# Atomic functions (AF) described in many books and hundreds of papers have been discovered in 1970s by Academician NAS of Ukraine Rvachev V.L. (https://ru.wikipedia.org/w/index.php?oldid=83948367) (author's teacher) and professor Rvachev V.A. and advanced by many followers, notably professor Kravchenko V.F. (https://ru.wikipedia.org/w/index.php?oldid=84521570), <NAME> (https://www.researchgate.net/profile/Hrvoje_Gotovac), <NAME> (https://www.researchgate.net/profile/Volodymyr_Kolodyazhny), <NAME> (https://www.researchgate.net/profile/Oleg_Kravchenko) as well as the author <NAME> (https://www.researchgate.net/profile/Sergei_Eremenko) [1-4] for a wide range of applications in mathematical physics, boundary value problems, statistics, radio-electronics, telecommunications, signal processing, and others.
# As per historical survey (https://www.researchgate.net/publication/308749839), some elements, analogs, subsets or Fourier transformations of AFs sometimes named differently (Fabius function, hat function, compactly supported smooth function) have been probably known since 1930s and rediscovered many times by scientists from different countries, including Fabius, W.Hilberg and others. However, the most comprehensive 50+ years’ theory development supported by many books, dissertations, hundreds of papers, lecture courses and multiple online resources have been performed by the schools of V.L. Rvachev, V.A. Rvachev and <NAME>.
# In 2017-2020, <NAME>, in papers "Atomic Strings and Fabric of Spacetime", "Atomic Solitons as a New Class of Solitons", "Atomic Machine Learning" and book "Soliton Nature" [1-8], has introduced <b>AString</b> atomic function as an integral and 'composing branch' of Atomic Function up(x): <font color=maroon>AString'(x) = AString(2x+1) - AString(2x-1) = up(x)</font>
# AString function, is a smooth solitonic kink function by joining of which on a periodic lattice it is possible to compose a straight-line resembling flat spacetime as well as to build 'solitonic atoms' composing different fields. It may lead to novel models of spacetime and quantized gravity where AString may describe Spacetime Quantum, or Spacetime Metriant. Also, representing of different fields via shift and stretches of AStrings and Atomic Functions may lead to unified theory where AString may describe some fundamental building block of quantum fields, like a string, elementary spacetime distortion or metriant.
# So, apart from traditional areas of AF applications in mathematical physics, radio-electronics and signal processing, AStrings and Atomic Functions may be expanded to Spacetime Physics, String theory, General and Special Relativity, Theory of Solitons, Lattice Physics, Quantized Gravity, Cosmology, Dark matter and Multiverse theories as well as Finite Element Methods, Nonarchimedean Computers, Atomic regression analysis, Atomic Kernels, Machine Learning and Artificial Intelligence.
# # <font color=teal>1. Atomic Function up(x) (introduced in 1971 by V.L.Rvachev and V.A.Rvachev)</font>
import numpy as np
import pylab as pl
pl.rcParams["figure.figsize"] = 9,6
# +
###################################################################
##This script calculates the values of Atomic Function up(x) (1971)
###################################################################
################### One Pulse of atomic function
def up1(x: float) -> float:
#Atomic function table
up_y = [0.5, 0.48, 0.460000017,0.440000421,0.420003478,0.400016184, 0.380053256, 0.360139056,
0.340308139, 0.320605107,0.301083436, 0.281802850, 0.262826445, 0.244218000, 0.226041554,
0.208361009, 0.191239338, 0.174736305, 0.158905389, 0.143991189, 0.129427260, 0.115840866,
0.103044024, 0.9110444278e-01, 0.798444445e-01, 0.694444445e-01, 0.598444445e-01,
0.510444877e-01, 0.430440239e-01, 0.358409663e-01, 0.294282603e-01, 0.237911889e-01,
0.189053889e-01, 0.147363055e-01, 0.112393379e-01, 0.836100883e-02, 0.604155412e-02,
0.421800000e-02, 0.282644445e-02, 0.180999032e-02, 0.108343562e-02, 0.605106267e-03,
0.308138660e-03, 0.139055523e-03, 0.532555251e-04, 0.161841328e-04, 0.347816874e-05,
0.420576116e-05, 0.167693347e-07, 0.354008603e-10, 0]
up_x = np.arange(0.5, 1.01, 0.01)
res = 0.
if ((x>=0.5) and (x<=1)):
for i in range(len(up_x) - 1):
if (up_x[i] >= x) and (x < up_x[i+1]):
N1 = 1 - (x - up_x[i])/0.01
res = N1 * up_y[i] + (1 - N1) * up_y[i+1]
return res
return res
############### Atomic Function Pulse with width, shift and scale #############
def upulse(t: float, a = 1., b = 0., c = 1., d = 0.) -> float:
x = (t - b)/a
res = 0.
if (x >= 0.5) and (x <= 1):
res = up1(x)
elif (x >= 0.0) and (x < 0.5):
res = 1 - up1(1 - x)
elif (x >= -1 and x <= -0.5):
res = up1(-x)
elif (x > -0.5) and (x < 0):
res = 1 - up1(1 + x)
res = d + res * c
return res
############### Atomic Function Applied to list with width, shift and scale #############
def up(x: list, a = 1., b = 0., c = 1., d = 0.) -> list:
res = []
for i in range(len(x)):
res.append(upulse(x[i], a, b, c, d))
return res
# -
x = np.arange(-2.0, 2.0, 0.01)
pl.title('Atomic Function up(x)')
pl.plot(x, up(x), label='Atomic Function')
pl.grid(True)
pl.show()
# # <font color=teal>2. Atomic String Function (AString) is an Integral and Composing Branch of Atomic Function up(x) (introduced in 2017 by <NAME>)</font>
# AString function is solitary kink function which simultaneously is integral and composing branch of atomic function up(x)
# ### <font color=maroon>AString'(x) = AString(2x+1) - AString(2x-1) = up(x)</font>
# +
############### Atomic String #############
def AString1(x: float) -> float:
res = 1 * (upulse(x/2.0 - 0.5) - 0.5)
return res
############### Atomic String Pulse with width, shift and scale #############
def AStringPulse(t: float, a = 1., b = 0., c = 1., d = 0.) -> float:
x = (t - b)/a
if (x < -1):
res = -0.5
elif (x > 1):
res = 0.5
else:
res = AString1(x)
res = d + res * c
return res
###### Atomic String Applied to list with width, shift and scale #############
def AString(x: list, a = 1., b = 0., c = 1., d = 0.) -> list:
res = []
for i in range(len(x)):
res.append(AStringPulse(x[i], a, b, c, d))
#res[i] = AStringPulse(x[i], a, b, c)
return res
###### Summation of two lists #############
def Sum(x1: list, x2: list) -> list:
res = []
for i in range(len(x1)):
res.append(x1[i] + x2[i])
return res
# -
x = np.arange(-2.0, 2.0, 0.01)
pl.title('Atomic String Function')
pl.plot(x, AString(x, 1.0, 0, 1, 0), label='Atomic String')
pl.grid(True)
pl.show()
# ## Atomic String, Atomic Function (AF) and AF Derivative plotted together
# +
x = np.arange(-2.0, 2.0, 0.01)
#This Calculates Derivative
dx = x[1] - x[0]
dydx = np.gradient(up(x), dx)
pl.plot(x, up(x), label='Atomic Function')
pl.plot(x, AString(x, 1.0, 0, 1, 0), linewidth=2, label='Atomic String Function')
pl.plot(x, dydx, '--', label='A-Function Derivative')
pl.title('Atomic and AString Functions')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# # <font color=teal>3. Properties of Atomic Function Up(x)</font>
# ## 3.1. Atomic Function Derivative expressed via Atomic Function itself
# Atomic Function Derivative can be exressed via Atomic Function itself - up'(x)= 2up(2x+1)-2up(2x-1) meaning the shape of pulses for derivative function can be represented by shifted and stratched Atomic Function itself - remarkable property
# ### <font color=maroon>up'(x)= 2up(2x+1)-2up(2x-1)</font>
# ### Atomic Function and its Derivative plotted together
# +
x = np.arange(-2.0, 2.0, 0.01)
pl.plot(x, up(x), label='Atomic Function', linewidth=2)
pl.plot(x, dydx, '--', label='Atomic Function Derivative', linewidth=1, color="Green")
pl.title('Atomic Function and Its Derivative')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# ## 3.2. Partition of Unity
# The Atomic Function pulses superposition set at points -2, -1, 0, +1, +2... can exactly represent a Unity (number 1):
# 1 = ... up(x-3) + up(x-2) + up(x-1) + up(x-0) + up(x+1) + up(x+2) + up(x+3) + ...
# ### <font color=maroon>1 = ... up(x-3) + up(x-2) + up(x-1) + up(x-0) + up(x+1) + up(x+2) + up(x+3) + ...</font>
x = np.arange(-2.0, 2.0, 0.01)
pl.plot(x, up(x, 1, -1), '--', linewidth=1, label='Atomic Function at x=-1')
pl.plot(x, up(x, 1, +0), '--', linewidth=1, label='Atomic Function at x=0')
pl.plot(x, up(x, 1, -1), '--', linewidth=1, label='Atomic Function at x=-1')
pl.plot(x, Sum(up(x, 1, -1), Sum(up(x), up(x, 1, 1))), linewidth=2, label='Atomic Function Compounding')
pl.title('Atomic Function Compounding represent 1')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# ## 3.3. Atomic Function (AF) is a 'finite', 'compactly supported', or 'solitary' function
# Like a Spline, Atomic Function (AF) 'compactly supported' not equal to zero only on section |x|<=1
# +
x = np.arange(-5.0, 5.0, 0.01)
pl.plot(x, up(x), label='Atomic Function', linewidth=2)
#pl.plot(x, dydx, '--', label='Atomic Function Derivative', linewidth=1, color="Green")
pl.title('Atomic Function is compactly supported')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# ## 3.4 Atomic Function is a non-analytical function (can not be represented by Taylor's series), but with known Fourier Transformation allowing to exactly calculate AF in certain points, with tabular representation provided in script above.
# # <font color=teal>4. Properties of Atomic String Function</font>
# ## 4.1. AString is not only Integral but also Composing Branch of Atomic Function
# ### <font color=maroon>AString'(x) = AString(2x+1) - AString(2x-1) = up(x)</font>
# Astring is a swing-like function - Integral of Atomic Function (AF) which can be expressed via AF itself:
# AString(x) = Integral(0,x)(Up(x)) = Up(x/2 - 1/2) - 1/2
# ### <font color=maroon>AString(x) = Integral(0,x)(Up(x)) = Up(x/2 - 1/2) - 1/2</font>
# ## 4.2. Atomic Function is a 'solitonic atom' composed from two opposite AStrings
# The concept of 'Solitonic Atoms' (bions) composed from opposite kinks is known in soliton theory [3,5].
# ### <font color=maroon>up(x) = AString(2x + 1) - AString(2x - 1)</font>
# +
######### Presentation of Atomic Function via Atomic Strings ##########
x = np.arange(-2.0, 2.0, 0.01)
pl.plot(x, AString(x, 1, 0, 1, 0), '--', linewidth=1, label='AString(x)')
pl.plot(x, AString(x, 0.5, -0.5, +1, 0), '--', linewidth=2, label='+AString(2x+1)')
pl.plot(x, AString(x, 0.5, +0.5, -1, 0), '--', linewidth=2, label='-AString(2x-1)')
#pl.plot(x, up(x, 1.0, 0, 1, 0), '--', linewidth=1, label='Atomic Function')
AS2 = Sum(AString(x, 0.5, -0.5, +1, 0), AString(x, 0.5, +0.5, -1, 0))
pl.plot(x, AS2, linewidth=3, label='Up(x) via Strings')
pl.title('Atomic Function as a Combination of AStrings')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# ## 4.3. AStrings and Atomic Solitons
# Solitonic mathematical properties of AString and Atomic Functions have been explored in author's paper [3] (<NAME>. Atomic solitons as a new class of solitons; 2018; https://www.researchgate.net/publication/329465767). They both satisfy differential equations with shifted arguments which introduce special kind of <b>nonlinearity</b> typical for all mathematical solitons.
# AString belong to the class of <b>Solitonic Kinks</b> similar to sine-Gordon, Frenkel-Kontorova, tanh and others. Unlike other kinks, AStrings are truly solitary (compactly-supported) and also have a unique property of composing of both straight-line and solitonic atoms on lattice resembling particle-like properties of solitons.
# Atomic Function up(x) is not actually a mathematical soliton, but a complex object composed from summation of two opposite AString kinks, and in solitonic terminology, is called 'solitonic atoms' (like bions).
# ## 4.4. All derivatives of AString can be represented via AString itself
# ### <font color=maroon>AString'(x) = AString(2x + 1) - AString(2x - 1)</font>
# It means AString is a smooth (infinitely divisible) function, with fractalic properties.
# ## 4.5. AString and Fabius Function
# Fabius Function https://en.wikipedia.org/wiki/Fabius_function, with unique property f'(x) = 2f(2x), published in 1966 but was probably known since 1935, is shifted and stretched AString function. Fabius function is not directly an integral of atomic function up(x).
# ### <font color=maroon>Fabius(x) = AString(2x - 1) + 0.5</font>
x = np.arange(-2, 2.0, 0.01)
pl.title('AString and Fabius Functions')
pl.plot(x, AString(x, 0.5, 0.5, 1, 0.5), label='Fabius Function')
pl.plot(x, AString(x, 1, 0, 1, 0), label='AString Function')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# ## 4.6. Partition of Line from Atomic String functions
# Combination/summation of Atomic Strings can exactly represent a straight line:
# x = ...Astring(x-2) + Astring(x-1) + AString(x) + Astring(x+1) + Astring(x+2)...
# ### <font color=maroon>x = ...Astring(x-2) + Astring(x-1) + AString(x) + Astring(x+1) + Astring(x+2)...</font>
# ### Partition based on AString function with width 1 and height 1
# +
x = np.arange(-3, 3, 0.01)
pl.plot(x, AString(x, 1, -1.0, 1, 0), '--', linewidth=1, label='AString 1')
pl.plot(x, AString(x, 1, +0.0, 1, 0), '--', linewidth=1, label='AString 2')
pl.plot(x, AString(x, 1, +1.0, 1, 0), '--', linewidth=1, label='AString 3')
AS2 = Sum(AString(x, 1, -1.0, 1, 0), AString(x, 1, +0.0, 1, 0))
AS3 = Sum(AS2, AString(x, 1, +1.0, 1, 0))
pl.plot(x, AS3, label='AStrings Sum', linewidth=2)
pl.title('Atomic Strings compose Line')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# ### Partition based on AString with certain width and height depending on a size of 'quanta'
# +
x = np.arange(-40.0, 40.0, 0.01)
width = 10.0
height = 10.0
#pl.plot(x, ABline (x, 1, 0), label='ABLine 1*x')
pl.plot(x, AString(x, width, -3*width/2, height, -3*width/2), '--', linewidth=1, label='AString 1')
pl.plot(x, AString(x, width, -1*width/2, height, -1*width/2), '--', linewidth=1, label='AString 2')
pl.plot(x, AString(x, width, +1*width/2, height, +1*width/2), '--', linewidth=1, label='AString 3')
pl.plot(x, AString(x, width, +3*width/2, height, +3*width/2), '--', linewidth=1, label='AString 4')
AS2 = Sum(AString(x, width, -3*width/2, height, -3*width/2), AString(x, width, -1*width/2, height, -1*width/2))
AS3 = Sum(AS2, AString(x, width,+1*width/2, height, +1*width/2))
AS4 = Sum(AS3, AString(x, width,+3*width/2, height, +3*width/2))
pl.plot(x, AS4, label='AStrings Joins', linewidth=2)
pl.title('Atomic Strings Combinations')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# # 5. Representing curved shapes via AStrings and Atomic Functions
# Shifts and stretches of Atomic adn AString functions allows reproducing curved surfaces (eq curved spacetime). Details are in author's papers "Atomic Strings and Fabric of Spacetime", "Atomic Solitons as a New Class of Solitons".
# +
x = np.arange(-50.0, 50.0, 0.1)
dx = x[1] - x[0]
CS6 = Sum(up(x, 5, -30, 5, 5), up(x, 15, 0, 15, 5))
CS6 = Sum(CS6, up(x, 10, +30, 10, 5))
pl.plot(x, CS6, label='Spacetime Density distribution')
IntC6 = np.cumsum(CS6)*dx/50
pl.plot(x, IntC6, label='Spacetime Shape (Geodesics)')
DerC6 = np.gradient(CS6, dx)
pl.plot(x, DerC6, label='Spacetime Curvature')
LightTrajectory = -10 -IntC6/5
pl.plot(x, LightTrajectory, label='Light Trajectory')
pl.title('Shape of Curved Spacetime model')
pl.legend(loc='best', numpoints=1)
pl.grid(True)
pl.show()
# -
# # <font color=teal>6. 'Soliton Nature' book</font>
# ## 6.1. AStrings and Atomic functions are also described in the book 'Soliton Nature'
# Soliton Nature book is easy-to-read, pictorial, interactive book which uses beautiful photography, video channel, and computer scripts in R and Python to demonstrate existing and explore new solitons – the magnificent and versatile energy concentration phenomenon of nature. New class of atomic solitons can be used to describe Higgs boson (‘the god particle’) fields, spacetime quanta and other fundamental building blocks of nature.
#pl.rcParams["figure.figsize"] = 16,12
book = pl.imread('BookSpread_small.png')
pl.imshow(book)
# ## 6.2. 'Soliton Nature' Video Channel, Book Trailer and Web Site
# Video channel https://www.youtube.com/channel/UCexT5iyczZH2HY1-jSafFeQ features amazing solitonic phenomena in nature - welcome to subscribe
# Book web site www.solitonnature.com contains book chapters and amazing video-gallery
# Book Trailers: https://www.youtube.com/watch?v=cZMZdW_3J84, https://www.youtube.com/watch?v=2lABLpIcevo, https://www.youtube.com/watch?v=hQ3zGFEnSWI
# ## 6.3. 'Soliton Nature' book in major bookstores around the globe
# - Amazon US https://www.amazon.com/gp/product/1951630777,
# - Amazon UK https://www.amazon.co.uk/Sergei-Eremenko/e/B082F3MQ4L,
# - Amazon Germany https://www.amazon.de/Sergei-Eremenko/e/B082F3MQ4L,
# - Amazon France https://www.amazon.fr/Soliton-Nature-Discover-Beautiful-Channel/dp/1951630777,
# - Google Books https://books.google.com.au/books/about/Soliton_Nature.html?id=d2zNDwAAQBAJ,
# - Kindle eBooks of your country, like https://www.amazon.com/Soliton-Nature-Discover-Beautiful-Channel-ebook/dp/B082B5PP6R.
# - Book web site www.solitonnature.com
# # <font color=teal>7. Online Source Code Repositories</font>
# This code is available on GitHub: https://solitonscientific.github.io/AtomicString/AFAString.html
# See also
#
# - https://github.com/SolitonScientific
#
# - https://solitonscientific.github.io/AtomicSoliton/AtomicSoliton.html
# - https://solitonscientific.github.io/AtomicString/AtomicString1.html
# - https://solitonscientific.github.io/AtomicMachineLearning/AtomicMachineLearning.html
#
# - https://notebooks.azure.com/Soliton/projects/AtomicString1
# - https://notebooks.azure.com/Soliton/projects/solitonnature
# - https://notebooks.azure.com/Soliton/projects/geosolitons
# # References
# 1. <NAME>. Atomic Strings and Fabric of Spacetime. Journal Achievements of Modern Radioelectronics, 2018. No.6. https://www.researchgate.net/publication/329455498
# 2. <NAME>. Atomic solitons as a new class of solitons. Journal Nonlinear World, No.6, Vol.16, 2018, p.39-63. DOI: 10.18127/j20700970-201806-06. https://www.researchgate.net/publication/329455498
# 3. <NAME>. Atomic solitons as a new class of solitons (English, with Russian Abstract). Journal Nonlinear World, No.6, Vol.16, 2018, p.39-63. DOI: 10.18127/j20700970-201806-06. https://www.researchgate.net/publication/329465767
# 4. <NAME>. Soliton Nature: Discover Beautiful Nature with 200 Images and Video Channel. ISBN: 978-1-951630-77-5. https://www.amazon.com/gp/product/1951630777; https://www.researchgate.net/publication/321228263;
# 5. Eremenko, S.Yu. Atomic Machine Learning. Journal Neurocomputers. 2018, No.3. https://www.researchgate.net/publication/322520539_Atomic_Machine_Learning
# 6. ResearchGate project: https://www.researchgate.net/project/Atomic-Strings-Quantum-of-Spacetime-and-Gravitation
# 7. ResearchGate project: https://www.researchgate.net/project/Atomic-String-and-Atomic-Function-New-Soliton-Candidates
# 8. ResearchGate project: https://www.researchgate.net/project/Atomic-Strings-Quantum-of-Spacetime-and-Gravitation
# <div align=right><i>By <font color=Teal><b><NAME></b></font>, PhD, Dr.Eng., Professor, Honorary Professor <br>
# https://www.researchgate.net/profile/Sergei_Eremenko <br>
# https://www.amazon.com/Sergei-Eremenko/e/B082F3MQ4L <br>
# https://www.linkedin.com/in/sergei-eremenko-3862079 <br>
# https://www.facebook.com/SergeiEremenko.Author/
# </i></div>
| AFAString.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_pytorch_p36
# language: python
# name: conda_pytorch_p36
# ---
# # Sentiment Analysis
#
# ## Updating a Model in SageMaker
#
# _Deep Learning Nanodegree Program | Deployment_
#
# ---
#
# In this notebook we will consider a situation in which a model that we constructed is no longer working as we intended. In particular, we will look at the XGBoost sentiment analysis model that we constructed earlier. In this case, however, we have some new data that our model doesn't seem to perform very well on. As a result, we will re-train our model and update an existing endpoint so that it uses our new model.
#
# This notebook starts by re-creating the XGBoost sentiment analysis model that was created in earlier notebooks. This means that you will have already seen the cells up to the end of Step 4. The new content in this notebook begins at Step 5.
#
# ## Instructions
#
# Some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this notebook. You will not need to modify the included code beyond what is requested. Sections that begin with '**TODO**' in the header indicate that you need to complete or implement some portion within them. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `# TODO: ...` comment. Please be sure to read the instructions carefully!
#
# In addition to implementing code, there will be questions for you to answer which relate to the task and your implementation. Each section where you will answer a question is preceded by a '**Question:**' header. Carefully read each question and provide your answer below the '**Answer:**' header by editing the Markdown cell.
#
# > **Note**: Code and Markdown cells can be executed using the **Shift+Enter** keyboard shortcut. In addition, a cell can be edited by typically clicking it (double-click for Markdown cells) or by pressing **Enter** while it is highlighted.
# ## Step 1: Downloading the data
#
# The dataset we are going to use is very popular among researchers in Natural Language Processing, usually referred to as the [IMDb dataset](http://ai.stanford.edu/~amaas/data/sentiment/). It consists of movie reviews from the website [imdb.com](http://www.imdb.com/), each labeled as either '**pos**itive', if the reviewer enjoyed the film, or '**neg**ative' otherwise.
#
# > Maas, <NAME>., et al. [Learning Word Vectors for Sentiment Analysis](http://ai.stanford.edu/~amaas/data/sentiment/). In _Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies_. Association for Computational Linguistics, 2011.
#
# We begin by using some Jupyter Notebook magic to download and extract the dataset.
# %mkdir ../data
# !wget -O ../data/aclImdb_v1.tar.gz http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz
# !tar -zxf ../data/aclImdb_v1.tar.gz -C ../data
# ## Step 2: Preparing the data
#
# The data we have downloaded is split into various files, each of which contains a single review. It will be much easier going forward if we combine these individual files into two large files, one for training and one for testing.
# +
import os
import glob
def read_imdb_data(data_dir='../data/aclImdb'):
data = {}
labels = {}
for data_type in ['train', 'test']:
data[data_type] = {}
labels[data_type] = {}
for sentiment in ['pos', 'neg']:
data[data_type][sentiment] = []
labels[data_type][sentiment] = []
path = os.path.join(data_dir, data_type, sentiment, '*.txt')
files = glob.glob(path)
for f in files:
with open(f) as review:
data[data_type][sentiment].append(review.read())
# Here we represent a positive review by '1' and a negative review by '0'
labels[data_type][sentiment].append(1 if sentiment == 'pos' else 0)
assert len(data[data_type][sentiment]) == len(labels[data_type][sentiment]), \
"{}/{} data size does not match labels size".format(data_type, sentiment)
return data, labels
# -
data, labels = read_imdb_data()
print("IMDB reviews: train = {} pos / {} neg, test = {} pos / {} neg".format(
len(data['train']['pos']), len(data['train']['neg']),
len(data['test']['pos']), len(data['test']['neg'])))
# +
from sklearn.utils import shuffle
def prepare_imdb_data(data, labels):
"""Prepare training and test sets from IMDb movie reviews."""
#Combine positive and negative reviews and labels
data_train = data['train']['pos'] + data['train']['neg']
data_test = data['test']['pos'] + data['test']['neg']
labels_train = labels['train']['pos'] + labels['train']['neg']
labels_test = labels['test']['pos'] + labels['test']['neg']
#Shuffle reviews and corresponding labels within training and test sets
data_train, labels_train = shuffle(data_train, labels_train)
data_test, labels_test = shuffle(data_test, labels_test)
# Return a unified training data, test data, training labels, test labets
return data_train, data_test, labels_train, labels_test
# -
train_X, test_X, train_y, test_y = prepare_imdb_data(data, labels)
print("IMDb reviews (combined): train = {}, test = {}".format(len(train_X), len(test_X)))
train_X[100]
# ## Step 3: Processing the data
#
# Now that we have our training and testing datasets merged and ready to use, we need to start processing the raw data into something that will be useable by our machine learning algorithm. To begin with, we remove any html formatting that may appear in the reviews and perform some standard natural language processing in order to homogenize the data.
import nltk
nltk.download("stopwords")
from nltk.corpus import stopwords
from nltk.stem.porter import *
stemmer = PorterStemmer()
# +
import re
from bs4 import BeautifulSoup
def review_to_words(review):
text = BeautifulSoup(review, "html.parser").get_text() # Remove HTML tags
text = re.sub(r"[^a-zA-Z0-9]", " ", text.lower()) # Convert to lower case
words = text.split() # Split string into words
words = [w for w in words if w not in stopwords.words("english")] # Remove stopwords
words = [PorterStemmer().stem(w) for w in words] # stem
return words
# -
review_to_words(train_X[100])
# +
import pickle
cache_dir = os.path.join("../cache", "sentiment_analysis") # where to store cache files
os.makedirs(cache_dir, exist_ok=True) # ensure cache directory exists
def preprocess_data(data_train, data_test, labels_train, labels_test,
cache_dir=cache_dir, cache_file="preprocessed_data.pkl"):
"""Convert each review to words; read from cache if available."""
# If cache_file is not None, try to read from it first
cache_data = None
if cache_file is not None:
try:
with open(os.path.join(cache_dir, cache_file), "rb") as f:
cache_data = pickle.load(f)
print("Read preprocessed data from cache file:", cache_file)
except:
pass # unable to read from cache, but that's okay
# If cache is missing, then do the heavy lifting
if cache_data is None:
# Preprocess training and test data to obtain words for each review
#words_train = list(map(review_to_words, data_train))
#words_test = list(map(review_to_words, data_test))
words_train = [review_to_words(review) for review in data_train]
words_test = [review_to_words(review) for review in data_test]
# Write to cache file for future runs
if cache_file is not None:
cache_data = dict(words_train=words_train, words_test=words_test,
labels_train=labels_train, labels_test=labels_test)
with open(os.path.join(cache_dir, cache_file), "wb") as f:
pickle.dump(cache_data, f)
print("Wrote preprocessed data to cache file:", cache_file)
else:
# Unpack data loaded from cache file
words_train, words_test, labels_train, labels_test = (cache_data['words_train'],
cache_data['words_test'], cache_data['labels_train'], cache_data['labels_test'])
return words_train, words_test, labels_train, labels_test
# -
# Preprocess data
train_X, test_X, train_y, test_y = preprocess_data(train_X, test_X, train_y, test_y)
# ### Extract Bag-of-Words features
#
# For the model we will be implementing, rather than using the reviews directly, we are going to transform each review into a Bag-of-Words feature representation. Keep in mind that 'in the wild' we will only have access to the training set so our transformer can only use the training set to construct a representation.
# +
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.externals import joblib
# joblib is an enhanced version of pickle that is more efficient for storing NumPy arrays
def extract_BoW_features(words_train, words_test, vocabulary_size=5000,
cache_dir=cache_dir, cache_file="bow_features.pkl"):
"""Extract Bag-of-Words for a given set of documents, already preprocessed into words."""
# If cache_file is not None, try to read from it first
cache_data = None
if cache_file is not None:
try:
with open(os.path.join(cache_dir, cache_file), "rb") as f:
cache_data = joblib.load(f)
print("Read features from cache file:", cache_file)
except:
pass # unable to read from cache, but that's okay
# If cache is missing, then do the heavy lifting
if cache_data is None:
# Fit a vectorizer to training documents and use it to transform them
# NOTE: Training documents have already been preprocessed and tokenized into words;
# pass in dummy functions to skip those steps, e.g. preprocessor=lambda x: x
vectorizer = CountVectorizer(max_features=vocabulary_size,
preprocessor=lambda x: x, tokenizer=lambda x: x) # already preprocessed
features_train = vectorizer.fit_transform(words_train).toarray()
# Apply the same vectorizer to transform the test documents (ignore unknown words)
features_test = vectorizer.transform(words_test).toarray()
# NOTE: Remember to convert the features using .toarray() for a compact representation
# Write to cache file for future runs (store vocabulary as well)
if cache_file is not None:
vocabulary = vectorizer.vocabulary_
cache_data = dict(features_train=features_train, features_test=features_test,
vocabulary=vocabulary)
with open(os.path.join(cache_dir, cache_file), "wb") as f:
joblib.dump(cache_data, f)
print("Wrote features to cache file:", cache_file)
else:
# Unpack data loaded from cache file
features_train, features_test, vocabulary = (cache_data['features_train'],
cache_data['features_test'], cache_data['vocabulary'])
# Return both the extracted features as well as the vocabulary
return features_train, features_test, vocabulary
# -
# Extract Bag of Words features for both training and test datasets
train_X, test_X, vocabulary = extract_BoW_features(train_X, test_X)
len(train_X[100])
# ## Step 4: Classification using XGBoost
#
# Now that we have created the feature representation of our training (and testing) data, it is time to start setting up and using the XGBoost classifier provided by SageMaker.
#
# ### Writing the dataset
#
# The XGBoost classifier that we will be using requires the dataset to be written to a file and stored using Amazon S3. To do this, we will start by splitting the training dataset into two parts, the data we will train the model with and a validation set. Then, we will write those datasets to a file and upload the files to S3. In addition, we will write the test set input to a file and upload the file to S3. This is so that we can use SageMakers Batch Transform functionality to test our model once we've fit it.
# +
import pandas as pd
# Earlier we shuffled the training dataset so to make things simple we can just assign
# the first 10 000 reviews to the validation set and use the remaining reviews for training.
val_X = pd.DataFrame(train_X[:10000])
train_X = pd.DataFrame(train_X[10000:])
val_y = pd.DataFrame(train_y[:10000])
train_y = pd.DataFrame(train_y[10000:])
# -
# The documentation for the XGBoost algorithm in SageMaker requires that the saved datasets should contain no headers or index and that for the training and validation data, the label should occur first for each sample.
#
# For more information about this and other algorithms, the SageMaker developer documentation can be found on __[Amazon's website.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__
# First we make sure that the local directory in which we'd like to store the training and validation csv files exists.
data_dir = '../data/sentiment_update'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# +
pd.DataFrame(test_X).to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)
pd.concat([val_y, val_X], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([train_y, train_X], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
# +
# To save a bit of memory we can set text_X, train_X, val_X, train_y and val_y to None.
test_X = train_X = val_X = train_y = val_y = None
# -
# ### Uploading Training / Validation files to S3
#
# Amazon's S3 service allows us to store files that can be access by both the built-in training models such as the XGBoost model we will be using as well as custom models such as the one we will see a little later.
#
# For this, and most other tasks we will be doing using SageMaker, there are two methods we could use. The first is to use the low level functionality of SageMaker which requires knowing each of the objects involved in the SageMaker environment. The second is to use the high level functionality in which certain choices have been made on the user's behalf. The low level approach benefits from allowing the user a great deal of flexibility while the high level approach makes development much quicker. For our purposes we will opt to use the high level approach although using the low-level approach is certainly an option.
#
# Recall the method `upload_data()` which is a member of object representing our current SageMaker session. What this method does is upload the data to the default bucket (which is created if it does not exist) into the path described by the key_prefix variable. To see this for yourself, once you have uploaded the data files, go to the S3 console and look to see where the files have been uploaded.
#
# For additional resources, see the __[SageMaker API documentation](http://sagemaker.readthedocs.io/en/latest/)__ and in addition the __[SageMaker Developer Guide.](https://docs.aws.amazon.com/sagemaker/latest/dg/)__
# +
import sagemaker
session = sagemaker.Session() # Store the current SageMaker session
# S3 prefix (which folder will we use)
prefix = 'sentiment-update'
test_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
# -
# ### Creating the XGBoost model
#
# Now that the data has been uploaded it is time to create the XGBoost model. To begin with, we need to do some setup. At this point it is worth discussing what a model is in SageMaker. It is easiest to think of a model of comprising three different objects in the SageMaker ecosystem, which interact with one another.
#
# - Model Artifacts
# - Training Code (Container)
# - Inference Code (Container)
#
# The Model Artifacts are what you might think of as the actual model itself. For example, if you were building a neural network, the model artifacts would be the weights of the various layers. In our case, for an XGBoost model, the artifacts are the actual trees that are created during training.
#
# The other two objects, the training code and the inference code are then used the manipulate the training artifacts. More precisely, the training code uses the training data that is provided and creates the model artifacts, while the inference code uses the model artifacts to make predictions on new data.
#
# The way that SageMaker runs the training and inference code is by making use of Docker containers. For now, think of a container as being a way of packaging code up so that dependencies aren't an issue.
# +
from sagemaker import get_execution_role
# Our current execution role is require when creating the model as the training
# and inference code will need to access the model artifacts.
role = get_execution_role()
# +
# We need to retrieve the location of the container which is provided by Amazon for using XGBoost.
# As a matter of convenience, the training and inference code both use the same container.
from sagemaker.amazon.amazon_estimator import get_image_uri
container = get_image_uri(session.boto_region_name, 'xgboost')
# +
# First we create a SageMaker estimator object for our model.
xgb = sagemaker.estimator.Estimator(container, # The location of the container we wish to use
role, # What is our current IAM Role
train_instance_count=1, # How many compute instances
train_instance_type='ml.m4.xlarge', # What kind of compute instances
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
sagemaker_session=session)
# And then set the algorithm specific parameters.
xgb.set_hyperparameters(max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.8,
silent=0,
objective='binary:logistic',
early_stopping_rounds=10,
num_round=500)
# -
# ### Fit the XGBoost model
#
# Now that our model has been set up we simply need to attach the training and validation datasets and then ask SageMaker to set up the computation.
s3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='csv')
s3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='csv')
xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})
# ### Testing the model
#
# Now that we've fit our XGBoost model, it's time to see how well it performs. To do this we will use SageMakers Batch Transform functionality. Batch Transform is a convenient way to perform inference on a large dataset in a way that is not realtime. That is, we don't necessarily need to use our model's results immediately and instead we can peform inference on a large number of samples. An example of this in industry might be peforming an end of month report. This method of inference can also be useful to us as it means to can perform inference on our entire test set.
#
# To perform a Batch Transformation we need to first create a transformer objects from our trained estimator object.
xgb_transformer = xgb.transformer(instance_count = 1, instance_type = 'ml.m4.xlarge')
# Next we actually perform the transform job. When doing so we need to make sure to specify the type of data we are sending so that it is serialized correctly in the background. In our case we are providing our model with csv data so we specify `text/csv`. Also, if the test data that we have provided is too large to process all at once then we need to specify how the data file should be split up. Since each line is a single entry in our data set we tell SageMaker that it can split the input on each line.
xgb_transformer.transform(test_location, content_type='text/csv', split_type='Line')
# Currently the transform job is running but it is doing so in the background. Since we wish to wait until the transform job is done and we would like a bit of feedback we can run the `wait()` method.
xgb_transformer.wait()
# Now the transform job has executed and the result, the estimated sentiment of each review, has been saved on S3. Since we would rather work on this file locally we can perform a bit of notebook magic to copy the file to the `data_dir`.
# !aws s3 cp --recursive $xgb_transformer.output_path $data_dir
# The last step is now to read in the output from our model, convert the output to something a little more usable, in this case we want the sentiment to be either `1` (positive) or `0` (negative), and then compare to the ground truth labels.
predictions = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)
predictions = [round(num) for num in predictions.squeeze().values]
from sklearn.metrics import accuracy_score
accuracy_score(test_y, predictions)
# ## Step 5: Looking at New Data
#
# So now we have an XGBoost sentiment analysis model that we believe is working pretty well. As a result, we deployed it and we are using it in some sort of app.
#
# However, as we allow users to use our app we periodically record submitted movie reviews so that we can perform some quality control on our deployed model. Once we've accumulated enough reviews we go through them by hand and evaluate whether they are positive or negative (there are many ways you might do this in practice aside from by hand). The reason for doing this is so that we can check to see how well our model is doing.
# +
import new_data
new_X, new_Y = new_data.get_new_data()
# -
# **NOTE:** Part of the fun in this notebook is trying to figure out what exactly is happening with the new data, so try not to cheat by looking in the `new_data` module. Also, the `new_data` module assumes that the cache created earlier in Step 3 is still stored in `../cache/sentiment_analysis`.
# ### (TODO) Testing the current model
#
# Now that we've loaded the new data, let's check to see how our current XGBoost model performs on it.
#
# First, note that the data that has been loaded has already been pre-processed so that each entry in `new_X` is a list of words that have been processed using `nltk`. However, we have not yet constructed the bag of words encoding, which we will do now.
#
# First, we use the vocabulary that we constructed earlier using the original training data to construct a `CountVectorizer` which we will use to transform our new data into its bag of words encoding.
#
# **TODO:** Create the CountVectorizer object using the vocabulary created earlier and use it to transform the new data.
# +
# TODO: Create the CountVectorizer using the previously constructed vocabulary
# vectorizer = None
# Solution:
vectorizer = CountVectorizer(vocabulary=vocabulary,
preprocessor=lambda x: x, tokenizer=lambda x: x)
# TODO: Transform our new data set and store the transformed data in the variable new_XV
# new_XV = None
# Solution
new_XV = vectorizer.transform(new_X).toarray()
# -
# As a quick sanity check, we make sure that the length of each of our bag of words encoded reviews is correct. In particular, it must be the same size as the vocabulary which in our case is `5000`.
len(new_XV[100])
# Now that we've performed the data processing that is required by our model we can save it locally and then upload it to S3 so that we can construct a batch transform job in order to see how well our model is working.
#
# First, we save the data locally.
#
# **TODO:** Save the new data (after it has been transformed using the original vocabulary) to the local notebook instance.
# +
# TODO: Save the data contained in new_XV locally in the data_dir with the file name new_data.csv
# Solution:
pd.DataFrame(new_XV).to_csv(os.path.join(data_dir, 'new_data.csv'), header=False, index=False)
# -
# Next, we upload the data to S3.
#
# **TODO:** Upload the csv file created above to S3.
# +
# TODO: Upload the new_data.csv file contained in the data_dir folder to S3 and save the resulting
# URI as new_data_location
# new_data_location = None
# Solution:
new_data_location = session.upload_data(os.path.join(data_dir, 'new_data.csv'), key_prefix=prefix)
# -
# Then, once the new data has been uploaded to S3, we create and run the batch transform job to get our model's predictions about the sentiment of the new movie reviews.
#
# **TODO:** Using the `xgb_transformer` object that was created earlier (at the end of Step 4 to test the XGBoost model), transform the data located at `new_data_location`.
# +
# TODO: Using xgb_transformer, transform the new_data_location data. You may wish to **wait** until
# the batch transform job has finished.
# Solution:
xgb_transformer.transform(new_data_location, content_type='text/csv', split_type='Line')
xgb_transformer.wait()
# -
# As usual, we copy the results of the batch transform job to our local instance.
# !aws s3 cp --recursive $xgb_transformer.output_path $data_dir
# Read in the results of the batch transform job.
predictions = pd.read_csv(os.path.join(data_dir, 'new_data.csv.out'), header=None)
predictions = [round(num) for num in predictions.squeeze().values]
# And check the accuracy of our current model.
accuracy_score(new_Y, predictions)
# So it would appear that *something* has changed since our model is no longer (as) effective at determining the sentiment of a user provided review.
#
# In a real life scenario you would check a number of different things to see what exactly is going on. In our case, we are only going to check one and that is whether some aspect of the underlying distribution has changed. In other words, we want to see if the words that appear in our new collection of reviews matches the words that appear in the original training set. Of course, we want to narrow our scope a little bit so we will only look at the `5000` most frequently appearing words in each data set, or in other words, the vocabulary generated by each data set.
#
# Before doing that, however, let's take a look at some of the incorrectly classified reviews in the new data set.
#
# To start, we will deploy the original XGBoost model. We will then use the deployed model to infer the sentiment of some of the new reviews. This will also serve as a nice excuse to deploy our model so that we can mimic a real life scenario where we have a model that has been deployed and is being used in production.
#
# **TODO:** Deploy the XGBoost model.
# +
# TODO: Deploy the model that was created earlier. Recall that the object name is 'xgb'.
# xgb_predictor = None
# Solution:
xgb_predictor = xgb.deploy(initial_instance_count = 1, instance_type = 'ml.m4.xlarge')
# -
# ### Diagnose the problem
#
# Now that we have our deployed "production" model, we can send some of our new data to it and filter out some of the incorrectly classified reviews.
# +
from sagemaker.predictor import csv_serializer
# We need to tell the endpoint what format the data we are sending is in so that SageMaker can perform the serialization.
xgb_predictor.content_type = 'text/csv'
xgb_predictor.serializer = csv_serializer
# -
# It will be useful to look at a few different examples of incorrectly classified reviews so we will start by creating a *generator* which we will use to iterate through some of the new reviews and find ones that are incorrect.
#
# **NOTE:** Understanding what Python generators are isn't really required for this module. The reason we use them here is so that we don't have to iterate through all of the new reviews, searching for incorrectly classified samples.
def get_sample(in_X, in_XV, in_Y):
for idx, smp in enumerate(in_X):
res = round(float(xgb_predictor.predict(in_XV[idx])))
if res != in_Y[idx]:
yield smp, in_Y[idx]
gn = get_sample(new_X, new_XV, new_Y)
# At this point, `gn` is the *generator* which generates samples from the new data set which are not classified correctly. To get the *next* sample we simply call the `next` method on our generator.
print(next(gn))
# After looking at a few examples, maybe we decide to look at the most frequently appearing `5000` words in each data set, the original training data set and the new data set. The reason for looking at this might be that we expect the frequency of use of different words to have changed, maybe there is some new slang that has been introduced or some other artifact of popular culture that has changed the way that people write movie reviews.
#
# To do this, we start by fitting a `CountVectorizer` to the new data.
new_vectorizer = CountVectorizer(max_features=5000,
preprocessor=lambda x: x, tokenizer=lambda x: x)
new_vectorizer.fit(new_X)
# Now that we have this new `CountVectorizor` object, we can check to see if the corresponding vocabulary has changed between the two data sets.
original_vocabulary = set(vocabulary.keys())
new_vocabulary = set(new_vectorizer.vocabulary_.keys())
# We can look at the words that were in the original vocabulary but not in the new vocabulary.
print(original_vocabulary - new_vocabulary)
# And similarly, we can look at the words that are in the new vocabulary but which were not in the original vocabulary.
print(new_vocabulary - original_vocabulary)
# These words themselves don't tell us much, however if one of these words occured with a large frequency, that might tell us something. In particular, we wouldn't really expect any of the words above to appear with too much frequency.
#
# **Question** What exactly is going on here. Not only what (if any) words appear with a larger than expected frequency but also, what does this mean? What has changed about the world that our original model no longer takes into account?
#
# **NOTE:** This is meant to be a very open ended question. To investigate you may need more cells than the one provided below. Also, there isn't really a *correct* answer, this is meant to be an opportunity to explore the data.
# ### (TODO) Build a new model
#
# Supposing that we believe something has changed about the underlying distribution of the words that our reviews are made up of, we need to create a new model. This way our new model will take into account whatever it is that has changed.
#
# To begin with, we will use the new vocabulary to create a bag of words encoding of the new data. We will then use this data to train a new XGBoost model.
#
# **NOTE:** Because we believe that the underlying distribution of words has changed it should follow that the original vocabulary that we used to construct a bag of words encoding of the reviews is no longer valid. This means that we need to be careful with our data. If we send an bag of words encoded review using the *original* vocabulary we should not expect any sort of meaningful results.
#
# In particular, this means that if we had deployed our XGBoost model like we did in the Web App notebook then we would need to implement this vocabulary change in the Lambda function as well.
new_XV = new_vectorizer.transform(new_X).toarray()
# And a quick check to make sure that the newly encoded reviews have the correct length, which should be the size of the new vocabulary which we created.
len(new_XV[0])
# Now that we have our newly encoded, newly collected data, we can split it up into a training and validation set so that we can train a new XGBoost model. As usual, we first split up the data, then save it locally and then upload it to S3.
# +
import pandas as pd
# Earlier we shuffled the training dataset so to make things simple we can just assign
# the first 10 000 reviews to the validation set and use the remaining reviews for training.
new_val_X = pd.DataFrame(new_XV[:10000])
new_train_X = pd.DataFrame(new_XV[10000:])
new_val_y = pd.DataFrame(new_Y[:10000])
new_train_y = pd.DataFrame(new_Y[10000:])
# -
# In order to save some memory we will effectively delete the `new_X` variable. Remember that this contained a list of reviews and each review was a list of words. Note that once this cell has been executed you will need to read the new data in again if you want to work with it.
new_X = None
# Next we save the new training and validation sets locally. Note that we overwrite the training and validation sets used earlier. This is mostly because the amount of space that we have available on our notebook instance is limited. Of course, you can increase this if you'd like but to do so may increase the cost of running the notebook instance.
# +
pd.DataFrame(new_XV).to_csv(os.path.join(data_dir, 'new_data.csv'), header=False, index=False)
pd.concat([new_val_y, new_val_X], axis=1).to_csv(os.path.join(data_dir, 'new_validation.csv'), header=False, index=False)
pd.concat([new_train_y, new_train_X], axis=1).to_csv(os.path.join(data_dir, 'new_train.csv'), header=False, index=False)
# -
# Now that we've saved our data to the local instance, we can safely delete the variables to save on memory.
new_val_y = new_val_X = new_train_y = new_train_X = new_XV = None
# Lastly, we make sure to upload the new training and validation sets to S3.
#
# **TODO:** Upload the new data as well as the new training and validation data sets to S3.
# +
# TODO: Upload the new data and the new validation.csv and train.csv files in the data_dir directory to S3.
# new_data_location = None
# new_val_location = None
# new_train_location = None
# Solution:
new_data_location = session.upload_data(os.path.join(data_dir, 'new_data.csv'), key_prefix=prefix)
new_val_location = session.upload_data(os.path.join(data_dir, 'new_validation.csv'), key_prefix=prefix)
new_train_location = session.upload_data(os.path.join(data_dir, 'new_train.csv'), key_prefix=prefix)
# -
# Once our new training data has been uploaded to S3, we can create a new XGBoost model that will take into account the changes that have occured in our data set.
#
# **TODO:** Create a new XGBoost estimator object.
# +
# TODO: First, create a SageMaker estimator object for our model.
# new_xgb = None
# Solution:
new_xgb = sagemaker.estimator.Estimator(container, # The location of the container we wish to use
role, # What is our current IAM Role
train_instance_count=1, # How many compute instances
train_instance_type='ml.m4.xlarge', # What kind of compute instances
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
sagemaker_session=session)
# TODO: Then set the algorithm specific parameters. You may wish to use the same parameters that were
# used when training the original model.
# Solution:
new_xgb.set_hyperparameters(max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.8,
silent=0,
objective='binary:logistic',
early_stopping_rounds=10,
num_round=500)
# -
# Once the model has been created, we can train it with our new data.
#
# **TODO:** Train the new XGBoost model.
# +
# TODO: First, make sure that you create s3 input objects so that SageMaker knows where to
# find the training and validation data.
s3_new_input_train = None
s3_new_input_validation = None
# Solution:
s3_new_input_train = sagemaker.s3_input(s3_data=new_train_location, content_type='csv')
s3_new_input_validation = sagemaker.s3_input(s3_data=new_val_location, content_type='csv')
# +
# TODO: Using the new validation and training data, 'fit' your new model.
# Solution:
new_xgb.fit({'train': s3_new_input_train, 'validation': s3_new_input_validation})
# -
# ### (TODO) Check the new model
#
# So now we have a new XGBoost model that we believe more accurately represents the state of the world at this time, at least in how it relates to the sentiment analysis problem that we are working on. The next step is to double check that our model is performing reasonably.
#
# To do this, we will first test our model on the new data.
#
# **Note:** In practice this is a pretty bad idea. We already trained our model on the new data, so testing it shouldn't really tell us much. In fact, this is sort of a textbook example of leakage. We are only doing it here so that we have a numerical baseline.
#
# **Question:** How might you address the leakage problem?
# First, we create a new transformer based on our new XGBoost model.
#
# **TODO:** Create a transformer object from the newly created XGBoost model.
# +
# TODO: Create a transformer object from the new_xgb model
# new_xgb_transformer = None
# Solution:
new_xgb_transformer = new_xgb.transformer(instance_count = 1, instance_type = 'ml.m4.xlarge')
# -
# Next we test our model on the new data.
#
# **TODO:** Use the transformer object to transform the new data (stored in the `new_data_location` variable)
# +
# TODO: Using new_xgb_transformer, transform the new_data_location data. You may wish to
# 'wait' for the transform job to finish.
# Solution:
new_xgb_transformer.transform(new_data_location, content_type='text/csv', split_type='Line')
new_xgb_transformer.wait()
# -
# Copy the results to our local instance.
# !aws s3 cp --recursive $new_xgb_transformer.output_path $data_dir
# And see how well the model did.
predictions = pd.read_csv(os.path.join(data_dir, 'new_data.csv.out'), header=None)
predictions = [round(num) for num in predictions.squeeze().values]
accuracy_score(new_Y, predictions)
# As expected, since we trained the model on this data, our model performs pretty well. So, we have reason to believe that our new XGBoost model is a "better" model.
#
# However, before we start changing our deployed model, we should first make sure that our new model isn't too different. In other words, if our new model performed really poorly on the original test data then this might be an indication that something else has gone wrong.
#
# To start with, since we got rid of the variable that stored the original test reviews, we will read them in again from the cache that we created in Step 3. Note that we need to make sure that we read in the original test data after it has been pre-processed with `nltk` but before it has been bag of words encoded. This is because we need to use the new vocabulary instead of the original one.
# +
cache_data = None
with open(os.path.join(cache_dir, "preprocessed_data.pkl"), "rb") as f:
cache_data = pickle.load(f)
print("Read preprocessed data from cache file:", "preprocessed_data.pkl")
test_X = cache_data['words_test']
test_Y = cache_data['labels_test']
# Here we set cache_data to None so that it doesn't occupy memory
cache_data = None
# -
# Once we've loaded the original test reviews, we need to create a bag of words encoding of them using the new vocabulary that we created, based on the new data.
#
# **TODO:** Transform the original test data using the new vocabulary.
# +
# TODO: Use the new_vectorizer object that you created earlier to transform the test_X data.
# test_X = None
# Solution:
test_X = new_vectorizer.transform(test_X).toarray()
# -
# Now that we have correctly encoded the original test data, we can write it to the local instance, upload it to S3 and test it.
pd.DataFrame(test_X).to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)
test_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)
new_xgb_transformer.transform(test_location, content_type='text/csv', split_type='Line')
new_xgb_transformer.wait()
# !aws s3 cp --recursive $new_xgb_transformer.output_path $data_dir
predictions = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)
predictions = [round(num) for num in predictions.squeeze().values]
accuracy_score(test_Y, predictions)
# It would appear that our new XGBoost model is performing quite well on the old test data. This gives us some indication that our new model should be put into production and replace our original model.
# ## Step 6: (TODO) Updating the Model
#
# So we have a new model that we'd like to use instead of one that is already deployed. Furthermore, we are assuming that the model that is already deployed is being used in some sort of application. As a result, what we want to do is update the existing endpoint so that it uses our new model.
#
# Of course, to do this we need to create an endpoint configuration for our newly created model.
#
# First, note that we can access the name of the model that we created above using the `model_name` property of the transformer. The reason for this is that in order for the transformer to create a batch transform job it needs to first create the model object inside of SageMaker. Since we've sort of already done this we should take advantage of it.
new_xgb_transformer.model_name
# Next, we create an endpoint configuration using the low level approach of creating the dictionary object which describes the endpoint configuration we want.
#
# **TODO:** Using the low level approach, create a new endpoint configuration. Don't forget that it needs a name and that the name needs to be unique. If you get stuck, try looking at the Boston Housing Low Level Deployment tutorial notebook.
# +
from time import gmtime, strftime
# TODO: Give our endpoint configuration a name. Remember, it needs to be unique.
# new_xgb_endpoint_config_name = None
# Solution:
new_xgb_endpoint_config_name = "sentiment-update-xgboost-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# TODO: Using the SageMaker Client, construct the endpoint configuration.
# new_xgb_endpoint_config_info = None
# Solution:
new_xgb_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = new_xgb_endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": new_xgb_transformer.model_name,
"VariantName": "XGB-Model"
}])
# -
# Once the endpoint configuration has been constructed, it is a straightforward matter to ask SageMaker to update the existing endpoint so that it uses the new endpoint configuration.
#
# Of note here is that SageMaker does this in such a way that there is no downtime. Essentially, SageMaker deploys the new model and then updates the original endpoint so that it points to the newly deployed model. After that, the original model is shut down. This way, whatever app is using our endpoint won't notice that we've changed the model that is being used.
#
# **TODO:** Use the SageMaker Client to update the endpoint that you deployed earlier.
# +
# TODO: Update the xgb_predictor.endpoint so that it uses new_xgb_endpoint_config_name.
# Solution:
session.sagemaker_client.update_endpoint(EndpointName=xgb_predictor.endpoint, EndpointConfigName=new_xgb_endpoint_config_name)
# -
# And, as is generally the case with SageMaker requests, this is being done in the background so if we want to wait for it to complete we need to call the appropriate method.
session.wait_for_endpoint(xgb_predictor.endpoint)
# ## Step 7: Delete the Endpoint
#
# Of course, since we are done with the deployed endpoint we need to make sure to shut it down, otherwise we will continue to be charged for it.
xgb_predictor.delete_endpoint()
# ## Some Additional Questions
#
# This notebook is a little different from the other notebooks in this module. In part, this is because it is meant to be a little bit closer to the type of problem you may face in a real world scenario. Of course, this problem is a very easy one with a prescribed solution, but there are many other interesting questions that we did not consider here and that you may wish to consider yourself.
#
# For example,
# - What other ways could the underlying distribution change?
# - Is it a good idea to re-train the model using only the new data?
# - What would change if the quantity of new data wasn't large. Say you only received 500 samples?
#
# ## Optional: Clean up
#
# The default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.
# +
# First we will remove all of the files contained in the data_dir directory
# !rm $data_dir/*
# And then we delete the directory itself
# !rmdir $data_dir
# Similarly we will remove the files in the cache_dir directory and the directory itself
# !rm $cache_dir/*
# !rmdir $cache_dir
# -
| Mini-Projects/IMDB Sentiment Analysis - XGBoost (Updating a Model) - Solution.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Developing an AI application
#
# Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
#
# In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
#
# <img src='assets/Flowers.png' width=500px>
#
# The project is broken down into multiple steps:
#
# * Load and preprocess the image dataset
# * Train the image classifier on your dataset
# * Use the trained classifier to predict image content
#
# We'll lead you through each part which you'll implement in Python.
#
# When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
#
# First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
# +
# Imports here
import numpy as np
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
import matplotlib.pyplot as plt
from torchvision import datasets, transforms, models
from collections import OrderedDict
from PIL import Image
import os
# -
# ## Load the data
#
# Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
#
# The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
#
# The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
#
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
image_path = (test_dir + '/74/' + 'image_01191.jpg')
save_dir='checkpoints/'
# +
if(os.path.exists(save_dir)):
print ("Directory for save the model %s already exists" % save_dir)
save_dir=save_dir+'checkpoint.pth'
else:
try:
os.mkdir(save_dir)
except OSError:
print ("Creation of the directory %s failed, we use the root directory" % save_dir)
save_dir='checkpoint.pth'
else:
print ("Successfully created the directory %s for save the model" % save_dir)
save_dir=save_dir+'checkpoint.pth'
# +
# TODO: Define your transforms for the training, validation, and testing sets
#data
data_transforms = transforms.Compose([transforms.RandomRotation(30),transforms.RandomResizedCrop(224),transforms.RandomHorizontalFlip(),transforms.ToTensor(),transforms.Normalize([0.485, 0.456, 0.406],[0.229, 0.224, 0.225])])
image_datasets = datasets.ImageFolder(train_dir, transform=data_transforms)
image_data = torch.utils.data.DataLoader(image_datasets, batch_size=64, shuffle=True)
#test
testval_transforms = transforms.Compose([transforms.Resize(256),transforms.CenterCrop(224),transforms.ToTensor(), transforms.Normalize([0.485, 0.456, 0.406],[0.229, 0.224, 0.225])])
image_testset = datasets.ImageFolder(test_dir, transform=testval_transforms)
image_test = torch.utils.data.DataLoader(image_testset, batch_size=64, shuffle=True)
#validation
val_transforms = transforms.Compose([transforms.Resize(256),transforms.CenterCrop(224),transforms.ToTensor(),transforms.Normalize([0.485, 0.456, 0.406],[0.229, 0.224, 0.225])])
image_valset = datasets.ImageFolder(valid_dir, transform=val_transforms)
image_val = torch.utils.data.DataLoader(image_valset, batch_size=64, shuffle=True)
# -
# ### Label mapping
#
# You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
# +
import json
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
#print(cat_to_name)
# -
# # Building and training the classifier
#
# Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
#
# We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
#
# * Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
# * Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
# * Train the classifier layers using backpropagation using the pre-trained network to get the features
# * Track the loss and accuracy on the validation set to determine the best hyperparameters
#
# We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
#
# When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
#
# One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
# GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
#
# **Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
epochs=4
learning_rate=0.001
print_every=10
hidden_sizes = [10200, 1020] # +10% total flowers - +10% --- use 7500 on pc
output_size = 102 #total flowers
arch='vgg16' #'mobilenet_v2' #vgg16
# +
def define_model():
if arch=="vgg16":
model = models.vgg16(pretrained=True)
input_size = 25088 #32768
else:
model = models.mobilenet_v2(pretrained=True)
input_size = 1280
#freeze parameters - less memory used
for param in model.parameters():
param.requires_grad = False
classifier = nn.Sequential(OrderedDict([
('dropout',nn.Dropout(0.5)),
('fc1', nn.Linear(input_size, hidden_sizes[0])),
('relu1', nn.ReLU()),
('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),
('relu2', nn.ReLU()),
('fc3', nn.Linear(hidden_sizes[1], output_size)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier=classifier
return model
model=define_model()
#model= model.share_memory()
model = model.cuda()
# +
#no reload
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), learning_rate)
# +
#no reload
def cal_accuracy(mod, data):
loss = 0
accuracy = 0
data_len=len(data)
for i, (inputs,labels) in enumerate(data):
inputs, labels = inputs.to('cuda') , labels.to('cuda')
mod.to('cuda')
with torch.no_grad():
outputs = mod.forward(inputs)
loss = criterion(outputs,labels)
ps = torch.exp(outputs).data
equality = (labels.data == ps.max(1)[1])
accuracy += equality.type_as(torch.FloatTensor()).mean()
loss = loss / data_len
accuracy = accuracy /data_len
return loss, accuracy
# +
#no reload
def training():
print('start training')
model.to('cuda')
#model.share_memory()
step=0
for epo in range(epochs):
running_loss=0
#take the inputs and labels for the trainload
for vai_int,(inputs,labels) in enumerate(image_data):
step+=1
inputs, labels = inputs.to('cuda'), labels.to('cuda')
optimizer.zero_grad()
outputs=model.forward(inputs)
loss= criterion(outputs,labels)
loss.backward()
optimizer.step()
#print('still working ')
running_loss+=loss.item()
if step % print_every == 0:
model.eval()
#print('end validation model')
val_loss,accuracy = cal_accuracy(model, image_val)
print("Step nro: {} ".format(step),
"Epoch: {}/{} ".format(1+epo, epochs),
"Loss: {:.4f} ".format(running_loss),
"Validation Loss {:.4f} ".format(val_loss),
"Accuracy {:.4f} ".format(accuracy))
running_loss = 0
#torch.cuda.empty_cache()
training()
print('end training')
# -
# ## Testing your network
#
# It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
# +
# TODO: Do validation on the test set
#no reload
def testing():
correctos = 0
total = 0
model.eval()
model.to('cuda')
with torch.no_grad():
for inputs, labels in image_test:
inputs, labels = inputs.to('cuda'), labels.to('cuda')
outputs = model(inputs)
aux , prediction = torch.max(outputs.data, 1)
total += labels.size(0)
tensor= (prediction == labels.data).sum()
correctos+=tensor.item()
accuracy=100 * correctos / total
print('Total: {} - Correct: {} - Accuracy: {:.2f}% '.format(total,correctos,accuracy))
testing()
# -
# ## Save the checkpoint
#
# Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
#
# ```model.class_to_idx = image_datasets['train'].class_to_idx```
#
# Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
# +
# TODO: Save the checkpoint
#no reload
model.class_to_idx = image_datasets.class_to_idx
model_state={
'learning_rate':learning_rate,
'epochs':epochs,
'hidden_sizes':hidden_sizes,
'output_size':output_size,
'state_dict':model.state_dict(),
'class_to_idx':model.class_to_idx,
'arch':arch
}
torch.save(model_state, save_dir)
# -
# ## Loading the checkpoint
#
# At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# +
# TODO: Write a function that loads a checkpoint and rebuilds the model
state_model = torch.load(save_dir)
learning_rate=state_model['learning_rate']
epochs=state_model['epochs']
hidden_sizes=state_model['hidden_sizes']
output_size=state_model['output_size']
arch=state_model['arch']
model=define_model()
model = model.cuda()
model.class_to_idx=state_model['class_to_idx']
model.load_state_dict(state_model['state_dict'])
print('model load')
# -
# # Inference for classification
#
# Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
#
# ```python
# probs, classes = predict(image_path, model)
# print(probs)
# print(classes)
# > [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
# > ['70', '3', '45', '62', '55']
# ```
#
# First you'll need to handle processing the input image such that it can be used in your network.
#
# ## Image Preprocessing
#
# You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
#
# First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
#
# Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
#
# As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
#
# And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
# +
def process_image(image):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
img = Image.open(image)
image_transforms = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
])
img = image_transforms(img)
return img
# show a image
processed_image = process_image(image_path)
print('image processed')
# -
# To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
# +
def imshow(image, ax=None, title=None):
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
imshow(processed_image)
# -
# ## Class Prediction
#
# Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
#
# To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
#
# Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
#
# ```python
# probs, classes = predict(image_path, model)
# print(probs)
# print(classes)
# > [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
# > ['70', '3', '45', '62', '55']
# ```
# +
def predict(top_k=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
model.eval()
model.cpu()
img = process_image(image_path)
img = img.unsqueeze_(0)
img = img.float()
with torch.no_grad():
output = model.forward(img)
probs, classes = torch.topk(output,top_k)
probs = probs.exp()
idx_to_class = {val: key for key, val in model.class_to_idx.items()}
top_n = [idx_to_class[each] for each in classes.cpu().numpy()[0]]
return probs, top_n
probs, classes = predict()
labels = []
for index in classes:
labels.append(cat_to_name[str(index)])
print('Name of the given image: ', labels[0])
probs=probs[0]
for name, prob in zip(labels, probs):
print("Name of class and probability {}: {:6f}".format(name, prob))
print(probs)
print(classes)
# -
# ## Sanity Checking
#
# Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
#
# <img src='assets/inference_example.png' width=300px>
#
# You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
# +
# TODO: Display an image along with the top 5 classes
def sanity_checking():
plt.rcParams["figure.figsize"] = (3,3)
plt.rcParams.update({'font.size': 12})
# Showing actual image
#image_path = (test_dir + '/37/' + 'image_03783.jpg')
probs, classes = predict()
image_to_show = process_image(image_path)
image = imshow(image_to_show, ax = plt)
image.axis('off')
image.title(cat_to_name[str(classes[0])])
image.show()
# Showing Top Classes
labels = []
for class_index in classes:
labels.append(cat_to_name[str(class_index)])
y_pos = np.arange(len(labels))
probs = probs[0]
plt.barh(y_pos, probs, align='center', color='red')
plt.yticks(y_pos, labels)
plt.xlabel('Probability')
plt.title('Top Classes')
plt.show()
sanity_checking()
# -
| Image Classifier Project.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] colab_type="text" id="_CMW13uMBlpk"
# # はじめに
#
# Chainer チュートリアルへようこそ。
#
# このチュートリアルは、機械学習やディープラーニングの仕組みや使い方を理解したい**大学学部生**以上の方に向けて書かれたオンライン学習資料です。
#
# 機械学習の勉強を進めるために必要な数学の知識から、Python というプログラミング言語を用いたコーディングの基本、機械学習・ディープラーニングの基礎的な理論、画像認識や自然言語処理などに機械学習を応用する方法に至るまで、幅広いトピックを解説しています。
#
# 機械学習を学び始めようとすると、ある程度、線形代数や確率統計といった数学の知識から、何らかのプログラミング言語が使えることなどが必要となってきます。
# しかし、そういった数学やプログラミングの全てに精通していなければ機械学習について学び始められないかというと、必ずしもそうではありません。
#
# 本チュートリアルでは、機械学習やディープラーニングに興味を持った方が、まず必要になる最低限の数学とプログラミングの知識から学び始められるように、資料を充実させています。
#
# そのため、できる限りこのサイト以外の教科書や資料を探さなくても、**このサイトだけで機械学習・ディープラーニングに入門できる**ことを目指して、作られています。初学者の方が「何から学び始めればいいのか」と迷うことなく学習を始められることを目指したサイトです。
#
# また、本チュートリアルの特徴として、資料の中に登場するコードが、Google Colaboratory というサービスを利用することで**そのままブラウザ上で実行できるようになっている**という点があります。
#
# ブラウザだけでコードを書き、実行して、結果を確認することができれば、説明に使われたサンプルコードを実行して結果を確かめるために、手元のコンピュータで環境構築を行う必要がなくなります。
#
# 本章ではまず、この **Google Colaboratory** というサービスの利用方法を説明します。
# + [markdown] colab_type="text" id="TK3cXCQuBlpm"
# ## 必要なもの
#
# - Google アカウント(お持ちでない場合は、こちらからお作りください:[Google アカウントの作成](https://accounts.google.com/signup))
# - ウェブブラウザ( Google Colaboratory はほとんどの主要なブラウザで動作します。PC 版の Chrome と Firefox では動作が検証されています。)
# + [markdown] colab_type="text" id="D3QflLv0qdiy"
# ## Google Colaboratory の基本
#
# Google Colaboratory(以下 Colab )は、クラウド上で [Jupyter Notebook](https://jupyter.org/) 環境を提供する Google のウェブサービスです。Jupyter Notebook はブラウザ上で主に以下のようなことが可能なオープンソースのウェブアプリケーションであり、データ分析の現場や研究、教育などで広く用いられています。
#
# - プログラムを実行と、その結果の確認
# - Markdown と呼ばれる文章を記述するためのマークアップ言語を使った、メモや解説などの記述の追加
#
# Colab では無料で GPU も使用することができますが、そのランタイムは**最大 12 時間**で消えてしまうため、長時間を要する処理などは別途環境を用意する必要があります。
# 学びはじめのうちは、数分から数時間程度で終わる処理がほとんどであるため、気にする必要はありませんが、本格的に使っていく場合は有料のクラウドサービスを利用するなどして、環境を整えるようにしましょう。
#
# 以降では、その基本的な使い方を説明します。
# + [markdown] colab_type="text" id="SNVvaFdwBlpn"
# ### Colab を開く
#
# まずは以下のURLにアクセスして、ブラウザで Colab を開いてください。
#
# [https://colab.research.google.com/](https://colab.research.google.com/)
#
# 「Colaboratory へようこそ」というタイトルの Jupyter Notebook が表示されます。
#
# 次に、タイトルの下にある 「ファイル」 から、「Python 3 の新しいノートブック」 を選択し、まっさらな Jupyter Notebook を作成しましょう。
#
# 
#
# Google アカウントにまだログインしていなかった場合は、以下のようなメッセージが表示されます。
#
# 
#
# その場合は、「ログイン」 をクリックして、Google アカウントでログインしてください。
#
# ログインが完了すると、以下のような画面が表示され、準備完了です。
# もうすでに Python を使ったプログラミングを開始する準備が整っています。
#
# 
# + [markdown] colab_type="text" id="SWEJAKWMBlpo"
# ### Open in Colab ボタン
#
# このチュートリアルの一部の章には、`Open in Colab` と書かれた以下のようなボタンがページ上部に設置されています。
#
# [](https://colab.research.google.com/github/kandalva/tutorials/blob/master/ja/01_Welcome_to_Chainer_Tutorial_ja.ipynb)
#
# このボタンを押すと、ブラウザで見ている資料が、Colab 上で Jupyter Notebook として開かれます。
# すると、チュートリアルの中で説明に用いられているコードを、**実際に実行して結果を確認することができます。**
#
# それでは、早速上のボタンか、このページの上部に配置されている `Open in Colab` ボタンを押して、このページを Colab で開いてください。
# すると、`Playground モード` という編集不可な状態でノートブックが Colab 上で開かれます。
# そこで、下図の位置にある `ドライブにコピー` というボタンを押して、自分の Google Drive 上にこのノートブックをコピーしてください。
# このボタンを押すと、コピーされたノートブックが自動的に開き、以降は内容に編集を加えたり、コードを実行したりすることができます。
#
# 
#
# この
#
# 1. `Open in Colab` から Colab へ移動
# 2. 自分のドライブへノートブックをコピーする
# 3. コードを実行しながら解説を読んでいく
#
# という手順が、本チュートリアルサイトのおすすめの利用方法です。
# + [markdown] colab_type="text" id="k4ak2UP9Blpp"
# ## Colab の基本的な使い方
#
# Colab 上の Jupyter Notebook を以降、単に**ノートブック**と呼びます。
#
# ノートブックは、複数の**セル**と呼ばれるブロックを持つことができます。
# 新しいノートブックを作った直後では、何も書かれていないセルが一つだけ存在している状態になっています。
# セルの内側のどこかをクリックすると、そのセルを選択することができます。
#
# セルには、**コードセル**と**テキストセル**の 2 種類があります。
# **コードセル** は Python のコードを書き込み、実行するためのセルであり、**テキストセル**は、Markdown 形式で文章を書くためのセルです。
#
# それぞれのセルタイプについてもう少し詳しく説明をします。
# + [markdown] colab_type="text" id="9hYkVljaBlpq"
# ### コードセル
#
# コードセルは、Python のコードを書き込み、実行することができるセルです。
# 実行するには、コードセルを選択した状態で、`Ctrl + Enter` または `Shift + Enter` を押します。
# 試しに、下のセルを選択して、`Ctrl + Enter` を押してみてください。
# + colab={"base_uri": "https://localhost:8080/", "height": 35} colab_type="code" id="EaOJalpbBlpr" outputId="40291477-aa22-4151-da78-d2ae6d2eb627"
print('Hello world!')
# + [markdown] colab_type="text" id="8QsnHDylXQrb"
# すぐ下に、Hello world! という文字列が表示されました。
# 上のセルに書き込まれているのは Python のコードで、与えられた文字列を表示する関数である `print()` に、`'Hello world!'` という文字列を渡しています。
# これを今実行したため、その結果が下に表示されています。
#
# プログラミング言語の Python については、[次の章](https://tutorials.chainer.org/ja/02_Basics_of_Python.html) でより詳しく解説します。
# + [markdown] colab_type="text" id="f7vtQ2SmBlpx"
# ### テキストセル
#
# テキストセルでは、Markdown 形式で記述された文章を扱います。
# 試しに、このセルを**ダブルクリック**してみてください。
# テキストセルが編集モードになり、Markdown 形式で文章を装飾するための、先程までは表示されていなかった記号が見えるようになります。
#
# その状態で `Shift + Enter` を押してみましょう。
#
# もとのレンダリングされた文章の表示に戻ります。
# + [markdown] colab_type="text" id="wEwqOW9bBlpy"
# ### Colab から Google Drive を使う
#
# Google Drive というオンラインストレージサービスを Colab で開いたノートブックから利用することができます。
# ノートブック中でコードを実行して作成したファイルなどを保存したり、逆に Google Drive 上に保存されているデータを読み込んだりすることができます。
#
# Colab 上のノートブックから Google Drive を使うには、Colab 専用のツールを使って、`/content/drive` というパスに現在ログイン中の Google アカウントが持っている Google Drive のスペースをマウントします。
# + colab={} colab_type="code" id="TI3-V_gN3Ekr"
from google.colab import drive
drive.mount('/content/drive')
# + [markdown] colab_type="text" id="zllU5vanBlp2"
# このノートブックを Colab で開いてから初めて上のコードセルを実行した場合は、以下のようなメッセージが表示されます。
#
# 
#
# 指示に従って表示されているURLへアクセスしてください。
# すると、「アカウントの選択」と書かれたページに飛び、すでにログイン済みの場合はログイン中の Google アカウントのアイコンやメールアドレスが表示されています。
# 利用したいアカウントをクリックして、次に進んで下さい。
# すると次に、`Google Drive File Stream が Google アカウントへのアクセスをリクエストしています` と書かれたページに飛びます。
#
# 
#
# 右下に「許可」と書かれたボタンが見えます。
# こちらをクリックしてください。
# すると以下のように認証コードが記載されたページへ移動します。
#
# 
#
# (この画像では認証コード部分をぼかしています)
# このコードを選択してコピーするか、右側にあるアイコンをクリックしてコピーしてください。
#
# 元のノートブックへ戻り、`Enter your authorization code:` というメッセージの下にある空欄に、先程コピーした認証コードを貼り付けて、Enter キーを押してください。
#
# **Mounted at /content/drive** と表示されたら、準備は完了です。
#
# 以下のセルを実行して、自分の Google Drive が Colab からアクセス可能になっていることを確認してください。
# + colab={"base_uri": "https://localhost:8080/", "height": 35} colab_type="code" id="osShvuIQ3GFy" outputId="e244c570-af51-4af8-a9f1-5a68da0fa41b"
# 'My Drive'の表記が出ていればマウントがうまく行われています。
# !ls 'drive/'
# + [markdown] colab_type="text" id="DbvFPwpova8M"
# 上のセルで実行しているのは Python のコードではありません。
# Jupyter Notebook では、コードセル中で `!` が先頭に付いている行は特別に解釈されます。`!ls` は、次に続くディレクトリの中にあるファイルまたはディレクトリの一覧を表示せよ、という意味です([注釈1](#note1))。
# + [markdown] colab_type="text" id="jZNTuBQ54BSu"
# ### Colab の便利なショートカット
#
# Colab を使用中に、セルのタイプの変更やセルの複製・追加などの操作をする場合は、メニューから該当する項目を選ぶ方法以外に、キーボードショートカットを利用する方法もあります。
#
# 下記によく使う**ショートカットキー**をまとめておきます。
# 多くのショートカットキーは**二段階**になっており、まず `Ctrl + M` を押してから、それぞれの機能によって異なるコマンドを入力する形になっています。
#
# | 説明 | コマンド |
# | -------------------- | ------------- |
# | Markdownモードへ変更 | Ctrl + M → M |
# | Codeモードへ変更 | Ctrl + M → Y |
# | セルの実行 | Shift + Enter |
# | セルを上に追加 | Ctrl + M → A |
# | セルを下に追加 | Ctrl + M → B |
# | セルのコピー | Ctrl + M → C |
# | セルの貼り付け | Ctrl + M → V |
# | セルの消去 | Ctrl + M → D |
# | コメントアウト | Ctrl + / |
#
# コメントアウトとは、コード中で実行時に無視したい行やコメントを選択した状態で行う操作です。
# Python では、`#` の後に続く文字列は全て、コメントとして無視され、実行時に評価されることはありません。
# + [markdown] colab_type="text" id="44vOyaBKEk3m"
# ### GPU を使用する
#
# Colab では GPU を無料で使用することができます。
# 初期設定では GPU を使用しない設定となっているため、GPU を使用する場合は設定を変更する必要があります。
#
# GPU を使用する場合は、画面上部のタブの中の 「Runtime」 (または「ランタイム」) をクリックし、「Change runtime type」 (または「ランタイムのタイプを変更」)を選択します。
#
# そして、下記の画像の様に 「Hardware accelerator」 (または「ハードウェアアクセラレータ」)を GPU に変更します。
#
# 
#
# これで Colab 上で GPU を使用できるようになりました。
#
#
# + [markdown] colab_type="text" id="QoQHVO6rva8O"
# これで、チュートリアルの本編に入っていく準備が完了しました。次の章では、Python というプログラミング言語の基本について解説します。
# + [markdown] colab_type="text" id="rot1jrxLy47Y"
# <hr />
# <div class="alert alert-info">
# **注釈 1**
#
# `ls` はシェルコマンドの 1 つです。
#
# [▲上へ戻る](#ref_note1)
# </div>
#
| ja/01_Welcome_to_Chainer_Tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# formats: ipynb,.pct.py:percent
# text_representation:
# extension: .py
# format_name: percent
# format_version: '1.3'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %% [markdown]
# # Manipulating GPflow models
#
# One of the key ingredients in GPflow is the model class, which enables you to carefully control parameters. This notebook shows how some of these parameter control features work, and how to build your own model with GPflow. First we'll look at:
#
# - how to view models and parameters
# - how to set parameter values
# - how to constrain parameters (for example, variance > 0)
# - how to fix model parameters
# - how to apply priors to parameters
# - how to optimize models
#
# Then we'll show how to build a simple logistic regression model, demonstrating the ease of the parameter framework.
#
# GPy users should feel right at home, but there are some small differences.
#
# First, let's deal with the usual notebook boilerplate and make a simple GP regression model. See [Basic (Gaussian likelihood) GP regression model](../basics/regression.ipynb) for specifics of the model; we just want some parameters to play with.
# %%
import numpy as np
import gpflow
import tensorflow_probability as tfp
from gpflow.utilities import print_summary, set_trainable, to_default_float
# %% [markdown]
# We begin by creating a very simple GP regression model:
# %%
# generate toy data
np.random.seed(1)
X = np.random.rand(20, 1)
Y = np.sin(12 * X) + 0.66 * np.cos(25 * X) + np.random.randn(20, 1) * 0.01
m = gpflow.models.GPR((X, Y), kernel=gpflow.kernels.Matern32() + gpflow.kernels.Linear())
# %% [markdown]
# ## Viewing, getting, and setting parameters
# You can display the state of the model in a terminal by using `print_summary(m)`. You can change the display format using the `fmt` keyword argument, e.g. `'html'`. In a notebook, you can also use `fmt='notebook'` or set the default printing format as `notebook`:
# %%
print_summary(m, fmt="notebook")
# %%
gpflow.config.set_default_summary_fmt("notebook")
# %% [markdown]
# This model has four parameters. The kernel is made of the sum of two parts. The first (counting from zero) is a Matern32 kernel that has a variance parameter and a lengthscales parameter; the second is a linear kernel that has only a variance parameter. There is also a parameter that controls the variance of the noise, as part of the likelihood.
#
# All the model variables have been initialized at `1.0`. You can access individual parameters in the same way that you display the state of the model in a terminal; for example, to see all the parameters that are part of the likelihood, run:
# %%
print_summary(m.likelihood)
# %% [markdown]
# This gets more useful with more complex models!
# %% [markdown]
# To set the value of a parameter, just use `assign()`:
# %%
m.kernel.kernels[0].lengthscales.assign(0.5)
m.likelihood.variance.assign(0.01)
print_summary(m, fmt="notebook")
# %% [markdown]
# ## Constraints and trainable variables
#
# GPflow helpfully creates an unconstrained representation of all the variables. In the previous example, all the variables are constrained positively (see the **transform** column in the table); the unconstrained representation is given by $\alpha = \log(\exp(\theta)-1)$. The `trainable_parameters` property returns the constrained values:
# %%
m.trainable_parameters
# %% [markdown]
# Each parameter has an `unconstrained_variable` attribute that enables you to access the unconstrained value as a TensorFlow `Variable`.
# %%
p = m.kernel.kernels[0].lengthscales
p.unconstrained_variable
# %% [markdown]
# You can also check the unconstrained value as follows:
# %%
p.transform.inverse(p)
# %% [markdown]
# Constraints are handled by the Bijector classes from the `tensorflow_probability` package. You might prefer to use the constraint $\alpha = \log(\theta)$; this is easily done by replacing the parameter with one that has a different `transform` attribute (here we make sure to copy all other attributes across from the old parameter; this is not necessary when there is no `prior` and the `trainable` state is still the default of `True`):
# %%
old_parameter = m.kernel.kernels[0].lengthscales
new_parameter = gpflow.Parameter(
old_parameter,
trainable=old_parameter.trainable,
prior=old_parameter.prior,
name=old_parameter.name.split(":")[0], # tensorflow is weird and adds ':0' to the name
transform=tfp.bijectors.Exp(),
)
m.kernel.kernels[0].lengthscales = new_parameter
# %% [markdown]
# Though the lengthscale itself remains the same, the unconstrained lengthscale has changed:
# %%
p.transform.inverse(p)
# %% [markdown]
# You can also change the `transform` attribute in place:
# %%
m.kernel.kernels[0].variance.transform = tfp.bijectors.Exp()
# %%
print_summary(m, fmt="notebook")
# %% [markdown]
# ## Changing whether a parameter will be trained in optimization
#
# Another helpful feature is the ability to fix parameters. To do this, simply set the `trainable` attribute to `False`; this is shown in the **trainable** column of the representation, and the corresponding variable is removed from the free state.
# %%
set_trainable(m.kernel.kernels[1].variance, False)
print_summary(m)
# %%
m.trainable_parameters
# %% [markdown]
# To unfix a parameter, just set the `trainable` attribute to `True` again.
# %%
set_trainable(m.kernel.kernels[1].variance, True)
print_summary(m)
# %% [markdown]
# **NOTE:** If you want to recursively change the `trainable` status of an object that *contains* parameters, you **must** use the `set_trainable()` utility function.
#
# A module (e.g. a model, kernel, likelihood, ... instance) does not have a `trainable` attribute:
# %%
try:
m.kernel.trainable
except AttributeError:
print(f"{m.kernel.__class__.__name__} does not have a trainable attribute")
# %%
set_trainable(m.kernel, False)
print_summary(m)
# %% [markdown]
# ## Priors
#
# You can set priors in the same way as transforms and trainability, by using `tensorflow_probability` distribution objects. Let's set a Gamma prior on the variance of the Matern32 kernel.
# %%
k = gpflow.kernels.Matern32()
k.variance.prior = tfp.distributions.Gamma(to_default_float(2), to_default_float(3))
print_summary(k)
# %%
m.kernel.kernels[0].variance.prior = tfp.distributions.Gamma(
to_default_float(2), to_default_float(3)
)
print_summary(m)
# %% [markdown]
# ## Optimization
#
# To optimize your model, first create an instance of an optimizer (in this case, `gpflow.optimizers.Scipy`), which has optional arguments that are passed to `scipy.optimize.minimize` (we minimize the negative log likelihood). Then, call the `minimize` method of that optimizer, with your model as the optimization target. Variables that have priors are maximum a priori (MAP) estimated, that is, we add the log prior to the log likelihood, and otherwise use Maximum Likelihood.
# %%
opt = gpflow.optimizers.Scipy()
opt.minimize(m.training_loss, variables=m.trainable_variables)
# %% [markdown]
# ## Building new models
#
# To build new models, you'll need to inherit from `gpflow.models.BayesianModel`.
# Parameters are instantiated with `gpflow.Parameter`.
# You might also be interested in `gpflow.Module` (a subclass of `tf.Module`), which acts as a 'container' for `Parameter`s (for example, kernels are `gpflow.Module`s).
#
# In this very simple demo, we'll implement linear multiclass classification.
#
# There are two parameters: a weight matrix and a bias (offset). You can use
# Parameter objects directly, like any TensorFlow tensor.
#
# The training objective depends on the type of model; it may be possible to
# implement the exact (log)marginal likelihood, or only a lower bound to the
# log marginal likelihood (ELBO). You need to implement this as the
# `maximum_log_likelihood_objective` method. The `BayesianModel` parent class
# provides a `log_posterior_density` method that returns the
# `maximum_log_likelihood_objective` plus the sum of the log-density of any priors
# on hyperparameters, which can be used for MCMC.
# GPflow provides mixin classes that define a `training_loss` method
# that returns the negative of (maximum likelihood objective + log prior
# density) for MLE/MAP estimation to be passed to optimizer's `minimize`
# method. Models that derive from `InternalDataTrainingLossMixin` are expected to store the data internally, and their `training_loss` does not take any arguments and can be passed directly to `minimize`.
# Models that take data as an argument to their `maximum_log_likelihood_objective` method derive from `ExternalDataTrainingLossMixin`, which provides a `training_loss_closure` to take the data and return the appropriate closure for `optimizer.minimize`.
# This is also discussed in the [GPflow with TensorFlow 2 notebook](../intro_to_gpflow2.ipynb).
# %%
import tensorflow as tf
class LinearMulticlass(gpflow.models.BayesianModel, gpflow.models.InternalDataTrainingLossMixin):
# The InternalDataTrainingLossMixin provides the training_loss method.
# (There is also an ExternalDataTrainingLossMixin for models that do not encapsulate data.)
def __init__(self, X, Y, name=None):
super().__init__(name=name) # always call the parent constructor
self.X = X.copy() # X is a NumPy array of inputs
self.Y = Y.copy() # Y is a 1-of-k (one-hot) representation of the labels
self.num_data, self.input_dim = X.shape
_, self.num_classes = Y.shape
# make some parameters
self.W = gpflow.Parameter(np.random.randn(self.input_dim, self.num_classes))
self.b = gpflow.Parameter(np.random.randn(self.num_classes))
# ^^ You must make the parameters attributes of the class for
# them to be picked up by the model. i.e. this won't work:
#
# W = gpflow.Parameter(... <-- must be self.W
def maximum_log_likelihood_objective(self):
p = tf.nn.softmax(
tf.matmul(self.X, self.W) + self.b
) # Parameters can be used like a tf.Tensor
return tf.reduce_sum(tf.math.log(p) * self.Y) # be sure to return a scalar
# %% [markdown]
# ...and that's it. Let's build a really simple demo to show that it works.
# %%
np.random.seed(123)
X = np.vstack(
[
np.random.randn(10, 2) + [2, 2],
np.random.randn(10, 2) + [-2, 2],
np.random.randn(10, 2) + [2, -2],
]
)
Y = np.repeat(np.eye(3), 10, 0)
import matplotlib.pyplot as plt
plt.style.use("ggplot")
# %matplotlib inline
plt.rcParams["figure.figsize"] = (12, 6)
_ = plt.scatter(X[:, 0], X[:, 1], 100, np.argmax(Y, 1), lw=2, cmap=plt.cm.viridis)
# %%
m = LinearMulticlass(X, Y)
m
# %%
opt = gpflow.optimizers.Scipy()
opt.minimize(m.training_loss, variables=m.trainable_variables)
# %%
xx, yy = np.mgrid[-4:4:200j, -4:4:200j]
X_test = np.vstack([xx.flatten(), yy.flatten()]).T
f_test = np.dot(X_test, m.W.read_value()) + m.b.read_value()
p_test = np.exp(f_test)
p_test /= p_test.sum(1)[:, None]
# %%
plt.figure(figsize=(12, 6))
for i in range(3):
plt.contour(xx, yy, p_test[:, i].reshape(200, 200), [0.5], colors="k", linewidths=1)
_ = plt.scatter(X[:, 0], X[:, 1], 100, np.argmax(Y, 1), lw=2, cmap=plt.cm.viridis)
# %% [markdown]
# That concludes the new model example and this notebook. You might want to see for yourself that the `LinearMulticlass` model and its parameters have all the functionality demonstrated here. You could also add some priors and run Hamiltonian Monte Carlo using the HMC optimizer `gpflow.train.HMC` and its `sample` method. See [Markov Chain Monte Carlo (MCMC)](../advanced/mcmc.ipynb) for more information on running the sampler.
| doc/source/notebooks/understanding/models.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [default]
# language: python
# name: python3
# ---
# + slideshow={"slide_type": "-"}
"""This area sets up the Jupyter environment.
Please do not modify anything in this cell.
"""
import os
import sys
# Add project to PYTHONPATH for future use
sys.path.insert(1, os.path.join(sys.path[0], '..'))
# Import miscellaneous modules
from IPython.core.display import display, HTML
# Set CSS styling
with open('../admin/custom.css', 'r') as f:
style = """<style>\n{}\n</style>""".format(f.read())
display(HTML(style))
# + [markdown] slideshow={"slide_type": "slide"}
# # Outline
#
# <div class="alert alert-warning">
# The following notebook will go through the basics of **supervised learning**.
# </div>
#
# In supervised learning we assume that our data consist of **input - output** pairs. A learning algorithm analyses the data and produces a function, or model, we can use to infer *outputs* given unseen future *inputs*.
#
# Below we can see a simplified illustration of the supervised learning problem.
#
# Pairs of inputs $\mathbf{x}$ and outputs $y$ constitutes our training examples, where the inputs are sampled from a probability distribution. A pair $(\mathbf{x}, y)$ is related by an *unknown* target function $f$ governed by a conditional probability distribution. The ultimate goal of supervised learning is to learn a function $g$ which approximates $f$ well.
#
# The particular approximation $g$ we pick is called a hypothesis. A learning algorithm is responsible for picking the most appropriate hypothesis from a hypothesis set. The decision between which hypothesis to pick is done by looking at the *data* and typically involves an error function which measures how good a hypothesis may be.
#
# <img src="resources/supervised-learning.png" alt="Supervised Learning" width="700" />
#
# When our learning algorithm has picked a good hypothesis, we can feed it new and unseen samples to produce output estimates.
#
# The name of the data typically differ depending on which area you are from.
#
# The **input** variables are commonly known as:
#
# - covariates
# - predictors
# - features
#
# The **output** variables are commonly known as:
#
# - variates
# - targets
# - labels
# -
# ## Linear Models: Regression
#
# For now we will focus on one of the simplest supervised learning problems: *linear regression*.
#
# A linear regression model learns a real-valued function where one or more dependent output variable(s) *depend* linearly on one or more independent input variable(s). Geometrically, this real-valued function can be interpreted as a hyperplane which we attempt to fit to our data.
#
#
# ### Motivation
#
# * Allows us to investigate the relationship between two or more variables statistically
# * Can be thought of as a building block of artificial neural networks
# * A solution can be found analytically or using data-driven optimisation
# * Basic introduction to supervised learning
# * Introduces you to the Python programming language and Jupyter notebook usage
#
# <img src="https://imgs.xkcd.com/comics/machine_learning.png" alt="xkcd" width="300" />
# ## Notation
#
# This notebook will use the following notation:
#
# * A (training) dataset has $N$ input - output pairs: $(\mathbf{x}_i, y_i)$, where $i$ signifies the $i$th example
# * Each input $\mathbf{x}_i$ is a $d$ dimensional column vector: $\mathbf{x}_i \in \mathbb{R}^d$
# * For this notebook we will assume the output to be univariate: $y \in \mathbb{R}$
#
# Keep in mind that additional notation will be introduced as we continue through the notebooks.
# # Example: Income vs. Education
#
# In the following example we will load data from a CSV file and use to estimate a linear model between an `Education index` and a `Income index`.
#
# * **input** $\rightarrow$ Scalar metric indicating level of education
# * **output** $\rightarrow$ Scalar metric indication level of income
#
# <div class="alert alert-info">
# <strong>In the follow code snippets we will:</strong>
# <ul>
# <li>Load data from a CSV file</li>
# <li>Plot the data</li>
# </ul>
# </div>
# First, let's begin by importing a selection of Python package that will prove useful for the rest of this Jupyter notebook.
# +
# Plots will be show inside the notebook
# %matplotlib notebook
import matplotlib.pyplot as plt
# NumPy is a package for manipulating N-dimensional array objects
import numpy as np
# Pandas is a data analysis package
import pandas as pd
import problem_unittests as tests
# -
# With Pandas we can load the aforementioned CSV data.
# +
# Load data and print the first n = 5 rows
# URL: http://www-bcf.usc.edu/~gareth/ISL/Income1.csv
DATA_URL = './resources/Income1.csv'
data = pd.read_csv(DATA_URL, index_col=0)
print(data.head(n=5))
# Put the second (education index) and third (income index) row in a NumPy array
X_data = data['Education'].values
y_data = data['Income'].values
# -
# With the data loaded we can plot it as a scatter plot using matplotlib.
# +
plt.figure()
plt.scatter(X_data, y_data, label='Training data')
plt.title('Education vs. Income')
plt.xlabel('Education index')
plt.ylabel('Income index')
plt.grid(linestyle='dotted')
plt.legend()
plt.show()
# -
# ## Modelling
#
# As previously mentioned, we will be using a linear model. That is, the output will be a linear combination of the input plus a bias or intercept:
#
# $$
# \begin{equation*}
# g(\mathbf{x}) = b + \sum_{j=1}^{d}w_jx_j
# \end{equation*}
# $$
#
# Keep in mind that in this problem there is only a single independent variable $\mathbf{x}$, which means the above can be simplified to: $g(x) = b + wx$, where $b$ is the intercept and $w$ is the slope.
#
#
# ### Notational Simplifications
#
# To simplify notation, it is quite common to merge the bias $b$ with the weights $w_i$ to get a single weight vector $\mathbf{w} = (w_0, w_1, \ldots, w_d)^\intercal$, where $w_0 = b$. Consequently, an extra dimension must be prepended to the input vector, i.e. $\mathbf{x} = (1, x_1, \ldots, x_d)^\intercal$.
#
# With this simplification the linear model can be written as:
#
# $$
# \begin{equation*}
# g(\mathbf{x}) = \sum_{j=1}^{d}w_jx_j
# \end{equation*}
# $$
#
#
# #### Matrix Form
#
# The above model takes a single input $\mathbf{x}$ and produces a single output prediction. We can take this one step further by putting all of the input examples in a single matrix called the *design matrix* $\mathbf{X}$. This matrix consists of one (training) example per row.
#
# <br class="math" />
# $$
# \begin{equation*}
# \mathbf{X} =
# \begin{bmatrix}
# 1 & \mathbf{x}_{11} & \cdots & \mathbf{x}_{1d} \\
# \vdots & \vdots & \ddots & \vdots \\
# 1 & \mathbf{x}_{N1} & \cdots & \mathbf{x}_{Nd}
# \end{bmatrix} =
# \left[ \begin{array}{c} \mathbf{x}_{1}^\intercal \\ \vdots\\ \mathbf{x}_{N}^\intercal\end{array} \right]
# \end{equation*}
# $$
# <br class="math" />
#
# With the design matrix, predictions can be done by matrix multiplication:
#
# <br class="math" />
# $$
# \begin{equation*}
# \hat{\mathbf{y}} = \mathbf{X}\mathbf{w} =
# \begin{bmatrix}
# 1 & \mathbf{x}_{11} & \cdots & \mathbf{x}_{1d} \\
# \vdots & \vdots & \ddots & \vdots \\
# 1 & \mathbf{x}_{N1} & \cdots & \mathbf{x}_{Nd}
# \end{bmatrix}
# \left[ \begin{array}{c} \mathbf{w}_{0} \\ \mathbf{w}_{1} \\ \vdots\\ \mathbf{w}_{d}\end{array} \right] =
# \left[ \begin{array}{c} y_{1} \\ y_{2} \\ \vdots\\ y_{N}\end{array} \right]
# \end{equation*}
# $$
# <br class="math" />
# ## Defining an Error Function
#
# To measure how well our hypothesis, i.e. a particular set of weights, approximates the unknown target function $f$ we will have to come up with an error function. This quantification, which we will call $J$, goes by several different names:
#
# * Cost
# * Energy
# * Error
# * Loss
# * Objective
#
# We will be using *squared error*: $(g(\mathbf{x}) - f(\mathbf{x}))^2$ to measure how well our hypothesis approximates $f$. Seeing as we do not have access to $f$ we will instead compute an in-sample squared error over all our training data. This measure is commonly known as *mean squared error* (MSE):
#
# $$
# \begin{equation*}
# J(\mathbf{w}) =
# \frac{1}{N}\sum_{i=1}^{N}(g(\mathbf{x}_i) - y_i)^2 =
# \frac{1}{N}\sum_{i=1}^{N}(\mathbf{w}^\intercal \mathbf{x}_i - y_i)^2 =
# \frac{1}{N}\lVert \mathbf{X}\mathbf{w} - \mathbf{y} \rVert^2
# \end{equation*}
# $$
#
# A simple analogy is to think of mean squared error as a set of springs, one per training example. The objective of the learning algorithm is to balance the learned hyperplane by attempting to push it as close as we can to each of the training samples. Thus, the futher the training sample is to our hyperplane, the stronger the force is on a particular spring.
#
# <img src="resources/mse.png" alt="MSE Springs" width="300" />
# ### Minimising the Error Function in Matrix Form
#
# Now, to get a good approximation, we need to select weights $\mathbf{w}$ so that the error $J(\mathbf{w})$ is minimised. This is commonly called *ordinary least squares* or OLS. There are several ways to do this, for example, gradient descent, however, for now we will simply take the derivative of $J(\mathbf{w})$ with respect to $\mathbf{w}$ and
# then equate it to zero to get the closed-form solution.
#
# First though, we need to expand the mean squared error representation so that we can differentiate it. The constant $\frac{1}{N}$ has been removed as it will not impact the selected weights.
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# J(\mathbf{w}) &= \lVert \mathbf{X}\mathbf{w} -
# \mathbf{y}\rVert^2 \\
# & = (\mathbf{X}\mathbf{w} - \mathbf{y})^\intercal(\mathbf{X}\mathbf{w} -
# \mathbf{y}) \\
# & = ((\mathbf{X}\mathbf{w})^\intercal - \mathbf{y}^\intercal)(\mathbf{X}
# \mathbf{w} - \mathbf{y}) \\
# & = (\mathbf{X}\mathbf{w})^\intercal \mathbf{X}\mathbf{w} -
# (\mathbf{X}\mathbf{w})^\intercal \mathbf{y} - \mathbf{y}^\intercal(\mathbf{X}
# \mathbf{w}) + \mathbf{y}^\intercal\mathbf{y} \\
# & = \mathbf{w}^\intercal\mathbf{X}^\intercal\mathbf{X}\mathbf{w} -
# 2(\mathbf{X}\mathbf{w})^\intercal \mathbf{y} + \mathbf{y}^\intercal\mathbf{y} \\
# & = \mathbf{w}^\intercal\mathbf{X}^\intercal\mathbf{X}\mathbf{w} -
# 2\mathbf{y}^\intercal\mathbf{X}\mathbf{w} + \mathbf{y}^\intercal\mathbf{y}
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# <div class="alert alert-warning">
# Before we move on, here are some useful properties for matrix differentiation:
# <ul>
# <li>$\frac{\partial \mathbf{w}^\intercal\mathbf{A}\mathbf{w}}{\partial \mathbf{w}} = 2\mathbf{A}^\intercal\mathbf{w}$</li>
# </ul>
# <ul>
# <li>$\frac{\partial \mathbf{B}\mathbf{w}}{\partial \mathbf{w}} = \mathbf{B}^\intercal$</li>
# </ul>
# </div>
#
# Let $A = \mathbf{X}^\intercal\mathbf{X}$ and $B = 2\mathbf{y}^\intercal\mathbf{X}$. Substitute and differentiate:
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# \frac{\partial J(\mathbf{w})}{\partial \mathbf{w}}
# &= \frac{\partial}{\partial \mathbf{w}}
# (\mathbf{w}^\intercal\mathbf{X}^\intercal\mathbf{X}\mathbf{w} -
# 2\mathbf{y}^\intercal\mathbf{X}\mathbf{w} +
# \mathbf{y}^\intercal\mathbf{y}) \\
# &= \frac{\partial}{\partial \mathbf{w}}
# (\mathbf{w}^\intercal A \mathbf{w} -
# B\mathbf{w} +
# \mathbf{y}^\intercal\mathbf{y}) \\
# &= 2\mathbf{A}^\intercal\mathbf{w} - \mathbf{B}^\intercal + 0
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# Now, let's replace $\mathbf{A}$ and $\mathbf{B}$:
#
# <br class="math" />
# $$
# \begin{equation*}
# \frac{\partial J(\mathbf{w})}{\partial \mathbf{w}}
# = 2\mathbf{X}^\intercal\mathbf{X}\mathbf{w} - 2\mathbf{X}^\intercal\mathbf{y}
# \end{equation*}
# $$
# <br class="math" />
#
# Finally, let's throw away constant terms, equate to zero, and solve for $\mathbf{w}$:
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# \frac{\partial J(\mathbf{w})}{\partial \mathbf{w}}
# &= 0 \\
# \mathbf{X}^\intercal\mathbf{X}\mathbf{w} - \mathbf{X}^\intercal\mathbf{y} &= 0 \\
# \mathbf{X}^\intercal\mathbf{X}\mathbf{w} &= \mathbf{X}^\intercal\mathbf{y} \\
# \mathbf{w} &= (\mathbf{X}^\intercal\mathbf{X})^{-1}\mathbf{X}^\intercal\mathbf{y}
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# And there we have it, the closed-form solution for ordinary least squares.
#
# Notice how we have to compute the inverse of a matrix. This means that $\mathbf{X}^\intercal\mathbf{X}$ must be non-singular, however, there are ways to circumvent this issue, for example, by using the Moore-Penrose pseudoinverse instead: `numpy.linalg.pinv()`.
# ## Using the Closed-Form Solution
#
# To use the closed-form solution we derived above to solve the `income` vs. `education` problem we require a few things, namely:
#
# * The design matrix $\mathbf{X}$
# * A column vector of ground truths $\mathbf{y}$
# * A function that takes the two aforementioned matrices and evaluates the closed-form solution to get a set of weights $\mathbf{w}$
#
# The last two requirements will have to be implemented by you.
#
# <div class="alert alert-info">
# <strong>In the follow code snippet we will:</strong>
# <ul>
# <li>Create the design matrix $\mathbf{X}$</li>
# </ul>
# </div>
# +
def build_X(x_data):
"""Return design matrix given an array of N samples with d dimensions.
"""
# Create matrix Ax1 if d = 1
if x_data.ndim == 1:
x_data = np.expand_dims(x_data, axis=1)
# Find the number of samples and dimensions
nb_samples = x_data.shape[0]
nb_dimensions = x_data.shape[1]
# Create Nxd+1 matrix filled with ones
_X = np.ones((nb_samples, nb_dimensions + 1))
# Paste in the data we have in the new matrix
_X[:nb_samples, 1:nb_dimensions + 1] = x_data
return _X
# Test and see that the design matrix was built correctly
tests.test_build_x(build_X)
# -
# ### Task I: Build y
#
# The second component we require is the vector $\mathbf{y}$. This is a column vector over all the ground truths or target values in our training dataset. For completeness, it has the following form:
#
# <br class="math" />
# $$
# \begin{equation*}
# \mathbf{y} = \left[ \begin{array}{c} y_{1} \\ y_{2} \\ \vdots\\ y_{N}\end{array} \right]
# \end{equation*}
# $$
# <br class="math" />
#
# <div class="alert alert-success">
# **Task**: Build the $\mathbf{y}$ vector shown above. Use the previous code snippet as a reference for your implementation.
# </div>
# + slideshow={"slide_type": "subslide"}
def build_y(y_data):
"""Return a column vector containing the target values y.
"""
# Make a copy of the argument that we can work on
_y = y_data.copy()
# Create y matrix Nx1
# Return result
return _y
### Do *not* modify the following line ###
# Test and see that the y vector was built correctly
tests.test_build_y(build_y)
# -
# ### Task II: Implement Closed-Form Solution
#
# Now that we have both the design matrix $\mathbf{X}$ and the vector of target values $\mathbf{y}$ we can fit a linear model using the closed-form solution we derived before. Remember all of we have to do is implement the following expression:
#
# $$
# \begin{equation*}
# \mathbf{w} = (\mathbf{X}^\intercal\mathbf{X})^{-1}\mathbf{X}^\intercal\mathbf{y}
# \end{equation*}
# $$
#
# Please refer to the following sources for how to utilise the various functions in NumPy when implementing your solution:
#
# * How to perform matrix multiplication in NumPy [np.dot()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html)
# * How to compute the inverse of a matrix in NumPy [np.linalg.inv()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.inv.html) or [np.linalg.pinv()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.pinv.html)
# * How to transpose a NumPy array [X.T](https://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.T.html)
#
# <div class="alert alert-success">
# **Task**: Implement a function that evaluates the closed-form solution given a design matrix $\mathbf{X}$ and target vector $\mathbf{y}$.
# </div>
# +
def compute_weights(X, y):
"""Return a vector of weights found by the derived closed-form solution.
"""
weights = None
# Implement closed-form solution here
return weights
### Do *not* modify the following line ###
# Test and see that the weights are calculated correctly
tests.test_compute_theta(compute_weights)
# -
# ### Task III: Learn a Linear Regression Model
#
# We have now implemeted all of the necessary building blocks:
#
# * `build_X()` : Used to build the design matrix $\mathbf{X}$
# * `build_y()` : Used to build the vector of target values $\mathbf{y}$
# * `compute_weights` : Used to fit a linear model to the data using the solution we derived above
#
# After we have estimated $\mathbf{w}$ we can perform predictions on unseen data by computing: $\hat{\mathbf{y}} = \mathbf{X}\mathbf{w}$.
#
# <div class="alert alert-success">
# **Task**: Learn the weights $\mathbf{w}$ given the building blocks we have implemented.
# </div>
# +
# Build design matrix (TASK)
X = None
# Build y vector (TASK)
y = None
# Learn linear model (TASK)
W = None
# -
# <div class="alert alert-info">
# <strong>In the follow code snippet we will:</strong>
# <ul>
# <li>Print the weights we learned</li>
# <li>Plot the hyperplane (line in our case because $d=1$) that $\mathbf{w}$ represents</li>
# </ul>
# </div>
# +
# Print weights
print('The learned linear model looks like this:')
print('Y = {:.3f} x + {:.3f}'.format(W[1, 0], W[0, 0]))
# Plot hyperplane and training data
xs = np.linspace(X_data.min(), X_data.max(), num=50)
ys = np.dot(build_X(xs), W)
plt.figure()
plt.scatter(X_data, y_data, label='Training data')
plt.plot(xs, ys, color='Red', linewidth=1, label='Fit')
plt.title('Education vs. Income')
plt.xlabel('Education index')
plt.ylabel('Income index')
plt.grid(linestyle='dotted')
plt.legend()
plt.show()
# -
# ## Critical Analysis
#
# Albeit easy to derive and easy to use, our closed-form solution has a few shortcomings:
#
# * Requires matrix inversion
# * Very computationally expensive
# * Not ideal for distributed computing
# * Issues become apparant when the number of features $d$ and number of samples $N$ begin to grow
# * Depending on the size of the dataset it might be difficult / infeasible to fit all of it in memory
#
# To tackle these issues we will attempt to solve the linear regression problem using an iterative optimisation method called **gradient descent**.
# # Gradient Descent
#
# <div class="alert alert-warning">
# In artificial neural network literature one can see several different symbols in use to signify the error function. For example, in addition to $J$ there is also $E$ (error), $L$ (loss), $C$ (cost), and even $err$. The rest of this notebook will use the symbol $E$ instead of $J$.
# </div>
#
# Gradient descent is an iterative optimisation algorithm. In general, it works by taking the derivative of an error function $E(\mathbf{w})$ with respect to the parameters $\mathbf{w}$ of the model, and then alter the parameters in the direction of the *negative* gradient.
#
# This can be summarised as: $\mathbf{w}(k+1)\leftarrow\mathbf{w}(k) - \eta\frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}$, where $\mathbf{w}(k)$ signifies the state of the model parameters at iteration $k$, and $\eta$ is known as the *learning rate* and decides how much the parameters should change with each application of the rule.
#
# This *update rule* is repeated until convergence or until the maximum number of iterations has been reached.
#
# **With gradient descent we can**:
#
# * Reduce memory issues by only working on parts of the data at a time
# * Distribute the computation among several computational nodes. This enables distributed computing and parallelisation which allows us to exploit new architectures such as GPUs, FPGAs, and ASICs
# * Gradient descent is a heavily use *type* of algorithm that opens the door for models such as artificial neural networks
# ## Digression: A Different Perspective
#
# Linear models, such as linear regression, can be represented as artifical neural networks. An illustration of this can be seen below:
#
# <img src="resources/linear-regression-net.png" alt="Linear regression as an artificial neural network" width="300" />
#
# As before, the input $\mathbf{x} \in \mathbb{R}^d$ and the input is integrated via a linear combination plus a bias. The integrated value is activated by an activation function $\sigma$, which for our linear regression model is defined as $\sigma(x) = x$.
#
# In other words, $\hat{y}$ is defined as $\sigma(\mathbf{X}\mathbf{w})$, which simplifies to $\mathbf{X}\mathbf{w}$ because the activation function used for linear regression is the identity function. In artificial neural network terminology we would typically say that the activation function is *linear*.
# ## Learning with Gradient Descent
#
# As we saw above, learning with gradient descent is easy. All we have to do is apply an *update rule* a set number of iterations until we are satisfied with the resulting weights. The update rule can be be seen below:
#
# $$
# \begin{equation*}
# \mathbf{w}(k+1)\leftarrow\mathbf{w}(k) - \eta\frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}
# \end{equation*}
# $$
#
# In words, the weights for the next iteration $k+1$ is the weights of the current iteration $k$ plus the *negative* gradient $\frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}$ scaled by the learning rate $\eta$. In other words, for each iteration in gradient descent we adjust the weights we have with respect to the gradient of the error function $E(\mathbf{w})$.
#
# An illustration of how this could look like with the mean squared error function can be seen below:
#
# <img src="resources/error-grad.png" alt="MSE gradient" width="300" />
#
# The current state of several different weight states are signified by red dots, while the arrow points in the negative gradient direction. The optimal weight state is found at the minima, which yields the lowest amount of error.
# ### Finding the Gradient
#
# To finalise the update rule we need to find: $\frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}$. This, of course, depends on the form of $E(\mathbf{w})$.
#
# The squared error for a single sample $\mathbf{x}_i$ in the training dataset is defined as:
#
# $E(\mathbf{w}) = (\hat{y}_i - y_i)^2$
#
# where $\hat{y}_i=\sigma(g)$ and $g(\mathbf{x})=\mathbf{w}^\intercal \mathbf{x}_i$.
#
# To simplify the derivation we will scale the squared error by halving it; this will not change the optimal solution:
#
# $E(\mathbf{w}) = \frac{1}{2}(\hat{y}_i - y_i)^2$
#
# Let's now attempt to find the derivative we need:
#
# <br class="math" />
# $$
# \begin{equation*}
# \frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}
# = \frac{\partial}{\partial\mathbf{w}}( \frac{1}{2}(\hat{y}_i - y_i)^2)
# \end{equation*}
# $$
# <br class="math" />
#
# Seeing as $\hat{y}$ is dependent on $\mathbf{w}$ we will need to use the chain rule of calculus.
#
# <div class="alert alert-warning">
# Let $a(b) = \frac{1}{2}(b)^2$ and $b(\mathbf{w}) = (\hat{y}_i - y_i)$, then $\frac{\partial a}{\partial\mathbf{w}}=\frac{\partial a}{\partial b}\frac{\partial b}{\partial\mathbf{w}}$.
# </div>
#
# Therefore:
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# \frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}
# &= \frac{\partial a}{\partial b}\frac{\partial b}{\partial\mathbf{w}} \\
# &= (\hat{y}_i - y_i)\frac{\partial}{\partial\mathbf{w}}(\hat{y}_i - y_i) \\
# &= (\hat{y}_i - y_i)((\frac{\partial}{\partial\mathbf{w}}\hat{y}_i) - (\frac{\partial}{\partial\mathbf{w}}y_i)) \\
# &= (\hat{y}_i - y_i)((\frac{\partial}{\partial\mathbf{w}}\hat{y}_i) - 0) \\
# &= (\hat{y}_i - y_i)\frac{\partial}{\partial\mathbf{w}}\hat{y}_i \\
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# Keep in mind that:
#
# * $\hat{y}_i=\sigma(g)$
# * $g(\mathbf{x})=\mathbf{w}^\intercal \mathbf{x}_i$.
#
# For now, let's replace $\hat{y}$ with $\sigma(g)$:
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# \frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}
# &= (\hat{y}_i - y_i)\frac{\partial}{\partial\mathbf{w}}\sigma(g)
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# Again we have to use the chain rule.
#
# <div class="alert alert-warning">
# Let $a(b) = \sigma(b)$ and $b(\mathbf{w}) = (\mathbf{w}^\intercal \mathbf{x}_i)$, then $\frac{\partial a}{\partial\mathbf{w}}=\frac{\partial a}{\partial b}\frac{\partial b}{\partial\mathbf{w}}$.
# </div>
#
# <br class="math" />
# $$
# \begin{equation*}
# \begin{aligned}
# \frac{\partial E(\mathbf{w})}{\partial\mathbf{w}}
# &= (\hat{y}_i - y_i)\frac{\partial a}{\partial b}\frac{\partial b}{\partial\mathbf{w}} \\
# &= (\hat{y}_i - y_i)\sigma '(g)\mathbf{x}_i
# \end{aligned}
# \end{equation*}
# $$
# <br class="math" />
#
# Thus, the update rule for gradient descent, regardless of activation function, is defined as:
#
# <br class="math" />
# $$
# \begin{equation*}
# \mathbf{w}(k+1) \leftarrow \mathbf{w}(k) - \eta((\hat{y}_i - y_i)\sigma '(g)\mathbf{x}_i)
# \end{equation*}
# $$
# <br class="math" />
#
# Seeing as we're doing linear regression, we know that activation function is linear, i.e. $\sigma(x)=x$, where $\sigma'(x)=1$. So the final update rule will look like this:
#
# <div class="alert alert-info">
# $$
# \begin{equation*}
# \begin{aligned}
# \mathbf{w}(k+1) &\leftarrow \mathbf{w}(k) - \eta((\hat{y}_i - y_i)\mathbf{x}_i) \\
# &\leftarrow \mathbf{w}(k) - \eta((\mathbf{w}^\intercal \mathbf{x}_i - y_i)\mathbf{x}_i)
# \end{aligned}
# \end{equation*}
# $$
# </div>
#
# Note that this updates the weights using only a single input example. This is generally called *stochastic* gradient descent. Typically the amount we adjust by is taken over a *batch*, i.e. subset, of examples.
#
# For completeness, the update rule above can be defined over a set of $m$ samples like so:
#
# $$
# \begin{equation*}
# \mathbf{w}(k+1) \leftarrow \mathbf{w}(k) - \eta\frac{1}{m}\sum_{i=i}^{m}(\mathbf{w}^\intercal \mathbf{x}_i - y_i)\mathbf{x}_i
# \end{equation*}
# $$
# ## Gradient Descent with Keras
#
# Thankfully, when using gradient descent we do not need to derive and implement it ourselves as there are many programming libraries out there that can do automatic differentiation for us.
#
# In this and future notebooks we will be using the Python library [Keras](https://keras.io/). This is a high-level library for building and training artificial neural networks running on either [TensorFlow](https://www.tensorflow.org/) or [Theano](http://deeplearning.net/software/theano/). We will be able to leverage Keras when creating our linear regression model with gradient descent because linear models can be interpreted as artificial neural networks.
#
# <div class="alert alert-info">
# <strong>In the following code snippets we will:</strong>
# <ul>
# <li>Create a linear regression model for the `Income` vs. `Education` problem in Keras</li>
# <li>Train the model using (stochastic) gradient descent</li>
# </ul>
# </div>
# Let's start by importing the modules we need from Keras as well as some additional ones we will use during training.
# +
import time
# A library for easily displaying progress meters
import tqdm
# Contains all built-in optimisation tools in Keras, such as stochastic gradient descent
from keras import optimizers
# An input "layer" and a densely-connected neural network layer
from keras.layers import Input, Dense
# Model is an API that wraps our linear regression model
from keras.models import Model
# -
# The input to our model is a single scalar value (`Education`). The output is also a single scalar value (`Income`).
# +
# There is only a *single* feature
input_X = Input(shape=(1,))
# The output of the model is a single value
output_y = Dense(units=1, use_bias=True)(input_X)
# We give the input and output to our Model API
model = Model(inputs=input_X, outputs=output_y)
# Print a summary of the model
model.summary()
# -
# Notice in the print above how the fully-connected layer `Dense()` has two *trainable* parameters. One is the weight (slope), while the second is the bias (intercept). Keras adds bias units by default, but it can be turned off by setting `use_bias=False`.
#
# The next thing we have to do in Keras is to set up an *optimiser* (sometimes called *solver*). There are many [alternatives](https://keras.io/optimizers/) to select from, however, we will settle for the stochastic gradient descent algorithm we discussed earlier.
# +
#
# Start by setting some user options
#
# Learning rate (set very small so we can clearly see the training progress)
lr = 0.0001
# Number of times to apply the update rule
nb_iterations = 100
# Number of samples to include each iteration (used to compute gradients)
nb_samples = 30
# Create optimiser using Keras
sgd = optimizers.SGD(lr=lr)
# Add the optimiser to our model, make it optimise mean squared error
model.compile(optimizer=sgd, loss='mean_squared_error')
# -
# Now that both the model definition and the optimiser is set up we can start training. Training using the Keras model API is done by calling the `fit()` method.
#
# Don't worry too much if this code is a bit too much right now. We will get much more experience with using Keras throughout the upcoming notebooks.
#
# While training the model, a plot is continuously updated to display the fitted line.
# +
fig, ax = plt.subplots(1,1)
# Perform `nb_iterations` update rule applications
for i in tqdm.tqdm(np.arange(nb_iterations)):
# Learn by calling the `fit` method
model.fit(X_data, y_data,
batch_size=nb_samples,
epochs=1,
verbose=0)
# Make a plot of the data and the current fit
xs = np.linspace(X_data.min(), X_data.max(), num=50)
ys = model.predict(xs)
ax.clear()
ax.scatter(X_data, y_data, label='Training data')
ax.plot(xs, ys, color='Red', linewidth=1, label='Fit')
ax.set_xlabel('Education index')
ax.set_ylabel('Income index')
ax.grid(linestyle='dotted')
ax.legend()
fig.canvas.draw()
time.sleep(0.05)
# -
| 1-regression/1-linear-regression.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Mixup data augmentation
# + hide_input=true
from fastai.gen_doc.nbdoc import *
from fastai.callbacks.mixup import *
from fastai.vision import *
# -
# ## What is mixup?
# This module contains the implementation of a data augmentation technique called [mixup](https://arxiv.org/abs/1710.09412). It is extremely efficient at regularizing models in computer vision (we used it to get our time to train CIFAR10 to 94% on one GPU to 6 minutes).
#
# As the name kind of suggests, the authors of the mixup article propose training the model on mixes of the training set images. For example, suppose we’re training on CIFAR10. Instead of feeding the model the raw images, we take two images (not necessarily from the same class) and make a linear combination of them: in terms of tensors, we have:
#
# `new_image = t * image1 + (1-t) * image2`
#
# where t is a float between 0 and 1. The target we assign to that new image is the same combination of the original targets:
#
# `new_target = t * target1 + (1-t) * target2`
#
# assuming the targets are one-hot encoded (which isn’t the case in PyTorch usually). And it's as simple as that.
#
# 
#
# Dog or cat? The right answer here is 70% dog and 30% cat!
#
# As the picture above shows, it’s a bit hard for the human eye to make sense of images obtained in this way (although we do see the shapes of a dog and a cat). However, it somehow makes a lot of sense to the model, which trains more efficiently. One important side note is that when training with mixup, the final loss (training or validation) will be higher than when training without it, even when the accuracy is far better: a model trained like this will make predictions that are a bit less confident.
# ## Basic Training
# To test this method, we first create a [`simple_cnn`](/layers.html#simple_cnn) and train it like we did with [`basic_train`](/basic_train.html#basic_train) so we can compare its results with a network trained with mixup.
path = untar_data(URLs.MNIST_SAMPLE)
data = ImageDataBunch.from_folder(path)
model = simple_cnn((3,16,16,2))
learn = Learner(data, model, metrics=[accuracy])
learn.fit(8)
# ## Mixup implementation in the library
# In the original article, the authors suggest four things:
#
# 1. Create two separate dataloaders, and draw a batch from each at every iteration to mix them up
# 2. Draw a value for t following a beta distribution with a parameter alpha (0.4 is suggested in their article)
# 3. Mix up the two batches with the same value t
# 4. Use one-hot encoded targets
#
# This module's implementation is based on these suggestions, and modified where experimental results suggested changes that would improve performance.
# The authors suggest using the beta distribution with parameters alpha=0.4. (In general, the beta distribution has two parameters, but in this case they're going to be equal.) Why do they suggest this? Well, with the parameters they suggest, the beta distribution looks like this:
#
# 
#
# meaning that there's a very high probability of picking values close to 0 or 1 (in which case the mixed up image is mostly from only one category) and then a somewhat constant, much smaller probability of picking something in the middle (notice that 0.33 is nearly as likely as 0.5, for instance).
#
# While this works very well, it’s not the fastest way, and this is the first suggestion we adjust. The unnecessary slowdown with this approach comes from drawing two different batches at every iteration, which means loading twice the number of images and additionally applying any other data augmentation functions to them. To avoid this, we apply mixup on a batch with a shuffled version of itself: this way, the images mixed up are still different.
#
# Using the same value of `t` for the whole batch is another suggestion we modify. In our experiments, we noticed that the model trained faster if we drew a different `t` for every image in the batch. (Both options got to the same result in terms of accuracy, it’s just that one arrived there more slowly.)
#
# Finally, notice that with this strategy we might create duplicate images: let’s say we are mixing `image0` with `image1` and `image1` with `image0`, and that we draw `t=0.1` for the first mix and `t=0.9` for the second. Then
#
# `image0 * 0.1 + shuffle0 * (1-0.1) = image0 * 0.1 + image1 * 0.9`
#
# and
#
# `image1 * 0.9 + shuffle1 * (1-0.9) = image1 * 0.9 + image0 * 0.1`
#
# will be the same. Of course we have to be a bit unlucky for this to happen, but in practice, we saw a drop in accuracy when we didn't remove duplicates. To avoid this, the trick is to replace the vector of `t` we drew with:
#
# `t = max(t, 1-t)`
#
# The beta distribution with the two parameters equal is symmetric in any case, and this way we ensure that the largest coefficient is always near the first image (the non-shuffled batch).
# ## Adding mixup to the mix
# We now add [`MixUpCallback`](/callbacks.mixup.html#MixUpCallback) to our Learner so that it modifies our input and target accordingly. The [`mixup`](/train.html#mixup) function does this for us behind the scenes, along with a few other tweaks described below:
# + hide_input=false
model = simple_cnn((3,16,16,2))
learner = Learner(data, model, metrics=[accuracy]).mixup()
learner.fit(8)
# -
# Training with mixup improves the best accuracy. Note that the validation loss is higher than without mixup, because the model makes less confident predictions: without mixup, most predictions are very close to 0. or 1. (in terms of probability) whereas the model with mixup makes predictions that are more nuanced. Before using mixup, make sure you know whether it's more important to optimize lower loss or better accuracy.
# + hide_input=true
show_doc(MixUpCallback)
# -
# Create a [`Callback`](/callback.html#Callback) for mixup on `learn` with a parameter `alpha` for the beta distribution. `stack_x` and `stack_y` determine whether we stack our inputs/targets with the vector lambda drawn or do the linear combination. (In general, we stack the inputs or outputs when they correspond to categories or classes and do the linear combination otherwise.)
# ### Callback methods
# You don't call these yourself - they're called by fastai's [`Callback`](/callback.html#Callback) system automatically to enable the class's functionality.
# + hide_input=true
show_doc(MixUpCallback.on_batch_begin)
# -
# Draws a vector of lambda following a beta distribution with `self.alpha` and operates the mixup on `last_input` and `last_target` according to `self.stack_x` and `self.stack_y`.
# ## Dealing with the loss
# We often have to modify the loss so that it is compatible with mixup. PyTorch was very careful to avoid one-hot encoding targets when possible, so it seems a bit of a drag to undo this. Fortunately for us, if the loss is a classic [cross-entropy](https://pytorch.org/docs/stable/nn.html#torch.nn.functional.cross_entropy), we have
#
# `loss(output, new_target) = t * loss(output, target1) + (1-t) * loss(output, target2)`
#
# so we don’t one-hot encode anything and instead just compute those two losses and find the linear combination.
#
# The following class is used to adapt the loss for mixup. Note that the [`mixup`](/train.html#mixup) function will use it to change the `Learner.loss_func` if necessary.
# + hide_input=true
show_doc(MixUpLoss, title_level=3)
# -
# ## Undocumented Methods - Methods moved below this line will intentionally be hidden
# + hide_input=true
show_doc(MixUpLoss.forward)
# -
| docs_src/callbacks.mixup.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <!--NOTEBOOK_HEADER-->
# *This notebook contains material from [Controlling Natural Watersheds](https://jckantor.github.io/Controlling-Natural-Watersheds);
# content is available [on Github](https://github.com/jckantor/Controlling-Natural-Watersheds.git).*
# <!--NAVIGATION-->
# < [Rainy River Flows](http://nbviewer.jupyter.org/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.05-Rainy_River_Flows.ipynb) | [Contents](toc.ipynb) | [USGS Surface Water Daily Data](http://nbviewer.jupyter.org/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.07-USGS_Surface_Water_Daily_Data.ipynb) ><p><a href="https://colab.research.google.com/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.06-Namakan_Lake_Outflows.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://raw.githubusercontent.com/jckantor/Controlling-Natural-Watersheds/master/notebooks/A.06-Namakan_Lake_Outflows.ipynb"><img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
# # Namakan Lake Outflows
# The purpose of this notebook is to create a data series for the outflows from Namakan Lake. The data series is written to a file `NL_outflow.pkl` that can be read into other notebooks using `NL = pd.read_pickle('../data/NL_outflow.pkl')`.
#
# The data series is constructed using data provided by <NAME> of the International Joint Commission.
# ## Read Data
#
# +
# Display graphics inline with the notebook
# %matplotlib notebook
# Standard Python modules
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import os
import datetime
import requests
# Modules to display images and data tables
from IPython.display import Image
from IPython.core.display import display
NL_outflow = pd.read_excel('../data/Namakan_Outflows.xls',index_col=0,header=3)['Stn 47']
# +
plt.figure(figsize=(10,5))
plt.hold(True)
NL_outflow.plot()
plt.hold(False)
plt.xlabel('Year')
plt.ylabel('Level [meters]')
plt.grid()
# -
RLLevelFlow = pd.read_csv('../data/RLLevelFlow.csv',index_col=0,parse_dates=True)
RL_inflow = RLLevelFlow['Inflow']
# +
plt.figure(figsize=(10,5))
plt.hold(True)
NL_outflow.plot()
RL_inflow.plot()
plt.hold(False)
plt.xlabel('Year')
plt.ylabel('Level [meters]')
plt.grid()
# +
flows = pd.concat([RL_inflow,NL_outflow],axis=1).dropna()
plt.figure(figsize=(10,5))
plt.hold(True)
flows['Inflow'].plot()
flows['Stn 47'].plot()
# +
plt.figure(figsize=(10,5))
plt.hold(True)
plt.plot(flows.ix['1970':'1999','Inflow'],flows.ix['1970':'1999','Stn 47'],'.',ms=5,color='b',alpha=0.6)
plt.plot(flows.ix['2000':,'Inflow'],flows.ix['2000':,'Stn 47'],'.',ms=5,color='r',alpha=0.6)
plt.xlim(0,2000)
plt.ylim(0,1000)
plt.grid()
plt.title('Namakan Outflow vs Rainy Lake Inflow, Daily for 1970-2014')
plt.ylabel('Namakan Outflow [cubic meters/sec]')
plt.xlabel('Rainy Lake Inflow [cubic meters/sec]')
# -
q = (flows['Stn 47']/flows['Inflow'])
plt.ylim(0,1)
q.plot(makers='.')
plt.figure(figsize=(10,5))
q = q[q > 0]
q = q[q < 1]
q.hist(bins=100,normed=True)
plt.xlim(0,1)
print q.mean()
# ## Data Reconciliation
NL.to_pickle('../data/NL.pkl')
# <!--NAVIGATION-->
# < [Rainy River Flows](http://nbviewer.jupyter.org/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.05-Rainy_River_Flows.ipynb) | [Contents](toc.ipynb) | [USGS Surface Water Daily Data](http://nbviewer.jupyter.org/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.07-USGS_Surface_Water_Daily_Data.ipynb) ><p><a href="https://colab.research.google.com/github/jckantor/Controlling-Natural-Watersheds/blob/master/notebooks/A.06-Namakan_Lake_Outflows.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://raw.githubusercontent.com/jckantor/Controlling-Natural-Watersheds/master/notebooks/A.06-Namakan_Lake_Outflows.ipynb"><img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
| notebooks/A.06-Namakan_Lake_Outflows.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# **Percentiles** For example, let's consider the sizes of the five largest continents – Africa, Antarctica, Asia, North America, and South America – rounded to the nearest million square miles.
import numpy as np
sizes = np.array([12, 17, 6, 9, 7])
sizes
# The `80th` percentile is the smallest value that is at least as large as `80%` of the elements of sizes
#
# **Step 1**: sort the list in ascending order <br/>
# **Step 2**: grasp `80%` of the elements from left to right
sorted_sizes = np.sort(sizes)
sorted_sizes
number_of_elements = 0.8*(len(sizes)-1)
number_of_elements
# `80th` percentile is at index `3th` (round down) or the number `12`
sorted_sizes[3]
# `80th` percentile is at index `4th` (round up) or the number `17`
sorted_sizes[4]
# **Handling with floating rank**
number_of_elements = 0.7*(len(sizes)-1)
number_of_elements
# round it up, becomes index `3th`; then `70th` percentile is at number `12`
sorted_sizes[3]
# **Interpoate ("linear" approach) with floating rank**
# **Step 1**: Determine the elements at the calculated rank using fomular `r=p(n-1)`; `70th` is at r=0.7*(5-1)=2.8; Example, rank `2.8` means that positions of elements `2th` and `3th` which are `9` and `12`, respectively
# **Step 2**: Take the difference between these two elements and multiply it by the fractional portion of the rank. For our example, this is: `(12 – 9)0.8 = 2.4`. <br/>
# **Step 3**: Take the lower-ranked value in **Step 1** and add the value from **Step 2** to obtain the interpolated value for the percentile. For our example, that value is `9 + 2.4 = 11.4`.
# **Usig numpy and pandas**
np.percentile(sizes, 80, interpolation='linear')
np.percentile(sizes, 70, interpolation='linear')
# +
import pandas as pd
my_data = {
"Size": sizes
}
df = pd.DataFrame(my_data)
df
# -
df["Size"].quantile(0.8, interpolation='linear')
df["Size"].quantile(0.7, interpolation='linear')
# **Other example**
import pandas as pd
scores_and_sections = pd.read_csv('scores_by_section.csv')
scores_and_sections
scores_and_sections['Midterm'].hist(bins=np.arange(-0.5, 25.6, 1))
scores_and_sections['Midterm'].quantile(0.85)
# **Quantiles**
scores_and_sections['Midterm'].quantile(0.25)
scores_and_sections['Midterm'].quantile(0.50)
scores_and_sections['Midterm'].quantile(0.75)
scores_and_sections['Midterm'].quantile(1)
scores_and_sections['Midterm'].max()
# **Bootstrap** We study the `Total Compensation` column
df = pd.read_csv("san_francisco_2015.csv")
df
# we will focus our attention on those who had at least the equivalent of a half-time job for the whole year. At a minimum wage of about `$10` per hour, and `20` hours per week for `52` weeks, that's a salary of about `$10,000`.
df = df.loc[df["Salaries"] > 10000]
df
# Visualize the histogram
my_bins = np.arange(0, 700000, 25000)
df['Total Compensation'].hist(bins=my_bins)
# **Compute the median**
pop_median = df['Total Compensation'].median()
pop_median
df['Total Compensation'].quantile(0.50)
# **Now we estimate this value using bootstrap (resampling)**
# +
my_bins = np.arange(0, 700000, 25000)
our_sample = df.sample(500, replace=False)
our_sample['Total Compensation'].hist(bins=my_bins)
# -
est_median = our_sample['Total Compensation'].median()
est_median
our_sample['Total Compensation'].quantile(0.50)
# The sample size is large. By the law of averages, the distribution of the sample resembles that of the population, and consequently the sample median is not very far from the population median (though of course it is not exactly the same).
# So now we have one estimate of the parameter. But had the sample come out differently, the estimate would have had a different value. We would like to be able to quantify the amount by which the estimate could vary across samples. That measure of variability will help us measure how accurately we can estimate the parameter.
# ## Bootstrap method
# * Treat the original sample as if it were the population.
# * Draw from the sample, at random with replacement, the same number of times as the original sample size.
resample_1 = our_sample.sample(frac=1.0, replace=True)
resample_1['Total Compensation'].hist(bins=my_bins)
# Compute the median of the new sample
resample_1['Total Compensation'].median()
resample_2 = our_sample.sample(frac=1.0, replace=True)
resampled_median_2 = resample_2['Total Compensation'].median()
resampled_median_2
# Resamnpling for `5,000` times
bstrap_medians = []
for i in range(1, 5000+1):
one_resample = our_sample.sample(frac=1.0, replace=True)
one_median = one_resample['Total Compensation'].median()
bstrap_medians.append(one_median)
# +
my_median_data = {
"Median": bstrap_medians
}
median_df = pd.DataFrame(my_median_data)
median_df
# -
median_df.hist()
# +
import matplotlib.pyplot as plt
plt.hist(bstrap_medians)
plt.xlabel("Median")
plt.ylabel("Frequency")
plt.show()
# +
plt.hist(bstrap_medians, zorder=1)
plt.xlabel("Median")
plt.ylabel("Frequency")
plt.scatter(pop_median, 0, color='red', s=30, zorder=2);
plt.show()
# -
# Let's find out the middle `95%` of the resampled medians contains the red dot
left = median_df.quantile(0.025)
left
right = median_df.quantile(0.975)
right
# **The population median of `$110,305` is between these two numbers. The interval and the population median are shown on the histogram below.**
# +
plt.hist(median_values, zorder=1)
plt.xlabel("Median")
plt.ylabel("Frequency")
plt.plot([left, right], [0, 0], color='yellow', lw=3, zorder=2)
plt.scatter(pop_median, 0, color='red', s=30, zorder=3);
plt.show()
# -
# So, the "middle 95%" interval of estimates captured the parameter in our example
# **Let repeat the processs 100 times to see how frequently the interval contains the parameter**. We will store all left and right ends per simulation.
def bootstrap_sample(our_sample):
bstrap_medians = []
for i in range(1, 5000+1):
one_resample = our_sample.sample(frac=1.0, replace=True)
one_median = one_resample['Total Compensation'].median()
bstrap_medians.append(one_median)
return bstrap_medians
# +
left_ends = []
right_ends = []
for i in range(1, 100+1):
our_sample = df.sample(500, replace=False)
bstrap_medians = bootstrap_sample(our_sample)
my_median_data = {
"Median": bstrap_medians
}
median_df = pd.DataFrame(my_median_data)
left = median_df['Median'].quantile(0.025)
right = median_df['Median'].quantile(0.975)
left_ends.append(left)
right_ends.append(right)
# +
my_left_right = {
"Left": left_ends,
"Right": right_ends
}
left_right_df = pd.DataFrame(my_left_right)
left_right_df
# -
good_experiments = left_right_df[(left_right_df["Left"] < pop_median) & (left_right_df["Right"] > pop_median)]
good_experiments
# +
for i in np.arange(100):
left = left_right_df.at[i, "Left"]
right = left_right_df.at[i, "Right"]
plt.plot([left, right], [i, i], color='gold')
plt.plot([pop_median, pop_median], [0, 100], color='red', lw=2)
plt.xlabel('Median (dollars)')
plt.ylabel('Replication')
plt.title('Population Median and Intervals of Estimates')
plt.show()
# -
# In other words, this process of estimation captures the parameter about `92%` of the time.
| 1-Lessons/Lesson17/OriginalPowerpoint/.ipynb_checkpoints/bootstrap-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src="images/strathsdr_banner.png" align="left">
# # Hardware Accelerated Spectrum Analysis on RFSoC
# ----
#
# <div class="alert alert-box alert-info">
# Please use Jupyter Labs http://board_ip_address/lab for this notebook.
# </div>
#
# This notebook presents a flexible hardware accelerated Spectrum Analyzer Module for the Zynq UltraScale+ RFSoC. The Spectrum Analyzer Module was developed by the [University of Strathclyde](https://github.com/strath-sdr).
#
# ## Table of Contents
# * [Introduction](#introduction)
# * [Hardware Setup](#hardware-setup)
# * [Software Setup](#software-setup)
# * [Simple Tone Generation](#simple-tone-generation)
# * [The Spectrum Analyzer](#the-spectrum-analyzer)
# * [A Simple Example](#a-simple-example)
# * [Conclusion](#conclusion)
#
# ## References
# * [Xilinx, Inc, "USP RF Data Converter: LogiCORE IP Product Guide", PG269, v2.3, June 2020](https://www.xilinx.com/support/documentation/ip_documentation/usp_rf_data_converter/v2_3/pg269-rf-data-converter.pdf)
#
# ## Revision History
# * **v1.0** | 12/02/2021 | Spectrum analyzer notebook
# * **v1.1** | 15/04/2021 | Update spectral resolution and minimum bandwidth with new value
# ----
# ## Introduction <a class="anchor" id="introduction"></a>
# The Zynq RFSoC contains high frequency samplers known as RF Data Converters (RF DCs). The RF DCs are tightly coupled with the Programmable Logic (PL), creating a high-throughput, low-latency path between the FPGA and analogue world. The Spectrum Analyzer Module employs the RF Analogue-to-Digital Converters (RF ADCs) to receive RF time domain signals. The received data is manipulated using spectral pre-processing techniques in the PL, to prepare it for frequency domain analysis and visualisation in the Processing System (PS).
#
# A significant portion of the design has been implemented in the RFSoC's PL to prevent the PS from applying highly computational arithemtic. [Figure 1](#fig-1) presents a simple diagram illustrating the system overview for one spectrum analyzer channel. There is a Spectrum Analyzer Module for each available RF ADC channel in the design. The Spectrum Analyzers are also interfaced to their very own flexible decimator, allowing different sample rates to be configured for each channel.
# <a class="anchor" id="fig-1"></a>
# <figure>
# <img src='images/spectrum_analyser_overview.png' height='50%' width='50%'/>
# <figcaption><b>Figure 1: The RFSoC Spectrum Analyzer system overview.</b></figcaption>
# </figure>
# ### Hardware Setup <a class="anchor" id="hardware-setup"></a>
# Your ZCU111 development board can host four Spectrum Analyzer Modules. To setup your board for this demonstration, you can connect each channel in loopback as shown in [Figure 2](#fig-2), or connect an antenna to one of the ADC channels.
#
# Don't worry if you don't have an antenna. The default loopback configuration will still be very interesting and is connected as follows:
# * Channel 0: DAC4 (Tile 229 Block 0) to ADC0 (Tile 224 Block 0)
# * Channel 1: DAC5 (Tile 229 Block 1) to ADC1 (Tile 224 Block 1)
# * Channel 2: DAC6 (Tile 229 Block 2) to ADC2 (Tile 225 Block 0)
# * Channel 3: DAC7 (Tile 229 Block 3) to ADC3 (Tile 225 Block 1)
#
# There has been several XM500 board revisions, and some contain different silkscreen and labels for the ADCs and DACs. Use the image below for further guidance and pay attention to the associated Tile and Block.
#
# <a class="anchor" id="fig-2"></a>
# <figure>
# <img src='images/zcu111_setup.png' height='50%' width='50%'/>
# <figcaption><b>Figure 2: ZCU111 and XM500 development board setup in loopback mode.</b></figcaption>
# </figure>
#
# If you have chosen to use an antenna, **do not** attach your antenna to any SMA interfaces labelled DAC.
#
# <div class="alert alert-box alert-danger">
# <b>Caution:</b>
# In this demonstration, we generate tones using the RFSoC development board. Your device should be setup in loopback mode. You should understand that the RFSoC platform can also transmit RF signals wirelessly. Remember that unlicensed wireless transmission of RF signals may be illegal in your geographical location. Radio signals may also interfere with nearby devices, such as pacemakers and emergency radio equipment. Note that it is also illegal to intercept and decode particular RF signals. If you are unsure, please seek professional support.
# </div>
# ### Software Setup <a class="anchor" id="software-setup"></a>
# We're nearly finished setting up the demonstration system. The majority of the libraries used by the spectrum analyzer design are contained inside the RFSoC-SAM software package. We only need to run a few code cells to initialise the software environment.
#
# The primary module for loading the Spectrum Analyzer design is contained inside `rfsoc_sam.overlay`. The class we are interested in using is `Overlay()`. During initialisation the class downloads the Spectrum Analyzer bitstream to the PL and configures the RF DCs and FPGA IP cores contained in our system. This process may take around a minute to complete.
#
# **Run** the code cell below to load the RFSoC-SAM Overlay class.
# +
from rfsoc_sam.overlay import Overlay
sam = Overlay()
# -
# When the RFSoC-SAM Overlay class is initialising, the setup script will also program the LMK and LMX low-jitter clock chips on the ZCU111 to 122.8MHz and 409.6MHz respectively.
#
# Lets now initialise the analyzer, and setup user control. The initialisation process takes around 2 minutes.
analyzer = sam.spectrum_analyzer()
# ----
# ## Simple Tone Generation <a class="anchor" id="simple-tone-generation"></a>
# A simple amplitude controller is required to generate tones using the RF Digital-to-Analogue Converters (RF DACs). We use tone generation in this demonstration to provide a signal for the user to inspect when using the Spectrum Analyzer Module.
#
# Run the code cell below to reveal a widget, which can be used to control the transmission frequency and amplitude.
analyzer.children[2]
# ## The Spectrum Analyzer <a class="anchor" id="the-spectrum-analyzer"></a>
# We will now explore the hardware accelerated Spectrum Analyzer Module. It is worthwhile noting the analyzers capabilities below:
#
# * The analyzer is capable of inspecting 1638.4MHz of bandwidth.
# * It can achieve a maximum spectral resolution of 0.244140625kHz.
# * The bandwidth is adjustable between 1638.4MHz and 1.6MHz.
# * The range of inspection is between 0 to 4096MHz using higher order Nyquist techniques.
analyzer.children[1]
# The Spectrum Analyzer Module contains a hardware accelerated FFT core, which can convert the RF sampled signal to the frequency domain using a range of different FFT lengths, $N = 64$ upto $N = 8192$. The frequency domain signal is further manipulated using a custom floating point processor to obtain the representative Power Spectral Density (PSD) or Power Spectrum. Furthermore, a hardware accelerated decibel (dB) converter is also used to condition the frequency domain signal for visual analysis.
#
# Through the loopback connection, you should be able to use the Spectrum Analyzer Module to locate the tone you previously generated using the tone generator. If you have an antenna connected to your board, try and locate signals of interest using the Spectrum Analyzer's control widgets.
# ### A Simple Example <a class="anchor" id="a-simple-example"></a>
# If you would like to enable stimulus for the spectrum analyzer, you can use your mobile phone to create WiFi traffic. Follow the steps below to create an interesting WiFi spectrum to visualise.
# * Connect your mobile phone to an access point that uses WiFi.
# * Then configure the spectrum analyzer for a centre frequency of 2400MHz and a decimation factor of 16.
# * Switch on the spectrum analyzer and spectrogram.
# * Use your phone to stream a video, or music. This will create WiFi traffic for inspection.
# * Place your phone close to the RF ADC ports of the spectrum analyzer.
#
# You should see a similar output as given in the [Figure 3](#fig-3) below.
#
# <a class="anchor" id="fig-3"></a>
# <figure>
# <img src='images/wifi_example.jpg' height='50%' width='50%'/>
# <figcaption><b>Figure 3: Capturing a WiFi signal using the Spectrum Analyser Module.</b></figcaption>
# </figure>
# ## Conclusion <a class="anchor" id="conclusion"></a>
# This notebook has presented a hardware accelerated Spectrum Analyzer Module for the ZCU111 development board.
| boards/ZCU111/rfsoc_sam/notebooks/rfsoc_spectrum_analysis.ipynb |
// ---
// jupyter:
// jupytext:
// text_representation:
// extension: .java
// format_name: light
// format_version: '1.5'
// jupytext_version: 1.14.4
// kernelspec:
// display_name: Java
// language: java
// name: java
// ---
// # Create your first deep learning neural network
//
// ## Introduction
//
// This is the first of our [beginner tutorial series](https://github.com/awslabs/djl/tree/master/jupyter/tutorial) that will take you through creating, training, and running inference on a neural network. In this tutorial, you will learn how to use the built-in `Block` to create your first neural network - a Multilayer Perceptron.
//
// ## Neural Network
//
// A neural network is a black box function. Instead of coding this function yourself, you provide many sample input/output pairs for this function. Then, we try to train the network to learn how to match the behavior of the function given only these input/output pairs. A better model with more data can more accurately match the function.
//
// ## Multilayer Perceptron
//
// A Multilayer Perceptron (MLP) is one of the simplest deep learning networks. The MLP has an input layer which contains your input data, an output layer which is produced by the network and contains the data the network is supposed to be learning, and some number of hidden layers. The example below contains an input of size 3, a single hidden layer of size 3, and an output of size 2. The number and sizes of the hidden layers are determined through experimentation but more layers enable the network to represent more complicated functions. Between each pair of layers is a linear operation (sometimes called a FullyConnected operation because each number in the input connected to each number in the output by a matrix multiplication). Not pictured, there is also a non-linear activation function after each linear operation. For more information, see [Multilayer Perceptron](https://en.wikipedia.org/wiki/Multilayer_perceptron).
//
// 
//
//
// ## Step 1: Setup development environment
//
// ### Installation
//
// This tutorial requires the installation of the Java Jupyter Kernel. To install the kernel, see the [Jupyter README](https://github.com/awslabs/djl/blob/master/jupyter/README.md).
// +
// Add the snapshot repository to get the DJL snapshot artifacts
// // %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/
// Add the maven dependencies
// %maven ai.djl:api:0.4.0
// %maven org.slf4j:slf4j-api:1.7.26
// %maven org.slf4j:slf4j-simple:1.7.26
// See https://github.com/awslabs/djl/blob/master/mxnet/mxnet-engine/README.md
// for more MXNet library selection options
// %maven ai.djl.mxnet:mxnet-native-auto:1.6.0
// -
import ai.djl.*;
import ai.djl.nn.*;
import ai.djl.nn.core.*;
import ai.djl.training.*;
// ## Step 2: Determine your input and output size
//
// The MLP model uses a one dimensional vector as the input and the output. You should determine the appropriate size of this vector based on your input data and what you will use the output of the model for. In a later tutorial, we will use this model for Mnist image classification.
//
// Our input vector will have size `28x28` because the input images have a height and width of 28 and it takes only a single number to represent each pixel. For a color image, you would need to further multiply this by `3` for the RGB channels.
//
// Our output vector has size `10` because there are `10` possible classes for each image.
long inputSize = 28*28;
long outputSize = 10;
// ## Step 3: Create a **SequentialBlock**
//
// ### NDArray
//
// The core data type used for working with Deep Learning is the [NDArray](https://javadoc.djl.ai/api/0.4.0/index.html?ai/djl/ndarray/NDArray.html). An NDArray represents a multidimensional, fixed-size homogeneous array. It has very similar behavior to the Numpy python package with the addition of efficient computing. We also have a helper class, the [NDList](https://javadoc.djl.ai/api/0.4.0/index.html?ai/djl/ndarray/NDList.html) which is a list of NDArrays which can have different sizes and data types.
//
// ### Block API
//
// In DJL, [Blocks](https://javadoc.djl.ai/api/0.4.0/index.html?ai/djl/nn/Block.html) serve a purpose similar to functions that convert an input `NDList` to an output `NDList`. They can represent single operations, parts of a neural network, and even the whole neural network. What makes blocks special is that they contain a number of parameters that are used in their function and are trained during deep learning. As these parameters are trained, the function represented by the blocks get more and more accurate.
//
// When building these block functions, the easiest way is to use composition. Similar to how functions are built by calling other functions, blocks can be built by combining other blocks. We refer to the containing block as the parent and the sub-blocks as the children.
//
//
// We provide several helpers to make it easy to build common block composition structures. For the MLP we will use the [SequentialBlock](https://javadoc.djl.ai/api/0.4.0/index.html?ai/djl/nn/SequentialBlock.html), a container block whose children form a chain of blocks where each child block feeds its output to the next child block in a sequence.
//
SequentialBlock block = new SequentialBlock();
// ## Step 4: Add blocks to SequentialBlock
//
// An MLP is organized into several layers. Each layer is composed of a [Linear Block](https://javadoc.djl.ai/api/0.4.0/index.html?ai/djl/nn/core/Linear.html) and a non-linear activation function. If we just had two linear blocks in a row, it would be the same as a combined linear block ($f(x) = W_2(W_1x) = (W_2W_1)x = W_{combined}x$). An activation is used to intersperse between the linear blocks to allow them to represent non-linear functions. We will use the popular [ReLU](https://javadoc.djl.ai/api/0.4.0/ai/djl/nn/Activation.html#reluBlock--) as our activation function.
//
// The first layer and last layers have fixed sizes depending on your desired input and output size. However, you are free to choose the number and sizes of the middle layers in the network. We will create a smaller MLP with two middle layers that gradually decrease the size. Typically, you would experiment with different values to see what works the best on your data set.
// +
block.add(Blocks.batchFlattenBlock(inputSize));
block.add(Linear.builder().setOutChannels(128).build());
block.add(Activation::relu);
block.add(Linear.builder().setOutChannels(64).build());
block.add(Activation::relu);
block.add(Linear.builder().setOutChannels(outputSize).build());
block
// -
// ## Summary
//
// Now that you've successfully created your first neural network, you can use this network to train your model.
//
// Next chapter: [Train your first model](train_your_first_model.ipynb)
//
// You can find the complete source code for this tutorial in the [model zoo](https://github.com/awslabs/djl/blob/master/model-zoo/src/main/java/ai/djl/basicmodelzoo/basic/Mlp.java).
| jupyter/tutorial/create_your_first_network.ipynb |
# ---
# jupyter:
# jupytext:
# split_at_heading: true
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
#export
from fastai2.basics import *
from fastai2.vision.all import *
# +
#default_exp vision.gan
#default_cls_lvl 3
# -
#hide
from nbdev.showdoc import *
# # GAN
#
# > Basic support for [Generative Adversial Networks](https://arxiv.org/abs/1406.2661)
# GAN stands for [Generative Adversarial Nets](https://arxiv.org/pdf/1406.2661.pdf) and were invented by <NAME>. The concept is that we train two models at the same time: a generator and a critic. The generator will try to make new images similar to the ones in a dataset, and the critic will try to classify real images from the ones the generator does. The generator returns images, the critic a single number (usually a probability, 0. for fake images and 1. for real ones).
#
# We train them against each other in the sense that at each step (more or less), we:
# 1. Freeze the generator and train the critic for one step by:
# - getting one batch of true images (let's call that `real`)
# - generating one batch of fake images (let's call that `fake`)
# - have the critic evaluate each batch and compute a loss function from that; the important part is that it rewards positively the detection of real images and penalizes the fake ones
# - update the weights of the critic with the gradients of this loss
#
#
# 2. Freeze the critic and train the generator for one step by:
# - generating one batch of fake images
# - evaluate the critic on it
# - return a loss that rewards posisitively the critic thinking those are real images
# - update the weights of the generator with the gradients of this loss
# > Note: The fastai library provides support for training GANs through the GANTrainer, but doesn't include more than basic models.
# ## Wrapping the modules
#export
class GANModule(Module):
"Wrapper around a `generator` and a `critic` to create a GAN."
def __init__(self, generator=None, critic=None, gen_mode=False):
if generator is not None: self.generator=generator
if critic is not None: self.critic =critic
store_attr(self, 'gen_mode')
def forward(self, *args):
return self.generator(*args) if self.gen_mode else self.critic(*args)
def switch(self, gen_mode=None):
"Put the module in generator mode if `gen_mode`, in critic mode otherwise."
self.gen_mode = (not self.gen_mode) if gen_mode is None else gen_mode
# This is just a shell to contain the two models. When called, it will either delegate the input to the `generator` or the `critic` depending of the value of `gen_mode`.
show_doc(GANModule.switch)
# By default (leaving `gen_mode` to `None`), this will put the module in the other mode (critic mode if it was in generator mode and vice versa).
#export
@delegates(ConvLayer.__init__)
def basic_critic(in_size, n_channels, n_features=64, n_extra_layers=0, norm_type=NormType.Batch, **kwargs):
"A basic critic for images `n_channels` x `in_size` x `in_size`."
layers = [ConvLayer(n_channels, n_features, 4, 2, 1, norm_type=None, **kwargs)]
cur_size, cur_ftrs = in_size//2, n_features
layers += [ConvLayer(cur_ftrs, cur_ftrs, 3, 1, norm_type=norm_type, **kwargs) for _ in range(n_extra_layers)]
while cur_size > 4:
layers.append(ConvLayer(cur_ftrs, cur_ftrs*2, 4, 2, 1, norm_type=norm_type, **kwargs))
cur_ftrs *= 2 ; cur_size //= 2
init = kwargs.get('init', nn.init.kaiming_normal_)
layers += [init_default(nn.Conv2d(cur_ftrs, 1, 4, padding=0), init), Flatten()]
return nn.Sequential(*layers)
#export
class AddChannels(Module):
"Add `n_dim` channels at the end of the input."
def __init__(self, n_dim): self.n_dim=n_dim
def forward(self, x): return x.view(*(list(x.shape)+[1]*self.n_dim))
#export
@delegates(ConvLayer.__init__)
def basic_generator(out_size, n_channels, in_sz=100, n_features=64, n_extra_layers=0, **kwargs):
"A basic generator from `in_sz` to images `n_channels` x `out_size` x `out_size`."
cur_size, cur_ftrs = 4, n_features//2
while cur_size < out_size: cur_size *= 2; cur_ftrs *= 2
layers = [AddChannels(2), ConvLayer(in_sz, cur_ftrs, 4, 1, transpose=True, **kwargs)]
cur_size = 4
while cur_size < out_size // 2:
layers.append(ConvLayer(cur_ftrs, cur_ftrs//2, 4, 2, 1, transpose=True, **kwargs))
cur_ftrs //= 2; cur_size *= 2
layers += [ConvLayer(cur_ftrs, cur_ftrs, 3, 1, 1, transpose=True, **kwargs) for _ in range(n_extra_layers)]
layers += [nn.ConvTranspose2d(cur_ftrs, n_channels, 4, 2, 1, bias=False), nn.Tanh()]
return nn.Sequential(*layers)
# +
critic = basic_critic(64, 3)
generator = basic_generator(64, 3)
tst = GANModule(critic=critic, generator=generator)
real = torch.randn(2, 3, 64, 64)
real_p = tst(real)
test_eq(real_p.shape, [2,1])
tst.switch() #tst is now in generator mode
noise = torch.randn(2, 100)
fake = tst(noise)
test_eq(fake.shape, real.shape)
tst.switch() #tst is back in critic mode
fake_p = tst(fake)
test_eq(fake_p.shape, [2,1])
# +
#export
_conv_args = dict(act_cls = partial(nn.LeakyReLU, negative_slope=0.2), norm_type=NormType.Spectral)
def _conv(ni, nf, ks=3, stride=1, self_attention=False, **kwargs):
if self_attention: kwargs['xtra'] = SelfAttention(nf)
return ConvLayer(ni, nf, ks=ks, stride=stride, **_conv_args, **kwargs)
# -
#export
@delegates(ConvLayer)
def DenseResBlock(nf, norm_type=NormType.Batch, **kwargs):
"Resnet block of `nf` features. `conv_kwargs` are passed to `conv_layer`."
return SequentialEx(ConvLayer(nf, nf, norm_type=norm_type, **kwargs),
ConvLayer(nf, nf, norm_type=norm_type, **kwargs),
MergeLayer(dense=True))
#export
def gan_critic(n_channels=3, nf=128, n_blocks=3, p=0.15):
"Critic to train a `GAN`."
layers = [
_conv(n_channels, nf, ks=4, stride=2),
nn.Dropout2d(p/2),
DenseResBlock(nf, **_conv_args)]
nf *= 2 # after dense block
for i in range(n_blocks):
layers += [
nn.Dropout2d(p),
_conv(nf, nf*2, ks=4, stride=2, self_attention=(i==0))]
nf *= 2
layers += [
ConvLayer(nf, 1, ks=4, bias=False, padding=0, norm_type=NormType.Spectral, act_cls=None),
Flatten()]
return nn.Sequential(*layers)
#export
class GANLoss(GANModule):
"Wrapper around `crit_loss_func` and `gen_loss_func`"
def __init__(self, gen_loss_func, crit_loss_func, gan_model):
super().__init__()
store_attr(self, 'gen_loss_func,crit_loss_func,gan_model')
def generator(self, output, target):
"Evaluate the `output` with the critic then uses `self.gen_loss_func`"
fake_pred = self.gan_model.critic(output)
self.gen_loss = self.gen_loss_func(fake_pred, output, target)
return self.gen_loss
def critic(self, real_pred, input):
"Create some `fake_pred` with the generator from `input` and compare them to `real_pred` in `self.crit_loss_func`."
fake = self.gan_model.generator(input.requires_grad_(False)).requires_grad_(True)
fake_pred = self.gan_model.critic(fake)
self.crit_loss = self.crit_loss_func(real_pred, fake_pred)
return self.crit_loss
# In generator mode, this loss function expects the `output` of the generator and some `target` (a batch of real images). It will evaluate if the generator successfully fooled the critic using `gen_loss_func`. This loss function has the following signature
# ```
# def gen_loss_func(fake_pred, output, target):
# ```
# to be able to combine the output of the critic on `output` (which the first argument `fake_pred`) with `output` and `target` (if you want to mix the GAN loss with other losses for instance).
#
# In critic mode, this loss function expects the `real_pred` given by the critic and some `input` (the noise fed to the generator). It will evaluate the critic using `crit_loss_func`. This loss function has the following signature
# ```
# def crit_loss_func(real_pred, fake_pred):
# ```
# where `real_pred` is the output of the critic on a batch of real images and `fake_pred` is generated from the noise using the generator.
#export
class AdaptiveLoss(Module):
"Expand the `target` to match the `output` size before applying `crit`."
def __init__(self, crit): self.crit = crit
def forward(self, output, target):
return self.crit(output, target[:,None].expand_as(output).float())
#export
def accuracy_thresh_expand(y_pred, y_true, thresh=0.5, sigmoid=True):
"Compute accuracy after expanding `y_true` to the size of `y_pred`."
if sigmoid: y_pred = y_pred.sigmoid()
return ((y_pred>thresh).byte()==y_true[:,None].expand_as(y_pred).byte()).float().mean()
# ## Callbacks for GAN training
#export
def set_freeze_model(m, rg):
for p in m.parameters(): p.requires_grad_(rg)
#export
class GANTrainer(Callback):
"Handles GAN Training."
run_after = TrainEvalCallback
def __init__(self, switch_eval=False, clip=None, beta=0.98, gen_first=False, show_img=True):
store_attr(self, 'switch_eval,clip,gen_first,show_img')
self.gen_loss,self.crit_loss = AvgSmoothLoss(beta=beta),AvgSmoothLoss(beta=beta)
def _set_trainable(self):
train_model = self.generator if self.gen_mode else self.critic
loss_model = self.generator if not self.gen_mode else self.critic
set_freeze_model(train_model, True)
set_freeze_model(loss_model, False)
if self.switch_eval:
train_model.train()
loss_model.eval()
def begin_fit(self):
"Initialize smootheners."
self.generator,self.critic = self.model.generator,self.model.critic
self.gen_mode = self.gen_first
self.switch(self.gen_mode)
self.crit_losses,self.gen_losses = [],[]
self.gen_loss.reset() ; self.crit_loss.reset()
#self.recorder.no_val=True
#self.recorder.add_metric_names(['gen_loss', 'disc_loss'])
#self.imgs,self.titles = [],[]
def begin_validate(self):
"Switch in generator mode for showing results."
self.switch(gen_mode=True)
def begin_batch(self):
"Clamp the weights with `self.clip` if it's not None, set the correct input/target."
if self.training and self.clip is not None:
for p in self.critic.parameters(): p.data.clamp_(-self.clip, self.clip)
if not self.gen_mode:
(self.learn.xb,self.learn.yb) = (self.yb,self.xb)
def after_batch(self):
"Record `last_loss` in the proper list."
if not self.training: return
if self.gen_mode:
self.gen_loss.accumulate(self.learn)
self.gen_losses.append(self.gen_loss.value)
self.last_gen = to_detach(self.pred)
else:
self.crit_loss.accumulate(self.learn)
self.crit_losses.append(self.crit_loss.value)
def begin_epoch(self):
"Put the critic or the generator back to eval if necessary."
self.switch(self.gen_mode)
#def after_epoch(self):
# "Show a sample image."
# if not hasattr(self, 'last_gen') or not self.show_img: return
# data = self.learn.data
# img = self.last_gen[0]
# norm = getattr(data,'norm',False)
# if norm and norm.keywords.get('do_y',False): img = data.denorm(img)
# img = data.train_ds.y.reconstruct(img)
# self.imgs.append(img)
# self.titles.append(f'Epoch {epoch}')
# pbar.show_imgs(self.imgs, self.titles)
# return add_metrics(last_metrics, [getattr(self.smoothenerG,'smooth',None),getattr(self.smoothenerC,'smooth',None)])
def switch(self, gen_mode=None):
"Switch the model and loss function, if `gen_mode` is provided, in the desired mode."
self.gen_mode = (not self.gen_mode) if gen_mode is None else gen_mode
self._set_trainable()
self.model.switch(gen_mode)
self.loss_func.switch(gen_mode)
# > Warning: The GANTrainer is useless on its own, you need to complete it with one of the following switchers
#export
class FixedGANSwitcher(Callback):
"Switcher to do `n_crit` iterations of the critic then `n_gen` iterations of the generator."
run_after = GANTrainer
def __init__(self, n_crit=1, n_gen=1): store_attr(self, 'n_crit,n_gen')
def begin_train(self): self.n_c,self.n_g = 0,0
def after_batch(self):
"Switch the model if necessary."
if not self.training: return
if self.learn.gan_trainer.gen_mode:
self.n_g += 1
n_iter,n_in,n_out = self.n_gen,self.n_c,self.n_g
else:
self.n_c += 1
n_iter,n_in,n_out = self.n_crit,self.n_g,self.n_c
target = n_iter if isinstance(n_iter, int) else n_iter(n_in)
if target == n_out:
self.learn.gan_trainer.switch()
self.n_c,self.n_g = 0,0
#export
class AdaptiveGANSwitcher(Callback):
"Switcher that goes back to generator/critic when the loss goes below `gen_thresh`/`crit_thresh`."
run_after = GANTrainer
def __init__(self, gen_thresh=None, critic_thresh=None):
store_attr(self, 'gen_thresh,critic_thresh')
def after_batch(self):
"Switch the model if necessary."
if not self.training: return
if self.gan_trainer.gen_mode:
if self.gen_thresh is None or self.loss < self.gen_thresh: self.gan_trainer.switch()
else:
if self.critic_thresh is None or self.loss < self.critic_thresh: self.gan_trainer.switch()
#export
class GANDiscriminativeLR(Callback):
"`Callback` that handles multiplying the learning rate by `mult_lr` for the critic."
run_after = GANTrainer
def __init__(self, mult_lr=5.): self.mult_lr = mult_lr
def begin_batch(self):
"Multiply the current lr if necessary."
if not self.learn.gan_trainer.gen_mode and self.training:
self.learn.opt.set_hyper('lr', self.learn.opt.hypers[0]['lr']*self.mult_lr)
def after_batch(self):
"Put the LR back to its value if necessary."
if not self.learn.gan_trainer.gen_mode: self.learn.opt.set_hyper('lr', self.learn.opt.hypers[0]['lr']/self.mult_lr)
# ## GAN data
#export
class InvisibleTensor(TensorBase):
def show(self, ctx=None, **kwargs): return ctx
#export
def generate_noise(fn, size=100): return cast(torch.randn(size), InvisibleTensor)
#export
@typedispatch
def show_batch(x:InvisibleTensor, y:TensorImage, samples, ctxs=None, max_n=10, nrows=None, ncols=None, figsize=None, **kwargs):
if ctxs is None: ctxs = get_grid(min(len(samples), max_n), nrows=nrows, ncols=ncols, figsize=figsize)
ctxs = show_batch[object](x, y, samples, ctxs=ctxs, max_n=max_n, **kwargs)
return ctxs
#export
@typedispatch
def show_results(x:InvisibleTensor, y:TensorImage, samples, outs, ctxs=None, max_n=10, nrows=None, ncols=None, figsize=None, **kwargs):
if ctxs is None: ctxs = get_grid(min(len(samples), max_n), nrows=nrows, ncols=ncols, add_vert=1, figsize=figsize)
ctxs = [b.show(ctx=c, **kwargs) for b,c,_ in zip(outs.itemgot(0),ctxs,range(max_n))]
return ctxs
bs = 128
size = 64
dblock = DataBlock(blocks = (TransformBlock, ImageBlock),
get_x = generate_noise,
get_items = get_image_files,
splitter = IndexSplitter([]),
item_tfms=Resize(size, method=ResizeMethod.Crop),
batch_tfms = Normalize.from_stats(torch.tensor([0.5,0.5,0.5]), torch.tensor([0.5,0.5,0.5])))
path = untar_data(URLs.LSUN_BEDROOMS)
dls = dblock.dataloaders(path, path=path, bs=bs)
dls.show_batch(max_n=16)
# ## GAN Learner
#export
def gan_loss_from_func(loss_gen, loss_crit, weights_gen=None):
"Define loss functions for a GAN from `loss_gen` and `loss_crit`."
def _loss_G(fake_pred, output, target, weights_gen=weights_gen):
ones = fake_pred.new_ones(fake_pred.shape[0])
weights_gen = ifnone(weights_gen, (1.,1.))
return weights_gen[0] * loss_crit(fake_pred, ones) + weights_gen[1] * loss_gen(output, target)
def _loss_C(real_pred, fake_pred):
ones = real_pred.new_ones (real_pred.shape[0])
zeros = fake_pred.new_zeros(fake_pred.shape[0])
return (loss_crit(real_pred, ones) + loss_crit(fake_pred, zeros)) / 2
return _loss_G, _loss_C
#export
def _tk_mean(fake_pred, output, target): return fake_pred.mean()
def _tk_diff(real_pred, fake_pred): return real_pred.mean() - fake_pred.mean()
# +
#export
@delegates()
class GANLearner(Learner):
"A `Learner` suitable for GANs."
def __init__(self, dls, generator, critic, gen_loss_func, crit_loss_func, switcher=None, gen_first=False,
switch_eval=True, show_img=True, clip=None, cbs=None, metrics=None, **kwargs):
gan = GANModule(generator, critic)
loss_func = GANLoss(gen_loss_func, crit_loss_func, gan)
if switcher is None: switcher = FixedGANSwitcher(n_crit=5, n_gen=1)
trainer = GANTrainer(clip=clip, switch_eval=switch_eval, show_img=show_img)
cbs = L(cbs) + L(trainer, switcher)
metrics = L(metrics) + L(*LossMetrics('gen_loss,crit_loss'))
super().__init__(dls, gan, loss_func=loss_func, cbs=cbs, metrics=metrics, **kwargs)
@classmethod
def from_learners(cls, gen_learn, crit_learn, switcher=None, weights_gen=None, **kwargs):
"Create a GAN from `learn_gen` and `learn_crit`."
losses = gan_loss_from_func(gen_learn.loss_func, crit_learn.loss_func, weights_gen=weights_gen)
return cls(gen_learn.dls, gen_learn.model, crit_learn.model, *losses, switcher=switcher, **kwargs)
@classmethod
def wgan(cls, dls, generator, critic, switcher=None, clip=0.01, switch_eval=False, **kwargs):
"Create a WGAN from `data`, `generator` and `critic`."
return cls(dls, generator, critic, _tk_mean, _tk_diff, switcher=switcher, clip=clip, switch_eval=switch_eval, **kwargs)
GANLearner.from_learners = delegates(to=GANLearner.__init__)(GANLearner.from_learners)
GANLearner.wgan = delegates(to=GANLearner.__init__)(GANLearner.wgan)
# -
from fastai2.callback.all import *
generator = basic_generator(64, n_channels=3, n_extra_layers=1)
critic = basic_critic (64, n_channels=3, n_extra_layers=1, act_cls=partial(nn.LeakyReLU, negative_slope=0.2))
learn = GANLearner.wgan(dls, generator, critic, opt_func = RMSProp)
learn.recorder.train_metrics=True
learn.recorder.valid_metrics=False
#slow
learn.fit(1, 2e-4, wd=0.)
learn.show_results(max_n=9, ds_idx=0)
# ## Export -
#hide
from nbdev.export import notebook2script
notebook2script()
| nbs/24_vision.gan.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
# # Kernel Density Estimate of Species Distributions
# This shows an example of a neighbors-based query (in particular a kernel
# density estimate) on geospatial data, using a Ball Tree built upon the
# Haversine distance metric -- i.e. distances over points in latitude/longitude.
# The dataset is provided by Phillips et. al. (2006).
# If available, the example uses
# `basemap <https://matplotlib.org/basemap/>`_
# to plot the coast lines and national boundaries of South America.
#
# This example does not perform any learning over the data
# (see `sphx_glr_auto_examples_applications_plot_species_distribution_modeling.py` for
# an example of classification based on the attributes in this dataset). It
# simply shows the kernel density estimate of observed data points in
# geospatial coordinates.
#
# The two species are:
#
# - `"Bradypus variegatus"
# <http://www.iucnredlist.org/apps/redlist/details/3038/0>`_ ,
# the Brown-throated Sloth.
#
# - `"Microryzomys minutus"
# <http://www.iucnredlist.org/details/13408/0>`_ ,
# also known as the Forest Small Rice Rat, a rodent that lives in Peru,
# Colombia, Ecuador, Peru, and Venezuela.
#
# ## References
#
# * `"Maximum entropy modeling of species geographic distributions"
# <http://rob.schapire.net/papers/ecolmod.pdf>`_
# <NAME>, <NAME>, <NAME> - Ecological Modelling,
# 190:231-259, 2006.
#
# +
# Author: <NAME> <<EMAIL>>
#
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_species_distributions
from sklearn.neighbors import KernelDensity
# if basemap is available, we'll use it.
# otherwise, we'll improvise later...
try:
from mpl_toolkits.basemap import Basemap
basemap = True
except ImportError:
basemap = False
def construct_grids(batch):
"""Construct the map grid from the batch object
Parameters
----------
batch : Batch object
The object returned by :func:`fetch_species_distributions`
Returns
-------
(xgrid, ygrid) : 1-D arrays
The grid corresponding to the values in batch.coverages
"""
# x,y coordinates for corner cells
xmin = batch.x_left_lower_corner + batch.grid_size
xmax = xmin + (batch.Nx * batch.grid_size)
ymin = batch.y_left_lower_corner + batch.grid_size
ymax = ymin + (batch.Ny * batch.grid_size)
# x coordinates of the grid cells
xgrid = np.arange(xmin, xmax, batch.grid_size)
# y coordinates of the grid cells
ygrid = np.arange(ymin, ymax, batch.grid_size)
return (xgrid, ygrid)
# Get matrices/arrays of species IDs and locations
data = fetch_species_distributions()
species_names = ['Bradypus Variegatus', 'Microryzomys Minutus']
Xtrain = np.vstack([data['train']['dd lat'],
data['train']['dd long']]).T
ytrain = np.array([d.decode('ascii').startswith('micro')
for d in data['train']['species']], dtype='int')
Xtrain *= np.pi / 180. # Convert lat/long to radians
# Set up the data grid for the contour plot
xgrid, ygrid = construct_grids(data)
X, Y = np.meshgrid(xgrid[::5], ygrid[::5][::-1])
land_reference = data.coverages[6][::5, ::5]
land_mask = (land_reference > -9999).ravel()
xy = np.vstack([Y.ravel(), X.ravel()]).T
xy = xy[land_mask]
xy *= np.pi / 180.
# Plot map of South America with distributions of each species
fig = plt.figure()
fig.subplots_adjust(left=0.05, right=0.95, wspace=0.05)
for i in range(2):
plt.subplot(1, 2, i + 1)
# construct a kernel density estimate of the distribution
print(" - computing KDE in spherical coordinates")
kde = KernelDensity(bandwidth=0.04, metric='haversine',
kernel='gaussian', algorithm='ball_tree')
kde.fit(Xtrain[ytrain == i])
# evaluate only on the land: -9999 indicates ocean
Z = np.full(land_mask.shape[0], -9999, dtype='int')
Z[land_mask] = np.exp(kde.score_samples(xy))
Z = Z.reshape(X.shape)
# plot contours of the density
levels = np.linspace(0, Z.max(), 25)
plt.contourf(X, Y, Z, levels=levels, cmap=plt.cm.Reds)
if basemap:
print(" - plot coastlines using basemap")
m = Basemap(projection='cyl', llcrnrlat=Y.min(),
urcrnrlat=Y.max(), llcrnrlon=X.min(),
urcrnrlon=X.max(), resolution='c')
m.drawcoastlines()
m.drawcountries()
else:
print(" - plot coastlines from coverage")
plt.contour(X, Y, land_reference,
levels=[-9998], colors="k",
linestyles="solid")
plt.xticks([])
plt.yticks([])
plt.title(species_names[i])
plt.show()
| 3_ml_start_knn_examples/plot_species_kde.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="3CA0tdmlIKk4" colab_type="text"
# # Flocculation Design Challenge
#
# Learn how to use the AguaClara code distribution and python to design a flocculator!
# The [AguaClara code documentation](https://aguaclara.github.io/aguaclara/index.html) will be helpful as you search for useful functions.
#
# 30 points total
# * 4 for style (define variables, comments in code, clear names, answers in sentences)
# * 26 for questions
# + id="rQ40v4xNxLRW" colab_type="code" outputId="1da61da3-9f5f-4352-fffb-d136b9ba7f07" executionInfo={"status": "ok", "timestamp": 1569503070498, "user_tz": 240, "elapsed": 7651, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AAuE7mDmYNDq6ij0468RSHe1goXE_t9gbSPdq5OAsU4-ejQ=s64", "userId": "08369668289863895493"}} colab={"base_uri": "https://localhost:8080/", "height": 513}
# !pip install aguaclara
# + id="wufnKCzBHPC5" colab_type="code" colab={}
from aguaclara.core.units import unit_registry as u
import aguaclara as ac
import numpy as np
import matplotlib.pyplot as plt
# + [markdown] id="BVUZ6Hkl8xRh" colab_type="text"
# # Velocity gradients and flow geometry
#
#
# ### 1) (2 points)
#
# Coagulant is injected in the center a long straight pipe. The pipe is 12 inches Nominal Diameter schedule 40 PVC and the flow rate is 120 L/s at $10^{\circ}C$. What distance is required for the coagulant to be completely mixed with the water in the pipe? Note that this estimate is based on the time required for an eddy to traverse the diameter of the pipe and that a safety factor of order 3 * $\pi$/2 would be reasonable. The eddy is assumed to not travel in a straight line and thus it more likely travels a distance of $\pi$/2 an the factor of 3 is an additional safety factor.Include this safety factor in the calculations. See the [(equation for pipe mixing)](https://aguaclara.github.io/Textbook/Rapid_Mix/RM_Derivations.html?highlight=energy%20dissipation#equation-rapid-mix-rm-derivations-42).
#
# * 1 point for correct friction factor
# * 1 point for correct distance
# + id="1DAxoEF09MNd" colab_type="code" colab={}
# + [markdown] id="mburwR8_939o" colab_type="text"
# ### 2) (1 point)
#
# What is the residence time in this mixing zone?
#
# * 1 point for correct answer
# + id="7p958wsl97dP" colab_type="code" colab={}
# + [markdown] id="7o7iBw1L-G-m" colab_type="text"
# ### 3) (1 point)
#
# How much head loss from wall shear will have occurred in the pipe in the distance measured in the previous problem? This analysis reveals how little energy is required to blend the coagulant with the raw water.
#
# * 1 point for correct answer
# + id="3G_lIWpK-Net" colab_type="code" colab={}
# + [markdown] id="sNgXh70UAbil" colab_type="text"
# ### 4) (1 point)
#
# What is the [Camp Stein velocity gradient](https://aguaclara.github.io/Textbook/Rapid_Mix/RM_Intro.html?highlight=camp%20stein#id15) in this pipe flow?
#
# * 1 point for correct answer
#
# + id="Yvv15jbGAexg" colab_type="code" colab={}
# + [markdown] id="jHaRWU6XAiOe" colab_type="text"
# ### 5) (2 points)
#
# What is the $G\theta$ for this mixing zone and how does it compare with the $G\theta$ recommended for [mechanical mixing units](https://aguaclara.github.io/Textbook/Rapid_Mix/RM_Intro.html#maximum-velocity-gradients)?
#
# * 1 for correct Gt
# * 1 point for comparison
#
# + id="d_lPTfo6Ap6Q" colab_type="code" colab={}
# + [markdown] id="ERhG_WGwCEg4" colab_type="text"
# ### 6) (1 point)
#
# What is the velocity gradient at the wall of the pipe? This will make it apparent that the velocity gradient is far from constant
#
# * 1 point for correct answer
# + id="vNVDpZ6PCFba" colab_type="code" colab={}
# + [markdown] id="F_V3xcPgCNeZ" colab_type="text"
# ### 7) (2 points)
#
# Suppose we insert a [flat plate oriented with the flat surface facing the flow](https://aguaclara.github.io/Textbook/Rapid_Mix/RM_Derivations.html?highlight=flat%20plate#behind-a-flat-plate) inside the pipe. Let the width of the plate be 0.5 cm so it is small enough that it doesn't significantly increase the velocity in the pipe. What is the maximum velocity gradient downstream of the plate? You may neglect the fact that the velocity in the center of the turbulent pipe flow is slightly higher than the average velocity.
#
# * 1 for correct Ratio
# * 1 for correct velocity gradient
# + id="s_1GJKRMCQJo" colab_type="code" colab={}
# + [markdown] id="Oo7xIHrSCW4Y" colab_type="text"
# ### 8) (1 point)
# What happens to the velocity gradient if a narrower flat plate is used? Does the maximum velocity gradient increase or decrease? Just look at the equation to answer this!
#
# * 1 point for correct answer
# + [markdown] id="60KdrJ-YCXeg" colab_type="text"
#
# + [markdown] id="CwAM-0xkEEe8" colab_type="text"
# # Flocculation model
#
# ### 1) (2 points)
# How far will two kaolin clay particles (density of 2650 $\frac{kg}{m^3}$) with a diameter of 5 $\mu m$ travel relative to each if they are in a uniform velocity gradient of 100 Hz for 400 s and separated (in the direction of the velocity gradient) by their average separation distance based on a turbidity of 0.5 NTU?
#
# We have defined NTU as a unit based on the concentration of clay in the aguaclara code base. You can derive these simple equations yourself or find them in the text. Note that in a uniform velocity gradient $\bar G = G_{CS}$. The [floc model code documentation](https://aguaclara.github.io/aguaclara/research/floc_model.html) and the [floc model chapter](https://aguaclara.github.io/Textbook/Flocculation/Floc_Model.html) will be helpful. The relative displacement caused simply by the deformation of the fluid is impressive!
#
# * 1 point for correct separation distance
# * 1 point for correct travel distance
# + id="bccfOxr-EHlX" colab_type="code" colab={}
# + [markdown] id="AyGEfBI2EOqn" colab_type="text"
# ### 2) (2 points)
#
# How much volume is "[cleared](https://aguaclara.github.io/Textbook/Flocculation/Floc_Model.html#equation-flocculation-floc-model-8)" by these particles divided by the volume occupied by the particles? This ratio is essentially how many times these particles should have collided in the 400 s.
#
# This analysis illustrates why 1 NTU is a practical limit for flocculation. Assuming that we don't want to apply so much coagulant that the clay particles are completely covered with coagulant, then some fraction of the collisions will be ineffective. Thus at 1 NTU a Gtheta of 40,000 might only cause one successful collision.
#
# * 1 for correct cleared volume
# * 1 for correct occupied volume
# + id="NzbU8xbxERPC" colab_type="code" colab={}
# + [markdown] id="dOkUG4liEc2w" colab_type="text"
# # Flocculator design
#
# Below we design a flocculator using [ac.Flocculator](https://aguaclara.github.io/aguaclara/design/floc.html) in the aguaclara distribution version. We will use the default settings for this design except change the flow rate to 60 L/s. The available inputs (and their default values) that you can change are shown in the [documentation](https://aguaclara.github.io/aguaclara/design/floc.html). You can change any of these parameters by including their keywords in the function call.
#
# See the [current cad drawing of a flocculator and entrance tank](https://cad.onshape.com/documents/c3a8ce032e33ebe875b9aab4/w/de9ad5474448b34f33fef097/e/08f41d8bdd9a9c90ab396f8a).
# + id="nl1iJikiEh_x" colab_type="code" outputId="88a90b63-7d18-46fb-8aed-a7909a8223f7" executionInfo={"status": "ok", "timestamp": 1569504425210, "user_tz": 240, "elapsed": 246, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AAuE7mDmYNDq6ij0468RSHe1goXE_t9gbSPdq5OAsU4-ejQ=s64", "userId": "08369668289863895493"}} colab={"base_uri": "https://localhost:8080/", "height": 204}
flow=60 * u.L/u.s
myF = ac.Flocculator(q=flow)
print('The number of channels is', myF.chan_n)
print('The channel length is',myF.chan_l)
print('The channel width is',ac.round_sig_figs(myF.chan_w,2))
print('The spacing between baffles is',ac.round_sig_figs(myF.baffle_s,2))
print('The number of obstacles per baffle is', myF.obstacle_n)
print('The velocity gradient is', ac.round_sig_figs(myF.vel_grad_avg,2))
print('The residence time (not counting the effect of head loss) is',ac.round_sig_figs(myF.retention_time,2))
print('The maximum distance between flow expansions is', ac.round_sig_figs(myF.expansion_h_max,2))
print('The drain diameter is', myF.drain_pipe.size)
print('The Gt is',myF.gt)
print('The length of the first channel occupied by the entrance tank is',myF.ent_l)
# + [markdown] id="QJVXHh9uJc-h" colab_type="text"
# ## Calculations and analysis
#
# ### 1) (2 points)
#
# How many expansions are there in total? Estimate this based on the spacing and flocculator size. You will have to account for the entrance tank that occupies volume in the first flocculator channel.
#
# * 2 points for correct answer
# + id="YkiAXqGbJgfH" colab_type="code" colab={}
# + [markdown] id="WCv6bN7AKQGL" colab_type="text"
# ### 2) (2 points)
# What is the head loss per expansion? (Calculate this head loss using the minor loss equation) You can use the BAFFLE_K that is defined in the flocculator class.
#
# * 1 point for correct velocity
# * 1 point for correct headloss
# + id="hakmCBhlKSpA" colab_type="code" colab={}
# + [markdown] id="excnCsR7Kdfv" colab_type="text"
# ### 3) (1 point)
# What is the total head loss of all of the expansions? Compare this with the target head loss of 40 cm.
#
# * 1 point for correct answer
# + id="TIdvFS75Kf7a" colab_type="code" colab={}
# + [markdown] id="Ffp3QLYSKxOk" colab_type="text"
# ### 4) (5 points)
# Change the design temperature over a range that would be applicable in Ithaca (0 to 30 degC) for a flocculator design of your choice. What happens as the temperature increases? Plot the following:
# * residence time
# * velocity gradient
# * baffle spacing
# * number of channels
# * channel width
#
# all as functions of temperature. Explain WHY these design changes occur.
#
# Hints:
# * I suggest creating about 50 designs
# * create a numpy array of flocculator objects.
# ```
# MyFn = 50
# myFs =np.empty(MyFn, dtype=type(myF))
# ```
# * create empty numpy arrays with the correct units for each parameter that you want to plot
# * use a single for loop to cycle through each design and extract the parameters that you want to plot from the flocculator objects (MyF) and place those values in the arrays that you created.
#
# Points
# * 1 for each Graph
#
# Make sure that each graph has correct axis labels with units!
# + id="5nYovZ_2K2Oy" colab_type="code" colab={}
# + [markdown] id="xmJa3qG1Nf1Q" colab_type="text"
# The water becomes more viscous as it gets colder. Thus it becomes more difficult to deform. Given that we are limiting the amount of energy that we are willing to use, we have to compensate by deforming the fluid more slowly. Thus if we hold the amount of energy available as a constant, then the velocity gradient decreases as the temperature decreases and the residence time increases. The number of channels increases as the temperature drops because the design ran up against the maximum channel width constraint as the flocculator volume increased.
# + [markdown] id="PVXiojzANjXM" colab_type="text"
# ### 5) (1 point)
# When designing a flocculator how should you select the design temperature?
#
# * 1 point for correct answer
# + [markdown] id="4m45FS6JNmeI" colab_type="text"
#
# + [markdown] id="0N9YaAjjIIMW" colab_type="text"
# ### 6) (4 points)
# Here at Cornell and in Honduras we have been experimenting with flocculators that have a $G\theta$ of 20,000 and a head loss of 50 cm for use in Honduras where the minimum temperature is about 15 $^\circ C$. Create a design for an entrance tank and flocculator with these inputs and flow rates of 10 L/s and 100 L/s. Previously in this design challenge you designed a flocculator. The AguaClara entrance tank is incorporated into the first channel of the flocculator. In object oriented programming this is handled by creating an object that is an [entrance tank flocculator assembly](https://aguaclara.github.io/aguaclara/design/ent_floc.html) that contains the entrance tank with an LFOM and the flocculator. This higher level assembly is able to optimize the width of the flocculator channels to best accommodate the flocculator and the entrance tank.
#
# Hints...
#
# * Place the two flows in an array and use a for loop to cycle through the two designs and print the design outputs.
# * Create a design: `myfastetf = ac.EntTankFloc(q=myq, floc = ac.Flocculator(q=myq, gt=20000, temp=15 * u.degC, hl = 50 * u.cm))`
# * At minimum you need to print the width and length of the flocculator! You might be curious about how other values have changed too.
# * Notice how we can set the values for a sub assembly inside the list of inputs for the assembly.
#
#
# Given these designs would you recommend that we change our plant layout to allow a single channel flocculator?
# List as many design implications as you can think of for this potential change. Check out the [current cad drawing](https://cad.onshape.com/documents/c3a8ce032e33ebe875b9aab4/w/de9ad5474448b34f33fef097/e/08f41d8bdd9a9c90ab396f8a) and explain why the plan view area of the plant would change and identify what else would need to change if the flocculator only had one channel. How is the entrance tank drained to remove accumulated solids? Do these designs make sense? As of 9/26/2019 the channel width for the low flow was not constructable.
#
# * 2 for designs
# * 2 for recommendation
# + [markdown] id="WyrWyNMnQg9r" colab_type="text"
# The entrance tank/flocculator would be less costly and the plan view area of the plant would be decreased because there wouldn't be so much wasted space at the end of the flocculator.
# The entrance tank has a drain for solids that accumulate and now that drain would be far from the central drain channel. It isn't clear how that drain would be connected because it will contain large heavy solids that could easily clog a horizontal drain pipe.
# + [markdown] id="3729SVJP8de0" colab_type="text"
#
# + [markdown] id="JkbUM5hHTW6w" colab_type="text"
# # Musings on the design for a horizontal flow flocculator
#
# The calculations and musings below are to get you thinking about what happens as we design larger water treatment plants!
#
# The maximum flow for a vertical flow flocculator is...
#
# $$Q = \frac{W_{Max}H_e}{\Pi_{HS_{Min}} } \left( \frac{2 H_e g^2 h_e^2}{K(G\theta)^2 \nu} \right)^\frac{1}{3}$$
# + id="jscBzneZTcuu" colab_type="code" outputId="7f80e5c4-b289-45ae-b363-e88b72600a2b" executionInfo={"status": "ok", "timestamp": 1569503071086, "user_tz": 240, "elapsed": 8172, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AAuE7mDmYNDq6ij0468RSHe1goXE_t9gbSPdq5OAsU4-ejQ=s64", "userId": "08369668289863895493"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
W_Max = 1.08 * u.m
K= 2.56
Ratio_H_S = 3
h_L = 40*u.cm
H_Min = 2*u.m
Gtheta = 3.7e4
T_Des = 15 * u.degC
#Find General Design Parameters
nu = ac.viscosity_kinematic(T_Des)
Q = (W_Max * H_Min/Ratio_H_S * ((2*H_Min*u.gravity**2*h_L**2)/(K*Gtheta**2 * nu))**(1/3)).to(u.L/u.s)
print('The maximum flow rate for a vertical flow flocculator is',Q)
# + [markdown] id="vpJexw4Q5sSy" colab_type="text"
#
# The minimum flow for a horizontal flow flocculator is...
#
#
# $$Q = W_{Min}S_{Min} \left( \frac{2 H_e g^2 h_e^2}{K(G\theta)^2 \nu} \right)^\frac{1}{3}$$
#
# * Here W is the width of flow which is the depth of water.
# * He is the distance between flow expansions which is the width of the channel
# + id="nOzDscp6vIEa" colab_type="code" outputId="1993bd20-5aa8-40d3-d69b-cdddc49258f4" executionInfo={"status": "ok", "timestamp": 1569503071088, "user_tz": 240, "elapsed": 8168, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AAuE7mDmYNDq6ij0468RSHe1goXE_t9gbSPdq5OAsU4-ejQ=s64", "userId": "08369668289863895493"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
# Now find the minimum flow for a horizontal flow flocculator given the constraint that the spacing must be 0.5 m for the masons
W_Min = 2*u.m
S_Min = 0.5 * u.m
H_Min = Ratio_H_S*S_Min
Q = (W_Min * S_Min * ((2*H_Min*u.gravity**2*h_L**2)/(K*Gtheta**2 * nu))**(1/3)).to(u.L/u.s)
print('The minimum flow rate for a horizontal flow flocculator is',Q)
# + [markdown] id="XJvowwb27AK_" colab_type="text"
# There is a gap in flow between our maximum vertical flow and minimum horizontal flow. We can bridge this gap by either increasing the depth of the vertical flow flocculator or decreasing the depth of the horizontal flow flocculator. We can solve the previous equation for W_Min given the flow rate required to find horizontal flow depth solutions that will work for flows between 180 and 220 L/s.
#
# We need to consult with the implementation partner to see what they would regard as the optimal depth for the flocculator tank. It is possible that we could increase the depth and provide an access ladder into the flocculator. This would allow us to minimize the plan view area required.
#
# Presumably the total wall area should be minimized to reduce construction costs. We need to determine if the minimum cost is at an H/S ratio of 3 or at an H/S ratio of 6. The optimal solution might depend on whether the walls that serve as baffles are equal or lower in cost to the walls used to create channels. This design process will need to include an exploration of possible materials and fabrication methods for the baffles. The baffles don't experience significant hydrostatic pressure and thus they must primarily be strong enough to resist being pushed by a human during construction or maintenance.
#
#
| DC/Floc_DC.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] deletable=true editable=true
# # Self-Driving Car Engineer Nanodegree
#
# ## Deep Learning
#
# ## Project: Build a Traffic Sign Recognition Classifier
#
# In this notebook, a template is provided for you to implement your functionality in stages, which is required to successfully complete this project. If additional code is required that cannot be included in the notebook, be sure that the Python code is successfully imported and included in your submission if necessary.
#
# > **Note**: Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You can then export the notebook by using the menu above and navigating to \n",
# "**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
#
# In addition to implementing code, there is a writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) that can be used to guide the writing process. Completing the code template and writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/481/view) for this project.
#
# The [rubric](https://review.udacity.com/#!/rubrics/481/view) contains "Stand Out Suggestions" for enhancing the project beyond the minimum requirements. The stand out suggestions are optional. If you decide to pursue the "stand out suggestions", you can include the code in this Ipython notebook and also discuss the results in the writeup file.
#
#
# >**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.
# + [markdown] deletable=true editable=true
# ---
# ## 1. Load The CIFAR10 Data
# + deletable=true editable=true
# Load pickled data
import pickle
from keras.datasets import cifar10
from sklearn.model_selection import train_test_split
(X_train_temp, y_train_temp), (X_test, y_test) = cifar10.load_data()
# y_train.shape is 2d, (50000, 1). While Keras is smart enough to handle this
# it's a good idea to flatten the array.
y_train_temp = y_train_temp.reshape(-1)
y_test = y_test.reshape(-1)
X_train, X_valid, y_train, y_valid = train_test_split(X_train_temp, y_train_temp, test_size=0.33, random_state=0)
assert(len(X_train) == len(y_train))
assert(len(X_valid) == len(y_valid))
assert(len(X_test) == len(y_test))
print("Loading done!")
# + [markdown] deletable=true editable=true
# ---
#
# ## Step 1: Dataset Summary & Exploration
#
# The pickled data is a dictionary with 4 key/value pairs:
#
# - `'features'` is a 4D array containing raw pixel data of the traffic sign images, (num examples, width, height, channels).
# - `'labels'` is a 1D array containing the label/class id of the traffic sign. The file `signnames.csv` contains id -> name mappings for each id.
# - `'sizes'` is a list containing tuples, (width, height) representing the original width and height the image.
# - `'coords'` is a list containing tuples, (x1, y1, x2, y2) representing coordinates of a bounding box around the sign in the image. **THESE COORDINATES ASSUME THE ORIGINAL IMAGE. THE PICKLED DATA CONTAINS RESIZED VERSIONS (32 by 32) OF THESE IMAGES**
#
# Complete the basic data summary below. Use python, numpy and/or pandas methods to calculate the data summary rather than hard coding the results. For example, the [pandas shape method](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.shape.html) might be useful for calculating some of the summary results.
# + [markdown] deletable=true editable=true
# ## 2. Provide a Basic Summary of the Data Set Using Python, Numpy and/or Pandas
# + deletable=true editable=true
### Replace each question mark with the appropriate value.
### Use python, pandas or numpy methods rather than hard coding the results
import numpy as np
# Number of training examples
n_train = len(X_train)
# Number of testing examples.
n_test = len(X_test)
# Number of validation examples
n_valid = len(X_valid)
# TODO: What's the shape of an traffic sign image?
image_shape = X_train[0].shape
# TODO: How many unique classes/labels there are in the dataset.
n_classes = np.unique(y_train).size
print("Number of training examples =", n_train)
print("Number of validation examples =", n_valid)
print("Number of testing examples =", n_test)
print("Image data shape =", image_shape)
print("Number of classes =", n_classes)
# + [markdown] deletable=true editable=true
# ## 3. Include an exploratory visualization of the dataset
# + [markdown] deletable=true editable=true
# Visualize the German Traffic Signs Dataset using the pickled file(s). This is open ended, suggestions include: plotting traffic sign images, plotting the count of each sign, etc.
#
# The [Matplotlib](http://matplotlib.org/) [examples](http://matplotlib.org/examples/index.html) and [gallery](http://matplotlib.org/gallery.html) pages are a great resource for doing visualizations in Python.
#
# **NOTE:** It's recommended you start with something simple first. If you wish to do more, come back to it after you've completed the rest of the sections.
# + deletable=true editable=true
import matplotlib.pyplot as plt
import random
import numpy as np
import csv
import pandas as pd
# Visualizations will be shown in the notebook.
# %matplotlib inline
def show_sample(features, labels, histogram = 1, sample_num = 1, sample_index = -1, color_map ='brg'):
if histogram == 1 :
col_num = 2
#Create training sample + histogram plot
f, axarr = plt.subplots(sample_num+1, col_num, figsize=(col_num*4,(sample_num+1)*3))
else:
if sample_num <= 4:
col_num = sample_num
else:
col_num = 4
if sample_num%col_num == 0:
row_num = int(sample_num/col_num)
else:
row_num = int(sample_num/col_num)+1
if sample_num == 1:
#Create training sample plot
f, ax = plt.subplots(row_num, col_num)
else:
#Create training sample plot
f, axarr = plt.subplots(row_num, col_num, figsize=(col_num*4,(row_num+1)*2))
signnames = pd.read_csv('signnames.csv')
index = sample_index - 1
for i in range(0, sample_num, 1):
if sample_index < -1:
index = random.randint(0, len(features))
else:
index = index + 1
if histogram == 1 :
image = features[index].squeeze()
axarr[i,0].set_title('%s' % signnames.iloc[labels[index], 1])
axarr[i,0].imshow(image,color_map)
hist,bins = np.histogram(image.flatten(),256, normed =1 )
cdf = hist.cumsum()
cdf_normalized = cdf * hist.max()/ cdf.max()
axarr[i,1].plot(cdf_normalized, color = 'b')
axarr[i,1].hist(image.flatten(),256, normed =1, color = 'r')
axarr[i,1].legend(('cdf','histogram'), loc = 'upper left')
axarr[i,0].axis('off')
axarr[sample_num,0].axis('off')
axarr[sample_num,1].axis('off')
else:
image = features[index].squeeze()
if row_num > 1:
axarr[int(i/col_num),i%col_num].set_title('%s' % signnames.iloc[labels[index], 1])
axarr[int(i/col_num),i%col_num].imshow(image,color_map)
axarr[int(i/col_num),i%col_num].axis('off')
axarr[int(i/col_num),i%col_num].axis('off')
axarr[int(i/col_num),i%col_num].axis('off')
elif sample_num == 1:
ax.set_title('%s' % signnames.iloc[labels[index], 1])
ax.imshow(image,color_map)
ax.axis('off')
ax.axis('off')
ax.axis('off')
else:
axarr[i%col_num].set_title('%s' % signnames.iloc[labels[index], 1])
axarr[i%col_num].imshow(image,color_map)
axarr[i%col_num].axis('off')
axarr[i%col_num].axis('off')
axarr[i%col_num].axis('off')
# Tweak spacing to prevent clipping of title labels
f.tight_layout()
plt.show()
def show_training_dataset_histogram(labels_train,labels_valid,labels_test):
fig, ax = plt.subplots(figsize=(15,5))
temp = [labels_train,labels_valid,labels_test]
n_classes = np.unique(y_train).size
# the histogram of the training data
n, bins, patches = ax.hist(temp, n_classes, label=["Train","Valid","Test"])
ax.set_xlabel('Classes')
ax.set_ylabel('Number of occurence')
ax.set_title(r'Histogram of the data sets')
ax.legend(bbox_to_anchor=(1.01, 1), loc="upper left")
plt.show()
show_training_dataset_histogram(y_train,y_valid,y_test)
show_sample(X_train, y_train, sample_num = 6)
# + [markdown] deletable=true editable=true
# ----
#
# ## Step 2: Design and Test a Model Architecture
#
# Design and implement a deep learning model that learns to recognize traffic signs. Train and test your model on the [German Traffic Sign Dataset](http://benchmark.ini.rub.de/?section=gtsrb&subsection=dataset).
#
# The LeNet-5 implementation shown in the [classroom](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) at the end of the CNN lesson is a solid starting point. You'll have to change the number of classes and possibly the preprocessing, but aside from that it's plug and play!
#
# With the LeNet-5 solution from the lecture, you should expect a validation set accuracy of about 0.89. To meet specifications, the validation set accuracy will need to be at least 0.93. It is possible to get an even higher accuracy, but 0.93 is the minimum for a successful project submission.
#
# There are various aspects to consider when thinking about this problem:
#
# - Neural network architecture (is the network over or underfitting?)
# - Play around preprocessing techniques (normalization, rgb to grayscale, etc)
# - Number of examples per label (some have more than others).
# - Generate fake data.
#
# Here is an example of a [published baseline model on this problem](http://yann.lecun.com/exdb/publis/pdf/sermanet-ijcnn-11.pdf). It's not required to be familiar with the approach used in the paper but, it's good practice to try to read papers like these.
# + [markdown] deletable=true editable=true
# ## 4. Augment the Data Set
# + deletable=true editable=true
import cv2
from tqdm import tqdm
from sklearn.utils import shuffle
def random_transform_image(dataset, index):
# Hyperparameters
# Values inspired from <NAME> and <NAME> Paper : Traffic Sign Recognition with Multi-Scale Convolutional Networks
Scale_change_max = 0.1
Translation_max = 2 #pixels
Rotation_max = 15 #degrees
Brightness_max = 0.1
# Generate random transformation values
trans_x = np.random.uniform(-Translation_max,Translation_max)
trans_y = np.random.uniform(-Translation_max,Translation_max)
angle = np.random.uniform(-Rotation_max,Rotation_max)
scale = np.random.uniform(1-Scale_change_max,1+Scale_change_max)
bright = np.random.uniform(-Brightness_max,Brightness_max)
#Brightness
#create white image
white_img = 255*np.ones((32,32,3), np.uint8)
black_img = np.zeros((32,32,3), np.uint8)
if bright >= 0:
img = cv2.addWeighted(dataset[index].squeeze(),1-bright,white_img,bright,0)
else:
img = cv2.addWeighted(dataset[index].squeeze(),bright+1,black_img,bright*-1,0)
# Scale
img = cv2.resize(img,None,fx=scale, fy=scale, interpolation = cv2.INTER_CUBIC)
# Get image shape afeter scaling
rows,cols,chan = img.shape
# Pad with zeroes before rotation if image shape is less than 32*32*3
if rows < 32:
offset = int((32-img.shape[0])/2)
# If shape is an even number
if img.shape[0] %2 == 0:
img = cv2.copyMakeBorder(img,offset,offset,offset,offset,cv2.BORDER_CONSTANT,value=[0,0,0])
else:
img = cv2.copyMakeBorder(img,offset,offset+1,offset+1,offset,cv2.BORDER_CONSTANT,value=[0,0,0])
# Update image shape after padding
rows,cols,chan = img.shape
# Rotate
M = cv2.getRotationMatrix2D((cols/2,rows/2),angle,1)
img = cv2.warpAffine(img,M,(cols,rows))
# Translation
M = np.float32([[1,0,trans_x],[0,1,trans_y]])
img = cv2.warpAffine(img,M,(cols,rows))
# Crop centered if image shape is greater than 32*32*3
if rows > 32:
offset = int((img.shape[0]-32)/2)
img = img[offset: 32 + offset, offset: 32 + offset]
return img
# Parameters
# Max example number per class
num_example_per_class = np.bincount(y_train)
min_example_num = max(num_example_per_class)
for i in range(len(num_example_per_class)):
# Update number of examples by class
num_example_per_class = np.bincount(y_train)
# If the class lacks examples...
if num_example_per_class[i] < min_example_num:
# Locate where pictures of this class are located in the training set..
pictures = np.array(np.where(y_train == i)).T
# Compute the number of pictures to be generated
num_example_to_generate = min_example_num - num_example_per_class[i]
# Compute the number of iteration necessary on the real data
num_iter = int( num_example_to_generate/len(pictures) ) + 1
# Compute the pool of real data necessary to fill the classes
if num_iter == 1 :
num_pictures = num_example_to_generate
else:
num_pictures = len(pictures)
# # Limit the number of iteration to 10
# num_iter = min(num_iter, 10)
# Create empty list
more_X = []
more_y = []
for k in range(num_iter):
# if we are in the last iteration, num_pictures is adjusted to fit the min_example_num
if (k == num_iter - 1) and (num_iter > 1):
num_pictures = min_example_num - num_iter * len(pictures)
# For each pictures of this class, generate 1 more synthetic image
pbar = tqdm(range(num_pictures), desc='Iter {:>2}/{}'.format(i+1, len(num_example_per_class)), unit='examples')
for j in pbar:
# Append the transformed picture
more_X.append(random_transform_image(X_train,pictures[j]))
# Append the class number
more_y.append(i)
# Append the synthetic images to the training set
X_train = np.append(X_train, np.array(more_X), axis=0)
y_train = np.append(y_train, np.array(more_y), axis=0)
print("New training feature shape",X_train.shape)
print("New training label shape",y_train.shape)
print("Data augmentation done!")
# + [markdown] deletable=true editable=true
# ## 5. Show a sample of the augmented dataset
# + deletable=true editable=true
# Visualization
show_training_dataset_histogram(y_train,y_valid,y_test)
show_sample(X_train, y_train, histogram = 0, sample_num = 8, sample_index = 35000)
# + [markdown] deletable=true editable=true
# ## 6. Pre-process functions
# + deletable=true editable=true
import cv2
from numpy import newaxis
def equalize_Y_histogram(features):
images = []
for image in features:
# Convert RGB to YUV
temp = cv2.cvtColor(image, cv2.COLOR_BGR2YUV);
# Equalize Y histogram in order to get better contrast accross the dataset
temp[:,:,0] = cv2.equalizeHist(temp[:,:,0])
# Convert back YUV to RGB
temp = cv2.cvtColor(temp, cv2.COLOR_YUV2BGR)
images.append(temp)
return np.array(images)
def CLAHE_contrast_normalization(features):
images = []
for image in features:
# create a CLAHE object
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(4,4))
temp = clahe.apply(image)
images.append(temp)
return np.array(images)
def convert_to_grayscale(features):
gray_images = []
for image in features:
# Convert RGB to grayscale
temp = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
gray_images.append(temp)
return np.array(gray_images)
def normalize_grayscale(image_data):
"""
Normalize the image data with Min-Max scaling to a range of [0.1, 0.9]
:param image_data: The image data to be normalized
:return: Normalized image data
"""
a = 0.1
b = 0.9
image_data_norm = a + ((image_data - np.amin(image_data))*(b-a))/(np.amax(image_data) - np.amin(image_data))
return image_data_norm
# + [markdown] deletable=true editable=true
# ## 7. Show a sample of the preprocess functions outputs
# + deletable=true editable=true
index = 255
X_temp1 = convert_to_grayscale(X_train)
X_temp2 = CLAHE_contrast_normalization(X_temp1)
X_temp3 = normalize_grayscale(X_temp2)
show_sample(X_train, y_train, histogram = 1, sample_num = 1, sample_index = index)
show_sample(X_temp1, y_train, histogram = 1, sample_num = 1, sample_index = index, color_map ='gray')
show_sample(X_temp2, y_train, histogram = 1, sample_num = 1, sample_index = index, color_map ='gray')
print(X_temp2[index])
print(X_temp3[index])
# + [markdown] deletable=true editable=true
# ## 8. Preprocess the Dataset
# + deletable=true editable=true
#Preprocessing pipeline
print('Preprocessing training features...')
X_train = convert_to_grayscale(X_train)
X_train = CLAHE_contrast_normalization(X_train)
X_train = normalize_grayscale(X_train)
X_train = X_train[..., newaxis]
print("Processed shape =", X_train.shape)
print('Preprocessing validation features...')
X_valid = convert_to_grayscale(X_valid)
X_valid = CLAHE_contrast_normalization(X_valid)
X_valid = normalize_grayscale(X_valid)
X_valid = X_valid[..., newaxis]
print("Processed shape =", X_valid.shape)
print('Preprocessing test features...')
X_test = convert_to_grayscale(X_test)
X_test = CLAHE_contrast_normalization(X_test)
X_test = normalize_grayscale(X_test)
X_test = X_test[..., newaxis]
print("Processed shape =", X_test.shape)
# Shuffle the training dataset
X_train, y_train = shuffle(X_train, y_train)
print("Pre-processing done!")
# + [markdown] deletable=true editable=true
# ## 9. Model Architecture
#
# [//]: # (Image References)
# [image1]: ./examples/architecture.png "Conv Net Architecture"
#
# ![alt text][image1]
#
# | Layer | Description | Input | Output |
# |:-------------:|:---------------------------------------------:|:-----------------:|:---------------------------:|
# | Input | 32x32x1 Grayscale image | Image | Convolution 1 |
# | Convolution 1 | 1x1 stride, valid padding, outputs 28x28x100 | Input | RELU |
# | RELU 1 | | Convolution 1 | Max Pooling 1 |
# | Max pooling 1 | 2x2 stride, outputs 14x14x100 | RELU 1 | Convolution 2, Max Pooling 3|
# | Convolution 2 | 1x1 stride, valid padding, outputs 10x10x200 | Max pooling 1 | RELU 2 |
# | RELU 2 | | Convolution 2 | Max pooling 2 |
# | Max pooling 2 | 2x2 stride, outputs 5x5x200 | RELU 2 | Flatten 2 |
# | Max pooling 3 | 2x2 stride, outputs 7x7x100 | Max pooling 1 | Flatten 1 |
# | Flatten 1 | Input = 7x7x100, Output = 4900 | Max pooling 3 | Concatenate 1 |
# | Flatten 2 | Input = 5x5x200, Output = 5000 | Max pooling 2 | Concatenate 1 |
# | Concatenate 1 | Input1 = 4900, Input1 = 5000, Output = 9900 | Max pooling 2 and 3 |Fully connected |
# | Fully connected | Fully Connected. Input = 9900, Output = 100 | Concatenate 1 | Dropout |
# | Dropout | Keep prob = 0.75 | Fully connected | Softmax |
# | Softmax | Fully Connected. Input = 100, Output = 43 | Dropout | Probabilities |
# + deletable=true editable=true
import tensorflow as tf
from tensorflow.contrib.layers import flatten
def model(x, keep_prob):
# Arguments used for tf.truncated_normal, randomly defines variables for the weights and biases for each layer
mu = 0
sigma = 0.1
# Network Parameters
n_classes = 10 # MNIST total classes (0-9 digits)
filter_size = 5
# Store layers weight & bias
weights = {
'wc1' : tf.Variable(tf.truncated_normal([filter_size, filter_size, 1, 100], mean = mu, stddev = sigma)),
'wc2' : tf.Variable(tf.truncated_normal([filter_size, filter_size, 100, 200], mean = mu, stddev = sigma)),
'wfc1': tf.Variable(tf.truncated_normal([9900, 100], mean = mu, stddev = sigma)),
'out' : tf.Variable(tf.truncated_normal([100, n_classes], mean = mu, stddev = sigma))}
biases = {
'bc1' : tf.Variable(tf.zeros([100])),
'bc2' : tf.Variable(tf.zeros([200])),
'bfc1': tf.Variable(tf.zeros([100])),
'out' : tf.Variable(tf.zeros([n_classes]))}
def conv2d(x, W, b, strides=1., padding='SAME'):
x = tf.nn.conv2d(x, W, strides=[1, strides, strides, 1], padding=padding)
x = tf.nn.bias_add(x, b)
return tf.nn.relu(x)
def maxpool2d(x, k=2, padding='SAME'):
return tf.nn.max_pool(x, ksize=[1, k, k, 1], strides=[1, k, k, 1], padding=padding)
# Layer 1: Convolution 1 - 32*32*1 to 28*28*100
conv1 = conv2d(x, weights['wc1'], biases['bc1'], padding='VALID')
# Max Pool - 28*28*100 to 14*14*100
conv1 = maxpool2d(conv1, k=2)
# Layer 2: Convolution 2 - 14*14*100 to 10*10*200
conv2 = conv2d(conv1, weights['wc2'], biases['bc2'], padding='VALID')
# Max Pool - 10*10*200 to 5*5*200
conv2 = maxpool2d(conv2, k=2)
#Fork second max pool - 14*14*100 to 7*7*100
conv1 = maxpool2d(conv1, k=2)
#Flatten conv1. Input = 7*7*100, Output = 4900
conv1 = tf.contrib.layers.flatten(conv1)
# Flatten conv2. Input = 5x5x200. Output = 5000.
conv2 = tf.contrib.layers.flatten(conv2)
# Concatenate
flat = tf.concat(1,[conv1,conv2])
# Layer 3 : Fully Connected. Input = 9900. Output = 100.
fc1 = tf.add(tf.matmul(flat, weights['wfc1']), biases['bfc1'])
fc1 = tf.nn.relu(fc1)
fc1 = tf.nn.dropout(fc1, keep_prob)
# Layer 4: Fully Connected. Input = 100. Output = n_classes.
logits = tf.add(tf.matmul(fc1, weights['out']), biases['out'])
return logits
# + [markdown] deletable=true editable=true
# ## 10. Train, Validate and Test the Model
# + [markdown] deletable=true editable=true
# A validation set can be used to assess how well the model is performing. A low accuracy on the training and validation
# sets imply underfitting. A high accuracy on the training set but low accuracy on the validation set implies overfitting.
# + deletable=true editable=true
### Train your model here.
### Calculate and report the accuracy on the training and validation set.
### Once a final model architecture is selected,
### the accuracy on the test set should be calculated and reported as well.
### Feel free to use as many code cells as needed.
#Hyperparameters
EPOCHS = 100 #Max EPOCH number, if ever early stopping doesn't kick in
BATCH_SIZE = 256 #Max batch size
rate = 0.001 #Base learning rate
keep_probability = 0.75 #Keep probability for dropout..
max_iter_wo_improvmnt = 3000 #For early stopping
# + [markdown] deletable=true editable=true
# ## 11. Features and Labels
#
# `x` is a placeholder for a batch of input images.
# `y` is a placeholder for a batch of output labels.
# + deletable=true editable=true
#Declare placeholder tensors
x = tf.placeholder(tf.float32, (None, 32, 32, 1))
y = tf.placeholder(tf.int32, (None))
keep_prob = tf.placeholder(tf.float32)
one_hot_y = tf.one_hot(y, n_classes)
# + [markdown] deletable=true editable=true
# ## 12. Training Pipeline
# Create a training pipeline that uses the model to classify German Traffic Sign Benchmarks data.
# + deletable=true editable=true
logits = model(x, keep_prob)
probabilities = tf.nn.softmax(logits)
cross_entropy = tf.nn.softmax_cross_entropy_with_logits(logits, one_hot_y)
loss_operation = tf.reduce_mean(cross_entropy)
optimizer = tf.train.AdamOptimizer(learning_rate = rate)
training_operation = optimizer.minimize(loss_operation)
# + [markdown] deletable=true editable=true
# ## 13. Model Evaluation
# Evaluate how well the loss and accuracy of the model for a given dataset.
# + deletable=true editable=true
correct_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(one_hot_y, 1))
accuracy_operation = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
saver = tf.train.Saver()
def evaluate(X_data, y_data):
num_examples = len(X_data)
total_accuracy = 0
sess = tf.get_default_session()
for offset in range(0, num_examples, BATCH_SIZE):
batch_x, batch_y = X_data[offset:offset+BATCH_SIZE], y_data[offset:offset+BATCH_SIZE]
accuracy = sess.run(accuracy_operation, feed_dict={x: batch_x, y: batch_y, keep_prob: 1.0})
total_accuracy += (accuracy * len(batch_x))
return total_accuracy / num_examples
# + [markdown] deletable=true editable=true
# ## 14. Train the Model
# Run the training data through the training pipeline to train the model.
#
# Before each epoch, shuffle the training set.
#
# After each epoch, measure the loss and accuracy of the validation set.
#
# Save the model after training.
# + deletable=true editable=true
from sklearn.utils import shuffle
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
num_examples = len(X_train)
# Max iteration number without improvement
max_interation_num_wo_improv = 1000
print("Training...")
iteration = 0
best_valid_accuracy = 0
best_accuracy_iter = 0
stop = 0
print()
for i in range(EPOCHS):
X_train, y_train = shuffle(X_train, y_train)
for offset in range(0, num_examples, BATCH_SIZE):
iteration = iteration + 1
end = offset + BATCH_SIZE
batch_x, batch_y = X_train[offset:end], y_train[offset:end]
sess.run(training_operation, feed_dict={x: batch_x, y: batch_y, keep_prob: keep_probability})
# After 10 Epochs, for every 200 iterations validation accuracy is checked
if (iteration % 200 == 0 and i > 10):
validation_accuracy = evaluate(X_valid, y_valid)
if validation_accuracy > best_valid_accuracy:
best_valid_accuracy = validation_accuracy
best_accuracy_iter = iteration
saver = tf.train.Saver()
saver.save(sess, './best_model')
print("Improvement found, model saved!")
stop = 0
# Stopping criteria : if not improvement since 1000 iterations stop training
if (iteration - best_accuracy_iter) > max_iter_wo_improvmnt:
print("Stopping criteria met..")
stop = 1
validation_accuracy = evaluate(X_valid, y_valid)
print("EPOCH {} ...".format(i+1))
print("Validation Accuracy = {:.3f}".format(validation_accuracy))
print()
if stop == 1:
break
# saver.save(sess, './lenet')
# print("Model saved")
# + [markdown] deletable=true editable=true
# ## 15. Evaluate accuracy of the different data sets
# + deletable=true editable=true
### Load the images and plot them here.
### Feel free to use as many code cells as needed.
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('.'))
print("Evaluating..")
train_accuracy = evaluate(X_train, y_train)
print("Train Accuracy = {:.3f}".format(train_accuracy))
valid_accuracy = evaluate(X_valid, y_valid)
print("Valid Accuracy = {:.3f}".format(valid_accuracy))
test_accuracy = evaluate(X_test, y_test)
print("Test Accuracy = {:.3f}".format(test_accuracy))
# + [markdown] deletable=true editable=true
# ---
#
# ## Step 3: Test a Model on New Images
#
# To give yourself more insight into how your model is working, download at least five pictures of German traffic signs from the web and use your model to predict the traffic sign type.
#
# You may find `signnames.csv` useful as it contains mappings from the class id (integer) to the actual sign name.
# + [markdown] deletable=true editable=true
# ## 16. Load and Show the Images
# + deletable=true editable=true
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
test_images = os.listdir('traffic-signs-data/web_found_signs/')
X_web = []
for file in test_images:
image = mpimg.imread('traffic-signs-data/web_found_signs/' + file)
plt.imshow(image)
plt.show()
print("Loaded ", file)
X_web.append(image)
X_web = np.array(X_web)
# Preprocess images
print('Preprocessing features...')
X_web = equalize_Y_histogram(X_web)
X_web = convert_to_grayscale(X_web)
X_web = normalize_grayscale(X_web)
X_web = X_web[..., newaxis]
print("Processed shape =", X_web.shape)
# + [markdown] deletable=true editable=true
# ## 17. Predict the Sign Type for Each Image
# + deletable=true editable=true
### Run the predictions here and use the model to output the prediction for each image.
### Make sure to pre-process the images with the same pre-processing pipeline used earlier.
### Feel free to use as many code cells as needed.
import tensorflow as tf
# hardcoded..
y_web = [9,22,2,18,1,17,4,10,38,4,4,23]
#We have to set the keep probability to 1.0 in the model..
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('.'))
logits_web = sess.run(tf.argmax(logits,1), feed_dict={x: X_web, keep_prob: 1.0})
print("Prediction =", logits_web)
# show_sample(X_web, logits_web, histogram = 0, sample_num = len(test_images), sample_index = 0, color_map = 'gray')
#Number of column to show
sample_num = len(test_images)
col_num = 4
if sample_num%col_num == 0:
row_num = int(sample_num/col_num)
else:
row_num = int(sample_num/col_num)+1
#Create training sample plot
f, axarr = plt.subplots(row_num, col_num, figsize=(col_num*4,(row_num+1)*2))
signnames = pd.read_csv('signnames.csv')
for i in range(0, sample_num, 1):
image = X_web[i].squeeze()
if logits_web[i] != y_web[i]:
color_str = 'red'
else:
color_str = 'green'
title_str = 'Predicted : %s \n Real: %s' % (signnames.iloc[logits_web[i], 1],signnames.iloc[y_web[i], 1])
axarr[int(i/col_num),i%col_num].set_title(title_str, color = color_str)
axarr[int(i/col_num),i%col_num].imshow(image,'gray')
axarr[int(i/col_num),i%col_num].axis('off')
axarr[int(i/col_num),i%col_num].axis('off')
axarr[int(i/col_num),i%col_num].axis('off')
f.tight_layout()
plt.show()
# + [markdown] deletable=true editable=true
# ## 18. Analyze Performance
# + deletable=true editable=true
### Calculate the accuracy for these 5 new images.
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('.'))
test_accuracy = evaluate(X_web, y_web)
print("Web images Accuracy = {:.3f}".format(test_accuracy))
# + [markdown] deletable=true editable=true
# ## 19. Output Top 5 Softmax Probabilities For Each Image Found on the Web
# + [markdown] deletable=true editable=true
# For each of the new images, print out the model's softmax probabilities to show the **certainty** of the model's predictions (limit the output to the top 5 probabilities for each image). [`tf.nn.top_k`](https://www.tensorflow.org/versions/r0.12/api_docs/python/nn.html#top_k) could prove helpful here.
#
# The example below demonstrates how tf.nn.top_k can be used to find the top k predictions for each image.
#
# `tf.nn.top_k` will return the values and indices (class ids) of the top k predictions. So if k=3, for each sign, it'll return the 3 largest probabilities (out of a possible 43) and the correspoding class ids.
#
# Take this numpy array as an example. The values in the array represent predictions. The array contains softmax probabilities for five candidate images with six possible classes. `tk.nn.top_k` is used to choose the three classes with the highest probability:
#
# ```
# # (5, 6) array
# a = np.array([[ 0.24879643, 0.07032244, 0.12641572, 0.34763842, 0.07893497,
# 0.12789202],
# [ 0.28086119, 0.27569815, 0.08594638, 0.0178669 , 0.18063401,
# 0.15899337],
# [ 0.26076848, 0.23664738, 0.08020603, 0.07001922, 0.1134371 ,
# 0.23892179],
# [ 0.11943333, 0.29198961, 0.02605103, 0.26234032, 0.1351348 ,
# 0.16505091],
# [ 0.09561176, 0.34396535, 0.0643941 , 0.16240774, 0.24206137,
# 0.09155967]])
# ```
#
# Running it through `sess.run(tf.nn.top_k(tf.constant(a), k=3))` produces:
#
# ```
# TopKV2(values=array([[ 0.34763842, 0.24879643, 0.12789202],
# [ 0.28086119, 0.27569815, 0.18063401],
# [ 0.26076848, 0.23892179, 0.23664738],
# [ 0.29198961, 0.26234032, 0.16505091],
# [ 0.34396535, 0.24206137, 0.16240774]]), indices=array([[3, 0, 5],
# [0, 1, 4],
# [0, 5, 1],
# [1, 3, 5],
# [1, 4, 3]], dtype=int32))
# ```
#
# Looking just at the first row we get `[ 0.34763842, 0.24879643, 0.12789202]`, you can confirm these are the 3 largest probabilities in `a`. You'll also notice `[3, 0, 5]` are the corresponding indices.
# + deletable=true editable=true
### Print out the top five softmax probabilities for the predictions on the German traffic sign images found on the web.
### Feel free to use as many code cells as needed.
import matplotlib.gridspec as gridspec
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('.'))
softmax_prob = sess.run(tf.nn.top_k(probabilities,k = 5), feed_dict={x: X_web, keep_prob: 1.0})
signnames = pd.read_csv('signnames.csv')
for i in range(len(test_images)):
plt.figure(figsize = (6,2))
gs = gridspec.GridSpec(1, 2,width_ratios=[2,3])
plt.subplot(gs[0])
plt.imshow(X_web[i].squeeze(),cmap="gray")
plt.axis('off')
plt.subplot(gs[1])
plt.barh(6-np.arange(5),softmax_prob[0][i], align='center')
if logits_web[i] != y_web[i]:
color_str = 'red'
else:
color_str = 'green'
for i_label in range(5):
temp_string = "%.1f %% : %s" % (softmax_prob[0][i][i_label]*100, str(signnames.iloc[softmax_prob[1][i][i_label], 1]))
plt.text(softmax_prob[0][i][0]*1.1,6-i_label-.15, temp_string, color = color_str)
plt.show()
# + [markdown] deletable=true editable=true
# ---
#
# ## Step 4: Visualize the Neural Network's State with Test Images
#
# This Section is not required to complete but acts as an additional excersise for understaning the output of a neural network's weights. While neural networks can be a great learning device they are often referred to as a black box. We can understand what the weights of a neural network look like better by plotting their feature maps. After successfully training your neural network you can see what it's feature maps look like by plotting the output of the network's weight layers in response to a test stimuli image. From these plotted feature maps, it's possible to see what characteristics of an image the network finds interesting. For a sign, maybe the inner network feature maps react with high activation to the sign's boundary outline or to the contrast in the sign's painted symbol.
#
# Provided for you below is the function code that allows you to get the visualization output of any tensorflow weight layer you want. The inputs to the function should be a stimuli image, one used during training or a new one you provided, and then the tensorflow variable name that represents the layer's state during the training process, for instance if you wanted to see what the [LeNet lab's](https://classroom.udacity.com/nanodegrees/nd013/parts/fbf77062-5703-404e-b60c-95b78b2f3f9e/modules/6df7ae49-c61c-4bb2-a23e-6527e69209ec/lessons/601ae704-1035-4287-8b11-e2c2716217ad/concepts/d4aca031-508f-4e0b-b493-e7b706120f81) feature maps looked like for it's second convolutional layer you could enter conv2 as the tf_activation variable.
#
# For an example of what feature map outputs look like, check out NVIDIA's results in their paper [End-to-End Deep Learning for Self-Driving Cars](https://devblogs.nvidia.com/parallelforall/deep-learning-self-driving-cars/) in the section Visualization of internal CNN State. NVIDIA was able to show that their network's inner weights had high activations to road boundary lines by comparing feature maps from an image with a clear path to one without. Try experimenting with a similar test to show that your trained network's weights are looking for interesting features, whether it's looking at differences in feature maps from images with or without a sign, or even what feature maps look like in a trained network vs a completely untrained one on the same sign image.
#
# <figure>
# <img src="visualize_cnn.png" width="380" alt="Combined Image" />
# <figcaption>
# <p></p>
# <p style="text-align: center;"> Your output should look something like this (above)</p>
# </figcaption>
# </figure>
# <p></p>
#
# + deletable=true editable=true
### Visualize your network's feature maps here.
### Feel free to use as many code cells as needed.
# image_input: the test image being fed into the network to produce the feature maps
# tf_activation: should be a tf variable name used during your training procedure that represents the calculated state of a specific weight layer
# activation_min/max: can be used to view the activation contrast in more detail, by default matplot sets min and max to the actual min and max values of the output
# plt_num: used to plot out multiple different weight feature map sets on the same block, just extend the plt number for each new feature map entry
def outputFeatureMap(image_input, tf_activation, activation_min=-1, activation_max=-1 ,plt_num=1):
# Here make sure to preprocess your image_input in a way your network expects
# with size, normalization, ect if needed
# image_input =
# Note: x should be the same name as your network's tensorflow data placeholder variable
# If you get an error tf_activation is not defined it maybe having trouble accessing the variable from inside a function
activation = tf_activation.eval(session=sess,feed_dict={x : image_input})
featuremaps = activation.shape[3]
plt.figure(plt_num, figsize=(15,15))
for featuremap in range(featuremaps):
plt.subplot(6,8, featuremap+1) # sets the number of feature maps to show on each row and column
plt.title('FeatureMap ' + str(featuremap)) # displays the feature map number
if activation_min != -1 & activation_max != -1:
plt.imshow(activation[0,:,:, featuremap], interpolation="nearest", vmin =activation_min, vmax=activation_max, cmap="gray")
elif activation_max != -1:
plt.imshow(activation[0,:,:, featuremap], interpolation="nearest", vmax=activation_max, cmap="gray")
elif activation_min !=-1:
plt.imshow(activation[0,:,:, featuremap], interpolation="nearest", vmin=activation_min, cmap="gray")
else:
plt.imshow(activation[0,:,:, featuremap], interpolation="nearest", cmap="gray")
# + [markdown] deletable=true editable=true
# ### Question 9
#
# Discuss how you used the visual output of your trained network's feature maps to show that it had learned to look for interesting characteristics in traffic sign images
#
# + [markdown] deletable=true editable=true
# **Answer:**
# + [markdown] deletable=true editable=true
# > **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \n",
# "**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
# + [markdown] deletable=true editable=true
# ### Project Writeup
#
# Once you have completed the code implementation, document your results in a project writeup using this [template](https://github.com/udacity/CarND-Traffic-Sign-Classifier-Project/blob/master/writeup_template.md) as a guide. The writeup can be in a markdown or pdf file.
| CarND-Traffic-Sign-Classifier-P2/Traffic_Sign_ClassifierCIfar10.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Using autograd to calculate the gradient of a log-likelihood
#
# It is straightforward to use the automatic differentiation library [autograd](https://github.com/HIPS/autograd) to take the derivative of log-likelihoods defined in pints. Below is an example of how to do this.
#
# WARNING: We currently find this method of caculating model sensitivities to be quite slow for most time-series models, and so do not recommended it for use.
# +
import matplotlib.pyplot as plt
import pints
import pints.toy as toy
import numpy as np
import warnings
try:
import autograd.numpy as np
from autograd.scipy.integrate import odeint
from autograd.builtins import tuple
from autograd import grad
except ImportError:
print("""This example requires autograd, which is not a pints dependency.
If you see this warning, try `pip install autograd`""")
exit(0)
from timeit import repeat
# -
# We begin be defining a model, identical to the [Fitzhugh Nagumo](https://pints.readthedocs.io/en/latest/toy/fitzhugh_nagumo_model.html) toy model implemented in pints. The corresponding toy model in pints has its `evaluateS1()` method defined, so we can compare the results using automatic differentiation.
class AutoGradFitzhughNagumoModel(pints.ForwardModel):
def simulate(self, parameters, times):
y0 = np.array([-1, 1], dtype=float)
def rhs(y, t, p):
V, R = y
a, b, c = p
dV_dt = (V - V**3 / 3 + R) * c
dR_dt = (V - a + b * R) / -c
return np.array([dV_dt, dR_dt])
return odeint(rhs, y0, times, tuple((parameters,)))
def n_parameters(self):
return 3
def n_outputs(self):
return 2
# Now we wrap an existing pints likelihood class, and use the `autograd.grad` function to calculate the gradient of the given log-likelihood
# +
class AutoGradLogLikelihood(pints.ProblemLogLikelihood):
def __init__(self, likelihood):
self.likelihood = likelihood
f = lambda x: self.likelihood(x)
self.likelihood_grad = grad(f)
def __call__(self, x):
return self.likelihood(x)
def evaluateS1(self, x):
values = self.likelihood(x)
gradient = self.likelihood_grad(x)
return values, gradient
def n_parameters(self):
return self.likelihood.n_parameters()
autograd_model = AutoGradFitzhughNagumoModel()
pints_model = pints.toy.FitzhughNagumoModel()
# -
# Now create some toy data and ensure that the new model gives the same output as the toy model in pints
# +
# Create some toy data
real_parameters = np.array(pints_model.suggested_parameters(), dtype='float64')
times = pints_model.suggested_times()
pints_values = pints_model.simulate(real_parameters, times)
autograd_values = autograd_model.simulate(real_parameters, times)
plt.figure()
plt.plot(times, autograd_values)
plt.plot(times, pints_values)
plt.show()
# -
# Add some noise to the values, and then create log-likelihoods using both the new model, and the pints model
# +
noise = 0.1
values = pints_values + np.random.normal(0, noise, pints_values.shape)
# Create an object with links to the model and time series
autograd_problem = pints.MultiOutputProblem(autograd_model, times, values)
pints_problem = pints.MultiOutputProblem(pints_model, times, values)
# Create a log-likelihood function
autograd_log_likelihood = pints.GaussianKnownSigmaLogLikelihood(autograd_problem, noise)
autograd_likelihood = AutoGradLogLikelihood(autograd_log_likelihood)
pints_log_likelihood = pints.GaussianKnownSigmaLogLikelihood(pints_problem, noise)
# -
# We can calculate the gradients of both likelihood functions at the given parameters to make sure that they are the same
autograd_likelihood.evaluateS1(real_parameters)
pints_log_likelihood.evaluateS1(real_parameters)
# Now we'll time both functions. You can see that the function using `autgrad` is significantly slower than the in-built `evaluateS1` function for the pints model. For reference, this function uses forward-mode sensitivity calculation using the symbolic Jacobian of the model.
# +
statement = 'autograd_likelihood.evaluateS1(real_parameters)'
setup = 'from __main__ import autograd_likelihood, real_parameters'
time_taken = min(repeat(stmt=statement, setup=setup, number=1, repeat=5))
'Elapsed time: {:.0f} ms'.format(1000. * time_taken)
# +
statement = 'pints_log_likelihood.evaluateS1(real_parameters)'
setup = 'from __main__ import pints_log_likelihood, real_parameters'
time_taken = min(repeat(stmt=statement, setup=setup, number=1, repeat=5))
'Elapsed time: {:.0f} ms'.format(1000. * time_taken)
| examples/toy/automatic-differentiation-using-autograd.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Universal concepts, preamble & glossary
#
# ## Types of Machine Learning
# There are three fundamental types of problems that machine learning algorithms are trying to solve.
#
# ### Supervised
# This is where we have some examples of a given input and its output and we are trying to model that function based on the data.
#
# #### Regression
# The output of our model is a number or vector of numbers which can take any real value. An example is trying to predict the price of a house based on a number of features such as number of rooms, number of windows, etc.
#
#
# #### Classification
# The output of our model, based on the inputs, is the probability of the input belonging to a class. An example is having an input image and outputting if it is a hotdog or not.
#
# ### Unsupervised
# There is no input-output relationship which we are trying to model. Rather, we want to find some hidden structure in the data. One type is clustering in which you are trying to group your data. A use case could be segmenting your customers by their interests so you can target them with different material.
#
# ### Reinforcement Learning
# In reinforcement learning, we have an agent, which takes in observations from an environment and takes actions based on those observations to maximize a reward function. This is inspired by pavlovian conditioning which has been shown to be the method that mammals learn by.
# ## Datapoint
#
# Here a single data point, $x^{(1)}$ is represented as a row vector where each column is a different **feature**.
#
# For example, if each training example is a house, then its vector of features may include elements for its price, no. rooms, no. windows etc.
#
# ### $X^{(1)} = \begin{bmatrix} x^{(1)}_1 & x^{(1)}_2 & \dots & x^{(1)}_{n-1}& x^{(1)}_n \end{bmatrix}$
#
# ## Design Matrix
# The **design matrix**, **X** contains all of our training data. Each row represents a certain example. There are $m$ training examples. Each row represents a different feature. There are $n$ features. Hence the design matrix has dimensions of $n$ by $m$.
#
# ### $Design \ matrix,\ X = \begin{bmatrix} \dots & x^{(1)} &\dots \\ & \vdots & \\ \dots & x^{(m)} & \dots \end{bmatrix} = \begin{bmatrix} x_{11} \dots x_{1n} \\ \vdots \ddots \vdots \\ x_{m1} \dots x_{mn} \end{bmatrix} \in m \times n$
# ## Hypothesis
# The hypothesis, $h$ is the output of your model. It is your current prediction of the mapping from input to output.
# ## Loss/cost function
#
#
# For our algorithms to learn, we need a way to evaluate their current performance, so that we can determine how to improve. We can mathematically define when our algorithm is performing well by evaluating an appropriate objective function. We usually try to minimise a function which indicates the error in our hypothesis (how bad our model is). We will represent the loss of our models with the symbol $J$. The cost function is dependent on as many dimensions as we have parameters (which are relevant to that loss function). Changing these parameters moves us around parameter space, in which the cost varies. Varying different parameters will have varying influence on how the cost changes - as such, some are more important to optimise.
#
# #### Mean Squared Error Loss
# MSE loss is the average over all training points of the squared error between your hypothesis and the label. The factor of $\frac{1}{2}$ is often included to cancel with the power of 2 when differentiated so that no constants are present.
#
# ### $ J =\frac{1}{2m} \sum_{i=1}^{m}(h^{(i)} - y^{(i)})^2$
#
# #### Binary Cross Entropy loss
# BCE loss is used to calculate error for classification tasks.
#
# ### $ J = \sum_{i=1}^{m} - y^{(i)} \cdot \text{log}(h^{(i)}) + (1-y^{(i)}) \cdot \text{log}(1-h^{(i)})$
#
# In classification tasks, for each class the label of a datapoint can only take binary values of 0 or 1; i.e. it *is* a member of that class or it *is not* a member of that class, and the output is usually a *confidence* value $\in [0, 1]$.
# When , $y, = 0$ the first term is 'turned off' and the second term
#
# #### Kullback-Leibler Divergence
# The KL divergence is a metric that quantifies the difference between two probability distributions, $p$ & $q$. It is used frequently in machine learning to measure the information lost when we try to represent a probability distribution in a different way (e.g. after reconstructing it from an encoding).
#
# ### $D_{KL}(p||q) = \sum_{i=1}^{m} p(x_i)\cdot (\text{log }p(x_i) - \text{log }q(x_i)) = \sum_{i=1}^{m} p(x_i)\cdot \text{log } \frac{p(x_i)}{q(x_i)}$
#
# ##### Practically, for a normal distribution, the KL-divergence can be evaluated as : $D_{KL} = \sum_i (\sigma^2 + $
#
# For a single datapoint, $x$, the KL divergence tests how similar the log probabilities of that value are and weights that difference by the value of the probability of sampling that $x$ from $p(x)$. The weighting $p(x)$ of the log difference makes the KL divergence different depending on which arrangement you compare the probability distributions in.
#
# Consider:
# - It takes large values when the sampled probabilities for the same values are more different, and the weighting probability distribution $p(x)$ is larger.
# - It takes a value of zero where the weighting probability distribution is zero.
# - Where the
# - The aim is often to minimise the KL divergence (the information difference between two probability distributions).
# ## Gradient Descent and the learning rate
# Gradient descent is the most popular optimization strategy currently used in machine learning. It has proven to be very effective even when there are millions of parameters to optimize as in the case of deep learning.<br>
# Lets say we have a function
# ### $J = f(x, y;\theta)$
# which we are trying to minimize by finding optimal values of $\theta$.<br>
#
# Gradient descent works by moving the weights that control a model in a direction that most decreases the cost. What is this direction? The gradient of a function at a point is a vector pointing in the direction which it increases fastest locally. So the direction which most *decreases* the cost function, is the negative gradient - in this case, it will be an $n+1$ (features and bias) dimensional vector containing the partial derivatives of the cost with respect to each of these model parameters.
#
# The negative gradient tells us the correct direction to move each weight in, but not the ideal size of the step.
#
# If we move the parameters by the value of the negative gradient, there is a chance that they may jump straight over the minima, perhaps to a point where the gradient is even higher! This can happen because the gradient can be greater than the distance of the parameter from its optimal position. This causes divergence, of the model parameters, instead of convergence.
# So, in gradient descent, we iteratively update the weights *proportionally* to the negative gradient at their local position. This proportionality constant, which the gradient is multiplied by to get the step size, is called the **learning rate**. The learning rate should be large enough so that the algorithm converges at a suitable rate, but small enough enough to ensure that it does not diverge.
#
# At a minima, the parameter should stabilise because the step size is proportional to the gradient, which will be zero.
#
# We can utilize the gradient descent strategy only if $J$ is a differentiable function.<br>
# We first start by initializing $\theta$ randomly. We then calculate $J$ and the derivative of $J$ w.r.t $\theta$.
# Once we have $\frac{\partial J}{\partial\theta}$, we update $\theta$ using the following update rule:
# ### $\theta := \theta - \alpha\frac{\partial J}{\partial\theta}$
#
# While it is important to understand the equations, it is equally important to have an intuitive understanding of what is going on. What we are doing when we calulate the partial derivative of $J$ w.r.t $\theta_i$ is we are finding out how a small increase in $\theta_i$ affects $J$. If this leads to an increase in $J$ we decrease our $\theta_i$ as we are trying to reduce $J$. If it leads to a decrease in $J$, we increase our $\theta_i$. This explains the negative sign in the update rule. <br>
#
# This can easily be visualised in the case where we have two parameters. We have a surface and we are trying to find the lowest point on this surface which corresponds to the lowest value of $J$. We start at a random point on the surface and interatively calculate the direction of greatest ascent ($\frac{\partial J}{\partial\theta}$) before taking a step in the opposite direction. We scale our steps by a factor of $\alpha$. We can set different values for $\alpha$ at runtime and we will get different results. If our $\alpha$ is too high, we will overshoot the minima and if it is too low, we will take too long to get to the minima.
# 
#
#
#
# ### Pseudo code for SGD
# #### Randomly select a batch of datapoints from the training set to train on.
# #### Make a prediction of the output for those
# #### Evaluate the model's cost for these predictions
# #### Find rate of change of cost wrt model parameters
# #### Update the model parameters proportionately to the negative gradients found above, according to:
#
# ### $ \theta\ \dot{=}\ \theta - \alpha \frac{\partial J}{\partial \theta}$
#
# #### Repeat for defined number of epochs
# ## Embedding
#
# #### One-hot
#
# If we have a classification problem, instead of having a word as a label, we can have a $K$-dimensional vector, where $K$ is the number of classes, and each element of that vector is zero except for one element that represents the true class label. This is a one-hot encoding. This is a way of representing a label numberically, using the same number of elements as there are classes.
#
# #### Embedding
#
# For one-hot encoding, each different possible label is a mutually orthogonal unit vector. All possible class labels make up a $K$-dimensional basis of vectors. This means that to be able to reach the whole range of our output space, we need a $K$- dimensional output from our model.
# Alternatively, class vectors can be embedded into a lower dimensional subspace, where less than $K$-dimensional vectors can be the output of our model. This embedding is not binary (discrete), and now has a continuous range not limited within the range $[0, 1]$.
#
# Imagine you are training a model to predict the next word in a sentance. You don't want your ouput space to have as many dimensions as you have predictable words in your corpus. So instead, you can embed these words into a low dimensional subspace where each of them is a vector. Similar words will be closer to each other and vector algebra can be done on these vectors.
#
#
# 
| Universal concepts, preamble & glossary.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Linear algebra in Python with NumPy
#
# In this lab, you will have the opportunity to remember some basic concepts about linear algebra and how to use them in Python.
#
# Numpy is one of the most used libraries in Python for arrays manipulation. It adds to Python a set of functions that allows us to operate on large multidimensional arrays with just a few lines. So forget about writing nested loops for adding matrices! With NumPy, this is as simple as adding numbers.
#
# Let us import the `numpy` library and assign the alias `np` for it. We will follow this convention in almost every notebook in this course, and you'll see this in many resources outside this course as well.
import numpy as np # The swiss knife of the data scientist.
# ## Defining lists and numpy arrays
alist = [1, 2, 3, 4, 5] # Define a python list. It looks like an np array
narray = np.array([1, 2, 3, 4]) # Define a numpy array
# Note the difference between a Python list and a NumPy array.
# +
print(alist)
print(narray)
print(type(alist))
print(type(narray))
# -
# ## Algebraic operators on NumPy arrays vs. Python lists
#
# One of the common beginner mistakes is to mix up the concepts of NumPy arrays and Python lists. Just observe the next example, where we add two objects of the two mentioned types. Note that the '+' operator on NumPy arrays perform an element-wise addition, while the same operation on Python lists results in a list concatenation. Be careful while coding. Knowing this can save many headaches.
print(narray + narray)
print(alist + alist)
# It is the same as with the product operator, `*`. In the first case, we scale the vector, while in the second case, we concatenate three times the same list.
print(narray * 3)
print(alist * 3)
# Be aware of the difference because, within the same function, both types of arrays can appear.
# Numpy arrays are designed for numerical and matrix operations, while lists are for more general purposes.
# ## Matrix or Array of Arrays
#
# In linear algebra, a matrix is a structure composed of n rows by m columns. That means each row must have the same number of columns. With NumPy, we have two ways to create a matrix:
# * Creating an array of arrays using `np.array` (recommended).
# * Creating a matrix using `np.matrix` (still available but might be removed soon).
#
# NumPy arrays or lists can be used to initialize a matrix, but the resulting matrix will be composed of NumPy arrays only.
# +
npmatrix1 = np.array([narray, narray, narray]) # Matrix initialized with NumPy arrays
npmatrix2 = np.array([alist, alist, alist]) # Matrix initialized with lists
npmatrix3 = np.array([narray, [1, 1, 1, 1], narray]) # Matrix initialized with both types
print(npmatrix1)
print(npmatrix2)
print(npmatrix3)
# -
# However, when defining a matrix, be sure that all the rows contain the same number of elements. Otherwise, the linear algebra operations could lead to unexpected results.
#
# Analyze the following two examples:
# +
# Example 1:
okmatrix = np.array([[1, 2], [3, 4]]) # Define a 2x2 matrix
print(okmatrix) # Print okmatrix
print(okmatrix * 2) # Print a scaled version of okmatrix
# +
# Example 2:
badmatrix = np.array([[1, 2], [3, 4], [5, 6, 7]]) # Define a matrix. Note the third row contains 3 elements
print(badmatrix) # Print the malformed matrix
print(badmatrix * 2) # It is supposed to scale the whole matrix
# -
# ## Scaling and translating matrices
#
# Now that you know how to build correct NumPy arrays and matrices, let us see how easy it is to operate with them in Python using the regular algebraic operators like + and -.
#
# Operations can be performed between arrays and arrays or between arrays and scalars.
# Scale by 2 and translate 1 unit the matrix
result = okmatrix * 2 + 1 # For each element in the matrix, multiply by 2 and add 1
print(result)
# +
# Add two sum compatible matrices
result1 = okmatrix + okmatrix
print(result1)
# Subtract two sum compatible matrices. This is called the difference vector
result2 = okmatrix - okmatrix
print(result2)
# -
# The product operator `*` when used on arrays or matrices indicates element-wise multiplications.
# Do not confuse it with the dot product.
result = okmatrix * okmatrix # Multiply each element by itself
print(result)
# ## Transpose a matrix
#
# In linear algebra, the transpose of a matrix is an operator that flips a matrix over its diagonal, i.e., the transpose operator switches the row and column indices of the matrix producing another matrix. If the original matrix dimension is n by m, the resulting transposed matrix will be m by n.
#
# **T** denotes the transpose operations with NumPy matrices.
matrix3x2 = np.array([[1, 2], [3, 4], [5, 6]]) # Define a 3x2 matrix
print('Original matrix 3 x 2')
print(matrix3x2)
print('Transposed matrix 2 x 3')
print(matrix3x2.T)
# However, note that the transpose operation does not affect 1D arrays.
nparray = np.array([1, 2, 3, 4]) # Define an array
print('Original array')
print(nparray)
print('Transposed array')
print(nparray.T)
# perhaps in this case you wanted to do:
nparray = np.array([[1, 2, 3, 4]]) # Define a 1 x 4 matrix. Note the 2 level of square brackets
print('Original array')
print(nparray)
print('Transposed array')
print(nparray.T)
# ## Get the norm of a nparray or matrix
#
# In linear algebra, the norm of an n-dimensional vector $\vec a$ is defined as:
#
# $$ norm(\vec a) = ||\vec a|| = \sqrt {\sum_{i=1}^{n} a_i ^ 2}$$
#
# Calculating the norm of vector or even of a matrix is a general operation when dealing with data. Numpy has a set of functions for linear algebra in the subpackage **linalg**, including the **norm** function. Let us see how to get the norm a given array or matrix:
# +
nparray1 = np.array([1, 2, 3, 4]) # Define an array
norm1 = np.linalg.norm(nparray1)
nparray2 = np.array([[1, 2], [3, 4]]) # Define a 2 x 2 matrix. Note the 2 level of square brackets
norm2 = np.linalg.norm(nparray2)
print(norm1)
print(norm2)
# -
# Note that without any other parameter, the norm function treats the matrix as being just an array of numbers.
# However, it is possible to get the norm by rows or by columns. The **axis** parameter controls the form of the operation:
# * **axis=0** means get the norm of each column
# * **axis=1** means get the norm of each row.
# +
nparray2 = np.array([[1, 1], [2, 2], [3, 3]]) # Define a 3 x 2 matrix.
normByCols = np.linalg.norm(nparray2, axis=0) # Get the norm for each column. Returns 2 elements
normByRows = np.linalg.norm(nparray2, axis=1) # get the norm for each row. Returns 3 elements
print(normByCols)
print(normByRows)
# -
# However, there are more ways to get the norm of a matrix in Python.
# For that, let us see all the different ways of defining the dot product between 2 arrays.
# ## The dot product between arrays: All the flavors
#
# The dot product or scalar product or inner product between two vectors $\vec a$ and $\vec b$ of the same size is defined as:
# $$\vec a \cdot \vec b = \sum_{i=1}^{n} a_i b_i$$
#
# The dot product takes two vectors and returns a single number.
# +
nparray1 = np.array([0, 1, 2, 3]) # Define an array
nparray2 = np.array([4, 5, 6, 7]) # Define an array
flavor1 = np.dot(nparray1, nparray2) # Recommended way
print(flavor1)
flavor2 = np.sum(nparray1 * nparray2) # Ok way
print(flavor2)
flavor3 = nparray1 @ nparray2 # Geeks way
print(flavor3)
# As you never should do: # Noobs way
flavor4 = 0
for a, b in zip(nparray1, nparray2):
flavor4 += a * b
print(flavor4)
# -
# **We strongly recommend using np.dot, since it is the only method that accepts arrays and lists without problems**
# +
norm1 = np.dot(np.array([1, 2]), np.array([3, 4])) # Dot product on nparrays
norm2 = np.dot([1, 2], [3, 4]) # Dot product on python lists
print(norm1, '=', norm2 )
# -
# Finally, note that the norm is the square root of the dot product of the vector with itself. That gives many options to write that function:
#
# $$ norm(\vec a) = ||\vec a|| = \sqrt {\sum_{i=1}^{n} a_i ^ 2} = \sqrt {a \cdot a}$$
#
# ## Sums by rows or columns
#
# Another general operation performed on matrices is the sum by rows or columns.
# Just as we did for the function norm, the **axis** parameter controls the form of the operation:
# * **axis=0** means to sum the elements of each column together.
# * **axis=1** means to sum the elements of each row together.
# +
nparray2 = np.array([[1, -1], [2, -2], [3, -3]]) # Define a 3 x 2 matrix.
sumByCols = np.sum(nparray2, axis=0) # Get the sum for each column. Returns 2 elements
sumByRows = np.sum(nparray2, axis=1) # get the sum for each row. Returns 3 elements
np.sum()
print('Sum by columns: ')
print(sumByCols)
print('Sum by rows:')
print(sumByRows)
# -
# ## Get the mean by rows or columns
#
# As with the sums, one can get the **mean** by rows or columns using the **axis** parameter. Just remember that the mean is the sum of the elements divided by the length of the vector
# $$ mean(\vec a) = \frac {{\sum_{i=1}^{n} a_i }}{n}$$
# +
nparray2 = np.array([[1, -1], [2, -2], [3, -3]]) # Define a 3 x 2 matrix. Chosen to be a matrix with 0 mean
mean = np.mean(nparray2) # Get the mean for the whole matrix
meanByCols = np.mean(nparray2, axis=0) # Get the mean for each column. Returns 2 elements
meanByRows = np.mean(nparray2, axis=1) # get the mean for each row. Returns 3 elements
print('Matrix mean: ')
print(mean)
print('Mean by columns: ')
print(meanByCols)
print('Mean by rows:')
print(meanByRows)
# -
# ## Center the columns of a matrix
#
# Centering the attributes of a data matrix is another essential preprocessing step. Centering a matrix means to remove the column mean to each element inside the column. The sum by columns of a centered matrix is always 0.
#
# With NumPy, this process is as simple as this:
# +
nparray2 = np.array([[1, 1], [2, 2], [3, 3]]) # Define a 3 x 2 matrix.
nparrayCentered = nparray2 - np.mean(nparray2, axis=0) # Remove the mean for each column
print('Original matrix')
print(nparray2)
print('Centered by columns matrix')
print(nparrayCentered)
print('New mean by column')
print(nparrayCentered.mean(axis=0))
# -
# **Warning:** This process does not apply for row centering. In such cases, consider transposing the matrix, centering by columns, and then transpose back the result.
#
# See the example below:
# +
nparray2 = np.array([[1, 3], [2, 4], [3, 5]]) # Define a 3 x 2 matrix.
nparrayCentered = nparray2.T - np.mean(nparray2, axis=1) # Remove the mean for each row
nparrayCentered = nparrayCentered.T # Transpose back the result
print('Original matrix')
print(nparray2)
print('Centered by columns matrix')
print(nparrayCentered)
print('New mean by rows')
print(nparrayCentered.mean(axis=1))
# -
# Note that some operations can be performed using static functions like `np.sum()` or `np.mean()`, or by using the inner functions of the array
# +
nparray2 = np.array([[1, 3], [2, 4], [3, 5]]) # Define a 3 x 2 matrix.
mean1 = np.mean(nparray2) # Static way
mean2 = nparray2.mean() # Dinamic way
print(mean1, ' == ', mean2)
# -
# Even if they are equivalent, we recommend the use of the static way always.
#
# **Congratulations! You have successfully reviewed vector and matrix operations with Numpy!**
| NLP_C1_W3_lecture_nb_01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: xaitools
# language: python
# name: xaitools
# ---
# # ASE2021 Hands-on Exercise
#
# Below are interactive hands-on exercises for model-agnostic techniques for generating local explanations.
# First, we need to load necesarry libraries as well as preparing datasets.
#
# +
## Load Data and preparing datasets
# Import for Load Data
from os import listdir
from os.path import isfile, join
import pandas as pd
# Import for Split Data into Training and Testing Samples
from sklearn.model_selection import train_test_split
train_dataset = pd.read_csv(("../../datasets/lucene-2.9.0.csv"), index_col = 'File')
test_dataset = pd.read_csv(("../../datasets/lucene-3.0.0.csv"), index_col = 'File')
outcome = 'RealBug'
features = ['OWN_COMMIT', 'Added_lines', 'CountClassCoupled', 'AvgLine', 'RatioCommentToCode']
# commits - # of commits that modify the file of interest
# Added lines - # of added lines of code
# Count class coupled - # of classes that interact or couple with the class of interest
# LOC - # of lines of code
# RatioCommentToCode - The ratio of lines of comments to lines of code
# process outcome to 0 and 1
train_dataset[outcome] = pd.Categorical(train_dataset[outcome])
train_dataset[outcome] = train_dataset[outcome].cat.codes
test_dataset[outcome] = pd.Categorical(test_dataset[outcome])
test_dataset[outcome] = test_dataset[outcome].cat.codes
X_train = train_dataset.loc[:, features]
X_test = test_dataset.loc[:, features]
y_train = train_dataset.loc[:, outcome]
y_test = test_dataset.loc[:, outcome]
class_labels = ['Clean', 'Defective']
X_train.columns = features
X_test.columns = features
training_data = pd.concat([X_train, y_train], axis=1)
testing_data = pd.concat([X_test, y_test], axis=1)
# -
# Then, we construct a Random Forests model as a predictive model to be explained.
#
# **(1) Please construct a Random Forests model using the code cell below.**
#
#
# `````{admonition} Tips
# :class: tip
# ````
#
# our_rf_model = RandomForestClassifier(random_state=0)
# our_rf_model.fit(X_train, y_train)
#
# ````
# `````
# +
from sklearn.ensemble import RandomForestClassifier
# Please fit your Random Forests model here!
# -
# ## LIME
#
# **LIME** (i.e., Local Interpretable Model-agnostic
# Explanations) {cite}`ribeiro2016should` is a model-agnostic technique that
# mimics the behaviour of the black-box model to generate the explanations
# of the predictions of the black-box model. Given a black-box model and
# an instance to explain, LIME performs 4 key steps to generate an
# instance explanation as follows:
#
# - First, LIME randomly generates instances surrounding the instance of
# interest.
#
# - Second, LIME uses the black-box model to generate predictions of the
# generated random instances.
#
# - Third, LIME constructs a local regression model using the generated
# random instances and their generated predictions from the black-box
# model.
#
# - Finally, the coefficients of the regression model indicate the
# contribution of each metric on the prediction of the instance of
# interest according to the black-box model.
#
# **(2) Please use LIME to explain the prediction of *DocumentsWriter.java* that is generated from your Random Forests model.**
#
# `````{admonition} Tips
# :class: tip
# ````
#
# # LIME Step 1 - Construct an explainer
# our_lime_explainer = lime.lime_tabular.LimeTabularExplainer(
# training_data = X_train.values,
# mode = 'classification',
# training_labels = y_train,
# feature_names = features,
# class_names = class_labels,
# discretize_continuous = True)
#
# # LIME Step 2 - Use the constructed explainer with the predict function
# # of your predictive model to explain any instance
# lime_local_explanation_of_an_instance = lime_explainer.explain_instance(
# data_row = X_test.loc['FileName.py', :],
# predict_fn = our_rf_model.predict_proba,
# num_features = 5,
# top_labels = 1)
#
# # Please use the code below to visualise the generated LIME explanation.
# lime_local_explanation_of_an_instance.show_in_notebook()
#
# ````
# `````
# + tags=[]
# Import for LIME
import lime
import lime.lime_tabular
file_to_be_explained = 'src/java/org/apache/lucene/index/DocumentsWriter.java'
print(f'Explaining {file_to_be_explained} with LIME')
# LIME Step 1 - Construct an explainer
# LIME Step 2 - Use the constructed explainer with the predict function of your predictive model to explain any instance
# visualise the generated LIME explanation
# -
# ## SHAP
#
# **SHAP** (Shapley values) {cite}`lundberg2018consistentshap` is a model-agnostic technique that generate the explanations of the black-box model based on game theory.
#
#
# **(2) Please use LIME to explain the prediction of *DocumentsWriter.java* that is generated from your Random Forests model.**
#
# `````{admonition} Tips
# :class: tip
# ````
#
# # SHAP Step 1 - Construct an explainer with the predict function
# # of your predictive model
# our_shap_explainer = shap.KernelExplainer(our_rf_model.predict, X_test)
#
# # SHAP Step 2 - Generate the SHAP explanation of an instance to be explained
# shap_explanations_of_an_instance = our_shap_explainer.shap_values(X_test.iloc[file_to_be_explained_idx, :])
#
# # Please use the code below to visualise the generated SHAP explanation (Force plot).
# shap.initjs()
# shap.force_plot(our_shap_explainer.expected_value,
# shap_explanations_of_instances,
# X_test.iloc[file_to_be_explained_idx,:])
#
# ````
# `````
# + tags=[]
# Import libraries for SHAP
import subprocess
import sys
import importlib
import numpy
import shap
file_to_be_explained = 'src/java/org/apache/lucene/index/DocumentsWriter.java'
file_to_be_explained_idx = list(X_test.index).index(file_to_be_explained)
# SHAP Step 1 - Construct an explainer with the predict function
# SHAP Step 2 - Generate the SHAP explanation of an instance to be explained
# visualise the generated SHAP explanation
# -
# ## PyExplainer
#
# **PyExplainer** {cite}`pornprasit2021pyexplainer` is a rule-based model-agnostic technique that utilises a local rule-based regression model to learn the associations between the characteristics of the synthetic instances and the predictions from the black-box model. Given a black-box model and an instance to explain, PyExplainer performs four key steps to generate an instance explanation as follows:
#
# - First, PyExplainer generates synthetic neighbors around the instance to be explained using the crossover
# and mutation techniques
#
# - Second, PyExplainer obtains the predictions of the synthetic neighbors from the black-box model
#
# - Third, PyExplainer builds a local rule-based regression model
#
# - Finally, PyExplainer generates an explanation from the local model for the instance to be explained
#
# **(3) Please use PyExplainer to explain the prediction of *DocumentsWriter.java* that is generated from your Random Forests model.**
#
# `````{admonition} Tips
# :class: tip
# ````
# import numpy as np
# np.random.seed(0)
#
# # PyExplainer Step 1 - Construct a PyExplainer
# our_pyexplainer = PyExplainer(X_train = X_train,
# y_train = y_train,
# indep = X_train.columns,
# dep = outcome,
# blackbox_model = rf_model)
#
# # PyExplainer Step 2 - Generate the rule-based explanation of an instance to be explained
# pyexplainer_explanation_of_an_instance = our_pyexplainer.explain(
# X_explain = X_test.loc[file_to_be_explained,:].to_frame().transpose(),
# y_explain = pd.Series(bool(y_test.loc[file_to_be_explained]),
# index = [file_to_be_explained],
# name = outcome),
# search_function = 'crossoverinterpolation',
# max_iter=1000,
# max_rules=20,
# random_state=0,
# reuse_local_model=True)
#
# # Please use the code below to visualise the generated PyExplainer explanation (What-If interactive visualisation).
# our_pyexplainer.visualise(pyexplainer_explanation_of_an_instance, title="Why this file is defect-introducing ?")
#
# ````
# `````
# +
# Import for PyExplainer
from pyexplainer.pyexplainer_pyexplainer import PyExplainer
file_to_be_explained = 'src/java/org/apache/lucene/index/DocumentsWriter.java'
# PyExplainer Step 1 - Construct a PyExplainer
# PyExplainer Step 2 - Generate the rule-based explanation of an instance to be explained
# visualise the generated rule-based PyExplainer explanation
# -
#
# All of the above explanations are the property-contrast explanation within a file (https://xai4se.github.io/xai/theory-of-explanations.html).
# In fact, model-agnostic techniques can be used to generate other types of explanations, e.g., Object-contrast (i.e., the differences of explanations between two objects).
#
# **(4) Please use LIME to generate the object-contrast explanations between *DocumentsWriter.java* and *TestStringIntern.java*.**
#
# +
# Import for LIME
import lime
import lime.lime_tabular
file_to_be_explained = 'src/java/org/apache/lucene/index/DocumentsWriter.java'
another_file_to_be_explained = 'src/test/org/apache/lucene/util/TestStringIntern.java'
print(f'Generating the object-contrast explanations between {file_to_be_explained} and {another_file_to_be_explained} with LIME')
# LIME Step 1 - Construct an explainer
# LIME Step 2 - Use the constructed explainer with the predict function of your predictive model to explain the two instances
# visualise the generated LIME explanation - (DocumentsWriter.java)
# -
# visualise the generated LIME explanation - (TestStringIntern.java)
| docs/tutorials/hands-on-exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Recommendations with MovieTweetings: Most Popular Recommendation
#
# Now that you have created the necessary columns we will be using throughout the rest of the lesson on creating recommendations, let's get started with the first of our recommendations.
#
# To get started, read in the libraries and the two datasets you will be using throughout the lesson using the code below.
#
# +
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tests as t
# %matplotlib inline
# Read in the datasets
movies = pd.read_csv('movies_clean.csv')
reviews = pd.read_csv('reviews_clean.csv')
del movies['Unnamed: 0']
del reviews['Unnamed: 0']
# -
# #### Part I: How To Find The Most Popular Movies?
#
# For this notebook, we have a single task. The task is that no matter the user, we need to provide a list of the recommendations based on simply the most popular items.
#
# For this task, we will consider what is "most popular" based on the following criteria:
#
# * A movie with the highest average rating is considered best
# * With ties, movies that have more ratings are better
# * A movie must have a minimum of 5 ratings to be considered among the best movies
# * If movies are tied in their average rating and number of ratings, the ranking is determined by the movie that is the most recent rating
#
# With these criteria, the goal for this notebook is to take a **user_id** and provide back the **n_top** recommendations. Use the function below as the scaffolding that will be used for all the future recommendations as well.
# +
movie_rating = reviews.groupby('movie_id')['rating']
avg_rating=movie_rating.mean() # highest average rating
num_rating=movie_rating.count() # number of rating
last_rating= pd.DataFrame(reviews.groupby('movie_id')['date'].max())
last_rating.columns=["last_rating"]
# -
rating_count_df = pd.DataFrame({'avg_rating': avg_rating, 'num_rating': num_rating})
rating_count_df = rating_count_df.join(last_rating)
# merge with the movies dataset
movie_recs = movies.set_index('movie_id').join(rating_count_df)
ranked_movies=movie_recs.sort_values(['avg_rating','num_rating','last_rating'],ascending=[False,False,False])
ranked_movies= ranked_movies[ranked_movies['num_rating']>4]
ranked_movies.head()
# +
def create_ranked_df(movies, reviews):
'''
INPUT
movies - the movies dataframe
reviews - the reviews dataframe
OUTPUT
ranked_movies - a dataframe with movies that are sorted by highest avg rating, more reviews,
then time, and must have more than 4 ratings
'''
# Pull the average ratings and number of ratings for each movie
movie_ratings = reviews.groupby('movie_id')['rating']
avg_ratings = movie_ratings.mean()
num_ratings = movie_ratings.count()
last_rating = pd.DataFrame(reviews.groupby('movie_id').max()['date'])
last_rating.columns = ['last_rating']
# Add Dates
rating_count_df = pd.DataFrame({'avg_rating': avg_ratings, 'num_ratings': num_ratings})
rating_count_df = rating_count_df.join(last_rating)
# merge with the movies dataset
movie_recs = movies.set_index('movie_id').join(rating_count_df)
# sort by top avg rating and number of ratings
ranked_movies = movie_recs.sort_values(['avg_rating', 'num_ratings', 'last_rating'], ascending=False)
# for edge cases - subset the movie list to those with only 5 or more reviews
ranked_movies = ranked_movies[ranked_movies['num_ratings'] > 4]
return ranked_movies
def popular_recommendations(user_id, n_top, ranked_movies):
'''
INPUT:
user_id - the user_id (str) of the individual you are making recommendations for
n_top - an integer of the number recommendations you want back
ranked_movies - a pandas dataframe of the already ranked movies based on avg rating, count, and time
OUTPUT:
top_movies - a list of the n_top recommended movies by movie title in order best to worst
'''
top_movies = list(ranked_movies['movie'][:n_top])
return top_movies
# -
# Usint the three criteria above, you should be able to put together the above function. If you feel confident in your solution, check the results of your function against our solution. On the next page, you can see a walkthrough and you can of course get the solution by looking at the solution notebook available in this workspace.
# +
# Top 20 movies recommended for id 1
ranked_movies = create_ranked_df(movies, reviews) # only run this once - it is not fast
recs_20_for_1 = popular_recommendations('1', 20, ranked_movies)
# Top 5 movies recommended for id 53968
recs_5_for_53968 = popular_recommendations('53968', 5, ranked_movies)
# Top 100 movies recommended for id 70000
recs_100_for_70000 = popular_recommendations('70000', 100, ranked_movies)
# Top 35 movies recommended for id 43
recs_35_for_43 = popular_recommendations('43', 35, ranked_movies)
# -
# Usint the three criteria above, you should be able to put together the above function. If you feel confident in your solution, check the results of your function against our solution. On the next page, you can see a walkthrough and you can of course get the solution by looking at the solution notebook available in this workspace.
# +
### You Should Not Need To Modify Anything In This Cell
# check 1
assert t.popular_recommendations('1', 20, ranked_movies) == recs_20_for_1, "The first check failed..."
# check 2
assert t.popular_recommendations('53968', 5, ranked_movies) == recs_5_for_53968, "The second check failed..."
# check 3
assert t.popular_recommendations('70000', 100, ranked_movies) == recs_100_for_70000, "The third check failed..."
# check 4
assert t.popular_recommendations('43', 35, ranked_movies) == recs_35_for_43, "The fourth check failed..."
print("If you got here, looks like you are good to go! Nice job!")
# -
# **Notice:** This wasn't the only way we could have determined the "top rated" movies. You can imagine that in keeping track of trending news or trending social events, you would likely want to create a time window from the current time, and then pull the articles in the most recent time frame. There are always going to be some subjective decisions to be made.
#
# If you find that no one is paying any attention to your most popular recommendations, then it might be time to find a new way to recommend, which is what the next parts of the lesson should prepare us to do!
#
# ### Part II: Adding Filters
#
# Now that you have created a function to give back the **n_top** movies, let's make it a bit more robust. Add arguments that will act as filters for the movie **year** and **genre**.
#
# Use the cells below to adjust your existing function to allow for **year** and **genre** arguments as **lists** of **strings**. Then your ending results are filtered to only movies within the lists of provided years and genres (as `or` conditions). If no list is provided, there should be no filter applied.
#
# You can adjust other necessary inputs as necessary to retrieve the final results you are looking for!
def popular_recs_filtered(user_id,n_top,ranked_movies,years=None,genres =None):
'''
INPUT:
user_id - user id of customer for recommendation
n_top - number of movies you want to recommend
ranked movies - previously generated movie ranking based on average movie rating, number of ratings, and recency.
years - a list of strings for movie release year
genres - list of strings for movie genre type
OUTPUT:
top_movies- recommended n_top movie titles in the order, best to worst.
'''
if years is not None:
ranked_movies= ranked_movies[ranked_movies.date.isin(years)]
if genres is not None:
num_genre_match = ranked_movies[genres].sum(axis=1)
ranked_movies = ranked_movies.loc[num_genre_match > 0, :]
# return a list of recommended movies
top_choice=list(ranked_movies['movie'][:n_top])
return top_choice
def popular_recs_filtered(user_id, n_top, ranked_movies, years=None, genres=None):
'''
INPUT:
user_id - the user_id (str) of the individual you are making recommendations for
n_top - an integer of the number recommendations you want back
ranked_movies - a pandas dataframe of the already ranked movies based on avg rating, count, and time
years - a list of strings with years of movies
genres - a list of strings with genres of movies
OUTPUT:
top_movies - a list of the n_top recommended movies by movie title in order best to worst
'''
# Filter movies based on year and genre
if years is not None:
ranked_movies = ranked_movies[ranked_movies['date'].isin(years)]
if genres is not None:
num_genre_match = ranked_movies[genres].sum(axis=1)
ranked_movies = ranked_movies.loc[num_genre_match > 0, :]
# create top movies list
top_movies = list(ranked_movies['movie'][:n_top])
return top_movies
# +
# Top 20 movies recommended for id 1 with years=['2015', '2016', '2017', '2018'], genres=['History']
recs_20_for_1_filtered = popular_recs_filtered('1', 20, ranked_movies, years=['2015', '2016', '2017', '2018'], genres=['History'])
# Top 5 movies recommended for id 53968 with no genre filter but years=['2015', '2016', '2017', '2018']
recs_5_for_53968_filtered = popular_recs_filtered('53968', 5, ranked_movies, years=['2015', '2016', '2017', '2018'])
# Top 100 movies recommended for id 70000 with no year filter but genres=['History', 'News']
recs_100_for_70000_filtered = popular_recs_filtered('70000', 100, ranked_movies, genres=['History', 'News'])
# +
### You Should Not Need To Modify Anything In This Cell
# check 1
assert t.popular_recs_filtered('1', 20, ranked_movies, years=['2015', '2016', '2017', '2018'], genres=['History']) == recs_20_for_1_filtered, "The first check failed..."
# check 2
assert t.popular_recs_filtered('53968', 5, ranked_movies, years=['2015', '2016', '2017', '2018']) == recs_5_for_53968_filtered, "The second check failed..."
# check 3
assert t.popular_recs_filtered('70000', 100, ranked_movies, genres=['History', 'News']) == recs_100_for_70000_filtered, "The third check failed..."
print("If you got here, looks like you are good to go! Nice job!")
# -
| Movie Recommendation/Most_Popular_Recommendations.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Erasmus+ ICCT project (2018-1-SI01-KA203-047081)
# Toggle cell visibility
from IPython.display import HTML
tag = HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide()
} else {
$('div.input').show()
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<sup>Promijeni vidljivost <a href="javascript:code_toggle()">ovdje</a>.</sup>''')
display(tag)
# Hide the code completely
# from IPython.display import HTML
# tag = HTML('''<style>
# div.input {
# display:none;
# }
# </style>''')
# display(tag)
# -
# %matplotlib notebook
import matplotlib.pyplot as plt
import numpy as np
import sympy as sym
import scipy.signal as signal
from ipywidgets import widgets, interact
import control as cn
# ## Geometrijsko mjesto korijena (Root locus)
#
# Geometrijsko mjesto korijena (Root locus) je graf položaja polova sustava zatvorene petlje u odnosu na određeni parametar (uobičajeno je to pojačanje). Može se pokazati da krivulje počinju u polovima otvorene petlje, a završavaju u nulama otvorene petlje (ili u beskonačnosti). Položaj polova sustava zatvorene petlje daje indikaciju stabilnosti sustava, a ukazuje i na druga svojstva odziva sustava poput prekoračenja, vremena porasta i vremena smirivanja.
#
# ---
#
# ### Kako koristiti ovaj interaktivni primjer?
# 1. Kliknite na gumb *P0*, *P1*, *I0* ili *I1* za odabir između sljedećih objekata: proporcija nultog, prvog ili drugog reda ili integral nultog ili prvog reda. Prijenosna funkcija objekta P0 je $k_p$ (u ovom primjeru $k_p=2$), objekta P1 $\frac{k_p}{\tau s+1}$ (u ovom primjeru $k_p=1$ and $\tau=2$), objekta I0 $\frac{k_i}{s}$ (u ovom primjeru $k_i=\frac{1}{10}$) i objakta I1 $\frac{k_i}{s(\tau s +1}$ (u ovom primjeru $k_i=1$ i $\tau=10$).
# 2. Kliknite na gumb *P*, *PI*, *PD* ili *PID* za odabir između proporcionalnog, proporcionalno-integracijskog, proporcionalno-derivacijskog ili proporcionalno-integracijsko-derivacijskog tipa algoritma upravljanja.
# 3. Pomičite klizače da biste promijenili vrijednosti proporcionalnog ($K_p$), integracijskog ($T_i$) i derivacijskog ($T_d$) koeficijenta PID regulacije.
# 4. Pomičite klizač $t_{max}$ za promjenu maksimalne vrijednosti vremena na osi x.
# +
A = 10
a=0.1
s, P, I, D = sym.symbols('s, P, I, D')
obj = 1/(A*s)
PID = P + P/(I*s) + P*D*s#/(a*D*s+1)
system = obj*PID/(1+obj*PID)
num = [sym.fraction(system.factor())[0].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system.factor())[0], gen=s)))]
den = [sym.fraction(system.factor())[1].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system.factor())[1], gen=s)))]
system_func_open = obj*PID
num_open = [sym.fraction(system_func_open.factor())[0].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func_open.factor())[0], gen=s)))]
den_open = [sym.fraction(system_func_open.factor())[1].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func_open.factor())[1], gen=s)))]
# make figure
fig = plt.figure(figsize=(9.8, 4),num='Geometrijsko mjesto korijena')
plt.subplots_adjust(wspace=0.3)
# add axes
ax = fig.add_subplot(121)
ax.grid(which='both', axis='both', color='lightgray')
ax.set_title('Vremenski odziv')
ax.set_xlabel('t [s]')
ax.set_ylabel('ulaz, izlaz')
rlocus = fig.add_subplot(122)
# plot step function and responses (initalisation)
input_plot, = ax.plot([],[],'C0', lw=1, label='ulaz')
response_plot, = ax.plot([],[], 'C1', lw=2, label='izlaz')
ax.legend()
rlocus_plot, = rlocus.plot([], [], 'r')
plt.show()
system_open = None
system_close = None
def update_plot(KP, TI, TD, Time_span):
global num, den, num_open, den_open
global system_open, system_close
num_temp = [float(i.subs(P,KP).subs(I,TI).subs(D,TD)) for i in num]
den_temp = [float(i.subs(P,KP).subs(I,TI).subs(D,TD)) for i in den]
system = signal.TransferFunction(num_temp, den_temp)
system_close = system
num_temp_open = [float(i.subs(P,KP).subs(I,TI).subs(D,TD)) for i in num_open]
den_temp_open = [float(i.subs(P,KP).subs(I,TI).subs(D,TD)) for i in den_open]
system_open = signal.TransferFunction(num_temp_open, den_temp_open)
rlocus.clear()
r, k, xlim, ylim = cn.root_locus_modified(system_open, Plot=False)
# r, k = cn.root_locus(system_open, Plot=False)
#rlocus.scatter(r)
#plot closed loop poles and zeros
poles = np.roots(system.den)
rlocus.plot(np.real(poles), np.imag(poles), 'kx')
zeros = np.roots(system.num)
if zeros.size > 0:
rlocus.plot(np.real(zeros), np.imag(zeros), 'ko', alpha=0.5)
# plot open loop poles and zeros
poles = np.roots(system_open.den)
rlocus.plot(np.real(poles), np.imag(poles), 'x', alpha=0.5)
zeros = np.roots(system_open.num)
if zeros.size > 0:
rlocus.plot(np.real(zeros), np.imag(zeros), 'o')
#plot root locus
for index, col in enumerate(r.T):
rlocus.plot(np.real(col), np.imag(col), 'b', alpha=0.5)
rlocus.set_title('Geometrijsko mjesto korijena')
rlocus.set_xlabel('Re')
rlocus.set_ylabel('Im')
rlocus.grid(which='both', axis='both', color='lightgray')
rlocus.axhline(linewidth=.3, color='g')
rlocus.axvline(linewidth=.3, color='g')
rlocus.set_ylim(ylim)
rlocus.set_xlim(xlim)
time = np.linspace(0, Time_span, 300)
u = np.ones_like(time)
u[0] = 0
time, response = signal.step(system, T=time)
response_plot.set_data(time, response)
input_plot.set_data(time, u)
ax.set_ylim([min([np.min(u), min(response),-.1]),min(100,max([max(response)*1.05, 1, 1.05*np.max(u)]))])
ax.set_xlim([-0.1,max(time)])
plt.show()
controller_ = PID
object_ = obj
def calc_tf():
global num, den, controller_, object_, num_open, den_open
system_func = object_*controller_/(1+object_*controller_)
num = [sym.fraction(system_func.factor())[0].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func.factor())[0], gen=s)))]
den = [sym.fraction(system_func.factor())[1].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func.factor())[1], gen=s)))]
system_func_open = object_*controller_
num_open = [sym.fraction(system_func_open.factor())[0].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func_open.factor())[0], gen=s)))]
den_open = [sym.fraction(system_func_open.factor())[1].expand().coeff(s, i) for i in reversed(range(1+sym.degree(sym.fraction(system_func_open.factor())[1], gen=s)))]
update_plot(Kp_widget.value, Ti_widget.value, Td_widget.value, time_span_widget.value)
def transfer_func(controller_type):
global controller_
proportional = P
integral = P/(I*s)
differential = P*D*s/(a*D*s+1)
if controller_type =='P':
controller_func = proportional
Kp_widget.disabled=False
Ti_widget.disabled=True
Td_widget.disabled=True
elif controller_type =='PI':
controller_func = proportional+integral
Kp_widget.disabled=False
Ti_widget.disabled=False
Td_widget.disabled=True
elif controller_type == 'PD':
controller_func = proportional+differential
Kp_widget.disabled=False
Ti_widget.disabled=True
Td_widget.disabled=False
else:
controller_func = proportional+integral+differential
Kp_widget.disabled=False
Ti_widget.disabled=False
Td_widget.disabled=False
controller_ = controller_func
calc_tf()
def transfer_func_obj(object_type):
global object_
if object_type == 'P0':
object_ = 2
elif object_type == 'P1':
object_ = 1/(2*s+1)
elif object_type == 'I0':
object_ = 1/(10*s)
elif object_type == 'I1':
object_ = 1/(s*(10*s+1))
calc_tf()
style = {'description_width': 'initial'}
def buttons_controller_clicked(event):
controller = buttons_controller.options[buttons_controller.index]
transfer_func(controller)
buttons_controller = widgets.ToggleButtons(
options=['P', 'PI', 'PD', 'PID'],
description='Odaberite tip algoritma upravljanja:',
disabled=False,
style=style)
buttons_controller.observe(buttons_controller_clicked)
def buttons_object_clicked(event):
object_ = buttons_object.options[buttons_object.index]
transfer_func_obj(object_)
buttons_object = widgets.ToggleButtons(
options=['P0', 'P1', 'I0', 'I1'],
description='Odaberite objekt:',
disabled=False,
style=style)
buttons_object.observe(buttons_object_clicked)
Kp_widget = widgets.FloatLogSlider(value=.5,min=-3,max=2.1,step=.001,description=r'\(K_p\)',
disabled=False,continuous_update=True,orientation='horizontal',readout=True,readout_format='.3f')
Ti_widget = widgets.FloatLogSlider(value=1.,min=-3,max=1.8,step=.001,description=r'\(T_{i} \)',
disabled=False,continuous_update=True,orientation='horizontal',readout=True,readout_format='.3f')
Td_widget = widgets.FloatLogSlider(value=1.,min=-3,max=1.8,step=.001,description=r'\(T_{d} \)',
disabled=False,continuous_update=True,orientation='horizontal',readout=True,readout_format='.3f')
time_span_widget = widgets.FloatSlider(value=10.,min=.5,max=50.,step=0.1,description=r'\(t_{max} \)',
disabled=False,continuous_update=True,orientation='horizontal',readout=True,readout_format='.1f')
transfer_func(buttons_controller.options[buttons_controller.index])
transfer_func_obj(buttons_object.options[buttons_object.index])
display(buttons_object)
display(buttons_controller)
interact(update_plot, KP=Kp_widget, TI=Ti_widget, TD=Td_widget, Time_span=time_span_widget);
| ICCT_hr/examples/02/.ipynb_checkpoints/TD-18-Geometrijsko_mjesto_korijena-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:torch] *
# language: python
# name: conda-env-torch-py
# ---
# +
import numpy as np
import torch
from torch import nn
from sklearn import datasets
import matplotlib.pyplot as plt
w0 = 0.125
b0 = 5.
x_range = [-20, 60]
def load_dataset(n=150, n_tst=150):
np.random.seed(43)
def s(x):
g = (x - x_range[0]) / (x_range[1] - x_range[0])
return 3 * (0.25 + g**2.)
x = (x_range[1] - x_range[0]) * np.random.rand(n) + x_range[0]
eps = np.random.randn(n) * s(x)
y = (w0 * x * (1. + np.sin(x)) + b0) + eps
y = (y - y.mean()) / y.std()
idx = np.argsort(x)
x = x[idx]
y = y[idx]
return y[:, None], x[:, None]
y, x = load_dataset()
# -
plt.plot(x,y,'.')
plt.show()
len(x)
X = torch.tensor(x, dtype=torch.float)
Y = torch.tensor(y, dtype=torch.float)
# - First we’ll model a neural network $g_{\theta}(x)$ with maximum likelihood estimation. Where we assume a Gaussian likelihood.
# $$\begin{equation}
# y \sim \mathcal{N}(g_{\theta}(x), \sigma^2)
# \end{equation}$$
# $$ \begin{equation}\hat{\theta}_{\text{MLE}} = \text{argmax}_\theta \prod_i^nP(y_i|\theta) \end{equation}$$
class MaximumLikelihood(nn.Module):
def __init__(self):
super().__init__()
self.out = nn.Sequential(
nn.Linear(1, 20),
nn.ReLU(),
nn.Linear(20, 1)
)
def forward(self, x):
return self.out(x)
epochs = 200
m = MaximumLikelihood()
optim = torch.optim.Adam(m.parameters(), lr=0.01)
for epoch in range(epochs):
optim.zero_grad()
y_pred = m(X)
loss = (0.5 * (y_pred - Y)**2).mean()
loss.backward()
optim.step()
m.eval()
y_estimate = m(X)
plt.figure(figsize=(10, 5))
plt.plot(x,y, 'b.', alpha=0.8)
plt.plot(x, y_estimate.detach().numpy(), 'r', alpha=0.6)
plt.show()
# - We are able to predict the expectation of $y$, but we are not able to make a statement about the uncertainty of our predictions.
# - In variational inference, we accept that we cannot obtain the true posterior $P(y|x)$, but we try to approximate this distribution with another distribution $Q_{\theta}(y)$, where $\theta$ are the variational parameters. This distribution we call a variational distribution.
# - If we choose a factorized (diagonal) Gaussian variational distribution, we define a function $g_{\theta}: x \mapsto \mu, \sigma$. The function $g_{\theta}$ will be a neural network that predicts the variational parameters.
# - The total model can thus be described as:
# $$ \begin{equation}P(y) = \mathcal{N}(0, 1) \end{equation}$$
# where we set a unit Gaussian prior $P(y)$.
# - from now on we will generalize to a notation that is often used. We’ll extend $y|x$ to any (latent) stochastic variable $Z$.
# --------------------------------
# - Variational inference is done by maximizing the ELBO:
# $$ \begin{equation}\text{argmax}_{Z} = E_{Z \sim Q}[\underbrace{\log P(D|Z)}_{\text{likelihood}}] - D_{KL}(Q(Z)||\underbrace{P(Z)}_{\text{prior}}) \label{eq:elbo} \end{equation}$$
# - Let's rewrite this ELBO definition so that it is more clear how we can use it to optimize the model.
# $$E_{Z \sim Q}[\log P(D|Z)] + E_{Z \sim Q}[ \frac{P(Z)}{Q(Z)}]dZ$$
# $$E_{Z \sim Q}[\log P(D|Z)] + E_{Z \sim Q}[\log P(Z) - \log Q(Z)]$$
# #### Monte Carlo ELBO and reparameterization trick
# - Deriving those expectations maybe not possible, thus we can get estimates of the true expectation by taking samples from $Q(Z)$ and average over those results.
# - If we start taking samples from a $Q(Z)$ we leave the deterministic world, and the gradient can not flow through the model anymore. We avoid this problem by reparameterizing the samples from the distribution.
# - Instead of sampling directly from the variational distribution, $z \sim Q(\mu, \sigma^2)$, we sample from a unit gaussian and recreate samples from the variational distribution. Now the stochasticity of $\epsilon$ is external and will not prevent the flow of gradients.
class VI(nn.Module):
def __init__(self):
super().__init__()
self.q_mu = nn.Sequential(
nn.Linear(1, 20),
nn.ReLU(),
nn.Linear(20, 10),
nn.ReLU(),
nn.Linear(10, 1)
)
self.q_log_var = nn.Sequential(
nn.Linear(1, 20),
nn.ReLU(),
nn.Linear(20, 10),
nn.ReLU(),
nn.Linear(10, 1)
)
def reparameterize(self, mu, log_var):
sigma = torch.exp(0.5 * log_var) + 1e-5
epsilon = torch.randn_like(sigma)
return mu + sigma * epsilon
def forward(self, x):
mu = self.q_mu(x)
log_var = self.q_log_var(x)
return self.reparameterize(mu, log_var), mu, log_var
# $$ \begin{aligned} \log p(\mathbf{y}|\mathbf{X, \mu, \sigma}) &= \sum_{i=1}^N \log N(y_i;\mathbf{\mu,\sigma^2}) \\ &= \sum_{i=1}^N \log \frac{1}{\sqrt{2\pi\sigma^2_e}}\exp (-\frac{(y_i - \mathbf{\mu})^2}{2\sigma^2_e}) \\ &= -\frac{N}{2}\log 2\pi\sigma^2_e - \sum_{i=1}^N \frac{(y_i-\mathbf{\mu)^2}}{2\sigma^2_e} \end{aligned}$$
def ll_gaussian(y, mu, log_var): #log-likelihood of gaussian
sigma = torch.exp(0.5 * log_var)
return -0.5 * torch.log(2 * np.pi * sigma**2) - (1 / (2 * sigma**2))* (y-mu)**2
def elbo(y_pred, y, mu, log_var):
# likelihood of observing y given Variational mu and sigma
likelihood = ll_gaussian(y, mu, log_var)
# prior probability of y_pred
log_prior = ll_gaussian(y_pred, 0, torch.log(torch.tensor(1.)))
# variational probability of y_pred
log_p_q = ll_gaussian(y_pred, mu, log_var)
# by taking the mean we approximate the expectation
return (likelihood + log_prior - log_p_q).mean()
def det_loss(y_pred, y, mu, log_var):
return -elbo(y_pred, y, mu, log_var)
# +
epochs = 1500
m = VI()
optim = torch.optim.Adam(m.parameters(), lr=0.005)
for epoch in range(epochs):
optim.zero_grad()
y_pred, mu, log_var = m(X)
loss = det_loss(y_pred, Y, mu, log_var)
loss.backward()
optim.step()
# +
# draw samples from Q(theta)
with torch.no_grad():
y_pred = torch.cat([m(X)[0] for _ in range(1000)], dim=1)
# Get some quantiles
q1, mu, q2 = np.quantile(y_pred, [0.05, 0.5, 0.95], axis=1)
plt.figure(figsize=(10, 5))
plt.scatter(X, Y, s=10)
plt.plot(X, mu, 'r', alpha=0.6)
plt.fill_between(X.flatten(), q1, q2, alpha=0.2)
# -
# #### Analytical KL-divergence
# - Above we have implemented ELBO by sampling from the variational posterior. It turns out that for the KL-divergence term, this isn’t necessary as there is an analytical solution:
# $$D_{KL}(Q(Z)||P(Z)) = \frac{1}{2}\sum_{i=1}^n(1+\log \sigma_i^2 - \mu_i^2 - \sigma_i^2)$$
# - For the likelihood term, we did implement Guassian log likelihood, this term can also be replaced with a similar loss functions. For Gaussian likelihood we can use squared mean error loss.
# - We can simplify the loss function as defined below:
# ```
# def det_loss(y, y_pred, mu, log_var):
# reconstruction_error = (0.5 * (y - y_pred)**2).sum()
# kl_divergence = (-0.5 * torch.sum(1 + log_var - mu**2 - log_var.exp()))
#
# return (reconstruction_error + kl_divergence).sum()
# ```
# #### Aleatoric and epistemic uncertainty
# - In the example above we were able to model the *aleatoric uncertainty*.
# - This is the *inherent variance* in the data which we have to accept because the underlaying data generation process is stochastic in nature. (e.g. throwing the dice, cards you get in a poker game)
# - Aleatory can have two flavors, being *homoscedastic* and *heteroscedastic*.
# - homoscedastic: For example in the model definition of linear regression $y = X \beta + \epsilon$ we incorporate $\epsilon$ for the noise in the data. In linear regression, $\epsilon$ is not dependent on $X$ and is therefore assumed to be constant.
# - heteroscedastic: If the aleatoric uncertainty is dependent on $X$, we speak of heteroscedastic uncertainty.
# - Epistemic uncertainty can be reduced by designing new model, acquiring more data, etc.
# - In the above example data has inherent noise which can't be reduced but when we generate model ensemble using dropout and measure only the *model inconsistency*, that epistemic uncertainty is low as shown in below. (because most of the uncertainty comes from aleatoric uncertainty in this case)
# <img src=attachment:image.png width=500>
| deep_learning/uncertainty-deep-learning-master/06. Aleatoric-epistemic uncertainty (toy example).ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # Introduction
#
# Welcome! In this set of tutorials you will learn about image classification using quantized neural networks (QNNs), and what kind of computations take place.
#
# ## What We Are Trying To Do
#
# For the purposes of this tutorial, we will view the QNN as a gray box. We will put in an image, do some operations, and get out a *classification result* which tells us what the QNN thinks this image is. The twenty-thousand feet view of how this goes is something like this:
#
# 1. We put in an image in the form of pixels, i.e. an array of numbers.
# 2. We multiply those pixel values (numbers) with some other numbers, which are the neural network weights, add them together, and perform some other simple operations.
# 4. We will repeat step 2 a couple of times with different weights.
# 3. At the end, we will obtain an array of numbers, one number for each class that the QNN knows about. The class with the largest number is the QNN's best guess on what the image is, the second largest is the second best guess, and so on.
#
# We won't concern ourselves with *where* the weights come from -- this tutorial will simply provide you with several pre-trained QNNs for that purpose. If you'd like to know more about neural networks in general, [here](https://github.com/stephencwelch/Neural-Networks-Demystified) is a popular tutorial in Jupyter Notebook form with accompanying YouTube videos, alongside countless other resources on the Internet.
#
# ## OK, Let's Do It!
#
# We'll start with a classical example in neural networks: classifying 28x28 grayscale images of digits (0 to 9). Let's load an image and see what it looks like first.
# +
from PIL import Image
from matplotlib.pyplot import imshow
import numpy as np
# load image using PIL
img = Image.open("7.png")
# convert to black and white
img = img.convert("L")
# convert to numpy array
img = np.asarray(img)
# display
% matplotlib inline
imshow(img, cmap='gray')
# -
# Looks like a seven to me, but to get a useful reminder of what images look like to a computer by default, let's have a look at the numpy array itself:
print(img.shape)
img
# It's all just numbers in a 28x28 array! Now let's see what the neural network says about this data. We will start by loading the QNN from the file it is stored in which is a [Python Pickle](https://wiki.python.org/moin/UsingPickle).
# +
from QNN.layers import *
import pickle
qnn = pickle.load(open("mnist-w1a1.pickle", "rb"))
qnn
# -
# As you can see, the QNN consists of several *layers*. The QNN we loaded seems to contain four types of layers: BipolarThresholding, FullyConnected, ScaleShift and Softmax. We will cover what all these do in more detail later on. Right now, let's just see if it works! The QNN module that we just imported contains a function called predict:
# get the predictions array
res = predict(qnn, img)
# return the index of the largest prediction
winner_ind = np.argmax(res)
# the sum of the output values add up to 1 due to softmax,
# so we can interpret them as probabilities
winner_prob = 100 * res[winner_ind]
print(res)
print("The QNN predicts this is a %d with %f percent probability" % (winner_ind, winner_prob))
# And our first image classification with a QNN is a success! In the following section, we will take a closer look at the computation that is taking place inside the .execute() functions for this network, and later we will cover more advanced types of networks.
| 0-basics.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Data Gathering for Power Analysis
#
# We've previously done power analyses for the Growth Team based on registration data from 2018. In this case, we want an updated analysis with more wikis, in order to understand how expanding to additional wikis will impact our statistical power.
#
# This work is tracked in [T250120](https://phabricator.wikimedia.org/T250120).
#
# The task asks to compare four groups of wikis:
#
# * Original target wikis: Czech, Korean, Arabic, Vietnamese
# * Current set: Czech, Korean, Arabic, Vietnamese, Ukrainian, Hungarian, Armenian, Basque
# * Adding just French: Czech, Korean, Arabic, Vietnamese, Ukrainian, Hungarian, Armenian, Basque, French
# * Adding our next set: Czech, Korean, Arabic, Vietnamese, Ukrainian, Hungarian, Armenian, Basque, French, Polish, Persian, Swedish, Danish, Indonesian, Italian, Portuguese.
#
# The first part of this is to gather registration, activation, and retention numbers for those wikis, for both desktop and mobile registrations.
# +
import datetime as dt
import pandas as pd
import numpy as np
from wmfdata import hive
# +
## Configuration variables
## Original target, then the next four, then the additional set
wikis = ['cswiki', 'kowiki', 'viwiki', 'arwiki',
'ukwiki', 'huwiki', 'hywiki', 'euwiki',
'frwiki', 'plwiki', 'fawiki', 'svwiki',
'dawiki', 'idwiki', 'itwiki', 'ptwiki']
## Activity tends to follow a yearly cycle, so let's use that.
start_date = '2019-01-01'
end_date = '2020-01-01'
## The mediawiki_history snapshot that we'll be using
snapshot = '2020-03'
# -
activity_query = '''
WITH regs AS (
SELECT wiki_db, event_user_id,
date_format(event_user_creation_timestamp, "yyyy-MM-01") as reg_month
FROM wmf.mediawiki_history
WHERE snapshot = "{snapshot}"
AND event_entity = "user"
AND event_type = "create"
AND event_user_is_created_by_self = TRUE
AND size(event_user_is_bot_by_historical) = 0
AND wiki_db IN ({wiki_list})
AND event_user_creation_timestamp >= "{start_time}"
AND event_user_creation_timestamp < "{end_time}"
),
mobile_data AS (
SELECT wiki AS wiki_db,
event.userid AS user_id,
IF(event.displaymobile, 'mobile', 'desktop') AS platform
FROM event_sanitized.serversideaccountcreation
WHERE year = 2019
AND wiki IN ({wiki_list})
AND event.isselfmade = true
),
edits AS (
SELECT wiki_db, event_user_id,
SUM(IF(unix_timestamp(event_timestamp) -
unix_timestamp(event_user_creation_timestamp) < 86400 , 1, 0)) AS activation_edits,
SUM(IF(unix_timestamp(event_timestamp) - unix_timestamp(event_user_creation_timestamp)
BETWEEN 86400 AND 15*86400, 1, 0)) AS retention_edits
FROM wmf.mediawiki_history
WHERE snapshot = "{snapshot}"
AND event_entity = "revision"
AND event_type = "create"
AND wiki_db IN ({wiki_list})
AND event_user_creation_timestamp >= "{start_time}"
AND event_user_creation_timestamp < "{end_time}"
AND SIZE(event_user_is_bot_by_historical) = 0
GROUP BY wiki_db, event_user_id
)
SELECT regs.wiki_db,
regs.event_user_id AS user_id,
regs.reg_month,
mobile_data.platform,
coalesce(edits.activation_edits, 0) AS activation_edits,
coalesce(edits.retention_edits, 0) AS retention_edits
FROM regs
JOIN mobile_data
ON regs.wiki_db = mobile_data.wiki_db
AND regs.event_user_id = mobile_data.user_id
LEFT JOIN edits
ON regs.wiki_db = edits.wiki_db
AND regs.event_user_id = edits.event_user_id
'''
# Grab user activity data:
user_activity = hive.run(activity_query.format(
snapshot = snapshot,
wiki_list = ','.join('"{}"'.format(w) for w in wikis),
start_time = start_date,
end_time = end_date
))
# Add boolean flags for whether a user is activated or retained:
user_activity['is_activated'] = user_activity['activation_edits'] > 0
user_activity['is_retained'] = user_activity['is_activated'] & (user_activity['retention_edits'] > 0)
# Aggregate per wiki, platform, and month of registration counts of registrations, activations, and retentions:
registrations_agg = (user_activity.groupby(['wiki_db', 'reg_month', 'platform'])
.agg({'user_id' : 'count'})
.rename(columns = {'user_id' : 'n_registered'}))
activations_agg = (user_activity.loc[user_activity['is_activated'] == True]
.groupby(['wiki_db', 'reg_month', 'platform'])
.agg({'user_id' : 'count'})
.rename(columns = {'user_id' : 'n_activated'}))
retentions_agg = (user_activity.loc[user_activity['is_retained'] == True]
.groupby(['wiki_db', 'reg_month', 'platform'])
.agg({'user_id' : 'count'})
.rename(columns = {'user_id' : 'n_retained'}))
# Merge the three aggregations to combine:
full_data = (registrations_agg.merge(activations_agg, how = 'left', left_index = True, right_index = True)
.merge(retentions_agg, how = 'left', left_index = True, right_index = True)
.fillna(0).reset_index())
# +
## Calculate activation and retention proportions
full_data['prop_activated'] = full_data['n_activated'] / full_data['n_registered']
full_data['prop_retained'] = full_data['n_retained'] / full_data['n_activated']
# -
# Aggregate over the whole year and calculate monthly averages:
fullyear_agg = (full_data.groupby(['wiki_db', 'platform'])
.agg({'n_registered' : 'mean', 'n_activated' : 'mean', 'n_retained' : 'mean',
'prop_activated' : 'mean', 'prop_retained' : 'mean'})
.reset_index())
# Write the resulting dataframe out as a TSV for import into R.
fullyear_agg.to_csv('datasets/aggregate_statistics.tsv',
header = True, index = False, sep = '\t')
| 01_data_gathering.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="view-in-github" colab_type="text"
# <a href="https://colab.research.google.com/github/mherbert93/DS-Unit-2-Linear-Models/blob/master/module3-ridge-regression/LS_DS_213_assignment.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# + [markdown] id="fq9wNXC4OJiI" colab_type="text"
# Lambda School Data Science
#
# *Unit 2, Sprint 1, Module 3*
#
# ---
# + [markdown] colab_type="text" id="7IXUfiQ2UKj6"
# # Ridge Regression
#
# ## Assignment
#
# We're going back to our other **New York City** real estate dataset. Instead of predicting apartment rents, you'll predict property sales prices.
#
# But not just for condos in Tribeca...
#
# - [x] Use a subset of the data where `BUILDING_CLASS_CATEGORY` == `'01 ONE FAMILY DWELLINGS'` and the sale price was more than 100 thousand and less than 2 million.
# - [x] Do train/test split. Use data from January — March 2019 to train. Use data from April 2019 to test.
# - [x] Do one-hot encoding of categorical features.
# - [x] Do feature selection with `SelectKBest`.
# - [x] Fit a ridge regression model with multiple features. Use the `normalize=True` parameter (or do [feature scaling](https://scikit-learn.org/stable/modules/preprocessing.html) beforehand — use the scaler's `fit_transform` method with the train set, and the scaler's `transform` method with the test set)
# - [x] Get mean absolute error for the test set.
# - [x] As always, commit your notebook to your fork of the GitHub repo.
#
# The [NYC Department of Finance](https://www1.nyc.gov/site/finance/taxes/property-rolling-sales-data.page) has a glossary of property sales terms and NYC Building Class Code Descriptions. The data comes from the [NYC OpenData](https://data.cityofnewyork.us/browse?q=NYC%20calendar%20sales) portal.
#
#
# ## Stretch Goals
#
# Don't worry, you aren't expected to do all these stretch goals! These are just ideas to consider and choose from.
#
# - [ ] Add your own stretch goal(s) !
# - [ ] Instead of `Ridge`, try `LinearRegression`. Depending on how many features you select, your errors will probably blow up! 💥
# - [x] Instead of `Ridge`, try [`RidgeCV`](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.RidgeCV.html).
# - [ ] Learn more about feature selection:
# - ["Permutation importance"](https://www.kaggle.com/dansbecker/permutation-importance)
# - [scikit-learn's User Guide for Feature Selection](https://scikit-learn.org/stable/modules/feature_selection.html)
# - [mlxtend](http://rasbt.github.io/mlxtend/) library
# - scikit-learn-contrib libraries: [boruta_py](https://github.com/scikit-learn-contrib/boruta_py) & [stability-selection](https://github.com/scikit-learn-contrib/stability-selection)
# - [_Feature Engineering and Selection_](http://www.feat.engineering/) by Kuhn & Johnson.
# - [ ] Try [statsmodels](https://www.statsmodels.org/stable/index.html) if you’re interested in more inferential statistical approach to linear regression and feature selection, looking at p values and 95% confidence intervals for the coefficients.
# - [ ] Read [_An Introduction to Statistical Learning_](http://faculty.marshall.usc.edu/gareth-james/ISL/ISLR%20Seventh%20Printing.pdf), Chapters 1-3, for more math & theory, but in an accessible, readable way.
# - [ ] Try [scikit-learn pipelines](https://scikit-learn.org/stable/modules/compose.html).
# + colab_type="code" id="o9eSnDYhUGD7" colab={}
# %%capture
import sys
# If you're on Colab:
if 'google.colab' in sys.modules:
DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'
# !pip install category_encoders==2.*
# If you're working locally:
else:
DATA_PATH = '../data/'
# Ignore this Numpy warning when using Plotly Express:
# FutureWarning: Method .ptp is deprecated and will be removed in a future version. Use numpy.ptp instead.
import warnings
warnings.filterwarnings(action='ignore', category=FutureWarning, module='numpy')
# + colab_type="code" id="QJBD4ruICm1m" colab={}
import pandas as pd
import pandas_profiling
# Read New York City property sales data
df = pd.read_csv(DATA_PATH+'condos/NYC_Citywide_Rolling_Calendar_Sales.csv')
# Change column names: replace spaces with underscores
df.columns = [col.replace(' ', '_') for col in df]
# SALE_PRICE was read as strings.
# Remove symbols, convert to integer
df['SALE_PRICE'] = (
df['SALE_PRICE']
.str.replace('$','')
.str.replace('-','')
.str.replace(',','')
.astype(int)
)
# + id="oIE711g7OJiW" colab_type="code" colab={}
# BOROUGH is a numeric column, but arguably should be a categorical feature,
# so convert it from a number to a string
df['BOROUGH'] = df['BOROUGH'].astype(str)
# + id="uEQe6U1HOJib" colab_type="code" colab={}
# Reduce cardinality for NEIGHBORHOOD feature
# Get a list of the top 10 neighborhoods
top10 = df['NEIGHBORHOOD'].value_counts()[:10].index
# At locations where the neighborhood is NOT in the top 10,
# replace the neighborhood with 'OTHER'
df.loc[~df['NEIGHBORHOOD'].isin(top10), 'NEIGHBORHOOD'] = 'OTHER'
# + id="MuzpHnr2OJif" colab_type="code" outputId="fae47f41-9bca-49a9-9f50-ddaeecfe396f" colab={"base_uri": "https://localhost:8080/", "height": 469}
df.head()
# + id="4pbdYV3lOv-b" colab_type="code" colab={}
df = df[(df['BUILDING_CLASS_CATEGORY'] == '01 ONE FAMILY DWELLINGS') & ((df['SALE_PRICE'] > 100000) & (df['SALE_PRICE'] < 2000000))]
# + id="p3O3SINBPXHi" colab_type="code" outputId="729b9ce4-8ae0-42a3-ce47-6b27d7167245" colab={"base_uri": "https://localhost:8080/", "height": 135}
df.head(1)
# + id="Dr_wjaeCbYDm" colab_type="code" outputId="405af31e-4f9c-47d0-ad14-86f9d63703ae" colab={"base_uri": "https://localhost:8080/", "height": 421}
df.isnull().sum()
# + id="bydd57_lbdM0" colab_type="code" outputId="3352d00a-60f3-4998-8105-6926c77b8df3" colab={"base_uri": "https://localhost:8080/", "height": 35}
df.shape
# + id="NtWGnBrwbfje" colab_type="code" colab={}
df.drop(['EASE-MENT', 'APARTMENT_NUMBER'], axis=1, inplace=True) #all values in this column are null, so drop them
# + id="ttzSRL5YPdjt" colab_type="code" colab={}
df['SALE_DATE'] = pd.to_datetime(df['SALE_DATE'], infer_datetime_format=True) #convert to datatime format
train = df[(df['SALE_DATE'] >= '2019-01-01') & (df['SALE_DATE'] < '2019-04-01')]
test = df[(df['SALE_DATE'] >= '2019-04-01') & (df['SALE_DATE'] < '2019-06-01')]
# + id="bIOxRdwxT4SB" colab_type="code" outputId="770b02bb-8338-4e50-e806-da669d735864" colab={"base_uri": "https://localhost:8080/", "height": 238}
train.describe(exclude='number')
# + id="h-QOekLivpJO" colab_type="code" outputId="a343ea20-123e-4f67-f425-7e97b20f74d2" colab={"base_uri": "https://localhost:8080/", "height": 300}
train.describe(include='number')
# + id="b5fTLVPJUFGR" colab_type="code" colab={}
#remove columns that have high cardinality
train = train.drop(['ADDRESS', 'LAND_SQUARE_FEET', 'SALE_DATE'], axis=1)
test = test.drop(['ADDRESS', 'LAND_SQUARE_FEET', 'SALE_DATE'], axis=1)
# + id="XMkS55OSXZUd" colab_type="code" colab={}
target = 'SALE_PRICE'
features = train.columns.drop([target])
# + id="ejs8eznaQxm5" colab_type="code" colab={}
X_train = train[features]
X_test = test[features]
Y_train = train['SALE_PRICE']
Y_test = test['SALE_PRICE']
import category_encoders as ce
encoder = ce.OneHotEncoder(use_cat_names=True)
X_train = encoder.fit_transform(X_train)
X_test = encoder.transform(X_test)
# + id="obSfs3bKX-oK" colab_type="code" outputId="25fd2788-41d5-4bdc-c579-11f82c83025a" colab={"base_uri": "https://localhost:8080/", "height": 244}
X_train.head()
# + id="rcpB4rDuYhII" colab_type="code" outputId="6994fff7-ffd5-42ee-bce8-2e866472b989" colab={"base_uri": "https://localhost:8080/", "height": 35}
X_train.shape
# + id="Wc0wtU6FZpUs" colab_type="code" colab={}
from sklearn.feature_selection import SelectKBest, f_regression
selector = SelectKBest(score_func=f_regression, k=9)
X_train_selected = selector.fit_transform(X_train, Y_train)
X_test_selected = selector.transform(X_test)
# + id="IS_3VU1hbIrC" colab_type="code" outputId="60a2d711-c907-45e2-ddfb-172a5cb2fa95" colab={"base_uri": "https://localhost:8080/", "height": 201}
selected_mask = selector.get_support()
all_names = X_train.columns
selected_names = all_names[selected_mask]
unselected_names = all_names[~selected_mask]
print('Features selected:')
for name in selected_names:
print(name)
# + id="BBMSxJ_zTGC5" colab_type="code" outputId="61c4734b-55eb-4fcd-d00c-03a8d653fcac" colab={"base_uri": "https://localhost:8080/", "height": 1000}
import warnings
warnings.filterwarnings("ignore")
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_absolute_error
from sklearn.linear_model import RidgeCV
previous_mae = 1000000000
previous_features = 100000000
diff = 100
alphas = [0.0001, 0.001, 0.01, 0.1, 1.0, 10.0, 100.0]
for k in range(1, len(X_train.columns)+1):
selector = SelectKBest(score_func=f_regression, k=k)
X_train_selected = selector.fit_transform(X_train, Y_train)
X_test_selected = selector.transform(X_test)
ridge = RidgeCV(alphas=alphas, normalize=True, cv=10)
ridge.fit(X_train_selected, Y_train)
prediction = ridge.predict(X_test_selected)
mae = mean_absolute_error(Y_test, prediction)
print(f'Test Mean Absolute Error: ${mae:,.0f} \n')
print("Alpha value is: ", ridge.alpha_)
print(f'{k} features')
selected_mask = selector.get_support()
all_names = X_train.columns
selected_names = all_names[selected_mask]
unselected_names = all_names[~selected_mask]
print('Features selected:')
for name in selected_names:
print(name)
# + id="xBAnyOdPj_x3" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 54} outputId="9d575cb0-9397-42fe-fc11-27981a1bc0c0"
from sklearn.linear_model import Ridge
selector = SelectKBest(score_func=f_regression, k=12)
X_train_selected = selector.fit_transform(X_train, Y_train)
X_test_selected = selector.transform(X_test)
ridge = Ridge(alpha=0.001, normalize=True) #use alpha value as determined from above
ridge.fit(X_train_selected, Y_train)
prediction = ridge.predict(X_test_selected)
mae = mean_absolute_error(Y_test, prediction)
print(f'Test Mean Absolute Error: ${mae:,.0f} \n')
# + id="r301sDMfkCAV" colab_type="code" colab={}
| module3-ridge-regression/LS_DS_213_assignment.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: mirror1
# language: python
# name: mirror1
# ---
# # MLFlow Pre-packaged Model Server AB Test Deployment
# In this example we will build two models with MLFlow and we will deploy them as an A/B test deployment. The reason this is powerful is because it allows you to deploy a new model next to the old one, distributing a percentage of traffic. These deployment strategies are quite simple using Seldon, and can be extended to shadow deployments, multi-armed-bandits, etc.
# ## Tutorial Overview
#
# This tutorial will follow closely break down in the following sections:
#
# 1. Train the MLFlow elastic net wine example
#
# 2. Deploy your trained model leveraging our pre-packaged MLFlow model server
#
# 3. Test the deployed MLFlow model by sending requests
#
# 4. Deploy your second model as an A/B test
#
# 5. Visualise and monitor the performance of your models using Seldon Analytics
#
# It will follow closely our talk at the [Spark + AI Summit 2019 on Seldon and MLflow](https://www.youtube.com/watch?v=D6eSfd9w9eA).
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# ## Dependencies
#
# For this example to work you must be running Seldon 0.3.2 or above - you can follow our [getting started guide for this](https://docs.seldon.io/projects/seldon-core/en/latest/workflow/install.html).
#
# In regards to other dependencies, make sure you have installed:
#
# * Helm v2.13.1+
# * kubectl v1.14+
# * Python 3.6+
# * MLFlow 1.1.0
# * pygmentize
#
# We will also take this chance to load the Python dependencies we will use through the tutorial:
# -
import pandas as pd
import numpy as np
from seldon_core.seldon_client import SeldonClient
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# #### Let's get started! 🚀🔥
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# ## 1. Train the first MLFlow Elastic Net Wine example
#
# For our example, we will use the elastic net wine example from [MLflow's tutorial](https://www.mlflow.org/docs/latest/tutorial.html).
# -
# ### MLproject
#
# As any other MLflow project, it is defined by its `MLproject` file:
# !pygmentize -l yaml MLproject
# We can see that this project uses Conda for the environment and that it's defined in the `conda.yaml` file:
# !pygmentize conda.yaml
# Lastly, we can also see that the training will be performed by the `train.py` file, which receives two parameters `alpha` and `l1_ratio`:
# !pygmentize train.py
# ### Dataset
#
# We will use the wine quality dataset.
# Let's load it to see what's inside:
data = pd.read_csv("wine-quality.csv")
data.head()
# ### Training
#
# We've set up our MLflow project and our dataset is ready, so we are now good to start training.
# MLflow allows us to train our model with the following command:
#
# ``` bash
# $ mlflow run . -P alpha=... -P l1_ratio=...
# ```
#
# On each run, `mlflow` will set up the Conda environment defined by the `conda.yaml` file and will run the training commands defined in the `MLproject` file.
# !mlflow run . -P alpha=0.5 -P l1_ratio=0.5
# Each of these commands will create a new run which can be visualised through the MLFlow dashboard as per the screenshot below.
#
# 
#
# Each of these models can actually be found on the `mlruns` folder:
# !tree -L 1 mlruns/0
# ### MLmodel
#
# Inside each of these folders, MLflow stores the parameters we used to train our model, any metric we logged during training, and a snapshot of our model.
# If we look into one of them, we can see the following structure:
# !tree mlruns/0/$(ls mlruns/0 | head -1)
# In particular, we are interested in the `MLmodel` file stored under `artifacts/model`:
# !pygmentize -l yaml mlruns/0/$(ls mlruns/0 | head -1)/artifacts/model/MLmodel
# This file stores the details of how the model was stored.
# With this information (plus the other files in the folder), we are able to load the model back.
# Seldon's MLflow server will use this information to serve this model.
#
# Now we should upload our newly trained model into a public Google Bucket or S3 bucket.
# We have already done this to make it simpler, which you will be able to find at `gs://seldon-models/mlflow/model-a`.
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# ## 2. Deploy your model using the Pre-packaged Moldel Server for MLFlow
#
# Once you have a Kubernetes Cluster running with [Seldon](https://docs.seldon.io/projects/seldon-core/en/latest/workflow/install.html) and [Ambassador](https://docs.seldon.io/projects/seldon-core/en/latest/workflow/install.html#install-ambassador) running we can deploy our trained MLFlow model.
# For this we have to create a Seldon definition of the model server definition, which we will break down further below.
#
# We will be using the model we updated to our google bucket (gs://seldon-models/mlflow/elasticnet_wine), but you can use your model if you uploaded it to a public bucket.
# -
# !pygmentize mlflow-model-server-seldon-config.yaml
# Once we write our configuration file, we are able to deploy it to our cluster by running it with our command
# !kubectl apply -f mlflow-model-server-seldon-config.yaml
# Once it's created we just wait until it's deployed.
#
# It will basically download the image for the pre-packaged MLFlow model server, and initialise it with the model we specified above.
#
# You can check the status of the deployment with the following command:
# !kubectl rollout status deployment.apps/mlflow-deployment-mlflow-deployment-dag-77efeb1
# Once it's deployed, we should see a "succcessfully rolled out" message above. We can now test it!
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# ## 3. Test the deployed MLFlow model by sending requests
# Now that our model is deployed in Kubernetes, we are able to send any requests.
# -
# We will first need the URL that is currently available through Ambassador.
#
# If you are running this locally, you should be able to reach it through localhost, in this case we can use port 80.
# !kubectl get svc | grep ambassador
# Now we will select the first datapoint in our dataset to send to the model.
x_0 = data.drop(["quality"], axis=1).values[:1]
print(list(x_0[0]))
# We can try sending a request first using curl:
# !curl -X POST -H 'Content-Type: application/json' \
# -d "{'data': {'names': [], 'ndarray': [[7.0, 0.27, 0.36, 20.7, 0.045, 45.0, 170.0, 1.001, 3.0, 0.45, 8.8]]}}" \
# http://localhost:80/seldon/default/mlflow-deployment/api/v0.1/predictions
# We can also send the request by using our python client
# +
from seldon_core.seldon_client import SeldonClient
import math
import numpy as np
import subprocess
HOST = "localhost" # Add the URL you found above
port = "80" # Make sure you use the port above
batch = x_0
payload_type = "ndarray"
sc = SeldonClient(
gateway="ambassador",
gateway_endpoint=HOST + ":" + port)
client_prediction = sc.predict(
data=batch,
deployment_name="mlflow-deployment",
names=[],
payload_type=payload_type)
print(client_prediction.response)
# + [markdown] toc-hr-collapsed=true toc-nb-collapsed=true
# ## 4. Deploy your second model as an A/B test
#
# Now that we have a model in production, it's possible to deploy a second model as an A/B test.
# Our model will also be an Elastic Net model but using a different set of parameters.
# We can easily train it by leveraging MLflow:
# -
# !mlflow run . -P alpha=0.75 -P l1_ratio=0.2
# As we did before, we will now need to upload our model to a cloud bucket.
# To speed things up, we already have done so and the second model is now accessible in `gs://seldon-models/mlflow/model-b`.
# ### A/B test
#
# We will deploy our second model as an A/B test.
# In particular, we will redirect 20% of the traffic to the new model.
#
# This can be done by simply adding a `traffic` attribute on our `SeldonDeployment` spec:
# !pygmentize ab-test-mlflow-model-server-seldon-config.yaml
# And similar to the model above, we only need to run the following to deploy it:
# !kubectl apply -f ab-test-mlflow-model-server-seldon-config.yaml
# We can check that the models have been deployed and are running with the following command.
#
# We should now see the "a-" model and the "b-" models.
# !kubectl get pods
# ## 5. Visualise and monitor the performance of your models using Seldon Analytics
#
# This section is optional, but by following the instructions you will be able to visualise the performance of both models as per the chart below.
#
# In order for this example to work you need to install and run the [Grafana Analytics package for Seldon Core](https://docs.seldon.io/projects/seldon-core/en/latest/analytics/analytics.html#helm-analytics-chart).
#
# For this we can access the URL with the command below, it will request an admin and password which by default are set to the following:
# * Username: admin
# * Password: <PASSWORD>
#
# You can access the grafana dashboard through the port provided below:
# !kubectl get svc grafana-prom -o jsonpath='{.spec.ports[0].nodePort}'
# Now that we have both models running in our Kubernetes cluster, we can analyse their performance using Seldon Core's integration with Prometheus and Grafana.
# To do so, we will iterate over the training set (which can be found in `wine-quality.csv`), making a request and sending the feedback of the prediction.
#
# Since the `/feedback` endpoint requires a `reward` signal (i.e. the higher the better), we will simulate one as:
#
# $$
# R(x_{n})
# = \begin{cases}
# \frac{1}{(y_{n} - f(x_{n}))^{2}} &, y_{n} \neq f(x_{n}) \\
# 500 &, y_{n} = f(x_{n})
# \end{cases}
# $$
#
# , where $R(x_{n})$ is the reward for input point $x_{n}$, $f(x_{n})$ is our trained model and $y_{n}$ is the actual value.
# +
def _get_reward(y, y_pred):
if y == y_pred:
return 500
return 1 / np.square(y - y_pred)
def _test_row(row):
input_features = row[:-1]
feature_names = input_features.index.to_list()
X = input_features.values.reshape(1, -1)
y = row[-1].reshape(1, -1)
# Note that we are re-using the SeldonClient defined previously
r = sc.predict(
deployment_name="mlflow-deployment",
data=X,
names=feature_names)
y_pred = r.response.data.tensor.values
reward = _get_reward(y, y_pred)
sc.feedback(
deployment_name="mlflow-deployment",
prediction_request=r.request,
prediction_response=r.response,
reward=reward)
return reward[0]
data.apply(_test_row, axis=1)
# -
# You should now be able to see Seldon's pre-built Grafana dashboard.
# 
# In bottom of the dashboard you can see the following charts:
#
# - On the left: the requests per second, which shows the different traffic breakdown we specified.
# - On the center: the reward, where you can see how model `a` outperforms model `b` by a large margin.
# - On the right, the latency for each one of them.
#
# You are able to add your own custom metrics, and try out other more complex deployments by following further guides at https://docs.seldon.io/projects/seldon-core/en/latest/workflow/README.html
| examples/models/mlflow_server_ab_test_ambassador/mlflow_server_ab_test_ambassador.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import matplotlib.pyplot as plt
import equations as eq
import sympy as sp
from solvers import *
# %matplotlib inline
# -
# To build the discrete operator
#
# \begin{equation}
# A = a(x, y)\partial_x^2 + b(x, y)\partial_y^2 + \alpha(x, y)\partial_x + \beta(x, y)\partial_y + q(x, y) \partial_x\partial_y
# \end{equation}
#
# with the exact solution
#
# \begin{equation}
# u(x, y) = u_{\sf exact}
# \end{equation}
#
# fill coefficients and exact solution in the following template
def my_favourite_equation(L_x, L_y):
x, y = sp.symbols('x, y', real=True)
a = .0
b = .0
alpha = .0
beta = .0
q = .0
exact = .0
return eq.construct_equation(a, b, alpha, beta, q, exact, x, y, L_x, L_y)
# Here $L_x, L_y$ define the physical space $x, y \in \left[0, L_x\right]\times\left[0, L_y\right]$.
# As an example we consider the following equation
#
# \begin{equation}
# \cosh(x y)\partial^2_x + \left[1 + \cos(\pi x^2 y)\right]^2 \partial_y^2 + \exp(x)\partial_x + \exp(y)\partial_y + (1-x)(1-y)\partial_x\partial_y,
# \end{equation}
#
# \begin{equation}
# u_{\sf exact} = x + y + 3x^3
# \end{equation}
def equation_1633(L_x, L_y):
x, y = sp.symbols('x, y', real=True)
a = sp.cosh(x*y)
b = (1 + sp.cos(sp.pi*x**2*y))**2
alpha = sp.exp(x)
beta = sp.exp(y)
q = (1-x)*(1-y)
exact = x + y + 3*x**3
return eq.construct_equation(a, b, alpha, beta, q, exact, x, y, L_x, L_y)
# When equation is defined we pass it to the function that construct ``coo_matrix``
# +
J = 5
n_x = n_y = 2**5
h = 2**-5
A, rhs, exact = eq.construct_matrix(equation_1633, 'Dirichlet', 1, 1, n_x, n_y)
# -
# We convert ``coo_matrix`` to dense format of ``numpy``
A = A.toarray()
# Now, we can run available solvers.
# BiCGSTAB (scipy)
solution, E_CG = BICGSTAB(A, rhs, tol=1e-15, verbose=True, write=True)
np.linalg.norm(solution - exact)
# Gauss-Seidel
solution, E_GS = GS(A, rhs, tol=h**2, verbose=True, write=True)
np.linalg.norm(solution - exact)
# Belief propagation split solver (see the article for details)
solution, E_split = split_BP_solver(A, rhs, tol=h**2, verbose=True, write=True)
np.linalg.norm(solution - exact)
solution_1, E_BP = GaBP(A, rhs, tol=h**2, verbose=True, write=True)
np.linalg.norm(solution - exact)
# For other available solvers see the notebook that reproduces figures from the article.
# Now it is possible to access convergence history. For example for the split solver
plt.plot(np.log(E_split))
# or for the Gaussian belief propagation solver
plt.plot(np.log(E_BP))
| template for your equations.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Performance metrics
#
# In this notebook we return to the **supervised learning** algorithms we trained in the last notebook, but dive deeper into how to evaluate them. As we saw, **classification** and **regression** each have their own performance metrics. Classification itself can be sub-divided into **binary** and **multi-class** classification with a set of metrics for each.
# The data and code-base in this notebook is very similar to the last notebook. However the exercises have changed to reflect the topic of this chapter.
#
# We use a bank marketing data, which has demographic and activity data about bank customers, as well as information about previous attempts to contact them for a marketing campain. The target `y` is binary and indicates whether the client signed up for a term deposit or not. Let's load the data again. You can read more about the data [here](https://archive.ics.uci.edu/ml/datasets/Bank+Marketing).
# +
import pandas as pd
import numpy as np
bank = pd.read_csv("data/bank-full.csv", sep = ";")
bank.head()
# -
# Since numeric and categorical features are often pre-processed differently, we will create variables that store the names of each to make it easier to refer to them later.
# +
num_cols = bank.select_dtypes(['integer', 'float']).columns
cat_cols = bank.select_dtypes(['object']).drop(columns = "y").columns
print("Numeric columns are {}.".format(", ".join(num_cols)))
print("Categorical columns are {}.".format(", ".join(cat_cols)))
# -
# As usual before we can proceed to machine learning, we need to get the data ready. And since we're doing supervised learning, we need to set aside a test data set to later be evaluate the model. So let's begin by splitting the data.
# +
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(bank.drop(columns = "y"), bank["y"],
test_size = 0.15, random_state = 42)
# -
X_train = X_train.reset_index(drop = True)
X_test = X_test.reset_index(drop = True)
print(f"Training data has {X_train.shape[0]} rows.")
print(f"Test data has {X_test.shape[0]} rows.")
# Before we begin our journey of trying out different algorithms in `sklearn` we do need to encode our categorical features.
# +
from sklearn.preprocessing import OneHotEncoder
onehoter = OneHotEncoder(sparse = False, drop = "first")
onehoter.fit(X_train[cat_cols])
onehot_cols = onehoter.get_feature_names(cat_cols)
X_train_onehot = pd.DataFrame(onehoter.transform(X_train[cat_cols]), columns = onehot_cols)
X_test_onehot = pd.DataFrame(onehoter.transform(X_test[cat_cols]), columns = onehot_cols)
# -
# Some algorithms we're going to use (such as decision tree) won't require that we normalize our numeric features, but most will. Not doing so won't break the algorithm, but just as we saw in the case of k-means, it will skew the results. So let's Z-normalize our numeric features now.
# +
from sklearn.preprocessing import StandardScaler
znormalizer = StandardScaler()
znormalizer.fit(X_train[num_cols])
X_train_norm = pd.DataFrame(znormalizer.transform(X_train[num_cols]), columns = num_cols)
X_test_norm = pd.DataFrame(znormalizer.transform(X_test[num_cols]), columns = num_cols)
X_train_norm.head()
# -
# We now join our numeric features and our one-hot-encoded categorical features into one data set that we pass to the decision tree classifier.
# +
X_train_featurized = X_train_onehot # add one-hot-encoded columns
X_test_featurized = X_test_onehot # add one-hot-encoded columns
X_train_featurized[num_cols] = X_train_norm # add numeric columns
X_test_featurized[num_cols] = X_test_norm # add numeric columns
del X_train_norm, X_test_norm, X_train_onehot, X_test_onehot
print("Featurized training data has {} rows and {} columns.".format(*X_train_featurized.shape))
print("Featurized test data has {} rows and {} columns.".format(*X_test_featurized.shape))
# -
# ## Decision tree classifier
#
# With our data ready, we can now train a decision tree classifier. There is a lot of detail that we leave for another time, but the common pattern to all the supervised learning algorithm is what we want to call attention to here:
#
# 1. We create an **instance** of the algorithm, along with any settings we want to use. Here we instantiate a `DecisionTreeClassifier` and specify `max_depth = 10`.
# 1. We train the algorithm on the training data by calling the `fit` method.
# 1. Once the model is trained, we obtain prediction by calling the `predict` method.
# +
from sklearn.tree import DecisionTreeClassifier
dtree = DecisionTreeClassifier(max_depth = 5)
dtree.fit(X_train_featurized, y_train)
y_hat_train = dtree.predict(X_train_featurized)
y_hat_test = dtree.predict(X_test_featurized)
# -
# If we wish to evaluate the model, we only need to predict for the test data, but in our case we predict both for the training and test data so we can show the effect of overfitting or underfitting.
# +
from sklearn.metrics import accuracy_score
acc_train = accuracy_score(y_train, y_hat_train) * 100
acc_test = accuracy_score(y_test, y_hat_test) * 100
print("Accuracy on the training data: {:.0f}%.".format(acc_train))
print("Accuracy on the test data: {:.0f}%.".format(acc_test))
# -
# ### Exercise
#
# - Find the counts of positive and negative classes for the target variable.
# - Based on the counts, would you recommend looking using **accuracy** to measure the model's performance?
# - Check out what other performance metrics are available in `sklearn.metrics` that could be relevant to evaluating this model. Choose one and report the value.
# ### End of exercise
# ## k-nearest neighbor classifier
#
# A model trained using the k-nearest neighbor algorithm on the other hand is very different. It doesn't have a tree structure. Instead it labels a new data point by finding the $k$ points nearest to it and looking up what their labels are, and letting the new data's label be the same as whatever label the majority of its neighbors have. Optionally, we can let closer neighbors influence the vote more than more distant neighbors.
# +
from sklearn.neighbors import KNeighborsClassifier
knnb = KNeighborsClassifier()
knnb.fit(X_train_featurized, y_train)
y_hat_train = knnb.predict(X_train_featurized)
y_hat_test = knnb.predict(X_test_featurized)
# -
# If we wish to evaluate the model, we only need to predict for the test data, but in our case we predict both for the training and test data so we can show the effect of overfitting or underfitting.
# +
acc_train = accuracy_score(y_train, y_hat_train) * 100
acc_test = accuracy_score(y_test, y_hat_test) * 100
print("Accuracy on the training data: {:.0f}%.".format(acc_train))
print("Accuracy on the test data: {:.0f}%.".format(acc_test))
# -
# ### Exercise
#
# - Get **precision** and **recall** for the model we trained above. Note that by default, the corresponding functions in `sklearn.metrics` expect the positive label to be the integer 1. For us, the positive label is the string `yes`, so we need to use `pos_label = 'yes'`.
# - Instead of calling the `predict` method to get predictions, call the `predict_proba` method to get the probability $P(Y_i = 1)$ for each row.
# - Change your the threshold from 0.50 (default) to 0.75 and based on this new threshold obtain hard predictions from the soft predictions we got in the last step.
# - Obtain **precision** and **recall** once more (now that we changed the threshold).
# - How did increasing the threshold change precision and recall.
# ### End of exercise
# ## Logistic regression classifier
#
# The logistic regression algorithm is another popular classifier. Careful here: even though it has the word **regression** in it, logistic regression is a **classification** algorithm, not a **regression** algorithm. A model trained using logistic regression predicts new classes using an **equation**. This makes logistic regression very efficient. In fact, once you have your trained model, you can pull out the equation's **coefficients** and implement it even in SQL: in just one query, although if we have a lot of features it could be nasty query!
# +
from sklearn.linear_model import LogisticRegression
logit = LogisticRegression(max_iter = 5000)
logit.fit(X_train_featurized, y_train)
y_hat_train = logit.predict(X_train_featurized)
y_hat_test = logit.predict(X_test_featurized)
# -
# Let's look at precision and recall on the training and test data.
# +
from sklearn.metrics import precision_score, recall_score
precision_train = precision_score(y_train, y_hat_train, pos_label = 'yes') * 100
precision_test = precision_score(y_test, y_hat_test, pos_label = 'yes') * 100
recall_train = recall_score(y_train, y_hat_train, pos_label = 'yes') * 100
recall_test = recall_score(y_test, y_hat_test, pos_label = 'yes') * 100
print("Precision = {:.0f}% and recall = {:.0f}% on the training data.".format(precision_train, recall_train))
print("Precision = {:.0f}% and recall = {:.0f}% on the test data.".format(precision_test, recall_test))
# -
# ## SVM classifier
#
# It looks like so far logistic regression performs better than the other two models right off the bat. So it might be time to compare its performance against one of the more advanced algorithms. Let's train an SVM model. SVM stands for support vector machines and before neural networks and deep learning started making a comeback in the last few years, SVMs were considered state of the art. As you will notice from running the next line, SVMs are also very compute-heavy.
#
# Note that by default, SVMs are **hard classifiers**, but by specifying `probability = True` we can get it to return soft predictions. Unfortunately, this comes at an added computational cost. But we need the soft predictions for later so we can plot an ROC plot.
# +
from sklearn.svm import SVC
svmc = SVC(probability = True)
svmc.fit(X_train_featurized, y_train)
y_hat_train = svmc.predict(X_train_featurized)
y_hat_test = svmc.predict(X_test_featurized)
# -
# ### Exercise
#
# Obtain precision and recall for the SVM classifier we trained above and compare them to the ones we got from training the logistic regression. What are your conclusion?
# ### End of exercise
# ## Comparing classifiers using the ROC curve and AUC
#
# So we trained so far four binary classification models. Having to look at precision and recall to determine which is best might be a little tedious, especially since in some cases there are trade-offs involved. Not to mention that the precision and recall metrics, as we saw in an earlier exercise, depend on a threshold. By default the threshold is set to 0.50, but we can change that and if we do we get new values for precision and recall, which means we have to go back to comparing our models again.
#
# This is when the ROC plot can be helpful. Unlike precision and recall, ROC is not a single metric but a graph. The ROC curve illustrates the trade-off that happens as we change our threshold from zero to 1. The closer the ROC curve comes to the top-left corner of the plot (the $(0, 1) point$, the better the classifier. If we have two classifiers $A$ and $B$, and the ROC curve of $A$ is higher than that of $B$ **at every point**, then this classifier $A$ outperforms $B$ **regardless of what threshold we choose**.
# +
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
def plot_roc(models, model_names):
plt.figure(0, figsize = [8, 7]).clf()
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0, 1])
plt.ylim([0, 1])
plt.ylabel('True Positive Rate')
plt.xlabel('False Positive Rate')
for ii, model in enumerate(models):
y_prob_test = model.predict_proba(X_test_featurized)[:, 1]
fpr, tpr, threshold = roc_curve(y_test, y_prob_test, pos_label = "yes")
roc_auc = auc(fpr, tpr)
fpr, tpr, threshold = roc_curve(y_test, y_prob_test, pos_label = "yes")
roc_auc = auc(fpr, tpr)
plt.plot(fpr, tpr, label = "{} AUC = {:0.2f}".format(model_names[ii], roc_auc))
plt.legend(loc = 'lower right');
# -
# An ROC curve for a single classifer is not very useful, but it's when we want to compare multiple classifiers that the ROC curve can save us a lot of time. Since creating an ROC curve can be tedious, the above cell has a function that does the hard work. When we call the function, all we need to do is give it the models we trained, in a list, and corresponding labels for each. In addition to plotting the ROC curve, the plot will also show the AUC (area under the ROC curve). The closer the AUC is to 1, the better the model.
plot_roc([logit, knnb, dtree, svmc], ['logistic', 'k-nearest-nb', 'decision-tree', 'SVM'])
# ## Multi-class classification
#
# We saw quite a few examples of training binary classification models. Now we're going to see examples of **multi-class classification**, namely when the number of classes is more than 2. As it turns out, multi-class classification isn't really that special. For example, one approach would be to build many classifiers, each one of each is going to distinguish one of the classes from the rest of them. This is referred to as **one-vs-all** or **one-vs-rest**. To get a prediction, we let each model predict with **soft predictions** and we predict the class to be whichever class obtained the highest probability.
#
# However, this can be very inefficient when the number of classes is high, and in some use-cases such as image classification the number of classes can be in the hundreds of thousands! As we will see in future lectures, neural networks can train multi-class classifier using a single model, which is a far superior approach than one-vs-rest.
#
# We train our multi-class classifier to predict the `job` column in the data. But because there are too many classes, we first reduce the number of classes by combining some of them. To do that, we use a remapping dictionary and pass it to the `replace` method of the `DataFrame`. We then change the target to be this new variable and drop the one-hot-encoded features related to `job` from the training and test data (otherwise we would be using `job` to predict `job` and we don't need ML to do that!).
# +
remap = {'entrepreneur': 'white-collar', 'housemaid': 'blue-collar', 'admin.': 'white-collar',
'management': 'white-collar', 'self-employed': 'self-emp', 'services': 'self-emp',
'student': 'unemployed', 'technician': 'blue-collar', 'unknown': 'self-emp'}
y_train = X_train['job'].replace(remap)
edu_onehot_cols = X_train_featurized.filter(like = 'job').columns
print(edu_onehot_cols)
X_train_featurized = X_train_featurized.drop(columns = edu_onehot_cols)
y_test = X_test['job'].replace(remap)
X_test_featurized = X_test_featurized.drop(columns = edu_onehot_cols)
y_train.value_counts(normalize = True)
# -
# ## Multi-class logistic regression
#
# In many cases, we can run exact same code with run to train the binary classifier, and train a multi-class classifier instead. **Logistic regressoion** is a great example to try our multi-class classification on. The reason is that logistic regression is very efficient and is able to train a true multi-class classifier, and not one-vs-rest.
# +
from sklearn.linear_model import LogisticRegression
logit = LogisticRegression(max_iter = 5000)
logit.fit(X_train_featurized, y_train)
y_hat_train = logit.predict(X_train_featurized)
y_hat_test = logit.predict(X_test_featurized)
# -
# Multi-class classification adds its own set of complications when it comes to model evaluation. But let's begin with the easy part: **accuracy is still accuracy**. In other words, whether we predict two classes or many, accuracy is still the precentage of correct predictions.
# +
acc_train = accuracy_score(y_train, y_hat_train) * 100
acc_test = accuracy_score(y_test, y_hat_test) * 100
print("Accuracy on the training data: {:.0f}%.".format(acc_train))
print("Accuracy on the test data: {:.0f}%.".format(acc_test))
# -
# But just as in the binary case, if the data has class imbalance, accuracy may paint too rosy a picture. One solution here is to use a **weighted accuracy** where the weights are chosen to give more importance to the classes we wish to emphasize. For example, in the next cell, we assgin `retired` and `unemployed` to have 10 times more weight than the remaining categories. We then measure weighted accuracy using the `sample_weight` argument.
# +
is_retired_or_self = y_test.isin(['retired', 'unemployed'])
y_test_wt = is_retired_or_self * 100 + ~is_retired_or_self * 1
acc_test_wt = accuracy_score(y_test, y_hat_test, sample_weight = y_test_wt) * 100
print("Weighted cccuracy on the test data: {:.0f}%.".format(acc_test_wt))
# -
# There are two things to note here:
# - A weighted accuracy measure as computed above does not really have too much value in terms of explainability. They mostly serve us to have a measure to tune our models with. In other words, we can use something like the weighted accuracy as defined above to evaluate a few different models and see which one has the best performance. But the exact value of the weighted accuracy isn't something particularly noteworthy since our choice of weights wasn't something we put a lot of thought into.
# - We can use weights at the time we **evaluate** a model, like we did in the above example, but we can also use weights at the time we **train** a model. Many algorithms such as `LogisticRegression` have an argument usally named `class_weight` which allows you to assign higher weights to certain classes so you can over-emphasize them **during training**. This way the model can focus on improving its predictions for those classes at the expense of the other classes.
# What about measures like **precision** and **recall**? They still apply in a multi-class classification setting, but now we need to calculate precision and recall **for each class** and then average them out. To calculate precison and recall for each class, we use a **one-vs-rest** approach. To average out the values for each class, we can use a simple average (called a **macro average** here) which gives each class equal weights, or we can weigh classes by their sample size (called **support** in the results below) and caluclate a **weighted average**.
#
# Of course we don't need to do any of that manually (phew!): we can use the `classification_report` function for that.
# +
from sklearn.metrics import classification_report
cl_report_train = classification_report(y_train, y_hat_train, zero_division = 0)
cl_report_test = classification_report(y_test, y_hat_test, zero_division = 0)
print("For training data:\n")
print(cl_report_train)
print("=====================================================\n")
print("For test data:\n")
print(cl_report_test)
# -
# One last word of caution about multi-class classification. We saw in the above example that `LogisticRegression` can easily and rather efficiently accomodate multi-class classification as well. However, not all algorithms are as generous! For example, the `SVC` algorithm we used in our binary classification example can also be used for multi-class classification, but at a great cost: as [explained here](https://scikit-learn.org/stable/modules/svm.html#multi-class-classification) `SVC` uses **one-vs-one** for multi-class classification. In other words, it builds a separate classifier to predict one class vs another class. If we have $m$ classes, we have $m \choose 2$ = $\frac{m!}{(m-2)!2!}$ (the left-hand-side reads **$m$ choose 2**) classifiers to build, which for even small values of $m$ can quickly get out of control. So it's important to read the documentation of each classifier to be aware of these limits.
# ## Linear regression regressor
# So far we've only seen classification algorithms. So it's time to change course and take a look at regression algorithms. For that we need to find a numeric target. We can use the `duration` column in the data as our target.
# +
y_train = X_train_featurized['duration']
X_train_featurized = X_train_featurized.drop(columns = 'duration')
y_test = X_test_featurized['duration']
X_test_featurized = X_test_featurized.drop(columns = 'duration')
# -
# Other than changing the target from categorical to numeric, we don't have to do things very differently from before. The training and predicting part of the code remain very similar.
# +
from sklearn.linear_model import LinearRegression
linreg = LinearRegression()
linreg.fit(X_train_featurized, y_train)
y_hat_train = linreg.predict(X_train_featurized)
y_hat_test = linreg.predict(X_test_featurized)
# -
# We've almost reached the end of the notebook and are just starting to talk about regression. This is because regression algorithms are more straight-forward. None of the topics we covered when we talked about classifications are really relevant in regression. Instead, we have a short list of metrics that can be used to tell us how close the prediction comes to the actual value (**root mean squared error** or **mean absolute error**) or by how much we were able to reduce our **uncertainty** (variability) about the target by modeling it using the **featuers** ($R^2$ and **adjusted $R^2$).
# +
from sklearn.metrics import mean_squared_error
rmse_train = mean_squared_error(y_train, y_hat_train) ** 0.5
rmse_test = mean_squared_error(y_test, y_hat_test) ** 0.5
print("RMSE on the training data: {:5.5f}.".format(rmse_train))
print("RMSE on the test data: {:5.5f}.".format(rmse_test))
# -
# ### Exercise
#
# - Find the MAE (mean absolute error) of the model trained above. How does it compare to the RMSE?
# - Find the $R^2$ (coefficient of determination) of the model trained above. How would you interpret this number?
# - Find the correlation between the predicted and actual values.
# - Show the distribution of the errors using `displot` in the `seaborn` package. What does the distribution suggest about the errors?
# ### End of exercise
# Of course there is always more we can be looking at if we want to get in the weeds. And model evaluation at the end of the day is similar to EDA (exploratory data analysis) in that you have a standard set of checks, but then you can get creative depending on what you're trying to answer. As an example, let's say we are wondering how our confidence about the prediction for `duration` depends on `marital`. To answer this we need to quantify what we mean by "confidence". That's simple: if our prediction is good then error should be low. So our confidence about the prediction can be measured using the standard deviation of the error. In other words, we can compute the standard deviation of the error grouped by `marital` to answer our question:
X_test['error'] = y_test - y_hat_test # compute the error
X_test['error'].groupby(X_test['marital']).std()
# The above example shows the importance of having some ida **ahead-of-time** of what metrics should be used to evaluate the model. Whether it's one or a few standard metrics or some pre-defined custom metric, clarifying it ahead of time can save us some time and **prevent** us from having to go fishing for the best performance metric **after training**. In fact, doing so can be dangerous and result in over-fitting. In future classes, we learn that if we need to do this properly, in addition to the training and test set, we should also be using a **validation set**.
| lesson_9.ipynb |
# +
"""
17. How to compute the mean squared error on a truth and predicted series?
"""
"""
Difficulty Level: L2
"""
"""
Compute the mean squared error of truth and pred series.
"""
"""
Input
"""
"""
truth = pd.Series(range(10))
pred = pd.Series(range(10)) + np.random.random(10)
"""
# Input
truth = pd.Series(range(10))
pred = pd.Series(range(10)) + np.random.random(10)
# Solution
np.mean((truth-pred)**2)
| pset_pandas_ext/101problems/solutions/nb/p17.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ---
# layout: post
# title: "Entropy와 Gini계수"
# author: "<NAME>"
# categories: Data분석
# tags: [DecisionTree, 의사결정나무, 불순도, Entropy와, Gini, 엔트로피, 지니계수, InformationGain, information]
# image: 03_entropy_gini.png
# ---
# ## **목적**
# - 지난번 포스팅에 ensemble 모델에 관하여 이야기하면서 약한 모형으로 의사결정나무를 많이 사용하는 것을 알 수 있었습니다. 이번에는 의사결정 나무를 만들기 위하여 사용되는 Entropy와 gini index에 대해서 알아보도록 하겠습니다.
# <br/>
# <br/>
#
# ### **트리 구축의 원칙**
# 
# > 출처 : https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011
# - 결정 트리를 구축할 때는 Occamm의 면도날처럼 데이터의 특성을 가장 잘 반영하는 간단한 가설을 먼저 채택하도록 되어있습니다. 어떻게 간단하고 합리적인 트리를 만들 수 있을 지 알아보겠습니다.
# <br>
# <br>
# ---
#
# ### **1. 결정 트리**
# 의사결정나무를 효율적으로 만들기 위해서는 변수의 기준에 따라 불순도/불확실성을 낮추는 방식으로 선택하여 만들게 됩니다.<br>
# 이에 불순도(Impurity) / 불확실성(Uncertainty)를 감소하는 것을 Information gain이라고 하며 이것을 최소화시키기 위하여 Gini Index와 Entropy라는 개념을 사용하게 되고 의사결정 나무의 종류에 따라 다르게 쓰입니다.<br>
# sklearn에서 default로 쓰이는 건 gini계수이며 이는 CART(Classificatioin And Regression Tree)에 쓰입니다.<br>
# ID3 그리고 이것을 개선한 C4.5, C5.0에서는 Entropy를 계산한다고 합니다. <br>
# CART tree는 항상 2진 분류를 하는 방식으로 나타나며, Entropy 혹은 Entropy 기반으로 계산되는 Information gain으로 계산되며 다중 분리가 됩니다. <br>
#
# - Gini계수와 Entropy 모두 높을수록 불순도가 높아져 분류를 하기 어렵습니다. <br>
# 
#
# |비 고|ID3|C4.5, C5|CART|
# |:---:|:---:|:---:|:---:|
# |평가지수|Entropy|Information gain|Gini Index(범주), 분산의 차이(수치)|
# |분리방식|다지분리|다지분리(범주) 및 이진분리(수치)|항상2진 분리|
# |비고|수치형 데이터 못 다룸|||
#
# <br>
# <br>
# > 출처/참고자료 : https://ko.wikipedia.org/wiki/%EA%B2%B0%EC%A0%95_%ED%8A%B8%EB%A6%AC_%ED%95%99%EC%8A%B5%EB%B2%95 <br>
# > 출처/참고자료 : https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=trashx&logNo=60099037740 <br>
# > 출처/참고자료 : https://ratsgo.github.io/machine%20learning/2017/03/26/tree/
# ---
#
# ### **1. Gini Index**
# 일단 sklearn의 DecisionTreeClassifier의 default 값인 Gini 계수에 대해서 먼저 설명하겠습니다. <br>
# 우선 Gini index의 공식입니다. <br>
#
# - 영역의 데이터 비율을 제곱하여 더한 값을 1에서 빼주게 된다.<br>
# 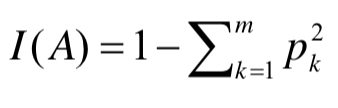 <br>
# <br>
# - 두개 영역 이상이 되면 비율의 제곱의 비율을 곱하여 1에서 빼주게 된다.<br>
# 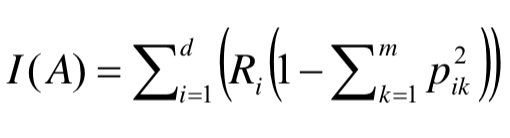
# > 출처 : https://soobarkbar.tistory.com/17
#
# <br>
#
# - 최대값을 보게되면 1 - ( (1/2)^2 + (1/2)^2 ) = 0.5
# - 최소값을 보게되면 1 - ( 1^2 + 0^2 ) = 0
# +
import os
import sys
import warnings
import math
import random
import numpy as np
import pandas as pd
import scipy
from sklearn import tree
from sklearn.tree import DecisionTreeClassifier
import matplotlib as mpl
from matplotlib import pyplot as plt
from plotnine import *
import graphviz
from sklearn.preprocessing import OneHotEncoder
# %matplotlib inline
warnings.filterwarnings("ignore")
# -
tennis = pd.read_csv("data/tennis.csv", index_col = "Day")
tennis
# - 위와 같은 데이터가 있다고 할 때, 우리는 어떤 요인이 가장 확실한(불확실성이 적은) 변수일지 생각을 하고 트리를 만들어야합니다.
# <br>
# <br>
#
# 아무것도 나누지 않았을 때 gini계수를 구하는 함수를 만든 후 얼마인지 출력해보겠습니다
def get_unique_dict(df) :
return {x : list(df[x].unique()) for x in ["Outlook", "Temperature", "Humidity", "Wind"]}
def get_gini(df, y_col) :
Ys = df[y_col].value_counts()
total_row = len(df)
return 1 - np.sum([np.square(len(df[df[y_col] == y]) / total_row) for y in Ys.index])
def gini_split(df, y_col, col, feature) :
r1 = len(df[df[col] == feature])
Y1 = dict(df[df[col] == feature][y_col].value_counts())
r2 = len(df[df[col] != feature])
Y2 = dict(df[df[col] != feature][y_col].value_counts())
ratio = r1 / (r1 + r2)
gi1 = 1 - np.sum([np.square(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) for x, y in Y1.items()])
gi2 = 1 - np.sum([np.square(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) for x, y in Y2.items()])
return (ratio * gi1) + ((1-ratio) * gi2)
# 어떤 기준으로 나누었을 때 gini계수를 구하는 함수를 만들어 예시로 Outlook이 Sunny일 때 gini 계수를 구해보겠습니다.
get_gini(tennis, "PlayTennis")
# 아무것도 나누지 않았을 때보다, Sunny로 나누었을 때 gini계수가 줄어드는 것을 볼 수 있습니다.<br>
# 이 때 이 차이값을 Information gain(정보획득)이라고 합니다. 그리고 정보획득량이 많은 쪽을 선택하여 트리의 구조를 만들기 시작합니다.
split_point = ["Outlook", "Sunny"]
print("{}, {} 기준 split 후 gini 계수 : {}".format(*split_point, gini_split(tennis, "PlayTennis", *split_point)))
print("information gain : {}".format(get_gini(tennis, "PlayTennis") - gini_split(tennis, "PlayTennis", *split_point)))
# - 이제 모든 변수에 대해서 각각의 gini계수를 구하여 정보획득량이 많은, 즉 gini계수가 적은 변수를 선정하여 트리를 만들어갑니다.
y_col = "PlayTennis"
unique_dict = get_unique_dict(tennis)
unique_dict
[f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis, y_col, idx, v)}" for idx, val in unique_dict.items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis, y_col, idx, v)] for idx, val in unique_dict.items() for v in val], columns = ["cat1", "cat2", "gini"])
print(gini_df.iloc[gini_df["gini"].argmax()])
print(gini_df.iloc[gini_df["gini"].argmin()])
# ---
# 임의로 x, y좌표를 생성하여 정보들이 얼마나 흩어져있는지 확인해보겠습니다.
def generate_xy(df, split_col = None, split_value = None) :
if split_col == None :
return df.assign(x = [random.random() for _ in range(len(df))], y = [random.random() for _ in range(len(df))])
else :
tmp_ = df[df[split_col] == split_value]
tmp__ = df[df[split_col] != split_value]
return pd.concat([tmp_.assign(x = [random.random() / 2 for _ in range(len(tmp_))], y = [random.random() for _ in range(len(tmp_))]),
tmp__.assign(x = [(random.random() / 2) + 0.5 for _ in range(len(tmp__))], y = [random.random() for _ in range(len(tmp__))])] )
# - 아무런 기준을 두지 않았을 때는 정보를 구분할 수 있는 정보가 없습니다.
p = (
ggplot(data = generate_xy(tennis), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/1.jpg")
# 
# - Outlook이 Overcast로 나누었을 때, Yes 4개가 확실히 구분되는 것을 볼 수 있습니다.
split_list = ["Outlook", "Overcast"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/2.jpg")
# 
# - 정보획득량이 가장 큰 Temperature가 Mild로 나누었을 때입니다.
split_list = ["Temperature", "Mild"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/3.jpg")
# 
# - Outlook이 Sunny, Rain으로 각각 나누었을 때입니다.
split_list = ["Outlook", "Sunny"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save(filename = "../assets/img/2021-06-01-Entropy/4.jpg")
# 
split_list = ["Outlook", "Rain"]
p = (
ggplot(data = generate_xy(tennis, *split_list), mapping = aes(x = "x", y = "y", color = y_col)) +
geom_point() +
geom_vline(xintercept = 0.5, color = "red", alpha = 0.7) +
theme_bw()
)
p.save("../assets/img/2021-06-01-Entropy/5.jpg")
# 
# #### **실제 tree 모델과 비교하기 위하여 OneHotEncoding 후 트리모형을 돌려보도록 하겠습니다.
cols = ["Outlook", "Temperature", "Humidity", "Wind"]
oe = OneHotEncoder()
Xs = pd.get_dummies(tennis[cols])
Ys = tennis[y_col]
dt_gini = DecisionTreeClassifier(criterion="gini")
dt_gini.fit(Xs, Ys)
def save_graphviz(grp, grp_num) :
p = graphviz.Source(grp)
p.save(filename = f"../assets/img/2021-06-01-Entropy/{grp_num}")
p.render(filename = f"../assets/img/2021-06-01-Entropy/{grp_num}", format = "jpg")
grp = tree.export_graphviz(dt_gini, out_file = None, feature_names=Xs.columns,
class_names=Ys.unique(),
filled=True)
save_graphviz(grp, 6)
# 
# #### **실제로 이 순서가 맞는지 확인해보겠습니다**
get_gini(tennis, "PlayTennis")
gini_df.iloc[gini_df["gini"].argmin()]
tennis_node1 = tennis[tennis["Outlook"] != "Overcast"]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_node1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_node1).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_node1, y_col, idx, v)] for idx, val in get_unique_dict(tennis_node1).items() for v in val], columns = ["cat1", "cat2", "gini"])
print("")
print("gini index : {}".format(get_gini(tennis_node1, y_col)))
print(gini_df.iloc[gini_df["gini"].argmin()])
tennis_node2 = tennis[(tennis["Outlook"] != "Overcast") & (tennis["Humidity"] == "High")]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_node2, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_node2).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_node2, y_col, idx, v)] for idx, val in get_unique_dict(tennis_node2).items() for v in val], columns = ["cat1", "cat2", "gini"])
print("")
print("gini index : {}".format(get_gini(tennis_node2, y_col)))
gini_df.iloc[gini_df["gini"].argmin()]
# #### - gini계수가 0이면 가장 끝쪽에 있는 terminal node가 됩니다.(데이터가 많으면 overfitting을 막기위하여 가지치기 컨셉이 활용됩니다)
tennis_ter1 = tennis[tennis["Outlook"] == "Overcast"]
[print(f"col : {idx}, split_feature : {v} : gini_index = {gini_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
gini_df = pd.DataFrame([[idx, v, gini_split(tennis_ter1, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "gini"])
gini_df.iloc[gini_df["gini"].argmin()]
# ---
#
# ### **2. Entropy**
# 다음은 ID3, C4.5 등 트리에서 정보획득량을 측정하기 위해 쓰이는 Entropy입니다.<br>
# 우선 Entropy의 공식입니다. <br>
#
# - 영역의 데이터 비율을 제곱하여 더한 값을 1에서 빼주게 된다.<br>
# 
# <br>
max_entropy = (-1 * ((0.5*np.log2(0.5)) + (0.5*np.log2(0.5))))
min_entropy = (-1 * ((1*np.log2(1))))
print(f"Entropy의 최대값 : {max_entropy}")
print(f"Entropy의 최대값 : {min_entropy}")
tennis
def get_entropy(df, y_col) :
Ys = df[y_col].value_counts()
total_row = len(df)
(-1 * ((0.5*np.log2(0.5)) + (0.5*np.log2(0.5))))
return -1 * np.sum([(len(df[df[y_col] == y]) / total_row) * np.log2(len(df[df[y_col] == y]) / total_row) for y in Ys.index])
get_entropy(tennis, y_col)
def entropy_split(df, y_col, col, feature) :
r1 = len(df[df[col] == feature])
Y1 = dict(df[df[col] == feature][y_col].value_counts())
r2 = len(df[df[col] != feature])
Y2 = dict(df[df[col] != feature][y_col].value_counts())
ratio = r1 / (r1 + r2)
ent1 = np.sum([(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) * np.log2(len(df[(df[col] == feature) & (df[y_col] == x)]) / r1) for x, y in Y1.items()])
ent2 = np.sum([(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) * np.log2(len(df[(df[col] != feature) & (df[y_col] == x)]) / r2) for x, y in Y2.items()])
return -1 * ((ratio * ent1) + ((1-ratio) * ent2))
entropy_split(tennis, "PlayTennis", "Outlook", "Sunny")
# Entropy 역시 gini index와 똑같은 개념으로 아무것도 나누지 않았을 때보다, Sunny로 나누었을 때 줄어드는 것을 볼 수 있습니다.<br>
# 이 때 차이값(Information gain)을 이용하여 트리를 만들면 ID3, C4.5 등의 트리 구조를 만들게 됩니다.
[f"col : {idx}, split_feature : {v} : Entropy = {entropy_split(tennis, y_col, idx, v)}" for idx, val in get_unique_dict(tennis).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in unique_dict.items() for v in val], columns = ["cat1", "cat2", "entropy"])
print(entropy_df.iloc[entropy_df["entropy"].argmin()])
print(entropy_df.iloc[gini_df["gini"].argmax()])
# #### **실제 tree 모델과 비교하기 위하여 OneHotEncoding 후 트리모형을 돌려보도록 하겠습니다.**
dt_entropy = DecisionTreeClassifier(criterion="entropy")
dt_entropy.fit(Xs, Ys)
grp = tree.export_graphviz(dt_entropy, out_file = None, feature_names=Xs.columns,
class_names=Ys.unique(),
filled=True)
save_graphviz(grp, 7)
# 
# #### **실제로 이 순서가 맞는지 확인해보겠습니다**
get_entropy(tennis, "PlayTennis")
entropy_df.iloc[entropy_df["entropy"].argmin()]
tennis_ter1 = tennis[tennis["Outlook"] == "Overcast"]
[print(f"col : {idx}, split_feature : {v} : entropy = {entropy_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "entropy"])
entropy_df.iloc[entropy_df["entropy"].argmin()]
tennis_ter1 = tennis[tennis["Outlook"] != "Overcast"]
[print(f"col : {idx}, split_feature : {v} : entropy = {entropy_split(tennis_ter1, y_col, idx, v)}") for idx, val in get_unique_dict(tennis_ter1).items() for v in val]
entropy_df = pd.DataFrame([[idx, v, entropy_split(tennis, y_col, idx, v)] for idx, val in get_unique_dict(tennis_ter1).items() for v in val], columns = ["cat1", "cat2", "entropy"])
entropy_df.iloc[entropy_df["entropy"].argmin()]
# ---
# ### **마지막으로 gini index와 entropy를 활용한 tree가 어떻게 노드가 나뉘었는지 보고 포스팅 마치겠습니다.**
#  
# <br>
# <br>
#
# ---
#
# <br>
#
# - code : [https://github.com/Chanjun-kim/Chanjun-kim.github.io/blob/main/_ipynb/2021-06-01-Entropy.ipynb](https://github.com/Chanjun-kim/Chanjun-kim.github.io/blob/main/_ipynb/2021-06-01-Entropy.ipynb) <br>
# - 참고 자료 : [https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011](https://m.blog.naver.com/PostView.naver?isHttpsRedirect=true&blogId=ehdrndd&logNo=221158124011)
| _ipynb/2021-06-01-Entropy.ipynb |
-- -*- coding: utf-8 -*-
-- ---
-- jupyter:
-- jupytext:
-- text_representation:
-- extension: .hs
-- format_name: light
-- format_version: '1.5'
-- jupytext_version: 1.14.4
-- kernelspec:
-- display_name: Haskell
-- language: haskell
-- name: haskell
-- ---
-- # 5. List comprehensions
-- ### 5.1 Basic concepts
-- $\displaystyle \big\{x2 \mid x\in\{1 . . 5\} \big\}$
--
-- $\{1,4,9,16,25\}$
[x^2 | x <- [1..5]]
-- 표현식 `x <- [1..5]` 에서 식 `[1..5]`가 제너레이터(generator)에 해당됨.
[(x,y) | x <- [1,2,3], y <- [4,5]]
[(x,y) | y <- [4,5], x <- [1,2,3]]
[(x,y) | x <- [1,2,3], y <- [1,2,3]]
:type concat
concat :: [[a]] -> [a]
concat x2 = [x | x1 <- x2, x <- x1]
-- concat xss = [x | xs <- xss, x <- xs]
concat [[1,2],[3,4,5],[6,7,8,9]]
firsts :: [(a,b)] -> [a]
firsts ps = [x | (x, _) <- ps]
firsts [(1,'a'), (2,'b'), (3,'c')] -- [1,2,3]
length :: [a] -> Int
length xs = sum [1 | _ <- xs]
length [1,2,3,4,5] -- 5
xs = [1,2,3,4,5]
[1 | _ <- xs]
sum [1, 2, 3, 4, 5]
-- ### 5.2 Guards (조건)
[x | x <- [1..10], even x]
[x | x <- [1..10], odd x]
-- 표현식 `even x` 와 `odd x` 는 가드라고 부른다.
factors :: Int -> [Int]
factors n = [x | x <- [1..n], n `mod` x == 0]
factors 15 -- [1,3,5,15]
factors 7 -- [1,7]
prime :: Int -> Bool
prime n = factors n == [1, n]
prime 15
prime 7
primes' :: Int -> [Int]
primes' t = [x | x <- [2..t], prime x]
primes' 40
find :: Eq a => a -> [(a, b)] -> [b]
find k t = [v | (k',v) <- t, k == k']
find 'a' [('a',1), ('b',2), ('c',3), ('b',4)] -- [1]
find 'b' [('a',1), ('b',2), ('c',3), ('b',4)] -- [2,4]
find 'c' [('a',1), ('b',2), ('c',3), ('b',4)] -- [3]
find 'd' [('a',1), ('b',2), ('c',3), ('b',4)] -- []
find' :: Eq b => b -> [(a, b)] -> [a]
find' t k = [v | (v, t') <- k, t == t']
find' 1 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['a']
find' 2 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['b']
find' 3 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['c']
find' 4 [('a',1), ('b',2), ('c',3), ('b',4)] -- ['d']
--
find''
find'' :: Eq a => a -> [(a,(b,c))] -> [(b,c)]
find'' k t = [v | (k',(v)) <- t, k == k']
find'' 'a' [('a',(1,2)), ('b',(3,4)), ('c',(5,6)), ('b',(7,8))] -- ['a']
-- 결론은
-- #### [처음 식 리스트 중 하나를 이용한 값(표현식) | 처음 식 리스트 중 하나 <- 처음 식, 조건] 구조로 이루어짐.
-- ### 5.3 The `zip` function
zip ['a','b','c'] [1,2,3,4]
:type zip
pairs :: [a] -> [(a, a)]
pairs ns = zip ns (tail ns)
xs = [1,2,3,4]
tail xs
zip [1,2,3] [2,3,4]
pairs [1,2,3,4] -- [(1,2),(2,3),(3,4)]
-- * `[1,2,3,4,5,6,7,8]` 을 인접한 수끼리 묶은 튜플 리스트를 만들고 싶다면
-- `zip [1,2,3,4,5,6,7] [2,3,4,5,6,7,8]`
zip [1,2,3,4,5,6,7] [2,3,4,5,6,7,8]
sorted :: Ord a => [a] -> Bool
sorted xs = and [x <= y | (x,y) <- pairs xs]
sorted [1,2,3,4] -- True
sorted [1,3,2,4] -- False
:type and
and [True, False]
sorted :: Ord a => [a] -> [a]
sorted xs = [if x > y then x else y | (x,y) <- pairs xs]
sorted [1,2,3,4] -- True
sorted [1,3,2,4] -- False
positions :: Eq a => a -> [a] -> [Int] -- zip a b 사용
positions x xs = [i | (x', i) <- zip xs [0..], x == x']
zip [True, False, True, False] [0..]
positions False [True, False, True, False] -- [1,3]
-- ### 5.4 String comprehensions
("abc" :: String) == (['a','b','c'] :: [Char])
"abcde" !! 2
take 3 "abcde"
length "abcde"
-- +
import Data.Char (isLower)
lowers :: String -> Int
lowers cs = length [c | c <- cs, isLower c]
-- -
lowers "Haskell" -- 6
lowers "LaTeX" -- 2
count :: Char -> String -> Int
count c cs = sum [1 | c' <- cs, c == c']
count' c cs = length [c' | c' <- cs, c == c']
count 's' "Mississippi"
count' 's' "Mississippi"
-- ### 5.5 The Caesar cipher
| PiHchap05.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Self-Driving Car Engineer Nanodegree
#
#
# ## Project: **Finding Lane Lines on the Road**
# ***
# In this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip "raw-lines-example.mp4" (also contained in this repository) to see what the output should look like after using the helper functions below.
#
# Once you have a result that looks roughly like "raw-lines-example.mp4", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.
#
# In addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.
#
# ---
# Let's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the "play" button above) to display the image.
#
# **Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the "Kernel" menu above and selecting "Restart & Clear Output".**
#
# ---
# **The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**
#
# ---
#
# <figure>
# <img src="examples/line-segments-example.jpg" width="380" alt="Combined Image" />
# <figcaption>
# <p></p>
# <p style="text-align: center;"> Your output should look something like this (above) after detecting line segments using the helper functions below </p>
# </figcaption>
# </figure>
# <p></p>
# <figure>
# <img src="examples/laneLines_thirdPass.jpg" width="380" alt="Combined Image" />
# <figcaption>
# <p></p>
# <p style="text-align: center;"> Your goal is to connect/average/extrapolate line segments to get output like this</p>
# </figcaption>
# </figure>
# **Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
# ## Import Packages
#importing some useful packages
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from ipywidgets import widgets
import numpy as np
import os
import cv2
# %matplotlib inline
# ## Read in an Image
# +
#reading in an image
image = mpimg.imread('test_images/solidWhiteRight.jpg')
#printing out some stats and plotting
print('This image is:', type(image), 'with dimensions:', image.shape)
plt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')
# -
# ## Ideas for Lane Detection Pipeline
# **Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**
#
# `cv2.inRange()` for color selection
# `cv2.fillPoly()` for regions selection
# `cv2.line()` to draw lines on an image given endpoints
# `cv2.addWeighted()` to coadd / overlay two images
# `cv2.cvtColor()` to grayscale or change color
# `cv2.imwrite()` to output images to file
# `cv2.bitwise_and()` to apply a mask to an image
#
# **Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**
# ## Helper Functions
# Below are some helper functions to help get you started. They should look familiar from the lesson!
# +
import math
class FindLanes(object):
def __init__(self):
left_lane = [0,0,0,0]
right_lane = [0,0,0,0]
def grayscale(self, img):
"""Applies the Grayscale transform
This will return an image with only one color channel
but NOTE: to see the returned image as grayscale
(assuming your grayscaled image is called 'gray')
you should call plt.imshow(gray, cmap='gray')"""
return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# Or use BGR2GRAY if you read an image with cv2.imread()
# return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
def canny(self, img, low_threshold, high_threshold):
"""Applies the Canny transform"""
return cv2.Canny(img, low_threshold, high_threshold)
def gaussian_blur(self, img, kernel_size):
"""Applies a Gaussian Noise kernel"""
return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)
def region_of_interest(self, img, vertices):
"""
Applies an image mask.
Only keeps the region of the image defined by the polygon
formed from `vertices`. The rest of the image is set to black.
`vertices` should be a numpy array of integer points.
"""
#defining a blank mask to start with
mask = np.zeros_like(img)
#defining a 3 channel or 1 channel color to fill the mask with depending on the input image
if len(img.shape) > 2:
channel_count = img.shape[2] # i.e. 3 or 4 depending on your image
ignore_mask_color = (255,) * channel_count
else:
ignore_mask_color = 255
#filling pixels inside the polygon defined by "vertices" with the fill color
cv2.fillPoly(mask, vertices, ignore_mask_color)
#returning the image only where mask pixels are nonzero
masked_image = cv2.bitwise_and(img, mask)
return masked_image
def draw_lines(self, img, lines, color=[255, 0, 0], thickness=2, interp_tol=10):
"""
NOTE: this is the function you might want to use as a starting point once you want to
average/extrapolate the line segments you detect to map out the full
extent of the lane (going from the result shown in raw-lines-example.mp4
to that shown in P1_example.mp4).
Think about things like separating line segments by their
slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left
line vs. the right line. Then, you can average the position of each of
the lines and extrapolate to the top and bottom of the lane.
This function draws `lines` with `color` and `thickness`.
Lines are drawn on the image inplace (mutates the image).
If you want to make the lines semi-transparent, think about combining
this function with the weighted_img() function below
"""
"""
Left Lane: > 0.0
Right Lane: < 0.0
"""
right_slopes = []
left_slopes = []
right_intercepts = []
left_intercepts = []
x_min_interp = 0
y_max = img.shape[0]
y_min_left = img.shape[0] + 1
y_min_right = img.shape[0] + 1
y_min_interp = 320 + 15 #top of ROI plus some offset for aesthetics
for line in lines:
for x1,y1,x2,y2 in line:
current_slope = (y2-y1)/(x2-x1)
# For each detected line, seperate lines into left and right lanes.
# Calculate the current slope and intercept and keep a history for averaging.
if current_slope < 0.0 and current_slope > -math.inf:
right_slopes.append(current_slope)
right_intercepts.append(y1 - current_slope*x1)
y_min_right = min(y_min_right, y1, y2)
if current_slope > 0.0 and current_slope < math.inf:
left_slopes.append(current_slope)
left_intercepts.append(y1 - current_slope*x1)
y_min_left = min(y_min_left, y1, y2)
# Calculate the average of the slopes, intercepts, x_min and x_max
# Interpolate the average line to the end of the region of interest (using equation of slopes)
if len(left_slopes) > 0:
ave_left_slope = sum(left_slopes) / len(left_slopes)
ave_intercept = sum(left_intercepts) / len(left_intercepts)
x_min=int((y_min_left - ave_intercept)/ ave_left_slope)
x_max = int((y_max - ave_intercept)/ ave_left_slope)
x_min_interp = int(((x_min*y_min_interp) - (x_min*y_max) - (x_max*y_min_interp) + (x_max*y_min_left))/(y_min_left - y_max))
self.left_lane = [x_min_interp, y_min_interp, x_max, y_max]
# Draw the left lane line
cv2.line(img, (self.left_lane[0], self.left_lane[1]), (self.left_lane[2], self.left_lane[3]), [255, 0, 0], 12)
if len(right_slopes) > 0:
ave_right_slope = sum(right_slopes) / len(right_slopes)
ave_intercept = sum(right_intercepts) / len(right_intercepts)
x_min = int((y_min_right - ave_intercept)/ ave_right_slope)
x_max = int((y_max - ave_intercept)/ ave_right_slope)
x_min_interp = int(((x_min*y_min_interp) - (x_min*y_max) - (x_max*y_min_interp) + (x_max*y_min_right))/(y_min_right - y_max))
self.right_lane = [x_min_interp, y_min_interp, x_max, y_max]
# Draw the right lane line
cv2.line(img, (self.right_lane[0], self.right_lane[1]), (self.right_lane[2], self.right_lane[3]), [255, 0, 0], 12)
def hough_lines(self, img, rho, theta, threshold, min_line_len, max_line_gap):
"""
`img` should be the output of a Canny transform.
Returns an image with hough lines drawn.
"""
lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)
line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)
self.draw_lines(line_img, lines)
return line_img
# Python 3 has support for cool math symbols.
def weighted_img(self, img, initial_img, α=0.8, β=1., γ=0.):
"""
`img` is the output of the hough_lines(), An image with lines drawn on it.
Should be a blank image (all black) with lines drawn on it.
`initial_img` should be the image before any processing.
The result image is computed as follows:
initial_img * α + img * β + γ
NOTE: initial_img and img must be the same shape!
"""
return cv2.addWeighted(initial_img, α, img, β, γ)
# -
# ## Test Images
#
# Build your pipeline to work on the images in the directory "test_images"
# **You should make sure your pipeline works well on these images before you try the videos.**
import os
os.listdir("test_images/")
# ## Build a Lane Finding Pipeline
#
#
# Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.
#
# Try tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.
# +
# TODO: Build your pipeline that will draw lane lines on the test_images
# then save them to the test_images_output directory.
# Load test images
dir = "test_images/"
out_dir = "test_images_output/"
input = os.listdir(dir)
# Define globals
kernel_size = 3
canny_thresh = [75,150]
rho = 2
theta = np.pi/180
threshold = 90
min_line_length = 20
max_line_gap = 20
fl = FindLanes()
# Setup pipeline
def process_image(image):
# Preprocess image
gray = fl.grayscale(image)
plt.imsave(out_dir + "gray.jpg", gray, cmap="gray")
blur = fl.gaussian_blur(gray, kernel_size)
plt.imsave(out_dir + "blur.jpg", blur, cmap="gray")
# Find edges
edges = fl.canny(blur, canny_thresh[0], canny_thresh[1])
plt.imsave(out_dir + "edges.jpg", edges, cmap="gray")
# Region of interest
verts = np.array([[(100,image.shape[0]),(450, 320), (500, 320), (image.shape[1],image.shape[0])]], dtype=np.int32)
masked_edges = fl.region_of_interest(edges, verts)
plt.imsave(out_dir + "masked_edges.jpg", masked_edges, cmap="gray")
# Draw hough lines
lines_image = fl.hough_lines(masked_edges, rho, theta, threshold, min_line_length, max_line_gap)
plt.imsave(out_dir + "lines.jpg", lines_image, cmap="gray")
result = fl.weighted_img(lines_image, image)
plt.imsave(out_dir + "final.jpg", result, cmap="gray")
return result
# Jupyter notebook validation error -----
# widgets.interact(process_image, canny_thresh=widgets.IntRangeSlider(min=0, max=255, step=1, value=canny_thresh))
# -----
for test_image in input:
image = mpimg.imread(dir + test_image)
processed = process_image(image)
plt.imshow(processed)
plt.show()
# -
# ## Test on Videos
#
# You know what's cooler than drawing lanes over images? Drawing lanes over video!
#
# We can test our solution on two provided videos:
#
# `solidWhiteRight.mp4`
#
# `solidYellowLeft.mp4`
#
# **Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**
#
# **If you get an error that looks like this:**
# ```
# NeedDownloadError: Need ffmpeg exe.
# You can download it by calling:
# imageio.plugins.ffmpeg.download()
# ```
# **Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**
# Import everything needed to edit/save/watch video clips
from moviepy.editor import VideoFileClip
from IPython.display import HTML
# +
# Define globals
# def process_image(image):
# # NOTE: The output you return should be a color image (3 channel) for processing video below
# # TODO: put your pipeline here,
# # you should return the final output (image where lines are drawn on lanes)
# -
# Let's try the one with the solid white lane on the right first ...
white_output = 'test_videos_output/solidWhiteRight.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4").subclip(0,5)
clip1 = VideoFileClip("test_videos/solidWhiteRight.mp4")
white_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!
# %time white_clip.write_videofile(white_output, audio=False)
# Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(white_output))
# ## Improve the draw_lines() function
#
# **At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video "P1_example.mp4".**
#
# **Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**
# Now for the one with the solid yellow lane on the left. This one's more tricky!
yellow_output = 'test_videos_output/solidYellowLeft.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)
clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')
yellow_clip = clip2.fl_image(process_image)
# %time yellow_clip.write_videofile(yellow_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(yellow_output))
# ## Writeup and Submission
#
# If you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.
#
# ## Optional Challenge
#
# Try your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!
challenge_output = 'test_videos_output/challenge.mp4'
## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video
## To do so add .subclip(start_second,end_second) to the end of the line below
## Where start_second and end_second are integer values representing the start and end of the subclip
## You may also uncomment the following line for a subclip of the first 5 seconds
##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)
clip3 = VideoFileClip('test_videos/challenge.mp4')
challenge_clip = clip3.fl_image(process_image)
# %time challenge_clip.write_videofile(challenge_output, audio=False)
HTML("""
<video width="960" height="540" controls>
<source src="{0}">
</video>
""".format(challenge_output))
| P1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Contrasts Overview
import numpy as np
import statsmodels.api as sm
# This document is based heavily on this excellent resource from UCLA http://www.ats.ucla.edu/stat/r/library/contrast_coding.htm
# A categorical variable of K categories, or levels, usually enters a regression as a sequence of K-1 dummy variables. This amounts to a linear hypothesis on the level means. That is, each test statistic for these variables amounts to testing whether the mean for that level is statistically significantly different from the mean of the base category. This dummy coding is called Treatment coding in R parlance, and we will follow this convention. There are, however, different coding methods that amount to different sets of linear hypotheses.
#
# In fact, the dummy coding is not technically a contrast coding. This is because the dummy variables add to one and are not functionally independent of the model's intercept. On the other hand, a set of *contrasts* for a categorical variable with `k` levels is a set of `k-1` functionally independent linear combinations of the factor level means that are also independent of the sum of the dummy variables. The dummy coding is not wrong *per se*. It captures all of the coefficients, but it complicates matters when the model assumes independence of the coefficients such as in ANOVA. Linear regression models do not assume independence of the coefficients and thus dummy coding is often the only coding that is taught in this context.
#
# To have a look at the contrast matrices in Patsy, we will use data from UCLA ATS. First let's load the data.
# #### Example Data
import pandas as pd
url = 'https://stats.idre.ucla.edu/stat/data/hsb2.csv'
hsb2 = pd.read_table(url, delimiter=",")
hsb2.head(10)
# It will be instructive to look at the mean of the dependent variable, write, for each level of race ((1 = Hispanic, 2 = Asian, 3 = African American and 4 = Caucasian)).
hsb2.groupby('race')['write'].mean()
# #### Treatment (Dummy) Coding
# Dummy coding is likely the most well known coding scheme. It compares each level of the categorical variable to a base reference level. The base reference level is the value of the intercept. It is the default contrast in Patsy for unordered categorical factors. The Treatment contrast matrix for race would be
from patsy.contrasts import Treatment
levels = [1,2,3,4]
contrast = Treatment(reference=0).code_without_intercept(levels)
print(contrast.matrix)
# Here we used `reference=0`, which implies that the first level, Hispanic, is the reference category against which the other level effects are measured. As mentioned above, the columns do not sum to zero and are thus not independent of the intercept. To be explicit, let's look at how this would encode the `race` variable.
hsb2.race.head(10)
print(contrast.matrix[hsb2.race-1, :][:20])
sm.categorical(hsb2.race.values)
# This is a bit of a trick, as the `race` category conveniently maps to zero-based indices. If it does not, this conversion happens under the hood, so this will not work in general but nonetheless is a useful exercise to fix ideas. The below illustrates the output using the three contrasts above
from statsmodels.formula.api import ols
mod = ols("write ~ C(race, Treatment)", data=hsb2)
res = mod.fit()
print(res.summary())
# We explicitly gave the contrast for race; however, since Treatment is the default, we could have omitted this.
# ### Simple Coding
# Like Treatment Coding, Simple Coding compares each level to a fixed reference level. However, with simple coding, the intercept is the grand mean of all the levels of the factors. Patsy does not have the Simple contrast included, but you can easily define your own contrasts. To do so, write a class that contains a code_with_intercept and a code_without_intercept method that returns a patsy.contrast.ContrastMatrix instance
# +
from patsy.contrasts import ContrastMatrix
def _name_levels(prefix, levels):
return ["[%s%s]" % (prefix, level) for level in levels]
class Simple(object):
def _simple_contrast(self, levels):
nlevels = len(levels)
contr = -1./nlevels * np.ones((nlevels, nlevels-1))
contr[1:][np.diag_indices(nlevels-1)] = (nlevels-1.)/nlevels
return contr
def code_with_intercept(self, levels):
contrast = np.column_stack((np.ones(len(levels)),
self._simple_contrast(levels)))
return ContrastMatrix(contrast, _name_levels("Simp.", levels))
def code_without_intercept(self, levels):
contrast = self._simple_contrast(levels)
return ContrastMatrix(contrast, _name_levels("Simp.", levels[:-1]))
# -
hsb2.groupby('race')['write'].mean().mean()
contrast = Simple().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Simple)", data=hsb2)
res = mod.fit()
print(res.summary())
# ### Sum (Deviation) Coding
# Sum coding compares the mean of the dependent variable for a given level to the overall mean of the dependent variable over all the levels. That is, it uses contrasts between each of the first k-1 levels and level k In this example, level 1 is compared to all the others, level 2 to all the others, and level 3 to all the others.
from patsy.contrasts import Sum
contrast = Sum().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Sum)", data=hsb2)
res = mod.fit()
print(res.summary())
# This corresponds to a parameterization that forces all the coefficients to sum to zero. Notice that the intercept here is the grand mean where the grand mean is the mean of means of the dependent variable by each level.
hsb2.groupby('race')['write'].mean().mean()
# ### Backward Difference Coding
# In backward difference coding, the mean of the dependent variable for a level is compared with the mean of the dependent variable for the prior level. This type of coding may be useful for a nominal or an ordinal variable.
from patsy.contrasts import Diff
contrast = Diff().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Diff)", data=hsb2)
res = mod.fit()
print(res.summary())
# For example, here the coefficient on level 1 is the mean of `write` at level 2 compared with the mean at level 1. Ie.,
res.params["C(race, Diff)[D.1]"]
hsb2.groupby('race').mean()["write"][2] - \
hsb2.groupby('race').mean()["write"][1]
# ### Helmert Coding
# Our version of Helmert coding is sometimes referred to as Reverse Helmert Coding. The mean of the dependent variable for a level is compared to the mean of the dependent variable over all previous levels. Hence, the name 'reverse' being sometimes applied to differentiate from forward Helmert coding. This comparison does not make much sense for a nominal variable such as race, but we would use the Helmert contrast like so:
from patsy.contrasts import Helmert
contrast = Helmert().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(race, Helmert)", data=hsb2)
res = mod.fit()
print(res.summary())
# To illustrate, the comparison on level 4 is the mean of the dependent variable at the previous three levels taken from the mean at level 4
grouped = hsb2.groupby('race')
grouped.mean()["write"][4] - grouped.mean()["write"][:3].mean()
# As you can see, these are only equal up to a constant. Other versions of the Helmert contrast give the actual difference in means. Regardless, the hypothesis tests are the same.
k = 4
1./k * (grouped.mean()["write"][k] - grouped.mean()["write"][:k-1].mean())
k = 3
1./k * (grouped.mean()["write"][k] - grouped.mean()["write"][:k-1].mean())
# ### Orthogonal Polynomial Coding
# The coefficients taken on by polynomial coding for `k=4` levels are the linear, quadratic, and cubic trends in the categorical variable. The categorical variable here is assumed to be represented by an underlying, equally spaced numeric variable. Therefore, this type of encoding is used only for ordered categorical variables with equal spacing. In general, the polynomial contrast produces polynomials of order `k-1`. Since `race` is not an ordered factor variable let's use `read` as an example. First we need to create an ordered categorical from `read`.
hsb2['readcat'] = np.asarray(pd.cut(hsb2.read, bins=3))
hsb2.groupby('readcat').mean()['write']
from patsy.contrasts import Poly
levels = hsb2.readcat.unique().tolist()
contrast = Poly().code_without_intercept(levels)
print(contrast.matrix)
mod = ols("write ~ C(readcat, Poly)", data=hsb2)
res = mod.fit()
print(res.summary())
# As you can see, readcat has a significant linear effect on the dependent variable `write` but not a significant quadratic or cubic effect.
| examples/notebooks/contrasts.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# <small><i>This notebook was prepared by [<NAME>](http://donnemartin.com). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).</i></small>
# # Challenge Notebook
# ## Problem: Implement a hash table with set, get, and remove methods.
#
# * [Constraints](#Constraints)
# * [Test Cases](#Test-Cases)
# * [Algorithm](#Algorithm)
# * [Code](#Code)
# * [Unit Test](#Unit-Test)
# * [Solution Notebook](#Solution-Notebook)
# ## Constraints
#
# * For simplicity, are the keys integers only?
# * Yes
# * For collision resolution, can we use chaining?
# * Yes
# * Do we have to worry about load factors?
# * No
# ## Test Cases
#
# * get on an empty hash table index
# * set on an empty hash table index
# * set on a non empty hash table index
# * set on a key that already exists
# * remove on a key with an entry
# * remove on a key without an entry
# ## Algorithm
#
# Refer to the [Solution Notebook](http://nbviewer.ipython.org/github/donnemartin/interactive-coding-challenges/blob/master/arrays_strings/hash_map/hash_map_solution.ipynb). If you are stuck and need a hint, the solution notebook's algorithm discussion might be a good place to start.
# ## Code
# +
class Item(object):
def __init__(self, key, value):
# TODO: Implement me
pass
class HashTable(object):
def __init__(self, size):
# TODO: Implement me
pass
def hash_function(self, key):
# TODO: Implement me
pass
def set(self, key, value):
# TODO: Implement me
pass
def get(self, key):
# TODO: Implement me
pass
def remove(self, key):
# TODO: Implement me
pass
# -
# ## Unit Test
#
#
# **The following unit test is expected to fail until you solve the challenge.**
# +
# # %load test_hash_map.py
from nose.tools import assert_equal
class TestHashMap(object):
# TODO: It would be better if we had unit tests for each
# method in addition to the following end-to-end test
def test_end_to_end(self):
hash_table = HashTable(10)
print("Test: get on an empty hash table index")
assert_equal(hash_table.get(0), None)
print("Test: set on an empty hash table index")
hash_table.set(0, 'foo')
assert_equal(hash_table.get(0), 'foo')
hash_table.set(1, 'bar')
assert_equal(hash_table.get(1), 'bar')
print("Test: set on a non empty hash table index")
hash_table.set(10, 'foo2')
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), 'foo2')
print("Test: set on a key that already exists")
hash_table.set(10, 'foo3')
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), 'foo3')
print("Test: remove on a key that already exists")
hash_table.remove(10)
assert_equal(hash_table.get(0), 'foo')
assert_equal(hash_table.get(10), None)
print("Test: remove on a key that doesn't exist")
hash_table.remove(-1)
print('Success: test_end_to_end')
def main():
test = TestHashMap()
test.test_end_to_end()
if __name__ == '__main__':
main()
# -
# ## Solution Notebook
#
# Review the [Solution Notebook](http://nbviewer.ipython.org/github/donnemartin/interactive-coding-challenges/blob/master/arrays_strings/hash_map/hash_map_solution.ipynb) for a discussion on algorithms and code solutions.
| arrays_strings/hash_map/hash_map_challenge.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.6.9 64-bit (''elfi36'': conda)'
# language: python
# name: python36964bitelfi36conda8bec451552304612bd355a97e3742bbb
# ---
# # Adaptive distance
#
# [ABC](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#approximate-bayesian-computation) provides means to sample an approximate posterior distribution over unknown parameters based on comparison between observed and simulated data.
# This comparison is often based on distance between features that summarise the data and are informative about the parameter values.
#
# Here we assume that the summaries calculated based on observed and simulated data are compared based on weighted distance with weight $w_i=1/\sigma_i$ calculated based on their standard deviation $\sigma_i$.
# This ensures that the selected summaries to have an equal contribution in the distance between observed and simulated data.
#
# This notebook studies [adaptive distance](https://projecteuclid.org/euclid.ba/1460641065) [SMC-ABC](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#sequential-monte-carlo-abc) where $\sigma_i$ and $w_i$ are recalculated between SMC iterations as proposed in [[1](#Reference)].
import numpy as np
import scipy.stats as ss
import matplotlib.pyplot as plt
# %matplotlib inline
import elfi
# ## Example 1:
#
# Assume we have an unknown parameter with prior distribution $\theta\sim U(0,50)$ and two simulator outputs $S_1\sim N(\theta, 1)$ and $S_2\sim N(\theta, 100)$ whose observed values are 20.
def simulator(mu, batch_size=1, random_state=None):
batches_mu = np.asarray(mu).reshape((-1,1))
obs_1 = ss.norm.rvs(loc=batches_mu, scale=1, random_state=random_state).reshape((-1,1))
obs_2 = ss.norm.rvs(loc=batches_mu, scale=100, random_state=random_state).reshape((-1,1))
return np.hstack((obs_1, obs_2))
observed_data = np.array([20,20])[None,:]
# Here the simulator outputs are both informative about the unknown model parameter, but $S_2$ has more observation noise than $S_1$. We do not calculate separate summaries in this example, but compare observed and simulated data based on these two variables.
#
# Euclidean distance between observed and simulated outputs or summaries can be used to find parameter values that could produce the observed data. Here we describe dependencies between the unknown parameter value and observed distances as an ELFI model `m` and sample the approximate posterior distribution with the [rejection sampler](https://elfi.readthedocs.io/en/latest/usage/tutorial.html#inference-with-rejection-sampling).
m = elfi.new_model()
theta = elfi.Prior(ss.uniform, 0, 50, model=m)
sim = elfi.Simulator(simulator, theta, observed=observed_data)
d = elfi.Distance('euclidean', sim)
rej = elfi.Rejection(d, batch_size=10000, seed=123)
# Let us sample 100 parameters with `quantile=0.01`. This means that we sample 10000 candidate parameters from the prior distribution and take the 100 parameters that produce simulated data closest to the observed data.
sample = rej.sample(100, quantile=0.01)
sample
plt.hist(sample.samples_array,range=(0,50),bins=20)
plt.xlabel('theta');
# The approximate posterior sample is concentrated around $\theta=20$ as expected in this example. However the sample distribution is much wider than we would observe in case the sample was selected based on $S_1$ alone.
#
# Now let us test adaptive distance in the same example.
#
# First we switch the distance node `d` to an adaptive distance node and initialise adaptive distance SMC-ABC. Initialisation is identical to the rejection sampler, and here we use the same batch size and seed as earlier, so that the methods are presented with the exact same candidate parameters.
d.become(elfi.AdaptiveDistance(sim))
ada_smc = elfi.AdaptiveDistanceSMC(d, batch_size=10000, seed=123)
# Since this is an iterative method, we must decide both sample size (`n_samples`) and how many populations are sampled (`rounds`). In addition we can decide the $\alpha$ quantile (`quantile`) used in estimation.
#
# Each population with `n_samples` parameter values is sampled as follows: 1. `n_samples/quantile` parameters are sampled from the current proposal distribution with acceptance threshold determined based on the previous population and 2. the distance measure is updated based on the observed sample and `n_samples` with the smallest distance are selected as the new population. The first population is sampled from the prior distribution and all samples are accepted in step 1.
#
# Here we sample one population with `quantile=0.01`. This means that the total simulation count will be the same as with the rejection sampler, but now the distance function is updated based on the 10000 simulated observations, and the 100 parameters included in the posterior sample are selected based on the new distance measure.
sample_ada = ada_smc.sample(100, 1, quantile=0.01)
sample_ada
plt.hist(sample_ada.samples_array,range=(0,50),bins=20)
plt.xlabel('theta');
# We see that the posterior distribution over unknown parameter values is narrower than in the previous example. This is because the simulator outputs are now normalised based on their estimated standard deviation.
#
# We can see $w_1$ and $w_2$:
sample_ada.adaptive_distance_w
# ## Example 2:
#
# This is the normal distribution example presented in [[1](#Reference)].
#
# Here we have an unknown parameter with prior distribution $\theta\sim N(0,100)$ and two simulator outputs $S_1\sim N(\theta, 0.1)$ and $S_2\sim N(1, 1)$ whose observed values are 0.
def simulator(mu, batch_size=1, random_state=None):
batches_mu = np.asarray(mu).reshape((-1,1))
obs_1 = ss.norm.rvs(loc=batches_mu, scale=0.1, random_state=random_state).reshape((-1,1))
obs_2 = ss.norm.rvs(loc=1, scale=1, size=batch_size, random_state=random_state).reshape((-1,1))
return np.hstack((obs_1, obs_2))
observed_data = np.array([0,0])[None,:]
# $S_1$ is now informative and $S_2$ uninformative about the unknown parameter value, and we note that between the two output variables, $S_1$ has larger variance under the prior predictive distribution. This means that normalisation estimated based on output data observed in the initial round or based on a separate sample would not work well in this example.
#
# Let us define a new model and initialise adaptive distance SMC-ABC.
m = elfi.new_model()
theta = elfi.Prior(ss.norm, 0, 100, model=m)
sim = elfi.Simulator(simulator, theta, observed=observed_data)
d = elfi.AdaptiveDistance(sim)
ada_smc = elfi.AdaptiveDistanceSMC(d, batch_size=2000, seed=123)
# Next we sample 1000 parameter values in 5 rounds with the default `quantile=0.5` which is recommended in sequential estimation [[1](#Reference)]:
sample_ada = ada_smc.sample(1000, 5)
sample_ada
plt.hist(sample_ada.samples_array, range=(-25,25), bins=20)
plt.xlabel(theta);
# The sample distribution is concentrated around $\theta=0$ but wider than could be expected. However we can continue the iterative estimation process. Here we sample two more populations:
sample_ada = ada_smc.sample(1000, 2)
sample_ada
plt.hist(sample_ada.samples_array, range=(-25,25), bins=20)
plt.xlabel('theta');
# We observe that the sample mean is now closer to zero and the sample distribution is narrower.
#
# Let us examine $w_1$ and $w_2$:
sample_ada.adaptive_distance_w
# We can see that $w_2$ (second column) is constant across iterations whereas $w_1$ increases as the method learns more about possible parameter values and the proposal distribution becomes more concentrated around $\theta=0$.
#
# ## Notes
#
# The adaptive distance SMC-ABC method demonstrated in this notebook normalises simulator outputs or summaries calculated based on simulator output based on their estimated standard deviation under the proposal distribution in each iteration. This ensures that all outputs or summaries have an equal contribution to the distance between simulated and observed data in all iterations.
#
# It is important to note that the method does not evaluate whether outputs or summaries are needed or informative. In both examples studied in this notebook, results would improve if inference was carried out based on $S_1$ alone. Hence one should choose the summaries used in adaptive distance SMC-ABC with the usual care. ELFI tools that aid in the selection process are discussed in the diagnostics notebook available [here](https://github.com/elfi-dev/notebooks/tree/master).
# ## Reference
# [1] <NAME> (2017). Adapting the ABC Distance Function. Bayesian Analysis 12(1): 289-309, 2017. https://projecteuclid.org/euclid.ba/1460641065
| adaptive_distance.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img style="float: left; margin: 30px 15px 15px 15px;" src="https://pngimage.net/wp-content/uploads/2018/06/logo-iteso-png-5.png" width="300" height="500" />
#
#
# ### <font color='navy'> Simulación de procesos financieros.
#
# **Nombres:** <NAME> y <NAME>
#
# **Fecha:** 22 de Febrero del 2021
#
# **Expediente** : if721470
# **Profesor:** <NAME>.
#
# ### LINK DE GITHUB:
#
# # Tarea 4: Clase 7. Ejemplos Simulación
# ## Enunciado de tarea
# > # 1
# Como ejemplo simple de una simulación de Monte Carlo, considere calcular la probabilidad de una suma particular del lanzamiento de tres dados (cada dado tiene valores del uno al seis). Además cada dado tiene las siguientes carácterísticas: el primer dado no está cargado (distribución uniforme todos son equiprobables); el segundo y tercer dado están cargados basados en una distribución binomial con parámetros (`n=5, p=0.5` y `n=5, p=0.2`). Calcule la probabilidad de que la suma resultante sea 7, 14 o 18.
#
#
# > # 2 Ejercicio de aplicación- Cafetería Central
#
# Premisas para la simulación:
# - Negocio de alimentos que vende bebidas y alimentos.
# - Negocio dentro del ITESO.
# - Negocio en cafetería central.
# - Tipo de clientes (hombres y mujeres).
# - Rentabilidad del 60%.
#
# ## Objetivo
# Realizar una simulación estimado el tiempo medio que se tardaran los clientes en ser atendidos entre el horario de 6:30 a 1 pm. Además saber el consumo.
# **Analizar supuestos y limitantes**
#
# ## Supuestos en simulación
# Clasificación de clientes:
# - Mujer = 1 $\longrightarrow$ aleatorio < 0.5
# - Hombre = 0 $\longrightarrow$ aleatorio $\geq$ 0.5.
#
# Condiciones iniciales:
# - Todas las distrubuciones de probabilidad se supondrán uniformes.
# - Tiempo de simulación: 6:30 am - 1:30pm $\longrightarrow$ T = 7 horas = 25200 seg.
# - Tiempo de llegada hasta ser atendido: Min=5seg, Max=30seg.
# - Tiempo que tardan los clientes en ser atendidos:
# - Mujer: Min = 1 min= 60seg, Max = 5 min = 300 seg
# - Hombre: Min = 40 seg, Max = 2 min= 120 seg
# - Consumo según el tipo de cliente:
# - Mujer: Min = 30 pesos, Max = 100 pesos
# - Hombre: Min = 20 pesos, Max = 80 pesos
#
# Responder las siguientes preguntas basados en los datos del problema:
# 1. ¿Cuáles fueron los gastos de los hombres y las mujeres en 5 días de trabajo?.
# 2. ¿Cuál fue el consumo promedio de los hombres y mujeres?
# 3. ¿Cuál fue el número de personas atendidas por día?
# 4. ¿Cuál fue el tiempo de atención promedio?
# 5. ¿Cuánto fue la ganancia promedio de la cafetería en 5 días de trabajo y su respectiva rentabilidad?
# ### Ejercicio 1:
# Como ejemplo simple de una simulación de Monte Carlo, considere calcular la probabilidad de una suma particular del lanzamiento de tres dados (cada dado tiene valores del uno al seis). Además cada dado tiene las siguientes carácterísticas: el primer dado no está cargado (distribución uniforme todos son equiprobables); el segundo y tercer dado están cargados basados en una distribución binomial con parámetros (`n=5, p=0.5` y `n=5, p=0.2`). Calcule la probabilidad de que la suma resultante sea 7, 14 o 18.
#
#
# +
# SOLUCION CRISTINA
# -
# Código de solución
.
.
.
.
# +
# SOLUCION DAYANA
# +
# Código de solución
import numpy as np
import scipy.stats as st
def suma1():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 7:
return True
else:
return False
def suma2():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 14:
return True
else:
return False
def suma3():
d1_no_caragado = np.random.randint(1,7)
d2_cargado = st.binom(n = 5, p=0.5).rvs(size = 1)
d3_cargado = st.binom(n = 5, p=0.2).rvs(size = 1)
suma_dados = d1_no_caragado + d2_cargado + d3_cargado
if suma_dados == 18:
return True
else:
return False
N=100
dado_1=[suma1( ) for i in range(N)]
dado_2=[suma2( ) for i in range(N)]
dado_3=[suma3( ) for i in range(N)]
dado_1.count(True)/N, dado_2.count(True)/N, dado_3.count(True)/N
# -
# ### Ejercicio 2 de aplicación- Cafetería Central
#
# Premisas para la simulación:
# - Negocio de alimentos que vende bebidas y alimentos.
# - Negocio dentro del ITESO.
# - Negocio en cafetería central.
# - Tipo de clientes (hombres y mujeres).
# - Rentabilidad del 60%.
#
# #### Objetivo
# Realizar una simulación estimado el tiempo medio que se tardaran los clientes en ser atendidos entre el horario de 6:30 a 1 pm. Además saber el consumo.
# **Analizar supuestos y limitantes**
#
# #### Supuestos en simulación
# Clasificación de clientes:
# - Mujer = 1 $\longrightarrow$ aleatorio < 0.5
# - Hombre = 0 $\longrightarrow$ aleatorio $\geq$ 0.5.
#
# Condiciones iniciales:
# - Todas las distrubuciones de probabilidad se supondrán uniformes.
# - Tiempo de simulación: 6:30 am - 1:30pm $\longrightarrow$ T = 7 horas = 25200 seg.
# - Tiempo de llegada hasta ser atendido: Min=5seg, Max=30seg.
# - Tiempo que tardan los clientes en ser atendidos:
# - Mujer: Min = 1 min= 60seg, Max = 5 min = 300 seg
# - Hombre: Min = 40 seg, Max = 2 min= 120 seg
# - Consumo según el tipo de cliente:
# - Mujer: Min = 30 pesos, Max = 100 pesos
# - Hombre: Min = 20 pesos, Max = 80 pesos
#
# Responder las siguientes preguntas basados en los datos del problema:
# 1. ¿Cuáles fueron los gastos de los hombres y las mujeres en 5 días de trabajo?.
# 2. ¿Cuál fue el consumo promedio de los hombres y mujeres?
# 3. ¿Cuál fue el número de personas atendidas por día?
# 4. ¿Cuál fue el tiempo de atención promedio?
# 5. ¿Cuánto fue la ganancia promedio de la cafetería en 5 días de trabajo y su respectiva rentabilidad?
################## Datos del problema
d = 5
T =25200
T_at_min = 5; T_at_max = 30
T_mujer_min =60; T_mujer_max = 300
T_hombre_min = 40; T_hombre_max = 120
C_mujer_min = 30; C_mujer_max = 100
C_hombre_min = 20; C_hombre_max = 80
# +
# SOLUCION CRISTINA
# -
# Código de solución
.
.
.
.
# +
# SOLUCION DAYANA
# -
# Código de solución
.
.
.
.
| TAREA_4. VazquezVargas_Cristina_NavarroValencia_Dayana.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Sense and Move
#
# In this notebook, let's put all of what we've learned together and see what happens to an initial probability distribution as a robot goes trough cycles of sensing then moving then sensing then moving, and so on! Recall that each time a robot senses (in this case a red or green color)it gains information about its environment, and everytime it moves, it loses some information due to motion uncertainty.
#
#
# <img src='images/sense_move.png' width=50% height=50% />
#
# First let's include our usual resource imports and display function.
# importing resources
import matplotlib.pyplot as plt
import numpy as np
# A helper function for visualizing a distribution.
def display_map(grid, bar_width=1):
if(len(grid) > 0):
x_labels = range(len(grid))
plt.bar(x_labels, height=grid, width=bar_width, color='b')
plt.xlabel('Grid Cell')
plt.ylabel('Probability')
plt.ylim(0, 1) # range of 0-1 for probability values
plt.title('Probability of the robot being at each cell in the grid')
plt.xticks(np.arange(min(x_labels), max(x_labels)+1, 1))
plt.show()
else:
print('Grid is empty')
# ### QUIZ: Given the list motions=[1,1], compute the posterior distribution if the robot first senses red, then moves right one, then senses green, then moves right again, starting with a uniform prior distribution, `p`.
#
# `motions=[1,1]` mean that the robot moves right one cell and then right again. You are given the initial variables and the complete `sense` and `move` function, below.
# +
# given initial variables
p=[0.2, 0.2, 0.2, 0.2, 0.2]
# the color of each grid cell in the 1D world
world=['green', 'red', 'red', 'green', 'green']
# Z, the sensor reading ('red' or 'green')
measurements = ['red', 'green']
pHit = 0.6
pMiss = 0.2
motions = [1,1]
pExact = 0.8
pOvershoot = 0.1
pUndershoot = 0.1
# You are given the complete sense function
def sense(p, Z):
''' Takes in a current probability distribution, p, and a sensor reading, Z.
Returns a *normalized* distribution after the sensor measurement has been made, q.
This should be accurate whether Z is 'red' or 'green'. '''
q=[]
# loop through all grid cells
for i in range(len(p)):
# check if the sensor reading is equal to the color of the grid cell
# if so, hit = 1
# if not, hit = 0
hit = (Z == world[i])
q.append(p[i] * (hit * pHit + (1-hit) * pMiss))
# sum up all the components
s = sum(q)
# divide all elements of q by the sum to normalize
for i in range(len(p)):
q[i] = q[i] / s
return q
# The complete move function
def move(p, U):
q=[]
# iterate through all values in p
for i in range(len(p)):
# use the modulo operator to find the new location for a p value
# this finds an index that is shifted by the correct amount
index = (i-U) % len(p)
nextIndex = (index+1) % len(p)
prevIndex = (index-1) % len(p)
s = pExact * p[index]
s = s + pOvershoot * p[nextIndex]
s = s + pUndershoot * p[prevIndex]
# append the correct, modified value of p to q
q.append(s)
return q
## TODO: Compute the posterior distribution if the robot first senses red, then moves
## right one, then senses green, then moves right again, starting with a uniform prior distribution.
for idx in range(len(motions)):
p = sense(p, measurements[idx])
p = move(p, motions[idx])
## print/display that distribution
print(p)
display_map(p)
# -
# ### Clarification about Entropy
#
# The video mentions that entropy will go down after the update step and that entropy will go up after the measurement step.
#
# In general, **entropy measures the amount of uncertainty**. Since the update step increases uncertainty, then entropy should increase. The measurement step decreases uncertainty, so entropy should decrease.
#
# Let's look at our current example where the robot could be at five different positions. The maximum uncertainty occurs when all positions have equal probabilities $[0.2, 0.2, 0.2, 0.2, 0.2]$
#
# Following the formula $$\text{Entropy} = \Sigma (-p \times log(p))$$we get $$-5 \times (.2)\times log(0.2) = 0.699$$
#
# Taking a measurement should decrease uncertainty and thus decrease entropy. Let's say after taking a measurement, the probabilities become <span class="mathquill">[0.05, 0.05, 0.05, 0.8, 0.05]</span>. Now the entropy decreased to 0.338. Hence a measurement step should decrease entropy whereas an update step should increase entropy.
| 4_2_Robot_Localization/9_1. Sense and Move, exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Fairness part of the Workshop
# Analyze fairness of a dataset with different techniques
# Imports
import pandas as pd
from IPython.display import display
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
# # Loading data
#
# We are first going to work with data from the Lending Club Dataset, a dataset of around 800k lending club users. This dataset doesn't have information about race or gender, so we will artificially create a "race" column to test our fairness metrics. Let's create a heavily unfair dataset: 80% of the users that were given a loan will be of race "1", and only 10% of the users who were not given a loan will be of race "1".
# +
# Data management
df = pd.read_csv('../data/loans_data.csv')
# Subset
df = df.sample(frac=0.1)
display(df)
# Random vector to add noise to protected class
p_nochange=0.9
random_vec = np.random.choice([0,1], size=len(df), p=[p_nochange,1-p_nochange])
print(len(df), sum(random_vec))
# Add protected class
df['protected_class'] = df['loan_status']^random_vec
print(df[['protected_class','loan_status']])
# +
quant_cols = ['loan_amnt', 'int_rate', 'annual_inc', 'dti', 'delinq_2yrs', 'fico_range_low',\
'inq_last_6mths', 'mths_since_last_delinq', 'mths_since_last_record', 'open_acc', 'pub_rec',\
'revol_bal', 'revol_util', 'total_acc', 'acc_now_delinq', 'tot_coll_amt',\
'tot_cur_bal', 'tax_liens', 'total_bal_ex_mort', 'total_bc_limit', 'total_il_high_credit_limit',\
'age_of_cr_line', 'installment','protected_class']
cat_cols = ['grade','emp_length', 'home_ownership','verification_status', 'term', 'initial_list_status',\
'disbursement_method', 'application_type']
other_cols = ['zip_code']
response_col = 'loan_status'
# Get train and test
df_x = pd.get_dummies(df[quant_cols+cat_cols], drop_first=False, columns=cat_cols)
df_y = df[response_col]
x_train, x_test, y_train, y_test = train_test_split(df_x, df_y, test_size=0.3, random_state=42)
# -
# ## Fitting our model
#
# Let's fit a Random Forest to our data. Because we artificially added a biased "protected class" column, our classifier will not be fair.
# +
# Fit model
model = RandomForestClassifier(n_estimators=25, max_depth=None, #class_weight='balanced_subsample', \
random_state=42).fit(x_train, y_train)
# -
# ## Getting predictions
# +
# Get predictions
preds_test = model.predict(x_test)
acc_train = model.score(x_train, y_train)
acc_test = model.score(x_test, y_test)
print(acc_train)
print(acc_test)
# -
# # Statistical Parity
#
# We will first test our model's predictions with statistical parity, a simple fairness measure that is easy to compute.
#
# ## What is statistical parity?
#
# This metric measures the difference between the probability of positive decisions for the protected group and the probability of positive decisions for ghe unprotected group. Mathematically:
# $$Sp = P(d=1|G=0) - P(d=1|G=1)$$
#
# This can be easily approximated with our data by calculating the proportion of positive decisions amongst people from race "0" and substracting the proportion of positive decisions amongst people from race "1":
#
# $$Sp = \frac{ \text{# people with positive decision and race 0}} { \text{ # people from race 0} } - \frac{ \text{# people with positive decision and race 1}} { \text{ # people from race 1}}$$
# Let's code a simple function that will calculate this for our dataset. In the next cell, complete the function `evaluate_statistical_parity` to perform the calculation above. The function definition and docstring will guide you.
# Statistical parity function
def evaluate_statistical_parity(predictions, protected_class_array):
"""Function to calculate statistical parity.
Parameters
----------
predictions (numpy array): binary decision labels outputted by our trained model.
protected_class_array (numpy array): boolean mask where protected rows are marked True
Returns
-------
bias (float): statistical parity bias
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
prob_g = np.sum(predictions & protected_class_array) / np.sum(protected_class_array)
prob_not_g = np.sum(predictions & ~protected_class_array) / np.sum(~protected_class_array)
bias = np.abs(prob_g - prob_not_g)
return bias
# # Conditional Parity
#
# Statistical parity is a simple measure, and it gives a fast overview on our model's fairness. However, it disregards important aspects of our dataset, such as the values of the features of each row. We could have a situation where the statistical parity measure tells us that we are giving loans to 20% of people from race 0 and 20% of people from race 1, which would be fair, but those 20% from race 0 are random, while the 20% from race 1 are people from developed countries. Our model would be hiding another layer of unfairness: we are not giving loans equally to people from race 1.
#
# We can use conditional parity to detect these types of imbalances. Conditional parity allows us to test for unfairness in a similar way as Statistical Parity, but conditioning on another feature (for example, country of origin). The equation is:
#
# $$Cp = P(d=1|G=0, L=l) - P(d=1|G=1, L=l)$$
#
# Again, this can be easily calculated by counting the number of positive outcome cases in por both protected groups, but this time only looking at the people that fulfill our conditional constraint (L=l)
# Conditional parity function
def evaluate_conditional_parity(predictions, protected_class_array, condition_array):
"""Function to calculate Conditional statistical parity.
Parameters
----------
predictions (numpy array): binary (decision) labels for X
protected_class_array (numpy array): boolean array where protected rows are marked True
condition_array (numpy array): boolean array that indicates conditional status
Returns
-------
bias (float): conditional parity bias
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
prob_g = np.sum(predictions & condition_array & protected_class_array) / np.sum(predictions & protected_class_array)
prob_not_g = np.sum(predictions & condition_array & ~protected_class_array) / np.sum(predictions & ~protected_class_array)
bias = np.abs(prob_g - prob_not_g)
return bias
# Evaluate statistical and conditional parity
stat_parity = evaluate_statistical_parity([bool(x) for x in preds_test], ~x_test['protected_class'].apply(lambda x: bool(x)))
cond_parity = evaluate_conditional_parity([bool(x) for x in preds_test], ~x_test['protected_class'].apply(lambda x: bool(x)), x_test['loan_amnt']>10000)
print(stat_parity)
print(cond_parity)
# # False Positive (Negative) Error Rate Balance
#
# The previous measures don't take into account the real labels of each observation; they only consider the predictions. The measure of fairness proposed here controls for equal poportions of false positives/false negatives in protected and unprotected classes. This measure is ideal in cases where committing mistakes disproportionately for different protected groups can bring negative outcomes.
#
# We will again code these measures as they are rather easy to understand. The function definition below will guide you through the process.
# +
# False positive and false negative rates
def evaluate_false_negative_rate(predictions, protected, y):
"""evaluate fnr
Parameters
----------
predictions (numpy array): binary (decision) labels for X predicted by our model
protected (numpy array): boolean mask where protected rows are marked True or 1
y (numpy array): boolean array that marks ground truth
Note:
FNR: FN / CP where FN=(predictions==0) & (y==1) CN = (y==1)
Returns
-------
bias (float)
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
cond_pos_protected = np.sum((y==1) & protected)
cond_pos_not_protected = np.sum((y==1) & ~protected)
if cond_pos_protected == 0:
return 'No Condition Positive in Protected'
if cond_pos_not_protected == 0:
return 'No Condition Positive in Not Protected'
false_neg_protected = np.sum((y==1) & (predictions==0) & protected)
false_neg_not_protected = np.sum((y==1) & (predictions==0) & ~protected)
fnr_g = false_neg_protected / cond_pos_protected
fnr_not_g = false_neg_not_protected / cond_pos_not_protected
bias = np.abs(fnr_g - fnr_not_g)
return bias
def evaluate_false_positive_rate(predictions, protected, y):
"""evaluate fpr
Parameters
----------
predictions (numpy array): binary (decision) labels for X predicted by our model
protected (numpy array): boolean mask where protected rows are marked True or 1
y (numpy array): boolean array that marks ground truth
Note:
FPR: FP / CN where FP=(predictions==1) & (y==0) CN = (y==0)
Returns
-------
bias (float)
"""
# --------------
# --------------
# Your code here
# --------------
# --------------
cond_neg_protected = np.sum((y==0) & protected)
cond_neg_not_protected = np.sum((y==0) & ~protected)
if cond_neg_protected == 0:
return 'No Condition Negative in Protected'
if cond_neg_not_protected == 0:
return 'No Condition Negative in Not Protected'
false_pos_protected = np.sum((y==0) & predictions & protected)
false_pos_not_protected = np.sum((y==0) & predictions & ~protected)
fpr_g = false_pos_protected / cond_neg_protected
fpr_not_g = false_pos_not_protected / cond_neg_not_protected
bias = np.abs(fpr_g - fpr_not_g)
return bias
# +
# Test FPR and FNR on this dataset
fnr = evaluate_false_negative_rate(x_test, preds_test, ~x_test['protected_class'], y_test)
fpr = evaluate_false_positive_rate(x_test, preds_test, ~x_test['protected_class'], y_test)
print(fpr)
print(fnr)
# -
# As we can see, the values of FPR and FNR are significantly higher than expected, showing that our dataset is clearly unfair.
# ## Other Fairness metrics
#
# We have coded and tested some basic Fairness metrics, but there are multiple other metrics that can be used, depending on the situation. Some of them are:
#
# **Predictive parity:**
# The fraction of correct positive predictions should be the same for protected and unprotected groups.
# $$P(Y=1|d=1, G=m) = P(Y=1|d=1, G=f)$$
#
#
# **Equalized odds:**
# Applicants with a good actual credit scope and applicants with a bad actual credit
# score should have a similar classification, regardless of the value of the protected class.
# $$P(d=1|Y=i, G=m) = P(d=1|Y=i, G=f), i\in \{0,1\}$$
#
#
# **Overall accuracy equality:**
# Both protected and unprotected groups have equal prediction accuracy.
# $$P(d=Y, G=m) = P(d=Y, G=f)$$
#
#
# **Treatment Equality:**
# Looks at ratio of errors a classifier makes instead of its accuracy. Satisfied if both protected and unprotected groups have equal ratio of false negatives and false positives.
#
# ## Fairness concepts
# - **Fairness through unawareness:**
# No sensitive attributes used in the decision making process.
# - **Fairness through awareness:**
# Similar individuals should have similar classification.
# - **Disparate impact:**
# Exists when decision outcomes disproportionately benefits or hurts individuals of a certain group.
# - **Disparate treatment:**
# Decision changes when protected feature changes.
# - **Disparate mistreatment:**
# Missclassification rates are different for people of different protected groups
#
# We refer the reader to http://fairware.cs.umass.edu/papers/Verma.pdf for more information.
# # Creating a Fair Model
#
# Once we have characterized and measured the fairness of the model, we might want to build a model that avoids discrimination given a protected class. As there are multiple ways to define fairness, there are also multiple ways to build a fair classifier, depending on what notion we want to emphasize.
#
# Some options are:
# - Preprocessing the data to remove biases, and training normal classifiers on that data
# - Training the classifier and post-processing the predictions to accomodate our measures of fairness
# - Training a modified classifier with clear constraints that enforce fairness
#
# We will exemplify the Optimized Preprocessing technique, published by our very own Flavio Calmon.
#
# ![../optimized.PNG]
# ### Census Income dataset
#
# The previous dataset was thorough and complex enough to demonstrate interpretability techniques, but as it is an anonymized dataset, it has little to no information on sensitive features. We will switch to another dataset for this part that is more suited to analyzing fairness techniques, as it possesses information on gender and race.
#
# This dataset is called the **Census Income dataset**, and it associates features of working adults to **whether or not they make more than $50k/yr**. It is extracted from the 1994 Census database, and contains **48842 observations** with a mix of continuous and categorical features (14 in total).
#
# List of features:
# - **age:** continuous.
# - **workclass:** categorical.
# - **education:** categorical.
# - **education-num:** continuous.
# - **marital-status:** categorical.
# - **relationship:** categorical.
# - **race:** categorical.
# - **sex:** categorical.
# - **capital-gain:** continuous.
# - **capital-loss:** continuous.
# - **hours-per-week:** continuous.
# - **fnlwgt:** (final weight) continuous.
# - **native-country:** categorical.
#
# Response: binary, corresponding to >50K (1) or <=50K (0).
#
#
# #### Reference:
# <NAME>, "Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid", Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, 1996
#
# +
# Imports
from aif360.algorithms.preprocessing import OptimPreproc
from sklearn.preprocessing import StandardScaler
from aif360.algorithms.preprocessing.optim_preproc_helpers.data_preproc_functions import load_preproc_data_german
from aif360.algorithms.preprocessing.optim_preproc_helpers.distortion_functions import get_distortion_german
from aif360.algorithms.preprocessing.optim_preproc_helpers.opt_tools import OptTools
from aif360.datasets import BinaryLabelDataset
from aif360.datasets import AdultDataset, GermanDataset, CompasDataset
from aif360.metrics import BinaryLabelDatasetMetric
from aif360.metrics import ClassificationMetric
from aif360.metrics.utils import compute_boolean_conditioning_vector
from aif360.algorithms.preprocessing.optim_preproc import OptimPreproc
from aif360.algorithms.preprocessing.optim_preproc_helpers.data_preproc_functions\
import load_preproc_data_adult, load_preproc_data_german, load_preproc_data_compas
from aif360.algorithms.preprocessing.optim_preproc_helpers.distortion_functions\
import get_distortion_adult, get_distortion_german, get_distortion_compas
from aif360.algorithms.preprocessing.optim_preproc_helpers.opt_tools import OptTools
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score
from IPython.display import Markdown, display
import matplotlib.pyplot as plt
# +
# Load German dataset with 'sex' as protected attribute
# privileged_groups = [{'sex': 1}]
# unprivileged_groups = [{'sex': 0}]
# dataset_orig = load_preproc_data_german(['sex'])
# optim_options = {
# "distortion_fun": get_distortion_german,
# "epsilon": 0.05,
# "clist": [0.99, 1.99, 2.99],
# "dlist": [.1, 0.05, 0]
# }
privileged_groups = [{'sex': 1}]
unprivileged_groups = [{'sex': 0}]
dataset_orig = load_preproc_data_adult(['sex'])
optim_options = {
"distortion_fun": get_distortion_adult,
"epsilon": 0.05,
"clist": [0.99, 1.99, 2.99],
"dlist": [.1, 0.05, 0]
}
# Split into train and test
dataset_orig_train, dataset_orig_test = dataset_orig.split([0.7], shuffle=True)
# -
print('Training Dataset shape:',dataset_orig_train.features.shape)
# print('Favorable and unfavorable labels:',dataset_orig_train.favorable_label, dataset_orig_train.unfavorable_label)
print('Protected attribute names:',dataset_orig_train.protected_attribute_names)
print('Privileged and unprivileged protected attribute values:' ,dataset_orig_train.privileged_protected_attributes,
dataset_orig_train.unprivileged_protected_attributes)
print('Dataset feature names:',dataset_orig_train.feature_names)
# +
# Extract data from AIF360 Data object
# We define a scaler to normalize our data
scale_orig = StandardScaler()
# We get our training numpy arrays
x_train = scale_orig.fit_transform(dataset_orig_train.features) #This fit_transform scales our data feature-wise.
y_train = (dataset_orig_train.labels.ravel()-2)*-1
y_train = dataset_orig_train.labels.ravel()
# And our testing arrays
x_test = scale_orig.transform(dataset_orig_test.features) # Here, we only transform, as we can't use the testing set to define the scaling factors.
y_test = (dataset_orig_test.labels.ravel()-2)*-1
y_test = dataset_orig_test.labels.ravel()
# +
# Train classifier on original data
rf_model = RandomForestClassifier(n_estimators=25,
max_depth=None,
random_state=42).fit(x_train, y_train)
rf_model.fit(x_train, y_train)
# +
# Getting accuracy and fairness metrics on test
acc_orig = rf_model.score(x_test, y_test)
print('Accuracy on test with original data:', acc_orig)
print(dataset_orig_test.feature_names)
predictions = rf_model.predict(x_test)>0.5
protected_class_array = dataset_orig_test.features[:,1]==1 # Here, we're taking the column corresponding to 'sex' and we are transforming it into a boolean array
statistical_parity_orig = evaluate_statistical_parity(predictions, protected_class_array)
fpr_orig = evaluate_false_positive_rate(predictions, protected_class_array, y_test)
fnr_orig = evaluate_false_negative_rate(predictions, protected_class_array, y_test)
print(statistical_parity_orig, fpr_orig, fnr_orig)
# metric_test_bef = compute_metrics(dataset_transf_test, dataset_transf_test_pred,
# unprivileged_groups, privileged_groups, disp=disp)
# bal_acc_arr_transf.append(metric_test_bef["Balanced accuracy"])
# avg_odds_diff_arr_transf.append(metric_test_bef["Average odds difference"])
# disp_imp_arr_transf.append(metric_test_bef["Disparate impact"])
# -
# ### Now, let's apply a dataset transformation to increase fairness !
# +
# Instantiate OptimizedDataPreprocessing module from AIF360
OP = OptimPreproc(OptTools, optim_options,
unprivileged_groups = unprivileged_groups,
privileged_groups = privileged_groups)
# Fit the module to the training data, effectively creating the mapping from original data to transformed, fair data
OP = OP.fit(dataset_orig_train)
# +
# Transform training data and align features
dataset_transf_train = OP.transform(dataset_orig_train, transform_Y=True)
dataset_transf_train = dataset_orig_train.align_datasets(dataset_transf_train)
# Same with test data
dataset_transf_test = OP.transform(dataset_orig_test, transform_Y = True)
dataset_transf_test = dataset_orig_test.align_datasets(dataset_transf_test)
# +
# Again, we have to get our training numpy arrays, this time on the TRANSFORMED training data
x_train_transf = scale_orig.fit_transform(dataset_transf_train.features)
y_train_transf = (dataset_transf_train.labels.ravel()-2)*-1
y_train_transf = dataset_transf_train.labels.ravel()
# And our testing arrays, on the TRANSFORMED test data
x_test_transf = scale_orig.transform(dataset_transf_test.features) # Here, we only transform, as we can't use the testing set to define the scaling factors.
y_test_transf = (dataset_transf_test.labels.ravel()-2)*-1
y_test_transf = dataset_transf_test.labels.ravel()
# -
# Train same classifier on TRANSFORMED data
rf_model_transf = RandomForestClassifier(n_estimators=25,
max_depth=None,
random_state=42).fit(x_train_transf, y_train_transf)
rf_model_transf.fit(x_train_transf, y_train_transf)
# +
# Getting accuracy and fairness metrics on TRANSFORMED test set
acc_transf = rf_model_transf.score(x_test_transf, y_test_transf)
print('Accuracy on test with original data (we should expect a bit less than before):', acc_transf)
predictions_transf = rf_model_transf.predict(x_test_transf)>0.5
protected_class_array_transf = dataset_orig_test.features[:,1]==1
statistical_parity_transf = evaluate_statistical_parity(predictions_transf, protected_class_array_transf)
fpr_transf = evaluate_false_positive_rate(predictions_transf, protected_class_array_transf, y_test_transf)
fnr_transf = evaluate_false_negative_rate(predictions_transf, protected_class_array_transf, y_test_transf)
print(statistical_parity_transf, fpr_transf, fnr_transf)
# +
# Compare the Results
# +
# +
# Predictions and fairness metrics on transformed test set
# -
dataset_transf_test_pred = dataset_transf_test.copy(deepcopy=True)
X_test = scale_transf.transform(dataset_transf_test_pred.features)
y_test = dataset_transf_test_pred.labels
dataset_transf_test_pred.scores = lmod.predict_proba(X_test)[:,pos_ind].reshape(-1,1)
# +
# Results
# -
Disparate impact
Average odds difference
Balanced accuracy
# # Conclusion
#
# We have analyzed particular fairness metrics and observed their behavior on an artificial dataset. It is important to remember that Fairness has multiple definitions, each one approriate for analyzing a specific situation. Statistical notions of fairness as described above are easy to measure. However, it is important to keep in mind that statistical definitions are insufficient in some cases (for example, when similarity has to be taken into account). Moreover, most valuable statistical metrics assume availability of actual, verified outcomes. While such outcomes are available for the training data, it is unclear whether the real classified data always conforms to the same distribution.
# # Appendix: extra resources
#
# ## Interesting Fairness analysis tools
# - Pymetrics audit-ai (https://github.com/pymetrics/audit-ai)
# - fairness metrics github (https://github.com/megantosh/fairness_measures_code)
# - fairness-comparison github (https://github.com/algofairness/fairness-comparison)
# - IBM AIF360 (https://github.com/IBM/AIF360, https://arxiv.org/pdf/1810.01943.pdf)
# - Themis ML (https://themis-ml.readthedocs.io/en/latest/)
# - FairML (https://github.com/adebayoj/fairml)
# - BlackBoxAuditing (https://github.com/algofairness/BlackBoxAuditing)
#
# ## Interesting papers
# - Learning Fair Representations (seminal paper) http://proceedings.mlr.press/v28/zemel13.pdf
# - Optimized Data Pre-Processing for Discrimination Prevention (by <NAME>) https://arxiv.org/pdf/1704.03354.pdf
# - Fairness Definitions Explained http://fairware.cs.umass.edu/papers/Verma.pdf
# - From parity to Preference-based notions of fairness https://arxiv.org/abs/1707.00010
# - Certifying and removing disparate impact https://arxiv.org/pdf/1412.3756.pdf
# - Learning Classification without Disparate Mistreatment https://arxiv.org/pdf/1610.08452.pdf
# - Fairness Constraints: Mechanisms for Fair Classification https://arxiv.org/abs/1507.05259
# - Fairness GAN https://arxiv.org/pdf/1805.09910.pdf
# - Adversarial Debiasing https://arxiv.org/pdf/1801.07593.pdf
# - Classification with Fairness Constraints: A Meta-Algorithm with Provable Guarantees https://arxiv.org/pdf/1806.06055.pdf
#
#
#
#
# +
# add IBM AIF360 examples if time
# +
# German Loan Dataset
import aif360
# +
from aif360.algorithms.preprocessing import DisparateImpactRemover
from aif360.datasets import AdultDataset
from aif360.metrics import BinaryLabelDatasetMetric
protected = 'sex'
# ad = AdultDataset(protected_attribute_names=[protected],
# privileged_classes=[['Male']], categorical_features=[],
# features_to_keep=['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week'])
data = GermanDataset()
print(data.feature_names)
aif360.algorithms.preprocessing.OptimPreproc(optimizer, optim_options, unprivileged_groups, privileged_groups, verbose=False, seed=None)
['age', 'sex', 'credit_history=Delay', 'credit_history=None/Paid', 'credit_history=Other', 'savings=500+', 'savings=<500', 'savings=Unknown/None', 'employment=1-4 years', 'employment=4+ years', 'employment=Unemployed']
# +
import urllib.request
url1 = 'https://archive.ics.uci.edu/ml/machine-learning-databases/statlog/german/german.data'
url2 = 'https://archive.ics.uci.edu/ml/machine-learning-databases/statlog/german/german.doc'
urllib.request.urlretrieve(url1,'C:/Users/Camilo/Anaconda3/lib/site-packages/aif360/data/raw/german/german.data')
urllib.request.urlretrieve(url2,'C:/Users/Camilo/Anaconda3/lib/site-packages/aif360/data/raw/german/german.doc')
# -
| notebooks/cf_fairness.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: dl
# language: python
# name: dl
# ---
# # Skip-gram Word2Vec
#
# In this notebook, I'll lead you through using PyTorch to implement the [Word2Vec algorithm](https://en.wikipedia.org/wiki/Word2vec) using the skip-gram architecture. By implementing this, you'll learn about embedding words for use in natural language processing. This will come in handy when dealing with things like machine translation.
#
# ## Readings
#
# Here are the resources I used to build this notebook. I suggest reading these either beforehand or while you're working on this material.
#
# * A really good [conceptual overview](http://mccormickml.com/2016/04/19/word2vec-tutorial-the-skip-gram-model/) of Word2Vec from <NAME>
# * [First Word2Vec paper](https://arxiv.org/pdf/1301.3781.pdf) from Mikolov et al.
# * [Neural Information Processing Systems, paper](http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf) with improvements for Word2Vec also from Mikolov et al.
#
# ---
# ## Word embeddings
#
# When you're dealing with words in text, you end up with tens of thousands of word classes to analyze; one for each word in a vocabulary. Trying to one-hot encode these words is massively inefficient because most values in a one-hot vector will be set to zero. So, the matrix multiplication that happens in between a one-hot input vector and a first, hidden layer will result in mostly zero-valued hidden outputs.
#
# To solve this problem and greatly increase the efficiency of our networks, we use what are called **embeddings**. Embeddings are just a fully connected layer like you've seen before. We call this layer the embedding layer and the weights are embedding weights. We skip the multiplication into the embedding layer by instead directly grabbing the hidden layer values from the weight matrix. We can do this because the multiplication of a one-hot encoded vector with a matrix returns the row of the matrix corresponding the index of the "on" input unit.
#
# <img src='assets/lookup_matrix.png' width=50%>
#
# Instead of doing the matrix multiplication, we use the weight matrix as a lookup table. We encode the words as integers, for example "heart" is encoded as 958, "mind" as 18094. Then to get hidden layer values for "heart", you just take the 958th row of the embedding matrix. This process is called an **embedding lookup** and the number of hidden units is the **embedding dimension**.
#
# There is nothing magical going on here. The embedding lookup table is just a weight matrix. The embedding layer is just a hidden layer. The lookup is just a shortcut for the matrix multiplication. The lookup table is trained just like any weight matrix.
#
# Embeddings aren't only used for words of course. You can use them for any model where you have a massive number of classes. A particular type of model called **Word2Vec** uses the embedding layer to find vector representations of words that contain semantic meaning.
# ---
# ## Word2Vec
#
# The Word2Vec algorithm finds much more efficient representations by finding vectors that represent the words. These vectors also contain semantic information about the words.
#
# <img src="assets/context_drink.png" width=40%>
#
# Words that show up in similar **contexts**, such as "coffee", "tea", and "water" will have vectors near each other. Different words will be further away from one another, and relationships can be represented by distance in vector space.
#
#
# There are two architectures for implementing Word2Vec:
# >* CBOW (Continuous Bag-Of-Words) and
# * Skip-gram
#
# <img src="assets/word2vec_architectures.png" width=60%>
#
# In this implementation, we'll be using the **skip-gram architecture** with **negative sampling** because it performs better than CBOW and trains faster with negative sampling. Here, we pass in a word and try to predict the words surrounding it in the text. In this way, we can train the network to learn representations for words that show up in similar contexts.
# ---
# ## Loading Data
#
# Next, we'll ask you to load in data and place it in the `data` directory
#
# 1. Load the [text8 dataset](https://s3.amazonaws.com/video.udacity-data.com/topher/2018/October/5bbe6499_text8/text8.zip); a file of cleaned up *Wikipedia article text* from <NAME>.
# 2. Place that data in the `data` folder in the home directory.
# 3. Then you can extract it and delete the archive, zip file to save storage space.
#
# After following these steps, you should have one file in your data directory: `data/text8`.
# +
# read in the extracted text file
with open('data/text8') as f:
text = f.read()
# print out the first 100 characters
print(text[:100])
# -
# ## Pre-processing
#
# Here I'm fixing up the text to make training easier. This comes from the `utils.py` file. The `preprocess` function does a few things:
# >* It converts any punctuation into tokens, so a period is changed to ` <PERIOD> `. In this data set, there aren't any periods, but it will help in other NLP problems.
# * It removes all words that show up five or *fewer* times in the dataset. This will greatly reduce issues due to noise in the data and improve the quality of the vector representations.
# * It returns a list of words in the text.
#
# This may take a few seconds to run, since our text file is quite large. If you want to write your own functions for this stuff, go for it!
# +
import utils
# get list of words
words = utils.preprocess(text)
print(words[:30])
# -
# print some stats about this word data
print("Total words in text: {}".format(len(words)))
print("Unique words: {}".format(len(set(words)))) # `set` removes any duplicate words
# ### Dictionaries
#
# Next, I'm creating two dictionaries to convert words to integers and back again (integers to words). This is again done with a function in the `utils.py` file. `create_lookup_tables` takes in a list of words in a text and returns two dictionaries.
# >* The integers are assigned in descending frequency order, so the most frequent word ("the") is given the integer 0 and the next most frequent is 1, and so on.
#
# Once we have our dictionaries, the words are converted to integers and stored in the list `int_words`.
# +
vocab_to_int, int_to_vocab = utils.create_lookup_tables(words)
int_words = [vocab_to_int[word] for word in words]
print(int_words[:30])
# -
# ## Subsampling
#
# Words that show up often such as "the", "of", and "for" don't provide much context to the nearby words. If we discard some of them, we can remove some of the noise from our data and in return get faster training and better representations. This process is called subsampling by Mikolov. For each word $w_i$ in the training set, we'll discard it with probability given by
#
# $$ P(w_i) = 1 - \sqrt{\frac{t}{f(w_i)}} $$
#
# where $t$ is a threshold parameter and $f(w_i)$ is the frequency of word $w_i$ in the total dataset.
#
# > Implement subsampling for the words in `int_words`. That is, go through `int_words` and discard each word given the probablility $P(w_i)$ shown above. Note that $P(w_i)$ is the probability that a word is discarded. Assign the subsampled data to `train_words`.
# +
from collections import Counter
import random
import numpy as np
threshold = 1e-5
word_counts = Counter(int_words)
#print(list(word_counts.items())[0]) # dictionary of int_words, how many times they appear
total_count = len(int_words)
freqs = {word: count/total_count for word, count in word_counts.items()}
p_drop = {word: 1 - np.sqrt(threshold/freqs[word]) for word in word_counts}
# discard some frequent words, according to the subsampling equation
# create a new list of words for training
train_words = [word for word in int_words if random.random() < (1 - p_drop[word])]
print(train_words[:30])
# -
# ## Making batches
# Now that our data is in good shape, we need to get it into the proper form to pass it into our network. With the skip-gram architecture, for each word in the text, we want to define a surrounding _context_ and grab all the words in a window around that word, with size $C$.
#
# From [Mikolov et al.](https://arxiv.org/pdf/1301.3781.pdf):
#
# "Since the more distant words are usually less related to the current word than those close to it, we give less weight to the distant words by sampling less from those words in our training examples... If we choose $C = 5$, for each training word we will select randomly a number $R$ in range $[ 1: C ]$, and then use $R$ words from history and $R$ words from the future of the current word as correct labels."
#
# > **Exercise:** Implement a function `get_target` that receives a list of words, an index, and a window size, then returns a list of words in the window around the index. Make sure to use the algorithm described above, where you chose a random number of words to from the window.
#
# Say, we have an input and we're interested in the idx=2 token, `741`:
# ```
# [5233, 58, 741, 10571, 27349, 0, 15067, 58112, 3580, 58, 10712]
# ```
#
# For `R=2`, `get_target` should return a list of four values:
# ```
# [5233, 58, 10571, 27349]
# ```
def get_target(words, idx, window_size=5):
''' Get a list of words in a window around an index. '''
R = np.random.randint(1, window_size+1)
start = idx - R if (idx - R) > 0 else 0
stop = idx + R
target_words = words[start:idx] + words[idx+1:stop+1]
return list(target_words)
# +
# test your code!
# run this cell multiple times to check for random window selection
int_text = [i for i in range(10)]
print('Input: ', int_text)
idx=5 # word index of interest
target = get_target(int_text, idx=idx, window_size=5)
print('Target: ', target) # you should get some indices around the idx
# -
# ### Generating Batches
#
# Here's a generator function that returns batches of input and target data for our model, using the `get_target` function from above. The idea is that it grabs `batch_size` words from a words list. Then for each of those batches, it gets the target words in a window.
def get_batches(words, batch_size, window_size=5):
''' Create a generator of word batches as a tuple (inputs, targets) '''
n_batches = len(words)//batch_size
# only full batches
words = words[:n_batches*batch_size]
for idx in range(0, len(words), batch_size):
x, y = [], []
batch = words[idx:idx+batch_size]
for ii in range(len(batch)):
batch_x = batch[ii]
batch_y = get_target(batch, ii, window_size)
y.extend(batch_y)
x.extend([batch_x]*len(batch_y))
yield x, y
# +
int_text = [i for i in range(20)]
x,y = next(get_batches(int_text, batch_size=4, window_size=5))
print('x\n', x)
print('y\n', y)
# -
# ---
# ## Validation
#
# Here, I'm creating a function that will help us observe our model as it learns. We're going to choose a few common words and few uncommon words. Then, we'll print out the closest words to them using the cosine similarity:
#
# <img src="assets/two_vectors.png" width=30%>
#
# $$
# \mathrm{similarity} = \cos(\theta) = \frac{\vec{a} \cdot \vec{b}}{|\vec{a}||\vec{b}|}
# $$
#
#
# We can encode the validation words as vectors $\vec{a}$ using the embedding table, then calculate the similarity with each word vector $\vec{b}$ in the embedding table. With the similarities, we can print out the validation words and words in our embedding table semantically similar to those words. It's a nice way to check that our embedding table is grouping together words with similar semantic meanings.
def cosine_similarity(embedding, valid_size=16, valid_window=100, device='cpu'):
""" Returns the cosine similarity of validation words with words in the embedding matrix.
Here, embedding should be a PyTorch embedding module.
"""
# Here we're calculating the cosine similarity between some random words and
# our embedding vectors. With the similarities, we can look at what words are
# close to our random words.
# sim = (a . b) / |a||b|
embed_vectors = embedding.weight
# magnitude of embedding vectors, |b|
magnitudes = embed_vectors.pow(2).sum(dim=1).sqrt().unsqueeze(0)
# pick N words from our ranges (0,window) and (1000,1000+window). lower id implies more frequent
valid_examples = np.array(random.sample(range(valid_window), valid_size//2))
valid_examples = np.append(valid_examples,
random.sample(range(1000,1000+valid_window), valid_size//2))
valid_examples = torch.LongTensor(valid_examples).to(device)
valid_vectors = embedding(valid_examples)
similarities = torch.mm(valid_vectors, embed_vectors.t())/magnitudes
return valid_examples, similarities
# ---
# # SkipGram model
#
# Define and train the SkipGram model.
# > You'll need to define an [embedding layer](https://pytorch.org/docs/stable/nn.html#embedding) and a final, softmax output layer.
#
# An Embedding layer takes in a number of inputs, importantly:
# * **num_embeddings** – the size of the dictionary of embeddings, or how many rows you'll want in the embedding weight matrix
# * **embedding_dim** – the size of each embedding vector; the embedding dimension
#
# Below is an approximate diagram of the general structure of our network.
# <img src="assets/skip_gram_arch.png" width=60%>
#
# >* The input words are passed in as batches of input word tokens.
# * This will go into a hidden layer of linear units (our embedding layer).
# * Then, finally into a softmax output layer.
#
# We'll use the softmax layer to make a prediction about the context words by sampling, as usual.
# ---
# ## Negative Sampling
#
# For every example we give the network, we train it using the output from the softmax layer. That means for each input, we're making very small changes to millions of weights even though we only have one true example. This makes training the network very inefficient. We can approximate the loss from the softmax layer by only updating a small subset of all the weights at once. We'll update the weights for the correct example, but only a small number of incorrect, or noise, examples. This is called ["negative sampling"](http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf).
#
# There are two modifications we need to make. First, since we're not taking the softmax output over all the words, we're really only concerned with one output word at a time. Similar to how we use an embedding table to map the input word to the hidden layer, we can now use another embedding table to map the hidden layer to the output word. Now we have two embedding layers, one for input words and one for output words. Secondly, we use a modified loss function where we only care about the true example and a small subset of noise examples.
#
# $$
# - \large \log{\sigma\left(u_{w_O}\hspace{0.001em}^\top v_{w_I}\right)} -
# \sum_i^N \mathbb{E}_{w_i \sim P_n(w)}\log{\sigma\left(-u_{w_i}\hspace{0.001em}^\top v_{w_I}\right)}
# $$
#
# This is a little complicated so I'll go through it bit by bit. $u_{w_O}\hspace{0.001em}^\top$ is the embedding vector for our "output" target word (transposed, that's the $^\top$ symbol) and $v_{w_I}$ is the embedding vector for the "input" word. Then the first term
#
# $$\large \log{\sigma\left(u_{w_O}\hspace{0.001em}^\top v_{w_I}\right)}$$
#
# says we take the log-sigmoid of the inner product of the output word vector and the input word vector. Now the second term, let's first look at
#
# $$\large \sum_i^N \mathbb{E}_{w_i \sim P_n(w)}$$
#
# This means we're going to take a sum over words $w_i$ drawn from a noise distribution $w_i \sim P_n(w)$. The noise distribution is basically our vocabulary of words that aren't in the context of our input word. In effect, we can randomly sample words from our vocabulary to get these words. $P_n(w)$ is an arbitrary probability distribution though, which means we get to decide how to weight the words that we're sampling. This could be a uniform distribution, where we sample all words with equal probability. Or it could be according to the frequency that each word shows up in our text corpus, the unigram distribution $U(w)$. The authors found the best distribution to be $U(w)^{3/4}$, empirically.
#
# Finally, in
#
# $$\large \log{\sigma\left(-u_{w_i}\hspace{0.001em}^\top v_{w_I}\right)},$$
#
# we take the log-sigmoid of the negated inner product of a noise vector with the input vector.
#
# <img src="assets/neg_sampling_loss.png" width=50%>
#
# To give you an intuition for what we're doing here, remember that the sigmoid function returns a probability between 0 and 1. The first term in the loss pushes the probability that our network will predict the correct word $w_O$ towards 1. In the second term, since we are negating the sigmoid input, we're pushing the probabilities of the noise words towards 0.
import torch
from torch import nn
import torch.optim as optim
class SkipGramNeg(nn.Module):
def __init__(self, n_vocab, n_embed, noise_dist=None):
super().__init__()
self.n_vocab = n_vocab
self.n_embed = n_embed
self.noise_dist = noise_dist
# define embedding layers for input and output words
self.in_embed = nn.Embedding(n_vocab, n_embed)
self.out_embed = nn.Embedding(n_vocab, n_embed)
# Initialize both embedding tables with uniform distribution
self.in_embed.weight.data.uniform_(-1, 1)
self.out_embed.weight.data.uniform_(-1, 1)
def forward_input(self, input_words):
# return input vector embeddings
input_vectors = self.in_embed(input_words)
return input_vectors
def forward_output(self, output_words):
# return output vector embeddings
output_vectors = self.out_embed(output_words)
return output_vectors
def forward_noise(self, batch_size, n_samples):
""" Generate noise vectors with shape (batch_size, n_samples, n_embed)"""
if self.noise_dist is None:
# Sample words uniformly
noise_dist = torch.ones(self.n_vocab)
else:
noise_dist = self.noise_dist
# Sample words from our noise distribution
noise_words = torch.multinomial(noise_dist,
batch_size * n_samples,
replacement=True)
device = "cuda" if model.out_embed.weight.is_cuda else "cpu"
noise_words = noise_words.to(device)
## TODO: get the noise embeddings
# reshape the embeddings so that they have dims (batch_size, n_samples, n_embed)
noise_vectors = self.out_embed(noise_words).view(batch_size, n_samples, self.n_embed)
return noise_vectors
class NegativeSamplingLoss(nn.Module):
def __init__(self):
super().__init__()
def forward(self, input_vectors, output_vectors, noise_vectors):
batch_size, embed_size = input_vectors.shape
# Input vectors should be a batch of column vectors
input_vectors = input_vectors.view(batch_size, embed_size, 1)
# Output vectors should be a batch of row vectors
output_vectors = output_vectors.view(batch_size, 1, embed_size)
# bmm = batch matrix multiplication
# correct log-sigmoid loss
out_loss = torch.bmm(output_vectors, input_vectors).sigmoid().log()
out_loss = out_loss.squeeze()
# incorrect log-sigmoid loss
noise_loss = torch.bmm(noise_vectors.neg(), input_vectors).sigmoid().log()
noise_loss = noise_loss.squeeze().sum(1) # sum the losses over the sample of noise vectors
# negate and sum correct and noisy log-sigmoid losses
# return average batch loss
return -(out_loss + noise_loss).mean()
# ### Training
#
# Below is our training loop, and I recommend that you train on GPU, if available.
# +
device = 'cuda' if torch.cuda.is_available() else 'cpu'
# Get our noise distribution
# Using word frequencies calculated earlier in the notebook
word_freqs = np.array(sorted(freqs.values(), reverse=True))
unigram_dist = word_freqs/word_freqs.sum()
noise_dist = torch.from_numpy(unigram_dist**(0.75)/np.sum(unigram_dist**(0.75)))
# instantiating the model
embedding_dim = 300
model = SkipGramNeg(len(vocab_to_int), embedding_dim, noise_dist=noise_dist).to(device)
# using the loss that we defined
criterion = NegativeSamplingLoss()
optimizer = optim.Adam(model.parameters(), lr=0.003)
print_every = 1500
steps = 0
epochs = 5
# train for some number of epochs
for e in range(epochs):
# get our input, target batches
for input_words, target_words in get_batches(train_words, 512):
steps += 1
inputs, targets = torch.LongTensor(input_words), torch.LongTensor(target_words)
inputs, targets = inputs.to(device), targets.to(device)
# input, outpt, and noise vectors
input_vectors = model.forward_input(inputs)
output_vectors = model.forward_output(targets)
noise_vectors = model.forward_noise(inputs.shape[0], 5)
# negative sampling loss
loss = criterion(input_vectors, output_vectors, noise_vectors)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# loss stats
if steps % print_every == 0:
print("Epoch: {}/{}".format(e+1, epochs))
print("Loss: ", loss.item()) # avg batch loss at this point in training
valid_examples, valid_similarities = cosine_similarity(model.in_embed, device=device)
_, closest_idxs = valid_similarities.topk(6)
valid_examples, closest_idxs = valid_examples.to('cpu'), closest_idxs.to('cpu')
for ii, valid_idx in enumerate(valid_examples):
closest_words = [int_to_vocab[idx.item()] for idx in closest_idxs[ii]][1:]
print(int_to_vocab[valid_idx.item()] + " | " + ', '.join(closest_words))
print("...\n")
# -
# ## Visualizing the word vectors
#
# Below we'll use T-SNE to visualize how our high-dimensional word vectors cluster together. T-SNE is used to project these vectors into two dimensions while preserving local stucture. Check out [this post from <NAME>](http://colah.github.io/posts/2014-10-Visualizing-MNIST/) to learn more about T-SNE and other ways to visualize high-dimensional data.
# +
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
# -
# getting embeddings from the embedding layer of our model, by name
embeddings = model.in_embed.weight.to('cpu').data.numpy()
viz_words = 380
tsne = TSNE()
embed_tsne = tsne.fit_transform(embeddings[:viz_words, :])
fig, ax = plt.subplots(figsize=(16, 16))
for idx in range(viz_words):
plt.scatter(*embed_tsne[idx, :], color='steelblue')
plt.annotate(int_to_vocab[idx], (embed_tsne[idx, 0], embed_tsne[idx, 1]), alpha=0.7)
| word2vec-embeddings/Negative_Sampling_Exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # TV Script Generation
# In this project, you'll generate your own [Simpsons](https://en.wikipedia.org/wiki/The_Simpsons) TV scripts using RNNs. You'll be using part of the [Simpsons dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data) of scripts from 27 seasons. The Neural Network you'll build will generate a new TV script for a scene at [Moe's Tavern](https://simpsonswiki.com/wiki/Moe's_Tavern).
# ## Get the Data
# The data is already provided for you. You'll be using a subset of the original dataset. It consists of only the scenes in Moe's Tavern. This doesn't include other versions of the tavern, like "Moe's Cavern", "Flaming Moe's", "Uncle Moe's Family Feed-Bag", etc..
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
data_dir = './data/simpsons/moes_tavern_lines.txt'
text = helper.load_data(data_dir)
# Ignore notice, since we don't use it for analysing the data
text = text[81:]
# -
# ## Explore the Data
# Play around with `view_sentence_range` to view different parts of the data.
# +
view_sentence_range = (0, 10)
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import numpy as np
print('Dataset Stats')
print('Roughly the number of unique words: {}'.format(len({word: None for word in text.split()})))
scenes = text.split('\n\n')
print('Number of scenes: {}'.format(len(scenes)))
sentence_count_scene = [scene.count('\n') for scene in scenes]
print('Average number of sentences in each scene: {}'.format(np.average(sentence_count_scene)))
sentences = [sentence for scene in scenes for sentence in scene.split('\n')]
print('Number of lines: {}'.format(len(sentences)))
word_count_sentence = [len(sentence.split()) for sentence in sentences]
print('Average number of words in each line: {}'.format(np.average(word_count_sentence)))
print()
print('The sentences {} to {}:'.format(*view_sentence_range))
print('\n'.join(text.split('\n')[view_sentence_range[0]:view_sentence_range[1]]))
# -
# ## Implement Preprocessing Functions
# The first thing to do to any dataset is preprocessing. Implement the following preprocessing functions below:
# - Lookup Table
# - Tokenize Punctuation
#
# ### Lookup Table
# To create a word embedding, you first need to transform the words to ids. In this function, create two dictionaries:
# - Dictionary to go from the words to an id, we'll call `vocab_to_int`
# - Dictionary to go from the id to word, we'll call `int_to_vocab`
#
# Return these dictionaries in the following tuple `(vocab_to_int, int_to_vocab)`
# +
import numpy as np
import problem_unittests as tests
def create_lookup_tables(text):
"""
Create lookup tables for vocabulary
:param text: The text of tv scripts split into words
:return: A tuple of dicts (vocab_to_int, int_to_vocab)
"""
# TODO: Implement Function
vocab = set(text)
vocab_to_int = {word: i for i,word in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
print(vocab_to_int)
print("\n")
print(int_to_vocab)
return vocab_to_int, int_to_vocab
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_create_lookup_tables(create_lookup_tables)
# -
# ### Tokenize Punctuation
# We'll be splitting the script into a word array using spaces as delimiters. However, punctuations like periods and exclamation marks make it hard for the neural network to distinguish between the word "bye" and "bye!".
#
# Implement the function `token_lookup` to return a dict that will be used to tokenize symbols like "!" into "||Exclamation_Mark||". Create a dictionary for the following symbols where the symbol is the key and value is the token:
# - Period ( . )
# - Comma ( , )
# - Quotation Mark ( " )
# - Semicolon ( ; )
# - Exclamation mark ( ! )
# - Question mark ( ? )
# - Left Parentheses ( ( )
# - Right Parentheses ( ) )
# - Dash ( -- )
# - Return ( \n )
#
# This dictionary will be used to token the symbols and add the delimiter (space) around it. This separates the symbols as it's own word, making it easier for the neural network to predict on the next word. Make sure you don't use a token that could be confused as a word. Instead of using the token "dash", try using something like "||dash||".
# +
def token_lookup():
"""
Generate a dict to turn punctuation into a token.
:return: Tokenize dictionary where the key is the punctuation and the value is the token
"""
# TODO: Implement Function
punc = {'.': '||period||',
',': '||comma||',
'"': '||quotation_mark||',
';': '||semicolon||',
'!': '||exclamation_mark||',
'?': '||question_mark||',
'(': '||left_parentheses',
')': '||right_parentheses',
'--': '||dash||',
'\n': '||return||'}
return punc
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_tokenize(token_lookup)
# -
# ## Preprocess all the data and save it
# Running the code cell below will preprocess all the data and save it to file.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Preprocess Training, Validation, and Testing Data
helper.preprocess_and_save_data(data_dir, token_lookup, create_lookup_tables)
# # Check Point
# This is your first checkpoint. If you ever decide to come back to this notebook or have to restart the notebook, you can start from here. The preprocessed data has been saved to disk.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import helper
import numpy as np
import problem_unittests as tests
int_text, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
# -
# ## Build the Neural Network
# You'll build the components necessary to build a RNN by implementing the following functions below:
# - get_inputs
# - get_init_cell
# - get_embed
# - build_rnn
# - build_nn
# - get_batches
#
# ### Check the Version of TensorFlow and Access to GPU
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from distutils.version import LooseVersion
import warnings
import tensorflow as tf
# Check TensorFlow Version
assert LooseVersion(tf.__version__) >= LooseVersion('1.3'), 'Please use TensorFlow version 1.3 or newer'
print('TensorFlow Version: {}'.format(tf.__version__))
# Check for a GPU
if not tf.test.gpu_device_name():
warnings.warn('No GPU found. Please use a GPU to train your neural network.')
else:
print('Default GPU Device: {}'.format(tf.test.gpu_device_name()))
# -
# ### Input
# Implement the `get_inputs()` function to create TF Placeholders for the Neural Network. It should create the following placeholders:
# - Input text placeholder named "input" using the [TF Placeholder](https://www.tensorflow.org/api_docs/python/tf/placeholder) `name` parameter.
# - Targets placeholder
# - Learning Rate placeholder
#
# Return the placeholders in the following tuple `(Input, Targets, LearningRate)`
# +
def get_inputs():
"""
Create TF Placeholders for input, targets, and learning rate.
:return: Tuple (input, targets, learning rate)
"""
# TODO: Implement Function
inputs = tf.placeholder(tf.int32, [None, None], name="input")
targets = tf.placeholder(tf.int32, [None, None], name="targets")
learning_rate = tf.placeholder(tf.float32, name="learning_rate")
return inputs, targets, learning_rate
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_inputs(get_inputs)
# -
# ### Build RNN Cell and Initialize
# Stack one or more [`BasicLSTMCells`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/BasicLSTMCell) in a [`MultiRNNCell`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell).
# - The Rnn size should be set using `rnn_size`
# - Initalize Cell State using the MultiRNNCell's [`zero_state()`](https://www.tensorflow.org/api_docs/python/tf/contrib/rnn/MultiRNNCell#zero_state) function
# - Apply the name "initial_state" to the initial state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)
#
# Return the cell and initial state in the following tuple `(Cell, InitialState)`
# +
def get_init_cell(batch_size, rnn_size):
"""
Create an RNN Cell and initialize it.
:param batch_size: Size of batches
:param rnn_size: Size of RNNs
:return: Tuple (cell, initialize state)
"""
# TODO: Implement Function
layers = 2
cells = []
for i in range(layers):
lstm = tf.contrib.rnn.BasicLSTMCell(rnn_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=0.7)
cells.append(drop)
cell = tf.contrib.rnn.MultiRNNCell(cells)
initial_state = tf.identity(cell.zero_state(batch_size, tf.float32), "initial_state")
return cell, initial_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_init_cell(get_init_cell)
# -
# ### Word Embedding
# Apply embedding to `input_data` using TensorFlow. Return the embedded sequence.
# +
def get_embed(input_data, vocab_size, embed_dim):
"""
Create embedding for <input_data>.
:param input_data: TF placeholder for text input.
:param vocab_size: Number of words in vocabulary.
:param embed_dim: Number of embedding dimensions
:return: Embedded input.
"""
# TODO: Implement Function
return tf.contrib.layers.embed_sequence(input_data, vocab_size, embed_dim)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_embed(get_embed)
# -
# ### Build RNN
# You created a RNN Cell in the `get_init_cell()` function. Time to use the cell to create a RNN.
# - Build the RNN using the [`tf.nn.dynamic_rnn()`](https://www.tensorflow.org/api_docs/python/tf/nn/dynamic_rnn)
# - Apply the name "final_state" to the final state using [`tf.identity()`](https://www.tensorflow.org/api_docs/python/tf/identity)
#
# Return the outputs and final_state state in the following tuple `(Outputs, FinalState)`
# +
def build_rnn(cell, inputs):
"""
Create a RNN using a RNN Cell
:param cell: RNN Cell
:param inputs: Input text data
:return: Tuple (Outputs, Final State)
"""
# TODO: Implement Function
outputs, final_state = tf.nn.dynamic_rnn(cell, inputs, dtype=tf.float32)
final_state = tf.identity(final_state, "final_state")
return outputs, final_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_build_rnn(build_rnn)
# -
# ### Build the Neural Network
# Apply the functions you implemented above to:
# - Apply embedding to `input_data` using your `get_embed(input_data, vocab_size, embed_dim)` function.
# - Build RNN using `cell` and your `build_rnn(cell, inputs)` function.
# - Apply a fully connected layer with a linear activation and `vocab_size` as the number of outputs.
#
# Return the logits and final state in the following tuple (Logits, FinalState)
# +
def build_nn(cell, rnn_size, input_data, vocab_size, embed_dim):
"""
Build part of the neural network
:param cell: RNN cell
:param rnn_size: Size of rnns
:param input_data: Input data
:param vocab_size: Vocabulary size
:param embed_dim: Number of embedding dimensions
:return: Tuple (Logits, FinalState)
"""
# TODO: Implement Function
embed = get_embed(input_data, vocab_size, embed_dim)
outputs, final_state = build_rnn(cell, embed)
logits = tf.contrib.layers.fully_connected(outputs, vocab_size, activation_fn=None)
return logits, final_state
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_build_nn(build_nn)
# -
# ### Batches
# Implement `get_batches` to create batches of input and targets using `int_text`. The batches should be a Numpy array with the shape `(number of batches, 2, batch size, sequence length)`. Each batch contains two elements:
# - The first element is a single batch of **input** with the shape `[batch size, sequence length]`
# - The second element is a single batch of **targets** with the shape `[batch size, sequence length]`
#
# If you can't fill the last batch with enough data, drop the last batch.
#
# For example, `get_batches([1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20], 3, 2)` would return a Numpy array of the following:
# ```
# [
# # First Batch
# [
# # Batch of Input
# [[ 1 2], [ 7 8], [13 14]]
# # Batch of targets
# [[ 2 3], [ 8 9], [14 15]]
# ]
#
# # Second Batch
# [
# # Batch of Input
# [[ 3 4], [ 9 10], [15 16]]
# # Batch of targets
# [[ 4 5], [10 11], [16 17]]
# ]
#
# # Third Batch
# [
# # Batch of Input
# [[ 5 6], [11 12], [17 18]]
# # Batch of targets
# [[ 6 7], [12 13], [18 1]]
# ]
# ]
# ```
#
# Notice that the last target value in the last batch is the first input value of the first batch. In this case, `1`. This is a common technique used when creating sequence batches, although it is rather unintuitive.
# +
def get_batches(int_text, batch_size, seq_length):
"""
Return batches of input and target
:param int_text: Text with the words replaced by their ids
:param batch_size: The size of batch
:param seq_length: The length of sequence
:return: Batches as a Numpy array
"""
# TODO: Implement Function
chars_per_batch = batch_size * seq_length
num_of_batches = len(int_text) // chars_per_batch
inputs = np.array(int_text[:num_of_batches*chars_per_batch])
targets = np.array(int_text[1:num_of_batches*chars_per_batch] + [int_text[0]])
inputs = inputs.reshape(batch_size, -1)
targets = targets.reshape(batch_size, -1)
inputs = np.split(inputs, num_of_batches, axis=1)
targets = np.split(targets, num_of_batches, axis=1)
batches = np.array(list(zip(inputs, targets)))
batches.reshape(num_of_batches, 2, batch_size, seq_length)
return batches
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_batches(get_batches)
# -
# ## Neural Network Training
# ### Hyperparameters
# Tune the following parameters:
#
# - Set `num_epochs` to the number of epochs.
# - Set `batch_size` to the batch size.
# - Set `rnn_size` to the size of the RNNs.
# - Set `embed_dim` to the size of the embedding.
# - Set `seq_length` to the length of sequence.
# - Set `learning_rate` to the learning rate.
# - Set `show_every_n_batches` to the number of batches the neural network should print progress.
# +
# Number of Epochs
num_epochs = 70
# Batch Size
batch_size = 64
# RNN Size
rnn_size = 1000
# Embedding Dimension Size
embed_dim = 500
# Sequence Length
seq_length = 14
# Learning Rate
learning_rate = 0.001
# Show stats for every n number of batches
show_every_n_batches = 15
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
save_dir = './save'
# -
# ### Build the Graph
# Build the graph using the neural network you implemented.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
from tensorflow.contrib import seq2seq
train_graph = tf.Graph()
with train_graph.as_default():
vocab_size = len(int_to_vocab)
input_text, targets, lr = get_inputs()
input_data_shape = tf.shape(input_text)
cell, initial_state = get_init_cell(input_data_shape[0], rnn_size)
logits, final_state = build_nn(cell, rnn_size, input_text, vocab_size, embed_dim)
# Probabilities for generating words
probs = tf.nn.softmax(logits, name='probs')
# Loss function
cost = seq2seq.sequence_loss(
logits,
targets,
tf.ones([input_data_shape[0], input_data_shape[1]]))
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -1., 1.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
# -
# ## Train
# Train the neural network on the preprocessed data. If you have a hard time getting a good loss, check the [forums](https://discussions.udacity.com/) to see if anyone is having the same problem.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
batches = get_batches(int_text, batch_size, seq_length)
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(num_epochs):
state = sess.run(initial_state, {input_text: batches[0][0]})
for batch_i, (x, y) in enumerate(batches):
feed = {
input_text: x,
targets: y,
initial_state: state,
lr: learning_rate}
train_loss, state, _ = sess.run([cost, final_state, train_op], feed)
# Show every <show_every_n_batches> batches
if (epoch_i * len(batches) + batch_i) % show_every_n_batches == 0:
print('Epoch {:>3} Batch {:>4}/{} train_loss = {:.3f}'.format(
epoch_i,
batch_i,
len(batches),
train_loss))
# Save Model
saver = tf.train.Saver()
saver.save(sess, save_dir)
print('Model Trained and Saved')
# -
# ## Save Parameters
# Save `seq_length` and `save_dir` for generating a new TV script.
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# Save parameters for checkpoint
helper.save_params((seq_length, save_dir))
# # Checkpoint
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import tensorflow as tf
import numpy as np
import helper
import problem_unittests as tests
_, vocab_to_int, int_to_vocab, token_dict = helper.load_preprocess()
seq_length, load_dir = helper.load_params()
# -
# ## Implement Generate Functions
# ### Get Tensors
# Get tensors from `loaded_graph` using the function [`get_tensor_by_name()`](https://www.tensorflow.org/api_docs/python/tf/Graph#get_tensor_by_name). Get the tensors using the following names:
# - "input:0"
# - "initial_state:0"
# - "final_state:0"
# - "probs:0"
#
# Return the tensors in the following tuple `(InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)`
# +
def get_tensors(loaded_graph):
"""
Get input, initial state, final state, and probabilities tensor from <loaded_graph>
:param loaded_graph: TensorFlow graph loaded from file
:return: Tuple (InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor)
"""
# TODO: Implement Function
InputTensor = loaded_graph.get_tensor_by_name("input:0")
InitialStateTensor = loaded_graph.get_tensor_by_name("initial_state:0")
FinalStateTensor = loaded_graph.get_tensor_by_name("final_state:0")
ProbsTensor = loaded_graph.get_tensor_by_name("probs:0")
return InputTensor, InitialStateTensor, FinalStateTensor, ProbsTensor
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_get_tensors(get_tensors)
# -
# ### Choose Word
# Implement the `pick_word()` function to select the next word using `probabilities`.
# +
def pick_word(probabilities, int_to_vocab):
"""
Pick the next word in the generated text
:param probabilities: Probabilites of the next word
:param int_to_vocab: Dictionary of word ids as the keys and words as the values
:return: String of the predicted word
"""
# TODO: Implement Function
predict = np.random.choice(range(0, len(int_to_vocab)), size=1, p=probabilities)
return int_to_vocab[predict[0]]
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_pick_word(pick_word)
# -
# ## Generate TV Script
# This will generate the TV script for you. Set `gen_length` to the length of TV script you want to generate.
# +
gen_length = 200
# homer_simpson, moe_szyslak, or Barney_Gumble
prime_word = 'moe_szyslak'
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(load_dir + '.meta')
loader.restore(sess, load_dir)
# Get Tensors from loaded model
input_text, initial_state, final_state, probs = get_tensors(loaded_graph)
# Sentences generation setup
gen_sentences = [prime_word + ':']
prev_state = sess.run(initial_state, {input_text: np.array([[1]])})
# Generate sentences
for n in range(gen_length):
# Dynamic Input
dyn_input = [[vocab_to_int[word] for word in gen_sentences[-seq_length:]]]
dyn_seq_length = len(dyn_input[0])
# Get Prediction
probabilities, prev_state = sess.run(
[probs, final_state],
{input_text: dyn_input, initial_state: prev_state})
pred_word = pick_word(probabilities[0][dyn_seq_length-1], int_to_vocab)
gen_sentences.append(pred_word)
# Remove tokens
tv_script = ' '.join(gen_sentences)
for key, token in token_dict.items():
ending = ' ' if key in ['\n', '(', '"'] else ''
tv_script = tv_script.replace(' ' + token.lower(), key)
tv_script = tv_script.replace('\n ', '\n')
tv_script = tv_script.replace('( ', '(')
print(tv_script)
# -
# # The TV Script is Nonsensical
# It's ok if the TV script doesn't make any sense. We trained on less than a megabyte of text. In order to get good results, you'll have to use a smaller vocabulary or get more data. Luckily there's more data! As we mentioned in the beggining of this project, this is a subset of [another dataset](https://www.kaggle.com/wcukierski/the-simpsons-by-the-data). We didn't have you train on all the data, because that would take too long. However, you are free to train your neural network on all the data. After you complete the project, of course.
# # Submitting This Project
# When submitting this project, make sure to run all the cells before saving the notebook. Save the notebook file as "dlnd_tv_script_generation.ipynb" and save it as a HTML file under "File" -> "Download as". Include the "helper.py" and "problem_unittests.py" files in your submission.
| dlnd_tv_script_generation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] _cell_guid="455c5288-0893-4f5c-be8f-b70a3ba51231" _uuid="978e24e319343f241547ac0ae94868a678c7f2cc" endofcell="--"
# *This tutorial is part of the [Learn Machine Learning](https://www.kaggle.com/learn/machine-learning/) series. In this step, you will learn what data leakage is and how to prevent it.*
#
#
# # What is Data Leakage
# Data leakage is one of the most important issues for a data scientist to understand. If you don't know how to prevent it, leakage will come up frequently, and it will ruin your models in the most subtle and dangerous ways. Specifically, leakage causes a model to look accurate until you start making decisions with the model, and then the model becomes very inaccurate. This tutorial will show you what leakage is and how to avoid it.
#
# There are two main types of leakage: **Leaky Predictors** and a **Leaky Validation Strategies.**
#
# ## Leaky Predictors
# This occurs when your predictors include data that will not be available at the time you make predictions.
#
# For example, imagine you want to predict who will get sick with pneumonia. The top few rows of your raw data might look like this:
#
# | got_pneumonia | age | weight | male | took_antibiotic_medicine | ... |
# |:-------------:|:---:|:------:|:-----:|:------------------------:|-----|
# | False | 65 | 100 | False | False | ... |
# | False | 72 | 130 | True | False | ... |
# | True | 58 | 100 | False | True | ... |
# -
#
#
# People take antibiotic medicines after getting pneumonia in order to recover. So the raw data shows a strong relationship between those columns. But *took_antibiotic_medicine* is frequently changed **after** the value for *got_pneumonia* is determined. This is target leakage.
#
# The model would see that anyone who has a value of `False` for `took_antibiotic_medicine` didn't have pneumonia. Validation data comes from the same source, so the pattern will repeat itself in validation, and the model will have great validation (or cross-validation) scores. But the model will be very inaccurate when subsequently deployed in the real world.
#
# To prevent this type of data leakage, any variable updated (or created) after the target value is realized should be excluded. Because when we use this model to make new predictions, that data won't be available to the model.
#
# 
# --
# + [markdown] _cell_guid="cca14623-d55f-49ba-8907-501e9ac2acca" _uuid="e5a3cf9b1bd44f7d2c8e8672b9e8594150d30ad6"
# ## Leaky Validation Strategy
#
# A much different type of leak occurs when you aren't careful distinguishing training data from validation data. For example, this happens if you run preprocessing (like fitting the Imputer for missing values) before calling train_test_split. Validation is meant to be a measure of how the model does on data it hasn't considered before. You can corrupt this process in subtle ways if the validation data affects the preprocessing behavoir.. The end result? Your model will get very good validation scores, giving you great confidence in it, but perform poorly when you deploy it to make decisions.
#
#
# ## Preventing Leaky Predictors
# There is no single solution that universally prevents leaky predictors. It requires knowledge about your data, case-specific inspection and common sense.
#
# However, leaky predictors frequently have high statistical correlations to the target. So two tactics to keep in mind:
# * To screen for possible leaky predictors, look for columns that are statistically correlated to your target.
# * If you build a model and find it extremely accurate, you likely have a leakage problem.
#
# ## Preventing Leaky Validation Strategies
#
# If your validation is based on a simple train-test split, exclude the validation data from any type of *fitting*, including the fitting of preprocessing steps. This is easier if you use [scikit-learn Pipelines](https://www.kaggle.com/dansbecker/pipelines). When using cross-validation, it's even more critical that you use pipelines and do your preprocessing inside the pipeline.
#
# # Example
# We will use a small dataset about credit card applications, and we will build a model predicting which applications were accepted (stored in a variable called *card*). Here is a look at the data:
# + _cell_guid="29c264f4-3836-4b48-b8c7-828e7bec45a0" _uuid="b95201cc2da5de79c022ab8c7cdfe38c16723907"
import pandas as pd
data = pd.read_csv('../input/AER_credit_card_data.csv',
true_values = ['yes'],
false_values = ['no'])
print(data.head())
# + [markdown] _cell_guid="12d34c22-ad00-4e6c-9d0b-443ab54caf35" _uuid="60436a2d7d8e7b87ce891639a2727b77761ff08d"
# We can see with `data.shape` that this is a small dataset (1312 rows), so we should use cross-validation to ensure accurate measures of model quality
# + _cell_guid="29a27f97-44b1-408f-95fb-00a6f01ea93f" _uuid="46cad269244b866c179b00f7f9048b9dc29e9de9"
data.shape
# + _cell_guid="2e15597f-171e-4da0-8c52-96c42f073a36" _uuid="90ea96255857648ffd6e74f33bf7e23c1c3da467"
from sklearn.pipeline import make_pipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
y = data.card
X = data.drop(['card'], axis=1)
# Since there was no preprocessing, we didn't need a pipeline here. Used anyway as best practice
modeling_pipeline = make_pipeline(RandomForestClassifier())
cv_scores = cross_val_score(modeling_pipeline, X, y, scoring='accuracy')
print("Cross-val accuracy: %f" %cv_scores.mean())
# + [markdown] _cell_guid="722ccd83-3d23-4d4e-989e-a9f3a719b308" _uuid="d7faed322b2b993b56f18922dac556950bfa8190"
# With experience, you'll find that it's very rare to find models that are accurate 98% of the time. It happens, but it's rare enough that we should inspect the data more closely to see if it is target leakage.
#
# Here is a summary of the data, which you can also find under the data tab:
#
# - **card:** Dummy variable, 1 if application for credit card accepted, 0 if not
# - **reports:** Number of major derogatory reports
# - **age:** Age n years plus twelfths of a year
# - **income:** Yearly income (divided by 10,000)
# - **share:** Ratio of monthly credit card expenditure to yearly income
# - **expenditure:** Average monthly credit card expenditure
# - **owner:** 1 if owns their home, 0 if rent
# - **selfempl:** 1 if self employed, 0 if not.
# - **dependents:** 1 + number of dependents
# - **months:** Months living at current address
# - **majorcards:** Number of major credit cards held
# - **active:** Number of active credit accounts
#
# A few variables look suspicious. For example, does **expenditure** mean expenditure on this card or on cards used before appying?
#
# At this point, basic data comparisons can be very helpful:
# + _cell_guid="1778dd97-db7e-47e6-a8bd-bb48e6da6327" _uuid="f6e587b54c565c9ca7990da8ff74ec4252c4ae49"
expenditures_cardholders = data.expenditure[data.card]
expenditures_noncardholders = data.expenditure[~data.card]
print('Fraction of those who received a card with no expenditures: %.2f' \
%(( expenditures_cardholders == 0).mean()))
print('Fraction of those who received a card with no expenditures: %.2f' \
%((expenditures_noncardholders == 0).mean()))
# + [markdown] _cell_guid="6ce5490e-f5eb-4ab8-b6cd-9309c1b3d832" _uuid="a5120e2851e400a9d70018496ceb4523a680ca06"
# Everyone with `card == False` had no expenditures, while only 2% of those with `card == True` had no expenditures. It's not surprising that our model appeared to have a high accuracy. But this seems a data leak, where expenditures probably means *expenditures on the card they applied for.**.
#
# Since **share** is partially determined by **expenditure**, it should be excluded too. The variables **active**, **majorcards** are a little less clear, but from the description, they sound concerning. In most situations, it's better to be safe than sorry if you can't track down the people who created the data to find out more.
#
# We would run a model without leakage as follows:
# + _cell_guid="6364e8d5-bb75-4f57-b635-f43627b72d62" _uuid="85f3ba9371ddb581005e096825c603faeacb9ffd"
potential_leaks = ['expenditure', 'share', 'active', 'majorcards']
X2 = X.drop(potential_leaks, axis=1)
cv_scores = cross_val_score(modeling_pipeline, X2, y, scoring='accuracy')
print("Cross-val accuracy: %f" %cv_scores.mean())
# + [markdown] _cell_guid="2b471192-b646-4b66-87db-3e977424f090" _uuid="f52399825ef93cc40a46d28c931cb3238c4bc613"
# This accuracy is quite a bit lower, which on the one hand is disappointing. However, we can expect it to be right about 80% of the time when used on new applications, whereas the leaky model would likely do much worse then that (even in spite of it's higher apparent score in cross-validation.).
#
# # Conclusion
# Data leakage can be multi-million dollar mistake in many data science applications. Careful separation of training and validation data is a first step, and pipelines can help implement this separation. Leaking predictors are a more frequent issue, and leaking predictors are harder to track down. A combination of caution, common sense and data exploration can help identify leaking predictors so you remove them from your model.
#
# # Exercise
# Review the data in your ongoing project. Are there any predictors that may cause leakage? As a hint, most datasets from Kaggle competitions don't have these variables. Once you get past those carefully curated datasets, this becomes a common issue.
#
# Click **[here](https://www.kaggle.com/learn/machine-learning)** to return the main page for *Learning Machine Learning.*
| ML_Learning/kernel (4).ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.7.3 64-bit (''base'': conda)'
# language: python
# name: python37364bitbaseconda65b5f305a1974c36abb2297a98801d43
# ---
# # ```Linear_Support_Vector_Machine``` Example
# Import the modules. We are also going to import `make_blobs()` function just to create the dataset.
import matplotlib.pyplot as plt
from Ardi.ml import Linear_Support_Vector_Machine_2D
from sklearn.datasets import make_blobs
# Create the dataset.
X, y = make_blobs(n_samples=50, n_features=2, centers=2, cluster_std=1.05, random_state=80)
# Display the data using scatter plot, where the data with label of 0 are displayed in blue, and the rest are of labels 1.
plt.scatter(X[:,0], X[:,1], c=y, cmap='winter')
# Initialize the Support Vector Machine model with linear kernel. Refer to the documentation to see the available parameters.
svm = Linear_Support_Vector_Machine_2D(iterations=800)
# Load the data that we just created to the `svm` model.
svm.take_data_raw(X, y)
# If you want to load the data from a csv file instead, you can use the following code.
# +
#svm.take_data_csv('dataset_test/sepal_petal_length.csv')
# -
# The following code is used just to ensure that our features and labels have been loaded properly.
svm.X[:5]
svm.y[:5]
# Display the first 5 data. Keep in mind that the `svm` model automatically converts label 0 to -1 because that's just how an SVM works.
plt.scatter(X[:5,0], X[:5,1], c=y[:5], cmap='winter')
# How to train the model.
svm.train()
# The `bias` and `weights` term before and after training. Note that both `bias` and `weights` are initially just a random number.
print('svm.bias\t\t:', svm.bias)
print('svm.updated_bias\t:', svm.updated_bias)
print()
print('svm.weights\t\t:', svm.weights)
print('svm.updated_weights\t:', svm.updated_weights)
# Perdicting multiple samples.
svm.predict_multiple_samples(svm.X)
# How the error decrease looks like. Note that these error values are the sum of hinge loss and its regularization term.
svm.plot_errors(print_details=True)
# How the decision boundary looks like before training.
svm.visualize_before()
# How the decision boundary looks like after training.
svm.visualize_after()
| Documentation/Linear_Support_Vector_Machine_2D Example.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# ## Differential expression anlaysis of the TCGA breast cancer set
#
# This notebook can be run locally or on a remote cloud computer by clicking the badge below:
#
# [](https://mybinder.org/v2/gh/statisticalbiotechnology/cb2030/master?filepath=nb%2Ftesting%2Ftesting.ipynb)
#
# First we retrieve the breast cancer RNAseq data as well as the clinical classification of the sets from cbioportal.org.
#
# The gene expresion data is stored in the [DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) `brca`, and the adherent clinical information of the cancers and their patients is stored in the [DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) `brca_clin`. It can be woth exploring these data structures.
#
# + slideshow={"slide_type": "fragment"}
import pandas as pd
import seaborn as sns
import numpy as np
import tarfile
import gzip
from scipy.stats import ttest_ind
import sys
sys.path.append("..") # Read loacal modules for tcga access and qvalue calculations
import tcga_read as tcga
brca = tcga.get_expression_data("../../data/brca.tsv.gz", 'http://download.cbioportal.org/brca_tcga_pub2015.tar.gz',"data_RNA_Seq_v2_expression_median.txt")
brca_clin = tcga.get_clinical_data("../../data/brca_clin.tsv.gz", 'http://download.cbioportal.org/brca_tcga_pub2015.tar.gz',"data_clinical_sample.txt")
# + [markdown] slideshow={"slide_type": "slide"}
# Before any further analysis we clean our data. This includes removal of genes where no transcripts were found for any of the samples , i.e. their values are either [NaN](https://en.wikipedia.org/wiki/NaN) or zero.
#
# The data is also log transformed. It is generally assumed that expression values follow a log-normal distribution, and hence the log transformation implies that the new values follow a nomal distribution.
# + slideshow={"slide_type": "fragment"}
brca.dropna(axis=0, how='any', inplace=True)
brca = brca.loc[~(brca<=0.0).any(axis=1)]
brca = pd.DataFrame(data=np.log2(brca),index=brca.index,columns=brca.columns)
# + [markdown] slideshow={"slide_type": "slide"}
# We can get an overview of the expression data:
# + slideshow={"slide_type": "fragment"}
brca
# + [markdown] slideshow={"slide_type": "slide"}
# and the clinical data:
# + slideshow={"slide_type": "fragment"}
brca_clin
# + [markdown] slideshow={"slide_type": "slide"}
# ### Differential expression analysis
#
# The goal of the excercise is to determine which genes that are differentially expressed in so called tripple negative cancers as compared to other cancers. A breast cancer is triple negative when it does not express either [Progesterone receptors](https://en.wikipedia.org/wiki/Progesterone_receptor), [Estrogen receptors](https://en.wikipedia.org/wiki/Estrogen_receptor) or [Epidermal growth factor receptor 2](https://en.wikipedia.org/wiki/HER2/neu). Such cancers are known to behave different than other cancers, and are not amendable to regular [hormonal theraphies](https://en.wikipedia.org/wiki/Hormonal_therapy_(oncology)).
#
# We first create a vector of booleans, that track which cancers that are tripple negative. This will be needed as an input for subsequent significance estimation.
# + slideshow={"slide_type": "fragment"}
brca_clin.loc["3N"]= (brca_clin.loc["PR status by ihc"]=="Negative") & (brca_clin.loc["ER Status By IHC"]=="Negative") & (brca_clin.loc["IHC-HER2"]=="Negative")
tripple_negative_bool = (brca_clin.loc["3N"] == True)
# + [markdown] slideshow={"slide_type": "slide"}
# Next, for each transcript that has been measured, we calculate (1) log of the average Fold Change difference between tripple negative and other cancers, and (2) the significance of the difference between tripple negative and other cancers.
#
# An easy way to do so is by defining a separate function, `get_significance_two_groups(row)`, that can do such calculations for any row of the `brca` DataFrame, and subsequently we use the function `apply` for the function to execute on each row of the DataFrame. For the significance test we use a $t$ test, which is provided by the function [`ttest_ind`.](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.ttest_ind.html)
#
# This results in a new table with gene names and their $p$ values of differential concentration, and their fold changes.
# + slideshow={"slide_type": "fragment"}
def get_significance_two_groups(row):
log_fold_change = row[tripple_negative_bool].mean() - row[~tripple_negative_bool].mean() # Calculate the log Fold Change
p = ttest_ind(row[tripple_negative_bool],row[~tripple_negative_bool],equal_var=False)[1] # Calculate the significance
return [p,-np.log10(p),log_fold_change]
pvalues = brca.apply(get_significance_two_groups,axis=1,result_type="expand")
pvalues.rename(columns = {list(pvalues)[0]: 'p', list(pvalues)[1]: '-log_p', list(pvalues)[2]: 'log_FC'}, inplace = True)
# + [markdown] slideshow={"slide_type": "slide"}
# The resulting list can be further investigated.
# + slideshow={"slide_type": "fragment"}
pvalues
# + [markdown] slideshow={"slide_type": "slide"}
# A common way to illustrate the diffrential expression values are by plotting the negative log of the $p$ values, as a function of the mean [fold change](https://en.wikipedia.org/wiki/Fold_change) of each transcript. This is known as a [Volcano plot](https://en.wikipedia.org/wiki/Volcano_plot_(statistics)).
# + slideshow={"slide_type": "fragment"}
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("white")
sns.set_context("talk")
ax = sns.relplot(data=pvalues,x="log_FC",y="-log_p",aspect=1.5,height=6)
ax.set(xlabel="$log_2(TN/not TN)$", ylabel="$-log_{10}(p)$");
# + [markdown] slideshow={"slide_type": "fragment"}
# The regular interpretation of a Volcano plot is that the ges in the top left and the top right corner are the most interesting ones, as the have a large fold change between the conditions as well as being very significant.
# -
| nb/testing/testing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# 
#
# ## 一. Density Estimation 密度估计
#
#
# 假如要更为正式定义异常检测问题,首先我们有一组从 $x^{(1)}$ 到 $x^{(m)}$ m个样本,且这些样本均为正常的。我们将这些样本数据建立一个模型 p(x) , p(x) 表示为 x 的分布概率。
#
#
# 
#
#
# 那么假如我们的测试集 $x_{test}$ 概率 p 低于阈值 $\varepsilon$ ,那么则将其标记为异常。
#
#
# 异常检测的核心就在于找到一个概率模型,帮助我们知道一个样本落入正常样本中的概率,从而帮助我们区分正常和异常样本。高斯分布(Gaussian Distribution)模型就是异常检测算法最常使用的概率分布模型。
#
# ### 1. 高斯分布
#
#
# 假如 x 服从高斯分布,那么我们将表示为: $x\sim N(\mu,\sigma^2)$ 。其分布概率为:
#
# $$p(x;\mu,\sigma^2)=\frac{1}{\sqrt{2\pi}\sigma}exp(-\frac{(x-\mu)^2}{2\sigma^2})$$
#
# 其中 $\mu$ 为期望值(均值), $\sigma^2$ 为方差。
#
# 其中,期望值 $\mu$ 决定了其轴的位置,标准差 $\sigma$ 决定了分布的幅度宽窄。当 $\mu=0,\sigma=1$ 时的正态分布是标准正态分布。
#
# 
#
#
# 期望值:$$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
#
# 方差: $$\sigma^2=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)}^2$$
#
#
# 假如我们有一组 m 个无标签训练集,其中每个训练数据又有 n 个特征,那么这个训练集应该是 m 个 n 维向量构成的样本矩阵。
#
#
# 在概率论中,对有限个样本进行参数估计
#
# $$\mu_j = \frac{1}{m} \sum_{i=1}^{m}x_j^{(i)}\;\;\;,\;\;\; \delta^2_j = \frac{1}{m} \sum_{i=1}^{m}(x_j^{(i)}-\mu_j)^2$$
#
# 这里对参数 $\mu$ 和参数 $\delta^2$ 的估计就是二者的极大似然估计。
#
# 假定每一个特征 $x_{1}$ 到 $x_{n}$ 均服从正态分布,则其模型的概率为:
#
# $$
# \begin{align*}
# p(x)&=p(x_1;\mu_1,\sigma_1^2)p(x_2;\mu_2,\sigma_2^2) \cdots p(x_n;\mu_n,\sigma_n^2)\\
# &=\prod_{j=1}^{n}p(x_j;\mu_j,\sigma_j^2)\\
# &=\prod_{j=1}^{n} \frac{1}{\sqrt{2\pi}\sigma_{j}}exp(-\frac{(x_{j}-\mu_{j})^2}{2\sigma_{j}^2})
# \end{align*}
# $$
#
#
# 当 $p(x)<\varepsilon$时,$x$ 为异常样本。
#
# ### 2. 举例
#
# 假定我们有两个特征 $x_1$ 、 $x_2$ ,它们都服从于高斯分布,并且通过参数估计,我们知道了分布参数:
#
# 
#
# 则模型 $p(x)$ 能由如下的热力图反映,热力图越热的地方,是正常样本的概率越高,参数 $\varepsilon$ 描述了一个截断高度,当概率落到了截断高度以下(下图紫色区域所示),则为异常样本:
#
# 
#
# 将 $p(x)$ 投影到特征 $x_1$ 、$x_2$ 所在平面,下图紫色曲线就反映了 $\varepsilon$ 的投影,它是一条截断曲线,落在截断曲线以外的样本,都会被认为是异常样本:
#
# 
#
#
# ### 3. 算法评估
#
# 由于异常样本是非常少的,所以整个数据集是非常偏斜的,我们不能单纯的用预测准确率来评估算法优劣,所以用我们之前的查准率(Precision)和召回率(Recall)计算出 F 值进行衡量异常检测算法了。
#
# - 真阳性、假阳性、真阴性、假阴性
# - 查准率(Precision)与 召回率(Recall)
# - F1 Score
#
# 我们还有一个参数 $\varepsilon$ ,这个 $\varepsilon$ 是我们用来决定什么时候把一个样本当做是异常样本的阈值。我们应该试用多个不同的 $\varepsilon$ 值,选取一个使得 F 值最大的那个 $\varepsilon$ 。
#
#
#
#
# ----------------------------------------------------------------------------------------------------------------
#
#
#
# ## 二. Building an Anomaly Detection System
#
#
#
# ### 1. 有监督学习与异常检测
#
#
#
#
# |有监督学习| 异常检测|
# | :----------: | :---: |
# |数据分布均匀 |数据非常偏斜,异常样本数目远小于正常样本数目
# |可以根据对正样本的拟合来知道正样本的形态,从而预测新来的样本是否是正样本 |异常的类型不一,很难根据对现有的异常样本(即正样本)的拟合来判断出异常样本的形态|
#
#
# 下面的表格则展示了二者的一些应用场景:
#
# |有监督学习| 异常检测|
# | :----------: | :---: |
# |垃圾邮件检测| 故障检测|
# |天气预测(预测雨天、晴天、或是多云天气)| 某数据中心对于机器设备的监控|
# |癌症的分类| 制造业判断一个零部件是否异常|
#
# 
#
# 假如我们的数据看起来不是很服从高斯分布,可以通过对数、指数、幂等数学变换让其接近于高斯分布。
#
#
# ----------------------------------------------------------------------------------------------------------------
#
#
#
# ## 三. Multivariate Gaussian Distribution (Optional)
#
#
#
# ### 1. 多元高斯分布模型
#
#
# 
#
#
# 我们以数据中心的监控计算机为例子。 $x_1$ 是CPU的负载,$x_2$ 是内存的使用量。其正常样本如左图红色点所示。假如我们有一个异常的样本(图中左上角绿色点),在图中看很明显它并不是正常样本所在的范围。但是在计算概率 $p(x)$ 的时候,因为它在 $x_1$ 和 $x_2$ 的高斯分布都属于正常范围,所以该点并不会被判断为异常点。
#
# 这是因为在高斯分布中,它并不能察觉在蓝色椭圆处才是正常样本概率高的范围,其概率是通过圆圈逐渐向外减小。所以在同一个圆圈内,虽然在计算中概率是一样的,但是在实际上却往往有很大偏差。
#
# 所以我们开发了一种改良版的异常检测算法:多元高斯分布。
#
#
#
# 我们不将每一个特征值都分开进行高斯分布的计算,而是作为整个模型进行高斯分布的拟合。
#
# 其概率模型为: $$p(x;\mu,\Sigma)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$ (其中 $|\Sigma|$ 是 $\Sigma$ 的行列式,$\mu$ 表示样本均值,$\Sigma$ 表示样本协方差矩阵。)。
#
# 多元高斯分布模型的热力图如下:
#
#
# 
#
#
# $\Sigma$ 是一个协方差矩阵,所以它衡量的是方差。减小 $\Sigma$ 其宽度也随之减少,增大反之。
#
#
# 
#
#
# $\Sigma$ 中第一个数字是衡量 $x_1$ 的,假如减少第一个数字,则可从图中观察到 $x_1$ 的范围也随之被压缩,变成了一个椭圆。
#
#
# 
#
#
# 多元高斯分布还可以给数据的相关性建立模型。假如我们在非主对角线上改变数据(如图中间那副),则其图像会根据 $y=x$ 这条直线上进行高斯分布。
#
#
# 
#
#
# 反之亦然。
#
#
# 
#
#
# 改变 $\mu$ 的值则是改变其中心点的位置。
#
#
# ### 2. 参数估计
#
#
# 多元高斯分布模型的参数估计如下:
#
#
#
# $$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
#
# $$\Sigma=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)(x^{(i)}-\mu)^T}$$
#
#
#
# ### 3. 算法流程
#
#
# 采用了多元高斯分布的异常检测算法流程如下:
#
# 1. 选择一些足够反映异常样本的特征 $x_j$ 。
# 2. 对各个样本进行参数估计:
# $$\mu=\frac{1}{m}\sum_{i=1}^{m}{x^{(i)}}$$
# $$\Sigma=\frac{1}{m}\sum_{i=1}^{m}{(x^{(i)}-\mu)(x^{(i)}-\mu)^T}$$
# 3. 当新的样本 x 到来时,计算 $p(x)$ :
#
# $$p(x)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$
#
# 如果 $p(x)<\varepsilon $ ,则认为样本 x 是异常样本。
#
#
#
# ### 4. 多元高斯分布模型与一般高斯分布模型的差异
#
# 一般的高斯分布模型只是多元高斯分布模型的一个约束,它将多元高斯分布的等高线约束到了如下所示同轴分布(概率密度的等高线是沿着轴向的):
#
# 
#
#
# 当: $\Sigma=\left[ \begin{array}{ccc}\sigma_1^2 \\ & \sigma_2^2 \\ &&…\\&&&\sigma_n^2\end{array} \right]$ 的时候,此时的多元高斯分布即是原来的多元高斯分布。(因为只有主对角线方差,并没有其它斜率的变化)
#
#
# 对比
#
# ### 模型定义
#
# 一般高斯模型:
#
# $$
# \begin{align*}
# p(x)&=p(x_1;\mu_1,\sigma_1^2)p(x_2;\mu_2,\sigma_2^2) \cdots p(x_n;\mu_n,\sigma_n^2)\\
# &=\prod_{j=1}^{n}p(x_j;\mu_j,\sigma_j^2)\\
# &=\prod_{j=1}^{n} \frac{1}{\sqrt{2\pi}\sigma_{j}}exp(-\frac{(x_{j}-\mu_{j})^2}{2\sigma_{j}^2})
# \end{align*}
# $$
#
# 多元高斯模型:
#
#
# $$p(x)=\frac{1}{(2\pi)^{\frac{n}{2}}|\Sigma|^{\frac{1}{2}}}exp(-\frac{1}{2}(x-\mu)^T\Sigma^{-1}(x-\mu))$$
#
#
# ### 相关性
#
# 一般高斯模型:
#
# 需要手动创建一些特征来描述某些特征的相关性
#
# 多元高斯模型:
#
# 利用协方差矩阵$\Sigma$获得了各个特征相关性
#
#
# ### 复杂度
#
# 一般高斯模型:
#
# 计算复杂度低,适用于高维特征
#
# 多元高斯模型:
#
# 计算复杂
#
# ### 效果
#
#
# 一般高斯模型:
#
# 在样本数目 m 较小时也工作良好
#
# 多元高斯模型:
#
# 需要 $\Sigma$ 可逆,亦即需要 $m>n$ ,且各个特征不能线性相关,如不能存在 $x_2=3x_1$ 或者 $x_3=x_1+2x_2$
#
#
#
# 结论:**基于多元高斯分布模型的异常检测应用十分有限**。
#
# ----------------------------------------------------------------------------------------------------------------
#
#
# ## 四. Anomaly Detection 测试
#
#
# ### 1. Question 1
#
#
# For which of the following problems would anomaly detection be a suitable algorithm?
#
# A. Given a dataset of credit card transactions, identify unusual transactions to flag them as possibly fraudulent.
#
# B. Given data from credit card transactions, classify each transaction according to type of purchase (for example: food, transportation, clothing).
#
# C. Given an image of a face, determine whether or not it is the face of a particular famous individual.
#
# D. From a large set of primary care patient records, identify individuals who might have unusual health conditions.
#
# 解答:A、D
#
# A、D 才适合异常检测算法。
#
#
# ### 2. Question 2
#
# Suppose you have trained an anomaly detection system for fraud detection, and your system that flags anomalies when $p(x)$ is less than ε, and you find on the cross-validation set that it is missing many fradulent transactions (i.e., failing to flag them as anomalies). What should you do?
#
#
# A. Decrease $\varepsilon$
#
# B. Increase $\varepsilon$
#
# 解答:B
#
#
#
# ### 3. Question 3
#
# Suppose you are developing an anomaly detection system to catch manufacturing defects in airplane engines. You model uses
#
# $$p(x) = \prod_{j=1}^{n}p(x_{j};\mu_{j},\sigma_{j}^{2})$$
#
# You have two features $x_1$ = vibration intensity, and $x_2$ = heat generated. Both $x_1$ and $x_2$ take on values between 0 and 1 (and are strictly greater than 0), and for most "normal" engines you expect that $x_1 \approx x_2$. One of the suspected anomalies is that a flawed engine may vibrate very intensely even without generating much heat (large $x_1$, small $x_2$), even though the particular values of $x_1$ and $x_2$ may not fall outside their typical ranges of values. What additional feature $x_3$ should you create to capture these types of anomalies:
#
#
# A. $x_3 = \frac{x_1}{x_2}$
#
# B. $x_3 = x_1^2\times x_2^2$
#
# C. $x_3 = (x_1 + x_2)^2$
#
# D. $x_3 = x_1 \times x_2^2$
#
#
# 解答:A
#
# 假如特征量 $x_1$ 和 $x_2$ ,可建立特征量 $x_3=\frac{x_1}{x_2}$ 结合两者。
#
# ### 4. Question 4
#
# Which of the following are true? Check all that apply.
#
#
# A. When evaluating an anomaly detection algorithm on the cross validation set (containing some positive and some negative examples), classification accuracy is usually a good evaluation metric to use.
#
# B. When developing an anomaly detection system, it is often useful to select an appropriate numerical performance metric to evaluate the effectiveness of the learning algorithm.
#
# C. In a typical anomaly detection setting, we have a large number of anomalous examples, and a relatively small number of normal/non-anomalous examples.
#
# D. In anomaly detection, we fit a model p(x) to a set of negative (y=0) examples, without using any positive examples we may have collected of previously observed anomalies.
#
# 解答:B、D
#
#
# ### 5. Question 5
#
# You have a 1-D dataset $\begin{Bmatrix}
# x^{(i)},\cdots,x^{(m)}
# \end{Bmatrix}$ and you want to detect outliers in the dataset. You first plot the dataset and it looks like this:
#
# 
#
# Suppose you fit the gaussian distribution parameters $\mu_1$ and $\sigma_1^2$ to this dataset. Which of the following values for $\mu_1$ and $\sigma_1^2$ might you get?
#
# A. $\mu = -3$,$\sigma_1^2 = 4$
#
# B. $\mu = -6$,$\sigma_1^2 = 4$
#
# C. $\mu = -3$,$\sigma_1^2 = 2$
#
# D. $\mu = -6$,$\sigma_1^2 = 2$
#
#
# 解答:A
#
# 中心点在-3,在-3周围即(-4,-2)周围仍比较密集,所以 $\sigma_1=2$ 。
#
#
# ----------------------------------------------------------------------------------------------------------------
# > GitHub Repo:[Halfrost-Field](https://github.com/halfrost/Halfrost-Field)
# >
# > Follow: [halfrost · GitHub](https://github.com/halfrost)
# >
# > Source: [https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine\_Learning/Anomaly\_Detection.ipynb](https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine_Learning/Anomaly_Detection.ipynb)
| contents/Machine_Learning/Anomaly_Detection.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: SageMath 9.0
# language: sage
# name: sagemath
# ---
# # Euclidean Algorithm
#
# Here we explore one implementation of the Euclidean Algorithm. Although this may not be the most efficient impelmentation of the Euclidean Algorithm, it is the one I wrote. Finding the GCD is the easy part of this Algorithm, it is the back substitution that needs some creativity here. In this document, we will go through the process of developing an algorithm that implements the both sides of the Euclidean Algorithm.
#
# Here we look at an implementation of the Euclidean Algorithm with no back substitution, that is, it only returns the greatest common divisor of two numbers. Here we override the gcd function that is provided to us by SageMath, and create our own. Let's take a look.
# define the gcd function that will take in two parameters which must be integers.
def gcd(a,b):
# set the variable atemp to the maximum of the two numbers.
# Note that we take the absolute value of each of the numbers,
# as this does not change the gcd.
atemp = max(abs(a),abs(b))
# similar with btemp, but the minimum.
btemp = min(abs(a),abs(b))
# let a be the maximum of the two, and b be the minimum of the two,
# where both are now positive, if they were not before
a = atemp
b = btemp
# while b is non zero...
while b > 0:
# obtain the quotient of a/b...
quotient = floor(a/b)
# as well as the remainder of a/b...
remainder = a % b
# then, for the next step in the Euclidean Algorithm, set
# a to be the current value of b, and set b to be the remainder
# obtained from the previous step, continue this process untill
# the remainder is zero.
a = b
b = remainder
# return the quotient, as this will be the greatest common divisor.
return a
# Here we give this function a test. We test a few different numbers, as well as some large ones to demonstrate the speed and efficiency of the Euclidean Algorithm.
print(f"gcd(5,12)={gcd(5,12)}")
result = gcd(5^23,23^5)
print(f"gcd(5^23,23^5)={result}")
# Now we turn our attention to the problem of finding some linear combination of the two numbers that equals their gcd. To see how this is done, consider the following general Euclidean Algorithm:
# \begin{align*}
# a = q_1 b+r_1 \\
# b = q_2 r_1 + r_2 \\
# \vdots \\
# r_{n-2}=q_n r_{n-1}+r_n \\
# r_{n-1}=q_{n+1}r_n
# \end{align*}
# Where we know that $\gcd(a,b)=r_n$. We can then work our way back up from the second to last line, we can write
#
# $$ r_n=r_{n-2}-q_{n}r_{n-1} $$
# But, since
#
# $$r_{n-1}=r_{n-3}-q_{n-1}r_{n-2} $$
#
# We can write
#
# $$r_n=r_{n-2}-q_n(r_{n-3}-q_{n-1}r_{n-2}) =(1+q_nq_{n-1})r_{n-2}-q_nr_{n-3}$$
#
# And so on through until we reach the top level, and have some linear combination of $a$ and $b$. To implement this in an algorithm, we consider the following steps a computer might take.
#
# > first, let $x=1$ and $y=-q_n$
# > then $r_n=xr_{n-2}+yr_{n-1}$
# > then write $r_n=xr_{n-2}+y(r_{n-3}-q_{n-1}r_{n-2})=(x-q_{n-1}y)r_{n-2}+yr_{n-3}$
# > set $x=x-q_{n-2}y$ and $y=y$
# > then $r_n=xr_{n-2}+yr_{n-3}$
# > then write $r_n=x(r_{n-4}-q_{n-2}r_{n-3})+yr_{n-3}=xr_{n-4}+(y-q_{n-2}x)r_{n-3}$
# > set $x=x$ and $y=y-q_{n-2}x)$
# > then $r_n=xr_{n-4}+yr_{n-3}$
# > continue until we reach the top equation, then we will have a
# > combination of $a$ and $b$ that equals $r_n=gcd(a,b)$
# redefine our gcd function, similar as before, must have
# two intengers as input.
def gcd_full(a: int,b: int):
# store the original values of a and b into the
# variables aOrig and bOrig, to be used at the end
aOrig = a
bOrig = b
# let atemp be the maximum of the absolute values of a and b
# let btemp be the minimum
atemp = max(abs(a),abs(b))
btemp = min(abs(a),abs(b))
# reassign the max value to be a and the minimum value to be b
a = atemp
b = btemp
# create a list to hold all of the quotients, there is no need to
# save all the remainders, as they are not used in the pseudocode
# above
quotients = []
# same as before, go through and perform the steps of the Euclidean
# algorithm, only this time, save all of the quotients into the list
# that we just defined.
while b > 0:
quotient = floor(a/b)
remainder = a % b
a = b
b = remainder
quotients.append(quotient)
# let d, be the value of the gcd
d = a
# throw the very last equation (the one with no remainder)
# away, since we do not use it in the pseudocode above
quotients.pop()
# set x and y to their initial values
x = 1
y = -quotients.pop()
# set the count equal to one, this will be used to alternate which one of x and y
# we change in each step
count = 0
# follow the pseudocode above until there are no longer any quotients left
while len(quotients)!=0:
if count % 2 == 0:
x = x-quotients.pop()*y
if count % 2 == 1:
y = y-quotients.pop()*x
count = count + 1
# this part is a little messy (sorry about that), but test to see which linear
# combination of the original values of a and b give us the gcd, then
# return those values
if x*aOrig+y*bOrig==d:
return [d,x,y]
elif (-x)*aOrig+y*bOrig==d:
return [d,-x,y]
elif x*aOrig+(-y)*bOrig==d:
return [d,x,-y]
elif (-x)*aOrig+(-y)*bOrig==d:
return [d,-x,-y]
elif y*aOrig+x*bOrig==d:
return [d,y,x]
elif (-y)*aOrig+x*bOrig==d:
return [d,-y,x]
elif y*aOrig+(-x)*bOrig==d:
return [d,y,-x]
elif (-y)*aOrig+(-x)*bOrig==d:
return [d,-y,-x]
# Here we go through an example of both of the above methods.
print(gcd(-339348,5423493))
print(gcd_full(-339348,5423493))
# The output of the above cell tells us that
#
# $$ \gcd(-339348,5423493)=3 $$
#
# and that
#
# $$ -339348(-146412)+5423493(-9161)=3 $$
| Misc/Euclid.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (system-wide)
# language: python
# metadata:
# cocalc:
# description: Python 3 programming language
# priority: 100
# url: https://www.python.org/
# name: python3
# ---
# # Laboratory 14
# ## Full name:
# ## R#:
# ## HEX:
# ## Title of the notebook
# ## Date:
# ### Important Terminology:
# __Plotting Position:__ An empirical distribution, based on a random sample from a (possibly unknown) probability distribution, obtained by plotting the exceedance (or cumulative) probability of the sample distribution against the sample value. <br>
# The exceedance probability for a particular sample value is a function of sample size and the rank of the particular sample. For exceedance probabilities, the sample values are ranked from largest to smallest. The general expression in common use for plotting position is
#
# $$ P = \frac{m - b}{N + 1 -2b}\ $$
#
# where m is the ordered rank of a sample value, N is the sample size, and b is a constant between 0 and 1, depending on the plotting method.<br>
#
# 
#
# __*From:__<br>
# __*https://glossary.ametsoc.org/wiki/*__<br>
#
# __Let's work on example. First, import the necessary packages:__
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# __Read the "lab14_E1data.csv" file as a dataset:__
data = pd.read_csv("lab14_E1data.csv")
data
# __The dataset contains two sets of values: "Set1" and "Set2". Use descriptive functions to learn more the sets.__
# Let's check out set1 and set2
set1 = data['Set1']
set2 = data['Set2']
print(set1)
print(set2)
set1.describe()
set2.describe()
# __Remember the Weibull Plotting Position formula from last session. Use Weibull Plotting Position formula to plot set1 and set2 quantiles on the same graph.__<br>
# __Do they look different? How?__
def weibull_pp(sample): # Weibull plotting position function
# returns a list of plotting positions; sample must be a numeric list
weibull_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
weibull_pp.append((i+1)/(len(sample)+1)) #values from the gringorten formula
return weibull_pp
#Convert to numpy arrays
set1 = np.array(set1)
set2 = np.array(set2)
#Apply the weibull pp function
set1_wei = weibull_pp(set1)
set2_wei = weibull_pp(set2)
myfigure = matplotlib.pyplot.figure(figsize = (4,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='blue')
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='orange')
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set1 and Set2 based on Weibull Plotting Function")
matplotlib.pyplot.show()
# __Do they look different? How?__
# __Define functions for Gringorten, Cunnane, California, and Hazen Plotting Position Formulas. Overlay and Plot them all for set 1 and set2 on two different graphs.__<br>
def gringorten_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
gringorten_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
gringorten_pp.append((i+1-0.44)/(len(sample)+0.12)) #values from the gringorten formula
return gringorten_pp
set1_grin = gringorten_pp(set1)
set2_grin = gringorten_pp(set2)
def cunnane_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
cunnane_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
cunnane_pp.append((i+1-0.40)/(len(sample)+0.2)) #values from the cunnane formula
return cunnane_pp
set1_cun = cunnane_pp(set1)
set2_cun = cunnane_pp(set2)
def california_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
california_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
california_pp.append((i+1)/(len(sample))) #values from the cunnane formula
return california_pp
set1_cal = california_pp(set1)
set2_cal = california_pp(set2)
def hazen_pp(sample): # plotting position function
# returns a list of plotting positions; sample must be a numeric list
hazen_pp = [] # null list to return after fill
sample.sort() # sort the sample list in place
for i in range(0,len(sample),1):
hazen_pp.append((i+1-0.5)/(len(sample))) #values from the cunnane formula
return hazen_pp
set1_haz = hazen_pp(set1)
set2_haz = hazen_pp(set2)
myfigure = matplotlib.pyplot.figure(figsize = (12,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='blue',
marker ="^",
s = 50)
matplotlib.pyplot.scatter(set1_grin, set1 ,color ='red',
marker ="o",
s = 20)
matplotlib.pyplot.scatter(set1_cun, set1 ,color ='green',
marker ="s",
s = 20)
matplotlib.pyplot.scatter(set1_cal, set1 ,color ='yellow',
marker ="p",
s = 20)
matplotlib.pyplot.scatter(set1_haz, set1 ,color ='black',
marker ="*",
s = 20)
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set1 based on Weibull, Gringorton, Cunnane, California, and Hazen Plotting Functions")
matplotlib.pyplot.show()
myfigure = matplotlib.pyplot.figure(figsize = (12,8)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='blue',
marker ="^",
s = 50)
matplotlib.pyplot.scatter(set2_grin, set2 ,color ='red',
marker ="o",
s = 20)
matplotlib.pyplot.scatter(set2_cun, set2 ,color ='green',
marker ="s",
s = 20)
matplotlib.pyplot.scatter(set2_cal, set2 ,color ='yellow',
marker ="p",
s = 20)
matplotlib.pyplot.scatter(set2_haz, set2 ,color ='black',
marker ="*",
s = 20)
matplotlib.pyplot.xlabel("Density or Quantile Value")
matplotlib.pyplot.ylabel("Value")
matplotlib.pyplot.title("Quantile Plot for Set2 based on Weibull, Gringorton, Cunnane, California, and Hazen Plotting Functions")
matplotlib.pyplot.show()
# __Plot a histogram of Set1 with 10 bins.__<br>
# +
import matplotlib.pyplot as plt
myfigure = matplotlib.pyplot.figure(figsize = (10,5)) # generate a object from the figure class, set aspect ratio
set1 = data['Set1']
set1.plot.hist(grid=False, bins=10, rwidth=1,
color='navy')
plt.title('Histogram of Set1')
plt.xlabel('Value')
plt.ylabel('Counts')
plt.grid(axis='y',color='yellow', alpha=1)
# -
# __Plot a histogram of Set2 with 10 bins.__<br>
set2 = data['Set2']
set2.plot.hist(grid=False, bins=10, rwidth=1,
color='darkorange')
plt.title('Histogram of Set2')
plt.xlabel('Value')
plt.ylabel('Counts')
plt.grid(axis='y',color='yellow', alpha=1)
# __Plot a histogram of both Set1 and Set2 and discuss the differences.__<br>
fig, ax = plt.subplots()
data.plot.hist(density=False, ax=ax, title='Histogram: Set1 vs. Set2', bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# __The cool 'seaborn' package: Another way for plotting histograms and more!__<br>
#
import seaborn as sns
sns.displot(set1,color='navy', rug=True)
sns.displot(set2,color='darkorange', rug=True)
# ### Important Terminology:
# __Kernel Density Estimation (KDE):__ a non-parametric way to estimate the probability density function of a random variable. Kernel density estimation is a fundamental data smoothing problem where inferences about the population are made, based on a finite data sample. This can be useful if you want to visualize just the “shape” of some data, as a kind of continuous replacement for the discrete histogram.<br>
#
# __*From:__<br>
# __*https://en.wikipedia.org/wiki/Kernel_density_estimation*__<br>
# __*https://mathisonian.github.io/kde/* >> A SUPERCOOL Blog!__<br>
# __*https://www.youtube.com/watch?v=fJoR3QsfXa0* >> A Nice Intro to distplot in seaborn | Note that displot is pretty much the same thing!__<br>
#
#
#
sns.displot(set1,color='navy',kind='kde',rug=True)
sns.displot(set1,color='navy',kde=True)
sns.displot(set2,color='orange',kde=True)
# ### Important Terminology:
# __Empirical Cumulative Distribution Function (ECDF):__ the distribution function associated with the empirical measure of a sample. This cumulative distribution function is a step function that jumps up by 1/n at each of the n data points. Its value at any specified value of the measured variable is the fraction of observations of the measured variable that are less than or equal to the specified value. <br>
#
# __*From:__<br>
# __*https://en.wikipedia.org/wiki/Empirical_distribution_function*__<br>
sns.displot(set1,color='navy',kind='ecdf')
# __Fit a Normal distribution data model to both Set1 and Set2. Plot them seperately. Describe the fit.__<br>
# +
set1 = data['Set1']
set2 = data['Set2']
set1 = np.array(set1)
set2 = np.array(set2)
set1_wei = weibull_pp(set1)
set2_wei = weibull_pp(set2)
# Normal Quantile Function
import math
def normdist(x,mu,sigma):
argument = (x - mu)/(math.sqrt(2.0)*sigma)
normdist = (1.0 + math.erf(argument))/2.0
return normdist
# For set1
mu = set1.mean() # Fitted Model
sigma = set1.std()
x = []; ycdf = []
xlow = 0; xhigh = 1.2*max(set1) ; howMany = 100
xstep = (xhigh - xlow)/howMany
for i in range(0,howMany+1,1):
x.append(xlow + i*xstep)
yvalue = normdist(xlow + i*xstep,mu,sigma)
ycdf.append(yvalue)
# Fitting Data to Normal Data Model
# Now plot the sample values and plotting position
myfigure = matplotlib.pyplot.figure(figsize = (7,9)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set1_wei, set1 ,color ='navy')
matplotlib.pyplot.plot(ycdf, x, color ='gold',linewidth=3)
matplotlib.pyplot.xlabel("Quantile Value")
matplotlib.pyplot.ylabel("Set1 Value")
mytitle = "Normal Distribution Data Model sample mean = : " + str(mu)+ " sample variance =:" + str(sigma**2)
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# -
# For set2
mu = set2.mean() # Fitted Model
sigma = set2.std()
x = []; ycdf = []
xlow = 0; xhigh = 1.2*max(set2) ; howMany = 100
xstep = (xhigh - xlow)/howMany
for i in range(0,howMany+1,1):
x.append(xlow + i*xstep)
yvalue = normdist(xlow + i*xstep,mu,sigma)
ycdf.append(yvalue)
# Fitting Data to Normal Data Model
# Now plot the sample values and plotting position
myfigure = matplotlib.pyplot.figure(figsize = (7,9)) # generate a object from the figure class, set aspect ratio
matplotlib.pyplot.scatter(set2_wei, set2 ,color ='orange')
matplotlib.pyplot.plot(ycdf, x, color ='purple',linewidth=3)
matplotlib.pyplot.xlabel("Quantile Value")
matplotlib.pyplot.ylabel("Set2 Value")
mytitle = "Normal Distribution Data Model sample mean = : " + str(mu)+ " sample variance =:" + str(sigma**2)
matplotlib.pyplot.title(mytitle)
matplotlib.pyplot.show()
# __Since it was an appropriate fit, we can use the normal distrubation to generate another sample randomly from the same population. Use a histogram with the new generated sets and compare them visually.__<br>
mu1 = set1.mean()
sd1 = set1.std()
mu2 = set2.mean()
sd2 = set2.std()
set1_s = np.random.normal(mu1, sd1, 100)
set2_s = np.random.normal(mu2, sd2, 100)
# +
data_d = pd.DataFrame({'Set1s':set1_s,'Set2s':set2_s})
fig, ax = plt.subplots()
data_d.plot.hist(density=False, ax=ax, title='Histogram: Set1 samples vs. Set2 samples', bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# +
fig, ax = plt.subplots()
data_d.plot.hist(density=False, ax=ax, title='Histogram: Set1 and Set1 samples vs. Set2 and Set2 samples', bins=40)
data.plot.hist(density=False, ax=ax, bins=40)
ax.set_ylabel('Count')
ax.grid(axis='y')
# -
# __Use boxplots to compare the four sets. Discuss their differences.__<br>
fig = plt.figure(figsize =(10, 7))
plt.boxplot ([set1, set1_s, set2, set2_s],1, '')
plt.show()
# __The first pair and the second pair look similar while the two pairs look differnet, right? The question is how can we KNOW if two sets are truly (significantly) different or not?__<br>
# ### Exercise 1:
# - __Step1:Read the "lab14_E2data.csv" file as a dataset.__<br>
# - __Step2:Describe the dataset numerically (using descriptive functions) and in your own words.__<br>
# - __Step3:Plot histograms and compare the sets in the dataset. What do you infer from the histograms?__<br>
# - __Step3*: This is a bonus step | Use "seaborn" to plot histograms with KDE and rugs!__<br>
# - __Step4:Write appropriate functions for the Beard, Tukey, and Adamowski Plotting Position Formulas.__<br>
# - __Step5:Apply your functions for the Beard, Tukey, and Adamowski Plotting Position Formulas on both sets and make quantile plots.__<br>
# - __Step6:Use the Tukey Plotting Position Formula and fit a Normal and a LogNormal distribution data model. Plot them and visually assess which one provides a better fit for each set__<br>
# - __Step7:Use the best distribution data model and a create two sample sets (one for each set) with 100 values.__<br>
# - __Step8:Use boxplots and illustrate the differences and similarities between the sets. What do you infer from the boxplots?__<br>
# Step1:
#Step2:
# Step3:
#Step3*: Bonus Step
#Step4: Functions for the Beard, Tukey, and Adamowski Plotting Position Formulas
#Step 5:
#Step6:
#Step7:
#Step8:
| 1-Lessons/Lesson14/Lab14/.ipynb_checkpoints/Lab14_Class-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_pytorch_p36
# language: python
# name: conda_pytorch_p36
# ---
# # Predicting Boston Housing Prices
#
# ## Using XGBoost in SageMaker (Deploy)
#
# _Deep Learning Nanodegree Program | Deployment_
#
# ---
#
# As an introduction to using SageMaker's Low Level Python API we will look at a relatively simple problem. Namely, we will use the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html) to predict the median value of a home in the area of Boston Mass.
#
# The documentation reference for the API used in this notebook is the [SageMaker Developer's Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/)
#
# ## General Outline
#
# Typically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.
#
# 1. Download or otherwise retrieve the data.
# 2. Process / Prepare the data.
# 3. Upload the processed data to S3.
# 4. Train a chosen model.
# 5. Test the trained model (typically using a batch transform job).
# 6. Deploy the trained model.
# 7. Use the deployed model.
#
# In this notebook we will be skipping step 5, testing the model. We will still test the model but we will do so by first deploying it and then sending the test data to the deployed model.
# ## Step 0: Setting up the notebook
#
# We begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.
# +
# %matplotlib inline
import os
import time
from time import gmtime, strftime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import load_boston
import sklearn.model_selection
# -
# In addition to the modules above, we need to import the various bits of SageMaker that we will be using.
# +
import sagemaker
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
# This is an object that represents the SageMaker session that we are currently operating in. This
# object contains some useful information that we will need to access later such as our region.
session = sagemaker.Session()
# This is an object that represents the IAM role that we are currently assigned. When we construct
# and launch the training job later we will need to tell it what IAM role it should have. Since our
# use case is relatively simple we will simply assign the training job the role we currently have.
role = get_execution_role()
# -
# ## Step 1: Downloading the data
#
# Fortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.
boston = load_boston()
# ## Step 2: Preparing and splitting the data
#
# Given that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.
# +
# First we package up the input data and the target variable (the median value) as pandas dataframes. This
# will make saving the data to a file a little easier later on.
X_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)
Y_bos_pd = pd.DataFrame(boston.target)
# We split the dataset into 2/3 training and 1/3 testing sets.
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)
# Then we split the training set further into 2/3 training and 1/3 validation sets.
X_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)
# -
# ## Step 3: Uploading the training and validation files to S3
#
# When a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. We can use the SageMaker API to do this and hide some of the details.
#
# ### Save the data locally
#
# First we need to create the train and validation csv files which we will then upload to S3.
# This is our local data directory. We need to make sure that it exists.
data_dir = '../data/boston'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# +
# We use pandas to save our train and validation data to csv files. Note that we make sure not to include header
# information or an index as this is required by the built in algorithms provided by Amazon. Also, it is assumed
# that the first entry in each row is the target variable.
pd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
# -
# ### Upload to S3
#
# Since we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.
# +
prefix = 'boston-xgboost-deploy-ll'
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
# -
# ## Step 4: Train and construct the XGBoost model
#
# Now that we have the training and validation data uploaded to S3, we can construct a training job for our XGBoost model and build the model itself.
#
# ### Set up the training job
#
# First, we will set up and execute a training job for our model. To do this we need to specify some information that SageMaker will use to set up and properly execute the computation. For additional documentation on constructing a training job, see the [CreateTrainingJob API](https://docs.aws.amazon.com/sagemaker/latest/dg/API_CreateTrainingJob.html) reference.
# +
# We will need to know the name of the container that we want to use for training. SageMaker provides
# a nice utility method to construct this for us.
container = get_image_uri(session.boto_region_name, 'xgboost')
# We now specify the parameters we wish to use for our training job
training_params = {}
# We need to specify the permissions that this training job will have. For our purposes we can use
# the same permissions that our current SageMaker session has.
training_params['RoleArn'] = role
# Here we describe the algorithm we wish to use. The most important part is the container which
# contains the training code.
training_params['AlgorithmSpecification'] = {
"TrainingImage": container,
"TrainingInputMode": "File"
}
# We also need to say where we would like the resulting model artifacst stored.
training_params['OutputDataConfig'] = {
"S3OutputPath": "s3://" + session.default_bucket() + "/" + prefix + "/output"
}
# We also need to set some parameters for the training job itself. Namely we need to describe what sort of
# compute instance we wish to use along with a stopping condition to handle the case that there is
# some sort of error and the training script doesn't terminate.
training_params['ResourceConfig'] = {
"InstanceCount": 1,
"InstanceType": "ml.m4.xlarge",
"VolumeSizeInGB": 5
}
training_params['StoppingCondition'] = {
"MaxRuntimeInSeconds": 86400
}
# Next we set the algorithm specific hyperparameters. You may wish to change these to see what effect
# there is on the resulting model.
training_params['HyperParameters'] = {
"max_depth": "5",
"eta": "0.2",
"gamma": "4",
"min_child_weight": "6",
"subsample": "0.8",
"objective": "reg:linear",
"early_stopping_rounds": "10",
"num_round": "200"
}
# Now we need to tell SageMaker where the data should be retrieved from.
training_params['InputDataConfig'] = [
{
"ChannelName": "train",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": train_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
},
{
"ChannelName": "validation",
"DataSource": {
"S3DataSource": {
"S3DataType": "S3Prefix",
"S3Uri": val_location,
"S3DataDistributionType": "FullyReplicated"
}
},
"ContentType": "csv",
"CompressionType": "None"
}
]
# -
# ### Execute the training job
#
# Now that we've built the dict containing the training job parameters, we can ask SageMaker to execute the job.
# +
# First we need to choose a training job name. This is useful for if we want to recall information about our
# training job at a later date. Note that SageMaker requires a training job name and that the name needs to
# be unique, which we accomplish by appending the current timestamp.
training_job_name = "boston-xgboost-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
training_params['TrainingJobName'] = training_job_name
# And now we ask SageMaker to create (and execute) the training job
training_job = session.sagemaker_client.create_training_job(**training_params)
# -
# The training job has now been created by SageMaker and is currently running. Since we need the output of the training job, we may wish to wait until it has finished. We can do so by asking SageMaker to output the logs generated by the training job and continue doing so until the training job terminates.
session.logs_for_job(training_job_name, wait=True)
# ### Build the model
#
# Now that the training job has completed, we have some model artifacts which we can use to build a model. Note that here we mean SageMaker's definition of a model, which is a collection of information about a specific algorithm along with the artifacts which result from a training job.
# +
# We begin by asking SageMaker to describe for us the results of the training job. The data structure
# returned contains a lot more information than we currently need, try checking it out yourself in
# more detail.
training_job_info = session.sagemaker_client.describe_training_job(TrainingJobName=training_job_name)
model_artifacts = training_job_info['ModelArtifacts']['S3ModelArtifacts']
# +
# Just like when we created a training job, the model name must be unique
model_name = training_job_name + "-model"
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the xgboost container that we used for training
# can also be used for inference.
primary_container = {
"Image": container,
"ModelDataUrl": model_artifacts
}
# And lastly we construct the SageMaker model
model_info = session.sagemaker_client.create_model(
ModelName = model_name,
ExecutionRoleArn = role,
PrimaryContainer = primary_container)
# -
# ## Step 5: Test the trained model
#
# We will be skipping this step for now. We will still test our trained model but we are going to do it by using the deployed model, rather than setting up a batch transform job.
#
# ## Step 6: Create and deploy the endpoint
#
# Now that we have trained and constructed a model it is time to build the associated endpoint and deploy it. As in the earlier steps, we first need to construct the appropriate configuration.
# +
# As before, we need to give our endpoint configuration a name which should be unique
endpoint_config_name = "boston-xgboost-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": model_name,
"VariantName": "AllTraffic"
}])
# -
# And now that the endpoint configuration has been created we can deploy the endpoint itself.
#
# **NOTE:** When deploying a model you are asking SageMaker to launch an compute instance that will wait for data to be sent to it. As a result, this compute instance will continue to run until *you* shut it down. This is important to know since the cost of a deployed endpoint depends on how long it has been running for.
#
# In other words **If you are no longer using a deployed endpoint, shut it down!**
# +
# Again, we need a unique name for our endpoint
endpoint_name = "boston-xgboost-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = endpoint_config_name)
# -
# Just like when we created a training job, SageMaker is now requisitioning and launching our endpoint. Since we can't do much until the endpoint has been completely deployed we can wait for it to finish.
endpoint_dec = session.wait_for_endpoint(endpoint_name)
# ## Step 7: Use the model
#
# Now that our model is trained and deployed we can send test data to it and evaluate the results. Here, because our test data is so small, we can send it all using a single call to our endpoint. If our test dataset was larger we would need to split it up and send the data in chunks, making sure to accumulate the results.
# First we need to serialize the input data. In this case we want to send the test data as a csv and
# so we manually do this. Of course, there are many other ways to do this.
payload = [[str(entry) for entry in row] for row in X_test.values]
payload = '\n'.join([','.join(row) for row in payload])
# +
# This time we use the sagemaker runtime client rather than the sagemaker client so that we can invoke
# the endpoint that we created.
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = payload)
# We need to make sure that we deserialize the result of our endpoint call.
result = response['Body'].read().decode("utf-8")
Y_pred = np.fromstring(result, sep=',')
# -
# To see how well our model works we can create a simple scatter plot between the predicted and actual values. If the model was completely accurate the resulting scatter plot would look like the line $x=y$. As we can see, our model seems to have done okay but there is room for improvement.
plt.scatter(Y_test, Y_pred)
plt.xlabel("Median Price")
plt.ylabel("Predicted Price")
plt.title("Median Price vs Predicted Price")
# ## Delete the endpoint
#
# Since we are no longer using the deployed model we need to make sure to shut it down. Remember that you have to pay for the length of time that your endpoint is deployed so the longer it is left running, the more it costs.
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
# ## Optional: Clean up
#
# The default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.
# +
# First we will remove all of the files contained in the data_dir directory
# !rm $data_dir/*
# And then we delete the directory itself
# !rmdir $data_dir
# -
| sagemaker_deployment/Tutorials/Boston Housing - XGBoost (Deploy) - Low Level.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Generate and visualize toy data sets
# +
import zfit
import numpy as np
from scipy.stats import norm, expon
from matplotlib import pyplot as plt
zfit.settings.set_seed(10) # fix seed
bounds = (0, 10)
obs = zfit.Space('x', limits=bounds)
# true parameters for signal and background
truth_n_sig = 1000
Nsig = zfit.Parameter("Nsig", truth_n_sig)
mean_sig = zfit.Parameter("mean_sig", 5.0)
sigma_sig = zfit.Parameter("sigma_sig", 0.5)
sig_pdf = zfit.pdf.Gauss(obs=obs, mu=mean_sig, sigma=sigma_sig).create_extended(Nsig)
truth_n_bkg = 10000
Nbkg = zfit.Parameter("Nbkg", truth_n_bkg)
lambda_bkg = zfit.Parameter("lambda_bkg", -1/4.0)
bkg_pdf = zfit.pdf.Exponential(obs=obs, lambda_=lambda_bkg).create_extended(Nbkg)
truth_sig_t = (1.0,)
truth_bkg_t = (2.5, 2.0)
# make a data set
m_sig = sig_pdf.sample(truth_n_sig).numpy()
m_bkg = bkg_pdf.sample(truth_n_bkg).numpy()
m = np.concatenate([m_sig, m_bkg]).flatten()
# fill t variables
t_sig = expon(0, *truth_sig_t).rvs(truth_n_sig)
t_bkg = norm(*truth_bkg_t).rvs(truth_n_bkg)
t = np.concatenate([t_sig, t_bkg])
# cut out range (0, 10) in m, t
ma = (bounds[0] < t) & (t < bounds[1])
m = m[ma]
t = t[ma]
fig, ax = plt.subplots(1, 3, figsize=(16, 4.5))
ax[0].hist2d(m, t, bins=(50, 50))
ax[0].set_xlabel("m")
ax[0].set_ylabel("t")
ax[1].hist([m_bkg, m_sig], bins=50, stacked=True, label=("background", "signal"))
ax[1].set_xlabel("m")
ax[1].legend()
ax[2].hist((t[truth_n_sig:], t[:truth_n_sig]), bins=50, stacked=True, label=("background", "signal"))
ax[2].set_xlabel("t")
ax[2].legend();
sorter = np.argsort(m)
m = m[sorter]
t = t[sorter]
# -
# # Fit toy data set
# +
from zfit.loss import ExtendedUnbinnedNLL
from zfit.minimize import Minuit
tot_pdf = zfit.pdf.SumPDF([sig_pdf, bkg_pdf])
loss = ExtendedUnbinnedNLL(model=tot_pdf, data=zfit.data.Data.from_numpy(obs=obs, array=m))
minimizer = Minuit()
minimum = minimizer.minimize(loss=loss)
minimum.hesse()
print(minimum)
# -
# ## Visualize fitted model
# +
from utils import pltdist, plotfitresult
fig = plt.figure(figsize=(8, 5.5))
nbins = 80
pltdist(m, nbins, bounds)
plotfitresult(tot_pdf, bounds, nbins, label="total model", color="crimson")
plotfitresult(bkg_pdf, bounds, nbins, label="background", color="forestgreen")
plotfitresult(sig_pdf, bounds, nbins, label="signal", color="orange")
plt.xlabel("m")
plt.ylabel("number of events")
plt.legend();
# -
# ## Compute sWeights
# +
from hepstats.splot import compute_sweights
weights = compute_sweights(tot_pdf, m)
print("Sum of signal sWeights: ", np.sum(weights[Nsig]))
# +
fig, ax = plt.subplots(1, 2, figsize=(16, 4.5))
plt.sca(ax[0])
nbins = 40
plt.plot(m, weights[Nsig], label="$w_\\mathrm{sig}$")
plt.plot(m, weights[Nbkg], label="$w_\\mathrm{bkg}$")
plt.plot(m, weights[Nsig] + weights[Nbkg], "-k")
plt.axhline(0, color="0.5")
plt.legend()
plt.sca(ax[1])
plt.hist(t, bins=nbins, range=bounds, weights=weights[Nsig], label="weighted histogram")
plt.hist(t_sig, bins=nbins, range=bounds, histtype="step", label="true histogram")
t1 = np.linspace(*bounds, nbins)
tcdf = expon(0, 1).pdf(t1) * np.sum(weights[Nsig]) * (bounds[1] - bounds[0])/nbins
plt.plot(t1, tcdf, label="model with $\lambda_\\mathrm{sig}$")
plt.xlabel("t")
plt.legend();
# -
np.average(t, weights=weights[Nsig])
np.average(t_sig)
| notebooks/splots/splot_example_2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # "Non-Intrusive Load Monitoring - NILM"
# > "A source separation problem that can enable a better, smarter electric grid"
#
# - toc:true
# - branch: master
# - badges: false
# - comments: true
# - author: <NAME>
# - categories: [projects]
# ## Context
#
# I spent just over three years at the research arm of Tata Consultancy Services (then called the Innovation Labs) from 2011 to 2014 (before I took a sabbatical for my doctoral studies and eventually moved out). The experience of working on futuristic technology problems in an industrial environment was interesting. I had the opportunity to witness an idea grow from its budding stage to eventual proof of concept and further adoption.
#
# One research problem that occupied most of my time during this stint was *Source Separation*. The idea underlying the problem stems from the need to analyse the multiple sources that give rise to a multitude of effects on a single (or a small number of) measurements. One application problem that came out of it (which was in its infancy during that time, and is at various stages of implementation today): [Non-intrusive load monitoring (NILM)](https://en.wikipedia.org/wiki/Nonintrusive_load_monitoring).
#
# In this article, I will discuss my experiences working on this problem.
# ## An intro
#
# A couple of years or so back, EDF, the French electric giant, with whom I have a contract for my house's electricity, updated their app/interface with a possible option to follow our daily consumption profile. This followed the installation of the smart electric meters, known as `Linky`, in our apartment. While this did not provide me with insights that I did not already have, there was something more on offer. The application also allowed me to dive further into the categories of my consumption based on different groupings:
# 1. Refrigeration
# 1. Water heating
# 1. Electric heater
# 1. Cooking
# 1. Infotainment and Computers.
#
# The recent screen capture of this looks something like this (I redacted the amount and my address):
#
# <img src="./../images/nilm/nilm_edf.jpg" alt="NILM - EDF" width="300"/>
#
# The refrigerator consumed 9% of the total electricity consumption. And one can get a vague idea already about how much the other categories consumed. Given the furore over data privacy, I had to *unlock* these features by deliberately giving EDF the rights handle my data.
#
# By now, one would have got the idea of what NILM means:
# > Note: To disaggregate or distinguish between different electrical appliances and their power consumption using a single power meter data.
# Or, one could view NILM algorithms acting like a prism
#
# <img src="./../images/nilm/nilm_prism.png" alt="NILM - Prism" width="500"/>
#
# When I first started to work on the problem of NILM, the academic papers were all interested in a sampling rate of once in 1 second. And right in line, the open datasets were also developed to the same tune. For example, the [REDD](http://redd.csail.mit.edu/) dataset from (the then) MIT team with [<NAME>](http://zicokolter.com/) at the helm provided data in that sampling rate range. Subsequent open data sets followed suit with similar ones. And some, like the popular dataset from Ubicomp Lab at the University of Washington on [Kaggle](https://www.kaggle.com/c/belkin-energy-disaggregation-competition) (that featured in the Belkin competition), had an even higher frequency of operation.
#
# However, the *Linky* smart meters installed in my apartment collects data once in 15 minutes and maps more closely to my ventures just before I started my PhD. In this work, we explored the use of AMI type data for NILM. AMI stands for Advanced Metering Infrastructure, the type of metering companies were hoping to install in households (sampling the cumulated power consumption every 15 minutes or so). This is in contrast with the high-frequency data over which most of the academic research work were based on. While challenging, that seemed unrealistic and so we wrote this paper summarizing our then-ongoing efforts.
#
# [Springer Link behind paywall](https://link.springer.com/chapter/10.1007/978-3-319-04960-1_8) or perhaps more useful would be the [link to Pre-print](https://github.com/krishnans14/feedback-control/tree/master/files/sirs_14_final.pdf)
#
#
# ## Client Project
#
# In our initial work, we focused on using the open datasets (for lack of data from our side). Things changed due to a pilot project to implement NILM in the Netherlands for a startup client. The unfortunate thing was we started without any data to work with and limited assumptions. The IT team built a data handling infrastructure awaiting installation of sensors, but developing a machine learning algorithm without data was a cruel joke (we weren't even sure on what would be the sampling rate of data because our client was still discussing with their potential clients about it).
#
# So we decided to do what today is terms as [*Transfer Learning*](https://en.wikipedia.org/wiki/Transfer_learning) which (as per Wikipedia)
# > focuses on storing knowledge gained while solving one problem and applying it to a different but related problem.
#
# without actually knowing what the term *transfer learning*. We collected some statistics/pattern about the characteristics of various appliances from the open datasets available. Then we created a pseudo database for different appliances that could then be used for training when the data arrived.
#
# An extra problem that plagued the initial efforts were in obtaining appliance level signatures. The pilot was in households where the wiring of the appliances was well integrated into the walls and it was difficult to put plugs to tap them. This led to an extra aspect of *gamification* introduced to label data. The process was as follows:
#
#
# * Using the transfer learning-based database, our NILM algorithm will provide detection of appliances.
# * The inhabitants of the households will get notifications at the end of the day on these detections through a mobile App.
# * The user labels the detection (correct or wrong) based on their own knowledge.
# * The NILM algorithm trains a model specific to each household based on this labelling.
#
# Several aspects of the above process were in flux. For instance, we were exploring different algorithms that can perform NILM (note that back in 2013, this area was fresh and had limited success) or the *gamification* aspects were not clear (how much to trust the feedback, etc.). But with all the limitations, a pilot went forward and looked good. But everything also came to an abrupt end due to financial constraints at the startup.
#
# During the same period, several other startups, notably in the US were working on the NILM problem. Some of them are still active: [Bidgely](https://www.bidgely.com/), [Opower](https://en.wikipedia.org/wiki/Opower), and more (check this [2012 entry on Oliver Parson's blog](https://blog.oliverparson.co.uk/2012/05/nialm-in-industry.html) on companies working on NILM). Several other companies came in and disappeared as it always happens. I worked with one of them.
#
# ## NILM Techniques Explored
#
# It is not surprising that we tried several techniques to realize NILM. Further, unlike academic freedom, we had to work with a limited set of assumptions and hence the need to use customized techniques (which were of course not published). But here are some techniques that were published:
# * Bayesian Inference [IEEE link behind paywall](https://ieeexplore.ieee.org/abstract/document/6603710) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/esiot13_final.pdf)
# * Factor Graphs [IEEE link behind paywall](https://ieeexplore.ieee.org/abstract/document/6637447/) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/icacci13_final.pdf)
#
# Apart from the different techniques, we also presented a paper on the approach to use a mix of transfer learning and simulation to generate labelled data over which NILM algorithms could be tested: [ACM link behind paywall](https://dl.acm.org/doi/abs/10.1145/2559627.2559630) or [Preprint](https://github.com/krishnans14/feedback-control/tree/master/files/es4cps_final.pdf)
#
# I will discuss these techniques and those we explored in more details sometime in the future.
# ## Beyond household disaggregation
#
# Our explorations for the application of NILM went beyond the household energy disaggregation pilot project with the Dutch startup. The following were the other problems that were at explored:
# * Cost-savings for a large building
# * A large office building of an enterprise also contained a cafeteria serving hundreds of diners. The power to these were supplied from a single transformer. This means that the electricity tarrif paid by the enterprise was corresponding to a commercial establishment (the cafeteria) and not the workplace. The latter was much cheaper. We gave a proposal of how one could attempt to use NILM and obtain an estimation of the two entities and save cost in the electricity bill paid.
# * Condition monitoring of appliances or industrial equipments
# * Today, the central aspect of Industry 4.0 is the condition monitoring of equipment to perform predictive maintenance, so much so that even [Amazon is into it](https://krishnans14.github.io/feedback-control/musings/industry-watch/2020/12/17/Monitron-and-Predictive-Maintenance.html). A couple of proposals were floated in that direction back in 2013-14.
# * Disaggregation of load versus generation (with rooftop solar installation)
# * When the rooftop solar installations became popular, different countries took different approaches to their integration with the grid. In some countries, there were no restrictions on how an individual household decides to integrate solar panels with their own usage or to connect back to the grid. We floated ideas on how to use NILM on the smart meter data to estimate generation capacity in a household.
# * A related application was whether we can use a single smart meter to identify defects in a host of solar PV panels (say on rooftops or a farm).
# * Activity monitoring
# * A more contentious application of NILM was on activity monitoring. A [patent](https://patents.google.com/patent/WO2015124972A1/en) was filed in this application towards the fag end of my stay in the lab.
#
# These are the applications that I remember from the top of my head.
# ## A few words before the end
#
# Recently, I bumped into the [NILM workshop for 2020](http://nilmworkshop.org/2020/) organized online and came across their papers and the YouTube live videos.
#
# I would try to spend some more time into these papers and posters and synthesize some thoughts for a future post where I would also like to discuss the [NILMTK python module](https://github.com/nilmtk/nilmtk), which I had been itching to try.
#
# It would be disingenious not to acknowledge the contributions of co-workers/supervisors in the above endeavours, though one can see the presence of [<NAME>](https://in.linkedin.com/in/mgirishchandra), [<NAME>](https://ca.linkedin.com/in/goutam-yelluru-gopal-93549428) prominently in all the publications.
| _notebooks/2020-12-30-NILM.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# This is hamoye Data science track introduction to python for machine learning code snippets
#
# # Lesson 1 no code snippet
# # Lesson 2 Numpy array and vectorization
#
#
# convention for importing numpy
import numpy as np
# +
arr = [6, 7, 8, 9]
print(type(arr)) # prints <class 'list'>
a = np.array(arr)
print(type(a)) # prints <class 'numpy.ndarray'>
print(a.shape) # prints (4, ) - a is a 1d array with 4 items
print(a.dtype) # prints int64
# get the dimension of a with ndim
print(a.ndim) # print's 1
b = np.array([
[3, 4, 5], [4, 5, 8]
])
print(b) # prints [[3 4 5] [4 5 8]]
print(b.ndim) # prints 2 - b is a 2d array
print(b.shape) # prints (2, 3) - b a 2d array with 2 rows and 3 columns
# -
# There are also inbuilt functions that can be used to initialize numpy which include empty(), zeros(), ones(), full(), random.random()
# +
# a 2x3 array with random values
print(np.random.random((2, 3))) # = array([[0.60793904, 0.02881965, 0.73022145], [0.34183628, 0.63274067, 0.07945224]])
# a 2x3 array of zeros
print(np.zeros((2, 3))) # = array([[0., 0., 0.], [0., 0., 0.]])
# a 2x3 array of ones
print(np.ones((2, 3))) # = array([[1., 1., 1.], [1., 1., 1]])
# a 3x3 identity matrix
print(np.zeros(3))# = array([[1., 0., 0.], [0., 0., 1.]])
# +
# Intra operability of arrays and scalars
c = np.array([[9.0, 8.0, 7.0], [1.0, 2.0, 3.0]])
d = np.array([[4.0, 5.0, 6.0], [9.0, 8.0, 7.0]])
print(c + d) # prints [[13. 13. 13.] [10. 10. 10.]]
print(5/d) # prints [[1.25 1. 0.83333333] [0.55555556 0.625 0.71428571]]
print(c ** 2) # prints [[81. 64. 49.] [ 1. 4. 9.]]
# -
# Indexing with arrays and using arrays for data processing
# +
print(d[1, 0:2]) # prints [9. 8.]
e = np.array([[10, 11, 12], [13, 14, 15], [16, 17, 18], [19, 20, 21]])
#print(e.shape, e.ndim)
# list slicing
print(e[:3, :2]) # prints 3 rows and 2 columns
# -
# There are other advanced methods of indexing which are shown below
# +
# Integer indexing
print(e[[2, 0, 3, 1], [2, 1, 0, 2]]) # prints [18 11 19 15]
# boolean indexing meeting a specified condition
print(e[e>15]) # prints [16 17 18 19 20 21]
# -
# # Lesson 3
# # Pandas - so much more than a cute animal
# it is a series of one dimensional array
#
# convention for importing pandas
import pandas as pd
# +
days = pd.Series(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'])
days
# +
# using numpy array
list_days = np.array(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'])
numpy_days = pd.Series(list_days)
numpy_days
# -
# using strings as index
d = pd.Series(['Monday', 'Tuesday', 'Wodnesday', 'Thursday', 'Friday'], ['a', 'b', 'c', 'd', 'e'])
d
# +
# create series from dictionary
d1 = pd.Series({'a':'Monday', 'b':'Tuesday', 'c':'Wednesday', 'd':'Thursday', 'e':'Friday'})
d1
# -
# Series can be accessed using specified index
d1[0]
d1[1:]
d1['c']
# # Pandas DataFrame
#
pd.DataFrame() # prints an empty dataframe
# +
# Create a dataframe from a dictionary
df_dict ={"Country": ['Ethiopia', 'Kenya', 'Nigeria', 'Ghana', 'Uganda'],
"Capital": ['Addis Ababa', 'Nirobi', 'Abuja', 'Accra', 'Kampala'],
"Population": [100000, 80500, 150000, 40000, 50000],
"Age": [60, 80, 70, 67, 90]}
df = pd.DataFrame(df_dict, index = [2, 4, 6, 8, 10])
df
# +
# create a dataframe from a list
df_list = [["Ethiopia", "<NAME>", 100000, 60],
["Kenya", "Nirobi", 805000, 80],
['Nigeria', 'Abuja', 150000, 70],
['Ghana', 'Accra', 40000, 67],
['Uganda', 'Kampala', 50000, 90]]
df1 = pd.DataFrame(df_list, columns=['Country', 'Capital', 'Population', 'Age'], index=[i+1 for i in range(len(df_list))])
df1
# -
# # at, iat, iloc, loc are accessors used to retrieve data in DataFrame
# Select the row at the index 0
df.iloc[0]
# select the Capital column
df['Capital']
# Select row with index label 6
df.loc[6]
# select single value with at label 6
df.at[6, 'Country']
# select single value using iat
df.iat[3, 0]
# # Statistical analysis
# find the sum of population
df['Population'].sum()
df.mean()
df.std()
df.median()
df.describe()
df.info()
# # missing value
# +
df_dict2 = {'Name':['Dejene', 'Asibeh', 'Tenager', np.nan],
'Profession':['Researcher', 'Software Engineer', 'Doctor', 'Data Scientist'],
'Experience':[7, np.nan, 8, 10],
'Height': [np.nan, 175, 180, 150]}
new_df = pd.DataFrame(df_dict2)
new_df
# -
# check for cells with missing values as True
new_df.isnull()
# remove rows with missing values
new_df.dropna()
# # Data types and Data wrangling
#
# - Working with different types of data: text files, CSV, JSON objects, HTML and databases
#
# Pandas can connect to databases, get data with queries and save in a dataframe
# impoting pandas library
import pandas as pd
# +
url = url='https://github.com/WalePhenomenon/climate_change/blob/master/fuel_ferc1.csv?raw=true'
fuel_df = pd.read_csv(url, error_bad_lines=False)
fuel_df.to_csv('fuel_data.csv', index=False)
# -
fuel_data = pd.read_csv('fuel_data.csv')
fuel_data.head()
fuel_data.describe(include='all')
# shows the skewness of the fuel data in two decimal points
round(fuel_data.skew(), 2)
# shows the kurtios of the fuel data in two decimal points
round(fuel_data.kurt(), 2)
# the correlation of the fuel data
fuel_data.corr()
# # Check for missing values
fuel_data.isnull().sum()
# Use groupby to count the sum of each unique value in the fuel unit column
fuel_data['fuel_count']= fuel_data.groupby('fuel_unit')['fuel_unit'].count()
fuel_count
fuel_data[['fuel_unit']] = fuel_data[['fuel_unit']].fillna(value='mcf')
# Check if missing values have been filled
fuel_data.isnull().sum()
# Count the number of report year
fuel_data.groupby('report_year')['report_year'].count()
# The average fuel_cost_per_unit_delivered in each year
fuel_data.groupby('report_year')['fuel_cost_per_unit_delivered'].mean()
# Merging in Pandas can be likened to join operations in relational databases like SQL.
# Group by the fuel type code and print the first entries in all the groups formed
fuel_data.groupby('fuel_type_code_pudl').first()
# Split the fuel data into two groups and merge using different methods
fuel_df1 = fuel_data.iloc[0:19000].reset_index(drop=True)
fuel_df2 = fuel_data.iloc[19000:].reset_index(drop=True)
# check that the length of both dataframes sum to the expexted length
assert len(fuel_data) == (len(fuel_df1) + len(fuel_df2))
# an inner merge will lose rows that do not match in both dataframes
pd.merge(fuel_df1, fuel_df2, how='inner')
# outer merge returns all rows in both dataframes
pd.merge(fuel_df1, fuel_df2, how='outer')
# removes rows from the right dataframe that do not have a match with the left and keeps all rows from the left
pd.merge(fuel_df1, fuel_df2, how='left')
# Concatenation is performed with the concat() function
data_to_concat = pd.DataFrame(np.zeros(fuel_data.shape))
pd.concat([fuel_data, data_to_concat]).reset_index(drop=True)
# Duplicates are a common occurrence in datasets which alter the results of analysis
# check for duplicate rows
fuel_data.duplicated().any()
# # Data Visualization and Representation in Python
# - The Anscombe Quartet and the importance of visualizing data
# +
# Import plotting library
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(7, 4))
plt.xticks(rotation=90)
fuel_unit = pd.DataFrame({'unit': ['BBL', 'GAL', 'GRAMSU', 'KGU', 'MCF', 'MMBTU', 'MWDTH', 'MWHTH', 'TON'],
'count': [7998, 84, 464, 110, 11354, 180, 95, 100, 8958]})
sns.barplot(data=fuel_unit, x='unit', y='count')
plt.xlabel('Fuel Unit')
# -
# Because of the extreme range of the values for the fuel unit, we can plot the barchart by taking the logarithm of the y-axis as follows:
g = sns.barplot(data=fuel_unit, x='unit', y='count')
plt.xticks(rotation=90)
g.set_yscale('log')
g.set_ylim(1, 12000)
plt.xlabel('Fuel Unit')
# Select a sample of the dataset
sample_df = fuel_data.sample(n=50, random_state=4)
sns.regplot(x=sample_df['utility_id_ferc1'], y=sample_df['fuel_cost_per_mmbtu'], fit_reg=False)
# - Advanced plotting: Kerbel Density Estimate plots, box plots and violin plots
# box plot
sns.boxplot(x='fuel_type_code_pudl', y='utility_id_ferc1', palette=['m','g'], data=fuel_data)
# KDE plot
sns.kdeplot(sample_df['fuel_cost_per_unit_burned'], shade=True, color='b')
# A heatmap is a representation of data that uses a spectrum of colours to indicate different values. It gives quick summaries and identifies patterns especially in large datasets. Alternatively, heatmaps can be described as table visualisations where the colour of each cell relates the values. The image below is an example of a heatmap
sns.heatmap(sample_df.corr())
| SectionA.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # <NAME>
# ## Computer Vision and IOT Intern @TSF
# ### Prediction using Decision tree algorithm
# ### Dataset : Iris.csv (https://bit.ly/34SRn3b)
# **Algorithm**
#
# One of the most important considerations when choosing a machine learning algorithm is how interpretable it is. The ability to explain how an algorithm makes predictions is useful to not only you, but also to potential stakeholders. A very interpretable machine learning algorithm is a decision tree which you can think of as a series of questions designed to assign a class or predict a continuous value depending on the task. The example image is a decision tree designed for classification.
# +
# %matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn import tree
# -
df=pd.read_csv('Iris.csv')
df
df.info()
# +
features = ['SepalLengthCm','SepalWidthCm','PetalLengthCm','PetalWidthCm']
# Create features matrix
x = df.loc[:, features].values
# -
y=df.Species
x_train,x_test,y_train,y_test=train_test_split(x, y, random_state=0)
clf = DecisionTreeClassifier(max_depth = 2,
random_state = 0)
# +
clf.fit(x_train, y_train)
# -
clf.predict(x_test[0:1])
from sklearn import metrics
import seaborn as sns
score = clf.score(x_test, y_test)
print(score)
print(metrics.classification_report(y_test,clf.predict(x_test)))
# +
cm = metrics.confusion_matrix(y_test, clf.predict(x_test))
plt.figure(figsize=(7,7))
sns.heatmap(cm, annot=True,
fmt=".0f",
linewidths=.5,
square = True,
cmap = 'Blues');
plt.ylabel('Actual label', fontsize = 17);
plt.xlabel('Predicted label', fontsize = 17);
plt.title('Accuracy Score: {}'.format(score), size = 17);
plt.tick_params(labelsize= 15)
# +
# List of values to try for max_depth:
max_depth_range = list(range(1, 6))
# List to store the average RMSE for each value of max_depth:
accuracy = []
for depth in max_depth_range:
clf = DecisionTreeClassifier(max_depth = depth,
random_state = 0)
clf.fit(x_train, y_train)
score = clf.score(x_test, y_test)
accuracy.append(score)
# +
#ploting accuracy score depth wise
fig, ax = plt.subplots(nrows = 1, ncols = 1, figsize = (10,7));
ax.plot(max_depth_range,
accuracy,
lw=2,
color='k')
ax.set_xlim([1, 5])
ax.set_ylim([.50, 1.00])
ax.grid(True,
axis = 'both',
zorder = 0,
linestyle = ':',
color = 'k')
ax.tick_params(labelsize = 18)
ax.set_xticks([1,2,3,4,5])
ax.set_xlabel('max_depth', fontsize = 24)
ax.set_ylabel('Accuracy', fontsize = 24)
fig.tight_layout()
#fig.savefig('images/max_depth_vs_accuracy.png', dpi = 300)
# +
fig, axes = plt.subplots(nrows = 1, ncols = 1, figsize = (7,4), dpi = 150)
tree.plot_tree(clf);
# -
# Putting the feature names and class names into variables
fn = ['sepal length (cm)','sepal width (cm)','petal length (cm)','petal width (cm)']
cn = ['setosa', 'versicolor', 'virginica']
# +
fig, axes = plt.subplots(nrows = 1, ncols = 1, figsize = (7,4), dpi = 300)
tree.plot_tree(clf,
feature_names = fn,
class_names=cn,
filled = True);
#fig.savefig('images/plottreefncn.png')
# -
# ### Conclusion
# - **After Importing, Fit our dataset in our model, accuracy is 89.47%.**
#
# - **We can clearly see model performance by confusion matrix and classification report.**
#
# - **By ploting accuracy score depth wise graph, optimal depth for model is 3.**
# ### Thank You!
| .ipynb_checkpoints/Prediction_using_Decision Tree-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Titanic - Machine Learning from Disaster
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
train = pd.read_csv('train.csv')
test = pd.read_csv('test.csv')
# + [markdown] tags=[]
# ## Variables Dictionary
# -
# - Variable | Definition | Key
# - survival | Survival | 0 = No, 1 = Yes
# - pclass | Ticket class | 1 = 1st, 2 = 2nd, 3 = 3rd
# - sex | Sex
# - Age | Age in years
# - sibsp | # of siblings / spouses aboard the Titanic
# - parch | # of parents / children aboard the Titanic
# - ticket | Ticket number
# - fare | Passenger fare
# - cabin | Cabin number
# - embarked | Port of Embarkation | C = Cherbourg, Q = Queenstown, S = Southampton
# + [markdown] tags=[]
# ## EDA
# -
train.head()
train.shape
train.info()
train.describe()
# + [markdown] tags=[]
# #### Let's make buckets for our columns
#
# 1) Target = Survived
# 2) Numerical = Pclass, Age, SibSp, Parch, Fare
# 3) Categorical = Sex, Embarked
# 4) Not sure = Name, Ticket, Cabin
# + [markdown] tags=[]
# ### Univariate Analysis
# -
# When exploring our dataset and its features, we have many options available to us. We can explore each feature individually, or compare pairs of features, finding the correlation between. Let's start with some simple Univariate (one feature) analysis.
#
# Features can be of multiple types:
#
# - Nominal: is for mutual exclusive, but not ordered, categories.
# - Ordinal: is one where the order matters but not the difference between values.
# - Interval: is a measurement where the difference between two values is meaningful.
# - Ratio: has all the properties of an interval variable, and also has a clear definition of 0.0.
#
# There are multiple ways of manipulating each feature type, but for simplicity, we'll define only two feature types:
#
# - Numerical: any feature that contains numeric values.
# - Categorical: any feature that contains categories, or text.
#
fig, ax = plt.subplots(2, 4, figsize=(16, 8))
sns.countplot(ax=ax[0, 0], data=train, x='Survived')
sns.countplot(ax=ax[0, 1], x='Pclass', data=train)
sns.countplot(ax=ax[0, 2], x='Sex', data=train)
sns.histplot(ax=ax[0, 3], x=train['Age'])
sns.countplot(ax=ax[1, 0], x='SibSp', data=train)
sns.countplot(ax=ax[1, 1], x='Parch', data=train)
sns.histplot(ax=ax[1, 2], x=train['Fare'], bins=20)
sns.countplot(ax=ax[1, 3], x='Embarked', data=train)
# + [markdown] tags=[]
# ### Is there any missing values in our data?
# -
train.isnull().sum()
msno.matrix(train, figsize=(16, 8))
# - Age contain 177 missing values, since the distribution of age is right skewed we might consider fill the missing value with median.
# - Cabin contain 687 missing values, roughly 70-80 % missing, we might consider to drop it.
# - Embarked only contain 2 missing values, great we can fill it with most frequent values.
# ### Feature vs Target
def biplot(feature, bins='auto'):
fig, ax = plt.subplots(1, 2, figsize=(12, 6))
sns.countplot(ax=ax[0], x=feature, data=train)
sns.histplot(ax=ax[1], x=feature, hue='Survived', data=train, kde=True, bins=bins)
# #### 1. Pclass
train.Pclass.describe()
train.Pclass.value_counts()
biplot('Pclass')
# From plot above, we can see that first class have higher chance of survive, on the other hand the third class have higher chance not survive.
# #### 2. Name
train.Name.head()
# After looking at the Name columns, my thinking was: Is a person title affecting survive rate ?
# #### 3. Sex
train.Sex.describe()
train.Sex.value_counts()
biplot('Sex')
# Male are more likely to not survive, maybe they prioritized women first ?
# #### 4. Age
train.Age.describe()
train.Age.value_counts()
biplot('Age')
# - People in age around 20 and 40 are have the higher chance of surviving
# - Also we see that many children survived too
# #### 5. SibSp
train.SibSp.describe()
train.SibSp.value_counts()
biplot('SibSp')
# - Having 0 sibling or spouse have higher chance of not surviving, maybe they prioritized to save other people rather than themself ?
# - Having 1 sibling or spouse have higher chance of surviving, maybe they helping each other to survive ?
# - Having more than 1 sibling or spouse suprisingly have higher chance of not surviving, maybe they helping other people too after finish help their sibling or spouse ?
# #### 6. Parch
train.Parch.describe()
train.Parch.value_counts()
biplot('Parch')
# #### 7. Ticket
len(train.Ticket.unique())
train.Ticket.head()
# There are 681 unique values in Ticket I'm not sure how to encode that to reduce the dimensionality
# #### 8. Fare
train.Fare.head()
train.Fare.describe()
biplot('Fare', bins=10)
# Looks like cheaper fare is not really safe, and higher price having higher chance to survive
# #### 9. Cabin
train.Cabin.unique()[:10]
len(train.Cabin.unique())
train.Cabin.isnull().sum()
# There just too many missing values in Cabin column we might just drop it later
# #### 10. Embarked
train.Embarked.describe()
train.Embarked.value_counts()
biplot('Embarked')
# Maybe cheaper fare are people from Southampton and that's decrease the chance of surviving
sns.histplot(x='Fare', hue='Embarked', data=train, bins=10)
# Surely cheaper fare come from Southampton, now that explain it
# + [markdown] tags=[]
# ## Model Building
# + [markdown] tags=[]
# ### Selecting features and splitting data into features and target variable
# -
train.columns
X = train.drop(['PassengerId', 'Survived', 'Name', 'Ticket', 'Cabin'], axis=1)
y = train.Survived
X.head()
# ### Train test split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=42)
# ### Preprocessing
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder
# +
numerical_pipeline = Pipeline([
('imputer', SimpleImputer(strategy='median')),
('scaler', MinMaxScaler())
])
categorical_pipeline = Pipeline([
('imputer', SimpleImputer(strategy='most_frequent')),
('onehot', OneHotEncoder())
])
# -
from sklearn.compose import ColumnTransformer
preprocessor = ColumnTransformer([
('numeric', numerical_pipeline, ['SibSp', 'Parch', 'Age', 'Fare']),
('categoric', categorical_pipeline, ['Sex', 'Pclass', 'Embarked'])
])
# ### Pipeline
from sklearn.neighbors import KNeighborsClassifier
pipeline = Pipeline([
('prep', preprocessor),
('algo', KNeighborsClassifier())
])
pipeline.fit(X_train, y_train)
pipeline.score(X_test,y_test)
# ### GridSearch CV
from sklearn.model_selection import GridSearchCV
pipeline.get_params()
# +
parameters = {
'algo__n_neighbors': range(1, 51, 2),
'algo__weights' : ['uniform', 'distance'],
'algo__p' : [1, 2]
}
model = GridSearchCV(pipeline, parameters, cv=5, n_jobs=-1, verbose=1)
model.fit(X_train, y_train)
# +
# result = pd.DataFrame(model.cv_results_)
# +
# result.sort_values('rank_test_score').head()
# -
model.best_params_
model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test)
# ### Predicting Jack & Rose
data = [
[1, 'female', 17, 1, 1, 40, 'S'],
[3, 'male', 20, 0, 0, 8, 'S']
]
X_pred = pd.DataFrame(data, columns=X_train.columns, index=['Rose', 'Jack'])
X_pred
X_pred['Survived'] = model.predict(X_pred)
X_pred
# ### Save Model
import pickle
filename = 'knn_titanic.pkl'
pickle.dump(model, open(filename, 'wb'))
model.best_estimator_
# ## Predict test
test.head()
X_pred = test.drop(['PassengerId', 'Name', 'Ticket'], axis=1)
model.predict(X_pred)
pred = pd.DataFrame({
'PassengerId':test.PassengerId,
'Survived':model.predict(X_pred)
})
pred.head()
pred.to_csv('gender_submission.csv', index=False)
| Titanic Survival - KNN.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Python basics
# ***
# ### General comments
# The first step in every Python script is to load those packages that we'll use during the analysis. A package is a set of tools that are not included in the built-in Python tools.
#
# There are four packages that are commonly used and we will usually load:
# * __[NumPy](http://www.numpy.org/)__ is a fundamental package for scientific computing that includes N-dimensional array objects, linear algebra, Fourier transforms, random number capabilities... __NumPy__ uses a vector structure called *array*; data in an *array* must be always of the same nature, i.e., integer, floating point number, string... To import __NumPy__, use the following command:
# > ```Python
# import numpy as np
# ```
#
# * __[pandas](https://pandas.pydata.org/)__ is a pacakge that allows organizind data in a structure named *data frame*. *Data frames* resemble the usual Excel table, in the sense that columns represent variables and rows represent samples. All the elements of a column (variable) must be of the same nature (integer, string...), but different columns may differ in the type of data they contain. As Excel talbes, a _data frame_ has an index and heading that identifies rows and columns, respectively, that allow us to search for specific values. To import __pandas__, use the following command:
# > ```Python
# import pandas as pd
# ```
#
# * __[matplotlib](https://matplotlib.org/)__ is a package designed to plot graphs similar to those in Matlab. To import __matplotlib__, you need the following commands:
# > ```Python
# import matplotlib.pyplot as plt
# # %matplotlib inline
# plt.style.use('seaborn-whitegrid')
# ```
#
# * __[SciPy](https://www.scipy.org/)__ contains several numerical tools that are efficient and easty to apply, e.g., numerical integration and optimization. We will not load the complete set of tools in __SciPy__, but those we need:
# > ```Python
# from scipy.stats import genextreme
# from scipy.optimize import curve_fit
# ```
#
# * [__os__](https://docs.python.org/3.4/library/os.html) is a package that allows us to change the working directory, create new directories, list the files contained in a directory, etc. To import it:
# > ```Python
# import os
# ```
# +
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
# %matplotlib inline
plt.style.use('seaborn-whitegrid')
from scipy.stats import genextreme
from scipy.optimize import curve_fit
import os
# -
# In case you need to install some of those packages, you'll need to do the following (example to install SciPy):<br>
# * Launch Anaconda Prompt<br>
# * Type `conda install scipy` + `Enter`<br>
#
# We're going to install a variable inspector to be able to check the existing objects in our analysis:<br>
# * Launch Anaconda Prompt<br>
# * Type:
# > `pip install jupyter_contrib_nbextensions` + `Enter`<br>
# `jupyter contrib nbextension install --user` + `Enter`<br>
# `jupyter nbextension enable varInspector/main` + `Enter`<br>
# ### Basic data structures in Python
# **Lists**<br>
# Lists are a data structure that can contain data of any type (integer, float, strings...) in a single object. Lists are mutable, meaning that we can modify the values inside a list after its declaration.
# create a list
a = [1, 'hello', 1.5]
# extract a value from the list
# modify one of the values in the list
# **Tuples**<br>
# Tuples are a data structure similar to lists because they can also contain data of any type. Contrary to lists, tuples can no be modified after declared.
# create a lista
b = (2, 'red', np.nan)
# extract a value from the tuple
# modify one of the values in the tuple
# **Arrays**<br>
# This is a specific structure of the package *NumPy* that allows us to work with vectores and matrices, and perform calculations upon them easily. All the values in an array must be of the same data type.
# create an array from the list 'a'
# create an array
c = np.array([1.5, 2.1, 4.5])
# extract values from the array
# invert the array
# modify a value in the array
# calculate the mean of the array
# **Pandas: _series_ and _data frames_**<br>
# _Pandas_ is a package suitable for working with bidimensional (_data frames_) or unidimensional (_series_) tables. Pandas' structures use the tools in *NumPy* to perform easily several tasks with the table. In _Pandas_, all the data contained in a column of the table must be of the same type; different columns may have different types of data.
# create a 'data frame' with name, age and weight
d = [['Peter', 36, 71],
['Laura', 40, 58],
['John', 25, 65]]
d = pd.DataFrame(data=d, columns=['name', 'age', 'weight'])
d
# a column in a data frame is a series
# calculate the mean of the dataframe
# **Dictionaries**<br>
# A dictionary can store several data structures (from those above mentioned) in a single object. We need to set a _key_ to access any of the data structures included in the dictionary.
# crear un diccionario que contenga todos los datos anteriormente creados
# siendo la clave el tipo de estructura
# create a dictionary that contains all the data structures previously created
# in this example, the key will be the type of structure
e = {'list': a,
'tuple': b,
'array': c,
'dataframe': d}
# extract one of the structures from the dictionary
| G1448-Hydrology/Precipitation/notebooks/Python_basics(incomplete).ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# #### New to Plotly?
# Plotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).
# <br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).
# <br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!
#
# #### Version Check
# Note: graph size attributes are available in version <b>1.9.2+</b><br>
# Run `pip install plotly --upgrade` to update your Plotly version
import plotly
plotly.__version__
# ### Adjusting Height, Width, & Margins###
# +
import plotly.plotly as py
import plotly.graph_objs as go
data = [
go.Scatter(
x=[0, 1, 2, 3, 4, 5, 6, 7, 8],
y=[0, 1, 2, 3, 4, 5, 6, 7, 8]
)
]
layout = go.Layout(
autosize=False,
width=500,
height=500,
margin=go.layout.Margin(
l=50,
r=50,
b=100,
t=100,
pad=4
),
paper_bgcolor='#7f7f7f',
plot_bgcolor='#c7c7c7'
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='size-margins')
# -
# ### Automatically Adjust Margins
# Set [automargin](https://plot.ly/python/reference/#layout-xaxis-automargin) to `True` and Plotly will automatically increase the margin size to prevent ticklabels from being cut off or overlapping with axis titles.
# +
import plotly.plotly as py
import plotly.graph_objs as go
data = [
go.Bar(
x=['Apples', 'Oranges', 'Watermelon', 'Pears'],
y=[3, 2, 1, 4]
)
]
layout = go.Layout(
autosize=False,
width=500,
height=500,
yaxis=go.layout.YAxis(
title='Y-axis Title',
ticktext=['Very long label','long label','3','label'],
tickvals=[1, 2, 3, 4],
tickmode='array',
automargin=True,
titlefont=dict(size=30),
),
paper_bgcolor='#7f7f7f',
plot_bgcolor='#c7c7c7'
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='automargin')
# -
# ### Dash Example
# [Dash](https://plot.ly/products/dash/) is an Open Source Python library which can help you convert plotly figures into a reactive, web-based application. Below is a simple example of a dashboard created using Dash. Its [source code](https://github.com/plotly/simple-example-chart-apps/tree/master/dash-graphsizeplot) can easily be deployed to a PaaS.
from IPython.display import IFrame
IFrame(src= "https://dash-simple-apps.plotly.host/dash-graphsizeplot/", width="100%", height="650px", frameBorder="0")
from IPython.display import IFrame
IFrame(src= "https://dash-simple-apps.plotly.host/dash-graphsizeplot/code", width="100%", height=500, frameBorder="0")
# #### Reference
# See https://plot.ly/python/reference/#layout for more information and chart attribute options!
# +
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
# ! pip install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'sizing.ipynb', 'python/setting-graph-size/', 'Setting Graph Size',
'How to manipulate the graph size in Python with Plotly.',
title = 'Setting Graph Size',
name = 'Setting Graph Size',
has_thumbnail='true', thumbnail='thumbnail/sizing.png',
language='python', order=2,
display_as='file_settings',
ipynb= '~notebook_demo/133')
| _posts/python-v3/fundamentals/sizing/sizing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <img src='https://wizardsourcer.com/wp-content/uploads/2019/03/Stackoverflow.png' width="400"></img>
#
# ## <center><h1> A Data Analysis using Stackoverflow’s 2019 and 2020 Annual Developer Survey - Colombia.</h1></center>
#
# > ## By <NAME>
#
# ## Each stage of the CRISP-DM process. These stages are:
#
#
# * [1. Business Understanding](#section1)
# * [2. Data Understanding.](#section2)
# * [3. Prepare Data.](#section3)
# * [4. Data Modeling.](#section4)
# * [5. Evaluate the Results.](#section5)
#
#
# <a id="section1"></a>
# ### <font color="#004D7F"> 1. Business Understanding </font>
# In this notebook, I will be exploring the 2019 and 2020 Stack Overflow results in order to glean some information on professional developers and what to focus on to have the best career in Colombia.
#
# There will be three questions I will seek to answer in order to get this information:
#
# 1. What Programming Languages are most used to work and Which Programming Languages are most Required in Stackoverflow survey data of 2019 and 2020 ?
# 2. How does Programming Languages used at work relates with Programming Languages, People Wants to Learn in Colombua According to Stackoverflow survey data of 2019 and 2020?
# 3. What are the most wanted Programming Languages in Colombia According to Stackoverflow survey data of 2019 and 2020?
# <a id="section2"></a>
# ### <font color="#004D7F"> 2. Data understanding </font>
#
# In order to gain some understanding of the data. I have to do these steps:
# * Handle categorical and missing data
# * Analyze, Model, and Visualize
# +
# import python libraries to handle datasets
import numpy as np
import pandas as pd
from collections import Counter
#make plots
import matplotlib.pyplot as plt
import seaborn as sns
#ignore warnings
import warnings
warnings.filterwarnings("ignore")
# to display graphs in jupyter notebook
# %matplotlib inline
# to visualise al the columns in the dataframe
pd.pandas.set_option('display.max_columns', None)
# -
# Loading Stackoverflow survey data of 2019 and 2020
# Ref link: https://insights.stackoverflow.com/survey
df_2019 = pd.read_csv('survey_results_public_2019.csv')
df_2020 = pd.read_csv('survey_results_public_2020.csv')
df_2019.head()
df_2020.head()
# <a id="section3"></a>
# ### <font color="#004D7F"> 3. Prepare Data </font>
#
rows, columns = df_2019.shape[0], df_2019.shape[1]
print(f'Number of rows and columns of 2019 Developer Survey: {rows}, {columns}')
rows, columns = df_2020.shape[0], df_2020.shape[1]
print(f'Number of rows and columns of 2020 Developer Survey: {rows}, {columns}')
#precentage missing values: 2019 Developer Survey
print(df_2019.isnull().sum()/df_2019.shape[0])
#precentage missing values: 2020 Developer Survey
print(df_2020.isnull().sum()/df_2020.shape[0])
#dtype check 2019 Developer Survey
df_2019.dtypes
#dtype check 2020 Developer Survey
df_2020.dtypes
#dist 2019 Developer Survey
df_2019.hist();
#dist 2020 Developer Survey
df_2020.hist();
#decribe 2019 Developer Survey
df_2019.describe()
#decribe 2020 Developer Survey
df_2020.describe()
#Missing Values in Data - Imput
def imput (x):
columns=x.columns.tolist()
del_ = []
for i in range(len(x.isnull().sum())):
if x.isnull().sum()[i]/x.shape[0] < 0.35:
if x[columns[i]].dtypes == object:
x[columns[i]].fillna(x[columns[i]].mode()[0] , inplace = True)
else:
x[columns[i]].fillna(x[columns[i]].median() , inplace = True)
else:
del_.append(columns[i])
x.drop(del_, axis = 1, inplace = True)
#Missing Values in Data - Imput
imput(df_2019)
imput(df_2020)
#precentage missing values: 2019 Developer Survey
print(df_2019.isnull().sum()/df_2019.shape[0])
#precentage missing values: 2020 Developer Survey
print(df_2020.isnull().sum()/df_2020.shape[0])
#
#
# ### What Programming Languages are most used to work and Which Programming Languages are most Required in Stackoverflow survey data of 2019 and 2020 ?
# +
#First make copy of dataframe and then Filter a dataframe by country
def filter_country(df, column_filter, country, column1,column2 ):
'''
Filter a dataframe by country(i.e. why i use dropna to drop all other country excpet Colombia)
Returns filtred dataframe
'''
df_copy = df
df_copy = df_copy[df_copy[column_filter] == country].dropna(subset=[column1, column2])
return df_copy
# Filtering the dataframe
col_2019 = filter_country(df_2019, 'Country', 'Colombia', 'LanguageWorkedWith', 'LanguageDesireNextYear')
col_2020 = filter_country(df_2020, 'Country', 'Colombia', 'LanguageWorkedWith', 'LanguageDesireNextYear')
# -
col2019, col2020 = col_2019.shape[0], col_2020.shape[0]
print(f'Nro 2019 and 2020: {col2019}, {col2020}' )
# +
def split_column(df, column):
'''
Split column by ;,
Returns a splited series.
'''
df_copy = df
columnSeries = df_copy[column].apply(lambda x: x.split(';'))
return columnSeries
# Splitting the dataframe by columns.
worked_languages_2019 = split_column(col_2019, 'LanguageWorkedWith')
wanted_languages_2019 = split_column(col_2019, 'LanguageDesireNextYear')
worked_languages_2020= split_column(col_2020, 'LanguageWorkedWith')
wanted_languages_2020 = split_column(col_2020, 'LanguageDesireNextYear')
# +
#Just Flating a nested list
def flat(array_list):
'''
Flat a nested list,
Returns a flat list.
'''
object_list = []
for row in array_list:
for obj in row:
object_list.append(obj.strip())
return object_list
# Flatting nested list objects.
list_worked_languages_2019 = flat(worked_languages_2019)
list_wanted_languages_2019 = flat(wanted_languages_2019)
list_worked_languages_2020 = flat(worked_languages_2020)
list_wanted_languages_2020 = flat(wanted_languages_2020)
# +
def list_of_group(data_list, year):
'''
Group by count to a list,
Returns a result dict
'''
grouped_list = dict(Counter(data_list))
grouped_dict = [{'Programming Language':key, 'Count': value, 'Year': year} for key, value in grouped_list.items()]
return grouped_dict
# Grouping the list and creating a dict.
dict_worked_languages_2019 = list_of_group(list_worked_languages_2019, '2019')
dict_wanted_languages_2019 = list_of_group(list_wanted_languages_2019, '2019')
dict_worked_languages_2020 = list_of_group(list_worked_languages_2020, '2020')
dict_wanted_languages_2020 = list_of_group(list_wanted_languages_2020, '2020')
# -
# <a id="section4"></a>
# ### <font color="#004D7F"> 4. Data Modeling </font>
#
# +
#Ref: https://stackoverflow.com/questions/23668427/pandas-three-way-joining-multiple-dataframes-on-columns
def create_dataframe(data_dicts):
'''
Create two dataframes and append them,
Returns a appended dataframe.
'''
df1 = pd.DataFrame(data_dicts[0])
df2 = pd.DataFrame(data_dicts[1])
df = df1.append(df2)
return df
worked_languages = create_dataframe([dict_worked_languages_2019, dict_worked_languages_2020])
wanted_languages = create_dataframe([dict_wanted_languages_2019, dict_wanted_languages_2020])
# +
#Adding Percentage to worked_languages and wanted_languages dataframe
def percentage(df, column):
'''
Scale data,
Returns data scaled.
'''
df_copy = df
series = []
for val in df_copy[column].unique():
series.append(df_copy[df_copy[column] == val]['Count'] /
df_copy[df_copy[column] == val]['Count'].sum())
joined = pd.Series()
for i_series in series:
joined = joined.append(i_series)
return joined
worked_languages['Percentage'] = percentage(worked_languages, 'Year')
wanted_languages['Percentage'] = percentage(wanted_languages, 'Year')
# +
# Get the top 12 languages
top_12_work = worked_languages.sort_values(by=['Percentage'], ascending=False).head(12)['Programming Language'].unique()
top_12_want = wanted_languages.sort_values(by=['Percentage'], ascending=False).head(12)['Programming Language'].unique()
worked_chart = worked_languages[worked_languages['Programming Language'].isin(top_12_work)]
wanted_chart = wanted_languages[wanted_languages['Programming Language'].isin(top_12_want)]
# +
plt.figure(figsize=(20,10))
sns.barplot(x = 'Programming Language',
y = 'Percentage',
hue = 'Year',
data = worked_chart.sort_values(by='Percentage', ascending=False))
plt.xlabel("Programming Languages", fontsize = 14)
plt.ylabel("Percentage", fontsize = 14)
plt.legend(title_fontsize='40')
plt.title('Most common Programming Languages used in Colombia', size = 16)
plt.show()
# -
# ## Evaluate the Results
#
# * The Highest Rate of percentage among all Programming Languages JavaScript have highest growth rate in 2019 which is around 18% whereas this percentage is drop significantly in 2020 about 2% in Colombia.
#
# ### How does Programming Languages used at work relates with Programming Languages, People Wants to Learn in Colombia According to Stackoverflow survey data of 2019 and 2020?
# ### Prepare Data
# +
row = []
for j,k in list(zip(worked_languages_2019, wanted_languages_2019)):
for i in j:
row.append({
'Worked_Programming_Languages': i,
'Wanted_Programming_Languages': Counter(k)
})
programming_language_transition = pd.DataFrame(row).groupby('Worked_Programming_Languages')\
.agg({'Wanted_Programming_Languages': 'sum'}).reset_index()
# -
# ### Data Modeling
# +
for row in programming_language_transition['Worked_Programming_Languages']:
programming_language_transition[row] = 0
for index, row in programming_language_transition.iterrows():
try:
total = sum([value for key, value in dict(row['Wanted_Programming_Languages']).items()])
for key, value in dict(row['Wanted_Programming_Languages']).items():
programming_language_transition[key].loc[index] = (value / total)
except:
continue
# +
prlt = programming_language_transition.drop('Wanted_Programming_Languages', axis=1)\
.set_index('Worked_Programming_Languages')
plt.figure(figsize=(20, 10))
sns.heatmap(prlt, cmap = "Reds")
plt.title('Programming Languages Heatmap', size = 20)
plt.ylabel('')
plt.show()
# -
# ### Evaluate the Results
#
# * With this Graph we can have some insights and they are:
# * `JavaScript` is highly correlated with every Programming Language (except: Elixir, Erlang and Scala) and `HTML/CSS` also have same trends.
# * There are around 27 Programming Language which has nearly no correlation among them with anyone.
# * `JavaScript` has strongest correlation with `Clojure` (i.e. about `25% to 30%`).
#
# ### What are the most wanted Programming Languages in Colombia According to Stackoverflow survey data of 2019 and 2020?
# +
plt.figure(figsize=(16,10))
sns.barplot(x = 'Programming Language',
y = 'Percentage',
hue = 'Year',
data = wanted_chart.sort_values(by='Percentage', ascending=False))
plt.title('Most wanted Programming Languages used in Colombia', size = 16)
plt.xlabel("Programming Languages", fontsize = 12)
plt.ylabel("Percentage", fontsize = 12)
plt.legend(title_fontsize='40')
plt.show()
# -
# ### Evaluate the Results
#
# * Most of the programming languages that have appeared as programming languages most used at work, also appeared in the ranking of most wanted programming languages, this show us that are many people wanting to learn these languages.
| Project - Stack Overflow Developer Survey.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# + [markdown] slideshow={"slide_type": "slide"}
# # Lecture 15, Further topics and current research in optimization
# + [markdown] slideshow={"slide_type": "slide"}
# ## Black box surrogate-based global optimization
# + [markdown] slideshow={"slide_type": "subslide"}
# In the current course, all the models have been based on algebraic equations.
#
# However, in many cases, you do not have algebraic equations describing the problem, but instead you have a software or a piece of code that can calculate the values for you.
# + [markdown] slideshow={"slide_type": "fragment"}
# In many cases like this, you need to treat the model as a *"black box"*, which means that you only know what goes in and what comes out.
# + [markdown] slideshow={"slide_type": "fragment"}
# Your method is going to have to be intelligent in how to figure out which solutions to evaluate and which not.
# + [markdown] slideshow={"slide_type": "subslide"}
# In addition, these models may be highly nonconvex and, thus, you are going to have to use *global optimization methods*.
#
# The methods described in this course are so-called local optimization methods. Local optimization methdos are highly efficient in finding a local minimum of a problem, but they cannot guarantee global optimum.
# + [markdown] slideshow={"slide_type": "fragment"}
# Global optimization methods need to have some strategy for searching as much as possible of the search space.
# + [markdown] slideshow={"slide_type": "fragment"}
# In global optimization, there is the so-called **exploration vs. exploitation** ratio. Exploitation means that the method is basically acting as a local optimization method to find the nearest local optimum and exploration means that the method uses some strategy to try to find other local optima.
# + [markdown] slideshow={"slide_type": "fragment"}
# So-called soft-computing methdos are very popular, although others also exist.
# + [markdown] slideshow={"slide_type": "subslide"}
# Finally, these black box models are often *computationally expensive*, which means that you need to use a so-called surrogate to save function calls to the black box model.
# + [markdown] slideshow={"slide_type": "fragment"}
# In practice, this means that there is a clever way of
# 1. deciding whether to evaluate a solution with the black box model or the surrogate model, and
# 2. when to update the surrogate with solutions calculated using the black-box.
# + [markdown] slideshow={"slide_type": "fragment"}
# Usual surrogates are neural networks, radial basis functions and Kriging models.
# -
# E.g., a recent survey by a PhD student of mine: http://link.springer.com/article/10.1007/s00158-015-1226-z#/page-1
# + [markdown] slideshow={"slide_type": "slide"}
# ## Connecting "Big Data" and optimization
# ### Also called prescriptive analytics
# + [markdown] slideshow={"slide_type": "subslide"}
# Sometimes, the model of the problem is not based on an algebraic model, nor a computer program, but instead you have (e.g., measured) data about the phenomena concerning the problem.
# + [markdown] slideshow={"slide_type": "fragment"}
# **This raises completely new kind of problems.**
# + [markdown] slideshow={"slide_type": "fragment"}
# Dealing with "Big Data", you have to deal with the four v:s:
# * volume:
# * the data is actually big and you need to have specific tools for accessing it
# * in addition, one needs to figure out what is the relevant data
# * variety:
# * the data is in completely different formats and you may have to deal with all of them (e.g., video, spread sheets, natural language),
# * velocity:
# * the data is constantly changing and more data is being gathered,
# * veracity:
# * the data is bad and untrusworthy,
# * there is a lot of missing data.
# + [markdown] slideshow={"slide_type": "fragment"}
# Also, in this case, one often needs machine learning techiques to first make sense of the data and then to optimize based on that information gathered.
# + [markdown] slideshow={"slide_type": "fragment"}
# **In TIES583 the students can make their own project that deals with data and optimization**
#
# The course will be starting right after this course!
#
# Please register at https://korppi.jyu.fi/kotka/course/student/generalCourseInfo.jsp?course=192670.
# + [markdown] slideshow={"slide_type": "fragment"}
# E.g., a recent paper at http://ieeexplore.ieee.org/xpls/abs_all.jsp?arnumber=6879615&tag=1
# + [markdown] slideshow={"slide_type": "slide"}
# ## Multiobjective optimization and decision support systems
# + [markdown] slideshow={"slide_type": "subslide"}
# ** The whole point of optimization is to support decision making! **
# + [markdown] slideshow={"slide_type": "fragment"}
# However,
# * most decision problems have multiple conflicting objectives, and
# * human beings are not rational decision makers.
# + [markdown] slideshow={"slide_type": "fragment"}
# First item needs methods to deal with multiple objectives.
#
# There are still a lot of unresolved questions in how the decision makers interact with optimization and, also, in just how to compute Pareto optimal solutions for complicated problems.
# + [markdown] slideshow={"slide_type": "fragment"}
# Second item needs a completely separate type of research.
#
# In fact, it has been shown that most of the decision making that humans do, is dictated by feelings.
#
# Thus, one needs to take into account human beings as complete beings.
#
# ** This is studied in behavioural operations research**
# + [markdown] slideshow={"slide_type": "fragment"}
# Multiobjective optimization e.g., in a recent paper by <NAME> and others http://dx.doi.org/10.1007/s11573-015-0786-0
#
# Behavioral aspects have been studied e.g., in a recent paper http://www.sciencedirect.com/science/article/pii/S0167487015001427
# + [markdown] slideshow={"slide_type": "slide"}
# ## Dealing with risk
# + [markdown] slideshow={"slide_type": "subslide"}
# ** Almost all real-life decisions include risk!**
# + [markdown] slideshow={"slide_type": "fragment"}
# How to deal with this risk, is a active research topic in optimization.
#
# Basically, there are two competing underlying approaches:
# 1. scenario-based approaches, where the possible states involving the decision problem are modelled as different scenarios and
# 2. probabilistic (and similar like fuzzy) approaches, where the possible states are modelled using a distribution (or similar).
# + [markdown] slideshow={"slide_type": "fragment"}
# There are also different risk measures that can be taken into account.
# + [markdown] slideshow={"slide_type": "fragment"}
# For example, there is one paper by <NAME> et al (incl. the lecturer) http://www.nrcresearchpress.com/doi/pdf/10.1139/cjfr-2014-0443, where the uncertainty is modelled using scenarios, but the twist is that there is a possibility of measuring the states, which removes a part or all of the uncertainty.
| Lecture 15, Further topics and current research topics in optimization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Ch `12`: Concept `01`
# ### Ranking by neural network
# +
import tensorflow as tf
import numpy as np
import random
# %matplotlib inline
import matplotlib.pyplot as plt
# +
n_features = 2
def get_data():
data_a = np.random.rand(10, n_features) + 1
data_b = np.random.rand(10, n_features)
plt.scatter(data_a[:, 0], data_a[:, 1], c='r', marker='x')
plt.scatter(data_b[:, 0], data_b[:, 1], c='g', marker='o')
plt.show()
return data_a, data_b
def get_data2():
data_a = np.asarray([[0.1, 0.9], [0.1, 0.8]])
data_b = np.asarray([[0.4,0.05], [0.45, 0.1]])
plt.scatter(data_a[:, 0], data_a[:, 1], c='r', marker='x')
plt.scatter(data_b[:, 0], data_b[:, 1], c='g', marker='o')
plt.xlim([0, 0.5])
plt.ylim([0, 1])
plt.axes().set_aspect('equal')
plt.show()
return data_a, data_b
data_a, data_b = get_data()
# -
n_hidden = 10
# +
with tf.name_scope("input"):
x1 = tf.placeholder(tf.float32, [None, n_features], name="x1")
x2 = tf.placeholder(tf.float32, [None, n_features], name="x2")
dropout_keep_prob = tf.placeholder(tf.float32, name='dropout_prob')
with tf.name_scope("hidden_layer"):
with tf.name_scope("weights"):
w1 = tf.Variable(tf.random_normal([n_features, n_hidden]), name="w1")
tf.summary.histogram("w1", w1)
b1 = tf.Variable(tf.random_normal([n_hidden]), name="b1")
tf.summary.histogram("b1", b1)
with tf.name_scope("output"):
h1 = tf.nn.dropout(tf.nn.relu(tf.matmul(x1,w1) + b1), keep_prob=dropout_keep_prob)
tf.summary.histogram("h1", h1)
h2 = tf.nn.dropout(tf.nn.relu(tf.matmul(x2, w1) + b1), keep_prob=dropout_keep_prob)
tf.summary.histogram("h2", h2)
with tf.name_scope("output_layer"):
with tf.name_scope("weights"):
w2 = tf.Variable(tf.random_normal([n_hidden, 1]), name="w2")
tf.summary.histogram("w2", w2)
b2 = tf.Variable(tf.random_normal([1]), name="b2")
tf.summary.histogram("b2", b2)
with tf.name_scope("output"):
s1 = tf.matmul(h1, w2) + b2
s2 = tf.matmul(h2, w2) + b2
# +
with tf.name_scope("loss"):
s12 = s1 - s2
s12_flat = tf.reshape(s12, [-1])
pred = tf.sigmoid(s12)
lable_p = tf.sigmoid(-tf.ones_like(s12))
cross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=tf.zeros_like(s12_flat), logits=s12_flat + 1)
loss = tf.reduce_mean(cross_entropy)
tf.summary.scalar("loss", loss)
with tf.name_scope("train_op"):
train_op = tf.train.AdamOptimizer(0.001).minimize(loss)
# -
sess = tf.InteractiveSession()
summary_op = tf.summary.merge_all()
writer = tf.summary.FileWriter("tb_files", sess.graph)
init = tf.global_variables_initializer()
sess.run(init)
for epoch in range(0, 10000):
loss_val, _ = sess.run([loss, train_op], feed_dict={x1:data_a, x2:data_b, dropout_keep_prob:0.5})
if epoch % 100 == 0 :
summary_result = sess.run(summary_op, feed_dict={x1:data_a, x2:data_b, dropout_keep_prob:1})
writer.add_summary(summary_result, epoch)
# print("Epoch {}: Loss {}".format(epoch, loss_val))
grid_size = 10
data_test = []
for y in np.linspace(0., 1., num=grid_size):
for x in np.linspace(0., 1., num=grid_size):
data_test.append([x, y])
# +
def visualize_results(data_test):
plt.figure()
scores_test = sess.run(s1, feed_dict={x1:data_test, dropout_keep_prob:1})
scores_img = np.reshape(scores_test, [grid_size, grid_size])
plt.imshow(scores_img, origin='lower')
plt.colorbar()
# -
visualize_results(data_test)
| ch12_rank/Concept01_ranknet.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Detecting and mitigating racial bias in income estimation
#
# The goal of this tutorial is to introduce the basic functionality of AI Fairness 360 to an interested developer who may not have a background in bias detection and mitigation.
#
# *Note: This demo is very similar to the [Credit Scoring Tutorial](tutorial_credit_scoring.ipynb). It is meant as an alternative introduction using a different dataset and mitigation algorithm.*
#
# ### Biases and Machine Learning
# A machine learning model makes predictions of an outcome for a particular instance. (Given an instance of a loan application, predict if the applicant will repay the loan.) The model makes these predictions based on a training dataset, where many other instances (other loan applications) and actual outcomes (whether they repaid) are provided. Thus, a machine learning algorithm will attempt to find patterns, or generalizations, in the training dataset to use when a prediction for a new instance is needed. (For example, one pattern it might discover is "if a person has salary > USD 40K and has outstanding debt < USD 5, they will repay the loan".) In many domains this technique, called supervised machine learning, has worked very well.
#
# However, sometimes the patterns that are found may not be desirable or may even be illegal. For example, a loan repay model may determine that age plays a significant role in the prediction of repayment because the training dataset happened to have better repayment for one age group than for another. This raises two problems: 1) the training dataset may not be representative of the true population of people of all age groups, and 2) even if it is representative, it is illegal to base any decision on a applicant's age, regardless of whether this is a good prediction based on historical data.
#
# AI Fairness 360 is designed to help address this problem with _fairness metrics_ and _bias mitigators_. Fairness metrics can be used to check for bias in machine learning workflows. Bias mitigators can be used to overcome bias in the workflow to produce a more fair outcome.
#
# The loan scenario describes an intuitive example of illegal bias. However, not all undesirable bias in machine learning is illegal it may also exist in more subtle ways. For example, a loan company may want a diverse portfolio of customers across all income levels, and thus, will deem it undesirable if they are making more loans to high income levels over low income levels. Although this is not illegal or unethical, it is undesirable for the company's strategy.
#
# As these two examples illustrate, a bias detection and/or mitigation toolkit needs to be tailored to the particular bias of interest. More specifically, it needs to know the attribute or attributes, called _protected attributes_, that are of interest: race is one example of a _protected attribute_ and age is a second.
#
# ### The Machine Learning Workflow
# To understand how bias can enter a machine learning model, we first review the basics of how a model is created in a supervised machine learning process.
#
#
#
# 
#
#
#
#
#
#
#
#
# First, the process starts with a _training dataset_, which contains a sequence of instances, where each instance has two components: the features and the correct prediction for those features. Next, a machine learning algorithm is trained on this training dataset to produce a machine learning model. This generated model can be used to make a prediction when given a new instance. A second dataset with features and correct predictions, called a _test dataset_, is used to assess the accuracy of the model.
# Since this test dataset is the same format as the training dataset, a set of instances of features and prediction pairs, often these two datasets derive from the same initial dataset. A random partitioning algorithm is used to split the initial dataset into training and test datasets.
#
# Bias can enter the system in any of the three steps above. The training data set may be biased in that its outcomes may be biased towards particular kinds of instances. The algorithm that creates the model may be biased in that it may generate models that are weighted towards particular features in the input. The test data set may be biased in that it has expectations on correct answers that may be biased. These three points in the machine learning process represent points for testing and mitigating bias. In AI Fairness 360 codebase, we call these points _pre-processing_, _in-processing_, and _post-processing_.
#
# ### AI Fairness 360
# We are now ready to utilize AI Fairness 360 (`aif360`) to detect and mitigate bias. We will use the Adult Census Income dataset, splitting it into a training and test dataset. We will look for bias in the creation of a machine learning model to predict if an individual's annual income exceeds $50,000 based on various personal attributes. The protected attribute will be "race", with "1" (white) and "0" (not white) being the values for the privileged and unprivileged groups, respectively.
# For this first tutorial, we will check for bias in the initial training data, mitigate the bias, and recheck. More sophisticated machine learning workflows are given in the author tutorials and demo notebooks in the codebase.
#
# Here are the steps involved
# #### Step 1: Write import statements
# #### Step 2: Set bias detection options, load dataset, and split between train and test
# #### Step 3: Compute fairness metric on original training dataset
# #### Step 4: Mitigate bias by transforming the original dataset
# #### Step 5: Compute fairness metric on transformed training dataset
#
# ### Step 1 Import Statements
# As with any Python program, the first step will be to import the necessary packages. Below we import several components from the aif360 package. We import a custom version of the AdultDataset with certain features binned, metrics to check for bias, and classes related to the algorithm we will use to mitigate bias. We also import some other non-aif360 useful packages.
# +
import sys
sys.path.append("../")
import numpy as np
from aif360.metrics import BinaryLabelDatasetMetric
from aif360.algorithms.preprocessing.optim_preproc import OptimPreproc
from aif360.algorithms.preprocessing.optim_preproc_helpers.data_preproc_functions\
import load_preproc_data_adult
from aif360.algorithms.preprocessing.optim_preproc_helpers.distortion_functions\
import get_distortion_adult
from aif360.algorithms.preprocessing.optim_preproc_helpers.opt_tools import OptTools
from IPython.display import Markdown, display
# -
np.random.seed(1)
# ### Step 2 Load dataset, specifying protected attribute, and split dataset into train and test
# In Step 2 we load the initial dataset, setting the protected attribute to be race. We then splits the original dataset into training and testing datasets. Although we will use only the training dataset in this tutorial, a normal workflow would also use a test dataset for assessing the efficacy (accuracy, fairness, etc.) during the development of a machine learning model. Finally, we set two variables (to be used in Step 3) for the privileged (1) and unprivileged (0) values for the race attribute. These are key inputs for detecting and mitigating bias, which will be Step 3 and Step 4.
# +
dataset_orig = load_preproc_data_adult(['race'])
dataset_orig_train, dataset_orig_test = dataset_orig.split([0.7], shuffle=True)
privileged_groups = [{'race': 1}] # White
unprivileged_groups = [{'race': 0}] # Not white
# -
# ### Step 3 Compute fairness metric on original training dataset
# Now that we've identified the protected attribute 'race' and defined privileged and unprivileged values, we can use aif360 to detect bias in the dataset. One simple test is to compare the percentage of favorable results for the privileged and unprivileged groups, subtracting the former percentage from the latter. A negative value indicates less favorable outcomes for the unprivileged groups. This is implemented in the method called mean_difference on the BinaryLabelDatasetMetric class. The code below performs this check and displays the output:
metric_orig_train = BinaryLabelDatasetMetric(dataset_orig_train,
unprivileged_groups=unprivileged_groups,
privileged_groups=privileged_groups)
display(Markdown("#### Original training dataset"))
print("Difference in mean outcomes between unprivileged and privileged groups = %f" % metric_orig_train.mean_difference())
# ### Step 4 Mitigate bias by transforming the original dataset
# The previous step showed that the privileged group was getting 10.5% more positive outcomes in the training dataset. Since this is not desirable, we are going to try to mitigate this bias in the training dataset. As stated above, this is called _pre-processing_ mitigation because it happens before the creation of the model.
#
# AI Fairness 360 implements several pre-processing mitigation algorithms. We will choose the Optimized Preprocess algorithm [1], which is implemented in "OptimPreproc" class in the "aif360.algorithms.preprocessing" directory. This algorithm will transform the dataset to have more equity in positive outcomes on the protected attribute for the privileged and unprivileged groups.
#
# The algorithm requires some tuning parameters, which are set in the optim_options variable and passed as an argument along with some other parameters, including the 2 variables containg the unprivileged and privileged groups defined in Step 3.
#
# We then call the fit and transform methods to perform the transformation, producing a newly transformed training dataset (dataset_transf_train). Finally, we ensure alignment of features between the transformed and the original dataset to enable comparisons.
#
# [1] Optimized Pre-Processing for Discrimination Prevention, NIPS 2017, <NAME>, <NAME>, <NAME>, <NAME>, and <NAME>
# +
optim_options = {
"distortion_fun": get_distortion_adult,
"epsilon": 0.05,
"clist": [0.99, 1.99, 2.99],
"dlist": [.1, 0.05, 0]
}
OP = OptimPreproc(OptTools, optim_options)
OP = OP.fit(dataset_orig_train)
dataset_transf_train = OP.transform(dataset_orig_train, transform_Y=True)
dataset_transf_train = dataset_orig_train.align_datasets(dataset_transf_train)
# -
# ### Step 5 Compute fairness metric on transformed dataset
# Now that we have a transformed dataset, we can check how effective it was in removing bias by using the same metric we used for the original training dataset in Step 3. Once again, we use the function mean_difference in the BinaryLabelDatasetMetric class:
metric_transf_train = BinaryLabelDatasetMetric(dataset_transf_train,
unprivileged_groups=unprivileged_groups,
privileged_groups=privileged_groups)
display(Markdown("#### Transformed training dataset"))
print("Difference in mean outcomes between unprivileged and privileged groups = %f" % metric_transf_train.mean_difference())
# We see the mitigation step was very effective, the difference in mean outcomes is now -0.051074. So we went from a 10.5% advantage for the privileged group to a 5.1% advantage for the privileged group — a reduction in more than half!
# ### Summary
# The purpose of this tutorial is to give a new user to bias detection and mitigation a gentle introduction to some of the functionality of AI Fairness 360. A more complete use case would take the next step and see how the transformed dataset impacts the accuracy and fairness of a trained model. This is implemented in the demo notebook in the examples directory of toolkit, called demo_optim_data_preproc.ipynb. I highly encourage readers to view that notebook as it is generalization and extension of this simple tutorial.
#
# There are many metrics one can use to detect the pressence of bias. AI Fairness 360 provides many of them for your use. Since it is not clear which of these metrics to use, we also provide some guidance. Likewise, there are many different bias mitigation algorithms one can employ, many of which are in AI Fairness 360. Other tutorials will demonstrate the use of some of these metrics and mitigations algorithms.
#
# As mentioned earlier, both fairness metrics and mitigation algorithms can be performed at various stages of the machine learning pipeline. We recommend checking for bias as often as possible, using as many metrics are relevant for the application domain. We also recommend incorporating bias detection in an automated continous integration pipeline to ensure bias awareness as a software project evolves.
| models/AIF360/examples/demo_optim_preproc_adult.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <p><font size="6"><b>01 - Pandas: Data Structures </b></font></p>
#
#
# > *© 2016-2018, <NAME> and <NAME> (<mailto:<EMAIL>>, <mailto:<EMAIL>>). Licensed under [CC BY 4.0 Creative Commons](http://creativecommons.org/licenses/by/4.0/)*
#
# ---
# + run_control={"frozen": false, "read_only": false}
import pandas as pd
# + run_control={"frozen": false, "read_only": false}
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# -
# # The pandas data structures: `DataFrame` and `Series`
#
# Pandas provides two fundamental data objects, for 1D (``Series``) and 2D data (``DataFrame``).
# ## One-dimensional data: `Series`
#
# A Series is a basic holder for **one-dimensional labeled data**. It can be created much as a NumPy array is created:
# + run_control={"frozen": false, "read_only": false}
s = pd.Series([0.1, 0.2, 0.3, 0.4])
s
# -
# ### Attributes of a Series: `index` and `values`
#
# The series also has an **index**, which by default is the numbers *0* through *N - 1*:
# + run_control={"frozen": false, "read_only": false}
s.index
# -
# You can access the underlying numpy array representation with the `.values` attribute:
# + run_control={"frozen": false, "read_only": false}
s.values
# -
# We can access series values via the index, just like for NumPy arrays:
# + run_control={"frozen": false, "read_only": false}
s[0]
# -
# Unlike the NumPy array, though, this index can be something other than integers:
# + run_control={"frozen": false, "read_only": false}
s2 = pd.Series(np.arange(4), index=['a', 'b', 'c', 'd'])
s2
# + run_control={"frozen": false, "read_only": false}
s2['c']
# -
# ### Pandas Series versus dictionaries
# In this way, a ``Series`` object can be thought of as similar to an ordered dictionary mapping one typed value to another typed value.
#
# In fact, it's possible to construct a series directly from a Python dictionary:
# + run_control={"frozen": false, "read_only": false}
pop_dict = {'Germany': 81.3,
'Belgium': 11.3,
'France': 64.3,
'United Kingdom': 64.9,
'Netherlands': 16.9}
population = pd.Series(pop_dict)
population
# -
# We can index the populations like a dict as expected ...
# + run_control={"frozen": false, "read_only": false}
population['France']
# -
# ... but with the power of numpy arrays. Many things you can do with numpy arrays, can also be applied on DataFrames / Series.
#
# Eg element-wise operations:
# + run_control={"frozen": false, "read_only": false}
population * 1000
# -
# ## Two-dimensional data: `DataFrame`
# A `DataFrame` is a **tabular data structure** (2D object to hold labelled data) comprised of rows and columns, akin to a spreadsheet, database table, or R's data.frame object. You can think of it as multiple Series objects which share the same index.
#
# <img align="left" width=50% src="../img/schema-dataframe.svg">
# For the examples here, we are going to create a small DataFrame with some data about a few countries.
#
# When creating a DataFrame manually, a common way to do this is from dictionary of arrays or lists:
# + run_control={"frozen": false, "read_only": false}
data = {'country': ['Belgium', 'France', 'Germany', 'Netherlands', 'United Kingdom'],
'population': [11.3, 64.3, 81.3, 16.9, 64.9],
'area': [30510, 671308, 357050, 41526, 244820],
'capital': ['Brussels', 'Paris', 'Berlin', 'Amsterdam', 'London']}
countries = pd.DataFrame(data)
countries
# -
# In practice, you will of course often import your data from an external source (text file, excel, database, ..), which we will see later.
#
# Note that in the IPython notebook, the dataframe will display in a rich HTML view.
# You access a Series representing a column in the data, using typical `[]` indexing syntax and the column name:
# + run_control={"frozen": false, "read_only": false}
countries['area']
# -
# ### Attributes of the DataFrame
#
# The DataFrame has a built-in concept of named rows and columns, the **`index`** and **`columns`** attributes:
# + run_control={"frozen": false, "read_only": false}
countries.index
# -
# By default, the index is the numbers *0* through *N - 1*
# + run_control={"frozen": false, "read_only": false}
countries.columns
# -
# To check the data types of the different columns:
# + run_control={"frozen": false, "read_only": false}
countries.dtypes
# -
# An overview of that information can be given with the `info()` method:
# + run_control={"frozen": false, "read_only": false}
countries.info()
# -
# A DataFrame has also a `values` attribute, but attention: when you have heterogeneous data, all values will be upcasted:
# + run_control={"frozen": false, "read_only": false}
countries.values
# -
# <div class="alert alert-info">
#
# **NumPy** provides
#
# <ul>
# <li>multi-dimensional, homogeneously typed arrays (single data type!)</li>
# </ul>
# <br>
#
# **Pandas** provides
#
# <ul>
# <li>2D, heterogeneous data structure (multiple data types!)</li>
# <li>labeled (named) row and column index</li>
# </ul>
#
#
# </div>
# ## Some useful methods on these data structures
# Exploration of the Series and DataFrame is essential (check out what you're dealing with).
# + run_control={"frozen": false, "read_only": false}
countries.head() # Top rows
# + run_control={"frozen": false, "read_only": false}
countries.tail() # Bottom rows
# -
# The ``describe`` method computes summary statistics for each numerical column:
# + run_control={"frozen": false, "read_only": false}
countries.describe()
# -
# **Sort**ing your data **by** a specific column is another important first-check:
# + run_control={"frozen": false, "read_only": false}
countries.sort_values(by='population')
# -
# The **`plot`** method can be used to quickly visualize the data in different ways:
# + run_control={"frozen": false, "read_only": false}
countries.plot()
# -
# However, for this dataset, it does not say that much:
# + run_control={"frozen": false, "read_only": false}
countries['population'].plot(kind='barh')
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>You can play with the `kind` keyword of the `plot` function in the figure above: 'line', 'bar', 'hist', 'density', 'area', 'pie', 'scatter', 'hexbin', 'box'</li>
# </ul>
# </div>
# # Importing and exporting data
# A wide range of input/output formats are natively supported by pandas:
#
# * CSV, text
# * SQL database
# * Excel
# * HDF5
# * json
# * html
# * pickle
# * sas, stata
# * Parquet
# * ...
# + run_control={"frozen": false, "read_only": false}
# pd.read_
# + run_control={"frozen": false, "read_only": false}
# countries.to_
# -
# <div class="alert alert-info">
#
#
# **Note: I/O interface**
#
#
# <ul>
# <li>All readers are `pd.read_...`</li>
# <li>All writers are `DataFrame.to_...` </li>
# </ul>
#
#
# </div>
# # Application on a real dataset
# Throughout the pandas notebooks, many of exercises will use the titanic dataset. This dataset has records of all the passengers of the Titanic, with characteristics of the passengers (age, class, etc. See below), and an indication whether they survived the disaster.
#
#
# The available metadata of the titanic data set provides the following information:
#
# VARIABLE | DESCRIPTION
# ------ | --------
# survival | Survival (0 = No; 1 = Yes)
# pclass | Passenger Class (1 = 1st; 2 = 2nd; 3 = 3rd)
# name | Name
# sex | Sex
# age | Age
# sibsp | Number of Siblings/Spouses Aboard
# parch | Number of Parents/Children Aboard
# ticket | Ticket Number
# fare | Passenger Fare
# cabin | Cabin
# embarked | Port of Embarkation (C = Cherbourg; Q = Queenstown; S = Southampton)
#
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>Read the CVS file (available at `../data/titanic.csv`) into a pandas DataFrame. Call the result `df`.</li>
# </ul>
# </div>
# + clear_cell=true run_control={"frozen": false, "read_only": false}
# # %load _solutions/pandas_01_data_structures1.py
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>Quick exploration: show the first 5 rows of the DataFrame.</li>
# </ul>
# </div>
# + clear_cell=true run_control={"frozen": false, "read_only": false}
# # %load _solutions/pandas_01_data_structures2.py
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>How many records (i.e. rows) has the titanic dataset?</li>
# </ul>
# </div>
# + clear_cell=true run_control={"frozen": false, "read_only": false}
# # %load _solutions/pandas_01_data_structures3.py
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>Select the 'Age' column (remember: we can use the [] indexing notation and the column label).</li>
# </ul>
# </div>
# + clear_cell=true
# # %load _solutions/pandas_01_data_structures4.py
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>Make a box plot of the Fare column.</li>
# </ul>
# </div>
# + clear_cell=true
# # %load _solutions/pandas_01_data_structures5.py
# -
# <div class="alert alert-success">
# <b>EXERCISE</b>:
#
# <ul>
# <li>Sort the rows of the DataFrame by 'Age' column, with the oldest passenger at the top. Check the help of the `sort_values` function and find out how to sort from the largest values to the lowest values</li>
# </ul>
# </div>
# + clear_cell=true
# # %load _solutions/pandas_01_data_structures6.py
# -
# ---
# # Acknowledgement
#
#
# > This notebook is partly based on material of <NAME> (https://github.com/jakevdp/OsloWorkshop2014).
#
| Day_1_Scientific_Python/pandas/pandas_01_data_structures.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Income prediction
#
# Recall that we have done a homework of data exploration on 'income.csv' to master the knowledge of Exploratory Data Analysis. In this homework, you are required to predict whether a person's income is high or low according to his relevant information including his age, education, occupation, race and so on.
#
#
# The attribute information is:
#
# - **income**: the label of this dataset, belongs to \[high, low\]
# - **age**: the age of a person, a continuous variable.
# - **work_class**: work class, belongs to \[Private, Self-emp-not-inc, Self-emp-inc, Federal-gov, Local-gov, State-gov, Without-pay, Never-worked\].
# - **education**: belongs to \[Bachelors, Some-college, 11th, HS-grad, Prof-school, Assoc-acdm, Assoc-voc, 9th, 7th-8th, 12th, - Masters, 1st-4th, 10th, Doctorate, 5th-6th, Preschool\].
# - **education_degree**: the education level of a person, an ordinal number variable.
# - **marital_status**: marital status, belongs to \[Married-civ-spouse, Divorced, Never-married, Separated, Widowed, Married-spouse-absent, Married-AF-spouse\].
# - **job**: occupation, belongs to \[Tech-support, Craft-repair, Other-service, Sales, Exec-managerial, Prof-specialty, Handlers-cleaners, Machine-op-inspct, Adm-clerical, Farming-fishing, Transport-moving, Priv-house-serv, Protective-serv, Armed-Forces\].
# - **relationship**: belongs to \[Wife, Own-child, Husband, Not-in-family, Other-relative, Unmarried\].
# - **race**: belongs to \[White, Asian-Pac-Islander, Amer-Indian-Eskimo, Other, Black\].
# - **sex**: belongs to \[Female, Male\].
# - **capital_gain**: capital gain, a continuous variable.
# - **capital_loss**: capital loss, a continuous variable.
# - **hours_per_week**: how long a person works every week, a continuous variable.
# - **birthplace**: belongs to \[United-States, Cambodia, England, Puerto-Rico, Canada, Germany, Outlying-US(Guam-USVI-etc), India, - Japan, Greece, South, China, Cuba, Iran, Honduras, Philippines, Italy, Poland, Jamaica, Vietnam, Mexico, Portugal, Ireland, France, Dominican-Republic, Laos, Ecuador, Taiwan, Haiti, Columbia, Hungary, Guatemala, Nicaragua, Scotland, Thailand, Yugoslavia, El-Salvador, Trinadad&Tobago, Peru, Hong, Holand-Netherlands\].
# Specifically, you are required to **fill the blanks of this notebook** based on your results. In this assignment, you will analyze how different features, models and hyper-parameters influence the performance.
# ## 1. Load Data
# +
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import sklearn
# %matplotlib inline
# # %config InlineBackend.figure_format = 'svg'
# -
df = pd.read_csv('income.csv')
# ## 2. Exploratory Data Analysis
# ### Take a brief look at the data using `head()`
df.head()
# ### Observe the basic statistical information of continuous attributes
df.describe() # only describe the continuous variables
# ### Count the NaN values
df.isnull().sum() ### before
# ### Remove NaN values due to small proportion to the whole dataset
df = df.dropna()
df.isnull().sum() ### after
# ### Pick out categorical and continuous variables
df.info()
# ### Observe categorical attributes
for col in df.select_dtypes([np.object]).columns:
print('{}: {}\n'.format(col, df[col].unique()))
# ### Merge values of similar semantics
df.education.replace({
'Preschool': 'dropout',
'10th': 'dropout',
'11th': 'dropout',
'12th': 'dropout',
'1st-4th': 'dropout',
'5th-6th': 'dropout',
'7th-8th': 'dropout',
'9th': 'dropout',
'HS-Grad': 'HighGrad',
'HS-grad': 'HighGrad',
'Some-colloge': 'CommunityCollege',
'Assoc-acdm': 'CommunityCollege',
'Assoc-voc': 'CommunityCollege',
'Prof-school': 'Masters',
}, inplace=True)
# ## 3. Classification Models
# +
from sklearn.model_selection import train_test_split
from sklearn import metrics
# tentatively take 3 numerical attributes for convenience
X = df[['education_degree', 'age', 'hours_per_week']].values
Y = df[['income']].values
# train, test split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.3, random_state=88, stratify=Y)
# -
# ### KNN
# +
## Example: Use KNN to predict income
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=50)
# change the shape of Y_train to (n_samples, ) using `.ravel()`
knn.fit(X_train, Y_train.ravel())
knn_pred = knn.predict(X_test)
print('The accuracy of the KNN is', metrics.accuracy_score(knn_pred, Y_test))
# -
# ### Hyper-parameter tuning with `GridSearchCV()`
# +
from sklearn.model_selection import GridSearchCV
param_grid = {'n_neighbors': np.arange(30, 70)}
knn = KNeighborsClassifier()
knn_cv = GridSearchCV(knn, param_grid, cv=5)
# change the shape of Y_train to (n_samples, ) using `.ravel()`
knn_cv.fit(X_train, Y_train.ravel())
print(knn_cv.best_params_)
print(knn_cv.best_score_)
# -
# ### Your Tasks
#
# As far as you can see, we have built a KNN classification model and select the best hyper-parameters with `GridSearchCV()`. In this task, you are asked to build your own models using `scikit-learn` APIs.
#
# **Question 1 [10pts]**. Build a `Logistic Regression` model on training data and calculate accuracy over testing data.
#
# **Question 2 [10pts]**. Build a `Decision Tree` model on training data and calculate accuracy over testing data.
#
# **Question 3 [20pts]**. Use graphviz to visualize the decision tree of Question 2, and use a proper tool to visualize the decision boundary of the decision tree.
#
# **Question 4 [10pts]**. Build a `Random Forest` model with your customized parameters on training data and calculate accuracy over testing data.
#
# **Question 5 [20pts]**. For `Random Forest`, use `GridSearchCV()` to find the **optimal** hyper-parameter combination over:
# - `n_estimator`: the number of trees in the forest
# - `max_depth`: the maximum depth of the tree
# - `max_leaf_nodes`: grow trees with ``max_leaf_nodes`` in best-first fashion.
#
# You should specify your own sets of values for these hyper-parameters. What's more, you are required to print the importance of each features of the dataset.
#
# (*tip: using the `feature_importances_` attributes of the `RandomForestClassifier()` as we have learned in class*)
#
# **Question 6 [10pts]**. Build a `AdaBoost` model on training data and calculate accuracy over testing data.
# +
# Question 1: Build a `Logistic Regression` model on training data and calculate accuracy over testing data.
from sklearn.linear_model import LogisticRegression
Lr = LogisticRegression(solver='liblinear')
# change the shape of Y_train to (n_samples, ) using `.ravel()`
Lr.fit(X_train, Y_train.ravel())
Lr_pred = Lr.predict(X_test)
# print the accuracy (we can also use different kinds of solver to find the optimal one for this task)
print('The accuracy of the Logistic Regression is', metrics.accuracy_score(Lr_pred, Y_test))
# +
# Question 2: Build a `Decision Tree` model on training data and calculate accuracy over testing data.
from sklearn import tree
Tree = tree.DecisionTreeClassifier(criterion='gini')
# train the model on the reaining set
Tree.fit(X_train,Y_train.ravel())
# use the model to predict the values on test set
Tree_pred = Tree.predict(X_test)
# print the accuracy (we can also use different kinds criterion for this task - 'gini' & ''entropy)
print('The accuracy of the Decision Tree is', metrics.accuracy_score(Tree_pred, Y_test))
# +
# Question 3: Use graphviz to visualize the decision tree of Question 2, and use a proper tool to visualize the decision boundary of the decision tree.
# # !pip install graphviz
# # !pip install IPython
# # !pip install pydotplus
import graphviz
from IPython.display import Image
from sklearn import tree
import pydotplus
# There are two versions, I because of environment problems, I cannot visualize it, so I keep 2 versions
# versoin 1
tree.export_graphviz(Tree)
# versoin 2
# dot_data = tree.export_graphviz(Tree, out_file=None, #Tree is the classifier in Question #2
# feature_names=df.income, #name of corresponding features
# class_names=df.capital_gain #name of corresponding classes
# filled=True, rounded=True,
# special_characters=True)
# # defining the graph (maybe there are some problem with environment, and I have changed the
# graph = pydotplus.graph_from_dot_data(dot_data)
# graph.write_png('example.png') #save the image
# Image(graph.create_png())
# +
# Question 4: Build a `Random Forest` model with your customized parameters on training data and calculate accuracy over testing data.
from sklearn.ensemble import RandomForestClassifier
RF = RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100, n_jobs=1,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
# training the model
RF.fit(X_train, Y_train.ravel())
# making predictions
RF_pred = RF.predict(X_test)
# print the accuracy (we can also use different combinition of parameters: criterion<gini...> & min_samples_leaf...)
print('The accuracy of the Random Forest is', metrics.accuracy_score(RF_pred, Y_test))
# +
# Question 5: Hyper-parameter serach over Random Forest and print feature importance list.
# Search round 1
# Below are the initial round of training
# We need to change the range of parameters step by step, to find the optimal ones
# just like binary search, we need to narrow down the range gradually
from sklearn.model_selection import GridSearchCV
param_set = {
'n_estimators': range(90, 110, 5),
'max_depth': range(10,21,3),
'max_leaf_nodes': range(45,55,5),
}
# Gsearch = GridSearchCV( RF, param_grid = param_set, scoring='roc_auc', cv=5 )
RF = RandomForestClassifier()
Gsearch = GridSearchCV( RF, param_grid = param_set, cv=5 )
Gsearch.fit(X_train, Y_train.ravel())
# Gsearch.grid_scores_, gsearch.best_params_, gsearch.best_score_
def print_best_score(gsearch,param_set):
# print best score
print("Best score: %0.3f" % gsearch.best_score_)
print("Best parameters set:")
# print the parameters
best_parameters = gsearch.best_estimator_.get_params()
for param_name in sorted(param_set.keys()):
print("\t%s: %r" % (param_name, best_parameters[param_name]))
print_best_score(Gsearch,param_set)
# Output log
# Best score: 0.795
# Best parameters set:
# max_depth: 16
# max_leaf_nodes: 45
# n_estimators: 105
# +
# Another round of training
# I just did 2 rounds of searching, beacuase of my limited computing resources
# each round of training takes about 50 mins on my PC
# the method & strategy is clear, so the rest is not difficult, I will not further carry them out, due to the poor computing capacity
from sklearn.model_selection import GridSearchCV
param_set = {
'n_estimators': range(100, 111, 1),
'max_depth': range(13,22,1),
'max_leaf_nodes': range(35,45,2),
}
# Gsearch = GridSearchCV( RF, param_grid = param_set, scoring='roc_auc', cv=5 )
RF = RandomForestClassifier()
Gsearch = GridSearchCV( RF, param_grid = param_set, cv=5 )
Gsearch.fit(X_train, Y_train.ravel())
# Gsearch.grid_scores_, gsearch.best_params_, gsearch.best_score_
def print_best_score(gsearch,param_set):
# best score has improved 0.1%, compared with round #
print("Best score: %0.3f" % gsearch.best_score_)
print("Best parameters set:")
# parameters
best_parameters = gsearch.best_estimator_.get_params()
for param_name in sorted(param_set.keys()):
print("\t%s: %r" % (param_name, best_parameters[param_name]))
print_best_score(Gsearch,param_set)
# each time, we can check, if the output parameters are on the border of the range, if so, we need to expand the range in this direction
# Best score: 0.796
# Best parameters set:
# max_depth: 19
# max_leaf_nodes: 35
# n_estimators: 105
# +
# Question 6: Build a `AdaBoost` model on training data and calculate accuracy over testing data.
from sklearn.ensemble import AdaBoostClassifier
# here the hyper-parameter is n_estimator, ew may as well take 100
Ada = AdaBoostClassifier(n_estimators=100, random_state=0)
Ada.fit(X_train, Y_train.ravel())
Ada_pred = Ada.predict(X_test)
# to improve performance, we can use loop to find the optimal parameter
print('The accuracy of the Ada Boost is', metrics.accuracy_score(Ada_pred, Y_test))
# -
# ## 4. Feature Engineering
#
# Before you start this part, we recommend you to read this [article](https://www.cnblogs.com/jasonfreak/p/5448385.html)
# ### Using `LabelEncoder()`: map categorical features to [0, C)
# +
from sklearn.preprocessing import LabelEncoder
encoded_df = df.apply(LabelEncoder().fit_transform)
encoded_df.head()
# -
# ### Using `pandas.get_dummies()`: map categorical features into one-hot encoding
# +
cols = list(set(df.select_dtypes([np.object]).columns) - set(['income']))
onehot_df = pd.get_dummies(df, columns=cols)
onehot_df.head()
# -
# The aforementioned machine learning models are built upon **3 distinct attributes** (`education_degree`, `age` and `hours_per_week`) with **10 more attributes unused**. You are required to utilize those unused columns using the feature engineering methods introduced above to address this issue.]
#
# **Question 7 [20pts]**. Compare the performance (accuracy) of different algorithms and different preprocessing methods on the dataset. Specifically, please fill the blanks in the table below:
#
# | Alg. | Original 3 columns | All columns with `LabelEncoder` | All columns with `OneHot` |
# | :---: | :----: | :----: | :----: |
# | Logistic Regression |  |  |  |
# | Decision Tree |  |  |  |
# | Random Forest |  |  |  |
# | AdaBoost |  |  |  |
# +
# Question 7: Compare the performance (accuracy) of different algorithms and different preprocessing methods on the dataset
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import tree
import warnings
warnings.filterwarnings('ignore')
encoded_feat = encoded_df.drop(columns=['income']).values
encoded_labl = encoded_df[['income']].values
encoded_X_train, encoded_X_test, encoded_Y_train, encoded_Y_test = train_test_split(encoded_feat, encoded_labl, test_size=0.3)
onehot_feat = onehot_df.drop(columns=['income']).values
onehot_labl = onehot_df[['income']].values
onehot_X_train, onehot_X_test, onehot_Y_train, onehot_Y_test = train_test_split(onehot_feat, onehot_labl, test_size=0.3)
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.3)
LR = LogisticRegression()
Tree = tree.DecisionTreeClassifier(criterion='gini')
RF = RandomForestClassifier()
Ada = AdaBoostClassifier()
models = [LR, Tree, RF, Ada]
model_names = ['LR', 'Tree', 'RF', 'Ada']
dataset = [[encoded_X_train, encoded_X_test, encoded_Y_train, encoded_Y_test], [onehot_X_train, onehot_X_test, onehot_Y_train, onehot_Y_test], [X_train, X_test, Y_train, Y_test]]
for index in range(len(models)):
model = models[index]
name = model_names[index]
for data in dataset:
model.fit(data[0], data[2])
model_pred = model.predict(data[1])
print('The accuracy of {} is'.format(name), metrics.accuracy_score(model_pred, data[3]))
print("The 3 accuracy are encoded, onehot and original in order.\n. To avoid filling in the table, I print them above")
# -
| DataScience/homework/hw7_tree&forest/hw7-tree&forest.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Keras `IMDB` dataset.
# * This is a dataset of 25,000 movies reviews from IMDB, labeled by sentiment (positive/negative). Reviews have been preprocessed, and each review is encoded as a list of word indexes (integers).
import tensorflow as tf
import numpy as np
from tensorflow.keras import datasets
from tensorflow import keras
dir(datasets)
# > Loading the data.
imdb = datasets.imdb.load_data(num_words=10000)
(X_train, y_train),(X_test, y_test) = imdb
print(X_train[0])
y_train[:2]
# > Let's join the train and test sets.
X = np.concatenate([X_train, X_test])
y = np.concatenate([y_train, y_test])
X, y
# > So the `X_train[0]` is just a list of integers that doesn't make sense to a human for now, but we can say it
# is a positive review about the movie according to the label we get.
word_indices = datasets.imdb.get_word_index()
word_indices
# > Let's create a function that decords integers lists into sentences.
word_indices_reversed = dict([(value, key) for (key, value) in word_indices.items()])
word_indices_reversed
def decord(sent):
INDEX_FROM=3
return " ".join([word_indices_reversed.get(i - INDEX_FROM, '#') for i in sent[0]])+"..."
decord([X[1]])
# > Lets create a function that will encode a given sentence to `word_embedings_list`.
def encode(sent):
pass
# > "Data preparation".
# > We want to preapare the sentences to have a same width. This is sometimes called `pad_sequencing` we are just make all sentences to have the same width by trancating long sentencs and appending 0 to shorter sentences.
def vectorize(sequences, dim=10000):
res = np.zeros((len(sequences), dim))
for i, seq in enumerate(sequences):
res[i, seq] = 1
return res
X_data = vectorize(X)
X_data[0], len(X_data[0]), len(X_data[1])
# > Converting the `X_data` and `y` to tensorflow_tensors.
X_tensors = tf.convert_to_tensor(X_data)
y_tensors = tf.convert_to_tensor(y)
y_tensors, X_tensors, y_tensors.shape, X_tensors.shape
# > Creating a `Functional NN`
# ### `CNN` for sentiment classification.
vocabulary_size = len(word_indices_reversed)
model = keras.Sequential([
keras.layers.Embedding(vocabulary_size,
100,
trainable= False,
input_length = 10000
),
keras.layers.Conv1D(128, 5, activation='relu'),
keras.layers.GlobalMaxPooling1D(),
keras.layers.Dense(1, activation="sigmoid")
])
model.compile(
loss = keras.losses.BinaryCrossentropy(from_logits=False),
metrics=["acc"]
)
model.summary()
model.fit(
X_tensors, y_tensors, epochs=2, validation_split=.3, batch_size=256
)
predictions = model.predict(X_tensors[:5])
# +
predictions= tf.squeeze(tf.round(predictions))
predictions, y_tensors[:5]
# -
| tf-rnn/01_IMDB_dataset/.ipynb_checkpoints/02_Sentiment_Analysis_IMDB_CNN-checkpoint.ipynb |
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.6.0
# language: julia
# name: julia-1.6
# ---
# ## Background
#
# [**Symbolics.jl**](https://github.com/JuliaSymbolics/Symbolics.jl) is a fast and modern Computer Algebra System (CAS) written in the Julia Programming Language. It is an integral part of the [SciML](https://sciml.ai/) ecosystem of differential equation solvers and scientific machine learning packages. While **Symbolics.jl** is primarily designed for modern scientific computing (e.g., auto-differentiation, machine learning), it is a powerful CAS and can also be useful for *classic* scientific computing. One such application is using the *perturbation* theory to solve algebraic and differential equations.
#
# Perturbation methods are a collection of techniques to solve intractable problems that generally don't have a closed solution but depend on a tunable parameter and have closed or easy solutions for some values of the parameter. The main idea is to assume a solution as a power series in the tunable parameter (say $ϵ$), such that $ϵ = 0$ corresponds to an easy solution.
#
# We will discuss the general steps of the perturbation methods to solve algebraic (this tutorial) and differential equations (*Mixed Symbolic/Numerical Methods for Perturbation Theory - Differential Equations*).
#
# The hallmark of the perturbation method is the generation of long and convoluted intermediate equations, which are subjected to algorithmic and mechanical manipulations. Therefore, these problems are well suited for CAS. In fact, CAS softwares have been used to help with the perturbation calculations since the early 1970s.
#
# In this tutorial our goal is to show how to use a mix of symbolic manipulations (**Symbolics.jl**) and numerical methods (**DifferentialEquations.jl**) to solve simple perturbation problems.
#
# ## Solving the Quintic
#
# We start with the "hello world!" analog of the perturbation problems, solving the quintic (fifth-order) equations. We want to find a real valued $x$ such that $x^5 + x = 1$. According to the Abel's theorem, a general quintic equation does not have a closed form solution. Of course, we can easily solve this equation numerically; for example, by using the Newton's method. We use the following implementation of the Newton's method:
# +
using Symbolics, SymbolicUtils
function solve_newton(f, x, x₀; abstol=1e-8, maxiter=50)
xₙ = Float64(x₀)
fₙ₊₁ = x - f / Symbolics.derivative(f, x)
for i = 1:maxiter
xₙ₊₁ = substitute(fₙ₊₁, Dict(x => xₙ))
if abs(xₙ₊₁ - xₙ) < abstol
return xₙ₊₁
else
xₙ = xₙ₊₁
end
end
return xₙ₊₁
end
# -
# In this code, `Symbolics.derivative(eq, x)` does exactly what it names implies: it calculates the symbolic derivative of `eq` (a **Symbolics.jl** expression) with respect to `x` (a **Symbolics.jl** variable). We use `Symbolics.substitute(eq, D)` to evaluate the update formula by substituting variables or sub-expressions (defined in a dictionary `D`) in `eq`. It should be noted that `substitute` is the workhorse of our code and will be used multiple times in the rest of these tutorials. `solve_newton` is written with simplicity and clarity, and not performance, in mind but suffices for our purpose.
#
# Let's go back to our quintic. We can define a Symbolics variable as `@variables x` and then solve the equation `solve_newton(x^5 + x - 1, x, 1.0)` (here, `x₀ = 0` is our first guess). The answer is 0.7549. Now, let's see how we can solve the same problem using the perturbation methods.
#
# We introduce a tuning parameter $\epsilon$ into our equation: $x^5 + \epsilon x = 1$. If $\epsilon = 1$, we get our original problem. For $\epsilon = 0$, the problem transforms to an easy one: $x^5 = 1$ which has an exact real solution $x = 1$ (and four complex solutions which we ignore here). We expand $x$ as a power series on $\epsilon$:
#
# $$
# x(\epsilon) = a_0 + a_1 \epsilon + a_2 \epsilon^2 + O(\epsilon^3)
# \,.
# $$
#
# $a_0$ is the solution of the easy equation, therefore $a_0 = 1$. Substituting into the original problem,
#
# $$
# (a_0 + a_1 \epsilon + a_2 \epsilon^2)^5 + \epsilon (a_0 + a_1 \epsilon + a_2 \epsilon^2) - 1 = 0
# \,.
# $$
#
# Expanding the equations, we get
# $$
# \epsilon (1 + 5 a_1) + \epsilon^2 (a_1 + 5 a_2 + 10 a1_2) + 𝑂(\epsilon^3) = 0
# \,.
# $$
#
# This equation should hold for each power of $\epsilon$. Therefore,
#
# $$
# 1 + 5 a_1 = 0
# \,,
# $$
#
# and
#
# $$
# a_1 + 5 a_2 + 10 a_1^2 = 0
# \,.
# $$
#
# This system of equations does not initially seem to be linear because of the presence of terms like $10 a_1^2$, but upon closer inspection is found to be in fact linear (this is a feature of the perturbation methods). In addition, the system is in a triangular form, meaning the first equation depends only on $a_1$, the second one on $a_1$ and $a_2$, such that we can replace the result of $a_1$ from the first one into the second equation and remove the non-linear term. We solve the first equation to get $a_1 = -\frac{1}{5}$. Substituting in the second one and solve for $a_2$:
#
# $$
# a_2 = \frac{(-\frac{1}{5} + 10(-(\frac{1}{5})²)}{5} = -\frac{1}{25}
# \,.
# $$
#
# Finally,
#
# $$
# x(\epsilon) = 1 - \frac{\epsilon}{5} - \frac{\epsilon^2}{25} + O(\epsilon^3)
# \,.
# $$
#
# Solving the original problem, $x(1) = 0.76$, compared to 0.7548 calculated numerically. We can improve the accuracy by including more terms in the expansion of $x$. However, the calculations, while straightforward, become messy and intractable to do manually very quickly. This is why a CAS is very helpful to solve perturbation problems.
#
# Now, let's see how we can do these calculations in Julia. Let $n$ be the order of the expansion. We start by defining the symbolic variables:
n = 2
@variables ϵ a[1:n]
# Then, we define
x = 1 + a[1]*ϵ + a[2]*ϵ^2
# The next step is to substitute `x` in the problem equation
eq = x^5 + ϵ*x - 1
# The expanded form of `eq` is
expand(eq)
# We need a way to get the coefficients of different powers of `ϵ`. Function `collect_powers(eq, x, ns)` returns the powers of variable `x` in expression `eq`. Argument `ns` is the range of the powers.
function collect_powers(eq, x, ns; max_power=100)
eq = substitute(expand(eq), Dict(x^j => 0 for j=last(ns)+1:max_power))
eqs = []
for i in ns
powers = Dict(x^j => (i==j ? 1 : 0) for j=1:last(ns))
push!(eqs, substitute(eq, powers))
end
eqs
end
# To return the coefficients of $ϵ$ and $ϵ^2$ in `eq`, we can write
eqs = collect_powers(eq, ϵ, 1:2)
# A few words on how `collect_powers` works, It uses `substitute` to find the coefficient of a given power of `x` by passing a `Dict` with all powers of `x` set to 0, except the target power which is set to 1. For example, the following expression returns the coefficient of `ϵ^2` in `eq`,
substitute(expand(eq), Dict(
ϵ => 0,
ϵ^2 => 1,
ϵ^3 => 0,
ϵ^4 => 0,
ϵ^5 => 0,
ϵ^6 => 0,
ϵ^7 => 0,
ϵ^8 => 0)
)
# Back to our problem. Having the coefficients of the powers of `ϵ`, we can set each equation in `eqs` to 0 (remember, we rearrange the problem such that `eq` is 0) and solve the system of linear equations to find the numerical values of the coefficients. **Symbolics.jl** has a function `Symbolics.solve_for` that can solve systems of linear equations. However, the presence of higher order terms in `eqs` prevents `Symbolics.solve_for(eqs .~ 0, a)` from workings properly. Instead, we can exploit the fact that our system is in a triangular form and start by solving `eqs[1]` for `a₁` and then substitute this in `eqs[2]` and solve for `a₂` (as continue the same process for higher order terms). This *cascading* process is done by function `solve_coef(eqs, ps)`:
function solve_coef(eqs, ps)
vals = Dict()
for i = 1:length(ps)
eq = substitute(eqs[i], vals)
vals[ps[i]] = Symbolics.solve_for(eq ~ 0, ps[i])
end
vals
end
# Here, `eqs` is an array of expressions (assumed to be equal to 0) and `ps` is an array of variables. The result is a dictionary of *variable* => *value* pairs. We apply `solve_coef` to `eqs` to get the numerical values of the parameters:
solve_coef(eqs, a)
# Finally, we substitute back the values of `a` in the definition of `x` as a function of `𝜀`. Note that `𝜀` is a number (usually Float64), whereas `ϵ` is a symbolic variable.
X = 𝜀 -> 1 + a[1]*𝜀 + a[2]*𝜀^2
# Therefore, the solution to our original problem becomes `X(1)`, which is equal to 0.76. We can use larger values of `n` to improve the accuracy of estimations.
#
# | n | x |
# |---|----------------|
# |1 |0.8 |
# |2 |0.76|
# |3 |0.752|
# |4 |0.752|
# |5 |0.7533|
# |6 |0.7543|
# |7 |0.7548|
# |8 |0.7550|
#
# Remember the numerical value is 0.7549. The two functions `collect_powers` and `solve_coef(eqs, a)` are used in all the examples in this and the next tutorial.
#
# ## Solving the Kepler's Equation
#
# Historically, the perturbation methods were first invented to solve orbital calculations of the Moon and the planets. In homage to this history, our second example has a celestial theme. Our goal is solve the Kepler's equation:
#
# $$
# E - e\sin(E) = M
# \,.
# $$
#
# where $e$ is the *eccentricity* of the elliptical orbit, $M$ is the *mean anomaly*, and $E$ (unknown) is the *eccentric anomaly* (the angle between the position of a planet in an elliptical orbit and the point of periapsis). This equation is central to solving two-body Keplerian orbits.
#
# Similar to the first example, it is easy to solve this problem using the Newton's method. For example, let $e = 0.01671$ (the eccentricity of the Earth) and $M = \pi/2$. We have `solve_newton(x - e*sin(x) - M, x, M)` equals to 1.5875 (compared to π/2 = 1.5708). Now, we try to solve the same problem using the perturbation techniques (see function `test_kepler`).
#
# For $e = 0$, we get $E = M$. Therefore, we can use $e$ as our perturbation parameter. For consistency with other problems, we also rename $e$ to $\epsilon$ and $E$ to $x$.
#
# From here on, we use the helper function `def_taylor` to define Taylor's series by calling it as `x = def_taylor(ϵ, a, 1)`, where the arguments are, respectively, the perturbation variable, an array of coefficients (starting from the coefficient of $\epsilon^1$), and an optional constant term.
def_taylor(x, ps) = sum([a*x^i for (i,a) in enumerate(ps)])
def_taylor(x, ps, p₀) = p₀ + def_taylor(x, ps)
# We start by defining the variables (assuming `n = 3`):
n = 3
@variables ϵ M a[1:n]
x = def_taylor(ϵ, a, M)
# We further simplify by substituting `sin` with its power series using the `expand_sin` helper function:
expand_sin(x, n) = sum([(isodd(k) ? -1 : 1)*(-x)^(2k-1)/factorial(2k-1) for k=1:n])
# To test,
expand_sin(0.1, 10) ≈ sin(0.1)
# The problem equation is
eq = x - ϵ * expand_sin(x, n) - M
# We follow the same process as the first example. We collect the coefficients of the powers of `ϵ`
eqs = collect_powers(eq, ϵ, 1:n)
# and then solve for `a`:
vals = solve_coef(eqs, a)
# Finally, we substitute `vals` back in `x`:
x′ = substitute(x, vals)
X = (𝜀, 𝑀) -> substitute(x′, Dict(ϵ => 𝜀, M => 𝑀))
X(0.01671, π/2)
# The result is 1.5876, compared to the numerical value of 1.5875. It is customary to order `X` based on the powers of `𝑀` instead of `𝜀`. We can calculate this series as `collect_powers(sol, M, 0:3)
# `. The result (after manual cleanup) is
#
# ```
# (1 + 𝜀 + 𝜀^2 + 𝜀^3)*𝑀
# - (𝜀 + 4*𝜀^2 + 10*𝜀^3)*𝑀^3/6
# + (𝜀 + 16*𝜀^2 + 91*𝜀^3)*𝑀^5/120
# ```
#
# Comparing the formula to the one for 𝐸 in the [Wikipedia article on the Kepler's equation](https://en.wikipedia.org/wiki/Kepler%27s_equation):
#
# $$
# E = \frac{1}{1-\epsilon}M
# -\frac{\epsilon}{(1-\epsilon)^4} \frac{M^3}{3!} + \frac{(9\epsilon^2
# + \epsilon)}{(1-\epsilon)^7}\frac{M^5}{5!}\cdots
# $$
#
# The first deviation is in the coefficient of $\epsilon^3 M^5$.
| notebook/perturbation/01-perturbation_algebraic.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# #!pip install -U tf-nightly-2.0-preview
# -
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
def plot_series(time, series, format="-", start=0, end=None, label=None):
plt.plot(time[start:end], series[start:end], format, label=label)
plt.xlabel("Time")
plt.ylabel("Value")
if label:
plt.legend(fontsize=14)
plt.grid(True)
# Trend and Seasonality
def trend(time, slope=0):
return slope * time
# Let's create a time series that just trends upward:
# +
time = np.arange(4 * 365 + 1)
baseline = 10
series = trend(time, 0.1)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
# -
# Now let's generate a time series with a seasonal pattern:
# +
def seasonal_pattern(season_time):
"""Just an arbitrary pattern, you can change it if you wish"""
return np.where(season_time < 0.4,
np.cos(season_time * 2 * np.pi),
1 / np.exp(3 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
"""Repeats the same pattern at each period"""
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
# +
baseline = 10
amplitude = 40
series = seasonality(time, period=365, amplitude=amplitude)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
# -
# Now let's create a time series with both trend and seasonality:
# +
slope = 0.05
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
# -
# NOISE
# In practice few real-life time series have such a smooth signal. They usually have some noise, and the signal-to-noise ratio can sometimes be very low. Let's generate some white noise:
def white_noise(time, noise_level=1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
# +
noise_level = 5
noise = white_noise(time, noise_level, seed=42)
plt.figure(figsize=(10, 6))
plot_series(time, noise)
plt.show()
# -
# Now let's add this white noise to the time series:
# +
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
# -
# All right, this looks realistic enough for now. Let's try to forecast it. We will split it into two periods: the training period and the validation period (in many cases, you would also want to have a test period). The split will be at time step 1000.
split_time = 1000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
def autocorrelation(time, amplitude, seed=None):
rnd = np.random.RandomState(seed)
φ1 = 0.5
φ2 = -0.1
ar = rnd.randn(len(time) + 50)
ar[:50] = 100
for step in range(50, len(time) + 50):
ar[step] += φ1 * ar[step - 50]
ar[step] += φ2 * ar[step - 33]
return ar[50:] * amplitude
def autocorrelation(time, amplitude, seed=None):
rnd = np.random.RandomState(seed)
φ = 0.8
ar = rnd.randn(len(time) + 1)
for step in range(1, len(time) + 1):
ar[step] += φ * ar[step - 1]
return ar[1:] * amplitude
series = autocorrelation(time, 10, seed=42)
plot_series(time[:200], series[:200])
plt.show()
series = autocorrelation(time, 10, seed=42) + trend(time, 2)
plot_series(time[:200], series[:200])
plt.show()
series = autocorrelation(time, 10, seed=42) + seasonality(time, period=50, amplitude=150) + trend(time, 2)
plot_series(time[:200], series[:200])
plt.show()
series = autocorrelation(time, 10, seed=42) + seasonality(time, period=50, amplitude=150) + trend(time, 2)
series2 = autocorrelation(time, 5, seed=42) + seasonality(time, period=50, amplitude=2) + trend(time, -1) + 550
series[200:] = series2[200:]
#series += noise(time, 30)
plot_series(time[:300], series[:300])
plt.show()
def impulses(time, num_impulses, amplitude=1, seed=None):
rnd = np.random.RandomState(seed)
impulse_indices = rnd.randint(len(time), size=10)
series = np.zeros(len(time))
for index in impulse_indices:
series[index] += rnd.rand() * amplitude
return series
series = impulses(time, 10, seed=42)
plot_series(time, series)
plt.show()
def autocorrelation(source, φs):
ar = source.copy()
max_lag = len(φs)
for step, value in enumerate(source):
for lag, φ in φs.items():
if step - lag > 0:
ar[step] += φ * ar[step - lag]
return ar
signal = impulses(time, 10, seed=42)
series = autocorrelation(signal, {1: 0.99})
plot_series(time, series)
plt.plot(time, signal, "k-")
plt.show()
signal = impulses(time, 10, seed=42)
series = autocorrelation(signal, {1: 0.70, 50: 0.2})
plot_series(time, series)
plt.plot(time, signal, "k-")
plt.show()
series_diff1 = series[1:] - series[:-1]
plot_series(time[1:], series_diff1)
# +
from pandas.plotting import autocorrelation_plot
autocorrelation_plot(series)
# +
from statsmodels.tsa.arima_model import ARIMA
model = ARIMA(series, order=(5, 1, 0))
model_fit = model.fit(disp=0)
print(model_fit.summary())
# -
root = r'D:\Users\Arkady\Verint\Coursera_2019_Tensorflow_Specialization\Course4_Sequences_TimeSeries_Prediction'
fpath = root + '/tmp/sunspots.csv'
# +
# #!wget --no-check-certificate \
# # https://storage.googleapis.com/laurencemoroney-blog.appspot.com/horse-or-human.zip \
# # -O /tmp/horse-or-human.zip
#import os
#import zipfile
#local_zip = '/tmp/horse-or-human.zip'
#zip_ref = zipfile.ZipFile(local_zip, 'r')
#zip_ref.extractall('/tmp/horse-or-human')
# to upload file from local computer to colab
#import pandas as pd
#from google.colab import files
#uploaded = files.upload()
# -
import pandas as pd
df = pd.read_csv(fpath, parse_dates=["Date"], index_col="Date")
series = df["Monthly Mean Total Sunspot Number"].asfreq("1M")
series.head()
series.plot(figsize=(12, 5))
series["1995-01-01":].plot()
series.diff(1).plot()
plt.axis([0, 100, -50, 50])
# +
from pandas.plotting import autocorrelation_plot
autocorrelation_plot(series)
# -
autocorrelation_plot(series.diff(1)[1:])
autocorrelation_plot(series.diff(1)[1:].diff(11 * 12)[11*12+1:])
plt.axis([0, 500, -0.1, 0.1])
autocorrelation_plot(series.diff(1)[1:])
plt.axis([0, 50, -0.1, 0.1])
116.7 - 104.3
[series.autocorr(lag) for lag in range(1, 50)]
filepath_or_buffer = fpath
pd.read_csv(filepath_or_buffer, sep=',', delimiter=None, header='infer', names=None, index_col=None, usecols=None, squeeze=False, prefix=None, mangle_dupe_cols=True, dtype=None, engine=None, converters=None, true_values=None, false_values=None, skipinitialspace=False, skiprows=None, skipfooter=0, nrows=None, na_values=None, keep_default_na=True, na_filter=True, verbose=False, skip_blank_lines=True, parse_dates=False, infer_datetime_format=False, keep_date_col=False, date_parser=None, dayfirst=False, iterator=False, chunksize=None, compression='infer', thousands=None, decimal=b'.', lineterminator=None, quotechar='"', quoting=0, doublequote=True, escapechar=None, comment=None, encoding=None, dialect=None, tupleize_cols=None, error_bad_lines=True, warn_bad_lines=True, delim_whitespace=False, low_memory=True, memory_map=False, float_precision=None)
#Read a comma-separated values (csv) file into DataFrame.
# +
from pandas.plotting import autocorrelation_plot
series_diff = series
for lag in range(50):
series_diff = series_diff[1:] - series_diff[:-1]
autocorrelation_plot(series_diff)
# +
import pandas as pd
series_diff1 = pd.Series(series[1:] - series[:-1])
autocorrs = [series_diff1.autocorr(lag) for lag in range(1, 60)]
plt.plot(autocorrs)
plt.show()
# -
| legacy/arkady TF legacy/TF_2020_course4_week1_notebook1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### X lines of Python
#
# # Wedge model
#
# This is part of [an Agile blog series](http://ageo.co/xlines00) called **x lines of Python**.
#
# We start with the usual preliminaries.
import matplotlib.pyplot as plt
import numpy as np
# ## Make an earth model
#
# We'll start off with an earth model --- an array of 'cells', each of which has some rock properties.
#
# Line 1 sets up some basic variables, then in line 2 I've used a little matrix-forming trick, `np.tri(m, n, k)`, which creates an *m* × *n* matrix with ones below the *k*th diagonal, and zeros above it. The `dtype` specification just makes sure we end up with integers, which we need later for the indexing trick.
#
# Then line 3 just sets every row above `depth//3` (the `//` is integer division, because NumPy prefers integers for indexing arrays), to 0.
length, depth = 40, 100
model = 1 + np.tri(depth, length, -depth//3, dtype=int)
model[:depth//3,:] = 0
# We'll have a quick look with some very basic plotting commands.
plt.imshow(model, cmap='viridis', aspect=0.2)
plt.show()
model[60]
# Now we can make some Vp-rho pairs (rock 0, rock 1, rock 2) and select from those with `np.take`. This works like `vlookup` in Excel --- it says "read this array, `model` in this case, in which the values *i* are like 0, 1, ... n, and give me the *i*th element from this other array, `rocks` in this case.
rocks = np.array([[2700, 2750], # Vp, rho
[2400, 2450],
[2800, 3000]])
# **Edit:** I was using `np.take` here, but ['fancy indexing'](http://docs.scipy.org/doc/numpy/user/basics.indexing.html) is shorter and more intuitive. We are just going to index `rocks` using the integers in `model`. That is, if `model` has a `1`, we take the second element, `[2400, 2450]`, from `rocks`. We'll end up with an array containing the rocks corresponding to each element of `earth`.
earth = rocks[model]
# Now apply `np.product` to those Vp-rho pairs to get impedance at every sample.
#
# This might look a bit magical, but we're just telling Python to apply the function `product()` to every set of numbers it encounters on the last axis (index `-1`). The array `earth` has shape (100, 40, 2), so you can think of it as a 100 row x 40 column 'section' in which each 'sample' is occupied by a Vp-rho pair. That pair is in the last axis. So product, which just takes a bunch of numbers and multiplies them, will return the impedance (the product of Vp and rho) at each sample location. We'll end up with a new 100 x 40 'section' with impedance at every sample.
imp = np.apply_along_axis(np.product, -1, earth)
# We could have saved a step by taking from `np.product(rocks, axis=1)` but I like the elegance of having an earth model with a set of rock properties at each sample location. That's how I think about the earth --- and it's similar to the concept of a geocellular model.
# ## Model seismic reflections
#
# Now we have an earth model — giving us acoustic impedance everywhere in this 2D grid — we define a function to compute reflection coefficients for every trace.
#
# I love this indexing trick though I admit it looks weird the first time you see it. It's easier to appreciate for a 1D array. Let's look at the differences:
#
# >>> a = np.array([1,1,1,2,2,2,3,3,3])
# >>> a[1:] - a[:-1]
# array([0, 0, 1, 0, 0, 1, 0, 0])
#
# This is equivalent to:
#
# >>> np.diff(a, axis=0)
#
# But I prefer to spell it out so it's analogous to the sum on the denominator.
# +
rc = (imp[1:,:] - imp[:-1,:]) / (imp[1:,:] + imp[:-1,:])
plt.imshow(rc, cmap='Greys', aspect=0.2)
plt.show()
# -
# We'll use a wavelet function from [`bruges`](https://github.com/agile-geoscience/bruges). This is not cheating! Well, I don't think it is... we could use `scipy.signal.ricker` but I can't figure out how to convert frequency into the 'width' parameter that function wants. Using the Ricker from `bruges` keeps things a bit simpler.
# +
import bruges
w = bruges.filters.ricker(duration=0.100, dt=0.001, f=40)
# -
# Let's make sure it looks OK:
plt.plot(w)
plt.show()
# Now one more application of `apply_along_axis`. We could use a loop to step over the traces, but the rule of thumb in Python is "if you are using a loop, you're doing it wrong.". So, we'll use `apply_along_axis`.
#
# It looks a bit more complicated this time, because we can't just pass a function like we did with `product` before. We want to pass in some more things, not just the trace that `apply_along_axis` is going to send it. So we use Python's 'unnamed function creator', `lambda` (in keeping with all things called `lambda`, it's a bad name that no-one can quite explain).
# +
synth = np.apply_along_axis(lambda t: np.convolve(t, w, mode='same'),
axis=0,
arr=rc)
plt.imshow(synth, cmap="Greys", aspect=0.2)
plt.show()
# -
# That's it! And it only needed 9 lines of Python! Not incldung boring old imports and plotting stuff.
#
# Here they are so you can count them:
length, depth = 40, 100
model = 1 + np.tri(depth, length, -depth//3)
model[:depth//3,:] = 0
rocks = np.array([[2700, 2750], [2400, 2450], [2800, 3000]])
earth = np.take(rocks, model.astype(int), axis=0)
imp = np.apply_along_axis(np.product, -1, earth)
rc = (imp[1:,:] - imp[:-1,:]) / (imp[1:,:] + imp[:-1,:])
w = bruges.filters.ricker(duration=0.100, dt=0.001, f=40)
synth = np.apply_along_axis(lambda t: np.convolve(t, w, mode='same'), axis=0, arr=rc)
# <hr />
#
# <div>
# <img src="https://avatars1.githubusercontent.com/u/1692321?s=50"><p style="text-align:center">© Agile Geoscience 2016</p>
# </div>
| notebooks/00_Synthetic_wedge_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <h1> Repeatable splitting </h1>
#
# In this notebook, we will explore the impact of different ways of creating machine learning datasets.
#
# <p>
#
# Repeatability is important in machine learning. If you do the same thing now and 5 minutes from now and get different answers, then it makes experimentation difficult. In other words, you will find it difficult to gauge whether a change you made has resulted in an improvement or not.
from google.cloud import bigquery
# <h3> Create a simple machine learning model </h3>
#
# The dataset that we will use is <a href="https://bigquery.cloud.google.com/table/bigquery-samples:airline_ontime_data.flights">a BigQuery public dataset</a> of airline arrival data. Click on the link, and look at the column names. Switch to the Details tab to verify that the number of records is 70 million, and then switch to the Preview tab to look at a few rows.
# <p>
# We want to predict the arrival delay of an airline based on the departure delay. The model that we will use is a zero-bias linear model:
# $$ delay_{arrival} = \alpha * delay_{departure} $$
# <p>
# To train the model is to estimate a good value for $\alpha$.
# <p>
# One approach to estimate alpha is to use this formula:
# $$ \alpha = \frac{\sum delay_{departure} delay_{arrival} }{ \sum delay_{departure}^2 } $$
# Because we'd like to capture the idea that this relationship is different for flights from New York to Los Angeles vs. flights from Austin to Indianapolis (shorter flight, less busy airports), we'd compute a different $alpha$ for each airport-pair. For simplicity, we'll do this model only for flights between Denver and Los Angeles.
# <h2> Naive random split (not repeatable) </h2>
compute_alpha = """
#standardSQL
SELECT
SAFE_DIVIDE(SUM(arrival_delay * departure_delay), SUM(departure_delay * departure_delay)) AS alpha
FROM
(
SELECT RAND() AS splitfield,
arrival_delay,
departure_delay
FROM
`bigquery-samples.airline_ontime_data.flights`
WHERE
departure_airport = 'DEN' AND arrival_airport = 'LAX'
)
WHERE
splitfield < 0.8
"""
results = bigquery.Client().query(compute_alpha).to_dataframe()
alpha = results['alpha'][0]
print(alpha)
# <h3> What is wrong with calculating RMSE on the training and test data as follows? </h3>
compute_rmse = """
#standardSQL
SELECT
dataset,
SQRT(AVG((arrival_delay - ALPHA * departure_delay)*(arrival_delay - ALPHA * departure_delay))) AS rmse,
COUNT(arrival_delay) AS num_flights
FROM (
SELECT
IF (RAND() < 0.8, 'train', 'eval') AS dataset,
arrival_delay,
departure_delay
FROM
`bigquery-samples.airline_ontime_data.flights`
WHERE
departure_airport = 'DEN'
AND arrival_airport = 'LAX' )
GROUP BY
dataset
"""
bigquery.Client().query(compute_rmse.replace('ALPHA', str(alpha))).to_dataframe()
# Hint:
# * Are you really getting the same training data in the compute_rmse query as in the compute_alpha query?
# * Do you get the same answers each time you rerun the compute_alpha and compute_rmse blocks?
# <h3> How do we correctly train and evaluate? </h3>
# <br/>
# Here's the right way to compute the RMSE using the actual training and held-out (evaluation) data. Note how much harder this feels.
#
# Although the calculations are now correct, the experiment is still not repeatable.
#
# Try running it several times; do you get the same answer?
train_and_eval_rand = """
#standardSQL
WITH
alldata AS (
SELECT
IF (RAND() < 0.8,
'train',
'eval') AS dataset,
arrival_delay,
departure_delay
FROM
`bigquery-samples.airline_ontime_data.flights`
WHERE
departure_airport = 'DEN'
AND arrival_airport = 'LAX' ),
training AS (
SELECT
SAFE_DIVIDE( SUM(arrival_delay * departure_delay) , SUM(departure_delay * departure_delay)) AS alpha
FROM
alldata
WHERE
dataset = 'train' )
SELECT
MAX(alpha) AS alpha,
dataset,
SQRT(AVG((arrival_delay - alpha * departure_delay)*(arrival_delay - alpha * departure_delay))) AS rmse,
COUNT(arrival_delay) AS num_flights
FROM
alldata,
training
GROUP BY
dataset
"""
bigquery.Client().query(train_and_eval_rand).to_dataframe()
# <h2> Using HASH of date to split the data </h2>
#
# Let's split by date and train.
compute_alpha = """
#standardSQL
SELECT
SAFE_DIVIDE(SUM(arrival_delay * departure_delay), SUM(departure_delay * departure_delay)) AS alpha
FROM
`bigquery-samples.airline_ontime_data.flights`
WHERE
departure_airport = 'DEN' AND arrival_airport = 'LAX'
AND ABS(MOD(FARM_FINGERPRINT(date), 10)) < 8
"""
results = bigquery.Client().query(compute_alpha).to_dataframe()
alpha = results['alpha'][0]
print(alpha)
# We can now use the alpha to compute RMSE. Because the alpha value is repeatable, we don't need to worry that the alpha in the compute_rmse will be different from the alpha computed in the compute_alpha.
compute_rmse = """
#standardSQL
SELECT
IF(ABS(MOD(FARM_FINGERPRINT(date), 10)) < 8, 'train', 'eval') AS dataset,
SQRT(AVG((arrival_delay - ALPHA * departure_delay)*(arrival_delay - ALPHA * departure_delay))) AS rmse,
COUNT(arrival_delay) AS num_flights
FROM
`bigquery-samples.airline_ontime_data.flights`
WHERE
departure_airport = 'DEN'
AND arrival_airport = 'LAX'
GROUP BY
dataset
"""
print(bigquery.Client().query(compute_rmse.replace('ALPHA', str(alpha))).to_dataframe().head())
# Note also that the RMSE on the evaluation dataset more from the RMSE on the training dataset when we do the split correctly. This should be expected; in the RAND() case, there was leakage between training and evaluation datasets, because there is high correlation between flights on the same day.
# <p>
# This is one of the biggest dangers with doing machine learning splits the wrong way -- <b> you will develop a false sense of confidence in how good your model is! </b>
# Copyright 2018 Google Inc.
# Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
| courses/machine_learning/deepdive/02_generalization/repeatable_splitting.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Анализ вариантов обслуживания автобусов на основе имитационной модели
# +
from load_packages import load_packages
load_packages()
# +
from packages.bas_models.src.bus_generator import BusGenerator
generator = BusGenerator([0.4, 0.7])
buses = generator.generate(1)
flights = 10
probs = [
[0.0,0.05],
[0.1,0.05],
[0.2,0.11],
[0.3,0.11],
[0.4,0.15],
[0.5,0.15],
[0.6,0.19],
[0.7,0.19],
[0.8,0.23],
[0.9,0.23],
[1.0,0.7]]
# +
from packages.bas_models.src.model import Model
def arrange_statistics(model: Model, timing: [int]):
statistics = dict()
for time in timing:
statistics[time] = get_statistics(model, time)
return statistics
def get_statistics(model: Model, days: int) -> int:
result = 0
for day in range(0, days):
buses = model.run(flights)
result += sum(list(map(lambda x: x.flights, buses)))
model.reset_buses()
return result / days
# +
import numpy as np
import matplotlib.pyplot as plt
def show_bar(keys, values, precision: int = 3):
keys = list(map(lambda x: str(x), keys))
values = list(map(lambda x: round(x, precision), values))
bars = plt.bar(keys, values)
autolabel(bars)
plt.show()
def autolabel(bars, xpos='center'):
xpos = xpos.lower()
ha = {'center': 'center', 'right': 'left', 'left': 'right'}
offset = {'center': 0.5, 'right': 0.57, 'left': 0.43}
for bar in bars:
height = bar.get_height()
plt.text(bar.get_x() + bar.get_width()*offset[xpos], 1.01*height,
'{}'.format(height), ha=ha[xpos], va='bottom')
# -
# # Модель без ремонта мелких поломок
# +
from packages.bas_models.src.nonstop_model import NonstopModel
model = NonstopModel(buses)
statistics = arrange_statistics(model, [1,2,3,4,5,10,20,90])
show_bar(list(statistics.keys()), list(statistics.values()))
# +
from bus_models.statistics_with_probs import StatisticsWithProbs
from packages.bas_models.src.nonstop_model import NonstopModel
stats_with_probs = StatisticsWithProbs(NonstopModel([]), 90, 10)
statistics = stats_with_probs.arrange_statistics(probs)
show_bar(list(statistics.keys()), list(statistics.values()), 2)
# -
# # Модель с ремонтом мелких поломок
# +
from packages.bas_models.src.repair_model import RepairModel
model = RepairModel(buses)
statistics = arrange_statistics(model, [1,2,3,4,5,10,20,90])
show_bar(list(statistics.keys()), list(statistics.values()))
# +
from bus_models.statistics_with_probs import StatisticsWithProbs
from packages.bas_models.src.repair_model import RepairModel
stats_with_probs = StatisticsWithProbs(RepairModel([]), 90, 10)
statistics = stats_with_probs.arrange_statistics(probs)
show_bar(list(statistics.keys()), list(statistics.values()), 2)
# -
# # Вывод
#
# В данных условиях модель без ремонта мелких поломок лучше подходит чем модель с ремонтом мелких поломок... Есть идея - если вероятность полной поломки автобуса
| examples/bus_models.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# This example shows how to use a `GridInterpolationKernel` module on an `ExactGP` model. This regression module is designed for when the inputs of the function you're modeling are one-dimensional.
#
# The use of inducing points allows for scaling up the training data by making computational complexity linear instead of cubic.
#
# Function to be modeled is y=sin(4*pi*x)
#
# GridInterpolationKernel exploits the regular grid structure of linspace for Toeplitz covariances.
#
# This notebook doesn't use cuda, in general we recommend GPU use if possible and most of our notebooks utilize cuda as well.
#
# Kernel interpolation for scalable structured Gaussian processes (KISS-GP) was introduced in this paper:
# http://proceedings.mlr.press/v37/wilson15.pdf
# +
import math
import torch
import gpytorch
from matplotlib import pyplot as plt
from torch import nn, optim
from torch.autograd import Variable
from gpytorch.kernels import RBFKernel, GridInterpolationKernel
from gpytorch.means import ConstantMean
from gpytorch.likelihoods import GaussianLikelihood
from gpytorch.random_variables import GaussianRandomVariable
# Make plots inline
# %matplotlib inline
# -
# Training points are in [0,1] every 1/999
train_x = Variable(torch.linspace(0, 1, 1000))
# Function to model is sin(4*pi*x)
# Gaussian noise from N(0,0.04)
train_y = Variable(torch.sin(train_x.data * (4 * math.pi)) + torch.randn(train_x.size()) * 0.2)
# +
# We use exact GP inference for regression
class GPRegressionModel(gpytorch.models.ExactGP):
def __init__(self, train_x, train_y, likelihood):
super(GPRegressionModel, self).__init__(train_x, train_y, likelihood)
self.mean_module = ConstantMean(constant_bounds=[-1e-5,1e-5])
# Put a grid interpolation kernel over the RBF kernel
self.base_covar_module = RBFKernel(log_lengthscale_bounds=(-5, 6))
self.covar_module = GridInterpolationKernel(self.base_covar_module, grid_size=400,
grid_bounds=[(0, 1)])
# Register kernel lengthscale as parameter
self.register_parameter('log_outputscale', nn.Parameter(torch.Tensor([0])), bounds=(-5,6))
def forward(self,x):
mean_x = self.mean_module(x)
covar_x = self.covar_module(x)
covar_x = covar_x.mul(self.log_outputscale.exp())
return GaussianRandomVariable(mean_x, covar_x)
# The likelihood output is a Gaussian with predictive mean and variance
likelihood = GaussianLikelihood()
# Initialize our model
model = GPRegressionModel(train_x.data, train_y.data, likelihood)
# +
# Find optimal model hyperparameters
model.train()
likelihood.train()
# Use the adam optimizer
optimizer = torch.optim.Adam([
{'params': model.parameters()}, # Includes GaussianLikelihood parameters
], lr=0.1)
# "Loss" for GPs - the marginal log likelihood
mll = gpytorch.mlls.ExactMarginalLogLikelihood(likelihood, model)
training_iterations = 30
for i in range(training_iterations):
# Zero backprop gradients
optimizer.zero_grad()
# Get output from model
output = model(train_x)
# Calc loss and backprop derivatives
loss = -mll(output, train_y)
loss.backward()
print('Iter %d/%d - Loss: %.3f' % (i + 1, training_iterations, loss.data[0]))
optimizer.step()
# +
# Put model & likelihood into eval mode
model.eval()
likelihood.eval()
# Initalize plot
f, observed_ax = plt.subplots(1, 1, figsize=(4, 3))
# Test points every 0.02 in [0,1] inclusive
test_x = Variable(torch.linspace(0, 1, 51))
# To make the predictions as accurate as possible, we're going to use lots of iterations of Conjugate Gradients
# This ensures that the matrix solves are as accurate as possible
with gpytorch.settings.max_cg_iterations(100):
observed_pred = likelihood(model(test_x))
# Define plotting function
def ax_plot(ax, rand_var, title):
# Get lower and upper predictive bounds
lower, upper = rand_var.confidence_region()
# Plot the training data as black stars
ax.plot(train_x.data.numpy(), train_y.data.numpy(), 'k*')
# Plot predictive means as blue line
ax.plot(test_x.data.numpy(), rand_var.mean().data.numpy(), 'b')
# Plot confidence bounds as lightly shaded region
ax.fill_between(test_x.data.numpy(), lower.data.numpy(), upper.data.numpy(), alpha=0.5)
ax.set_ylim([-3, 3])
ax.legend(['Observed Data', 'Mean', 'Confidence'])
ax.set_title(title)
ax_plot(observed_ax, observed_pred, 'Observed Values (Likelihood)')
| examples/kissgp_gp_regression.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] tags=[]
# # Fetching supplimentary model input from the Planetary Computer
#
# This notebook produces additional input layers for the training data used in the [sentinel 1 flood detection](https://www.drivendata.org/competitions/81/detect-flood-water/) competition run by DrivenData. If fetches JRC Global Surface Water and NASADEM elevation data from the Planetary Computer (PC) STAC API and creates pixel-aligned chips that match what will be used in the evaluation process for the competition.
#
# The notebook will iterate through chip paths and query the PC STAC API for the `nasadem` and `jrc-gsw` Collections. It then creates a set of GeoTiffs by "coregistering" the raster data with the chip GeoTIFF, so that all of the additional input layers have the same CRS, bounds, and resolution as the chip. These additional layers are then saved alongside the training chip.
# +
from dataclasses import dataclass
import os
from tempfile import TemporaryDirectory
from typing import List, Any, Dict
from shapely.geometry import box, mapping
import rasterio
from rasterio.warp import reproject, Resampling
import pyproj
from osgeo import gdal
from pystac_client import Client
import planetary_computer as pc
# -
# #### Extract training chips
#
# Download the `flood-train-images.tgz` file from [competition Data Download page](https://www.drivendata.org/competitions/81/detect-flood-water/data/) and upload it to the Hub in the same directory as this notebook.
#
# Then run:
# !tar -xvf flood-train-images.tgz
# to uncompress this. Afterwards you should see an `train_features` directory containing all of the training chips ending in `.tif`.
#
# Use this directory to define the location of the chips, or if you have already uncompressed the chips elsewhere set the location here:
TRAINING_DATA_DIR = "train_features"
# #### Gather chip paths
#
# These chip paths will be used later in the notebook to process the chips. These paths should be to only one GeoTIFF per chip; for example, if both `VV.tif` and `VH.tif` are available for a chip, use only one of these paths. The GeoTIFFs at these paths will be read to get the bounds, CRS and resolution that will be used to fetch auxiliary input data. These can be relative paths. The auxiliary input data will be saved in the same directory as the GeoTIFF files at these paths.
chip_paths = []
for file_name in os.listdir(TRAINING_DATA_DIR):
if file_name.endswith("_vv.tif"):
chip_paths.append(os.path.join(TRAINING_DATA_DIR, file_name))
print(f"{len(chip_paths)} chips found.")
# #### Create the STAC API client
#
# This will be used in the methods below to query the PC STAC API.
STAC_API = "https://planetarycomputer.microsoft.com/api/stac/v1"
catalog = Client.open(STAC_API)
# #### Define functions and classes
# Define a `ChipInfo` dataclass to encapsulate the required data for the target chip. This includes geospatial information that will be used to coregister the incoming jrc-gsw and nasadem data.
# +
@dataclass
class ChipInfo:
"""
Holds information about a training chip, including geospatial info for coregistration
"""
path: str
prefix: str
crs: Any
shape: List[int]
transform: List[float]
bounds: rasterio.coords.BoundingBox
footprint: Dict[str, Any]
def get_footprint(bounds, crs):
"""Gets a GeoJSON footprint (in epsg:4326) from rasterio bounds and CRS"""
transformer = pyproj.Transformer.from_crs(crs, "epsg:4326", always_xy=True)
minx, miny = transformer.transform(bounds.left, bounds.bottom)
maxx, maxy = transformer.transform(bounds.right, bounds.top)
return mapping(box(minx, miny, maxx, maxy))
def get_chip_info(chip_path):
"""Gets chip info from a GeoTIFF file"""
with rasterio.open(chip_path) as ds:
chip_crs = ds.crs
chip_shape = ds.shape
chip_transform = ds.transform
chip_bounds = ds.bounds
# Use the first part of the chip filename as a prefix
prefix = os.path.basename(chip_path).split("_")[0]
return ChipInfo(
path=chip_path,
prefix=prefix,
crs=chip_crs,
shape=chip_shape,
transform=chip_transform,
bounds=chip_bounds,
footprint=get_footprint(chip_bounds, chip_crs),
)
# -
# This method reprojects coregisters raster data to the bounds, CRS and resolution described by the ChipInfo.
def reproject_to_chip(
chip_info, input_path, output_path, resampling=Resampling.nearest
):
"""
Reproject a raster at input_path to chip_info, saving to output_path.
Use Resampling.nearest for classification rasters. Otherwise use something
like Resampling.bilinear for continuous data.
"""
with rasterio.open(input_path) as src:
kwargs = src.meta.copy()
kwargs.update(
{
"crs": chip_info.crs,
"transform": chip_info.transform,
"width": chip_info.shape[1],
"height": chip_info.shape[0],
"driver": "GTiff",
}
)
with rasterio.open(output_path, "w", **kwargs) as dst:
for i in range(1, src.count + 1):
reproject(
source=rasterio.band(src, i),
destination=rasterio.band(dst, i),
src_transform=src.transform,
src_crs=src.crs,
dst_transform=chip_info.transform,
dst_crs=chip_info.crs,
resampling=Resampling.nearest,
)
# This method will take in a set of items and a asset key and write a [VRT](https://gdal.org/drivers/raster/vrt.html) using signed HREFs. This is useful when there's multiple results from the query, so we can treat the resulting rasters as a single set of raster data. It uses the `planetary_computer.sign` method to sign the HREFs with a SAS token generated by the PC [Data Auth API](https://planetarycomputer.microsoft.com/docs/concepts/sas/).
def write_vrt(items, asset_key, dest_path):
"""Write a VRT with hrefs extracted from a list of items for a specific asset."""
hrefs = [pc.sign(item.assets[asset_key].href) for item in items]
vsi_hrefs = [f"/vsicurl/{href}" for href in hrefs]
gdal.BuildVRT(dest_path, vsi_hrefs).FlushCache()
# This method ties it all together - for a given `ChipInfo`, Collection, and Asset, write an auxiliary input chip with the given file name.
def create_chip_aux_file(
chip_info, collection_id, asset_key, file_name, resampling=Resampling.nearest
):
"""
Write an auxiliary chip file.
The auxiliary chip file includes chip_info for the Collection and Asset, and is
saved in the same directory as the original chip with the given file_name.
"""
output_path = os.path.join(
os.path.dirname(chip_info.path), f"{chip_info.prefix}_{file_name}"
)
search = catalog.search(collections=[collection_id], intersects=chip_info.footprint)
items = list(search.get_items())
with TemporaryDirectory() as tmp_dir:
vrt_path = os.path.join(tmp_dir, "source.vrt")
write_vrt(items, asset_key, vrt_path)
reproject_to_chip(chip_info, vrt_path, output_path, resampling=resampling)
return output_path
# #### Configurate the auxiliary input files that we will generate.
# Define a set of parameters to pass into create_chip_aux_file
aux_file_params = [
("nasadem", "elevation", "nasadem.tif", Resampling.bilinear),
("jrc-gsw", "extent", "jrc-gsw-extent.tif", Resampling.nearest),
("jrc-gsw", "occurrence", "jrc-gsw-occurrence.tif", Resampling.nearest),
("jrc-gsw", "recurrence", "jrc-gsw-recurrence.tif", Resampling.nearest),
("jrc-gsw", "seasonality", "jrc-gsw-seasonality.tif", Resampling.nearest),
("jrc-gsw", "transitions", "jrc-gsw-transitions.tif", Resampling.nearest),
("jrc-gsw", "change", "jrc-gsw-change.tif", Resampling.nearest),
]
# #### Generate auxiliary input chips for NASADEM and JRC
# + tags=[]
# Iterate over the chips and generate all aux input files.
count = len(chip_paths)
for i, chip_path in enumerate(chip_paths):
print(f"({i+1} of {count}) {chip_path}")
chip_info = get_chip_info(chip_path)
for collection_id, asset_key, file_name, resampling_method in aux_file_params:
print(f" ... Creating chip data for {collection_id} {asset_key}")
create_chip_aux_file(
chip_info, collection_id, asset_key, file_name, resampling=resampling_method
)
# -
| generate_auxiliary_input.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %matplotlib inline
#
# # Recognizing hand-written digits
#
# This example shows how scikit-learn can be used to recognize images of
# hand-written digits, from 0-9.
#
# +
# Author: <NAME> <gael dot varoquaux at normalesup dot org>
# License: BSD 3 clause
# Standard scientific Python imports
import matplotlib.pyplot as plt
# Import datasets, classifiers and performance metrics
from sklearn import datasets, svm, metrics
from sklearn.model_selection import train_test_split
# -
# ## Digits dataset
#
# The digits dataset consists of 8x8
# pixel images of digits. The ``images`` attribute of the dataset stores
# 8x8 arrays of grayscale values for each image. We will use these arrays to
# visualize the first 4 images. The ``target`` attribute of the dataset stores
# the digit each image represents and this is included in the title of the 4
# plots below.
#
# Note: if we were working from image files (e.g., 'png' files), we would load
# them using :func:`matplotlib.pyplot.imread`.
#
#
# +
digits = datasets.load_digits()
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, label in zip(axes, digits.images, digits.target):
ax.set_axis_off()
ax.imshow(image, cmap=plt.cm.gray_r, interpolation="nearest")
ax.set_title("Training: %i" % label)
# -
# ## Classification
#
# To apply a classifier on this data, we need to flatten the images, turning
# each 2-D array of grayscale values from shape ``(8, 8)`` into shape
# ``(64,)``. Subsequently, the entire dataset will be of shape
# ``(n_samples, n_features)``, where ``n_samples`` is the number of images and
# ``n_features`` is the total number of pixels in each image.
#
# We can then split the data into train and test subsets and fit a support
# vector classifier on the train samples. The fitted classifier can
# subsequently be used to predict the value of the digit for the samples
# in the test subset.
#
#
# +
# flatten the images
n_samples = len(digits.images)
data = digits.images.reshape((n_samples, -1))
# Create a classifier: a support vector classifier
clf = svm.SVC(gamma=0.001)
# Split data into 50% train and 50% test subsets
X_train, X_test, y_train, y_test = train_test_split(
data, digits.target, test_size=0.5, shuffle=False
)
# Learn the digits on the train subset
clf.fit(X_train, y_train)
# Predict the value of the digit on the test subset
predicted = clf.predict(X_test)
# -
# Below we visualize the first 4 test samples and show their predicted
# digit value in the title.
#
#
_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))
for ax, image, prediction in zip(axes, X_test, predicted):
ax.set_axis_off()
image = image.reshape(8, 8)
ax.imshow(image, cmap=plt.cm.gray_r, interpolation="nearest")
ax.set_title(f"Prediction: {prediction}")
# :func:`~sklearn.metrics.classification_report` builds a text report showing
# the main classification metrics.
#
#
print(
f"Classification report for classifier {clf}:\n"
f"{metrics.classification_report(y_test, predicted)}\n"
)
# We can also plot a `confusion matrix <confusion_matrix>` of the
# true digit values and the predicted digit values.
#
#
# +
disp = metrics.ConfusionMatrixDisplay.from_predictions(y_test, predicted)
disp.figure_.suptitle("Confusion Matrix")
print(f"Confusion matrix:\n{disp.confusion_matrix}")
plt.show()
| exercises/deep_learning/.ipynb_checkpoints/plot_digits_classification-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# ## Classification of the Palmer penguins data
#
# This document is part of the showcase, where I replicate the same brief and simple analyses with different tools.
#
# This particular file focuses on simple classification of the Palmer penguins data from the tidytuesday project.
#
# The data can be found in <https://github.com/rfordatascience/tidytuesday/tree/master/data/2020/2020-07-28>. They consist of one documents: *penguins.csv* contains information and measurements about some penguins.
#
# For the specific analysis I will use **Python** and **scikit-learn** (plus **Jupyter notebook**).
#
# We start by loading the packages:
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
# and the dataset:
penguins = pd.read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-07-28/penguins.csv')
# We can have a look at the schema of the data:
penguins.info(verbose=True)
# and the summary statistics:
penguins.describe()
# Our main goal is to try and build a model that classifies the species of the penguins based on their other characteristics.
#
# We start by checking for missing values:
sns.heatmap(penguins.isnull(), cbar=False)
penguins.isna().sum()
# The plot shows that 5 of the features contain missing data (11 regard sex and 2 for each of the penguin measurements). In addition there are 2 penguins for which we have neither sex information nor the measurements, and 9 additional penguins for which we are only missing the sex.
#
# In practice, since this is a small amount of missing data we could drop all of them, but for the purposes of this showcase, we are going to drop only those that are missing the majority of information and impute the ones that are missing only the sex (later).
penguins = penguins.dropna(axis = 0, thresh=4, how = "all")
penguins.isna().sum()
# Now we can plot the classes:
penguins['species'].value_counts().plot.bar(color=['red', 'green', 'blue'])
# This is not very bad for a small and simple dataset like this, but once again for this showcase we are going to try and balance the classes (later).
#
# We can also plot the scatterplots, conditional distributions and boxplots and also check the individual correlations (for the continuous features).
#
# We exclude the year each penguin was recorded.
penguins = penguins.drop(columns='year')
sns.pairplot(penguins, hue="species")
# There are some pretty clear patterns patterns, so we are going to use all the features.
#
# First, we are going to split the data into features and label:
species = penguins.pop('species')
# Now we can split the dataset to training and testing:
X_train, X_test, y_train, y_test = train_test_split(penguins, species, test_size=0.2, random_state=1, stratify=species)
# The first model we are going to use is a support vector machine. We define the model and the pre-processing steps:
# * Switch all nominal predictors to one-hot encoding
# * k nearest neighbor imputation for the sex feature
# * Normalize all numeric predictors
# * Apply the classifier
# +
from sklearn.pipeline import Pipeline
from sklearn import svm
from sklearn.impute import KNNImputer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler
pipe_svc = Pipeline([ ('onehot', OneHotEncoder(handle_unknown='ignore', sparse=False)), ('imputer', KNNImputer(n_neighbors=2, weights="uniform")), ('scaler', StandardScaler(with_mean=False)), ('svc', svm.SVC())])
# -
# We fit the training data into the pipeline:
pipe_svc.fit(X_train, y_train)
# We can check the predictions of the testing data:
pipe_svc.score(X_test, y_test)
# And finally, we can check some metrics:
# +
from sklearn.metrics import classification_report
y_pred_svc = pipe_svc.predict(X_test)
print(classification_report(y_test, y_pred_svc))
# -
# We are also going to fit a simple decision tree model. We specify a new pipeline with the new algorithm and the same pre-processing steps:
from sklearn import tree
pipe_tree = Pipeline([ ('onehot', OneHotEncoder(handle_unknown='ignore', sparse=False)), ('imputer', KNNImputer(n_neighbors=2, weights="uniform")), ('scaler', StandardScaler(with_mean=False)), ('dec_tree', tree.DecisionTreeClassifier())])
# And fit on the training data data:
pipe_tree.fit(X_train, y_train)
# And once again test on the testing data:
pipe_tree.score(X_test, y_test)
# And check on the metrics:
y_pred_tree = pipe_tree.predict(X_test)
print(classification_report(y_test, y_pred_tree))
# In most aspects the support vector machine classifier performed significantly better, though it struggled with classifying the Chinstrap penguins.
| PalmerPenguins_Python_scikit-learn.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
#
# # Regular Grid Variogram in Python for Engineers and Geoscientists
#
# ## with GSLIB's GAMV Program Converted to Python
#
# ### <NAME>, Associate Professor, University of Texas at Austin
#
#
# #### Contacts: [Twitter/@GeostatsGuy](https://twitter.com/geostatsguy) | [GitHub/GeostatsGuy](https://github.com/GeostatsGuy) | [www.michaelpyrcz.com](http://michaelpyrcz.com) | [GoogleScholar](https://scholar.google.com/citations?user=QVZ20eQAAAAJ&hl=en&oi=ao) | [Book](https://www.amazon.com/Geostatistical-Reservoir-Modeling-Michael-Pyrcz/dp/0199731446)
#
# This is a tutorial for / demonstration of **Irregularly Sampled Variogram Calculation in Python with GSLIB's GAMV program translated to Python, wrappers and reimplementations of other GSLIB: Geostatistical Library methods** (Deutsch and Journel, 1997).
#
# This exercise demonstrates the semivariogram calculation method in Python with wrappers and reimplimentation of GSLIB methods. The steps include:
#
# 1. generate a 2D model with sequential Gaussian simulation
# 2. sample from the simulation
# 3. calculate and visualize experimental semivariograms
#
# To accomplish this I have provide wrappers or reimplementation in Python for the following GSLIB methods:
#
# 1. sgsim - sequantial Gaussian simulation limited to 2D and unconditional
# 2. hist - histograms plots reimplemented with GSLIB parameters using python methods
# 3. locmap - location maps reimplemented with GSLIB parameters using python methods
# 4. pixelplt - pixel plots reimplemented with GSLIB parameters using python methods
# 5. locpix - my modification of GSLIB to superimpose a location map on a pixel plot reimplemented with GSLIB parameters using Python methods
# 5. affine - affine correction adjust the mean and standard deviation of a feature reimplemented with GSLIB parameters using Python methods
#
# I have also started to translate the GSLIB support subfunctions to Python. Stay tuned.
#
# The GSLIB source and executables are available at http://www.statios.com/Quick/gslib.html. For the reference on using GSLIB check out the User Guide, GSLIB: Geostatistical Software Library and User's Guide by <NAME> and <NAME>. Overtime, more of the GSLIB programs will be translated to Python and there will be no need to have the executables. For this workflow you will need sgsim.exe from GSLIB.com for windows and Mac OS executables from https://github.com/GeostatsGuy/GSLIB_MacOS.
#
# I did this to allow people to use these GSLIB functions that are extremely robust in Python. Also this should be a bridge to allow so many familar with GSLIB to work in Python as a kept the parameterization and displays consistent with GSLIB. The wrappers are simple functions declared below that write the parameter files, run the GSLIB executable in the working directory and load and visualize the output in Python. This will be included on GitHub for anyone to try it out https://github.com/GeostatsGuy/.
#
# This was my first effort to translate the GSLIB Fortran to Python. It was pretty easy so I'll start translating other critical GSLIB functions. I've completed NSCORE, DECLUS, GAM and now GAMV as of now.
#
# #### Load the required libraries
#
# The following code loads the required libraries.
import os # to set current working directory
import numpy as np # arrays and matrix math
import pandas as pd # DataFrames
import matplotlib.pyplot as plt # plotting
# If you get a package import error, you may have to first install some of these packages. This can usually be accomplished by opening up a command window on Windows and then typing 'python -m pip install [package-name]'. More assistance is available with the respective package docs.
#
# #### Declare functions
#
# Here are the wrappers and reimplementations of GSLIB method along with two utilities to load GSLIB's Geo-EAS from data files into DataFrames and 2D Numpy arrays. These are used in the testing workflow.
# +
# Some GeostatsPy Functions - by <NAME>, maintained at https://git.io/fNgR7.
# A set of functions to provide access to GSLIB in Python.
# GSLIB executables: nscore.exe, declus.exe, gam.exe, gamv.exe, vmodel.exe, kb2d.exe & sgsim.exe must be in the working directory
import pandas as pd
import os
import numpy as np
import matplotlib.pyplot as plt
import random as rand
image_type = 'tif'; dpi = 600
# utility to convert GSLIB Geo-EAS files to a 1D or 2D numpy ndarray for use with Python methods
def GSLIB2ndarray(data_file,kcol,nx,ny):
colArray = []
if ny > 1:
array = np.ndarray(shape=(ny,nx),dtype=float,order='F')
else:
array = np.zeros(nx)
with open(data_file) as myfile: # read first two lines
head = [next(myfile) for x in range(2)]
line2 = head[1].split()
ncol = int(line2[0]) # get the number of columns
for icol in range(0, ncol): # read over the column names
head = [next(myfile) for x in range(1)]
if icol == kcol:
col_name = head[0].split()[0]
if ny > 1:
for iy in range(0,ny):
for ix in range(0,nx):
head = [next(myfile) for x in range(1)]
array[ny-1-iy][ix] = head[0].split()[kcol]
else:
for ix in range(0,nx):
head = [next(myfile) for x in range(1)]
array[ix] = head[0].split()[kcol]
return array,col_name
# utility to convert GSLIB Geo-EAS files to a pandas DataFrame for use with Python methods
def GSLIB2Dataframe(data_file):
colArray = []
with open(data_file) as myfile: # read first two lines
head = [next(myfile) for x in range(2)]
line2 = head[1].split()
ncol = int(line2[0])
for icol in range(0, ncol):
head = [next(myfile) for x in range(1)]
colArray.append(head[0].split()[0])
data = np.loadtxt(myfile, skiprows = 0)
df = pd.DataFrame(data)
df.columns = colArray
return df
# histogram, reimplemented in Python of GSLIB hist with MatPlotLib methods, displayed and as image file
def hist(array,xmin,xmax,log,cumul,bins,weights,xlabel,title,fig_name):
plt.figure(figsize=(8,6))
cs = plt.hist(array, alpha = 0.2, color = 'red', edgecolor = 'black', bins=bins, range = [xmin,xmax], weights = weights, log = log, cumulative = cumul)
plt.title(title)
plt.xlabel(xlabel); plt.ylabel('Frequency')
plt.savefig(fig_name + '.' + image_type,dpi=dpi)
plt.show()
return
# histogram, reimplemented in Python of GSLIB hist with MatPlotLib methods (version for subplots)
def hist_st(array,xmin,xmax,log,cumul,bins,weights,xlabel,title):
cs = plt.hist(array, alpha = 0.2, color = 'red', edgecolor = 'black', bins=bins, range = [xmin,xmax], weights = weights, log = log, cumulative = cumul)
plt.title(title)
plt.xlabel(xlabel); plt.ylabel('Frequency')
return
# location map, reimplemention in Python of GSLIB locmap with MatPlotLib methods
def locmap(df,xcol,ycol,vcol,xmin,xmax,ymin,ymax,vmin,vmax,title,xlabel,ylabel,vlabel,cmap,fig_name):
ixy = 0
plt.figure(figsize=(8,6))
im = plt.scatter(df[xcol],df[ycol],s=None, c=df[vcol], marker=None, cmap=cmap, norm=None, vmin=vmin, vmax=vmax, alpha=0.8, linewidths=0.8, edgecolors="black")
plt.title(title)
plt.xlim(xmin,xmax)
plt.ylim(ymin,ymax)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
cbar = plt.colorbar(im, orientation = 'vertical',ticks=np.linspace(vmin,vmax,10))
cbar.set_label(vlabel, rotation=270, labelpad=20)
plt.savefig(fig_name + '.' + image_type,dpi=dpi)
plt.show()
return im
# location map, reimplemention in Python of GSLIB locmap with MatPlotLib methods (version for subplots)
def locmap_st(df,xcol,ycol,vcol,xmin,xmax,ymin,ymax,vmin,vmax,title,xlabel,ylabel,vlabel,cmap):
ixy = 0
im = plt.scatter(df[xcol],df[ycol],s=None, c=df[vcol], marker=None, cmap=cmap, norm=None, vmin=vmin, vmax=vmax, alpha=0.8, linewidths=0.8, verts=None, edgecolors="black")
plt.title(title)
plt.xlim(xmin,xmax)
plt.ylim(ymin,ymax)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
cbar = plt.colorbar(im, orientation = 'vertical',ticks=np.linspace(vmin,vmax,10))
cbar.set_label(vlabel, rotation=270, labelpad=20)
return im
# pixel plot, reimplemention in Python of GSLIB pixelplt with MatPlotLib methods
def pixelplt(array,xmin,xmax,ymin,ymax,step,vmin,vmax,title,xlabel,ylabel,vlabel,cmap,fig_name):
print(str(step))
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax, ymin, -1*step))
plt.figure(figsize=(8,6))
im = plt.contourf(xx,yy,array,cmap=cmap,vmin=vmin,vmax=vmax,levels=np.linspace(vmin,vmax,100))
plt.title(title)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
cbar = plt.colorbar(im,orientation = 'vertical',ticks=np.linspace(vmin,vmax,10))
cbar.set_label(vlabel, rotation=270, labelpad=20)
plt.savefig(fig_name + '.' + image_type,dpi=dpi)
plt.show()
return im
# pixel plot, reimplemention in Python of GSLIB pixelplt with MatPlotLib methods(version for subplots)
def pixelplt_st(array,xmin,xmax,ymin,ymax,step,vmin,vmax,title,xlabel,ylabel,vlabel,cmap):
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax, ymin, -1*step))
ixy = 0
x = [];y = []; v = [] # use dummy since scatter plot controls legend min and max appropriately and contour does not!
cs = plt.contourf(xx,yy,array,cmap=cmap,vmin=vmin,vmax=vmax,levels = np.linspace(vmin,vmax,100))
im = plt.scatter(x,y,s=None, c=v, marker=None,cmap=cmap, vmin=vmin, vmax=vmax, alpha=0.8, linewidths=0.8, edgecolors="black")
plt.title(title)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
plt.clim(vmin,vmax)
cbar = plt.colorbar(im, orientation = 'vertical')
cbar.set_label(vlabel, rotation=270, labelpad=20)
return cs
# pixel plot and location map, reimplementation in Python of a GSLIB MOD with MatPlotLib methods
def locpix(array,xmin,xmax,ymin,ymax,step,vmin,vmax,df,xcol,ycol,vcol,title,xlabel,ylabel,vlabel,cmap,fig_name):
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax, ymin, -1*step))
ixy = 0
plt.figure(figsize=(8,6))
cs = plt.contourf(xx, yy, array, cmap=cmap,vmin=vmin, vmax=vmax,levels = np.linspace(vmin,vmax,100))
im = plt.scatter(df[xcol],df[ycol],s=None, c=df[vcol], marker=None, cmap=cmap, vmin=vmin, vmax=vmax, alpha=0.8, linewidths=0.8, edgecolors="black")
plt.title(title)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
plt.xlim(xmin,xmax)
plt.ylim(ymin,ymax)
cbar = plt.colorbar(orientation = 'vertical')
cbar.set_label(vlabel, rotation=270, labelpad=20)
plt.savefig(fig_name + '.' + image_type,dpi=dpi)
plt.show()
return cs
# pixel plot and location map, reimplementation in Python of a GSLIB MOD with MatPlotLib methods(version for subplots)
def locpix_st(array,xmin,xmax,ymin,ymax,step,vmin,vmax,df,xcol,ycol,vcol,title,xlabel,ylabel,vlabel,cmap):
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax, ymin, -1*step))
ixy = 0
cs = plt.contourf(xx, yy, array, cmap=cmap,vmin=vmin, vmax=vmax,levels = np.linspace(vmin,vmax,100))
im = plt.scatter(df[xcol],df[ycol],s=None, c=df[vcol], marker=None, cmap=cmap, vmin=vmin, vmax=vmax, alpha=0.8, linewidths=0.8, edgecolors="black")
plt.title(title)
plt.xlabel(xlabel)
plt.ylabel(ylabel)
plt.xlim(xmin,xmax)
plt.ylim(ymin,ymax)
cbar = plt.colorbar(orientation = 'vertical')
cbar.set_label(vlabel, rotation=270, labelpad=20)
# affine distribution correction reimplemented in Python with numpy methods
def affine(array,tmean,tstdev):
mean = np.average(array)
stdev = np.std(array)
array = (tstdev/stdev)*(array - mean) + tmean
return(array)
def make_variogram(nug,nst,it1,cc1,azi1,hmaj1,hmin1,it2=1,cc2=0,azi2=0,hmaj2=0,hmin2=0):
if cc2 == 0:
nst = 1
var = dict([('nug', nug), ('nst', nst), ('it1', it1),('cc1', cc1),('azi1', azi1),('hmaj1', hmaj1), ('hmin1', hmin1),
('it2', it2),('cc2', cc2),('azi2', azi2),('hmaj2', hmaj2), ('hmin2', hmin2)])
if nug + cc1 + cc2 != 1:
print('\x1b[0;30;41m make_variogram Warning: sill does not sum to 1.0, do not use in simulation \x1b[0m')
if cc1 < 0 or cc2 < 0 or nug < 0 or hmaj1 < 0 or hmaj2 < 0 or hmin1 < 0 or hmin2 < 0:
print('\x1b[0;30;41m make_variogram Warning: contributions and ranges must be all positive \x1b[0m')
if hmaj1 < hmin1 or hmaj2 < hmin2:
print('\x1b[0;30;41m make_variogram Warning: major range should be greater than minor range \x1b[0m')
return var
# sequential Gaussian simulation, 2D unconditional wrapper for sgsim from GSLIB (.exe must be in working directory)
def GSLIB_sgsim_2d_uncond(nreal,nx,ny,hsiz,seed,var,output_file):
import os
import numpy as np
nug = var['nug']
nst = var['nst']; it1 = var['it1']; cc1 = var['cc1']; azi1 = var['azi1']; hmaj1 = var['hmaj1']; hmin1 = var['hmin1']
it2 = var['it2']; cc2 = var['cc2']; azi2 = var['azi2']; hmaj2 = var['hmaj2']; hmin2 = var['hmin2']
max_range = max(hmaj1,hmaj2)
hmn = hsiz * 0.5
hctab = int(max_range/hsiz)*2 + 1
sim_array = np.random.rand(nx,ny)
file = open("sgsim.par", "w")
file.write(" Parameters for SGSIM \n")
file.write(" ******************** \n")
file.write(" \n")
file.write("START OF PARAMETER: \n")
file.write("none -file with data \n")
file.write("1 2 0 3 5 0 - columns for X,Y,Z,vr,wt,sec.var. \n")
file.write("-1.0e21 1.0e21 - trimming limits \n")
file.write("0 -transform the data (0=no, 1=yes) \n")
file.write("none.trn - file for output trans table \n")
file.write("1 - consider ref. dist (0=no, 1=yes) \n")
file.write("none.dat - file with ref. dist distribution \n")
file.write("1 0 - columns for vr and wt \n")
file.write("-4.0 4.0 - zmin,zmax(tail extrapolation) \n")
file.write("1 -4.0 - lower tail option, parameter \n")
file.write("1 4.0 - upper tail option, parameter \n")
file.write("0 -debugging level: 0,1,2,3 \n")
file.write("nonw.dbg -file for debugging output \n")
file.write(str(output_file) + " -file for simulation output \n")
file.write(str(nreal) + " -number of realizations to generate \n")
file.write(str(nx) + " " + str(hmn) + " " + str(hsiz) + " \n")
file.write(str(ny) + " " + str(hmn) + " " + str(hsiz) + " \n")
file.write("1 0.0 1.0 - nz zmn zsiz \n")
file.write(str(seed) + " -random number seed \n")
file.write("0 8 -min and max original data for sim \n")
file.write("12 -number of simulated nodes to use \n")
file.write("0 -assign data to nodes (0=no, 1=yes) \n")
file.write("1 3 -multiple grid search (0=no, 1=yes),num \n")
file.write("0 -maximum data per octant (0=not used) \n")
file.write(str(max_range) + " " + str(max_range) + " 1.0 -maximum search (hmax,hmin,vert) \n")
file.write(str(azi1) + " 0.0 0.0 -angles for search ellipsoid \n")
file.write(str(hctab) + " " + str(hctab) + " 1 -size of covariance lookup table \n")
file.write("0 0.60 1.0 -ktype: 0=SK,1=OK,2=LVM,3=EXDR,4=COLC \n")
file.write("none.dat - file with LVM, EXDR, or COLC variable \n")
file.write("4 - column for secondary variable \n")
file.write(str(nst) + " " + str(nug) + " -nst, nugget effect \n")
file.write(str(it1) + " " + str(cc1) + " " +str(azi1) + " 0.0 0.0 -it,cc,ang1,ang2,ang3\n")
file.write(" " + str(hmaj1) + " " + str(hmin1) + " 1.0 - a_hmax, a_hmin, a_vert \n")
file.write(str(it2) + " " + str(cc2) + " " +str(azi2) + " 0.0 0.0 -it,cc,ang1,ang2,ang3\n")
file.write(" " + str(hmaj2) + " " + str(hmin2) + " 1.0 - a_hmax, a_hmin, a_vert \n")
file.close()
os.system('"sgsim.exe sgsim.par"')
sim_array = GSLIB2ndarray(output_file,0,nx,ny)
return(sim_array[0])
# extract regular spaced samples from a model
def regular_sample(array,xmin,xmax,ymin,ymax,step,mx,my,name):
x = []; y = []; v = []; iix = 0; iiy = 0;
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax, ymin, -1*step))
iiy = 0
for iy in range(0,ny):
if iiy >= my:
iix = 0
for ix in range(0,nx):
if iix >= mx:
x.append(xx[ix,iy]);y.append(yy[ix,iy]); v.append(array[ix,iy])
iix = 0; iiy = 0
iix = iix + 1
iiy = iiy + 1
df = pd.DataFrame(np.c_[x,y,v],columns=['X', 'Y', name])
return(df)
def random_sample(array,xmin,xmax,ymin,ymax,step,nsamp,name):
import random as rand
x = []; y = []; v = []; iix = 0; iiy = 0;
xx, yy = np.meshgrid(np.arange(xmin, xmax, step),np.arange(ymax-1, ymin-1, -1*step))
ny = xx.shape[0]
nx = xx.shape[1]
sample_index = rand.sample(range((nx)*(ny)), nsamp)
for isamp in range(0,nsamp):
iy = int(sample_index[isamp]/ny)
ix = sample_index[isamp] - iy*nx
x.append(xx[iy,ix])
y.append(yy[iy,ix])
v.append(array[iy,ix])
df = pd.DataFrame(np.c_[x,y,v],columns=['X', 'Y', name])
return(df)
# -
# Here's the GAMV program translated to Python. Note: it was simplified to run just one experimental semivariogram at a time (in a simgle direction) and only for 2D datasets. I have applied Numba to speedup the required double loop over the data.
# +
import math # for trig and constants
from numba import jit # for precompile speed up of loops with NumPy ndarrays
# GSLIB's GAMV program (Deutsch and Journel, 1998) converted from the original Fortran to Python
# by <NAME>, the University of Texas at Austin (Jan, 2019)
# Note simplified for 2D, semivariogram only and one direction at a time
def gamv(df,xcol,ycol,vcol,tmin,tmax,xlag,xltol,nlag,azm,atol,bandwh,isill):
# Parameters - consistent with original GSLIB
# df - DataFrame with the spatial data, xcol, ycol, vcol coordinates and property columns
# tmin, tmax - property trimming limits
# xlag, xltol - lag distance and lag distance tolerance
# nlag - number of lags to calculate
# azm, atol - azimuth and azimuth tolerance
# bandwh - horizontal bandwidth / maximum distance offset orthogonal to azimuth
# isill - 1 for standardize sill
# Load the data
df_extract = df.loc[(df[vcol] >= tmin) & (df[vcol] <= tmax)] # trim values outside tmin and tmax
nd = len(df_extract)
x = df_extract[xcol].values
y = df_extract[ycol].values
vr = df_extract[vcol].values
# Summary statistics for the data after trimming
avg = vr.mean()
stdev = vr.std()
sills = stdev**2.0
ssq = sills
vrmin = vr.min()
vrmax = vr.max()
#print('Number of Data ' + str(nd) +', Average ' + str(avg) + ' Variance ' + str(sills))
# Define the distance tolerance if it isn't already:
if xltol < 0.0: xltol = 0.5 * xlag
# Loop over combinatorial of data pairs to calculate the variogram
dis, vario, npp = variogram_loop(x,y,vr,xlag,xltol,nlag,azm,atol,bandwh)
# Standardize sill to one by dividing all variogram values by the variance
for il in range(0,nlag+2):
if isill == 1:
vario[il] = vario[il] / sills
# Apply 1/2 factor to go from variogram to semivariogram
vario[il] = 0.5 * vario[il]
# END - return variogram model information
return dis, vario, npp
@jit(nopython=True) # all NumPy array operations included in this function for precompile with NumBa
def variogram_loop(x,y,vr,xlag,xltol,nlag,azm,atol,bandwh):
# Allocate the needed memory:
nvarg = 1
mxdlv = nlag + 2 # in gamv the npp etc. arrays go to nlag + 2
dis = np.zeros(mxdlv)
lag = np.zeros(mxdlv)
vario = np.zeros(mxdlv)
hm = np.zeros(mxdlv)
tm = np.zeros(mxdlv)
hv = np.zeros(mxdlv)
npp = np.zeros(mxdlv)
ivtail = np.zeros(nvarg + 2)
ivhead = np.zeros(nvarg + 2)
ivtype = np.ones(nvarg + 2)
ivtail[0] = 0; ivhead[0] = 0; ivtype[0] = 0;
EPSLON = 1.0e-20
nd = len(x)
# The mathematical azimuth is measured counterclockwise from EW and
# not clockwise from NS as the conventional azimuth is:
azmuth = (90.0-azm)*math.pi/180.0
uvxazm = math.cos(azmuth)
uvyazm = math.sin(azmuth)
if atol <= 0.0:
csatol = math.cos(45.0*math.pi/180.0)
else:
csatol = math.cos(atol*math.pi/180.0)
# Initialize the arrays for each direction, variogram, and lag:
nsiz = nlag+2
dismxs = ((float(nlag) + 0.5 - EPSLON) * xlag) ** 2
# MAIN LOOP OVER ALL PAIRS:
for i in range(0,nd):
for j in range(0,nd):
# Definition of the lag corresponding to the current pair:
dx = x[j] - x[i]
dy = y[j] - y[i]
dxs = dx*dx
dys = dy*dy
hs = dxs + dys
if hs <= dismxs:
if hs < 0.0:
hs = 0.0
h = np.sqrt(hs)
# Determine which lag this is and skip if outside the defined distance
# tolerance:
if h <= EPSLON:
lagbeg = 0
lagend = 0
else:
lagbeg = -1
lagend = -1
for ilag in range(1,nlag+1):
if h >= (xlag*float(ilag-1)-xltol) and h <= (xlag*float(ilag-1)+xltol): # reduced to -1
if lagbeg < 0:
lagbeg = ilag
lagend = ilag
if lagend >= 0:
# Definition of the direction corresponding to the current pair. All
# directions are considered (overlapping of direction tolerance cones
# is allowed):
# Check for an acceptable azimuth angle:
dxy = np.sqrt(max((dxs+dys),0.0))
if dxy < EPSLON:
dcazm = 1.0
else:
dcazm = (dx*uvxazm+dy*uvyazm)/dxy
# Check the horizontal bandwidth criteria (maximum deviation
# perpendicular to the specified direction azimuth):
band = uvxazm*dy - uvyazm*dx
# Apply all the previous checks at once to avoid a lot of nested if statements
if (abs(dcazm) >= csatol) and (abs(band) <= bandwh):
# Check whether or not an omni-directional variogram is being computed:
omni = False
if atol >= 90.0: omni = True
# For this variogram, sort out which is the tail and the head value:
iv = 0 # hardcoded just one varioigram
it = ivtype[iv]
if dcazm >= 0.0:
vrh = vr[i]
vrt = vr[j]
if omni:
vrtpr = vr[i]
vrhpr = vr[j]
else:
vrh = vr[j]
vrt = vr[i]
if omni:
vrtpr = vr[j]
vrhpr = vr[i]
# Reject this pair on the basis of missing values:
# Data was trimmed at the beginning
# The Semivariogram (all other types of measures are removed for now)
for il in range(lagbeg,lagend+1):
npp[il] = npp[il] + 1
dis[il] = dis[il] + h
tm[il] = tm[il] + vrt
hm[il] = hm[il] + vrh
vario[il] = vario[il] + ((vrh-vrt)*(vrh-vrt))
if(omni):
npp[il] = npp[il] + 1.0
dis[il] = dis[il] + h
tm[il] = tm[il] + vrtpr
hm[il] = hm[il] + vrhpr
vario[il] = vario[il] + ((vrhpr-vrtpr)*(vrhpr-vrtpr))
# Get average values for gam, hm, tm, hv, and tv, then compute
# the correct "variogram" measure:
for il in range(0,nlag+2):
i = il
if npp[i] > 0:
rnum = npp[i]
dis[i] = dis[i] / (rnum)
vario[i] = vario[i] / (rnum)
hm[i] = hm[i] / (rnum)
tm[i] = tm[i] / (rnum)
return dis, vario, npp
# -
# Here's a simple test of the GAMV code with visualizations to check the results including the gridded data pixelplt, histogram and experimental semivariograms in 4 directions.
#
# #### Set the working directory
#
# I always like to do this so I don't lose files and to simplify subsequent read and writes (avoid including the full address each time). Also, in this case make sure to place the required (see above) GSLIB executables in this directory or a location identified in the environmental variable *Path*.
os.chdir("c:/PGE337") # set the working directory
# You will have to update the part in quotes with your own working directory and the format is different on a Mac (e.g. "~/PGE").
#
# ##### Make a 2D spatial model
#
# The following are the basic parameters for the demonstration. This includes the number of cells in the 2D regular grid, the cell size (step) and the x and y min and max along with the color scheme.
#
# Then we make a single realization of a Gausian distributed feature over the specified 2D grid and then apply affine correction to ensure we have a reasonable mean and spread for our feature's distribution, assumed to be Porosity (e.g. no negative values) while retaining the Gaussian distribution. Any transform could be applied at this point. We are keeping this workflow simple. *This is our truth model that we will sample*.
#
# The parameters of *GSLIB_sgsim_2d_uncond* are (nreal,nx,ny,hsiz,seed,hrange1,hrange2,azi,output_file). nreal is the number of realizations, nx and ny are the number of cells in x and y, hsiz is the cell siz, seed is the random number seed, hrange and hrange2 are the variogram ranges in major and minor directions respectively, azi is the azimuth of the primary direction of continuity (0 is aligned with Y axis) and output_file is a GEO_DAS file with the simulated realization. The ouput is the 2D numpy array of the simulation along with the name of the property.
# +
nx = 100; ny = 100; cell_size = 10 # grid number of cells and cell size
xmin = 0.0; ymin = 0.0; # grid origin
xmax = xmin + nx * cell_size; ymax = ymin + ny * cell_size # calculate the extent of model
seed = 74073 # random number seed for stochastic simulation
vario = make_variogram(0.0,nst=1,it1=1,cc1=1.0,azi1=0,hmaj1=500,hmin1=500)
mean = 10.0; stdev = 2.0 # Porosity mean and standard deviation
#cmap = plt.cm.RdYlBu
vmin = 4; vmax = 16; cmap = plt.cm.plasma # color min and max and using the plasma color map
# calculate a stochastic realization with standard normal distribution
sim = GSLIB_sgsim_2d_uncond(1,nx,ny,cell_size,seed,vario,"Por")
sim = affine(sim,mean,stdev) # correct the distribution to a target mean and standard deviation.
sampling_ncell = 10 # sample every 10th node from the model
#samples = regular_sample(sim,xmin,xmax,ymin,ymax,sampling_ncell,30,30,'Realization')
#samples_cluster = samples.drop([80,79,78,73,72,71,70,65,64,63,61,57,56,54,53,47,45,42]) # this removes specific rows (samples)
#samples_cluster = samples_cluster.reset_index(drop=True) # we reset and remove the index (it is not sequential anymore)
samples = random_sample(sim,xmin,xmax,ymin,ymax,cell_size,100,"Por")
locpix(sim,xmin,xmax,ymin,ymax,cell_size,vmin,vmax,samples,'X','Y','Por','Porosity Realization and Regular Samples','X(m)','Y(m)','Porosity (%)',cmap,"Por_Samples")
# -
# Below I calculate the isotropic, 000 and 090 directional experimental semivariograms from our sample set. Then the variograms are visualized together on the same variogram plot.
# +
tmin = -9999.; tmax = 9999.
lagiso, varioiso, nppiso = gamv(samples,'X','Y','Por',tmin,tmax,xlag = 100,xltol = 50,nlag = 10,azm = 0,atol = 90.0,bandwh = 9999,isill = 1)
lag000, vario000, npp000 = gamv(samples,'X','Y','Por',tmin,tmax,xlag = 100,xltol = 50,nlag = 10,azm = 0,atol = 22.5,bandwh = 9999,isill = 1)
lag090, vario090, npp090 = gamv(samples,'X','Y','Por',tmin,tmax,xlag = 100,xltol = 50,nlag = 10,azm = 90,atol = 22.5,bandwh = 9999,isill = 1)
plt.subplot(121)
locpix_st(sim,xmin,xmax,ymin,ymax,cell_size,vmin,vmax,samples,'X','Y','Por','Porosity Realization and Random Samples','X(m)','Y(m)','Porosity (%)',cmap)
plt.subplot(122)
plt.scatter(lagiso,varioiso,s=nppiso/len(samples),marker='x',color = 'black',label = 'Iso')
plt.scatter(lag000,vario000,s=npp000/len(samples),marker='o',color = 'blue',label = '000')
plt.scatter(lag090,vario090,s=npp090/len(samples),marker='o',color = 'green',label = '090')
plt.plot([0,1000],[1.0,1.0],color = 'black')
plt.xlabel('Lag Distance(m)')
plt.ylabel('Semivariogram')
plt.title('Iregular Samples Experimental Variograms')
plt.ylim(0,1.5)
plt.xlim(0,1000)
handles, labels = plt.gca().get_legend_handles_labels()
plt.gca().legend(handles[::], labels[::])
plt.subplots_adjust(left=0.0, bottom=0.0, right=2.0, top=1.0, wspace=0.3, hspace=0.3)
plt.show()
# -
# Here's the gamv program without the numba acceleration, just incase that is helpful. Warning it is quite slow with more than a couple hundred data samples.
# +
import math
# GSLIB's GAM program (Deutsch and Journel, 1998) converted from the original Fortran to Python
# by <NAME>, the University of Texas at Austin (Jan, 2019)
def gamv(df,xcol,ycol,vcol,tmin,tmax,xlag,xltol,nlag,azm,atol,isill):
# Parameters - consistent with original GSLIB
# array - 2D gridded data / model
# tmin, tmax - property trimming limits
# xsiz, ysiz - grid cell extents in x and y directions
# ixd, iyd - lag offset in grid cells
# nlag - number of lags to calculate
# isill - 1 for standardize sill
#
nvarg = 1 # for mulitple variograms repeat the program
nxy = nx*ny
mxdlv = nlag + 2 # in gamv the npp etc. arrays go to nlag + 2
dip = 0.0; dtol = 1.0 # hard code for 2D for now
EPSLON = 1.0e-20
bandwh = 1.0e20
bandwd = 1.0e20
# Allocate the needed memory:
dis = np.zeros(mxdlv)
lag = np.zeros(mxdlv)
vario = np.zeros(mxdlv)
hm = np.zeros(mxdlv)
tm = np.zeros(mxdlv)
hv = np.zeros(mxdlv)
npp = np.zeros(mxdlv)
ivtail = np.zeros(nvarg + 2)
ivhead = np.zeros(nvarg + 2)
ivtype = np.ones(nvarg + 2)
ivtail[0] = 0; ivhead[0] = 0; ivtype[0] = 0;
# Load the data
df_extract = df.loc[(df[vcol] >= tmin) & (df[vcol] <= tmax)] # trim values outside tmin and tmax
nd = len(df_extract)
x = df_extract[xcol]
y = df_extract[ycol]
vr = df_extract[vcol]
# Summary statistics for the data after trimming
avg = vr.mean()
stdev = vr.std()
sills = stdev**2.0
ssq = sills
vrmin = vr.min()
vrmax = vr.max()
#print('Number of Data ' + str(nd) +', Average ' + str(avg) + ' Variance ' + str(sills))
# Define the distance tolerance if it isn't already:
if xltol < 0.0: xltol = 0.5 * xlag
# Removed loop over directions
# The mathematical azimuth is measured counterclockwise from EW and
# not clockwise from NS as the conventional azimuth is:
azmuth = (90.0-azm)*math.pi/180.0
uvxazm = math.cos(azmuth)
uvyazm = math.sin(azmuth)
if atol <= 0.0:
csatol = math.cos(45.0*math.pi/180.0)
else:
csatol = math.cos(atol*math.pi/180.0)
# The declination is measured positive down from vertical (up) rather
# than negative down from horizontal:
declin = (90.0-dip)*math.pi/180.0
uvzdec = math.cos(declin)
uvhdec = math.sin(declin)
if dtol <= 0.0:
csdtol = math.cos(45.0*math.pi/180.0)
else:
csdtol = math.cos(dtol*math.pi/180.0)
# Initialize the arrays for each direction, variogram, and lag:
nsiz = nlag+2
dismxs = ((float(nlag) + 0.5 - EPSLON) * xlag) ** 2
# MAIN LOOP OVER ALL PAIRS:
irepo = max(1,min((nd/10),1000))
for i in range(0,nd):
# if((int(i/irepo)*irepo) == i):
# print( ' currently on seed point ' + str(i) + ' of '+ str(nd))
for j in range(0,nd):
# Definition of the lag corresponding to the current pair:
dx = x[j] - x[i]
dy = y[j] - y[i]
dxs = dx*dx
dys = dy*dy
hs = dxs + dys
if hs <= dismxs:
if hs < 0.0:
hs = 0.0
h = np.sqrt(hs)
# Determine which lag this is and skip if outside the defined distance
# tolerance:
if h <= EPSLON:
lagbeg = 0
lagend = 0
else:
lagbeg = -1
lagend = -1
for ilag in range(1,nlag+1):
if h >= (xlag*float(ilag-1)-xltol) and h <= (xlag*float(ilag-1)+xltol): # reduced to -1
if lagbeg < 0:
lagbeg = ilag
lagend = ilag
if lagend >= 0:
# Definition of the direction corresponding to the current pair. All
# directions are considered (overlapping of direction tolerance cones
# is allowed):
# Check for an acceptable azimuth angle:
dxy = np.sqrt(max((dxs+dys),0.0))
if dxy < EPSLON:
dcazm = 1.0
else:
dcazm = (dx*uvxazm+dy*uvyazm)/dxy
# Check the horizontal bandwidth criteria (maximum deviation
# perpendicular to the specified direction azimuth):
band = uvxazm*dy - uvyazm*dx
# Apply all the previous checks at once to avoid a lot of nested if statements
if (abs(dcazm) >= csatol) and (abs(band) <= bandwh):
# Check whether or not an omni-directional variogram is being computed:
omni = False
if atol >= 90.0: omni = True
# For this variogram, sort out which is the tail and the head value:
iv = 0 # hardcoded just one varioigram
it = ivtype[iv]
if dcazm >= 0.0:
vrh = vr[i]
vrt = vr[j]
if omni:
vrtpr = vr[i]
vrhpr = vr[j]
else:
vrh = vr[j]
vrt = vr[i]
if omni:
vrtpr = vr[j]
vrhpr = vr[i]
# Reject this pair on the basis of missing values:
# Data was trimmed at the beginning
# The Semivariogram:
for il in range(lagbeg,lagend+1):
npp[il] = npp[il] + 1
dis[il] = dis[il] + h
tm[il] = tm[il] + vrt
hm[il] = hm[il] + vrh
vario[il] = vario[il] + ((vrh-vrt)*(vrh-vrt))
if(omni):
npp[il] = npp[il] + 1.0
dis[il] = dis[il] + h
tm[il] = tm[il] + vrtpr
hm[il] = hm[il] + vrhpr
vario[il] = vario[il] + ((vrhpr-vrtpr)*(vrhpr-vrtpr))
# Get average values for gam, hm, tm, hv, and tv, then compute
# the correct "variogram" measure:
for il in range(0,nlag+2):
i = il
if npp[i] > 0:
rnum = npp[i]
dis[i] = dis[i] / (rnum)
vario[i] = vario[i] / (rnum)
hm[i] = hm[i] / (rnum)
tm[i] = tm[i] / (rnum)
# Attempt to standardize:
if isill == 1:
vario[i] = vario[i] / sills
# semivariogram
vario[i] = 0.5 * vario[i]
return dis, vario, npp
# -
# I hope you find this code and demonstration useful. I'm always happy to discuss geostatistics, statistical modeling, uncertainty modeling and machine learning,
#
# *Michael*
#
# **<NAME>**, Ph.D., P.Eng. Associate Professor The Hildebrand Department of Petroleum and Geosystems Engineering, Bureau of Economic Geology, The Jackson School of Geosciences, The University of Texas at Austin
# On Twitter I'm the **GeostatsGuy** and on YouTube my lectures are on the channel, **GeostatsGuy Lectures**.
| examples/Gamv.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
# # Данные для тестов
def malanchev_dataset(inliers=2**10, outliers=2**5, seed=0):
rng = np.random.default_rng(seed)
x = np.concatenate([rng.uniform([0, 0], [0.5, 0.5], (inliers, 2)),
rng.normal([1, 1], 0.1, (outliers, 2)),
rng.normal([0, 1], 0.1, (outliers, 2)),
rng.normal([1, 0], 0.1, (outliers, 2))])
return x
plt.scatter(*malanchev_dataset().T);
# # Оценка средней длины пути в дереве
# +
def _average_path_length(n):
"""
Average path length formula.
"""
return 2.0 * (np.log(n - 1.0) + np.euler_gamma) - 2.0 * (n - 1.0) / n
def average_path_length(n):
"""
Average path length computation.
Parameters
----------
n
Either array of tree depths to computer average path length of
or one tree depth scalar.
Returns
-------
Average path length.
"""
if np.isscalar(n):
if n <= 1:
apl = 0
elif n == 2:
apl = 1
else:
apl = _average_path_length(n)
else:
n = np.asarray(n)
apl = np.zeros_like(n)
apl[n > 1] = _average_path_length(n[n > 1])
apl[n == 2] = 1
return apl
# -
# # Изоляционный лес
# +
class IsolationForest:
def __init__(self, trees=100, subsamples=256, depth=None, seed=0):
self.subsamples = subsamples
self.trees = trees
self.depth = depth
self.seedseq = np.random.SeedSequence(seed)
self.rng = np.random.default_rng(seed)
self.estimators = []
self.n = 0
def fit(self, data):
n = data.shape[0]
self.n = n
self.subsamples = self.subsamples if n > self.subsamples else n
self.depth = self.depth or int(np.ceil(np.log2(self.subsamples)))
self.estimators = [None] * self.trees
seeds = self.seedseq.spawn(self.trees)
for i in range(self.trees):
subs = self.rng.choice(n, self.subsamples)
gen = IsolationForestGenerator(data[subs, :], self.depth, seeds[i])
self.estimators[i] = gen.pine
return self
def mean_paths(self, data):
means = np.zeros(data.shape[0])
for ti in range(self.trees):
path = self.estimators[ti].paths(data)
means += path
means /= self.trees
return means
def scores(self, data):
means = self.mean_paths(data)
return - 2 ** (-means / average_path_length(self.subsamples))
class Tree:
def __init__(self, features, selectors, values):
self.features = features
self.len = selectors.shape[0]
# Two complementary arrays.
# Selectors select feature to branch on.
self.selectors = selectors
# Values either set the deciding feature value or set the closing path length
self.values = values
def _get_one_path(self, key):
i = 1
while 2 * i < self.selectors.shape[0]:
f = self.selectors[i]
if f < 0:
break
if key[f] <= self.values[i]:
i = 2 * i
else:
i = 2 * i + 1
return self.values[i]
def paths(self, x):
n = x.shape[0]
paths = np.empty(n)
for i in range(n):
paths[i] = self._get_one_path(x[i, :])
return paths
class IsolationForestGenerator:
def __init__(self, sample, depth, seed=0):
self.depth = depth
self.features = sample.shape[1]
self.length = 1 << (depth + 1)
self.rng = np.random.default_rng(seed)
self.selectors = np.full(self.length, -1, dtype=np.int32)
self.values = np.full(self.length, 0, dtype=np.float64)
self._populate(1, sample)
self.pine = Tree(self.features, self.selectors, self.values)
def _populate(self, i, sample):
if sample.shape[0] == 1:
self.values[i] = np.floor(np.log2(i))
return
if self.length <= 2 * i:
self.values[i] = np.floor(np.log2(i)) + \
average_path_length(sample.shape[0])
return
selector = self.rng.integers(self.features)
self.selectors[i] = selector
minval = np.min(sample[:, selector])
maxval = np.max(sample[:, selector])
if minval == maxval:
self.selectors[i] = -1
self.values[i] = np.floor(np.log2(i)) + \
average_path_length(sample.shape[0])
return
value = self.rng.uniform(minval, maxval)
self.values[i] = value
self._populate(2 * i, sample[sample[:, selector] <= value])
self._populate(2 * i + 1, sample[sample[:, selector] > value])
# -
# # Пример
# +
data = malanchev_dataset()
isoforest = IsolationForest(trees=100, subsamples=16, depth=4)
isoforest.fit(data)
scores = isoforest.scores(data)
sorting = np.argsort(scores)
plt.scatter(*data[sorting[:96]].T, color='C1', label='anomaly')
plt.scatter(*data[sorting[96:]].T, color='C0', label='regular')
plt.legend()
pass
# -
# # Тесты
data = malanchev_dataset(inliers=2**13)
# %%time
isoforest = IsolationForest(trees=200, subsamples=1024, depth=10)
isoforest.fit(data)
pass
# %%time
scores = isoforest.scores(data)
# Если посчитать для 10e6 точек (в часах):
48 / 2**13 * 10e6 / 60.0 / 60.0
| j07_casestudy/j07_isoforest_python_naive.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] nbsphinx="hidden"
# # Random Signals
#
# *This jupyter notebook is part of a [collection of notebooks](../index.ipynb) on various topics of Digital Signal Processing.
# -
# ## Introduction
#
# Random signals are signals whose values are not (or only to a limited extend) predictable. Frequently used alternative terms are
#
# * stochastic signals
# * non-deterministic signals
#
# Random signals play an important role in various fields of signal processing and communications. This is due to the fact that only random signals carry information. A signal which is observed by a receiver has to be unknown to some degree in order to represent novel [information](https://en.wikipedia.org/wiki/Information).
#
# Random signals are often classified as useful/desired and disturbing/interfering signals. For instance
#
# * useful signals: data, speech, music, images, ...
# * disturbing signals: thermal noise at a resistor, amplifier noise, quantization noise, ...
#
# Practical signals are frequently modeled as a combination of useful signals and additive noise.
#
# As the values of a random signal cannot be foreseen, the properties of random signals are described by the their statistical characteristics. One measure is for instance the average value of a random signal.
# **Example - Random Signals**
#
# The following audio examples illustrate the characteristics of some deterministic and random signals. Lower the volume of your headphones or loudspeakers before playing back the examples.
#
# 1. Cosine signal
#
# <audio src="./cosine.wav" controls>Your browser does not support the audio element.</audio>[./cosine.wav](./cosine.wav)
# 2. Noise
#
# <audio src="./noise.wav" controls>Your browser does not support the audio element.</audio>[./noise.wav](./noise.wav)
# 3. Cosine signal superpositioned by noise
#
# <audio src="./cosine_noise.wav" controls>Your browser does not support the audio element.</audio>[./cosine_noise.wav](./cosine_noise.wav)
# 4. Speech signal
#
# <audio src="../data/speech.wav" controls>Your browser does not support the audio element.</audio>[../data/speech.wav](../data/speech.wav)
# 5. Speech signal superpositioned by noise
#
# <audio src="./speech_noise.wav" controls>Your browser does not support the audio element.</audio>[./speech_noise.wav](./speech_noise.wav)
# **Excercise**
#
# * Which example can be considered as deterministic, random signal or combination of both?
#
# Solution: The cosine signal is the only deterministic signal. Noise and speech are random signals, as their samples can not (or only to a limited extend) be predicted from previous samples. The superposition of the cosine and noise signals is a combination of a deterministic and a random signal.
# ### Processing of Random Signals
#
# In contrary to the assumption of deterministic signals in traditional signal processing, [statistical signal processing](https://en.wikipedia.org/wiki/Statistical_signal_processing) treats signals explicitly as random signals. Two prominent application examples involving random signals are
# #### Measurement of physical quantities
#
# The measurement of physical quantities is often subject to additive noise and distortions. The additive noise models e.g. the sensor noise. The distortions, by e.g. the transmission properties of an amplifier, may be modeled by a system.
#
# 
#
# $\mathcal{H}$ denotes an arbitrary (not necessarily LTI) system. The aim of statistical signal processing is to estimate the physical quantity from the observed sensor data, given some knowledge on the disturbing system and the statistical properties of the noise.
# #### Communication channel
#
# In communications engineering a message is sent over a channel distorting the signal by e.g. multipath propagation. Additive noise is present at the receiver due to background and amplifier noise.
#
# 
#
# The aim of statistical signal processing is to estimate the send message from the received message, given some knowledge on the disturbing system and the statistical properties of the noise.
# ### Random Processes
#
# A random process is a [stochastic process](https://en.wikipedia.org/wiki/Stochastic_process) which generates an ensemble of random signals. A random process
#
# * provides a mathematical model for an ensemble of random signals and
# * generates different sample functions with specific common properties.
#
# It is important to differentiate between an
#
# * *ensemble*: collection of all possible signals of a random process and an
# * *sample function*: one specific random signal.
#
# An example for a random process is speech produced by humans. Here the ensemble is composed from the speech signals produced by all humans on earth, one particular speech signal produced by one person at a specific time is a sample function.
# **Example - Sample functions of a random process**
#
# The following example shows sample functions of a continuous amplitude real-valued random process. All sample functions have the same properties with respect to certain statistical measures.
# +
# %matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
N = 5 # number of sample functions
# draw N sample functions from a random process
np.random.seed(0)
x = np.random.normal(size=(N, 32))
# plot sample functions
fig = plt.figure(figsize=(10, 12))
for n in range(N):
plt.subplot(N, 1, n+1)
plt.tight_layout()
plt.stem(x[n,:], basefmt='k-')
plt.title('Sample Function %d' %n)
plt.xlabel(r'$k$')
plt.ylabel(r'$x_%d[k]$' %n)
plt.axis([-1, 32, -3, 3])
plt.grid()
# -
# **Exercise**
#
# * What is different, what is common between the sample functions?
#
# Solution: You may have observed that the amplitude values of the individual sample functions $x_n[k]$ differ for a fixed time instant $k$. However, the sample functions seem to share some common properties. For instance, positive and negative values seem to occur with approximately the same probability.
# ### Properties of Random Processes and Random Signals
#
# It was already argued above, that it is not meaningful to describe a random signal by the amplitude values of a particular sample function. Instead, random signals are characterized by specific statistical measures. In statistical signal processing it is common to use
#
# * amplitude distributions and
# * ensemble averages/moments
#
# for this purpose. These measures will be introduced in the remainder.
| Lectures_Advanced-DSP/random_signals/introduction.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Regularization
#
# Welcome to the second assignment of this week. Deep Learning models have so much flexibility and capacity that **overfitting can be a serious problem**, if the training dataset is not big enough. Sure it does well on the training set, but the learned network **doesn't generalize to new examples** that it has never seen!
#
# **You will learn to:** Use regularization in your deep learning models.
#
# Let's get started!
# ## Table of Contents
# - [1 - Packages](#1)
# - [2 - Problem Statement](#2)
# - [3 - Loading the Dataset](#3)
# - [4 - Non-Regularized Model](#4)
# - [5 - L2 Regularization](#5)
# - [Exercise 1 - compute_cost_with_regularization](#ex-1)
# - [Exercise 2 - backward_propagation_with_regularization](#ex-2)
# - [6 - Dropout](#6)
# - [6.1 - Forward Propagation with Dropout](#6-1)
# - [Exercise 3 - forward_propagation_with_dropout](#ex-3)
# - [6.2 - Backward Propagation with Dropout](#6-2)
# - [Exercise 4 - backward_propagation_with_dropout](#ex-4)
# - [7 - Conclusions](#7)
# <a name='1'></a>
# ## 1 - Packages
# +
# import packages
import numpy as np
import matplotlib.pyplot as plt
import sklearn
import sklearn.datasets
import scipy.io
from reg_utils import sigmoid, relu, plot_decision_boundary, initialize_parameters, load_2D_dataset, predict_dec
from reg_utils import compute_cost, predict, forward_propagation, backward_propagation, update_parameters
from testCases import *
from public_tests import *
# %matplotlib inline
plt.rcParams['figure.figsize'] = (7.0, 4.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# %load_ext autoreload
# %autoreload 2
# -
# <a name='2'></a>
# ## 2 - Problem Statement
# You have just been hired as an AI expert by the French Football Corporation. They would like you to recommend positions where France's goal keeper should kick the ball so that the French team's players can then hit it with their head.
#
# <img src="images/field_kiank.png" style="width:600px;height:350px;">
#
# <caption><center><font color='purple'><b>Figure 1</b>: Football field. The goal keeper kicks the ball in the air, the players of each team are fighting to hit the ball with their head </font></center></caption>
#
#
# They give you the following 2D dataset from France's past 10 games.
# <a name='3'></a>
# ## 3 - Loading the Dataset
train_X, train_Y, test_X, test_Y = load_2D_dataset()
# Each dot corresponds to a position on the football field where a football player has hit the ball with his/her head after the French goal keeper has shot the ball from the left side of the football field.
# - If the dot is blue, it means the French player managed to hit the ball with his/her head
# - If the dot is red, it means the other team's player hit the ball with their head
#
# **Your goal**: Use a deep learning model to find the positions on the field where the goalkeeper should kick the ball.
# **Analysis of the dataset**: This dataset is a little noisy, but it looks like a diagonal line separating the upper left half (blue) from the lower right half (red) would work well.
#
# You will first try a non-regularized model. Then you'll learn how to regularize it and decide which model you will choose to solve the French Football Corporation's problem.
# <a name='4'></a>
# ## 4 - Non-Regularized Model
#
# You will use the following neural network (already implemented for you below). This model can be used:
# - in *regularization mode* -- by setting the `lambd` input to a non-zero value. We use "`lambd`" instead of "`lambda`" because "`lambda`" is a reserved keyword in Python.
# - in *dropout mode* -- by setting the `keep_prob` to a value less than one
#
# You will first try the model without any regularization. Then, you will implement:
# - *L2 regularization* -- functions: "`compute_cost_with_regularization()`" and "`backward_propagation_with_regularization()`"
# - *Dropout* -- functions: "`forward_propagation_with_dropout()`" and "`backward_propagation_with_dropout()`"
#
# In each part, you will run this model with the correct inputs so that it calls the functions you've implemented. Take a look at the code below to familiarize yourself with the model.
def model(X, Y, learning_rate = 0.3, num_iterations = 30000, print_cost = True, lambd = 0, keep_prob = 1):
"""
Implements a three-layer neural network: LINEAR->RELU->LINEAR->RELU->LINEAR->SIGMOID.
Arguments:
X -- input data, of shape (input size, number of examples)
Y -- true "label" vector (1 for blue dot / 0 for red dot), of shape (output size, number of examples)
learning_rate -- learning rate of the optimization
num_iterations -- number of iterations of the optimization loop
print_cost -- If True, print the cost every 10000 iterations
lambd -- regularization hyperparameter, scalar
keep_prob - probability of keeping a neuron active during drop-out, scalar.
Returns:
parameters -- parameters learned by the model. They can then be used to predict.
"""
grads = {}
costs = [] # to keep track of the cost
m = X.shape[1] # number of examples
layers_dims = [X.shape[0], 20, 3, 1]
# Initialize parameters dictionary.
parameters = initialize_parameters(layers_dims)
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID.
if keep_prob == 1:
a3, cache = forward_propagation(X, parameters)
elif keep_prob < 1:
a3, cache = forward_propagation_with_dropout(X, parameters, keep_prob)
# Cost function
if lambd == 0:
cost = compute_cost(a3, Y)
else:
cost = compute_cost_with_regularization(a3, Y, parameters, lambd)
# Backward propagation.
assert (lambd == 0 or keep_prob == 1) # it is possible to use both L2 regularization and dropout,
# but this assignment will only explore one at a time
if lambd == 0 and keep_prob == 1:
grads = backward_propagation(X, Y, cache)
elif lambd != 0:
grads = backward_propagation_with_regularization(X, Y, cache, lambd)
elif keep_prob < 1:
grads = backward_propagation_with_dropout(X, Y, cache, keep_prob)
# Update parameters.
parameters = update_parameters(parameters, grads, learning_rate)
# Print the loss every 10000 iterations
if print_cost and i % 10000 == 0:
print("Cost after iteration {}: {}".format(i, cost))
if print_cost and i % 1000 == 0:
costs.append(cost)
# plot the cost
plt.plot(costs)
plt.ylabel('cost')
plt.xlabel('iterations (x1,000)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters
# Let's train the model without any regularization, and observe the accuracy on the train/test sets.
parameters = model(train_X, train_Y)
print ("On the training set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
# The train accuracy is 94.8% while the test accuracy is 91.5%. This is the **baseline model** (you will observe the impact of regularization on this model). Run the following code to plot the decision boundary of your model.
plt.title("Model without regularization")
axes = plt.gca()
axes.set_xlim([-0.75,0.40])
axes.set_ylim([-0.75,0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
# The non-regularized model is obviously overfitting the training set. It is fitting the noisy points! Lets now look at two techniques to reduce overfitting.
# <a name='5'></a>
# ## 5 - L2 Regularization
#
# The standard way to avoid overfitting is called **L2 regularization**. It consists of appropriately modifying your cost function, from:
# $$J = -\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)} \tag{1}$$
# To:
# $$J_{regularized} = \small \underbrace{-\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)} }_\text{cross-entropy cost} + \underbrace{\frac{1}{m} \frac{\lambda}{2} \sum\limits_l\sum\limits_k\sum\limits_j W_{k,j}^{[l]2} }_\text{L2 regularization cost} \tag{2}$$
#
# Let's modify your cost and observe the consequences.
#
# <a name='ex-1'></a>
# ### Exercise 1 - compute_cost_with_regularization
# Implement `compute_cost_with_regularization()` which computes the cost given by formula (2). To calculate $\sum\limits_k\sum\limits_j W_{k,j}^{[l]2}$ , use :
# ```python
# np.sum(np.square(Wl))
# ```
# Note that you have to do this for $W^{[1]}$, $W^{[2]}$ and $W^{[3]}$, then sum the three terms and multiply by $ \frac{1}{m} \frac{\lambda}{2} $.
# + deletable=false nbgrader={"cell_type": "code", "checksum": "88e54417c158ef5260e3107ab846463e", "grade": false, "grade_id": "cell-02a896d283f479aa", "locked": false, "schema_version": 3, "solution": true, "task": false}
# GRADED FUNCTION: compute_cost_with_regularization
def compute_cost_with_regularization(A3, Y, parameters, lambd):
"""
Implement the cost function with L2 regularization. See formula (2) above.
Arguments:
A3 -- post-activation, output of forward propagation, of shape (output size, number of examples)
Y -- "true" labels vector, of shape (output size, number of examples)
parameters -- python dictionary containing parameters of the model
Returns:
cost - value of the regularized loss function (formula (2))
"""
m = Y.shape[1]
W1 = parameters["W1"]
W2 = parameters["W2"]
W3 = parameters["W3"]
cross_entropy_cost = compute_cost(A3, Y) # This gives you the cross-entropy part of the cost
#(≈ 1 lines of code)
# L2_regularization_cost =
# YOUR CODE STARTS HERE
L2_regularization_cost = lambd/(2*m)*(np.sum(np.square(W1))+np.sum(np.square(W2))+np.sum(np.square(W3)))
# YOUR CODE ENDS HERE
cost = cross_entropy_cost + L2_regularization_cost
return cost
# + deletable=false editable=false nbgrader={"cell_type": "code", "checksum": "c8efc715a4d6127a214a1b9f97e9f4cb", "grade": true, "grade_id": "cell-8a99b24d8ecfe0c3", "locked": true, "points": 10, "schema_version": 3, "solution": false, "task": false}
A3, t_Y, parameters = compute_cost_with_regularization_test_case()
cost = compute_cost_with_regularization(A3, t_Y, parameters, lambd=0.1)
print("cost = " + str(cost))
compute_cost_with_regularization_test(compute_cost_with_regularization)
# -
# Of course, because you changed the cost, you have to change backward propagation as well! All the gradients have to be computed with respect to this new cost.
#
# <a name='ex-2'></a>
# ### Exercise 2 - backward_propagation_with_regularization
# Implement the changes needed in backward propagation to take into account regularization. The changes only concern dW1, dW2 and dW3. For each, you have to add the regularization term's gradient ($\frac{d}{dW} ( \frac{1}{2}\frac{\lambda}{m} W^2) = \frac{\lambda}{m} W$).
# + deletable=false nbgrader={"cell_type": "code", "checksum": "eb2dfa385aa47fe2e2edf5c6821618e6", "grade": false, "grade_id": "cell-c6f6ed3630e04d4b", "locked": false, "schema_version": 3, "solution": true, "task": false}
# GRADED FUNCTION: backward_propagation_with_regularization
def backward_propagation_with_regularization(X, Y, cache, lambd):
"""
Implements the backward propagation of our baseline model to which we added an L2 regularization.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- "true" labels vector, of shape (output size, number of examples)
cache -- cache output from forward_propagation()
lambd -- regularization hyperparameter, scalar
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
m = X.shape[1]
(Z1, A1, W1, b1, Z2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
#(≈ 1 lines of code)
# dW3 = 1./m * np.dot(dZ3, A2.T) + None
# YOUR CODE STARTS HERE
dW3 = 1. / m * np.dot(dZ3, A2.T) + (lambd / m) * W3
# YOUR CODE ENDS HERE
db3 = 1. / m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
#(≈ 1 lines of code)
# dW2 = 1./m * np.dot(dZ2, A1.T) + None
# YOUR CODE STARTS HERE
dW2 = 1. / m * np.dot(dZ2, A1.T) + (lambd / m) * W2
# YOUR CODE ENDS HERE
db2 = 1. / m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
#(≈ 1 lines of code)
# dW1 = 1./m * np.dot(dZ1, X.T) + None
# YOUR CODE STARTS HERE
dW1 = 1. / m * np.dot(dZ1, X.T) + (lambd / m) * W1
# YOUR CODE ENDS HERE
db1 = 1. / m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3,"dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
# + deletable=false editable=false nbgrader={"cell_type": "code", "checksum": "bd8e0024ad54c2facd2fb8e11d21d2a0", "grade": true, "grade_id": "cell-9826510f7bfdd0f8", "locked": true, "points": 10, "schema_version": 3, "solution": false, "task": false}
t_X, t_Y, cache = backward_propagation_with_regularization_test_case()
grads = backward_propagation_with_regularization(t_X, t_Y, cache, lambd = 0.7)
print ("dW1 = \n"+ str(grads["dW1"]))
print ("dW2 = \n"+ str(grads["dW2"]))
print ("dW3 = \n"+ str(grads["dW3"]))
backward_propagation_with_regularization_test(backward_propagation_with_regularization)
# -
# Let's now run the model with L2 regularization $(\lambda = 0.7)$. The `model()` function will call:
# - `compute_cost_with_regularization` instead of `compute_cost`
# - `backward_propagation_with_regularization` instead of `backward_propagation`
parameters = model(train_X, train_Y, lambd = 0.7)
print ("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
# Congrats, the test set accuracy increased to 93%. You have saved the French football team!
#
# You are not overfitting the training data anymore. Let's plot the decision boundary.
plt.title("Model with L2-regularization")
axes = plt.gca()
axes.set_xlim([-0.75,0.40])
axes.set_ylim([-0.75,0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
# **Observations**:
# - The value of $\lambda$ is a hyperparameter that you can tune using a dev set.
# - L2 regularization makes your decision boundary smoother. If $\lambda$ is too large, it is also possible to "oversmooth", resulting in a model with high bias.
#
# **What is L2-regularization actually doing?**:
#
# L2-regularization relies on the assumption that a model with small weights is simpler than a model with large weights. Thus, by penalizing the square values of the weights in the cost function you drive all the weights to smaller values. It becomes too costly for the cost to have large weights! This leads to a smoother model in which the output changes more slowly as the input changes.
#
# <br>
# <font color='blue'>
#
# **What you should remember:** the implications of L2-regularization on:
# - The cost computation:
# - A regularization term is added to the cost.
# - The backpropagation function:
# - There are extra terms in the gradients with respect to weight matrices.
# - Weights end up smaller ("weight decay"):
# - Weights are pushed to smaller values.
# <a name='6'></a>
# ## 6 - Dropout
#
# Finally, **dropout** is a widely used regularization technique that is specific to deep learning.
# **It randomly shuts down some neurons in each iteration.** Watch these two videos to see what this means!
#
# <!--
# To understand drop-out, consider this conversation with a friend:
# - Friend: "Why do you need all these neurons to train your network and classify images?".
# - You: "Because each neuron contains a weight and can learn specific features/details/shape of an image. The more neurons I have, the more featurse my model learns!"
# - Friend: "I see, but are you sure that your neurons are learning different features and not all the same features?"
# - You: "Good point... Neurons in the same layer actually don't talk to each other. It should be definitly possible that they learn the same image features/shapes/forms/details... which would be redundant. There should be a solution."
# !-->
#
#
# <center>
# <video width="620" height="440" src="images/dropout1_kiank.mp4" type="video/mp4" controls>
# </video>
# </center>
# <br>
# <caption><center><font color='purple'><b>Figure 2 </b>: <b>Drop-out on the second hidden layer.</b> <br> At each iteration, you shut down (= set to zero) each neuron of a layer with probability $1 - keep\_prob$ or keep it with probability $keep\_prob$ (50% here). The dropped neurons don't contribute to the training in both the forward and backward propagations of the iteration. </font></center></caption>
#
# <center>
# <video width="620" height="440" src="images/dropout2_kiank.mp4" type="video/mp4" controls>
# </video>
# </center>
#
# <caption><center><font color='purple'><b>Figure 3</b>:<b> Drop-out on the first and third hidden layers. </b><br> $1^{st}$ layer: we shut down on average 40% of the neurons. $3^{rd}$ layer: we shut down on average 20% of the neurons. </font></center></caption>
#
#
# When you shut some neurons down, you actually modify your model. The idea behind drop-out is that at each iteration, you train a different model that uses only a subset of your neurons. With dropout, your neurons thus become less sensitive to the activation of one other specific neuron, because that other neuron might be shut down at any time.
#
# <a name='6-1'></a>
# ### 6.1 - Forward Propagation with Dropout
#
# <a name='ex-3'></a>
# ### Exercise 3 - forward_propagation_with_dropout
#
# Implement the forward propagation with dropout. You are using a 3 layer neural network, and will add dropout to the first and second hidden layers. We will not apply dropout to the input layer or output layer.
#
# **Instructions**:
# You would like to shut down some neurons in the first and second layers. To do that, you are going to carry out 4 Steps:
# 1. In lecture, we dicussed creating a variable $d^{[1]}$ with the same shape as $a^{[1]}$ using `np.random.rand()` to randomly get numbers between 0 and 1. Here, you will use a vectorized implementation, so create a random matrix $D^{[1]} = [d^{[1](1)} d^{[1](2)} ... d^{[1](m)}] $ of the same dimension as $A^{[1]}$.
# 2. Set each entry of $D^{[1]}$ to be 1 with probability (`keep_prob`), and 0 otherwise.
#
# **Hint:** Let's say that keep_prob = 0.8, which means that we want to keep about 80% of the neurons and drop out about 20% of them. We want to generate a vector that has 1's and 0's, where about 80% of them are 1 and about 20% are 0.
# This python statement:
# `X = (X < keep_prob).astype(int)`
#
# is conceptually the same as this if-else statement (for the simple case of a one-dimensional array) :
#
# ```
# for i,v in enumerate(x):
# if v < keep_prob:
# x[i] = 1
# else: # v >= keep_prob
# x[i] = 0
# ```
# Note that the `X = (X < keep_prob).astype(int)` works with multi-dimensional arrays, and the resulting output preserves the dimensions of the input array.
#
# Also note that without using `.astype(int)`, the result is an array of booleans `True` and `False`, which Python automatically converts to 1 and 0 if we multiply it with numbers. (However, it's better practice to convert data into the data type that we intend, so try using `.astype(int)`.)
#
# 3. Set $A^{[1]}$ to $A^{[1]} * D^{[1]}$. (You are shutting down some neurons). You can think of $D^{[1]}$ as a mask, so that when it is multiplied with another matrix, it shuts down some of the values.
# 4. Divide $A^{[1]}$ by `keep_prob`. By doing this you are assuring that the result of the cost will still have the same expected value as without drop-out. (This technique is also called inverted dropout.)
# + deletable=false nbgrader={"cell_type": "code", "checksum": "249ddfb0abac7c799948d3e600db7a4c", "grade": false, "grade_id": "cell-a81658747a0683be", "locked": false, "schema_version": 3, "solution": true, "task": false}
# GRADED FUNCTION: forward_propagation_with_dropout
def forward_propagation_with_dropout(X, parameters, keep_prob = 0.5):
"""
Implements the forward propagation: LINEAR -> RELU + DROPOUT -> LINEAR -> RELU + DROPOUT -> LINEAR -> SIGMOID.
Arguments:
X -- input dataset, of shape (2, number of examples)
parameters -- python dictionary containing your parameters "W1", "b1", "W2", "b2", "W3", "b3":
W1 -- weight matrix of shape (20, 2)
b1 -- bias vector of shape (20, 1)
W2 -- weight matrix of shape (3, 20)
b2 -- bias vector of shape (3, 1)
W3 -- weight matrix of shape (1, 3)
b3 -- bias vector of shape (1, 1)
keep_prob - probability of keeping a neuron active during drop-out, scalar
Returns:
A3 -- last activation value, output of the forward propagation, of shape (1,1)
cache -- tuple, information stored for computing the backward propagation
"""
np.random.seed(1)
# retrieve parameters
W1 = parameters["W1"]
b1 = parameters["b1"]
W2 = parameters["W2"]
b2 = parameters["b2"]
W3 = parameters["W3"]
b3 = parameters["b3"]
# LINEAR -> RELU -> LINEAR -> RELU -> LINEAR -> SIGMOID
Z1 = np.dot(W1, X) + b1
A1 = relu(Z1)
#(≈ 4 lines of code) # Steps 1-4 below correspond to the Steps 1-4 described above.
# D1 = # Step 1: initialize matrix D1 = np.random.rand(..., ...)
# D1 = # Step 2: convert entries of D1 to 0 or 1 (using keep_prob as the threshold)
# A1 = # Step 3: shut down some neurons of A1
# A1 = # Step 4: scale the value of neurons that haven't been shut down
# YOUR CODE STARTS HERE
D1 = np.random.rand(A1.shape[0], A1.shape[1]) # Step 1: initialize matrix D1 = np.random.rand(..., ...)
D1 = (D1 < keep_prob) # Step 2: convert entries of D1 to 0 or 1 (using keep_prob as the threshold)
A1 = A1 * D1 # Step 3: shut down some neurons of A1
A1 = A1 / keep_prob # Step 4: scale the value of neurons that haven't been shut down
# YOUR CODE ENDS HERE
Z2 = np.dot(W2, A1) + b2
A2 = relu(Z2)
#(≈ 4 lines of code)
# D2 = # Step 1: initialize matrix D2 = np.random.rand(..., ...)
# D2 = # Step 2: convert entries of D2 to 0 or 1 (using keep_prob as the threshold)
# A2 = # Step 3: shut down some neurons of A2
# A2 = # Step 4: scale the value of neurons that haven't been shut down
# YOUR CODE STARTS HERE
D2 = np.random.rand(A2.shape[0], A2.shape[1]) # Step 1: initialize matrix D2 = np.random.rand(..., ...)
D2 = (D2 < keep_prob) # Step 2: convert entries of D2 to 0 or 1 (using keep_prob as the threshold)
A2 = A2 * D2 # Step 3: shut down some neurons of A2
A2 = A2 / keep_prob # Step 4: scale the value of neurons that haven't been shut down
# YOUR CODE ENDS HERE
Z3 = np.dot(W3, A2) + b3
A3 = sigmoid(Z3)
cache = (Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3)
return A3, cache
# + deletable=false editable=false nbgrader={"cell_type": "code", "checksum": "caec66931ac05dbe474596e75f3a14cd", "grade": true, "grade_id": "cell-be6195c629f586bf", "locked": true, "points": 20, "schema_version": 3, "solution": false, "task": false}
t_X, parameters = forward_propagation_with_dropout_test_case()
A3, cache = forward_propagation_with_dropout(t_X, parameters, keep_prob=0.7)
print ("A3 = " + str(A3))
forward_propagation_with_dropout_test(forward_propagation_with_dropout)
# -
# <a name='6-2'></a>
# ### 6.2 - Backward Propagation with Dropout
#
# <a name='ex-4'></a>
# ### Exercise 4 - backward_propagation_with_dropout
# Implement the backward propagation with dropout. As before, you are training a 3 layer network. Add dropout to the first and second hidden layers, using the masks $D^{[1]}$ and $D^{[2]}$ stored in the cache.
#
# **Instruction**:
# Backpropagation with dropout is actually quite easy. You will have to carry out 2 Steps:
# 1. You had previously shut down some neurons during forward propagation, by applying a mask $D^{[1]}$ to `A1`. In backpropagation, you will have to shut down the same neurons, by reapplying the same mask $D^{[1]}$ to `dA1`.
# 2. During forward propagation, you had divided `A1` by `keep_prob`. In backpropagation, you'll therefore have to divide `dA1` by `keep_prob` again (the calculus interpretation is that if $A^{[1]}$ is scaled by `keep_prob`, then its derivative $dA^{[1]}$ is also scaled by the same `keep_prob`).
#
# + deletable=false nbgrader={"cell_type": "code", "checksum": "ee4145889a9c078fcf6aef51aceb3ba9", "grade": false, "grade_id": "cell-5b97731b540b0b87", "locked": false, "schema_version": 3, "solution": true, "task": false}
# GRADED FUNCTION: backward_propagation_with_dropout
def backward_propagation_with_dropout(X, Y, cache, keep_prob):
"""
Implements the backward propagation of our baseline model to which we added dropout.
Arguments:
X -- input dataset, of shape (2, number of examples)
Y -- "true" labels vector, of shape (output size, number of examples)
cache -- cache output from forward_propagation_with_dropout()
keep_prob - probability of keeping a neuron active during drop-out, scalar
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
m = X.shape[1]
(Z1, D1, A1, W1, b1, Z2, D2, A2, W2, b2, Z3, A3, W3, b3) = cache
dZ3 = A3 - Y
dW3 = 1./m * np.dot(dZ3, A2.T)
db3 = 1./m * np.sum(dZ3, axis=1, keepdims=True)
dA2 = np.dot(W3.T, dZ3)
#(≈ 2 lines of code)
# dA2 = # Step 1: Apply mask D2 to shut down the same neurons as during the forward propagation
# dA2 = # Step 2: Scale the value of neurons that haven't been shut down
# YOUR CODE STARTS HERE
dA2 = dA2 * D2 # Step 1: Apply mask D2 to shut down the same neurons as during the forward propagation
dA2 = dA2 / keep_prob # Step 2: Scale the value of neurons that haven't been shut down
# YOUR CODE ENDS HERE
dZ2 = np.multiply(dA2, np.int64(A2 > 0))
dW2 = 1./m * np.dot(dZ2, A1.T)
db2 = 1./m * np.sum(dZ2, axis=1, keepdims=True)
dA1 = np.dot(W2.T, dZ2)
#(≈ 2 lines of code)
# dA1 = # Step 1: Apply mask D1 to shut down the same neurons as during the forward propagation
# dA1 = # Step 2: Scale the value of neurons that haven't been shut down
# YOUR CODE STARTS HERE
dA1 = dA1 * D1 # Step 1: Apply mask D1 to shut down the same neurons as during the forward propagation
dA1 = dA1 / keep_prob # Step 2: Scale the value of neurons that haven't been shut down
# YOUR CODE ENDS HERE
dZ1 = np.multiply(dA1, np.int64(A1 > 0))
dW1 = 1./m * np.dot(dZ1, X.T)
db1 = 1./m * np.sum(dZ1, axis=1, keepdims=True)
gradients = {"dZ3": dZ3, "dW3": dW3, "db3": db3,"dA2": dA2,
"dZ2": dZ2, "dW2": dW2, "db2": db2, "dA1": dA1,
"dZ1": dZ1, "dW1": dW1, "db1": db1}
return gradients
# + deletable=false editable=false nbgrader={"cell_type": "code", "checksum": "c30bb7a9f59c7d421c8627d5d9252b29", "grade": true, "grade_id": "cell-958c189ce5b16569", "locked": true, "points": 10, "schema_version": 3, "solution": false, "task": false}
t_X, t_Y, cache = backward_propagation_with_dropout_test_case()
gradients = backward_propagation_with_dropout(t_X, t_Y, cache, keep_prob=0.8)
print ("dA1 = \n" + str(gradients["dA1"]))
print ("dA2 = \n" + str(gradients["dA2"]))
backward_propagation_with_dropout_test(backward_propagation_with_dropout)
# -
# Let's now run the model with dropout (`keep_prob = 0.86`). It means at every iteration you shut down each neurons of layer 1 and 2 with 14% probability. The function `model()` will now call:
# - `forward_propagation_with_dropout` instead of `forward_propagation`.
# - `backward_propagation_with_dropout` instead of `backward_propagation`.
# +
parameters = model(train_X, train_Y, keep_prob = 0.86, learning_rate = 0.3)
print ("On the train set:")
predictions_train = predict(train_X, train_Y, parameters)
print ("On the test set:")
predictions_test = predict(test_X, test_Y, parameters)
# -
# Dropout works great! The test accuracy has increased again (to 95%)! Your model is not overfitting the training set and does a great job on the test set. The French football team will be forever grateful to you!
#
# Run the code below to plot the decision boundary.
plt.title("Model with dropout")
axes = plt.gca()
axes.set_xlim([-0.75,0.40])
axes.set_ylim([-0.75,0.65])
plot_decision_boundary(lambda x: predict_dec(parameters, x.T), train_X, train_Y)
# **Note**:
# - A **common mistake** when using dropout is to use it both in training and testing. You should use dropout (randomly eliminate nodes) only in training.
# - Deep learning frameworks like [tensorflow](https://www.tensorflow.org/api_docs/python/tf/nn/dropout), [PaddlePaddle](http://doc.paddlepaddle.org/release_doc/0.9.0/doc/ui/api/trainer_config_helpers/attrs.html), [keras](https://keras.io/layers/core/#dropout) or [caffe](http://caffe.berkeleyvision.org/tutorial/layers/dropout.html) come with a dropout layer implementation. Don't stress - you will soon learn some of these frameworks.
#
# <font color='blue'>
#
# **What you should remember about dropout:**
# - Dropout is a regularization technique.
# - You only use dropout during training. Don't use dropout (randomly eliminate nodes) during test time.
# - Apply dropout both during forward and backward propagation.
# - During training time, divide each dropout layer by keep_prob to keep the same expected value for the activations. For example, if keep_prob is 0.5, then we will on average shut down half the nodes, so the output will be scaled by 0.5 since only the remaining half are contributing to the solution. Dividing by 0.5 is equivalent to multiplying by 2. Hence, the output now has the same expected value. You can check that this works even when keep_prob is other values than 0.5.
# <a name='7'></a>
# ## 7 - Conclusions
# **Here are the results of our three models**:
#
# <table>
# <tr>
# <td>
# <b>model</b>
# </td>
# <td>
# <b>train accuracy</b>
# </td>
# <td>
# <b>test accuracy</b>
# </td>
# </tr>
# <td>
# 3-layer NN without regularization
# </td>
# <td>
# 95%
# </td>
# <td>
# 91.5%
# </td>
# <tr>
# <td>
# 3-layer NN with L2-regularization
# </td>
# <td>
# 94%
# </td>
# <td>
# 93%
# </td>
# </tr>
# <tr>
# <td>
# 3-layer NN with dropout
# </td>
# <td>
# 93%
# </td>
# <td>
# 95%
# </td>
# </tr>
# </table>
# Note that regularization hurts training set performance! This is because it limits the ability of the network to overfit to the training set. But since it ultimately gives better test accuracy, it is helping your system.
# Congratulations for finishing this assignment! And also for revolutionizing French football. :-)
# <font color='blue'>
#
# **What we want you to remember from this notebook**:
# - Regularization will help you reduce overfitting.
# - Regularization will drive your weights to lower values.
# - L2 regularization and Dropout are two very effective regularization techniques.
| C02W01/1.2 Regularization/Regularization.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# # Carving Unit Tests
#
# So far, we have always generated _system input_, i.e. data that the program as a whole obtains via its input channels. If we are interested in testing only a small set of functions, having to go through the system can be very inefficient. This chapter introduces a technique known as _carving_, which, given a system test, automatically extracts a set of _unit tests_ that replicate the calls seen during the unit test. The key idea is to _record_ such calls such that we can _replay_ them later – as a whole or selectively. On top, we also explore how to synthesize API grammars from carved unit tests; this means that we can _synthesize API tests without having to write a grammar at all._
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"}
# **Prerequisites**
#
# * Carving makes use of dynamic traces of function calls and variables, as introduced in the [chapter on configuration fuzzing](ConfigurationFuzzer.ipynb).
# * Using grammars to test units was introduced in the [chapter on API fuzzing](APIFuzzer.ipynb).
# + slideshow={"slide_type": "skip"}
import bookutils
# + slideshow={"slide_type": "skip"}
import APIFuzzer
# + [markdown] slideshow={"slide_type": "slide"}
# ## Synopsis
# <!-- Automatically generated. Do not edit. -->
#
# To [use the code provided in this chapter](Importing.ipynb), write
#
# ```python
# >>> from fuzzingbook.Carver import <identifier>
# ```
#
# and then make use of the following features.
#
#
# This chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such "carving" mechanisms have the potential to run tests much faster.
#
# ### Recording Calls
#
# The `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:
#
# ```python
# >>> with CallCarver() as carver:
# >>> y = my_sqrt(2)
# >>> y = my_sqrt(4)
# ```
# After execution, `called_functions()` lists the names of functions encountered:
#
# ```python
# >>> carver.called_functions()
# ['my_sqrt', '__exit__']
# ```
# The `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).
#
# ```python
# >>> carver.arguments('my_sqrt')
# [[('x', 2)], [('x', 4)]]
# ```
# Complex arguments are properly serialized, such that they can be easily restored.
#
# ### Synthesizing Calls
#
# While such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.
#
# The `CallGrammarMiner` class turns a list of carved executions into a grammar.
#
# ```python
# >>> my_sqrt_miner = CallGrammarMiner(carver)
# >>> my_sqrt_grammar = my_sqrt_miner.mine_call_grammar()
# >>> my_sqrt_grammar
# {'<start>': ['<call>'],
# '<call>': ['<my_sqrt>'],
# '<my_sqrt-x>': ['4', '2'],
# '<my_sqrt>': ['my_sqrt(<my_sqrt-x>)']}
# ```
# This grammar can be used to synthesize calls.
#
# ```python
# >>> fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)
# >>> fuzzer.fuzz()
# 'my_sqrt(2)'
# ```
# These calls can be executed in isolation, effectively extracting unit tests from system tests:
#
# ```python
# >>> eval(fuzzer.fuzz())
# 2.0
# ```
#
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## System Tests vs Unit Tests
#
# Remember the URL grammar introduced for [grammar fuzzing](Grammars.ipynb)? With such a grammar, we can happily test a Web browser again and again, checking how it reacts to arbitrary page requests.
#
# Let us define a very simple "web browser" that goes and downloads the content given by the URL.
# + slideshow={"slide_type": "skip"}
import urllib.parse
# + slideshow={"slide_type": "fragment"}
def webbrowser(url):
"""Download the http/https resource given by the URL"""
import requests # Only import if needed
r = requests.get(url)
return r.text
# + [markdown] slideshow={"slide_type": "subslide"}
# Let us apply this on [fuzzingbook.org](https://www.fuzzingbook.org/) and measure the time, using the [Timer class](Timer.ipynb):
# + slideshow={"slide_type": "skip"}
from Timer import Timer
# + slideshow={"slide_type": "fragment"}
with Timer() as webbrowser_timer:
fuzzingbook_contents = webbrowser(
"http://www.fuzzingbook.org/html/Fuzzer.html")
print("Downloaded %d bytes in %.2f seconds" %
(len(fuzzingbook_contents), webbrowser_timer.elapsed_time()))
# + slideshow={"slide_type": "fragment"}
fuzzingbook_contents[:100]
# + [markdown] slideshow={"slide_type": "subslide"}
# A full webbrowser, of course, would also render the HTML content. We can achieve this using these commands (but we don't, as we do not want to replicate the entire Web page here):
#
#
# ```python
# from IPython.display import HTML, display
# HTML(fuzzingbook_contents)
# ```
# + [markdown] slideshow={"slide_type": "subslide"}
# Having to start a whole browser (or having it render a Web page) again and again means lots of overhead, though – in particular if we want to test only a subset of its functionality. In particular, after a change in the code, we would prefer to test only the subset of functions that is affected by the change, rather than running the well-tested functions again and again.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "fragment"}
# Let us assume we change the function that takes care of parsing the given URL and decomposing it into the individual elements – the scheme ("http"), the network location (`"www.fuzzingbook.com"`), or the path (`"/html/Fuzzer.html"`). This function is named `urlparse()`:
# + slideshow={"slide_type": "skip"}
from urllib.parse import urlparse
# + slideshow={"slide_type": "fragment"}
urlparse('https://www.fuzzingbook.com/html/Carver.html')
# + [markdown] slideshow={"slide_type": "subslide"}
# You see how the individual elements of the URL – the _scheme_ (`"http"`), the _network location_ (`"www.fuzzingbook.com"`), or the path (`"//html/Carver.html"`) are all properly identified. Other elements (like `params`, `query`, or `fragment`) are empty, because they were not part of our input.
# + [markdown] slideshow={"slide_type": "fragment"}
# The interesting thing is that executing only `urlparse()` is orders of magnitude faster than running all of `webbrowser()`. Let us measure the factor:
# + slideshow={"slide_type": "subslide"}
runs = 1000
with Timer() as urlparse_timer:
for i in range(runs):
urlparse('https://www.fuzzingbook.com/html/Carver.html')
avg_urlparse_time = urlparse_timer.elapsed_time() / 1000
avg_urlparse_time
# + [markdown] slideshow={"slide_type": "fragment"}
# Compare this to the time required by the webbrowser
# + slideshow={"slide_type": "fragment"}
webbrowser_timer.elapsed_time()
# + [markdown] slideshow={"slide_type": "fragment"}
# The difference in time is huge:
# + slideshow={"slide_type": "fragment"}
webbrowser_timer.elapsed_time() / avg_urlparse_time
# + [markdown] slideshow={"slide_type": "subslide"}
# Hence, in the time it takes to run `webbrowser()` once, we can have _tens of thousands_ of executions of `urlparse()` – and this does not even take into account the time it takes the browser to render the downloaded HTML, to run the included scripts, and whatever else happens when a Web page is loaded. Hence, strategies that allow us to test at the _unit_ level are very promising as they can save lots of overhead.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Carving Unit Tests
#
# Testing methods and functions at the unit level requires a very good understanding of the individual units to be tested as well as their interplay with other units. Setting up an appropriate infrastructure and writing unit tests by hand thus is demanding, yet rewarding. There is, however, an interesting alternative to writing unit tests by hand. The technique of _carving_ automatically _converts system tests into unit tests_ by means of recording and replaying function calls:
#
# 1. During a system test (given or generated), we _record_ all calls into a function, including all arguments and other variables the function reads.
# 2. From these, we synthesize a self-contained _unit test_ that reconstructs the function call with all arguments.
# 3. This unit test can be executed (replayed) at any time with high efficiency.
#
# In the remainder of this chapter, let us explore these steps.
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Recording Calls
#
# Our first challenge is to record function calls together with their arguments. (In the interest of simplicity, we restrict ourself to arguments, ignoring any global variables or other non-arguments that are read by the function.) To record calls and arguments, we use the mechanism [we introduced for coverage](Coverage.ipynb): By setting up a tracer function, we track all calls into individual functions, also saving their arguments. Just like `Coverage` objects, we want to use `Carver` objects to be able to be used in conjunction with the `with` statement, such that we can trace a particular code block:
#
# ```python
# with Carver() as carver:
# function_to_be_traced()
# c = carver.calls()
# ```
#
# The initial definition supports this construct:
# + [markdown] slideshow={"slide_type": "subslide"}
# \todo{Get tracker from [dynamic invariants](DynamicInvariants.ipynb)}
# + slideshow={"slide_type": "skip"}
import sys
# + slideshow={"slide_type": "subslide"}
class Carver(object):
def __init__(self, log=False):
self._log = log
self.reset()
def reset(self):
self._calls = {}
# Start of `with` block
def __enter__(self):
self.original_trace_function = sys.gettrace()
sys.settrace(self.traceit)
return self
# End of `with` block
def __exit__(self, exc_type, exc_value, tb):
sys.settrace(self.original_trace_function)
# + [markdown] slideshow={"slide_type": "subslide"}
# The actual work takes place in the `traceit()` method, which records all calls in the `_calls` attribute. First, we define two helper functions:
# + slideshow={"slide_type": "skip"}
import inspect
# + slideshow={"slide_type": "fragment"}
def get_qualified_name(code):
"""Return the fully qualified name of the current function"""
name = code.co_name
module = inspect.getmodule(code)
if module is not None:
name = module.__name__ + "." + name
return name
# + slideshow={"slide_type": "subslide"}
def get_arguments(frame):
"""Return call arguments in the given frame"""
# When called, all arguments are local variables
local_variables = frame.f_locals.copy()
arguments = [(var, frame.f_locals[var])
for var in local_variables]
arguments.reverse() # Want same order as call
return arguments
# + slideshow={"slide_type": "subslide"}
class CallCarver(Carver):
def add_call(self, function_name, arguments):
"""Add given call to list of calls"""
if function_name not in self._calls:
self._calls[function_name] = []
self._calls[function_name].append(arguments)
# Tracking function: Record all calls and all args
def traceit(self, frame, event, arg):
if event != "call":
return None
code = frame.f_code
function_name = code.co_name
qualified_name = get_qualified_name(code)
arguments = get_arguments(frame)
self.add_call(function_name, arguments)
if qualified_name != function_name:
self.add_call(qualified_name, arguments)
if self._log:
print(simple_call_string(function_name, arguments))
return None
# + [markdown] slideshow={"slide_type": "subslide"}
# Finally, we need some convenience functions to access the calls:
# + slideshow={"slide_type": "subslide"}
class CallCarver(CallCarver):
def calls(self):
"""Return a dictionary of all calls traced."""
return self._calls
def arguments(self, function_name):
"""Return a list of all arguments of the given function
as (VAR, VALUE) pairs.
Raises an exception if the function was not traced."""
return self._calls[function_name]
def called_functions(self, qualified=False):
"""Return all functions called."""
if qualified:
return [function_name for function_name in self._calls.keys()
if function_name.find('.') >= 0]
else:
return [function_name for function_name in self._calls.keys()
if function_name.find('.') < 0]
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Recording my_sqrt()
# + [markdown] slideshow={"slide_type": "fragment"}
# Let's try out our new `Carver` class – first on a very simple function:
# + slideshow={"slide_type": "skip"}
from Intro_Testing import my_sqrt
# + slideshow={"slide_type": "fragment"}
with CallCarver() as sqrt_carver:
my_sqrt(2)
my_sqrt(4)
# + [markdown] slideshow={"slide_type": "fragment"}
# We can retrieve all calls seen...
# + slideshow={"slide_type": "fragment"}
sqrt_carver.calls()
# + slideshow={"slide_type": "subslide"}
sqrt_carver.called_functions()
# + [markdown] slideshow={"slide_type": "fragment"}
# ... as well as the arguments of a particular function:
# + slideshow={"slide_type": "fragment"}
sqrt_carver.arguments("my_sqrt")
# + [markdown] slideshow={"slide_type": "fragment"}
# We define a convenience function for nicer printing of these lists:
# + slideshow={"slide_type": "fragment"}
def simple_call_string(function_name, argument_list):
"""Return function_name(arg[0], arg[1], ...) as a string"""
return function_name + "(" + \
", ".join([var + "=" + repr(value)
for (var, value) in argument_list]) + ")"
# + slideshow={"slide_type": "subslide"}
for function_name in sqrt_carver.called_functions():
for argument_list in sqrt_carver.arguments(function_name):
print(simple_call_string(function_name, argument_list))
# + [markdown] slideshow={"slide_type": "fragment"}
# This is a syntax we can directly use to invoke `my_sqrt()` again:
# + slideshow={"slide_type": "fragment"}
eval("my_sqrt(x=2)")
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Carving urlparse()
# + [markdown] slideshow={"slide_type": "fragment"}
# What happens if we apply this to `webbrowser()`?
# + slideshow={"slide_type": "fragment"}
with CallCarver() as webbrowser_carver:
webbrowser("http://www.example.com")
# + [markdown] slideshow={"slide_type": "fragment"}
# We see that retrieving a URL from the Web requires quite some functionality:
# + slideshow={"slide_type": "fragment"}
function_list = webbrowser_carver.called_functions(qualified=True)
len(function_list)
# + slideshow={"slide_type": "fragment"}
print(function_list[:50])
# + [markdown] slideshow={"slide_type": "fragment"}
# Among several other functions, we also have a call to `urlparse()`:
# + slideshow={"slide_type": "subslide"}
urlparse_argument_list = webbrowser_carver.arguments("urllib.parse.urlparse")
urlparse_argument_list
# + [markdown] slideshow={"slide_type": "subslide"}
# Again, we can convert this into a well-formatted call:
# + slideshow={"slide_type": "fragment"}
urlparse_call = simple_call_string("urlparse", urlparse_argument_list[0])
urlparse_call
# + [markdown] slideshow={"slide_type": "fragment"}
# Again, we can re-execute this call:
# + slideshow={"slide_type": "fragment"}
eval(urlparse_call)
# + [markdown] slideshow={"slide_type": "fragment"}
# We now have successfully carved the call to `urlparse()` out of the `webbrowser()` execution.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Replaying Calls
# + [markdown] slideshow={"slide_type": "subslide"}
# Replaying calls in their entirety and in all generality is tricky, as there are several challenges to be addressed. These include:
#
# 1. We need to be able to _access_ individual functions. If we access a function by name, the name must be in scope. If the name is not visible (for instance, because it is a name internal to the module), we must make it visible.
#
# 2. Any _resources_ accessed outside of arguments must be recorded and reconstructed for replay as well. This can be difficult if variables refer to external resources such as files or network resources.
#
# 3. _Complex objects_ must be reconstructed as well.
# + [markdown] slideshow={"slide_type": "subslide"}
# These constraints make carving hard or even impossible if the function to be tested interacts heavily with its environment. To illustrate these issues, consider the `email.parser.parse()` method that is invoked in `webbrowser()`:
# + slideshow={"slide_type": "fragment"}
email_parse_argument_list = webbrowser_carver.arguments("email.parser.parse")
# + [markdown] slideshow={"slide_type": "fragment"}
# Calls to this method look like this:
# + slideshow={"slide_type": "fragment"}
email_parse_call = simple_call_string(
"email.parser.Parser.parse",
email_parse_argument_list[0])
email_parse_call
# + [markdown] slideshow={"slide_type": "fragment"}
# We see that `email.parser.Parser.parse()` is part of a `email.parser.Parser` object (`self`) and it gets a `StringIO` object (`fp`). Both are non-primitive values. How could we possibly reconstruct them?
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Serializing Objects
#
# The answer to the problem of complex objects lies in creating a _persistent_ representation that can be _reconstructed_ at later points in time. This process is known as _serialization_; in Python, it is also known as _pickling_. The `pickle` module provides means to create a serialized representation of an object. Let us apply this on the `email.parser.Parser` object we just found:
# + slideshow={"slide_type": "skip"}
import pickle
# + slideshow={"slide_type": "fragment"}
email_parse_argument_list
# + slideshow={"slide_type": "fragment"}
parser_object = email_parse_argument_list[0][2][1]
parser_object
# + slideshow={"slide_type": "subslide"}
pickled = pickle.dumps(parser_object)
pickled
# + [markdown] slideshow={"slide_type": "fragment"}
# From this string representing the serialized `email.parser.Parser` object, we can recreate the Parser object at any time:
# + slideshow={"slide_type": "fragment"}
unpickled_parser_object = pickle.loads(pickled)
unpickled_parser_object
# + [markdown] slideshow={"slide_type": "fragment"}
# The serialization mechanism allows us to produce a representation for all objects passed as parameters (assuming they can be pickled, that is). We can now extend the `simple_call_string()` function such that it automatically pickles objects. Additionally, we set it up such that if the first parameter is named `self` (i.e., it is a class method), we make it a method of the `self` object.
# + slideshow={"slide_type": "subslide"}
def call_value(value):
value_as_string = repr(value)
if value_as_string.find('<') >= 0:
# Complex object
value_as_string = "pickle.loads(" + repr(pickle.dumps(value)) + ")"
return value_as_string
# + slideshow={"slide_type": "subslide"}
def call_string(function_name, argument_list):
"""Return function_name(arg[0], arg[1], ...) as a string, pickling complex objects"""
if len(argument_list) > 0:
(first_var, first_value) = argument_list[0]
if first_var == "self":
# Make this a method call
method_name = function_name.split(".")[-1]
function_name = call_value(first_value) + "." + method_name
argument_list = argument_list[1:]
return function_name + "(" + \
", ".join([var + "=" + call_value(value)
for (var, value) in argument_list]) + ")"
# + [markdown] slideshow={"slide_type": "fragment"}
# Let us apply the extended `call_string()` method to create a call for `email.parser.parse()`, including pickled objects:
# + slideshow={"slide_type": "subslide"}
call = call_string("email.parser.Parser.parse", email_parse_argument_list[0])
print(call)
# + [markdown] slideshow={"slide_type": "fragment"}
# With this call involving the pickled object, we can now re-run the original call and obtain a valid result:
# + slideshow={"slide_type": "skip"}
import email
# + slideshow={"slide_type": "fragment"}
eval(call)
# + [markdown] slideshow={"slide_type": "subslide"}
# ### All Calls
#
# So far, we have seen only one call of `webbrowser()`. How many of the calls within `webbrowser()` can we actually carve and replay? Let us try this out and compute the numbers.
# + slideshow={"slide_type": "skip"}
import traceback
# + slideshow={"slide_type": "skip"}
import enum
import socket
# + slideshow={"slide_type": "fragment"}
all_functions = set(webbrowser_carver.called_functions(qualified=True))
call_success = set()
run_success = set()
# + slideshow={"slide_type": "subslide"}
exceptions_seen = set()
for function_name in webbrowser_carver.called_functions(qualified=True):
for argument_list in webbrowser_carver.arguments(function_name):
try:
call = call_string(function_name, argument_list)
call_success.add(function_name)
result = eval(call)
run_success.add(function_name)
except Exception as exc:
exceptions_seen.add(repr(exc))
# print("->", call, file=sys.stderr)
# traceback.print_exc()
# print("", file=sys.stderr)
continue
# + slideshow={"slide_type": "subslide"}
print("%d/%d calls (%.2f%%) successfully created and %d/%d calls (%.2f%%) successfully ran" % (
len(call_success), len(all_functions), len(
call_success) * 100 / len(all_functions),
len(run_success), len(all_functions), len(run_success) * 100 / len(all_functions)))
# + [markdown] slideshow={"slide_type": "fragment"}
# About a quarter of the calls succeed. Let us take a look into some of the error messages we get:
# + slideshow={"slide_type": "subslide"}
for i in range(10):
print(list(exceptions_seen)[i])
# + [markdown] slideshow={"slide_type": "subslide"}
# We see that:
#
# * **A large majority of calls could be converted into call strings.** If this is not the case, this is mostly due to having unserialized objects being passed.
# * **About a quarter of the calls could be executed.** The error messages for the failing runs are varied; the most frequent being that some internal name is invoked that is not in scope.
# + [markdown] slideshow={"slide_type": "fragment"}
# Our carving mechanism should be taken with a grain of salt: We still do not cover the situation where external variables and values (such as global variables) are being accessed, and the serialization mechanism cannot recreate external resources. Still, if the function of interest falls among those that _can_ be carved and replayed, we can very effectively re-run its calls with their original arguments.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Mining API Grammars from Carved Calls
#
# So far, we have used carved calls to replay exactly the same invocations as originally encountered. However, we can also _mutate_ carved calls to effectively fuzz APIs with previously recorded arguments.
#
# The general idea is as follows:
#
# 1. First, we record all calls of a specific function from a given execution of the program.
# 2. Second, we create a grammar that incorporates all these calls, with separate rules for each argument and alternatives for each value found; this allows us to produce calls that arbitrarily _recombine_ these arguments.
#
# Let us explore these steps in the following sections.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### From Calls to Grammars
#
# Let us start with an example. The `power(x, y)` function returns $x^y$; it is but a wrapper around the equivalent `math.pow()` function. (Since `power()` is defined in Python, we can trace it – in contrast to `math.pow()`, which is implemented in C.)
# + slideshow={"slide_type": "skip"}
import math
# + slideshow={"slide_type": "fragment"}
def power(x, y):
return math.pow(x, y)
# + [markdown] slideshow={"slide_type": "fragment"}
# Let us invoke `power()` while recording its arguments:
# + slideshow={"slide_type": "fragment"}
with CallCarver() as power_carver:
z = power(1, 2)
z = power(3, 4)
# + slideshow={"slide_type": "fragment"}
power_carver.arguments("power")
# + [markdown] slideshow={"slide_type": "subslide"}
# From this list of recorded arguments, we could now create a grammar for the `power()` call, with `x` and `y` expanding into the values seen:
# + slideshow={"slide_type": "skip"}
from Grammars import START_SYMBOL, is_valid_grammar, new_symbol, extend_grammar
# + slideshow={"slide_type": "fragment"}
POWER_GRAMMAR = {
"<start>": ["power(<x>, <y>)"],
"<x>": ["1", "3"],
"<y>": ["2", "4"]
}
assert is_valid_grammar(POWER_GRAMMAR)
# + [markdown] slideshow={"slide_type": "fragment"}
# When fuzzing with this grammar, we then get arbitrary combinations of `x` and `y`; aiming for coverage will ensure that all values are actually tested at least once:
# + slideshow={"slide_type": "skip"}
from GrammarCoverageFuzzer import GrammarCoverageFuzzer
# + slideshow={"slide_type": "subslide"}
power_fuzzer = GrammarCoverageFuzzer(POWER_GRAMMAR)
[power_fuzzer.fuzz() for i in range(5)]
# + [markdown] slideshow={"slide_type": "fragment"}
# What we need is a method to automatically convert the arguments as seen in `power_carver` to the grammar as seen in `POWER_GRAMMAR`. This is what we define in the next section.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### A Grammar Miner for Calls
#
# We introduce a class `CallGrammarMiner`, which, given a `Carver`, automatically produces a grammar from the calls seen. To initialize, we pass the carver object:
# + slideshow={"slide_type": "fragment"}
class CallGrammarMiner(object):
def __init__(self, carver, log=False):
self.carver = carver
self.log = log
# + [markdown] slideshow={"slide_type": "subslide"}
# #### Initial Grammar
#
# The initial grammar produces a single call. The possible `<call>` expansions are to be constructed later:
# + slideshow={"slide_type": "skip"}
import copy
# + slideshow={"slide_type": "fragment"}
class CallGrammarMiner(CallGrammarMiner):
CALL_SYMBOL = "<call>"
def initial_grammar(self):
return extend_grammar(
{START_SYMBOL: [self.CALL_SYMBOL],
self.CALL_SYMBOL: []
})
# + slideshow={"slide_type": "subslide"}
m = CallGrammarMiner(power_carver)
initial_grammar = m.initial_grammar()
initial_grammar
# + [markdown] slideshow={"slide_type": "subslide"}
# #### A Grammar from Arguments
#
# Let us start by creating a grammar from a list of arguments. The method `mine_arguments_grammar()` creates a grammar for the arguments seen during carving, such as these:
# + slideshow={"slide_type": "fragment"}
arguments = power_carver.arguments("power")
arguments
# + [markdown] slideshow={"slide_type": "fragment"}
# The `mine_arguments_grammar()` method iterates through the variables seen and creates a mapping `variables` of variable names to a set of values seen (as strings, going through `call_value()`). In a second step, it then creates a grammar with a rule for each variable name, expanding into the values seen.
# + slideshow={"slide_type": "subslide"}
class CallGrammarMiner(CallGrammarMiner):
def var_symbol(self, function_name, var, grammar):
return new_symbol(grammar, "<" + function_name + "-" + var + ">")
def mine_arguments_grammar(self, function_name, arguments, grammar):
var_grammar = {}
variables = {}
for argument_list in arguments:
for (var, value) in argument_list:
value_string = call_value(value)
if self.log:
print(var, "=", value_string)
if value_string.find("<") >= 0:
var_grammar["<langle>"] = ["<"]
value_string = value_string.replace("<", "<langle>")
if var not in variables:
variables[var] = set()
variables[var].add(value_string)
var_symbols = []
for var in variables:
var_symbol = self.var_symbol(function_name, var, grammar)
var_symbols.append(var_symbol)
var_grammar[var_symbol] = list(variables[var])
return var_grammar, var_symbols
# + slideshow={"slide_type": "subslide"}
m = CallGrammarMiner(power_carver)
var_grammar, var_symbols = m.mine_arguments_grammar(
"power", arguments, initial_grammar)
# + slideshow={"slide_type": "fragment"}
var_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# The additional return value `var_symbols` is a list of argument symbols in the call:
# + slideshow={"slide_type": "fragment"}
var_symbols
# + [markdown] slideshow={"slide_type": "subslide"}
# #### A Grammar from Calls
#
# To get the grammar for a single function (`mine_function_grammar()`), we add a call to the function:
# + slideshow={"slide_type": "subslide"}
class CallGrammarMiner(CallGrammarMiner):
def function_symbol(self, function_name, grammar):
return new_symbol(grammar, "<" + function_name + ">")
def mine_function_grammar(self, function_name, grammar):
arguments = self.carver.arguments(function_name)
if self.log:
print(function_name, arguments)
var_grammar, var_symbols = self.mine_arguments_grammar(
function_name, arguments, grammar)
function_grammar = var_grammar
function_symbol = self.function_symbol(function_name, grammar)
if len(var_symbols) > 0 and var_symbols[0].find("-self") >= 0:
# Method call
function_grammar[function_symbol] = [
var_symbols[0] + "." + function_name + "(" + ", ".join(var_symbols[1:]) + ")"]
else:
function_grammar[function_symbol] = [
function_name + "(" + ", ".join(var_symbols) + ")"]
if self.log:
print(function_symbol, "::=", function_grammar[function_symbol])
return function_grammar, function_symbol
# + slideshow={"slide_type": "subslide"}
m = CallGrammarMiner(power_carver)
function_grammar, function_symbol = m.mine_function_grammar(
"power", initial_grammar)
function_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# The additionally returned `function_symbol` holds the name of the function call just added:
# + slideshow={"slide_type": "fragment"}
function_symbol
# + [markdown] slideshow={"slide_type": "subslide"}
# #### A Grammar from all Calls
#
# Let us now repeat the above for all function calls seen during carving. To this end, we simply iterate over all function calls seen:
# + slideshow={"slide_type": "fragment"}
power_carver.called_functions()
# + slideshow={"slide_type": "subslide"}
class CallGrammarMiner(CallGrammarMiner):
def mine_call_grammar(self, function_list=None, qualified=False):
grammar = self.initial_grammar()
fn_list = function_list
if function_list is None:
fn_list = self.carver.called_functions(qualified=qualified)
for function_name in fn_list:
if function_list is None and (function_name.startswith("_") or function_name.startswith("<")):
continue # Internal function
# Ignore errors with mined functions
try:
function_grammar, function_symbol = self.mine_function_grammar(
function_name, grammar)
except:
if function_list is not None:
raise
if function_symbol not in grammar[self.CALL_SYMBOL]:
grammar[self.CALL_SYMBOL].append(function_symbol)
grammar.update(function_grammar)
assert is_valid_grammar(grammar)
return grammar
# + [markdown] slideshow={"slide_type": "subslide"}
# The method `mine_call_grammar()` is the one that clients can and should use – first for mining...
# + slideshow={"slide_type": "fragment"}
m = CallGrammarMiner(power_carver)
power_grammar = m.mine_call_grammar()
power_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# ...and then for fuzzing:
# + slideshow={"slide_type": "fragment"}
power_fuzzer = GrammarCoverageFuzzer(power_grammar)
[power_fuzzer.fuzz() for i in range(5)]
# + [markdown] slideshow={"slide_type": "subslide"}
# With this, we have successfully extracted a grammar from a recorded execution; in contrast to "simple" carving, our grammar allows us to _recombine_ arguments and thus to fuzz at the API level.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Fuzzing Web Functions
#
# Let us now apply our grammar miner on a larger API – the `urlparse()` function we already encountered during carving.
# + slideshow={"slide_type": "fragment"}
with CallCarver() as webbrowser_carver:
webbrowser("https://www.fuzzing<EMAIL>")
webbrowser("http://www.example.com")
# + [markdown] slideshow={"slide_type": "fragment"}
# We can mine a grammar from the calls encountered:
# + slideshow={"slide_type": "fragment"}
m = CallGrammarMiner(webbrowser_carver)
webbrowser_grammar = m.mine_call_grammar()
# + [markdown] slideshow={"slide_type": "fragment"}
# This is a rather large grammar:
# + slideshow={"slide_type": "fragment"}
call_list = webbrowser_grammar['<call>']
len(call_list)
# + slideshow={"slide_type": "subslide"}
print(call_list[:20])
# + [markdown] slideshow={"slide_type": "fragment"}
# Here's the rule for the `urlsplit()` function:
# + slideshow={"slide_type": "fragment"}
webbrowser_grammar["<urlsplit>"]
# + [markdown] slideshow={"slide_type": "fragment"}
# Here are the arguments. Note that although we only passed `http://www.fuzzingbook.org` as a parameter, we also see the `https:` variant. That is because opening the `http:` URL automatically redirects to the `https:` URL, which is then also processed by `urlsplit()`.
# + slideshow={"slide_type": "fragment"}
webbrowser_grammar["<urlsplit-url>"]
# + [markdown] slideshow={"slide_type": "subslide"}
# There also is some variation in the `scheme` argument:
# + slideshow={"slide_type": "fragment"}
webbrowser_grammar["<urlsplit-scheme>"]
# + [markdown] slideshow={"slide_type": "fragment"}
# If we now apply a fuzzer on these rules, we systematically cover all variations of arguments seen, including, of course, combinations not seen during carving. Again, we are fuzzing at the API level here.
# + slideshow={"slide_type": "subslide"}
urlsplit_fuzzer = GrammarCoverageFuzzer(
webbrowser_grammar, start_symbol="<urlsplit>")
for i in range(5):
print(urlsplit_fuzzer.fuzz())
# + [markdown] slideshow={"slide_type": "fragment"}
# Just as seen with carving, running tests at the API level is orders of magnitude faster than executing system tests. Hence, this calls for means to fuzz at the method level:
# + slideshow={"slide_type": "skip"}
from urllib.parse import urlsplit
# + slideshow={"slide_type": "skip"}
from Timer import Timer
# + slideshow={"slide_type": "subslide"}
with Timer() as urlsplit_timer:
urlsplit('http://www.fuzzingbook.org/', 'http', True)
urlsplit_timer.elapsed_time()
# + slideshow={"slide_type": "fragment"}
with Timer() as webbrowser_timer:
webbrowser("http://www.fuzzingbook.org")
webbrowser_timer.elapsed_time()
# + slideshow={"slide_type": "fragment"}
webbrowser_timer.elapsed_time() / urlsplit_timer.elapsed_time()
# + [markdown] slideshow={"slide_type": "subslide"}
# But then again, the caveats encountered during carving apply, notably the requirement to recreate the original function environment. If we also alter or recombine arguments, we get the additional risk of _violating an implicit precondition_ – that is, invoking a function with arguments the function was never designed for. Such _false alarms_, resulting from incorrect invocations rather than incorrect implementations, must then be identified (typically manually) and wed out (for instance, by altering or constraining the grammar). The huge speed gains at the API level, however, may well justify this additional investment.
# + [markdown] slideshow={"slide_type": "slide"}
# ## Synopsis
#
# This chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such "carving" mechanisms have the potential to run tests much faster.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Recording Calls
#
# The `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:
# + slideshow={"slide_type": "fragment"}
with CallCarver() as carver:
y = my_sqrt(2)
y = my_sqrt(4)
# + [markdown] slideshow={"slide_type": "fragment"}
# After execution, `called_functions()` lists the names of functions encountered:
# + slideshow={"slide_type": "fragment"}
carver.called_functions()
# + [markdown] slideshow={"slide_type": "fragment"}
# The `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).
# + slideshow={"slide_type": "subslide"}
carver.arguments('my_sqrt')
# + [markdown] slideshow={"slide_type": "fragment"}
# Complex arguments are properly serialized, such that they can be easily restored.
# + [markdown] slideshow={"slide_type": "subslide"}
# ### Synthesizing Calls
#
# While such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.
# + [markdown] slideshow={"slide_type": "fragment"}
# The `CallGrammarMiner` class turns a list of carved executions into a grammar.
# + slideshow={"slide_type": "subslide"}
my_sqrt_miner = CallGrammarMiner(carver)
my_sqrt_grammar = my_sqrt_miner.mine_call_grammar()
my_sqrt_grammar
# + [markdown] slideshow={"slide_type": "fragment"}
# This grammar can be used to synthesize calls.
# + slideshow={"slide_type": "fragment"}
fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)
fuzzer.fuzz()
# + [markdown] slideshow={"slide_type": "fragment"}
# These calls can be executed in isolation, effectively extracting unit tests from system tests:
# + slideshow={"slide_type": "fragment"}
eval(fuzzer.fuzz())
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Lessons Learned
#
# * _Carving_ allows for effective replay of function calls recorded during a system test.
# * A function call can be _orders of magnitude faster_ than a system invocation.
# * _Serialization_ allows to create persistent representations of complex objects.
# * Functions that heavily interact with their environment and/or access external resources are difficult to carve.
# * From carved calls, one can produce API grammars that arbitrarily combine carved arguments.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Next Steps
#
# In the next chapter, we will discuss [how to reduce failure-inducing inputs](Reducer.ipynb).
# + [markdown] slideshow={"slide_type": "slide"}
# ## Background
#
# Carving was invented by Elbaum et al. \cite{Elbaum2006} and originally implemented for Java. In this chapter, we follow several of their design choices (including recording and serializing method arguments only).
#
# The combination of carving and fuzzing at the API level is described in \cite{Kampmann2018}.
# + [markdown] button=false new_sheet=true run_control={"read_only": false} slideshow={"slide_type": "slide"}
# ## Exercises
#
# ### Exercise 1: Carving for Regression Testing
#
# So far, during carving, we only have looked into reproducing _calls_, but not into actually checking the _results_ of these calls. This is important for _regression testing_ – i.e. checking whether a change to code does not impede existing functionality. We can build this by recording not only _calls_, but also _return values_ – and then later compare whether the same calls result in the same values. This may not work on all occasions; values that depend on time, randomness, or other external factors may be different. Still, for functionality that abstracts from these details, checking that nothing has changed is an important part of testing.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"} solution="hidden" solution2="hidden" solution2_first=true solution_first=true
# Our aim is to design a class `ResultCarver` that extends `CallCarver` by recording both calls and return values.
#
# In a first step, create a `traceit()` method that also tracks return values by extending the `traceit()` method. The `traceit()` event type is `"return"` and the `arg` parameter is the returned value. Here is a prototype that only prints out the returned values:
# + slideshow={"slide_type": "subslide"}
class ResultCarver(CallCarver):
def traceit(self, frame, event, arg):
if event == "return":
if self._log:
print("Result:", arg)
super().traceit(frame, event, arg)
# Need to return traceit function such that it is invoked for return
# events
return self.traceit
# + slideshow={"slide_type": "subslide"}
with ResultCarver(log=True) as result_carver:
my_sqrt(2)
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# #### Part 1: Store function results
#
# Extend the above code such that results are _stored_ in a way that associates them with the currently returning function (or method). To this end, you need to keep track of the _current stack of called functions_.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"} solution="hidden" solution2="hidden"
# **Solution.** Here's a solution, building on the above:
# + slideshow={"slide_type": "skip"} solution2="hidden"
class ResultCarver(CallCarver):
def reset(self):
super().reset()
self._call_stack = []
self._results = {}
def add_result(self, function_name, arguments, result):
key = simple_call_string(function_name, arguments)
self._results[key] = result
def traceit(self, frame, event, arg):
if event == "call":
code = frame.f_code
function_name = code.co_name
qualified_name = get_qualified_name(code)
self._call_stack.append(
(function_name, qualified_name, get_arguments(frame)))
if event == "return":
result = arg
(function_name, qualified_name, arguments) = self._call_stack.pop()
self.add_result(function_name, arguments, result)
if function_name != qualified_name:
self.add_result(qualified_name, arguments, result)
if self._log:
print(
simple_call_string(
function_name,
arguments),
"=",
result)
# Keep on processing current calls
super().traceit(frame, event, arg)
# Need to return traceit function such that it is invoked for return
# events
return self.traceit
# + slideshow={"slide_type": "skip"} solution2="hidden"
with ResultCarver(log=True) as result_carver:
my_sqrt(2)
result_carver._results
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"} solution="hidden" solution2="hidden" solution2_first=true solution_first=true
# #### Part 2: Access results
#
# Give it a method `result()` that returns the value recorded for that particular function name and result:
#
# ```python
# class ResultCarver(CallCarver):
# def result(self, function_name, argument):
# """Returns the result recorded for function_name(argument"""
# ```
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** This is mostly done in the code for part 1:
# + slideshow={"slide_type": "skip"} solution2="hidden"
class ResultCarver(ResultCarver):
def result(self, function_name, argument):
key = simple_call_string(function_name, arguments)
return self._results[key]
# + [markdown] slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# #### Part 3: Produce assertions
#
# For the functions called during `webbrowser()` execution, create a set of _assertions_ that check whether the result returned is still the same. Test this for `urllib.parse.urlparse()` and `urllib.parse.urlsplit()`.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "skip"} solution="hidden" solution2="hidden"
# **Solution.** Not too hard now:
# + slideshow={"slide_type": "skip"} solution2="hidden"
with ResultCarver() as webbrowser_result_carver:
webbrowser("http://www.example.com")
# + slideshow={"slide_type": "skip"} solution2="hidden"
for function_name in ["urllib.parse.urlparse", "urllib.parse.urlsplit"]:
for arguments in webbrowser_result_carver.arguments(function_name):
try:
call = call_string(function_name, arguments)
result = webbrowser_result_carver.result(function_name, arguments)
print("assert", call, "==", call_value(result))
except Exception:
continue
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# We can run these assertions:
# + slideshow={"slide_type": "skip"} solution2="hidden"
from urllib.parse import SplitResult, ParseResult, urlparse, urlsplit
# + slideshow={"slide_type": "skip"} solution2="hidden"
assert urlparse(
url='http://www.example.com',
scheme='',
allow_fragments=True) == ParseResult(
scheme='http',
netloc='www.example.com',
path='',
params='',
query='',
fragment='')
assert urlsplit(
url='http://www.example.com',
scheme='',
allow_fragments=True) == SplitResult(
scheme='http',
netloc='www.example.com',
path='',
query='',
fragment='')
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# We can now add these carved tests to a _regression test suite_ which would be run after every change to ensure that the functionality of `urlparse()` and `urlsplit()` is not changed.
# + [markdown] button=false new_sheet=false run_control={"read_only": false} slideshow={"slide_type": "subslide"} solution2="hidden" solution2_first=true
# ### Exercise 2: Abstracting Arguments
#
# When mining an API grammar from executions, set up an abstraction scheme to widen the range of arguments to be used during testing. If the values for an argument, all conform to some type `T`. abstract it into `<T>`. For instance, if calls to `foo(1)`, `foo(2)`, `foo(3)` have been seen, the grammar should abstract its calls into `foo(<int>)`, with `<int>` being appropriately defined.
#
# Do this for a number of common types: integers, positive numbers, floating-point numbers, host names, URLs, mail addresses, and more.
# + [markdown] slideshow={"slide_type": "skip"} solution2="hidden"
# **Solution.** Left to the reader.
| docs/beta/notebooks/Carver.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="Cb4espuLKJiA"
# ##### Copyright 2021 The TensorFlow Authors.
# + cellView="form" id="DjZQV2njKJ3U"
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] id="mTL0TERThT6z"
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/tutorials/audio/transfer_learning_audio"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/audio/transfer_learning_audio.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/audio/transfer_learning_audio.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/tutorials/audio/transfer_learning_audio.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# <td>
# <a href="https://tfhub.dev/google/yamnet/1"><img src="https://www.tensorflow.org/images/hub_logo_32px.png" />See TF Hub model</a>
# </td>
# </table>
# + [markdown] id="K2madPFAGHb3"
# # Transfer Learning with YAMNet for environmental sound classification
#
# [YAMNet](https://tfhub.dev/google/yamnet/1) is an audio event classifier that can predict audio events from [521 classes](https://github.com/tensorflow/models/blob/master/research/audioset/yamnet/yamnet_class_map.csv), like laughter, barking, or a siren.
#
# In this tutorial you will learn how to:
#
# - Load and use the YAMNet model for inference.
# - Build a new model using the YAMNet embeddings to classify cat and dog sounds.
# - Evaluate and export your model.
#
# + [markdown] id="5Mdp2TpBh96Y"
# ## Import TensorFlow and other libraries
#
# + [markdown] id="zCcKYqu_hvKe"
# Start by installing [TensorFlow I/O](https://www.tensorflow.org/io), which will make it easier for you to load audio files off disk.
# + id="urBpRWDHTHHU"
# !pip install tensorflow_io
# + id="7l3nqdWVF-kC"
import os
from IPython import display
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
import tensorflow_io as tfio
# + [markdown] id="v9ZhybCnt_bM"
# ## About YAMNet
#
# YAMNet is an audio event classifier that takes audio waveform as input and makes independent predictions for each of 521 audio events from the [AudioSet](https://research.google.com/audioset/) ontology.
#
# Internally, the model extracts "frames" from the audio signal and processes batches of these frames. This version of the model uses frames that are 0.96s long and extracts one frame every 0.48s.
#
# The model accepts a 1-D float32 Tensor or NumPy array containing a waveform of arbitrary length, represented as mono 16 kHz samples in the range `[-1.0, +1.0]`. This tutorial contains code to help you convert a `.wav` file into the correct format.
#
# The model returns 3 outputs, including the class scores, embeddings (which you will use for transfer learning), and the log mel spectrogram. You can find more details [here](https://tfhub.dev/google/yamnet/1), and this tutorial will walk you through using these in practice.
#
# One specific use of YAMNet is as a high-level feature extractor: the `1024-D` embedding output of YAMNet can be used as the input features of another shallow model which can then be trained on a small amount of data for a particular task. This allows the quick creation of specialized audio classifiers without requiring a lot of labeled data and without having to train a large model end-to-end.
#
# You will use YAMNet's embeddings output for transfer learning and train one or more [Dense](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense) layers on top of this.
#
# First, you will try the model and see the results of classifying audio. You will then construct the data pre-processing pipeline.
#
# ### Loading YAMNet from TensorFlow Hub
#
# You are going to use YAMNet from [Tensorflow Hub](https://tfhub.dev/) to extract the embeddings from the sound files.
#
# Loading a model from TensorFlow Hub is straightforward: choose the model, copy its URL and use the `load` function.
#
# Note: to read the documentation of the model, you can use the model url in your browser.
# + id="06CWkBV5v3gr"
yamnet_model_handle = 'https://tfhub.dev/google/yamnet/1'
yamnet_model = hub.load(yamnet_model_handle)
# + [markdown] id="GmrPJ0GHw9rr"
# With the model loaded and following the [models's basic usage tutorial](https://www.tensorflow.org/hub/tutorials/yamnet) you'll download a sample wav file and run the inference.
#
# + id="C5i6xktEq00P"
testing_wav_file_name = tf.keras.utils.get_file('miaow_16k.wav',
'https://storage.googleapis.com/audioset/miaow_16k.wav',
cache_dir='./',
cache_subdir='test_data')
print(testing_wav_file_name)
# + [markdown] id="mBm9y9iV2U_-"
# You will need a function to load the audio files. They will also be used later when working with the training data.
#
# Note: The returned `wav_data` from `load_wav_16k_mono` is already normalized to values in `[-1.0, 1.0]` (as stated in the model's [documentation](https://tfhub.dev/google/yamnet/1)).
# + id="Xwc9Wrdg2EtY"
# Util functions for loading audio files and ensure the correct sample rate
@tf.function
def load_wav_16k_mono(filename):
""" read in a waveform file and convert to 16 kHz mono """
file_contents = tf.io.read_file(filename)
wav, sample_rate = tf.audio.decode_wav(
file_contents,
desired_channels=1)
wav = tf.squeeze(wav, axis=-1)
sample_rate = tf.cast(sample_rate, dtype=tf.int64)
wav = tfio.audio.resample(wav, rate_in=sample_rate, rate_out=16000)
return wav
# + id="FRqpjkwB0Jjw"
testing_wav_data = load_wav_16k_mono(testing_wav_file_name)
_ = plt.plot(testing_wav_data)
# Play the audio file.
display.Audio(testing_wav_data,rate=16000)
# + [markdown] id="6z6rqlEz20YB"
# ### Load the class mapping
#
# It's important to load the class names that YAMNet is able to recognize. The mapping file is present at `yamnet_model.class_map_path()`, in the `csv` format.
# + id="6Gyj23e_3Mgr"
class_map_path = yamnet_model.class_map_path().numpy().decode('utf-8')
class_names =list(pd.read_csv(class_map_path)['display_name'])
for name in class_names[:20]:
print(name)
print('...')
# + [markdown] id="5xbycDnT40u0"
# ### Run inference
#
# YAMNet provides frame-level class-scores (i.e., 521 scores for every frame). In order to determine clip-level predictions, the scores can be aggregated per-class across frames (e.g., using mean or max aggregation). This is done below by `scores_np.mean(axis=0)`. Finally, in order to find the top-scored class at the clip-level, we take the maximum of the 521 aggregated scores.
#
# + id="NT0otp-A4Y3u"
scores, embeddings, spectrogram = yamnet_model(testing_wav_data)
class_scores = tf.reduce_mean(scores, axis=0)
top_class = tf.argmax(class_scores)
infered_class = class_names[top_class]
print(f'The main sound is: {infered_class}')
print(f'The embeddings shape: {embeddings.shape}')
# + [markdown] id="YBaLNg5H5IWa"
# Note: The model correctly inferred an animal sound. Your goal is to increase accuracy for specific classes. Also, notice that the the model generated 13 embeddings, 1 per frame.
# + [markdown] id="fmthELBg1A2-"
# ## ESC-50 dataset
#
# The [ESC-50 dataset](https://github.com/karolpiczak/ESC-50#repository-content), well described [here](https://www.karolpiczak.com/papers/Piczak2015-ESC-Dataset.pdf), is a labeled collection of 2000 environmental audio recordings (each 5 seconds long). The data consists of 50 classes, with 40 examples per class.
#
# Next, you will download and extract it.
#
# + id="MWobqK8JmZOU"
_ = tf.keras.utils.get_file('esc-50.zip',
'https://github.com/karoldvl/ESC-50/archive/master.zip',
cache_dir='./',
cache_subdir='datasets',
extract=True)
# + [markdown] id="qcruxiuX1cO5"
# ### Explore the data
#
# The metadata for each file is specified in the csv file at `./datasets/ESC-50-master/meta/esc50.csv`
#
# and all the audio files are in `./datasets/ESC-50-master/audio/`
#
# You will create a pandas dataframe with the mapping and use that to have a clearer view of the data.
#
# + id="jwmLygPrMAbH"
esc50_csv = './datasets/ESC-50-master/meta/esc50.csv'
base_data_path = './datasets/ESC-50-master/audio/'
pd_data = pd.read_csv(esc50_csv)
pd_data.head()
# + [markdown] id="7d4rHBEQ2QAU"
# ### Filter the data
#
# Given the data on the dataframe, you will apply some transformations:
#
# - filter out rows and use only the selected classes (dog and cat). If you want to use any other classes, this is where you can choose them.
# - change the filename to have the full path. This will make loading easier later.
# - change targets to be within a specific range. In this example, dog will remain 0, but cat will become 1 instead of its original value of 5.
# + id="tFnEoQjgs14I"
my_classes = ['dog', 'cat']
map_class_to_id = {'dog':0, 'cat':1}
filtered_pd = pd_data[pd_data.category.isin(my_classes)]
class_id = filtered_pd['category'].apply(lambda name: map_class_to_id[name])
filtered_pd = filtered_pd.assign(target=class_id)
full_path = filtered_pd['filename'].apply(lambda row: os.path.join(base_data_path, row))
filtered_pd = filtered_pd.assign(filename=full_path)
filtered_pd.head(10)
# + [markdown] id="BkDcBS-aJdCz"
# ### Load the audio files and retrieve embeddings
#
# Here you'll apply the `load_wav_16k_mono` and prepare the wav data for the model.
#
# When extracting embeddings from the wav data, you get an array of shape `(N, 1024)` where `N` is the number of frames that YAMNet found (one for every 0.48 seconds of audio).
# + [markdown] id="AKDT5RomaDKO"
# Your model will use each frame as one input so you need to to create a new column that has one frame per row. You also need to expand the labels and fold column to proper reflect these new rows.
#
# The expanded fold column keeps the original value. You cannot mix frames because, when doing the splits, you might end with parts of the same audio on different splits and that would make our validation and test steps less effective.
# + id="u5Rq3_PyKLtU"
filenames = filtered_pd['filename']
targets = filtered_pd['target']
folds = filtered_pd['fold']
main_ds = tf.data.Dataset.from_tensor_slices((filenames, targets, folds))
main_ds.element_spec
# + id="rsEfovDVAHGY"
def load_wav_for_map(filename, label, fold):
return load_wav_16k_mono(filename), label, fold
main_ds = main_ds.map(load_wav_for_map)
main_ds.element_spec
# + id="k0tG8DBNAHcE"
# applies the embedding extraction model to a wav data
def extract_embedding(wav_data, label, fold):
''' run YAMNet to extract embedding from the wav data '''
scores, embeddings, spectrogram = yamnet_model(wav_data)
num_embeddings = tf.shape(embeddings)[0]
return (embeddings,
tf.repeat(label, num_embeddings),
tf.repeat(fold, num_embeddings))
# extract embedding
main_ds = main_ds.map(extract_embedding).unbatch()
main_ds.element_spec
# + [markdown] id="ZdfPIeD0Qedk"
# ### Split the data
#
# You will use the `fold` column to split the dataset into train, validation and test.
#
# The fold values are so that files from the same original wav file are keep on the same split, you can find more information on the [paper](https://www.karolpiczak.com/papers/Piczak2015-ESC-Dataset.pdf) describing the dataset.
#
# The last step is to remove the `fold` column from the dataset since we're not going to use it anymore on the training process.
#
# + id="1ZYvlFiVsffC"
cached_ds = main_ds.cache()
train_ds = cached_ds.filter(lambda embedding, label, fold: fold < 4)
val_ds = cached_ds.filter(lambda embedding, label, fold: fold == 4)
test_ds = cached_ds.filter(lambda embedding, label, fold: fold == 5)
# remove the folds column now that it's not needed anymore
remove_fold_column = lambda embedding, label, fold: (embedding, label)
train_ds = train_ds.map(remove_fold_column)
val_ds = val_ds.map(remove_fold_column)
test_ds = test_ds.map(remove_fold_column)
train_ds = train_ds.cache().shuffle(1000).batch(32).prefetch(tf.data.AUTOTUNE)
val_ds = val_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
test_ds = test_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
# + [markdown] id="v5PaMwvtcAIe"
# ## Create your model
#
# You did most of the work!
# Next, define a very simple Sequential Model to start with -- one hiden layer and 2 outputs to recognize cats and dogs.
#
# + id="JYCE0Fr1GpN3"
my_model = tf.keras.Sequential([
tf.keras.layers.Input(shape=(1024), dtype=tf.float32,
name='input_embedding'),
tf.keras.layers.Dense(512, activation='relu'),
tf.keras.layers.Dense(len(my_classes))
], name='my_model')
my_model.summary()
# + id="l1qgH35HY0SE"
my_model.compile(loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer="adam",
metrics=['accuracy'])
callback = tf.keras.callbacks.EarlyStopping(monitor='loss',
patience=3,
restore_best_weights=True)
# + id="T3sj84eOZ3pk"
history = my_model.fit(train_ds,
epochs=20,
validation_data=val_ds,
callbacks=callback)
# + [markdown] id="OAbraYKYpdoE"
# Lets run the evaluate method on the test data just to be sure there's no overfitting.
# + id="H4Nh5nec3Sky"
loss, accuracy = my_model.evaluate(test_ds)
print("Loss: ", loss)
print("Accuracy: ", accuracy)
# + [markdown] id="cid-qIrIpqHS"
# You did it!
# + [markdown] id="nCKZonrJcXab"
# ## Test your model
#
# Next, try your model on the embedding from the previous test using YAMNet only.
#
# + id="79AFpA3_ctCF"
scores, embeddings, spectrogram = yamnet_model(testing_wav_data)
result = my_model(embeddings).numpy()
infered_class = my_classes[result.mean(axis=0).argmax()]
print(f'The main sound is: {infered_class}')
# + [markdown] id="k2yleeev645r"
# ## Save a model that can directly take a wav file as input
#
# Your model works when you give it the embeddings as input.
#
# In a real situation you'll want to give it the sound data directly.
#
# To do that you will combine YAMNet with your model into one single model that you can export for other applications.
#
# To make it easier to use the model's result, the final layer will be a `reduce_mean` operation. When using this model for serving, as you will see bellow, you will need the name of the final layer. If you don't define one, TF will auto define an incremental one that makes it hard to test as it will keep changing everytime you train the model. When using a raw tf operation you can't assign a name to it. To address this issue, you'll create a custom layer that just apply `reduce_mean` and you will call it 'classifier'.
#
# + id="QUVCI2Suunpw"
class ReduceMeanLayer(tf.keras.layers.Layer):
def __init__(self, axis=0, **kwargs):
super(ReduceMeanLayer, self).__init__(**kwargs)
self.axis = axis
def call(self, input):
return tf.math.reduce_mean(input, axis=self.axis)
# + id="zE_Npm0nzlwc"
saved_model_path = './dogs_and_cats_yamnet'
input_segment = tf.keras.layers.Input(shape=(), dtype=tf.float32, name='audio')
embedding_extraction_layer = hub.KerasLayer(yamnet_model_handle,
trainable=False, name='yamnet')
_, embeddings_output, _ = embedding_extraction_layer(input_segment)
serving_outputs = my_model(embeddings_output)
serving_outputs = ReduceMeanLayer(axis=0, name='classifier')(serving_outputs)
serving_model = tf.keras.Model(input_segment, serving_outputs)
serving_model.save(saved_model_path, include_optimizer=False)
# + id="y-0bY5FMme1C"
tf.keras.utils.plot_model(serving_model)
# + [markdown] id="btHQDN9mqxM_"
# Load your saved model to verify that it works as expected.
# + id="KkYVpJS72WWB"
reloaded_model = tf.saved_model.load(saved_model_path)
# + [markdown] id="4BkmvvNzq49l"
# And for the final test: given some sound data, does your model return the correct result?
# + id="xeXtD5HO28y-"
reloaded_results = reloaded_model(testing_wav_data)
cat_or_dog = my_classes[tf.argmax(reloaded_results)]
print(f'The main sound is: {cat_or_dog}')
# + [markdown] id="ZRrOcBYTUgwn"
# If you want to try your new model on a serving setup, you can use the 'serving_default' signature.
# + id="ycC8zzDSUG2s"
serving_results = reloaded_model.signatures['serving_default'](testing_wav_data)
cat_or_dog = my_classes[tf.argmax(serving_results['classifier'])]
print(f'The main sound is: {cat_or_dog}')
# + [markdown] id="da7blblCHs8c"
# ## (Optional) Some more testing
#
# The model is ready.
#
# Let's compare it to YAMNet on the test dataset.
# + id="vDf5MASIIN1z"
test_pd = filtered_pd.loc[filtered_pd['fold'] == 5]
row = test_pd.sample(1)
filename = row['filename'].item()
print(filename)
waveform = load_wav_16k_mono(filename)
print(f'Waveform values: {waveform}')
_ = plt.plot(waveform)
display.Audio(waveform, rate=16000)
# + id="eYUzFxYJIcE1"
# Run the model, check the output.
scores, embeddings, spectrogram = yamnet_model(waveform)
class_scores = tf.reduce_mean(scores, axis=0)
top_class = tf.argmax(class_scores)
infered_class = class_names[top_class]
top_score = class_scores[top_class]
print(f'[YAMNet] The main sound is: {infered_class} ({top_score})')
reloaded_results = reloaded_model(waveform)
your_top_class = tf.argmax(reloaded_results)
your_infered_class = my_classes[your_top_class]
class_probabilities = tf.nn.softmax(reloaded_results, axis=-1)
your_top_score = class_probabilities[your_top_class]
print(f'[Your model] The main sound is: {your_infered_class} ({your_top_score})')
# + [markdown] id="g8Tsym8Rq-0V"
# ## Next steps
#
# You just created a model that can classify sounds from dogs or cats. With the same idea and proper data you could, for example, build a bird recognizer based on their singing.
#
# Let us know what you come up with! Share your project with us on social media.
#
| site/en/tutorials/audio/transfer_learning_audio.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] id="X24v90qcPhq2" colab_type="text"
# # Blood Cell Classification
# #### <NAME>
# Using AI, we are able to train the programs to perform a task without us expliciting programming it; instead, we train the program to train itself by letting the program play games with itself. The program below uses a convoluted neutron network to train itself to classify what type of white blood cell it actually is. The convoluted neutron network imitate the structure of neurons that process images in the brain and use techniques to reduce neuron count, as well as maintaining positional relationships in the data by processing the data through multiple layers. The four different white blood cells that the program is training to classify are Eosinophil, Lymphocyte, Monocyte, and Neutrophil. Eosinophil make up 2 to 4 percent of white blood cells(WBC) which excretes acids to combat parasites; Lymphocyte make up 20 to 30 percent of WBCs which migrates in and out of blood; Monocytes make up 2 to 8 percent of WBCs which enter peripheral tissues to become tissue macrophages which can engulf large particles and pathogens; and Neutrophil make up 50 to 70 percent of WBCs and their cytoplasm is packed with pale granules containing lysosomal enzymes and bacteria-killing compounds. In the future, this program can help hasten the mandatory blood tests.
# + id="iHtq_5lrPhq3" colab_type="code" colab={} executionInfo={"status": "ok", "timestamp": 1598673330677, "user_tz": 240, "elapsed": 611, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
# %matplotlib inline
# + id="Uha77-GhPhq7" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 368} executionInfo={"status": "error", "timestamp": 1598673333459, "user_tz": 240, "elapsed": 3385, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}} outputId="9c9e7113-5391-49da-aa99-6b4920420eda"
import numpy as np
from keras.models import Sequential, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input
from keras.layers import Conv2D, GlobalAveragePooling2D, LeakyReLU
from keras.utils import np_utils
from keras.optimizers import adam, SGD, rmsprop
from keras.applications import MobileNet
from string import ascii_uppercase
import matplotlib.pyplot as plt
from pandas_ml import ConfusionMatrix
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
from PIL import Image
from glob import glob
import cv2
# + [markdown] id="9UmS3MpcPhq-" colab_type="text"
# # Data Preparation
#
# * Uploading and formatting images for training and testing the convoluted neutron network*
#
# + id="lUfauRpIPhq_" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333452, "user_tz": 240, "elapsed": 3376, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
# TODO
classes = []
for x in glob("data/train/*"):
classes.append(x[11:])
num_classes = len(classes)
print(classes)
# + id="uK-Npmi7PhrB" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333453, "user_tz": 240, "elapsed": 3376, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
read_img = lambda path: cv2.resize(cv2.imread(path), (224, 224))
#TODO load in dataset for training
x_train = []
y_train = []
label = 0
for folder in glob("data/train/*"):
for img in glob(folder + "/*"):
x_train.append(read_img(img))
y_train.append(label)
label += 1
x_train = np.asarray(x_train)
y_train = np.asarray(y_train)
x_test = []
y_test = []
label = 0
for folder in glob("data/test/*"):
for img in glob(folder + "/*"):
x_test.append(read_img(img))
y_test.append(label)
label += 1
x_test = np.asarray(x_test)
y_test = np.asarray(y_test)
x_train, y_train = shuffle(x_train, y_train)
# Converts labels for train and test set to one hot encodings
y_train = np_utils.to_categorical(y_train, num_classes)
y_test = np_utils.to_categorical(y_test, num_classes)
x_train.shape, y_train.shape, x_test.shape, y_test.shape
# + [markdown] id="jKgtjnYLPhrE" colab_type="text"
# # A collection of image of White Blood Cells
# + id="Wgh-SUg5PhrE" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333453, "user_tz": 240, "elapsed": 3375, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
w=x_train.shape[1]
h=x_train.shape[2]
fig=plt.figure(figsize=(8, 8))
columns = 4
rows = 5
for i in range(1, columns*rows +1):
img = np.random.randint(10, size=(h,w))
fig.add_subplot(rows, columns, i)
plt.imshow(x_train[i])
# + id="j0diWrKzPhrG" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333454, "user_tz": 240, "elapsed": 3375, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
#TODO
model_name = "Blood CNN.h5"
load_checkpoint = False
# + id="FqNWoOIqPhrI" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333454, "user_tz": 240, "elapsed": 3374, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
#Load existing model
if load_checkpoint:
model = load_model(model_name)
#Create new model
else:
model_base = MobileNet(include_top=False,input_shape=x_train.shape[1:])
model = Sequential()
model.add(model_base)
model.add(GlobalAveragePooling2D())
model.add(Dropout(0,5))
model.add(Dense(num_classes,activation='softmax'))
model.summary()
# + id="duJVlu9APhrM" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333455, "user_tz": 240, "elapsed": 3374, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
#TODO
opt = SGD(lr=0.01)
model.compile(loss='categorical_crossentropy',
optimizer=opt,
metrics=['accuracy'])
# + [markdown] id="oCel4p4xPhrO" colab_type="text"
# # Training the Program
# + id="xT1bVBVAPhrP" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333455, "user_tz": 240, "elapsed": 3373, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
while True:
model.fit(x_train, y_train,
batch_size= 16, epochs=1, verbose=1)
model.save(model_name)
# + [markdown] id="nNqWY5XcPhrR" colab_type="text"
# # Evaluation and Testing
# + [markdown] id="fYEoFAicPhrS" colab_type="text"
# # Loss vs Accuracy
# + id="vsN8kGF5PhrS" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333456, "user_tz": 240, "elapsed": 3373, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
score = model.evaluate(x_test, y_test, verbose=0)
"Loss: %s, Accuracy: %s" % (score[0], score[1])
# + [markdown] id="EUDa6id0PhrV" colab_type="text"
# # Proof of Concept
# + id="5QBpychPPhrW" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333456, "user_tz": 240, "elapsed": 3372, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
i = 5
plt.imshow(x_test[i])
prediction = model.predict(np.expand_dims(x_test[i], axis=0))
"Expected: %s, Predicted: %s" % (classes[y_test[i].argmax()], classes[prediction.argmax()])
# + [markdown] id="Bk3ZVkBlPhrZ" colab_type="text"
# # The Confusion Matrix
# * Where is the program making the most mistakes in its classification*
# + id="Z08TnbN_PhrZ" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333457, "user_tz": 240, "elapsed": 3372, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
ConfusionMatrix([classes[one_hot.argmax()] for one_hot in y_test], [classes[pred.argmax()] for pred in model.predict(x_test)]).plot()
# + id="TxjtiDOxPhrc" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333457, "user_tz": 240, "elapsed": 3370, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
# + id="IZMnOl7sPhre" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333458, "user_tz": 240, "elapsed": 3370, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
# + id="8bc8a8ujPhrg" colab_type="code" colab={} executionInfo={"status": "aborted", "timestamp": 1598673333458, "user_tz": 240, "elapsed": 3369, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "14769114701246385476"}}
| Neural Network Classifier.ipynb |
# +
# Зависимости
import pandas as pd
# import numpy as np
# import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.svm import SVR, SVC
from sklearn.metrics import mean_squared_error, f1_score
# -
# Генерируем уникальный seed
my_code = "Рахматуллаев и Тимуров"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Читаем данные из файла
example_data = pd.read_csv("datasets/Fish.csv")
example_data.head()
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(example_data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(example_data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# +
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
num_columns = ['Weight', 'Length1', 'Length2', 'Length3', 'Height', 'Width']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), num_columns)], remainder='passthrough')
ct.fit(train)
# -
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_test = pd.DataFrame(ct.transform(test))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = num_columns + ['Species']
sc_train.columns = column_names
sc_test.columns = column_names
sc_val.columns = column_names
sc_train
# +
# Задание №1 - анализ метода опорных векторов в задаче регрессии
# https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVR.html#sklearn.svm.SVR
# kernel : {'linear', 'poly', 'rbf', 'sigmoid', 'precomputed'}, default='rbf'
# Только для kernel = 'poly' : degreeint, default=3
# +
# Выбираем 4 числовых переменных, три их них будут предикторами, одна - зависимой переменной
n = 4
labels = random.sample(num_columns, n)
y_label = labels[0]
x_labels = labels[1:]
print(x_labels)
print(y_label)
# +
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
# -
x_train
# Создайте 4 модели с различными ядрами: 'linear', 'poly', 'rbf', 'sigmoid'.
# Решите получившуюся задачу регрессии с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметр регуляризации C : float, default=1.0
# Укажите, какая модель решает задачу лучше других.
r_model_1 = SVR(kernel='linear', C=0.8)
r_model_2 = SVR(kernel='poly', degree=3, C=1.0)
r_model_3 = SVR(kernel='rbf', C=1.0)
r_model_4 = SVR(kernel='sigmoid', C=0.6)
r_models = []
r_models.append(r_model_1)
r_models.append(r_model_2)
r_models.append(r_model_3)
r_models.append(r_model_4)
# Обучаем модели
for model in r_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке
mses = []
for model in r_models:
val_pred = model.predict(x_val)
mse = mean_squared_error(y_val, val_pred)
mses.append(mse)
print(mse)
# Выбираем лучшую модель
i_min = mses.index(min(mses))
best_r_model = r_models[i_min]
best_r_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_r_model.predict(x_test)
mse = mean_squared_error(y_test, test_pred)
print(mse)
# +
# Задание №2 - анализ метода опорных векторов в задаче классификации
# https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC
# kernel : {'linear', 'poly', 'rbf', 'sigmoid', 'precomputed'}, default='rbf'
# Только для kernel = 'poly' : degreeint, default=3
# +
# Выбираем 2 числовых переменных, которые будут параметрами элементов набора данных
# Метка класса всегда 'Species'
n = 2
x_labels = random.sample(num_columns, n)
y_label = 'Species'
print(x_labels)
print(y_label)
# +
# Отберем необходимые параметры
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_label]
y_test = sc_test[y_label]
y_val = sc_val[y_label]
# -
x_train
# Создайте 4 модели с различными ядрами: 'linear', 'poly', 'rbf', 'sigmoid'.
# Решите получившуюся задачу регрессии с помощью созданных моделей и сравните их эффективность.
# При необходимости применяйте параметр регуляризации C : float, default=1.0
# Укажите, какая модель решает задачу лучше других.
c_model = SVC()
c_model_1 = SVC(kernel='linear', C=0.8)
c_model_2 = SVC(kernel='poly', degree=3, C=1.0)
c_model_3 = SVC(kernel='rbf', C=1.0)
c_model_4 = SVC(kernel='sigmoid', C=0.6)
c_models = []
c_models.append(c_model_1)
c_models.append(c_model_2)
c_models.append(c_model_3)
c_models.append(c_model_4)
# Обучаем модели
for model in c_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
f1s = []
for model in c_models:
val_pred = model.predict(x_val)
f1 = f1_score(y_val, val_pred, average='weighted')
f1s.append(f1)
print(f1)
# Выбираем лучшую модель
i_min = f1s.index(min(f1s))
best_c_model = c_models[i_min]
best_c_model.get_params()
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_c_model.predict(x_test)
f1 = f1_score(y_test, test_pred, average='weighted')
print(f1)
| 2021 Весенний семестр/Практическое задание 2/ПЗ-2_Рахматуллаев.Ж.Ж_Тимуров.У.Т._ИСТ-18-2/ПЗ-2_Рахматуллаев.Ж.Ж_Тимуров.У.Т._ИСТ-18-2.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
Test params: 2, 2, 0.05, 1, 0.5
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/ipykernel_launcher.py:18: FutureWarning: 'argmin' is deprecated. Use 'idxmin' instead. The behavior of 'argmin' will be corrected to return the positional minimum in the future. Use 'series.values.argmin' to get the position of the minimum now.
test-merror 0.116666 for 29 rounds
Test params: 2, 2, 0.05, 1, 1
test-merror 0.1085652 for 116 rounds
Test params: 2, 2, 0.05, 2, 0.5
test-merror 0.1046756 for 132 rounds
Test params: 2, 2, 0.05, 2, 1
test-merror 0.1182866 for 17 rounds
Test params: 2, 2, 0.1, 1, 0.5
test-merror 0.1004634 for 87 rounds
Test params: 2, 2, 0.1, 1, 1
test-merror 0.1030542 for 95 rounds
Test params: 2, 2, 0.1, 2, 0.5
test-merror 0.1017588 for 100 rounds
Test params: 2, 2, 0.1, 2, 1
test-merror 0.1150464 for 27 rounds
Test params: 2, 2, 0.15, 1, 0.5
test-merror 0.1007872 for 66 rounds
Test params: 2, 2, 0.15, 1, 1
test-merror 0.1011102 for 85 rounds
Test params: 2, 2, 0.15, 2, 0.5
test-merror 0.1166668 for 11 rounds
Test params: 2, 2, 0.15, 2, 1
test-merror 0.0998146 for 93 rounds
Test params: 2, 2, 0.2, 1, 0.5
test-merror 0.1017596 for 38 rounds
Test params: 2, 2, 0.2, 1, 1
test-merror 0.1001388 for 75 rounds
Test params: 2, 2, 0.2, 2, 0.5
test-merror 0.0962486 for 74 rounds
Test params: 2, 2, 0.2, 2, 1
test-merror 0.0972226 for 82 rounds
Test params: 2, 4, 0.05, 1, 0.5
test-merror 0.1049984 for 31 rounds
Test params: 2, 4, 0.05, 1, 1
test-merror 0.1066198 for 34 rounds
Test params: 2, 4, 0.05, 2, 0.5
test-merror 0.0981958 for 108 rounds
Test params: 2, 4, 0.05, 2, 1
test-merror 0.105648 for 64 rounds
Test params: 2, 4, 0.1, 1, 0.5
test-merror 0.093983 for 68 rounds
Test params: 2, 4, 0.1, 1, 1
test-merror 0.0975456 for 42 rounds
Test params: 2, 4, 0.1, 2, 0.5
test-merror 0.100463 for 44 rounds
Test params: 2, 4, 0.1, 2, 1
test-merror 0.0965744 for 65 rounds
Test params: 2, 4, 0.15, 1, 0.5
test-merror 0.0933316 for 49 rounds
Test params: 2, 4, 0.15, 1, 1
test-merror 0.0962502 for 40 rounds
Test params: 2, 4, 0.15, 2, 0.5
test-merror 0.094306 for 54 rounds
Test params: 2, 4, 0.15, 2, 1
test-merror 0.0949542 for 41 rounds
Test params: 2, 4, 0.2, 1, 0.5
test-merror 0.0952776 for 24 rounds
Test params: 2, 4, 0.2, 1, 1
test-merror 0.0956006 for 31 rounds
Test params: 2, 4, 0.2, 2, 0.5
test-merror 0.0985182 for 34 rounds
Test params: 2, 4, 0.2, 2, 1
test-merror 0.0981946 for 28 rounds
Test params: 2, 6, 0.05, 1, 0.5
test-merror 0.1014342 for 30 rounds
Test params: 2, 6, 0.05, 1, 1
test-merror 0.1040268 for 20 rounds
Test params: 2, 6, 0.05, 2, 0.5
test-merror 0.1040276 for 39 rounds
Test params: 2, 6, 0.05, 2, 1
test-merror 0.1014362 for 58 rounds
Test params: 2, 6, 0.1, 1, 0.5
test-merror 0.0956018 for 59 rounds
Test params: 2, 6, 0.1, 1, 1
test-merror 0.095602 for 40 rounds
Test params: 2, 6, 0.1, 2, 0.5
test-merror 0.0985198 for 40 rounds
Test params: 2, 6, 0.1, 2, 1
test-merror 0.103704 for 18 rounds
Test params: 2, 6, 0.15, 1, 0.5
test-merror 0.0965732 for 30 rounds
Test params: 2, 6, 0.15, 1, 1
test-merror 0.0949526 for 31 rounds
Test params: 2, 6, 0.15, 2, 0.5
test-merror 0.0975456 for 33 rounds
Test params: 2, 6, 0.15, 2, 1
test-merror 0.1017588 for 25 rounds
Test params: 2, 6, 0.2, 1, 0.5
test-merror 0.0962482 for 21 rounds
Test params: 2, 6, 0.2, 1, 1
test-merror 0.093334 for 25 rounds
Test params: 2, 6, 0.2, 2, 0.5
test-merror 0.097548 for 36 rounds
Test params: 2, 6, 0.2, 2, 1
test-merror 0.097222 for 39 rounds
Test params: 6, 2, 0.05, 1, 0.5
test-merror 0.116666 for 29 rounds
Test params: 2, 2, 0.05, 1, 1
test-merror 0.1085652 for 116 rounds
Test params: 2, 2, 0.05, 2, 0.5
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
<ipython-input-37-f47408f9405e> in <module>()
14 bst = xgb.train(param, dtrain, num_round)
15
---> 16 cv = xgb.cv(param, dtrain, 999, nfold=5, early_stopping_rounds=10)
17 mean = cv['test-merror-mean'].min()
18 boost_rounds = cv['test-merror-mean'].argmin()
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/xgboost/training.pyc in cv(params, dtrain, num_boost_round, nfold, stratified, folds, metrics, obj, feval, maximize, early_stopping_rounds, fpreproc, as_pandas, verbose_eval, show_stdv, seed, callbacks, shuffle)
405 for fold in cvfolds:
406 fold.update(i, obj)
--> 407 res = aggcv([f.eval(i, feval) for f in cvfolds])
408
409 for key, mean, std in res:
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/xgboost/training.pyc in eval(self, iteration, feval)
220 def eval(self, iteration, feval):
221 """"Evaluate the CVPack for one iteration."""
--> 222 return self.bst.eval_set(self.watchlist, iteration, feval)
223
224
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/xgboost/core.pyc in eval_set(self, evals, iteration, feval)
949 if not isinstance(d[1], STRING_TYPES):
950 raise TypeError('expected string, got {}'.format(type(d[1]).__name__))
--> 951 self._validate_features(d[0])
952
953 dmats = c_array(ctypes.c_void_p, [d[0].handle for d in evals])
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/xgboost/core.pyc in _validate_features(self, data)
1271 else:
1272 # Booster can't accept data with different feature names
-> 1273 if self.feature_names != data.feature_names:
1274 dat_missing = set(self.feature_names) - set(data.feature_names)
1275 my_missing = set(data.feature_names) - set(self.feature_names)
/Users/fanxu/anaconda2/envs/my-rdkit-env/lib/python2.7/site-packages/xgboost/core.pyc in feature_names(self)
629 """
630 if self._feature_names is None:
--> 631 return ['f{0}'.format(i) for i in range(self.num_col())]
632 else:
633 return self._feature_names
KeyboardInterrupt:
| fangli/Untitled.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Project 5 : Classification
# ## Instructions
#
# ### Description
#
# Practice classification on the Titanic dataset.
#
# ### Grading
#
# For grading purposes, we will clear all outputs from all your cells and then run them all from the top. Please test your notebook in the same fashion before turning it in.
#
# ### Submitting Your Solution
#
# To submit your notebook, first clear all the cells (this won't matter too much this time, but for larger data sets in the future, it will make the file smaller). Then use the File->Download As->Notebook to obtain the notebook file. Finally, submit the notebook file on Canvas.
#
# +
import pandas as pd
import numpy as np
import sklearn as sk
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('ggplot')
# %matplotlib inline
# %config InlineBackend.figure_format = 'retina'
# -
# ### Introduction
#
# On April 15, 1912, the largest passenger liner ever made collided with an iceberg during her maiden voyage. When the Titanic sank it killed 1502 out of 2224 passengers and crew. This sensational tragedy shocked the international community and led to better safety regulations for ships. One of the reasons that the shipwreck resulted in such loss of life was that there were not enough lifeboats for the passengers and crew. Although there was some element of luck involved in surviving the sinking, some groups of people were more likely to survive than others.
#
# Intro Videos:
# https://www.youtube.com/watch?v=3lyiZMeTKIo
# and
# https://www.youtube.com/watch?v=ItjXTieWKyI
#
# The `titanic_data.csv` file contains data for `887` of the real Titanic passengers. Each row represents one person. The columns describe different attributes about the person including whether they survived (`0=No`), their age, their passenger-class (`1=1st Class, Upper`), gender, and the fare they paid (£s*). For more on the currency: http://www.statisticalconsultants.co.nz/blog/titanic-fare-data.html
#
# We are going to try to see if there are correlations between the feature data provided (find a best subset of features) and passenger survival.
# ### Problem 1: Load and understand the data (35 points)
#
# #### Your task (some of this is the work you completed for L14 - be sure to copy that work into here as needed)
# Conduct some preprocessing steps to explore the following and provide code/answers in the below cells:
# 1. Load the `titanic_data.csv` file into a pandas dataframe
# 2. Explore the data provided (e.g., looking at statistics using describe(), value_counts(), histograms, scatter plots of various features, etc.)
# 3. What are the names of feature columns that appear to be usable for learning?
# 4. What is the name of the column that appears to represent our target?
# 5. Formulate a hypothesis about the relationship between given feature data and the target
# 6. How did Pclass affect passenngers' chances of survival?
# 7. What is the age distribution of survivors?
# Step 1. Load the `titanic_data.csv` file into a pandas dataframe
boat = pd.read_csv("titanic_data.csv")
# +
# Step 2. Explore the data provided (e.g., looking at statistics using describe(), value_counts(), histograms, scatter plots of various features, etc.)
print("Headers: " + str(list(boat)))
print("\nTotal Number of Survivors: " + str(boat["Survived"].value_counts()[1]))
print("Survival Rate: " + str(342 / 757))
children = boat[boat["Age"] < 18]
print("\nNumber of Children: " + str(len(children["Age"])))
print("Surviving Children: " + str(children["Survived"].value_counts()[1]))
print("Survival Rate: " + str(65 / 130 * 100) + "%")
print("\nNumber of adults: " + str(887 - len(children["Age"])))
print("Surviving Adults: " + str(boat["Survived"].value_counts()[1] - children["Survived"].value_counts()[1]))
print("Survival Rate: " + str(277 / 757 * 100) + "%")
boat.describe()
print('\nSurvived Col:')
for i in range(10):
print(boat['Survived'][i])
# -
# ---
#
# **Edit this cell to provide answers to the following steps:**
#
# ---
#
# Step 3. What are the names of feature columns that appear to be usable for learning?
#
# Age, Pclass, Survived, Sex, Age, Parents/Children Aboard
#
# Step 4. What is the name of the column that appears to represent our target?
#
# Survived (Binary true false classifications)
#
# Step 5. Formulate a hypothesis about the relationship between given feature data and the target
#
# General survival rate will be higher in children
# +
#Step 6. How did Pclass affect passenngers' chances of survival?
#Show your work with a bar plot, dataframe selection, or visual of your choice.
classSums = [0,0,0,0]
for x in range(len(boat)):
classSums[boat['Pclass'][x]] += 1
for x in range(1,len(classSums)):
print(f"Class {x} had a survival rate of: {classSums[x] / 887}")
# +
#Step 7. What is the age distribution of survivors?
#Show your work with a dataframe operation and/or histogram plot.
ages = []
for x in range(len(boat)):
if boat['Survived'][x]:
ages.extend([boat['Age'][x]])
plt.title('Surviver Age Distrobution')
plt.xlabel("Age")
plt.hist(ages)
plt.show()
plt.title('Overall Age Distrobution')
plt.xlabel("Age")
plt.hist(boat['Age'])
plt.show()
# -
# ### Problem 2: transform the data (10 points)
# The `Sex` column is categorical, meaning its data are separable into groups, but not numerical. To be able to work with this data, we need numbers, so you task is to transform the `Sex` column into numerical data with pandas' `get_dummies` feature and remove the original categorical `Sex` column.
boat['Sex']= pd.get_dummies(boat['Sex'])
# ### Problem 3: Classification (30 points)
# Now that the data is transformed, we want to run various classification experiments on it. The first is `K Nearest Neighbors`, which you will conduct by:
#
# 1. Define input and target data by creating lists of dataframe columns (e.g., inputs = ['Pclass', etc.)
# 2. Split the data into training and testing sets with `train_test_split()`
# 3. Create a `KNeighborsClassifier` using `5` neighbors at first (you can experiment with this parameter)
# 4. Train your model by passing the training dataset to `fit()`
# 5. Calculate predicted target values(y_hat) by passing the testing dataset to `predict()`
# 6. Print the accuracy of the model with `score()`
#
# ** Note: If you get a python warning as you use the Y, trainY, or testY vector in some of the function calls about "DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, )", you can look up how to use trainY.values.ravel() or trainY.values.flatten() or another function, etc.
# +
#inputs = Pclass, Age, Sex, Fare
#inputs = pd.concat([boat["Pclass"], boat["Age"],boat["Pclass"], boat["Fare"]], axis=1)
#target = Surviaval
#target = boat["Survived"]
#inputs.describe()
# -
from sklearn.model_selection import train_test_split
train , test =train_test_split(boat)
train.describe()
# +
from sklearn.neighbors import KNeighborsClassifier
k = 5
model = KNeighborsClassifier(k)
model.fit(train[["Pclass", "Age", "Pclass", "Fare"]], train["Survived"])
yhat = model.predict(train[["Pclass", "Age", "Pclass", "Fare"]])
model.score(test[["Pclass", "Age", "Pclass", "Fare"]], test['Survived'])
# -
# ### Problem 4: Cross validation, classification report (15 points)
# - Using the concepts from the 17-model_selection slides and the [`cross_val_score`](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_val_score.html) function from scikit-learn, estimate the f-score ([`f1-score`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html#sklearn.metrics.f1_score) (you can use however many folds you wish). To get `cross_val_score` to use `f1-score` rather than the default accuracy measure, you will need to set the `scoring` parameter and use a scorer object created via [`make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html#sklearn.metrics.make_scorer). Since this has a few parts to it, let me just give you that parameter: ```scorerVar = make_scorer(f1_score, pos_label=1)```
#
# - Using the concepts from the end of the 14-classification slides, output a confusion matrix.
#
# - Also, output a classification report [`classification_report`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html) from sklearn.metrics showing more of the metrics: precision, recall, f1-score for both of our classes.
# +
from sklearn import metrics
from sklearn.metrics import confusion_matrix, f1_score, classification_report, make_scorer
from sklearn import model_selection
scorerVar = make_scorer(f1_score, pos_label=1)
scores = cross_val_score(model, boat[["Pclass", "Age", "Pclass", "Fare"]], boat["Survived"],cv = 5, scoring = scorerVar)
print(scores.mean())
print(confusion_matrix(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
print(classification_report(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
# -
# ### Problem 5: Logistic Regression (15 points)
#
# Now, repeat the above experiment using the [`LogisticRegression`](http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) model in scikit-learn, and output:
#
# - The fit accuracy (using the `score` method of the model)
# - The f-score (using the [`cross_val_score`](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_val_score.html) function)
# - The confusion matrix
# - The precision, recall, and f-measure for the 1 class (you can just print the results of the [`classification_report`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html) function from sklearn.metrics)
# +
from sklearn.linear_model import LogisticRegression
#create a model object
model = LogisticRegression()
#train our model
model.fit(train[["Pclass", "Age", "Pclass", "Fare"]], train["Survived"])
#evaluate the model
yhat = model.predict(train[["Pclass", "Age", "Pclass", "Fare"]])
score = model.score(test[["Pclass", "Age", "Pclass", "Fare"]], test['Survived'])
print(f"model score: {score}")
#setup to get f-score and cv
scorerVar = make_scorer(f1_score, pos_label=1)
scores = cross_val_score(model, boat[["Pclass", "Age", "Pclass", "Fare"]], boat["Survived"],cv = 5, scoring = scorerVar)
print(f"Cross Validation f1_score: {scores.mean()}")
#confusion matrix
print("Confusion Matrix")
print(confusion_matrix(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
#classification report
print("\nClassification Report")
print(classification_report(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
# -
# ### Problem 6: Support Vector Machines (15 points)
# Now, repeat the above experiment using the using a Support Vector classifier [`SVC`](http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html) with default parameters (RBF kernel) model in scikit-learn, and output:
#
# - The fit accuracy (using the `score` method of the model)
# - The f-score (using the [`cross_val_score`](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_val_score.html) function)
# - The confusion matrix
# - The precision, recall, and f-measure for the 1 class (you can just print the results of the [`classification_report`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html) function from sklearn.metrics)
# +
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
#create a model object
model = SVC()
#train our model
model.fit(train[["Pclass", "Age", "Pclass", "Fare"]], train["Survived"])
#evaluate the model
yhat = model.predict(train[["Pclass", "Age", "Pclass", "Fare"]])
score = model.score(test[["Pclass", "Age", "Pclass", "Fare"]], test['Survived'])
print(f"model score: {score}")
#setup to get f-score and cv
scorerVar = make_scorer(f1_score, pos_label=1)
scores = cross_val_score(model, boat[["Pclass", "Age", "Pclass", "Fare"]], boat["Survived"],cv = 5, scoring = scorerVar)
print(f"Cross Validation f1_score: {scores.mean()}")
#confusion matrix
print("Confusion Matrix")
print(confusion_matrix(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
#classification report
print("\nClassification Report")
print(classification_report(test['Survived'], model.predict(test[["Pclass", "Age", "Pclass", "Fare"]])))
# -
# ### Problem 7: Comparision and Discussion (5 points)
# Edit this cell to provide a brief discussion (3-5 sentances at most):
# 1. What was the model/algorithm that performed best for you?
#
# Logistic Regression performed the best
#
# 2. What feaures and parameters were used to achieve that performance?
#
# Tweaking SVM to use a linear kernal also worked just as well as Logistical Regression
#
# 3. What insights did you gain from your experimentation about the predictive power of this dataset and did it match your original hypothesis about the relationship between given feature data and the target?
#
# Age really did not effect survival rates. I was really surpised by this!
# ### Questionnaire
# 1) How long did you spend on this assignment?
# <br>~2hrs<br>
# 2) What did you like about it? What did you not like about it?
# <br>The breadth of classification measures<br>
# 3) Did you find any errors or is there anything you would like changed?
# <br>Nope<br>
| python/examples/05-classify-new.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # SVI Part III: ELBO Gradient Estimators
#
# ## Setup
#
# We've defined a Pyro model with observations ${\bf x}$ and latents ${\bf z}$ of the form $p_{\theta}({\bf x}, {\bf z}) = p_{\theta}({\bf x}|{\bf z}) p_{\theta}({\bf z})$. We've also defined a Pyro guide (i.e. a variational distribution) of the form $q_{\phi}({\bf z})$. Here ${\theta}$ and $\phi$ are variational parameters for the model and guide, respectively. (In particular these are _not_ random variables that call for a Bayesian treatment).
#
# We'd like to maximize the log evidence $\log p_{\theta}({\bf x})$ by maximizing the ELBO (the evidence lower bound) given by
#
# $${\rm ELBO} \equiv \mathbb{E}_{q_{\phi}({\bf z})} \left [
# \log p_{\theta}({\bf x}, {\bf z}) - \log q_{\phi}({\bf z})
# \right]$$
#
# To do this we're going to take (stochastic) gradient steps on the ELBO in the parameter space $\{ \theta, \phi \}$ (see references [1,2] for early work on this approach). So we need to be able to compute unbiased estimates of
#
# $$\nabla_{\theta,\phi} {\rm ELBO} = \nabla_{\theta,\phi}\mathbb{E}_{q_{\phi}({\bf z})} \left [
# \log p_{\theta}({\bf x}, {\bf z}) - \log q_{\phi}({\bf z})
# \right]$$
#
# How do we do this for general stochastic functions `model()` and `guide()`? To simplify notation let's generalize our discussion a bit and ask how we can compute gradients of expectations of an arbitrary cost function $f({\bf z})$. Let's also drop any distinction between $\theta$ and $\phi$. So we want to compute
#
# $$\nabla_{\phi}\mathbb{E}_{q_{\phi}({\bf z})} \left [
# f_{\phi}({\bf z}) \right]$$
#
# Let's start with the easiest case.
#
# ## Easy Case: Reparameterizable Random Variables
#
# Suppose that we can reparameterize things such that
#
# $$\mathbb{E}_{q_{\phi}({\bf z})} \left [f_{\phi}({\bf z}) \right]
# =\mathbb{E}_{q({\bf \epsilon})} \left [f_{\phi}(g_{\phi}({\bf \epsilon})) \right]$$
#
# Crucially we've moved all the $\phi$ dependence inside of the exectation; $q({\bf \epsilon})$ is a fixed distribution with no dependence on $\phi$. This kind of reparameterization can be done for many distributions (e.g. the normal distribution); see reference [3] for a discussion. In this case we can pass the gradient straight through the expectation to get
#
# $$\nabla_{\phi}\mathbb{E}_{q({\bf \epsilon})} \left [f_{\phi}(g_{\phi}({\bf \epsilon})) \right]=
# \mathbb{E}_{q({\bf \epsilon})} \left [\nabla_{\phi}f_{\phi}(g_{\phi}({\bf \epsilon})) \right]$$
#
# Assuming $f(\cdot)$ and $g(\cdot)$ are sufficiently smooth, we can now get unbiased estimates of the gradient of interest by taking a Monte Carlo estimate of this expectation.
#
# ## Tricky Case: Non-reparameterizable Random Variables
#
# What if we can't do the above reparameterization? Unfortunately this is the case for many distributions of interest, for example all discrete distributions. In this case our estimator takes a bit more complicated form.
#
# We begin by expanding the gradient of interest as
#
# $$\nabla_{\phi}\mathbb{E}_{q_{\phi}({\bf z})} \left [
# f_{\phi}({\bf z}) \right]=
# \nabla_{\phi} \int d{\bf z} \; q_{\phi}({\bf z}) f_{\phi}({\bf z})$$
#
# and use the chain rule to write this as
#
# $$ \int d{\bf z} \; \left \{ (\nabla_{\phi} q_{\phi}({\bf z})) f_{\phi}({\bf z}) + q_{\phi}({\bf z})(\nabla_{\phi} f_{\phi}({\bf z}))\right \} $$
#
# At this point we run into a problem. We know how to generate samples from $q(\cdot)$—we just run the guide forward—but $\nabla_{\phi} q_{\phi}({\bf z})$ isn't even a valid probability density. So we need to massage this formula so that it's in the form of an expectation w.r.t. $q(\cdot)$. This is easily done using the identity
#
# $$ \nabla_{\phi} q_{\phi}({\bf z}) =
# q_{\phi}({\bf z})\nabla_{\phi} \log q_{\phi}({\bf z})$$
#
# which allows us to rewrite the gradient of interest as
#
# $$\mathbb{E}_{q_{\phi}({\bf z})} \left [
# (\nabla_{\phi} \log q_{\phi}({\bf z})) f_{\phi}({\bf z}) + \nabla_{\phi} f_{\phi}({\bf z})\right]$$
#
# This form of the gradient estimator—variously known as the REINFORCE estimator or the score function estimator or the likelihood ratio estimator—is amenable to simple Monte Carlo estimation.
#
# Note that one way to package this result (which is covenient for implementation) is to introduce a surrogate loss function
#
# $${\rm surrogate \;loss} \equiv
# \log q_{\phi}({\bf z}) \overline{f_{\phi}({\bf z})} + f_{\phi}({\bf z})$$
#
# Here the bar indicates that the term is held constant (i.e. it is not to be differentiated w.r.t. $\phi$). To get a (single-sample) Monte Carlo gradient estimate, we sample the latent random variables, compute the surrogate loss, and differentiate. The result is an unbiased estimate of $\nabla_{\phi}\mathbb{E}_{q_{\phi}({\bf z})} \left [
# f_{\phi}({\bf z}) \right]$. In equations:
#
# $$\nabla_{\phi} {\rm ELBO} = \mathbb{E}_{q_{\phi}({\bf z})} \left [
# \nabla_{\phi} ({\rm surrogate \; loss}) \right]$$
#
# ## Variance or Why I Wish I Was Doing MLE Deep Learning
#
# We now have a general recipe for an unbiased gradient estimator of expectations of cost functions. Unfortunately, in the more general case where our $q(\cdot)$ includes non-reparameterizable random variables, this estimator tends to have high variance. Indeed in many cases of interest the variance is so high that the estimator is effectively unusable. So we need strategies to reduce variance (for a discussion see reference [4]). We're going to pursue two strategies. The first strategy takes advantage of the particular structure of the cost function $f(\cdot)$. The second strategy effectively introduces a way to reduce variance by using information from previous estimates of
# $\mathbb{E}_{q_{\phi}({\bf z})} [ f_{\phi}({\bf z})]$. As such it is somewhat analogous to using momentum in stochastic gradient descent.
#
# ### Reducing Variance via Dependency Structure
#
# In the above discussion we stuck to a general cost function $f_{\phi}({\bf z})$. We could continue in this vein (the approach we're about to discuss is applicable in the general case) but for concreteness let's zoom back in. In the case of stochastic variational inference, we're interested in a particular cost function of the form <br/><br/>
#
# $$\log p_{\theta}({\bf x} | {\rm Pa}_p ({\bf x})) +
# \sum_i \log p_{\theta}({\bf z}_i | {\rm Pa}_p ({\bf z}_i))
# - \sum_i \log q_{\phi}({\bf z}_i | {\rm Pa}_q ({\bf z}_i))$$
#
# where we've broken the log ratio $\log p_{\theta}({\bf x}, {\bf z})/q_{\phi}({\bf z})$ into an observation log likelihood piece and a sum over the different latent random variables $\{{\bf z}_i \}$. We've also introduced the notation
# ${\rm Pa}_p (\cdot)$ and ${\rm Pa}_q (\cdot)$ to denote the parents of a given random variable in the model and in the guide, respectively. (The reader might worry what the appropriate notion of dependency would be in the case of general stochastic functions; here we simply mean regular ol' dependency within a single execution trace). The point is that different terms in the cost function have different dependencies on the random variables $\{ {\bf z}_i \}$ and this is something we can leverage.
#
# To make a long story short, for any non-reparameterizable latent random variable ${\bf z}_i$ the surrogate loss is going to have a term
#
# $$\log q_{\phi}({\bf z}_i) \overline{f_{\phi}({\bf z})} $$
#
# It turns out that we can remove some of the terms in $\overline{f_{\phi}({\bf z})}$ and still get an unbiased gradient estimator; furthermore, doing so will generally decrease the variance. In particular (see reference [4] for details) we can remove any terms in $\overline{f_{\phi}({\bf z})}$ that are not downstream of the latent variable ${\bf z}_i$ (downstream w.r.t. to the dependency structure of the guide).
#
# In Pyro, all of this logic is taken care of automatically by the `SVI` class. In particular as long as we switch on `trace_graph=True`, Pyro will keep track of the dependency structure within the execution traces of the model and guide and construct a surrogate loss that has all the unnecessary terms removed:
#
# ```python
# svi = SVI(model, guide, optimizer, "ELBO", trace_graph=True)
# ```
#
# Note that leveraging this dependency information takes extra computations, so `trace_graph=True` should only be invoked in the case where your model has non-reparameterizable random variables.
#
#
# ### Aside: Dependency tracking in Pyro
#
# Finally, a word about dependency tracking. Tracking dependency within a stochastic function that includes arbitrary Python code is a bit tricky. The approach currently implemented in Pyro is analogous to the one used in WebPPL (cf. reference [5]). Briefly, a conservative notion of dependency is used that relies on sequential ordering. If random variable ${\bf z}_2$ follows ${\bf z}_1$ in a given stochastic function then ${\bf z}_2$ _may be_ dependent on ${\bf z}_1$ and therefore _is_ assumed to be dependent. To mitigate the overly coarse conclusions that can be drawn by this kind of dependency tracking, Pyro includes constructs for declaring things as independent, namely `irange` and `iarange` ([see the previous tutorial](svi_part_ii.html)). For use cases with non-reparameterizable variables, it is therefore important for the user to make use of these constructs (when applicable) to take full advantage of the variance reduction provided by `SVI`. In some cases it may also pay to consider reordering random variables within a stochastic function (if possible). It's also worth noting that we expect to add finer notions of dependency tracking in a future version of Pyro.
#
# ### Reducing Variance with Data-Dependent Baselines
#
# The second strategy for reducing variance in our ELBO gradient estimator goes under the name of baselines (see e.g. reference [6]). It actually makes use of the same bit of math that underlies the variance reduction strategy discussed above, except now instead of removing terms we're going to add terms. Basically, instead of removing terms with zero expectation that tend to _contribute_ to the variance, we're going to add specially chosen terms with zero expectation that work to _reduce_ the variance. As such, this is a control variate strategy.
#
# In more detail, the idea is to take advantage of the fact that for any constant $b$, the following identity holds
#
# $$\mathbb{E}_{q_{\phi}({\bf z})} \left [\nabla_{\phi}
# (\log q_{\phi}({\bf z}) \times b) \right]=0$$
#
# This follows since $q(\cdot)$ is normalized:
#
# $$\mathbb{E}_{q_{\phi}({\bf z})} \left [\nabla_{\phi}
# \log q_{\phi}({\bf z}) \right]=
# \int \!d{\bf z} \; q_{\phi}({\bf z}) \nabla_{\phi}
# \log q_{\phi}({\bf z})=
# \int \! d{\bf z} \; \nabla_{\phi} q_{\phi}({\bf z})=
# \nabla_{\phi} \int \! d{\bf z} \; q_{\phi}({\bf z})=\nabla_{\phi} 1 = 0$$
#
# What this means is that we can replace any term
#
# $$\log q_{\phi}({\bf z}_i) \overline{f_{\phi}({\bf z})} $$
#
# in our surrogate loss with
#
# $$\log q_{\phi}({\bf z}_i) \left(\overline{f_{\phi}({\bf z})}-b\right) $$
#
# Doing so doesn't affect the mean of our gradient estimator but it does affect the variance. If we choose $b$ wisely, we can hope to reduce the variance. In fact, $b$ need not be a constant: it can depend on any of the random choices upstream (or sidestream) of ${\bf z}_i$.
#
# #### Baselines in Pyro
#
# There are several ways the user can instruct Pyro to use baselines in the context of stochastic variational inference. Since baselines can be attached to any non-reparameterizable random variable, the current baseline interface is at the level of the `pyro.sample` statement. In particular the baseline interface makes use of an argument `baseline`, which is a dictionary that specifies baseline options. Note that it only makes sense to specify baselines for sample statements within the guide (and not in the model).
#
# ##### Decaying Average Baseline
#
# The simplest baseline is constructed from a running average of recent samples of $\overline{f_{\phi}({\bf z})}$. In Pyro this kind of baseline can be invoked as follows
#
# ```python
# z = pyro.sample("z", dist.bernoulli, ...,
# baseline={'use_decaying_avg_baseline': True,
# 'baseline_beta': 0.95})
# ```
#
# The optional argument `baseline_beta` specifies the decay rate of the decaying average (default value: `0.90`).
#
# #### Neural Baselines
#
# In some cases a decaying average baseline works well. In others using a baseline that depends on upstream randomness is crucial for getting good variance reduction. A powerful approach for constructing such a baseline is to use a neural network that can be adapted during the course of learning. Pyro provides two ways to specify such a baseline (for an extended example see the [AIR tutorial](air.html)).
#
# First the user needs to decide what inputs the baseline is going to consume (e.g. the current datapoint under consideration or the previously sampled random variable). Then the user needs to construct a `nn.Module` that encapsulates the baseline computation. This might look something like
#
# ```python
# class BaselineNN(nn.Module):
# def __init__(self, dim_input, dim_hidden):
# super(BaselineNN, self).__init__()
# self.linear = nn.Linear(dim_input, dim_hidden)
# # ... finish initialization ...
#
# def forward(self, x):
# hidden = self.linear(x)
# # ... do more computations ...
# return baseline
# ```
#
# Then, assuming the BaselineNN object `baseline_module` has been initialized somewhere else, in the guide we'll have something like
#
# ```python
# def guide(x): # here x is the current mini-batch of data
# pyro.module("my_baseline", baseline_module, tags="baseline")
# # ... other computations ...
# z = pyro.sample("z", dist.bernoulli, ...,
# baseline={'nn_baseline': baseline_module,
# 'nn_baseline_input': x})
# ```
#
# Here the argument `nn_baseline` tells Pyro which `nn.Module` to use to construct the baseline. On the backend the argument `nn_baseline_input` is fed into the forward method of the module to compute the baseline $b$. Note that the baseline module needs to be registered with Pyro with a `pyro.module` call so that Pyro is aware of the trainable parameters within the module.
#
# Under the hood Pyro constructs a loss of the form
#
# $${\rm baseline\; loss} \equiv\left(\overline{f_{\phi}({\bf z})} - b \right)^2$$
#
# which is used to adapt the parameters of the neural network. There's no theorem that suggests this is the optimal loss function to use in this context (it's not), but in practice it can work pretty well. Just as for the decaying average baseline, the idea is that a baseline that can track the mean $\overline{f_{\phi}({\bf z})}$ will help reduce the variance. Under the hood `SVI` takes one step on the baseline loss in conjunction with a step on the ELBO.
#
# Note that the module `baseline_module` has been tagged with the string `"baseline"` above; this has the effect of tagging all parameters inside of `baseline_module` with the parameter tag `"baseline"`. This gives the user a convenient handle for controlling how the baseline parameters are optimized. For example, if the user wants the baseline parameters to have a larger learning rate (usually a good idea) an appropriate optimizer could be constructed as follows:
#
# ```python
# def per_param_args(module_name, param_name, tags):
# if 'baseline' in tags:
# return {"lr": 0.010}
# else:
# return {"lr": 0.001}
#
# optimizer = optim.Adam(per_param_args)
# ```
#
# Note that in order for the overall procedure to be correct the baseline parameters should only be optimized through the baseline loss. Similarly the model and guide parameters should only be optimized through the ELBO. To ensure that this is the case under the hood `SVI` detaches the baseline $b$ that enters the ELBO from the autograd graph. Also, since the inputs to the neural baseline may depend on the parameters of the model and guide, the inputs are also detached from the autograd graph before they are fed into the neural network.
#
# Finally, there is an alternate way for the user to specify a neural baseline. Simply use the argument `baseline_value`:
#
# ```python
# b = # do baseline computation
# z = pyro.sample("z", dist.bernoulli, ...,
# baseline={'baseline_value': b})
# ```
#
# This works as above, except in this case it's the user's responsibility to make sure that any autograd tape connecting $b$ to the parameters of the model and guide has been cut. Or to say the same thing in language more familiar to PyTorch users, any inputs to $b$ that depend on $\theta$ or $\phi$ need to be detached from the autograd graph with `detach()` statements.
#
# #### A complete example with baselines
#
# Recall that in the [first SVI tutorial](svi_part_i.html) we considered a bernoulli-beta model for coin flips. Because the beta random variable is non-reparameterizable, the corresponding ELBO gradients are quite noisy. In that context we dealt with this problem by dialing up the number of Monte Carlo samples used to form the estimator. This isn't necessarily a bad approach, but it can be an expensive one.
# Here we showcase how a simple decaying average baseline can reduce the variance. While we're at it, we also use `iarange` to write our model in a fully vectorized manner.
#
# Instead of directly comparing gradient variances, we're going to see how many steps it takes for SVI to converge. Recall that for this particular model (because of conjugacy) we can compute the exact posterior. So to assess the utility of baselines in this context, we setup the following simple experiment. We initialize the guide at a specified set of variational parameters. We then do SVI until the variational parameters have gotten to within a fixed tolerance of the parameters of the exact posterior. We do this both with and without the decaying average baseline. We then compare the number of gradient steps we needed in the two cases. Here's the complete code:
#
# (_Since apart from the use of_ `iarange` _and_ `use_decaying_avg_baseline`, _this code is very similar to the code in parts I and II of the SVI tutorial, we're not going to go through the code line by line._)
# +
from __future__ import print_function
import numpy as np
import torch
from torch.autograd import Variable
import pyro
import pyro.distributions as dist
import pyro.optim as optim
from pyro.infer import SVI
import sys
def param_abs_error(name, target):
return torch.sum(torch.abs(target - pyro.param(name))).data.numpy()[0]
class BernoulliBetaExample(object):
def __init__(self):
# the two hyperparameters for the beta prior
self.alpha0 = Variable(torch.Tensor([10.0]))
self.beta0 = Variable(torch.Tensor([10.0]))
# the dataset consists of six 1s and four 0s
self.data = Variable(torch.zeros(10,1))
self.data[0:6, 0].data = torch.ones(6)
self.n_data = self.data.size(0)
# compute the alpha parameter of the exact beta posterior
self.alpha_n = self.alpha0 + self.data.sum()
# compute the beta parameter of the exact beta posterior
self.beta_n = self.beta0 - self.data.sum() + Variable(torch.Tensor([self.n_data]))
# for convenience compute the logs
self.log_alpha_n = torch.log(self.alpha_n)
self.log_beta_n = torch.log(self.beta_n)
def setup(self):
# initialize values of the two variational parameters
# set to be quite close to the true values
# so that the experiment doesn't take too long
self.log_alpha_q_0 = Variable(torch.Tensor([np.log(15.0)]), requires_grad=True)
self.log_beta_q_0 = Variable(torch.Tensor([np.log(15.0)]), requires_grad=True)
def model(self, use_decaying_avg_baseline):
# sample `latent_fairness` from the beta prior
f = pyro.sample("latent_fairness", dist.beta, self.alpha0, self.beta0)
# use iarange to indicate that the observations are
# conditionally independent given f and get vectorization
with pyro.iarange("data_iarange"):
# observe all ten datapoints using the bernoulli likelihood
pyro.observe("obs", dist.bernoulli, self.data, f)
def guide(self, use_decaying_avg_baseline):
# register the two variational parameters with pyro
log_alpha_q = pyro.param("log_alpha_q", self.log_alpha_q_0)
log_beta_q = pyro.param("log_beta_q", self.log_beta_q_0)
alpha_q, beta_q = torch.exp(log_alpha_q), torch.exp(log_beta_q)
# sample f from the beta variational distribution
baseline_dict = {'use_decaying_avg_baseline': use_decaying_avg_baseline,
'baseline_beta': 0.90}
# note that the baseline_dict specifies whether we're using
# decaying average baselines or not
pyro.sample("latent_fairness", dist.beta, alpha_q, beta_q,
baseline=baseline_dict)
def do_inference(self, use_decaying_avg_baseline, tolerance=0.05):
# clear the param store in case we're in a REPL
pyro.clear_param_store()
# initialize the variational parameters for this run
self.setup()
# setup the optimizer and the inference algorithm
optimizer = optim.Adam({"lr": .0008, "betas": (0.93, 0.999)})
svi = SVI(self.model, self.guide, optimizer, loss="ELBO", trace_graph=True)
print("Doing inference with use_decaying_avg_baseline=%s" % use_decaying_avg_baseline)
# do up to 10000 steps of inference
for k in range(10000):
svi.step(use_decaying_avg_baseline)
if k % 100 == 0:
print('.', end='')
sys.stdout.flush()
# compute the distance to the parameters of the true posterior
alpha_error = param_abs_error("log_alpha_q", self.log_alpha_n)
beta_error = param_abs_error("log_beta_q", self.log_beta_n)
# stop inference early if we're close to the true posterior
if alpha_error < tolerance and beta_error < tolerance:
break
print("\nDid %d steps of inference." % k)
print(("Final absolute errors for the two variational parameters " +
"(in log space) were %.4f & %.4f") % (alpha_error, beta_error))
# do the experiment
bbe = BernoulliBetaExample()
bbe.do_inference(use_decaying_avg_baseline=True)
bbe.do_inference(use_decaying_avg_baseline=False)
# -
# **Sample output:**
# ```
# Doing inference with use_decaying_avg_baseline=True
# ...........
# Did 2070 steps of inference.
# Final absolute errors for the two variational parameters (in log space) were 0.0500 & 0.0443
# Doing inference with use_decaying_avg_baseline=False
# .....................
# Did 4159 steps of inference.
# Final absolute errors for the two variational parameters (in log space) were 0.0500 & 0.0306
# ```
# For this particular run we can see that baselines roughly halved the number of steps of SVI we needed to do. The results are stochastic and will vary from run to run, but this is an encouraging result. For certain model and guide pairs, baselines can provide an even bigger win.
# ## References
#
# [1] `Automated Variational Inference in Probabilistic Programming`,
# <br/>
# <NAME>, <NAME>
#
# [2] `Black Box Variational Inference`,<br/>
# <NAME>, <NAME>, <NAME>
#
# [3] `Auto-Encoding Variational Bayes`,<br/>
# <NAME>, <NAME>
#
# [4] `Gradient Estimation Using Stochastic Computation Graphs`,
# <br/>
# <NAME>, <NAME>, <NAME>, <NAME>
#
# [5] `Deep Amortized Inference for Probabilistic Programs`
# <br/>
# <NAME>, <NAME>, <NAME>
#
# [6] `Neural Variational Inference and Learning in Belief Networks`
# <br/>
# <NAME>, <NAME>
| tutorial/source/svi_part_iii.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Portfolio Exercise: Starbucks
# <br>
#
# <img src="https://opj.ca/wp-content/uploads/2018/02/New-Starbucks-Logo-1200x969.jpg" width="200" height="200">
# <br>
# <br>
#
# #### Background Information
#
# The dataset you will be provided in this portfolio exercise was originally used as a take-home assignment provided by Starbucks for their job candidates. The data for this exercise consists of about 120,000 data points split in a 2:1 ratio among training and test files. In the experiment simulated by the data, an advertising promotion was tested to see if it would bring more customers to purchase a specific product priced at $10. Since it costs the company 0.15 to send out each promotion, it would be best to limit that promotion only to those that are most receptive to the promotion. Each data point includes one column indicating whether or not an individual was sent a promotion for the product, and one column indicating whether or not that individual eventually purchased that product. Each individual also has seven additional features associated with them, which are provided abstractly as V1-V7.
#
# #### Optimization Strategy
#
# Your task is to use the training data to understand what patterns in V1-V7 to indicate that a promotion should be provided to a user. Specifically, your goal is to maximize the following metrics:
#
# * **Incremental Response Rate (IRR)**
#
# IRR depicts how many more customers purchased the product with the promotion, as compared to if they didn't receive the promotion. Mathematically, it's the ratio of the number of purchasers in the promotion group to the total number of customers in the purchasers group (_treatment_) minus the ratio of the number of purchasers in the non-promotional group to the total number of customers in the non-promotional group (_control_).
#
# $$ IRR = \frac{purch_{treat}}{cust_{treat}} - \frac{purch_{ctrl}}{cust_{ctrl}} $$
#
#
# * **Net Incremental Revenue (NIR)**
#
# NIR depicts how much is made (or lost) by sending out the promotion. Mathematically, this is 10 times the total number of purchasers that received the promotion minus 0.15 times the number of promotions sent out, minus 10 times the number of purchasers who were not given the promotion.
#
# $$ NIR = (10\cdot purch_{treat} - 0.15 \cdot cust_{treat}) - 10 \cdot purch_{ctrl}$$
#
# For a full description of what Starbucks provides to candidates see the [instructions available here](https://drive.google.com/open?id=18klca9Sef1Rs6q8DW4l7o349r8B70qXM).
#
# Below you can find the training data provided. Explore the data and different optimization strategies.
#
# #### How To Test Your Strategy?
#
# When you feel like you have an optimization strategy, complete the `promotion_strategy` function to pass to the `test_results` function.
# From past data, we know there are four possible outomes:
#
# Table of actual promotion vs. predicted promotion customers:
#
# <table>
# <tr><th></th><th colspan = '2'>Actual</th></tr>
# <tr><th>Predicted</th><th>Yes</th><th>No</th></tr>
# <tr><th>Yes</th><td>I</td><td>II</td></tr>
# <tr><th>No</th><td>III</td><td>IV</td></tr>
# </table>
#
# The metrics are only being compared for the individuals we predict should obtain the promotion – that is, quadrants I and II. Since the first set of individuals that receive the promotion (in the training set) receive it randomly, we can expect that quadrants I and II will have approximately equivalent participants.
#
# Comparing quadrant I to II then gives an idea of how well your promotion strategy will work in the future.
#
# Get started by reading in the data below. See how each variable or combination of variables along with a promotion influences the chance of purchasing. When you feel like you have a strategy for who should receive a promotion, test your strategy against the test dataset used in the final `test_results` function.
# +
# Load in packages
from itertools import combinations
from test_results import test_results, score
import numpy as np
import pandas as pd
import scipy as sp
import seaborn as sns
import sklearn as sk
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_score
import matplotlib.pyplot as plt
import seaborn as sb
# %matplotlib inline
# Load in the train data and inspecting the first few rows
train_data = pd.read_csv('./training.csv')
train_data.head()
# -
# Inspecting the data
train_data.info()
# Checking for any null-values
train_data[train_data.isnull()].sum()
# Checking distribution of promotion
prom_dist = train_data.groupby('Promotion')['purchase'].value_counts()
prom_dist
# +
# Calculating and printing group counts
customer_total = train_data.shape[0]
customer_control = train_data.query('Promotion == "No"').shape[0]
customer_treatment = train_data.query('Promotion == "Yes"').shape[0]
purchase_total = train_data.query('purchase == 1').shape[0]
purchase_control = train_data.query('Promotion == "No" and purchase == 1').shape[0]
purchase_treatment = train_data.query('Promotion == "Yes" and purchase == 1').shape[0]
print('Customer count:', customer_total)
print('Control group count:', customer_control)
print('Treatment group count:', customer_treatment)
print('Total purchase count:', purchase_total)
print('Control purchase count:', purchase_control)
print('Total treatment count:', purchase_treatment)
# -
# Calculating Incremental Response Rate (IRR)
irr = (purchase_treatment / customer_treatment) - (purchase_control / customer_control)
print('IRR:',irr)
# Calculating Net Incremental Revenue (NIR)
nir = 10*purchase_treatment - 0.15*customer_treatment - 10*purchase_control
print('NIR:', nir)
# ### Hypothesis test for IRR value
#
# Null Hypothesis (H0): IRR <= 0;
# Alternate Hypothesis (H1): IRR != 0
#
# alpha = 0.05
#
# Bonferroni Correction = alpha / number of measures = 0.025
#
#
# +
# Checking IRR, simulate outcomes under null and compare to observed outcome
n_trials = 200000
p_null = train_data['purchase'].mean()
sim_control = np.random.binomial(customer_control, p_null, n_trials)
sim_treatment = np.random.binomial(customer_treatment, p_null, n_trials)
samples = (sim_treatment / customer_treatment) - (sim_control / customer_control)
p_val = (samples >= irr).mean()
# Conclusion of the experiment
print('The p-value for the test on IRR is {}. Therefore we reject the null hypothesis that IRR = 0.' .format(p_val))
# -
# ### Hypothesis test for NIR value
#
# H0: NIR = 0;
# H1: NIR != 0
#
# alpha = 0.05
#
# Bonferroni Correction = alpha / number of measures = 0.025
#
# +
# Checking NIR, simulate outcomes under null and compare to observed outcome
n_trials = 200000
p_null = train_data['purchase'].mean()
sim_control = np.random.binomial(customer_control, p_null, n_trials)
sim_treatment = np.random.binomial(customer_treatment, p_null, n_trials)
samples = 10*sim_treatment - 0.15*customer_treatment - 10*sim_control
p_val = (samples >= nir).mean()
# Conclusion of the experiment
print('The p-value for the test on NIR is {}. Therefore we reject the null hypothesis that NIR = 0.' .format(p_val))
# -
# ### Building promotion strategy model
# +
# Creating X and y variables
X = train_data[['V1', 'V2', 'V3', 'V4', 'V5', 'V6', 'V7']]
y = train_data['purchase'].values
# Scaling X
scaler = StandardScaler()
X = scaler.fit_transform(X)
# Perform train test split in 2:1 ratio
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.67, test_size=0.33, random_state=55)
# +
# Build a pipeline, using n_jobs = -1 to improve processing speeds
pipeline = Pipeline([('clf', RandomForestClassifier(n_jobs=-1, class_weight='balanced'))])
# Checking pipeline parameters
pipeline.get_params().keys()
# -
# Hyperparameter tuning, using precision as scoring method
parameters = {'clf__n_estimators': [50,100,200],
'clf__max_depth': [3,4,5]}
# +
# Noted it costs the company 0.15 to send out each promotion and it would be best to limit
# that promotion only to those that are most receptive to the promotion.
# Therefore we want to minimise false positives (ie we are seeking higher Precision, which will be the used metric)
# Also noted that higher the purchase_treatment (true positives), the higher the IRR and NRR.
# passing grid search object
cv = GridSearchCV(pipeline, param_grid = parameters, scoring ='precision')
# +
# Training grid search model
cv.fit(X_train, y_train)
# Predict on test data
y_pred = cv.predict(X_test)
# -
# Evaluating the model
class_report = classification_report(y_test, y_pred)
# +
# Confusion matrix
conf_matrix = confusion_matrix(y_test,y_pred)
index = ['No','Yes']
columns = ['No','Yes']
cm_df = pd.DataFrame(conf_matrix, columns, index)
sns.heatmap(cm_df,annot=True, cmap='Blues',fmt='g')
plt.plot()
plt.xlabel('Predicted label')
plt.ylabel('True label')
plt.title('Confusion Matrix');
# -
# Printing confusion matrix to check the above chart
confusion_matrix(y_test,y_pred)
def promotion_strategy(df):
'''
INPUT
df - a dataframe with *only* the columns V1 - V7 (same as train_data)
OUTPUT
promotion_df - np.array with the values
'Yes' or 'No' related to whether or not an
individual should recieve a promotion
should be the length of df.shape[0]
Ex:
INPUT: df
V1 V2 V3 V4 V5 V6 V7
2 30 -1.1 1 1 3 2
3 32 -0.6 2 3 2 2
2 30 0.13 1 1 4 2
OUTPUT: promotion
array(['Yes', 'Yes', 'No'])
indicating the first two users would recieve the promotion and
the last should not.
'''
# Scaling dataframe using the above scaler
df = scaler.transform(df)
# Predict on the data frame
purchases = cv.predict(df)
promotion = np.where(purchases == 1, 'Yes','No')
return promotion
# +
# This will test your results, and provide you back some information
# on how well your promotion_strategy will work in practice
test_results(promotion_strategy)
| Starbucks.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Environment and RL Agent Controller for a Thermostat
#
# ```
# Author: <NAME>
# Github: mpettis
# Twitter: @mtpettis
# Date: 2020-04-27
# ```
#
# This is a toy example of a room with a heater. When the heater is off, the temperature will decay to 0.0, and when it is on, it will rise to 1.0. The decay and rise is not instantaneous, but has exponential decay behavior in time given by the following formula:
#
# temperature[i + 1] = heater[i] + (temperature[i] - heater[i]) * exp(-1/tau)
#
# Where:
#
# temperature[i] is the temperature at timestep i (between 0 and 1).
# heater[i] is the applied heater, 0 when not applied, 1 when applied.
# tau is the characteristic heat decay constant.
#
# So, when the heater is off, the temperature will decay towards 0, and when the heater is on, it will rise towards 1. When the heater is toggled on/off, it will drift towards 1/0.
#
# Here is a sample plot of what the temperature response looks like when the heater is on for a while, then off for a while. You will see the characteristic rise and decay of the temperature to the response.
# +
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import math
## Compute the response for a given action and current temperature
def respond(action, current_temp, tau):
return action + (current_temp - action) * math.exp(-1.0/tau)
## Actions of a series of on, then off
sAction = pd.Series(np.array([1,1,1,1,1,1,1,1,1,1,0,0,0,0,0,0,0,0,0,0]))
sResponse = np.zeros(sAction.size)
## Update the response with the response to the action
for i in range(sAction.size):
## Get last response
if i == 0:
last_response = 0
else:
last_response = sResponse[i - 1]
sResponse[i] = respond(sAction[i], last_response, 3.0)
## Assemble and plot
df = pd.DataFrame(list(zip(sAction, sResponse)), columns=['action', 'response'])
df.plot()
# -
# ## Goal and Reward
# The goal here is to make an agent that will take actions that will keep the temperature between 0.4 and 0.6.
#
# We make a reward function to reflect our goal. When the temperature is between 0.4 and 0.6, we set the reward as 0.0. When the temperature is outside of this band, we set the reward to be the negative distance the temperature is from its closest band. So if the temperature is 0.1, then the reward is -(0.4 - 0.1) = -0.3, and if it is 0.8, then the reward is -(0.8 - 0.6) = -0.2.
#
# Let's chart the reward vs. temperature to show what is meant:
# +
def reward(temp):
delta = abs(temp - 0.5)
if delta < 0.1:
return 0.0
else:
return -delta + 0.1
temps = [x * 0.01 for x in range(100)]
rewards = [reward(x) for x in temps]
fig=plt.figure(figsize=(12, 4))
plt.scatter(temps, rewards)
plt.xlabel('Temperature')
plt.ylabel('Reward')
plt.title('Reward vs. Temperature')
# -
# # Environment Setup
#
# The environment responds to actions. It is what keeps track of the temperature state of the room, returns the reward for being in that temperature state, and tells you if the episode is over or not (in this case, we just set a max episode length that can happen).
#
# Here is the gist of the flow:
#
# - Create an environment by calling `Environment.create()`, see below, telling it to use the class you created for this (here, the ThermostatEnvironment) and the max timesteps per episode. The enviroment is assigned to the name `environment`.
# - Initialize the environment `environment` by calling `environment.reset()`. This will do stuff, most importantly, it will initialize the `timestep` attribute to 0.
# - When you want to take an action on the current state of the environment, you will call `environment.execute(<action-value>)`. If you want to have the heater off, you call `environment.execute(0)`, and if you want to have the heater on, you call `environment.execute(1)`.
# - What the `execute()` call returns is a tuple with 3 entries:
# - __state__. In this case, the state is the current temperature that results from taking the action. If you turn on the heater, the temperature will rise from the previous state, and if the heater was turned off, the temperature will fall from the previous state. This should be kept as a numpy array, even though it seems like overkill with a single value for the state coming back. For more complex examples beyond this thermostat, there will be more than 1 component to the state.
# - __terminal__. This is a True/False value. It is True if the episode terminated. In this case, that will happen once you exceed the max number of steps you have set. Otherwise, it will be False, which lets the agent know that it can take further steps.
# - __reward__. This is the reward for taking the action you took.
#
# Below, to train the agent, you will have the agent take actions on the environment, and the environment will return these signals so that the agent can self-train to optimize its reward.
# +
###-----------------------------------------------------------------------------
## Imports
from tensorforce.environments import Environment
from tensorforce.agents import Agent
###-----------------------------------------------------------------------------
### Environment definition
class ThermostatEnvironment(Environment):
"""This class defines a simple thermostat environment. It is a room with
a heater, and when the heater is on, the room temperature will approach
the max heater temperature (usually 1.0), and when off, the room will
decay to a temperature of 0.0. The exponential constant that determines
how fast it approaches these temperatures over timesteps is tau.
"""
def __init__(self):
## Some initializations. Will eventually parameterize this in the constructor.
self.tau = 3.0
self.current_temp = np.random.random(size=(1,))
super().__init__()
def states(self):
return dict(type='float', shape=(1,))
def actions(self):
"""Action 0 means no heater, temperature approaches 0.0. Action 1 means
the heater is on and the room temperature approaches 1.0.
"""
return dict(type='int', num_values=2)
# Optional, should only be defined if environment has a natural maximum
# episode length
def max_episode_timesteps(self):
return super().max_episode_timesteps()
# Optional
def close(self):
super().close()
def reset(self):
"""Reset state.
"""
# state = np.random.random(size=(1,))
self.timestep = 0
self.current_temp = np.random.random(size=(1,))
return self.current_temp
def response(self, action):
"""Respond to an action. When the action is 1, the temperature
exponentially decays approaches 1.0. When the action is 0,
the current temperature decays towards 0.0.
"""
return action + (self.current_temp - action) * math.exp(-1.0 / self.tau)
def reward_compute(self):
""" The reward here is 0 if the current temp is between 0.4 and 0.6,
else it is distance the temp is away from the 0.4 or 0.6 boundary.
Return the value within the numpy array, not the numpy array.
"""
delta = abs(self.current_temp - 0.5)
if delta < 0.1:
return 0.0
else:
return -delta[0] + 0.1
def execute(self, actions):
## Check the action is either 0 or 1 -- heater on or off.
assert actions == 0 or actions == 1
## Increment timestamp
self.timestep += 1
## Update the current_temp
self.current_temp = self.response(actions)
## Compute the reward
reward = self.reward_compute()
## The only way to go terminal is to exceed max_episode_timestamp.
## terminal == False means episode is not done
## terminal == True means it is done.
terminal = False
if self.timestep > self.max_episode_timesteps():
terminal = True
return self.current_temp, terminal, reward
###-----------------------------------------------------------------------------
### Create the environment
### - Tell it the environment class
### - Set the max timestamps that can happen per episode
environment = environment = Environment.create(
environment=ThermostatEnvironment,
max_episode_timesteps=100)
# -
# # Agent setup
#
# Here we configure a type of agent to learn against this environment. There are many agent configurations to choose from, which we will not cover here. We will not discuss what type of agent to choose here -- we will just take a basic agent to train.
agent = Agent.create(
agent='tensorforce', environment=environment, update=64,
objective='policy_gradient', reward_estimation=dict(horizon=1)
)
# # Check: Untrained Agent Performance
#
# Let's see how the untrained agent performs on the environment. The red horizontal lines are the target bands for the temperature.
#
# The agent doesn't take actions to try and get the temperature within the bands. It either initializes a policy to the heater always off or always on.
# +
### Initialize
environment.reset()
## Creation of the environment via Environment.create() creates
## a wrapper class around the original Environment defined here.
## That wrapper mainly keeps track of the number of timesteps.
## In order to alter the attributes of your instance of the original
## class, like to set the initial temp to a custom value, like here,
## you need to access the `environment` member of this wrapped class.
## That is why you see the way to set the current_temp like below.
environment.environment.current_temp = np.array([0.5])
states = environment.environment.current_temp
internals = agent.initial_internals()
terminal = False
### Run an episode
temp = [environment.environment.current_temp[0]]
while not terminal:
actions, internals = agent.act(states=states, internals=internals, evaluation=True)
states, terminal, reward = environment.execute(actions=actions)
temp += [states[0]]
### Plot the run
plt.figure(figsize=(12, 4))
ax=plt.subplot()
ax.set_ylim([0.0, 1.0])
plt.plot(range(len(temp)), temp)
plt.hlines(y=0.4, xmin=0, xmax=99, color='r')
plt.hlines(y=0.6, xmin=0, xmax=99, color='r')
plt.xlabel('Timestep')
plt.ylabel('Temperature')
plt.title('Temperature vs. Timestep')
plt.show()
# -
# # Train the agent
#
# Here we train the agent against episodes of interacting with the environment.
# Train for 200 episodes
for _ in range(200):
states = environment.reset()
terminal = False
while not terminal:
actions = agent.act(states=states)
states, terminal, reward = environment.execute(actions=actions)
agent.observe(terminal=terminal, reward=reward)
# # Check: Trained Agent Performance
#
# You can plainly see that this is toggling the heater on/off to keep the temperature within the target band!
# +
### Initialize
environment.reset()
## Creation of the environment via Environment.create() creates
## a wrapper class around the original Environment defined here.
## That wrapper mainly keeps track of the number of timesteps.
## In order to alter the attributes of your instance of the original
## class, like to set the initial temp to a custom value, like here,
## you need to access the `environment` member of this wrapped class.
## That is why you see the way to set the current_temp like below.
environment.environment.current_temp = np.array([1.0])
states = environment.environment.current_temp
internals = agent.initial_internals()
terminal = False
### Run an episode
temp = [environment.environment.current_temp[0]]
while not terminal:
actions, internals = agent.act(states=states, internals=internals, evaluation=True)
states, terminal, reward = environment.execute(actions=actions)
temp += [states[0]]
### Plot the run
plt.figure(figsize=(12, 4))
ax=plt.subplot()
ax.set_ylim([0.0, 1.0])
plt.plot(range(len(temp)), temp)
plt.hlines(y=0.4, xmin=0, xmax=99, color='r')
plt.hlines(y=0.6, xmin=0, xmax=99, color='r')
plt.xlabel('Timestep')
plt.ylabel('Temperature')
plt.title('Temperature vs. Timestep')
plt.show()
| examples/temperature-controller.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: computationalPhysics
# language: python
# name: conda-env-computationalPhysics-py
# ---
# <h1>A Simple Model of a ballon in a fluid of Uniform Density</h1>
# <h2><NAME></h2>
# <h3>Introduction</h3>
# Here I present two simple models of a balloon in a confined space. In the first the balloon is acted upon by gravity and a buoyant force. Additionally, there is an effective infintite in magnitude and infintesimal in time normal force applied to the ballon at the boundaries of some user defined volume. The volume containes two fluids, each with a different (but uniform) density. The second model is simialar to the first; however, it may contain much more complex density perturbations throughout, and an additional "wind" force is included in all three spacial dimensions. This model demonstrates how density perturbations may be used as approximations of soft constraint boundaries.
# +
import numpy as np
import scipy as sp
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.ticker import AutoMinorLocator
from mplEasyAnimate import animation
from tqdm import tqdm
import time
import integrators as intgs
from helper import display_video, make_animation
from IPython.display import HTML
plt.rc('text', usetex = True)
plt.rc('font', family='serif')
# -
g = 9.8 # m s^-2
# <h3>Balloon & Helper Objects</h3>
# Here I define a balloon class which will store the ballons position and velocity. This object will also loose energy when reflected off of a boundary. The function
# $$
# c_{r}(P) = -40e^{-4.5P} + 0.86
# $$
# empirically determined <a href=http://isjos.org/JoP/vol3iss2/Papers/JoPv3i2-2COR.pdf>here</a>, is used to calculate the coefficient of restitution for the balloon in one atmosphere of pressure. At a boundary with axis $i$ the new velocity along axis $i$ is then given as
# $$
# v_{i, f} = -c_{r}(P) \cdot v_{i, 0}
# $$
class balloonObj:
def __init__(self, radius, rho=0.164, r0=[0, 0, 0], v0=[0, 0, 0], a1=1, a2=1, a3=1):
self.radius = radius
self.rho = rho # kg m^-3
self.volume = ((4/3)*np.pi*self.radius**3)/1000 # m^3
self.ppos = None
self.pos = r0 # m
self.mass = self.volume*self.rho # kg
self.velocity = v0
P = 1 #atm
self.a1 = a1
self.a2 = a2
self.a3 = a3
self.cr = -40*np.exp(-4.5*P)+0.86 #http://isjos.org/JoP/vol3iss2/Papers/JoPv3i2-2COR.pdf
def reflect(self, axis):
self.velocity[axis] = -self.cr * self.velocity[axis]
# Next we define a "helper object" -- confinment -- which simply is used to store the bounds of the rectangular cuboid the balloon is confined within. This confinment object is responsible for reporting if a collision has happened, and what axis that collison is along.
class confinment:
def __init__(self, bounds):
# [[xmin, xmax], [ymin, ymax], [zmin, zmax]]
self.bounds = bounds
def check_x(self, x):
if self.bounds[0][0] < x < self.bounds[0][1]:
return False
else:
return True
def check_y(self, y):
if self.bounds[1][0] < y < self.bounds[1][1]:
return False
else:
return True
def check_z(self, z):
if self.bounds[2][0] < z < self.bounds[2][1]:
return False
else:
return True
def check_for_collision(self, pos):
if self.check_x(pos[0]) and self.check_y(pos[1]) and self.check_z(pos[2]):
return True
else:
return False
# Finally we define an object to coordinate the integration. The "worldIntegrator" takes some model, some balloon, some confiner, and an integration scheme to use. It will then allow the user to step the system through time.
class wordIntegrator:
def __init__(self, confiner, obj, model, method=intgs.rk4, upper_density=1.18, lower_density=1.18):
self.object = obj
self.method = method
self.model = model
self.confiner = confiner
self.clock = 0
self.step = 0
self.upper_density = upper_density
self.lower_density = lower_density
def get_rho(self, ypos):
if ypos <= self.confiner.bounds[1][1]/2:
return self.lower_density # kg m^-3
else:
return self.upper_density # kg m^-3
def get_args(self):
args = dict()
args['m'] = self.object.mass
args['V'] = self.object.volume
args['a1'] = self.object.a1
args['a2'] = self.object.a2
args['a3'] = self.object.a3
args['rho_air'] = self.get_rho(self.object.pos[1])
return args
def timeEvolve(self, dt):
"""
Desc:
Incriment system by time step dt
"""
cx = self.confiner.check_x(self.object.pos[0])
cy = self.confiner.check_y(self.object.pos[1])
cz = self.confiner.check_z(self.object.pos[2])
if cx:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(0)
if cy:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(1)
if cz:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(2)
cI = list(self.object.pos) + list(self.object.velocity)
nI = self.method(self.model, cI, self.clock, dt, self.get_args())
self.object.ppos = self.object.pos
self.object.pos = nI[:3]
self.object.velocity = nI[3:]
self.step += 1
self.clock += dt
# <h3>Model</h3>
# We develop a three--dimensional model to describe the system, the model is given as
# $$
# \frac{dx}{dt} = v_{x} \\
# \frac{dy}{dt} = v_{y}\\
# \frac{dz}{dt} = v_{z} \\
# $$
# with the velocity components being given by
# $$
# \frac{dv_{x}}{dt} = 0 \\
# \frac{dv_{y}}{dt} = -mg+gV\rho_{c} \\
# \frac{dv_{z}}{dt} = 0 \\
# $$
# Initially we had hoped to include quadradic drag in three dimensions into this model; however, this proved infeasible for this stage of this project. Future work will aim to include quadradic drag into the model.
#
# The force in the $y$ direction is given as the sum of the weight of the ballon and the weight of the displaced fluid. This model of buoyancy assumes the density of the fluid over the height of object is a constant. A more complex, and physically representative manner of estimating the boyant force may be desirable in future given that the balloon traverses density boundary. However, the method presented here acts as an effective 1st order estimate.
def bouyModel(I, t, args):
# 0 1 2 3 4 5
# [x, y, z, vx, vy, vz]
dIdt = np.zeros(6)
dIdt[0] = I[3]
dIdt[1] = I[4]
dIdt[2] = I[5]
# Weight # Boyant Force
dIdt[4] = (-args['m']*g) + g*args['V']*(args['rho_air'])
return dIdt
# <h3>Integration</h3>
# I integrate the model with a balloon density of 1 kg m$^{-3}$ over 1000 seconds, with a time step of 0.01 seconds. I have set up the densities of the volume such that the ballon is more dense than the top half of the volume, and less dense than the bottom half of the volume. This should result in an soft boundary which the balloon tends to stay within at the intersection of the two regions.
# +
balloon = balloonObj(0.31, v0=[1.5, 0, 0], r0=[1, 4.5, 1], rho=1)
confiner = confinment([[-5, 5], [0, 10], [-5, 5]])
world = wordIntegrator(confiner, balloon, bouyModel, upper_density=0.5, lower_density=2)
pos = list()
vel = list()
dt = 0.01
time_vals = np.arange(0, 1000, dt)
for t in time_vals:
world.timeEvolve(dt)
pos.append(world.object.pos)
vel.append(world.object.velocity)
pos = np.array(pos)
vel = np.array(vel)
# -
# <h3>Data Visualization</h3>
# I defined a helper function to set the style of all plots in a consistent manner
def setup_plot(xBounds=False, yBounds=False, yBV = [0, 10], xBV = [-5, 5]):
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
if yBounds:
ax.axhline(y=yBV[0], color='gray', alpha=0.5)
ax.axhline(y=yBV[1], color='gray', alpha=0.5)
if xBounds:
ax.axvline(x=xBV[0], color='gray', alpha=0.5)
ax.axvline(x=xBV[1], color='gray', alpha=0.5)
ax.xaxis.set_minor_locator(AutoMinorLocator())
ax.yaxis.set_minor_locator(AutoMinorLocator())
ax.tick_params(which='both', labelsize=17, direction='in', top=True, right=True)
ax.tick_params(which='major', length=10, width=1)
ax.tick_params(which='minor', length=5, width=1)
return fig, ax
# First we investigate the x-y postition of the balloon from the integration above. Note how the balloon looses energy on impact with the wall (related to its velocity before impact through the calculated coefficient of restitution). However also note that the ballon reverses velocity in the y direction without interacting with the hard boundary. This is a demonstration that the pressure difference may act as a soft boundary (i.e. the balloon can pass through it but will eventually be forced back the way it came).
#
# Because of the energy loss to reflection off the x bounds the ''wavelength'' of the oscillation shortens with time, this can be more clearly seen in the animation presented below this cell.
# +
fig, ax = setup_plot(xBounds=True, yBounds=True)
ax.plot(pos[:, 0], pos[:, 1], 'k')
ax.set_xlabel('$x$ [m]', fontsize=20)
ax.set_ylabel('$y$ [m]', fontsize=20)
plt.show()
# -
make_animation(pos, 'BallBouncing.mp4', plt, AutoMinorLocator, step=500)
# +
import io
import base64
from IPython.display import HTML
import os
if not os.path.exists('BallBouncing.mp4'):
raise IOError('ERROR! Animation has not been generated to the local directory yet!')
video = io.open('BallBouncing.mp4', 'r+b').read()
encoded = base64.b64encode(video)
HTML(data='''<video alt="test" controls>
<source src="data:video/mp4;base64,{0}" type="video/mp4" />
</video>'''.format(encoded.decode('ascii')))
# -
# Looking at just the x position vs time we see linear sections connected with discontinuities at the bounds as we would expect to see given there are no forces acting on the ballon in the x direction.
# +
fig, ax = setup_plot(yBounds=True, yBV=[-5, 5])
ax.plot(time_vals, pos[:, 0], 'k')
ax.set_xlabel('Time [s]', fontsize=20)
ax.set_ylabel('$x$ [m]', fontsize=20)
plt.show()
# -
# We equally see what we might expect to see in the y direction, the balloon osscillating around the pressure boundary
# +
fig, ax = setup_plot(yBounds=True)
ax.plot(time_vals, pos[:, 1], 'k')
ax.set_xlabel('Time [s]', fontsize=20)
ax.set_ylabel('$y$ [m]', fontsize=20)
plt.show()
# +
fig, ax = setup_plot()
ax.plot(time_vals, vel[:, 0], 'k')
ax.set_xlabel('Time [s]', fontsize=20)
ax.set_ylabel(r'$v_{x}$ [m s$^{-1}$]', fontsize=20)
plt.show()
# +
fig, ax = setup_plot()
ax.plot(time_vals, vel[:, 1], 'k')
ax.set_xlabel('Time [s]', fontsize=20)
ax.set_ylabel(r'$v_{y}$ [m s$^{-1}$]', fontsize=20)
plt.show()
# -
# <h3>Wind & 3 Dimensions</h3>
#
# The model I have presented so far is relatively boring in all but the y directions. It is possible to plot all three spacial dimensions here; however, given there are no forces in either the x or z directions, that does not hold much interest beyond the 2D situations I have presented here. Below I present an updated model containing an extra "wind" force in both the x and z axis. It should be noted that this is a contrived force; however, because of the implimentation, may model a somewhat accurate situation.
#
# This more complex model also describes the density based on a function.
def bouyModel_wind(I, t, args):
# 0 1 2 3 4 5
# [x, y, z, vx, vy, vz]
dIdt = np.zeros(6)
dIdt[0] = I[3]
dIdt[1] = I[4]
dIdt[2] = I[5]
dIdt[3] = args['wind'][0](I, t)
# Weight # Boyant Force # Wind Force
dIdt[4] = (-args['m']*g) + g*args['V']*(args['rho_air']) + args['wind'][1](I, t)
dIdt[5] = args['wind'][2](I, t)
return dIdt
class wordIntegrator_wind:
def __init__(self, confiner, obj, model, method=intgs.rk4,
density_func=lambda y, ty: 1.18, wind_0=lambda x, t: 0,
wind_1 = lambda y, t: 0, wind_2=lambda z, t: 0):
self.object = obj
self.method = method
self.model = model
self.confiner = confiner
self.clock = 0
self.step = 0
self.get_rho = density_func
self.wind = (wind_0, wind_1, wind_2)
def get_args(self):
args = dict()
args['m'] = self.object.mass
args['V'] = self.object.volume
args['a1'] = self.object.a1
args['a2'] = self.object.a2
args['a3'] = self.object.a3
args['rho_air'] = self.get_rho(self.object.pos[1], self.confiner.bounds[1][1])
args['wind'] = self.wind
return args
def timeEvolve(self, dt):
cx = self.confiner.check_x(self.object.pos[0])
cy = self.confiner.check_y(self.object.pos[1])
cz = self.confiner.check_z(self.object.pos[2])
if cx:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(0)
if cy:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(1)
if cz:
if self.object.ppos is not None:
self.object.pos = self.object.ppos
self.object.reflect(2)
cI = list(self.object.pos) + list(self.object.velocity)
nI = self.method(self.model, cI, self.clock, dt, self.get_args())
self.object.ppos = self.object.pos
self.object.pos = nI[:3]
self.object.velocity = nI[3:]
self.step += 1
self.clock += dt
# I define both a density function
# $$
# \rho_{air}(y) = 5\sin(\ln(y^{5}))
# $$
# and a function describing wind in the x-direction
# $$
# F_{w,x}(x, t) = \frac{0.01\sin(x)}{0.005961t+0.01}
# $$
# These are then passed into the new word Integrator. The effects of the wind pushing the balloon to one side are clear.
def density(y, ty):
return 5*np.sin(np.log(y**5))
def xwind(I, t):
return 0.01*np.sin(I[0])/(0.005960*t+0.01)
balloon = balloonObj(0.31, v0=[1.5, 0, 0], r0=[1, 4.5, 1], rho=1)
confiner = confinment([[-5, 5], [0, 10], [-5, 5]])
world = wordIntegrator_wind(confiner, balloon, bouyModel_wind, density_func=density, wind_0=xwind)
pos = list()
vel = list()
dt = 0.01
time_vals = np.arange(0, 1000, dt)
for t in time_vals:
world.timeEvolve(dt)
pos.append(world.object.pos)
vel.append(world.object.velocity)
pos = np.array(pos)
vel = np.array(vel)
# +
fig, ax = setup_plot(xBounds=True, yBounds=True)
ax.plot(pos[:, 0], pos[:, 1], 'k')
ax.set_xlabel('$x$ [m]', fontsize=20)
ax.set_ylabel('$y$ [m]', fontsize=20)
plt.show()
# -
# Finally we will look at 3D. I define the same initial condtitions for integration as above, except I also give the balloon an initial z velocity of
# $$
# v_{z} = -1 \text{ m s}^{-1}
# $$
# I then plot this in 3D below. If one changes the z velocity so that it approaches 0 it is clear how the motion collapses into one plane
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
balloon = balloonObj(0.31, v0=[1.5, 0, -1], r0=[1, 4.5, 1], rho=1)
confiner = confinment([[-5, 5], [0, 10], [-5, 5]])
world = wordIntegrator_wind(confiner, balloon, bouyModel_wind, density_func=density, wind_0=xwind)
pos = list()
vel = list()
dt = 0.1
time_vals = np.arange(0, 1000, dt)
for t in time_vals:
world.timeEvolve(dt)
pos.append(world.object.pos)
vel.append(world.object.velocity)
pos = np.array(pos)
vel = np.array(vel)
# +
fig = plt.figure(figsize=(10, 7))
ax = fig.add_subplot(111, projection='3d')
ax.plot(pos[:, 0], pos[:, 1], pos[:, 2], 'k')
ax.set_zlim(-5, 5)
ax.set_xlim(-5, 5)
ax.set_ylim(0, 10)
ax.set_xlabel('$x$ [m]', fontsize=20)
ax.set_ylabel('$y$ [m]', fontsize=20)
ax.set_zlabel('$z$ [m]', fontsize=20)
plt.show()
| BallonInAFluid/BoudreauxNotebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # SciPy
# SciPy is a collection of mathematical algorithms and convenience functions. In this this notebook there are just a few examples of the features that are most important to us. But if you want to see all that SciPy has to offer, have a look at the [official documentation](https://docs.scipy.org/doc/scipy/reference/).
#
# Since SciPy has several sublibraries, it is commom practice to import just the one we are going to use, as you'll in the following examples.
import numpy as np
import matplotlib as mpl # ignore this for now
import matplotlib.pyplot as plt # ignore this for now
# # Interpolation
# There are several general interpolation facilities available in SciPy, for data in 1, 2, and higher dimensions. First, let's generate some sample data.
# +
x = np.linspace(0, 10, num=11, endpoint=True)
y = np.cos(-x**2/9.0)
plt.scatter(x,y)
# -
# The `interp1d` funtions grabs data points and **returns a *function***. The default interpolation method is the linear interpolation, but there are several to choose from.
# +
from scipy.interpolate import interp1d
f1 = interp1d(x, y) # linear is the default
f2 = interp1d(x, y, kind='cubic') # cubic splines
f3 = interp1d(x, y, kind='nearest') # grab the nearest value
f4 = interp1d(x, y, kind='previous') # hold last value
f5 = interp1d(x, y, kind='next') # grab the next value
# +
print(f1(4))
print(f2(4))
print(f1(4.6))
print(f2(4.6))
# -
# Now that we have the interpolated function, lets generate a tighter grid in the x axis and plot the resulto of the different interpolation methods.
xnew = np.linspace(0, 10, num=101, endpoint=True)
xnew
plt.plot(x, y, 'o', xnew, f1(xnew), '-', xnew, f2(xnew), '--', xnew, f3(xnew), '-.')
plt.legend(['data', 'linear', 'cubic', 'nearest'], loc='best')
plt.show()
# The `interpolate` sublibrary also has interpolation methods for multivariate data and has **integration with pandas**. Have a look at the documentation.
# # Definite Integrals
# The function `quad` is provided to integrate a function of one variable between two points. This functions has 2 outputs, the first one is the computed integral value and the second is an estimate of the absolute error.
# +
import scipy.integrate as integrate
def my_func(x):
return x**2
integrate.quad(my_func, 0, 2)
# -
# The `quad` functions also allows for infinite limits.
#
# $$
# \int_{-\infty}^{\infty} e^{-x^{2}}dx
# $$
# +
def my_func(x):
return np.exp(-x**2)
integrate.quad(my_func, -np.inf, np.inf)
# -
# SciPy's `integrate` library also has functions for double and triple integrals. Check them out in the documentations.
# # Optimization
# The `scipy.optimize` package provides several commonly used optimization algorithms. Here we are going to use just one to illustrate.
#
# Consider that you have 3 assets available. Their expected returns, risks (standard-deviations) and betas are on the table bellow and $\rho$ is the correlation matrix of the returns.
#
# | Asset | Return | Risk | Beta |
# |-------|--------|------|------|
# |A |3% | 10% | 0.5 |
# |B |3.5% | 11% | 1.2 |
# |C |5% | 15% | 1.8 |
#
# $$
# \rho =
# \begin{bmatrix}
# 1 & 0.3 & -0.6 \\
# 0.3 & 1 & 0 \\
# -0.6 & 0 & 1
# \end{bmatrix}
# $$
#
# Use the `minimize` function to find the weights of each asset that maximizes it's Sharpe index.
# +
retu = np.array([0.03, 0.035, 0.05])
risk = np.array([0.10, 0.11, 0.15])
beta = np.array([0.5, 1.2, 1.8])
corr = np.array([[1, 0.3, -0.6],
[0.3, 1, 0],
[-0.6, 0, 1]])
def port_return(w):
return retu.dot(w)
def port_risk(w):
covar = np.diag(risk).dot(corr).dot(np.diag(risk))
return (w.dot(covar).dot(w))**0.5
def port_sharpe(w):
return -1*(port_return(w) / port_risk(w)) # The -1 is because we want to MINIMIZE the negative of the Sharpe
def port_weight(w):
return w.sum()
# -
# When declaring an optimization problem with inequality restrictions, they have the form of:
#
# $$
# \begin{align*}
# \min_{w} & f\left(w\right)\\
# s.t. & g\left(w\right)\geq0
# \end{align*}
# $$
# +
from scipy.optimize import minimize
eq_cons = {'type': 'eq',
'fun' : lambda w: port_weight(w) - 1}
w0 = np.array([1, 0, 0])
res = minimize(port_sharpe, w0, method='SLSQP', constraints=eq_cons, options={'ftol': 1e-9, 'disp': True})
# -
res.x
res.x.sum()
-1*res.fun
# # Linear Algebra (again)
# `scipy.linalg` contains all the functions in `numpy.linalg` plus some more advanced ones.
# +
from scipy import linalg as la
A = np.array([[1,3,5],[2,5,1],[2,3,8]])
la.inv(A)
# -
# Matrix and vector **norms** can also be computed with SciPy. A wide range of norm definitions are available using different parameters to the order argument of `linalg.norm`.
A = np.array([[1, 2], [3, 4]])
print(la.norm(A)) # frobenius norm is the default.
print(la.norm(A, 1)) # L1 norm (max column sum)
print(la.norm(A, np.inf)) # L inf norm (max row sum)
# Some more advanced matrix decompositions are also available, like the **Schur Decomposition**
la.schur(A)
# Some notable matrices can also be created, like block **diagonal matrices**.
# +
A = np.array([[1, 0],
[0, 1]])
B = np.array([[3, 4, 5],
[6, 7, 8]])
C = np.array([[7]])
la.block_diag(A, B, C)
# -
# # Solving Linear Systems
#
#
# $$
# \begin{align}
# x+3y+5 & =10\\
# 2x+5y+z & =8\\
# 2x+3y+8z & =3
# \end{align}
# $$
#
# The system above can be written with matrix notation as $AX=B$ and we know we can find the solution by doing $X=A^{-1}B$, but inverting a matrix is computationally expensive. When solving big linear system it is advised to use the `solve` method.
A = np.array([[1, 3, 5], [2, 5, 1], [2, 3, 8]])
B = np.array([[10], [8], [3]])
# Lets check the time that it takes to solve the system in both ways...
la.inv(A).dot(B)
la.solve(A, B)
# let's try with a bigger matrix
import numpy.random as rnd
A = rnd.random((1000, 1000))
B = rnd.random((1000, 1))
# %%timeit
la.inv(A).dot(B)
# %%timeit
la.solve(A, B)
| fhnotebooks/Introduction to Python/Section 03 - SciPy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# <div style="color:#777777;background-color:#ffffff;font-size:12px;text-align:right;">
# prepared by <NAME> (QuSoft@Riga) | November 07, 2018
# </div>
# <table><tr><td><i> I have some macros here. If there is a problem with displaying mathematical formulas, please run me to load these macros.</i></td></td></table>
# $ \newcommand{\bra}[1]{\langle #1|} $
# $ \newcommand{\ket}[1]{|#1\rangle} $
# $ \newcommand{\braket}[2]{\langle #1|#2\rangle} $
# $ \newcommand{\inner}[2]{\langle #1,#2\rangle} $
# $ \newcommand{\biginner}[2]{\left\langle #1,#2\right\rangle} $
# $ \newcommand{\mymatrix}[2]{\left( \begin{array}{#1} #2\end{array} \right)} $
# $ \newcommand{\myvector}[1]{\mymatrix{c}{#1}} $
# $ \newcommand{\myrvector}[1]{\mymatrix{r}{#1}} $
# $ \newcommand{\mypar}[1]{\left( #1 \right)} $
# $ \newcommand{\mybigpar}[1]{ \Big( #1 \Big)} $
# $ \newcommand{\sqrttwo}{\frac{1}{\sqrt{2}}} $
# $ \newcommand{\dsqrttwo}{\dfrac{1}{\sqrt{2}}} $
# $ \newcommand{\onehalf}{\frac{1}{2}} $
# $ \newcommand{\donehalf}{\dfrac{1}{2}} $
# $ \newcommand{\hadamard}{ \mymatrix{rr}{ \sqrttwo & \sqrttwo \\ \sqrttwo & -\sqrttwo }} $
# $ \newcommand{\vzero}{\myvector{1\\0}} $
# $ \newcommand{\vone}{\myvector{0\\1}} $
# $ \newcommand{\vhadamardzero}{\myvector{ \sqrttwo \\ \sqrttwo } } $
# $ \newcommand{\vhadamardone}{ \myrvector{ \sqrttwo \\ -\sqrttwo } } $
# $ \newcommand{\myarray}[2]{ \begin{array}{#1}#2\end{array}} $
# $ \newcommand{\X}{ \mymatrix{cc}{0 & 1 \\ 1 & 0} } $
# $ \newcommand{\Z}{ \mymatrix{rr}{1 & 0 \\ 0 & -1} } $
# $ \newcommand{\Htwo}{ \mymatrix{rrrr}{ \frac{1}{2} & \frac{1}{2} & \frac{1}{2} & \frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & \frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} & \frac{1}{2} } } $
# $ \newcommand{\CNOT}{ \mymatrix{cccc}{1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 1 \\ 0 & 0 & 1 & 0} } $
# $ \newcommand{\norm}[1]{ \left\lVert #1 \right\rVert } $
# <h2>Quantum State</h2>
#
# The overall probability must be 1 when we observe a quantum system.
#
# For example, the following vectors <u>cannot</u> be a valid quantum state:
#
# $$
# \myvector{ \frac{1}{2} \\ \frac{1}{2} }
# \mbox{ and }
# \myvector{ \frac{\sqrt{3}}{2} \\ \frac{1}{\sqrt{2}} }.
# $$
#
# For the first vector: the probabilities of observing the states $\ket{0} $ and $ \ket{1} $ are $ \frac{1}{4} $.
#
# So, the overall probability of getting a result is $ \frac{1}{4} + \frac{1}{4} = \frac{1}{2} $, which is less than 1.
#
# For the second vector: the probabilities of observing the states $\ket{0} $ and $ \ket{1} $ are respectively $ \frac{3}{4} $ and $ \frac{1}{2} $.
#
# So, the overall probability of getting a result is $ \frac{3}{4} + \frac{1}{2} = \frac{5}{4} $, which is greater than 1.
# <font color="blue"><b>The summation of amplitude squares must be 1 for a valid quantum state.</b></font>
# <font color="blue"><b>In other words, a quantum state can be represented by a vector having length 1, and vice versa.</b></font>
#
# <i>The summation of amplitude squares gives the square of the length of vector.
#
# But, this summation is 1, and its sqaure root is also 1. So, we directly use the term <u>length</u> in the defintion.</i>
#
# We represent a quantum state as $ \ket{u} $ instead of $ u $.
#
# Remember the relation between the length and inner product: $ \norm{u} = \sqrt{\inner{u}{u}} $.
#
# In quantum computation, we use almost the same notation for the inner product: $ \braket{u}{u}$.
#
# $ \norm{ \ket{u} } = \sqrt{ \braket{u}{u} } = 1 $, or equivalently $ \braket{u}{u} = 1 $.
# <h3> Task 1 </h3>
#
# Let $a$ and $b$ be real numbers.
#
# If the folllowing vectors are valid quantum states, then what can be the values of $a$ and $b$?
#
# $$
# \ket{v} = \myrvector{a \\ -0.1 \\ -0.3 \\ 0.4 \\ 0.5}
# ~~~~~ \mbox{and} ~~~~~
# \ket{u} = \myrvector{ \frac{1}{\sqrt{2}} \\ \frac{1}{\sqrt{b}} \\ -\frac{1}{\sqrt{3}} }.
# $$
#
# your code is here or you may find the values by hand (in mind)
#
# <a href="..\bronze-solutions\B46_Quantum_State_Solutions.ipynb#task1">click for our solution</a>
# <h3> Quantum Operators </h3>
#
# Once the quantum state is defined, the definition of quantum operator is very easy.
#
# <font color="blue"><b>Any length preserving matrix is a quantum operator, and vice versa.</b></font>
# <h3> Task 2</h3>
#
# Remember Hadamard operator:
#
# $$
# H = \hadamard.
# $$
#
# Let's randomly create a 2-dimensional quantum state, and test whether Hadamard operator preserves the length or not.
#
# Write a function that returns a randomly created 2-dimensional quantum state:
# <ul>
# <li> Pick a random value between 0 and 100 </li>
# <li> Divide it by 100</li>
# <li> Take sqaure root of it</li>
# <li> Randomly determine its sign ($+$ or $-$)</li>
# <li> This is the first entry of the vector </li>
# <li> Find an appropriate value for the second entry </li>
# <li> Randomly determine its sign ($+$ or $-$)</li>
# </ul>
#
# Write a function that determines whether a given vector is a valid quantum state or not.
#
# (Due to precision problem, the summation of squares may not be exactly 1 but very close to 1, e.g., 0.9999999999999998.)
#
# Repeat 10 times:
# <ul>
# <li> Randomly create a quantum state </li>
# <li> Multiply Hadamard matrix with the randomly created quantum state </li>
# <li> Check whether the result quantum state is valid </li>
# </ul>
#
# your solution is here
#
# <a href="..\bronze-solutions\B46_Quantum_State_Solutions.ipynb#task2">click for our solution</a>
| community/awards/teach_me_quantum_2018/bronze/bronze/B46_Quantum_State.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Part 9 - Intro to Encrypted Programs
#
# Believe it or not, it is possible to compute with encrypted data. In other words, it's possible to run a program where **ALL of the variables** in the program are **encrypted**!
#
# In this tutorial, we're going to walk through very basic tools of encrypted computation. In particular, we're going to focus on one popular approach called Secure Multi-Party Computation. In this lesson, we'll learn how to build an encrypted calculator which can perform calculations on encrypted numbers.
#
# Authors:
# - <NAME> - Twitter: [@iamtrask](https://twitter.com/iamtrask)
# - <NAME> - GitHub: [@LaRiffle](https://github.com/LaRiffle)
#
# References:
# - <NAME> - [Blog](https://mortendahl.github.io) - Twitter: [@mortendahlcs](https://twitter.com/mortendahlcs)
# # Step 1: Encryption Using Secure Multi-Party Computation
#
# SMPC is at first glance a rather strange form of "encryption". Instead of using a public/private key to encrypt a variable, each value is split into multiple `shares`, each of which operates like a private key. Typically, these `shares` will be distributed amongst 2 or more _owners_. Thus, in order to decrypt the variable, all owners must agree to allow the decryption. In essence, everyone has a private key.
#
# ### Encrypt()
#
# So, let's say we wanted to "encrypt" a variable `x`, we could do so in the following way.
#
# > Encryption doesn't use floats or real numbers but happens in a mathematical space called [integer quotient ring](http://mathworld.wolfram.com/QuotientRing.html) which is basically the integers between `0` and `Q-1`, where `Q` is prime and "big enough" so that the space can contain all the numbers that we use in our experiments. In practice, given a value `x` integer, we do `x % Q` to fit in the ring. (That's why we avoid using number `x' > Q`).
Q = 1234567891011
x = 25
# +
import random
def encrypt(x):
share_a = random.randint(-Q,Q)
share_b = random.randint(-Q,Q)
share_c = (x - share_a - share_b) % Q
return (share_a, share_b, share_c)
# -
encrypt(x)
# As you can see here, we have split our variable `x` into 3 different shares, which could be sent to 3 different owners.
#
# ### Decrypt()
#
# If we wanted to decrypt these 3 shares, we could simply sum them together and take the modulus of the result (mod Q)
def decrypt(*shares):
return sum(shares) % Q
a,b,c = encrypt(25)
decrypt(a, b, c)
# Importantly, notice that if we try to decrypt with only two shares, the decryption does not work!
decrypt(a, b)
# Thus, we need all of the owners to participate in order to decrypt the value. It is in this way that the `shares` act like private keys, all of which must be present in order to decrypt a value.
# # Step 2: Basic Arithmetic Using SMPC
#
# However, the truly extraordinary property of Secure Multi-Party Computation is the ability to perform computation **while the variables are still encrypted**. Let's demonstrate simple addition below.
x = encrypt(25)
y = encrypt(5)
def add(x, y):
z = list()
# the first worker adds their shares together
z.append((x[0] + y[0]) % Q)
# the second worker adds their shares together
z.append((x[1] + y[1]) % Q)
# the third worker adds their shares together
z.append((x[2] + y[2]) % Q)
return z
decrypt(*add(x,y))
# ### Success!!!
#
# And there you have it! If each worker (separately) adds their shares together, then the resulting shares will decrypt to the correct value (25 + 5 == 30).
#
# As it turns out, SMPC protocols exist which can allow this encrypted computation for the following operations:
# - addition (which we've just seen)
# - multiplication
# - comparison
#
# and using these basic underlying primitives, we can perform arbitrary computation!!!
#
# In the next section, we're going to learn how to use the PySyft library to perform these operations!
# # Step 3: SMPC Using PySyft
#
# In the previous sections, we outlined some basic intuitions around SMPC is supposed to work. However, in practice we don't want to have to hand-write all of the primitive operations ourselves when writing our encrypted programs. So, in this section we're going to walk through the basics of how to do encrypted computation using PySyft. In particular, we're going to focus on how to do the 3 primitives previously mentioned: addition, multiplication, and comparison.
#
# First, we need to create a few Virtual Workers (which hopefully you're now familiar with given our previous tutorials).
# +
import torch
import syft as sy
hook = sy.TorchHook(torch)
bob = sy.VirtualWorker(hook, id="bob")
alice = sy.VirtualWorker(hook, id="alice")
bill = sy.VirtualWorker(hook, id="bill")
# -
# ### Basic Encryption/Decryption
#
# Encryption is as simple as taking any PySyft tensor and calling .share(). Decryption is as simple as calling .get() on the shared variable
x = torch.tensor([25])
x
encrypted_x = x.share(bob, alice, bill)
encrypted_x.get()
# ### Introspecting the Encrypted Values
#
# If we look closer at Bob, Alice, and Bill's workers, we can see the shares that get created!
list(bob._tensors.values())
x = torch.tensor([25]).share(bob, alice, bill)
# Bob's share
bobs_share = list(bob._tensors.values())[0]
bobs_share
# Alice's share
alices_share = list(alice._tensors.values())[0]
alices_share
# Bill's share
bills_share = list(bill._tensors.values())[0]
bills_share
# And if we wanted to, we could decrypt these values using the SAME approach we talked about earlier!!!
(bobs_share + alices_share + bills_share)
# As you can see, when we called `.share()` it simply split the value into 3 shares and sent one share to each of the parties!
# # Encrypted Arithmetic
#
# And now you see that we can perform arithmetic on the underlying values! The API is constructed so that we can simply perform arithmetic like we would normal PyTorch tensors.
x = torch.tensor([25]).share(bob,alice)
y = torch.tensor([5]).share(bob,alice)
z = x + y
z.get()
z = x - y
z.get()
# # Encrypted Multiplication
#
# For multiplication we need an additional party who is responsible for consistently generating random numbers (and not colluding with any of the other parties). We call this person a "crypto provider". For all intensive purposes, the crypto provider is just an additional VirtualWorker, but it's important to acknowledge that the crypto provider is not an "owner" in that he/she doesn't own shares but is someone who needs to be trusted to not collude with any of the existing shareholders.
crypto_provider = sy.VirtualWorker(hook, id="crypto_provider")
x = torch.tensor([25]).share(bob,alice, crypto_provider=crypto_provider)
y = torch.tensor([5]).share(bob,alice, crypto_provider=crypto_provider)
# +
# multiplication
z = x * y
z.get()
# -
# You can also do matrix multiplication
x = torch.tensor([[1, 2],[3,4]]).share(bob,alice, crypto_provider=crypto_provider)
y = torch.tensor([[2, 0],[0,2]]).share(bob,alice, crypto_provider=crypto_provider)
# +
# matrix multiplication
z = x.mm(y)
z.get()
# -
# # Encrypted comparison
# It is also possible to private comparisons between private values. We rely here on the SecureNN protocol, the details of which can be found [here](https://eprint.iacr.org/2018/442.pdf). The result of the comparison is also a private shared tensor.
x = torch.tensor([25]).share(bob,alice, crypto_provider=crypto_provider)
y = torch.tensor([5]).share(bob,alice, crypto_provider=crypto_provider)
z = x > y
z.get()
z = x <= y
z.get()
z = x == y
z.get()
z = x == y + 20
z.get()
# You can also perform max operations
x = torch.tensor([2, 3, 4, 1]).share(bob,alice, crypto_provider=crypto_provider)
x.max().get()
x = torch.tensor([[2, 3], [4, 1]]).share(bob,alice, crypto_provider=crypto_provider)
max_values = x.max(dim=0)
max_values.get()
# # Congratulations!!! - Time to Join the Community!
#
# Congratulations on completing this notebook tutorial! If you enjoyed this and would like to join the movement toward privacy preserving, decentralized ownership of AI and the AI supply chain (data), you can do so in the following ways!
#
# ### Star PySyft on GitHub
#
# The easiest way to help our community is just by starring the Repos! This helps raise awareness of the cool tools we're building.
#
# - [Star PySyft](https://github.com/OpenMined/PySyft)
#
# ### Join our Slack!
#
# The best way to keep up to date on the latest advancements is to join our community! You can do so by filling out the form at [http://slack.openmined.org](http://slack.openmined.org)
#
# ### Join a Code Project!
#
# The best way to contribute to our community is to become a code contributor! At any time you can go to PySyft GitHub Issues page and filter for "Projects". This will show you all the top level Tickets giving an overview of what projects you can join! If you don't want to join a project, but you would like to do a bit of coding, you can also look for more "one off" mini-projects by searching for GitHub issues marked "good first issue".
#
# - [PySyft Projects](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3AProject)
# - [Good First Issue Tickets](https://github.com/OpenMined/PySyft/issues?q=is%3Aopen+is%3Aissue+label%3A%22good+first+issue%22)
#
# ### Donate
#
# If you don't have time to contribute to our codebase, but would still like to lend support, you can also become a Backer on our Open Collective. All donations go toward our web hosting and other community expenses such as hackathons and meetups!
#
# [OpenMined's Open Collective Page](https://opencollective.com/openmined)
| examples/tutorials/Part 09 - Intro to Encrypted Programs.ipynb |